hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
cb43138ea6cf564da1e6a0ae5404c2c8ef9b7aa9 | 20,189 | ipynb | Jupyter Notebook | docs/source/examples/Visualizing Frame Dragging in Kerr Spacetime using EinsteinPy!.ipynb | bibek22/einsteinpy | 78bf5d942cbb12393852f8e4d7a8426f1ffe6f23 | [
"MIT"
] | null | null | null | docs/source/examples/Visualizing Frame Dragging in Kerr Spacetime using EinsteinPy!.ipynb | bibek22/einsteinpy | 78bf5d942cbb12393852f8e4d7a8426f1ffe6f23 | [
"MIT"
] | null | null | null | docs/source/examples/Visualizing Frame Dragging in Kerr Spacetime using EinsteinPy!.ipynb | bibek22/einsteinpy | 78bf5d942cbb12393852f8e4d7a8426f1ffe6f23 | [
"MIT"
] | null | null | null | 140.201389 | 17,124 | 0.896032 | [
[
[
"# Visualizing Frame Dragging in Kerr Spacetime\n\n### Importing required modules",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\nfrom einsteinpy.coordinates.utils import four_position, stacked_vec\nfrom einsteinpy.geodesic import Geodesic\nfrom einsteinpy.metric import Kerr",
"_____no_output_____"
]
],
[
[
"### Setting up metric and intial conditions",
"_____no_output_____"
]
],
[
[
"# Metric Parameters\nM, a = 1.989e30, 0.3\n # Kerr Metric Object\nms_cov = Kerr(coords=\"BL\", M=M, a=a)\n\n# Initial conditions\nx_vec = np.array([49.95e8, np.pi / 2, np.pi])\nv_vec = np.array([0., 0., 0.])\nt = 0.\n\n# Getting Position 4-Vector\nx_4vec = four_position(t, x_vec)\n# Calculating Schwarzschild Metric at x_4vec\nms_cov_mat = ms_cov.metric_covariant(x_4vec)\n\n# Getting stacked (Length-8) initial vector, containing 4-Pos and 4-Vel\ninit_vec = stacked_vec(ms_cov_mat, t, x_vec, v_vec, time_like=True)",
"_____no_output_____"
]
],
[
[
"### Calculating the geodesic",
"_____no_output_____"
]
],
[
[
"geod = Geodesic(metric=ms_cov, init_vec=init_vec, end_lambda=33932.90, step_size=1.2)\n\nans = geod.trajectory\n\nx, y = ans[:,1], ans[:,2]",
"_____no_output_____"
]
],
[
[
"### Plotting the geodesic",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nfig = plt.figure(figsize=(12,10))\nplt.scatter(x, y, s=0.2)\nplt.scatter(0, 0)\nplt.show()",
"_____no_output_____"
]
],
[
[
" As can be seen in the plot above, the test particle is dragged by the Kerr black hole.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb431c193431149aa1088aed266d478d94e3f1eb | 158,698 | ipynb | Jupyter Notebook | analysis/Phasing Probabilistic Model Analysis.ipynb | AbeelLab/phasm-benchmarks | 74486a6abe545333f0e360a63a8b3644424234c3 | [
"MIT"
] | 1 | 2017-07-18T10:11:27.000Z | 2017-07-18T10:11:27.000Z | analysis/Phasing Probabilistic Model Analysis.ipynb | lrvdijk/phasm-benchmarks | 74486a6abe545333f0e360a63a8b3644424234c3 | [
"MIT"
] | null | null | null | analysis/Phasing Probabilistic Model Analysis.ipynb | lrvdijk/phasm-benchmarks | 74486a6abe545333f0e360a63a8b3644424234c3 | [
"MIT"
] | null | null | null | 193.534146 | 55,538 | 0.855877 | [
[
[
"%matplotlib inline\n\nimport sys\nimport os\nimport json\nfrom glob import glob\nfrom collections import defaultdict, OrderedDict\n\nimport dinopy\nimport yaml\nimport matplotlib.pyplot as plt\nfrom matplotlib.ticker import MultipleLocator\nimport seaborn\nimport numpy\nimport pandas as pd\nimport networkx\nfrom scipy.special import binom\nfrom scipy import stats\nfrom IPython.display import Image, display\n\nfrom phasm.io import gfa\nfrom phasm.alignments import AlignmentType\nfrom phasm.assembly_graph import AssemblyGraph\nfrom phasm.bubbles import find_superbubbles\n\nBASE_DIR = os.path.realpath(os.path.join(os.getcwd(), '..'))\n\nwith open(os.path.join(BASE_DIR, \"config.yml\")) as f:\n config = yaml.load(f)\n \nseaborn.set_style('whitegrid')",
"_____no_output_____"
],
[
"spanning_read_stats = []\ncandidate_prob_stats = []\nbubble_map = defaultdict(dict)\nfor assembly, asm_config in config['assemblies'].items():\n parts = assembly.split('-')\n ploidy = int(parts[0].replace(\"ploidy\", \"\"))\n coverage = int(parts[1].replace(\"x\", \"\"))\n \n asm_folder = os.path.join(BASE_DIR, \"assemblies\", assembly)\n \n for debugdata in glob(\"{}/04_phase/component[0-9].bubblechain[0-9]-debugdata.json\".format(asm_folder)):\n print(debugdata)\n graphml = debugdata.replace(\"04_phase\", \"03_chain\").replace(\"-debugdata.json\", \".graphml\")\n g = AssemblyGraph(networkx.read_graphml(graphml))\n \n curr_bubble = None\n bubble_num = 0\n num_candidates = -1\n with open(debugdata) as f:\n for line in f:\n data = json.loads(line)\n if data['type'] == \"new_bubble\":\n curr_bubble = data\n bubble_map[ploidy, coverage][(data['entrance'], data['exit'])] = data\n \n if data['start_of_block'] == True:\n bubble_num = 1\n else:\n dist_between_bubbles = (\n min(e[2] for e in g.out_edges_iter(data['entrance'], data=g.edge_len))\n )\n spanning_read_stats.append({\n 'dist': dist_between_bubbles,\n 'spanning_reads': len(data['rel_read_info']),\n 'ploidy': ploidy\n })\n \n bubble_num += 1\n \n if data['type'] == \"candidate_set\":\n p_sr = data['p_sr']\n prior = data['prior']\n prob = 10**(p_sr + prior)\n entrance = curr_bubble['entrance']\n exit = curr_bubble['exit']\n candidate_prob_stats.append({\n 'bubble': (entrance, exit),\n 'bubble_num': bubble_num,\n 'candidate_prob': prob,\n 'ploidy': ploidy,\n 'coverage': coverage\n })",
"/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy2-60x-error-free/04_phase/component0.bubblechain0-debugdata.json\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy2-60x-error-free/04_phase/component1.bubblechain0-debugdata.json\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy3-60x-error-free/04_phase/component0.bubblechain0-debugdata.json\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy3-60x-error-free/04_phase/component1.bubblechain0-debugdata.json\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy4-60x-error-free/04_phase/component0.bubblechain0-debugdata.json\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy4-60x-error-free/04_phase/component1.bubblechain0-debugdata.json\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy6-60x-error-free/04_phase/component0.bubblechain0-debugdata.json\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy6-60x-error-free/04_phase/component1.bubblechain0-debugdata.json\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy8-60x-error-free/04_phase/component0.bubblechain0-debugdata.json\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy8-60x-error-free/04_phase/component1.bubblechain0-debugdata.json\n"
],
[
"srdf = pd.DataFrame(spanning_read_stats)\nsrdf['spanning_reads_norm'] = srdf['spanning_reads'] / srdf['ploidy']\ng = seaborn.JointGrid(x=\"dist\", y=\"spanning_reads_norm\", data=srdf, size=7)\nx_bin_size = 2500\ng.ax_marg_x.hist(srdf['dist'], alpha=0.6, bins=numpy.arange(0, srdf['dist'].max()+x_bin_size, x_bin_size))\ny_bin_size = 10\ng.ax_marg_y.hist(srdf['spanning_reads_norm'], alpha=0.6, orientation=\"horizontal\",\n bins=numpy.arange(0, srdf['spanning_reads_norm'].max()+y_bin_size, y_bin_size))\ng.plot_joint(seaborn.regplot)\ng.annotate(stats.pearsonr)\n\nseaborn.plt.suptitle(\"Number of spanning reads against the distance between two bubbles,\\n normalised for ploidy\")\nplt.ylim(ymin=0)\nplt.xlabel(\"Distance between two bubbles [bases]\")\nplt.ylabel(\"Number of spanning reads\")\nplt.subplots_adjust(top=0.9)\nplt.savefig(os.path.join(BASE_DIR, 'figures', 'spanning-reads.png'), transparent=True, dpi=256)\n\n",
"_____no_output_____"
],
[
"candidate_df = pd.DataFrame(candidate_prob_stats)\ncandidate_df.set_index('bubble')\nplt.figure()\nseaborn.distplot(candidate_df['candidate_prob'], kde=False, hist_kws={\"alpha\": 0.8})\nplt.title(\"Distribution of candidate extension relative likelihoods\")\nplt.xlabel(\"Relative likelihood of an extension\")\nplt.ylabel(\"Count\")\n# plt.xlim(xmax=1.0)\nplt.axvline(1e-3, linestyle='--', color='black')\nplt.savefig(os.path.join(BASE_DIR, 'figures', 'rel-likelihood-abs.png'), transparent=True, dpi=256)",
"_____no_output_____"
],
[
"grouped = candidate_df.groupby(['bubble', 'ploidy'])['candidate_prob']\nmax_probs = grouped.max()\n\nfor bubble, ploidy in grouped.groups.keys():\n candidate_df.loc[grouped.groups[bubble, ploidy], 'max_prob'] = max_probs[bubble, ploidy]\n \ncandidate_df['relative_prob'] = candidate_df['candidate_prob'] / candidate_df['max_prob']\ncandidate_df\n\nplt.figure()\nseaborn.distplot(candidate_df[candidate_df['relative_prob'] < 1.0]['relative_prob'], kde=False, hist_kws={\"alpha\": 0.8})\nplt.title(\"Distribution of relative probabilities for each candidate extension\\n\"\n \"at each superbubble\")\nplt.xlabel(r\"$RL[E|H]\\ /\\ \\omega$\")\nplt.ylabel(\"Count\")\nplt.savefig(os.path.join(BASE_DIR, \"figures\", \"rl-relative-dist.png\"), transparent=True, dpi=256)",
"_____no_output_____"
],
[
"c1, c2, c3, c4, c5 = seaborn.color_palette(n_colors=5)\npruning_stats = []\n\nfor assembly, asm_config in config['assemblies'].items():\n parts = assembly.split('-')\n ploidy = int(parts[0].replace(\"ploidy\", \"\"))\n coverage = int(parts[1].replace(\"x\", \"\"))\n \n if coverage != 60:\n continue\n \n asm_folder = os.path.join(BASE_DIR, \"assemblies\", assembly)\n \n for chain_num, graphml in enumerate(glob(\"{}/03_chain/component[0-9].bubblechain[0-9].graphml\".format(asm_folder))):\n print(graphml)\n \n # Calculate effect of pruning\n g = AssemblyGraph(networkx.read_graphml(graphml))\n \n bubbles = OrderedDict(find_superbubbles(g, report_nested=False))\n bubble_num = 0\n for i, bubble in enumerate(reversed(bubbles.items())):\n entrance, exit = bubble\n num_paths = len(list(networkx.all_simple_paths(g, entrance, exit)))\n \n if not bubble in bubble_map[ploidy, coverage]:\n continue\n \n bubble_data = bubble_map[ploidy, coverage][bubble]\n if bubble_data['start_of_block']:\n bubble_num = 1\n else:\n bubble_num += 1\n \n kappa = 0.0\n pruned = 0\n num_candidates_left = sys.maxsize\n while num_candidates_left > 500 and kappa < 1.0:\n kappa += 0.1\n num_candidates_left = len(\n candidate_df.query('(bubble == @bubble) and (ploidy == @ploidy) and (relative_prob >= @kappa)')\n )\n pruned = len(\n candidate_df.query('(bubble == @bubble) and (ploidy == @ploidy) and (relative_prob < @kappa)')\n )\n \n pruning_stats.append({\n 'ploidy': ploidy,\n 'coverage': coverage,\n 'bubble_num': bubble_num,\n 'pruned': pruned,\n 'kappa': kappa\n })\n ",
"/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy2-60x-error-free/03_chain/component0.bubblechain0.graphml\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy2-60x-error-free/03_chain/component1.bubblechain0.graphml\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy3-60x-error-free/03_chain/component0.bubblechain0.graphml\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy3-60x-error-free/03_chain/component1.bubblechain0.graphml\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy4-60x-error-free/03_chain/component0.bubblechain0.graphml\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy4-60x-error-free/03_chain/component1.bubblechain0.graphml\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy6-60x-error-free/03_chain/component0.bubblechain0.graphml\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy6-60x-error-free/03_chain/component1.bubblechain0.graphml\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy8-60x-error-free/03_chain/component0.bubblechain0.graphml\n/run/media/lucas/data/bioinformatics/thesis-data/assemblies/ploidy8-60x-error-free/03_chain/component1.bubblechain0.graphml\n"
],
[
"pruning_df = pd.DataFrame(pruning_stats)\nagg_df = pd.DataFrame(pruning_df.groupby(['bubble_num', 'kappa']).size().rename('counts'))\nagg_df.reset_index(level=agg_df.index.names, inplace=True)\nagg_df = agg_df.query('kappa <= 1.0')\n\n",
"_____no_output_____"
],
[
"sum_df = pd.DataFrame(agg_df.groupby('bubble_num')['counts'].sum()).reset_index()\nsum_df\n\nfor i in sum_df['bubble_num'].unique():\n agg_df.loc[agg_df['bubble_num'] == i, 'total'] = int(sum_df['counts'].loc[sum_df['bubble_num'] == i].values[0])\n \nagg_df['fraction'] = agg_df['counts'] / agg_df['total']\nagg_df",
"_____no_output_____"
],
[
"plt.figure()\ng = seaborn.factorplot(x=\"kappa\", y=\"fraction\", col=\"bubble_num\", \n kind=\"bar\", col_wrap=3, sharex=False, color=c1,\n data=agg_df.query('(bubble_num < 7) and (kappa <= 1.0)'))\nseaborn.plt.suptitle('The maximum pruning factor $\\kappa$ at different stages of the phasing process')\nplt.subplots_adjust(top=0.9, hspace=0.3)\n\nfor i, ax in enumerate(g.axes):\n ax.set_xlabel(\"$\\kappa$\")\n \n if i % 3 == 0:\n ax.set_ylabel(\"Fraction\")\n ax.set_title(\"Superbubble {}\".format(i+1))\n\nplt.savefig(os.path.join(BASE_DIR, 'figures', 'pruning.png'), transparent=True, dpi=256)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4320b9347455051b938cdca99490e143937e4f | 41,886 | ipynb | Jupyter Notebook | Seminario/WilliamSteveRodriguezVillamizar_Seminario-2.ipynb | wisrovi/03MAIR---Algoritmos-de-Optimizacion---2019 | a962bd3cb1b4180d3f2fc64cdc689f6334808750 | [
"MIT"
] | null | null | null | Seminario/WilliamSteveRodriguezVillamizar_Seminario-2.ipynb | wisrovi/03MAIR---Algoritmos-de-Optimizacion---2019 | a962bd3cb1b4180d3f2fc64cdc689f6334808750 | [
"MIT"
] | null | null | null | Seminario/WilliamSteveRodriguezVillamizar_Seminario-2.ipynb | wisrovi/03MAIR---Algoritmos-de-Optimizacion---2019 | a962bd3cb1b4180d3f2fc64cdc689f6334808750 | [
"MIT"
] | null | null | null | 31.707797 | 315 | 0.367354 | [
[
[
"<a href=\"https://colab.research.google.com/github/wisrovi/03MAIR---Algoritmos-de-Optimizacion---2019/blob/master/Seminario/WilliamSteveRodriguezVillamizar_Seminario-2.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Algoritmos de optimización - Seminario<br>\nNombre y Apellidos: <br>\nGithub: https://github.com/wisrovi/03MAIR---Algoritmos-de-Optimizacion---2019/blob/master/Seminario/WilliamSteveRodriguezVillamizar_Seminario-2.ipynb<br>\nProblema:\n\nColab: https://colab.research.google.com/github/wisrovi/03MAIR---Algoritmos-de-Optimizacion---2019/blob/master/Seminario/WilliamSteveRodriguezVillamizar_Seminario-2.ipynb\n> 1. Combinar cifras y operaciones <br>\n\nDescripción del problema:\n\nElección de grupos de población homogéneos\n\n• El problema consiste en analizar el siguiente problema y diseñar un algoritmo que lo resuelva.\n\n• Disponemos de las 9 cifras del 1 al 9 (excluimos el cero) y de los 4 signos básicos de las\noperaciones fundamentales: suma(+), resta(-), multiplicación(*) y división(/)\n\n• Debemos combinarlos alternativamente sin repetir ninguno de ellos para obtener una\ncantidad dada.\n\n....\n\n(*) La respuesta es obligatoria\n\n\n\n\n\n ",
"_____no_output_____"
],
[
"# Solución del problema usando algoritmos por recursividad y el algoritmo divide y venceras",
"_____no_output_____"
],
[
"(*)¿Cuantas posibilidades hay sin tener en cuenta las restricciones?<br>\n\n\n\n¿Cuantas posibilidades hay teniendo en cuenta todas las restricciones.\n\n\n",
"_____no_output_____"
],
[
"Respuesta",
"_____no_output_____"
],
[
"Debido a que el algoritmo genera en diversas ocasiones soluciones repetidas y no todas las combinaciones de numeros y signos son posibles, además de que cada valor ingresado tiene su própio número de soluciones único, siendo más facil generar soluciones para valores enteros que para valores flotantes.\n\nNo fue posible determinar una ecuación que entregara la cantidad de soluciones absoluta para todos los valores ingresados.\n\nPor ello se ha elaborado un algoritmo para buscar toda la cantidad de respuestas posibles para un número dado, para determinar cuantas soluciones se pueden generar para la respuesta solicituda (se verá más adelante en el código).\n\nEn la evaluacion del algoritmo se ha logrado generar hasta 700 soluciones posibles para la respuesta de un entero de valor 10 (siendo este valor el que mayor soluciones se ha logrado encontrar. \n",
"_____no_output_____"
],
[
"Modelo para el espacio de soluciones<br>\n(*) ¿Cual es la estructura de datos que mejor se adapta al problema? Argumentalo.(Es posible que hayas elegido una al principio y veas la necesidad de cambiar, arguentalo)\n",
"_____no_output_____"
],
[
"Respuesta",
"_____no_output_____"
],
[
"Como este problema no tiene un dataseet inicial sino un dato de ingreso y con este generar una solución al problema, \nsólo se puede decir que hay mejores soluciones para datos de entrada de tipo int menores a 100 que para datos float de cualquier valor\n\nEsto se evidencia debido a que tratando de evaluar el mayor numero de soluciones para un dato, se ha logrado minimo 250 soluciones diferentes para datos int\nmientras que para datos float sólo se han logrado 35 soluciones\n\nSin embargo si se pudiere usar una maquina con mayor GPU a la mia, \nla cantidad de soluciones debería aumentar de 35 a 100 y de 250 a 1000 para float e int respectivamente.",
"_____no_output_____"
],
[
"Según el modelo para el espacio de soluciones<br>\n(*)¿Cual es la función objetivo?\n\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"(*)¿Es un problema de maximización o minimización?",
"_____no_output_____"
],
[
"Respuesta: Minimizacion",
"_____no_output_____"
],
[
"## Implementaciones Básicas de código para fuerza bruta y algoritmo final",
"_____no_output_____"
]
],
[
[
"import random\nfrom sympy.parsing.sympy_parser import parse_expr\nimport numpy as np",
"_____no_output_____"
],
[
"Diccionario_signos = {\n \"suma\" : \"+\",\n \"resta\" : \"-\",\n \"multiplicacion\" : \"*\",\n \"division\" : \"/\"\n }\n\nLista_Numeros_Posibles = (1,\n 2,\n 3,\n 4,\n 5,\n 6,\n 7,\n 8,\n 9\n )",
"_____no_output_____"
],
[
"def BuscarExiste(lista, item):\n buscandoEncontrando = False\n for i in lista:\n if i == item:\n buscandoEncontrando = True\n break\n return buscandoEncontrando\n\ndef getNumeroAleatorio(listaNumeros = []):\n \"\"\"\n usamos recursividad para crear este set de datos\n \"\"\"\n if len(listaNumeros) < 5: \n numer = random.choice(Lista_Numeros_Posibles)\n if not BuscarExiste(listaNumeros, numer):\n listaNumeros.append(numer)\n getNumeroAleatorio(listaNumeros) \n return tuple(listaNumeros) \n\ndef getSignosAleatorios(listaSignos = []): \n \"\"\"\n usamos recursividad para crear este set de datos\n \"\"\"\n if len(listaSignos) < 4:\n sign = random.choice( list(Diccionario_signos.keys()) ) \n if not BuscarExiste(listaSignos, sign):\n listaSignos.append(sign)\n getSignosAleatorios(listaSignos)\n return tuple(listaSignos)\n\ndef HallarResultado(tuplaNumeros, tuplaSignos):\n expresion = \"\"\"%s %s %s %s %s %s %s %s %s\"\"\" %(\n str(tuplaNumeros[0]),\n Diccionario_signos[tuplaSignos[0]],\n str(tuplaNumeros[1]),\n Diccionario_signos[tuplaSignos[1]],\n str(tuplaNumeros[2]),\n Diccionario_signos[tuplaSignos[2]],\n str(tuplaNumeros[3]),\n Diccionario_signos[tuplaSignos[3]],\n str(tuplaNumeros[4])\n ) \n solucion = float(parse_expr(expresion))\n solucion = \"{0:.2f}\".format(solucion)\n solucion = float(solucion)\n return (expresion, solucion)\n\ndef HallarValorDeseado(valorDeseado, stepsMax=10000): \n for i in range(stepsMax):\n (expresion, solucion) = HallarResultado(getNumeroAleatorio([]), getSignosAleatorios([]))\n if valorDeseado == solucion:\n return (expresion, \"=\", valorDeseado) \n\ndef BuscarOperacionesPorListado(valoresDeseados):\n respuestas = []\n numerosNoEncontrados = []\n for valorDeseado in valoresDeseados:\n rta = HallarValorDeseado(valorDeseado) \n if rta is None:\n numerosNoEncontrados.append(valorDeseado)\n #print(\"None = \", valorDeseado )\n else:\n respuestas.append(rta)\n #print(rta)\n return respuestas, numerosNoEncontrados ",
"_____no_output_____"
]
],
[
[
"## Algoritmo Por fuerza bruta",
"_____no_output_____"
],
[
"Diseña un algoritmo para resolver el problema por fuerza bruta",
"_____no_output_____"
],
[
"Respuesta",
"_____no_output_____"
]
],
[
[
"#Buscando la cantidad máxima de soluciones posibles\nvalor = 5.25\ncantidadSolucionesBuscar = 500 #Número probado de soluciones diferentes encontradas 500 para int y 150 float\n\n\nvaloresDeseados = []\nfor i in range(cantidadSolucionesBuscar):\n valoresDeseados.append(valor)\n\n\nprint(\"Total numeros buscar respuesta: \", len(valoresDeseados))\nrespuestas, numerosNoEncontrados =BuscarOperacionesPorListado(valoresDeseados)\n\nprint(\"Numeros no encontrados: \")\nfor rta in numerosNoEncontrados:\n print(rta)\n pass\n\nrespuestas = sorted(set(respuestas))\nprint(\"Numeros con respuesta encontrada: \", len(respuestas) )\nfor rta in respuestas:\n #print(rta)\n pass",
"Total numeros buscar respuesta: 500\nNumeros no encontrados: \nNumeros con respuesta encontrada: 337\n"
],
[
"print(\"solución a hallar: \", valor)\nprint()\nprint(\"solución mejor (usando los numero más pequeños posibles): \" )\nprint(respuestas[0][0], respuestas[0][1], respuestas[0][2])",
"solución a hallar: 5.25\n\nsolución mejor (usando los numero más pequeños posibles): \n1 * 2 + 4 - 6 / 8 = 5.25\n"
]
],
[
[
"Calcula la complejidad del algoritmo por fuerza bruta",
"_____no_output_____"
],
[
"Respuesta",
"_____no_output_____"
],
[
"O(n log n)",
"_____no_output_____"
],
[
"## Algoritmo final (mejora al de fuerza bruta)",
"_____no_output_____"
],
[
"ok - (*)Diseña un algoritmo que mejore la complejidad del algortimo por fuerza bruta. Argumenta porque crees que mejora el algoritmo por fuerza bruta",
"_____no_output_____"
],
[
"Respuesta",
"_____no_output_____"
]
],
[
[
"valoresDeseados = [-3.5]\n\nprint(\"Total numeros buscar respuesta: \", len(valoresDeseados))\nrespuestas, numerosNoEncontrados =BuscarOperacionesPorListado(valoresDeseados)\n\nprint(\"solución a hallar: \", valoresDeseados[0])\nprint()\nprint(\"solución mejor (usando los numero más pequeños posibles): \" )\nprint(respuestas[0][0], respuestas[0][1], respuestas[0][2])",
"Total numeros buscar respuesta: 1\nsolución a hallar: -3.5\n\nsolución mejor (usando los numero más pequeños posibles): \n2 - 7 + 4 / 8 * 3 = -3.5\n"
],
[
"valoresDeseados = [-3.5, 2, 9, -9.5, 4, 1.3, 1,2,3,4,5,6,7,8,9]\n\nprint(\"Total numeros buscar respuesta: \", len(valoresDeseados))\nrespuestas, numerosNoEncontrados =BuscarOperacionesPorListado(valoresDeseados)\n\nprint(\"Numeros no encontrados: \")\nfor rta in numerosNoEncontrados:\n print(rta)\n pass\n\nprint(\"Numeros con respuesta encontrada: \")\nfor rta in respuestas:\n print(rta)\n pass",
"Total numeros buscar respuesta: 15\nNumeros no encontrados: \n1.3\nNumeros con respuesta encontrada: \n('3 * 5 / 6 - 8 + 2', '=', -3.5)\n('4 / 8 * 2 + 7 - 6', '=', 2)\n('9 + 6 - 4 / 2 * 3', '=', 9)\n('2 / 4 + 8 - 6 * 3', '=', -9.5)\n('7 - 3 * 4 + 9 / 1', '=', 4)\n('6 - 8 + 1 / 3 * 9', '=', 1)\n('5 + 2 * 3 / 6 - 4', '=', 2)\n('1 - 4 * 6 / 8 + 5', '=', 3)\n('4 / 2 - 7 + 1 * 9', '=', 4)\n('2 + 5 / 4 * 8 - 7', '=', 5)\n('8 - 4 + 3 / 9 * 6', '=', 6)\n('6 / 2 - 8 + 3 * 4', '=', 7)\n('8 * 3 / 6 - 1 + 5', '=', 8)\n('9 + 6 * 4 / 8 - 3', '=', 9)\n"
],
[
"valoresDeseados = range (-10, 11, 1)\n\nprint(\"Total numeros buscar respuesta: \", len(valoresDeseados))\nrespuestas, numerosNoEncontrados =BuscarOperacionesPorListado(valoresDeseados)\n\nprint(\"Numeros no encontrados: \")\nfor rta in numerosNoEncontrados:\n print(rta)\n pass\n\nrespuestas = sorted(set(respuestas))\nprint(\"Numeros con respuesta encontrada: \")\nfor rta in respuestas:\n print(rta)\n pass",
"Total numeros buscar respuesta: 21\nNumeros no encontrados: \nNumeros con respuesta encontrada: \n('1 - 5 + 6 / 4 * 8', '=', 8)\n('1 - 8 / 2 * 4 + 6', '=', -9)\n('2 - 7 / 1 + 4 * 3', '=', 7)\n('2 - 8 / 4 + 5 * 1', '=', 5)\n('3 + 5 - 6 / 4 * 8', '=', -4)\n('3 - 5 / 2 * 4 + 6', '=', -1)\n('3 - 9 + 2 / 4 * 6', '=', -3)\n('4 + 8 / 1 - 6 * 2', '=', 0)\n('4 - 9 + 3 / 2 * 6', '=', 4)\n('5 + 1 - 4 * 9 / 3', '=', -6)\n('5 + 4 * 9 / 6 - 2', '=', 9)\n('5 - 4 / 2 * 7 + 1', '=', -8)\n('5 - 6 + 8 * 1 / 4', '=', 1)\n('7 + 1 / 2 * 6 - 4', '=', 6)\n('7 + 2 / 4 * 6 - 8', '=', 2)\n('7 + 4 - 9 * 6 / 3', '=', -7)\n('8 - 7 / 1 * 2 + 4', '=', -2)\n('8 / 4 + 5 * 3 - 7', '=', 10)\n('9 + 2 / 1 - 3 * 7', '=', -10)\n('9 - 3 * 6 + 8 / 2', '=', -5)\n('9 - 4 / 2 * 7 + 8', '=', 3)\n"
],
[
"valoresDeseados = []\nvalores = np.arange(-10, 11, 0.05)\nfor valorDeseado in valores:\n valorDeseado = float(\"{0:.2f}\".format(valorDeseado))\n valoresDeseados.append(valorDeseado)\n\nprint(\"Total numeros buscar respuesta: \", len(valoresDeseados))\nrespuestas, numerosNoEncontrados =BuscarOperacionesPorListado(valoresDeseados)\nprint(\"Con rta: \",len(respuestas))\nprint(\"Sin rta: \",len(numerosNoEncontrados))\n\n\n\nprint(\"Numeros no encontrados: \")\nfor rta in numerosNoEncontrados:\n print(rta)\n pass\n\nprint(\"Numeros con respuesta encontrada: \")\nfor rta in respuestas:\n print(rta)\n pass\n",
"Total numeros buscar respuesta: 420\nCon rta: 162\nSin rta: 258\nNumeros no encontrados: \n-9.95\n-9.9\n-9.85\n-9.7\n-9.65\n-9.6\n-9.55\n-9.45\n-9.35\n-9.3\n-9.15\n-9.1\n-9.05\n-8.95\n-8.9\n-8.85\n-8.7\n-8.65\n-8.6\n-8.55\n-8.45\n-8.35\n-8.3\n-8.15\n-8.1\n-8.05\n-7.95\n-7.9\n-7.85\n-7.7\n-7.65\n-7.6\n-7.55\n-7.45\n-7.35\n-7.3\n-7.15\n-7.1\n-7.05\n-6.95\n-6.9\n-6.85\n-6.7\n-6.65\n-6.6\n-6.55\n-6.45\n-6.35\n-6.3\n-6.15\n-6.1\n-6.05\n-5.95\n-5.9\n-5.85\n-5.7\n-5.65\n-5.55\n-5.45\n-5.35\n-5.3\n-5.25\n-5.2\n-5.15\n-5.1\n-5.05\n-4.95\n-4.9\n-4.85\n-4.7\n-4.65\n-4.55\n-4.45\n-4.35\n-4.3\n-4.15\n-4.1\n-4.05\n-3.95\n-3.9\n-3.85\n-3.7\n-3.65\n-3.55\n-3.45\n-3.35\n-3.3\n-3.15\n-3.1\n-3.05\n-2.95\n-2.9\n-2.85\n-2.7\n-2.65\n-2.55\n-2.45\n-2.35\n-2.3\n-2.15\n-2.1\n-2.05\n-1.95\n-1.9\n-1.85\n-1.7\n-1.65\n-1.55\n-1.45\n-1.35\n-1.3\n-1.15\n-1.1\n-1.05\n-0.95\n-0.9\n-0.85\n-0.7\n-0.65\n-0.55\n-0.45\n-0.35\n-0.3\n-0.15\n-0.1\n-0.05\n0.05\n0.1\n0.15\n0.3\n0.35\n0.45\n0.55\n0.65\n0.7\n0.85\n0.9\n0.95\n1.05\n1.1\n1.15\n1.3\n1.35\n1.45\n1.55\n1.65\n1.7\n1.85\n1.9\n1.95\n2.05\n2.1\n2.15\n2.3\n2.35\n2.45\n2.55\n2.65\n2.7\n2.85\n2.9\n2.95\n3.05\n3.1\n3.15\n3.3\n3.35\n3.45\n3.55\n3.65\n3.7\n3.85\n3.9\n3.95\n4.05\n4.1\n4.15\n4.3\n4.35\n4.45\n4.55\n4.65\n4.7\n4.85\n4.9\n4.95\n5.05\n5.1\n5.15\n5.3\n5.35\n5.45\n5.55\n5.65\n5.7\n5.85\n5.9\n5.95\n6.05\n6.1\n6.15\n6.3\n6.35\n6.45\n6.55\n6.65\n6.7\n6.85\n6.9\n6.95\n7.05\n7.1\n7.15\n7.3\n7.35\n7.45\n7.55\n7.65\n7.7\n7.85\n7.9\n7.95\n8.05\n8.1\n8.15\n8.3\n8.35\n8.45\n8.55\n8.65\n8.7\n8.85\n8.9\n8.95\n9.05\n9.1\n9.15\n9.3\n9.35\n9.45\n9.55\n9.65\n9.7\n9.85\n9.9\n9.95\n10.05\n10.1\n10.15\n10.3\n10.35\n10.45\n10.55\n10.65\n10.7\n10.85\n10.9\n10.95\nNumeros con respuesta encontrada: \n('3 + 5 - 8 * 9 / 4', '=', -10.0)\n('1 / 5 + 6 - 2 * 8', '=', -9.8)\n('1 - 2 * 6 + 5 / 4', '=', -9.75)\n('8 - 9 * 2 + 3 / 6', '=', -9.5)\n('1 - 8 / 5 * 9 + 4', '=', -9.4)\n('3 / 4 + 8 - 9 * 2', '=', -9.25)\n('4 / 5 + 8 - 2 * 9', '=', -9.2)\n('2 - 6 * 8 / 4 + 1', '=', -9.0)\n('7 - 8 * 2 + 1 / 5', '=', -8.8)\n('9 + 1 / 4 - 3 * 6', '=', -8.75)\n('2 + 9 / 6 - 4 * 3', '=', -8.5)\n('4 - 2 * 7 + 8 / 5', '=', -8.4)\n('5 + 3 / 4 - 2 * 7', '=', -8.25)\n('4 / 5 - 2 * 8 + 7', '=', -8.2)\n('8 + 5 - 7 * 3 / 1', '=', -8.0)\n('2 - 6 * 9 / 5 + 1', '=', -7.8)\n('7 + 5 / 4 - 8 * 2', '=', -7.75)\n('7 - 5 * 3 + 2 / 4', '=', -7.5)\n('8 / 5 - 3 * 6 + 9', '=', -7.4)\n('3 / 4 + 6 - 2 * 7', '=', -7.25)\n('6 - 7 * 2 + 4 / 5', '=', -7.2)\n('4 + 5 - 8 / 3 * 6', '=', -7.0)\n('1 + 3 * 2 / 5 - 9', '=', -6.8)\n('8 - 7 / 4 * 9 + 1', '=', -6.75)\n('9 - 8 * 2 + 3 / 6', '=', -6.5)\n('8 / 5 - 7 * 2 + 6', '=', -6.4)\n('2 + 3 - 9 / 4 * 5', '=', -6.25)\n('1 + 4 - 7 / 5 * 8', '=', -6.2)\n('2 - 7 / 4 * 8 + 6', '=', -6.0)\n('1 + 3 * 2 / 5 - 8', '=', -5.8)\n('2 / 8 + 9 - 5 * 3', '=', -5.75)\n('2 * 1 / 5 - 9 + 3', '=', -5.6)\n('7 - 3 * 9 / 2 + 1', '=', -5.5)\n('2 - 8 * 9 / 5 + 7', '=', -5.4)\n('1 + 2 / 6 * 3 - 7', '=', -5.0)\n('3 - 9 + 6 * 1 / 5', '=', -4.8)\n('5 + 6 - 9 / 4 * 7', '=', -4.75)\n('3 + 2 / 5 - 1 * 8', '=', -4.6)\n('8 - 7 * 2 + 6 / 4', '=', -4.5)\n('8 / 5 + 3 - 9 * 1', '=', -4.4)\n('3 - 9 + 2 * 7 / 8', '=', -4.25)\n('9 / 5 + 2 - 8 * 1', '=', -4.2)\n('9 + 7 - 5 * 4 / 1', '=', -4.0)\n('3 - 1 * 8 + 6 / 5', '=', -3.8)\n('8 + 1 / 4 - 2 * 6', '=', -3.75)\n('6 * 2 / 5 + 1 - 7', '=', -3.6)\n('6 + 1 - 3 / 2 * 7', '=', -3.5)\n('4 + 7 - 8 / 5 * 9', '=', -3.4)\n('3 * 1 / 4 + 5 - 9', '=', -3.25)\n('6 - 8 / 5 * 7 + 2', '=', -3.2)\n('7 - 8 / 4 * 6 + 2', '=', -3.0)\n('1 + 7 - 6 / 5 * 9', '=', -2.8)\n('3 + 1 - 6 / 8 * 9', '=', -2.75)\n('2 / 5 * 6 + 4 - 9', '=', -2.6)\n('9 / 6 * 3 - 8 + 1', '=', -2.5)\n('2 / 5 * 4 + 3 - 7', '=', -2.4)\n('6 - 1 * 9 + 3 / 4', '=', -2.25)\n('9 / 5 + 8 - 6 * 2', '=', -2.2)\n('8 - 4 / 1 * 3 + 2', '=', -2.0)\n('9 - 3 * 4 + 6 / 5', '=', -1.8)\n('2 / 8 * 9 + 1 - 5', '=', -1.75)\n('7 / 5 - 9 + 3 * 2', '=', -1.6)\n('1 - 6 + 7 * 4 / 8', '=', -1.5)\n('3 / 5 * 1 + 6 - 8', '=', -1.4)\n('6 - 9 + 1 / 4 * 7', '=', -1.25)\n('7 - 9 * 1 + 4 / 5', '=', -1.2)\n('6 - 8 / 1 * 2 + 9', '=', -1.0)\n('2 * 3 - 8 + 6 / 5', '=', -0.8)\n('9 - 2 * 5 + 1 / 4', '=', -0.75)\n('7 * 1 - 8 + 2 / 5', '=', -0.6)\n('1 / 2 * 3 + 6 - 8', '=', -0.5)\n('1 * 2 + 8 / 5 - 4', '=', -0.4)\n('3 / 4 + 5 - 6 * 1', '=', -0.25)\n('2 + 9 - 8 / 5 * 7', '=', -0.2)\n('6 * 2 / 3 + 1 - 5', '=', 0.0)\n('4 + 3 / 5 * 7 - 8', '=', 0.2)\n('2 - 4 + 3 / 8 * 6', '=', 0.25)\n('1 * 3 - 4 + 7 / 5', '=', 0.4)\n('1 + 7 - 6 * 5 / 4', '=', 0.5)\n('2 / 5 * 4 - 9 + 8', '=', 0.6)\n('1 - 9 + 7 / 4 * 5', '=', 0.75)\n('9 / 5 * 1 - 3 + 2', '=', 0.8)\n('3 - 4 * 2 + 6 / 1', '=', 1.0)\n('7 * 3 / 5 + 6 - 9', '=', 1.2)\n('6 - 7 + 9 / 8 * 2', '=', 1.25)\n('3 * 4 / 5 + 6 - 7', '=', 1.4)\n('3 / 6 - 7 + 2 * 4', '=', 1.5)\n('6 + 2 - 4 / 5 * 8', '=', 1.6)\n('3 / 4 - 8 * 1 + 9', '=', 1.75)\n('4 / 5 + 9 * 1 - 8', '=', 1.8)\n('6 * 4 / 8 + 1 - 2', '=', 2.0)\n('8 - 1 * 7 + 6 / 5', '=', 2.2)\n('2 / 8 - 5 + 7 * 1', '=', 2.25)\n('4 - 7 + 9 / 5 * 3', '=', 2.4)\n('3 / 6 * 9 - 4 + 2', '=', 2.5)\n('8 / 5 - 1 * 6 + 7', '=', 2.6)\n('8 + 6 - 5 * 9 / 4', '=', 2.75)\n('3 - 4 / 5 * 9 + 7', '=', 2.8)\n('2 / 6 * 3 - 7 + 9', '=', 3.0)\n('9 - 7 + 6 * 1 / 5', '=', 3.2)\n('9 / 4 * 1 + 7 - 6', '=', 3.25)\n('2 - 4 + 9 * 3 / 5', '=', 3.4)\n('5 * 3 / 6 - 1 + 2', '=', 3.5)\n('3 + 1 * 2 - 7 / 5', '=', 3.6)\n('5 + 2 / 8 * 7 - 3', '=', 3.75)\n('9 - 6 + 1 / 5 * 4', '=', 3.8)\n('9 - 4 * 3 / 2 + 1', '=', 4.0)\n('2 + 4 - 9 / 5 * 1', '=', 4.2)\n('7 - 5 + 2 * 9 / 8', '=', 4.25)\n('3 * 4 / 5 - 6 + 8', '=', 4.4)\n('5 * 3 / 6 + 9 - 7', '=', 4.5)\n('6 + 8 / 5 * 1 - 3', '=', 4.6)\n('7 * 1 / 4 + 9 - 6', '=', 4.75)\n('1 + 7 - 2 / 5 * 8', '=', 4.8)\n('3 * 4 / 2 - 9 + 8', '=', 5.0)\n('2 + 8 - 4 / 5 * 6', '=', 5.2)\n('5 / 4 * 9 + 1 - 7', '=', 5.25)\n('3 + 1 * 4 - 8 / 5', '=', 5.4)\n('2 + 4 * 9 / 8 - 1', '=', 5.5)\n('4 - 2 + 3 * 6 / 5', '=', 5.6)\n('9 - 5 + 1 * 7 / 4', '=', 5.75)\n('7 - 2 + 1 / 5 * 4', '=', 5.8)\n('5 * 2 - 7 + 3 / 1', '=', 6.0)\n('6 / 5 - 4 + 1 * 9', '=', 6.2)\n('6 - 7 / 4 * 5 + 9', '=', 6.25)\n('7 + 3 / 5 * 9 - 6', '=', 6.4)\n('6 + 9 / 2 * 1 - 4', '=', 6.5)\n('7 + 9 / 5 * 2 - 4', '=', 6.6)\n('6 / 8 * 1 + 9 - 3', '=', 6.75)\n('4 / 5 * 6 - 7 + 9', '=', 6.8)\n('6 / 3 - 4 + 1 * 9', '=', 7.0)\n('1 + 2 * 4 - 9 / 5', '=', 7.2)\n('2 / 8 * 5 - 1 + 7', '=', 7.25)\n('4 / 5 * 3 - 1 + 6', '=', 7.4)\n('1 - 4 + 7 / 2 * 3', '=', 7.5)\n('9 - 3 + 4 * 2 / 5', '=', 7.6)\n('9 + 1 - 3 * 6 / 8', '=', 7.75)\n('6 - 1 + 7 * 2 / 5', '=', 7.8)\n('1 - 5 + 4 / 3 * 9', '=', 8.0)\n('7 / 5 * 8 + 6 - 9', '=', 8.2)\n('7 / 8 * 6 + 5 - 2', '=', 8.25)\n('1 * 6 + 4 - 8 / 5', '=', 8.4)\n('9 + 3 - 1 * 7 / 2', '=', 8.5)\n('6 - 3 + 4 / 5 * 7', '=', 8.6)\n('2 - 1 / 4 * 5 + 8', '=', 8.75)\n('8 - 6 * 1 / 5 + 2', '=', 8.8)\n('6 / 3 * 2 + 9 - 4', '=', 9.0)\n('6 * 2 + 1 / 5 - 3', '=', 9.2)\n('2 + 1 * 8 - 3 / 4', '=', 9.25)\n('8 + 4 * 3 / 5 - 1', '=', 9.4)\n('2 - 3 + 7 * 9 / 6', '=', 9.5)\n('1 * 3 - 2 / 5 + 7', '=', 9.6)\n('3 / 4 * 9 + 5 - 2', '=', 9.75)\n('8 / 5 * 3 - 4 + 9', '=', 9.8)\n('6 / 2 * 3 + 8 - 7', '=', 10.0)\n('3 * 4 + 1 / 5 - 2', '=', 10.2)\n('6 * 7 / 8 + 9 - 4', '=', 10.25)\n('3 * 9 / 5 + 6 - 1', '=', 10.4)\n('6 / 4 - 1 + 5 * 2', '=', 10.5)\n('2 - 4 + 9 / 5 * 7', '=', 10.6)\n('6 - 2 + 9 * 3 / 4', '=', 10.75)\n('4 / 5 + 2 * 8 - 6', '=', 10.8)\n"
]
],
[
[
"Este algoritmo mejora al algoritmo de fuerza bruta debido a que al no tener que generar muchas soluciones para la misma entrada para luego elegir una, sino que se elije la qprimera que cumpla con lo solicitado, logrando así respuestas más rápidas.",
"_____no_output_____"
],
[
"(*)Calcula la complejidad del algoritmo ",
"_____no_output_____"
],
[
"Respuesta",
"_____no_output_____"
],
[
"O(n^2)",
"_____no_output_____"
],
[
"Según el problema (y tenga sentido), diseña un juego de datos de entrada aleatorios",
"_____no_output_____"
]
],
[
[
"sizeDataseet = 20\n\n\nvaloresDeseados = []\nfor valorDeseado in range(sizeDataseet):\n valorAzar = random.random()*10\n valorAzar = float(\"{0:.2f}\".format(valorAzar))\n valoresDeseados.append(valorAzar)\n\nprint(valoresDeseados)",
"[2.3, 9.82, 2.15, 8.72, 3.18, 4.42, 4.31, 2.75, 4.76, 1.19, 0.51, 7.58, 9.02, 2.12, 0.22, 6.34, 1.52, 8.96, 8.13, 5.09]\n"
]
],
[
[
"Aplica el algoritmo al juego de datos generado",
"_____no_output_____"
]
],
[
[
"print(\"Total numeros buscar respuesta: \", len(valoresDeseados))\nrespuestas, numerosNoEncontrados =BuscarOperacionesPorListado(valoresDeseados)\nprint(\"Con rta: \",len(respuestas))\nprint(\"Sin rta: \",len(numerosNoEncontrados))\n\n\n\nprint(\"Numeros no encontrados: \")\nfor rta in numerosNoEncontrados:\n print(rta)\n pass\n\nprint(\"Numeros con respuesta encontrada: \")\nfor rta in respuestas:\n print(rta)\n pass",
"Total numeros buscar respuesta: 20\nCon rta: 3\nSin rta: 17\nNumeros no encontrados: \n2.3\n9.82\n2.15\n8.72\n3.18\n4.42\n4.31\n4.76\n1.19\n0.51\n7.58\n9.02\n6.34\n1.52\n8.96\n8.13\n5.09\nNumeros con respuesta encontrada: \n('2 / 8 * 3 + 9 - 7', '=', 2.75)\n('4 - 3 + 9 * 1 / 8', '=', 2.12)\n('1 - 3 + 5 / 9 * 4', '=', 0.22)\n"
]
],
[
[
"## Conclusiones",
"_____no_output_____"
],
[
"Para este problema se ha usado recursividad y el algoritmo de divide y venceras para la solución del problema, pero usando algoritmos heuristicos seguro se podrá lograr obtener los mismos resultados en un tiempo menor",
"_____no_output_____"
],
[
"Las restricciones del problema hacen que el algoritmo le resulte dificil generar soluciones para valores float no multiplos de 0.25.... ",
"_____no_output_____"
],
[
"El algoritmo generado fue por métodos exactos, pero un ejercicio futuro sería usar algoritmos heuristicos para la solución. Incluso la posibilidad de generar los set de signos y set de numeros (ambos son el set de datos) mediante algoritmos geneticos, y comparar las soluciones.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cb432ed16ed91abb72ee401d41e0a7f92e08f87a | 20,481 | ipynb | Jupyter Notebook | courses/machine_learning/deepdive2/structured/labs/4c_keras_wide_and_deep_babyweight.ipynb | ecuriotto/training-data-analyst | 3da6b9a4f9715c6cf9653c2025ac071b15011111 | [
"Apache-2.0"
] | 4 | 2019-02-14T19:12:43.000Z | 2021-12-21T20:48:48.000Z | courses/machine_learning/deepdive2/structured/labs/4c_keras_wide_and_deep_babyweight.ipynb | jgamblegeorge/training-data-analyst | 9f9006e7b740f59798ac6016d55dd4e38a1e0528 | [
"Apache-2.0"
] | 9 | 2020-09-26T01:05:43.000Z | 2022-03-12T00:29:52.000Z | courses/machine_learning/deepdive2/structured/labs/4c_keras_wide_and_deep_babyweight.ipynb | jgamblegeorge/training-data-analyst | 9f9006e7b740f59798ac6016d55dd4e38a1e0528 | [
"Apache-2.0"
] | 2 | 2020-10-07T12:13:48.000Z | 2021-01-05T13:59:24.000Z | 36.185512 | 553 | 0.606269 | [
[
[
"# LAB 4c: Create Keras Wide and Deep model.\n\n**Learning Objectives**\n\n1. Set CSV Columns, label column, and column defaults\n1. Make dataset of features and label from CSV files\n1. Create input layers for raw features\n1. Create feature columns for inputs\n1. Create wide layer, deep dense hidden layers, and output layer\n1. Create custom evaluation metric\n1. Build wide and deep model tying all of the pieces together\n1. Train and evaluate\n\n\n## Introduction \nIn this notebook, we'll be using Keras to create a wide and deep model to predict the weight of a baby before it is born.\n\nWe'll start by defining the CSV column names, label column, and column defaults for our data inputs. Then, we'll construct a tf.data Dataset of features and the label from the CSV files and create inputs layers for the raw features. Next, we'll set up feature columns for the model inputs and build a wide and deep neural network in Keras. We'll create a custom evaluation metric and build our wide and deep model. Finally, we'll train and evaluate our model.\n\nEach learning objective will correspond to a __#TODO__ in this student lab notebook -- try to complete this notebook first and then review the [solution notebook](../solutions/4c_keras_wide_and_deep_babyweight.ipynb).",
"_____no_output_____"
],
[
"## Load necessary libraries",
"_____no_output_____"
]
],
[
[
"import datetime\nimport os\nimport shutil\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport tensorflow as tf\nprint(tf.__version__)",
"_____no_output_____"
]
],
[
[
"## Verify CSV files exist\n\nIn the seventh lab of this series [4a_sample_babyweight](../solutions/4a_sample_babyweight.ipynb), we sampled from BigQuery our train, eval, and test CSV files. Verify that they exist, otherwise go back to that lab and create them.",
"_____no_output_____"
]
],
[
[
"%%bash\nls *.csv",
"_____no_output_____"
],
[
"%%bash\nhead -5 *.csv",
"_____no_output_____"
]
],
[
[
"## Create Keras model",
"_____no_output_____"
],
[
"### Lab Task #1: Set CSV Columns, label column, and column defaults.\n\nNow that we have verified that our CSV files exist, we need to set a few things that we will be using in our input function.\n* `CSV_COLUMNS` are going to be our header names of our columns. Make sure that they are in the same order as in the CSV files\n* `LABEL_COLUMN` is the header name of the column that is our label. We will need to know this to pop it from our features dictionary.\n* `DEFAULTS` is a list with the same length as `CSV_COLUMNS`, i.e. there is a default for each column in our CSVs. Each element is a list itself with the default value for that CSV column.",
"_____no_output_____"
]
],
[
[
"# Determine CSV, label, and key columns\n# TODO: Create list of string column headers, make sure order matches.\nCSV_COLUMNS = [\"\"]\n\n# TODO: Add string name for label column\nLABEL_COLUMN = \"\"\n\n# Set default values for each CSV column as a list of lists.\n# Treat is_male and plurality as strings.\nDEFAULTS = []",
"_____no_output_____"
]
],
[
[
"### Lab Task #2: Make dataset of features and label from CSV files.\n\nNext, we will write an input_fn to read the data. Since we are reading from CSV files we can save ourself from trying to recreate the wheel and can use `tf.data.experimental.make_csv_dataset`. This will create a CSV dataset object. However we will need to divide the columns up into features and a label. We can do this by applying the map method to our dataset and popping our label column off of our dictionary of feature tensors.",
"_____no_output_____"
]
],
[
[
"def features_and_labels(row_data):\n \"\"\"Splits features and labels from feature dictionary.\n\n Args:\n row_data: Dictionary of CSV column names and tensor values.\n Returns:\n Dictionary of feature tensors and label tensor.\n \"\"\"\n label = row_data.pop(LABEL_COLUMN)\n\n return row_data, label # features, label\n\n\ndef load_dataset(pattern, batch_size=1, mode=tf.estimator.ModeKeys.EVAL):\n \"\"\"Loads dataset using the tf.data API from CSV files.\n\n Args:\n pattern: str, file pattern to glob into list of files.\n batch_size: int, the number of examples per batch.\n mode: tf.estimator.ModeKeys to determine if training or evaluating.\n Returns:\n `Dataset` object.\n \"\"\"\n # TODO: Make a CSV dataset\n dataset = tf.data.experimental.make_csv_dataset()\n\n # TODO: Map dataset to features and label\n dataset = dataset.map() # features, label\n\n # Shuffle and repeat for training\n if mode == tf.estimator.ModeKeys.TRAIN:\n dataset = dataset.shuffle(buffer_size=1000).repeat()\n\n # Take advantage of multi-threading; 1=AUTOTUNE\n dataset = dataset.prefetch(buffer_size=1)\n\n return dataset",
"_____no_output_____"
]
],
[
[
"### Lab Task #3: Create input layers for raw features.\n\nWe'll need to get the data read in by our input function to our model function, but just how do we go about connecting the dots? We can use Keras input layers [(tf.Keras.layers.Input)](https://www.tensorflow.org/api_docs/python/tf/keras/Input) by defining:\n* shape: A shape tuple (integers), not including the batch size. For instance, shape=(32,) indicates that the expected input will be batches of 32-dimensional vectors. Elements of this tuple can be None; 'None' elements represent dimensions where the shape is not known.\n* name: An optional name string for the layer. Should be unique in a model (do not reuse the same name twice). It will be autogenerated if it isn't provided.\n* dtype: The data type expected by the input, as a string (float32, float64, int32...)",
"_____no_output_____"
]
],
[
[
"def create_input_layers():\n \"\"\"Creates dictionary of input layers for each feature.\n\n Returns:\n Dictionary of `tf.Keras.layers.Input` layers for each feature.\n \"\"\"\n # TODO: Create dictionary of tf.keras.layers.Input for each dense feature\n deep_inputs = {}\n\n # TODO: Create dictionary of tf.keras.layers.Input for each sparse feature\n wide_inputs = {}\n\n inputs = {**wide_inputs, **deep_inputs}\n\n return inputs",
"_____no_output_____"
]
],
[
[
"### Lab Task #4: Create feature columns for inputs.\n\nNext, define the feature columns. `mother_age` and `gestation_weeks` should be numeric. The others, `is_male` and `plurality`, should be categorical. Remember, only dense feature columns can be inputs to a DNN.",
"_____no_output_____"
]
],
[
[
"def create_feature_columns(nembeds):\n \"\"\"Creates wide and deep dictionaries of feature columns from inputs.\n\n Args:\n nembeds: int, number of dimensions to embed categorical column down to.\n Returns:\n Wide and deep dictionaries of feature columns.\n \"\"\"\n # TODO: Create deep feature columns for numeric features\n deep_fc = {}\n\n # TODO: Create wide feature columns for categorical features\n wide_fc = {}\n\n # TODO: Bucketize the float fields. This makes them wide\n\n # TODO: Cross all the wide cols, have to do the crossing before we one-hot\n\n # TODO: Embed cross and add to deep feature columns\n\n return wide_fc, deep_fc",
"_____no_output_____"
]
],
[
[
"### Lab Task #5: Create wide and deep model and output layer.\n\nSo we've figured out how to get our inputs ready for machine learning but now we need to connect them to our desired output. Our model architecture is what links the two together. We need to create a wide and deep model now. The wide side will just be a linear regression or dense layer. For the deep side, let's create some hidden dense layers. All of this will end with a single dense output layer. This is regression so make sure the output layer activation is correct and that the shape is right.",
"_____no_output_____"
]
],
[
[
"def get_model_outputs(wide_inputs, deep_inputs, dnn_hidden_units):\n \"\"\"Creates model architecture and returns outputs.\n\n Args:\n wide_inputs: Dense tensor used as inputs to wide side of model.\n deep_inputs: Dense tensor used as inputs to deep side of model.\n dnn_hidden_units: List of integers where length is number of hidden\n layers and ith element is the number of neurons at ith layer.\n Returns:\n Dense tensor output from the model.\n \"\"\"\n # Hidden layers for the deep side\n layers = [int(x) for x in dnn_hidden_units]\n deep = deep_inputs\n\n # TODO: Create DNN model for the deep side\n deep_out =\n\n # TODO: Create linear model for the wide side\n wide_out =\n\n # Concatenate the two sides\n both = tf.keras.layers.concatenate(\n inputs=[deep_out, wide_out], name=\"both\")\n\n # TODO: Create final output layer\n\n return output",
"_____no_output_____"
]
],
[
[
"### Lab Task #6: Create custom evaluation metric.\n\nWe want to make sure that we have some useful way to measure model performance for us. Since this is regression, we would like to know the RMSE of the model on our evaluation dataset, however, this does not exist as a standard evaluation metric, so we'll have to create our own by using the true and predicted labels.",
"_____no_output_____"
]
],
[
[
"def rmse(y_true, y_pred):\n \"\"\"Calculates RMSE evaluation metric.\n\n Args:\n y_true: tensor, true labels.\n y_pred: tensor, predicted labels.\n Returns:\n Tensor with value of RMSE between true and predicted labels.\n \"\"\"\n # TODO: Calculate RMSE from true and predicted labels\n pass",
"_____no_output_____"
]
],
[
[
"### Lab Task #7: Build wide and deep model tying all of the pieces together.\n\nExcellent! We've assembled all of the pieces, now we just need to tie them all together into a Keras Model. This is NOT a simple feedforward model with no branching, side inputs, etc. so we can't use Keras' Sequential Model API. We're instead going to use Keras' Functional Model API. Here we will build the model using [tf.keras.models.Model](https://www.tensorflow.org/api_docs/python/tf/keras/Model) giving our inputs and outputs and then compile our model with an optimizer, a loss function, and evaluation metrics.",
"_____no_output_____"
]
],
[
[
"def build_wide_deep_model(dnn_hidden_units=[64, 32], nembeds=3):\n \"\"\"Builds wide and deep model using Keras Functional API.\n\n Returns:\n `tf.keras.models.Model` object.\n \"\"\"\n # Create input layers\n inputs = create_input_layers()\n\n # Create feature columns\n wide_fc, deep_fc = create_feature_columns(nembeds)\n\n # The constructor for DenseFeatures takes a list of numeric columns\n # The Functional API in Keras requires: LayerConstructor()(inputs)\n\n # TODO: Add wide and deep feature colummns\n wide_inputs = tf.keras.layers.DenseFeatures(\n feature_columns=#TODO, name=\"wide_inputs\")(inputs)\n deep_inputs = tf.keras.layers.DenseFeatures(\n feature_columns=#TODO, name=\"deep_inputs\")(inputs)\n\n # Get output of model given inputs\n output = get_model_outputs(wide_inputs, deep_inputs, dnn_hidden_units)\n\n # Build model and compile it all together\n model = tf.keras.models.Model(inputs=inputs, outputs=output)\n\n # TODO: Add custom eval metrics to list\n model.compile(optimizer=\"adam\", loss=\"mse\", metrics=[\"mse\"])\n\n return model\n\nprint(\"Here is our wide and deep architecture so far:\\n\")\nmodel = build_wide_deep_model()\nprint(model.summary())",
"_____no_output_____"
]
],
[
[
"We can visualize the wide and deep network using the Keras plot_model utility.",
"_____no_output_____"
]
],
[
[
"tf.keras.utils.plot_model(\n model=model, to_file=\"wd_model.png\", show_shapes=False, rankdir=\"LR\")",
"_____no_output_____"
]
],
[
[
"## Run and evaluate model",
"_____no_output_____"
],
[
"### Lab Task #8: Train and evaluate.\n\nWe've built our Keras model using our inputs from our CSV files and the architecture we designed. Let's now run our model by training our model parameters and periodically running an evaluation to track how well we are doing on outside data as training goes on. We'll need to load both our train and eval datasets and send those to our model through the fit method. Make sure you have the right pattern, batch size, and mode when loading the data. Also, don't forget to add the callback to TensorBoard.",
"_____no_output_____"
]
],
[
[
"TRAIN_BATCH_SIZE = 32\nNUM_TRAIN_EXAMPLES = 10000 * 5 # training dataset repeats, it'll wrap around\nNUM_EVALS = 5 # how many times to evaluate\n# Enough to get a reasonable sample, but not so much that it slows down\nNUM_EVAL_EXAMPLES = 10000\n\n# TODO: Load training dataset\ntrainds = load_dataset()\n\n# TODO: Load evaluation dataset\nevalds = load_dataset().take(count=NUM_EVAL_EXAMPLES // 1000)\n\nsteps_per_epoch = NUM_TRAIN_EXAMPLES // (TRAIN_BATCH_SIZE * NUM_EVALS)\n\nlogdir = os.path.join(\n \"logs\", datetime.datetime.now().strftime(\"%Y%m%d-%H%M%S\"))\ntensorboard_callback = tf.keras.callbacks.TensorBoard(\n log_dir=logdir, histogram_freq=1)\n\n# TODO: Fit model on training dataset and evaluate every so often\nhistory = model.fit()",
"_____no_output_____"
]
],
[
[
"### Visualize loss curve",
"_____no_output_____"
]
],
[
[
"# Plot\nnrows = 1\nncols = 2\nfig = plt.figure(figsize=(10, 5))\n\nfor idx, key in enumerate([\"loss\", \"rmse\"]):\n ax = fig.add_subplot(nrows, ncols, idx+1)\n plt.plot(history.history[key])\n plt.plot(history.history[\"val_{}\".format(key)])\n plt.title(\"model {}\".format(key))\n plt.ylabel(key)\n plt.xlabel(\"epoch\")\n plt.legend([\"train\", \"validation\"], loc=\"upper left\");",
"_____no_output_____"
]
],
[
[
"### Save the model",
"_____no_output_____"
]
],
[
[
"OUTPUT_DIR = \"babyweight_trained_wd\"\nshutil.rmtree(OUTPUT_DIR, ignore_errors=True)\nEXPORT_PATH = os.path.join(\n OUTPUT_DIR, datetime.datetime.now().strftime(\"%Y%m%d%H%M%S\"))\ntf.saved_model.save(\n obj=model, export_dir=EXPORT_PATH) # with default serving function\nprint(\"Exported trained model to {}\".format(EXPORT_PATH))",
"_____no_output_____"
],
[
"!ls $EXPORT_PATH",
"_____no_output_____"
]
],
[
[
"## Lab Summary: \nIn this lab, we started by defining the CSV column names, label column, and column defaults for our data inputs. Then, we constructed a tf.data Dataset of features and the label from the CSV files and created inputs layers for the raw features. Next, we set up feature columns for the model inputs and built a wide and deep neural network in Keras. We created a custom evaluation metric and built our wide and deep model. Finally, we trained and evaluated our model.",
"_____no_output_____"
],
[
"Copyright 2019 Google Inc. Licensed under the Apache License, Version 2.0 (the \"License\"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an \"AS IS\" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
cb432fbe296b89b2bbc26dad5442c189381bb7fb | 15,713 | ipynb | Jupyter Notebook | others/hive_command_note.ipynb | johnnychiuchiu/Machine-Learning | 0fd6fd2c08025134cf7d20b245c39f82d5453e14 | [
"MIT"
] | 13 | 2018-03-19T18:16:03.000Z | 2022-03-22T03:44:13.000Z | others/hive_command_note.ipynb | johnnychiuchiu/Machine-Learning | 0fd6fd2c08025134cf7d20b245c39f82d5453e14 | [
"MIT"
] | null | null | null | others/hive_command_note.ipynb | johnnychiuchiu/Machine-Learning | 0fd6fd2c08025134cf7d20b245c39f82d5453e14 | [
"MIT"
] | 7 | 2018-01-11T04:03:11.000Z | 2021-01-22T07:56:42.000Z | 25.262058 | 275 | 0.56291 | [
[
[
"# Hive Command Note",
"_____no_output_____"
],
[
"**Outline**\n\n* [Introduction](#intro)\n* [Syntax](#syntax)\n* [Reference](#refer)",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
],
[
"Hive is a data warehouse infrastructure tool to process structured data in Hadoop. It resides on top of Hadoop to \nsummarize Big Data, and makes querying and analyzing easy.",
"_____no_output_____"
],
[
"* **Access Hive**: in cmd, type *`hive`*\n* **Run hive script**: hive -f xxx.hql",
"_____no_output_____"
],
[
"> **Database in HIVE**",
"_____no_output_____"
],
[
"Each database is a collection of tables. \n[link](http://www.tutorialspoint.com/hive/hive_create_database.htm)",
"_____no_output_____"
]
],
[
[
"# create database\nCREATE DATABASE [IF NOT EXISTS] userdb;\n\n# show all the databases\nshow databases;\n\n# use a certain database, every table we create afterwards will be within the database\nuse databaseName;",
"_____no_output_____"
],
[
"# drop database\nDROP DATABASE IF EXISTS userdb;",
"_____no_output_____"
]
],
[
[
"> **Create Table**",
"_____no_output_____"
],
[
"1. employees.csv -> HDFS\n2. create table & load employees.csv\n3. drop employees table (Be careful that by dropping the table, HIVE will actually delete the original csv not just the table itself). Instead, we can create an external table. \n * External tables: if you drop them, data in hdfs will NOT be deleted.",
"_____no_output_____"
],
[
"**Data Types**\n* **Integers**\n * *TINYINT*—1 byte integer\n * *SMALLINT*—2 byte integer\n * *INT*—4 byte integer\n * *BIGINT*—8 byte integer\n* **Boolean type**\n * *BOOLEAN*—TRUE/FALSE\n* **Floating point numbers**\n * *FLOAT*—single precision\n * *DOUBLE*—Double precision\n* **Fixed point numbers**\n * *DECIMAL*—a fixed point value of user defined scale and precision\n* **String types**\n * *STRING*—sequence of characters in a specified character set\n * *VARCHAR*—sequence of characters in a specified character set with a maximum length\n * *CHAR*—sequence of characters in a specified character set with a defined length\n* **Date and time types**\n * *TIMESTAMP*— a specific point in time, up to nanosecond precision\n * *DATE*—a date\n* **Binary types**\n * *BINARY*—a sequence of bytes\n\n**Complex Types**\n* **Structs**: the elements within the type can be accessed using the DOT (.) notation. For example, for a column c of type STRUCT {a INT; b INT}, the a field is accessed by the expression c.a\n * format: `<first, second>`\n * access: mystruct.first \n* **Maps (key-value tuples)**: The elements are accessed using ['element name'] notation. For example in a map M comprising of a mapping from 'group' -> gid the gid value can be accessed using M['group']\n * format: index based\n * access: myarray[0]\n* **Arrays (indexable lists)**: The elements in the array have to be in the same type. Elements can be accessed using the [n] notation where n is an index (zero-based) into the array. For example, for an array A having the elements ['a', 'b', 'c'], A[1] retruns 'b'.\n * format: key based\n * access: myMap['KEY']",
"_____no_output_____"
],
[
"* **ROW FORMAT DELIMITED**: one row per line\n* **FIELDS TERMINATED BY ','**: split column by comma",
"_____no_output_____"
]
],
[
[
"# use external table in this example\nCREATE EXTERNAL TABLE movies(\n userid INT,\n movieid INT,\n rating INT,\n timestamp TIMESTAMP)\nROW FORMAT DELIMITED\nFIELDS TERMINATED BY '\\t';",
"_____no_output_____"
],
[
"CREATE TABLE myemployees( \n name STRING, \n salary FLOAT, \n subordinates ARRAY<STRING>, \n deductions MAP<STRING, FLOAT>, \n address STRUCT<street:STRING, city:STRING, state:STRING,zip:INT>)\nROW FORMAT DELIMITED # This line is telling Hive to expect the file to contain one row per line. So basically, we are telling Hive that when it finds a new line character that means is a new records.\nFIELDS TERMINATED BY ',' # split column by comma\nCOLLECTION ITEMS TERMINATED BY '#' # split the struct type item by `#`\nMAP KEYS TERMINATED BY '-' # split the map type column by `-`\nLINES TERMINATED BY '\\N'; # separate line by `\\N`",
"_____no_output_____"
]
],
[
[
"> **load file from hdfs into hive**",
"_____no_output_____"
],
[
"[StackOverFlow: Which is the difference between LOAD DATA INPATH and LOAD DATA LOCAL INPATH in HIVE](https://stackoverflow.com/questions/43204716/which-is-the-difference-between-load-data-inpath-and-load-data-local-inpath-in-h/43205970)",
"_____no_output_____"
]
],
[
[
"# load data into table movie. Noted that the path is hdfs path\n# noted that the original file in hdfs://hw5/ will be move to ''hdfs://wolf.xxx.ooo.edu:8000/user/hive/warehouse/jchiu.db/movie/u.data'' after this command\nLOAD DATA INPATH 'hw5/u.data' into table movie;\n\n# load data into table movie. Noted that the path is local path\n# LOCAL is identifier to specify the local path. It is optional.\n# when using LOCAL, the file is copied to the hive directory\nLOAD DATA LOCAL INPATH 'localpath' into table movie;\nLOAD DATA LOCAL INPATH '/home/public/course/recommendationEngine/u.data' into table movies;",
"_____no_output_____"
],
[
"# create an external table\nCREATE EXTERNAL TABLE myemployees",
"_____no_output_____"
],
[
"LOAD DATA INPATH '...' INTO TABLE employees",
"_____no_output_____"
]
],
[
[
"> **see column name; describe table**",
"_____no_output_____"
]
],
[
[
"# method 1\ndescribe database.tablename;\n\n# method 2\nuse database;\ndescribe tablename;",
"_____no_output_____"
]
],
[
[
"> **Query**",
"_____no_output_____"
]
],
[
[
"SELECT [ALL | DISTINCT] select_expr, select_expr, ... \nFROM table_reference \n[WHERE where_condition] \n[GROUP BY col_list] \n[HAVING having_condition] \n[ORDER BY col_list]] \n[LIMIT number];",
"_____no_output_____"
],
[
"select address.city from employees",
"_____no_output_____"
]
],
[
[
"> **show tables**",
"_____no_output_____"
]
],
[
[
"# if already use database, it'll show tables in this database; if not, it'll show all the tables\nshow tables;",
"_____no_output_____"
]
],
[
[
"> **drop tables**",
"_____no_output_____"
],
[
"[] means optional. When used, we don't need these.",
"_____no_output_____"
]
],
[
[
"DROP TABLE [IF EXISTS] table_name;",
"_____no_output_____"
]
],
[
[
"> **create view in hive**",
"_____no_output_____"
]
],
[
[
"CREATE VIEW [IF NOT EXISTS] emp_30000 AS\nSELECT * FROM employee\nWHERE salary>30000;",
"_____no_output_____"
]
],
[
[
"> **drop a view**",
"_____no_output_____"
]
],
[
[
"DROP VIEW view_name",
"_____no_output_____"
]
],
[
[
"> **join**",
"_____no_output_____"
],
[
"[tutorialspoint: hiveql join](https://www.tutorialspoint.com/hive/hiveql_joins.htm)\n\nSyntax-wise is essentially the same as SQL",
"_____no_output_____"
],
[
"> **hive built in aggregation functions**",
"_____no_output_____"
],
[
"[treasuredata: hive-aggregate-functions](https://docs.treasuredata.com/articles/hive-aggregate-functions)",
"_____no_output_____"
],
[
"> **hive built in operators**",
"_____no_output_____"
],
[
"[tutorialspoint: built-in operators](https://www.tutorialspoint.com/hive/hive_built_in_operators.htm)\n\ndeal with NULL/NA, equal...etc",
"_____no_output_____"
],
[
"> **writing data into the filesystem from queries**",
"_____no_output_____"
],
[
"[hive doc](https://cwiki.apache.org/confluence/display/Hive/LanguageManual+DML#LanguageManualDML-Writingdataintothefilesystemfromqueries)\n\n[Hive INSERT OVERWRITE DIRECTORY command output is not separated by a delimiter. Why?](https://stackoverflow.com/questions/16459790/hive-insert-overwrite-directory-command-output-is-not-separated-by-a-delimiter)\nThe discussion happened at 2013. Not sure if it's still valid or not.",
"_____no_output_____"
],
[
"* If LOCAL keyword is used, Hive will write data to the directory on the local file system.\n* Data written to the filesystem is serialized as text with columns separated by ^A and rows separated by newlines. If any of the columns are not of primitive type, then those columns are serialized to JSON format.",
"_____no_output_____"
]
],
[
[
"INSERT OVERWRITE [LOCAL] DIRECTORY directory1 \n SELECT ... FROM ...",
"_____no_output_____"
]
],
[
[
"* **STORED AS TEXTFILE**: Stored as plain text files. TEXTFILE is the default file format, unless the configuration parameter hive.default.fileformat has a different setting.",
"_____no_output_____"
]
],
[
[
"# in a newer hive version, this should work just fine\nINSERT OVERWRITE [LOCAL] DIRECTORY directory1 \nROW FORMAT DELIMITED\nFIELDS TERMINATED BY '\\t'\n SELECT ... FROM ...",
"_____no_output_____"
],
[
"# another way to work around this\n# concat_ws: concat column together as string\nINSERT OVERWRITE DIRECTORY '/user/hadoop/output' \n SELECT concat_ws(',', col1, col2) \n FROM graph_edges;",
"_____no_output_____"
]
],
[
[
"> **Create User Defined Fucntions (UDF)**",
"_____no_output_____"
],
[
"**Steps**\n* write in java\n* jar file\n* import jar file\n* use UDF as query",
"_____no_output_____"
],
[
"# Lab Material",
"_____no_output_____"
]
],
[
[
"### sample code from lab\n\nCREATE EXTERNAL TABLE employees(\nname STRING,\nsalary FLOAT)\nROW FORMAT DELIMITED\nFIELDS TERMINATED BY ’,’; \nLOAD DATA INPATH ‘employees.csv’ into table employees;\n\nCREATE DATABASE msia;\nSHOW DATABASES;\nDROP DATABASE msia;\nUSE msia;\nSHOW TABLES;\n\nCREATE TABLE employees(\nname STRING,\nsalary FLOAT,\nsubordinates ARRAY<STRING>, \ndeductions MAP<STRING, FLOAT>, \naddress STRUCT<street:STRING, city: STRING, state: STRING, zip: INT>); CREATE TABLE t (\ns STRING,\nf FLOAT,\na ARRAY<MAP<STRING, STRUCT<p1: INT, p2:INT> >);\nROW FORMAT DELIMITED\nFIELDS TERMINATED BY ’,’\nCOLLECTION ITEMS TERMINATED BY ’#’\nMAP KEYS TERMINATED BY ’-’\nLINES TERMINATED BY ’\\n’;\nLOAD DATA INPATH ’employees.csv’ into table employees;",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
],
[
"# <a id='refer'>Reference</a>",
"_____no_output_____"
],
[
"* [Tutorialspoint Hive Tutorial](https://www.tutorialspoint.com/hive/index.htm)\n* [Hive tutorial doc](https://cwiki.apache.org/confluence/display/Hive/Tutorial)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cb43348fbdac91b44c2c8a52ed20d9fc93f84dcc | 799,147 | ipynb | Jupyter Notebook | pyfolio/examples/sector_mapping_example.ipynb | NunoEdgarGFlowHub/pyfolio | 68efdcc2e2d0f140ddbc408a260c6318ac8b06d3 | [
"Apache-2.0"
] | 2 | 2016-08-30T02:16:05.000Z | 2019-03-15T07:27:53.000Z | pyfolio/examples/sector_mapping_example.ipynb | riverdarda/pyfolio | 145ac36fdf286cc01b79989aa90f40480b40cab7 | [
"Apache-2.0"
] | null | null | null | pyfolio/examples/sector_mapping_example.ipynb | riverdarda/pyfolio | 145ac36fdf286cc01b79989aa90f40480b40cab7 | [
"Apache-2.0"
] | 1 | 2016-12-04T16:22:07.000Z | 2016-12-04T16:22:07.000Z | 2,536.974603 | 678,368 | 0.948863 | [
[
[
"# Sector Mappings",
"_____no_output_____"
],
[
"To generate sector allocation plots in the positions tearsheet and PnL by sector in the round trips tearsheet, you must pass pyfolio a dictionary (or dict-like data struction) of symbol-sector mappings, where symbols are keys and sectors are values. `create_full_tearsheet` will also take symbol-sector mappings as keyword argument `sector_mappings`.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport pyfolio as pf\nimport gzip\nimport os\nimport pandas as pd",
"_____no_output_____"
],
[
"transactions = pd.read_csv(gzip.open('../tests/test_data/test_txn.csv.gz'),\n index_col=0, parse_dates=0)\npositions = pd.read_csv(gzip.open('../tests/test_data/test_pos.csv.gz'),\n index_col=0, parse_dates=0)\nreturns = pd.read_csv(gzip.open('../tests/test_data/test_returns.csv.gz'),\n index_col=0, parse_dates=0, header=None)[1]\ngross_lev = pd.read_csv(gzip.open('../tests/test_data/test_gross_lev.csv.gz'),\n index_col=0, parse_dates=0, header=None)[1]",
"_____no_output_____"
],
[
"returns.index = returns.index.tz_localize(\"UTC\")\npositions.index = positions.index.tz_localize(\"UTC\")\ntransactions.index = transactions.index.tz_localize(\"UTC\")\ngross_lev.index = gross_lev.index.tz_localize(\"UTC\")\n",
"_____no_output_____"
],
[
"positions.head(2)",
"_____no_output_____"
],
[
"sect_map = {'COST': 'Consumer Goods', \n 'INTC': 'Technology', \n 'CERN': 'Healthcare', \n 'GPS': 'Technology',\n 'MMM': 'Construction', \n 'DELL': 'Technology', \n 'AMD': 'Technology'}",
"_____no_output_____"
],
[
"pf.create_position_tear_sheet(returns, positions, gross_lev=gross_lev, sector_mappings=sect_map)\n",
"\n\nTop 10 long positions of all time (and max%)\n['COST' 'DELL' 'CERN' 'MMM' 'INTC' 'AMD' 'GPS']\n[ 0.9 0.857 0.835 0.821 0.786 0.758 0.622]\n\n\nTop 10 short positions of all time (and max%)\n['AMD' 'DELL' 'CERN' 'MMM' 'GPS' 'INTC' 'COST']\n[-0.301 -0.266 -0.255 -0.226 -0.201 -0.185 -0.164]\n\n\nTop 10 positions of all time (and max%)\n['COST' 'DELL' 'CERN' 'MMM' 'INTC' 'AMD' 'GPS']\n[ 0.9 0.857 0.835 0.821 0.786 0.758 0.622]\n\n\nAll positions ever held\n['COST' 'DELL' 'CERN' 'MMM' 'INTC' 'AMD' 'GPS']\n[ 0.9 0.857 0.835 0.821 0.786 0.758 0.622]\n\n\n"
],
[
"pf.create_round_trip_tear_sheet(positions, transactions, sector_mappings=sect_map)",
" duration pnl returns long\ncount 1430 1430.000000 1430.000000 1430\nmean 9 days 16:40:56.154545 45.737238 0.003543 0.523077\nstd 22 days 02:16:41.165898 1616.537844 0.031288 0.499642\nmin 0 days 00:00:00 -30697.460000 -0.218045 False\n25% 0 days 23:59:59 -5.773144 -0.011450 0\n50% 2 days 23:59:59 0.871629 0.003885 1\n75% 5 days 23:59:59 40.438366 0.018126 1\nmax 286 days 00:00:00 17835.869482 0.204385 True\nPercent of round trips profitable = 57.2%\nMean return per winning round trip = 0.02181\nMean return per losing round trip = -0.02108\nA decision is made every 1.053 days.\n0.9495 trading decisions per day.\n19.94 trading decisions per month.\n\nProfitability (PnL / PnL total) per name:\nsymbol\nCOST 0.398964\nINTC 0.382659\nCERN 0.323077\nMMM 0.221479\nGPS 0.049385\nAMD -0.064091\nDELL -0.311473\nName: pnl, dtype: float64\n\nProfitability (PnL / PnL total) per name:\nsymbol\nConsumer Goods 0.398964\nHealthcare 0.323077\nConstruction 0.221479\nTechnology 0.056480\nName: pnl, dtype: float64\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4334cc8aaef570e93b208f408eda3d9a2a5207 | 84,756 | ipynb | Jupyter Notebook | examples/loading_data.ipynb | hassan-jvd90/sktime | 7ef5e3db837100b96357346a451b26b6142e4639 | [
"BSD-3-Clause"
] | 1 | 2020-06-02T22:24:44.000Z | 2020-06-02T22:24:44.000Z | examples/loading_data.ipynb | sparkingdark/sktime | a9470b6bca4d3bc98f4ddaee191b19d359c66714 | [
"BSD-3-Clause"
] | 1 | 2020-11-20T13:51:20.000Z | 2020-11-20T13:51:20.000Z | examples/loading_data.ipynb | sparkingdark/sktime | a9470b6bca4d3bc98f4ddaee191b19d359c66714 | [
"BSD-3-Clause"
] | 3 | 2020-10-18T04:54:30.000Z | 2021-02-15T18:04:18.000Z | 30.932847 | 667 | 0.409682 | [
[
[
"# Loading and working with data in sktime\n\nPython provides a variety of useful ways to represent data, but NumPy arrays and pandas DataFrames are commonly used for data analysis. When using NumPy 2d-arrays or pandas DataFrames to analyze tabular data the rows are commony used to represent each instance (e.g. case or observation) of the data, while the columns are used to represent a given feature (e.g. variable or dimension) for an observation. Since timeseries data also has a time dimension for a given instance and feature, several alternative data formats could be used to represent this data, including nested pandas DataFrame structures, NumPy 3d-arrays, or multi-indexed pandas DataFrames. \n\nSktime is designed to work with timeseries data stored as nested pandas DataFrame objects. Similar to working with pandas DataFrames with tabular data, this allows instances to be represented by rows and the feature data for each dimension of a problem (e.g. variables or features) to be stored in the DataFrame columns. To accomplish this the timepoints for each instance-feature combination are stored in a single cell in the input Pandas DataFrame ([see Sktime pandas DataFrame format](#sktime_df_format) for more details). \n\nUsers can load or convert data into sktime's format in a variety of ways. Data can be loaded directly from a bespoke sktime file format (.ts) ([see Representing data with .ts files](#ts_files)) or supported file formats provided by [other existing data sources](#other_file_types) (such as Weka ARFF and .tsv). Sktime also provides functions to convert data to and from sktime's nested pandas DataFrame format and several other common ways for representing timeseries data using NumPy arrays or pandas DataFrames. [see Converting between sktime and alternative timeseries formats](#convert).\n\nThe rest of this sktime tutorial will provide a more detailed description of the sktime pandas DataFrame format, a brief description of the .ts file format, how to load data from other supported formats, and how to convert between other common ways of representing timeseries data in NumPy arrays or pandas DataFrames.\n\n<a id=\"sktime_df_format\"></a>\n## Sktime pandas DataFrame format\n\nThe core data structure for storing datasets in sktime is a _nested_ pandas DataFrame, where rows of the dataframe correspond to instances (cases or observations), and columns correspond to dimensions of the problem (features or variables). The multiple timepoints and their corresponding values for each instance-feature pair are stored as pandas Series object _nested_ within the applicable DataFrame cell.\n\nFor example, for a problem with n cases that each have data across c timeseries dimensions:\n\n DataFrame:\n index | dim_0 | dim_1 | ... | dim_c-1\n 0 | pd.Series | pd.Series | pd.Series | pd.Series\n 1 | pd.Series | pd.Series | pd.Series | pd.Series\n ... | ... | ... | ... | ...\n n | pd.Series | pd.Series | pd.Series | pd.Series\n\nRepresenting timeseries data in this way makes it easy to align the timeseries features for a given instance with non-timeseries information. For example, in a classification problem, it is easy to align the timeseries features for an observation with its (index-aligned) target class label:\n\n index | class_val\n 0 | int\n 1 | int\n ... | ...\n n | int\n\n\nWhile sktime's format uses pandas Series objects in its nested DataFrame structure, other data structures like NumPy arrays could be used to hold the timeseries values in each cell. However, the use of pandas Series objects helps to facilitate simple storage of sparse data and make it easy to accomodate series with non-integer timestamps (such as dates). \n\n\n<a id=\"ts_files\"></a>\n## The .ts file format\nOne common use case is to load locally stored data. To make this easy, the .ts file format has been created for representing problems in a standard format for use with sktime. \n\n### Representing data with .ts files\nA .ts file include two main parts:\n* header information\n* data\n\nThe header information is used to facilitate simple representation of the data through including metadata about the structure of the problem. The header contains the following:\n\n @problemName <problem name>\n @timeStamps <true/false>\n @univariate <true/false>\n @classLabel <true/false> <space delimited list of possible class values>\n @data\n\nThe data for the problem should begin after the @data tag. In the simplest case where @timestamps is false, values for a series are expressed in a comma-separated list and the index of each value is relative to its position in the list (0, 1, ..., m). An _instance_ may contain 1 to many dimensions, where instances are line-delimited and dimensions within an instance are colon (:) delimited. For example:\n\n 2,3,2,4:4,3,2,2\n 13,12,32,12:22,23,12,32\n 4,4,5,4:3,2,3,2\n\nThis example data has 3 _instances_, corresponding to the three lines shown above. Each instance has 2 _dimensions_ with 4 observations per dimension. For example, the intitial instance's first dimension has the timepoint values of 2, 3, 2, 4 and the second dimension has the values 4, 3, 2, 2.\n\nMissing readings can be specified using ?. For example, \n\n 2,?,2,4:4,3,2,2\n 13,12,32,12:22,23,12,32\n 4,4,5,4:3,2,3,2\n \nwould indicate the second timepoint value of the initial instance's first dimension is missing. \n\nAlternatively, for sparse datasets, readings can be specified by setting @timestamps to true in the header and representing the data with tuples in the form of (timestamp, value) just for the obser. For example, the first instance in the example above could be specified in this representation as:\n\n (0,2),(1,3)(2,2)(3,4):(0,4),(1,3),(2,2),(3,2)\n\nEquivalently, the sparser example\n\n 2,5,?,?,?,?,?,5,?,?,?,?,4\n\ncould be represented with just the non-missing timestamps as:\n\n (0,2),(0,5),(7,5),(12,4)\n\nWhen using the .ts file format to store data for timeseries classification problems, the class label for an instance should be specified in the last dimension and @classLabel should be set to true in the header information and be followed by the set of possible class values. For example, if a case consists of a single dimension and has a class value of 1 it would be specified as:\n\n 1,4,23,34:1\n\n\n### Loading from .ts file to pandas DataFrame\n\nA dataset can be loaded from a .ts file using the following method in sktime.utils.data_io.py:\n\n load_from_tsfile_to_dataframe(full_file_path_and_name, replace_missing_vals_with='NaN')\n\nThis can be demonstrated using the Arrow Head problem that is included in sktime under sktime/datasets/data",
"_____no_output_____"
]
],
[
[
"import os\n\nimport sktime\nfrom sktime.utils.data_io import load_from_tsfile_to_dataframe\n\nDATA_PATH = os.path.join(os.path.dirname(sktime.__file__), \"datasets/data\")\n\ntrain_x, train_y = load_from_tsfile_to_dataframe(\n os.path.join(DATA_PATH, \"ArrowHead/ArrowHead_TRAIN.ts\")\n)\ntest_x, test_y = load_from_tsfile_to_dataframe(\n os.path.join(DATA_PATH, \"ArrowHead/ArrowHead_TEST.ts\")\n)",
"_____no_output_____"
]
],
[
[
"Train and test partitions of the ArrowHead problem have been loaded into nested dataframes with an associated array of class values. As an example, below are the first 5 rows from the train_x and train_y:",
"_____no_output_____"
]
],
[
[
"train_x.head()",
"_____no_output_____"
],
[
"train_y[0:5]",
"_____no_output_____"
]
],
[
[
"<a id=\"other_file_types\"></a>\n## Loading other file formats\nResearchers who have made timeseries data available have used two other common formats, including:\n\n+ Weka ARFF files\n+ UCR .tsv files\n\n\n### Loading from Weka ARFF files\n\nIt is also possible to load data from Weka's attribute-relation file format (ARFF) files. Data for timeseries problems are made available in this format by researchers at the University of East Anglia (among others) at www.timeseriesclassification.com. The `load_from_arff_to_dataframe` method in `sktime.utils.data_io` supports reading data for both univariate and multivariate timeseries problems. \n\nThe univariate functionality is demonstrated below using data on the ArrowHead problem again (this time loading from ARFF file).",
"_____no_output_____"
]
],
[
[
"from sktime.utils.data_io import load_from_arff_to_dataframe\n\nX, y = load_from_arff_to_dataframe(\n os.path.join(DATA_PATH, \"ArrowHead/ArrowHead_TRAIN.arff\")\n)\nX.head()",
"_____no_output_____"
]
],
[
[
"The multivariate BasicMotions problem is used below to illustrate the ability to read multivariate timeseries data from ARFF files into the sktime format. ",
"_____no_output_____"
]
],
[
[
"X, y = load_from_arff_to_dataframe(\n os.path.join(DATA_PATH, \"BasicMotions/BasicMotions_TRAIN.arff\")\n)\nX.head()",
"_____no_output_____"
]
],
[
[
"### Loading from UCR .tsv Format Files\n\nA further option is to load data into sktime from tab separated value (.tsv) files. Researchers at the University of Riverside, California make a variety of timeseries data available in this format at https://www.cs.ucr.edu/~eamonn/time_series_data_2018. \n\nThe `load_from_ucr_tsv_to_dataframe` method in `sktime.utils.data_io` supports reading univariate problems. An example with ArrowHead is given below to demonstrate equivalence with loading from the .ts and ARFF file formats.",
"_____no_output_____"
]
],
[
[
"from sktime.utils.data_io import load_from_ucr_tsv_to_dataframe\n\nX, y = load_from_ucr_tsv_to_dataframe(\n os.path.join(DATA_PATH, \"ArrowHead/ArrowHead_TRAIN.tsv\")\n)\nX.head()",
"_____no_output_____"
]
],
[
[
"<a id=\"convert\"></a>\n## Converting between other NumPy and pandas formats\n\nIt is also possible to use data from sources other than .ts and .arff files by manually shaping the data into the format described above. \n\nFunctions to convert from and to these types to sktime's nested DataFrame format are provided in `sktime.utils.data_processing`\n\n### Using tabular data with sktime\n\nOne approach to representing timeseries data is a tabular DataFrame. As usual, each row represents an instance. In the tabular setting each timepoint of the univariate timeseries being measured for each instance are treated as feature and stored as a primitive data type in the DataFrame's cells. \n\nIn a univariate setting, where there are `n` instances of the series and each univariate timeseries has `t` timepoints, this would yield a pandas DataFrame with shape (n, t). In practice, this could be used to represent sensors measuring the same signal over time (features) on different machines (instances) or the same economic variable over time (features) for different countries (instances). \n\nThe function `from_2d_array_to_nested` converts a (n, t) tabular DataFrame to nested DataFrame with shape (n, 1). To convert from a nested DataFrame to a tabular array the function `from_nested_to_2d_array` can be used.\n\nThe example below uses 50 instances with 20 timepoints each. ",
"_____no_output_____"
]
],
[
[
"from numpy.random import default_rng\n\nfrom sktime.utils.data_processing import (\n from_2d_array_to_nested,\n from_nested_to_2d_array,\n is_nested_dataframe,\n)\n\nrng = default_rng()\nX_2d = rng.standard_normal((50, 20))\nprint(f\"The tabular data has the shape {X_2d.shape}\")",
"The tabular data has the shape (50, 20)\n"
]
],
[
[
"The `from_2d_array_to_nested` function makes it easy to convert this to a nested DataFrame.",
"_____no_output_____"
]
],
[
[
"X_nested = from_2d_array_to_nested(X_2d)\nprint(f\"X_nested is a nested DataFrame: {is_nested_dataframe(X_nested)}\")\nprint(f\"The cell contains a {type(X_nested.iloc[0,0])}.\")\nprint(f\"The nested DataFrame has shape {X_nested.shape}\")\nX_nested.head()",
"X_nested is a nested DataFrame: True\nThe cell contains a <class 'pandas.core.series.Series'>.\nThe nested DataFrame has shape (50, 1)\n"
]
],
[
[
"This nested DataFrame can also be converted back to a tabular DataFrame using easily. ",
"_____no_output_____"
]
],
[
[
"X_2d = from_nested_to_2d_array(X_nested)\nprint(f\"The tabular data has the shape {X_2d.shape}\")",
"The tabular data has the shape (50, 20)\n"
]
],
[
[
"### Using long-format data with sktime\n\nTimeseries data can also be represented in _long_ format where each row identifies the value for a single timepoint for a given dimension for a given instance. \n\nThis format may be encountered in a database where each row stores a single value measurement identified by several identification columns. For example, where `case_id` is an id to identify a specific instance in the data, `dimension_id` is an integer between 0 and d-1 for d dimensions in the data, `reading_id` is the index of timepoints for the associated `case_id` and `dimension_id`, and `value` is the actual value of the observation. E.g.:\n\n | case_id | dim_id | reading_id | value\n ------------------------------------------------\n 0 | int | int | int | double\n 1 | int | int | int | double\n 2 | int | int | int | double\n 3 | int | int | int | double\n \nSktime provides functions to convert to and from the long data format in `sktime.utils.data_processing`. \n\nThe `from_long_to_nested` function converts from a long format DataFrame to sktime's nested format (with assumptions made on how the data is initially formatted). Conversely, `from_nested_to_long` converts from a sktime nested DataFrame into a long format DataFrame. \n\n\nTo demonstrate this functionality the method below creates a dataset with a 50 instances (cases), 5 dimensions and 20 timepoints per dimension.",
"_____no_output_____"
]
],
[
[
"from sktime.utils.data_io import generate_example_long_table\n\nX = generate_example_long_table(num_cases=50, series_len=20, num_dims=5)\n\nX.head()",
"_____no_output_____"
],
[
"X.tail()",
"_____no_output_____"
]
],
[
[
"As shown below, applying the `from_long_to_nested` method returns a sktime-formatted dataset with individual dimensions represented by columns of the output dataframe.",
"_____no_output_____"
]
],
[
[
"from sktime.utils.data_processing import from_long_to_nested, from_nested_to_long\n\nX_nested = from_long_to_nested(X)\nX_nested.head()",
"_____no_output_____"
]
],
[
[
"As expected the result is a nested DataFrame and the cells include nested pandas Series objects. ",
"_____no_output_____"
]
],
[
[
"print(f\"X_nested is a nested DataFrame: {is_nested_dataframe(X_nested)}\")\nprint(f\"The cell contains a {type(X_nested.iloc[0,0])}.\")\nprint(f\"The nested DataFrame has shape {X_nested.shape}\")\nX_nested.iloc[0, 0].head()",
"X_nested is a nested DataFrame: True\nThe cell contains a <class 'pandas.core.series.Series'>.\nThe nested DataFrame has shape (50, 5)\n"
]
],
[
[
"As shown below, the `from_nested_to_long` function can be used to convert the resulting nested DataFrame (or any nested DataFrame) to a long format DataFrame. ",
"_____no_output_____"
]
],
[
[
"X_long = from_nested_to_long(\n X_nested,\n instance_column_name=\"case_id\",\n time_column_name=\"reading_id\",\n dimension_column_name=\"dim_id\",\n)\nX_long.head()",
"_____no_output_____"
],
[
"X_long.tail()",
"_____no_output_____"
]
],
[
[
"### Using multi-indexed pandas DataFrames\n\nPandas deprecated its Panel object in version 0.20.1. Since that time pandas has recommended representing 3-dimensional data using a multi-indexed DataFrame. \n\nStoring timeseries data in a Pandas multi-indexed DataFrame is a natural option since many timeseries problems include data over the instance, feature and time dimensions. \n\nSktime provides the functions `from_multi_index_to_nested` and `from_nested_to_multi_index` in `sktime.utils.data_processing` to easily convert between pandas multi-indexed DataFrames and sktime's nested DataFrame structure. \n\nThe example below illustrates how these functions can be used to convert to and from the nested structure given data with 50 instances, 5 features (columns) and 20 timepoints per feature. In the multi-indexed DataFrame a row represents a unique combination of the instance and timepoint indices. Therefore, the resulting multi-indexed DataFrame should have the shape (1000, 5). ",
"_____no_output_____"
]
],
[
[
"from sktime.utils.data_io import make_multi_index_dataframe\nfrom sktime.utils.data_processing import (\n from_multi_index_to_nested,\n from_nested_to_multi_index,\n)\n\nX_mi = make_multi_index_dataframe(n_instances=50, n_columns=5, n_timepoints=20)\n\nprint(f\"The multi-indexed DataFrame has shape {X_mi.shape}\")\nprint(f\"The multi-index names are {X_mi.index.names}\")\n\nX_mi.head()",
"The multi-indexed DataFrame has shape (1000, 5)\nThe multi-index names are ['case_id', 'reading_id']\n"
]
],
[
[
"The multi-indexed DataFrame can be easily converted to a nested DataFrame with shape (50, 5). Note that the conversion to the nested DataFrame has preserved the column names (it has also preserved the values of the instance index and the pandas Series objects nested in each cell have preserved the time index). ",
"_____no_output_____"
]
],
[
[
"X_nested = from_multi_index_to_nested(X_mi, instance_index=\"case_id\")\nprint(f\"X_nested is a nested DataFrame: {is_nested_dataframe(X_nested)}\")\nprint(f\"The cell contains a {type(X_nested.iloc[0,0])}.\")\nprint(f\"The nested DataFrame has shape {X_nested.shape}\")\nX_nested.head()",
"X_nested is a nested DataFrame: True\nThe cell contains a <class 'pandas.core.series.Series'>.\nThe nested DataFrame has shape (50, 5)\n"
]
],
[
[
"Nested DataFrames can also be converted to a multi-indexed Pandas DataFrame",
"_____no_output_____"
]
],
[
[
"X_mi = from_nested_to_multi_index(\n X_nested, instance_index=\"case_id\", time_index=\"reading_id\"\n)\nX_mi.head()",
"_____no_output_____"
]
],
[
[
"### Using NumPy 3d-arrays with sktime\n\nAnother common approach for representing timeseries data is to use a 3-dimensional NumPy array with shape (n_instances, n_columns, n_timepoints). \n\nSktime provides the functions `from_3d_numpy_to_nested` `from_nested_to_3d_numpy` in `sktime.utils.data_processing` to let users easily convert between NumPy 3d-arrays and nested pandas DataFrames. \n\nThis is demonstrated using a 3d-array with 50 instances, 5 features (columns) and 20 timepoints, resulting in a 3d-array with shape (50, 5, 20). ",
"_____no_output_____"
]
],
[
[
"from sktime.utils.data_processing import (\n from_3d_numpy_to_nested,\n from_multi_index_to_3d_numpy,\n from_nested_to_3d_numpy,\n)\n\nX_mi = make_multi_index_dataframe(n_instances=50, n_columns=5, n_timepoints=20)\nX_3d = from_multi_index_to_3d_numpy(\n X_mi, instance_index=\"case_id\", time_index=\"reading_id\"\n)\n\nprint(f\"The 3d-array has shape {X_3d.shape}\")",
"The 3d-array has shape (50, 5, 20)\n"
]
],
[
[
"The 3d-array can be easily converted to a nested DataFrame with shape (50, 5). Note that since NumPy array doesn't have indices, the instance index is the numerical range over the number of instances and the columns are automatically assigned. Users can optionally supply their own columns names via the columns_names parameter. ",
"_____no_output_____"
]
],
[
[
"X_nested = from_3d_numpy_to_nested(X_3d)\nprint(f\"X_nested is a nested DataFrame: {is_nested_dataframe(X_nested)}\")\nprint(f\"The cell contains a {type(X_nested.iloc[0,0])}.\")\nprint(f\"The nested DataFrame has shape {X_nested.shape}\")\nX_nested.head()",
"X_nested is a nested DataFrame: True\nThe cell contains a <class 'pandas.core.series.Series'>.\nThe nested DataFrame has shape (50, 5)\n"
]
],
[
[
"Nested DataFrames can also be converted to NumPy 3d-arrays. ",
"_____no_output_____"
]
],
[
[
"X_3d = from_nested_to_3d_numpy(X_nested)\nprint(f\"The resulting object is a {type(X_3d)}\")\nprint(f\"The shape of the 3d-array is {X_3d.shape}\")",
"The resulting object is a <class 'numpy.ndarray'>\nThe shape of the 3d-array is (50, 5, 20)\n"
]
],
[
[
"### Converting between NumPy 3d-arrays and pandas multi-indexed DataFrame\n\nAlthough an example is not provided here, sktime lets users convert data between NumPy 3d-arrays and a multi-indexed pandas DataFrame formats using the functions `from_3d_numpy_to_multi_index` and `from_multi_index_to_3d_numpy` in `sktime.utils.data_processing`. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb4335ec879f8121c6cffd70135015d5ee2728a7 | 43,876 | ipynb | Jupyter Notebook | jupyter/.ipynb_checkpoints/Train_model-checkpoint.ipynb | jahehalo/potential_of_restarts_hidden | 4aa3427adb452ae65e56268e64c412b2c626c7c4 | [
"CC-BY-4.0",
"MIT"
] | null | null | null | jupyter/.ipynb_checkpoints/Train_model-checkpoint.ipynb | jahehalo/potential_of_restarts_hidden | 4aa3427adb452ae65e56268e64c412b2c626c7c4 | [
"CC-BY-4.0",
"MIT"
] | null | null | null | jupyter/.ipynb_checkpoints/Train_model-checkpoint.ipynb | jahehalo/potential_of_restarts_hidden | 4aa3427adb452ae65e56268e64c412b2c626c7c4 | [
"CC-BY-4.0",
"MIT"
] | null | null | null | 44.634791 | 105 | 0.446121 | [
[
[
"import numpy as np\nimport pandas as pd\nimport os\nfrom glob import glob\n\nfrom sklearn.feature_selection import VarianceThreshold\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.ensemble import RandomForestRegressor\n\nfrom scipy.optimize import root_scalar\n\nfrom joblib import dump, load\n\nfrom expon_mixture import ExponMixture ",
"_____no_output_____"
],
[
"fits = pd.read_csv('./fits/expon_mix_2comp_fits.txt', index_col=0)",
"_____no_output_____"
],
[
"fits[fits['p1']<1.0].describe()",
"_____no_output_____"
],
[
"fits",
"_____no_output_____"
],
[
"df = pd.read_csv('./calculate_features/features_train.csv')\ndf.set_index('instance', inplace=True)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"merged_data = pd.concat([df, fits], join='inner', axis=1)",
"_____no_output_____"
],
[
"v2 = merged_data[merged_data['p1']<1.0]\ny = v2[['p1', 'lambda1', 'lambda2']]\nX = v2.drop(['p1', 'p2', 'lambda1', 'lambda2'], axis=1)",
"_____no_output_____"
],
[
"regr_rf = RandomForestRegressor()\nregr_rf.fit(X, y)",
"_____no_output_____"
],
[
"dump(regr_rf, 'rf_regressor.joblib') ",
"_____no_output_____"
],
[
"regr_rf = load('rf_regressor.joblib') ",
"_____no_output_____"
],
[
"regr_rf.predict(X)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb433a1ec70653fdbdef7c6c81f8ded2fef56e03 | 21,720 | ipynb | Jupyter Notebook | book/distributions/Binomial-Distribution.ipynb | willettk/stats-ds-book | 06bc751a7e82f73f9d7419f32fe5882ec5742f2f | [
"MIT"
] | 41 | 2020-08-18T12:14:43.000Z | 2022-03-31T16:37:17.000Z | book/distributions/Binomial-Distribution.ipynb | willettk/stats-ds-book | 06bc751a7e82f73f9d7419f32fe5882ec5742f2f | [
"MIT"
] | 7 | 2020-08-19T04:22:24.000Z | 2020-12-22T15:18:24.000Z | book/distributions/Binomial-Distribution.ipynb | willettk/stats-ds-book | 06bc751a7e82f73f9d7419f32fe5882ec5742f2f | [
"MIT"
] | 13 | 2020-08-19T02:57:47.000Z | 2022-03-03T15:24:07.000Z | 48.266667 | 6,884 | 0.770028 | [
[
[
"import numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"np.random.choice(['red','green',3,'dragon',5,6], size=5,p=[.1,.1,.1,.7,0,0])",
"_____no_output_____"
],
[
"thisroll = np.random.choice([1,2,3,4,5,6],3)\nthisroll",
"_____no_output_____"
],
[
"1.*(thisroll[0]==1)+1.*(thisroll[1]==1)+1.*(thisroll[2]==1)",
"_____no_output_____"
],
[
"1.*(thisroll[0]==1)",
"_____no_output_____"
],
[
"N_experiments = 100000\nN_roll = 9",
"_____no_output_____"
],
[
"data = np.random.choice([1,2,3,4,5,6],N_experiments*N_roll)\ndata = data.reshape(N_experiments,N_roll)",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"n_aces = 1.*(data[:,0]==1)+1.*(data[:,1]==1)+1.*(data[:,2]==1)+1.*(data[:,3]==1)+1.*(data[:,4]==1)+1.*(data[:,5]==1) +1.*(data[:,6]==1)+1.*(data[:,7]==1)+1.*(data[:,8]==1)",
"_____no_output_____"
],
[
"n_aces",
"_____no_output_____"
],
[
"mybins = np.arange(0,11)-.5\nk_array = np.arange(0,10)\nk_array",
"_____no_output_____"
],
[
"from scipy.stats import binom\nn = N_roll\np = 1./6\nrv = binom(n, p)",
"_____no_output_____"
],
[
"plt.hist(n_aces, bins=mybins, density=True)\nplt.vlines(k_array, 0, rv.pmf(k_array), colors='k', linestyles='-', lw=1, label='frozen pmf')",
"_____no_output_____"
],
[
"np.sum(n_aces==3)/N_experiments",
"_____no_output_____"
],
[
"import scipy.special",
"_____no_output_____"
],
[
"scipy.special.binom(9,3)",
"_____no_output_____"
],
[
"scipy.special.binom(9,3)*np.power(1./6,3)*np.power(5./6,6)",
"_____no_output_____"
],
[
"scipy.special.binom(9,2)*np.power(1./6,2)*np.power(5./6,7)",
"_____no_output_____"
],
[
"np.sum(n_aces==2)/N_experiments",
"_____no_output_____"
]
],
[
[
"## From scipy",
"_____no_output_____"
]
],
[
[
"from scipy.stats import binom",
"_____no_output_____"
],
[
"n, p = 50, 0.1",
"_____no_output_____"
],
[
"x = np.arange(0,n)\n# Fancy way\n#x = np.arange(binom.ppf(0.01, n, p), binom.ppf(0.99, n, p))",
"_____no_output_____"
],
[
"plt.scatter(x,binom.pmf(x,n,p))",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cb433e34fbe673d3353fd944b15d36e4a65756f8 | 26,420 | ipynb | Jupyter Notebook | topicnet/demos/RTL-WIKI-PREPROCESSING.ipynb | tkhirianov/TopicNet | 41318897d25b622da16f20d68fe9c315cd177b3c | [
"MIT"
] | 1 | 2021-04-25T21:38:48.000Z | 2021-04-25T21:38:48.000Z | topicnet/demos/RTL-WIKI-PREPROCESSING.ipynb | tkhirianov/TopicNet | 41318897d25b622da16f20d68fe9c315cd177b3c | [
"MIT"
] | null | null | null | topicnet/demos/RTL-WIKI-PREPROCESSING.ipynb | tkhirianov/TopicNet | 41318897d25b622da16f20d68fe9c315cd177b3c | [
"MIT"
] | null | null | null | 52.213439 | 7,130 | 0.65651 | [
[
[
"This notebook will show an example of text preprocessing applied to RTL-Wiki dataset.\n\nThis dataset was introduced in [1] and later recreated in [2]. You can download it in from http://139.18.2.164/mroeder/palmetto/datasets/rtl-wiki.tar.gz\n\n--------\n\n[1] \"Reading Tea Leaves: How Humans Interpret Topic Models\" (NIPS 2009)\n\n[2] \"Exploring the Space of Topic Coherence Measures\" (WSDM 2015)\n\n\n\n",
"_____no_output_____"
]
],
[
[
"# download corpus and unpack it:\n! wget http://139.18.2.164/mroeder/palmetto/datasets/rtl-wiki.tar.gz -O rtl-wiki.tar.gz\n! tar xzf rtl-wiki.tar.gz\n",
"--2020-01-25 23:56:21-- http://139.18.2.164/mroeder/palmetto/datasets/rtl-wiki.tar.gz\nConnecting to 139.18.2.164:80... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 176728881 (169M) [application/octet-stream]\nSaving to: ‘rtl-wiki.tar.gz’\n\nrtl-wiki.tar.gz 100%[===================>] 168,54M 12,5MB/s in 12s \n\n2020-01-25 23:56:34 (13,6 MB/s) - ‘rtl-wiki.tar.gz’ saved [176728881/176728881]\n\n"
]
],
[
[
"The corpus is a sample of 10000 articles from English Wikipedia in a MediaWiki markup format.\n\nHence, we need to strip specific wiki formatting. We advise using a `mwparserfromhell` fork optimized to deal with the English Wikipedia.\n\n```\ngit clone --branch images_and_interwiki https://github.com/bt2901/mwparserfromhell.git\n```",
"_____no_output_____"
]
],
[
[
"! git clone --branch images_and_interwiki https://github.com/bt2901/mwparserfromhell.git",
"Cloning into 'mwparserfromhell'...\nremote: Enumerating objects: 34, done.\u001b[K\nremote: Counting objects: 100% (34/34), done.\u001b[K\nremote: Compressing objects: 100% (25/25), done.\u001b[K\nremote: Total 5756 (delta 15), reused 19 (delta 9), pack-reused 5722\nReceiving objects: 100% (5756/5756), 1.75 MiB | 2.78 MiB/s, done.\nResolving deltas: 100% (3987/3987), done.\n"
]
],
[
[
"The Wikipedia dataset is too heterogenous. Building a good topic model here requires a lot of topics or a lot of documents.\n\nTo make collection more focused, we will filter out everything which isn't about people. We will use the following criteria to distinguish people and not-people:\n\n\n",
"_____no_output_____"
]
],
[
[
"import re\n\n# all infoboxes related to persons, according to https://en.wikipedia.org/wiki/Wikipedia:List_of_infoboxes\nperson_infoboxes = {'infobox magic: the gathering player', 'infobox architect', 'infobox mountaineer', 'infobox scientist', 'infobox chess biography', 'infobox racing driver', 'infobox saint', 'infobox snooker player', 'infobox figure skater', 'infobox theological work', 'infobox gaelic athletic association player', 'infobox professional wrestler', 'infobox noble', 'infobox pelotari', 'infobox native american leader', 'infobox pretender', 'infobox amateur wrestler', 'infobox college football player', 'infobox buddha', 'infobox cfl biography', 'infobox playboy playmate', 'infobox cyclist', 'infobox martial artist', 'infobox motorcycle rider', 'infobox motocross rider', 'infobox bandy biography', 'infobox video game player', 'infobox dancer', 'infobox nahua officeholder', 'infobox criminal', 'infobox squash player', 'infobox go player', 'infobox bullfighting career', 'infobox engineering career', 'infobox pirate', 'infobox latter day saint biography', 'infobox sumo wrestler', 'infobox youtube personality', 'infobox national hockey league coach', 'infobox rebbe', 'infobox football official', 'infobox aviator', 'infobox pharaoh', 'infobox classical composer', 'infobox fbi ten most wanted', 'infobox chef', 'infobox engineer', 'infobox nascar driver', 'infobox medical person', 'infobox jewish leader', 'infobox horseracing personality', 'infobox poker player', 'infobox economist', 'infobox peer', 'infobox war on terror detainee', 'infobox philosopher', 'infobox professional bowler', 'infobox champ car driver', 'infobox golfer', 'infobox le mans driver', 'infobox alpine ski racer', 'infobox boxer (amateur)', 'infobox bodybuilder', 'infobox college coach', 'infobox speedway rider', 'infobox skier', 'infobox medical details', 'infobox field hockey player', 'infobox badminton player', 'infobox sports announcer details', 'infobox academic', 'infobox f1 driver', 'infobox ncaa athlete', 'infobox biathlete', 'infobox comics creator', 'infobox rugby league biography', 'infobox fencer', 'infobox theologian', 'infobox religious biography', 'infobox egyptian dignitary', 'infobox curler', 'infobox racing driver series section', 'infobox afl biography', 'infobox speed skater', 'infobox climber', 'infobox rugby biography', 'infobox clergy', 'infobox equestrian', 'infobox member of the knesset', 'infobox pageant titleholder', 'infobox lacrosse player', 'infobox tennis biography', 'infobox gymnast', 'infobox sport wrestler', 'infobox sports announcer', 'infobox surfer', 'infobox darts player', 'infobox christian leader', 'infobox presenter', 'infobox gunpowder plotter', 'infobox table tennis player', 'infobox sailor', 'infobox astronaut', 'infobox handball biography', 'infobox volleyball biography', 'infobox spy', 'infobox wrc driver', 'infobox police officer', 'infobox swimmer', 'infobox netball biography', 'infobox model', 'infobox comedian', 'infobox boxer'}\n# is page included in a category with demography information?\ndemography_re = re.compile(\"([0-9]+ (deaths|births))|(living people)\")\n",
"_____no_output_____"
],
[
"dir_name = \"persons\"",
"_____no_output_____"
],
[
"! mkdir $dir_name",
"_____no_output_____"
],
[
"import glob\nfrom bs4 import BeautifulSoup\nfrom mwparserfromhell import mwparserfromhell\n\nfrom tqdm import tqdm_notebook as tqdm\n\nfor filename in tqdm(glob.glob(\"documents/*.html\")):\n doc_id = filename.partition(\"/\")[-1]\n doc_id = doc_id.rpartition(\".\")[0] + \".txt\"\n is_about_person = False\n with open(filename, \"r\") as f:\n soup = BeautifulSoup(\"\".join(f.readlines()))\n text = soup.findAll('textarea', id=\"wpTextbox1\")[0].contents[0]\n text = text.replace(\"&\", \"&\").replace('<', '<').replace('>', '>')\n wikicode = mwparserfromhell.parse(text)\n if dir_name == \"persons\":\n for node in wikicode.nodes:\n entry_type = str(type(node))\n if \"Wikilink\" in entry_type:\n special_link_name, _, cat_name = node.title.lower().strip().partition(\":\")\n if special_link_name == \"category\":\n if demography_re.match(cat_name):\n is_about_person = True\n if \"Template\" in entry_type:\n name = str(node.name).lower().strip()\n if name in person_infoboxes:\n is_about_person = True\n should_be_saved = is_about_person\n else:\n should_be_saved = True\n if should_be_saved:\n with open(f\"{dir_name}/{doc_id}\", \"w\") as f2:\n stripped_text = wikicode.strip_code()\n f2.write(stripped_text)\n",
"_____no_output_____"
]
],
[
[
"Now we have a folder `persons` which contains 1201 document. Let's take a look at the one of them:",
"_____no_output_____"
]
],
[
[
"! head $dir_name/Eusebius.txt",
"Eusebius of Caesarea\r\nEusebius of Caesarea (c. 263 – c. 339) (often called Eusebius Pamphili, \"Eusebius [the friend] of Pamphilus\") became the bishop of Caesarea Palaestina, the capital of Iudaea province, c 314. He is often referred to as the Father of Church History because of his work in recording the history of the early Christian church, especially Chronicle and Ecclesiastical History Wetterau, Bruce. World history. New York: Henry Holt and company. 1994. . \r\n\r\nBiography\r\n\r\nHis date and place of birth are unknown and little is known of his youth, however it is estimated that he was born in 265 Encyclopedia of the Early Church, Published in 1992, English Version, page 299 . He became acquainted with the presbyter Dorotheus in Antioch and probably received exegetical instruction from him. In 296 he was in Palestine and saw Constantine who visited the country with Diocletian. He was in Caesarea when Agapius was bishop and became friendly with Pamphilus of Caesarea, with whom he seems to have studied the text of the Bible, with the aid of Origen's Hexapla and commentaries collected by Pamphilus, in an attempt to prepare a correct version.\r\n\r\nIn 307, Pamphilus was imprisoned, but Eusebius continued their project. The resulting defence of Origen, in which they had collaborated, was finished by Eusebius after the death of Pamphilus and sent to the martyrs in the mines of Phaeno located in modern Jordan. Eusebius then seems to have gone to Tyre and later to Egypt, where he first suffered persecution.\r\n\r\nEusebius is next heard of as bishop of Caesarea Maritima. He succeeded Agapius, whose time of office is not certain, but Eusebius must have become bishop soon after 313. Nothing is known about the early years of his tenure. When the Council of Nicaea met in 325, Eusebius was prominent in its transactions. He was not naturally a spiritual leader or theologian, but as a very learned man and a famous author who enjoyed the special favour of the emperor, he came to the fore among the members of the council (traditionally given as 318 attendees). He presented a confession to the council which was rejected in favour of a more specifically anti-Arian creed from Palestine. This later creed became the basis of the council's major theological statement, the Nicene Creed. Bruce L. Shelley, Church History in Plain Language, (2nd ed. Dallas, TX: Word Publishing, 1995.), p.102. \r\n"
]
],
[
[
"We need to lemmatize texts, remove stopwords and extract informative ngramms.\n\nThere's no one \"correct\" way to do it, but the reasonable baseline is using well-known `nltk` library.",
"_____no_output_____"
]
],
[
[
"import nltk\nimport string\n\nimport pandas as pd\nfrom glob import glob\n\n",
"_____no_output_____"
],
[
"nltk.data.path.append('/home/evgenyegorov/nltk_data/')",
"_____no_output_____"
],
[
"files = glob(dir_name + '/*.txt')\n",
"_____no_output_____"
],
[
"data = []\nfor path in files:\n entry = {}\n entry['id'] = path.split('/')[-1].rpartition(\".\")[0]\n with open(path, 'r') as f:\n entry['raw_text'] = \" \".join(line.strip() for line in f.readlines())\n data.append(entry)\n\nwiki_texts = pd.DataFrame(data)",
"_____no_output_____"
],
[
"from tqdm import tqdm",
"_____no_output_____"
],
[
"\ntokenized_text = []\n\nfor text in tqdm(wiki_texts['raw_text'].values):\n tokens = nltk.wordpunct_tokenize(text.lower())\n tokenized_text.append(nltk.pos_tag(tokens))\nwiki_texts['tokenized'] = tokenized_text",
"100%|██████████| 1201/1201 [01:37<00:00, 12.29it/s]\n"
],
[
"from nltk.corpus import wordnet\n\ndef nltk2wn_tag(nltk_tag):\n if nltk_tag.startswith('J'):\n return wordnet.ADJ\n elif nltk_tag.startswith('V'):\n return wordnet.VERB\n elif nltk_tag.startswith('N'):\n return wordnet.NOUN\n elif nltk_tag.startswith('R'):\n return wordnet.ADV\n else: \n return ''",
"_____no_output_____"
],
[
"from nltk.stem import WordNetLemmatizer\nfrom nltk.corpus import stopwords\nstop = set(stopwords.words('english'))\n\nlemmatized_text = []\nwnl = WordNetLemmatizer()\nfor text in wiki_texts['tokenized'].values:\n lemmatized = [wnl.lemmatize(word,nltk2wn_tag(pos))\n if nltk2wn_tag(pos) != ''\n else wnl.lemmatize(word)\n for word, pos in text ]\n lemmatized = [word for word in lemmatized \n if word not in stop and word.isalpha()]\n lemmatized_text.append(lemmatized)\nwiki_texts['lemmatized'] = lemmatized_text",
"_____no_output_____"
]
],
[
[
"Ngrams are a powerful feature, and BigARTM is able to take advantage of it (the technical term is 'multimodal topic modeling': our topic model could model a lot of different features linked to a specific document, not just words).",
"_____no_output_____"
]
],
[
[
"from nltk.collocations import BigramAssocMeasures, BigramCollocationFinder\n\nbigram_measures = BigramAssocMeasures()\nfinder = BigramCollocationFinder.from_documents(wiki_texts['lemmatized'])\nfinder.apply_freq_filter(5)\nset_dict = set(finder.nbest(bigram_measures.pmi,32100)[100:])\ndocuments = wiki_texts['lemmatized']\nbigrams = []\nfor doc in documents:\n entry = ['_'.join([word_first, word_second])\n for word_first, word_second in zip(doc[:-1],doc[1:])\n if (word_first, word_second) in set_dict]\n bigrams.append(entry)\nwiki_texts['bigram'] = bigrams",
"_____no_output_____"
],
[
"from collections import Counter\n\ndef vowpalize_sequence(sequence):\n word_2_frequency = Counter(sequence)\n del word_2_frequency['']\n vw_string = ''\n for word in word_2_frequency:\n vw_string += word + \":\" + str(word_2_frequency[word]) + ' '\n return vw_string\n\n",
"_____no_output_____"
],
[
"vw_text = []\nfor index, data in wiki_texts.iterrows():\n vw_string = '' \n doc_id = data.id\n lemmatized = '@lemmatized ' + vowpalize_sequence(data.lemmatized)\n bigram = '@bigram ' + vowpalize_sequence(data.bigram)\n vw_string = ' |'.join([doc_id, lemmatized, bigram])\n vw_text.append(vw_string)\nwiki_texts['vw_text'] = vw_text\n\n",
"_____no_output_____"
]
],
[
[
"Vowpal Wabbit (\"wv\") is a text format which is a good fit for multimodal topic modeling. Here, we elected to store dataset in a Bag-of-Words format (for performance reasons), but VW could store everything as a sequence of words as well.\n\nIt looks like this:",
"_____no_output_____"
]
],
[
[
"wiki_texts['vw_text'].head().values[0]\n",
"_____no_output_____"
],
[
"wiki_texts[['id','raw_text', 'vw_text']].to_csv('./wiki_data.csv')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cb433ed59fab4496e639a25c3a7445eea0e37b81 | 23,449 | ipynb | Jupyter Notebook | multiclass/multiclass_test.ipynb | kesamet/examples | 6baef94cc7db99abf17ca3df7dae35fcc3b08d25 | [
"Apache-2.0"
] | 1 | 2021-07-23T02:21:08.000Z | 2021-07-23T02:21:08.000Z | multiclass/multiclass_test.ipynb | kesamet/examples | 6baef94cc7db99abf17ca3df7dae35fcc3b08d25 | [
"Apache-2.0"
] | 2 | 2021-04-05T13:25:11.000Z | 2021-04-09T03:50:38.000Z | multiclass/multiclass_test.ipynb | kesamet/examples | 6baef94cc7db99abf17ca3df7dae35fcc3b08d25 | [
"Apache-2.0"
] | 14 | 2021-03-31T06:58:11.000Z | 2022-01-18T09:02:11.000Z | 38.822848 | 120 | 0.486076 | [
[
[
"import pandas as pd\nimport numpy as np\n\nfrom sklearn import metrics\nfrom sklearn import preprocessing\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.preprocessing import OneHotEncoder, StandardScaler\n\nfrom bedrock_client.bedrock.analyzer.model_analyzer import ModelAnalyzer\nfrom bedrock_client.bedrock.analyzer import ModelTypes\nfrom bedrock_client.bedrock.api import BedrockApi\nfrom bedrock_client.bedrock.metrics.service import ModelMonitoringService\nimport logging",
"WARNING:root:No module named 'tensorflow': AdversarialDebiasing will be unavailable. To install, run:\npip install 'aif360[AdversarialDebiasing]'\n"
],
[
"def load_dataset(filepath, target):\n df = pd.read_csv(filepath)\n df['large_rings'] = (df['Rings'] > 10).astype(int)\n\n # Ensure nothing missing\n original_len = len(df)\n df.dropna(how=\"any\", axis=0, inplace=True)\n num_rows_dropped = original_len - len(df)\n if num_rows_dropped > 0:\n print(f\"Warning - dropped {num_rows_dropped} rows with NA data.\")\n\n y = df[target].values\n df.drop(target, axis=1, inplace=True)\n\n return df, y",
"_____no_output_____"
],
[
"def train_log_reg_model(X, y, seed=0, C=1, verbose=False):\n verbose and print('\\nTraining\\nScaling...')\n scaling = StandardScaler()\n X = scaling.fit_transform(X)\n\n verbose and print('Fitting...')\n verbose and print('C:', C)\n model = LogisticRegression(random_state=seed, C=C, max_iter=4000)\n model.fit(X, y)\n\n verbose and print('Chaining pipeline...')\n pipe = Pipeline([('scaling', scaling), ('model', model)])\n\n verbose and print('Training Done.')\n\n return pipe",
"_____no_output_____"
],
[
"def compute_log_metrics(pipe,\n x_test,\n y_test,\n y_test_onehot):\n test_prob = pipe.predict_proba(x_test)\n test_pred = pipe.predict(x_test)\n\n acc = metrics.accuracy_score(y_test, test_pred)\n precision = metrics.precision_score(y_test, test_pred, average='macro')\n recall = metrics.recall_score(y_test, test_pred, average='macro')\n f1_score = metrics.f1_score(y_test, test_pred, average='macro')\n roc_auc = metrics.roc_auc_score(y_test_onehot, test_prob, average='macro', multi_class='ovr')\n avg_prc = metrics.average_precision_score(y_test_onehot, test_prob, average='macro')\n print(\"\\nEvaluation\\n\"\n f\"\\tAccuracy = {acc:.4f}\\n\"\n f\"\\tPrecision (macro) = {precision:.4f}\\n\"\n f\"\\tRecall (macro) = {recall:.4f}\\n\"\n f\"\\tF1 score (macro) = {f1_score:.4f}\\n\"\n f\"\\tROC AUC (macro) = {roc_auc:.4f}\\n\"\n f\"\\tAverage precision (macro) = {avg_prc:.4f}\")\n\n\n # Bedrock Logger: captures model metrics\n bedrock = BedrockApi(logging.getLogger(__name__))\n \n # `log_chart_data` assumes binary classification\n # For multiclass labels, we can use a \"micro-average\" by \n # quantifying score on all classes jointly \n # See https://scikit-learn.org/stable/auto_examples/model_selection/plot_precision_recall.html\n # This will allow us to use the same `log_chart_data` method\n bedrock.log_chart_data(\n y_test_onehot.ravel().astype(int).tolist(), # list of int\n test_prob.ravel().astype(float).tolist() # list of float\n )\n\n bedrock.log_metric(\"Accuracy\", acc)\n bedrock.log_metric(\"Precision (macro)\", precision)\n bedrock.log_metric(\"Recall (macro)\", recall)\n bedrock.log_metric(\"F1 Score (macro)\", f1_score)\n bedrock.log_metric(\"ROC AUC (macro)\", roc_auc)\n bedrock.log_metric(\"Avg precision (macro)\", avg_prc)\n\n return test_prob, test_pred",
"_____no_output_____"
],
[
"x_train, y_train = load_dataset(\n filepath=\"data/abalone_train.csv\",\n target=\"Type\"\n)\nx_test, y_test = load_dataset(\n filepath=\"data/abalone_test.csv\",\n target=\"Type\"\n)",
"_____no_output_____"
],
[
"enc = OneHotEncoder(handle_unknown='ignore', sparse=False)\n# sklearn `roc_auc_score` and `average_precision_score` expects\n# binary label indicators with shape (n_samples, n_classes)\ny_train_onehot = enc.fit_transform(y_train.reshape(-1, 1))\ny_test_onehot = enc.fit_transform(y_test.reshape(-1, 1))\n\n# Convert target variable to numeric values\n# ModelMonitoringService.export_text expect both features\n# and inference to be numeric values\ny_train = np.argmax(y_train_onehot, axis=1)\ny_test = np.argmax(y_test_onehot, axis=1)",
"_____no_output_____"
],
[
"for value, category in enumerate(enc.categories_[0]):\n print(f'{category} : {value}')",
"F : 0\nI : 1\nM : 2\n"
],
[
"pipe = train_log_reg_model(x_train,\n y_train,\n seed=0,\n C=1e-1,\n verbose=True)",
"\nTraining\nScaling...\nFitting...\nC: 0.1\nChaining pipeline...\nTraining Done.\n"
],
[
"test_prob, test_pred = compute_log_metrics(pipe,\n x_test,\n y_test,\n y_test_onehot)\n# Ignore ERROR, this is for testing purposes",
"ERROR:__main__:BEDROCK API TOKEN not found\nERROR:__main__:BEDROCK API TOKEN not found\nERROR:__main__:BEDROCK API TOKEN not found\nERROR:__main__:BEDROCK API TOKEN not found\nERROR:__main__:BEDROCK API TOKEN not found\nERROR:__main__:BEDROCK API TOKEN not found\nERROR:__main__:BEDROCK API TOKEN not found\n"
],
[
"CONFIG_FAI = {\n 'large_rings': {\n 'privileged_attribute_values': [1],\n # privileged group name corresponding to values=[1]\n 'privileged_group_name': 'Large', \n 'unprivileged_attribute_values': [0],\n # unprivileged group name corresponding to values=[0]\n 'unprivileged_group_name': 'Small', \n }\n}\n\n# Train Shap model and calculate xafai metrics\nanalyzer = (\n ModelAnalyzer(pipe[1],\n model_name='logistic',\n model_type=ModelTypes.LINEAR)\n .train_features(x_train)\n .test_features(x_test)\n .fairness_config(CONFIG_FAI)\n .test_labels(y_test)\n .test_inference(test_pred)\n)\nanalyzer.analyze()",
"_____no_output_____"
],
[
"ModelMonitoringService.export_text(\n features=x_train.iteritems(), # assumes numeric values\n inference=test_pred.tolist(), # assumes numeric values\n)",
"_____no_output_____"
],
[
"for item in x_train.iteritems():\n print(item)",
"('LongestShell', 0 0.245\n1 0.620\n2 0.455\n3 0.480\n4 0.430\n ... \n2917 0.455\n2918 0.665\n2919 0.440\n2920 0.505\n2921 0.580\nName: LongestShell, Length: 2922, dtype: float64)\n('Diameter', 0 0.195\n1 0.510\n2 0.345\n3 0.355\n4 0.325\n ... \n2917 0.350\n2918 0.535\n2919 0.350\n2920 0.405\n2921 0.450\nName: Diameter, Length: 2922, dtype: float64)\n('Height', 0 0.060\n1 0.180\n2 0.105\n3 0.115\n4 0.115\n ... \n2917 0.140\n2918 0.225\n2919 0.135\n2920 0.140\n2921 0.120\nName: Height, Length: 2922, dtype: float64)\n('WholeWeight', 0 0.0950\n1 1.3315\n2 0.4005\n3 0.4725\n4 0.3865\n ... \n2917 0.5725\n2918 2.1835\n2919 0.4350\n2920 0.8750\n2921 0.8685\nName: WholeWeight, Length: 2922, dtype: float64)\n('ShuckedWeight', 0 0.0445\n1 0.5940\n2 0.1640\n3 0.2065\n4 0.1475\n ... \n2917 0.1965\n2918 0.7535\n2919 0.1815\n2920 0.2665\n2921 0.4180\nName: ShuckedWeight, Length: 2922, dtype: float64)\n('VisceraWeight', 0 0.0245\n1 0.2760\n2 0.0755\n3 0.1120\n4 0.1065\n ... \n2917 0.1325\n2918 0.3910\n2919 0.0830\n2920 0.1740\n2921 0.1475\nName: VisceraWeight, Length: 2922, dtype: float64)\n('ShellWeight', 0 0.0260\n1 0.3880\n2 0.1260\n3 0.1320\n4 0.1100\n ... \n2917 0.1750\n2918 0.8850\n2919 0.1250\n2920 0.2850\n2921 0.2605\nName: ShellWeight, Length: 2922, dtype: float64)\n('Rings', 0 4\n1 11\n2 8\n3 8\n4 11\n ..\n2917 10\n2918 27\n2919 12\n2920 12\n2921 8\nName: Rings, Length: 2922, dtype: int64)\n('large_rings', 0 0\n1 1\n2 0\n3 0\n4 1\n ..\n2917 0\n2918 1\n2919 1\n2920 1\n2921 0\nName: large_rings, Length: 2922, dtype: int64)\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb435b4753614b7501bca58671aaa89f448d6f89 | 19,404 | ipynb | Jupyter Notebook | 01_Getting_&_Knowing_Your_Data/Chipotle/Exercise_with_Solutions.ipynb | Nick-Kolowich/pandas_exercises | 93c846a81f35fdf645a80a2f3e797de8bd91767d | [
"BSD-3-Clause"
] | null | null | null | 01_Getting_&_Knowing_Your_Data/Chipotle/Exercise_with_Solutions.ipynb | Nick-Kolowich/pandas_exercises | 93c846a81f35fdf645a80a2f3e797de8bd91767d | [
"BSD-3-Clause"
] | null | null | null | 01_Getting_&_Knowing_Your_Data/Chipotle/Exercise_with_Solutions.ipynb | Nick-Kolowich/pandas_exercises | 93c846a81f35fdf645a80a2f3e797de8bd91767d | [
"BSD-3-Clause"
] | null | null | null | 23.52 | 195 | 0.422387 | [
[
[
"# Ex2 - Getting and Knowing your Data\n\nCheck out [Chipotle Exercises Video Tutorial](https://www.youtube.com/watch?v=lpuYZ5EUyS8&list=PLgJhDSE2ZLxaY_DigHeiIDC1cD09rXgJv&index=2) to watch a data scientist go through the exercises",
"_____no_output_____"
],
[
"This time we are going to pull data directly from the internet.\nSpecial thanks to: https://github.com/justmarkham for sharing the dataset and materials.\n\n### Step 1. Import the necessary libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"### Step 2. Import the dataset from this [address](https://raw.githubusercontent.com/justmarkham/DAT8/master/data/chipotle.tsv). ",
"_____no_output_____"
],
[
"### Step 3. Assign it to a variable called chipo.",
"_____no_output_____"
]
],
[
[
"url = 'https://raw.githubusercontent.com/justmarkham/DAT8/master/data/chipotle.tsv'\n \nchipo = pd.read_csv(url, sep = '\\t')",
"_____no_output_____"
]
],
[
[
"### Step 4. See the first 10 entries",
"_____no_output_____"
]
],
[
[
"chipo.head(10)",
"_____no_output_____"
]
],
[
[
"### Step 5. What is the number of observations in the dataset?",
"_____no_output_____"
]
],
[
[
"# Solution 1\n\nchipo.shape[0] # entries <= 4622 observations",
"_____no_output_____"
],
[
"# Solution 2\n\nchipo.info() # entries <= 4622 observations",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 4622 entries, 0 to 4621\nData columns (total 5 columns):\norder_id 4622 non-null int64\nquantity 4622 non-null int64\nitem_name 4622 non-null object\nchoice_description 3376 non-null object\nitem_price 4622 non-null object\ndtypes: int64(2), object(3)\nmemory usage: 180.6+ KB\n"
]
],
[
[
"### Step 6. What is the number of columns in the dataset?",
"_____no_output_____"
]
],
[
[
"chipo.shape[1]",
"_____no_output_____"
]
],
[
[
"### Step 7. Print the name of all the columns.",
"_____no_output_____"
]
],
[
[
"chipo.columns",
"_____no_output_____"
]
],
[
[
"### Step 8. How is the dataset indexed?",
"_____no_output_____"
]
],
[
[
"chipo.index",
"_____no_output_____"
]
],
[
[
"### Step 9. Which was the most-ordered item? ",
"_____no_output_____"
]
],
[
[
"c = chipo.groupby('item_name')\nc = c.sum()\nc = c.sort_values(['quantity'], ascending=False)\nc.head(1)",
"_____no_output_____"
]
],
[
[
"### Step 10. For the most-ordered item, how many items were ordered?",
"_____no_output_____"
]
],
[
[
"c = chipo.groupby('item_name')\nc = c.sum()\nc = c.sort_values(['quantity'], ascending=False)\nc.head(1)",
"_____no_output_____"
]
],
[
[
"### Step 11. What was the most ordered item in the choice_description column?",
"_____no_output_____"
]
],
[
[
"c = chipo.groupby('choice_description').sum()\nc = c.sort_values(['quantity'], ascending=False)\nc.head(1)\n# Diet Coke 159",
"_____no_output_____"
]
],
[
[
"### Step 12. How many items were orderd in total?",
"_____no_output_____"
]
],
[
[
"total_items_orders = chipo.quantity.sum()\ntotal_items_orders",
"_____no_output_____"
]
],
[
[
"### Step 13. Turn the item price into a float",
"_____no_output_____"
],
[
"#### Step 13.a. Check the item price type",
"_____no_output_____"
]
],
[
[
"chipo.item_price.dtype",
"_____no_output_____"
]
],
[
[
"#### Step 13.b. Create a lambda function and change the type of item price",
"_____no_output_____"
]
],
[
[
"dollarizer = lambda x: float(x[1:-1])\nchipo.item_price = chipo.item_price.apply(dollarizer)",
"_____no_output_____"
]
],
[
[
"#### Step 13.c. Check the item price type",
"_____no_output_____"
]
],
[
[
"chipo.item_price.dtype",
"_____no_output_____"
]
],
[
[
"### Step 14. How much was the revenue for the period in the dataset?",
"_____no_output_____"
]
],
[
[
"revenue = (chipo['quantity']* chipo['item_price']).sum()\n\nprint('Revenue was: $' + str(np.round(revenue,2)))",
"Revenue was: $39237.02\n"
]
],
[
[
"### Step 15. How many orders were made in the period?",
"_____no_output_____"
]
],
[
[
"orders = chipo.order_id.value_counts().count()\norders",
"_____no_output_____"
]
],
[
[
"### Step 16. What is the average revenue amount per order?",
"_____no_output_____"
]
],
[
[
"# Solution 1\n\nchipo['revenue'] = chipo['quantity'] * chipo['item_price']\norder_grouped = chipo.groupby(by=['order_id']).sum()\norder_grouped.mean()['revenue']",
"_____no_output_____"
],
[
"# Solution 2\n\nchipo.groupby(by=['order_id']).sum().mean()['revenue']",
"_____no_output_____"
]
],
[
[
"### Step 17. How many different items are sold?",
"_____no_output_____"
]
],
[
[
"chipo.item_name.value_counts().count()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb43613d030e70036e187fc3c73fe2bca508081c | 483,527 | ipynb | Jupyter Notebook | notebooks/Untitled.ipynb | mwang87/GNPS_ClassicalNetwork_GroupsComparison | 21aa04552b5cbdf7c0d9072e4c58d2075cf37485 | [
"MIT"
] | null | null | null | notebooks/Untitled.ipynb | mwang87/GNPS_ClassicalNetwork_GroupsComparison | 21aa04552b5cbdf7c0d9072e4c58d2075cf37485 | [
"MIT"
] | null | null | null | notebooks/Untitled.ipynb | mwang87/GNPS_ClassicalNetwork_GroupsComparison | 21aa04552b5cbdf7c0d9072e4c58d2075cf37485 | [
"MIT"
] | null | null | null | 1,469.68693 | 417,388 | 0.957841 | [
[
[
"from upsetplot import generate_counts\nfrom upsetplot import from_memberships",
"_____no_output_____"
],
[
"example = generate_counts()",
"_____no_output_____"
],
[
"example",
"_____no_output_____"
],
[
"example.to_dict()",
"_____no_output_____"
],
[
"import pandas as pd\nurl = \"https://gnps.ucsd.edu/ProteoSAFe/DownloadResultFile?task=62b32c4c5fd440a4802088da0d48b07c&file=clusterinfosummarygroup_attributes_withIDs_withcomponentID/\"\n\ndf = pd.read_csv(url, sep=\"\\t\")",
"_____no_output_____"
],
[
"df.columns.to_list()\n#df[\"ATTRIBUTE_OrganismID\"]",
"_____no_output_____"
],
[
"df[\"DefaultGroups\"].head().to_list()",
"_____no_output_____"
],
[
"import pandas as pd\nfrom upsetplot import generate_counts\nfrom upsetplot import from_memberships\nfrom upsetplot import plot\nfrom matplotlib import pyplot\nimport uuid\n\n\ngnps_task = \"8443d633eb2447538cb977a7b271eaed\"\nmetadata_column = \"ATTRIBUTE_Media\"\n\ndata_url = \"https://gnps.ucsd.edu/ProteoSAFe/DownloadResultFile?task={}&file=clusterinfosummarygroup_attributes_withIDs_withcomponentID/\".format(gnps_task)\ndata_df = pd.read_csv(data_url, sep=\"\\t\")",
"_____no_output_____"
],
[
"merged_terms = list(set(data_df[metadata_column].dropna()))\n\nterms_to_consider = set()\nfor term in merged_terms:\n terms_to_consider = terms_to_consider | set(term.split(\",\"))\n\nmetadata_terms = set(terms_to_consider)\nmetadata_terms = set(list(metadata_terms)[:3])\n\nmembership = []\nfor group_value in data_df[metadata_column].to_list():\n group_splits = set(group_value.split(\",\"))\n group_splits = list(group_splits & metadata_terms)\n membership.append(group_splits)\n\nupset_data_df = from_memberships(membership)\nplotting_object = plot(upset_data_df, subset_size=\"count\", sort_by=\"cardinality\", orientation=\"vertical\")\n\n#uuid_save = str(uuid.uuid4())\n#pyplot.savefig(\"./{}.png\".format(uuid_save))",
"_____no_output_____"
],
[
"plotting_object = plot(upset_data_df, subset_size=\"count\", sort_by=\"cardinality\", totals_plot_elements=2)",
"_____no_output_____"
],
[
"plotting_object = plot(upset_data_df, subset_size=\"count\", sort_by=\"cardinality\", totals_plot_elements=4)",
"_____no_output_____"
],
[
"plotting_object = plot(upset_data_df, subset_size=\"count\", sort_by=\"cardinality\", totals_plot_elements=10)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4367cdc4dd8995a42fe06907848a425a22e0ae | 60,739 | ipynb | Jupyter Notebook | ai-platform-unified/notebooks/official/custom/custom-tabular-bq-managed-dataset.ipynb | thehardikv/ai-platform-samples | 0050d12476bcbfdb99d9894a3755a97da5cd80fe | [
"Apache-2.0"
] | null | null | null | ai-platform-unified/notebooks/official/custom/custom-tabular-bq-managed-dataset.ipynb | thehardikv/ai-platform-samples | 0050d12476bcbfdb99d9894a3755a97da5cd80fe | [
"Apache-2.0"
] | null | null | null | ai-platform-unified/notebooks/official/custom/custom-tabular-bq-managed-dataset.ipynb | thehardikv/ai-platform-samples | 0050d12476bcbfdb99d9894a3755a97da5cd80fe | [
"Apache-2.0"
] | null | null | null | 39.186452 | 417 | 0.539044 | [
[
[
"# Copyright 2020 Google LLC\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Custom training and online prediction\n\n<table align=\"left\">\n <td>\n <a href=\"https://colab.research.google.com/github/GoogleCloudPlatform/ai-platform-samples/blob/master/ai-platform-unified/notebooks/official/custom-tabular-bq-managed-dataset.ipynb\">\n <img src=\"https://cloud.google.com/ml-engine/images/colab-logo-32px.png\" alt=\"Colab logo\"> Run in Colab\n </a>\n </td>\n <td>\n <a href=\"https://github.com/GoogleCloudPlatform/ai-platform-samples/blob/master/ai-platform-unified/notebooks/official/custom-tabular-bq-managed-dataset.ipynb\">\n <img src=\"https://cloud.google.com/ml-engine/images/github-logo-32px.png\" alt=\"GitHub logo\">\n View on GitHub\n </a>\n </td>\n</table>\n<br/><br/><br/>",
"_____no_output_____"
],
[
"## Overview\n\n\nThis tutorial demonstrates how to use the Vertex SDK for Python to train and deploy a custom tabular classification model for online prediction.",
"_____no_output_____"
],
[
"### Dataset\n\nThe dataset used for this tutorial is the penguins dataset from [BigQuery public datasets](https://cloud.google.com/bigquery/public-data). In this version of the dataset, you will use only the fields `culmen_length_mm`, `culmen_depth_mm`, `flipper_length_mm`, `body_mass_g` to predict the penguins species (`species`).",
"_____no_output_____"
],
[
"### Objective\n\nIn this notebook, you create a custom-trained model from a Python script in a Docker container using the Vertex SDK for Python, and then do a prediction on the deployed model by sending data. Alternatively, you can create custom-trained models using `gcloud` command-line tool, or online using the Cloud Console.\n\nThe steps performed include:\n\n- Create an Vertex AI custom job for training a model.\n- Train a TensorFlow model.\n- Deploy the `Model` resource to a serving `Endpoint` resource.\n- Make a prediction.\n- Undeploy the `Model` resource.",
"_____no_output_____"
],
[
"### Costs\n\nThis tutorial uses billable components of Google Cloud (GCP):\n\n* Vertex AI\n* Cloud Storage\n\nLearn about [Cloud Vertex AI\npricing](https://cloud.google.com/vertex-ai/pricing) and [Cloud Storage\npricing](https://cloud.google.com/storage/pricing), and use the [Pricing\nCalculator](https://cloud.google.com/products/calculator/)\nto generate a cost estimate based on your projected usage.",
"_____no_output_____"
],
[
"## Installation\n\nInstall the latest (preview) version of Vertex SDK for Python.",
"_____no_output_____"
]
],
[
[
"import os\n\n# The Google Cloud Notebook product has specific requirements\nIS_GOOGLE_CLOUD_NOTEBOOK = os.path.exists(\"/opt/deeplearning/metadata/env_version\")\n\n# Google Cloud Notebook requires dependencies to be installed with '--user'\nUSER_FLAG = \"\"\nif IS_GOOGLE_CLOUD_NOTEBOOK:\n USER_FLAG = \"--user\"",
"_____no_output_____"
],
[
"! pip3 install {USER_FLAG} --upgrade google-cloud-aiplatform",
"_____no_output_____"
]
],
[
[
"Install the latest GA version of *google-cloud-storage* library as well.",
"_____no_output_____"
]
],
[
[
"! pip3 install {USER_FLAG} -U google-cloud-storage",
"_____no_output_____"
]
],
[
[
"Install the latest GA version of *google-cloud-bigquery* library as well.",
"_____no_output_____"
]
],
[
[
"! pip3 install {USER_FLAG} -U \"google-cloud-bigquery[all]\"",
"_____no_output_____"
]
],
[
[
"### Restart the kernel\n\nOnce you've installed everything, you need to restart the notebook kernel so it can find the packages.",
"_____no_output_____"
]
],
[
[
"import os\n\nif not os.getenv(\"IS_TESTING\"):\n # Automatically restart kernel after installs\n import IPython\n\n app = IPython.Application.instance()\n app.kernel.do_shutdown(True)",
"_____no_output_____"
]
],
[
[
"## Before you begin\n\n### Select a GPU runtime\n\n**Make sure you're running this notebook in a GPU runtime if you have that option. In Colab, select \"Runtime --> Change runtime type > GPU\"**\n\n### Set up your Google Cloud project\n\n**The following steps are required, regardless of your notebook environment.**\n\n1. [Select or create a Google Cloud project](https://console.cloud.google.com/cloud-resource-manager). When you first create an account, you get a $300 free credit towards your compute/storage costs.\n\n2. [Make sure that billing is enabled for your project](https://cloud.google.com/billing/docs/how-to/modify-project).\n\n3. [Enable the Vertex AI API and Compute Engine API](https://console.cloud.google.com/flows/enableapi?apiid=aiplatform.googleapis.com,compute_component).\n\n4. If you are running this notebook locally, you will need to install the [Cloud SDK](https://cloud.google.com/sdk).\n\n5. Enter your project ID in the cell below. Then run the cell to make sure the\nCloud SDK uses the right project for all the commands in this notebook.\n\n**Note**: Jupyter runs lines prefixed with `!` as shell commands, and it interpolates Python variables prefixed with `$` into these commands.",
"_____no_output_____"
],
[
"#### Set your project ID\n\n**If you don't know your project ID**, you may be able to get your project ID using `gcloud`.",
"_____no_output_____"
]
],
[
[
"PROJECT_ID = \"\"\n\nif not os.getenv(\"IS_TESTING\"):\n # Get your Google Cloud project ID from gcloud\n shell_output=!gcloud config list --format 'value(core.project)' 2>/dev/null\n PROJECT_ID = shell_output[0]\n print(\"Project ID: \", PROJECT_ID)",
"_____no_output_____"
]
],
[
[
"Otherwise, set your project ID here.",
"_____no_output_____"
]
],
[
[
"if PROJECT_ID == \"\" or PROJECT_ID is None:\n PROJECT_ID = \"[your-project-id]\" # @param {type:\"string\"}",
"_____no_output_____"
]
],
[
[
"#### Timestamp\n\nIf you are in a live tutorial session, you might be using a shared test account or project. To avoid name collisions between users on resources created, you create a timestamp for each instance session, and append it onto the name of resources you create in this tutorial.",
"_____no_output_____"
]
],
[
[
"from datetime import datetime\n\nTIMESTAMP = datetime.now().strftime(\"%Y%m%d%H%M%S\")",
"_____no_output_____"
]
],
[
[
"### Authenticate your Google Cloud account\n\n**If you are using Google Cloud Notebooks**, your environment is already\nauthenticated. Skip this step.\n\n**If you are using Colab**, run the cell below and follow the instructions\nwhen prompted to authenticate your account via oAuth.\n\n**Otherwise**, follow these steps:\n\n1. In the Cloud Console, go to the [**Create service account key**\n page](https://console.cloud.google.com/apis/credentials/serviceaccountkey).\n\n2. Click **Create service account**.\n\n3. In the **Service account name** field, enter a name, and\n click **Create**.\n\n4. In the **Grant this service account access to project** section, click the **Role** drop-down list. Type \"Vertex AI\"\ninto the filter box, and select\n **Vertex AI Administrator**. Type \"Storage Object Admin\" into the filter box, and select **Storage Object Admin**.\n\n5. Click *Create*. A JSON file that contains your key downloads to your\nlocal environment.\n\n6. Enter the path to your service account key as the\n`GOOGLE_APPLICATION_CREDENTIALS` variable in the cell below and run the cell.",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\n\n# If you are running this notebook in Colab, run this cell and follow the\n# instructions to authenticate your GCP account. This provides access to your\n# Cloud Storage bucket and lets you submit training jobs and prediction\n# requests.\n\n# The Google Cloud Notebook product has specific requirements\nIS_GOOGLE_CLOUD_NOTEBOOK = os.path.exists(\"/opt/deeplearning/metadata/env_version\")\n\n# If on Google Cloud Notebooks, then don't execute this code\nif not IS_GOOGLE_CLOUD_NOTEBOOK:\n if \"google.colab\" in sys.modules:\n from google.colab import auth as google_auth\n\n google_auth.authenticate_user()\n\n # If you are running this notebook locally, replace the string below with the\n # path to your service account key and run this cell to authenticate your GCP\n # account.\n elif not os.getenv(\"IS_TESTING\"):\n %env GOOGLE_APPLICATION_CREDENTIALS ''",
"_____no_output_____"
]
],
[
[
"### Create a Cloud Storage bucket\n\n**The following steps are required, regardless of your notebook environment.**\n\nWhen you submit a training job using the Cloud SDK, you upload a Python package\ncontaining your training code to a Cloud Storage bucket. Vertex AI runs\nthe code from this package. In this tutorial, Vertex AI also saves the\ntrained model that results from your job in the same bucket. Using this model artifact, you can then\ncreate Vertex AI model and endpoint resources in order to serve\nonline predictions.\n\nSet the name of your Cloud Storage bucket below. It must be unique across all\nCloud Storage buckets.\n\nYou may also change the `REGION` variable, which is used for operations\nthroughout the rest of this notebook. Make sure to [choose a region where Vertex AI services are\navailable](https://cloud.google.com/vertex-ai/docs/general/locations#available_regions). You may\nnot use a Multi-Regional Storage bucket for training with Vertex AI.",
"_____no_output_____"
]
],
[
[
"BUCKET_NAME = \"\" # @param {type:\"string\"}\nREGION = \"us-central1\" # @param {type:\"string\"}",
"_____no_output_____"
],
[
"if BUCKET_NAME == \"\" or BUCKET_NAME is None or BUCKET_NAME == \"gs://[your-bucket-name]\":\n BUCKET_NAME = \"gs://\" + PROJECT_ID + \"aip-\" + TIMESTAMP",
"_____no_output_____"
]
],
[
[
"**Only if your bucket doesn't already exist**: Run the following cell to create your Cloud Storage bucket.",
"_____no_output_____"
]
],
[
[
"! gsutil mb -l $REGION $BUCKET_NAME",
"_____no_output_____"
]
],
[
[
"Finally, validate access to your Cloud Storage bucket by examining its contents:",
"_____no_output_____"
]
],
[
[
"! gsutil ls -al $BUCKET_NAME",
"_____no_output_____"
]
],
[
[
"### Set up variables\n\nNext, set up some variables used throughout the tutorial.",
"_____no_output_____"
],
[
"#### Import Vertex SDK for Python\n\nImport the Vertex SDK for Python into your Python environment and initialize it.",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\n\nfrom google.cloud import aiplatform\nfrom google.cloud.aiplatform import gapic as aip\n\naiplatform.init(project=PROJECT_ID, location=REGION, staging_bucket=BUCKET_NAME)",
"_____no_output_____"
]
],
[
[
"#### Set hardware accelerators\n\nYou can set hardware accelerators for both training and prediction.\n\nSet the variables `TRAIN_GPU/TRAIN_NGPU` and `DEPLOY_GPU/DEPLOY_NGPU` to use a container image supporting a GPU and the number of GPUs allocated to the virtual machine (VM) instance. For example, to use a GPU container image with 4 Nvidia Tesla K80 GPUs allocated to each VM, you would specify:\n\n (aip.AcceleratorType.NVIDIA_TESLA_K80, 4)\n\nSee the [locations where accelerators are available](https://cloud.google.com/vertex-ai/docs/general/locations#accelerators).\n\nOtherwise specify `(None, None)` to use a container image to run on a CPU.\n\nLearn [which accelerators are available in your region.](https://cloud.google.com/vertex-ai/docs/general/locations#accelerators)",
"_____no_output_____"
]
],
[
[
"TRAIN_GPU, TRAIN_NGPU = (aip.AcceleratorType.NVIDIA_TESLA_K80, 1)\n\nDEPLOY_GPU, DEPLOY_NGPU = (aip.AcceleratorType.NVIDIA_TESLA_K80, 1)",
"_____no_output_____"
]
],
[
[
"#### Set pre-built containers\n\nVertex AI provides pre-built containers to run training and prediction.\n\nFor the latest list, see [Pre-built containers for training](https://cloud.google.com/vertex-ai/docs/training/pre-built-containers) and [Pre-built containers for prediction](https://cloud.google.com/vertex-ai/docs/predictions/pre-built-containers)",
"_____no_output_____"
]
],
[
[
"TRAIN_VERSION = \"tf-gpu.2-4\"\nDEPLOY_VERSION = \"tf2-gpu.2-4\"\n\nTRAIN_IMAGE = \"gcr.io/cloud-aiplatform/training/{}:latest\".format(TRAIN_VERSION)\nDEPLOY_IMAGE = \"gcr.io/cloud-aiplatform/prediction/{}:latest\".format(DEPLOY_VERSION)\n\nprint(\"Training:\", TRAIN_IMAGE, TRAIN_GPU, TRAIN_NGPU)\nprint(\"Deployment:\", DEPLOY_IMAGE, DEPLOY_GPU, DEPLOY_NGPU)",
"_____no_output_____"
]
],
[
[
"#### Set machine types\n\nNext, set the machine types to use for training and prediction.\n\n- Set the variables `TRAIN_COMPUTE` and `DEPLOY_COMPUTE` to configure your compute resources for training and prediction.\n - `machine type`\n - `n1-standard`: 3.75GB of memory per vCPU\n - `n1-highmem`: 6.5GB of memory per vCPU\n - `n1-highcpu`: 0.9 GB of memory per vCPU\n - `vCPUs`: number of \\[2, 4, 8, 16, 32, 64, 96 \\]\n\n*Note: The following is not supported for training:*\n\n - `standard`: 2 vCPUs\n - `highcpu`: 2, 4 and 8 vCPUs\n\n*Note: You may also use n2 and e2 machine types for training and deployment, but they do not support GPUs*.\n\nLearn [which machine types are available](https://cloud.google.com/vertex-ai/docs/training/configure-compute) for training and [for prediction](https://cloud.google.com/vertex-ai/docs/predictions/configure-compute)",
"_____no_output_____"
]
],
[
[
"MACHINE_TYPE = \"n1-standard\"\n\nVCPU = \"4\"\nTRAIN_COMPUTE = MACHINE_TYPE + \"-\" + VCPU\nprint(\"Train machine type\", TRAIN_COMPUTE)\n\nMACHINE_TYPE = \"n1-standard\"\n\nVCPU = \"4\"\nDEPLOY_COMPUTE = MACHINE_TYPE + \"-\" + VCPU\nprint(\"Deploy machine type\", DEPLOY_COMPUTE)",
"_____no_output_____"
]
],
[
[
"# Tutorial\n\nNow you are ready to start creating your own custom-trained model with the penguins dataset.",
"_____no_output_____"
],
[
"## Prepare the data\n\nTo improve the convergence of our custom deep learning model, we need to normalize the data. To prepare for this, calculate the mean and standard deviation for each numeric column.\n\nThis will be passed to the training script to normalize the data before training.\nThis will also be used to normalize the testing data later in this notebook.",
"_____no_output_____"
]
],
[
[
"BQ_SOURCE = \"bq://bigquery-public-data.ml_datasets.penguins\"",
"_____no_output_____"
],
[
"import json\n\nimport numpy as np\n# Calculate mean and std across all rows\nfrom google.cloud import bigquery\n\nNA_VALUES = [\"NA\", \".\"]\n\n# Set up BigQuery clients\nbqclient = bigquery.Client()\n\n\n# Download a table\ndef download_table(bq_table_uri: str):\n # Remove bq:// prefix if present\n prefix = \"bq://\"\n if bq_table_uri.startswith(prefix):\n bq_table_uri = bq_table_uri[len(prefix) :]\n\n table = bigquery.TableReference.from_string(bq_table_uri)\n rows = bqclient.list_rows(\n table,\n )\n return rows.to_dataframe()\n\n\n# Remove NA values\ndef clean_dataframe(df):\n return df.replace(to_replace=NA_VALUES, value=np.NaN).dropna()\n\n\ndef calculate_mean_and_std(df):\n # Calculate mean and std for each applicable column\n mean_and_std = {}\n dtypes = list(zip(df.dtypes.index, map(str, df.dtypes)))\n # Normalize numeric columns.\n for column, dtype in dtypes:\n if dtype == \"float32\" or dtype == \"float64\":\n mean_and_std[column] = {\n \"mean\": df[column].mean(),\n \"std\": df[column].std(),\n }\n\n return mean_and_std\n\n\ndataframe = download_table(BQ_SOURCE)\ndataframe = clean_dataframe(dataframe)\nmean_and_std = calculate_mean_and_std(dataframe)\n\nprint(\"The mean and stds for each column are: \" + str(mean_and_std))\n\n# Write to a file\nMEAN_AND_STD_JSON_FILE = \"mean_and_std.json\"\n\nwith open(MEAN_AND_STD_JSON_FILE, \"w\") as outfile:\n json.dump(mean_and_std, outfile)\n\n# Save to the staging bucket\n! gsutil cp {MEAN_AND_STD_JSON_FILE} {BUCKET_NAME}",
"_____no_output_____"
]
],
[
[
"## Create a managed tabular dataset from BigQuery dataset\n\nYour first step in training a model is to create a managed dataset instance.",
"_____no_output_____"
]
],
[
[
"dataset = aiplatform.TabularDataset.create(\n display_name=\"sample-penguins\", bq_source=BQ_SOURCE\n)",
"_____no_output_____"
]
],
[
[
"## Train a model\n\nThere are two ways you can train a custom model using a container image:\n\n- **Use a Google Cloud prebuilt container**. If you use a prebuilt container, you will additionally specify a Python package to install into the container image. This Python package contains your code for training a custom model.\n\n- **Use your own custom container image**. If you use your own container, the container needs to contain your code for training a custom model.",
"_____no_output_____"
],
[
"### Define the command args for the training script\n\nPrepare the command-line arguments to pass to your training script.\n- `args`: The command line arguments to pass to the corresponding Python module. In this example, they will be:\n - `\"--epochs=\" + EPOCHS`: The number of epochs for training.\n - `\"--batch_size=\" + BATCH_SIZE`: The number of batch size for training.\n - `\"--distribute=\" + TRAIN_STRATEGY\"` : The training distribution strategy to use for single or distributed training.\n - `\"single\"`: single device.\n - `\"mirror\"`: all GPU devices on a single compute instance.\n - `\"multi\"`: all GPU devices on all compute instances.\n - `\"--mean_and_std_json_file=\" + FILE_PATH`: The file on Google Cloud Storage with pre-calculated means and standard deviations.",
"_____no_output_____"
]
],
[
[
"JOB_NAME = \"custom_job_\" + TIMESTAMP\nMODEL_DIR = \"{}/{}\".format(BUCKET_NAME, JOB_NAME)\n\nif not TRAIN_NGPU or TRAIN_NGPU < 2:\n TRAIN_STRATEGY = \"single\"\nelse:\n TRAIN_STRATEGY = \"mirror\"\n\nEPOCHS = 20\nBATCH_SIZE = 10\n\nCMDARGS = [\n \"--epochs=\" + str(EPOCHS),\n \"--batch_size=\" + str(BATCH_SIZE),\n \"--distribute=\" + TRAIN_STRATEGY,\n \"--mean_and_std_json_file=\" + f\"{BUCKET_NAME}/{MEAN_AND_STD_JSON_FILE}\",\n]",
"_____no_output_____"
]
],
[
[
"#### Training script\n\nIn the next cell, you will write the contents of the training script, `task.py`. In summary:\n\n- Loads the data from the BigQuery table using the BigQuery Python client library.\n- Loads the pre-calculated mean and standard deviation from the Google Cloud Storage bucket.\n- Builds a model using TF.Keras model API.\n- Compiles the model (`compile()`).\n- Sets a training distribution strategy according to the argument `args.distribute`.\n- Trains the model (`fit()`) with epochs and batch size according to the arguments `args.epochs` and `args.batch_size`\n- Get the directory where to save the model artifacts from the environment variable `AIP_MODEL_DIR`. This variable is set by the training service.\n- Saves the trained model (`save(MODEL_DIR)`) to the specified model directory.",
"_____no_output_____"
]
],
[
[
"%%writefile task.py\n\nimport argparse\nimport tensorflow as tf\nimport numpy as np\nimport os\n\nimport pandas as pd\nimport tensorflow as tf\n\nfrom google.cloud import bigquery\nfrom google.cloud import storage\n\n# Read environmental variables\ntraining_data_uri = os.environ[\"AIP_TRAINING_DATA_URI\"]\nvalidation_data_uri = os.environ[\"AIP_VALIDATION_DATA_URI\"]\ntest_data_uri = os.environ[\"AIP_TEST_DATA_URI\"]\n\n# Read args\nparser = argparse.ArgumentParser()\nparser.add_argument('--epochs', dest='epochs',\n default=10, type=int,\n help='Number of epochs.')\nparser.add_argument('--batch_size', dest='batch_size',\n default=10, type=int,\n help='Batch size.')\nparser.add_argument('--distribute', dest='distribute', type=str, default='single',\n help='Distributed training strategy.')\nparser.add_argument('--mean_and_std_json_file', dest='mean_and_std_json_file', type=str,\n help='GCS URI to the JSON file with pre-calculated column means and standard deviations.')\nargs = parser.parse_args()\n\ndef download_blob(bucket_name, source_blob_name, destination_file_name):\n \"\"\"Downloads a blob from the bucket.\"\"\"\n # bucket_name = \"your-bucket-name\"\n # source_blob_name = \"storage-object-name\"\n # destination_file_name = \"local/path/to/file\"\n\n storage_client = storage.Client()\n\n bucket = storage_client.bucket(bucket_name)\n\n # Construct a client side representation of a blob.\n # Note `Bucket.blob` differs from `Bucket.get_blob` as it doesn't retrieve\n # any content from Google Cloud Storage. As we don't need additional data,\n # using `Bucket.blob` is preferred here.\n blob = bucket.blob(source_blob_name)\n blob.download_to_filename(destination_file_name)\n\n print(\n \"Blob {} downloaded to {}.\".format(\n source_blob_name, destination_file_name\n )\n )\n\ndef extract_bucket_and_prefix_from_gcs_path(gcs_path: str):\n \"\"\"Given a complete GCS path, return the bucket name and prefix as a tuple.\n\n Example Usage:\n\n bucket, prefix = extract_bucket_and_prefix_from_gcs_path(\n \"gs://example-bucket/path/to/folder\"\n )\n\n # bucket = \"example-bucket\"\n # prefix = \"path/to/folder\"\n\n Args:\n gcs_path (str):\n Required. A full path to a Google Cloud Storage folder or resource.\n Can optionally include \"gs://\" prefix or end in a trailing slash \"/\".\n\n Returns:\n Tuple[str, Optional[str]]\n A (bucket, prefix) pair from provided GCS path. If a prefix is not\n present, a None will be returned in its place.\n \"\"\"\n if gcs_path.startswith(\"gs://\"):\n gcs_path = gcs_path[5:]\n if gcs_path.endswith(\"/\"):\n gcs_path = gcs_path[:-1]\n\n gcs_parts = gcs_path.split(\"/\", 1)\n gcs_bucket = gcs_parts[0]\n gcs_blob_prefix = None if len(gcs_parts) == 1 else gcs_parts[1]\n\n return (gcs_bucket, gcs_blob_prefix)\n\n# Download means and std\ndef download_mean_and_std(mean_and_std_json_file):\n \"\"\"Download mean and std for each column\"\"\"\n import json\n \n bucket, file_path = extract_bucket_and_prefix_from_gcs_path(mean_and_std_json_file)\n download_blob(bucket_name=bucket, source_blob_name=file_path, destination_file_name=file_path)\n \n with open(file_path, 'r') as file:\n return json.loads(file.read())\n \nmean_and_std = download_mean_and_std(args.mean_and_std_json_file)\n\n# Single Machine, single compute device\nif args.distribute == 'single':\n if tf.test.is_gpu_available():\n strategy = tf.distribute.OneDeviceStrategy(device=\"/gpu:0\")\n else:\n strategy = tf.distribute.OneDeviceStrategy(device=\"/cpu:0\")\n# Single Machine, multiple compute device\nelif args.distribute == 'mirror':\n strategy = tf.distribute.MirroredStrategy()\n# Multiple Machine, multiple compute device\nelif args.distribute == 'multi':\n strategy = tf.distribute.experimental.MultiWorkerMirroredStrategy()\n\n# Set up training variables\nLABEL_COLUMN = \"species\"\nUNUSED_COLUMNS = []\nNA_VALUES = [\"NA\", \".\"]\n\n# Possible categorical values\nSPECIES = ['Adelie Penguin (Pygoscelis adeliae)',\n 'Chinstrap penguin (Pygoscelis antarctica)',\n 'Gentoo penguin (Pygoscelis papua)']\nISLANDS = ['Dream', 'Biscoe', 'Torgersen']\nSEXES = ['FEMALE', 'MALE']\n\n# Set up BigQuery clients\nbqclient = bigquery.Client()\n\n# Download a table\ndef download_table(bq_table_uri: str):\n # Remove bq:// prefix if present\n prefix = \"bq://\"\n if bq_table_uri.startswith(prefix):\n bq_table_uri = bq_table_uri[len(prefix):]\n\n table = bigquery.TableReference.from_string(bq_table_uri)\n rows = bqclient.list_rows(\n table,\n )\n return rows.to_dataframe()\n\n\ndf_train = download_table(training_data_uri)\ndf_validation = download_table(validation_data_uri)\ndf_test = download_table(test_data_uri)\n\n# Remove NA values\ndef clean_dataframe(df):\n return df.replace(to_replace=NA_VALUES, value=np.NaN).dropna()\n\n\ndf_train = clean_dataframe(df_train)\n# df_validation = clean_dataframe(df_validation)\ndf_validation = clean_dataframe(df_validation)\n\n_CATEGORICAL_TYPES = {\n \"island\": pd.api.types.CategoricalDtype(categories=ISLANDS),\n \"species\": pd.api.types.CategoricalDtype(categories=SPECIES),\n \"sex\": pd.api.types.CategoricalDtype(categories=SEXES),\n}\n\n\ndef standardize(df, mean_and_std):\n \"\"\"Scales numerical columns using their means and standard deviation to get\n z-scores: the mean of each numerical column becomes 0, and the standard\n deviation becomes 1. This can help the model converge during training.\n\n Args:\n df: Pandas df\n\n Returns:\n Input df with the numerical columns scaled to z-scores\n \"\"\"\n dtypes = list(zip(df.dtypes.index, map(str, df.dtypes)))\n # Normalize numeric columns.\n for column, dtype in dtypes:\n if dtype == \"float32\":\n df[column] -= mean_and_std[column][\"mean\"]\n df[column] /= mean_and_std[column][\"std\"]\n return df\n\ndef preprocess(df):\n \"\"\"Converts categorical features to numeric. Removes unused columns.\n\n Args:\n df: Pandas df with raw data\n\n Returns:\n df with preprocessed data\n \"\"\"\n df = df.drop(columns=UNUSED_COLUMNS)\n\n # Drop rows with NaN's\n df = df.dropna()\n\n # Convert integer valued (numeric) columns to floating point\n numeric_columns = df.select_dtypes([\"int32\", \"float32\", \"float64\"]).columns\n df[numeric_columns] = df[numeric_columns].astype(\"float32\")\n\n # Convert categorical columns to numeric\n cat_columns = df.select_dtypes([\"object\"]).columns\n\n df[cat_columns] = df[cat_columns].apply(\n lambda x: x.astype(_CATEGORICAL_TYPES[x.name])\n )\n df[cat_columns] = df[cat_columns].apply(lambda x: x.cat.codes)\n return df\n\n\ndef convert_dataframe_to_dataset(\n df_train,\n df_validation,\n mean_and_std\n):\n df_train = preprocess(df_train)\n df_validation = preprocess(df_validation)\n\n df_train_x, df_train_y = df_train, df_train.pop(LABEL_COLUMN)\n df_validation_x, df_validation_y = df_validation, df_validation.pop(LABEL_COLUMN)\n\n # Join train_x and eval_x to normalize on overall means and standard\n # deviations. Then separate them again.\n all_x = pd.concat([df_train_x, df_validation_x], keys=[\"train\", \"eval\"])\n all_x = standardize(all_x, mean_and_std)\n df_train_x, df_validation_x = all_x.xs(\"train\"), all_x.xs(\"eval\")\n\n y_train = np.asarray(df_train_y).astype(\"float32\")\n y_validation = np.asarray(df_validation_y).astype(\"float32\")\n\n # Convert to numpy representation\n x_train = np.asarray(df_train_x)\n x_test = np.asarray(df_validation_x)\n\n # Convert to one-hot representation\n y_train = tf.keras.utils.to_categorical(y_train, num_classes=len(SPECIES))\n y_validation = tf.keras.utils.to_categorical(y_validation, num_classes=len(SPECIES))\n\n dataset_train = tf.data.Dataset.from_tensor_slices((x_train, y_train))\n dataset_validation = tf.data.Dataset.from_tensor_slices((x_test, y_validation))\n return (dataset_train, dataset_validation)\n\n# Create datasets\ndataset_train, dataset_validation = convert_dataframe_to_dataset(df_train, df_validation, mean_and_std)\n\n# Shuffle train set\ndataset_train = dataset_train.shuffle(len(df_train))\n\ndef create_model(num_features):\n # Create model\n Dense = tf.keras.layers.Dense\n model = tf.keras.Sequential(\n [\n Dense(\n 100,\n activation=tf.nn.relu,\n kernel_initializer=\"uniform\",\n input_dim=num_features,\n ),\n Dense(75, activation=tf.nn.relu),\n Dense(50, activation=tf.nn.relu),\n Dense(25, activation=tf.nn.relu),\n Dense(3, activation=tf.nn.softmax),\n ]\n )\n \n # Compile Keras model\n optimizer = tf.keras.optimizers.RMSprop(lr=0.001)\n model.compile(\n loss=\"categorical_crossentropy\", metrics=[\"accuracy\"], optimizer=optimizer\n )\n \n return model\n\n# Create the model\nwith strategy.scope():\n model = create_model(num_features=dataset_train._flat_shapes[0].dims[0].value)\n\n# Set up datasets\nNUM_WORKERS = strategy.num_replicas_in_sync\n# Here the batch size scales up by number of workers since\n# `tf.data.Dataset.batch` expects the global batch size.\nGLOBAL_BATCH_SIZE = args.batch_size * NUM_WORKERS\ndataset_train = dataset_train.batch(GLOBAL_BATCH_SIZE)\ndataset_validation = dataset_validation.batch(GLOBAL_BATCH_SIZE)\n\n# Train the model\nmodel.fit(dataset_train, epochs=args.epochs, validation_data=dataset_validation)\n\ntf.saved_model.save(model, os.environ[\"AIP_MODEL_DIR\"])\n\ndf_test.head()",
"_____no_output_____"
]
],
[
[
"### Train the model\n\nDefine your custom training job on Vertex AI.\n\nUse the `CustomTrainingJob` class to define the job, which takes the following parameters:\n\n- `display_name`: The user-defined name of this training pipeline.\n- `script_path`: The local path to the training script.\n- `container_uri`: The URI of the training container image.\n- `requirements`: The list of Python package dependencies of the script.\n- `model_serving_container_image_uri`: The URI of a container that can serve predictions for your model — either a prebuilt container or a custom container.\n\nUse the `run` function to start training, which takes the following parameters:\n\n- `args`: The command line arguments to be passed to the Python script.\n- `replica_count`: The number of worker replicas.\n- `model_display_name`: The display name of the `Model` if the script produces a managed `Model`.\n- `machine_type`: The type of machine to use for training.\n- `accelerator_type`: The hardware accelerator type.\n- `accelerator_count`: The number of accelerators to attach to a worker replica.\n\nThe `run` function creates a training pipeline that trains and creates a `Model` object. After the training pipeline completes, the `run` function returns the `Model` object.",
"_____no_output_____"
]
],
[
[
"job = aiplatform.CustomTrainingJob(\n display_name=JOB_NAME,\n script_path=\"task.py\",\n container_uri=TRAIN_IMAGE,\n requirements=[\"google-cloud-bigquery>=2.20.0\"],\n model_serving_container_image_uri=DEPLOY_IMAGE,\n)\n\nMODEL_DISPLAY_NAME = \"penguins-\" + TIMESTAMP\n\n# Start the training\nif TRAIN_GPU:\n model = job.run(\n dataset=dataset,\n model_display_name=MODEL_DISPLAY_NAME,\n bigquery_destination=f\"bq://{PROJECT_ID}\",\n args=CMDARGS,\n replica_count=1,\n machine_type=TRAIN_COMPUTE,\n accelerator_type=TRAIN_GPU.name,\n accelerator_count=TRAIN_NGPU,\n )\nelse:\n model = job.run(\n dataset=dataset,\n model_display_name=MODEL_DISPLAY_NAME,\n bigquery_destination=f\"bq://{PROJECT_ID}\",\n args=CMDARGS,\n replica_count=1,\n machine_type=TRAIN_COMPUTE,\n accelerator_count=0,\n )",
"_____no_output_____"
]
],
[
[
"### Deploy the model\n\nBefore you use your model to make predictions, you need to deploy it to an `Endpoint`. You can do this by calling the `deploy` function on the `Model` resource. This will do two things:\n\n1. Create an `Endpoint` resource for deploying the `Model` resource to.\n2. Deploy the `Model` resource to the `Endpoint` resource.\n\n\nThe function takes the following parameters:\n\n- `deployed_model_display_name`: A human readable name for the deployed model.\n- `traffic_split`: Percent of traffic at the endpoint that goes to this model, which is specified as a dictionary of one or more key/value pairs.\n - If only one model, then specify as **{ \"0\": 100 }**, where \"0\" refers to this model being uploaded and 100 means 100% of the traffic.\n - If there are existing models on the endpoint, for which the traffic will be split, then use `model_id` to specify as **{ \"0\": percent, model_id: percent, ... }**, where `model_id` is the model id of an existing model to the deployed endpoint. The percents must add up to 100.\n- `machine_type`: The type of machine to use for training.\n- `accelerator_type`: The hardware accelerator type.\n- `accelerator_count`: The number of accelerators to attach to a worker replica.\n- `starting_replica_count`: The number of compute instances to initially provision.\n- `max_replica_count`: The maximum number of compute instances to scale to. In this tutorial, only one instance is provisioned.\n\n### Traffic split\n\nThe `traffic_split` parameter is specified as a Python dictionary. You can deploy more than one instance of your model to an endpoint, and then set the percentage of traffic that goes to each instance.\n\nYou can use a traffic split to introduce a new model gradually into production. For example, if you had one existing model in production with 100% of the traffic, you could deploy a new model to the same endpoint, direct 10% of traffic to it, and reduce the original model's traffic to 90%. This allows you to monitor the new model's performance while minimizing the distruption to the majority of users.\n\n### Compute instance scaling\n\nYou can specify a single instance (or node) to serve your online prediction requests. This tutorial uses a single node, so the variables `MIN_NODES` and `MAX_NODES` are both set to `1`.\n\nIf you want to use multiple nodes to serve your online prediction requests, set `MAX_NODES` to the maximum number of nodes you want to use. Vertex AI autoscales the number of nodes used to serve your predictions, up to the maximum number you set. Refer to the [pricing page](https://cloud.google.com/vertex-ai/pricing#prediction-prices) to understand the costs of autoscaling with multiple nodes.\n\n### Endpoint\n\nThe method will block until the model is deployed and eventually return an `Endpoint` object. If this is the first time a model is deployed to the endpoint, it may take a few additional minutes to complete provisioning of resources.",
"_____no_output_____"
]
],
[
[
"DEPLOYED_NAME = \"penguins_deployed-\" + TIMESTAMP\n\nTRAFFIC_SPLIT = {\"0\": 100}\n\nMIN_NODES = 1\nMAX_NODES = 1\n\nif DEPLOY_GPU:\n endpoint = model.deploy(\n deployed_model_display_name=DEPLOYED_NAME,\n traffic_split=TRAFFIC_SPLIT,\n machine_type=DEPLOY_COMPUTE,\n accelerator_type=DEPLOY_GPU.name,\n accelerator_count=DEPLOY_NGPU,\n min_replica_count=MIN_NODES,\n max_replica_count=MAX_NODES,\n )\nelse:\n endpoint = model.deploy(\n deployed_model_display_name=DEPLOYED_NAME,\n traffic_split=TRAFFIC_SPLIT,\n machine_type=DEPLOY_COMPUTE,\n accelerator_type=DEPLOY_COMPUTE.name,\n accelerator_count=0,\n min_replica_count=MIN_NODES,\n max_replica_count=MAX_NODES,\n )",
"_____no_output_____"
]
],
[
[
"## Make an online prediction request\n\nSend an online prediction request to your deployed model.",
"_____no_output_____"
],
[
"### Prepare test data\n\nPrepare test data by normalizing it and converting categorical values to numeric values.\nThese values have to match what was just at training.\n\nIn this example, testing is done with the same dataset that was used for training. In practice, you will want to use a separate dataset to correctly verify your results.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom google.cloud import bigquery\n\nUNUSED_COLUMNS = []\nLABEL_COLUMN = \"species\"\n\n# Possible categorical values\nSPECIES = [\n \"Adelie Penguin (Pygoscelis adeliae)\",\n \"Chinstrap penguin (Pygoscelis antarctica)\",\n \"Gentoo penguin (Pygoscelis papua)\",\n]\nISLANDS = [\"Dream\", \"Biscoe\", \"Torgersen\"]\nSEXES = [\"FEMALE\", \"MALE\"]\n\n_CATEGORICAL_TYPES = {\n \"island\": pd.api.types.CategoricalDtype(categories=ISLANDS),\n \"species\": pd.api.types.CategoricalDtype(categories=SPECIES),\n \"sex\": pd.api.types.CategoricalDtype(categories=SEXES),\n}\n\n\ndef standardize(df, mean_and_std):\n \"\"\"Scales numerical columns using their means and standard deviation to get\n z-scores: the mean of each numerical column becomes 0, and the standard\n deviation becomes 1. This can help the model converge during training.\n\n Args:\n df: Pandas df\n\n Returns:\n Input df with the numerical columns scaled to z-scores\n \"\"\"\n dtypes = list(zip(df.dtypes.index, map(str, df.dtypes)))\n # Normalize numeric columns.\n for column, dtype in dtypes:\n if dtype == \"float32\":\n df[column] -= mean_and_std[column][\"mean\"]\n df[column] /= mean_and_std[column][\"std\"]\n return df\n\n\ndef preprocess(df, mean_and_std):\n \"\"\"Converts categorical features to numeric. Removes unused columns.\n\n Args:\n df: Pandas df with raw data\n\n Returns:\n df with preprocessed data\n \"\"\"\n df = df.drop(columns=UNUSED_COLUMNS)\n\n # Drop rows with NaN's\n df = df.dropna()\n\n # Convert integer valued (numeric) columns to floating point\n numeric_columns = df.select_dtypes([\"int32\", \"float32\", \"float64\"]).columns\n df[numeric_columns] = df[numeric_columns].astype(\"float32\")\n\n # Convert categorical columns to numeric\n cat_columns = df.select_dtypes([\"object\"]).columns\n\n df[cat_columns] = df[cat_columns].apply(\n lambda x: x.astype(_CATEGORICAL_TYPES[x.name])\n )\n df[cat_columns] = df[cat_columns].apply(lambda x: x.cat.codes)\n return df\n\n\ndef convert_dataframe_to_list(df, mean_and_std):\n df = preprocess(df, mean_and_std)\n\n df_x, df_y = df, df.pop(LABEL_COLUMN)\n\n # Normalize on overall means and standard deviations.\n df = standardize(df, mean_and_std)\n\n y = np.asarray(df_y).astype(\"float32\")\n\n # Convert to numpy representation\n x = np.asarray(df_x)\n\n # Convert to one-hot representation\n return x.tolist(), y.tolist()\n\n\nx_test, y_test = convert_dataframe_to_list(dataframe, mean_and_std)",
"_____no_output_____"
]
],
[
[
"### Send the prediction request\n\nNow that you have test data, you can use them to send a prediction request. Use the `Endpoint` object's `predict` function, which takes the following parameters:\n\n- `instances`: A list of penguin measurement instances. According to your custom model, each instance should be an array of numbers. This was prepared in the previous step.\n\nThe `predict` function returns a list, where each element in the list corresponds to the corresponding instance in the request. You will see in the output for each prediction:\n\n- Confidence level for the prediction (`predictions`), between 0 and 1, for each of the ten classes.\n\nYou can then run a quick evaluation on the prediction results:\n1. `np.argmax`: Convert each list of confidence levels to a label\n2. Compare the predicted labels to the actual labels\n3. Calculate `accuracy` as `correct/total`",
"_____no_output_____"
]
],
[
[
"predictions = endpoint.predict(instances=x_test)\ny_predicted = np.argmax(predictions.predictions, axis=1)\n\ncorrect = sum(y_predicted == np.array(y_test))\naccuracy = len(y_predicted)\nprint(\n f\"Correct predictions = {correct}, Total predictions = {accuracy}, Accuracy = {correct/accuracy}\"\n)",
"_____no_output_____"
]
],
[
[
"## Undeploy the model\n\nTo undeploy your `Model` resource from the serving `Endpoint` resource, use the endpoint's `undeploy` method with the following parameter:\n\n- `deployed_model_id`: The model deployment identifier returned by the endpoint service when the `Model` resource was deployed. You can retrieve the deployed models using the endpoint's `deployed_models` property.\n\nSince this is the only deployed model on the `Endpoint` resource, you can omit `traffic_split`.",
"_____no_output_____"
]
],
[
[
"deployed_model_id = endpoint.list_models()[0].id\nendpoint.undeploy(deployed_model_id=deployed_model_id)",
"_____no_output_____"
]
],
[
[
"# Cleaning up\n\nTo clean up all Google Cloud resources used in this project, you can [delete the Google Cloud project](https://cloud.google.com/resource-manager/docs/creating-managing-projects#shutting_down_projects) you used for the tutorial.\n\nOtherwise, you can delete the individual resources you created in this tutorial:\n\n- Training Job\n- Model\n- Endpoint\n- Cloud Storage Bucket",
"_____no_output_____"
]
],
[
[
"delete_training_job = True\ndelete_model = True\ndelete_endpoint = True\n\n# Warning: Setting this to true will delete everything in your bucket\ndelete_bucket = False\n\n# Delete the training job\njob.delete()\n\n# Delete the model\nmodel.delete()\n\n# Delete the endpoint\nendpoint.delete()\n\nif delete_bucket and \"BUCKET_NAME\" in globals():\n ! gsutil -m rm -r $BUCKET_NAME",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4372361a9096f7917a14340d8e0e662bd9c979 | 1,782 | ipynb | Jupyter Notebook | example/MyPython3.ipynb | nufeng1999/jupyter-MyPython-kernel | b459a48f082aed35eeb50945caf503b44911891d | [
"MIT"
] | null | null | null | example/MyPython3.ipynb | nufeng1999/jupyter-MyPython-kernel | b459a48f082aed35eeb50945caf503b44911891d | [
"MIT"
] | null | null | null | example/MyPython3.ipynb | nufeng1999/jupyter-MyPython-kernel | b459a48f082aed35eeb50945caf503b44911891d | [
"MIT"
] | null | null | null | 21.46988 | 96 | 0.461841 | [
[
[
"#%define M1 print(\"这是我的宏替换测试\")\n##%log:0\n#%define TEST\n#%if defined TEST:\nprint(\"this is test code.\")\n#%else:\nprint(\"------------------.\")\nprint(\"------------------.\")\nprint(\"------------------.\")\nprint(\"------------------.\")\nprint(\"------------------.\")\nprint(\"------------------.\")\nM1",
"_____no_output_____"
],
[
"import os\nimport sys\n# %cd \"h:\\Jupyter\\ipynb\\MyPython\"\nif os.getcwd() not in sys.path:\n\tsys.path",
"[MyPythonKernel125328] Info:The process :h:\\Jupyter\\ipynb\\MyPython\\tmpoguyf3ix.py\n[MyPythonKernel125328] Info:python3 h:\\Jupyter\\ipynb\\MyPython\\tmpoguyf3ix.py \n[MyPythonKernel125328] Info:The process end:8628\n"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
cb43732872ebf75d87ad65d8ec67835c3be84bee | 410,097 | ipynb | Jupyter Notebook | 6_survival_analysis/expression_eda.ipynb | jjc2718/mutation-fn | e6bab15109a79c7d42dcdbedb1b949a6289d8958 | [
"BSD-3-Clause"
] | null | null | null | 6_survival_analysis/expression_eda.ipynb | jjc2718/mutation-fn | e6bab15109a79c7d42dcdbedb1b949a6289d8958 | [
"BSD-3-Clause"
] | null | null | null | 6_survival_analysis/expression_eda.ipynb | jjc2718/mutation-fn | e6bab15109a79c7d42dcdbedb1b949a6289d8958 | [
"BSD-3-Clause"
] | null | null | null | 221.554295 | 95,724 | 0.891762 | [
[
[
"## Explore one-hit vs. two-hit samples in expression space",
"_____no_output_____"
]
],
[
[
"from pathlib import Path\nimport pickle as pkl\n\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nfrom sklearn.preprocessing import StandardScaler\n\nimport sys; sys.path.append('..')\nimport config as cfg\nfrom data_utilities import load_cnv_data\n\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"# park et al. geneset info\npark_loss_data = cfg.data_dir / 'park_loss_df.tsv'\npark_gain_data = cfg.data_dir / 'park_gain_df.tsv'\n\n# park et al. significant gene info\npark_loss_sig_data = cfg.data_dir / 'park_loss_df_sig_only.tsv'\npark_gain_sig_data = cfg.data_dir / 'park_gain_df_sig_only.tsv'\n\n# park et al. gene/cancer type predictions\npark_preds_dir = cfg.data_dir / 'park_genes_all_preds'\n\n# mutation and copy number data\npancancer_pickle = Path('/home/jake/research/mpmp/data/pancancer_data.pkl')\n\n# gene expression/rppa data files\ndata_type = 'gene expression'\nsubset_feats = 10000\ngene_expression_data_file = Path(\n '/home/jake/research/mpmp/data/tcga_expression_matrix_processed.tsv.gz'\n)\nrppa_data_file = Path(\n '/home/jake/research/mpmp/data/tcga_rppa_matrix_processed.tsv'\n)",
"_____no_output_____"
]
],
[
[
"### Load mutation info\n\nFor now, just use binary mutation status from the pancancer repo. In the future we could pull more granular info from MC3, but it would take some engineering of `1_get_mutation_counts` to do this for lots of genes.",
"_____no_output_____"
]
],
[
[
"park_loss_df = pd.read_csv(park_loss_data, sep='\\t', index_col=0)\npark_loss_df.head()",
"_____no_output_____"
],
[
"park_gain_df = pd.read_csv(park_gain_data, sep='\\t', index_col=0)\npark_gain_df.head()",
"_____no_output_____"
],
[
"with open(pancancer_pickle, 'rb') as f:\n pancancer_data = pkl.load(f)",
"_____no_output_____"
],
[
"# get (binary) mutation data\n# 1 = observed non-silent mutation in this gene for this sample, 0 otherwise\nmutation_df = pancancer_data[1]\nprint(mutation_df.shape)\nmutation_df.iloc[:5, :5]",
"(9074, 20938)\n"
]
],
[
[
"### Load copy number info\n\nGet copy loss/gain info directly from GISTIC \"thresholded\" output. This should be the same as (or very similar to) what the Park et al. study uses.",
"_____no_output_____"
]
],
[
[
"sample_freeze_df = pancancer_data[0]\ncopy_samples = set(sample_freeze_df.SAMPLE_BARCODE)\nprint(len(copy_samples))",
"9074\n"
],
[
"copy_loss_df, copy_gain_df = load_cnv_data(\n cfg.data_dir / 'pancan_GISTIC_threshold.tsv',\n copy_samples\n)\nprint(copy_loss_df.shape)\ncopy_loss_df.iloc[:5, :5]",
"(9068, 25128)\n"
],
[
"print(copy_gain_df.shape)\ncopy_gain_df.iloc[:5, :5]",
"(9068, 25128)\n"
],
[
"sample_freeze_df.head()",
"_____no_output_____"
]
],
[
[
"### Load expression data\n\nWe'll also standardize each feature, and subset to the top features by mean absolute deviation if `subset_feats` is set.",
"_____no_output_____"
]
],
[
[
"if data_type == 'gene expression':\n exp_df = pd.read_csv(gene_expression_data_file, sep='\\t', index_col=0)\nelif data_type == 'rppa':\n exp_df = pd.read_csv(rppa_data_file, sep='\\t', index_col=0)\n \nprint(exp_df.shape)\nexp_df.iloc[:5, :5]",
"(11060, 15369)\n"
],
[
"# standardize features first\nexp_df = pd.DataFrame(\n StandardScaler().fit_transform(exp_df),\n index=exp_df.index.copy(),\n columns=exp_df.columns.copy()\n)\nprint(exp_df.shape)\nexp_df.iloc[:5, :5]",
"(11060, 15369)\n"
],
[
"# subset to subset_feats features by mean absolute deviation\nif subset_feats is not None:\n mad_ranking = (\n exp_df.mad(axis=0)\n .sort_values(ascending=False)\n )\n top_feats = mad_ranking[:subset_feats].index.astype(str).values\n exp_mad_df = exp_df.reindex(top_feats, axis='columns')\nelse:\n exp_mad_df = exp_df\n \nprint(exp_mad_df.shape)\nexp_mad_df.iloc[:5, :5]",
"(11060, 10000)\n"
]
],
[
[
"### Get sample info and hit groups for gene/cancer type",
"_____no_output_____"
]
],
[
[
"def get_hits_for_gene_and_tissue(identifier, cancer_classification):\n \"\"\"Given a gene and tissue, load the relevant mutation/CNV information,\n and divide the samples into groups to compare survival.\n \"\"\"\n # get patient ids in given cancer type \n gene, tissue = identifier.split('_')\n tissue_ids = (sample_freeze_df\n .query('DISEASE == @tissue')\n .SAMPLE_BARCODE\n )\n \n # get mutation and copy status\n mutation_status = mutation_df.loc[tissue_ids, gene]\n if cancer_classification == 'TSG':\n copy_status = copy_loss_df.loc[tissue_ids, gene]\n elif cancer_classification == 'Oncogene':\n copy_status = copy_gain_df.loc[tissue_ids, gene]\n \n # get hit groups from mutation/CNV data\n two_hit_samples = (mutation_status & copy_status).astype(int)\n one_hit_samples = (mutation_status | copy_status).astype(int)\n \n return pd.DataFrame(\n {'group': one_hit_samples + two_hit_samples}\n )",
"_____no_output_____"
],
[
"identifier = 'ATRX_LGG'\ncancer_classification = 'Oncogene'\n\nsample_mut_df = get_hits_for_gene_and_tissue(identifier, cancer_classification)\n\n# make sure sample data overlaps exactly with expression data\noverlap_ixs = sample_mut_df.index.intersection(exp_mad_df.index)\nsample_mut_df = sample_mut_df.loc[overlap_ixs, :].copy()\nexp_mad_df = exp_mad_df.loc[overlap_ixs, :].copy()\n\n# add group info for legends\nsample_mut_df['group'] = sample_mut_df.group.map({\n 0: 'wild-type',\n 1: 'one-hit',\n 2: 'two-hit'\n})\n\nprint(sample_mut_df.shape)\nprint(sample_mut_df.group.unique())\nsample_mut_df.iloc[:5, :5]",
"(507, 1)\n['wild-type' 'two-hit' 'one-hit']\n"
]
],
[
[
"### Plot samples by hit group",
"_____no_output_____"
]
],
[
[
"from sklearn.decomposition import PCA\n\npca = PCA(n_components=2)\n\nX_proj_pca = pca.fit_transform(exp_mad_df)\n\nprint(X_proj_pca.shape)\nX_proj_pca[:5, :5]",
"(507, 2)\n"
],
[
"sns.set({'figure.figsize': (8, 6)})\n\nsns.scatterplot(x=X_proj_pca[:, 0],\n y=X_proj_pca[:, 1],\n hue=sample_mut_df.group,\n hue_order=['wild-type', 'one-hit', 'two-hit'])\n\nplt.title('PCA of {} {} features, colored by {} status'.format(\n subset_feats, data_type, identifier))\nplt.xlabel('PC1')\nplt.ylabel('PC2')",
"_____no_output_____"
],
[
"from umap import UMAP\n\nreducer = UMAP(n_components=2, random_state=42)\n\nX_proj_umap = reducer.fit_transform(exp_mad_df)\n\nprint(X_proj_umap.shape)\nX_proj_umap[:5, :5]",
"/home/jake/anaconda3/envs/mutation_fn/lib/python3.8/site-packages/tqdm/auto.py:22: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html\n from .autonotebook import tqdm as notebook_tqdm\n"
],
[
"sns.set({'figure.figsize': (8, 6)})\n\nsns.scatterplot(x=X_proj_umap[:, 0],\n y=X_proj_umap[:, 1],\n hue=sample_mut_df.group,\n hue_order=['wild-type', 'one-hit', 'two-hit'])\n\nplt.title('UMAP of {} {} features, colored by {} status'.format(\n subset_feats, data_type, identifier))\nplt.xlabel('UMAP1')\nplt.ylabel('UMAP2')",
"_____no_output_____"
]
],
[
[
"### Plot samples by hit group, using features selected by pan-cancer classifiers",
"_____no_output_____"
]
],
[
[
"coefs_file = Path(\n '/home/jake/research/mpmp/data/final_models/final_expression_all_merged_coefs.tsv'\n)\n\ncoefs_df = pd.read_csv(coefs_file, sep='\\t', index_col=0)\ncoefs_df.iloc[:5, :5]",
"/home/jake/anaconda3/envs/mutation_fn/lib/python3.8/site-packages/IPython/core/interactiveshell.py:3457: DtypeWarning: Columns (0) have mixed types.Specify dtype option on import or set low_memory=False.\n exec(code_obj, self.user_global_ns, self.user_ns)\n"
],
[
"gene, tissue = identifier.split('_')\n\ncoefs_gene = coefs_df.loc[:, gene]\ncoefs_gene = coefs_gene[(~coefs_gene.isna()) &\n (~(coefs_gene == 0.0)) &\n # get rid of log10_mut and cancer type covariates\n (coefs_gene.index.astype(str).str.isdigit())]\n\ncoefs_gene.index = coefs_gene.index.astype(str)\n\nprint(coefs_gene.shape)\ncoefs_gene.head()",
"(465,)\n"
],
[
"print(coefs_gene.index)\nprint(coefs_gene.index.isna().sum())",
"Index(['100134938', '10014', '10018', '10072', '10094', '10142', '10217',\n '10228', '10417', '10423',\n ...\n '9761', '9843', '9846', '9858', '9877', '9905', '9909', '9922', '9924',\n '9926'],\n dtype='object', length=465)\n0\n"
],
[
"exp_coefs_df = exp_df.loc[overlap_ixs, coefs_gene.index].copy()\n\nprint(exp_coefs_df.shape)\nexp_coefs_df.iloc[:5, :5]",
"(507, 465)\n"
],
[
"sns.set({'figure.figsize': (8, 6)})\n\npca = PCA(n_components=2)\nX_proj_pca = pca.fit_transform(exp_coefs_df)\n\nsns.scatterplot(x=X_proj_pca[:, 0],\n y=X_proj_pca[:, 1],\n hue=sample_mut_df.group,\n hue_order=['wild-type', 'one-hit', 'two-hit'])\n\nplt.title('PCA of non-zero {} features, colored by {} status'.format(\n data_type, identifier))\nplt.xlabel('PC1')\nplt.ylabel('PC2')",
"_____no_output_____"
],
[
"sns.set({'figure.figsize': (8, 6)})\n\nreducer = UMAP(n_components=2, random_state=42)\nX_proj_umap = reducer.fit_transform(exp_coefs_df)\n\nsns.scatterplot(x=X_proj_umap[:, 0],\n y=X_proj_umap[:, 1],\n hue=sample_mut_df.group,\n hue_order=['wild-type', 'one-hit', 'two-hit'])\n\nplt.title('UMAP of nonzero {} features, colored by {} status'.format(\n data_type, identifier))\nplt.xlabel('UMAP1')\nplt.ylabel('UMAP2')",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4374b328d4f817ed6256e31e176a84adcc870f | 4,567 | ipynb | Jupyter Notebook | 03-Loops_Condicionais_Metodos_Funcoes/03-While.ipynb | alineAssuncao/Python_Fundamentos_Analise_Dados | 872781f2cec24487b0f29f62afeb60650a451bfd | [
"MIT"
] | 1 | 2019-02-03T10:53:55.000Z | 2019-02-03T10:53:55.000Z | 03-Loops_Condicionais_Metodos_Funcoes/03-While.ipynb | alineAssuncao/Python_Fundamentos_Analise_Dados | 872781f2cec24487b0f29f62afeb60650a451bfd | [
"MIT"
] | null | null | null | 03-Loops_Condicionais_Metodos_Funcoes/03-While.ipynb | alineAssuncao/Python_Fundamentos_Analise_Dados | 872781f2cec24487b0f29f62afeb60650a451bfd | [
"MIT"
] | null | null | null | 20.853881 | 62 | 0.440552 | [
[
[
"# While",
"_____no_output_____"
]
],
[
[
"counter = 0\nwhile counter < 10:\n print(counter)\n counter = counter + 1",
"0\n1\n2\n3\n4\n5\n6\n7\n8\n9\n"
],
[
"# usando clausula else com o while\nx = 0\n\nwhile x < 10:\n print(\"o valor de x nesta iteração é: \", x)\n print(\" x ainda é menor que 10, somando 1 a x\")\n x += 1\nelse:\n print(\"Loop concluido!\")",
"o valor de x nesta iteração é: 0\n x ainda é menor que 10, somando 1 a x\no valor de x nesta iteração é: 1\n x ainda é menor que 10, somando 1 a x\no valor de x nesta iteração é: 2\n x ainda é menor que 10, somando 1 a x\no valor de x nesta iteração é: 3\n x ainda é menor que 10, somando 1 a x\no valor de x nesta iteração é: 4\n x ainda é menor que 10, somando 1 a x\no valor de x nesta iteração é: 5\n x ainda é menor que 10, somando 1 a x\no valor de x nesta iteração é: 6\n x ainda é menor que 10, somando 1 a x\no valor de x nesta iteração é: 7\n x ainda é menor que 10, somando 1 a x\no valor de x nesta iteração é: 8\n x ainda é menor que 10, somando 1 a x\no valor de x nesta iteração é: 9\n x ainda é menor que 10, somando 1 a x\nLoop concluido!\n"
]
],
[
[
"# Pass, break, continue",
"_____no_output_____"
]
],
[
[
"counter = 0\nwhile counter < 100:\n if counter == 4:\n break\n else:\n pass\n print(counter)\n counter += 1",
"0\n1\n2\n3\n"
],
[
"for verificador in \"Python\":\n if verificador == \"h\":\n continue\n print(verificador)",
"P\ny\nt\no\nn\n"
]
],
[
[
"# while e for juntos",
"_____no_output_____"
]
],
[
[
"for i in range(2,30):\n j = 2\n counter = 0\n while j < i:\n if i % j == 0:\n counter = 1\n j += 1\n else:\n j += 1\n if counter == 0:\n print(str(i) + \" é um número primo\")\n counter = 0\n else:\n counter = 0\n ",
"2 é um número primo\n3 é um número primo\n5 é um número primo\n7 é um número primo\n11 é um número primo\n13 é um número primo\n17 é um número primo\n19 é um número primo\n23 é um número primo\n29 é um número primo\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb439e2845c55c258a00daa76b0698a6f2223a63 | 13,489 | ipynb | Jupyter Notebook | Bokeh/03 - models.ipynb | prakash123mayank/Data-Science-45min-Intros | f9208e43d69f791f8611998b39238444e9a7b7ba | [
"Unlicense"
] | 1,406 | 2015-01-05T19:20:55.000Z | 2022-03-17T08:35:09.000Z | Bokeh/03 - models.ipynb | prakash123mayank/Data-Science-45min-Intros | f9208e43d69f791f8611998b39238444e9a7b7ba | [
"Unlicense"
] | 1 | 2019-07-27T11:53:24.000Z | 2019-10-02T19:34:32.000Z | Bokeh/03 - models.ipynb | prakash123mayank/Data-Science-45min-Intros | f9208e43d69f791f8611998b39238444e9a7b7ba | [
"Unlicense"
] | 495 | 2015-01-06T11:39:21.000Z | 2022-03-15T10:21:43.000Z | 27.528571 | 282 | 0.536882 | [
[
[
"<table style=\"float:left; border:none\">\n <tr style=\"border:none\">\n <td style=\"border:none\">\n <a href=\"http://bokeh.pydata.org/\"> \n <img \n src=\"http://bokeh.pydata.org/en/latest/_static/bokeh-transparent.png\" \n style=\"width:70px\"\n >\n </a> \n </td>\n <td style=\"border:none\">\n <h1>Bokeh Tutorial — <tt style=\"display:inline\">bokeh.models</tt> interface</h1>\n </td>\n </tr>\n</table>",
"_____no_output_____"
],
[
"## Models\n\nNYTimes interactive chart [Usain Bolt vs. 116 years of Olympic sprinters](http://www.nytimes.com/interactive/2012/08/05/sports/olympics/the-100-meter-dash-one-race-every-medalist-ever.html)",
"_____no_output_____"
],
[
"The first thing we need is to get the data. The data for this chart is located in the ``bokeh.sampledata`` module as a Pandas DataFrame. You can see the first ten rows below:?",
"_____no_output_____"
]
],
[
[
"from bokeh.sampledata.sprint import sprint\nsprint[:10]",
"_____no_output_____"
]
],
[
[
"Next we import some of the Bokeh models that need to be assembled to make a plot. At a minimum, we need to start with ``Plot``, the glyphs (``Circle`` and ``Text``) we want to display, as well as ``ColumnDataSource`` to hold the data and range obejcts to set the plot bounds. ",
"_____no_output_____"
]
],
[
[
"from bokeh.io import output_notebook, show\nfrom bokeh.models.glyphs import Circle, Text\nfrom bokeh.models import ColumnDataSource, Range1d, DataRange1d, Plot",
"_____no_output_____"
],
[
"output_notebook()",
"_____no_output_____"
]
],
[
[
"## Setting up Data",
"_____no_output_____"
]
],
[
[
"abbrev_to_country = {\n \"USA\": \"United States\",\n \"GBR\": \"Britain\",\n \"JAM\": \"Jamaica\",\n \"CAN\": \"Canada\",\n \"TRI\": \"Trinidad and Tobago\",\n \"AUS\": \"Australia\",\n \"GER\": \"Germany\",\n \"CUB\": \"Cuba\",\n \"NAM\": \"Namibia\",\n \"URS\": \"Soviet Union\",\n \"BAR\": \"Barbados\",\n \"BUL\": \"Bulgaria\",\n \"HUN\": \"Hungary\",\n \"NED\": \"Netherlands\",\n \"NZL\": \"New Zealand\",\n \"PAN\": \"Panama\",\n \"POR\": \"Portugal\",\n \"RSA\": \"South Africa\",\n \"EUA\": \"United Team of Germany\",\n}\n\ngold_fill = \"#efcf6d\"\ngold_line = \"#c8a850\"\nsilver_fill = \"#cccccc\"\nsilver_line = \"#b0b0b1\"\nbronze_fill = \"#c59e8a\"\nbronze_line = \"#98715d\"\n\nfill_color = { \"gold\": gold_fill, \"silver\": silver_fill, \"bronze\": bronze_fill }\nline_color = { \"gold\": gold_line, \"silver\": silver_line, \"bronze\": bronze_line }\n\ndef selected_name(name, medal, year):\n return name if medal == \"gold\" and year in [1988, 1968, 1936, 1896] else None\n\nt0 = sprint.Time[0]\n\nsprint[\"Abbrev\"] = sprint.Country\nsprint[\"Country\"] = sprint.Abbrev.map(lambda abbr: abbrev_to_country[abbr])\nsprint[\"Medal\"] = sprint.Medal.map(lambda medal: medal.lower())\nsprint[\"Speed\"] = 100.0/sprint.Time\nsprint[\"MetersBack\"] = 100.0*(1.0 - t0/sprint.Time)\nsprint[\"MedalFill\"] = sprint.Medal.map(lambda medal: fill_color[medal])\nsprint[\"MedalLine\"] = sprint.Medal.map(lambda medal: line_color[medal])\nsprint[\"SelectedName\"] = sprint[[\"Name\", \"Medal\", \"Year\"]].apply(tuple, axis=1).map(lambda args: selected_name(*args))\n\nsource = ColumnDataSource(sprint)",
"_____no_output_____"
]
],
[
[
"## Basic Plot with Glyphs",
"_____no_output_____"
]
],
[
[
"plot_options = dict(plot_width=800, plot_height=480, toolbar_location=None, \n outline_line_color=None, title = \"Usain Bolt vs. 116 years of Olympic sprinters\")",
"_____no_output_____"
],
[
"radius = dict(value=5, units=\"screen\")\nmedal_glyph = Circle(x=\"MetersBack\", y=\"Year\", radius=radius, fill_color=\"MedalFill\", \n line_color=\"MedalLine\", fill_alpha=0.5)\n\nathlete_glyph = Text(x=\"MetersBack\", y=\"Year\", x_offset=10, text=\"SelectedName\",\n text_align=\"left\", text_baseline=\"middle\", text_font_size=\"9pt\")\n\nno_olympics_glyph = Text(x=7.5, y=1942, text=[\"No Olympics in 1940 or 1944\"],\n text_align=\"center\", text_baseline=\"middle\",\n text_font_size=\"9pt\", text_font_style=\"italic\", text_color=\"silver\")\n",
"_____no_output_____"
],
[
"xdr = Range1d(start=sprint.MetersBack.max()+2, end=0) # +2 is for padding\nydr = DataRange1d(range_padding=0.05) \n\nplot = Plot(x_range=xdr, y_range=ydr, **plot_options)\nplot.add_glyph(source, medal_glyph)\nplot.add_glyph(source, athlete_glyph)\nplot.add_glyph(no_olympics_glyph)",
"_____no_output_____"
],
[
"show(plot)",
"_____no_output_____"
]
],
[
[
"## Adding Axes and Grids",
"_____no_output_____"
]
],
[
[
"from bokeh.models import Grid, LinearAxis, SingleIntervalTicker",
"_____no_output_____"
],
[
"xdr = Range1d(start=sprint.MetersBack.max()+2, end=0) # +2 is for padding\nydr = DataRange1d(range_padding=0.05) \n\nplot = Plot(x_range=xdr, y_range=ydr, **plot_options)\nplot.add_glyph(source, medal_glyph)\nplot.add_glyph(source, athlete_glyph)\nplot.add_glyph(no_olympics_glyph)",
"_____no_output_____"
],
[
"xticker = SingleIntervalTicker(interval=5, num_minor_ticks=0)\nxaxis = LinearAxis(ticker=xticker, axis_line_color=None, major_tick_line_color=None,\n axis_label=\"Meters behind 2012 Bolt\", axis_label_text_font_size=\"10pt\", \n axis_label_text_font_style=\"bold\")\nplot.add_layout(xaxis, \"below\")\n\nxgrid = Grid(dimension=0, ticker=xaxis.ticker, grid_line_dash=\"dashed\")\nplot.add_layout(xgrid)\n\nyticker = SingleIntervalTicker(interval=12, num_minor_ticks=0)\nyaxis = LinearAxis(ticker=yticker, major_tick_in=-5, major_tick_out=10)\nplot.add_layout(yaxis, \"right\")",
"_____no_output_____"
],
[
"show(plot)",
"_____no_output_____"
]
],
[
[
"## Adding a Hover Tool",
"_____no_output_____"
]
],
[
[
"from bokeh.models import HoverTool",
"_____no_output_____"
],
[
"tooltips = \"\"\"\n<div>\n <span style=\"font-size: 15px;\">@Name</span> \n <span style=\"font-size: 10px; color: #666;\">(@Abbrev)</span>\n</div>\n<div>\n <span style=\"font-size: 17px; font-weight: bold;\">@Time{0.00}</span> \n <span style=\"font-size: 10px; color: #666;\">@Year</span>\n</div>\n<div style=\"font-size: 11px; color: #666;\">@{MetersBack}{0.00} meters behind</div>\n\"\"\"",
"_____no_output_____"
],
[
"xdr = Range1d(start=sprint.MetersBack.max()+2, end=0) # +2 is for padding\nydr = DataRange1d(range_padding=0.05) \n\nplot = Plot(x_range=xdr, y_range=ydr, **plot_options)\nmedal = plot.add_glyph(source, medal_glyph) # we need this renderer to configure the hover tool\nplot.add_glyph(source, athlete_glyph)\nplot.add_glyph(no_olympics_glyph)\n\nxticker = SingleIntervalTicker(interval=5, num_minor_ticks=0)\nxaxis = LinearAxis(ticker=xticker, axis_line_color=None, major_tick_line_color=None,\n axis_label=\"Meters behind 2012 Bolt\", axis_label_text_font_size=\"10pt\", \n axis_label_text_font_style=\"bold\")\nplot.add_layout(xaxis, \"below\")\n\nxgrid = Grid(dimension=0, ticker=xaxis.ticker, grid_line_dash=\"dashed\")\nplot.add_layout(xgrid)\n\nyticker = SingleIntervalTicker(interval=12, num_minor_ticks=0)\nyaxis = LinearAxis(ticker=yticker, major_tick_in=-5, major_tick_out=10)\nplot.add_layout(yaxis, \"right\")",
"_____no_output_____"
],
[
"hover = HoverTool(tooltips=tooltips, renderers=[medal])\nplot.add_tools(hover)",
"_____no_output_____"
],
[
"show(plot)",
"_____no_output_____"
],
[
"from bubble_plot import get_1964_data\n\ndef get_plot():\n return Plot(\n x_range=Range1d(1, 9), y_range=Range1d(20, 100),\n title=\"\", plot_width=800, plot_height=400,\n outline_line_color=None, toolbar_location=None,\n )\n\ndf = get_1964_data()\ndf.head()",
"_____no_output_____"
],
[
"# EXERCISE: Add Circles to the plot from the data in `df`. \n# With `fertility` for the x coordinates, `life` for the y coordinates.\n\nplot = get_plot()\n\n\n",
"_____no_output_____"
],
[
"# EXERCISE: Color the circles by region_color & change the size of the color by population\n\n\n",
"_____no_output_____"
],
[
"# EXERCISE: Add axes and grid lines\n\n",
"_____no_output_____"
],
[
"# EXERCISE: Manually add a legend using Circle & Text. The color key is as follows \n\nregion_name_and_color = [\n ('America', '#3288bd'),\n ('East Asia & Pacific', '#99d594'),\n ('Europe & Central Asia', '#e6f598'),\n ('Middle East & North Africa', '#fee08b'),\n ('South Asia', '#fc8d59'),\n ('Sub-Saharan Africa', '#d53e4f')\n]",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb43a1025e2eb408d4f02c40063528e156439fbd | 83,046 | ipynb | Jupyter Notebook | tensorflow/practice/convnets/ImageRegularizationCIFAR10.ipynb | lisuizhe/ml-algorithm | af0755869657b4085de44d4ec95b8c5269d9ac1a | [
"Apache-2.0"
] | 3 | 2019-04-21T06:04:20.000Z | 2019-04-26T00:03:14.000Z | tensorflow/practice/convnets/ImageRegularizationCIFAR10.ipynb | lisuizhe/ml-algorithm | af0755869657b4085de44d4ec95b8c5269d9ac1a | [
"Apache-2.0"
] | null | null | null | tensorflow/practice/convnets/ImageRegularizationCIFAR10.ipynb | lisuizhe/ml-algorithm | af0755869657b4085de44d4ec95b8c5269d9ac1a | [
"Apache-2.0"
] | null | null | null | 432.53125 | 29,160 | 0.934855 | [
[
[
"%matplotlib inline\n\nimport tensorflow.keras as keras\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nfrom tensorflow.keras.datasets import cifar10\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"(x_train, y_train), (x_test, y_test) = cifar10.load_data()\n\nx_train = x_train.astype('float32') / 255\ny_train = np.eye(10)[y_train.astype('int32').flatten()]\nx_test = x_test.astype('float32') / 255\ny_test = np.eye(10)[y_test.astype('int32').flatten()]\n\nx_train, x_valid, y_train, y_valid = train_test_split(x_train, y_train, test_size=10000)",
"_____no_output_____"
],
[
"from tensorflow.keras.preprocessing.image import ImageDataGenerator\n\nfig = plt.figure(figsize=(9, 15))\nfig.subplots_adjust(left=0, right=1, bottom=0, top=0.5, hspace=0.05, wspace=0.05)\n\nfor i in range(5):\n ax = fig.add_subplot(1, 5, i+1, xticks=[], yticks=[])\n ax.imshow(x_train[i])",
"_____no_output_____"
],
[
"def normalize(x):\n max_x = np.max(x, axis=(0, 1), keepdims=True)\n min_x = np.min(x, axis=(0, 1), keepdims=True)\n return (x - min_x) / (max_x - min_x)",
"_____no_output_____"
],
[
"from tensorflow.keras.preprocessing.image import ImageDataGenerator\n\n# Global Contrast Normalization(GCN)\ngcn_whitening = ImageDataGenerator(samplewise_center=True, samplewise_std_normalization=True)\ngcn_whitening.fit(x_train)\n\nfig = plt.figure(figsize=(9, 15))\nfig.subplots_adjust(left=0, right=1, bottom=0, top=0.5, hspace=0.05, wspace=0.05)\n\nfor x_batch, y_batch in gcn_whitening.flow(x_train, y_train, batch_size=9, shuffle=False):\n for i in range(5):\n ax = fig.add_subplot(1, 5, i+1, xticks=[], yticks=[])\n ax.imshow(x_batch[i])\n break;",
"Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\n"
],
[
"# Zero-phase Component Analysis Whitening\nzca_whitening = ImageDataGenerator(zca_whitening=True)\nzca_whitening.fit(x_train)\n\nfig = plt.figure(figsize=(9, 15))\nfig.subplots_adjust(left=0, right=1, bottom=0, top=0.5, hspace=0.05, wspace=0.05)\n\nfor x_batch, y_batch in zca_whitening.flow(x_train, y_train, batch_size=9, shuffle=False):\n for i in range(5):\n ax = fig.add_subplot(1, 5, i+1, xticks=[], yticks=[])\n ax.imshow(x_batch[i])\n break;",
"C:\\Users\\suizh\\Anaconda3\\lib\\site-packages\\keras_preprocessing\\image\\image_data_generator.py:334: UserWarning: This ImageDataGenerator specifies `zca_whitening`, which overrides setting of `featurewise_center`.\n warnings.warn('This ImageDataGenerator specifies '\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\nClipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb43a7f34aa04f33515a3013ccb5ad51fa909e73 | 680,111 | ipynb | Jupyter Notebook | docs/source/notebooks/ODE_with_manual_gradients.ipynb | tandelDipak/pymc3 | e13a7ed8656ecea0fec9c5d5af5b1830315b44f9 | [
"Apache-2.0"
] | 1 | 2020-09-08T22:47:43.000Z | 2020-09-08T22:47:43.000Z | docs/source/notebooks/ODE_with_manual_gradients.ipynb | aseyboldt/pymc3 | 5b000c14f19042d5317fa31c0a9e5021190b40f4 | [
"Apache-2.0"
] | null | null | null | docs/source/notebooks/ODE_with_manual_gradients.ipynb | aseyboldt/pymc3 | 5b000c14f19042d5317fa31c0a9e5021190b40f4 | [
"Apache-2.0"
] | null | null | null | 602.935284 | 332,208 | 0.936325 | [
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport pymc3 as pm\nimport theano\n\nfrom scipy.integrate import odeint\nfrom theano import *\n\nTHEANO_FLAGS = \"optimizer=fast_compile\"",
"_____no_output_____"
]
],
[
[
"# Lotka-Volterra with manual gradients\n\nby [Sanmitra Ghosh](https://www.mrc-bsu.cam.ac.uk/people/in-alphabetical-order/a-to-g/sanmitra-ghosh/)",
"_____no_output_____"
],
[
"Mathematical models are used ubiquitously in a variety of science and engineering domains to model the time evolution of physical variables. These mathematical models are often described as ODEs that are characterised by model structure - the functions of the dynamical variables - and model parameters. However, for the vast majority of systems of practical interest it is necessary to infer both the model parameters and an appropriate model structure from experimental observations. This experimental data often appears to be scarce and incomplete. Furthermore, a large variety of models described as dynamical systems show traits of sloppiness (see [Gutenkunst et al., 2007](https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.0030189)) and have unidentifiable parameter combinations. The task of inferring model parameters and structure from experimental data is of paramount importance to reliably analyse the behaviour of dynamical systems and draw faithful predictions in light of the difficulties posit by their complexities. Moreover, any future model prediction should encompass and propagate variability and uncertainty in model parameters and/or structure. Thus, it is also important that the inference methods are equipped to quantify and propagate the aforementioned uncertainties from the model descriptions to model predictions. As a natural choice to handle uncertainty, at least in the parameters, Bayesian inference is increasingly used to fit ODE models to experimental data ([Mark Girolami, 2008](https://www.sciencedirect.com/science/article/pii/S030439750800501X)). However, due to some of the difficulties that I pointed above, fitting an ODE model using Bayesian inference is a challenging task. In this tutorial I am going to take up that challenge and will show how PyMC3 could be potentially used for this purpose. \n\nI must point out that model fitting (inference of the unknown parameters) is just one of many crucial tasks that a modeller has to complete in order to gain a deeper understanding of a physical process. However, success in this task is crucial and this is where PyMC3, and probabilistic programming (ppl) in general, is extremely useful. The modeller can take full advantage of the variety of samplers and distributions provided by PyMC3 to automate inference.\n\nIn this tutorial I will focus on the fitting exercise, that is estimating the posterior distribution of the parameters given some noisy experimental time series. ",
"_____no_output_____"
],
[
"## Bayesian inference of the parameters of an ODE\n\nI begin by first introducing the Bayesian framework for inference in a coupled non-linear ODE defined as \n$$\n\\frac{d X(t)}{dt}=\\boldsymbol{f}\\big(X(t),\\boldsymbol{\\theta}\\big),\n$$\nwhere $X(t)\\in\\mathbb{R}^K$ is the solution, at each time point, of the system composed of $K$ coupled ODEs - the state vector - and $\\boldsymbol{\\theta}\\in\\mathbb{R}^D$ is the parameter vector that we wish to infer. $\\boldsymbol{f}(\\cdot)$ is a non-linear function that describes the governing dynamics. Also, in case of an initial value problem, let the matrix $\\boldsymbol{X}(\\boldsymbol{\\theta}, \\mathbf{x_0})$ denote the solution of the above system of equations at some specified time points for the parameters $\\boldsymbol{\\theta}$ and initial conditions $\\mathbf{x_0}$.\n\nConsider a set of noisy experimental observations $\\boldsymbol{Y} \\in \\mathbb{R}^{T\\times K}$ observed at $T$ experimental time points for the $K$ states. We can obtain the likelihood $p(\\boldsymbol{Y}|\\boldsymbol{X})$, where I use the symbol $\\boldsymbol{X}:=\\boldsymbol{X}(\\boldsymbol{\\theta}, \\mathbf{x_0})$, and combine that with a prior distribution $p(\\boldsymbol{\\theta})$ on the parameters, using the Bayes theorem, to obtain the posterior distribution as\n$$\np(\\boldsymbol{\\theta}|\\boldsymbol{Y})=\\frac{1}{Z}p(\\boldsymbol{Y}|\\boldsymbol{X})p(\\boldsymbol{\\theta}),\n$$\nwhere $Z=\\int p(\\boldsymbol{Y}|\\boldsymbol{X})p(\\boldsymbol{\\theta}) d\\boldsymbol{\\theta} $ is the intractable marginal likelihood. Due to this intractability we resort to approximate inference and apply MCMC. \n\nFor this tutorial I have chosen two ODEs: \n1. The [__Lotka-Volterra predator prey model__ ](http://www.scholarpedia.org/article/Predator-prey_model)\n2. The [__Fitzhugh-Nagumo action potential model__](http://www.scholarpedia.org/article/FitzHugh-Nagumo_model)\n\nI will showcase two distinctive approaches (__NUTS__ and __SMC__ step methods), supported by PyMC3, for the estimation of unknown parameters in these models. ",
"_____no_output_____"
],
[
"## Lotka-Volterra predator prey model\n\n The Lotka Volterra model depicts an ecological system that is used to describe the interaction between a predator and prey species. This ODE given by\n $$\n \\begin{aligned}\n \t\\frac{d x}{dt} &=\\alpha x -\\beta xy \\\\\n \t\\frac{d y}{dt} &=-\\gamma y + \\delta xy,\n \\end{aligned}\n $$\n shows limit cycle behaviour and has often been used for benchmarking Bayesian inference methods. $\\boldsymbol{\\theta}=(\\alpha,\\beta,\\gamma,\\delta, x(0),y(0))$ is the set of unknown parameters that we wish to infer from experimental observations of the state vector $X(t)=(x(t),y(t))$ comprising the concentrations of the prey and the predator species respectively. $x(0), y(0)$ are the initial values of the states needed to solve the ODE, which are also treated as unknown quantities. The predator prey model was recently used to demonstrate the applicability of the NUTS sampler, and the Stan ppl in general, for inference in ODE models. I will closely follow [this](https://mc-stan.org/users/documentation/case-studies/lotka-volterra-predator-prey.html) Stan tutorial and thus I will setup this model and associated inference problem (including the data) exactly as was done for the Stan tutorial. Let me first write down the code to solve this ODE using the SciPy's `odeint`. Note that the methods in this tutorial is not limited or tied to `odeint`. Here I have chosen `odeint` to simply stay within PyMC3's dependencies (SciPy in this case). ",
"_____no_output_____"
]
],
[
[
"class LotkaVolterraModel:\n def __init__(self, y0=None):\n self._y0 = y0\n\n def simulate(self, parameters, times):\n alpha, beta, gamma, delta, Xt0, Yt0 = [x for x in parameters]\n\n def rhs(y, t, p):\n X, Y = y\n dX_dt = alpha * X - beta * X * Y\n dY_dt = -gamma * Y + delta * X * Y\n return dX_dt, dY_dt\n\n values = odeint(rhs, [Xt0, Yt0], times, (parameters,))\n return values\n\n\node_model = LotkaVolterraModel()",
"_____no_output_____"
]
],
[
[
"## Handling ODE gradients\n\nNUTS requires the gradient of the log of the target density w.r.t. the unknown parameters, $\\nabla_{\\boldsymbol{\\theta}}p(\\boldsymbol{\\theta}|\\boldsymbol{Y})$, which can be evaluated using the chain rule of differentiation as \n$$ \\nabla_{\\boldsymbol{\\theta}}p(\\boldsymbol{\\theta}|\\boldsymbol{Y}) = \\frac{\\partial p(\\boldsymbol{\\theta}|\\boldsymbol{Y})}{\\partial \\boldsymbol{X}}^T \\frac{\\partial \\boldsymbol{X}}{\\partial \\boldsymbol{\\theta}}.$$\n\nThe gradient of an ODE w.r.t. its parameters, the term $\\frac{\\partial \\boldsymbol{X}}{\\partial \\boldsymbol{\\theta}}$, can be obtained using local sensitivity analysis, although this is not the only method to obtain gradients. However, just like solving an ODE (a non-linear one to be precise) evaluation of the gradients can only be carried out using some sort of numerical method, say for example the famous Runge-Kutta method for non-stiff ODEs. PyMC3 uses Theano as the automatic differentiation engine and thus all models are implemented by stitching together available primitive operations (Ops) supported by Theano. Even to extend PyMC3 we need to compose models that can be expressed as symbolic combinations of Theano's Ops. However, if we take a step back and think about Theano then it is apparent that neither the ODE solution nor its gradient w.r.t. to the parameters can be expressed symbolically as combinations of Theano’s primitive Ops. Hence, from Theano’s perspective an ODE (and for that matter any other form of a non-linear differential equation) is a non-differentiable black-box function. However, one might argue that if a numerical method is coded up in Theano (using say the `scan` Op), then it is possible to symbolically express the numerical method that evaluates the ODE states, and then we can easily use Theano’s automatic differentiation engine to obtain the gradients as well by differentiating through the numerical solver itself. I like to point out that the former, obtaining the solution, is indeed possible this way but the obtained gradient would be error-prone. Additionally, this entails to a complete ‘re-inventing the wheel’ as one would have to implement decades old sophisticated numerical algorithms again from scratch in Theano. \n\nThus, in this tutorial I am going to present the alternative approach which consists of defining new [custom Theano Ops](http://deeplearning.net/software/theano_versions/dev/extending/extending_theano.html), extending Theano, that will wrap both the numerical solution and the vector-Matrix product, $ \\frac{\\partial p(\\boldsymbol{\\theta}|\\boldsymbol{Y})}{\\partial \\boldsymbol{X}}^T \\frac{\\partial \\boldsymbol{X}}{\\partial \\boldsymbol{\\theta}}$, often known as the _**vector-Jacobian product**_ (VJP) in automatic differentiation literature. I like to point out here that in the context of non-linear ODEs the term Jacobian is used to denote gradients of the ODE dynamics $\\boldsymbol{f}$ w.r.t. the ODE states $X(t)$. Thus, to avoid confusion, from now on I will use the term _**vector-sensitivity product**_ (VSP) to denote the same quantity that the term VJP denotes.\n\nI will start by introducing the forward sensitivity analysis.\n\n## ODE sensitivity analysis\n\nFor a coupled ODE system $\\frac{d X(t)}{dt} = \\boldsymbol{f}(X(t),\\boldsymbol{\\theta})$, the local sensitivity of the solution to a parameter is defined by how much the solution would change by changes in the parameter, i.e. the sensitivity of the the $k$-th state is simply put the time evolution of its graident w.r.t. the $d$-th parameter. This quantitiy, denoted as $Z_{kd}(t)$, is given by\n$$Z_{kd}(t)=\\frac{d }{d t} \\left\\{\\frac{\\partial X_k (t)}{\\partial \\theta_d}\\right\\} = \\sum_{i=1}^K \\frac{\\partial f_k}{\\partial X_i (t)}\\frac{\\partial X_i (t)}{\\partial \\theta_d} + \\frac{\\partial f_k}{\\partial \\theta_d}.$$\n\nUsing forward sensitivity analysis we can obtain both the state $X(t)$ and its derivative w.r.t the parameters, at each time point, as the solution to an initial value problem by augmenting the original ODE system with the sensitivity equations $Z_{kd}$. The augmented ODE system $\\big(X(t), Z(t)\\big)$ can then be solved together using a chosen numerical method. The augmented ODE system needs the initial values for the sensitivity equations. All of these should be set to zero except the ones where the sensitivity of a state w.r.t. its own initial value is sought, that is $ \\frac{\\partial X_k(t)}{\\partial X_k (0)} =1 $. Note that in order to solve this augmented system we have to embark in the tedious process of deriving $ \\frac{\\partial f_k}{\\partial X_i (t)}$, also known as the Jacobian of an ODE, and $\\frac{\\partial f_k}{\\partial \\theta_d}$ terms. Thankfully, many ODE solvers calculate these terms and solve the augmented system when asked for by the user. An example would be the [SUNDIAL CVODES solver suite](https://computation.llnl.gov/projects/sundials/cvodes). A Python wrapper for CVODES can be found [here](https://jmodelica.org/assimulo/). \n\nHowever, for this tutorial I would go ahead and derive the terms mentioned above, manually, and solve the Lotka-Volterra ODEs alongwith the sensitivites in the following code block. The functions `jac` and `dfdp` below calculate $ \\frac{\\partial f_k}{\\partial X_i (t)}$ and $\\frac{\\partial f_k}{\\partial \\theta_d}$ respectively for the Lotka-Volterra model. For conveniance I have transformed the sensitivity equation in a matrix form. Here I extended the solver code snippet above to include sensitivities when asked for.",
"_____no_output_____"
]
],
[
[
"n_states = 2\nn_odeparams = 4\nn_ivs = 2\n\n\nclass LotkaVolterraModel:\n def __init__(self, n_states, n_odeparams, n_ivs, y0=None):\n self._n_states = n_states\n self._n_odeparams = n_odeparams\n self._n_ivs = n_ivs\n self._y0 = y0\n\n def simulate(self, parameters, times):\n return self._simulate(parameters, times, False)\n\n def simulate_with_sensitivities(self, parameters, times):\n return self._simulate(parameters, times, True)\n\n def _simulate(self, parameters, times, sensitivities):\n alpha, beta, gamma, delta, Xt0, Yt0 = [x for x in parameters]\n\n def r(y, t, p):\n X, Y = y\n dX_dt = alpha * X - beta * X * Y\n dY_dt = -gamma * Y + delta * X * Y\n return dX_dt, dY_dt\n\n if sensitivities:\n\n def jac(y):\n X, Y = y\n ret = np.zeros((self._n_states, self._n_states))\n ret[0, 0] = alpha - beta * Y\n ret[0, 1] = -beta * X\n ret[1, 0] = delta * Y\n ret[1, 1] = -gamma + delta * X\n return ret\n\n def dfdp(y):\n X, Y = y\n ret = np.zeros(\n (self._n_states, self._n_odeparams + self._n_ivs)\n ) # except the following entries\n ret[\n 0, 0\n ] = X # \\frac{\\partial [\\alpha X - \\beta XY]}{\\partial \\alpha}, and so on...\n ret[0, 1] = -X * Y\n ret[1, 2] = -Y\n ret[1, 3] = X * Y\n\n return ret\n\n def rhs(y_and_dydp, t, p):\n y = y_and_dydp[0 : self._n_states]\n dydp = y_and_dydp[self._n_states :].reshape(\n (self._n_states, self._n_odeparams + self._n_ivs)\n )\n dydt = r(y, t, p)\n d_dydp_dt = np.matmul(jac(y), dydp) + dfdp(y)\n return np.concatenate((dydt, d_dydp_dt.reshape(-1)))\n\n y0 = np.zeros((2 * (n_odeparams + n_ivs)) + n_states)\n y0[6] = 1.0 # \\frac{\\partial [X]}{\\partial Xt0} at t==0, and same below for Y\n y0[13] = 1.0\n y0[0:n_states] = [Xt0, Yt0]\n result = odeint(rhs, y0, times, (parameters,), rtol=1e-6, atol=1e-5)\n values = result[:, 0 : self._n_states]\n dvalues_dp = result[:, self._n_states :].reshape(\n (len(times), self._n_states, self._n_odeparams + self._n_ivs)\n )\n return values, dvalues_dp\n else:\n values = odeint(r, [Xt0, Yt0], times, (parameters,), rtol=1e-6, atol=1e-5)\n return values\n\n\node_model = LotkaVolterraModel(n_states, n_odeparams, n_ivs)",
"_____no_output_____"
]
],
[
[
"For this model I have set the relative and absolute tolerances to $10^{-6}$ and $10^{-5}$ respectively, as was suggested in the Stan tutorial. This will produce sufficiently accurate solutions. Further reducing the tolerances will increase accuracy but at the cost of increasing the computational time. A thorough discussion on the choice and use of a numerical method for solving the ODE is out of the scope of this tutorial. However, I must point out that the inaccuracies of the ODE solver do affect the likelihood and as a result the inference. This is more so the case for stiff systems. I would recommend interested readers to this nice blog article where this effect is discussed thoroughly for a [cardiac ODE model](https://mirams.wordpress.com/2018/10/17/ode-errors-and-optimisation/). There is also an emerging area of uncertainty quantification that attacks the problem of noise arisng from impreciseness of numerical algorithms, [probabilistic numerics](http://probabilistic-numerics.org/). This is indeed an elegant framework to carry out inference while taking into account the errors coming from the numeric ODE solvers.\n\n## Custom ODE Op\n\nIn order to define the custom `Op` I have written down two `theano.Op` classes `ODEGradop`, `ODEop`. `ODEop` essentially wraps the ODE solution and will be called by PyMC3. The `ODEGradop` wraps the numerical VSP and this op is then in turn used inside the `grad` method in the `ODEop` to return the VSP. Note that we pass in two functions: `state`, `numpy_vsp` as arguments to respective Ops. I will define these functions later. These functions act as shims using which we connect the python code for numerical solution of sate and VSP to Theano and thus PyMC3.",
"_____no_output_____"
]
],
[
[
"class ODEGradop(theano.Op):\n def __init__(self, numpy_vsp):\n self._numpy_vsp = numpy_vsp\n\n def make_node(self, x, g):\n x = theano.tensor.as_tensor_variable(x)\n g = theano.tensor.as_tensor_variable(g)\n node = theano.Apply(self, [x, g], [g.type()])\n return node\n\n def perform(self, node, inputs_storage, output_storage):\n x = inputs_storage[0]\n\n g = inputs_storage[1]\n out = output_storage[0]\n out[0] = self._numpy_vsp(x, g) # get the numerical VSP\n\n\nclass ODEop(theano.Op):\n def __init__(self, state, numpy_vsp):\n self._state = state\n self._numpy_vsp = numpy_vsp\n\n def make_node(self, x):\n x = theano.tensor.as_tensor_variable(x)\n\n return theano.Apply(self, [x], [x.type()])\n\n def perform(self, node, inputs_storage, output_storage):\n x = inputs_storage[0]\n out = output_storage[0]\n\n out[0] = self._state(x) # get the numerical solution of ODE states\n\n def grad(self, inputs, output_grads):\n x = inputs[0]\n g = output_grads[0]\n\n grad_op = ODEGradop(self._numpy_vsp) # pass the VSP when asked for gradient\n grad_op_apply = grad_op(x, g)\n\n return [grad_op_apply]",
"_____no_output_____"
]
],
[
[
"I must point out that the way I have defined the custom ODE Ops above there is the possibility that the ODE is solved twice for the same parameter values, once for the states and another time for the VSP. To avoid this behaviour I have written a helper class which stops this double evaluation.",
"_____no_output_____"
]
],
[
[
"class solveCached:\n def __init__(self, times, n_params, n_outputs):\n\n self._times = times\n self._n_params = n_params\n self._n_outputs = n_outputs\n self._cachedParam = np.zeros(n_params)\n self._cachedSens = np.zeros((len(times), n_outputs, n_params))\n self._cachedState = np.zeros((len(times), n_outputs))\n\n def __call__(self, x):\n\n if np.all(x == self._cachedParam):\n state, sens = self._cachedState, self._cachedSens\n\n else:\n state, sens = ode_model.simulate_with_sensitivities(x, times)\n\n return state, sens\n\n\ntimes = np.arange(0, 21) # number of measurement points (see below)\ncached_solver = solveCached(times, n_odeparams + n_ivs, n_states)",
"_____no_output_____"
]
],
[
[
"### The ODE state & VSP evaluation\n\nMost ODE systems of practical interest will have multiple states and thus the output of the solver, which I have denoted so far as $\\boldsymbol{X}$, for a system with $K$ states solved on $T$ time points, would be a $T \\times K$-dimensional matrix. For the Lotka-Volterra model the columns of this matrix represent the time evolution of the individual species concentrations. I flatten this matrix to a $TK$-dimensional vector $vec(\\boldsymbol{X})$, and also rearrange the sensitivities accordingly to obtain the desired vector-matrix product. It is beneficial at this point to test the custom Op as described [here](http://deeplearning.net/software/theano_versions/dev/extending/extending_theano.html#how-to-test-it).",
"_____no_output_____"
]
],
[
[
"def state(x):\n State, Sens = cached_solver(np.array(x, dtype=np.float64))\n cached_solver._cachedState, cached_solver._cachedSens, cached_solver._cachedParam = (\n State,\n Sens,\n x,\n )\n return State.reshape((2 * len(State),))\n\n\ndef numpy_vsp(x, g):\n numpy_sens = cached_solver(np.array(x, dtype=np.float64))[1].reshape(\n (n_states * len(times), len(x))\n )\n return numpy_sens.T.dot(g)",
"_____no_output_____"
]
],
[
[
"## The Hudson's Bay Company data\n\nThe Lotka-Volterra predator prey model has been used previously to successfully explain the dynamics of natural populations of predators and prey, such as the lynx and snowshoe hare data of the Hudson's Bay Company. This is the same data (that was shared [here](https://github.com/stan-dev/example-models/tree/master/knitr/lotka-volterra)) used in the Stan example and thus I will use this data-set as the experimental observations $\\boldsymbol{Y}(t)$ to infer the parameters. ",
"_____no_output_____"
]
],
[
[
"Year = np.arange(1900, 1921, 1)\n# fmt: off\nLynx = np.array([4.0, 6.1, 9.8, 35.2, 59.4, 41.7, 19.0, 13.0, 8.3, 9.1, 7.4,\n 8.0, 12.3, 19.5, 45.7, 51.1, 29.7, 15.8, 9.7, 10.1, 8.6])\nHare = np.array([30.0, 47.2, 70.2, 77.4, 36.3, 20.6, 18.1, 21.4, 22.0, 25.4, \n 27.1, 40.3, 57.0, 76.6, 52.3, 19.5, 11.2, 7.6, 14.6, 16.2, 24.7])\n# fmt: on\nplt.figure(figsize=(15, 7.5))\nplt.plot(Year, Lynx, color=\"b\", lw=4, label=\"Lynx\")\nplt.plot(Year, Hare, color=\"g\", lw=4, label=\"Hare\")\nplt.legend(fontsize=15)\nplt.xlim([1900, 1920])\nplt.xlabel(\"Year\", fontsize=15)\nplt.ylabel(\"Concentrations\", fontsize=15)\nplt.xticks(Year, rotation=45)\nplt.title(\"Lynx (predator) - Hare (prey): oscillatory dynamics\", fontsize=25);",
"_____no_output_____"
]
],
[
[
"## The probablistic model\n\nI have now got all the ingredients needed in order to define the probabilistic model in PyMC3. As I have mentioned previously I will set up the probabilistic model with the exact same likelihood and priors used in the Stan example. The observed data is defined as follows:\n\n$$\\log (\\boldsymbol{Y(t)}) = \\log (\\boldsymbol{X(t)}) + \\eta(t),$$ \n\nwhere $\\eta(t)$ is assumed to be zero mean i.i.d Gaussian noise with an unknown standard deviation $\\sigma$, that needs to be estimated. The above multiplicative (on the natural scale) noise model encodes a lognormal distribution as the likelihood:\n\n$$\\boldsymbol{Y(t)} \\sim \\mathcal{L}\\mathcal{N}(\\log (\\boldsymbol{X(t)}), \\sigma^2).$$\n\nThe following priors are then placed on the parameters:\n\n$$\n\\begin{aligned}\nx(0), y(0) &\\sim \\mathcal{L}\\mathcal{N}(\\log(10),1),\\\\\n\\alpha, \\gamma &\\sim \\mathcal{N}(1,0.5),\\\\\n\\beta, \\delta &\\sim \\mathcal{N}(0.05,0.05),\\\\\n\\sigma &\\sim \\mathcal{L}\\mathcal{N}(-1,1).\n\\end{aligned}\n$$\n\nFor an intuitive explanation, which I am omitting for brevity, regarding the choice of priors as well as the likelihood model, I would recommend the Stan example mentioned above. The above probabilistic model is defined in PyMC3 below. Note that the flattened state vector is reshaped to match the data dimensionality.\n\nFinally, I use the `pm.sample` method to run NUTS by default and obtain $1500$ post warm-up samples from the posterior.",
"_____no_output_____"
]
],
[
[
"theano.config.exception_verbosity = \"high\"\ntheano.config.floatX = \"float64\"\n\n\n# Define the data matrix\nY = np.vstack((Hare, Lynx)).T\n\n# Now instantiate the theano custom ODE op\nmy_ODEop = ODEop(state, numpy_vsp)\n\n# The probabilistic model\nwith pm.Model() as LV_model:\n\n # Priors for unknown model parameters\n\n alpha = pm.Normal(\"alpha\", mu=1, sd=0.5)\n beta = pm.Normal(\"beta\", mu=0.05, sd=0.05)\n gamma = pm.Normal(\"gamma\", mu=1, sd=0.5)\n delta = pm.Normal(\"delta\", mu=0.05, sd=0.05)\n\n xt0 = pm.Lognormal(\"xto\", mu=np.log(10), sd=1)\n yt0 = pm.Lognormal(\"yto\", mu=np.log(10), sd=1)\n sigma = pm.Lognormal(\"sigma\", mu=-1, sd=1, shape=2)\n\n # Forward model\n all_params = pm.math.stack([alpha, beta, gamma, delta, xt0, yt0], axis=0)\n ode_sol = my_ODEop(all_params)\n forward = ode_sol.reshape(Y.shape)\n\n # Likelihood\n Y_obs = pm.Lognormal(\"Y_obs\", mu=pm.math.log(forward), sd=sigma, observed=Y)\n\n trace = pm.sample(1500, tune=1000, init=\"adapt_diag\")\ntrace[\"diverging\"].sum()",
"Auto-assigning NUTS sampler...\nInitializing NUTS using adapt_diag...\nMultiprocess sampling (2 chains in 2 jobs)\nNUTS: [sigma, yto, xto, delta, gamma, beta, alpha]\nSampling 2 chains, 0 divergences: 2%|▏ | 94/5000 [01:02<59:45, 1.37draws/s] /Users/demetri/anaconda3/envs/gsoc/lib/python3.6/site-packages/scipy/integrate/odepack.py:247: ODEintWarning: Excess work done on this call (perhaps wrong Dfun type). Run with full_output = 1 to get quantitative information.\n warnings.warn(warning_msg, ODEintWarning)\nSampling 2 chains, 0 divergences: 2%|▏ | 108/5000 [01:09<54:44, 1.49draws/s] /Users/demetri/anaconda3/envs/gsoc/lib/python3.6/site-packages/scipy/integrate/odepack.py:247: ODEintWarning: Excess work done on this call (perhaps wrong Dfun type). Run with full_output = 1 to get quantitative information.\n warnings.warn(warning_msg, ODEintWarning)\nSampling 2 chains, 0 divergences: 100%|██████████| 5000/5000 [12:57<00:00, 4.16draws/s] \nThe acceptance probability does not match the target. It is 0.6992852935132228, but should be close to 0.8. Try to increase the number of tuning steps.\n"
],
[
"with LV_model:\n pm.traceplot(trace);",
"_____no_output_____"
],
[
"import pandas as pd\n\nsummary = pm.summary(trace)\nSTAN_mus = [0.549, 0.028, 0.797, 0.024, 33.960, 5.949, 0.248, 0.252]\nSTAN_sds = [0.065, 0.004, 0.091, 0.004, 2.909, 0.533, 0.045, 0.044]\nsummary[\"STAN_mus\"] = pd.Series(np.array(STAN_mus), index=summary.index)\nsummary[\"STAN_sds\"] = pd.Series(np.array(STAN_sds), index=summary.index)\nsummary",
"/Users/demetri/Documents/GitHub/pymc3/pymc3/stats.py:991: FutureWarning: The join_axes-keyword is deprecated. Use .reindex or .reindex_like on the result to achieve the same functionality.\n axis=1, join_axes=[dforg.index])\n"
]
],
[
[
"These estimates are almost identical to those obtained in the Stan tutorial (see the last two columns above), which is what we can expect. Posterior predictives can be drawn as below. ",
"_____no_output_____"
]
],
[
[
"ppc_samples = pm.sample_posterior_predictive(trace, samples=1000, model=LV_model)[\"Y_obs\"]\nmean_ppc = ppc_samples.mean(axis=0)\nCriL_ppc = np.percentile(ppc_samples, q=2.5, axis=0)\nCriU_ppc = np.percentile(ppc_samples, q=97.5, axis=0)",
"/Users/demetri/Documents/GitHub/pymc3/pymc3/sampling.py:1078: UserWarning: samples parameter is smaller than nchains times ndraws, some draws and/or chains may not be represented in the returned posterior predictive sample\n warnings.warn(\"samples parameter is smaller than nchains times ndraws, some draws \"\n100%|██████████| 1000/1000 [00:10<00:00, 98.26it/s]\n"
],
[
"plt.figure(figsize=(15, 2 * (5)))\nplt.subplot(2, 1, 1)\nplt.plot(Year, Lynx, \"o\", color=\"b\", lw=4, ms=10.5)\nplt.plot(Year, mean_ppc[:, 1], color=\"b\", lw=4)\nplt.plot(Year, CriL_ppc[:, 1], \"--\", color=\"b\", lw=2)\nplt.plot(Year, CriU_ppc[:, 1], \"--\", color=\"b\", lw=2)\nplt.xlim([1900, 1920])\nplt.ylabel(\"Lynx conc\", fontsize=15)\nplt.xticks(Year, rotation=45)\nplt.subplot(2, 1, 2)\nplt.plot(Year, Hare, \"o\", color=\"g\", lw=4, ms=10.5, label=\"Observed\")\nplt.plot(Year, mean_ppc[:, 0], color=\"g\", lw=4, label=\"mean of ppc\")\nplt.plot(Year, CriL_ppc[:, 0], \"--\", color=\"g\", lw=2, label=\"credible intervals\")\nplt.plot(Year, CriU_ppc[:, 0], \"--\", color=\"g\", lw=2)\nplt.legend(fontsize=15)\nplt.xlim([1900, 1920])\nplt.xlabel(\"Year\", fontsize=15)\nplt.ylabel(\"Hare conc\", fontsize=15)\nplt.xticks(Year, rotation=45);",
"_____no_output_____"
]
],
[
[
"# Efficient exploration of the posterior landscape with SMC\n\nIt has been pointed out in several papers that the complex non-linear dynamics of an ODE results in a posterior landscape that is extremely difficult to navigate efficiently by many MCMC samplers. Thus, recently the curvature information of the posterior surface has been used to construct powerful geometrically aware samplers ([Mark Girolami and Ben Calderhead, 2011](https://rss.onlinelibrary.wiley.com/doi/epdf/10.1111/j.1467-9868.2010.00765.x)) that perform extremely well in ODE inference problems. Another set of ideas suggest breaking down a complex inference task into a sequence of simpler tasks. In essence the idea is to use sequential-importance-sampling to sample from an artificial sequence of increasingly complex distributions where the first in the sequence is a distribution that is easy to sample from, the prior, and the last in the sequence is the actual complex target distribution. The associated importance distribution is constructed by moving the set of particles sampled at the previous step using a Markov kernel, say for example the MH kernel. \n\nA simple way of building the sequence of distributions is to use a temperature $\\beta$, that is raised slowly from $0$ to $1$. Using this temperature variable $\\beta$ we can write down the annealed intermediate distribution as\n\n$$p_{\\beta}(\\boldsymbol{\\theta}|\\boldsymbol{y})\\propto p(\\boldsymbol{y}|\\boldsymbol{\\theta})^{\\beta} p(\\boldsymbol{\\theta}).$$\n\nSamplers that carry out sequential-importance-sampling from these artificial sequence of distributions, to avoid the difficult task of sampling directly from $p(\\boldsymbol{\\theta}|\\boldsymbol{y})$, are known as Sequential Monte Carlo (SMC) samplers ([P Del Moral et al., 2006](https://rss.onlinelibrary.wiley.com/doi/full/10.1111/j.1467-9868.2006.00553.x)). The performance of these samplers are sensitive to the choice of the temperature schedule, that is the set of user-defined increasing values of $\\beta$ between $0$ and $1$. Fortunately, PyMC3 provides a version of the SMC sampler ([Jianye Ching and Yi-Chu Chen, 2007](https://ascelibrary.org/doi/10.1061/%28ASCE%290733-9399%282007%29133%3A7%28816%29)) that automatically figures out this temperature schedule. Moreover, the PyMC3's SMC sampler does not require the gradient of the log target density. As a result it is extremely easy to use this sampler for inference in ODE models. In the next example I will apply this SMC sampler to estimate the parameters of the Fitzhugh-Nagumo model. ",
"_____no_output_____"
],
[
"## The Fitzhugh-Nagumo model\n\nThe Fitzhugh-Nagumo model given by\n$$\n\\begin{aligned}\n\\frac{dV}{dt}&=(V - \\frac{V^3}{3} + R)c\\\\\n\\frac{dR}{dt}&=\\frac{-(V-a+bR)}{c},\n\\end{aligned}\n$$\nconsisting of a membrane voltage variable $V(t)$ and a recovery variable $R(t)$ is a two-dimensional simplification of the [Hodgkin-Huxley](http://www.scholarpedia.org/article/Conductance-based_models) model of spike (action potential) generation in squid giant axons and where $a$, $b$, $c$ are the model parameters. This model produces a rich dynamics and as a result a complex geometry of the posterior surface that often leads to poor performance of many MCMC samplers. As a result this model was used to test the efficacy of the discussed geometric MCMC scheme and since then has been used to benchmark other novel MCMC methods. Following [Mark Girolami and Ben Calderhead, 2011](https://rss.onlinelibrary.wiley.com/doi/epdf/10.1111/j.1467-9868.2010.00765.x) I will also use artificially generated data from this model to setup the inference task for estimating $\\boldsymbol{\\theta}=(a,b,c)$.",
"_____no_output_____"
]
],
[
[
"class FitzhughNagumoModel:\n def __init__(self, times, y0=None):\n self._y0 = np.array([-1, 1], dtype=np.float64)\n self._times = times\n\n def _simulate(self, parameters, times):\n a, b, c = [float(x) for x in parameters]\n\n def rhs(y, t, p):\n V, R = y\n dV_dt = (V - V ** 3 / 3 + R) * c\n dR_dt = (V - a + b * R) / -c\n return dV_dt, dR_dt\n\n values = odeint(rhs, self._y0, times, (parameters,), rtol=1e-6, atol=1e-6)\n return values\n\n def simulate(self, x):\n return self._simulate(x, self._times)",
"_____no_output_____"
]
],
[
[
"## Simulated Data\n\nFor this example I am going to use simulated data that is I will generate noisy traces from the forward model defined above with parameters $\\theta$ set to $(0.2,0.2,3)$ respectively and corrupted by i.i.d Gaussian noise with a standard deviation $\\sigma=0.5$. The initial values are set to $V(0)=-1$ and $R(0)=1$ respectively. Again following [Mark Girolami and Ben Calderhead, 2011](https://rss.onlinelibrary.wiley.com/doi/epdf/10.1111/j.1467-9868.2010.00765.x) I will assume that the initial values are known. These parameter values pushes the model into the oscillatory regime.",
"_____no_output_____"
]
],
[
[
"n_states = 2\nn_times = 200\ntrue_params = [0.2, 0.2, 3.0]\nnoise_sigma = 0.5\nFN_solver_times = np.linspace(0, 20, n_times)\node_model = FitzhughNagumoModel(FN_solver_times)\nsim_data = ode_model.simulate(true_params)\nnp.random.seed(42)\nY_sim = sim_data + np.random.randn(n_times, n_states) * noise_sigma\nplt.figure(figsize=(15, 7.5))\nplt.plot(FN_solver_times, sim_data[:, 0], color=\"darkblue\", lw=4, label=r\"$V(t)$\")\nplt.plot(FN_solver_times, sim_data[:, 1], color=\"darkgreen\", lw=4, label=r\"$R(t)$\")\nplt.plot(FN_solver_times, Y_sim[:, 0], \"o\", color=\"darkblue\", ms=4.5, label=\"Noisy traces\")\nplt.plot(FN_solver_times, Y_sim[:, 1], \"o\", color=\"darkgreen\", ms=4.5)\nplt.legend(fontsize=15)\nplt.xlabel(\"Time\", fontsize=15)\nplt.ylabel(\"Values\", fontsize=15)\nplt.title(\"Fitzhugh-Nagumo Action Potential Model\", fontsize=25);",
"_____no_output_____"
]
],
[
[
"## Define a non-differentiable black-box op using Theano @as_op\n\nRemember that I told SMC sampler does not require gradients, this is by the way the case for other samplers such as the Metropolis-Hastings, Slice sampler that are also supported in PyMC3. For all these gradient-free samplers I will show a simple and quick way of wrapping the forward model i.e. the ODE solution in Theano. All we have to do is to simply to use the decorator `as_op` that converts a python function into a basic Theano Op. We also tell Theano using the `as_op` decorator that we have three parameters each being a Theano scalar. The output then is a Theano matrix whose columns are the state vectors.",
"_____no_output_____"
]
],
[
[
"import theano.tensor as tt\n\nfrom theano.compile.ops import as_op\n\n\n@as_op(itypes=[tt.dscalar, tt.dscalar, tt.dscalar], otypes=[tt.dmatrix])\ndef th_forward_model(param1, param2, param3):\n\n param = [param1, param2, param3]\n th_states = ode_model.simulate(param)\n\n return th_states",
"_____no_output_____"
]
],
[
[
"## Generative model\n\nSince I have corrupted the original traces with i.i.d Gaussian thus the likelihood is given by\n$$\\boldsymbol{Y} = \\prod_{i=1}^T \\mathcal{N}(\\boldsymbol{X}(t_i)), \\sigma^2\\mathbb{I}),$$ \nwhere $\\mathbb{I}\\in \\mathbb{R}^{K \\times K}$. We place a Gamma, Normal, Uniform prior on $(a,b,c)$ and a HalfNormal prior on $\\sigma$ as follows:\n$$\n\\begin{aligned}\n\ta & \\sim \\mathcal{Gamma}(2,1),\\\\\n\tb & \\sim \\mathcal{N}(0,1),\\\\\n\tc & \\sim \\mathcal{U}(0.1,1),\\\\\n\t\\sigma & \\sim \\mathcal{H}(1).\n\\end{aligned}\n$$\n\nNotice how I have used the `start` argument for this example. Just like `pm.sample` `pm.sample_smc` has a number of settings, but I found the default ones good enough for simple models such as this one.",
"_____no_output_____"
]
],
[
[
"draws = 1000\nwith pm.Model() as FN_model:\n\n a = pm.Gamma(\"a\", alpha=2, beta=1)\n b = pm.Normal(\"b\", mu=0, sd=1)\n c = pm.Uniform(\"c\", lower=0.1, upper=10)\n\n sigma = pm.HalfNormal(\"sigma\", sd=1)\n\n forward = th_forward_model(a, b, c)\n\n cov = np.eye(2) * sigma ** 2\n\n Y_obs = pm.MvNormal(\"Y_obs\", mu=forward, cov=cov, observed=Y_sim)\n\n startsmc = {v.name: np.random.uniform(1e-3, 2, size=draws) for v in FN_model.free_RVs}\n\n trace_FN = pm.sample_smc(draws, start=startsmc)",
"Sample initial stage: ...\nStage: 0 Beta: 0.009 Steps: 25\nStage: 1 Beta: 0.015 Steps: 8\nStage: 2 Beta: 0.020 Steps: 4\nStage: 3 Beta: 0.030 Steps: 13\nStage: 4 Beta: 0.049 Steps: 3\nStage: 5 Beta: 0.089 Steps: 10\nStage: 6 Beta: 0.178 Steps: 3\nStage: 7 Beta: 0.368 Steps: 8\nStage: 8 Beta: 0.782 Steps: 3\nStage: 9 Beta: 1.000 Steps: 7\n"
],
[
"pm.plot_posterior(trace_FN, kind=\"hist\", bins=30, color=\"seagreen\");",
"_____no_output_____"
]
],
[
[
"## Inference summary\n\nWith `pm.SMC`, do I get similar performance to geometric MCMC samplers (see [Mark Girolami and Ben Calderhead, 2011](https://rss.onlinelibrary.wiley.com/doi/epdf/10.1111/j.1467-9868.2010.00765.x))? I think so !",
"_____no_output_____"
]
],
[
[
"results = [\n pm.summary(trace_FN, [\"a\"]),\n pm.summary(trace_FN, [\"b\"]),\n pm.summary(trace_FN, [\"c\"]),\n pm.summary(trace_FN, [\"sigma\"]),\n]\nresults = pd.concat(results)\ntrue_params.append(noise_sigma)\nresults[\"True values\"] = pd.Series(np.array(true_params), index=results.index)\ntrue_params.pop()\nresults",
"_____no_output_____"
]
],
[
[
"## Reconstruction of the phase portrait\n\nIts good to check that we can reconstruct the (famous) pahse portrait for this model based on the obtained samples.",
"_____no_output_____"
]
],
[
[
"params = np.array([trace_FN.get_values(\"a\"), trace_FN.get_values(\"b\"), trace_FN.get_values(\"c\")]).T\nparams.shape\nnew_values = []\nfor ind in range(len(params)):\n ppc_sol = ode_model.simulate(params[ind])\n new_values.append(ppc_sol)\nnew_values = np.array(new_values)\nmean_values = np.mean(new_values, axis=0)\nplt.figure(figsize=(15, 7.5))\n\nplt.plot(\n mean_values[:, 0],\n mean_values[:, 1],\n color=\"black\",\n lw=4,\n label=\"Inferred (mean of sampled) phase portrait\",\n)\nplt.plot(\n sim_data[:, 0], sim_data[:, 1], \"--\", color=\"#ff7f0e\", lw=4, ms=6, label=\"True phase portrait\"\n)\nplt.legend(fontsize=15)\nplt.xlabel(r\"$V(t)$\", fontsize=15)\nplt.ylabel(r\"$R(t)$\", fontsize=15);",
"_____no_output_____"
]
],
[
[
"# Perspectives\n\n### Using some other ODE models\n\nI have tried to keep everything as general as possible. So, my custom ODE Op, the state and VSP evaluator as well as the cached solver are not tied to a specific ODE model. Thus, to use any other ODE model one only needs to implement a `simulate_with_sensitivities` method according to their own specific ODE model.\n\n### Other forms of differential equation (DDE, DAE, PDE)\n\nI hope the two examples have elucidated the applicability of PyMC3 in regards to fitting ODE models. Although ODEs are the most fundamental constituent of a mathematical model, there are indeed other forms of dynamical systems such as a delay differential equation (DDE), a differential algebraic equation (DAE) and the partial differential equation (PDE) whose parameter estimation is equally important. The SMC and for that matter any other non-gradient sampler supported by PyMC3 can be used to fit all these forms of differential equation, of course using the `as_op`. However, just like an ODE we can solve augmented systems of DDE/DAE along with their sensitivity equations. The sensitivity equations for a DDE and a DAE can be found in this recent paper, [C Rackauckas et al., 2018](https://arxiv.org/abs/1812.01892) (Equation 9 and 10). Thus we can easily apply NUTS sampler to these models.\n\n### Stan already supports ODEs\n\nWell there are many problems where I believe SMC sampler would be more suitable than NUTS and thus its good to have that option. \n\n### Model selection\n\nMost ODE inference literature since [Vladislav Vyshemirsky and Mark Girolami, 2008](https://academic.oup.com/bioinformatics/article/24/6/833/192524) recommend the usage of Bayes factor for the purpose of model selection/comparison. This involves the calculation of the marginal likelihood which is a much more nuanced topic and I would refrain from any discussion about that. Fortunately, the SMC sampler calculates the marginal likelihood as a by product so this can be used for obtaining Bayes factors. Follow PyMC3's other tutorials for further information regarding how to obtain the marginal likelihood after running the SMC sampler.\n\nSince we generally frame the ODE inference as a regression problem (along with the i.i.d measurement noise assumption in most cases) we can straight away use any of the supported information criterion, such as the widely available information criterion (WAIC), irrespective of what sampler is used for inference. See the PyMC3's API for further information regarding WAIC.\n\n### Other AD packages\n\nAlthough this is a slight digression nonetheless I would still like to point out my observations on this issue. The approach that I have presented here for embedding an ODE (also extends to DDE/DAE) as a custom Op can be trivially carried forward to other AD packages such as TensorFlow and PyTorch. I had been able to use TensorFlow's [py_func](https://www.tensorflow.org/api_docs/python/tf/py_func) to build a custom TensorFlow ODE Op and then use that in the [Edward](http://edwardlib.org/) ppl. I would recommend [this](https://pytorch.org/tutorials/advanced/numpy_extensions_tutorial.html) tutorial, for writing PyTorch extensions, to those who are interested in using the [Pyro](http://pyro.ai/) ppl.\n \n\n",
"_____no_output_____"
]
],
[
[
"%load_ext watermark\n%watermark -n -u -v -iv -w",
"pymc3 3.8\narviz 0.7.0\npandas 0.25.3\nseaborn 0.9.0\nnumpy 1.17.5\nlast updated: Wed Apr 22 2020 \n\nCPython 3.8.0\nIPython 7.11.0\nwatermark 2.0.2\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb43c107bca6bd4c67fe5ad33d2f1883a9c4151b | 451,492 | ipynb | Jupyter Notebook | notebooks/ch-algorithms/teleportation.ipynb | kifumi/platypus | 619f0842a207ba19088728895736f02d905d5aa2 | [
"Apache-2.0"
] | 12 | 2022-01-07T17:21:42.000Z | 2022-03-23T12:53:16.000Z | notebooks/ch-algorithms/teleportation.ipynb | kifumi/platypus | 619f0842a207ba19088728895736f02d905d5aa2 | [
"Apache-2.0"
] | 477 | 2022-01-06T17:05:03.000Z | 2022-03-31T15:46:10.000Z | notebooks/ch-algorithms/teleportation.ipynb | kifumi/platypus | 619f0842a207ba19088728895736f02d905d5aa2 | [
"Apache-2.0"
] | 21 | 2022-01-06T18:59:42.000Z | 2022-03-31T07:44:03.000Z | 399.90434 | 187,176 | 0.672269 | [
[
[
"# Quantum Teleportation",
"_____no_output_____"
],
[
"This notebook demonstrates quantum teleportation. We first use Qiskit's built-in simulators to test our quantum circuit, and then try it out on a real quantum computer.",
"_____no_output_____"
],
[
"## 1. Overview <a id='overview'></a>",
"_____no_output_____"
],
[
"Alice wants to send quantum information to Bob. Specifically, suppose she wants to send the qubit state\n$\\vert\\psi\\rangle = \\alpha\\vert0\\rangle + \\beta\\vert1\\rangle$. \nThis entails passing on information about $\\alpha$ and $\\beta$ to Bob.\n\nThere exists a theorem in quantum mechanics which states that you cannot simply make an exact copy of an unknown quantum state. This is known as the no-cloning theorem. As a result of this we can see that Alice can't simply generate a copy of $\\vert\\psi\\rangle$ and give the copy to Bob. We can only copy classical states (not superpositions).\n\nHowever, by taking advantage of two classical bits and an entangled qubit pair, Alice can transfer her state $\\vert\\psi\\rangle$ to Bob. We call this teleportation because, at the end, Bob will have $\\vert\\psi\\rangle$ and Alice won't anymore.",
"_____no_output_____"
],
[
"## 2. The Quantum Teleportation Protocol <a id='how'></a>\nTo transfer a quantum bit, Alice and Bob must use a third party (Telamon) to send them an entangled qubit pair. Alice then performs some operations on her qubit, sends the results to Bob over a classical communication channel, and Bob then performs some operations on his end to receive Alice’s qubit.\n\n\n\nWe will describe the steps on a quantum circuit below. Here, no qubits are actually ‘sent’, you’ll just have to imagine that part!\n\nFirst we set up our session:",
"_____no_output_____"
]
],
[
[
"# Do the necessary imports\nimport numpy as np\nfrom qiskit import QuantumCircuit, QuantumRegister, ClassicalRegister\nfrom qiskit import IBMQ, Aer, transpile\nfrom qiskit.visualization import plot_histogram, plot_bloch_multivector, array_to_latex\nfrom qiskit.extensions import Initialize\nfrom qiskit.result import marginal_counts\nfrom qiskit.quantum_info import random_statevector",
"_____no_output_____"
]
],
[
[
"and create our quantum circuit:",
"_____no_output_____"
]
],
[
[
"## SETUP\n# Protocol uses 3 qubits and 2 classical bits in 2 different registers\n\nqr = QuantumRegister(3, name=\"q\") # Protocol uses 3 qubits\ncrz = ClassicalRegister(1, name=\"crz\") # and 2 classical bits\ncrx = ClassicalRegister(1, name=\"crx\") # in 2 different registers\nteleportation_circuit = QuantumCircuit(qr, crz, crx)",
"_____no_output_____"
]
],
[
[
"#### Step 1\nA third party, Telamon, creates an entangled pair of qubits and gives one to Bob and one to Alice.\n\nThe pair Telamon creates is a special pair called a Bell pair. In quantum circuit language, the way to create a Bell pair between two qubits is to first transfer one of them to the X-basis ($|+\\rangle$ and $|-\\rangle$) using a Hadamard gate, and then to apply a CNOT gate onto the other qubit controlled by the one in the X-basis. ",
"_____no_output_____"
]
],
[
[
"def create_bell_pair(qc, a, b):\n \"\"\"Creates a bell pair in qc using qubits a & b\"\"\"\n qc.h(a) # Put qubit a into state |+>\n qc.cx(a,b) # CNOT with a as control and b as target",
"_____no_output_____"
],
[
"## SETUP\n# Protocol uses 3 qubits and 2 classical bits in 2 different registers\nqr = QuantumRegister(3, name=\"q\")\ncrz, crx = ClassicalRegister(1, name=\"crz\"), ClassicalRegister(1, name=\"crx\")\nteleportation_circuit = QuantumCircuit(qr, crz, crx)\n\n## STEP 1\n# In our case, Telamon entangles qubits q1 and q2\n# Let's apply this to our circuit:\ncreate_bell_pair(teleportation_circuit, 1, 2)\n# And view the circuit so far:\nteleportation_circuit.draw()",
"_____no_output_____"
]
],
[
[
"Let's say Alice owns $q_1$ and Bob owns $q_2$ after they part ways.\n\n#### Step 2 \n\nAlice applies a CNOT gate to $q_1$, controlled by $\\vert\\psi\\rangle$ (the qubit she is trying to send Bob). Then Alice applies a Hadamard gate to $|\\psi\\rangle$. In our quantum circuit, the qubit ($|\\psi\\rangle$) Alice is trying to send is $q_0$:",
"_____no_output_____"
]
],
[
[
"def alice_gates(qc, psi, a):\n qc.cx(psi, a)\n qc.h(psi)",
"_____no_output_____"
],
[
"## SETUP\n# Protocol uses 3 qubits and 2 classical bits in 2 different registers\nqr = QuantumRegister(3, name=\"q\")\ncrz, crx = ClassicalRegister(1, name=\"crz\"), ClassicalRegister(1, name=\"crx\")\nteleportation_circuit = QuantumCircuit(qr, crz, crx)\n\n## STEP 1\ncreate_bell_pair(teleportation_circuit, 1, 2)\n\n## STEP 2\nteleportation_circuit.barrier() # Use barrier to separate steps\nalice_gates(teleportation_circuit, 0, 1)\nteleportation_circuit.draw()",
"_____no_output_____"
]
],
[
[
"#### Step 3\n\nNext, Alice applies a measurement to both qubits that she owns, $q_1$ and $\\vert\\psi\\rangle$, and stores this result in two classical bits. She then sends these two bits to Bob.",
"_____no_output_____"
]
],
[
[
"def measure_and_send(qc, a, b):\n \"\"\"Measures qubits a & b and 'sends' the results to Bob\"\"\"\n qc.barrier()\n qc.measure(a,0)\n qc.measure(b,1)",
"_____no_output_____"
],
[
"## SETUP\n# Protocol uses 3 qubits and 2 classical bits in 2 different registers\nqr = QuantumRegister(3, name=\"q\")\ncrz, crx = ClassicalRegister(1, name=\"crz\"), ClassicalRegister(1, name=\"crx\")\nteleportation_circuit = QuantumCircuit(qr, crz, crx)\n\n## STEP 1\ncreate_bell_pair(teleportation_circuit, 1, 2)\n\n## STEP 2\nteleportation_circuit.barrier() # Use barrier to separate steps\nalice_gates(teleportation_circuit, 0, 1)\n\n## STEP 3\nmeasure_and_send(teleportation_circuit, 0 ,1)\nteleportation_circuit.draw()",
"_____no_output_____"
]
],
[
[
"#### Step 4\n\nBob, who already has the qubit $q_2$, then applies the following gates depending on the state of the classical bits:\n\n00 $\\rightarrow$ Do nothing\n\n01 $\\rightarrow$ Apply $X$ gate\n\n10 $\\rightarrow$ Apply $Z$ gate\n\n11 $\\rightarrow$ Apply $ZX$ gate\n\n(*Note that this transfer of information is purely classical*.)",
"_____no_output_____"
]
],
[
[
"# This function takes a QuantumCircuit (qc), integer (qubit)\n# and ClassicalRegisters (crz & crx) to decide which gates to apply\ndef bob_gates(qc, qubit, crz, crx):\n # Here we use c_if to control our gates with a classical\n # bit instead of a qubit\n qc.x(qubit).c_if(crx, 1) # Apply gates if the registers \n qc.z(qubit).c_if(crz, 1) # are in the state '1'",
"_____no_output_____"
],
[
"## SETUP\n# Protocol uses 3 qubits and 2 classical bits in 2 different registers\nqr = QuantumRegister(3, name=\"q\")\ncrz, crx = ClassicalRegister(1, name=\"crz\"), ClassicalRegister(1, name=\"crx\")\nteleportation_circuit = QuantumCircuit(qr, crz, crx)\n\n## STEP 1\ncreate_bell_pair(teleportation_circuit, 1, 2)\n\n## STEP 2\nteleportation_circuit.barrier() # Use barrier to separate steps\nalice_gates(teleportation_circuit, 0, 1)\n\n## STEP 3\nmeasure_and_send(teleportation_circuit, 0, 1)\n\n## STEP 4\nteleportation_circuit.barrier() # Use barrier to separate steps\nbob_gates(teleportation_circuit, 2, crz, crx)\nteleportation_circuit.draw()",
"_____no_output_____"
]
],
[
[
"And voila! At the end of this protocol, Alice's qubit has now teleported to Bob.",
"_____no_output_____"
],
[
"## 3. Simulating the Teleportation Protocol <a id='simulating'></a>",
"_____no_output_____"
],
[
"### 3.1 How Will We Test the Protocol on a Quantum Computer? <a id='testing'></a>",
"_____no_output_____"
],
[
"In this notebook, we will initialize Alice's qubit in a random state $\\vert\\psi\\rangle$ (`psi`). This state will be created using an `Initialize` gate on $|q_0\\rangle$. In this chapter we use the function `random_statevector` to choose `psi` for us, but feel free to set `psi` to any qubit state you want.",
"_____no_output_____"
]
],
[
[
"# Create random 1-qubit state\npsi = random_statevector(2)\n\n# Display it nicely\ndisplay(array_to_latex(psi, prefix=\"|\\\\psi\\\\rangle =\"))\n# Show it on a Bloch sphere\nplot_bloch_multivector(psi)",
"_____no_output_____"
]
],
[
[
"Let's create our initialization instruction to create $|\\psi\\rangle$ from the state $|0\\rangle$:",
"_____no_output_____"
]
],
[
[
"init_gate = Initialize(psi)\ninit_gate.label = \"init\"",
"_____no_output_____"
]
],
[
[
"(`Initialize` is technically not a gate since it contains a reset operation, and so is not reversible. We call it an 'instruction' instead). If the quantum teleportation circuit works, then at the end of the circuit the qubit $|q_2\\rangle$ will be in this state. We will check this using the statevector simulator.",
"_____no_output_____"
],
[
"### 3.2 Using the Simulated Statevector <a id='simulating-sv'></a>\n\nWe can use the Aer simulator to verify our qubit has been teleported.",
"_____no_output_____"
]
],
[
[
"## SETUP\nqr = QuantumRegister(3, name=\"q\") # Protocol uses 3 qubits\ncrz = ClassicalRegister(1, name=\"crz\") # and 2 classical registers\ncrx = ClassicalRegister(1, name=\"crx\")\nqc = QuantumCircuit(qr, crz, crx)\n\n## STEP 0\n# First, let's initialize Alice's q0\nqc.append(init_gate, [0])\nqc.barrier()\n\n## STEP 1\n# Now begins the teleportation protocol\ncreate_bell_pair(qc, 1, 2)\nqc.barrier()\n\n## STEP 2\n# Send q1 to Alice and q2 to Bob\nalice_gates(qc, 0, 1)\n\n## STEP 3\n# Alice then sends her classical bits to Bob\nmeasure_and_send(qc, 0, 1)\n\n## STEP 4\n# Bob decodes qubits\nbob_gates(qc, 2, crz, crx)\n\n# Display the circuit\nqc.draw()",
"_____no_output_____"
]
],
[
[
"We can see below, using the statevector obtained from the aer simulator, that the state of $|q_2\\rangle$ is the same as the state $|\\psi\\rangle$ we created above, while the states of $|q_0\\rangle$ and $|q_1\\rangle$ have been collapsed to either $|0\\rangle$ or $|1\\rangle$. The state $|\\psi\\rangle$ has been teleported from qubit 0 to qubit 2.",
"_____no_output_____"
]
],
[
[
"sim = Aer.get_backend('aer_simulator')\nqc.save_statevector()\nout_vector = sim.run(qc).result().get_statevector()\nplot_bloch_multivector(out_vector)",
"/usr/local/anaconda3/envs/terra-unstable/lib/python3.9/site-packages/qiskit/visualization/bloch.py:69: MatplotlibDeprecationWarning: \nThe M attribute was deprecated in Matplotlib 3.4 and will be removed two minor releases later. Use self.axes.M instead.\n x_s, y_s, _ = proj3d.proj_transform(xs3d, ys3d, zs3d, renderer.M)\n"
]
],
[
[
"You can run this cell a few times to make sure. You may notice that the qubits 0 & 1 change states, but qubit 2 is always in the state $|\\psi\\rangle$.",
"_____no_output_____"
],
[
"### 3.3 Using the Simulated Counts <a id='simulating-fc'></a>\n\nQuantum teleportation is designed to send qubits between two parties. We do not have the hardware to demonstrate this, but we can demonstrate that the gates perform the correct transformations on a single quantum chip. Here we again use the aer simulator to simulate how we might test our protocol.\n\nOn a real quantum computer, we would not be able to sample the statevector, so if we wanted to check our teleportation circuit is working, we need to do things slightly differently. The `Initialize` instruction first performs a reset, setting our qubit to the state $|0\\rangle$. It then applies gates to turn our $|0\\rangle$ qubit into the state $|\\psi\\rangle$:\n\n$$ |0\\rangle \\xrightarrow{\\text{Initialize gates}} |\\psi\\rangle $$\n\nSince all quantum gates are reversible, we can find the inverse of these gates using:",
"_____no_output_____"
]
],
[
[
"inverse_init_gate = init_gate.gates_to_uncompute()",
"_____no_output_____"
]
],
[
[
"This operation has the property:\n\n$$ |\\psi\\rangle \\xrightarrow{\\text{Inverse Initialize gates}} |0\\rangle $$\n\nTo prove the qubit $|q_0\\rangle$ has been teleported to $|q_2\\rangle$, if we do this inverse initialization on $|q_2\\rangle$, we expect to measure $|0\\rangle$ with certainty. We do this in the circuit below:",
"_____no_output_____"
]
],
[
[
"## SETUP\nqr = QuantumRegister(3, name=\"q\") # Protocol uses 3 qubits\ncrz = ClassicalRegister(1, name=\"crz\") # and 2 classical registers\ncrx = ClassicalRegister(1, name=\"crx\")\nqc = QuantumCircuit(qr, crz, crx)\n\n## STEP 0\n# First, let's initialize Alice's q0\nqc.append(init_gate, [0])\nqc.barrier()\n\n## STEP 1\n# Now begins the teleportation protocol\ncreate_bell_pair(qc, 1, 2)\nqc.barrier()\n\n## STEP 2\n# Send q1 to Alice and q2 to Bob\nalice_gates(qc, 0, 1)\n\n## STEP 3\n# Alice then sends her classical bits to Bob\nmeasure_and_send(qc, 0, 1)\n\n## STEP 4\n# Bob decodes qubits\nbob_gates(qc, 2, crz, crx)\n\n## STEP 5\n# reverse the initialization process\nqc.append(inverse_init_gate, [2])\n\n# Display the circuit\nqc.draw()",
"_____no_output_____"
]
],
[
[
"We can see the `inverse_init_gate` appearing, labelled 'disentangler' on the circuit diagram. Finally, we measure the third qubit and store the result in the third classical bit:",
"_____no_output_____"
]
],
[
[
"# Need to add a new ClassicalRegister\n# to see the result\ncr_result = ClassicalRegister(1)\nqc.add_register(cr_result)\nqc.measure(2,2)\nqc.draw()",
"_____no_output_____"
]
],
[
[
"and we run our experiment:",
"_____no_output_____"
]
],
[
[
"t_qc = transpile(qc, sim)\nt_qc.save_statevector()\ncounts = sim.run(t_qc).result().get_counts()\nqubit_counts = [marginal_counts(counts, [qubit]) for qubit in range(3)]\nplot_histogram(qubit_counts)",
"_____no_output_____"
]
],
[
[
"We can see we have a 100% chance of measuring $q_2$ (the purple bar in the histogram) in the state $|0\\rangle$. This is the expected result, and indicates the teleportation protocol has worked properly.",
"_____no_output_____"
],
[
"## 4. Understanding Quantum Teleportation <a id=\"understanding-qt\"></a>",
"_____no_output_____"
],
[
"As you have worked with the Quantum Teleportation's implementation, it is time to understand the mathematics behind the protocol.\n\n#### Step 1\n\nQuantum Teleportation begins with the fact that Alice needs to transmit $|\\psi\\rangle = \\alpha|0\\rangle + \\beta|1\\rangle$ (a random qubit) to Bob. She doesn't know the state of the qubit. For this, Alice and Bob take the help of a third party (Telamon). Telamon prepares a pair of entangled qubits for Alice and Bob. The entangled qubits could be written in Dirac Notation as:\n\n$$ |e \\rangle = \\frac{1}{\\sqrt{2}} (|00\\rangle + |11\\rangle) $$\n\nAlice and Bob each possess one qubit of the entangled pair (denoted as A and B respectively),\n\n$$|e\\rangle = \\frac{1}{\\sqrt{2}} (|0\\rangle_A |0\\rangle_B + |1\\rangle_A |1\\rangle_B) $$\n\nThis creates a three qubit quantum system where Alice has the first two qubits and Bob the last one.\n\n$$ \\begin{aligned}\n|\\psi\\rangle \\otimes |e\\rangle &= \\frac{1}{\\sqrt{2}} (\\alpha |0\\rangle \\otimes (|00\\rangle + |11\\rangle) + \\beta |1\\rangle \\otimes (|00\\rangle + |11\\rangle))\\\\\n&= \\frac{1}{\\sqrt{2}} (\\alpha|000\\rangle + \\alpha|011\\rangle + \\beta|100\\rangle + \\beta|111\\rangle) \n\\end{aligned}$$",
"_____no_output_____"
],
[
"#### Step 2\n\nNow according to the protocol Alice applies CNOT gate on her two qubits followed by Hadamard gate on the first qubit. This results in the state:\n\n$$ \n\\begin{aligned} &(H \\otimes I \\otimes I) (CNOT \\otimes I) (|\\psi\\rangle \\otimes |e\\rangle)\\\\\n&=(H \\otimes I \\otimes I) (CNOT \\otimes I) \\frac{1}{\\sqrt{2}} (\\alpha|000\\rangle + \\alpha|011\\rangle + \\beta|100\\rangle + \\beta|111\\rangle) \\\\\n&= (H \\otimes I \\otimes I) \\frac{1}{\\sqrt{2}} (\\alpha|000\\rangle + \\alpha|011\\rangle + \\beta|110\\rangle + \\beta|101\\rangle) \\\\\n&= \\frac{1}{2} (\\alpha(|000\\rangle + |011\\rangle + |100\\rangle + |111\\rangle) + \\beta(|010\\rangle + |001\\rangle - |110\\rangle - |101\\rangle)) \\\\\n\\end{aligned}\n$$\n\nWhich can then be separated and written as: \n\n$$\n\\begin{aligned}\n= \\frac{1}{2}(\n & \\phantom{+} |00\\rangle (\\alpha|0\\rangle + \\beta|1\\rangle) \\hphantom{\\quad )} \\\\\n & + |01\\rangle (\\alpha|1\\rangle + \\beta|0\\rangle) \\hphantom{\\quad )}\\\\[4pt]\n & + |10\\rangle (\\alpha|0\\rangle - \\beta|1\\rangle) \\hphantom{\\quad )}\\\\[4pt]\n & + |11\\rangle (\\alpha|1\\rangle - \\beta|0\\rangle) \\quad )\\\\\n\\end{aligned}\n$$",
"_____no_output_____"
],
[
"#### Step 3\n\nAlice measures the first two qubit (which she owns) and sends them as two classical bits to Bob. The result she obtains is always one of the four standard basis states $|00\\rangle, |01\\rangle, |10\\rangle,$ and $|11\\rangle$ with equal probability. \n\nOn the basis of her measurement, Bob's state will be projected to, \n$$\n|00\\rangle \\rightarrow (\\alpha|0\\rangle + \\beta|1\\rangle)\\\\\n|01\\rangle \\rightarrow (\\alpha|1\\rangle + \\beta|0\\rangle)\\\\\n|10\\rangle \\rightarrow (\\alpha|0\\rangle - \\beta|1\\rangle)\\\\\n|11\\rangle \\rightarrow (\\alpha|1\\rangle - \\beta|0\\rangle)\n$$",
"_____no_output_____"
],
[
"#### Step 4\n\nBob, on receiving the bits from Alice, knows he can obtain the original state $|\\psi\\rangle$ by applying appropriate transformations on his qubit that was once part of the entangled pair.\n\nThe transformations he needs to apply are:\n\n$$\n\\begin{array}{c c c}\n\\mbox{Bob's State} & \\mbox{Bits Received} & \\mbox{Gate Applied} \\\\\n(\\alpha|0\\rangle + \\beta|1\\rangle) & 00 & I \\\\\n(\\alpha|1\\rangle + \\beta|0\\rangle) & 01 & X \\\\\n(\\alpha|0\\rangle - \\beta|1\\rangle) & 10 & Z \\\\\n(\\alpha|1\\rangle - \\beta|0\\rangle) & 11 & ZX\n\\end{array}\n$$\n\nAfter this step Bob will have successfully reconstructed Alice's state.",
"_____no_output_____"
],
[
"## 5. Teleportation on a Real Quantum Computer <a id='real_qc'></a>",
"_____no_output_____"
],
[
"### 5.1 IBM hardware and Deferred Measurement <a id='deferred-measurement'></a>\n\nThe IBM quantum computers currently do not support instructions after measurements, meaning we cannot run the quantum teleportation in its current form on real hardware. Fortunately, this does not limit our ability to perform any computations due to the _deferred measurement principle_ discussed in chapter 4.4 of [1]. The principle states that any measurement can be postponed until the end of the circuit, i.e. we can move all the measurements to the end, and we should see the same results.\n\n\n\nAny benefits of measuring early are hardware related: If we can measure early, we may be able to reuse qubits, or reduce the amount of time our qubits are in their fragile superposition. In this example, the early measurement in quantum teleportation would have allowed us to transmit a qubit state without a direct quantum communication channel.\n\nWhile moving the gates allows us to demonstrate the \"teleportation\" circuit on real hardware, it should be noted that the benefit of the teleportation process (transferring quantum states via classical channels) is lost.\n\nLet us re-write the `bob_gates` function to `new_bob_gates`:",
"_____no_output_____"
]
],
[
[
"def new_bob_gates(qc, a, b, c):\n qc.cx(b, c)\n qc.cz(a, c)",
"_____no_output_____"
]
],
[
[
"And create our new circuit:",
"_____no_output_____"
]
],
[
[
"qc = QuantumCircuit(3,1)\n\n# First, let's initialize Alice's q0\nqc.append(init_gate, [0])\nqc.barrier()\n\n# Now begins the teleportation protocol\ncreate_bell_pair(qc, 1, 2)\nqc.barrier()\n# Send q1 to Alice and q2 to Bob\nalice_gates(qc, 0, 1)\nqc.barrier()\n# Alice sends classical bits to Bob\nnew_bob_gates(qc, 0, 1, 2)\n\n# We undo the initialization process\nqc.append(inverse_init_gate, [2])\n\n# See the results, we only care about the state of qubit 2\nqc.measure(2,0)\n\n# View the results:\nqc.draw()",
"_____no_output_____"
]
],
[
[
"### 5.2 Executing <a id='executing'></a> ",
"_____no_output_____"
]
],
[
[
"# First, see what devices we are allowed to use by loading our saved accounts\nIBMQ.load_account()\nprovider = IBMQ.get_provider(hub='ibm-q')",
"_____no_output_____"
],
[
"# get the least-busy backend at IBM and run the quantum circuit there\nfrom qiskit.providers.ibmq import least_busy\nfrom qiskit.tools.monitor import job_monitor\nbackend = least_busy(provider.backends(filters=lambda b: b.configuration().n_qubits >= 3 and\n not b.configuration().simulator and b.status().operational==True))\nt_qc = transpile(qc, backend, optimization_level=3)\njob = backend.run(t_qc)\njob_monitor(job) # displays job status under cell",
"Job Status: job has successfully run\n"
],
[
"# Get the results and display them\nexp_result = job.result()\nexp_counts = exp_result.get_counts(qc)\nprint(exp_counts)\nplot_histogram(exp_counts)",
"{'0': 849, '1': 175}\n"
]
],
[
[
"As we see here, there are a few results in which we measured $|1\\rangle$. These arise due to errors in the gates and the qubits. In contrast, our simulator in the earlier part of the notebook had zero errors in its gates, and allowed error-free teleportation.",
"_____no_output_____"
]
],
[
[
"print(f\"The experimental error rate : {exp_counts['1']*100/sum(exp_counts.values()):.3f}%\")",
"The experimental error rate : 17.090%\n"
]
],
[
[
"## 6. References <a id='references'></a>\n[1] M. Nielsen and I. Chuang, Quantum Computation and Quantum Information, Cambridge Series on Information and the Natural Sciences (Cambridge University Press, Cambridge, 2000).\n\n[2] Eleanor Rieffel and Wolfgang Polak, Quantum Computing: a Gentle Introduction (The MIT Press Cambridge England, Massachusetts, 2011).",
"_____no_output_____"
]
],
[
[
"import qiskit.tools.jupyter\n%qiskit_version_table",
"/usr/local/anaconda3/envs/terra-unstable/lib/python3.9/site-packages/qiskit/aqua/__init__.py:86: DeprecationWarning: The package qiskit.aqua is deprecated. It was moved/refactored to qiskit-terra For more information see <https://github.com/Qiskit/qiskit-aqua/blob/main/README.md#migration-guide>\n warn_package('aqua', 'qiskit-terra')\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb43c3a68ef837e0b8163b5a358ad99dbe2a64bb | 10,457 | ipynb | Jupyter Notebook | Dates4.ipynb | 4dsolutions/Python5 | 8d80753e823441a571b827d24d21577446409b52 | [
"MIT"
] | 11 | 2016-08-17T00:15:26.000Z | 2020-07-17T21:31:10.000Z | Dates4.ipynb | 4dsolutions/Python5 | 8d80753e823441a571b827d24d21577446409b52 | [
"MIT"
] | null | null | null | Dates4.ipynb | 4dsolutions/Python5 | 8d80753e823441a571b827d24d21577446409b52 | [
"MIT"
] | 5 | 2017-02-22T05:15:52.000Z | 2019-11-08T06:17:34.000Z | 16.893376 | 379 | 0.486947 | [
[
[
"#### PYT-DS SAISOFT\n\n<a data-flickr-embed=\"true\" href=\"https://www.flickr.com/photos/kirbyurner/27963484878/in/album-72157693427665102/\" title=\"Barry at Large\"><img src=\"https://farm1.staticflickr.com/969/27963484878_b38f0db42a_m.jpg\" width=\"240\" height=\"180\" alt=\"Barry at Large\"></a><script async src=\"//embedr.flickr.com/assets/client-code.js\" charset=\"utf-8\"></script>\n\n# DATA SCIENCE WITH PYTHON\n\n\n# Pandas Periods",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom pandas import Series, DataFrame, Period, Timestamp\nimport numpy as np",
"_____no_output_____"
],
[
"p = Period(2007, freq='A-DEC') # an interval from Jan 1 to Dec 31, 2007\np",
"_____no_output_____"
],
[
"p + 5",
"_____no_output_____"
],
[
"p - 2",
"_____no_output_____"
],
[
"p - Period(2000, freq='A-DEC')",
"_____no_output_____"
],
[
"rng_years = pd.period_range('1/1/2000', '1/1/2018', freq='Y')",
"_____no_output_____"
],
[
"rng_years",
"_____no_output_____"
],
[
"rng_months = pd.period_range('1/1/2000', '1/1/2018', freq='M')",
"_____no_output_____"
],
[
"rng_months",
"_____no_output_____"
],
[
"type(rng_months)",
"_____no_output_____"
],
[
"s = Series(np.random.randn(len(rng_years)), index=rng_years)\ns",
"_____no_output_____"
],
[
"s.index",
"_____no_output_____"
],
[
"rng_years",
"_____no_output_____"
],
[
"rng_years.asfreq('M', how='start')",
"_____no_output_____"
],
[
"rng_years.asfreq('M', how='end')",
"_____no_output_____"
],
[
"rng_years.asfreq('D', how='end')",
"_____no_output_____"
],
[
"rng_months.asfreq('D', how='end')",
"_____no_output_____"
],
[
"fiscal_year = Period('2007', freq='A-JUN')",
"_____no_output_____"
],
[
"fiscal_year.asfreq('M', 'start')",
"_____no_output_____"
],
[
"fiscal_year.asfreq('M', 'end')",
"_____no_output_____"
],
[
"fiscal_year.asfreq('D', 'start')",
"_____no_output_____"
],
[
"fiscal_year.asfreq('D', 'end')",
"_____no_output_____"
]
],
[
[
"## Quarterly Period Frequencies",
"_____no_output_____"
]
],
[
[
"p = Period('2018Q4', freq='Q-JAN')",
"_____no_output_____"
],
[
"p",
"_____no_output_____"
],
[
"p.asfreq('D', 'start')",
"_____no_output_____"
],
[
"p.asfreq('D', 'end')",
"_____no_output_____"
],
[
"p.asfreq('Y', 'start')",
"_____no_output_____"
],
[
"p.asfreq('M', 'start')",
"_____no_output_____"
],
[
"p.asfreq('M', 'start') + 3",
"_____no_output_____"
],
[
"p.asfreq('Q-JAN', 'start')",
"_____no_output_____"
],
[
"p.asfreq('Q-JAN', 'end')",
"_____no_output_____"
],
[
"p.asfreq('Q', 'end')",
"_____no_output_____"
]
],
[
[
"## Timestamps to Periods (and Back)",
"_____no_output_____"
]
],
[
[
"rng = pd.date_range('1/1/2018', periods=6, freq='M')",
"_____no_output_____"
],
[
"rng",
"_____no_output_____"
],
[
"ts = Series(np.random.randn(6), index=rng)",
"_____no_output_____"
],
[
"ts",
"_____no_output_____"
],
[
"pts = ts.to_period()",
"_____no_output_____"
],
[
"pts",
"_____no_output_____"
],
[
"rng = pd.date_range('1/28/2018', periods=10, freq='D')\nts2 = Series(np.random.randn(len(rng)), index=rng)",
"_____no_output_____"
],
[
"ts2",
"_____no_output_____"
],
[
"ts2.to_period('M')",
"_____no_output_____"
],
[
"pts",
"_____no_output_____"
],
[
"pts.to_timestamp(how='end')",
"_____no_output_____"
],
[
"pts.to_timestamp(how='start')",
"_____no_output_____"
],
[
"pts2 = ts2.to_period('M')",
"_____no_output_____"
],
[
"pts2",
"_____no_output_____"
],
[
"pts2.to_timestamp(how='end')",
"_____no_output_____"
],
[
"ts",
"_____no_output_____"
],
[
"%matplotlib inline\nts.plot(kind=\"bar\")",
"_____no_output_____"
],
[
"ts.plot(kind=\"line\")",
"_____no_output_____"
]
],
[
[
"### LAB CHALLENGE: \n\nUsing documentation (of course!) create a horizontal bar graph from the same data (ts).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb43dbf9df0b1b5bbfc8f3f1c77c61206c1f9595 | 702,788 | ipynb | Jupyter Notebook | Hands on/CS185C02_Sp21_ho10.ipynb | shahadeshubhu/CS-185C | 7c210fc4198d2dc6009d1c470ae292be05c62bf6 | [
"MIT"
] | null | null | null | Hands on/CS185C02_Sp21_ho10.ipynb | shahadeshubhu/CS-185C | 7c210fc4198d2dc6009d1c470ae292be05c62bf6 | [
"MIT"
] | null | null | null | Hands on/CS185C02_Sp21_ho10.ipynb | shahadeshubhu/CS-185C | 7c210fc4198d2dc6009d1c470ae292be05c62bf6 | [
"MIT"
] | null | null | null | 702,788 | 702,788 | 0.961603 | [
[
[
"<img src=\"https://pm1.narvii.com/5887/02b61b74eaec1060b56a3fcfed42ecc24a457a2e_hq.jpg\">\n\nIn this hands-on, we will use the Marvel dataset to practice using different plots to visualize distributions of values between groups. You are free to come up with you own questions and use one of the categorical plots to help answer each question. You are also free to build your own dataframe that contains a specific subset of the data to help you answer your questions. \n\nThe dataset is in https://raw.githubusercontent.com/csbfx/advpy122-data/master/marvel-wikia-data.csv\n\nData source: https://github.com/fivethirtyeight/data/tree/master/comic-characters",
"_____no_output_____"
],
[
"| Variable |\tDefinition |\n| :------- | :-\n|page_id|\tThe unique identifier for that characters page within the wikia\n|name|\tThe name of the character\n|urlslug|\tThe unique url within the wikia that takes you to the character\n|ID|\tThe identity status of the character (Secret Identity, Public identity, [on marvel only: No Dual Identity])\n|ALIGN|\tIf the character is Good, Bad or Neutral\n|EYE|\tEye color of the character\n|HAIR|\tHair color of the character\n|SEX|\tSex of the character (e.g. Male, Female, etc.)\n|GSM|\tIf the character is a gender or sexual minority (e.g. Homosexual characters, bisexual characters)\n|ALIVE|\tIf the character is alive or deceased\n|APPEARANCES|\tThe number of appareances of the character in comic books (as of Sep. 2, 2014. Number will become increasingly out of date as time goes on.)\n|FIRST APPEARANCE|\tThe month and year of the character's first appearance in a comic book, if available\n|YEAR|\tThe year of the character's first appearance in a comic book, if available",
"_____no_output_____"
],
[
"## Q1. How big is this dataset? \nUse pandas to find out the number of rows and columns.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport seaborn as sns\nfrom matplotlib import pyplot as plt\n%matplotlib inline\nmarvel = pd.read_csv(\"https://raw.githubusercontent.com/csbfx/advpy122-data/master/marvel-wikia-data.csv\")\nprint(\"rows \" + str(len(marvel)))\nprint(\"columns \" + str(len(marvel.columns)))",
"rows 16376\ncolumns 13\n"
]
],
[
[
"## Q2. Strip plots\nCome up with a question using this dataset and use a `strip` plot to help answer the question. State your question in a markdown cell. Recall that a `strip` plot is a categorical plot where one axis is category and the other is a continuous variable. Set the appropriate arguments to make the plot more readable. Be sure to include a meaning title for the plot.",
"_____no_output_____"
],
[
"**Show the distribution of appearances of characters based on how their alignment. What can you infer from the data?**",
"_____no_output_____"
]
],
[
[
"stripplt = sns.catplot(data= marvel, x=\"ALIGN\", y=\"APPEARANCES\", aspect = 1.5)\nstripplt.fig.suptitle(\"Appearances of characters based on their alignments\", y = 1)",
"_____no_output_____"
]
],
[
[
"Amongst all the characters, it is the good characters who've made more appearances showing the marvel theme where heroes or good characters eventually defeat bad characters or evil men who most likely might've been eventually killed off or have been defeated.",
"_____no_output_____"
],
[
"## Q3. Multiples of Strip plots\nCome up with a question using this dataset and use a strip plot that contains multiples (splitting the plot into multiples by a category that has two or more unique values) by using the `row` or `col` argument. State your question in a markdown cell. Recall that a strip plot is a categorical plot where one axis is category and the other is a continuous variable. Set the appropriate arguments to make the plot more readable. Be sure to include a meaning title for the plot.",
"_____no_output_____"
],
[
"**Create a strip plot pointing out the relationship between Marvel characters appearance like eyes and hair to the appearsnces they've made in comics**",
"_____no_output_____"
]
],
[
[
"marvel['EYE'] = marvel.EYE.str.replace(\" Eyes\",\"\")\nmstripplt = sns.catplot(data= marvel, x=\"EYE\", y=\"APPEARANCES\", row=\"HAIR\", hue=\"EYE\", aspect = 3.5)\nmstripplt.fig.suptitle(\"Physical characteristics of characters and their relationship with the appearances they've made\", y = 1)",
"_____no_output_____"
]
],
[
[
"## Q4. Swarm plot\nCome up with a question using this dataset and use a `swarm` plot to help answer the question. State your question in a markdown cell. Recall that a `swarm` plot is also a categorical plot where one axis is category and the other is a continuous variable. Set the appropriate arguments to make the plot more readable. Be sure to include a meaning title for the plot.",
"_____no_output_____"
],
[
"**What does the identity of a good character from marvel say about their chances of living or dying? create a swarm plot to illustrate your point**",
"_____no_output_____"
]
],
[
[
"marvel_good = marvel[marvel[\"ALIGN\"] == \"Good Characters\"]\nswarm = sns.catplot(data=marvel_good, x=\"ALIVE\", y= \"Year\", hue=\"ID\", kind= \"swarm\")\nswarm.fig.suptitle(\"Living status of good characters based on their identities\")",
"/usr/local/lib/python3.7/dist-packages/seaborn/categorical.py:1296: UserWarning: 66.3% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\n/usr/local/lib/python3.7/dist-packages/seaborn/categorical.py:1296: UserWarning: 16.6% of the points cannot be placed; you may want to decrease the size of the markers or use stripplot.\n warnings.warn(msg, UserWarning)\n"
]
],
[
[
"From the swarm plot, interesting readings can be found. Surprisingly it is characters with secret identites that have died rather than those with public identities. We'd expect good characters who hide their identities to have a higher chance of survival but that's not the case. ",
"_____no_output_____"
],
[
"## Q5. Box plots\nBox plot is one of the most commonly used plot for visualizing data distribution. We can convert the `swamp` plot into a `box` plot by simply changing the kind argument to `kind=\"box\"`. Convert the swarm plot that you created in Q4 with a boxplot here. Set the appropriate arguments to make the plot more readable. Be sure to include a meaning title for the plot.",
"_____no_output_____"
]
],
[
[
"box = sns.catplot(data=marvel_good, x=\"ALIVE\", y= \"Year\", hue=\"ID\", kind= \"box\")\nbox.fig.suptitle(\"Box plot for Living status of good characters based on their identities\")",
"_____no_output_____"
]
],
[
[
"## Q6. Violin plots\nCome up with a question using this dataset and use a `violin` plot to help answer the question. State your question in a markdown cell. Recall that a `violin` plot is also a categorical plot where one axis is category and the other is a continuous variable. Set the appropriate arguments to make the plot more readable. You might want to set setting `cut` to zero if the distribution spreads beyond the values of the data in the dataset. Be sure to include a meaning title for the plot.",
"_____no_output_____"
],
[
"**Create a violin plot which defines a relationship between characters of a gender or sexual minority and year when they made their first appearance. What do you conclude from the strip plot?**",
"_____no_output_____"
]
],
[
[
"stripplt = sns.catplot(data = marvel, x =\"GSM\", y = \"Year\", aspect = 2.5, kind= \"violin\", hue=\"ALIVE\")\nstripplt.fig.suptitle(\"Inclusion of GSM characters over the years\", y = 1)",
"_____no_output_____"
]
],
[
[
"From the given strip plot, we can conclude that Marvel started including characters of GSM very late from the 1990s.",
"_____no_output_____"
],
[
"## Bonus: \nBecause violin plots are symetrical, when we have only two categories we can put one on each side with `split = True`. Try to create a violin plot using the `split` parameter. You will need to come up with a dataframe using this dataset with data that has two categories. ",
"_____no_output_____"
]
],
[
[
"marvel\n",
"_____no_output_____"
],
[
"stripplt1 = sns.catplot(data = marvel, x =\"GSM\", y = \"Year\", aspect = 2.5, kind= \"violin\",hue=\"ALIVE\", split= True)\nstripplt1.fig.suptitle(\"Inclusion of GSM characters over the years\", y = 1)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cb43e15f2fdb45160a653b2f5b9882d3595f071c | 14,071 | ipynb | Jupyter Notebook | notebooks/3. CNN - Text.ipynb | aivizionz/DeepLearning-NLP | d14fabe20bbe58ed1c0b876387338e1a55d6c70c | [
"MIT"
] | 294 | 2017-07-22T20:13:59.000Z | 2022-03-04T15:55:54.000Z | notebooks/3. CNN - Text.ipynb | aivizionz/DeepLearning-NLP | d14fabe20bbe58ed1c0b876387338e1a55d6c70c | [
"MIT"
] | 1 | 2018-04-24T03:16:58.000Z | 2018-04-24T03:16:58.000Z | notebooks/3. CNN - Text.ipynb | aivizionz/DeepLearning-NLP | d14fabe20bbe58ed1c0b876387338e1a55d6c70c | [
"MIT"
] | 140 | 2017-07-27T10:20:46.000Z | 2022-01-20T15:57:50.000Z | 26.70019 | 123 | 0.536991 | [
[
[
"# Train convolutional network for sentiment analysis. \n\nBased on\n\"Convolutional Neural Networks for Sentence Classification\" by Yoon Kim\nhttp://arxiv.org/pdf/1408.5882v2.pdf\n\nFor `CNN-non-static` gets to 82.1% after 61 epochs with following settings:\nembedding_dim = 20 \nfilter_sizes = (3, 4)\nnum_filters = 3\ndropout_prob = (0.7, 0.8)\nhidden_dims = 100\n\nFor `CNN-rand` gets to 78-79% after 7-8 epochs with following settings:\nembedding_dim = 20 \nfilter_sizes = (3, 4)\nnum_filters = 150\ndropout_prob = (0.25, 0.5)\nhidden_dims = 150\n\nFor `CNN-static` gets to 75.4% after 7 epochs with following settings:\nembedding_dim = 100 \nfilter_sizes = (3, 4)\nnum_filters = 150\ndropout_prob = (0.25, 0.5)\nhidden_dims = 150\n\n* it turns out that such a small data set as \"Movie reviews with one\nsentence per review\" (Pang and Lee, 2005) requires much smaller network\nthan the one introduced in the original article:\n- embedding dimension is only 20 (instead of 300; 'CNN-static' still requires ~100)\n- 2 filter sizes (instead of 3)\n- higher dropout probabilities and\n- 3 filters per filter size is enough for 'CNN-non-static' (instead of 100)\n- embedding initialization does not require prebuilt Google Word2Vec data.\nTraining Word2Vec on the same \"Movie reviews\" data set is enough to \nachieve performance reported in the article (81.6%)\n\nAnother distinct difference is sliding MaxPooling window of length=2\ninstead of MaxPooling over whole feature map as in the article\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport data_helpers\nfrom w2v import train_word2vec\n\nfrom keras.models import Sequential, Model\nfrom keras.layers import Activation, Dense, Dropout, Embedding, Flatten, Input, Merge, Convolution1D, MaxPooling1D\n\nfrom sklearn.cross_validation import train_test_split\n\nnp.random.seed(2)",
"_____no_output_____"
],
[
"model_variation = 'CNN-rand' # CNN-rand | CNN-non-static | CNN-static\nprint('Model variation is %s' % model_variation)",
"_____no_output_____"
],
[
"# Model Hyperparameters\nsequence_length = 56\nembedding_dim = 20 \nfilter_sizes = (3, 4)\nnum_filters = 150\ndropout_prob = (0.25, 0.5)\nhidden_dims = 150",
"_____no_output_____"
],
[
"# Training parameters\nbatch_size = 32\nnum_epochs = 2",
"_____no_output_____"
],
[
"# Word2Vec parameters, see train_word2vec\nmin_word_count = 1 # Minimum word count \ncontext = 10 # Context window size ",
"_____no_output_____"
],
[
"print(\"Loading data...\")\nx, y, vocabulary, vocabulary_inv = data_helpers.load_data()",
"_____no_output_____"
],
[
"\nif model_variation=='CNN-non-static' or model_variation=='CNN-static':\n embedding_weights = train_word2vec(x, vocabulary_inv, embedding_dim, min_word_count, context)\n if model_variation=='CNN-static':\n x = embedding_weights[0][x]\nelif model_variation=='CNN-rand':\n embedding_weights = None\nelse:\n raise ValueError('Unknown model variation') ",
"_____no_output_____"
],
[
"data = np.append(x,y,axis = 1)",
"_____no_output_____"
],
[
"train, test = train_test_split(data, test_size = 0.15,random_state = 0)",
"_____no_output_____"
],
[
"X_test = test[:,:56]\nY_test = test[:,56:58]\n\n\nX_train = train[:,:56]\nY_train = train[:,56:58]\ntrain_rows = np.random.randint(0,X_train.shape[0],2500)\nX_train = X_train[train_rows]\nY_train = Y_train[train_rows]",
"_____no_output_____"
],
[
"print(\"Vocabulary Size: {:d}\".format(len(vocabulary)))\n",
"_____no_output_____"
],
[
"def initialize():\n \n global graph_in\n global convs\n \n graph_in = Input(shape=(sequence_length, embedding_dim))\n convs = []",
"_____no_output_____"
],
[
"#Buliding the first layer (Convolution Layer) of the network\ndef build_layer_1(filter_length):\n \n \n conv = Convolution1D(nb_filter=num_filters,\n filter_length=filter_length,\n border_mode='valid',\n activation='relu',\n subsample_length=1)(graph_in)\n return conv",
"_____no_output_____"
],
[
"#Adding a max pooling layer to the model(network)\ndef add_max_pooling(conv):\n \n pool = MaxPooling1D(pool_length=2)(conv)\n return pool",
"_____no_output_____"
],
[
"#Adding a flattening layer to the model(network), before adding a dense layer\ndef add_flatten(conv_or_pool):\n \n flatten = Flatten()(conv_or_pool)\n return flatten",
"_____no_output_____"
],
[
"def add_sequential(graph):\n \n #main sequential model\n model = Sequential()\n if not model_variation=='CNN-static':\n model.add(Embedding(len(vocabulary), embedding_dim, input_length=sequence_length,\n weights=embedding_weights))\n model.add(Dropout(dropout_prob[0], input_shape=(sequence_length, embedding_dim)))\n model.add(graph)\n model.add(Dense(2))\n model.add(Activation('sigmoid'))\n \n return model",
"_____no_output_____"
],
[
"#1.Convolution 2.Flatten\ndef one_layer_convolution():\n \n initialize()\n \n conv = build_layer_1(3)\n flatten = add_flatten(conv)\n \n convs.append(flatten)\n out = convs[0]\n\n graph = Model(input=graph_in, output=out)\n \n model = add_sequential(graph)\n model.compile(loss='binary_crossentropy', optimizer='rmsprop', metrics=['accuracy'])\n \n model.fit(X_train, Y_train, batch_size=batch_size,\n nb_epoch=1, validation_data=(X_test, Y_test))\n \n score = model.evaluate(X_test, Y_test, verbose=0)\n print('Test score:', score[0])\n print('Test accuracy:', score[1])",
"_____no_output_____"
],
[
"#1.Convolution 2.Max Pooling 3.Flatten\ndef two_layer_convolution():\n \n initialize()\n \n conv = build_layer_1(3)\n pool = add_max_pooling(conv)\n flatten = add_flatten(pool)\n \n convs.append(flatten)\n out = convs[0]\n\n graph = Model(input=graph_in, output=out)\n \n model = add_sequential(graph)\n model.compile(loss='binary_crossentropy', optimizer='rmsprop', metrics=['accuracy'])\n \n model.fit(X_train, Y_train, batch_size=batch_size,\n nb_epoch=num_epochs, validation_data=(X_test, Y_test))\n \n score = model.evaluate(X_test, Y_test, verbose=0)\n print('Test score:', score[0])\n print('Test accuracy:', score[1])",
"_____no_output_____"
],
[
"#1.Convolution 2.Max Pooling 3.Flatten 4.Convolution 5.Flatten\ndef three_layer_convolution():\n \n initialize()\n \n conv = build_layer_1(3)\n pool = add_max_pooling(conv)\n flatten = add_flatten(pool)\n \n convs.append(flatten)\n \n conv = build_layer_1(4)\n flatten = add_flatten(conv)\n \n convs.append(flatten)\n \n if len(filter_sizes)>1:\n out = Merge(mode='concat')(convs)\n else:\n out = convs[0]\n\n graph = Model(input=graph_in, output=out)\n \n model = add_sequential(graph)\n model.compile(loss='binary_crossentropy', optimizer='rmsprop', metrics=['accuracy'])\n \n model.fit(X_train, Y_train, batch_size=batch_size,\n nb_epoch=1, validation_data=(X_test, Y_test))\n \n score = model.evaluate(X_test, Y_test, verbose=0)\n print('Test score:', score[0])\n print('Test accuracy:', score[1])",
"_____no_output_____"
],
[
"#1.Convolution 2.Max Pooling 3.Flatten 4.Convolution 5.Max Pooling 6.Flatten\ndef four_layer_convolution():\n \n initialize()\n \n conv = build_layer_1(3)\n pool = add_max_pooling(conv)\n flatten = add_flatten(pool)\n \n convs.append(flatten)\n \n conv = build_layer_1(4)\n pool = add_max_pooling(conv)\n flatten = add_flatten(pool)\n \n convs.append(flatten)\n \n if len(filter_sizes)>1:\n out = Merge(mode='concat')(convs)\n else:\n out = convs[0]\n\n graph = Model(input=graph_in, output=out)\n \n model = add_sequential(graph)\n model.compile(loss='binary_crossentropy', optimizer='rmsprop', metrics=['accuracy'])\n \n model.fit(X_train, Y_train, batch_size=batch_size,\n nb_epoch=num_epochs, validation_data=(X_test, Y_test))\n \n score = model.evaluate(X_test, Y_test, verbose=0)\n print('Test score:', score[0])\n print('Test accuracy:', score[1])",
"_____no_output_____"
],
[
"%%time\n#1.Convolution 2.Flatten\none_layer_convolution()",
"_____no_output_____"
],
[
"%%time\n#1.Convolution 2.Max Pooling 3.Flatten\ntwo_layer_convolution()",
"_____no_output_____"
],
[
"%%time\n#1.Convolution 2.Max Pooling 3.Flatten 4.Convolution 5.Flatten\nthree_layer_convolution()",
"_____no_output_____"
],
[
"%%time\n#1.Convolution 2.Max Pooling 3.Flatten 4.Convolution 5.Max Pooling 6.Flatten\nfour_layer_convolution()",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb43e50e45368a3f99aafc86f7a0f96371099e03 | 254,907 | ipynb | Jupyter Notebook | MiscNotebookFiles/Testing.ipynb | ZYVE255/ebm-optimizer | 9b1cf6014f987ef4b8d65d4a5659c704b6ea15c4 | [
"MIT"
] | null | null | null | MiscNotebookFiles/Testing.ipynb | ZYVE255/ebm-optimizer | 9b1cf6014f987ef4b8d65d4a5659c704b6ea15c4 | [
"MIT"
] | 1 | 2019-05-25T12:17:19.000Z | 2019-05-25T12:17:19.000Z | MiscNotebookFiles/Testing.ipynb | ZYVE255/ebm-optimizer | 9b1cf6014f987ef4b8d65d4a5659c704b6ea15c4 | [
"MIT"
] | null | null | null | 88.355979 | 18,072 | 0.745091 | [
[
[
"#==========Imports==========\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport astropy.constants as const\nimport time\nfrom scipy import interpolate\nimport Zach_OPTIMIZER.EBMFunctions as opt\n\nimport Bell_EBM as ebm",
"_____no_output_____"
],
[
"#==========Set Up System==========\nplanet = ebm.Planet(rad=1.500*const.R_jup.value, mass=1.170*const.M_jup.value,\n Porb=1.09142030, a=0.02340*2*const.au.value, inc=83.37, vWind=5e3, nlat = 8, e=0.2)\nstar = ebm.Star(teff=6300., rad=1.59, mass=1.20)\nsystem = ebm.System(star, planet)",
"_____no_output_____"
],
[
"def CreateBaseline(star, planet, temporal=5000, spacial=32,orbit=2):\n _star = star\n _planet = planet\n _system = ebm.System(_star, _planet)\n \n Teq = _system.get_teq()\n T0 = np.ones_like(_system.planet.map.values)*Teq\n t0 = 0.\n t1 = t0+_system.planet.Porb*orbit\n dt = _system.planet.Porb/temporal\n baselineTimes, baselineMaps = _system.run_model(T0, t0, t1, dt, verbose=False, intermediates=False)\n if (planet.orbit.e != 0.):\n T0 = baselineMaps[-1]\n t0 = baselineTimes[-1]\n t1 = t0+system.planet.Porb\n dt = (system.planet.Porb)/1000.\n baselineTimes, baselineMaps = system.run_model(T0, t0, t1, dt, verbose=False, intermediates=True)\n \n baselineLightcurve = system.lightcurve(baselineTimes, baselineMaps, bolo=False, wav=4.5e-6)\n \n# phaseBaseline = system.get_phase(baselineTimes).flatten()\n# order = np.argsort(phaseBaseline)\n# baselineLightcurve = baselineLightcurve[order]\n# phaseBaseline = phaseBaseline[order]\n else:\n baselineLightcurve = system.lightcurve(bolo=False, wav=4.5e-6)\n \n return baselineTimes, baselineMaps, baselineLightcurve",
"_____no_output_____"
],
[
"blt, blm, blc = opt.CreateBaseline(star,planet)",
"_____no_output_____"
],
[
"plt.plot(blc)",
"_____no_output_____"
],
[
"def RunTests(star, planet, points, base):\n data = np.zeros(shape=(points.shape[0],4))\n _star = star\n _planet = planet\n _system = ebm.System(_star,_planet)\n for i in range(0, points.shape[0]):\n \n _star = star\n _planet = planet \n _planet.map = ebm.Map.Map(nlat=points[i,1])\n _system = ebm.System(_star, _planet)\n \n data[i,0] = points[i,0]\n data[i,1] = points[i,1]\n \n tInt = time.time()\n \n Teq = _system.get_teq()\n T0 = np.ones_like(_system.planet.map.values)*Teq\n t0 = 0.\n t1 = t0+_system.planet.Porb\n dt = _system.planet.Porb/points[i,0]\n testTimes, testMaps = system.run_model(T0, t0, t1, dt, verbose=False)\n if (_planet.orbit.e != 0):\n T0 = testMaps[-1]\n t0 = testTimes[-1]\n t1 = t0+_system.planet.Porb\n dt = system.planet.Porb/points[i,0]\n testTimes, testMaps = system.run_model(T0, t0, t1, dt, verbose=False, intermediates=True)\n testLightcurve = system.lightcurve(testTimes, testMaps, bolo=False, wav=4.5e-6)\n \n phaseTest = _system.get_phase(testTimes).flatten()\n order = np.argsort(phaseTest)\n testLightcurve = testLightcurve[order]\n phaseTest = phaseTest[order]\n testLightcurve = np.interp(base, phaseTest, testLightcurve)\n else:\n testLightcurve = system.lightcurve(bolo=False, wav=4.5e-6)\n\n tFin = time.time()\n \n data[i,3] = (1e6)*(np.amax(np.absolute(base - testLightcurve)))\n data[i,2] = (tFin - tInt)*(1e3)\n\n return testLightcurve, data\n",
"_____no_output_____"
],
[
"p = np.zeros(shape=((10),2))",
"_____no_output_____"
],
[
"p[:,0]=500\np[:,1]=8\np[9,0] = 500\np[9,1] = 8",
"_____no_output_____"
],
[
"p",
"_____no_output_____"
],
[
"lc, data = opt.RunTests(star,planet,p,blc,blt)",
"_____no_output_____"
],
[
"plt.plot(lc)",
"_____no_output_____"
],
[
"plt.plot(lc, c='g')\nplt.plot(blc, c='b')",
"_____no_output_____"
],
[
"plt.plot((blc-lc)*(1e6))",
"_____no_output_____"
],
[
"def Optimize(star, planet, error, verbose=False):\n _planet = planet\n _star = star\n aError = error\n\n #==========High Res Baseline Creation========== \n if (verbose == True): \n print(\"Starting baseline generation...\")\n \n tInt = time.time()\n blt, blm, blc = CreateBaseline(_star, _planet)\n tFin = time.time()\n \n if (verbose == True): \n print(\"Baseline generation complete; Time to Compute: \" + str(round(tFin-tInt,2)) + \"s\")\n\n #===========Initial data creationg================\n space_points = 5\n temp_points = 5\n data = np.zeros(shape=((space_points*temp_points),4))\n for i in range (0, temp_points):\n for j in range (0, space_points):\n data[(i*space_points)+j,0]= ((i+1)*250)+0\n data[(i*space_points)+j,1] = ((j+1)*4)+0\n if (verbose == True): \n print(\"First pass data points assigned\")\n\n #==================First pass testing Area======================\n if (verbose == True): \n print(\"Starting first pass...\")\n \n tInt = time.time()\n lc, data = RunTests(_star, _planet, data, blc)\n tFin = time.time()\n \n if (verbose == True): \n print(\"First pass finished : Time to compute: \" + str(round(tFin-tInt,2)) + \"s\")\n\n #=================First pass best point===================\n #print(data) #For debugging purposes \n if (verbose == True):\n print(\"Processing first pass data...\")\n iBest = None\n for i in range(0,space_points*temp_points):\n if (data[i,3]<=(aError*1.05)):\n if (iBest == None):\n iBest = i\n if(data[i,2] < data[iBest,2]):\n iBest = i\n \n #===========Second pass data creation================\n space_points = 5\n temp_points = 5\n dataDouble = np.zeros(shape=((space_points*temp_points),2))\n for i in range (0, temp_points):\n for j in range (0, space_points):\n dataDouble[(i*space_points)+j,0] = ((i)*50)+(data[iBest,0]-100)\n if (dataDouble[(i*space_points)+j,0]<100):\n dataDouble[(i*space_points)+j,0] = 100\n dataDouble[(i*space_points)+j,1] = ((j)*2)+(data[iBest,1]-4)\n if (dataDouble[(i*space_points)+j,1]<2):\n dataDouble[(i*space_points)+j,1] = 2\n if (verbose == True): \n print(\"Second pass data points assigned\")\n \n #==================Second pass testing Area======================\n if (verbose == True): \n print(\"Starting second pass...\")\n \n tInt = time.time()\n lc, dataDouble = RunTests(_star, _planet, dataDouble, blc)\n tFin = time.time()\n if (verbose == True): \n print(\"Second pass finished : Time to compute: \" + str(round(tFin-tInt,2)) + \"s\")\n \n #=================Finding best second pass point===================\n #print(data) #For debugging purposes \n if (verbose == True):\n print(\"Processing second pass data...\")\n iBest = None\n for i in range(0,space_points*temp_points):\n if (dataDouble[i,3]<=aError):\n if (iBest == None):\n iBest = i\n if(dataDouble[i,2] < dataDouble[iBest,2]):\n iBest = i\n\n if (iBest == None):\n print(\"No points match requested error\")\n else:\n print(\"Temporal: \" + str(dataDouble[iBest,0]) + \" Spacial: \" + str(dataDouble[iBest,1]))\n print(\"Time for compute: \" + str(round(dataDouble[iBest, 2],2)) +\"ms : Error: \" + str(round(dataDouble[iBest, 3],2)) + \"ppm\")\n print(\"Expected compute time @ 1,000,000 cycles: \" + str((round((dataDouble[iBest, 2]*1e3/60)/60,2))) + \" Hrs\")\n \n return dataDouble[iBest,0], dataDouble[iBest,1]\n# #print(data) #For debugging\n# #print(dataDouble) #For debugging\n# #=========Create Maps==================\n# if (verbose == True):\n# planet.map = ebm.Map.Map(nlat=dataDouble[iBest,1])\n# system = ebm.System(star, planet)\n\n# TotalTimeToCompute = 0.\n\n# Teq = system.get_teq()\n# T0 = np.ones_like(system.planet.map.values)*Teq\n# t0 = 0.\n# t1 = t0+system.planet.Porb*1\n# dt = system.planet.Porb/dataDouble[iBest,0]\n# times, maps, ttc = system.run_model_tester(T0, t0, t1, dt, verbose=False)\n# TotalTimeToCompute += ttc\n\n# if (planet.orbit.e != 0):\n# T0 = maps[-1]\n# t0 = times[-1]\n# t1 = t0+system.planet.Porb\n# dt = system.planet.Porb/dataDouble[iBest,0]\n# times, maps, ttc = system.run_model_tester(T0, t0, t1, dt, verbose=False, intermediates=True)\n# TotalTimeToCompute += ttc\n\n# testLightcurve = system.lightcurve(times, maps, bolo=False, wav=4.5e-6)\n\n# phaseTest = system.get_phase(times).flatten()\n# order = np.argsort(phaseTest)\n# testLightcurve = testLightcurve[order]\n# phaseTest = phaseTest[order]\n# testLightcurve = np.interp(phaseBaseline, phaseTest, testLightcurve)\n \n\n# plt.plot((baselineLightcurve)*1e6, lw=2, c='g')\n# plt.plot((testLightcurve)*1e6, lw=1, c='r')\n# plt.title(\"Lightcurves of baseline (green) compared to recommended values (red)\")\n# plt.show()\n\n",
"_____no_output_____"
],
[
"temp, space = opt.Optimize(star, planet, 100, verbose=True)",
"Starting baseline generation...\nBaseline generation complete; Time to Compute: 2.76s\nFirst pass data points assigned\nStarting first pass...\nFirst pass finished : Time to compute: 25.06s\nProcessing first pass data...\nSecond pass data points assigned\nStarting second pass...\nSecond pass finished : Time to compute: 3.04s\nProcessing second pass data...\nTemporal: 150.0 Spacial: 6.0\nTime for compute: 66.81ms : Error: 44.1ppm\nExpected compute time @ 1,000,000 cycles: 18.56 Hrs\n"
],
[
"phaseBaseline = system.get_phase(blt).flatten()\norder = np.argsort(phaseBaseline)\nbaselineLightcurve = blc[order]\nphaseBaseline = phaseBaseline[order]\n\n\n_star = star\n_planet = planet \n_planet.map = ebm.Map.Map(nlat=8)\n_system = ebm.System(_star, _planet)\n\ntInt = time.time()\n\nTeq = _system.get_teq()\nT0 = np.ones_like(_system.planet.map.values)*Teq\nt0 = 0.\nt1 = t0+_system.planet.Porb\ndt = _system.planet.Porb/500\ntestTimes, testMaps = system.run_model(T0, t0, t1, dt, verbose=False)\nif (_planet.orbit.e != 0):\n T0 = testMaps[-1]\n t0 = testTimes[-1]\n t1 = t0+_system.planet.Porb\n dt = system.planet.Porb/500\n testTimes, testMaps = system.run_model(T0, t0, t1, dt, verbose=False, intermediates=True)\n testLightcurve = system.lightcurve(testTimes, testMaps, bolo=False, wav=4.5e-6)\n \n testbeta = testLightcurve\n \n phaseTest = _system.get_phase(testTimes).flatten()\n order = np.argsort(phaseTest)\n testLightcurve = testLightcurve[order]\n testalpha = testLightcurve\n phaseTest = phaseTest[order]\n testLightcurve = np.interp(phaseBaseline, phaseTest, testLightcurve)\nelse:\n testLightcurve = system.lightcurve(bolo=False, wav=4.5e-6)\n\ntFin = time.time()\n\n",
"_____no_output_____"
],
[
"plt.plot(blc)",
"_____no_output_____"
],
[
"plt.plot(testbeta)",
"_____no_output_____"
],
[
"plt.plot(testalpha)",
"_____no_output_____"
],
[
"plt.plot(testLightcurve)",
"_____no_output_____"
],
[
"phaseTest",
"_____no_output_____"
],
[
"testTimes",
"_____no_output_____"
],
[
"blt",
"_____no_output_____"
],
[
"blt.shape",
"_____no_output_____"
],
[
"testTimes.shape",
"_____no_output_____"
],
[
"def RunTests(star, planet, points, base, basetimes, basemap):\n \"\"\"\n Runs several test of a system and returns time \n to compute and error as comapared to baseline for each test.\n \n Args:\n star (ebm.Star): The star to runs the tests on\n planet (ebm.Planet): The planet to run the tests on\n points (2darray (n by 2)): The array of points to be tested by the model, \n each point must contain [temporal, spacial], n points are provided\n base (ndarray): Baseline lightcurve as generated by the CreateBaseline function\n \n Return:\n ndarray: Latest tested lightcurve, mainly used for debugging purposes\n ndarray: (n by 4), n points of format [temporal, spacial, time_to_compute, error_in_ppm]\n \"\"\"\n \n data = np.zeros(shape=(points.shape[0],4))\n _star = star\n _planet = planet\n _system = ebm.System(_star,_planet)\n \n if (_planet.orbit.e != 0):\n phaseBaseline = _system.get_phase(basetimes).flatten()\n order = np.argsort(phaseBaseline)\n baselineLightcurve = base[order]\n phaseBaseline = phaseBaseline[order]\n \n for i in range(0, points.shape[0]):\n \n _star = star\n _planet = planet \n _planet.map = ebm.Map.Map(nlat=points[i,1])\n _system = ebm.System(_star, _planet)\n \n data[i,0] = points[i,0]\n data[i,1] = points[i,1]\n \n tInt = time.time()\n \n Teq = _system.get_teq()\n T0 = np.ones_like(_system.planet.map.values)*Teq\n t0 = 0.\n t1 = t0+_system.planet.Porb\n dt = _system.planet.Porb/points[i,0]\n testTimes, testMaps = _system.run_model(T0, t0, t1, dt, verbose=False)\n# if (_planet.orbit.e != 0):\n# T0 = testMaps[-1]\n# t0 = testTimes[-1]\n# t1 = t0+_system.planet.Porb\n# dt = _system.planet.Porb/points[i,0]\n# testTimes, testMaps = _system.run_model(T0, t0, t1, dt, verbose=False, intermediates=True)\n# testLightcurve = _system.lightcurve(testTimes, testMaps, bolo=False, wav=4.5e-6)\n \n# phaseTest = _system.get_phase(testTimes).flatten()\n# order = np.argsort(phaseTest)\n# testLightcurve = testLightcurve[order]\n# phaseTest = phaseTest[order]\n# testLightcurve = np.interp(phaseBaseline, phaseTest, testLightcurve)\n# else:\n# testLightcurve = _system.lightcurve(bolo=False, wav=4.5e-6)\n if (_planet.orbit.e != 0):\n T0 = testMaps[-1]\n t0 = testTimes[-1]\n t1 = t0+_system.planet.Porb\n dt = system.planet.Porb/points[i,0]\n testTimes, testMaps = _system.run_model(T0, t0, t1, dt, verbose=False, intermediates=True)\n testLightcurve = _system.lightcurve(testTimes, testMaps, bolo=False, wav=4.5e-6)\n\n #testbeta = testLightcurve\n\n phaseTest = _system.get_phase(testTimes).flatten()\n order = np.argsort(phaseTest)\n testLightcurve = testLightcurve[order]\n #testalpha = testLightcurve\n phaseTest = phaseTest[order]\n testLightcurve = np.interp(phaseBaseline, phaseTest, testLightcurve)\n else:\n testLightcurve = system.lightcurve(bolo=False, wav=4.5e-6)\n\n tFin = time.time()\n \n data[i,3] = (1e6)*(np.amax(np.absolute(base - testLightcurve)))\n data[i,2] = (tFin - tInt)*(1e3)\n\n return testLightcurve, data",
"_____no_output_____"
],
[
"light, ded = RunTests(star,planet,p,blc,blt,blm)",
"_____no_output_____"
],
[
"plt.plot(light)",
"_____no_output_____"
],
[
"phaseBaseline = system.get_phase(blt).flatten()\norder = np.argsort(phaseBaseline)\nbaselineLightcurve = blc[order]\nphaseBaseline = phaseBaseline[order]\n",
"_____no_output_____"
],
[
"#==========Imports==========\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport astropy.constants as const\nimport time\nfrom scipy import interpolate\nimport Zach_OPTIMIZER.EBMFunctions as opt\n\nimport Bell_EBM as ebm",
"_____no_output_____"
],
[
"\ndef RunTests(star, planet, points, base, basetimes):\n \"\"\"\n Runs several test of a system and returns time \n to compute and error as comapared to baseline for each test.\n \n Args:\n star (ebm.Star): The star to runs the tests on\n planet (ebm.Planet): The planet to run the tests on\n points (2darray (n by 2)): The array of points to be tested by the model, \n each point must contain [temporal, spacial], n points are provided\n base (ndarray): Baseline lightcurve as generated by the CreateBaseline function\n basetime (ndarray): Baseline times as generated by CreateBaseline function\n \n Return:\n ndarray: Latest tested lightcurve, mainly used for debugging purposes\n ndarray: (n by 4), n points of format [temporal, spacial, time_to_compute, error_in_ppm]\n \"\"\"\n \n data = np.zeros(shape=(points.shape[0],4))\n _star = star\n _planet = planet\n _system = ebm.System(_star,_planet)\n \n if (_planet.orbit.e != 0):\n phaseBaseline = _system.get_phase(basetimes).flatten()\n order = np.argsort(phaseBaseline)\n baselineLightcurve = base[order]\n phaseBaseline = phaseBaseline[order]\n \n for i in range(0, points.shape[0]):\n \n _star = star\n _planet = planet \n _planet.map = ebm.Map.Map(nlat=points[i,1])\n _system = ebm.System(_star, _planet)\n \n data[i,0] = points[i,0]\n data[i,1] = points[i,1]\n \n tInt = time.time()\n \n Teq = _system.get_teq()\n T0 = np.ones_like(_system.planet.map.values)*Teq\n t0 = 0.\n t1 = t0+_system.planet.Porb\n dt = _system.planet.Porb/points[i,0]\n testTimes, testMaps = _system.run_model(T0, t0, t1, dt, verbose=False)\n if (_planet.orbit.e != 0):\n T0 = testMaps[-1]\n t0 = testTimes[-1]\n t1 = t0+_system.planet.Porb\n dt = _system.planet.Porb/points[i,0]\n testTimes, testMaps = _system.run_model(T0, t0, t1, dt, verbose=False, intermediates=True)\n testLightcurve = _system.lightcurve(testTimes, testMaps, bolo=False, wav=4.5e-6)\n \n phaseTest = _system.get_phase(testTimes).flatten()\n order = np.argsort(phaseTest)\n testLightcurve = testLightcurve[order]\n phaseTest = phaseTest[order]\n testLightcurve = np.interp(phaseBaseline, phaseTest, testLightcurve)\n else:\n testLightcurve = _system.lightcurve(bolo=False, wav=4.5e-6)\n\n tFin = time.time()\n \n data[i,3] = (1e6)*(np.amax(np.absolute(base - testLightcurve)))\n data[i,2] = (tFin - tInt)*(1e3)\n\n return testLightcurve, data\n\n\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb43f4adac8d49c69bd4f9c38fda720509e1caa6 | 15,224 | ipynb | Jupyter Notebook | nltk-inverted-indexing/notebook2.ipynb | kapilkalra04/nlp-research | dcbff74796cc01bb4a49c031eae9643063f4e68d | [
"MIT"
] | null | null | null | nltk-inverted-indexing/notebook2.ipynb | kapilkalra04/nlp-research | dcbff74796cc01bb4a49c031eae9643063f4e68d | [
"MIT"
] | null | null | null | nltk-inverted-indexing/notebook2.ipynb | kapilkalra04/nlp-research | dcbff74796cc01bb4a49c031eae9643063f4e68d | [
"MIT"
] | null | null | null | 29.220729 | 1,714 | 0.490673 | [
[
[
"### STEPS:\n\n#### Pipeline - 1\n1. Tokenization\n1. Remove StopWords and Punctuation\n1. Stemming\n\n#### Pipeline - 2\n1. Tokenization\n1. POS Tagger\n1. Lemmatization\n\n***Remember to Deal With Everything in Lower Cases***",
"_____no_output_____"
]
],
[
[
"import nltk\nnltk.download('punkt') # For Tokenizing\nnltk.download('stopwords') # For Stopwords\nnltk.download('wordnet') # For Lemmatization\nnltk.download('averaged_perceptron_tagger') # For POS Tagging\nfrom nltk.stem import WordNetLemmatizer\nfrom nltk.stem.snowball import EnglishStemmer\nfrom nltk import pos_tag # POS Tagger\nfrom nltk.corpus import wordnet as wn\nimport string",
"[nltk_data] Downloading package punkt to\n[nltk_data] C:\\Users\\kapilkalra04\\AppData\\Roaming\\nltk_data...\n[nltk_data] Package punkt is already up-to-date!\n[nltk_data] Downloading package stopwords to\n[nltk_data] C:\\Users\\kapilkalra04\\AppData\\Roaming\\nltk_data...\n[nltk_data] Package stopwords is already up-to-date!\n[nltk_data] Downloading package wordnet to\n[nltk_data] C:\\Users\\kapilkalra04\\AppData\\Roaming\\nltk_data...\n[nltk_data] Package wordnet is already up-to-date!\n[nltk_data] Downloading package averaged_perceptron_tagger to\n[nltk_data] C:\\Users\\kapilkalra04\\AppData\\Roaming\\nltk_data...\n[nltk_data] Package averaged_perceptron_tagger is already up-to-\n[nltk_data] date!\n"
],
[
"input = [\n 'Industrial Disease',\n'Private Investigations',\n'So Far Away',\n'Twisting by the Pool',\n'Skateaway',\n'Walk of Life',\n'Romeo and Juliet',\n'Tunnel of Love',\n'Money for Nothing',\n'Sultans of Swing',\n'Stairway To Heaven',\n'Kashmir',\n'Achilles Last Stand',\n'Whole Lotta Love',\n'Immigrant Song',\n'Black Dog',\n'When The Levee Breaks',\n'Since I\\'ve Been Lovin\\' You',\n'Since I\\'ve Been Loving You',\n'Over the Hills and Far Away',\n'Dazed and Confused' \n]",
"_____no_output_____"
],
[
"len(input)",
"_____no_output_____"
]
],
[
[
"#### TOKENIZER",
"_____no_output_____"
]
],
[
[
"def tokenize(sentence):\n return nltk.tokenize.word_tokenize(sentence)",
"_____no_output_____"
],
[
"print input[17]\nprint tokenize(input[17]) # there a \"(\\')\" being tokenized we will remove it later",
"Since I've Been Lovin' You\n['Since', 'I', \"'ve\", 'Been', 'Lovin', \"'\", 'You']\n"
]
],
[
[
"#### POS TAGGER CONVERTER",
"_____no_output_____"
]
],
[
[
"def penn_to_wn(tag):\n \"\"\"\n Convert between the PennTreebank (pos_tag) tags to simple Wordnet tags\n \"\"\"\n if tag.startswith('J'):\n return wn.ADJ\n elif tag.startswith('N'):\n return wn.NOUN\n elif tag.startswith('R'):\n return wn.ADV\n elif tag.startswith('V'):\n return wn.VERB\n return None",
"_____no_output_____"
]
],
[
[
"#### LEMMATIZER",
"_____no_output_____"
]
],
[
[
"wnl = WordNetLemmatizer()",
"_____no_output_____"
],
[
"def lemmatize(word, pos=wn.NOUN):\n return wnl.lemmatize(word,pos=pos)",
"_____no_output_____"
],
[
"print input[3]\nprint tokenize(input[3])\nfor w in tokenize(input[3]):\n w.lower()\n print lemmatize(w)",
"Twisting by the Pool\n['Twisting', 'by', 'the', 'Pool']\nTwisting\nby\nthe\nPool\n"
]
],
[
[
"#### STEMMER",
"_____no_output_____"
]
],
[
[
"def stem(word):\n stemmer = EnglishStemmer()\n return stemmer.stem(word)",
"_____no_output_____"
],
[
"print input[3]\nprint tokenize(input[3])\nfor w in tokenize(input[3]):\n w.lower()\n print stem(w)",
"Twisting by the Pool\n['Twisting', 'by', 'the', 'Pool']\ntwist\nby\nthe\npool\n"
]
],
[
[
"#### STOPWORDS",
"_____no_output_____"
]
],
[
[
"stopwords = nltk.corpus.stopwords.words('English')\nprint stopwords",
"[u'i', u'me', u'my', u'myself', u'we', u'our', u'ours', u'ourselves', u'you', u\"you're\", u\"you've\", u\"you'll\", u\"you'd\", u'your', u'yours', u'yourself', u'yourselves', u'he', u'him', u'his', u'himself', u'she', u\"she's\", u'her', u'hers', u'herself', u'it', u\"it's\", u'its', u'itself', u'they', u'them', u'their', u'theirs', u'themselves', u'what', u'which', u'who', u'whom', u'this', u'that', u\"that'll\", u'these', u'those', u'am', u'is', u'are', u'was', u'were', u'be', u'been', u'being', u'have', u'has', u'had', u'having', u'do', u'does', u'did', u'doing', u'a', u'an', u'the', u'and', u'but', u'if', u'or', u'because', u'as', u'until', u'while', u'of', u'at', u'by', u'for', u'with', u'about', u'against', u'between', u'into', u'through', u'during', u'before', u'after', u'above', u'below', u'to', u'from', u'up', u'down', u'in', u'out', u'on', u'off', u'over', u'under', u'again', u'further', u'then', u'once', u'here', u'there', u'when', u'where', u'why', u'how', u'all', u'any', u'both', u'each', u'few', u'more', u'most', u'other', u'some', u'such', u'no', u'nor', u'not', u'only', u'own', u'same', u'so', u'than', u'too', u'very', u's', u't', u'can', u'will', u'just', u'don', u\"don't\", u'should', u\"should've\", u'now', u'd', u'll', u'm', u'o', u're', u've', u'y', u'ain', u'aren', u\"aren't\", u'couldn', u\"couldn't\", u'didn', u\"didn't\", u'doesn', u\"doesn't\", u'hadn', u\"hadn't\", u'hasn', u\"hasn't\", u'haven', u\"haven't\", u'isn', u\"isn't\", u'ma', u'mightn', u\"mightn't\", u'mustn', u\"mustn't\", u'needn', u\"needn't\", u'shan', u\"shan't\", u'shouldn', u\"shouldn't\", u'wasn', u\"wasn't\", u'weren', u\"weren't\", u'won', u\"won't\", u'wouldn', u\"wouldn't\"]\n"
]
],
[
[
"#### INDEXED DATABASES",
"_____no_output_____"
],
[
"#### Pipeline 1",
"_____no_output_____"
]
],
[
[
"db = {}",
"_____no_output_____"
],
[
"for sentence in input:\n words = tokenize(sentence)\n for word in words:\n word = word.lower()\n if word not in stopwords and word not in string.punctuation:\n root = stem(word)\n if db.has_key(root):\n db[root].append(sentence)\n else :\n db[root] = [sentence]",
"_____no_output_____"
],
[
"db",
"_____no_output_____"
]
],
[
[
"#### Pipeline 2",
"_____no_output_____"
]
],
[
[
"db2 = {}",
"_____no_output_____"
],
[
"for sentence in input:\n words = tokenize(sentence)\n tagged_sentence = pos_tag(words)\n for word, tag in tagged_sentence:\n word = word.lower()\n tag = penn_to_wn(tag)\n if tag in (wn.NOUN,wn.ADJ,wn.VERB,wn.ADV):\n root = lemmatize(word,tag)\n if db2.has_key(root):\n db2[root].append(sentence)\n else :\n db2[root] = [sentence]",
"_____no_output_____"
],
[
"db2",
"_____no_output_____"
]
],
[
[
"#### WE CAN OBSERVE THAT BY USING LEMMATIZATION WE PRESERVED THE MORPHOLOGY",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb43fcd50c1c6444fb35af169215edad3575b27a | 10,288 | ipynb | Jupyter Notebook | 11_estructura_de_openapi.ipynb | rogernavarro/py411 | 19984eb523162071440b1da39351be3b1e469ff1 | [
"MIT"
] | 2 | 2020-09-16T20:16:29.000Z | 2020-12-02T21:43:03.000Z | 11_estructura_de_openapi.ipynb | PythonistaMX/py411 | c77f9d74d7045e3e85eb7ef5d16f569d8dd466e0 | [
"MIT"
] | null | null | null | 11_estructura_de_openapi.ipynb | PythonistaMX/py411 | c77f9d74d7045e3e85eb7ef5d16f569d8dd466e0 | [
"MIT"
] | null | null | null | 21.613445 | 405 | 0.48853 | [
[
[
"[](https://www.pythonista.io)",
"_____no_output_____"
],
[
"# Esquema de *OpenAPI*.\n\nhttps://swagger.io/docs/specification/basic-structure/",
"_____no_output_____"
],
[
"## Estructura.",
"_____no_output_____"
],
[
"* Versión de *OpenAPI*.\n* Información (```info```).\n* Etiquetas (```tags```).\n* Servidores (```servers```).\n* Componentes (```components```).\n * Esquemas (```schemas```).\n * Cuerpos de petición (```requestBodies```)\n* Rutas (```paths```).",
"_____no_output_____"
],
[
"## Versión de *Open API*.",
"_____no_output_____"
],
[
"```yaml\nopenapi: <versión>\n```\n\nhttps://github.com/OAI/OpenAPI-Specification/blob/main/versions/3.0.3.md#versions",
"_____no_output_____"
],
[
"## información.",
"_____no_output_____"
],
[
"``` yaml\ninfo:\n description: <Descripción de la API>\n version: <Version de la API>\n title: <Título de la documentación de la API>\n termsOfService: <URL de los términos de servicio>\n contact:\n name: <Nombre del contacto>\n email: <Correo electrónico del contacto>\n url: <URL de referencia>\n license:\n nombre: <Nombre de la licencia>\n url: <URL de la licencia>\n externalDocs:\n description: <Descripción de documentos externos>\n url: <URL de la licencia>\n```\n\nhttps://swagger.io/docs/specification/api-general-info/",
"_____no_output_____"
],
[
"## Etiquetas:",
"_____no_output_____"
],
[
"```yaml\ntags:\n - name: <nombre de la etiqueta 1>\n description: <descripción de la etiqueta 1>\n - name: <nombre de la etiqueta 2>\n description: <descripción de la etiqueta 2>\n```\n\nhttps://swagger.io/docs/specification/grouping-operations-with-tags/",
"_____no_output_____"
],
[
"## Servidores:",
"_____no_output_____"
],
[
"``` yaml\nservers:\n - url: <URL del servidor 1>\n description: <descripción del servidor 1>\n - url: <URL del servidor 2\n description: <descripción del servidor 2>\n```",
"_____no_output_____"
],
[
"## Componentes.",
"_____no_output_____"
],
[
"https://swagger.io/docs/specification/components/",
"_____no_output_____"
],
[
"* Esquemas (*schemas*)\n* Cuerpos de peticiones (*requestBodies*)\n",
"_____no_output_____"
],
[
"``` yaml\ncomponents:\n requestBodies:\n - <esquema de peticion 1>\n - <esquema de peticion 2>\n schemas:\n - <esquema 1>\n - <esquema 2>\n parameters:\n - <parámetro 1>\n - <parámetro 2>\n responses:\n - <respuesta 1>\n - <respuesta 1>\n headers:\n - <encabezado 1>\n - <encabezado 2>\n examples:\n - <ejemplo 1>\n - <ejemplo 2>\n callbacks:\n - <URL 1>\n - <URL 2>\n```",
"_____no_output_____"
],
[
"## Rutas.",
"_____no_output_____"
],
[
"https://swagger.io/docs/specification/paths-and-operations/",
"_____no_output_____"
],
[
"```\n\"/<segmento 1>{<parámetro 1>}<segmento 2>{<parámetro 2>}\"\n```",
"_____no_output_____"
],
[
"**Ejemplos:**",
"_____no_output_____"
],
[
"* ```/api/{clave}```\n* ```/api/{clave}-{id}/mensajes```\n* ```/auth/logout```",
"_____no_output_____"
],
[
"``` yaml\npaths:\n <ruta 1>:\n <método 1>\n <metodo 2>\n <parameters>:\n <parámetro 1>\n <parámetro 2> \n``` ",
"_____no_output_____"
],
[
"### Parámetros.\n\nLos parámetros son datos obtenidos a partir de la ruta o de la consulta enviado en la petición.",
"_____no_output_____"
],
[
"``` yaml\nparameters:\n - name: <Nombre del parámetro>\n in: <Fuente>\n description: <Descripción del parámetro>\n required: <booleano>\n example: <Ejemplo del parámetro>\n schema:\n <esquema>\n```",
"_____no_output_____"
],
[
"### Métodos.",
"_____no_output_____"
],
[
"``` yaml\n<método>:\n tags:\n - <etiqueta 1>\n - <etiqueta 2>\n summary: <Resumen de la funcionalidad>\n description: <Descripción de la funcionalidad>\n parameters:\n - <parámetro 1>\n - <parámetro 2>\n responses:\n <código de estado 1>\n <código de estado 2>\n requestBody:\n <esquema de petición>\n```",
"_____no_output_____"
],
[
"### Códigos de estado.\n\n``` yaml\n<número de código de estado 1>:\n description: <Descripción de la funcionalidad>\n content:\n <tipo de aplicación>:\n <esquema del contenido de la respuesta>\n```",
"_____no_output_____"
],
[
"### Contenidos de respuesta.\n\nhttps://swagger.io/docs/specification/describing-responses/\n\nhttps://swagger.io/docs/specification/data-models/representing-xml/",
"_____no_output_____"
],
[
"## Esquemas.\n\nhttps://swagger.io/docs/specification/data-models/",
"_____no_output_____"
],
[
"## Tipos de datos.\n\nhttps://swagger.io/docs/specification/data-models/data-types/",
"_____no_output_____"
],
[
"### Tipo ```string```.\n\nhttps://swagger.io/docs/specification/data-models/data-types/#string",
"_____no_output_____"
],
[
"### Tipos ```number``` e ```integer```.\n\nhttps://swagger.io/docs/specification/data-models/data-types/#numbers",
"_____no_output_____"
],
[
"### Tipo ```boolean```.\n\nhttps://swagger.io/docs/specification/data-models/data-types/#boolean",
"_____no_output_____"
],
[
"### Tipo ```array```.\n\nhttps://swagger.io/docs/specification/data-models/data-types/#array",
"_____no_output_____"
],
[
"### Tipo ```object```.\n\nhttps://swagger.io/docs/specification/data-models/data-types/#object",
"_____no_output_____"
],
[
"## Enums.",
"_____no_output_____"
],
[
"``` yaml\ntype: <tipo>\nenum:\n - <elemento 1>\n - <elemento 2>\n```\n\nhttps://swagger.io/docs/specification/data-models/enums/",
"_____no_output_____"
],
[
"## Referencias.",
"_____no_output_____"
],
[
"```\n$ref: \"ruta\"\n```\n\nhttps://swagger.io/docs/specification/using-ref/",
"_____no_output_____"
],
[
"### Referencias dentro del documento.\n\n``` #/<nivel 1>/<nivel 2>/... /<nivel n>/<elemento> ```",
"_____no_output_____"
],
[
"## Ejemplos.\n\nhttps://swagger.io/docs/specification/adding-examples/",
"_____no_output_____"
],
[
"# ",
"_____no_output_____"
],
[
"<p style=\"text-align: center\"><a rel=\"license\" href=\"http://creativecommons.org/licenses/by/4.0/\"><img alt=\"Licencia Creative Commons\" style=\"border-width:0\" src=\"https://i.creativecommons.org/l/by/4.0/80x15.png\" /></a><br />Esta obra está bajo una <a rel=\"license\" href=\"http://creativecommons.org/licenses/by/4.0/\">Licencia Creative Commons Atribución 4.0 Internacional</a>.</p>\n<p style=\"text-align: center\">© José Luis Chiquete Valdivieso. 2022.</p>",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cb4401843d36e0254eebda72442870b5c33c053b | 323,229 | ipynb | Jupyter Notebook | notebooks/tensorflow/getting-started-keras.ipynb | wuby216/Around-ML | 5870a50fb098ab53f7cd2cee845b0b48154fea6e | [
"Apache-2.0"
] | null | null | null | notebooks/tensorflow/getting-started-keras.ipynb | wuby216/Around-ML | 5870a50fb098ab53f7cd2cee845b0b48154fea6e | [
"Apache-2.0"
] | null | null | null | notebooks/tensorflow/getting-started-keras.ipynb | wuby216/Around-ML | 5870a50fb098ab53f7cd2cee845b0b48154fea6e | [
"Apache-2.0"
] | null | null | null | 74.665974 | 24,012 | 0.537074 | [
[
[
"# Copyright 2019 Google LLC\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Getting started: Training and prediction with Keras in AI Platform\n\n<img src=\"https://storage.googleapis.com/cloud-samples-data/ml-engine/census/keras-tensorflow-cmle.png\" alt=\"Keras, TensorFlow, and AI Platform logos\" width=\"300px\">\n\n<table align=\"left\">\n <td>\n <a href=\"https://cloud.google.com/ml-engine/docs/tensorflow/getting-started-keras\">\n <img src=\"https://cloud.google.com/_static/images/cloud/icons/favicons/onecloud/super_cloud.png\"\n alt=\"Google Cloud logo\" width=\"32px\"> Read on cloud.google.com\n </a>\n </td>\n <td>\n <a href=\"https://colab.research.google.com/github/GoogleCloudPlatform/cloudml-samples/blob/master/notebooks/tensorflow/getting-started-keras.ipynb\">\n <img src=\"https://cloud.google.com/ml-engine/images/colab-logo-32px.png\" alt=\"Colab logo\"> Run in Colab\n </a>\n </td>\n <td>\n <a href=\"https://github.com/GoogleCloudPlatform/cloudml-samples/blob/master/notebooks/tensorflow/getting-started-keras.ipynb\">\n <img src=\"https://cloud.google.com/ml-engine/images/github-logo-32px.png\" alt=\"GitHub logo\">\n View on GitHub\n </a>\n </td>\n</table>",
"_____no_output_____"
],
[
"## Overview\n\nThis tutorial shows how to train a neural network on Cloud Machine Learning\nEngine using the Keras sequential API and how to serve predictions from that\nmodel.\n\nKeras is a high-level API for building and training deep learning models.\n[tf.keras](https://www.tensorflow.org/guide/keras) is TensorFlow’s\nimplementation of this API.\n\nThe first two parts of the tutorial walk through training a model on Cloud\nAI Platform using prewritten Keras code, deploying the trained model to Cloud ML\nEngine, and serving online predictions from the deployed model.\n\nThe last part of the tutorial digs into the training code used for this model and ensuring it's compatible with AI Platform. To learn more about building\nmachine learning models in Keras more generally, read [TensorFlow's Keras\ntutorials](https://www.tensorflow.org/tutorials/keras).",
"_____no_output_____"
],
[
"### Dataset\n\nThis tutorial uses the [United States Census Income\nDataset](https://archive.ics.uci.edu/ml/datasets/census+income) provided by the\n[UC Irvine Machine Learning\nRepository](https://archive.ics.uci.edu/ml/index.php). This dataset contains\ninformation about people from a 1994 Census database, including age, education,\nmarital status, occupation, and whether they make more than $50,000 a year.",
"_____no_output_____"
],
[
"### Objective\n\nThe goal is to train a deep neural network (DNN) using Keras that predicts\nwhether a person makes more than $50,000 a year (target label) based on other\nCensus information about the person (features).\n\nThis tutorial focuses more on using this model with AI Platform than on\nthe design of the model itself. However, it's always important to think about\npotential problems and unintended consequences when building machine learning\nsystems. See the [Machine Learning Crash Course exercise about\nfairness](https://developers.google.com/machine-learning/crash-course/fairness/programming-exercise)\nto learn about sources of bias in the Census dataset, as well as machine\nlearning fairness more generally.",
"_____no_output_____"
],
[
"### Costs\n\nThis tutorial uses billable components of Google Cloud Platform (GCP):\n\n* AI Platform\n* Cloud Storage\n\nLearn about [AI Platform\npricing](https://cloud.google.com/ml-engine/docs/pricing) and [Cloud Storage\npricing](https://cloud.google.com/storage/pricing), and use the [Pricing\nCalculator](https://cloud.google.com/products/calculator/)\nto generate a cost estimate based on your projected usage.",
"_____no_output_____"
],
[
"## Before you begin\n\nYou must do several things before you can train and deploy a model in Cloud ML\nEngine:\n\n* Set up your local development environment.\n* Set up a GCP project with billing and the necessary\n APIs enabled.\n* Authenticate your GCP account in this notebook.\n* Create a Cloud Storage bucket to store your training package and your\n trained model.\n",
"_____no_output_____"
],
[
"### Set up your local development environment\n\n**If you are using Colab or Cloud ML Notebooks**, your environment already meets\nall the requirements to run this notebook. You can skip this step.",
"_____no_output_____"
],
[
"**Otherwise**, make sure your environment meets this notebook's requirements.\nYou need the following:\n\n* The Google Cloud SDK\n* Git\n* Python 3\n* virtualenv\n* Jupyter notebook running in a virtual environment with Python 3\n\nThe Google Cloud guide to [Setting up a Python development\nenvironment](https://cloud.google.com/python/setup) and the [Jupyter\ninstallation guide](https://jupyter.org/install) provide detailed instructions\nfor meeting these requirements. The following steps provide a condensed set of\ninstructions:\n\n1. [Install and initialize the Cloud SDK.](https://cloud.google.com/sdk/docs/)\n\n2. [Install Python 3.](https://cloud.google.com/python/setup#installing_python)\n\n3. [Install\n virtualenv](https://cloud.google.com/python/setup#installing_and_using_virtualenv)\n and create a virtual environment that uses Python 3.\n\n4. Activate that environment and run `pip install jupyter` in a shell to install\n Jupyter.\n\n5. Run `jupyter notebook` in a shell to launch Jupyter.\n\n6. Open this notebook in the Jupyter Notebook Dashboard.",
"_____no_output_____"
],
[
"### Set up your GCP project\n\n**The following steps are required, regardless of your notebook environment.**\n\n1. [Select or create a GCP project.](https://console.cloud.google.com/cloud-resource-manager)\n\n2. [Make sure that billing is enabled for your project.](https://cloud.google.com/billing/docs/how-to/modify-project)\n\n3. [Enable the AI Platform and Compute Engine APIs.](https://console.cloud.google.com/flows/enableapi?apiid=ml.googleapis.com,compute_component)\n\n4. Enter your project ID in the cell below. Then run the cell to make sure the\nCloud SDK uses the right project for all the commands in this notebook.\n\n**Note**: Jupyter runs lines prefixed with `!` as shell commands, and it interpolates Python variables prefixed with `$` into these commands.",
"_____no_output_____"
]
],
[
[
"PROJECT_ID = \"<your-project-id>\" #@param {type:\"string\"}\n! gcloud config set project $PROJECT_ID",
"Updated property [core/project].\n"
]
],
[
[
"### Authenticate your GCP account\n\n**If you are using Cloud ML Notebooks**, your environment is already\nauthenticated. Skip this step.",
"_____no_output_____"
],
[
"**If you are using Colab**, run the cell below and follow the instructions\nwhen prompted to authenticate your account via oAuth.\n\n**Otherwise**, follow these steps:\n\n1. In the GCP Console, go to the [**Create service account key**\n page](https://console.cloud.google.com/apis/credentials/serviceaccountkey).\n\n2. From the **Service account** drop-down list, select **New service account**.\n\n3. In the **Service account name** field, enter a name.\n\n4. From the **Role** drop-down list, select\n **Machine Learning Engine > AI Platform Admin** and\n **Storage > Storage Object Admin**.\n\n5. Click *Create*. A JSON file that contains your key downloads to your\nlocal environment.\n\n6. Enter the path to your service account key as the\n`GOOGLE_APPLICATION_CREDENTIALS` variable in the cell below and run the cell.",
"_____no_output_____"
]
],
[
[
"import sys\n\n# If you are running this notebook in Colab, run this cell and follow the\n# instructions to authenticate your GCP account. This provides access to your\n# Cloud Storage bucket and lets you submit training jobs and prediction\n# requests.\n\nif 'google.colab' in sys.modules:\n from google.colab import auth as google_auth\n google_auth.authenticate_user()\n\n# If you are running this notebook locally, replace the string below with the\n# path to your service account key and run this cell to authenticate your GCP\n# account.\nelse:\n %env GOOGLE_APPLICATION_CREDENTIALS ''\n",
"_____no_output_____"
]
],
[
[
"### Create a Cloud Storage bucket\n\n**The following steps are required, regardless of your notebook environment.**\n\nWhen you submit a training job using the Cloud SDK, you upload a Python package\ncontaining your training code to a Cloud Storage bucket. AI Platform runs\nthe code from this package. In this tutorial, AI Platform also saves the\ntrained model that results from your job in the same bucket. You can then\ncreate an AI Platform model verison based on this output in order to serve\nonline predictions.\n\nSet the name of your Cloud Storage bucket below. It must be unique across all\nCloud Storage buckets. \n\nYou may also change the `REGION` variable, which is used for operations\nthroughout the rest of this notebook. Make sure to [choose a region where Cloud\nAI Platform services are\navailable](https://cloud.google.com/ml-engine/docs/tensorflow/regions). You may\nnot use a Multi-Regional Storage bucket for training with AI Platform.",
"_____no_output_____"
]
],
[
[
"BUCKET_NAME = \"<your-bucket-name>\" #@param {type:\"string\"}\nREGION = \"us-central1\" #@param {type:\"string\"}",
"_____no_output_____"
]
],
[
[
"**Only if your bucket doesn't already exist**: Run the following cell to create your Cloud Storage bucket.",
"_____no_output_____"
]
],
[
[
"! gsutil mb -l $REGION gs://$BUCKET_NAME",
"Creating gs://<your-bucket-name>/...\n"
]
],
[
[
"Finally, validate access to your Cloud Storage bucket by examining its contents:\n",
"_____no_output_____"
]
],
[
[
"! gsutil ls -al gs://$BUCKET_NAME",
"_____no_output_____"
]
],
[
[
"## Part 1. Quickstart for training in AI Platform\n\nThis section of the tutorial walks you through submitting a training job to Cloud\nAI Platform. This job runs sample code that uses Keras to train a deep neural\nnetwork on the United States Census data. It outputs the trained model as a\n[TensorFlow SavedModel\ndirectory](https://www.tensorflow.org/guide/saved_model#save_and_restore_models)\nin your Cloud Storage bucket.\n",
"_____no_output_____"
],
[
"### Get training code and dependencies\n\nFirst, download the training code and change the notebook's working directory:",
"_____no_output_____"
]
],
[
[
"# Clone the repository of AI Platform samples\n! git clone --depth 1 https://github.com/GoogleCloudPlatform/cloudml-samples\n\n# Set the working directory to the sample code directory\n%cd cloudml-samples/census/tf-keras",
"Cloning into 'cloudml-samples'...\nremote: Enumerating objects: 404, done.\u001b[K\nremote: Counting objects: 100% (404/404), done.\u001b[K\nremote: Compressing objects: 100% (333/333), done.\u001b[K\nremote: Total 404 (delta 110), reused 199 (delta 46), pack-reused 0\u001b[K\nReceiving objects: 100% (404/404), 22.67 MiB | 19.76 MiB/s, done.\nResolving deltas: 100% (110/110), done.\n/content/cloudml-samples/census/tf-keras\n"
]
],
[
[
"Notice that the training code is structured as a Python package in the\n`trainer/` subdirectory:",
"_____no_output_____"
]
],
[
[
"# `ls` shows the working directory's contents. The `p` flag adds trailing \n# slashes to subdirectory names. The `R` flag lists subdirectories recursively.\n! ls -pR",
".:\nREADME.md requirements.txt trainer/\n\n./trainer:\n__init__.py model.py task.py\tutil.py\n"
]
],
[
[
"Run the following cell to install Python dependencies needed to train the model locally. When you run the training job in AI Platform,\ndependencies are preinstalled based on the [runtime\nverison](https://cloud.google.com/ml-engine/docs/tensorflow/runtime-version-list)\nyou choose.",
"_____no_output_____"
]
],
[
[
"! pip install -r requirements.txt",
"Requirement already satisfied: numpy>=1.14.* in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 1)) (1.14.6)\nRequirement already satisfied: pandas>=0.22.* in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 2)) (0.22.0)\nRequirement already satisfied: six==1.11.* in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 3)) (1.11.0)\nRequirement already satisfied: tensorflow==1.13.* in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 4)) (1.13.1)\nRequirement already satisfied: pytz>=2011k in /usr/local/lib/python3.6/dist-packages (from pandas>=0.22.*->-r requirements.txt (line 2)) (2018.9)\nRequirement already satisfied: python-dateutil>=2 in /usr/local/lib/python3.6/dist-packages (from pandas>=0.22.*->-r requirements.txt (line 2)) (2.5.3)\nRequirement already satisfied: protobuf>=3.6.1 in /usr/local/lib/python3.6/dist-packages (from tensorflow==1.13.*->-r requirements.txt (line 4)) (3.7.0)\nRequirement already satisfied: keras-preprocessing>=1.0.5 in /usr/local/lib/python3.6/dist-packages (from tensorflow==1.13.*->-r requirements.txt (line 4)) (1.0.9)\nRequirement already satisfied: keras-applications>=1.0.6 in /usr/local/lib/python3.6/dist-packages (from tensorflow==1.13.*->-r requirements.txt (line 4)) (1.0.7)\nRequirement already satisfied: termcolor>=1.1.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow==1.13.*->-r requirements.txt (line 4)) (1.1.0)\nRequirement already satisfied: tensorflow-estimator<1.14.0rc0,>=1.13.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow==1.13.*->-r requirements.txt (line 4)) (1.13.0)\nRequirement already satisfied: grpcio>=1.8.6 in /usr/local/lib/python3.6/dist-packages (from tensorflow==1.13.*->-r requirements.txt (line 4)) (1.15.0)\nRequirement already satisfied: astor>=0.6.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow==1.13.*->-r requirements.txt (line 4)) (0.7.1)\nRequirement already satisfied: gast>=0.2.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow==1.13.*->-r requirements.txt (line 4)) (0.2.2)\nRequirement already satisfied: absl-py>=0.1.6 in /usr/local/lib/python3.6/dist-packages (from tensorflow==1.13.*->-r requirements.txt (line 4)) (0.7.1)\nRequirement already satisfied: tensorboard<1.14.0,>=1.13.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow==1.13.*->-r requirements.txt (line 4)) (1.13.1)\nRequirement already satisfied: wheel>=0.26 in /usr/local/lib/python3.6/dist-packages (from tensorflow==1.13.*->-r requirements.txt (line 4)) (0.33.1)\nRequirement already satisfied: setuptools in /usr/local/lib/python3.6/dist-packages (from protobuf>=3.6.1->tensorflow==1.13.*->-r requirements.txt (line 4)) (40.8.0)\nRequirement already satisfied: h5py in /usr/local/lib/python3.6/dist-packages (from keras-applications>=1.0.6->tensorflow==1.13.*->-r requirements.txt (line 4)) (2.8.0)\nRequirement already satisfied: mock>=2.0.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow-estimator<1.14.0rc0,>=1.13.0->tensorflow==1.13.*->-r requirements.txt (line 4)) (2.0.0)\nRequirement already satisfied: markdown>=2.6.8 in /usr/local/lib/python3.6/dist-packages (from tensorboard<1.14.0,>=1.13.0->tensorflow==1.13.*->-r requirements.txt (line 4)) (3.0.1)\nRequirement already satisfied: werkzeug>=0.11.15 in /usr/local/lib/python3.6/dist-packages (from tensorboard<1.14.0,>=1.13.0->tensorflow==1.13.*->-r requirements.txt (line 4)) (0.14.1)\nRequirement already satisfied: pbr>=0.11 in /usr/local/lib/python3.6/dist-packages (from mock>=2.0.0->tensorflow-estimator<1.14.0rc0,>=1.13.0->tensorflow==1.13.*->-r requirements.txt (line 4)) (5.1.3)\n"
]
],
[
[
"### Train your model locally\n\nBefore training on AI Platform, train the job locally to verify the file\nstructure and packaging is correct.\n\nFor a complex or resource-intensive job, you\nmay want to train locally on a small sample of your dataset to verify your code.\nThen you can run the job on AI Platform to train on the whole dataset.\n\nThis sample runs a relatively quick job on a small dataset, so the local\ntraining and the AI Platform job run the same code on the same data.\n\nRun the following cell to train a model locally:",
"_____no_output_____"
]
],
[
[
"# Explicitly tell `gcloud ml-engine local train` to use Python 3 \n! gcloud config set ml_engine/local_python $(which python3)\n\n# This is similar to `python -m trainer.task --job-dir local-training-output`\n# but it better replicates the AI Platform environment, especially for\n# distributed training (not applicable here).\n! gcloud ml-engine local train \\\n --package-path trainer \\\n --module-name trainer.task \\\n --job-dir local-training-output",
"Updated property [ml_engine/local_python].\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/ops/resource_variable_ops.py:435: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nColocations handled automatically by placer.\n2019-03-27 17:53:24.297156: I tensorflow/core/platform/profile_utils/cpu_utils.cc:94] CPU Frequency: 2200000000 Hz\n2019-03-27 17:53:24.297428: I tensorflow/compiler/xla/service/service.cc:150] XLA service 0x2441b80 executing computations on platform Host. Devices:\n2019-03-27 17:53:24.297464: I tensorflow/compiler/xla/service/service.cc:158] StreamExecutor device (0): <undefined>, <undefined>\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/ops/math_ops.py:3066: to_int32 (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.cast instead.\n\nEpoch 00001: LearningRateScheduler reducing learning rate to 0.02.\nEpoch 1/20\n254/254 [==============================] - 1s 5ms/step - loss: 0.5032 - acc: 0.7890 - val_loss: 0.4553 - val_acc: 0.8030\n\nEpoch 00002: LearningRateScheduler reducing learning rate to 0.015.\nEpoch 2/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3616 - acc: 0.8362 - val_loss: 0.3273 - val_acc: 0.8468\n\nEpoch 00003: LearningRateScheduler reducing learning rate to 0.0125.\nEpoch 3/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3411 - acc: 0.8450 - val_loss: 0.3294 - val_acc: 0.8447\n\nEpoch 00004: LearningRateScheduler reducing learning rate to 0.01125.\nEpoch 4/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3362 - acc: 0.8454 - val_loss: 0.3566 - val_acc: 0.8410\n\nEpoch 00005: LearningRateScheduler reducing learning rate to 0.010625.\nEpoch 5/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3325 - acc: 0.8469 - val_loss: 0.3264 - val_acc: 0.8507\n\nEpoch 00006: LearningRateScheduler reducing learning rate to 0.0103125.\nEpoch 6/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3316 - acc: 0.8471 - val_loss: 0.3228 - val_acc: 0.8515\n\nEpoch 00007: LearningRateScheduler reducing learning rate to 0.01015625.\nEpoch 7/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3297 - acc: 0.8475 - val_loss: 0.3308 - val_acc: 0.8452\n\nEpoch 00008: LearningRateScheduler reducing learning rate to 0.010078125.\nEpoch 8/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3287 - acc: 0.8479 - val_loss: 0.3449 - val_acc: 0.8394\n\nEpoch 00009: LearningRateScheduler reducing learning rate to 0.0100390625.\nEpoch 9/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3274 - acc: 0.8488 - val_loss: 0.3213 - val_acc: 0.8522\n\nEpoch 00010: LearningRateScheduler reducing learning rate to 0.01001953125.\nEpoch 10/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3256 - acc: 0.8488 - val_loss: 0.3302 - val_acc: 0.8508\n\nEpoch 00011: LearningRateScheduler reducing learning rate to 0.010009765625.\nEpoch 11/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3280 - acc: 0.8502 - val_loss: 0.3342 - val_acc: 0.8443\n\nEpoch 00012: LearningRateScheduler reducing learning rate to 0.010004882812500001.\nEpoch 12/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3283 - acc: 0.8502 - val_loss: 0.3511 - val_acc: 0.8506\n\nEpoch 00013: LearningRateScheduler reducing learning rate to 0.01000244140625.\nEpoch 13/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3258 - acc: 0.8498 - val_loss: 0.3450 - val_acc: 0.8491\n\nEpoch 00014: LearningRateScheduler reducing learning rate to 0.010001220703125.\nEpoch 14/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3272 - acc: 0.8500 - val_loss: 0.3220 - val_acc: 0.8524\n\nEpoch 00015: LearningRateScheduler reducing learning rate to 0.0100006103515625.\nEpoch 15/20\n254/254 [==============================] - 1s 3ms/step - loss: 0.3261 - acc: 0.8498 - val_loss: 0.3221 - val_acc: 0.8520\n\nEpoch 00016: LearningRateScheduler reducing learning rate to 0.01000030517578125.\nEpoch 16/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3239 - acc: 0.8509 - val_loss: 0.3232 - val_acc: 0.8480\n\nEpoch 00017: LearningRateScheduler reducing learning rate to 0.010000152587890625.\nEpoch 17/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3275 - acc: 0.8484 - val_loss: 0.3377 - val_acc: 0.8540\n\nEpoch 00018: LearningRateScheduler reducing learning rate to 0.010000076293945313.\nEpoch 18/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3242 - acc: 0.8490 - val_loss: 0.3275 - val_acc: 0.8522\n\nEpoch 00019: LearningRateScheduler reducing learning rate to 0.010000038146972657.\nEpoch 19/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3236 - acc: 0.8507 - val_loss: 0.3302 - val_acc: 0.8471\n\nEpoch 00020: LearningRateScheduler reducing learning rate to 0.010000019073486329.\nEpoch 20/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3259 - acc: 0.8490 - val_loss: 0.3218 - val_acc: 0.8518\nWARNING:tensorflow:This model was compiled with a Keras optimizer (<tensorflow.python.keras.optimizers.RMSprop object at 0x7fa74da22278>) but is being saved in TensorFlow format with `save_weights`. The model's weights will be saved, but unlike with TensorFlow optimizers in the TensorFlow format the optimizer's state will not be saved.\n\nConsider using a TensorFlow optimizer from `tf.train`.\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/keras/engine/network.py:1436: update_checkpoint_state (from tensorflow.python.training.checkpoint_management) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.train.CheckpointManager to manage checkpoints rather than manually editing the Checkpoint proto.\nWARNING:tensorflow:Model was compiled with an optimizer, but the optimizer is not from `tf.train` (e.g. `tf.train.AdagradOptimizer`). Only the serving graph was exported. The train and evaluate graphs were not added to the SavedModel.\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/saved_model/signature_def_utils_impl.py:205: build_tensor_info (from tensorflow.python.saved_model.utils_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nThis function will only be available through the v1 compatibility library as tf.compat.v1.saved_model.utils.build_tensor_info or tf.compat.v1.saved_model.build_tensor_info.\nINFO:tensorflow:Signatures INCLUDED in export for Classify: None\nINFO:tensorflow:Signatures INCLUDED in export for Regress: None\nINFO:tensorflow:Signatures INCLUDED in export for Predict: ['serving_default']\nINFO:tensorflow:Signatures INCLUDED in export for Train: None\nINFO:tensorflow:Signatures INCLUDED in export for Eval: None\nINFO:tensorflow:No assets to save.\nINFO:tensorflow:No assets to write.\nINFO:tensorflow:SavedModel written to: local-training-output/keras_export/1553709223/saved_model.pb\nModel exported to: local-training-output/keras_export/1553709223\n"
]
],
[
[
"### Train your model using AI Platform\n\nNext, submit a training job to AI Platform. This runs the training module\nin the cloud and exports the trained model to Cloud Storage.\n\nFirst, give your training job a name and choose a directory within your Cloud\nStorage bucket for saving intermediate and output files:",
"_____no_output_____"
]
],
[
[
"JOB_NAME = 'my_first_keras_job'\nJOB_DIR = 'gs://' + BUCKET_NAME + '/keras-job-dir'",
"_____no_output_____"
]
],
[
[
"Run the following command to package the `trainer/` directory, upload it to the\nspecified `--job-dir`, and instruct AI Platform to run the\n`trainer.task` module from that package.\n\nThe `--stream-logs` flag lets you view training logs in the cell below. You can\nalso see logs and other job details in the GCP Console.",
"_____no_output_____"
]
],
[
[
"! gcloud ml-engine jobs submit training $JOB_NAME \\\n --package-path trainer/ \\\n --module-name trainer.task \\\n --region $REGION \\\n --python-version 3.5 \\\n --runtime-version 1.13 \\\n --job-dir $JOB_DIR \\\n --stream-logs",
"Job [my_first_keras_job] submitted successfully.\nINFO\t2019-03-27 17:54:27 +0000\tservice\t\tValidating job requirements...\nINFO\t2019-03-27 17:54:27 +0000\tservice\t\tJob creation request has been successfully validated.\nINFO\t2019-03-27 17:54:27 +0000\tservice\t\tJob my_first_keras_job is queued.\nINFO\t2019-03-27 17:54:27 +0000\tservice\t\tWaiting for job to be provisioned.\nINFO\t2019-03-27 17:54:30 +0000\tservice\t\tWaiting for training program to start.\nINFO\t2019-03-27 17:56:09 +0000\tmaster-replica-0\t\tRunning task with arguments: --cluster={\"master\": [\"127.0.0.1:2222\"]} --task={\"type\": \"master\", \"index\": 0} --job={ \"package_uris\": [\"gs://<your-bucket-name>/keras-job-dir/packages/dcc159f40836cff74a27866227b327b0a8ccb5266194e76cff5368266b6d1cdd/trainer-0.0.0.tar.gz\"], \"python_module\": \"trainer.task\", \"region\": \"us-central1\", \"runtime_version\": \"1.13\", \"job_dir\": \"gs://<your-bucket-name>/keras-job-dir\", \"run_on_raw_vm\": true, \"python_version\": \"3.5\"}\nWARNING\t2019-03-27 17:56:09 +0000\tmaster-replica-0\t\tFrom /usr/local/lib/python3.5/dist-packages/tensorflow/python/framework/op_def_library.py:263: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.\nWARNING\t2019-03-27 17:56:09 +0000\tmaster-replica-0\t\tInstructions for updating:\nWARNING\t2019-03-27 17:56:09 +0000\tmaster-replica-0\t\tColocations handled automatically by placer.\nINFO\t2019-03-27 17:56:18 +0000\tmaster-replica-0\t\tRunning module trainer.task.\nINFO\t2019-03-27 17:56:18 +0000\tmaster-replica-0\t\tDownloading the package: gs://<your-bucket-name>/keras-job-dir/packages/dcc159f40836cff74a27866227b327b0a8ccb5266194e76cff5368266b6d1cdd/trainer-0.0.0.tar.gz\nINFO\t2019-03-27 17:56:18 +0000\tmaster-replica-0\t\tRunning command: gsutil -q cp gs://<your-bucket-name>/keras-job-dir/packages/dcc159f40836cff74a27866227b327b0a8ccb5266194e76cff5368266b6d1cdd/trainer-0.0.0.tar.gz trainer-0.0.0.tar.gz\nINFO\t2019-03-27 17:56:20 +0000\tmaster-replica-0\t\tInstalling the package: gs://<your-bucket-name>/keras-job-dir/packages/dcc159f40836cff74a27866227b327b0a8ccb5266194e76cff5368266b6d1cdd/trainer-0.0.0.tar.gz\nINFO\t2019-03-27 17:56:20 +0000\tmaster-replica-0\t\tRunning command: pip3 install --user --upgrade --force-reinstall --no-deps trainer-0.0.0.tar.gz\nINFO\t2019-03-27 17:56:22 +0000\tmaster-replica-0\t\tProcessing ./trainer-0.0.0.tar.gz\nINFO\t2019-03-27 17:56:22 +0000\tmaster-replica-0\t\tGenerating grammar tables from /usr/lib/python3.5/lib2to3/Grammar.txt\nINFO\t2019-03-27 17:56:22 +0000\tmaster-replica-0\t\tGenerating grammar tables from /usr/lib/python3.5/lib2to3/PatternGrammar.txt\nINFO\t2019-03-27 17:56:23 +0000\tmaster-replica-0\t\tBuilding wheels for collected packages: trainer\nINFO\t2019-03-27 17:56:23 +0000\tmaster-replica-0\t\t Building wheel for trainer (setup.py): started\nINFO\t2019-03-27 17:56:23 +0000\tmaster-replica-0\t\tGenerating grammar tables from /usr/lib/python3.5/lib2to3/Grammar.txt\nINFO\t2019-03-27 17:56:23 +0000\tmaster-replica-0\t\tGenerating grammar tables from /usr/lib/python3.5/lib2to3/PatternGrammar.txt\nINFO\t2019-03-27 17:56:23 +0000\tmaster-replica-0\t\t Building wheel for trainer (setup.py): finished with status 'done'\nINFO\t2019-03-27 17:56:23 +0000\tmaster-replica-0\t\t Stored in directory: /root/.cache/pip/wheels/0d/1b/db/f8e86b296734f0b137e17e5d34862f4ae4faf8388755c6272f\nINFO\t2019-03-27 17:56:23 +0000\tmaster-replica-0\t\tSuccessfully built trainer\nINFO\t2019-03-27 17:56:23 +0000\tmaster-replica-0\t\tInstalling collected packages: trainer\nINFO\t2019-03-27 17:56:23 +0000\tmaster-replica-0\t\tSuccessfully installed trainer-0.0.0\nINFO\t2019-03-27 17:56:23 +0000\tmaster-replica-0\t\tRunning command: pip3 install --user trainer-0.0.0.tar.gz\nINFO\t2019-03-27 17:56:24 +0000\tmaster-replica-0\t\tProcessing ./trainer-0.0.0.tar.gz\nINFO\t2019-03-27 17:56:24 +0000\tmaster-replica-0\t\tGenerating grammar tables from /usr/lib/python3.5/lib2to3/Grammar.txt\nINFO\t2019-03-27 17:56:24 +0000\tmaster-replica-0\t\tGenerating grammar tables from /usr/lib/python3.5/lib2to3/PatternGrammar.txt\nINFO\t2019-03-27 17:56:24 +0000\tmaster-replica-0\t\tBuilding wheels for collected packages: trainer\nINFO\t2019-03-27 17:56:24 +0000\tmaster-replica-0\t\t Building wheel for trainer (setup.py): started\nINFO\t2019-03-27 17:56:25 +0000\tmaster-replica-0\t\tGenerating grammar tables from /usr/lib/python3.5/lib2to3/Grammar.txt\nINFO\t2019-03-27 17:56:25 +0000\tmaster-replica-0\t\tGenerating grammar tables from /usr/lib/python3.5/lib2to3/PatternGrammar.txt\nINFO\t2019-03-27 17:56:25 +0000\tmaster-replica-0\t\t Building wheel for trainer (setup.py): finished with status 'done'\nINFO\t2019-03-27 17:56:25 +0000\tmaster-replica-0\t\t Stored in directory: /root/.cache/pip/wheels/0d/1b/db/f8e86b296734f0b137e17e5d34862f4ae4faf8388755c6272f\nINFO\t2019-03-27 17:56:25 +0000\tmaster-replica-0\t\tSuccessfully built trainer\nINFO\t2019-03-27 17:56:29 +0000\tmaster-replica-0\t\tInstalling collected packages: trainer\nINFO\t2019-03-27 17:56:29 +0000\tmaster-replica-0\t\t Found existing installation: trainer 0.0.0\nINFO\t2019-03-27 17:56:29 +0000\tmaster-replica-0\t\t Uninstalling trainer-0.0.0:\nINFO\t2019-03-27 17:56:29 +0000\tmaster-replica-0\t\t Successfully uninstalled trainer-0.0.0\nINFO\t2019-03-27 17:56:29 +0000\tmaster-replica-0\t\tSuccessfully installed trainer-0.0.0\nINFO\t2019-03-27 17:56:29 +0000\tmaster-replica-0\t\tRunning command: python3 -m trainer.task --job-dir gs://<your-bucket-name>/keras-job-dir\nWARNING\t2019-03-27 17:56:43 +0000\tmaster-replica-0\t\tFrom /usr/local/lib/python3.5/dist-packages/tensorflow/python/ops/resource_variable_ops.py:435: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.\nWARNING\t2019-03-27 17:56:43 +0000\tmaster-replica-0\t\tInstructions for updating:\nWARNING\t2019-03-27 17:56:43 +0000\tmaster-replica-0\t\tColocations handled automatically by placer.\nINFO\t2019-03-27 17:56:44 +0000\tmaster-replica-0\t\tYour CPU supports instructions that this TensorFlow binary was not compiled to use: AVX2 FMA\nINFO\t2019-03-27 17:56:44 +0000\tmaster-replica-0\t\tCPU Frequency: 2300000000 Hz\nINFO\t2019-03-27 17:56:44 +0000\tmaster-replica-0\t\tXLA service 0x4f15c40 executing computations on platform Host. Devices:\nINFO\t2019-03-27 17:56:44 +0000\tmaster-replica-0\t\t StreamExecutor device (0): <undefined>, <undefined>\nWARNING\t2019-03-27 17:56:44 +0000\tmaster-replica-0\t\tFrom /usr/local/lib/python3.5/dist-packages/tensorflow/python/ops/math_ops.py:3066: to_int32 (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nWARNING\t2019-03-27 17:56:44 +0000\tmaster-replica-0\t\tInstructions for updating:\nWARNING\t2019-03-27 17:56:44 +0000\tmaster-replica-0\t\tUse tf.cast instead.\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\tEpoch 00001: LearningRateScheduler reducing learning rate to 0.02.\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\tEpoch 1/20\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 1:06 - loss: 0.5855 - acc: 0.7891\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t 16/254 [>.............................] - ETA: 4s - loss: 3.8615 - acc: 0.7495 \b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t 34/254 [===>..........................] - ETA: 2s - loss: 3.1560 - acc: 0.7415\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t 52/254 [=====>........................] - ETA: 1s - loss: 2.2601 - acc: 0.7515\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t 69/254 [=======>......................] - ETA: 1s - loss: 1.8414 - acc: 0.7505\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t 87/254 [=========>....................] - ETA: 0s - loss: 1.5580 - acc: 0.7543\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t105/254 [===========>..................] - ETA: 0s - loss: 1.3674 - acc: 0.7613\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t121/254 [=============>................] - ETA: 0s - loss: 1.2418 - acc: 0.7677\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t141/254 [===============>..............] - ETA: 0s - loss: 1.1292 - acc: 0.7703\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t161/254 [==================>...........] - ETA: 0s - loss: 1.0420 - acc: 0.7741\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t179/254 [====================>.........] - ETA: 0s - loss: 0.9786 - acc: 0.7781\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t198/254 [======================>.......] - ETA: 0s - loss: 0.9222 - acc: 0.7832\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t218/254 [========================>.....] - ETA: 0s - loss: 0.8751 - acc: 0.7866\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t238/254 [===========================>..] - ETA: 0s - loss: 0.8347 - acc: 0.7886\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 4ms/step - loss: 0.8076 - acc: 0.7896 - val_loss: 0.4046 - val_acc: 0.8322\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\tEpoch 00002: LearningRateScheduler reducing learning rate to 0.015.\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\tEpoch 2/20\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 0s - loss: 0.3897 - acc: 0.8438\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:45 +0000\tmaster-replica-0\t\t 21/254 [=>............................] - ETA: 0s - loss: 0.3849 - acc: 0.8307\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t 40/254 [===>..........................] - ETA: 0s - loss: 0.3795 - acc: 0.8273\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t 61/254 [======>.......................] - ETA: 0s - loss: 0.3706 - acc: 0.8262\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t 80/254 [========>.....................] - ETA: 0s - loss: 0.3666 - acc: 0.8264\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t 99/254 [==========>...................] - ETA: 0s - loss: 0.3618 - acc: 0.8298\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t119/254 [=============>................] - ETA: 0s - loss: 0.3604 - acc: 0.8303\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t139/254 [===============>..............] - ETA: 0s - loss: 0.3622 - acc: 0.8294\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t158/254 [=================>............] - ETA: 0s - loss: 0.3593 - acc: 0.8310\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t177/254 [===================>..........] - ETA: 0s - loss: 0.3601 - acc: 0.8312\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t197/254 [======================>.......] - ETA: 0s - loss: 0.3598 - acc: 0.8309\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t216/254 [========================>.....] - ETA: 0s - loss: 0.3599 - acc: 0.8311\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t237/254 [==========================>...] - ETA: 0s - loss: 0.3597 - acc: 0.8312\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 3ms/step - loss: 0.3580 - acc: 0.8321 - val_loss: 0.3400 - val_acc: 0.8372\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\tEpoch 00003: LearningRateScheduler reducing learning rate to 0.0125.\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\tEpoch 3/20\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 1s - loss: 0.3455 - acc: 0.8203\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t 17/254 [=>............................] - ETA: 0s - loss: 0.3449 - acc: 0.8419\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t 36/254 [===>..........................] - ETA: 0s - loss: 0.3414 - acc: 0.8414\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t 55/254 [=====>........................] - ETA: 0s - loss: 0.3324 - acc: 0.8449\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t 74/254 [=======>......................] - ETA: 0s - loss: 0.3378 - acc: 0.8417\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t 91/254 [=========>....................] - ETA: 0s - loss: 0.3372 - acc: 0.8423\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:46 +0000\tmaster-replica-0\t\t107/254 [===========>..................] - ETA: 0s - loss: 0.3394 - acc: 0.8424\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t122/254 [=============>................] - ETA: 0s - loss: 0.3433 - acc: 0.8397\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t138/254 [===============>..............] - ETA: 0s - loss: 0.3426 - acc: 0.8398\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t158/254 [=================>............] - ETA: 0s - loss: 0.3421 - acc: 0.8412\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t175/254 [===================>..........] - ETA: 0s - loss: 0.3427 - acc: 0.8413\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t192/254 [=====================>........] - ETA: 0s - loss: 0.3417 - acc: 0.8418\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t209/254 [=======================>......] - ETA: 0s - loss: 0.3405 - acc: 0.8423\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t226/254 [=========================>....] - ETA: 0s - loss: 0.3409 - acc: 0.8424\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t247/254 [============================>.] - ETA: 0s - loss: 0.3412 - acc: 0.8428\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 3ms/step - loss: 0.3409 - acc: 0.8425 - val_loss: 0.3308 - val_acc: 0.8496\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\tEpoch 00004: LearningRateScheduler reducing learning rate to 0.01125.\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\tEpoch 4/20\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 0s - loss: 0.4057 - acc: 0.8203\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t 17/254 [=>............................] - ETA: 0s - loss: 0.3522 - acc: 0.8323\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t 34/254 [===>..........................] - ETA: 0s - loss: 0.3467 - acc: 0.8447\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t 54/254 [=====>........................] - ETA: 0s - loss: 0.3441 - acc: 0.8459\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t 71/254 [=======>......................] - ETA: 0s - loss: 0.3448 - acc: 0.8445\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t 91/254 [=========>....................] - ETA: 0s - loss: 0.3435 - acc: 0.8449\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t111/254 [============>.................] - ETA: 0s - loss: 0.3417 - acc: 0.8449\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t130/254 [==============>...............] - ETA: 0s - loss: 0.3392 - acc: 0.8462\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t149/254 [================>.............] - ETA: 0s - loss: 0.3386 - acc: 0.8461\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t167/254 [==================>...........] - ETA: 0s - loss: 0.3370 - acc: 0.8455\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:47 +0000\tmaster-replica-0\t\t188/254 [=====================>........] - ETA: 0s - loss: 0.3361 - acc: 0.8459\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t208/254 [=======================>......] - ETA: 0s - loss: 0.3349 - acc: 0.8458\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t228/254 [=========================>....] - ETA: 0s - loss: 0.3364 - acc: 0.8452\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t245/254 [===========================>..] - ETA: 0s - loss: 0.3367 - acc: 0.8453\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 3ms/step - loss: 0.3366 - acc: 0.8451 - val_loss: 0.3431 - val_acc: 0.8319\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\tEpoch 00005: LearningRateScheduler reducing learning rate to 0.010625.\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\tEpoch 5/20\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 0s - loss: 0.3805 - acc: 0.8281\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t 22/254 [=>............................] - ETA: 0s - loss: 0.3307 - acc: 0.8544\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t 41/254 [===>..........................] - ETA: 0s - loss: 0.3297 - acc: 0.8470\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t 59/254 [=====>........................] - ETA: 0s - loss: 0.3334 - acc: 0.8455\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t 74/254 [=======>......................] - ETA: 0s - loss: 0.3299 - acc: 0.8502\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t 87/254 [=========>....................] - ETA: 0s - loss: 0.3323 - acc: 0.8495\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t 97/254 [==========>...................] - ETA: 0s - loss: 0.3336 - acc: 0.8490\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t107/254 [===========>..................] - ETA: 0s - loss: 0.3344 - acc: 0.8495\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t120/254 [=============>................] - ETA: 0s - loss: 0.3367 - acc: 0.8472\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t136/254 [===============>..............] - ETA: 0s - loss: 0.3368 - acc: 0.8463\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t151/254 [================>.............] - ETA: 0s - loss: 0.3359 - acc: 0.8468\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t169/254 [==================>...........] - ETA: 0s - loss: 0.3373 - acc: 0.8449\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t187/254 [=====================>........] - ETA: 0s - loss: 0.3364 - acc: 0.8450\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t205/254 [=======================>......] - ETA: 0s - loss: 0.3348 - acc: 0.8449\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:48 +0000\tmaster-replica-0\t\t223/254 [=========================>....] - ETA: 0s - loss: 0.3345 - acc: 0.8451\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t242/254 [===========================>..] - ETA: 0s - loss: 0.3353 - acc: 0.8448\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 3ms/step - loss: 0.3339 - acc: 0.8453 - val_loss: 0.3486 - val_acc: 0.8504\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\tEpoch 00006: LearningRateScheduler reducing learning rate to 0.0103125.\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\tEpoch 6/20\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 0s - loss: 0.5068 - acc: 0.7891\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t 18/254 [=>............................] - ETA: 0s - loss: 0.3490 - acc: 0.8459\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t 38/254 [===>..........................] - ETA: 0s - loss: 0.3355 - acc: 0.8512\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t 58/254 [=====>........................] - ETA: 0s - loss: 0.3312 - acc: 0.8528\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t 78/254 [========>.....................] - ETA: 0s - loss: 0.3311 - acc: 0.8526\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t 99/254 [==========>...................] - ETA: 0s - loss: 0.3317 - acc: 0.8501\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t116/254 [============>.................] - ETA: 0s - loss: 0.3312 - acc: 0.8496\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t134/254 [==============>...............] - ETA: 0s - loss: 0.3326 - acc: 0.8500\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t153/254 [=================>............] - ETA: 0s - loss: 0.3326 - acc: 0.8499\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t169/254 [==================>...........] - ETA: 0s - loss: 0.3351 - acc: 0.8485\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t187/254 [=====================>........] - ETA: 0s - loss: 0.3351 - acc: 0.8481\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t204/254 [=======================>......] - ETA: 0s - loss: 0.3346 - acc: 0.8478\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t220/254 [========================>.....] - ETA: 0s - loss: 0.3345 - acc: 0.8482\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t237/254 [==========================>...] - ETA: 0s - loss: 0.3330 - acc: 0.8483\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 3ms/step - loss: 0.3317 - acc: 0.8483 - val_loss: 0.3297 - val_acc: 0.8402\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\tEpoch 00007: LearningRateScheduler reducing learning rate to 0.01015625.\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\tEpoch 7/20\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 0s - loss: 0.2934 - acc: 0.8906\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t 19/254 [=>............................] - ETA: 0s - loss: 0.3127 - acc: 0.8586\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:49 +0000\tmaster-replica-0\t\t 37/254 [===>..........................] - ETA: 0s - loss: 0.3242 - acc: 0.8554\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t 55/254 [=====>........................] - ETA: 0s - loss: 0.3300 - acc: 0.8482\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t 74/254 [=======>......................] - ETA: 0s - loss: 0.3345 - acc: 0.8486\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t 93/254 [=========>....................] - ETA: 0s - loss: 0.3329 - acc: 0.8465\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t111/254 [============>.................] - ETA: 0s - loss: 0.3327 - acc: 0.8466\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t128/254 [==============>...............] - ETA: 0s - loss: 0.3327 - acc: 0.8468\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t147/254 [================>.............] - ETA: 0s - loss: 0.3307 - acc: 0.8479\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t163/254 [==================>...........] - ETA: 0s - loss: 0.3310 - acc: 0.8484\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t183/254 [====================>.........] - ETA: 0s - loss: 0.3316 - acc: 0.8488\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t202/254 [======================>.......] - ETA: 0s - loss: 0.3335 - acc: 0.8477\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t219/254 [========================>.....] - ETA: 0s - loss: 0.3312 - acc: 0.8491\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t238/254 [===========================>..] - ETA: 0s - loss: 0.3312 - acc: 0.8489\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 3ms/step - loss: 0.3316 - acc: 0.8480 - val_loss: 0.3250 - val_acc: 0.8461\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\tEpoch 00008: LearningRateScheduler reducing learning rate to 0.010078125.\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\tEpoch 8/20\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 0s - loss: 0.3214 - acc: 0.8594\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t 19/254 [=>............................] - ETA: 0s - loss: 0.3520 - acc: 0.8425\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t 34/254 [===>..........................] - ETA: 0s - loss: 0.3456 - acc: 0.8447\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t 49/254 [====>.........................] - ETA: 0s - loss: 0.3347 - acc: 0.8492\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t 64/254 [======>.......................] - ETA: 0s - loss: 0.3296 - acc: 0.8503\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t 79/254 [========>.....................] - ETA: 0s - loss: 0.3293 - acc: 0.8502\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t 95/254 [==========>...................] - ETA: 0s - loss: 0.3288 - acc: 0.8502\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:50 +0000\tmaster-replica-0\t\t112/254 [============>.................] - ETA: 0s - loss: 0.3285 - acc: 0.8489\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t128/254 [==============>...............] - ETA: 0s - loss: 0.3307 - acc: 0.8480\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t144/254 [================>.............] - ETA: 0s - loss: 0.3294 - acc: 0.8484\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t161/254 [==================>...........] - ETA: 0s - loss: 0.3294 - acc: 0.8488\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t175/254 [===================>..........] - ETA: 0s - loss: 0.3289 - acc: 0.8494\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t190/254 [=====================>........] - ETA: 0s - loss: 0.3308 - acc: 0.8486\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t206/254 [=======================>......] - ETA: 0s - loss: 0.3302 - acc: 0.8491\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t221/254 [=========================>....] - ETA: 0s - loss: 0.3310 - acc: 0.8492\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t237/254 [==========================>...] - ETA: 0s - loss: 0.3327 - acc: 0.8483\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 3ms/step - loss: 0.3313 - acc: 0.8483 - val_loss: 0.3255 - val_acc: 0.8500\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\tEpoch 00009: LearningRateScheduler reducing learning rate to 0.0100390625.\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\tEpoch 9/20\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 0s - loss: 0.4250 - acc: 0.7969\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t 20/254 [=>............................] - ETA: 0s - loss: 0.3384 - acc: 0.8500\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t 35/254 [===>..........................] - ETA: 0s - loss: 0.3326 - acc: 0.8484\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t 51/254 [=====>........................] - ETA: 0s - loss: 0.3307 - acc: 0.8508\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t 68/254 [=======>......................] - ETA: 0s - loss: 0.3312 - acc: 0.8516\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t 85/254 [=========>....................] - ETA: 0s - loss: 0.3337 - acc: 0.8500\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t102/254 [===========>..................] - ETA: 0s - loss: 0.3346 - acc: 0.8494\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t118/254 [============>.................] - ETA: 0s - loss: 0.3358 - acc: 0.8485\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t134/254 [==============>...............] - ETA: 0s - loss: 0.3320 - acc: 0.8493\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:51 +0000\tmaster-replica-0\t\t150/254 [================>.............] - ETA: 0s - loss: 0.3319 - acc: 0.8489\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t168/254 [==================>...........] - ETA: 0s - loss: 0.3337 - acc: 0.8492\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t187/254 [=====================>........] - ETA: 0s - loss: 0.3332 - acc: 0.8481\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t206/254 [=======================>......] - ETA: 0s - loss: 0.3315 - acc: 0.8489\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t224/254 [=========================>....] - ETA: 0s - loss: 0.3311 - acc: 0.8490\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t242/254 [===========================>..] - ETA: 0s - loss: 0.3304 - acc: 0.8489\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 3ms/step - loss: 0.3314 - acc: 0.8485 - val_loss: 0.3236 - val_acc: 0.8520\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\tEpoch 00010: LearningRateScheduler reducing learning rate to 0.01001953125.\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\tEpoch 10/20\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 0s - loss: 0.2766 - acc: 0.8906\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t 18/254 [=>............................] - ETA: 0s - loss: 0.3305 - acc: 0.8507\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t 33/254 [==>...........................] - ETA: 0s - loss: 0.3337 - acc: 0.8487\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t 48/254 [====>.........................] - ETA: 0s - loss: 0.3414 - acc: 0.8439\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t 62/254 [======>.......................] - ETA: 0s - loss: 0.3370 - acc: 0.8448\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t 73/254 [=======>......................] - ETA: 0s - loss: 0.3332 - acc: 0.8472\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t 84/254 [========>.....................] - ETA: 0s - loss: 0.3283 - acc: 0.8501\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t 94/254 [==========>...................] - ETA: 0s - loss: 0.3290 - acc: 0.8503\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t105/254 [===========>..................] - ETA: 0s - loss: 0.3292 - acc: 0.8510\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t119/254 [=============>................] - ETA: 0s - loss: 0.3262 - acc: 0.8522\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t133/254 [==============>...............] - ETA: 0s - loss: 0.3295 - acc: 0.8505\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t148/254 [================>.............] - ETA: 0s - loss: 0.3296 - acc: 0.8493\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:52 +0000\tmaster-replica-0\t\t164/254 [==================>...........] - ETA: 0s - loss: 0.3290 - acc: 0.8492\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t181/254 [====================>.........] - ETA: 0s - loss: 0.3303 - acc: 0.8485\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t199/254 [======================>.......] - ETA: 0s - loss: 0.3291 - acc: 0.8492\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t217/254 [========================>.....] - ETA: 0s - loss: 0.3297 - acc: 0.8496\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t236/254 [==========================>...] - ETA: 0s - loss: 0.3295 - acc: 0.8496\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 4ms/step - loss: 0.3291 - acc: 0.8494 - val_loss: 0.3264 - val_acc: 0.8516\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\tEpoch 00011: LearningRateScheduler reducing learning rate to 0.010009765625.\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\tEpoch 11/20\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 0s - loss: 0.3474 - acc: 0.8594\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t 18/254 [=>............................] - ETA: 0s - loss: 0.3394 - acc: 0.8572\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t 37/254 [===>..........................] - ETA: 0s - loss: 0.3287 - acc: 0.8554\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t 56/254 [=====>........................] - ETA: 0s - loss: 0.3220 - acc: 0.8559\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t 74/254 [=======>......................] - ETA: 0s - loss: 0.3257 - acc: 0.8545\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t 92/254 [=========>....................] - ETA: 0s - loss: 0.3259 - acc: 0.8527\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t111/254 [============>.................] - ETA: 0s - loss: 0.3267 - acc: 0.8504\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t130/254 [==============>...............] - ETA: 0s - loss: 0.3266 - acc: 0.8497\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t147/254 [================>.............] - ETA: 0s - loss: 0.3260 - acc: 0.8510\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t166/254 [==================>...........] - ETA: 0s - loss: 0.3253 - acc: 0.8500\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t185/254 [====================>.........] - ETA: 0s - loss: 0.3249 - acc: 0.8502\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t204/254 [=======================>......] - ETA: 0s - loss: 0.3257 - acc: 0.8496\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t221/254 [=========================>....] - ETA: 0s - loss: 0.3275 - acc: 0.8496\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t241/254 [===========================>..] - ETA: 0s - loss: 0.3264 - acc: 0.8496\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 3ms/step - loss: 0.3270 - acc: 0.8494 - val_loss: 0.3246 - val_acc: 0.8499\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\tEpoch 00012: LearningRateScheduler reducing learning rate to 0.010004882812500001.\nINFO\t2019-03-27 17:56:53 +0000\tmaster-replica-0\t\tEpoch 12/20\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 0s - loss: 0.3179 - acc: 0.8516\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t 19/254 [=>............................] - ETA: 0s - loss: 0.3353 - acc: 0.8433\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t 39/254 [===>..........................] - ETA: 0s - loss: 0.3319 - acc: 0.8482\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t 59/254 [=====>........................] - ETA: 0s - loss: 0.3268 - acc: 0.8546\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t 79/254 [========>.....................] - ETA: 0s - loss: 0.3309 - acc: 0.8492\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t 99/254 [==========>...................] - ETA: 0s - loss: 0.3315 - acc: 0.8509\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t119/254 [=============>................] - ETA: 0s - loss: 0.3311 - acc: 0.8513\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t135/254 [==============>...............] - ETA: 0s - loss: 0.3284 - acc: 0.8524\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t153/254 [=================>............] - ETA: 0s - loss: 0.3270 - acc: 0.8516\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t172/254 [===================>..........] - ETA: 0s - loss: 0.3263 - acc: 0.8517\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t190/254 [=====================>........] - ETA: 0s - loss: 0.3263 - acc: 0.8513\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t206/254 [=======================>......] - ETA: 0s - loss: 0.3271 - acc: 0.8509\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t225/254 [=========================>....] - ETA: 0s - loss: 0.3274 - acc: 0.8505\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t242/254 [===========================>..] - ETA: 0s - loss: 0.3271 - acc: 0.8505\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 3ms/step - loss: 0.3276 - acc: 0.8500 - val_loss: 0.3452 - val_acc: 0.8444\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\tEpoch 00013: LearningRateScheduler reducing learning rate to 0.01000244140625.\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\tEpoch 13/20\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 0s - loss: 0.3248 - acc: 0.8516\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t 19/254 [=>............................] - ETA: 0s - loss: 0.3278 - acc: 0.8376\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t 38/254 [===>..........................] - ETA: 0s - loss: 0.3317 - acc: 0.8425\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:54 +0000\tmaster-replica-0\t\t 53/254 [=====>........................] - ETA: 0s - loss: 0.3286 - acc: 0.8464\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t 71/254 [=======>......................] - ETA: 0s - loss: 0.3204 - acc: 0.8524\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t 88/254 [=========>....................] - ETA: 0s - loss: 0.3232 - acc: 0.8511\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t105/254 [===========>..................] - ETA: 0s - loss: 0.3271 - acc: 0.8512\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t124/254 [=============>................] - ETA: 0s - loss: 0.3308 - acc: 0.8484\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t141/254 [===============>..............] - ETA: 0s - loss: 0.3316 - acc: 0.8480\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t157/254 [=================>............] - ETA: 0s - loss: 0.3294 - acc: 0.8488\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t171/254 [===================>..........] - ETA: 0s - loss: 0.3298 - acc: 0.8480\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t189/254 [=====================>........] - ETA: 0s - loss: 0.3304 - acc: 0.8482\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t206/254 [=======================>......] - ETA: 0s - loss: 0.3295 - acc: 0.8490\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t225/254 [=========================>....] - ETA: 0s - loss: 0.3315 - acc: 0.8477\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t242/254 [===========================>..] - ETA: 0s - loss: 0.3305 - acc: 0.8481\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 3ms/step - loss: 0.3301 - acc: 0.8485 - val_loss: 0.3439 - val_acc: 0.8439\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\tEpoch 00014: LearningRateScheduler reducing learning rate to 0.010001220703125.\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\tEpoch 14/20\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 1s - loss: 0.3098 - acc: 0.8906\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t 16/254 [>.............................] - ETA: 0s - loss: 0.3104 - acc: 0.8550\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t 33/254 [==>...........................] - ETA: 0s - loss: 0.3184 - acc: 0.8520\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t 49/254 [====>.........................] - ETA: 0s - loss: 0.3204 - acc: 0.8520\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t 64/254 [======>.......................] - ETA: 0s - loss: 0.3218 - acc: 0.8510\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t 80/254 [========>.....................] - ETA: 0s - loss: 0.3252 - acc: 0.8482\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:55 +0000\tmaster-replica-0\t\t 96/254 [==========>...................] - ETA: 0s - loss: 0.3242 - acc: 0.8476\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t112/254 [============>.................] - ETA: 0s - loss: 0.3230 - acc: 0.8488\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t129/254 [==============>...............] - ETA: 0s - loss: 0.3256 - acc: 0.8485\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t144/254 [================>.............] - ETA: 0s - loss: 0.3255 - acc: 0.8480\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t162/254 [==================>...........] - ETA: 0s - loss: 0.3251 - acc: 0.8479\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t177/254 [===================>..........] - ETA: 0s - loss: 0.3249 - acc: 0.8488\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t195/254 [======================>.......] - ETA: 0s - loss: 0.3263 - acc: 0.8482\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t211/254 [=======================>......] - ETA: 0s - loss: 0.3261 - acc: 0.8489\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t227/254 [=========================>....] - ETA: 0s - loss: 0.3257 - acc: 0.8497\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t242/254 [===========================>..] - ETA: 0s - loss: 0.3262 - acc: 0.8502\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 4ms/step - loss: 0.3265 - acc: 0.8503 - val_loss: 0.3399 - val_acc: 0.8413\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\tEpoch 00015: LearningRateScheduler reducing learning rate to 0.0100006103515625.\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\tEpoch 15/20\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 1s - loss: 0.2755 - acc: 0.8594\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t 14/254 [>.............................] - ETA: 0s - loss: 0.3182 - acc: 0.8482\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t 27/254 [==>...........................] - ETA: 0s - loss: 0.3285 - acc: 0.8466\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t 39/254 [===>..........................] - ETA: 0s - loss: 0.3268 - acc: 0.8496\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t 47/254 [====>.........................] - ETA: 0s - loss: 0.3321 - acc: 0.8499\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t 59/254 [=====>........................] - ETA: 0s - loss: 0.3231 - acc: 0.8538\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t 72/254 [=======>......................] - ETA: 0s - loss: 0.3262 - acc: 0.8516\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t 86/254 [=========>....................] - ETA: 0s - loss: 0.3273 - acc: 0.8483\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:56 +0000\tmaster-replica-0\t\t101/254 [==========>...................] - ETA: 0s - loss: 0.3294 - acc: 0.8476\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t117/254 [============>.................] - ETA: 0s - loss: 0.3276 - acc: 0.8488\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t131/254 [==============>...............] - ETA: 0s - loss: 0.3263 - acc: 0.8504\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t148/254 [================>.............] - ETA: 0s - loss: 0.3250 - acc: 0.8513\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t165/254 [==================>...........] - ETA: 0s - loss: 0.3260 - acc: 0.8511\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t182/254 [====================>.........] - ETA: 0s - loss: 0.3261 - acc: 0.8510\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t201/254 [======================>.......] - ETA: 0s - loss: 0.3259 - acc: 0.8504\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t218/254 [========================>.....] - ETA: 0s - loss: 0.3261 - acc: 0.8502\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t237/254 [==========================>...] - ETA: 0s - loss: 0.3266 - acc: 0.8506\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 4ms/step - loss: 0.3285 - acc: 0.8496 - val_loss: 0.3191 - val_acc: 0.8499\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\tEpoch 00016: LearningRateScheduler reducing learning rate to 0.01000030517578125.\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\tEpoch 16/20\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 0s - loss: 0.3200 - acc: 0.8281\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t 20/254 [=>............................] - ETA: 0s - loss: 0.3262 - acc: 0.8441\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t 40/254 [===>..........................] - ETA: 0s - loss: 0.3369 - acc: 0.8436\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t 59/254 [=====>........................] - ETA: 0s - loss: 0.3348 - acc: 0.8451\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t 79/254 [========>.....................] - ETA: 0s - loss: 0.3302 - acc: 0.8462\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t 98/254 [==========>...................] - ETA: 0s - loss: 0.3292 - acc: 0.8478\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t117/254 [============>.................] - ETA: 0s - loss: 0.3301 - acc: 0.8468\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t134/254 [==============>...............] - ETA: 0s - loss: 0.3275 - acc: 0.8482\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t151/254 [================>.............] - ETA: 0s - loss: 0.3253 - acc: 0.8492\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t167/254 [==================>...........] - ETA: 0s - loss: 0.3259 - acc: 0.8493\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:57 +0000\tmaster-replica-0\t\t185/254 [====================>.........] - ETA: 0s - loss: 0.3271 - acc: 0.8489\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t205/254 [=======================>......] - ETA: 0s - loss: 0.3292 - acc: 0.8473\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t224/254 [=========================>....] - ETA: 0s - loss: 0.3284 - acc: 0.8485\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t244/254 [===========================>..] - ETA: 0s - loss: 0.3271 - acc: 0.8495\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 3ms/step - loss: 0.3278 - acc: 0.8495 - val_loss: 0.3245 - val_acc: 0.8537\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\tEpoch 00017: LearningRateScheduler reducing learning rate to 0.010000152587890625.\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\tEpoch 17/20\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 0s - loss: 0.2615 - acc: 0.8828\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t 19/254 [=>............................] - ETA: 0s - loss: 0.3367 - acc: 0.8442\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t 37/254 [===>..........................] - ETA: 0s - loss: 0.3328 - acc: 0.8461\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t 55/254 [=====>........................] - ETA: 0s - loss: 0.3300 - acc: 0.8503\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t 71/254 [=======>......................] - ETA: 0s - loss: 0.3255 - acc: 0.8537\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t 88/254 [=========>....................] - ETA: 0s - loss: 0.3239 - acc: 0.8533\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t105/254 [===========>..................] - ETA: 0s - loss: 0.3245 - acc: 0.8519\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t122/254 [=============>................] - ETA: 0s - loss: 0.3234 - acc: 0.8515\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t139/254 [===============>..............] - ETA: 0s - loss: 0.3218 - acc: 0.8526\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t156/254 [=================>............] - ETA: 0s - loss: 0.3244 - acc: 0.8518\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t172/254 [===================>..........] - ETA: 0s - loss: 0.3267 - acc: 0.8508\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t188/254 [=====================>........] - ETA: 0s - loss: 0.3286 - acc: 0.8497\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t203/254 [======================>.......] - ETA: 0s - loss: 0.3278 - acc: 0.8500\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:58 +0000\tmaster-replica-0\t\t219/254 [========================>.....] - ETA: 0s - loss: 0.3263 - acc: 0.8503\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t236/254 [==========================>...] - ETA: 0s - loss: 0.3270 - acc: 0.8502\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 3ms/step - loss: 0.3270 - acc: 0.8500 - val_loss: 0.3272 - val_acc: 0.8521\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\tEpoch 00018: LearningRateScheduler reducing learning rate to 0.010000076293945313.\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\tEpoch 18/20\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 1s - loss: 0.3422 - acc: 0.8281\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t 16/254 [>.............................] - ETA: 0s - loss: 0.3106 - acc: 0.8506\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t 32/254 [==>...........................] - ETA: 0s - loss: 0.3128 - acc: 0.8508\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t 47/254 [====>.........................] - ETA: 0s - loss: 0.3246 - acc: 0.8477\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t 63/254 [======>.......................] - ETA: 0s - loss: 0.3241 - acc: 0.8508\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t 79/254 [========>.....................] - ETA: 0s - loss: 0.3233 - acc: 0.8495\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t 95/254 [==========>...................] - ETA: 0s - loss: 0.3282 - acc: 0.8479\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t111/254 [============>.................] - ETA: 0s - loss: 0.3282 - acc: 0.8470\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t126/254 [=============>................] - ETA: 0s - loss: 0.3266 - acc: 0.8486\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t145/254 [================>.............] - ETA: 0s - loss: 0.3273 - acc: 0.8484\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t163/254 [==================>...........] - ETA: 0s - loss: 0.3287 - acc: 0.8471\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t178/254 [====================>.........] - ETA: 0s - loss: 0.3267 - acc: 0.8485\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t194/254 [=====================>........] - ETA: 0s - loss: 0.3253 - acc: 0.8494\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t209/254 [=======================>......] - ETA: 0s - loss: 0.3248 - acc: 0.8497\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t224/254 [=========================>....] - ETA: 0s - loss: 0.3249 - acc: 0.8495\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t240/254 [===========================>..] - ETA: 0s - loss: 0.3254 - acc: 0.8494\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 4ms/step - loss: 0.3244 - acc: 0.8493 - val_loss: 0.3271 - val_acc: 0.8508\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\tEpoch 00019: LearningRateScheduler reducing learning rate to 0.010000038146972657.\nINFO\t2019-03-27 17:56:59 +0000\tmaster-replica-0\t\tEpoch 19/20\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 1s - loss: 0.4041 - acc: 0.8516\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t 16/254 [>.............................] - ETA: 0s - loss: 0.3522 - acc: 0.8467\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t 33/254 [==>...........................] - ETA: 0s - loss: 0.3445 - acc: 0.8492\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t 50/254 [====>.........................] - ETA: 0s - loss: 0.3336 - acc: 0.8500\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t 65/254 [======>.......................] - ETA: 0s - loss: 0.3331 - acc: 0.8520\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t 84/254 [========>.....................] - ETA: 0s - loss: 0.3301 - acc: 0.8520\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t102/254 [===========>..................] - ETA: 0s - loss: 0.3289 - acc: 0.8513\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t118/254 [============>.................] - ETA: 0s - loss: 0.3290 - acc: 0.8505\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t134/254 [==============>...............] - ETA: 0s - loss: 0.3310 - acc: 0.8502\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t154/254 [=================>............] - ETA: 0s - loss: 0.3283 - acc: 0.8517\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t173/254 [===================>..........] - ETA: 0s - loss: 0.3270 - acc: 0.8511\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t190/254 [=====================>........] - ETA: 0s - loss: 0.3266 - acc: 0.8501\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t201/254 [======================>.......] - ETA: 0s - loss: 0.3260 - acc: 0.8504\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t217/254 [========================>.....] - ETA: 0s - loss: 0.3281 - acc: 0.8502\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t231/254 [==========================>...] - ETA: 0s - loss: 0.3281 - acc: 0.8502\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t242/254 [===========================>..] - ETA: 0s - loss: 0.3270 - acc: 0.8508\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 4ms/step - loss: 0.3281 - acc: 0.8504 - val_loss: 0.3239 - val_acc: 0.8521\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\tEpoch 00020: LearningRateScheduler reducing learning rate to 0.010000019073486329.\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\tEpoch 20/20\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t 1/254 [..............................] - ETA: 0s - loss: 0.2766 - acc: 0.8672\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:00 +0000\tmaster-replica-0\t\t 20/254 [=>............................] - ETA: 0s - loss: 0.3260 - acc: 0.8504\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:01 +0000\tmaster-replica-0\t\t 37/254 [===>..........................] - ETA: 0s - loss: 0.3271 - acc: 0.8476\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:01 +0000\tmaster-replica-0\t\t 58/254 [=====>........................] - ETA: 0s - loss: 0.3302 - acc: 0.8479\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:01 +0000\tmaster-replica-0\t\t 79/254 [========>.....................] - ETA: 0s - loss: 0.3354 - acc: 0.8473\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:01 +0000\tmaster-replica-0\t\t 98/254 [==========>...................] - ETA: 0s - loss: 0.3344 - acc: 0.8465\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:01 +0000\tmaster-replica-0\t\t119/254 [=============>................] - ETA: 0s - loss: 0.3318 - acc: 0.8491\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:01 +0000\tmaster-replica-0\t\t139/254 [===============>..............] - ETA: 0s - loss: 0.3314 - acc: 0.8502\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:01 +0000\tmaster-replica-0\t\t156/254 [=================>............] - ETA: 0s - loss: 0.3316 - acc: 0.8502\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:01 +0000\tmaster-replica-0\t\t175/254 [===================>..........] - ETA: 0s - loss: 0.3317 - acc: 0.8510\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:01 +0000\tmaster-replica-0\t\t192/254 [=====================>........] - ETA: 0s - loss: 0.3309 - acc: 0.8508\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:01 +0000\tmaster-replica-0\t\t212/254 [========================>.....] - ETA: 0s - loss: 0.3296 - acc: 0.8507\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:01 +0000\tmaster-replica-0\t\t231/254 [==========================>...] - ETA: 0s - loss: 0.3282 - acc: 0.8509\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:01 +0000\tmaster-replica-0\t\t250/254 [============================>.] - ETA: 0s - loss: 0.3294 - acc: 0.8508\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\nINFO\t2019-03-27 17:57:01 +0000\tmaster-replica-0\t\t254/254 [==============================] - 1s 3ms/step - loss: 0.3294 - acc: 0.8508 - val_loss: 0.3282 - val_acc: 0.8519\nWARNING\t2019-03-27 17:57:03 +0000\tmaster-replica-0\t\tThis model was compiled with a Keras optimizer (<tensorflow.python.keras.optimizers.RMSprop object at 0x7f3904fa0518>) but is being saved in TensorFlow format with `save_weights`. The model's weights will be saved, but unlike with TensorFlow optimizers in the TensorFlow format the optimizer's state will not be saved.\nWARNING\t2019-03-27 17:57:03 +0000\tmaster-replica-0\t\t\nWARNING\t2019-03-27 17:57:03 +0000\tmaster-replica-0\t\tConsider using a TensorFlow optimizer from `tf.train`.\nWARNING\t2019-03-27 17:57:05 +0000\tmaster-replica-0\t\tFrom /usr/local/lib/python3.5/dist-packages/tensorflow/python/keras/engine/network.py:1436: update_checkpoint_state (from tensorflow.python.training.checkpoint_management) is deprecated and will be removed in a future version.\nWARNING\t2019-03-27 17:57:05 +0000\tmaster-replica-0\t\tInstructions for updating:\nWARNING\t2019-03-27 17:57:05 +0000\tmaster-replica-0\t\tUse tf.train.CheckpointManager to manage checkpoints rather than manually editing the Checkpoint proto.\nWARNING\t2019-03-27 17:57:05 +0000\tmaster-replica-0\t\tModel was compiled with an optimizer, but the optimizer is not from `tf.train` (e.g. `tf.train.AdagradOptimizer`). Only the serving graph was exported. The train and evaluate graphs were not added to the SavedModel.\nWARNING\t2019-03-27 17:57:10 +0000\tmaster-replica-0\t\tFrom /usr/local/lib/python3.5/dist-packages/tensorflow/python/saved_model/signature_def_utils_impl.py:205: build_tensor_info (from tensorflow.python.saved_model.utils_impl) is deprecated and will be removed in a future version.\nWARNING\t2019-03-27 17:57:10 +0000\tmaster-replica-0\t\tInstructions for updating:\nWARNING\t2019-03-27 17:57:10 +0000\tmaster-replica-0\t\tThis function will only be available through the v1 compatibility library as tf.compat.v1.saved_model.utils.build_tensor_info or tf.compat.v1.saved_model.build_tensor_info.\nINFO\t2019-03-27 17:57:10 +0000\tmaster-replica-0\t\tSignatures INCLUDED in export for Classify: None\nINFO\t2019-03-27 17:57:10 +0000\tmaster-replica-0\t\tSignatures INCLUDED in export for Eval: None\nINFO\t2019-03-27 17:57:10 +0000\tmaster-replica-0\t\tSignatures INCLUDED in export for Predict: ['serving_default']\nINFO\t2019-03-27 17:57:10 +0000\tmaster-replica-0\t\tSignatures INCLUDED in export for Regress: None\nINFO\t2019-03-27 17:57:10 +0000\tmaster-replica-0\t\tSignatures INCLUDED in export for Train: None\nINFO\t2019-03-27 17:57:10 +0000\tmaster-replica-0\t\tNo assets to save.\nINFO\t2019-03-27 17:57:10 +0000\tmaster-replica-0\t\tNo assets to write.\nINFO\t2019-03-27 17:57:10 +0000\tmaster-replica-0\t\tSavedModel written to: gs://<your-bucket-name>/keras-job-dir/keras_export/1553709421/saved_model.pb\nINFO\t2019-03-27 17:57:11 +0000\tmaster-replica-0\t\tModel exported to: gs://<your-bucket-name>/keras-job-dir/keras_export/1553709421\nINFO\t2019-03-27 17:57:11 +0000\tmaster-replica-0\t\tModule completed; cleaning up.\nINFO\t2019-03-27 17:57:11 +0000\tmaster-replica-0\t\tClean up finished.\nINFO\t2019-03-27 17:57:11 +0000\tmaster-replica-0\t\tTask completed successfully.\nendTime: '2019-03-27T18:01:46'\njobId: my_first_keras_job\nstartTime: '2019-03-27T17:55:34'\nstate: SUCCEEDED\n"
]
],
[
[
"## Part 2. Quickstart for online predictions in AI Platform\n\nThis section shows how to use AI Platform and your trained model from Part 1\nto predict a person's income bracket from other Census information about them.",
"_____no_output_____"
],
[
"### Create model and version resources in AI Platform\n\nTo serve online predictions using the model you trained and exported in Part 1,\ncreate a *model* resource in AI Platform and a *version* resource\nwithin it. The version resource is what actually uses your trained model to\nserve predictions. This structure lets you adjust and retrain your model many times and\norganize all the versions together in AI Platform. Learn more about [models\nand\nversions](https://cloud.google.com/ml-engine/docs/tensorflow/projects-models-versions-jobs).\n\nFirst, name and create the model resource:",
"_____no_output_____"
]
],
[
[
"MODEL_NAME = \"my_first_keras_model\"\n\n! gcloud ml-engine models create $MODEL_NAME \\\n --regions $REGION",
"Created AI Platform model [projects/<your-project-id>/models/my_first_keras_model].\n"
]
],
[
[
"Next, create the model version. The training job from Part 1 exported a timestamped\n[TensorFlow SavedModel\ndirectory](https://www.tensorflow.org/guide/saved_model#structure_of_a_savedmodel_directory)\nto your Cloud Storage bucket. AI Platform uses this directory to create a\nmodel version. Learn more about [SavedModel and Cloud ML\nEngine](https://cloud.google.com/ml-engine/docs/tensorflow/deploying-models).\n\nYou may be able to find the path to this directory in your training job's logs.\nLook for a line like:\n\n```\nModel exported to: gs://<your-bucket-name>/keras-job-dir/keras_export/1545439782\n```\n\nExecute the following command to identify your SavedModel directory and use it to create a model version resource:",
"_____no_output_____"
]
],
[
[
"MODEL_VERSION = \"v1\"\n\n# Get a list of directories in the `keras_export` parent directory\nKERAS_EXPORT_DIRS = ! gsutil ls $JOB_DIR/keras_export/\n\n# Pick the directory with the latest timestamp, in case you've trained\n# multiple times\nSAVED_MODEL_PATH = KERAS_EXPORT_DIRS[-1]\n\n# Create model version based on that SavedModel directory\n! gcloud ml-engine versions create $MODEL_VERSION \\\n --model $MODEL_NAME \\\n --runtime-version 1.13 \\\n --python-version 3.5 \\\n --framework tensorflow \\\n --origin $SAVED_MODEL_PATH",
"_____no_output_____"
]
],
[
[
"### Prepare input for prediction\n\nTo receive valid and useful predictions, you must preprocess input for prediction in the same way that training data was preprocessed. In a production\nsystem, you may want to create a preprocessing pipeline that can be used identically at training time and prediction time.\n\nFor this exercise, use the training package's data-loading code to select a random sample from the evaluation data. This data is in the form that was used to evaluate accuracy after each epoch of training, so it can be used to send test predictions without further preprocessing:",
"_____no_output_____"
]
],
[
[
"from trainer import util\n\n_, _, eval_x, eval_y = util.load_data()\n\nprediction_input = eval_x.sample(20)\nprediction_targets = eval_y[prediction_input.index]\n\nprediction_input",
"_____no_output_____"
]
],
[
[
"Notice that categorical fields, like `occupation`, have already been converted to integers (with the same mapping that was used for training). Numerical fields, like `age`, have been scaled to a\n[z-score](https://developers.google.com/machine-learning/crash-course/representation/cleaning-data). Some fields have been dropped from the original\ndata. Compare the prediction input with the raw data for the same examples:",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\n_, eval_file_path = util.download(util.DATA_DIR)\nraw_eval_data = pd.read_csv(eval_file_path,\n names=util._CSV_COLUMNS,\n na_values='?')\n\nraw_eval_data.iloc[prediction_input.index]",
"_____no_output_____"
]
],
[
[
"Export the prediction input to a newline-delimited JSON file:",
"_____no_output_____"
]
],
[
[
"import json\n\nwith open('prediction_input.json', 'w') as json_file:\n for row in prediction_input.values.tolist():\n json.dump(row, json_file)\n json_file.write('\\n')\n\n! cat prediction_input.json",
"[0.9012127751273994, 1.0, 1.525541514460902, 2.0, 9.0, 0.0, 4.0, -0.14479173735784842, -0.21713186390175285, -0.43754385253479555, 38.0]\n[-0.9221541171760282, 3.0, -0.4192650914017433, 4.0, 2.0, 3.0, 4.0, -0.14479173735784842, -0.21713186390175285, -0.03403923708700391, 38.0]\n[-1.2138928199445767, 3.0, -0.030303770229214273, 4.0, 10.0, 1.0, 4.0, -0.14479173735784842, -0.21713186390175285, 1.5799792247041626, 38.0]\n[-0.6304154144074798, 3.0, 0.35865755094331475, 4.0, 0.0, 3.0, 4.0, -0.14479173735784842, -0.21713186390175285, -0.6796466218034705, 38.0]\n[-1.5056315227131252, 3.0, -1.5861490549193304, 4.0, 7.0, 3.0, 0.0, -0.14479173735784842, -0.21713186390175285, -0.03403923708700391, 38.0]\n[-0.11987268456252011, 5.0, 0.35865755094331475, 2.0, 2.0, 0.0, 4.0, -0.14479173735784842, -0.21713186390175285, -0.8410484679825871, 38.0]\n[0.24480069389816542, 3.0, 1.525541514460902, 2.0, 9.0, 0.0, 4.0, -0.14479173735784842, -0.21713186390175285, 1.176474609256371, 6.0]\n[0.0989313425138912, 1.0, 1.525541514460902, 2.0, 9.0, 0.0, 4.0, 0.8868473744801746, -0.21713186390175285, -0.03403923708700391, 38.0]\n[0.39067004528243965, 5.0, -0.030303770229214273, 2.0, 4.0, 0.0, 4.0, -0.14479173735784842, -0.21713186390175285, 4.7273152251969375, 38.0]\n[1.1200168022038106, 3.0, 1.1365801932883728, 2.0, 12.0, 0.0, 4.0, -0.14479173735784842, -0.21713186390175285, -0.03403923708700391, 38.0]\n[-1.2868274956367138, 3.0, -0.030303770229214273, 4.0, 11.0, 3.0, 4.0, -0.14479173735784842, -0.21713186390175285, -1.6480576988781703, 38.0]\n[-0.7033500900996169, 3.0, -0.4192650914017433, 2.0, 7.0, 5.0, 4.0, -0.14479173735784842, 4.5022796885373735, -0.43754385253479555, 38.0]\n[0.5365393966667138, 3.0, 1.525541514460902, 4.0, 3.0, 4.0, 4.0, -0.14479173735784842, -0.21713186390175285, -0.03403923708700391, 38.0]\n[-1.3597621713288508, 3.0, -0.4192650914017433, 4.0, 6.0, 3.0, 2.0, -0.14479173735784842, -0.21713186390175285, -0.03403923708700391, 38.0]\n[0.5365393966667138, 3.0, 1.1365801932883728, 0.0, 11.0, 2.0, 2.0, -0.14479173735784842, -0.21713186390175285, -0.03403923708700391, 38.0]\n[1.338820829280222, 3.0, -0.4192650914017433, 2.0, 2.0, 0.0, 4.0, -0.14479173735784842, -0.21713186390175285, -0.03403923708700391, 38.0]\n[0.6824087480509881, 3.0, 1.1365801932883728, 0.0, 12.0, 3.0, 4.0, -0.14479173735784842, -0.21713186390175285, -0.03403923708700391, 38.0]\n[0.0259966668217541, 3.0, 1.1365801932883728, 2.0, 11.0, 0.0, 4.0, -0.14479173735784842, -0.21713186390175285, 0.3694653783607877, 38.0]\n[-0.4116113873310685, 3.0, -0.4192650914017433, 2.0, 11.0, 0.0, 4.0, -0.14479173735784842, -0.21713186390175285, 1.176474609256371, 38.0]\n[0.39067004528243965, 6.0, 1.1365801932883728, 4.0, 4.0, 1.0, 4.0, -0.14479173735784842, -0.21713186390175285, -0.03403923708700391, 38.0]\n"
]
],
[
[
"The `gcloud` command-line tool accepts newline-delimited JSON for online\nprediction, and this particular Keras model expects a flat list of\nnumbers for each input example.\n\nAI Platform requires a different format when you make online prediction requests to the REST API without using the `gcloud` tool. The way you structure\nyour model may also change how you must format data for prediction. Learn more\nabout [formatting data for online\nprediction](https://cloud.google.com/ml-engine/docs/tensorflow/prediction-overview#prediction_input_data).",
"_____no_output_____"
],
[
"### Submit the online prediction request",
"_____no_output_____"
],
[
"Use `gcloud` to submit your online prediction request.",
"_____no_output_____"
]
],
[
[
"! gcloud ml-engine predict \\\n --model $MODEL_NAME \\\n --version $MODEL_VERSION \\\n --json-instances prediction_input.json",
"DENSE_4\n[0.6854287385940552]\n[0.011786997318267822]\n[0.037236183881759644]\n[0.016223609447479248]\n[0.0012015104293823242]\n[0.23621389269828796]\n[0.6174039244651794]\n[0.9822691679000854]\n[0.3815768361091614]\n[0.6715215444564819]\n[0.001094043254852295]\n[0.43077391386032104]\n[0.22132840752601624]\n[0.004075437784194946]\n[0.22736871242523193]\n[0.4111979305744171]\n[0.27328649163246155]\n[0.6981356143951416]\n[0.3309604525566101]\n[0.20807647705078125]\n"
]
],
[
[
"Since the model's last layer uses a [sigmoid function](https://developers.google.com/machine-learning/glossary/#sigmoid_function) for its activation, outputs between 0 and 0.5 represent negative predictions (\"<=50K\") and outputs between 0.5 and 1 represent positive ones (\">50K\").\n\nDo the predicted income brackets match the actual ones? Run the following cell\nto see the true labels.",
"_____no_output_____"
]
],
[
[
"prediction_targets",
"_____no_output_____"
]
],
[
[
"## Part 3. Developing the Keras model from scratch\n\nAt this point, you have trained a machine learning model on AI Platform, deployed the trained model as a version resource on AI Platform, and received online predictions from the deployment. The next section walks through recreating the Keras code used to train your model. It covers the following parts of developing a machine learning model for use with AI Platform:\n\n* Downloading and preprocessing data\n* Designing and training the model\n* Visualizing training and exporting the trained model\n\nWhile this section provides more detailed insight to the tasks completed in previous parts, to learn more about using `tf.keras`, read [TensorFlow's guide to Keras](https://www.tensorflow.org/tutorials/keras). To learn more about structuring code as a training packge for AI Platform, read [Packaging a training application](https://cloud.google.com/ml-engine/docs/tensorflow/packaging-trainer) and reference the [complete training code](https://github.com/GoogleCloudPlatform/cloudml-samples/tree/master/census/tf-keras), which is structured as a Python package.",
"_____no_output_____"
],
[
"### Import libraries and define constants\n\nFirst, import Python libraries required for training:",
"_____no_output_____"
]
],
[
[
"import os\nfrom six.moves import urllib\nimport tempfile\n\nimport numpy as np\nimport pandas as pd\nimport tensorflow as tf\n\n# Examine software versions\nprint(__import__('sys').version)\nprint(tf.__version__)\nprint(tf.keras.__version__)",
"3.6.7 (default, Oct 22 2018, 11:32:17) \n[GCC 8.2.0]\n1.13.1\n2.2.4-tf\n"
]
],
[
[
"Then, define some useful constants:\n\n* Information for downloading training and evaluation data\n* Information required for Pandas to interpret the data and convert categorical fields into numeric features\n* Hyperparameters for training, such as learning rate and batch size",
"_____no_output_____"
]
],
[
[
"### For downloading data ###\n\n# Storage directory\nDATA_DIR = os.path.join(tempfile.gettempdir(), 'census_data')\n\n# Download options.\nDATA_URL = 'https://storage.googleapis.com/cloud-samples-data/ml-engine' \\\n '/census/data'\nTRAINING_FILE = 'adult.data.csv'\nEVAL_FILE = 'adult.test.csv'\nTRAINING_URL = '%s/%s' % (DATA_URL, TRAINING_FILE)\nEVAL_URL = '%s/%s' % (DATA_URL, EVAL_FILE)\n\n### For interpreting data ###\n\n# These are the features in the dataset.\n# Dataset information: https://archive.ics.uci.edu/ml/datasets/census+income\n_CSV_COLUMNS = [\n 'age', 'workclass', 'fnlwgt', 'education', 'education_num',\n 'marital_status', 'occupation', 'relationship', 'race', 'gender',\n 'capital_gain', 'capital_loss', 'hours_per_week', 'native_country',\n 'income_bracket'\n]\n\n_CATEGORICAL_TYPES = {\n 'workclass': pd.api.types.CategoricalDtype(categories=[\n 'Federal-gov', 'Local-gov', 'Never-worked', 'Private', 'Self-emp-inc',\n 'Self-emp-not-inc', 'State-gov', 'Without-pay'\n ]),\n 'marital_status': pd.api.types.CategoricalDtype(categories=[\n 'Divorced', 'Married-AF-spouse', 'Married-civ-spouse',\n 'Married-spouse-absent', 'Never-married', 'Separated', 'Widowed'\n ]),\n 'occupation': pd.api.types.CategoricalDtype([\n 'Adm-clerical', 'Armed-Forces', 'Craft-repair', 'Exec-managerial',\n 'Farming-fishing', 'Handlers-cleaners', 'Machine-op-inspct',\n 'Other-service', 'Priv-house-serv', 'Prof-specialty', 'Protective-serv',\n 'Sales', 'Tech-support', 'Transport-moving'\n ]),\n 'relationship': pd.api.types.CategoricalDtype(categories=[\n 'Husband', 'Not-in-family', 'Other-relative', 'Own-child', 'Unmarried',\n 'Wife'\n ]),\n 'race': pd.api.types.CategoricalDtype(categories=[\n 'Amer-Indian-Eskimo', 'Asian-Pac-Islander', 'Black', 'Other', 'White'\n ]),\n 'native_country': pd.api.types.CategoricalDtype(categories=[\n 'Cambodia', 'Canada', 'China', 'Columbia', 'Cuba', 'Dominican-Republic',\n 'Ecuador', 'El-Salvador', 'England', 'France', 'Germany', 'Greece',\n 'Guatemala', 'Haiti', 'Holand-Netherlands', 'Honduras', 'Hong', 'Hungary',\n 'India', 'Iran', 'Ireland', 'Italy', 'Jamaica', 'Japan', 'Laos', 'Mexico',\n 'Nicaragua', 'Outlying-US(Guam-USVI-etc)', 'Peru', 'Philippines', 'Poland',\n 'Portugal', 'Puerto-Rico', 'Scotland', 'South', 'Taiwan', 'Thailand',\n 'Trinadad&Tobago', 'United-States', 'Vietnam', 'Yugoslavia'\n ]),\n 'income_bracket': pd.api.types.CategoricalDtype(categories=[\n '<=50K', '>50K'\n ])\n}\n\n# This is the label (target) we want to predict.\n_LABEL_COLUMN = 'income_bracket'\n\n### Hyperparameters for training ###\n\n# This the training batch size\nBATCH_SIZE = 128\n\n# This is the number of epochs (passes over the full training data)\nNUM_EPOCHS = 20\n\n# Define learning rate.\nLEARNING_RATE = .01",
"_____no_output_____"
]
],
[
[
"### Download and preprocess data",
"_____no_output_____"
],
[
"#### Download the data\n\nNext, define functions to download training and evaluation data. These functions also fix minor irregularities in the data's formatting.",
"_____no_output_____"
]
],
[
[
"def _download_and_clean_file(filename, url):\n \"\"\"Downloads data from url, and makes changes to match the CSV format.\n \n The CSVs may use spaces after the comma delimters (non-standard) or include\n rows which do not represent well-formed examples. This function strips out\n some of these problems.\n \n Args:\n filename: filename to save url to\n url: URL of resource to download\n \"\"\"\n temp_file, _ = urllib.request.urlretrieve(url)\n with tf.gfile.Open(temp_file, 'r') as temp_file_object:\n with tf.gfile.Open(filename, 'w') as file_object:\n for line in temp_file_object:\n line = line.strip()\n line = line.replace(', ', ',')\n if not line or ',' not in line:\n continue\n if line[-1] == '.':\n line = line[:-1]\n line += '\\n'\n file_object.write(line)\n tf.gfile.Remove(temp_file)\n\n\ndef download(data_dir):\n \"\"\"Downloads census data if it is not already present.\n \n Args:\n data_dir: directory where we will access/save the census data\n \"\"\"\n tf.gfile.MakeDirs(data_dir)\n\n training_file_path = os.path.join(data_dir, TRAINING_FILE)\n if not tf.gfile.Exists(training_file_path):\n _download_and_clean_file(training_file_path, TRAINING_URL)\n\n eval_file_path = os.path.join(data_dir, EVAL_FILE)\n if not tf.gfile.Exists(eval_file_path):\n _download_and_clean_file(eval_file_path, EVAL_URL)\n\n return training_file_path, eval_file_path",
"_____no_output_____"
]
],
[
[
"Use those functions to download the data for training and verify that you have CSV files for training and evaluation: ",
"_____no_output_____"
]
],
[
[
"training_file_path, eval_file_path = download(DATA_DIR)\n\n# You should see 2 files: adult.data.csv and adult.test.csv\n!ls -l $DATA_DIR",
"total 5156\n-rw-r--r-- 1 root root 3518450 Mar 27 17:52 adult.data.csv\n-rw-r--r-- 1 root root 1758573 Mar 27 17:52 adult.test.csv\n"
]
],
[
[
"Next, load these files using Pandas and examine the data:",
"_____no_output_____"
]
],
[
[
"# This census data uses the value '?' for fields (column) that are missing data. \n# We use na_values to find ? and set it to NaN values.\n# https://pandas.pydata.org/pandas-docs/stable/generated/pandas.read_csv.html\n\ntrain_df = pd.read_csv(training_file_path, names=_CSV_COLUMNS, na_values='?')\neval_df = pd.read_csv(eval_file_path, names=_CSV_COLUMNS, na_values='?')\n\n# Here's what the data looks like before we preprocess the data.\ntrain_df.head()",
"_____no_output_____"
]
],
[
[
"#### Preprocess the data\n\nThe first preprocessing step removes certain features from the data and\nconverts categorical features to numerical values for use with Keras.\n\nLearn more about [feature engineering](https://developers.google.com/machine-learning/crash-course/representation/feature-engineering) and [bias in data](https://developers.google.com/machine-learning/crash-course/fairness/types-of-bias).",
"_____no_output_____"
]
],
[
[
"UNUSED_COLUMNS = ['fnlwgt', 'education', 'gender']\n\n\ndef preprocess(dataframe):\n \"\"\"Converts categorical features to numeric. Removes unused columns.\n \n Args:\n dataframe: Pandas dataframe with raw data\n \n Returns:\n Dataframe with preprocessed data\n \"\"\"\n dataframe = dataframe.drop(columns=UNUSED_COLUMNS)\n\n # Convert integer valued (numeric) columns to floating point\n numeric_columns = dataframe.select_dtypes(['int64']).columns\n dataframe[numeric_columns] = dataframe[numeric_columns].astype('float32')\n\n # Convert categorical columns to numeric\n cat_columns = dataframe.select_dtypes(['object']).columns\n dataframe[cat_columns] = dataframe[cat_columns].apply(lambda x: x.astype(\n _CATEGORICAL_TYPES[x.name]))\n dataframe[cat_columns] = dataframe[cat_columns].apply(lambda x: x.cat.codes)\n return dataframe\n\n\nprepped_train_df = preprocess(train_df)\nprepped_eval_df = preprocess(eval_df)",
"_____no_output_____"
]
],
[
[
"Run the following cell to see how preprocessing changed the data. Notice in particular that `income_bracket`, the label that you're training the model to predict, has changed from `<=50K` and `>50K` to `0` and `1`:",
"_____no_output_____"
]
],
[
[
"prepped_train_df.head()",
"_____no_output_____"
]
],
[
[
"Next, separate the data into features (\"x\") and labels (\"y\"), and reshape the label arrays into a format for use with `tf.data.Dataset` later:",
"_____no_output_____"
]
],
[
[
"# Split train and test data with labels.\n# The pop() method will extract (copy) and remove the label column from the dataframe\ntrain_x, train_y = prepped_train_df, prepped_train_df.pop(_LABEL_COLUMN)\neval_x, eval_y = prepped_eval_df, prepped_eval_df.pop(_LABEL_COLUMN)\n\n# Reshape label columns for use with tf.data.Dataset\ntrain_y = np.asarray(train_y).astype('float32').reshape((-1, 1))\neval_y = np.asarray(eval_y).astype('float32').reshape((-1, 1))",
"_____no_output_____"
]
],
[
[
"Scaling training data so each numerical feature column has a mean of 0 and a standard deviation of 1 [can improve your model](https://developers.google.com/machine-learning/crash-course/representation/cleaning-data).\n\nIn a production system, you may want to save the means and standard deviations from your training set and use them to perform an identical transformation on test data at prediction time. For convenience in this exercise, temporarily combine the training and evaluation data to scale all of them:",
"_____no_output_____"
]
],
[
[
"def standardize(dataframe):\n \"\"\"Scales numerical columns using their means and standard deviation to get\n z-scores: the mean of each numerical column becomes 0, and the standard\n deviation becomes 1. This can help the model converge during training.\n \n Args:\n dataframe: Pandas dataframe\n \n Returns:\n Input dataframe with the numerical columns scaled to z-scores\n \"\"\"\n dtypes = list(zip(dataframe.dtypes.index, map(str, dataframe.dtypes)))\n # Normalize numeric columns.\n for column, dtype in dtypes:\n if dtype == 'float32':\n dataframe[column] -= dataframe[column].mean()\n dataframe[column] /= dataframe[column].std()\n return dataframe\n\n\n# Join train_x and eval_x to normalize on overall means and standard\n# deviations. Then separate them again.\nall_x = pd.concat([train_x, eval_x], keys=['train', 'eval'])\nall_x = standardize(all_x)\ntrain_x, eval_x = all_x.xs('train'), all_x.xs('eval')",
"_____no_output_____"
]
],
[
[
"Finally, examine some of your fully preprocessed training data:",
"_____no_output_____"
]
],
[
[
"# Verify dataset features\n# Note how only the numeric fields (not categorical) have been standardized\ntrain_x.head()",
"_____no_output_____"
]
],
[
[
"### Design and train the model",
"_____no_output_____"
],
[
"#### Create training and validation datasets\n\nCreate an input function to convert features and labels into a\n[`tf.data.Dataset`](https://www.tensorflow.org/guide/datasets) for training or evaluation:",
"_____no_output_____"
]
],
[
[
"def input_fn(features, labels, shuffle, num_epochs, batch_size):\n \"\"\"Generates an input function to be used for model training.\n \n Args:\n features: numpy array of features used for training or inference\n labels: numpy array of labels for each example\n shuffle: boolean for whether to shuffle the data or not (set True for\n training, False for evaluation)\n num_epochs: number of epochs to provide the data for\n batch_size: batch size for training\n \n Returns:\n A tf.data.Dataset that can provide data to the Keras model for training or\n evaluation\n \"\"\"\n if labels is None:\n inputs = features\n else:\n inputs = (features, labels)\n dataset = tf.data.Dataset.from_tensor_slices(inputs)\n\n if shuffle:\n dataset = dataset.shuffle(buffer_size=len(features))\n\n # We call repeat after shuffling, rather than before, to prevent separate\n # epochs from blending together.\n dataset = dataset.repeat(num_epochs)\n dataset = dataset.batch(batch_size)\n return dataset",
"_____no_output_____"
]
],
[
[
"Next, create these training and evaluation datasets.Use the `NUM_EPOCHS`\nand `BATCH_SIZE` hyperparameters defined previously to define how the training\ndataset provides examples to the model during training. Set up the validation\ndataset to provide all its examples in one batch, for a single validation step\nat the end of each training epoch.",
"_____no_output_____"
]
],
[
[
"# Pass a numpy array by using DataFrame.values\ntraining_dataset = input_fn(features=train_x.values, \n labels=train_y, \n shuffle=True, \n num_epochs=NUM_EPOCHS, \n batch_size=BATCH_SIZE)\n\nnum_eval_examples = eval_x.shape[0]\n\n# Pass a numpy array by using DataFrame.values\nvalidation_dataset = input_fn(features=eval_x.values, \n labels=eval_y, \n shuffle=False, \n num_epochs=NUM_EPOCHS, \n batch_size=num_eval_examples) ",
"_____no_output_____"
]
],
[
[
"#### Design a Keras Model",
"_____no_output_____"
],
[
"Design your neural network using the [Keras Sequential API](https://www.tensorflow.org/guide/keras#sequential_model).\n\nThis deep neural network (DNN) has several hidden layers, and the last layer uses a sigmoid activation function to output a value between 0 and 1:\n\n* The input layer has 100 units using the ReLU activation function.\n* The hidden layer has 75 units using the ReLU activation function.\n* The hidden layer has 50 units using the ReLU activation function.\n* The hidden layer has 25 units using the ReLU activation function.\n* The output layer has 1 units using a sigmoid activation function.\n* The optimizer uses the binary cross-entropy loss function, which is appropriate for a binary classification problem like this one.\n\nFeel free to change these layers to try to improve the model:",
"_____no_output_____"
]
],
[
[
"def create_keras_model(input_dim, learning_rate):\n \"\"\"Creates Keras Model for Binary Classification.\n \n Args:\n input_dim: How many features the input has\n learning_rate: Learning rate for training\n \n Returns:\n The compiled Keras model (still needs to be trained)\n \"\"\"\n model = tf.keras.Sequential()\n model.add(\n tf.keras.layers.Dense(\n 100,\n activation=tf.nn.relu,\n kernel_initializer='uniform',\n input_shape=(input_dim,)))\n model.add(tf.keras.layers.Dense(75, activation=tf.nn.relu))\n model.add(tf.keras.layers.Dense(50, activation=tf.nn.relu))\n model.add(tf.keras.layers.Dense(25, activation=tf.nn.relu))\n # The single output node and Sigmoid activation makes this a Logistic\n # Regression.\n model.add(tf.keras.layers.Dense(1, activation=tf.nn.sigmoid))\n\n # Custom Optimizer:\n # https://www.tensorflow.org/api_docs/python/tf/train/RMSPropOptimizer\n optimizer = tf.keras.optimizers.RMSprop(\n lr=learning_rate, rho=0.9, epsilon=1e-08, decay=0.0)\n\n # Compile Keras model\n model.compile(\n loss='binary_crossentropy', optimizer=optimizer, metrics=['accuracy'])\n return model",
"_____no_output_____"
]
],
[
[
"Next, create the Keras model object and examine its structure:",
"_____no_output_____"
]
],
[
[
"num_train_examples, input_dim = train_x.shape\nprint('Number of features: {}'.format(input_dim))\nprint('Number of examples: {}'.format(num_train_examples))\n\nkeras_model = create_keras_model(\n input_dim=input_dim,\n learning_rate=LEARNING_RATE)\n\n# Take a detailed look inside the model\nkeras_model.summary()",
"Number of features: 11\nNumber of examples: 32561\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/ops/resource_variable_ops.py:435: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nColocations handled automatically by placer.\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense (Dense) (None, 100) 1200 \n_________________________________________________________________\ndense_1 (Dense) (None, 75) 7575 \n_________________________________________________________________\ndense_2 (Dense) (None, 50) 3800 \n_________________________________________________________________\ndense_3 (Dense) (None, 25) 1275 \n_________________________________________________________________\ndense_4 (Dense) (None, 1) 26 \n=================================================================\nTotal params: 13,876\nTrainable params: 13,876\nNon-trainable params: 0\n_________________________________________________________________\n"
]
],
[
[
"#### Train and evaluate the model\n\nDefine a learning rate decay to encourage model paramaters to make smaller\nchanges as training goes on:",
"_____no_output_____"
]
],
[
[
"# Setup Learning Rate decay.\nlr_decay = tf.keras.callbacks.LearningRateScheduler(\n lambda epoch: LEARNING_RATE + 0.02 * (0.5 ** (1 + epoch)),\n verbose=True)",
"_____no_output_____"
]
],
[
[
"Finally, train the model. Provide the appropriate `steps_per_epoch` for the\nmodel to train on the entire training dataset (with `BATCH_SIZE` examples per step) during each epoch. And instruct the model to calculate validation\naccuracy with one big validation batch at the end of each epoch.\n",
"_____no_output_____"
]
],
[
[
"history = keras_model.fit(training_dataset, \n epochs=NUM_EPOCHS, \n steps_per_epoch=int(num_train_examples/BATCH_SIZE), \n validation_data=validation_dataset, \n validation_steps=1, \n callbacks=[lr_decay],\n verbose=1)",
"WARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/ops/math_ops.py:3066: to_int32 (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.cast instead.\n\nEpoch 00001: LearningRateScheduler reducing learning rate to 0.02.\nEpoch 1/20\n254/254 [==============================] - 1s 5ms/step - loss: 0.6986 - acc: 0.7893 - val_loss: 0.3894 - val_acc: 0.8329\n\nEpoch 00002: LearningRateScheduler reducing learning rate to 0.015.\nEpoch 2/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3574 - acc: 0.8335 - val_loss: 0.3861 - val_acc: 0.8131\n\nEpoch 00003: LearningRateScheduler reducing learning rate to 0.0125.\nEpoch 3/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3422 - acc: 0.8404 - val_loss: 0.3304 - val_acc: 0.8445\n\nEpoch 00004: LearningRateScheduler reducing learning rate to 0.01125.\nEpoch 4/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3353 - acc: 0.8465 - val_loss: 0.3610 - val_acc: 0.8435\n\nEpoch 00005: LearningRateScheduler reducing learning rate to 0.010625.\nEpoch 5/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3315 - acc: 0.8457 - val_loss: 0.3288 - val_acc: 0.8445\n\nEpoch 00006: LearningRateScheduler reducing learning rate to 0.0103125.\nEpoch 6/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3294 - acc: 0.8475 - val_loss: 0.3331 - val_acc: 0.8489\n\nEpoch 00007: LearningRateScheduler reducing learning rate to 0.01015625.\nEpoch 7/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3296 - acc: 0.8476 - val_loss: 0.3296 - val_acc: 0.8508\n\nEpoch 00008: LearningRateScheduler reducing learning rate to 0.010078125.\nEpoch 8/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3287 - acc: 0.8486 - val_loss: 0.3254 - val_acc: 0.8494\n\nEpoch 00009: LearningRateScheduler reducing learning rate to 0.0100390625.\nEpoch 9/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3274 - acc: 0.8483 - val_loss: 0.3205 - val_acc: 0.8511\n\nEpoch 00010: LearningRateScheduler reducing learning rate to 0.01001953125.\nEpoch 10/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3274 - acc: 0.8493 - val_loss: 0.3233 - val_acc: 0.8483\n\nEpoch 00011: LearningRateScheduler reducing learning rate to 0.010009765625.\nEpoch 11/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3268 - acc: 0.8485 - val_loss: 0.3315 - val_acc: 0.8511\n\nEpoch 00012: LearningRateScheduler reducing learning rate to 0.010004882812500001.\nEpoch 12/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3258 - acc: 0.8500 - val_loss: 0.3328 - val_acc: 0.8502\n\nEpoch 00013: LearningRateScheduler reducing learning rate to 0.01000244140625.\nEpoch 13/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3278 - acc: 0.8488 - val_loss: 0.3196 - val_acc: 0.8536\n\nEpoch 00014: LearningRateScheduler reducing learning rate to 0.010001220703125.\nEpoch 14/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3261 - acc: 0.8508 - val_loss: 0.3355 - val_acc: 0.8384\n\nEpoch 00015: LearningRateScheduler reducing learning rate to 0.0100006103515625.\nEpoch 15/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3249 - acc: 0.8508 - val_loss: 0.3379 - val_acc: 0.8478\n\nEpoch 00016: LearningRateScheduler reducing learning rate to 0.01000030517578125.\nEpoch 16/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3277 - acc: 0.8485 - val_loss: 0.3253 - val_acc: 0.8524\n\nEpoch 00017: LearningRateScheduler reducing learning rate to 0.010000152587890625.\nEpoch 17/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3251 - acc: 0.8521 - val_loss: 0.3261 - val_acc: 0.8512\n\nEpoch 00018: LearningRateScheduler reducing learning rate to 0.010000076293945313.\nEpoch 18/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3287 - acc: 0.8494 - val_loss: 0.3232 - val_acc: 0.8543\n\nEpoch 00019: LearningRateScheduler reducing learning rate to 0.010000038146972657.\nEpoch 19/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3239 - acc: 0.8512 - val_loss: 0.3334 - val_acc: 0.8496\n\nEpoch 00020: LearningRateScheduler reducing learning rate to 0.010000019073486329.\nEpoch 20/20\n254/254 [==============================] - 1s 4ms/step - loss: 0.3279 - acc: 0.8504 - val_loss: 0.3174 - val_acc: 0.8523\n"
]
],
[
[
"### Visualize training and export the trained model",
"_____no_output_____"
],
[
"#### Visualize training\n\nImport `matplotlib` to visualize how the model learned over the training period.",
"_____no_output_____"
]
],
[
[
"! pip install matplotlib\n\nfrom matplotlib import pyplot as plt\n\n%matplotlib inline",
"Requirement already satisfied: matplotlib in /usr/local/lib/python3.6/dist-packages (3.0.3)\nRequirement already satisfied: numpy>=1.10.0 in /usr/local/lib/python3.6/dist-packages (from matplotlib) (1.14.6)\nRequirement already satisfied: python-dateutil>=2.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib) (2.5.3)\nRequirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib) (1.0.1)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib) (2.3.1)\nRequirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.6/dist-packages (from matplotlib) (0.10.0)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.6/dist-packages (from python-dateutil>=2.1->matplotlib) (1.11.0)\nRequirement already satisfied: setuptools in /usr/local/lib/python3.6/dist-packages (from kiwisolver>=1.0.1->matplotlib) (40.8.0)\n"
]
],
[
[
"Plot the model's loss (binary cross-entropy) and accuracy, as measured at the\nend of each training epoch:",
"_____no_output_____"
]
],
[
[
"# Visualize History for Loss.\nplt.title('Keras model loss')\nplt.plot(history.history['loss'])\nplt.plot(history.history['val_loss'])\nplt.ylabel('loss')\nplt.xlabel('epoch')\nplt.legend(['training', 'validation'], loc='upper right')\nplt.show()\n\n# Visualize History for Accuracy.\nplt.title('Keras model accuracy')\nplt.ylabel('accuracy')\nplt.xlabel('epoch')\nplt.plot(history.history['acc'])\nplt.plot(history.history['val_acc'])\nplt.legend(['training', 'validation'], loc='lower right')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Over time, loss decreases and accuracy increases. But do they converge to a\nstable level? Are there big differences between the training and validation\nmetrics (a sign of overfitting)?\n\nLearn about [how to improve your machine learning\nmodel](https://developers.google.com/machine-learning/crash-course/). Then, feel\nfree to adjust hyperparameters or the model architecture and train again.",
"_____no_output_____"
],
[
"#### Export the model for serving\n\nUse\n[tf.contrib.saved_model.save_keras_model](https://www.tensorflow.org/api_docs/python/tf/contrib/saved_model/save_keras_model) to export a TensorFlow SavedModel directory. This is the format that Cloud\nAI Platform requires when you [create a model version\nresource](https://cloud.google.com/ml-engine/docs/tensorflow/deploying-models#creating_a_model_version).\n\nSince not all optimizers can be exported to the SavedModel format, you may see\nwarnings during the export process. As long you successfully export a serving\ngraph, AI Platform can used the SavedModel to serve predictions.",
"_____no_output_____"
]
],
[
[
"# Export the model to a local SavedModel directory \nexport_path = tf.contrib.saved_model.save_keras_model(keras_model, 'keras_export')\nprint(\"Model exported to: \", export_path)",
"\nWARNING: The TensorFlow contrib module will not be included in TensorFlow 2.0.\nFor more information, please see:\n * https://github.com/tensorflow/community/blob/master/rfcs/20180907-contrib-sunset.md\n * https://github.com/tensorflow/addons\nIf you depend on functionality not listed there, please file an issue.\n\nWARNING:tensorflow:This model was compiled with a Keras optimizer (<tensorflow.python.keras.optimizers.RMSprop object at 0x7fc198c4e400>) but is being saved in TensorFlow format with `save_weights`. The model's weights will be saved, but unlike with TensorFlow optimizers in the TensorFlow format the optimizer's state will not be saved.\n\nConsider using a TensorFlow optimizer from `tf.train`.\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/keras/engine/network.py:1436: update_checkpoint_state (from tensorflow.python.training.checkpoint_management) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.train.CheckpointManager to manage checkpoints rather than manually editing the Checkpoint proto.\nWARNING:tensorflow:Model was compiled with an optimizer, but the optimizer is not from `tf.train` (e.g. `tf.train.AdagradOptimizer`). Only the serving graph was exported. The train and evaluate graphs were not added to the SavedModel.\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/saved_model/signature_def_utils_impl.py:205: build_tensor_info (from tensorflow.python.saved_model.utils_impl) is deprecated and will be removed in a future version.\nInstructions for updating:\nThis function will only be available through the v1 compatibility library as tf.compat.v1.saved_model.utils.build_tensor_info or tf.compat.v1.saved_model.build_tensor_info.\nINFO:tensorflow:Signatures INCLUDED in export for Classify: None\nINFO:tensorflow:Signatures INCLUDED in export for Regress: None\nINFO:tensorflow:Signatures INCLUDED in export for Predict: ['serving_default']\nINFO:tensorflow:Signatures INCLUDED in export for Train: None\nINFO:tensorflow:Signatures INCLUDED in export for Eval: None\nINFO:tensorflow:No assets to save.\nINFO:tensorflow:No assets to write.\nINFO:tensorflow:SavedModel written to: keras_export/1553710367/saved_model.pb\nModel exported to: b'keras_export/1553710367'\n"
]
],
[
[
"You may export a SavedModel directory to your local filesystem or to Cloud\nStorage, as long as you have the necessary permissions. In your current\nenvironment, you granted access to Cloud Storage by authenticating your GCP account and setting the `GOOGLE_APPLICATION_CREDENTIALS` environment variable. Cloud ML\nEngine training jobs can also export directly to Cloud Storage, because Cloud ML\nEngine service accounts [have access to Cloud Storage buckets in their own\nproject](https://cloud.google.com/ml-engine/docs/tensorflow/working-with-cloud-storage).\n\nTry exporting directly to Cloud Storage:",
"_____no_output_____"
]
],
[
[
"# Export the model to a SavedModel directory in Cloud Storage\nexport_path = tf.contrib.saved_model.save_keras_model(keras_model, JOB_DIR + '/keras_export')\nprint(\"Model exported to: \", export_path)",
"WARNING:tensorflow:This model was compiled with a Keras optimizer (<tensorflow.python.keras.optimizers.RMSprop object at 0x7fc198c4e400>) but is being saved in TensorFlow format with `save_weights`. The model's weights will be saved, but unlike with TensorFlow optimizers in the TensorFlow format the optimizer's state will not be saved.\n\nConsider using a TensorFlow optimizer from `tf.train`.\nWARNING:tensorflow:Model was compiled with an optimizer, but the optimizer is not from `tf.train` (e.g. `tf.train.AdagradOptimizer`). Only the serving graph was exported. The train and evaluate graphs were not added to the SavedModel.\nINFO:tensorflow:Signatures INCLUDED in export for Classify: None\nINFO:tensorflow:Signatures INCLUDED in export for Regress: None\nINFO:tensorflow:Signatures INCLUDED in export for Predict: ['serving_default']\nINFO:tensorflow:Signatures INCLUDED in export for Train: None\nINFO:tensorflow:Signatures INCLUDED in export for Eval: None\nINFO:tensorflow:No assets to save.\nINFO:tensorflow:No assets to write.\nINFO:tensorflow:SavedModel written to: gs://<your-bucket-name>/keras-job-dir/keras_export/1553710379/saved_model.pb\nModel exported to: b'gs://<your-bucket-name>/keras-job-dir/keras_export/1553710379'\n"
]
],
[
[
"You can now deploy this model to AI Platform and serve predictions by\nfollowing the steps from Part 2.",
"_____no_output_____"
],
[
"## Cleaning up\n\nTo clean up all GCP resources used in this project, you can [delete the GCP\nproject](https://cloud.google.com/resource-manager/docs/creating-managing-projects#shutting_down_projects) you used for the tutorial.\n\nAlternatively, you can clean up individual resources by running the following\ncommands:",
"_____no_output_____"
]
],
[
[
"# Delete model version resource\n! gcloud ml-engine versions delete $MODEL_VERSION --quiet --model $MODEL_NAME \n\n# Delete model resource\n! gcloud ml-engine models delete $MODEL_NAME --quiet\n\n# Delete Cloud Storage objects that were created\n! gsutil -m rm -r $JOB_DIR\n\n# If the training job is still running, cancel it\n! gcloud ml-engine jobs cancel $JOB_NAME --quiet --verbosity critical",
"Removing gs://<your-bucket-name>/keras-job-dir/keras_export/1553709421/#1553709423098988...\nRemoving gs://<your-bucket-name>/keras-job-dir/keras_export/#1553709422852130...\nRemoving gs://<your-bucket-name>/keras-job-dir/keras_export/1553709421/variables/checkpoint#1553709429999272...\nRemoving gs://<your-bucket-name>/keras-job-dir/keras_export/1553709421/variables/#1553709428368512...\nRemoving gs://<your-bucket-name>/keras-job-dir/keras_export/1553709421/assets/#1553709430948151...\nRemoving gs://<your-bucket-name>/keras-job-dir/keras_export/1553709421/variables/variables.data-00000-of-00001#1553709428975750...\nRemoving gs://<your-bucket-name>/keras-job-dir/keras_export/1553709421/assets/saved_model.json#1553709431121952...\nRemoving gs://<your-bucket-name>/keras-job-dir/keras_export/1553709421/variables/variables.index#1553709429461522...\nRemoving gs://<your-bucket-name>/keras-job-dir/keras_export/1553709421/saved_model.pb#1553709430502605...\nRemoving gs://<your-bucket-name>/keras-job-dir/keras_export/1553710379/#1553710381998179...\nRemoving gs://<your-bucket-name>/keras-job-dir/keras_export/1553710379/assets/#1553710395035632...\nRemoving gs://<your-bucket-name>/keras-job-dir/keras_export/1553710379/assets/saved_model.json#1553710395421499...\nRemoving gs://<your-bucket-name>/keras-job-dir/keras_export/1553710379/saved_model.pb#1553710394249816...\nRemoving gs://<your-bucket-name>/keras-job-dir/keras_export/1553710379/variables/#1553710390778836...\nRemoving gs://<your-bucket-name>/keras-job-dir/keras_export/1553710379/variables/checkpoint#1553710393369087...\nRemoving gs://<your-bucket-name>/keras-job-dir/keras_export/1553710379/variables/variables.data-00000-of-00001#1553710391609457...\nRemoving gs://<your-bucket-name>/keras-job-dir/keras_export/1553710379/variables/variables.index#1553710392464814...\nRemoving gs://<your-bucket-name>/keras-job-dir/packages/dcc159f40836cff74a27866227b327b0a8ccb5266194e76cff5368266b6d1cdd/trainer-0.0.0.tar.gz#1553709266664674...\n/ [18/18 objects] 100% Done \nOperation completed over 18 objects. \n"
]
],
[
[
"If your Cloud Storage bucket doesn't contain any other objects and you would like to delete it, run `gsutil rm -r gs://$BUCKET_NAME`.",
"_____no_output_____"
],
[
"## What's next?\n\n* View the [complete training\ncode](https://github.com/GoogleCloudPlatform/cloudml-samples/tree/master/census/tf-keras) used in this guide, which structures the code to accept custom\nhyperparameters as command-line flags.\n* Read about [packaging\ncode](https://cloud.google.com/ml-engine/docs/tensorflow/packaging-trainer) for an AI Platform training job.\n* Read about [deploying a\nmodel](https://cloud.google.com/ml-engine/docs/tensorflow/deploying-models) to serve predictions.",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb4408e37ebc72f09dc5bd03e4f4f6ab791d2f1d | 37,767 | ipynb | Jupyter Notebook | Convolutional Neural Networks/Course_2_Part_6_Lesson_3_Notebook.ipynb | ornob39/Tensor-Flow-in-Practice-Specialization | b955658ced6231ab19a1fc1197ae6616745bfe1d | [
"MIT"
] | null | null | null | Convolutional Neural Networks/Course_2_Part_6_Lesson_3_Notebook.ipynb | ornob39/Tensor-Flow-in-Practice-Specialization | b955658ced6231ab19a1fc1197ae6616745bfe1d | [
"MIT"
] | null | null | null | Convolutional Neural Networks/Course_2_Part_6_Lesson_3_Notebook.ipynb | ornob39/Tensor-Flow-in-Practice-Specialization | b955658ced6231ab19a1fc1197ae6616745bfe1d | [
"MIT"
] | null | null | null | 101.524194 | 22,210 | 0.767548 | [
[
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
],
[
"import os\n\nfrom tensorflow.keras import layers\nfrom tensorflow.keras import Model\n!wget --no-check-certificate \\\n https://storage.googleapis.com/mledu-datasets/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5 \\\n -O /tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5\n \nfrom tensorflow.keras.applications.inception_v3 import InceptionV3\n\nlocal_weights_file = '/tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5'\n\npre_trained_model = InceptionV3(input_shape = (150, 150, 3), \n include_top = False, \n weights = None)\n\npre_trained_model.load_weights(local_weights_file)\n\nfor layer in pre_trained_model.layers:\n layer.trainable = False\n \n# pre_trained_model.summary()\n\nlast_layer = pre_trained_model.get_layer('mixed7')\nprint('last layer output shape: ', last_layer.output_shape)\nlast_output = last_layer.output",
"--2020-08-19 11:42:40-- https://storage.googleapis.com/mledu-datasets/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5\nResolving storage.googleapis.com (storage.googleapis.com)... 74.125.20.128, 74.125.142.128, 74.125.195.128, ...\nConnecting to storage.googleapis.com (storage.googleapis.com)|74.125.20.128|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 87910968 (84M) [application/x-hdf]\nSaving to: ‘/tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5’\n\n\r /tmp/ince 0%[ ] 0 --.-KB/s \r /tmp/incep 9%[> ] 8.01M 28.2MB/s \r /tmp/incept 47%[========> ] 40.01M 58.0MB/s \r /tmp/incepti 76%[==============> ] 64.01M 65.0MB/s \r/tmp/inception_v3_w 100%[===================>] 83.84M 81.1MB/s in 1.0s \n\n2020-08-19 11:42:41 (81.1 MB/s) - ‘/tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5’ saved [87910968/87910968]\n\nlast layer output shape: (None, 7, 7, 768)\n"
],
[
"from tensorflow.keras.optimizers import RMSprop\n\n# Flatten the output layer to 1 dimension\nx = layers.Flatten()(last_output)\n# Add a fully connected layer with 1,024 hidden units and ReLU activation\nx = layers.Dense(1024, activation='relu')(x)\n# Add a dropout rate of 0.2\nx = layers.Dropout(0.2)(x) \n# Add a final sigmoid layer for classification\nx = layers.Dense (1, activation='sigmoid')(x) \n\nmodel = Model( pre_trained_model.input, x) \n\nmodel.compile(optimizer = RMSprop(lr=0.0001), \n loss = 'binary_crossentropy', \n metrics = ['accuracy'])\n",
"_____no_output_____"
],
[
"!wget --no-check-certificate \\\n https://storage.googleapis.com/mledu-datasets/cats_and_dogs_filtered.zip \\\n -O /tmp/cats_and_dogs_filtered.zip\n\nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator\n\nimport os\nimport zipfile\n\nlocal_zip = '//tmp/cats_and_dogs_filtered.zip'\n\nzip_ref = zipfile.ZipFile(local_zip, 'r')\n\nzip_ref.extractall('/tmp')\nzip_ref.close()\n\n# Define our example directories and files\nbase_dir = '/tmp/cats_and_dogs_filtered'\n\ntrain_dir = os.path.join( base_dir, 'train')\nvalidation_dir = os.path.join( base_dir, 'validation')\n\n\ntrain_cats_dir = os.path.join(train_dir, 'cats') # Directory with our training cat pictures\ntrain_dogs_dir = os.path.join(train_dir, 'dogs') # Directory with our training dog pictures\nvalidation_cats_dir = os.path.join(validation_dir, 'cats') # Directory with our validation cat pictures\nvalidation_dogs_dir = os.path.join(validation_dir, 'dogs')# Directory with our validation dog pictures\n\ntrain_cat_fnames = os.listdir(train_cats_dir)\ntrain_dog_fnames = os.listdir(train_dogs_dir)\n\n# Add our data-augmentation parameters to ImageDataGenerator\ntrain_datagen = ImageDataGenerator(rescale = 1./255.,\n rotation_range = 40,\n width_shift_range = 0.2,\n height_shift_range = 0.2,\n shear_range = 0.2,\n zoom_range = 0.2,\n horizontal_flip = True)\n\n# Note that the validation data should not be augmented!\ntest_datagen = ImageDataGenerator( rescale = 1.0/255. )\n\n# Flow training images in batches of 20 using train_datagen generator\ntrain_generator = train_datagen.flow_from_directory(train_dir,\n batch_size = 20,\n class_mode = 'binary', \n target_size = (150, 150)) \n\n# Flow validation images in batches of 20 using test_datagen generator\nvalidation_generator = test_datagen.flow_from_directory( validation_dir,\n batch_size = 20,\n class_mode = 'binary', \n target_size = (150, 150))",
"--2020-08-19 11:43:02-- https://storage.googleapis.com/mledu-datasets/cats_and_dogs_filtered.zip\nResolving storage.googleapis.com (storage.googleapis.com)... 74.125.28.128, 74.125.142.128, 74.125.195.128, ...\nConnecting to storage.googleapis.com (storage.googleapis.com)|74.125.28.128|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 68606236 (65M) [application/zip]\nSaving to: ‘/tmp/cats_and_dogs_filtered.zip’\n\n\r /tmp/cats 0%[ ] 0 --.-KB/s \r /tmp/cats_ 24%[===> ] 16.33M 81.6MB/s \r /tmp/cats_a 36%[======> ] 24.01M 59.9MB/s \r/tmp/cats_and_dogs_ 100%[===================>] 65.43M 120MB/s in 0.5s \n\n2020-08-19 11:43:03 (120 MB/s) - ‘/tmp/cats_and_dogs_filtered.zip’ saved [68606236/68606236]\n\nFound 2000 images belonging to 2 classes.\nFound 1000 images belonging to 2 classes.\n"
],
[
"history = model.fit(\n train_generator,\n validation_data = validation_generator,\n steps_per_epoch = 100,\n epochs = 20,\n validation_steps = 50,\n verbose = 2)",
"Epoch 1/20\n100/100 - 18s - loss: 0.3639 - accuracy: 0.8610 - val_loss: 0.0994 - val_accuracy: 0.9600\nEpoch 2/20\n100/100 - 17s - loss: 0.2290 - accuracy: 0.9130 - val_loss: 0.0987 - val_accuracy: 0.9590\nEpoch 3/20\n100/100 - 17s - loss: 0.2051 - accuracy: 0.9200 - val_loss: 0.1590 - val_accuracy: 0.9450\nEpoch 4/20\n100/100 - 17s - loss: 0.1928 - accuracy: 0.9275 - val_loss: 0.1050 - val_accuracy: 0.9640\nEpoch 5/20\n100/100 - 17s - loss: 0.1679 - accuracy: 0.9395 - val_loss: 0.1534 - val_accuracy: 0.9580\nEpoch 6/20\n100/100 - 17s - loss: 0.1496 - accuracy: 0.9485 - val_loss: 0.1191 - val_accuracy: 0.9690\nEpoch 7/20\n100/100 - 17s - loss: 0.1816 - accuracy: 0.9350 - val_loss: 0.1108 - val_accuracy: 0.9630\nEpoch 8/20\n100/100 - 18s - loss: 0.1605 - accuracy: 0.9445 - val_loss: 0.1108 - val_accuracy: 0.9650\nEpoch 9/20\n100/100 - 17s - loss: 0.1524 - accuracy: 0.9520 - val_loss: 0.1427 - val_accuracy: 0.9600\nEpoch 10/20\n100/100 - 17s - loss: 0.1440 - accuracy: 0.9530 - val_loss: 0.1324 - val_accuracy: 0.9650\nEpoch 11/20\n100/100 - 17s - loss: 0.1822 - accuracy: 0.9435 - val_loss: 0.1339 - val_accuracy: 0.9660\nEpoch 12/20\n100/100 - 17s - loss: 0.1378 - accuracy: 0.9500 - val_loss: 0.1213 - val_accuracy: 0.9680\nEpoch 13/20\n100/100 - 17s - loss: 0.1494 - accuracy: 0.9580 - val_loss: 0.1215 - val_accuracy: 0.9680\nEpoch 14/20\n100/100 - 17s - loss: 0.1353 - accuracy: 0.9600 - val_loss: 0.1363 - val_accuracy: 0.9650\nEpoch 15/20\n100/100 - 17s - loss: 0.1301 - accuracy: 0.9550 - val_loss: 0.1847 - val_accuracy: 0.9510\nEpoch 16/20\n100/100 - 17s - loss: 0.1347 - accuracy: 0.9575 - val_loss: 0.1345 - val_accuracy: 0.9700\nEpoch 17/20\n100/100 - 17s - loss: 0.1510 - accuracy: 0.9535 - val_loss: 0.1570 - val_accuracy: 0.9630\nEpoch 18/20\n100/100 - 17s - loss: 0.1417 - accuracy: 0.9515 - val_loss: 0.1193 - val_accuracy: 0.9750\nEpoch 19/20\n100/100 - 17s - loss: 0.1362 - accuracy: 0.9625 - val_loss: 0.1395 - val_accuracy: 0.9700\nEpoch 20/20\n100/100 - 17s - loss: 0.1341 - accuracy: 0.9610 - val_loss: 0.1287 - val_accuracy: 0.9690\n"
],
[
"import matplotlib.pyplot as plt\nacc = history.history['accuracy']\nval_acc = history.history['val_accuracy']\nloss = history.history['loss']\nval_loss = history.history['val_loss']\n\nepochs = range(len(acc))\n\nplt.plot(epochs, acc, 'r', label='Training accuracy')\nplt.plot(epochs, val_acc, 'b', label='Validation accuracy')\nplt.title('Training and validation accuracy')\nplt.legend(loc=0)\nplt.figure()\n\n\nplt.show()",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4419102e259d8afc55fda3483a5779b5b22017 | 57,490 | ipynb | Jupyter Notebook | notebook/side_try_pheno.ipynb | liangyy/ptrs-ukb | e63b3827b9873d580675f1ba9bc27e817d7ea380 | [
"MIT"
] | 7 | 2021-05-10T19:58:58.000Z | 2022-03-07T05:37:22.000Z | notebook/side_try_pheno.ipynb | liangyy/ptrs-ukb | e63b3827b9873d580675f1ba9bc27e817d7ea380 | [
"MIT"
] | null | null | null | notebook/side_try_pheno.ipynb | liangyy/ptrs-ukb | e63b3827b9873d580675f1ba9bc27e817d7ea380 | [
"MIT"
] | 3 | 2021-08-22T00:12:55.000Z | 2022-03-30T20:19:14.000Z | 139.538835 | 23,244 | 0.677909 | [
[
[
"import hail as hl\nfrom hail.plot import show\nfrom pprint import pprint\nimport matplotlib",
"_____no_output_____"
],
[
"pheno = (hl.import_table('/homes/yanyul/query_first_attempt_with_qc.csv', impute = True, delimiter = ',')\n .key_by('eid'))",
"2019-12-06 14:06:28 Hail: INFO: Reading table to impute column types\n2019-12-06 14:06:30 Hail: INFO: Finished type imputation\n Loading column 'eid' as type 'int32' (imputed)\n Loading column 'age_recruitment' as type 'int32' (imputed)\n Loading column 'sex' as type 'int32' (imputed)\n Loading column 'pc1' as type 'float64' (imputed)\n Loading column 'pc2' as type 'float64' (imputed)\n Loading column 'pc3' as type 'float64' (imputed)\n Loading column 'pc4' as type 'float64' (imputed)\n Loading column 'pc5' as type 'float64' (imputed)\n Loading column 'pc6' as type 'float64' (imputed)\n Loading column 'pc7' as type 'float64' (imputed)\n Loading column 'pc8' as type 'float64' (imputed)\n Loading column 'pc9' as type 'float64' (imputed)\n Loading column 'pc10' as type 'float64' (imputed)\n Loading column 'ethnicity_agg' as type 'int32' (imputed)\n Loading column 'meaning' as type 'str' (imputed)\n Loading column 'height' as type 'float64' (imputed)\n Loading column 'dbp' as type 'int32' (imputed)\n Loading column 'sbp' as type 'int32' (imputed)\n Loading column 'bmi' as type 'float64' (imputed)\n Loading column 'wbc' as type 'float64' (imputed)\n Loading column 'rbc' as type 'float64' (imputed)\n Loading column 'hb' as type 'float64' (imputed)\n Loading column 'ht' as type 'float64' (imputed)\n Loading column 'mcv' as type 'float64' (imputed)\n Loading column 'mch' as type 'float64' (imputed)\n Loading column 'mchc' as type 'float64' (imputed)\n Loading column 'platelet' as type 'float64' (imputed)\n Loading column 'lymphocyte' as type 'float64' (imputed)\n Loading column 'monocyte' as type 'float64' (imputed)\n Loading column 'neutrophil' as type 'float64' (imputed)\n Loading column 'eosinophil' as type 'float64' (imputed)\n Loading column 'basophil' as type 'float64' (imputed)\n"
],
[
"pheno.describe()",
"----------------------------------------\nGlobal fields:\n None\n----------------------------------------\nRow fields:\n 'eid': int32 \n 'age_recruitment': int32 \n 'sex': int32 \n 'pc1': float64 \n 'pc2': float64 \n 'pc3': float64 \n 'pc4': float64 \n 'pc5': float64 \n 'pc6': float64 \n 'pc7': float64 \n 'pc8': float64 \n 'pc9': float64 \n 'pc10': float64 \n 'ethnicity_agg': int32 \n 'meaning': str \n 'height': float64 \n 'dbp': int32 \n 'sbp': int32 \n 'bmi': float64 \n 'wbc': float64 \n 'rbc': float64 \n 'hb': float64 \n 'ht': float64 \n 'mcv': float64 \n 'mch': float64 \n 'mchc': float64 \n 'platelet': float64 \n 'lymphocyte': float64 \n 'monocyte': float64 \n 'neutrophil': float64 \n 'eosinophil': float64 \n 'basophil': float64 \n----------------------------------------\nKey: ['eid']\n----------------------------------------\n"
],
[
"pheno.show(width=100)",
"_____no_output_____"
],
[
"pheno.aggregate(hl.agg.counter(pheno.meaning)) #(hl.agg.counter(table.SuperPopulation)))",
"_____no_output_____"
],
[
"pheno.aggregate(hl.agg.stats(pheno.age_recruitment))",
"_____no_output_____"
],
[
"pheno.count()",
"_____no_output_____"
],
[
"p = hl.plot.histogram(pheno.pc10)\nshow(p)",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport numpy as np\nplt.figure(1)\nplt.plot(np.sin(np.linspace(0, 20, 100)))\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4419db1dcb9669f6d3cb6ddf8091c04d468126 | 5,725 | ipynb | Jupyter Notebook | work/COVIDvu-US.ipynb | VirusTrack/COVIDvu | ebe459deaa00cb1246ee46255a2ce437401bf31a | [
"BSD-3-Clause"
] | 12 | 2020-03-23T01:33:14.000Z | 2020-04-19T21:57:34.000Z | work/COVIDvu-US.ipynb | VirusTrack/COVIDvu | ebe459deaa00cb1246ee46255a2ce437401bf31a | [
"BSD-3-Clause"
] | 229 | 2020-03-22T15:33:06.000Z | 2020-11-20T05:09:33.000Z | work/COVIDvu-US.ipynb | VirusTrack/COVIDvu | ebe459deaa00cb1246ee46255a2ce437401bf31a | [
"BSD-3-Clause"
] | 6 | 2020-03-26T02:24:33.000Z | 2020-04-04T11:52:14.000Z | 22.53937 | 136 | 0.520873 | [
[
[
"# COVIDvu - US regions visualizer <img src='resources/American-flag.png' align = 'right'>",
"_____no_output_____"
],
[
"---\n## Runtime prerequisites",
"_____no_output_____"
]
],
[
[
"%%capture --no-stderr requirementsOutput\n\ndisplayRequirementsOutput = False\n\n%pip install -r requirements.txt\nfrom covidvu.utils import autoReloadCode; autoReloadCode()",
"_____no_output_____"
],
[
"if displayRequirementsOutput:\n requirementsOutput.show()",
"_____no_output_____"
]
],
[
[
"---\n## Pull latest datasets\n",
"_____no_output_____"
]
],
[
[
"%sx ./refreshdata local patch",
"_____no_output_____"
]
],
[
[
"---\n## Confirmed, deaths, recovered datasets",
"_____no_output_____"
]
],
[
[
"import os\n\nimport numpy as np\nimport pandas as pd\n\nfrom covidvu.cryostation import Cryostation\n\npd.options.mode.chained_assignment = None",
"_____no_output_____"
],
[
"databasePath = './database/virustrack.db'",
"_____no_output_____"
],
[
"storage = Cryostation(databasePath=databasePath)\nconfirmedCases = storage.timeSeriesFor(regionType = 'province',\n countryName = 'US',\n casesType = 'confirmed', disableProgressBar=False)\nconfirmedDeaths = storage.timeSeriesFor(regionType = 'province',\n countryName = 'US',\n casesType = 'deaths', disableProgressBar=False)",
"_____no_output_____"
]
],
[
[
"---\n## Cases by US state",
"_____no_output_____"
]
],
[
[
"from ipywidgets import fixed\nfrom ipywidgets import interact\nfrom ipywidgets import widgets\n\nfrom covidvu import visualize",
"_____no_output_____"
],
[
"statesUS = list(confirmedCases.columns)\nmultiState = widgets.SelectMultiple(\n options=statesUS,\n value=['New York'],\n description='State',\n disabled=False\n)\nlog = widgets.Checkbox(value=False, description='Log scale')",
"_____no_output_____"
]
],
[
[
"### Confirmed cases",
"_____no_output_____"
]
],
[
[
"interact(visualize.plotTimeSeriesInteractive, \n df=fixed(confirmedCases), \n selectedColumns=multiState, \n log=log,\n yLabel=fixed('Total confirmed cases'),\n title=fixed('COVID-19 total confirmed cases in US states')\n);",
"_____no_output_____"
],
[
"def viewTopStates(n):\n return pd.DataFrame(confirmedCases.iloc[-1,:].sort_values(ascending=False).iloc[1:n]).style.background_gradient(cmap=\"Reds\")",
"_____no_output_____"
],
[
"interact(viewTopStates, n=widgets.IntSlider(min=1, max=len(statesUS), step=1, value=5));",
"_____no_output_____"
]
],
[
[
"---\n## Cases by US region",
"_____no_output_____"
]
],
[
[
"regionsUS = list(confirmedCases.columns)\nmultiRegion = widgets.SelectMultiple(\n options=regionsUS,\n value=['New York'],\n description='State',\n disabled=False\n)",
"_____no_output_____"
],
[
"interact(visualize.plotTimeSeriesInteractive, \n df=fixed(confirmedCases), \n selectedColumns=multiRegion, \n log=log,\n yLabel=fixed('Total confirmed cases'),\n title=fixed('COVID-19 total confirmed cases in US regions')\n);",
"_____no_output_____"
]
],
[
[
"---\n© the COVIDvu Contributors. All rights reserved.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cb442783f0f7383747d9a0b6cd1416c930a610ec | 3,137 | ipynb | Jupyter Notebook | Week - 4/Functions in Python.ipynb | AshishJangra27/Data-Science-Specialization | 638e490fdd00e57fa8cdc7fcbc946307babd6d0a | [
"Apache-2.0"
] | null | null | null | Week - 4/Functions in Python.ipynb | AshishJangra27/Data-Science-Specialization | 638e490fdd00e57fa8cdc7fcbc946307babd6d0a | [
"Apache-2.0"
] | null | null | null | Week - 4/Functions in Python.ipynb | AshishJangra27/Data-Science-Specialization | 638e490fdd00e57fa8cdc7fcbc946307babd6d0a | [
"Apache-2.0"
] | null | null | null | 16.253886 | 77 | 0.43226 | [
[
[
"2^5 = 2*2*2*2*2",
"_____no_output_____"
],
[
"2**5",
"_____no_output_____"
],
[
"pow(2,5)",
"_____no_output_____"
],
[
"def power(num , pow_): \n n = 1\n for i in range(pow_):\n n = n * num\n print(n)\n return n",
"_____no_output_____"
],
[
"x = power(33,43)\n\nif (x%2 == 0):\n print('Even')\nelse:\n print(\"Odd\")",
"197742224185065366932324177968221745157026572653527148127098282337\nOdd\n"
],
[
"2*2*2*2*2",
"_____no_output_____"
],
[
"pow_ = 5\nnum = 2\n\n",
"32\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb442b90acbee7f157400385f8d396658321498d | 94,808 | ipynb | Jupyter Notebook | Projetos/trabalhoCovid.ipynb | quesia-araujo/Curso-IA | e0ec17c56be46a8670081392ae5ca71be0bd1b19 | [
"MIT"
] | null | null | null | Projetos/trabalhoCovid.ipynb | quesia-araujo/Curso-IA | e0ec17c56be46a8670081392ae5ca71be0bd1b19 | [
"MIT"
] | null | null | null | Projetos/trabalhoCovid.ipynb | quesia-araujo/Curso-IA | e0ec17c56be46a8670081392ae5ca71be0bd1b19 | [
"MIT"
] | null | null | null | 94,808 | 94,808 | 0.858335 | [
[
[
"import urllib.request\nimport os\nfrom PIL import Image,ImageStat\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport torch.optim as optim\nimport torchvision\nimport torch\nimport torch.nn as nn\nfrom torch.utils.data import DataLoader\nimport torchvision.transforms as transforms\nimport torch.functional as F\nimport torchsummary\nfrom sklearn.model_selection import StratifiedKFold",
"_____no_output_____"
]
],
[
[
"# Carregando os dados",
"_____no_output_____"
]
],
[
[
"main_dir = \"/content/drive/My Drive/COVID-19/X-Ray Image DataSet\"\nos.chdir(main_dir)",
"_____no_output_____"
],
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3aietf%3awg%3aoauth%3a2.0%3aoob&response_type=code&scope=email%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdocs.test%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive.photos.readonly%20https%3a%2f%2fwww.googleapis.com%2fauth%2fpeopleapi.readonly\n\nEnter your authorization code:\n··········\nMounted at /content/drive\n"
],
[
"!ls",
"Covid-19 No_findings Pneumonia\n"
],
[
"image_loader = lambda x: Image.open(x)\n",
"_____no_output_____"
]
],
[
[
"## Tratamento de Imagem",
"_____no_output_____"
]
],
[
[
"class ToRGB(object):\n def __call__(self,img):\n if img.mode == 'RGBA':\n r, g, b, a = img.split()\n return Image.merge('RGB', (r, g, b))\n if img.mode == 'L':\n rgb = img.convert('RGB')\n return rgb\n return img",
"_____no_output_____"
],
[
"class ToNorm(object):\n def __call__(self,img):\n mean = torch.mean(img)\n std = torch.std(img)\n return (img - mean)/std",
"_____no_output_____"
],
[
"transform = transforms.Compose([ToRGB(),\n transforms.Resize((256,256)),\n transforms.ToTensor(),\n ToNorm(), \n ])",
"_____no_output_____"
]
],
[
[
"## Carregando os dados em um dataloader",
"_____no_output_____"
]
],
[
[
"dataset = torchvision.datasets.DatasetFolder(main_dir,loader = image_loader,extensions = ('png','jpg','jpeg',) ,transform=transform )",
"_____no_output_____"
],
[
"dataset.classes",
"_____no_output_____"
],
[
"len(dataset)",
"_____no_output_____"
],
[
"dl = DataLoader(dataset,batch_size=32)",
"_____no_output_____"
],
[
"data,class_att = next(iter(dl))",
"_____no_output_____"
],
[
"grid_img = torchvision.utils.make_grid(data,nrow=5)",
"_____no_output_____"
],
[
"grid_img.shape",
"_____no_output_____"
],
[
"plt.imshow(grid_img.permute(1,2,0))",
"Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\n"
]
],
[
[
"# rede",
"_____no_output_____"
]
],
[
[
"def conv_block(ni, nf, size=3, stride=1):\n for_pad = lambda s: s if s > 2 else 3\n return nn.Sequential(\n nn.Conv2d(ni, nf, kernel_size=size, stride=stride, padding=(for_pad(size) - 1)//2, bias=False),\n nn.BatchNorm2d(nf),\n nn.LeakyReLU(negative_slope=0.1, inplace=True)\n )\n\ndef triple_conv(ni, nf):\n return nn.Sequential(\n conv_block(ni, nf),\n conv_block(ni, nf, size=1),\n conv_block(ni, nf)\n )\ndef maxpooling():\n return nn.MaxPool2d(2, stride=2)",
"_____no_output_____"
],
[
"nn.Module??",
"_____no_output_____"
]
],
[
[
"# teste\n",
"_____no_output_____"
]
],
[
[
"def conv_block(in_c, out_c, size=3, stride=1):\n for_pad = lambda size: size if size > 2 else 3\n return nn.Sequential(\n nn.Conv2d(in_c, out_c, kernel_size=size, stride=stride, padding=(for_pad(size) - 1) // 2, bias=False),\n nn.BatchNorm2d(out_c),\n nn.LeakyReLU(negative_slope=0.1, inplace=True),\n )\n\ndef last_conv_block(in_c, out_c, size=3, stride=1):\n for_pad = lambda size: size if size > 2 else 3\n return nn.Sequential(\n nn.Conv2d(in_c, out_c, kernel_size=size, stride=stride, padding=(for_pad(size) - 1) // 2, bias=False),\n nn.ReLU(inplace=True),\n nn.BatchNorm2d(out_c),\n )\n\ndef triple_conv(in_c, out_c):\n return nn.Sequential(\n conv_block(in_c, out_c),\n conv_block(out_c, in_c, size=1),\n conv_block(in_c, out_c)\n )\n\nclass DarkCovidNet(nn.Module):\n def __init__(self):\n super().__init__()\n #maxpooling\n self.maxp = nn.MaxPool2d(2, stride=2)\n #backbone \n self.block1 = conv_block(3,8)\n self.block2 = conv_block(8,16)\n self.block3 = triple_conv(16,32)\n self.block4 = triple_conv(32,64)\n self.block5 = triple_conv(64,128)\n self.block6 = triple_conv(128,256)\n self.block7 = conv_block(256,128, size=1)\n self.block8 = conv_block(128,256)\n self.block9 = last_conv_block(256,3)\n #classifier\n self.flatten = nn.Flatten()\n self.linear = nn.Linear(507,3)\n\n def forward(self, x):\n #backbone \n x = self.block1(x)\n x = self.maxp(x)\n x = self.block2(x)\n x = self.maxp(x)\n x = self.block3(x)\n x = self.maxp(x)\n x = self.block4(x)\n x = self.maxp(x)\n x = self.block5(x)\n x = self.maxp(x)\n x = self.block6(x)\n x = self.block7(x)\n x = self.block8(x)\n x = self.block9(x)\n #classifier\n x = self.flatten(x)\n x = self.linear(x)\n return x",
"_____no_output_____"
],
[
"model = DarkCovidNet()\n\ntorchsummary.summary(model,(3,256,256),device='cpu')\n\ndel model",
"----------------------------------------------------------------\n Layer (type) Output Shape Param #\n================================================================\n Conv2d-1 [-1, 8, 256, 256] 216\n BatchNorm2d-2 [-1, 8, 256, 256] 16\n LeakyReLU-3 [-1, 8, 256, 256] 0\n MaxPool2d-4 [-1, 8, 128, 128] 0\n Conv2d-5 [-1, 16, 128, 128] 1,152\n BatchNorm2d-6 [-1, 16, 128, 128] 32\n LeakyReLU-7 [-1, 16, 128, 128] 0\n MaxPool2d-8 [-1, 16, 64, 64] 0\n Conv2d-9 [-1, 32, 64, 64] 4,608\n BatchNorm2d-10 [-1, 32, 64, 64] 64\n LeakyReLU-11 [-1, 32, 64, 64] 0\n Conv2d-12 [-1, 16, 66, 66] 512\n BatchNorm2d-13 [-1, 16, 66, 66] 32\n LeakyReLU-14 [-1, 16, 66, 66] 0\n Conv2d-15 [-1, 32, 66, 66] 4,608\n BatchNorm2d-16 [-1, 32, 66, 66] 64\n LeakyReLU-17 [-1, 32, 66, 66] 0\n MaxPool2d-18 [-1, 32, 33, 33] 0\n Conv2d-19 [-1, 64, 33, 33] 18,432\n BatchNorm2d-20 [-1, 64, 33, 33] 128\n LeakyReLU-21 [-1, 64, 33, 33] 0\n Conv2d-22 [-1, 32, 35, 35] 2,048\n BatchNorm2d-23 [-1, 32, 35, 35] 64\n LeakyReLU-24 [-1, 32, 35, 35] 0\n Conv2d-25 [-1, 64, 35, 35] 18,432\n BatchNorm2d-26 [-1, 64, 35, 35] 128\n LeakyReLU-27 [-1, 64, 35, 35] 0\n MaxPool2d-28 [-1, 64, 17, 17] 0\n Conv2d-29 [-1, 128, 17, 17] 73,728\n BatchNorm2d-30 [-1, 128, 17, 17] 256\n LeakyReLU-31 [-1, 128, 17, 17] 0\n Conv2d-32 [-1, 64, 19, 19] 8,192\n BatchNorm2d-33 [-1, 64, 19, 19] 128\n LeakyReLU-34 [-1, 64, 19, 19] 0\n Conv2d-35 [-1, 128, 19, 19] 73,728\n BatchNorm2d-36 [-1, 128, 19, 19] 256\n LeakyReLU-37 [-1, 128, 19, 19] 0\n MaxPool2d-38 [-1, 128, 9, 9] 0\n Conv2d-39 [-1, 256, 9, 9] 294,912\n BatchNorm2d-40 [-1, 256, 9, 9] 512\n LeakyReLU-41 [-1, 256, 9, 9] 0\n Conv2d-42 [-1, 128, 11, 11] 32,768\n BatchNorm2d-43 [-1, 128, 11, 11] 256\n LeakyReLU-44 [-1, 128, 11, 11] 0\n Conv2d-45 [-1, 256, 11, 11] 294,912\n BatchNorm2d-46 [-1, 256, 11, 11] 512\n LeakyReLU-47 [-1, 256, 11, 11] 0\n Conv2d-48 [-1, 128, 13, 13] 32,768\n BatchNorm2d-49 [-1, 128, 13, 13] 256\n LeakyReLU-50 [-1, 128, 13, 13] 0\n Conv2d-51 [-1, 256, 13, 13] 294,912\n BatchNorm2d-52 [-1, 256, 13, 13] 512\n LeakyReLU-53 [-1, 256, 13, 13] 0\n Conv2d-54 [-1, 3, 13, 13] 6,912\n ReLU-55 [-1, 3, 13, 13] 0\n BatchNorm2d-56 [-1, 3, 13, 13] 6\n Flatten-57 [-1, 507] 0\n Linear-58 [-1, 3] 1,524\n================================================================\nTotal params: 1,167,586\nTrainable params: 1,167,586\nNon-trainable params: 0\n----------------------------------------------------------------\nInput size (MB): 0.75\nForward/backward pass size (MB): 37.53\nParams size (MB): 4.45\nEstimated Total Size (MB): 42.73\n----------------------------------------------------------------\n"
]
],
[
[
"# Treinamento",
"_____no_output_____"
]
],
[
[
"n = len(dataset)\nn_train = int(0.7*n)\nn_test = n - n_train",
"_____no_output_____"
],
[
"n_train ,n_test",
"_____no_output_____"
],
[
"ds_train,ds_test = torch.utils.data.random_split(dataset,(n_train,n_test))",
"_____no_output_____"
],
[
"len(ds_train)",
"_____no_output_____"
],
[
"x,y = next(iter(ds_train))",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
],
[
"skf = StratifiedKFold(n_splits=5)",
"_____no_output_____"
],
[
"for train, test in skf.split(x, y):\n print(train,test)",
"_____no_output_____"
],
[
"def data_split(dataset,lists):\n return [torch.utils.data.Subset(dataset, llist ) for llist in lists]",
"_____no_output_____"
],
[
"dl_train = torch.utils.data.DataLoader(ds_train,batch_size=32)\ndl_test = torch.utils.data.DataLoader(ds_test,batch_size=32)",
"_____no_output_____"
],
[
"import sklearn.metrics as metrics",
"_____no_output_____"
],
[
"def train_model(dl,model,opt,criterion,epochs,device):\n model.to(device)\n model.train()\n lloss = []\n for epoch in range(epochs):\n for x,y in dl:\n x = x.to(device)\n y = y.to(device)\n pred = model(x)\n loss = criterion(pred,y)\n loss.backward()\n opt.step()\n opt.zero_grad()\n lloss.append(loss.item())\n ",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score",
"_____no_output_____"
],
[
"lacc = []\ndevice = torch.device('cuda:0')\n#for train, test in skf.split(range(len(dataset)), dataset.targets):\n #ds_train,ds_test = data_split(ds,(train,test))\n \nmodel = DarkCovidNet()\n\ncriterion = nn.CrossEntropyLoss()\nopt = optim.Adam(model.parameters(),lr=1e-2)\n \ndl_train = torch.utils.data.DataLoader(ds_train,batch_size=32)\ndl_test = torch.utils.data.DataLoader(ds_test,batch_size=32)\n \ntrain_model(dl_train,model,opt,criterion,150,device)\n(acc,loss)=evaluate(dl_test,model,criterion)\nprint(\"accuracy:%4.3f loss:%4.3f\"%(acc,loss))\ntorch.save(model.state_dict(),'model.pth')\nlacc.append(acc)\nnp.mean(lacc),np.std(lacc)",
"accuracy:0.725 loss:1.951\n"
],
[
"def evaluate(dl,model,criterion):\n model.to(device)\n model.eval()\n lacc = []\n lloss = []\n with torch.no_grad():\n for x,y in dl:\n x = x.to(device)\n pred = model(x)\n loss = criterion(pred,y.to(device)) \n y_pred = pred.argmax(dim=1).cpu()\n acc = accuracy_score(y,y_pred)\n lacc.append(acc)\n lloss.append(loss.item())\n return np.mean(lacc),np.mean(lloss)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb442cc54d2644ff2b9fd2d87add05f099716734 | 40,249 | ipynb | Jupyter Notebook | rough_work/HW8 Q1.ipynb | tylerlum/math_318_assignments | 7116b5695c42a6c7af1efc4b518e0ffdbda94657 | [
"MIT"
] | null | null | null | rough_work/HW8 Q1.ipynb | tylerlum/math_318_assignments | 7116b5695c42a6c7af1efc4b518e0ffdbda94657 | [
"MIT"
] | null | null | null | rough_work/HW8 Q1.ipynb | tylerlum/math_318_assignments | 7116b5695c42a6c7af1efc4b518e0ffdbda94657 | [
"MIT"
] | null | null | null | 36.993566 | 65 | 0.550846 | [
[
[
"import numpy as np\nfrom tqdm import tqdm",
"_____no_output_____"
],
[
"p = 0.5\nN = 1000000",
"_____no_output_____"
],
[
"def run_test():\n nums = [0]\n for _ in range(N):\n if np.random.uniform(low=0, high=1) < p:\n nums.append(nums[-1] + 1)\n else:\n nums.append(nums[-1] - 1)\n freq = {}\n for num in nums:\n if num in freq:\n freq[num] += 1\n else:\n freq[num] = 1\n for x in freq:\n freq[x] /= len(nums)\n return freq",
"_____no_output_____"
],
[
"runs = 10000\ncounter = 0\nfor _ in tqdm(range(runs)):\n if run_test() > 1:\n counter += 1\nprint(f'Fraction return to origin = {counter / runs}')",
"100%|██████████| 10000/10000 [00:18<00:00, 540.40it/s]"
],
[
"print(f'Expected fraction return to origin = {2*(1-p)}')",
"Expected fraction return to origin = 0.6000000000000001\n"
],
[
"freq = run_test()",
"_____no_output_____"
],
[
"freq",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb443426b7694be7721853e099417e34ad524b0f | 44,062 | ipynb | Jupyter Notebook | Welcome_to_Colaboratory.ipynb | psuto/TensorFlow2ForDL | 7a2520fd6acfd83a4a5ae924076669b13ede8be8 | [
"Unlicense"
] | null | null | null | Welcome_to_Colaboratory.ipynb | psuto/TensorFlow2ForDL | 7a2520fd6acfd83a4a5ae924076669b13ede8be8 | [
"Unlicense"
] | null | null | null | Welcome_to_Colaboratory.ipynb | psuto/TensorFlow2ForDL | 7a2520fd6acfd83a4a5ae924076669b13ede8be8 | [
"Unlicense"
] | null | null | null | 148.858108 | 31,886 | 0.852004 | [
[
[
"<a href=\"https://colab.research.google.com/github/psuto/TensorFlow2ForDL/blob/main/Welcome_to_Colaboratory.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# New section",
"_____no_output_____"
],
[
"<p><img alt=\"Colaboratory logo\" height=\"45px\" src=\"/img/colab_favicon.ico\" align=\"left\" hspace=\"10px\" vspace=\"0px\"></p>\n\n<h1>What is Colaboratory?</h1>\n\nColaboratory, or 'Colab' for short, allows you to write and execute Python in your browser, with \n- Zero configuration required\n- Free access to GPUs\n- Easy sharing\n\nWhether you're a <strong>student</strong>, a <strong>data scientist</strong> or an <strong>AI researcher</strong>, Colab can make your work easier. Watch <a href=\"https://www.youtube.com/watch?v=inN8seMm7UI\">Introduction to Colab</a> to find out more, or just get started below!",
"_____no_output_____"
],
[
"## <strong>Getting started</strong>\n\nThe document that you are reading is not a static web page, but an interactive environment called a <strong>Colab notebook</strong> that lets you write and execute code.\n\nFor example, here is a <strong>code cell</strong> with a short Python script that computes a value, stores it in a variable and prints the result:",
"_____no_output_____"
]
],
[
[
"seconds_in_a_day = 24 * 60 * 60\nseconds_in_a_day",
"_____no_output_____"
]
],
[
[
"To execute the code in the above cell, select it with a click and then either press the play button to the left of the code, or use the keyboard shortcut 'Command/Ctrl+Enter'. To edit the code, just click the cell and start editing.\n\nVariables that you define in one cell can later be used in other cells:",
"_____no_output_____"
]
],
[
[
"seconds_in_a_week = 7 * seconds_in_a_day\nseconds_in_a_week",
"_____no_output_____"
]
],
[
[
"Colab notebooks allow you to combine <strong>executable code</strong> and <strong>rich text</strong> in a single document, along with <strong>images</strong>, <strong>HTML</strong>, <strong>LaTeX</strong> and more. When you create your own Colab notebooks, they are stored in your Google Drive account. You can easily share your Colab notebooks with co-workers or friends, allowing them to comment on your notebooks or even edit them. To find out more, see <a href=\"/notebooks/basic_features_overview.ipynb\">Overview of Colab</a>. To create a new Colab notebook you can use the File menu above, or use the following link: <a href=\"http://colab.research.google.com#create=true\">Create a new Colab notebook</a>.\n\nColab notebooks are Jupyter notebooks that are hosted by Colab. To find out more about the Jupyter project, see <a href=\"https://www.jupyter.org\">jupyter.org</a>.",
"_____no_output_____"
],
[
"## Data science\n\nWith Colab you can harness the full power of popular Python libraries to analyse and visualise data. The code cell below uses <strong>numpy</strong> to generate some random data, and uses <strong>matplotlib</strong> to visualise it. To edit the code, just click the cell and start editing.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom matplotlib import pyplot as plt\n\nys = 200 + np.random.randn(100)\nx = [x for x in range(len(ys))]\n\nplt.plot(x, ys, '-')\nplt.fill_between(x, ys, 195, where=(ys > 195), facecolor='g', alpha=0.6)\n\nplt.title(\"Sample Visualization\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"You can import your own data into Colab notebooks from your Google Drive account, including from spreadsheets, as well as from GitHub and many other sources. To find out more about importing data, and how Colab can be used for data science, see the links below under <a href=\"#working-with-data\">Working with data</a>.",
"_____no_output_____"
],
[
"## Machine learning\n\nWith Colab you can import an image dataset, train an image classifier on it, and evaluate the model, all in just <a href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/tutorials/quickstart/beginner.ipynb\">a few lines of code</a>. Colab notebooks execute code on Google's cloud servers, meaning you can leverage the power of Google hardware, including <a href=\"#using-accelerated-hardware\">GPUs and TPUs</a>, regardless of the power of your machine. All you need is a browser.",
"_____no_output_____"
],
[
"Colab is used extensively in the machine learning community with applications including:\n- Getting started with TensorFlow\n- Developing and training neural networks\n- Experimenting with TPUs\n- Disseminating AI research\n- Creating tutorials\n\nTo see sample Colab notebooks that demonstrate machine learning applications, see the <a href=\"#machine-learning-examples\">machine learning examples</a> below.",
"_____no_output_____"
],
[
"## More resources\n\n### Working with notebooks in Colab\n- [Overview of Colaboratory](/notebooks/basic_features_overview.ipynb)\n- [Guide to markdown](/notebooks/markdown_guide.ipynb)\n- [Importing libraries and installing dependencies](/notebooks/snippets/importing_libraries.ipynb)\n- [Saving and loading notebooks in GitHub](https://colab.research.google.com/github/googlecolab/colabtools/blob/master/notebooks/colab-github-demo.ipynb)\n- [Interactive forms](/notebooks/forms.ipynb)\n- [Interactive widgets](/notebooks/widgets.ipynb)\n- <img src=\"/img/new.png\" height=\"20px\" align=\"left\" hspace=\"4px\" alt=\"New\"></img>\n [TensorFlow 2 in Colab](/notebooks/tensorflow_version.ipynb)\n\n<a name=\"working-with-data\"></a>\n### Working with data\n- [Loading data: Drive, Sheets and Google Cloud Storage](/notebooks/io.ipynb) \n- [Charts: visualising data](/notebooks/charts.ipynb)\n- [Getting started with BigQuery](/notebooks/bigquery.ipynb)\n\n### Machine learning crash course\nThese are a few of the notebooks from Google's online machine learning course. See the <a href=\"https://developers.google.com/machine-learning/crash-course/\">full course website</a> for more.\n- [Intro to Pandas](/notebooks/mlcc/intro_to_pandas.ipynb)\n- [TensorFlow concepts](/notebooks/mlcc/tensorflow_programming_concepts.ipynb)\n- [First steps with TensorFlow](/notebooks/mlcc/first_steps_with_tensor_flow.ipynb)\n- [Intro to neural nets](/notebooks/mlcc/intro_to_neural_nets.ipynb)\n- [Intro to sparse data and embeddings](/notebooks/mlcc/intro_to_sparse_data_and_embeddings.ipynb)\n\n<a name=\"using-accelerated-hardware\"></a>\n### Using accelerated hardware\n- [TensorFlow with GPUs](/notebooks/gpu.ipynb)\n- [TensorFlow with TPUs](/notebooks/tpu.ipynb)",
"_____no_output_____"
],
[
"<a name=\"machine-learning-examples\"></a>\n\n## Machine learning examples\n\nTo see end-to-end examples of the interactive machine-learning analyses that Colaboratory makes possible, take a look at these tutorials using models from <a href=\"https://tfhub.dev\">TensorFlow Hub</a>.\n\nA few featured examples:\n\n- <a href=\"https://tensorflow.org/hub/tutorials/tf2_image_retraining\">Retraining an Image Classifier</a>: Build a Keras model on top of a pre-trained image classifier to distinguish flowers.\n- <a href=\"https://tensorflow.org/hub/tutorials/tf2_text_classification\">Text Classification</a>: Classify IMDB film reviews as either <em>positive</em> or <em>negative</em>.\n- <a href=\"https://tensorflow.org/hub/tutorials/tf2_arbitrary_image_stylization\">Style Transfer</a>: Use deep learning to transfer style between images.\n- <a href=\"https://tensorflow.org/hub/tutorials/retrieval_with_tf_hub_universal_encoder_qa\">Multilingual Universal Sentence Encoder Q&A</a>: Use a machine-learning model to answer questions from the SQuAD dataset.\n- <a href=\"https://tensorflow.org/hub/tutorials/tweening_conv3d\">Video Interpolation</a>: Predict what happened in a video between the first and the last frame.\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cb443b3deefb03479c491608d03e0ba7708b0ea3 | 245,081 | ipynb | Jupyter Notebook | remote_sensing/python/colab/0_Tutorials/ee_ImageCollection.ipynb | HNoorazar/Kirti | fb7108dac1190774bd90a527aaa8a3cb405f127d | [
"MIT"
] | null | null | null | remote_sensing/python/colab/0_Tutorials/ee_ImageCollection.ipynb | HNoorazar/Kirti | fb7108dac1190774bd90a527aaa8a3cb405f127d | [
"MIT"
] | null | null | null | remote_sensing/python/colab/0_Tutorials/ee_ImageCollection.ipynb | HNoorazar/Kirti | fb7108dac1190774bd90a527aaa8a3cb405f127d | [
"MIT"
] | null | null | null | 245,081 | 245,081 | 0.750291 | [
[
[
"<!--COURSE_INFORMATION-->\n<img align=\"left\" style=\"padding-right:10px;\" src=\"https://sitejerk.com/images/google-earth-logo-png-5.png\" width=5% >\n<img align=\"right\" style=\"padding-left:10px;\" src=\"https://colab.research.google.com/img/colab_favicon_256px.png\" width=6% >\n\n\n>> *This notebook is part of the free course [EEwPython](https://colab.research.google.com/github/csaybar/EEwPython/blob/master/index.ipynb); the content is available [on GitHub](https://github.com/csaybar/EEwPython)* and released under the [Apache 2.0 License](https://www.gnu.org/licenses/gpl-3.0.en.html). 99% of this material has been adapted from [Google Earth Engine Guides](https://developers.google.com/earth-engine/).",
"_____no_output_____"
],
[
"<!--NAVIGATION-->\n < [Image](2_eeImage.ipynb) | [Contents](index.ipynb) | [Geometry, Feature and FeatureCollection](4_features.ipynb)>\n\n<a href=\"https://colab.research.google.com/github/csaybar/EEwPython/blob/master/3_eeImageCollection.ipynb\"><img align=\"left\" src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open in Colab\" title=\"Open and Execute in Google Colaboratory\"></a>",
"_____no_output_____"
],
[
"<center>\n<h1>Google Earth Engine with Python </h1>\n<h2> ee.ImageCollection</h2>\n</center>\n<h2> Topics:</h2>\n\n1. ImageCollection Overview\n2. ImageCollection Information and Metadata\n3. Filtering and ImageCollection\n4. Mapping over an ImageCollection\n5. Reducing an ImageCollection\n6. Compositing and Mosaicking\n7. Iterating over an ImageCollection\n\n\nAn **ImageCollection** is a stack or time series of images. In addition to loading an **ImageCollection** using an Earth Engine collection ID, Earth Engine has methods to create image collections. The constructor **ee.ImageCollection()** or the convenience method **ee.ImageCollection.fromImages()** create image collections from lists of images. You can also create new image collections by merging existing collections.",
"_____no_output_____"
],
[
"## Connecting GEE with Google Services",
"_____no_output_____"
],
[
"- **Authenticate to Earth Engine**",
"_____no_output_____"
]
],
[
[
"!pip install earthengine-api #earth-engine Python API",
"Requirement already satisfied: earthengine-api in /usr/local/lib/python3.6/dist-packages (0.1.209)\nRequirement already satisfied: future in /usr/local/lib/python3.6/dist-packages (from earthengine-api) (0.16.0)\nRequirement already satisfied: google-cloud-storage in /usr/local/lib/python3.6/dist-packages (from earthengine-api) (1.16.2)\nRequirement already satisfied: google-auth>=1.4.1 in /usr/local/lib/python3.6/dist-packages (from earthengine-api) (1.4.2)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from earthengine-api) (1.12.0)\nRequirement already satisfied: google-api-python-client in /usr/local/lib/python3.6/dist-packages (from earthengine-api) (1.7.11)\nRequirement already satisfied: google-auth-httplib2>=0.0.3 in /usr/local/lib/python3.6/dist-packages (from earthengine-api) (0.0.3)\nRequirement already satisfied: httplib2<1dev,>=0.9.2 in /usr/local/lib/python3.6/dist-packages (from earthengine-api) (0.11.3)\nRequirement already satisfied: google-resumable-media<0.5.0dev,>=0.3.1 in /usr/local/lib/python3.6/dist-packages (from google-cloud-storage->earthengine-api) (0.4.1)\nRequirement already satisfied: google-cloud-core<2.0dev,>=1.0.0 in /usr/local/lib/python3.6/dist-packages (from google-cloud-storage->earthengine-api) (1.0.3)\nRequirement already satisfied: cachetools>=2.0.0 in /usr/local/lib/python3.6/dist-packages (from google-auth>=1.4.1->earthengine-api) (4.0.0)\nRequirement already satisfied: pyasn1-modules>=0.2.1 in /usr/local/lib/python3.6/dist-packages (from google-auth>=1.4.1->earthengine-api) (0.2.7)\nRequirement already satisfied: rsa>=3.1.4 in /usr/local/lib/python3.6/dist-packages (from google-auth>=1.4.1->earthengine-api) (4.0)\nRequirement already satisfied: uritemplate<4dev,>=3.0.0 in /usr/local/lib/python3.6/dist-packages (from google-api-python-client->earthengine-api) (3.0.1)\nRequirement already satisfied: google-api-core<2.0.0dev,>=1.14.0 in /usr/local/lib/python3.6/dist-packages (from google-cloud-core<2.0dev,>=1.0.0->google-cloud-storage->earthengine-api) (1.15.0)\nRequirement already satisfied: pyasn1<0.5.0,>=0.4.6 in /usr/local/lib/python3.6/dist-packages (from pyasn1-modules>=0.2.1->google-auth>=1.4.1->earthengine-api) (0.4.8)\nRequirement already satisfied: setuptools>=34.0.0 in /usr/local/lib/python3.6/dist-packages (from google-api-core<2.0.0dev,>=1.14.0->google-cloud-core<2.0dev,>=1.0.0->google-cloud-storage->earthengine-api) (42.0.2)\nRequirement already satisfied: googleapis-common-protos<2.0dev,>=1.6.0 in /usr/local/lib/python3.6/dist-packages (from google-api-core<2.0.0dev,>=1.14.0->google-cloud-core<2.0dev,>=1.0.0->google-cloud-storage->earthengine-api) (1.6.0)\nRequirement already satisfied: requests<3.0.0dev,>=2.18.0 in /usr/local/lib/python3.6/dist-packages (from google-api-core<2.0.0dev,>=1.14.0->google-cloud-core<2.0dev,>=1.0.0->google-cloud-storage->earthengine-api) (2.21.0)\nRequirement already satisfied: pytz in /usr/local/lib/python3.6/dist-packages (from google-api-core<2.0.0dev,>=1.14.0->google-cloud-core<2.0dev,>=1.0.0->google-cloud-storage->earthengine-api) (2018.9)\nRequirement already satisfied: protobuf>=3.4.0 in /usr/local/lib/python3.6/dist-packages (from google-api-core<2.0.0dev,>=1.14.0->google-cloud-core<2.0dev,>=1.0.0->google-cloud-storage->earthengine-api) (3.10.0)\nRequirement already satisfied: idna<2.9,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests<3.0.0dev,>=2.18.0->google-api-core<2.0.0dev,>=1.14.0->google-cloud-core<2.0dev,>=1.0.0->google-cloud-storage->earthengine-api) (2.8)\nRequirement already satisfied: urllib3<1.25,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from requests<3.0.0dev,>=2.18.0->google-api-core<2.0.0dev,>=1.14.0->google-cloud-core<2.0dev,>=1.0.0->google-cloud-storage->earthengine-api) (1.24.3)\nRequirement already satisfied: chardet<3.1.0,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests<3.0.0dev,>=2.18.0->google-api-core<2.0.0dev,>=1.14.0->google-cloud-core<2.0dev,>=1.0.0->google-cloud-storage->earthengine-api) (3.0.4)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.6/dist-packages (from requests<3.0.0dev,>=2.18.0->google-api-core<2.0.0dev,>=1.14.0->google-cloud-core<2.0dev,>=1.0.0->google-cloud-storage->earthengine-api) (2019.11.28)\n"
],
[
"!earthengine authenticate ",
"Running command using Cloud API. Set --no-use_cloud_api to go back to using the API\n\n/usr/local/lib/python3.6/dist-packages/IPython/utils/traitlets.py:5: UserWarning: IPython.utils.traitlets has moved to a top-level traitlets package.\n warn(\"IPython.utils.traitlets has moved to a top-level traitlets package.\")\nTo authorize access needed by Earth Engine, open the following URL in a web browser and follow the instructions. If the web browser does not start automatically, please manually browse the URL below.\n\n https://accounts.google.com/o/oauth2/auth?client_id=517222506229-vsmmajv00ul0bs7p89v5m89qs8eb9359.apps.googleusercontent.com&scope=https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fearthengine+https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdevstorage.full_control&redirect_uri=urn%3Aietf%3Awg%3Aoauth%3A2.0%3Aoob&response_type=code\n\nThe authorization workflow will generate a code, which you should paste in the box below. \nEnter verification code: 4/vgFoxnSFz9UAnqVcyHJAB-a9t_cv9zzj4BcIFl6zN1gXDCBQ6Jbwu9Q\n\nSuccessfully saved authorization token.\n"
]
],
[
[
"- **Authenticate to Google Drive (OPTIONAL)**",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"_____no_output_____"
]
],
[
[
"- **Authenticate to Google Cloud (OPTIONAL)**",
"_____no_output_____"
]
],
[
[
"from google.colab import auth\nauth.authenticate_user()",
"_____no_output_____"
]
],
[
[
"## Testing the software setup\n\n\n",
"_____no_output_____"
]
],
[
[
"# Earth Engine Python API\nimport ee\nee.Initialize()",
"_____no_output_____"
],
[
"import folium\n\n# Define the URL format used for Earth Engine generated map tiles.\nEE_TILES = 'https://earthengine.googleapis.com/map/{mapid}/{{z}}/{{x}}/{{y}}?token={token}'\n\nprint('Folium version: ' + folium.__version__)",
"Folium version: 0.8.3\n"
],
[
"# @title Mapdisplay: Display GEE objects using folium.\n\ndef Mapdisplay(center, dicc, Tiles=\"OpensTreetMap\",zoom_start=10):\n '''\n :param center: Center of the map (Latitude and Longitude).\n :param dicc: Earth Engine Geometries or Tiles dictionary\n :param Tiles: Mapbox Bright,Mapbox Control Room,Stamen Terrain,Stamen Toner,stamenwatercolor,cartodbpositron.\n :zoom_start: Initial zoom level for the map.\n :return: A folium.Map object.\n '''\n mapViz = folium.Map(location=center,tiles=Tiles, zoom_start=zoom_start)\n for k,v in dicc.items():\n if ee.image.Image in [type(x) for x in v.values()]:\n folium.TileLayer(\n tiles = EE_TILES.format(**v),\n attr = 'Google Earth Engine',\n overlay =True,\n name = k\n ).add_to(mapViz)\n else:\n folium.GeoJson(\n data = v,\n name = k\n ).add_to(mapViz)\n mapViz.add_child(folium.LayerControl())\n return mapViz",
"_____no_output_____"
]
],
[
[
"# 1. Image Collection Overview\n\nAn `ImageCollection` is a **stack or time series of images**. In addition to loading an `ImageCollection` using an Earth Engine collection ID, Earth Engine has methods to create image collections. The constructor `ee.ImageCollection()` or the convenience method `ee.ImageCollection.fromImages()` create image collections from lists of images. You can also create new image collections by merging existing collections. For example:\n",
"_____no_output_____"
]
],
[
[
"# Create arbitrary constant images.\nconstant1 = ee.Image(1)\nconstant2 = ee.Image(2)\n\n# Create a collection by giving a list to the constructor.\ncollectionFromConstructor = ee.ImageCollection([constant1, constant2])\nprint('collectionFromConstructor: ')\ncollectionFromConstructor.getInfo()",
"collectionFromConstructor: \n"
],
[
"# Create a collection with fromImages().\ncollectionFromImages = ee.ImageCollection.fromImages([ee.Image(3), ee.Image(4)])\nprint('collectionFromImages: ')\ncollectionFromImages.getInfo()",
"collectionFromImages: \n"
],
[
"# Merge two collections.\nmergedCollection = collectionFromConstructor.merge(collectionFromImages)\nprint('mergedCollection: ')\nmergedCollection.getInfo()",
"mergedCollection: \n"
],
[
"# Create a toy FeatureCollection\nfeatures = ee.FeatureCollection(\n [ee.Feature(None, {'foo': 1}), ee.Feature(None, {'foo': 2})])\n\n# Create an ImageCollection from the FeatureCollection\n# by mapping a function over the FeatureCollection.\nimages = features.map(lambda feature:ee.Image(ee.Number(feature.get('foo'))))\n\n# Print the resultant collection.\nprint('Image collection: ')\nimages.getInfo()",
"Image collection: \n"
]
],
[
[
"Note that in this example an `ImageCollection` is created by mapping a function that returns an `Image` over a `FeatureCollection`. Learn more about mapping in the [Mapping over an ImageCollection section](https://developers.google.com/earth-engine/ic_mapping). Learn more about feature collections from the [FeatureCollection section](https://developers.google.com/earth-engine/feature_collections).",
"_____no_output_____"
],
[
"# 2. ImageCollection Information and Metadata\n\nAs with Images, there are a variety of ways to get information about an ImageCollection. The collection can be printed directly to the console, but the console printout is **limited to 5000 elements**. Collections larger than 5000 images will need to be filtered before printing. Printing a large collection will be correspondingly slower. The following example shows various ways of getting information about image collections programmatically.",
"_____no_output_____"
]
],
[
[
"# Load a Landsat 8 ImageCollection for a single path-row.\ncollection = ee.ImageCollection('LANDSAT/LC08/C01/T1_TOA')\\\n .filter(ee.Filter.eq('WRS_PATH', 44))\\\n .filter(ee.Filter.eq('WRS_ROW', 34))\\\n .filterDate('2014-03-01', '2014-08-01')\nprint('Collection: ')\ncollection.getInfo()",
"Collection: \n"
],
[
"# Get the number of images.\ncount = collection.size()\nprint('Count: ', count.getInfo())",
"Count: 9\n"
],
[
"from datetime import datetime as dt\n# Get the date range of images in the collection.\nrango = collection.reduceColumns(ee.Reducer.minMax(), [\"system:time_start\"])\n\n# Passing numeric date to standard\ninit_date = ee.Date(rango.get('min')).getInfo()['value']/1000.\ninit_date_f = dt.utcfromtimestamp(init_date).strftime('%Y-%m-%d %H:%M:%S')\n\nlast_date = ee.Date(rango.get('max')).getInfo()['value']/1000.\nlast_date_f = dt.utcfromtimestamp(last_date).strftime('%Y-%m-%d %H:%M:%S')\n\nprint('Date range: ',init_date_f,' - ',last_date_f)",
"Date range: 2014-03-18 18:46:32 - 2014-07-24 18:45:57\n"
],
[
"# Get statistics for a property of the images in the collection.\nsunStats = collection.aggregate_stats('SUN_ELEVATION')\nprint('Sun elevation statistics: ')\nsunStats.getInfo()",
"Sun elevation statistics: \n"
],
[
"# Sort by a cloud cover property, get the least cloudy image.\nimage = ee.Image(collection.sort('CLOUD_COVER').first())\nprint('Least cloudy image: ', )\nimage.getInfo()",
"Least cloudy image: \n"
],
[
"# Limit the collection to the 10 most recent images.\nrecent = collection.sort('system:time_start', False).limit(10)\nprint('Recent images: ')\n# recent.getInfo()",
"Recent images: \n"
]
],
[
[
"# 3. Filtering an ImageCollection\n\nAs illustrated in the Get Started section and the ImageCollection Information section, Earth Engine provides a variety of convenience methods for filtering image collections. Specifically, many common use cases are handled by **imageCollection.filterDate()**, and **imageCollection.filterBounds()**. For general purpose filtering, use **imageCollection.filter()** with an **ee.Filter** as an argument. The following example demonstrates both convenience methods and **filter()** to identify and remove images with bad registration from an **ImageCollection**.",
"_____no_output_____"
]
],
[
[
"# Load Landsat 5 data, filter by date and bounds.\ncollection = ee.ImageCollection('LANDSAT/LT05/C01/T2').filterDate('1987-01-01', '1990-05-01').filterBounds(ee.Geometry.Point(25.8544, -18.08874))\n\n# Also filter the collection by the IMAGE_QUALITY property.\nfiltered = collection.filterMetadata(name = 'IMAGE_QUALITY', operator = 'equals', value = 9)\n\n# Create two composites to check the effect of filtering by IMAGE_QUALITY.\nbadComposite = ee.Algorithms.Landsat.simpleComposite(collection = collection, percentile = 75, cloudScoreRange = 3)\ngoodComposite = ee.Algorithms.Landsat.simpleComposite(collection = filtered, percentile = 75, cloudScoreRange = 3)\n\ndicc = {\n 'Bad composite' : badComposite.getMapId({'bands': ['B3', 'B2', 'B1'], 'gain': 3.5}),\n 'Good composite': goodComposite.getMapId({'bands': ['B3', 'B2', 'B1'], 'gain': 3.5})\n}\n\n# Display the results\ncenter = [-18.08874, 25.8544]\nMapdisplay(center, dicc, zoom_start= 13)",
"_____no_output_____"
],
[
"dicc = {\n 'Bad composite' : badComposite.getMapId({'bands': ['B3', 'B2', 'B1'], 'gain': 3.5}) # ,\n # 'Good composite': goodComposite.getMapId({'bands': ['B3', 'B2', 'B1'], 'gain': 3.5})\n}\n\n# Display the results\ncenter = [-18.08874, 25.8544]\nMapdisplay(center, dicc, zoom_start= 13)",
"_____no_output_____"
]
],
[
[
"# 4. Mapping over an ImageCollection\n\nTo apply a function to every Image in an ImageCollection use imageCollection.map(). The only argument to map() is a function which takes one parameter: an ee.Image. For example, the following code adds a timestamp band to every image in the collection.",
"_____no_output_____"
]
],
[
[
"from pprint import pprint\n# Load a Landsat 8 collection for a single path-row.\ncollection = ee.ImageCollection('LANDSAT/LC08/C01/T1_TOA').filter(ee.Filter.eq('WRS_PATH', 44)).filter(ee.Filter.eq('WRS_ROW', 34))\n\n# This function adds a band representing the image timestamp.\ndef addTime(image):\n\treturn image.addBands(image.metadata('system:time_start'))\n\n# Map the function over the collection and display the result.\npprint(collection.map(addTime).limit(3).getInfo())",
"_____no_output_____"
]
],
[
[
"Note that in the predefined function, the **metadata()** method is used to create a new Image from the value of a property. As discussed in the *Reducing* and *Compositing* sections, having that time band is useful for the linear modeling of change and for making composites.\n\nThe mapped function is limited in the operations it can perform. Specifically, it can’t modify variables outside the function; it can’t print anything; it can’t use JavaScript ‘if’ or ‘for’ statements. However, you can use **ee.Algorithms.If()** to perform conditional operations in a mapped function.",
"_____no_output_____"
]
],
[
[
"# Load a Landsat 8 collection for a single path-row.\ncollection = ee.ImageCollection('LANDSAT/LC8_L1T_TOA').filter(ee.Filter.eq('WRS_PATH', 44)).filter(ee.Filter.eq('WRS_ROW', 34))\n\n# This function uses a conditional statement to return the image if\n# the solar elevation > 40 degrees. Otherwise it returns a zero image.\ndef conditional(image):\n\treturn ee.Algorithms.If(ee.Number(image.get('SUN_ELEVATION')).gt(40), image, ee.Image(0))\n\n# Map the function over the collection, convert to a List and print the result.\nprint('Expand this to see the result: ')\npprint(collection.map(conditional).limit(3).getInfo())",
"_____no_output_____"
]
],
[
[
"Inspect the list of images in the output ImageCollection and note that the when the condition evaluated by the **If()** algorithm is true, the output contains a constant image. Although this demonstrates a server-side conditional function (learn more about client vs. server in Earth Engine on this page), avoid **If()** in general and use filters instead.",
"_____no_output_____"
],
[
"# 5. Reducing an ImageCollection\n\nTo composite images in an **ImageCollection**, use **imageCollection.reduce()**. This will composite all the images in the collection to a single image representing, for example, the min, max, mean or standard deviation of the images. (See the Reducers section for more information about reducers). For example, to create a median value image from a collection:",
"_____no_output_____"
]
],
[
[
"# Load a Landsat 8 collection for a single path-row.\ncollection = ee.ImageCollection('LANDSAT/LC08/C01/T1_TOA').filter(ee.Filter.eq('WRS_PATH', 44)).filter(ee.Filter.eq('WRS_ROW', 34)).filterDate('2014-01-01', '2015-01-01')\n\n# Compute a median image and display.\nmedian = collection.median()\n\ndicc = {\n 'median' : median.getMapId({'bands': ['B4', 'B3', 'B2'], 'max': 0.3})\n}\n\n# Display the results\ncenter = [37.7726, -122.3578]\nMapdisplay(center, dicc, zoom_start= 12)",
"_____no_output_____"
]
],
[
[
"At each location in the output image, in each band, the pixel value is the median of all unmasked pixels in the input imagery (the images in the collection). In the previous example, median() is a convenience method for the following call:",
"_____no_output_____"
]
],
[
[
"# Reduce the collection with a median reducer.\nmedian = collection.reduce(ee.Reducer.median())\n\n# Display the median image.\ndicc = {'also median' : median.getMapId({'bands': ['B4_median', 'B3_median', 'B2_median'], 'max': 0.3})}\n\n# Display the results\ncenter = [37.7726, -122.3578]\nMapdisplay(center, dicc, zoom_start= 12)",
"_____no_output_____"
]
],
[
[
"Note that the band names differ as a result of using reduce() instead of the convenience method. Specifically, the names of the reducer have been appended to the band names.\n\nMore complex reductions are also possible using reduce(). For example, to compute the long term linear trend over a collection, use one of the linear regression reducers. The following code computes the linear trend of MODIS Enhanced Vegetation Index (EVI).",
"_____no_output_____"
]
],
[
[
"# This function adds a band representing the image timestamp.\ndef addTime(image):\n return image.addBands(image.metadata('system:time_start').divide(1000 * 60 * 60 * 24 * 365))\n\n# Load a MODIS collection, filter to several years of 16 day mosaics, and map the time band function over it.\ncollection = ee.ImageCollection('MODIS/006/MYD13A1').filterDate('2004-01-01', '2010-10-31').map(addTime)\n\n# Select the bands to model with the independent variable first.\n# Compute the linear trend over time.\ntrend = collection.select(['system:time_start', 'EVI']).reduce(ee.Reducer.linearFit())\n\n# Display the trend with increasing slopes in green, decreasing in red.\ndicc = {\n 'EVI trend' : trend.getMapId({'min': 0, 'max': [-100, 100, 10000], 'bands': ['scale', 'scale', 'offset']})\n}\n\n# Display the results\ncenter = [39.436, -96.943]\nMapdisplay(center, dicc, zoom_start= 5)",
"_____no_output_____"
]
],
[
[
"Note that the output of the reduction in this example is a two banded image with one band for the slope of a linear regression (**scale**) and one band for the intercept (**offset**). Explore the API documentation to see a list of the reducers that are available to reduce an **ImageCollection** to a single Image. See the **ImageCollection.reduce()** section for more information about reducing image collections.\n\n",
"_____no_output_____"
],
[
"# 6. Compositing and Mosaicking\n\nIn general, compositing refers to the process of combining spatially overlapping images into a single image based on an aggregation function. Mosaicking refers to the process of spatially assembling image datasets to produce a spatially continuous image. In Earth Engine, these terms are used interchangeably, though both compositing and mosaicking are supported. For example, consider the task of compositing multiple images in the same location. For example, using one National Agriculture Imagery Program (NAIP) Digital Orthophoto Quarter Quadrangle (DOQQ) at different times, the following example demonstrates making a maximum value composite:",
"_____no_output_____"
]
],
[
[
"# Load three NAIP quarter quads in the same location, different times.\nnaip2004_2012 = ee.ImageCollection('USDA/NAIP/DOQQ')\\\n .filterBounds(ee.Geometry.Point(-71.08841, 42.39823))\\\n .filterDate('2004-07-01', '2012-12-31')\\\n .select(['R', 'G', 'B'])\n\n# Temporally composite the images with a maximum value function.\ncomposite = naip2004_2012.max()\ncenter = [42.3712, -71.12532]\nMapdisplay(center, {'max value composite':composite.getMapId()},zoom_start=12)",
"_____no_output_____"
]
],
[
[
"Consider the need to mosaic four different DOQQs at the same time, but different locations. The following example demonstrates that using **imageCollection.mosaic()**:\n\n",
"_____no_output_____"
]
],
[
[
"# Load four 2012 NAIP quarter quads, different locations.\nnaip2012 = ee.ImageCollection('USDA/NAIP/DOQQ')\\\n .filterBounds(ee.Geometry.Rectangle(-71.17965, 42.35125, -71.08824, 42.40584))\\\n .filterDate('2012-01-01', '2012-12-31')\n\n# Spatially mosaic the images in the collection and display.\nmosaic = naip2012.mosaic()\ncenter = [42.3712,-71.12532]\nMapdisplay(center,{'spatial mosaic':mosaic.getMapId()},zoom_start=12)",
"_____no_output_____"
]
],
[
[
"Note that there is some overlap in the DOQQs in the previous example. The **mosaic()** method composites overlapping images according to their order in the collection (last on top). To control the source of pixels in a mosaic (or a composite), use image masks. For example, the following uses thresholds on spectral indices to mask the image data in a mosaic:\n\n",
"_____no_output_____"
]
],
[
[
"# Load a NAIP quarter quad, display.\nnaip = ee.Image('USDA/NAIP/DOQQ/m_4207148_nw_19_1_20120710')\n\n# Create the NDVI and NDWI spectral indices.\nndvi = naip.normalizedDifference(['N', 'R'])\nndwi = naip.normalizedDifference(['G', 'N'])\n\n# Create some binary images from thresholds on the indices.\n# This threshold is designed to detect bare land.\nbare1 = ndvi.lt(0.2).And(ndwi.lt(0.3))\n# This detects bare land with lower sensitivity. It also detects shadows.\nbare2 = ndvi.lt(0.2).And(ndwi.lt(0.8));\n\n# Define visualization parameters for the spectral indices.\nndviViz = {'min': -1, 'max': 1, 'palette': ['FF0000', '00FF00']}\nndwiViz = {'min': 0.5, 'max': 1, 'palette': ['00FFFF', '0000FF']}\n\n# Mask and mosaic visualization images. The last layer is on top.\nmosaic = ee.ImageCollection([\n # NDWI > 0.5 is water. Visualize it with a blue palette.\n ndwi.updateMask(ndwi.gte(0.5)).visualize(**ndwiViz),\n # NDVI > 0.2 is vegetation. Visualize it with a green palette.\n ndvi.updateMask(ndvi.gte(0.2)).visualize(**ndviViz),\n # Visualize bare areas with shadow (bare2 but not bare1) as gray.\n bare2.updateMask(bare2.And(bare1.Not())).visualize(**{'palette': ['AAAAAA']}),\n # Visualize the other bare areas as white.\n bare1.updateMask(bare1).visualize(**{'palette': ['FFFFFF']}),\n]).mosaic()\n\ncenter = [42.3443, -71.0915]\ndicc = {'NAIP DOQQ':naip.getMapId(),\n 'Visualization mosaic':mosaic.getMapId()}\nMapdisplay(center,dicc,zoom_start=14) ",
"_____no_output_____"
]
],
[
[
"To make a composite which maximizes an arbitrary band in the input, use **imageCollection.qualityMosaic()**. The **qualityMosaic()** method sets each pixel in the composite based on which image in the collection has a maximum value for the specified band. For example, the following code demonstrates making a greenest pixel composite and a recent value composite:",
"_____no_output_____"
]
],
[
[
"# This function masks clouds in Landsat 8 imagery.\ndef maskClouds(img):\n scored = ee.Algorithms.Landsat.simpleCloudScore(img)\n return img.updateMask(scored.select(['cloud']).lt(20))\n\n\n# This function masks clouds and adds quality bands to Landsat 8 images.\ndef addQualityBands(img):\n return maskClouds(img).addBands(img.normalizedDifference(['B5', 'B4']))\\\n .addBands(img.metadata('system:time_start')) # time in days\n\n\n# Load a 2014 Landsat 8 ImageCollection.\n# Map the cloud masking and quality band function over the collection.\ncollection = ee.ImageCollection('LANDSAT/LC08/C01/T1_TOA')\\\n .filterDate('2014-06-01', '2014-12-31')\\\n .map(addQualityBands)\n\n# Create a cloud-free, most recent value composite.\nrecentValueComposite = collection.qualityMosaic('system:time_start')\n\n# Create a greenest pixel composite.\ngreenestPixelComposite = collection.qualityMosaic('nd')\n\n\n# Create a cloudy image in the collection.\ncloudy = ee.Image('LANDSAT/LC08/C01/T1_TOA/LC08_044034_20140825');\n\n# Display the results.\ncenter = [37.8239, -122.374] # San Francisco Bay\nvizParams = {'bands': ['B5', 'B4', 'B3'], 'min': 0, 'max': 0.4}\ndicc = {'recent value composite':recentValueComposite.getMapId(vizParams),\n 'greenest pixel composite':greenestPixelComposite.getMapId(vizParams),\n 'cloudy':cloudy.getMapId(vizParams)}\n\nMapdisplay(center,dicc,zoom_start=12)",
"_____no_output_____"
]
],
[
[
"# 7. Iterating over an ImageCollection\n\nAlthough `map()` applies a function to every image in a collection, the function visits every image in the collection independently. For example, suppose you want to compute a cumulative anomaly ($A_t$) at time t from a time series. To obtain a recursively defined series of the form $A_t = f(Image_t, A_{t-1})$, mapping won't work because the function (f) depends on the previous result ($A{t-1}$). \n\nFor example, suppose you want to compute a series of cumulative Normalized Difference Vegetation Index (NDVI) anomaly images relative to a baseline. Let $A_0 = 0$ and $f(Image_t, A_{t-1}) = Image_t + A_{t-1}$ where $A_{t-1}$ is the cumulative anomaly up to time $t-1$ and Image_t is the anomaly at time t. Use **imageCollection.iterate()** to make this recursively defined ImageCollection. In the following example, the function **accumulate()** takes two parameters: an image in the collection, and a list of all the previous outputs. With each call to **iterate()**, the anomaly is added to the running sum and the result is added to the list. The final result is passed to the **ImageCollection** constructor to get a new sequence of images:\n",
"_____no_output_____"
]
],
[
[
"# Load MODIS EVI imagery.\ncollection = ee.ImageCollection('MODIS/006/MYD13A1').select('EVI');\n\n# Define reference conditions from the first 10 years of data.\nreference = collection.filterDate('2001-01-01', '2010-12-31')\\\n .sort('system:time_start', False) # Sort chronologically in descending order.\n\n# Compute the mean of the first 10 years.\nmean = reference.mean()\n\n# Compute anomalies by subtracting the 2001-2010 mean from each image in a\n# collection of 2011-2014 images. Copy the date metadata over to the\n# computed anomaly images in the new collection.\nseries = collection.filterDate('2011-01-01', '2014-12-31')\\\n .map(lambda img: img.subtract(mean).set('system:time_start', img.get('system:time_start')))\n\n# Display cumulative anomalies.\ncenter = [40.2,-100.811]\nvizParams = {'min': -60000, 'max': 60000, 'palette': ['FF0000', '000000', '00FF00']}\ndicc = {'EVI anomaly':series.sum().getMapId(vizParams)}\nMapdisplay(center,dicc,zoom_start=5)",
"_____no_output_____"
],
[
"# Get the timestamp from the most recent image in the reference collection.\ntime0 = reference.first().get('system:time_start')\n\n# Use imageCollection.iterate() to make a collection of cumulative anomaly over time.\n# The initial value for iterate() is a list of anomaly images already processed.\n# The first anomaly image in the list is just 0, with the time0 timestamp.\nfirst = ee.List([ee.Image(0).set('system:time_start', time0).select([0], ['EVI'])]) # Rename the first band 'EVI'.\n\n# This is a function to pass to Iterate().\n# As anomaly images are computed, add them to the list.\ndef accumulate(img,lista):\n # Get the latest cumulative anomaly image from the end of the list with\n # get(-1). Since the type of the list argument to the function is unknown,\n # it needs to be cast to a List. Since the return type of get() is unknown,\n # cast it to Image.\n img = ee.Image(img)\n previous = ee.Image(ee.List(lista).get(-1))\n # Add the current anomaly to make a new cumulative anomaly image.\n added = img.add(previous)\\\n .set('system:time_start', img.get('system:time_start')) # Propagate metadata to the new image.\n # Return the list with the cumulative anomaly inserted.\n return ee.List(lista).add(added)\n\n# Create an ImageCollection of cumulative anomaly images by iterating.\n# Since the return type of iterate is unknown, it needs to be cast to a List.\ncumulative = ee.ImageCollection(ee.List(series.iterate(accumulate, first)))",
"_____no_output_____"
],
[
"def stackCollection(collection):\n # Create an initial image.\n first = ee.Image(collection.first()).select([])\n\n # Write a function that appends a band to an image.\n def appendBands(image, previous):\n return ee.Image(previous).addBands(image)\n return ee.Image(collection.iterate(appendBands, first))",
"_____no_output_____"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\n# Chart some interesting locations.\npt1 = ee.Geometry.Point(116.4647, 40.1054)\n\n# ee.ImageCollection to ee.Image\nimg_cumulative = stackCollection(cumulative)\n\n\nseries = img_cumulative.reduceRegions(collection=pt1,\n reducer=ee.Reducer.mean(),\n scale=500)\n\ndic_series = series.getInfo()\nEVI_anom = np.array(list(dic_series['features'][0]['properties'].values()))\n\nplt.plot(EVI_anom)\nplt.show()",
"_____no_output_____"
]
],
[
[
"<!--NAVIGATION-->\n < [Image](2_eeImage.ipynb) | [Contents](index.ipynb) | [Geometry, Feature and FeatureCollection](4_features.ipynb)>\n\n<a href=\"https://colab.research.google.com/github/csaybar/EEwPython/blob/master/3_eeImageCollection.ipynb\"><img align=\"left\" src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open in Colab\" title=\"Open and Execute in Google Colaboratory\"></a>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb443d798d616b7dd7ee08aa85a5a8e7ed4a77f2 | 17,502 | ipynb | Jupyter Notebook | Transfer Learning/ClassifyFlowers_DL (TransferLearning_MobileNet).ipynb | a-shah8/DeepLearning | 8bc29643f4e2d50d78740f1500da70e20499efe7 | [
"MIT"
] | 1 | 2021-01-26T05:11:53.000Z | 2021-01-26T05:11:53.000Z | Transfer Learning/ClassifyFlowers_DL (TransferLearning_MobileNet).ipynb | a-shah8/DeepLearning | 8bc29643f4e2d50d78740f1500da70e20499efe7 | [
"MIT"
] | null | null | null | Transfer Learning/ClassifyFlowers_DL (TransferLearning_MobileNet).ipynb | a-shah8/DeepLearning | 8bc29643f4e2d50d78740f1500da70e20499efe7 | [
"MIT"
] | null | null | null | 27.780952 | 107 | 0.567421 | [
[
[
"## Libraries",
"_____no_output_____"
]
],
[
[
"### Uncomment the next two lines to,\n### install tensorflow_hub and tensorflow datasets\n\n#!pip install tensorflow_hub\n#!pip install tensorflow_datasets",
"_____no_output_____"
],
[
"import numpy as np\nimport tensorflow as tf\nimport matplotlib.pyplot as plt\n\nimport tensorflow_hub as hub\nimport tensorflow_datasets as tfds\n\nfrom tensorflow.keras import layers",
"_____no_output_____"
]
],
[
[
"### Download and Split data into Train and Validation",
"_____no_output_____"
]
],
[
[
"def get_data():\n (train_set, validation_set), info = tfds.load(\n 'tf_flowers', \n with_info=True, \n as_supervised=True, \n split=['train[:70%]', 'train[70%:]'],\n )\n \n return train_set, validation_set, info\n\ntrain_set, validation_set, info = get_data()",
"_____no_output_____"
],
[
"num_examples = info.splits['train'].num_examples\nnum_classes = info.features['label'].num_classes\n\nprint('Total Number of Classes: {}'.format(num_classes))\nprint('Total Number of Training Images: {}'.format(len(train_set)))\nprint('Total Number of Validation Images: {} \\n'.format(len(validation_set)))",
"_____no_output_____"
],
[
"img_shape = 224\nbatch_size = 32\n\ndef format_image(image, label):\n image = tf.image.resize(image, (img_shape, img_shape))/255.0\n return image, label\n\ntrain_batches = train_set.shuffle(num_examples//4).map(format_image).batch(batch_size).prefetch(1)\nvalidation_batches = validation_set.map(format_image).batch(batch_size).prefetch(1)",
"_____no_output_____"
]
],
[
[
"### Getting MobileNet model's learned features",
"_____no_output_____"
]
],
[
[
"def get_mobilenet_features():\n URL = \"https://tfhub.dev/google/tf2-preview/mobilenet_v2/feature_vector/4\"\n global img_shape\n feature_extractor = hub.KerasLayer(URL, input_shape=(img_shape, img_shape,3))\n \n return feature_extractor",
"_____no_output_____"
],
[
"### Freezing the layers of transferred model (MobileNet)\nfeature_extractor = get_mobilenet_features()\nfeature_extractor.trainable = False",
"_____no_output_____"
]
],
[
[
"## Deep Learning Model - Transfer Learning using MobileNet",
"_____no_output_____"
]
],
[
[
"def create_transfer_learned_model(feature_extractor):\n \n global num_classes\n model = tf.keras.Sequential([\n feature_extractor, \n layers.Dense(num_classes, activation='softmax')\n ])\n\n model.compile(\n optimizer='adam', \n loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True), \n metrics=['accuracy'])\n\n model.summary()\n \n return model",
"_____no_output_____"
]
],
[
[
"### Training the last classification layer of the model",
"_____no_output_____"
],
[
"Achieved Validation Accuracy: 90.10% (significant improvement over simple architecture)",
"_____no_output_____"
]
],
[
[
"epochs = 6\nmodel = create_transfer_learned_model(feature_extractor)\nhistory = model.fit(train_batches,\n epochs=epochs,\n validation_data=validation_batches)",
"_____no_output_____"
]
],
[
[
"### Plotting Accuracy and Loss Curves",
"_____no_output_____"
]
],
[
[
"def create_plots(history):\n acc = history.history['accuracy']\n val_acc = history.history['val_accuracy']\n\n loss = history.history['loss']\n val_loss = history.history['val_loss']\n\n global epochs\n epochs_range = range(epochs)\n\n plt.figure(figsize=(8, 8))\n plt.subplot(1, 2, 1)\n plt.plot(epochs_range, acc, label='Training Accuracy')\n plt.plot(epochs_range, val_acc, label='Validation Accuracy')\n plt.legend(loc='lower right')\n plt.title('Training and Validation Accuracy')\n\n plt.subplot(1, 2, 2)\n plt.plot(epochs_range, loss, label='Training Loss')\n plt.plot(epochs_range, val_loss, label='Validation Loss')\n plt.legend(loc='upper right')\n plt.title('Training and Validation Loss')\n plt.show()\n \ncreate_plots(history)",
"_____no_output_____"
]
],
[
[
"### Prediction",
"_____no_output_____"
]
],
[
[
"def predict():\n \n global train_batches, info\n \n image_batch, label_batch = next(iter(train_batches.take(1)))\n image_batch = image_batch.numpy()\n label_batch = label_batch.numpy()\n\n predicted_batch = model.predict(image_batch)\n predicted_batch = tf.squeeze(predicted_batch).numpy()\n\n class_names = np.array(info.features['label'].names)\n predicted_ids = np.argmax(predicted_batch, axis=-1)\n predicted_class_names = class_names[predicted_ids]\n \n return image_batch, label_batch, predicted_ids, predicted_class_names",
"_____no_output_____"
],
[
"image_batch, label_batch, predicted_ids, predicted_class_names = predict()\nprint(\"Labels: \", label_batch)\nprint(\"Predicted labels: \", predicted_ids)",
"_____no_output_____"
],
[
"def plot_figures():\n \n global image_batch, predicted_ids, label_batch\n plt.figure(figsize=(10,9))\n for n in range(30):\n plt.subplot(6,5,n+1)\n plt.subplots_adjust(hspace = 0.3)\n plt.imshow(image_batch[n])\n color = \"blue\" if predicted_ids[n] == label_batch[n] else \"red\"\n plt.title(predicted_class_names[n].title(), color=color)\n plt.axis('off')\n _ = plt.suptitle(\"Model predictions (blue: correct, red: incorrect)\")\n \nplot_figures()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb44429fa7610e95c0549cb893cac5ce4efaaf1e | 35,391 | ipynb | Jupyter Notebook | International debt.ipynb | monisha-anila/Data-Camp- | f87c59c1af7e642e1df50152341a3a6d1347ac84 | [
"MIT"
] | null | null | null | International debt.ipynb | monisha-anila/Data-Camp- | f87c59c1af7e642e1df50152341a3a6d1347ac84 | [
"MIT"
] | null | null | null | International debt.ipynb | monisha-anila/Data-Camp- | f87c59c1af7e642e1df50152341a3a6d1347ac84 | [
"MIT"
] | null | null | null | 32.952514 | 503 | 0.477607 | [
[
[
"## 1. The World Bank's international debt data\n<p>It's not that we humans only take debts to manage our necessities. A country may also take debt to manage its economy. For example, infrastructure spending is one costly ingredient required for a country's citizens to lead comfortable lives. <a href=\"https://www.worldbank.org\">The World Bank</a> is the organization that provides debt to countries.</p>\n<p>In this notebook, we are going to analyze international debt data collected by The World Bank. The dataset contains information about the amount of debt (in USD) owed by developing countries across several categories. We are going to find the answers to questions like: </p>\n<ul>\n<li>What is the total amount of debt that is owed by the countries listed in the dataset?</li>\n<li>Which country owns the maximum amount of debt and what does that amount look like?</li>\n<li>What is the average amount of debt owed by countries across different debt indicators?</li>\n</ul>\n<p><img src=\"https://assets.datacamp.com/production/project_754/img/image.jpg\" alt></p>\n<p>The first line of code connects us to the <code>international_debt</code> database where the table <code>international_debt</code> is residing. Let's first <code>SELECT</code> <em>all</em> of the columns from the <code>international_debt</code> table. Also, we'll limit the output to the first ten rows to keep the output clean.</p>",
"_____no_output_____"
]
],
[
[
"%%sql\npostgresql:///international_debt\nSELECT * FROM international_debt\nLIMIT 10;\n ",
"10 rows affected.\n"
]
],
[
[
"## 2. Finding the number of distinct countries\n<p>From the first ten rows, we can see the amount of debt owed by <em>Afghanistan</em> in the different debt indicators. But we do not know the number of different countries we have on the table. There are repetitions in the country names because a country is most likely to have debt in more than one debt indicator. </p>\n<p>Without a count of unique countries, we will not be able to perform our statistical analyses holistically. In this section, we are going to extract the number of unique countries present in the table. </p>",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT COUNT(DISTINCT country_name) AS total_distinct_countries \nFROM international_debt;",
" * postgresql:///international_debt\n1 rows affected.\n"
]
],
[
[
"## 3. Finding out the distinct debt indicators\n<p>We can see there are a total of 124 countries present on the table. As we saw in the first section, there is a column called <code>indicator_name</code> that briefly specifies the purpose of taking the debt. Just beside that column, there is another column called <code>indicator_code</code> which symbolizes the category of these debts. Knowing about these various debt indicators will help us to understand the areas in which a country can possibly be indebted to. </p>",
"_____no_output_____"
]
],
[
[
"%%sql\n\nSELECT DISTINCT indicator_code AS distinct_debt_indicators\nFROM international_debt\nORDER BY distinct_debt_indicators;\n",
" * postgresql:///international_debt\n25 rows affected.\n"
]
],
[
[
"## 4. Totaling the amount of debt owed by the countries\n<p>As mentioned earlier, the financial debt of a particular country represents its economic state. But if we were to project this on an overall global scale, how will we approach it?</p>\n<p>Let's switch gears from the debt indicators now and find out the total amount of debt (in USD) that is owed by the different countries. This will give us a sense of how the overall economy of the entire world is holding up.</p>",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT \n ROUND(SUM(debt)/1000000, 2) AS total_debt\nFROM international_debt; ",
" * postgresql:///international_debt\n1 rows affected.\n"
]
],
[
[
"## 5. Country with the highest debt\n<p>\"Human beings cannot comprehend very large or very small numbers. It would be useful for us to acknowledge that fact.\" - <a href=\"https://en.wikipedia.org/wiki/Daniel_Kahneman\">Daniel Kahneman</a>. That is more than <em>3 million <strong>million</strong></em> USD, an amount which is really hard for us to fathom. </p>\n<p>Now that we have the exact total of the amounts of debt owed by several countries, let's now find out the country that owns the highest amount of debt along with the amount. <strong>Note</strong> that this debt is the sum of different debts owed by a country across several categories. This will help to understand more about the country in terms of its socio-economic scenarios. We can also find out the category in which the country owns its highest debt. But we will leave that for now. </p>",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT \n country_name, \n SUM(debt) AS total_debt\nFROM international_debt\nGROUP BY country_name\nORDER BY total_debt DESC\nLIMIT 1;",
" * postgresql:///international_debt\n1 rows affected.\n"
]
],
[
[
"## 6. Average amount of debt across indicators\n<p>So, it was <em>China</em>. A more in-depth breakdown of China's debts can be found <a href=\"https://datatopics.worldbank.org/debt/ids/country/CHN\">here</a>. </p>\n<p>We now have a brief overview of the dataset and a few of its summary statistics. We already have an idea of the different debt indicators in which the countries owe their debts. We can dig even further to find out on an average how much debt a country owes? This will give us a better sense of the distribution of the amount of debt across different indicators.</p>",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT \n indicator_code AS debt_indicator,\n indicator_name,\n AVG(debt) AS average_debt\nFROM international_debt\nGROUP BY debt_indicator,indicator_name\nORDER BY average_debt DESC\nLIMIT 10;",
" * postgresql:///international_debt\n10 rows affected.\n"
]
],
[
[
"## 7. The highest amount of principal repayments\n<p>We can see that the indicator <code>DT.AMT.DLXF.CD</code> tops the chart of average debt. This category includes repayment of long term debts. Countries take on long-term debt to acquire immediate capital. More information about this category can be found <a href=\"https://datacatalog.worldbank.org/principal-repayments-external-debt-long-term-amt-current-us-0\">here</a>. </p>\n<p>An interesting observation in the above finding is that there is a huge difference in the amounts of the indicators after the second one. This indicates that the first two indicators might be the most severe categories in which the countries owe their debts.</p>\n<p>We can investigate this a bit more so as to find out which country owes the highest amount of debt in the category of long term debts (<code>DT.AMT.DLXF.CD</code>). Since not all the countries suffer from the same kind of economic disturbances, this finding will allow us to understand that particular country's economic condition a bit more specifically. </p>",
"_____no_output_____"
]
],
[
[
"%%sql\nSELECT \n country_name, \n indicator_name\nFROM international_debt\nWHERE debt = (SELECT \n MAX(debt)\n FROM international_debt\n LIMIT 1);",
" * postgresql:///international_debt\n1 rows affected.\n"
]
],
[
[
"## 8. The most common debt indicator\n<p>China has the highest amount of debt in the long-term debt (<code>DT.AMT.DLXF.CD</code>) category. This is verified by <a href=\"https://data.worldbank.org/indicator/DT.AMT.DLXF.CD?end=2018&most_recent_value_desc=true\">The World Bank</a>. It is often a good idea to verify our analyses like this since it validates that our investigations are correct. </p>\n<p>We saw that long-term debt is the topmost category when it comes to the average amount of debt. But is it the most common indicator in which the countries owe their debt? Let's find that out. </p>",
"_____no_output_____"
]
],
[
[
"%%sql\n\nSELECT indicator_code,\n COUNT(indicator_code) AS indicator_count\nFROM international_debt\nGROUP BY indicator_code\nORDER BY indicator_count DESC,indicator_code DESC\nLIMIT 20;\n\n\n\n",
" * postgresql:///international_debt\n20 rows affected.\n"
]
],
[
[
"## 9. Other viable debt issues and conclusion\n<p>There are a total of six debt indicators in which all the countries listed in our dataset have taken debt. The indicator <code>DT.AMT.DLXF.CD</code> is also there in the list. So, this gives us a clue that all these countries are suffering from a common economic issue. But that is not the end of the story, a part of the story rather. </p>\n<p>Let's change tracks from <code>debt_indicator</code>s now and focus on the amount of debt again. Let's find out the maximum amount of debt across the indicators along with the respective country names. With this, we will be in a position to identify the other plausible economic issues a country might be going through. By the end of this section, we will have found out the debt indicators in which a country owes its highest debt. </p>\n<p>In this notebook, we took a look at debt owed by countries across the globe. We extracted a few summary statistics from the data and unraveled some interesting facts and figures. We also validated our findings to make sure the investigations are correct.</p>",
"_____no_output_____"
]
],
[
[
"%%sql\n\nSELECT country_name,\n indicator_code,\n MAX(debt) AS maximum_debt\nFROM international_debt\nGROUP BY country_name,indicator_code\nORDER BY maximum_debt DESC\nLIMIT 10;",
" * postgresql:///international_debt\n10 rows affected.\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4451007062d9a7490a4df9a6f8d3b32f4e2980 | 171,151 | ipynb | Jupyter Notebook | P1.ipynb | shahriar-sust13/CarND-LaneLines-P1 | 0b37708f4e2507744cd88dec9d7281dfb4935a4c | [
"MIT"
] | null | null | null | P1.ipynb | shahriar-sust13/CarND-LaneLines-P1 | 0b37708f4e2507744cd88dec9d7281dfb4935a4c | [
"MIT"
] | null | null | null | P1.ipynb | shahriar-sust13/CarND-LaneLines-P1 | 0b37708f4e2507744cd88dec9d7281dfb4935a4c | [
"MIT"
] | null | null | null | 197.862428 | 116,972 | 0.859189 | [
[
[
"# Self-Driving Car Engineer Nanodegree\n\n\n## Project: **Finding Lane Lines on the Road** \n***\nIn this project, you will use the tools you learned about in the lesson to identify lane lines on the road. You can develop your pipeline on a series of individual images, and later apply the result to a video stream (really just a series of images). Check out the video clip \"raw-lines-example.mp4\" (also contained in this repository) to see what the output should look like after using the helper functions below. \n\nOnce you have a result that looks roughly like \"raw-lines-example.mp4\", you'll need to get creative and try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video \"P1_example.mp4\". Ultimately, you would like to draw just one line for the left side of the lane, and one for the right.\n\nIn addition to implementing code, there is a brief writeup to complete. The writeup should be completed in a separate file, which can be either a markdown file or a pdf document. There is a [write up template](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) that can be used to guide the writing process. Completing both the code in the Ipython notebook and the writeup template will cover all of the [rubric points](https://review.udacity.com/#!/rubrics/322/view) for this project.\n\n---\nLet's have a look at our first image called 'test_images/solidWhiteRight.jpg'. Run the 2 cells below (hit Shift-Enter or the \"play\" button above) to display the image.\n\n**Note: If, at any point, you encounter frozen display windows or other confounding issues, you can always start again with a clean slate by going to the \"Kernel\" menu above and selecting \"Restart & Clear Output\".**\n\n---",
"_____no_output_____"
],
[
"**The tools you have are color selection, region of interest selection, grayscaling, Gaussian smoothing, Canny Edge Detection and Hough Tranform line detection. You are also free to explore and try other techniques that were not presented in the lesson. Your goal is piece together a pipeline to detect the line segments in the image, then average/extrapolate them and draw them onto the image for display (as below). Once you have a working pipeline, try it out on the video stream below.**\n\n---\n\n<figure>\n <img src=\"examples/line-segments-example.jpg\" width=\"380\" alt=\"Combined Image\" />\n <figcaption>\n <p></p> \n <p style=\"text-align: center;\"> Your output should look something like this (above) after detecting line segments using the helper functions below </p> \n </figcaption>\n</figure>\n <p></p> \n<figure>\n <img src=\"examples/laneLines_thirdPass.jpg\" width=\"380\" alt=\"Combined Image\" />\n <figcaption>\n <p></p> \n <p style=\"text-align: center;\"> Your goal is to connect/average/extrapolate line segments to get output like this</p> \n </figcaption>\n</figure>",
"_____no_output_____"
],
[
"**Run the cell below to import some packages. If you get an `import error` for a package you've already installed, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.** ",
"_____no_output_____"
],
[
"## Import Packages",
"_____no_output_____"
]
],
[
[
"#importing some useful packages\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\nimport numpy as np\nimport cv2\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## Read in an Image",
"_____no_output_____"
]
],
[
[
"#reading in an image\nimage = mpimg.imread('test_images/solidWhiteRight.jpg')\n\n#printing out some stats and plotting\nprint('This image is:', type(image), 'with dimensions:', image.shape)\nplt.imshow(image) # if you wanted to show a single color channel image called 'gray', for example, call as plt.imshow(gray, cmap='gray')",
"This image is: <class 'numpy.ndarray'> with dimensions: (540, 960, 3)\n"
]
],
[
[
"## Ideas for Lane Detection Pipeline",
"_____no_output_____"
],
[
"**Some OpenCV functions (beyond those introduced in the lesson) that might be useful for this project are:**\n\n`cv2.inRange()` for color selection \n`cv2.fillPoly()` for regions selection \n`cv2.line()` to draw lines on an image given endpoints \n`cv2.addWeighted()` to coadd / overlay two images\n`cv2.cvtColor()` to grayscale or change color\n`cv2.imwrite()` to output images to file \n`cv2.bitwise_and()` to apply a mask to an image\n\n**Check out the OpenCV documentation to learn about these and discover even more awesome functionality!**",
"_____no_output_____"
],
[
"## Helper Functions",
"_____no_output_____"
],
[
"Below are some helper functions to help get you started. They should look familiar from the lesson!",
"_____no_output_____"
]
],
[
[
"import math\n\ndef grayscale(img):\n \"\"\"Applies the Grayscale transform\n This will return an image with only one color channel\n but NOTE: to see the returned image as grayscale\n (assuming your grayscaled image is called 'gray')\n you should call plt.imshow(gray, cmap='gray')\"\"\"\n return cv2.cvtColor(img, cv2.COLOR_RGB2GRAY)\n # Or use BGR2GRAY if you read an image with cv2.imread()\n # return cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)\n \ndef canny(img, low_threshold, high_threshold):\n \"\"\"Applies the Canny transform\"\"\"\n return cv2.Canny(img, low_threshold, high_threshold)\n\ndef gaussian_blur(img, kernel_size):\n \"\"\"Applies a Gaussian Noise kernel\"\"\"\n return cv2.GaussianBlur(img, (kernel_size, kernel_size), 0)\n\ndef region_of_interest(img, vertices):\n \"\"\"\n Applies an image mask.\n \n Only keeps the region of the image defined by the polygon\n formed from `vertices`. The rest of the image is set to black.\n \"\"\"\n #defining a blank mask to start with\n mask = np.zeros_like(img) \n \n #defining a 3 channel or 1 channel color to fill the mask with depending on the input image\n if len(img.shape) > 2:\n channel_count = img.shape[2] # i.e. 3 or 4 depending on your image\n ignore_mask_color = (255,) * channel_count\n else:\n ignore_mask_color = 255\n \n #filling pixels inside the polygon defined by \"vertices\" with the fill color \n cv2.fillPoly(mask, vertices, ignore_mask_color)\n \n #returning the image only where mask pixels are nonzero\n masked_image = cv2.bitwise_and(img, mask)\n return masked_image\n\ndef get_dist(a, b):\n x = (a[0]-b[0])*(a[0]-b[0]) + (a[1]-b[1])*(a[1]-b[1]);\n return x;\n\ndef get_full_line(a, b):\n y = 540;\n x = ( (a[0]-b[0])*(y-a[1]) )/(a[1]-b[1]) + a[0];\n point1 = ( int(round(x)), int(round(y)) );\n y = 320;\n x = ( (a[0]-b[0])*(y-a[1]) )/(a[1]-b[1]) + a[0];\n point2 = ( int(round(x)), int(round(y)) );\n return point1, point2;\n \n\ndef draw_lines(img, lines, color=[255, 0, 0], thickness=10):\n \"\"\"\n NOTE: this is the function you might want to use as a starting point once you want to \n average/extrapolate the line segments you detect to map out the full\n extent of the lane (going from the result shown in raw-lines-example.mp4\n to that shown in P1_example.mp4). \n \n Think about things like separating line segments by their \n slope ((y2-y1)/(x2-x1)) to decide which segments are part of the left\n line vs. the right line. Then, you can average the position of each of \n the lines and extrapolate to the top and bottom of the lane.\n \n This function draws `lines` with `color` and `thickness`. \n Lines are drawn on the image inplace (mutates the image).\n If you want to make the lines semi-transparent, think about combining\n this function with the weighted_img() function below\n \"\"\"\n \n imshape = img.shape;\n height = imshape[0];\n width = imshape[1];\n current_dist = 0;\n \n for line in lines:\n for x1, y1, x2, y2 in line:\n if( 2*x1<width and 2*x2<width ):\n if( get_dist((x1, y1), (x2, y2))>current_dist ):\n current_dist = get_dist((x1, y1), (x2, y2));\n point1 = (x1, y1);\n point2 = (x2, y2);\n \n point1, point2 = get_full_line(point1, point2);\n \n cv2.line(img, point1, point2, color, thickness);\n \n current_dist = 0;\n for line in lines:\n for x1, y1, x2, y2 in line:\n if( 2*x1>width and 2*x2>width ):\n if( get_dist((x1, y1), (x2, y2))>current_dist ):\n current_dist = get_dist((x1, y1), (x2, y2));\n point1 = (x1, y1);\n point2 = (x2, y2);\n \n point1, point2 = get_full_line(point1, point2);\n \n cv2.line(img, point1, point2, color, thickness);\n \ndef hough_lines(img, rho, theta, threshold, min_line_len, max_line_gap):\n \"\"\"\n `img` should be the output of a Canny transform.\n \n Returns an image with hough lines drawn.\n \"\"\"\n lines = cv2.HoughLinesP(img, rho, theta, threshold, np.array([]), minLineLength=min_line_len, maxLineGap=max_line_gap)\n line_img = np.zeros((img.shape[0], img.shape[1], 3), dtype=np.uint8)\n draw_lines(line_img, lines)\n return line_img\n\n# Python 3 has support for cool math symbols.\n\ndef weighted_img(img, initial_img, α=0.8, β=1., γ=0.):\n \"\"\"\n `img` is the output of the hough_lines(), An image with lines drawn on it.\n Should be a blank image (all black) with lines drawn on it.\n \n `initial_img` should be the image before any processing.\n \n The result image is computed as follows:\n \n initial_img * α + img * β + γ\n NOTE: initial_img and img must be the same shape!\n \"\"\"\n return cv2.addWeighted(initial_img, α, img, β, γ)",
"_____no_output_____"
]
],
[
[
"## Test Images\n\nBuild your pipeline to work on the images in the directory \"test_images\" \n**You should make sure your pipeline works well on these images before you try the videos.**",
"_____no_output_____"
]
],
[
[
"import os\nos.listdir(\"test_images/\")",
"_____no_output_____"
]
],
[
[
"## Build a Lane Finding Pipeline\n\n",
"_____no_output_____"
],
[
"Build the pipeline and run your solution on all test_images. Make copies into the `test_images_output` directory, and you can use the images in your writeup report.\n\nTry tuning the various parameters, especially the low and high Canny thresholds as well as the Hough lines parameters.",
"_____no_output_____"
]
],
[
[
"def make_line(a, b):\n return (a[0], a[1], b[0], b[1]);\n '''\n line = {};\n line.x1 = a[0];\n line.y1 = a[1];\n line.x2 = b[0];\n line.y2 = b[1];\n return line;\n '''",
"_____no_output_____"
],
[
"def get_lines(vertices):\n lines = [];\n n = len(vertices);\n for i in range(0, n):\n lines.append( make_line(vertices[i], vertices[(i+1)%n]) );\n return lines;",
"_____no_output_____"
],
[
"def find_lane_line(image):\n gray_image = grayscale(image);\n blur_image = gaussian_blur(gray_image, 5);\n canny_edge = canny(blur_image, 50, 100);\n imshape = image.shape;\n #print(\"--->\", imshape[0], imshape[1]);\n vertices = [(0, imshape[0]), (imshape[1], imshape[0]), (500, 325), (470, 325)];\n polygon = np.array([vertices], dtype=np.int32);\n masked_image = region_of_interest(canny_edge, polygon);\n #lines = get_lines(vertices);\n #draw_lines(masked_image, lines);\n rho = 2; # distance resolution in pixels of the Hough grid\n theta = np.pi/180; # angular resolution in radians of the Hough grid\n threshold = 10; # minimum number of votes (intersections in Hough grid cell)\n min_line_len = 10; #minimum number of pixels making up a line\n max_line_gap = 20; # maximum gap in pixels between connectable line segments\n hough_image = hough_lines(masked_image, rho, theta, threshold, min_line_len, max_line_gap);\n processed = weighted_img(hough_image, image);\n return processed;",
"_____no_output_____"
],
[
"# TODO: Build your pipeline that will draw lane lines on the test_images\n# then save them to the test_imaes = os.lisdir(\"test_images/\");\n\ninames = os.listdir(\"test_images/\");\n\nfor iname in inames:\n image = mpimg.imread('test_images/'+iname);\n new_image = find_lane_line(image);\n #plt.imshow(new_image, cmap='Greys_r');\n mpimg.imsave('test_images_output/'+iname, new_image);\n #break;",
"_____no_output_____"
]
],
[
[
"## Test on Videos\n\nYou know what's cooler than drawing lanes over images? Drawing lanes over video!\n\nWe can test our solution on two provided videos:\n\n`solidWhiteRight.mp4`\n\n`solidYellowLeft.mp4`\n\n**Note: if you get an import error when you run the next cell, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.**\n\n**If you get an error that looks like this:**\n```\nNeedDownloadError: Need ffmpeg exe. \nYou can download it by calling: \nimageio.plugins.ffmpeg.download()\n```\n**Follow the instructions in the error message and check out [this forum post](https://discussions.udacity.com/t/project-error-of-test-on-videos/274082) for more troubleshooting tips across operating systems.**",
"_____no_output_____"
]
],
[
[
"# Import everything needed to edit/save/watch video clips\nimport imageio\nfrom moviepy.editor import VideoFileClip\nfrom IPython.display import HTML",
"_____no_output_____"
],
[
"def process_image(image):\n # NOTE: The output you return should be a color image (3 channel) for processing video below\n # TODO: put your pipeline here,\n # you should return the final output (image where lines are drawn on lanes)\n result = find_lane_line(image);\n return result",
"_____no_output_____"
]
],
[
[
"Let's try the one with the solid white lane on the right first ...",
"_____no_output_____"
]
],
[
[
"white_output = 'test_videos_output/solidWhiteRight.mp4'\n## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video\n## To do so add .subclip(start_second,end_second) to the end of the line below\n## Where start_second and end_second are integer values representing the start and end of the subclip\n## You may also uncomment the following line for a subclip of the first 5 seconds\n##clip1 = VideoFileClip(\"test_videos/solidWhiteRight.mp4\").subclip(0,5)\nclip1 = VideoFileClip(\"test_videos/solidWhiteRight.mp4\")\nwhite_clip = clip1.fl_image(process_image) #NOTE: this function expects color images!!\n%time white_clip.write_videofile(white_output, audio=False)",
"[MoviePy] >>>> Building video test_videos_output/solidWhiteRight.mp4\n[MoviePy] Writing video test_videos_output/solidWhiteRight.mp4\n"
]
],
[
[
"Play the video inline, or if you prefer find the video in your filesystem (should be in the same directory) and play it in your video player of choice.",
"_____no_output_____"
]
],
[
[
"HTML(\"\"\"\n<video width=\"960\" height=\"540\" controls>\n <source src=\"{0}\">\n</video>\n\"\"\".format(white_output))",
"_____no_output_____"
]
],
[
[
"## Improve the draw_lines() function\n\n**At this point, if you were successful with making the pipeline and tuning parameters, you probably have the Hough line segments drawn onto the road, but what about identifying the full extent of the lane and marking it clearly as in the example video (P1_example.mp4)? Think about defining a line to run the full length of the visible lane based on the line segments you identified with the Hough Transform. As mentioned previously, try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video \"P1_example.mp4\".**\n\n**Go back and modify your draw_lines function accordingly and try re-running your pipeline. The new output should draw a single, solid line over the left lane line and a single, solid line over the right lane line. The lines should start from the bottom of the image and extend out to the top of the region of interest.**",
"_____no_output_____"
],
[
"Now for the one with the solid yellow lane on the left. This one's more tricky!",
"_____no_output_____"
]
],
[
[
"yellow_output = 'test_videos_output/solidYellowLeft.mp4'\n## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video\n## To do so add .subclip(start_second,end_second) to the end of the line below\n## Where start_second and end_second are integer values representing the start and end of the subclip\n## You may also uncomment the following line for a subclip of the first 5 seconds\n##clip2 = VideoFileClip('test_videos/solidYellowLeft.mp4').subclip(0,5)\nclip2 = VideoFileClip('test_videos/solidYellowLeft.mp4')\nyellow_clip = clip2.fl_image(process_image)\n%time yellow_clip.write_videofile(yellow_output, audio=False)",
"[MoviePy] >>>> Building video test_videos_output/solidYellowLeft.mp4\n[MoviePy] Writing video test_videos_output/solidYellowLeft.mp4\n"
],
[
"HTML(\"\"\"\n<video width=\"960\" height=\"540\" controls>\n <source src=\"{0}\">\n</video>\n\"\"\".format(yellow_output))",
"_____no_output_____"
]
],
[
[
"## Writeup and Submission\n\nIf you're satisfied with your video outputs, it's time to make the report writeup in a pdf or markdown file. Once you have this Ipython notebook ready along with the writeup, it's time to submit for review! Here is a [link](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) to the writeup template file.\n",
"_____no_output_____"
],
[
"## Optional Challenge\n\nTry your lane finding pipeline on the video below. Does it still work? Can you figure out a way to make it more robust? If you're up for the challenge, modify your pipeline so it works with this video and submit it along with the rest of your project!",
"_____no_output_____"
]
],
[
[
"challenge_output = 'test_videos_output/challenge.mp4'\n## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video\n## To do so add .subclip(start_second,end_second) to the end of the line below\n## Where start_second and end_second are integer values representing the start and end of the subclip\n## You may also uncomment the following line for a subclip of the first 5 seconds\n##clip3 = VideoFileClip('test_videos/challenge.mp4').subclip(0,5)\nclip3 = VideoFileClip('test_videos/challenge.mp4')\nchallenge_clip = clip3.fl_image(process_image)\n%time challenge_clip.write_videofile(challenge_output, audio=False)",
"[MoviePy] >>>> Building video test_videos_output/challenge.mp4\n[MoviePy] Writing video test_videos_output/challenge.mp4\n"
],
[
"HTML(\"\"\"\n<video width=\"960\" height=\"540\" controls>\n <source src=\"{0}\">\n</video>\n\"\"\".format(challenge_output))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cb4453cb7db7037c4ab250ed093c86658c5fbb8d | 12,295 | ipynb | Jupyter Notebook | 2. Deep Learning Frameworks/2.1 Introduction - Theano.ipynb | jianhuayang/deep-learning-keras-tensorflow | cfa82c4b2ce03db6f33f5bf290fec3ad6dfee4d9 | [
"MIT"
] | 2,482 | 2016-10-25T19:29:02.000Z | 2022-03-28T06:40:14.000Z | 2. Deep Learning Frameworks/2.1 Introduction - Theano.ipynb | jianhuayang/deep-learning-keras-tensorflow | cfa82c4b2ce03db6f33f5bf290fec3ad6dfee4d9 | [
"MIT"
] | 21 | 2017-02-17T12:19:44.000Z | 2021-04-28T18:10:43.000Z | 2. Deep Learning Frameworks/2.1 Introduction - Theano.ipynb | jianhuayang/deep-learning-keras-tensorflow | cfa82c4b2ce03db6f33f5bf290fec3ad6dfee4d9 | [
"MIT"
] | 1,171 | 2016-10-27T15:13:18.000Z | 2022-03-17T09:52:15.000Z | 17.052705 | 165 | 0.457096 | [
[
[
"Theano \n===\nA language in a language",
"_____no_output_____"
],
[
"Dealing with weights matrices and gradients can be tricky and sometimes not trivial.\nTheano is a great framework for handling vectors, matrices and high dimensional tensor algebra. \nMost of this tutorial will refer to Theano however TensorFlow is another great framework capable of providing an incredible abstraction for complex algebra.\nMore on TensorFlow in the next chapters.",
"_____no_output_____"
]
],
[
[
"import theano\nimport theano.tensor as T",
"_____no_output_____"
]
],
[
[
"Symbolic variables\n==========",
"_____no_output_____"
],
[
"Theano has it's own variables and functions, defined the following",
"_____no_output_____"
]
],
[
[
"x = T.scalar()",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
]
],
[
[
"Variables can be used in expressions",
"_____no_output_____"
]
],
[
[
"y = 3*(x**2) + 1",
"_____no_output_____"
]
],
[
[
"y is an expression now ",
"_____no_output_____"
],
[
"Result is symbolic as well",
"_____no_output_____"
]
],
[
[
"type(y)\ny.shape",
"_____no_output_____"
]
],
[
[
"#####printing",
"_____no_output_____"
],
[
"As we are about to see, normal printing isn't the best when it comes to theano",
"_____no_output_____"
]
],
[
[
"print(y)",
"Elemwise{add,no_inplace}.0\n"
],
[
"theano.pprint(y)",
"_____no_output_____"
],
[
"theano.printing.debugprint(y)",
"Elemwise{add,no_inplace} [id A] '' \n |Elemwise{mul,no_inplace} [id B] '' \n | |TensorConstant{3} [id C]\n | |Elemwise{pow,no_inplace} [id D] '' \n | |<TensorType(float64, scalar)> [id E]\n | |TensorConstant{2} [id F]\n |TensorConstant{1} [id G]\n"
]
],
[
[
"Evaluating expressions\n============\n\nSupply a `dict` mapping variables to values",
"_____no_output_____"
]
],
[
[
"y.eval({x: 2})",
"_____no_output_____"
]
],
[
[
"Or compile a function",
"_____no_output_____"
]
],
[
[
"f = theano.function([x], y)",
"_____no_output_____"
],
[
"f(2)",
"_____no_output_____"
]
],
[
[
"Other tensor types\n==========",
"_____no_output_____"
]
],
[
[
"X = T.vector()\nX = T.matrix()\nX = T.tensor3()\nX = T.tensor4()",
"_____no_output_____"
]
],
[
[
"Automatic differention\n============\n- Gradients are free!",
"_____no_output_____"
]
],
[
[
"x = T.scalar()\ny = T.log(x)",
"_____no_output_____"
],
[
"gradient = T.grad(y, x)\nprint(gradient)\nprint(gradient.eval({x: 2}))\nprint((2 * gradient))",
"Elemwise{true_div}.0\n0.5\nElemwise{mul,no_inplace}.0\n"
]
],
[
[
"# Shared Variables\n\n- Symbolic + Storage",
"_____no_output_____"
]
],
[
[
"import numpy as np\nx = theano.shared(np.zeros((2, 3), dtype=theano.config.floatX))",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
]
],
[
[
"We can get and set the variable's value",
"_____no_output_____"
]
],
[
[
"values = x.get_value()\nprint(values.shape)\nprint(values)",
"(2, 3)\n[[ 0. 0. 0.]\n [ 0. 0. 0.]]\n"
],
[
"x.set_value(values)",
"_____no_output_____"
]
],
[
[
"Shared variables can be used in expressions as well",
"_____no_output_____"
]
],
[
[
"(x + 2) ** 2",
"_____no_output_____"
]
],
[
[
"Their value is used as input when evaluating",
"_____no_output_____"
]
],
[
[
"((x + 2) ** 2).eval()",
"_____no_output_____"
],
[
"theano.function([], (x + 2) ** 2)()",
"_____no_output_____"
]
],
[
[
"# Updates\n\n- Store results of function evalution\n- `dict` mapping shared variables to new values",
"_____no_output_____"
]
],
[
[
"count = theano.shared(0)\nnew_count = count + 1\nupdates = {count: new_count}\n\nf = theano.function([], count, updates=updates)",
"_____no_output_____"
],
[
"f()",
"_____no_output_____"
],
[
"f()",
"_____no_output_____"
],
[
"f()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cb44542362ce41dd3b84bcf0a3a8aeae04e001ae | 131 | ipynb | Jupyter Notebook | .ipynb_checkpoints/basic_usage-checkpoint.ipynb | rgilman33/obs-tower | 895faff27f0bfcd7beb3f0f53047467c567106c5 | [
"Apache-2.0"
] | null | null | null | .ipynb_checkpoints/basic_usage-checkpoint.ipynb | rgilman33/obs-tower | 895faff27f0bfcd7beb3f0f53047467c567106c5 | [
"Apache-2.0"
] | 2 | 2021-10-12T22:04:31.000Z | 2021-10-12T22:50:08.000Z | .ipynb_checkpoints/basic_usage-checkpoint.ipynb | rgilman33/obs-tower | 895faff27f0bfcd7beb3f0f53047467c567106c5 | [
"Apache-2.0"
] | null | null | null | 32.75 | 75 | 0.885496 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
cb445bb3a7599a6ae4f442c5bf72b63ccb583db6 | 2,277 | ipynb | Jupyter Notebook | notebooks/02 - FingerTapping.ipynb | gkroussos/pdkit | 59b92a4fa624ad1050eeb18c00436694bea0580c | [
"MIT"
] | 21 | 2018-03-14T11:47:47.000Z | 2022-02-10T19:58:36.000Z | notebooks/02 - FingerTapping.ipynb | gkroussos/pdkit | 59b92a4fa624ad1050eeb18c00436694bea0580c | [
"MIT"
] | 23 | 2018-03-11T15:47:22.000Z | 2021-03-20T14:54:53.000Z | notebooks/02 - FingerTapping.ipynb | gkroussos/pdkit | 59b92a4fa624ad1050eeb18c00436694bea0580c | [
"MIT"
] | 14 | 2018-03-11T15:40:20.000Z | 2022-01-31T02:57:43.000Z | 19.461538 | 102 | 0.53184 | [
[
[
"# FingerTapping",
"_____no_output_____"
]
],
[
[
"import pdkit",
"_____no_output_____"
],
[
"# Read a finger tapping data file\nfilename = './data/2458/FT_-_ONE_TARGET_-_LEFT_HAND_2016-10-14_10-36-02_-_f73512ccfc_-_2458.csv'",
"_____no_output_____"
],
[
"# Use the Finger Tapping Time Series to load the data file\nts = pdkit.FingerTappingTimeSeries().load(filename)",
"_____no_output_____"
],
[
"# Instance to the finger Tapping Processor\nftp = pdkit.FingerTappingProcessor()",
"_____no_output_____"
],
[
"# Example how to get the mean alternate distance of the finger tapping tests (in pixels)\nmatd, total_time = ftp.mean_alnt_target_distance(ts)\nprint(matd)",
"10.686484959607274\n"
],
[
"# Example how to get the kinesia scores (the number of key taps)\nks, total_time = ftp.kinesia_scores(ts)\nprint(ks)",
"248\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4474f34cc77b44fb283a0a4a557b3e149f4b73 | 155,475 | ipynb | Jupyter Notebook | .ipynb_checkpoints/SSA_slow-checkpoint.ipynb | SystemsBiologyUniandes/PyEcoLib | 3c46a34af51e29a2d5cca1f894606bbc9738f7a0 | [
"MIT"
] | 1 | 2020-12-31T06:37:14.000Z | 2020-12-31T06:37:14.000Z | .ipynb_checkpoints/SSA_slow-checkpoint.ipynb | SystemsBiologyUniandes/PyEcoLib | 3c46a34af51e29a2d5cca1f894606bbc9738f7a0 | [
"MIT"
] | null | null | null | .ipynb_checkpoints/SSA_slow-checkpoint.ipynb | SystemsBiologyUniandes/PyEcoLib | 3c46a34af51e29a2d5cca1f894606bbc9738f7a0 | [
"MIT"
] | 1 | 2021-12-19T19:48:20.000Z | 2021-12-19T19:48:20.000Z | 280.640794 | 65,384 | 0.90585 | [
[
[
"import math\nimport numpy as np\nimport pandas as pd\nfrom matplotlib import pyplot as plt\nfrom scipy.stats import bayes_mvs as bayesest\nimport os \nimport time\n\n\nfrom szsimulator import Szsimulator\n\n%matplotlib inline",
"_____no_output_____"
],
[
"mean_size = 3 # micron\ndoubling_time = 18 #min\ntmax = 180 #min\nsample_time = 2 #min\ndiv_steps = 10 \nncells = 1000",
"_____no_output_____"
],
[
"gr = np.log(2)/doubling_time \nkd = div_steps*gr/(mean_size)",
"_____no_output_____"
],
[
"\nncells = 2000\nsampling_time = sample_time \nrprom = 10 # RNA mean concentration\npprom = 1000 # prot mean concentration \ngammar = 5*gr # RNA Active degradation rate\nkr = rprom*(gr+gammar) # RNA transcription rate\nkp = pprom*gr/rprom # Protein translation rate\n\npop = np.zeros([ncells,6])\n\nindexes = np.int(tmax/sampling_time)\nrarray = np.zeros([ncells,indexes])\nparray = np.zeros([ncells,indexes])\ntarray = np.zeros([indexes])\nszarray = np.zeros([ncells,indexes])\ncellindex = 0\nindexref = 0\nstart = time.time()\nfor cell in pop:\n if ncells > 100:\n if cellindex/ncells > indexref:\n print(str(np.int(100*cellindex/ncells))+\"%\")\n indexref += 0.1\n #Initialize the simulator\n sim = Szsimulator(tmax = tmax, sample_time = sample_time, ncells=1, gr = gr, k = kd, steps = div_steps)\n #_______________\n #Example of a direct SSA simulation\n cell[0] = mean_size #Initial size\n cell[1] = mean_size*rprom #Initial RNA number\n cell[2] = mean_size*pprom #Initial Protein number\n cell[3] = (1/gr)*np.log(1-(gr/(kr*cell[0]))*np.log(np.random.rand())) #time to thenext rna creation\n cell[4] = -np.log(np.random.rand())/(gammar*cell[1]) #time to the next rna degradation\n cell[5] = -np.log(np.random.rand())/(kp*cell[1]) #time to next protein creation\n t=0\n reactions=[[0,1,0,0,0,0],[0,-1,0,0,0,0],[0,0,1,0,0,0]] #Reactions (RNA creation, RNA active degradation, Protein creation)\n nextt = 0\n index = 0\n ndiv = 0\n while t<tmax: #iterating over time\n nr = cell[1]\n nprot = cell[2]\n sz = cell[0]\n tnextarr = [cell[3],cell[4],cell[5]]\n tau = np.min(tnextarr)\n cell += reactions[np.argmin(tnextarr)]\n #------------------\n sim.simulate(tmax=tau,export = False) #Simulate size dynamics for that given time\n #--------------------\n cell[0] = sim.get_sz(0) #Taking the cell size after that simulation\n if sim.get_ndiv(0) > ndiv: #Check if cell got divided\n cell[1] = np.random.binomial(nr,0.5) # RNA segregated binomially\n cell[2] = np.random.binomial(nprot,0.5) # Protein segregated binomially\n ndiv += 1 # New number of divisions\n nr = cell[1] #Refreshing RNA number\n nprot = cell[2] #Refreshing Protein number\n sz = cell[0] #Refreshing size number\n cell[3] = (1/gr)*np.log(1-(gr/(kr*cell[0]))*np.log(np.random.rand())) #time to thenext rna creation\n cell[4] = -np.log(np.random.rand())/(gammar*cell[1]) #time to the next rna degradation\n cell[5] = -np.log(np.random.rand())/(kp*cell[1]) #time to next protein creation \n t+=tau\n if t > nextt and index<len(tarray): #storing data\n rarray[cellindex,index] = nr/sz # RNA concentration\n parray[cellindex,index] = nprot/sz # Protein concentration\n szarray[cellindex,index] = sz # Cell size\n tarray[index] = t # Time\n index += 1\n nextt += sampling_time\n cellindex += 1 \nprint('It took', np.int(time.time()-start), 'seconds.')",
"0%\n10%\n20%\n30%\n40%\n50%\n60%\n70%\n80%\n90%\nIt took 2835.5868401527405 seconds.\n"
],
[
"data=pd.DataFrame(np.transpose(np.array(szarray)))\nind=0\nnewcol=[]\nfor name in data.columns:\n newcol.append(\"mom\"+str(ind))\n ind+=1\ndata.columns=newcol\nmnszarray=[]\ncvszarray=[]\nerrcv2sz=[]\nerrmnsz=[]\nfor m in range(len(data)):\n szs=data.loc[m, :].values.tolist()\n mean_cntr, var_cntr, std_cntr = bayesest(szs,alpha=0.95)\n mnszarray.append(mean_cntr[0])\n errmnsz.append(mean_cntr[1][1]-mean_cntr[0])\n cvszarray.append(var_cntr[0]/mean_cntr[0]**2)\n errv=(var_cntr[1][1]-var_cntr[0])/mean_cntr[0]**2+2*(mean_cntr[1][1]-mean_cntr[0])*var_cntr[0]/mean_cntr[0]**3\n errcv2sz.append(errv)\n\ndata['time'] = tarray\ndata['Mean_sz'] = mnszarray\ndata['Error_mean'] = errmnsz\ndata['sz_CV2'] = cvszarray\ndata['Error_CV2'] = errcv2sz\nif not os.path.exists('./data/SSA'):\n os.makedirs('./data/SSA')\ndata.to_csv(\"./data/SSA/szsim.csv\")",
"_____no_output_____"
],
[
"tmax=9*doubling_time\ndt=0.0001*doubling_time\nlamb=1\na=gr\nnsteps=div_steps\nk=kd\n\nv0=mean_size\n#psz1=[]\nndivs=10\nt=0\nbigdeltat=0.1\nsteps=int(np.floor(tmax/dt))\nu=np.zeros([ndivs,nsteps])#(DIVS,STEPS)\nu[0]=np.zeros(nsteps)\nu[0][0]=1#P_00\nallmeandivs4=[]#average divisions along the time\nallvardiv4=[] # variace of pn along the time\nallmeansz4=[]\nallvarsz4=[]\ntime4=[]#time array\nyenvol=[]\nxenvol=[]\nstart=0\ncount=int(np.floor(tmax/(dt*1000)))-1\ncount2=0\nstart = time.time()\nfor l in range(steps): \n utemp=u\n for n in range(len(utemp)):#n=divs, \n for m in range(len(utemp[n])):#m=steps\n if (m==0):#m=steps\n if(n==0):#n=divs\n dun=-k*v0**lamb*np.exp(lamb*a*t)*(utemp[0][0])\n u[n][m]+=dun*dt\n else:\n arg=lamb*(a*t-n*np.log(2))\n dun=k*v0**lamb*np.exp(arg)*((2**lamb)*utemp[n-1][len(utemp[n])-1]-utemp[n][0])\n u[n][m]+=dun*dt\n elif(m==len(utemp[n])-1):\n if(n==len(utemp)-1):\n arg=lamb*(a*t-n*np.log(2))\n dun=k*v0**lamb*np.exp(arg)*(utemp[n][len(utemp[n])-2])\n u[n][m]+=dun*dt\n else:\n arg=lamb*(a*t-n*np.log(2))\n dun=k*v0**lamb*np.exp(arg)*(utemp[n][m-1]-utemp[n][m])\n u[n][m]+=dun*dt\n else:\n arg=lamb*(a*t-n*np.log(2))\n dun=k*v0**lamb*np.exp(arg)*(utemp[n][m-1]-utemp[n][m])\n u[n][m]+=dun*dt\n t+=dt\n count=count+1\n if count==int(np.floor(tmax/(dt*1000))):\n time4.append(t/doubling_time)\n mean=0\n for n in range(len(utemp)):\n pnval=np.sum(u[n])\n mean+=n*pnval\n allmeandivs4.append(mean/mean_size)\n var=0\n for n in range(len(utemp)):#divs\n pnval=np.sum(u[n])\n var+=(n-mean)**2*pnval\n allvardiv4.append(np.sqrt(var))\n pn=np.zeros(ndivs)\n sizen=np.zeros(ndivs)\n meansz=0\n for ll in range(len(utemp)):\n pnltemp=np.sum(u[ll])#prob of n divs\n pn[ll]=pnltemp#\n sizen[ll]=np.exp(a*t)/2**ll#\n meansz+=pnltemp*v0*np.exp(a*t)/2**ll\n allmeansz4.append(meansz)\n varsz=0\n for ll in range(len(utemp)):\n pnltemp=np.sum(u[ll])\n varsz+=(v0*np.exp(a*t)/2**ll-meansz)**2*pnltemp\n allvarsz4.append(varsz)\n count=0\n count2+=1\n if(count2==100):\n print(str(int(100*t/tmax))+\"%\")\n count2=0\nprint('It took', np.int(time.time()-start), 'seconds.')",
"9%\n19%\n29%\n39%\n49%\n59%\n69%\n79%\n88%\n98%\nIt took 46 seconds.\n"
],
[
"fig, ax = plt.subplots(1,2, figsize=(12,4))\n#ax[0].plot(tarray,mnszarray)\nax[0].fill_between(np.array(tarray)/doubling_time,np.array(mnszarray)-np.array(errmnsz),np.array(mnszarray)+np.array(errmnsz),\n alpha=1, edgecolor='#4db8ff', facecolor='#4db8ff',linewidth=0,label='SSA')\n#ax[1].plot(tarray,cvszarray)\nax[1].fill_between(np.array(tarray)/doubling_time,np.array(cvszarray)-np.array(errcv2sz),np.array(cvszarray)+np.array(errcv2sz),\n alpha=1, edgecolor='#4db8ff', facecolor='#4db8ff',linewidth=0)\nax[0].plot(np.array(time4),np.array(allmeansz4),lw=2,c='#006599',label=\"Numerical\")\nax[1].plot(np.array(time4),np.array(allvarsz4)/np.array(allmeansz4)**2,lw=2,c='#006599')\n\nax[0].set_ylabel(\"$s$ ($\\mu$m)\",size=20)\nax[1].set_ylabel(\"$C_V^2(s)$\",size=20)\nax[0].set_xlabel(r\"$t/\\tau$\",size=20)\nax[1].set_xlabel(r\"$t/\\tau$\",size=20)\nax[0].set_ylim([1,1.2*np.max(mnszarray)])\nax[1].set_ylim([0,1.2*np.max(cvszarray)])\nfor l in [0,1]:\n ax[l].set_xlim([0,tmax/doubling_time])\n taqui=np.arange(0,(tmax+1)/doubling_time,step=1)\n ax[l].set_xticks(np.array(taqui))\n ax[l].grid()\n ax[l].tick_params(axis='x', labelsize=15)\n ax[l].tick_params(axis='y', labelsize=15)\n for axis in ['bottom','left']:\n ax[l].spines[axis].set_linewidth(2)\n ax[l].tick_params(axis='both', width=2,length=6)\n for axis in ['top','right']:\n ax[l].spines[axis].set_linewidth(0)\n ax[l].tick_params(axis='both', width=0,length=6)\nplt.subplots_adjust(hspace=0.3,wspace=0.3)\ntaqui=np.arange(0,0.15,step=0.02)\nax[1].set_yticks(np.array(taqui))\nax[0].legend(fontsize=15)\nif not os.path.exists('./figures/SSA'):\n os.makedirs('./figures/SSA')\nplt.savefig('./figures/SSA/size_statistics.svg',bbox_inches='tight')\nplt.savefig('./figures/SSA/size_statistics.png',bbox_inches='tight')",
"_____no_output_____"
],
[
"data=pd.DataFrame(np.transpose(np.array(rarray)))\nind=0\nnewcol=[]\nfor name in data.columns:\n newcol.append(\"mom\"+str(ind))\n ind+=1\ndata.columns=newcol\nmnrnaarray=[]\ncvrnaarray=[]\nerrcv2rna=[]\nerrmnrna=[]\nfor m in range(len(data)):\n rnas=data.loc[m, :].values.tolist()\n mean_cntr, var_cntr, std_cntr = bayesest(rnas,alpha=0.95)\n mnrnaarray.append(mean_cntr[0])\n errmnrna.append(mean_cntr[1][1]-mean_cntr[0])\n cvrnaarray.append(var_cntr[0]/mean_cntr[0]**2)\n errv=(var_cntr[1][1]-var_cntr[0])/mean_cntr[0]**2+2*(mean_cntr[1][1]-mean_cntr[0])*var_cntr[0]/mean_cntr[0]**3\n errcv2rna.append(errv)\n\ndata['time'] = tarray\ndata['Mean_RNA'] = mnrnaarray\ndata['Error_mean'] = errmnrna\ndata['RNA_CV2'] = cvrnaarray\ndata['Error_CV2'] = errcv2rna\nif not os.path.exists('./data/SSA'):\n os.makedirs('./data/SSA')\ndata.to_csv(\"./data/SSA/RNAsim.csv\")",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1,2, figsize=(12,4))\nax[0].plot(np.array(tarray)/doubling_time,mnrnaarray,c=\"#BD0025\")\nax[0].fill_between(np.array(tarray)/doubling_time,np.array(mnrnaarray)-np.array(errmnrna),np.array(mnrnaarray)+np.array(errmnrna),\n alpha=1, edgecolor='#FF3333', facecolor='#FF3333',linewidth=0)\nax[1].plot(np.array(tarray)/doubling_time,cvrnaarray,c=\"#BD0025\")\nax[1].fill_between(np.array(tarray)/doubling_time,np.array(cvrnaarray)-np.array(errcv2rna),np.array(cvrnaarray)+np.array(errcv2rna),\n alpha=1, edgecolor='#FF3333', facecolor='#FF3333',linewidth=0)\n\n\nax[0].set_ylabel(\"RNA\",size=20)\nax[1].set_ylabel(\"$C_V^2(r)$\",size=20)\nax[0].set_xlabel(r\"$t/\\tau$\",size=20)\nax[1].set_xlabel(r\"$t/\\tau$\",size=20)\nax[0].set_ylim([0,1.2*np.max(mnrnaarray)])\nax[1].set_ylim([0,1.2*np.max(cvrnaarray)])\nfor l in [0,1]:\n ax[l].set_xlim([0,tmax/doubling_time])\n taqui=np.arange(0,(tmax+1)/doubling_time,step=1)\n ax[l].set_xticks(np.array(taqui))\n ax[l].grid()\n ax[l].tick_params(axis='x', labelsize=15)\n ax[l].tick_params(axis='y', labelsize=15)\n for axis in ['bottom','left']:\n ax[l].spines[axis].set_linewidth(2)\n ax[l].tick_params(axis='both', width=2,length=6)\n for axis in ['top','right']:\n ax[l].spines[axis].set_linewidth(0)\n ax[l].tick_params(axis='both', width=0,length=6)\nplt.subplots_adjust(hspace=0.3,wspace=0.3)\ntaqui=np.arange(0,1.2*np.max(cvrnaarray),step=np.round(.2*np.max(cvrnaarray),2))\nax[1].set_yticks(np.array(taqui))\nif not os.path.exists('./figures/SSA'):\n os.makedirs('./figures/SSA')\nplt.savefig('./figures/SSA/rna_statistics.svg',bbox_inches='tight')\nplt.savefig('./figures/SSA/rna_statistics.png',bbox_inches='tight')",
"_____no_output_____"
],
[
"data=pd.DataFrame(np.transpose(np.array(parray)))\nind=0\nnewcol=[]\nfor name in data.columns:\n newcol.append(\"mom\"+str(ind))\n ind+=1\ndata.columns=newcol\nmnprotarray=[]\ncvprotarray=[]\nerrcv2prot=[]\nerrmnprot=[]\nfor m in range(len(data)):\n rnas=data.loc[m, :].values.tolist()\n mean_cntr, var_cntr, std_cntr = bayesest(rnas,alpha=0.95)\n mnprotarray.append(mean_cntr[0])\n errmnprot.append(mean_cntr[1][1]-mean_cntr[0])\n cvprotarray.append(var_cntr[0]/mean_cntr[0]**2)\n errv=(var_cntr[1][1]-var_cntr[0])/mean_cntr[0]**2+2*(mean_cntr[1][1]-mean_cntr[0])*var_cntr[0]/mean_cntr[0]**3\n errcv2prot.append(errv)\n\ndata['time'] = tarray\ndata['Mean_prot'] = mnrnaarray\ndata['Error_mean'] = errmnrna\ndata['prot_CV2'] = cvrnaarray\ndata['Error_CV2'] = errcv2rna\nif not os.path.exists('./data/SSA'):\n os.makedirs('./data/SSA')\ndata.to_csv(\"./data/SSA/protsim.csv\")",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1,2, figsize=(12,4))\nax[0].plot(np.array(tarray)/doubling_time,mnprotarray,c=\"#3BB000\")\nax[0].fill_between(np.array(tarray)/doubling_time,np.array(mnprotarray)-np.array(errmnprot),np.array(mnprotarray)+np.array(errmnprot),\n alpha=1, edgecolor='#4BE000', facecolor='#4BE000',linewidth=0)\nax[1].plot(np.array(tarray)/doubling_time,cvprotarray,c=\"#3BB000\")\nax[1].fill_between(np.array(tarray)/doubling_time,np.array(cvprotarray)-np.array(errcv2prot),np.array(cvprotarray)+np.array(errcv2prot),\n alpha=1, edgecolor='#4BE000', facecolor='#4BE000',linewidth=0)\n\n\nax[0].set_ylabel(\"Protein\",size=20)\nax[1].set_ylabel(\"$C_V^2(p)$\",size=20)\nax[0].set_xlabel(r\"$t/\\tau$\",size=20)\nax[1].set_xlabel(r\"$t/\\tau$\",size=20)\nax[0].set_ylim([0,1.2*np.max(mnprotarray)])\nax[1].set_ylim([0,1.2*np.max(cvprotarray)])\nfor l in [0,1]:\n ax[l].set_xlim([0,tmax/doubling_time])\n taqui=np.arange(0,(tmax+1)/doubling_time,step=1)\n ax[l].set_xticks(np.array(taqui))\n ax[l].grid()\n ax[l].tick_params(axis='x', labelsize=15)\n ax[l].tick_params(axis='y', labelsize=15)\n for axis in ['bottom','left']:\n ax[l].spines[axis].set_linewidth(2)\n ax[l].tick_params(axis='both', width=2,length=6)\n for axis in ['top','right']:\n ax[l].spines[axis].set_linewidth(0)\n ax[l].tick_params(axis='both', width=0,length=6)\nplt.subplots_adjust(hspace=0.3,wspace=0.5)\ntaqui=np.arange(0,1.2*np.max(cvprotarray),step=np.round(.2*np.max(cvprotarray),4))\nax[1].set_yticks(np.array(taqui))\nif not os.path.exists('./figures'):\n os.makedirs('./figures')\nif not os.path.exists('./figures/SSA'):\n os.makedirs('./figures/SSA')\nplt.savefig('./figures/SSA/prot_statistics.svg',bbox_inches='tight')\nplt.savefig('./figures/SSA/prot_statistics.png',bbox_inches='tight')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb4477adfabc315d479d32e2a56cea738e51f66b | 19,877 | ipynb | Jupyter Notebook | notebooks/01-text-data.ipynb | ankitaguhaoakland/ml-workshop-advanced | f4475f34ecbd5c010a2ce32ee0b43afcafeee5a0 | [
"MIT"
] | 1 | 2021-04-09T21:14:39.000Z | 2021-04-09T21:14:39.000Z | notebooks/01-text-data.ipynb | ankitaguhaoakland/ml-workshop-advanced | f4475f34ecbd5c010a2ce32ee0b43afcafeee5a0 | [
"MIT"
] | null | null | null | notebooks/01-text-data.ipynb | ankitaguhaoakland/ml-workshop-advanced | f4475f34ecbd5c010a2ce32ee0b43afcafeee5a0 | [
"MIT"
] | null | null | null | 20.576605 | 129 | 0.526689 | [
[
[
"# Text Data in scikit-learn",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport sklearn\n\nsklearn.set_config(display='diagram')",
"_____no_output_____"
],
[
"from pathlib import Path\nimport tarfile\nfrom urllib import request\n\ndata_path = Path(\"data\")\nextracted_path = Path(\"data\") / \"train\"\nimdb_path = data_path / \"aclImdbmini.tar.gz\"\n\ndef untar_imdb():\n if extracted_path.exists():\n print(\"imdb dataset already extracted\")\n return\n with tarfile.open(imdb_path, \"r\") as tar_f:\n tar_f.extractall(data_path)\n \n# This may take some time to run since it will download and extracted\nuntar_imdb()",
"_____no_output_____"
]
],
[
[
"## CountVectorizer",
"_____no_output_____"
]
],
[
[
"sample_text = [\"Can we go to the hill? I finished my homework.\",\n \"The hill is very tall. Please be careful\"]",
"_____no_output_____"
],
[
"from sklearn.feature_extraction.text import CountVectorizer",
"_____no_output_____"
],
[
"vect = CountVectorizer()\nvect.fit(sample_text)\n\nvect.get_feature_names()",
"_____no_output_____"
],
[
"X = vect.transform(sample_text)\nX",
"_____no_output_____"
],
[
"X.toarray()",
"_____no_output_____"
]
],
[
[
"### Bag of words",
"_____no_output_____"
]
],
[
[
"sample_text",
"_____no_output_____"
],
[
"X_inverse = vect.inverse_transform(X)",
"_____no_output_____"
],
[
"X_inverse[0]",
"_____no_output_____"
],
[
"X_inverse[1]",
"_____no_output_____"
]
],
[
[
"## Loading text data with scikit-learn",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import load_files\n\nreviews_train = load_files(extracted_path, categories=[\"neg\", \"pos\"])\nraw_text_train, raw_y_train = reviews_train.data, reviews_train.target\nraw_text_train = [doc.replace(b\"<br />\", b\" \") for doc in raw_text_train]",
"_____no_output_____"
],
[
"import numpy as np\nnp.unique(raw_y_train)",
"_____no_output_____"
],
[
"np.bincount(raw_y_train)",
"_____no_output_____"
],
[
"len(raw_text_train)",
"_____no_output_____"
],
[
"raw_text_train[5]",
"_____no_output_____"
]
],
[
[
"## Split dataset",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\ntext_train, text_test, y_train, y_test = train_test_split(\n raw_text_train, raw_y_train, stratify=raw_y_train, random_state=0)",
"_____no_output_____"
]
],
[
[
"### Transform training data",
"_____no_output_____"
]
],
[
[
"vect = CountVectorizer()\nX_train = vect.fit_transform(text_train)",
"_____no_output_____"
],
[
"len(text_train)",
"_____no_output_____"
],
[
"X_train",
"_____no_output_____"
]
],
[
[
"### Transform testing set",
"_____no_output_____"
]
],
[
[
"len(text_test)",
"_____no_output_____"
],
[
"X_test = vect.transform(text_test)",
"_____no_output_____"
],
[
"X_test",
"_____no_output_____"
]
],
[
[
"### Extract feature names",
"_____no_output_____"
]
],
[
[
"feature_names = vect.get_feature_names()",
"_____no_output_____"
],
[
"feature_names[10000:10020]",
"_____no_output_____"
],
[
"feature_names[::3000]",
"_____no_output_____"
]
],
[
[
"### Linear model for classification",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\nlr = LogisticRegression(solver='liblinear', random_state=42).fit(X_train, y_train)",
"_____no_output_____"
],
[
"lr.score(X_test, y_test)",
"_____no_output_____"
],
[
"def plot_important_features(coef, feature_names, top_n=20, ax=None, rotation=40):\n if ax is None:\n ax = plt.gca()\n feature_names = np.asarray(feature_names)\n coef = coef.reshape(-1)\n inds = np.argsort(coef)\n low = inds[:top_n]\n high = inds[-top_n:]\n important = np.hstack([low, high])\n myrange = range(len(important))\n colors = ['red'] * top_n + ['blue'] * top_n\n \n ax.bar(myrange, coef[important], color=colors)\n ax.set_xticks(myrange)\n ax.set_xticklabels(feature_names[important], rotation=rotation, ha=\"right\")\n ax.set_xlim(-.7, 2 * top_n)\n ax.set_frame_on(False)",
"_____no_output_____"
],
[
"feature_names = vect.get_feature_names()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(15, 6))\nplot_important_features(lr.coef_, feature_names, top_n=20, ax=ax)",
"_____no_output_____"
]
],
[
[
"## Exercise 1\n\n1. Train a `sklearn.ensemble.RandomForestClassifier` on the training set, `X_train` and `y_train`.\n2. Evalute the accuracy on the test set.\n3. What are the top 20 important features accourind go `feature_importances_` of the random forst.",
"_____no_output_____"
]
],
[
[
"# %load solutions/01-ex01-solutions.py",
"_____no_output_____"
]
],
[
[
"## CountVectorizer Options",
"_____no_output_____"
]
],
[
[
"sample_text = [\"Can we go to the hill? I finished my homework.\",\n \"The hill is very tall. Please be careful\"]",
"_____no_output_____"
],
[
"vect = CountVectorizer()\nvect.fit(sample_text)\nvect.get_feature_names()",
"_____no_output_____"
]
],
[
[
"### Stop words",
"_____no_output_____"
]
],
[
[
"vect = CountVectorizer(stop_words='english')\nvect.fit(sample_text)\nvect.get_feature_names()",
"_____no_output_____"
],
[
"from sklearn.feature_extraction.text import ENGLISH_STOP_WORDS\nprint(list(ENGLISH_STOP_WORDS))",
"_____no_output_____"
]
],
[
[
"### Max features",
"_____no_output_____"
]
],
[
[
"vect = CountVectorizer(max_features=4, stop_words='english')\nvect.fit(sample_text)\nvect.get_feature_names()",
"_____no_output_____"
]
],
[
[
"### Min frequency on the imdb dataset\n\nWith `min_df=1` (default)",
"_____no_output_____"
]
],
[
[
"X_train.shape",
"_____no_output_____"
]
],
[
[
"With `min_df=4`",
"_____no_output_____"
]
],
[
[
"vect = CountVectorizer(min_df=4)\nX_train_min_df_4 = vect.fit_transform(text_train)",
"_____no_output_____"
],
[
"X_train_min_df_4.shape",
"_____no_output_____"
],
[
"lr_df_4 = LogisticRegression(solver='liblinear', random_state=42).fit(X_train_min_df_4, y_train)",
"_____no_output_____"
],
[
"X_test_min_df_4 = vect.transform(text_test)",
"_____no_output_____"
]
],
[
[
"#### Scores with different min frequencies",
"_____no_output_____"
]
],
[
[
"lr_df_4.score(X_test_min_df_4, y_test)",
"_____no_output_____"
],
[
"lr.score(X_test, y_test)",
"_____no_output_____"
]
],
[
[
"## Pipelines and Vectorizers",
"_____no_output_____"
]
],
[
[
"from sklearn.pipeline import Pipeline\n\nlog_reg = Pipeline([\n ('vectorizer', CountVectorizer()),\n ('classifier', LogisticRegression(random_state=42, solver='liblinear'))\n])",
"_____no_output_____"
],
[
"log_reg",
"_____no_output_____"
],
[
"text_train[:2]",
"_____no_output_____"
],
[
"log_reg.fit(text_train, y_train)",
"_____no_output_____"
],
[
"log_reg.score(text_train, y_train)",
"_____no_output_____"
],
[
"log_reg.score(text_test, y_test)",
"_____no_output_____"
]
],
[
[
"## Exercise 2\n\n1. Create a pipeline with a `CountVectorizer` with `min_df=5` and `stop_words='english'` and a `RandomForestClassifier`.\n2. What is the score of the random forest on the test dataset?",
"_____no_output_____"
]
],
[
[
"# %load solutions/01-ex02-solutions.py",
"_____no_output_____"
]
],
[
[
"## Bigrams\n\n`CountVectorizer` takes a `ngram_range` parameter",
"_____no_output_____"
]
],
[
[
"sample_text",
"_____no_output_____"
],
[
"cv = CountVectorizer(ngram_range=(1, 1)).fit(sample_text)\nprint(\"Vocabulary size:\", len(cv.vocabulary_))\nprint(\"Vocabulary:\", cv.get_feature_names())",
"_____no_output_____"
],
[
"cv = CountVectorizer(ngram_range=(2, 2)).fit(sample_text)\nprint(\"Vocabulary size:\", len(cv.vocabulary_))\nprint(\"Vocabulary:\")\nprint(cv.get_feature_names())",
"_____no_output_____"
],
[
"cv = CountVectorizer(ngram_range=(1, 2)).fit(sample_text)\nprint(\"Vocabulary size:\", len(cv.vocabulary_))\nprint(\"Vocabulary:\")\nprint(cv.get_feature_names())",
"_____no_output_____"
]
],
[
[
"## n-grams with stop words",
"_____no_output_____"
]
],
[
[
"cv_n_gram = CountVectorizer(ngram_range=(1, 2), min_df=4, stop_words=\"english\")\ncv_n_gram.fit(text_train)",
"_____no_output_____"
],
[
"len(cv_n_gram.vocabulary_)",
"_____no_output_____"
],
[
"print(cv_n_gram.get_feature_names()[::2000])",
"_____no_output_____"
],
[
"pipe_cv_n_gram = Pipeline([\n ('vectorizer', cv_n_gram),\n ('classifier', LogisticRegression(random_state=42, solver='liblinear'))\n])",
"_____no_output_____"
],
[
"pipe_cv_n_gram.fit(text_train, y_train)",
"_____no_output_____"
],
[
"pipe_cv_n_gram.score(text_test, y_test)",
"_____no_output_____"
],
[
"feature_names = pipe_cv_n_gram['vectorizer'].get_feature_names()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(15, 6))\nplot_important_features(pipe_cv_n_gram['classifier'].coef_.ravel(), feature_names, top_n=20, ax=ax)",
"_____no_output_____"
]
],
[
[
"## Tf-idf rescaling",
"_____no_output_____"
]
],
[
[
"sample_text",
"_____no_output_____"
],
[
"from sklearn.feature_extraction.text import TfidfVectorizer",
"_____no_output_____"
],
[
"tfidvect = TfidfVectorizer().fit(sample_text)\ntfid_trans = tfidvect.transform(sample_text)",
"_____no_output_____"
],
[
"tfid_trans.toarray()",
"_____no_output_____"
]
],
[
[
"## Train on the imdb dataset",
"_____no_output_____"
]
],
[
[
"log_reg_tfid = Pipeline([\n ('vectorizer', TfidfVectorizer(ngram_range=(1, 2), min_df=4,\n stop_words=\"english\")),\n ('classifier', LogisticRegression(random_state=42, solver='liblinear'))\n])",
"_____no_output_____"
],
[
"log_reg_tfid.fit(text_train, y_train)",
"_____no_output_____"
],
[
"log_reg_tfid.score(text_test, y_test)",
"_____no_output_____"
]
],
[
[
"## Exercise 3\n\n0. Load data from `fetch_20newsgroups`:\n\n```python\nfrom sklearn.datasets import fetch_20newsgroups\ncategories = [\n 'alt.atheism',\n 'sci.space',\n]\nremove = ('headers', 'footers', 'quotes')\n\ndata_train = fetch_20newsgroups(subset='train', categories=categories,\n remove=remove)\n\ndata_test = fetch_20newsgroups(subset='test', categories=categories,\n remove=remove)\n\nX_train, y_train = data_train.data, data_train.target\nX_test, y_test = data_test.data, data_test.target\n```\n\n1. How many samples are there in the training dataset and test dataset?\n1. Construct a pipeline with a `TfidfVectorizer` and `LogisticRegression`.\n1. Evalute the pipeline on the test set.\n1. Plot the feature importances using `plot_important_features`.",
"_____no_output_____"
]
],
[
[
"# %load solutions/01-ex03-solutions.py",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb447c8bb652689598f49cf269fc0911e841b6be | 9,191 | ipynb | Jupyter Notebook | tensorflow/day3/practice/P_03_06_iris_multi_classificataion_callback1.ipynb | daludaluking/LG_AI_all_in_one- | e0855af811deb1e5cf1695430bd52a8eb3d48827 | [
"Apache-2.0"
] | null | null | null | tensorflow/day3/practice/P_03_06_iris_multi_classificataion_callback1.ipynb | daludaluking/LG_AI_all_in_one- | e0855af811deb1e5cf1695430bd52a8eb3d48827 | [
"Apache-2.0"
] | null | null | null | tensorflow/day3/practice/P_03_06_iris_multi_classificataion_callback1.ipynb | daludaluking/LG_AI_all_in_one- | e0855af811deb1e5cf1695430bd52a8eb3d48827 | [
"Apache-2.0"
] | null | null | null | 9,191 | 9,191 | 0.675117 | [
[
[
"import tensorflow as tf\n\nimport matplotlib.pyplot as plt\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import LabelEncoder\n\nimport pandas as pd\nimport math\n\ngpus = tf.config.experimental.list_physical_devices('GPU')\nif gpus:\n try:\n # Currently, memory growth needs to be the same across GPUs\n for gpu in gpus:\n tf.config.experimental.set_memory_growth(gpu, True)\n logical_gpus = tf.config.experimental.list_logical_devices('GPU')\n print(len(gpus), \"Physical GPUs,\", len(logical_gpus), \"Logical GPUs\")\n except RuntimeError as e:\n # Memory growth must be set before GPUs have been initialized\n print(e)",
"1 Physical GPUs, 1 Logical GPUs\n"
],
[
"\ndef step_decay(epoch):\n\tinitial_lrate = 0.001\n\tdrop = 0.98\n\tepochs_drop = 50.0\n\tlrate = initial_lrate * math.pow(drop, math.floor((1+epoch)/epochs_drop))\n\treturn lrate\n\n",
"_____no_output_____"
],
[
"\n# 데이터 입력\ndf = pd.read_csv('../dataset/iris.csv', names = [\"sepal_length\", \"sepal_width\", \"petal_length\", \"petal_width\", \"species\"])\n\n# 데이터 분류\ndataset=df.copy()\n\n# 데이터 분류\nY_obj=dataset.pop(\"species\")\nX=dataset.copy()\n\n# 문자열을 숫자로 변환\nY_encoded=pd.get_dummies(Y_obj)\n",
"_____no_output_____"
],
[
"\n# 전체 데이터에서 학습 데이터와 테스트 데이터(0.1)로 구분\nX_train1, X_test, Y_train1, Y_test = train_test_split(X, Y_encoded, test_size=0.3,shuffle=True, stratify=Y_encoded) ## shuffle=True로 하면 데이터를 섞어서 나눔\n## 학습 셋에서 학습과 검증 데이터(0.2)로 구분\nX_train, X_valid, Y_train, Y_valid = train_test_split(X_train1, Y_train1, test_size=0.2, shuffle=True, stratify=Y_train1) ## shuffle=True로 하면 데이터를 섞어서 나눔\n\n",
"_____no_output_____"
],
[
"\n# 모델의 설정\nactivation=tf.keras.activations.sigmoid\ninput_Layer = tf.keras.layers.Input(shape=(4,))\nx = tf.keras.layers.Dense(16, activation=activation,)(input_Layer)\nx = tf.keras.layers.Dense(12, activation=activation)(x)\nOut_Layer= tf.keras.layers.Dense(3, activation='softmax')(x)\nmodel = tf.keras.models.Model(inputs=[input_Layer], outputs=[Out_Layer])\nmodel.summary()\n",
"Model: \"model\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) [(None, 4)] 0 \n_________________________________________________________________\ndense (Dense) (None, 16) 80 \n_________________________________________________________________\ndense_1 (Dense) (None, 12) 204 \n_________________________________________________________________\ndense_2 (Dense) (None, 3) 39 \n=================================================================\nTotal params: 323\nTrainable params: 323\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"\n# 모델 컴파일\nmodel.compile(loss=tf.keras.losses.categorical_crossentropy,\n optimizer=tf.keras.optimizers.SGD(learning_rate=0.001),\n metrics=[tf.keras.metrics.categorical_accuracy])\n\nmodelpath=\"./best_model/{epoch:02d}-{val_loss:.4f}.h5\"\nclabacks_list =[tf.keras.callbacks.EarlyStopping(monitor='val_loss', patience=100),\n tf.keras.callbacks.ModelCheckpoint(filepath=modelpath, monitor='val_loss', verbose=1, save_best_only=True),\n tf.keras.callbacks.LearningRateScheduler(step_decay, verbose=1)]\n\n## model fit은 histoy를 반환한다. 훈련중의 발생하는 모든 정보를 담고 있는 딕셔너리.\nresult=model.fit(X_train, Y_train, epochs=50, batch_size=50, validation_data=(X_valid,Y_valid),callbacks=clabacks_list) # validation_data=(X_valid,Y_valid)을 추가하여 학습시 검증을 해줌.\n## histoy는 딕셔너리이므로 keys()를 통해 출력의 key(카테고리)를 확인하여 무엇을 받고 있는지 확인.\nprint(result.history.keys())\n",
"_____no_output_____"
],
[
"\n### result에서 loss와 val_loss의 key를 가지는 값들만 추출\nloss = result.history['loss']\nval_loss = result.history['val_loss']\n### loss와 val_loss를 그래프화\nepochs = range(1, len(loss) + 1)\nplt.subplot(211) ## 2x1 개의 그래프 중에 1번째\nplt.plot(epochs, loss, 'bo', label='Training loss')\nplt.plot(epochs, val_loss, 'b', label='Validation loss')\nplt.title('Training and validation loss')\nplt.xlabel('Epochs')\nplt.ylabel('Loss')\nplt.legend()\n\n### history에서 binary_accuracy와 val_binary_accuracy key를 가지는 값들만 추출\nacc = result.history['categorical_accuracy']\nval_acc = result.history['val_categorical_accuracy']\n\n### binary_accuracy와 val_binary_accuracy key를 그래프화\nplt.subplot(212) ## 2x1 개의 그래프 중에 2번째\nplt.plot(epochs, acc, 'ro', label='Training acc')\nplt.plot(epochs, val_acc, 'r', label='Validation acc')\nplt.title('Training and validation accuracy')\nplt.xlabel('Epochs')\nplt.ylabel('Accuracy')\nplt.legend()\n\n\n# model.evalueate를 통해 테스트 데이터로 정확도 확인하기.\n## model.evaluate(X_test, Y_test)의 리턴값은 [loss, binary_acuuracy ] -> 위 model.compile에서 metrics=[ keras.metrics.binary_accuracy]옵션을 주어서 binary acuuracy 출력됨.\nprint(\"\\n Test Accuracy: %.4f\" % (model.evaluate(X_test, Y_test)[1]))\n\n## 그래프 띄우기\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cb44826877f1ff760cc45046df4be91662b7ad5f | 74,601 | ipynb | Jupyter Notebook | Natural Language Processing/Transformer for Translation.ipynb | NiloyPurkait/cognitive-libraries | 424f0d778d120b780d794a66a5eb509355674937 | [
"BSD-3-Clause"
] | null | null | null | Natural Language Processing/Transformer for Translation.ipynb | NiloyPurkait/cognitive-libraries | 424f0d778d120b780d794a66a5eb509355674937 | [
"BSD-3-Clause"
] | null | null | null | Natural Language Processing/Transformer for Translation.ipynb | NiloyPurkait/cognitive-libraries | 424f0d778d120b780d794a66a5eb509355674937 | [
"BSD-3-Clause"
] | null | null | null | 36.390732 | 457 | 0.520288 | [
[
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Transformer model for language Translation",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/tutorials/text/transformer\">\n <img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />\n View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/tutorials/text/transformer.ipynb\">\n <img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />\n Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs/blob/master/site/en/tutorials/text/transformer.ipynb\">\n <img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />\n View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/docs/site/en/tutorials/text/transformer.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"This tutorial trains a <a href=\"https://arxiv.org/abs/1706.03762\" class=\"external\">Transformer model</a> to translate Portuguese to English. This is an advanced example that assumes knowledge of [text generation](text_generation.ipynb) and [attention](nmt_with_attention.ipynb).\n\nThe core idea behind the Transformer model is *self-attention*—the ability to attend to different positions of the input sequence to compute a representation of that sequence. Transformer creates stacks of self-attention layers and is explained below in the sections *Scaled dot product attention* and *Multi-head attention*.\n\nA transformer model handles variable-sized input using stacks of self-attention layers instead of [RNNs](text_classification_rnn.ipynb) or [CNNs](../images/intro_to_cnns.ipynb). This general architecture has a number of advantages:\n\n* It make no assumptions about the temporal/spatial relationships across the data. This is ideal for processing a set of objects (for example, [StarCraft units](https://deepmind.com/blog/alphastar-mastering-real-time-strategy-game-starcraft-ii/#block-8)).\n* Layer outputs can be calculated in parallel, instead of a series like an RNN.\n* Distant items can affect each other's output without passing through many RNN-steps, or convolution layers (see [Scene Memory Transformer](https://arxiv.org/pdf/1903.03878.pdf) for example).\n* It can learn long-range dependencies. This is a challenge in many sequence tasks.\n\nThe downsides of this architecture are:\n\n* For a time-series, the output for a time-step is calculated from the *entire history* instead of only the inputs and current hidden-state. This _may_ be less efficient. \n* If the input *does* have a temporal/spatial relationship, like text, some positional encoding must be added or the model will effectively see a bag of words. \n\nAfter training the model in this notebook, you will be able to input a Portuguese sentence and return the English translation.\n\n<img src=\"https://www.tensorflow.org/images/tutorials/transformer/attention_map_portuguese.png\" width=\"800\" alt=\"Attention heatmap\">",
"_____no_output_____"
]
],
[
[
"!pip install -q tfds-nightly",
"_____no_output_____"
],
[
"import tensorflow_datasets as tfds\nimport tensorflow as tf\n\nimport time\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Setup input pipeline",
"_____no_output_____"
],
[
"Use [TFDS](https://www.tensorflow.org/datasets) to load the [Portugese-English translation dataset](https://github.com/neulab/word-embeddings-for-nmt) from the [TED Talks Open Translation Project](https://www.ted.com/participate/translate).\n\nThis dataset contains approximately 50000 training examples, 1100 validation examples, and 2000 test examples.",
"_____no_output_____"
]
],
[
[
"examples, metadata = tfds.load('ted_hrlr_translate/pt_to_en', with_info=True,\n as_supervised=True)\ntrain_examples, val_examples = examples['train'], examples['validation']",
"_____no_output_____"
]
],
[
[
"Create a custom subwords tokenizer from the training dataset. ",
"_____no_output_____"
]
],
[
[
"tokenizer_en = tfds.deprecated.text.SubwordTextEncoder.build_from_corpus(\n (en.numpy() for pt, en in train_examples), target_vocab_size=2**13)\n\ntokenizer_pt = tfds.deprecated.text.SubwordTextEncoder.build_from_corpus(\n (pt.numpy() for pt, en in train_examples), target_vocab_size=2**13)",
"_____no_output_____"
],
[
"sample_string = 'Transformer is awesome.'\n\ntokenized_string = tokenizer_en.encode(sample_string)\nprint ('Tokenized string is {}'.format(tokenized_string))\n\noriginal_string = tokenizer_en.decode(tokenized_string)\nprint ('The original string: {}'.format(original_string))\n\nassert original_string == sample_string",
"_____no_output_____"
]
],
[
[
"The tokenizer encodes the string by breaking it into subwords if the word is not in its dictionary.",
"_____no_output_____"
]
],
[
[
"for ts in tokenized_string:\n print ('{} ----> {}'.format(ts, tokenizer_en.decode([ts])))",
"_____no_output_____"
],
[
"BUFFER_SIZE = 20000\nBATCH_SIZE = 64",
"_____no_output_____"
]
],
[
[
"Add a start and end token to the input and target. ",
"_____no_output_____"
]
],
[
[
"def encode(lang1, lang2):\n lang1 = [tokenizer_pt.vocab_size] + tokenizer_pt.encode(\n lang1.numpy()) + [tokenizer_pt.vocab_size+1]\n\n lang2 = [tokenizer_en.vocab_size] + tokenizer_en.encode(\n lang2.numpy()) + [tokenizer_en.vocab_size+1]\n \n return lang1, lang2",
"_____no_output_____"
]
],
[
[
"You want to use `Dataset.map` to apply this function to each element of the dataset. `Dataset.map` runs in graph mode.\n\n* Graph tensors do not have a value. \n* In graph mode you can only use TensorFlow Ops and functions. \n\nSo you can't `.map` this function directly: You need to wrap it in a `tf.py_function`. The `tf.py_function` will pass regular tensors (with a value and a `.numpy()` method to access it), to the wrapped python function.",
"_____no_output_____"
]
],
[
[
"def tf_encode(pt, en):\n result_pt, result_en = tf.py_function(encode, [pt, en], [tf.int64, tf.int64])\n result_pt.set_shape([None])\n result_en.set_shape([None])\n\n return result_pt, result_en",
"_____no_output_____"
]
],
[
[
"Note: To keep this example small and relatively fast, drop examples with a length of over 40 tokens.",
"_____no_output_____"
]
],
[
[
"MAX_LENGTH = 40",
"_____no_output_____"
],
[
"def filter_max_length(x, y, max_length=MAX_LENGTH):\n return tf.logical_and(tf.size(x) <= max_length,\n tf.size(y) <= max_length)",
"_____no_output_____"
],
[
"train_dataset = train_examples.map(tf_encode)\ntrain_dataset = train_dataset.filter(filter_max_length)\n# cache the dataset to memory to get a speedup while reading from it.\ntrain_dataset = train_dataset.cache()\ntrain_dataset = train_dataset.shuffle(BUFFER_SIZE).padded_batch(BATCH_SIZE)\ntrain_dataset = train_dataset.prefetch(tf.data.experimental.AUTOTUNE)\n\n\nval_dataset = val_examples.map(tf_encode)\nval_dataset = val_dataset.filter(filter_max_length).padded_batch(BATCH_SIZE)",
"_____no_output_____"
],
[
"pt_batch, en_batch = next(iter(val_dataset))\npt_batch, en_batch",
"_____no_output_____"
]
],
[
[
"## Positional encoding\n\nSince this model doesn't contain any recurrence or convolution, positional encoding is added to give the model some information about the relative position of the words in the sentence. \n\nThe positional encoding vector is added to the embedding vector. Embeddings represent a token in a d-dimensional space where tokens with similar meaning will be closer to each other. But the embeddings do not encode the relative position of words in a sentence. So after adding the positional encoding, words will be closer to each other based on the *similarity of their meaning and their position in the sentence*, in the d-dimensional space.\n\nSee the notebook on [positional encoding](https://github.com/tensorflow/examples/blob/master/community/en/position_encoding.ipynb) to learn more about it. The formula for calculating the positional encoding is as follows:\n\n$$\\Large{PE_{(pos, 2i)} = sin(pos / 10000^{2i / d_{model}})} $$\n$$\\Large{PE_{(pos, 2i+1)} = cos(pos / 10000^{2i / d_{model}})} $$",
"_____no_output_____"
]
],
[
[
"def get_angles(pos, i, d_model):\n angle_rates = 1 / np.power(10000, (2 * (i//2)) / np.float32(d_model))\n return pos * angle_rates",
"_____no_output_____"
],
[
"def positional_encoding(position, d_model):\n angle_rads = get_angles(np.arange(position)[:, np.newaxis],\n np.arange(d_model)[np.newaxis, :],\n d_model)\n \n # apply sin to even indices in the array; 2i\n angle_rads[:, 0::2] = np.sin(angle_rads[:, 0::2])\n \n # apply cos to odd indices in the array; 2i+1\n angle_rads[:, 1::2] = np.cos(angle_rads[:, 1::2])\n \n pos_encoding = angle_rads[np.newaxis, ...]\n \n return tf.cast(pos_encoding, dtype=tf.float32)",
"_____no_output_____"
],
[
"pos_encoding = positional_encoding(50, 512)\nprint (pos_encoding.shape)\n\nplt.pcolormesh(pos_encoding[0], cmap='RdBu')\nplt.xlabel('Depth')\nplt.xlim((0, 512))\nplt.ylabel('Position')\nplt.colorbar()\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Masking",
"_____no_output_____"
],
[
"Mask all the pad tokens in the batch of sequence. It ensures that the model does not treat padding as the input. The mask indicates where pad value `0` is present: it outputs a `1` at those locations, and a `0` otherwise.",
"_____no_output_____"
]
],
[
[
"def create_padding_mask(seq):\n seq = tf.cast(tf.math.equal(seq, 0), tf.float32)\n \n # add extra dimensions to add the padding\n # to the attention logits.\n return seq[:, tf.newaxis, tf.newaxis, :] # (batch_size, 1, 1, seq_len)",
"_____no_output_____"
],
[
"x = tf.constant([[7, 6, 0, 0, 1], [1, 2, 3, 0, 0], [0, 0, 0, 4, 5]])\ncreate_padding_mask(x)",
"_____no_output_____"
]
],
[
[
"The look-ahead mask is used to mask the future tokens in a sequence. In other words, the mask indicates which entries should not be used.\n\nThis means that to predict the third word, only the first and second word will be used. Similarly to predict the fourth word, only the first, second and the third word will be used and so on.",
"_____no_output_____"
]
],
[
[
"def create_look_ahead_mask(size):\n mask = 1 - tf.linalg.band_part(tf.ones((size, size)), -1, 0)\n return mask # (seq_len, seq_len)",
"_____no_output_____"
],
[
"x = tf.random.uniform((1, 3))\ntemp = create_look_ahead_mask(x.shape[1])\ntemp",
"_____no_output_____"
]
],
[
[
"## Scaled dot product attention",
"_____no_output_____"
],
[
"<img src=\"https://www.tensorflow.org/images/tutorials/transformer/scaled_attention.png\" width=\"500\" alt=\"scaled_dot_product_attention\">\n\nThe attention function used by the transformer takes three inputs: Q (query), K (key), V (value). The equation used to calculate the attention weights is:\n\n$$\\Large{Attention(Q, K, V) = softmax_k(\\frac{QK^T}{\\sqrt{d_k}}) V} $$\n\nThe dot-product attention is scaled by a factor of square root of the depth. This is done because for large values of depth, the dot product grows large in magnitude pushing the softmax function where it has small gradients resulting in a very hard softmax. \n\nFor example, consider that `Q` and `K` have a mean of 0 and variance of 1. Their matrix multiplication will have a mean of 0 and variance of `dk`. Hence, *square root of `dk`* is used for scaling (and not any other number) because the matmul of `Q` and `K` should have a mean of 0 and variance of 1, and you get a gentler softmax.\n\nThe mask is multiplied with -1e9 (close to negative infinity). This is done because the mask is summed with the scaled matrix multiplication of Q and K and is applied immediately before a softmax. The goal is to zero out these cells, and large negative inputs to softmax are near zero in the output.",
"_____no_output_____"
]
],
[
[
"def scaled_dot_product_attention(q, k, v, mask):\n \"\"\"Calculate the attention weights.\n q, k, v must have matching leading dimensions.\n k, v must have matching penultimate dimension, i.e.: seq_len_k = seq_len_v.\n The mask has different shapes depending on its type(padding or look ahead) \n but it must be broadcastable for addition.\n \n Args:\n q: query shape == (..., seq_len_q, depth)\n k: key shape == (..., seq_len_k, depth)\n v: value shape == (..., seq_len_v, depth_v)\n mask: Float tensor with shape broadcastable \n to (..., seq_len_q, seq_len_k). Defaults to None.\n \n Returns:\n output, attention_weights\n \"\"\"\n\n matmul_qk = tf.matmul(q, k, transpose_b=True) # (..., seq_len_q, seq_len_k)\n \n # scale matmul_qk\n dk = tf.cast(tf.shape(k)[-1], tf.float32)\n scaled_attention_logits = matmul_qk / tf.math.sqrt(dk)\n\n # add the mask to the scaled tensor.\n if mask is not None:\n scaled_attention_logits += (mask * -1e9) \n\n # softmax is normalized on the last axis (seq_len_k) so that the scores\n # add up to 1.\n attention_weights = tf.nn.softmax(scaled_attention_logits, axis=-1) # (..., seq_len_q, seq_len_k)\n\n output = tf.matmul(attention_weights, v) # (..., seq_len_q, depth_v)\n\n return output, attention_weights",
"_____no_output_____"
]
],
[
[
"As the softmax normalization is done on K, its values decide the amount of importance given to Q.\n\nThe output represents the multiplication of the attention weights and the V (value) vector. This ensures that the words you want to focus on are kept as-is and the irrelevant words are flushed out.",
"_____no_output_____"
]
],
[
[
"def print_out(q, k, v):\n temp_out, temp_attn = scaled_dot_product_attention(\n q, k, v, None)\n print ('Attention weights are:')\n print (temp_attn)\n print ('Output is:')\n print (temp_out)",
"_____no_output_____"
],
[
"np.set_printoptions(suppress=True)\n\ntemp_k = tf.constant([[10,0,0],\n [0,10,0],\n [0,0,10],\n [0,0,10]], dtype=tf.float32) # (4, 3)\n\ntemp_v = tf.constant([[ 1,0],\n [ 10,0],\n [ 100,5],\n [1000,6]], dtype=tf.float32) # (4, 2)\n\n# This `query` aligns with the second `key`,\n# so the second `value` is returned.\ntemp_q = tf.constant([[0, 10, 0]], dtype=tf.float32) # (1, 3)\nprint_out(temp_q, temp_k, temp_v)",
"_____no_output_____"
],
[
"# This query aligns with a repeated key (third and fourth), \n# so all associated values get averaged.\ntemp_q = tf.constant([[0, 0, 10]], dtype=tf.float32) # (1, 3)\nprint_out(temp_q, temp_k, temp_v)",
"_____no_output_____"
],
[
"# This query aligns equally with the first and second key, \n# so their values get averaged.\ntemp_q = tf.constant([[10, 10, 0]], dtype=tf.float32) # (1, 3)\nprint_out(temp_q, temp_k, temp_v)",
"_____no_output_____"
]
],
[
[
"Pass all the queries together.",
"_____no_output_____"
]
],
[
[
"temp_q = tf.constant([[0, 0, 10], [0, 10, 0], [10, 10, 0]], dtype=tf.float32) # (3, 3)\nprint_out(temp_q, temp_k, temp_v)",
"_____no_output_____"
]
],
[
[
"## Multi-head attention",
"_____no_output_____"
],
[
"<img src=\"https://www.tensorflow.org/images/tutorials/transformer/multi_head_attention.png\" width=\"500\" alt=\"multi-head attention\">\n\n\nMulti-head attention consists of four parts:\n* Linear layers and split into heads.\n* Scaled dot-product attention.\n* Concatenation of heads.\n* Final linear layer.",
"_____no_output_____"
],
[
"Each multi-head attention block gets three inputs; Q (query), K (key), V (value). These are put through linear (Dense) layers and split up into multiple heads. \n\nThe `scaled_dot_product_attention` defined above is applied to each head (broadcasted for efficiency). An appropriate mask must be used in the attention step. The attention output for each head is then concatenated (using `tf.transpose`, and `tf.reshape`) and put through a final `Dense` layer.\n\nInstead of one single attention head, Q, K, and V are split into multiple heads because it allows the model to jointly attend to information at different positions from different representational spaces. After the split each head has a reduced dimensionality, so the total computation cost is the same as a single head attention with full dimensionality.",
"_____no_output_____"
]
],
[
[
"class MultiHeadAttention(tf.keras.layers.Layer):\n def __init__(self, d_model, num_heads):\n super(MultiHeadAttention, self).__init__()\n self.num_heads = num_heads\n self.d_model = d_model\n \n assert d_model % self.num_heads == 0\n \n self.depth = d_model // self.num_heads\n \n self.wq = tf.keras.layers.Dense(d_model)\n self.wk = tf.keras.layers.Dense(d_model)\n self.wv = tf.keras.layers.Dense(d_model)\n \n self.dense = tf.keras.layers.Dense(d_model)\n \n def split_heads(self, x, batch_size):\n \"\"\"Split the last dimension into (num_heads, depth).\n Transpose the result such that the shape is (batch_size, num_heads, seq_len, depth)\n \"\"\"\n x = tf.reshape(x, (batch_size, -1, self.num_heads, self.depth))\n return tf.transpose(x, perm=[0, 2, 1, 3])\n \n def call(self, v, k, q, mask):\n batch_size = tf.shape(q)[0]\n \n q = self.wq(q) # (batch_size, seq_len, d_model)\n k = self.wk(k) # (batch_size, seq_len, d_model)\n v = self.wv(v) # (batch_size, seq_len, d_model)\n \n q = self.split_heads(q, batch_size) # (batch_size, num_heads, seq_len_q, depth)\n k = self.split_heads(k, batch_size) # (batch_size, num_heads, seq_len_k, depth)\n v = self.split_heads(v, batch_size) # (batch_size, num_heads, seq_len_v, depth)\n \n # scaled_attention.shape == (batch_size, num_heads, seq_len_q, depth)\n # attention_weights.shape == (batch_size, num_heads, seq_len_q, seq_len_k)\n scaled_attention, attention_weights = scaled_dot_product_attention(\n q, k, v, mask)\n \n scaled_attention = tf.transpose(scaled_attention, perm=[0, 2, 1, 3]) # (batch_size, seq_len_q, num_heads, depth)\n\n concat_attention = tf.reshape(scaled_attention, \n (batch_size, -1, self.d_model)) # (batch_size, seq_len_q, d_model)\n\n output = self.dense(concat_attention) # (batch_size, seq_len_q, d_model)\n \n return output, attention_weights",
"_____no_output_____"
]
],
[
[
"Create a `MultiHeadAttention` layer to try out. At each location in the sequence, `y`, the `MultiHeadAttention` runs all 8 attention heads across all other locations in the sequence, returning a new vector of the same length at each location.",
"_____no_output_____"
]
],
[
[
"temp_mha = MultiHeadAttention(d_model=512, num_heads=8)\ny = tf.random.uniform((1, 60, 512)) # (batch_size, encoder_sequence, d_model)\nout, attn = temp_mha(y, k=y, q=y, mask=None)\nout.shape, attn.shape",
"_____no_output_____"
]
],
[
[
"## Point wise feed forward network",
"_____no_output_____"
],
[
"Point wise feed forward network consists of two fully-connected layers with a ReLU activation in between.",
"_____no_output_____"
]
],
[
[
"def point_wise_feed_forward_network(d_model, dff):\n return tf.keras.Sequential([\n tf.keras.layers.Dense(dff, activation='relu'), # (batch_size, seq_len, dff)\n tf.keras.layers.Dense(d_model) # (batch_size, seq_len, d_model)\n ])",
"_____no_output_____"
],
[
"sample_ffn = point_wise_feed_forward_network(512, 2048)\nsample_ffn(tf.random.uniform((64, 50, 512))).shape",
"_____no_output_____"
]
],
[
[
"## Encoder and decoder",
"_____no_output_____"
],
[
"<img src=\"https://www.tensorflow.org/images/tutorials/transformer/transformer.png\" width=\"600\" alt=\"transformer\">",
"_____no_output_____"
],
[
"The transformer model follows the same general pattern as a standard [sequence to sequence with attention model](nmt_with_attention.ipynb). \n\n* The input sentence is passed through `N` encoder layers that generates an output for each word/token in the sequence.\n* The decoder attends on the encoder's output and its own input (self-attention) to predict the next word. ",
"_____no_output_____"
],
[
"### Encoder layer\n\nEach encoder layer consists of sublayers:\n\n1. Multi-head attention (with padding mask) \n2. Point wise feed forward networks. \n\nEach of these sublayers has a residual connection around it followed by a layer normalization. Residual connections help in avoiding the vanishing gradient problem in deep networks.\n\nThe output of each sublayer is `LayerNorm(x + Sublayer(x))`. The normalization is done on the `d_model` (last) axis. There are N encoder layers in the transformer.",
"_____no_output_____"
]
],
[
[
"class EncoderLayer(tf.keras.layers.Layer):\n def __init__(self, d_model, num_heads, dff, rate=0.1):\n super(EncoderLayer, self).__init__()\n\n self.mha = MultiHeadAttention(d_model, num_heads)\n self.ffn = point_wise_feed_forward_network(d_model, dff)\n\n self.layernorm1 = tf.keras.layers.LayerNormalization(epsilon=1e-6)\n self.layernorm2 = tf.keras.layers.LayerNormalization(epsilon=1e-6)\n \n self.dropout1 = tf.keras.layers.Dropout(rate)\n self.dropout2 = tf.keras.layers.Dropout(rate)\n \n def call(self, x, training, mask):\n\n attn_output, _ = self.mha(x, x, x, mask) # (batch_size, input_seq_len, d_model)\n attn_output = self.dropout1(attn_output, training=training)\n out1 = self.layernorm1(x + attn_output) # (batch_size, input_seq_len, d_model)\n \n ffn_output = self.ffn(out1) # (batch_size, input_seq_len, d_model)\n ffn_output = self.dropout2(ffn_output, training=training)\n out2 = self.layernorm2(out1 + ffn_output) # (batch_size, input_seq_len, d_model)\n \n return out2",
"_____no_output_____"
],
[
"sample_encoder_layer = EncoderLayer(512, 8, 2048)\n\nsample_encoder_layer_output = sample_encoder_layer(\n tf.random.uniform((64, 43, 512)), False, None)\n\nsample_encoder_layer_output.shape # (batch_size, input_seq_len, d_model)",
"_____no_output_____"
]
],
[
[
"### Decoder layer\n\nEach decoder layer consists of sublayers:\n\n1. Masked multi-head attention (with look ahead mask and padding mask)\n2. Multi-head attention (with padding mask). V (value) and K (key) receive the *encoder output* as inputs. Q (query) receives the *output from the masked multi-head attention sublayer.*\n3. Point wise feed forward networks\n\nEach of these sublayers has a residual connection around it followed by a layer normalization. The output of each sublayer is `LayerNorm(x + Sublayer(x))`. The normalization is done on the `d_model` (last) axis.\n\nThere are N decoder layers in the transformer.\n\nAs Q receives the output from decoder's first attention block, and K receives the encoder output, the attention weights represent the importance given to the decoder's input based on the encoder's output. In other words, the decoder predicts the next word by looking at the encoder output and self-attending to its own output. See the demonstration above in the scaled dot product attention section.",
"_____no_output_____"
]
],
[
[
"class DecoderLayer(tf.keras.layers.Layer):\n def __init__(self, d_model, num_heads, dff, rate=0.1):\n super(DecoderLayer, self).__init__()\n\n self.mha1 = MultiHeadAttention(d_model, num_heads)\n self.mha2 = MultiHeadAttention(d_model, num_heads)\n\n self.ffn = point_wise_feed_forward_network(d_model, dff)\n \n self.layernorm1 = tf.keras.layers.LayerNormalization(epsilon=1e-6)\n self.layernorm2 = tf.keras.layers.LayerNormalization(epsilon=1e-6)\n self.layernorm3 = tf.keras.layers.LayerNormalization(epsilon=1e-6)\n \n self.dropout1 = tf.keras.layers.Dropout(rate)\n self.dropout2 = tf.keras.layers.Dropout(rate)\n self.dropout3 = tf.keras.layers.Dropout(rate)\n \n \n def call(self, x, enc_output, training, \n look_ahead_mask, padding_mask):\n # enc_output.shape == (batch_size, input_seq_len, d_model)\n\n attn1, attn_weights_block1 = self.mha1(x, x, x, look_ahead_mask) # (batch_size, target_seq_len, d_model)\n attn1 = self.dropout1(attn1, training=training)\n out1 = self.layernorm1(attn1 + x)\n \n attn2, attn_weights_block2 = self.mha2(\n enc_output, enc_output, out1, padding_mask) # (batch_size, target_seq_len, d_model)\n attn2 = self.dropout2(attn2, training=training)\n out2 = self.layernorm2(attn2 + out1) # (batch_size, target_seq_len, d_model)\n \n ffn_output = self.ffn(out2) # (batch_size, target_seq_len, d_model)\n ffn_output = self.dropout3(ffn_output, training=training)\n out3 = self.layernorm3(ffn_output + out2) # (batch_size, target_seq_len, d_model)\n \n return out3, attn_weights_block1, attn_weights_block2",
"_____no_output_____"
],
[
"sample_decoder_layer = DecoderLayer(512, 8, 2048)\n\nsample_decoder_layer_output, _, _ = sample_decoder_layer(\n tf.random.uniform((64, 50, 512)), sample_encoder_layer_output, \n False, None, None)\n\nsample_decoder_layer_output.shape # (batch_size, target_seq_len, d_model)",
"_____no_output_____"
]
],
[
[
"### Encoder\n\nThe `Encoder` consists of:\n1. Input Embedding\n2. Positional Encoding\n3. N encoder layers\n\nThe input is put through an embedding which is summed with the positional encoding. The output of this summation is the input to the encoder layers. The output of the encoder is the input to the decoder.",
"_____no_output_____"
]
],
[
[
"class Encoder(tf.keras.layers.Layer):\n def __init__(self, num_layers, d_model, num_heads, dff, input_vocab_size,\n maximum_position_encoding, rate=0.1):\n super(Encoder, self).__init__()\n\n self.d_model = d_model\n self.num_layers = num_layers\n \n self.embedding = tf.keras.layers.Embedding(input_vocab_size, d_model)\n self.pos_encoding = positional_encoding(maximum_position_encoding, \n self.d_model)\n \n \n self.enc_layers = [EncoderLayer(d_model, num_heads, dff, rate) \n for _ in range(num_layers)]\n \n self.dropout = tf.keras.layers.Dropout(rate)\n \n def call(self, x, training, mask):\n\n seq_len = tf.shape(x)[1]\n \n # adding embedding and position encoding.\n x = self.embedding(x) # (batch_size, input_seq_len, d_model)\n x *= tf.math.sqrt(tf.cast(self.d_model, tf.float32))\n x += self.pos_encoding[:, :seq_len, :]\n\n x = self.dropout(x, training=training)\n \n for i in range(self.num_layers):\n x = self.enc_layers[i](x, training, mask)\n \n return x # (batch_size, input_seq_len, d_model)",
"_____no_output_____"
],
[
"sample_encoder = Encoder(num_layers=2, d_model=512, num_heads=8, \n dff=2048, input_vocab_size=8500,\n maximum_position_encoding=10000)\ntemp_input = tf.random.uniform((64, 62), dtype=tf.int64, minval=0, maxval=200)\n\nsample_encoder_output = sample_encoder(temp_input, training=False, mask=None)\n\nprint (sample_encoder_output.shape) # (batch_size, input_seq_len, d_model)",
"_____no_output_____"
]
],
[
[
"### Decoder",
"_____no_output_____"
],
[
" The `Decoder` consists of:\n1. Output Embedding\n2. Positional Encoding\n3. N decoder layers\n\nThe target is put through an embedding which is summed with the positional encoding. The output of this summation is the input to the decoder layers. The output of the decoder is the input to the final linear layer.",
"_____no_output_____"
]
],
[
[
"class Decoder(tf.keras.layers.Layer):\n def __init__(self, num_layers, d_model, num_heads, dff, target_vocab_size,\n maximum_position_encoding, rate=0.1):\n super(Decoder, self).__init__()\n\n self.d_model = d_model\n self.num_layers = num_layers\n \n self.embedding = tf.keras.layers.Embedding(target_vocab_size, d_model)\n self.pos_encoding = positional_encoding(maximum_position_encoding, d_model)\n \n self.dec_layers = [DecoderLayer(d_model, num_heads, dff, rate) \n for _ in range(num_layers)]\n self.dropout = tf.keras.layers.Dropout(rate)\n \n def call(self, x, enc_output, training, \n look_ahead_mask, padding_mask):\n\n seq_len = tf.shape(x)[1]\n attention_weights = {}\n \n x = self.embedding(x) # (batch_size, target_seq_len, d_model)\n x *= tf.math.sqrt(tf.cast(self.d_model, tf.float32))\n x += self.pos_encoding[:, :seq_len, :]\n \n x = self.dropout(x, training=training)\n\n for i in range(self.num_layers):\n x, block1, block2 = self.dec_layers[i](x, enc_output, training,\n look_ahead_mask, padding_mask)\n \n attention_weights['decoder_layer{}_block1'.format(i+1)] = block1\n attention_weights['decoder_layer{}_block2'.format(i+1)] = block2\n \n # x.shape == (batch_size, target_seq_len, d_model)\n return x, attention_weights",
"_____no_output_____"
],
[
"sample_decoder = Decoder(num_layers=2, d_model=512, num_heads=8, \n dff=2048, target_vocab_size=8000,\n maximum_position_encoding=5000)\ntemp_input = tf.random.uniform((64, 26), dtype=tf.int64, minval=0, maxval=200)\n\noutput, attn = sample_decoder(temp_input, \n enc_output=sample_encoder_output, \n training=False,\n look_ahead_mask=None, \n padding_mask=None)\n\noutput.shape, attn['decoder_layer2_block2'].shape",
"_____no_output_____"
]
],
[
[
"## Create the Transformer",
"_____no_output_____"
],
[
"Transformer consists of the encoder, decoder and a final linear layer. The output of the decoder is the input to the linear layer and its output is returned.",
"_____no_output_____"
]
],
[
[
"class Transformer(tf.keras.Model):\n def __init__(self, num_layers, d_model, num_heads, dff, input_vocab_size, \n target_vocab_size, pe_input, pe_target, rate=0.1):\n super(Transformer, self).__init__()\n\n self.encoder = Encoder(num_layers, d_model, num_heads, dff, \n input_vocab_size, pe_input, rate)\n\n self.decoder = Decoder(num_layers, d_model, num_heads, dff, \n target_vocab_size, pe_target, rate)\n\n self.final_layer = tf.keras.layers.Dense(target_vocab_size)\n \n def call(self, inp, tar, training, enc_padding_mask, \n look_ahead_mask, dec_padding_mask):\n\n enc_output = self.encoder(inp, training, enc_padding_mask) # (batch_size, inp_seq_len, d_model)\n \n # dec_output.shape == (batch_size, tar_seq_len, d_model)\n dec_output, attention_weights = self.decoder(\n tar, enc_output, training, look_ahead_mask, dec_padding_mask)\n \n final_output = self.final_layer(dec_output) # (batch_size, tar_seq_len, target_vocab_size)\n \n return final_output, attention_weights",
"_____no_output_____"
],
[
"sample_transformer = Transformer(\n num_layers=2, d_model=512, num_heads=8, dff=2048, \n input_vocab_size=8500, target_vocab_size=8000, \n pe_input=10000, pe_target=6000)\n\ntemp_input = tf.random.uniform((64, 38), dtype=tf.int64, minval=0, maxval=200)\ntemp_target = tf.random.uniform((64, 36), dtype=tf.int64, minval=0, maxval=200)\n\nfn_out, _ = sample_transformer(temp_input, temp_target, training=False, \n enc_padding_mask=None, \n look_ahead_mask=None,\n dec_padding_mask=None)\n\nfn_out.shape # (batch_size, tar_seq_len, target_vocab_size)",
"_____no_output_____"
]
],
[
[
"## Set hyperparameters",
"_____no_output_____"
],
[
"To keep this example small and relatively fast, the values for *num_layers, d_model, and dff* have been reduced. \n\nThe values used in the base model of transformer were; *num_layers=6*, *d_model = 512*, *dff = 2048*. See the [paper](https://arxiv.org/abs/1706.03762) for all the other versions of the transformer.\n\nNote: By changing the values below, you can get the model that achieved state of the art on many tasks.",
"_____no_output_____"
]
],
[
[
"num_layers = 4\nd_model = 128\ndff = 512\nnum_heads = 8\n\ninput_vocab_size = tokenizer_pt.vocab_size + 2\ntarget_vocab_size = tokenizer_en.vocab_size + 2\ndropout_rate = 0.1",
"_____no_output_____"
]
],
[
[
"## Optimizer",
"_____no_output_____"
],
[
"Use the Adam optimizer with a custom learning rate scheduler according to the formula in the [paper](https://arxiv.org/abs/1706.03762).\n\n$$\\Large{lrate = d_{model}^{-0.5} * min(step{\\_}num^{-0.5}, step{\\_}num * warmup{\\_}steps^{-1.5})}$$\n",
"_____no_output_____"
]
],
[
[
"class CustomSchedule(tf.keras.optimizers.schedules.LearningRateSchedule):\n def __init__(self, d_model, warmup_steps=4000):\n super(CustomSchedule, self).__init__()\n \n self.d_model = d_model\n self.d_model = tf.cast(self.d_model, tf.float32)\n\n self.warmup_steps = warmup_steps\n \n def __call__(self, step):\n arg1 = tf.math.rsqrt(step)\n arg2 = step * (self.warmup_steps ** -1.5)\n \n return tf.math.rsqrt(self.d_model) * tf.math.minimum(arg1, arg2)",
"_____no_output_____"
],
[
"learning_rate = CustomSchedule(d_model)\n\noptimizer = tf.keras.optimizers.Adam(learning_rate, beta_1=0.9, beta_2=0.98, \n epsilon=1e-9)",
"_____no_output_____"
],
[
"temp_learning_rate_schedule = CustomSchedule(d_model)\n\nplt.plot(temp_learning_rate_schedule(tf.range(40000, dtype=tf.float32)))\nplt.ylabel(\"Learning Rate\")\nplt.xlabel(\"Train Step\")",
"_____no_output_____"
]
],
[
[
"## Loss and metrics",
"_____no_output_____"
],
[
"Since the target sequences are padded, it is important to apply a padding mask when calculating the loss.",
"_____no_output_____"
]
],
[
[
"loss_object = tf.keras.losses.SparseCategoricalCrossentropy(\n from_logits=True, reduction='none')",
"_____no_output_____"
],
[
"def loss_function(real, pred):\n mask = tf.math.logical_not(tf.math.equal(real, 0))\n loss_ = loss_object(real, pred)\n\n mask = tf.cast(mask, dtype=loss_.dtype)\n loss_ *= mask\n \n return tf.reduce_sum(loss_)/tf.reduce_sum(mask)",
"_____no_output_____"
],
[
"train_loss = tf.keras.metrics.Mean(name='train_loss')\ntrain_accuracy = tf.keras.metrics.SparseCategoricalAccuracy(\n name='train_accuracy')",
"_____no_output_____"
]
],
[
[
"## Training and checkpointing",
"_____no_output_____"
]
],
[
[
"transformer = Transformer(num_layers, d_model, num_heads, dff,\n input_vocab_size, target_vocab_size, \n pe_input=input_vocab_size, \n pe_target=target_vocab_size,\n rate=dropout_rate)",
"_____no_output_____"
],
[
"def create_masks(inp, tar):\n # Encoder padding mask\n enc_padding_mask = create_padding_mask(inp)\n \n # Used in the 2nd attention block in the decoder.\n # This padding mask is used to mask the encoder outputs.\n dec_padding_mask = create_padding_mask(inp)\n \n # Used in the 1st attention block in the decoder.\n # It is used to pad and mask future tokens in the input received by \n # the decoder.\n look_ahead_mask = create_look_ahead_mask(tf.shape(tar)[1])\n dec_target_padding_mask = create_padding_mask(tar)\n combined_mask = tf.maximum(dec_target_padding_mask, look_ahead_mask)\n \n return enc_padding_mask, combined_mask, dec_padding_mask",
"_____no_output_____"
]
],
[
[
"Create the checkpoint path and the checkpoint manager. This will be used to save checkpoints every `n` epochs.",
"_____no_output_____"
]
],
[
[
"checkpoint_path = \"./checkpoints/train\"\n\nckpt = tf.train.Checkpoint(transformer=transformer,\n optimizer=optimizer)\n\nckpt_manager = tf.train.CheckpointManager(ckpt, checkpoint_path, max_to_keep=5)\n\n# if a checkpoint exists, restore the latest checkpoint.\nif ckpt_manager.latest_checkpoint:\n ckpt.restore(ckpt_manager.latest_checkpoint)\n print ('Latest checkpoint restored!!')",
"_____no_output_____"
]
],
[
[
"The target is divided into tar_inp and tar_real. tar_inp is passed as an input to the decoder. `tar_real` is that same input shifted by 1: At each location in `tar_input`, `tar_real` contains the next token that should be predicted.\n\nFor example, `sentence` = \"SOS A lion in the jungle is sleeping EOS\"\n\n`tar_inp` = \"SOS A lion in the jungle is sleeping\"\n\n`tar_real` = \"A lion in the jungle is sleeping EOS\"\n\nThe transformer is an auto-regressive model: it makes predictions one part at a time, and uses its output so far to decide what to do next. \n\nDuring training this example uses teacher-forcing (like in the [text generation tutorial](./text_generation.ipynb)). Teacher forcing is passing the true output to the next time step regardless of what the model predicts at the current time step.\n\nAs the transformer predicts each word, *self-attention* allows it to look at the previous words in the input sequence to better predict the next word.\n\nTo prevent the model from peeking at the expected output the model uses a look-ahead mask.",
"_____no_output_____"
]
],
[
[
"EPOCHS = 20",
"_____no_output_____"
],
[
"# The @tf.function trace-compiles train_step into a TF graph for faster\n# execution. The function specializes to the precise shape of the argument\n# tensors. To avoid re-tracing due to the variable sequence lengths or variable\n# batch sizes (the last batch is smaller), use input_signature to specify\n# more generic shapes.\n\ntrain_step_signature = [\n tf.TensorSpec(shape=(None, None), dtype=tf.int64),\n tf.TensorSpec(shape=(None, None), dtype=tf.int64),\n]\n\[email protected](input_signature=train_step_signature)\ndef train_step(inp, tar):\n tar_inp = tar[:, :-1]\n tar_real = tar[:, 1:]\n \n enc_padding_mask, combined_mask, dec_padding_mask = create_masks(inp, tar_inp)\n \n with tf.GradientTape() as tape:\n predictions, _ = transformer(inp, tar_inp, \n True, \n enc_padding_mask, \n combined_mask, \n dec_padding_mask)\n loss = loss_function(tar_real, predictions)\n\n gradients = tape.gradient(loss, transformer.trainable_variables) \n optimizer.apply_gradients(zip(gradients, transformer.trainable_variables))\n \n train_loss(loss)\n train_accuracy(tar_real, predictions)",
"_____no_output_____"
]
],
[
[
"Portuguese is used as the input language and English is the target language.",
"_____no_output_____"
]
],
[
[
"for epoch in range(EPOCHS):\n start = time.time()\n \n train_loss.reset_states()\n train_accuracy.reset_states()\n \n # inp -> portuguese, tar -> english\n for (batch, (inp, tar)) in enumerate(train_dataset):\n train_step(inp, tar)\n \n if batch % 50 == 0:\n print ('Epoch {} Batch {} Loss {:.4f} Accuracy {:.4f}'.format(\n epoch + 1, batch, train_loss.result(), train_accuracy.result()))\n \n if (epoch + 1) % 5 == 0:\n ckpt_save_path = ckpt_manager.save()\n print ('Saving checkpoint for epoch {} at {}'.format(epoch+1,\n ckpt_save_path))\n \n print ('Epoch {} Loss {:.4f} Accuracy {:.4f}'.format(epoch + 1, \n train_loss.result(), \n train_accuracy.result()))\n\n print ('Time taken for 1 epoch: {} secs\\n'.format(time.time() - start))",
"_____no_output_____"
]
],
[
[
"## Evaluate",
"_____no_output_____"
],
[
"The following steps are used for evaluation:\n\n* Encode the input sentence using the Portuguese tokenizer (`tokenizer_pt`). Moreover, add the start and end token so the input is equivalent to what the model is trained with. This is the encoder input.\n* The decoder input is the `start token == tokenizer_en.vocab_size`.\n* Calculate the padding masks and the look ahead masks.\n* The `decoder` then outputs the predictions by looking at the `encoder output` and its own output (self-attention).\n* Select the last word and calculate the argmax of that.\n* Concatentate the predicted word to the decoder input as pass it to the decoder.\n* In this approach, the decoder predicts the next word based on the previous words it predicted.\n\nNote: The model used here has less capacity to keep the example relatively faster so the predictions maybe less right. To reproduce the results in the paper, use the entire dataset and base transformer model or transformer XL, by changing the hyperparameters above.",
"_____no_output_____"
]
],
[
[
"def evaluate(inp_sentence):\n start_token = [tokenizer_pt.vocab_size]\n end_token = [tokenizer_pt.vocab_size + 1]\n \n # inp sentence is portuguese, hence adding the start and end token\n inp_sentence = start_token + tokenizer_pt.encode(inp_sentence) + end_token\n encoder_input = tf.expand_dims(inp_sentence, 0)\n \n # as the target is english, the first word to the transformer should be the\n # english start token.\n decoder_input = [tokenizer_en.vocab_size]\n output = tf.expand_dims(decoder_input, 0)\n \n for i in range(MAX_LENGTH):\n enc_padding_mask, combined_mask, dec_padding_mask = create_masks(\n encoder_input, output)\n \n # predictions.shape == (batch_size, seq_len, vocab_size)\n predictions, attention_weights = transformer(encoder_input, \n output,\n False,\n enc_padding_mask,\n combined_mask,\n dec_padding_mask)\n \n # select the last word from the seq_len dimension\n predictions = predictions[: ,-1:, :] # (batch_size, 1, vocab_size)\n\n predicted_id = tf.cast(tf.argmax(predictions, axis=-1), tf.int32)\n \n # return the result if the predicted_id is equal to the end token\n if predicted_id == tokenizer_en.vocab_size+1:\n return tf.squeeze(output, axis=0), attention_weights\n \n # concatentate the predicted_id to the output which is given to the decoder\n # as its input.\n output = tf.concat([output, predicted_id], axis=-1)\n\n return tf.squeeze(output, axis=0), attention_weights",
"_____no_output_____"
],
[
"def plot_attention_weights(attention, sentence, result, layer):\n fig = plt.figure(figsize=(16, 8))\n \n sentence = tokenizer_pt.encode(sentence)\n \n attention = tf.squeeze(attention[layer], axis=0)\n \n for head in range(attention.shape[0]):\n ax = fig.add_subplot(2, 4, head+1)\n \n # plot the attention weights\n ax.matshow(attention[head][:-1, :], cmap='viridis')\n\n fontdict = {'fontsize': 10}\n \n ax.set_xticks(range(len(sentence)+2))\n ax.set_yticks(range(len(result)))\n \n ax.set_ylim(len(result)-1.5, -0.5)\n \n ax.set_xticklabels(\n ['<start>']+[tokenizer_pt.decode([i]) for i in sentence]+['<end>'], \n fontdict=fontdict, rotation=90)\n \n ax.set_yticklabels([tokenizer_en.decode([i]) for i in result \n if i < tokenizer_en.vocab_size], \n fontdict=fontdict)\n \n ax.set_xlabel('Head {}'.format(head+1))\n \n plt.tight_layout()\n plt.show()",
"_____no_output_____"
],
[
"def translate(sentence, plot=''):\n result, attention_weights = evaluate(sentence)\n \n predicted_sentence = tokenizer_en.decode([i for i in result \n if i < tokenizer_en.vocab_size]) \n\n print('Input: {}'.format(sentence))\n print('Predicted translation: {}'.format(predicted_sentence))\n \n if plot:\n plot_attention_weights(attention_weights, sentence, result, plot)",
"_____no_output_____"
],
[
"translate(\"este é um problema que temos que resolver.\")\nprint (\"Real translation: this is a problem we have to solve .\")",
"_____no_output_____"
],
[
"translate(\"os meus vizinhos ouviram sobre esta ideia.\")\nprint (\"Real translation: and my neighboring homes heard about this idea .\")",
"_____no_output_____"
],
[
"translate(\"vou então muito rapidamente partilhar convosco algumas histórias de algumas coisas mágicas que aconteceram.\")\nprint (\"Real translation: so i 'll just share with you some stories very quickly of some magical things that have happened .\")",
"_____no_output_____"
]
],
[
[
"You can pass different layers and attention blocks of the decoder to the `plot` parameter.",
"_____no_output_____"
]
],
[
[
"translate(\"este é o primeiro livro que eu fiz.\", plot='decoder_layer4_block2')\nprint (\"Real translation: this is the first book i've ever done.\")",
"_____no_output_____"
]
],
[
[
"## Summary\n\nIn this tutorial, you learned about positional encoding, multi-head attention, the importance of masking and how to create a transformer.\n\nTry using a different dataset to train the transformer. You can also create the base transformer or transformer XL by changing the hyperparameters above. You can also use the layers defined here to create [BERT](https://arxiv.org/abs/1810.04805) and train state of the art models. Futhermore, you can implement beam search to get better predictions.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb44b75614763e129223762ce47c9c1a7809a051 | 10,828 | ipynb | Jupyter Notebook | research/bayesian_deeplab/deeplab_demo.ipynb | omegafragger/models | 6518e3e78d898398aa7c19c8cfe7133a859e60e6 | [
"Apache-2.0"
] | null | null | null | research/bayesian_deeplab/deeplab_demo.ipynb | omegafragger/models | 6518e3e78d898398aa7c19c8cfe7133a859e60e6 | [
"Apache-2.0"
] | null | null | null | research/bayesian_deeplab/deeplab_demo.ipynb | omegafragger/models | 6518e3e78d898398aa7c19c8cfe7133a859e60e6 | [
"Apache-2.0"
] | null | null | null | 31.114943 | 185 | 0.555504 | [
[
[
"# DeepLab Demo\n\nThis demo will demostrate the steps to run deeplab semantic segmentation model on sample input images.",
"_____no_output_____"
]
],
[
[
"#@title Imports\n\nimport os\nfrom io import BytesIO\nimport tarfile\nimport tempfile\nfrom six.moves import urllib\n\nfrom matplotlib import gridspec\nfrom matplotlib import pyplot as plt\nimport numpy as np\nfrom PIL import Image\n\nimport tensorflow as tf",
"_____no_output_____"
],
[
"#@title Helper methods\n\n\nclass DeepLabModel(object):\n \"\"\"Class to load deeplab model and run inference.\"\"\"\n\n INPUT_TENSOR_NAME = 'ImageTensor:0'\n OUTPUT_TENSOR_NAME = 'SemanticPredictions:0'\n INPUT_SIZE = 513\n FROZEN_GRAPH_NAME = 'frozen_inference_graph'\n\n def __init__(self, tarball_path):\n \"\"\"Creates and loads pretrained deeplab model.\"\"\"\n self.graph = tf.Graph()\n\n graph_def = None\n # Extract frozen graph from tar archive.\n tar_file = tarfile.open(tarball_path)\n for tar_info in tar_file.getmembers():\n if self.FROZEN_GRAPH_NAME in os.path.basename(tar_info.name):\n file_handle = tar_file.extractfile(tar_info)\n graph_def = tf.GraphDef.FromString(file_handle.read())\n break\n\n tar_file.close()\n\n if graph_def is None:\n raise RuntimeError('Cannot find inference graph in tar archive.')\n\n with self.graph.as_default():\n tf.import_graph_def(graph_def, name='')\n\n self.sess = tf.Session(graph=self.graph)\n\n def run(self, image):\n \"\"\"Runs inference on a single image.\n\n Args:\n image: A PIL.Image object, raw input image.\n\n Returns:\n resized_image: RGB image resized from original input image.\n seg_map: Segmentation map of `resized_image`.\n \"\"\"\n width, height = image.size\n resize_ratio = 1.0 * self.INPUT_SIZE / max(width, height)\n target_size = (int(resize_ratio * width), int(resize_ratio * height))\n resized_image = image.convert('RGB').resize(target_size, Image.ANTIALIAS)\n batch_seg_map = self.sess.run(\n self.OUTPUT_TENSOR_NAME,\n feed_dict={self.INPUT_TENSOR_NAME: [np.asarray(resized_image)]})\n seg_map = batch_seg_map[0]\n return resized_image, seg_map\n\n\ndef create_pascal_label_colormap():\n \"\"\"Creates a label colormap used in PASCAL VOC segmentation benchmark.\n\n Returns:\n A Colormap for visualizing segmentation results.\n \"\"\"\n colormap = np.zeros((256, 3), dtype=int)\n ind = np.arange(256, dtype=int)\n\n for shift in reversed(range(8)):\n for channel in range(3):\n colormap[:, channel] |= ((ind >> channel) & 1) << shift\n ind >>= 3\n\n return colormap\n\n\ndef label_to_color_image(label):\n \"\"\"Adds color defined by the dataset colormap to the label.\n\n Args:\n label: A 2D array with integer type, storing the segmentation label.\n\n Returns:\n result: A 2D array with floating type. The element of the array\n is the color indexed by the corresponding element in the input label\n to the PASCAL color map.\n\n Raises:\n ValueError: If label is not of rank 2 or its value is larger than color\n map maximum entry.\n \"\"\"\n if label.ndim != 2:\n raise ValueError('Expect 2-D input label')\n\n colormap = create_pascal_label_colormap()\n\n if np.max(label) >= len(colormap):\n raise ValueError('label value too large.')\n\n return colormap[label]\n\n\ndef vis_segmentation(image, seg_map):\n \"\"\"Visualizes input image, segmentation map and overlay view.\"\"\"\n plt.figure(figsize=(15, 5))\n grid_spec = gridspec.GridSpec(1, 4, width_ratios=[6, 6, 6, 1])\n\n plt.subplot(grid_spec[0])\n plt.imshow(image)\n plt.axis('off')\n plt.title('input image')\n\n plt.subplot(grid_spec[1])\n seg_image = label_to_color_image(seg_map).astype(np.uint8)\n plt.imshow(seg_image)\n plt.axis('off')\n plt.title('segmentation map')\n\n plt.subplot(grid_spec[2])\n plt.imshow(image)\n plt.imshow(seg_image, alpha=0.7)\n plt.axis('off')\n plt.title('segmentation overlay')\n\n unique_labels = np.unique(seg_map)\n ax = plt.subplot(grid_spec[3])\n plt.imshow(\n FULL_COLOR_MAP[unique_labels].astype(np.uint8), interpolation='nearest')\n ax.yaxis.tick_right()\n plt.yticks(range(len(unique_labels)), LABEL_NAMES[unique_labels])\n plt.xticks([], [])\n ax.tick_params(width=0.0)\n plt.grid('off')\n plt.show()\n\n\nLABEL_NAMES = np.asarray([\n 'background', 'aeroplane', 'bicycle', 'bird', 'boat', 'bottle', 'bus',\n 'car', 'cat', 'chair', 'cow', 'diningtable', 'dog', 'horse', 'motorbike',\n 'person', 'pottedplant', 'sheep', 'sofa', 'train', 'tv'\n])\n\nFULL_LABEL_MAP = np.arange(len(LABEL_NAMES)).reshape(len(LABEL_NAMES), 1)\nFULL_COLOR_MAP = label_to_color_image(FULL_LABEL_MAP)",
"_____no_output_____"
],
[
"#@title Select and download models {display-mode: \"form\"}\n\nMODEL_NAME = 'mobilenetv2_coco_voctrainaug' # @param ['mobilenetv2_coco_voctrainaug', 'mobilenetv2_coco_voctrainval', 'xception_coco_voctrainaug', 'xception_coco_voctrainval']\n\n_DOWNLOAD_URL_PREFIX = 'http://download.tensorflow.org/models/'\n_MODEL_URLS = {\n 'mobilenetv2_coco_voctrainaug':\n 'deeplabv3_mnv2_pascal_train_aug_2018_01_29.tar.gz',\n 'mobilenetv2_coco_voctrainval':\n 'deeplabv3_mnv2_pascal_trainval_2018_01_29.tar.gz',\n 'xception_coco_voctrainaug':\n 'deeplabv3_pascal_train_aug_2018_01_04.tar.gz',\n 'xception_coco_voctrainval':\n 'deeplabv3_pascal_trainval_2018_01_04.tar.gz',\n}\n_TARBALL_NAME = 'deeplab_model.tar.gz'\n\nmodel_dir = tempfile.mkdtemp()\ntf.gfile.MakeDirs(model_dir)\n\ndownload_path = os.path.join(model_dir, _TARBALL_NAME)\nprint('downloading model, this might take a while...')\nurllib.request.urlretrieve(_DOWNLOAD_URL_PREFIX + _MODEL_URLS[MODEL_NAME],\n download_path)\nprint('download completed! loading DeepLab model...')\n\nMODEL = DeepLabModel(download_path)\nprint('model loaded successfully!')",
"_____no_output_____"
]
],
[
[
"## Run on sample images\n\nSelect one of sample images (leave `IMAGE_URL` empty) or feed any internet image\nurl for inference.\n\nNote that we are using single scale inference in the demo for fast computation,\nso the results may slightly differ from the visualizations in\n[README](https://github.com/tensorflow/models/blob/master/research/deeplab/README.md),\nwhich uses multi-scale and left-right flipped inputs.",
"_____no_output_____"
]
],
[
[
"#@title Run on sample images {display-mode: \"form\"}\n\nSAMPLE_IMAGE = 'image1' # @param ['image1', 'image2', 'image3']\nIMAGE_URL = '' #@param {type:\"string\"}\n\n_SAMPLE_URL = ('https://github.com/tensorflow/models/blob/master/research/'\n 'deeplab/g3doc/img/%s.jpg?raw=true')\n\n\ndef run_visualization(url):\n \"\"\"Inferences DeepLab model and visualizes result.\"\"\"\n try:\n f = urllib.request.urlopen(url)\n jpeg_str = f.read()\n orignal_im = Image.open(BytesIO(jpeg_str))\n except IOError:\n print('Cannot retrieve image. Please check url: ' + url)\n return\n\n print('running deeplab on image %s...' % url)\n resized_im, seg_map = MODEL.run(orignal_im)\n\n vis_segmentation(resized_im, seg_map)\n\n\nimage_url = IMAGE_URL or _SAMPLE_URL % SAMPLE_IMAGE\nrun_visualization(image_url)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cb44c02777a61f074fce3ad026c98fb4aa86bd96 | 229,525 | ipynb | Jupyter Notebook | 0_Dataset.ipynb | calincan2000/CVND-Image-Captioning-PyTorch- | d277ef8be766552f08112c806620eec457c5175c | [
"MIT"
] | 1 | 2021-12-02T08:22:40.000Z | 2021-12-02T08:22:40.000Z | 0_Dataset.ipynb | calincan2000/CVND-Image-Captioning-PyTorch- | d277ef8be766552f08112c806620eec457c5175c | [
"MIT"
] | null | null | null | 0_Dataset.ipynb | calincan2000/CVND-Image-Captioning-PyTorch- | d277ef8be766552f08112c806620eec457c5175c | [
"MIT"
] | null | null | null | 1,326.734104 | 224,456 | 0.956088 | [
[
[
"# Computer Vision Nanodegree\n\n## Project: Image Captioning\n\n---\n\nThe Microsoft **C**ommon **O**bjects in **CO**ntext (MS COCO) dataset is a large-scale dataset for scene understanding. The dataset is commonly used to train and benchmark object detection, segmentation, and captioning algorithms. \n\n\n\nYou can read more about the dataset on the [website](http://cocodataset.org/#home) or in the [research paper](https://arxiv.org/pdf/1405.0312.pdf).\n\nIn this notebook, you will explore this dataset, in preparation for the project.\n\n## Step 1: Initialize the COCO API\n\nWe begin by initializing the [COCO API](https://github.com/cocodataset/cocoapi) that you will use to obtain the data.",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\nsys.path.append('/opt/cocoapi/PythonAPI')\nfrom pycocotools.coco import COCO\n\n# initialize COCO API for instance annotations\ndataDir = '/opt/cocoapi'\ndataType = 'val2014'\ninstances_annFile = os.path.join(dataDir, 'annotations/instances_{}.json'.format(dataType))\ncoco = COCO(instances_annFile)\n\n# initialize COCO API for caption annotations\ncaptions_annFile = os.path.join(dataDir, 'annotations/captions_{}.json'.format(dataType))\ncoco_caps = COCO(captions_annFile)\n\n# get image ids \nids = list(coco.anns.keys())",
"loading annotations into memory...\nDone (t=6.53s)\ncreating index...\nindex created!\nloading annotations into memory...\nDone (t=0.97s)\ncreating index...\nindex created!\n"
]
],
[
[
"## Step 2: Plot a Sample Image\n\nNext, we plot a random image from the dataset, along with its five corresponding captions. Each time you run the code cell below, a different image is selected. \n\nIn the project, you will use this dataset to train your own model to generate captions from images!",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport skimage.io as io\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\n# pick a random image and obtain the corresponding URL\nann_id = np.random.choice(ids)\nimg_id = coco.anns[ann_id]['image_id']\nimg = coco.loadImgs(img_id)[0]\nurl = img['coco_url']\n\n# print URL and visualize corresponding image\nprint(url)\nI = io.imread(url)\nplt.axis('off')\nplt.imshow(I)\nplt.show()\n\n# load and display captions\nannIds = coco_caps.getAnnIds(imgIds=img['id']);\nanns = coco_caps.loadAnns(annIds)\ncoco_caps.showAnns(anns)",
"http://images.cocodataset.org/val2014/COCO_val2014_000000429913.jpg\n"
]
],
[
[
"## Step 3: What's to Come!\n\nIn this project, you will use the dataset of image-caption pairs to train a CNN-RNN model to automatically generate images from captions. You'll learn more about how to design the architecture in the next notebook in the sequence (**1_Preliminaries.ipynb**).\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb44c5bf9a285f2bee85643cb7caf227c38b8c0a | 31,404 | ipynb | Jupyter Notebook | train.ipynb | sinamehrabi/Montreal_tv_channels | 3624a4e8b25a0878692b3ca06284f9d0d4712183 | [
"MIT"
] | null | null | null | train.ipynb | sinamehrabi/Montreal_tv_channels | 3624a4e8b25a0878692b3ca06284f9d0d4712183 | [
"MIT"
] | null | null | null | train.ipynb | sinamehrabi/Montreal_tv_channels | 3624a4e8b25a0878692b3ca06284f9d0d4712183 | [
"MIT"
] | null | null | null | 34.134783 | 246 | 0.419055 | [
[
[
"## Import libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nfrom sklearn import preprocessing\nfrom sklearn.preprocessing import MinMaxScaler, StandardScaler\nfrom sklearn.model_selection import ShuffleSplit\nfrom sklearn.metrics import mean_absolute_error, r2_score\nfrom sklearn.ensemble import RandomForestRegressor\nimport scipy.stats",
"_____no_output_____"
]
],
[
[
"## Read data",
"_____no_output_____"
]
],
[
[
"train_df = pd.read_csv('data.csv') #read original train data",
"_____no_output_____"
]
],
[
[
"## Functions\n",
"_____no_output_____"
]
],
[
[
"def drop_columns(df, column_names):\n \"\"\"\n \n df: input dataframe\n column_names: list of column's name\n return: dataframe with dropped columns\n \n \"\"\"\n new_df = df.copy(deep=True)\n new_df.drop(column_names, axis=1, inplace=True)\n return new_df\n\n",
"_____no_output_____"
]
],
[
[
"# Preprocessing",
"_____no_output_____"
],
[
"### 1- First discover our data generally",
"_____no_output_____"
]
],
[
[
"train_df.head(-5)",
"_____no_output_____"
]
],
[
[
"#### we have 20 features and 616656 samples",
"_____no_output_____"
]
],
[
[
"train_df.shape",
"_____no_output_____"
]
],
[
[
"### Check number of unique values of features",
"_____no_output_____"
]
],
[
[
"for col in train_df:\n print(\"column:{}\".format(str(col)) + \" ---------> \" + str(len(train_df[col].unique())))",
"column:Unnamed: 0 ---------> 616656\ncolumn:Episode ---------> 6687\ncolumn:Station ---------> 24\ncolumn:Channel Type ---------> 2\ncolumn:Season ---------> 4\ncolumn:Year ---------> 4\ncolumn:Date ---------> 877\ncolumn:Day of week ---------> 7\ncolumn:Start_time ---------> 138322\ncolumn:End_time ---------> 138334\ncolumn:Length ---------> 39\ncolumn:Name of show ---------> 6687\ncolumn:Name of episode ---------> 86557\ncolumn:Genre ---------> 27\ncolumn:First time or rerun ---------> 2\ncolumn:# of episode in the season ---------> 2\ncolumn:Movie? ---------> 2\ncolumn:Game of the Canadiens during episode? ---------> 2\ncolumn:Market Share_total ---------> 545\ncolumn:Temperature in Montreal during episode ---------> 11829\n"
]
],
[
[
"## features with NaN samples",
"_____no_output_____"
]
],
[
[
"train_df.columns[train_df.isna().any()].tolist()",
"_____no_output_____"
]
],
[
[
"## 2- pirmary feature selection",
"_____no_output_____"
],
[
"### Episode and Name of show are same features! then choose one of them",
"_____no_output_____"
]
],
[
[
"# check equality of two columns\nassert sum(train_df['Episode'] == train_df['Name of show'][:]) == train_df.shape[0] , \"Columns are not same\"",
"_____no_output_____"
]
],
[
[
"### difference of end time and start time are high correlated with Length (using Pearson Correlation Coefficient about 99.5%), also we have NaN values in start time and end time, then we can remove start time and end time features from data",
"_____no_output_____"
]
],
[
[
"df = train_df.copy()\ndf = df[df['Start_time'].notna()]\ndf['Start_time'] = pd.to_datetime(df['Start_time'])\ndf['End_time'] = pd.to_datetime(df['End_time'])\ndf['Time_diff'] = (df['End_time'] - df['Start_time'])\ndf['Time_diff'] = df['Time_diff'].dt.seconds /3600\n\nprint(scipy.stats.pearsonr(df[\"Time_diff\"],df[\"Length\"] ))\ndel df",
"(0.9949798563110538, 0.0)\n"
]
],
[
[
"### Convert Date feature to Month and Day",
"_____no_output_____"
]
],
[
[
"train_df['Date'] = pd.to_datetime(train_df['Date'])\ntrain_df['Month'] = train_df.Date.dt.month\ntrain_df['Day'] = train_df.Date.dt.day",
"_____no_output_____"
]
],
[
[
"### 36% of a \"Name of episode\" feature is NaN and we can not use interpolate or some other approach...we can drop it!",
"_____no_output_____"
]
],
[
[
"100 * train_df[(train_df['Name of episode'].isnull())].shape[0]/len(train_df) ",
"_____no_output_____"
]
],
[
[
"### Conclution: These features should be dropped",
"_____no_output_____"
]
],
[
[
"column_names = ['Unnamed: 0','Date', 'Start_time', 'End_time','Name of show', 'Name of episode']",
"_____no_output_____"
]
],
[
[
"### We have NaN values on \"Temperature in Montreal during episode\" features..I choose linear Interpolate to fill NaN values",
"_____no_output_____"
]
],
[
[
"train_df['Temperature in Montreal during episode'].interpolate(inplace=True)",
"_____no_output_____"
]
],
[
[
"## Label Encoding with simple label encoder",
"_____no_output_____"
]
],
[
[
"temp_train_df = drop_columns(train_df, column_names)\n# temp_test_df = drop_columns(test_df, column_names)\n\n\ntrain_target_df = temp_train_df['Market Share_total']\ntrain_df = temp_train_df.copy(deep=True)\ntrain_df.drop(['Market Share_total'], axis=1, inplace=True)\n\n# test_target_df = new_test_df['Market Share_total']\n# test_df = new_test_df.copy(deep=True)\n# test_df.drop(['Market Share_total'], axis=1, inplace=True)\n\nle = preprocessing.LabelEncoder()\n\nfor item in train_df.loc[:, ~train_df.columns.isin(['Temperature in Montreal during episode','Year', 'Length', 'Month', 'Day'])]:\n \n train_df[item] = le.fit_transform(train_df[item]) + 1",
"_____no_output_____"
]
],
[
[
"### Normalize our data",
"_____no_output_____"
]
],
[
[
"scaler = StandardScaler()\nNormalized_train_arr = scaler.fit_transform(train_df)\nNormalized_train_target_arr = scaler.fit_transform(train_target_df.values.reshape(-1,1))",
"_____no_output_____"
]
],
[
[
"## Use shuffle split train and test data: 70% for train and 30% for validation data\n## Choose RandomForest Regressor model with 12 estimator for our data\n## Metrics are R square and MAE",
"_____no_output_____"
]
],
[
[
"ss = ShuffleSplit(n_splits=5, test_size=0.3, random_state=0)\npreds = []\nreals = []\nr2_scores_list = []\nmae_list = []\npcc = []\nspc = []\n\nfor train_index, val_index in ss.split(Normalized_train_arr):\n \n train_X = Normalized_train_arr[train_index]\n train_Y = Normalized_train_target_arr[train_index]\n \n validation_X = Normalized_train_arr[val_index]\n validation_y = Normalized_train_target_arr[val_index]\n \n regr = RandomForestRegressor(n_estimators=12, random_state=0, n_jobs=-1)\n regr.fit(train_X, train_Y)\n pred_y = regr.predict(validation_X)\n # Model Metrics Calculation\n r2_scores_list.append(regr.score(validation_X, validation_y))\n mae_list.append(mean_absolute_error(scaler.inverse_transform(validation_y), scaler.inverse_transform(pred_y.reshape(-1,1))))\n # Pearson Correlation Coefficient and Spearman Correlation Coefficient Calculations\n pcc.append(scipy.stats.pearsonr(pred_y, validation_y.ravel())[0])\n spc.append(scipy.stats.spearmanr(pred_y, validation_y.ravel())[0])\n ",
"/home/cna/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:18: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples,), for example using ravel().\n/home/cna/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:18: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples,), for example using ravel().\n/home/cna/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:18: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples,), for example using ravel().\n/home/cna/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:18: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples,), for example using ravel().\n/home/cna/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:18: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples,), for example using ravel().\n"
]
],
[
[
"### Calculate Mean value of model metrics and statistical metrics",
"_____no_output_____"
]
],
[
[
"print(\"R Square mean value: \", str(np.mean(r2_scores_list)))\nprint(\"MAE mean value: \", str(np.mean(mae_list)))\nprint(\"Pearson Correlation Coefficient: \", str(np.mean(pcc)))\nprint(\"Spearman Correlation Coefficient: \", str(np.mean(spc)))",
"R Square mean value: 0.8566461292615359\nMAE mean value: 1.10838557054771\nPearson Correlation Coefficient: 0.9258654084141197\nSpearman Correlation Coefficient: 0.8056042127167841\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb44d0043907d1729cb25d89bfd93ce874573f83 | 16,198 | ipynb | Jupyter Notebook | lqr/python_src/simple.ipynb | YoshimitsuMatsutaIe/abc_2022 | 9c6fb487c7ec22fdc57cc1eb0abec4c9786ad995 | [
"MIT"
] | null | null | null | lqr/python_src/simple.ipynb | YoshimitsuMatsutaIe/abc_2022 | 9c6fb487c7ec22fdc57cc1eb0abec4c9786ad995 | [
"MIT"
] | null | null | null | lqr/python_src/simple.ipynb | YoshimitsuMatsutaIe/abc_2022 | 9c6fb487c7ec22fdc57cc1eb0abec4c9786ad995 | [
"MIT"
] | null | null | null | 37.757576 | 7,498 | 0.671935 | [
[
[
"# 自分で実装",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport scipy as sp\nfrom scipy import linalg as LA",
"_____no_output_____"
]
],
[
[
"リカッチ代数方程式\n$$\nPA+A^{\\top}P-PBR^{-1}B^{\\top}P+Q=0\n$$\nの正定解$P$を求める. ",
"_____no_output_____"
]
],
[
[
"A = np.array([\n [1.1, 2],\n [-0.3, -1],\n])\nB = np.array([\n [1, 2],\n [0.847, 3],\n])\nQ = np.diag([10.0, 10.0])\nR = np.diag([1., 1.])\n\nP_solver = LA.solve_continuous_are(A, B, Q, R)\nP_solver # ソルバを使って求めた解",
"_____no_output_____"
]
],
[
[
"# リカッチの非線形行列微分方程式を解いて求める\n[教科書](#1)のp.160にある方法.力技.こちらは簡単. ",
"_____no_output_____"
]
],
[
[
"def solve_care(A, B, Q, R):\n P = np.zeros_like(A)\n P_ = P.copy()\n \n invR = np.linalg.inv(R)\n \n while True:\n P += (P @ A + A.T @ P - P @ B @ invR @ B.T @ P + Q) * 0.001\n if np.abs(P - P_).max() < 1e-12:\n break\n P_ = P.copy()\n \n return P\n\n\nP = solve_care(A, B, Q, R)\nP",
"_____no_output_____"
]
],
[
[
"scipyの結果と比較",
"_____no_output_____"
]
],
[
[
"P - P_solver",
"_____no_output_____"
]
],
[
[
"ほぼ同じ.",
"_____no_output_____"
],
[
"****\n## 有本・ポッターの方法\n[教科書](#1)のp.160にある方法. \nまずはハミルトン行列$H$をつくる. \n$$\nH = \\begin{bmatrix}\nA & -BR^{-1}B^{\\top}\\\\\n-Q & -A^{\\top}\n\\end{bmatrix}\n$$",
"_____no_output_____"
]
],
[
[
"H = np.block([\n [A, -B @ np.linalg.inv(R) @ B.T],\n [-Q, -A.T],\n])\nH",
"_____no_output_____"
]
],
[
[
"ハミルトン行列の固有値,固有ベクトルを求める. ",
"_____no_output_____"
]
],
[
[
"L, W = np.linalg.eig(H) # 固有値と固有ベクトル\n\nplt.scatter(np.real(L), np.imag(L), label=\"eigvals\")\nplt.legend(); plt.grid()\nplt.xlabel(\"Re\"); plt.ylabel(\"Im\")",
"_____no_output_____"
]
],
[
[
"最適極は左半面にあるもの全てである.\n今回は-11.86...と-2.73...の2つが最適極となる. \n$i$番目の最適極の固有ベクトル$\\omega_i$を2つに分割する. \n$$\n\\omega_i = \\begin{bmatrix}\nu_i\\\\\nv_i\n\\end{bmatrix}\n$$",
"_____no_output_____"
]
],
[
[
"L",
"_____no_output_____"
],
[
"W",
"_____no_output_____"
],
[
"# -11.16...について\nw1 = W[:, 0:1]\nu1 = w1[:2, :]\nv1 = w1[2:, :]\n\n# -2.13...について\nw2 = W[:, 1:2]\nu2 = w2[:2, :]\nv2 = w2[2:, :]",
"_____no_output_____"
]
],
[
[
"リカッチ解は次式となる. \n$$\nP = [v_1, v_2, ...,v_n][u_1, u_2, ...,u_n]^{-1}\n$$",
"_____no_output_____"
]
],
[
[
"U = np.block([u1, u2])\nV = np.block([v1, v2])\nP_ap = V @ np.linalg.inv(U)\nP_ap",
"_____no_output_____"
]
],
[
[
"ソルバー解と比較",
"_____no_output_____"
]
],
[
[
"P_ap - P_solver",
"_____no_output_____"
]
],
[
[
"以上を1つの関数にまとめたもの⇓",
"_____no_output_____"
]
],
[
[
"def arimoto_potter(A, B, Q, R):\n \"\"\"有本・ポッターの方法\"\"\"\n n = len(A)\n H = np.block([\n [A, -B @ np.linalg.inv(R) @ B.T],\n [-Q, -A.T],\n ]) # ハミルトン行列\n \n L, W = np.linalg.eig(H) # 固有値と固有ベクトル\n U, V = [], []\n for i, l in enumerate(L):\n if np.real(l) < 0: # 固有値が負の最適極の場合\n U.append(W[:n, i:i+1])\n V.append(W[n:, i:i+1])\n U = np.concatenate(U, axis=1)\n V = np.concatenate(V, axis=1)\n \n return V @ np.linalg.inv(U)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb44ddf9617742107d8d4964fdb834778530fd65 | 893,022 | ipynb | Jupyter Notebook | 11 Pandas Teil 3/Viele Dateien.ipynb | edzardschade/Edzard_CAS | 42c5db16e506e1fd8653d49e7509b8d2b59353e7 | [
"MIT"
] | null | null | null | 11 Pandas Teil 3/Viele Dateien.ipynb | edzardschade/Edzard_CAS | 42c5db16e506e1fd8653d49e7509b8d2b59353e7 | [
"MIT"
] | null | null | null | 11 Pandas Teil 3/Viele Dateien.ipynb | edzardschade/Edzard_CAS | 42c5db16e506e1fd8653d49e7509b8d2b59353e7 | [
"MIT"
] | null | null | null | 301.289474 | 382,320 | 0.918509 | [
[
[
"# Viele Dateien",
"_____no_output_____"
],
[
"**Inhalt:** Massenverarbeitung von gescrapten Zeitreihen\n\n**Nötige Skills:** Daten explorieren, Time+Date Basics\n\n**Lernziele:**\n- Pandas in Kombination mit Scraping\n- Öffnen und zusammenfügen von vielen Dateien (Glob)\n- Umstrukturierung von Dataframes (Pivot)\n- Plotting Level 4 (Small Multiples)",
"_____no_output_____"
],
[
"## Das Beispiel",
"_____no_output_____"
],
[
"Wir interessieren uns in diesem Notebook für Krypto-Coins.\n\nDie Webseite https://coinmarketcap.com/ führt Marktdaten zu den hundert wichtigsten Coins auf.\n\nMit einem einfachen Scraper werden wir diese Daten beschaffen und rudimentär analysieren.\n\nDer Pfad zum Projektordner heisst `dataprojects/Krypto/`",
"_____no_output_____"
],
[
"## Vorbereitung",
"_____no_output_____"
]
],
[
[
"import requests\nfrom bs4 import BeautifulSoup",
"_____no_output_____"
],
[
"import pandas as pd",
"_____no_output_____"
],
[
"import numpy as np",
"_____no_output_____"
],
[
"import re",
"_____no_output_____"
],
[
"import glob",
"_____no_output_____"
],
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## Scraper",
"_____no_output_____"
]
],
[
[
"path = 'dataprojects/Krypto/'",
"_____no_output_____"
]
],
[
[
"### Liste von allen Kryptowährungen",
"_____no_output_____"
],
[
"Zuerst kucken wir auf der Seite, welches die 100 grössten Kryptowährungen sind, und laden uns Namen und Links derselbigen.",
"_____no_output_____"
]
],
[
[
"base_url = 'https://coinmarketcap.com/'",
"_____no_output_____"
],
[
"response = requests.get(base_url)\ndoc = BeautifulSoup(response.text, \"html.parser\")",
"_____no_output_____"
],
[
"currencies = doc.find_all('a', class_='currency-name-container link-secondary')",
"_____no_output_____"
],
[
"currencies[0]",
"_____no_output_____"
],
[
"len(currencies)",
"_____no_output_____"
],
[
"currency_list = []",
"_____no_output_____"
],
[
"for currency in currencies:\n this_currency = {}\n this_currency['name'] = currency.text\n this_currency['link'] = currency['href']\n currency_list.append(this_currency)",
"_____no_output_____"
],
[
"df_currencies = pd.DataFrame(currency_list)",
"_____no_output_____"
],
[
"df_currencies.head(2)",
"_____no_output_____"
],
[
"df_currencies['link'] = df_currencies['link'].str.extract('/currencies/(.+)/')",
"_____no_output_____"
],
[
"df_currencies.head(2)",
"_____no_output_____"
],
[
"df_currencies.to_csv(path + 'currencies.csv', index=False)",
"_____no_output_____"
]
],
[
[
"### Daten von den einzelnen Währungen",
"_____no_output_____"
],
[
"Zuerst testen wir mit einer Probewährung aus, wie wir an die Informationen kommen.",
"_____no_output_____"
]
],
[
[
"base_url = 'https://coinmarketcap.com/currencies/bitcoin/historical-data/?start=20171015&end=20181015'",
"_____no_output_____"
],
[
"response = requests.get(base_url)\ndoc = BeautifulSoup(response.text, \"html.parser\")",
"_____no_output_____"
],
[
"days = doc.find_all('tr', class_='text-right')",
"_____no_output_____"
],
[
"days_list = []",
"_____no_output_____"
],
[
"cells = days[0].find_all('td')",
"_____no_output_____"
],
[
"cells",
"_____no_output_____"
],
[
"this_day = {}",
"_____no_output_____"
],
[
"this_day['date'] = cells[0].text\nthis_day['open'] = cells[1].text\nthis_day['high'] = cells[2].text\nthis_day['low'] = cells[3].text\nthis_day['close'] = cells[4].text\nthis_day['volume'] = cells[5].text\nthis_day['marketcap'] = cells[6].text",
"_____no_output_____"
],
[
"this_day",
"_____no_output_____"
],
[
"for day in days:\n this_day = {}\n cells = day.find_all('td')\n this_day['date'] = cells[0].text\n this_day['open'] = cells[1].text\n this_day['high'] = cells[2].text\n this_day['low'] = cells[3].text\n this_day['close'] = cells[4].text\n this_day['volume'] = cells[5].text\n this_day['marketcap'] = cells[6].text\n days_list.append(this_day)",
"_____no_output_____"
],
[
"df = pd.DataFrame(days_list)",
"_____no_output_____"
],
[
"df.head(2)",
"_____no_output_____"
]
],
[
[
"Nun wenden wir den Scraper auf alle Währungen an",
"_____no_output_____"
]
],
[
[
"df_currencies = pd.read_csv(path + 'currencies.csv')",
"_____no_output_____"
],
[
"df_currencies.head(2)",
"_____no_output_____"
],
[
"len(df_currencies)",
"_____no_output_____"
],
[
"currencies = df_currencies.to_dict(orient='records')",
"_____no_output_____"
],
[
"url_start = 'https://coinmarketcap.com/currencies/'\nurl_end = '/historical-data/?start=20171015&end=20181015'",
"_____no_output_____"
],
[
"for currency in currencies:\n print ('working on: ' + currency['name'])\n \n url = url_start + currency['link'] + url_end\n response = requests.get(url)\n doc = BeautifulSoup(response.text, \"html.parser\")\n \n days = doc.find_all('tr', class_='text-right')\n days_list = []\n \n this_day = {}\n for day in days:\n this_day = {}\n cells = day.find_all('td')\n this_day['date'] = cells[0].text\n this_day['open'] = cells[1].text\n this_day['high'] = cells[2].text\n this_day['low'] = cells[3].text\n this_day['close'] = cells[4].text\n this_day['volume'] = cells[5].text\n this_day['marketcap'] = cells[6].text\n days_list.append(this_day)\n \n df = pd.DataFrame(days_list)\n filename = currency['name'] + '.csv'\n df.to_csv(path + 'data/' + filename, index=False)\n \nprint('Done')",
"_____no_output_____"
]
],
[
[
"Am Ende haben wir eine Liste von Dateien: Zu jeder Kryptowährung existiert eine Tabelle mit den Marktdaten über den definierten Zeitraum.\n\nDie Daten sind im Unterordner `data/` abgelegt.",
"_____no_output_____"
],
[
"## Daten analysieren",
"_____no_output_____"
],
[
"### Einlesen",
"_____no_output_____"
],
[
"Wir starten damit, dass wir das Verzeichnis durchsuchen, in dem alle Kryptowährungs-Daten abgelegt sind.\n\nDazu benutzen wir `glob`, ein praktisches Tool aus der Standard Library: https://docs.python.org/3/library/glob.html",
"_____no_output_____"
]
],
[
[
"filenames = glob.glob(path + 'data/*.csv') # wir machen eine Liste mit den Filenames ",
"_____no_output_____"
],
[
"len(filenames)",
"_____no_output_____"
],
[
"filenames[0:2]",
"_____no_output_____"
]
],
[
[
"Mit Glob haben wir nun eine Liste mit den Dateinamen erstellt.\n\nNun lesen wir jede einzelne Datei aus der Liste ein.\n\nSo, dass wir als Ergebnis eine Liste von Dataframes erhalten.",
"_____no_output_____"
]
],
[
[
"dfs = []",
"_____no_output_____"
],
[
"dfs = [pd.read_csv(filename) for filename in filenames] # Listcomprehension",
"_____no_output_____"
],
[
"dfs[0].head(2)",
"_____no_output_____"
]
],
[
[
"Die einzelnen Dataframes in der Liste enthalten die Marktdaten. Doch sie enthalten selbst keine Information darüber, zu welcher Kryptowährung die Daten gehören. Wir führen zu dem Zweck in jedes Dataframe noch eine zusätzliche Spalte hinzu mit dem Namen der Währung. ",
"_____no_output_____"
]
],
[
[
"for df, filename in zip(dfs, filenames): #zip ==> führt den code für die in Klammern aufgefüheten df gleichz. aus\n df['currency'] = filename\n df['currency'] = df['currency'].str.extract('/data/(.+).csv')",
"_____no_output_____"
],
[
"dfs[0].head(2)",
"_____no_output_____"
]
],
[
[
"Nun fügen wir alle Dataframes zu einem einzigen, sehr langen Dataframe zusammen.\n\nDazu benutzen wir die Funktion `pd.concat()`: https://pandas.pydata.org/pandas-docs/stable/generated/pandas.concat.html",
"_____no_output_____"
]
],
[
[
"df_all = pd.concat(dfs, ignore_index=True) #ignore_index=True ==> die alte Indexierung soll überschrieben werden.\n#concat führt listen untereinaner zusammen",
"_____no_output_____"
],
[
"df_all.shape",
"_____no_output_____"
],
[
"df_all.head(2)",
"_____no_output_____"
],
[
"df_all.tail(2)",
"_____no_output_____"
],
[
"df_all.dtypes",
"_____no_output_____"
]
],
[
[
"Wir haben nun ein ellenlanges Dataframe. What next?",
"_____no_output_____"
],
[
"### Arrangieren",
"_____no_output_____"
],
[
"Das hängt davon ab, was wir mit den Daten genau tun wollen.\n\nEine Option wäre: die verschiedenen Währungen miteinander zu vergleichen. Und zwar anhand der Schlusskurse.\n\nDazu müssen wir das Dataframe leicht umstellen, mit `pivot()`: https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.pivot.html",
"_____no_output_____"
]
],
[
[
"df_pivoted = df_all.pivot(index='date', columns='currency', values='close')",
"_____no_output_____"
],
[
"df_pivoted.shape",
"_____no_output_____"
],
[
"df_pivoted.head(2)",
"_____no_output_____"
],
[
"df_pivoted.tail(2)",
"_____no_output_____"
],
[
"df_pivoted.rename_axis(None, inplace=True)",
"_____no_output_____"
]
],
[
[
"Nun verfügen wir über einen Index, bei dem eine Zeile jeweils einem einzigartigen Zeitpunkt entspricht.\n\nUm damit zu arbeiten, verwandeln wir den Text in der Indexspalte in ein echtes Datum des Typs datetime.",
"_____no_output_____"
]
],
[
[
"df_pivoted.index = pd.to_datetime(df_pivoted.index, format=\"%b %d, %Y\")",
"_____no_output_____"
],
[
"df_pivoted.sort_index(inplace=True)",
"_____no_output_____"
],
[
"df_pivoted.head(2)",
"_____no_output_____"
]
],
[
[
"Wir haben nun ein sauber formatiertes Dataframe. Mit hundert Spalten, die für jede Kryptowährung, sofern sie zum betreffenden Zeitpunkt existierte, einen Handelskurs enthält.\n\nDie nächste Frage ist: Wie vergleichen wir diese Kurse? Was sagt es aus, wenn eine Währung an einem bestimmten Tag zu 0,1976 USD gehandelt wurde und eine andere zu 18,66 USD?",
"_____no_output_____"
],
[
"### Vergleichbarkeit herstellen",
"_____no_output_____"
],
[
"Diverse Dinge würden sich hier anbieten:\n- zB `pct_change()` um die Veränderungen in den Kursen zu analysieren\n- oder eine indexierte Zeitreihe, die an einem bestimmten Tag bei 100 beginnt\n\nWir wählen die zweite Variante. Und speichern dazu die erste Zeile separat ab.",
"_____no_output_____"
]
],
[
[
"row_0 = df_pivoted.iloc[0]\nrow_0",
"_____no_output_____"
]
],
[
[
"Dann teilen wir jede einzelne Zeile im Dataframe durch die erste Zeile. Und speichern als neues DF ab.",
"_____no_output_____"
]
],
[
[
"df_pivoted_100 = df_pivoted.apply(lambda row: row / row_0 * 100, axis=1)",
"_____no_output_____"
]
],
[
[
"Das neue Dataframe ist nun indexiert auf 100. Alle Währungen starten am gleichen Punkt...",
"_____no_output_____"
]
],
[
[
"df_pivoted_100.head(5)",
"_____no_output_____"
],
[
"df_pivoted_100.tail(1)",
"_____no_output_____"
]
],
[
[
"... und enden an einem bestimmten Punkt. Anhand dieses Punktes können wir die relative Entwicklung ablesen.",
"_____no_output_____"
]
],
[
[
"s_last = df_pivoted_100.iloc[-1]",
"_____no_output_____"
]
],
[
[
"Welche zehn Kryptowährungen am meisten Wert zugelegt haben...",
"_____no_output_____"
]
],
[
[
"s_last.sort_values(ascending=False).head(10)",
"_____no_output_____"
]
],
[
[
"... und welche am meisten Wert verloren haben.",
"_____no_output_____"
]
],
[
[
"s_last.sort_values(ascending=False, na_position='first').tail(10)",
"_____no_output_____"
]
],
[
[
"Und so sieht die Performance aller Währungen aus:",
"_____no_output_____"
]
],
[
[
"df_pivoted_100.plot(figsize=(10,6), legend=False)",
"_____no_output_____"
]
],
[
[
"Wow, das sind ziemlich viele Linien!",
"_____no_output_____"
],
[
"# Plotting Level 4",
"_____no_output_____"
],
[
"Wie wir diesen Chart etwas auseinandernehmen können, lernen wir hier.\n\nEine Gelegenheit, zu sehen, wie man die matplotlib-Funktionen direkt benutzen kann.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"import matplotlib.dates as mdates\nimport matplotlib.ticker as ticker",
"_____no_output_____"
]
],
[
[
"Und eine neue Art kennenlernen, wie man einen Plot erstellt.",
"_____no_output_____"
],
[
"### Ein Plot",
"_____no_output_____"
],
[
"Starten wir zuerst mal mit einem Plots: Bitcoin.\n\nWir müssen uns dazu zuerst zwei Dinge basteln:\n1. Eine \"figure\", also eine Abbildung\n1. Einen \"subplot\", also der Plot selbst",
"_____no_output_____"
]
],
[
[
"# Wir erstellen beide Dinge in einem Atemzug\nfig, ax = plt.subplots(figsize=(10,6))\n\n# Und füllen den Plot jetzt mit Inhalt:\ndf_pivoted_100['Bitcoin'].plot(title=\"Bitcoin\", ax=ax)",
"_____no_output_____"
]
],
[
[
"### Zwei Plots",
"_____no_output_____"
],
[
"Als nächstes Plotten wir zwei Währungen auf derselben Figure: Bitcoin und Ethereum.\n\nWir müssen uns dazu erneut zwei Dinge basteln:\n1. Eine \"figure\", also eine Abbildung\n1. Diverse \"subplots\" für die jeweiligen Währungen\n\nDazu formatieren wir jetzt die x-Achse etwas speziell.",
"_____no_output_____"
]
],
[
[
"# Zuerst kreieren wir nur die Figure\nfig = plt.figure(figsize=(12,3))\n\n# Danach die einzelnen Subplots\nax1 = fig.add_subplot(1, 2, 1) # total 1 Zeile, total 2 Spalten, Subplot Nr. 1\nax2 = fig.add_subplot(1, 2, 2) # total 1 Zeile, total 2 Spalten, Subplot Nr. 2\n\n# Und schliesslich füllen wir die Subplots mit Inhalt\ndf_pivoted_100['Bitcoin'].plot(title=\"Bitcoin\", ax=ax1)\ndf_pivoted_100['Ethereum'].plot(title=\"Ethereum\", ax=ax2)\n\n# Hier formatieren wir die x-Achse für Plot 1\nax1.xaxis.set_major_locator(mdates.MonthLocator())\nax1.xaxis.set_major_formatter(mdates.DateFormatter('%m'))\nax1.xaxis.set_minor_locator(ticker.NullLocator())\n\n# Hier formatieren wir die x-Achse für Plot 2\nax2.xaxis.set_major_locator(mdates.MonthLocator())\nax2.xaxis.set_major_formatter(mdates.DateFormatter('%m'))\nax2.xaxis.set_minor_locator(ticker.NullLocator())",
"_____no_output_____"
]
],
[
[
"Einige Angaben dazu, wie man Zeitachsen formatieren kann, gibt es hier:\n- TickLocators: https://matplotlib.org/examples/ticks_and_spines/tick-locators.html\n- TickFormatters: https://matplotlib.org/gallery/ticks_and_spines/tick-formatters.html",
"_____no_output_____"
],
[
"### Sehr viele Plots",
"_____no_output_____"
],
[
"Nun plotten wir sämtliche Währungen auf einmal. Wie viele sind es?",
"_____no_output_____"
]
],
[
[
"anzahl_charts = s_last.notnull().sum()\nanzahl_charts",
"_____no_output_____"
]
],
[
[
"Wir sortieren unsere Liste der Währungen etwas:",
"_____no_output_____"
]
],
[
[
"sortierte_waehrungen = s_last[s_last.notnull()].sort_values(ascending=False)\nsortierte_waehrungen.head(2)",
"_____no_output_____"
]
],
[
[
"Und wiederholen dann wiederum dasselbe Vorgehen wie vorher.",
"_____no_output_____"
]
],
[
[
"sortierte_waehrungen.index",
"_____no_output_____"
],
[
"# Eine Abbildung, die gross genug ist\nfig = plt.figure(figsize=(15,22))\n\n# Und nun, für jede einzelne Währung:\nfor i, waehrung in enumerate(sortierte_waehrungen.index): #der enumerate-Funktion indexiert die Plots\n \n # einen Subplot kreieren ...\n ax = fig.add_subplot(11, 6, i + 1)\n\n # ... und mit Inhalt füllen\n df_pivoted_100[waehrung].plot(title=waehrung, ax=ax)\n \n # Auf Ticks verzichten wir hier ganz\n ax.xaxis.set_major_locator(ticker.NullLocator())\n ax.xaxis.set_minor_locator(ticker.NullLocator())",
"_____no_output_____"
]
],
[
[
"Falls wir zusätzlich noch wollen, dass jeder Plot dieselbe y-Achse hat:",
"_____no_output_____"
]
],
[
[
"# Eine Abbildung, die gross genug ist\nfig = plt.figure(figsize=(15,22))\n\n# Und nun, für jede einzelne Währung:\nfor i, waehrung in enumerate(sortierte_waehrungen.index):\n \n # einen Subplot kreieren ...\n ax = fig.add_subplot(11, 6, i + 1)\n\n # ... und mit Inhalt füllen\n df_pivoted_100[waehrung].plot(title=waehrung, ax=ax)\n \n # Auf Ticks verzichten wir hier ganz\n ax.xaxis.set_major_locator(ticker.NullLocator())\n ax.xaxis.set_minor_locator(ticker.NullLocator())\n \n # Hier setzen wir eine einheitliche y-Achse (und schalten sie aus)\n ax.set_ylim([0, 25000])\n ax.yaxis.set_major_locator(ticker.NullLocator())",
"_____no_output_____"
]
],
[
[
"### Aber es geht auch einfacher...",
"_____no_output_____"
],
[
"Ha! Nachdem wir nun alles Manuell zusammengebastelt haben, mit Matplotlib, hier die gute Nachricht:\n\n*Wir können das mit wenigen Codezeilen auch direkt aus der Pandas-Plot()-Funktion haben :-)*",
"_____no_output_____"
]
],
[
[
"axes = df_pivoted_100[sortierte_waehrungen.index].plot(subplots=True,layout=(22, 3), sharey=True, figsize=(15,22))\n\naxes[0,0].xaxis.set_major_locator(ticker.NullLocator())\naxes[0,0].xaxis.set_minor_locator(ticker.NullLocator())",
"_____no_output_____"
]
],
[
[
"# Übung",
"_____no_output_____"
],
[
"Hier schauen wir uns nicht mehr die Handelskurse, sondern die Handelsvolumen an! Also: Wie viel von den einzelnen Kryptowährungen an einem bestimmten Tag gekfauft und verkauft wurde (gemessen in USD).",
"_____no_output_____"
],
[
"Schauen Sie sich nochmals das Dataframe `df_all` an, das wir im Verlauf des Notebooks erstellt haben - es enthält alle Informationen, die wir brauchen, ist aber noch relativ unstrukturiert.",
"_____no_output_____"
],
[
"Welche Spalte interessiert uns? Müssen wir noch etwas daran machen?",
"_____no_output_____"
],
[
"### Daten arrangieren",
"_____no_output_____"
],
[
"Unternehmen Sie die nötigen Schritte, um mit der Spalte arbeiten zu können. Sie sollten am Ende eine Spalte haben, die nicht mehr als Object, sondern als Float formatiert ist.\n\nTipp: Speichern Sie alle Modifikationen in einer neuen Spalte ab, damit das Original unverändert bleibt.",
"_____no_output_____"
],
[
"Nun wollen wir die Daten umgliedern:\n- Für jedes Datum wollen wir eine Zeile\n- Für jede Kryptowährung eine Spalte\n- Wir interessieren uns für die Handelsvolumen",
"_____no_output_____"
],
[
"Formatieren Sie die Werte in der Index-Spalte als Datetime-Objekte und sortieren Sie das Dataframe nach Datum.",
"_____no_output_____"
],
[
"### Analyse",
"_____no_output_____"
],
[
"Wir machen in dieser Sektion einige einfache Auswertungen und repetieren einige Befehle, u.a. aus dem Time Series Sheet.",
"_____no_output_____"
],
[
"**Top-10**: Welches waren, im Schnitt, die zehn meistgehandelten Währungen? Liste und Chart.",
"_____no_output_____"
],
[
"Welches waren die zehn Währungen, bei denen das Volumen in absoluten Zahlen am meisten geschwankt ist? (Standardabweichung)",
"_____no_output_____"
],
[
"Sieht so aus, als wären es dieselben zehn Währungen.\n\nKönnen wir angeben, welche von ihnen relativ die grössten Schwankungen hatten, also im Vergleich zum Handelsvolumen?",
"_____no_output_____"
],
[
"**Bitcoin vs Ethereum**",
"_____no_output_____"
],
[
"Erstellen Sie einen Chart mit dem wöchentlichen Umsatztotal von Bitcoin und Ethereum!",
"_____no_output_____"
],
[
"In welchem der letzten 12 Monate wurde insgesamt am meisten mit Bitcoin gehandelt? Mit Ethereum?",
"_____no_output_____"
],
[
"Wie viel Bitcoin und Ethereum wird im Durchschnitt an den sieben Wochentagen gehandelt? Barchart.",
"_____no_output_____"
],
[
"**Small Multiples**: Hier erstellen wir einen Plot, ähnlich wie oben",
"_____no_output_____"
],
[
"Kreieren Sie zuerst eine Liste von Währungen:\n- Alle Währungen, die am letzten Handelstag einen Eintrag haben\n- Sortiert in absteigender Reihenfolge nach dem Handelsvolumen\n- Wir wählen nur die zehn grössten aus",
"_____no_output_____"
],
[
"Und jetzt: Small Multiples plotten! Überlegen Sie sich:\n- Wie viele Subplots braucht es, wie sollen sie angeordnet sein?\n- Wie gross muss die Abbildung insgesamt sein?\n- Was ist eine sinnvolle Einstellung für die Y-Achse?\n\n(Sie können die Matplotlib-Funktionalität dafür nutzen oder direkt Pandas-plot()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cb44e717151ad8ee53927973595c441d9d0b0389 | 1,600 | ipynb | Jupyter Notebook | work/dbs.ipynb | nmfc2003/pyspark-setup-demo | a5797e0188723483b5ae663c2738ca7a792bb8c2 | [
"MIT"
] | null | null | null | work/dbs.ipynb | nmfc2003/pyspark-setup-demo | a5797e0188723483b5ae663c2738ca7a792bb8c2 | [
"MIT"
] | null | null | null | work/dbs.ipynb | nmfc2003/pyspark-setup-demo | a5797e0188723483b5ae663c2738ca7a792bb8c2 | [
"MIT"
] | null | null | null | 17.582418 | 55 | 0.505 | [
[
[
"# PySpark task Notebook\n1. Run PostgreSQL ddl script\n2. Load CSV Data files\n3. Write Data to PostgreSQL source db\n4. Analyze Data with Spark SQL\n\n_Prepared by: [Noam Marianne]",
"_____no_output_____"
],
[
"### Run PostgreSQL Script\nRun the PostgreSQL sql script",
"_____no_output_____"
]
],
[
[
"# ! pip install psycopg2-binary --upgrade --quiet",
"_____no_output_____"
],
[
"%run -i 'dbs_ddl.py'",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cb44edd9e9df8cae4325c558f14c401f870cb813 | 36,345 | ipynb | Jupyter Notebook | general-models/General_Model_3_Create_the_Model--Multi_layer.ipynb | janetrbarclay/gw-general-models | 6f004d055dc73fe97a4545e19690b30d45a68379 | [
"CC0-1.0"
] | 4 | 2018-12-20T15:07:09.000Z | 2019-06-26T09:05:11.000Z | general-models/General_Model_3_Create_the_Model--Multi_layer.ipynb | janetrbarclay/gw-general-models | 6f004d055dc73fe97a4545e19690b30d45a68379 | [
"CC0-1.0"
] | 3 | 2018-05-10T15:53:07.000Z | 2021-06-28T15:11:10.000Z | general-models/General_Model_3_Create_the_Model--Multi_layer.ipynb | janetrbarclay/gw-general-models | 6f004d055dc73fe97a4545e19690b30d45a68379 | [
"CC0-1.0"
] | 5 | 2018-02-14T18:04:10.000Z | 2020-02-25T03:41:46.000Z | 33.715213 | 927 | 0.570065 | [
[
[
"# Create a general MODFLOW model from the NHDPlus dataset",
"_____no_output_____"
],
[
"Project specific variables are imported in the model_spec.py and gen_mod_dict.py files that must be included in the notebook directory. The first first includes pathnames to data sources that will be different for each user. The second file includes a dictionary of model-specific information such as cell size, default hydraulic parameter values, and scenario defintion (e.g. include bedrock, number of layers, etc.). There are examples in the repository. Run the following cells up to the \"Run to here\" cell to get a pull-down menu of models in the model_dict. Then, without re-running that cell, run all the remaining cells. Re-running the following cell would re-set the model to the first one in the list, which you probably don't want. If you use the notebook option to run all cells below, it runs the cell you're in, so if you use that option, move to the next cell (below the pull-down menu of models) first.",
"_____no_output_____"
]
],
[
[
"__author__ = 'Jeff Starn'\n%matplotlib notebook\nfrom model_specs import *\nfrom gen_mod_dict import *\nimport os\nimport sys\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport flopy as fp\nimport pandas as pd\nimport gdal\ngdal.UseExceptions()\nimport shutil\n# from model_specs import *\n# from gen_mod_dict import *\n\nfrom ipywidgets import interact, Dropdown\nfrom IPython.display import display",
"_____no_output_____"
],
[
"for key, value in model_dict.items():\n md = key\n ms = model_dict[md]\n print('trying {}'.format(md))\n try:\n pass\n except:\n pass",
"_____no_output_____"
],
[
"models = list(model_dict.keys())\nmodels.sort()\nmodel_area = Dropdown(\n options=models,\n description='Model:',\n background_color='cyan',\n border_color='black',\n border_width=2)\ndisplay(model_area)",
"_____no_output_____"
]
],
[
[
"### Run to here to initiate notebook",
"_____no_output_____"
],
[
"First time using this notebook in this session (before restarting the notebook), run the cells up to this point. Then select your model from the dropdown list above. Move your cursor to this cell and use the toolbar menu Cell --> Run All Below. After the first time, if you want to run another model, select your model and start running from this cell--you don't need to re-run the cells from the beginning.",
"_____no_output_____"
],
[
"## Preliminary stuff",
"_____no_output_____"
]
],
[
[
"md = model_area.value\nms = model_dict[md]\nprint('The model being processed is {}\\n'.format(md))",
"_____no_output_____"
]
],
[
[
"Set pathnames and create workspace directories for geographic data (from Notebook 1) and this model.",
"_____no_output_____"
]
],
[
[
"geo_ws = os.path.join(proj_dir, ms['ws'])\nmodel_ws = os.path.join(geo_ws, scenario_dir)\narray_pth = os.path.join(model_ws, 'arrays')\n \ntry:\n shutil.rmtree(array_pth)\nexcept:\n pass\n\ntry:\n shutil.rmtree(model_ws)\nexcept:\n pass\n\nos.makedirs(model_ws)\n\nhead_file_name = '{}.hds'.format(md)\nhead_file_pth = os.path.join(model_ws, head_file_name)\n\nprint (model_ws)",
"_____no_output_____"
]
],
[
[
"Replace entries from the default K_dict with the model specific K values from model_dict if they exist. ",
"_____no_output_____"
]
],
[
[
"for key, value in K_dict.items():\n if key in ms.keys():\n K_dict[key] = ms[key]",
"_____no_output_____"
]
],
[
[
"Replace entries from the default rock_riv_dict with the model specific values from model_dict if they exist. rock_riv_dict has various attributes of bedrock and stream geometry.",
"_____no_output_____"
]
],
[
[
"for key, value in rock_riv_dict.items():\n if key in ms.keys():\n rock_riv_dict[key] = ms[key]",
"_____no_output_____"
]
],
[
[
"Assign values to variables used in this notebook using rock_riv_dict",
"_____no_output_____"
]
],
[
[
"min_thk = rock_riv_dict['min_thk'] \nstream_width = rock_riv_dict['stream_width'] \nstream_bed_thk = rock_riv_dict['stream_bed_thk']\nriver_depth = rock_riv_dict['river_depth'] \nbedrock_thk = rock_riv_dict['bedrock_thk']\nstream_bed_kadjust = rock_riv_dict['stream_bed_kadjust']",
"_____no_output_____"
]
],
[
[
"## Read the information for a model domain processed using Notebook 1",
"_____no_output_____"
],
[
"Read the model_grid data frame from a csv file. Extract grid dimensions and ibound array.",
"_____no_output_____"
]
],
[
[
"model_file = os.path.join(geo_ws, 'model_grid.csv')\nmodel_grid = pd.read_csv(model_file, index_col='node_num', na_values=['nan', hnoflo])\n\nNROW = model_grid.row.max() + 1\nNCOL = model_grid.col.max() + 1\nnum_cells = NROW * NCOL\n\nibound = model_grid.ibound.reshape(NROW, NCOL)\ninactive = (ibound == 0)",
"_____no_output_____"
]
],
[
[
"## Translate geologic information into hydrologic properties",
"_____no_output_____"
]
],
[
[
"# # old geology used in general models prior to 4/5/2016\n# coarse_deposits = (model_grid.coarse_flag == 2)\n# coarse_is_1 = coarse_deposits.reshape(NROW, NCOL)",
"_____no_output_____"
]
],
[
[
"This version replaces Soller's Surfmat with the Quaternary Atlas. Look-up table for coarse deposits (zone = 1) from Dick Yager's new_unit. All other categories are lumped with fine deposits (zone = 0).\n* alluvium = 1\n* ice contact = 9\n* lacustrine coarse = 11\n* outwash = 17",
"_____no_output_____"
],
[
"Create a dictionary that maps the K_dict from gen_mod_dict to zone numbers (key=zone number, value=entry in K_dict). Make sure these correspond with the correct units. If you're using the defaults, it is correct.",
"_____no_output_____"
]
],
[
[
"zone_dict = {0 : 'K_fine', 1 : 'K_coarse', 2 : 'K_lakes', 3 : 'K_bedrock'}",
"_____no_output_____"
]
],
[
[
"Perform the mapping from zone number to K to create the Kh1d array.",
"_____no_output_____"
]
],
[
[
"zones1d = np.zeros(( NROW, NCOL ), dtype=np.int32)\n\nqa = model_grid.qu_atlas.reshape( NROW, NCOL )\nzones1d[qa == 1] = 1\nzones1d[qa == 9] = 1\nzones1d[qa == 11] = 1\nzones1d[qa == 17] = 1\n\nla = model_grid.lake.reshape( NROW, NCOL )\nzones1d[la == 1] = 2\n\nKh1d = np.zeros(( NROW, NCOL ), dtype=np.float32)\n\nfor key, val in zone_dict.items():\n Kh1d[zones1d == key] = K_dict[val]\n \nmodel_grid['K0'] = Kh1d.ravel()",
"_____no_output_____"
]
],
[
[
"## Process boundary condition information",
"_____no_output_____"
],
[
"Create a dictionary of stream information for the drain or river package.\nRiver package input also needs the elevation of the river bed. Don't use both packages. The choice is made by commenting/uncommenting sections of the modflow function. Replace segment_len (segment length) with the conductance. The river package has not been tested.",
"_____no_output_____"
]
],
[
[
"drn_flag = (model_grid.stage != np.nan) & (model_grid.ibound == 1)\ndrn_data = model_grid.loc[drn_flag, ['lay', 'row', 'col', 'stage', 'segment_len', 'K0']]\ndrn_data.columns = ['k', 'i', 'j', 'stage', 'segment_len', 'K0']\ndcond = drn_data.K0 *stream_bed_kadjust* drn_data.segment_len * stream_width / stream_bed_thk\ndrn_data['segment_len'] = dcond\ndrn_data.rename(columns={'segment_len' : 'cond'}, inplace=True)\ndrn_data.drop('K0', axis=1, inplace=True)\ndrn_data.dropna(axis='index', inplace=True)\ndrn_data.insert(drn_data.shape[1], 'iface', 6)\ndrn_recarray = drn_data.to_records(index=False)\ndrn_dict = {0 : drn_recarray}",
"_____no_output_____"
],
[
"riv_flag = (model_grid.stage != np.nan) & (model_grid.ibound == 1)\nriv_data = model_grid.loc[riv_flag, ['lay', 'row', 'col', 'stage', 'segment_len', \n 'reach_intermit', 'K0']]\nriv_data.columns = ['k', 'i', 'j', 'stage', 'segment_len', 'rbot', 'K0']\nriv_data[['rbot']] = riv_data.stage - river_depth\nrcond = riv_data.K0 * stream_bed_kadjust* riv_data.segment_len * stream_width / stream_bed_thk\nriv_data['segment_len'] = rcond\nriv_data.rename(columns={'segment_len' : 'rcond'}, inplace=True)\nriv_data.drop('K0', axis=1, inplace=True)\nriv_data.dropna(axis='index', inplace=True)\nriv_data.insert(riv_data.shape[1], 'iface', 6)\nriv_recarray = riv_data.to_records(index=False)\nriv_dict = {0 : riv_recarray}",
"_____no_output_____"
]
],
[
[
"Create a dictionary of information for the general-head boundary package.\nSimilar to the above cell. Not tested.",
"_____no_output_____"
]
],
[
[
"if model_grid.ghb.sum() > 0:\n ghb_flag = model_grid.ghb == 1\n ghb_data = model_grid.loc[ghb_flag, ['lay', 'row', 'col', 'top', 'segment_len', 'K0']]\n ghb_data.columns = ['k', 'i', 'j', 'stage', 'segment_len', 'K0']\n gcond = ghb_data.K0 * L * L / stream_bed_thk\n ghb_data['segment_len'] = gcond\n ghb_data.rename(columns={'segment_len' : 'cond'}, inplace=True)\n ghb_data.drop('K0', axis=1, inplace=True)\n ghb_data.dropna(axis='index', inplace=True)\n ghb_data.insert(ghb_data.shape[1], 'iface', 6)\n ghb_recarray = ghb_data.to_records(index=False)\n ghb_dict = {0 : ghb_recarray}",
"_____no_output_____"
]
],
[
[
"### Create 1-layer model to get initial top-of-aquifer on which to drape subsequent layering",
"_____no_output_____"
],
[
"Get starting heads from top elevations. The top is defined as the model-cell-mean NED elevation except in streams, where it is interpolated between MaxElevSmo and MinElevSmo in the NHD (called 'stage' in model_grid). Make them a little higher than land so that drains don't accidentally go dry too soon.",
"_____no_output_____"
]
],
[
[
"top = model_grid.top.reshape(NROW, NCOL)\nstrt = top * 1.05",
"_____no_output_____"
]
],
[
[
"Modify the bedrock surface, ensuring that it is always at least min_thk below the top elevation. This calculation will be revisited for the multi-layer case.",
"_____no_output_____"
]
],
[
[
"bedrock = model_grid.bedrock_el.reshape(NROW, NCOL)\nthk = top - bedrock\nthk[thk < min_thk] = min_thk\nbot = top - thk",
"_____no_output_____"
]
],
[
[
"## Create recharge array",
"_____no_output_____"
],
[
"This version replaces the Wolock/Yager recharge grid with the GWRP SWB grid.",
"_____no_output_____"
]
],
[
[
"## used in general models prior to 4/5/2016\n# rech = model_grid.recharge.reshape(NROW, NCOL)",
"_____no_output_____"
]
],
[
[
"Replace rech array with\n* calculate total recharge for the model domain\n* calculate areas of fine and coarse deposits\n* apportion recharge according to the ratio specified in gen_mod_dict.py\n* write the values to an array",
"_____no_output_____"
]
],
[
[
"r_swb = model_grid.swb.reshape(NROW, NCOL) / 365.25\n\nrech_ma = np.ma.MaskedArray(r_swb, mask=inactive)\ncoarse_ma = np.ma.MaskedArray(zones1d != 0, mask=inactive)\nfine_ma = np.ma.MaskedArray(zones1d == 0, mask=inactive)\n\ntotal_rech = rech_ma.sum()\nAf = fine_ma.sum()\nAc = coarse_ma.sum()\nRf = total_rech / (rech_fact * Ac + Af)\nRc = rech_fact * Rf\n\nrech = np.zeros_like(r_swb)\nrech[zones1d != 0] = Rc\nrech[zones1d == 0] = Rf",
"_____no_output_____"
]
],
[
[
"## Define a function to create and run MODFLOW",
"_____no_output_____"
]
],
[
[
"def modflow(md, mfpth, model_ws, nlay=1, top=top, strt=strt, nrow=NROW, ncol=NCOL, botm=bedrock, \n ibound=ibound, hk=Kh1d, rech=rech, stream_dict=drn_dict, delr=L, delc=L, \n hnoflo=hnoflo, hdry=hdry, iphdry=1):\n\n strt_dir = os.getcwd()\n os.chdir(model_ws)\n\n ml = fp.modflow.Modflow(modelname=md, exe_name=mfpth, version='mfnwt', \n external_path='arrays') \n\n # add packages (DIS has to come before either BAS or the flow package)\n dis = fp.modflow.ModflowDis(ml, nlay=nlay, nrow=NROW, ncol=NCOL, nper=1, delr=L, delc=L, \n laycbd=0, top=top, botm=botm, perlen=1.E+05, nstp=1, tsmult=1, \n steady=True, itmuni=4, lenuni=2, extension='dis', \n unitnumber=11) \n\n bas = fp.modflow.ModflowBas(ml, ibound=ibound, strt=strt, ifrefm=True, \n ixsec=False, ichflg=False, stoper=None, hnoflo=hnoflo, extension='bas', \n unitnumber=13)\n\n upw = fp.modflow.ModflowUpw(ml, laytyp=1, layavg=0, chani=1.0, layvka=1, laywet=0, ipakcb=53, \n hdry=hdry, iphdry=iphdry, hk=hk, hani=1.0, vka=1.0, ss=1e-05, \n sy=0.15, vkcb=0.0, noparcheck=False, extension='upw', \n unitnumber=31)\n\n rch = fp.modflow.ModflowRch(ml, nrchop=3, ipakcb=53, rech=rech, irch=1, \n extension='rch', unitnumber=19)\n\n drn = fp.modflow.ModflowDrn(ml, ipakcb=53, stress_period_data=drn_dict, \n dtype=drn_dict[0].dtype,\n extension='drn', unitnumber=21, options=['NOPRINT', 'AUX IFACE'])\n\n riv = fp.modflow.ModflowRiv(ml, ipakcb=53, stress_period_data=riv_dict, \n dtype=riv_dict[0].dtype,\n extension='riv', unitnumber=18, options=['NOPRINT', 'AUX IFACE'])\n\n if GHB:\n ghb = fp.modflow.ModflowGhb(ml, ipakcb=53, stress_period_data=ghb_dict, \n dtype=ghb_dict[0].dtype,\n extension='ghb', unitnumber=23, options=['NOPRINT', 'AUX IFACE'])\n\n oc = fp.modflow.ModflowOc(ml, ihedfm=0, iddnfm=0, chedfm=None, cddnfm=None, cboufm=None, \n compact=True, stress_period_data={(0, 0): ['save head', 'save budget']}, \n extension=['oc', 'hds', 'ddn', 'cbc'], unitnumber=[14, 51, 52, 53])\n\n# nwt = fp.modflow.ModflowNwt(ml, headtol=0.0001, fluxtol=500, maxiterout=1000, \n# thickfact=1e-05, linmeth=2, iprnwt=1, ibotav=0, options='COMPLEX')\n\n nwt = fp.modflow.ModflowNwt(ml, headtol=0.0001, fluxtol=500, maxiterout=100, thickfact=1e-05, \n linmeth=2, iprnwt=1, ibotav=1, options='SPECIFIED', dbdtheta =0.80, \n dbdkappa = 0.00001, dbdgamma = 0.0, momfact = 0.10, backflag = 1, \n maxbackiter=30, backtol=1.05, backreduce=0.4, iacl=2, norder=1, \n level=3, north=7, iredsys=1, rrctols=0.0,idroptol=1, epsrn=1.0E-3,\n hclosexmd= 1.0e-4, mxiterxmd=200)\n \n ml.write_input()\n ml.remove_package('RIV')\n \n ml.write_input()\n success, output = ml.run_model(silent=True)\n os.chdir(strt_dir)\n if success:\n print(\" Your {:0d} layer model ran successfully\".format(nlay))\n else:\n print(\" Your {:0d} layer model didn't work\".format(nlay))",
"_____no_output_____"
]
],
[
[
"## Run 1-layer MODFLOW",
"_____no_output_____"
],
[
"Use the function to run MODFLOW for 1 layer to getting approximate top-of-aquifer elevation",
"_____no_output_____"
]
],
[
[
"modflow(md, mfpth, model_ws, nlay=1, top=top, strt=strt, nrow=NROW, ncol=NCOL, botm=bot, ibound=ibound, \n hk=Kh1d, rech=rech, stream_dict=drn_dict, delr=L, delc=L, hnoflo=hnoflo, hdry=hdry, iphdry=0)",
"_____no_output_____"
]
],
[
[
"Read the head file and calculate new layer top (wt) and bottom (bot) elevations based on the estimated\nwater table (wt) being the top of the top layer. Divide the surficial layer into NLAY equally thick layers between wt and the bedrock surface elevation (as computed using minimum surficial thickness). ",
"_____no_output_____"
]
],
[
[
"hdobj = fp.utils.HeadFile(head_file_pth)\nheads1 = hdobj.get_data(kstpkper=(0, 0))\nheads1[heads1 == hnoflo] = np.nan\nheads1[heads1 <= hdry] = np.nan\nheads1 = heads1[0, :, :]\nhdobj = None",
"_____no_output_____"
]
],
[
[
"## Create layering using the scenario in gen_mod_dict",
"_____no_output_____"
],
[
"Make new model with (possibly) multiple layers. If there are dry cells in the 1 layer model, they are converted to NaN (not a number). The minimum function in the first line returns NaN if the element of either input arrays is NaN. In that case, replace NaN in modeltop with the top elevation. The process is similar to the 1 layer case. Thickness is estimated based on modeltop and bedrock and is constrained to be at least min_thk (set in gen_mod_dict.py). This thickness is divided into num_surf_layers number of layers. The cumulative thickness of these layers is the distance from the top of the model to the bottom of the layers. This 3D array of distances (the same for each layer) is subtracted from modeltop.",
"_____no_output_____"
]
],
[
[
"modeltop = np.minimum(heads1, top)\nnan = np.isnan(heads1)\nmodeltop[nan] = top[nan]\nthk = modeltop - bedrock\nthk[thk < min_thk] = min_thk\n\nNLAY = num_surf_layers\nlay_extrude = np.ones((NLAY, NROW, NCOL))\nlay_thk = lay_extrude * thk / NLAY\nbot = modeltop - np.cumsum(lay_thk, axis=0)",
"_____no_output_____"
]
],
[
[
"Using the estimated water table as the new top-of-aquifer elevations sometimes leads to the situation, in usually a very small number of cells, that the drain elevation is below the bottom of the cell. The following procedure resets the bottom elevation to one meter below the drain elevation if that is the case. ",
"_____no_output_____"
]
],
[
[
"stg = model_grid.stage.fillna(1.E+30, inplace=False)\ntmpdrn = (lay_extrude * stg.reshape(NROW, NCOL)).ravel()\ntmpbot = bot.ravel()\nindex = np.less(tmpdrn, tmpbot)\ntmpbot[index] = tmpdrn[index] - 1.0\nbot = tmpbot.reshape(NLAY, NROW, NCOL)",
"_____no_output_____"
]
],
[
[
"* If add_bedrock = True in gen_mod_dict.py, add a layer to the bottom and increment NLAY by 1.\n* Assign the new bottom-most layer an elevation equal to the elevation of the bottom of the lowest surficial layer minus bedrock_thk, which is specified in rock_riv_dict (in gen_mod_dict.py).\n* Concatenate the new bottom-of-bedrock-layer to the bottom of the surficial bottom array.\n* Compute the vertical midpoint of each cell. Make an array (bedrock_index) that is True if the bedrock surface is higher than the midpoint and False if it is not.\n* lay_extrude replaces the old lay_extrude to account for the new bedrock layer. It is not used in this cell, but is used later to extrude other arrays.",
"_____no_output_____"
]
],
[
[
"sol_thk = model_grid.soller_thk.reshape(NROW, NCOL)\ntmp = top - sol_thk\nbedrock_4_K = bedrock.copy()\nbedrock_4_K[bedrock > top] = tmp[bedrock > top]\n\nif add_bedrock:\n NLAY = num_surf_layers + 1\n lay_extrude = np.ones((NLAY, NROW, NCOL))\n bed_bot = bot[-1:,:,:] - bedrock_thk\n bot = np.concatenate((bot, bed_bot), axis=0)\n\n mids = bot + thk / NLAY / 2\n bedrock_index = mids < bedrock_4_K\n bedrock_index[-1:,:,:] = True\n\nelif not add_bedrock:\n print(' no bedrock')\n pass\n\nelse:\n print(' add_bedrock variable needs to True or False')",
"_____no_output_____"
]
],
[
[
"Extrude all arrays to NLAY number of layers. Create a top-of-aquifer elevation (fake_top) that is higher (20% in this case) than the simulated 1-layer water table because in doing this approximation, some stream elevations end up higher than top_of_aquifer and thus do not operate as drains. The fake_top shouldn't affect model computations if it is set high enough because the model uses convertible (confined or unconfined) layers.",
"_____no_output_____"
]
],
[
[
"fake_top = (modeltop * 1.2).astype(np.float32)\nstrt = (lay_extrude * modeltop * 1.05).astype(np.float32)\nibound = (lay_extrude * ibound).astype(np.int16)",
"_____no_output_____"
]
],
[
[
"Perform the mapping from zone number to K to create the Kh3d array.",
"_____no_output_____"
]
],
[
[
"zones3d = np.zeros(( NLAY, NROW, NCOL ), dtype=np.int32)\n\nqa = model_grid.qu_atlas.reshape(NROW, NCOL)\nqa3d = (lay_extrude * qa).astype(np.int32)\n\nzones3d[qa3d == 1] = 1\nzones3d[qa3d == 9] = 1\nzones3d[qa3d == 11] = 1\nzones3d[qa3d == 17] = 1\n\nif add_bedrock:\n zones3d[bedrock_index] = 3\n\nla = model_grid.lake.reshape(NROW, NCOL)\nzones3d[0, la == 1] = 2\n\nKh3d = np.zeros(( NLAY, NROW, NCOL ), dtype=np.float32)\n\nfor key, val in zone_dict.items():\n Kh3d[zones3d == key] = K_dict[val]",
"_____no_output_____"
]
],
[
[
"Run MODFLOW again using the new layer definitions. The difference from the first run is that the top-of-aquifer elevation is the 1-layer water table rather than land surface, and of course, the number of surficial layers and/or the presence of a bedrock layer is different. ",
"_____no_output_____"
]
],
[
[
"modflow(md, mfpth, model_ws, nlay=NLAY, top=fake_top, strt=strt, nrow=NROW, ncol=NCOL, \n botm=bot, ibound=ibound, hk=Kh3d, rech=rech, stream_dict=drn_dict, delr=L, \n delc=L, hnoflo=hnoflo, hdry=hdry, iphdry=1)",
"_____no_output_____"
]
],
[
[
"Read the new head array",
"_____no_output_____"
]
],
[
[
"hdobj = fp.utils.HeadFile(head_file_pth)\nheads = hdobj.get_data()\nhdobj = None",
"_____no_output_____"
]
],
[
[
"Make a 2D array of the heads in the highest active cells and call it the water_table",
"_____no_output_____"
]
],
[
[
"heads[heads == hnoflo] = np.nan\nheads[heads <= hdry] = np.nan\nhin = np.argmax(np.isfinite(heads), axis=0)\nrow, col = np.indices((hin.shape))\nwater_table = heads[hin, row, col]\n\nwater_table_ma = np.ma.MaskedArray(water_table, inactive)",
"_____no_output_____"
]
],
[
[
"Save the head array to a geotiff file.",
"_____no_output_____"
]
],
[
[
"data = water_table_ma\n\nsrc_pth = os.path.join(geo_ws, 'ibound.tif')\nsrc = gdal.Open(src_pth)\n\ndst_pth = os.path.join(model_ws, 'pre-heads.tif')\ndriver = gdal.GetDriverByName('GTiff')\ndst = driver.CreateCopy(dst_pth, src, 0)\n\nband = dst.GetRasterBand(1)\nband.WriteArray(data)\nband.SetNoDataValue(np.nan)\n\ndst = None\nsrc = None",
"_____no_output_____"
]
],
[
[
"Save the heads and K from the upper-most layer to model_grid.csv",
"_____no_output_____"
]
],
[
[
"model_grid['pre_cal_heads'] = water_table_ma.ravel()\nmodel_grid['pre_cal_K'] = Kh3d[0,:,:].ravel()\n\nif add_bedrock:\n model_grid['thk'] = model_grid.top - bot[-1,:,:].ravel() + bedrock_thk\nelse:\n model_grid['thk'] = model_grid.top - bot[-1,:,:].ravel()\n\nmodel_grid['thkR'] = model_grid.thk / model_grid.recharge\n \nmodel_grid.to_csv(os.path.join(model_ws, 'model_grid.csv'))",
"_____no_output_____"
]
],
[
[
"Save zone array for use in calibration.",
"_____no_output_____"
]
],
[
[
"zone_file = os.path.join(model_ws, 'zone_array.npz')\nnp.savez(zone_file, zone=zones3d)",
"_____no_output_____"
]
],
[
[
"Plot a cross-section to see what the layers look like. Change row_to_plot to see other rows. Columns could be easily added.",
"_____no_output_____"
]
],
[
[
"def calc_error(top, head, obs_type):\n # an offset of 1 is used to eliminate counting heads that\n # are within 1 m of their target as errors.\n # count topo and hydro errors\n t = top < (head - err_tol)\n h = top > (head + err_tol)\n\n tmp_df = pd.DataFrame({'head':head, 'ot':obs_type, 't':t, 'h':h})\n\n tmp = tmp_df.groupby('ot').sum()\n h_e_ = tmp.loc['hydro', 'h']\n t_e_ = tmp.loc['topo', 't']\n result = np.array([h_e_, t_e_])\n return result",
"_____no_output_____"
],
[
"hydro, topo = calc_error(model_grid.top, water_table.ravel(), model_grid.obs_type)\nnum_hydro = model_grid.obs_type.value_counts()['hydro']\nnum_topo = model_grid.obs_type.value_counts()['topo']\nnum_cells = num_hydro + num_topo\nhydro = hydro / num_hydro\ntopo = topo / num_topo",
"_____no_output_____"
],
[
"def ma2(data2D):\n return np.ma.MaskedArray(data2D, mask=inactive)\n\ndef ma3(data3D):\n return np.ma.MaskedArray(data3D, mask=(ibound == 0))\n\nrow_to_plot = NROW / 2\nxplot = np.linspace( L / 2, NCOL * L - L / 2, NCOL)\n\nmKh = ma3(Kh3d)\nmtop = ma2(top)\nmbed = ma2(bedrock)\nmbot = ma3(bot)\n\ncolors = ['green', 'red', 'gray']\n\nfig = plt.figure(figsize=(8,8))\n\nax1 = plt.subplot2grid((3, 1), (0, 0), rowspan=2)\n\nax1.plot(xplot, mtop[row_to_plot, ], label='land surface', color='black', lw=0.5)\nax1.plot(xplot, water_table_ma[row_to_plot, ], label='water table', color='blue', lw=1.)\nax1.fill_between(xplot, mtop[row_to_plot, ], mbot[0, row_to_plot, :], alpha=0.25, \n color='blue', label='layer 1', lw=0.75)\nfor lay in range(NLAY-1):\n label = 'layer {}'.format(lay+2)\n ax1.fill_between(xplot, mbot[lay, row_to_plot, :], mbot[lay+1, row_to_plot, :], label=label, \n color=colors[lay], alpha=0.250, lw=0.75)\nax1.plot(xplot, mbed[row_to_plot, :], label='bedrock (Soller)', color='red', linestyle='dotted', lw=1.5)\nax1.plot(xplot, mbot[-1, row_to_plot, :], color='black', linestyle='solid', lw=0.5)\nax1.legend(loc=0, frameon=False, fontsize=10, ncol=3)#, bbox_to_anchor=(1.0, 0.5))\nax1.set_ylabel('Altitude, in meters')\nax1.set_xticklabels('')\nax1.set_title('Default section along row {}, {} model, weight {:0.1f}\\nK fine = {:0.1f} K coarse = {:0.1f}\\\n K bedrock = {:0.1f}\\nFraction dry drains {:0.2f} Fraction flooded cells {:0.2f}'.format(row_to_plot, \\\n md, 1, K_dict['K_fine'], K_dict['K_coarse'], K_dict['K_bedrock'], hydro, topo))\n\nax2 = plt.subplot2grid((3, 1), (2, 0))\nax2.fill_between(xplot, 0, mKh[0, row_to_plot, :], alpha=0.25, color='blue', \n label='layer 1', lw=0.75, step='mid')\nax2.set_xlabel('Distance in meters')\nax2.set_yscale('log')\nax2.set_ylabel('Hydraulic conductivity\\n in layer 1, in meters / day')\n\nline = '{}_{}_xs.png'.format(md, scenario_dir)\nfig_name = os.path.join(model_ws, line)\nplt.savefig(fig_name)",
"_____no_output_____"
],
[
"t = top < (water_table - err_tol)\nh = top > (water_table + err_tol)\n\nmt = np.ma.MaskedArray(t.reshape(NROW, NCOL), model_grid.obs_type != 'topo')\nmh = np.ma.MaskedArray(h.reshape(NROW, NCOL), model_grid.obs_type != 'hydro')",
"_____no_output_____"
],
[
"from matplotlib import colors\n\ncmap = colors.ListedColormap(['0.50', 'red'])\ncmap2 = colors.ListedColormap(['blue'])\n\nback = np.ma.MaskedArray(ibound[0,:,:], ibound[0,:,:] == 0)\nfig, ax = plt.subplots(1,2)\nax[0].imshow(back, cmap=cmap2, alpha=0.2)\nim0 = ax[0].imshow(mh, cmap=cmap, interpolation='None')\nax[0].axhline(row_to_plot)\n# fig.colorbar(im0, ax=ax[0])\nax[1].imshow(back, cmap=cmap2, alpha=0.2)\nim1 = ax[1].imshow(mt, cmap=cmap, interpolation='None')\nax[1].axhline(row_to_plot)\n# fig.colorbar(im1, ax=ax[1])\nfig.suptitle('Default model errors (in red) along row {}, {} model, weight {:0.1f}\\nK fine = {:0.1f} K coarse = {:0.1f}\\\n K bedrock = {:0.1f}\\nFraction dry drains {:0.2f} Fraction flooded cells {:0.2f}'.format(row_to_plot, \\\n md, 1.0, K_dict['K_fine'], K_dict['K_coarse'], K_dict['K_bedrock'], hydro, topo))\n\n# fig.subplots_adjust(left=None, bottom=None, right=None, top=None,\n# wspace=None, hspace=None)\n\nfig.set_size_inches(6, 6)\n\n# line = '{}_{}_error_map_cal.png'.format(md, scenario_dir)\nline = '{}_{}_error_map.png'.format(md, scenario_dir) #csc\nfig_name = os.path.join(model_ws, line)\nplt.savefig(fig_name)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cb44f28e26930f2299a4fd0db235358297458390 | 23,312 | ipynb | Jupyter Notebook | Data_Modeling/Creating_Denormalized_Tables.ipynb | annadutkiewicz/Data_Engineering_Nanodegree | 976ca0f8586669313826535594a9ceb4bd5099ee | [
"MIT"
] | null | null | null | Data_Modeling/Creating_Denormalized_Tables.ipynb | annadutkiewicz/Data_Engineering_Nanodegree | 976ca0f8586669313826535594a9ceb4bd5099ee | [
"MIT"
] | null | null | null | Data_Modeling/Creating_Denormalized_Tables.ipynb | annadutkiewicz/Data_Engineering_Nanodegree | 976ca0f8586669313826535594a9ceb4bd5099ee | [
"MIT"
] | null | null | null | 31.545332 | 313 | 0.496525 | [
[
[
"# Lesson 2 Exercise 2: Creating Denormalized Tables\n\n<img src=\"images/postgresSQLlogo.png\" width=\"250\" height=\"250\">",
"_____no_output_____"
],
[
"## Walk through the basics of modeling data from normalized from to denormalized form. We will create tables in PostgreSQL, insert rows of data, and do simple JOIN SQL queries to show how these multiple tables can work together. \n\n#### Where you see ##### you will need to fill in code. This exercise will be more challenging than the last. Use the information provided to create the tables and write the insert statements.\n\n#### Remember the examples shown are simple, but imagine these situations at scale with large datasets, many users, and the need for quick response time. \n\nNote: __Do not__ click the blue Preview button in the lower task bar",
"_____no_output_____"
],
[
"### Import the library \nNote: An error might popup after this command has exectuted. If it does read it careful before ignoring. ",
"_____no_output_____"
]
],
[
[
"import psycopg2",
"_____no_output_____"
]
],
[
[
"### Create a connection to the database, get a cursor, and set autocommit to true",
"_____no_output_____"
]
],
[
[
"try: \n conn = psycopg2.connect(\"host=127.0.0.1 dbname=studentdb user=student password=student\")\nexcept psycopg2.Error as e: \n print(\"Error: Could not make connection to the Postgres database\")\n print(e)\ntry: \n cur = conn.cursor()\nexcept psycopg2.Error as e: \n print(\"Error: Could not get cursor to the Database\")\n print(e)\nconn.set_session(autocommit=True)",
"_____no_output_____"
]
],
[
[
"#### Let's start with our normalized (3NF) database set of tables we had in the last exercise, but we have added a new table `sales`. \n\n`Table Name: transactions2 \ncolumn 0: transaction Id\ncolumn 1: Customer Name\ncolumn 2: Cashier Id\ncolumn 3: Year `\n\n`Table Name: albums_sold\ncolumn 0: Album Id\ncolumn 1: Transaction Id\ncolumn 3: Album Name` \n\n`Table Name: employees\ncolumn 0: Employee Id\ncolumn 1: Employee Name `\n\n`Table Name: sales\ncolumn 0: Transaction Id\ncolumn 1: Amount Spent\n`\n<img src=\"images/table16.png\" width=\"450\" height=\"450\"> <img src=\"images/table15.png\" width=\"450\" height=\"450\"> <img src=\"images/table17.png\" width=\"350\" height=\"350\"> <img src=\"images/table18.png\" width=\"350\" height=\"350\">\n",
"_____no_output_____"
],
[
"### TO-DO: Add all Create statements for all Tables and Insert data into the tables",
"_____no_output_____"
]
],
[
[
"# TO-DO: Add all Create statements for all tables\ntry: \n cur.execute(\"CREATE TABLE IF NOT EXISTS transactions2 (transaction_id int, \\\n customer_name varchar, cashier_id int, \\\n year int);\")\nexcept psycopg2.Error as e: \n print(\"Error: Issue creating table\")\n print (e)\n\ntry: \n cur.execute(\"CREATE TABLE IF NOT EXISTS employees (employee_id int, \\\n employee_name varchar);\")\nexcept psycopg2.Error as e: \n print(\"Error: Issue creating table\")\n print (e)\n\ntry: \n cur.execute(\"CREATE TABLE IF NOT EXISTS albums_sold (album_id int, transaction_id int, \\\n album_name varchar);\")\nexcept psycopg2.Error as e: \n print(\"Error: Issue creating table\")\n print (e)\n\ntry: \n cur.execute(\"CREATE TABLE IF NOT EXISTS sales (transaction_id int, amount_spent int);\")\nexcept psycopg2.Error as e: \n print(\"Error: Issue creating table\")\n print (e)\n \n# TO-DO: Insert data into the tables \n \n \n \ntry: \n cur.execute(\"INSERT INTO transactions2 (transaction_id, customer_name, cashier_id, year) \\\n VALUES (%s, %s, %s, %s)\", \\\n (1, \"Amanda\", 1, 2000))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e)\n\ntry: \n cur.execute(\"INSERT INTO transactions2 (transaction_id, customer_name, cashier_id, year) \\\n VALUES (%s, %s, %s, %s)\", \\\n (2, \"Toby\", 1, 2000))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e)\n \ntry: \n cur.execute(\"INSERT INTO transactions2 (transaction_id, customer_name, cashier_id, year) \\\n VALUES (%s, %s, %s, %s)\", \\\n (3, \"Max\", 2, 2018))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e)\n \ntry: \n cur.execute(\"INSERT INTO albums_sold (album_id, transaction_id, album_name) \\\n VALUES (%s, %s, %s)\", \\\n (1, 1, \"Rubber Soul\"))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e)\n\ntry: \n cur.execute(\"INSERT INTO albums_sold (album_id, transaction_id, album_name) \\\n VALUES (%s, %s, %s)\", \\\n (2, 1, \"Let It Be\"))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e)\n \ntry: \n cur.execute(\"INSERT INTO albums_sold (album_id, transaction_id, album_name) \\\n VALUES (%s, %s, %s)\", \\\n (3, 2, \"My Generation\"))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e)\n \ntry: \n cur.execute(\"INSERT INTO albums_sold (album_id, transaction_id, album_name) \\\n VALUES (%s, %s, %s)\", \\\n (4, 3, \"Meet the Beatles\"))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e)\n\ntry: \n cur.execute(\"INSERT INTO albums_sold (album_id, transaction_id, album_name) \\\n VALUES (%s, %s, %s)\", \\\n (5, 3, \"Help!\"))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e)\n\ntry: \n cur.execute(\"INSERT INTO employees (employee_id, employee_name) \\\n VALUES (%s, %s)\", \\\n (1, \"Sam\"))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e)\n\ntry: \n cur.execute(\"INSERT INTO employees (employee_id, employee_name) \\\n VALUES (%s, %s)\", \\\n (2, \"Bob\"))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e) \n \ntry: \n cur.execute(\"INSERT INTO sales (transaction_id, amount_spent) \\\n VALUES (%s, %s)\", \\\n (1, 40))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e) \n \ntry: \n cur.execute(\"INSERT INTO sales (transaction_id, amount_spent) \\\n VALUES (%s, %s)\", \\\n (2, 19))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e) \n\ntry: \n cur.execute(\"INSERT INTO sales (transaction_id, amount_spent) \\\n VALUES (%s, %s)\", \\\n (3, 45))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e) ",
"_____no_output_____"
]
],
[
[
"#### TO-DO: Confirm using the Select statement the data were added correctly",
"_____no_output_____"
]
],
[
[
"print(\"Table: transactions2\\n\")\ntry: \n cur.execute(\"SELECT * FROM transactions2;\")\nexcept psycopg2.Error as e: \n print(\"Error: select *\")\n print (e)\n\nrow = cur.fetchone()\nwhile row:\n print(row)\n row = cur.fetchone()\n\nprint(\"\\nTable: albums_sold\\n\")\ntry: \n cur.execute(\"SELECT * FROM albums_sold;\")\nexcept psycopg2.Error as e: \n print(\"Error: select *\")\n print (e)\n\nrow = cur.fetchone()\nwhile row:\n print(row)\n row = cur.fetchone()\n\nprint(\"\\nTable: employees\\n\")\ntry: \n cur.execute(\"SELECT * FROM employees;\")\nexcept psycopg2.Error as e: \n print(\"Error: select *\")\n print (e)\n\nrow = cur.fetchone()\nwhile row:\n print(row)\n row = cur.fetchone()\n \nprint(\"\\nTable: sales\\n\")\ntry: \n cur.execute(\"SELECT * FROM sales;\")\nexcept psycopg2.Error as e: \n print(\"Error: select *\")\n print (e)\n\nrow = cur.fetchone()\nwhile row:\n print(row)\n row = cur.fetchone()",
"Table: transactions2\n\n(1, 'Amanda', 1, 2000)\n(2, 'Toby', 1, 2000)\n(3, 'Max', 2, 2018)\n\nTable: albums_sold\n\n(1, 1, 'Rubber Soul')\n(2, 1, 'Let It Be')\n(3, 2, 'My Generation')\n(4, 3, 'Meet the Beatles')\n(5, 3, 'Help!')\n\nTable: employees\n\n(1, 'Sam')\n(2, 'Bob')\n\nTable: sales\n\n(1, 40)\n(2, 19)\n(3, 45)\n"
]
],
[
[
"### Let's say you need to do a query that gives:\n\n`transaction_id\n customer_name\n cashier name\n year \n albums sold\n amount sold` \n\n### TO-DO: Complete the statement below to perform a 3 way `JOIN` on the 4 tables you have created. ",
"_____no_output_____"
]
],
[
[
"try: \n cur.execute(\"SELECT transactions2.transaction_id, customer_name, employees.employee_name, \\\n year, albums_sold.album_name, sales.amount_spent\\\n FROM ((transactions2 JOIN employees ON \\\n transactions2.cashier_id = employees.employee_id) JOIN \\\n albums_sold ON albums_sold.transaction_id=transactions2.transaction_id) JOIN\\\n sales ON transactions2.transaction_id=sales.transaction_id;\")\n \nexcept psycopg2.Error as e: \n print(\"Error: select *\")\n print (e)\n\nrow = cur.fetchone()\nwhile row:\n print(row)\n row = cur.fetchone()",
"(1, 'Amanda', 'Sam', 2000, 'Rubber Soul', 40)\n(1, 'Amanda', 'Sam', 2000, 'Let It Be', 40)\n(2, 'Toby', 'Sam', 2000, 'My Generation', 19)\n(3, 'Max', 'Bob', 2018, 'Meet the Beatles', 45)\n(3, 'Max', 'Bob', 2018, 'Help!', 45)\n"
]
],
[
[
"#### Great we were able to get the data we wanted.\n\n### But, we had to perform a 3 way `JOIN` to get there. While it's great we had that flexibility, we need to remember that `JOINS` are slow and if we have a read heavy workload that required low latency queries we want to reduce the number of `JOINS`. Let's think about denormalizing our normalized tables.",
"_____no_output_____"
],
[
"### With denormalization you want to think about the queries you are running and how to reduce the number of JOINS even if that means duplicating data. The following are the queries you need to run.",
"_____no_output_____"
],
[
"#### Query 1 : `select transaction_id, customer_name, amount_spent FROM <min number of tables>` \nIt should generate the amount spent on each transaction \n#### Query 2: `select cashier_name, SUM(amount_spent) FROM <min number of tables> GROUP BY cashier_name` \nIt should generate the total sales by cashier ",
"_____no_output_____"
],
[
"### Query 1: `select transaction_id, customer_name, amount_spent FROM <min number of tables>`\n\nOne way to do this would be to do a JOIN on the `sales` and `transactions2` table but we want to minimize the use of `JOINS`. \n\nTo reduce the number of tables, first add `amount_spent` to the `transactions` table so that you will not need to do a JOIN at all. \n\n`Table Name: transactions \ncolumn 0: transaction Id\ncolumn 1: Customer Name\ncolumn 2: Cashier Id\ncolumn 3: Year\ncolumn 4: amount_spent`\n\n<img src=\"images/table19.png\" width=\"450\" height=\"450\">\n",
"_____no_output_____"
],
[
"### TO-DO: Add the tables as part of the denormalization process",
"_____no_output_____"
]
],
[
[
"# TO-DO: Create all tables\ntry: \n cur.execute(\"CREATE TABLE IF NOT EXISTS transactions (transaction_id int, \\\n customer_name varchar, cashier_id int, \\\n year int, amount_spent int);\")\nexcept psycopg2.Error as e: \n print(\"Error: Issue creating table\")\n print (e)\n\n\n\n#Insert data into all tables \n \ntry: \n cur.execute(\"INSERT INTO transactions (transaction_id, customer_name, cashier_id, year, amount_spent) \\\n VALUES (%s, %s, %s, %s, %s)\", \\\n (1, \"Amanda\", 1, 2000, 40))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e)\n \ntry: \n cur.execute(\"INSERT INTO transactions (transaction_id, customer_name, cashier_id, year, amount_spent) \\\n VALUES (%s, %s, %s, %s, %s)\", \\\n (2, \"Toby\", 1, 2000, 19))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e)\n \ntry: \n cur.execute(\"INSERT INTO transactions (transaction_id, customer_name, cashier_id, year, amount_spent) \\\n VALUES (%s, %s, %s, %s, %s)\", \\\n (3, \"Max\", 2, 2018, 45))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e)",
"_____no_output_____"
]
],
[
[
"### Now you should be able to do a simplifed query to get the information you need. No `JOIN` is needed.",
"_____no_output_____"
]
],
[
[
"try: \n cur.execute(\"SELECT transaction_id, customer_name, amount_spent FROM transactions;\")\n \nexcept psycopg2.Error as e: \n print(\"Error: select *\")\n print (e)\n\nrow = cur.fetchone()\nwhile row:\n print(row)\n row = cur.fetchone()",
"(1, 'Amanda', 40)\n(2, 'Toby', 19)\n(3, 'Max', 45)\n"
]
],
[
[
"#### Your output for the above cell should be the following:\n(1, 'Amanda', 40)<br>\n(2, 'Toby', 19)<br>\n(3, 'Max', 45)",
"_____no_output_____"
],
[
"### Query 2: `select cashier_name, SUM(amount_spent) FROM <min number of tables> GROUP BY cashier_name` \n\nTo avoid using any `JOINS`, first create a new table with just the information we need. \n\n`Table Name: cashier_sales\ncol: Transaction Id\nCol: Cashier Name\nCol: Cashier Id\ncol: Amount_Spent\n`\n\n<img src=\"images/table20.png\" width=\"350\" height=\"350\">\n\n### TO-DO: Create a new table with just the information you need.",
"_____no_output_____"
]
],
[
[
"try: \n cur.execute(\"CREATE TABLE IF NOT EXISTS cashier_sales (transaction_id int, cashier_name varchar, \\\n cashier_id int, amount_spent int);\")\nexcept psycopg2.Error as e: \n print(\"Error: Issue creating table\")\n print (e)\n\n\n#Insert into all tables \n \ntry: \n cur.execute(\"INSERT INTO cashier_sales (transaction_id, cashier_name, cashier_id, amount_spent) \\\n VALUES (%s, %s, %s, %s)\", \\\n (1, \"Sam\", 1, 40 ))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e)\n\ntry: \n cur.execute(\"INSERT INTO cashier_sales (transaction_id, cashier_name, cashier_id, amount_spent) \\\n VALUES (%s, %s, %s, %s)\", \\\n (2, \"Sam\", 1, 19 ))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e)\n\ntry: \n cur.execute(\"INSERT INTO cashier_sales (transaction_id, cashier_name, cashier_id, amount_spent) \\\n VALUES (%s, %s, %s, %s)\", \\\n (3, \"Max\", 2, 45))\nexcept psycopg2.Error as e: \n print(\"Error: Inserting Rows\")\n print (e)",
"_____no_output_____"
]
],
[
[
"### Run the query",
"_____no_output_____"
]
],
[
[
"try: \n cur.execute(\"select cashier_name, SUM(amount_spent) FROM cashier_sales GROUP BY cashier_name;\")\n \nexcept psycopg2.Error as e: \n print(\"Error: select *\")\n print (e)\n\nrow = cur.fetchone()\nwhile row:\n print(row)\n row = cur.fetchone()",
"('Sam', 59)\n('Max', 45)\n"
]
],
[
[
"#### Your output for the above cell should be the following:\n('Sam', 59)<br>\n('Max', 45)\n",
"_____no_output_____"
],
[
"#### We have successfully taken normalized table and denormalized them inorder to speed up our performance and allow for simplier queries to be executed. ",
"_____no_output_____"
],
[
"### Drop the tables",
"_____no_output_____"
]
],
[
[
"try: \n cur.execute(\"DROP table albums_sold\")\nexcept psycopg2.Error as e: \n print(\"Error: Dropping table\")\n print (e)\ntry: \n cur.execute(\"DROP table employees\")\nexcept psycopg2.Error as e: \n print(\"Error: Dropping table\")\n print (e)\ntry: \n cur.execute(\"DROP table transactions\")\nexcept psycopg2.Error as e: \n print(\"Error: Dropping table\")\n print (e)\ntry: \n cur.execute(\"DROP table transactions2\")\nexcept psycopg2.Error as e: \n print(\"Error: Dropping table\")\n print (e)\ntry: \n cur.execute(\"DROP table sales\")\nexcept psycopg2.Error as e: \n print(\"Error: Dropping table\")\n print (e)\ntry: \n cur.execute(\"DROP table cashier_sales\")\nexcept psycopg2.Error as e: \n print(\"Error: Dropping table\")\n print (e)",
"_____no_output_____"
]
],
[
[
"### And finally close your cursor and connection. ",
"_____no_output_____"
]
],
[
[
"cur.close()\nconn.close()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4516f8342f8e871c6ef90738580e950104dc21 | 17,627 | ipynb | Jupyter Notebook | notebook/.ipynb_checkpoints/advanced_tutorial-checkpoint.ipynb | stevehamwu/EmotionCauseExtraction | b5a160f35f7b03bf3730b6885096dbc5f958df8b | [
"MIT"
] | 3 | 2022-02-07T12:08:38.000Z | 2022-03-28T04:26:39.000Z | notebook/.ipynb_checkpoints/advanced_tutorial-checkpoint.ipynb | stevehamwu/EmotionCauseExtraction | b5a160f35f7b03bf3730b6885096dbc5f958df8b | [
"MIT"
] | null | null | null | notebook/.ipynb_checkpoints/advanced_tutorial-checkpoint.ipynb | stevehamwu/EmotionCauseExtraction | b5a160f35f7b03bf3730b6885096dbc5f958df8b | [
"MIT"
] | null | null | null | 151.956897 | 6,534 | 0.672434 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\nAdvanced: Making Dynamic Decisions and the Bi-LSTM CRF\n======================================================\n\nDynamic versus Static Deep Learning Toolkits\n--------------------------------------------\n\nPytorch is a *dynamic* neural network kit. Another example of a dynamic\nkit is `Dynet <https://github.com/clab/dynet>`__ (I mention this because\nworking with Pytorch and Dynet is similar. If you see an example in\nDynet, it will probably help you implement it in Pytorch). The opposite\nis the *static* tool kit, which includes Theano, Keras, TensorFlow, etc.\nThe core difference is the following:\n\n* In a static toolkit, you define\n a computation graph once, compile it, and then stream instances to it.\n* In a dynamic toolkit, you define a computation graph *for each\n instance*. It is never compiled and is executed on-the-fly\n\nWithout a lot of experience, it is difficult to appreciate the\ndifference. One example is to suppose we want to build a deep\nconstituent parser. Suppose our model involves roughly the following\nsteps:\n\n* We build the tree bottom up\n* Tag the root nodes (the words of the sentence)\n* From there, use a neural network and the embeddings\n of the words to find combinations that form constituents. Whenever you\n form a new constituent, use some sort of technique to get an embedding\n of the constituent. In this case, our network architecture will depend\n completely on the input sentence. In the sentence \"The green cat\n scratched the wall\", at some point in the model, we will want to combine\n the span $(i,j,r) = (1, 3, \\text{NP})$ (that is, an NP constituent\n spans word 1 to word 3, in this case \"The green cat\").\n\nHowever, another sentence might be \"Somewhere, the big fat cat scratched\nthe wall\". In this sentence, we will want to form the constituent\n$(2, 4, NP)$ at some point. The constituents we will want to form\nwill depend on the instance. If we just compile the computation graph\nonce, as in a static toolkit, it will be exceptionally difficult or\nimpossible to program this logic. In a dynamic toolkit though, there\nisn't just 1 pre-defined computation graph. There can be a new\ncomputation graph for each instance, so this problem goes away.\n\nDynamic toolkits also have the advantage of being easier to debug and\nthe code more closely resembling the host language (by that I mean that\nPytorch and Dynet look more like actual Python code than Keras or\nTheano).\n\nBi-LSTM Conditional Random Field Discussion\n-------------------------------------------\n\nFor this section, we will see a full, complicated example of a Bi-LSTM\nConditional Random Field for named-entity recognition. The LSTM tagger\nabove is typically sufficient for part-of-speech tagging, but a sequence\nmodel like the CRF is really essential for strong performance on NER.\nFamiliarity with CRF's is assumed. Although this name sounds scary, all\nthe model is is a CRF but where an LSTM provides the features. This is\nan advanced model though, far more complicated than any earlier model in\nthis tutorial. If you want to skip it, that is fine. To see if you're\nready, see if you can:\n\n- Write the recurrence for the viterbi variable at step i for tag k.\n- Modify the above recurrence to compute the forward variables instead.\n- Modify again the above recurrence to compute the forward variables in\n log-space (hint: log-sum-exp)\n\nIf you can do those three things, you should be able to understand the\ncode below. Recall that the CRF computes a conditional probability. Let\n$y$ be a tag sequence and $x$ an input sequence of words.\nThen we compute\n\n\\begin{align}P(y|x) = \\frac{\\exp{(\\text{Score}(x, y)})}{\\sum_{y'} \\exp{(\\text{Score}(x, y')})}\\end{align}\n\nWhere the score is determined by defining some log potentials\n$\\log \\psi_i(x,y)$ such that\n\n\\begin{align}\\text{Score}(x,y) = \\sum_i \\log \\psi_i(x,y)\\end{align}\n\nTo make the partition function tractable, the potentials must look only\nat local features.\n\nIn the Bi-LSTM CRF, we define two kinds of potentials: emission and\ntransition. The emission potential for the word at index $i$ comes\nfrom the hidden state of the Bi-LSTM at timestep $i$. The\ntransition scores are stored in a $|T|x|T|$ matrix\n$\\textbf{P}$, where $T$ is the tag set. In my\nimplementation, $\\textbf{P}_{j,k}$ is the score of transitioning\nto tag $j$ from tag $k$. So:\n\n\\begin{align}\\text{Score}(x,y) = \\sum_i \\log \\psi_\\text{EMIT}(y_i \\rightarrow x_i) + \\log \\psi_\\text{TRANS}(y_{i-1} \\rightarrow y_i)\\end{align}\n\n\\begin{align}= \\sum_i h_i[y_i] + \\textbf{P}_{y_i, y_{i-1}}\\end{align}\n\nwhere in this second expression, we think of the tags as being assigned\nunique non-negative indices.\n\nIf the above discussion was too brief, you can check out\n`this <http://www.cs.columbia.edu/%7Emcollins/crf.pdf>`__ write up from\nMichael Collins on CRFs.\n\nImplementation Notes\n--------------------\n\nThe example below implements the forward algorithm in log space to\ncompute the partition function, and the viterbi algorithm to decode.\nBackpropagation will compute the gradients automatically for us. We\ndon't have to do anything by hand.\n\nThe implementation is not optimized. If you understand what is going on,\nyou'll probably quickly see that iterating over the next tag in the\nforward algorithm could probably be done in one big operation. I wanted\nto code to be more readable. If you want to make the relevant change,\nyou could probably use this tagger for real tasks.\n\n",
"_____no_output_____"
]
],
[
[
"# Author: Robert Guthrie\n\nimport torch\nimport torch.autograd as autograd\nimport torch.nn as nn\nimport torch.optim as optim\n\ntorch.manual_seed(1)",
"_____no_output_____"
]
],
[
[
"Helper functions to make the code more readable.\n\n",
"_____no_output_____"
]
],
[
[
"def to_scalar(var):\n # returns a python float\n return var.view(-1).data.tolist()[0]\n\n\ndef argmax(vec):\n # return the argmax as a python int\n _, idx = torch.max(vec, 1)\n return to_scalar(idx)\n\n\ndef prepare_sequence(seq, to_ix):\n idxs = [to_ix[w] for w in seq]\n tensor = torch.LongTensor(idxs)\n return autograd.Variable(tensor)\n\n\n# Compute log sum exp in a numerically stable way for the forward algorithm\ndef log_sum_exp(vec):\n max_score = vec[0, argmax(vec)]\n max_score_broadcast = max_score.view(1, -1).expand(1, vec.size()[1])\n return max_score + \\\n torch.log(torch.sum(torch.exp(vec - max_score_broadcast)))",
"_____no_output_____"
]
],
[
[
"Create model\n\n",
"_____no_output_____"
]
],
[
[
"class BiLSTM_CRF(nn.Module):\n\n def __init__(self, vocab_size, tag_to_ix, embedding_dim, hidden_dim):\n super(BiLSTM_CRF, self).__init__()\n self.embedding_dim = embedding_dim\n self.hidden_dim = hidden_dim\n self.vocab_size = vocab_size\n self.tag_to_ix = tag_to_ix\n self.tagset_size = len(tag_to_ix)\n\n self.word_embeds = nn.Embedding(vocab_size, embedding_dim)\n self.lstm = nn.LSTM(embedding_dim, hidden_dim // 2,\n num_layers=1, bidirectional=True)\n\n # Maps the output of the LSTM into tag space.\n self.hidden2tag = nn.Linear(hidden_dim, self.tagset_size)\n\n # Matrix of transition parameters. Entry i,j is the score of\n # transitioning *to* i *from* j.\n self.transitions = nn.Parameter(\n torch.randn(self.tagset_size, self.tagset_size))\n\n # These two statements enforce the constraint that we never transfer\n # to the start tag and we never transfer from the stop tag \n self.transitions.data[tag_to_ix[START_TAG], :] = -10000\n self.transitions.data[:, tag_to_ix[STOP_TAG]] = -10000\n\n self.hidden = self.init_hidden()\n\n def init_hidden(self):\n return (autograd.Variable(torch.randn(2, 1, self.hidden_dim // 2)),\n autograd.Variable(torch.randn(2, 1, self.hidden_dim // 2)))\n\n def _forward_alg(self, feats):\n # Do the forward algorithm to compute the partition function\n init_alphas = torch.Tensor(1, self.tagset_size).fill_(-10000.)\n # START_TAG has all of the score.\n init_alphas[0][self.tag_to_ix[START_TAG]] = 0.\n\n # Wrap in a variable so that we will get automatic backprop\n forward_var = autograd.Variable(init_alphas)\n\n # Iterate through the sentence\n for feat in feats:\n alphas_t = [] # The forward variables at this timestep\n for next_tag in range(self.tagset_size):\n # broadcast the emission score: it is the same regardless of\n # the previous tag\n emit_score = feat[next_tag].view(\n 1, -1).expand(1, self.tagset_size)\n # the ith entry of trans_score is the score of transitioning to\n # next_tag from i\n trans_score = self.transitions[next_tag].view(1, -1)\n # The ith entry of next_tag_var is the value for the\n # edge (i -> next_tag) before we do log-sum-exp\n next_tag_var = forward_var + trans_score + emit_score\n # The forward variable for this tag is log-sum-exp of all the\n # scores.\n alphas_t.append(log_sum_exp(next_tag_var))\n forward_var = torch.cat(alphas_t).view(1, -1)\n terminal_var = forward_var + self.transitions[self.tag_to_ix[STOP_TAG]]\n alpha = log_sum_exp(terminal_var)\n return alpha\n\n def _get_lstm_features(self, sentence):\n self.hidden = self.init_hidden()\n embeds = self.word_embeds(sentence).view(len(sentence), 1, -1)\n lstm_out, self.hidden = self.lstm(embeds, self.hidden)\n lstm_out = lstm_out.view(len(sentence), self.hidden_dim)\n lstm_feats = self.hidden2tag(lstm_out)\n return lstm_feats\n\n def _score_sentence(self, feats, tags):\n # Gives the score of a provided tag sequence\n score = autograd.Variable(torch.Tensor([0]))\n tags = torch.cat([torch.LongTensor([self.tag_to_ix[START_TAG]]), tags])\n for i, feat in enumerate(feats):\n score = score + \\\n self.transitions[tags[i + 1], tags[i]] + feat[tags[i + 1]]\n score = score + self.transitions[self.tag_to_ix[STOP_TAG], tags[-1]]\n return score\n\n def _viterbi_decode(self, feats):\n backpointers = []\n\n # Initialize the viterbi variables in log space\n init_vvars = torch.Tensor(1, self.tagset_size).fill_(-10000.)\n init_vvars[0][self.tag_to_ix[START_TAG]] = 0\n\n # forward_var at step i holds the viterbi variables for step i-1\n forward_var = autograd.Variable(init_vvars)\n for feat in feats:\n bptrs_t = [] # holds the backpointers for this step\n viterbivars_t = [] # holds the viterbi variables for this step\n\n for next_tag in range(self.tagset_size):\n # next_tag_var[i] holds the viterbi variable for tag i at the\n # previous step, plus the score of transitioning\n # from tag i to next_tag.\n # We don't include the emission scores here because the max\n # does not depend on them (we add them in below)\n next_tag_var = forward_var + self.transitions[next_tag]\n best_tag_id = argmax(next_tag_var)\n bptrs_t.append(best_tag_id)\n viterbivars_t.append(next_tag_var[0][best_tag_id])\n # Now add in the emission scores, and assign forward_var to the set\n # of viterbi variables we just computed\n forward_var = (torch.cat(viterbivars_t) + feat).view(1, -1)\n backpointers.append(bptrs_t)\n\n # Transition to STOP_TAG\n terminal_var = forward_var + self.transitions[self.tag_to_ix[STOP_TAG]]\n best_tag_id = argmax(terminal_var)\n path_score = terminal_var[0][best_tag_id]\n\n # Follow the back pointers to decode the best path.\n best_path = [best_tag_id]\n for bptrs_t in reversed(backpointers):\n best_tag_id = bptrs_t[best_tag_id]\n best_path.append(best_tag_id)\n # Pop off the start tag (we dont want to return that to the caller)\n start = best_path.pop()\n assert start == self.tag_to_ix[START_TAG] # Sanity check\n best_path.reverse()\n return path_score, best_path\n\n def neg_log_likelihood(self, sentence, tags):\n feats = self._get_lstm_features(sentence)\n forward_score = self._forward_alg(feats)\n gold_score = self._score_sentence(feats, tags)\n return forward_score - gold_score\n\n def forward(self, sentence): # dont confuse this with _forward_alg above.\n # Get the emission scores from the BiLSTM\n lstm_feats = self._get_lstm_features(sentence)\n\n # Find the best path, given the features.\n score, tag_seq = self._viterbi_decode(lstm_feats)\n return score, tag_seq",
"_____no_output_____"
]
],
[
[
"Run training\n\n",
"_____no_output_____"
]
],
[
[
"START_TAG = \"<START>\"\nSTOP_TAG = \"<STOP>\"\nEMBEDDING_DIM = 5\nHIDDEN_DIM = 4\n\n# Make up some training data\ntraining_data = [(\n \"the wall street journal reported today that apple corporation made money\".split(),\n \"B I I I O O O B I O O\".split()\n), (\n \"georgia tech is a university in georgia\".split(),\n \"B I O O O O B\".split()\n)]\n\nword_to_ix = {}\nfor sentence, tags in training_data:\n for word in sentence:\n if word not in word_to_ix:\n word_to_ix[word] = len(word_to_ix)\n\ntag_to_ix = {\"B\": 0, \"I\": 1, \"O\": 2, START_TAG: 3, STOP_TAG: 4}\n\nmodel = BiLSTM_CRF(len(word_to_ix), tag_to_ix, EMBEDDING_DIM, HIDDEN_DIM)\noptimizer = optim.SGD(model.parameters(), lr=0.01, weight_decay=1e-4)\n\n# Check predictions before training\nprecheck_sent = prepare_sequence(training_data[0][0], word_to_ix)\nprecheck_tags = torch.LongTensor([tag_to_ix[t] for t in training_data[0][1]])\nprint(model(precheck_sent))\n\n# Make sure prepare_sequence from earlier in the LSTM section is loaded\nfor epoch in range(\n 300): # again, normally you would NOT do 300 epochs, it is toy data\n for sentence, tags in training_data:\n # Step 1. Remember that Pytorch accumulates gradients.\n # We need to clear them out before each instance\n model.zero_grad()\n\n # Step 2. Get our inputs ready for the network, that is,\n # turn them into Variables of word indices.\n sentence_in = prepare_sequence(sentence, word_to_ix)\n targets = torch.LongTensor([tag_to_ix[t] for t in tags])\n\n # Step 3. Run our forward pass.\n neg_log_likelihood = model.neg_log_likelihood(sentence_in, targets)\n\n # Step 4. Compute the loss, gradients, and update the parameters by\n # calling optimizer.step()\n neg_log_likelihood.backward()\n optimizer.step()\n\n# Check predictions after training\nprecheck_sent = prepare_sequence(training_data[0][0], word_to_ix)\nprint(model(precheck_sent))\n# We got it!",
"_____no_output_____"
]
],
[
[
"Exercise: A new loss function for discriminative tagging\n--------------------------------------------------------\n\nIt wasn't really necessary for us to create a computation graph when\ndoing decoding, since we do not backpropagate from the viterbi path\nscore. Since we have it anyway, try training the tagger where the loss\nfunction is the difference between the Viterbi path score and the score\nof the gold-standard path. It should be clear that this function is\nnon-negative and 0 when the predicted tag sequence is the correct tag\nsequence. This is essentially *structured perceptron*.\n\nThis modification should be short, since Viterbi and score\\_sentence are\nalready implemented. This is an example of the shape of the computation\ngraph *depending on the training instance*. Although I haven't tried\nimplementing this in a static toolkit, I imagine that it is possible but\nmuch less straightforward.\n\nPick up some real data and do a comparison!\n\n\n",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb451f44ceccc3a87438df086e560e288e35acca | 21,183 | ipynb | Jupyter Notebook | assignment2/NetworkVisualization.ipynb | AnthonyNg404/Deep-Learning | ef1dafaa1d07e9c9b574ba1722a7954c16ef463d | [
"Unlicense"
] | null | null | null | assignment2/NetworkVisualization.ipynb | AnthonyNg404/Deep-Learning | ef1dafaa1d07e9c9b574ba1722a7954c16ef463d | [
"Unlicense"
] | null | null | null | assignment2/NetworkVisualization.ipynb | AnthonyNg404/Deep-Learning | ef1dafaa1d07e9c9b574ba1722a7954c16ef463d | [
"Unlicense"
] | null | null | null | 41.535294 | 736 | 0.605816 | [
[
[
"# Network Visualization (PyTorch)\n\nIn this notebook we will explore the use of *image gradients* for generating new images.\n\nWhen training a model, we define a loss function which measures our current unhappiness with the model's performance; we then use backpropagation to compute the gradient of the loss with respect to the model parameters, and perform gradient descent on the model parameters to minimize the loss.\n\nHere we will do something slightly different. We will start from a convolutional neural network model which has been pretrained to perform image classification on the ImageNet dataset. We will use this model to define a loss function which quantifies our current unhappiness with our image, then use backpropagation to compute the gradient of this loss with respect to the pixels of the image. We will then keep the model fixed, and perform gradient descent *on the image* to synthesize a new image which minimizes the loss.\n\nIn this notebook we will explore three techniques for image generation:\n\n1. **Saliency Maps**: Saliency maps are a quick way to tell which part of the image influenced the classification decision made by the network.\n2. **Fooling Images**: We can perturb an input image so that it appears the same to humans, but will be misclassified by the pretrained network.\n3. **Class Visualization**: We can synthesize an image to maximize the classification score of a particular class; this can give us some sense of what the network is looking for when it classifies images of that class.\n\nThis notebook uses **PyTorch**;",
"_____no_output_____"
]
],
[
[
"import torch\nimport torchvision\nimport torchvision.transforms as T\nimport random\n\nimport numpy as np\nfrom scipy.ndimage.filters import gaussian_filter1d\nimport matplotlib.pyplot as plt\nfrom deeplearning.image_utils import SQUEEZENET_MEAN, SQUEEZENET_STD\nfrom PIL import Image\n\nfrom deeplearning.network_visualization import compute_saliency_maps, make_fooling_image, update_class_visulization\n\n%matplotlib inline\nplt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots\nplt.rcParams['image.interpolation'] = 'nearest'\nplt.rcParams['image.cmap'] = 'gray'\n\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
]
],
[
[
"### Helper Functions\n\nOur pretrained model was trained on images that had been preprocessed by subtracting the per-color mean and dividing by the per-color standard deviation. We define a few helper functions for performing and undoing this preprocessing. You don't need to do anything in this cell.",
"_____no_output_____"
]
],
[
[
"def preprocess(img, size=224):\n transform = T.Compose([\n T.Resize(size),\n T.ToTensor(),\n T.Normalize(mean=SQUEEZENET_MEAN.tolist(),\n std=SQUEEZENET_STD.tolist()),\n T.Lambda(lambda x: x[None]),\n ])\n return transform(img)\n\ndef deprocess(img, should_rescale=True):\n transform = T.Compose([\n T.Lambda(lambda x: x[0]),\n T.Normalize(mean=[0, 0, 0], std=(1.0 / SQUEEZENET_STD).tolist()),\n T.Normalize(mean=(-SQUEEZENET_MEAN).tolist(), std=[1, 1, 1]),\n T.Lambda(rescale) if should_rescale else T.Lambda(lambda x: x),\n T.ToPILImage(),\n ])\n return transform(img)\n\ndef rescale(x):\n low, high = x.min(), x.max()\n x_rescaled = (x - low) / (high - low)\n return x_rescaled\n \ndef blur_image(X, sigma=1):\n X_np = X.cpu().clone().numpy()\n X_np = gaussian_filter1d(X_np, sigma, axis=2)\n X_np = gaussian_filter1d(X_np, sigma, axis=3)\n X.copy_(torch.Tensor(X_np).type_as(X))\n return X\n\ndef rel_error(x, y):\n return torch.max(torch.abs(x - y) / (torch.maximum(torch.tensor(1e-8), torch.abs(x) + torch.abs(y))))",
"_____no_output_____"
]
],
[
[
"# Pretrained Model\n\nFor all of our image generation experiments, we will start with a convolutional neural network which was pretrained to perform image classification on ImageNet. We can use any model here, but for the purposes of this assignment we will use SqueezeNet [1], which achieves accuracies comparable to AlexNet but with a significantly reduced parameter count and computational complexity.\n\nUsing SqueezeNet rather than AlexNet or VGG or ResNet means that we can easily perform all image generation experiments on CPU.\n\n[1] Iandola et al, \"SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and < 0.5MB model size\", arXiv 2016",
"_____no_output_____"
]
],
[
[
"# Download and load the pretrained SqueezeNet model.\n# model = torchvision.models.squeezenet1_1(pretrained=True)\nmodel = torch.load('squeezenet1_1.pt')\n\n# We don't want to train the model, so tell PyTorch not to compute gradients\n# with respect to model parameters.\nfor param in model.parameters():\n param.requires_grad = False\n \nreference_data = torch.load('network_visualization_check.pt')",
"_____no_output_____"
]
],
[
[
"## Load some ImageNet images\nWe have provided a few example images from the validation set of the ImageNet ILSVRC 2012 Classification dataset. To download these images, change to `deeplearning/datasets/` and run `get_imagenet_val.sh`.\n\nSince they come from the validation set, our pretrained model did not see these images during training.\n\nRun the following cell to visualize some of these images, along with their ground-truth labels.",
"_____no_output_____"
]
],
[
[
"from deeplearning.data_utils import load_imagenet_val\nX, y, class_names = load_imagenet_val(num=5)\n\nplt.figure(figsize=(12, 6))\nfor i in range(5):\n plt.subplot(1, 5, i + 1)\n plt.imshow(X[i])\n plt.title(class_names[y[i]])\n plt.axis('off')\nplt.gcf().tight_layout()",
"_____no_output_____"
]
],
[
[
"# Saliency Maps\nUsing this pretrained model, we will compute class saliency maps as described in Section 3.1 of [2].\n\nA **saliency map** tells us the degree to which each pixel in the image affects the classification score for that image. To compute it, we compute the gradient of the unnormalized score corresponding to the correct class (which is a scalar) with respect to the pixels of the image. If the image has shape `(3, H, W)` then this gradient will also have shape `(3, H, W)`; for each pixel in the image, this gradient tells us the amount by which the classification score will change if the pixel changes by a small amount. To compute the saliency map, we take the absolute value of this gradient, then take the maximum value over the 3 input channels; the final saliency map thus has shape `(H, W)` and all entries are nonnegative.\n\n[2] Karen Simonyan, Andrea Vedaldi, and Andrew Zisserman. \"Deep Inside Convolutional Networks: Visualising\nImage Classification Models and Saliency Maps\", ICLR Workshop 2014.",
"_____no_output_____"
],
[
"### Hint: PyTorch `gather` method\nRecall in Assignment 1 you needed to select one element from each row of a matrix; if `s` is an numpy array of shape `(N, C)` and `y` is a numpy array of shape `(N,`) containing integers `0 <= y[i] < C`, then `s[np.arange(N), y]` is a numpy array of shape `(N,)` which selects one element from each element in `s` using the indices in `y`.\n\nIn PyTorch you can perform the same operation using the `gather()` method. If `s` is a PyTorch Tensor or Variable of shape `(N, C)` and `y` is a PyTorch Tensor or Variable of shape `(N,)` containing longs in the range `0 <= y[i] < C`, then\n\n`s.gather(1, y.view(-1, 1)).squeeze()`\n\nwill be a PyTorch Tensor (or Variable) of shape `(N,)` containing one entry from each row of `s`, selected according to the indices in `y`.\n\nrun the following cell to see an example.\n\nYou can also read the documentation for [the gather method](http://pytorch.org/docs/torch.html#torch.gather)\nand [the squeeze method](http://pytorch.org/docs/torch.html#torch.squeeze).",
"_____no_output_____"
]
],
[
[
"# Example of using gather to select one entry from each row in PyTorch\ndef gather_example():\n N, C = 4, 5\n s = torch.randn(N, C)\n y = torch.LongTensor([1, 2, 1, 3])\n print(s)\n print(y)\n print(s.gather(1, y.view(-1, 1)).squeeze())\ngather_example()",
"_____no_output_____"
]
],
[
[
"**Now implement the saliency map in the `compute_saliency_maps` function in the file `deeplearning/network_visualization.py`.** \n\nOnce you have tested the implementation, run the following to visualize some class saliency maps on our example images from the ImageNet validation set:",
"_____no_output_____"
]
],
[
[
"def show_saliency_maps(X, y):\n # Convert X and y from numpy arrays to Torch Tensors\n X_tensor = torch.cat([preprocess(Image.fromarray(x)) for x in X], dim=0)\n y_tensor = torch.LongTensor(y)\n\n # Compute saliency maps for images in X\n saliency = compute_saliency_maps(X_tensor, y_tensor, model)\n\n # Convert the saliency map from Torch Tensor to numpy array and show images\n # and saliency maps together.\n saliency = saliency.numpy()\n N = X.shape[0]\n for i in range(N):\n plt.subplot(2, N, i + 1)\n plt.imshow(X[i])\n plt.axis('off')\n plt.title(class_names[y[i]])\n plt.subplot(2, N, N + i + 1)\n plt.imshow(saliency[i], cmap=plt.cm.hot)\n plt.axis('off')\n plt.gcf().set_size_inches(12, 5)\n plt.show()\n\nshow_saliency_maps(X, y)",
"_____no_output_____"
]
],
[
[
"# Fooling Images\nWe can also use image gradients to generate \"fooling images\" as discussed in [3]. Given an image and a target class, we can perform gradient **ascent** over the image to maximize the target class, stopping when the network classifies the image as the target class.\n\n**Implement the fooling image generation in the `make_fooling_image` function in the file `deeplearning/network_visualization.py`.**\n\n[3] Szegedy et al, \"Intriguing properties of neural networks\", ICLR 2014\n\nRun the following cell to generate a fooling image:",
"_____no_output_____"
]
],
[
[
"target_y = reference_data['fooling_input_y']\nX_tensor = reference_data['fooling_input_X']\nX_fooling = make_fooling_image(X_tensor, target_y, model)\n\nscores = model(X_fooling)\nassert target_y == scores.data.max(dim=1)[1][0], 'The model is not fooled!'",
"_____no_output_____"
]
],
[
[
"After generating a fooling image, run the following cell to visualize the original image, the fooling image, as well as the difference between them.",
"_____no_output_____"
]
],
[
[
"idx = 0\ntarget_y = 6\n\nX_fooling_np = deprocess(X_fooling.clone())\nX_fooling_np = np.asarray(X_fooling_np).astype(np.uint8)\n\nplt.subplot(1, 4, 1)\nplt.imshow(X[idx])\nplt.title(class_names[y[idx]])\nplt.axis('off')\n\nplt.subplot(1, 4, 2)\nplt.imshow(X_fooling_np)\nplt.title(class_names[target_y])\nplt.axis('off')\n\nplt.subplot(1, 4, 3)\nX_pre = preprocess(Image.fromarray(X[idx]))\ndiff = np.asarray(deprocess(X_fooling - X_pre, should_rescale=False))\nplt.imshow(diff)\nplt.title('Difference')\nplt.axis('off')\n\nplt.subplot(1, 4, 4)\ndiff = np.asarray(deprocess(10 * (X_fooling - X_pre), should_rescale=False))\nplt.imshow(diff)\nplt.title('Magnified difference (10x)')\nplt.axis('off')\n\nplt.gcf().set_size_inches(12, 5)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Class visualization\nBy starting with a random noise image and performing gradient ascent on a target class, we can generate an image that the network will recognize as the target class. This idea was first presented in [2]; [3] extended this idea by suggesting several regularization techniques that can improve the quality of the generated image.\n\nConcretely, let $I$ be an image and let $y$ be a target class. Let $s_y(I)$ be the score that a convolutional network assigns to the image $I$ for class $y$; note that these are raw unnormalized scores, not class probabilities. We wish to generate an image $I^*$ that achieves a high score for the class $y$ by solving the problem\n\n$$\nI^* = \\arg\\max_I s_y(I) - R(I)\n$$\n\nwhere $R$ is a (possibly implicit) regularizer (note the sign of $R(I)$ in the argmax: we want to minimize this regularization term). We can solve this optimization problem using gradient ascent, computing gradients with respect to the generated image. We will use (explicit) L2 regularization of the form\n\n$$\nR(I) = \\lambda \\|I\\|_2^2\n$$\n\n**and** implicit regularization as suggested by [3] by periodically blurring the generated image. We can solve this problem using gradient ascent on the generated image.\n\n**Complete the implementation of the `update_class_visulization` function in the file `deeplearning/network_visualization.py`.**\n\n\n[2] Karen Simonyan, Andrea Vedaldi, and Andrew Zisserman. \"Deep Inside Convolutional Networks: Visualising\nImage Classification Models and Saliency Maps\", ICLR Workshop 2014.\n\n[3] Yosinski et al, \"Understanding Neural Networks Through Deep Visualization\", ICML 2015 Deep Learning Workshop",
"_____no_output_____"
],
[
"Once you have test the implementation in the cell above, run the following cell to generate an image of a Tarantula:",
"_____no_output_____"
]
],
[
[
"def jitter(X, ox, oy):\n \"\"\"\n Helper function to randomly jitter an image.\n\n Inputs\n - X: PyTorch Tensor of shape (N, C, H, W)\n - ox, oy: Integers giving number of pixels to jitter along W and H axes\n\n Returns: A new PyTorch Tensor of shape (N, C, H, W)\n \"\"\"\n if ox != 0:\n left = X[:, :, :, :-ox]\n right = X[:, :, :, -ox:]\n X = torch.cat([right, left], dim=3)\n if oy != 0:\n top = X[:, :, :-oy]\n bottom = X[:, :, -oy:]\n X = torch.cat([bottom, top], dim=2)\n return X\n\ndef create_class_visualization(target_y, model, **kwargs):\n \"\"\"\n Generate an image to maximize the score of target_y under a pretrained model.\n\n Inputs:\n - target_y: Integer in the range [0, 1000) giving the index of the class\n - model: A pretrained CNN that will be used to generate the image\n\n Keyword arguments:\n - l2_reg: Strength of L2 regularization on the image\n - learning_rate: How big of a step to take\n - num_iterations: How many iterations to use\n - blur_every: How often to blur the image as an implicit regularizer\n - max_jitter: How much to gjitter the image as an implicit regularizer\n - show_every: How often to show the intermediate result\n \"\"\"\n l2_reg = kwargs.pop('l2_reg', 1e-3)\n learning_rate = kwargs.pop('learning_rate', 25)\n num_iterations = kwargs.pop('num_iterations', 100)\n blur_every = kwargs.pop('blur_every', 10)\n max_jitter = kwargs.pop('max_jitter', 16)\n show_every = kwargs.pop('show_every', 25)\n\n # Randomly initialize the image as a PyTorch Tensor\n img = torch.randn(1, 3, 224, 224).mul_(1.0)\n for t in range(num_iterations):\n # Randomly jitter the image a bit; this gives slightly nicer results\n ox, oy = random.randint(0, max_jitter), random.randint(0, max_jitter)\n img.copy_(jitter(img, ox, oy))\n img.copy_(update_class_visulization(model, target_y, l2_reg, learning_rate, img).data)\n\n # Undo the random jitter\n img.copy_(jitter(img, -ox, -oy))\n\n # As regularizer, clamp and periodically blur the image\n for c in range(3):\n lo = float(-SQUEEZENET_MEAN[c] / SQUEEZENET_STD[c])\n hi = float((1.0 - SQUEEZENET_MEAN[c]) / SQUEEZENET_STD[c])\n img[:, c].clamp_(min=lo, max=hi)\n if t % blur_every == 0:\n blur_image(img, sigma=0.5)\n\n # Periodically show the image\n if t == 0 or (t + 1) % show_every == 0 or t == num_iterations - 1:\n plt.imshow(deprocess(img.clone().cpu()))\n class_name = class_names[target_y]\n plt.title('%s\\nIteration %d / %d' % (class_name, t + 1, num_iterations))\n plt.gcf().set_size_inches(4, 4)\n plt.axis('off')\n plt.show()\n\n return deprocess(img.cpu())",
"_____no_output_____"
],
[
"target_y = 76 # Tarantula\n# target_y = 78 # Tick\n# target_y = 187 # Yorkshire Terrier\n# target_y = 683 # Oboe\n# target_y = 366 # Gorilla\n# target_y = 604 # Hourglass\nout = create_class_visualization(target_y, model)",
"_____no_output_____"
]
],
[
[
"Try out your class visualization on other classes! You should also feel free to play with various hyperparameters to try and improve the quality of the generated image, but this is not required.",
"_____no_output_____"
]
],
[
[
"# target_y = 78 # Tick\n# target_y = 187 # Yorkshire Terrier\n# target_y = 683 # Oboe\n# target_y = 366 # Gorilla\n# target_y = 604 # Hourglass\ntarget_y = np.random.randint(1000)\nprint(class_names[target_y])\nX = create_class_visualization(target_y, model)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4527d15330e91ebb5739c0f61f71313510803a | 1,165 | ipynb | Jupyter Notebook | Python-Programming/Teaching-Material/some-practice-questions.ipynb | vivekparasharr/Learn-Programming | 1ae07ef5143bff3c504978e1d375698820f59af0 | [
"MIT"
] | null | null | null | Python-Programming/Teaching-Material/some-practice-questions.ipynb | vivekparasharr/Learn-Programming | 1ae07ef5143bff3c504978e1d375698820f59af0 | [
"MIT"
] | null | null | null | Python-Programming/Teaching-Material/some-practice-questions.ipynb | vivekparasharr/Learn-Programming | 1ae07ef5143bff3c504978e1d375698820f59af0 | [
"MIT"
] | null | null | null | 16.180556 | 46 | 0.449785 | [
[
[
"Print the fibonacci series: 1 1 2 3 5 8 ",
"_____no_output_____"
]
],
[
[
"a, b = 0, 1\nwhile b < 10:\n print(b)\n a, b = b, a+b",
"1\n1\n2\n3\n5\n8\n"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
cb452e2f870f4a815e7f032f64f6039b38b4ea1f | 39,805 | ipynb | Jupyter Notebook | sparse_logistic_regression/Sparse_Logistic_Regression.ipynb | ymalitsky/graal | c0366e96a0e6ad8463a03ab94db66ebeab0fc55e | [
"MIT"
] | 1 | 2022-02-03T16:49:33.000Z | 2022-02-03T16:49:33.000Z | sparse_logistic_regression/Sparse_Logistic_Regression.ipynb | ymalitsky/graal | c0366e96a0e6ad8463a03ab94db66ebeab0fc55e | [
"MIT"
] | null | null | null | sparse_logistic_regression/Sparse_Logistic_Regression.ipynb | ymalitsky/graal | c0366e96a0e6ad8463a03ab94db66ebeab0fc55e | [
"MIT"
] | null | null | null | 109.054795 | 29,868 | 0.833413 | [
[
[
"## Sparse logistic regression\n$\\newcommand{\\n}[1]{\\left\\|#1 \\right\\|}$ \n$\\newcommand{\\R}{\\mathbb R} $ \n$\\newcommand{\\N}{\\mathbb N} $ \n$\\newcommand{\\Z}{\\mathbb Z} $ \n$\\newcommand{\\lr}[1]{\\left\\langle #1\\right\\rangle}$\n\nWe want to minimize\n$$\\min_x J(x) := \\sum_{i=1}^m \\log\\bigl(1+\\exp (-b_i\\lr{a_i, x})\\bigr) + \\gamma \\n{x}_1$$\nwhere $(a_i, b_i)\\in \\R^n\\times \\{-1,1\\}$ is the training set and $\\gamma >0$. We can rewrite the objective as \n$J(x) = \\tilde f(Kx)+g(x)$,\nwhere $$\\tilde f(y)=\\sum_{i=1}^{} \\log (1+\\exp(y_i)), \\quad K = -b*A \\in \\R^{m\\times n}, \\quad g(x) = \\gamma \\n{x}_1$$\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport scipy.linalg as LA\nimport scipy.sparse as spr\nimport scipy.sparse.linalg as spr_LA\nfrom time import perf_counter\nfrom sklearn import datasets",
"_____no_output_____"
],
[
"filename = \"data/a9a\"\n#filename = \"data/real-sim.bz2\"\n#filename = \"data/rcv1_train.binary.bz2\"\n#filename = \"data/kdda.t.bz2\"\n\nA, b = datasets.load_svmlight_file(filename)\nm, n = A.shape\n\nprint(\"The dataset {}. The dimensions: m={}, n={}\".format(filename[5:], m, n))\n\n\n# define all ingredients for sparse logistic regression\ngamma = 0.005 * LA.norm(A.T.dot(b), np.inf)\nK = (A.T.multiply(-b)).T.tocsr()\n\n# find the norm of K^T K\nL = spr_LA.svds(K, k=1, return_singular_vectors=False)**2\n\n# starting point\nx0 = np.zeros(n)\n\n# stepsize\nss = 4/L\n\n\ng = lambda x: gamma*LA.norm(x,1)\nprox_g = lambda x, rho: x + np.clip(-x, -rho*gamma, rho*gamma)\n\n\nf = lambda x: np.log(1. + np.exp(x)).sum()\n\ndef df(x):\n exp_x = np.exp(x)\n return exp_x/(1.+exp_x)\n\ndh = lambda x, Kx: K.T.dot(df(Kx))\n\n# residual\nres = lambda x: LA.norm(x-prox_g(x-dh(x,K.dot(x)), 1))\n\n# energy\nJ = lambda x, Kx: f(Kx)+g(x)",
"The dataset a9a. The dimensions: m=32561, n=123\n"
],
[
"### Algorithms\n\ndef prox_grad(x1, s=1, numb_iter=100):\n \"\"\"\n Implementation of the proximal gradient method.\n\n x1: array, a starting point\n s: positive number, a stepsize\n numb_iter: positive integer, number of iterations\n\n Returns an array of energy values, computed in each iteration, and the\n argument x_k after numb_iter iterations\n \"\"\"\n begin = perf_counter()\n x = x1.copy()\n Kx = K.dot(x)\n values = [J(x, Kx)]\n dhx = dh(x,Kx)\n\n for i in range(numb_iter):\n #x = prox_g(x - s * dh(x, Kx), s)\n x = prox_g(x - s * dhx, s)\n Kx = K.dot(x)\n dhx = dh(x,Kx)\n values.append(J(x, Kx))\n\n end = perf_counter()\n\n print(\"Time execution of prox-grad:\", end - begin)\n return np.array(values), x\n\n\ndef fista(x1, s=1, numb_iter=100):\n \"\"\"\n Implementation of the FISTA.\n\n x1: array, a starting point\n s: positive number, a stepsize\n numb_iter: positive integer, number of iterations\n\n Returns an array of energy values, computed in each iteration, and the\n argument x_k after numb_iter iterations\n \"\"\"\n begin = perf_counter()\n x, y = x1.copy(), x1.copy()\n t = 1.\n Ky = K.dot(y)\n values = [J(y,Ky)]\n\n for i in range(numb_iter):\n x1 = prox_g(y - s * dh(y, Ky), s)\n t1 = 0.5 * (1 + np.sqrt(1 + 4 * t**2))\n y = x1 + (t - 1) / t1 * (x1 - x)\n x, t = x1, t1\n Ky = K.dot(y)\n values.append(J(y, Ky))\n\n end = perf_counter()\n\n print(\"Time execution of FISTA:\", end - begin)\n return np.array(values), x\n\n\ndef adaptive_graal(x1, numb_iter=100):\n \"\"\"\n Implementation of the adaptive GRAAL.\n\n x1: array, a starting point\n numb_iter: positive integer, number of iterations\n\n Returns an array of energy values, computed in each iteration, and the\n argument x_k after numb_iter iterations\n \"\"\"\n begin = perf_counter()\n phi = 1.5\n x, x_ = x1.copy(), x1.copy()\n x0 = x + np.random.randn(x.shape[0]) * 1e-9\n Kx = K.dot(x)\n dhx = dh(x, Kx)\n la = phi / 2 * LA.norm(x - x0) / LA.norm(dhx - dh(x0, K.dot(x0)))\n rho = 1. / phi + 1. / phi**2\n values = [J(x, Kx)]\n th = 1\n\n for i in range(numb_iter):\n x1 = prox_g(x_ - la * dhx, la)\n Kx1 = K.dot(x1)\n dhx1 = dh(x1, Kx1)\n\n n1 = LA.norm(x1 - x)**2\n n2 = LA.norm(dhx1 - dhx)**2\n\n n1_div_n2 = n1/n2 if n2 != 0 else la*10\n la1 = min(rho * la, 0.25 * phi * th / la * (n1_div_n2))\n x_ = ((phi - 1) * x1 + x_) / phi\n th = phi * la1 / la\n x, la, dhx = x1, la1, dhx1\n values.append(J(x1, Kx1))\n\n end = perf_counter()\n\n print(\"Time execution of aGRAAL:\", end - begin)\n return values, x, x_\n",
"_____no_output_____"
]
],
[
[
"Run the algorithms. It might take some time, if the dataset and/or the number of iterations are huge",
"_____no_output_____"
]
],
[
[
"N = 10000\nans1 = prox_grad(x0, ss, numb_iter=N)\nans2 = fista(x0, ss, numb_iter=N)\nans3 = adaptive_graal(x0, numb_iter=N)\n\nx1, x2, x3 = ans1[1], ans2[1], ans3[1]\nx1, x3 = ans1[1], ans3[1]\nprint(\"Residuals:\", [res(x) for x in [x1, x2, x3]])",
"Time execution of prox-grad: 68.03384687801008\nTime execution of FISTA: 68.20270787499612\nTime execution of aGRAAL: 70.1420427080011\nResiduals: [4.084755911350097e-05, 6.0765061701526585e-06, 7.583397935460372e-10]\n"
]
],
[
[
"Plot the results",
"_____no_output_____"
]
],
[
[
"values = [ans1[0], ans2[0], ans3[0]]\nlabels = [\"PGM\", \"FISTA\", \"aGRAAL\"]\nlinestyles = [':', \"--\", \"-\"]\ncolors = ['b', 'g', '#FFD700']\n\nv_min = min([min(v) for v in values])\nplt.figure(figsize=(6,4))\nfor i,v in enumerate(values):\n plt.plot(v - v_min, color=colors[i], label=labels[i], linestyle=linestyles[i])\nplt.yscale('log')\nplt.xlabel(u'iterations, k')\nplt.ylabel('$J(x^k)-J_{_*}$')\nplt.legend()\n#plt.savefig('figures/a9a.pdf', bbox_inches='tight')\nplt.show()\n\n\nplt.clf()\n",
"_____no_output_____"
],
[
"np.max(spr_LA.eigsh(K.T.dot(K))[0])",
"_____no_output_____"
],
[
"L",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb453b0918a493cf8831733315e07fbf20c0b24e | 719 | ipynb | Jupyter Notebook | test/ipynb/groovy/DomClassesSupport.ipynb | ssadedin/beakerx | 34479b07d2dfdf1404692692f483faf0251632c3 | [
"Apache-2.0"
] | 1,491 | 2017-03-30T03:05:05.000Z | 2022-03-27T04:26:02.000Z | test/ipynb/groovy/DomClassesSupport.ipynb | ssadedin/beakerx | 34479b07d2dfdf1404692692f483faf0251632c3 | [
"Apache-2.0"
] | 3,268 | 2015-01-01T00:10:26.000Z | 2017-05-05T18:59:41.000Z | test/ipynb/groovy/DomClassesSupport.ipynb | ssadedin/beakerx | 34479b07d2dfdf1404692692f483faf0251632c3 | [
"Apache-2.0"
] | 287 | 2017-04-03T01:30:06.000Z | 2022-03-17T06:09:15.000Z | 17.536585 | 53 | 0.520167 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
cb453e7d36f17c0d7d738e809b8fcb78f4e80f6c | 6,871 | ipynb | Jupyter Notebook | 04 Python Teil 2/05 Dateien.ipynb | edzardschade/Edzard_CAS | 42c5db16e506e1fd8653d49e7509b8d2b59353e7 | [
"MIT"
] | null | null | null | 04 Python Teil 2/05 Dateien.ipynb | edzardschade/Edzard_CAS | 42c5db16e506e1fd8653d49e7509b8d2b59353e7 | [
"MIT"
] | null | null | null | 04 Python Teil 2/05 Dateien.ipynb | edzardschade/Edzard_CAS | 42c5db16e506e1fd8653d49e7509b8d2b59353e7 | [
"MIT"
] | null | null | null | 22.454248 | 137 | 0.484791 | [
[
[
"# Dateien",
"_____no_output_____"
],
[
"## Eine Textdatei lesen und ihren Inhalt ausgeben",
"_____no_output_____"
]
],
[
[
"# Wir öffnen die Datei lesen.txt zum Lesen (\"r\") und speichern ihren Inhalt in die Variable file\nfile = open(\"lesen.txt\", \"r\")\n\n# Wir gehen alle Zeilen nacheinander durch\n# In der txt-Datei stehen für uns nicht sichtbare Zeilenumbruchszeichen, durch die jeweils das Ende einer Zeile markiert ist\nfor line in file:\n # Eine Zeile ohne Zeilenumbruch ausgeben\n print(line.strip())\nfile.close()",
"Hallo Leute.\nIch bin eine wunderschöne weitere Zeile.\nJa, das ist wirklich toll!\n"
],
[
"file = open(\"gemischtertext.txt\", \"r\")\n\n",
"_____no_output_____"
]
],
[
[
"## In eine Textdatei schreiben",
"_____no_output_____"
]
],
[
[
"# Wir öffnen eine Datei zum Schreiben (\"w\": write)\nfile = open(\"schreiben.txt\", \"w\")\n\nstudents = [\"Max\", \"Monika\", \"hgcvhgv\", \"Erik\", \"Franziska\"]\n\n# Wir loopen mit einer for-Schleife durch die Liste students\nfor student in students:\n # Mit der write-Methode schreiben wir den aktuellen String student und einen Zeilenumbruch in das file-Objekt\n file.write(student + \"\\n\")\n\n# Abschließend müssen wir die Datei wieder schließen\nfile.close()",
"_____no_output_____"
]
],
[
[
"## Dateien öffnen mit with\nWenn wir Dateien mit einer with-Konstruktion öffnen, dann brauchen wir sie nicht mehr explizit mit der close()-Methode schließen.",
"_____no_output_____"
]
],
[
[
"with open(\"lesen.txt\", \"r\") as file:\n for line in file:\n print(line)",
"Hallo Leute.\n\nIch bin eine wunderschöne weitere Zeile.\n"
]
],
[
[
"## CSV-Datei lesen\ncsv steht für comma separated values. Auch solche csv-Dateien können wir mit Python auslesen.",
"_____no_output_____"
]
],
[
[
"with open(\"datei.csv\") as file:\n for line in file:\n data = line.strip().split(\";\")\n print(data[0] + \": \" + data[1])",
"Muenchen: 1800000\nBerlin: 3000000\nBudapest: 2000000\n"
]
],
[
[
"## CSV-Datei lesen (und Daten überspringen)\n\nIn dieser Lektion lernst du:\n\n- Wie du eine CSV-Datei einliest, und Zeilen überspringen kannst.",
"_____no_output_____"
]
],
[
[
"with open(\"datei.csv\") as file:\n for line in file:\n data = line.strip().split(\";\")\n \n if int(data[1]) < 2000000:\n continue\n \n if data[2] == \"BUD\":\n continue\n \n print(data)\n \n #if data[2] == \"BER\" or data[2] == \"BUD\":\n # print(data[2])\n # print(data)",
"['Berlin', '3000000', 'BER']\n"
]
],
[
[
"## Übung\n- Besorgt euch die datei https://data.stadt-zuerich.ch/dataset/pd-stapo-hundenamen/resource/8bf2127d-c354-4834-8590-9666cbd6e160\n- Ihr findet sie auch im Ordner 20151001_hundenamen.csv\n- Findet heraus wie oft der Hundename \"Aaron\" zwischen 2000 - 2012 gebraucht wurde. ",
"_____no_output_____"
]
],
[
[
"n = \"1975\"\nprint(int(n) < 1990)",
"True\n"
],
[
"jahre = [\"Year\", \"1990\", \"1992\"]\n\nfor jahr in jahre:\n if jahr == \"Year\":\n continue\n print(int(jahr))",
"1990\n1992\n"
],
[
"### Euer code hier",
"_____no_output_____"
],
[
"with open(\"20151001_hundenamen.csv\", \"r\") as file:\n for line in file:\n anzahl=0\n data = line.strip().split(\",\")\n if data[1] == \"GEBURTSJAHR_HUND\":\n continue\n \n if data[0] == '\"Aaron\"' and int(data[1]) >= 2000 and int(data[1]) < 2013:\n anzahl = anzahl + 1\n \nprint(anzahl)",
"0\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cb453f325570cfc0da1308984dde2253157898b0 | 56,527 | ipynb | Jupyter Notebook | Tutorial 2.ipynb | Junyoungpark/2021-Spring-IE437 | 4c10fddda71d5f44fa8bf51f61e54f0a04673d57 | [
"MIT"
] | null | null | null | Tutorial 2.ipynb | Junyoungpark/2021-Spring-IE437 | 4c10fddda71d5f44fa8bf51f61e54f0a04673d57 | [
"MIT"
] | null | null | null | Tutorial 2.ipynb | Junyoungpark/2021-Spring-IE437 | 4c10fddda71d5f44fa8bf51f61e54f0a04673d57 | [
"MIT"
] | null | null | null | 200.450355 | 31,768 | 0.904081 | [
[
[
"from functools import partial\n\nimport torch\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom scipy.optimize import minimize\n\nfrom src.utils import generate_y\nfrom src.MLP import MLP",
"_____no_output_____"
]
],
[
[
"## Tutorial for `Type2` problem\n\nThe usage of `type 1` problems are prominent. However it is often true that without constraints on optimization varaibles $x$ that the optimal solution might be trivial.\nAs an concrete example, let's consider the following optimization problem:\n$$ \\max_{x} x^2 $$\n\nWe can visually inspect the solution of above optimization problem. Let's look around the following figure",
"_____no_output_____"
]
],
[
[
"xs = np.linspace(-10.0, 10.0, 1000)\nys = xs ** 2\nfig, ax = plt.subplots(1,1)\nax.grid()\n_ = ax.plot(xs, ys)",
"_____no_output_____"
]
],
[
[
"As you can notice, as we push the solution $x$ toward $\\infty$ or $-\\infty$, the objective function $x^2$ increase. We call this scenarios as 'the problem is unbounded'. Even in the practical setup, we will confront such cases easily. Moreover to make the optimization problem 'practically' useful, we can consider the constrain on to the optimization problem. We firstly consider the following type of constraint optimziation problem.",
"_____no_output_____"
],
[
"### Solve optimization problem with box constraints\n\n$$\n\\begin{aligned}\n \\min_{x} &\\, f(x) \\\\\n \\text{s.t.} &\\, x_{min} \\leq x \\leq x_{max} \\\\\n\\end{aligned}\n$$\n\nThis type of optimization problem is to handle the box constraint on optimization variable $x$. The $\\text{s.t.}$ is abbreviation of '**s**uch **t**hat'.\n\n#### Box constraint\n\n$$x_{min} \\leq x \\leq x_{max}$$\n\nThe box constraint indicates that the solution of optmization problem $x^*$ must be larger or equal to $x_{min}$ and smaller or equal to $x_{max}$. The box constraint is a special case of general constraints. Mathemacically to solve the optimziation problems, either the box constraint and general linear/non linear constraints can be handled in the same manner. However most of the off-the-shelf solvers explictly consider the box constrains as a different arguments from the other constraints. \n\n> For instance, `scipy` solvers consider the box constrains as `bounds`.\n\nFor now, let's investigate the box constraint first and reserve some room for the general constraint. ",
"_____no_output_____"
],
[
"## `scipy.optimize` pacakge\n\n`scipy.optimize` is a package that implements various types of optimziation solvers. Most importantly, they offer nice python interface of the solvers so that you can setup your own optmization problem with few lines of codes. We will also solve the optimization problems with a variant of QP solver of this package.\n\nEspecially, `scipy.optmize.minize` function is powerful in practice. you can 'some-how-magically' optimize your own function. Even in the case your function is not analytically differntiable.\n\n> In such case, `scipy.optmize.minize` employs numerical methods to estimate the jacobian and hessian. As a cost of emplyoing numerical methods, the optmization procedure will be slower.\n\n### Interface of `scipy.optimize.minimize`\n\n`scipy.optimize.minimize` majorly requires following arguments:\n\n1. `fun`: the objective function that you want to optmize\n2. `x0`: the initial solution. You can set arbitarily unless it doesn't violate constraints\n3. `jac`: (optional) the method for computing the jacobian of the objective function\n4. `hessian`: (optional) the method for computing the hessian of the objective function\n5. `bounds`: (optional) box constraints\n6. `constraints`: (optional) linear/non-linear constraints\n\nwhen `jac` and `hessian` is not specified, the `scipy.optmize.minize` function estimate the jacobian and hessian of the objective function numerically.",
"_____no_output_____"
],
[
"## Implementing `fun`, `jac`, `hessian`\n\nOur primiary interest is to bind the `torch` module and `scipy.optimize.minimize`. Since pytorch is automatic differentiation tool, we can compute the `jac` and `hessian` efficiently.",
"_____no_output_____"
]
],
[
[
"def objective(x, model):\n # Note that we will not use the pytorch's automatic differentiation functionality\n # while computing objective\n with torch.no_grad(): \n torch_x = torch.from_numpy(x).view(-1,1).float()\n y = model(torch_x)\n y = y.sum().numpy()\n return y\n\ndef jac(x, model):\n torch_x = torch.from_numpy(x).view(-1,1).float()\n jac = torch.autograd.functional.jacobian(model, torch_x).numpy()\n return jac",
"_____no_output_____"
],
[
"m = MLP(1, 1, num_neurons=[128, 128])\nm.load_state_dict(torch.load('./model.pt'))",
"_____no_output_____"
],
[
"lb, ub = -3.0, 2.0 # declare lower and upper bound of optmization variable\n\nx_init = np.random.uniform(lb, ub) \nx0 = np.ones(1) * x_init\nx0_tensor = torch.ones(1,1) * x_init\ny0 = m(x0_tensor).detach()",
"_____no_output_____"
],
[
"b = (lb, ub)\nbnds = (b,)\n\nsoln_nn = minimize(partial(objective, model=m), \n x0, \n method='SLSQP', \n bounds=bnds, \n jac=partial(jac, model=m))",
"_____no_output_____"
]
],
[
[
"## Visualize optimization result",
"_____no_output_____"
]
],
[
[
"x_min, x_max = -4.0, 4.0\n\nxs_linspace = torch.linspace(-4, 4, 2000).view(-1, 1)\nys_linspace = generate_y(xs_linspace)",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(1, 1, figsize=(10, 5))\naxes.grid()\naxes.plot(xs_linspace, ys_linspace, label='Ground truth')\n\nys_pred = m(xs_linspace).detach()\naxes.plot(xs_linspace, ys_pred, label='Ground truth')\naxes.fill_between(np.linspace(lb, ub, 100),\n ys_linspace.min(),\n ys_linspace.max(),\n color='grey', \n alpha=0.3,\n label='constraint region')\naxes.scatter(x_init, y0, label='Opt start', \n c='green', marker='*', s=100.0)\n\naxes.scatter(soln_nn.x, soln_nn.fun, label='NN opt', c='green')\naxes.legend()\naxes.set_xlabel(\"input\")\naxes.set_ylabel(\"y value\")\nplt.show()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cb4544037352e4132dd4ef6537b497a01809d072 | 4,061 | ipynb | Jupyter Notebook | ipynb/Eswatini.ipynb | RobertRosca/oscovida.github.io | d609949076e3f881e38ec674ecbf0887e9a2ec25 | [
"CC-BY-4.0"
] | null | null | null | ipynb/Eswatini.ipynb | RobertRosca/oscovida.github.io | d609949076e3f881e38ec674ecbf0887e9a2ec25 | [
"CC-BY-4.0"
] | null | null | null | ipynb/Eswatini.ipynb | RobertRosca/oscovida.github.io | d609949076e3f881e38ec674ecbf0887e9a2ec25 | [
"CC-BY-4.0"
] | null | null | null | 28.398601 | 162 | 0.508742 | [
[
[
"# Eswatini\n\n* Homepage of project: https://oscovida.github.io\n* [Execute this Jupyter Notebook using myBinder](https://mybinder.org/v2/gh/oscovida/binder/master?filepath=ipynb/Eswatini.ipynb)",
"_____no_output_____"
]
],
[
[
"import datetime\nimport time\n\nstart = datetime.datetime.now()\nprint(f\"Notebook executed on: {start.strftime('%d/%m/%Y %H:%M:%S%Z')} {time.tzname[time.daylight]}\")",
"_____no_output_____"
],
[
"%config InlineBackend.figure_formats = ['svg']\nfrom oscovida import *",
"_____no_output_____"
],
[
"overview(\"Eswatini\");",
"_____no_output_____"
],
[
"# load the data\ncases, deaths, region_label = get_country_data(\"Eswatini\")\n\n# compose into one table\ntable = compose_dataframe_summary(cases, deaths)\n\n# show tables with up to 500 rows\npd.set_option(\"max_rows\", 500)\n\n# display the table\ntable",
"_____no_output_____"
]
],
[
[
"# Explore the data in your web browser\n\n- If you want to execute this notebook, [click here to use myBinder](https://mybinder.org/v2/gh/oscovida/binder/master?filepath=ipynb/Eswatini.ipynb)\n- and wait (~1 to 2 minutes)\n- Then press SHIFT+RETURN to advance code cell to code cell\n- See http://jupyter.org for more details on how to use Jupyter Notebook",
"_____no_output_____"
],
[
"# Acknowledgements:\n\n- Johns Hopkins University provides data for countries\n- Robert Koch Institute provides data for within Germany\n- Open source and scientific computing community for the data tools\n- Github for hosting repository and html files\n- Project Jupyter for the Notebook and binder service\n- The H2020 project Photon and Neutron Open Science Cloud ([PaNOSC](https://www.panosc.eu/))\n\n--------------------",
"_____no_output_____"
]
],
[
[
"print(f\"Download of data from Johns Hopkins university: cases at {fetch_cases_last_execution()} and \"\n f\"deaths at {fetch_deaths_last_execution()}.\")",
"_____no_output_____"
],
[
"# to force a fresh download of data, run \"clear_cache()\"",
"_____no_output_____"
],
[
"print(f\"Notebook execution took: {datetime.datetime.now()-start}\")\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb454abc56f464082abd3bfd95bb2e0519fafa97 | 18,203 | ipynb | Jupyter Notebook | tutorials/beginner/ibis.ipynb | fugue-project/tutorials | a485db6c223588eb2d462f61e813a9ce3d6df9ee | [
"Apache-2.0"
] | 28 | 2020-06-26T19:43:26.000Z | 2022-03-27T23:01:28.000Z | tutorials/beginner/ibis.ipynb | fugue-project/tutorials | a485db6c223588eb2d462f61e813a9ce3d6df9ee | [
"Apache-2.0"
] | 50 | 2020-09-28T08:12:06.000Z | 2022-01-26T06:16:54.000Z | tutorials/beginner/ibis.ipynb | fugue-project/tutorials | a485db6c223588eb2d462f61e813a9ce3d6df9ee | [
"Apache-2.0"
] | 7 | 2020-10-28T19:27:44.000Z | 2021-11-16T06:25:00.000Z | 29.171474 | 594 | 0.501071 | [
[
[
"# Ibis Integration (Experimental)\n\nThe [Ibis project](https://ibis-project.org/docs/) tries to bridge the gap between local Python and [various backends](https://ibis-project.org/docs/backends/index.html) including distributed systems such as Spark and Dask. The main idea is to create a pythonic interface to express SQL semantic, so the expression is agnostic to the backends.\n\nThe design idea is very aligned with Fugue. But please notice there are a few key differences:\n\n* **Fugue supports both pythonic APIs and SQL**, and the choice should be determined by particular cases or users' preferences. On the other hand, Ibis focuses on the pythonic expression of SQL and perfectionizes it.\n* **Fugue supports SQL and non-SQL semantic for data transformation.** Besides SQL, another important option is [Fugue Transform](introduction.html#fugue-transform). The Fugue transformers can wrap complicated Python/Pandas logic and apply them distributedly on dataframes. A typical example is distributed model inference, the inference part has to be done by Python, it can be easily achieved by a transformer, but the data preparation may be done nicely by SQL or Ibis.\n* **Fugue and Ibis are on different abstraction layers.** Ibis is nice to construct single SQL statements to accomplish single tasks. Even it involves multiple tables and multiple steps, its final step is either outputting one table or inserting one table into a database. On the other hand, Fugue workflow is to orchestrate these tasks. For example, it can read a table, do the first transformation and save to a file, then do the second transformation and print. Each transformation may be done using Ibis, but loading, saving and printing and the orchestration can be done by Fugue.\n\nThis is also why Ibis can be a very nice option for Fugue users to build their pipelines. For people who prefer pythonic APIs, they can keep all the logic in Python with the help of Ibis. Although Fugue has its own functional API similar to Ibis, the programming interface of Ibis is really elegant. It usually helps users write less but more expressive code to achieve the same thing.\n\n## Hello World\n\nIn this example, we try to achieve this SQL semantic:\n\n```sql\nSELECT a, a+1 AS b FROM\n (SELECT a FROM tb1 UNION SELECT a FROM tb2)\n```",
"_____no_output_____"
]
],
[
[
"from ibis import BaseBackend, literal\nimport ibis.expr.types as ir\n\ndef ibis_func(backend:BaseBackend) -> ir.TableExpr:\n tb1 = backend.table(\"tb1\")\n tb2 = backend.table(\"tb2\")\n tb3 = tb1.union(tb2)\n return tb3.mutate(b=tb3.a+literal(1))",
"_____no_output_____"
]
],
[
[
"Now let's test with the pandas backend",
"_____no_output_____"
]
],
[
[
"import ibis\nimport pandas as pd\n\ncon = ibis.pandas.connect({\n \"tb1\": pd.DataFrame([[0]], columns=[\"a\"]),\n \"tb2\": pd.DataFrame([[1]], columns=[\"a\"])\n})\nibis_func(con).execute()",
"_____no_output_____"
]
],
[
[
"Now let's make this a part of Fugue",
"_____no_output_____"
]
],
[
[
"from fugue import FugueWorkflow\nfrom fugue_ibis import run_ibis\n\ndag = FugueWorkflow()\ndf1 = dag.df([[0]], \"a:long\")\ndf2 = dag.df([[1]], \"a:long\")\ndf3 = run_ibis(ibis_func, tb1=df1, tb2=df2)\ndf3.show()",
"_____no_output_____"
]
],
[
[
"Now let's run on Pandas",
"_____no_output_____"
]
],
[
[
"dag.run()",
"PandasDataFrame\na:long|b:long\n------+------\n0 |1 \n1 |2 \nTotal count: 2\n\n"
]
],
[
[
"Now let's run on Dask",
"_____no_output_____"
]
],
[
[
"import fugue_dask\n\ndag.run(\"dask\")",
"DaskDataFrame\na:long|b:long\n------+------\n0 |1 \n1 |2 \nTotal count: 2\n\n"
]
],
[
[
"Now let's run on DuckDB",
"_____no_output_____"
]
],
[
[
"import fugue_duckdb\n\ndag.run(\"duck\")",
"PandasDataFrame\na:long|b:long\n------+------\n0 |1 \n1 |2 \nTotal count: 2\n\n"
]
],
[
[
"For each different execution engine, Ibis will also run on the correspondent backend.\n\n## A deeper integration\n\nThe above approach needs a function taking in an Ibis backend and returning a `TableExpr`. The following is another approach that simpler and more elegant.",
"_____no_output_____"
]
],
[
[
"from fugue_ibis import as_ibis, as_fugue\n\ndag = FugueWorkflow()\ntb1 = as_ibis(dag.df([[0]], \"a:long\"))\ntb2 = as_ibis(dag.df([[1]], \"a:long\"))\ntb3 = tb1.union(tb2)\ndf3 = as_fugue(tb3.mutate(b=tb3.a+literal(1)))\ndf3.show()\n\ndag.run()",
"PandasDataFrame\na:long|b:long\n------+------\n0 |1 \n1 |2 \nTotal count: 2\n\n"
]
],
[
[
"Alternatively, you can treat `as_ibis` and `as_fugue` as class methods. This is more convenient to use, but it's a bit magical. This is achieved by adding these two methods using `setattr` to the correspondent classes. This patching-like design pattern is widely used by Ibis.",
"_____no_output_____"
]
],
[
[
"import fugue_ibis # must import\n\ndag = FugueWorkflow()\ntb1 = dag.df([[0]], \"a:long\").as_ibis()\ntb2 = dag.df([[1]], \"a:long\").as_ibis()\ntb3 = tb1.union(tb2)\ndf3 = tb3.mutate(b=tb3.a+literal(1)).as_fugue()\ndf3.show()\n\ndag.run()",
"PandasDataFrame\na:long|b:long\n------+------\n0 |1 \n1 |2 \nTotal count: 2\n\n"
]
],
[
[
"By importing `fugue_ibis`, the two methods were automatically added.\n\nIt's up to the users which way to go. The first approach (`run_ibis`) is the best to separate Ibis logic, as you can see, it is also great for unit testing. The second approach is elegant, but you will have to unit test the code with the logic before and after the conversions. The third approach is the most intuitive, but it's a bit magical.\n\n## Z-Score\n\nNow, let's consider a practical example. We want to use Fugue to compute z-score of a dataframe, partitioning should be an option. The reason to implement it on Fugue level is that the compute becomes scale agnostic and framework agnostic.",
"_____no_output_____"
]
],
[
[
"from fugue import WorkflowDataFrame\nfrom fugue_ibis import as_ibis, as_fugue\n\ndef z_score(df:WorkflowDataFrame, input_col:str, output_col:str) -> WorkflowDataFrame:\n by = df.partition_spec.partition_by\n idf = as_ibis(df)\n col = idf[input_col] \n if len(by)==0:\n return as_fugue(idf.mutate(**{output_col:(col - col.mean())/col.std()}))\n agg = idf.group_by(by).aggregate(mean_=col.mean(), std_=col.std())\n j = idf.inner_join(agg, by)[idf, ((idf[input_col]-agg.mean_)/agg.std_).name(output_col)]\n return as_fugue(j)",
"_____no_output_____"
]
],
[
[
"Now, generate testing data",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nnp.random.seed(0)\npdf = pd.DataFrame(dict(\n a=np.random.choice([\"a\",\"b\"], 100),\n b=np.random.choice([\"c\",\"d\"], 100),\n c=np.random.rand(100),\n))\n\npdf[\"expected1\"] = (pdf.c - pdf.c.mean())/pdf.c.std()\npdf = pdf.groupby([\"a\", \"b\"]).apply(lambda tdf: tdf.assign(expected2=(tdf.c - tdf.c.mean())/tdf.c.std())).reset_index(drop=True)",
"_____no_output_____"
]
],
[
[
"And here is the final code.",
"_____no_output_____"
]
],
[
[
"dag = FugueWorkflow()\ndf = z_score(dag.df(pdf), \"c\", \"z1\")\ndf = z_score(df.partition_by(\"a\", \"b\"), \"c\", \"z2\")\ndf.show()\n\ndag.run()",
"PandasDataFrame\na:str|b:str|c:double |expected1:double |expected2:double |z1:double |z2:double \n-----+-----+-------------+------------------+------------------+-------------+------------\na |c |0.84640867247|1.144636809499835 |1.577770556350802 |1.14463680949|1.5777705563\n | |11278 | | |98345 |508022 \na |c |0.69947927531|0.6163095213101546|1.0316513450169476|0.61630952131|1.0316513450\n | |75043 | | |01543 |169478 \na |c |0.81379781970|1.0273750242983348|1.4565598691775665|1.02737502429|1.4565598691\n | |24772 | | |83344 |77567 \na |c |0.39650574084|-0.473119748536303|-0.094465490044129|-0.4731197485|-0.094465490\n | |698464 |06 |49 |3630345 |0441295 \na |c |0.58127287263|0.1912640955118427|0.5922921108698395|0.19126409551|0.5922921108\n | |58587 |9 | |18424 |698396 \na |c |0.29828232595|-0.826310542423581|-0.459550319315429|-0.8263105424|-0.459550319\n | |603077 |5 |6 |235819 |3154297 \na |c |0.57432524884|0.1662818979021328|0.5664686140318282|0.16628189790|0.5664686140\n | |95788 |8 | |21325 |318284 \na |c |0.43141843543|-0.347581023054070|0.0353008712338056|-0.3475810230|0.0353008712\n | |397396 |96 |9 |540714 |33805695 \na |c |0.43586492526|-0.331592378438154|0.0518279486404278|-0.3315923784|0.0518279486\n | |56268 |5 |1 |3815487 |4042782 \na |c |0.14944830465|-1.361486459469925|-1.012748793586346|-1.3614864594|-1.012748793\n | |799375 |9 |2 |699263 |5863462 \n"
]
],
[
[
"## Consistency issues\n\nIbis as of 2.0.0 can have different behaviors on different backends. Here are some examples from the common discrepencies between pandas and SQL.",
"_____no_output_____"
]
],
[
[
"# pandas drops null keys on group (by default), SQL doesn't\n\ndag = FugueWorkflow()\ndf = dag.df([[\"a\",1],[None,2]], \"a:str,b:int\").as_ibis()\ndf.groupby([\"a\"]).aggregate(s=df.b.sum()).as_fugue().show()\n\ndag.run()\ndag.run(\"duckdb\")\n",
"PandasDataFrame\na:str|s:long\n-----+------\na |1 \nTotal count: 1\n\nPandasDataFrame\na:str|s:long\n-----+------\na |1 \nNULL |2 \nTotal count: 2\n\n"
],
[
"# pandas joins on NULLs, SQL doesn't\n\ndag = FugueWorkflow()\ndf1 = dag.df([[\"a\",1],[None,2]], \"a:str,b:int\").as_ibis()\ndf2 = dag.df([[\"a\",1],[None,2]], \"a:str,c:int\").as_ibis()\ndf1.inner_join(df2, [\"a\"])[df1, df2.c].as_fugue().show()\n\ndag.run()\ndag.run(\"duckdb\")",
"PandasDataFrame\na:str|b:int|c:int\n-----+-----+-----\na |1 |1 \nNULL |2 |2 \nTotal count: 2\n\nPandasDataFrame\na:str|b:int|c:int\n-----+-----+-----\na |1 |1 \nTotal count: 1\n\n"
]
],
[
[
"Since Ibis integration is experimental, we rely on Ibis to achieve consistent behaviors. If you have any Ibis specific question please also consider asking in [Ibis issues](https://github.com/ibis-project/ibis/issues).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cb454b8d808912fec8a73b9e486f56b9fdb46852 | 446,325 | ipynb | Jupyter Notebook | demos/Init_Image.ipynb | DazhiZhong/clipit | 799c4a9267615f8de15bec11c0cff54aa7162d4e | [
"MIT"
] | null | null | null | demos/Init_Image.ipynb | DazhiZhong/clipit | 799c4a9267615f8de15bec11c0cff54aa7162d4e | [
"MIT"
] | null | null | null | demos/Init_Image.ipynb | DazhiZhong/clipit | 799c4a9267615f8de15bec11c0cff54aa7162d4e | [
"MIT"
] | null | null | null | 705.094787 | 223,645 | 0.94295 | [
[
[
"<a href=\"https://colab.research.google.com/github/dribnet/clipit/blob/master/demos/Init_Image.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"#Pixray Init Image demo\nUse a starting image for better control.\n\n<br>\nGreat White Shark by <a href=\"https://twitter.com/ViktorAlm/status/1428112740128894981\">Viktor Alm</a>",
"_____no_output_____"
]
],
[
[
"#@title Setup\n\n#@markdown Please execute this cell by pressing the _Play_ button \n#@markdown on the left. For setup,\n#@markdown **you need to run this cell,\n#@markdown then choose Runtime -> Restart Runtime from the menu,\n#@markdown and then run the cell again**. It should remind you to\n#@markdown do this after the first run.\n\n#@markdown Setup can take 5-10 minutes, but once it is complete it usually does not need to be repeated\n#@markdown until you close the window.\n\n#@markdown **Note**: This installs the software on the Colab \n#@markdown notebook in the cloud and not on your computer.\n\n# https://stackoverflow.com/a/56727659/1010653\n\n# Add a gpu check\n# (this can get better over time)\nfrom google.colab import output\n\nnvidia_output = !nvidia-smi --query-gpu=memory.total --format=noheader,nounits,csv\ngpu_memory = int(nvidia_output[0])\nif gpu_memory < 14000:\n output.eval_js('new Audio(\"https://upload.wikimedia.org/wikipedia/commons/0/05/Beep-09.ogg\").play()')\n warning_string = f\"--> GPU check: ONLY {gpu_memory} MiB available: WARNING, THIS IS PROBABLY NOT ENOUGH <--\"\n print(warning_string)\n output.eval_js('alert(\"Warning - low GPU (see message)\")')\nelse:\n print(f\"GPU check: {gpu_memory} MiB available: this should be fine\")\n\nfrom IPython.utils import io\nwith io.capture_output() as captured:\n !git clone https://github.com/openai/CLIP\n # !pip install taming-transformers\n !git clone https://github.com/CompVis/taming-transformers.git\n !rm -Rf pixray\n !git clone https://github.com/dribnet/pixray\n !pip install ftfy regex tqdm omegaconf pytorch-lightning\n !pip install kornia\n !pip install imageio-ffmpeg \n !pip install einops\n !pip install torch-optimizer\n !pip install easydict\n !pip install braceexpand\n !pip install git+https://github.com/pvigier/perlin-numpy\n\n # ClipDraw deps\n !pip install svgwrite\n !pip install svgpathtools\n !pip install cssutils\n !pip install numba\n !pip install torch-tools\n !pip install visdom\n\n !git clone https://github.com/BachiLi/diffvg\n %cd diffvg\n # !ls\n !git submodule update --init --recursive\n !python setup.py install\n %cd ..\n\noutput.clear()\nimport sys\nsys.path.append(\"pixray\")\n\nresult_msg = \"setup complete\"\nimport IPython\nimport os\nif not os.path.isfile(\"first_init_complete\"):\n # put stuff in here that should only happen once\n !mkdir -p models\n os.mknod(\"first_init_complete\")\n result_msg = \"Please choose Runtime -> Restart Runtime from the menu, and then run Setup again\"\n\njs_code = f'''\ndocument.querySelector(\"#output-area\").appendChild(document.createTextNode(\"{result_msg}\"));\n'''\njs_code += '''\nfor (rule of document.styleSheets[0].cssRules){\n if (rule.selectorText=='body') break\n}\nrule.style.fontSize = '30px'\n'''\ndisplay(IPython.display.Javascript(js_code))",
"_____no_output_____"
],
[
"#@title Settings\n\n#@markdown Enter a description of what you want to draw - I usually add #pixelart to the prompt.\n#@markdown The drawer can also be swapped with VQGAN. init_image can be any image but best to\n#@markdown try to match the aspect ratio. Palette is optional and usually with an init_image\n#@markdown fewer iterations are necessary.<br>\n\nprompts = \"The Matrix. #pixelart #gameart\" #@param {type:\"string\"}\n\naspect = \"widescreen\" #@param [\"widescreen\", \"square\"]\n\ninit_image = \"https://cdn.mos.cms.futurecdn.net/D8VcfmEXTEUW99vtdexD3o.jpg\" #@param {type:\"string\"}\n\ndrawer = \"pixel\" #@param [\"vqgan\", \"pixel\"]\n\npixel_scale = 0.75#@param {type:\"number\"}\n\npixel_type = \"rect\" #@param [\"rect\", \"rectshift\", \"hex\", \"tri\", \"diamond\"]\n\nuse_palette = \"white->black->neon green->black->neon blue\" #@param {type:\"string\"}\n\niterations = 100#@param {type:\"number\"}\n\n\n#@markdown When you have the settings you want, press the play button on the left.\n#@markdown The system will save these and start generating images below.\n\n#@markdown When that is done you can change these\n#@markdown settings and see if you get different results. Or if you get\n#@markdown impatient, just select \"Runtime -> Interrupt Execution\".\n#@markdown Note that the first time you run it may take a bit longer\n#@markdown as nessary files are downloaded.\n\n\n#@markdown\n#@markdown *Advanced: you can also edit this cell and add add additional\n#@markdown settings, combining settings from different notebooks.*\n\n\n# Simple setup\nimport pixray\n\n# these are good settings for pixeldraw\npixray.reset_settings()\npixray.add_settings(prompts=prompts, aspect=aspect)\npixray.add_settings(quality=\"better\", scale=3)\npixray.add_settings(drawer=drawer, iterations=iterations)\nif drawer == \"pixel\":\n pixray.add_settings(pixel_scale=pixel_scale, pixel_type=pixel_type)\nif len(use_palette) > 0:\n pixray.add_settings(target_palette=use_palette)\n pixray.add_settings(smoothness=1.0, smoothness_type='log')\n pixray.add_settings(saturation=0.75)\nif len(init_image) > 0:\n pixray.add_settings(init_image=init_image)\n\n#### YOU CAN ADD YOUR OWN CUSTOM SETTING HERE ####\n# this is the example of how to run longer with less frequent display\n# pixray.add_settings(iterations=500, display_every=50)\npixray.add_settings(display_clear=True)\n\nsettings = pixray.apply_settings()\npixray.do_init(settings)\npixray.do_run(settings)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cb454d8dd625ccebead625136d4e6d7fb933b605 | 531,436 | ipynb | Jupyter Notebook | examples/rosenbrock.ipynb | paulthebaker/kombine | 9d558e666bc30ee2dda8061513046887a0af1e9e | [
"MIT"
] | null | null | null | examples/rosenbrock.ipynb | paulthebaker/kombine | 9d558e666bc30ee2dda8061513046887a0af1e9e | [
"MIT"
] | null | null | null | examples/rosenbrock.ipynb | paulthebaker/kombine | 9d558e666bc30ee2dda8061513046887a0af1e9e | [
"MIT"
] | null | null | null | 2,280.841202 | 372,256 | 0.962823 | [
[
[
"import numpy as np\n\nimport kombine",
"_____no_output_____"
]
],
[
[
"Import some cool visualization stuff.",
"_____no_output_____"
]
],
[
[
"from matplotlib import pyplot as plt\nimport corner\nimport prism\n\n%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\n\nprism.inline_ipynb()",
"_____no_output_____"
]
],
[
[
"# 2-D Rosenbrock Target Distribution",
"_____no_output_____"
],
[
"Define the target function.",
"_____no_output_____"
]
],
[
[
"ndim = 2\n\ndef rosenbrock(x):\n return -np.sum((1.0 - x[:-1])*(1.0 - x[:-1]) + 100.0*(x[1:] - x[:-1]*x[:-1])*(x[1:] - x[:-1]*x[:-1]))",
"_____no_output_____"
]
],
[
[
"Create a uniformly distributed ensemble and burn it in.",
"_____no_output_____"
]
],
[
[
"nwalkers = 500 \nsampler = kombine.Sampler(nwalkers, ndim, rosenbrock)\n\np0 = np.random.uniform(-10, 10, size=(nwalkers, ndim))\np, post, q = sampler.burnin(p0)",
"_____no_output_____"
]
],
[
[
"See what burnin did.",
"_____no_output_____"
]
],
[
[
"prism.corner(sampler.chain)",
"_____no_output_____"
]
],
[
[
"Generate some more samples.",
"_____no_output_____"
]
],
[
[
"p, post, q = sampler.run_mcmc(500)",
"_____no_output_____"
],
[
"fig, [ax1, ax2, ax3] = plt.subplots(1, 3, figsize=(15, 5))\n\nax1.plot(sampler.acceptance_fraction, 'k', alpha=.5, label=\"Mean Acceptance Rate\");\n\nfor p, ax in zip(range(sampler.dim), [ax2, ax3]):\n ax.plot(sampler.chain[..., p], alpha=0.1)\n\nax1.legend(loc='lower right');",
"_____no_output_____"
]
],
[
[
"Plot independent samples.",
"_____no_output_____"
]
],
[
[
"acls = np.ceil(2/np.mean(sampler.acceptance[-500:], axis=0) - 1).astype(int)\n\nind_samps = np.concatenate([sampler.chain[-500::acl, c].reshape(-1, 2) for c, acl in enumerate(acls)])\nprint(\"{} independent samples collected with a mean ACL of {}.\".format(len(ind_samps), np.mean(acls)))\n\ncorner.corner(ind_samps, plot_density=False, plot_contours=False);",
"105262 independent samples collected with a mean ACL of 2.478.\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb455141902bc2e214c5834f7778a50c35f62ea8 | 37,422 | ipynb | Jupyter Notebook | site/ja/tutorials/images/classification.ipynb | justaverygoodboy/docs-l10n | 8d4857750f2b5e8e6889acbb4b1e2f98ad7ce34e | [
"Apache-2.0"
] | null | null | null | site/ja/tutorials/images/classification.ipynb | justaverygoodboy/docs-l10n | 8d4857750f2b5e8e6889acbb4b1e2f98ad7ce34e | [
"Apache-2.0"
] | null | null | null | site/ja/tutorials/images/classification.ipynb | justaverygoodboy/docs-l10n | 8d4857750f2b5e8e6889acbb4b1e2f98ad7ce34e | [
"Apache-2.0"
] | null | null | null | 28.031461 | 429 | 0.483112 | [
[
[
"##### Copyright 2018 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Image classification",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/tutorials/images/classification\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs-l10n/blob/master/site/ja/tutorials/images/classification.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs-l10n/blob/master/site/ja/tutorials/images/classification.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/docs-l10n/site/ja/tutorials/images/classification.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"Note: これらのドキュメントは私たちTensorFlowコミュニティが翻訳したものです。コミュニティによる 翻訳は**ベストエフォート**であるため、この翻訳が正確であることや[英語の公式ドキュメント](https://www.tensorflow.org/?hl=en)の 最新の状態を反映したものであることを保証することはできません。 この翻訳の品質を向上させるためのご意見をお持ちの方は、GitHubリポジトリ[tensorflow/docs](https://github.com/tensorflow/docs)にプルリクエストをお送りください。 コミュニティによる翻訳やレビューに参加していただける方は、 [[email protected] メーリングリスト](https://groups.google.com/a/tensorflow.org/forum/#!forum/docs-ja)にご連絡ください。",
"_____no_output_____"
],
[
"このチュートリアルでは、画像から猫または犬を分類する方法を示します。 `tf.keras.Sequential` モデルを使用して画像分類器を構築し、 `tf.keras.preprocessing.image.ImageDataGenerator` を使用してデータをロードします。このチュートリアルでは、以下のコンセプトにしたがって、実践的な経験と感覚を養います。\n\n* `tf.keras.preprocessing.image.ImageDataGenerator` クラスを使用して _データ入力パイプライン_ を構築し、モデルで使用するディスク上のデータを効率的に処理します。\n* _過学習(Overfitting)_ —過学習を識別および防止する方法。\n* _データ拡張(Data Augmentation)_ および _ドロップアウト(dropout)_ —データパイプラインおよび画像分類モデルに組み込むコンピュータービジョンタスクの過学習と戦うための重要なテクニック。\n\nこのチュートリアルは、基本的な機械学習のワークフローに従います。\n\n1. データの調査及び理解\n2. 入力パイプラインの構築\n3. モデルの構築\n4. モデルの学習\n5. モデルのテスト\n6. モデルの改善とプロセスの繰り返し",
"_____no_output_____"
],
[
"## パッケージのインポート",
"_____no_output_____"
],
[
"まずは必要なパッケージをインポートすることから始めましょう。 `os`パッケージはファイルとディレクトリ構造を読み込み、 NumPy は python リストの numpy 配列への変換と必要な行列演算の実行、 `matplotlib.pyplot` はグラフの描画や学習データおよび検証データに含まれる画像の表示、に利用します。",
"_____no_output_____"
],
[
"モデルの構築に必要な TensorFlow と Keras クラスをインポートします。",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf",
"_____no_output_____"
],
[
"from tensorflow.keras.models import Sequential\nfrom tensorflow.keras.layers import Dense, Conv2D, Flatten, Dropout, MaxPooling2D\nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator\n\nimport os\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## データの読み込み",
"_____no_output_____"
],
[
"データセットのダウンロードから始めます。このチュートリアルでは、 Kaggle の <a href=\"https://www.kaggle.com/c/dogs-vs-cats/data\" target=\"_blank\">Dogs vs Cats</a> データセットをフィルタリングしたバージョンを使用します。データセットのアーカイブバージョンをダウンロードし、\"/tmp/\"ディレクトリに保存します。",
"_____no_output_____"
]
],
[
[
"_URL = 'https://storage.googleapis.com/mledu-datasets/cats_and_dogs_filtered.zip'\n\npath_to_zip = tf.keras.utils.get_file('cats_and_dogs.zip', origin=_URL, extract=True)\n\nPATH = os.path.join(os.path.dirname(path_to_zip), 'cats_and_dogs_filtered')",
"_____no_output_____"
]
],
[
[
"データセットのディレクトリ構造は次のとおりです:\n\n<pre>\n<b>cats_and_dogs_filtered</b>\n|__ <b>train</b>\n |______ <b>cats</b>: [cat.0.jpg, cat.1.jpg, cat.2.jpg ....]\n |______ <b>dogs</b>: [dog.0.jpg, dog.1.jpg, dog.2.jpg ...]\n|__ <b>validation</b>\n |______ <b>cats</b>: [cat.2000.jpg, cat.2001.jpg, cat.2002.jpg ....]\n |______ <b>dogs</b>: [dog.2000.jpg, dog.2001.jpg, dog.2002.jpg ...]\n</pre>",
"_____no_output_____"
],
[
"データの内容を抽出した後、学習および検証セットのための適切なファイルパスで変数を設定します。",
"_____no_output_____"
]
],
[
[
"train_dir = os.path.join(PATH, 'train')\nvalidation_dir = os.path.join(PATH, 'validation')",
"_____no_output_____"
],
[
"train_cats_dir = os.path.join(train_dir, 'cats') # 学習用の猫画像のディレクトリ\ntrain_dogs_dir = os.path.join(train_dir, 'dogs') # 学習用の犬画像のディレクトリ\nvalidation_cats_dir = os.path.join(validation_dir, 'cats') # 検証用の猫画像のディレクトリ\nvalidation_dogs_dir = os.path.join(validation_dir, 'dogs') # 検証用の犬画像のディレクトリ",
"_____no_output_____"
]
],
[
[
"### データの理解",
"_____no_output_____"
],
[
"学習および検証ディレクトリの中にある猫と犬の画像の数を見てみましょう:",
"_____no_output_____"
]
],
[
[
"num_cats_tr = len(os.listdir(train_cats_dir))\nnum_dogs_tr = len(os.listdir(train_dogs_dir))\n\nnum_cats_val = len(os.listdir(validation_cats_dir))\nnum_dogs_val = len(os.listdir(validation_dogs_dir))\n\ntotal_train = num_cats_tr + num_dogs_tr\ntotal_val = num_cats_val + num_dogs_val",
"_____no_output_____"
],
[
"print('total training cat images:', num_cats_tr)\nprint('total training dog images:', num_dogs_tr)\n\nprint('total validation cat images:', num_cats_val)\nprint('total validation dog images:', num_dogs_val)\nprint(\"--\")\nprint(\"Total training images:\", total_train)\nprint(\"Total validation images:\", total_val)",
"_____no_output_____"
]
],
[
[
"便宜上、データセットの前処理およびネットワークの学習中に使用する変数を設定します。",
"_____no_output_____"
]
],
[
[
"batch_size = 128\nepochs = 15\nIMG_HEIGHT = 150\nIMG_WIDTH = 150",
"_____no_output_____"
]
],
[
[
"## データの準備",
"_____no_output_____"
],
[
"モデルにデータを送る前に、画像を適切に前処理された浮動小数点テンソルにフォーマットします。\n\n1.ディスクから画像を読み取ります。\n2.これらの画像のコンテンツをデコードし、RGB値にしたがって適切なグリッド形式に変換します。\n3.それらを浮動小数点テンソルに変換します。\n4.ニューラルネットワークは小さな入力値を扱う方が適しているため、テンソルを0〜255の値から0〜1の値にリスケーリングします。\n\n幸い、これらすべてのタスクは、 `tf.keras` によって提供される `ImageDataGenerator` クラスで実行できます。この `ImageDataGenerator` はディスクから画像を読み取り、適切なテンソルに前処理を行います。さらに、これらの画像をテンソルのバッチに変換するジェネレータをセットアップします。これは、ネットワーク学習時に便利です。",
"_____no_output_____"
]
],
[
[
"train_image_generator = ImageDataGenerator(rescale=1./255) # 学習データのジェネレータ\nvalidation_image_generator = ImageDataGenerator(rescale=1./255) # 検証データのジェネレータ",
"_____no_output_____"
]
],
[
[
"学習および検証画像のジェネレータを定義したのち、 `flow_from_directory` メソッドはディスクから画像をロードし、リスケーリングを適用し、画像を必要な大きさにリサイズします。",
"_____no_output_____"
]
],
[
[
"train_data_gen = train_image_generator.flow_from_directory(batch_size=batch_size,\n directory=train_dir,\n shuffle=True,\n target_size=(IMG_HEIGHT, IMG_WIDTH),\n class_mode='binary')",
"_____no_output_____"
],
[
"val_data_gen = validation_image_generator.flow_from_directory(batch_size=batch_size,\n directory=validation_dir,\n target_size=(IMG_HEIGHT, IMG_WIDTH),\n class_mode='binary')",
"_____no_output_____"
]
],
[
[
"### 学習用画像の可視化",
"_____no_output_____"
],
[
"学習用のジェネレータから画像バッチを抽出して可視化します。(この例では32個の画像を抽出し、そのうち5つを `matplotlib` で描画します。)",
"_____no_output_____"
]
],
[
[
"sample_training_images, _ = next(train_data_gen)",
"_____no_output_____"
]
],
[
[
" `next` 関数はデータセットからバッチを返します。 `next` 関数の返り値は `(x_train、y_train)` の形式で、 `x_train` は学習用の特徴量、 `y_train` はそのラベルです。ラベルを破棄して、学習用画像の可視化のみを行います。",
"_____no_output_____"
]
],
[
[
"# この関数は、1行5列のグリッド形式で画像をプロットし、画像は各列に配置されます。\ndef plotImages(images_arr):\n fig, axes = plt.subplots(1, 5, figsize=(20,20))\n axes = axes.flatten()\n for img, ax in zip( images_arr, axes):\n ax.imshow(img)\n ax.axis('off')\n plt.tight_layout()\n plt.show()",
"_____no_output_____"
],
[
"plotImages(sample_training_images[:5])",
"_____no_output_____"
]
],
[
[
"## モデルの構築",
"_____no_output_____"
],
[
"モデルはmax pooling層を伴う3つの畳み込みブロックからなります。さらに `relu` 活性化関数によるアクティベーションを伴う512ユニットの全結合層があります。モデルは、シグモイド活性化関数による2値分類に基づいてクラスに属する確率を出力します。",
"_____no_output_____"
]
],
[
[
"model = Sequential([\n Conv2D(16, 3, padding='same', activation='relu', input_shape=(IMG_HEIGHT, IMG_WIDTH ,3)),\n MaxPooling2D(),\n Conv2D(32, 3, padding='same', activation='relu'),\n MaxPooling2D(),\n Conv2D(64, 3, padding='same', activation='relu'),\n MaxPooling2D(),\n Flatten(),\n Dense(512, activation='relu'),\n Dense(1, activation='sigmoid')\n])",
"_____no_output_____"
]
],
[
[
"### モデルのコンパイル\nこのチュートリアルでは、 *ADAM* オプティマイザーと *binary cross entropy* 損失関数を選択します。各学習エポックの学習と検証の精度を表示するために、`metrics` 引数を渡します。",
"_____no_output_____"
]
],
[
[
"model.compile(optimizer='adam',\n loss='binary_crossentropy',\n metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"### モデルの概要\n\nすべてのネットワークのレイヤーを見るには、モデルの `summary` メソッドを利用します:",
"_____no_output_____"
]
],
[
[
"model.summary()",
"_____no_output_____"
]
],
[
[
"### モデルの学習",
"_____no_output_____"
],
[
"`ImageDataGenerator` クラスの `fit_generator` メソッドを使用して、ネットワークを学習します。",
"_____no_output_____"
]
],
[
[
"history = model.fit_generator(\n train_data_gen,\n steps_per_epoch=total_train // batch_size,\n epochs=epochs,\n validation_data=val_data_gen,\n validation_steps=total_val // batch_size\n)",
"_____no_output_____"
]
],
[
[
"### 学習結果の可視化",
"_____no_output_____"
],
[
"ネットワークを学習した後、結果を可視化します。",
"_____no_output_____"
]
],
[
[
"acc = history.history['accuracy']\nval_acc = history.history['val_accuracy']\n\nloss = history.history['loss']\nval_loss = history.history['val_loss']\n\nepochs_range = range(epochs)\n\nplt.figure(figsize=(8, 8))\nplt.subplot(1, 2, 1)\nplt.plot(epochs_range, acc, label='Training Accuracy')\nplt.plot(epochs_range, val_acc, label='Validation Accuracy')\nplt.legend(loc='lower right')\nplt.title('Training and Validation Accuracy')\n\nplt.subplot(1, 2, 2)\nplt.plot(epochs_range, loss, label='Training Loss')\nplt.plot(epochs_range, val_loss, label='Validation Loss')\nplt.legend(loc='upper right')\nplt.title('Training and Validation Loss')\nplt.show()",
"_____no_output_____"
]
],
[
[
"プロットからわかるように、学習セットの精度と検証セットの精度は大幅に外れており、モデルは検証セットで約70%の精度しか達成していません。\n\n何がうまくいかなかったかを見て、モデル全体のパフォーマンスを向上してみましょう。",
"_____no_output_____"
],
[
"## 過学習",
"_____no_output_____"
],
[
"上記のプロットでは、学習セットの精度は時間とともに直線的に向上していますが、検証セットの精度は学習プロセスの中で約70%あたりで頭打ちになっています。そして、学習と検証の精度の違いが顕著です。これは *過学習* のサインです。\n\n学習サンプルが少ない場合、モデルは学習サンプルに含まれるノイズや不要な詳細から学習してしまい、これによって新しいサンプルに対するモデルの性能に悪影響を与えることがあります。この現象は、過学習として知られています。過学習とは、モデルが新しいデータセットに対して汎化するのが難しい状態をいいます。\n\n学習プロセスにおいて過学習に対抗する手段はいくつかあります。このチュートリアルでは、*データ拡張(data Augmentation)* を使用し、さらにモデルに *ドロップアウト(dropout)* を追加します。",
"_____no_output_____"
],
[
"## データ拡張(Data augmentation)",
"_____no_output_____"
],
[
"過学習は一般に、学習サンプルが少ない場合に発生します。この問題を解決する方法の1つは、十分な数の学習サンプルが含まれるようにデータセットを拡張することです。データ拡張は、既存の学習サンプルに対してランダムな変換を行い、データセットとして利用できそうな画像を生成するアプローチをとります。このデータ拡張の目的は、学習中にモデルがまったくおなじ画像を2回利用しないようにすることです。これによってモデルをデータのより多くの特徴を利用し、より汎化することができます。\n\n`tf.keras` においては、このデータ拡張を `ImageDataGenerator` クラスを使用して実装します。データセットに対するさまざまな変換を指定することによって、学習プロセス中にそれが適用されます。",
"_____no_output_____"
],
[
"### データの拡張と可視化",
"_____no_output_____"
],
[
"最初に、ランダムな水平反転による拡張をデータセットに適用し、それぞれの画像が変換後にどのように見えるかを確認します。",
"_____no_output_____"
],
[
"### 水平反転の適用",
"_____no_output_____"
],
[
" このデータ拡張を適用するためには、 `ImageDataGenerator` クラスの引数として `horizontal_flip` を渡し、 `True`を設定します。",
"_____no_output_____"
]
],
[
[
"image_gen = ImageDataGenerator(rescale=1./255, horizontal_flip=True)",
"_____no_output_____"
],
[
"train_data_gen = image_gen.flow_from_directory(batch_size=batch_size,\n directory=train_dir,\n shuffle=True,\n target_size=(IMG_HEIGHT, IMG_WIDTH))",
"_____no_output_____"
]
],
[
[
"学習サンプルから1つのサンプル画像を取得する作業を5回繰り返して、おなじ画像に5回データ拡張が適用されるようにします。",
"_____no_output_____"
]
],
[
[
"augmented_images = [train_data_gen[0][0][0] for i in range(5)]",
"_____no_output_____"
],
[
"# 上で学習用画像の可視化のために定義、使用されたおなじカスタムプロット関数を再利用する\nplotImages(augmented_images)",
"_____no_output_____"
]
],
[
[
"### 画像のランダムな回転",
"_____no_output_____"
],
[
"回転のデータ拡張を利用して学習用サンプルをランダムに左右45度の範囲で回転させてみましょう。",
"_____no_output_____"
]
],
[
[
"image_gen = ImageDataGenerator(rescale=1./255, rotation_range=45)",
"_____no_output_____"
],
[
"train_data_gen = image_gen.flow_from_directory(batch_size=batch_size,\n directory=train_dir,\n shuffle=True,\n target_size=(IMG_HEIGHT, IMG_WIDTH))\n\naugmented_images = [train_data_gen[0][0][0] for i in range(5)]",
"_____no_output_____"
],
[
"plotImages(augmented_images)",
"_____no_output_____"
]
],
[
[
"### ズームによるデータ拡張の適用",
"_____no_output_____"
],
[
"データセットにズームによるデータ拡張を適用して、画像をランダムに最大50%拡大します。",
"_____no_output_____"
]
],
[
[
"image_gen = ImageDataGenerator(rescale=1./255, zoom_range=0.5)",
"_____no_output_____"
],
[
"train_data_gen = image_gen.flow_from_directory(batch_size=batch_size,\n directory=train_dir,\n shuffle=True,\n target_size=(IMG_HEIGHT, IMG_WIDTH))\n\naugmented_images = [train_data_gen[0][0][0] for i in range(5)]",
"_____no_output_____"
],
[
"plotImages(augmented_images)",
"_____no_output_____"
]
],
[
[
"### すべてのデータ拡張を同時に利用する",
"_____no_output_____"
],
[
"ここまでで紹介したすべてのデータ拡張機能を適用します。ここでは、学習用画像に対して、リスケール、45度の回転、幅シフト、高さシフト、水平反転、ズームを適用しました。",
"_____no_output_____"
]
],
[
[
"image_gen_train = ImageDataGenerator(\n rescale=1./255,\n rotation_range=45,\n width_shift_range=.15,\n height_shift_range=.15,\n horizontal_flip=True,\n zoom_range=0.5\n )",
"_____no_output_____"
],
[
"train_data_gen = image_gen_train.flow_from_directory(batch_size=batch_size,\n directory=train_dir,\n shuffle=True,\n target_size=(IMG_HEIGHT, IMG_WIDTH),\n class_mode='binary')",
"_____no_output_____"
]
],
[
[
"これらのデータ拡張がデータセットにランダムに適用されたときに、一つの画像に対して5回の個別の適用を行った際にそれぞれどのように見えるかを可視化します。",
"_____no_output_____"
]
],
[
[
"augmented_images = [train_data_gen[0][0][0] for i in range(5)]\nplotImages(augmented_images)",
"_____no_output_____"
]
],
[
[
"### 検証データジェネレータの構築",
"_____no_output_____"
],
[
"一般に、データ拡張は学習サンプルのみに適用します。今回は、 `ImageDataGenerator` を使用して検証画像に対してリスケールのみを実施し、バッチに変換します。",
"_____no_output_____"
]
],
[
[
"image_gen_val = ImageDataGenerator(rescale=1./255)",
"_____no_output_____"
],
[
"val_data_gen = image_gen_val.flow_from_directory(batch_size=batch_size,\n directory=validation_dir,\n target_size=(IMG_HEIGHT, IMG_WIDTH),\n class_mode='binary')",
"_____no_output_____"
]
],
[
[
"## ドロップアウト(dropout)",
"_____no_output_____"
],
[
"過学習を避けるもう一つの方法は、ネットワークに *ドロップアウト* を導入することです。これは、ネットワークにおいて重みを小さくする正則化の方式で、これによって重みの値の分布がより規則的になり、少ない学習データに対する過学習を減らすことができます。ドロップアウトはこのチュートリアルで利用される正則化手法の一つです。\n\nドロップアウトをレイヤーに適用すると、学習プロセス中に適用されたレイヤーのうちランダムに出力ユニットをドロップアウト(ゼロに設定)します。ドロップアウトは、入力値として0.1、0.2、0.4といった形式の小数をとります。これは、適用されたレイヤーからランダムに出力単位の10%、20%、または40%をドロップアウトすることを意味します。\n\n特定のレイヤーに0.1ドロップアウトを適用すると、各学習エポックにおいて出力ユニットの10%がランダムに0にされます。\n\nこの新しいドロップアウト機能を使用したネットワークアーキテクチャを作成し、異なる畳み込みレイヤーや全接続レイヤーに適用してみましょう。",
"_____no_output_____"
],
[
"## ドロップアウトを追加した新しいネットワークの構築",
"_____no_output_____"
],
[
"ここでは、ドロップアウトを最初と最後の max pool 層に適用します。ドロップアウトを適用すると、各学習エポック中にニューロンの20%がランダムにゼロに設定されます。これにより、学習データセットに対する過学習を避けることができます。",
"_____no_output_____"
]
],
[
[
"model_new = Sequential([\n Conv2D(16, 3, padding='same', activation='relu', \n input_shape=(IMG_HEIGHT, IMG_WIDTH ,3)),\n MaxPooling2D(),\n Dropout(0.2),\n Conv2D(32, 3, padding='same', activation='relu'),\n MaxPooling2D(),\n Conv2D(64, 3, padding='same', activation='relu'),\n MaxPooling2D(),\n Dropout(0.2),\n Flatten(),\n Dense(512, activation='relu'),\n Dense(1, activation='sigmoid')\n])",
"_____no_output_____"
]
],
[
[
"### モデルのコンパイル",
"_____no_output_____"
],
[
"ネットワークにドロップアウトを導入した後、モデルをコンパイルし、レイヤーの概要を表示します。",
"_____no_output_____"
]
],
[
[
"model_new.compile(optimizer='adam',\n loss='binary_crossentropy',\n metrics=['accuracy'])\n\nmodel_new.summary()",
"_____no_output_____"
]
],
[
[
"### モデルの学習",
"_____no_output_____"
],
[
"学習サンプルにデータ拡張を導入し、ネットワークにドロップアウトを追加した後、この新しいネットワークを学習します:",
"_____no_output_____"
]
],
[
[
"history = model_new.fit_generator(\n train_data_gen,\n steps_per_epoch=total_train // batch_size,\n epochs=epochs,\n validation_data=val_data_gen,\n validation_steps=total_val // batch_size\n)",
"_____no_output_____"
]
],
[
[
"### モデルの可視化",
"_____no_output_____"
],
[
"学習後に新しいモデルを可視化すると、過学習が前回よりも大幅に少ないことがわかります。より多くのエポックでモデルを学習すると、精度はさらに向上するはずです。",
"_____no_output_____"
]
],
[
[
"acc = history.history['accuracy']\nval_acc = history.history['val_accuracy']\n\nloss = history.history['loss']\nval_loss = history.history['val_loss']\n\nepochs_range = range(epochs)\n\nplt.figure(figsize=(8, 8))\nplt.subplot(1, 2, 1)\nplt.plot(epochs_range, acc, label='Training Accuracy')\nplt.plot(epochs_range, val_acc, label='Validation Accuracy')\nplt.legend(loc='lower right')\nplt.title('Training and Validation Accuracy')\n\nplt.subplot(1, 2, 2)\nplt.plot(epochs_range, loss, label='Training Loss')\nplt.plot(epochs_range, val_loss, label='Validation Loss')\nplt.legend(loc='upper right')\nplt.title('Training and Validation Loss')\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
cb455a701986600123b47eb18c80315834a176c9 | 3,738 | ipynb | Jupyter Notebook | content/lessons/08/Now-You-Code/NYC2-Shopping-List.ipynb | MahopacHS/spring2019-Christian64Aguilar | 2e9ac4a5245d459f6d086c61ad3c0540db39b981 | [
"MIT"
] | null | null | null | content/lessons/08/Now-You-Code/NYC2-Shopping-List.ipynb | MahopacHS/spring2019-Christian64Aguilar | 2e9ac4a5245d459f6d086c61ad3c0540db39b981 | [
"MIT"
] | null | null | null | content/lessons/08/Now-You-Code/NYC2-Shopping-List.ipynb | MahopacHS/spring2019-Christian64Aguilar | 2e9ac4a5245d459f6d086c61ad3c0540db39b981 | [
"MIT"
] | null | null | null | 51.205479 | 644 | 0.61985 | [
[
[
"# Now You Code 2: Shopping List\n\nWrite a program to input a list of grocery items for your shopping list and then writes them to a file a line at a time. The program should keep asking you to enter grocery items until you type `'done'`.\n\nAfter you complete the list, the program should then load the file and read the list back to you by printing each item out.\n\nSample Run:\n\n```\nLet's make a shopping list. Type 'done' when you're finished:\nEnter Item: milk\nEnter Item: cheese\nEnter Item: eggs\nEnter Item: beer\nEnter Item: apples\nEnter Item: done\nYour shopping list:\nmilk\ncheese\neggs\nbeer\napples\n```\n",
"_____no_output_____"
],
[
"## Step 1: Problem Analysis\n\nInputs:\n\nOutputs:\n\nAlgorithm (Steps in Program):\n\n",
"_____no_output_____"
]
],
[
[
"filename = \"NYC2-shopping-list.txt\"\n## Step 2: write code here \ndef addtolist(item):\n add= list.append(newitem)\n print(\"added {} now I have {}items\".format(newitem,len(lista)))\n file=open('NYC2-shopping-list.txt','a')\n file.write(item+\"\\n\")",
"_____no_output_____"
]
],
[
[
"## Step 3: Refactoring\n\nRefactor the part of your program which reads the shopping list from the file into a separate user-defined function called `readShoppingList` \nre-write your program to use this function.\n\n## ReadShoppingList function\n\nInputs:\n\nOutputs:\n\nAlgorithm (Steps in Program):\n\n\n\n",
"_____no_output_____"
]
],
[
[
"## Step 4: Write program again with refactored code.\ndef readShoppingList():\n shoppingList = [NYC2-shopping-list.txt]\n # todo read shopping list here\n \n return shoppingList\n\n\n# TODO Main code here\n\n",
"_____no_output_____"
]
],
[
[
"## Step 5: Questions\n\n1. Is the refactored code in step 4 easier to read? Why or why not? no because it requires more steps while step one didnt need much at all.\n\n2. Explain how this program could be refarctored further (there's one thing that's obvious). by using the split function. \n\n3. Describe how this program can be modified to support multiple shopping lists? by creating more files and having the program open multiple files \n",
"_____no_output_____"
],
[
"## Reminder of Evaluation Criteria\n\n1. What the problem attempted (analysis, code, and answered questions) ?\n2. What the problem analysis thought out? (does the program match the plan?)\n3. Does the code execute without syntax error?\n4. Does the code solve the intended problem?\n5. Is the code well written? (easy to understand, modular, and self-documenting, handles errors)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb45611cbfff0b5cc302e78bdd66f1f3d31b2e08 | 93,833 | ipynb | Jupyter Notebook | docs/site/tutorials/model_training_walkthrough.ipynb | realdoug/tensorflow-swift | 2bc8e308c74c0e9d1c0d97a066d033b4458eea4d | [
"CC-BY-4.0"
] | null | null | null | docs/site/tutorials/model_training_walkthrough.ipynb | realdoug/tensorflow-swift | 2bc8e308c74c0e9d1c0d97a066d033b4458eea4d | [
"CC-BY-4.0"
] | null | null | null | docs/site/tutorials/model_training_walkthrough.ipynb | realdoug/tensorflow-swift | 2bc8e308c74c0e9d1c0d97a066d033b4458eea4d | [
"CC-BY-4.0"
] | null | null | null | 66.266243 | 22,846 | 0.709772 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
cb45642fe58f9499f9f83626ad7d526d5c6101e4 | 77,640 | ipynb | Jupyter Notebook | week03_lm/homework.ipynb | waytobehigh/nlp_course | 3529616f17fd4f854e79ae32d87934af689d88e1 | [
"MIT"
] | 2 | 2019-10-31T15:06:52.000Z | 2020-04-08T18:51:59.000Z | week03_lm/homework.ipynb | waytobehigh/nlp_course | 3529616f17fd4f854e79ae32d87934af689d88e1 | [
"MIT"
] | null | null | null | week03_lm/homework.ipynb | waytobehigh/nlp_course | 3529616f17fd4f854e79ae32d87934af689d88e1 | [
"MIT"
] | null | null | null | 87.138047 | 22,572 | 0.798905 | [
[
[
"### Homework: going neural (6 pts)\n\nWe've checked out statistical approaches to language models in the last notebook. Now let's go find out what deep learning has to offer.\n\n<img src='https://raw.githubusercontent.com/yandexdataschool/nlp_course/master/resources/expanding_mind_lm_kn_3.png' width=300px>\n\nWe're gonna use the same dataset as before, except this time we build a language model that's character-level, not word level.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"Working on character level means that we don't need to deal with large vocabulary or missing words. Heck, we can even keep uppercase words in text! The downside, however, is that all our sequences just got a lot longer.\n\nHowever, we still need special tokens:\n* Begin Of Sequence (__BOS__) - this token is at the start of each sequence. We use it so that we always have non-empty input to our neural network. $P(x_t) = P(x_1 | BOS)$\n* End Of Sequence (__EOS__) - you guess it... this token is at the end of each sequence. The catch is that it should __not__ occur anywhere else except at the very end. If our model produces this token, the sequence is over.\n",
"_____no_output_____"
]
],
[
[
"BOS, EOS = ' ', '\\n'\n\ndata = pd.read_json(\"./arxivData.json\")\nlines = data.apply(lambda row: (row['title'] + ' ; ' + row['summary'])[:512], axis=1) \\\n .apply(lambda line: BOS + line.replace(EOS, ' ') + EOS) \\\n .tolist()\n\n# if you missed the seminar, download data here - https://yadi.sk/d/_nGyU2IajjR9-w",
"_____no_output_____"
]
],
[
[
"Our next step is __building char-level vocabulary__. Put simply, you need to assemble a list of all unique tokens in the dataset.",
"_____no_output_____"
]
],
[
[
"# get all unique characters from lines (including capital letters and symbols)\ntokens = set(''.join(lines))\n\ntokens = sorted(tokens)\nn_tokens = len(tokens)\nprint ('n_tokens = ',n_tokens)\nassert 100 < n_tokens < 150\nassert BOS in tokens, EOS in tokens",
"n_tokens = 136\n"
]
],
[
[
"We can now assign each character with it's index in tokens list. This way we can encode a string into a TF-friendly integer vector.",
"_____no_output_____"
]
],
[
[
"# dictionary of character -> its identifier (index in tokens list)\ntoken_to_id = {token: id for id, token in enumerate(tokens)}",
"_____no_output_____"
],
[
"assert len(tokens) == len(token_to_id), \"dictionaries must have same size\"\nfor i in range(n_tokens):\n assert token_to_id[tokens[i]] == i, \"token identifier must be it's position in tokens list\"\n\nprint(\"Seems alright!\")",
"Seems alright!\n"
]
],
[
[
"Our final step is to assemble several strings in a integet matrix `[batch_size, text_length]`. \n\nThe only problem is that each sequence has a different length. We can work around that by padding short sequences with extra _EOS_ or cropping long sequences. Here's how it works:",
"_____no_output_____"
]
],
[
[
"def to_matrix(lines, max_len=None, pad=token_to_id[EOS], dtype='int32'):\n \"\"\"Casts a list of lines into tf-digestable matrix\"\"\"\n max_len = max_len or max(map(len, lines))\n lines_ix = np.zeros([len(lines), max_len], dtype) + pad\n for i in range(len(lines)):\n line_ix = list(map(token_to_id.get, lines[i][:max_len]))\n lines_ix[i, :len(line_ix)] = line_ix\n return lines_ix",
"_____no_output_____"
],
[
"#Example: cast 4 random names to matrices, pad with zeros\ndummy_lines = [\n ' abc\\n',\n ' abacaba\\n',\n ' abc1234567890\\n',\n]\nprint(to_matrix(dummy_lines))\n\n",
"[[ 1 66 67 68 0 0 0 0 0 0 0 0 0 0 0]\n [ 1 66 67 66 68 66 67 66 0 0 0 0 0 0 0]\n [ 1 66 67 68 18 19 20 21 22 23 24 25 26 17 0]]\n"
]
],
[
[
"### Neural Language Model\n\nJust like for N-gram LMs, we want to estimate probability of text as a joint probability of tokens (symbols this time).\n\n$$P(X) = \\prod_t P(x_t \\mid x_0, \\dots, x_{t-1}).$$ \n\nInstead of counting all possible statistics, we want to train a neural network with parameters $\\theta$ that estimates the conditional probabilities:\n\n$$ P(x_t \\mid x_0, \\dots, x_{t-1}) \\approx p(x_t \\mid x_0, \\dots, x_{t-1}, \\theta) $$\n\n\nBut before we optimize, we need to define our neural network. Let's start with a fixed-window (aka convolutional) architecture:\n\n<img src='https://raw.githubusercontent.com/yandexdataschool/nlp_course/master/resources/fixed_window_lm.jpg' width=400px>\n",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport keras, keras.layers as L\nsess = tf.InteractiveSession()",
"Using TensorFlow backend.\n"
],
[
"class FixedWindowLanguageModel:\n def __init__(self, n_tokens=n_tokens, emb_size=16, hid_size=64):\n \"\"\" \n A fixed window model that looks on at least 5 previous symbols.\n \n Note: fixed window LM is effectively performing a convolution over a sequence of words.\n This convolution only looks on current and previous words.\n Such convolution can be represented as a sequence of 2 operations:\n - pad input vectors by {strides * (filter_size - 1)} zero vectors on the \"left\", do not pad right\n - perform regular convolution with {filter_size} and {strides}\n \n You can stack several convolutions at once\n \"\"\"\n \n #YOUR CODE - create layers/variables and any metadata you want, e.g. self.emb = L.Embedding(...)\n \n self.emb = L.Embedding(input_dim=n_tokens, output_dim=emb_size)\n self.conv1 = L.Convolution1D(filters=hid_size, kernel_size=5,\n padding='causal', name='conv1')\n self.conv2 = L.Convolution1D(filters=n_tokens, kernel_size=5,\n padding='causal', name='conv2')\n self.activation = L.Activation('relu')\n \n #END OF YOUR CODE\n \n self.prefix_ix = tf.placeholder('int32', [None, None])\n self.next_token_probs = tf.nn.softmax(self(self.prefix_ix)[:, -1])\n \n def __call__(self, input_ix):\n \"\"\"\n compute language model logits given input tokens\n :param input_ix: batch of sequences with token indices, tf tensor: int32[batch_size, sequence_length]\n :returns: pre-softmax linear outputs of language model [batch_size, sequence_length, n_tokens]\n these outputs will be used as logits to compute P(x_t | x_0, ..., x_{t - 1})\n \"\"\"\n \n embedding = self.emb(input_ix)\n conv1 = self.conv1(embedding)\n conv1 = self.activation(conv1)\n conv2 = self.conv2(conv1)\n \n return conv2\n \n def get_possible_next_tokens(self, prefix=BOS, temperature=1.0, max_len=100, sess=sess):\n \"\"\" :returns: probabilities of next token, dict {token : prob} for all tokens \"\"\"\n probs = sess.run(self.next_token_probs, {self.prefix_ix: to_matrix([prefix])})[0]\n return dict(zip(tokens, probs))\n ",
"_____no_output_____"
],
[
"window_lm = FixedWindowLanguageModel()",
"_____no_output_____"
],
[
"dummy_input_ix = tf.constant(to_matrix(dummy_lines))\ndummy_lm_out = window_lm(dummy_input_ix)\n# note: tensorflow and keras layers only create variables after they're first applied (called)\n\nsess.run(tf.global_variables_initializer())\ndummy_logits = sess.run(dummy_lm_out)\n\nassert dummy_logits.shape == (len(dummy_lines), max(map(len, dummy_lines)), n_tokens), \"please check output shape\"\nassert np.all(np.isfinite(dummy_logits)), \"inf/nan encountered\"\nassert not np.allclose(dummy_logits.sum(-1), 1), \"please predict linear outputs, don't use softmax (maybe you've just got unlucky)\"",
"_____no_output_____"
],
[
"# test for lookahead\ndummy_input_ix_2 = tf.constant(to_matrix([line[:3] + 'e' * (len(line) - 3) for line in dummy_lines]))\ndummy_lm_out_2 = window_lm(dummy_input_ix_2)\ndummy_logits_2 = sess.run(dummy_lm_out_2)\nassert np.allclose(dummy_logits[:, :3] - dummy_logits_2[:, :3], 0), \"your model's predictions depend on FUTURE tokens. \" \\\n \" Make sure you don't allow any layers to look ahead of current token.\" \\\n \" You can also get this error if your model is not deterministic (e.g. dropout). Disable it for this test.\"",
"_____no_output_____"
]
],
[
[
"We can now tune our network's parameters to minimize categorical crossentropy over training dataset $D$:\n\n$$ L = {\\frac1{|D|}} \\sum_{X \\in D} \\sum_{x_i \\in X} - \\log p(x_t \\mid x_1, \\dots, x_{t-1}, \\theta) $$\n\nAs usual with with neural nets, this optimization is performed via stochastic gradient descent with backprop. One can also note that minimizing crossentropy is equivalent to minimizing model __perplexity__, KL-divergence or maximizng log-likelihood.",
"_____no_output_____"
]
],
[
[
"def compute_lengths(input_ix, eos_ix=token_to_id[EOS]):\n \"\"\" compute length of each line in input ix (incl. first EOS), int32 vector of shape [batch_size] \"\"\"\n count_eos = tf.cumsum(tf.to_int32(tf.equal(input_ix, eos_ix)), axis=1, exclusive=True)\n lengths = tf.reduce_sum(tf.to_int32(tf.equal(count_eos, 0)), axis=1)\n return lengths\n\nprint('matrix:\\n', dummy_input_ix.eval())\nprint('lengths:', compute_lengths(dummy_input_ix).eval())",
"matrix:\n [[ 1 66 67 68 0 0 0 0 0 0 0 0 0 0 0]\n [ 1 66 67 66 68 66 67 66 0 0 0 0 0 0 0]\n [ 1 66 67 68 18 19 20 21 22 23 24 25 26 17 0]]\nlengths: [ 5 9 15]\n"
],
[
"input_ix = tf.placeholder('int32', [None, None])\n\nlogits = window_lm(input_ix[:, :-1])\nreference_answers = input_ix[:, 1:]\n\n# Your task: implement loss function as per formula above\n# your loss should only be computed on actual tokens, excluding padding\n# predicting actual tokens and first EOS do count. Subsequent EOS-es don't\n# you will likely need to use compute_lengths and/or tf.sequence_mask to get it right.\n\nlengths = compute_lengths(input_ix)\nmask = tf.to_float(tf.sequence_mask(lengths, tf.shape(input_ix)[1])[:, 1:])\nloss = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=reference_answers, logits=logits)\nloss = tf.reduce_sum(loss * mask) / tf.reduce_sum(mask) \n\n# operation to update network weights\ntrain_step = tf.train.AdamOptimizer().minimize(loss)",
"_____no_output_____"
],
[
"loss_1 = sess.run(loss, {input_ix: to_matrix(dummy_lines, max_len=50)})\nloss_2 = sess.run(loss, {input_ix: to_matrix(dummy_lines, max_len=100)})\nassert (np.ndim(loss_1) == 0) and (0 < loss_1 < 100), \"loss must be a positive scalar\"\nassert np.allclose(loss_1, loss_2), 'do not include AFTER first EOS into loss. '\\\n 'Hint: use tf.sequence_mask. Beware +/-1 errors. And be careful when averaging!'",
"_____no_output_____"
]
],
[
[
"### Training loop\n\nNow let's train our model on minibatches of data",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\ntrain_lines, dev_lines = train_test_split(lines, test_size=0.25, random_state=42)\n\nsess.run(tf.global_variables_initializer())\nbatch_size = 256\nscore_dev_every = 250\ntrain_history, dev_history = [], []",
"_____no_output_____"
],
[
"def score_lines(dev_lines, batch_size):\n \"\"\" computes average loss over the entire dataset \"\"\"\n dev_loss_num, dev_loss_len = 0., 0.\n for i in range(0, len(dev_lines), batch_size):\n batch_ix = to_matrix(dev_lines[i: i + batch_size])\n dev_loss_num += sess.run(loss, {input_ix: batch_ix}) * len(batch_ix)\n dev_loss_len += len(batch_ix)\n return dev_loss_num / dev_loss_len\n\ndef generate(lm, prefix=BOS, temperature=1.0, max_len=100):\n \"\"\"\n Samples output sequence from probability distribution obtained by lm\n :param temperature: samples proportionally to lm probabilities ^ temperature\n if temperature == 0, always takes most likely token. Break ties arbitrarily.\n \"\"\"\n while True:\n token_probs = lm.get_possible_next_tokens(prefix)\n tokens, probs = zip(*token_probs.items())\n if temperature == 0:\n next_token = tokens[np.argmax(probs)]\n else:\n probs = np.array([p ** (1. / temperature) for p in probs])\n probs /= sum(probs)\n next_token = np.random.choice(tokens, p=probs)\n \n prefix += next_token\n if next_token == EOS or len(prefix) > max_len: break\n return prefix\n\nif len(dev_history) == 0:\n dev_history.append((0, score_lines(dev_lines, batch_size)))\n print(\"Before training:\", generate(window_lm, 'Bridging'))",
"Before training: BridgingY\"öŁfcμF}'[GÜàβáLWÜμ'ρ{1xZYεL(S}+8V#!ô|4`,ü.\"e(7;My.χ\"èDÖlμaõ|00KÉó93/(n+A4ŁW<R>RS!4èFM%q:A°É\n"
],
[
"from IPython.display import clear_output\nfrom random import sample\nfrom tqdm import trange\n\nfor i in trange(len(train_history), 5000):\n batch = to_matrix(sample(train_lines, batch_size))\n loss_i, _ = sess.run([loss, train_step], {input_ix: batch})\n train_history.append((i, loss_i))\n \n if (i + 1) % 50 == 0:\n clear_output(True)\n plt.scatter(*zip(*train_history), alpha=0.1, label='train_loss')\n if len(dev_history):\n plt.plot(*zip(*dev_history), color='red', label='dev_loss')\n plt.legend(); plt.grid(); plt.show()\n print(\"Generated examples (tau=0.5):\")\n for j in range(3):\n print(generate(window_lm, temperature=0.5))\n \n if (i + 1) % score_dev_every == 0:\n print(\"Scoring dev...\")\n dev_history.append((i, score_lines(dev_lines, batch_size)))\n print('#%i Dev loss: %.3f' % dev_history[-1])\n",
"_____no_output_____"
],
[
"assert np.mean(train_history[:10], axis=0)[1] > np.mean(train_history[-10:], axis=0)[1], \"The model didn't converge.\"\nprint(\"Final dev loss:\", dev_history[-1][-1])\n\nfor i in range(10):\n print(generate(window_lm, temperature=0.5))",
"Final dev loss: 1.577635495837142\n Search in the sements are of the model method space of a propose a novel method for stocally sentati\n Acture and compution training a sumplitical structure of the are is a propose sets of the the the fu\n The relater stoch as the method foun and maching cansumed to the problem with a noural networks wher\n Evolution of a structure is the detical contration of the and as agence able of the annomation from \n Application of semails classification and problems of the enterent deep neural network (CNN) and met\n Datasion poselation of the segmentation for exploit the fields for distrate of a monsider be often g\n Matrix Space of the context and event in the search to componition of the object to dead for constru\n Set of shown go use of convolutional Networks ; Tho subleation ; The recormation problems a large fo\n In this paper work of adgation of the conved of the problem of the Semband for state the explodica\n A Botification and explorical sefference models of the sequence the problems for $n$ convertation wi\n"
]
],
[
[
"### RNN Language Models\n\nFixed-size architectures are reasonably good when capturing short-term dependencies, but their design prevents them from capturing any signal outside their window. We can mitigate this problem by using a __recurrent neural network__:\n\n$$ h_0 = \\vec 0 ; \\quad h_{t+1} = RNN(x_t, h_t) $$\n\n$$ p(x_t \\mid x_0, \\dots, x_{t-1}, \\theta) = dense_{softmax}(h_{t-1}) $$\n\nSuch model processes one token at a time, left to right, and maintains a hidden state vector between them. Theoretically, it can learn arbitrarily long temporal dependencies given large enough hidden size.\n\n<img src='https://raw.githubusercontent.com/yandexdataschool/nlp_course/master/resources/rnn_lm.jpg' width=480px>",
"_____no_output_____"
]
],
[
[
"class RNNLanguageModel:\n def __init__(self, n_tokens=n_tokens, emb_size=16, hid_size=256):\n \"\"\" \n Build a recurrent language model.\n You are free to choose anything you want, but the recommended architecture is\n - token embeddings\n - one or more LSTM/GRU layers with hid size\n - linear layer to predict logits\n \"\"\"\n \n # YOUR CODE - create layers/variables/etc\n \n self.emb = L.Embedding(n_tokens, emb_size)\n self.lstm = L.LSTM(hid_size, return_sequences=True)\n self.linear = L.Dense(n_tokens)\n \n #END OF YOUR CODE\n \n \n self.prefix_ix = tf.placeholder('int32', [None, None])\n self.next_token_probs = tf.nn.softmax(self(self.prefix_ix)[:, -1])\n \n def __call__(self, input_ix):\n \"\"\"\n compute language model logits given input tokens\n :param input_ix: batch of sequences with token indices, tf tensor: int32[batch_size, sequence_length]\n :returns: pre-softmax linear outputs of language model [batch_size, sequence_length, n_tokens]\n these outputs will be used as logits to compute P(x_t | x_0, ..., x_{t - 1})\n \"\"\"\n \n embedding = self.emb(input_ix)\n lstm = self.lstm(embedding)\n linear = self.linear(lstm)\n \n return linear\n \n def get_possible_next_tokens(self, prefix=BOS, temperature=1.0, max_len=100, sess=sess):\n \"\"\" :returns: probabilities of next token, dict {token : prob} for all tokens \"\"\"\n probs = sess.run(self.next_token_probs, {self.prefix_ix: to_matrix([prefix])})[0]\n return dict(zip(tokens, probs))\n ",
"_____no_output_____"
],
[
"rnn_lm = RNNLanguageModel()",
"_____no_output_____"
],
[
"dummy_input_ix = tf.constant(to_matrix(dummy_lines))\ndummy_lm_out = rnn_lm(dummy_input_ix)\n# note: tensorflow and keras layers only create variables after they're first applied (called)\n\nsess.run(tf.global_variables_initializer())\ndummy_logits = sess.run(dummy_lm_out)\n\nassert dummy_logits.shape == (len(dummy_lines), max(map(len, dummy_lines)), n_tokens), \"please check output shape\"\nassert np.all(np.isfinite(dummy_logits)), \"inf/nan encountered\"\nassert not np.allclose(dummy_logits.sum(-1), 1), \"please predict linear outputs, don't use softmax (maybe you've just got unlucky)\"",
"_____no_output_____"
],
[
"# test for lookahead\ndummy_input_ix_2 = tf.constant(to_matrix([line[:3] + 'e' * (len(line) - 3) for line in dummy_lines]))\ndummy_lm_out_2 = rnn_lm(dummy_input_ix_2)\ndummy_logits_2 = sess.run(dummy_lm_out_2)\nassert np.allclose(dummy_logits[:, :3] - dummy_logits_2[:, :3], 0), \"your model's predictions depend on FUTURE tokens. \" \\\n \" Make sure you don't allow any layers to look ahead of current token.\" \\\n \" You can also get this error if your model is not deterministic (e.g. dropout). Disable it for this test.\"",
"_____no_output_____"
]
],
[
[
"### RNN training\n\nOur RNN language model should optimize the same loss function as fixed-window model. But there's a catch. Since RNN recurrently multiplies gradients through many time-steps, gradient values may explode, [breaking](https://raw.githubusercontent.com/yandexdataschool/nlp_course/master/resources/nan.jpg) your model.\nThe common solution to that problem is to clip gradients either [individually](https://www.tensorflow.org/versions/r1.1/api_docs/python/tf/clip_by_value) or [globally](https://www.tensorflow.org/versions/r1.1/api_docs/python/tf/clip_by_global_norm).\n\nYour task here is to prepare tensorflow graph that would minimize the same loss function. If you encounter large loss fluctuations during training, please add gradient clipping using urls above.\n\n_Note: gradient clipping is not exclusive to RNNs. Convolutional networks with enough depth often suffer from the same issue._",
"_____no_output_____"
]
],
[
[
"input_ix = tf.placeholder('int32', [None, None])\n\nlogits = rnn_lm(input_ix[:, :-1])\nreference_answers = input_ix[:, 1:]\n\n# Copy the loss function and train step from the fixed-window model training\nlengths = compute_lengths(input_ix)\nmask = tf.to_float(tf.sequence_mask(lengths, tf.shape(input_ix)[1])[:, 1:])\nloss = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=reference_answers, logits=logits)\nloss = tf.reduce_sum(loss * mask) / tf.reduce_sum(mask) \n\n# and the train step\ntrain_step = tf.train.AdamOptimizer().minimize(loss)",
"_____no_output_____"
],
[
"loss_1 = sess.run(loss, {input_ix: to_matrix(dummy_lines, max_len=50)})\nloss_2 = sess.run(loss, {input_ix: to_matrix(dummy_lines, max_len=100)})\nassert (np.ndim(loss_1) == 0) and (0 < loss_1 < 100), \"loss must be a positive scalar\"\nassert np.allclose(loss_1, loss_2), 'do not include AFTER first EOS into loss. Hint: use tf.sequence_mask. Be careful when averaging!'",
"_____no_output_____"
]
],
[
[
"### RNN: Training loop",
"_____no_output_____"
]
],
[
[
"sess.run(tf.global_variables_initializer())\nbatch_size = 128\nscore_dev_every = 250\ntrain_history, dev_history = [], []\n\ndev_history.append((0, score_lines(dev_lines, batch_size)))",
"_____no_output_____"
],
[
"for i in trange(len(train_history), 5000):\n batch = to_matrix(sample(train_lines, batch_size))\n loss_i, _ = sess.run([loss, train_step], {input_ix: batch})\n train_history.append((i, loss_i))\n \n if (i + 1) % 50 == 0:\n clear_output(True)\n plt.scatter(*zip(*train_history), alpha=0.1, label='train_loss')\n if len(dev_history):\n plt.plot(*zip(*dev_history), color='red', label='dev_loss')\n plt.legend(); plt.grid(); plt.show()\n print(\"Generated examples (tau=0.5):\")\n for j in range(3):\n print(generate(rnn_lm, temperature=0.5))\n \n if (i + 1) % score_dev_every == 0:\n print(\"Scoring dev...\")\n dev_history.append((i, score_lines(dev_lines, batch_size)))\n print('#%i Dev loss: %.3f' % dev_history[-1])\n",
"_____no_output_____"
],
[
"assert np.mean(train_history[:10]) > np.mean(train_history[-10:]), \"The model didn't converge.\"\nprint(\"Final dev loss:\", dev_history[-1][-1])\nfor i in range(10):\n print(generate(rnn_lm, temperature=0.5))",
"Final dev loss: 1.1499693006189857\n Fast Classification for Machine Learning ; This paper presents a set of a compositional information \n Scalable Convolutional Neural Networks ; In this paper we present a novel approach that are introduc\n Regression from a Generative Semantic Algorithm for Statistical Deep Networks ; The problem of discr\n A Constraint Optimal Transformation of Scale Transition ; This paper presents on the distributions o\n A Convergence and Classification of Action Resparse Selection ; Specific methods are a problems of a\n A Diagnostic Approach for Automatic Convex Algorithms ; Computational accurate content of the proble\n Semantics for Network for the adversarial networks ; Described from the first sentence of the weak m\n Sward Computer Neural Networks ; We show that popular image segmentation from a simple methods combi\n The Set Samples Based Model for Generation ; We present a new consistency set of sentence in the con\n Appearance Systems ; In this paper we propose a novel approaches to the interest capturing systems a\n"
]
],
[
[
"### Bonus quest: Ultimate Language Model\n\nSo you've learned the building blocks of neural language models, you can now build the ultimate monster: \n* Make it char-level, word level or maybe use sub-word units like [bpe](https://github.com/rsennrich/subword-nmt);\n* Combine convolutions, recurrent cells, pre-trained embeddings and all the black magic deep learning has to offer;\n * Use strides to get larger window size quickly. Here's a [scheme](https://storage.googleapis.com/deepmind-live-cms/documents/BlogPost-Fig2-Anim-160908-r01.gif) from google wavenet.\n* Train on large data. Like... really large. Try [1 Billion Words](http://www.statmt.org/lm-benchmark/1-billion-word-language-modeling-benchmark-r13output.tar.gz) benchmark;\n* Use training schedules to speed up training. Start with small length and increase over time; Take a look at [one cycle](https://medium.com/@nachiket.tanksale/finding-good-learning-rate-and-the-one-cycle-policy-7159fe1db5d6) for learning rate;\n\n_You are NOT required to submit this assignment. Please make sure you don't miss your deadline because of it :)_",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb458f44503af1a146ecbe5a00368f08157fcb20 | 5,547 | ipynb | Jupyter Notebook | MDOF_identification_tutorial.ipynb | itomac/Extended_Morlet-Wave | e7f84368f8253ac815b02b67e2f69d36174a5bc9 | [
"MIT"
] | null | null | null | MDOF_identification_tutorial.ipynb | itomac/Extended_Morlet-Wave | e7f84368f8253ac815b02b67e2f69d36174a5bc9 | [
"MIT"
] | null | null | null | MDOF_identification_tutorial.ipynb | itomac/Extended_Morlet-Wave | e7f84368f8253ac815b02b67e2f69d36174a5bc9 | [
"MIT"
] | null | null | null | 27.597015 | 272 | 0.524067 | [
[
[
"# Identification of modal parametes using extended Morlet-Wave method from MDOF system\nver. 0.1",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom morlet_wave import *",
"_____no_output_____"
]
],
[
[
"**Steps required by the user:**\n1. Load Impulse Response Functions as a numpy array of the shape: \\\n`x[(number_of_samples, measure_points)]`\n2. Define sampling frequency: \\\neg. `fs = 1024` S/s\n3. Estimate natural frequencies: \\\neg. `nat_freq_est = np.array([315, 860, 1667]) * 2*np.pi` \\\nunit of natural frequencies is [rad/s]. In case of noisy signals it is important to estimate natural frequency as accurate as it is possible.",
"_____no_output_____"
]
],
[
[
"# x = \n# fs = \n# nat_freq_est =",
"_____no_output_____"
]
],
[
[
"**Set parameters of the method:**\n1. Set time spread parameters. One can set any two set parameters, but according to the author these three sets should be used:\n * `tsprd = (5, 10)`\n * `tsprd = (7, 14)` <- default\n * `tsprd = (10, 20)`\n2. Set range of morlet-wave function cycles, default value:\n * `ncycl = (30, 300)` <- default",
"_____no_output_____"
]
],
[
[
"tsprd = (5, 10)\nncycl = (30, 300)",
"_____no_output_____"
]
],
[
[
"Defined container to store identified modal parameters:",
"_____no_output_____"
]
],
[
[
"data = {\n \"zeta\" : [],\n \"omega\": [],\n \"X\" : []\n}",
"_____no_output_____"
]
],
[
[
"Following cell does the identification. It iterates along all measurement spots and natural frquencies and stores data in container `data`.\\\n*Note:* in case of very noisy data, if identified natural frequencies varies significantly from estimated, then estimated natural frequencies can be used for identification, but calling method for frequency identification: ```detect_frequency(use_estimated=True)``` ",
"_____no_output_____"
]
],
[
[
"measure_points = x.shape[1]\nnat_freq = nat_freq_est.size\nfor i in range(measure_points):\n zeta = []\n omega = []\n X = []\n for j in range(nat_freq):\n print(\"i, j: \", i, j)\n if j == 0:\n freq = (nat_freq_est[0], nat_freq_est[1])\n elif j == nat_freq-1:\n freq = (nat_freq_est[-1], nat_freq_est[-2])\n elif np.abs(nat_freq_est[j]-nat_freq_est[j+1]) < np.abs(nat_freq_est[j]-nat_freq_est[j-1]):\n freq = (nat_freq_est[j], nat_freq_est[j+1])\n else:\n freq = (nat_freq_est[j], nat_freq_est[j-1])\n \n sys = ExtendedMW(fs, x[i,], freq, tsprd, ncycl)\n sys.detect_frequency()\n sys.detect_damp()\n sys.estimate(True)\n sys.detect_amplitude(True)\n \n zeta.append(sys.zeta)\n omega.append(sys.omega)\n X.append(sys.X * np.exp(1j * sys.phi))\n del sys\n data[\"zeta\"].append(zeta)\n data[\"omega\"].append(omega)\n data[\"X\"].append(X)",
"_____no_output_____"
]
],
[
[
"Calculate modeshapes from the identified amplitudes and phases.",
"_____no_output_____"
]
],
[
[
"psi = np.sign(np.sin(np.angle(beam[\"X\"])))*np.abs(beam[\"X\"])\nprint(psi)",
"_____no_output_____"
]
],
[
[
"Plot the mode shapes:",
"_____no_output_____"
]
],
[
[
"m = np.linspace(1, measure_points, measure_points)\nfor i in range(nat_freq):\n y = np.zeros(measure_points)\n m = np.linspace(1, measure_points, measure_points)\n m[np.isnan(psi[:, i])] = np.nan\n y[np.invert(np.isnan(psi[:, i]))] = psi[np.invert(np.isnan(psi[:, i])), i]\n print(m, y)\n plt.plot(m, y/np.max(np.abs(y)))\nplt.grid(True)\nplt.xticks(np.linspace(1, measure_points, measure_points));",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb45afff19f4d964018bc4cad8d2a710d438dede | 19,732 | ipynb | Jupyter Notebook | 01/Alapok.ipynb | TamasPoloskei/BME-VEMA | 542725bf78e9ad0962018c1cf9ff40c860f8e1f0 | [
"MIT"
] | null | null | null | 01/Alapok.ipynb | TamasPoloskei/BME-VEMA | 542725bf78e9ad0962018c1cf9ff40c860f8e1f0 | [
"MIT"
] | 1 | 2018-11-20T14:17:52.000Z | 2018-11-20T14:17:52.000Z | 01/Alapok.ipynb | TamasPoloskei/BME-VEMA | 542725bf78e9ad0962018c1cf9ff40c860f8e1f0 | [
"MIT"
] | null | null | null | 23.744886 | 665 | 0.512923 | [
[
[
"# Bevezetés\n\n### Python programozási nyelv\n\nA Python egy open-source (OS), interpretált, általános célú programozási nyelv (vagy script-nyelv).\n\n**Tulajdonságai:**\n- Objektum orientált\n- Interpretált\n - Nem szükséges fordítani (mint a pl a *C++*-t), elég csak beírni a parancsot, és már futtatható is a kód\n - Alkalmas teszi számítások gyors-prototipizálására\n - Cserébe lassú\n- Open-source:\n - Ingyenes\n - Folyamatosan karban tartott \n - Széles körben felhasznált iparban és akadémiában is\n - Nagy \"Community\", sok segédlettel, fórummal (pl.: [stackoverflow](https://stackoverflow.com/questions/tagged/python)) \n- Moduláris:\n - Rengetek feladatra létezik \"*package*\" (pl.: numerikus számításokra *numpy*, szimbolikus számításokra *sympy*, táblázatfájl-kezelésre *CSV*)\n - Csak azt kell behívni, amire szükségünk van\n - Ismerni kell a *package* ekoszisztémát, mik léteznek, mi mire jó, stb...\n- Sok IDE (*Integrated Development Environment*) létezik:\n - Alapvetően shell (terminál) alapú\n - Notebook: **_jupyter notebook_**, *jupyter lab*\n - Szövegszerkesztő: *Spyder*, *VS Code* (ingyenes/open source - ezek tartalmaznak *Debugger*-t is)\n - Fizetős szövegszerkeszők (lista nem teljes): *Visual Studio*, *PyCharm*, stb...\n \n### Jupyter notebook működése (+ Python kernel):\n\nLegfontosabb tudnivalók:\n\n- Csak egy *front-end*, ami kommunikál egy *kernel*-lel (ez a kernel menüben választható).\n- Két mód létezik:\n - Command mode (cellaműveleteket lehet végezni)\n - Edit mode (szövegbevitel cellába)\n- Command mode (`ESC` billentyű lenyomásával érhető el, kék csík a cella kijelölése esetén):\n - Notebook mentése: `s`\n - Cellák hozzáadása: `b` az aktuális cella alá, `a` az aktuális cella fölé\n - Cella törlése: kétszer egymás után a `d` billentyű lenyomása\n - Cella törlésének visszavonása: `z`\n - Cella másolás: `c`, kivágás: `x`, beillesztés az aktuális cella alá: `v`\n - Számozás bekapcsolása a cella soraira: `l` (kis L), vagy `Shift + l` az összes cellára\n - Cellamódok: futtatandó kód: `y`, nyers kód (nem futtatható): `r`, markdown (formázott szöveg): `m` \n- Edit mode (`Enter` billenytű lenyomásával érhető el, zöld szín):\n - Sor \"kikommentelése\"/\"visszaállítása\": `Ctrl + /`\n - Több kurzor lehelyezése: `Ctrl + Bal egérgomb` \n - Téglalap-szerű kijelölés (rectangular selection): `Alt + Bal egérgomb` \"húzása\" (dragging)\n- Közös\n - Cella futtatása, majd cellaléptetés: `Shift + Enter` (ez létrehoz egy új cellát, ha nincs hova lépnie)\n - Cella futtatása cellaléptetés nélkül: `Ctrl + Enter` \n\n**Jupyter notebook help-jének előhozása**: *Edit mode*-ban `h` lenyomásával \n**Python help**: Kurzorral a függvény nevén állva `Shift + Tab` vagy egy cellába `?\"fv_név\"` beírása és futtatása",
"_____no_output_____"
],
[
"# Python bevezető",
"_____no_output_____"
],
[
"## Alapműveletek (Shift/Ctrl + Enter-rel futtassuk)",
"_____no_output_____"
]
],
[
[
"17 + 7 #Összeadás",
"_____no_output_____"
],
[
"333 - 7 #Kivonás",
"_____no_output_____"
],
[
"11 * 22 #Szorzás",
"_____no_output_____"
],
[
"7/9 #Osztás (ez nem egész (int) lesz: külön típus float)",
"_____no_output_____"
],
[
"0.3-0.1-0.2 # float: számábrázolási hiba lehet!!",
"_____no_output_____"
],
[
"2**3 # Hatványozás (** és NEM ^!)",
"_____no_output_____"
],
[
"2**(0.5) # Gyökvönás hatványozás segítségével",
"_____no_output_____"
],
[
"5e-3 #normálalak e segítségével (vagy 5E-3)",
"_____no_output_____"
]
],
[
[
"Néhány alapművelet működik szövegre is",
"_____no_output_____"
]
],
[
[
"'str1_' + 'str2_' #Összeadás",
"_____no_output_____"
],
[
"2 * 'str2_' #Szorzás",
"_____no_output_____"
]
],
[
[
"## Összetettebb függvények",
"_____no_output_____"
]
],
[
[
"sin(2) #szinusz",
"_____no_output_____"
]
],
[
[
"Összetettebb függvények már nincsenek a python alapnyelvben - ilyenkor szükséges behívni külső csomagokat, pl a **math** csomagot",
"_____no_output_____"
]
],
[
[
"import math",
"_____no_output_____"
],
[
"sin(2) # ez így továbbra sem létezik",
"_____no_output_____"
],
[
"math.sin(2)",
"_____no_output_____"
],
[
"# Több parancs együttes beírásakor nem látszik, csak az utolsó sor kimenete: print függvény alkalmazása!\nprint(math.sqrt(2))\nprint(math.tan(2))\nprint(math.atan(2))",
"1.4142135623730951\n-2.185039863261519\n1.1071487177940904\n"
],
[
"# Kimenet el is rejthető a ; segítségével (\"suppress output\")\n1+1;",
"_____no_output_____"
]
],
[
[
"Amennyiben szükséges, definiálhatunk mi is saját változókat az `=` jellel. \nMegjegyzés: a `=` értékadó függvénynek nincs kimenete",
"_____no_output_____"
]
],
[
[
"a=2\nb=3\nc=4.0 # automatikus típusadás",
"_____no_output_____"
],
[
"(a+b*c)**a # a legáltalánosabb típus lesz a kimenet (int < float)",
"_____no_output_____"
],
[
"# Fontos, hogy igyekezzük kerülni védett változó neveket! ILYET NE!\nmath.sqrt = 1\nmath.sqrt(2)",
"_____no_output_____"
]
],
[
[
"Ha véletlenül ilyet teszünk, akkor érdemes újraindítani a *kernel* a fent látható körkörös nyíllal, vagy a *Kernel* $\\rightarrow$ *Restart* segítségével",
"_____no_output_____"
],
[
"## Függvények\n\nSzerkezet:\n```python\ndef function(*arguments):\n instruction1\n instruction2\n ...\n return result\n```\n\nA függvény alá tartozó utasításokat tabulátoros behúzással (indent) kell beírni (nincs `{}` zárójel, vagy `end`). A függvény neve után jönnek az argumentumok majd kettősponttal `:` jelezzük, hogy hol kezdődik a függvény.",
"_____no_output_____"
]
],
[
[
"def foo(x):\n return 3*x\n\ndef bar(x,y):\n a = x+y**2\n return 2*a + 4",
"_____no_output_____"
],
[
"print(foo(3))\nprint(foo(3.))\nprint(foo('szöveg_'))\n\nprint(bar(3,4.))",
"9\n9.0\nszöveg_szöveg_szöveg_\n42.0\n"
]
],
[
[
"Lehetséges úgynevezett anonim függvényeket (*anonymous function* vagy *lambda function*) is létrehozni, amely gyors módja az egyszerű, egysoros függvények létrehozására:\n\n```python\nlambda arguments: instruction\n```\n\nEz akár egy változóhoz is hozzárendelhető, mint egy szám vagy string.",
"_____no_output_____"
]
],
[
[
"double = lambda x : x*2\nmultiply = lambda x,y : x*y",
"_____no_output_____"
],
[
"print(double(3))\nprint(multiply(10,3))",
"6\n30\n"
]
],
[
[
"## Listák\n\nPythonban egyszerűen létrehozhatóak listák, amelyekbe bármilyen adattípust tárolhatunk. A lista indexelése 0-tól indul",
"_____no_output_____"
]
],
[
[
"lista = [1,2,3,4,\"valami\",[1.0,4]]",
"_____no_output_____"
],
[
"print(lista[0]) # lista 1. eleme\nprint(lista[3]) # lista 4. eleme\nprint(lista[-1]) # negatív számokkal hátulról indexeljük a listát, és (-1)-től indul\nprint(lista[-2]) # lista utolsó előtti eleme",
"1\n4\n[1.0, 4]\nvalami\n"
],
[
"print(lista[1:-1]) # egyszerre több elem [inkluzív:exklúzív módon]\nprint(lista[1:2]) # egyszerre több elem [inkluzív:exklúzív módon]\nprint(lista[2:]) # lista utolsó elemét is figyelembe vesszük",
"[2, 3, 4, 'valami']\n[2]\n[3, 4, 'valami', [1.0, 4]]\n"
]
],
[
[
"## Vezérlési szerkezetek (Control Flow) - csak a legfontosabbak",
"_____no_output_____"
],
[
"### if-then-else\n\n```python\nif condition:\n instruction1\nelif condition2:\n instruction2\nelse:\n intsturction3\n```",
"_____no_output_____"
]
],
[
[
"a=4\nif a<=3:\n print('\"a\" nem nagyobb, mint 3')\nelif a>=10:\n print('\"a\" nem kisebb, mint 10')\nelse:\n print('\"a\" nagyobb mint 3, de kisebb mint 10')",
"\"a\" nagyobb mint 3, de kisebb mint 10\n"
]
],
[
[
"### for ciklus (for loop)\n```python\nfor i in array:\n instruction\n```",
"_____no_output_____"
]
],
[
[
"for i in range(3):\n print(i)\n \nprint()\n\nfor (i,elem) in enumerate(lista):\n print('lista ',i,'. eleme: ',elem,sep='') # több elem printelése egyszerr, szeparátor = ''\n",
"0\n1\n2\n\nlista 0. eleme: 1\nlista 1. eleme: 2\nlista 2. eleme: 3\nlista 3. eleme: 4\nlista 4. eleme: valami\nlista 5. eleme: [1.0, 4]\n"
]
],
[
[
"## Listák gyors létrehozása (List comprehension)",
"_____no_output_____"
]
],
[
[
"lista2 = [3*i**2 for i in range(2,5)] # range: 2,3,4\nlista2",
"_____no_output_____"
],
[
"lista3 = list(range(10))\nlista3",
"_____no_output_____"
],
[
"myfun = lambda x: 3*x**2\n\nlista4 = [myfun(i) for i in range(2,10) if i%3 != 0] # ha i nem osztható 3-al\nlista4",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb45ba7e73baa129af3915fa60ec5e8463dc9f75 | 509,015 | ipynb | Jupyter Notebook | Siamese-networks-medium.ipynb | codyhex/Facial-Similarity-with-Siamese-Networks-in-Pytorch | cc4f29f51b2c0e0a437443353ca01c8099226a16 | [
"MIT"
] | 884 | 2017-07-21T00:53:32.000Z | 2022-03-31T02:45:02.000Z | Siamese-networks-medium.ipynb | codyhex/Facial-Similarity-with-Siamese-Networks-in-Pytorch | cc4f29f51b2c0e0a437443353ca01c8099226a16 | [
"MIT"
] | 42 | 2017-08-28T18:25:37.000Z | 2022-03-18T03:23:31.000Z | Siamese-networks-medium.ipynb | codyhex/Facial-Similarity-with-Siamese-Networks-in-Pytorch | cc4f29f51b2c0e0a437443353ca01c8099226a16 | [
"MIT"
] | 279 | 2017-07-22T01:18:38.000Z | 2022-03-15T11:35:36.000Z | 563.693245 | 56,624 | 0.943086 | [
[
[
"# One Shot Learning with Siamese Networks\n\nThis is the jupyter notebook that accompanies",
"_____no_output_____"
],
[
"## Imports\nAll the imports are defined here",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport torchvision\nimport torchvision.datasets as dset\nimport torchvision.transforms as transforms\nfrom torch.utils.data import DataLoader,Dataset\nimport matplotlib.pyplot as plt\nimport torchvision.utils\nimport numpy as np\nimport random\nfrom PIL import Image\nimport torch\nfrom torch.autograd import Variable\nimport PIL.ImageOps \nimport torch.nn as nn\nfrom torch import optim\nimport torch.nn.functional as F",
"_____no_output_____"
]
],
[
[
"## Helper functions\nSet of helper functions",
"_____no_output_____"
]
],
[
[
"def imshow(img,text=None,should_save=False):\n npimg = img.numpy()\n plt.axis(\"off\")\n if text:\n plt.text(75, 8, text, style='italic',fontweight='bold',\n bbox={'facecolor':'white', 'alpha':0.8, 'pad':10})\n plt.imshow(np.transpose(npimg, (1, 2, 0)))\n plt.show() \n\ndef show_plot(iteration,loss):\n plt.plot(iteration,loss)\n plt.show()",
"_____no_output_____"
]
],
[
[
"## Configuration Class\nA simple class to manage configuration",
"_____no_output_____"
]
],
[
[
"class Config():\n training_dir = \"./data/faces/training/\"\n testing_dir = \"./data/faces/testing/\"\n train_batch_size = 64\n train_number_epochs = 100",
"_____no_output_____"
]
],
[
[
"## Custom Dataset Class\nThis dataset generates a pair of images. 0 for geniune pair and 1 for imposter pair",
"_____no_output_____"
]
],
[
[
"class SiameseNetworkDataset(Dataset):\n \n def __init__(self,imageFolderDataset,transform=None,should_invert=True):\n self.imageFolderDataset = imageFolderDataset \n self.transform = transform\n self.should_invert = should_invert\n \n def __getitem__(self,index):\n img0_tuple = random.choice(self.imageFolderDataset.imgs)\n #we need to make sure approx 50% of images are in the same class\n should_get_same_class = random.randint(0,1) \n if should_get_same_class:\n while True:\n #keep looping till the same class image is found\n img1_tuple = random.choice(self.imageFolderDataset.imgs) \n if img0_tuple[1]==img1_tuple[1]:\n break\n else:\n while True:\n #keep looping till a different class image is found\n \n img1_tuple = random.choice(self.imageFolderDataset.imgs) \n if img0_tuple[1] !=img1_tuple[1]:\n break\n\n img0 = Image.open(img0_tuple[0])\n img1 = Image.open(img1_tuple[0])\n img0 = img0.convert(\"L\")\n img1 = img1.convert(\"L\")\n \n if self.should_invert:\n img0 = PIL.ImageOps.invert(img0)\n img1 = PIL.ImageOps.invert(img1)\n\n if self.transform is not None:\n img0 = self.transform(img0)\n img1 = self.transform(img1)\n \n return img0, img1 , torch.from_numpy(np.array([int(img1_tuple[1]!=img0_tuple[1])],dtype=np.float32))\n \n def __len__(self):\n return len(self.imageFolderDataset.imgs)",
"_____no_output_____"
]
],
[
[
"## Using Image Folder Dataset",
"_____no_output_____"
]
],
[
[
"folder_dataset = dset.ImageFolder(root=Config.training_dir)",
"_____no_output_____"
],
[
"siamese_dataset = SiameseNetworkDataset(imageFolderDataset=folder_dataset,\n transform=transforms.Compose([transforms.Resize((100,100)),\n transforms.ToTensor()\n ])\n ,should_invert=False)",
"_____no_output_____"
]
],
[
[
"## Visualising some of the data\nThe top row and the bottom row of any column is one pair. The 0s and 1s correspond to the column of the image.\n1 indiciates dissimilar, and 0 indicates similar.",
"_____no_output_____"
]
],
[
[
"vis_dataloader = DataLoader(siamese_dataset,\n shuffle=True,\n num_workers=8,\n batch_size=8)\ndataiter = iter(vis_dataloader)\n\n\nexample_batch = next(dataiter)\nconcatenated = torch.cat((example_batch[0],example_batch[1]),0)\nimshow(torchvision.utils.make_grid(concatenated))\nprint(example_batch[2].numpy())",
"_____no_output_____"
]
],
[
[
"## Neural Net Definition\nWe will use a standard convolutional neural network",
"_____no_output_____"
]
],
[
[
"class SiameseNetwork(nn.Module):\n def __init__(self):\n super(SiameseNetwork, self).__init__()\n self.cnn1 = nn.Sequential(\n nn.ReflectionPad2d(1),\n nn.Conv2d(1, 4, kernel_size=3),\n nn.ReLU(inplace=True),\n nn.BatchNorm2d(4),\n \n nn.ReflectionPad2d(1),\n nn.Conv2d(4, 8, kernel_size=3),\n nn.ReLU(inplace=True),\n nn.BatchNorm2d(8),\n\n\n nn.ReflectionPad2d(1),\n nn.Conv2d(8, 8, kernel_size=3),\n nn.ReLU(inplace=True),\n nn.BatchNorm2d(8),\n\n\n )\n\n self.fc1 = nn.Sequential(\n nn.Linear(8*100*100, 500),\n nn.ReLU(inplace=True),\n\n nn.Linear(500, 500),\n nn.ReLU(inplace=True),\n\n nn.Linear(500, 5))\n\n def forward_once(self, x):\n output = self.cnn1(x)\n output = output.view(output.size()[0], -1)\n output = self.fc1(output)\n return output\n\n def forward(self, input1, input2):\n output1 = self.forward_once(input1)\n output2 = self.forward_once(input2)\n return output1, output2",
"_____no_output_____"
]
],
[
[
"## Contrastive Loss",
"_____no_output_____"
]
],
[
[
"class ContrastiveLoss(torch.nn.Module):\n \"\"\"\n Contrastive loss function.\n Based on: http://yann.lecun.com/exdb/publis/pdf/hadsell-chopra-lecun-06.pdf\n \"\"\"\n\n def __init__(self, margin=2.0):\n super(ContrastiveLoss, self).__init__()\n self.margin = margin\n\n def forward(self, output1, output2, label):\n euclidean_distance = F.pairwise_distance(output1, output2, keepdim = True)\n loss_contrastive = torch.mean((1-label) * torch.pow(euclidean_distance, 2) +\n (label) * torch.pow(torch.clamp(self.margin - euclidean_distance, min=0.0), 2))\n\n\n return loss_contrastive",
"_____no_output_____"
]
],
[
[
"## Training Time!",
"_____no_output_____"
]
],
[
[
"train_dataloader = DataLoader(siamese_dataset,\n shuffle=True,\n num_workers=8,\n batch_size=Config.train_batch_size)",
"_____no_output_____"
],
[
"net = SiameseNetwork().cuda()\ncriterion = ContrastiveLoss()\noptimizer = optim.Adam(net.parameters(),lr = 0.0005 )",
"_____no_output_____"
],
[
"counter = []\nloss_history = [] \niteration_number= 0",
"_____no_output_____"
],
[
"for epoch in range(0,Config.train_number_epochs):\n for i, data in enumerate(train_dataloader,0):\n img0, img1 , label = data\n img0, img1 , label = img0.cuda(), img1.cuda() , label.cuda()\n optimizer.zero_grad()\n output1,output2 = net(img0,img1)\n loss_contrastive = criterion(output1,output2,label)\n loss_contrastive.backward()\n optimizer.step()\n if i %10 == 0 :\n print(\"Epoch number {}\\n Current loss {}\\n\".format(epoch,loss_contrastive.item()))\n iteration_number +=10\n counter.append(iteration_number)\n loss_history.append(loss_contrastive.item())\nshow_plot(counter,loss_history)",
"Epoch number 0\n Current loss 2.0378851890563965\n\nEpoch number 1\n Current loss 2.8077077865600586\n\nEpoch number 2\n Current loss 1.4288808107376099\n\nEpoch number 3\n Current loss 1.273261547088623\n\nEpoch number 4\n Current loss 1.1692599058151245\n\nEpoch number 5\n Current loss 1.0952510833740234\n\nEpoch number 6\n Current loss 1.2235523462295532\n\nEpoch number 7\n Current loss 1.1519213914871216\n\nEpoch number 8\n Current loss 1.1717911958694458\n\nEpoch number 9\n Current loss 1.1029901504516602\n\nEpoch number 10\n Current loss 1.1344356536865234\n\nEpoch number 11\n Current loss 1.17781400680542\n\nEpoch number 12\n Current loss 1.129368543624878\n\nEpoch number 13\n Current loss 1.1467115879058838\n\nEpoch number 14\n Current loss 1.163444995880127\n\nEpoch number 15\n Current loss 1.1398472785949707\n\nEpoch number 16\n Current loss 1.1301088333129883\n\nEpoch number 17\n Current loss 1.0887219905853271\n\nEpoch number 18\n Current loss 1.140424370765686\n\nEpoch number 19\n Current loss 1.0663241147994995\n\nEpoch number 20\n Current loss 1.08632230758667\n\nEpoch number 21\n Current loss 1.1327917575836182\n\nEpoch number 22\n Current loss 1.1568373441696167\n\nEpoch number 23\n Current loss 1.070037841796875\n\nEpoch number 24\n Current loss 1.1004447937011719\n\nEpoch number 25\n Current loss 1.1675760746002197\n\nEpoch number 26\n Current loss 1.107505202293396\n\nEpoch number 27\n Current loss 1.164154052734375\n\nEpoch number 28\n Current loss 1.1572291851043701\n\nEpoch number 29\n Current loss 1.0625216960906982\n\nEpoch number 30\n Current loss 1.0926334857940674\n\nEpoch number 31\n Current loss 1.1182385683059692\n\nEpoch number 32\n Current loss 1.1999294757843018\n\nEpoch number 33\n Current loss 1.0879995822906494\n\nEpoch number 34\n Current loss 1.1144282817840576\n\nEpoch number 35\n Current loss 1.1479668617248535\n\nEpoch number 36\n Current loss 1.1063014268875122\n\nEpoch number 37\n Current loss 1.1686123609542847\n\nEpoch number 38\n Current loss 1.1108856201171875\n\nEpoch number 39\n Current loss 1.0782051086425781\n\nEpoch number 40\n Current loss 1.0159437656402588\n\nEpoch number 41\n Current loss 1.0994377136230469\n\nEpoch number 42\n Current loss 1.101317286491394\n\nEpoch number 43\n Current loss 1.068577527999878\n\nEpoch number 44\n Current loss 1.109604835510254\n\nEpoch number 45\n Current loss 1.1685329675674438\n\nEpoch number 46\n Current loss 1.1148097515106201\n\nEpoch number 47\n Current loss 1.125104308128357\n\nEpoch number 48\n Current loss 1.123309850692749\n\nEpoch number 49\n Current loss 1.1074304580688477\n\nEpoch number 50\n Current loss 1.0960524082183838\n\nEpoch number 51\n Current loss 1.1231727600097656\n\nEpoch number 52\n Current loss 1.0602667331695557\n\nEpoch number 53\n Current loss 1.1933255195617676\n\nEpoch number 54\n Current loss 1.0731847286224365\n\nEpoch number 55\n Current loss 1.1229645013809204\n\nEpoch number 56\n Current loss 1.1323256492614746\n\nEpoch number 57\n Current loss 1.1582694053649902\n\nEpoch number 58\n Current loss 1.087444543838501\n\nEpoch number 59\n Current loss 1.112156867980957\n\nEpoch number 60\n Current loss 1.1236321926116943\n\nEpoch number 61\n Current loss 1.089324951171875\n\nEpoch number 62\n Current loss 1.151943325996399\n\nEpoch number 63\n Current loss 1.0736372470855713\n\nEpoch number 64\n Current loss 1.0705881118774414\n\nEpoch number 65\n Current loss 1.0880753993988037\n\nEpoch number 66\n Current loss 1.0668013095855713\n\nEpoch number 67\n Current loss 1.0845987796783447\n\nEpoch number 68\n Current loss 1.117438554763794\n\nEpoch number 69\n Current loss 1.1109552383422852\n\nEpoch number 70\n Current loss 1.0705968141555786\n\nEpoch number 71\n Current loss 1.0489232540130615\n\nEpoch number 72\n Current loss 1.0667352676391602\n\nEpoch number 73\n Current loss 1.1204578876495361\n\nEpoch number 74\n Current loss 1.0965818166732788\n\nEpoch number 75\n Current loss 1.1749546527862549\n\nEpoch number 76\n Current loss 1.1329829692840576\n\nEpoch number 77\n Current loss 1.1027050018310547\n\nEpoch number 78\n Current loss 1.0796157121658325\n\nEpoch number 79\n Current loss 1.0907905101776123\n\nEpoch number 80\n Current loss 1.0403797626495361\n\nEpoch number 81\n Current loss 1.1375505924224854\n\nEpoch number 82\n Current loss 1.143746256828308\n\nEpoch number 83\n Current loss 1.0821889638900757\n\nEpoch number 84\n Current loss 1.1218210458755493\n\nEpoch number 85\n Current loss 1.08035147190094\n\nEpoch number 86\n Current loss 1.0216319561004639\n\nEpoch number 87\n Current loss 1.0868322849273682\n\nEpoch number 88\n Current loss 1.1195446252822876\n\nEpoch number 89\n Current loss 1.1785082817077637\n\nEpoch number 90\n Current loss 1.0771337747573853\n\nEpoch number 91\n Current loss 1.0936869382858276\n\nEpoch number 92\n Current loss 1.1433212757110596\n\nEpoch number 93\n Current loss 1.153044581413269\n\nEpoch number 94\n Current loss 1.103136420249939\n\nEpoch number 95\n Current loss 1.1112598180770874\n\nEpoch number 96\n Current loss 1.0496973991394043\n\nEpoch number 97\n Current loss 1.0448846817016602\n\nEpoch number 98\n Current loss 1.1437416076660156\n\nEpoch number 99\n Current loss 1.1138417720794678\n\n"
]
],
[
[
"## Some simple testing\nThe last 3 subjects were held out from the training, and will be used to test. The Distance between each image pair denotes the degree of similarity the model found between the two images. Less means it found more similar, while higher values indicate it found them to be dissimilar.",
"_____no_output_____"
]
],
[
[
"folder_dataset_test = dset.ImageFolder(root=Config.testing_dir)\nsiamese_dataset = SiameseNetworkDataset(imageFolderDataset=folder_dataset_test,\n transform=transforms.Compose([transforms.Resize((100,100)),\n transforms.ToTensor()\n ])\n ,should_invert=False)\n\ntest_dataloader = DataLoader(siamese_dataset,num_workers=6,batch_size=1,shuffle=True)\ndataiter = iter(test_dataloader)\nx0,_,_ = next(dataiter)\n\nfor i in range(10):\n _,x1,label2 = next(dataiter)\n concatenated = torch.cat((x0,x1),0)\n \n output1,output2 = net(Variable(x0).cuda(),Variable(x1).cuda())\n euclidean_distance = F.pairwise_distance(output1, output2)\n imshow(torchvision.utils.make_grid(concatenated),'Dissimilarity: {:.2f}'.format(euclidean_distance.item()))\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb45bc1d7adf23b328795b77d04e4d04c0c4362a | 9,542 | ipynb | Jupyter Notebook | ML/NaiveBayes.ipynb | Hacky-DH/learn | 8035d7e237a18e3a958e633767a3061789165ac0 | [
"MIT"
] | 3 | 2019-11-11T03:03:47.000Z | 2019-11-11T13:54:18.000Z | ML/NaiveBayes.ipynb | Hacky-DH/learn | 8035d7e237a18e3a958e633767a3061789165ac0 | [
"MIT"
] | 1 | 2020-05-26T13:21:54.000Z | 2020-05-26T13:21:54.000Z | ML/NaiveBayes.ipynb | Hacky-DH/learn | 8035d7e237a18e3a958e633767a3061789165ac0 | [
"MIT"
] | null | null | null | 29.269939 | 777 | 0.492559 | [
[
[
"# Naive Bayes\n$$ \\begin{split} \\mathop{argmax}_{c_k}p(y=c_k|x) &= \\mathop{argmax}_{c_k}p(y=c_k)p(x|y=c_k) \\\\\n& \\left( due to: p(y=c_k|x) = \\frac{p(y=c_k)p(x|y=c_k)}{p(x)} \\right) \\\\\n&= \\mathop{argmax}_{c_k}p(y=c_k)\\prod_jp(x^{(j)}|y=c_k) \\end{split} $$\nUse Maximum Likelihood Estimate(MLE) to evaluate $ p(y=c_k)$ and $ p(x^{(j)}|y=c_k) $ in datasets.\n$$ \\hat{p}(y=c_k) = \\frac{\\sum_i I(y_i=c_k)}{N} \\\\\n\\hat{p}(x^{(j)}=a_j|y=c_k) = \\frac{\\sum_i I(x_i^{(j)}=a_j,y=c_k)}{I(y_i=c_k)}\n$$\nBayesian estimation add $ \\lambda $ on numerator and denominator in MLE.",
"_____no_output_____"
],
[
"# Naive Bayes in Scikit-learn\nClassifiers: GaussianNB, MultinomialNB, BernoulliNB\n\n## Documents Classification\nUse TF-IDF(Term Frequency and Inverse Document Frequency) of term in documents as feature\n$$ TF-IDF = TF*IDF \\\\\nTF(t) = \\frac {\\text{Number of times term t appears in a document}}{\\text{Total number of terms in the document}}\\\\\nIDF(t) = log_e\\frac {\\text{Total number of documents}}{\\text{Number of documents with term t in it + 1}} $$\nBag of Words",
"_____no_output_____"
],
[
"### TfidfVectorizer\nsklearn.feature_extraction.text.TfidfVectorizer(stop_words, token_pattern, max_df)",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction.text import TfidfVectorizer",
"_____no_output_____"
],
[
"vect = TfidfVectorizer()",
"_____no_output_____"
],
[
"documents=[\n 'my dog has flea problems help please',\n 'maybe not take him to dog park stupid',\n 'my dalmation is so cute I love him',\n 'stop posting stupid worthless garbage',\n 'mr licks ate my steak how to stop him',\n 'quit buying worthlsess dog food stupid',\n]\ntargets=[0,1,0,1,0,1] # 0 normal, 1 insult",
"_____no_output_____"
],
[
"tf_matrix = vect.fit_transform(documents)",
"_____no_output_____"
],
[
"# all unique words\nwords = vect.get_feature_names()\nprint(len(words), words)",
"32 ['ate', 'buying', 'cute', 'dalmation', 'dog', 'flea', 'food', 'garbage', 'has', 'help', 'him', 'how', 'is', 'licks', 'love', 'maybe', 'mr', 'my', 'not', 'park', 'please', 'posting', 'problems', 'quit', 'so', 'steak', 'stop', 'stupid', 'take', 'to', 'worthless', 'worthlsess']\n"
],
[
"# words id\nprint(len(vect.vocabulary_), vect.vocabulary_)",
"32 {'my': 17, 'dog': 4, 'has': 8, 'flea': 5, 'problems': 22, 'help': 9, 'please': 20, 'maybe': 15, 'not': 18, 'take': 28, 'him': 10, 'to': 29, 'park': 19, 'stupid': 27, 'dalmation': 3, 'is': 12, 'so': 24, 'cute': 2, 'love': 14, 'stop': 26, 'posting': 21, 'worthless': 30, 'garbage': 7, 'mr': 16, 'licks': 13, 'ate': 0, 'steak': 25, 'how': 11, 'quit': 23, 'buying': 1, 'worthlsess': 31, 'food': 6}\n"
],
[
"tfidf = tf_matrix.toarray()\nprint(tfidf.shape, tfidf[0])",
"(6, 32) [0. 0. 0. 0. 0.2836157 0.40966432\n 0. 0. 0.40966432 0.40966432 0. 0.\n 0. 0. 0. 0. 0. 0.2836157\n 0. 0. 0.40966432 0. 0.40966432 0.\n 0. 0. 0. 0. 0. 0.\n 0. 0. ]\n"
]
],
[
[
"### CountVectorizer",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction.text import CountVectorizer",
"_____no_output_____"
],
[
"c_vect = CountVectorizer()\nc_matrix = c_vect.fit_transform(documents)",
"_____no_output_____"
],
[
"print(c_vect.get_feature_names())",
"['ate', 'buying', 'cute', 'dalmation', 'dog', 'flea', 'food', 'garbage', 'has', 'help', 'him', 'how', 'is', 'licks', 'love', 'maybe', 'mr', 'my', 'not', 'park', 'please', 'posting', 'problems', 'quit', 'so', 'steak', 'stop', 'stupid', 'take', 'to', 'worthless', 'worthlsess']\n"
],
[
"c_matrix.toarray()",
"_____no_output_____"
],
[
"# default ngram_range is (1, 1), token_pattern=’(?u)\\b\\w\\w+\\b’\nc_vect_ngram = CountVectorizer(ngram_range=(1, 2))\nc_matrix_ngram = c_vect_ngram.fit_transform(documents)\nprint(c_vect_ngram.get_feature_names())",
"['ate', 'ate my', 'buying', 'buying worthlsess', 'cute', 'cute love', 'dalmation', 'dalmation is', 'dog', 'dog food', 'dog has', 'dog park', 'flea', 'flea problems', 'food', 'food stupid', 'garbage', 'has', 'has flea', 'help', 'help please', 'him', 'him to', 'how', 'how to', 'is', 'is so', 'licks', 'licks ate', 'love', 'love him', 'maybe', 'maybe not', 'mr', 'mr licks', 'my', 'my dalmation', 'my dog', 'my steak', 'not', 'not take', 'park', 'park stupid', 'please', 'posting', 'posting stupid', 'problems', 'problems help', 'quit', 'quit buying', 'so', 'so cute', 'steak', 'steak how', 'stop', 'stop him', 'stop posting', 'stupid', 'stupid worthless', 'take', 'take him', 'to', 'to dog', 'to stop', 'worthless', 'worthless garbage', 'worthlsess', 'worthlsess dog']\n"
]
],
[
[
"### MultinomialNB",
"_____no_output_____"
]
],
[
[
"from sklearn.naive_bayes import MultinomialNB",
"_____no_output_____"
],
[
"clf = MultinomialNB(alpha=0.001).fit(tf_matrix, targets)",
"_____no_output_____"
],
[
"test_vect = TfidfVectorizer(vocabulary=vect.vocabulary_)\ntest_features = test_vect.fit_transform([documents[3]])\npredicted_labels = clf.predict(test_features)",
"_____no_output_____"
],
[
"from sklearn import metrics\nprint(metrics.accuracy_score([targets[3]], predicted_labels))",
"1.0\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cb45d416ee553ce7ac06a6028731dc7f386555bb | 87,845 | ipynb | Jupyter Notebook | examples/crop-type-classification/CropTypeClassification_Workflow.ipynb | Gnilliw/eo-learn | dc3f52b3ad96741081afc39e14903f45714d0868 | [
"MIT"
] | null | null | null | examples/crop-type-classification/CropTypeClassification_Workflow.ipynb | Gnilliw/eo-learn | dc3f52b3ad96741081afc39e14903f45714d0868 | [
"MIT"
] | null | null | null | examples/crop-type-classification/CropTypeClassification_Workflow.ipynb | Gnilliw/eo-learn | dc3f52b3ad96741081afc39e14903f45714d0868 | [
"MIT"
] | null | null | null | 42.998042 | 712 | 0.607342 | [
[
[
"# How To: Crop type classification for Austria\n\nThis example notebook shows the steps towards constructing an automated machine learning pipeline for crop type identification in an area of interest in Austria. Along the pipeline, two different approaches are applied and compared. The first one, the LightGBM, represents a state-of-the-art machine learning algorithm. The second one is a Temporal Convolutional Neural Network architecture from the field of deep learning. The prediction is performed on a time-series of Sentinel-2 scenes from 2018. The example notebook will lead you through the whole process of creating the pipeline, with details provided at each step (see **Overview**). \n\n## Before start\nEnjoying the functionality of eo-learn and the simplicity of this example workflow is preceded by the unavoidable setup of an adequate working environment. But trust us, it's worth it! And we'll take you by the hand.\n### Requirements\n#### Sentinel Hub account\nTo run the example you'll need a Sentinel Hub account. If you do not have one yet, you can create a free trial account at the [Sentinel Hub webpage](https://services.sentinel-hub.com/oauth/subscription). If you are a researcher you can even apply for a free non-commercial account at the [ESA OSEO page](https://earth.esa.int/aos/OSEO).\n\nOnce you have the account set up, login to [Sentinel Hub Configurator](https://apps.sentinel-hub.com/configurator/). By default you will already have the default configuration with an **instance ID** (alpha numeric code of length 36). For this tutorial we recommend that you create a new configuration (`Add new configuration`) and set the configuration to be based on **Python scripts template**. Such configuration will already contain all layers used in a more general Land Use/ Land Cover (LULC) example which are adopted for this example. Otherwise you will have to define the layers for your configuration yourself.\n\nOne layer you have to define yourself is your \"MY-S2-L2A-BANDS\" layer. Therefore you: \n* log in to your Sentinel Hub account\n* go to `Configuration Utility` and access your newly created `LULC` configuration\n* here you choose `+ Add new layer` \n* your Layer name is `MY-S2-L2A-BANDS`\n* in red letters you are requested to _! Please select predefined product or enter your processing script_ - so you better do...\n * to set your custom script you copy/paste `return [B02,B03,B04,B05,B06,B07,B08,B8A,B11,B12]` into the `Custom script editor` and click `</> Set Custom Script`\n * You just told Sentinel Hub which bands you want to download in the following. Now, go on and `Save` your own layer\n\nAfter you have prepared the configuration please put configuration's **instance ID** into `sentinelhub` package's configuration file following the [configuration instructions](http://sentinelhub-py.readthedocs.io/en/latest/configure.html). For Processing API request you also need to obtain and set your `oauth` client id and secret. You can do this either manually or by using the respective variables in the configuration section of the following workflow.\n\n#### Sentinel Hub Python package\nThe [Sentinel Hub Python package](https://sentinelhub-py.readthedocs.io/en/latest/) allows users to make OGC (WMS and WCS) web requests to download and process satellite images within your Python scripts. It supports Sentinel-2 L1C and L2A, Sentinel-1, Landsat 8, MODIS and DEM data source.\n#### eo-learn library\nBetween the acquisition of a satellite image and actionable information, there is a large processing effort. [eo-learn](https://eo-learn.readthedocs.io/en/latest/index.html) as a collection of modular Python sub-packages allows easy and quick pro-cessing of spatio-temporal data to prototype, build and automate these required large scale EO workflows for AOIs of any size. It also directly enables the application of state-of-the-art tools for computer vision, machine learning and deep learning packages in Python to the data. Especially for non-experts to the field of remote sensing and machine learning it makes extraction of valuable information from satellite imagery easier and more comfortable.\n#### Additional packages\nIn addition to the previous packages the installation of the packages [keras](https://keras.io/), [tensor flow](https://www.tensorflow.org/) and [ipywidgets](https://ipywidgets.readthedocs.io/en/latest/) is required.\n\n## Overview\nWith the help of the eo-learn library, the entire classification process can be executed in 4 processing blocks, i.e. `EOWorkflows`.\n1. Ground truth data\n2. EO data\n3. Feature engineering - Crop type grouping - Sampling\n4. Prediction\n\n**In more detail the notebook is structured as follows:**\n\n\nI. Imports\n\nII. Configurations\n### Part I\n1. BBox-Splitter\n - Plot AOI and give the extent\n - Create BBoxes\n - Visualize the selection\n2. Add ground truth data\n - Create EOPatches and add LPIS + area ratio\n3. Add EO data\n - Choose EO features\n - Clean EOPatch list\n4. Feature/ label engineering and Sampling\n - Data visualization\n - Resampling, Interpolation, LPIS data preparation\n - Sampling\n \n### Part II\n\n6. Prediction\n - Set up and train LightGBM model\n - Set up and train TempCNN model\n - Model validation and evaluation\n - Prediction\n - Visualization of the results\n7. Next steps\n\nNow, after the setup you are curious what is next and can't wait to get your hands dirty? Well, let's get started!",
"_____no_output_____"
],
[
"# Imports\nLets start with some necessary imports.",
"_____no_output_____"
]
],
[
[
"# set module directory to system path\nimport sys, os\nMAIN_FOLDER = os.getcwd()\nimport_path = os.path.join(MAIN_FOLDER, 'Tasks')\nif import_path not in sys.path:\n sys.path.append(import_path)",
"_____no_output_____"
],
[
"# Built-in modules\nimport math\nimport shutil\nimport itertools\nfrom datetime import timedelta\n\n\n# Basics of Python data handling and visualization\nimport pandas as pd\nimport geopandas as gpd\nimport numpy as np\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\nfrom matplotlib.colors import LinearSegmentedColormap\nfrom shapely.geometry import Polygon\nfrom tqdm.notebook import tqdm\n\n\n# Imports from eo-learn, sentinelhub-py, and perceptive-sentinel\nfrom sentinelhub import CRS, BBoxSplitter, MimeType\n\nfrom eolearn.core import LinearWorkflow, FeatureType, SaveTask, OverwritePermission, LoadTask\nfrom eolearn.core import EOPatch, EOTask, CreateEOPatchTask, ZipFeatureTask, MapFeatureTask\nfrom eolearn.geometry import VectorToRaster, ErosionTask\nfrom eolearn.io import SentinelHubInputTask, ExportToTiff\nfrom eolearn.mask import get_s2_pixel_cloud_detector, AddCloudMaskTask, AddValidDataMaskTask\nfrom eolearn.features import SimpleFilterTask, LinearInterpolation\nfrom eolearn.features import NormalizedDifferenceIndexTask, EuclideanNormTask\n\n# Machine learning\nimport lightgbm as lgb\nimport joblib\nfrom sklearn.preprocessing import StandardScaler, OneHotEncoder\nfrom sklearn.model_selection import train_test_split\nfrom sklearn import metrics\n\n# Deep Learning\nfrom keras.models import Sequential, load_model\nfrom keras.layers import Dense, Flatten, Dropout\nfrom keras.layers.convolutional import Conv1D, MaxPooling1D\nfrom keras.utils import to_categorical\n\n# Notebook specific classes and functions\nfrom CropTypeClassTasks import CleanLPIS, GroupLPIS, LPISCLASS, ConcatenateData, SamplingTaskTask\nfrom CropTypeClassTasks import train_test_split_eopatches, train_test_split_eopatch\nfrom CropTypeClassTasks import plot_confusion_matrix, PredictPatch, AddAreaRatio, FixLPIS, masking\nfrom CropTypeClassTasks import AddGeopediaVectorFeature, Sen2CorValidData, ValidDataFractionPredicate",
"_____no_output_____"
]
],
[
[
"# Configurations\nIn this part you can define your configurations. The basic configurations are set for an example running smoothly.\n## Configuration file customization",
"_____no_output_____"
]
],
[
[
"# In case you put the credentials into the configuration file by hand you can leave this unchanged\nINSTANCE_ID = ''\nCLIENT_ID = ''\nCLIENT_SECRET = ''",
"_____no_output_____"
],
[
"config = SHConfig()\n\nif CLIENT_ID and CLIENT_SECRET and INSTANCE_ID:\n config.instance_id = INSTANCE_ID\n config.sh_client_id = CLIENT_ID\n config.sh_client_secret = CLIENT_SECRET\n\nif config.sh_client_id == '' or config.sh_client_secret == '' or config.instance_id == '':\n print(\"Warning! To use Sentinel Hub services, please provide the credentials (client ID and client secret).\")",
"_____no_output_____"
]
],
[
[
"## Workflow configuration",
"_____no_output_____"
]
],
[
[
"# define in- and output folders\noutput_path = os.path.join(MAIN_FOLDER, 'Output')\ngeneral_data_path = os.path.join(MAIN_FOLDER, 'GeneralData')\npatch_path = os.path.join(MAIN_FOLDER, 'Output', 'EOPatches')\nthresLPIS_path = os.path.join(MAIN_FOLDER, 'Output', 'EOPatches_Low_LPIS_Thres')\nsamples_path = os.path.join(MAIN_FOLDER, 'Output', 'Samples')\nmodels_path = os.path.join(MAIN_FOLDER, 'Output', 'Models')\npredictions_path = os.path.join(MAIN_FOLDER, 'Output', 'Predictions')\n\n# For reference colormap\nlpisclass_cmap = mpl.colors.ListedColormap([entry.color for entry in LPISCLASS])\nlpisclass_norm = mpl.colors.BoundaryNorm(np.arange(-0.5, 26, 1), lpisclass_cmap.N)\nclass_names = [entry.class_name for entry in LPISCLASS]\nclass_ids = [entry.id for entry in LPISCLASS]\n\n\n### 1. BBox-Splitter\n## Plot AOI and give extent\nINPUT_FILE = os.path.join(general_data_path, 'Area_AOI.geojson') # Geojson or Shapefile of the area of interest\naustria = os.path.join(general_data_path, 'Area_Austria.geojson') # Geojson of austrian national borders\ncrs = CRS.UTM_33N # wanted coordinate System of the AOI\n\n\n### 2. Add ground truth data\n## Create EOPatches and add LPIS + area ratio\nyear = '2018' # year of interest\nlayerID_dict = {'2018': 2647, '2017': 2034, '2016': 2033} # Layer IDs of Geopedia Layer\nlayerID = layerID_dict[year] # Layer ID for Austria of year set by \"year\"\npatch_list = os.listdir(patch_path) # List of created EOPatches names\n\n\n### 3. Add EO data\n## Choose EO features\nmaxcloud = 0.8 # maximum cloudcoverage of sentinel tiles used for download\ndatafrac = 0.7 # keep only frames with valid data fraction over x%\n## Clean EOPatch list\nlpis_thres = 0.13 # Patches with less than x% of LPIS coverage are excluded in the following\n## Add EO data\ntime_interval = [f'{year}-01-01', f'{year}-09-30'] # the start and end date for downloading satellite data\n\n\n### 4. Feature and label engineering\n## Feature concatenation, interpolation and LPIS data preparation\nday_range = 8 # the realtime range of valid satellite images is resampled to a x day equidistant range \n## Prepare LPIS data\ngrouping_id = 'basic' # define grouping id (has to be identical to endings of the two grouping files)\n# File that contains LPIS Crop ID and wanted groups - Colums of shape: CROP_ID, english, slovenian, latin, GROUP_1\nlpis_to_group_file = os.path.join(general_data_path, 'at_lpis_{}_crop_to_group_mapping_{}.csv'.format(year, grouping_id))\n# File that contains the wanted groups and their ID - Colums of shape: GROUP_1, GROUP_1_ID\ncrop_group_file = os.path.join(general_data_path, 'crop_group_1_definition_{}.csv'.format(grouping_id))\n\n\n### 5. Sampling\n## Sampling per EOPatch\npixel_thres = 1000 # Pixel thresold necessary for a class to be considered in sampling\nsamp_class = 500 # take x samples per class per EOPatch\n## Combine samples and split into train and test data\ntest_ratio = 4 # take every xth patch for testing\nfeatures_dict = 'FEATURES_SAMPLED' # name of the dictionary where the sample features are saved\nlabels_dict = 'LPIS_class_{}_ERODED_SAMPLED'.format(grouping_id) # name of the dictionary where the sample labels are saved",
"_____no_output_____"
]
],
[
[
"# 1. From AOI to BBox",
"_____no_output_____"
],
[
"## Plot AOI and give extent\nSpotlight on your \"INPUT_FILE\" configuration! This is where you have the possibility to easily adapt the workflow to your needs. **Take your pick** and replace the AOI file in the _General data_ folder. Either shapefile or geojson formatted version of your AOI is split into smaller patches by `eo-learn`. The total number of patches depends on the AOIs size. Automated splitting is supposed to create patches of size 10 x 10 km. ",
"_____no_output_____"
]
],
[
[
"# define Area of interest\naoi = gpd.read_file(INPUT_FILE) # read AOI file\n\naoi_shape = aoi.geometry.values[-1] # get aoi shape\n\n# define BBox-Splitter split values\nShapeVal_a = round(aoi_shape.bounds[2] - aoi_shape.bounds[0])\nShapeVal_b = round(aoi_shape.bounds[3] - aoi_shape.bounds[1])\n\nSplitVal_a = max(1, int(ShapeVal_a/1e4))\nSplitVal_b = max(1, int(ShapeVal_b/1e4))\n\n# Give extent of AOI + grid count and plot AOI\nprint('The extent of the AOI is {}m x {}m, so it is split into a grid of {} x {}.'.format(ShapeVal_a, \n ShapeVal_b, \n SplitVal_a, \n SplitVal_b))\n\naoi.plot()\nplt.axis('off');",
"_____no_output_____"
]
],
[
[
"## Create BBoxes\nThe simple patch polygons are transformed into bounding boxes suitable for serving as geometrical EOPatch frame. ",
"_____no_output_____"
]
],
[
[
"# split area of interest into an appropriate number of BBoxes\nbbox_splitter = BBoxSplitter([aoi_shape], crs, (SplitVal_a, SplitVal_b))\n\nbbox_list = np.array(bbox_splitter.get_bbox_list()) # get list of BBox geometries\ninfo_list = np.array(bbox_splitter.get_info_list()) # get list of x (column) and y(row) indices\n\nprint('Each bounding box also has some info how it was created.\\nExample:\\n'\n 'bbox: {}\\ninfo: {}\\n'.format(bbox_list[0].__repr__(), info_list[0]))",
"_____no_output_____"
]
],
[
[
"## Visualize the selection\nFirst visualize the GeoDataFrame of the bounding boxes",
"_____no_output_____"
]
],
[
[
"# create GeoDataFrame of BBoxes\ngeometry = [Polygon(bbox.get_polygon()) for bbox in bbox_list] # get geometry from bbox_list for creating GeoSeries\nidxs_x = [info['index_x'] for info in info_list] # get column index for naming EOPatch\nidxs_y = [info['index_y'] for info in info_list] # get row index for naming EOPatch\n\ngdf = gpd.GeoDataFrame({'index_x': idxs_x, 'index_y': idxs_y},\n crs={'init': CRS.ogc_string(crs)},\n geometry=geometry)\n\nshapefile_name = os.path.join(output_path, 'BBoxes.shp')\ngdf.to_file(shapefile_name)\n\ngdf.head()",
"_____no_output_____"
]
],
[
[
"Second visualize the split AOI with reference to Austrian national borders",
"_____no_output_____"
]
],
[
[
"# Plot AOI overview\naustria_gdf = gpd.read_file(austria)\n\nfontdict = {'family': 'monospace', 'weight': 'normal', 'size': 11}\n\n# if bboxes have all same size, estimate offset\nxl, yl, xu, yu = gdf.geometry[0].bounds\nxoff, yoff = (xu - xl) / 3, (yu - yl) / 5\n\n# main figure\nfig, ax = plt.subplots(figsize=(20, 20))\ngdf.plot(ax=ax,facecolor='w',edgecolor='r',alpha=0.5,linewidth=5)\naoi.plot(ax=ax, facecolor='w',edgecolor='k',alpha=0.5)\naustria_gdf.plot(ax=ax, facecolor='w',edgecolor='b',alpha=0.5)\nax.set_title('Test Area Splitted');\nplt.axis('off')\n\n# sub figure\na = plt.axes([0.2, 0.6, .2, .2])\ngdf.plot(ax=a, facecolor='w',edgecolor='r',alpha=0.5, linewidth=3)\naoi.plot(ax=a, facecolor='w',edgecolor='k',alpha=0.5, linewidth=3)\nplt.xticks([])\nplt.yticks([])",
"_____no_output_____"
]
],
[
[
"# 2. Add ground truth data",
"_____no_output_____"
],
[
"## Create EOPatches and add LPIS data + area ratio\nNow it's time to create `EOPatches` and start filling them with data.\n\n#### Add data\n* At first you transform your basic geometric frames into proper `EOPatches`. You can then fill these handy data containers endlessly. \n* As a start you add your ground truth data that is later used as a reference to validate your prediction results. Here, you use Austrian LPIS data containing agricultural information on the field-level. In the case of this example you download your 2018 data in vector format automatically from [Geopedia](http://portal.geopedia.world/) using Sentinel-Hub tasks. For further observation you can also download the complete and free dataset for Austria [here](https://www.data.gv.at/katalog/dataset?q=INVEKOS+Schl%C3%A4ge&sort=score+desc%2C+metadata_modified+desc). \n* Additionally a ratio value is added showing the percentage of the agricultural area in the respective `EOPatch`. The importance of this ratio will become apparent in the following steps.\n\nAn `EOPatch` is created and manipulated using `EOTasks`. Due to the potentially large number of `EOPatches`, automation of the processing pipeline is absolutely crucial. Therefore `EOTasks` are chained in an `EOWorkflow`. In this example the final workflow is executed on all patches, which are saved to the specified directory.",
"_____no_output_____"
],
[
"### Set up your 1. EOWorkflow - Ground truth data\nThe `EOTasks` need to be put in some order and executed one by one. This can be achieved by manually executing the tasks, or more conveniently, defining an `EOWorkflow` which does this for you.\nAn `EOWorkflow` can be linear or more complex, but it should be acyclic. Here we will use the linear case of the EOWorkflow, available as `LinearWorkflow`",
"_____no_output_____"
]
],
[
[
"# TASK FOR CREATING EOPATCH\ncreate = CreateEOPatchTask()\n\n# TASK FOR ADDING LPIS DATA FROM GEOPEDIA\n# here you can choose the year of interest\n# also you have to set the corresponding Geopedialayer-ID\nadd_lpis = AddGeopediaVectorFeature((FeatureType.VECTOR_TIMELESS, 'LPIS_{}'.format(year)),\n layer=layerID, year_filter=None, drop_duplicates=True)\n\n# TASK FOR ADDING AN AREA RATIO\n# the area ratio indicates the EOPatches proportion of LPIS coverage\narea_ratio = AddAreaRatio((FeatureType.VECTOR_TIMELESS, 'LPIS_{}'.format(year)),\n (FeatureType.SCALAR_TIMELESS, 'FIELD_AREA_RATIO'))\n\n# TASK FOR SAVING TO OUTPUT\nsave = SaveTask(patch_path, overwrite_permission=OverwritePermission.OVERWRITE_PATCH)\n\n# define the workflow\nworkflow = LinearWorkflow(create, \n add_lpis, \n area_ratio, \n save)",
"_____no_output_____"
]
],
[
[
"### Run your first EOWorkflow",
"_____no_output_____"
]
],
[
[
"# execute workflow\npbar = tqdm(total=len(bbox_list))\nfor idx, bbox in enumerate(bbox_list):\n bbox = bbox_splitter.bbox_list[idx]\n info = bbox_splitter.info_list[idx]\n patch_name = f'eopatch_{idx}_col-{info[\"index_x\"]}_row-{info[\"index_y\"]}'\n workflow.execute({create:{'bbox':bbox}, save:{'eopatch_folder':patch_name}})\n pbar.update(1)",
"_____no_output_____"
]
],
[
[
"Visualize the added vector data for one example EOPatch",
"_____no_output_____"
]
],
[
[
"eopatch_name = 'eopatch_0_col-0_row-0' # get the name of the first newly created EOPatch\neopatch = EOPatch.load(os.path.join(patch_path, eopatch_name))\n\n# plot vector data\nprint('Plotting LPIS vector data of eopatch: {}'.format(eopatch_name))\nfig, ax = plt.subplots(figsize=(20, 20))\nLPIS = eopatch.vector_timeless['LPIS_{}'.format(year)]\nLPIS.plot(column='SNAR_BEZEI', ax=ax, categorical=True)\nax.set_aspect('auto')\nax.set_xticks(ticks=[])\nax.set_yticks(ticks=[])\n\ndel eopatch",
"_____no_output_____"
]
],
[
[
"As you can see, the crop types in your AOI are very diverse. Each colour stands for one of the over 200 LPIS classes.",
"_____no_output_____"
],
[
"# 3. Add EO data",
"_____no_output_____"
],
[
"## Choose EO features\nNow, it's time to add Sentinel-2 data to the EOPatches. You are lucky to be using `eo-learn`, as this is simply done by setting up another single EOWorkflow including only a single EOTask for adding your satellite images. The remaining tasks allow you to create extensive valid data masks and useful indices using a ridiculously small amount of code.\n\nIn detail you add:\n* L2A bands [B02,B03,B04,B05,B06,B07,B08,B8A,B11,B12]\n* Sen2cor's scene classification map and snow probability map\n* SentinelHub's cloud probability map and cloud mask\n* A mask of validity, based on acquired data from Sentinel and cloud coverage.\n\n 1. IS_DATA == True\n 2. CLOUD_MASK == 0 (1 indicates that pixel was identified to be covered with cloud)\n \n\n* Filter out time frames with < 70 % valid coverage (no clouds)\n* Calculate and add NDVI, NDWI, NORM for helping the algorithm to detect relationships between the spectral bands.\n* Prepare following Workflow by adding two features",
"_____no_output_____"
],
[
"### Set up your 2. EOWorkflow - EO data",
"_____no_output_____"
]
],
[
[
"# TASK TO LOAD EXISTING EOPATCH\nload = LoadTask(patch_path)\n\n# TASK TO ADD SENTINEL 2 LEVEL 2A DATA\n# Here also a simple filter of cloudy scenes is done. A detailed cloud cover \n# detection is performed within the next steps\n\n# Using SH we will download the following data:\n# * L2A bands: B02, B03, B04, B05, B06, B07, B08, B8A, B11, and B12\n# * sen2cor's scene classification: SCL\n# * s2cloudless' cloud mask: CLM\nband_names = ['B02','B03','B04','B05','B06','B07','B08','B8A','B11','B12']\nadd_l2a = SentinelHubInputTask(\n bands_feature=(FeatureType.DATA, 'MY-S2-L2A-BANDS'),\n bands = band_names,\n resolution=10,\n maxcc=maxcloud,\n time_difference=timedelta(minutes=120),\n data_source=DataSource.SENTINEL2_L2A,\n additional_data=[(FeatureType.MASK, 'dataMask', 'IS_DATA'),\n (FeatureType.MASK, 'SCL'),\n (FeatureType.MASK, 'CLM')],\n config=config,\n max_threads=5\n)\n\n# create valid data masks\nscl_valid_classes = [2, 4, 5, 6, 7]\n\n# TASKs FOR ADDING L2A and L1C VALID DATA MASKS\n# convert cloudmask to validmask\nadd_clm_valid =MapFeatureTask((FeatureType.MASK, 'CLM'),\n (FeatureType.MASK, 'CLM_VALID'),\n np.logical_not)\n# combine IS_DATA and CLM_VALID\nadd_l1c_valmask = ZipFeatureTask({FeatureType.MASK: ['IS_DATA', 'CLM_VALID']}, \n (FeatureType.MASK, 'L1C_VALID'),\n np.logical_and)\n# combine IS_DATA and SCL (using an erosion radius of 6 and a dilation radius of 22 pixel for SCL classes)\nadd_l2a_valmask = AddValidDataMaskTask(Sen2CorValidData(scl_valid_classes, 6, 22), 'L2A_VALID')\n# combine all validmasks\nadd_valmask = ZipFeatureTask({FeatureType.MASK: ['L1C_VALID', 'L2A_VALID']}, \n (FeatureType.MASK, 'VALID_DATA'),\n np.logical_and)\n\n# TASK TO FILTER OUT SCENES INCLUDING TOO MANY UNVALID PIXEL\n# keep frames with > x % valid coverage\nvalid_data_predicate = ValidDataFractionPredicate(datafrac)\nfilter_task = SimpleFilterTask((FeatureType.MASK, 'VALID_DATA'), valid_data_predicate)\n\n# TASK FOR CALCULATING INDICES\n# NDVI = Normalized Difference Vegetation Index\n# NDWI = Normalized Difference Water Index\n# NORM = Euclidean Norm\nndvi = NormalizedDifferenceIndexTask((FeatureType.DATA, 'MY-S2-L2A-BANDS'), \n (FeatureType.DATA, 'NDVI'),\n [6, 2])\nndwi = NormalizedDifferenceIndexTask((FeatureType.DATA, 'MY-S2-L2A-BANDS'), \n (FeatureType.DATA, 'NDWI'),\n [1, 6])\nnorm = EuclideanNormTask((FeatureType.DATA, 'MY-S2-L2A-BANDS'), (FeatureType.DATA, 'NORM'))\n\n# TASK FOR SAVING TO OUTPUT\nsave = SaveTask(patch_path, compress_level=1, overwrite_permission=OverwritePermission.OVERWRITE_PATCH)\n\nworkflow = LinearWorkflow(load, \n add_l2a, \n add_clm_valid, \n add_l1c_valmask, \n add_l2a_valmask, \n add_valmask, \n filter_task, \n ndvi, \n ndwi, \n norm, \n save)",
"_____no_output_____"
]
],
[
[
"## Clean EOPatch list\nMost likely, along with this innovative workflow, you are pushing humankind forward with other processes on your machine. Therefore you do not want to waste your ressources on EOPatches containing very little agricultural area. Before running your already set up EOWorkflow, clean your EOPatch list.\n\nRemember the earlier calculated LPIS ratio? From here on you only keep EOPatches containing more than 13% agricultural area. The irrelevant ones are moved to the sidetrack. If you want to use EOPatches more extensively covered with agricultural area simply increase your \"lpis_thres\" configuration.",
"_____no_output_____"
]
],
[
[
"# in GeoDataFrame label patches with certain thresold either as to do (1) or not to do (0)\ngdf[f'far{year}'] = -2.0\nfor idx, row in gdf.iterrows():\n patch_name = os.path.join(patch_path, f'eopatch_{idx}_col-{row.index_x}_row-{row.index_y}')\n eop = EOPatch.load(str(patch_name), lazy_loading=True)\n gdf.loc[idx, f'far{year}'] = eop.scalar_timeless['FIELD_AREA_RATIO'][0]\n \ngdf[f'todo{year}'] = (gdf[f'far{year}'] > lpis_thres) * 1\ngdf.to_file(shapefile_name)\n\n# move EOPatch folders with LPIS coverage beneath thresold into seperate folder\nmove = []\npatch_list_delete = gpd.read_file(shapefile_name)\npatch_list_delete = patch_list_delete[patch_list_delete[f'todo{year}'] == 0] # identify EOPatches with insufficient LPIS thresold\n\n# create list including names of the identified EOPatches\nfor idx in patch_list_delete.index:\n info = bbox_splitter.info_list[idx]\n patch_name = f'eopatch_{idx}_col-{info[\"index_x\"]}_row-{info[\"index_y\"]}'\n move.append(patch_name)\n \nprint('EOPatches moved to sidetrack: ' + str([patch_name for patch_name in move]))\n\n# move identified EOPatches to alternative folder\nfor patch_name in move:\n shutil.move(os.path.join(patch_path, patch_name), os.path.join(thresLPIS_path, patch_name))\n\npatch_list = os.listdir(patch_path) # update patch_list",
"_____no_output_____"
]
],
[
[
"### Run second EOWorkflow\n* Set up EOWorkflow? **Check!**\n* Ignored irrelevant EOPatches? **Check!**\n\nThen go ahead and run your EOWorkflow on the basis of your \"time_interval\" configuration!",
"_____no_output_____"
]
],
[
[
"# execute workflow and save the names of those that failed\nfailed = []\n\npbar = tqdm(total=len(patch_list))\nfor patch_name in patch_list:\n # add EO data if possible\n try:\n workflow.execute({load: {'eopatch_folder': patch_name},\n add_l2a: {'time_interval': time_interval},\n add_sc_feature: {'data': {}},\n add_rl_feature: {'data': {}},\n save: {'eopatch_folder': patch_name}})\n # append EOPatch name to list for further investigation\n except Exception as ex:\n print(f'Failed {patch_name} with {ex}')\n failed.append(patch_name)\n\n pbar.update()\n",
"_____no_output_____"
]
],
[
[
"# 4. Feature/ label engineering and Sampling\nThe classifier you are using for the following prediction is very picky when it comes to the format of the input data. To feed your thoughtfully compiled data to the algorithm it needs some preparation.",
"_____no_output_____"
],
[
"## Data visualization\n\nNow, after all necessary data is added let's load a single EOPatch and look at the structure. By executing \n```\nEOPatch.load(os.path.join(patch_path, 'eopatch_0_col-0_row-0'))\n```\n\nYou obtain the following structure:\n\n```\nEOPatch(\n data: {\n MY-S2-L2A-BANDS: numpy.ndarray(shape=(39, 1028, 1033, 10), dtype=float32)\n NDVI: numpy.ndarray(shape=(39, 1028, 1033, 1), dtype=float32)\n NDWI: numpy.ndarray(shape=(39, 1028, 1033, 1), dtype=float32)\n NORM: numpy.ndarray(shape=(39, 1028, 1033, 1), dtype=float32)\n }\n mask: {\n CLM: numpy.ndarray(shape=(39, 1028, 1033, 1), dtype=bool)\n CLM_VALID: numpy.ndarray(shape=(39, 1028, 1033, 1), dtype=bool)\n IS_DATA: numpy.ndarray(shape=(39, 1028, 1033, 1), dtype=bool)\n L1C_VALID: numpy.ndarray(shape=(39, 1028, 1033, 1), dtype=bool)\n L2A_VALID: numpy.ndarray(shape=(39, 1028, 1033, 1), dtype=bool)\n SCL: numpy.ndarray(shape=(39, 1028, 1033, 1), dtype=int32)\n VALID_DATA: numpy.ndarray(shape=(39, 1028, 1033, 1), dtype=bool)\n }\n scalar: {}\n label: {}\n vector: {}\n data_timeless: {}\n mask_timeless: {}\n scalar_timeless: {\n FIELD_AREA_RATIO: numpy.ndarray(shape=(1,), dtype=float64)\n }\n label_timeless: {}\n vector_timeless: {\n LPIS_2018: geopandas.GeoDataFrame(columns=['geometry', 'FS_KENNUNG', 'SL_FLAECHE', 'ID', 'SNAR_BEZEI', 'DateImported'], length=4091, crs=EPSG:32633)\n }\n meta_info: {\n maxcc: 0.8\n service_type: 'wcs'\n size_x: '10m'\n size_y: '10m'\n time_difference: datetime.timedelta(seconds=7200)\n time_interval: ['2018-01-01', '2018-09-30']\n }\n bbox: BBox(((420862.3179607267, 5329537.336315366), (431194.28800678457, 5339817.792378783)), crs=CRS('32633'))\n timestamp: [datetime.datetime(2018, 1, 6, 10, 4, 51), ..., datetime.datetime(2018, 9, 28, 10, 0, 24)], length=39\n)\n```\n\nAs you can see your EO data and indices are stored in `data.FeatureType` your valid data masks in `mask.FeatureType` and your ground truth data in `vector_timeless.FeatureType`\n\n\nIt is possible to access various EOPatch content via calls like:\n```\neopatch.timestamp\neopatch.vector_timeless['LPIS_2018']\neopatch.data['NDVI'][0]\neopatch.data['MY-S2-L2A-BANDS'][5][..., [3, 2, 1]]\n\n```",
"_____no_output_____"
],
[
"### Plot RGB image\nIn order to get a quick and realistic overview of your AOI you plot the true color image of one EOPatch",
"_____no_output_____"
]
],
[
[
"eopatch_name = 'eopatch_0_col-0_row-0' # get the name of the first newly created EOPatch\neopatch = EOPatch.load(os.path.join(patch_path, eopatch_name), lazy_loading=True)\n\nfig, ax = plt.subplots(figsize=(20, 20))\nplt.imshow(np.clip(eopatch.data['MY-S2-L2A-BANDS'][0][..., [2, 1, 0]] * 3.5, 0, 1))\nplt.xticks([])\nplt.yticks([])\nax.set_aspect('auto')",
"_____no_output_____"
]
],
[
[
"### Plot mean NDVI\nPlot the time-wise mean of NDVI for the whole region. Filter out clouds in the mean calculation.",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(figsize=(20, 20))\nndvi = eopatch.data['NDVI']\nmask = eopatch.mask['VALID_DATA']\nndvi[~mask] = np.nan\nndvi_mean = np.nanmean(ndvi, axis=0).squeeze()\nim = ax.imshow(ndvi_mean, vmin=0, vmax=0.8, cmap=plt.get_cmap('YlGn'))\nax.set_xticks([])\nax.set_yticks([])\nax.set_aspect('auto')\n\ncb = fig.colorbar(im, ax=ax, orientation='horizontal', pad=0.01, aspect=100)\ncb.ax.tick_params(labelsize=20)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Plot all masks\nTo see how the valid data masks look like and work together, you can compare them to a regular RGB image. For demonstration reasons a timeframe is selected which contains cloud-covered area.",
"_____no_output_____"
]
],
[
[
"tidx = 1\nplt.figure(figsize=(20,20))\n\nplt.subplot(331)\nplt.imshow(np.clip(eopatch.data['MY-S2-L2A-BANDS'][tidx][..., [2,1,0]] * 3.5,0,1))\nplt.xticks([])\nplt.yticks([])\nplt.title('MY-S2-L2A-BANDS - RGB')\n\nplt.subplot(332)\nplt.imshow(eopatch.mask['IS_DATA'][tidx].squeeze(), vmin=0, vmax=1, cmap='gray')\nplt.xticks([])\nplt.yticks([])\nplt.title('IS_DATA - Data availability')\n\nplt.subplot(333)\nplt.imshow(eopatch.mask['CLM'][tidx].squeeze(), vmin=0, vmax=1, cmap='gray')\nplt.xticks([])\nplt.yticks([])\nplt.title('CLM - Cloudmask')\n\nplt.subplot(334)\nplt.imshow(eopatch.mask['L1C_VALID'][tidx].squeeze(), vmin=0, vmax=1, cmap='gray')\nplt.xticks([])\nplt.yticks([])\nplt.title('L1C_VALID - L1C valid data mask')\n\nplt.subplot(335)\nplt.imshow(eopatch.mask['L2A_VALID'][tidx].squeeze(), vmin=0, vmax=1, cmap='gray')\nplt.xticks([])\nplt.yticks([])\nplt.title('L2A_VALID - L2A valid data mask')\n\nplt.subplot(336)\nplt.imshow(eopatch.mask['SCL'][tidx].squeeze(), cmap='jet')\nplt.xticks([])\nplt.yticks([])\nplt.title('SCL - Sen2Cor scene classification map')\n\nplt.subplot(338)\nplt.imshow(eopatch.mask['VALID_DATA'][tidx].squeeze(), vmin=0, vmax=1, cmap='gray')\nplt.xticks([])\nplt.yticks([])\nplt.title('VALID_DATA - Combined valid data mask')",
"_____no_output_____"
]
],
[
[
"As you can see invalid pixel from the different cloud masks and Sen2Cor scene classification map are combined. For SCL the classes: \n* 1 SC_SATURATED_DEFECTIVE \n* 3 SC_CLOUD_SHADOW \n* 8 SC_CLOUD_MEDIUM_PROBABILITY \n* 9 CLOUD_HIGH_PROBABILITY \n* 10 THIN_CIRRUS \n* 11 SNOW \n\nare considered as invalid.",
"_____no_output_____"
],
[
"### Plot spatial mean NDVI timeseries\nPlot the mean of NDVI over all pixels in a single patch throughout the year. Filter out clouds in the mean calculation.",
"_____no_output_____"
]
],
[
[
"ndvi_series = eopatch.data['NDVI']\ntime = np.array(eopatch.timestamp)\nmask = eopatch.mask['VALID_DATA']\nt, w, h, _ = ndvi_series.shape\n\nndvi_clean = ndvi_series.copy()\nndvi_clean[~mask] = np.nan # set values of invalid pixels to NaN's\n\n# Calculate means, remove NaN's from means\nndvi_mean = np.nanmean(ndvi_series.reshape(t, w * h).squeeze(), axis=1)\nndvi_mean_clean = np.nanmean(ndvi_clean.reshape(t, w * h).squeeze(), axis=1)\ntime_clean = time[~np.isnan(ndvi_mean_clean)]\nndvi_mean_clean = ndvi_mean_clean[~np.isnan(ndvi_mean_clean)]\n\nfig, ax = plt.subplots(figsize=(20, 5))\nplt.plot(time_clean, ndvi_mean_clean, 's-', label = 'Mean NDVI with cloud cleaning')\nplt.plot(time, ndvi_mean, 'o-', label='Mean NDVI without cloud cleaning')\nplt.xlabel('Time', fontsize=15)\nplt.ylabel('Mean NDVI over patch', fontsize=15)\nplt.xticks(fontsize=15)\nplt.yticks(fontsize=15)\nplt.legend(loc=2, prop={'size': 15});\nax.set_aspect('auto')\n\ndel eopatch # delete eopatch variable to enable further processing",
"_____no_output_____"
]
],
[
[
"The time series displayed looks very fragmented for the temporal resolution of the Sentinel 2 data to be so hyped, right?\n\nThis is what you get if you choose to keep timeframes with valid data fraction over 70% only. You set the value in your \"datafrac\" configuration. If you expect a nice overview of vegetation growing stages, reality kicks in and gives you mostly cloudy conditions in the first months of the year.\nThe good thing about being picky about the validity of your timeframes is reduced data volume. Invalid frames contain no additional value for your later analysis anyways.",
"_____no_output_____"
],
[
"## Resampling - Interpolation - LPIS data preparation - Sampling\n### Feature concatenation and interpolation\n* For easier handling of the data you concatenate MY-S2-L2A-BANDS, NDVI, NDWI, NORM info into a single feature called FEATURES\n* Perform temporal interpolation (filling gaps and resampling to the same dates) by:\n * creating a linear interpolation task in the temporal dimension\n * providing the cloud mask to tell the interpolating function which values to update\n * using only timeframes from a timerange all EOPatches have in common (from earliest date to latest date)\n\n### LPIS data preparation\n* From scratch, LPIS data is divided into 200 different crop type classes. As the classification is based on spectral signatures, those have to be distinctive. 200 classes are obviously too detailed for achieving accurate prediction results. Therefore you group these classes into reasonable groups also based on similar spectral characteristics using the two CSV files from the \"General data\" folder. The basic grouping defines 14 groups namely: Grass, Maize, Orchards, Peas, Potatoes, Pumpkins, Soybean, Summer cereals, Sunflower, Vegetables, Vine-yards, Winter cereals, Winter rape, Other. This grouping turned out to perform best in classification.\n* After the grouping, the data set stored in vector format is converted into a raster format. Thus, each EO pixel can be assigned to a crop type value. All polygons belonging to one of the classes are separately burned to the raster mask.\n* In order to get rid of artifacts with a width of 1 pixel, and mixed pixels at the edges between polygons of different classes you perform an erosion. That means a buffer of 1 pixel (10m) size is applied to each individual field in the border area.\n\n### Sampling\nBy a spatial sampling of the EOPatches you randomly take a subset of pixels from a patch to use in the machine learning training and testing. Here you only want to consider classes that are represented to a certain quantity of pixels.\n* Remember your \"pixel_tres\" configuration - a threshold of 1000 pixel is necessary for a class to be considered in sampling\n* Remember your \"samp_class\" configuration - 500 pixel per class per EOPatch are sampled",
"_____no_output_____"
]
],
[
[
"# for linear interpolation find earliest and latest overlapping dates\n\n# list EOPatches\neopatches = []\n\npatch_list = os.listdir(patch_path)\n\nfor i in patch_list:\n eopatches.append(EOPatch.load(os.path.join(patch_path, i), lazy_loading=True))\n\neopatches = np.array(eopatches)\n\n# identify earliest date\ntimelist = []\nfor eopatch in eopatches:\n timelist.append(eopatch.timestamp[0])\nmindate = str(max(timelist).date())\nprint('Earliest date: ' + str(max(timelist)))\n\n# identify latest date\ntimelist = []\nfor eopatch in eopatches:\n timelist.append(eopatch.timestamp[-1])\nmaxdate = str(min(timelist).date())\nprint('Latest date: ' + str(min(timelist)))",
"_____no_output_____"
]
],
[
[
"### Set up your 3. EOWorkflow - Feature engineering/ Crop type grouping/ Sampling",
"_____no_output_____"
]
],
[
[
"# TASK FOR LOADING EXISTING EOPATCHES\nload = LoadTask(patch_path)\n\n# TASK FOR CONCATENATION\n# bands and indices are concatenated into one features dictionary\nconcatenate = ConcatenateData('FEATURES', ['MY-S2-L2A-BANDS','NDVI','NDWI','NORM'])\n\n# TASK FOR LINEAR INTERPOLATION\n# linear interpolation of full time-series and date resampling\nresample_range = (mindate, maxdate, day_range)\nlinear_interp = LinearInterpolation(\n 'FEATURES', # name of field to interpolate\n mask_feature=(FeatureType.MASK, 'VALID_DATA'), # mask to be used in interpolation\n copy_features=[(FeatureType.VECTOR_TIMELESS, 'LPIS_{}'.format(year))], # features to keep\n resample_range=resample_range, # set the resampling range\n bounds_error=False # extrapolate with NaN's\n)\n\n# TASK TO FIX AUSTRIAN LPIS DATA\n# on the basis of the wrongly defined column \"SNAR_BEZEI\" \n# a column \"SNAR_BEZEI_NAME\" is added which defines the LPIS class\nfixlpis = FixLPIS(feature='LPIS_{}'.format(year), country='Austria')\n\n# TASK FOR GROUPING LPIS INTO WANTED CLASSES\n# on the basis of the two grouping files an individual crop type grouping can be applied\n# for changes these files have to be adapted\ngrouplpis = GroupLPIS(year=year, lpis_to_group_file=lpis_to_group_file, crop_group_file=crop_group_file)\n\n# TASK FOR CONVERTING LPIS DATA FROM VECTOR TO RASTER FORMAT\n# multiple rasterized layers appling different crop type groupings can be stored in an EOPatch\nvtr = VectorToRaster(\n vector_input=(FeatureType.VECTOR_TIMELESS, 'LPIS_{}'.format(year)), \n raster_feature=(FeatureType.MASK_TIMELESS, 'LPIS_class_{}'.format(grouping_id)),\n values_column='GROUP_1_ID',\n raster_shape=(FeatureType.DATA, 'FEATURES'),\n no_data_value=0)\n\n# TASK FOR EROSION\n# erode each class of the reference map\nerosion = ErosionTask(mask_feature=(FeatureType.MASK_TIMELESS,'LPIS_class_{}'.format(grouping_id),\n 'LPIS_class_{}_ERODED'.format(grouping_id)), disk_radius=1)\n\n# TASK FOR SPATIAL SAMPLING\n# evenly sample about pixels from patches\nspatial_sampling = SamplingTaskTask(grouping_id, pixel_thres, samp_class)\n\n# TASK FOR SAVING TO OUTPUT\nsave = SaveTask(patch_path, overwrite_permission=OverwritePermission.OVERWRITE_PATCH)\n\n# define the workflow\nworkflow = LinearWorkflow(load, \n concatenate, \n linear_interp, \n fixlpis, \n grouplpis, \n vtr, \n erosion, \n spatial_sampling,\n save)",
"_____no_output_____"
]
],
[
[
"### Run third EOWorkflow",
"_____no_output_____"
]
],
[
[
"pbar = tqdm(total=len(patch_list))\nfor patch_name in patch_list:\n extra_param = {load: {'eopatch_folder': patch_name},\n grouplpis: {'col_cropN_lpis': 'SNAR_BEZEI_NAME',\n 'col_cropN_lpistogroup': 'CROP_ID'},\n save: {'eopatch_folder': patch_name}}\n\n workflow.execute(extra_param)\n pbar.update(1)",
"_____no_output_____"
]
],
[
[
"### EOPatch data visualization\n\nNow, after all the data is transformed and sampled let's load the single EOPatch again and look at the structure. By executing \n```\nEOPatch.load(os.path.join(patch_path, 'eopatch_0_col-0_row-0')\n```\n\nYou obtain the following structure:\n\n\n```\nEOPatch(\n data: {\n FEATURES: numpy.ndarray(shape=(31, 1033, 1040, 13), dtype=float64)\n FEATURES_SAMPLED: numpy.ndarray(shape=(31, 6000, 1, 13), dtype=float64)\n }\n mask: {}\n scalar: {}\n label: {}\n vector: {}\n data_timeless: {}\n mask_timeless: {\n LPIS_class_basic: numpy.ndarray(shape=(1033, 1040, 1), dtype=uint8)\n LPIS_class_basic_ERODED: numpy.ndarray(shape=(1033, 1040, 1), dtype=uint8)\n LPIS_class_basic_ERODED_SAMPLED: numpy.ndarray(shape=(6000, 1, 1), dtype=uint8)\n }\n scalar_timeless: {}\n label_timeless: {}\n vector_timeless: {\n LPIS_2018: geopandas.GeoDataFrame(columns=['geometry', 'FS_KENNUNG', 'SL_FLAECHE', 'ID', 'SNAR_BEZEI', 'DateImported', 'SNAR_BEZEI_NAME', 'CROP_ID', 'english', 'slovenian', 'latin', 'GROUP_1', 'GROUP_1_original', 'GROUP_1_ID'], length=4140, crs=epsg:32633)\n }\n meta_info: {}\n bbox: BBox(((420717.14926283853, 5329441.919254168), (431121.7036578405, 5339770.083848184)), crs=EPSG:32633)\n timestamp: [datetime.datetime(2018, 1, 29, 0, 0), ..., datetime.datetime(2018, 9, 26, 0, 0)], length=31\n)\n```\n\nThings have changed, haven't they?\n\nYour 10 spectral bands and 3 indices are combined in `FEATURES` and the randomly sampled pixels are stored in `FEATURES_SAMPLED`. After filtering, your valid data masks have been deleted and your eroded and sampled reference data is available in practical raster format as `mask_timeless.FeatureType`.",
"_____no_output_____"
],
[
"## Combine samples and split into train and test data\nAs you performed the spatial sampling for each patch separately you have to combine the samples. But first you have to assign your EOPatches either to the training or validation dataset. In this case you take one in four EOPatches for testing.\n\nOnly classes present in both train and test dataset are considered in the classification.\n\nThe sampled features and labels are loaded and reshaped into $n \\times m$, where $n$ represents the number of training pixels, and $m = f \\times t$ the number of all features, with $f$ the size of bands and band combinations (in this example 13) and $t$ the length of the resampled time-series (in this example 34)\n\nTerminology: In data science features are commonly refered to as \"X\" and labels as \"y\"",
"_____no_output_____"
]
],
[
[
"patch_list = os.listdir(patch_path) # update patch list\n\n# combine EOPatches to one dataset\neopatches = []\nfor i in patch_list:\n eopatches.append(EOPatch.load(os.path.join(patch_path, i), lazy_loading=True))\n\neopatches = np.array(eopatches)\n\n# depending on the number of EOPatches adjust test_ratio if necessary and split into test and train data accordingly\nif len(patch_list) == 1:\n # split combined dataset into train and test data\n X_train, X_test, y_train, y_test, n_timesteps, n_features = train_test_split_eopatch(eopatches, \n features_dict, \n labels_dict)\n\nelif len(patch_list) < 4:\n test_ratio = 3\n # split combined dataset into train and test data\n X_train, X_test, y_train, y_test, n_timesteps, n_features = train_test_split_eopatches(eopatches, \n test_ratio, \n features_dict, \n labels_dict)\n\nelse:\n # split combined dataset into train and test data\n X_train, X_test, y_train, y_test, n_timesteps, n_features = train_test_split_eopatches(eopatches, \n test_ratio, \n features_dict, \n labels_dict)\n\n# mask out labels that are not in both train and test data and also mask out samples where features include NaN values\nX_train, X_test, y_train, y_test = masking(X_train, X_test, y_train, y_test)\n\ntotal_samp_count = X_train.shape[0] + X_test.shape[0]\nprint('From your {} EOPatch(es) at total of {} samples were taken. '\n 'This sampling dataset includes {} training and {} test samples.'.format(len(patch_list), \n total_samp_count, \n X_train.shape[0], \n X_test.shape[0]))",
"_____no_output_____"
]
],
[
[
"### Plot sample distribution",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(20, 15))\n\ny_ids_train, y_counts_train = np.unique(y_train, return_counts=True)\n\nplt.subplot(2, 1, 1)\nplt.bar(range(len(y_ids_train)), y_counts_train)\nplt.xticks(range(len(y_ids_train)), [class_names[i] for i in y_ids_train], rotation=90, fontsize=20);\nplt.yticks(fontsize=20)\nplt.grid(True)\nplt.title('Training samples', size=20)\n\ny_ids_test, y_counts_test = np.unique(y_test, return_counts=True)\n\nplt.subplot(2, 1, 2)\nplt.bar(range(len(y_ids_test)), y_counts_test)\nplt.xticks(range(len(y_ids_test)), [class_names[i] for i in y_ids_test], rotation=90, fontsize=20);\nplt.yticks(fontsize=20)\nplt.grid(True)\nplt.title('Test samples', size=20)\n\nfig.subplots_adjust(wspace=0, hspace=1)",
"_____no_output_____"
]
],
[
[
"As you can see you have managed to generate a well balanced dataset. In both your 3/4 training and 1/4 test dataset no group is under or over represented, which provides a reasonable basis for the following classification.",
"_____no_output_____"
],
[
"### Scaling and one-hot-encoding\nIn the following you want to feed your samples into two different algorithms. To guarantee equivalent conditions for both models you need scaled features and one-hot-endcoded labels.",
"_____no_output_____"
]
],
[
[
"# scale features\nscaler = StandardScaler()\nX_train = scaler.fit_transform(X_train)\nX_test = scaler.transform(X_test)\nX_train = np.reshape(X_train, (-1,n_timesteps,n_features))\nX_test = np.reshape(X_test, (-1,n_timesteps,n_features))\n# save feature related scaling properties\njoblib.dump(scaler, os.path.join(samples_path, \n 'Scaler_{}.bin'.format(grouping_id)), \n compress=True)\n\n# labels one- hot-encoding\ny_train = y_train.reshape(-1, 1)\ny_test = y_test.reshape(-1, 1)\nenc = OneHotEncoder(sparse=False)\nenc.fit(np.array(class_ids).reshape(-1, 1))\ny_train = enc.transform(y_train)\ny_test = enc.transform(y_test)\nlabel_count = y_train.shape[1]",
"_____no_output_____"
]
],
[
[
"## Save or load samples (optional)\nYou can choose to save your samples for later applications. For entering the upcoming part of prediction, this is not necessary.",
"_____no_output_____"
]
],
[
[
"# save samples(optional)\nnp.save(os.path.join(samples_path, 'X_train_{}'.format(grouping_id)), X_train)\n\nnp.save(os.path.join(samples_path, 'X_test_{}'.format(grouping_id)), X_test)\n\nnp.save(os.path.join(samples_path, 'y_train_{}'.format(grouping_id)), y_train)\n\nnp.save(os.path.join(samples_path, 'y_test_{}'.format(grouping_id)), y_test)",
"_____no_output_____"
],
[
"# load samples(optional)\nX_train = np.load(os.path.join(samples_path, 'X_train_{}.npy'.format(grouping_id)))\n\nX_test = np.load(os.path.join(samples_path, 'X_test_{}.npy'.format(grouping_id)))\n\ny_train = np.load(os.path.join(samples_path, 'y_train_{}.npy'.format(grouping_id)))\n\ny_test = np.load(os.path.join(samples_path, 'y_test_{}.npy'.format(grouping_id)))",
"_____no_output_____"
]
],
[
[
"# 6. Prediction\nCongrats, you've mastered the heavy preprocessing steps! Now, this is where the magic of Machine and Deep Learning happens. \n\nState-of-the-art [LightGBM](https://github.com/Microsoft/LightGBM) is used as a ML model. It is a fast, distributed, high-performance gradient boosting framework based on decision tree algorithms, used for many ML tasks.\n\nAs novel competitors, [TempCNN](https://www.mdpi.com/2072-4292/11/5/523/htm#sec4-remotesensing-11-00523) DL architectures are entering the game. \nSo far Convolutional Neural Networks were mainly and successfully applied for image and language recognition tasks. Modifying the convolutional filters of the architectures the Temporal CNN is supposed to exploit the temporal information of satellite image time series.",
"_____no_output_____"
],
[
"## Set up and train LightGBM model\nThe [default hyper-parameters](https://lightgbm.readthedocs.io/en/latest/Parameters.html) are used in this example. For more info on [parameter tuning](https://lightgbm.readthedocs.io/en/latest/Parameters-Tuning.html), check the documentation of the package.",
"_____no_output_____"
]
],
[
[
"%%time\n\n# Set up training classes\nrev_y_train = [np.argmax(y, axis=None, out=None) for y in y_train]\nrev_y_train_unique = np.unique(rev_y_train)\n\n# reshape features from count-timeframes-features to timeframes-count-features\na, b, c = X_train.shape\nX_train_lgbm = X_train.reshape(a,b * c)\n\n\n# Set up the LightGBM model\nmodel_lgbm = lgb.LGBMClassifier(\n objective='multiclass', \n num_class=len(rev_y_train_unique), \n metric='multi_logloss'\n)\n\n# Train the model\nmodel_lgbm.fit(X_train_lgbm, rev_y_train)\n\n# Save the model\njoblib.dump(model_lgbm, os.path.join(models_path, 'model_lgbm_CropTypeClass_{}.pkl'.format(grouping_id)))",
"_____no_output_____"
]
],
[
[
"## Set up and train TempCNN model\nIn this example an approved architecture from the scientific paper linked above is adopted.",
"_____no_output_____"
]
],
[
[
"%%time\n\n# Set up the TempCNN architecture\nmodel_tcnn = Sequential()\nmodel_tcnn.add(Conv1D(filters=5, kernel_size=3, activation='relu', input_shape=(n_timesteps,n_features)))\nmodel_tcnn.add(Dropout(0.5))\nmodel_tcnn.add(Conv1D(filters=5, kernel_size=3, activation='relu', input_shape=(n_timesteps,n_features)))\nmodel_tcnn.add(Dropout(0.5))\nmodel_tcnn.add(Conv1D(filters=5, kernel_size=3, activation='relu', input_shape=(n_timesteps,n_features)))\nmodel_tcnn.add(Dropout(0.5))\nmodel_tcnn.add(Flatten())\nmodel_tcnn.add(Dense(256, activation='relu'))\nmodel_tcnn.add(Dense(label_count, activation='softmax'))\nmodel_tcnn.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])\n\n# Train the model\nmodel_tcnn.fit(X_train, \n y_train, \n validation_data=(X_test, y_test), \n epochs=10, \n batch_size=32, \n verbose=1, \n shuffle=True)\n\n# Save the model\nmodel_tcnn.save(os.path.join(models_path, 'model_tcnn_CropTypeClass_{}.h5'.format(grouping_id)))",
"_____no_output_____"
]
],
[
[
"## Validation and evaluation\nValidation of the models is a crucial step in data science. All models are wrong, but some are less wrong than others, so model evaluation is important.\n\nIn order to validate the models, we use the training set to predict the classes, and then compare the predicted set of labels to the \"ground truth\".\n\nThe validation is performed by evaluating various metrics, such as accuracy, precision, recall, $F_1$ score, some of which are nicely described [in this blog post](https://medium.com/greyatom/performance-metrics-for-classification-problems-in-machine-learning-part-i-b085d432082b)",
"_____no_output_____"
],
[
"Get the overall accuracy (OA) and the weighted $F_1$ score",
"_____no_output_____"
]
],
[
[
"# reshape features from count-timeframes-features to timeframes-count-features\n# and set up training classes\nd, e, f = X_test.shape\nX_test_lgbm = X_test.reshape(d, e * f)\nrev_y_test = [np.argmax(y, axis=None, out=None) for y in y_test]\n\n# Load the models\nmodel_lgbm = joblib.load(os.path.join(models_path, 'model_lgbm_CropTypeClass_{}.pkl'.format(grouping_id)))\nmodel_tcnn = load_model(os.path.join(models_path, 'model_tcnn_CropTypeClass_{}.h5'.format(grouping_id)))\n\n# get overall accuracy and weighted F1-score for LightGBM\npy_test_lgbm = model_lgbm.predict(X_test_lgbm)\nprint('Classification accuracy LightGBM {:.1f}%'.format(100 * metrics.accuracy_score(rev_y_test, py_test_lgbm)))\nprint('Classification F1-score LightGBM {:.1f}%'.format(100 * metrics.f1_score(rev_y_test, py_test_lgbm, average='weighted')))\n\npy_test_tcnn = model_tcnn.predict_classes(X_test)\nprint('Classification accuracy TempCNN {:.1f}%'.format(100 * metrics.accuracy_score(rev_y_test, py_test_tcnn)))\nprint('Classification F1-score TempCNN {:.1f}%'.format(100 * metrics.f1_score(rev_y_test, py_test_tcnn, average='weighted')))",
"_____no_output_____"
]
],
[
[
"$F_1$ score, precision, and recall for each class separately",
"_____no_output_____"
]
],
[
[
"# LightGBM: F1-score, precision, and recall for each class separately\nclass_labels = np.unique(rev_y_test)\nclass_names = [entry.class_name for entry in LPISCLASS]\n\nf1_scores = metrics.f1_score(rev_y_test, py_test_lgbm, labels=class_labels, average=None)\nrecall = metrics.recall_score(rev_y_test, py_test_lgbm, labels=class_labels, average=None)\nprecision = metrics.precision_score(rev_y_test, py_test_lgbm, labels=class_labels, average=None) \n\nprint('LightGBM:')\nprint(' Class = F1 | Recall | Precision')\nprint(' --------------------------------------------------')\nfor idx, croptype in enumerate([class_names[idx] for idx in class_labels]):\n print(' * {0:20s} = {1:2.1f} | {2:2.1f} | {3:2.1f}'.format(croptype, \n f1_scores[idx] * 100, \n recall[idx] * 100, \n precision[idx] * 100))\n\n# TempCNN: F1-score, precision, and recall for each class separately\nclass_names = [entry.class_name for entry in LPISCLASS]\n\nf1_scores = metrics.f1_score(rev_y_test, py_test_tcnn, labels=class_labels, average=None)\nrecall = metrics.recall_score(rev_y_test, py_test_tcnn, labels=class_labels, average=None)\nprecision = metrics.precision_score(rev_y_test, py_test_tcnn, labels=class_labels, average=None) \n\nprint('TempCNN:')\nprint(' Class = F1 | Recall | Precision')\nprint(' --------------------------------------------------')\nfor idx, croptype in enumerate([class_names[idx] for idx in class_labels]):\n print(' * {0:20s} = {1:2.1f} | {2:2.1f} | {3:2.1f}'.format(croptype, \n f1_scores[idx] * 100, \n recall[idx] * 100, \n precision[idx] * 100))",
"_____no_output_____"
]
],
[
[
"### Plot the standard Confusion Matrix for LightGBM",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(20, 20))\nconf_matrix_gbm = metrics.confusion_matrix(rev_y_test, py_test_lgbm)\nplot_confusion_matrix(conf_matrix_gbm,\n classes=[name for idx, name in enumerate(class_names) if idx in class_labels],\n normalize=True,\n ylabel='Truth (CROPS)',\n xlabel='Predicted (LightGBM)',\n title='Confusion matrix');",
"_____no_output_____"
]
],
[
[
"### Plot the standard Confusion Matrix for TempCNN",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(20, 20))\nconf_matrix_gbm = metrics.confusion_matrix(rev_y_test, py_test_tcnn)\nplot_confusion_matrix(conf_matrix_gbm,\n classes=[name for idx, name in enumerate(class_names) if idx in class_labels],\n normalize=True,\n ylabel='Truth (CROPS)',\n xlabel='Predicted (TempCNN)',\n title='Confusion matrix');",
"_____no_output_____"
]
],
[
[
"The validation of the models shows that for most of the groups both perform very well. However, there seem to be differences in their confusion for certain classes:\n* In this specific case orchards might catch your attention mostly. LightGBM performs worse than TempCNN. But more interesting than the overall accuracy is, that LightGBM classifies actual orchards as grass a lot (low recall) while, no other class is mistaken as orchards (high precision). In contrast TempCNN recognizes actual orchards well (high recall) but identifies acutal grass as orchards frequently (lower precision). Generally, confusion with grass class is not surprising, as there is a lot of it between the individual trees.\n* There is also poor performance received for potatoes in both models as their cultivation practices are quite similar to peas.\n* Poor performance for the group Other is expectable in consequence of its diverse class composition",
"_____no_output_____"
],
[
"### Most important features\n\nThe LightGBM model contains the information about feature importances. Let's check which features are most important for classification.",
"_____no_output_____"
]
],
[
[
"eopatch_name = 'eopatch_0_col-0_row-0' # get the name of the first newly created EOPatch\neopatch = EOPatch.load(os.path.join(patch_path, eopatch_name), lazy_loading=True)\n\ntimeframe_count = eopatch.data['FEATURES'].shape[0]\nfeatures_count = eopatch.data['FEATURES'].shape[3]\n\ndel eopatch\n\nz = model_lgbm.feature_importances_.reshape((timeframe_count, features_count))\n\nfnames = ['B02','B03','B04','B05','B06','B07','B08','B8A','B11','B12','NDVI','NDWI','NORM']\n\nfig = plt.figure(figsize=(15, 15))\nax = plt.gca()\n\n# plot the importances\nim = ax.imshow(z, aspect=0.25)\nplt.xticks(range(len(fnames)), fnames, rotation=45, fontsize=20)\nplt.yticks(range(timeframe_count), ['T{}'.format(i) for i in range(timeframe_count)], fontsize=20)\nplt.xlabel('Bands and band related features', fontsize=20)\nplt.ylabel('Time frames', fontsize=15)\nplt.ylim(top=-0.5, bottom=timeframe_count - 0.5)\nax.xaxis.tick_top()\nax.xaxis.set_label_position('top')\n\n\nfig.subplots_adjust(wspace=0, hspace=0)\n\ncb = fig.colorbar(im, ax=[ax], orientation='horizontal', pad=0.01, aspect=100)\ncb.ax.tick_params(labelsize=20)\ncb.set_label('Feature importance', fontsize=15)",
"_____no_output_____"
]
],
[
[
"As you can see, the most important features for LightGBM are recorded within the main growth period. Here different growing stages can be detected that constitute certain crop types. ",
"_____no_output_____"
],
[
"## Prediction\n\nNow that both models have been validated, the remaining thing is to predict the whole AOI. As LightGBM receives higher overall accurays it is used for further predictions. If you are interested in a specific crop group TempCNN is outperforming LightGBM simply change the following configuration.",
"_____no_output_____"
]
],
[
[
"# swap commentation for using a different model\n\n# model = load_model(os.path.join(models_path, 'model_tcnn_CropTypeClass_{}.h5'.format(grouping_id))) # load TempCNN model\nmodel = joblib.load(os.path.join(models_path, 'model_lgbm_CropTypeClass_{}.pkl'.format(grouping_id))) # load LightGBM model\n\n# load respective feature scaler\nscaler = joblib.load(os.path.join(samples_path, 'Scaler_{}.bin'.format(grouping_id)))",
"_____no_output_____"
]
],
[
[
"In the following you define a workflow to make a prediction on the existing EOPatches. The EOTask accepts the features and the names for the labels. In addition you export GeoTIFF images of the prediction to easily access your visual results.",
"_____no_output_____"
],
[
"### Set up your 4. EOWorklow - Prediction",
"_____no_output_____"
]
],
[
[
"# TASK TO LOAD EXISTING EOPATCHES\nload = LoadTask(patch_path)\n\n# TASK FOR PREDICTION\npredict = PredictPatch(model, (FeatureType.DATA, 'FEATURES'), 'LBL_GBM', scaler)\n\n# TASK TO EXPORT TIFF\nexport_tiff = ExportToTiff((FeatureType.MASK_TIMELESS, 'LBL_GBM'))\ntiff_location = predictions_path\nif not os.path.isdir(tiff_location):\n os.makedirs(tiff_location)\n \n# TASK FOR SAVING\nsave = SaveTask(patch_path, overwrite_permission=OverwritePermission.OVERWRITE_PATCH)\n\n\nworkflow = LinearWorkflow(load,\n predict,\n export_tiff,\n save)",
"_____no_output_____"
]
],
[
[
"### Run fourth EOWorkflow",
"_____no_output_____"
]
],
[
[
"patch_list = os.listdir(patch_path) # update patch list\n# execute workflow\npbar = tqdm(total=len(patch_list))\nfor patch_name in patch_list:\n extra_param = {load: {'eopatch_folder': patch_name}, \n export_tiff: {'filename': '{}/prediction_{}.tiff'.format(predictions_path, patch_name)}, \n save: {'eopatch_folder': patch_name}}\n\n workflow.execute(extra_param)\n pbar.update()",
"_____no_output_____"
]
],
[
[
"### EOPatch data visualization\n\nFinishing the last processing step, let's have a look at the final EOPatch by executing\n```\nEOPatch.load(os.path.join(patch_path, 'eopatch_0_col-0_row-0')\n```\n\nYou obtain the following structure which is extended by your predicted data stored as `LBL_GBM` in `mask_timeless.FeatureType`:\n\n\n```\nEOPatch(\n data: {\n FEATURES: numpy.ndarray(shape=(34, 1028, 1033, 13), dtype=float64)\n FEATURES_SAMPLED: numpy.ndarray(shape=(34, 6000, 1, 13), dtype=float64)\n }\n mask: {}\n scalar: {}\n label: {}\n vector: {}\n data_timeless: {}\n mask_timeless: {\n LBL_GBM: numpy.ndarray(shape=(1028, 1033, 1), dtype=int64)\n LPIS_class_basic: numpy.ndarray(shape=(1028, 1033, 1), dtype=uint8)\n LPIS_class_basic_ERODED: numpy.ndarray(shape=(1028, 1033, 1), dtype=uint8)\n LPIS_class_basic_ERODED_SAMPLED: numpy.ndarray(shape=(6000, 1, 1), dtype=uint8)\n }\n scalar_timeless: {}\n label_timeless: {}\n vector_timeless: {\n LPIS_2018: geopandas.GeoDataFrame(columns=['geometry', 'FS_KENNUNG', 'SL_FLAECHE', 'ID', 'SNAR_BEZEI', 'DateImported', 'SNAR_BEZEI_NAME', 'CROP_ID', 'english', 'slovenian', 'latin', 'GROUP_1', 'GROUP_1_original', 'GROUP_1_ID'], length=4091, crs=EPSG:32633)\n }\n meta_info: {}\n bbox: BBox(((420862.3179607267, 5329537.336315366), (431194.28800678457, 5339817.792378783)), crs=CRS('32633'))\n timestamp: [datetime.datetime(2018, 1, 6, 0, 0), ..., datetime.datetime(2018, 9, 27, 0, 0)], length=34\n)\n```",
"_____no_output_____"
],
[
"## Visualization of the results",
"_____no_output_____"
],
[
"### Visualize predicted EOPatch data",
"_____no_output_____"
]
],
[
[
"eopatch_name = 'eopatch_0_col-0_row-0' # get the name of the first newly created EOPatch\neopatch = EOPatch.load(os.path.join(patch_path, eopatch_name), lazy_loading=True)\n\n# update colormap\ncb_classes = np.unique(np.unique(eopatch.mask_timeless['LBL_GBM']))\ncustom_cmap = mpl.colors.ListedColormap([lpisclass_cmap.colors[i] for i in cb_classes])\ncustom_norm = mpl.colors.BoundaryNorm(np.arange(-0.5, len(cb_classes), 1), custom_cmap.N)\n\n# mask prediction - exclude pixel with no LPIS reference\nlabels = np.array(eopatch.mask_timeless['LPIS_class_{}'.format(grouping_id)])\nmask = labels == 0\nlabelspred = np.array(eopatch.mask_timeless['LBL_GBM'])\nLBL = np.ma.masked_array(labelspred, mask)\n\n# plot figure\nfig, ax = plt.subplots(figsize=(20, 20))\nim = ax.imshow(LBL.squeeze(), cmap=lpisclass_cmap, norm=lpisclass_norm)\nax.set_xticks([])\nax.set_yticks([])\nax.set_aspect('auto')\n\nfig.subplots_adjust(wspace=0, hspace=0)\n\n# plot colorbar\ncb = fig.colorbar(mpl.cm.ScalarMappable(norm=custom_norm, cmap=custom_cmap), \n orientation=\"horizontal\", \n pad=0.01, \n aspect=100)\ncb.ax.tick_params(labelsize=20)\ncb.set_ticks(range(len(cb_classes)))\ncb.ax.set_xticklabels([class_names[i] for i in cb_classes], rotation=90, fontsize=15)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Compare ground truth and prediction",
"_____no_output_____"
]
],
[
[
"# mask prediction - exclude pixel with no LPIS reference\nlabels = np.array(eopatch.mask_timeless['LPIS_class_{}'.format(grouping_id)])\nmask = labels == 0\nlabelspred = np.array(eopatch.mask_timeless['LBL_GBM'])\nLBL = np.ma.masked_array(labelspred, mask)\n\nfig, axes = plt.subplots(2,2,figsize=(20, 10))\n\n# plot prediction\nax1 = plt.subplot(121)\nim = ax1.imshow(LBL.squeeze(), cmap=lpisclass_cmap, norm=lpisclass_norm)\nplt.title('Prediction')\nax1.set_xticks([])\nax1.set_yticks([])\nax1.set_aspect('auto')\n\n# plot ground truth\nax2 = plt.subplot(122)\nim = ax2.imshow(labels.squeeze(), cmap=lpisclass_cmap, norm=lpisclass_norm)\nplt.title('Ground truth')\nax2.set_xticks([])\nax2.set_yticks([])\nax2.set_aspect('auto')\n\naxlist=[ax1,ax2]\n\nfig.subplots_adjust(wspace=0, hspace=0)\n\n# plot colorbar\ncb = fig.colorbar(mpl.cm.ScalarMappable(norm=custom_norm, cmap=custom_cmap), \n ax = axlist,\n orientation=\"horizontal\", \n pad=0.01, \n aspect=100)\ncb.ax.tick_params(labelsize=20)\ncb.set_ticks(range(len(cb_classes)))\ncb.ax.set_xticklabels([class_names[i] for i in cb_classes], rotation=90, fontsize=15)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Close-up comparison",
"_____no_output_____"
]
],
[
[
"# create red-green colormap\ncolors = [(0, 1, 0), (1, 0, 0)] # G -> R\ncmap_name = 'my_list'\ncm = LinearSegmentedColormap.from_list(\n cmap_name, colors)\n\n\nfig = plt.figure(figsize=(20, 20))\n\ninspect_size = 100\n\nw, h = labels.squeeze().shape\n\nw_min = np.random.choice(range(w - inspect_size))\nh_min = np.random.choice(range(h - inspect_size))\n\nax = plt.subplot(2, 2, 1)\nplt.imshow(labels.squeeze()[w_min: w_min + inspect_size, h_min : h_min + inspect_size],\n cmap=lpisclass_cmap, norm=lpisclass_norm)\nplt.xticks([])\nplt.yticks([])\nax.set_aspect('auto')\nplt.title('Ground truth', fontsize=20)\n\nax = plt.subplot(2, 2, 2)\nplt.imshow(LBL.squeeze()[w_min: w_min + inspect_size, h_min: h_min + inspect_size],\n cmap=lpisclass_cmap, norm=lpisclass_norm)\nplt.xticks([])\nplt.yticks([])\nax.set_aspect('auto')\nplt.title('Prediction', fontsize=20)\n\nax = plt.subplot(2, 2, 3)\nmask = LBL.squeeze() != labels.squeeze()\nplt.imshow(mask[w_min: w_min + inspect_size, h_min: h_min + inspect_size], cmap=cm)\nplt.xticks([])\nplt.yticks([]);\nax.set_aspect('auto')\nplt.title('Difference', fontsize=20)\n\nax = plt.subplot(2, 2, 4)\nimage = np.clip(eopatch.data['FEATURES'][8][..., [2, 1, 0]] * 3.5, 0, 1)\nplt.imshow(image[w_min: w_min + inspect_size, h_min: h_min + inspect_size])\nplt.xticks([])\nplt.yticks([]);\nax.set_aspect('auto')\nplt.title('True Color', fontsize=20)\n\nfig.subplots_adjust(wspace=0.1, hspace=0.1)",
"_____no_output_____"
]
],
[
[
"As you can probably see in the randomly chosen section of the AOI there are certain patterns of misclassified pixels:\n* There are complete fields mistaken as another crop group. In this case the algorithm got confused because of similar spectral characteristics. You already got an overview of the frequency and combination of those incidents in the evaluation part above.\n* Misclassified single pixels are usually located at the border of the respective fields. Here the \"mixed-pixel-problem\" impacts the prediction results. For the modeling these pixels were excluded, as they may include spectral reflectance values of different vegetation types and thereby confuse the algorithm.",
"_____no_output_____"
],
[
"# Next steps\n\nNow, after your first successful classification you are hooked? But the region around Wels in Austria was surprisingly not your actual AOI or you want to try other vegetation groupings? Then here are some suggestions on how you could proceed:\n* **Customize configurations**\n\n The notebook offers various possibilities to change parameters and evaluate their effects. Simply enter the configuration section in the beginning and modify e.g. cloudcover thresholds or your sampling strategy.\n\n\n* **Change the AOI within Austria**\n\n This would be the simplest case to apply. You just have to place a Shapefile or Geojson of your own AOI in the location of the \"Area_AOI.geojson\" from the example. The size and shape of the included polygon are irrelevant. \n \n \n* **Try alternative crop groupings**\n\n In order to regroup the LPIS classes you need to have a closer look at the two CSV files in the `GeneralData` folder. \n * `at_lpis_2018_crop_to_group_mapping_basic.csv`: Here you can assign LPIS classes to different crop groups.\\\n *_CROP_ID_* represents the respective LPIS class\\\n *_GROUP_1_* represents the respective group you want a class in\n * `crop_group_1_definition_basic.csv`: Here you can combine or separate individual crop groups by assigning the respective ID.\\\n *_GROUP_1_* again represents the groups\\\n *_GROUP_1_ID_* represents the respective numeric ID\n \n \n* **Apply the notebook to another country**\n\n Another country means different AOI plus different LPIS classes. \n * The first requires no additional effort. Change your AOI file and run the processes. EO data is downloaded and processed exactly as in the example. \n * But when it comes to the ground truth data, this is were things get tricky as you additionally need to customize the CSV grouping files for your specific country",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb45e7106b62b7c74b762216074d6c43a67a1378 | 2,206 | ipynb | Jupyter Notebook | Decorator Order.ipynb | tylerdave/ipython-notebooks | 8e69e653f8f57cc5d5b5adf5f27b4d3238802340 | [
"MIT"
] | null | null | null | Decorator Order.ipynb | tylerdave/ipython-notebooks | 8e69e653f8f57cc5d5b5adf5f27b4d3238802340 | [
"MIT"
] | null | null | null | Decorator Order.ipynb | tylerdave/ipython-notebooks | 8e69e653f8f57cc5d5b5adf5f27b4d3238802340 | [
"MIT"
] | null | null | null | 22.742268 | 59 | 0.446056 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
cb45ea2f96f5cb1378e1c65c6b189c243809e41f | 3,483 | ipynb | Jupyter Notebook | critique-DataCats-Eric-Lance.ipynb | chapman-cs510-2016f/cw-06-eric_lance | 2e1446ab614ed5e9484fb20e6286c643448da42b | [
"MIT"
] | null | null | null | critique-DataCats-Eric-Lance.ipynb | chapman-cs510-2016f/cw-06-eric_lance | 2e1446ab614ed5e9484fb20e6286c643448da42b | [
"MIT"
] | null | null | null | critique-DataCats-Eric-Lance.ipynb | chapman-cs510-2016f/cw-06-eric_lance | 2e1446ab614ed5e9484fb20e6286c643448da42b | [
"MIT"
] | null | null | null | 59.033898 | 521 | 0.705426 | [
[
[
"# Classwork 6\n### Critique another group's classwork 5\n\n### Group Eric-Lance",
"_____no_output_____"
],
[
"#### Is it clear how the code is organized? \nWe, the people of Datacats, believe this module is very well organized and easy to follow. We can clearly see what each function does, where each function lies, i.e. nothing looks confusing or out of place. There are comments inbetween different steps, which makes understanding the different parts of the code easy to follow. Overall, the organization is good.\n\n#### Is the code properly documented with both docstrings and supplementary comments according to industry standards? \nYes, the module has correctly documented docstrings, both for the module docstring and the function docstrings. As stated before, there are comments throughout the code which makes the algorithms easy to follow. One critique we feel is the docstrings themselves could contain more information. \nThe Module Docstring, we feel is fine.\nThe Function Docstring for the init, could use the wording \"the user needs to input\" or use the words \"parameter 1, parameter 2\" so if someone who was reading the docstring did not know how to use the function, they will have a better clerification of how to use the function, and which parameter input goes to which variable. We're not sure if it's a necessity, but it could be something to think about when writing programs where users not as familiar with cloding, needs to figure out how to use a module.\nThe other Function Docstrings were taken from the original abscplane class, which is straightforward, so we think they're stated well, but we suppose it could use some more elaboration on the function. Once again, not sure if it's needed or necessary, but could be something to think.\n\n#### Can you follow the algorithm of the code, i.e., what it is doing, and how? \nIt is very easy to follow. We loved how you guys made the xstep and ystep variable a part of the class' initial variables, so it can be used again in different functions. We liked how you built the plane, the coding of it is very concise, short, and saved a lot of steps, in comparison to what we did. Very straightforward and simple, which is good.\n\n#### Do you see any suggestions for how to improve the code? Discuss your critique with the members of the other group.\nProbaby the biggest suggestion for you guys is to put the test functions in a different module/file. This way, the code does not look unnecessarily long or cluttered. Although the test functions themselves don't interfere with code itself, we think asthetically wise, it would make your module look better. \nAlthough we will say we liked how you programmed your test functions. It is very good. ",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown"
]
] |
cb45ea469145d5d39b668f727a7f2cd3c672a4d4 | 17,456 | ipynb | Jupyter Notebook | notebooks/building_production_ml_systems/labs/4b_streaming_data_inference_vertex.ipynb | Jonathanpro/asl-ml-immersion | c461aa215339a6816810dfef5a92a6e375f9bc66 | [
"Apache-2.0"
] | 11 | 2021-09-08T05:39:02.000Z | 2022-03-25T14:35:22.000Z | notebooks/building_production_ml_systems/labs/4b_streaming_data_inference_vertex.ipynb | Jonathanpro/asl-ml-immersion | c461aa215339a6816810dfef5a92a6e375f9bc66 | [
"Apache-2.0"
] | 118 | 2021-08-28T03:09:44.000Z | 2022-03-31T00:38:44.000Z | notebooks/building_production_ml_systems/labs/4b_streaming_data_inference_vertex.ipynb | Jonathanpro/asl-ml-immersion | c461aa215339a6816810dfef5a92a6e375f9bc66 | [
"Apache-2.0"
] | 110 | 2021-09-02T15:01:35.000Z | 2022-03-31T12:32:48.000Z | 37.539785 | 553 | 0.635713 | [
[
[
"# Working with Streaming Data\n\nLearning Objectives\n 1. Learn how to process real-time data for ML models using Cloud Dataflow\n 2. Learn how to serve online predictions using real-time data\n\n## Introduction\n\nIt can be useful to leverage real time data in a machine learning model when making a prediction. However, doing so requires setting up a streaming data pipeline which can be non-trivial. \n\nTypically you will have the following:\n - A series of IoT devices generating and sending data from the field in real-time (in our case these are the taxis)\n - A messaging bus to that receives and temporarily stores the IoT data (in our case this is Cloud Pub/Sub)\n - A streaming processing service that subscribes to the messaging bus, windows the messages and performs data transformations on each window (in our case this is Cloud Dataflow)\n - A persistent store to keep the processed data (in our case this is BigQuery)\n\nThese steps happen continuously and in real-time, and are illustrated by the blue arrows in the diagram below. \n\nOnce this streaming data pipeline is established, we need to modify our model serving to leverage it. This simply means adding a call to the persistent store (BigQuery) to fetch the latest real-time data when a prediction request comes in. This flow is illustrated by the red arrows in the diagram below. \n\n<img src='../assets/taxi_streaming_data.png' width='80%'>\n\n\nIn this lab we will address how to process real-time data for machine learning models. We will use the same data as our previous 'taxifare' labs, but with the addition of `trips_last_5min` data as an additional feature. This is our proxy for real-time traffic.\n\n",
"_____no_output_____"
]
],
[
[
"!pip install --upgrade apache-beam[gcp]",
"_____no_output_____"
]
],
[
[
"Restart the kernel before proceeding further (On the Notebook menu - Kernel - Restart Kernel).",
"_____no_output_____"
]
],
[
[
"import os\nimport shutil\n\nimport numpy as np\nimport tensorflow as tf\nfrom google import api_core\nfrom google.cloud import aiplatform, bigquery\nfrom google.protobuf import json_format\nfrom google.protobuf.struct_pb2 import Value\nfrom matplotlib import pyplot as plt\nfrom tensorflow import keras\nfrom tensorflow.keras.callbacks import TensorBoard\nfrom tensorflow.keras.layers import Dense, DenseFeatures\nfrom tensorflow.keras.models import Sequential\n\nprint(tf.__version__)",
"_____no_output_____"
],
[
"# Change below if necessary\nPROJECT = !gcloud config get-value project # noqa: E999\nPROJECT = PROJECT[0]\nBUCKET = PROJECT\nREGION = \"us-central1\"\n\n%env PROJECT=$PROJECT\n%env BUCKET=$BUCKET\n%env REGION=$REGION",
"_____no_output_____"
],
[
"%%bash\ngcloud config set project $PROJECT\ngcloud config set ai/region $REGION",
"_____no_output_____"
]
],
[
[
"## Re-train our model with `trips_last_5min` feature\n\nIn this lab, we want to show how to process real-time data for training and prediction. So, we need to retrain our previous model with this additional feature. Go through the notebook `4a_streaming_data_training.ipynb`. Open and run the notebook to train and save a model. This notebook is very similar to what we did in the Introduction to Tensorflow module but note the added feature for `trips_last_5min` in the model and the dataset.",
"_____no_output_____"
],
[
"## Simulate Real Time Taxi Data\n\nSince we don’t actually have real-time taxi data we will synthesize it using a simple python script. The script publishes events to Google Cloud Pub/Sub.\n\nInspect the `iot_devices.py` script in the `taxicab_traffic` folder. It is configured to send about 2,000 trip messages every five minutes with some randomness in the frequency to mimic traffic fluctuations. These numbers come from looking at the historical average of taxi ride frequency in BigQuery. \n\nIn production this script would be replaced with actual taxis with IoT devices sending trip data to Cloud Pub/Sub. \n\nTo execute the `iot_devices.py` script, launch a terminal and navigate to the `asl-ml-immersion/notebooks/building_production_ml_systems/solutions` directory. Then run the following two commands.",
"_____no_output_____"
],
[
"```bash\nPROJECT_ID=$(gcloud config get-value project)\npython3 ./taxicab_traffic/iot_devices.py --project=$PROJECT_ID\n```",
"_____no_output_____"
],
[
"You will see new messages being published every 5 seconds. **Keep this terminal open** so it continues to publish events to the Pub/Sub topic. If you open [Pub/Sub in your Google Cloud Console](https://console.cloud.google.com/cloudpubsub/topic/list), you should be able to see a topic called `taxi_rides`.",
"_____no_output_____"
],
[
"## Create a BigQuery table to collect the processed data\n\nIn the next section, we will create a dataflow pipeline to write processed taxifare data to a BigQuery Table, however that table does not yet exist. Execute the following commands to create a BigQuery dataset called `taxifare` and a table within that dataset called `traffic_realtime`. ",
"_____no_output_____"
]
],
[
[
"bq = bigquery.Client()\n\ndataset = bigquery.Dataset(bq.dataset(\"taxifare\"))\ntry:\n bq.create_dataset(dataset) # will fail if dataset already exists\n print(\"Dataset created.\")\nexcept api_core.exceptions.Conflict:\n print(\"Dataset already exists.\")",
"_____no_output_____"
]
],
[
[
"Next, we create a table called `traffic_realtime` and set up the schema.",
"_____no_output_____"
]
],
[
[
"dataset = bigquery.Dataset(bq.dataset(\"taxifare\"))\n\ntable_ref = dataset.table(\"traffic_realtime\")\nSCHEMA = [\n bigquery.SchemaField(\"trips_last_5min\", \"INTEGER\", mode=\"REQUIRED\"),\n bigquery.SchemaField(\"time\", \"TIMESTAMP\", mode=\"REQUIRED\"),\n]\ntable = bigquery.Table(table_ref, schema=SCHEMA)\n\ntry:\n bq.create_table(table)\n print(\"Table created.\")\nexcept api_core.exceptions.Conflict:\n print(\"Table already exists.\")",
"_____no_output_____"
]
],
[
[
"## Launch Streaming Dataflow Pipeline\n\nNow that we have our taxi data being pushed to Pub/Sub, and our BigQuery table set up, let’s consume the Pub/Sub data using a streaming DataFlow pipeline.\n\nThe pipeline is defined in `./taxicab_traffic/streaming_count.py`. Open that file and inspect it. \n\nThere are 5 transformations being applied:\n - Read from PubSub\n - Window the messages\n - Count number of messages in the window\n - Format the count for BigQuery\n - Write results to BigQuery\n\n**TODO:** Open the file ./taxicab_traffic/streaming_count.py and find the TODO there. Specify a sliding window that is 5 minutes long, and gets recalculated every 15 seconds. Hint: Reference the [beam programming guide](https://beam.apache.org/documentation/programming-guide/#windowing) for guidance. To check your answer reference the solution. \n\nFor the second transform, we specify a sliding window that is 5 minutes long, and recalculate values every 15 seconds. \n\nIn a new terminal, launch the dataflow pipeline using the command below. You can change the `BUCKET` variable, if necessary. Here it is assumed to be your `PROJECT_ID`.",
"_____no_output_____"
],
[
"```bash\nPROJECT_ID=$(gcloud config get-value project)\nREGION=$(gcloud config get-value ai/region)\nBUCKET=$PROJECT_ID # change as necessary \npython3 ./taxicab_traffic/streaming_count.py \\\n --input_topic taxi_rides \\\n --runner=DataflowRunner \\\n --project=$PROJECT_ID \\\n --region=$REGION \\\n --temp_location=gs://$BUCKET/dataflow_streaming\n```",
"_____no_output_____"
],
[
"Once you've submitted the command above you can examine the progress of that job in the [Dataflow section of Cloud console](https://console.cloud.google.com/dataflow). ",
"_____no_output_____"
],
[
"## Explore the data in the table",
"_____no_output_____"
],
[
"After a few moments, you should also see new data written to your BigQuery table as well. \n\nRe-run the query periodically to observe new data streaming in! You should see a new row every 15 seconds. ",
"_____no_output_____"
]
],
[
[
"%%bigquery\nSELECT\n *\nFROM\n `taxifare.traffic_realtime`\nORDER BY\n time DESC\nLIMIT 10",
"_____no_output_____"
]
],
[
[
"## Make predictions from the new data\n\nIn the rest of the lab, we'll referece the model we trained and deployed from the previous labs, so make sure you have run the code in the `4a_streaming_data_training.ipynb` notebook. \n\nThe `add_traffic_last_5min` function below will query the `traffic_realtime` table to find the most recent traffic information and add that feature to our instance for prediction.",
"_____no_output_____"
],
[
"**Exercise.** Complete the code in the function below. Write a SQL query that will return the most recent entry in `traffic_realtime` and add it to the instance.",
"_____no_output_____"
]
],
[
[
"# TODO 2a. Write a function to take most recent entry in `traffic_realtime`\n# table and add it to instance.\ndef add_traffic_last_5min(instance):\n bq = bigquery.Client()\n query_string = \"\"\"\n TODO: Your code goes here\n \"\"\"\n trips = bq.query(query_string).to_dataframe()[\"trips_last_5min\"][0]\n instance['traffic_last_5min'] = # TODO: Your code goes here.\n return instance",
"_____no_output_____"
]
],
[
[
"The `traffic_realtime` table is updated in realtime using Cloud Pub/Sub and Dataflow so, if you run the cell below periodically, you should see the `traffic_last_5min` feature added to the instance and change over time. ",
"_____no_output_____"
]
],
[
[
"add_traffic_last_5min(\n instance={\n \"dayofweek\": 4,\n \"hourofday\": 13,\n \"pickup_longitude\": -73.99,\n \"pickup_latitude\": 40.758,\n \"dropoff_latitude\": 41.742,\n \"dropoff_longitude\": -73.07,\n }\n)",
"_____no_output_____"
]
],
[
[
"Finally, we'll use the python api to call predictions on an instance, using the realtime traffic information in our prediction. Just as above, you should notice that our resulting predicitons change with time as our realtime traffic information changes as well.",
"_____no_output_____"
],
[
"**Exercise.** Complete the code below to call prediction on an instance incorporating realtime traffic info. You should\n- use the function `add_traffic_last_5min` to add the most recent realtime traffic data to the prediction instance\n- call prediction on your model for this realtime instance and save the result as a variable called `response`\n- parse the json of `response` to print the predicted taxifare cost",
"_____no_output_____"
],
[
"Copy the `ENDPOINT_RESOURCENAME` from the deployment in the previous lab to the beginning of the block below.",
"_____no_output_____"
]
],
[
[
"# TODO 2b. Write code to call prediction on instance using realtime traffic\n# info. Hint: Look at this sample\n# https://github.com/googleapis/python-aiplatform/blob/master/samples/snippets/predict_custom_trained_model_sample.py\n\n# TODO: Copy the `ENDPOINT_RESOURCENAME` from the deployment in the previous\n# lab.\nENDPOINT_RESOURCENAME = \"\"\n\napi_endpoint = f\"{REGION}-aiplatform.googleapis.com\"\n\n# The AI Platform services require regional API endpoints.\nclient_options = {\"api_endpoint\": api_endpoint}\n# Initialize client that will be used to create and send requests.\n# This client only needs to be created once, and can be reused for multiple\n# requests.\nclient = aiplatform.gapic.PredictionServiceClient(client_options=client_options)\n\ninstance = {\n \"dayofweek\": 4,\n \"hourofday\": 13,\n \"pickup_longitude\": -73.99,\n \"pickup_latitude\": 40.758,\n \"dropoff_latitude\": 41.742,\n \"dropoff_longitude\": -73.07,\n}\n\n# The format of each instance should conform to the deployed model's\n# prediction input schema.\ninstance_dict = # TODO: Your code goes here.\n\ninstance = json_format.ParseDict(instance, Value())\ninstances = [instance]\nresponse = # TODO: Your code goes here.\n\n# The predictions are a google.protobuf.Value representation of the model's\n# predictions.\nprint(\" prediction:\",\n # TODO: Your code goes here.\n)",
"_____no_output_____"
]
],
[
[
"## Cleanup",
"_____no_output_____"
],
[
"In order to avoid ongoing charges, when you are finished with this lab, you can delete your Dataflow job of that job from the [Dataflow section of Cloud console](https://console.cloud.google.com/dataflow).\n\nAn endpoint with a model deployed to it incurs ongoing charges, as there must be at least one replica defined (the `min-replica-count` parameter is at least 1). In order to stop incurring charges, you can click on the endpoint on the [Endpoints page of the Cloud Console](https://console.cloud.google.com/vertex-ai/endpoints) and un-deploy your model.",
"_____no_output_____"
],
[
"Copyright 2021 Google Inc. Licensed under the Apache License, Version 2.0 (the \"License\"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an \"AS IS\" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cb45ff2e0f013c4519f7a1f89bdc251fd9d91722 | 36,391 | ipynb | Jupyter Notebook | advanced_functionality/pytorch_bring_your_own_gan/build_gan_with_pytorch.ipynb | npalm/amazon-sagemaker-examples | 5b9c3ed1f82efa2826402492971b9904696f57df | [
"Apache-2.0"
] | 2,610 | 2020-10-01T14:14:53.000Z | 2022-03-31T18:02:31.000Z | advanced_functionality/pytorch_bring_your_own_gan/build_gan_with_pytorch.ipynb | niklas-palm/amazon-sagemaker-examples | 84f451a193f2c2ee92ad3f91e36148547e948f16 | [
"Apache-2.0"
] | 1,959 | 2020-09-30T20:22:42.000Z | 2022-03-31T23:58:37.000Z | advanced_functionality/pytorch_bring_your_own_gan/build_gan_with_pytorch.ipynb | niklas-palm/amazon-sagemaker-examples | 84f451a193f2c2ee92ad3f91e36148547e948f16 | [
"Apache-2.0"
] | 2,052 | 2020-09-30T22:11:46.000Z | 2022-03-31T23:02:51.000Z | 39.172228 | 802 | 0.564892 | [
[
[
"# Build GAN (Generative Adversarial Networks) with PyTorch and SageMaker",
"_____no_output_____"
],
[
"### About GAN\n\nGenerative Adversarial Network (GAN) i is a generative machine learning model, which is widely used in advertising, games, entertainment, media, pharmaceuticals and other industries. It can be used to create fictional characters and scenes, simulate facial aging, and change image styles, and produce chemical formulas and so on.\n\nGAN was proposed by Ian Goodfellow in 2014, it is a deep neural network architecture consisting of a generative network and a discriminant network. The generation network generates \"fake\" data and tries to deceive the discrimination network; the discrimination network authenticates the generated data and tries to correctly identify all \"fake\" data. In the process of training iterations, the two networks continue to evolve and confront until they reach an equilibrium state (reference: Nash equilibrium), the discriminant network can no longer recognize \"fake\" data, and the training ends.\n\nThis example will lead you to build a GAN model leveraging the PyTorch framework, introducing GAN from the perspective of engineering practice, and opening a new and interesting AI/ML experience in generative models.",
"_____no_output_____"
],
[
"### Environment setup\nUpgrade packages",
"_____no_output_____"
]
],
[
[
"!pip install --upgrade pip sagemaker awscli boto3 numpy ipywidgets\n!pip install Pillow==7.1.2",
"_____no_output_____"
]
],
[
[
"Create folders",
"_____no_output_____"
]
],
[
[
"!mkdir -p data src tmp",
"_____no_output_____"
]
],
[
[
"### Download data\nThere are many public datasets on the Internet, which are very helpful for machine learning engineering and scientific research, such as algorithm study and evaluation. We will use MNIST dataset, which is a handwritten digits dataset, we will use it to train a GAN model, and eventually generate some fake \"handwritten\" digits.",
"_____no_output_____"
]
],
[
[
"!aws s3 cp --recursive s3://sagemaker-sample-files/datasets/image/MNIST/pytorch/ ./data",
"_____no_output_____"
]
],
[
[
"### Data preparation",
"_____no_output_____"
],
[
"PyTorch framework has a torchvision.datasets package, which provides access to a number of datasets, you may use the following commands to read MNIST pre-downloaded dataset from local storage, for later use.\n",
"_____no_output_____"
]
],
[
[
"from torchvision import datasets\n\ndataroot = './data'\ntrainset = datasets.MNIST(root=dataroot, train=True, download=False)\ntestset = datasets.MNIST(root=dataroot, train=False, download=False)\nprint(trainset)\nprint(testset)",
"_____no_output_____"
]
],
[
[
"SageMaker SDK will create a default Amazon S3 bucket for you to access various files and data, that you may need in the machine learning engineering lifecycle. We can get the name of this bucket through the default_bucket method of the sagemaker.session.Session class in the SageMaker SDK.",
"_____no_output_____"
]
],
[
[
"from sagemaker.session import Session\n\nsess = Session()\n\n# S3 bucket for saving code and model artifacts.\n# Feel free to specify a different bucket here if you wish.\nbucket = sess.default_bucket()\nprefix = 'byos-pytorch-gan'\n\n# Location to save your custom code in tar.gz format.\ns3_custom_code_upload_location = f's3://{bucket}/{prefix}/customcode'\n\n# Location where results of model training are saved.\ns3_model_artifacts_location = f's3://{bucket}/{prefix}/artifacts/'",
"_____no_output_____"
]
],
[
[
"The SageMaker SDK provides tools for operating AWS services. For example, the S3Downloader class is used to download objects in S3, and the S3Uploader is used to upload local files to S3. You will upload the dataset files to Amazon S3 for model training. During model training, we do not download data from the Internet to avoid network latency caused by fetching data from the Internet, and at the same time avoiding possible security risks due to direct access to the Internet.\n",
"_____no_output_____"
]
],
[
[
"import os\nfrom sagemaker.s3 import S3Uploader as s3up\n\ns3_data_location = s3up.upload(os.path.join(dataroot, \"MNIST\"), f\"s3://{bucket}/{prefix}/data/mnist\")",
"_____no_output_____"
]
],
[
[
"### Training\n\n\n",
"_____no_output_____"
],
[
"DCGAN (Deep Convolutional Generative Adversarial Networks) is a variant of the GAN families. This architecture essentially leverages Deep Convolutional Neural Networks to generate images belonging to a given distribution from noisy data using the Generator-Discriminator framework.",
"_____no_output_____"
]
],
[
[
"%%writefile src/train.py\n\nfrom __future__ import print_function\nimport argparse\nimport json\nimport logging\nimport os\nimport sys\nimport random\n\nimport torch\nimport torch.nn as nn\nimport torch.nn.parallel\nimport torch.nn.functional as F\n\nimport torch.optim as optim\nimport torch.backends.cudnn as cudnn\nimport torch.utils.data\nimport torchvision.datasets as dset\nimport torchvision.transforms as transforms\nimport torchvision.utils as vutils\n\n\ncudnn.benchmark = True\n\nlogger = logging.getLogger(__name__)\nlogger.setLevel(logging.DEBUG)\nlogger.addHandler(logging.StreamHandler(sys.stdout))\n\n\nclass Generator(nn.Module):\n def __init__(self, *, nz, nc, ngf, ngpu=1):\n super(Generator, self).__init__()\n self.ngpu = ngpu\n self.main = nn.Sequential(\n # input is Z, going into a convolution\n nn.ConvTranspose2d( nz, ngf * 8, 4, 1, 0, bias=False),\n nn.BatchNorm2d(ngf * 8),\n nn.ReLU(True),\n # state size. (ngf*8) x 4 x 4\n nn.ConvTranspose2d(ngf * 8, ngf * 4, 4, 2, 1, bias=False),\n nn.BatchNorm2d(ngf * 4),\n nn.ReLU(True),\n # state size. (ngf*4) x 8 x 8\n nn.ConvTranspose2d(ngf * 4, ngf * 2, 4, 2, 1, bias=False),\n nn.BatchNorm2d(ngf * 2),\n nn.ReLU(True),\n # state size. (ngf*2) x 16 x 16\n nn.ConvTranspose2d(ngf * 2, ngf, 4, 2, 1, bias=False),\n nn.BatchNorm2d(ngf),\n nn.ReLU(True),\n # state size. (ngf) x 32 x 32\n nn.ConvTranspose2d( ngf, nc, 4, 2, 1, bias=False),\n nn.Tanh()\n # state size. (nc) x 64 x 64\n )\n\n def forward(self, input):\n if input.is_cuda and self.ngpu > 1:\n output = nn.parallel.data_parallel(self.main, input, range(self.ngpu))\n else:\n output = self.main(input)\n return output\n\n def save(self, path, *, filename=None, device='cpu'):\n # recommended way from http://pytorch.org/docs/master/notes/serialization.html\n self.to(device)\n if not filename is None:\n path = os.path.join(path, filename)\n torch.save(self.state_dict(), path)\n\n def load(self, path, *, filename=None):\n if not filename is None:\n path = os.path.join(path, filename)\n with open(path, 'rb') as f:\n self.load_state_dict(torch.load(f))\n\n \nclass Discriminator(nn.Module):\n def __init__(self, *, nc, ndf, ngpu=1):\n super(Discriminator, self).__init__()\n self.ngpu = ngpu\n self.main = nn.Sequential(\n # input is (nc) x 64 x 64\n nn.Conv2d(nc, ndf, 4, 2, 1, bias=False),\n nn.LeakyReLU(0.2, inplace=True),\n # state size. (ndf) x 32 x 32\n nn.Conv2d(ndf, ndf * 2, 4, 2, 1, bias=False),\n nn.BatchNorm2d(ndf * 2),\n nn.LeakyReLU(0.2, inplace=True),\n # state size. (ndf*2) x 16 x 16\n nn.Conv2d(ndf * 2, ndf * 4, 4, 2, 1, bias=False),\n nn.BatchNorm2d(ndf * 4),\n nn.LeakyReLU(0.2, inplace=True),\n # state size. (ndf*4) x 8 x 8\n nn.Conv2d(ndf * 4, ndf * 8, 4, 2, 1, bias=False),\n nn.BatchNorm2d(ndf * 8),\n nn.LeakyReLU(0.2, inplace=True),\n # state size. (ndf*8) x 4 x 4\n nn.Conv2d(ndf * 8, 1, 4, 1, 0, bias=False),\n nn.Sigmoid()\n )\n\n def forward(self, input):\n if input.is_cuda and self.ngpu > 1:\n output = nn.parallel.data_parallel(self.main, input, range(self.ngpu))\n else:\n output = self.main(input)\n\n return output.view(-1, 1).squeeze(1)\n\n def save(self, path, *, filename=None, device='cpu'):\n # recommended way from http://pytorch.org/docs/master/notes/serialization.html\n self.to(device)\n if not filename is None:\n path = os.path.join(path, filename)\n torch.save(self.state_dict(), path)\n\n\n def load(self, path, *, filename=None):\n if not filename is None:\n path = os.path.join(path, filename)\n with open(path, 'rb') as f:\n self.load_state_dict(torch.load(f))\n \n \nclass DCGAN(object):\n \"\"\"\n A wrapper class for Generator and Discriminator,\n 'train_step' method is for single batch training.\n \"\"\"\n\n fixed_noise = None\n criterion = None\n device = None\n netG = None\n netD = None\n optimizerG = None\n optimizerD = None\n nz = None\n nc = None\n ngf = None\n ndf = None\n real_cpu = None\n \n def __init__(self, *, batch_size, nz, nc, ngf, ndf, device, weights_init,\n learning_rate, betas, real_label, fake_label):\n\n super(DCGAN, self).__init__()\n\n import torch\n \n self.nz = nz\n self.nc = nc\n self.ngf = ngf\n self.ndf = ndf\n \n self.real_label = real_label\n self.fake_label = fake_label\n \n self.fixed_noise = torch.randn(batch_size, nz, 1, 1, device=device)\n self.criterion = nn.BCELoss()\n self.device = device\n \n self.netG = Generator(nz=nz, nc=nc, ngf=ngf).to(device)\n # print(netG)\n self.netD = Discriminator(nc=nc, ndf=ndf).to(device)\n # print(netD)\n \n self.netG.apply(weights_init)\n self.netD.apply(weights_init)\n \n # setup optimizer\n self.optimizerG = optim.Adam(self.netG.parameters(), lr=learning_rate, betas=betas)\n self.optimizerD = optim.Adam(self.netD.parameters(), lr=learning_rate, betas=betas)\n\n\n def train_step(self, data, *, epoch, epochs):\n import torch\n\n ############################\n # (1) Update D network: maximize log(D(x)) + log(1 - D(G(z)))\n ###########################\n # train with real\n self.netD.zero_grad()\n self.real_cpu = data[0]\n real = data[0].to(self.device)\n batch_size = real.size(0)\n label = torch.full((batch_size,), self.real_label, device=self.device)\n \n output = self.netD(real).view(-1)\n errD_real = self.criterion(output, label)\n errD_real.backward()\n D_x = output.mean().item()\n\n\n # train with fake\n noise = torch.randn(batch_size, self.nz, 1, 1, device=self.device)\n fake = self.netG(noise)\n label.fill_(self.fake_label)\n output = self.netD(fake.detach()).view(-1)\n errD_fake = self.criterion(output, label)\n errD_fake.backward()\n D_G_z1 = output.mean().item()\n errD = errD_real + errD_fake\n self.optimizerD.step()\n \n\n ############################\n # (2) Update G network: maximize log(D(G(z)))\n ###########################\n self.netG.zero_grad()\n label.fill_(self.real_label) # fake labels are real for generator cost\n output = self.netD(fake).view(-1)\n errG = self.criterion(output, label)\n errG.backward()\n D_G_z2 = output.mean().item()\n self.optimizerG.step()\n\n\n return errG.item(), errD.item(), D_x, D_G_z1, D_G_z2\n \n\n \n# custom weights initialization called on netG and netD\ndef weights_init(m):\n classname = m.__class__.__name__\n if classname.find('Conv') != -1:\n torch.nn.init.normal_(m.weight, 0.0, 0.02)\n elif classname.find('BatchNorm') != -1:\n torch.nn.init.normal_(m.weight, 1.0, 0.02)\n torch.nn.init.zeros_(m.bias)\n\n\ndef log_batch(epoch, epochs, batch, batches, errD, errG, D_x, D_G_z1, D_G_z2, *, log_interval=10, output_dir):\n\n if batch % log_interval == 0:\n logger.info(f\"Epoch[{epoch}/{epochs}], Batch[{batch}/{batches}], \" +\n f\"Loss_D: {errD:.4}, Loss_G: {errG:.4}, D(x): {D_x:.4}, D(G(z)): {D_G_z1:.4}/{D_G_z2:.4}\")\n\n\n\ndef get_device(use_cuda):\n import torch\n\n device = \"cpu\"\n num_gpus = 0\n \n if torch.cuda.is_available():\n if use_cuda:\n device = \"cuda\"\n torch.cuda.set_device(0)\n num_gpus = torch.cuda.device_count()\n else:\n logger.debug(\"WARNING: You have a CUDA device, so you should probably run with --cuda 1\")\n\n logger.debug(f\"Number of gpus available: {num_gpus}\")\n \n return device, num_gpus\n\n\ndef train(dataloader, hps, test_batch_size,\n device, model_dir, output_dir, seed, log_interval):\n \n epochs = hps['epochs']\n batch_size = hps['batch-size']\n nz = hps['nz']\n ngf = hps['ngf']\n ndf = hps['ndf']\n learning_rate = hps['learning-rate']\n beta1 = hps['beta1']\n \n dcgan = DCGAN(batch_size=batch_size, nz=nz, nc=1, ngf=ngf, ndf=ndf,\n device=device, weights_init=weights_init, learning_rate=learning_rate,\n betas=(beta1, 0.999), real_label=1, fake_label=0)\n\n for epoch in range(epochs):\n batches = len(dataloader)\n for batch, data in enumerate(dataloader, 0):\n errG, errD, D_x, D_G_z1, D_G_z2 = dcgan.train_step(data,\n epoch=epoch, epochs=epochs)\n \n log_batch(epoch, epochs, batch, batches, errD, errG,\n D_x, D_G_z1, D_G_z2, log_interval=log_interval, output_dir=output_dir)\n\n \n save_model(model_dir, dcgan.netG)\n\n return\n\n\ndef save_model(model_dir, model):\n logger.info(\"Saving the model.\")\n model.save(model_dir, filename=\"model.pth\")\n\n \ndef load_model(model_dir, device=None):\n logger.info(\"Loading the model.\")\n if device is None:\n device = get_training_device_name(1)\n\n netG.load(model_dir, filename=\"model.pth\", device=device)\n\n return netG\n\n\ndef parse_args():\n # Training settings\n parser = argparse.ArgumentParser(description='PyTorch Example')\n \n parser.add_argument('--batch-size', type=int, default=1000, metavar='N',\n help='input batch size (default: 1000)')\n parser.add_argument('--test-batch-size', type=int, default=1000, metavar='N',\n help='input batch size for testing (default: 1000)')\n parser.add_argument('--seed', type=int, default=None, metavar='S',\n help='random seed')\n parser.add_argument('--log-interval', type=int, default=10, metavar='N',\n help='how many batches to wait before logging training status')\n\n parser.add_argument('--save-model', action='store_true', default=False,\n help='For Saving the current Model')\n\n parser.add_argument('--model-dir', type=str, default=os.environ.get('SM_MODEL_DIR', None))\n \n parser.add_argument('--cuda', type=int, default=1)\n parser.add_argument('--num-gpus', type=int, default=os.environ.get('SM_NUM_GPUS', None))\n \n parser.add_argument('--pin-memory', type=bool, default=os.environ.get('SM_PIN_MEMORY', False))\n\n parser.add_argument('--data-dir', required=False, default=None, help='path to data dir')\n parser.add_argument('--workers', type=int, help='number of data loading workers', default=2)\n parser.add_argument('--output-dir', default=os.environ.get('SM_OUTPUT_DATA_DIR', None), help='folder to output images and model checkpoints')\n parser.add_argument('--hps', default=os.environ.get('SM_HPS', None), help='Hyperparameters')\n \n return parser.parse_known_args()\n\n\ndef get_datasets(*, dataroot='/opt/ml/input/data', classes=None):\n\n dataset = dset.MNIST(root=dataroot,\n transform=transforms.Compose([\n transforms.Resize(64),\n transforms.ToTensor(),\n transforms.Normalize((0.5,), (0.5,)),\n ]))\n\n return dataset\n \n\nif __name__ == '__main__':\n args, unknown = parse_args()\n \n # get training options\n hps = json.loads(args.hps)\n\n try:\n os.makedirs(args.output_dir)\n except OSError:\n pass\n\n if args.seed is None:\n random_seed = random.randint(1, 10000)\n logger.debug(f\"Generated Random Seed: {random_seed}\")\n cudnn.benchmark = True\n else:\n logger.debug(f\"Provided Random Seed: {args.seed}\")\n random_seed = args.seed\n cudnn.deterministic = True\n cudnn.benchmark = False\n \n random.seed(random_seed)\n torch.manual_seed(random_seed)\n\n pin_memory=args.pin_memory\n num_workers = int(args.workers)\n \n device, num_gpus = get_device(args.cuda)\n \n if device == 'cuda':\n num_workers = 1\n pin_memory = True\n\n \n if args.data_dir is None:\n input_dir = os.environ.get('SM_INPUT_DIR', None)\n if input_dir is None and str(args.dataset).lower() != 'fake':\n raise ValueError(f\"`--data-dir` parameter is required for dataset \\\"{args.dataset}\\\"\")\n\n dataroot = input_dir + \"/data\"\n else:\n dataroot = args.data_dir\n\n dataset = get_datasets(dataroot=dataroot)\n\n assert dataset\n dataloader = torch.utils.data.DataLoader(dataset, batch_size=args.batch_size,\n shuffle=True, num_workers=num_workers, pin_memory=pin_memory)\n\n \n train(dataloader, hps, args.test_batch_size, device, args.model_dir, args.output_dir, args.seed, args.log_interval)",
"_____no_output_____"
]
],
[
[
"Per sagemaker.get_execution_role() method, the notebook can get the role pre-assigned to the notebook instance. This role will be used to obtain training resources, such as downloading training framework images, allocating Amazon EC2 instances, and so on.",
"_____no_output_____"
]
],
[
[
"from sagemaker import get_execution_role\n\n# IAM execution role that gives SageMaker access to resources in your AWS account.\n# We can use the SageMaker Python SDK to get the role from our notebook environment. \nrole = get_execution_role()",
"_____no_output_____"
]
],
[
[
"The hyperparameters, that used in the model training tasks, can be defined in the notebook so that it is separated from the algorithm and training code. The hyperparameters are passed in when the training task is created and dynamically combined with the training task.",
"_____no_output_____"
]
],
[
[
"import json\n\nhps = {\n 'seed': 0,\n 'learning-rate': 0.0002,\n 'epochs': 18,\n 'pin-memory': 1,\n 'beta1': 0.5,\n 'nz': 100,\n 'ngf': 28,\n 'ndf': 28,\n 'batch-size': 128,\n 'log-interval': 20,\n }\n\n\nstr_hps = json.dumps(hps, indent = 4)\nprint(str_hps)",
"_____no_output_____"
]
],
[
[
"```PyTorch``` class from sagemaker.pytorch package, is an estimator for PyTorch framework, it can be used to create and execute training tasks, as well as to deploy trained models. In the parameter list, ``instance_type`` is used to specify the instance type, such as CPU or GPU instances. The directory containing training script and the model code are specified by ``source_dir``, and the training script file name must be clearly defined by ``entry_point``. These parameters will be passed to the training task along with other parameters, and they determine the environment settings of the training task.",
"_____no_output_____"
]
],
[
[
"from sagemaker.pytorch import PyTorch\n\nestimator = PyTorch(role=role,\n entry_point='train.py',\n source_dir='./src',\n output_path=s3_model_artifacts_location,\n code_location=s3_custom_code_upload_location,\n instance_count=1,\n instance_type='ml.g4dn.2xlarge',\n framework_version='1.5.0',\n py_version='py3',\n hyperparameters=hps,\n )",
"_____no_output_____"
]
],
[
[
"Please pay special attention to the ``train_use_spot_instances`` parameter. The value of ``True`` means that you want to use SPOT instances first. Since machine learning training usually requires a large amount of computing resources to run for a long time, leveraging SPOT instances can help you control your cost. The SPOT instances may save cost up to 90% of the on-demand instances, depending on the instance type, region, and time, the actual price might be different.",
"_____no_output_____"
],
[
"You have created a PyTorch object, and you can use it to fit pre-uploaded data on Amazon S3. The following command will initiate the training task, and the training data will be imported into the training environment in the form of an input channel named **MNIST**. When the training task starts, the training data was already downloaded from S3 to the local file system of the training instance, and the training script ```train.py``` will load the data from the local disk afterwards.",
"_____no_output_____"
]
],
[
[
"# Start training\nestimator.fit({\"MNIST\": s3_data_location}, wait=False)",
"_____no_output_____"
]
],
[
[
"Depending on the training instance you choose, the training process may last from tens of minutes to several hours. It is recommended to set the ``wait`` parameter to ``False``, this option will detach the notebook from the training task. In scenarios with long training time and many training logs, it can prevent the notebook context from being lost due to network interruption or session timeout. After the notebook detached from the training task, the output will be temporarily invisible. You can execute the following code, and the notebook will obtain and resume the previous training session.",
"_____no_output_____"
]
],
[
[
"%%time\nfrom sagemaker.estimator import Estimator\n\n# Attaching previous training session\ntraining_job_name = estimator.latest_training_job.name\nattached_estimator = Estimator.attach(training_job_name)",
"_____no_output_____"
]
],
[
[
"Since the model was designed to leverage the GPU power to accelerate training, it will be much faster than training tasks on CPU instances. For example, the p3.2xlarge instance will take about 15 minutes, while the c5.xlarge instance may take more than 6 hours. The current model does not support distributed and parallel training, so multi-instance and multi-CPU/GPU will not bring extra benefits in training speed boosting.\n\nWhen the training completes, the trained model will be uploaded to S3. The upload location is specified by the `output_path` parameter provided when creating the `PyTorch` object.",
"_____no_output_____"
],
[
"### Model verification",
"_____no_output_____"
],
[
"You will download the trained model from Amazon S3 to the local file system of the instance where the notebook is located. The following code will load the model, and then generate a picture with a random number as input, then display picture.\n",
"_____no_output_____"
]
],
[
[
"from sagemaker.s3 import S3Downloader as s3down\n\n!mkdir -p ./tmp\n\nmodel_url = attached_estimator.model_data\ns3down.download(model_url, './tmp')",
"_____no_output_____"
],
[
"!tar -zxf tmp/model.tar.gz -C ./tmp",
"_____no_output_____"
]
],
[
[
"Execute the following instructions to load the trained model, and generate a set of \"handwritten\" digitals.",
"_____no_output_____"
]
],
[
[
"def generate_fake_handwriting(model, *, num_images, nz, device=None):\n\n import torch\n import torchvision.utils as vutils\n from io import BytesIO\n from PIL import Image\n \n\n z = torch.randn(num_images, nz, 1, 1, device=device)\n fake = model(z)\n\n imgio = BytesIO()\n vutils.save_image(fake.detach(), imgio, normalize=True, format=\"PNG\")\n img = Image.open(imgio)\n \n return img\n\n\ndef load_model(path, *, model_cls=None, params=None, filename=None, device=None, strict=True):\n\n import os\n import torch\n \n model_pt_path = path\n if not filename is None:\n model_pt_path = os.path.join(path, filename)\n \n if device is None:\n device = 'cpu'\n \n if not model_cls is None:\n model = model_cls(**params)\n model.load_state_dict(torch.load(model_pt_path, map_location=torch.device(device)), strict=strict)\n else:\n model = torch.jit.load(model_pt_path, map_location=torch.device(device))\n\n model.to(device)\n \n return model\n",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport torch\nfrom src.train import Generator\n\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\n\nparams = {'nz': hps['nz'], 'nc': 1, 'ngf': hps['ngf']}\nmodel = load_model(\"./tmp/model.pth\", model_cls=Generator, params=params, device=device, strict=False)\nimg = generate_fake_handwriting(model, num_images=64, nz=hps['nz'], device=device)\n\nplt.imshow(np.asarray(img))",
"_____no_output_____"
]
],
[
[
"### Clean up\n\nRun the following commandline in a terminal, to remove files generated by this notebook from S3 and local storage\n",
"_____no_output_____"
]
],
[
[
"import os\n\nprint(f\"aws s3 rm --recursive s3://{bucket}/{prefix}\")\nprint(f\"rm -rf {os.path.abspath(dataroot)}\")\n",
"_____no_output_____"
]
],
[
[
"### Conclusion",
"_____no_output_____"
],
[
"The PyTorch framework, as one of the most popular deep learning framework, is being widely recognised and applied, has become one of the de facto mainstream frameworks.\n\nAmazon SageMaker is tightly integrated with a variety of AWS services, such as Amazon EC2 instances of various types and sizes, Amazon S3, Amazon ECR, etc., providing an end-to-end, consistent machine learning experience for all framework practitioners. Amazon SageMaker continues to support mainstream machine learning frameworks, including PyTorch. Machine learning algorithms and models developed with PyTorch can be easily transplanted to Amazon SageMaker environment, by using Amazon SageMaker's fully managed Jupyter Notebook, SPOT training instances, Amazon Elastic Container Registry, SageMaker SDK, and so on, the complexity of machine learning engineering and infrastracture operation are simplified, productivity and efficiency are improved, operation and maintenance costs reduced.\n\nDCGAN is a landmark in the field of generative confrontation networks, and it is the cornerstone of many complex generative confrontation networks today. We will explore some of the most recent and interesting variants of GAN in later exmaples.\n\nI believe that through the introduction and engineering practice of this example, it will be helpful for you to understand the principles and engineering methods for GAN in general.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb4611c3550dca621dc6978ddc035cb38c051c09 | 204,089 | ipynb | Jupyter Notebook | example3/CFD generation Vortex Patch.ipynb | mleprovost/LowRankVortex.jl | 8eedffb54a94ec0376c9e3f05f42e56b7d775a11 | [
"MIT"
] | 5 | 2022-02-11T19:20:16.000Z | 2022-03-14T05:18:36.000Z | example3/CFD generation Vortex Patch.ipynb | mleprovost/LowRankVortex.jl | 8eedffb54a94ec0376c9e3f05f42e56b7d775a11 | [
"MIT"
] | null | null | null | example3/CFD generation Vortex Patch.ipynb | mleprovost/LowRankVortex.jl | 8eedffb54a94ec0376c9e3f05f42e56b7d775a11 | [
"MIT"
] | null | null | null | 86.441762 | 10,151 | 0.72831 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
cb4613f124e027c54057e6127b679bcb9da41416 | 898 | ipynb | Jupyter Notebook | rocksetta-examples/z00-template-notebook.ipynb | shankar96/TensorFlow | bead51575338b0e15ebec7da54234a4ba6b4a58c | [
"MIT"
] | 58 | 2016-01-31T19:32:09.000Z | 2021-11-16T19:33:41.000Z | rocksetta-examples/z00-template-notebook.ipynb | shankar96/TensorFlow | bead51575338b0e15ebec7da54234a4ba6b4a58c | [
"MIT"
] | 4 | 2016-01-23T13:27:09.000Z | 2018-12-18T20:30:11.000Z | rocksetta-examples/z00-template-notebook.ipynb | shankar96/TensorFlow | bead51575338b0e15ebec7da54234a4ba6b4a58c | [
"MIT"
] | 34 | 2016-01-04T04:02:10.000Z | 2021-11-16T19:32:36.000Z | 17.96 | 84 | 0.534521 | [
[
[
"#Enter a bash command below and click run to activate\n!",
"_____no_output_____"
],
[
"#Enter a python file to load and edit. Changes will be automatically saved!\n%load ",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
cb462b61971e6d42678f8017f73384d8ca1974fd | 4,608 | ipynb | Jupyter Notebook | BBC-news-data/STAGE 3 - Abstractive Summary (Encoder-Decoder).ipynb | navyamehta/economic-data-NLP | 66ad4849561585ea008c2676a5cbfd1455e72d7a | [
"MIT"
] | null | null | null | BBC-news-data/STAGE 3 - Abstractive Summary (Encoder-Decoder).ipynb | navyamehta/economic-data-NLP | 66ad4849561585ea008c2676a5cbfd1455e72d7a | [
"MIT"
] | 6 | 2020-11-13T18:54:54.000Z | 2022-02-10T02:28:29.000Z | BBC-news-data/STAGE 3 - Abstractive Summary (Encoder-Decoder).ipynb | navyamehta/data-sentiment-NLP | 66ad4849561585ea008c2676a5cbfd1455e72d7a | [
"MIT"
] | null | null | null | 31.346939 | 300 | 0.614149 | [
[
[
"We attempt to build an encoder-decoder system for abstractive text summarization with a Bahdanau attention layer. 100-D GloVe Embeddings are used to initialize the encoder-decoder design, with LSTM and CNN architectures tested for the intermediary layers. The overarching idea of the design is:",
"_____no_output_____"
],
[
"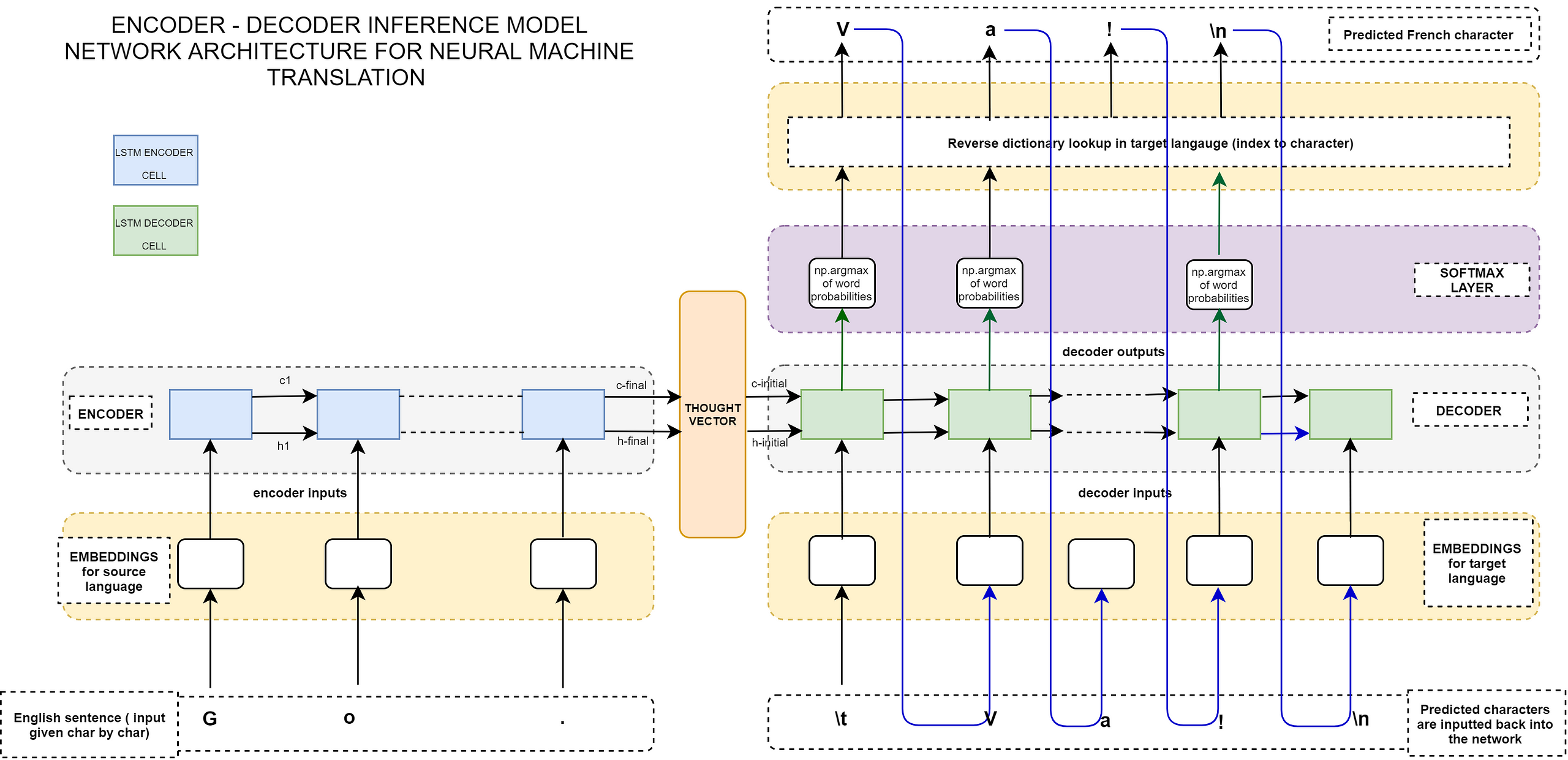",
"_____no_output_____"
]
],
[
[
"from attention import AttentionLayer\nimport pandas as pd\nimport numpy as np\nimport scipy as sp\nimport re\nfrom bs4 import BeautifulSoup\nimport tensorflow as tf\nfrom tensorflow.keras.layers import LSTM, TimeDistributed, Dense, Bidirectional, Input, Embedding, Dropout\nfrom tensorflow.keras.models import Model\nimport os\nimport collections\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport nltk.stem\nfrom nltk.corpus import stopwords",
"_____no_output_____"
],
[
"lstmdim = 500\ntf.keras.backend.clear_session()\n#Encoding Segment\ntextinput = Input(shape=(MAX_TEXT_LEN,))\ntextembed = Embedding(len(texttoken.word_index)+1, lstmdim, trainable=True, mask_zero=True)(textinput)\nencout1, _, _, _, _ = Bidirectional(LSTM(lstmdim, return_sequences=True, return_state=True))(textembed)\nencout1 = Dropout(0.1)(encout1)\n_, enc_h, enc_c = LSTM(lstmdim, return_sequences=True, return_state=True)(encout1)\n#Decoding Segment\nsumminput = Input(shape=(None,))\ndecembed_layer = Embedding(len(summtoken.word_index)+1, lstmdim, trainable=True)\nsummembed = decembed_layer(summinput)\ndeclstm_layer = LSTM(lstmdim, return_sequences=True, return_state=True)\ndecout, _, _ = declstm_layer(summembed, initial_state=[enc_h, enc_c])\ndecdense_layer = Dense(len(summtoken.word_index)+1, activation=\"softmax\")\npreds = decdense_layer(decout)\nmdl = Model(inputs=[textinput, summinput], outputs=preds)\nmdl.compile(optimizer=\"adam\", loss=\"sparse_categorical_crossentropy\")\nmdl.summary()",
"_____no_output_____"
],
[
"check = tf.keras.callbacks.ModelCheckpoint(\"newsbbcseq2seq.h5\", save_best_only=True, monitor=\"val_loss\", verbose=True)\nhist = mdl.fit([trainX, trainY[:,:-1]], trainY.reshape(trainY.shape[0], trainY.shape[1], 1)[:,1:], epochs=10, callbacks=[check],\n batch_size=16, verbose=True,\n validation_data=([testX, testY[:,:-1]], testY.reshape(testY.shape[0], testY.shape[1], 1)[:,1:]))",
"_____no_output_____"
],
[
"#Run-Time Model Graphs\nencode_model = Model(inputs=textinput, outputs=[enc_h, enc_c])\ndec_h = Input(shape=(lstmdim,))\ndec_c = Input(shape=(lstmdim,))\ndecinput = Input(shape=(None,))\ndecembed = decembed_layer(decinput)\noutput, new_h, new_c = declstm_layer(decembed, initial_state=[dec_h, dec_c])\noutput = decdense_layer(output)\ndecode_model = Model(inputs=[decinput, dec_h, dec_c], outputs=[output, new_h, new_c])",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cb462c69e151baaf077fcafe1ba3eef84d01b049 | 107,090 | ipynb | Jupyter Notebook | quantization/linear_uniform_characteristic.ipynb | mgrubisic/digital-signal-processing-lecture | 7098b958639eb5cfcabd110d26ddd30ff8444e0a | [
"MIT"
] | 2 | 2018-12-29T19:13:49.000Z | 2020-05-25T09:53:21.000Z | quantization/linear_uniform_characteristic.ipynb | mgrubisic/digital-signal-processing-lecture | 7098b958639eb5cfcabd110d26ddd30ff8444e0a | [
"MIT"
] | null | null | null | quantization/linear_uniform_characteristic.ipynb | mgrubisic/digital-signal-processing-lecture | 7098b958639eb5cfcabd110d26ddd30ff8444e0a | [
"MIT"
] | 3 | 2020-10-17T07:48:22.000Z | 2022-03-17T06:28:58.000Z | 459.613734 | 49,414 | 0.926221 | [
[
[
"# Quantization of Signals\n\n*This jupyter notebook is part of a [collection of notebooks](../index.ipynb) on various topics of Digital Signal Processing. Please direct questions and suggestions to [[email protected]](mailto:[email protected]).*",
"_____no_output_____"
],
[
"## Characteristic of a Linear Uniform Quantizer\n\nThe characteristics of a quantizer depend on the mapping functions $f(\\cdot)$, $g(\\cdot)$ and the rounding operation $\\lfloor \\cdot \\rfloor$ introduced in the [previous section](introduction.ipynb). A linear quantizer bases on linear mapping functions $f(\\cdot)$ and $g(\\cdot)$. A uniform quantizer splits the mapped input signal into quantization steps of equal size. Quantizers can be described by their nonlinear in-/output characteristic $x_Q[k] = \\mathcal{Q} \\{ x[k] \\}$, where $\\mathcal{Q} \\{ \\cdot \\}$ denotes the quantization process. For linear uniform quantization it is common to differentiate between two characteristic curves, the so called mid-tread and mid-rise. Both are introduced in the following.",
"_____no_output_____"
],
[
"### Mid-Tread Characteristic Curve\n\nThe in-/output relation of the mid-tread quantizer is given as\n\n\\begin{equation}\nx_Q[k] = Q \\cdot \\underbrace{\\left\\lfloor \\frac{x[k]}{Q} + \\frac{1}{2} \\right\\rfloor}_{index}\n\\end{equation}\n\nwhere $Q$ denotes the constant quantization step size and $\\lfloor \\cdot \\rfloor$ the [floor function](https://en.wikipedia.org/wiki/Floor_and_ceiling_functions) which maps a real number to the largest integer not greater than its argument. Without restricting $x[k]$ in amplitude, the resulting quantization indexes are [countable infinite](https://en.wikipedia.org/wiki/Countable_set). For a finite number of quantization indexes, the input signal has to be restricted to a minimal/maximal amplitude $x_\\text{min} < x[k] < x_\\text{max}$ before quantization. The resulting quantization characteristic of a linear uniform mid-tread quantizer is shown below\n\n\n\nThe term mid-tread is due to the fact that small values $|x[k]| < \\frac{Q}{2}$ are mapped to zero.",
"_____no_output_____"
],
[
"#### Example - Mid-tread quantization of a sine signal\n\nThe quantization of one period of a sine signal $x[k] = A \\cdot \\sin[\\Omega_0\\,k]$ by a mid-tread quantizer is simulated. $A$ denotes the amplitude of the signal, $x_\\text{min} = -1$ and $x_\\text{max} = 1$ are the smallest and largest output values of the quantizer, respectively.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nA = 1.2 # amplitude of signal\nQ = 1/10 # quantization stepsize\nN = 2000 # number of samples\n\ndef uniform_midtread_quantizer(x, Q):\n # limiter\n x = np.copy(x)\n idx = np.where(np.abs(x) >= 1)\n x[idx] = np.sign(x[idx])\n # linear uniform quantization\n xQ = Q * np.floor(x/Q + 1/2)\n \n return xQ\n\ndef plot_signals(x, xQ):\n e = xQ - x\n plt.figure(figsize=(10,6))\n plt.plot(x, label=r'signal $x[k]$')\n plt.plot(xQ, label=r'quantized signal $x_Q[k]$')\n plt.plot(e, label=r'quantization error $e[k]$')\n plt.xlabel(r'$k$')\n plt.axis([0, N, -1.1*A, 1.1*A])\n plt.legend()\n plt.grid()\n\n# generate signal\nx = A * np.sin(2*np.pi/N * np.arange(N))\n# quantize signal\nxQ = uniform_midtread_quantizer(x, Q)\n# plot signals\nplot_signals(x, xQ)",
"_____no_output_____"
]
],
[
[
"**Exercise**\n\n* Change the quantization stepsize `Q` and the amplitude `A` of the signal. Which effect does this have on the quantization error?\n\nSolution: The smaller the quantization step size, the smaller the quantization error is for $|x[k]| < 1$. Note, the quantization error is not bounded for $|x[k]| > 1$ due to the clipping of the signal $x[k]$.",
"_____no_output_____"
],
[
"### Mid-Rise Characteristic Curve\n\nThe in-/output relation of the mid-rise quantizer is given as\n\n\\begin{equation}\nx_Q[k] = Q \\cdot \\Big( \\underbrace{\\left\\lfloor\\frac{ x[k] }{Q}\\right\\rfloor}_{index} + \\frac{1}{2} \\Big)\n\\end{equation}\n\nwhere $\\lfloor \\cdot \\rfloor$ denotes the floor function. The quantization characteristic of a linear uniform mid-rise quantizer is illustrated below\n\n\n\nThe term mid-rise copes for the fact that $x[k] = 0$ is not mapped to zero. Small positive/negative values around zero are mapped to $\\pm \\frac{Q}{2}$.",
"_____no_output_____"
],
[
"#### Example - Mid-rise quantization of a sine signal\n\nThe previous example is now reevaluated using the mid-rise characteristic",
"_____no_output_____"
]
],
[
[
"A = 1.2 # amplitude of signal\nQ = 1/10 # quantization stepsize\nN = 2000 # number of samples\n\ndef uniform_midrise_quantizer(x, Q):\n # limiter\n x = np.copy(x)\n idx = np.where(np.abs(x) >= 1)\n x[idx] = np.sign(x[idx])\n # linear uniform quantization\n xQ = Q * (np.floor(x/Q) + .5)\n \n return xQ\n\n# generate signal\nx = A * np.sin(2*np.pi/N * np.arange(N))\n# quantize signal\nxQ = uniform_midrise_quantizer(x, Q)\n# plot signals\nplot_signals(x, xQ)",
"_____no_output_____"
]
],
[
[
"**Exercise**\n\n* What are the differences between the mid-tread and the mid-rise characteristic curves for the given example?\n\nSolution: The mid-tread and the mid-rise quantization of the sine signal differ for signal values smaller than half of the quantization interval. Mid-tread has a representation of $x[k] = 0$ while this is not the case for the mid-rise quantization.",
"_____no_output_____"
],
[
"**Copyright**\n\nThis notebook is provided as [Open Educational Resource](https://en.wikipedia.org/wiki/Open_educational_resources). Feel free to use the notebook for your own purposes. The text is licensed under [Creative Commons Attribution 4.0](https://creativecommons.org/licenses/by/4.0/), the code of the IPython examples under the [MIT license](https://opensource.org/licenses/MIT). Please attribute the work as follows: *Sascha Spors, Digital Signal Processing - Lecture notes featuring computational examples, 2016-2018*.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cb4638d9d26691685c71a7c23c65ae734e9aa932 | 8,983 | ipynb | Jupyter Notebook | 5-4Tensorboard可视化.ipynb | LZY2006/Tensorflow-1.x-jupyter-notebooks | 4d658251732b3826ca47b730a56bbf6f6952930b | [
"MIT"
] | 2 | 2021-01-23T00:50:29.000Z | 2021-04-28T02:20:31.000Z | 5-4Tensorboard可视化.ipynb | LZY2006/Tensorflow-1.x-jupyter-notebooks | 4d658251732b3826ca47b730a56bbf6f6952930b | [
"MIT"
] | null | null | null | 5-4Tensorboard可视化.ipynb | LZY2006/Tensorflow-1.x-jupyter-notebooks | 4d658251732b3826ca47b730a56bbf6f6952930b | [
"MIT"
] | 1 | 2021-10-31T10:39:10.000Z | 2021-10-31T10:39:10.000Z | 36.967078 | 259 | 0.555048 | [
[
[
"import tensorflow as tf\nfrom tensorflow.examples.tutorials.mnist import input_data\nfrom tensorflow.contrib.tensorboard.plugins import projector",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\h5py\\__init__.py:34: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\n"
],
[
"# 载入数据集\nmnist = input_data.read_data_sets(r\"C:\\Users\\zdwxx\\Downloads\\Compressed\\MNIST_data\", one_hot=True)\n\n# 运行次数\nmax_steps = 550 * 21\n\n# 图片数量\nimage_num = 3000\n\n# 定义会话\nsess = tf.Session()\n\n# 文件路径\nDIR = \"C:/Tensorflow/\"\n\n# 载入图片\nembedding = tf.Variable(tf.stack(mnist.test.images[:image_num]), \n trainable=False, name=\"embedding\")\n\n# 定义一个参数概要\ndef varible_summaries(var):\n \n with tf.name_scope(\"summary\"):\n \n mean = tf.reduce_mean(var)\n tf.summary.scalar(\"mean\", mean) # 平均值\n with tf.name_scope(\"stddev\"):\n stddev = tf.sqrt(tf.reduce_mean(tf.square(var - mean)))\n tf.summary.scalar(\"stddev\", stddev) # 标准差\n tf.summary.scalar(\"max\", tf.reduce_max(var)) #最大值\n tf.summary.scalar(\"min\", tf.reduce_min(var)) # 最小值\n tf.summary.histogram(\"histogram\", var) # 直方图\n\n# 命名空间\nwith tf.name_scope(\"input\"):\n # 定义两个placeholder\n x = tf.placeholder(tf.float32, [None, 784], name=\"x-input\")\n y = tf.placeholder(tf.float32, [None, 10], name=\"y-input\") \n\n# 显示图片\nwith tf.name_scope(\"input_reshape\"):\n \n image_shaped_input = tf.reshape(x, [-1, 28, 28, 1])\n tf.summary.image(\"input\", image_shaped_input, 10)\n \n\nwith tf.name_scope(\"layer\"):\n #创建一个简单的神经网络\n with tf.name_scope(\"wights1\"):\n W1 = tf.Variable(tf.truncated_normal([784, 500], stddev=0.1), name=\"W1\")\n varible_summaries(W1)\n \n with tf.name_scope(\"biases1\"):\n b1 = tf.Variable(tf.zeros([500]) + 0.1, name=\"b1\")\n varible_summaries(b1)\n \n# with tf.name_scope(\"wx_plus_b1\"):\n# wx_plus_b1 = tf.matmul(x, W1) + b1\n \n with tf.name_scope(\"L1\"):\n L1 = tf.nn.tanh(tf.matmul(x, W1) + b1)\n \n with tf.name_scope(\"wights2\"):\n W2 = tf.Variable(tf.truncated_normal([500, 10], stddev=0.1), name=\"W2\")\n varible_summaries(W2)\n \n with tf.name_scope(\"biases2\"):\n b2 = tf.Variable(tf.zeros([10]) + 0.1, name=\"b2\")\n varible_summaries(b2)\n \n with tf.name_scope(\"wx_plus_b2\"):\n wx_plus_b2 = tf.matmul(L1, W2) + b2\n \n with tf.name_scope(\"softmax\"):\n prediction = tf.nn.softmax(wx_plus_b2) # 预测值\n\n# 二次代价函数\n\n# loss = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(labels=y, logits=prediction))\nwith tf.name_scope(\"loss\"):\n loss = loss = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(labels=y, logits=prediction))\n tf.summary.scalar(\"loss\", loss)\n \n# 梯度下降法\nwith tf.name_scope(\"train\"):\n train_step = tf.train.GradientDescentOptimizer(0.2).minimize(loss)\n\n# 初始化变量\ninit = tf.global_variables_initializer()\nsess.run(init)\n\nwith tf.name_scope(\"accuracy\"):\n # 结果存放在一个布尔型列表中\n with tf.name_scope(\"correct_prediction\"):\n correct_prediction = tf.equal(tf.argmax(y, 1), tf.argmax(prediction, 1)) #argmax返回1维张量中最大的值所在的位置\n # 求准确率\n with tf.name_scope(\"accuracy\"):\n accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))#cast转换类型,True->1.0, False->0.0\n tf.summary.scalar(\"accuracy\", accuracy)\n\n# 产生 metadata文件 \nif tf.gfile.Exists(DIR + \"projector/projector/metadata.tsv\"):\n tf.gfile.DeleteRecursively(DIR + \"projector/projector/metadata.tsv\")\n\nwith open(DIR + \"projector/projector/metadata.tsv\", \"w\") as f:\n lables = sess.run(tf.argmax(mnist.test.labels[:], 1))\n for i in range(image_num):\n f.write(str(lables[i]) + \"\\n\")\n\n# 合并所有的summary\nmerged = tf.summary.merge_all()\n\n\nprojector_writer = tf.summary.FileWriter(DIR + \"projector/projector\", sess.graph)\nsaver = tf.train.Saver()\nconfig = projector.ProjectorConfig()\nembed = config.embeddings.add()\nembed.tensor_name = embedding.name\nembed.metadata_path = DIR + \"projector/projector/metadata.tsv\"\nembed.sprite.image_path = DIR + \"projector/data/mnist_10k_sprite.png\"\nembed.sprite.single_image_dim.extend([28, 28])\nprojector.visualize_embeddings(projector_writer, config)\n \n\nfor i in range(max_steps):\n\n batch_xs, batch_ys = mnist.train.next_batch(100) #类似于read,一次读取100张图片\n run_options = tf.RunOptions(trace_level=tf.RunOptions.FULL_TRACE)\n run_metadata = tf.RunMetadata()\n summary = sess.run([train_step, merged], feed_dict={x : batch_xs, y : batch_ys})[1]\n projector_writer.add_run_metadata(run_metadata, \"step%03d\" % i)\n projector_writer.add_summary(summary, i)\n \n if i % 550 == 0:\n acc = sess.run(accuracy, feed_dict={x:mnist.test.images, y:mnist.test.labels})\n print(\"第\", i, \"个周期\", \"准确率是\", acc)\n\nsaver.save(sess, DIR + \"projector/projector/a_model.ckpt\")\nprojector_writer.close()\nsess.close()",
"Extracting C:\\Users\\zdwxx\\Downloads\\Compressed\\MNIST_data\\train-images-idx3-ubyte.gz\nExtracting C:\\Users\\zdwxx\\Downloads\\Compressed\\MNIST_data\\train-labels-idx1-ubyte.gz\nExtracting C:\\Users\\zdwxx\\Downloads\\Compressed\\MNIST_data\\t10k-images-idx3-ubyte.gz\nExtracting C:\\Users\\zdwxx\\Downloads\\Compressed\\MNIST_data\\t10k-labels-idx1-ubyte.gz\nWARNING:tensorflow:From <ipython-input-2-bbab3714fffc>:81: softmax_cross_entropy_with_logits (from tensorflow.python.ops.nn_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\n\nFuture major versions of TensorFlow will allow gradients to flow\ninto the labels input on backprop by default.\n\nSee tf.nn.softmax_cross_entropy_with_logits_v2.\n\n第 0 个周期 准确率是 0.0913\n第 550 个周期 准确率是 0.8161\n第 1100 个周期 准确率是 0.8317\n第 1650 个周期 准确率是 0.8383\n第 2200 个周期 准确率是 0.9278\n第 2750 个周期 准确率是 0.9331\n第 3300 个周期 准确率是 0.9383\n第 3850 个周期 准确率是 0.941\n第 4400 个周期 准确率是 0.9433\n第 4950 个周期 准确率是 0.9455\n第 5500 个周期 准确率是 0.9466\n第 6050 个周期 准确率是 0.9485\n第 6600 个周期 准确率是 0.9499\n第 7150 个周期 准确率是 0.9506\n第 7700 个周期 准确率是 0.953\n第 8250 个周期 准确率是 0.9539\n第 8800 个周期 准确率是 0.9557\n第 9350 个周期 准确率是 0.9562\n第 9900 个周期 准确率是 0.9569\n第 10450 个周期 准确率是 0.9575\n第 11000 个周期 准确率是 0.9589\n"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
cb463aced5f75b356684b1b189270504a8516f86 | 116,435 | ipynb | Jupyter Notebook | legacy/unidad_5/03_C.ipynb | LVFerrero/diseno_sci_sfw | d782c0b6ccd5dbfd7c59fd2ac408207470e22301 | [
"BSD-3-Clause"
] | 1 | 2020-10-15T12:48:48.000Z | 2020-10-15T12:48:48.000Z | legacy/unidad_5/03_C.ipynb | LVFerrero/diseno_sci_sfw | d782c0b6ccd5dbfd7c59fd2ac408207470e22301 | [
"BSD-3-Clause"
] | null | null | null | legacy/unidad_5/03_C.ipynb | LVFerrero/diseno_sci_sfw | d782c0b6ccd5dbfd7c59fd2ac408207470e22301 | [
"BSD-3-Clause"
] | null | null | null | 130.094972 | 34,764 | 0.87989 | [
[
[
"\n# Diseño de software para cómputo científico\n\n----\n\n## Unidad 5: Integración con lenguajes de alto nivel con bajo nivel.\n",
"_____no_output_____"
],
[
"## Agenda de la Unidad 5\n\n- JIT (Numba)\n- Cython.\n- Integración de Python con FORTRAN.\n- **Integración de Python con C.**\n",
"_____no_output_____"
],
[
"## Recapitulando\n\n- Escribimos el código Python.\n- Pasamos todo a numpy.\n- Hicimos profile.\n- Paralelisamos (joblib/dask).\n- Hicimos profile.\n- Usamos Numba.\n- Hicimos profile.\n- Si podemos elegir el lenguaje: Cython\n - Si no podemos elegir el lenguaje y vamos a hacer cómputo numérico FORTRAN.\n- Si no podemos elegir vamos con C/C++/Rust/lo-que-sea.\n",
"_____no_output_____"
],
[
"## Ctypes\n\n- Permite usar bibliotecas existentes en otros idiomas escribiendo envoltorios **simples** en Python.\n- Viene con Python.\n- Puede ser un poco **Dificil** de usar.\n- Es una herramienta ideal para comper Python",
"_____no_output_____"
],
[
"### Ejemplo para Ctypes 1/2\n\nEl código C que usaremos en este tutorial está diseñado para ser lo más simple posible mientras demuestra los conceptos que estamos cubriendo. Es más un \"ejemplo de juguete\" y no pretende ser útil por sí solo. Estas son las funciones que utilizaremos:\n\n```c\nint simple_function(void) {\n static int counter = 0;\n counter++;\n return counter;\n}\n```\n\n- `simple_function` simplemente devuelve números de conteo. \n- Cada vez que se llama en incrementos de contador y devuelve ese valor.",
"_____no_output_____"
],
[
"### Ejemplo para Ctypes 2/2\n\n```c\nvoid add_one_to_string(char *input) {\n int ii = 0;\n for (; ii < strlen(input); ii++) {\n input[ii]++;\n }\n}\n```\n\n- Agrega uno a cada carácter en una matriz de caracteres que se pasa. \n- Usaremos esto para hablar sobre las cadenas inmutables de Python y cómo solucionarlas cuando sea necesario.\n\nEstos ejemplos estan guardadoe en `clibc1.c`, y se compilan con:\n\n```bash\ngcc -c -Wall -Werror -fpic clib1.c # crea el código objeto\ngcc -shared -o libclib1.so clib1.o # crea el .so\n```\n",
"_____no_output_____"
],
[
"## Llamando a una función simple",
"_____no_output_____"
]
],
[
[
"import ctypes\n\n# Load the shared library into c types.\nlibc = ctypes.CDLL(\"ctypes/libclib1.so\")",
"_____no_output_____"
],
[
"counter = libc.simple_function()\ncounter",
"_____no_output_____"
]
],
[
[
"## Cadenas inmutables en Python con Ctypes",
"_____no_output_____"
]
],
[
[
"print(\"Calling C function which tries to modify Python string\")\noriginal_string = \"starting string\"\nprint(\"Before:\", original_string)\n\n# This call does not change value, even though it tries!\nlibc.add_one_to_string(original_string)\n\nprint(\"After: \", original_string)",
"Calling C function which tries to modify Python string\nBefore: starting string\nAfter: starting string\n"
]
],
[
[
"- Como notarán esto **no anda**.\n- El `original_string` no está disponible en la función C en absoluto al hacer esto. \n- La función C modificó alguna otra memoria, no la cadena. \n- La función C no solo no hace lo que desea, sino que también modifica la memoria que no debería, lo que genera posibles problemas de corrupción de memoria.\n- Si queremos que la función C tenga acceso a la cadena, necesitamos hacer un poco de trabajo de serialización.",
"_____no_output_____"
],
[
"## Cadenas inmutables en Python con Ctypes\n\n- Necesitamos convertir la cadena original a bytes usando `str.encode,` y luego pasar esto al constructor para un `ctypes.string_buffer`. \n- Los String_buffers son mutables y se pasan a C como `char *`.",
"_____no_output_____"
]
],
[
[
"# The ctypes string buffer IS mutable, however.\nprint(\"Calling C function with mutable buffer this time\")\n\n# Need to encode the original to get bytes for string_buffer\nmutable_string = ctypes.create_string_buffer(str.encode(original_string))\n\nprint(\"Before:\", mutable_string.value)\nlibc.add_one_to_string(mutable_string) # Works!\nprint(\"After: \", mutable_string.value)",
"Calling C function with mutable buffer this time\nBefore: b'starting string'\nAfter: b'tubsujoh!tusjoh'\n"
]
],
[
[
"## Especificación de firmas de funciones en ctypes\n\n- Como vimos anteriormente, podemos especificar el tipo de retorno si es necesario.\n- Podemos hacer una especificación similar de los parámetros de la función. \n- Además, proporcionar una firma de función le permite a Python verificar que está pasando los parámetros correctos cuando llama a una función C, de lo contrario, pueden suceder cosas **malas**.\n\nPara especificar el tipo de retorno de una función, hayque obtener el bjeto de la función y establecer la propiedad `restype`:\n\n```python\nlibc.func.restype = ctypes.POINTER(ctypes.c_char)\n```\n\ny para especificar las firmas\n\n```python\nlibc.func.argtypes = [ctypes.POINTER(ctypes.c_char), ]\n```",
"_____no_output_____"
],
[
"## Escribir una interfaz Python en C\n\nVamos a \"envolver\" función de biblioteca C `fputs()`:\n\n```C\nint fputs (const char *, FILE *)\n```\n\n- Esta función toma dos argumentos:\n 1. `const char *` es una matriz de caracteres.\n 2. `FILE *` es un puntero a un stream de archivo.\n- `fputs()` escribe la matriz de caracteres en el archivo especificado y devuelve un valor no negativo, si la operación es exitosa, este valor indicará el número de bytes escritos en el archivo. \n- Si hay un error, entonces devuelve `EOF`. ",
"_____no_output_____"
],
[
"## Escribir la función C para `fputs()`\n\nEste es un programa básico de C que usa fputs() para escribir una cadena en una secuencia de archivos:\n\n```C\n#include <stdio.h>\n#include <stdlib.h>\n#include <unistd.h>\n\nint main() {\n FILE *fp = fopen(\"write.txt\", \"w\");\n fputs(\"Real Python!\", fp);\n fclose(fp);\n return 0;\n}\n```",
"_____no_output_____"
],
[
"## Envolviendo `fputs()`\n\nEl siguiente bloque de código muestra la versión final envuelta de su código C:\n\n```C\n#include <Python.h>\n\nstatic PyObject *method_fputs(PyObject *self, PyObject *args) {\n char *str, *filename = NULL;\n int bytes_copied = -1;\n\n /* Parse arguments */\n if(!PyArg_ParseTuple(args, \"ss\", &str, &filename)) {\n return NULL;\n }\n\n FILE *fp = fopen(filename, \"w\");\n bytes_copied = fputs(str, fp);\n fclose(fp);\n\n return PyLong_FromLong(bytes_copied);\n}\n```\n\nEste fragmento de código hace referencia a tres estructuras de objetos que se definen en `Python.h`:\n`PyObject`, `PyArg_ParseTuple()` y `PyLong_FromLong()`",
"_____no_output_____"
],
[
"## `PyObject`\n\n- `PyObject` es una estructura de objetos que utiliza para definir tipos de objetos para Python. \n- Todos los demás tipos de objetos Python son extensiones de este tipo.\n- Establecer el tipo de retorno de la función anterior como `PyObject` define los campos comunes que requiere Python para reconocer esto como un tipo válido.\n\nEche otro vistazo a las primeras líneas de su código C:\n\n```C\nstatic PyObject *method_fputs(PyObject *self, PyObject *args) {\n char *str, *filename = NULL;\n int bytes_copied = -1;\n ...\n``` \n\nEn la línea 2, declara los tipos de argumento que desea recibir de su código Python:\n\n- `char *str` es la cadena que desea escribir en la secuencia del archivo.\n- `char *filename` es el nombre del archivo para escribir.",
"_____no_output_____"
],
[
"## `PyArg_ParseTuple()`\n\n`PyArg_ParseTuple()` transforma los argumentos que recibirá de su programa Python en variables locales:\n\n```C\nstatic PyObject *method_fputs(PyObject *self, PyObject *args) {\n char *str, *filename = NULL;\n int bytes_copied = -1;\n \n if(!PyArg_ParseTuple(args, \"ss\", &str, &filename)) {\n return NULL;\n }\n ...\n``` \n\n`PyArg_ParseTuple()` toma los siguientes argumentos:\n\n- `args` de tipo `PyObject`.\n- `\"ss\"` especifica el tipo de datos de los argumentos a analizar.\n- `&str` y `&filename` son punteros a variables locales a las que se asignarán los valores analizados.\n\n`PyArg_ParseTuple()` retorna `false` frente a un error.",
"_____no_output_____"
],
[
"## `fputs()` y `PyLongFromLon()`\n\n```C\nstatic PyObject *method_fputs(PyObject *self, PyObject *args) {\n char *str, *filename = NULL;\n int bytes_copied = -1;\n if(!PyArg_ParseTuple(args, \"ss\", &str, &filename)) {\n return NULL;\n }\n\n FILE *fp = fopen(filename, \"w\");\n bytes_copied = fputs(str, fp);\n fclose(fp);\n\n return PyLong_FromLong(bytes_copied);\n}\n\n``` \n\n- Las llamadas a `fputs()` fueron explicadas anteriormente, la única diferencia es que las variables utilizadas son las que provienen de `*args` y almacenadas localmente.\n- Finalmente `PyLong_FromLong()` retorna un `PyLongObject`, que representa objecto entero en Python.",
"_____no_output_____"
],
[
"## Módulo de extensión\n\nYa se escribió el código que constituye la funcionalidad principal de su módulo de extensión Python C. \n- Sin embargo queda escribir las definiciones de su módulo y los métodos que contiene, de esta manera:\n\n```C\nstatic PyMethodDef FputsMethods[] = {\n {\"fputs\", method_fputs, METH_VARARGS, \"Python interface for fputs C library function\"},\n {NULL, NULL, 0, NULL}\n};\n\n\nstatic struct PyModuleDef fputsmodule = {\n PyModuleDef_HEAD_INIT,\n \"fputs\",\n \"Python interface for the fputs C library function\",\n -1,\n FputsMethods\n};\n```",
"_____no_output_____"
],
[
"## `PyMethodDef`\n\n\n- `PyMethodDef` informa al intérprete de Python sobre ello los métodos definidos en el módulo\n- Idealmente, habrá más de un método en la. Es por eso que necesita definir una matriz de estructuras:\n\n```C\nstatic PyMethodDef FputsMethods[] = {\n {\"fputs\", method_fputs, METH_VARARGS, \"Python interface for fputs C library function\"},\n {NULL, NULL, 0, NULL}\n};\n```\n\nCada miembro individual de la estructura contiene la siguiente información:\n- `fputs` es el nombre que el usuario escribiría para invocar esta función en particular desde Python.\n- `method_fputs` es el nombre de la función C a invocar.\n- `METH_VARARGS` indica que la función aceptará dos argumentos de tipo\n`PyObject *`:\n - `self` es el objeto del módulo.\n - `args` es una tupla que contiene los argumentos de la función (descomprimibles `PyArg_ParseTuple()`.\n- La cadena final es un valor para representar el docstring.",
"_____no_output_____"
],
[
"### `PyModuleDef`\n\nDefine un módulo Python (un archivo `.py`) en C.\n\n```C\nstatic struct PyModuleDef fputsmodule = {\n PyModuleDef_HEAD_INIT, \"fputs\",\n \"Interface for the fputs C function\", -1, FputsMethods};```\n\nHay un total de 9 miembros en esta estructura, pero el bloque de código anterior, inicializa los siguientes cinco:\n\n- `PyModuleDef_HEAD_INIT` es la clase \"base\" del módulo (normalmente esto siempre es igual).\n- `\"fputs\"` nombre del módulo.\n- La cadena es la documentación del módulo.\n- `-1` cantidad de memoria necesaria para almacenar el estado del programa. Es útil cuando su módulo se utiliza en múltiples subinterpretadores, y puede tener los siguientes valores:\n - Un valor negativo indica que este módulo no tiene soporte para subinterpretadores.\n - Un valor no negativo permite la reinicialización del módulo. También especifica el requisito de memoria que se asignará en cada sesión de subinterpretador.\n- `FputsMethods` es tabla de métodos. ",
"_____no_output_____"
],
[
"## Inicializando el módulo\n\n- Ahora que ha definido la extensión Python C y las estructuras de métodos, es hora de ponerlas en uso. \n- Cuando un programa Python importa su módulo por primera vez, llamará a `PyInit_fputs()`:\n\n```C\nPyMODINIT_FUNC PyInit_fputs(void) {\n return PyModule_Create(&fputsmodule);\n}\n``` \n\n`PyMODINIT_FUNC hace 3 cosas implícitamente`\n\n- Establece implícitamente el tipo de retorno de la función como PyObject *.\n- Declara cualquier enlace especial.\n- Declara la función como \"C\" externa. En caso de que esté usando C++, le dice al compilador de C ++ que no haga cambios de nombre en los símbolos.\n\n`PyModule_Create()` devolverá un nuevo objeto de módulo de tipo `PyObject *`.",
"_____no_output_____"
],
[
"## Poniendo todo junto - Qué pasa cuando importamos el módulo?\n\n",
"_____no_output_____"
],
[
"## Poniendo todo junto - Qué retorna cuando se importa el módulo?\n\n",
"_____no_output_____"
],
[
"## Poniendo todo junto - Qué sucede cuando llamamos a `fputs.fputs()`\n\n",
"_____no_output_____"
],
[
"## Empaquetado con `distutils` \n\n```python\nfrom distutils.core import setup, Extension\n\ndef main():\n setup(name=\"fputs\",\n ext_modules=[Extension(\"fputs\", [\"fputsmodule.c\"])],\n ...)\n\nif __name__ == \"__main__\":\n main()\n \n```\n\nPara instalar:\n\n```bash\n$ python3 setup.py install \n```\n\nPara compilar\n\n```bash\n$ python setup.py build_ext --inplace\n```\n\nSi se quiere especificar el compilador\n\n```bash\n$ CC=gcc python3 setup.py install\n```",
"_____no_output_____"
],
[
"## Usando la extensión",
"_____no_output_____"
]
],
[
[
"import sys; sys.path.insert(0, \"./c_extensions\")\nimport fputs",
"_____no_output_____"
],
[
"fputs?",
"_____no_output_____"
],
[
"fputs.fputs?",
"_____no_output_____"
],
[
"fputs.fputs(\"Hola mundo!\", \"salida.txt\")",
"_____no_output_____"
],
[
"with open(\"salida.txt\") as fp: \n print(fp.read())",
"Hola mundo!\n"
]
],
[
[
"## Raising Exceptions\n\n- Si desea lanzar excepciones de Python desde C, puede usar la API de Python para hacerlo. \n- Algunas de las funciones proporcionadas por la API de Python para generar excepciones son las siguientes:\n\n- `PyErr_SetString(PyObject *type, const char *message)`\n- `PyErr_Format(PyObject *type, const char *format)`\n- `PyErr_SetObject(PyObject *type, PyObject *value)`\n\nTodas las exceptions de Python estan definidas en las API.",
"_____no_output_____"
],
[
"## Raising Exceptions\n\n\n```C\nstatic PyObject *method_fputs(PyObject *self, PyObject *args) {\n char *str, *filename = NULL;\n int bytes_copied = -1;\n\n /* Parse arguments */\n if(!PyArg_ParseTuple(args, \"ss\", &str, &fd)) {\n return NULL;\n }\n if (strlen(str) <= 0) {\n PyErr_SetString(PyExc_ValueError, \"String length must be greater than 0\");\n return NULL;\n }\n\n fp = fopen(filename, \"w\");\n bytes_copied = fputs(str, fp);\n fclose(fp);\n\n return PyLong_FromLong(bytes_copied);\n}\n```",
"_____no_output_____"
],
[
"## Raising Custom Exceptions\n\nPara crear y usar excepción personalizada, se debe agregarla instancia de módulo:\n\n```C\nstatic PyObject *StringTooShortError = NULL;\n\nPyMODINIT_FUNC PyInit_fputs(void) {\n /* Assign module value */\n PyObject *module = PyModule_Create(&fputsmodule);\n /* Initialize new exception object */\n StringTooShortError = PyErr_NewException(\"fputs.StringTooShortError\", NULL, NULL);\n /* Add exception object to your module */\n PyModule_AddObject(module, \"StringTooShortError\", StringTooShortError);\n\n return module;\n}\nstatic PyObject *method_fputs(PyObject *self, PyObject *args) {\n ...\n if (strlen(str) <=0 10) {\n /* Passing custom exception */\n PyErr_SetString(StringTooShortError, \"String length must be greater than 0\");\n return NULL;}\n ...\n}\n \n```",
"_____no_output_____"
],
[
"## Referencias\n\n- https://docs.python.org/3.8/library/ctypes.html\n- https://dbader.org/blog/python-ctypes-tutorial\n- https://realpython.com/build-python-c-extension-module/",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cb463ea05128085bc0c4de47cdcafc4115a1f914 | 123,622 | ipynb | Jupyter Notebook | intro_python/Intro_Python_Resbaz_2021.ipynb | drakeasberry/2021_Resbaz_Python | b3e9602d15cba61bc81854025f65632d13bf984b | [
"MIT"
] | 2 | 2021-05-17T17:01:05.000Z | 2021-12-25T07:09:48.000Z | intro_python/Intro_Python_Resbaz_2021.ipynb | drakeasberry/2021_Resbaz_Python | b3e9602d15cba61bc81854025f65632d13bf984b | [
"MIT"
] | null | null | null | intro_python/Intro_Python_Resbaz_2021.ipynb | drakeasberry/2021_Resbaz_Python | b3e9602d15cba61bc81854025f65632d13bf984b | [
"MIT"
] | 31 | 2021-05-15T17:13:27.000Z | 2021-05-18T16:27:53.000Z | 29.932688 | 642 | 0.573943 | [
[
[
"# An Introduction to Python using Jupyter Notebooks\n<a id='toc'></a>\n## Table of Contents:\n### Introduction\n* [Python programs are plain text files](#python-programs)\n* [Use the Jupyter Notebook for editing and running Python](#jn-editing-python) \n* [How are Jupyter Notebooks stored](#how-its-stored)\n* [What you need to know](#need-to-know)\n* [The Notebook has Control and Edit modes](#notebook-modes)\n* [Use the keyboard and mouse to select and edit cells](#keyboard-mouse)\n * [Practice: Run your first Jupyter Notebook cells](#prac-jupyter)\n\n### Using Markdown\n* [The Notebook will turn Markdown into pretty-printed documentation](#markdown)\n* [How to use Markdown](#how-to-markdown)\n* [Markdown Exercises](#md-exercises)\n* [Markdown Exercise Soultions](#md-solutions)\n\n### Introduction to Python 1: Data\n* [Intro to Python 1: Prerequisites](#python-1)\n* [Programming with Python](#python-introduction) \n * [What is Python and why would I use it?](#python-introduction)\n * [Special Characters](#python-sp-char)\n * [Variables](#variables)\n * [Practice](#prac-variable)\n * [Variables can be used in calculations](#variable-calc)\n * [Data Types](#data-types)\n * [Practice with Strings](#prac-strings)\n * [Practice with Numerics](#numbers)\n * [Practice with Booleans](#booleans)\n * [Python \"Type\" function](#py-type)\n * [Lists](#py-lists)\n * [Tuples](#py-tuples)\n * [Differences between lists and tuples](#lists-vs-tuples)\n * [Sets](#py-sets)\n * [Dictionaries](#py-dictionaries)\n* [Python Statements](#py-statements)\n * [Conditionals](#py-conditionals)\n * [Loops](#py-loops)\n * [For Loops](#for-loops)\n * [While Loops](#while-loops)\n* [Pandas: Working with Existing Data](#pandas)\n * [Pandas: Importing Data](#read-data)\n * [Pandas: Manipulating Data](#manipulate-data)\n * [Pandas: Writing Data](#write-data)\n * [Pandas: Working with more than file](#all-countries)\n * [Pandas: Slicing and selecting values](#slicing)\n\n* Python I Exercises\n * [Problem 5: Assigning variables and printing values](#prob-variable)\n * [Problem 6: Print your first and last name](#py-concatenate)\n * [Problem 7: What variable type do I have?](#py-data-type)\n * [Problem 8: Creating and Working with Lists](#prob-lists)\n * [Problem 9: Creating and Accessing Dictionaries](#prob-dictionaries)\n * [Problem 10: Writing Conditional If/Else Statements](#prob-if-else)\n * [Problem 11: Reverse the string using a for loop](#prob-str-reverse-loop)\n * [Problem 12: Looping through Dictionaries](#prob-dict-loop)\n * [Problem 13: Checking assumptions about your data](#prob-unique)\n * [Problem 14: Slice and save summary statistics](#summary-stats)\n* [Python I Exercise Soultions](#py1-solutions)\n\n### Introduction to Python 2: A Tool for Programming\n* [Intro to Python 2: Prerequisites](#python-2)\n* [Setup if you are joining in for Python II](#python-2-setup)\n* [Functions:](#functions)\n * [Why Use Functions?](#why-functions)\n * [Let's revist the reverse string and turn it into a function](#str-reverse-func)\n * [Let's look at a real world example of where constants could be used in functions](#temp-func)\n* [Scripting](#scripting)\n* Python II Exercises\n* [Python II Exercise Soultions](#py2-solutions)\n\n### Common Errors\n* [Common Errors](#errors)\n\n",
"_____no_output_____"
],
[
"<a id='python-programs'></a>\n### Python programs are plain text files\n[Table of Contents](#toc)\n* They have the `.py` extension to let everyone (including the operating system) \n know it is a Python program.\n * This is convention, not a requirement.\n* It's common to write them using a text editor but we are going to use a [Jupyter Notebook](http://jupyter.org/).\n* There is a bit of extra setup, but it is well worth it because Jupyter Notebooks provide code completion\n and other helpful features such as markdown integration. This means you can take notes in this notebook while we are working throughout the session.\n* There are some pitfalls that can also cause confusion if we are unaware of them. While code generally runs from top to bottom, a Jupyter Notebook allows you to run items out of sequence. The order of code blocks running order will appear as a number to the left of the code text field.\n* Notebook files have the extension `.ipynb` to distinguish them from plain-text Python programs.\n\n<a id='jn-editing-python'></a>\n### Use the Jupyter Notebook for editing and running Python\n[Table of Contents](#toc)\n* The [Anaconda package manager](http://www.anaconda.com) is an automated way to install the Jupyter notebook.\n * See [the setup instructions]({{ site.github.url }}/setup/) for Anaconda installation \n instructions.\n* It also installs all the extra libraries it needs to run.\n* Once you have installed Python and the Jupyter Notebook requirements, open a shell and type:\n\n> `jupyter notebook`\n\n* This will start a Jupyter Notebook server and open your default web browser. \n* The server runs locally on your machine only and does not use an internet connection.\n* The server sends messages to your browser.\n* The server does the work and the web browser renders the notebook.\n* You can type code into the browser and see the result when the web page talks to the server.\n* This has several advantages:\n\t- You can easily type, edit, and copy and paste blocks of code.\n\t- Tab completion allows you to easily access the names of things you are \n using and learn more about them.\n\t- It allows you to annotate your code with links, different sized text, bullets, \n etc to make it more accessible to you and your collaborators.\n\t- It allows you to display figures next to the code that produces them to \n tell a complete story of the analysis.\n - **Note: This will modify and delete files on your local machine.**\n* The notebook is stored as JSON but can be saved as a .py file if you would\n like to run it from the bash shell or a python interpreter.\n* Just like a webpage, the saved notebook looks different to what you see when \n it gets rendered by your browser.\n \n<a id='how-its-stored'></a>\n### How are Jupyter Notebooks Stored\n[Table of Contents](#toc)\n* The notebook file is stored in a format called JSON.\n* Just like a webpage, what's saved looks different from what you see in your browser.\n* But this format allows Jupyter to mix software (in several languages) with documentation and graphics, all in one file.\n\n<a id='need-to-know'></a>\n### What you need to know for today's lesson\n[Table of Contents](#toc)\n**Jupyter Notebook options when running locally:**\n\n\n**Jupyter Notebook options when running in Binder:**\n\n\n* Commands are only run when you tell them to run. Some lessons require you to run their code in order.\n* The File menu has an option called \"Revert to Checkpoint\". Use that to reset your file in case you delete something on accident.\n* The Kernel menu has an options to restart the interpreter and clear the output. \n* The Run button will send the code in the selected cell to the interpreter.\n* The command pallate function will show you and let you set hotkeys.\n* Saving to browser storage is the button with a cloud and downward facing arrow. Click on this button frequently\nto save progress as we go.\n* Restoring from browser storage is the button with a cloud and upward facing arrow. Click on this button if you\nare disconnected or Binder quits working after you have refreshed the page. This will load your previously save work.\n\n<a id='notebook-modes'></a>\n### The Notebook has Control and Edit modes.\n[Table of Contents](#toc)\n* Open a new notebook from the dropdown menu in the top right corner of the file browser page.\n* Each notebook contains one or more cells of various types.\n\n> ## Code vs. Markdown\n>\n> We often use the term \"code\" to mean \"the source code of software written in a language such as Python\".\n> A \"code cell\" in a Jupyter Notebook contains software code or that which is for the computer to read.\n> A \"markdown cell\" is one that contains ordinary prose written for human beings to read.\n\n* If you press `esc` and `return` keys alternately, the outer border of your code cell will change from blue to green.\n * The difference in color can be subtle, but indicate different modes of you notebook.\n * <span style='color:blue'>Blue</span> is the command mode while <span style='color:green'>Green</span> is the\n edit mode.\n* If you use the \"esc\" key to make the surrounding box blue (enter into command mode) and then press the \"H\" key, a\n list of all the shortcut keys will appear.\n* When in command mode (esc/blue),\n * The `B key` will make a new cell below the currently selected cell.\n * The `A key` will make one above.\n * The `X key` will delete the current cell.\n* There are lots of shortcuts you can try out and most actions can be done with the menus at the top of the page if you forget the shortcuts.\n* If you remember the `esc` and `H` shortcuts, you will be able to find all the tools you need to work in a notebook.\n\n<a id='keyboard-mouse'></a>\n### Use the keyboard and mouse to select and edit cells.\n[Table of Contents](#toc)\n* Pressing the `return key turns the surrounding box green to signal edit mode and allows you type in the cell.\n* Because we want to be able to write many lines of code in a single cell, pressing the `return` key when the\nborder is green moves the cursor to the next line in the cell just like in a text editor.\n* We need some other way to tell the Notebook we want to run what's in the cell.\n* Pressing the `shift` and the `return` keys together will execute the contents of the cell.\n* Notice that the `return` and `shift` keys on the right of the keyboard are right next to each other.",
"_____no_output_____"
],
[
"<a id='prac-jupyter'></a>\n### Practice: Running Jupyter Notebook Cell\n[Table of Contents](#toc)",
"_____no_output_____"
]
],
[
[
"# Find the shortcut in the command pallate and run this cell.\nmessage = \"run me first\"",
"_____no_output_____"
]
],
[
[
"If you ran the above cell correctly, there should be a number **1** inside the square brackets to the left of the cell. **Note:** the number will increase everytime you run the cell.",
"_____no_output_____"
]
],
[
[
"# Run this cell and see what the output is. \nprint(message)",
"_____no_output_____"
]
],
[
[
"**If the output beneath the cell looks like this:**\n```python\nrun me first\n```\nThen you have run the cells in the correct order and received the expected output. Why did we get this output?\n\n**If the output beneath the cell looks like this:**\n```python\n---------------------------------------------------------------------------\nNameError Traceback (most recent call last)\n<ipython-input-1-a4525a899574> in <module>\n 1 # Run this cell and see what the output is.\n----> 2 print(message)\n\nNameError: name 'message' is not defined\n```\n\nThen you have received an error. Read the error message to see what went wrong. Here we have a `NameError`because the computer does not know what the variable `message` is. We need to go back to the first code cell and run it correctly first to define the variable `message`. Then we should be able to run the second code cell and receive the first output (prints the string we assigned to the variable `message`).\n\n**Getting Error Messages**:\nError messages are commonplace for anyone writing code. You should expect to get them frequently and learn how to\ninterpret them as best as possible. Some languages give more descriptive error messages than others, but in both\ncases you are likely to find the answer with a quick Google search.",
"_____no_output_____"
],
[
"## Using Markdown\n<a id='markdown'></a>\n### The Notebook will turn Markdown into pretty-printed documentation.\n[Table of Contents](#toc)\n* Notebooks can also render [Markdown][markdown].\n * A simple plain-text format for writing lists, links and other things that might go into a web page.\n * Equivalently, a subset of HTML that looks like what you would send in an old-fashioned email.\n* Turn the current cell into a Markdown cell by entering the command mode (esc/blue) and press the `M key`.\n* `In [ ]:` will disappear to show it is no longer a code cell and you will be able to write in Markdown.\n* Turn the current cell into a Code cell by entering the command mode (esc/blue) and press the `Y key`.",
"_____no_output_____"
],
[
"<a id='how-to-markdown'></a>\n### How to use Markdown\n[Table of Contents](#toc)\n<div class=\"row\">\n <div class=\"col-md-6\" markdown=\"1\">",
"_____no_output_____"
],
[
"**The asterisk is a special character in markdown. It will create a bulleted list.**\n```markdown\nMarkdown syntax to produce output below.\n\n* Use asterisks\n* to create\n* bullet lists.\n```\n\n* Use asterisks \n* to create \n* bulleted lists. ",
"_____no_output_____"
],
[
"**But what happens when I want to use asterisk in my text. We can use another special, the back slash `\\`, also known as\n an escape charatcer. Place the back slash before any markdown special character without a space to use the special character in your text.**\n```markdown\nMarkdown syntax to produce output below.\n\n\\* Use asterisks\n\\* to create\n\\* bullet lists.\n```\n\n\\* Use asterisks \n\\* to create \n\\* bullet lists.\n\nNote: Escape characters can change depending on the language you are writing in.",
"_____no_output_____"
],
[
"**Use can use numbers to create a numbered list:**\n```markdown\nMarkdown syntax to produce numbered lists.\n1. Use numbers\n1. to create\n1. numbered lists.\n```\n\n1. Use numbers\n1. to create\n1. numbered lists.\n\nNote: That we did not have to type numbers in order but markdown still converted correctly in output. This is nice\nbecause it saves us time when we modify or edit lists later because we do not have to renumber the entire list.",
"_____no_output_____"
],
[
"**Using differnt Headings to keep consistency through document:**\n```markdown\nMarkdown syntax to produce headings.\n\n# A Level-1 Heading\n## A Level-2 Heading\n### A Level-3 Heading\n```\n\nPrint version of the three lines of markdown code from above.\n# A Level-1 Heading\n## A Level-2 Heading\n### A Level-3 Heading",
"_____no_output_____"
],
[
"**Line breaks don't matter. But blank lines create new paragraphs.**\n```markdown\n**Markdown syntax:**\n\nLine breaks\ndo not matter. _(accomplished by pressing the return key once)_\n\nSometimes though we want to include a line break without starting a new paragraph. We can accomplish this by including two spaces at the end of the line.\n\nHere is the first line. \nThe second line is on the second line but in same paragraph (no blank line).\n```\n\n**Print version of markdown code from above:**\n\nLine breaks\ndon't matter. _(accomplished by pressing the return key once)_\n\nSometimes though we want to include a line break without starting a new paragraph. We can accomplish this by including two spaces at the end of the line.\n\nHere is the first line. \nThe second line is on the second line but in same paragraph (no blank line).",
"_____no_output_____"
],
[
"**Creating links in markdown:**\n\nThe information inside the `[...]` is what the user will see and the information inside the `(...)` is the pointer or url that the link will take the user to.\n ```markdown\n**Markdown Syntax:**\n\n[Create links](http://software-carpentry.org) with the following syntax `[...](...)`.\nOr use [named links][data_carpentry].\n\n_Notice the line below only defines the link and is not in printed output. Double click on the cell below this one if you don't believe me._\n[data_carpentry]: http://datacarpentry.org \n```",
"_____no_output_____"
],
[
"**Output of markdown syntax:**\n\n[Create links](http://software-carpentry.org) with `[...](...)`. \nOr use [named links][data_carpentry].\n\n[data_carpentry]: http://datacarpentry.org",
"_____no_output_____"
],
[
"<a id='md-exercises'></a>\n## Markdown Exercises\n[Table of Contents](#toc)",
"_____no_output_____"
],
[
"### Creating Lists in Markdown\n<a id='md-exercises-p01'></a>\n**Problem 1: Creating Lists** Create a nested list in a Markdown cell in a notebook that looks like this:\n\n1. Get funding.\n1. Do work.\n * Design experiment.\n * Collect data.\n * Analyze.\n1. Write up.\n1. Publish.\n\n**Hint:**_Double click this cell to see the answer._\n\n[Solution](#md-solutions-p01)",
"_____no_output_____"
],
[
"<a id='md-exercises-p02'></a>\n### Math anyone?\n**Problem 2: Math in python** What is displayed when a Python cell in a notebook that contains several calculations is executed? For example, what happens when this cell is executed?",
"_____no_output_____"
]
],
[
[
"7 * 3",
"_____no_output_____"
]
],
[
[
"What is displayed when a Python cell in a notebook that contains several calculations is executed? For example, what happens when this cell is executed?",
"_____no_output_____"
]
],
[
[
"7 * 3\n\n2 + 1\n\n6 * 7 + 12",
"_____no_output_____"
]
],
[
[
"[Solution](#md-solutions-p02)\n\n<a id='md-exercises-p03'></a>\n**Problem 3: Math in markdown** Change an Existing Cell from Code to Markdown\n\nWhat happens if you write some Python in a code cell and then you switch it to a Markdown cell? For example, put the following in a code cell.\n\n1. Run the cell below with `shift + return` to be sure that it works as a code cell. _Hint: it should give you the\nsame result as **Problem 2**_.\n1. Select the cell below and use `escape + M` to switch the cell to Markdown and run it again with `shift + return`. What happened and how might this be useful?",
"_____no_output_____"
]
],
[
[
"7 * 3 \n2 + 1\n\nx = 6 * 7 + 12\nprint(x)",
"_____no_output_____"
]
],
[
[
"Print statements can help us find errors or unexpected results from our code. They allow us to check our assumptions.\n Does the computer have stored what we think it does?\n\nThis could also be useful if you wanted to show what the code generating your document looks like. Think code reviews,\ncolleagues, advisors, etc.\n\n[Solution](#md-solutions-p03)",
"_____no_output_____"
],
[
"<a id='md-exercises-p04'></a>\n**Problem 4:** Equations\n \nStandard Markdown (such as we’re using for these notes) won’t render equations, but the Notebook will. \n\n`$\\Sigma_{i=1}^{N} 2^{-i} \\approx 1$`\n",
"_____no_output_____"
],
[
"Think about the following questions:\n1. What will it display?\n1. What do you think the underscore `_` does?\n1. What do you think the circumflex `^` does?\n1. What do you think the dollar sign `$` does?\n\nChange the Code cell below containing the equation to a Markdown cell and run it.",
"_____no_output_____"
]
],
[
[
"$\\Sigma_{i=1}^{N} 2^{-i} \\approx 1$",
"_____no_output_____"
]
],
[
[
"**Note:** If you received a <span style='color:red'> SyntaxError</span>, then you need to change the cell to a Markdown\ncell and rerun.\n\n[Solution](#md-solutions-p04)",
"_____no_output_____"
],
[
"<a id='md-solutions'></a>\n## Markdown Exercise Solutions\n[Table of Contents](#toc)",
"_____no_output_____"
],
[
"<a id='md-solutions-p01'></a>\n### Problem 1: Creating Lists\nThis challenge integrates both the numbered list and bullet list. Note that the bullet list is tabbed over to create the nesting necesary for the list.\n```markdown\n**Type the following in your Markdown cell:**\n1. Get funding.\n1. Do work.\n * Design experiment.\n * Collect data.\n * Analyze.\n1. Write up.\n1. Publish.\n```\n[Back to Problem](#md-exercises-p01)",
"_____no_output_____"
],
[
"<a id='md-solutions-p02'></a>\n### Problem 2: Math in python\nThe output of running the code cell is 54 because 6 multiplied by 7 is 42 and 42 plus 12 equals 54. This equation was stored as a variable called `x` and the last line executed was `print(x)`, which simply prints out the value of variable `x` at the current time. However, it still did all the other mathematical equations `7*3` and `2+1`, but it did not print them out because we did not ask the computer to do so.\n\n[Back to Problem](#md-exercises-p02)",
"_____no_output_____"
],
[
"<a id='md-solutions-p03'></a>\n### Problem 3: Math in markdown\nIn step 1, The output of running the code cell is 54 because 6 multiplied by 7 is 42 and 42 plus 12 equals 54. This\nequation was stored as a variable called `x` and the last line executed was `print(x)`, which simply prints out the\nvalue of variable `x` at the current time. However, it still did all the other mathematical equations `7*3` and\n`2+1`, but it did not print them out because we did not store the value and ask the computer to print them.\n\nThe Python code gets treated like markdown text. The lines appear as if they are part of one contiguous paragraph.\nThis could be useful to temporarily turn on and off cells in notebooks that get used for multiple purposes. It is\nalso useful when you want to show the code you have written rather than the output of the code execution.\n```markdown\n7*3\n2+1\nx = 6 * 7 + 12 \nprint(x)\n```\n\n[Back to Problem](#md-exercises-p03)",
"_____no_output_____"
],
[
"<a id='md-solutions-p04'></a>\n### Problem 4: Equations\n`$\\Sigma_{i=1}^{N} 2^{-i} \\approx 1$`\n\n$\\Sigma_{i=1}^{N} 2^{-i} \\approx 1$\n\nThe notebook shows the equation as it would be rendered from latex equation syntax. The dollar sign,`$`, is used to tell markdown that the text in between is a latex equation. If you are not familiar with latex, the underscore, `_`, is used for subscripts and the circumflex, `^`, is used for superscripts. A pair of curly braces, `{` and `}`, is used to group text together so that the statement `i=1` becomes the the subscript and `N` becomes the superscript. Similarly, `-i` is in curly braces to make the whole statement the superscript for `2`. `\\sum` and `\\approx` are latex commands for “sum over” and “approximate” symbols.\n\n[anaconda]: https://docs.continuum.io/anaconda/install\n\n[markdown]: https://en.wikipedia.org/wiki/Markdown\n\n**A common error is to forgot to run the cell as markdown.** The python interpreter does not know what to do with the \\$. Syntax errors generally mean that the user has entered something incorrectly (check for typos before assuming the line of code is wrong altogether.\n\n```markdown\n File \"<ipython-input-1-a80a20b3c603>\", line 1\n $\\Sigma_{i=1}^{N} 2^{-i} \\approx 1$\n ^\nSyntaxError: invalid syntax\n```\n\n[Back to Problem](#md-exercises-p04)",
"_____no_output_____"
],
[
"<a id='python-1'></a>\n# Intro to Python I: Data\n[Table of Contents](#toc)\n\n\n**Prerequisites:** None\n\nThis workshop will help researchers with no prior programming experience learn how to utilize Python to analyze research data. You will learn how to open data files in Python, complete basic data manipulation tasks and save your work without compromising original data. Oftentimes, researchers find themselves needing to do the same task with different data and you will gain basic experience on how Python can help you make more efficient use of your time.\n\n**Learning Objectives:**\n1. Clean/manipulate data\n1. Automate repetitive tasks\n\n**Learning Outcomes:** you will be able to…\n1. read data into Pandas dataframe\n1. use Pandas to manipulate data\n1. save work to a datafile useable in other programs needed by researcher\n1. write if/else statements\n1. build for and while loops",
"_____no_output_____"
],
[
"<a id='python-introduction'></a>\n## Programming with Python\n[Table of Contents](#toc)\n### What is Python and why would I use it?\n\nA programming language is a way of writing commands so that an interpreter or compiler can turn them into machine\ninstructions. Python is just one of many different programming languages.\n\nEven if you are not using Python in your work, you can use Python to learn the fundamentals of programming that will apply across languages.\n\n**We like using Python in workshops for lots of reasons:**\n* It is widely used in science\n* It's easy to read and write\n* There is a huge supporting community - lots of ways to learn and get help\n* This Jupyter Notebook. Not a lot of languages have this kind of thing (name comes from Julia, Python, and R).\n\n<a id='python-sp-char'></a>\n### Special Characters\n[Table of Contents](#toc)\n\nWe have already worked with special characters in markdown. Similarly, python uses certain special characters as part of its syntax. **Note:** special characters are not consistent across languages so make sure you familiarize yourself with the special characters in the languages in which you write code.\n\n**Python Special Characters:**\n\n* `[` : left `square bracket`\n* `]` : right `square bracket`\n* `(` : left `paren` (parentheses)\n* `)` : right `paren` (parentheses)\n* `{` : left `curly brace`\n* `}` : right `curly brace`\n* `<` : left `angle bracket`\n* `>` : right `angle bracket`\n* `-` `dash` (not hyphen. Minus only when used in an equation or formula)\n* `\"` : `double quote`\n* `'` : `single quote` (apostrophe)\n\n<a id='variables'></a>\n### Variables\n[Table of Contents](#toc)\n\nVariables are used to store information in the computer that can later be referenced, manipulated and/or used by our programs. Important things to remember about variables include:\n* We store values inside variables. \n* We can refer to variables in other parts of our programs.\n* In Python, the variable is created when a value is assigned to it.\n * Values are assigned to variable names using the equals sign `=`. \n* A variable can hold two types of things. \n * Basic data types. For descriptions and details [(See Data Types)](#data-types) \n * Objects - ways to structure data and code. In Python, all variables are objects.\n* Variable naming convention:\n * Cannot start with a digit\n * Cannot contain spaces, quotation marks, or other punctuation\n * Using a descriptive name can make the code easier to read **(You will thank yourself later)**\n \n<a id='prac-variable'></a>\n### Practice\n[Table of Contents](#toc)",
"_____no_output_____"
]
],
[
[
"# What is happening in this code python cell\nage = 34\nfirst_name = 'Drake'",
"_____no_output_____"
]
],
[
[
"In the cell above, Python assigns an age (in this example 34) to a variable `age` and a name (Drake) in quotation marks to a variable `first_name`.\n\nIf you want to see the stored value of a variable in python, you can display the value by using the print command \n`print()` with the variable name placed inside the parenthesis.",
"_____no_output_____"
]
],
[
[
"# what is the current value stored in the variable `age`\nprint(age)",
"_____no_output_____"
]
],
[
[
"**Write a print statement to show the value of variable `first_name` in the code cell below.**",
"_____no_output_____"
]
],
[
[
"# Print out the current value stored in the variable `first_name``\n",
"_____no_output_____"
]
],
[
[
"<a id='prob-variable'></a>\n### Problem 5: Assigning variables and printing values\n[Table of Contents](#toc)\n\n1. Create two new variables called `age` and `first_name` with your own age and name\n2. Print each variable out to dispaly it's value\n",
"_____no_output_____"
],
[
"**Extra Credit:** Combine values in a single print command by separating them with commas",
"_____no_output_____"
]
],
[
[
"# Insert your variable values into the print statement below\nprint(<insert variable here>, 'is', <insert variable here>, 'years old.')",
"_____no_output_____"
]
],
[
[
"The `print` command automatically puts a single space between items to separate them and wraps around to a new line at the end.\n[Solution](#prob-variable-sol)",
"_____no_output_____"
],
[
"<a id='variable-calc'></a>\n## Variables can be used in calculations.\n[Table of Contents](#toc)\n\n\n* We can use variables in calculations just as if they were values.\n * Remember, we assigned **our own age** to `age` a few lines ago.",
"_____no_output_____"
]
],
[
[
"age = age + 3\nprint('My age in three years:', age)",
"_____no_output_____"
]
],
[
[
"* This now sets our age value **our current age + 3 years**. \n* We can also add strings together, but it works a bit differently. When you add strings together it is called **concatenating**.",
"_____no_output_____"
]
],
[
[
"name = \"Sonoran\"\nfull_name = name + \" Desert\"\nprint(full_name)",
"_____no_output_____"
]
],
[
[
"* Notice how I included a space in the quotes before \"Desert\". If we hadn't, we would have had \"SonoranDesert\"\n* Can we subtract, multiply, or divide strings?",
"_____no_output_____"
],
[
"<a id='py-concatenate'></a>\n## Problem 6: Printing your first and last name\n[Table of Contents](#toc)\n\nIn the code cell below, create a new variable called last_name with your own last name.\nCreate a second new variable called full_name that is a combination of your first and last name.\n\n",
"_____no_output_____"
]
],
[
[
"# Print full name",
"_____no_output_____"
]
],
[
[
"[Solution](#py-concatenate-sol)\n<a id='data-types'></a>\n### Data Types\n[Table of Contents](#toc)\n\n**Some data types you will find in almost every language include:**",
"_____no_output_____"
],
[
"| Data Type| Abbreviation | Type of Information | Examples |\n| :-| :-| :-| :-|\n| Strings | str | characters, words, sentences or paragraphs| 'a' 'b' 'c' 'abc' '0' '3' ';' '?'|\n| Integers | int | whole numbers | 1 2 3 100 10000 -100 |\n| Floating point or Float | float | decimals | 10.0 56.9 -3.765 |\n| Booleans | bool | logical test | True, False |",
"_____no_output_____"
],
[
"<a id='strings'></a>\n### Strings\n[Table of Contents](#toc)\n\nOne or more characters strung together and enclosed in quotes (single or double): \"Hello World!\"",
"_____no_output_____"
]
],
[
[
"greeting = \"Hello World!\"\nprint(\"The greeting is:\", greeting)",
"_____no_output_____"
],
[
"greeting = 'Hello World!'\nprint('The greeting is:', greeting)",
"_____no_output_____"
]
],
[
[
"#### Need to use single quotes in your string?\nUse double quotes to make your string.",
"_____no_output_____"
]
],
[
[
"greeting = \"Hello 'World'!\"\nprint(\"The greeting is:\", greeting)",
"_____no_output_____"
]
],
[
[
"#### Need to use both?",
"_____no_output_____"
]
],
[
[
"greeting1 = \"'Hello'\"\ngreeting2 = '\"World\"!'\nprint(\"The greeting is:\", greeting1, greeting2)",
"_____no_output_____"
]
],
[
[
"#### Concatenation",
"_____no_output_____"
]
],
[
[
"bear = \"wild\"\ndown = \"cats\"\nprint(bear + down)",
"_____no_output_____"
]
],
[
[
"Why aren't `greeting`, `greeting1`, `greeting2`, `bear`, or `down` enclosed in quotes in the statements above?\n\n<a id='prac-strings'></a>\n### Practice: Strings\n[Table of Contents](#toc)",
"_____no_output_____"
],
[
"#### Use an index to get a single character from a string.\n * The characters (individual letters, numbers, and so on) in a string are ordered.\n * For example, the string ‘AB’ is not the same as ‘BA’. Because of this ordering, we can treat the string as a list of characters.\n * Each position in the string (first, second, etc.) is given a number. This number is called an index or sometimes a subscript.\n * Indices are numbered from 0. \n * Use the position’s index in square brackets to get the character at that position.",
"_____no_output_____"
]
],
[
[
"# String : H e l i u m \n# Index Location: 0 1 2 3 4 5\n\natom_name = 'helium'\nprint(atom_name[0], atom_name[3])",
"_____no_output_____"
]
],
[
[
"<a id='numbers'></a>\n### Numbers\n[Table of Contents](#toc)\n\n* Numbers are stored as numbers (no quotes) and are either integers (whole) or real numbers (decimal). \n* In programming, numbers with decimal precision are called floating-point, or float.\n* Floats use more processing than integers so use them wisely!\n* Floats and integers come in various sizes but Python switches between them transparently.",
"_____no_output_____"
]
],
[
[
"my_integer = 10\nmy_float = 10.99998\nmy_value = my_integer\n\nprint(\"My numeric value:\", my_value)",
"_____no_output_____"
]
],
[
[
"<a id='py-type'></a>\n### Using Python built-in type() function\n[Table of Contents](#toc)\n\n\nIf you are not sure of what your variables' types are, you can call a python function called `type()` in the same manner as you used `print()` function.\nPython is an object-oriented language, so any defined variable has a type. Default common types are **str, int, float, list and tuple.** We will cover [list](#py-list) and [tuple](#py-tuple) later.",
"_____no_output_____"
]
],
[
[
"print(\"Type:\", type(age))\nprint(\"Type:\", type(first_name))",
"_____no_output_____"
],
[
"# Print out datatype of variables\nprint(\"my_value Type:\", type(my_value))\nprint(\"my_float Type:\", type(my_float))",
"_____no_output_____"
]
],
[
[
"<a id='booleans'></a>\n### Boolean\n[Table of Contents](#toc)\n\n* Boolean values are binary, meaning they can only either true or false.\n* In python True and False (no quotes) are boolean values",
"_____no_output_____"
]
],
[
[
"is_true = True\nis_false = False\n\nprint(\"My true boolean variable:\", is_true)\nprint(\"Type:\", type(is_false))",
"_____no_output_____"
]
],
[
[
"<a id='py-data-type'></a>\n### Problem 7: What variable type do I have? \n[Table of Contents](#toc)\n\nsize = '1024' \nWhat data type is `size`? Use some of the python you have learned to provide proof of your answer.\n<ol style=\"list-style-type:lower-alpha\">\n <li>float</li>\n <li>string</li>\n <li>integer</li>\n <li>boolean</li>\n</ol>",
"_____no_output_____"
]
],
[
[
"# Write your explanation as a comment and write the python code that outputs support for your answer.\n",
"_____no_output_____"
]
],
[
[
"[Solution](#py-data-type)\n<a id='py-data-structures'></a>\n## Data Structures\n[Table of Contents](#toc)\n\nPython has many objects that can be used to structure data including:\n\n| Object | Data Structure | Mutable |\n| :- | :- | :- |\n| List | collections of values held together in brackets | Mutable |\n| Tuple | collection of grouped values held together in parentheses | Immutable |\n| Set | collections of unique values held together in curly braces | Mutable |\n| Dictionary | collections of keys & values held together in curly braces | Mutable |",
"_____no_output_____"
],
[
"<a id='py-lists'></a>\n### Lists\n[Table of Contents](#toc)\n\nLists are collections of values held together in brackets: ",
"_____no_output_____"
]
],
[
[
"list_of_characters = ['a', 'b', 'c'] \nprint(list_of_characters)",
"_____no_output_____"
]
],
[
[
"<a id='prob-lists'></a>\n### Problem 8: Creating and Working with Lists\n[Table of Contents](#toc)\n\n1. Create a new list called list_of_numbers with four numbers in it.",
"_____no_output_____"
]
],
[
[
"# Print out the list of numbers you created\n",
"_____no_output_____"
]
],
[
[
"* Just like strings, we can access any value in the list by it's position in the list.\n* **IMPORTANT:** Indexes start at 0\n ~~~\n list: ['a', 'b', 'c', 'd']\n index location: 0 1 2 3\n ~~~",
"_____no_output_____"
]
],
[
[
"# Print out the second value in the list list_of_numbers\n",
"_____no_output_____"
]
],
[
[
"2. Once you have created a list you can add more items to it with the append method",
"_____no_output_____"
]
],
[
[
"# Append a number to your list_of_numbers\n",
"_____no_output_____"
]
],
[
[
"[Solution](#prob-lists-sol)\n#### Aside: Sizes of data structures\n\nTo determine how large (how many values/entries/elements/etc.) any Python data structure has, use the `len()` function",
"_____no_output_____"
]
],
[
[
"len(list_of_numbers)",
"_____no_output_____"
]
],
[
[
"Note that you cannot compute the length of a numeric variable:",
"_____no_output_____"
]
],
[
[
"len(age)",
"_____no_output_____"
]
],
[
[
"This will give an error: `TypeError: object of type 'int' has no len()`",
"_____no_output_____"
],
[
"However, `len()` can compute the lengths of strings",
"_____no_output_____"
]
],
[
[
"# Get the length of the string\nprint(len('this is a sentence'))",
"_____no_output_____"
],
[
"# You can also get the lengths of strings in a list\nlist_of_strings = [\"Python is Awesome!\", \"Look! I'm programming.\", \"E = mc^2\"]\n\n# This will get the length of \"Look! I'm programming.\"\nprint(len(list_of_strings[1]))",
"_____no_output_____"
]
],
[
[
"<a id='py-tuples'></a>\n### Tuples\n[Table of Contents](#toc)\n\nTuples are like a List, but **cannot be changed (immutable).**\n\nTuples can be used to represent any collection of data. They work well for things like coordinates. Notice below that\n tuples are surrounded by parentheses `()` rather than square brackets `[]` that were used for lists.",
"_____no_output_____"
]
],
[
[
"tuple_of_x_y_coordinates = (3, 4)\nprint (tuple_of_x_y_coordinates)",
"_____no_output_____"
]
],
[
[
"Tuples can have any number of values",
"_____no_output_____"
]
],
[
[
"coordinates = (1, 7, 38, 9, 0)\nprint (coordinates)\n\nicecream_flavors = (\"strawberry\", \"vanilla\", \"chocolate\")\nprint (icecream_flavors)",
"_____no_output_____"
]
],
[
[
"... and any types of values.\n\nOnce created, you `cannot add more items to a tuple` (but you can add items to a list). If we try to append, like we did with lists, we get an error",
"_____no_output_____"
]
],
[
[
"icecream_flavors.append('bubblegum')",
"_____no_output_____"
]
],
[
[
"<a id='lists-vs-tuples'></a>\n### The Difference Between Lists and Tuples\n[Table of Contents](#toc)\n\nLists are good for manipulating data sets. It's easy for the computer to add, remove and sort items. Sorted tuples are easier to search and index. This happens because tuples reserve entire blocks of memory to make finding specific locations easier while lists use addressing and force the computer to step through the whole list.\n\nLet's say you want to get to the last item. The tuple can calculate the location because:\n\n(address)=(size of data)×(index of the item)+(original address)\n\nThis is how zero indexing works. The computer can do the calculation and jump directly to the address. The list would need to go through every item in the list to get there.\n\nNow lets say you wanted to remove the third item. Removing it from the tuple requires it to be resized and copied. Python would even make you do this manually. Removing the third item in the list is as simple as making the second item point to the fourth. Python makes this as easy as calling a method on the tuple object.",
"_____no_output_____"
],
[
"<a id='py-sets'></a>\n### Sets\n[Table of Contents](#toc)\n\nSets are similar to lists and tuples, but can only contain unique values and are held inside curly braces.\n\nFor example a list could contain multiple exact values",
"_____no_output_____"
]
],
[
[
"# In the gapminder data that we will use, we will have data entries for the continents\n# of each country in the dataset\nmy_list = ['Africa', 'Europe', 'North America', 'Africa', 'Europe', 'North America']\nprint(\"my_list is\", my_list)\n\n# A set would only allow for unique values to be held\nmy_set = {'Africa', 'Europe', 'North America', 'Africa', 'Europe', 'North America'}\nprint(\"my_set is\", my_set)",
"_____no_output_____"
]
],
[
[
"Just like lists, you can append to a set using the add() method.",
"_____no_output_____"
]
],
[
[
"my_set.add('Asia')\n\n# Now let's try to append one that is in:\nmy_set.add('Europe')",
"_____no_output_____"
]
],
[
[
"What will the print statements show now in the code cell below?",
"_____no_output_____"
]
],
[
[
"print(\"my_list is\", my_list)\nprint(\"my_set is\", my_set)",
"_____no_output_____"
]
],
[
[
"<a id='py-dictionaries'></a>\n### Dictionaries\n[Table of Contents](#toc)\n\n* Dictionaries are collections of things that you can lookup like in a real dictionary:\n* Dictionarys can organized into key and value pairs separated by commas (like lists) and surrounded by curly braces.\n * E.g. {key1: value1, key2: value2}\n * We call each association a \"key-value pair\". ",
"_____no_output_____"
]
],
[
[
"dictionary_of_definitions = {\"aardvark\" : \"The aardvark is a medium-sized, burrowing, nocturnal mammal native to \"\n \"Africa.\",\n \"boat\" : \"A boat is a thing that floats on water\"}",
"_____no_output_____"
]
],
[
[
"We can find the definition of aardvark by giving the dictionary the \"key\" to the definition we want in brackets.\n\nIn this case the key is the word we want to lookup",
"_____no_output_____"
]
],
[
[
"print(\"The definition of aardvark is:\", dictionary_of_definitions[\"aardvark\"]) ",
"_____no_output_____"
],
[
"# Print out the definition of a boat\n",
"_____no_output_____"
]
],
[
[
"Just like lists and sets, you can add to dictionaries by doing the following:",
"_____no_output_____"
]
],
[
[
"dictionary_of_definitions['ocean'] = \"An ocean is a very large expanse of sea, in particular each of the main areas into which the sea is divided geographically.\"\nprint(dictionary_of_definitions)",
"_____no_output_____"
]
],
[
[
"<a id='prob-dictionaries'></a>\n### Problem 9: Creating and Accessing Dictionaries\n[Table of Contents](#toc)\n\n1. Create a dictionary called `zoo` with at least three animal types with a different count for each animal (How many\n animals of that type are found at the zoo).\n1. `print` out the count of the second animal in your dictionary \n",
"_____no_output_____"
]
],
[
[
"# Zoo Dictionary\n",
"_____no_output_____"
]
],
[
[
"[Solution](#prob-dictionaries-sol)\n<a id='py-statements'></a>\n## Statements\n[Table of Contents](#toc)\n\nOK great. Now what can we do with all of this? \n\nWe can plug everything together with a bit of logic and python language to make a program that can do things like:\n\n* process data (data wrangling or manipulation)\n* parse files\n* data analysis",
"_____no_output_____"
],
[
"What kind of logic are we talking about?\n\nWe are talking about something called a \"logical structure\" which starts at the top (first line) and reads down the page in order\n\nIn python a logical structure are often composed of statements. Statements are powerful operators that control the\nflow of your script. There are two main types of statements:\n\n* conditionals (if, while)\n* loops (for, while)\n",
"_____no_output_____"
],
[
"<a id='py-conditionals'></a>\n### Conditionals\n[Table of Contents](#toc)\n\nConditionals are how we make a decision in the program.\nIn python, conditional statements are called if/else statements.\n\n* If statement use boolean values to define flow.\n * If something is True, do this. Else, do this\n * While something is True, do some process.\n \n**Building if/else statements in Python:**\n1. Start first line with `if` \n1. Then `some-condition` must be a logical test that can be evaulated as True or False\n1. End the first line with `:` \n1. Indent the next line(s) with `tab` or `4 spaces` (Jupyter does the indent automatically!)\n 1. `do-things`: give python commands to execute\n1. End the statement with `else` and `:` (notice that if and else are in the same indent) \n1. Indent the next line(s) with `tab` or `4 spaces` (Jupyter does the indent automatically!)\n 1. `do-different-things`: give python commands to execute",
"_____no_output_____"
],
[
"### Comparison operators:\n\n`==` equality \n`!=` not equal \n`>` greater than \n`>=` greater than or equal to \n`<` less than \n`<=` less than or equal to ",
"_____no_output_____"
]
],
[
[
"weight = 3.56\n\nif weight >= 2:\n print(weight,'is greater than or equal to 2')\nelse:\n print(weight,'is less than 2')",
"_____no_output_____"
]
],
[
[
"### Membership operators:\n\n`in` check to see if data is **present** in some collection \n`not in` check to see if data is **absent** from some collection",
"_____no_output_____"
]
],
[
[
"groceries=['bread', 'tomato', 'hot sauce', 'cheese']\n\nif 'basil' in groceries: \n print('Will buy basil')\nelse:\n print(\"Don't need basil\")",
"_____no_output_____"
],
[
"# this is the variable that holds the current condition of it_is_daytime \n# which is True or False \nit_is_daytime = True \n\n# if/else statement that evaluates current value of it_is_daytime variable\nif it_is_daytime:\n print (\"Have a nice day.\")\nelse:\n print (\"Have a nice night.\")\n \n# before running this cell\n# what will happen if we change it_is_daytime to True?\n# what will happen if we change it_is_daytime to False?",
"_____no_output_____"
]
],
[
[
"* Often if/else statement use a comparison between two values to determine True or False\n* These comparisons use \"comparison operators\" such as ==, >, and <.\n* \\>= and <= can be used if you need the comparison to be inclusive.\n* **NOTE**: Two equal signs `==` is used to compare values, while one equals sign `=` is used to assign a value\n * E.g.\n \n 1 > 2 is False<br/>\n 2 > 2 is False<br/>\n 2 >= 2 is True<br/>\n 'abc' == 'abc' is True",
"_____no_output_____"
]
],
[
[
"user_name = \"Ben\"\n\nif user_name == \"Marnee\":\n print (\"Marnee likes to program in Python.\")\nelse:\n print (\"We do not know who you are.\")",
"_____no_output_____"
]
],
[
[
"* What if a condition has more than two choices? Does it have to use a boolean?\n* Python if-statments will let you do that with elif\n* `elif` stands for \"else if\"\n ",
"_____no_output_____"
]
],
[
[
"if user_name == \"Marnee\":\n print (\"Marnee likes to program in Python.\")\nelif user_name == \"Ben\":\n print (\"Ben likes maps.\")\nelif user_name == \"Brian\":\n print (\"Brian likes plant genomes\")\nelse:\n print (\"We do not know who you are\")\n \n# for each possibility of user_name we have an if or else-if statment to check the \n# value of the name and print a message accordingly.",
"_____no_output_____"
]
],
[
[
"What does the following statement print?\n\n my_num = 42\n my_num = 8 + my_num\n new_num = my_num / 2\n if new_num >= 30:\n print(\"Greater than thirty\")\n elif my_num == 25:\n print(\"Equals 25\")\n elif new_num <= 30:\n print(\"Less than thirty\")\n else:\n print(\"Unknown\")",
"_____no_output_____"
],
[
"<a id='prob-if-else'></a>\n### Problem 10: Writing Conditional If/Else Statements\n[Table of Contents](#toc)\n\nCheck to see if you have more than three entries in the `zoo` dictionary you created earlier. If you do, print \"more than three animals\". If you don't, print \"three or less animals\"",
"_____no_output_____"
]
],
[
[
"# write an if/else statement\n",
"_____no_output_____"
]
],
[
[
"Can you modify your code above to tell the user that they have exactly three animals in the dictionary?",
"_____no_output_____"
]
],
[
[
"# Modify conditional to include exactly three as potential output",
"_____no_output_____"
]
],
[
[
"[Solution](#prob-if-else-sol)\n<a id='py-loops'></a>\n### Loops\n[Table of Contents](#toc)\n\nLoops tell a program to do the same thing over and over again until a certain condition is met. \n* In python two main loop types: \n * For loops\n * While loops",
"_____no_output_____"
],
[
"<a id='for-loops'></a>\n### For Loops\n[Table of Contents](#toc)\n\nA for loop executes the same command through each value in a collection.\n\nBuilding blocks of a for loop:\n\n> `for` each-item `in` variable `:` \n>> `do-something`\n\n**Building for loops in python:**\n1. Start the first line with `for` \n1. `each-item` is an arbitrary name for each item in the variable/list. \n1. Use `in` to indicate the variable that hold the collection of information \n1. End the first line with `:` \n1. indent the following line(s) with `tab` or `4 spaces` (Jupyter does the indent automatically!)\n 1. `do-something` give python commands to execute",
"_____no_output_____"
],
[
"In the example below, `number` is our `each-item` and the `print()` command is our `do-something`.",
"_____no_output_____"
]
],
[
[
"# Running this cell will give a NameError because it has not be defined yet.\nprint(number)",
"_____no_output_____"
],
[
"# Run this cell and see if you figure out what this for loop does\nfor number in range(10): # does not include 10! \n print(number)",
"_____no_output_____"
]
],
[
[
"#### LOOPING a set number of times\nWe can do this with the function `range()`. Range automatically creates a list of numbers in a specified range.\n\nIn the example above, we have a list of 10 numbers starting with 0 and increasing by one until we have 10 numbers. In the example below, we get the same end result although we have give two numbers to `range()`. In the example below we have given the start and end points of the range. **Note: Do not forget about python's zero-indexing**",
"_____no_output_____"
]
],
[
[
"# What will be printed\nfor number in range(0,10):\n print(number)",
"_____no_output_____"
],
[
"# What will be printed\nfor number in range(1,11):\n print(number)",
"_____no_output_____"
],
[
"# What will be printed\nfor number in range(10,0, -1):\n print(number)",
"_____no_output_____"
],
[
"# Change the code from the cell above so that python prints 9 to 0 in descending order\n",
"_____no_output_____"
],
[
"# This loop prints in each iteration of the loop which shows us a value for each of the 10 runs.\ntotal = 0 # global variable\n\nfor i in range(10):\n total=total+i \n print(total) ",
"_____no_output_____"
],
[
"# This loop prints the value for last value after the 10 runs have occured.\ntotal=0\n\nfor i in range(10):\n total=total+i\n\nprint(total)",
"_____no_output_____"
]
],
[
[
"#### Saving Time\nLooping can save you lots of time. We will look at a simple example to see how it works with lists, but imagine if your list was 100 items long. You do not want to write 100 individual print commands, do you?",
"_____no_output_____"
]
],
[
[
"# LOOPING over a collection\n# LIST\n\n# If I want to print a list of fruits, I could write out each print statment like this:\nprint(\"apple\")\nprint(\"banana\")\nprint(\"mango\")\n\n# or I could create a list of fruit\n# loop over the list\n# and print each item in the list\nlist_of_fruit = [\"apple\", \"banana\", \"mango\"]\n\n# this is how we write the loop\n# \"fruit\" here is a variable that will hold each item in the list, the fruit, as we loop\n# over the items in the list\nprint (\">>looping>>\")\nfor fruit in list_of_fruit:\n print (fruit)",
"_____no_output_____"
]
],
[
[
"#### Creating New Data\nYou can also use loops to create new datasets as well. In the cell below, we use a mathematical operator to create a new list `data_2` where each value is double that of the value in the original list `data`.",
"_____no_output_____"
]
],
[
[
"data = [35,45,60,1.5,40,50]\ndata_2 = []\n\nfor i in data:\n data_2.append(i*2)\n \nprint(data_2)",
"_____no_output_____"
]
],
[
[
"<a id='prob-str-reverse-loop'></a>\n### Problem 11: Reverse the string using a for loop\n[Table of Contents](#toc)\n\nThere are many ways to reverse a string. I want to challenge you to use a for loop. The goal is to practice how to build a for loop (use multiple print statements) to help you understand what is happening in each step.",
"_____no_output_____"
]
],
[
[
"string = \"waterfall\"\nreversed_string = \"\"\n\n# For loop reverses the string given as input\n\n\n# Print out the both the original and reversed strings",
"_____no_output_____"
]
],
[
[
"**Extra Credit: Accomplish the same task (reverse a string) with out using a for loop.** _Hint: the reversing range example above gives you a clue AND Google always has an answer!_",
"_____no_output_____"
]
],
[
[
"# Reversing the string can be done by writing only one more line\nstring = \"waterfall\"\n",
"_____no_output_____"
]
],
[
[
"We can loop over collections of things like lists or dictionaries or we can create a looping structure.",
"_____no_output_____"
]
],
[
[
"# LOOPING over a collection\n# DICTIONARY\n\n# We can do the same thing with a dictionary and each association in the dictionary\n\nfruit_price = {\"apple\" : 0.10, \"banana\" : 0.50, \"mango\" : 0.75}\nfor key, value in fruit_price.items():\n print (\"%s price is %s\" % (key, value))",
"_____no_output_____"
]
],
[
[
"[Solution](#prob-str-reverse-loop-sol)\n<a id='prob-dict-loop'></a>\n### Problem 12: Looping through Dictionaries\n[Table of Contents](#toc)\n\n1. For each entry in your `zoo` dictionary, print that key",
"_____no_output_____"
]
],
[
[
"# print only dictionary keys using a for loop\n",
"_____no_output_____"
]
],
[
[
"2. For each entry in your zoo dictionary, print that value",
"_____no_output_____"
]
],
[
[
"# print only dictionary values using a for loop\n",
"_____no_output_____"
]
],
[
[
"3. Can you print both the key and its associated value using a for loop?",
"_____no_output_____"
]
],
[
[
"# print dictionary keys and values using a single for loop\n",
"_____no_output_____"
]
],
[
[
"[Solution](#prob-dict-loop-sol)\n<a id='while-loops'></a>\n### While Loops\n[Table of Contents](#toc)\n\nSimilar to if statements, while loops use a boolean test to either continue looping or break out of the loop.",
"_____no_output_____"
]
],
[
[
"# While Loops\nmy_num = 10\n\nwhile my_num > 0:\n print(\"My number\", my_num)\n my_num = my_num - 1\nprint('My value is no longer greater than zero and I have exited the \"while\" loop as a result.')",
"_____no_output_____"
]
],
[
[
"NOTE: While loops can be dangerous, because if you forget to include an operation that modifies the variable being\ntested (above, we're subtracting 1 at the end of each loop), it will continue to run forever and your script will never finish.",
"_____no_output_____"
],
[
"That is it. With just these data types, structures and logic, you can build a program. We will write program with functions in [Python II: A tool for programming](#python-2)",
"_____no_output_____"
],
[
"<a id='pandas'></a>\n## Pandas: Working with Existing Data\n[Table of Contents](#toc)\n\nThus far, we have been creating our own data as we go along and you are probably thinking \"How in the world can this save me time?\" This next section is going to help you learn how to import data that you already have. [Pandas](https://pandas.pydata.org/docs/) is a python package that is great for doing data manipulation.\n\n<a id='read-data'></a>\n### Pandas: Importing Data\n[Table of Contents](#toc)\n\n**Importing packages:** Pandas is a package that is written for python but is not part of the base python install. In\n order to use these add on packages, we must first import them. This is conventionally the first thing you do in a\n script. If I were building a script using Jupyter Notebooks, I generally do all the importing of packages I need for\n the entire notebook in the first code cell.",
"_____no_output_____"
]
],
[
[
"# Import packages\nimport pandas",
"_____no_output_____"
]
],
[
[
"**Note:** pandas is a long name and you will generally find a shortened version of the name in online help resources. As such, we will use the same convention in this workshop. It only requires a small modification to the import statement.",
"_____no_output_____"
]
],
[
[
"# Import packages\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"Now that we have access to pandas at our disposal we are ready to import some data. We will be working a freely available dataset called [gapminder](https://www.gapminder.org/). The first data set we are going to look at is called `Afghanistan_Raw`. ",
"_____no_output_____"
]
],
[
[
"# import from excel spreadsheet\nafghanistan_xlsx = pd.read_excel('gapminder_data/Afghanistan_Raw.xlsx')\n",
"_____no_output_____"
],
[
"# import from csv file\nafghanistan_csv = pd.read_csv('gapminder_data/Afghanistan_Raw.csv')",
"_____no_output_____"
]
],
[
[
"The cell above assigns a `variable` to a pandas dataframe. \n\nTo create a pandas dataframe: \n1. We use `pd` to tell python that we want to use the pandas package that we imported. \n1. We use `.read_excel()` or `.read_csv()` to tell pandas what type of file format we are giving it. \n1. We have given the `relative path` to the file in parentheses.\n\n**Relative paths** are your best friend when you want your code to be easily moved or shared with collaborators. They\n use your current position in the computer's file structure as the starting point.\n* If you work on a script with relative paths on your work computer, email it to yourself and try to continue working\n on your personal home computer, it should work because the usernames may be different but are bypassed and\n computer's file structure are the same from the directory in which we are working.\n\n* The `current working directory` is where the Jupyter Notebook is stored unless you manually change it.\n\n#### Project Directory\n\nIntro_Python_Resbaz_2021.ipynb\n ├── array_vs_list.png \n ├── gapminder_data \n │ ├── Afghanistan_Raw.csv \n │ ├── Afghanistan_Raw.xlsx\n │ ├── Afghanistan_Fixed.csv\n │ └── gapminder_by_country\n ├── jn_binder_options.png \n ├── jn_options.png \n ├── Intro_Python_Resbaz_2021.ipynb\n └── scripting_practice.ipynb\n\n**Absolute paths** can be useful if the thing you are trying to access is never going to move. They start at the root of the computer's file structure and work out to the file's location. **Note: this includes the computer's username.**\n* If you work on a script with absolute paths on your work computer, email it to yourself and try to continue working on your personal home computer, it will fail because the usernames and computer's file structure are different.\n\n * My absolute path (work): /Users/**drakeasberry**/Desktop/2021_Resbaz_Python/intro_python\n * My absolute path (home): /Users/**drake**/Desktop/2021_Resbaz_Python/intro_python",
"_____no_output_____"
]
],
[
[
"print('This is the excel file:\\n\\n', afghanistan_xlsx)\nprint('\\nThis is the csv file:\\n\\n', afghanistan_csv)",
"_____no_output_____"
]
],
[
[
"This prints out each file separatly and I have added a few line break `\\n` just to make it a little easier read when it is printed. However, these may still feel unfamiliar and hard to read for you or your colleagues. If we do not include the data varibale inside the `print()`, then pandas will render a formatted table that is more visually pleasing. Let's look at the difference.",
"_____no_output_____"
]
],
[
[
"# Use print to label the output, but let pandas render the table\nprint('This is the excel file:')\nafghanistan_xlsx",
"_____no_output_____"
],
[
"# Use print to label the output, but let pandas render the table\nprint('This is the csv file:')\nafghanistan_csv",
"_____no_output_____"
]
],
[
[
"<a id='manipulate-data'></a>\n### Pandas: Manipulating Data\n[Table of Contents](#toc)\n\nAs you can see above, both ways of importing data have produced the same results. The type of data file you use is a personal choice, but not one that should be taken for granted. Microsoft Excel is licensed product and not everyone may have access to open `.xlsx` files whereas a `.csv`file is a comma separated values document that can be read by many free text editors. `.csv` files are also genereally smaller than the same information stored in a `.xlsx` file. My preferred choice is using `.csv` files due to smaller size and easier accessibility. ",
"_____no_output_____"
]
],
[
[
"afghanistan_csv.country.unique()",
"_____no_output_____"
],
[
"# Drop all rows with no values\nafghanistan_csv.dropna(how='all')",
"_____no_output_____"
],
[
"# What prints now and why?\nafghanistan_csv",
"_____no_output_____"
],
[
"# If we want to save the operations we need to store it in a variable (we will overwrite the existing one here)\nafghanistan_csv = afghanistan_csv.dropna(how='all')\nafghanistan_csv",
"_____no_output_____"
],
[
"# we will store a new dataframe called df to save some typing\n# we will subset the data to only rows that have a country name\ndf = afghanistan_csv.dropna(subset=['country'])\ndf",
"_____no_output_____"
],
[
"df = df.rename(columns={'pop':'population'})",
"_____no_output_____"
],
[
"# We are only expecting Afghanistan to be the only country in this file\n# Let's check our assumptions\ndf.country.unique()",
"_____no_output_____"
]
],
[
[
"<a id='prob-unique'></a>\n### Problem 13: Checking assumptions about your data\n[Table of Contents](#toc)\n\nYou can use df.info() to general idea about the data and then you can investigate the remaining columns to see if the\ndata is as you expect.",
"_____no_output_____"
]
],
[
[
"# this will give a quick overview of the data frame to give you an idea of where to start looks\n",
"_____no_output_____"
],
[
"# Hint: Check your assumptions about values dataframe\n",
"_____no_output_____"
]
],
[
[
"[Solution](#prob-unique-sol)\nOur investigation has showed us that some of data has errors but probably still useful if we correct them.\n* The year column is being read as a float instead of an object (we will not be doing mathematics on years)\n* The year column still has a missing value\n* The population column is being read as an object instead of an integer (we may want to do mathematics on population)\n* The continent column has a typo `Asiaa` and `tbd`\n\nLet's see if we can fix these issues together.",
"_____no_output_____"
]
],
[
[
"# Let's fix the typos in continent column\ndf = df.replace(to_replace =[\"Asiaa\", \"tbd\"], value =\"Asia\")\ndf",
"_____no_output_____"
],
[
"# Let's take a closer look at year column by sorting\ndf.sort_values(by='year')",
"_____no_output_____"
]
],
[
[
"By sorting the dataframe based on year, we can see that the years are incrementing by 5 years. We can also deduce that the year 1982 is missing. \nDepending on the data, you will have to make a decision as the researcher:\n* Are you confident that you can say that you have replaced the value correctly and the rest of the data is good?\n* Do you delete the data based on the fact that it had missing data?\n\nIn this case, we are going to replace the missing value with 1982 because we believe it is the right thing to do in this particular case.\n\n**Note:** In general, you should be very selective on replacing missing values. ",
"_____no_output_____"
]
],
[
[
"df['year'] = df['year'].fillna(1982)\ndf",
"_____no_output_____"
],
[
"# Finally, let's fix the datatypes of columns\ndf = df.astype({\"year\": int, \"population\": int})\ndf",
"_____no_output_____"
],
[
"# Let's check to see if it is working the way we think it is\ndf.info()",
"_____no_output_____"
]
],
[
[
"<a id='write-data'></a>\n### Pandas: Writing Data\n[Table of Contents](#toc)\n\nNow that we have made all the changes necessary, we should save our corrected datafram as a new file.",
"_____no_output_____"
]
],
[
[
"# Save file with changes we made\ndf.to_csv('gapminder_data/Afghanistan_Fixed.csv')",
"_____no_output_____"
]
],
[
[
"<a id='all-countries'></a>\n### Pandas: Working with more than file\n[Table of Contents](#toc)",
"_____no_output_____"
]
],
[
[
"#Import pandas library using an alias\nimport pandas as pd\n# Import glob library which allows us to use regular expressions to select multiple files\nimport glob",
"_____no_output_____"
],
[
"# Let's see where we are within the computer's directory structure\n# The exclamation point allows us to utilize a bash command in the notebook\n!pwd",
"_____no_output_____"
],
[
"# Let's see what files and folders are in our current location\n!ls",
"_____no_output_____"
],
[
"# Let's see what files and folders are in the gapminder_data directory\n!ls gapminder_data/",
"_____no_output_____"
],
[
"# Let's see what files and folders are in the gapminder_data/gapminder_by_country directory\n!ls gapminder_data/gapminder_by_country/",
"_____no_output_____"
]
],
[
[
"We worked with one file `Afghanistan` in the previous section, now we will combine everything we have seen to work with all the countries data that we have.\n\n1. Find files in `gapminder_data/gapminder_by_country/` \n1. Get all filenames into a list \n1. Remove `country.cc.txt`\n1. For loop to append file lines into a pandas dataframe \n1. Add column names from `country.cc.txt`",
"_____no_output_____"
]
],
[
[
"# glob.glob will match files in the current directory based on a pattern\ncountries = sorted(glob.glob('gapminder_data/gapminder_by_country/*.cc.txt'))\nlen(countries)",
"_____no_output_____"
],
[
"# Remove header item from item of files\n# If you try to run this cell more than once, you will get an error \n# because the item does not exist once it has been removed after the first execution of this cell\ncountries.remove('gapminder_data/gapminder_by_country/country.cc.txt')",
"_____no_output_____"
],
[
"# Check the length of the list to ensure the item was correctly removed\nlen(countries)",
"_____no_output_____"
],
[
"# creating dataframe from a for loop:\ndf = pd.DataFrame()\n\n# Go through each of 142 files and append until all countries are in one dataframe\nfor country in countries:\n c=pd.read_csv(country,sep='\\t',header=None)\n df=df.append(c,ignore_index=True)\n\n# Import header and store as list\nheader = pd.read_csv('gapminder_data/gapminder_by_country/country.cc.txt', sep='\\t')\ncolumn_names = list(header.columns)\n\n# Add header to dataframe created with the loop\ndf.columns = column_names",
"_____no_output_____"
],
[
"# Gives us number of rows and columns\ndf.shape",
"_____no_output_____"
],
[
"# Get summary statistics\ndf.describe()",
"_____no_output_____"
],
[
"# Do you remember how to change column types\n",
"_____no_output_____"
],
[
"# Solution\n# Do you remember how to change column types\ndf = df.astype({\"year\": int, \"pop\": int})\ndf.describe()",
"_____no_output_____"
]
],
[
[
"Save to summary of the dataframe `to_csv`, create a NEW file name, otherwise will overwrite the files we downloaded!",
"_____no_output_____"
]
],
[
[
"df.describe().to_csv('gapminder_summ_stats.csv')",
"_____no_output_____"
],
[
"ls",
"_____no_output_____"
]
],
[
[
"<a id='slicing'></a>\n### Pandas: Slicing and selecting values\n[Table of Contents](#toc)\n\n<div class=\"alert alert-block alert-success\">\n<b>Pandas Dataframe:</b> \n- 2-dimensional representation of a table \n- Series is the data-structure Pandas use to represent a column.\n</div>\n\n\nBecause it's 2 dimensional, we have to specify which rows and which columns we want to select.",
"_____no_output_____"
]
],
[
[
"# see the first 5 rows in the dataframe by default, but you can use any number in parentheses to see more or less\ndf.head()",
"_____no_output_____"
]
],
[
[
"**`.loc[]` to select values by the name**\n\n\n**`.loc[a:b,i:j]`**, where \n a and b are the rows\n i and j are the columns",
"_____no_output_____"
],
[
"Need to set index first:",
"_____no_output_____"
]
],
[
[
"df=df.set_index('country')\ndf",
"_____no_output_____"
],
[
"# this returns all the rows and columns where the index is Brazil\ndf.loc['Brazil']",
"_____no_output_____"
],
[
"# this returns all the rows and columns where the index is Brazil through Ecuador (alphabetically)\ndf.loc['Brazil':'Ecuador']",
"_____no_output_____"
],
[
"# this returns all the rows where the index is Brazil through Ecuador (alphabetically), but only includes the columns\n# between year and lifeExp (moving from left to right across the dataframe)\ndf.loc['Brazil':'Ecuador','year':'lifeExp']",
"_____no_output_____"
],
[
"# this returns all the rows where the index is Brazil or Ecuador, but only includes the columns\n# between year and lifeExp (moving from left to right across the dataframe)\ndf.loc[['Brazil','Ecuador'],'year':'lifeExp']",
"_____no_output_____"
]
],
[
[
"**`.iloc[]` to select values by the index**\n\n**`.iloc[a:b,i:j]`**, where \n a and b are the indexes of rows \n i and j are the indexes of columns",
"_____no_output_____"
]
],
[
[
"# this returns rows 10 through 16 and all but the last column (gdpPercap)\ndf.iloc[9:16,:-1]",
"_____no_output_____"
]
],
[
[
"**Observation:** \n```\n-3:-1, omits the final index (column gdpPercap) in the range provided, while a named slice includes the final element.\n ```",
"_____no_output_____"
]
],
[
[
"# this returns rows 10 and 17 and all but the columns (continent and lifeExp)\ndf.iloc[[9,16],-3:-1]\n",
"_____no_output_____"
],
[
"# this also returns rows 10 and 17 and all but the columns (continent and lifeExp)\ndf.iloc[[9,16],2:4]",
"_____no_output_____"
]
],
[
[
"<a id='summary-stats'></a>\n### Problem 14: Slice and save summary statistics\n[Table of Contents](#toc)\n\nSelect two countries of your interest. Slice the `df` to select only these countries. Then, obtain summary statistics by country, and save to a file.",
"_____no_output_____"
]
],
[
[
"# pick two countries to subset and save file with a descriptive name\n",
"_____no_output_____"
]
],
[
[
"[Solution](#summary-stats-sol)\n<a id='py1-solutions'></a>\n## Python I: Problem Solutions\n[Table of Contents](#toc)\n\n<a id='prob-variable-sol'></a>\n### Problem 5: Assigning variables and printing values\n1. Create two new variables called `age` and `first_name` with your own age and name\n2. Print each variable out to dispaly it's value\n[Back to Problem](#prob-variable)",
"_____no_output_____"
]
],
[
[
"age = '<your age>'\nfirst_name = '<your first name>'\nprint(age)\nprint(first_name)",
"_____no_output_____"
]
],
[
[
"**Extra Credit:** You can also combine values in a single print command by separating them with commas",
"_____no_output_____"
]
],
[
[
"# Insert your variable values into the print statement below\nprint(first_name, 'is', age, 'years old')",
"_____no_output_____"
]
],
[
[
"Correct Output:\nIf you received this output, then you correctly assigned new variables and combined them correctly in the print statment. The information represented between `<>` should reflect your personal information at this point.\n```markdown\n<your age>\n<your first name>\n<your first name> is <your age> years old\n```\n\nIf you received this output, then you forget to assign new variables.\n```markdown\n34\nDrake\nDrake is 34 years old\n```\n\nIf you received this output, then you correctly assigned new variables but mixed up the order in the combined print statment.\n```markdown\n<your age>\n<your first name>\n<your age> is <your first name> years old\n```",
"_____no_output_____"
],
[
"<a id='py-concatenate-sol'></a>\n### Problem 6: Printing your first and last name\n\nIn the code cell below, create a new variable called last_name with your own last name.\nCreate a second new variable called full_name that is a combination of your first and last name.\n\n",
"_____no_output_____"
]
],
[
[
"# Print full name\nfirst_name = 'Drake'\nlast_name = 'Asberry'\n\nprint(first_name, last_name)",
"_____no_output_____"
]
],
[
[
"[Back to Problem](#py-concatenate-sol)\n<a id='py-data-type-sol'></a>\n### Problem 7: What variable type do I have? \n\nsize = '1024' \nWhat data type is `size`? Use some of the python you have learned to provide proof of your answer.\n<ol style=\"list-style-type:lower-alpha\">\n <li>float</li>\n <li>string</li>\n <li>integer</li>\n <li>boolean</li>\n</ol>",
"_____no_output_____"
]
],
[
[
"# Write your explanation as a comment and write the python code that outputs support for your answer.\nsize = '1024'\nprint(type(size), \"is a string because when we stored the variable, we wrapped it in single quotes ''. Python \"\n \"understood this to be a string instead of an integer as a result.\")",
"_____no_output_____"
]
],
[
[
"[Back to Problem](#py-data-type)\n<a id='prob-lists-sol'></a>\n### Problem 8: Creating and Working with Lists\n\n1. Create a new list called list_of_numbers with four numbers in it.",
"_____no_output_____"
]
],
[
[
"# Print out the list of numbers you created\nlist_of_numbers = [0, 1, 2, 3]\nprint(list_of_numbers)",
"_____no_output_____"
],
[
"# Print out the second value in the list list_of_numbers\nprint(list_of_characters[1])",
"_____no_output_____"
]
],
[
[
"2. Once you have created a list you can add more items to it with the append method",
"_____no_output_____"
]
],
[
[
"# Append a number to your list\nlist_of_numbers.append(5)\nprint(list_of_numbers)",
"_____no_output_____"
]
],
[
[
"[Back to Problem](#prob-lists)\n### Problem 9: Creating and Accessing Dictionaries\n\n1. Create a dictionary called `zoo` with at least three animal types with a different count for each animal.\n1. `print` out the count of the second animal in your dictionary \n",
"_____no_output_____"
]
],
[
[
"# Zoo Dictionary\nzoo = {'bears':25, 'lions':19, 'monkeys':67}\nprint(zoo['lions'])",
"_____no_output_____"
]
],
[
[
"[Back to Problem](#prob-dictionaries)\n<a id='prob-if-else-sol'></a>\n### Problem 10: Writing Conditional If/Else Statements\nCheck to see if you have more than three entries in the `zoo` dictionary you created earlier. If you do, print \"more than three animals\". If you don't, print \"three or less animals\"",
"_____no_output_____"
]
],
[
[
"# write an if/else statement\nif len(zoo) > 3:\n print(\"more than three animals\")\nelse:\n print(\"three or less animals\")",
"_____no_output_____"
]
],
[
[
"Can you modify your code above to tell the user that they have exactly three animals in the dictionary?",
"_____no_output_____"
]
],
[
[
"# Modify conditional to include exactly three as potential output\nif len(zoo) > 3:\n print(\"more than three animals\")\nelif len(zoo) < 3:\n print(\"less than three animals\")\nelse:\n print(\"exactly three animals\")",
"_____no_output_____"
]
],
[
[
"[Back to Problem](#prob-if-else)\n<a id='prob-str-reverse-loop-sol'></a>\n### Problem 11: Reversing Strings\n\nThere are many ways to reverse a string. I want to challenge you to use a for loop. The goal is to practice how to build a for loop (use multiple print statements) to help you understand what is happening in each step.",
"_____no_output_____"
]
],
[
[
"string = \"waterfall\"\nreversed_string = \"\"\n\nfor char in string:\n #print(reversed_string)\n reversed_string = char + reversed_string\n #print(char)\n #print(reversed_string)\n\nprint('The original string was:', string)\nprint('The reversed string is:', reversed_string)",
"_____no_output_____"
]
],
[
[
"**Extra Credit: Accomplish the same task (reverse a string) with out using a for loop.** _Hint: the reversing range example above gives you a clue AND Google always has an answer!_",
"_____no_output_____"
]
],
[
[
"string = \"waterfall\"\nprint(string[::-1])",
"_____no_output_____"
]
],
[
[
"[Back to Problem](#prob-str-reverse-loop)\n<a id='prob-dict-loop'></a>\n### Problem 12: Looping through Dictionaries\n[Table of Contents](#toc)\n\n1. For each entry in your `zoo` dictionary, print that key",
"_____no_output_____"
]
],
[
[
"# print only dictionary keys using a for loop\nfor key in zoo.keys():\n print(key)",
"_____no_output_____"
]
],
[
[
"2. For each entry in your zoo dictionary, print that value",
"_____no_output_____"
]
],
[
[
"# print only dictionary values using a for loop\nfor value in zoo.values():\n print(value)",
"_____no_output_____"
]
],
[
[
"3. Can you print both the key and its associated value using a for loop?",
"_____no_output_____"
]
],
[
[
"# print dictionary keys and values using a single for loop\nfor key, value in zoo.items():\n print(key,value)",
"_____no_output_____"
]
],
[
[
"[Back to Problem](#prob-dict-loop)\n<a id='prob-unique'></a>\n### Problem 13: Checking assumptions about your data\n[Table of Contents](#toc)\n\nYou can use df.info() to general idea about the data and then you can investigate the remaining columns to see if the\ndata is as you expect.",
"_____no_output_____"
]
],
[
[
"# this will give a quick overview of the data frame to give you an idea of where to start looks\nprint('total rows in dataframe:', len(df))\ndf.info()",
"_____no_output_____"
],
[
"# Hint: Check your assumptions about values dataframe\ndf.year.unique()\ncolumns = list(df.columns)\nfor column in columns:\n unique_val = eval('df.' + column + '.unique()')\n print(column, ':\\nunique values:\\n', unique_val, '\\n\\n')",
"_____no_output_____"
]
],
[
[
"[Back to Problem](#prob-unique)\n<a id='summary-stats-sol'></a>\n### Problem 14: Slice and save summary statistics\n\nSelect two countries of your interest. Slice the `df` to select only these countries. Then, obtain summary statistics by country, and save to a file.",
"_____no_output_____"
]
],
[
[
"# My Solution\nmy_countries = df.loc[['China','Germany'],'pop':]\nmy_countries.describe().to_csv('china_germany_summ_stats.csv')\nmy_countries",
"_____no_output_____"
]
],
[
[
"[Back to Problem](#summary-stats)\n<a id='python-2'></a>\n## Intro to Python II: A Tool for Programming \n[Table of Contents](#toc)\n \n**Prerequisites:** Intro to Python 1: Data OR knowledge of another programming language \n\nThis workshop will help attendees build on previous knowledge of Python or other programming language in order to harness the powers of Python to make your computer work for you. You will learn how to write their own Python functions, save their code as scripts that can be called from future projects and build a workflow to chain multiple scripts together.\n\n**Learning Objectives:**\n1. Understand the syntax of python functions\n1. Understand the basics of scripting in python\n1. Understand data analysis cycles\n\n**Learning Outcomes:** you will be able to…\n1. Write your own functions\n1. Save code as a script\n1. Build a workflow",
"_____no_output_____"
],
[
"<a id='python-2-setup'></a>\n## Setup if you are joining in for Python II\n[Table of Contents](#toc)\n\n**Run the next three code cells to have the data you need to work with in this section.**",
"_____no_output_____"
]
],
[
[
"# import libraries \nimport pandas as pd",
"_____no_output_____"
],
[
"# Create a dictionary with rainfall, temperature and pressure\ndata={'rainfall_inches':[1.34,1.56,4.33],\n 'temperature_F':[75,80,96],\n 'pressure_psi':[10,2,35]}\ndata",
"_____no_output_____"
],
[
"string = \"waterfall\"\nprint(string[::-1])",
"_____no_output_____"
]
],
[
[
"<a id='functions'></a>\n## Functions:\n[Table of Contents](#toc)\n\nCreate your own functions, especially if you need to make the same operation many times. This will make you code cleaner.\n\n* Functions are known by many names in other languages. Most commonly methods and subroutines.\n* A function has a contract that guarantees certain output based on certain input(s)\n* Variables get passed into the function\n* The function then preforms actions based on the variables that are passed\n* A new value is returned from the function\n\nIn python we are able to define a function with `def`. First you define the function and later you call the defined function. \n\nHere we define a function that we will call \"add_two_numbers\"\n* def add_two_numbers():",
"_____no_output_____"
]
],
[
[
"# this defines our function\ndef add_two_numbers():\n answer = 50 + 15\n return answer",
"_____no_output_____"
],
[
"# this calls the function and stores in the variable `x`\nx = add_two_numbers()\nx",
"_____no_output_____"
]
],
[
[
"That function seems a little silly because we could just add 50 and 15 easier than defining a function to do it for us. However, imagine 50 was some constant that we need to add observations to. Now we could rewrite the function to accept an observation to add to our constant of 50.",
"_____no_output_____"
]
],
[
[
"# this defines our function\n# the \"num1\" inside the parentheses means it is expecting us to pass a value to the function when we call it\ndef add_to_constant(num1):\n answer = 50 + num1\n return answer",
"_____no_output_____"
],
[
"# this calls the function and stores in the variable `y`\n# the value we want to pass goes inside the parentheses in the call\ny = add_to_constant(10)\ny",
"_____no_output_____"
]
],
[
[
"Change the value that you pass to the function to see how it works.\n<a id='why-functions'></a>\n### Why Use Functions?\n[Table of Contents](#toc)\n\nFunctions let us break down our programs into smaller bits that can be reused and tested.\n\nHuman beings can only keep a few items in working memory at a time. we can only understand larger/more complicated ideas\n by understanding smaller pieces and combining them. Functions serve the same purpose in programs. We encapsulate\n complexity so that we can treat it as a single “thing” and this enables reusablility. Write code one time, but use\n many times in our program or programs.\n\n1. Testability\n\n* Imagine a really big program with lots of lines of code. There is a problem somewhere in the code because you are\nnot getting the results you expect.\n\n* How do you find the problem in your code?\n * If your program is composed of lots of small functions that only do one thing then you can test each function individually.\n\n2. Reusability\n\n* Imagine a really big program with lots of lines of code. There is a section of code you want to use in a different part of the program.\n\n* How do you reuse that part of the code?\n * If you just have one big program then you have to copy and paste that bit of code where you want it to go, but\n if that bit was a function, you could just use that function again.\n\n3. Writing cleaner code\n\n* Always keep both of these concepts in mind when writing programs.\n* Write small functions that do one thing.\n* Never have one giant function that does a million things.\n* A well written script is composed of lots of functions that do one thing.",
"_____no_output_____"
],
[
"<a id='str-reverse-func'></a>\n### Let's revist the reverse string and turn it into a function\n[Table of Contents](#toc)",
"_____no_output_____"
]
],
[
[
"# Create the function\ndef reverse_text(string):\n \"\"\"Function to reverse text in strings.\n \n \"\"\"\n result=string[::-1]\n return result",
"_____no_output_____"
],
[
"# Call the function and pass a string as input\nreverse_text(\"waterfall\")",
"_____no_output_____"
],
[
"# you can also pass a variable to function\noriginal='pool'\nreverse_text(original)",
"_____no_output_____"
]
],
[
[
"This may seem trivial, but we could use a function like to ask a user for a word that they would like to see written\nin reverse. Each time the input is given, we would run the same code to return the reversed spelling of the word they\n gave us.\n\n<a id='temp-func'></a>\n### Let's look at a real world example of where constants could be used in functions\n[Table of Contents](#toc)",
"_____no_output_____"
]
],
[
[
"# Create the function\ndef convert_temp(temperature,unit):\n \"\"\"Function to convert temperature from F to C, and vice-versa.\n Need temperature (integer or float) and unit (string, uppercase F or C)\n \n \"\"\"\n t=int(temperature)\n u=str(unit)\n \n if u == 'C':\n fahr=(9/5*t)+32\n print('{}C is {}F'.format(t,int(fahr)))\n \n elif u == 'F': # or else:\n celsius=(t-32)*5/9\n print('{}F is {}C'.format(t,int(celsius)))",
"_____no_output_____"
],
[
"convert_temp(85,'C')",
"_____no_output_____"
],
[
"# Using the question mark following the function name, we see information about the function and how we might use it\nconvert_temp?",
"_____no_output_____"
],
[
"# will demonstrate this depending on time\n\ndef convert_temp2():\n \"\"\"Function to convert temperature from F to C, and vice-versa.\n User input.\n \n \"\"\"\n t=int(input('Enter temperature:'))\n u=str(input('Enter unit (F or C):'))\n \n if u == 'C':\n fahr=9/5*t+32\n return '{}C is {}F'.format(t,int(fahr))\n \n elif u == 'F':\n celsius=(t-32)*5/9\n return '{}F is {}C'.format(t,int(celsius))\n \n else:\n return \"Don't know how to convert...\"",
"_____no_output_____"
],
[
"convert_temp2()",
"_____no_output_____"
],
[
"convert_temp2()",
"_____no_output_____"
],
[
"convert_temp2()",
"_____no_output_____"
]
],
[
[
"<a id='scripting'></a>\n## Scripting\n[Table of Contents](#toc)\n",
"_____no_output_____"
],
[
"For this section we are going to open the other Jupyter Notebook found in our repository to ensure we are starting\nwith a clean slate.\n\n1. Save your progress in the current notebook and you may want download a copy for your records as well which can be\ndone using the `File` menu.\n1. `Go to File > Open > scripting_practice.ipynb` to open the notebook.",
"_____no_output_____"
],
[
"<a id='errors'></a>\n## Common Errors\n[Table of Contents](#toc)\n\n",
"_____no_output_____"
],
[
"### Help yourself",
"_____no_output_____"
]
],
[
[
"help(print)",
"_____no_output_____"
],
[
"help(len)",
"_____no_output_____"
],
[
"?len",
"_____no_output_____"
],
[
"?data",
"_____no_output_____"
],
[
"dir(data)",
"_____no_output_____"
]
],
[
[
"```\nhelp(your_data_object)\ndir(your_data_object) \n```",
"_____no_output_____"
],
[
"### Variable errors",
"_____no_output_____"
]
],
[
[
"# need to create/define a variable before using it\nchocolate_cake",
"_____no_output_____"
],
[
"# this also includes mispellings...\nfirst_name='Nathalia'",
"_____no_output_____"
],
[
"firt_name",
"_____no_output_____"
]
],
[
[
"### Syntax errors",
"_____no_output_____"
]
],
[
[
"# Syntax errors: when you forget to close a ) \n## EOF - end of file\n## means that the end of your source code was reached before all code blocks were completed\nprint(len(first_name)",
"_____no_output_____"
],
[
"print(len(first_name))",
"_____no_output_____"
],
[
"# Syntax errors: when you forgot a , \ntires=4\nprint('My car has'tires,' tires')",
"_____no_output_____"
],
[
"# Syntax errors: forgot to close a quote ' in a string\n## EOL = end of line\nprint('My car has',tires,' tires)",
"_____no_output_____"
],
[
"tires=4\nprint('My car has',tires,' tires')",
"_____no_output_____"
],
[
"# Syntax errors: when you forget the colon at the end of a line\ndata=[1,2,3,4]\n\nfor i in data\n print(i**2)",
"_____no_output_____"
],
[
"# Indentation errors: forgot to indent\nfor i in data:\nprint(i**2)",
"_____no_output_____"
],
[
"for i in data:\n print(i**2)",
"_____no_output_____"
]
],
[
[
"### Index errors",
"_____no_output_____"
]
],
[
[
"groceries=['banana','cheese','bread']",
"_____no_output_____"
],
[
"groceries[3]",
"_____no_output_____"
]
],
[
[
"### Character in strings are IMMUTABLE",
"_____no_output_____"
]
],
[
[
"fruit='mango'",
"_____no_output_____"
],
[
"fruit[3]",
"_____no_output_____"
],
[
"fruit[3]='G'",
"_____no_output_____"
]
],
[
[
"### Item in list is MUTABLE",
"_____no_output_____"
]
],
[
[
"fruits=['mango','cherry']",
"_____no_output_____"
],
[
"fruits[1]",
"_____no_output_____"
],
[
"fruits[1]='apple'",
"_____no_output_____"
],
[
"fruits",
"_____no_output_____"
]
],
[
[
"### Character in item of a list is IMMUTABLE",
"_____no_output_____"
]
],
[
[
"fruits[1]",
"_____no_output_____"
],
[
"fruits[1][2]",
"_____no_output_____"
],
[
"fruits[1][2]='P'",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cb46439bde03b5a9582e833c00b2cfaaedd50b01 | 737 | ipynb | Jupyter Notebook | pset_loops/dict_loops/tests/nb/test2.ipynb | mottaquikarim/pydev-psets | 9749e0d216ee0a5c586d0d3013ef481cc21dee27 | [
"MIT"
] | 5 | 2019-04-08T20:05:37.000Z | 2019-12-04T20:48:45.000Z | pset_loops/dict_loops/tests/nb/test2.ipynb | mottaquikarim/pydev-psets | 9749e0d216ee0a5c586d0d3013ef481cc21dee27 | [
"MIT"
] | 8 | 2019-04-15T15:16:05.000Z | 2022-02-12T10:33:32.000Z | pset_loops/dict_loops/tests/nb/test2.ipynb | mottaquikarim/pydev-psets | 9749e0d216ee0a5c586d0d3013ef481cc21dee27 | [
"MIT"
] | 2 | 2019-04-10T00:14:42.000Z | 2020-02-26T20:35:21.000Z | 17.97561 | 46 | 0.462687 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
cb4654e0efccff50515220c7dcf410dd90ebafeb | 2,294 | ipynb | Jupyter Notebook | tutorials/W0D4_Calculus/student/W0D4_Outro.ipynb | ofou/course-content | 04cc6450ee20c57c7832f86da6826e516d2daeed | [
"CC-BY-4.0",
"BSD-3-Clause"
] | null | null | null | tutorials/W0D4_Calculus/student/W0D4_Outro.ipynb | ofou/course-content | 04cc6450ee20c57c7832f86da6826e516d2daeed | [
"CC-BY-4.0",
"BSD-3-Clause"
] | null | null | null | tutorials/W0D4_Calculus/student/W0D4_Outro.ipynb | ofou/course-content | 04cc6450ee20c57c7832f86da6826e516d2daeed | [
"CC-BY-4.0",
"BSD-3-Clause"
] | null | null | null | 27.638554 | 275 | 0.605057 | [
[
[
"<a href=\"https://colab.research.google.com/github/NeuromatchAcademy/course-content/blob/master/tutorials/W0D3_LinearAlgebra/student/W0D3_Outro.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>[](https://kaggle.com/kernels/welcome?src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W0D3_LinearAlgebra/student/W0D3_Outro.ipynb)",
"_____no_output_____"
],
[
"# Outro\n",
"_____no_output_____"
],
[
"**Our 2021 Sponsors, including Presenting Sponsor Facebook Reality Labs**\n\n<p align='center'><img src='https://github.com/NeuromatchAcademy/widgets/blob/master/sponsors.png?raw=True'/></p>",
"_____no_output_____"
],
[
"## Daily survey\n\nDon't forget to complete your reflections and content check in the daily survey! Please be patient after logging in as there is\na small delay before you will be redirected to the survey.\n\n<a href=\"https://portal.neuromatchacademy.org/api/redirect/to/bdd6ad66-3e51-49e0-91b4-be4158f55bd7\"><img src=\"../static/button.png\" alt=\"button link to survey\" style=\"width:410px\"></a>\n",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cb465cfcc0219484d1b503fe04398773730e83f3 | 1,581 | ipynb | Jupyter Notebook | svm/svm-base.ipynb | lybenson/ML | a9cdc04f01c3d1d266bf5e578cfdb3b1073b3279 | [
"MIT"
] | null | null | null | svm/svm-base.ipynb | lybenson/ML | a9cdc04f01c3d1d266bf5e578cfdb3b1073b3279 | [
"MIT"
] | null | null | null | svm/svm-base.ipynb | lybenson/ML | a9cdc04f01c3d1d266bf5e578cfdb3b1073b3279 | [
"MIT"
] | null | null | null | 23.597015 | 288 | 0.605313 | [
[
[
"# 支持向量机(SVM)\n\n支持向量机(英语:support vector machine,常简称为SVM)是在分类与回归分析中分析数据的监督式学习模型与相关的学习算法。给定一组训练实例,每个训练实例被标记为属于两个类别中的一个或另一个,SVM训练算法创建一个将新的实例分配给两个类别之一的模型,使其成为非概率二元线性分类器。SVM模型是将实例表示为空间中的点,这样映射就使得单独类别的实例被尽可能宽的明显的间隔分开。然后,将新的实例映射到同一空间,并基于它们落在间隔的哪一侧来预测所属类别。\n\n假设某些给定的数据点各自属于两个类之一,而目标是确定新数据点将在哪个类中。对于支持向量机来说,数据点被视为 ${\\displaystyle p}$ 维向量,而我们想知道是否可以用 ${\\displaystyle (p-1)}$ 维超平面来分开这些点。这就是所谓的**线性分类器**。可能有许多超平面可以把数据分类。最佳超平面的一个合理选择是以最大间隔把两个类分开的超平面。因此,我们要选择能够让到每边最近的数据点的距离最大化的超平面。如果存在这样的超平面,则称为**最大间隔超平面**,而其定义的线性分类器被称为最大间隔分类器,或者叫做最佳稳定性感知器。\n\n\n一个线性分类器的学习目标便是要在n维的数据空间中找到一个超平面(hyper plane),这个超平面的方程可以表示为\n\n$\nw^Tx + b = 0\n$\n\n\nhard margin\n\nsoft margin",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
cb466537fb6c937cd6ddceb47e8e1429e30cb15e | 21,590 | ipynb | Jupyter Notebook | notebooks/Chapter_11/03_Code_Breaking.ipynb | choldgraf/prob140 | 0750fc62fb114220035278ed2161e4b82ddca15f | [
"MIT"
] | null | null | null | notebooks/Chapter_11/03_Code_Breaking.ipynb | choldgraf/prob140 | 0750fc62fb114220035278ed2161e4b82ddca15f | [
"MIT"
] | null | null | null | notebooks/Chapter_11/03_Code_Breaking.ipynb | choldgraf/prob140 | 0750fc62fb114220035278ed2161e4b82ddca15f | [
"MIT"
] | null | null | null | 39.326047 | 496 | 0.555628 | [
[
[
"# HIDDEN\nfrom datascience import *\nfrom prob140 import *\nimport numpy as np\nimport matplotlib.pyplot as plt\nplt.style.use('fivethirtyeight')\n%matplotlib inline\nimport math\nfrom scipy import stats\nfrom scipy import misc\nfrom itertools import permutations",
"_____no_output_____"
],
[
"# HIDDEN\n# The alphabet\nalph = make_array('a', 'd', 't')",
"_____no_output_____"
],
[
"# HIDDEN\n# Decode atdt using all possible decoders\nx1 = [['a', 't', 'd', 't'], ['a','d','t','d'], ['d','t','a','t']]\nx2 = [['d','a','t','a'], ['t','d','a','d'], ['t','a','d','a']]\ndecoded = x1+x2",
"_____no_output_____"
],
[
"# HIDDEN\ndecoding = Table().with_columns(\n 'Decoder', list(permutations(alph)),\n 'atdt Decoded', decoded\n)",
"_____no_output_____"
],
[
"# HIDDEN\n# Make bigram transition matrix\n# Data from Peter Norvig's bigram table\n\naa = 1913489177\ndd = 6513992572\ntt = 19222971337\nad = 23202347740\nda = 23279747379\nat = 80609883139\nta = 42344542093\ndt = 10976756096\ntd = 3231292348\n\nrow1 = make_array(aa, ad, at)/sum([aa, ad, at])\nrow2 = make_array(da, dd, dt)/sum([da, dd, dt])\nrow3 = make_array(ta, td, tt)/sum([ta, td, tt])\nrows = np.append(np.append(row1, row2), row3)\n",
"_____no_output_____"
],
[
"# HIDDEN\nbigrams = MarkovChain.from_table(Table().states(alph).transition_probability(rows))",
"_____no_output_____"
]
],
[
[
"## Code Breaking ##",
"_____no_output_____"
],
[
"While it is interesting that many Markov Chains are reversible, the examples that we have seen so far haven't explained what we get by reversing a chain. After all, if it looks the same running forwards as it does backwards, why not just run it forwards? Why bother with reversibility?\n\nIt turns out that reversing Markov Chains can help solve a class of problems that are intractable by other methods. In this section we present an example of how such problems arise. In the next section we discuss a solution.",
"_____no_output_____"
],
[
"### Assumptions ###\nPeople have long been fascinated by encryption and decryption, well before cybersecurity became part of our lives. Decoding encrypted information can be complex and computation intensive. Reversed Markov Chains can help us in this task.\n\nTo get a sense of one approach to solving such problems, and of the extent of the task, let's try to decode a short piece of text that has been encoded using a simple code called a *substituion code*. Text is written in an *alphabet*, which you can think of as a set of letters and punctuation. In a substitution code, each letter of the alphabet is simply replaced by another in such a way that the code is just a permutation of the alphabet.\n\nTo decode a message encrypted by a substitution code, you have to *invert* the permutation that was used. In other words, you have to apply a permutation to the *coded* message in order to recover the original text. We will call this permutation the *decoder*.\n\nTo decode a textual message, we have to make some assumptions. For example, it helps to know the language in which the message was written, and what combinations of letters are common in that language. For example, suppose we try to decode a message that was written in English and then encrypted. If our decoding process ends up with \"words\" like zzxtf and tbgdgaa, we might want to try a different way.\n\nSo we need data about which sequences of letters are common. Such data are now increasingly easy to gather; see for example this [web page](http://norvig.com/ngrams/) by [Peter Norvig](http://norvig.com), a Director of Research at Google. ",
"_____no_output_____"
],
[
"### Decoding a Message ###\nLet's see how we can use such an approach to decode a message. For simplicity, suppose our alphabet consists of only three letters: a, d, and t. Now suppose we get the coded message atdt. We believe it's an English word. How can we go about decoding it in a manner that can be replicated by a computer for other words too?\n\nAs a first step, we will write down all 3! = 6 possible permutations of the letters in the alphabet and use each one to decode the message. The table `decoding` contains all the results. Each entry in the `Decoder` column is a permutation that we will apply to our coded text atdt. The permutation determines which letters we will use as substitutes in our decoding process. \n\nTo see how to do this, start by keeping the alphabet in \"alphabetical\" order in your head: 'a', 'd', 't'. Now look at the rows of the table.\n\n- The decoder in the first row is ['a', 'd', 't']. This decoder simply leaves the letters unchanged; atdt gets decoded as atdt. \n$$\n\\text{Decoder ['a', 'd', 't']: } ~~~ a \\to a, ~~~ d \\to d, ~~~ t \\to t\n$$\n\n- The decoder in the second row is ['a', 't', 'd']. This keeps the first letter of the alphabet 'a' unchanged, but replaces the second letter 'd' by 't' and the third letter 't' by 'd'.\n$$\n\\text{Decoder ['a', 't', 'd']: } ~~~ a \\to a, ~~~ d \\to t, ~~~ t \\to d\n$$\nSo atdt gets decoded as adtd.\n\nYou can read the rest of the table in the same way. \n\nNotice that in each decoded message, a letter appears twice, at indices 1 and 3. That's the letter being used to decode t in atdt. A feature of substitution codes is that each letter *original* is coded by a letter *code*, with the same letter *code* being used every time the letter *original* appears in the text. So the decoder must have the same feature.",
"_____no_output_____"
]
],
[
[
"decoding",
"_____no_output_____"
]
],
[
[
"Which one of these decoders should we use? To make this decision, we have to know something about the frequency of letter transitions in English. Our goal will be to pick the decoder according to the frequency of the decoded word.\n\nWe have put together some data on the frequency of the different *bigrams*, or two-letter combinations, in English. Here is a transition matrix called `bigrams` that is a gross simplification of available information about bigrams in English; we used Peter Norvig's bigrams table and restricted it to our three-letter alphabet. The row corresponding to the letter 'a' assumes that about 2% of the bigrams that start with 'a' are 'aa', about 22% are 'ad', and the remaining 76% are 'at'. \n\nIt makes sense that the 'aa' transitions are rare; we don't use words like aardvark very often. Even 2% seems large until you remember that it is the proportion of 'aa' transitions only among transitions 'aa', 'ad', and 'at', because we have restricted the alphabet. If you look at its proportion among all $26\\times26$ bigrams, that will be much lower.",
"_____no_output_____"
]
],
[
[
"bigrams",
"_____no_output_____"
]
],
[
[
"Now think of the true text as a path of a Markov Chain that has this transition matrix. An interesting historical note is that this is what Markov did when he first came up with the process that now bears his name – he analyzed the transitions between vowels and consonants in *Eugene Onegin*, Alexander Pushkin's novel written in verse.\n\nIf the true text is tada, then we can think of the sequence tada as the path of a Markov chain. Its probability can be calculated at $P(t)P(t, a)P(a, d)P(d, a)$. We will give each decoder a score based on this probability. Higher scores correspond to better decoders.\n\nTo assign the score, we assume that all three letters are equally likely to start the path. For three common letters in the alphabet, this won't be far from the truth. That means the probability of each path will start with a factor of 1/3, which we can ignore because all we are trying to do is rank all the probabilities. We will just calculate $P(t, a)P(a, d)P(d, a)$ which is about 8%. \n\nAccording to our `decoding` table above, tada is the result we get by applying the decoder ['t', 'd', 'a'] to our data atdt. For now, we will say that *the score of this decoder, given the data*, is 8%. Later we will introduce more formal calculations and terminology.",
"_____no_output_____"
]
],
[
[
"# score of decoder ['t', 'd', 'a']\n0.653477 * 0.219458 * 0.570995",
"_____no_output_____"
]
],
[
[
"To automate such calcuations we can use the `prob_of_path` method. Remember that its first argument is the initial state, and the second argument is a list or array consisting of the remaining states in sequence.",
"_____no_output_____"
]
],
[
[
"bigrams.prob_of_path('t', ['a', 'd', 'a'])",
"_____no_output_____"
]
],
[
[
"Should we decide that our message atdt should be decoded as tada? Perhaps, if we think that 8% is a high likelihood. But what if some other possible decoder has a higher likelihood? In that case it would be natural to prefer that one.\n\nSo we are going to need the probabilities of each of the six \"decoded\" paths.\n\nLet's define a function `score` that will take a list or array of characters and return the probability of the corresponding path using the `bigrams` transition matrix. In our example, this is the same as returning the score of the corresponding decoder.",
"_____no_output_____"
]
],
[
[
"def score(x):\n return bigrams.prob_of_path(x[0], x[1:])",
"_____no_output_____"
]
],
[
[
"Here are the results in decreasing order of score. There is a clear winner: the decoder ['d', 't', 'a'] corresponding to the message 'data' has more than twice the score of any other decoder.",
"_____no_output_____"
]
],
[
[
"decoding = decoding.with_column('Score of Decoder', decoding.apply(score, 1))\ndecoding.sort('Score of Decoder', descending=True)",
"_____no_output_____"
]
],
[
[
"### The Size of the Problem ###\nWhat we have been able to do with an alphabet of three characters becomes daunting when the alphabet is larger. The 52 lower case and upper case letters, along with a space character and all the punctuations, form an alphabet of around 70 characters. That gives us 70! different decoders to consider. In theory, we have to find the likelihood of each of these 70! candidates and sort them.\n\nHere is the number 70!. That's a lot of decoders. Our computing system can't handle that many, and other systems will have the same problem.",
"_____no_output_____"
]
],
[
[
"math.factorial(70)",
"_____no_output_____"
]
],
[
[
"One potential solution is to sample at random from these 70! possible decoders and just pick from among the sampled permutations. But how should we draw from 70! items? It's not a good idea to choose uniform random permutations of the alphabet, as those are unlikely to get us quickly to the desired solution. \n\nWhat we would really like our sampling procedure to do is to choose good decoders with high probability. A good decoder is one that generates text that has higher probability than text produced by almost all other decoders. In other words, a good decoder has higher likelihood than other decoders, given the data.\n\nYou can write down this likelihood using Bayes' Rule. Let $S$ represent the space of all possible permutations; if the alphabet has $N$ characters, then $S$ has $N!$ elements. For any randomly picked permutation $j$, the likelihood of that decoder given the data is:\n\n\\begin{align*}\n\\text{Likelihood of } j \\text{ given the encoded text}\n&= \\frac{\\frac{1}{N!} P(\\text{encoded text} \\mid \\text{decoder = }j)}\n{ {\\sum_{i \\in S} } \\frac{1}{N!} P(\\text{encoded text} \\mid \\text{decoder = }i)} \\\\ \\\\\n&=\\frac{P(\\text{encoded text} \\mid \\text{decoder = }j)}\n{ {\\sum_{i \\in S} } P(\\text{encoded text} \\mid \\text{decoder = }i)}\n\\end{align*}\n\nFor the given encoded text, the denominator is the normalizing constant that makes all the likelihoods sum to 1. It appears in the likelihood of every decoder. In our example with the three-letter alphabet, we ignored it because we could figure out the numerators for all six decoders and just compare them. The numerator was what we called the *score* of the decoder.\n\nEven when the alphabet is large, for any particular decoder $j$ we can find the numerator by multiplying transition probabilities sequentially, as we did in our example. But with a large alphabet we can't do this for all possible decoders, so we can't list all possible scores and we can't add them all up. Therefore we don't know the denominator of the likelihoods, not even upto a decent approximation.\n\nWhat we need now is a method that helps us draw from a probability distribution even when we don't know the normalizing constant. That is what Markov Chain Monte Carlo helps us to do.",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cb466ebf93e8c26ac67359459cb4c0116acedafa | 58,421 | ipynb | Jupyter Notebook | chatbot_tutorial.ipynb | vndee/pytorch-vi | 454f309e856f186504ba8e087601b51ab3738ac1 | [
"MIT"
] | 1 | 2019-08-19T07:52:21.000Z | 2019-08-19T07:52:21.000Z | chatbot_tutorial.ipynb | vndee/pytorch-vi | 454f309e856f186504ba8e087601b51ab3738ac1 | [
"MIT"
] | null | null | null | chatbot_tutorial.ipynb | vndee/pytorch-vi | 454f309e856f186504ba8e087601b51ab3738ac1 | [
"MIT"
] | null | null | null | 47.496748 | 753 | 0.527978 | [
[
[
"<a href=\"https://colab.research.google.com/github/vndee/pytorch-vi/blob/master/chatbot_tutorial.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"## CHATBOT\n**Tác giả**: [Matthew Inkawhich](https://github.com/MatthewInkawhich)\n\nTrong hướng dẫn này chúng ta sẽ khám phá một ứng dụng thú vị của mô hình seq2seq. Chúng ta sẽ huấn luyện một chatbot đơn giản sử dụng data là lời thoại trong phim từ [Cornell Movie-Dialogs Corpus](https://www.cs.cornell.edu/~cristian/Cornell_Movie-Dialogs_Corpus.html)\n\nCác mô hình có khả năng đàm thoại là một mảng nghiên cứu đang rất được chú ý của trí tuệ nhân tạo. Chatbot có thể tìm thấy trong rất nhiều sản phẩm tiện ích như bộ phận chăm sóc khách hàng hoặc các dịch vụ tư vấn online. Nhưng con bot này thường thuộc dạng retrieval-based (dựa trên truy xuất), đó là các mô hình mà câu trả lời đã được định sẵn cho mỗi loại câu hỏi nhất định. Dạy một cỗ máy để nó có khả năng đàm thoại với con người một cách tự nhiên vẫn là một bài toán khó và còn xa để đi đến lời giải. Gần đây, đi theo sự bùng nổ của học sâu, các mô hình sinh mạnh mẽ như Google's Neural Conversational Model đã tạo ra một bước nhảy vọt ấn tượng. Trong bài hướng dẫn này, chúng ta sẽ hiện thực một kiểu mô hình sinh như vậy với PyTorch.\n\n\n\n```\n> hello?\nBot: hello .\n> where am I?\nBot: you re in a hospital .\n> who are you?\nBot: i m a lawyer .\n> how are you doing?\nBot: i m fine .\n> are you my friend?\nBot: no .\n> you're under arrest\nBot: i m trying to help you !\n> i'm just kidding\nBot: i m sorry .\n> where are you from?\nBot: san francisco .\n> it's time for me to leave\nBot: i know .\n> goodbye\nBot: goodbye .\n```\n\n### Các phần chính:\n- Load và tiền xử lý [Cornell Movie-Dialogs Corpus](https://www.cs.cornell.edu/~cristian/Cornell_Movie-Dialogs_Corpus.html) dataset.\n- Hiện thực mô hình seq2seq với Luong's attention.\n- Phối hợp huấn luyện mô hình encoder-decoder với mini-batches.\n- Hiện thực thuật toán decoding bằng tìm kiếm tham lam.\n- Tương tác với mô hình đã huấn luyện.\n\n### Lời cảm ơn:\nCode trong bài viết này được mượn từ các project mã nguồn mở sau:\n- Yuan-Kuei Wu’s pytorch-chatbot implementation: https://github.com/ywk991112/pytorch-chatbot\n- Sean Robertson’s practical-pytorch seq2seq-translation example: https://github.com/spro/practical-pytorch/tree/master/seq2seq-translation\n- FloydHub’s Cornell Movie Corpus preprocessing code: https://github.com/floydhub/textutil-preprocess-cornell-movie-corpus",
"_____no_output_____"
],
[
"## Chuẩn bị\nĐầu tiên chúng ta cần tải dữ liệu tại [đây](https://www.cs.cornell.edu/~cristian/Cornell_Movie-Dialogs_Corpus.html) và giải nén.",
"_____no_output_____"
]
],
[
[
"!wget --header 'Host: www.cs.cornell.edu' --user-agent 'Mozilla/5.0 (X11; Ubuntu; Linux x86_64; rv:66.0) Gecko/20100101 Firefox/66.0' --header 'Accept: text/html,application/xhtml+xml,application/xml;q=0.9,*/*;q=0.8' --header 'Accept-Language: en-US,en;q=0.5' --header 'Upgrade-Insecure-Requests: 1' 'http://www.cs.cornell.edu/~cristian/data/cornell_movie_dialogs_corpus.zip' --output-document 'cornell_movie_dialogs_corpus.zip'\n!unzip cornell_movie_dialogs_corpus.zip",
"--2019-05-22 05:32:16-- http://www.cs.cornell.edu/~cristian/data/cornell_movie_dialogs_corpus.zip\nResolving www.cs.cornell.edu (www.cs.cornell.edu)... 132.236.207.20\nConnecting to www.cs.cornell.edu (www.cs.cornell.edu)|132.236.207.20|:80... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 9916637 (9.5M) [application/zip]\nSaving to: ‘cornell_movie_dialogs_corpus.zip’\n\ncornell_movie_dialo 100%[===================>] 9.46M 1.13MB/s in 8.4s \n\n2019-05-22 05:32:30 (1.13 MB/s) - ‘cornell_movie_dialogs_corpus.zip’ saved [9916637/9916637]\n\nArchive: cornell_movie_dialogs_corpus.zip\n creating: cornell movie-dialogs corpus/\n inflating: cornell movie-dialogs corpus/.DS_Store \n creating: __MACOSX/\n creating: __MACOSX/cornell movie-dialogs corpus/\n inflating: __MACOSX/cornell movie-dialogs corpus/._.DS_Store \n inflating: cornell movie-dialogs corpus/chameleons.pdf \n inflating: __MACOSX/cornell movie-dialogs corpus/._chameleons.pdf \n inflating: cornell movie-dialogs corpus/movie_characters_metadata.txt \n inflating: cornell movie-dialogs corpus/movie_conversations.txt \n inflating: cornell movie-dialogs corpus/movie_lines.txt \n inflating: cornell movie-dialogs corpus/movie_titles_metadata.txt \n inflating: cornell movie-dialogs corpus/raw_script_urls.txt \n inflating: cornell movie-dialogs corpus/README.txt \n inflating: __MACOSX/cornell movie-dialogs corpus/._README.txt \n"
],
[
"!ls cornell\\ movie-dialogs\\ corpus",
"chameleons.pdf\t\t movie_lines.txt\t\t README.txt\nmovie_characters_metadata.txt movie_titles_metadata.txt\nmovie_conversations.txt raw_script_urls.txt\n"
]
],
[
[
"Import một số thư viện hỗ trợ:",
"_____no_output_____"
]
],
[
[
"from __future__ import absolute_import\nfrom __future__ import division\nfrom __future__ import print_function\nfrom __future__ import unicode_literals\n\nimport torch\nfrom torch.jit import script, trace\nimport torch.nn as nn\nfrom torch import optim\nimport torch.nn.functional as F\nimport csv\nimport random\nimport re\nimport os\nimport unicodedata\nimport codecs\nfrom io import open\nimport itertools\nimport math\n\nUSE_CUDA = torch.cuda.is_available()\ndevice = torch.device(\"cuda\" if USE_CUDA else \"cpu\")",
"_____no_output_____"
]
],
[
[
"## Load và tiền xử lý dữ liệu\nBước tiếp theo chúng ta cần tổ chức lại dữ liệu. Cornell Movie-Dialogs Corpus là một tập dữ liệu lớn gồm các đoạn hội thoại của các nhân vật trong phim.\n- 220,579 đoạn hội thoại của 10,292 cặp nhân vật.\n- 9,035 nhân vật từ 617 bộ phim.\n- 304,713 cách diễn đạt.\n\nTập dữ liệu này rất lớn và phân tán, đa dạng trong phong cách ngôn ngữ, thời gian, địa điểm cũng như ý nghĩa. Chúng ta hi vọng mô hình của mình sẽ đủ tốt để làm việc với nhiều cách nói hay truy vấn khác nhau.\nTrước hết, hãy xem một vài dòng từ dữ liệu gốc, xem chúng ta có gì ở đây.",
"_____no_output_____"
]
],
[
[
"corpus_name = 'cornell movie-dialogs corpus'\n\ndef printLines(file, n=10):\n with open(file, 'rb') as datafile:\n lines = datafile.readlines()\n for line in lines[:n]:\n print(line)\n\nprintLines(os.path.join(corpus_name, 'movie_lines.txt'))",
"b'L1045 +++$+++ u0 +++$+++ m0 +++$+++ BIANCA +++$+++ They do not!\\n'\nb'L1044 +++$+++ u2 +++$+++ m0 +++$+++ CAMERON +++$+++ They do to!\\n'\nb'L985 +++$+++ u0 +++$+++ m0 +++$+++ BIANCA +++$+++ I hope so.\\n'\nb'L984 +++$+++ u2 +++$+++ m0 +++$+++ CAMERON +++$+++ She okay?\\n'\nb\"L925 +++$+++ u0 +++$+++ m0 +++$+++ BIANCA +++$+++ Let's go.\\n\"\nb'L924 +++$+++ u2 +++$+++ m0 +++$+++ CAMERON +++$+++ Wow\\n'\nb\"L872 +++$+++ u0 +++$+++ m0 +++$+++ BIANCA +++$+++ Okay -- you're gonna need to learn how to lie.\\n\"\nb'L871 +++$+++ u2 +++$+++ m0 +++$+++ CAMERON +++$+++ No\\n'\nb'L870 +++$+++ u0 +++$+++ m0 +++$+++ BIANCA +++$+++ I\\'m kidding. You know how sometimes you just become this \"persona\"? And you don\\'t know how to quit?\\n'\nb'L869 +++$+++ u0 +++$+++ m0 +++$+++ BIANCA +++$+++ Like my fear of wearing pastels?\\n'\n"
]
],
[
[
"Để thuận tiện, chúng ta sẽ tổ chức lại dữ liệu theo một format mỗi dòng trong file sẽ được tách ra bởi dấu tab cho một câu hỏi và một câu trả lời.\n\nPhía dưới chúng ta sẽ cần một số phương thức để phân tích dữ liệu từ file movie_lines.tx\n- `loadLines': Tách mỗi dòng dữ liệu thành một đối tượng dictionary trong python gồm các thuộc tính (lineID, characterID, movieID, character, text).\n-`loadConversations`: Nhóm các thuộc tính của từng dòng trong `loadLines` thành một đoạn hội thoại dựa trên movie_conversations.txt.\n- `extractSentencePairs`: Trích xuất một cặp câu trong đoạn hội thoại.",
"_____no_output_____"
]
],
[
[
"# Splits each line of the file into a dictionary of fields\ndef loadLines(fileName, fields):\n lines = {}\n with open(fileName, 'r', encoding='iso-8859-1') as f:\n for line in f:\n values = line.split(\" +++$+++ \")\n # Extract fields\n lineObj = {}\n for i, field in enumerate(fields):\n lineObj[field] = values[i]\n lines[lineObj['lineID']] = lineObj\n return lines\n\n\n# Groups fields of lines from `loadLines` into conversations based on *movie_conversations.txt*\ndef loadConversations(fileName, lines, fields):\n conversations = []\n with open(fileName, 'r', encoding='iso-8859-1') as f:\n for line in f:\n values = line.split(\" +++$+++ \")\n # Extract fields\n convObj = {}\n for i, field in enumerate(fields):\n convObj[field] = values[i]\n # Convert string to list (convObj[\"utteranceIDs\"] == \"['L598485', 'L598486', ...]\")\n lineIds = eval(convObj[\"utteranceIDs\"])\n # Reassemble lines\n convObj[\"lines\"] = []\n for lineId in lineIds:\n convObj[\"lines\"].append(lines[lineId])\n conversations.append(convObj)\n return conversations\n\n\n# Extracts pairs of sentences from conversations\ndef extractSentencePairs(conversations):\n qa_pairs = []\n for conversation in conversations:\n # Iterate over all the lines of the conversation\n for i in range(len(conversation[\"lines\"]) - 1): # We ignore the last line (no answer for it)\n inputLine = conversation[\"lines\"][i][\"text\"].strip()\n targetLine = conversation[\"lines\"][i+1][\"text\"].strip()\n # Filter wrong samples (if one of the lists is empty)\n if inputLine and targetLine:\n qa_pairs.append([inputLine, targetLine])\n return qa_pairs\n",
"_____no_output_____"
]
],
[
[
"Bây giờ chúng ta sẽ gọi các phương thức ở trên để tạo ra một file dữ liệu mới tên là formatted_movie_lines.txt.",
"_____no_output_____"
]
],
[
[
"# Define path to new file\ndatafile = os.path.join(corpus_name, 'formatted_movie_lines.txt')\n\ndelimiter = '\\t'\n# Unescape the delimiter\ndelimiter = str(codecs.decode(delimiter, 'unicode_escape'))\n\n# Initialize lines dict, conversations list, and field ids\nlines = {}\nconversations = []\nMOVIE_LINES_FIELDS = [\"lineID\", \"characterID\", \"movieID\", \"character\", \"text\"]\nMOVIE_CONVERSATIONS_FIELDS = [\"character1ID\", \"character2ID\", \"movieID\", \"utteranceIDs\"]\n\n# Load lines and process conversations\nprint(\"\\nProcessing corpus...\")\nlines = loadLines(os.path.join(corpus_name, \"movie_lines.txt\"), MOVIE_LINES_FIELDS)\nprint(\"\\nLoading conversations...\")\nconversations = loadConversations(os.path.join(corpus_name, \"movie_conversations.txt\"),\n lines, MOVIE_CONVERSATIONS_FIELDS)\n\n# Write new csv file\nprint(\"\\nWriting newly formatted file...\")\nwith open(datafile, 'w', encoding='utf-8') as outputfile:\n writer = csv.writer(outputfile, delimiter=delimiter, lineterminator='\\n')\n for pair in extractSentencePairs(conversations):\n writer.writerow(pair)\n\n# Print a sample of lines\nprint(\"\\nSample lines from file:\")\nprintLines(datafile)",
"\nProcessing corpus...\n\nLoading conversations...\n\nWriting newly formatted file...\n\nSample lines from file:\nb\"Can we make this quick? Roxanne Korrine and Andrew Barrett are having an incredibly horrendous public break- up on the quad. Again.\\tWell, I thought we'd start with pronunciation, if that's okay with you.\\n\"\nb\"Well, I thought we'd start with pronunciation, if that's okay with you.\\tNot the hacking and gagging and spitting part. Please.\\n\"\nb\"Not the hacking and gagging and spitting part. Please.\\tOkay... then how 'bout we try out some French cuisine. Saturday? Night?\\n\"\nb\"You're asking me out. That's so cute. What's your name again?\\tForget it.\\n\"\nb\"No, no, it's my fault -- we didn't have a proper introduction ---\\tCameron.\\n\"\nb\"Cameron.\\tThe thing is, Cameron -- I'm at the mercy of a particularly hideous breed of loser. My sister. I can't date until she does.\\n\"\nb\"The thing is, Cameron -- I'm at the mercy of a particularly hideous breed of loser. My sister. I can't date until she does.\\tSeems like she could get a date easy enough...\\n\"\nb'Why?\\tUnsolved mystery. She used to be really popular when she started high school, then it was just like she got sick of it or something.\\n'\nb\"Unsolved mystery. She used to be really popular when she started high school, then it was just like she got sick of it or something.\\tThat's a shame.\\n\"\nb'Gosh, if only we could find Kat a boyfriend...\\tLet me see what I can do.\\n'\n"
]
],
[
[
"### Đọc và cắt dữ liệu\nSau khi đã tổ chức lại dữ liệu, chúng ta cần tạo một từ điển các từ dùng trong tập dữ liệu và đọc các cặp câu truy vấn - phản hồi vào bộ nhớ.\n\nChú ý rằng chúng ta xem một câu là một chuỗi liên tiếp các **từ**, không có một ánh xạ ngầm nào của nó ở một không gian số học rời rạc. Do đó chúng ta cần phải tạo một hàm ánh xạ sao cho mỗi từ riêng biệt chỉ có duy nhất một giá trị chỉ số đại diện chính là vị trí của nó trong từ điển.\n\nĐể làm điều đó chúng ta định nghĩa lớp `Voc`, nơi sẽ lưu một dictionary ánh xạ **từ** sang **chỉ số**, một dictionary ánh xạ ngược **chỉ số** sang **từ**, một biến đếm cho mỗi từ và một biến đếm tổng số các từ. Lớp `Voc` cũng cung cắp các phương thức để thêm một từ vào từ điển (`addWord`), thêm tất cả các từ trong một câu (`addSentence`) và lược bỏ (trimming) các từ không thường gặp. Chúng ta sẽ nói về trimming sau:\n",
"_____no_output_____"
]
],
[
[
"# Default word tokens\nPAD_token = 0 # Used for padding short sentences\nSOS_token = 1 # Start-of-sentence token\nEOS_token = 2 # End-of-sentence token\n\nclass Voc:\n def __init__(self, name):\n self.name = name\n self.trimmed = False\n self.word2index = {}\n self.word2count = {}\n self.index2word = {PAD_token: \"PAD\", SOS_token: \"SOS\", EOS_token: \"EOS\"}\n self.num_words = 3 # Count SOS, EOS, PAD\n\n def addSentence(self, sentence):\n for word in sentence.split(' '):\n self.addWord(word)\n\n def addWord(self, word):\n if word not in self.word2index:\n self.word2index[word] = self.num_words\n self.word2count[word] = 1\n self.index2word[self.num_words] = word\n self.num_words += 1\n else:\n self.word2count[word] += 1\n\n # Remove words below a certain count threshold\n def trim(self, min_count):\n if self.trimmed:\n return\n self.trimmed = True\n\n keep_words = []\n\n for k, v in self.word2count.items():\n if v >= min_count:\n keep_words.append(k)\n\n print('keep_words {} / {} = {:.4f}'.format(\n len(keep_words), len(self.word2index), len(keep_words) / len(self.word2index)\n ))\n\n # Reinitialize dictionaries\n self.word2index = {}\n self.word2count = {}\n self.index2word = {PAD_token: \"PAD\", SOS_token: \"SOS\", EOS_token: \"EOS\"}\n self.num_words = 3 # Count default tokens\n\n for word in keep_words:\n self.addWord(word)",
"_____no_output_____"
]
],
[
[
"Trước khi đưa vào huấn luyện ta cần một số thao tác tiền xử lý dữ liệu. Đầu tiên, chúng ta cần chuyển đổi các chuỗi Unicode thành ASCII sử dụng `unicodeToAscii`. Tiếp theo phải chuyển tất cả các kí tự thành chữ viết thường và lược bỏ các kí tự không ở trong bảng chữ cái ngoại trừ một số dấu câu (`normalizedString`). Cuối cùng để giúp quá trình huấn luyện nhanh chóng hội tụ chúng ta sẽ lọc ra các câu có độ dài lớn hơn ngưỡng `MAX_LENGTH` (`filterPairs`).",
"_____no_output_____"
]
],
[
[
"MAX_LENGTH = 10 # Maximum sentence length to consider\n\n# Turn a Unicode string to plain ASCII, thanks to\n# https://stackoverflow.com/a/518232/2809427\ndef unicodeToAscii(s):\n return ''.join(\n c for c in unicodedata.normalize('NFD', s)\n if unicodedata.category(c) != 'Mn'\n )\n\n# Lowercase, trim, and remove non-letter characters\ndef normalizeString(s):\n s = unicodeToAscii(s.lower().strip())\n s = re.sub(r\"([.!?])\", r\" \\1\", s)\n s = re.sub(r\"[^a-zA-Z.!?]+\", r\" \", s)\n s = re.sub(r\"\\s+\", r\" \", s).strip()\n return s\n\n# Read query/response pairs and return a voc object\ndef readVocs(datafile, corpus_name):\n print(\"Reading lines...\")\n # Read the file and split into lines\n lines = open(datafile, encoding='utf-8').\\\n read().strip().split('\\n')\n # Split every line into pairs and normalize\n pairs = [[normalizeString(s) for s in l.split('\\t')] for l in lines]\n voc = Voc(corpus_name)\n return voc, pairs\n\n# Returns True iff both sentences in a pair 'p' are under the MAX_LENGTH threshold\ndef filterPair(p):\n # Input sequences need to preserve the last word for EOS token\n return len(p[0].split(' ')) < MAX_LENGTH and len(p[1].split(' ')) < MAX_LENGTH\n\n# Filter pairs using filterPair condition\ndef filterPairs(pairs):\n return [pair for pair in pairs if filterPair(pair)]\n\n# Using the functions defined above, return a populated voc object and pairs list\ndef loadPrepareData(corpus_name, datafile, save_dir):\n print(\"Start preparing training data ...\")\n voc, pairs = readVocs(datafile, corpus_name)\n print(\"Read {!s} sentence pairs\".format(len(pairs)))\n pairs = filterPairs(pairs)\n print(\"Trimmed to {!s} sentence pairs\".format(len(pairs)))\n print(\"Counting words...\")\n for pair in pairs:\n voc.addSentence(pair[0])\n voc.addSentence(pair[1])\n print(\"Counted words:\", voc.num_words)\n return voc, pairs\n\n\n# Load/Assemble voc and pairs\nsave_dir = os.path.join(\"save\")\nvoc, pairs = loadPrepareData(corpus_name, datafile, save_dir)\n# Print some pairs to validate\nprint(\"\\npairs:\")\nfor pair in pairs[:10]:\n print(pair)",
"Start preparing training data ...\nReading lines...\nRead 221282 sentence pairs\nTrimmed to 64271 sentence pairs\nCounting words...\nCounted words: 18008\n\npairs:\n['there .', 'where ?']\n['you have my word . as a gentleman', 'you re sweet .']\n['hi .', 'looks like things worked out tonight huh ?']\n['you know chastity ?', 'i believe we share an art instructor']\n['have fun tonight ?', 'tons']\n['well no . . .', 'then that s all you had to say .']\n['then that s all you had to say .', 'but']\n['but', 'you always been this selfish ?']\n['do you listen to this crap ?', 'what crap ?']\n['what good stuff ?', 'the real you .']\n"
]
],
[
[
"Một chiến thuật khác để giúp mô hình học nhanh hơn đó là lược bỏ các từ hiếm gặp trong dữ liệu. Việc này giúp làm giảm đi độ khó của bài toán, và do đó mô hình sẽ hội tụ nhanh hơn. Chúng ta sẽ làm điều này bằng 2 bước.\n- Lược bỏ các từ với tần suất xuất hiện ít hơn `MIN_COUNT` sử dụng phương thức `voc.trim`.\n- Lược bỏ các cặp câu hội thoại có chứa từ bị cắt ở bước trên.\n",
"_____no_output_____"
]
],
[
[
"MIN_COUNT = 3 # Minimum word count threshold for trimming\n\ndef trimRareWords(voc, pairs, MIN_COUNT):\n # Trim words used under the MIN_COUNT from the voc\n voc.trim(MIN_COUNT)\n # Filter out pairs with trimmed words\n keep_pairs = []\n for pair in pairs:\n input_sentence = pair[0]\n output_sentence = pair[1]\n keep_input = True\n keep_output = True\n # Check input sentence\n for word in input_sentence.split(' '):\n if word not in voc.word2index:\n keep_input = False\n break\n # Check output sentence\n for word in output_sentence.split(' '):\n if word not in voc.word2index:\n keep_output = False\n break\n\n # Only keep pairs that do not contain trimmed word(s) in their input or output sentence\n if keep_input and keep_output:\n keep_pairs.append(pair)\n\n print(\"Trimmed from {} pairs to {}, {:.4f} of total\".format(len(pairs), len(keep_pairs), len(keep_pairs) / len(pairs)))\n return keep_pairs\n\n\n# Trim voc and pairs\npairs = trimRareWords(voc, pairs, MIN_COUNT)",
"keep_words 7823 / 18005 = 0.4345\nTrimmed from 64271 pairs to 53165, 0.8272 of total\n"
]
],
[
[
"## Chuẩn bị dữ liệu cho mô hình\n\nMặc dù ở trên chúng ta đã làm rất nhiều thứ để có một bộ dữ liệu tốt gồm các cặp câu hội thoại, từ điển. Nhưng mô hình của chúng ta luôn mong đợi dữ liệu vào của nó phải là numerical torch tensor. Cách để chuyển dữ liệu dạng này thành tensor có thể tìm thấy ở bài viết [seq2seq translation tutorial](https://pytorch.org/tutorials/intermediate/seq2seq_translation_tutorial.html). Trong bài viết này chúng ta chỉ dùng batch size bằng 1, tất cả những gì chúng ta phải làm là chuyển tất cả các từ trong một cặp câu thành chỉ số tương ứng của nó trong từ điển và đưa vào mô hình huấn luyện.\n\nTuy nhiên, nếu muốn quá trình huấn luyện nhanh hơn và tận dụng được khả năng tính toán song song của GPU chúng ta nên huấn luyện theo mini-batches.\n\nSử dụng mini-batches thì cần phải chú ý rằng các câu trong một batch có thể sẽ có độ dài không giống nhau. Vì vậy chúng ta nên đặt số chiều của các tensor batch cố định là (max_length, batch_size). Các câu có độ dài nhỏ hơn max_length sẽ được thêm zero padding phía sau kí tự EOS_token (kí tự kết thúc câu).\n\nMột vấn đề khác đặt ra là nếu chúng ta chuyển tất cả các từ của một cặp câu vào một batch tensor, lúc này tensor của chúng ta sẽ có kích thước là (max_length, batch_size). Tuy nhiên cái chúng ta cần là một tensor với kích thước (batch_size, max_length) và lúc đó cần phải hiện thực thêm một phướng thức để chuyển vị ma trận. Thay vì rườm ra như vậy, chúng ta sẽ thực hiện việc chuyển vị đó ngay từ trong hàm `zeroPadding`.\n\n\n",
"_____no_output_____"
]
],
[
[
"def indexesFromSentence(voc, sentence):\n return [voc.word2index[word] for word in sentence.split(' ')] + [EOS_token]\n\ndef zeroPadding(l, fillvalue=PAD_token):\n return list(itertools.zip_longest(*l, fillvalue=fillvalue))\n\ndef binaryMatrix(l, value=PAD_token):\n m = []\n for i, seq in enumerate(l):\n m.append([])\n for token in seq:\n if token == PAD_token:\n m[i].append(0)\n else:\n m[i].append(1)\n \n return m\n\n# Returns padded input sequene tensor and lengths\ndef inputVar(l, voc):\n indexes_batch = [indexesFromSentence(voc, sentence) for sentence in l]\n lengths = torch.tensor([len(indexes) for indexes in indexes_batch])\n padList = zeroPadding(indexes_batch)\n padVar = torch.LongTensor(padList)\n return padVar, lengths\n\n# Returns padded target sequence tensor, padding mask, and max target length\ndef outputVar(l, voc):\n indexes_batch = [indexesFromSentence(voc, sentence) for sentence in l]\n max_target_len = max([len(indexes) for indexes in indexes_batch])\n padList = zeroPadding(indexes_batch)\n mask = binaryMatrix(padList)\n mask = torch.ByteTensor(mask)\n padVar = torch.LongTensor(padList)\n return padVar, mask, max_target_len\n\ndef batch2TrainData(voc, pair_batch):\n pair_batch.sort(key=lambda x: len(x[0].split(' ')), reverse=True)\n input_batch, output_batch = [], []\n for pair in pair_batch:\n input_batch.append(pair[0])\n output_batch.append(pair[1])\n \n inp, lengths = inputVar(input_batch, voc)\n output, mask, max_target_len = outputVar(output_batch, voc)\n return inp, lengths, output, mask, max_target_len\n\n# Example for validation\nsmall_batch_size = 5\nbatches = batch2TrainData(voc, [random.choice(pairs) for _ in range(small_batch_size)])\ninput_variable, lengths, target_variable, mask, max_target_len = batches\n\nprint('input_variable:', input_variable)\nprint('lengths:', lengths)\nprint('target_variable:', target_variable)\nprint('mask:', mask)\nprint('max_target_len:', max_target_len)",
"input_variable: tensor([[ 112, 726, 5, 242, 48],\n [ 12, 3610, 37, 188, 1626],\n [ 130, 4, 53, 45, 4],\n [ 4, 758, 606, 140, 2],\n [ 34, 1205, 6, 4, 0],\n [ 7, 4, 2, 2, 0],\n [ 197, 2, 0, 0, 0],\n [ 117, 0, 0, 0, 0],\n [ 4, 0, 0, 0, 0],\n [ 2, 0, 0, 0, 0]])\nlengths: tensor([10, 7, 6, 6, 4])\ntarget_variable: tensor([[ 27, 25, 7, 167, 25],\n [ 14, 296, 118, 6, 112],\n [ 67, 66, 70, 2, 94],\n [123, 25, 606, 0, 117],\n [ 21, 296, 6, 0, 4],\n [ 22, 66, 2, 0, 2],\n [ 4, 2, 0, 0, 0],\n [ 2, 0, 0, 0, 0]])\nmask: tensor([[1, 1, 1, 1, 1],\n [1, 1, 1, 1, 1],\n [1, 1, 1, 1, 1],\n [1, 1, 1, 0, 1],\n [1, 1, 1, 0, 1],\n [1, 1, 1, 0, 1],\n [1, 1, 0, 0, 0],\n [1, 0, 0, 0, 0]], dtype=torch.uint8)\nmax_target_len: 8\n"
]
],
[
[
"##Định nghĩa mô hình\n###Mô hình Seq2Seq\n\nBộ não chatbot của chúng ta là một mô hình sequence-to-sequence (seq2seq). Mục tiêu của mô hình seq2seq là nhận một chuỗi đầu vào và dự đoán chuỗi đầu ra dựa trên mô mô hình cố định.\n\n[Sutskever và các cộng sự](https://arxiv.org/abs/1409.3215) đã đề xuất một phương pháp dựa trên hai mô hình mạng nơ-ron hồi quy (RNN) có thể giải quyết được bài toán này. Một RNN hoạt động như một encoder (bộ mã hóa), encoder có nhiệm vụ mã hóa chuỗi đầu vào thành một context vector (vector ngữ cảnh). Trên lý thuyết, context vector (layer cuối cùng của RNN) sẽ chứa các thông tin ngữ nghĩa của chuỗi đầu vào. RNN thứ hai là decoder (bộ giải mã), nó dùng context vector của encoder để dự đoán chuỗi đầu ra tương ứng.\n\n\n\n*Nguồn ảnh: https://jeddy92.github.io/JEddy92.github.io/ts_seq2seq_intro/*\n\n###Encoder\n\nBộ mã hóa sử dụng mạng nơ-ron hồi quy (encoder RNN) duyệt qua từng token của chuỗi đầu vào, tại mỗi thời điểm xuất ra một \"output\" vector và một \"hidden state\" vector. Hidden state vector sau đó sẽ được dùng để tính hidden state vector tại thời điểm tiếp theo như trong ý tưởng cơ bản của RNN. Mạng encoder sẽ cố gắn g chuyển đổi những cái gì nó nhìn thấy trong chuỗi đầu vào bao gồm cả ngữ cảnh và ngữ nghĩa thành một tập hợp các điểm trong một không gian nhiều chiều, nơi decoder nhìn vào để giải mã chuỗi đầu ra có ý nghĩa.\n\nTrái tim của encoder là multi-layered Gate Recurrent Unit, được đề xuất bởi [Cho và các cộng sư](https://arxiv.org/pdf/1406.1078v3.pdf) vào năm 2014. Chúng ta sẽ dùng dạng hai chiều của GRU, đồng nghĩa với việc có 2 mạng RNN độc lập: một đọc chuỗi đầu vào theo một thứ tự từ trái sáng phải, một từ phải sang trái.\n\n\n\n*Nguồn ảnh: https://colah.github.io/posts/2015-09-NN-Types-FP/*\n\nChú ý rằng `embedding` layer được dùng để mã hóa từng từ trong câu văn đầu vào thành một vector trong không gian ngữ nghĩa của nó.\n\nCuối cùng, nếu đưa một batch dữ liệu vào RNN, chúng ta cần phải \"unpack\" zeros padding xung quanh của từng chuỗi. \n\n####Các bước tính toán\n1. Chuyển word index thành embedding vector.\n2. Đóng gói các câu thành một các batch.\n3. Đưa từng batch qua GRU để tính toán.\n4. Unpack padding.\n5. Cộng tất cả các output của GRU hai chiều.\n6. Trả về kết quả và hidden state cuối cùng.\n\n####Input:\n- `input_seq`: batch of input sentences, kích thước (max_length, batch_size)\n- `input_lengths`: Danh sách chứa độ dài câu tương ứng với từng câu trong batch, kích thước (batch_size)\n- `hidden`: hidden state, kích thước (n_layers * num_directions, batch_size, hidden_size)\n\n####Output:\n- `output`: Layer của cuối cùng của GRU, kích thước (max_length, batch_size, hidden_size)\n- `hidden`: cập nhật hidden state từ GRU, kích thước (n_layers * num_directions, batch_size, hidden_size)",
"_____no_output_____"
]
],
[
[
"class EncoderRNN(nn.Module):\n def __init__(self, hidden_size, embedding, n_layers=1, dropout=0):\n super(EncoderRNN, self).__init__()\n self.n_layers = n_layers\n self.hidden_size = hidden_size\n self.embedding = embedding\n \n # Initialize GRU; the input_size and hidden_size params are both set to \n # 'hidden_size' because our input size is a word embedding with number \n # of features == hidden_size\n self.gru = nn.GRU(hidden_size, hidden_size, n_layers,\n dropout=(0 if n_layers == 1 else dropout), bidirectional=True)\n \n def forward(self, input_seq, input_lengths, hidden=None):\n # Convert word indexes to embedding vector\n embedded = self.embedding(input_seq)\n \n # Pack padded batch of sequences for RNN module\n packed = nn.utils.rnn.pack_padded_sequence(embedded, input_lengths)\n \n # Forward pass through GRU\n outputs, hidden = self.gru(packed, hidden)\n \n # Unpack padding\n outputs, _ = nn.utils.rnn.pad_packed_sequence(outputs)\n \n # Sum bidirectional GRU outputs\n output = outputs[:, :, :self.hidden_size] + outputs[:, :, self.hidden_size:]\n \n # Return output and final hidden state\n return outputs, hidden",
"_____no_output_____"
]
],
[
[
"###Decoder\nBộ giải mã RNN sẽ sinh ra chuỗi đầu ra theo từng token. Nó sử dụng context vector của encoder và hidden state để sinh từ tiếp theo trong chuỗi đầu ra cho đến khi gặp phải EOS_token (kí hiệu kết thúc câu). Một vấn đề với bài toán seq2seq truyền thống đó là nếu chỉ dùng context vector và hidden state thì sẽ bị mất mát thông tin, đặc biệt là với những câu dài.\n\nĐể đối phó với điều đó, [Bahdanau](https://arxiv.org/abs/1409.0473) đã đề xuất một phương pháp gọi là cơ chế attention. Cơ chế này cho phép decoder đặt sự chú ý lên một vài điểm nhất định trong câu thay vì nhìn các từ với mức độ quan trọng y như nhau. \n\nAttention được tính toán dựa vào hidden state hiện tại của decoder và kết quả của encoder. Bộ trọng số của attention có cùng kích thước với chuồi đầu vào.\n\n\n\n[Luong](https://arxiv.org/abs/1508.04025) attention là một phiên bản cải tiến với ý tưởng \"Global attention\". Sự khác biệt là với \"Global attention\" chúng ta sẽ nhìn tất cả các hidden state của encoder, thay vì chỉ nhìn hidden state cuối cùng của encoder như của Bahdanau. Một khác biệt nữa là \"global attention\" tính dựa trên duy nhất hidden state hiện tại của decoder chứ không như phiên bản của Bahdanau cần phải tính qua hidden state tại các bước trước đó.\n\n\n\nTrong đó: $h_{t}$ là hidden state hiện tại của decoder và $h_{s}$ là toàn bộ hidden state của encoder.\n\nNhìn chung, global attention có thể tổng hợp như hình bên dưới.\n\n",
"_____no_output_____"
]
],
[
[
"# Luong attention layer\nclass Attn(nn.Module):\n def __init__(self, method, hidden_size):\n super(Attn, self).__init__()\n self.method = method\n if self.method not in ['dot', 'general', 'concat']:\n raise ValueError(self.method, 'is not an appropriate attention method.')\n self.hidden_size = hidden_size\n \n if self.method == 'general':\n self.attn = nn.Linear(self.hidden_size, hidden_size)\n \n elif self.method == 'concat':\n self.attn = nn.Linear(self.hidden_size * 2, hidden_size)\n self.v = nn.Parameter(torch.FloatTensor(hidden_size))\n \n def dot_score(self, hidden, encoder_output):\n return torch.sum(hidden * encoder_ouput, dim=2)\n \n def general_score(self, hidden, encoder_output):\n energy = self.attn(encoder_output)\n return torch.sum(hidden * energy, dim=2)\n \n def concat_score(self, hidden, encoder_outputs):\n energy = self.attn(torch.cat((hidden.expand(encoder_output.size(0), -1, -1),\n encoder_ouputs), 2)).tanh()\n \n return torch.sum(self.v * energy, dim=2)\n \n def forward(self, hidden, encoder_outputs):\n # Calculate the attention weights (energies) based on the given method\n if self.method == 'general':\n attn_energies = self.general_score(hidden, encoder_outputs)\n elif self.method == 'concat':\n attn_energies = self.concat_score(hidden, encoder_outputs)\n elif self.method == 'dot':\n attn_energies = self.dot_score(hidden, encoder_outputs)\n \n # Transpose max_length and batch_size dimensions\n attn_energies = attn_energies.t()\n \n # Return the softmax normalized probability scores (with added dimension)\n return F.softmax(attn_energies, dim=1).unsqueeze(1)",
"_____no_output_____"
]
],
[
[
"####Các bước tính toán\n1. Lấy embedding vector của từ hiện tại\n2. Đưa dữ liệu qua GRU hai chiều để tính toán\n3. Tính trọng số attention từ output của GRU\n4. Nhân trọng số của attention của encoder output để có được trọng số mới của context vector.\n5. Nối (concat) context vector và GRU hidden state như trong công thức của Luong attention.\n6. Dự đoán từ tiếp theo dựa trên Luong attention\n7. Trả về kết quả và hidden state cuối cùng\n\n####Inputs:\n- `input_step`: Một step là một đơn vị thời gian, kích thước (1, batch_size)\n- `last_hidden`: hidden layer cuối của GRU, kích thước (n_layers * num_directión, batch_size, hidden_size)\n- `encoder_outputs`: encoder output, kích thước (max_length, batch_size, hidden_size)\n\n####Outputs:\n- `output`: softmax normalized tensor, kích thước (batch_size, voc.num_words)\n- `hidden`: hidden state cuối của GRU, kích thước (n_layers * num_directions, batch_size, hidden_size)",
"_____no_output_____"
]
],
[
[
"class LuongAttnDecoderRNN(nn.Module):\n def __init__(self, attn_model, embedding, hidden_size, output_size, n_layers=1, dropout=0.1):\n super(LuongAttnDecoderRNN, self).__init__()\n\n # Keep for reference\n self.attn_model = attn_model\n self.hidden_size = hidden_size\n self.output_size = output_size\n self.n_layers = n_layers\n self.dropout = dropout\n\n # Define layers\n self.embedding = embedding\n self.embedding_dropout = nn.Dropout(dropout)\n self.gru = nn.GRU(hidden_size, hidden_size, n_layers, dropout=(0 if n_layers == 1 else dropout))\n self.concat = nn.Linear(hidden_size * 2, hidden_size)\n self.out = nn.Linear(hidden_size, output_size)\n\n self.attn = Attn(attn_model, hidden_size)\n\n def forward(self, input_step, last_hidden, encoder_outputs):\n # Note: we run this one step (word) at a time\n # Get embedding of current input word\n embedded = self.embedding(input_step)\n embedded = self.embedding_dropout(embedded)\n \n # Forward through unidirectional GRU\n rnn_output, hidden = self.gru(embedded, last_hidden)\n \n # Calculate attention weights from the current GRU output\n attn_weights = self.attn(rnn_output, encoder_outputs)\n \n # Multiply attention weights to encoder outputs to get new \"weighted sum\" context vector\n context = attn_weights.bmm(encoder_outputs.transpose(0, 1))\n \n # Concatenate weighted context vector and GRU output using Luong eq. 5\n rnn_output = rnn_output.squeeze(0)\n context = context.squeeze(1)\n concat_input = torch.cat((rnn_output, context), 1)\n concat_output = torch.tanh(self.concat(concat_input))\n \n # Predict next word using Luong eq. 6\n output = self.out(concat_output)\n output = F.softmax(output, dim=1)\n \n # Return output and final hidden state\n return output, hidden",
"_____no_output_____"
]
],
[
[
"##Huấn luyện\n###Masked loss\nVì chúng ta đang làm việc với batch of padded sentences, cho nên không thể dễ dàng để tính loss cho tất cả các thành phần của tensor. Chúng ta định nghĩa hàm `maskNLLLoss` để tính loss dựa trên output của decoder. Kết quả trả về là trung bình negative log likelihood của các thành phần trong tensor (mỗi thành phần là một câu).\n",
"_____no_output_____"
]
],
[
[
"def maskNLLLoss(inp, target, mask):\n nTotal = mask.sum()\n crossEntropy = -troch.log(torch.gather(inp, 1, target.view(-1, 1)).squeeze(1))\n loss = crossEntropy.masked_selected(mask).mean()\n loss = loss.to(device)\n return loss, nTotal.item()",
"_____no_output_____"
]
],
[
[
"###Training\nHàm `train` hiện thực thuật toán huấn luyện cho một lần lặp.\nChúng ta sẽ dùng một vài kỹ thuật để quá trình training diễn ra tốt hơn:\n- **Teacher forcing**: Kỹ thuật này cho phép với một xác suất được quy định sẵn `teacher_forcing_ratio`, decoder sẽ dùng target word tại thời điểm hiện tại để dự đoán từ tiếp theo thay vì dùng từ được dự đoán bởi decoder tại thời điểm hiện tại.\n- **Gradient clipping**: Đây là một kỹ thuật thường dùng để đối phố với \"exploding gradient\". Kỹ thuật này đơn giản là chặn giá trị gradient ở một ngưỡng trên, không để nó trở nên quá lớn.\n\n\n*Nguồn ảnh: Goodfellow et al. Deep Learning. 2016. https://www.deeplearningbook.org/*\n\n####Các bước tính toán\n1. Đưa toàn bộ batch vào encoder đê tính toán.\n2. Khởi tạo input cho decoder bằng SOS_token và hidden state bằng với hidden state cuối cùng của encoder.\n3. Đưa chuỗi input qua decoder.\n4. If teacher_forcing: gán input tại thời điểm tiếp theo của decoder bằng nhãn đúng của từ dự đoán hiện tại, ngược lại gán bằng từ được decoder dự đoán tại thời điểm hiện tại.\n5. Tính loss\n6. Thực hiện giải thuật lan truyền ngược.\n7. Clip gradients.\n8. Cập nhật trọng số encoder và decoder.\n\n",
"_____no_output_____"
]
],
[
[
"def train(input_variable, lengths, target_variable, mask, max_target_len, encoder, decoder, embedding,\n encoder_optimizer, decoder_optimizer, batch_size, clip, max_length=MAX_LENGTH):\n\n # Zero gradients\n encoder_optimizer.zero_grad()\n decoder_optimizer.zero_grad()\n\n # Set device options\n input_variable = input_variable.to(device)\n lengths = lengths.to(device)\n target_variable = target_variable.to(device)\n mask = mask.to(device)\n\n # Initialize variables\n loss = 0\n print_losses = []\n n_totals = 0\n\n # Forward pass through encoder\n encoder_outputs, encoder_hidden = encoder(input_variable, lengths)\n\n # Create initial decoder input (start with SOS tokens for each sentence)\n decoder_input = torch.LongTensor([[SOS_token for _ in range(batch_size)]])\n decoder_input = decoder_input.to(device)\n\n # Set initial decoder hidden state to the encoder's final hidden state\n decoder_hidden = encoder_hidden[:decoder.n_layers]\n\n # Determine if we are using teacher forcing this iteration\n use_teacher_forcing = True if random.random() < teacher_forcing_ratio else False\n\n # Forward batch of sequences through decoder one time step at a time\n if use_teacher_forcing:\n for t in range(max_target_len):\n decoder_output, decoder_hidden = decoder(\n decoder_input, decoder_hidden, encoder_outputs\n )\n # Teacher forcing: next input is current target\n decoder_input = target_variable[t].view(1, -1)\n # Calculate and accumulate loss\n mask_loss, nTotal = maskNLLLoss(decoder_output, target_variable[t], mask[t])\n loss += mask_loss\n print_losses.append(mask_loss.item() * nTotal)\n n_totals += nTotal\n else:\n for t in range(max_target_len):\n decoder_output, decoder_hidden = decoder(\n decoder_input, decoder_hidden, encoder_outputs\n )\n # No teacher forcing: next input is decoder's own current output\n _, topi = decoder_output.topk(1)\n decoder_input = torch.LongTensor([[topi[i][0] for i in range(batch_size)]])\n decoder_input = decoder_input.to(device)\n # Calculate and accumulate loss\n mask_loss, nTotal = maskNLLLoss(decoder_output, target_variable[t], mask[t])\n loss += mask_loss\n print_losses.append(mask_loss.item() * nTotal)\n n_totals += nTotal\n\n # Perform backpropatation\n loss.backward()\n\n # Clip gradients: gradients are modified in place\n _ = nn.utils.clip_grad_norm_(encoder.parameters(), clip)\n _ = nn.utils.clip_grad_norm_(decoder.parameters(), clip)\n\n # Adjust model weights\n encoder_optimizer.step()\n decoder_optimizer.step()\n\n return sum(print_losses) / n_totals",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cb46723e494f13325fa9aa2786eabc0c117ff135 | 19,263 | ipynb | Jupyter Notebook | Intraday_dist_impact.ipynb | shubhaguha/airlab-retail | bafa80b410decf12f14fe60f0bbd0c66daf003f4 | [
"MIT"
] | 12 | 2020-12-07T19:50:59.000Z | 2021-08-21T11:49:21.000Z | Intraday_dist_impact.ipynb | shubhaguha/airlab-retail | bafa80b410decf12f14fe60f0bbd0c66daf003f4 | [
"MIT"
] | null | null | null | Intraday_dist_impact.ipynb | shubhaguha/airlab-retail | bafa80b410decf12f14fe60f0bbd0c66daf003f4 | [
"MIT"
] | 4 | 2020-11-04T10:21:25.000Z | 2021-08-23T17:36:12.000Z | 109.448864 | 14,452 | 0.867726 | [
[
[
"import torch.nn as nn\nimport torch\nfrom retail import retail\nimport torch.distributions as d\nimport torch.nn.functional as F\nimport numpy as np\nimport seaborn as sns",
"_____no_output_____"
]
],
[
[
"We define the arguments used by our store. We also define the intra-day distribution as being sampled by a uniform distribution.",
"_____no_output_____"
]
],
[
[
"# Define main store arguments as well as sampler size\nn_customers = 2500\nn_buckets = 4\nmonte_carlo_size = 100\nstore_args= {'assortment_size': 1000, 'bucket_cov': torch.eye(n_buckets)/100, 'seed' : 1066,\n 'max_stock': 1000, 'forecastVariance' :0., 'horizon': 100, 'lead_time': 1}\nbucketDist = d.uniform.Uniform(0,1)",
"_____no_output_____"
],
[
"#Create the list of the average daily reward for each customer distribution for the chosen policy\nsummed_rewards_policy = []\nfor i in range(monte_carlo_size):\n sub_rewards = []\n done = False\n #Generate the store and its customer repartition throughout the day\n torch.manual_seed(i)\n sampled = bucketDist.sample((n_buckets,))\n sample_bucket_customers = (n_customers*sampled/sampled.sum()).round()\n store = retail.StoreEnv(**store_args, bucket_customers = sample_bucket_customers)\n while not (done):\n #Compute the order according to the policy \n customers = sample_bucket_customers.max()\n p = store.forecast.squeeze()\n std = torch.sqrt(customers*p+(1-p))\n order = F.relu(3*std+store.forecast.squeeze()*customers-store.get_full_inventory_position()).round()\n # Step the environment and get its observation\n obs = store.step(order.numpy())\n # Store reward for the specific time step\n sub_rewards.append(obs[1])\n done = obs[2]\n #Append average reward of this customer repartition to the list of rewards\n summed_rewards_policy.append(torch.stack(sub_rewards).mean())",
"_____no_output_____"
],
[
"sns.kdeplot(torch.stack(summed_rewards_policy).numpy())",
"_____no_output_____"
]
],
[
[
"We observe a skewed distribution, where one distribution yields very high results. \n\nLet's investigate this distribution - it corresponds to seed 49.",
"_____no_output_____"
]
],
[
[
"torch.manual_seed(49)\nsampled = bucketDist.sample((n_buckets,))\nsample_bucket_customers = (n_customers*sampled/sampled.sum()).round()\n\n\nsample_bucket_customers",
"_____no_output_____"
]
],
[
[
"All customers are coming at the end of the day. This is actually a realistic distribution, as patrons usually visit the store after their working hours. ",
"_____no_output_____"
]
],
[
[
"np.savetxt(\"imbalance_result.csv\", torch.stack(summed_rewards_policy).numpy(), delimiter=\",\")\n",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb468141483ae3d80abfaef71486539c99323557 | 12,531 | ipynb | Jupyter Notebook | content/lessons/10/Class-Coding-Lab/CCL-Dictionaries.ipynb | hvpuzzan-su/intern-project | b65e8d2cd43e21c3fdc61b7bbb8e81f5c9148f08 | [
"MIT"
] | null | null | null | content/lessons/10/Class-Coding-Lab/CCL-Dictionaries.ipynb | hvpuzzan-su/intern-project | b65e8d2cd43e21c3fdc61b7bbb8e81f5c9148f08 | [
"MIT"
] | null | null | null | content/lessons/10/Class-Coding-Lab/CCL-Dictionaries.ipynb | hvpuzzan-su/intern-project | b65e8d2cd43e21c3fdc61b7bbb8e81f5c9148f08 | [
"MIT"
] | null | null | null | 29.835714 | 1,016 | 0.539622 | [
[
[
"# In-Class Coding Lab: Dictionaries\n\nThe goals of this lab are to help you understand:\n\n - How to use Python Dictionaries\n - Basic Dictionary methods\n - Dealing with Key errors\n - How to use lists of Dictionaries\n - How to encode / decode python dictionaries to json.\n \n## Dictionaries are Key-Value Pairs.\n\nThe **key** is unique for each Python dictionary object and is always of type `str`. The **value** stored under the key can be any Python type. \n\nThis example creates a `stock` variable with two keys `symbol` and `name`. We access the dictionary key with `['keyname']`.",
"_____no_output_____"
]
],
[
[
"stock = {} # empty dictionary\nstock['symbol'] = 'AAPL'\nstock['name'] = 'Apple Computer'\nprint(stock)\nprint(stock['symbol'])\nprint(stock['name'])",
"_____no_output_____"
]
],
[
[
"While Python lists are best suited for storing multiple values of the same type ( like grades ), Python dictionaries are best suited for storing hybrid values, or values with multiple attributes.\n\nIn the example above we created an empty dictionary `{}` then assigned keys `symbol` and `name` as part of individual assignment statements.\n\nWe can also build the dictionary in a single statement, like this:",
"_____no_output_____"
]
],
[
[
"stock = { 'name' : 'Apple Computer', 'symbol' : 'AAPL', 'value' : 125.6 }\nprint(stock)\nprint(\"%s (%s) has a value of $%.2f\" %(stock['name'], stock['symbol'], stock['value']))",
"_____no_output_____"
]
],
[
[
"## Dictionaries are mutable\n\nThis means we can change their value. We can add and remove keys and update the value of keys. This makes dictionaries quite useful for storing data.",
"_____no_output_____"
]
],
[
[
"# let's add 2 new keys\nprint(\"Before changes\", stock)\nstock['low'] = 119.85\nstock['high'] = 127.0\n\n# and update the value key\nstock['value'] = 126.25\nprint(\"After change\", stock)\n",
"_____no_output_____"
]
],
[
[
"## Now you Try It!\n\nCreate a python dictionary called `car` with the following keys `make`, `model` and `price`. Set appropriate values and print out the dictionary.\n",
"_____no_output_____"
]
],
[
[
"# TODO: Write code here \nstock = {}\nstock['make'] = 'Acura'\nstock['model'] = 'MDX'\nstock['price'] = '$50,000'\nprint(stock)\nprint(stock['make'])\nprint(stock['model'])\nprint(stock['price'])",
"{'make': 'Acura', 'model': 'MDX', 'price': '$50,000'}\nAcura\nMDX\n$50,000\n"
]
],
[
[
"## What Happens when the key is not there?\n\nLet's go back to our stock example. What happens when we try to read a key not present in the dictionary? \n\nThe answer is that Python will report a `KeyError` ",
"_____no_output_____"
]
],
[
[
"print( stock['change'] )",
"_____no_output_____"
]
],
[
[
"No worries. We know how to handle run-time errors in Python... use `try except` !!!",
"_____no_output_____"
]
],
[
[
"try:\n print( stock['change'] )\nexcept KeyError:\n print(\"The key 'change' does not exist!\")",
"The key 'change' does not exist!\n"
]
],
[
[
"## Avoiding KeyError\n\nYou can avoid `KeyError` using the `get()` dictionary method. This method will return a default value when the key does not exist.\n\nThe first argument to `get()` is the key to get, the second argument is the value to return when the key does not exist. ",
"_____no_output_____"
]
],
[
[
"print(stock.get('name','no key'))\nprint(stock.get('change', 'no key'))",
"no key\nno key\n"
]
],
[
[
"## Now You try It!\n\nWrite a program to ask the user to input a key for the `stock` variable. \n\nIf the key exists, print the value, otherwise print 'Key does not exist'",
"_____no_output_____"
]
],
[
[
"# TODO: write code here\nstock = {}\nstock = input('Enter key')\ntry:\n print(stock.get('stock'))\nexcept KeyError:\n print('key does not exist')\n",
"Enter keystock\n"
]
],
[
[
"## Enumerating keys and values\n\nYou can enumerate keys and values easily, using the `keys()` and `values()` methods:",
"_____no_output_____"
]
],
[
[
"print(\"KEYS\")\nfor k in stock.keys():\n print(k)\n \nprint(\"VALUES\")\nfor v in stock.values():\n print(v)",
"KEYS\nmake\nmodel\nprice\nVALUES\nAcura\nMDX\n$50,000\n"
]
],
[
[
"## List of Dictionary\n\nThe List of Dictionary object in Python allows us to create useful in-memory data structures. It's one of the features of Python that sets it apart from other programming languages. \n\nLet's use it to build a portfolio (list of 4 stocks).",
"_____no_output_____"
]
],
[
[
"portfolio = [\n { 'symbol' : 'AAPL', 'name' : 'Apple Computer Corp.', 'value': 136.66 },\n { 'symbol' : 'AMZN', 'name' : 'Amazon.com, Inc.', 'value': 845.24 },\n { 'symbol' : 'MSFT', 'name' : 'Microsoft Corporation', 'value': 64.62 },\n { 'symbol' : 'TSLA', 'name' : 'Tesla, Inc.', 'value': 257.00 } \n]\n\nprint(\"first stock\", portfolio[0]) \nprint(\"name of first stock\", portfolio[0]['name']) \nprint(\"last stock\", portfolio[-1]) \nprint(\"value of 2nd stock\", portfolio[1]['value']) ",
"first stock {'symbol': 'AAPL', 'name': 'Apple Computer Corp.', 'value': 136.66}\nname of first stock Apple Computer Corp.\nlast stock {'symbol': 'TSLA', 'name': 'Tesla, Inc.', 'value': 257.0}\nvalue of 2nd stock 845.24\n"
]
],
[
[
"## Putting It All Together\n\nWrite a program to build out your personal stock portfolio.\n\n```\n1. Start with an empty list, called portfolio\n2. loop\n3. create a new stock dictionary\n3. input a stock symbol, or type 'QUIT' to print portfolio\n4. if symbol equals 'QUIT' exit loop\n5. add symbol value to stock dictionary under 'symbol' key\n6. input stock value as float\n7. add stock value to stock dictionary under 'value key\n8. append stock variable to portfolio list variable\n9. time to print the portfolio: for each stock in the portfolio\n10. print stock symbol and stock value, like this \"AAPL $136.66\"\n```",
"_____no_output_____"
]
],
[
[
"portfolio = []\nwhile True:\n stock = {}\n symbol = input(\"Enter a stock symbol, or type \\'Quit\\' to print portfolio.\")\n if symbol == 'Quit':\n break\n stock['symbol'] = symbol\n value = float(input('Enter the value of the stock.'))\n stock['value'] = value\n portfolio.append(stock)\nfor stock in portfolio:\n print(stock['symbol'],stock['value'],)",
"Enter a stock symbol, or type 'Quit' to print portfolio.Quit\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb46817a41eea89bd27794e07d176de199e17591 | 250,187 | ipynb | Jupyter Notebook | kaggle/machine-learning-with-a-heart/Lab4.ipynb | xR86/ml-stuff | 2a1b79408897171b78032ff2531ab6f8b18be6c4 | [
"MIT"
] | 3 | 2018-12-11T03:03:15.000Z | 2020-02-11T19:38:07.000Z | kaggle/machine-learning-with-a-heart/Lab4.ipynb | xR86/ml-stuff | 2a1b79408897171b78032ff2531ab6f8b18be6c4 | [
"MIT"
] | 6 | 2017-05-31T20:58:32.000Z | 2021-02-16T23:13:15.000Z | kaggle/machine-learning-with-a-heart/Lab4.ipynb | xR86/ml-stuff | 2a1b79408897171b78032ff2531ab6f8b18be6c4 | [
"MIT"
] | null | null | null | 62.111966 | 93,296 | 0.630632 | [
[
[
"# Tema 4.1 <a class=\"tocSkip\">",
"_____no_output_____"
],
[
"# Imports",
"_____no_output_____"
]
],
[
[
"import math\n\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport graphviz\n\nimport sklearn.tree\nimport sklearn.neighbors\nimport sklearn.naive_bayes\nimport sklearn.svm\nimport sklearn.metrics\nimport sklearn.preprocessing\nimport sklearn.model_selection",
"_____no_output_____"
]
],
[
[
"# Data\nhttps://www.drivendata.org/competitions/54/machine-learning-with-a-heart/page/109/\n- Numeric\n - slope\\_of\\_peak\\_exercise\\_st\\_segment (int, semi-categorical, 1-3)\n - resting\\_blood\\_pressure (int)\n - chest\\_pain\\_type (int, semi-categorical, 1-4)\n - num\\_major\\_vessels (int, semi-categorical, 0-3)\n - resting\\_ekg\\_results (int, semi-categorical, 0-2)\n - serum\\_cholesterol\\_mg\\_per\\_dl (int)\n - oldpeak\\_eq\\_st\\_depression (float)\n - age (int)\n - max\\_heart\\_rate\\_achieved (int)\n- Categorical\n - thal\n - normal\n - fixed\\_defect\n - reversible\\_defect\n - fasting\\_blood\\_sugar\\_gt\\_120\\_mg\\_per\\_dl (blood sugar > 120)\n - 0\n - 1\n - sex\n - 0 (f)\n - 1 (m)\n - exercise\\_induced\\_angina \n - 0\n - 1\n",
"_____no_output_____"
]
],
[
[
"features = pd.read_csv('train_values.csv')\nlabels = pd.read_csv('train_labels.csv')",
"_____no_output_____"
],
[
"features.head()",
"_____no_output_____"
],
[
"labels.head()",
"_____no_output_____"
],
[
"FEATURES = ['slope_of_peak_exercise_st_segment', \n 'thal',\n 'resting_blood_pressure', \n 'chest_pain_type', \n 'num_major_vessels', \n 'fasting_blood_sugar_gt_120_mg_per_dl',\n 'resting_ekg_results', \n 'serum_cholesterol_mg_per_dl', \n 'oldpeak_eq_st_depression', \n 'sex',\n 'age', \n 'max_heart_rate_achieved', \n 'exercise_induced_angina']\n\nLABEL = 'heart_disease_present'\n\nEXPLANATIONS = {'slope_of_peak_exercise_st_segment' : 'Quality of Blood Flow to the Heart',\n 'thal' : 'Thallium Stress Test Measuring Blood Flow to the Heart',\n 'resting_blood_pressure' : 'Resting Blood Pressure', \n 'chest_pain_type' : 'Chest Pain Type (1-4)',\n 'num_major_vessels' : 'Major Vessels (0-3) Colored by Flourosopy',\n 'fasting_blood_sugar_gt_120_mg_per_dl' : 'Fasting Blood Sugar > 120 mg/dl',\n 'resting_ekg_results' : 'Resting Electrocardiographic Results (0-2)',\n 'serum_cholesterol_mg_per_dl' : 'Serum Cholesterol in mg/dl',\n 'oldpeak_eq_st_depression' : 'Exercise vs. Rest\\nA Measure of Abnormality in Electrocardiograms',\n 'age' : 'Age (years)',\n 'sex' : 'Sex (m/f)',\n 'max_heart_rate_achieved' : 'Maximum Heart Rate Achieved (bpm)',\n 'exercise_induced_angina' : 'Exercise-Induced Chest Pain (yes/no)'}\n\nNUMERICAL_FEATURES = ['slope_of_peak_exercise_st_segment', \n 'resting_blood_pressure', \n 'chest_pain_type', \n 'num_major_vessels', \n 'resting_ekg_results', \n 'serum_cholesterol_mg_per_dl', \n 'oldpeak_eq_st_depression', \n 'age', \n 'max_heart_rate_achieved']\n\nCATEGORICAL_FEATURES = ['thal', \n 'fasting_blood_sugar_gt_120_mg_per_dl', \n 'sex', \n 'exercise_induced_angina']\n\nCATEGORICAL_FEATURE_VALUES = {'thal' : [[0, 1, 2], ['Normal', \n 'Fixed Defect', \n 'Reversible Defect']], \n 'fasting_blood_sugar_gt_120_mg_per_dl' : [[0, 1], ['No', 'Yes']],\n 'sex' : [[0, 1], ['F', 'M']], \n 'exercise_induced_angina' : [[0, 1], ['No', 'Yes']]}\n\nSEMI_CATEGORICAL_FEATURES = ['slope_of_peak_exercise_st_segment',\n 'chest_pain_type',\n 'num_major_vessels',\n 'resting_ekg_results']\n\nSEMI_CATEGORICAL_FEATURE_LIMITS = {'slope_of_peak_exercise_st_segment' : [1, 3],\n 'chest_pain_type' : [1, 4],\n 'num_major_vessels' : [0, 3],\n 'resting_ekg_results' : [0, 2]}\n\nLABEL_VALUES = [[0, 1], ['No', 'Yes']]\n",
"_____no_output_____"
],
[
"for feature in CATEGORICAL_FEATURES:\n if len(CATEGORICAL_FEATURE_VALUES[feature][0]) > 2:\n \n onehot_feature = pd.get_dummies(features[feature])\n \n feature_index = features.columns.get_loc(feature)\n features.drop(feature, axis=1, inplace=True)\n \n onehot_feature.columns = [f'{feature}={feature_value}' for feature_value in onehot_feature.columns]\n for colname in onehot_feature.columns[::-1]:\n features.insert(feature_index, colname, onehot_feature[colname])",
"_____no_output_____"
],
[
"features.head()",
"_____no_output_____"
],
[
"x = features.values[:,1:].astype(int)\ny = labels.values[:,-1].astype(int)\n\nprint('x =\\n', x)\nprint('y =\\n', y)",
"x =\n [[ 1 0 1 ... 45 170 0]\n [ 2 0 1 ... 54 158 0]\n [ 1 0 1 ... 77 162 1]\n ...\n [ 2 0 0 ... 64 131 1]\n [ 1 0 1 ... 48 175 0]\n [ 1 0 1 ... 54 163 0]]\ny =\n [0 0 1 1 0 0 1 1 1 0 0 0 1 0 1 1 0 1 0 1 0 0 0 0 0 0 0 0 1 0 0 1 1 0 1 0 0\n 0 0 1 0 0 0 0 0 1 0 0 1 0 1 0 1 1 1 0 1 1 0 1 0 1 1 0 0 1 1 0 0 1 0 0 0 0\n 1 1 0 1 1 1 0 1 1 1 0 1 0 1 0 0 0 1 0 1 0 0 1 0 0 0 0 0 1 0 0 0 0 1 0 1 0\n 0 1 1 0 0 0 1 1 1 1 0 0 0 1 0 0 0 1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 1 0 0 1\n 1 1 0 0 1 1 1 1 0 0 0 1 0 1 1 1 0 0 0 1 0 0 1 1 1 1 0 1 1 1 0 0]\n"
],
[
"stratified_kflod_validator = sklearn.model_selection.StratifiedKFold(n_splits=5, shuffle=True)\n\nstratified_kflod_validator",
"_____no_output_____"
]
],
[
[
"# Decision Trees",
"_____no_output_____"
]
],
[
[
"tree_mean_acc = 0\ntree_score_df = pd.DataFrame(columns = ['Fold', 'Accuracy', 'Precision', 'Recall'])\n\nfor fold_ind, (train_indices, test_indices) in enumerate(stratified_kflod_validator.split(x, y), 1):\n \n x_train, x_test = x[train_indices], x[test_indices]\n y_train, y_test = y[train_indices], y[test_indices]\n \n dec_tree = sklearn.tree.DecisionTreeClassifier(min_samples_split = 5)\n dec_tree.fit(x_train, y_train)\n \n acc = dec_tree.score(x_test, y_test)\n tree_mean_acc += acc\n \n y_pred = dec_tree.predict(x_test)\n precision = sklearn.metrics.precision_score(y_test, y_pred)\n recall = sklearn.metrics.recall_score(y_test, y_pred)\n \n tree_score_df.loc[fold_ind] = [f'{fold_ind}', \n f'{acc*100:.2f} %', \n f'{precision*100:.2f} %', \n f'{recall*100:.2f} %']\n \n tree_plot_data = sklearn.tree.export_graphviz(dec_tree, out_file = None,\n feature_names = features.columns[1:], \n class_names = [f'{labels.columns[1]}={label_value}' \n for label_value \n in LABEL_VALUES[1]],\n filled = True, \n rounded = True, \n special_characters = True) \n graph = graphviz.Source(tree_plot_data) \n graph.render(f'Fold {fold_ind}')\n \nnext_ind = len(tree_score_df) + 1\n\nmean_acc = tree_score_df['Accuracy'].apply(lambda n: float(n[:-2])).mean()\nmean_prec = tree_score_df['Precision'].apply(lambda n: float(n[:-2])).mean()\nmean_rec = tree_score_df['Recall'].apply(lambda n: float(n[:-2])).mean()\n\ntree_score_df.loc[next_ind] = ['Avg', f'{mean_acc:.2f} %', f'{mean_prec:.2f} %', f'{mean_rec:.2f} %']\ntree_score_df",
"_____no_output_____"
]
],
[
[
"# KNN",
"_____no_output_____"
]
],
[
[
"# TODO Normalize\n\nknn_mean_score_df = pd.DataFrame(columns = ['k', 'Avg. Accuracy', 'Avg. Precision', 'Avg. Recall'])\n\nnormalized_x = sklearn.preprocessing.normalize(x) # No improvement over un-normalized data.\n\nmean_accs = []\nfor k in list(range(1, 10)) + [math.ceil(len(features) * step) for step in [0.1, 0.2, 0.3, 0.4, 0.5]]:\n \n knn_score_df = pd.DataFrame(columns = ['Fold', 'Accuracy', 'Precision', 'Recall'])\n\n mean_acc = 0\n for fold_ind, (train_indices, test_indices) in enumerate(stratified_kflod_validator.split(x, y), 1):\n\n x_train, x_test = normalized_x[train_indices], normalized_x[test_indices]\n y_train, y_test = y[train_indices], y[test_indices]\n\n knn = sklearn.neighbors.KNeighborsClassifier(n_neighbors = k)\n knn.fit(x_train, y_train)\n\n acc = knn.score(x_test, y_test)\n mean_acc += acc\n \n y_pred = knn.predict(x_test)\n precision = sklearn.metrics.precision_score(y_test, y_pred)\n recall = sklearn.metrics.recall_score(y_test, y_pred)\n\n knn_score_df.loc[fold_ind] = [f'{fold_ind}', \n f'{acc*100:.2f} %',\n f'{precision*100:.2f} %', \n f'{recall*100:.2f} %']\n\n next_ind = len(knn_score_df) + 1\n \n mean_acc = knn_score_df['Accuracy'].apply(lambda n: float(n[:-2])).mean()\n mean_prec = knn_score_df['Precision'].apply(lambda n: float(n[:-2])).mean()\n mean_rec = knn_score_df['Recall'].apply(lambda n: float(n[:-2])).mean()\n \n knn_score_df.loc[next_ind] = ['Avg', \n f'{acc*100:.2f} %',\n f'{precision*100:.2f} %', \n f'{recall*100:.2f} %']\n \n knn_mean_score_df.loc[k] = [k, \n f'{mean_acc:.2f} %', \n f'{mean_prec:.2f} %', \n f'{mean_rec:.2f} %']\n\n# print(f'k = {k}')\n# print(knn_score_df)\n# print()\n \nbest_accuracy = knn_mean_score_df.sort_values(by = ['Avg. Accuracy']).iloc[-1]\nprint('Best avg. accuracy is', best_accuracy['Avg. Accuracy'], 'for k =', best_accuracy['k'], '.')\nknn_mean_score_df.sort_values(by = ['Avg. Accuracy'])",
"Best avg. accuracy is 62.78 % for k = 90 .\n"
]
],
[
[
"# Naive Bayes",
"_____no_output_____"
]
],
[
[
"nb_classifier_types = [sklearn.naive_bayes.GaussianNB,\n sklearn.naive_bayes.MultinomialNB,\n sklearn.naive_bayes.ComplementNB,\n sklearn.naive_bayes.BernoulliNB]\n\nnb_mean_score_df = pd.DataFrame(columns = ['Type', 'Avg. Accuracy', 'Avg. Precision', 'Avg. Recall'])\n\nfor nb_classifier_type in nb_classifier_types:\n \n nb_score_df = pd.DataFrame(columns = ['Fold', 'Accuracy', 'Precision', 'Recall'])\n\n mean_acc = 0\n for fold_ind, (train_indices, test_indices) in enumerate(stratified_kflod_validator.split(x, y), 1):\n\n x_train, x_test = x[train_indices], x[test_indices]\n y_train, y_test = y[train_indices], y[test_indices]\n\n nb = nb_classifier_type()\n nb.fit(x_train, y_train)\n\n acc = nb.score(x_test, y_test)\n mean_acc += acc\n \n y_pred = nb.predict(x_test)\n precision = sklearn.metrics.precision_score(y_test, y_pred)\n recall = sklearn.metrics.recall_score(y_test, y_pred)\n\n nb_score_df.loc[fold_ind] = [f'{fold_ind}', \n f'{acc*100:.2f} %', \n f'{precision*100:.2f} %', \n f'{recall*100:.2f} %']\n\n next_ind = len(nb_score_df) + 1\n \n mean_acc = nb_score_df['Accuracy'].apply(lambda n: float(n[:-2])).mean()\n mean_prec = nb_score_df['Precision'].apply(lambda n: float(n[:-2])).mean()\n mean_rec = nb_score_df['Recall'].apply(lambda n: float(n[:-2])).mean()\n \n nb_score_df.loc[next_ind] = ['Avg', \n f'{mean_acc:.2f} %', \n f'{mean_prec:.2f} %', \n f'{mean_rec:.2f} %']\n \n nb_mean_score_df.loc[len(nb_mean_score_df) + 1] = [nb_classifier_type.__name__, \n f'{mean_acc:.2f} %', \n f'{mean_prec:.2f} %', \n f'{mean_rec:.2f} %']\n\n print(nb_classifier_type.__name__)\n print()\n print(nb_score_df)\n print()\n \nnb_mean_score_df.sort_values(by = ['Avg. Accuracy'])",
"GaussianNB\n\n Fold Accuracy Precision Recall\n1 1 86.11 % 82.35 % 87.50 %\n2 2 77.78 % 72.22 % 81.25 %\n3 3 88.89 % 87.50 % 87.50 %\n4 4 77.78 % 72.22 % 81.25 %\n5 5 83.33 % 81.25 % 81.25 %\n6 Avg 82.78 % 79.11 % 83.75 %\n\nMultinomialNB\n\n Fold Accuracy Precision Recall\n1 1 83.33 % 91.67 % 68.75 %\n2 2 80.56 % 73.68 % 87.50 %\n3 3 77.78 % 78.57 % 68.75 %\n4 4 72.22 % 68.75 % 68.75 %\n5 5 75.00 % 68.42 % 81.25 %\n6 Avg 77.78 % 76.22 % 75.00 %\n\nComplementNB\n\n Fold Accuracy Precision Recall\n1 1 91.67 % 88.24 % 93.75 %\n2 2 75.00 % 68.42 % 81.25 %\n3 3 69.44 % 69.23 % 56.25 %\n4 4 77.78 % 70.00 % 87.50 %\n5 5 80.56 % 76.47 % 81.25 %\n6 Avg 78.89 % 74.47 % 80.00 %\n\nBernoulliNB\n\n Fold Accuracy Precision Recall\n1 1 69.44 % 69.23 % 56.25 %\n2 2 86.11 % 86.67 % 81.25 %\n3 3 80.56 % 80.00 % 75.00 %\n4 4 80.56 % 71.43 % 93.75 %\n5 5 77.78 % 72.22 % 81.25 %\n6 Avg 78.89 % 75.91 % 77.50 %\n\n"
]
],
[
[
"# SVM",
"_____no_output_____"
]
],
[
[
"svm_classifier_type = sklearn.svm.SVC\n\n# Avg.\n# Args -> acc / prec / rec\n#\n# kernel: linear -> 78.89 % 78.31 % 73.75 %\n# kernel: linear, C: 0.1 -> 84.44 % 88.54 % 75.00 %\n#\n# * No improvement for larger C.\n#\n# kernel: poly, max_iter: 1 -> 46.67 % 34.67 % 21.25 %\n# kernel: poly, max_iter: 10 -> 57.22 % 51.27 % 66.25 %\n# kernel: poly, max_iter: 100 -> 61.67 % 60.18 % 40.00 %\n# kernel: poly, max_iter: 100, coef0: 1 -> 62.22 % 62.19 % 41.25 %\n#\n# * No improvement for more iters.\n# * No improvement for larger C.\n# * No improvement for higher degree.\n# * No improvement for different coef0.\n#\n# kernel: rbf, max_iter: 10 -> 48.89 % 46.07 % 72.50 %\n# kernel: rbf, max_iter: 100 -> 60.00 % 74.00 % 17.50 %\n# kernel: rbf, max_iter: 1000 -> 60.56 % 78.33 % 15.00 %\n\n\nargs = {'kernel': 'linear', 'C': 0.1}\n\nsvm_score_df = pd.DataFrame(columns = ['Type', 'Accuracy', 'Precision', 'Recall'])\n\n# normalized_x = sklearn.preprocessing.normalize(x)\n\nmean_acc = 0\nfor fold_ind, (train_indices, test_indices) in enumerate(stratified_kflod_validator.split(x, y), 1):\n\n x_train, x_test = x[train_indices], x[test_indices]\n y_train, y_test = y[train_indices], y[test_indices]\n\n svm = svm_classifier_type(**args, gamma = 'scale', cache_size = 256)\n svm.fit(x_train, y_train)\n\n acc = svm.score(x_test, y_test)\n mean_acc += acc\n\n y_pred = svm.predict(x_test)\n precision = sklearn.metrics.precision_score(y_test, y_pred)\n recall = sklearn.metrics.recall_score(y_test, y_pred)\n\n svm_score_df.loc[fold_ind] = [f'{fold_ind}', \n f'{acc*100:.2f} %', \n f'{precision*100:.2f} %', \n f'{recall*100:.2f} %']\n\nnext_ind = len(svm_score_df) + 1\n\nmean_acc = svm_score_df['Accuracy'].apply(lambda n: float(n[:-2])).mean()\nmean_prec = svm_score_df['Precision'].apply(lambda n: float(n[:-2])).mean()\nmean_rec = svm_score_df['Recall'].apply(lambda n: float(n[:-2])).mean()\n\nsvm_score_df.loc[next_ind] = ['Avg', \n f'{mean_acc:.2f} %', \n f'{mean_prec:.2f} %', \n f'{mean_rec:.2f} %']\n\nprint(svm_score_df)",
" Type Accuracy Precision Recall\n1 1 75.00 % 81.82 % 56.25 %\n2 2 91.67 % 88.24 % 93.75 %\n3 3 86.11 % 82.35 % 87.50 %\n4 4 77.78 % 75.00 % 75.00 %\n5 5 80.56 % 80.00 % 75.00 %\n6 Avg 82.22 % 81.48 % 77.50 %\n"
]
],
[
[
"# Shallow Neural Nets",
"_____no_output_____"
],
[
"## Import deps",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\nfrom sklearn.model_selection import train_test_split\n\nimport keras\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Dropout, Activation, Flatten\nfrom keras.layers import Conv2D, MaxPooling2D\n\n\nfrom keras.layers import Input, Dense, Conv2D, MaxPooling2D, Dropout, Flatten, BatchNormalization, LeakyReLU",
"Using TensorFlow backend.\n"
]
],
[
[
"## Import data",
"_____no_output_____"
]
],
[
[
"features = pd.read_csv('train_values.csv')\nlabels = pd.read_csv('train_labels.csv')\n\nprint(labels.head())\nfeatures.head()",
" patient_id heart_disease_present\n0 0z64un 0\n1 ryoo3j 0\n2 yt1s1x 1\n3 l2xjde 1\n4 oyt4ek 0\n"
],
[
"FEATURES = ['slope_of_peak_exercise_st_segment', \n 'thal',\n 'resting_blood_pressure', \n 'chest_pain_type', \n 'num_major_vessels', \n 'fasting_blood_sugar_gt_120_mg_per_dl',\n 'resting_ekg_results', \n 'serum_cholesterol_mg_per_dl', \n 'oldpeak_eq_st_depression', \n 'sex',\n 'age', \n 'max_heart_rate_achieved', \n 'exercise_induced_angina']\n\nLABEL = 'heart_disease_present'\n\nEXPLANATIONS = {'slope_of_peak_exercise_st_segment' : 'Quality of Blood Flow to the Heart',\n 'thal' : 'Thallium Stress Test Measuring Blood Flow to the Heart',\n 'resting_blood_pressure' : 'Resting Blood Pressure', \n 'chest_pain_type' : 'Chest Pain Type (1-4)',\n 'num_major_vessels' : 'Major Vessels (0-3) Colored by Flourosopy',\n 'fasting_blood_sugar_gt_120_mg_per_dl' : 'Fasting Blood Sugar > 120 mg/dl',\n 'resting_ekg_results' : 'Resting Electrocardiographic Results (0-2)',\n 'serum_cholesterol_mg_per_dl' : 'Serum Cholesterol in mg/dl',\n 'oldpeak_eq_st_depression' : 'Exercise vs. Rest\\nA Measure of Abnormality in Electrocardiograms',\n 'age' : 'Age (years)',\n 'sex' : 'Sex (m/f)',\n 'max_heart_rate_achieved' : 'Maximum Heart Rate Achieved (bpm)',\n 'exercise_induced_angina' : 'Exercise-Induced Chest Pain (yes/no)'}\n\nNUMERICAL_FEATURES = ['slope_of_peak_exercise_st_segment', \n 'resting_blood_pressure', \n 'chest_pain_type', \n 'num_major_vessels', \n 'resting_ekg_results', \n 'serum_cholesterol_mg_per_dl', \n 'oldpeak_eq_st_depression', \n 'age', \n 'max_heart_rate_achieved']\n\nCATEGORICAL_FEATURES = ['thal', \n 'fasting_blood_sugar_gt_120_mg_per_dl', \n 'sex', \n 'exercise_induced_angina']\n\nCATEGORICAL_FEATURE_VALUES = {'thal' : [[0, 1, 2], ['Normal', \n 'Fixed Defect', \n 'Reversible Defect']], \n 'fasting_blood_sugar_gt_120_mg_per_dl' : [[0, 1], ['No', 'Yes']],\n 'sex' : [[0, 1], ['F', 'M']], \n 'exercise_induced_angina' : [[0, 1], ['No', 'Yes']]}\n\nSEMI_CATEGORICAL_FEATURES = ['slope_of_peak_exercise_st_segment',\n 'chest_pain_type',\n 'num_major_vessels',\n 'resting_ekg_results']\n\nSEMI_CATEGORICAL_FEATURE_LIMITS = {'slope_of_peak_exercise_st_segment' : [1, 3],\n 'chest_pain_type' : [1, 4],\n 'num_major_vessels' : [0, 3],\n 'resting_ekg_results' : [0, 2]}\n\nLABEL_VALUES = [[0, 1], ['No', 'Yes']]\n",
"_____no_output_____"
],
[
"for feature in CATEGORICAL_FEATURES:\n if len(CATEGORICAL_FEATURE_VALUES[feature][0]) > 2:\n \n onehot_feature = pd.get_dummies(features[feature])\n \n feature_index = features.columns.get_loc(feature)\n features.drop(feature, axis=1, inplace=True)\n \n onehot_feature.columns = ['%s=%s' % (feature, feature_value) for feature_value in onehot_feature.columns]\n for colname in onehot_feature.columns[::-1]:\n features.insert(feature_index, colname, onehot_feature[colname])",
"_____no_output_____"
],
[
"x = features.values[:,1:].astype(int)\ny = labels.values[:,-1].astype(int)\n\nprint('x =\\n', x)\nprint('y =\\n', y)",
"x =\n [[ 1 0 1 ... 45 170 0]\n [ 2 0 1 ... 54 158 0]\n [ 1 0 1 ... 77 162 1]\n ...\n [ 2 0 0 ... 64 131 1]\n [ 1 0 1 ... 48 175 0]\n [ 1 0 1 ... 54 163 0]]\ny =\n [0 0 1 1 0 0 1 1 1 0 0 0 1 0 1 1 0 1 0 1 0 0 0 0 0 0 0 0 1 0 0 1 1 0 1 0 0\n 0 0 1 0 0 0 0 0 1 0 0 1 0 1 0 1 1 1 0 1 1 0 1 0 1 1 0 0 1 1 0 0 1 0 0 0 0\n 1 1 0 1 1 1 0 1 1 1 0 1 0 1 0 0 0 1 0 1 0 0 1 0 0 0 0 0 1 0 0 0 0 1 0 1 0\n 0 1 1 0 0 0 1 1 1 1 0 0 0 1 0 0 0 1 1 0 0 0 1 1 0 1 1 0 0 1 0 0 1 1 0 0 1\n 1 1 0 0 1 1 1 1 0 0 0 1 0 1 1 1 0 0 0 1 0 0 1 1 1 1 0 1 1 1 0 0]\n"
],
[
"# for fold_ind, (train_indices, test_indices) in enumerate(stratified_kflod_validator.split(x, y), 1):\n\n# x_train, x_test = x[train_indices], x[test_indices]\n# y_train, y_test = y[train_indices], y[test_indices]",
"_____no_output_____"
],
[
"x_train, x_test, y_train, y_test = \\\n train_test_split(x, y, test_size=0.2, random_state=42)\n\nprint(x_train.shape, x_test.shape)\nprint(y_train.shape, y_test.shape)",
"(144, 15) (36, 15)\n(144,) (36,)\n"
]
],
[
[
"## Define model",
"_____no_output_____"
]
],
[
[
"input_shape = (1,15)\nnum_classes = 2\n\nprint(x.shape)\nprint(y.shape)\n\nprint(x[:1])\nprint(y[:1])",
"(180, 15)\n(180,)\n[[ 1 0 1 0 128 2 0 0 2 308 0 1 45 170 0]]\n[0]\n"
]
],
[
[
"### Architecture 0 - Inflating Dense 120-225, 0.5 Dropout, Batch Norm, Sigmoid Classification",
"_____no_output_____"
]
],
[
[
"arch_cnt = 'arch-0-3'\n\nmodel = Sequential()\nmodel.add(\n Dense(120, input_dim=15, kernel_initializer='normal',\n # kernel_regularizer=keras.regularizers.l2(0.001), # pierd 0.2 acc\n activation='relu'))\nmodel.add(Dropout(0.5))\nmodel.add(Dense(225, input_dim=15, kernel_initializer='normal', activation='relu'))\n# model.add(LeakyReLU(alpha=0.1))\nmodel.add(BatchNormalization(axis = 1))\nmodel.add(Dense(1, kernel_initializer='normal', activation='sigmoid'))\n\n\nmodel.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])\n\nmodel.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_19 (Dense) (None, 120) 1920 \n_________________________________________________________________\ndropout_7 (Dropout) (None, 120) 0 \n_________________________________________________________________\ndense_20 (Dense) (None, 225) 27225 \n_________________________________________________________________\nbatch_normalization_7 (Batch (None, 225) 900 \n_________________________________________________________________\ndense_21 (Dense) (None, 1) 226 \n=================================================================\nTotal params: 30,271\nTrainable params: 29,821\nNon-trainable params: 450\n_________________________________________________________________\n"
],
[
"%%time\n\n# earlystop_cb = keras.callbacks.EarlyStopping(\n# monitor='val_loss',\n# patience=5, restore_best_weights=True,\n# verbose=1)\nreduce_lr_cb = keras.callbacks.ReduceLROnPlateau(\n monitor='val_loss', factor=0.05,\n patience=5, min_lr=0.001,\n verbose=1)\n\n# es_cb = keras.callbacks.EarlyStopping(\n# monitor='val_loss',\n# min_delta=0.1,\n# patience=7,\n# verbose=1,\n# mode='auto'\n# )\n# 'restore_best_weights' in dir(keras.callbacks.EarlyStopping()) # FALSE = library is not up-to-date\n\ntb_cb = keras.callbacks.TensorBoard(log_dir='./tensorboard/%s' % arch_cnt, histogram_freq=0, \n write_graph=True, write_images=True)\n\n\nepochs = 50\nbatch_size = 32\n\nmodel.fit(\n x_train, y_train,\n batch_size=batch_size,\n epochs=epochs,\n verbose=1,\n \n shuffle=False,\n validation_data=(x_test, y_test),\n callbacks=[reduce_lr_cb, es_cb, tb_cb]\n)",
"Train on 144 samples, validate on 36 samples\nEpoch 1/50\n144/144 [==============================] - 0s 2ms/step - loss: 0.6969 - acc: 0.5417 - val_loss: 0.6813 - val_acc: 0.5556\nEpoch 2/50\n144/144 [==============================] - 0s 71us/step - loss: 0.7291 - acc: 0.5417 - val_loss: 0.6876 - val_acc: 0.5556\nEpoch 3/50\n144/144 [==============================] - 0s 81us/step - loss: 0.7537 - acc: 0.5000 - val_loss: 0.7145 - val_acc: 0.4444\nEpoch 4/50\n144/144 [==============================] - 0s 138us/step - loss: 0.6780 - acc: 0.5556 - val_loss: 0.7250 - val_acc: 0.4444\nEpoch 5/50\n144/144 [==============================] - 0s 80us/step - loss: 0.7757 - acc: 0.4444 - val_loss: 0.7068 - val_acc: 0.4444\nEpoch 6/50\n144/144 [==============================] - 0s 106us/step - loss: 0.6985 - acc: 0.5556 - val_loss: 0.6883 - val_acc: 0.4167\n\nEpoch 00006: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 7/50\n144/144 [==============================] - 0s 84us/step - loss: 0.7108 - acc: 0.5556 - val_loss: 0.6835 - val_acc: 0.5278\nEpoch 8/50\n144/144 [==============================] - 0s 115us/step - loss: 0.6764 - acc: 0.5903 - val_loss: 0.6797 - val_acc: 0.5556\nEpoch 00008: early stopping\nCPU times: user 2.04 s, sys: 47.7 ms, total: 2.09 s\nWall time: 2.04 s\n"
],
[
"score = model.evaluate(x_test, y_test, verbose=0)\nprint('Test loss:', score[0])\nprint('Test accuracy:', score[1])",
"Test loss: 0.6797179447280036\nTest accuracy: 0.5555555555555556\n"
]
],
[
[
"### Architecture 1 - **`Deflating Dense 225-112`**, 0.5 Dropout, Batch Norm, Sigmoid Classification",
"_____no_output_____"
]
],
[
[
"arch_cnt = 'arch-1'\n\nmodel = Sequential()\nmodel.add(\n Dense(225, input_dim=15, kernel_initializer='normal',\n # kernel_regularizer=keras.regularizers.l2(0.001), # pierd 0.2 acc\n activation='relu'))\nmodel.add(Dropout(0.5))\nmodel.add(Dense(112, input_dim=15, kernel_initializer='normal', activation='relu'))\n# model.add(LeakyReLU(alpha=0.1))\nmodel.add(BatchNormalization(axis = 1))\nmodel.add(Dense(1, kernel_initializer='normal', activation='sigmoid'))\n\n\nmodel.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])\n\nmodel.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_4 (Dense) (None, 225) 3600 \n_________________________________________________________________\ndropout_2 (Dropout) (None, 225) 0 \n_________________________________________________________________\ndense_5 (Dense) (None, 112) 25312 \n_________________________________________________________________\nbatch_normalization_2 (Batch (None, 112) 448 \n_________________________________________________________________\ndense_6 (Dense) (None, 1) 113 \n=================================================================\nTotal params: 29,473\nTrainable params: 29,249\nNon-trainable params: 224\n_________________________________________________________________\n"
],
[
"%%time\n\n# earlystop_cb = keras.callbacks.EarlyStopping(\n# monitor='val_loss',\n# patience=5, restore_best_weights=True,\n# verbose=1)\nreduce_lr_cb = keras.callbacks.ReduceLROnPlateau(\n monitor='val_loss', factor=0.05,\n patience=7, min_lr=0.001,\n verbose=1)\n\ntb_cb = keras.callbacks.TensorBoard(log_dir='./tensorboard/%s' % arch_cnt, histogram_freq=0, \n write_graph=True, write_images=True)\n\n\nepochs = 50\nbatch_size = 32\n\nmodel.fit(\n x_train, y_train,\n batch_size=batch_size,\n epochs=epochs,\n verbose=1,\n \n shuffle=False,\n validation_data=(x_test, y_test),\n callbacks=[reduce_lr_cb, tb_cb]\n # callbacks=[earlystop_cb, reduce_lr_cb]\n)",
"Train on 144 samples, validate on 36 samples\nEpoch 1/50\n144/144 [==============================] - 0s 2ms/step - loss: 0.7093 - acc: 0.5069 - val_loss: 0.6825 - val_acc: 0.5833\nEpoch 2/50\n144/144 [==============================] - 0s 73us/step - loss: 0.6929 - acc: 0.5694 - val_loss: 0.6799 - val_acc: 0.7500\nEpoch 3/50\n144/144 [==============================] - 0s 80us/step - loss: 0.6967 - acc: 0.5069 - val_loss: 0.6785 - val_acc: 0.6111\nEpoch 4/50\n144/144 [==============================] - 0s 81us/step - loss: 0.6565 - acc: 0.6042 - val_loss: 0.6805 - val_acc: 0.5278\nEpoch 5/50\n144/144 [==============================] - 0s 75us/step - loss: 0.7139 - acc: 0.4861 - val_loss: 0.6793 - val_acc: 0.5000\nEpoch 6/50\n144/144 [==============================] - 0s 71us/step - loss: 0.6951 - acc: 0.5833 - val_loss: 0.6761 - val_acc: 0.5278\nEpoch 7/50\n144/144 [==============================] - 0s 88us/step - loss: 0.6469 - acc: 0.6181 - val_loss: 0.6785 - val_acc: 0.4722\nEpoch 8/50\n144/144 [==============================] - 0s 99us/step - loss: 0.6730 - acc: 0.6042 - val_loss: 0.6817 - val_acc: 0.4444\nEpoch 9/50\n144/144 [==============================] - 0s 97us/step - loss: 0.6321 - acc: 0.6389 - val_loss: 0.6897 - val_acc: 0.5000\nEpoch 10/50\n144/144 [==============================] - 0s 77us/step - loss: 0.6615 - acc: 0.5694 - val_loss: 0.6838 - val_acc: 0.4722\nEpoch 11/50\n144/144 [==============================] - 0s 115us/step - loss: 0.6357 - acc: 0.6458 - val_loss: 0.6758 - val_acc: 0.4444\nEpoch 12/50\n144/144 [==============================] - 0s 70us/step - loss: 0.6651 - acc: 0.5556 - val_loss: 0.6755 - val_acc: 0.4722\nEpoch 13/50\n144/144 [==============================] - 0s 83us/step - loss: 0.6498 - acc: 0.6111 - val_loss: 0.6762 - val_acc: 0.4167\nEpoch 14/50\n144/144 [==============================] - 0s 77us/step - loss: 0.6679 - acc: 0.5903 - val_loss: 0.6807 - val_acc: 0.5000\nEpoch 15/50\n144/144 [==============================] - 0s 75us/step - loss: 0.6756 - acc: 0.5972 - val_loss: 0.6921 - val_acc: 0.4722\nEpoch 16/50\n144/144 [==============================] - 0s 64us/step - loss: 0.6558 - acc: 0.6458 - val_loss: 0.7126 - val_acc: 0.5000\nEpoch 17/50\n144/144 [==============================] - 0s 85us/step - loss: 0.6583 - acc: 0.5764 - val_loss: 0.7052 - val_acc: 0.5000\nEpoch 18/50\n144/144 [==============================] - 0s 97us/step - loss: 0.6644 - acc: 0.5972 - val_loss: 0.7022 - val_acc: 0.5000\nEpoch 19/50\n144/144 [==============================] - 0s 67us/step - loss: 0.6542 - acc: 0.6181 - val_loss: 0.6827 - val_acc: 0.5000\n\nEpoch 00019: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 20/50\n144/144 [==============================] - 0s 60us/step - loss: 0.6295 - acc: 0.6528 - val_loss: 0.6693 - val_acc: 0.4722\nEpoch 21/50\n144/144 [==============================] - 0s 88us/step - loss: 0.6372 - acc: 0.6458 - val_loss: 0.6694 - val_acc: 0.5278\nEpoch 22/50\n144/144 [==============================] - 0s 86us/step - loss: 0.6372 - acc: 0.6528 - val_loss: 0.6713 - val_acc: 0.5278\nEpoch 23/50\n144/144 [==============================] - 0s 126us/step - loss: 0.6302 - acc: 0.6667 - val_loss: 0.6722 - val_acc: 0.5278\nEpoch 24/50\n144/144 [==============================] - 0s 95us/step - loss: 0.6399 - acc: 0.6389 - val_loss: 0.6669 - val_acc: 0.5278\nEpoch 25/50\n144/144 [==============================] - 0s 72us/step - loss: 0.6367 - acc: 0.6875 - val_loss: 0.6621 - val_acc: 0.5833\nEpoch 26/50\n144/144 [==============================] - 0s 104us/step - loss: 0.6440 - acc: 0.6111 - val_loss: 0.6633 - val_acc: 0.5833\nEpoch 27/50\n144/144 [==============================] - 0s 81us/step - loss: 0.6119 - acc: 0.6875 - val_loss: 0.6646 - val_acc: 0.6111\nEpoch 28/50\n144/144 [==============================] - 0s 103us/step - loss: 0.6463 - acc: 0.6458 - val_loss: 0.6685 - val_acc: 0.6111\nEpoch 29/50\n144/144 [==============================] - 0s 80us/step - loss: 0.6254 - acc: 0.6528 - val_loss: 0.6906 - val_acc: 0.5278\nEpoch 30/50\n144/144 [==============================] - 0s 89us/step - loss: 0.6357 - acc: 0.6667 - val_loss: 0.6953 - val_acc: 0.5000\nEpoch 31/50\n144/144 [==============================] - 0s 130us/step - loss: 0.6132 - acc: 0.7014 - val_loss: 0.6744 - val_acc: 0.5556\nEpoch 32/50\n144/144 [==============================] - 0s 98us/step - loss: 0.5881 - acc: 0.6806 - val_loss: 0.6497 - val_acc: 0.6389\nEpoch 33/50\n144/144 [==============================] - 0s 75us/step - loss: 0.6306 - acc: 0.6597 - val_loss: 0.6354 - val_acc: 0.6111\nEpoch 34/50\n144/144 [==============================] - 0s 76us/step - loss: 0.5916 - acc: 0.6458 - val_loss: 0.6416 - val_acc: 0.6389\nEpoch 35/50\n144/144 [==============================] - 0s 82us/step - loss: 0.6336 - acc: 0.6389 - val_loss: 0.6559 - val_acc: 0.5833\nEpoch 36/50\n144/144 [==============================] - 0s 79us/step - loss: 0.6226 - acc: 0.6667 - val_loss: 0.6697 - val_acc: 0.6111\nEpoch 37/50\n144/144 [==============================] - 0s 83us/step - loss: 0.6108 - acc: 0.6875 - val_loss: 0.6779 - val_acc: 0.5278\nEpoch 38/50\n144/144 [==============================] - 0s 100us/step - loss: 0.6044 - acc: 0.6667 - val_loss: 0.6830 - val_acc: 0.5278\nEpoch 39/50\n144/144 [==============================] - 0s 92us/step - loss: 0.5959 - acc: 0.6806 - val_loss: 0.6889 - val_acc: 0.5556\nEpoch 40/50\n144/144 [==============================] - 0s 80us/step - loss: 0.6000 - acc: 0.7153 - val_loss: 0.7006 - val_acc: 0.5278\n\nEpoch 00040: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 41/50\n144/144 [==============================] - 0s 96us/step - loss: 0.5673 - acc: 0.7014 - val_loss: 0.7056 - val_acc: 0.5278\nEpoch 42/50\n144/144 [==============================] - 0s 73us/step - loss: 0.6208 - acc: 0.6111 - val_loss: 0.6841 - val_acc: 0.5556\nEpoch 43/50\n144/144 [==============================] - 0s 90us/step - loss: 0.5972 - acc: 0.7083 - val_loss: 0.6565 - val_acc: 0.6944\nEpoch 44/50\n144/144 [==============================] - 0s 89us/step - loss: 0.6086 - acc: 0.7014 - val_loss: 0.6394 - val_acc: 0.6667\nEpoch 45/50\n144/144 [==============================] - 0s 88us/step - loss: 0.5894 - acc: 0.6806 - val_loss: 0.6096 - val_acc: 0.6389\nEpoch 46/50\n144/144 [==============================] - 0s 87us/step - loss: 0.5571 - acc: 0.7569 - val_loss: 0.6087 - val_acc: 0.6389\nEpoch 47/50\n144/144 [==============================] - 0s 144us/step - loss: 0.5807 - acc: 0.6875 - val_loss: 0.6171 - val_acc: 0.6389\nEpoch 48/50\n144/144 [==============================] - 0s 110us/step - loss: 0.5782 - acc: 0.6736 - val_loss: 0.6319 - val_acc: 0.6944\nEpoch 49/50\n144/144 [==============================] - 0s 99us/step - loss: 0.6167 - acc: 0.6458 - val_loss: 0.6380 - val_acc: 0.6944\nEpoch 50/50\n144/144 [==============================] - 0s 120us/step - loss: 0.5846 - acc: 0.6806 - val_loss: 0.6483 - val_acc: 0.6944\nCPU times: user 2.14 s, sys: 50.7 ms, total: 2.19 s\nWall time: 2.05 s\n"
],
[
"score = model.evaluate(x_test, y_test, verbose=0)\nprint('Test loss:', score[0])\nprint('Test accuracy:', score[1])",
"Test loss: 0.6483488314681582\nTest accuracy: 0.6944444444444444\n"
]
],
[
[
"### Architecture 2 - Deflating Dense 225-112, 0.5 Dropout, Batch Norm, Sigmoid Classification, **`HE Initialization`**",
"_____no_output_____"
]
],
[
[
"arch_cnt = 'arch-2'\n\nmodel = Sequential()\nmodel.add(\n Dense(225, input_dim=15, kernel_initializer='he_uniform',\n kernel_regularizer=keras.regularizers.l2(0.001), # pierd 0.2 acc\n activation='relu'))\nmodel.add(Dropout(0.5))\nmodel.add(Dense(112, input_dim=15, kernel_initializer='he_uniform', activation='relu'))\n# model.add(LeakyReLU(alpha=0.1))\nmodel.add(BatchNormalization(axis = 1))\nmodel.add(Dense(1, kernel_initializer='normal', activation='sigmoid'))\n\n\nmodel.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])\n\nmodel.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_7 (Dense) (None, 225) 3600 \n_________________________________________________________________\ndropout_3 (Dropout) (None, 225) 0 \n_________________________________________________________________\ndense_8 (Dense) (None, 112) 25312 \n_________________________________________________________________\nbatch_normalization_3 (Batch (None, 112) 448 \n_________________________________________________________________\ndense_9 (Dense) (None, 1) 113 \n=================================================================\nTotal params: 29,473\nTrainable params: 29,249\nNon-trainable params: 224\n_________________________________________________________________\n"
],
[
"%%time\n\n# earlystop_cb = keras.callbacks.EarlyStopping(\n# monitor='val_loss',\n# patience=5, restore_best_weights=True,\n# verbose=1)\nreduce_lr_cb = keras.callbacks.ReduceLROnPlateau(\n monitor='val_loss', factor=0.05,\n patience=7, min_lr=0.001,\n verbose=1)\n\ntb_cb = keras.callbacks.TensorBoard(log_dir='./tensorboard/%s' % arch_cnt, histogram_freq=0, \n write_graph=True, write_images=True)\n\n\nepochs = 50\nbatch_size = 32\n\nmodel.fit(\n x_train, y_train,\n batch_size=batch_size,\n epochs=epochs,\n verbose=1,\n \n shuffle=False,\n validation_data=(x_test, y_test),\n callbacks=[reduce_lr_cb, tb_cb]\n # callbacks=[earlystop_cb, reduce_lr_cb]\n)",
"Train on 144 samples, validate on 36 samples\nEpoch 1/50\n144/144 [==============================] - 0s 2ms/step - loss: 1.1839 - acc: 0.4792 - val_loss: 1.1616 - val_acc: 0.4167\nEpoch 2/50\n144/144 [==============================] - 0s 105us/step - loss: 1.1813 - acc: 0.4583 - val_loss: 1.1539 - val_acc: 0.4167\nEpoch 3/50\n144/144 [==============================] - 0s 79us/step - loss: 1.1809 - acc: 0.3958 - val_loss: 1.1394 - val_acc: 0.4444\nEpoch 4/50\n144/144 [==============================] - 0s 104us/step - loss: 1.1154 - acc: 0.5347 - val_loss: 1.1291 - val_acc: 0.4444\nEpoch 5/50\n144/144 [==============================] - 0s 80us/step - loss: 1.1076 - acc: 0.5417 - val_loss: 1.1173 - val_acc: 0.4444\nEpoch 6/50\n144/144 [==============================] - 0s 112us/step - loss: 1.0832 - acc: 0.5764 - val_loss: 1.1076 - val_acc: 0.4444\nEpoch 7/50\n144/144 [==============================] - 0s 81us/step - loss: 1.0518 - acc: 0.6458 - val_loss: 1.0964 - val_acc: 0.4444\nEpoch 8/50\n144/144 [==============================] - 0s 80us/step - loss: 1.0829 - acc: 0.5417 - val_loss: 1.0868 - val_acc: 0.4444\nEpoch 9/50\n144/144 [==============================] - 0s 91us/step - loss: 1.0699 - acc: 0.5278 - val_loss: 1.0821 - val_acc: 0.4444\nEpoch 10/50\n144/144 [==============================] - 0s 116us/step - loss: 1.0633 - acc: 0.5833 - val_loss: 1.0795 - val_acc: 0.4444\nEpoch 11/50\n144/144 [==============================] - 0s 136us/step - loss: 1.0887 - acc: 0.5139 - val_loss: 1.0843 - val_acc: 0.4722\nEpoch 12/50\n144/144 [==============================] - 0s 96us/step - loss: 1.0217 - acc: 0.6111 - val_loss: 1.0854 - val_acc: 0.4444\nEpoch 13/50\n144/144 [==============================] - 0s 113us/step - loss: 1.0282 - acc: 0.6042 - val_loss: 1.0917 - val_acc: 0.4167\nEpoch 14/50\n144/144 [==============================] - 0s 92us/step - loss: 1.0108 - acc: 0.5694 - val_loss: 1.0987 - val_acc: 0.4167\nEpoch 15/50\n144/144 [==============================] - 0s 113us/step - loss: 0.9797 - acc: 0.6597 - val_loss: 1.1029 - val_acc: 0.4444\nEpoch 16/50\n144/144 [==============================] - 0s 82us/step - loss: 1.0015 - acc: 0.5764 - val_loss: 1.1065 - val_acc: 0.4444\nEpoch 17/50\n144/144 [==============================] - 0s 97us/step - loss: 0.9799 - acc: 0.6181 - val_loss: 1.1098 - val_acc: 0.4444\n\nEpoch 00017: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 18/50\n144/144 [==============================] - 0s 83us/step - loss: 0.9983 - acc: 0.5764 - val_loss: 1.1117 - val_acc: 0.4444\nEpoch 19/50\n144/144 [==============================] - 0s 85us/step - loss: 0.9640 - acc: 0.6458 - val_loss: 1.1135 - val_acc: 0.4444\nEpoch 20/50\n144/144 [==============================] - 0s 124us/step - loss: 0.9538 - acc: 0.6181 - val_loss: 1.1163 - val_acc: 0.4444\nEpoch 21/50\n144/144 [==============================] - 0s 117us/step - loss: 0.9511 - acc: 0.6528 - val_loss: 1.1067 - val_acc: 0.4444\nEpoch 22/50\n144/144 [==============================] - 0s 70us/step - loss: 0.9432 - acc: 0.6319 - val_loss: 1.1034 - val_acc: 0.4444\nEpoch 23/50\n144/144 [==============================] - 0s 77us/step - loss: 0.9707 - acc: 0.5556 - val_loss: 1.0927 - val_acc: 0.4444\nEpoch 24/50\n144/144 [==============================] - 0s 108us/step - loss: 0.9418 - acc: 0.5903 - val_loss: 1.0725 - val_acc: 0.4722\nEpoch 25/50\n144/144 [==============================] - 0s 93us/step - loss: 0.9269 - acc: 0.6528 - val_loss: 1.0518 - val_acc: 0.4444\nEpoch 26/50\n144/144 [==============================] - 0s 97us/step - loss: 0.9566 - acc: 0.5903 - val_loss: 1.0330 - val_acc: 0.4444\nEpoch 27/50\n144/144 [==============================] - 0s 83us/step - loss: 0.9245 - acc: 0.6250 - val_loss: 1.0146 - val_acc: 0.4722\nEpoch 28/50\n144/144 [==============================] - 0s 66us/step - loss: 0.8902 - acc: 0.6597 - val_loss: 1.0130 - val_acc: 0.4722\nEpoch 29/50\n144/144 [==============================] - 0s 84us/step - loss: 0.9109 - acc: 0.6181 - val_loss: 1.0215 - val_acc: 0.4444\nEpoch 30/50\n144/144 [==============================] - 0s 89us/step - loss: 0.8991 - acc: 0.6389 - val_loss: 1.0251 - val_acc: 0.4444\nEpoch 31/50\n144/144 [==============================] - 0s 94us/step - loss: 0.8714 - acc: 0.6528 - val_loss: 1.0364 - val_acc: 0.4722\nEpoch 32/50\n144/144 [==============================] - 0s 87us/step - loss: 0.8695 - acc: 0.6736 - val_loss: 1.0542 - val_acc: 0.4444\nEpoch 33/50\n144/144 [==============================] - 0s 118us/step - loss: 0.8849 - acc: 0.6667 - val_loss: 1.0607 - val_acc: 0.4444\nEpoch 34/50\n144/144 [==============================] - 0s 77us/step - loss: 0.8582 - acc: 0.6944 - val_loss: 1.0637 - val_acc: 0.4444\nEpoch 35/50\n144/144 [==============================] - 0s 69us/step - loss: 0.8696 - acc: 0.6736 - val_loss: 1.0548 - val_acc: 0.4444\n\nEpoch 00035: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 36/50\n144/144 [==============================] - 0s 74us/step - loss: 0.8510 - acc: 0.6389 - val_loss: 1.0397 - val_acc: 0.4444\nEpoch 37/50\n144/144 [==============================] - 0s 91us/step - loss: 0.8735 - acc: 0.6319 - val_loss: 1.0156 - val_acc: 0.4722\nEpoch 38/50\n144/144 [==============================] - 0s 78us/step - loss: 0.8834 - acc: 0.6250 - val_loss: 0.9912 - val_acc: 0.4722\nEpoch 39/50\n144/144 [==============================] - 0s 81us/step - loss: 0.8446 - acc: 0.6736 - val_loss: 0.9728 - val_acc: 0.4444\nEpoch 40/50\n144/144 [==============================] - 0s 138us/step - loss: 0.8724 - acc: 0.6111 - val_loss: 0.9568 - val_acc: 0.4722\nEpoch 41/50\n144/144 [==============================] - 0s 66us/step - loss: 0.8606 - acc: 0.6389 - val_loss: 0.9440 - val_acc: 0.5000\nEpoch 42/50\n144/144 [==============================] - 0s 74us/step - loss: 0.8378 - acc: 0.6181 - val_loss: 0.9356 - val_acc: 0.5000\nEpoch 43/50\n144/144 [==============================] - 0s 68us/step - loss: 0.8190 - acc: 0.6597 - val_loss: 0.9292 - val_acc: 0.5000\nEpoch 44/50\n144/144 [==============================] - 0s 82us/step - loss: 0.8186 - acc: 0.6944 - val_loss: 0.9218 - val_acc: 0.5000\nEpoch 45/50\n144/144 [==============================] - 0s 77us/step - loss: 0.8526 - acc: 0.6389 - val_loss: 0.9180 - val_acc: 0.4722\nEpoch 46/50\n144/144 [==============================] - 0s 93us/step - loss: 0.8269 - acc: 0.6181 - val_loss: 0.9148 - val_acc: 0.4722\nEpoch 47/50\n144/144 [==============================] - 0s 81us/step - loss: 0.8286 - acc: 0.6111 - val_loss: 0.9087 - val_acc: 0.4444\nEpoch 48/50\n144/144 [==============================] - 0s 67us/step - loss: 0.8373 - acc: 0.6458 - val_loss: 0.9028 - val_acc: 0.4444\nEpoch 49/50\n144/144 [==============================] - 0s 72us/step - loss: 0.8445 - acc: 0.5833 - val_loss: 0.8974 - val_acc: 0.4444\nEpoch 50/50\n144/144 [==============================] - 0s 73us/step - loss: 0.8133 - acc: 0.6319 - val_loss: 0.8961 - val_acc: 0.4444\nCPU times: user 2.22 s, sys: 91.4 ms, total: 2.31 s\nWall time: 2.16 s\n"
],
[
"score = model.evaluate(x_test, y_test, verbose=0)\nprint('Test loss:', score[0])\nprint('Test accuracy:', score[1])",
"Test loss: 0.8960549301571317\nTest accuracy: 0.4444444444444444\n"
]
],
[
[
"### Architecture 3 - Deflating Dense 225-112, 0.5 Dropout, Batch Norm, Sigmoid Classification, **`L2 = 1e^-4`**",
"_____no_output_____"
]
],
[
[
"arch_cnt = 'arch-3-4'\n\nmodel = Sequential()\nmodel.add(\n Dense(225, input_dim=15, kernel_initializer='normal',\n kernel_regularizer=keras.regularizers.l2(0.0001), # pierd 0.2 acc\n activation='relu'))\nmodel.add(Dropout(0.5))\nmodel.add(\n Dense(112, input_dim=15, kernel_initializer='normal',\n kernel_regularizer=keras.regularizers.l2(0.0001), # pierd 0.2 acc\n activation='relu'))\n# model.add(LeakyReLU(alpha=0.1))\nmodel.add(BatchNormalization(axis = 1))\nmodel.add(Dense(1, kernel_initializer='normal', activation='sigmoid'))\n\n\nmodel.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])\n\nmodel.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_31 (Dense) (None, 225) 3600 \n_________________________________________________________________\ndropout_11 (Dropout) (None, 225) 0 \n_________________________________________________________________\ndense_32 (Dense) (None, 112) 25312 \n_________________________________________________________________\nbatch_normalization_11 (Batc (None, 112) 448 \n_________________________________________________________________\ndense_33 (Dense) (None, 1) 113 \n=================================================================\nTotal params: 29,473\nTrainable params: 29,249\nNon-trainable params: 224\n_________________________________________________________________\n"
],
[
"%%time\n\n# earlystop_cb = keras.callbacks.EarlyStopping(\n# monitor='val_loss',\n# patience=5, restore_best_weights=True,\n# verbose=1)\nreduce_lr_cb = keras.callbacks.ReduceLROnPlateau(\n monitor='val_loss', factor=0.05,\n patience=7, min_lr=0.001,\n verbose=1)\n\ntb_cb = keras.callbacks.TensorBoard(log_dir='./tensorboard/%s' % arch_cnt, histogram_freq=0, \n write_graph=True, write_images=True)\n\n\nepochs = 50\nbatch_size = 32\n\nmodel.fit(\n x_train, y_train,\n batch_size=batch_size,\n epochs=epochs,\n verbose=1,\n \n shuffle=False,\n validation_data=(x_test, y_test),\n callbacks=[reduce_lr_cb, tb_cb]\n # callbacks=[earlystop_cb, reduce_lr_cb]\n)",
"Train on 144 samples, validate on 36 samples\nEpoch 1/50\n144/144 [==============================] - 0s 2ms/step - loss: 0.6725 - acc: 0.6528 - val_loss: 0.7009 - val_acc: 0.5000\nEpoch 2/50\n144/144 [==============================] - 0s 90us/step - loss: 0.7097 - acc: 0.5417 - val_loss: 0.6948 - val_acc: 0.6111\nEpoch 3/50\n144/144 [==============================] - 0s 105us/step - loss: 0.7187 - acc: 0.5069 - val_loss: 0.6937 - val_acc: 0.6111\nEpoch 4/50\n144/144 [==============================] - 0s 88us/step - loss: 0.6837 - acc: 0.6042 - val_loss: 0.6939 - val_acc: 0.6111\nEpoch 5/50\n144/144 [==============================] - 0s 108us/step - loss: 0.6921 - acc: 0.5694 - val_loss: 0.7035 - val_acc: 0.4722\nEpoch 6/50\n144/144 [==============================] - 0s 115us/step - loss: 0.6707 - acc: 0.5972 - val_loss: 0.7112 - val_acc: 0.4444\nEpoch 7/50\n144/144 [==============================] - 0s 119us/step - loss: 0.6915 - acc: 0.5764 - val_loss: 0.7157 - val_acc: 0.4444\nEpoch 8/50\n144/144 [==============================] - 0s 133us/step - loss: 0.6749 - acc: 0.5903 - val_loss: 0.7153 - val_acc: 0.4444\nEpoch 9/50\n144/144 [==============================] - 0s 167us/step - loss: 0.6569 - acc: 0.6389 - val_loss: 0.7225 - val_acc: 0.4444\nEpoch 10/50\n144/144 [==============================] - 0s 110us/step - loss: 0.6670 - acc: 0.6319 - val_loss: 0.7271 - val_acc: 0.4444\n\nEpoch 00010: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 11/50\n144/144 [==============================] - 0s 88us/step - loss: 0.6593 - acc: 0.6042 - val_loss: 0.7341 - val_acc: 0.4444\nEpoch 12/50\n144/144 [==============================] - 0s 97us/step - loss: 0.6468 - acc: 0.6181 - val_loss: 0.7384 - val_acc: 0.4444\nEpoch 13/50\n144/144 [==============================] - 0s 105us/step - loss: 0.6363 - acc: 0.6944 - val_loss: 0.7379 - val_acc: 0.4444\nEpoch 14/50\n144/144 [==============================] - 0s 96us/step - loss: 0.6457 - acc: 0.6250 - val_loss: 0.7422 - val_acc: 0.4444\nEpoch 15/50\n144/144 [==============================] - 0s 91us/step - loss: 0.6465 - acc: 0.6458 - val_loss: 0.7480 - val_acc: 0.4722\nEpoch 16/50\n144/144 [==============================] - 0s 98us/step - loss: 0.6247 - acc: 0.6458 - val_loss: 0.7744 - val_acc: 0.4722\nEpoch 17/50\n144/144 [==============================] - 0s 96us/step - loss: 0.6359 - acc: 0.6597 - val_loss: 0.8232 - val_acc: 0.4444\n\nEpoch 00017: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 18/50\n144/144 [==============================] - 0s 81us/step - loss: 0.6326 - acc: 0.6806 - val_loss: 0.8864 - val_acc: 0.4722\nEpoch 19/50\n144/144 [==============================] - 0s 108us/step - loss: 0.6292 - acc: 0.6250 - val_loss: 0.9086 - val_acc: 0.4444\nEpoch 20/50\n144/144 [==============================] - 0s 117us/step - loss: 0.6426 - acc: 0.6458 - val_loss: 0.9152 - val_acc: 0.4722\nEpoch 21/50\n144/144 [==============================] - 0s 122us/step - loss: 0.6155 - acc: 0.6389 - val_loss: 0.9242 - val_acc: 0.4722\nEpoch 22/50\n144/144 [==============================] - 0s 116us/step - loss: 0.6173 - acc: 0.6667 - val_loss: 0.9376 - val_acc: 0.4444\nEpoch 23/50\n144/144 [==============================] - 0s 95us/step - loss: 0.6576 - acc: 0.6042 - val_loss: 0.9239 - val_acc: 0.4722\nEpoch 24/50\n144/144 [==============================] - 0s 95us/step - loss: 0.6005 - acc: 0.6944 - val_loss: 0.8889 - val_acc: 0.4722\n\nEpoch 00024: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 25/50\n144/144 [==============================] - 0s 105us/step - loss: 0.6153 - acc: 0.6944 - val_loss: 0.8436 - val_acc: 0.4722\nEpoch 26/50\n144/144 [==============================] - 0s 90us/step - loss: 0.5863 - acc: 0.7153 - val_loss: 0.8369 - val_acc: 0.4722\nEpoch 27/50\n144/144 [==============================] - 0s 91us/step - loss: 0.6248 - acc: 0.6667 - val_loss: 0.8485 - val_acc: 0.4722\nEpoch 28/50\n144/144 [==============================] - 0s 115us/step - loss: 0.6113 - acc: 0.6667 - val_loss: 0.8225 - val_acc: 0.4722\nEpoch 29/50\n144/144 [==============================] - 0s 104us/step - loss: 0.5913 - acc: 0.7083 - val_loss: 0.8296 - val_acc: 0.4722\nEpoch 30/50\n144/144 [==============================] - 0s 98us/step - loss: 0.6350 - acc: 0.6528 - val_loss: 0.8113 - val_acc: 0.5000\nEpoch 31/50\n144/144 [==============================] - 0s 91us/step - loss: 0.6476 - acc: 0.6736 - val_loss: 0.7497 - val_acc: 0.5000\n\nEpoch 00031: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 32/50\n144/144 [==============================] - 0s 144us/step - loss: 0.5939 - acc: 0.7083 - val_loss: 0.7081 - val_acc: 0.5000\nEpoch 33/50\n144/144 [==============================] - 0s 84us/step - loss: 0.6042 - acc: 0.6667 - val_loss: 0.6770 - val_acc: 0.5833\nEpoch 34/50\n144/144 [==============================] - 0s 135us/step - loss: 0.6072 - acc: 0.6667 - val_loss: 0.6625 - val_acc: 0.6111\nEpoch 35/50\n144/144 [==============================] - 0s 92us/step - loss: 0.5964 - acc: 0.7153 - val_loss: 0.6697 - val_acc: 0.6111\nEpoch 36/50\n144/144 [==============================] - 0s 105us/step - loss: 0.6051 - acc: 0.6806 - val_loss: 0.6875 - val_acc: 0.5833\nEpoch 37/50\n144/144 [==============================] - 0s 95us/step - loss: 0.5794 - acc: 0.7014 - val_loss: 0.7249 - val_acc: 0.5278\nEpoch 38/50\n144/144 [==============================] - 0s 101us/step - loss: 0.5880 - acc: 0.6944 - val_loss: 0.7636 - val_acc: 0.5000\nEpoch 39/50\n144/144 [==============================] - 0s 99us/step - loss: 0.6396 - acc: 0.6736 - val_loss: 0.7869 - val_acc: 0.5000\nEpoch 40/50\n144/144 [==============================] - 0s 114us/step - loss: 0.5761 - acc: 0.7083 - val_loss: 0.7492 - val_acc: 0.5278\nEpoch 41/50\n144/144 [==============================] - 0s 127us/step - loss: 0.5895 - acc: 0.6458 - val_loss: 0.7077 - val_acc: 0.6111\n\nEpoch 00041: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 42/50\n144/144 [==============================] - 0s 109us/step - loss: 0.6104 - acc: 0.6667 - val_loss: 0.6913 - val_acc: 0.6111\nEpoch 43/50\n144/144 [==============================] - 0s 103us/step - loss: 0.5877 - acc: 0.6736 - val_loss: 0.6822 - val_acc: 0.6389\nEpoch 44/50\n144/144 [==============================] - 0s 102us/step - loss: 0.5405 - acc: 0.7361 - val_loss: 0.6942 - val_acc: 0.6111\nEpoch 45/50\n144/144 [==============================] - 0s 91us/step - loss: 0.6039 - acc: 0.7014 - val_loss: 0.6933 - val_acc: 0.6389\nEpoch 46/50\n144/144 [==============================] - 0s 129us/step - loss: 0.5611 - acc: 0.7153 - val_loss: 0.6480 - val_acc: 0.6667\nEpoch 47/50\n144/144 [==============================] - 0s 92us/step - loss: 0.5768 - acc: 0.7222 - val_loss: 0.6161 - val_acc: 0.6667\nEpoch 48/50\n144/144 [==============================] - 0s 122us/step - loss: 0.5598 - acc: 0.7361 - val_loss: 0.6042 - val_acc: 0.6667\nEpoch 49/50\n144/144 [==============================] - 0s 114us/step - loss: 0.5568 - acc: 0.7708 - val_loss: 0.5950 - val_acc: 0.6667\nEpoch 50/50\n144/144 [==============================] - 0s 98us/step - loss: 0.6006 - acc: 0.6944 - val_loss: 0.5939 - val_acc: 0.6667\nCPU times: user 3.43 s, sys: 101 ms, total: 3.54 s\nWall time: 3.38 s\n"
],
[
"score = model.evaluate(x_test, y_test, verbose=0)\nprint('Test loss:', score[0])\nprint('Test accuracy:', score[1])",
"Test loss: 0.5939429865943061\nTest accuracy: 0.6666666666666666\n"
]
],
[
[
"### Architecture 3 - Deflating Dense 225-112, 0.5 Dropout, Batch Norm, Sigmoid Classification, **`L2 = 1e^-3`**",
"_____no_output_____"
]
],
[
[
"arch_cnt = 'arch-3-3'\n\nmodel = Sequential()\nmodel.add(\n Dense(225, input_dim=15, kernel_initializer='normal',\n kernel_regularizer=keras.regularizers.l2(0.001), # pierd 0.2 acc\n activation='relu'))\nmodel.add(Dropout(0.5))\nmodel.add(\n Dense(112, input_dim=15, kernel_initializer='normal',\n kernel_regularizer=keras.regularizers.l2(0.001), # pierd 0.2 acc\n activation='relu'))\n# model.add(LeakyReLU(alpha=0.1))\nmodel.add(BatchNormalization(axis = 1))\nmodel.add(Dense(1, kernel_initializer='normal', activation='sigmoid'))\n\n\nmodel.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])\n\nmodel.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_28 (Dense) (None, 225) 3600 \n_________________________________________________________________\ndropout_10 (Dropout) (None, 225) 0 \n_________________________________________________________________\ndense_29 (Dense) (None, 112) 25312 \n_________________________________________________________________\nbatch_normalization_10 (Batc (None, 112) 448 \n_________________________________________________________________\ndense_30 (Dense) (None, 1) 113 \n=================================================================\nTotal params: 29,473\nTrainable params: 29,249\nNon-trainable params: 224\n_________________________________________________________________\n"
],
[
"%%time\n\n# earlystop_cb = keras.callbacks.EarlyStopping(\n# monitor='val_loss',\n# patience=5, restore_best_weights=True,\n# verbose=1)\nreduce_lr_cb = keras.callbacks.ReduceLROnPlateau(\n monitor='val_loss', factor=0.05,\n patience=7, min_lr=0.001,\n verbose=1)\n\ntb_cb = keras.callbacks.TensorBoard(log_dir='./tensorboard/%s' % arch_cnt, histogram_freq=0, \n write_graph=True, write_images=True)\n\n\nepochs = 50\nbatch_size = 32\n\nmodel.fit(\n x_train, y_train,\n batch_size=batch_size,\n epochs=epochs,\n verbose=1,\n \n shuffle=False,\n validation_data=(x_test, y_test),\n callbacks=[reduce_lr_cb, tb_cb]\n # callbacks=[earlystop_cb, reduce_lr_cb]\n)",
"Train on 144 samples, validate on 36 samples\nEpoch 1/50\n144/144 [==============================] - 0s 3ms/step - loss: 0.7692 - acc: 0.5417 - val_loss: 0.7977 - val_acc: 0.4167\nEpoch 2/50\n144/144 [==============================] - 0s 105us/step - loss: 0.7819 - acc: 0.4792 - val_loss: 0.7888 - val_acc: 0.4167\nEpoch 3/50\n144/144 [==============================] - 0s 98us/step - loss: 0.7486 - acc: 0.5972 - val_loss: 0.7901 - val_acc: 0.4167\nEpoch 4/50\n144/144 [==============================] - 0s 89us/step - loss: 0.8262 - acc: 0.4514 - val_loss: 0.8170 - val_acc: 0.4167\nEpoch 5/50\n144/144 [==============================] - 0s 89us/step - loss: 0.7510 - acc: 0.5139 - val_loss: 0.8423 - val_acc: 0.4167\nEpoch 6/50\n144/144 [==============================] - 0s 111us/step - loss: 0.7443 - acc: 0.5278 - val_loss: 0.8328 - val_acc: 0.4167\nEpoch 7/50\n144/144 [==============================] - 0s 112us/step - loss: 0.7216 - acc: 0.5833 - val_loss: 0.8259 - val_acc: 0.4167\nEpoch 8/50\n144/144 [==============================] - 0s 129us/step - loss: 0.6948 - acc: 0.6250 - val_loss: 0.8210 - val_acc: 0.4167\nEpoch 9/50\n144/144 [==============================] - 0s 93us/step - loss: 0.7271 - acc: 0.5694 - val_loss: 0.8228 - val_acc: 0.4167\n\nEpoch 00009: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 10/50\n144/144 [==============================] - 0s 81us/step - loss: 0.6597 - acc: 0.6875 - val_loss: 0.8274 - val_acc: 0.4444\nEpoch 11/50\n144/144 [==============================] - 0s 93us/step - loss: 0.7129 - acc: 0.6042 - val_loss: 0.8302 - val_acc: 0.4444\nEpoch 12/50\n144/144 [==============================] - 0s 115us/step - loss: 0.7131 - acc: 0.5903 - val_loss: 0.8399 - val_acc: 0.4444\nEpoch 13/50\n144/144 [==============================] - 0s 119us/step - loss: 0.6937 - acc: 0.6389 - val_loss: 0.8475 - val_acc: 0.4444\nEpoch 14/50\n144/144 [==============================] - 0s 110us/step - loss: 0.7281 - acc: 0.6111 - val_loss: 0.8439 - val_acc: 0.4444\nEpoch 15/50\n144/144 [==============================] - 0s 89us/step - loss: 0.7205 - acc: 0.5625 - val_loss: 0.8283 - val_acc: 0.4444\nEpoch 16/50\n144/144 [==============================] - 0s 85us/step - loss: 0.6667 - acc: 0.6875 - val_loss: 0.8356 - val_acc: 0.4444\n\nEpoch 00016: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 17/50\n144/144 [==============================] - 0s 83us/step - loss: 0.7025 - acc: 0.6389 - val_loss: 0.8482 - val_acc: 0.4444\nEpoch 18/50\n144/144 [==============================] - 0s 130us/step - loss: 0.6816 - acc: 0.6250 - val_loss: 0.8651 - val_acc: 0.4444\nEpoch 19/50\n144/144 [==============================] - 0s 114us/step - loss: 0.6924 - acc: 0.6181 - val_loss: 0.8572 - val_acc: 0.4444\nEpoch 20/50\n144/144 [==============================] - 0s 117us/step - loss: 0.7176 - acc: 0.5347 - val_loss: 0.8635 - val_acc: 0.4722\nEpoch 21/50\n144/144 [==============================] - 0s 92us/step - loss: 0.6687 - acc: 0.6458 - val_loss: 0.8664 - val_acc: 0.4722\nEpoch 22/50\n144/144 [==============================] - 0s 102us/step - loss: 0.6501 - acc: 0.6458 - val_loss: 0.8912 - val_acc: 0.4444\nEpoch 23/50\n144/144 [==============================] - 0s 126us/step - loss: 0.6612 - acc: 0.6597 - val_loss: 0.9411 - val_acc: 0.4444\n\nEpoch 00023: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 24/50\n144/144 [==============================] - 0s 104us/step - loss: 0.6831 - acc: 0.6111 - val_loss: 0.9712 - val_acc: 0.4444\nEpoch 25/50\n144/144 [==============================] - 0s 106us/step - loss: 0.6770 - acc: 0.6597 - val_loss: 0.9774 - val_acc: 0.4444\nEpoch 26/50\n144/144 [==============================] - 0s 84us/step - loss: 0.6666 - acc: 0.6667 - val_loss: 0.9166 - val_acc: 0.4444\nEpoch 27/50\n144/144 [==============================] - 0s 117us/step - loss: 0.6062 - acc: 0.7431 - val_loss: 0.8773 - val_acc: 0.4722\nEpoch 28/50\n144/144 [==============================] - 0s 136us/step - loss: 0.6729 - acc: 0.6181 - val_loss: 0.8392 - val_acc: 0.4722\nEpoch 29/50\n144/144 [==============================] - 0s 83us/step - loss: 0.6451 - acc: 0.6458 - val_loss: 0.8356 - val_acc: 0.4722\nEpoch 30/50\n144/144 [==============================] - 0s 101us/step - loss: 0.6248 - acc: 0.6875 - val_loss: 0.8405 - val_acc: 0.4722\n\nEpoch 00030: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 31/50\n144/144 [==============================] - 0s 117us/step - loss: 0.6316 - acc: 0.7014 - val_loss: 0.8406 - val_acc: 0.4722\nEpoch 32/50\n144/144 [==============================] - 0s 101us/step - loss: 0.6501 - acc: 0.6597 - val_loss: 0.8330 - val_acc: 0.4722\nEpoch 33/50\n144/144 [==============================] - 0s 100us/step - loss: 0.6490 - acc: 0.6528 - val_loss: 0.7997 - val_acc: 0.5000\nEpoch 34/50\n144/144 [==============================] - 0s 89us/step - loss: 0.6211 - acc: 0.7431 - val_loss: 0.7778 - val_acc: 0.5278\nEpoch 35/50\n144/144 [==============================] - 0s 81us/step - loss: 0.6794 - acc: 0.6181 - val_loss: 0.7906 - val_acc: 0.5278\nEpoch 36/50\n144/144 [==============================] - 0s 103us/step - loss: 0.6693 - acc: 0.6806 - val_loss: 0.7985 - val_acc: 0.5278\nEpoch 37/50\n144/144 [==============================] - 0s 89us/step - loss: 0.6580 - acc: 0.6528 - val_loss: 0.8106 - val_acc: 0.5000\nEpoch 38/50\n144/144 [==============================] - 0s 124us/step - loss: 0.6284 - acc: 0.6667 - val_loss: 0.8280 - val_acc: 0.5000\nEpoch 39/50\n144/144 [==============================] - 0s 80us/step - loss: 0.6384 - acc: 0.6597 - val_loss: 0.8437 - val_acc: 0.5000\nEpoch 40/50\n144/144 [==============================] - 0s 72us/step - loss: 0.6360 - acc: 0.6111 - val_loss: 0.8712 - val_acc: 0.4722\nEpoch 41/50\n144/144 [==============================] - 0s 127us/step - loss: 0.6752 - acc: 0.6111 - val_loss: 0.9283 - val_acc: 0.4722\n\nEpoch 00041: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 42/50\n144/144 [==============================] - 0s 159us/step - loss: 0.6466 - acc: 0.6458 - val_loss: 0.9391 - val_acc: 0.4722\nEpoch 43/50\n144/144 [==============================] - 0s 115us/step - loss: 0.6189 - acc: 0.6667 - val_loss: 0.9385 - val_acc: 0.4722\nEpoch 44/50\n144/144 [==============================] - 0s 110us/step - loss: 0.6068 - acc: 0.6944 - val_loss: 0.9006 - val_acc: 0.4722\nEpoch 45/50\n144/144 [==============================] - 0s 79us/step - loss: 0.6409 - acc: 0.6875 - val_loss: 0.9075 - val_acc: 0.5000\nEpoch 46/50\n144/144 [==============================] - 0s 159us/step - loss: 0.6519 - acc: 0.6528 - val_loss: 0.8841 - val_acc: 0.5000\nEpoch 47/50\n144/144 [==============================] - 0s 118us/step - loss: 0.6561 - acc: 0.6806 - val_loss: 0.8750 - val_acc: 0.5000\nEpoch 48/50\n144/144 [==============================] - 0s 88us/step - loss: 0.6223 - acc: 0.7014 - val_loss: 0.8971 - val_acc: 0.5000\n\nEpoch 00048: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 49/50\n144/144 [==============================] - 0s 93us/step - loss: 0.6592 - acc: 0.6319 - val_loss: 0.8518 - val_acc: 0.5000\nEpoch 50/50\n144/144 [==============================] - 0s 107us/step - loss: 0.5995 - acc: 0.6944 - val_loss: 0.8409 - val_acc: 0.5000\nCPU times: user 3.56 s, sys: 77.9 ms, total: 3.64 s\nWall time: 3.49 s\n"
],
[
"score = model.evaluate(x_test, y_test, verbose=0)\nprint('Test loss:', score[0])\nprint('Test accuracy:', score[1])",
"Test loss: 0.8408659166759915\nTest accuracy: 0.5\n"
]
],
[
[
"### Architecture 3 - Deflating Dense 225-112, 0.5 Dropout, Batch Norm, Sigmoid Classification, **`L2 = 1e^-2`**",
"_____no_output_____"
]
],
[
[
"arch_cnt = 'arch-3-2'\n\nmodel = Sequential()\nmodel.add(\n Dense(225, input_dim=15, kernel_initializer='normal',\n kernel_regularizer=keras.regularizers.l2(0.01), # pierd 0.2 acc\n activation='relu'))\nmodel.add(Dropout(0.5))\nmodel.add(\n Dense(112, input_dim=15, kernel_initializer='normal',\n kernel_regularizer=keras.regularizers.l2(0.01), # pierd 0.2 acc\n activation='relu'))\n# model.add(LeakyReLU(alpha=0.1))\nmodel.add(BatchNormalization(axis = 1))\nmodel.add(Dense(1, kernel_initializer='normal', activation='sigmoid'))\n\n\nmodel.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])\n\nmodel.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_25 (Dense) (None, 225) 3600 \n_________________________________________________________________\ndropout_9 (Dropout) (None, 225) 0 \n_________________________________________________________________\ndense_26 (Dense) (None, 112) 25312 \n_________________________________________________________________\nbatch_normalization_9 (Batch (None, 112) 448 \n_________________________________________________________________\ndense_27 (Dense) (None, 1) 113 \n=================================================================\nTotal params: 29,473\nTrainable params: 29,249\nNon-trainable params: 224\n_________________________________________________________________\n"
],
[
"%%time\n\n# earlystop_cb = keras.callbacks.EarlyStopping(\n# monitor='val_loss',\n# patience=5, restore_best_weights=True,\n# verbose=1)\nreduce_lr_cb = keras.callbacks.ReduceLROnPlateau(\n monitor='val_loss', factor=0.05,\n patience=7, min_lr=0.001,\n verbose=1)\n\ntb_cb = keras.callbacks.TensorBoard(log_dir='./tensorboard/%s' % arch_cnt, histogram_freq=0, \n write_graph=True, write_images=True)\n\n\nepochs = 50\nbatch_size = 32\n\nmodel.fit(\n x_train, y_train,\n batch_size=batch_size,\n epochs=epochs,\n verbose=1,\n \n shuffle=False,\n validation_data=(x_test, y_test),\n callbacks=[reduce_lr_cb, tb_cb]\n # callbacks=[earlystop_cb, reduce_lr_cb]\n)",
"Train on 144 samples, validate on 36 samples\nEpoch 1/50\n144/144 [==============================] - 0s 2ms/step - loss: 1.3970 - acc: 0.6111 - val_loss: 1.3253 - val_acc: 0.5833\nEpoch 2/50\n144/144 [==============================] - 0s 78us/step - loss: 1.3548 - acc: 0.5069 - val_loss: 1.2600 - val_acc: 0.6389\nEpoch 3/50\n144/144 [==============================] - 0s 134us/step - loss: 1.2752 - acc: 0.5417 - val_loss: 1.2089 - val_acc: 0.5278\nEpoch 4/50\n144/144 [==============================] - 0s 95us/step - loss: 1.1935 - acc: 0.5833 - val_loss: 1.1714 - val_acc: 0.5000\nEpoch 5/50\n144/144 [==============================] - 0s 103us/step - loss: 1.1574 - acc: 0.5347 - val_loss: 1.1544 - val_acc: 0.4444\nEpoch 6/50\n144/144 [==============================] - 0s 85us/step - loss: 1.1429 - acc: 0.5000 - val_loss: 1.1270 - val_acc: 0.4722\nEpoch 7/50\n144/144 [==============================] - 0s 93us/step - loss: 1.0410 - acc: 0.6458 - val_loss: 1.0989 - val_acc: 0.4444\nEpoch 8/50\n144/144 [==============================] - 0s 104us/step - loss: 1.0218 - acc: 0.6181 - val_loss: 1.0568 - val_acc: 0.5000\nEpoch 9/50\n144/144 [==============================] - 0s 96us/step - loss: 0.9996 - acc: 0.6458 - val_loss: 1.0285 - val_acc: 0.5000\nEpoch 10/50\n144/144 [==============================] - 0s 94us/step - loss: 0.9516 - acc: 0.6736 - val_loss: 1.0158 - val_acc: 0.5000\nEpoch 11/50\n144/144 [==============================] - 0s 85us/step - loss: 0.9309 - acc: 0.6528 - val_loss: 1.0130 - val_acc: 0.5000\nEpoch 12/50\n144/144 [==============================] - 0s 113us/step - loss: 0.9125 - acc: 0.6528 - val_loss: 1.0007 - val_acc: 0.5000\nEpoch 13/50\n144/144 [==============================] - 0s 83us/step - loss: 0.9088 - acc: 0.6111 - val_loss: 0.9850 - val_acc: 0.5000\nEpoch 14/50\n144/144 [==============================] - 0s 93us/step - loss: 0.9277 - acc: 0.6181 - val_loss: 0.9572 - val_acc: 0.4722\nEpoch 15/50\n144/144 [==============================] - 0s 93us/step - loss: 0.8736 - acc: 0.6458 - val_loss: 0.9331 - val_acc: 0.4444\nEpoch 16/50\n144/144 [==============================] - 0s 104us/step - loss: 0.9136 - acc: 0.5694 - val_loss: 0.9267 - val_acc: 0.4444\nEpoch 17/50\n144/144 [==============================] - 0s 124us/step - loss: 0.8606 - acc: 0.6042 - val_loss: 0.9140 - val_acc: 0.4444\nEpoch 18/50\n144/144 [==============================] - 0s 157us/step - loss: 0.8596 - acc: 0.6875 - val_loss: 0.9056 - val_acc: 0.4444\nEpoch 19/50\n144/144 [==============================] - 0s 111us/step - loss: 0.8568 - acc: 0.6389 - val_loss: 0.8979 - val_acc: 0.4444\nEpoch 20/50\n144/144 [==============================] - 0s 114us/step - loss: 0.8296 - acc: 0.6736 - val_loss: 0.8832 - val_acc: 0.4722\nEpoch 21/50\n144/144 [==============================] - 0s 99us/step - loss: 0.8324 - acc: 0.6458 - val_loss: 0.8805 - val_acc: 0.4444\nEpoch 22/50\n144/144 [==============================] - 0s 100us/step - loss: 0.8512 - acc: 0.6111 - val_loss: 0.8644 - val_acc: 0.5000\nEpoch 23/50\n144/144 [==============================] - 0s 87us/step - loss: 0.8416 - acc: 0.6319 - val_loss: 0.8458 - val_acc: 0.5000\nEpoch 24/50\n144/144 [==============================] - 0s 90us/step - loss: 0.8018 - acc: 0.6389 - val_loss: 0.8344 - val_acc: 0.5556\nEpoch 25/50\n144/144 [==============================] - 0s 88us/step - loss: 0.8188 - acc: 0.6736 - val_loss: 0.8333 - val_acc: 0.5278\nEpoch 26/50\n144/144 [==============================] - 0s 81us/step - loss: 0.7958 - acc: 0.6597 - val_loss: 0.8315 - val_acc: 0.5278\nEpoch 27/50\n144/144 [==============================] - 0s 86us/step - loss: 0.8079 - acc: 0.6597 - val_loss: 0.8293 - val_acc: 0.5000\nEpoch 28/50\n144/144 [==============================] - 0s 111us/step - loss: 0.8112 - acc: 0.6250 - val_loss: 0.8219 - val_acc: 0.5278\nEpoch 29/50\n144/144 [==============================] - 0s 86us/step - loss: 0.7719 - acc: 0.6667 - val_loss: 0.8144 - val_acc: 0.5556\nEpoch 30/50\n144/144 [==============================] - 0s 98us/step - loss: 0.7737 - acc: 0.6319 - val_loss: 0.8104 - val_acc: 0.5556\nEpoch 31/50\n144/144 [==============================] - 0s 85us/step - loss: 0.7806 - acc: 0.6458 - val_loss: 0.8084 - val_acc: 0.5556\nEpoch 32/50\n144/144 [==============================] - 0s 85us/step - loss: 0.7911 - acc: 0.6319 - val_loss: 0.8180 - val_acc: 0.5000\nEpoch 33/50\n144/144 [==============================] - 0s 90us/step - loss: 0.7738 - acc: 0.6458 - val_loss: 0.8365 - val_acc: 0.4444\nEpoch 34/50\n144/144 [==============================] - 0s 119us/step - loss: 0.7785 - acc: 0.6250 - val_loss: 0.8442 - val_acc: 0.4444\nEpoch 35/50\n144/144 [==============================] - 0s 107us/step - loss: 0.7921 - acc: 0.6250 - val_loss: 0.8281 - val_acc: 0.4444\nEpoch 36/50\n144/144 [==============================] - 0s 109us/step - loss: 0.7810 - acc: 0.6250 - val_loss: 0.8109 - val_acc: 0.5000\nEpoch 37/50\n144/144 [==============================] - 0s 98us/step - loss: 0.7913 - acc: 0.5903 - val_loss: 0.7972 - val_acc: 0.5278\nEpoch 38/50\n144/144 [==============================] - 0s 125us/step - loss: 0.7589 - acc: 0.6806 - val_loss: 0.7809 - val_acc: 0.6111\nEpoch 39/50\n144/144 [==============================] - 0s 112us/step - loss: 0.7842 - acc: 0.6458 - val_loss: 0.7754 - val_acc: 0.5833\nEpoch 40/50\n144/144 [==============================] - 0s 77us/step - loss: 0.7520 - acc: 0.6806 - val_loss: 0.7724 - val_acc: 0.5833\nEpoch 41/50\n144/144 [==============================] - 0s 97us/step - loss: 0.7662 - acc: 0.6597 - val_loss: 0.7766 - val_acc: 0.6111\nEpoch 42/50\n144/144 [==============================] - 0s 73us/step - loss: 0.7877 - acc: 0.6944 - val_loss: 0.7872 - val_acc: 0.5556\nEpoch 43/50\n144/144 [==============================] - 0s 100us/step - loss: 0.7609 - acc: 0.6597 - val_loss: 0.8043 - val_acc: 0.4722\nEpoch 44/50\n144/144 [==============================] - 0s 79us/step - loss: 0.7633 - acc: 0.6458 - val_loss: 0.8180 - val_acc: 0.4722\nEpoch 45/50\n144/144 [==============================] - 0s 75us/step - loss: 0.7369 - acc: 0.6875 - val_loss: 0.8321 - val_acc: 0.5000\nEpoch 46/50\n144/144 [==============================] - 0s 111us/step - loss: 0.7548 - acc: 0.6458 - val_loss: 0.8193 - val_acc: 0.4444\nEpoch 47/50\n144/144 [==============================] - 0s 136us/step - loss: 0.7429 - acc: 0.6319 - val_loss: 0.8042 - val_acc: 0.5278\n\nEpoch 00047: ReduceLROnPlateau reducing learning rate to 0.001.\nEpoch 48/50\n144/144 [==============================] - 0s 79us/step - loss: 0.7433 - acc: 0.6181 - val_loss: 0.7955 - val_acc: 0.5278\nEpoch 49/50\n144/144 [==============================] - 0s 84us/step - loss: 0.7543 - acc: 0.6389 - val_loss: 0.7788 - val_acc: 0.5556\nEpoch 50/50\n144/144 [==============================] - 0s 91us/step - loss: 0.7357 - acc: 0.6250 - val_loss: 0.7693 - val_acc: 0.5833\nCPU times: user 3.09 s, sys: 102 ms, total: 3.19 s\nWall time: 3.01 s\n"
],
[
"score = model.evaluate(x_test, y_test, verbose=0)\nprint('Test loss:', score[0])\nprint('Test accuracy:', score[1])",
"Test loss: 0.7693172958162096\nTest accuracy: 0.5833333333333334\n"
]
],
[
[
"### Architecture 3 - Deflating Dense 225-112, 0.5 Dropout, Batch Norm, Sigmoid Classification, **`L2 = 1e^-1`**",
"_____no_output_____"
]
],
[
[
"arch_cnt = 'arch-3-1'\n\nmodel = Sequential()\nmodel.add(\n Dense(225, input_dim=15, kernel_initializer='normal',\n kernel_regularizer=keras.regularizers.l2(0.1), # pierd 0.2 acc\n activation='relu'))\nmodel.add(Dropout(0.5))\nmodel.add(\n Dense(112, input_dim=15, kernel_initializer='normal',\n kernel_regularizer=keras.regularizers.l2(0.1), # pierd 0.2 acc\n activation='relu'))\n# model.add(LeakyReLU(alpha=0.1))\nmodel.add(BatchNormalization(axis = 1))\nmodel.add(Dense(1, kernel_initializer='normal', activation='sigmoid'))\n\n\nmodel.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])\n\nmodel.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_22 (Dense) (None, 225) 3600 \n_________________________________________________________________\ndropout_8 (Dropout) (None, 225) 0 \n_________________________________________________________________\ndense_23 (Dense) (None, 112) 25312 \n_________________________________________________________________\nbatch_normalization_8 (Batch (None, 112) 448 \n_________________________________________________________________\ndense_24 (Dense) (None, 1) 113 \n=================================================================\nTotal params: 29,473\nTrainable params: 29,249\nNon-trainable params: 224\n_________________________________________________________________\n"
],
[
"%%time\n\n# earlystop_cb = keras.callbacks.EarlyStopping(\n# monitor='val_loss',\n# patience=5, restore_best_weights=True,\n# verbose=1)\nreduce_lr_cb = keras.callbacks.ReduceLROnPlateau(\n monitor='val_loss', factor=0.05,\n patience=7, min_lr=0.001,\n verbose=1)\n\ntb_cb = keras.callbacks.TensorBoard(log_dir='./tensorboard/%s' % arch_cnt, histogram_freq=0, \n write_graph=True, write_images=True)\n\n\nepochs = 50\nbatch_size = 32\n\nmodel.fit(\n x_train, y_train,\n batch_size=batch_size,\n epochs=epochs,\n verbose=1,\n \n shuffle=False,\n validation_data=(x_test, y_test),\n callbacks=[reduce_lr_cb, tb_cb]\n # callbacks=[earlystop_cb, reduce_lr_cb]\n)",
"Train on 144 samples, validate on 36 samples\nEpoch 1/50\n144/144 [==============================] - 0s 2ms/step - loss: 7.4829 - acc: 0.5486 - val_loss: 6.8780 - val_acc: 0.4444\nEpoch 2/50\n144/144 [==============================] - 0s 94us/step - loss: 6.5561 - acc: 0.5764 - val_loss: 6.0230 - val_acc: 0.4444\nEpoch 3/50\n144/144 [==============================] - 0s 90us/step - loss: 5.7389 - acc: 0.5208 - val_loss: 5.2785 - val_acc: 0.4444\nEpoch 4/50\n144/144 [==============================] - 0s 109us/step - loss: 5.0140 - acc: 0.5903 - val_loss: 4.6354 - val_acc: 0.4722\nEpoch 5/50\n144/144 [==============================] - 0s 107us/step - loss: 4.4265 - acc: 0.5556 - val_loss: 4.0890 - val_acc: 0.4444\nEpoch 6/50\n144/144 [==============================] - 0s 104us/step - loss: 3.9032 - acc: 0.5833 - val_loss: 3.6264 - val_acc: 0.4444\nEpoch 7/50\n144/144 [==============================] - 0s 98us/step - loss: 3.4811 - acc: 0.5208 - val_loss: 3.2298 - val_acc: 0.4444\nEpoch 8/50\n144/144 [==============================] - 0s 79us/step - loss: 3.0772 - acc: 0.6111 - val_loss: 2.8905 - val_acc: 0.4722\nEpoch 9/50\n144/144 [==============================] - 0s 123us/step - loss: 2.7821 - acc: 0.5486 - val_loss: 2.6020 - val_acc: 0.5278\nEpoch 10/50\n144/144 [==============================] - 0s 100us/step - loss: 2.5395 - acc: 0.5417 - val_loss: 2.3626 - val_acc: 0.4722\nEpoch 11/50\n144/144 [==============================] - 0s 100us/step - loss: 2.2919 - acc: 0.5625 - val_loss: 2.1634 - val_acc: 0.4444\nEpoch 12/50\n144/144 [==============================] - 0s 128us/step - loss: 2.0781 - acc: 0.5764 - val_loss: 1.9884 - val_acc: 0.4444\nEpoch 13/50\n144/144 [==============================] - 0s 88us/step - loss: 1.9107 - acc: 0.5903 - val_loss: 1.8417 - val_acc: 0.4444\nEpoch 14/50\n144/144 [==============================] - 0s 84us/step - loss: 1.7622 - acc: 0.5417 - val_loss: 1.7146 - val_acc: 0.4444\nEpoch 15/50\n144/144 [==============================] - 0s 95us/step - loss: 1.6451 - acc: 0.5972 - val_loss: 1.6169 - val_acc: 0.4444\nEpoch 16/50\n144/144 [==============================] - 0s 120us/step - loss: 1.5554 - acc: 0.5694 - val_loss: 1.5401 - val_acc: 0.4167\nEpoch 17/50\n144/144 [==============================] - 0s 116us/step - loss: 1.4397 - acc: 0.6111 - val_loss: 1.4735 - val_acc: 0.4444\nEpoch 18/50\n144/144 [==============================] - 0s 104us/step - loss: 1.4129 - acc: 0.5139 - val_loss: 1.4041 - val_acc: 0.4444\nEpoch 19/50\n144/144 [==============================] - 0s 90us/step - loss: 1.3058 - acc: 0.5903 - val_loss: 1.3251 - val_acc: 0.4444\nEpoch 20/50\n144/144 [==============================] - 0s 81us/step - loss: 1.2432 - acc: 0.6250 - val_loss: 1.2571 - val_acc: 0.4444\nEpoch 21/50\n144/144 [==============================] - 0s 97us/step - loss: 1.1539 - acc: 0.6597 - val_loss: 1.2116 - val_acc: 0.4722\nEpoch 22/50\n144/144 [==============================] - 0s 107us/step - loss: 1.1573 - acc: 0.6042 - val_loss: 1.1877 - val_acc: 0.4444\nEpoch 23/50\n144/144 [==============================] - 0s 93us/step - loss: 1.1166 - acc: 0.5833 - val_loss: 1.1431 - val_acc: 0.4722\nEpoch 24/50\n144/144 [==============================] - 0s 84us/step - loss: 1.0777 - acc: 0.6042 - val_loss: 1.1021 - val_acc: 0.4444\nEpoch 25/50\n144/144 [==============================] - 0s 90us/step - loss: 0.9982 - acc: 0.6250 - val_loss: 1.0659 - val_acc: 0.4722\nEpoch 26/50\n144/144 [==============================] - 0s 120us/step - loss: 1.0227 - acc: 0.6319 - val_loss: 1.0479 - val_acc: 0.4444\nEpoch 27/50\n144/144 [==============================] - 0s 103us/step - loss: 0.9950 - acc: 0.5833 - val_loss: 1.0425 - val_acc: 0.4722\nEpoch 28/50\n144/144 [==============================] - 0s 101us/step - loss: 0.9684 - acc: 0.6458 - val_loss: 1.0459 - val_acc: 0.4444\nEpoch 29/50\n144/144 [==============================] - 0s 94us/step - loss: 0.9391 - acc: 0.6042 - val_loss: 1.0335 - val_acc: 0.4444\nEpoch 30/50\n144/144 [==============================] - 0s 90us/step - loss: 0.9301 - acc: 0.6667 - val_loss: 0.9903 - val_acc: 0.4722\nEpoch 31/50\n144/144 [==============================] - 0s 109us/step - loss: 0.8917 - acc: 0.6319 - val_loss: 0.9629 - val_acc: 0.4444\nEpoch 32/50\n144/144 [==============================] - 0s 85us/step - loss: 0.8565 - acc: 0.6319 - val_loss: 0.9693 - val_acc: 0.4444\nEpoch 33/50\n144/144 [==============================] - 0s 122us/step - loss: 0.8659 - acc: 0.6736 - val_loss: 0.9631 - val_acc: 0.4444\nEpoch 34/50\n144/144 [==============================] - 0s 98us/step - loss: 0.8961 - acc: 0.5833 - val_loss: 0.9642 - val_acc: 0.4444\nEpoch 35/50\n144/144 [==============================] - 0s 114us/step - loss: 0.8323 - acc: 0.6528 - val_loss: 0.9652 - val_acc: 0.4444\nEpoch 36/50\n144/144 [==============================] - 0s 108us/step - loss: 0.8590 - acc: 0.5972 - val_loss: 0.9168 - val_acc: 0.4722\nEpoch 37/50\n144/144 [==============================] - 0s 87us/step - loss: 0.8576 - acc: 0.6458 - val_loss: 0.8841 - val_acc: 0.4722\nEpoch 38/50\n144/144 [==============================] - 0s 84us/step - loss: 0.8472 - acc: 0.5972 - val_loss: 0.8537 - val_acc: 0.4444\nEpoch 39/50\n144/144 [==============================] - 0s 111us/step - loss: 0.8216 - acc: 0.6458 - val_loss: 0.8376 - val_acc: 0.5556\nEpoch 40/50\n144/144 [==============================] - 0s 127us/step - loss: 0.8348 - acc: 0.5903 - val_loss: 0.8320 - val_acc: 0.6667\nEpoch 41/50\n144/144 [==============================] - 0s 97us/step - loss: 0.7779 - acc: 0.6528 - val_loss: 0.8274 - val_acc: 0.6111\nEpoch 42/50\n144/144 [==============================] - 0s 95us/step - loss: 0.8031 - acc: 0.6181 - val_loss: 0.8249 - val_acc: 0.5556\nEpoch 43/50\n144/144 [==============================] - 0s 93us/step - loss: 0.8192 - acc: 0.6042 - val_loss: 0.8221 - val_acc: 0.5278\nEpoch 44/50\n144/144 [==============================] - 0s 73us/step - loss: 0.8374 - acc: 0.5903 - val_loss: 0.8187 - val_acc: 0.5556\nEpoch 45/50\n144/144 [==============================] - 0s 77us/step - loss: 0.7896 - acc: 0.5903 - val_loss: 0.8259 - val_acc: 0.4722\nEpoch 46/50\n144/144 [==============================] - 0s 83us/step - loss: 0.8045 - acc: 0.5833 - val_loss: 0.8416 - val_acc: 0.4444\nEpoch 47/50\n144/144 [==============================] - 0s 97us/step - loss: 0.7633 - acc: 0.6806 - val_loss: 0.8655 - val_acc: 0.4444\nEpoch 48/50\n144/144 [==============================] - 0s 87us/step - loss: 0.8088 - acc: 0.6250 - val_loss: 0.8355 - val_acc: 0.4444\nEpoch 49/50\n144/144 [==============================] - 0s 119us/step - loss: 0.7518 - acc: 0.6597 - val_loss: 0.8255 - val_acc: 0.4722\nEpoch 50/50\n144/144 [==============================] - 0s 110us/step - loss: 0.7830 - acc: 0.6389 - val_loss: 0.8165 - val_acc: 0.4722\nCPU times: user 3.01 s, sys: 70.6 ms, total: 3.08 s\nWall time: 2.89 s\n"
],
[
"score = model.evaluate(x_test, y_test, verbose=0)\nprint('Test loss:', score[0])\nprint('Test accuracy:', score[1])",
"Test loss: 0.8165454334682889\nTest accuracy: 0.4722222222222222\n"
]
],
[
[
"---",
"_____no_output_____"
],
[
"# Ensemble Methods",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## Bagging Strategies",
"_____no_output_____"
],
[
"### Random Forests",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier",
"_____no_output_____"
],
[
"# x_train, x_test, y_train, y_test",
"_____no_output_____"
],
[
"\nclf = RandomForestClassifier(n_estimators=100, max_depth=2, random_state=0)",
"_____no_output_____"
],
[
"clf.fit(x_train, y_train)",
"_____no_output_____"
],
[
"print(clf.feature_importances_)",
"[0.04117183 0.00496237 0.2124435 0.11033686 0.03552996 0.12413992\n 0.09559455 0. 0.00413384 0.03759457 0.05501764 0.02024867\n 0.03323923 0.07819483 0.14739222]\n"
],
[
"print(clf.predict(x_test))",
"[0 0 0 1 0 1 0 0 1 1 1 1 1 0 0 0 0 0 1 1 0 0 1 0 0 1 0 0 0 1 1 0 1 0 0 0]\n"
],
[
"# make predictions for test data\ny_pred = clf.predict(x_test)\npredictions = [round(value) for value in y_pred]",
"_____no_output_____"
],
[
"# evaluate predictions\naccuracy = accuracy_score(y_test, predictions)\nprint(\"Accuracy: %.2f%%\" % (accuracy * 100.0))",
"Accuracy: 83.33%\n"
]
],
[
[
"### ExtraTrees",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import ExtraTreesClassifier",
"_____no_output_____"
],
[
"# x_train, x_test, y_train, y_test",
"_____no_output_____"
],
[
"\nclf = ExtraTreesClassifier(n_estimators=100, max_depth=2, random_state=0)",
"_____no_output_____"
],
[
"clf.fit(x_train, y_train)",
"_____no_output_____"
],
[
"print(clf.feature_importances_)",
"[0.03595362 0.00066439 0.19711 0.21279172 0.00303455 0.06847946\n 0.10905054 0.00071238 0.00961193 0.00356046 0.03753535 0.04054296\n 0.01259753 0.06053845 0.20781666]\n"
],
[
"print(clf.predict(x_test))",
"[0 0 0 1 0 1 0 0 1 1 1 1 1 0 0 0 0 0 1 1 0 0 1 0 0 1 1 0 0 1 1 0 1 0 0 0]\n"
],
[
"# make predictions for test data\ny_pred = clf.predict(x_test)\npredictions = [round(value) for value in y_pred]",
"_____no_output_____"
],
[
"# evaluate predictions\naccuracy = accuracy_score(y_test, predictions)\nprint(\"Accuracy: %.2f%%\" % (accuracy * 100.0))",
"Accuracy: 80.56%\n"
],
[
"fig = plt.figure(figsize=(10,5))\n\nplot_learning_curves(x_train, y_train, x_test, y_test, clf)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Stacking Strategies",
"_____no_output_____"
],
[
"### SuperLearner",
"_____no_output_____"
],
[
"## Boosting Strategies",
"_____no_output_____"
],
[
"### xgboost",
"_____no_output_____"
]
],
[
[
"# import xgboost as xgb\nfrom xgboost import XGBClassifier\nfrom sklearn.metrics import accuracy_score",
"_____no_output_____"
],
[
"# x_train, x_test, y_train, y_test",
"_____no_output_____"
],
[
"model = XGBClassifier()\nmodel.fit(x_train, y_train)",
"_____no_output_____"
],
[
"print(model)",
"XGBClassifier(base_score=0.5, booster='gbtree', colsample_bylevel=1,\n colsample_bytree=1, gamma=0, learning_rate=0.1, max_delta_step=0,\n max_depth=3, min_child_weight=1, missing=None, n_estimators=100,\n n_jobs=1, nthread=None, objective='binary:logistic', random_state=0,\n reg_alpha=0, reg_lambda=1, scale_pos_weight=1, seed=None,\n silent=True, subsample=1)\n"
],
[
"# make predictions for test data\ny_pred = model.predict(x_test)\npredictions = [round(value) for value in y_pred]",
"/usr/local/lib/python3.5/dist-packages/sklearn/preprocessing/label.py:151: DeprecationWarning: The truth value of an empty array is ambiguous. Returning False, but in future this will result in an error. Use `array.size > 0` to check that an array is not empty.\n if diff:\n"
],
[
"# evaluate predictions\naccuracy = accuracy_score(y_test, predictions)\nprint(\"Accuracy: %.2f%%\" % (accuracy * 100.0))",
"Accuracy: 86.11%\n"
]
],
[
[
"---",
"_____no_output_____"
],
[
"# Bibliography",
"_____no_output_____"
],
[
"+ https://medium.com/@datalesdatales/why-you-should-be-plotting-learning-curves-in-your-next-machine-learning-project-221bae60c53\n\n+ https://slideplayer.com/slide/4684120/15/images/6/Outline+Bias%2FVariance+Tradeoff+Ensemble+methods+that+minimize+variance.jpg\n + https://slideplayer.com/slide/4684120/\n\n+ plot confusion matrix\n+ http://rasbt.github.io/mlxtend/user_guide/plotting/plot_learning_curves/\n+ https://machinelearningmastery.com/roc-curves-and-precision-recall-curves-for-classification-in-python/\n+ \n\n\n+ http://docs.h2o.ai/h2o-tutorials/latest-stable/tutorials/ensembles-stacking/index.html\n",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cb4689c2955240b0969e100736a790f2a2726815 | 148,042 | ipynb | Jupyter Notebook | module1-join-and-reshape-data/LS_DS_121_Join_and_Reshape_Data.ipynb | mikvikpik/DS-Unit-1-Sprint-2-Data-Wrangling-and-Storytelling | 1fe9eaa78293abb6b01c1ca4a3f8a62a89f76398 | [
"MIT"
] | null | null | null | module1-join-and-reshape-data/LS_DS_121_Join_and_Reshape_Data.ipynb | mikvikpik/DS-Unit-1-Sprint-2-Data-Wrangling-and-Storytelling | 1fe9eaa78293abb6b01c1ca4a3f8a62a89f76398 | [
"MIT"
] | null | null | null | module1-join-and-reshape-data/LS_DS_121_Join_and_Reshape_Data.ipynb | mikvikpik/DS-Unit-1-Sprint-2-Data-Wrangling-and-Storytelling | 1fe9eaa78293abb6b01c1ca4a3f8a62a89f76398 | [
"MIT"
] | null | null | null | 32.479596 | 320 | 0.334891 | [
[
[
"<a href=\"https://colab.research.google.com/github/mikvikpik/DS-Unit-1-Sprint-2-Data-Wrangling-and-Storytelling/blob/master/module1-join-and-reshape-data/LS_DS_121_Join_and_Reshape_Data.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"# used for machine learning when doing suggestions for purchasing",
"_____no_output_____"
]
],
[
[
"_Lambda School Data Science_\n\n# Join and Reshape datasets\n\nObjectives\n- concatenate data with pandas\n- merge data with pandas\n- understand tidy data formatting\n- melt and pivot data with pandas\n\nLinks\n- [Pandas Cheat Sheet](https://github.com/pandas-dev/pandas/blob/master/doc/cheatsheet/Pandas_Cheat_Sheet.pdf)\n- [Tidy Data](https://en.wikipedia.org/wiki/Tidy_data)\n - Combine Data Sets: Standard Joins\n - Tidy Data\n - Reshaping Data\n- Python Data Science Handbook\n - [Chapter 3.6](https://jakevdp.github.io/PythonDataScienceHandbook/03.06-concat-and-append.html), Combining Datasets: Concat and Append\n - [Chapter 3.7](https://jakevdp.github.io/PythonDataScienceHandbook/03.07-merge-and-join.html), Combining Datasets: Merge and Join\n - [Chapter 3.8](https://jakevdp.github.io/PythonDataScienceHandbook/03.08-aggregation-and-grouping.html), Aggregation and Grouping\n - [Chapter 3.9](https://jakevdp.github.io/PythonDataScienceHandbook/03.09-pivot-tables.html), Pivot Tables\n \nReference\n- Pandas Documentation: [Reshaping and Pivot Tables](https://pandas.pydata.org/pandas-docs/stable/reshaping.html)\n- Modern Pandas, Part 5: [Tidy Data](https://tomaugspurger.github.io/modern-5-tidy.html)",
"_____no_output_____"
],
[
"## Download data\n\nWe’ll work with a dataset of [3 Million Instacart Orders, Open Sourced](https://tech.instacart.com/3-million-instacart-orders-open-sourced-d40d29ead6f2)!",
"_____no_output_____"
]
],
[
[
"!wget https://s3.amazonaws.com/instacart-datasets/instacart_online_grocery_shopping_2017_05_01.tar.gz",
"--2019-06-03 21:52:12-- https://s3.amazonaws.com/instacart-datasets/instacart_online_grocery_shopping_2017_05_01.tar.gz\nResolving s3.amazonaws.com (s3.amazonaws.com)... 52.216.230.93\nConnecting to s3.amazonaws.com (s3.amazonaws.com)|52.216.230.93|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 205548478 (196M) [application/x-gzip]\nSaving to: ‘instacart_online_grocery_shopping_2017_05_01.tar.gz’\n\ninstacart_online_gr 100%[===================>] 196.03M 66.8MB/s in 2.9s \n\n2019-06-03 21:52:15 (66.8 MB/s) - ‘instacart_online_grocery_shopping_2017_05_01.tar.gz’ saved [205548478/205548478]\n\n"
],
[
"!tar --gunzip --extract --verbose --file=instacart_online_grocery_shopping_2017_05_01.tar.gz",
"instacart_2017_05_01/\ninstacart_2017_05_01/._aisles.csv\ninstacart_2017_05_01/aisles.csv\ninstacart_2017_05_01/._departments.csv\ninstacart_2017_05_01/departments.csv\ninstacart_2017_05_01/._order_products__prior.csv\ninstacart_2017_05_01/order_products__prior.csv\ninstacart_2017_05_01/._order_products__train.csv\ninstacart_2017_05_01/order_products__train.csv\ninstacart_2017_05_01/._orders.csv\ninstacart_2017_05_01/orders.csv\ninstacart_2017_05_01/._products.csv\ninstacart_2017_05_01/products.csv\n"
],
[
"%cd instacart_2017_05_01",
"/content/instacart_2017_05_01\n"
],
[
"!ls -lh *.csv",
"-rw-r--r-- 1 502 staff 2.6K May 2 2017 aisles.csv\n-rw-r--r-- 1 502 staff 270 May 2 2017 departments.csv\n-rw-r--r-- 1 502 staff 551M May 2 2017 order_products__prior.csv\n-rw-r--r-- 1 502 staff 24M May 2 2017 order_products__train.csv\n-rw-r--r-- 1 502 staff 104M May 2 2017 orders.csv\n-rw-r--r-- 1 502 staff 2.1M May 2 2017 products.csv\n"
]
],
[
[
"# Join Datasets",
"_____no_output_____"
],
[
"## Goal: Reproduce this example\n\nThe first two orders for user id 1:",
"_____no_output_____"
]
],
[
[
"from IPython.display import display, Image\nurl = 'https://cdn-images-1.medium.com/max/1600/1*vYGFQCafJtGBBX5mbl0xyw.png'\nexample = Image(url=url, width=600)\n\ndisplay(example) #important to refer to what we are creating",
"_____no_output_____"
]
],
[
[
"## Load data\n\nHere's a list of all six CSV filenames",
"_____no_output_____"
]
],
[
[
"!ls -lh *.csv",
"-rw-r--r-- 1 502 staff 2.6K May 2 2017 aisles.csv\n-rw-r--r-- 1 502 staff 270 May 2 2017 departments.csv\n-rw-r--r-- 1 502 staff 551M May 2 2017 order_products__prior.csv\n-rw-r--r-- 1 502 staff 24M May 2 2017 order_products__train.csv\n-rw-r--r-- 1 502 staff 104M May 2 2017 orders.csv\n-rw-r--r-- 1 502 staff 2.1M May 2 2017 products.csv\n"
]
],
[
[
"For each CSV\n- Load it with pandas\n- Look at the dataframe's shape\n- Look at its head (first rows)\n- `display(example)`\n- Which columns does it have in common with the example we want to reproduce?",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"### aisles",
"_____no_output_____"
]
],
[
[
"aisles = pd.read_csv('aisles.csv')\nprint(aisles.shape)\naisles.head()",
"(134, 2)\n"
],
[
"display(example)\n# aisles.csv not found or needed",
"_____no_output_____"
]
],
[
[
"### departments",
"_____no_output_____"
]
],
[
[
"!head departments.csv",
"department_id,department\n1,frozen\n2,other\n3,bakery\n4,produce\n5,alcohol\n6,international\n7,beverages\n8,pets\n9,dry goods pasta\n"
],
[
"departments = pd.read_csv('departments.csv')\nprint(departments.shape)\ndepartments.head()",
"(21, 2)\n"
],
[
"display(example)\n# department.csv not needed either because not found",
"_____no_output_____"
]
],
[
[
"### order_products__prior",
"_____no_output_____"
]
],
[
[
"# order_products__prior has doublle underscore in second underscore\norder_products__prior = pd.read_csv('order_products__prior.csv')\nprint(order_products__prior.shape)\norder_products__prior.head()",
"(32434489, 4)\n"
],
[
"display(example)\n# need order_id, product_id, add_to_cart_order, from order_products__prior",
"_____no_output_____"
],
[
"!free -m\n#shows memory used in bang notation",
" total used free shared buff/cache available\nMem: 13022 2400 7490 0 3131 12325\nSwap: 0 0 0\n"
],
[
"order_products__prior.groupby('order_id')\n#creates a new dataframe, groupby object\n#order_products__prior.groupby('order_id')['product_id'].count()\n#shows total count of each item\n#.mean() shows average count of ordered item",
"_____no_output_____"
]
],
[
[
"### order_products__train",
"_____no_output_____"
]
],
[
[
"order_products__train = pd.read_csv('order_products__train.csv')\nprint(order_products__train.shape)\norder_products__train.head()",
"(1384617, 4)\n"
],
[
"display(example)\n# need order_id, product_id, add_to_cart_order, from order_products__train\n# IMPORTANT - same data from order_products__prior\n# beware of overwrite",
"_____no_output_____"
]
],
[
[
"### orders",
"_____no_output_____"
]
],
[
[
"orders = pd.read_csv('orders.csv')\nprint(orders.shape)\norders.head()\n# best dataframe to start with",
"(3421083, 7)\n"
],
[
"display(example)\n# has order_id, order_number, order_dow, order_hour_of_day, from orders\n# useful to match up for merge as index to match rows on",
"_____no_output_____"
]
],
[
[
"### products",
"_____no_output_____"
]
],
[
[
"products = pd.read_csv('products.csv')\nprint(products.shape)\nproducts.head()",
"(49688, 4)\n"
],
[
"display(example)\n# need product_id, product_name, from products",
"_____no_output_____"
]
],
[
[
"## Concatenate order_products__prior and order_products__train",
"_____no_output_____"
]
],
[
[
"order_products__prior.shape",
"_____no_output_____"
],
[
"order_products__train.shape",
"_____no_output_____"
],
[
"# pd.concat used to add onto from \norder_products = pd.concat([order_products__prior, order_products__train])\norder_products.shape",
"_____no_output_____"
],
[
"# assert is used to simple test for usability\n# refactoring is to make the dataframe more readable and precise\n# no error means True statement\nassert len(order_products) == len(order_products__prior) + len(order_products__train)",
"_____no_output_____"
],
[
"# Filter order products to get as close as we can to example table\n# condition used to filter \n# this shows True/False for condition\norder_products['order_id'] == 2539329",
"_____no_output_____"
],
[
"# shows data of what condition is True\norder_products[order_products['order_id'] == 2539329]",
"_____no_output_____"
],
[
"import numpy as np",
"_____no_output_____"
],
[
"order_products[np.logical_or((order_products['order_id'] == 2539329), (order_products['order_id'] == 2398795))]",
"_____no_output_____"
],
[
"#condition = (orders['user_id' == 1]) & (orders['order_number'] <= 2)\n#\n#columns = [\n# 'user_id',\n# 'order_id',\n# 'order_number',\n# 'order_dow',\n# 'order_hour_of_day'\n#]\n#\n#subset = orders.loc[conditions, columns]\n#subset",
"_____no_output_____"
]
],
[
[
"## Get a subset of orders — the first two orders for user id 1",
"_____no_output_____"
],
[
"From `orders` dataframe:\n- user_id\n- order_id\n- order_number\n- order_dow\n- order_hour_of_day",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"## Merge dataframes",
"_____no_output_____"
],
[
"Merge the subset from `orders` with columns from `order_products`",
"_____no_output_____"
]
],
[
[
"# merging dataframes is the most important of today\nhelp(pd.merge)",
"_____no_output_____"
]
],
[
[
"Merge with columns from `products`",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"# Reshape Datasets",
"_____no_output_____"
],
[
"## Why reshape data?\n\n#### Some libraries prefer data in different formats\n\nFor example, the Seaborn data visualization library prefers data in \"Tidy\" format often (but not always).\n\n> \"[Seaborn will be most powerful when your datasets have a particular organization.](https://seaborn.pydata.org/introduction.html#organizing-datasets) This format ia alternately called “long-form” or “tidy” data and is described in detail by Hadley Wickham. The rules can be simply stated:\n\n> - Each variable is a column\n- Each observation is a row\n\n> A helpful mindset for determining whether your data are tidy is to think backwards from the plot you want to draw. From this perspective, a “variable” is something that will be assigned a role in the plot.\"\n\n#### Data science is often about putting square pegs in round holes\n\nHere's an inspiring [video clip from _Apollo 13_](https://www.youtube.com/watch?v=ry55--J4_VQ): “Invent a way to put a square peg in a round hole.” It's a good metaphor for data wrangling!",
"_____no_output_____"
],
[
"## Hadley Wickham's Examples\n\nFrom his paper, [Tidy Data](http://vita.had.co.nz/papers/tidy-data.html)",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport pandas as pd\nimport numpy as np\nimport seaborn as sns\n\ntable1 = pd.DataFrame(\n [[np.nan, 2],\n [16, 11], \n [3, 1]],\n index=['John Smith', 'Jane Doe', 'Mary Johnson'], \n columns=['treatmenta', 'treatmentb'])\n\ntable2 = table1.T",
"_____no_output_____"
]
],
[
[
"\"Table 1 provides some data about an imaginary experiment in a format commonly seen in the wild. \n\nThe table has two columns and three rows, and both rows and columns are labelled.\"",
"_____no_output_____"
]
],
[
[
"table1",
"_____no_output_____"
]
],
[
[
"\"There are many ways to structure the same underlying data. \n\nTable 2 shows the same data as Table 1, but the rows and columns have been transposed. The data is the same, but the layout is different.\"",
"_____no_output_____"
]
],
[
[
"table2",
"_____no_output_____"
]
],
[
[
"\"Table 3 reorganises Table 1 to make the values, variables and obserations more clear.\n\nTable 3 is the tidy version of Table 1. Each row represents an observation, the result of one treatment on one person, and each column is a variable.\"\n\n| name | trt | result |\n|--------------|-----|--------|\n| John Smith | a | - |\n| Jane Doe | a | 16 |\n| Mary Johnson | a | 3 |\n| John Smith | b | 2 |\n| Jane Doe | b | 11 |\n| Mary Johnson | b | 1 |",
"_____no_output_____"
],
[
"## Table 1 --> Tidy\n\nWe can use the pandas `melt` function to reshape Table 1 into Tidy format.",
"_____no_output_____"
]
],
[
[
"table1 = table1.reset_index()\ntable1",
"_____no_output_____"
],
[
"# id_vars is the unit of observation\n# when calling melt pick id_vars very carefully\n\ntidy = table1.melt(id_vars='index')\ntidy",
"_____no_output_____"
],
[
"tidy = tidy.rename(columns={\n 'index': 'name',\n 'variable': 'trt',\n 'value': 'result'\n})\n\ntidy",
"_____no_output_____"
],
[
"tidy.trt = tidy.trt.str.replace('treatment', '')\n\ntidy",
"_____no_output_____"
]
],
[
[
"## Table 2 --> Tidy",
"_____no_output_____"
]
],
[
[
"tidy = table2.reset_index().melt(id_vars = 'index')\ntidy",
"_____no_output_____"
],
[
"tidy = tidy[['variable', 'index', 'value']]\ntidy",
"_____no_output_____"
],
[
"tidy = tidy.rename(columns = {\n 'variable': 'name',\n 'index': 'trt',\n 'value': 'result'\n})\n\ntidy",
"_____no_output_____"
],
[
"tidy.trt = tidy.trt.str.replace('treatment', '')\n\ntidy",
"_____no_output_____"
]
],
[
[
"## Tidy --> Table 1\n\nThe `pivot_table` function is the inverse of `melt`.",
"_____no_output_____"
]
],
[
[
"tidy = tidy.pivot_table(index='name', columns='trt', values='result')\n\ntidy",
"_____no_output_____"
]
],
[
[
"## Tidy --> Table 2",
"_____no_output_____"
]
],
[
[
"tidy = tidy.T\ntidy",
"_____no_output_____"
]
],
[
[
"# Seaborn example\n\nThe rules can be simply stated:\n\n- Each variable is a column\n- Each observation is a row\n\nA helpful mindset for determining whether your data are tidy is to think backwards from the plot you want to draw. From this perspective, a “variable” is something that will be assigned a role in the plot.\"",
"_____no_output_____"
]
],
[
[
"sns.catplot(x='trt', y='result', col='name', \n kind='bar', data=tidy, height=2);",
"_____no_output_____"
]
],
[
[
"## Now with Instacart data",
"_____no_output_____"
]
],
[
[
"products = pd.read_csv('products.csv')\n\norder_products = pd.concat([pd.read_csv('order_products__prior.csv'), \n pd.read_csv('order_products__train.csv')])\n\norders = pd.read_csv('orders.csv')",
"_____no_output_____"
]
],
[
[
"## Goal: Reproduce part of this example\n\nInstead of a plot with 50 products, we'll just do two — the first products from each list\n- Half And Half Ultra Pasteurized\n- Half Baked Frozen Yogurt",
"_____no_output_____"
]
],
[
[
"from IPython.display import display, Image\nurl = 'https://cdn-images-1.medium.com/max/1600/1*wKfV6OV-_1Ipwrl7AjjSuw.png'\nexample = Image(url=url, width=600)\n\ndisplay(example)",
"_____no_output_____"
]
],
[
[
"So, given a `product_name` we need to calculate its `order_hour_of_day` pattern.",
"_____no_output_____"
],
[
"## Subset and Merge\n\nOne challenge of performing a merge on this data is that the `products` and `orders` datasets do not have any common columns that we can merge on. Due to this we will have to use the `order_products` dataset to provide the columns that we will use to perform the merge.",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"## 4 ways to reshape and plot",
"_____no_output_____"
],
[
"### 1. value_counts",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"### 2. crosstab",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"### 3. Pivot Table",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"### 4. melt",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"# Assignment\n\n## Join Data Section\n\nThese are the top 10 most frequently ordered products. How many times was each ordered? \n\n1. Banana\n2. Bag of Organic Bananas\n3. Organic Strawberries\n4. Organic Baby Spinach \n5. Organic Hass Avocado\n6. Organic Avocado\n7. Large Lemon \n8. Strawberries\n9. Limes \n10. Organic Whole Milk\n\nFirst, write down which columns you need and which dataframes have them.\n\nNext, merge these into a single dataframe.\n\nThen, use pandas functions from the previous lesson to get the counts of the top 10 most frequently ordered products.\n\n## Reshape Data Section\n\n- Replicate the lesson code\n- Complete the code cells we skipped near the beginning of the notebook\n- Table 2 --> Tidy\n- Tidy --> Table 2\n- Load seaborn's `flights` dataset by running the cell below. Then create a pivot table showing the number of passengers by month and year. Use year for the index and month for the columns. You've done it right if you get 112 passengers for January 1949 and 432 passengers for December 1960.",
"_____no_output_____"
]
],
[
[
"# Join Data Section Assignment\n# Need 'product_name', 'product_id' from 'products'\n# Need products_list dataframe merged to orders\n# Then need sum of produce_list frequency. Each time they were ordered.\n# creating products_list from products\nproducts_list = products[['product_name', 'product_id']]\nprint(products_list.shape)\nproducts_list.head()",
"(49688, 2)\n"
],
[
"# merge products list to orders by product_id\nproducts_list = pd.merge(products_list, order_products, how='left', on='product_id')\nprint(products_list.shape)\nproducts_list.head(20)",
"(33819109, 5)\n"
],
[
"# create list of only shopping_list\nshopping_list = ['Banana', 'Bag of Organic Bananas', 'Organic Strawberries',\n 'Organic Baby Spinach', 'Organic Hass Avocado', 'Organic Avocado',\n 'Large Lemon', 'Strawberries', 'Limes', 'Organic Whole Milk']\n# filter products_list by shopping_list\nproducts_list = products_list.drop(['add_to_cart_order','reordered'], axis=1)\nproducts_list.head()",
"_____no_output_____"
],
[
"# created dataframe of only shopping_list items included\nproducts_list = products_list[products_list['product_name'].isin(shopping_list)] \nprint(products_list.shape)\nproducts_list.head()",
"(2418314, 3)\n"
],
[
"# counts of each item ordered\nproducts_list['product_name'].value_counts()",
"_____no_output_____"
],
[
"# Reshape Data Section\n# Load flights\n# Pivot using .T\n# Use year for the index and month for the columns. \n# You've done it right if you get 112 passengers for January 1949 and 432 passengers for December 1960.",
"_____no_output_____"
],
[
"flights = sns.load_dataset('flights')",
"_____no_output_____"
],
[
"flights = flights.T\nflights",
"_____no_output_____"
]
],
[
[
"## Join Data Stretch Challenge\n\nThe [Instacart blog post](https://tech.instacart.com/3-million-instacart-orders-open-sourced-d40d29ead6f2) has a visualization of \"**Popular products** purchased earliest in the day (green) and latest in the day (red).\" \n\nThe post says,\n\n> \"We can also see the time of day that users purchase specific products.\n\n> Healthier snacks and staples tend to be purchased earlier in the day, whereas ice cream (especially Half Baked and The Tonight Dough) are far more popular when customers are ordering in the evening.\n\n> **In fact, of the top 25 latest ordered products, the first 24 are ice cream! The last one, of course, is a frozen pizza.**\"\n\nYour challenge is to reproduce the list of the top 25 latest ordered popular products.\n\nWe'll define \"popular products\" as products with more than 2,900 orders.\n\n## Reshape Data Stretch Challenge\n\n_Try whatever sounds most interesting to you!_\n\n- Replicate more of Instacart's visualization showing \"Hour of Day Ordered\" vs \"Percent of Orders by Product\"\n- Replicate parts of the other visualization from [Instacart's blog post](https://tech.instacart.com/3-million-instacart-orders-open-sourced-d40d29ead6f2), showing \"Number of Purchases\" vs \"Percent Reorder Purchases\"\n- Get the most recent order for each user in Instacart's dataset. This is a useful baseline when [predicting a user's next order](https://www.kaggle.com/c/instacart-market-basket-analysis)\n- Replicate parts of the blog post linked at the top of this notebook: [Modern Pandas, Part 5: Tidy Data](https://tomaugspurger.github.io/modern-5-tidy.html)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb4689e58726aa8ef12814734fc8aae875e8ccd3 | 6,283 | ipynb | Jupyter Notebook | Final_KNN_B&E.ipynb | beny2000/HeartDiseaseClassifier | a4b7f02893a95246060ded9d4112e8c2b5255f75 | [
"Unlicense"
] | 4 | 2020-11-18T01:58:18.000Z | 2020-11-18T02:02:45.000Z | Final_KNN_B&E.ipynb | DelaneySteve/HeartDiseaseClassifier | a4b7f02893a95246060ded9d4112e8c2b5255f75 | [
"Unlicense"
] | null | null | null | Final_KNN_B&E.ipynb | DelaneySteve/HeartDiseaseClassifier | a4b7f02893a95246060ded9d4112e8c2b5255f75 | [
"Unlicense"
] | 3 | 2020-11-18T01:58:20.000Z | 2021-02-25T11:29:43.000Z | 33.59893 | 243 | 0.514245 | [
[
[
"<a href=\"https://colab.research.google.com/github/beny2000/HeartDiseaseClassifier/blob/main/Final_KNN_B%26E.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"#imports\nimport sys\nimport pandas as pd\nimport numpy as np\nimport sklearn\nimport matplotlib\nimport keras\nimport math\nfrom sklearn import preprocessing\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.preprocessing import MinMaxScaler\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.preprocessing import RobustScaler\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.preprocessing import scale\nimport pickle\n\n!pip install anvil-uplink\nimport anvil.server\nanvil.server.connect(\"2ZZFEIO7RSWJ2NSXVOIJU7NK-PUMVCYIB7OL5ZPSL\")\n\n",
"Requirement already satisfied: anvil-uplink in /usr/local/lib/python3.6/dist-packages (0.3.34)\nRequirement already satisfied: ws4py in /usr/local/lib/python3.6/dist-packages (from anvil-uplink) (0.5.1)\nRequirement already satisfied: argparse in /usr/local/lib/python3.6/dist-packages (from anvil-uplink) (1.4.0)\nRequirement already satisfied: future in /usr/local/lib/python3.6/dist-packages (from anvil-uplink) (0.16.0)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from anvil-uplink) (1.15.0)\nConnecting to wss://anvil.works/uplink\nAnvil websocket open\nConnected to \"Default environment (dev)\" as SERVER\n"
]
],
[
[
"KNN\n",
"_____no_output_____"
]
],
[
[
"#read data\nemily = pd.read_csv('heart.csv')\ndata=emily\n\n#split into features and labels\nX = data.iloc[:,:-1].values\ny = data.iloc[:,13].values\n\n#split into test & train\nx_train, x_test, y_train, y_test = train_test_split(X,y, test_size=0.2)\n\n#preprocess data\nscaler = RobustScaler().fit(X)\nx_Train = scaler.transform(x_train)\nx_Test = scaler.transform(x_test)\n\n#build model\nmodel = KNeighborsClassifier(n_neighbors=5)\nmodel.fit(x_Train, y_train)\n\n#save model\npickle.dump(model, open('model_new.pickle', 'wb'))",
"_____no_output_____"
],
[
"#Check model performance on the test set\nprint('Accuracy of K-NN classifier on training set: {:.2f}'\n .format(model.score(x_Train, y_train)))\nprint('Accuracy of K-NN classifier on test set: {:.2f}'\n .format(model.score(x_Test, y_test)))\nprint(\"Pediction for\", x_Test[0], model.predict([x_Test[0]]), \"Correct prediction:\", y_test[0])",
"Accuracy of K-NN classifier on training set: 0.89\nAccuracy of K-NN classifier on test set: 0.82\nPediction for [ 0.44444444 -1. -0.5 0.75 1.05511811 0.\n -1. -0.21538462 1. 0.125 0. 0.\n 1. ] [0] Correct prediction: 0\n"
],
[
"#Make predict using model\r\n\r\n#[age, sex, cp, trestbps, chol, fbs, restecg, thalach, exang, oldpeak, slope, ca, thal]\r\n#input = [51,\t0,\t0,\t130,\t305,\t0,\t1,\t142,\t1,\t1.2,\t1,\t0,\t3] #0\r\n#input = [63,\t1,\t3,\t145,\t233,\t1,\t0,\t150,\t0,\t2.3,\t0,\t0,\t1] #1\r\n\r\[email protected]\r\ndef knnpredict(input):\r\n #loads model and makes prediction on input data\r\n\r\n model = pickle.load(open('model_new.pickle', 'rb'))\r\n input_scaled = scaler.transform([np.array(input)])\r\n\r\n return model.predict(input_scaled)[0], model.score(x_Test, y_test)\r\n",
"_____no_output_____"
],
[
"anvil.server.wait_forever()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cb468e0f33f4da28dd56e35000834673b9d0e623 | 138,806 | ipynb | Jupyter Notebook | examples/resampling/GlobalCS.ipynb | NaIwo/multi-imbalance | 237c5842b27a58edfdfb88073faa0021eb243348 | [
"MIT"
] | 63 | 2019-08-12T09:12:53.000Z | 2022-03-29T10:39:37.000Z | examples/resampling/GlobalCS.ipynb | NaIwo/multi-imbalance | 237c5842b27a58edfdfb88073faa0021eb243348 | [
"MIT"
] | 31 | 2019-08-15T20:24:05.000Z | 2022-03-21T11:54:05.000Z | examples/resampling/GlobalCS.ipynb | NaIwo/multi-imbalance | 237c5842b27a58edfdfb88073faa0021eb243348 | [
"MIT"
] | 6 | 2020-12-19T22:39:47.000Z | 2021-07-09T02:24:53.000Z | 775.452514 | 134,387 | 0.951335 | [
[
[
"Unzip datasets and prepare data:",
"_____no_output_____"
]
],
[
[
"from collections import Counter\n\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom sklearn.decomposition import PCA\n\nfrom multi_imbalance.datasets import load_datasets\nfrom multi_imbalance.resampling.global_cs import GlobalCS\nfrom multi_imbalance.utils.data import construct_flat_2pc_df\n\n%matplotlib inline\nsns.set_style('darkgrid')\n\ndf = load_datasets()['new_ecoli']\nX, y = df['data'], df['target']\nprint(X[:5])\nprint(y[:5])",
"[[0.49 0.29 0.48 0.5 0.56 0.24 0.35]\n [0.07 0.4 0.48 0.5 0.54 0.35 0.44]\n [0.56 0.4 0.48 0.5 0.49 0.37 0.46]\n [0.59 0.49 0.48 0.5 0.52 0.45 0.36]\n [0.23 0.32 0.48 0.5 0.55 0.25 0.35]]\n[0 0 0 0 0]\n"
]
],
[
[
"Resample data using Global CS algorithm",
"_____no_output_____"
]
],
[
[
"clf = GlobalCS()\nresampled_X, resampled_y = clf.fit_resample(X, y)",
"_____no_output_____"
]
],
[
[
"Compare results by plotting data in 2 dimensions",
"_____no_output_____"
]
],
[
[
"n = len(Counter(y).keys())\np = sns.color_palette(\"husl\", n)\n\npca = PCA(n_components=2)\npca.fit(X)\n\nfig, axs = plt.subplots(ncols=2, nrows=2)\nfig.set_size_inches( 16, 10)\naxs = axs.flatten()\n\naxs[1].set_title(\"Base\")\nsns.countplot(y, ax=axs[0], palette=p)\nX = pca.transform(X)\ndf = construct_flat_2pc_df(X, y)\nsns.scatterplot(x='x1', y='x2', hue='y', style='y', data=df, alpha=0.7, ax=axs[1], legend='full', palette=p)\n\n\naxs[3].set_title(\"MDO\")\nsns.countplot(resampled_y, ax=axs[2],palette=p)\nresampled_X = pca.transform(resampled_X)\ndf = construct_flat_2pc_df(resampled_X, resampled_y)\nsns.scatterplot(x='x1', y='x2', hue='y', style='y', data=df, alpha=0.7, ax=axs[3], legend='full', palette=p)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb46942750c6471ba3f31da6b7bee96afc467164 | 17,992 | ipynb | Jupyter Notebook | notebooks/207-vision-paddlegan-superresolution/207-vision-paddlegan-superresolution.ipynb | zzk0/openvino_notebooks | 8303642d77960e247a04ff571e733fae1c8bfba4 | [
"Apache-2.0"
] | 1 | 2022-03-25T10:35:54.000Z | 2022-03-25T10:35:54.000Z | notebooks/207-vision-paddlegan-superresolution/207-vision-paddlegan-superresolution.ipynb | zzk0/openvino_notebooks | 8303642d77960e247a04ff571e733fae1c8bfba4 | [
"Apache-2.0"
] | null | null | null | notebooks/207-vision-paddlegan-superresolution/207-vision-paddlegan-superresolution.ipynb | zzk0/openvino_notebooks | 8303642d77960e247a04ff571e733fae1c8bfba4 | [
"Apache-2.0"
] | null | null | null | 32.071301 | 694 | 0.596876 | [
[
[
"# Super Resolution with PaddleGAN and OpenVINO\n\nThis notebook demonstrates converting the RealSR (real-world super-resolution) model from [PaddlePaddle/PaddleGAN](https://github.com/PaddlePaddle/PaddleGAN) to OpenVINO's Intermediate Representation (IR) format, and shows inference results on both the PaddleGAN and IR models.\n\nFor more information about the various PaddleGAN superresolution models, see [PaddleGAN's documentation](https://github.com/PaddlePaddle/PaddleGAN/blob/develop/docs/en_US/tutorials/single_image_super_resolution.md). For more information about RealSR, see the [research paper](https://openaccess.thecvf.com/content_CVPRW_2020/papers/w31/Ji_Real-World_Super-Resolution_via_Kernel_Estimation_and_Noise_Injection_CVPRW_2020_paper.pdf) from CVPR 2020.\n\nThis notebook works best with small images (up to 800x600).",
"_____no_output_____"
],
[
"## Imports",
"_____no_output_____"
]
],
[
[
"import sys\nimport time\nimport warnings\nfrom pathlib import Path\n\nimport cv2\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport paddle\nfrom IPython.display import HTML, FileLink, ProgressBar, clear_output, display\nfrom IPython.display import Image as DisplayImage\nfrom PIL import Image\nfrom openvino.runtime import Core, PartialShape\nfrom paddle.static import InputSpec\nfrom ppgan.apps import RealSRPredictor\n\nsys.path.append(\"../utils\")\nfrom notebook_utils import NotebookAlert",
"_____no_output_____"
]
],
[
[
"## Settings",
"_____no_output_____"
]
],
[
[
"# The filenames of the downloaded and converted models\nMODEL_NAME = \"paddlegan_sr\"\nMODEL_DIR = Path(\"model\")\nOUTPUT_DIR = Path(\"output\")\nOUTPUT_DIR.mkdir(exist_ok=True)\n\nmodel_path = MODEL_DIR / MODEL_NAME\nir_path = model_path.with_suffix(\".xml\")\nonnx_path = model_path.with_suffix(\".onnx\")",
"_____no_output_____"
]
],
[
[
"## Inference on PaddlePaddle Model\n\n### Investigate PaddleGAN Model\n\nThe [PaddleGAN documentation](https://github.com/PaddlePaddle/PaddleGAN) explains to run the model with `sr.run()`. Let's see what that function does, and check other relevant functions that are called from that function. Adding `??` to the methods shows the docstring and source code.",
"_____no_output_____"
]
],
[
[
"# Running this cell will download the model weights if they have not been downloaded before\n# This may take a while\nsr = RealSRPredictor()",
"_____no_output_____"
],
[
"sr.run??",
"_____no_output_____"
],
[
"sr.run_image??",
"_____no_output_____"
],
[
"sr.norm??",
"_____no_output_____"
],
[
"sr.denorm??",
"_____no_output_____"
]
],
[
[
"The run checks whether the input is an image or a video. For an image, it loads the image as an RGB image, normalizes it, and converts it to a Paddle tensor. It is propagated to the network by calling `self.model()` and then \"denormalized\". The normalization function simply divides all image values by 255. This converts an image with integer values in the range of 0 to 255 to an image with floating point values in the range of 0 to 1. The denormalization function transforms the output from network shape (C,H,W) to image shape (H,W,C). It then clips the image values between 0 and 255, and converts the image to a standard RGB image with integer values in the range of 0 to 255.\n\nTo get more information about the model, we can check what it looks like with `sr.model??`. ",
"_____no_output_____"
]
],
[
[
"# sr.model??",
"_____no_output_____"
]
],
[
[
"### Do Inference\n\nTo show inference on the PaddlePaddle model, set PADDLEGAN_INFERENCE to True in the cell below. Performing inference may take some time.",
"_____no_output_____"
]
],
[
[
"# Set PADDLEGAN_INFERENCE to True to show inference on the PaddlePaddle model.\n# This may take a long time, especially for larger images.\n#\nPADDLEGAN_INFERENCE = False\nif PADDLEGAN_INFERENCE:\n # load the input image and convert to tensor with input shape\n IMAGE_PATH = Path(\"data/coco_tulips.jpg\")\n image = cv2.cvtColor(cv2.imread(str(IMAGE_PATH)), cv2.COLOR_BGR2RGB)\n input_image = image.transpose(2, 0, 1)[None, :, :, :] / 255\n input_tensor = paddle.to_tensor(input_image.astype(np.float32))\n if max(image.shape) > 400:\n NotebookAlert(\n f\"This image has shape {image.shape}. Doing inference will be slow \"\n \"and the notebook may stop responding. Set PADDLEGAN_INFERENCE to False \"\n \"to skip doing inference on the PaddlePaddle model.\",\n \"warning\",\n )",
"_____no_output_____"
],
[
"if PADDLEGAN_INFERENCE:\n # Do inference, and measure how long it takes\n print(f\"Start superresolution inference for {IMAGE_PATH.name} with shape {image.shape}...\")\n start_time = time.perf_counter()\n sr.model.eval()\n with paddle.no_grad():\n result = sr.model(input_tensor)\n end_time = time.perf_counter()\n duration = end_time - start_time\n result_image = (\n (result.numpy().squeeze() * 255).clip(0, 255).astype(\"uint8\").transpose((1, 2, 0))\n )\n print(f\"Superresolution image shape: {result_image.shape}\")\n print(f\"Inference duration: {duration:.2f} seconds\")\n plt.imshow(result_image);",
"_____no_output_____"
]
],
[
[
"## Convert PaddleGAN Model to ONNX and OpenVINO IR\n\nTo convert the PaddlePaddle model to OpenVINO IR, we first convert the model to ONNX, and then convert the ONNX model to the IR format.\n\n### Convert PaddlePaddle Model to ONNX",
"_____no_output_____"
]
],
[
[
"# Ignore PaddlePaddle warnings:\n# The behavior of expression A + B has been unified with elementwise_add(X, Y, axis=-1)\nwarnings.filterwarnings(\"ignore\")\nsr.model.eval()\n# ONNX export requires an input shape in this format as parameter\nx_spec = InputSpec([None, 3, 299, 299], \"float32\", \"x\")\npaddle.onnx.export(sr.model, str(model_path), input_spec=[x_spec], opset_version=13)",
"_____no_output_____"
]
],
[
[
"### Convert ONNX Model to OpenVINO IR",
"_____no_output_____"
]
],
[
[
"## Uncomment the command below to show Model Optimizer help, which shows the possible arguments for Model Optimizer\n# ! mo --help",
"_____no_output_____"
],
[
"if not ir_path.exists():\n print(\"Exporting ONNX model to IR... This may take a few minutes.\")\n ! mo --input_model $onnx_path --input_shape \"[1,3,299,299]\" --model_name $MODEL_NAME --output_dir \"$MODEL_DIR\" --data_type \"FP16\" --log_level \"CRITICAL\"",
"_____no_output_____"
]
],
[
[
"## Do Inference on IR Model",
"_____no_output_____"
]
],
[
[
"# Read network and get input and output names\nie = Core()\nmodel = ie.read_model(model=ir_path)\ninput_layer = next(iter(model.inputs))",
"_____no_output_____"
],
[
"# Load and show image\nIMAGE_PATH = Path(\"data/coco_tulips.jpg\")\nimage = cv2.cvtColor(cv2.imread(str(IMAGE_PATH)), cv2.COLOR_BGR2RGB)\nif max(image.shape) > 800:\n NotebookAlert(\n f\"This image has shape {image.shape}. The notebook works best with images with \"\n \"a maximum side of 800x600. Larger images may work well, but inference may \"\n \"be slow\",\n \"warning\",\n )\nplt.imshow(image)",
"_____no_output_____"
],
[
"# Reshape network to image size\nmodel.reshape({input_layer.any_name: PartialShape([1, 3, image.shape[0], image.shape[1]])})\n# Load network to the CPU device (this may take a few seconds)\ncompiled_model = ie.compile_model(model=model, device_name=\"CPU\")\noutput_layer = next(iter(compiled_model.outputs))",
"_____no_output_____"
],
[
"# Convert image to network input shape and divide pixel values by 255\n# See \"Investigate PaddleGAN model\" section\ninput_image = image.transpose(2, 0, 1)[None, :, :, :] / 255\nstart_time = time.perf_counter()\n# Do inference\nir_result = compiled_model([input_image])[output_layer]\nend_time = time.perf_counter()\nduration = end_time - start_time\nprint(f\"Inference duration: {duration:.2f} seconds\")",
"_____no_output_____"
],
[
"# Get result array in CHW format\nresult_array = ir_result.squeeze()\n# Convert array to image with same method as PaddleGAN:\n# Multiply by 255, clip values between 0 and 255, convert to HWC INT8 image\n# See \"Investigate PaddleGAN model\" section\nimage_super = (result_array * 255).clip(0, 255).astype(\"uint8\").transpose((1, 2, 0))\n# Resize image with bicubic upsampling for comparison\nimage_bicubic = cv2.resize(image, tuple(image_super.shape[:2][::-1]), interpolation=cv2.INTER_CUBIC)",
"_____no_output_____"
],
[
"plt.imshow(image_super)",
"_____no_output_____"
]
],
[
[
"### Show Animated GIF\n\nTo visualize the difference between the bicubic image and the superresolution image, we create an imated gif that switches between both versions.",
"_____no_output_____"
]
],
[
[
"result_pil = Image.fromarray(image_super)\nbicubic_pil = Image.fromarray(image_bicubic)\ngif_image_path = OUTPUT_DIR / Path(IMAGE_PATH.stem + \"_comparison.gif\")\nfinal_image_path = OUTPUT_DIR / Path(IMAGE_PATH.stem + \"_super.png\")\n\nresult_pil.save(\n fp=str(gif_image_path),\n format=\"GIF\",\n append_images=[bicubic_pil],\n save_all=True,\n duration=1000,\n loop=0,\n)\n\nresult_pil.save(fp=str(final_image_path), format=\"png\")\nDisplayImage(open(gif_image_path, \"rb\").read(), width=1920 // 2)",
"_____no_output_____"
]
],
[
[
"### Create Comparison Video\n\nCreate a video with a \"slider\", showing the bicubic image to the right and the superresolution image on the left. \n\nFor the video, the superresolution and bicubic image are resized to half the original width and height, to improve processing speed. This gives an indication of the superresolution effect. The video is saved as an .avi video. You can click on the link to download the video, or open it directly from the images directory, and play it locally.",
"_____no_output_____"
]
],
[
[
"FOURCC = cv2.VideoWriter_fourcc(*\"MJPG\")\nIMAGE_PATH = Path(IMAGE_PATH)\nresult_video_path = OUTPUT_DIR / Path(f\"{IMAGE_PATH.stem}_comparison_paddlegan.avi\")\nvideo_target_height, video_target_width = (\n image_super.shape[0] // 2,\n image_super.shape[1] // 2,\n)\n\nout_video = cv2.VideoWriter(\n str(result_video_path),\n FOURCC,\n 90,\n (video_target_width, video_target_height),\n)\n\nresized_result_image = cv2.resize(image_super, (video_target_width, video_target_height))[\n :, :, (2, 1, 0)\n]\nresized_bicubic_image = cv2.resize(image_bicubic, (video_target_width, video_target_height))[\n :, :, (2, 1, 0)\n]\n\nprogress_bar = ProgressBar(total=video_target_width)\nprogress_bar.display()\n\nfor i in range(2, video_target_width):\n # Create a frame where the left part (until i pixels width) contains the\n # superresolution image, and the right part (from i pixels width) contains\n # the bicubic image\n comparison_frame = np.hstack(\n (\n resized_result_image[:, :i, :],\n resized_bicubic_image[:, i:, :],\n )\n )\n\n # create a small black border line between the superresolution\n # and bicubic part of the image\n comparison_frame[:, i - 1 : i + 1, :] = 0\n out_video.write(comparison_frame)\n progress_bar.progress = i\n progress_bar.update()\nout_video.release()\nclear_output()\n\nvideo_link = FileLink(result_video_path)\nvideo_link.html_link_str = \"<a href='%s' download>%s</a>\"\ndisplay(HTML(f\"The video has been saved to {video_link._repr_html_()}\"))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cb4699fdffc57913732930e602fe63fecec44f91 | 57,908 | ipynb | Jupyter Notebook | demo/full_demo.ipynb | joaopalmeiro/b2-presentation | 6d0ffa5874ecb6071ca23f358253d8a4b375ab49 | [
"MIT"
] | null | null | null | demo/full_demo.ipynb | joaopalmeiro/b2-presentation | 6d0ffa5874ecb6071ca23f358253d8a4b375ab49 | [
"MIT"
] | null | null | null | demo/full_demo.ipynb | joaopalmeiro/b2-presentation | 6d0ffa5874ecb6071ca23f358253d8a4b375ab49 | [
"MIT"
] | null | null | null | 221.869732 | 52,002 | 0.663 | [
[
[
"# Imports",
"_____no_output_____"
]
],
[
[
"from datetime import datetime\n\nfrom b2 import B2",
"_____no_output_____"
]
],
[
[
"# B2 kick-off and data loading",
"_____no_output_____"
],
[
"`fire_earlier.csv` and `fire_later.csv` are samples of the \"[1.88 Million US Wildfires](https://www.kaggle.com/rtatman/188-million-us-wildfires)\" dataset made available on Kaggle by Rachael Tatman.",
"_____no_output_____"
]
],
[
[
"b2 = B2()\n\ndata = b2.from_file(\"./data/fire_earlier.csv\")\n# data = b2.from_file(\"./data/fire_later.csv\")",
"_____no_output_____"
],
[
"data[\"DT\"] = data.apply(\n lambda ts: datetime.fromtimestamp(ts).replace(microsecond=0), \"DISCOVERY_DATE\"\n)\ndata[\"YEAR\"] = data.apply(lambda d: d.year, \"DT\")\ndata[\"MINUTE\"] = data.apply(lambda d: d.minute, \"DT\")\n\nb2.show_profile(data) # Update the dashboard columns",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
],
[
"data.num_rows",
"_____no_output_____"
]
],
[
[
"# B2 in action",
"_____no_output_____"
]
],
[
[
"states = data.group(\"STATE\")\nstates.vis()",
"_____no_output_____"
],
[
"# 🟡 01:47 🟡\nCAUSE_DESCR_data_dist = data.group('CAUSE_DESCR')\nCAUSE_DESCR_data_dist.vis()",
"_____no_output_____"
],
[
"large_fires = data.where(\"FIRE_SIZE\", lambda x: x > 1000)\ntime_vs_size = large_fires.select([\"DISCOVERY_TIME\", \"FIRE_SIZE\"])\ntime_vs_size.vis()",
"_____no_output_____"
],
[
"# 🟡 01:48 🟡\nYEAR_data_dist = data.group('YEAR')\nYEAR_data_dist.vis()",
"_____no_output_____"
],
[
"%%reactive\n\ndata.get_filtered_data().head(10)",
"_____no_output_____"
],
[
"%%reactive\n\nfiltered_locs = data.get_filtered_data().select([\"LATITUDE\", \"LONGITUDE\"])\nfiltered_locs.plot_heatmap(zoom_start=3, radius=6)",
"_____no_output_____"
],
[
"# 🔵 02:02 🔵\n# b2.sel([{\"CAUSE_DESCR_data_dist\": {\"CAUSE_DESCR\": [\"Lightning\"]}}])\nb2.sel([{\"CAUSE_DESCR_data_dist\": {\"CAUSE_DESCR\": [\"Debris Burning\"]}}])",
"_____no_output_____"
],
[
"# 🟠 02:15 🟠\n## Current snapshot queries:\n# data.where('CAUSE_DESCR', b2.are.contained_in(['Debris Burning'])).group('STATE')\n# data.group('CAUSE_DESCR')\n# data.where('CAUSE_DESCR', b2.are.contained_in(['Debris Burning'])).where('FIRE_SIZE', lambda x: x > 1000).select(['DISCOVERY_TIME', 'FIRE_SIZE'])\n# data.where('CAUSE_DESCR', b2.are.contained_in(['Debris Burning'])).group('YEAR')\nfrom IPython.display import HTML, display\ndisplay(HTML(\"\"\"<div><svg class=\"marks\" width=\"677\" height=\"173\" viewBox=\"0 0 677 173\" version=\"1.1\" xmlns=\"http://www.w3.org/2000/svg\" xmlns:xlink=\"http://www.w3.org/1999/xlink\"><rect width=\"677\" height=\"173\" style=\"fill: white;\"></rect><g transform=\"translate(52,10)\"><g class=\"mark-group role-frame root\"><g transform=\"translate(0,0)\"><path class=\"background\" d=\"M0.5,0.5h620v120h-620Z\" style=\"fill: none; stroke: #ddd;\"></path><g><g class=\"mark-group role-axis\"><g transform=\"translate(0.5,0.5)\"><path class=\"background\" d=\"M0,0h0v0h0Z\" style=\"pointer-events: none; fill: none;\"></path><g><g class=\"mark-rule role-axis-grid\" style=\"pointer-events: none;\"><line transform=\"translate(0,120)\" x2=\"620\" y2=\"0\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,90)\" x2=\"620\" y2=\"0\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,60)\" x2=\"620\" y2=\"0\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,30)\" x2=\"620\" y2=\"0\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,0)\" x2=\"620\" y2=\"0\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line></g></g><path class=\"foreground\" d=\"\" style=\"pointer-events: none; display: none; fill: none;\"></path></g></g><g class=\"mark-group role-axis\"><g transform=\"translate(0.5,120.5)\"><path class=\"background\" d=\"M0,0h0v0h0Z\" style=\"pointer-events: none; fill: none;\"></path><g><g class=\"mark-rule role-axis-tick\" style=\"pointer-events: none;\"><line transform=\"translate(10,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(30,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(50,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(70,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(90,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(110,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(130,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(150,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(170,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(190,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(210,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(230,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(250,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(270,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(290,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(310,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(330,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(350,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(370,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(390,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(410,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(430,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(450,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(470,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(490,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(510,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(530,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(550,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(570,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(590,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(610,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line></g><g class=\"mark-text role-axis-label\" style=\"pointer-events: none;\"><text text-anchor=\"end\" transform=\"translate(9.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">GA</text><text text-anchor=\"end\" transform=\"translate(29.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">MS</text><text text-anchor=\"end\" transform=\"translate(49.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">NY</text><text text-anchor=\"end\" transform=\"translate(69.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">WV</text><text text-anchor=\"end\" transform=\"translate(89.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">AZ</text><text text-anchor=\"end\" transform=\"translate(109.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">OK</text><text text-anchor=\"end\" transform=\"translate(129.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">CA</text><text text-anchor=\"end\" transform=\"translate(149.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">WI</text><text text-anchor=\"end\" transform=\"translate(169.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">ID</text><text text-anchor=\"end\" transform=\"translate(189.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">CO</text><text text-anchor=\"end\" transform=\"translate(209.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">MI</text><text text-anchor=\"end\" transform=\"translate(229.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">UT</text><text text-anchor=\"end\" transform=\"translate(249.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">OR</text><text text-anchor=\"end\" transform=\"translate(269.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">NV</text><text text-anchor=\"end\" transform=\"translate(289.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">MT</text><text text-anchor=\"end\" transform=\"translate(309.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">NM</text><text text-anchor=\"end\" transform=\"translate(329.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">WA</text><text text-anchor=\"end\" transform=\"translate(349.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">AL</text><text text-anchor=\"end\" transform=\"translate(369.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">ME</text><text text-anchor=\"end\" transform=\"translate(389.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">KY</text><text text-anchor=\"end\" transform=\"translate(409.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">PA</text><text text-anchor=\"end\" transform=\"translate(429.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">WY</text><text text-anchor=\"end\" transform=\"translate(449.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">SC</text><text text-anchor=\"end\" transform=\"translate(469.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">NC</text><text text-anchor=\"end\" transform=\"translate(489.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">TN</text><text text-anchor=\"end\" transform=\"translate(509.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">VA</text><text text-anchor=\"end\" transform=\"translate(529.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">SD</text><text text-anchor=\"end\" transform=\"translate(549.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">TX</text><text text-anchor=\"end\" transform=\"translate(569.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">FL</text><text text-anchor=\"end\" transform=\"translate(589.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">KS</text><text text-anchor=\"end\" transform=\"translate(609.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">ND</text></g><g class=\"mark-rule role-axis-domain\" style=\"pointer-events: none;\"><line transform=\"translate(0,0)\" x2=\"620\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line></g><g class=\"mark-text role-axis-title\" style=\"pointer-events: none;\"><text text-anchor=\"middle\" transform=\"translate(310,35.859375)\" style=\"font-family: sans-serif; font-size: 11px; font-weight: bold; fill: #000; opacity: 1;\">STATE</text></g></g><path class=\"foreground\" d=\"\" style=\"pointer-events: none; display: none; fill: none;\"></path></g></g><g class=\"mark-group role-axis\"><g transform=\"translate(0.5,0.5)\"><path class=\"background\" d=\"M0,0h0v0h0Z\" style=\"pointer-events: none; fill: none;\"></path><g><g class=\"mark-rule role-axis-tick\" style=\"pointer-events: none;\"><line transform=\"translate(0,120)\" x2=\"-5\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,90)\" x2=\"-5\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,60)\" x2=\"-5\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,30)\" x2=\"-5\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,0)\" x2=\"-5\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line></g><g class=\"mark-text role-axis-label\" style=\"pointer-events: none;\"><text text-anchor=\"end\" transform=\"translate(-7,123)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">0</text><text text-anchor=\"end\" transform=\"translate(-7,93)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">500</text><text text-anchor=\"end\" transform=\"translate(-7,63)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">1,000</text><text text-anchor=\"end\" transform=\"translate(-7,33)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">1,500</text><text text-anchor=\"end\" transform=\"translate(-7,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">2,000</text></g><g class=\"mark-rule role-axis-domain\" style=\"pointer-events: none;\"><line transform=\"translate(0,120)\" x2=\"0\" y2=\"-120\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line></g><g class=\"mark-text role-axis-title\" style=\"pointer-events: none;\"><text text-anchor=\"middle\" transform=\"translate(-35.423828125,60) rotate(-90) translate(0,-2)\" style=\"font-family: sans-serif; font-size: 11px; font-weight: bold; fill: #000; opacity: 1;\">count</text></g></g><path class=\"foreground\" d=\"\" style=\"pointer-events: none; display: none; fill: none;\"></path></g></g><g class=\"mark-rect role-mark layer_0_marks\"><path d=\"M341,117.48h18v2.519999999999996h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M81,105.66000000000001h18v14.33999999999999h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M121,108.17999999999999h18v11.820000000000007h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M181,112.98h18v7.019999999999996h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M561,119.94000000000001h18v0.05999999999998806h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M1,10.559999999999995h18v109.44h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M161,112.5h18v7.5h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M581,119.94000000000001h18v0.05999999999998806h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M381,117.84h18v2.1599999999999966h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M361,117.48h18v2.519999999999996h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M201,114.42h18v5.579999999999998h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M21,72.17999999999999h18v47.82000000000001h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M281,115.92h18v4.079999999999998h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M461,118.86h18v1.1400000000000006h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M601,119.94000000000001h18v0.05999999999998806h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M301,116.58h18v3.4200000000000017h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M261,115.8h18v4.200000000000003h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M41,102.6h18v17.400000000000006h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M101,106.8h18v13.200000000000003h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M241,114.9h18v5.099999999999994h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M401,118.08h18v1.9200000000000017h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M441,118.56h18v1.4399999999999977h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M521,119.88h18v0.12000000000000455h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M481,119.34h18v0.6599999999999966h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M541,119.88h18v0.12000000000000455h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M221,114.78h18v5.219999999999999h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M501,119.64h18v0.35999999999999943h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M321,117.36h18v2.6400000000000006h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M141,111.17999999999999h18v8.820000000000007h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M61,102.6h18v17.400000000000006h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M421,118.14h18v1.8599999999999994h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M341,119.03999999999999h18v0.960000000000008h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M81,119.34h18v0.6599999999999966h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M181,119.76h18v0.23999999999999488h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M1,64.80000000000001h18v55.19999999999999h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M161,119.88h18v0.12000000000000455h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M381,119.16h18v0.8400000000000034h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M361,118.98h18v1.019999999999996h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M201,117.96h18v2.0400000000000063h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M21,105.36h18v14.64h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M281,119.94000000000001h18v0.05999999999998806h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M461,119.88h18v0.12000000000000455h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M301,119.88h18v0.12000000000000455h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M41,119.16h18v0.8400000000000034h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M101,119.76h18v0.23999999999999488h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M401,119.46000000000001h18v0.539999999999992h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M441,119.88h18v0.12000000000000455h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M321,119.94000000000001h18v0.05999999999998806h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M141,117.06h18v2.9399999999999977h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M61,113.88h18v6.1200000000000045h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path><path d=\"M421,119.88h18v0.12000000000000455h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path></g><g class=\"mark-rect role-mark layer_1_marks\" style=\"pointer-events: none;\"></g></g><path class=\"foreground\" d=\"\" style=\"display: none; fill: none;\"></path></g></g></g></svg><svg class=\"marks\" width=\"317\" height=\"237\" viewBox=\"0 0 317 237\" version=\"1.1\" xmlns=\"http://www.w3.org/2000/svg\" xmlns:xlink=\"http://www.w3.org/1999/xlink\"><rect width=\"317\" height=\"237\" style=\"fill: white;\"></rect><g transform=\"translate(52,5)\"><g class=\"mark-group role-frame root\"><g transform=\"translate(0,0)\"><path class=\"background\" d=\"M0.5,0.5h260v120h-260Z\" style=\"fill: none; stroke: #ddd;\"></path><g><g class=\"mark-group role-axis\"><g transform=\"translate(0.5,0.5)\"><path class=\"background\" d=\"M0,0h0v0h0Z\" style=\"pointer-events: none; fill: none;\"></path><g><g class=\"mark-rule role-axis-grid\" style=\"pointer-events: none;\"><line transform=\"translate(0,120)\" x2=\"260\" y2=\"0\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,83)\" x2=\"260\" y2=\"0\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,45)\" x2=\"260\" y2=\"0\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,8)\" x2=\"260\" y2=\"0\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line></g></g><path class=\"foreground\" d=\"\" style=\"pointer-events: none; display: none; fill: none;\"></path></g></g><g class=\"mark-group role-axis\"><g transform=\"translate(0.5,120.5)\"><path class=\"background\" d=\"M0,0h0v0h0Z\" style=\"pointer-events: none; fill: none;\"></path><g><g class=\"mark-rule role-axis-tick\" style=\"pointer-events: none;\"><line transform=\"translate(10,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(30,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(50,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(70,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(90,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(110,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(130,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(150,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(170,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(190,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(210,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(230,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(250,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line></g><g class=\"mark-text role-axis-label\" style=\"pointer-events: none;\"><text text-anchor=\"end\" transform=\"translate(9.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">Debris Burning</text><text text-anchor=\"end\" transform=\"translate(29.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">Arson</text><text text-anchor=\"end\" transform=\"translate(49.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">Lightning</text><text text-anchor=\"end\" transform=\"translate(69.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">Miscellaneous</text><text text-anchor=\"end\" transform=\"translate(89.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">Equipment Use</text><text text-anchor=\"end\" transform=\"translate(109.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">Missing/Undefined</text><text text-anchor=\"end\" transform=\"translate(129.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">Smoking</text><text text-anchor=\"end\" transform=\"translate(149.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">Campfire</text><text text-anchor=\"end\" transform=\"translate(169.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">Children</text><text text-anchor=\"end\" transform=\"translate(189.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">Railroad</text><text text-anchor=\"end\" transform=\"translate(209.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">Powerline</text><text text-anchor=\"end\" transform=\"translate(229.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">Fireworks</text><text text-anchor=\"end\" transform=\"translate(249.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">Structure</text></g><g class=\"mark-rule role-axis-domain\" style=\"pointer-events: none;\"><line transform=\"translate(0,0)\" x2=\"260\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line></g><g class=\"mark-text role-axis-title\" style=\"pointer-events: none;\"><text text-anchor=\"middle\" transform=\"translate(130,104.0771484375)\" style=\"font-family: sans-serif; font-size: 11px; font-weight: bold; fill: #000; opacity: 1;\">CAUSE_DESCR</text></g></g><path class=\"foreground\" d=\"\" style=\"pointer-events: none; display: none; fill: none;\"></path></g></g><g class=\"mark-group role-axis\"><g transform=\"translate(0.5,0.5)\"><path class=\"background\" d=\"M0,0h0v0h0Z\" style=\"pointer-events: none; fill: none;\"></path><g><g class=\"mark-rule role-axis-tick\" style=\"pointer-events: none;\"><line transform=\"translate(0,120)\" x2=\"-5\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,83)\" x2=\"-5\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,45)\" x2=\"-5\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,8)\" x2=\"-5\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line></g><g class=\"mark-text role-axis-label\" style=\"pointer-events: none;\"><text text-anchor=\"end\" transform=\"translate(-7,123)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">0</text><text text-anchor=\"end\" transform=\"translate(-7,85.5)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">500</text><text text-anchor=\"end\" transform=\"translate(-7,48)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">1,000</text><text text-anchor=\"end\" transform=\"translate(-7,10.5)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">1,500</text></g><g class=\"mark-rule role-axis-domain\" style=\"pointer-events: none;\"><line transform=\"translate(0,120)\" x2=\"0\" y2=\"-120\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line></g><g class=\"mark-text role-axis-title\" style=\"pointer-events: none;\"><text text-anchor=\"middle\" transform=\"translate(-35.423828125,60) rotate(-90) translate(0,-2)\" style=\"font-family: sans-serif; font-size: 11px; font-weight: bold; fill: #000; opacity: 1;\">count</text></g></g><path class=\"foreground\" d=\"\" style=\"pointer-events: none; display: none; fill: none;\"></path></g></g><g class=\"mark-rect role-mark layer_0_marks\"><path d=\"M21,32.25000000000001h18v87.75h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M141,108.89999999999999h18v11.100000000000009h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M161,109.35h18v10.650000000000006h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M1,11.25h18v108.75h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M81,98.775h18v21.224999999999994h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M221,118.425h18v1.5750000000000028h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M41,64.05h18v55.95h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M61,79.95h18v40.05h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M101,103.27499999999999h18v16.72500000000001h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M201,117.6h18v2.4000000000000057h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M181,114.45h18v5.549999999999997h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M121,107.1h18v12.900000000000006h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M241,119.625h18v0.375h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path></g><g class=\"mark-rect role-mark layer_1_marks\" style=\"pointer-events: none;\"><path d=\"M1,11.25h18v108.75h-18Z\" style=\"fill: #fdae6b; opacity: 0.5;\"></path></g></g><path class=\"foreground\" d=\"\" style=\"display: none; fill: none;\"></path></g></g></g></svg><svg class=\"marks\" width=\"264\" height=\"165\" viewBox=\"0 0 264 165\" version=\"1.1\" xmlns=\"http://www.w3.org/2000/svg\" xmlns:xlink=\"http://www.w3.org/1999/xlink\"><rect width=\"264\" height=\"165\" style=\"fill: white;\"></rect><defs><clipPath id=\"clip83\"><rect x=\"0\" y=\"0\" width=\"200\" height=\"120\"></rect></clipPath><clipPath id=\"clip84\"><rect x=\"0\" y=\"0\" width=\"200\" height=\"120\"></rect></clipPath></defs><g transform=\"translate(58,8)\"><g class=\"mark-group role-frame root\"><g transform=\"translate(0,0)\"><path class=\"background\" d=\"M0.5,0.5h200v120h-200Z\" style=\"fill: none; stroke: #ddd;\"></path><g><g class=\"mark-group role-axis\"><g transform=\"translate(0.5,120.5)\"><path class=\"background\" d=\"M0,0h0v0h0Z\" style=\"pointer-events: none; fill: none;\"></path><g><g class=\"mark-rule role-axis-grid\" style=\"pointer-events: none;\"><line transform=\"translate(0,-120)\" x2=\"0\" y2=\"120\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(29,-120)\" x2=\"0\" y2=\"120\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(57,-120)\" x2=\"0\" y2=\"120\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(86,-120)\" x2=\"0\" y2=\"120\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(114,-120)\" x2=\"0\" y2=\"120\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(143,-120)\" x2=\"0\" y2=\"120\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(171,-120)\" x2=\"0\" y2=\"120\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(200,-120)\" x2=\"0\" y2=\"120\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line></g></g><path class=\"foreground\" d=\"\" style=\"pointer-events: none; display: none; fill: none;\"></path></g></g><g class=\"mark-group role-axis\"><g transform=\"translate(0.5,0.5)\"><path class=\"background\" d=\"M0,0h0v0h0Z\" style=\"pointer-events: none; fill: none;\"></path><g><g class=\"mark-rule role-axis-grid\" style=\"pointer-events: none;\"><line transform=\"translate(0,120)\" x2=\"200\" y2=\"0\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,70)\" x2=\"200\" y2=\"0\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,20)\" x2=\"200\" y2=\"0\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line></g></g><path class=\"foreground\" d=\"\" style=\"pointer-events: none; display: none; fill: none;\"></path></g></g><g class=\"mark-group role-axis\"><g transform=\"translate(0.5,120.5)\"><path class=\"background\" d=\"M0,0h0v0h0Z\" style=\"pointer-events: none; fill: none;\"></path><g><g class=\"mark-rule role-axis-tick\" style=\"pointer-events: none;\"><line transform=\"translate(0,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(29,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(57,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(86,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(114,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(143,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(171,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(200,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line></g><g class=\"mark-text role-axis-label\" style=\"pointer-events: none;\"><text text-anchor=\"start\" transform=\"translate(0,15)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">1,000</text><text text-anchor=\"middle\" transform=\"translate(28.57142857142857,15)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 0;\">1,200</text><text text-anchor=\"middle\" transform=\"translate(57.14285714285714,15)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">1,400</text><text text-anchor=\"middle\" transform=\"translate(85.71428571428571,15)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 0;\">1,600</text><text text-anchor=\"middle\" transform=\"translate(114.28571428571428,15)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">1,800</text><text text-anchor=\"middle\" transform=\"translate(142.85714285714286,15)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 0;\">2,000</text><text text-anchor=\"middle\" transform=\"translate(171.42857142857142,15)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">2,200</text><text text-anchor=\"end\" transform=\"translate(200,15)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 0;\">2,400</text></g><g class=\"mark-rule role-axis-domain\" style=\"pointer-events: none;\"><line transform=\"translate(0,0)\" x2=\"200\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line></g><g class=\"mark-text role-axis-title\" style=\"pointer-events: none;\"><text text-anchor=\"middle\" transform=\"translate(100,30)\" style=\"font-family: sans-serif; font-size: 11px; font-weight: bold; fill: #000; opacity: 1;\">DISCOVERY_TIME</text></g></g><path class=\"foreground\" d=\"\" style=\"pointer-events: none; display: none; fill: none;\"></path></g></g><g class=\"mark-group role-axis\"><g transform=\"translate(0.5,0.5)\"><path class=\"background\" d=\"M0,0h0v0h0Z\" style=\"pointer-events: none; fill: none;\"></path><g><g class=\"mark-rule role-axis-tick\" style=\"pointer-events: none;\"><line transform=\"translate(0,120)\" x2=\"-5\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,70)\" x2=\"-5\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,20)\" x2=\"-5\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line></g><g class=\"mark-text role-axis-label\" style=\"pointer-events: none;\"><text text-anchor=\"end\" transform=\"translate(-7,123)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">0</text><text text-anchor=\"end\" transform=\"translate(-7,72.99999999999999)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">10,000</text><text text-anchor=\"end\" transform=\"translate(-7,22.999999999999996)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">20,000</text></g><g class=\"mark-rule role-axis-domain\" style=\"pointer-events: none;\"><line transform=\"translate(0,120)\" x2=\"0\" y2=\"-120\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line></g><g class=\"mark-text role-axis-title\" style=\"pointer-events: none;\"><text text-anchor=\"middle\" transform=\"translate(-41.0390625,60) rotate(-90) translate(0,-2)\" style=\"font-family: sans-serif; font-size: 11px; font-weight: bold; fill: #000; opacity: 1;\">FIRE_SIZE</text></g></g><path class=\"foreground\" d=\"\" style=\"pointer-events: none; display: none; fill: none;\"></path></g></g><g class=\"mark-rect role-mark brush_brush_bg\" clip-path=\"url(#clip83)\"><path d=\"M0,0h0v0h0Z\" style=\"fill: #333; fill-opacity: 0.125;\"></path></g><g class=\"mark-symbol role-mark marks\"><path transform=\"translate(161.42857142857144,77.49999999999999)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(30.28571428571429,114.655)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(63.28571428571429,112.64)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(175.71428571428572,109.47)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(75.14285714285714,107.05)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(85.71428571428571,2.38799999999999)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(47.14285714285714,81.5)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(115.99999999999999,112.89)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(14.285714285714285,84.62)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(100.42857142857142,62.725)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(30.28571428571429,107.73400000000001)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(36.42857142857142,100)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(49.28571428571429,105.50750000000001)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(129.85714285714286,114.095)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(60.14285714285714,45)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(57.14285714285714,101.47)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(30.714285714285715,112.35000000000001)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(75.71428571428571,98.855)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(74.28571428571429,113.5)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(75.71428571428571,111.88499999999999)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(186.42857142857144,103.92)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(58.285714285714285,114.195)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(89.71428571428571,92.5)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(88,19.810000000000002)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(102.14285714285714,110.405)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(114.71428571428572,109.36)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(142.85714285714286,105.285)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(57.14285714285714,113.25)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(121.14285714285715,102.65)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(0,114)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(74.71428571428571,112.5)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(119.28571428571428,108.75)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(28.57142857142857,15.395000000000003)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #9FB3C8; stroke-width: 2; opacity: 0.5;\"></path><path transform=\"translate(102.14285714285714,110.405)\" d=\"M2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,-2.7386127875258306,0A2.7386127875258306,2.7386127875258306,0,1,1,2.7386127875258306,0\" style=\"fill: none; stroke: #003E6B; stroke-width: 2; opacity: 0.5;\"></path></g><g class=\"mark-rect role-mark brush_brush\" clip-path=\"url(#clip84)\"><path d=\"M0,0h0v0h0Z\" style=\"fill: none;\"></path></g></g><path class=\"foreground\" d=\"\" style=\"display: none; fill: none;\"></path></g></g></g></svg><svg class=\"marks\" width=\"81\" height=\"175\" viewBox=\"0 0 81 175\" version=\"1.1\" xmlns=\"http://www.w3.org/2000/svg\" xmlns:xlink=\"http://www.w3.org/1999/xlink\"><rect width=\"81\" height=\"175\" style=\"fill: white;\"></rect><defs><clipPath id=\"clip85\"><rect x=\"0\" y=\"0\" width=\"20\" height=\"120\"></rect></clipPath><clipPath id=\"clip86\"><rect x=\"0\" y=\"0\" width=\"20\" height=\"120\"></rect></clipPath></defs><g transform=\"translate(52,5)\"><g class=\"mark-group role-frame root\"><g transform=\"translate(0,0)\"><path class=\"background\" d=\"M0.5,0.5h20v120h-20Z\" style=\"fill: none; stroke: #ddd;\"></path><g><g class=\"mark-group role-axis\"><g transform=\"translate(0.5,0.5)\"><path class=\"background\" d=\"M0,0h0v0h0Z\" style=\"pointer-events: none; fill: none;\"></path><g><g class=\"mark-rule role-axis-grid\" style=\"pointer-events: none;\"><line transform=\"translate(0,120)\" x2=\"20\" y2=\"0\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,72)\" x2=\"20\" y2=\"0\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,24)\" x2=\"20\" y2=\"0\" style=\"fill: none; stroke: #ddd; stroke-width: 1; opacity: 1;\"></line></g></g><path class=\"foreground\" d=\"\" style=\"pointer-events: none; display: none; fill: none;\"></path></g></g><g class=\"mark-group role-axis\"><g transform=\"translate(0.5,120.5)\"><path class=\"background\" d=\"M0,0h0v0h0Z\" style=\"pointer-events: none; fill: none;\"></path><g><g class=\"mark-rule role-axis-tick\" style=\"pointer-events: none;\"><line transform=\"translate(10,0)\" x2=\"0\" y2=\"5\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line></g><g class=\"mark-text role-axis-label\" style=\"pointer-events: none;\"><text text-anchor=\"end\" transform=\"translate(9.5,7) rotate(270) translate(0,3)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">1970</text></g><g class=\"mark-rule role-axis-domain\" style=\"pointer-events: none;\"><line transform=\"translate(0,0)\" x2=\"20\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line></g><g class=\"mark-text role-axis-title\" style=\"pointer-events: none;\"><text text-anchor=\"middle\" transform=\"translate(10,42.4609375)\" style=\"font-family: sans-serif; font-size: 11px; font-weight: bold; fill: #000; opacity: 1;\">YEAR</text></g></g><path class=\"foreground\" d=\"\" style=\"pointer-events: none; display: none; fill: none;\"></path></g></g><g class=\"mark-group role-axis\"><g transform=\"translate(0.5,0.5)\"><path class=\"background\" d=\"M0,0h0v0h0Z\" style=\"pointer-events: none; fill: none;\"></path><g><g class=\"mark-rule role-axis-tick\" style=\"pointer-events: none;\"><line transform=\"translate(0,120)\" x2=\"-5\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,72)\" x2=\"-5\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line><line transform=\"translate(0,24)\" x2=\"-5\" y2=\"0\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line></g><g class=\"mark-text role-axis-label\" style=\"pointer-events: none;\"><text text-anchor=\"end\" transform=\"translate(-7,123)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">0</text><text text-anchor=\"end\" transform=\"translate(-7,75)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">2,000</text><text text-anchor=\"end\" transform=\"translate(-7,26.999999999999993)\" style=\"font-family: sans-serif; font-size: 10px; fill: #000; opacity: 1;\">4,000</text></g><g class=\"mark-rule role-axis-domain\" style=\"pointer-events: none;\"><line transform=\"translate(0,120)\" x2=\"0\" y2=\"-120\" style=\"fill: none; stroke: #888; stroke-width: 1; opacity: 1;\"></line></g><g class=\"mark-text role-axis-title\" style=\"pointer-events: none;\"><text text-anchor=\"middle\" transform=\"translate(-35.423828125,60) rotate(-90) translate(0,-2)\" style=\"font-family: sans-serif; font-size: 11px; font-weight: bold; fill: #000; opacity: 1;\">count</text></g></g><path class=\"foreground\" d=\"\" style=\"pointer-events: none; display: none; fill: none;\"></path></g></g><g class=\"mark-rect role-mark brush_brush_bg\" clip-path=\"url(#clip85)\"><path d=\"M0,0h0v0h0Z\" style=\"fill: #333; fill-opacity: 0.125;\"></path></g><g class=\"mark-rect role-mark layer_0_marks\"><path d=\"M1,0h18v120h-18Z\" style=\"fill: #9FB3C8; opacity: 0.5;\"></path><path d=\"M1,85.19999999999999h18v34.80000000000001h-18Z\" style=\"fill: #003E6B; opacity: 0.5;\"></path></g><g class=\"mark-rect role-mark layer_1_marks\" style=\"pointer-events: none;\"></g><g class=\"mark-rect role-mark brush_brush\" clip-path=\"url(#clip86)\"><path d=\"M0,0h0v0h0Z\" style=\"fill: none;\"></path></g></g><path class=\"foreground\" d=\"\" style=\"display: none; fill: none;\"></path></g></g></g></svg><div>\"\"\"))",
"_____no_output_____"
],
[
"# 🔵 02:21 🔵\n# b2.sel([{\"time_vs_size\": {\"DISCOVERY_TIME\": [1007, 2400]}}, {\"time_vs_size\": {\"FIRE_SIZE\": [0, 9800]}}, {\"CAUSE_DESCR_data_dist\": {\"CAUSE_DESCR\": [\"Smoking\"]}}])",
"_____no_output_____"
],
[
"# 🟡 02:23 🟡\ndata['DISCOVERY_DATE_bin'] = data.apply(lambda x: 'null' if b2.np.isnan(x) else int(x/300.0) * 300.0, 'DISCOVERY_DATE')\nDISCOVERY_DATE_data_dist = data.group('DISCOVERY_DATE_bin')\n# DISCOVERY_DATE_data_dist.vis()\nDISCOVERY_DATE_data_dist.vis(mark=\"line\", x_type=\"temporal\")",
"_____no_output_____"
],
[
"minute = data.group(\"MINUTE\")\nminute.vis(mark=\"line\", x_type=\"ordinal\")",
"_____no_output_____"
],
[
"help(DISCOVERY_DATE_data_dist.vis)",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cb46a3a7abe7b7cc08f43fa441a77be298909f18 | 14 | ipynb | Jupyter Notebook | hello_world/mnist_sigit_classification.ipynb | sumeetkr/deeplearning_tutorials | 924a91e1cf0fb8a2a6143b084976a939f4c9840a | [
"MIT"
] | null | null | null | hello_world/mnist_sigit_classification.ipynb | sumeetkr/deeplearning_tutorials | 924a91e1cf0fb8a2a6143b084976a939f4c9840a | [
"MIT"
] | null | null | null | hello_world/mnist_sigit_classification.ipynb | sumeetkr/deeplearning_tutorials | 924a91e1cf0fb8a2a6143b084976a939f4c9840a | [
"MIT"
] | null | null | null | 7 | 13 | 0.642857 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
cb46a5259055a8939a1198540696df1abdeb654a | 86,069 | ipynb | Jupyter Notebook | lstmwithimdb.ipynb | wjnnora/LSTM-Com-Dataset-IMDB | c54c7be4871e8087ff0fafc97100889d1d6fdb60 | [
"MIT"
] | null | null | null | lstmwithimdb.ipynb | wjnnora/LSTM-Com-Dataset-IMDB | c54c7be4871e8087ff0fafc97100889d1d6fdb60 | [
"MIT"
] | null | null | null | lstmwithimdb.ipynb | wjnnora/LSTM-Com-Dataset-IMDB | c54c7be4871e8087ff0fafc97100889d1d6fdb60 | [
"MIT"
] | null | null | null | 113.998675 | 21,396 | 0.844346 | [
[
[
"# Treinando LSTMs com o dataset IMDB",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport keras\nfrom keras.models import Sequential\nfrom keras.layers import Dense, LSTM, Conv1D, MaxPool1D, Dropout, Embedding\nfrom keras.preprocessing import sequence\nfrom keras.callbacks import EarlyStopping\nimport matplotlib.pyplot as plt\nimport sys\nimport warnings",
"_____no_output_____"
],
[
"# Desativa os avisos\nif not sys.warnoptions:\n warnings.simplefilter(\"ignore\")",
"_____no_output_____"
],
[
"# Carrega o dataset contendo as 5000 palavras mais utilizadas do dataset\n(x_train, y_train), (x_test, y_test) = keras.datasets.imdb.load_data(num_words = 5000)",
"_____no_output_____"
],
[
"# Separando os dados de treino\nx_aux, y_aux = x_test, y_test\nx_test = x_aux[:15000]\ny_test = y_aux[:15000]\nx_validation = x_aux[15000:]\ny_validation = y_aux[15000:]\n\ndel x_aux, y_aux",
"_____no_output_____"
],
[
"# Exibindo a quantidade de listas dentro do dataset\nprint(\"X train: \", len(x_train))\nprint(\"Y train: \", len(y_train))\nprint(\"X test: \", len(x_test))\nprint(\"Y test: \", len(y_test))\nprint(\"X validation: \", len(x_validation))\nprint(\"Y validation: \", len(y_validation))",
"X train: 25000\nY train: 25000\nX test: 15000\nY test: 15000\nX validation: 10000\nY validation: 10000\n"
],
[
"# Exibindo o comprimento das 10 primeiras listas do dataset\nfor l, t in zip(x_train[:10], y_train[:10]):\n print(f\"Comprimento lista : {len(l)} - Target: {t}\")",
"Comprimento lista : 218 - Target: 1\nComprimento lista : 189 - Target: 0\nComprimento lista : 141 - Target: 0\nComprimento lista : 550 - Target: 1\nComprimento lista : 147 - Target: 0\nComprimento lista : 43 - Target: 0\nComprimento lista : 123 - Target: 1\nComprimento lista : 562 - Target: 0\nComprimento lista : 233 - Target: 1\nComprimento lista : 130 - Target: 0\n"
]
],
[
[
"Para o treinamento será necessário deixar todas as listas no mesmo tamanho.",
"_____no_output_____"
]
],
[
[
"# Truncando as sequências de entrada de treinamento e teste para ficarem todas no mesmo tamanho\n# O modelo aprenderá que valores 0 não possuem informação\nx_train = sequence.pad_sequences(x_train, maxlen=500)\nx_test = sequence.pad_sequences(x_test, maxlen=500)\nx_validation = sequence.pad_sequences(x_validation, maxlen=500)",
"_____no_output_____"
],
[
"# Verificando novamente as 10 primeiras listas do dataset de treino\nfor x, y in zip(x_train[:10], y_train[:10]):\n print(f\"Comprimento da lista: {len(x)} - Target: {y}\")",
"Comprimento da lista: 500 - Target: 1\nComprimento da lista: 500 - Target: 0\nComprimento da lista: 500 - Target: 0\nComprimento da lista: 500 - Target: 1\nComprimento da lista: 500 - Target: 0\nComprimento da lista: 500 - Target: 0\nComprimento da lista: 500 - Target: 1\nComprimento da lista: 500 - Target: 0\nComprimento da lista: 500 - Target: 1\nComprimento da lista: 500 - Target: 0\n"
]
],
[
[
"Agora sim está adequado para o treinamento.",
"_____no_output_____"
],
[
"## Definindo algumas funções ",
"_____no_output_____"
]
],
[
[
"def exibir_performance_dados_teste(loss, accuracy):\n print(\"Loss: %.2f\" % (loss * 100))\n print(\"Accuracy: %.2f\" % (accuracy * 100))",
"_____no_output_____"
],
[
"def exibir_evolucao_treino_validacao(train_hist, train, validation):\n plt.plot(train_hist.history[train])\n plt.plot(train_hist.history[validation])\n plt.title(\"Histórico do treinamento\")\n plt.ylabel(train)\n plt.xlabel(\"epoch\")\n plt.legend(['train', 'validation'], loc = 'upper left')\n plt.show()",
"_____no_output_____"
]
],
[
[
"# Criando vários modelos",
"_____no_output_____"
],
[
"## LSTM",
"_____no_output_____"
]
],
[
[
"# Criando o primeiro modelo\nmodel1 = Sequential()\n\n# Parâmetros:\n# input_dim = tamanho do vocabulário do dataset. Nesse caso, 5000.\n# output_dim = tamanho do vetor para representar cada palavra.\n# input_length = tamanho de cada vetor de saída.\nmodel1.add(Embedding(input_dim = 5000, output_dim = 64, input_length = 500))\n\n# Camada LSTM com 100 neurônios\nmodel1.add(LSTM(100))\n\n# Camada de saída com um neurônio e função sigmoid\nmodel1.add(Dense(1, activation = 'sigmoid'))",
"_____no_output_____"
],
[
"# Compilando o modelo\nmodel1.compile(loss = keras.losses.binary_crossentropy, optimizer = 'adam', metrics = ['accuracy'])",
"_____no_output_____"
],
[
"# Verificando a arquitetura da rede\nmodel1.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding (Embedding) (None, 500, 64) 320000 \n_________________________________________________________________\nlstm (LSTM) (None, 100) 66000 \n_________________________________________________________________\ndense (Dense) (None, 1) 101 \n=================================================================\nTotal params: 386,101\nTrainable params: 386,101\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"%time\n\n# Treinando o modelo\nhist = model1.fit(x_train, y_train, validation_data=(x_validation, y_validation), batch_size = 128, epochs = 10)",
"CPU times: user 9 µs, sys: 1e+03 ns, total: 10 µs\nWall time: 20 µs\nEpoch 1/10\n196/196 [==============================] - 8s 39ms/step - loss: 0.5168 - accuracy: 0.7442 - val_loss: 0.3653 - val_accuracy: 0.8452\nEpoch 2/10\n196/196 [==============================] - 7s 37ms/step - loss: 0.3466 - accuracy: 0.8603 - val_loss: 0.3242 - val_accuracy: 0.8710\nEpoch 3/10\n196/196 [==============================] - 7s 37ms/step - loss: 0.2494 - accuracy: 0.9022 - val_loss: 0.3101 - val_accuracy: 0.8673\nEpoch 4/10\n196/196 [==============================] - 7s 37ms/step - loss: 0.2076 - accuracy: 0.9226 - val_loss: 0.2967 - val_accuracy: 0.8836\nEpoch 5/10\n196/196 [==============================] - 7s 37ms/step - loss: 0.1868 - accuracy: 0.9306 - val_loss: 0.3059 - val_accuracy: 0.8771\nEpoch 6/10\n196/196 [==============================] - 7s 37ms/step - loss: 0.1650 - accuracy: 0.9398 - val_loss: 0.3351 - val_accuracy: 0.8644\nEpoch 7/10\n196/196 [==============================] - 7s 37ms/step - loss: 0.1466 - accuracy: 0.9456 - val_loss: 0.4005 - val_accuracy: 0.8700\nEpoch 8/10\n196/196 [==============================] - 7s 37ms/step - loss: 0.1446 - accuracy: 0.9469 - val_loss: 0.3587 - val_accuracy: 0.8679\nEpoch 9/10\n196/196 [==============================] - 7s 37ms/step - loss: 0.1431 - accuracy: 0.9477 - val_loss: 0.3753 - val_accuracy: 0.8604\nEpoch 10/10\n196/196 [==============================] - 7s 37ms/step - loss: 0.1583 - accuracy: 0.9395 - val_loss: 0.3555 - val_accuracy: 0.8663\n"
],
[
"# Avaliando o modelo\nloss, accuracy = model1.evaluate(x_test, y_test)\n\nexibir_performance_dados_teste(loss, accuracy)",
"469/469 [==============================] - 5s 10ms/step - loss: 0.3839 - accuracy: 0.8585\nLoss: 38.39\nAccuracy: 85.85\n"
],
[
"# Exibindo a evolução dos dados de treino e validação\nexibir_evolucao_treino_validacao(hist, 'accuracy', 'val_accuracy')",
"_____no_output_____"
]
],
[
[
"## LSTM com técnica de regulização Dropout",
"_____no_output_____"
],
[
"O objetivo do segundo modelo é melhorar a generalização do modelo aplicando a técnica de regularização Dropout.",
"_____no_output_____"
]
],
[
[
"# Criando o modelo\nmodel2 = Sequential()\n\n# Parâmetros:\n# input_dim = tamanho do vocabulário do dataset. Nesse caso, 5000.\n# output_dim = tamanho do vetor para representar cada palavra.\n# input_length = tamanho de cada vetor de saída.\nmodel2.add(Embedding(input_dim = 5000, output_dim = 64, input_length = 500))\n\n# Camada Dropout com 25% de drop\nmodel2.add(Dropout(0.25))\n\n# Camada LSTM com 100 neurônios\nmodel2.add(LSTM(100))\n\n# Camada Dropout com 25% de drop\nmodel2.add(Dropout(0.25))\n\n# Última camada com 1 neurônio de saída e função de ativação sigmoid\nmodel2.add(Dense(1, activation = 'sigmoid'))",
"_____no_output_____"
],
[
"# Compilando o modelo\nmodel2.compile(loss = 'binary_crossentropy', optimizer = 'adam', metrics = ['accuracy'])",
"_____no_output_____"
],
[
"# Visualizando a arquitetura da rede\nmodel2.summary()",
"Model: \"sequential_2\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding_2 (Embedding) (None, 500, 64) 320000 \n_________________________________________________________________\ndropout_2 (Dropout) (None, 500, 64) 0 \n_________________________________________________________________\nlstm_2 (LSTM) (None, 100) 66000 \n_________________________________________________________________\ndropout_3 (Dropout) (None, 100) 0 \n_________________________________________________________________\ndense_2 (Dense) (None, 1) 101 \n=================================================================\nTotal params: 386,101\nTrainable params: 386,101\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"%time\n\n# Treinando o modelo\nhist = model2.fit(x_train, y_train, validation_data=(x_validation, y_validation), batch_size = 128, epochs = 10)",
"CPU times: user 12 µs, sys: 0 ns, total: 12 µs\nWall time: 25.5 µs\nEpoch 1/10\n196/196 [==============================] - 8s 39ms/step - loss: 0.5290 - accuracy: 0.7239 - val_loss: 0.3568 - val_accuracy: 0.8521\nEpoch 2/10\n196/196 [==============================] - 7s 38ms/step - loss: 0.3222 - accuracy: 0.8644 - val_loss: 0.3058 - val_accuracy: 0.8823\nEpoch 3/10\n196/196 [==============================] - 7s 37ms/step - loss: 0.2460 - accuracy: 0.9027 - val_loss: 0.3028 - val_accuracy: 0.8728\nEpoch 4/10\n196/196 [==============================] - 7s 37ms/step - loss: 0.2157 - accuracy: 0.9192 - val_loss: 0.2970 - val_accuracy: 0.8834\nEpoch 5/10\n196/196 [==============================] - 7s 37ms/step - loss: 0.1877 - accuracy: 0.9298 - val_loss: 0.3144 - val_accuracy: 0.8633\nEpoch 6/10\n196/196 [==============================] - 7s 38ms/step - loss: 0.1672 - accuracy: 0.9389 - val_loss: 0.3327 - val_accuracy: 0.8611\nEpoch 7/10\n196/196 [==============================] - 7s 38ms/step - loss: 0.1648 - accuracy: 0.9388 - val_loss: 0.4198 - val_accuracy: 0.8687\nEpoch 8/10\n196/196 [==============================] - 7s 37ms/step - loss: 0.1385 - accuracy: 0.9487 - val_loss: 0.3859 - val_accuracy: 0.8711\nEpoch 9/10\n196/196 [==============================] - 7s 38ms/step - loss: 0.2914 - accuracy: 0.8750 - val_loss: 0.3920 - val_accuracy: 0.8320\nEpoch 10/10\n196/196 [==============================] - 7s 38ms/step - loss: 0.2150 - accuracy: 0.9165 - val_loss: 0.4142 - val_accuracy: 0.8118\n"
],
[
"# Avaliando o modelo\nloss, accuracy = model2.evaluate(x_test, y_test)\n\nexibir_performance_dados_teste(loss, accuracy)",
"469/469 [==============================] - 5s 10ms/step - loss: 0.4350 - accuracy: 0.8008\nLoss: 43.50\nAccuracy: 80.08\n"
],
[
"# Exibindo a evolução dos dados de treino e validação\nexibir_evolucao_treino_validacao(hist, 'accuracy', 'val_accuracy')",
"_____no_output_____"
]
],
[
[
"## LSTM como CNN e Técnicas de regularização",
"_____no_output_____"
],
[
"O objetivo do terceiro modelo é aplicar uma camada de CNN e mais uma técnica de regularização",
"_____no_output_____"
]
],
[
[
"# Criando o modelo\nmodel3 = Sequential()\n\n# Parâmetros:\n# input_dim = tamanho do vocabulário do dataset. Nesse caso, 5000.\n# output_dim = tamanho do vetor para representar cada palavra.\n# input_length = tamanho de cada vetor de saída.\nmodel3.add(Embedding(input_dim = 5000, output_dim = 64, input_length = 500))\n\n# Camada de convolução\nmodel3.add(Conv1D(filters = 64, kernel_size = 3, padding = 'same', activation = 'relu'))\n\n# Camada de pooling\nmodel3.add(MaxPool1D(pool_size = 2))\n\n# Camada de Dropout com 25%\nmodel3.add(Dropout(0.25))\n\n# Camada LSTM com 100 neurônios, 1 camada de dropout antes e uma depois, ambas com 25% de drop\n#model3.add(LSTM(100, dropout = 0.25, recurrent_dropout = 0.25))\nmodel3.add(LSTM(100))\n\n# Camada de Dropout com 25%\nmodel3.add(Dropout(0.25))\n\n# Última camada com 1 neurônio de saída e função de ativação sigmoid\nmodel3.add(Dense(units = 1, activation = 'sigmoid'))",
"_____no_output_____"
],
[
"# Compilando o modelo\nmodel3.compile(loss = 'binary_crossentropy', optimizer = 'adam', metrics = ['accuracy'])",
"_____no_output_____"
],
[
"# Exibindo a arquitetura da rede\nmodel3.summary()",
"Model: \"sequential_5\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding_5 (Embedding) (None, 500, 64) 320000 \n_________________________________________________________________\nconv1d_2 (Conv1D) (None, 500, 64) 12352 \n_________________________________________________________________\nmax_pooling1d_2 (MaxPooling1 (None, 250, 64) 0 \n_________________________________________________________________\ndropout_6 (Dropout) (None, 250, 64) 0 \n_________________________________________________________________\nlstm_5 (LSTM) (None, 100) 66000 \n_________________________________________________________________\ndropout_7 (Dropout) (None, 100) 0 \n_________________________________________________________________\ndense_5 (Dense) (None, 1) 101 \n=================================================================\nTotal params: 398,453\nTrainable params: 398,453\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# Definindo o EarlyStopping e treinando o modelo\nmonitor = EarlyStopping(monitor = 'val_loss', min_delta=1e-1, patience = 5, verbose = 3, mode = 'auto')\n\nhist = model3.fit(x_train,\n y_train,\n validation_data = (x_validation, y_validation),\n callbacks = [monitor],\n batch_size = 64, \n epochs = 100)",
"Epoch 1/100\n391/391 [==============================] - 9s 24ms/step - loss: 0.4131 - accuracy: 0.7997 - val_loss: 0.2752 - val_accuracy: 0.8867\nEpoch 2/100\n391/391 [==============================] - 9s 23ms/step - loss: 0.2475 - accuracy: 0.9058 - val_loss: 0.2694 - val_accuracy: 0.8883\nEpoch 3/100\n391/391 [==============================] - 9s 23ms/step - loss: 0.1888 - accuracy: 0.9303 - val_loss: 0.3321 - val_accuracy: 0.8775\nEpoch 4/100\n391/391 [==============================] - 9s 23ms/step - loss: 0.1561 - accuracy: 0.9431 - val_loss: 0.2961 - val_accuracy: 0.8831\nEpoch 5/100\n391/391 [==============================] - 9s 22ms/step - loss: 0.1310 - accuracy: 0.9548 - val_loss: 0.3410 - val_accuracy: 0.8841\nEpoch 6/100\n391/391 [==============================] - 9s 23ms/step - loss: 0.1097 - accuracy: 0.9617 - val_loss: 0.3864 - val_accuracy: 0.8709\nEpoch 00006: early stopping\n"
],
[
"# Avaliando o modelo\nloss, accuracy = model3.evaluate(x_test, y_test)\n\nexibir_performance_dados_teste(loss, accuracy)",
"469/469 [==============================] - 3s 7ms/step - loss: 0.4061 - accuracy: 0.8651\nLoss: 40.61\nAccuracy: 86.51\n"
],
[
"#xibindo a evolução do modelo\nexibir_evolucao_treino_validacao(hist, 'accuracy', 'val_accuracy')",
"_____no_output_____"
]
],
[
[
"Com 86,51% de acurácia nos dados de teste, o último modelo (model3) foi o que apresentou o melhor resultado. Utilizando EarlyStopping, consegui obter o melhor modelo com apenas 6 epochs.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.