repo_id
stringlengths 15
86
| file_path
stringlengths 28
180
| content
stringlengths 1
1.75M
| __index_level_0__
int64 0
0
|
|---|---|---|---|
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/frugalscore/frugalscore.py | # Copyright 2022 The HuggingFace Datasets Authors and the current metric script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""FrugalScore metric."""
import torch
from transformers import AutoModelForSequenceClassification, AutoTokenizer, Trainer, TrainingArguments
import datasets
_CITATION = """\
@article{eddine2021frugalscore,
title={FrugalScore: Learning Cheaper, Lighter and Faster Evaluation Metrics for Automatic Text Generation},
author={Eddine, Moussa Kamal and Shang, Guokan and Tixier, Antoine J-P and Vazirgiannis, Michalis},
journal={arXiv preprint arXiv:2110.08559},
year={2021}
}
"""
_DESCRIPTION = """\
FrugalScore is a reference-based metric for NLG models evaluation. It is based on a distillation approach that allows to learn a fixed, low cost version of any expensive NLG metric, while retaining most of its original performance.
"""
_KWARGS_DESCRIPTION = """
Calculates how good are predictions given some references, using certain scores.
Args:
predictions (list of str): list of predictions to score. Each predictions
should be a string.
references (list of str): list of reference for each prediction. Each
reference should be a string.
batch_size (int): the batch size for predictions.
max_length (int): maximum sequence length.
device (str): either gpu or cpu
Returns:
scores (list of int): list of scores.
Examples:
>>> frugalscore = datasets.load_metric("frugalscore")
>>> results = frugalscore.compute(predictions=['hello there', 'huggingface'], references=['hello world', 'hugging face'])
>>> print([round(s, 3) for s in results["scores"]])
[0.631, 0.645]
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class FRUGALSCORE(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Value("string"),
"references": datasets.Value("string"),
}
),
homepage="https://github.com/moussaKam/FrugalScore",
)
def _download_and_prepare(self, dl_manager):
if self.config_name == "default":
checkpoint = "moussaKam/frugalscore_tiny_bert-base_bert-score"
else:
checkpoint = self.config_name
self.model = AutoModelForSequenceClassification.from_pretrained(checkpoint)
self.tokenizer = AutoTokenizer.from_pretrained(checkpoint)
def _compute(
self,
predictions,
references,
batch_size=32,
max_length=128,
device=None,
):
"""Returns the scores"""
assert len(predictions) == len(
references
), "predictions and references should have the same number of sentences."
if device is not None:
assert device in ["gpu", "cpu"], "device should be either gpu or cpu."
else:
device = "gpu" if torch.cuda.is_available() else "cpu"
training_args = TrainingArguments(
"trainer",
fp16=(device == "gpu"),
per_device_eval_batch_size=batch_size,
report_to="all",
no_cuda=(device == "cpu"),
log_level="warning",
)
dataset = {"sentence1": predictions, "sentence2": references}
raw_datasets = datasets.Dataset.from_dict(dataset)
def tokenize_function(data):
return self.tokenizer(
data["sentence1"], data["sentence2"], max_length=max_length, truncation=True, padding=True
)
tokenized_datasets = raw_datasets.map(tokenize_function, batched=True)
tokenized_datasets.remove_columns(["sentence1", "sentence2"])
trainer = Trainer(self.model, training_args, tokenizer=self.tokenizer)
predictions = trainer.predict(tokenized_datasets)
return {"scores": list(predictions.predictions.squeeze(-1))}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/precision/README.md | # Metric Card for Precision
## Metric Description
Precision is the fraction of correctly labeled positive examples out of all of the examples that were labeled as positive. It is computed via the equation:
Precision = TP / (TP + FP)
where TP is the True positives (i.e. the examples correctly labeled as positive) and FP is the False positive examples (i.e. the examples incorrectly labeled as positive).
## How to Use
At minimum, precision takes as input a list of predicted labels, `predictions`, and a list of output labels, `references`.
```python
>>> precision_metric = datasets.load_metric("precision")
>>> results = precision_metric.compute(references=[0, 1], predictions=[0, 1])
>>> print(results)
{'precision': 1.0}
```
### Inputs
- **predictions** (`list` of `int`): Predicted class labels.
- **references** (`list` of `int`): Actual class labels.
- **labels** (`list` of `int`): The set of labels to include when `average` is not set to `'binary'`. If `average` is `None`, it should be the label order. Labels present in the data can be excluded, for example to calculate a multiclass average ignoring a majority negative class. Labels not present in the data will result in 0 components in a macro average. For multilabel targets, labels are column indices. By default, all labels in `predictions` and `references` are used in sorted order. Defaults to None.
- **pos_label** (`int`): The class to be considered the positive class, in the case where `average` is set to `binary`. Defaults to 1.
- **average** (`string`): This parameter is required for multiclass/multilabel targets. If set to `None`, the scores for each class are returned. Otherwise, this determines the type of averaging performed on the data. Defaults to `'binary'`.
- 'binary': Only report results for the class specified by `pos_label`. This is applicable only if the classes found in `predictions` and `references` are binary.
- 'micro': Calculate metrics globally by counting the total true positives, false negatives and false positives.
- 'macro': Calculate metrics for each label, and find their unweighted mean. This does not take label imbalance into account.
- 'weighted': Calculate metrics for each label, and find their average weighted by support (the number of true instances for each label). This alters `'macro'` to account for label imbalance. This option can result in an F-score that is not between precision and recall.
- 'samples': Calculate metrics for each instance, and find their average (only meaningful for multilabel classification).
- **sample_weight** (`list` of `float`): Sample weights Defaults to None.
- **zero_division** (): Sets the value to return when there is a zero division. Defaults to .
- 0: Returns 0 when there is a zero division.
- 1: Returns 1 when there is a zero division.
- 'warn': Raises warnings and then returns 0 when there is a zero division.
### Output Values
- **precision**(`float` or `array` of `float`): Precision score or list of precision scores, depending on the value passed to `average`. Minimum possible value is 0. Maximum possible value is 1. Higher values indicate that fewer negative examples were incorrectly labeled as positive, which means that, generally, higher scores are better.
Output Example(s):
```python
{'precision': 0.2222222222222222}
```
```python
{'precision': array([0.66666667, 0.0, 0.0])}
```
#### Values from Popular Papers
### Examples
Example 1-A simple binary example
```python
>>> precision_metric = datasets.load_metric("precision")
>>> results = precision_metric.compute(references=[0, 1, 0, 1, 0], predictions=[0, 0, 1, 1, 0])
>>> print(results)
{'precision': 0.5}
```
Example 2-The same simple binary example as in Example 1, but with `pos_label` set to `0`.
```python
>>> precision_metric = datasets.load_metric("precision")
>>> results = precision_metric.compute(references=[0, 1, 0, 1, 0], predictions=[0, 0, 1, 1, 0], pos_label=0)
>>> print(round(results['precision'], 2))
0.67
```
Example 3-The same simple binary example as in Example 1, but with `sample_weight` included.
```python
>>> precision_metric = datasets.load_metric("precision")
>>> results = precision_metric.compute(references=[0, 1, 0, 1, 0], predictions=[0, 0, 1, 1, 0], sample_weight=[0.9, 0.5, 3.9, 1.2, 0.3])
>>> print(results)
{'precision': 0.23529411764705882}
```
Example 4-A multiclass example, with different values for the `average` input.
```python
>>> predictions = [0, 2, 1, 0, 0, 1]
>>> references = [0, 1, 2, 0, 1, 2]
>>> results = precision_metric.compute(predictions=predictions, references=references, average='macro')
>>> print(results)
{'precision': 0.2222222222222222}
>>> results = precision_metric.compute(predictions=predictions, references=references, average='micro')
>>> print(results)
{'precision': 0.3333333333333333}
>>> results = precision_metric.compute(predictions=predictions, references=references, average='weighted')
>>> print(results)
{'precision': 0.2222222222222222}
>>> results = precision_metric.compute(predictions=predictions, references=references, average=None)
>>> print([round(res, 2) for res in results['precision']])
[0.67, 0.0, 0.0]
```
## Limitations and Bias
[Precision](https://huggingface.co/metrics/precision) and [recall](https://huggingface.co/metrics/recall) are complementary and can be used to measure different aspects of model performance -- using both of them (or an averaged measure like [F1 score](https://huggingface.co/metrics/F1) to better represent different aspects of performance. See [Wikipedia](https://en.wikipedia.org/wiki/Precision_and_recall) for more information.
## Citation(s)
```bibtex
@article{scikit-learn,
title={Scikit-learn: Machine Learning in {P}ython},
author={Pedregosa, F. and Varoquaux, G. and Gramfort, A. and Michel, V.
and Thirion, B. and Grisel, O. and Blondel, M. and Prettenhofer, P.
and Weiss, R. and Dubourg, V. and Vanderplas, J. and Passos, A. and
Cournapeau, D. and Brucher, M. and Perrot, M. and Duchesnay, E.},
journal={Journal of Machine Learning Research},
volume={12},
pages={2825--2830},
year={2011}
}
```
## Further References
- [Wikipedia -- Precision and recall](https://en.wikipedia.org/wiki/Precision_and_recall)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/precision/precision.py | # Copyright 2020 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Precision metric."""
from sklearn.metrics import precision_score
import datasets
_DESCRIPTION = """
Precision is the fraction of correctly labeled positive examples out of all of the examples that were labeled as positive. It is computed via the equation:
Precision = TP / (TP + FP)
where TP is the True positives (i.e. the examples correctly labeled as positive) and FP is the False positive examples (i.e. the examples incorrectly labeled as positive).
"""
_KWARGS_DESCRIPTION = """
Args:
predictions (`list` of `int`): Predicted class labels.
references (`list` of `int`): Actual class labels.
labels (`list` of `int`): The set of labels to include when `average` is not set to `'binary'`. If `average` is `None`, it should be the label order. Labels present in the data can be excluded, for example to calculate a multiclass average ignoring a majority negative class. Labels not present in the data will result in 0 components in a macro average. For multilabel targets, labels are column indices. By default, all labels in `predictions` and `references` are used in sorted order. Defaults to None.
pos_label (`int`): The class to be considered the positive class, in the case where `average` is set to `binary`. Defaults to 1.
average (`string`): This parameter is required for multiclass/multilabel targets. If set to `None`, the scores for each class are returned. Otherwise, this determines the type of averaging performed on the data. Defaults to `'binary'`.
- 'binary': Only report results for the class specified by `pos_label`. This is applicable only if the classes found in `predictions` and `references` are binary.
- 'micro': Calculate metrics globally by counting the total true positives, false negatives and false positives.
- 'macro': Calculate metrics for each label, and find their unweighted mean. This does not take label imbalance into account.
- 'weighted': Calculate metrics for each label, and find their average weighted by support (the number of true instances for each label). This alters `'macro'` to account for label imbalance. This option can result in an F-score that is not between precision and recall.
- 'samples': Calculate metrics for each instance, and find their average (only meaningful for multilabel classification).
sample_weight (`list` of `float`): Sample weights Defaults to None.
zero_division (`int` or `string`): Sets the value to return when there is a zero division. Defaults to 'warn'.
- 0: Returns 0 when there is a zero division.
- 1: Returns 1 when there is a zero division.
- 'warn': Raises warnings and then returns 0 when there is a zero division.
Returns:
precision (`float` or `array` of `float`): Precision score or list of precision scores, depending on the value passed to `average`. Minimum possible value is 0. Maximum possible value is 1. Higher values indicate that fewer negative examples were incorrectly labeled as positive, which means that, generally, higher scores are better.
Examples:
Example 1-A simple binary example
>>> precision_metric = datasets.load_metric("precision")
>>> results = precision_metric.compute(references=[0, 1, 0, 1, 0], predictions=[0, 0, 1, 1, 0])
>>> print(results)
{'precision': 0.5}
Example 2-The same simple binary example as in Example 1, but with `pos_label` set to `0`.
>>> precision_metric = datasets.load_metric("precision")
>>> results = precision_metric.compute(references=[0, 1, 0, 1, 0], predictions=[0, 0, 1, 1, 0], pos_label=0)
>>> print(round(results['precision'], 2))
0.67
Example 3-The same simple binary example as in Example 1, but with `sample_weight` included.
>>> precision_metric = datasets.load_metric("precision")
>>> results = precision_metric.compute(references=[0, 1, 0, 1, 0], predictions=[0, 0, 1, 1, 0], sample_weight=[0.9, 0.5, 3.9, 1.2, 0.3])
>>> print(results)
{'precision': 0.23529411764705882}
Example 4-A multiclass example, with different values for the `average` input.
>>> predictions = [0, 2, 1, 0, 0, 1]
>>> references = [0, 1, 2, 0, 1, 2]
>>> results = precision_metric.compute(predictions=predictions, references=references, average='macro')
>>> print(results)
{'precision': 0.2222222222222222}
>>> results = precision_metric.compute(predictions=predictions, references=references, average='micro')
>>> print(results)
{'precision': 0.3333333333333333}
>>> results = precision_metric.compute(predictions=predictions, references=references, average='weighted')
>>> print(results)
{'precision': 0.2222222222222222}
>>> results = precision_metric.compute(predictions=predictions, references=references, average=None)
>>> print([round(res, 2) for res in results['precision']])
[0.67, 0.0, 0.0]
"""
_CITATION = """
@article{scikit-learn,
title={Scikit-learn: Machine Learning in {P}ython},
author={Pedregosa, F. and Varoquaux, G. and Gramfort, A. and Michel, V.
and Thirion, B. and Grisel, O. and Blondel, M. and Prettenhofer, P.
and Weiss, R. and Dubourg, V. and Vanderplas, J. and Passos, A. and
Cournapeau, D. and Brucher, M. and Perrot, M. and Duchesnay, E.},
journal={Journal of Machine Learning Research},
volume={12},
pages={2825--2830},
year={2011}
}
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Precision(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Sequence(datasets.Value("int32")),
"references": datasets.Sequence(datasets.Value("int32")),
}
if self.config_name == "multilabel"
else {
"predictions": datasets.Value("int32"),
"references": datasets.Value("int32"),
}
),
reference_urls=["https://scikit-learn.org/stable/modules/generated/sklearn.metrics.precision_score.html"],
)
def _compute(
self,
predictions,
references,
labels=None,
pos_label=1,
average="binary",
sample_weight=None,
zero_division="warn",
):
score = precision_score(
references,
predictions,
labels=labels,
pos_label=pos_label,
average=average,
sample_weight=sample_weight,
zero_division=zero_division,
)
return {"precision": float(score) if score.size == 1 else score}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/squad/README.md | # Metric Card for SQuAD
## Metric description
This metric wraps the official scoring script for version 1 of the [Stanford Question Answering Dataset (SQuAD)](https://huggingface.co/datasets/squad).
SQuAD is a reading comprehension dataset, consisting of questions posed by crowdworkers on a set of Wikipedia articles, where the answer to every question is a segment of text, or span, from the corresponding reading passage, or the question might be unanswerable.
## How to use
The metric takes two files or two lists of question-answers dictionaries as inputs : one with the predictions of the model and the other with the references to be compared to:
```python
from datasets import load_metric
squad_metric = load_metric("squad")
results = squad_metric.compute(predictions=predictions, references=references)
```
## Output values
This metric outputs a dictionary with two values: the average exact match score and the average [F1 score](https://huggingface.co/metrics/f1).
```
{'exact_match': 100.0, 'f1': 100.0}
```
The range of `exact_match` is 0-100, where 0.0 means no answers were matched and 100.0 means all answers were matched.
The range of `f1` is 0-1 -- its lowest possible value is 0, if either the precision or the recall is 0, and its highest possible value is 1.0, which means perfect precision and recall.
### Values from popular papers
The [original SQuAD paper](https://nlp.stanford.edu/pubs/rajpurkar2016squad.pdf) reported an F1 score of 51.0% and an Exact Match score of 40.0%. They also report that human performance on the dataset represents an F1 score of 90.5% and an Exact Match score of 80.3%.
For more recent model performance, see the [dataset leaderboard](https://paperswithcode.com/dataset/squad).
## Examples
Maximal values for both exact match and F1 (perfect match):
```python
from datasets import load_metric
squad_metric = load_metric("squad")
predictions = [{'prediction_text': '1976', 'id': '56e10a3be3433e1400422b22'}]
references = [{'answers': {'answer_start': [97], 'text': ['1976']}, 'id': '56e10a3be3433e1400422b22'}]
results = squad_metric.compute(predictions=predictions, references=references)
results
{'exact_match': 100.0, 'f1': 100.0}
```
Minimal values for both exact match and F1 (no match):
```python
from datasets import load_metric
squad_metric = load_metric("squad")
predictions = [{'prediction_text': '1999', 'id': '56e10a3be3433e1400422b22'}]
references = [{'answers': {'answer_start': [97], 'text': ['1976']}, 'id': '56e10a3be3433e1400422b22'}]
results = squad_metric.compute(predictions=predictions, references=references)
results
{'exact_match': 0.0, 'f1': 0.0}
```
Partial match (2 out of 3 answers correct) :
```python
from datasets import load_metric
squad_metric = load_metric("squad")
predictions = [{'prediction_text': '1976', 'id': '56e10a3be3433e1400422b22'}, {'prediction_text': 'Beyonce', 'id': '56d2051ce7d4791d0090260b'}, {'prediction_text': 'climate change', 'id': '5733b5344776f419006610e1'}]
references = [{'answers': {'answer_start': [97], 'text': ['1976']}, 'id': '56e10a3be3433e1400422b22'}, {'answers': {'answer_start': [233], 'text': ['Beyoncé and Bruno Mars']}, 'id': '56d2051ce7d4791d0090260b'}, {'answers': {'answer_start': [891], 'text': ['climate change']}, 'id': '5733b5344776f419006610e1'}]
results = squad_metric.compute(predictions=predictions, references=references)
results
{'exact_match': 66.66666666666667, 'f1': 66.66666666666667}
```
## Limitations and bias
This metric works only with datasets that have the same format as [SQuAD v.1 dataset](https://huggingface.co/datasets/squad).
The SQuAD dataset does contain a certain amount of noise, such as duplicate questions as well as missing answers, but these represent a minority of the 100,000 question-answer pairs. Also, neither exact match nor F1 score reflect whether models do better on certain types of questions (e.g. who questions) or those that cover a certain gender or geographical area -- carrying out more in-depth error analysis can complement these numbers.
## Citation
@inproceedings{Rajpurkar2016SQuAD10,
title={SQuAD: 100, 000+ Questions for Machine Comprehension of Text},
author={Pranav Rajpurkar and Jian Zhang and Konstantin Lopyrev and Percy Liang},
booktitle={EMNLP},
year={2016}
}
## Further References
- [The Stanford Question Answering Dataset: Background, Challenges, Progress (blog post)](https://rajpurkar.github.io/mlx/qa-and-squad/)
- [Hugging Face Course -- Question Answering](https://huggingface.co/course/chapter7/7)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/squad/evaluate.py | """ Official evaluation script for v1.1 of the SQuAD dataset. """
import argparse
import json
import re
import string
import sys
from collections import Counter
def normalize_answer(s):
"""Lower text and remove punctuation, articles and extra whitespace."""
def remove_articles(text):
return re.sub(r"\b(a|an|the)\b", " ", text)
def white_space_fix(text):
return " ".join(text.split())
def remove_punc(text):
exclude = set(string.punctuation)
return "".join(ch for ch in text if ch not in exclude)
def lower(text):
return text.lower()
return white_space_fix(remove_articles(remove_punc(lower(s))))
def f1_score(prediction, ground_truth):
prediction_tokens = normalize_answer(prediction).split()
ground_truth_tokens = normalize_answer(ground_truth).split()
common = Counter(prediction_tokens) & Counter(ground_truth_tokens)
num_same = sum(common.values())
if num_same == 0:
return 0
precision = 1.0 * num_same / len(prediction_tokens)
recall = 1.0 * num_same / len(ground_truth_tokens)
f1 = (2 * precision * recall) / (precision + recall)
return f1
def exact_match_score(prediction, ground_truth):
return normalize_answer(prediction) == normalize_answer(ground_truth)
def metric_max_over_ground_truths(metric_fn, prediction, ground_truths):
scores_for_ground_truths = []
for ground_truth in ground_truths:
score = metric_fn(prediction, ground_truth)
scores_for_ground_truths.append(score)
return max(scores_for_ground_truths)
def evaluate(dataset, predictions):
f1 = exact_match = total = 0
for article in dataset:
for paragraph in article["paragraphs"]:
for qa in paragraph["qas"]:
total += 1
if qa["id"] not in predictions:
message = "Unanswered question " + qa["id"] + " will receive score 0."
print(message, file=sys.stderr)
continue
ground_truths = [x["text"] for x in qa["answers"]]
prediction = predictions[qa["id"]]
exact_match += metric_max_over_ground_truths(exact_match_score, prediction, ground_truths)
f1 += metric_max_over_ground_truths(f1_score, prediction, ground_truths)
exact_match = 100.0 * exact_match / total
f1 = 100.0 * f1 / total
return {"exact_match": exact_match, "f1": f1}
if __name__ == "__main__":
expected_version = "1.1"
parser = argparse.ArgumentParser(description="Evaluation for SQuAD " + expected_version)
parser.add_argument("dataset_file", help="Dataset file")
parser.add_argument("prediction_file", help="Prediction File")
args = parser.parse_args()
with open(args.dataset_file) as dataset_file:
dataset_json = json.load(dataset_file)
if dataset_json["version"] != expected_version:
print(
"Evaluation expects v-" + expected_version + ", but got dataset with v-" + dataset_json["version"],
file=sys.stderr,
)
dataset = dataset_json["data"]
with open(args.prediction_file) as prediction_file:
predictions = json.load(prediction_file)
print(json.dumps(evaluate(dataset, predictions)))
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/squad/squad.py | # Copyright 2020 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" SQuAD metric. """
import datasets
from .evaluate import evaluate
_CITATION = """\
@inproceedings{Rajpurkar2016SQuAD10,
title={SQuAD: 100, 000+ Questions for Machine Comprehension of Text},
author={Pranav Rajpurkar and Jian Zhang and Konstantin Lopyrev and Percy Liang},
booktitle={EMNLP},
year={2016}
}
"""
_DESCRIPTION = """
This metric wrap the official scoring script for version 1 of the Stanford Question Answering Dataset (SQuAD).
Stanford Question Answering Dataset (SQuAD) is a reading comprehension dataset, consisting of questions posed by
crowdworkers on a set of Wikipedia articles, where the answer to every question is a segment of text, or span,
from the corresponding reading passage, or the question might be unanswerable.
"""
_KWARGS_DESCRIPTION = """
Computes SQuAD scores (F1 and EM).
Args:
predictions: List of question-answers dictionaries with the following key-values:
- 'id': id of the question-answer pair as given in the references (see below)
- 'prediction_text': the text of the answer
references: List of question-answers dictionaries with the following key-values:
- 'id': id of the question-answer pair (see above),
- 'answers': a Dict in the SQuAD dataset format
{
'text': list of possible texts for the answer, as a list of strings
'answer_start': list of start positions for the answer, as a list of ints
}
Note that answer_start values are not taken into account to compute the metric.
Returns:
'exact_match': Exact match (the normalized answer exactly match the gold answer)
'f1': The F-score of predicted tokens versus the gold answer
Examples:
>>> predictions = [{'prediction_text': '1976', 'id': '56e10a3be3433e1400422b22'}]
>>> references = [{'answers': {'answer_start': [97], 'text': ['1976']}, 'id': '56e10a3be3433e1400422b22'}]
>>> squad_metric = datasets.load_metric("squad")
>>> results = squad_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'exact_match': 100.0, 'f1': 100.0}
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Squad(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": {"id": datasets.Value("string"), "prediction_text": datasets.Value("string")},
"references": {
"id": datasets.Value("string"),
"answers": datasets.features.Sequence(
{
"text": datasets.Value("string"),
"answer_start": datasets.Value("int32"),
}
),
},
}
),
codebase_urls=["https://rajpurkar.github.io/SQuAD-explorer/"],
reference_urls=["https://rajpurkar.github.io/SQuAD-explorer/"],
)
def _compute(self, predictions, references):
pred_dict = {prediction["id"]: prediction["prediction_text"] for prediction in predictions}
dataset = [
{
"paragraphs": [
{
"qas": [
{
"answers": [{"text": answer_text} for answer_text in ref["answers"]["text"]],
"id": ref["id"],
}
for ref in references
]
}
]
}
]
score = evaluate(dataset=dataset, predictions=pred_dict)
return score
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/bertscore/README.md | # Metric Card for BERT Score
## Metric description
BERTScore is an automatic evaluation metric for text generation that computes a similarity score for each token in the candidate sentence with each token in the reference sentence. It leverages the pre-trained contextual embeddings from [BERT](https://huggingface.co/bert-base-uncased) models and matches words in candidate and reference sentences by cosine similarity.
Moreover, BERTScore computes precision, recall, and F1 measure, which can be useful for evaluating different language generation tasks.
## How to use
BERTScore takes 3 mandatory arguments : `predictions` (a list of string of candidate sentences), `references` (a list of strings or list of list of strings of reference sentences) and either `lang` (a string of two letters indicating the language of the sentences, in [ISO 639-1 format](https://en.wikipedia.org/wiki/List_of_ISO_639-1_codes)) or `model_type` (a string specififying which model to use, according to the BERT specification). The default behavior of the metric is to use the suggested model for the target language when one is specified, otherwise to use the `model_type` indicated.
```python
from datasets import load_metric
bertscore = load_metric("bertscore")
predictions = ["hello there", "general kenobi"]
references = ["hello there", "general kenobi"]
results = bertscore.compute(predictions=predictions, references=references, lang="en")
```
BERTScore also accepts multiple optional arguments:
`num_layers` (int): The layer of representation to use. The default is the number of layers tuned on WMT16 correlation data, which depends on the `model_type` used.
`verbose` (bool): Turn on intermediate status update. The default value is `False`.
`idf` (bool or dict): Use idf weighting; can also be a precomputed idf_dict.
`device` (str): On which the contextual embedding model will be allocated on. If this argument is `None`, the model lives on `cuda:0` if cuda is available.
`nthreads` (int): Number of threads used for computation. The default value is `4`.
`rescale_with_baseline` (bool): Rescale BERTScore with the pre-computed baseline. The default value is `False`.
`batch_size` (int): BERTScore processing batch size, at least one of `model_type` or `lang`. `lang` needs to be specified when `rescale_with_baseline` is `True`.
`baseline_path` (str): Customized baseline file.
`use_fast_tokenizer` (bool): `use_fast` parameter passed to HF tokenizer. The default value is `False`.
## Output values
BERTScore outputs a dictionary with the following values:
`precision`: The [precision](https://huggingface.co/metrics/precision) for each sentence from the `predictions` + `references` lists, which ranges from 0.0 to 1.0.
`recall`: The [recall](https://huggingface.co/metrics/recall) for each sentence from the `predictions` + `references` lists, which ranges from 0.0 to 1.0.
`f1`: The [F1 score](https://huggingface.co/metrics/f1) for each sentence from the `predictions` + `references` lists, which ranges from 0.0 to 1.0.
`hashcode:` The hashcode of the library.
### Values from popular papers
The [original BERTScore paper](https://openreview.net/pdf?id=SkeHuCVFDr) reported average model selection accuracies (Hits@1) on WMT18 hybrid systems for different language pairs, which ranged from 0.004 for `en<->tr` to 0.824 for `en<->de`.
For more recent model performance, see the [metric leaderboard](https://paperswithcode.com/paper/bertscore-evaluating-text-generation-with).
## Examples
Maximal values with the `distilbert-base-uncased` model:
```python
from datasets import load_metric
bertscore = load_metric("bertscore")
predictions = ["hello world", "general kenobi"]
references = ["hello world", "general kenobi"]
results = bertscore.compute(predictions=predictions, references=references, model_type="distilbert-base-uncased")
print(results)
{'precision': [1.0, 1.0], 'recall': [1.0, 1.0], 'f1': [1.0, 1.0], 'hashcode': 'distilbert-base-uncased_L5_no-idf_version=0.3.10(hug_trans=4.10.3)'}
```
Partial match with the `bert-base-uncased` model:
```python
from datasets import load_metric
bertscore = load_metric("bertscore")
predictions = ["hello world", "general kenobi"]
references = ["goodnight moon", "the sun is shining"]
results = bertscore.compute(predictions=predictions, references=references, model_type="distilbert-base-uncased")
print(results)
{'precision': [0.7380737066268921, 0.5584042072296143], 'recall': [0.7380737066268921, 0.5889028906822205], 'f1': [0.7380737066268921, 0.5732481479644775], 'hashcode': 'bert-base-uncased_L5_no-idf_version=0.3.10(hug_trans=4.10.3)'}
```
## Limitations and bias
The [original BERTScore paper](https://openreview.net/pdf?id=SkeHuCVFDr) showed that BERTScore correlates well with human judgment on sentence-level and system-level evaluation, but this depends on the model and language pair selected.
Furthermore, not all languages are supported by the metric -- see the [BERTScore supported language list](https://github.com/google-research/bert/blob/master/multilingual.md#list-of-languages) for more information.
Finally, calculating the BERTScore metric involves downloading the BERT model that is used to compute the score-- the default model for `en`, `roberta-large`, takes over 1.4GB of storage space and downloading it can take a significant amount of time depending on the speed of your internet connection. If this is an issue, choose a smaller model; for instance `distilbert-base-uncased` is 268MB. A full list of compatible models can be found [here](https://docs.google.com/spreadsheets/d/1RKOVpselB98Nnh_EOC4A2BYn8_201tmPODpNWu4w7xI/edit#gid=0).
## Citation
```bibtex
@inproceedings{bert-score,
title={BERTScore: Evaluating Text Generation with BERT},
author={Tianyi Zhang* and Varsha Kishore* and Felix Wu* and Kilian Q. Weinberger and Yoav Artzi},
booktitle={International Conference on Learning Representations},
year={2020},
url={https://openreview.net/forum?id=SkeHuCVFDr}
}
```
## Further References
- [BERTScore Project README](https://github.com/Tiiiger/bert_score#readme)
- [BERTScore ICLR 2020 Poster Presentation](https://iclr.cc/virtual_2020/poster_SkeHuCVFDr.html)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/bertscore/bertscore.py | # Copyright 2020 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" BERTScore metric. """
import functools
from contextlib import contextmanager
import bert_score
from packaging import version
import datasets
@contextmanager
def filter_logging_context():
def filter_log(record):
return False if "This IS expected if you are initializing" in record.msg else True
logger = datasets.utils.logging.get_logger("transformers.modeling_utils")
logger.addFilter(filter_log)
try:
yield
finally:
logger.removeFilter(filter_log)
_CITATION = """\
@inproceedings{bert-score,
title={BERTScore: Evaluating Text Generation with BERT},
author={Tianyi Zhang* and Varsha Kishore* and Felix Wu* and Kilian Q. Weinberger and Yoav Artzi},
booktitle={International Conference on Learning Representations},
year={2020},
url={https://openreview.net/forum?id=SkeHuCVFDr}
}
"""
_DESCRIPTION = """\
BERTScore leverages the pre-trained contextual embeddings from BERT and matches words in candidate and reference
sentences by cosine similarity.
It has been shown to correlate with human judgment on sentence-level and system-level evaluation.
Moreover, BERTScore computes precision, recall, and F1 measure, which can be useful for evaluating different language
generation tasks.
See the project's README at https://github.com/Tiiiger/bert_score#readme for more information.
"""
_KWARGS_DESCRIPTION = """
BERTScore Metrics with the hashcode from a source against one or more references.
Args:
predictions (list of str): Prediction/candidate sentences.
references (list of str or list of list of str): Reference sentences.
lang (str): Language of the sentences; required (e.g. 'en').
model_type (str): Bert specification, default using the suggested
model for the target language; has to specify at least one of
`model_type` or `lang`.
num_layers (int): The layer of representation to use,
default using the number of layers tuned on WMT16 correlation data.
verbose (bool): Turn on intermediate status update.
idf (bool or dict): Use idf weighting; can also be a precomputed idf_dict.
device (str): On which the contextual embedding model will be allocated on.
If this argument is None, the model lives on cuda:0 if cuda is available.
nthreads (int): Number of threads.
batch_size (int): Bert score processing batch size,
at least one of `model_type` or `lang`. `lang` needs to be
specified when `rescale_with_baseline` is True.
rescale_with_baseline (bool): Rescale bertscore with pre-computed baseline.
baseline_path (str): Customized baseline file.
use_fast_tokenizer (bool): `use_fast` parameter passed to HF tokenizer. New in version 0.3.10.
Returns:
precision: Precision.
recall: Recall.
f1: F1 score.
hashcode: Hashcode of the library.
Examples:
>>> predictions = ["hello there", "general kenobi"]
>>> references = ["hello there", "general kenobi"]
>>> bertscore = datasets.load_metric("bertscore")
>>> results = bertscore.compute(predictions=predictions, references=references, lang="en")
>>> print([round(v, 2) for v in results["f1"]])
[1.0, 1.0]
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class BERTScore(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
homepage="https://github.com/Tiiiger/bert_score",
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Value("string", id="sequence"),
"references": datasets.Sequence(datasets.Value("string", id="sequence"), id="references"),
}
),
codebase_urls=["https://github.com/Tiiiger/bert_score"],
reference_urls=[
"https://github.com/Tiiiger/bert_score",
"https://arxiv.org/abs/1904.09675",
],
)
def _compute(
self,
predictions,
references,
lang=None,
model_type=None,
num_layers=None,
verbose=False,
idf=False,
device=None,
batch_size=64,
nthreads=4,
all_layers=False,
rescale_with_baseline=False,
baseline_path=None,
use_fast_tokenizer=False,
):
get_hash = bert_score.utils.get_hash
scorer = bert_score.BERTScorer
if version.parse(bert_score.__version__) >= version.parse("0.3.10"):
get_hash = functools.partial(get_hash, use_fast_tokenizer=use_fast_tokenizer)
scorer = functools.partial(scorer, use_fast_tokenizer=use_fast_tokenizer)
elif use_fast_tokenizer:
raise ImportWarning(
"To use a fast tokenizer, the module `bert-score>=0.3.10` is required, and the current version of `bert-score` doesn't match this condition.\n"
'You can install it with `pip install "bert-score>=0.3.10"`.'
)
if model_type is None:
assert lang is not None, "either lang or model_type should be specified"
model_type = bert_score.utils.lang2model[lang.lower()]
if num_layers is None:
num_layers = bert_score.utils.model2layers[model_type]
hashcode = get_hash(
model=model_type,
num_layers=num_layers,
idf=idf,
rescale_with_baseline=rescale_with_baseline,
use_custom_baseline=baseline_path is not None,
)
with filter_logging_context():
if not hasattr(self, "cached_bertscorer") or self.cached_bertscorer.hash != hashcode:
self.cached_bertscorer = scorer(
model_type=model_type,
num_layers=num_layers,
batch_size=batch_size,
nthreads=nthreads,
all_layers=all_layers,
idf=idf,
device=device,
lang=lang,
rescale_with_baseline=rescale_with_baseline,
baseline_path=baseline_path,
)
(P, R, F) = self.cached_bertscorer.score(
cands=predictions,
refs=references,
verbose=verbose,
batch_size=batch_size,
)
output_dict = {
"precision": P.tolist(),
"recall": R.tolist(),
"f1": F.tolist(),
"hashcode": hashcode,
}
return output_dict
def add_batch(self, predictions=None, references=None, **kwargs):
"""Add a batch of predictions and references for the metric's stack."""
# References can be strings or lists of strings
# Let's change strings to lists of strings with one element
if references is not None:
references = [[ref] if isinstance(ref, str) else ref for ref in references]
super().add_batch(predictions=predictions, references=references, **kwargs)
def add(self, prediction=None, reference=None, **kwargs):
"""Add one prediction and reference for the metric's stack."""
# References can be strings or lists of strings
# Let's change strings to lists of strings with one element
if isinstance(reference, str):
reference = [reference]
super().add(prediction=prediction, reference=reference, **kwargs)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/mean_iou/README.md | # Metric Card for Mean IoU
## Metric Description
IoU (Intersection over Union) is the area of overlap between the predicted segmentation and the ground truth divided by the area of union between the predicted segmentation and the ground truth.
For binary (two classes) or multi-class segmentation, the *mean IoU* of the image is calculated by taking the IoU of each class and averaging them.
## How to Use
The Mean IoU metric takes two numeric arrays as input corresponding to the predicted and ground truth segmentations:
```python
>>> import numpy as np
>>> mean_iou = datasets.load_metric("mean_iou")
>>> predicted = np.array([[2, 2, 3], [8, 2, 4], [3, 255, 2]])
>>> ground_truth = np.array([[1, 2, 2], [8, 2, 1], [3, 255, 1]])
>>> results = mean_iou.compute(predictions=predicted, references=ground_truth, num_labels=10, ignore_index=255)
```
### Inputs
**Mandatory inputs**
- `predictions` (`List[ndarray]`): List of predicted segmentation maps, each of shape (height, width). Each segmentation map can be of a different size.
- `references` (`List[ndarray]`): List of ground truth segmentation maps, each of shape (height, width). Each segmentation map can be of a different size.
- `num_labels` (`int`): Number of classes (categories).
- `ignore_index` (`int`): Index that will be ignored during evaluation.
**Optional inputs**
- `nan_to_num` (`int`): If specified, NaN values will be replaced by the number defined by the user.
- `label_map` (`dict`): If specified, dictionary mapping old label indices to new label indices.
- `reduce_labels` (`bool`): Whether or not to reduce all label values of segmentation maps by 1. Usually used for datasets where 0 is used for background, and background itself is not included in all classes of a dataset (e.g. ADE20k). The background label will be replaced by 255. The default value is `False`.
### Output Values
The metric returns a dictionary with the following elements:
- `mean_iou` (`float`): Mean Intersection-over-Union (IoU averaged over all categories).
- `mean_accuracy` (`float`): Mean accuracy (averaged over all categories).
- `overall_accuracy` (`float`): Overall accuracy on all images.
- `per_category_accuracy` (`ndarray` of shape `(num_labels,)`): Per category accuracy.
- `per_category_iou` (`ndarray` of shape `(num_labels,)`): Per category IoU.
The values of all of the scores reported range from from `0.0` (minimum) and `1.0` (maximum).
Output Example:
```python
{'mean_iou': 0.47750000000000004, 'mean_accuracy': 0.5916666666666666, 'overall_accuracy': 0.5263157894736842, 'per_category_iou': array([0. , 0. , 0.375, 0.4 , 0.5 , 0. , 0.5 , 1. , 1. , 1. ]), 'per_category_accuracy': array([0. , 0. , 0.75 , 0.66666667, 1. , 0. , 0.5 , 1. , 1. , 1. ])}
```
#### Values from Popular Papers
The [leaderboard for the CityScapes dataset](https://paperswithcode.com/sota/semantic-segmentation-on-cityscapes) reports a Mean IOU ranging from 64 to 84; that of [ADE20k](https://paperswithcode.com/sota/semantic-segmentation-on-ade20k) ranges from 30 to a peak of 59.9, indicating that the dataset is more difficult for current approaches (as of 2022).
### Examples
```python
>>> from datasets import load_metric
>>> import numpy as np
>>> mean_iou = load_metric("mean_iou")
>>> # suppose one has 3 different segmentation maps predicted
>>> predicted_1 = np.array([[1, 2], [3, 4], [5, 255]])
>>> actual_1 = np.array([[0, 3], [5, 4], [6, 255]])
>>> predicted_2 = np.array([[2, 7], [9, 2], [3, 6]])
>>> actual_2 = np.array([[1, 7], [9, 2], [3, 6]])
>>> predicted_3 = np.array([[2, 2, 3], [8, 2, 4], [3, 255, 2]])
>>> actual_3 = np.array([[1, 2, 2], [8, 2, 1], [3, 255, 1]])
>>> predictions = [predicted_1, predicted_2, predicted_3]
>>> references = [actual_1, actual_2, actual_3]
>>> results = mean_iou.compute(predictions=predictions, references=references, num_labels=10, ignore_index=255, reduce_labels=False)
>>> print(results) # doctest: +NORMALIZE_WHITESPACE
{'mean_iou': 0.47750000000000004, 'mean_accuracy': 0.5916666666666666, 'overall_accuracy': 0.5263157894736842, 'per_category_iou': array([0. , 0. , 0.375, 0.4 , 0.5 , 0. , 0.5 , 1. , 1. , 1. ]), 'per_category_accuracy': array([0. , 0. , 0.75 , 0.66666667, 1. , 0. , 0.5 , 1. , 1. , 1. ])}
```
## Limitations and Bias
Mean IOU is an average metric, so it will not show you where model predictions differ from the ground truth (i.e. if there are particular regions or classes that the model does poorly on). Further error analysis is needed to gather actional insights that can be used to inform model improvements.
## Citation(s)
```bibtex
@software{MMSegmentation_Contributors_OpenMMLab_Semantic_Segmentation_2020,
author = {{MMSegmentation Contributors}},
license = {Apache-2.0},
month = {7},
title = {{OpenMMLab Semantic Segmentation Toolbox and Benchmark}},
url = {https://github.com/open-mmlab/mmsegmentation},
year = {2020}
}"
```
## Further References
- [Wikipedia article - Jaccard Index](https://en.wikipedia.org/wiki/Jaccard_index)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/mean_iou/mean_iou.py | # Copyright 2022 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Mean IoU (Intersection-over-Union) metric."""
from typing import Dict, Optional
import numpy as np
import datasets
_DESCRIPTION = """
IoU is the area of overlap between the predicted segmentation and the ground truth divided by the area of union
between the predicted segmentation and the ground truth. For binary (two classes) or multi-class segmentation,
the mean IoU of the image is calculated by taking the IoU of each class and averaging them.
"""
_KWARGS_DESCRIPTION = """
Args:
predictions (`List[ndarray]`):
List of predicted segmentation maps, each of shape (height, width). Each segmentation map can be of a different size.
references (`List[ndarray]`):
List of ground truth segmentation maps, each of shape (height, width). Each segmentation map can be of a different size.
num_labels (`int`):
Number of classes (categories).
ignore_index (`int`):
Index that will be ignored during evaluation.
nan_to_num (`int`, *optional*):
If specified, NaN values will be replaced by the number defined by the user.
label_map (`dict`, *optional*):
If specified, dictionary mapping old label indices to new label indices.
reduce_labels (`bool`, *optional*, defaults to `False`):
Whether or not to reduce all label values of segmentation maps by 1. Usually used for datasets where 0 is used for background,
and background itself is not included in all classes of a dataset (e.g. ADE20k). The background label will be replaced by 255.
Returns:
`Dict[str, float | ndarray]` comprising various elements:
- *mean_iou* (`float`):
Mean Intersection-over-Union (IoU averaged over all categories).
- *mean_accuracy* (`float`):
Mean accuracy (averaged over all categories).
- *overall_accuracy* (`float`):
Overall accuracy on all images.
- *per_category_accuracy* (`ndarray` of shape `(num_labels,)`):
Per category accuracy.
- *per_category_iou* (`ndarray` of shape `(num_labels,)`):
Per category IoU.
Examples:
>>> import numpy as np
>>> mean_iou = datasets.load_metric("mean_iou")
>>> # suppose one has 3 different segmentation maps predicted
>>> predicted_1 = np.array([[1, 2], [3, 4], [5, 255]])
>>> actual_1 = np.array([[0, 3], [5, 4], [6, 255]])
>>> predicted_2 = np.array([[2, 7], [9, 2], [3, 6]])
>>> actual_2 = np.array([[1, 7], [9, 2], [3, 6]])
>>> predicted_3 = np.array([[2, 2, 3], [8, 2, 4], [3, 255, 2]])
>>> actual_3 = np.array([[1, 2, 2], [8, 2, 1], [3, 255, 1]])
>>> predicted = [predicted_1, predicted_2, predicted_3]
>>> ground_truth = [actual_1, actual_2, actual_3]
>>> results = mean_iou.compute(predictions=predicted, references=ground_truth, num_labels=10, ignore_index=255, reduce_labels=False)
>>> print(results) # doctest: +NORMALIZE_WHITESPACE
{'mean_iou': 0.47750000000000004, 'mean_accuracy': 0.5916666666666666, 'overall_accuracy': 0.5263157894736842, 'per_category_iou': array([0. , 0. , 0.375, 0.4 , 0.5 , 0. , 0.5 , 1. , 1. , 1. ]), 'per_category_accuracy': array([0. , 0. , 0.75 , 0.66666667, 1. , 0. , 0.5 , 1. , 1. , 1. ])}
"""
_CITATION = """\
@software{MMSegmentation_Contributors_OpenMMLab_Semantic_Segmentation_2020,
author = {{MMSegmentation Contributors}},
license = {Apache-2.0},
month = {7},
title = {{OpenMMLab Semantic Segmentation Toolbox and Benchmark}},
url = {https://github.com/open-mmlab/mmsegmentation},
year = {2020}
}"""
def intersect_and_union(
pred_label,
label,
num_labels,
ignore_index: bool,
label_map: Optional[Dict[int, int]] = None,
reduce_labels: bool = False,
):
"""Calculate intersection and Union.
Args:
pred_label (`ndarray`):
Prediction segmentation map of shape (height, width).
label (`ndarray`):
Ground truth segmentation map of shape (height, width).
num_labels (`int`):
Number of categories.
ignore_index (`int`):
Index that will be ignored during evaluation.
label_map (`dict`, *optional*):
Mapping old labels to new labels. The parameter will work only when label is str.
reduce_labels (`bool`, *optional*, defaults to `False`):
Whether or not to reduce all label values of segmentation maps by 1. Usually used for datasets where 0 is used for background,
and background itself is not included in all classes of a dataset (e.g. ADE20k). The background label will be replaced by 255.
Returns:
area_intersect (`ndarray`):
The intersection of prediction and ground truth histogram on all classes.
area_union (`ndarray`):
The union of prediction and ground truth histogram on all classes.
area_pred_label (`ndarray`):
The prediction histogram on all classes.
area_label (`ndarray`):
The ground truth histogram on all classes.
"""
if label_map is not None:
for old_id, new_id in label_map.items():
label[label == old_id] = new_id
# turn into Numpy arrays
pred_label = np.array(pred_label)
label = np.array(label)
if reduce_labels:
label[label == 0] = 255
label = label - 1
label[label == 254] = 255
mask = label != ignore_index
mask = np.not_equal(label, ignore_index)
pred_label = pred_label[mask]
label = np.array(label)[mask]
intersect = pred_label[pred_label == label]
area_intersect = np.histogram(intersect, bins=num_labels, range=(0, num_labels - 1))[0]
area_pred_label = np.histogram(pred_label, bins=num_labels, range=(0, num_labels - 1))[0]
area_label = np.histogram(label, bins=num_labels, range=(0, num_labels - 1))[0]
area_union = area_pred_label + area_label - area_intersect
return area_intersect, area_union, area_pred_label, area_label
def total_intersect_and_union(
results,
gt_seg_maps,
num_labels,
ignore_index: bool,
label_map: Optional[Dict[int, int]] = None,
reduce_labels: bool = False,
):
"""Calculate Total Intersection and Union, by calculating `intersect_and_union` for each (predicted, ground truth) pair.
Args:
results (`ndarray`):
List of prediction segmentation maps, each of shape (height, width).
gt_seg_maps (`ndarray`):
List of ground truth segmentation maps, each of shape (height, width).
num_labels (`int`):
Number of categories.
ignore_index (`int`):
Index that will be ignored during evaluation.
label_map (`dict`, *optional*):
Mapping old labels to new labels. The parameter will work only when label is str.
reduce_labels (`bool`, *optional*, defaults to `False`):
Whether or not to reduce all label values of segmentation maps by 1. Usually used for datasets where 0 is used for background,
and background itself is not included in all classes of a dataset (e.g. ADE20k). The background label will be replaced by 255.
Returns:
total_area_intersect (`ndarray`):
The intersection of prediction and ground truth histogram on all classes.
total_area_union (`ndarray`):
The union of prediction and ground truth histogram on all classes.
total_area_pred_label (`ndarray`):
The prediction histogram on all classes.
total_area_label (`ndarray`):
The ground truth histogram on all classes.
"""
total_area_intersect = np.zeros((num_labels,), dtype=np.float64)
total_area_union = np.zeros((num_labels,), dtype=np.float64)
total_area_pred_label = np.zeros((num_labels,), dtype=np.float64)
total_area_label = np.zeros((num_labels,), dtype=np.float64)
for result, gt_seg_map in zip(results, gt_seg_maps):
area_intersect, area_union, area_pred_label, area_label = intersect_and_union(
result, gt_seg_map, num_labels, ignore_index, label_map, reduce_labels
)
total_area_intersect += area_intersect
total_area_union += area_union
total_area_pred_label += area_pred_label
total_area_label += area_label
return total_area_intersect, total_area_union, total_area_pred_label, total_area_label
def mean_iou(
results,
gt_seg_maps,
num_labels,
ignore_index: bool,
nan_to_num: Optional[int] = None,
label_map: Optional[Dict[int, int]] = None,
reduce_labels: bool = False,
):
"""Calculate Mean Intersection and Union (mIoU).
Args:
results (`ndarray`):
List of prediction segmentation maps, each of shape (height, width).
gt_seg_maps (`ndarray`):
List of ground truth segmentation maps, each of shape (height, width).
num_labels (`int`):
Number of categories.
ignore_index (`int`):
Index that will be ignored during evaluation.
nan_to_num (`int`, *optional*):
If specified, NaN values will be replaced by the number defined by the user.
label_map (`dict`, *optional*):
Mapping old labels to new labels. The parameter will work only when label is str.
reduce_labels (`bool`, *optional*, defaults to `False`):
Whether or not to reduce all label values of segmentation maps by 1. Usually used for datasets where 0 is used for background,
and background itself is not included in all classes of a dataset (e.g. ADE20k). The background label will be replaced by 255.
Returns:
`Dict[str, float | ndarray]` comprising various elements:
- *mean_iou* (`float`):
Mean Intersection-over-Union (IoU averaged over all categories).
- *mean_accuracy* (`float`):
Mean accuracy (averaged over all categories).
- *overall_accuracy* (`float`):
Overall accuracy on all images.
- *per_category_accuracy* (`ndarray` of shape `(num_labels,)`):
Per category accuracy.
- *per_category_iou* (`ndarray` of shape `(num_labels,)`):
Per category IoU.
"""
total_area_intersect, total_area_union, total_area_pred_label, total_area_label = total_intersect_and_union(
results, gt_seg_maps, num_labels, ignore_index, label_map, reduce_labels
)
# compute metrics
metrics = {}
all_acc = total_area_intersect.sum() / total_area_label.sum()
iou = total_area_intersect / total_area_union
acc = total_area_intersect / total_area_label
metrics["mean_iou"] = np.nanmean(iou)
metrics["mean_accuracy"] = np.nanmean(acc)
metrics["overall_accuracy"] = all_acc
metrics["per_category_iou"] = iou
metrics["per_category_accuracy"] = acc
if nan_to_num is not None:
metrics = {metric: np.nan_to_num(metric_value, nan=nan_to_num) for metric, metric_value in metrics.items()}
return metrics
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class MeanIoU(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
# 1st Seq - height dim, 2nd - width dim
{
"predictions": datasets.Sequence(datasets.Sequence(datasets.Value("uint16"))),
"references": datasets.Sequence(datasets.Sequence(datasets.Value("uint16"))),
}
),
reference_urls=[
"https://github.com/open-mmlab/mmsegmentation/blob/71c201b1813267d78764f306a297ca717827c4bf/mmseg/core/evaluation/metrics.py"
],
)
def _compute(
self,
predictions,
references,
num_labels: int,
ignore_index: bool,
nan_to_num: Optional[int] = None,
label_map: Optional[Dict[int, int]] = None,
reduce_labels: bool = False,
):
iou_result = mean_iou(
results=predictions,
gt_seg_maps=references,
num_labels=num_labels,
ignore_index=ignore_index,
nan_to_num=nan_to_num,
label_map=label_map,
reduce_labels=reduce_labels,
)
return iou_result
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/mse/README.md | # Metric Card for MSE
## Metric Description
Mean Squared Error(MSE) represents the average of the squares of errors -- i.e. the average squared difference between the estimated values and the actual values.
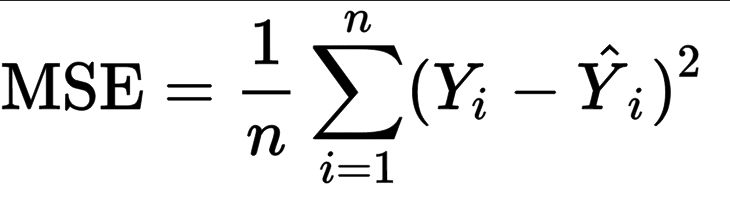
## How to Use
At minimum, this metric requires predictions and references as inputs.
```python
>>> mse_metric = datasets.load_metric("mse")
>>> predictions = [2.5, 0.0, 2, 8]
>>> references = [3, -0.5, 2, 7]
>>> results = mse_metric.compute(predictions=predictions, references=references)
```
### Inputs
Mandatory inputs:
- `predictions`: numeric array-like of shape (`n_samples,`) or (`n_samples`, `n_outputs`), representing the estimated target values.
- `references`: numeric array-like of shape (`n_samples,`) or (`n_samples`, `n_outputs`), representing the ground truth (correct) target values.
Optional arguments:
- `sample_weight`: numeric array-like of shape (`n_samples,`) representing sample weights. The default is `None`.
- `multioutput`: `raw_values`, `uniform_average` or numeric array-like of shape (`n_outputs,`), which defines the aggregation of multiple output values. The default value is `uniform_average`.
- `raw_values` returns a full set of errors in case of multioutput input.
- `uniform_average` means that the errors of all outputs are averaged with uniform weight.
- the array-like value defines weights used to average errors.
- `squared` (`bool`): If `True` returns MSE value, if `False` returns RMSE (Root Mean Squared Error). The default value is `True`.
### Output Values
This metric outputs a dictionary, containing the mean squared error score, which is of type:
- `float`: if multioutput is `uniform_average` or an ndarray of weights, then the weighted average of all output errors is returned.
- numeric array-like of shape (`n_outputs,`): if multioutput is `raw_values`, then the score is returned for each output separately.
Each MSE `float` value ranges from `0.0` to `1.0`, with the best value being `0.0`.
Output Example(s):
```python
{'mse': 0.5}
```
If `multioutput="raw_values"`:
```python
{'mse': array([0.41666667, 1. ])}
```
#### Values from Popular Papers
### Examples
Example with the `uniform_average` config:
```python
>>> from datasets import load_metric
>>> mse_metric = load_metric("mse")
>>> predictions = [2.5, 0.0, 2, 8]
>>> references = [3, -0.5, 2, 7]
>>> results = mse_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'mse': 0.375}
```
Example with `squared = True`, which returns the RMSE:
```python
>>> from datasets import load_metric
>>> mse_metric = load_metric("mse")
>>> predictions = [2.5, 0.0, 2, 8]
>>> references = [3, -0.5, 2, 7]
>>> rmse_result = mse_metric.compute(predictions=predictions, references=references, squared=False)
>>> print(rmse_result)
{'mse': 0.6123724356957945}
```
Example with multi-dimensional lists, and the `raw_values` config:
```python
>>> from datasets import load_metric
>>> mse_metric = load_metric("mse", "multilist")
>>> predictions = [[0.5, 1], [-1, 1], [7, -6]]
>>> references = [[0, 2], [-1, 2], [8, -5]]
>>> results = mse_metric.compute(predictions=predictions, references=references, multioutput='raw_values')
>>> print(results)
{'mse': array([0.41666667, 1. ])}
"""
```
## Limitations and Bias
MSE has the disadvantage of heavily weighting outliers -- given that it squares them, this results in large errors weighing more heavily than small ones. It can be used alongside [MAE](https://huggingface.co/metrics/mae), which is complementary given that it does not square the errors.
## Citation(s)
```bibtex
@article{scikit-learn,
title={Scikit-learn: Machine Learning in {P}ython},
author={Pedregosa, F. and Varoquaux, G. and Gramfort, A. and Michel, V.
and Thirion, B. and Grisel, O. and Blondel, M. and Prettenhofer, P.
and Weiss, R. and Dubourg, V. and Vanderplas, J. and Passos, A. and
Cournapeau, D. and Brucher, M. and Perrot, M. and Duchesnay, E.},
journal={Journal of Machine Learning Research},
volume={12},
pages={2825--2830},
year={2011}
}
```
```bibtex
@article{willmott2005advantages,
title={Advantages of the mean absolute error (MAE) over the root mean square error (RMSE) in assessing average model performance},
author={Willmott, Cort J and Matsuura, Kenji},
journal={Climate research},
volume={30},
number={1},
pages={79--82},
year={2005}
}
```
## Further References
- [Mean Squared Error - Wikipedia](https://en.wikipedia.org/wiki/Mean_squared_error)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/mse/mse.py | # Copyright 2022 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""MSE - Mean Squared Error Metric"""
from sklearn.metrics import mean_squared_error
import datasets
_CITATION = """\
@article{scikit-learn,
title={Scikit-learn: Machine Learning in {P}ython},
author={Pedregosa, F. and Varoquaux, G. and Gramfort, A. and Michel, V.
and Thirion, B. and Grisel, O. and Blondel, M. and Prettenhofer, P.
and Weiss, R. and Dubourg, V. and Vanderplas, J. and Passos, A. and
Cournapeau, D. and Brucher, M. and Perrot, M. and Duchesnay, E.},
journal={Journal of Machine Learning Research},
volume={12},
pages={2825--2830},
year={2011}
}
"""
_DESCRIPTION = """\
Mean Squared Error(MSE) is the average of the square of difference between the predicted
and actual values.
"""
_KWARGS_DESCRIPTION = """
Args:
predictions: array-like of shape (n_samples,) or (n_samples, n_outputs)
Estimated target values.
references: array-like of shape (n_samples,) or (n_samples, n_outputs)
Ground truth (correct) target values.
sample_weight: array-like of shape (n_samples,), default=None
Sample weights.
multioutput: {"raw_values", "uniform_average"} or array-like of shape (n_outputs,), default="uniform_average"
Defines aggregating of multiple output values. Array-like value defines weights used to average errors.
"raw_values" : Returns a full set of errors in case of multioutput input.
"uniform_average" : Errors of all outputs are averaged with uniform weight.
squared : bool, default=True
If True returns MSE value, if False returns RMSE (Root Mean Squared Error) value.
Returns:
mse : mean squared error.
Examples:
>>> mse_metric = datasets.load_metric("mse")
>>> predictions = [2.5, 0.0, 2, 8]
>>> references = [3, -0.5, 2, 7]
>>> results = mse_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'mse': 0.375}
>>> rmse_result = mse_metric.compute(predictions=predictions, references=references, squared=False)
>>> print(rmse_result)
{'mse': 0.6123724356957945}
If you're using multi-dimensional lists, then set the config as follows :
>>> mse_metric = datasets.load_metric("mse", "multilist")
>>> predictions = [[0.5, 1], [-1, 1], [7, -6]]
>>> references = [[0, 2], [-1, 2], [8, -5]]
>>> results = mse_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'mse': 0.7083333333333334}
>>> results = mse_metric.compute(predictions=predictions, references=references, multioutput='raw_values')
>>> print(results) # doctest: +NORMALIZE_WHITESPACE
{'mse': array([0.41666667, 1. ])}
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Mse(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(self._get_feature_types()),
reference_urls=[
"https://scikit-learn.org/stable/modules/generated/sklearn.metrics.mean_squared_error.html"
],
)
def _get_feature_types(self):
if self.config_name == "multilist":
return {
"predictions": datasets.Sequence(datasets.Value("float")),
"references": datasets.Sequence(datasets.Value("float")),
}
else:
return {
"predictions": datasets.Value("float"),
"references": datasets.Value("float"),
}
def _compute(self, predictions, references, sample_weight=None, multioutput="uniform_average", squared=True):
mse = mean_squared_error(
references, predictions, sample_weight=sample_weight, multioutput=multioutput, squared=squared
)
return {"mse": mse}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/competition_math/README.md | # Metric Card for Competition MATH
## Metric description
This metric is used to assess performance on the [Mathematics Aptitude Test of Heuristics (MATH) dataset](https://huggingface.co/datasets/competition_math).
It first canonicalizes the inputs (e.g., converting `1/2` to `\\frac{1}{2}`) and then computes accuracy.
## How to use
This metric takes two arguments:
`predictions`: a list of predictions to score. Each prediction is a string that contains natural language and LaTeX.
`references`: list of reference for each prediction. Each reference is a string that contains natural language and LaTeX.
```python
>>> from datasets import load_metric
>>> math = load_metric("competition_math")
>>> references = ["\\frac{1}{2}"]
>>> predictions = ["1/2"]
>>> results = math.compute(references=references, predictions=predictions)
```
N.B. To be able to use Competition MATH, you need to install the `math_equivalence` dependency using `pip install git+https://github.com/hendrycks/math.git`.
## Output values
This metric returns a dictionary that contains the [accuracy](https://huggingface.co/metrics/accuracy) after canonicalizing inputs, on a scale between 0.0 and 1.0.
### Values from popular papers
The [original MATH dataset paper](https://arxiv.org/abs/2103.03874) reported accuracies ranging from 3.0% to 6.9% by different large language models.
More recent progress on the dataset can be found on the [dataset leaderboard](https://paperswithcode.com/sota/math-word-problem-solving-on-math).
## Examples
Maximal values (full match):
```python
>>> from datasets import load_metric
>>> math = load_metric("competition_math")
>>> references = ["\\frac{1}{2}"]
>>> predictions = ["1/2"]
>>> results = math.compute(references=references, predictions=predictions)
>>> print(results)
{'accuracy': 1.0}
```
Minimal values (no match):
```python
>>> from datasets import load_metric
>>> math = load_metric("competition_math")
>>> references = ["\\frac{1}{2}"]
>>> predictions = ["3/4"]
>>> results = math.compute(references=references, predictions=predictions)
>>> print(results)
{'accuracy': 0.0}
```
Partial match:
```python
>>> from datasets import load_metric
>>> math = load_metric("competition_math")
>>> references = ["\\frac{1}{2}","\\frac{3}{4}"]
>>> predictions = ["1/5", "3/4"]
>>> results = math.compute(references=references, predictions=predictions)
>>> print(results)
{'accuracy': 0.5}
```
## Limitations and bias
This metric is limited to datasets with the same format as the [Mathematics Aptitude Test of Heuristics (MATH) dataset](https://huggingface.co/datasets/competition_math), and is meant to evaluate the performance of large language models at solving mathematical problems.
N.B. The MATH dataset also assigns levels of difficulty to different problems, so disagregating model performance by difficulty level (similarly to what was done in the [original paper](https://arxiv.org/abs/2103.03874) can give a better indication of how a given model does on a given difficulty of math problem, compared to overall accuracy.
## Citation
```bibtex
@article{hendrycksmath2021,
title={Measuring Mathematical Problem Solving With the MATH Dataset},
author={Dan Hendrycks
and Collin Burns
and Saurav Kadavath
and Akul Arora
and Steven Basart
and Eric Tang
and Dawn Song
and Jacob Steinhardt},
journal={arXiv preprint arXiv:2103.03874},
year={2021}
}
```
## Further References
- [MATH dataset](https://huggingface.co/datasets/competition_math)
- [MATH leaderboard](https://paperswithcode.com/sota/math-word-problem-solving-on-math)
- [MATH paper](https://arxiv.org/abs/2103.03874)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/competition_math/competition_math.py | # Copyright 2020 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Accuracy metric for the Mathematics Aptitude Test of Heuristics (MATH) dataset."""
import math_equivalence # From: git+https://github.com/hendrycks/math.git
import datasets
_CITATION = """\
@article{hendrycksmath2021,
title={Measuring Mathematical Problem Solving With the MATH Dataset},
author={Dan Hendrycks
and Collin Burns
and Saurav Kadavath
and Akul Arora
and Steven Basart
and Eric Tang
and Dawn Song
and Jacob Steinhardt},
journal={arXiv preprint arXiv:2103.03874},
year={2021}
}
"""
_DESCRIPTION = """\
This metric is used to assess performance on the Mathematics Aptitude Test of Heuristics (MATH) dataset.
It first canonicalizes the inputs (e.g., converting "1/2" to "\\frac{1}{2}") and then computes accuracy.
"""
_KWARGS_DESCRIPTION = r"""
Calculates accuracy after canonicalizing inputs.
Args:
predictions: list of predictions to score. Each prediction
is a string that contains natural language and LaTex.
references: list of reference for each prediction. Each
reference is a string that contains natural language
and LaTex.
Returns:
accuracy: accuracy after canonicalizing inputs
(e.g., converting "1/2" to "\\frac{1}{2}")
Examples:
>>> metric = datasets.load_metric("competition_math")
>>> results = metric.compute(references=["\\frac{1}{2}"], predictions=["1/2"])
>>> print(results)
{'accuracy': 1.0}
"""
@datasets.utils.file_utils.add_end_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class CompetitionMathMetric(datasets.Metric):
"""Accuracy metric for the MATH dataset."""
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Value("string"),
"references": datasets.Value("string"),
}
),
# Homepage of the metric for documentation
homepage="https://github.com/hendrycks/math",
# Additional links to the codebase or references
codebase_urls=["https://github.com/hendrycks/math"],
)
def _compute(self, predictions, references):
"""Returns the scores"""
n_correct = 0.0
for i, j in zip(predictions, references):
n_correct += 1.0 if math_equivalence.is_equiv(i, j) else 0.0
accuracy = n_correct / len(predictions)
return {
"accuracy": accuracy,
}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/xtreme_s/README.md | # Metric Card for XTREME-S
## Metric Description
The XTREME-S metric aims to evaluate model performance on the Cross-lingual TRansfer Evaluation of Multilingual Encoders for Speech (XTREME-S) benchmark.
This benchmark was designed to evaluate speech representations across languages, tasks, domains and data regimes. It covers 102 languages from 10+ language families, 3 different domains and 4 task families: speech recognition, translation, classification and retrieval.
## How to Use
There are two steps: (1) loading the XTREME-S metric relevant to the subset of the benchmark being used for evaluation; and (2) calculating the metric.
1. **Loading the relevant XTREME-S metric** : the subsets of XTREME-S are the following: `mls`, `voxpopuli`, `covost2`, `fleurs-asr`, `fleurs-lang_id`, `minds14` and `babel`. More information about the different subsets can be found on the [XTREME-S benchmark page](https://huggingface.co/datasets/google/xtreme_s).
```python
>>> from datasets import load_metric
>>> xtreme_s_metric = datasets.load_metric('xtreme_s', 'mls')
```
2. **Calculating the metric**: the metric takes two inputs :
- `predictions`: a list of predictions to score, with each prediction a `str`.
- `references`: a list of lists of references for each translation, with each reference a `str`.
```python
>>> references = ["it is sunny here", "paper and pen are essentials"]
>>> predictions = ["it's sunny", "paper pen are essential"]
>>> results = xtreme_s_metric.compute(predictions=predictions, references=references)
```
It also has two optional arguments:
- `bleu_kwargs`: a `dict` of keywords to be passed when computing the `bleu` metric for the `covost2` subset. Keywords can be one of `smooth_method`, `smooth_value`, `force`, `lowercase`, `tokenize`, `use_effective_order`.
- `wer_kwargs`: optional dict of keywords to be passed when computing `wer` and `cer`, which are computed for the `mls`, `fleurs-asr`, `voxpopuli`, and `babel` subsets. Keywords are `concatenate_texts`.
## Output values
The output of the metric depends on the XTREME-S subset chosen, consisting of a dictionary that contains one or several of the following metrics:
- `accuracy`: the proportion of correct predictions among the total number of cases processed, with a range between 0 and 1 (see [accuracy](https://huggingface.co/metrics/accuracy) for more information). This is returned for the `fleurs-lang_id` and `minds14` subsets.
- `f1`: the harmonic mean of the precision and recall (see [F1 score](https://huggingface.co/metrics/f1) for more information). Its range is 0-1 -- its lowest possible value is 0, if either the precision or the recall is 0, and its highest possible value is 1.0, which means perfect precision and recall. It is returned for the `minds14` subset.
- `wer`: Word error rate (WER) is a common metric of the performance of an automatic speech recognition system. The lower the value, the better the performance of the ASR system, with a WER of 0 being a perfect score (see [WER score](https://huggingface.co/metrics/wer) for more information). It is returned for the `mls`, `fleurs-asr`, `voxpopuli` and `babel` subsets of the benchmark.
- `cer`: Character error rate (CER) is similar to WER, but operates on character instead of word. The lower the CER value, the better the performance of the ASR system, with a CER of 0 being a perfect score (see [CER score](https://huggingface.co/metrics/cer) for more information). It is returned for the `mls`, `fleurs-asr`, `voxpopuli` and `babel` subsets of the benchmark.
- `bleu`: the BLEU score, calculated according to the SacreBLEU metric approach. It can take any value between 0.0 and 100.0, inclusive, with higher values being better (see [SacreBLEU](https://huggingface.co/metrics/sacrebleu) for more details). This is returned for the `covost2` subset.
### Values from popular papers
The [original XTREME-S paper](https://arxiv.org/pdf/2203.10752.pdf) reported average WERs ranging from 9.2 to 14.6, a BLEU score of 20.6, an accuracy of 73.3 and F1 score of 86.9, depending on the subsets of the dataset tested on.
## Examples
For the `mls` subset (which outputs `wer` and `cer`):
```python
>>> from datasets import load_metric
>>> xtreme_s_metric = datasets.load_metric('xtreme_s', 'mls')
>>> references = ["it is sunny here", "paper and pen are essentials"]
>>> predictions = ["it's sunny", "paper pen are essential"]
>>> results = xtreme_s_metric.compute(predictions=predictions, references=references)
>>> print({k: round(v, 2) for k, v in results.items()})
{'wer': 0.56, 'cer': 0.27}
```
For the `covost2` subset (which outputs `bleu`):
```python
>>> from datasets import load_metric
>>> xtreme_s_metric = datasets.load_metric('xtreme_s', 'covost2')
>>> references = ["bonjour paris", "il est necessaire de faire du sport de temps en temp"]
>>> predictions = ["bonjour paris", "il est important de faire du sport souvent"]
>>> results = xtreme_s_metric.compute(predictions=predictions, references=references)
>>> print({k: round(v, 2) for k, v in results.items()})
{'bleu': 31.65}
```
For the `fleurs-lang_id` subset (which outputs `accuracy`):
```python
>>> from datasets import load_metric
>>> xtreme_s_metric = datasets.load_metric('xtreme_s', 'fleurs-lang_id')
>>> references = [0, 1, 0, 0, 1]
>>> predictions = [0, 1, 1, 0, 0]
>>> results = xtreme_s_metric.compute(predictions=predictions, references=references)
>>> print({k: round(v, 2) for k, v in results.items()})
{'accuracy': 0.6}
```
For the `minds14` subset (which outputs `f1` and `accuracy`):
```python
>>> from datasets import load_metric
>>> xtreme_s_metric = datasets.load_metric('xtreme_s', 'minds14')
>>> references = [0, 1, 0, 0, 1]
>>> predictions = [0, 1, 1, 0, 0]
>>> results = xtreme_s_metric.compute(predictions=predictions, references=references)
>>> print({k: round(v, 2) for k, v in results.items()})
{'f1': 0.58, 'accuracy': 0.6}
```
## Limitations and bias
This metric works only with datasets that have the same format as the [XTREME-S dataset](https://huggingface.co/datasets/google/xtreme_s).
While the XTREME-S dataset is meant to represent a variety of languages and tasks, it has inherent biases: it is missing many languages that are important and under-represented in NLP datasets.
It also has a particular focus on read-speech because common evaluation benchmarks like CoVoST-2 or LibriSpeech evaluate on this type of speech, which results in a mismatch between performance obtained in a read-speech setting and a more noisy setting (in production or live deployment, for instance).
## Citation
```bibtex
@article{conneau2022xtreme,
title={XTREME-S: Evaluating Cross-lingual Speech Representations},
author={Conneau, Alexis and Bapna, Ankur and Zhang, Yu and Ma, Min and von Platen, Patrick and Lozhkov, Anton and Cherry, Colin and Jia, Ye and Rivera, Clara and Kale, Mihir and others},
journal={arXiv preprint arXiv:2203.10752},
year={2022}
}
```
## Further References
- [XTREME-S dataset](https://huggingface.co/datasets/google/xtreme_s)
- [XTREME-S github repository](https://github.com/google-research/xtreme)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/xtreme_s/xtreme_s.py | # Copyright 2022 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" XTREME-S benchmark metric. """
from typing import List
from packaging import version
from sklearn.metrics import f1_score
import datasets
from datasets.config import PY_VERSION
if PY_VERSION < version.parse("3.8"):
import importlib_metadata
else:
import importlib.metadata as importlib_metadata
# TODO(Patrick/Anton)
_CITATION = """\
"""
_DESCRIPTION = """\
XTREME-S is a benchmark to evaluate universal cross-lingual speech representations in many languages.
XTREME-S covers four task families: speech recognition, classification, speech-to-text translation and retrieval.
"""
_KWARGS_DESCRIPTION = """
Compute XTREME-S evaluation metric associated to each XTREME-S dataset.
Args:
predictions: list of predictions to score.
Each translation should be tokenized into a list of tokens.
references: list of lists of references for each translation.
Each reference should be tokenized into a list of tokens.
bleu_kwargs: optional dict of keywords to be passed when computing 'bleu'.
Keywords include Dict can be one of 'smooth_method', 'smooth_value', 'force', 'lowercase',
'tokenize', 'use_effective_order'.
wer_kwargs: optional dict of keywords to be passed when computing 'wer' and 'cer'.
Keywords include 'concatenate_texts'.
Returns: depending on the XTREME-S task, one or several of:
"accuracy": Accuracy - for 'fleurs-lang_id', 'minds14'
"f1": F1 score - for 'minds14'
"wer": Word error rate - for 'mls', 'fleurs-asr', 'voxpopuli', 'babel'
"cer": Character error rate - for 'mls', 'fleurs-asr', 'voxpopuli', 'babel'
"bleu": BLEU score according to the `sacrebleu` metric - for 'covost2'
Examples:
>>> xtreme_s_metric = datasets.load_metric('xtreme_s', 'mls') # 'mls', 'voxpopuli', 'fleurs-asr' or 'babel'
>>> references = ["it is sunny here", "paper and pen are essentials"]
>>> predictions = ["it's sunny", "paper pen are essential"]
>>> results = xtreme_s_metric.compute(predictions=predictions, references=references)
>>> print({k: round(v, 2) for k, v in results.items()})
{'wer': 0.56, 'cer': 0.27}
>>> xtreme_s_metric = datasets.load_metric('xtreme_s', 'covost2')
>>> references = ["bonjour paris", "il est necessaire de faire du sport de temps en temp"]
>>> predictions = ["bonjour paris", "il est important de faire du sport souvent"]
>>> results = xtreme_s_metric.compute(predictions=predictions, references=references)
>>> print({k: round(v, 2) for k, v in results.items()})
{'bleu': 31.65}
>>> xtreme_s_metric = datasets.load_metric('xtreme_s', 'fleurs-lang_id')
>>> references = [0, 1, 0, 0, 1]
>>> predictions = [0, 1, 1, 0, 0]
>>> results = xtreme_s_metric.compute(predictions=predictions, references=references)
>>> print({k: round(v, 2) for k, v in results.items()})
{'accuracy': 0.6}
>>> xtreme_s_metric = datasets.load_metric('xtreme_s', 'minds14')
>>> references = [0, 1, 0, 0, 1]
>>> predictions = [0, 1, 1, 0, 0]
>>> results = xtreme_s_metric.compute(predictions=predictions, references=references)
>>> print({k: round(v, 2) for k, v in results.items()})
{'f1': 0.58, 'accuracy': 0.6}
"""
_CONFIG_NAMES = ["fleurs-asr", "mls", "voxpopuli", "babel", "covost2", "fleurs-lang_id", "minds14"]
SENTENCE_DELIMITER = ""
try:
from jiwer import transforms as tr
_jiwer_available = True
except ImportError:
_jiwer_available = False
if _jiwer_available and version.parse(importlib_metadata.version("jiwer")) < version.parse("2.3.0"):
class SentencesToListOfCharacters(tr.AbstractTransform):
def __init__(self, sentence_delimiter: str = " "):
self.sentence_delimiter = sentence_delimiter
def process_string(self, s: str):
return list(s)
def process_list(self, inp: List[str]):
chars = []
for sent_idx, sentence in enumerate(inp):
chars.extend(self.process_string(sentence))
if self.sentence_delimiter is not None and self.sentence_delimiter != "" and sent_idx < len(inp) - 1:
chars.append(self.sentence_delimiter)
return chars
cer_transform = tr.Compose(
[tr.RemoveMultipleSpaces(), tr.Strip(), SentencesToListOfCharacters(SENTENCE_DELIMITER)]
)
elif _jiwer_available:
cer_transform = tr.Compose(
[
tr.RemoveMultipleSpaces(),
tr.Strip(),
tr.ReduceToSingleSentence(SENTENCE_DELIMITER),
tr.ReduceToListOfListOfChars(),
]
)
else:
cer_transform = None
def simple_accuracy(preds, labels):
return float((preds == labels).mean())
def f1_and_simple_accuracy(preds, labels):
return {
"f1": float(f1_score(y_true=labels, y_pred=preds, average="macro")),
"accuracy": simple_accuracy(preds, labels),
}
def bleu(
preds,
labels,
smooth_method="exp",
smooth_value=None,
force=False,
lowercase=False,
tokenize=None,
use_effective_order=False,
):
# xtreme-s can only have one label
labels = [[label] for label in labels]
preds = list(preds)
try:
import sacrebleu as scb
except ImportError:
raise ValueError(
"sacrebleu has to be installed in order to apply the bleu metric for covost2."
"You can install it via `pip install sacrebleu`."
)
if version.parse(scb.__version__) < version.parse("1.4.12"):
raise ImportWarning(
"To use `sacrebleu`, the module `sacrebleu>=1.4.12` is required, and the current version of `sacrebleu` doesn't match this condition.\n"
'You can install it with `pip install "sacrebleu>=1.4.12"`.'
)
references_per_prediction = len(labels[0])
if any(len(refs) != references_per_prediction for refs in labels):
raise ValueError("Sacrebleu requires the same number of references for each prediction")
transformed_references = [[refs[i] for refs in labels] for i in range(references_per_prediction)]
output = scb.corpus_bleu(
preds,
transformed_references,
smooth_method=smooth_method,
smooth_value=smooth_value,
force=force,
lowercase=lowercase,
use_effective_order=use_effective_order,
**({"tokenize": tokenize} if tokenize else {}),
)
return {"bleu": output.score}
def wer_and_cer(preds, labels, concatenate_texts, config_name):
try:
from jiwer import compute_measures
except ImportError:
raise ValueError(
f"jiwer has to be installed in order to apply the wer metric for {config_name}."
"You can install it via `pip install jiwer`."
)
if concatenate_texts:
wer = compute_measures(labels, preds)["wer"]
cer = compute_measures(labels, preds, truth_transform=cer_transform, hypothesis_transform=cer_transform)["wer"]
return {"wer": wer, "cer": cer}
else:
def compute_score(preds, labels, score_type="wer"):
incorrect = 0
total = 0
for prediction, reference in zip(preds, labels):
if score_type == "wer":
measures = compute_measures(reference, prediction)
elif score_type == "cer":
measures = compute_measures(
reference, prediction, truth_transform=cer_transform, hypothesis_transform=cer_transform
)
incorrect += measures["substitutions"] + measures["deletions"] + measures["insertions"]
total += measures["substitutions"] + measures["deletions"] + measures["hits"]
return incorrect / total
return {"wer": compute_score(preds, labels, "wer"), "cer": compute_score(preds, labels, "cer")}
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class XtremeS(datasets.Metric):
def _info(self):
if self.config_name not in _CONFIG_NAMES:
raise KeyError(f"You should supply a configuration name selected in {_CONFIG_NAMES}")
pred_type = "int64" if self.config_name in ["fleurs-lang_id", "minds14"] else "string"
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{"predictions": datasets.Value(pred_type), "references": datasets.Value(pred_type)}
),
codebase_urls=[],
reference_urls=[],
format="numpy",
)
def _compute(self, predictions, references, bleu_kwargs=None, wer_kwargs=None):
bleu_kwargs = bleu_kwargs if bleu_kwargs is not None else {}
wer_kwargs = wer_kwargs if wer_kwargs is not None else {}
if self.config_name == "fleurs-lang_id":
return {"accuracy": simple_accuracy(predictions, references)}
elif self.config_name == "minds14":
return f1_and_simple_accuracy(predictions, references)
elif self.config_name == "covost2":
smooth_method = bleu_kwargs.pop("smooth_method", "exp")
smooth_value = bleu_kwargs.pop("smooth_value", None)
force = bleu_kwargs.pop("force", False)
lowercase = bleu_kwargs.pop("lowercase", False)
tokenize = bleu_kwargs.pop("tokenize", None)
use_effective_order = bleu_kwargs.pop("use_effective_order", False)
return bleu(
preds=predictions,
labels=references,
smooth_method=smooth_method,
smooth_value=smooth_value,
force=force,
lowercase=lowercase,
tokenize=tokenize,
use_effective_order=use_effective_order,
)
elif self.config_name in ["fleurs-asr", "mls", "voxpopuli", "babel"]:
concatenate_texts = wer_kwargs.pop("concatenate_texts", False)
return wer_and_cer(predictions, references, concatenate_texts, self.config_name)
else:
raise KeyError(f"You should supply a configuration name selected in {_CONFIG_NAMES}")
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/roc_auc/README.md | # Metric Card for ROC AUC
## Metric Description
This metric computes the area under the curve (AUC) for the Receiver Operating Characteristic Curve (ROC). The return values represent how well the model used is predicting the correct classes, based on the input data. A score of `0.5` means that the model is predicting exactly at chance, i.e. the model's predictions are correct at the same rate as if the predictions were being decided by the flip of a fair coin or the roll of a fair die. A score above `0.5` indicates that the model is doing better than chance, while a score below `0.5` indicates that the model is doing worse than chance.
This metric has three separate use cases:
- **binary**: The case in which there are only two different label classes, and each example gets only one label. This is the default implementation.
- **multiclass**: The case in which there can be more than two different label classes, but each example still gets only one label.
- **multilabel**: The case in which there can be more than two different label classes, and each example can have more than one label.
## How to Use
At minimum, this metric requires references and prediction scores:
```python
>>> roc_auc_score = datasets.load_metric("roc_auc")
>>> refs = [1, 0, 1, 1, 0, 0]
>>> pred_scores = [0.5, 0.2, 0.99, 0.3, 0.1, 0.7]
>>> results = roc_auc_score.compute(references=refs, prediction_scores=pred_scores)
>>> print(round(results['roc_auc'], 2))
0.78
```
The default implementation of this metric is the **binary** implementation. If employing the **multiclass** or **multilabel** use cases, the keyword `"multiclass"` or `"multilabel"` must be specified when loading the metric:
- In the **multiclass** case, the metric is loaded with:
```python
>>> roc_auc_score = datasets.load_metric("roc_auc", "multiclass")
```
- In the **multilabel** case, the metric is loaded with:
```python
>>> roc_auc_score = datasets.load_metric("roc_auc", "multilabel")
```
See the [Examples Section Below](#examples_section) for more extensive examples.
### Inputs
- **`references`** (array-like of shape (n_samples,) or (n_samples, n_classes)): Ground truth labels. Expects different inputs based on use case:
- binary: expects an array-like of shape (n_samples,)
- multiclass: expects an array-like of shape (n_samples,)
- multilabel: expects an array-like of shape (n_samples, n_classes)
- **`prediction_scores`** (array-like of shape (n_samples,) or (n_samples, n_classes)): Model predictions. Expects different inputs based on use case:
- binary: expects an array-like of shape (n_samples,)
- multiclass: expects an array-like of shape (n_samples, n_classes). The probability estimates must sum to 1 across the possible classes.
- multilabel: expects an array-like of shape (n_samples, n_classes)
- **`average`** (`str`): Type of average, and is ignored in the binary use case. Defaults to `'macro'`. Options are:
- `'micro'`: Calculates metrics globally by considering each element of the label indicator matrix as a label. Only works with the multilabel use case.
- `'macro'`: Calculate metrics for each label, and find their unweighted mean. This does not take label imbalance into account.
- `'weighted'`: Calculate metrics for each label, and find their average, weighted by support (i.e. the number of true instances for each label).
- `'samples'`: Calculate metrics for each instance, and find their average. Only works with the multilabel use case.
- `None`: No average is calculated, and scores for each class are returned. Only works with the multilabels use case.
- **`sample_weight`** (array-like of shape (n_samples,)): Sample weights. Defaults to None.
- **`max_fpr`** (`float`): If not None, the standardized partial AUC over the range [0, `max_fpr`] is returned. Must be greater than `0` and less than or equal to `1`. Defaults to `None`. Note: For the multiclass use case, `max_fpr` should be either `None` or `1.0` as ROC AUC partial computation is not currently supported for `multiclass`.
- **`multi_class`** (`str`): Only used for multiclass targets, in which case it is required. Determines the type of configuration to use. Options are:
- `'ovr'`: Stands for One-vs-rest. Computes the AUC of each class against the rest. This treats the multiclass case in the same way as the multilabel case. Sensitive to class imbalance even when `average == 'macro'`, because class imbalance affects the composition of each of the 'rest' groupings.
- `'ovo'`: Stands for One-vs-one. Computes the average AUC of all possible pairwise combinations of classes. Insensitive to class imbalance when `average == 'macro'`.
- **`labels`** (array-like of shape (n_classes,)): Only used for multiclass targets. List of labels that index the classes in `prediction_scores`. If `None`, the numerical or lexicographical order of the labels in `prediction_scores` is used. Defaults to `None`.
### Output Values
This metric returns a dict containing the `roc_auc` score. The score is a `float`, unless it is the multilabel case with `average=None`, in which case the score is a numpy `array` with entries of type `float`.
The output therefore generally takes the following format:
```python
{'roc_auc': 0.778}
```
In contrast, though, the output takes the following format in the multilabel case when `average=None`:
```python
{'roc_auc': array([0.83333333, 0.375, 0.94444444])}
```
ROC AUC scores can take on any value between `0` and `1`, inclusive.
#### Values from Popular Papers
### <a name="examples_section"></a>Examples
Example 1, the **binary** use case:
```python
>>> roc_auc_score = datasets.load_metric("roc_auc")
>>> refs = [1, 0, 1, 1, 0, 0]
>>> pred_scores = [0.5, 0.2, 0.99, 0.3, 0.1, 0.7]
>>> results = roc_auc_score.compute(references=refs, prediction_scores=pred_scores)
>>> print(round(results['roc_auc'], 2))
0.78
```
Example 2, the **multiclass** use case:
```python
>>> roc_auc_score = datasets.load_metric("roc_auc", "multiclass")
>>> refs = [1, 0, 1, 2, 2, 0]
>>> pred_scores = [[0.3, 0.5, 0.2],
... [0.7, 0.2, 0.1],
... [0.005, 0.99, 0.005],
... [0.2, 0.3, 0.5],
... [0.1, 0.1, 0.8],
... [0.1, 0.7, 0.2]]
>>> results = roc_auc_score.compute(references=refs,
... prediction_scores=pred_scores,
... multi_class='ovr')
>>> print(round(results['roc_auc'], 2))
0.85
```
Example 3, the **multilabel** use case:
```python
>>> roc_auc_score = datasets.load_metric("roc_auc", "multilabel")
>>> refs = [[1, 1, 0],
... [1, 1, 0],
... [0, 1, 0],
... [0, 0, 1],
... [0, 1, 1],
... [1, 0, 1]]
>>> pred_scores = [[0.3, 0.5, 0.2],
... [0.7, 0.2, 0.1],
... [0.005, 0.99, 0.005],
... [0.2, 0.3, 0.5],
... [0.1, 0.1, 0.8],
... [0.1, 0.7, 0.2]]
>>> results = roc_auc_score.compute(references=refs,
... prediction_scores=pred_scores,
... average=None)
>>> print([round(res, 2) for res in results['roc_auc'])
[0.83, 0.38, 0.94]
```
## Limitations and Bias
## Citation
```bibtex
@article{doi:10.1177/0272989X8900900307,
author = {Donna Katzman McClish},
title ={Analyzing a Portion of the ROC Curve},
journal = {Medical Decision Making},
volume = {9},
number = {3},
pages = {190-195},
year = {1989},
doi = {10.1177/0272989X8900900307},
note ={PMID: 2668680},
URL = {https://doi.org/10.1177/0272989X8900900307},
eprint = {https://doi.org/10.1177/0272989X8900900307}
}
```
```bibtex
@article{10.1023/A:1010920819831,
author = {Hand, David J. and Till, Robert J.},
title = {A Simple Generalisation of the Area Under the ROC Curve for Multiple Class Classification Problems},
year = {2001},
issue_date = {November 2001},
publisher = {Kluwer Academic Publishers},
address = {USA},
volume = {45},
number = {2},
issn = {0885-6125},
url = {https://doi.org/10.1023/A:1010920819831},
doi = {10.1023/A:1010920819831},
journal = {Mach. Learn.},
month = {oct},
pages = {171–186},
numpages = {16},
keywords = {Gini index, AUC, error rate, ROC curve, receiver operating characteristic}
}
```
```bibtex
@article{scikit-learn,
title={Scikit-learn: Machine Learning in {P}ython},
author={Pedregosa, F. and Varoquaux, G. and Gramfort, A. and Michel, V. and Thirion, B. and Grisel, O. and Blondel, M. and Prettenhofer, P. and Weiss, R. and Dubourg, V. and Vanderplas, J. and Passos, A. and Cournapeau, D. and Brucher, M. and Perrot, M. and Duchesnay, E.},
journal={Journal of Machine Learning Research},
volume={12},
pages={2825--2830},
year={2011}
}
```
## Further References
This implementation is a wrapper around the [Scikit-learn implementation](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.roc_auc_score.html). Much of the documentation here was adapted from their existing documentation, as well.
The [Guide to ROC and AUC](https://youtu.be/iCZJfO-7C5Q) video from the channel Data Science Bits is also very informative.
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/roc_auc/roc_auc.py | # Copyright 2020 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Accuracy metric."""
from sklearn.metrics import roc_auc_score
import datasets
_DESCRIPTION = """
This metric computes the area under the curve (AUC) for the Receiver Operating Characteristic Curve (ROC). The return values represent how well the model used is predicting the correct classes, based on the input data. A score of `0.5` means that the model is predicting exactly at chance, i.e. the model's predictions are correct at the same rate as if the predictions were being decided by the flip of a fair coin or the roll of a fair die. A score above `0.5` indicates that the model is doing better than chance, while a score below `0.5` indicates that the model is doing worse than chance.
This metric has three separate use cases:
- binary: The case in which there are only two different label classes, and each example gets only one label. This is the default implementation.
- multiclass: The case in which there can be more than two different label classes, but each example still gets only one label.
- multilabel: The case in which there can be more than two different label classes, and each example can have more than one label.
"""
_KWARGS_DESCRIPTION = """
Args:
- references (array-like of shape (n_samples,) or (n_samples, n_classes)): Ground truth labels. Expects different input based on use case:
- binary: expects an array-like of shape (n_samples,)
- multiclass: expects an array-like of shape (n_samples,)
- multilabel: expects an array-like of shape (n_samples, n_classes)
- prediction_scores (array-like of shape (n_samples,) or (n_samples, n_classes)): Model predictions. Expects different inputs based on use case:
- binary: expects an array-like of shape (n_samples,)
- multiclass: expects an array-like of shape (n_samples, n_classes)
- multilabel: expects an array-like of shape (n_samples, n_classes)
- average (`str`): Type of average, and is ignored in the binary use case. Defaults to 'macro'. Options are:
- `'micro'`: Calculates metrics globally by considering each element of the label indicator matrix as a label. Only works with the multilabel use case.
- `'macro'`: Calculate metrics for each label, and find their unweighted mean. This does not take label imbalance into account.
- `'weighted'`: Calculate metrics for each label, and find their average, weighted by support (i.e. the number of true instances for each label).
- `'samples'`: Calculate metrics for each instance, and find their average. Only works with the multilabel use case.
- `None`: No average is calculated, and scores for each class are returned. Only works with the multilabels use case.
- sample_weight (array-like of shape (n_samples,)): Sample weights. Defaults to None.
- max_fpr (`float`): If not None, the standardized partial AUC over the range [0, `max_fpr`] is returned. Must be greater than `0` and less than or equal to `1`. Defaults to `None`. Note: For the multiclass use case, `max_fpr` should be either `None` or `1.0` as ROC AUC partial computation is not currently supported for `multiclass`.
- multi_class (`str`): Only used for multiclass targets, where it is required. Determines the type of configuration to use. Options are:
- `'ovr'`: Stands for One-vs-rest. Computes the AUC of each class against the rest. This treats the multiclass case in the same way as the multilabel case. Sensitive to class imbalance even when `average == 'macro'`, because class imbalance affects the composition of each of the 'rest' groupings.
- `'ovo'`: Stands for One-vs-one. Computes the average AUC of all possible pairwise combinations of classes. Insensitive to class imbalance when `average == 'macro'`.
- labels (array-like of shape (n_classes,)): Only used for multiclass targets. List of labels that index the classes in
`prediction_scores`. If `None`, the numerical or lexicographical order of the labels in
`prediction_scores` is used. Defaults to `None`.
Returns:
roc_auc (`float` or array-like of shape (n_classes,)): Returns array if in multilabel use case and `average='None'`. Otherwise, returns `float`.
Examples:
Example 1:
>>> roc_auc_score = datasets.load_metric("roc_auc")
>>> refs = [1, 0, 1, 1, 0, 0]
>>> pred_scores = [0.5, 0.2, 0.99, 0.3, 0.1, 0.7]
>>> results = roc_auc_score.compute(references=refs, prediction_scores=pred_scores)
>>> print(round(results['roc_auc'], 2))
0.78
Example 2:
>>> roc_auc_score = datasets.load_metric("roc_auc", "multiclass")
>>> refs = [1, 0, 1, 2, 2, 0]
>>> pred_scores = [[0.3, 0.5, 0.2],
... [0.7, 0.2, 0.1],
... [0.005, 0.99, 0.005],
... [0.2, 0.3, 0.5],
... [0.1, 0.1, 0.8],
... [0.1, 0.7, 0.2]]
>>> results = roc_auc_score.compute(references=refs, prediction_scores=pred_scores, multi_class='ovr')
>>> print(round(results['roc_auc'], 2))
0.85
Example 3:
>>> roc_auc_score = datasets.load_metric("roc_auc", "multilabel")
>>> refs = [[1, 1, 0],
... [1, 1, 0],
... [0, 1, 0],
... [0, 0, 1],
... [0, 1, 1],
... [1, 0, 1]]
>>> pred_scores = [[0.3, 0.5, 0.2],
... [0.7, 0.2, 0.1],
... [0.005, 0.99, 0.005],
... [0.2, 0.3, 0.5],
... [0.1, 0.1, 0.8],
... [0.1, 0.7, 0.2]]
>>> results = roc_auc_score.compute(references=refs, prediction_scores=pred_scores, average=None)
>>> print([round(res, 2) for res in results['roc_auc']])
[0.83, 0.38, 0.94]
"""
_CITATION = """\
@article{doi:10.1177/0272989X8900900307,
author = {Donna Katzman McClish},
title ={Analyzing a Portion of the ROC Curve},
journal = {Medical Decision Making},
volume = {9},
number = {3},
pages = {190-195},
year = {1989},
doi = {10.1177/0272989X8900900307},
note ={PMID: 2668680},
URL = {https://doi.org/10.1177/0272989X8900900307},
eprint = {https://doi.org/10.1177/0272989X8900900307}
}
@article{10.1023/A:1010920819831,
author = {Hand, David J. and Till, Robert J.},
title = {A Simple Generalisation of the Area Under the ROC Curve for Multiple Class Classification Problems},
year = {2001},
issue_date = {November 2001},
publisher = {Kluwer Academic Publishers},
address = {USA},
volume = {45},
number = {2},
issn = {0885-6125},
url = {https://doi.org/10.1023/A:1010920819831},
doi = {10.1023/A:1010920819831},
journal = {Mach. Learn.},
month = {oct},
pages = {171–186},
numpages = {16},
keywords = {Gini index, AUC, error rate, ROC curve, receiver operating characteristic}
}
@article{scikit-learn,
title={Scikit-learn: Machine Learning in {P}ython},
author={Pedregosa, F. and Varoquaux, G. and Gramfort, A. and Michel, V. and Thirion, B. and Grisel, O. and Blondel, M. and Prettenhofer, P. and Weiss, R. and Dubourg, V. and Vanderplas, J. and Passos, A. and Cournapeau, D. and Brucher, M. and Perrot, M. and Duchesnay, E.},
journal={Journal of Machine Learning Research},
volume={12},
pages={2825--2830},
year={2011}
}
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class ROCAUC(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"prediction_scores": datasets.Sequence(datasets.Value("float")),
"references": datasets.Value("int32"),
}
if self.config_name == "multiclass"
else {
"references": datasets.Sequence(datasets.Value("int32")),
"prediction_scores": datasets.Sequence(datasets.Value("float")),
}
if self.config_name == "multilabel"
else {
"references": datasets.Value("int32"),
"prediction_scores": datasets.Value("float"),
}
),
reference_urls=["https://scikit-learn.org/stable/modules/generated/sklearn.metrics.roc_auc_score.html"],
)
def _compute(
self,
references,
prediction_scores,
average="macro",
sample_weight=None,
max_fpr=None,
multi_class="raise",
labels=None,
):
return {
"roc_auc": roc_auc_score(
references,
prediction_scores,
average=average,
sample_weight=sample_weight,
max_fpr=max_fpr,
multi_class=multi_class,
labels=labels,
)
}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/wer/README.md | # Metric Card for WER
## Metric description
Word error rate (WER) is a common metric of the performance of an automatic speech recognition (ASR) system.
The general difficulty of measuring the performance of ASR systems lies in the fact that the recognized word sequence can have a different length from the reference word sequence (supposedly the correct one). The WER is derived from the [Levenshtein distance](https://en.wikipedia.org/wiki/Levenshtein_distance), working at the word level.
This problem is solved by first aligning the recognized word sequence with the reference (spoken) word sequence using dynamic string alignment. Examination of this issue is seen through a theory called the power law that states the correlation between [perplexity](https://huggingface.co/metrics/perplexity) and word error rate (see [this article](https://www.cs.cmu.edu/~roni/papers/eval-metrics-bntuw-9802.pdf) for further information).
Word error rate can then be computed as:
`WER = (S + D + I) / N = (S + D + I) / (S + D + C)`
where
`S` is the number of substitutions,
`D` is the number of deletions,
`I` is the number of insertions,
`C` is the number of correct words,
`N` is the number of words in the reference (`N=S+D+C`).
## How to use
The metric takes two inputs: references (a list of references for each speech input) and predictions (a list of transcriptions to score).
```python
from datasets import load_metric
wer = load_metric("wer")
wer_score = wer.compute(predictions=predictions, references=references)
```
## Output values
This metric outputs a float representing the word error rate.
```
print(wer_score)
0.5
```
This value indicates the average number of errors per reference word.
The **lower** the value, the **better** the performance of the ASR system, with a WER of 0 being a perfect score.
### Values from popular papers
This metric is highly dependent on the content and quality of the dataset, and therefore users can expect very different values for the same model but on different datasets.
For example, datasets such as [LibriSpeech](https://huggingface.co/datasets/librispeech_asr) report a WER in the 1.8-3.3 range, whereas ASR models evaluated on [Timit](https://huggingface.co/datasets/timit_asr) report a WER in the 8.3-20.4 range.
See the leaderboards for [LibriSpeech](https://paperswithcode.com/sota/speech-recognition-on-librispeech-test-clean) and [Timit](https://paperswithcode.com/sota/speech-recognition-on-timit) for the most recent values.
## Examples
Perfect match between prediction and reference:
```python
from datasets import load_metric
wer = load_metric("wer")
predictions = ["hello world", "good night moon"]
references = ["hello world", "good night moon"]
wer_score = wer.compute(predictions=predictions, references=references)
print(wer_score)
0.0
```
Partial match between prediction and reference:
```python
from datasets import load_metric
wer = load_metric("wer")
predictions = ["this is the prediction", "there is an other sample"]
references = ["this is the reference", "there is another one"]
wer_score = wer.compute(predictions=predictions, references=references)
print(wer_score)
0.5
```
No match between prediction and reference:
```python
from datasets import load_metric
wer = load_metric("wer")
predictions = ["hello world", "good night moon"]
references = ["hi everyone", "have a great day"]
wer_score = wer.compute(predictions=predictions, references=references)
print(wer_score)
1.0
```
## Limitations and bias
WER is a valuable tool for comparing different systems as well as for evaluating improvements within one system. This kind of measurement, however, provides no details on the nature of translation errors and further work is therefore required to identify the main source(s) of error and to focus any research effort.
## Citation
```bibtex
@inproceedings{woodard1982,
author = {Woodard, J.P. and Nelson, J.T.,
year = {1982},
journal = Ẅorkshop on standardisation for speech I/O technology, Naval Air Development Center, Warminster, PA},
title = {An information theoretic measure of speech recognition performance}
}
```
```bibtex
@inproceedings{morris2004,
author = {Morris, Andrew and Maier, Viktoria and Green, Phil},
year = {2004},
month = {01},
pages = {},
title = {From WER and RIL to MER and WIL: improved evaluation measures for connected speech recognition.}
}
```
## Further References
- [Word Error Rate -- Wikipedia](https://en.wikipedia.org/wiki/Word_error_rate)
- [Hugging Face Tasks -- Automatic Speech Recognition](https://huggingface.co/tasks/automatic-speech-recognition)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/wer/wer.py | # Copyright 2021 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" Word Error Ratio (WER) metric. """
from jiwer import compute_measures
import datasets
_CITATION = """\
@inproceedings{inproceedings,
author = {Morris, Andrew and Maier, Viktoria and Green, Phil},
year = {2004},
month = {01},
pages = {},
title = {From WER and RIL to MER and WIL: improved evaluation measures for connected speech recognition.}
}
"""
_DESCRIPTION = """\
Word error rate (WER) is a common metric of the performance of an automatic speech recognition system.
The general difficulty of measuring performance lies in the fact that the recognized word sequence can have a different length from the reference word sequence (supposedly the correct one). The WER is derived from the Levenshtein distance, working at the word level instead of the phoneme level. The WER is a valuable tool for comparing different systems as well as for evaluating improvements within one system. This kind of measurement, however, provides no details on the nature of translation errors and further work is therefore required to identify the main source(s) of error and to focus any research effort.
This problem is solved by first aligning the recognized word sequence with the reference (spoken) word sequence using dynamic string alignment. Examination of this issue is seen through a theory called the power law that states the correlation between perplexity and word error rate.
Word error rate can then be computed as:
WER = (S + D + I) / N = (S + D + I) / (S + D + C)
where
S is the number of substitutions,
D is the number of deletions,
I is the number of insertions,
C is the number of correct words,
N is the number of words in the reference (N=S+D+C).
This value indicates the average number of errors per reference word. The lower the value, the better the
performance of the ASR system with a WER of 0 being a perfect score.
"""
_KWARGS_DESCRIPTION = """
Compute WER score of transcribed segments against references.
Args:
references: List of references for each speech input.
predictions: List of transcriptions to score.
concatenate_texts (bool, default=False): Whether to concatenate all input texts or compute WER iteratively.
Returns:
(float): the word error rate
Examples:
>>> predictions = ["this is the prediction", "there is an other sample"]
>>> references = ["this is the reference", "there is another one"]
>>> wer = datasets.load_metric("wer")
>>> wer_score = wer.compute(predictions=predictions, references=references)
>>> print(wer_score)
0.5
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class WER(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Value("string", id="sequence"),
"references": datasets.Value("string", id="sequence"),
}
),
codebase_urls=["https://github.com/jitsi/jiwer/"],
reference_urls=[
"https://en.wikipedia.org/wiki/Word_error_rate",
],
)
def _compute(self, predictions=None, references=None, concatenate_texts=False):
if concatenate_texts:
return compute_measures(references, predictions)["wer"]
else:
incorrect = 0
total = 0
for prediction, reference in zip(predictions, references):
measures = compute_measures(reference, prediction)
incorrect += measures["substitutions"] + measures["deletions"] + measures["insertions"]
total += measures["substitutions"] + measures["deletions"] + measures["hits"]
return incorrect / total
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/cuad/README.md | # Metric Card for CUAD
## Metric description
This metric wraps the official scoring script for version 1 of the [Contract Understanding Atticus Dataset (CUAD)](https://huggingface.co/datasets/cuad), which is a corpus of more than 13,000 labels in 510 commercial legal contracts that have been manually labeled to identify 41 categories of important clauses that lawyers look for when reviewing contracts in connection with corporate transactions.
The CUAD metric computes several scores: [Exact Match](https://huggingface.co/metrics/exact_match), [F1 score](https://huggingface.co/metrics/f1), Area Under the Precision-Recall Curve, [Precision](https://huggingface.co/metrics/precision) at 80% [recall](https://huggingface.co/metrics/recall) and Precision at 90% recall.
## How to use
The CUAD metric takes two inputs :
`predictions`, a list of question-answer dictionaries with the following key-values:
- `id`: the id of the question-answer pair as given in the references.
- `prediction_text`: a list of possible texts for the answer, as a list of strings depending on a threshold on the confidence probability of each prediction.
`references`: a list of question-answer dictionaries with the following key-values:
- `id`: the id of the question-answer pair (the same as above).
- `answers`: a dictionary *in the CUAD dataset format* with the following keys:
- `text`: a list of possible texts for the answer, as a list of strings.
- `answer_start`: a list of start positions for the answer, as a list of ints.
Note that `answer_start` values are not taken into account to compute the metric.
```python
from datasets import load_metric
cuad_metric = load_metric("cuad")
predictions = [{'prediction_text': ['The seller:', 'The buyer/End-User: Shenzhen LOHAS Supply Chain Management Co., Ltd.'], 'id': 'LohaCompanyltd_20191209_F-1_EX-10.16_11917878_EX-10.16_Supply Agreement__Parties'}]
references = [{'answers': {'answer_start': [143, 49], 'text': ['The seller:', 'The buyer/End-User: Shenzhen LOHAS Supply Chain Management Co., Ltd.']}, 'id': 'LohaCompanyltd_20191209_F-1_EX-10.16_11917878_EX-10.16_Supply Agreement__Parties'}]
results = cuad_metric.compute(predictions=predictions, references=references)
```
## Output values
The output of the CUAD metric consists of a dictionary that contains one or several of the following metrics:
`exact_match`: The normalized answers that exactly match the reference answer, with a range between 0.0 and 1.0 (see [exact match](https://huggingface.co/metrics/exact_match) for more information).
`f1`: The harmonic mean of the precision and recall (see [F1 score](https://huggingface.co/metrics/f1) for more information). Its range is between 0.0 and 1.0 -- its lowest possible value is 0, if either the precision or the recall is 0, and its highest possible value is 1.0, which means perfect precision and recall.
`aupr`: The Area Under the Precision-Recall curve, with a range between 0.0 and 1.0, with a higher value representing both high recall and high precision, and a low value representing low values for both. See the [Wikipedia article](https://en.wikipedia.org/wiki/Receiver_operating_characteristic#Area_under_the_curve) for more information.
`prec_at_80_recall`: The fraction of true examples among the predicted examples at a recall rate of 80%. Its range is between 0.0 and 1.0. For more information, see [precision](https://huggingface.co/metrics/precision) and [recall](https://huggingface.co/metrics/recall).
`prec_at_90_recall`: The fraction of true examples among the predicted examples at a recall rate of 90%. Its range is between 0.0 and 1.0.
### Values from popular papers
The [original CUAD paper](https://arxiv.org/pdf/2103.06268.pdf) reports that a [DeBERTa model](https://huggingface.co/microsoft/deberta-base) attains
an AUPR of 47.8%, a Precision at 80% Recall of 44.0%, and a Precision at 90% Recall of 17.8% (they do not report F1 or Exact Match separately).
For more recent model performance, see the [dataset leaderboard](https://paperswithcode.com/dataset/cuad).
## Examples
Maximal values :
```python
from datasets import load_metric
cuad_metric = load_metric("cuad")
predictions = [{'prediction_text': ['The seller:', 'The buyer/End-User: Shenzhen LOHAS Supply Chain Management Co., Ltd.'], 'id': 'LohaCompanyltd_20191209_F-1_EX-10.16_11917878_EX-10.16_Supply Agreement__Parties'}]
references = [{'answers': {'answer_start': [143, 49], 'text': ['The seller:', 'The buyer/End-User: Shenzhen LOHAS Supply Chain Management Co., Ltd.']}, 'id': 'LohaCompanyltd_20191209_F-1_EX-10.16_11917878_EX-10.16_Supply Agreement__Parties'}]
results = cuad_metric.compute(predictions=predictions, references=references)
print(results)
{'exact_match': 100.0, 'f1': 100.0, 'aupr': 0.0, 'prec_at_80_recall': 1.0, 'prec_at_90_recall': 1.0}
```
Minimal values:
```python
from datasets import load_metric
cuad_metric = load_metric("cuad")
predictions = [{'prediction_text': ['The Company appoints the Distributor as an exclusive distributor of Products in the Market, subject to the terms and conditions of this Agreement.'], 'id': 'LIMEENERGYCO_09_09_1999-EX-10-DISTRIBUTOR AGREEMENT__Exclusivity_0'}]
references = [{'answers': {'answer_start': [143], 'text': 'The seller'}, 'id': 'LIMEENERGYCO_09_09_1999-EX-10-DISTRIBUTOR AGREEMENT__Exclusivity_0'}]
results = cuad_metric.compute(predictions=predictions, references=references)
print(results)
{'exact_match': 0.0, 'f1': 0.0, 'aupr': 0.0, 'prec_at_80_recall': 0, 'prec_at_90_recall': 0}
```
Partial match:
```python
from datasets import load_metric
cuad_metric = load_metric("cuad")
predictions = [{'prediction_text': ['The seller:', 'The buyer/End-User: Shenzhen LOHAS Supply Chain Management Co., Ltd.'], 'id': 'LohaCompanyltd_20191209_F-1_EX-10.16_11917878_EX-10.16_Supply Agreement__Parties'}]
predictions = [{'prediction_text': ['The Company appoints the Distributor as an exclusive distributor of Products in the Market, subject to the terms and conditions of this Agreement.', 'The buyer/End-User: Shenzhen LOHAS Supply Chain Management Co., Ltd.'], 'id': 'LohaCompanyltd_20191209_F-1_EX-10.16_11917878_EX-10.16_Supply Agreement__Parties'}]
results = cuad_metric.compute(predictions=predictions, references=references)
print(results)
{'exact_match': 100.0, 'f1': 50.0, 'aupr': 0.0, 'prec_at_80_recall': 0, 'prec_at_90_recall': 0}
```
## Limitations and bias
This metric works only with datasets that have the same format as the [CUAD dataset](https://huggingface.co/datasets/cuad). The limitations of the biases of this dataset are not discussed, but could exhibit annotation bias given the homogeneity of annotators for this dataset.
In terms of the metric itself, the accuracy of AUPR has been debated because its estimates are quite noisy and because of the fact that reducing the Precision-Recall Curve to a single number ignores the fact that it is about the tradeoffs between the different systems or performance points plotted and not the performance of an individual system. Reporting the original F1 and exact match scores is therefore useful to ensure a more complete representation of system performance.
## Citation
```bibtex
@article{hendrycks2021cuad,
title={CUAD: An Expert-Annotated NLP Dataset for Legal Contract Review},
author={Dan Hendrycks and Collin Burns and Anya Chen and Spencer Ball},
journal={arXiv preprint arXiv:2103.06268},
year={2021}
}
```
## Further References
- [CUAD dataset homepage](https://www.atticusprojectai.org/cuad-v1-performance-announcements)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/cuad/cuad.py | # Copyright 2020 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" CUAD metric. """
import datasets
from .evaluate import evaluate
_CITATION = """\
@article{hendrycks2021cuad,
title={CUAD: An Expert-Annotated NLP Dataset for Legal Contract Review},
author={Dan Hendrycks and Collin Burns and Anya Chen and Spencer Ball},
journal={arXiv preprint arXiv:2103.06268},
year={2021}
}
"""
_DESCRIPTION = """
This metric wrap the official scoring script for version 1 of the Contract
Understanding Atticus Dataset (CUAD).
Contract Understanding Atticus Dataset (CUAD) v1 is a corpus of more than 13,000 labels in 510
commercial legal contracts that have been manually labeled to identify 41 categories of important
clauses that lawyers look for when reviewing contracts in connection with corporate transactions.
"""
_KWARGS_DESCRIPTION = """
Computes CUAD scores (EM, F1, AUPR, Precision@80%Recall, and Precision@90%Recall).
Args:
predictions: List of question-answers dictionaries with the following key-values:
- 'id': id of the question-answer pair as given in the references (see below)
- 'prediction_text': list of possible texts for the answer, as a list of strings
depending on a threshold on the confidence probability of each prediction.
references: List of question-answers dictionaries with the following key-values:
- 'id': id of the question-answer pair (see above),
- 'answers': a Dict in the CUAD dataset format
{
'text': list of possible texts for the answer, as a list of strings
'answer_start': list of start positions for the answer, as a list of ints
}
Note that answer_start values are not taken into account to compute the metric.
Returns:
'exact_match': Exact match (the normalized answer exactly match the gold answer)
'f1': The F-score of predicted tokens versus the gold answer
'aupr': Area Under the Precision-Recall curve
'prec_at_80_recall': Precision at 80% recall
'prec_at_90_recall': Precision at 90% recall
Examples:
>>> predictions = [{'prediction_text': ['The seller:', 'The buyer/End-User: Shenzhen LOHAS Supply Chain Management Co., Ltd.'], 'id': 'LohaCompanyltd_20191209_F-1_EX-10.16_11917878_EX-10.16_Supply Agreement__Parties'}]
>>> references = [{'answers': {'answer_start': [143, 49], 'text': ['The seller:', 'The buyer/End-User: Shenzhen LOHAS Supply Chain Management Co., Ltd.']}, 'id': 'LohaCompanyltd_20191209_F-1_EX-10.16_11917878_EX-10.16_Supply Agreement__Parties'}]
>>> cuad_metric = datasets.load_metric("cuad")
>>> results = cuad_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'exact_match': 100.0, 'f1': 100.0, 'aupr': 0.0, 'prec_at_80_recall': 1.0, 'prec_at_90_recall': 1.0}
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class CUAD(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": {
"id": datasets.Value("string"),
"prediction_text": datasets.features.Sequence(datasets.Value("string")),
},
"references": {
"id": datasets.Value("string"),
"answers": datasets.features.Sequence(
{
"text": datasets.Value("string"),
"answer_start": datasets.Value("int32"),
}
),
},
}
),
codebase_urls=["https://www.atticusprojectai.org/cuad"],
reference_urls=["https://www.atticusprojectai.org/cuad"],
)
def _compute(self, predictions, references):
pred_dict = {prediction["id"]: prediction["prediction_text"] for prediction in predictions}
dataset = [
{
"paragraphs": [
{
"qas": [
{
"answers": [{"text": answer_text} for answer_text in ref["answers"]["text"]],
"id": ref["id"],
}
for ref in references
]
}
]
}
]
score = evaluate(dataset=dataset, predictions=pred_dict)
return score
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/cuad/evaluate.py | """ Official evaluation script for CUAD dataset. """
import argparse
import json
import re
import string
import sys
import numpy as np
IOU_THRESH = 0.5
def get_jaccard(prediction, ground_truth):
remove_tokens = [".", ",", ";", ":"]
for token in remove_tokens:
ground_truth = ground_truth.replace(token, "")
prediction = prediction.replace(token, "")
ground_truth, prediction = ground_truth.lower(), prediction.lower()
ground_truth, prediction = ground_truth.replace("/", " "), prediction.replace("/", " ")
ground_truth, prediction = set(ground_truth.split(" ")), set(prediction.split(" "))
intersection = ground_truth.intersection(prediction)
union = ground_truth.union(prediction)
jaccard = len(intersection) / len(union)
return jaccard
def normalize_answer(s):
"""Lower text and remove punctuation, articles and extra whitespace."""
def remove_articles(text):
return re.sub(r"\b(a|an|the)\b", " ", text)
def white_space_fix(text):
return " ".join(text.split())
def remove_punc(text):
exclude = set(string.punctuation)
return "".join(ch for ch in text if ch not in exclude)
def lower(text):
return text.lower()
return white_space_fix(remove_articles(remove_punc(lower(s))))
def compute_precision_recall(predictions, ground_truths, qa_id):
tp, fp, fn = 0, 0, 0
substr_ok = "Parties" in qa_id
# first check if ground truth is empty
if len(ground_truths) == 0:
if len(predictions) > 0:
fp += len(predictions) # false positive for each one
else:
for ground_truth in ground_truths:
assert len(ground_truth) > 0
# check if there is a match
match_found = False
for pred in predictions:
if substr_ok:
is_match = get_jaccard(pred, ground_truth) >= IOU_THRESH or ground_truth in pred
else:
is_match = get_jaccard(pred, ground_truth) >= IOU_THRESH
if is_match:
match_found = True
if match_found:
tp += 1
else:
fn += 1
# now also get any fps by looping through preds
for pred in predictions:
# Check if there's a match. if so, don't count (don't want to double count based on the above)
# but if there's no match, then this is a false positive.
# (Note: we get the true positives in the above loop instead of this loop so that we don't double count
# multiple predictions that are matched with the same answer.)
match_found = False
for ground_truth in ground_truths:
assert len(ground_truth) > 0
if substr_ok:
is_match = get_jaccard(pred, ground_truth) >= IOU_THRESH or ground_truth in pred
else:
is_match = get_jaccard(pred, ground_truth) >= IOU_THRESH
if is_match:
match_found = True
if not match_found:
fp += 1
precision = tp / (tp + fp) if tp + fp > 0 else np.nan
recall = tp / (tp + fn) if tp + fn > 0 else np.nan
return precision, recall
def process_precisions(precisions):
"""
Processes precisions to ensure that precision and recall don't both get worse.
Assumes the list precision is sorted in order of recalls
"""
precision_best = precisions[::-1]
for i in range(1, len(precision_best)):
precision_best[i] = max(precision_best[i - 1], precision_best[i])
precisions = precision_best[::-1]
return precisions
def get_aupr(precisions, recalls):
processed_precisions = process_precisions(precisions)
aupr = np.trapz(processed_precisions, recalls)
if np.isnan(aupr):
return 0
return aupr
def get_prec_at_recall(precisions, recalls, recall_thresh):
"""Assumes recalls are sorted in increasing order"""
processed_precisions = process_precisions(precisions)
prec_at_recall = 0
for prec, recall in zip(processed_precisions, recalls):
if recall >= recall_thresh:
prec_at_recall = prec
break
return prec_at_recall
def exact_match_score(prediction, ground_truth):
return normalize_answer(prediction) == normalize_answer(ground_truth)
def metric_max_over_ground_truths(metric_fn, predictions, ground_truths):
score = 0
for pred in predictions:
for ground_truth in ground_truths:
score = metric_fn(pred, ground_truth)
if score == 1: # break the loop when one prediction matches the ground truth
break
if score == 1:
break
return score
def evaluate(dataset, predictions):
f1 = exact_match = total = 0
precisions = []
recalls = []
for article in dataset:
for paragraph in article["paragraphs"]:
for qa in paragraph["qas"]:
total += 1
if qa["id"] not in predictions:
message = "Unanswered question " + qa["id"] + " will receive score 0."
print(message, file=sys.stderr)
continue
ground_truths = [x["text"] for x in qa["answers"]]
prediction = predictions[qa["id"]]
precision, recall = compute_precision_recall(prediction, ground_truths, qa["id"])
precisions.append(precision)
recalls.append(recall)
if precision == 0 and recall == 0:
f1 += 0
else:
f1 += 2 * (precision * recall) / (precision + recall)
exact_match += metric_max_over_ground_truths(exact_match_score, prediction, ground_truths)
precisions = [x for _, x in sorted(zip(recalls, precisions))]
recalls.sort()
f1 = 100.0 * f1 / total
exact_match = 100.0 * exact_match / total
aupr = get_aupr(precisions, recalls)
prec_at_90_recall = get_prec_at_recall(precisions, recalls, recall_thresh=0.9)
prec_at_80_recall = get_prec_at_recall(precisions, recalls, recall_thresh=0.8)
return {
"exact_match": exact_match,
"f1": f1,
"aupr": aupr,
"prec_at_80_recall": prec_at_80_recall,
"prec_at_90_recall": prec_at_90_recall,
}
if __name__ == "__main__":
parser = argparse.ArgumentParser(description="Evaluation for CUAD")
parser.add_argument("dataset_file", help="Dataset file")
parser.add_argument("prediction_file", help="Prediction File")
args = parser.parse_args()
with open(args.dataset_file) as dataset_file:
dataset_json = json.load(dataset_file)
dataset = dataset_json["data"]
with open(args.prediction_file) as prediction_file:
predictions = json.load(prediction_file)
print(json.dumps(evaluate(dataset, predictions)))
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/perplexity/README.md | # Metric Card for Perplexity
## Metric Description
Given a model and an input text sequence, perplexity measures how likely the model is to generate the input text sequence. This can be used in two main ways:
1. to evaluate how well the model has learned the distribution of the text it was trained on
- In this case, the model input should be the trained model to be evaluated, and the input texts should be the text that the model was trained on.
2. to evaluate how well a selection of text matches the distribution of text that the input model was trained on
- In this case, the model input should be a trained model, and the input texts should be the text to be evaluated.
## Intended Uses
Any language generation task.
## How to Use
The metric takes a list of text as input, as well as the name of the model used to compute the metric:
```python
from datasets import load_metric
perplexity = load_metric("perplexity")
results = perplexity.compute(input_texts=input_texts, model_id='gpt2')
```
### Inputs
- **model_id** (str): model used for calculating Perplexity. NOTE: Perplexity can only be calculated for causal language models.
- This includes models such as gpt2, causal variations of bert, causal versions of t5, and more (the full list can be found in the AutoModelForCausalLM documentation here: https://huggingface.co/docs/transformers/master/en/model_doc/auto#transformers.AutoModelForCausalLM )
- **input_texts** (list of str): input text, each separate text snippet is one list entry.
- **batch_size** (int): the batch size to run texts through the model. Defaults to 16.
- **add_start_token** (bool): whether to add the start token to the texts, so the perplexity can include the probability of the first word. Defaults to True.
- **device** (str): device to run on, defaults to 'cuda' when available
### Output Values
This metric outputs a dictionary with the perplexity scores for the text input in the list, and the average perplexity.
If one of the input texts is longer than the max input length of the model, then it is truncated to the max length for the perplexity computation.
```
{'perplexities': [8.182524681091309, 33.42122268676758, 27.012239456176758], 'mean_perplexity': 22.871995608011883}
```
This metric's range is 0 and up. A lower score is better.
#### Values from Popular Papers
### Examples
Calculating perplexity on input_texts defined here:
```python
perplexity = datasets.load_metric("perplexity")
input_texts = ["lorem ipsum", "Happy Birthday!", "Bienvenue"]
results = perplexity.compute(model_id='gpt2',
add_start_token=False,
input_texts=input_texts)
print(list(results.keys()))
>>>['perplexities', 'mean_perplexity']
print(round(results["mean_perplexity"], 2))
>>>78.22
print(round(results["perplexities"][0], 2))
>>>11.11
```
Calculating perplexity on input_texts loaded in from a dataset:
```python
perplexity = datasets.load_metric("perplexity")
input_texts = datasets.load_dataset("wikitext",
"wikitext-2-raw-v1",
split="test")["text"][:50]
input_texts = [s for s in input_texts if s!='']
results = perplexity.compute(model_id='gpt2',
input_texts=input_texts)
print(list(results.keys()))
>>>['perplexities', 'mean_perplexity']
print(round(results["mean_perplexity"], 2))
>>>60.35
print(round(results["perplexities"][0], 2))
>>>81.12
```
## Limitations and Bias
Note that the output value is based heavily on what text the model was trained on. This means that perplexity scores are not comparable between models or datasets.
## Citation
```bibtex
@article{jelinek1977perplexity,
title={Perplexity—a measure of the difficulty of speech recognition tasks},
author={Jelinek, Fred and Mercer, Robert L and Bahl, Lalit R and Baker, James K},
journal={The Journal of the Acoustical Society of America},
volume={62},
number={S1},
pages={S63--S63},
year={1977},
publisher={Acoustical Society of America}
}
```
## Further References
- [Hugging Face Perplexity Blog Post](https://huggingface.co/docs/transformers/perplexity)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/perplexity/perplexity.py | # Copyright 2022 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Perplexity Metric."""
import numpy as np
import torch
from torch.nn import CrossEntropyLoss
from transformers import AutoModelForCausalLM, AutoTokenizer
import datasets
from datasets import logging
_CITATION = """\
"""
_DESCRIPTION = """
Perplexity (PPL) is one of the most common metrics for evaluating language models.
It is defined as the exponentiated average negative log-likelihood of a sequence.
For more information, see https://huggingface.co/docs/transformers/perplexity
"""
_KWARGS_DESCRIPTION = """
Args:
model_id (str): model used for calculating Perplexity
NOTE: Perplexity can only be calculated for causal language models.
This includes models such as gpt2, causal variations of bert,
causal versions of t5, and more (the full list can be found
in the AutoModelForCausalLM documentation here:
https://huggingface.co/docs/transformers/master/en/model_doc/auto#transformers.AutoModelForCausalLM )
input_texts (list of str): input text, each separate text snippet
is one list entry.
batch_size (int): the batch size to run texts through the model. Defaults to 16.
add_start_token (bool): whether to add the start token to the texts,
so the perplexity can include the probability of the first word. Defaults to True.
device (str): device to run on, defaults to 'cuda' when available
Returns:
perplexity: dictionary containing the perplexity scores for the texts
in the input list, as well as the mean perplexity. If one of the input texts is
longer than the max input length of the model, then it is truncated to the
max length for the perplexity computation.
Examples:
Example 1:
>>> perplexity = datasets.load_metric("perplexity")
>>> input_texts = ["lorem ipsum", "Happy Birthday!", "Bienvenue"]
>>> results = perplexity.compute(model_id='gpt2',
... add_start_token=False,
... input_texts=input_texts) # doctest:+ELLIPSIS
>>> print(list(results.keys()))
['perplexities', 'mean_perplexity']
>>> print(round(results["mean_perplexity"], 2))
78.22
>>> print(round(results["perplexities"][0], 2))
11.11
Example 2:
>>> perplexity = datasets.load_metric("perplexity")
>>> input_texts = datasets.load_dataset("wikitext",
... "wikitext-2-raw-v1",
... split="test")["text"][:50]
>>> input_texts = [s for s in input_texts if s!='']
>>> results = perplexity.compute(model_id='gpt2',
... input_texts=input_texts) # doctest:+ELLIPSIS
>>> print(list(results.keys()))
['perplexities', 'mean_perplexity']
>>> print(round(results["mean_perplexity"], 2))
60.35
>>> print(round(results["perplexities"][0], 2))
81.12
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Perplexity(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"input_texts": datasets.Value("string"),
}
),
reference_urls=["https://huggingface.co/docs/transformers/perplexity"],
)
def _compute(self, input_texts, model_id, batch_size: int = 16, add_start_token: bool = True, device=None):
if device is not None:
assert device in ["gpu", "cpu", "cuda"], "device should be either gpu or cpu."
if device == "gpu":
device = "cuda"
else:
device = "cuda" if torch.cuda.is_available() else "cpu"
model = AutoModelForCausalLM.from_pretrained(model_id)
model = model.to(device)
tokenizer = AutoTokenizer.from_pretrained(model_id)
# if batch_size > 1 (which generally leads to padding being required), and
# if there is not an already assigned pad_token, assign an existing
# special token to also be the padding token
if tokenizer.pad_token is None and batch_size > 1:
existing_special_tokens = list(tokenizer.special_tokens_map_extended.values())
# check that the model already has at least one special token defined
assert (
len(existing_special_tokens) > 0
), "If batch_size > 1, model must have at least one special token to use for padding. Please use a different model or set batch_size=1."
# assign one of the special tokens to also be the pad token
tokenizer.add_special_tokens({"pad_token": existing_special_tokens[0]})
if add_start_token:
# leave room for <BOS> token to be added:
assert (
tokenizer.bos_token is not None
), "Input model must already have a BOS token if using add_start_token=True. Please use a different model, or set add_start_token=False"
max_tokenized_len = model.config.max_length - 1
else:
max_tokenized_len = model.config.max_length
encodings = tokenizer(
input_texts,
add_special_tokens=False,
padding=True,
truncation=True,
max_length=max_tokenized_len,
return_tensors="pt",
return_attention_mask=True,
).to(device)
encoded_texts = encodings["input_ids"]
attn_masks = encodings["attention_mask"]
# check that each input is long enough:
if add_start_token:
assert torch.all(torch.ge(attn_masks.sum(1), 1)), "Each input text must be at least one token long."
else:
assert torch.all(
torch.ge(attn_masks.sum(1), 2)
), "When add_start_token=False, each input text must be at least two tokens long. Run with add_start_token=True if inputting strings of only one token, and remove all empty input strings."
ppls = []
loss_fct = CrossEntropyLoss(reduction="none")
for start_index in logging.tqdm(range(0, len(encoded_texts), batch_size)):
end_index = min(start_index + batch_size, len(encoded_texts))
encoded_batch = encoded_texts[start_index:end_index]
attn_mask = attn_masks[start_index:end_index]
if add_start_token:
bos_tokens_tensor = torch.tensor([[tokenizer.bos_token_id]] * encoded_batch.size(dim=0)).to(device)
encoded_batch = torch.cat([bos_tokens_tensor, encoded_batch], dim=1)
attn_mask = torch.cat(
[torch.ones(bos_tokens_tensor.size(), dtype=torch.int64).to(device), attn_mask], dim=1
)
labels = encoded_batch
with torch.no_grad():
out_logits = model(encoded_batch, attention_mask=attn_mask).logits
shift_logits = out_logits[..., :-1, :].contiguous()
shift_labels = labels[..., 1:].contiguous()
shift_attention_mask_batch = attn_mask[..., 1:].contiguous()
perplexity_batch = torch.exp2(
(loss_fct(shift_logits.transpose(1, 2), shift_labels) * shift_attention_mask_batch).sum(1)
/ shift_attention_mask_batch.sum(1)
)
ppls += perplexity_batch.tolist()
return {"perplexities": ppls, "mean_perplexity": np.mean(ppls)}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/xnli/README.md | # Metric Card for XNLI
## Metric description
The XNLI metric allows to evaluate a model's score on the [XNLI dataset](https://huggingface.co/datasets/xnli), which is a subset of a few thousand examples from the [MNLI dataset](https://huggingface.co/datasets/glue/viewer/mnli) that have been translated into a 14 different languages, some of which are relatively low resource such as Swahili and Urdu.
As with MNLI, the task is to predict textual entailment (does sentence A imply/contradict/neither sentence B) and is a classification task (given two sentences, predict one of three labels).
## How to use
The XNLI metric is computed based on the `predictions` (a list of predicted labels) and the `references` (a list of ground truth labels).
```python
from datasets import load_metric
xnli_metric = load_metric("xnli")
predictions = [0, 1]
references = [0, 1]
results = xnli_metric.compute(predictions=predictions, references=references)
```
## Output values
The output of the XNLI metric is simply the `accuracy`, i.e. the proportion of correct predictions among the total number of cases processed, with a range between 0 and 1 (see [accuracy](https://huggingface.co/metrics/accuracy) for more information).
### Values from popular papers
The [original XNLI paper](https://arxiv.org/pdf/1809.05053.pdf) reported accuracies ranging from 59.3 (for `ur`) to 73.7 (for `en`) for the BiLSTM-max model.
For more recent model performance, see the [dataset leaderboard](https://paperswithcode.com/dataset/xnli).
## Examples
Maximal values:
```python
>>> from datasets import load_metric
>>> xnli_metric = load_metric("xnli")
>>> predictions = [0, 1]
>>> references = [0, 1]
>>> results = xnli_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'accuracy': 1.0}
```
Minimal values:
```python
>>> from datasets import load_metric
>>> xnli_metric = load_metric("xnli")
>>> predictions = [1, 0]
>>> references = [0, 1]
>>> results = xnli_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'accuracy': 0.0}
```
Partial match:
```python
>>> from datasets import load_metric
>>> xnli_metric = load_metric("xnli")
>>> predictions = [1, 0, 1]
>>> references = [1, 0, 0]
>>> results = xnli_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'accuracy': 0.6666666666666666}
```
## Limitations and bias
While accuracy alone does give a certain indication of performance, it can be supplemented by error analysis and a better understanding of the model's mistakes on each of the categories represented in the dataset, especially if they are unbalanced.
While the XNLI dataset is multilingual and represents a diversity of languages, in reality, cross-lingual sentence understanding goes beyond translation, given that there are many cultural differences that have an impact on human sentiment annotations. Since the XNLI dataset was obtained by translation based on English sentences, it does not capture these cultural differences.
## Citation
```bibtex
@InProceedings{conneau2018xnli,
author = "Conneau, Alexis
and Rinott, Ruty
and Lample, Guillaume
and Williams, Adina
and Bowman, Samuel R.
and Schwenk, Holger
and Stoyanov, Veselin",
title = "XNLI: Evaluating Cross-lingual Sentence Representations",
booktitle = "Proceedings of the 2018 Conference on Empirical Methods
in Natural Language Processing",
year = "2018",
publisher = "Association for Computational Linguistics",
location = "Brussels, Belgium",
}
```
## Further References
- [XNI Dataset GitHub](https://github.com/facebookresearch/XNLI)
- [HuggingFace Tasks -- Text Classification](https://huggingface.co/tasks/text-classification)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/xnli/xnli.py | # Copyright 2020 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" XNLI benchmark metric. """
import datasets
_CITATION = """\
@InProceedings{conneau2018xnli,
author = "Conneau, Alexis
and Rinott, Ruty
and Lample, Guillaume
and Williams, Adina
and Bowman, Samuel R.
and Schwenk, Holger
and Stoyanov, Veselin",
title = "XNLI: Evaluating Cross-lingual Sentence Representations",
booktitle = "Proceedings of the 2018 Conference on Empirical Methods
in Natural Language Processing",
year = "2018",
publisher = "Association for Computational Linguistics",
location = "Brussels, Belgium",
}
"""
_DESCRIPTION = """\
XNLI is a subset of a few thousand examples from MNLI which has been translated
into a 14 different languages (some low-ish resource). As with MNLI, the goal is
to predict textual entailment (does sentence A imply/contradict/neither sentence
B) and is a classification task (given two sentences, predict one of three
labels).
"""
_KWARGS_DESCRIPTION = """
Computes XNLI score which is just simple accuracy.
Args:
predictions: Predicted labels.
references: Ground truth labels.
Returns:
'accuracy': accuracy
Examples:
>>> predictions = [0, 1]
>>> references = [0, 1]
>>> xnli_metric = datasets.load_metric("xnli")
>>> results = xnli_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'accuracy': 1.0}
"""
def simple_accuracy(preds, labels):
return (preds == labels).mean()
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Xnli(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Value("int64" if self.config_name != "sts-b" else "float32"),
"references": datasets.Value("int64" if self.config_name != "sts-b" else "float32"),
}
),
codebase_urls=[],
reference_urls=[],
format="numpy",
)
def _compute(self, predictions, references):
return {"accuracy": simple_accuracy(predictions, references)}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/rouge/README.md | # Metric Card for ROUGE
## Metric Description
ROUGE, or Recall-Oriented Understudy for Gisting Evaluation, is a set of metrics and a software package used for evaluating automatic summarization and machine translation software in natural language processing. The metrics compare an automatically produced summary or translation against a reference or a set of references (human-produced) summary or translation.
Note that ROUGE is case insensitive, meaning that upper case letters are treated the same way as lower case letters.
This metrics is a wrapper around the [Google Research reimplementation of ROUGE](https://github.com/google-research/google-research/tree/master/rouge)
## How to Use
At minimum, this metric takes as input a list of predictions and a list of references:
```python
>>> rouge = datasets.load_metric('rouge')
>>> predictions = ["hello there", "general kenobi"]
>>> references = ["hello there", "general kenobi"]
>>> results = rouge.compute(predictions=predictions,
... references=references)
>>> print(list(results.keys()))
['rouge1', 'rouge2', 'rougeL', 'rougeLsum']
>>> print(results["rouge1"])
AggregateScore(low=Score(precision=1.0, recall=1.0, fmeasure=1.0), mid=Score(precision=1.0, recall=1.0, fmeasure=1.0), high=Score(precision=1.0, recall=1.0, fmeasure=1.0))
>>> print(results["rouge1"].mid.fmeasure)
1.0
```
### Inputs
- **predictions** (`list`): list of predictions to score. Each prediction
should be a string with tokens separated by spaces.
- **references** (`list`): list of reference for each prediction. Each
reference should be a string with tokens separated by spaces.
- **rouge_types** (`list`): A list of rouge types to calculate. Defaults to `['rouge1', 'rouge2', 'rougeL', 'rougeLsum']`.
- Valid rouge types:
- `"rouge1"`: unigram (1-gram) based scoring
- `"rouge2"`: bigram (2-gram) based scoring
- `"rougeL"`: Longest common subsequence based scoring.
- `"rougeLSum"`: splits text using `"\n"`
- See [here](https://github.com/huggingface/datasets/issues/617) for more information
- **use_aggregator** (`boolean`): If True, returns aggregates. Defaults to `True`.
- **use_stemmer** (`boolean`): If `True`, uses Porter stemmer to strip word suffixes. Defaults to `False`.
### Output Values
The output is a dictionary with one entry for each rouge type in the input list `rouge_types`. If `use_aggregator=False`, each dictionary entry is a list of Score objects, with one score for each sentence. Each Score object includes the `precision`, `recall`, and `fmeasure`. E.g. if `rouge_types=['rouge1', 'rouge2']` and `use_aggregator=False`, the output is:
```python
{'rouge1': [Score(precision=1.0, recall=0.5, fmeasure=0.6666666666666666), Score(precision=1.0, recall=1.0, fmeasure=1.0)], 'rouge2': [Score(precision=0.0, recall=0.0, fmeasure=0.0), Score(precision=1.0, recall=1.0, fmeasure=1.0)]}
```
If `rouge_types=['rouge1', 'rouge2']` and `use_aggregator=True`, the output is of the following format:
```python
{'rouge1': AggregateScore(low=Score(precision=1.0, recall=1.0, fmeasure=1.0), mid=Score(precision=1.0, recall=1.0, fmeasure=1.0), high=Score(precision=1.0, recall=1.0, fmeasure=1.0)), 'rouge2': AggregateScore(low=Score(precision=1.0, recall=1.0, fmeasure=1.0), mid=Score(precision=1.0, recall=1.0, fmeasure=1.0), high=Score(precision=1.0, recall=1.0, fmeasure=1.0))}
```
The `precision`, `recall`, and `fmeasure` values all have a range of 0 to 1.
#### Values from Popular Papers
### Examples
An example without aggregation:
```python
>>> rouge = datasets.load_metric('rouge')
>>> predictions = ["hello goodbye", "ankh morpork"]
>>> references = ["goodbye", "general kenobi"]
>>> results = rouge.compute(predictions=predictions,
... references=references)
>>> print(list(results.keys()))
['rouge1', 'rouge2', 'rougeL', 'rougeLsum']
>>> print(results["rouge1"])
[Score(precision=0.5, recall=0.5, fmeasure=0.5), Score(precision=0.0, recall=0.0, fmeasure=0.0)]
```
The same example, but with aggregation:
```python
>>> rouge = datasets.load_metric('rouge')
>>> predictions = ["hello goodbye", "ankh morpork"]
>>> references = ["goodbye", "general kenobi"]
>>> results = rouge.compute(predictions=predictions,
... references=references,
... use_aggregator=True)
>>> print(list(results.keys()))
['rouge1', 'rouge2', 'rougeL', 'rougeLsum']
>>> print(results["rouge1"])
AggregateScore(low=Score(precision=0.0, recall=0.0, fmeasure=0.0), mid=Score(precision=0.25, recall=0.25, fmeasure=0.25), high=Score(precision=0.5, recall=0.5, fmeasure=0.5))
```
The same example, but only calculating `rouge_1`:
```python
>>> rouge = datasets.load_metric('rouge')
>>> predictions = ["hello goodbye", "ankh morpork"]
>>> references = ["goodbye", "general kenobi"]
>>> results = rouge.compute(predictions=predictions,
... references=references,
... rouge_types=['rouge_1'],
... use_aggregator=True)
>>> print(list(results.keys()))
['rouge1']
>>> print(results["rouge1"])
AggregateScore(low=Score(precision=0.0, recall=0.0, fmeasure=0.0), mid=Score(precision=0.25, recall=0.25, fmeasure=0.25), high=Score(precision=0.5, recall=0.5, fmeasure=0.5))
```
## Limitations and Bias
See [Schluter (2017)](https://aclanthology.org/E17-2007/) for an in-depth discussion of many of ROUGE's limits.
## Citation
```bibtex
@inproceedings{lin-2004-rouge,
title = "{ROUGE}: A Package for Automatic Evaluation of Summaries",
author = "Lin, Chin-Yew",
booktitle = "Text Summarization Branches Out",
month = jul,
year = "2004",
address = "Barcelona, Spain",
publisher = "Association for Computational Linguistics",
url = "https://www.aclweb.org/anthology/W04-1013",
pages = "74--81",
}
```
## Further References
- This metrics is a wrapper around the [Google Research reimplementation of ROUGE](https://github.com/google-research/google-research/tree/master/rouge) | 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/rouge/rouge.py | # Copyright 2020 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" ROUGE metric from Google Research github repo. """
# The dependencies in https://github.com/google-research/google-research/blob/master/rouge/requirements.txt
import absl # noqa: F401 # Here to have a nice missing dependency error message early on
import nltk # noqa: F401 # Here to have a nice missing dependency error message early on
import numpy # noqa: F401 # Here to have a nice missing dependency error message early on
import six # noqa: F401 # Here to have a nice missing dependency error message early on
from rouge_score import rouge_scorer, scoring
import datasets
_CITATION = """\
@inproceedings{lin-2004-rouge,
title = "{ROUGE}: A Package for Automatic Evaluation of Summaries",
author = "Lin, Chin-Yew",
booktitle = "Text Summarization Branches Out",
month = jul,
year = "2004",
address = "Barcelona, Spain",
publisher = "Association for Computational Linguistics",
url = "https://www.aclweb.org/anthology/W04-1013",
pages = "74--81",
}
"""
_DESCRIPTION = """\
ROUGE, or Recall-Oriented Understudy for Gisting Evaluation, is a set of metrics and a software package used for
evaluating automatic summarization and machine translation software in natural language processing.
The metrics compare an automatically produced summary or translation against a reference or a set of references (human-produced) summary or translation.
Note that ROUGE is case insensitive, meaning that upper case letters are treated the same way as lower case letters.
This metrics is a wrapper around Google Research reimplementation of ROUGE:
https://github.com/google-research/google-research/tree/master/rouge
"""
_KWARGS_DESCRIPTION = """
Calculates average rouge scores for a list of hypotheses and references
Args:
predictions: list of predictions to score. Each prediction
should be a string with tokens separated by spaces.
references: list of reference for each prediction. Each
reference should be a string with tokens separated by spaces.
rouge_types: A list of rouge types to calculate.
Valid names:
`"rouge{n}"` (e.g. `"rouge1"`, `"rouge2"`) where: {n} is the n-gram based scoring,
`"rougeL"`: Longest common subsequence based scoring.
`"rougeLSum"`: rougeLsum splits text using `"\n"`.
See details in https://github.com/huggingface/datasets/issues/617
use_stemmer: Bool indicating whether Porter stemmer should be used to strip word suffixes.
use_aggregator: Return aggregates if this is set to True
Returns:
rouge1: rouge_1 (precision, recall, f1),
rouge2: rouge_2 (precision, recall, f1),
rougeL: rouge_l (precision, recall, f1),
rougeLsum: rouge_lsum (precision, recall, f1)
Examples:
>>> rouge = datasets.load_metric('rouge')
>>> predictions = ["hello there", "general kenobi"]
>>> references = ["hello there", "general kenobi"]
>>> results = rouge.compute(predictions=predictions, references=references)
>>> print(list(results.keys()))
['rouge1', 'rouge2', 'rougeL', 'rougeLsum']
>>> print(results["rouge1"])
AggregateScore(low=Score(precision=1.0, recall=1.0, fmeasure=1.0), mid=Score(precision=1.0, recall=1.0, fmeasure=1.0), high=Score(precision=1.0, recall=1.0, fmeasure=1.0))
>>> print(results["rouge1"].mid.fmeasure)
1.0
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Rouge(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Value("string", id="sequence"),
"references": datasets.Value("string", id="sequence"),
}
),
codebase_urls=["https://github.com/google-research/google-research/tree/master/rouge"],
reference_urls=[
"https://en.wikipedia.org/wiki/ROUGE_(metric)",
"https://github.com/google-research/google-research/tree/master/rouge",
],
)
def _compute(self, predictions, references, rouge_types=None, use_aggregator=True, use_stemmer=False):
if rouge_types is None:
rouge_types = ["rouge1", "rouge2", "rougeL", "rougeLsum"]
scorer = rouge_scorer.RougeScorer(rouge_types=rouge_types, use_stemmer=use_stemmer)
if use_aggregator:
aggregator = scoring.BootstrapAggregator()
else:
scores = []
for ref, pred in zip(references, predictions):
score = scorer.score(ref, pred)
if use_aggregator:
aggregator.add_scores(score)
else:
scores.append(score)
if use_aggregator:
result = aggregator.aggregate()
else:
result = {}
for key in scores[0]:
result[key] = [score[key] for score in scores]
return result
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/chrf/README.md | # Metric Card for chrF(++)
## Metric Description
ChrF and ChrF++ are two MT evaluation metrics that use the F-score statistic for character n-gram matches. ChrF++ additionally includes word n-grams, which correlate more strongly with direct assessment. We use the implementation that is already present in sacrebleu.
While this metric is included in sacreBLEU, the implementation here is slightly different from sacreBLEU in terms of the required input format. Here, the length of the references and hypotheses lists need to be the same, so you may need to transpose your references compared to sacrebleu's required input format. See https://github.com/huggingface/datasets/issues/3154#issuecomment-950746534
See the [sacreBLEU README.md](https://github.com/mjpost/sacreBLEU#chrf--chrf) for more information.
## How to Use
At minimum, this metric requires a `list` of predictions and a `list` of `list`s of references:
```python
>>> prediction = ["The relationship between cats and dogs is not exactly friendly.", "a good bookshop is just a genteel black hole that knows how to read."]
>>> reference = [["The relationship between dogs and cats is not exactly friendly.", ], ["A good bookshop is just a genteel Black Hole that knows how to read."]]
>>> chrf = datasets.load_metric("chrf")
>>> results = chrf.compute(predictions=prediction, references=reference)
>>> print(results)
{'score': 84.64214891738334, 'char_order': 6, 'word_order': 0, 'beta': 2}
```
### Inputs
- **`predictions`** (`list` of `str`): The predicted sentences.
- **`references`** (`list` of `list` of `str`): The references. There should be one reference sub-list for each prediction sentence.
- **`char_order`** (`int`): Character n-gram order. Defaults to `6`.
- **`word_order`** (`int`): Word n-gram order. If equals to 2, the metric is referred to as chrF++. Defaults to `0`.
- **`beta`** (`int`): Determine the importance of recall w.r.t precision. Defaults to `2`.
- **`lowercase`** (`bool`): If `True`, enables case-insensitivity. Defaults to `False`.
- **`whitespace`** (`bool`): If `True`, include whitespaces when extracting character n-grams. Defaults to `False`.
- **`eps_smoothing`** (`bool`): If `True`, applies epsilon smoothing similar to reference chrF++.py, NLTK, and Moses implementations. If `False`, takes into account effective match order similar to sacreBLEU < 2.0.0. Defaults to `False`.
### Output Values
The output is a dictionary containing the following fields:
- **`'score'`** (`float`): The chrF (chrF++) score.
- **`'char_order'`** (`int`): The character n-gram order.
- **`'word_order'`** (`int`): The word n-gram order. If equals to `2`, the metric is referred to as chrF++.
- **`'beta'`** (`int`): Determine the importance of recall w.r.t precision.
The output is formatted as below:
```python
{'score': 61.576379378113785, 'char_order': 6, 'word_order': 0, 'beta': 2}
```
The chrF(++) score can be any value between `0.0` and `100.0`, inclusive.
#### Values from Popular Papers
### Examples
A simple example of calculating chrF:
```python
>>> prediction = ["The relationship between cats and dogs is not exactly friendly.", "a good bookshop is just a genteel black hole that knows how to read."]
>>> reference = [["The relationship between dogs and cats is not exactly friendly.", ], ["A good bookshop is just a genteel Black Hole that knows how to read."]]
>>> chrf = datasets.load_metric("chrf")
>>> results = chrf.compute(predictions=prediction, references=reference)
>>> print(results)
{'score': 84.64214891738334, 'char_order': 6, 'word_order': 0, 'beta': 2}
```
The same example, but with the argument `word_order=2`, to calculate chrF++ instead of chrF:
```python
>>> prediction = ["The relationship between cats and dogs is not exactly friendly.", "a good bookshop is just a genteel black hole that knows how to read."]
>>> reference = [["The relationship between dogs and cats is not exactly friendly.", ], ["A good bookshop is just a genteel Black Hole that knows how to read."]]
>>> chrf = datasets.load_metric("chrf")
>>> results = chrf.compute(predictions=prediction,
... references=reference,
... word_order=2)
>>> print(results)
{'score': 82.87263732906315, 'char_order': 6, 'word_order': 2, 'beta': 2}
```
The same chrF++ example as above, but with `lowercase=True` to normalize all case:
```python
>>> prediction = ["The relationship between cats and dogs is not exactly friendly.", "a good bookshop is just a genteel black hole that knows how to read."]
>>> reference = [["The relationship between dogs and cats is not exactly friendly.", ], ["A good bookshop is just a genteel Black Hole that knows how to read."]]
>>> chrf = datasets.load_metric("chrf")
>>> results = chrf.compute(predictions=prediction,
... references=reference,
... word_order=2,
... lowercase=True)
>>> print(results)
{'score': 92.12853119829202, 'char_order': 6, 'word_order': 2, 'beta': 2}
```
## Limitations and Bias
- According to [Popović 2017](https://www.statmt.org/wmt17/pdf/WMT70.pdf), chrF+ (where `word_order=1`) and chrF++ (where `word_order=2`) produce scores that correlate better with human judgements than chrF (where `word_order=0`) does.
## Citation
```bibtex
@inproceedings{popovic-2015-chrf,
title = "chr{F}: character n-gram {F}-score for automatic {MT} evaluation",
author = "Popovi{\'c}, Maja",
booktitle = "Proceedings of the Tenth Workshop on Statistical Machine Translation",
month = sep,
year = "2015",
address = "Lisbon, Portugal",
publisher = "Association for Computational Linguistics",
url = "https://aclanthology.org/W15-3049",
doi = "10.18653/v1/W15-3049",
pages = "392--395",
}
@inproceedings{popovic-2017-chrf,
title = "chr{F}++: words helping character n-grams",
author = "Popovi{\'c}, Maja",
booktitle = "Proceedings of the Second Conference on Machine Translation",
month = sep,
year = "2017",
address = "Copenhagen, Denmark",
publisher = "Association for Computational Linguistics",
url = "https://aclanthology.org/W17-4770",
doi = "10.18653/v1/W17-4770",
pages = "612--618",
}
@inproceedings{post-2018-call,
title = "A Call for Clarity in Reporting {BLEU} Scores",
author = "Post, Matt",
booktitle = "Proceedings of the Third Conference on Machine Translation: Research Papers",
month = oct,
year = "2018",
address = "Belgium, Brussels",
publisher = "Association for Computational Linguistics",
url = "https://www.aclweb.org/anthology/W18-6319",
pages = "186--191",
}
```
## Further References
- See the [sacreBLEU README.md](https://github.com/mjpost/sacreBLEU#chrf--chrf) for more information on this implementation. | 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/chrf/chrf.py | # Copyright 2021 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" Chrf(++) metric as available in sacrebleu. """
import sacrebleu as scb
from packaging import version
from sacrebleu import CHRF
import datasets
_CITATION = """\
@inproceedings{popovic-2015-chrf,
title = "chr{F}: character n-gram {F}-score for automatic {MT} evaluation",
author = "Popovi{\'c}, Maja",
booktitle = "Proceedings of the Tenth Workshop on Statistical Machine Translation",
month = sep,
year = "2015",
address = "Lisbon, Portugal",
publisher = "Association for Computational Linguistics",
url = "https://aclanthology.org/W15-3049",
doi = "10.18653/v1/W15-3049",
pages = "392--395",
}
@inproceedings{popovic-2017-chrf,
title = "chr{F}++: words helping character n-grams",
author = "Popovi{\'c}, Maja",
booktitle = "Proceedings of the Second Conference on Machine Translation",
month = sep,
year = "2017",
address = "Copenhagen, Denmark",
publisher = "Association for Computational Linguistics",
url = "https://aclanthology.org/W17-4770",
doi = "10.18653/v1/W17-4770",
pages = "612--618",
}
@inproceedings{post-2018-call,
title = "A Call for Clarity in Reporting {BLEU} Scores",
author = "Post, Matt",
booktitle = "Proceedings of the Third Conference on Machine Translation: Research Papers",
month = oct,
year = "2018",
address = "Belgium, Brussels",
publisher = "Association for Computational Linguistics",
url = "https://www.aclweb.org/anthology/W18-6319",
pages = "186--191",
}
"""
_DESCRIPTION = """\
ChrF and ChrF++ are two MT evaluation metrics. They both use the F-score statistic for character n-gram matches,
and ChrF++ adds word n-grams as well which correlates more strongly with direct assessment. We use the implementation
that is already present in sacrebleu.
The implementation here is slightly different from sacrebleu in terms of the required input format. The length of
the references and hypotheses lists need to be the same, so you may need to transpose your references compared to
sacrebleu's required input format. See https://github.com/huggingface/datasets/issues/3154#issuecomment-950746534
See the README.md file at https://github.com/mjpost/sacreBLEU#chrf--chrf for more information.
"""
_KWARGS_DESCRIPTION = """
Produces ChrF(++) scores for hypotheses given reference translations.
Args:
predictions (list of str): The predicted sentences.
references (list of list of str): The references. There should be one reference sub-list for each prediction sentence.
char_order (int): Character n-gram order. Defaults to `6`.
word_order (int): Word n-gram order. If equals to `2`, the metric is referred to as chrF++. Defaults to `0`.
beta (int): Determine the importance of recall w.r.t precision. Defaults to `2`.
lowercase (bool): if `True`, enables case-insensitivity. Defaults to `False`.
whitespace (bool): If `True`, include whitespaces when extracting character n-grams.
eps_smoothing (bool): If `True`, applies epsilon smoothing similar
to reference chrF++.py, NLTK and Moses implementations. If `False`,
it takes into account effective match order similar to sacreBLEU < 2.0.0. Defaults to `False`.
Returns:
'score' (float): The chrF (chrF++) score,
'char_order' (int): The character n-gram order,
'word_order' (int): The word n-gram order. If equals to 2, the metric is referred to as chrF++,
'beta' (int): Determine the importance of recall w.r.t precision
Examples:
Example 1--a simple example of calculating chrF:
>>> prediction = ["The relationship between cats and dogs is not exactly friendly.", "a good bookshop is just a genteel black hole that knows how to read."]
>>> reference = [["The relationship between dogs and cats is not exactly friendly."], ["A good bookshop is just a genteel Black Hole that knows how to read."]]
>>> chrf = datasets.load_metric("chrf")
>>> results = chrf.compute(predictions=prediction, references=reference)
>>> print(results)
{'score': 84.64214891738334, 'char_order': 6, 'word_order': 0, 'beta': 2}
Example 2--the same example, but with the argument word_order=2, to calculate chrF++ instead of chrF:
>>> prediction = ["The relationship between cats and dogs is not exactly friendly.", "a good bookshop is just a genteel black hole that knows how to read."]
>>> reference = [["The relationship between dogs and cats is not exactly friendly."], ["A good bookshop is just a genteel Black Hole that knows how to read."]]
>>> chrf = datasets.load_metric("chrf")
>>> results = chrf.compute(predictions=prediction,
... references=reference,
... word_order=2)
>>> print(results)
{'score': 82.87263732906315, 'char_order': 6, 'word_order': 2, 'beta': 2}
Example 3--the same chrF++ example as above, but with `lowercase=True` to normalize all case:
>>> prediction = ["The relationship between cats and dogs is not exactly friendly.", "a good bookshop is just a genteel black hole that knows how to read."]
>>> reference = [["The relationship between dogs and cats is not exactly friendly."], ["A good bookshop is just a genteel Black Hole that knows how to read."]]
>>> chrf = datasets.load_metric("chrf")
>>> results = chrf.compute(predictions=prediction,
... references=reference,
... word_order=2,
... lowercase=True)
>>> print(results)
{'score': 92.12853119829202, 'char_order': 6, 'word_order': 2, 'beta': 2}
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class ChrF(datasets.Metric):
def _info(self):
if version.parse(scb.__version__) < version.parse("1.4.12"):
raise ImportWarning(
"To use `sacrebleu`, the module `sacrebleu>=1.4.12` is required, and the current version of `sacrebleu` doesn't match this condition.\n"
'You can install it with `pip install "sacrebleu>=1.4.12"`.'
)
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
homepage="https://github.com/mjpost/sacreBLEU#chrf--chrf",
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Value("string", id="sequence"),
"references": datasets.Sequence(datasets.Value("string", id="sequence"), id="references"),
}
),
codebase_urls=["https://github.com/mjpost/sacreBLEU#chrf--chrf"],
reference_urls=[
"https://github.com/m-popovic/chrF",
],
)
def _compute(
self,
predictions,
references,
char_order: int = CHRF.CHAR_ORDER,
word_order: int = CHRF.WORD_ORDER,
beta: int = CHRF.BETA,
lowercase: bool = False,
whitespace: bool = False,
eps_smoothing: bool = False,
):
references_per_prediction = len(references[0])
if any(len(refs) != references_per_prediction for refs in references):
raise ValueError("Sacrebleu requires the same number of references for each prediction")
transformed_references = [[refs[i] for refs in references] for i in range(references_per_prediction)]
sb_chrf = CHRF(char_order, word_order, beta, lowercase, whitespace, eps_smoothing)
output = sb_chrf.corpus_score(predictions, transformed_references)
return {
"score": output.score,
"char_order": output.char_order,
"word_order": output.word_order,
"beta": output.beta,
}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/recall/README.md | # Metric Card for Recall
## Metric Description
Recall is the fraction of the positive examples that were correctly labeled by the model as positive. It can be computed with the equation:
Recall = TP / (TP + FN)
Where TP is the number of true positives and FN is the number of false negatives.
## How to Use
At minimum, this metric takes as input two `list`s, each containing `int`s: predictions and references.
```python
>>> recall_metric = datasets.load_metric('recall')
>>> results = recall_metric.compute(references=[0, 1], predictions=[0, 1])
>>> print(results)
["{'recall': 1.0}"]
```
### Inputs
- **predictions** (`list` of `int`): The predicted labels.
- **references** (`list` of `int`): The ground truth labels.
- **labels** (`list` of `int`): The set of labels to include when `average` is not set to `binary`, and their order when average is `None`. Labels present in the data can be excluded in this input, for example to calculate a multiclass average ignoring a majority negative class, while labels not present in the data will result in 0 components in a macro average. For multilabel targets, labels are column indices. By default, all labels in y_true and y_pred are used in sorted order. Defaults to None.
- **pos_label** (`int`): The class label to use as the 'positive class' when calculating the recall. Defaults to `1`.
- **average** (`string`): This parameter is required for multiclass/multilabel targets. If None, the scores for each class are returned. Otherwise, this determines the type of averaging performed on the data. Defaults to `'binary'`.
- `'binary'`: Only report results for the class specified by `pos_label`. This is applicable only if the target labels and predictions are binary.
- `'micro'`: Calculate metrics globally by counting the total true positives, false negatives, and false positives.
- `'macro'`: Calculate metrics for each label, and find their unweighted mean. This does not take label imbalance into account.
- `'weighted'`: Calculate metrics for each label, and find their average weighted by support (the number of true instances for each label). This alters `'macro'` to account for label imbalance. Note that it can result in an F-score that is not between precision and recall.
- `'samples'`: Calculate metrics for each instance, and find their average (only meaningful for multilabel classification).
- **sample_weight** (`list` of `float`): Sample weights Defaults to `None`.
- **zero_division** (): Sets the value to return when there is a zero division. Defaults to .
- `'warn'`: If there is a zero division, the return value is `0`, but warnings are also raised.
- `0`: If there is a zero division, the return value is `0`.
- `1`: If there is a zero division, the return value is `1`.
### Output Values
- **recall**(`float`, or `array` of `float`, for multiclass targets): Either the general recall score, or the recall scores for individual classes, depending on the values input to `labels` and `average`. Minimum possible value is 0. Maximum possible value is 1. A higher recall means that more of the positive examples have been labeled correctly. Therefore, a higher recall is generally considered better.
Output Example(s):
```python
{'recall': 1.0}
```
```python
{'recall': array([1., 0., 0.])}
```
This metric outputs a dictionary with one entry, `'recall'`.
#### Values from Popular Papers
### Examples
Example 1-A simple example with some errors
```python
>>> recall_metric = datasets.load_metric('recall')
>>> results = recall_metric.compute(references=[0, 0, 1, 1, 1], predictions=[0, 1, 0, 1, 1])
>>> print(results)
{'recall': 0.6666666666666666}
```
Example 2-The same example as Example 1, but with `pos_label=0` instead of the default `pos_label=1`.
```python
>>> recall_metric = datasets.load_metric('recall')
>>> results = recall_metric.compute(references=[0, 0, 1, 1, 1], predictions=[0, 1, 0, 1, 1], pos_label=0)
>>> print(results)
{'recall': 0.5}
```
Example 3-The same example as Example 1, but with `sample_weight` included.
```python
>>> recall_metric = datasets.load_metric('recall')
>>> sample_weight = [0.9, 0.2, 0.9, 0.3, 0.8]
>>> results = recall_metric.compute(references=[0, 0, 1, 1, 1], predictions=[0, 1, 0, 1, 1], sample_weight=sample_weight)
>>> print(results)
{'recall': 0.55}
```
Example 4-A multiclass example, using different averages.
```python
>>> recall_metric = datasets.load_metric('recall')
>>> predictions = [0, 2, 1, 0, 0, 1]
>>> references = [0, 1, 2, 0, 1, 2]
>>> results = recall_metric.compute(predictions=predictions, references=references, average='macro')
>>> print(results)
{'recall': 0.3333333333333333}
>>> results = recall_metric.compute(predictions=predictions, references=references, average='micro')
>>> print(results)
{'recall': 0.3333333333333333}
>>> results = recall_metric.compute(predictions=predictions, references=references, average='weighted')
>>> print(results)
{'recall': 0.3333333333333333}
>>> results = recall_metric.compute(predictions=predictions, references=references, average=None)
>>> print(results)
{'recall': array([1., 0., 0.])}
```
## Limitations and Bias
## Citation(s)
```bibtex
@article{scikit-learn, title={Scikit-learn: Machine Learning in {P}ython}, author={Pedregosa, F. and Varoquaux, G. and Gramfort, A. and Michel, V. and Thirion, B. and Grisel, O. and Blondel, M. and Prettenhofer, P. and Weiss, R. and Dubourg, V. and Vanderplas, J. and Passos, A. and Cournapeau, D. and Brucher, M. and Perrot, M. and Duchesnay, E.}, journal={Journal of Machine Learning Research}, volume={12}, pages={2825--2830}, year={2011}
```
## Further References
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/recall/recall.py | # Copyright 2020 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Recall metric."""
from sklearn.metrics import recall_score
import datasets
_DESCRIPTION = """
Recall is the fraction of the positive examples that were correctly labeled by the model as positive. It can be computed with the equation:
Recall = TP / (TP + FN)
Where TP is the true positives and FN is the false negatives.
"""
_KWARGS_DESCRIPTION = """
Args:
- **predictions** (`list` of `int`): The predicted labels.
- **references** (`list` of `int`): The ground truth labels.
- **labels** (`list` of `int`): The set of labels to include when `average` is not set to `binary`, and their order when average is `None`. Labels present in the data can be excluded in this input, for example to calculate a multiclass average ignoring a majority negative class, while labels not present in the data will result in 0 components in a macro average. For multilabel targets, labels are column indices. By default, all labels in y_true and y_pred are used in sorted order. Defaults to None.
- **pos_label** (`int`): The class label to use as the 'positive class' when calculating the recall. Defaults to `1`.
- **average** (`string`): This parameter is required for multiclass/multilabel targets. If None, the scores for each class are returned. Otherwise, this determines the type of averaging performed on the data. Defaults to `'binary'`.
- `'binary'`: Only report results for the class specified by `pos_label`. This is applicable only if the target labels and predictions are binary.
- `'micro'`: Calculate metrics globally by counting the total true positives, false negatives, and false positives.
- `'macro'`: Calculate metrics for each label, and find their unweighted mean. This does not take label imbalance into account.
- `'weighted'`: Calculate metrics for each label, and find their average weighted by support (the number of true instances for each label). This alters `'macro'` to account for label imbalance. Note that it can result in an F-score that is not between precision and recall.
- `'samples'`: Calculate metrics for each instance, and find their average (only meaningful for multilabel classification).
- **sample_weight** (`list` of `float`): Sample weights Defaults to `None`.
- **zero_division** (): Sets the value to return when there is a zero division. Defaults to .
- `'warn'`: If there is a zero division, the return value is `0`, but warnings are also raised.
- `0`: If there is a zero division, the return value is `0`.
- `1`: If there is a zero division, the return value is `1`.
Returns:
- **recall** (`float`, or `array` of `float`): Either the general recall score, or the recall scores for individual classes, depending on the values input to `labels` and `average`. Minimum possible value is 0. Maximum possible value is 1. A higher recall means that more of the positive examples have been labeled correctly. Therefore, a higher recall is generally considered better.
Examples:
Example 1-A simple example with some errors
>>> recall_metric = datasets.load_metric('recall')
>>> results = recall_metric.compute(references=[0, 0, 1, 1, 1], predictions=[0, 1, 0, 1, 1])
>>> print(results)
{'recall': 0.6666666666666666}
Example 2-The same example as Example 1, but with `pos_label=0` instead of the default `pos_label=1`.
>>> recall_metric = datasets.load_metric('recall')
>>> results = recall_metric.compute(references=[0, 0, 1, 1, 1], predictions=[0, 1, 0, 1, 1], pos_label=0)
>>> print(results)
{'recall': 0.5}
Example 3-The same example as Example 1, but with `sample_weight` included.
>>> recall_metric = datasets.load_metric('recall')
>>> sample_weight = [0.9, 0.2, 0.9, 0.3, 0.8]
>>> results = recall_metric.compute(references=[0, 0, 1, 1, 1], predictions=[0, 1, 0, 1, 1], sample_weight=sample_weight)
>>> print(results)
{'recall': 0.55}
Example 4-A multiclass example, using different averages.
>>> recall_metric = datasets.load_metric('recall')
>>> predictions = [0, 2, 1, 0, 0, 1]
>>> references = [0, 1, 2, 0, 1, 2]
>>> results = recall_metric.compute(predictions=predictions, references=references, average='macro')
>>> print(results)
{'recall': 0.3333333333333333}
>>> results = recall_metric.compute(predictions=predictions, references=references, average='micro')
>>> print(results)
{'recall': 0.3333333333333333}
>>> results = recall_metric.compute(predictions=predictions, references=references, average='weighted')
>>> print(results)
{'recall': 0.3333333333333333}
>>> results = recall_metric.compute(predictions=predictions, references=references, average=None)
>>> print(results)
{'recall': array([1., 0., 0.])}
"""
_CITATION = """
@article{scikit-learn, title={Scikit-learn: Machine Learning in {P}ython}, author={Pedregosa, F. and Varoquaux, G. and Gramfort, A. and Michel, V. and Thirion, B. and Grisel, O. and Blondel, M. and Prettenhofer, P. and Weiss, R. and Dubourg, V. and Vanderplas, J. and Passos, A. and Cournapeau, D. and Brucher, M. and Perrot, M. and Duchesnay, E.}, journal={Journal of Machine Learning Research}, volume={12}, pages={2825--2830}, year={2011}
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Recall(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Sequence(datasets.Value("int32")),
"references": datasets.Sequence(datasets.Value("int32")),
}
if self.config_name == "multilabel"
else {
"predictions": datasets.Value("int32"),
"references": datasets.Value("int32"),
}
),
reference_urls=["https://scikit-learn.org/stable/modules/generated/sklearn.metrics.recall_score.html"],
)
def _compute(
self,
predictions,
references,
labels=None,
pos_label=1,
average="binary",
sample_weight=None,
zero_division="warn",
):
score = recall_score(
references,
predictions,
labels=labels,
pos_label=pos_label,
average=average,
sample_weight=sample_weight,
zero_division=zero_division,
)
return {"recall": float(score) if score.size == 1 else score}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/wiki_split/README.md | # Metric Card for WikiSplit
## Metric description
WikiSplit is the combination of three metrics: [SARI](https://huggingface.co/metrics/sari), [exact match](https://huggingface.co/metrics/exact_match) and [SacreBLEU](https://huggingface.co/metrics/sacrebleu).
It can be used to evaluate the quality of sentence splitting approaches, which require rewriting a long sentence into two or more coherent short sentences, e.g. based on the [WikiSplit dataset](https://huggingface.co/datasets/wiki_split).
## How to use
The WIKI_SPLIT metric takes three inputs:
`sources`: a list of source sentences, where each sentence should be a string.
`predictions`: a list of predicted sentences, where each sentence should be a string.
`references`: a list of lists of reference sentences, where each sentence should be a string.
```python
>>> from datasets import load_metric
>>> wiki_split = load_metric("wiki_split")
>>> sources = ["About 95 species are currently accepted ."]
>>> predictions = ["About 95 you now get in ."]
>>> references= [["About 95 species are currently known ."]]
>>> results = wiki_split.compute(sources=sources, predictions=predictions, references=references)
```
## Output values
This metric outputs a dictionary containing three scores:
`sari`: the [SARI](https://huggingface.co/metrics/sari) score, whose range is between `0.0` and `100.0` -- the higher the value, the better the performance of the model being evaluated, with a SARI of 100 being a perfect score.
`sacrebleu`: the [SacreBLEU](https://huggingface.co/metrics/sacrebleu) score, which can take any value between `0.0` and `100.0`, inclusive.
`exact`: the [exact match](https://huggingface.co/metrics/exact_match) score, which represents the sum of all of the individual exact match scores in the set, divided by the total number of predictions in the set. It ranges from `0.0` to `100`, inclusive. Here, `0.0` means no prediction/reference pairs were matches, while `100.0` means they all were.
```python
>>> print(results)
{'sari': 21.805555555555557, 'sacrebleu': 14.535768424205482, 'exact': 0.0}
```
### Values from popular papers
This metric was initially used by [Rothe et al.(2020)](https://arxiv.org/pdf/1907.12461.pdf) to evaluate the performance of different split-and-rephrase approaches on the [WikiSplit dataset](https://huggingface.co/datasets/wiki_split). They reported a SARI score of 63.5, a SacreBLEU score of 77.2, and an EXACT_MATCH score of 16.3.
## Examples
Perfect match between prediction and reference:
```python
>>> from datasets import load_metric
>>> wiki_split = load_metric("wiki_split")
>>> sources = ["About 95 species are currently accepted ."]
>>> predictions = ["About 95 species are currently accepted ."]
>>> references= [["About 95 species are currently accepted ."]]
>>> results = wiki_split.compute(sources=sources, predictions=predictions, references=references)
>>> print(results)
{'sari': 100.0, 'sacrebleu': 100.00000000000004, 'exact': 100.0
```
Partial match between prediction and reference:
```python
>>> from datasets import load_metric
>>> wiki_split = load_metric("wiki_split")
>>> sources = ["About 95 species are currently accepted ."]
>>> predictions = ["About 95 you now get in ."]
>>> references= [["About 95 species are currently known ."]]
>>> results = wiki_split.compute(sources=sources, predictions=predictions, references=references)
>>> print(results)
{'sari': 21.805555555555557, 'sacrebleu': 14.535768424205482, 'exact': 0.0}
```
No match between prediction and reference:
```python
>>> from datasets import load_metric
>>> wiki_split = load_metric("wiki_split")
>>> sources = ["About 95 species are currently accepted ."]
>>> predictions = ["Hello world ."]
>>> references= [["About 95 species are currently known ."]]
>>> results = wiki_split.compute(sources=sources, predictions=predictions, references=references)
>>> print(results)
{'sari': 14.047619047619046, 'sacrebleu': 0.0, 'exact': 0.0}
```
## Limitations and bias
This metric is not the official metric to evaluate models on the [WikiSplit dataset](https://huggingface.co/datasets/wiki_split). It was initially proposed by [Rothe et al.(2020)](https://arxiv.org/pdf/1907.12461.pdf), whereas the [original paper introducing the WikiSplit dataset (2018)](https://aclanthology.org/D18-1080.pdf) uses different metrics to evaluate performance, such as corpus-level [BLEU](https://huggingface.co/metrics/bleu) and sentence-level BLEU.
## Citation
```bibtex
@article{rothe2020leveraging,
title={Leveraging pre-trained checkpoints for sequence generation tasks},
author={Rothe, Sascha and Narayan, Shashi and Severyn, Aliaksei},
journal={Transactions of the Association for Computational Linguistics},
volume={8},
pages={264--280},
year={2020},
publisher={MIT Press}
}
```
## Further References
- [WikiSplit dataset](https://huggingface.co/datasets/wiki_split)
- [WikiSplit paper (Botha et al., 2018)](https://aclanthology.org/D18-1080.pdf)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/wiki_split/wiki_split.py | # Copyright 2020 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" WIKI_SPLIT metric."""
import re
import string
from collections import Counter
import sacrebleu
import sacremoses
from packaging import version
import datasets
_CITATION = """
@inproceedings{xu-etal-2016-optimizing,
title = {Optimizing Statistical Machine Translation for Text Simplification},
authors={Xu, Wei and Napoles, Courtney and Pavlick, Ellie and Chen, Quanze and Callison-Burch, Chris},
journal = {Transactions of the Association for Computational Linguistics},
volume = {4},
year={2016},
url = {https://www.aclweb.org/anthology/Q16-1029},
pages = {401--415
},
@inproceedings{post-2018-call,
title = "A Call for Clarity in Reporting {BLEU} Scores",
author = "Post, Matt",
booktitle = "Proceedings of the Third Conference on Machine Translation: Research Papers",
month = oct,
year = "2018",
address = "Belgium, Brussels",
publisher = "Association for Computational Linguistics",
url = "https://www.aclweb.org/anthology/W18-6319",
pages = "186--191",
}
"""
_DESCRIPTION = """\
WIKI_SPLIT is the combination of three metrics SARI, EXACT and SACREBLEU
It can be used to evaluate the quality of machine-generated texts.
"""
_KWARGS_DESCRIPTION = """
Calculates sari score (between 0 and 100) given a list of source and predicted
sentences, and a list of lists of reference sentences. It also computes the BLEU score as well as the exact match score.
Args:
sources: list of source sentences where each sentence should be a string.
predictions: list of predicted sentences where each sentence should be a string.
references: list of lists of reference sentences where each sentence should be a string.
Returns:
sari: sari score
sacrebleu: sacrebleu score
exact: exact score
Examples:
>>> sources=["About 95 species are currently accepted ."]
>>> predictions=["About 95 you now get in ."]
>>> references=[["About 95 species are currently known ."]]
>>> wiki_split = datasets.load_metric("wiki_split")
>>> results = wiki_split.compute(sources=sources, predictions=predictions, references=references)
>>> print(results)
{'sari': 21.805555555555557, 'sacrebleu': 14.535768424205482, 'exact': 0.0}
"""
def normalize_answer(s):
"""Lower text and remove punctuation, articles and extra whitespace."""
def remove_articles(text):
regex = re.compile(r"\b(a|an|the)\b", re.UNICODE)
return re.sub(regex, " ", text)
def white_space_fix(text):
return " ".join(text.split())
def remove_punc(text):
exclude = set(string.punctuation)
return "".join(ch for ch in text if ch not in exclude)
def lower(text):
return text.lower()
return white_space_fix(remove_articles(remove_punc(lower(s))))
def compute_exact(a_gold, a_pred):
return int(normalize_answer(a_gold) == normalize_answer(a_pred))
def compute_em(predictions, references):
scores = [any(compute_exact(ref, pred) for ref in refs) for pred, refs in zip(predictions, references)]
return (sum(scores) / len(scores)) * 100
def SARIngram(sgrams, cgrams, rgramslist, numref):
rgramsall = [rgram for rgrams in rgramslist for rgram in rgrams]
rgramcounter = Counter(rgramsall)
sgramcounter = Counter(sgrams)
sgramcounter_rep = Counter()
for sgram, scount in sgramcounter.items():
sgramcounter_rep[sgram] = scount * numref
cgramcounter = Counter(cgrams)
cgramcounter_rep = Counter()
for cgram, ccount in cgramcounter.items():
cgramcounter_rep[cgram] = ccount * numref
# KEEP
keepgramcounter_rep = sgramcounter_rep & cgramcounter_rep
keepgramcountergood_rep = keepgramcounter_rep & rgramcounter
keepgramcounterall_rep = sgramcounter_rep & rgramcounter
keeptmpscore1 = 0
keeptmpscore2 = 0
for keepgram in keepgramcountergood_rep:
keeptmpscore1 += keepgramcountergood_rep[keepgram] / keepgramcounter_rep[keepgram]
# Fix an alleged bug [2] in the keep score computation.
# keeptmpscore2 += keepgramcountergood_rep[keepgram] / keepgramcounterall_rep[keepgram]
keeptmpscore2 += keepgramcountergood_rep[keepgram]
# Define 0/0=1 instead of 0 to give higher scores for predictions that match
# a target exactly.
keepscore_precision = 1
keepscore_recall = 1
if len(keepgramcounter_rep) > 0:
keepscore_precision = keeptmpscore1 / len(keepgramcounter_rep)
if len(keepgramcounterall_rep) > 0:
# Fix an alleged bug [2] in the keep score computation.
# keepscore_recall = keeptmpscore2 / len(keepgramcounterall_rep)
keepscore_recall = keeptmpscore2 / sum(keepgramcounterall_rep.values())
keepscore = 0
if keepscore_precision > 0 or keepscore_recall > 0:
keepscore = 2 * keepscore_precision * keepscore_recall / (keepscore_precision + keepscore_recall)
# DELETION
delgramcounter_rep = sgramcounter_rep - cgramcounter_rep
delgramcountergood_rep = delgramcounter_rep - rgramcounter
delgramcounterall_rep = sgramcounter_rep - rgramcounter
deltmpscore1 = 0
deltmpscore2 = 0
for delgram in delgramcountergood_rep:
deltmpscore1 += delgramcountergood_rep[delgram] / delgramcounter_rep[delgram]
deltmpscore2 += delgramcountergood_rep[delgram] / delgramcounterall_rep[delgram]
# Define 0/0=1 instead of 0 to give higher scores for predictions that match
# a target exactly.
delscore_precision = 1
if len(delgramcounter_rep) > 0:
delscore_precision = deltmpscore1 / len(delgramcounter_rep)
# ADDITION
addgramcounter = set(cgramcounter) - set(sgramcounter)
addgramcountergood = set(addgramcounter) & set(rgramcounter)
addgramcounterall = set(rgramcounter) - set(sgramcounter)
addtmpscore = 0
for addgram in addgramcountergood:
addtmpscore += 1
# Define 0/0=1 instead of 0 to give higher scores for predictions that match
# a target exactly.
addscore_precision = 1
addscore_recall = 1
if len(addgramcounter) > 0:
addscore_precision = addtmpscore / len(addgramcounter)
if len(addgramcounterall) > 0:
addscore_recall = addtmpscore / len(addgramcounterall)
addscore = 0
if addscore_precision > 0 or addscore_recall > 0:
addscore = 2 * addscore_precision * addscore_recall / (addscore_precision + addscore_recall)
return (keepscore, delscore_precision, addscore)
def SARIsent(ssent, csent, rsents):
numref = len(rsents)
s1grams = ssent.split(" ")
c1grams = csent.split(" ")
s2grams = []
c2grams = []
s3grams = []
c3grams = []
s4grams = []
c4grams = []
r1gramslist = []
r2gramslist = []
r3gramslist = []
r4gramslist = []
for rsent in rsents:
r1grams = rsent.split(" ")
r2grams = []
r3grams = []
r4grams = []
r1gramslist.append(r1grams)
for i in range(0, len(r1grams) - 1):
if i < len(r1grams) - 1:
r2gram = r1grams[i] + " " + r1grams[i + 1]
r2grams.append(r2gram)
if i < len(r1grams) - 2:
r3gram = r1grams[i] + " " + r1grams[i + 1] + " " + r1grams[i + 2]
r3grams.append(r3gram)
if i < len(r1grams) - 3:
r4gram = r1grams[i] + " " + r1grams[i + 1] + " " + r1grams[i + 2] + " " + r1grams[i + 3]
r4grams.append(r4gram)
r2gramslist.append(r2grams)
r3gramslist.append(r3grams)
r4gramslist.append(r4grams)
for i in range(0, len(s1grams) - 1):
if i < len(s1grams) - 1:
s2gram = s1grams[i] + " " + s1grams[i + 1]
s2grams.append(s2gram)
if i < len(s1grams) - 2:
s3gram = s1grams[i] + " " + s1grams[i + 1] + " " + s1grams[i + 2]
s3grams.append(s3gram)
if i < len(s1grams) - 3:
s4gram = s1grams[i] + " " + s1grams[i + 1] + " " + s1grams[i + 2] + " " + s1grams[i + 3]
s4grams.append(s4gram)
for i in range(0, len(c1grams) - 1):
if i < len(c1grams) - 1:
c2gram = c1grams[i] + " " + c1grams[i + 1]
c2grams.append(c2gram)
if i < len(c1grams) - 2:
c3gram = c1grams[i] + " " + c1grams[i + 1] + " " + c1grams[i + 2]
c3grams.append(c3gram)
if i < len(c1grams) - 3:
c4gram = c1grams[i] + " " + c1grams[i + 1] + " " + c1grams[i + 2] + " " + c1grams[i + 3]
c4grams.append(c4gram)
(keep1score, del1score, add1score) = SARIngram(s1grams, c1grams, r1gramslist, numref)
(keep2score, del2score, add2score) = SARIngram(s2grams, c2grams, r2gramslist, numref)
(keep3score, del3score, add3score) = SARIngram(s3grams, c3grams, r3gramslist, numref)
(keep4score, del4score, add4score) = SARIngram(s4grams, c4grams, r4gramslist, numref)
avgkeepscore = sum([keep1score, keep2score, keep3score, keep4score]) / 4
avgdelscore = sum([del1score, del2score, del3score, del4score]) / 4
avgaddscore = sum([add1score, add2score, add3score, add4score]) / 4
finalscore = (avgkeepscore + avgdelscore + avgaddscore) / 3
return finalscore
def normalize(sentence, lowercase: bool = True, tokenizer: str = "13a", return_str: bool = True):
# Normalization is requried for the ASSET dataset (one of the primary
# datasets in sentence simplification) to allow using space
# to split the sentence. Even though Wiki-Auto and TURK datasets,
# do not require normalization, we do it for consistency.
# Code adapted from the EASSE library [1] written by the authors of the ASSET dataset.
# [1] https://github.com/feralvam/easse/blob/580bba7e1378fc8289c663f864e0487188fe8067/easse/utils/preprocessing.py#L7
if lowercase:
sentence = sentence.lower()
if tokenizer in ["13a", "intl"]:
if version.parse(sacrebleu.__version__).major >= 2:
normalized_sent = sacrebleu.metrics.bleu._get_tokenizer(tokenizer)()(sentence)
else:
normalized_sent = sacrebleu.TOKENIZERS[tokenizer]()(sentence)
elif tokenizer == "moses":
normalized_sent = sacremoses.MosesTokenizer().tokenize(sentence, return_str=True, escape=False)
elif tokenizer == "penn":
normalized_sent = sacremoses.MosesTokenizer().penn_tokenize(sentence, return_str=True)
else:
normalized_sent = sentence
if not return_str:
normalized_sent = normalized_sent.split()
return normalized_sent
def compute_sari(sources, predictions, references):
if not (len(sources) == len(predictions) == len(references)):
raise ValueError("Sources length must match predictions and references lengths.")
sari_score = 0
for src, pred, refs in zip(sources, predictions, references):
sari_score += SARIsent(normalize(src), normalize(pred), [normalize(sent) for sent in refs])
sari_score = sari_score / len(predictions)
return 100 * sari_score
def compute_sacrebleu(
predictions,
references,
smooth_method="exp",
smooth_value=None,
force=False,
lowercase=False,
use_effective_order=False,
):
references_per_prediction = len(references[0])
if any(len(refs) != references_per_prediction for refs in references):
raise ValueError("Sacrebleu requires the same number of references for each prediction")
transformed_references = [[refs[i] for refs in references] for i in range(references_per_prediction)]
output = sacrebleu.corpus_bleu(
predictions,
transformed_references,
smooth_method=smooth_method,
smooth_value=smooth_value,
force=force,
lowercase=lowercase,
use_effective_order=use_effective_order,
)
return output.score
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class WikiSplit(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Value("string", id="sequence"),
"references": datasets.Sequence(datasets.Value("string", id="sequence"), id="references"),
}
),
codebase_urls=[
"https://github.com/huggingface/transformers/blob/master/src/transformers/data/metrics/squad_metrics.py",
"https://github.com/cocoxu/simplification/blob/master/SARI.py",
"https://github.com/tensorflow/tensor2tensor/blob/master/tensor2tensor/utils/sari_hook.py",
"https://github.com/mjpost/sacreBLEU",
],
reference_urls=[
"https://www.aclweb.org/anthology/Q16-1029.pdf",
"https://github.com/mjpost/sacreBLEU",
"https://en.wikipedia.org/wiki/BLEU",
"https://towardsdatascience.com/evaluating-text-output-in-nlp-bleu-at-your-own-risk-e8609665a213",
],
)
def _compute(self, sources, predictions, references):
result = {}
result.update({"sari": compute_sari(sources=sources, predictions=predictions, references=references)})
result.update({"sacrebleu": compute_sacrebleu(predictions=predictions, references=references)})
result.update({"exact": compute_em(predictions=predictions, references=references)})
return result
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/mae/README.md | # Metric Card for MAE
## Metric Description
Mean Absolute Error (MAE) is the mean of the magnitude of difference between the predicted and actual numeric values:
## How to Use
At minimum, this metric requires predictions and references as inputs.
```python
>>> mae_metric = datasets.load_metric("mae")
>>> predictions = [2.5, 0.0, 2, 8]
>>> references = [3, -0.5, 2, 7]
>>> results = mae_metric.compute(predictions=predictions, references=references)
```
### Inputs
Mandatory inputs:
- `predictions`: numeric array-like of shape (`n_samples,`) or (`n_samples`, `n_outputs`), representing the estimated target values.
- `references`: numeric array-like of shape (`n_samples,`) or (`n_samples`, `n_outputs`), representing the ground truth (correct) target values.
Optional arguments:
- `sample_weight`: numeric array-like of shape (`n_samples,`) representing sample weights. The default is `None`.
- `multioutput`: `raw_values`, `uniform_average` or numeric array-like of shape (`n_outputs,`), which defines the aggregation of multiple output values. The default value is `uniform_average`.
- `raw_values` returns a full set of errors in case of multioutput input.
- `uniform_average` means that the errors of all outputs are averaged with uniform weight.
- the array-like value defines weights used to average errors.
### Output Values
This metric outputs a dictionary, containing the mean absolute error score, which is of type:
- `float`: if multioutput is `uniform_average` or an ndarray of weights, then the weighted average of all output errors is returned.
- numeric array-like of shape (`n_outputs,`): if multioutput is `raw_values`, then the score is returned for each output separately.
Each MAE `float` value ranges from `0.0` to `1.0`, with the best value being 0.0.
Output Example(s):
```python
{'mae': 0.5}
```
If `multioutput="raw_values"`:
```python
{'mae': array([0.5, 1. ])}
```
#### Values from Popular Papers
### Examples
Example with the `uniform_average` config:
```python
>>> from datasets import load_metric
>>> mae_metric = load_metric("mae")
>>> predictions = [2.5, 0.0, 2, 8]
>>> references = [3, -0.5, 2, 7]
>>> results = mae_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'mae': 0.5}
```
Example with multi-dimensional lists, and the `raw_values` config:
```python
>>> from datasets import load_metric
>>> mae_metric = datasets.load_metric("mae", "multilist")
>>> predictions = [[0.5, 1], [-1, 1], [7, -6]]
>>> references = [[0, 2], [-1, 2], [8, -5]]
>>> results = mae_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'mae': 0.75}
>>> results = mae_metric.compute(predictions=predictions, references=references, multioutput='raw_values')
>>> print(results)
{'mae': array([0.5, 1. ])}
```
## Limitations and Bias
One limitation of MAE is that the relative size of the error is not always obvious, meaning that it can be difficult to tell a big error from a smaller one -- metrics such as Mean Absolute Percentage Error (MAPE) have been proposed to calculate MAE in percentage terms.
Also, since it calculates the mean, MAE may underestimate the impact of big, but infrequent, errors -- metrics such as the Root Mean Square Error (RMSE) compensate for this by taking the root of error values.
## Citation(s)
```bibtex
@article{scikit-learn,
title={Scikit-learn: Machine Learning in {P}ython},
author={Pedregosa, F. and Varoquaux, G. and Gramfort, A. and Michel, V.
and Thirion, B. and Grisel, O. and Blondel, M. and Prettenhofer, P.
and Weiss, R. and Dubourg, V. and Vanderplas, J. and Passos, A. and
Cournapeau, D. and Brucher, M. and Perrot, M. and Duchesnay, E.},
journal={Journal of Machine Learning Research},
volume={12},
pages={2825--2830},
year={2011}
}
```
```bibtex
@article{willmott2005advantages,
title={Advantages of the mean absolute error (MAE) over the root mean square error (RMSE) in assessing average model performance},
author={Willmott, Cort J and Matsuura, Kenji},
journal={Climate research},
volume={30},
number={1},
pages={79--82},
year={2005}
}
```
## Further References
- [Mean Absolute Error - Wikipedia](https://en.wikipedia.org/wiki/Mean_absolute_error)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/mae/mae.py | # Copyright 2022 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""MAE - Mean Absolute Error Metric"""
from sklearn.metrics import mean_absolute_error
import datasets
_CITATION = """\
@article{scikit-learn,
title={Scikit-learn: Machine Learning in {P}ython},
author={Pedregosa, F. and Varoquaux, G. and Gramfort, A. and Michel, V.
and Thirion, B. and Grisel, O. and Blondel, M. and Prettenhofer, P.
and Weiss, R. and Dubourg, V. and Vanderplas, J. and Passos, A. and
Cournapeau, D. and Brucher, M. and Perrot, M. and Duchesnay, E.},
journal={Journal of Machine Learning Research},
volume={12},
pages={2825--2830},
year={2011}
}
"""
_DESCRIPTION = """\
Mean Absolute Error (MAE) is the mean of the magnitude of difference between the predicted and actual
values.
"""
_KWARGS_DESCRIPTION = """
Args:
predictions: array-like of shape (n_samples,) or (n_samples, n_outputs)
Estimated target values.
references: array-like of shape (n_samples,) or (n_samples, n_outputs)
Ground truth (correct) target values.
sample_weight: array-like of shape (n_samples,), default=None
Sample weights.
multioutput: {"raw_values", "uniform_average"} or array-like of shape (n_outputs,), default="uniform_average"
Defines aggregating of multiple output values. Array-like value defines weights used to average errors.
"raw_values" : Returns a full set of errors in case of multioutput input.
"uniform_average" : Errors of all outputs are averaged with uniform weight.
Returns:
mae : mean absolute error.
If multioutput is "raw_values", then mean absolute error is returned for each output separately. If multioutput is "uniform_average" or an ndarray of weights, then the weighted average of all output errors is returned.
MAE output is non-negative floating point. The best value is 0.0.
Examples:
>>> mae_metric = datasets.load_metric("mae")
>>> predictions = [2.5, 0.0, 2, 8]
>>> references = [3, -0.5, 2, 7]
>>> results = mae_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'mae': 0.5}
If you're using multi-dimensional lists, then set the config as follows :
>>> mae_metric = datasets.load_metric("mae", "multilist")
>>> predictions = [[0.5, 1], [-1, 1], [7, -6]]
>>> references = [[0, 2], [-1, 2], [8, -5]]
>>> results = mae_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'mae': 0.75}
>>> results = mae_metric.compute(predictions=predictions, references=references, multioutput='raw_values')
>>> print(results)
{'mae': array([0.5, 1. ])}
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Mae(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(self._get_feature_types()),
reference_urls=[
"https://scikit-learn.org/stable/modules/generated/sklearn.metrics.mean_absolute_error.html"
],
)
def _get_feature_types(self):
if self.config_name == "multilist":
return {
"predictions": datasets.Sequence(datasets.Value("float")),
"references": datasets.Sequence(datasets.Value("float")),
}
else:
return {
"predictions": datasets.Value("float"),
"references": datasets.Value("float"),
}
def _compute(self, predictions, references, sample_weight=None, multioutput="uniform_average"):
mae_score = mean_absolute_error(references, predictions, sample_weight=sample_weight, multioutput=multioutput)
return {"mae": mae_score}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/seqeval/README.md | # Metric Card for seqeval
## Metric description
seqeval is a Python framework for sequence labeling evaluation. seqeval can evaluate the performance of chunking tasks such as named-entity recognition, part-of-speech tagging, semantic role labeling and so on.
## How to use
Seqeval produces labelling scores along with its sufficient statistics from a source against one or more references.
It takes two mandatory arguments:
`predictions`: a list of lists of predicted labels, i.e. estimated targets as returned by a tagger.
`references`: a list of lists of reference labels, i.e. the ground truth/target values.
It can also take several optional arguments:
`suffix` (boolean): `True` if the IOB tag is a suffix (after type) instead of a prefix (before type), `False` otherwise. The default value is `False`, i.e. the IOB tag is a prefix (before type).
`scheme`: the target tagging scheme, which can be one of [`IOB1`, `IOB2`, `IOE1`, `IOE2`, `IOBES`, `BILOU`]. The default value is `None`.
`mode`: whether to count correct entity labels with incorrect I/B tags as true positives or not. If you want to only count exact matches, pass `mode="strict"` and a specific `scheme` value. The default is `None`.
`sample_weight`: An array-like of shape (n_samples,) that provides weights for individual samples. The default is `None`.
`zero_division`: Which value to substitute as a metric value when encountering zero division. Should be one of [`0`,`1`,`"warn"`]. `"warn"` acts as `0`, but the warning is raised.
```python
>>> from datasets import load_metric
>>> seqeval = load_metric('seqeval')
>>> predictions = [['O', 'O', 'B-MISC', 'I-MISC', 'I-MISC', 'I-MISC', 'O'], ['B-PER', 'I-PER', 'O']]
>>> references = [['O', 'O', 'O', 'B-MISC', 'I-MISC', 'I-MISC', 'O'], ['B-PER', 'I-PER', 'O']]
>>> results = seqeval.compute(predictions=predictions, references=references)
```
## Output values
This metric returns a dictionary with a summary of scores for overall and per type:
Overall:
`accuracy`: the average [accuracy](https://huggingface.co/metrics/accuracy), on a scale between 0.0 and 1.0.
`precision`: the average [precision](https://huggingface.co/metrics/precision), on a scale between 0.0 and 1.0.
`recall`: the average [recall](https://huggingface.co/metrics/recall), on a scale between 0.0 and 1.0.
`f1`: the average [F1 score](https://huggingface.co/metrics/f1), which is the harmonic mean of the precision and recall. It also has a scale of 0.0 to 1.0.
Per type (e.g. `MISC`, `PER`, `LOC`,...):
`precision`: the average [precision](https://huggingface.co/metrics/precision), on a scale between 0.0 and 1.0.
`recall`: the average [recall](https://huggingface.co/metrics/recall), on a scale between 0.0 and 1.0.
`f1`: the average [F1 score](https://huggingface.co/metrics/f1), on a scale between 0.0 and 1.0.
### Values from popular papers
The 1995 "Text Chunking using Transformation-Based Learning" [paper](https://aclanthology.org/W95-0107) reported a baseline recall of 81.9% and a precision of 78.2% using non Deep Learning-based methods.
More recently, seqeval continues being used for reporting performance on tasks such as [named entity detection](https://www.mdpi.com/2306-5729/6/8/84/htm) and [information extraction](https://ieeexplore.ieee.org/abstract/document/9697942/).
## Examples
Maximal values (full match) :
```python
>>> from datasets import load_metric
>>> seqeval = load_metric('seqeval')
>>> predictions = [['O', 'O', 'B-MISC', 'I-MISC', 'I-MISC', 'I-MISC', 'O'], ['B-PER', 'I-PER', 'O']]
>>> references = [['O', 'O', 'B-MISC', 'I-MISC', 'I-MISC', 'I-MISC', 'O'], ['B-PER', 'I-PER', 'O']]
>>> results = seqeval.compute(predictions=predictions, references=references)
>>> print(results)
{'MISC': {'precision': 1.0, 'recall': 1.0, 'f1': 1.0, 'number': 1}, 'PER': {'precision': 1.0, 'recall': 1.0, 'f1': 1.0, 'number': 1}, 'overall_precision': 1.0, 'overall_recall': 1.0, 'overall_f1': 1.0, 'overall_accuracy': 1.0}
```
Minimal values (no match):
```python
>>> from datasets import load_metric
>>> seqeval = load_metric('seqeval')
>>> predictions = [['O', 'B-MISC', 'I-MISC'], ['B-PER', 'I-PER', 'O']]
>>> references = [['B-MISC', 'O', 'O'], ['I-PER', '0', 'I-PER']]
>>> results = seqeval.compute(predictions=predictions, references=references)
>>> print(results)
{'MISC': {'precision': 0.0, 'recall': 0.0, 'f1': 0.0, 'number': 1}, 'PER': {'precision': 0.0, 'recall': 0.0, 'f1': 0.0, 'number': 2}, '_': {'precision': 0.0, 'recall': 0.0, 'f1': 0.0, 'number': 1}, 'overall_precision': 0.0, 'overall_recall': 0.0, 'overall_f1': 0.0, 'overall_accuracy': 0.0}
```
Partial match:
```python
>>> from datasets import load_metric
>>> seqeval = load_metric('seqeval')
>>> predictions = [['O', 'O', 'B-MISC', 'I-MISC', 'I-MISC', 'I-MISC', 'O'], ['B-PER', 'I-PER', 'O']]
>>> references = [['O', 'O', 'O', 'B-MISC', 'I-MISC', 'I-MISC', 'O'], ['B-PER', 'I-PER', 'O']]
>>> results = seqeval.compute(predictions=predictions, references=references)
>>> print(results)
{'MISC': {'precision': 0.0, 'recall': 0.0, 'f1': 0.0, 'number': 1}, 'PER': {'precision': 1.0, 'recall': 1.0, 'f1': 1.0, 'number': 1}, 'overall_precision': 0.5, 'overall_recall': 0.5, 'overall_f1': 0.5, 'overall_accuracy': 0.8}
```
## Limitations and bias
seqeval supports following IOB formats (short for inside, outside, beginning) : `IOB1`, `IOB2`, `IOE1`, `IOE2`, `IOBES`, `IOBES` (only in strict mode) and `BILOU` (only in strict mode).
For more information about IOB formats, refer to the [Wikipedia page](https://en.wikipedia.org/wiki/Inside%E2%80%93outside%E2%80%93beginning_(tagging)) and the description of the [CoNLL-2000 shared task](https://aclanthology.org/W02-2024).
## Citation
```bibtex
@inproceedings{ramshaw-marcus-1995-text,
title = "Text Chunking using Transformation-Based Learning",
author = "Ramshaw, Lance and
Marcus, Mitch",
booktitle = "Third Workshop on Very Large Corpora",
year = "1995",
url = "https://www.aclweb.org/anthology/W95-0107",
}
```
```bibtex
@misc{seqeval,
title={{seqeval}: A Python framework for sequence labeling evaluation},
url={https://github.com/chakki-works/seqeval},
note={Software available from https://github.com/chakki-works/seqeval},
author={Hiroki Nakayama},
year={2018},
}
```
## Further References
- [README for seqeval at GitHub](https://github.com/chakki-works/seqeval)
- [CoNLL-2000 shared task](https://www.clips.uantwerpen.be/conll2002/ner/bin/conlleval.txt)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/seqeval/seqeval.py | # Copyright 2020 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" seqeval metric. """
import importlib
from typing import List, Optional, Union
from seqeval.metrics import accuracy_score, classification_report
import datasets
_CITATION = """\
@inproceedings{ramshaw-marcus-1995-text,
title = "Text Chunking using Transformation-Based Learning",
author = "Ramshaw, Lance and
Marcus, Mitch",
booktitle = "Third Workshop on Very Large Corpora",
year = "1995",
url = "https://www.aclweb.org/anthology/W95-0107",
}
@misc{seqeval,
title={{seqeval}: A Python framework for sequence labeling evaluation},
url={https://github.com/chakki-works/seqeval},
note={Software available from https://github.com/chakki-works/seqeval},
author={Hiroki Nakayama},
year={2018},
}
"""
_DESCRIPTION = """\
seqeval is a Python framework for sequence labeling evaluation.
seqeval can evaluate the performance of chunking tasks such as named-entity recognition, part-of-speech tagging, semantic role labeling and so on.
This is well-tested by using the Perl script conlleval, which can be used for
measuring the performance of a system that has processed the CoNLL-2000 shared task data.
seqeval supports following formats:
IOB1
IOB2
IOE1
IOE2
IOBES
See the [README.md] file at https://github.com/chakki-works/seqeval for more information.
"""
_KWARGS_DESCRIPTION = """
Produces labelling scores along with its sufficient statistics
from a source against one or more references.
Args:
predictions: List of List of predicted labels (Estimated targets as returned by a tagger)
references: List of List of reference labels (Ground truth (correct) target values)
suffix: True if the IOB prefix is after type, False otherwise. default: False
scheme: Specify target tagging scheme. Should be one of ["IOB1", "IOB2", "IOE1", "IOE2", "IOBES", "BILOU"].
default: None
mode: Whether to count correct entity labels with incorrect I/B tags as true positives or not.
If you want to only count exact matches, pass mode="strict". default: None.
sample_weight: Array-like of shape (n_samples,), weights for individual samples. default: None
zero_division: Which value to substitute as a metric value when encountering zero division. Should be on of 0, 1,
"warn". "warn" acts as 0, but the warning is raised.
Returns:
'scores': dict. Summary of the scores for overall and per type
Overall:
'accuracy': accuracy,
'precision': precision,
'recall': recall,
'f1': F1 score, also known as balanced F-score or F-measure,
Per type:
'precision': precision,
'recall': recall,
'f1': F1 score, also known as balanced F-score or F-measure
Examples:
>>> predictions = [['O', 'O', 'B-MISC', 'I-MISC', 'I-MISC', 'I-MISC', 'O'], ['B-PER', 'I-PER', 'O']]
>>> references = [['O', 'O', 'O', 'B-MISC', 'I-MISC', 'I-MISC', 'O'], ['B-PER', 'I-PER', 'O']]
>>> seqeval = datasets.load_metric("seqeval")
>>> results = seqeval.compute(predictions=predictions, references=references)
>>> print(list(results.keys()))
['MISC', 'PER', 'overall_precision', 'overall_recall', 'overall_f1', 'overall_accuracy']
>>> print(results["overall_f1"])
0.5
>>> print(results["PER"]["f1"])
1.0
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Seqeval(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
homepage="https://github.com/chakki-works/seqeval",
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Sequence(datasets.Value("string", id="label"), id="sequence"),
"references": datasets.Sequence(datasets.Value("string", id="label"), id="sequence"),
}
),
codebase_urls=["https://github.com/chakki-works/seqeval"],
reference_urls=["https://github.com/chakki-works/seqeval"],
)
def _compute(
self,
predictions,
references,
suffix: bool = False,
scheme: Optional[str] = None,
mode: Optional[str] = None,
sample_weight: Optional[List[int]] = None,
zero_division: Union[str, int] = "warn",
):
if scheme is not None:
try:
scheme_module = importlib.import_module("seqeval.scheme")
scheme = getattr(scheme_module, scheme)
except AttributeError:
raise ValueError(f"Scheme should be one of [IOB1, IOB2, IOE1, IOE2, IOBES, BILOU], got {scheme}")
report = classification_report(
y_true=references,
y_pred=predictions,
suffix=suffix,
output_dict=True,
scheme=scheme,
mode=mode,
sample_weight=sample_weight,
zero_division=zero_division,
)
report.pop("macro avg")
report.pop("weighted avg")
overall_score = report.pop("micro avg")
scores = {
type_name: {
"precision": score["precision"],
"recall": score["recall"],
"f1": score["f1-score"],
"number": score["support"],
}
for type_name, score in report.items()
}
scores["overall_precision"] = overall_score["precision"]
scores["overall_recall"] = overall_score["recall"]
scores["overall_f1"] = overall_score["f1-score"]
scores["overall_accuracy"] = accuracy_score(y_true=references, y_pred=predictions)
return scores
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/sari/README.md | # Metric Card for SARI
## Metric description
SARI (***s**ystem output **a**gainst **r**eferences and against the **i**nput sentence*) is a metric used for evaluating automatic text simplification systems.
The metric compares the predicted simplified sentences against the reference and the source sentences. It explicitly measures the goodness of words that are added, deleted and kept by the system.
SARI can be computed as:
`sari = ( F1_add + F1_keep + P_del) / 3`
where
`F1_add` is the n-gram F1 score for add operations
`F1_keep` is the n-gram F1 score for keep operations
`P_del` is the n-gram precision score for delete operations
The number of n grams, `n`, is equal to 4, as in the original paper.
This implementation is adapted from [Tensorflow's tensor2tensor implementation](https://github.com/tensorflow/tensor2tensor/blob/master/tensor2tensor/utils/sari_hook.py).
It has two differences with the [original GitHub implementation](https://github.com/cocoxu/simplification/blob/master/SARI.py):
1) It defines 0/0=1 instead of 0 to give higher scores for predictions that match a target exactly.
2) It fixes an [alleged bug](https://github.com/cocoxu/simplification/issues/6) in the keep score computation.
## How to use
The metric takes 3 inputs: sources (a list of source sentence strings), predictions (a list of predicted sentence strings) and references (a list of lists of reference sentence strings)
```python
from datasets import load_metric
sari = load_metric("sari")
sources=["About 95 species are currently accepted."]
predictions=["About 95 you now get in."]
references=[["About 95 species are currently known.","About 95 species are now accepted.","95 species are now accepted."]]
sari_score = sari.compute(sources=sources, predictions=predictions, references=references)
```
## Output values
This metric outputs a dictionary with the SARI score:
```
print(sari_score)
{'sari': 26.953601953601954}
```
The range of values for the SARI score is between 0 and 100 -- the higher the value, the better the performance of the model being evaluated, with a SARI of 100 being a perfect score.
### Values from popular papers
The [original paper that proposes the SARI metric](https://aclanthology.org/Q16-1029.pdf) reports scores ranging from 26 to 43 for different simplification systems and different datasets. They also find that the metric ranks all of the simplification systems and human references in the same order as the human assessment used as a comparison, and that it correlates reasonably with human judgments.
More recent SARI scores for text simplification can be found on leaderboards for datasets such as [TurkCorpus](https://paperswithcode.com/sota/text-simplification-on-turkcorpus) and [Newsela](https://paperswithcode.com/sota/text-simplification-on-newsela).
## Examples
Perfect match between prediction and reference:
```python
from datasets import load_metric
sari = load_metric("sari")
sources=["About 95 species are currently accepted ."]
predictions=["About 95 species are currently accepted ."]
references=[["About 95 species are currently accepted ."]]
sari_score = sari.compute(sources=sources, predictions=predictions, references=references)
print(sari_score)
{'sari': 100.0}
```
Partial match between prediction and reference:
```python
from datasets import load_metric
sari = load_metric("sari")
sources=["About 95 species are currently accepted ."]
predictions=["About 95 you now get in ."]
references=[["About 95 species are currently known .","About 95 species are now accepted .","95 species are now accepted ."]]
sari_score = sari.compute(sources=sources, predictions=predictions, references=references)
print(sari_score)
{'sari': 26.953601953601954}
```
## Limitations and bias
SARI is a valuable measure for comparing different text simplification systems as well as one that can assist the iterative development of a system.
However, while the [original paper presenting SARI](https://aclanthology.org/Q16-1029.pdf) states that it captures "the notion of grammaticality and meaning preservation", this is a difficult claim to empirically validate.
## Citation
```bibtex
@inproceedings{xu-etal-2016-optimizing,
title = {Optimizing Statistical Machine Translation for Text Simplification},
authors={Xu, Wei and Napoles, Courtney and Pavlick, Ellie and Chen, Quanze and Callison-Burch, Chris},
journal = {Transactions of the Association for Computational Linguistics},
volume = {4},
year={2016},
url = {https://www.aclweb.org/anthology/Q16-1029},
pages = {401--415},
}
```
## Further References
- [NLP Progress -- Text Simplification](http://nlpprogress.com/english/simplification.html)
- [Hugging Face Hub -- Text Simplification Models](https://huggingface.co/datasets?filter=task_ids:text-simplification)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/sari/sari.py | # Copyright 2020 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" SARI metric."""
from collections import Counter
import sacrebleu
import sacremoses
from packaging import version
import datasets
_CITATION = """\
@inproceedings{xu-etal-2016-optimizing,
title = {Optimizing Statistical Machine Translation for Text Simplification},
authors={Xu, Wei and Napoles, Courtney and Pavlick, Ellie and Chen, Quanze and Callison-Burch, Chris},
journal = {Transactions of the Association for Computational Linguistics},
volume = {4},
year={2016},
url = {https://www.aclweb.org/anthology/Q16-1029},
pages = {401--415},
}
"""
_DESCRIPTION = """\
SARI is a metric used for evaluating automatic text simplification systems.
The metric compares the predicted simplified sentences against the reference
and the source sentences. It explicitly measures the goodness of words that are
added, deleted and kept by the system.
Sari = (F1_add + F1_keep + P_del) / 3
where
F1_add: n-gram F1 score for add operation
F1_keep: n-gram F1 score for keep operation
P_del: n-gram precision score for delete operation
n = 4, as in the original paper.
This implementation is adapted from Tensorflow's tensor2tensor implementation [3].
It has two differences with the original GitHub [1] implementation:
(1) Defines 0/0=1 instead of 0 to give higher scores for predictions that match
a target exactly.
(2) Fixes an alleged bug [2] in the keep score computation.
[1] https://github.com/cocoxu/simplification/blob/master/SARI.py
(commit 0210f15)
[2] https://github.com/cocoxu/simplification/issues/6
[3] https://github.com/tensorflow/tensor2tensor/blob/master/tensor2tensor/utils/sari_hook.py
"""
_KWARGS_DESCRIPTION = """
Calculates sari score (between 0 and 100) given a list of source and predicted
sentences, and a list of lists of reference sentences.
Args:
sources: list of source sentences where each sentence should be a string.
predictions: list of predicted sentences where each sentence should be a string.
references: list of lists of reference sentences where each sentence should be a string.
Returns:
sari: sari score
Examples:
>>> sources=["About 95 species are currently accepted ."]
>>> predictions=["About 95 you now get in ."]
>>> references=[["About 95 species are currently known .","About 95 species are now accepted .","95 species are now accepted ."]]
>>> sari = datasets.load_metric("sari")
>>> results = sari.compute(sources=sources, predictions=predictions, references=references)
>>> print(results)
{'sari': 26.953601953601954}
"""
def SARIngram(sgrams, cgrams, rgramslist, numref):
rgramsall = [rgram for rgrams in rgramslist for rgram in rgrams]
rgramcounter = Counter(rgramsall)
sgramcounter = Counter(sgrams)
sgramcounter_rep = Counter()
for sgram, scount in sgramcounter.items():
sgramcounter_rep[sgram] = scount * numref
cgramcounter = Counter(cgrams)
cgramcounter_rep = Counter()
for cgram, ccount in cgramcounter.items():
cgramcounter_rep[cgram] = ccount * numref
# KEEP
keepgramcounter_rep = sgramcounter_rep & cgramcounter_rep
keepgramcountergood_rep = keepgramcounter_rep & rgramcounter
keepgramcounterall_rep = sgramcounter_rep & rgramcounter
keeptmpscore1 = 0
keeptmpscore2 = 0
for keepgram in keepgramcountergood_rep:
keeptmpscore1 += keepgramcountergood_rep[keepgram] / keepgramcounter_rep[keepgram]
# Fix an alleged bug [2] in the keep score computation.
# keeptmpscore2 += keepgramcountergood_rep[keepgram] / keepgramcounterall_rep[keepgram]
keeptmpscore2 += keepgramcountergood_rep[keepgram]
# Define 0/0=1 instead of 0 to give higher scores for predictions that match
# a target exactly.
keepscore_precision = 1
keepscore_recall = 1
if len(keepgramcounter_rep) > 0:
keepscore_precision = keeptmpscore1 / len(keepgramcounter_rep)
if len(keepgramcounterall_rep) > 0:
# Fix an alleged bug [2] in the keep score computation.
# keepscore_recall = keeptmpscore2 / len(keepgramcounterall_rep)
keepscore_recall = keeptmpscore2 / sum(keepgramcounterall_rep.values())
keepscore = 0
if keepscore_precision > 0 or keepscore_recall > 0:
keepscore = 2 * keepscore_precision * keepscore_recall / (keepscore_precision + keepscore_recall)
# DELETION
delgramcounter_rep = sgramcounter_rep - cgramcounter_rep
delgramcountergood_rep = delgramcounter_rep - rgramcounter
delgramcounterall_rep = sgramcounter_rep - rgramcounter
deltmpscore1 = 0
deltmpscore2 = 0
for delgram in delgramcountergood_rep:
deltmpscore1 += delgramcountergood_rep[delgram] / delgramcounter_rep[delgram]
deltmpscore2 += delgramcountergood_rep[delgram] / delgramcounterall_rep[delgram]
# Define 0/0=1 instead of 0 to give higher scores for predictions that match
# a target exactly.
delscore_precision = 1
if len(delgramcounter_rep) > 0:
delscore_precision = deltmpscore1 / len(delgramcounter_rep)
# ADDITION
addgramcounter = set(cgramcounter) - set(sgramcounter)
addgramcountergood = set(addgramcounter) & set(rgramcounter)
addgramcounterall = set(rgramcounter) - set(sgramcounter)
addtmpscore = 0
for addgram in addgramcountergood:
addtmpscore += 1
# Define 0/0=1 instead of 0 to give higher scores for predictions that match
# a target exactly.
addscore_precision = 1
addscore_recall = 1
if len(addgramcounter) > 0:
addscore_precision = addtmpscore / len(addgramcounter)
if len(addgramcounterall) > 0:
addscore_recall = addtmpscore / len(addgramcounterall)
addscore = 0
if addscore_precision > 0 or addscore_recall > 0:
addscore = 2 * addscore_precision * addscore_recall / (addscore_precision + addscore_recall)
return (keepscore, delscore_precision, addscore)
def SARIsent(ssent, csent, rsents):
numref = len(rsents)
s1grams = ssent.split(" ")
c1grams = csent.split(" ")
s2grams = []
c2grams = []
s3grams = []
c3grams = []
s4grams = []
c4grams = []
r1gramslist = []
r2gramslist = []
r3gramslist = []
r4gramslist = []
for rsent in rsents:
r1grams = rsent.split(" ")
r2grams = []
r3grams = []
r4grams = []
r1gramslist.append(r1grams)
for i in range(0, len(r1grams) - 1):
if i < len(r1grams) - 1:
r2gram = r1grams[i] + " " + r1grams[i + 1]
r2grams.append(r2gram)
if i < len(r1grams) - 2:
r3gram = r1grams[i] + " " + r1grams[i + 1] + " " + r1grams[i + 2]
r3grams.append(r3gram)
if i < len(r1grams) - 3:
r4gram = r1grams[i] + " " + r1grams[i + 1] + " " + r1grams[i + 2] + " " + r1grams[i + 3]
r4grams.append(r4gram)
r2gramslist.append(r2grams)
r3gramslist.append(r3grams)
r4gramslist.append(r4grams)
for i in range(0, len(s1grams) - 1):
if i < len(s1grams) - 1:
s2gram = s1grams[i] + " " + s1grams[i + 1]
s2grams.append(s2gram)
if i < len(s1grams) - 2:
s3gram = s1grams[i] + " " + s1grams[i + 1] + " " + s1grams[i + 2]
s3grams.append(s3gram)
if i < len(s1grams) - 3:
s4gram = s1grams[i] + " " + s1grams[i + 1] + " " + s1grams[i + 2] + " " + s1grams[i + 3]
s4grams.append(s4gram)
for i in range(0, len(c1grams) - 1):
if i < len(c1grams) - 1:
c2gram = c1grams[i] + " " + c1grams[i + 1]
c2grams.append(c2gram)
if i < len(c1grams) - 2:
c3gram = c1grams[i] + " " + c1grams[i + 1] + " " + c1grams[i + 2]
c3grams.append(c3gram)
if i < len(c1grams) - 3:
c4gram = c1grams[i] + " " + c1grams[i + 1] + " " + c1grams[i + 2] + " " + c1grams[i + 3]
c4grams.append(c4gram)
(keep1score, del1score, add1score) = SARIngram(s1grams, c1grams, r1gramslist, numref)
(keep2score, del2score, add2score) = SARIngram(s2grams, c2grams, r2gramslist, numref)
(keep3score, del3score, add3score) = SARIngram(s3grams, c3grams, r3gramslist, numref)
(keep4score, del4score, add4score) = SARIngram(s4grams, c4grams, r4gramslist, numref)
avgkeepscore = sum([keep1score, keep2score, keep3score, keep4score]) / 4
avgdelscore = sum([del1score, del2score, del3score, del4score]) / 4
avgaddscore = sum([add1score, add2score, add3score, add4score]) / 4
finalscore = (avgkeepscore + avgdelscore + avgaddscore) / 3
return finalscore
def normalize(sentence, lowercase: bool = True, tokenizer: str = "13a", return_str: bool = True):
# Normalization is requried for the ASSET dataset (one of the primary
# datasets in sentence simplification) to allow using space
# to split the sentence. Even though Wiki-Auto and TURK datasets,
# do not require normalization, we do it for consistency.
# Code adapted from the EASSE library [1] written by the authors of the ASSET dataset.
# [1] https://github.com/feralvam/easse/blob/580bba7e1378fc8289c663f864e0487188fe8067/easse/utils/preprocessing.py#L7
if lowercase:
sentence = sentence.lower()
if tokenizer in ["13a", "intl"]:
if version.parse(sacrebleu.__version__).major >= 2:
normalized_sent = sacrebleu.metrics.bleu._get_tokenizer(tokenizer)()(sentence)
else:
normalized_sent = sacrebleu.TOKENIZERS[tokenizer]()(sentence)
elif tokenizer == "moses":
normalized_sent = sacremoses.MosesTokenizer().tokenize(sentence, return_str=True, escape=False)
elif tokenizer == "penn":
normalized_sent = sacremoses.MosesTokenizer().penn_tokenize(sentence, return_str=True)
else:
normalized_sent = sentence
if not return_str:
normalized_sent = normalized_sent.split()
return normalized_sent
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Sari(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"sources": datasets.Value("string", id="sequence"),
"predictions": datasets.Value("string", id="sequence"),
"references": datasets.Sequence(datasets.Value("string", id="sequence"), id="references"),
}
),
codebase_urls=[
"https://github.com/cocoxu/simplification/blob/master/SARI.py",
"https://github.com/tensorflow/tensor2tensor/blob/master/tensor2tensor/utils/sari_hook.py",
],
reference_urls=["https://www.aclweb.org/anthology/Q16-1029.pdf"],
)
def _compute(self, sources, predictions, references):
if not (len(sources) == len(predictions) == len(references)):
raise ValueError("Sources length must match predictions and references lengths.")
sari_score = 0
for src, pred, refs in zip(sources, predictions, references):
sari_score += SARIsent(normalize(src), normalize(pred), [normalize(sent) for sent in refs])
sari_score = sari_score / len(predictions)
return {"sari": 100 * sari_score}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/coval/README.md | # Metric Card for COVAL
## Metric description
CoVal is a coreference evaluation tool for the [CoNLL](https://huggingface.co/datasets/conll2003) and [ARRAU](https://catalog.ldc.upenn.edu/LDC2013T22) datasets which implements of the common evaluation metrics including MUC [Vilain et al, 1995](https://aclanthology.org/M95-1005.pdf), B-cubed [Bagga and Baldwin, 1998](https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.34.2578&rep=rep1&type=pdf), CEAFe [Luo et al., 2005](https://aclanthology.org/H05-1004.pdf), LEA [Moosavi and Strube, 2016](https://aclanthology.org/P16-1060.pdf) and the averaged CoNLL score (the average of the F1 values of MUC, B-cubed and CEAFe).
CoVal code was written by [`@ns-moosavi`](https://github.com/ns-moosavi), with some parts borrowed from [Deep Coref](https://github.com/clarkkev/deep-coref/blob/master/evaluation.py). The test suite is taken from the [official CoNLL code](https://github.com/conll/reference-coreference-scorers/), with additions by [`@andreasvc`](https://github.com/andreasvc) and file parsing developed by Leo Born.
## How to use
The metric takes two lists of sentences as input: one representing `predictions` and `references`, with the sentences consisting of words in the CoNLL format (see the [Limitations and bias](#Limitations-and-bias) section below for more details on the CoNLL format).
```python
from datasets import load_metric
coval = load_metric('coval')
words = ['bc/cctv/00/cctv_0005 0 0 Thank VBP (TOP(S(VP* thank 01 1 Xu_li * (V*) * -',
... 'bc/cctv/00/cctv_0005 0 1 you PRP (NP*) - - - Xu_li * (ARG1*) (ARG0*) (116)',
... 'bc/cctv/00/cctv_0005 0 2 everyone NN (NP*) - - - Xu_li * (ARGM-DIS*) * (116)',
... 'bc/cctv/00/cctv_0005 0 3 for IN (PP* - - - Xu_li * (ARG2* * -',
... 'bc/cctv/00/cctv_0005 0 4 watching VBG (S(VP*)))) watch 01 1 Xu_li * *) (V*) -',
... 'bc/cctv/00/cctv_0005 0 5 . . *)) - - - Xu_li * * * -']
references = [words]
predictions = [words]
results = coval.compute(predictions=predictions, references=references)
```
It also has several optional arguments:
`keep_singletons`: After extracting all mentions of key or system file mentions whose corresponding coreference chain is of size one are considered as singletons. The default evaluation mode will include singletons in evaluations if they are included in the key or the system files. By setting `keep_singletons=False`, all singletons in the key and system files will be excluded from the evaluation.
`NP_only`: Most of the recent coreference resolvers only resolve NP mentions and leave out the resolution of VPs. By setting the `NP_only` option, the scorer will only evaluate the resolution of NPs.
`min_span`: By setting `min_span`, the scorer reports the results based on automatically detected minimum spans. Minimum spans are determined using the [MINA algorithm](https://arxiv.org/pdf/1906.06703.pdf).
## Output values
The metric outputs a dictionary with the following key-value pairs:
`mentions`: number of mentions, ranges from 0-1
`muc`: MUC metric, which expresses performance in terms of recall and precision, ranging from 0-1.
`bcub`: B-cubed metric, which is the averaged precision of all items in the distribution, ranging from 0-1.
`ceafe`: CEAFe (Constrained Entity Alignment F-Measure) is computed by aligning reference and system entities with the constraint that a reference entity is aligned with at most one reference entity. It ranges from 0-1
`lea`: LEA is a Link-Based Entity-Aware metric which, for each entity, considers how important the entity is and how well it is resolved. It ranges from 0-1.
`conll_score`: averaged CoNLL score (the average of the F1 values of `muc`, `bcub` and `ceafe`), ranging from 0 to 100.
### Values from popular papers
Given that many of the metrics returned by COVAL come from different sources, is it hard to cite reference values for all of them.
The CoNLL score is used to track progress on different datasets such as the [ARRAU corpus](https://paperswithcode.com/sota/coreference-resolution-on-the-arrau-corpus) and [CoNLL 2012](https://paperswithcode.com/sota/coreference-resolution-on-conll-2012).
## Examples
Maximal values
```python
from datasets import load_metric
coval = load_metric('coval')
words = ['bc/cctv/00/cctv_0005 0 0 Thank VBP (TOP(S(VP* thank 01 1 Xu_li * (V*) * -',
... 'bc/cctv/00/cctv_0005 0 1 you PRP (NP*) - - - Xu_li * (ARG1*) (ARG0*) (116)',
... 'bc/cctv/00/cctv_0005 0 2 everyone NN (NP*) - - - Xu_li * (ARGM-DIS*) * (116)',
... 'bc/cctv/00/cctv_0005 0 3 for IN (PP* - - - Xu_li * (ARG2* * -',
... 'bc/cctv/00/cctv_0005 0 4 watching VBG (S(VP*)))) watch 01 1 Xu_li * *) (V*) -',
... 'bc/cctv/00/cctv_0005 0 5 . . *)) - - - Xu_li * * * -']
references = [words]
predictions = [words]
results = coval.compute(predictions=predictions, references=references)
print(results)
{'mentions/recall': 1.0, 'mentions/precision': 1.0, 'mentions/f1': 1.0, 'muc/recall': 1.0, 'muc/precision': 1.0, 'muc/f1': 1.0, 'bcub/recall': 1.0, 'bcub/precision': 1.0, 'bcub/f1': 1.0, 'ceafe/recall': 1.0, 'ceafe/precision': 1.0, 'ceafe/f1': 1.0, 'lea/recall': 1.0, 'lea/precision': 1.0, 'lea/f1': 1.0, 'conll_score': 100.0}
```
## Limitations and bias
This wrapper of CoVal currently only works with [CoNLL line format](https://huggingface.co/datasets/conll2003), which has one word per line with all the annotation for this word in column separated by spaces:
| Column | Type | Description |
|:-------|:----------------------|:--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|
| 1 | Document ID | This is a variation on the document filename |
| 2 | Part number | Some files are divided into multiple parts numbered as 000, 001, 002, ... etc. |
| 3 | Word number | |
| 4 | Word | This is the token as segmented/tokenized in the Treebank. Initially the *_skel file contain the placeholder [WORD] which gets replaced by the actual token from the Treebank which is part of the OntoNotes release. |
| 5 | Part-of-Speech | |
| 6 | Parse bit | This is the bracketed structure broken before the first open parenthesis in the parse, and the word/part-of-speech leaf replaced with a *. The full parse can be created by substituting the asterix with the "([pos] [word])" string (or leaf) and concatenating the items in the rows of that column. |
| 7 | Predicate lemma | The predicate lemma is mentioned for the rows for which we have semantic role information. All other rows are marked with a "-". |
| 8 | Predicate Frameset ID | This is the PropBank frameset ID of the predicate in Column 7. |
| 9 | Word sense | This is the word sense of the word in Column 3. |
| 10 | Speaker/Author | This is the speaker or author name where available. Mostly in Broadcast Conversation and Web Log data. |
| 11 | Named Entities | These columns identifies the spans representing various named entities. |
| 12:N | Predicate Arguments | There is one column each of predicate argument structure information for the predicate mentioned in Column 7. |
| N | Coreference | Coreference chain information encoded in a parenthesis structure. |
## Citations
```bibtex
@InProceedings{moosavi2019minimum,
author = { Nafise Sadat Moosavi, Leo Born, Massimo Poesio and Michael Strube},
title = {Using Automatically Extracted Minimum Spans to Disentangle Coreference Evaluation from Boundary Detection},
year = {2019},
booktitle = {Proceedings of the 57th Annual Meeting of
the Association for Computational Linguistics (Volume 1: Long Papers)},
publisher = {Association for Computational Linguistics},
address = {Florence, Italy},
}
```
```bibtex
@inproceedings{10.3115/1072399.1072405,
author = {Vilain, Marc and Burger, John and Aberdeen, John and Connolly, Dennis and Hirschman, Lynette},
title = {A Model-Theoretic Coreference Scoring Scheme},
year = {1995},
isbn = {1558604022},
publisher = {Association for Computational Linguistics},
address = {USA},
url = {https://doi.org/10.3115/1072399.1072405},
doi = {10.3115/1072399.1072405},
booktitle = {Proceedings of the 6th Conference on Message Understanding},
pages = {45–52},
numpages = {8},
location = {Columbia, Maryland},
series = {MUC6 ’95}
}
```
```bibtex
@INPROCEEDINGS{Bagga98algorithmsfor,
author = {Amit Bagga and Breck Baldwin},
title = {Algorithms for Scoring Coreference Chains},
booktitle = {In The First International Conference on Language Resources and Evaluation Workshop on Linguistics Coreference},
year = {1998},
pages = {563--566}
}
```
```bibtex
@INPROCEEDINGS{Luo05oncoreference,
author = {Xiaoqiang Luo},
title = {On coreference resolution performance metrics},
booktitle = {In Proc. of HLT/EMNLP},
year = {2005},
pages = {25--32},
publisher = {URL}
}
```
```bibtex
@inproceedings{moosavi-strube-2016-coreference,
title = "Which Coreference Evaluation Metric Do You Trust? A Proposal for a Link-based Entity Aware Metric",
author = "Moosavi, Nafise Sadat and
Strube, Michael",
booktitle = "Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers)",
month = aug,
year = "2016",
address = "Berlin, Germany",
publisher = "Association for Computational Linguistics",
url = "https://www.aclweb.org/anthology/P16-1060",
doi = "10.18653/v1/P16-1060",
pages = "632--642",
}
```
## Further References
- [CoNLL 2012 Task Description](http://www.conll.cemantix.org/2012/data.html): for information on the format (section "*_conll File Format")
- [CoNLL Evaluation details](https://github.com/ns-moosavi/coval/blob/master/conll/README.md)
- [Hugging Face - Neural Coreference Resolution (Neuralcoref)](https://huggingface.co/coref/)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/coval/coval.py | # Copyright 2020 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" CoVal metric. """
import coval # From: git+https://github.com/ns-moosavi/coval.git # noqa: F401
from coval.conll import reader, util
from coval.eval import evaluator
import datasets
logger = datasets.logging.get_logger(__name__)
_CITATION = """\
@InProceedings{moosavi2019minimum,
author = { Nafise Sadat Moosavi, Leo Born, Massimo Poesio and Michael Strube},
title = {Using Automatically Extracted Minimum Spans to Disentangle Coreference Evaluation from Boundary Detection},
year = {2019},
booktitle = {Proceedings of the 57th Annual Meeting of
the Association for Computational Linguistics (Volume 1: Long Papers)},
publisher = {Association for Computational Linguistics},
address = {Florence, Italy},
}
@inproceedings{10.3115/1072399.1072405,
author = {Vilain, Marc and Burger, John and Aberdeen, John and Connolly, Dennis and Hirschman, Lynette},
title = {A Model-Theoretic Coreference Scoring Scheme},
year = {1995},
isbn = {1558604022},
publisher = {Association for Computational Linguistics},
address = {USA},
url = {https://doi.org/10.3115/1072399.1072405},
doi = {10.3115/1072399.1072405},
booktitle = {Proceedings of the 6th Conference on Message Understanding},
pages = {45–52},
numpages = {8},
location = {Columbia, Maryland},
series = {MUC6 ’95}
}
@INPROCEEDINGS{Bagga98algorithmsfor,
author = {Amit Bagga and Breck Baldwin},
title = {Algorithms for Scoring Coreference Chains},
booktitle = {In The First International Conference on Language Resources and Evaluation Workshop on Linguistics Coreference},
year = {1998},
pages = {563--566}
}
@INPROCEEDINGS{Luo05oncoreference,
author = {Xiaoqiang Luo},
title = {On coreference resolution performance metrics},
booktitle = {In Proc. of HLT/EMNLP},
year = {2005},
pages = {25--32},
publisher = {URL}
}
@inproceedings{moosavi-strube-2016-coreference,
title = "Which Coreference Evaluation Metric Do You Trust? A Proposal for a Link-based Entity Aware Metric",
author = "Moosavi, Nafise Sadat and
Strube, Michael",
booktitle = "Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers)",
month = aug,
year = "2016",
address = "Berlin, Germany",
publisher = "Association for Computational Linguistics",
url = "https://www.aclweb.org/anthology/P16-1060",
doi = "10.18653/v1/P16-1060",
pages = "632--642",
}
"""
_DESCRIPTION = """\
CoVal is a coreference evaluation tool for the CoNLL and ARRAU datasets which
implements of the common evaluation metrics including MUC [Vilain et al, 1995],
B-cubed [Bagga and Baldwin, 1998], CEAFe [Luo et al., 2005],
LEA [Moosavi and Strube, 2016] and the averaged CoNLL score
(the average of the F1 values of MUC, B-cubed and CEAFe)
[Denis and Baldridge, 2009a; Pradhan et al., 2011].
This wrapper of CoVal currently only work with CoNLL line format:
The CoNLL format has one word per line with all the annotation for this word in column separated by spaces:
Column Type Description
1 Document ID This is a variation on the document filename
2 Part number Some files are divided into multiple parts numbered as 000, 001, 002, ... etc.
3 Word number
4 Word itself This is the token as segmented/tokenized in the Treebank. Initially the *_skel file contain the placeholder [WORD] which gets replaced by the actual token from the Treebank which is part of the OntoNotes release.
5 Part-of-Speech
6 Parse bit This is the bracketed structure broken before the first open parenthesis in the parse, and the word/part-of-speech leaf replaced with a *. The full parse can be created by substituting the asterix with the "([pos] [word])" string (or leaf) and concatenating the items in the rows of that column.
7 Predicate lemma The predicate lemma is mentioned for the rows for which we have semantic role information. All other rows are marked with a "-"
8 Predicate Frameset ID This is the PropBank frameset ID of the predicate in Column 7.
9 Word sense This is the word sense of the word in Column 3.
10 Speaker/Author This is the speaker or author name where available. Mostly in Broadcast Conversation and Web Log data.
11 Named Entities These columns identifies the spans representing various named entities.
12:N Predicate Arguments There is one column each of predicate argument structure information for the predicate mentioned in Column 7.
N Coreference Coreference chain information encoded in a parenthesis structure.
More informations on the format can be found here (section "*_conll File Format"): http://www.conll.cemantix.org/2012/data.html
Details on the evaluation on CoNLL can be found here: https://github.com/ns-moosavi/coval/blob/master/conll/README.md
CoVal code was written by @ns-moosavi.
Some parts are borrowed from https://github.com/clarkkev/deep-coref/blob/master/evaluation.py
The test suite is taken from https://github.com/conll/reference-coreference-scorers/
Mention evaluation and the test suite are added by @andreasvc.
Parsing CoNLL files is developed by Leo Born.
"""
_KWARGS_DESCRIPTION = """
Calculates coreference evaluation metrics.
Args:
predictions: list of sentences. Each sentence is a list of word predictions to score in the CoNLL format.
Each prediction is a word with its annotations as a string made of columns joined with spaces.
Only columns 4, 5, 6 and the last column are used (word, POS, Pars and coreference annotation)
See the details on the format in the description of the metric.
references: list of sentences. Each sentence is a list of word reference to score in the CoNLL format.
Each reference is a word with its annotations as a string made of columns joined with spaces.
Only columns 4, 5, 6 and the last column are used (word, POS, Pars and coreference annotation)
See the details on the format in the description of the metric.
keep_singletons: After extracting all mentions of key or system files,
mentions whose corresponding coreference chain is of size one,
are considered as singletons. The default evaluation mode will include
singletons in evaluations if they are included in the key or the system files.
By setting 'keep_singletons=False', all singletons in the key and system files
will be excluded from the evaluation.
NP_only: Most of the recent coreference resolvers only resolve NP mentions and
leave out the resolution of VPs. By setting the 'NP_only' option, the scorer will only evaluate the resolution of NPs.
min_span: By setting 'min_span', the scorer reports the results based on automatically detected minimum spans.
Minimum spans are determined using the MINA algorithm.
Returns:
'mentions': mentions
'muc': MUC metric [Vilain et al, 1995]
'bcub': B-cubed [Bagga and Baldwin, 1998]
'ceafe': CEAFe [Luo et al., 2005]
'lea': LEA [Moosavi and Strube, 2016]
'conll_score': averaged CoNLL score (the average of the F1 values of MUC, B-cubed and CEAFe)
Examples:
>>> coval = datasets.load_metric('coval')
>>> words = ['bc/cctv/00/cctv_0005 0 0 Thank VBP (TOP(S(VP* thank 01 1 Xu_li * (V*) * -',
... 'bc/cctv/00/cctv_0005 0 1 you PRP (NP*) - - - Xu_li * (ARG1*) (ARG0*) (116)',
... 'bc/cctv/00/cctv_0005 0 2 everyone NN (NP*) - - - Xu_li * (ARGM-DIS*) * (116)',
... 'bc/cctv/00/cctv_0005 0 3 for IN (PP* - - - Xu_li * (ARG2* * -',
... 'bc/cctv/00/cctv_0005 0 4 watching VBG (S(VP*)))) watch 01 1 Xu_li * *) (V*) -',
... 'bc/cctv/00/cctv_0005 0 5 . . *)) - - - Xu_li * * * -']
>>> references = [words]
>>> predictions = [words]
>>> results = coval.compute(predictions=predictions, references=references)
>>> print(results) # doctest:+ELLIPSIS
{'mentions/recall': 1.0,[...] 'conll_score': 100.0}
"""
def get_coref_infos(
key_lines, sys_lines, NP_only=False, remove_nested=False, keep_singletons=True, min_span=False, doc="dummy_doc"
):
key_doc_lines = {doc: key_lines}
sys_doc_lines = {doc: sys_lines}
doc_coref_infos = {}
key_nested_coref_num = 0
sys_nested_coref_num = 0
key_removed_nested_clusters = 0
sys_removed_nested_clusters = 0
key_singletons_num = 0
sys_singletons_num = 0
key_clusters, singletons_num = reader.get_doc_mentions(doc, key_doc_lines[doc], keep_singletons)
key_singletons_num += singletons_num
if NP_only or min_span:
key_clusters = reader.set_annotated_parse_trees(key_clusters, key_doc_lines[doc], NP_only, min_span)
sys_clusters, singletons_num = reader.get_doc_mentions(doc, sys_doc_lines[doc], keep_singletons)
sys_singletons_num += singletons_num
if NP_only or min_span:
sys_clusters = reader.set_annotated_parse_trees(sys_clusters, key_doc_lines[doc], NP_only, min_span)
if remove_nested:
nested_mentions, removed_clusters = reader.remove_nested_coref_mentions(key_clusters, keep_singletons)
key_nested_coref_num += nested_mentions
key_removed_nested_clusters += removed_clusters
nested_mentions, removed_clusters = reader.remove_nested_coref_mentions(sys_clusters, keep_singletons)
sys_nested_coref_num += nested_mentions
sys_removed_nested_clusters += removed_clusters
sys_mention_key_cluster = reader.get_mention_assignments(sys_clusters, key_clusters)
key_mention_sys_cluster = reader.get_mention_assignments(key_clusters, sys_clusters)
doc_coref_infos[doc] = (key_clusters, sys_clusters, key_mention_sys_cluster, sys_mention_key_cluster)
if remove_nested:
logger.info(
"Number of removed nested coreferring mentions in the key "
f"annotation: {key_nested_coref_num}; and system annotation: {sys_nested_coref_num}"
)
logger.info(
"Number of resulting singleton clusters in the key "
f"annotation: {key_removed_nested_clusters}; and system annotation: {sys_removed_nested_clusters}"
)
if not keep_singletons:
logger.info(
f"{key_singletons_num:d} and {sys_singletons_num:d} singletons are removed from the key and system "
"files, respectively"
)
return doc_coref_infos
def evaluate(key_lines, sys_lines, metrics, NP_only, remove_nested, keep_singletons, min_span):
doc_coref_infos = get_coref_infos(key_lines, sys_lines, NP_only, remove_nested, keep_singletons, min_span)
output_scores = {}
conll = 0
conll_subparts_num = 0
for name, metric in metrics:
recall, precision, f1 = evaluator.evaluate_documents(doc_coref_infos, metric, beta=1)
if name in ["muc", "bcub", "ceafe"]:
conll += f1
conll_subparts_num += 1
output_scores.update({f"{name}/recall": recall, f"{name}/precision": precision, f"{name}/f1": f1})
logger.info(
name.ljust(10),
f"Recall: {recall * 100:.2f}",
f" Precision: {precision * 100:.2f}",
f" F1: {f1 * 100:.2f}",
)
if conll_subparts_num == 3:
conll = (conll / 3) * 100
logger.info(f"CoNLL score: {conll:.2f}")
output_scores.update({"conll_score": conll})
return output_scores
def check_gold_parse_annotation(key_lines):
has_gold_parse = False
for line in key_lines:
if not line.startswith("#"):
if len(line.split()) > 6:
parse_col = line.split()[5]
if not parse_col == "-":
has_gold_parse = True
break
else:
break
return has_gold_parse
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Coval(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Sequence(datasets.Value("string")),
"references": datasets.Sequence(datasets.Value("string")),
}
),
codebase_urls=["https://github.com/ns-moosavi/coval"],
reference_urls=[
"https://github.com/ns-moosavi/coval",
"https://www.aclweb.org/anthology/P16-1060",
"http://www.conll.cemantix.org/2012/data.html",
],
)
def _compute(
self, predictions, references, keep_singletons=True, NP_only=False, min_span=False, remove_nested=False
):
allmetrics = [
("mentions", evaluator.mentions),
("muc", evaluator.muc),
("bcub", evaluator.b_cubed),
("ceafe", evaluator.ceafe),
("lea", evaluator.lea),
]
if min_span:
has_gold_parse = util.check_gold_parse_annotation(references)
if not has_gold_parse:
raise NotImplementedError("References should have gold parse annotation to use 'min_span'.")
# util.parse_key_file(key_file)
# key_file = key_file + ".parsed"
score = evaluate(
key_lines=references,
sys_lines=predictions,
metrics=allmetrics,
NP_only=NP_only,
remove_nested=remove_nested,
keep_singletons=keep_singletons,
min_span=min_span,
)
return score
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/meteor/README.md | # Metric Card for METEOR
## Metric description
METEOR (Metric for Evaluation of Translation with Explicit ORdering) is a machine translation evaluation metric, which is calculated based on the harmonic mean of precision and recall, with recall weighted more than precision.
METEOR is based on a generalized concept of unigram matching between the machine-produced translation and human-produced reference translations. Unigrams can be matched based on their surface forms, stemmed forms, and meanings. Once all generalized unigram matches between the two strings have been found, METEOR computes a score for this matching using a combination of unigram-precision, unigram-recall, and a measure of fragmentation that is designed to directly capture how well-ordered the matched words in the machine translation are in relation to the reference.
## How to use
METEOR has two mandatory arguments:
`predictions`: a list of predictions to score. Each prediction should be a string with tokens separated by spaces.
`references`: a list of references for each prediction. Each reference should be a string with tokens separated by spaces.
It also has several optional parameters:
`alpha`: Parameter for controlling relative weights of precision and recall. The default value is `0.9`.
`beta`: Parameter for controlling shape of penalty as a function of fragmentation. The default value is `3`.
`gamma`: The relative weight assigned to fragmentation penalty. The default is `0.5`.
Refer to the [METEOR paper](https://aclanthology.org/W05-0909.pdf) for more information about parameter values and ranges.
```python
>>> from datasets import load_metric
>>> meteor = load_metric('meteor')
>>> predictions = ["It is a guide to action which ensures that the military always obeys the commands of the party"]
>>> references = ["It is a guide to action that ensures that the military will forever heed Party commands"]
>>> results = meteor.compute(predictions=predictions, references=references)
```
## Output values
The metric outputs a dictionary containing the METEOR score. Its values range from 0 to 1.
### Values from popular papers
The [METEOR paper](https://aclanthology.org/W05-0909.pdf) does not report METEOR score values for different models, but it does report that METEOR gets an R correlation value of 0.347 with human evaluation on the Arabic data and 0.331 on the Chinese data.
## Examples
Maximal values :
```python
>>> from datasets import load_metric
>>> meteor = load_metric('meteor')
>>> predictions = ["It is a guide to action which ensures that the military always obeys the commands of the party"]
>>> references = ["It is a guide to action which ensures that the military always obeys the commands of the party"]
>>> results = meteor.compute(predictions=predictions, references=references)
>>> print(round(results['meteor'], 2))
1.0
```
Minimal values:
```python
>>> from datasets import load_metric
>>> meteor = load_metric('meteor')
>>> predictions = ["It is a guide to action which ensures that the military always obeys the commands of the party"]
>>> references = ["Hello world"]
>>> results = meteor.compute(predictions=predictions, references=references)
>>> print(round(results['meteor'], 2))
0.0
```
Partial match:
```python
>>> from datasets import load_metric
>>> meteor = load_metric('meteor')
>>> predictions = ["It is a guide to action which ensures that the military always obeys the commands of the party"]
>>> references = ["It is a guide to action that ensures that the military will forever heed Party commands"]
>>> results = meteor.compute(predictions=predictions, references=references)
>>> print(round(results['meteor'], 2))
0.69
```
## Limitations and bias
While the correlation between METEOR and human judgments was measured for Chinese and Arabic and found to be significant, further experimentation is needed to check its correlation for other languages.
Furthermore, while the alignment and matching done in METEOR is based on unigrams, using multiple word entities (e.g. bigrams) could contribute to improving its accuracy -- this has been proposed in [more recent publications](https://www.cs.cmu.edu/~alavie/METEOR/pdf/meteor-naacl-2010.pdf) on the subject.
## Citation
```bibtex
@inproceedings{banarjee2005,
title = {{METEOR}: An Automatic Metric for {MT} Evaluation with Improved Correlation with Human Judgments},
author = {Banerjee, Satanjeev and Lavie, Alon},
booktitle = {Proceedings of the {ACL} Workshop on Intrinsic and Extrinsic Evaluation Measures for Machine Translation and/or Summarization},
month = jun,
year = {2005},
address = {Ann Arbor, Michigan},
publisher = {Association for Computational Linguistics},
url = {https://www.aclweb.org/anthology/W05-0909},
pages = {65--72},
}
```
## Further References
- [METEOR -- Wikipedia](https://en.wikipedia.org/wiki/METEOR)
- [METEOR score -- NLTK](https://www.nltk.org/_modules/nltk/translate/meteor_score.html)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/meteor/meteor.py | # Copyright 2020 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" METEOR metric. """
import importlib.metadata
import numpy as np
from nltk.translate import meteor_score
from packaging import version
import datasets
NLTK_VERSION = version.parse(importlib.metadata.version("nltk"))
if NLTK_VERSION >= version.Version("3.6.4"):
from nltk import word_tokenize
_CITATION = """\
@inproceedings{banarjee2005,
title = {{METEOR}: An Automatic Metric for {MT} Evaluation with Improved Correlation with Human Judgments},
author = {Banerjee, Satanjeev and Lavie, Alon},
booktitle = {Proceedings of the {ACL} Workshop on Intrinsic and Extrinsic Evaluation Measures for Machine Translation and/or Summarization},
month = jun,
year = {2005},
address = {Ann Arbor, Michigan},
publisher = {Association for Computational Linguistics},
url = {https://www.aclweb.org/anthology/W05-0909},
pages = {65--72},
}
"""
_DESCRIPTION = """\
METEOR, an automatic metric for machine translation evaluation
that is based on a generalized concept of unigram matching between the
machine-produced translation and human-produced reference translations.
Unigrams can be matched based on their surface forms, stemmed forms,
and meanings; furthermore, METEOR can be easily extended to include more
advanced matching strategies. Once all generalized unigram matches
between the two strings have been found, METEOR computes a score for
this matching using a combination of unigram-precision, unigram-recall, and
a measure of fragmentation that is designed to directly capture how
well-ordered the matched words in the machine translation are in relation
to the reference.
METEOR gets an R correlation value of 0.347 with human evaluation on the Arabic
data and 0.331 on the Chinese data. This is shown to be an improvement on
using simply unigram-precision, unigram-recall and their harmonic F1
combination.
"""
_KWARGS_DESCRIPTION = """
Computes METEOR score of translated segments against one or more references.
Args:
predictions: list of predictions to score. Each prediction
should be a string with tokens separated by spaces.
references: list of reference for each prediction. Each
reference should be a string with tokens separated by spaces.
alpha: Parameter for controlling relative weights of precision and recall. default: 0.9
beta: Parameter for controlling shape of penalty as a function of fragmentation. default: 3
gamma: Relative weight assigned to fragmentation penalty. default: 0.5
Returns:
'meteor': meteor score.
Examples:
>>> meteor = datasets.load_metric('meteor')
>>> predictions = ["It is a guide to action which ensures that the military always obeys the commands of the party"]
>>> references = ["It is a guide to action that ensures that the military will forever heed Party commands"]
>>> results = meteor.compute(predictions=predictions, references=references)
>>> print(round(results["meteor"], 4))
0.6944
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Meteor(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Value("string", id="sequence"),
"references": datasets.Value("string", id="sequence"),
}
),
codebase_urls=["https://github.com/nltk/nltk/blob/develop/nltk/translate/meteor_score.py"],
reference_urls=[
"https://www.nltk.org/api/nltk.translate.html#module-nltk.translate.meteor_score",
"https://en.wikipedia.org/wiki/METEOR",
],
)
def _download_and_prepare(self, dl_manager):
import nltk
nltk.download("wordnet")
if NLTK_VERSION >= version.Version("3.6.5"):
nltk.download("punkt")
if NLTK_VERSION >= version.Version("3.6.6"):
nltk.download("omw-1.4")
def _compute(self, predictions, references, alpha=0.9, beta=3, gamma=0.5):
if NLTK_VERSION >= version.Version("3.6.5"):
scores = [
meteor_score.single_meteor_score(
word_tokenize(ref), word_tokenize(pred), alpha=alpha, beta=beta, gamma=gamma
)
for ref, pred in zip(references, predictions)
]
else:
scores = [
meteor_score.single_meteor_score(ref, pred, alpha=alpha, beta=beta, gamma=gamma)
for ref, pred in zip(references, predictions)
]
return {"meteor": np.mean(scores)}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/super_glue/README.md | # Metric Card for SuperGLUE
## Metric description
This metric is used to compute the SuperGLUE evaluation metric associated to each of the subsets of the [SuperGLUE dataset](https://huggingface.co/datasets/super_glue).
SuperGLUE is a new benchmark styled after GLUE with a new set of more difficult language understanding tasks, improved resources, and a new public leaderboard.
## How to use
There are two steps: (1) loading the SuperGLUE metric relevant to the subset of the dataset being used for evaluation; and (2) calculating the metric.
1. **Loading the relevant SuperGLUE metric** : the subsets of SuperGLUE are the following: `boolq`, `cb`, `copa`, `multirc`, `record`, `rte`, `wic`, `wsc`, `wsc.fixed`, `axb`, `axg`.
More information about the different subsets of the SuperGLUE dataset can be found on the [SuperGLUE dataset page](https://huggingface.co/datasets/super_glue) and on the [official dataset website](https://super.gluebenchmark.com/).
2. **Calculating the metric**: the metric takes two inputs : one list with the predictions of the model to score and one list of reference labels. The structure of both inputs depends on the SuperGlUE subset being used:
Format of `predictions`:
- for `record`: list of question-answer dictionaries with the following keys:
- `idx`: index of the question as specified by the dataset
- `prediction_text`: the predicted answer text
- for `multirc`: list of question-answer dictionaries with the following keys:
- `idx`: index of the question-answer pair as specified by the dataset
- `prediction`: the predicted answer label
- otherwise: list of predicted labels
Format of `references`:
- for `record`: list of question-answers dictionaries with the following keys:
- `idx`: index of the question as specified by the dataset
- `answers`: list of possible answers
- otherwise: list of reference labels
```python
from datasets import load_metric
super_glue_metric = load_metric('super_glue', 'copa')
predictions = [0, 1]
references = [0, 1]
results = super_glue_metric.compute(predictions=predictions, references=references)
```
## Output values
The output of the metric depends on the SuperGLUE subset chosen, consisting of a dictionary that contains one or several of the following metrics:
`exact_match`: A given predicted string's exact match score is 1 if it is the exact same as its reference string, and is 0 otherwise. (See [Exact Match](https://huggingface.co/metrics/exact_match) for more information).
`f1`: the harmonic mean of the precision and recall (see [F1 score](https://huggingface.co/metrics/f1) for more information). Its range is 0-1 -- its lowest possible value is 0, if either the precision or the recall is 0, and its highest possible value is 1.0, which means perfect precision and recall.
`matthews_correlation`: a measure of the quality of binary and multiclass classifications (see [Matthews Correlation](https://huggingface.co/metrics/matthews_correlation) for more information). Its range of values is between -1 and +1, where a coefficient of +1 represents a perfect prediction, 0 an average random prediction and -1 an inverse prediction.
### Values from popular papers
The [original SuperGLUE paper](https://arxiv.org/pdf/1905.00537.pdf) reported average scores ranging from 47 to 71.5%, depending on the model used (with all evaluation values scaled by 100 to make computing the average possible).
For more recent model performance, see the [dataset leaderboard](https://super.gluebenchmark.com/leaderboard).
## Examples
Maximal values for the COPA subset (which outputs `accuracy`):
```python
from datasets import load_metric
super_glue_metric = load_metric('super_glue', 'copa') # any of ["copa", "rte", "wic", "wsc", "wsc.fixed", "boolq", "axg"]
predictions = [0, 1]
references = [0, 1]
results = super_glue_metric.compute(predictions=predictions, references=references)
print(results)
{'accuracy': 1.0}
```
Minimal values for the MultiRC subset (which outputs `pearson` and `spearmanr`):
```python
from datasets import load_metric
super_glue_metric = load_metric('super_glue', 'multirc')
predictions = [{'idx': {'answer': 0, 'paragraph': 0, 'question': 0}, 'prediction': 0}, {'idx': {'answer': 1, 'paragraph': 2, 'question': 3}, 'prediction': 1}]
references = [1,0]
results = super_glue_metric.compute(predictions=predictions, references=references)
print(results)
{'exact_match': 0.0, 'f1_m': 0.0, 'f1_a': 0.0}
```
Partial match for the COLA subset (which outputs `matthews_correlation`)
```python
from datasets import load_metric
super_glue_metric = load_metric('super_glue', 'axb')
references = [0, 1]
predictions = [1,1]
results = super_glue_metric.compute(predictions=predictions, references=references)
print(results)
{'matthews_correlation': 0.0}
```
## Limitations and bias
This metric works only with datasets that have the same format as the [SuperGLUE dataset](https://huggingface.co/datasets/super_glue).
The dataset also includes Winogender, a subset of the dataset that is designed to measure gender bias in coreference resolution systems. However, as noted in the SuperGLUE paper, this subset has its limitations: *"It offers only positive predictive value: A poor bias score is clear evidence that a model exhibits gender bias, but a good score does not mean that the model is unbiased.[...] Also, Winogender does not cover all forms of social bias, or even all forms of gender. For instance, the version of the data used here offers no coverage of gender-neutral they or non-binary pronouns."
## Citation
```bibtex
@article{wang2019superglue,
title={Super{GLUE}: A Stickier Benchmark for General-Purpose Language Understanding Systems},
author={Wang, Alex and Pruksachatkun, Yada and Nangia, Nikita and Singh, Amanpreet and Michael, Julian and Hill, Felix and Levy, Omer and Bowman, Samuel R},
journal={arXiv preprint arXiv:1905.00537},
year={2019}
}
```
## Further References
- [SuperGLUE benchmark homepage](https://super.gluebenchmark.com/)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/super_glue/super_glue.py | # Copyright 2020 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""The SuperGLUE benchmark metric."""
from sklearn.metrics import f1_score, matthews_corrcoef
import datasets
from .record_evaluation import evaluate as evaluate_record
_CITATION = """\
@article{wang2019superglue,
title={SuperGLUE: A Stickier Benchmark for General-Purpose Language Understanding Systems},
author={Wang, Alex and Pruksachatkun, Yada and Nangia, Nikita and Singh, Amanpreet and Michael, Julian and Hill, Felix and Levy, Omer and Bowman, Samuel R},
journal={arXiv preprint arXiv:1905.00537},
year={2019}
}
"""
_DESCRIPTION = """\
SuperGLUE (https://super.gluebenchmark.com/) is a new benchmark styled after
GLUE with a new set of more difficult language understanding tasks, improved
resources, and a new public leaderboard.
"""
_KWARGS_DESCRIPTION = """
Compute SuperGLUE evaluation metric associated to each SuperGLUE dataset.
Args:
predictions: list of predictions to score. Depending on the SuperGlUE subset:
- for 'record': list of question-answer dictionaries with the following keys:
- 'idx': index of the question as specified by the dataset
- 'prediction_text': the predicted answer text
- for 'multirc': list of question-answer dictionaries with the following keys:
- 'idx': index of the question-answer pair as specified by the dataset
- 'prediction': the predicted answer label
- otherwise: list of predicted labels
references: list of reference labels. Depending on the SuperGLUE subset:
- for 'record': list of question-answers dictionaries with the following keys:
- 'idx': index of the question as specified by the dataset
- 'answers': list of possible answers
- otherwise: list of reference labels
Returns: depending on the SuperGLUE subset:
- for 'record':
- 'exact_match': Exact match between answer and gold answer
- 'f1': F1 score
- for 'multirc':
- 'exact_match': Exact match between answer and gold answer
- 'f1_m': Per-question macro-F1 score
- 'f1_a': Average F1 score over all answers
- for 'axb':
'matthews_correlation': Matthew Correlation
- for 'cb':
- 'accuracy': Accuracy
- 'f1': F1 score
- for all others:
- 'accuracy': Accuracy
Examples:
>>> super_glue_metric = datasets.load_metric('super_glue', 'copa') # any of ["copa", "rte", "wic", "wsc", "wsc.fixed", "boolq", "axg"]
>>> predictions = [0, 1]
>>> references = [0, 1]
>>> results = super_glue_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'accuracy': 1.0}
>>> super_glue_metric = datasets.load_metric('super_glue', 'cb')
>>> predictions = [0, 1]
>>> references = [0, 1]
>>> results = super_glue_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'accuracy': 1.0, 'f1': 1.0}
>>> super_glue_metric = datasets.load_metric('super_glue', 'record')
>>> predictions = [{'idx': {'passage': 0, 'query': 0}, 'prediction_text': 'answer'}]
>>> references = [{'idx': {'passage': 0, 'query': 0}, 'answers': ['answer', 'another_answer']}]
>>> results = super_glue_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'exact_match': 1.0, 'f1': 1.0}
>>> super_glue_metric = datasets.load_metric('super_glue', 'multirc')
>>> predictions = [{'idx': {'answer': 0, 'paragraph': 0, 'question': 0}, 'prediction': 0}, {'idx': {'answer': 1, 'paragraph': 2, 'question': 3}, 'prediction': 1}]
>>> references = [0, 1]
>>> results = super_glue_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'exact_match': 1.0, 'f1_m': 1.0, 'f1_a': 1.0}
>>> super_glue_metric = datasets.load_metric('super_glue', 'axb')
>>> references = [0, 1]
>>> predictions = [0, 1]
>>> results = super_glue_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'matthews_correlation': 1.0}
"""
def simple_accuracy(preds, labels):
return float((preds == labels).mean())
def acc_and_f1(preds, labels, f1_avg="binary"):
acc = simple_accuracy(preds, labels)
f1 = float(f1_score(y_true=labels, y_pred=preds, average=f1_avg))
return {
"accuracy": acc,
"f1": f1,
}
def evaluate_multirc(ids_preds, labels):
"""
Computes F1 score and Exact Match for MultiRC predictions.
"""
question_map = {}
for id_pred, label in zip(ids_preds, labels):
question_id = f'{id_pred["idx"]["paragraph"]}-{id_pred["idx"]["question"]}'
pred = id_pred["prediction"]
if question_id in question_map:
question_map[question_id].append((pred, label))
else:
question_map[question_id] = [(pred, label)]
f1s, ems = [], []
for question, preds_labels in question_map.items():
question_preds, question_labels = zip(*preds_labels)
f1 = f1_score(y_true=question_labels, y_pred=question_preds, average="macro")
f1s.append(f1)
em = int(sum(pred == label for pred, label in preds_labels) == len(preds_labels))
ems.append(em)
f1_m = float(sum(f1s) / len(f1s))
em = sum(ems) / len(ems)
f1_a = float(f1_score(y_true=labels, y_pred=[id_pred["prediction"] for id_pred in ids_preds]))
return {"exact_match": em, "f1_m": f1_m, "f1_a": f1_a}
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class SuperGlue(datasets.Metric):
def _info(self):
if self.config_name not in [
"boolq",
"cb",
"copa",
"multirc",
"record",
"rte",
"wic",
"wsc",
"wsc.fixed",
"axb",
"axg",
]:
raise KeyError(
"You should supply a configuration name selected in "
'["boolq", "cb", "copa", "multirc", "record", "rte", "wic", "wsc", "wsc.fixed", "axb", "axg",]'
)
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(self._get_feature_types()),
codebase_urls=[],
reference_urls=[],
format="numpy" if not self.config_name == "record" and not self.config_name == "multirc" else None,
)
def _get_feature_types(self):
if self.config_name == "record":
return {
"predictions": {
"idx": {
"passage": datasets.Value("int64"),
"query": datasets.Value("int64"),
},
"prediction_text": datasets.Value("string"),
},
"references": {
"idx": {
"passage": datasets.Value("int64"),
"query": datasets.Value("int64"),
},
"answers": datasets.Sequence(datasets.Value("string")),
},
}
elif self.config_name == "multirc":
return {
"predictions": {
"idx": {
"answer": datasets.Value("int64"),
"paragraph": datasets.Value("int64"),
"question": datasets.Value("int64"),
},
"prediction": datasets.Value("int64"),
},
"references": datasets.Value("int64"),
}
else:
return {
"predictions": datasets.Value("int64"),
"references": datasets.Value("int64"),
}
def _compute(self, predictions, references):
if self.config_name == "axb":
return {"matthews_correlation": matthews_corrcoef(references, predictions)}
elif self.config_name == "cb":
return acc_and_f1(predictions, references, f1_avg="macro")
elif self.config_name == "record":
dataset = [
{
"qas": [
{"id": ref["idx"]["query"], "answers": [{"text": ans} for ans in ref["answers"]]}
for ref in references
]
}
]
predictions = {pred["idx"]["query"]: pred["prediction_text"] for pred in predictions}
return evaluate_record(dataset, predictions)[0]
elif self.config_name == "multirc":
return evaluate_multirc(predictions, references)
elif self.config_name in ["copa", "rte", "wic", "wsc", "wsc.fixed", "boolq", "axg"]:
return {"accuracy": simple_accuracy(predictions, references)}
else:
raise KeyError(
"You should supply a configuration name selected in "
'["boolq", "cb", "copa", "multirc", "record", "rte", "wic", "wsc", "wsc.fixed", "axb", "axg",]'
)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/super_glue/record_evaluation.py | """
Official evaluation script for ReCoRD v1.0.
(Some functions are adopted from the SQuAD evaluation script.)
"""
import argparse
import json
import re
import string
import sys
from collections import Counter
def normalize_answer(s):
"""Lower text and remove punctuation, articles and extra whitespace."""
def remove_articles(text):
return re.sub(r"\b(a|an|the)\b", " ", text)
def white_space_fix(text):
return " ".join(text.split())
def remove_punc(text):
exclude = set(string.punctuation)
return "".join(ch for ch in text if ch not in exclude)
def lower(text):
return text.lower()
return white_space_fix(remove_articles(remove_punc(lower(s))))
def f1_score(prediction, ground_truth):
prediction_tokens = normalize_answer(prediction).split()
ground_truth_tokens = normalize_answer(ground_truth).split()
common = Counter(prediction_tokens) & Counter(ground_truth_tokens)
num_same = sum(common.values())
if num_same == 0:
return 0
precision = 1.0 * num_same / len(prediction_tokens)
recall = 1.0 * num_same / len(ground_truth_tokens)
f1 = (2 * precision * recall) / (precision + recall)
return f1
def exact_match_score(prediction, ground_truth):
return normalize_answer(prediction) == normalize_answer(ground_truth)
def metric_max_over_ground_truths(metric_fn, prediction, ground_truths):
scores_for_ground_truths = []
for ground_truth in ground_truths:
score = metric_fn(prediction, ground_truth)
scores_for_ground_truths.append(score)
return max(scores_for_ground_truths)
def evaluate(dataset, predictions):
f1 = exact_match = total = 0
correct_ids = []
for passage in dataset:
for qa in passage["qas"]:
total += 1
if qa["id"] not in predictions:
message = f'Unanswered question {qa["id"]} will receive score 0.'
print(message, file=sys.stderr)
continue
ground_truths = [x["text"] for x in qa["answers"]]
prediction = predictions[qa["id"]]
_exact_match = metric_max_over_ground_truths(exact_match_score, prediction, ground_truths)
if int(_exact_match) == 1:
correct_ids.append(qa["id"])
exact_match += _exact_match
f1 += metric_max_over_ground_truths(f1_score, prediction, ground_truths)
exact_match = exact_match / total
f1 = f1 / total
return {"exact_match": exact_match, "f1": f1}, correct_ids
if __name__ == "__main__":
expected_version = "1.0"
parser = argparse.ArgumentParser("Official evaluation script for ReCoRD v1.0.")
parser.add_argument("data_file", help="The dataset file in JSON format.")
parser.add_argument("pred_file", help="The model prediction file in JSON format.")
parser.add_argument("--output_correct_ids", action="store_true", help="Output the correctly answered query IDs.")
args = parser.parse_args()
with open(args.data_file) as data_file:
dataset_json = json.load(data_file)
if dataset_json["version"] != expected_version:
print(
f'Evaluation expects v-{expected_version}, but got dataset with v-{dataset_json["version"]}',
file=sys.stderr,
)
dataset = dataset_json["data"]
with open(args.pred_file) as pred_file:
predictions = json.load(pred_file)
metrics, correct_ids = evaluate(dataset, predictions)
if args.output_correct_ids:
print(f"Output {len(correct_ids)} correctly answered question IDs.")
with open("correct_ids.json", "w") as f:
json.dump(correct_ids, f)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/pearsonr/pearsonr.py | # Copyright 2021 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Pearson correlation coefficient metric."""
from scipy.stats import pearsonr
import datasets
_DESCRIPTION = """
Pearson correlation coefficient and p-value for testing non-correlation.
The Pearson correlation coefficient measures the linear relationship between two datasets. The calculation of the p-value relies on the assumption that each dataset is normally distributed. Like other correlation coefficients, this one varies between -1 and +1 with 0 implying no correlation. Correlations of -1 or +1 imply an exact linear relationship. Positive correlations imply that as x increases, so does y. Negative correlations imply that as x increases, y decreases.
The p-value roughly indicates the probability of an uncorrelated system producing datasets that have a Pearson correlation at least as extreme as the one computed from these datasets.
"""
_KWARGS_DESCRIPTION = """
Args:
predictions (`list` of `int`): Predicted class labels, as returned by a model.
references (`list` of `int`): Ground truth labels.
return_pvalue (`boolean`): If `True`, returns the p-value, along with the correlation coefficient. If `False`, returns only the correlation coefficient. Defaults to `False`.
Returns:
pearsonr (`float`): Pearson correlation coefficient. Minimum possible value is -1. Maximum possible value is 1. Values of 1 and -1 indicate exact linear positive and negative relationships, respectively. A value of 0 implies no correlation.
p-value (`float`): P-value, which roughly indicates the probability of an The p-value roughly indicates the probability of an uncorrelated system producing datasets that have a Pearson correlation at least as extreme as the one computed from these datasets. Minimum possible value is 0. Maximum possible value is 1. Higher values indicate higher probabilities.
Examples:
Example 1-A simple example using only predictions and references.
>>> pearsonr_metric = datasets.load_metric("pearsonr")
>>> results = pearsonr_metric.compute(predictions=[10, 9, 2.5, 6, 4], references=[1, 2, 3, 4, 5])
>>> print(round(results['pearsonr'], 2))
-0.74
Example 2-The same as Example 1, but that also returns the `p-value`.
>>> pearsonr_metric = datasets.load_metric("pearsonr")
>>> results = pearsonr_metric.compute(predictions=[10, 9, 2.5, 6, 4], references=[1, 2, 3, 4, 5], return_pvalue=True)
>>> print(sorted(list(results.keys())))
['p-value', 'pearsonr']
>>> print(round(results['pearsonr'], 2))
-0.74
>>> print(round(results['p-value'], 2))
0.15
"""
_CITATION = """
@article{2020SciPy-NMeth,
author = {Virtanen, Pauli and Gommers, Ralf and Oliphant, Travis E. and
Haberland, Matt and Reddy, Tyler and Cournapeau, David and
Burovski, Evgeni and Peterson, Pearu and Weckesser, Warren and
Bright, Jonathan and {van der Walt}, St{\'e}fan J. and
Brett, Matthew and Wilson, Joshua and Millman, K. Jarrod and
Mayorov, Nikolay and Nelson, Andrew R. J. and Jones, Eric and
Kern, Robert and Larson, Eric and Carey, C J and
Polat, Ilhan and Feng, Yu and Moore, Eric W. and
{VanderPlas}, Jake and Laxalde, Denis and Perktold, Josef and
Cimrman, Robert and Henriksen, Ian and Quintero, E. A. and
Harris, Charles R. and Archibald, Anne M. and
Ribeiro, Antonio H. and Pedregosa, Fabian and
{van Mulbregt}, Paul and {SciPy 1.0 Contributors}},
title = {{{SciPy} 1.0: Fundamental Algorithms for Scientific
Computing in Python}},
journal = {Nature Methods},
year = {2020},
volume = {17},
pages = {261--272},
adsurl = {https://rdcu.be/b08Wh},
doi = {10.1038/s41592-019-0686-2},
}
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Pearsonr(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Value("float"),
"references": datasets.Value("float"),
}
),
reference_urls=["https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.pearsonr.html"],
)
def _compute(self, predictions, references, return_pvalue=False):
if return_pvalue:
results = pearsonr(references, predictions)
return {"pearsonr": results[0], "p-value": results[1]}
else:
return {"pearsonr": float(pearsonr(references, predictions)[0])}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/pearsonr/README.md | # Metric Card for Pearson Correlation Coefficient (pearsonr)
## Metric Description
Pearson correlation coefficient and p-value for testing non-correlation.
The Pearson correlation coefficient measures the linear relationship between two datasets. The calculation of the p-value relies on the assumption that each dataset is normally distributed. Like other correlation coefficients, this one varies between -1 and +1 with 0 implying no correlation. Correlations of -1 or +1 imply an exact linear relationship. Positive correlations imply that as x increases, so does y. Negative correlations imply that as x increases, y decreases.
The p-value roughly indicates the probability of an uncorrelated system producing datasets that have a Pearson correlation at least as extreme as the one computed from these datasets.
## How to Use
This metric takes a list of predictions and a list of references as input
```python
>>> pearsonr_metric = datasets.load_metric("pearsonr")
>>> results = pearsonr_metric.compute(predictions=[10, 9, 2.5, 6, 4], references=[1, 2, 3, 4, 5])
>>> print(round(results['pearsonr']), 2)
['-0.74']
```
### Inputs
- **predictions** (`list` of `int`): Predicted class labels, as returned by a model.
- **references** (`list` of `int`): Ground truth labels.
- **return_pvalue** (`boolean`): If `True`, returns the p-value, along with the correlation coefficient. If `False`, returns only the correlation coefficient. Defaults to `False`.
### Output Values
- **pearsonr**(`float`): Pearson correlation coefficient. Minimum possible value is -1. Maximum possible value is 1. Values of 1 and -1 indicate exact linear positive and negative relationships, respectively. A value of 0 implies no correlation.
- **p-value**(`float`): P-value, which roughly indicates the probability of an The p-value roughly indicates the probability of an uncorrelated system producing datasets that have a Pearson correlation at least as extreme as the one computed from these datasets. Minimum possible value is 0. Maximum possible value is 1. Higher values indicate higher probabilities.
Like other correlation coefficients, this one varies between -1 and +1 with 0 implying no correlation. Correlations of -1 or +1 imply an exact linear relationship. Positive correlations imply that as x increases, so does y. Negative correlations imply that as x increases, y decreases.
Output Example(s):
```python
{'pearsonr': -0.7}
```
```python
{'p-value': 0.15}
```
#### Values from Popular Papers
### Examples
Example 1-A simple example using only predictions and references.
```python
>>> pearsonr_metric = datasets.load_metric("pearsonr")
>>> results = pearsonr_metric.compute(predictions=[10, 9, 2.5, 6, 4], references=[1, 2, 3, 4, 5])
>>> print(round(results['pearsonr'], 2))
-0.74
```
Example 2-The same as Example 1, but that also returns the `p-value`.
```python
>>> pearsonr_metric = datasets.load_metric("pearsonr")
>>> results = pearsonr_metric.compute(predictions=[10, 9, 2.5, 6, 4], references=[1, 2, 3, 4, 5], return_pvalue=True)
>>> print(sorted(list(results.keys())))
['p-value', 'pearsonr']
>>> print(round(results['pearsonr'], 2))
-0.74
>>> print(round(results['p-value'], 2))
0.15
```
## Limitations and Bias
As stated above, the calculation of the p-value relies on the assumption that each data set is normally distributed. This is not always the case, so verifying the true distribution of datasets is recommended.
## Citation(s)
```bibtex
@article{2020SciPy-NMeth,
author = {Virtanen, Pauli and Gommers, Ralf and Oliphant, Travis E. and
Haberland, Matt and Reddy, Tyler and Cournapeau, David and
Burovski, Evgeni and Peterson, Pearu and Weckesser, Warren and
Bright, Jonathan and {van der Walt}, St{\'e}fan J. and
Brett, Matthew and Wilson, Joshua and Millman, K. Jarrod and
Mayorov, Nikolay and Nelson, Andrew R. J. and Jones, Eric and
Kern, Robert and Larson, Eric and Carey, C J and
Polat, {\.I}lhan and Feng, Yu and Moore, Eric W. and
{VanderPlas}, Jake and Laxalde, Denis and Perktold, Josef and
Cimrman, Robert and Henriksen, Ian and Quintero, E. A. and
Harris, Charles R. and Archibald, Anne M. and
Ribeiro, Ant{\^o}nio H. and Pedregosa, Fabian and
{van Mulbregt}, Paul and {SciPy 1.0 Contributors}},
title = {{{SciPy} 1.0: Fundamental Algorithms for Scientific
Computing in Python}},
journal = {Nature Methods},
year = {2020},
volume = {17},
pages = {261--272},
adsurl = {https://rdcu.be/b08Wh},
doi = {10.1038/s41592-019-0686-2},
}
```
## Further References
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/mahalanobis/README.md | # Metric Card for Mahalanobis Distance
## Metric Description
Mahalonobis distance is the distance between a point and a distribution (as opposed to the distance between two points), making it the multivariate equivalent of the Euclidean distance.
It is often used in multivariate anomaly detection, classification on highly imbalanced datasets and one-class classification.
## How to Use
At minimum, this metric requires two `list`s of datapoints:
```python
>>> mahalanobis_metric = datasets.load_metric("mahalanobis")
>>> results = mahalanobis_metric.compute(reference_distribution=[[0, 1], [1, 0]], X=[[0, 1]])
```
### Inputs
- `X` (`list`): data points to be compared with the `reference_distribution`.
- `reference_distribution` (`list`): data points from the reference distribution that we want to compare to.
### Output Values
`mahalanobis` (`array`): the Mahalonobis distance for each data point in `X`.
```python
>>> print(results)
{'mahalanobis': array([0.5])}
```
#### Values from Popular Papers
*N/A*
### Example
```python
>>> mahalanobis_metric = datasets.load_metric("mahalanobis")
>>> results = mahalanobis_metric.compute(reference_distribution=[[0, 1], [1, 0]], X=[[0, 1]])
>>> print(results)
{'mahalanobis': array([0.5])}
```
## Limitations and Bias
The Mahalanobis distance is only able to capture linear relationships between the variables, which means it cannot capture all types of outliers. Mahalanobis distance also fails to faithfully represent data that is highly skewed or multimodal.
## Citation
```bibtex
@inproceedings{mahalanobis1936generalized,
title={On the generalized distance in statistics},
author={Mahalanobis, Prasanta Chandra},
year={1936},
organization={National Institute of Science of India}
}
```
```bibtex
@article{de2000mahalanobis,
title={The Mahalanobis distance},
author={De Maesschalck, Roy and Jouan-Rimbaud, Delphine and Massart, D{\'e}sir{\'e} L},
journal={Chemometrics and intelligent laboratory systems},
volume={50},
number={1},
pages={1--18},
year={2000},
publisher={Elsevier}
}
```
## Further References
-[Wikipedia -- Mahalanobis Distance](https://en.wikipedia.org/wiki/Mahalanobis_distance)
-[Machine Learning Plus -- Mahalanobis Distance](https://www.machinelearningplus.com/statistics/mahalanobis-distance/)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/mahalanobis/mahalanobis.py | # Copyright 2021 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Mahalanobis metric."""
import numpy as np
import datasets
_DESCRIPTION = """
Compute the Mahalanobis Distance
Mahalonobis distance is the distance between a point and a distribution.
And not between two distinct points. It is effectively a multivariate equivalent of the Euclidean distance.
It was introduced by Prof. P. C. Mahalanobis in 1936
and has been used in various statistical applications ever since
[source: https://www.machinelearningplus.com/statistics/mahalanobis-distance/]
"""
_CITATION = """\
@article{de2000mahalanobis,
title={The mahalanobis distance},
author={De Maesschalck, Roy and Jouan-Rimbaud, Delphine and Massart, D{\'e}sir{\'e} L},
journal={Chemometrics and intelligent laboratory systems},
volume={50},
number={1},
pages={1--18},
year={2000},
publisher={Elsevier}
}
"""
_KWARGS_DESCRIPTION = """
Args:
X: List of datapoints to be compared with the `reference_distribution`.
reference_distribution: List of datapoints from the reference distribution we want to compare to.
Returns:
mahalanobis: The Mahalonobis distance for each datapoint in `X`.
Examples:
>>> mahalanobis_metric = datasets.load_metric("mahalanobis")
>>> results = mahalanobis_metric.compute(reference_distribution=[[0, 1], [1, 0]], X=[[0, 1]])
>>> print(results)
{'mahalanobis': array([0.5])}
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Mahalanobis(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"X": datasets.Sequence(datasets.Value("float", id="sequence"), id="X"),
}
),
)
def _compute(self, X, reference_distribution):
# convert to numpy arrays
X = np.array(X)
reference_distribution = np.array(reference_distribution)
# Assert that arrays are 2D
if len(X.shape) != 2:
raise ValueError("Expected `X` to be a 2D vector")
if len(reference_distribution.shape) != 2:
raise ValueError("Expected `reference_distribution` to be a 2D vector")
if reference_distribution.shape[0] < 2:
raise ValueError(
"Expected `reference_distribution` to be a 2D vector with more than one element in the first dimension"
)
# Get mahalanobis distance for each prediction
X_minus_mu = X - np.mean(reference_distribution)
cov = np.cov(reference_distribution.T)
try:
inv_covmat = np.linalg.inv(cov)
except np.linalg.LinAlgError:
inv_covmat = np.linalg.pinv(cov)
left_term = np.dot(X_minus_mu, inv_covmat)
mahal_dist = np.dot(left_term, X_minus_mu.T).diagonal()
return {"mahalanobis": mahal_dist}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/cer/README.md | # Metric Card for CER
## Metric description
Character error rate (CER) is a common metric of the performance of an automatic speech recognition (ASR) system. CER is similar to Word Error Rate (WER), but operates on character instead of word.
Character error rate can be computed as:
`CER = (S + D + I) / N = (S + D + I) / (S + D + C)`
where
`S` is the number of substitutions,
`D` is the number of deletions,
`I` is the number of insertions,
`C` is the number of correct characters,
`N` is the number of characters in the reference (`N=S+D+C`).
## How to use
The metric takes two inputs: references (a list of references for each speech input) and predictions (a list of transcriptions to score).
```python
from datasets import load_metric
cer = load_metric("cer")
cer_score = cer.compute(predictions=predictions, references=references)
```
## Output values
This metric outputs a float representing the character error rate.
```
print(cer_score)
0.34146341463414637
```
The **lower** the CER value, the **better** the performance of the ASR system, with a CER of 0 being a perfect score.
However, CER's output is not always a number between 0 and 1, in particular when there is a high number of insertions (see [Examples](#Examples) below).
### Values from popular papers
This metric is highly dependent on the content and quality of the dataset, and therefore users can expect very different values for the same model but on different datasets.
Multilingual datasets such as [Common Voice](https://huggingface.co/datasets/common_voice) report different CERs depending on the language, ranging from 0.02-0.03 for languages such as French and Italian, to 0.05-0.07 for English (see [here](https://github.com/speechbrain/speechbrain/tree/develop/recipes/CommonVoice/ASR/CTC) for more values).
## Examples
Perfect match between prediction and reference:
```python
from datasets import load_metric
cer = load_metric("cer")
predictions = ["hello world", "good night moon"]
references = ["hello world", "good night moon"]
cer_score = cer.compute(predictions=predictions, references=references)
print(cer_score)
0.0
```
Partial match between prediction and reference:
```python
from datasets import load_metric
cer = load_metric("cer")
predictions = ["this is the prediction", "there is an other sample"]
references = ["this is the reference", "there is another one"]
cer_score = cer.compute(predictions=predictions, references=references)
print(cer_score)
0.34146341463414637
```
No match between prediction and reference:
```python
from datasets import load_metric
cer = load_metric("cer")
predictions = ["hello"]
references = ["gracias"]
cer_score = cer.compute(predictions=predictions, references=references)
print(cer_score)
1.0
```
CER above 1 due to insertion errors:
```python
from datasets import load_metric
cer = load_metric("cer")
predictions = ["hello world"]
references = ["hello"]
cer_score = cer.compute(predictions=predictions, references=references)
print(cer_score)
1.2
```
## Limitations and bias
CER is useful for comparing different models for tasks such as automatic speech recognition (ASR) and optic character recognition (OCR), especially for multilingual datasets where WER is not suitable given the diversity of languages. However, CER provides no details on the nature of translation errors and further work is therefore required to identify the main source(s) of error and to focus any research effort.
Also, in some cases, instead of reporting the raw CER, a normalized CER is reported where the number of mistakes is divided by the sum of the number of edit operations (`I` + `S` + `D`) and `C` (the number of correct characters), which results in CER values that fall within the range of 0–100%.
## Citation
```bibtex
@inproceedings{morris2004,
author = {Morris, Andrew and Maier, Viktoria and Green, Phil},
year = {2004},
month = {01},
pages = {},
title = {From WER and RIL to MER and WIL: improved evaluation measures for connected speech recognition.}
}
```
## Further References
- [Hugging Face Tasks -- Automatic Speech Recognition](https://huggingface.co/tasks/automatic-speech-recognition)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/cer/test_cer.py | # Copyright 2021 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import unittest
from cer import CER
cer = CER()
class TestCER(unittest.TestCase):
def test_cer_case_senstive(self):
refs = ["White House"]
preds = ["white house"]
# S = 2, D = 0, I = 0, N = 11, CER = 2 / 11
char_error_rate = cer.compute(predictions=preds, references=refs)
self.assertTrue(abs(char_error_rate - 0.1818181818) < 1e-6)
def test_cer_whitespace(self):
refs = ["were wolf"]
preds = ["werewolf"]
# S = 0, D = 0, I = 1, N = 9, CER = 1 / 9
char_error_rate = cer.compute(predictions=preds, references=refs)
self.assertTrue(abs(char_error_rate - 0.1111111) < 1e-6)
refs = ["werewolf"]
preds = ["weae wolf"]
# S = 1, D = 1, I = 0, N = 8, CER = 0.25
char_error_rate = cer.compute(predictions=preds, references=refs)
self.assertTrue(abs(char_error_rate - 0.25) < 1e-6)
# consecutive whitespaces case 1
refs = ["were wolf"]
preds = ["were wolf"]
# S = 0, D = 0, I = 0, N = 9, CER = 0
char_error_rate = cer.compute(predictions=preds, references=refs)
self.assertTrue(abs(char_error_rate - 0.0) < 1e-6)
# consecutive whitespaces case 2
refs = ["were wolf"]
preds = ["were wolf"]
# S = 0, D = 0, I = 0, N = 9, CER = 0
char_error_rate = cer.compute(predictions=preds, references=refs)
self.assertTrue(abs(char_error_rate - 0.0) < 1e-6)
def test_cer_sub(self):
refs = ["werewolf"]
preds = ["weaewolf"]
# S = 1, D = 0, I = 0, N = 8, CER = 0.125
char_error_rate = cer.compute(predictions=preds, references=refs)
self.assertTrue(abs(char_error_rate - 0.125) < 1e-6)
def test_cer_del(self):
refs = ["werewolf"]
preds = ["wereawolf"]
# S = 0, D = 1, I = 0, N = 8, CER = 0.125
char_error_rate = cer.compute(predictions=preds, references=refs)
self.assertTrue(abs(char_error_rate - 0.125) < 1e-6)
def test_cer_insert(self):
refs = ["werewolf"]
preds = ["wereolf"]
# S = 0, D = 0, I = 1, N = 8, CER = 0.125
char_error_rate = cer.compute(predictions=preds, references=refs)
self.assertTrue(abs(char_error_rate - 0.125) < 1e-6)
def test_cer_equal(self):
refs = ["werewolf"]
char_error_rate = cer.compute(predictions=refs, references=refs)
self.assertEqual(char_error_rate, 0.0)
def test_cer_list_of_seqs(self):
refs = ["werewolf", "I am your father"]
char_error_rate = cer.compute(predictions=refs, references=refs)
self.assertEqual(char_error_rate, 0.0)
refs = ["werewolf", "I am your father", "doge"]
preds = ["werxwolf", "I am your father", "doge"]
# S = 1, D = 0, I = 0, N = 28, CER = 1 / 28
char_error_rate = cer.compute(predictions=preds, references=refs)
self.assertTrue(abs(char_error_rate - 0.03571428) < 1e-6)
def test_correlated_sentences(self):
refs = ["My hovercraft", "is full of eels"]
preds = ["My hovercraft is full", " of eels"]
# S = 0, D = 0, I = 2, N = 28, CER = 2 / 28
# whitespace at the front of " of eels" will be strip during preporcessing
# so need to insert 2 whitespaces
char_error_rate = cer.compute(predictions=preds, references=refs, concatenate_texts=True)
self.assertTrue(abs(char_error_rate - 0.071428) < 1e-6)
def test_cer_unicode(self):
refs = ["我能吞下玻璃而不伤身体"]
preds = [" 能吞虾玻璃而 不霜身体啦"]
# S = 3, D = 2, I = 0, N = 11, CER = 5 / 11
char_error_rate = cer.compute(predictions=preds, references=refs)
self.assertTrue(abs(char_error_rate - 0.4545454545) < 1e-6)
refs = ["我能吞下玻璃", "而不伤身体"]
preds = ["我 能 吞 下 玻 璃", "而不伤身体"]
# S = 0, D = 5, I = 0, N = 11, CER = 5 / 11
char_error_rate = cer.compute(predictions=preds, references=refs)
self.assertTrue(abs(char_error_rate - 0.454545454545) < 1e-6)
refs = ["我能吞下玻璃而不伤身体"]
char_error_rate = cer.compute(predictions=refs, references=refs)
self.assertFalse(char_error_rate, 0.0)
def test_cer_empty(self):
refs = [""]
preds = ["Hypothesis"]
with self.assertRaises(ValueError):
cer.compute(predictions=preds, references=refs)
if __name__ == "__main__":
unittest.main()
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/cer/cer.py | # Copyright 2021 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" Character Error Ratio (CER) metric. """
from typing import List
import jiwer
import jiwer.transforms as tr
from packaging import version
import datasets
from datasets.config import PY_VERSION
if PY_VERSION < version.parse("3.8"):
import importlib_metadata
else:
import importlib.metadata as importlib_metadata
SENTENCE_DELIMITER = ""
if version.parse(importlib_metadata.version("jiwer")) < version.parse("2.3.0"):
class SentencesToListOfCharacters(tr.AbstractTransform):
def __init__(self, sentence_delimiter: str = " "):
self.sentence_delimiter = sentence_delimiter
def process_string(self, s: str):
return list(s)
def process_list(self, inp: List[str]):
chars = []
for sent_idx, sentence in enumerate(inp):
chars.extend(self.process_string(sentence))
if self.sentence_delimiter is not None and self.sentence_delimiter != "" and sent_idx < len(inp) - 1:
chars.append(self.sentence_delimiter)
return chars
cer_transform = tr.Compose(
[tr.RemoveMultipleSpaces(), tr.Strip(), SentencesToListOfCharacters(SENTENCE_DELIMITER)]
)
else:
cer_transform = tr.Compose(
[
tr.RemoveMultipleSpaces(),
tr.Strip(),
tr.ReduceToSingleSentence(SENTENCE_DELIMITER),
tr.ReduceToListOfListOfChars(),
]
)
_CITATION = """\
@inproceedings{inproceedings,
author = {Morris, Andrew and Maier, Viktoria and Green, Phil},
year = {2004},
month = {01},
pages = {},
title = {From WER and RIL to MER and WIL: improved evaluation measures for connected speech recognition.}
}
"""
_DESCRIPTION = """\
Character error rate (CER) is a common metric of the performance of an automatic speech recognition system.
CER is similar to Word Error Rate (WER), but operates on character instead of word. Please refer to docs of WER for further information.
Character error rate can be computed as:
CER = (S + D + I) / N = (S + D + I) / (S + D + C)
where
S is the number of substitutions,
D is the number of deletions,
I is the number of insertions,
C is the number of correct characters,
N is the number of characters in the reference (N=S+D+C).
CER's output is not always a number between 0 and 1, in particular when there is a high number of insertions. This value is often associated to the percentage of characters that were incorrectly predicted. The lower the value, the better the
performance of the ASR system with a CER of 0 being a perfect score.
"""
_KWARGS_DESCRIPTION = """
Computes CER score of transcribed segments against references.
Args:
references: list of references for each speech input.
predictions: list of transcribtions to score.
concatenate_texts: Whether or not to concatenate sentences before evaluation, set to True for more accurate result.
Returns:
(float): the character error rate
Examples:
>>> predictions = ["this is the prediction", "there is an other sample"]
>>> references = ["this is the reference", "there is another one"]
>>> cer = datasets.load_metric("cer")
>>> cer_score = cer.compute(predictions=predictions, references=references)
>>> print(cer_score)
0.34146341463414637
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class CER(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Value("string", id="sequence"),
"references": datasets.Value("string", id="sequence"),
}
),
codebase_urls=["https://github.com/jitsi/jiwer/"],
reference_urls=[
"https://en.wikipedia.org/wiki/Word_error_rate",
"https://sites.google.com/site/textdigitisation/qualitymeasures/computingerrorrates",
],
)
def _compute(self, predictions, references, concatenate_texts=False):
if concatenate_texts:
return jiwer.compute_measures(
references,
predictions,
truth_transform=cer_transform,
hypothesis_transform=cer_transform,
)["wer"]
incorrect = 0
total = 0
for prediction, reference in zip(predictions, references):
measures = jiwer.compute_measures(
reference,
prediction,
truth_transform=cer_transform,
hypothesis_transform=cer_transform,
)
incorrect += measures["substitutions"] + measures["deletions"] + measures["insertions"]
total += measures["substitutions"] + measures["deletions"] + measures["hits"]
return incorrect / total
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/glue/README.md | # Metric Card for GLUE
## Metric description
This metric is used to compute the GLUE evaluation metric associated to each [GLUE dataset](https://huggingface.co/datasets/glue).
GLUE, the General Language Understanding Evaluation benchmark is a collection of resources for training, evaluating, and analyzing natural language understanding systems.
## How to use
There are two steps: (1) loading the GLUE metric relevant to the subset of the GLUE dataset being used for evaluation; and (2) calculating the metric.
1. **Loading the relevant GLUE metric** : the subsets of GLUE are the following: `sst2`, `mnli`, `mnli_mismatched`, `mnli_matched`, `qnli`, `rte`, `wnli`, `cola`,`stsb`, `mrpc`, `qqp`, and `hans`.
More information about the different subsets of the GLUE dataset can be found on the [GLUE dataset page](https://huggingface.co/datasets/glue).
2. **Calculating the metric**: the metric takes two inputs : one list with the predictions of the model to score and one lists of references for each translation.
```python
from datasets import load_metric
glue_metric = load_metric('glue', 'sst2')
references = [0, 1]
predictions = [0, 1]
results = glue_metric.compute(predictions=predictions, references=references)
```
## Output values
The output of the metric depends on the GLUE subset chosen, consisting of a dictionary that contains one or several of the following metrics:
`accuracy`: the proportion of correct predictions among the total number of cases processed, with a range between 0 and 1 (see [accuracy](https://huggingface.co/metrics/accuracy) for more information).
`f1`: the harmonic mean of the precision and recall (see [F1 score](https://huggingface.co/metrics/f1) for more information). Its range is 0-1 -- its lowest possible value is 0, if either the precision or the recall is 0, and its highest possible value is 1.0, which means perfect precision and recall.
`pearson`: a measure of the linear relationship between two datasets (see [Pearson correlation](https://huggingface.co/metrics/pearsonr) for more information). Its range is between -1 and +1, with 0 implying no correlation, and -1/+1 implying an exact linear relationship. Positive correlations imply that as x increases, so does y, whereas negative correlations imply that as x increases, y decreases.
`spearmanr`: a nonparametric measure of the monotonicity of the relationship between two datasets(see [Spearman Correlation](https://huggingface.co/metrics/spearmanr) for more information). `spearmanr` has the same range as `pearson`.
`matthews_correlation`: a measure of the quality of binary and multiclass classifications (see [Matthews Correlation](https://huggingface.co/metrics/matthews_correlation) for more information). Its range of values is between -1 and +1, where a coefficient of +1 represents a perfect prediction, 0 an average random prediction and -1 an inverse prediction.
The `cola` subset returns `matthews_correlation`, the `stsb` subset returns `pearson` and `spearmanr`, the `mrpc` and `qqp` subsets return both `accuracy` and `f1`, and all other subsets of GLUE return only accuracy.
### Values from popular papers
The [original GLUE paper](https://huggingface.co/datasets/glue) reported average scores ranging from 58 to 64%, depending on the model used (with all evaluation values scaled by 100 to make computing the average possible).
For more recent model performance, see the [dataset leaderboard](https://paperswithcode.com/dataset/glue).
## Examples
Maximal values for the MRPC subset (which outputs `accuracy` and `f1`):
```python
from datasets import load_metric
glue_metric = load_metric('glue', 'mrpc') # 'mrpc' or 'qqp'
references = [0, 1]
predictions = [0, 1]
results = glue_metric.compute(predictions=predictions, references=references)
print(results)
{'accuracy': 1.0, 'f1': 1.0}
```
Minimal values for the STSB subset (which outputs `pearson` and `spearmanr`):
```python
from datasets import load_metric
glue_metric = load_metric('glue', 'stsb')
references = [0., 1., 2., 3., 4., 5.]
predictions = [-10., -11., -12., -13., -14., -15.]
results = glue_metric.compute(predictions=predictions, references=references)
print(results)
{'pearson': -1.0, 'spearmanr': -1.0}
```
Partial match for the COLA subset (which outputs `matthews_correlation`)
```python
from datasets import load_metric
glue_metric = load_metric('glue', 'cola')
references = [0, 1]
predictions = [1, 1]
results = glue_metric.compute(predictions=predictions, references=references)
results
{'matthews_correlation': 0.0}
```
## Limitations and bias
This metric works only with datasets that have the same format as the [GLUE dataset](https://huggingface.co/datasets/glue).
While the GLUE dataset is meant to represent "General Language Understanding", the tasks represented in it are not necessarily representative of language understanding, and should not be interpreted as such.
Also, while the GLUE subtasks were considered challenging during its creation in 2019, they are no longer considered as such given the impressive progress made since then. A more complex (or "stickier") version of it, called [SuperGLUE](https://huggingface.co/datasets/super_glue), was subsequently created.
## Citation
```bibtex
@inproceedings{wang2019glue,
title={{GLUE}: A Multi-Task Benchmark and Analysis Platform for Natural Language Understanding},
author={Wang, Alex and Singh, Amanpreet and Michael, Julian and Hill, Felix and Levy, Omer and Bowman, Samuel R.},
note={In the Proceedings of ICLR.},
year={2019}
}
```
## Further References
- [GLUE benchmark homepage](https://gluebenchmark.com/)
- [Fine-tuning a model with the Trainer API](https://huggingface.co/course/chapter3/3?)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/glue/glue.py | # Copyright 2020 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" GLUE benchmark metric. """
from scipy.stats import pearsonr, spearmanr
from sklearn.metrics import f1_score, matthews_corrcoef
import datasets
_CITATION = """\
@inproceedings{wang2019glue,
title={{GLUE}: A Multi-Task Benchmark and Analysis Platform for Natural Language Understanding},
author={Wang, Alex and Singh, Amanpreet and Michael, Julian and Hill, Felix and Levy, Omer and Bowman, Samuel R.},
note={In the Proceedings of ICLR.},
year={2019}
}
"""
_DESCRIPTION = """\
GLUE, the General Language Understanding Evaluation benchmark
(https://gluebenchmark.com/) is a collection of resources for training,
evaluating, and analyzing natural language understanding systems.
"""
_KWARGS_DESCRIPTION = """
Compute GLUE evaluation metric associated to each GLUE dataset.
Args:
predictions: list of predictions to score.
Each translation should be tokenized into a list of tokens.
references: list of lists of references for each translation.
Each reference should be tokenized into a list of tokens.
Returns: depending on the GLUE subset, one or several of:
"accuracy": Accuracy
"f1": F1 score
"pearson": Pearson Correlation
"spearmanr": Spearman Correlation
"matthews_correlation": Matthew Correlation
Examples:
>>> glue_metric = datasets.load_metric('glue', 'sst2') # 'sst2' or any of ["mnli", "mnli_mismatched", "mnli_matched", "qnli", "rte", "wnli", "hans"]
>>> references = [0, 1]
>>> predictions = [0, 1]
>>> results = glue_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'accuracy': 1.0}
>>> glue_metric = datasets.load_metric('glue', 'mrpc') # 'mrpc' or 'qqp'
>>> references = [0, 1]
>>> predictions = [0, 1]
>>> results = glue_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'accuracy': 1.0, 'f1': 1.0}
>>> glue_metric = datasets.load_metric('glue', 'stsb')
>>> references = [0., 1., 2., 3., 4., 5.]
>>> predictions = [0., 1., 2., 3., 4., 5.]
>>> results = glue_metric.compute(predictions=predictions, references=references)
>>> print({"pearson": round(results["pearson"], 2), "spearmanr": round(results["spearmanr"], 2)})
{'pearson': 1.0, 'spearmanr': 1.0}
>>> glue_metric = datasets.load_metric('glue', 'cola')
>>> references = [0, 1]
>>> predictions = [0, 1]
>>> results = glue_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'matthews_correlation': 1.0}
"""
def simple_accuracy(preds, labels):
return float((preds == labels).mean())
def acc_and_f1(preds, labels):
acc = simple_accuracy(preds, labels)
f1 = float(f1_score(y_true=labels, y_pred=preds))
return {
"accuracy": acc,
"f1": f1,
}
def pearson_and_spearman(preds, labels):
pearson_corr = float(pearsonr(preds, labels)[0])
spearman_corr = float(spearmanr(preds, labels)[0])
return {
"pearson": pearson_corr,
"spearmanr": spearman_corr,
}
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Glue(datasets.Metric):
def _info(self):
if self.config_name not in [
"sst2",
"mnli",
"mnli_mismatched",
"mnli_matched",
"cola",
"stsb",
"mrpc",
"qqp",
"qnli",
"rte",
"wnli",
"hans",
]:
raise KeyError(
"You should supply a configuration name selected in "
'["sst2", "mnli", "mnli_mismatched", "mnli_matched", '
'"cola", "stsb", "mrpc", "qqp", "qnli", "rte", "wnli", "hans"]'
)
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Value("int64" if self.config_name != "stsb" else "float32"),
"references": datasets.Value("int64" if self.config_name != "stsb" else "float32"),
}
),
codebase_urls=[],
reference_urls=[],
format="numpy",
)
def _compute(self, predictions, references):
if self.config_name == "cola":
return {"matthews_correlation": matthews_corrcoef(references, predictions)}
elif self.config_name == "stsb":
return pearson_and_spearman(predictions, references)
elif self.config_name in ["mrpc", "qqp"]:
return acc_and_f1(predictions, references)
elif self.config_name in ["sst2", "mnli", "mnli_mismatched", "mnli_matched", "qnli", "rte", "wnli", "hans"]:
return {"accuracy": simple_accuracy(predictions, references)}
else:
raise KeyError(
"You should supply a configuration name selected in "
'["sst2", "mnli", "mnli_mismatched", "mnli_matched", '
'"cola", "stsb", "mrpc", "qqp", "qnli", "rte", "wnli", "hans"]'
)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/squad_v2/README.md | # Metric Card for SQuAD v2
## Metric description
This metric wraps the official scoring script for version 2 of the [Stanford Question Answering Dataset (SQuAD)](https://huggingface.co/datasets/squad_v2).
SQuAD is a reading comprehension dataset, consisting of questions posed by crowdworkers on a set of Wikipedia articles, where the answer to every question is a segment of text, or span, from the corresponding reading passage, or the question might be unanswerable.
SQuAD 2.0 combines the 100,000 questions in SQuAD 1.1 with over 50,000 unanswerable questions written adversarially by crowdworkers to look similar to answerable ones. To do well on SQuAD2.0, systems must not only answer questions when possible, but also determine when no answer is supported by the paragraph and abstain from answering.
## How to use
The metric takes two files or two lists - one representing model predictions and the other the references to compare them to.
*Predictions* : List of triple for question-answers to score with the following key-value pairs:
* `'id'`: the question-answer identification field of the question and answer pair
* `'prediction_text'` : the text of the answer
* `'no_answer_probability'` : the probability that the question has no answer
*References*: List of question-answers dictionaries with the following key-value pairs:
* `'id'`: id of the question-answer pair (see above),
* `'answers'`: a list of Dict {'text': text of the answer as a string}
* `'no_answer_threshold'`: the probability threshold to decide that a question has no answer.
```python
from datasets import load_metric
squad_metric = load_metric("squad_v2")
results = squad_metric.compute(predictions=predictions, references=references)
```
## Output values
This metric outputs a dictionary with 13 values:
* `'exact'`: Exact match (the normalized answer exactly match the gold answer) (see the `exact_match` metric (forthcoming))
* `'f1'`: The average F1-score of predicted tokens versus the gold answer (see the [F1 score](https://huggingface.co/metrics/f1) metric)
* `'total'`: Number of scores considered
* `'HasAns_exact'`: Exact match (the normalized answer exactly match the gold answer)
* `'HasAns_f1'`: The F-score of predicted tokens versus the gold answer
* `'HasAns_total'`: How many of the questions have answers
* `'NoAns_exact'`: Exact match (the normalized answer exactly match the gold answer)
* `'NoAns_f1'`: The F-score of predicted tokens versus the gold answer
* `'NoAns_total'`: How many of the questions have no answers
* `'best_exact'` : Best exact match (with varying threshold)
* `'best_exact_thresh'`: No-answer probability threshold associated to the best exact match
* `'best_f1'`: Best F1 score (with varying threshold)
* `'best_f1_thresh'`: No-answer probability threshold associated to the best F1
The range of `exact_match` is 0-100, where 0.0 means no answers were matched and 100.0 means all answers were matched.
The range of `f1` is 0-1 -- its lowest possible value is 0, if either the precision or the recall is 0, and its highest possible value is 1.0, which means perfect precision and recall.
The range of `total` depends on the length of predictions/references: its minimal value is 0, and maximal value is the total number of questions in the predictions and references.
### Values from popular papers
The [SQuAD v2 paper](https://arxiv.org/pdf/1806.03822.pdf) reported an F1 score of 66.3% and an Exact Match score of 63.4%.
They also report that human performance on the dataset represents an F1 score of 89.5% and an Exact Match score of 86.9%.
For more recent model performance, see the [dataset leaderboard](https://paperswithcode.com/dataset/squad).
## Examples
Maximal values for both exact match and F1 (perfect match):
```python
from datasets import load_metric
squad_v2_ metric = load_metric("squad_v2")
predictions = [{'prediction_text': '1976', 'id': '56e10a3be3433e1400422b22', 'no_answer_probability': 0.}]
references = [{'answers': {'answer_start': [97], 'text': ['1976']}, 'id': '56e10a3be3433e1400422b22'}]
results = squad_v2_metric.compute(predictions=predictions, references=references)
results
{'exact': 100.0, 'f1': 100.0, 'total': 1, 'HasAns_exact': 100.0, 'HasAns_f1': 100.0, 'HasAns_total': 1, 'best_exact': 100.0, 'best_exact_thresh': 0.0, 'best_f1': 100.0, 'best_f1_thresh': 0.0}
```
Minimal values for both exact match and F1 (no match):
```python
from datasets import load_metric
squad_metric = load_metric("squad_v2")
predictions = [{'prediction_text': '1999', 'id': '56e10a3be3433e1400422b22', 'no_answer_probability': 0.}]
references = [{'answers': {'answer_start': [97], 'text': ['1976']}, 'id': '56e10a3be3433e1400422b22'}]
results = squad_v2_metric.compute(predictions=predictions, references=references)
results
{'exact': 0.0, 'f1': 0.0, 'total': 1, 'HasAns_exact': 0.0, 'HasAns_f1': 0.0, 'HasAns_total': 1, 'best_exact': 0.0, 'best_exact_thresh': 0.0, 'best_f1': 0.0, 'best_f1_thresh': 0.0}
```
Partial match (2 out of 3 answers correct) :
```python
from datasets import load_metric
squad_metric = load_metric("squad_v2")
predictions = [{'prediction_text': '1976', 'id': '56e10a3be3433e1400422b22', 'no_answer_probability': 0.}, {'prediction_text': 'Beyonce', 'id': '56d2051ce7d4791d0090260b', 'no_answer_probability': 0.}, {'prediction_text': 'climate change', 'id': '5733b5344776f419006610e1', 'no_answer_probability': 0.}]
references = [{'answers': {'answer_start': [97], 'text': ['1976']}, 'id': '56e10a3be3433e1400422b22'}, {'answers': {'answer_start': [233], 'text': ['Beyoncé and Bruno Mars']}, 'id': '56d2051ce7d4791d0090260b'}, {'answers': {'answer_start': [891], 'text': ['climate change']}, 'id': '5733b5344776f419006610e1'}]
results = squad_v2_metric.compute(predictions=predictions, references=references)
results
{'exact': 66.66666666666667, 'f1': 66.66666666666667, 'total': 3, 'HasAns_exact': 66.66666666666667, 'HasAns_f1': 66.66666666666667, 'HasAns_total': 3, 'best_exact': 66.66666666666667, 'best_exact_thresh': 0.0, 'best_f1': 66.66666666666667, 'best_f1_thresh': 0.0}
```
## Limitations and bias
This metric works only with the datasets in the same format as the [SQuAD v.2 dataset](https://huggingface.co/datasets/squad_v2).
The SQuAD datasets do contain a certain amount of noise, such as duplicate questions as well as missing answers, but these represent a minority of the 100,000 question-answer pairs. Also, neither exact match nor F1 score reflect whether models do better on certain types of questions (e.g. who questions) or those that cover a certain gender or geographical area -- carrying out more in-depth error analysis can complement these numbers.
## Citation
```bibtex
@inproceedings{Rajpurkar2018SQuAD2,
title={Know What You Don't Know: Unanswerable Questions for SQuAD},
author={Pranav Rajpurkar and Jian Zhang and Percy Liang},
booktitle={ACL 2018},
year={2018}
}
```
## Further References
- [The Stanford Question Answering Dataset: Background, Challenges, Progress (blog post)](https://rajpurkar.github.io/mlx/qa-and-squad/)
- [Hugging Face Course -- Question Answering](https://huggingface.co/course/chapter7/7)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/squad_v2/evaluate.py | """Official evaluation script for SQuAD version 2.0.
In addition to basic functionality, we also compute additional statistics and
plot precision-recall curves if an additional na_prob.json file is provided.
This file is expected to map question ID's to the model's predicted probability
that a question is unanswerable.
"""
import argparse
import collections
import json
import os
import re
import string
import sys
import numpy as np
ARTICLES_REGEX = re.compile(r"\b(a|an|the)\b", re.UNICODE)
OPTS = None
def parse_args():
parser = argparse.ArgumentParser("Official evaluation script for SQuAD version 2.0.")
parser.add_argument("data_file", metavar="data.json", help="Input data JSON file.")
parser.add_argument("pred_file", metavar="pred.json", help="Model predictions.")
parser.add_argument(
"--out-file", "-o", metavar="eval.json", help="Write accuracy metrics to file (default is stdout)."
)
parser.add_argument(
"--na-prob-file", "-n", metavar="na_prob.json", help="Model estimates of probability of no answer."
)
parser.add_argument(
"--na-prob-thresh",
"-t",
type=float,
default=1.0,
help='Predict "" if no-answer probability exceeds this (default = 1.0).',
)
parser.add_argument(
"--out-image-dir", "-p", metavar="out_images", default=None, help="Save precision-recall curves to directory."
)
parser.add_argument("--verbose", "-v", action="store_true")
if len(sys.argv) == 1:
parser.print_help()
sys.exit(1)
return parser.parse_args()
def make_qid_to_has_ans(dataset):
qid_to_has_ans = {}
for article in dataset:
for p in article["paragraphs"]:
for qa in p["qas"]:
qid_to_has_ans[qa["id"]] = bool(qa["answers"]["text"])
return qid_to_has_ans
def normalize_answer(s):
"""Lower text and remove punctuation, articles and extra whitespace."""
def remove_articles(text):
return ARTICLES_REGEX.sub(" ", text)
def white_space_fix(text):
return " ".join(text.split())
def remove_punc(text):
exclude = set(string.punctuation)
return "".join(ch for ch in text if ch not in exclude)
def lower(text):
return text.lower()
return white_space_fix(remove_articles(remove_punc(lower(s))))
def get_tokens(s):
if not s:
return []
return normalize_answer(s).split()
def compute_exact(a_gold, a_pred):
return int(normalize_answer(a_gold) == normalize_answer(a_pred))
def compute_f1(a_gold, a_pred):
gold_toks = get_tokens(a_gold)
pred_toks = get_tokens(a_pred)
common = collections.Counter(gold_toks) & collections.Counter(pred_toks)
num_same = sum(common.values())
if len(gold_toks) == 0 or len(pred_toks) == 0:
# If either is no-answer, then F1 is 1 if they agree, 0 otherwise
return int(gold_toks == pred_toks)
if num_same == 0:
return 0
precision = 1.0 * num_same / len(pred_toks)
recall = 1.0 * num_same / len(gold_toks)
f1 = (2 * precision * recall) / (precision + recall)
return f1
def get_raw_scores(dataset, preds):
exact_scores = {}
f1_scores = {}
for article in dataset:
for p in article["paragraphs"]:
for qa in p["qas"]:
qid = qa["id"]
gold_answers = [t for t in qa["answers"]["text"] if normalize_answer(t)]
if not gold_answers:
# For unanswerable questions, only correct answer is empty string
gold_answers = [""]
if qid not in preds:
print(f"Missing prediction for {qid}")
continue
a_pred = preds[qid]
# Take max over all gold answers
exact_scores[qid] = max(compute_exact(a, a_pred) for a in gold_answers)
f1_scores[qid] = max(compute_f1(a, a_pred) for a in gold_answers)
return exact_scores, f1_scores
def apply_no_ans_threshold(scores, na_probs, qid_to_has_ans, na_prob_thresh):
new_scores = {}
for qid, s in scores.items():
pred_na = na_probs[qid] > na_prob_thresh
if pred_na:
new_scores[qid] = float(not qid_to_has_ans[qid])
else:
new_scores[qid] = s
return new_scores
def make_eval_dict(exact_scores, f1_scores, qid_list=None):
if not qid_list:
total = len(exact_scores)
return collections.OrderedDict(
[
("exact", 100.0 * sum(exact_scores.values()) / total),
("f1", 100.0 * sum(f1_scores.values()) / total),
("total", total),
]
)
else:
total = len(qid_list)
return collections.OrderedDict(
[
("exact", 100.0 * sum(exact_scores[k] for k in qid_list) / total),
("f1", 100.0 * sum(f1_scores[k] for k in qid_list) / total),
("total", total),
]
)
def merge_eval(main_eval, new_eval, prefix):
for k in new_eval:
main_eval[f"{prefix}_{k}"] = new_eval[k]
def plot_pr_curve(precisions, recalls, out_image, title):
plt.step(recalls, precisions, color="b", alpha=0.2, where="post")
plt.fill_between(recalls, precisions, step="post", alpha=0.2, color="b")
plt.xlabel("Recall")
plt.ylabel("Precision")
plt.xlim([0.0, 1.05])
plt.ylim([0.0, 1.05])
plt.title(title)
plt.savefig(out_image)
plt.clf()
def make_precision_recall_eval(scores, na_probs, num_true_pos, qid_to_has_ans, out_image=None, title=None):
qid_list = sorted(na_probs, key=lambda k: na_probs[k])
true_pos = 0.0
cur_p = 1.0
cur_r = 0.0
precisions = [1.0]
recalls = [0.0]
avg_prec = 0.0
for i, qid in enumerate(qid_list):
if qid_to_has_ans[qid]:
true_pos += scores[qid]
cur_p = true_pos / float(i + 1)
cur_r = true_pos / float(num_true_pos)
if i == len(qid_list) - 1 or na_probs[qid] != na_probs[qid_list[i + 1]]:
# i.e., if we can put a threshold after this point
avg_prec += cur_p * (cur_r - recalls[-1])
precisions.append(cur_p)
recalls.append(cur_r)
if out_image:
plot_pr_curve(precisions, recalls, out_image, title)
return {"ap": 100.0 * avg_prec}
def run_precision_recall_analysis(main_eval, exact_raw, f1_raw, na_probs, qid_to_has_ans, out_image_dir):
if out_image_dir and not os.path.exists(out_image_dir):
os.makedirs(out_image_dir)
num_true_pos = sum(1 for v in qid_to_has_ans.values() if v)
if num_true_pos == 0:
return
pr_exact = make_precision_recall_eval(
exact_raw,
na_probs,
num_true_pos,
qid_to_has_ans,
out_image=os.path.join(out_image_dir, "pr_exact.png"),
title="Precision-Recall curve for Exact Match score",
)
pr_f1 = make_precision_recall_eval(
f1_raw,
na_probs,
num_true_pos,
qid_to_has_ans,
out_image=os.path.join(out_image_dir, "pr_f1.png"),
title="Precision-Recall curve for F1 score",
)
oracle_scores = {k: float(v) for k, v in qid_to_has_ans.items()}
pr_oracle = make_precision_recall_eval(
oracle_scores,
na_probs,
num_true_pos,
qid_to_has_ans,
out_image=os.path.join(out_image_dir, "pr_oracle.png"),
title="Oracle Precision-Recall curve (binary task of HasAns vs. NoAns)",
)
merge_eval(main_eval, pr_exact, "pr_exact")
merge_eval(main_eval, pr_f1, "pr_f1")
merge_eval(main_eval, pr_oracle, "pr_oracle")
def histogram_na_prob(na_probs, qid_list, image_dir, name):
if not qid_list:
return
x = [na_probs[k] for k in qid_list]
weights = np.ones_like(x) / float(len(x))
plt.hist(x, weights=weights, bins=20, range=(0.0, 1.0))
plt.xlabel("Model probability of no-answer")
plt.ylabel("Proportion of dataset")
plt.title(f"Histogram of no-answer probability: {name}")
plt.savefig(os.path.join(image_dir, f"na_prob_hist_{name}.png"))
plt.clf()
def find_best_thresh(preds, scores, na_probs, qid_to_has_ans):
num_no_ans = sum(1 for k in qid_to_has_ans if not qid_to_has_ans[k])
cur_score = num_no_ans
best_score = cur_score
best_thresh = 0.0
qid_list = sorted(na_probs, key=lambda k: na_probs[k])
for i, qid in enumerate(qid_list):
if qid not in scores:
continue
if qid_to_has_ans[qid]:
diff = scores[qid]
else:
if preds[qid]:
diff = -1
else:
diff = 0
cur_score += diff
if cur_score > best_score:
best_score = cur_score
best_thresh = na_probs[qid]
return 100.0 * best_score / len(scores), best_thresh
def find_all_best_thresh(main_eval, preds, exact_raw, f1_raw, na_probs, qid_to_has_ans):
best_exact, exact_thresh = find_best_thresh(preds, exact_raw, na_probs, qid_to_has_ans)
best_f1, f1_thresh = find_best_thresh(preds, f1_raw, na_probs, qid_to_has_ans)
main_eval["best_exact"] = best_exact
main_eval["best_exact_thresh"] = exact_thresh
main_eval["best_f1"] = best_f1
main_eval["best_f1_thresh"] = f1_thresh
def main():
with open(OPTS.data_file) as f:
dataset_json = json.load(f)
dataset = dataset_json["data"]
with open(OPTS.pred_file) as f:
preds = json.load(f)
if OPTS.na_prob_file:
with open(OPTS.na_prob_file) as f:
na_probs = json.load(f)
else:
na_probs = {k: 0.0 for k in preds}
qid_to_has_ans = make_qid_to_has_ans(dataset) # maps qid to True/False
has_ans_qids = [k for k, v in qid_to_has_ans.items() if v]
no_ans_qids = [k for k, v in qid_to_has_ans.items() if not v]
exact_raw, f1_raw = get_raw_scores(dataset, preds)
exact_thresh = apply_no_ans_threshold(exact_raw, na_probs, qid_to_has_ans, OPTS.na_prob_thresh)
f1_thresh = apply_no_ans_threshold(f1_raw, na_probs, qid_to_has_ans, OPTS.na_prob_thresh)
out_eval = make_eval_dict(exact_thresh, f1_thresh)
if has_ans_qids:
has_ans_eval = make_eval_dict(exact_thresh, f1_thresh, qid_list=has_ans_qids)
merge_eval(out_eval, has_ans_eval, "HasAns")
if no_ans_qids:
no_ans_eval = make_eval_dict(exact_thresh, f1_thresh, qid_list=no_ans_qids)
merge_eval(out_eval, no_ans_eval, "NoAns")
if OPTS.na_prob_file:
find_all_best_thresh(out_eval, preds, exact_raw, f1_raw, na_probs, qid_to_has_ans)
if OPTS.na_prob_file and OPTS.out_image_dir:
run_precision_recall_analysis(out_eval, exact_raw, f1_raw, na_probs, qid_to_has_ans, OPTS.out_image_dir)
histogram_na_prob(na_probs, has_ans_qids, OPTS.out_image_dir, "hasAns")
histogram_na_prob(na_probs, no_ans_qids, OPTS.out_image_dir, "noAns")
if OPTS.out_file:
with open(OPTS.out_file, "w") as f:
json.dump(out_eval, f)
else:
print(json.dumps(out_eval, indent=2))
if __name__ == "__main__":
OPTS = parse_args()
if OPTS.out_image_dir:
import matplotlib
matplotlib.use("Agg")
import matplotlib.pyplot as plt
main()
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/squad_v2/squad_v2.py | # Copyright 2020 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" SQuAD v2 metric. """
import datasets
from .evaluate import (
apply_no_ans_threshold,
find_all_best_thresh,
get_raw_scores,
make_eval_dict,
make_qid_to_has_ans,
merge_eval,
)
_CITATION = """\
@inproceedings{Rajpurkar2016SQuAD10,
title={SQuAD: 100, 000+ Questions for Machine Comprehension of Text},
author={Pranav Rajpurkar and Jian Zhang and Konstantin Lopyrev and Percy Liang},
booktitle={EMNLP},
year={2016}
}
"""
_DESCRIPTION = """
This metric wrap the official scoring script for version 2 of the Stanford Question
Answering Dataset (SQuAD).
Stanford Question Answering Dataset (SQuAD) is a reading comprehension dataset, consisting of questions posed by
crowdworkers on a set of Wikipedia articles, where the answer to every question is a segment of text, or span,
from the corresponding reading passage, or the question might be unanswerable.
SQuAD2.0 combines the 100,000 questions in SQuAD1.1 with over 50,000 unanswerable questions
written adversarially by crowdworkers to look similar to answerable ones.
To do well on SQuAD2.0, systems must not only answer questions when possible, but also
determine when no answer is supported by the paragraph and abstain from answering.
"""
_KWARGS_DESCRIPTION = """
Computes SQuAD v2 scores (F1 and EM).
Args:
predictions: List of triple for question-answers to score with the following elements:
- the question-answer 'id' field as given in the references (see below)
- the text of the answer
- the probability that the question has no answer
references: List of question-answers dictionaries with the following key-values:
- 'id': id of the question-answer pair (see above),
- 'answers': a list of Dict {'text': text of the answer as a string}
no_answer_threshold: float
Probability threshold to decide that a question has no answer.
Returns:
'exact': Exact match (the normalized answer exactly match the gold answer)
'f1': The F-score of predicted tokens versus the gold answer
'total': Number of score considered
'HasAns_exact': Exact match (the normalized answer exactly match the gold answer)
'HasAns_f1': The F-score of predicted tokens versus the gold answer
'HasAns_total': Number of score considered
'NoAns_exact': Exact match (the normalized answer exactly match the gold answer)
'NoAns_f1': The F-score of predicted tokens versus the gold answer
'NoAns_total': Number of score considered
'best_exact': Best exact match (with varying threshold)
'best_exact_thresh': No-answer probability threshold associated to the best exact match
'best_f1': Best F1 (with varying threshold)
'best_f1_thresh': No-answer probability threshold associated to the best F1
Examples:
>>> predictions = [{'prediction_text': '1976', 'id': '56e10a3be3433e1400422b22', 'no_answer_probability': 0.}]
>>> references = [{'answers': {'answer_start': [97], 'text': ['1976']}, 'id': '56e10a3be3433e1400422b22'}]
>>> squad_v2_metric = datasets.load_metric("squad_v2")
>>> results = squad_v2_metric.compute(predictions=predictions, references=references)
>>> print(results)
{'exact': 100.0, 'f1': 100.0, 'total': 1, 'HasAns_exact': 100.0, 'HasAns_f1': 100.0, 'HasAns_total': 1, 'best_exact': 100.0, 'best_exact_thresh': 0.0, 'best_f1': 100.0, 'best_f1_thresh': 0.0}
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class SquadV2(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": {
"id": datasets.Value("string"),
"prediction_text": datasets.Value("string"),
"no_answer_probability": datasets.Value("float32"),
},
"references": {
"id": datasets.Value("string"),
"answers": datasets.features.Sequence(
{"text": datasets.Value("string"), "answer_start": datasets.Value("int32")}
),
},
}
),
codebase_urls=["https://rajpurkar.github.io/SQuAD-explorer/"],
reference_urls=["https://rajpurkar.github.io/SQuAD-explorer/"],
)
def _compute(self, predictions, references, no_answer_threshold=1.0):
no_answer_probabilities = {p["id"]: p["no_answer_probability"] for p in predictions}
dataset = [{"paragraphs": [{"qas": references}]}]
predictions = {p["id"]: p["prediction_text"] for p in predictions}
qid_to_has_ans = make_qid_to_has_ans(dataset) # maps qid to True/False
has_ans_qids = [k for k, v in qid_to_has_ans.items() if v]
no_ans_qids = [k for k, v in qid_to_has_ans.items() if not v]
exact_raw, f1_raw = get_raw_scores(dataset, predictions)
exact_thresh = apply_no_ans_threshold(exact_raw, no_answer_probabilities, qid_to_has_ans, no_answer_threshold)
f1_thresh = apply_no_ans_threshold(f1_raw, no_answer_probabilities, qid_to_has_ans, no_answer_threshold)
out_eval = make_eval_dict(exact_thresh, f1_thresh)
if has_ans_qids:
has_ans_eval = make_eval_dict(exact_thresh, f1_thresh, qid_list=has_ans_qids)
merge_eval(out_eval, has_ans_eval, "HasAns")
if no_ans_qids:
no_ans_eval = make_eval_dict(exact_thresh, f1_thresh, qid_list=no_ans_qids)
merge_eval(out_eval, no_ans_eval, "NoAns")
find_all_best_thresh(out_eval, predictions, exact_raw, f1_raw, no_answer_probabilities, qid_to_has_ans)
return dict(out_eval)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/ter/README.md | # Metric Card for TER
## Metric Description
TER (Translation Edit Rate, also called Translation Error Rate) is a metric to quantify the edit operations that a hypothesis requires to match a reference translation. We use the implementation that is already present in [sacrebleu](https://github.com/mjpost/sacreBLEU#ter), which in turn is inspired by the [TERCOM implementation](https://github.com/jhclark/tercom).
The implementation here is slightly different from sacrebleu in terms of the required input format. The length of the references and hypotheses lists need to be the same, so you may need to transpose your references compared to sacrebleu's required input format. See [this github issue](https://github.com/huggingface/datasets/issues/3154#issuecomment-950746534).
See the README.md file at https://github.com/mjpost/sacreBLEU#ter for more information.
## How to Use
This metric takes, at minimum, predicted sentences and reference sentences:
```python
>>> predictions = ["does this sentence match??",
... "what about this sentence?",
... "What did the TER metric user say to the developer?"]
>>> references = [["does this sentence match", "does this sentence match!?!"],
... ["wHaT aBoUt ThIs SeNtEnCe?", "wHaT aBoUt ThIs SeNtEnCe?"],
... ["Your jokes are...", "...TERrible"]]
>>> ter = datasets.load_metric("ter")
>>> results = ter.compute(predictions=predictions,
... references=references,
... case_sensitive=True)
>>> print(results)
{'score': 150.0, 'num_edits': 15, 'ref_length': 10.0}
```
### Inputs
This metric takes the following as input:
- **`predictions`** (`list` of `str`): The system stream (a sequence of segments).
- **`references`** (`list` of `list` of `str`): A list of one or more reference streams (each a sequence of segments).
- **`normalized`** (`boolean`): If `True`, applies basic tokenization and normalization to sentences. Defaults to `False`.
- **`ignore_punct`** (`boolean`): If `True`, applies basic tokenization and normalization to sentences. Defaults to `False`.
- **`support_zh_ja_chars`** (`boolean`): If `True`, tokenization/normalization supports processing of Chinese characters, as well as Japanese Kanji, Hiragana, Katakana, and Phonetic Extensions of Katakana. Only applies if `normalized = True`. Defaults to `False`.
- **`case_sensitive`** (`boolean`): If `False`, makes all predictions and references lowercase to ignore differences in case. Defaults to `False`.
### Output Values
This metric returns the following:
- **`score`** (`float`): TER score (num_edits / sum_ref_lengths * 100)
- **`num_edits`** (`int`): The cumulative number of edits
- **`ref_length`** (`float`): The cumulative average reference length
The output takes the following form:
```python
{'score': ter_score, 'num_edits': num_edits, 'ref_length': ref_length}
```
The metric can take on any value `0` and above. `0` is a perfect score, meaning the predictions exactly match the references and no edits were necessary. Higher scores are worse. Scores above 100 mean that the cumulative number of edits, `num_edits`, is higher than the cumulative length of the references, `ref_length`.
#### Values from Popular Papers
### Examples
Basic example with only predictions and references as inputs:
```python
>>> predictions = ["does this sentence match??",
... "what about this sentence?"]
>>> references = [["does this sentence match", "does this sentence match!?!"],
... ["wHaT aBoUt ThIs SeNtEnCe?", "wHaT aBoUt ThIs SeNtEnCe?"]]
>>> ter = datasets.load_metric("ter")
>>> results = ter.compute(predictions=predictions,
... references=references,
... case_sensitive=True)
>>> print(results)
{'score': 62.5, 'num_edits': 5, 'ref_length': 8.0}
```
Example with `normalization = True`:
```python
>>> predictions = ["does this sentence match??",
... "what about this sentence?"]
>>> references = [["does this sentence match", "does this sentence match!?!"],
... ["wHaT aBoUt ThIs SeNtEnCe?", "wHaT aBoUt ThIs SeNtEnCe?"]]
>>> ter = datasets.load_metric("ter")
>>> results = ter.compute(predictions=predictions,
... references=references,
... normalized=True,
... case_sensitive=True)
>>> print(results)
{'score': 57.14285714285714, 'num_edits': 6, 'ref_length': 10.5}
```
Example ignoring punctuation and capitalization, and everything matches:
```python
>>> predictions = ["does this sentence match??",
... "what about this sentence?"]
>>> references = [["does this sentence match", "does this sentence match!?!"],
... ["wHaT aBoUt ThIs SeNtEnCe?", "wHaT aBoUt ThIs SeNtEnCe?"]]
>>> ter = datasets.load_metric("ter")
>>> results = ter.compute(predictions=predictions,
... references=references,
... ignore_punct=True,
... case_sensitive=False)
>>> print(results)
{'score': 0.0, 'num_edits': 0, 'ref_length': 8.0}
```
Example ignoring punctuation and capitalization, but with an extra (incorrect) sample:
```python
>>> predictions = ["does this sentence match??",
... "what about this sentence?",
... "What did the TER metric user say to the developer?"]
>>> references = [["does this sentence match", "does this sentence match!?!"],
... ["wHaT aBoUt ThIs SeNtEnCe?", "wHaT aBoUt ThIs SeNtEnCe?"],
... ["Your jokes are...", "...TERrible"]]
>>> ter = datasets.load_metric("ter")
>>> results = ter.compute(predictions=predictions,
... references=references,
... ignore_punct=True,
... case_sensitive=False)
>>> print(results)
{'score': 100.0, 'num_edits': 10, 'ref_length': 10.0}
```
## Limitations and Bias
## Citation
```bibtex
@inproceedings{snover-etal-2006-study,
title = "A Study of Translation Edit Rate with Targeted Human Annotation",
author = "Snover, Matthew and
Dorr, Bonnie and
Schwartz, Rich and
Micciulla, Linnea and
Makhoul, John",
booktitle = "Proceedings of the 7th Conference of the Association for Machine Translation in the Americas: Technical Papers",
month = aug # " 8-12",
year = "2006",
address = "Cambridge, Massachusetts, USA",
publisher = "Association for Machine Translation in the Americas",
url = "https://aclanthology.org/2006.amta-papers.25",
pages = "223--231",
}
@inproceedings{post-2018-call,
title = "A Call for Clarity in Reporting {BLEU} Scores",
author = "Post, Matt",
booktitle = "Proceedings of the Third Conference on Machine Translation: Research Papers",
month = oct,
year = "2018",
address = "Belgium, Brussels",
publisher = "Association for Computational Linguistics",
url = "https://www.aclweb.org/anthology/W18-6319",
pages = "186--191",
}
```
## Further References
- See [the sacreBLEU github repo](https://github.com/mjpost/sacreBLEU#ter) for more information.
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/ter/ter.py | # Copyright 2021 The HuggingFace Datasets Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
""" TER metric as available in sacrebleu. """
import sacrebleu as scb
from packaging import version
from sacrebleu import TER
import datasets
_CITATION = """\
@inproceedings{snover-etal-2006-study,
title = "A Study of Translation Edit Rate with Targeted Human Annotation",
author = "Snover, Matthew and
Dorr, Bonnie and
Schwartz, Rich and
Micciulla, Linnea and
Makhoul, John",
booktitle = "Proceedings of the 7th Conference of the Association for Machine Translation in the Americas: Technical Papers",
month = aug # " 8-12",
year = "2006",
address = "Cambridge, Massachusetts, USA",
publisher = "Association for Machine Translation in the Americas",
url = "https://aclanthology.org/2006.amta-papers.25",
pages = "223--231",
}
@inproceedings{post-2018-call,
title = "A Call for Clarity in Reporting {BLEU} Scores",
author = "Post, Matt",
booktitle = "Proceedings of the Third Conference on Machine Translation: Research Papers",
month = oct,
year = "2018",
address = "Belgium, Brussels",
publisher = "Association for Computational Linguistics",
url = "https://www.aclweb.org/anthology/W18-6319",
pages = "186--191",
}
"""
_DESCRIPTION = """\
TER (Translation Edit Rate, also called Translation Error Rate) is a metric to quantify the edit operations that a
hypothesis requires to match a reference translation. We use the implementation that is already present in sacrebleu
(https://github.com/mjpost/sacreBLEU#ter), which in turn is inspired by the TERCOM implementation, which can be found
here: https://github.com/jhclark/tercom.
The implementation here is slightly different from sacrebleu in terms of the required input format. The length of
the references and hypotheses lists need to be the same, so you may need to transpose your references compared to
sacrebleu's required input format. See https://github.com/huggingface/datasets/issues/3154#issuecomment-950746534
See the README.md file at https://github.com/mjpost/sacreBLEU#ter for more information.
"""
_KWARGS_DESCRIPTION = """
Produces TER scores alongside the number of edits and reference length.
Args:
predictions (list of str): The system stream (a sequence of segments).
references (list of list of str): A list of one or more reference streams (each a sequence of segments).
normalized (boolean): If `True`, applies basic tokenization and normalization to sentences. Defaults to `False`.
ignore_punct (boolean): If `True`, applies basic tokenization and normalization to sentences. Defaults to `False`.
support_zh_ja_chars (boolean): If `True`, tokenization/normalization supports processing of Chinese characters,
as well as Japanese Kanji, Hiragana, Katakana, and Phonetic Extensions of Katakana.
Only applies if `normalized = True`. Defaults to `False`.
case_sensitive (boolean): If `False`, makes all predictions and references lowercase to ignore differences in case. Defaults to `False`.
Returns:
'score' (float): TER score (num_edits / sum_ref_lengths * 100)
'num_edits' (int): The cumulative number of edits
'ref_length' (float): The cumulative average reference length
Examples:
Example 1:
>>> predictions = ["does this sentence match??",
... "what about this sentence?",
... "What did the TER metric user say to the developer?"]
>>> references = [["does this sentence match", "does this sentence match!?!"],
... ["wHaT aBoUt ThIs SeNtEnCe?", "wHaT aBoUt ThIs SeNtEnCe?"],
... ["Your jokes are...", "...TERrible"]]
>>> ter = datasets.load_metric("ter")
>>> results = ter.compute(predictions=predictions,
... references=references,
... case_sensitive=True)
>>> print(results)
{'score': 150.0, 'num_edits': 15, 'ref_length': 10.0}
Example 2:
>>> predictions = ["does this sentence match??",
... "what about this sentence?"]
>>> references = [["does this sentence match", "does this sentence match!?!"],
... ["wHaT aBoUt ThIs SeNtEnCe?", "wHaT aBoUt ThIs SeNtEnCe?"]]
>>> ter = datasets.load_metric("ter")
>>> results = ter.compute(predictions=predictions,
... references=references,
... case_sensitive=True)
>>> print(results)
{'score': 62.5, 'num_edits': 5, 'ref_length': 8.0}
Example 3:
>>> predictions = ["does this sentence match??",
... "what about this sentence?"]
>>> references = [["does this sentence match", "does this sentence match!?!"],
... ["wHaT aBoUt ThIs SeNtEnCe?", "wHaT aBoUt ThIs SeNtEnCe?"]]
>>> ter = datasets.load_metric("ter")
>>> results = ter.compute(predictions=predictions,
... references=references,
... normalized=True,
... case_sensitive=True)
>>> print(results)
{'score': 57.14285714285714, 'num_edits': 6, 'ref_length': 10.5}
Example 4:
>>> predictions = ["does this sentence match??",
... "what about this sentence?"]
>>> references = [["does this sentence match", "does this sentence match!?!"],
... ["wHaT aBoUt ThIs SeNtEnCe?", "wHaT aBoUt ThIs SeNtEnCe?"]]
>>> ter = datasets.load_metric("ter")
>>> results = ter.compute(predictions=predictions,
... references=references,
... ignore_punct=True,
... case_sensitive=False)
>>> print(results)
{'score': 0.0, 'num_edits': 0, 'ref_length': 8.0}
Example 5:
>>> predictions = ["does this sentence match??",
... "what about this sentence?",
... "What did the TER metric user say to the developer?"]
>>> references = [["does this sentence match", "does this sentence match!?!"],
... ["wHaT aBoUt ThIs SeNtEnCe?", "wHaT aBoUt ThIs SeNtEnCe?"],
... ["Your jokes are...", "...TERrible"]]
>>> ter = datasets.load_metric("ter")
>>> results = ter.compute(predictions=predictions,
... references=references,
... ignore_punct=True,
... case_sensitive=False)
>>> print(results)
{'score': 100.0, 'num_edits': 10, 'ref_length': 10.0}
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Ter(datasets.Metric):
def _info(self):
if version.parse(scb.__version__) < version.parse("1.4.12"):
raise ImportWarning(
"To use `sacrebleu`, the module `sacrebleu>=1.4.12` is required, and the current version of `sacrebleu` doesn't match this condition.\n"
'You can install it with `pip install "sacrebleu>=1.4.12"`.'
)
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
homepage="http://www.cs.umd.edu/~snover/tercom/",
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Value("string", id="sequence"),
"references": datasets.Sequence(datasets.Value("string", id="sequence"), id="references"),
}
),
codebase_urls=["https://github.com/mjpost/sacreBLEU#ter"],
reference_urls=[
"https://github.com/jhclark/tercom",
],
)
def _compute(
self,
predictions,
references,
normalized: bool = False,
ignore_punct: bool = False,
support_zh_ja_chars: bool = False,
case_sensitive: bool = False,
):
references_per_prediction = len(references[0])
if any(len(refs) != references_per_prediction for refs in references):
raise ValueError("Sacrebleu requires the same number of references for each prediction")
transformed_references = [[refs[i] for refs in references] for i in range(references_per_prediction)]
sb_ter = TER(
normalized=normalized,
no_punct=ignore_punct,
asian_support=support_zh_ja_chars,
case_sensitive=case_sensitive,
)
output = sb_ter.corpus_score(predictions, transformed_references)
return {"score": output.score, "num_edits": output.num_edits, "ref_length": output.ref_length}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/code_eval/README.md | # Metric Card for Code Eval
## Metric description
The CodeEval metric estimates the pass@k metric for code synthesis.
It implements the evaluation harness for the HumanEval problem solving dataset described in the paper ["Evaluating Large Language Models Trained on Code"](https://arxiv.org/abs/2107.03374).
## How to use
The Code Eval metric calculates how good are predictions given a set of references. Its arguments are:
`predictions`: a list of candidates to evaluate. Each candidate should be a list of strings with several code candidates to solve the problem.
`references`: a list with a test for each prediction. Each test should evaluate the correctness of a code candidate.
`k`: number of code candidates to consider in the evaluation. The default value is `[1, 10, 100]`.
`num_workers`: the number of workers used to evaluate the candidate programs (The default value is `4`).
`timeout`: The maximum time taken to produce a prediction before it is considered a "timeout". The default value is `3.0` (i.e. 3 seconds).
```python
from datasets import load_metric
code_eval = load_metric("code_eval")
test_cases = ["assert add(2,3)==5"]
candidates = [["def add(a,b): return a*b", "def add(a, b): return a+b"]]
pass_at_k, results = code_eval.compute(references=test_cases, predictions=candidates, k=[1, 2])
```
N.B.
This metric exists to run untrusted model-generated code. Users are strongly encouraged not to do so outside of a robust security sandbox. Before running this metric and once you've taken the necessary precautions, you will need to set the `HF_ALLOW_CODE_EVAL` environment variable. Use it at your own risk:
```python
import os
os.environ["HF_ALLOW_CODE_EVAL"] = "1"`
```
## Output values
The Code Eval metric outputs two things:
`pass_at_k`: a dictionary with the pass rates for each k value defined in the arguments.
`results`: a dictionary with granular results of each unit test.
### Values from popular papers
The [original CODEX paper](https://arxiv.org/pdf/2107.03374.pdf) reported that the CODEX-12B model had a pass@k score of 28.8% at `k=1`, 46.8% at `k=10` and 72.3% at `k=100`. However, since the CODEX model is not open source, it is hard to verify these numbers.
## Examples
Full match at `k=1`:
```python
from datasets import load_metric
code_eval = load_metric("code_eval")
test_cases = ["assert add(2,3)==5"]
candidates = [["def add(a, b): return a+b"]]
pass_at_k, results = code_eval.compute(references=test_cases, predictions=candidates, k=[1])
print(pass_at_k)
{'pass@1': 1.0}
```
No match for k = 1:
```python
from datasets import load_metric
code_eval = load_metric("code_eval")
test_cases = ["assert add(2,3)==5"]
candidates = [["def add(a,b): return a*b"]]
pass_at_k, results = code_eval.compute(references=test_cases, predictions=candidates, k=[1])
print(pass_at_k)
{'pass@1': 0.0}
```
Partial match at k=1, full match at k=2:
```python
from datasets import load_metric
code_eval = load_metric("code_eval")
test_cases = ["assert add(2,3)==5"]
candidates = [["def add(a, b): return a+b", "def add(a,b): return a*b"]]
pass_at_k, results = code_eval.compute(references=test_cases, predictions=candidates, k=[1, 2])
print(pass_at_k)
{'pass@1': 0.5, 'pass@2': 1.0}
```
## Limitations and bias
As per the warning included in the metric code itself:
> This program exists to execute untrusted model-generated code. Although it is highly unlikely that model-generated code will do something overtly malicious in response to this test suite, model-generated code may act destructively due to a lack of model capability or alignment. Users are strongly encouraged to sandbox this evaluation suite so that it does not perform destructive actions on their host or network. For more information on how OpenAI sandboxes its code, see the accompanying paper. Once you have read this disclaimer and taken appropriate precautions, uncomment the following line and proceed at your own risk:
More information about the limitations of the code can be found on the [Human Eval Github repository](https://github.com/openai/human-eval).
## Citation
```bibtex
@misc{chen2021evaluating,
title={Evaluating Large Language Models Trained on Code},
author={Mark Chen and Jerry Tworek and Heewoo Jun and Qiming Yuan \
and Henrique Ponde de Oliveira Pinto and Jared Kaplan and Harri Edwards \
and Yuri Burda and Nicholas Joseph and Greg Brockman and Alex Ray \
and Raul Puri and Gretchen Krueger and Michael Petrov and Heidy Khlaaf \
and Girish Sastry and Pamela Mishkin and Brooke Chan and Scott Gray \
and Nick Ryder and Mikhail Pavlov and Alethea Power and Lukasz Kaiser \
and Mohammad Bavarian and Clemens Winter and Philippe Tillet \
and Felipe Petroski Such and Dave Cummings and Matthias Plappert \
and Fotios Chantzis and Elizabeth Barnes and Ariel Herbert-Voss \
and William Hebgen Guss and Alex Nichol and Alex Paino and Nikolas Tezak \
and Jie Tang and Igor Babuschkin and Suchir Balaji and Shantanu Jain \
and William Saunders and Christopher Hesse and Andrew N. Carr \
and Jan Leike and Josh Achiam and Vedant Misra and Evan Morikawa \
and Alec Radford and Matthew Knight and Miles Brundage and Mira Murati \
and Katie Mayer and Peter Welinder and Bob McGrew and Dario Amodei \
and Sam McCandlish and Ilya Sutskever and Wojciech Zaremba},
year={2021},
eprint={2107.03374},
archivePrefix={arXiv},
primaryClass={cs.LG}
}
```
## Further References
- [Human Eval Github repository](https://github.com/openai/human-eval)
- [OpenAI Codex website](https://openai.com/blog/openai-codex/)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/code_eval/code_eval.py | # Copyright 2020 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""The CodeEval metric estimates the pass@k metric for code synthesis.
This is an evaluation harness for the HumanEval problem solving dataset
described in the paper "Evaluating Large Language Models Trained on Code"
(https://arxiv.org/abs/2107.03374)."""
import itertools
import os
from collections import Counter, defaultdict
from concurrent.futures import ThreadPoolExecutor, as_completed
import numpy as np
import datasets
from .execute import check_correctness
_CITATION = """\
@misc{chen2021evaluating,
title={Evaluating Large Language Models Trained on Code},
author={Mark Chen and Jerry Tworek and Heewoo Jun and Qiming Yuan \
and Henrique Ponde de Oliveira Pinto and Jared Kaplan and Harri Edwards \
and Yuri Burda and Nicholas Joseph and Greg Brockman and Alex Ray \
and Raul Puri and Gretchen Krueger and Michael Petrov and Heidy Khlaaf \
and Girish Sastry and Pamela Mishkin and Brooke Chan and Scott Gray \
and Nick Ryder and Mikhail Pavlov and Alethea Power and Lukasz Kaiser \
and Mohammad Bavarian and Clemens Winter and Philippe Tillet \
and Felipe Petroski Such and Dave Cummings and Matthias Plappert \
and Fotios Chantzis and Elizabeth Barnes and Ariel Herbert-Voss \
and William Hebgen Guss and Alex Nichol and Alex Paino and Nikolas Tezak \
and Jie Tang and Igor Babuschkin and Suchir Balaji and Shantanu Jain \
and William Saunders and Christopher Hesse and Andrew N. Carr \
and Jan Leike and Josh Achiam and Vedant Misra and Evan Morikawa \
and Alec Radford and Matthew Knight and Miles Brundage and Mira Murati \
and Katie Mayer and Peter Welinder and Bob McGrew and Dario Amodei \
and Sam McCandlish and Ilya Sutskever and Wojciech Zaremba},
year={2021},
eprint={2107.03374},
archivePrefix={arXiv},
primaryClass={cs.LG}
}
"""
_DESCRIPTION = """\
This metric implements the evaluation harness for the HumanEval problem solving dataset
described in the paper "Evaluating Large Language Models Trained on Code"
(https://arxiv.org/abs/2107.03374).
"""
_KWARGS_DESCRIPTION = """
Calculates how good are predictions given some references, using certain scores
Args:
predictions: list of candidates to evaluate. Each candidates should be a list
of strings with several code candidates to solve the problem.
references: a list with a test for each prediction. Each test should evaluate the
correctness of a code candidate.
k: number of code candidates to consider in the evaluation (Default: [1, 10, 100])
num_workers: number of workers used to evaluate the canidate programs (Default: 4).
timeout:
Returns:
pass_at_k: dict with pass rates for each k
results: dict with granular results of each unittest
Examples:
>>> code_eval = datasets.load_metric("code_eval")
>>> test_cases = ["assert add(2,3)==5"]
>>> candidates = [["def add(a,b): return a*b", "def add(a, b): return a+b"]]
>>> pass_at_k, results = code_eval.compute(references=test_cases, predictions=candidates, k=[1, 2])
>>> print(pass_at_k)
{'pass@1': 0.5, 'pass@2': 1.0}
"""
_WARNING = """
################################################################################
!!!WARNING!!!
################################################################################
The "code_eval" metric executes untrusted model-generated code in Python.
Although it is highly unlikely that model-generated code will do something
overtly malicious in response to this test suite, model-generated code may act
destructively due to a lack of model capability or alignment.
Users are strongly encouraged to sandbox this evaluation suite so that it
does not perform destructive actions on their host or network. For more
information on how OpenAI sandboxes its code, see the paper "Evaluating Large
Language Models Trained on Code" (https://arxiv.org/abs/2107.03374).
Once you have read this disclaimer and taken appropriate precautions,
set the environment variable HF_ALLOW_CODE_EVAL="1". Within Python you can to this
with:
>>> import os
>>> os.environ["HF_ALLOW_CODE_EVAL"] = "1"
################################################################################\
"""
_LICENSE = """The MIT License
Copyright (c) OpenAI (https://openai.com)
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the "Software"), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in
all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN
THE SOFTWARE."""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class CodeEval(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
# This is the description that will appear on the metrics page.
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
# This defines the format of each prediction and reference
features=datasets.Features(
{
"predictions": datasets.Sequence(datasets.Value("string")),
"references": datasets.Value("string"),
}
),
homepage="https://github.com/openai/human-eval",
codebase_urls=["https://github.com/openai/human-eval"],
reference_urls=["https://github.com/openai/human-eval"],
license=_LICENSE,
)
def _compute(self, predictions, references, k=[1, 10, 100], num_workers=4, timeout=3.0):
"""Returns the scores"""
if os.getenv("HF_ALLOW_CODE_EVAL", 0) != "1":
raise ValueError(_WARNING)
if os.name == "nt":
raise NotImplementedError("This metric is currently not supported on Windows.")
with ThreadPoolExecutor(max_workers=num_workers) as executor:
futures = []
completion_id = Counter()
n_samples = 0
results = defaultdict(list)
for task_id, (candidates, test_case) in enumerate(zip(predictions, references)):
for candidate in candidates:
test_program = candidate + "\n" + test_case
args = (test_program, timeout, task_id, completion_id[task_id])
future = executor.submit(check_correctness, *args)
futures.append(future)
completion_id[task_id] += 1
n_samples += 1
for future in as_completed(futures):
result = future.result()
results[result["task_id"]].append((result["completion_id"], result))
total, correct = [], []
for result in results.values():
result.sort()
passed = [r[1]["passed"] for r in result]
total.append(len(passed))
correct.append(sum(passed))
total = np.array(total)
correct = np.array(correct)
ks = k
pass_at_k = {f"pass@{k}": estimate_pass_at_k(total, correct, k).mean() for k in ks if (total >= k).all()}
return pass_at_k, results
def estimate_pass_at_k(num_samples, num_correct, k):
"""Estimates pass@k of each problem and returns them in an array."""
def estimator(n: int, c: int, k: int) -> float:
"""Calculates 1 - comb(n - c, k) / comb(n, k)."""
if n - c < k:
return 1.0
return 1.0 - np.prod(1.0 - k / np.arange(n - c + 1, n + 1))
if isinstance(num_samples, int):
num_samples_it = itertools.repeat(num_samples, len(num_correct))
else:
assert len(num_samples) == len(num_correct)
num_samples_it = iter(num_samples)
return np.array([estimator(int(n), int(c), k) for n, c in zip(num_samples_it, num_correct)])
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/code_eval/execute.py | # Copyright 2020 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# This code is adapted from OpenAI's release
# https://github.com/openai/human-eval/blob/master/human_eval/execution.py
import contextlib
import faulthandler
import io
import multiprocessing
import os
import platform
import signal
import tempfile
def check_correctness(check_program, timeout, task_id, completion_id):
"""
Evaluates the functional correctness of a completion by running the test
suite provided in the problem.
:param completion_id: an optional completion ID so we can match
the results later even if execution finishes asynchronously.
"""
manager = multiprocessing.Manager()
result = manager.list()
p = multiprocessing.Process(target=unsafe_execute, args=(check_program, result, timeout))
p.start()
p.join(timeout=timeout + 1)
if p.is_alive():
p.kill()
if not result:
result.append("timed out")
return {
"task_id": task_id,
"passed": result[0] == "passed",
"result": result[0],
"completion_id": completion_id,
}
def unsafe_execute(check_program, result, timeout):
with create_tempdir():
# These system calls are needed when cleaning up tempdir.
import os
import shutil
rmtree = shutil.rmtree
rmdir = os.rmdir
chdir = os.chdir
# Disable functionalities that can make destructive changes to the test.
reliability_guard()
# Run program.
try:
exec_globals = {}
with swallow_io():
with time_limit(timeout):
exec(check_program, exec_globals)
result.append("passed")
except TimeoutException:
result.append("timed out")
except BaseException as e:
result.append(f"failed: {e}")
# Needed for cleaning up.
shutil.rmtree = rmtree
os.rmdir = rmdir
os.chdir = chdir
@contextlib.contextmanager
def time_limit(seconds):
def signal_handler(signum, frame):
raise TimeoutException("Timed out!")
signal.setitimer(signal.ITIMER_REAL, seconds)
signal.signal(signal.SIGALRM, signal_handler)
try:
yield
finally:
signal.setitimer(signal.ITIMER_REAL, 0)
@contextlib.contextmanager
def swallow_io():
stream = WriteOnlyStringIO()
with contextlib.redirect_stdout(stream):
with contextlib.redirect_stderr(stream):
with redirect_stdin(stream):
yield
@contextlib.contextmanager
def create_tempdir():
with tempfile.TemporaryDirectory() as dirname:
with chdir(dirname):
yield dirname
class TimeoutException(Exception):
pass
class WriteOnlyStringIO(io.StringIO):
"""StringIO that throws an exception when it's read from"""
def read(self, *args, **kwargs):
raise OSError
def readline(self, *args, **kwargs):
raise OSError
def readlines(self, *args, **kwargs):
raise OSError
def readable(self, *args, **kwargs):
"""Returns True if the IO object can be read."""
return False
class redirect_stdin(contextlib._RedirectStream): # type: ignore
_stream = "stdin"
@contextlib.contextmanager
def chdir(root):
if root == ".":
yield
return
cwd = os.getcwd()
os.chdir(root)
try:
yield
except BaseException as exc:
raise exc
finally:
os.chdir(cwd)
def reliability_guard(maximum_memory_bytes=None):
"""
This disables various destructive functions and prevents the generated code
from interfering with the test (e.g. fork bomb, killing other processes,
removing filesystem files, etc.)
WARNING
This function is NOT a security sandbox. Untrusted code, including, model-
generated code, should not be blindly executed outside of one. See the
Codex paper for more information about OpenAI's code sandbox, and proceed
with caution.
"""
if maximum_memory_bytes is not None:
import resource
resource.setrlimit(resource.RLIMIT_AS, (maximum_memory_bytes, maximum_memory_bytes))
resource.setrlimit(resource.RLIMIT_DATA, (maximum_memory_bytes, maximum_memory_bytes))
if not platform.uname().system == "Darwin":
resource.setrlimit(resource.RLIMIT_STACK, (maximum_memory_bytes, maximum_memory_bytes))
faulthandler.disable()
import builtins
builtins.exit = None
builtins.quit = None
import os
os.environ["OMP_NUM_THREADS"] = "1"
os.kill = None
os.system = None
os.putenv = None
os.remove = None
os.removedirs = None
os.rmdir = None
os.fchdir = None
os.setuid = None
os.fork = None
os.forkpty = None
os.killpg = None
os.rename = None
os.renames = None
os.truncate = None
os.replace = None
os.unlink = None
os.fchmod = None
os.fchown = None
os.chmod = None
os.chown = None
os.chroot = None
os.fchdir = None
os.lchflags = None
os.lchmod = None
os.lchown = None
os.getcwd = None
os.chdir = None
import shutil
shutil.rmtree = None
shutil.move = None
shutil.chown = None
import subprocess
subprocess.Popen = None # type: ignore
__builtins__["help"] = None
import sys
sys.modules["ipdb"] = None
sys.modules["joblib"] = None
sys.modules["resource"] = None
sys.modules["psutil"] = None
sys.modules["tkinter"] = None
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/exact_match/README.md | # Metric Card for Exact Match
## Metric Description
A given predicted string's exact match score is 1 if it is the exact same as its reference string, and is 0 otherwise.
- **Example 1**: The exact match score of prediction "Happy Birthday!" is 0, given its reference is "Happy New Year!".
- **Example 2**: The exact match score of prediction "The Colour of Magic (1983)" is 1, given its reference is also "The Colour of Magic (1983)".
The exact match score of a set of predictions is the sum of all of the individual exact match scores in the set, divided by the total number of predictions in the set.
- **Example**: The exact match score of the set {Example 1, Example 2} (above) is 0.5.
## How to Use
At minimum, this metric takes as input predictions and references:
```python
>>> from datasets import load_metric
>>> exact_match_metric = load_metric("exact_match")
>>> results = exact_match_metric.compute(predictions=predictions, references=references)
```
### Inputs
- **`predictions`** (`list` of `str`): List of predicted texts.
- **`references`** (`list` of `str`): List of reference texts.
- **`regexes_to_ignore`** (`list` of `str`): Regex expressions of characters to ignore when calculating the exact matches. Defaults to `None`. Note: the regex changes are applied before capitalization is normalized.
- **`ignore_case`** (`bool`): If `True`, turns everything to lowercase so that capitalization differences are ignored. Defaults to `False`.
- **`ignore_punctuation`** (`bool`): If `True`, removes punctuation before comparing strings. Defaults to `False`.
- **`ignore_numbers`** (`bool`): If `True`, removes all digits before comparing strings. Defaults to `False`.
### Output Values
This metric outputs a dictionary with one value: the average exact match score.
```python
{'exact_match': 100.0}
```
This metric's range is 0-100, inclusive. Here, 0.0 means no prediction/reference pairs were matches, while 100.0 means they all were.
#### Values from Popular Papers
The exact match metric is often included in other metrics, such as SQuAD. For example, the [original SQuAD paper](https://nlp.stanford.edu/pubs/rajpurkar2016squad.pdf) reported an Exact Match score of 40.0%. They also report that the human performance Exact Match score on the dataset was 80.3%.
### Examples
Without including any regexes to ignore:
```python
>>> exact_match = datasets.load_metric("exact_match")
>>> refs = ["the cat", "theater", "YELLING", "agent007"]
>>> preds = ["cat?", "theater", "yelling", "agent"]
>>> results = exact_match.compute(references=refs, predictions=preds)
>>> print(round(results["exact_match"], 1))
25.0
```
Ignoring regexes "the" and "yell", as well as ignoring case and punctuation:
```python
>>> exact_match = datasets.load_metric("exact_match")
>>> refs = ["the cat", "theater", "YELLING", "agent007"]
>>> preds = ["cat?", "theater", "yelling", "agent"]
>>> results = exact_match.compute(references=refs, predictions=preds, regexes_to_ignore=["the ", "yell"], ignore_case=True, ignore_punctuation=True)
>>> print(round(results["exact_match"], 1))
50.0
```
Note that in the example above, because the regexes are ignored before the case is normalized, "yell" from "YELLING" is not deleted.
Ignoring "the", "yell", and "YELL", as well as ignoring case and punctuation:
```python
>>> exact_match = datasets.load_metric("exact_match")
>>> refs = ["the cat", "theater", "YELLING", "agent007"]
>>> preds = ["cat?", "theater", "yelling", "agent"]
>>> results = exact_match.compute(references=refs, predictions=preds, regexes_to_ignore=["the ", "yell", "YELL"], ignore_case=True, ignore_punctuation=True)
>>> print(round(results["exact_match"], 1))
75.0
```
Ignoring "the", "yell", and "YELL", as well as ignoring case, punctuation, and numbers:
```python
>>> exact_match = datasets.load_metric("exact_match")
>>> refs = ["the cat", "theater", "YELLING", "agent007"]
>>> preds = ["cat?", "theater", "yelling", "agent"]
>>> results = exact_match.compute(references=refs, predictions=preds, regexes_to_ignore=["the ", "yell", "YELL"], ignore_case=True, ignore_punctuation=True, ignore_numbers=True)
>>> print(round(results["exact_match"], 1))
100.0
```
An example that includes sentences:
```python
>>> exact_match = datasets.load_metric("exact_match")
>>> refs = ["The cat sat on the mat.", "Theaters are great.", "It's like comparing oranges and apples."]
>>> preds = ["The cat sat on the mat?", "Theaters are great.", "It's like comparing apples and oranges."]
>>> results = exact_match.compute(references=refs, predictions=preds)
>>> print(round(results["exact_match"], 1))
33.3
```
## Limitations and Bias
This metric is limited in that it outputs the same score for something that is completely wrong as for something that is correct except for a single character. In other words, there is no award for being *almost* right.
## Citation
## Further References
- Also used in the [SQuAD metric](https://github.com/huggingface/datasets/tree/main/metrics/squad)
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/exact_match/exact_match.py | # Copyright 2020 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Exact Match metric."""
import re
import string
import numpy as np
import datasets
_DESCRIPTION = """
Returns the rate at which the input predicted strings exactly match their references, ignoring any strings input as part of the regexes_to_ignore list.
"""
_KWARGS_DESCRIPTION = """
Args:
predictions: List of predicted texts.
references: List of reference texts.
regexes_to_ignore: List, defaults to None. Regex expressions of characters to
ignore when calculating the exact matches. Note: these regexes are removed
from the input data before the changes based on the options below (e.g. ignore_case,
ignore_punctuation, ignore_numbers) are applied.
ignore_case: Boolean, defaults to False. If true, turns everything
to lowercase so that capitalization differences are ignored.
ignore_punctuation: Boolean, defaults to False. If true, removes all punctuation before
comparing predictions and references.
ignore_numbers: Boolean, defaults to False. If true, removes all punctuation before
comparing predictions and references.
Returns:
exact_match: Dictionary containing exact_match rate. Possible values are between 0.0 and 100.0, inclusive.
Examples:
>>> exact_match = datasets.load_metric("exact_match")
>>> refs = ["the cat", "theater", "YELLING", "agent007"]
>>> preds = ["cat?", "theater", "yelling", "agent"]
>>> results = exact_match.compute(references=refs, predictions=preds)
>>> print(round(results["exact_match"], 1))
25.0
>>> exact_match = datasets.load_metric("exact_match")
>>> refs = ["the cat", "theater", "YELLING", "agent007"]
>>> preds = ["cat?", "theater", "yelling", "agent"]
>>> results = exact_match.compute(references=refs, predictions=preds, regexes_to_ignore=["the ", "yell"], ignore_case=True, ignore_punctuation=True)
>>> print(round(results["exact_match"], 1))
50.0
>>> exact_match = datasets.load_metric("exact_match")
>>> refs = ["the cat", "theater", "YELLING", "agent007"]
>>> preds = ["cat?", "theater", "yelling", "agent"]
>>> results = exact_match.compute(references=refs, predictions=preds, regexes_to_ignore=["the ", "yell", "YELL"], ignore_case=True, ignore_punctuation=True)
>>> print(round(results["exact_match"], 1))
75.0
>>> exact_match = datasets.load_metric("exact_match")
>>> refs = ["the cat", "theater", "YELLING", "agent007"]
>>> preds = ["cat?", "theater", "yelling", "agent"]
>>> results = exact_match.compute(references=refs, predictions=preds, regexes_to_ignore=["the ", "yell", "YELL"], ignore_case=True, ignore_punctuation=True, ignore_numbers=True)
>>> print(round(results["exact_match"], 1))
100.0
>>> exact_match = datasets.load_metric("exact_match")
>>> refs = ["The cat sat on the mat.", "Theaters are great.", "It's like comparing oranges and apples."]
>>> preds = ["The cat sat on the mat?", "Theaters are great.", "It's like comparing apples and oranges."]
>>> results = exact_match.compute(references=refs, predictions=preds)
>>> print(round(results["exact_match"], 1))
33.3
"""
_CITATION = """
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class ExactMatch(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Value("string", id="sequence"),
"references": datasets.Value("string", id="sequence"),
}
),
reference_urls=[],
)
def _compute(
self,
predictions,
references,
regexes_to_ignore=None,
ignore_case=False,
ignore_punctuation=False,
ignore_numbers=False,
):
if regexes_to_ignore is not None:
for s in regexes_to_ignore:
predictions = np.array([re.sub(s, "", x) for x in predictions])
references = np.array([re.sub(s, "", x) for x in references])
else:
predictions = np.asarray(predictions)
references = np.asarray(references)
if ignore_case:
predictions = np.char.lower(predictions)
references = np.char.lower(references)
if ignore_punctuation:
repl_table = string.punctuation.maketrans("", "", string.punctuation)
predictions = np.char.translate(predictions, table=repl_table)
references = np.char.translate(references, table=repl_table)
if ignore_numbers:
repl_table = string.digits.maketrans("", "", string.digits)
predictions = np.char.translate(predictions, table=repl_table)
references = np.char.translate(references, table=repl_table)
score_list = predictions == references
return {"exact_match": np.mean(score_list) * 100}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/f1/README.md | # Metric Card for F1
## Metric Description
The F1 score is the harmonic mean of the precision and recall. It can be computed with the equation:
F1 = 2 * (precision * recall) / (precision + recall)
## How to Use
At minimum, this metric requires predictions and references as input
```python
>>> f1_metric = datasets.load_metric("f1")
>>> results = f1_metric.compute(predictions=[0, 1], references=[0, 1])
>>> print(results)
["{'f1': 1.0}"]
```
### Inputs
- **predictions** (`list` of `int`): Predicted labels.
- **references** (`list` of `int`): Ground truth labels.
- **labels** (`list` of `int`): The set of labels to include when `average` is not set to `'binary'`, and the order of the labels if `average` is `None`. Labels present in the data can be excluded, for example to calculate a multiclass average ignoring a majority negative class. Labels not present in the data will result in 0 components in a macro average. For multilabel targets, labels are column indices. By default, all labels in `predictions` and `references` are used in sorted order. Defaults to None.
- **pos_label** (`int`): The class to be considered the positive class, in the case where `average` is set to `binary`. Defaults to 1.
- **average** (`string`): This parameter is required for multiclass/multilabel targets. If set to `None`, the scores for each class are returned. Otherwise, this determines the type of averaging performed on the data. Defaults to `'binary'`.
- 'binary': Only report results for the class specified by `pos_label`. This is applicable only if the classes found in `predictions` and `references` are binary.
- 'micro': Calculate metrics globally by counting the total true positives, false negatives and false positives.
- 'macro': Calculate metrics for each label, and find their unweighted mean. This does not take label imbalance into account.
- 'weighted': Calculate metrics for each label, and find their average weighted by support (the number of true instances for each label). This alters `'macro'` to account for label imbalance. This option can result in an F-score that is not between precision and recall.
- 'samples': Calculate metrics for each instance, and find their average (only meaningful for multilabel classification).
- **sample_weight** (`list` of `float`): Sample weights Defaults to None.
### Output Values
- **f1**(`float` or `array` of `float`): F1 score or list of f1 scores, depending on the value passed to `average`. Minimum possible value is 0. Maximum possible value is 1. Higher f1 scores are better.
Output Example(s):
```python
{'f1': 0.26666666666666666}
```
```python
{'f1': array([0.8, 0.0, 0.0])}
```
This metric outputs a dictionary, with either a single f1 score, of type `float`, or an array of f1 scores, with entries of type `float`.
#### Values from Popular Papers
### Examples
Example 1-A simple binary example
```python
>>> f1_metric = datasets.load_metric("f1")
>>> results = f1_metric.compute(references=[0, 1, 0, 1, 0], predictions=[0, 0, 1, 1, 0])
>>> print(results)
{'f1': 0.5}
```
Example 2-The same simple binary example as in Example 1, but with `pos_label` set to `0`.
```python
>>> f1_metric = datasets.load_metric("f1")
>>> results = f1_metric.compute(references=[0, 1, 0, 1, 0], predictions=[0, 0, 1, 1, 0], pos_label=0)
>>> print(round(results['f1'], 2))
0.67
```
Example 3-The same simple binary example as in Example 1, but with `sample_weight` included.
```python
>>> f1_metric = datasets.load_metric("f1")
>>> results = f1_metric.compute(references=[0, 1, 0, 1, 0], predictions=[0, 0, 1, 1, 0], sample_weight=[0.9, 0.5, 3.9, 1.2, 0.3])
>>> print(round(results['f1'], 2))
0.35
```
Example 4-A multiclass example, with different values for the `average` input.
```python
>>> predictions = [0, 2, 1, 0, 0, 1]
>>> references = [0, 1, 2, 0, 1, 2]
>>> results = f1_metric.compute(predictions=predictions, references=references, average="macro")
>>> print(round(results['f1'], 2))
0.27
>>> results = f1_metric.compute(predictions=predictions, references=references, average="micro")
>>> print(round(results['f1'], 2))
0.33
>>> results = f1_metric.compute(predictions=predictions, references=references, average="weighted")
>>> print(round(results['f1'], 2))
0.27
>>> results = f1_metric.compute(predictions=predictions, references=references, average=None)
>>> print(results)
{'f1': array([0.8, 0. , 0. ])}
```
## Limitations and Bias
## Citation(s)
```bibtex
@article{scikit-learn,
title={Scikit-learn: Machine Learning in {P}ython},
author={Pedregosa, F. and Varoquaux, G. and Gramfort, A. and Michel, V.
and Thirion, B. and Grisel, O. and Blondel, M. and Prettenhofer, P.
and Weiss, R. and Dubourg, V. and Vanderplas, J. and Passos, A. and
Cournapeau, D. and Brucher, M. and Perrot, M. and Duchesnay, E.},
journal={Journal of Machine Learning Research},
volume={12},
pages={2825--2830},
year={2011}
}
```
## Further References | 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/f1/f1.py | # Copyright 2020 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""F1 metric."""
from sklearn.metrics import f1_score
import datasets
_DESCRIPTION = """
The F1 score is the harmonic mean of the precision and recall. It can be computed with the equation:
F1 = 2 * (precision * recall) / (precision + recall)
"""
_KWARGS_DESCRIPTION = """
Args:
predictions (`list` of `int`): Predicted labels.
references (`list` of `int`): Ground truth labels.
labels (`list` of `int`): The set of labels to include when `average` is not set to `'binary'`, and the order of the labels if `average` is `None`. Labels present in the data can be excluded, for example to calculate a multiclass average ignoring a majority negative class. Labels not present in the data will result in 0 components in a macro average. For multilabel targets, labels are column indices. By default, all labels in `predictions` and `references` are used in sorted order. Defaults to None.
pos_label (`int`): The class to be considered the positive class, in the case where `average` is set to `binary`. Defaults to 1.
average (`string`): This parameter is required for multiclass/multilabel targets. If set to `None`, the scores for each class are returned. Otherwise, this determines the type of averaging performed on the data. Defaults to `'binary'`.
- 'binary': Only report results for the class specified by `pos_label`. This is applicable only if the classes found in `predictions` and `references` are binary.
- 'micro': Calculate metrics globally by counting the total true positives, false negatives and false positives.
- 'macro': Calculate metrics for each label, and find their unweighted mean. This does not take label imbalance into account.
- 'weighted': Calculate metrics for each label, and find their average weighted by support (the number of true instances for each label). This alters `'macro'` to account for label imbalance. This option can result in an F-score that is not between precision and recall.
- 'samples': Calculate metrics for each instance, and find their average (only meaningful for multilabel classification).
sample_weight (`list` of `float`): Sample weights Defaults to None.
Returns:
f1 (`float` or `array` of `float`): F1 score or list of f1 scores, depending on the value passed to `average`. Minimum possible value is 0. Maximum possible value is 1. Higher f1 scores are better.
Examples:
Example 1-A simple binary example
>>> f1_metric = datasets.load_metric("f1")
>>> results = f1_metric.compute(references=[0, 1, 0, 1, 0], predictions=[0, 0, 1, 1, 0])
>>> print(results)
{'f1': 0.5}
Example 2-The same simple binary example as in Example 1, but with `pos_label` set to `0`.
>>> f1_metric = datasets.load_metric("f1")
>>> results = f1_metric.compute(references=[0, 1, 0, 1, 0], predictions=[0, 0, 1, 1, 0], pos_label=0)
>>> print(round(results['f1'], 2))
0.67
Example 3-The same simple binary example as in Example 1, but with `sample_weight` included.
>>> f1_metric = datasets.load_metric("f1")
>>> results = f1_metric.compute(references=[0, 1, 0, 1, 0], predictions=[0, 0, 1, 1, 0], sample_weight=[0.9, 0.5, 3.9, 1.2, 0.3])
>>> print(round(results['f1'], 2))
0.35
Example 4-A multiclass example, with different values for the `average` input.
>>> predictions = [0, 2, 1, 0, 0, 1]
>>> references = [0, 1, 2, 0, 1, 2]
>>> results = f1_metric.compute(predictions=predictions, references=references, average="macro")
>>> print(round(results['f1'], 2))
0.27
>>> results = f1_metric.compute(predictions=predictions, references=references, average="micro")
>>> print(round(results['f1'], 2))
0.33
>>> results = f1_metric.compute(predictions=predictions, references=references, average="weighted")
>>> print(round(results['f1'], 2))
0.27
>>> results = f1_metric.compute(predictions=predictions, references=references, average=None)
>>> print(results)
{'f1': array([0.8, 0. , 0. ])}
"""
_CITATION = """
@article{scikit-learn,
title={Scikit-learn: Machine Learning in {P}ython},
author={Pedregosa, F. and Varoquaux, G. and Gramfort, A. and Michel, V.
and Thirion, B. and Grisel, O. and Blondel, M. and Prettenhofer, P.
and Weiss, R. and Dubourg, V. and Vanderplas, J. and Passos, A. and
Cournapeau, D. and Brucher, M. and Perrot, M. and Duchesnay, E.},
journal={Journal of Machine Learning Research},
volume={12},
pages={2825--2830},
year={2011}
}
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class F1(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Sequence(datasets.Value("int32")),
"references": datasets.Sequence(datasets.Value("int32")),
}
if self.config_name == "multilabel"
else {
"predictions": datasets.Value("int32"),
"references": datasets.Value("int32"),
}
),
reference_urls=["https://scikit-learn.org/stable/modules/generated/sklearn.metrics.f1_score.html"],
)
def _compute(self, predictions, references, labels=None, pos_label=1, average="binary", sample_weight=None):
score = f1_score(
references, predictions, labels=labels, pos_label=pos_label, average=average, sample_weight=sample_weight
)
return {"f1": float(score) if score.size == 1 else score}
| 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/matthews_correlation/README.md | # Metric Card for Matthews Correlation Coefficient
## Metric Description
The Matthews correlation coefficient is used in machine learning as a
measure of the quality of binary and multiclass classifications. It takes
into account true and false positives and negatives and is generally
regarded as a balanced measure which can be used even if the classes are of
very different sizes. The MCC is in essence a correlation coefficient value
between -1 and +1. A coefficient of +1 represents a perfect prediction, 0
an average random prediction and -1 an inverse prediction. The statistic
is also known as the phi coefficient. [source: Wikipedia]
## How to Use
At minimum, this metric requires a list of predictions and a list of references:
```python
>>> matthews_metric = datasets.load_metric("matthews_correlation")
>>> results = matthews_metric.compute(references=[0, 1], predictions=[0, 1])
>>> print(results)
{'matthews_correlation': 1.0}
```
### Inputs
- **`predictions`** (`list` of `int`s): Predicted class labels.
- **`references`** (`list` of `int`s): Ground truth labels.
- **`sample_weight`** (`list` of `int`s, `float`s, or `bool`s): Sample weights. Defaults to `None`.
### Output Values
- **`matthews_correlation`** (`float`): Matthews correlation coefficient.
The metric output takes the following form:
```python
{'matthews_correlation': 0.54}
```
This metric can be any value from -1 to +1, inclusive.
#### Values from Popular Papers
### Examples
A basic example with only predictions and references as inputs:
```python
>>> matthews_metric = datasets.load_metric("matthews_correlation")
>>> results = matthews_metric.compute(references=[1, 3, 2, 0, 3, 2],
... predictions=[1, 2, 2, 0, 3, 3])
>>> print(results)
{'matthews_correlation': 0.5384615384615384}
```
The same example as above, but also including sample weights:
```python
>>> matthews_metric = datasets.load_metric("matthews_correlation")
>>> results = matthews_metric.compute(references=[1, 3, 2, 0, 3, 2],
... predictions=[1, 2, 2, 0, 3, 3],
... sample_weight=[0.5, 3, 1, 1, 1, 2])
>>> print(results)
{'matthews_correlation': 0.09782608695652174}
```
The same example as above, with sample weights that cause a negative correlation:
```python
>>> matthews_metric = datasets.load_metric("matthews_correlation")
>>> results = matthews_metric.compute(references=[1, 3, 2, 0, 3, 2],
... predictions=[1, 2, 2, 0, 3, 3],
... sample_weight=[0.5, 1, 0, 0, 0, 1])
>>> print(results)
{'matthews_correlation': -0.25}
```
## Limitations and Bias
*Note any limitations or biases that the metric has.*
## Citation
```bibtex
@article{scikit-learn,
title={Scikit-learn: Machine Learning in {P}ython},
author={Pedregosa, F. and Varoquaux, G. and Gramfort, A. and Michel, V.
and Thirion, B. and Grisel, O. and Blondel, M. and Prettenhofer, P.
and Weiss, R. and Dubourg, V. and Vanderplas, J. and Passos, A. and
Cournapeau, D. and Brucher, M. and Perrot, M. and Duchesnay, E.},
journal={Journal of Machine Learning Research},
volume={12},
pages={2825--2830},
year={2011}
}
```
## Further References
- This Hugging Face implementation uses [this scikit-learn implementation](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.matthews_corrcoef.html) | 0 |
hf_public_repos/datasets/metrics | hf_public_repos/datasets/metrics/matthews_correlation/matthews_correlation.py | # Copyright 2021 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Matthews Correlation metric."""
from sklearn.metrics import matthews_corrcoef
import datasets
_DESCRIPTION = """
Compute the Matthews correlation coefficient (MCC)
The Matthews correlation coefficient is used in machine learning as a
measure of the quality of binary and multiclass classifications. It takes
into account true and false positives and negatives and is generally
regarded as a balanced measure which can be used even if the classes are of
very different sizes. The MCC is in essence a correlation coefficient value
between -1 and +1. A coefficient of +1 represents a perfect prediction, 0
an average random prediction and -1 an inverse prediction. The statistic
is also known as the phi coefficient. [source: Wikipedia]
"""
_KWARGS_DESCRIPTION = """
Args:
predictions (list of int): Predicted labels, as returned by a model.
references (list of int): Ground truth labels.
sample_weight (list of int, float, or bool): Sample weights. Defaults to `None`.
Returns:
matthews_correlation (dict containing float): Matthews correlation.
Examples:
Example 1, a basic example with only predictions and references as inputs:
>>> matthews_metric = datasets.load_metric("matthews_correlation")
>>> results = matthews_metric.compute(references=[1, 3, 2, 0, 3, 2],
... predictions=[1, 2, 2, 0, 3, 3])
>>> print(round(results['matthews_correlation'], 2))
0.54
Example 2, the same example as above, but also including sample weights:
>>> matthews_metric = datasets.load_metric("matthews_correlation")
>>> results = matthews_metric.compute(references=[1, 3, 2, 0, 3, 2],
... predictions=[1, 2, 2, 0, 3, 3],
... sample_weight=[0.5, 3, 1, 1, 1, 2])
>>> print(round(results['matthews_correlation'], 2))
0.1
Example 3, the same example as above, but with sample weights that cause a negative correlation:
>>> matthews_metric = datasets.load_metric("matthews_correlation")
>>> results = matthews_metric.compute(references=[1, 3, 2, 0, 3, 2],
... predictions=[1, 2, 2, 0, 3, 3],
... sample_weight=[0.5, 1, 0, 0, 0, 1])
>>> print(round(results['matthews_correlation'], 2))
-0.25
"""
_CITATION = """\
@article{scikit-learn,
title={Scikit-learn: Machine Learning in {P}ython},
author={Pedregosa, F. and Varoquaux, G. and Gramfort, A. and Michel, V.
and Thirion, B. and Grisel, O. and Blondel, M. and Prettenhofer, P.
and Weiss, R. and Dubourg, V. and Vanderplas, J. and Passos, A. and
Cournapeau, D. and Brucher, M. and Perrot, M. and Duchesnay, E.},
journal={Journal of Machine Learning Research},
volume={12},
pages={2825--2830},
year={2011}
}
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class MatthewsCorrelation(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Value("int32"),
"references": datasets.Value("int32"),
}
),
reference_urls=[
"https://scikit-learn.org/stable/modules/generated/sklearn.metrics.matthews_corrcoef.html"
],
)
def _compute(self, predictions, references, sample_weight=None):
return {
"matthews_correlation": float(matthews_corrcoef(references, predictions, sample_weight=sample_weight)),
}
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_metric_common.py | # Copyright 2020 HuggingFace Inc.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import doctest
import glob
import importlib
import inspect
import os
import re
from contextlib import contextmanager
from functools import wraps
from unittest.mock import patch
import numpy as np
import pytest
from absl.testing import parameterized
import datasets
from datasets import load_metric
from .utils import for_all_test_methods, local, slow
# mark all tests as integration
pytestmark = pytest.mark.integration
REQUIRE_FAIRSEQ = {"comet"}
_has_fairseq = importlib.util.find_spec("fairseq") is not None
UNSUPPORTED_ON_WINDOWS = {"code_eval"}
_on_windows = os.name == "nt"
REQUIRE_TRANSFORMERS = {"bertscore", "frugalscore", "perplexity"}
_has_transformers = importlib.util.find_spec("transformers") is not None
def skip_if_metric_requires_fairseq(test_case):
@wraps(test_case)
def wrapper(self, metric_name):
if not _has_fairseq and metric_name in REQUIRE_FAIRSEQ:
self.skipTest('"test requires Fairseq"')
else:
test_case(self, metric_name)
return wrapper
def skip_if_metric_requires_transformers(test_case):
@wraps(test_case)
def wrapper(self, metric_name):
if not _has_transformers and metric_name in REQUIRE_TRANSFORMERS:
self.skipTest('"test requires transformers"')
else:
test_case(self, metric_name)
return wrapper
def skip_on_windows_if_not_windows_compatible(test_case):
@wraps(test_case)
def wrapper(self, metric_name):
if _on_windows and metric_name in UNSUPPORTED_ON_WINDOWS:
self.skipTest('"test not supported on Windows"')
else:
test_case(self, metric_name)
return wrapper
def get_local_metric_names():
metrics = [metric_dir.split(os.sep)[-2] for metric_dir in glob.glob("./metrics/*/")]
return [{"testcase_name": x, "metric_name": x} for x in metrics if x != "gleu"] # gleu is unfinished
@parameterized.named_parameters(get_local_metric_names())
@for_all_test_methods(
skip_if_metric_requires_fairseq, skip_if_metric_requires_transformers, skip_on_windows_if_not_windows_compatible
)
@local
class LocalMetricTest(parameterized.TestCase):
INTENSIVE_CALLS_PATCHER = {}
metric_name = None
@pytest.mark.filterwarnings("ignore:metric_module_factory is deprecated:FutureWarning")
@pytest.mark.filterwarnings("ignore:load_metric is deprecated:FutureWarning")
def test_load_metric(self, metric_name):
doctest.ELLIPSIS_MARKER = "[...]"
metric_module = importlib.import_module(
datasets.load.metric_module_factory(os.path.join("metrics", metric_name)).module_path
)
metric = datasets.load.import_main_class(metric_module.__name__, dataset=False)
# check parameters
parameters = inspect.signature(metric._compute).parameters
self.assertTrue(all(p.kind != p.VAR_KEYWORD for p in parameters.values())) # no **kwargs
# run doctest
with self.patch_intensive_calls(metric_name, metric_module.__name__):
with self.use_local_metrics():
try:
results = doctest.testmod(metric_module, verbose=True, raise_on_error=True)
except doctest.UnexpectedException as e:
raise e.exc_info[1] # raise the exception that doctest caught
self.assertEqual(results.failed, 0)
self.assertGreater(results.attempted, 1)
@slow
def test_load_real_metric(self, metric_name):
doctest.ELLIPSIS_MARKER = "[...]"
metric_module = importlib.import_module(
datasets.load.metric_module_factory(os.path.join("metrics", metric_name)).module_path
)
# run doctest
with self.use_local_metrics():
results = doctest.testmod(metric_module, verbose=True, raise_on_error=True)
self.assertEqual(results.failed, 0)
self.assertGreater(results.attempted, 1)
@contextmanager
def patch_intensive_calls(self, metric_name, module_name):
if metric_name in self.INTENSIVE_CALLS_PATCHER:
with self.INTENSIVE_CALLS_PATCHER[metric_name](module_name):
yield
else:
yield
@contextmanager
def use_local_metrics(self):
def load_local_metric(metric_name, *args, **kwargs):
return load_metric(os.path.join("metrics", metric_name), *args, **kwargs)
with patch("datasets.load_metric") as mock_load_metric:
mock_load_metric.side_effect = load_local_metric
yield
@classmethod
def register_intensive_calls_patcher(cls, metric_name):
def wrapper(patcher):
patcher = contextmanager(patcher)
cls.INTENSIVE_CALLS_PATCHER[metric_name] = patcher
return patcher
return wrapper
# Metrics intensive calls patchers
# --------------------------------
@LocalMetricTest.register_intensive_calls_patcher("bleurt")
def patch_bleurt(module_name):
import tensorflow.compat.v1 as tf
from bleurt.score import Predictor
tf.flags.DEFINE_string("sv", "", "") # handle pytest cli flags
class MockedPredictor(Predictor):
def predict(self, input_dict):
assert len(input_dict["input_ids"]) == 2
return np.array([1.03, 1.04])
# mock predict_fn which is supposed to do a forward pass with a bleurt model
with patch("bleurt.score._create_predictor") as mock_create_predictor:
mock_create_predictor.return_value = MockedPredictor()
yield
@LocalMetricTest.register_intensive_calls_patcher("bertscore")
def patch_bertscore(module_name):
import torch
def bert_cos_score_idf(model, refs, *args, **kwargs):
return torch.tensor([[1.0, 1.0, 1.0]] * len(refs))
# mock get_model which is supposed to do download a bert model
# mock bert_cos_score_idf which is supposed to do a forward pass with a bert model
with patch("bert_score.scorer.get_model"), patch(
"bert_score.scorer.bert_cos_score_idf"
) as mock_bert_cos_score_idf:
mock_bert_cos_score_idf.side_effect = bert_cos_score_idf
yield
@LocalMetricTest.register_intensive_calls_patcher("comet")
def patch_comet(module_name):
def load_from_checkpoint(model_path):
class Model:
def predict(self, data, *args, **kwargs):
assert len(data) == 2
scores = [0.19, 0.92]
return scores, sum(scores) / len(scores)
return Model()
# mock load_from_checkpoint which is supposed to do download a bert model
# mock load_from_checkpoint which is supposed to do download a bert model
with patch("comet.download_model") as mock_download_model:
mock_download_model.return_value = None
with patch("comet.load_from_checkpoint") as mock_load_from_checkpoint:
mock_load_from_checkpoint.side_effect = load_from_checkpoint
yield
def test_seqeval_raises_when_incorrect_scheme():
metric = load_metric(os.path.join("metrics", "seqeval"))
wrong_scheme = "ERROR"
error_message = f"Scheme should be one of [IOB1, IOB2, IOE1, IOE2, IOBES, BILOU], got {wrong_scheme}"
with pytest.raises(ValueError, match=re.escape(error_message)):
metric.compute(predictions=[], references=[], scheme=wrong_scheme)
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_metadata_util.py | import re
import sys
import tempfile
import unittest
from pathlib import Path
import pytest
import yaml
from huggingface_hub import DatasetCard, DatasetCardData
from datasets.config import METADATA_CONFIGS_FIELD
from datasets.utils.metadata import MetadataConfigs
def _dedent(string: str) -> str:
indent_level = min(re.search("^ +", t).end() if t.startswith(" ") else 0 for t in string.splitlines())
return "\n".join([line[indent_level:] for line in string.splitlines() if indent_level < len(line)])
README_YAML = """\
---
language:
- zh
- en
task_ids:
- sentiment-classification
---
# Begin of markdown
Some cool dataset card
"""
README_EMPTY_YAML = """\
---
---
# Begin of markdown
Some cool dataset card
"""
README_NO_YAML = """\
# Begin of markdown
Some cool dataset card
"""
README_METADATA_CONFIG_INCORRECT_FORMAT = f"""\
---
{METADATA_CONFIGS_FIELD}:
data_dir: v1
drop_labels: true
---
"""
README_METADATA_SINGLE_CONFIG = f"""\
---
{METADATA_CONFIGS_FIELD}:
- config_name: custom
data_dir: v1
drop_labels: true
---
"""
README_METADATA_TWO_CONFIGS_WITH_DEFAULT_FLAG = f"""\
---
{METADATA_CONFIGS_FIELD}:
- config_name: v1
data_dir: v1
drop_labels: true
- config_name: v2
data_dir: v2
drop_labels: false
default: true
---
"""
README_METADATA_TWO_CONFIGS_WITH_DEFAULT_NAME = f"""\
---
{METADATA_CONFIGS_FIELD}:
- config_name: custom
data_dir: custom
drop_labels: true
- config_name: default
data_dir: data
drop_labels: false
---
"""
EXPECTED_METADATA_SINGLE_CONFIG = {"custom": {"data_dir": "v1", "drop_labels": True}}
EXPECTED_METADATA_TWO_CONFIGS_DEFAULT_FLAG = {
"v1": {"data_dir": "v1", "drop_labels": True},
"v2": {"data_dir": "v2", "drop_labels": False, "default": True},
}
EXPECTED_METADATA_TWO_CONFIGS_DEFAULT_NAME = {
"custom": {"data_dir": "custom", "drop_labels": True},
"default": {"data_dir": "data", "drop_labels": False},
}
@pytest.fixture
def data_dir_with_two_subdirs(tmp_path):
data_dir = tmp_path / "data_dir_with_two_configs_in_metadata"
cats_data_dir = data_dir / "cats"
cats_data_dir.mkdir(parents=True)
dogs_data_dir = data_dir / "dogs"
dogs_data_dir.mkdir(parents=True)
with open(cats_data_dir / "cat.jpg", "wb") as f:
f.write(b"this_is_a_cat_image_bytes")
with open(dogs_data_dir / "dog.jpg", "wb") as f:
f.write(b"this_is_a_dog_image_bytes")
return str(data_dir)
class TestMetadataUtils(unittest.TestCase):
def test_metadata_dict_from_readme(self):
with tempfile.TemporaryDirectory() as tmp_dir:
path = Path(tmp_dir) / "README.md"
with open(path, "w+") as readme_file:
readme_file.write(README_YAML)
dataset_card_data = DatasetCard.load(path).data
self.assertDictEqual(
dataset_card_data.to_dict(), {"language": ["zh", "en"], "task_ids": ["sentiment-classification"]}
)
with open(path, "w+") as readme_file:
readme_file.write(README_EMPTY_YAML)
if (
sys.platform != "win32"
): # there is a bug on windows, see https://github.com/huggingface/huggingface_hub/issues/1546
dataset_card_data = DatasetCard.load(path).data
self.assertDictEqual(dataset_card_data.to_dict(), {})
with open(path, "w+") as readme_file:
readme_file.write(README_NO_YAML)
dataset_card_data = DatasetCard.load(path).data
self.assertEqual(dataset_card_data.to_dict(), {})
def test_from_yaml_string(self):
valid_yaml_string = _dedent(
"""\
annotations_creators:
- found
language_creators:
- found
language:
- en
license:
- unknown
multilinguality:
- monolingual
pretty_name: Test Dataset
size_categories:
- 10K<n<100K
source_datasets:
- extended|other-yahoo-webscope-l6
task_categories:
- question-answering
task_ids:
- open-domain-qa
"""
)
assert DatasetCardData(**yaml.safe_load(valid_yaml_string)).to_dict()
valid_yaml_with_optional_keys = _dedent(
"""\
annotations_creators:
- found
language_creators:
- found
language:
- en
license:
- unknown
multilinguality:
- monolingual
pretty_name: Test Dataset
size_categories:
- 10K<n<100K
source_datasets:
- extended|other-yahoo-webscope-l6
task_categories:
- text-classification
task_ids:
- multi-class-classification
paperswithcode_id:
- squad
configs:
- en
train-eval-index:
- config: en
task: text-classification
task_id: multi_class_classification
splits:
train_split: train
eval_split: test
col_mapping:
text: text
label: target
metrics:
- type: accuracy
name: Accuracy
extra_gated_prompt: |
By clicking on “Access repository” below, you also agree to ImageNet Terms of Access:
[RESEARCHER_FULLNAME] (the "Researcher") has requested permission to use the ImageNet database (the "Database") at Princeton University and Stanford University. In exchange for such permission, Researcher hereby agrees to the following terms and conditions:
1. Researcher shall use the Database only for non-commercial research and educational purposes.
extra_gated_fields:
Company: text
Country: text
I agree to use this model for non-commerical use ONLY: checkbox
"""
)
assert DatasetCardData(**yaml.safe_load(valid_yaml_with_optional_keys)).to_dict()
@pytest.mark.parametrize(
"readme_content, expected_metadata_configs_dict, expected_default_config_name",
[
(README_METADATA_SINGLE_CONFIG, EXPECTED_METADATA_SINGLE_CONFIG, None),
(README_METADATA_TWO_CONFIGS_WITH_DEFAULT_FLAG, EXPECTED_METADATA_TWO_CONFIGS_DEFAULT_FLAG, "v2"),
(README_METADATA_TWO_CONFIGS_WITH_DEFAULT_NAME, EXPECTED_METADATA_TWO_CONFIGS_DEFAULT_NAME, "default"),
],
)
def test_metadata_configs_dataset_card_data(
readme_content, expected_metadata_configs_dict, expected_default_config_name
):
with tempfile.TemporaryDirectory() as tmp_dir:
path = Path(tmp_dir) / "README.md"
with open(path, "w+") as readme_file:
readme_file.write(readme_content)
dataset_card_data = DatasetCard.load(path).data
metadata_configs_dict = MetadataConfigs.from_dataset_card_data(dataset_card_data)
assert metadata_configs_dict == expected_metadata_configs_dict
assert metadata_configs_dict.get_default_config_name() == expected_default_config_name
def test_metadata_configs_incorrect_yaml():
with tempfile.TemporaryDirectory() as tmp_dir:
path = Path(tmp_dir) / "README.md"
with open(path, "w+") as readme_file:
readme_file.write(README_METADATA_CONFIG_INCORRECT_FORMAT)
dataset_card_data = DatasetCard.load(path).data
with pytest.raises(ValueError):
_ = MetadataConfigs.from_dataset_card_data(dataset_card_data)
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_warnings.py | import pytest
from datasets import inspect_metric, list_metrics, load_metric
@pytest.fixture
def mock_emitted_deprecation_warnings(monkeypatch):
monkeypatch.setattr("datasets.utils.deprecation_utils._emitted_deprecation_warnings", set())
# Used by list_metrics
@pytest.fixture
def mock_hfh(monkeypatch):
class MetricMock:
def __init__(self, metric_id):
self.id = metric_id
class HfhMock:
_metrics = [MetricMock(metric_id) for metric_id in ["accuracy", "mse", "precision", "codeparrot/apps_metric"]]
def list_metrics(self):
return self._metrics
monkeypatch.setattr("datasets.inspect.huggingface_hub", HfhMock())
@pytest.mark.parametrize(
"func, args", [(load_metric, ("metrics/mse",)), (list_metrics, ()), (inspect_metric, ("metrics/mse", "tmp_path"))]
)
def test_metric_deprecation_warning(func, args, mock_emitted_deprecation_warnings, mock_hfh, tmp_path):
if "tmp_path" in args:
args = tuple(arg if arg != "tmp_path" else tmp_path for arg in args)
with pytest.warns(FutureWarning, match="https://huggingface.co/docs/evaluate"):
func(*args)
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_file_utils.py | import os
from pathlib import Path
from unittest.mock import patch
import pytest
import zstandard as zstd
from datasets.download.download_config import DownloadConfig
from datasets.utils.file_utils import (
OfflineModeIsEnabled,
cached_path,
fsspec_get,
fsspec_head,
ftp_get,
ftp_head,
get_from_cache,
http_get,
http_head,
)
FILE_CONTENT = """\
Text data.
Second line of data."""
FILE_PATH = "file"
@pytest.fixture(scope="session")
def zstd_path(tmp_path_factory):
path = tmp_path_factory.mktemp("data") / (FILE_PATH + ".zstd")
data = bytes(FILE_CONTENT, "utf-8")
with zstd.open(path, "wb") as f:
f.write(data)
return path
@pytest.fixture
def tmpfs_file(tmpfs):
with open(os.path.join(tmpfs.local_root_dir, FILE_PATH), "w") as f:
f.write(FILE_CONTENT)
return FILE_PATH
@pytest.mark.parametrize("compression_format", ["gzip", "xz", "zstd"])
def test_cached_path_extract(compression_format, gz_file, xz_file, zstd_path, tmp_path, text_file):
input_paths = {"gzip": gz_file, "xz": xz_file, "zstd": zstd_path}
input_path = input_paths[compression_format]
cache_dir = tmp_path / "cache"
download_config = DownloadConfig(cache_dir=cache_dir, extract_compressed_file=True)
extracted_path = cached_path(input_path, download_config=download_config)
with open(extracted_path) as f:
extracted_file_content = f.read()
with open(text_file) as f:
expected_file_content = f.read()
assert extracted_file_content == expected_file_content
@pytest.mark.parametrize("default_extracted", [True, False])
@pytest.mark.parametrize("default_cache_dir", [True, False])
def test_extracted_datasets_path(default_extracted, default_cache_dir, xz_file, tmp_path, monkeypatch):
custom_cache_dir = "custom_cache"
custom_extracted_dir = "custom_extracted_dir"
custom_extracted_path = tmp_path / "custom_extracted_path"
if default_extracted:
expected = ("downloads" if default_cache_dir else custom_cache_dir, "extracted")
else:
monkeypatch.setattr("datasets.config.EXTRACTED_DATASETS_DIR", custom_extracted_dir)
monkeypatch.setattr("datasets.config.EXTRACTED_DATASETS_PATH", str(custom_extracted_path))
expected = custom_extracted_path.parts[-2:] if default_cache_dir else (custom_cache_dir, custom_extracted_dir)
filename = xz_file
download_config = (
DownloadConfig(extract_compressed_file=True)
if default_cache_dir
else DownloadConfig(cache_dir=tmp_path / custom_cache_dir, extract_compressed_file=True)
)
extracted_file_path = cached_path(filename, download_config=download_config)
assert Path(extracted_file_path).parent.parts[-2:] == expected
def test_cached_path_local(text_file):
# absolute path
text_file = str(Path(text_file).resolve())
assert cached_path(text_file) == text_file
# relative path
text_file = str(Path(__file__).resolve().relative_to(Path(os.getcwd())))
assert cached_path(text_file) == text_file
def test_cached_path_missing_local(tmp_path):
# absolute path
missing_file = str(tmp_path.resolve() / "__missing_file__.txt")
with pytest.raises(FileNotFoundError):
cached_path(missing_file)
# relative path
missing_file = "./__missing_file__.txt"
with pytest.raises(FileNotFoundError):
cached_path(missing_file)
def test_get_from_cache_fsspec(tmpfs_file):
output_path = get_from_cache(f"tmp://{tmpfs_file}")
with open(output_path) as f:
output_file_content = f.read()
assert output_file_content == FILE_CONTENT
@patch("datasets.config.HF_DATASETS_OFFLINE", True)
def test_cached_path_offline():
with pytest.raises(OfflineModeIsEnabled):
cached_path("https://huggingface.co")
@patch("datasets.config.HF_DATASETS_OFFLINE", True)
def test_http_offline(tmp_path_factory):
filename = tmp_path_factory.mktemp("data") / "file.html"
with pytest.raises(OfflineModeIsEnabled):
http_get("https://huggingface.co", temp_file=filename)
with pytest.raises(OfflineModeIsEnabled):
http_head("https://huggingface.co")
@patch("datasets.config.HF_DATASETS_OFFLINE", True)
def test_ftp_offline(tmp_path_factory):
filename = tmp_path_factory.mktemp("data") / "file.html"
with pytest.raises(OfflineModeIsEnabled):
ftp_get("ftp://huggingface.co", temp_file=filename)
with pytest.raises(OfflineModeIsEnabled):
ftp_head("ftp://huggingface.co")
@patch("datasets.config.HF_DATASETS_OFFLINE", True)
def test_fsspec_offline(tmp_path_factory):
filename = tmp_path_factory.mktemp("data") / "file.html"
with pytest.raises(OfflineModeIsEnabled):
fsspec_get("s3://huggingface.co", temp_file=filename)
with pytest.raises(OfflineModeIsEnabled):
fsspec_head("s3://huggingface.co")
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/README.md | ## Add Dummy data test
**Important** In order to pass the `load_dataset_<dataset_name>` test, dummy data is required for all possible config names.
First we distinguish between datasets scripts that
- A) have no config class and
- B) have a config class
For A) the dummy data folder structure, will always look as follows:
- ``dummy/<version>/dummy_data.zip``, *e.g.* ``cosmos_qa/dummy/0.1.0/dummy_data.zip``.
For B) the dummy data folder structure, will always look as follows:
- ``dummy/<config_name>/<version>/dummy_data.zip``, *e.g.* ``squad/dummy/plain-text/1.0.0/dummy_data.zip``.
Now the difficult part is to create the correct `dummy_data.zip` file.
**Important** When checking the dummy folder structure of already added datasets, always unzip ``dummy_data.zip``. If a folder ``dummy_data`` is found next to ``dummy_data.zip``, it is probably an old version and should be deleted. The tests only take the ``dummy_data.zip`` file into account.
Here we have to pay close attention to the ``_split_generators(self, dl_manager)`` function of the dataset script in question.
There are three general possibilties:
1) The ``dl_manager.download_and_extract()`` is given a **single path variable** of type `str` as its argument. In this case the file `dummy_data.zip` should unzip to the following structure:
``os.path.join("dummy_data", <additional-paths-as-defined-in-split-generations>)`` *e.g.* for ``sentiment140``, the unzipped ``dummy_data.zip`` has the following dir structure ``dummy_data/testdata.manual.2009.06.14.csv`` and ``dummy_data/training.1600000.processed.noemoticon.csv``.
**Note** if there are no ``<additional-paths-as-defined-in-split-generations>``, then ``dummy_data`` should be the name of the single file. An example for this is the ``crime-and-punishment`` dataset script.
2) The ``dl_manager.download_and_extract()`` is given a **dictionary of paths** of type `str` as its argument. In this case the file `dummy_data.zip` should unzip to the following structure:
``os.path.join("dummy_data", <value_of_dict>.split('/')[-1], <additional-paths-as-defined-in-split-generations>)`` *e.g.* for ``squad``, the unzipped ``dummy_data.zip`` has the following dir structure ``dummy_data/dev-v1.1.json``, etc...
**Note** if ``<value_of_dict>`` is a zipped file then the dummy data folder structure should contain the exact name of the zipped file and the following extracted folder structure. The file `dummy_data.zip` should **never** itself contain a zipped file since the dummy data is not unzipped by the ``MockDownloadManager`` during testing. *E.g.* check the dummy folder structure of ``hansards`` where the folders have to be named ``*.tar`` or the structure of ``wiki_split`` where the folders have to be named ``*.zip``.
3) The ``dl_manager.download_and_extract()`` is given a **dictionary of lists of paths** of type `str` as its argument. This is a very special case and has been seen only for the dataset ``ensli``. In this case the values are simply flattened and the dummy folder structure is the same as in 2).
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_info.py | import os
import pytest
import yaml
from datasets.features.features import Features, Value
from datasets.info import DatasetInfo, DatasetInfosDict
@pytest.mark.parametrize(
"files",
[
["full:README.md", "dataset_infos.json"],
["empty:README.md", "dataset_infos.json"],
["dataset_infos.json"],
["full:README.md"],
],
)
def test_from_dir(files, tmp_path_factory):
dataset_infos_dir = tmp_path_factory.mktemp("dset_infos_dir")
if "full:README.md" in files:
with open(dataset_infos_dir / "README.md", "w") as f:
f.write("---\ndataset_info:\n dataset_size: 42\n---")
if "empty:README.md" in files:
with open(dataset_infos_dir / "README.md", "w") as f:
f.write("")
# we want to support dataset_infos.json for backward compatibility
if "dataset_infos.json" in files:
with open(dataset_infos_dir / "dataset_infos.json", "w") as f:
f.write('{"default": {"dataset_size": 42}}')
dataset_infos = DatasetInfosDict.from_directory(dataset_infos_dir)
assert dataset_infos
assert dataset_infos["default"].dataset_size == 42
@pytest.mark.parametrize(
"dataset_info",
[
DatasetInfo(),
DatasetInfo(
description="foo",
features=Features({"a": Value("int32")}),
builder_name="builder",
config_name="config",
version="1.0.0",
splits=[{"name": "train"}],
download_size=42,
),
],
)
def test_dataset_info_dump_and_reload(tmp_path, dataset_info: DatasetInfo):
tmp_path = str(tmp_path)
dataset_info.write_to_directory(tmp_path)
reloaded = DatasetInfo.from_directory(tmp_path)
assert dataset_info == reloaded
assert os.path.exists(os.path.join(tmp_path, "dataset_info.json"))
def test_dataset_info_to_yaml_dict():
dataset_info = DatasetInfo(
description="foo",
citation="bar",
homepage="https://foo.bar",
license="CC0",
features=Features({"a": Value("int32")}),
post_processed={},
supervised_keys=(),
task_templates=[],
builder_name="builder",
config_name="config",
version="1.0.0",
splits=[{"name": "train", "num_examples": 42}],
download_checksums={},
download_size=1337,
post_processing_size=442,
dataset_size=1234,
size_in_bytes=1337 + 442 + 1234,
)
dataset_info_yaml_dict = dataset_info._to_yaml_dict()
assert sorted(dataset_info_yaml_dict) == sorted(DatasetInfo._INCLUDED_INFO_IN_YAML)
for key in DatasetInfo._INCLUDED_INFO_IN_YAML:
assert key in dataset_info_yaml_dict
assert isinstance(dataset_info_yaml_dict[key], (list, dict, int, str))
dataset_info_yaml = yaml.safe_dump(dataset_info_yaml_dict)
reloaded = yaml.safe_load(dataset_info_yaml)
assert dataset_info_yaml_dict == reloaded
def test_dataset_info_to_yaml_dict_empty():
dataset_info = DatasetInfo()
dataset_info_yaml_dict = dataset_info._to_yaml_dict()
assert dataset_info_yaml_dict == {}
@pytest.mark.parametrize(
"dataset_infos_dict",
[
DatasetInfosDict(),
DatasetInfosDict({"default": DatasetInfo()}),
DatasetInfosDict({"my_config_name": DatasetInfo()}),
DatasetInfosDict(
{
"default": DatasetInfo(
description="foo",
features=Features({"a": Value("int32")}),
builder_name="builder",
config_name="config",
version="1.0.0",
splits=[{"name": "train"}],
download_size=42,
)
}
),
DatasetInfosDict(
{
"v1": DatasetInfo(dataset_size=42),
"v2": DatasetInfo(dataset_size=1337),
}
),
],
)
def test_dataset_infos_dict_dump_and_reload(tmp_path, dataset_infos_dict: DatasetInfosDict):
tmp_path = str(tmp_path)
dataset_infos_dict.write_to_directory(tmp_path)
reloaded = DatasetInfosDict.from_directory(tmp_path)
# the config_name of the dataset_infos_dict take over the attribute
for config_name, dataset_info in dataset_infos_dict.items():
dataset_info.config_name = config_name
# the yaml representation doesn't include fields like description or citation
# so we just test that we can recover what we can from the yaml
dataset_infos_dict[config_name] = DatasetInfo._from_yaml_dict(dataset_info._to_yaml_dict())
assert dataset_infos_dict == reloaded
if dataset_infos_dict:
assert os.path.exists(os.path.join(tmp_path, "README.md"))
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_experimental.py | import unittest
import warnings
from datasets.utils import experimental
@experimental
def dummy_function():
return "success"
class TestExperimentalFlag(unittest.TestCase):
def test_experimental_warning(self):
with warnings.catch_warnings(record=True) as w:
warnings.simplefilter("always")
self.assertEqual(dummy_function(), "success")
self.assertEqual(len(w), 1)
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_filelock.py | import os
import time
import pytest
from datasets.utils.filelock import FileLock, Timeout
def test_filelock(tmpdir):
lock1 = FileLock(str(tmpdir / "foo.lock"))
lock2 = FileLock(str(tmpdir / "foo.lock"))
timeout = 0.01
with lock1.acquire():
with pytest.raises(Timeout):
_start = time.time()
lock2.acquire(timeout)
assert time.time() - _start > timeout
def test_long_filename(tmpdir):
filename = "a" * 1000 + ".lock"
lock1 = FileLock(str(tmpdir / filename))
assert lock1._lock_file.endswith(".lock")
assert not lock1._lock_file.endswith(filename)
assert len(os.path.basename(lock1._lock_file)) <= 255
lock2 = FileLock(tmpdir / filename)
with lock1.acquire():
with pytest.raises(Timeout):
lock2.acquire(0)
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_iterable_dataset.py | from copy import deepcopy
from itertools import chain, islice
import numpy as np
import pandas as pd
import pyarrow as pa
import pyarrow.compute as pc
import pytest
from datasets import Dataset, load_dataset
from datasets.combine import concatenate_datasets, interleave_datasets
from datasets.features import (
ClassLabel,
Features,
Image,
Value,
)
from datasets.formatting import get_format_type_from_alias
from datasets.info import DatasetInfo
from datasets.iterable_dataset import (
ArrowExamplesIterable,
BufferShuffledExamplesIterable,
CyclingMultiSourcesExamplesIterable,
ExamplesIterable,
FilteredExamplesIterable,
FormattingConfig,
HorizontallyConcatenatedMultiSourcesExamplesIterable,
IterableDataset,
MappedExamplesIterable,
RandomlyCyclingMultiSourcesExamplesIterable,
SelectColumnsIterable,
ShuffledDataSourcesArrowExamplesIterable,
ShuffledDataSourcesExamplesIterable,
ShufflingConfig,
SkipExamplesIterable,
StepExamplesIterable,
TakeExamplesIterable,
TypedExamplesIterable,
VerticallyConcatenatedMultiSourcesExamplesIterable,
_BaseExamplesIterable,
_batch_arrow_tables,
_batch_to_examples,
_convert_to_arrow,
_examples_to_batch,
)
from .utils import (
assert_arrow_memory_doesnt_increase,
is_rng_equal,
require_dill_gt_0_3_2,
require_not_windows,
require_pyspark,
require_tf,
require_torch,
)
DEFAULT_N_EXAMPLES = 20
DEFAULT_BATCH_SIZE = 4
DEFAULT_FILEPATH = "file.txt"
SAMPLE_DATASET_IDENTIFIER = "lhoestq/test" # has dataset script
def generate_examples_fn(**kwargs):
kwargs = kwargs.copy()
n = kwargs.pop("n", DEFAULT_N_EXAMPLES)
filepaths = kwargs.pop("filepaths", None)
for filepath in filepaths or [DEFAULT_FILEPATH]:
if filepaths is not None:
kwargs["filepath"] = filepath
for i in range(n):
yield f"{filepath}_{i}", {"id": i, **kwargs}
def generate_tables_fn(**kwargs):
kwargs = kwargs.copy()
n = kwargs.pop("n", DEFAULT_N_EXAMPLES)
batch_size = kwargs.pop("batch_size", DEFAULT_BATCH_SIZE)
filepaths = kwargs.pop("filepaths", None)
for filepath in filepaths or [DEFAULT_FILEPATH]:
buffer = []
batch_idx = 0
if filepaths is not None:
kwargs["filepath"] = filepath
for i in range(n):
buffer.append({"id": i, **kwargs})
if len(buffer) == batch_size:
yield f"{filepath}_{batch_idx}", pa.Table.from_pylist(buffer)
buffer = []
batch_idx += 1
yield batch_idx, pa.Table.from_pylist(buffer)
@pytest.fixture
def dataset():
ex_iterable = ExamplesIterable(generate_examples_fn, {})
return IterableDataset(ex_iterable, info=DatasetInfo(description="dummy"), split="train")
@pytest.fixture
def dataset_with_several_columns():
ex_iterable = ExamplesIterable(
generate_examples_fn,
{"filepath": ["data0.txt", "data1.txt", "data2.txt"], "metadata": {"sources": ["https://foo.bar"]}},
)
return IterableDataset(ex_iterable, info=DatasetInfo(description="dummy"), split="train")
@pytest.fixture
def arrow_file(tmp_path_factory, dataset: IterableDataset):
filename = str(tmp_path_factory.mktemp("data") / "file.arrow")
Dataset.from_generator(dataset.__iter__).map(cache_file_name=filename)
return filename
################################
#
# Utilities tests
#
################################
@pytest.mark.parametrize("batch_size", [1, 2, 3, 9, 10, 11, 20])
@pytest.mark.parametrize("drop_last_batch", [False, True])
def test_convert_to_arrow(batch_size, drop_last_batch):
examples = [{"foo": i} for i in range(10)]
full_table = pa.Table.from_pylist(examples)
num_rows = len(full_table) if not drop_last_batch else len(full_table) // batch_size * batch_size
num_batches = (num_rows // batch_size) + 1 if num_rows % batch_size else num_rows // batch_size
subtables = list(
_convert_to_arrow(
[(i, example) for i, example in enumerate(examples)],
batch_size=batch_size,
drop_last_batch=drop_last_batch,
)
)
assert len(subtables) == num_batches
if drop_last_batch:
assert all(len(subtable) == batch_size for _, subtable in subtables)
else:
assert all(len(subtable) == batch_size for _, subtable in subtables[:-1])
assert len(subtables[-1][1]) <= batch_size
if num_rows > 0:
reloaded = pa.concat_tables([subtable for _, subtable in subtables])
assert full_table.slice(0, num_rows).to_pydict() == reloaded.to_pydict()
@pytest.mark.parametrize(
"tables",
[
[pa.table({"foo": range(10)})],
[pa.table({"foo": range(0, 5)}), pa.table({"foo": range(5, 10)})],
[pa.table({"foo": [i]}) for i in range(10)],
],
)
@pytest.mark.parametrize("batch_size", [1, 2, 3, 9, 10, 11, 20])
@pytest.mark.parametrize("drop_last_batch", [False, True])
def test_batch_arrow_tables(tables, batch_size, drop_last_batch):
full_table = pa.concat_tables(tables)
num_rows = len(full_table) if not drop_last_batch else len(full_table) // batch_size * batch_size
num_batches = (num_rows // batch_size) + 1 if num_rows % batch_size else num_rows // batch_size
subtables = list(
_batch_arrow_tables(
[(i, table) for i, table in enumerate(tables)], batch_size=batch_size, drop_last_batch=drop_last_batch
)
)
assert len(subtables) == num_batches
if drop_last_batch:
assert all(len(subtable) == batch_size for _, subtable in subtables)
else:
assert all(len(subtable) == batch_size for _, subtable in subtables[:-1])
assert len(subtables[-1][1]) <= batch_size
if num_rows > 0:
reloaded = pa.concat_tables([subtable for _, subtable in subtables])
assert full_table.slice(0, num_rows).to_pydict() == reloaded.to_pydict()
################################
#
# _BaseExampleIterable tests
#
################################
def test_examples_iterable():
ex_iterable = ExamplesIterable(generate_examples_fn, {})
expected = list(generate_examples_fn())
assert next(iter(ex_iterable)) == expected[0]
assert list(ex_iterable) == expected
assert ex_iterable.iter_arrow is None
def test_examples_iterable_with_kwargs():
ex_iterable = ExamplesIterable(generate_examples_fn, {"filepaths": ["0.txt", "1.txt"], "split": "train"})
expected = list(generate_examples_fn(filepaths=["0.txt", "1.txt"], split="train"))
assert list(ex_iterable) == expected
assert all("split" in ex for _, ex in ex_iterable)
assert sorted({ex["filepath"] for _, ex in ex_iterable}) == ["0.txt", "1.txt"]
def test_examples_iterable_shuffle_data_sources():
ex_iterable = ExamplesIterable(generate_examples_fn, {"filepaths": ["0.txt", "1.txt"]})
ex_iterable = ex_iterable.shuffle_data_sources(np.random.default_rng(40))
expected = list(generate_examples_fn(filepaths=["1.txt", "0.txt"])) # shuffle the filepaths
assert list(ex_iterable) == expected
def test_examples_iterable_shuffle_shards_and_metadata():
def gen(filepaths, all_metadata):
for i, (filepath, metadata) in enumerate(zip(filepaths, all_metadata)):
yield i, {"filepath": filepath, "metadata": metadata}
ex_iterable = ExamplesIterable(
gen,
{
"filepaths": [f"{i}.txt" for i in range(100)],
"all_metadata": [{"id": str(i)} for i in range(100)],
},
)
ex_iterable = ex_iterable.shuffle_data_sources(np.random.default_rng(42))
out = list(ex_iterable)
filepaths_ids = [x["filepath"].split(".")[0] for _, x in out]
metadata_ids = [x["metadata"]["id"] for _, x in out]
assert filepaths_ids == metadata_ids, "entangled lists of shards/metadata should be shuffled the same way"
def test_arrow_examples_iterable():
ex_iterable = ArrowExamplesIterable(generate_tables_fn, {})
expected = sum([pa_table.to_pylist() for _, pa_table in generate_tables_fn()], [])
assert next(iter(ex_iterable))[1] == expected[0]
assert [example for _, example in ex_iterable] == expected
expected = list(generate_tables_fn())
assert list(ex_iterable.iter_arrow()) == expected
def test_arrow_examples_iterable_with_kwargs():
ex_iterable = ArrowExamplesIterable(generate_tables_fn, {"filepaths": ["0.txt", "1.txt"], "split": "train"})
expected = sum(
[pa_table.to_pylist() for _, pa_table in generate_tables_fn(filepaths=["0.txt", "1.txt"], split="train")], []
)
assert [example for _, example in ex_iterable] == expected
assert all("split" in ex for _, ex in ex_iterable)
assert sorted({ex["filepath"] for _, ex in ex_iterable}) == ["0.txt", "1.txt"]
expected = list(generate_tables_fn(filepaths=["0.txt", "1.txt"], split="train"))
assert list(ex_iterable.iter_arrow()) == expected
def test_arrow_examples_iterable_shuffle_data_sources():
ex_iterable = ArrowExamplesIterable(generate_tables_fn, {"filepaths": ["0.txt", "1.txt"]})
ex_iterable = ex_iterable.shuffle_data_sources(np.random.default_rng(40))
expected = sum(
[pa_table.to_pylist() for _, pa_table in generate_tables_fn(filepaths=["1.txt", "0.txt"])], []
) # shuffle the filepaths
assert [example for _, example in ex_iterable] == expected
expected = list(generate_tables_fn(filepaths=["1.txt", "0.txt"]))
assert list(ex_iterable.iter_arrow()) == expected
@pytest.mark.parametrize("seed", [42, 1337, 101010, 123456])
def test_buffer_shuffled_examples_iterable(seed):
n, buffer_size = 100, 30
generator = np.random.default_rng(seed)
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n})
ex_iterable = BufferShuffledExamplesIterable(base_ex_iterable, buffer_size=buffer_size, generator=generator)
rng = deepcopy(generator)
expected_indices_used_for_shuffling = list(
islice(BufferShuffledExamplesIterable._iter_random_indices(rng, buffer_size=buffer_size), n - buffer_size)
)
# indices to pick in the shuffle buffer should all be in the right range
assert all(0 <= index_to_pick < buffer_size for index_to_pick in expected_indices_used_for_shuffling)
# it should be random indices
assert expected_indices_used_for_shuffling != list(range(buffer_size))
# The final order of examples is the result of a shuffle buffer.
all_examples = list(generate_examples_fn(n=n))
# We create a buffer and we pick random examples from it.
buffer, rest = all_examples[:buffer_size], all_examples[buffer_size:]
expected = []
for i, index_to_pick in enumerate(expected_indices_used_for_shuffling):
expected.append(buffer[index_to_pick])
# The picked examples are directly replaced by the next examples from the iterable.
buffer[index_to_pick] = rest.pop(0)
# Once we have reached the end of the iterable, we shuffle the buffer and return the remaining examples.
rng.shuffle(buffer)
expected += buffer
assert next(iter(ex_iterable)) == expected[0]
assert list(ex_iterable) == expected
assert sorted(ex_iterable) == sorted(all_examples)
def test_cycling_multi_sources_examples_iterable():
ex_iterable1 = ExamplesIterable(generate_examples_fn, {"text": "foo"})
ex_iterable2 = ExamplesIterable(generate_examples_fn, {"text": "bar"})
ex_iterable = CyclingMultiSourcesExamplesIterable([ex_iterable1, ex_iterable2])
expected = list(chain(*zip(generate_examples_fn(text="foo"), generate_examples_fn(text="bar"))))
# The cycling stops as soon as one iterable is out of examples (here ex_iterable1), so the last sample from ex_iterable2 is unecessary
expected = expected[:-1]
assert next(iter(ex_iterable)) == expected[0]
assert list(ex_iterable) == expected
assert all((x["id"], x["text"]) == (i // 2, "bar" if i % 2 else "foo") for i, (_, x) in enumerate(ex_iterable))
@pytest.mark.parametrize("probabilities", [None, (0.5, 0.5), (0.9, 0.1)])
def test_randomly_cycling_multi_sources_examples_iterable(probabilities):
seed = 42
generator = np.random.default_rng(seed)
ex_iterable1 = ExamplesIterable(generate_examples_fn, {"text": "foo"})
ex_iterable2 = ExamplesIterable(generate_examples_fn, {"text": "bar"})
ex_iterable = RandomlyCyclingMultiSourcesExamplesIterable(
[ex_iterable1, ex_iterable2], generator=generator, probabilities=probabilities
)
# The source used randomly changes at each example. It stops when one of the iterators is empty.
rng = deepcopy(generator)
iterators = (generate_examples_fn(text="foo"), generate_examples_fn(text="bar"))
indices_iterator = RandomlyCyclingMultiSourcesExamplesIterable._iter_random_indices(
rng, len(iterators), p=probabilities
)
expected = []
lengths = [len(list(ex_iterable1)), len(list(ex_iterable2))]
for i in indices_iterator:
if lengths[0] == 0 or lengths[1] == 0:
break
for key, example in iterators[i]:
expected.append((key, example))
lengths[i] -= 1
break
else:
break
assert next(iter(ex_iterable)) == expected[0]
assert list(ex_iterable) == expected
@pytest.mark.parametrize(
"n, func, batched, batch_size",
[
(3, lambda x: {"id+1": x["id"] + 1}, False, None), # just add 1 to the id
(3, lambda x: {"id+1": [x["id"][0] + 1]}, True, 1), # same with bs=1
(5, lambda x: {"id+1": [i + 1 for i in x["id"]]}, True, 10), # same with bs=10
(25, lambda x: {"id+1": [i + 1 for i in x["id"]]}, True, 10), # same with bs=10
(5, lambda x: {"id+1": [i + 1 for i in x["id"]]}, True, None), # same with bs=None
(5, lambda x: {"id+1": [i + 1 for i in x["id"]]}, True, -1), # same with bs<=0
(3, lambda x: {k: v * 2 for k, v in x.items()}, True, 1), # make a duplicate of each example
],
)
def test_mapped_examples_iterable(n, func, batched, batch_size):
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n})
ex_iterable = MappedExamplesIterable(base_ex_iterable, func, batched=batched, batch_size=batch_size)
all_examples = [x for _, x in generate_examples_fn(n=n)]
if batched is False:
expected = [{**x, **func(x)} for x in all_examples]
else:
# For batched map we have to format the examples as a batch (i.e. in one single dictionary) to pass the batch to the function
all_transformed_examples = []
# If batch_size is None or <=0, we use the whole dataset as a single batch
if batch_size is None or batch_size <= 0:
batch_size = len(all_examples)
for batch_offset in range(0, len(all_examples), batch_size):
examples = all_examples[batch_offset : batch_offset + batch_size]
batch = _examples_to_batch(examples)
transformed_batch = func(batch)
all_transformed_examples.extend(_batch_to_examples(transformed_batch))
expected = _examples_to_batch(all_examples)
expected.update(_examples_to_batch(all_transformed_examples))
expected = list(_batch_to_examples(expected))
assert next(iter(ex_iterable))[1] == expected[0]
assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
"n, func, batched, batch_size",
[
(3, lambda x: {"id+1": x["id"] + 1}, False, None), # just add 1 to the id
(3, lambda x: {"id+1": [x["id"][0] + 1]}, True, 1), # same with bs=1
(5, lambda x: {"id+1": [i + 1 for i in x["id"]]}, True, 10), # same with bs=10
(25, lambda x: {"id+1": [i + 1 for i in x["id"]]}, True, 10), # same with bs=10
(5, lambda x: {"id+1": [i + 1 for i in x["id"]]}, True, None), # same with bs=None
(5, lambda x: {"id+1": [i + 1 for i in x["id"]]}, True, -1), # same with bs<=0
(3, lambda x: {k: v * 2 for k, v in x.items()}, True, 1), # make a duplicate of each example
],
)
def test_mapped_examples_iterable_drop_last_batch(n, func, batched, batch_size):
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n})
ex_iterable = MappedExamplesIterable(
base_ex_iterable, func, batched=batched, batch_size=batch_size, drop_last_batch=True
)
all_examples = [x for _, x in generate_examples_fn(n=n)]
is_empty = False
if batched is False:
# `drop_last_batch` has no effect here
expected = [{**x, **func(x)} for x in all_examples]
else:
# For batched map we have to format the examples as a batch (i.e. in one single dictionary) to pass the batch to the function
all_transformed_examples = []
# If batch_size is None or <=0, we use the whole dataset as a single batch
if batch_size is None or batch_size <= 0:
batch_size = len(all_examples)
for batch_offset in range(0, len(all_examples), batch_size):
examples = all_examples[batch_offset : batch_offset + batch_size]
if len(examples) < batch_size: # ignore last batch
break
batch = _examples_to_batch(examples)
transformed_batch = func(batch)
all_transformed_examples.extend(_batch_to_examples(transformed_batch))
all_examples = all_examples if n % batch_size == 0 else all_examples[: n // batch_size * batch_size]
if all_examples:
expected = _examples_to_batch(all_examples)
expected.update(_examples_to_batch(all_transformed_examples))
expected = list(_batch_to_examples(expected))
else:
is_empty = True
if not is_empty:
assert next(iter(ex_iterable))[1] == expected[0]
assert [x for _, x in ex_iterable] == expected
else:
with pytest.raises(StopIteration):
next(iter(ex_iterable))
@pytest.mark.parametrize(
"n, func, batched, batch_size",
[
(3, lambda x, index: {"id+idx": x["id"] + index}, False, None), # add the index to the id
(
25,
lambda x, indices: {"id+idx": [i + j for i, j in zip(x["id"], indices)]},
True,
10,
), # add the index to the id
(5, lambda x, indices: {"id+idx": [i + j for i, j in zip(x["id"], indices)]}, True, None), # same with bs=None
(5, lambda x, indices: {"id+idx": [i + j for i, j in zip(x["id"], indices)]}, True, -1), # same with bs<=0
],
)
def test_mapped_examples_iterable_with_indices(n, func, batched, batch_size):
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n})
ex_iterable = MappedExamplesIterable(
base_ex_iterable, func, batched=batched, batch_size=batch_size, with_indices=True
)
all_examples = [x for _, x in generate_examples_fn(n=n)]
if batched is False:
expected = [{**x, **func(x, idx)} for idx, x in enumerate(all_examples)]
else:
# For batched map we have to format the examples as a batch (i.e. in one single dictionary) to pass the batch to the function
all_transformed_examples = []
# If batch_size is None or <=0, we use the whole dataset as a single batch
if batch_size is None or batch_size <= 0:
batch_size = len(all_examples)
for batch_offset in range(0, len(all_examples), batch_size):
examples = all_examples[batch_offset : batch_offset + batch_size]
batch = _examples_to_batch(examples)
indices = list(range(batch_offset, batch_offset + len(examples)))
transformed_batch = func(batch, indices)
all_transformed_examples.extend(_batch_to_examples(transformed_batch))
expected = _examples_to_batch(all_examples)
expected.update(_examples_to_batch(all_transformed_examples))
expected = list(_batch_to_examples(expected))
assert next(iter(ex_iterable))[1] == expected[0]
assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
"n, func, batched, batch_size, remove_columns",
[
(3, lambda x: {"id+1": x["id"] + 1}, False, None, ["extra_column"]), # just add 1 to the id
(25, lambda x: {"id+1": [i + 1 for i in x["id"]]}, True, 10, ["extra_column"]), # same with bs=10
(
50,
lambda x: {"foo": ["bar"] * np.random.default_rng(x["id"][0]).integers(0, 10)},
True,
8,
["extra_column", "id"],
), # make a duplicate of each example
(5, lambda x: {"id+1": [i + 1 for i in x["id"]]}, True, None, ["extra_column"]), # same with bs=None
(5, lambda x: {"id+1": [i + 1 for i in x["id"]]}, True, -1, ["extra_column"]), # same with bs<=0
],
)
def test_mapped_examples_iterable_remove_columns(n, func, batched, batch_size, remove_columns):
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n, "extra_column": "foo"})
ex_iterable = MappedExamplesIterable(
base_ex_iterable, func, batched=batched, batch_size=batch_size, remove_columns=remove_columns
)
all_examples = [x for _, x in generate_examples_fn(n=n)]
columns_to_remove = remove_columns if isinstance(remove_columns, list) else [remove_columns]
if batched is False:
expected = [{**{k: v for k, v in x.items() if k not in columns_to_remove}, **func(x)} for x in all_examples]
else:
# For batched map we have to format the examples as a batch (i.e. in one single dictionary) to pass the batch to the function
all_transformed_examples = []
# If batch_size is None or <=0, we use the whole dataset as a single batch
if batch_size is None or batch_size <= 0:
batch_size = len(all_examples)
for batch_offset in range(0, len(all_examples), batch_size):
examples = all_examples[batch_offset : batch_offset + batch_size]
batch = _examples_to_batch(examples)
transformed_batch = func(batch)
all_transformed_examples.extend(_batch_to_examples(transformed_batch))
expected = {k: v for k, v in _examples_to_batch(all_examples).items() if k not in columns_to_remove}
expected.update(_examples_to_batch(all_transformed_examples))
expected = list(_batch_to_examples(expected))
assert next(iter(ex_iterable))[1] == expected[0]
assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
"n, func, batched, batch_size, fn_kwargs",
[
(3, lambda x, y=0: {"id+y": x["id"] + y}, False, None, None),
(3, lambda x, y=0: {"id+y": x["id"] + y}, False, None, {"y": 3}),
(25, lambda x, y=0: {"id+y": [i + y for i in x["id"]]}, True, 10, {"y": 3}),
(5, lambda x, y=0: {"id+y": [i + y for i in x["id"]]}, True, None, {"y": 3}), # same with bs=None
(5, lambda x, y=0: {"id+y": [i + y for i in x["id"]]}, True, -1, {"y": 3}), # same with bs<=0
],
)
def test_mapped_examples_iterable_fn_kwargs(n, func, batched, batch_size, fn_kwargs):
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n})
ex_iterable = MappedExamplesIterable(
base_ex_iterable, func, batched=batched, batch_size=batch_size, fn_kwargs=fn_kwargs
)
all_examples = [x for _, x in generate_examples_fn(n=n)]
if fn_kwargs is None:
fn_kwargs = {}
if batched is False:
expected = [{**x, **func(x, **fn_kwargs)} for x in all_examples]
else:
# For batched map we have to format the examples as a batch (i.e. in one single dictionary) to pass the batch to the function
all_transformed_examples = []
# If batch_size is None or <=0, we use the whole dataset as a single batch
if batch_size is None or batch_size <= 0:
batch_size = len(all_examples)
for batch_offset in range(0, len(all_examples), batch_size):
examples = all_examples[batch_offset : batch_offset + batch_size]
batch = _examples_to_batch(examples)
transformed_batch = func(batch, **fn_kwargs)
all_transformed_examples.extend(_batch_to_examples(transformed_batch))
expected = _examples_to_batch(all_examples)
expected.update(_examples_to_batch(all_transformed_examples))
expected = list(_batch_to_examples(expected))
assert next(iter(ex_iterable))[1] == expected[0]
assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
"n, func, batched, batch_size, input_columns",
[
(3, lambda id_: {"id+1": id_ + 1}, False, None, ["id"]), # just add 1 to the id
(25, lambda ids_: {"id+1": [i + 1 for i in ids_]}, True, 10, ["id"]), # same with bs=10
(5, lambda ids_: {"id+1": [i + 1 for i in ids_]}, True, None, ["id"]), # same with bs=None
(5, lambda ids_: {"id+1": [i + 1 for i in ids_]}, True, -1, ["id"]), # same with bs<=0
],
)
def test_mapped_examples_iterable_input_columns(n, func, batched, batch_size, input_columns):
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n})
ex_iterable = MappedExamplesIterable(
base_ex_iterable, func, batched=batched, batch_size=batch_size, input_columns=input_columns
)
all_examples = [x for _, x in generate_examples_fn(n=n)]
columns_to_input = input_columns if isinstance(input_columns, list) else [input_columns]
if batched is False:
expected = [{**x, **func(*[x[col] for col in columns_to_input])} for x in all_examples]
else:
# For batched map we have to format the examples as a batch (i.e. in one single dictionary) to pass the batch to the function
all_transformed_examples = []
# If batch_size is None or <=0, we use the whole dataset as a single batch
if batch_size is None or batch_size <= 0:
batch_size = len(all_examples)
for batch_offset in range(0, len(all_examples), batch_size):
examples = all_examples[batch_offset : batch_offset + batch_size]
batch = _examples_to_batch(examples)
transformed_batch = func(*[batch[col] for col in columns_to_input])
all_transformed_examples.extend(_batch_to_examples(transformed_batch))
expected = _examples_to_batch(all_examples)
expected.update(_examples_to_batch(all_transformed_examples))
expected = list(_batch_to_examples(expected))
assert next(iter(ex_iterable))[1] == expected[0]
assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
"n, func, batched, batch_size",
[
(3, lambda t: t.append_column("id+1", pc.add(t["id"], 1)), False, None), # just add 1 to the id
(3, lambda t: t.append_column("id+1", pc.add(t["id"], 1)), True, 1), # same with bs=1
(5, lambda t: t.append_column("id+1", pc.add(t["id"], 1)), True, 10), # same with bs=10
(25, lambda t: t.append_column("id+1", pc.add(t["id"], 1)), True, 10), # same with bs=10
(5, lambda t: t.append_column("id+1", pc.add(t["id"], 1)), True, None), # same with bs=None
(5, lambda t: t.append_column("id+1", pc.add(t["id"], 1)), True, -1), # same with bs<=0
(3, lambda t: pa.concat_tables([t] * 2), True, 1), # make a duplicate of each example
],
)
def test_mapped_examples_iterable_arrow_format(n, func, batched, batch_size):
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n})
ex_iterable = MappedExamplesIterable(
base_ex_iterable,
func,
batched=batched,
batch_size=batch_size,
formatting=FormattingConfig(format_type="arrow"),
)
all_examples = [x for _, x in generate_examples_fn(n=n)]
if batched is False:
expected = [func(pa.Table.from_pylist([x])).to_pylist()[0] for x in all_examples]
else:
expected = []
# If batch_size is None or <=0, we use the whole dataset as a single batch
if batch_size is None or batch_size <= 0:
batch_size = len(all_examples)
for batch_offset in range(0, len(all_examples), batch_size):
examples = all_examples[batch_offset : batch_offset + batch_size]
batch = pa.Table.from_pylist(examples)
expected.extend(func(batch).to_pylist())
assert next(iter(ex_iterable))[1] == expected[0]
assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
"n, func, batched, batch_size",
[
(3, lambda t: t.append_column("id+1", pc.add(t["id"], 1)), False, None), # just add 1 to the id
(3, lambda t: t.append_column("id+1", pc.add(t["id"], 1)), True, 1), # same with bs=1
(5, lambda t: t.append_column("id+1", pc.add(t["id"], 1)), True, 10), # same with bs=10
(25, lambda t: t.append_column("id+1", pc.add(t["id"], 1)), True, 10), # same with bs=10
(5, lambda t: t.append_column("id+1", pc.add(t["id"], 1)), True, None), # same with bs=None
(5, lambda t: t.append_column("id+1", pc.add(t["id"], 1)), True, -1), # same with bs<=0
(3, lambda t: pa.concat_tables([t] * 2), True, 1), # make a duplicate of each example
],
)
def test_mapped_examples_iterable_drop_last_batch_and_arrow_format(n, func, batched, batch_size):
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n})
ex_iterable = MappedExamplesIterable(
base_ex_iterable,
func,
batched=batched,
batch_size=batch_size,
drop_last_batch=True,
formatting=FormattingConfig(format_type="arrow"),
)
all_examples = [x for _, x in generate_examples_fn(n=n)]
is_empty = False
if batched is False:
# `drop_last_batch` has no effect here
expected = [func(pa.Table.from_pylist([x])).to_pylist()[0] for x in all_examples]
else:
all_transformed_examples = []
# If batch_size is None or <=0, we use the whole dataset as a single batch
if batch_size is None or batch_size <= 0:
batch_size = len(all_examples)
for batch_offset in range(0, len(all_examples), batch_size):
examples = all_examples[batch_offset : batch_offset + batch_size]
if len(examples) < batch_size: # ignore last batch
break
batch = pa.Table.from_pylist(examples)
out = func(batch)
all_transformed_examples.extend(
out.to_pylist()
) # we don't merge with input since they're arrow tables and not dictionaries
all_examples = all_examples if n % batch_size == 0 else all_examples[: n // batch_size * batch_size]
if all_examples:
expected = all_transformed_examples
else:
is_empty = True
if not is_empty:
assert next(iter(ex_iterable))[1] == expected[0]
assert [x for _, x in ex_iterable] == expected
else:
with pytest.raises(StopIteration):
next(iter(ex_iterable))
@pytest.mark.parametrize(
"n, func, batched, batch_size",
[
(
3,
lambda t, index: t.append_column("id+idx", pc.add(t["id"], index)),
False,
None,
), # add the index to the id
(
25,
lambda t, indices: t.append_column("id+idx", pc.add(t["id"], indices)),
True,
10,
), # add the index to the id
(5, lambda t, indices: t.append_column("id+idx", pc.add(t["id"], indices)), True, None), # same with bs=None
(5, lambda t, indices: t.append_column("id+idx", pc.add(t["id"], indices)), True, -1), # same with bs<=0
],
)
def test_mapped_examples_iterable_with_indices_and_arrow_format(n, func, batched, batch_size):
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n})
ex_iterable = MappedExamplesIterable(
base_ex_iterable,
func,
batched=batched,
batch_size=batch_size,
with_indices=True,
formatting=FormattingConfig(format_type="arrow"),
)
all_examples = [x for _, x in generate_examples_fn(n=n)]
if batched is False:
expected = [func(pa.Table.from_pylist([x]), i).to_pylist()[0] for i, x in enumerate(all_examples)]
else:
expected = []
# If batch_size is None or <=0, we use the whole dataset as a single batch
if batch_size is None or batch_size <= 0:
batch_size = len(all_examples)
for batch_offset in range(0, len(all_examples), batch_size):
examples = all_examples[batch_offset : batch_offset + batch_size]
batch = pa.Table.from_pylist(examples)
expected.extend(func(batch, list(range(batch_offset, batch_offset + len(batch)))).to_pylist())
assert next(iter(ex_iterable))[1] == expected[0]
assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
"n, func, batched, batch_size, remove_columns",
[
(
3,
lambda t: t.append_column("id+1", pc.add(t["id"], 1)),
False,
None,
["extra_column"],
), # just add 1 to the id
(25, lambda t: t.append_column("id+1", pc.add(t["id"], 1)), True, 10, ["extra_column"]), # same with bs=10
(
50,
lambda t: pa.table({"foo": ["bar"] * np.random.default_rng(t["id"][0].as_py()).integers(0, 10)}),
True,
8,
["extra_column", "id"],
), # make a duplicate of each example
(5, lambda t: t.append_column("id+1", pc.add(t["id"], 1)), True, None, ["extra_column"]), # same with bs=None
(5, lambda t: t.append_column("id+1", pc.add(t["id"], 1)), True, -1, ["extra_column"]), # same with bs<=0
],
)
def test_mapped_examples_iterable_remove_columns_arrow_format(n, func, batched, batch_size, remove_columns):
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n, "extra_column": "foo"})
ex_iterable = MappedExamplesIterable(
base_ex_iterable,
func,
batched=batched,
batch_size=batch_size,
remove_columns=remove_columns,
formatting=FormattingConfig(format_type="arrow"),
)
all_examples = [x for _, x in generate_examples_fn(n=n)]
columns_to_remove = remove_columns if isinstance(remove_columns, list) else [remove_columns]
if batched is False:
expected = [
{**{k: v for k, v in func(pa.Table.from_pylist([x])).to_pylist()[0].items() if k not in columns_to_remove}}
for x in all_examples
]
else:
expected = []
# If batch_size is None or <=0, we use the whole dataset as a single batch
if batch_size is None or batch_size <= 0:
batch_size = len(all_examples)
for batch_offset in range(0, len(all_examples), batch_size):
examples = all_examples[batch_offset : batch_offset + batch_size]
batch = pa.Table.from_pylist(examples)
expected.extend(
[{k: v for k, v in x.items() if k not in columns_to_remove} for x in func(batch).to_pylist()]
)
assert next(iter(ex_iterable))[1] == expected[0]
assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
"n, func, batched, batch_size, fn_kwargs",
[
(3, lambda t, y=0: t.append_column("id+idx", pc.add(t["id"], y)), False, None, None),
(3, lambda t, y=0: t.append_column("id+idx", pc.add(t["id"], y)), False, None, {"y": 3}),
(25, lambda t, y=0: t.append_column("id+idx", pc.add(t["id"], y)), True, 10, {"y": 3}),
(5, lambda t, y=0: t.append_column("id+idx", pc.add(t["id"], y)), True, None, {"y": 3}), # same with bs=None
(5, lambda t, y=0: t.append_column("id+idx", pc.add(t["id"], y)), True, -1, {"y": 3}), # same with bs<=0
],
)
def test_mapped_examples_iterable_fn_kwargs_and_arrow_format(n, func, batched, batch_size, fn_kwargs):
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n})
ex_iterable = MappedExamplesIterable(
base_ex_iterable,
func,
batched=batched,
batch_size=batch_size,
fn_kwargs=fn_kwargs,
formatting=FormattingConfig(format_type="arrow"),
)
all_examples = [x for _, x in generate_examples_fn(n=n)]
if fn_kwargs is None:
fn_kwargs = {}
if batched is False:
expected = [func(pa.Table.from_pylist([x]), **fn_kwargs).to_pylist()[0] for x in all_examples]
else:
expected = []
# If batch_size is None or <=0, we use the whole dataset as a single batch
if batch_size is None or batch_size <= 0:
batch_size = len(all_examples)
for batch_offset in range(0, len(all_examples), batch_size):
examples = all_examples[batch_offset : batch_offset + batch_size]
batch = pa.Table.from_pylist(examples)
expected.extend(func(batch, **fn_kwargs).to_pylist())
assert next(iter(ex_iterable))[1] == expected[0]
assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
"n, func, batched, batch_size, input_columns",
[
(3, lambda id_: pa.table({"id+1": pc.add(id_, 1)}), False, None, ["id"]), # just add 1 to the id
(25, lambda ids_: pa.table({"id+1": pc.add(ids_, 1)}), True, 10, ["id"]), # same with bs=10
(5, lambda ids_: pa.table({"id+1": pc.add(ids_, 1)}), True, None, ["id"]), # same with bs=None
(5, lambda ids_: pa.table({"id+1": pc.add(ids_, 1)}), True, -1, ["id"]), # same with bs<=0
],
)
def test_mapped_examples_iterable_input_columns_and_arrow_format(n, func, batched, batch_size, input_columns):
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n})
ex_iterable = MappedExamplesIterable(
base_ex_iterable,
func,
batched=batched,
batch_size=batch_size,
input_columns=input_columns,
formatting=FormattingConfig(format_type="arrow"),
)
all_examples = [x for _, x in generate_examples_fn(n=n)]
columns_to_input = input_columns if isinstance(input_columns, list) else [input_columns]
if batched is False:
expected = [
func(*[pa.Table.from_pylist([x])[col] for col in columns_to_input]).to_pylist()[0] for x in all_examples
]
else:
expected = []
# If batch_size is None or <=0, we use the whole dataset as a single batch
if batch_size is None or batch_size <= 0:
batch_size = len(all_examples)
for batch_offset in range(0, len(all_examples), batch_size):
examples = all_examples[batch_offset : batch_offset + batch_size]
batch = pa.Table.from_pylist(examples)
expected.extend(func(*[batch[col] for col in columns_to_input]).to_pylist())
assert next(iter(ex_iterable))[1] == expected[0]
assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
"n, func, batched, batch_size",
[
(3, lambda x: x["id"] % 2 == 0, False, None), # keep even number
(3, lambda x: [x["id"][0] % 2 == 0], True, 1), # same with bs=1
(25, lambda x: [i % 2 == 0 for i in x["id"]], True, 10), # same with bs=10
(5, lambda x: [i % 2 == 0 for i in x["id"]], True, None), # same with bs=None
(5, lambda x: [i % 2 == 0 for i in x["id"]], True, -1), # same with bs<=0
(3, lambda x: False, False, None), # return 0 examples
(3, lambda x: [False] * len(x["id"]), True, 10), # same with bs=10
],
)
def test_filtered_examples_iterable(n, func, batched, batch_size):
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n})
ex_iterable = FilteredExamplesIterable(base_ex_iterable, func, batched=batched, batch_size=batch_size)
all_examples = [x for _, x in generate_examples_fn(n=n)]
if batched is False:
expected = [x for x in all_examples if func(x)]
else:
# For batched filter we have to format the examples as a batch (i.e. in one single dictionary) to pass the batch to the function
expected = []
# If batch_size is None or <=0, we use the whole dataset as a single batch
if batch_size is None or batch_size <= 0:
batch_size = len(all_examples)
for batch_offset in range(0, len(all_examples), batch_size):
examples = all_examples[batch_offset : batch_offset + batch_size]
batch = _examples_to_batch(examples)
mask = func(batch)
expected.extend([x for x, to_keep in zip(examples, mask) if to_keep])
if expected:
assert next(iter(ex_iterable))[1] == expected[0]
assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
"n, func, batched, batch_size",
[
(3, lambda x, index: index % 2 == 0, False, None), # keep even number
(25, lambda x, indices: [idx % 2 == 0 for idx in indices], True, 10), # same with bs=10
(5, lambda x, indices: [idx % 2 == 0 for idx in indices], True, None), # same with bs=None
(5, lambda x, indices: [idx % 2 == 0 for idx in indices], True, -1), # same with bs<=0
],
)
def test_filtered_examples_iterable_with_indices(n, func, batched, batch_size):
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n})
ex_iterable = FilteredExamplesIterable(
base_ex_iterable, func, batched=batched, batch_size=batch_size, with_indices=True
)
all_examples = [x for _, x in generate_examples_fn(n=n)]
if batched is False:
expected = [x for idx, x in enumerate(all_examples) if func(x, idx)]
else:
# For batched filter we have to format the examples as a batch (i.e. in one single dictionary) to pass the batch to the function
expected = []
# If batch_size is None or <=0, we use the whole dataset as a single batch
if batch_size is None or batch_size <= 0:
batch_size = len(all_examples)
for batch_offset in range(0, len(all_examples), batch_size):
examples = all_examples[batch_offset : batch_offset + batch_size]
batch = _examples_to_batch(examples)
indices = list(range(batch_offset, batch_offset + len(examples)))
mask = func(batch, indices)
expected.extend([x for x, to_keep in zip(examples, mask) if to_keep])
assert next(iter(ex_iterable))[1] == expected[0]
assert [x for _, x in ex_iterable] == expected
@pytest.mark.parametrize(
"n, func, batched, batch_size, input_columns",
[
(3, lambda id_: id_ % 2 == 0, False, None, ["id"]), # keep even number
(25, lambda ids_: [i % 2 == 0 for i in ids_], True, 10, ["id"]), # same with bs=10
(3, lambda ids_: [i % 2 == 0 for i in ids_], True, None, ["id"]), # same with bs=None
(3, lambda ids_: [i % 2 == 0 for i in ids_], True, None, ["id"]), # same with bs=None
],
)
def test_filtered_examples_iterable_input_columns(n, func, batched, batch_size, input_columns):
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n})
ex_iterable = FilteredExamplesIterable(
base_ex_iterable, func, batched=batched, batch_size=batch_size, input_columns=input_columns
)
all_examples = [x for _, x in generate_examples_fn(n=n)]
columns_to_input = input_columns if isinstance(input_columns, list) else [input_columns]
if batched is False:
expected = [x for x in all_examples if func(*[x[col] for col in columns_to_input])]
else:
# For batched filter we have to format the examples as a batch (i.e. in one single dictionary) to pass the batch to the function
expected = []
# If batch_size is None or <=0, we use the whole dataset as a single batch
if batch_size is None or batch_size <= 0:
batch_size = len(all_examples)
for batch_offset in range(0, len(all_examples), batch_size):
examples = all_examples[batch_offset : batch_offset + batch_size]
batch = _examples_to_batch(examples)
mask = func(*[batch[col] for col in columns_to_input])
expected.extend([x for x, to_keep in zip(examples, mask) if to_keep])
assert next(iter(ex_iterable))[1] == expected[0]
assert [x for _, x in ex_iterable] == expected
def test_skip_examples_iterable():
total, count = 10, 2
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": total})
skip_ex_iterable = SkipExamplesIterable(base_ex_iterable, n=count)
expected = list(generate_examples_fn(n=total))[count:]
assert list(skip_ex_iterable) == expected
assert (
skip_ex_iterable.shuffle_data_sources(np.random.default_rng(42)) is skip_ex_iterable
), "skip examples makes the shards order fixed"
def test_take_examples_iterable():
total, count = 10, 2
base_ex_iterable = ExamplesIterable(generate_examples_fn, {"n": total})
take_ex_iterable = TakeExamplesIterable(base_ex_iterable, n=count)
expected = list(generate_examples_fn(n=total))[:count]
assert list(take_ex_iterable) == expected
assert (
take_ex_iterable.shuffle_data_sources(np.random.default_rng(42)) is take_ex_iterable
), "skip examples makes the shards order fixed"
def test_vertically_concatenated_examples_iterable():
ex_iterable1 = ExamplesIterable(generate_examples_fn, {"label": 10})
ex_iterable2 = ExamplesIterable(generate_examples_fn, {"label": 5})
concatenated_ex_iterable = VerticallyConcatenatedMultiSourcesExamplesIterable([ex_iterable1, ex_iterable2])
expected = [x for _, x in ex_iterable1] + [x for _, x in ex_iterable2]
assert [x for _, x in concatenated_ex_iterable] == expected
def test_vertically_concatenated_examples_iterable_with_different_columns():
# having different columns is supported
# Though iterable datasets fill the missing data with nulls
ex_iterable1 = ExamplesIterable(generate_examples_fn, {"label": 10})
ex_iterable2 = ExamplesIterable(generate_examples_fn, {})
concatenated_ex_iterable = VerticallyConcatenatedMultiSourcesExamplesIterable([ex_iterable1, ex_iterable2])
expected = [x for _, x in ex_iterable1] + [x for _, x in ex_iterable2]
assert [x for _, x in concatenated_ex_iterable] == expected
def test_vertically_concatenated_examples_iterable_shuffle_data_sources():
ex_iterable1 = ExamplesIterable(generate_examples_fn, {"label": 10})
ex_iterable2 = ExamplesIterable(generate_examples_fn, {"label": 5})
concatenated_ex_iterable = VerticallyConcatenatedMultiSourcesExamplesIterable([ex_iterable1, ex_iterable2])
rng = np.random.default_rng(42)
shuffled_ex_iterable = concatenated_ex_iterable.shuffle_data_sources(rng)
# make sure the list of examples iterables is shuffled, and each examples iterable is shuffled
expected = [x for _, x in ex_iterable2.shuffle_data_sources(rng)] + [
x for _, x in ex_iterable1.shuffle_data_sources(rng)
]
assert [x for _, x in shuffled_ex_iterable] == expected
def test_horizontally_concatenated_examples_iterable():
ex_iterable1 = ExamplesIterable(generate_examples_fn, {"label1": 10})
ex_iterable2 = ExamplesIterable(generate_examples_fn, {"label2": 5})
concatenated_ex_iterable = HorizontallyConcatenatedMultiSourcesExamplesIterable([ex_iterable1, ex_iterable2])
with pytest.raises(ValueError): # column "id" is duplicated -> raise an error
list(concatenated_ex_iterable)
ex_iterable2 = MappedExamplesIterable(ex_iterable2, lambda x: x, remove_columns=["id"])
concatenated_ex_iterable = HorizontallyConcatenatedMultiSourcesExamplesIterable([ex_iterable1, ex_iterable2])
expected = [{**x, **y} for (_, x), (_, y) in zip(ex_iterable1, ex_iterable2)]
assert [x for _, x in concatenated_ex_iterable] == expected
assert (
concatenated_ex_iterable.shuffle_data_sources(np.random.default_rng(42)) is concatenated_ex_iterable
), "horizontally concatenated examples makes the shards order fixed"
@pytest.mark.parametrize(
"ex_iterable",
[
ExamplesIterable(generate_examples_fn, {}),
ShuffledDataSourcesExamplesIterable(generate_examples_fn, {}, np.random.default_rng(42)),
SelectColumnsIterable(ExamplesIterable(generate_examples_fn, {}), ["id"]),
StepExamplesIterable(ExamplesIterable(generate_examples_fn, {}), 2, 0),
CyclingMultiSourcesExamplesIterable([ExamplesIterable(generate_examples_fn, {})]),
VerticallyConcatenatedMultiSourcesExamplesIterable([ExamplesIterable(generate_examples_fn, {})]),
HorizontallyConcatenatedMultiSourcesExamplesIterable([ExamplesIterable(generate_examples_fn, {})]),
RandomlyCyclingMultiSourcesExamplesIterable(
[ExamplesIterable(generate_examples_fn, {})], np.random.default_rng(42)
),
MappedExamplesIterable(ExamplesIterable(generate_examples_fn, {}), lambda x: x),
MappedExamplesIterable(ArrowExamplesIterable(generate_tables_fn, {}), lambda x: x),
FilteredExamplesIterable(ExamplesIterable(generate_examples_fn, {}), lambda x: True),
FilteredExamplesIterable(ArrowExamplesIterable(generate_tables_fn, {}), lambda x: True),
BufferShuffledExamplesIterable(ExamplesIterable(generate_examples_fn, {}), 10, np.random.default_rng(42)),
SkipExamplesIterable(ExamplesIterable(generate_examples_fn, {}), 10),
TakeExamplesIterable(ExamplesIterable(generate_examples_fn, {}), 10),
TypedExamplesIterable(
ExamplesIterable(generate_examples_fn, {}), Features({"id": Value("int32")}), token_per_repo_id={}
),
],
)
def test_no_iter_arrow(ex_iterable: _BaseExamplesIterable):
assert ex_iterable.iter_arrow is None
@pytest.mark.parametrize(
"ex_iterable",
[
ArrowExamplesIterable(generate_tables_fn, {}),
ShuffledDataSourcesArrowExamplesIterable(generate_tables_fn, {}, np.random.default_rng(42)),
SelectColumnsIterable(ArrowExamplesIterable(generate_tables_fn, {}), ["id"]),
# StepExamplesIterable(ArrowExamplesIterable(generate_tables_fn, {}), 2, 0), # not implemented
# CyclingMultiSourcesExamplesIterable([ArrowExamplesIterable(generate_tables_fn, {})]), # not implemented
VerticallyConcatenatedMultiSourcesExamplesIterable([ArrowExamplesIterable(generate_tables_fn, {})]),
# HorizontallyConcatenatedMultiSourcesExamplesIterable([ArrowExamplesIterable(generate_tables_fn, {})]), # not implemented
# RandomlyCyclingMultiSourcesExamplesIterable([ArrowExamplesIterable(generate_tables_fn, {})], np.random.default_rng(42)), # not implemented
MappedExamplesIterable(
ExamplesIterable(generate_examples_fn, {}), lambda t: t, formatting=FormattingConfig(format_type="arrow")
),
MappedExamplesIterable(
ArrowExamplesIterable(generate_tables_fn, {}),
lambda t: t,
formatting=FormattingConfig(format_type="arrow"),
),
FilteredExamplesIterable(
ExamplesIterable(generate_examples_fn, {}),
lambda t: True,
formatting=FormattingConfig(format_type="arrow"),
),
FilteredExamplesIterable(
ArrowExamplesIterable(generate_tables_fn, {}),
lambda t: True,
formatting=FormattingConfig(format_type="arrow"),
),
# BufferShuffledExamplesIterable(ArrowExamplesIterable(generate_tables_fn, {}), 10, np.random.default_rng(42)), # not implemented
# SkipExamplesIterable(ArrowExamplesIterable(generate_tables_fn, {}), 10), # not implemented
# TakeExamplesIterable(ArrowExamplesIterable(generate_tables_fn, {}), 10), # not implemented
TypedExamplesIterable(
ArrowExamplesIterable(generate_tables_fn, {}), Features({"id": Value("int32")}), token_per_repo_id={}
),
],
)
def test_iter_arrow(ex_iterable: _BaseExamplesIterable):
assert ex_iterable.iter_arrow is not None
key, pa_table = next(ex_iterable.iter_arrow())
assert isinstance(pa_table, pa.Table)
############################
#
# IterableDataset tests
#
############################
def test_iterable_dataset():
dataset = IterableDataset(ExamplesIterable(generate_examples_fn, {}))
expected = [x for _, x in generate_examples_fn()]
assert next(iter(dataset)) == expected[0]
assert list(dataset) == expected
def test_iterable_dataset_from_generator():
data = [
{"col_1": "0", "col_2": 0, "col_3": 0.0},
{"col_1": "1", "col_2": 1, "col_3": 1.0},
{"col_1": "2", "col_2": 2, "col_3": 2.0},
{"col_1": "3", "col_2": 3, "col_3": 3.0},
]
def gen():
yield from data
dataset = IterableDataset.from_generator(gen)
assert isinstance(dataset, IterableDataset)
assert list(dataset) == data
def test_iterable_dataset_from_generator_with_shards():
def gen(shard_names):
for shard_name in shard_names:
for i in range(10):
yield {"shard_name": shard_name, "i": i}
shard_names = [f"data{shard_idx}.txt" for shard_idx in range(4)]
dataset = IterableDataset.from_generator(gen, gen_kwargs={"shard_names": shard_names})
assert isinstance(dataset, IterableDataset)
assert dataset.n_shards == len(shard_names)
def test_iterable_dataset_from_file(dataset: IterableDataset, arrow_file: str):
with assert_arrow_memory_doesnt_increase():
dataset_from_file = IterableDataset.from_file(arrow_file)
expected_features = dataset._resolve_features().features
assert dataset_from_file.features.type == expected_features.type
assert dataset_from_file.features == expected_features
assert isinstance(dataset_from_file, IterableDataset)
assert list(dataset_from_file) == list(dataset)
@require_not_windows
@require_dill_gt_0_3_2
@require_pyspark
def test_from_spark_streaming():
import pyspark
spark = pyspark.sql.SparkSession.builder.master("local[*]").appName("pyspark").getOrCreate()
data = [
("0", 0, 0.0),
("1", 1, 1.0),
("2", 2, 2.0),
("3", 3, 3.0),
]
df = spark.createDataFrame(data, "col_1: string, col_2: int, col_3: float")
dataset = IterableDataset.from_spark(df)
assert isinstance(dataset, IterableDataset)
results = []
for ex in dataset:
results.append(ex)
assert results == [
{"col_1": "0", "col_2": 0, "col_3": 0.0},
{"col_1": "1", "col_2": 1, "col_3": 1.0},
{"col_1": "2", "col_2": 2, "col_3": 2.0},
{"col_1": "3", "col_2": 3, "col_3": 3.0},
]
@require_not_windows
@require_dill_gt_0_3_2
@require_pyspark
def test_from_spark_streaming_features():
import PIL.Image
import pyspark
spark = pyspark.sql.SparkSession.builder.master("local[*]").appName("pyspark").getOrCreate()
data = [(0, np.arange(4 * 4 * 3).reshape(4, 4, 3).tolist())]
df = spark.createDataFrame(data, "idx: int, image: array<array<array<int>>>")
features = Features({"idx": Value("int64"), "image": Image()})
dataset = IterableDataset.from_spark(
df,
features=features,
)
assert isinstance(dataset, IterableDataset)
results = []
for ex in dataset:
results.append(ex)
assert len(results) == 1
isinstance(results[0]["image"], PIL.Image.Image)
@require_torch
def test_iterable_dataset_torch_integration():
ex_iterable = ExamplesIterable(generate_examples_fn, {})
dataset = IterableDataset(ex_iterable)
import torch.utils.data
assert isinstance(dataset, torch.utils.data.IterableDataset)
assert isinstance(dataset, IterableDataset)
assert dataset._ex_iterable is ex_iterable
@require_torch
def test_iterable_dataset_torch_picklable():
import pickle
ex_iterable = ExamplesIterable(generate_examples_fn, {})
dataset = IterableDataset(ex_iterable, formatting=FormattingConfig(format_type="torch"))
reloaded_dataset = pickle.loads(pickle.dumps(dataset))
import torch.utils.data
assert isinstance(reloaded_dataset, IterableDataset)
assert isinstance(reloaded_dataset, torch.utils.data.IterableDataset)
assert reloaded_dataset._formatting.format_type == "torch"
assert len(list(dataset)) == len(list(reloaded_dataset))
@require_torch
def test_iterable_dataset_with_format_torch():
ex_iterable = ExamplesIterable(generate_examples_fn, {})
dataset = IterableDataset(ex_iterable)
from torch.utils.data import DataLoader
dataloader = DataLoader(dataset)
assert len(list(dataloader)) == len(list(ex_iterable))
@require_torch
def test_iterable_dataset_torch_dataloader_parallel():
from torch.utils.data import DataLoader
ex_iterable = ExamplesIterable(generate_examples_fn, {})
dataset = IterableDataset(ex_iterable)
dataloader = DataLoader(dataset, num_workers=2, batch_size=None)
result = list(dataloader)
expected = [example for _, example in ex_iterable]
assert len(result) == len(expected)
assert {str(x) for x in result} == {str(x) for x in expected}
@require_torch
@pytest.mark.filterwarnings("ignore:This DataLoader will create:UserWarning")
@pytest.mark.parametrize("n_shards, num_workers", [(2, 1), (2, 2), (3, 2), (2, 3)])
def test_sharded_iterable_dataset_torch_dataloader_parallel(n_shards, num_workers):
from torch.utils.data import DataLoader
ex_iterable = ExamplesIterable(generate_examples_fn, {"filepaths": [f"{i}.txt" for i in range(n_shards)]})
dataset = IterableDataset(ex_iterable)
dataloader = DataLoader(dataset, batch_size=None, num_workers=num_workers)
result = list(dataloader)
expected = [example for _, example in ex_iterable]
assert len(result) == len(expected)
assert {str(x) for x in result} == {str(x) for x in expected}
@require_torch
@pytest.mark.integration
@pytest.mark.parametrize("num_workers", [1, 2])
def test_iterable_dataset_from_hub_torch_dataloader_parallel(num_workers, tmp_path):
from torch.utils.data import DataLoader
dataset = load_dataset(SAMPLE_DATASET_IDENTIFIER, cache_dir=str(tmp_path), streaming=True, split="train")
dataloader = DataLoader(dataset, batch_size=None, num_workers=num_workers)
result = list(dataloader)
assert len(result) == 2
@pytest.mark.parametrize("batch_size", [4, 5])
@pytest.mark.parametrize("drop_last_batch", [False, True])
def test_iterable_dataset_iter_batch(batch_size, drop_last_batch):
n = 25
dataset = IterableDataset(ExamplesIterable(generate_examples_fn, {"n": n}))
all_examples = [ex for _, ex in generate_examples_fn(n=n)]
expected = []
for i in range(0, len(all_examples), batch_size):
if len(all_examples[i : i + batch_size]) < batch_size and drop_last_batch:
continue
expected.append(_examples_to_batch(all_examples[i : i + batch_size]))
assert next(iter(dataset.iter(batch_size, drop_last_batch=drop_last_batch))) == expected[0]
assert list(dataset.iter(batch_size, drop_last_batch=drop_last_batch)) == expected
def test_iterable_dataset_info():
info = DatasetInfo(description="desc", citation="@article{}", size_in_bytes=42)
ex_iterable = ExamplesIterable(generate_examples_fn, {})
dataset = IterableDataset(ex_iterable, info=info)
assert dataset.info == info
assert dataset.description == info.description
assert dataset.citation == info.citation
assert dataset.size_in_bytes == info.size_in_bytes
def test_iterable_dataset_set_epoch(dataset: IterableDataset):
assert dataset._epoch == 0
dataset.set_epoch(42)
assert dataset._epoch == 42
@pytest.mark.parametrize("seed", [None, 42, 1337])
@pytest.mark.parametrize("epoch", [None, 0, 1, 10])
def test_iterable_dataset_set_epoch_of_shuffled_dataset(dataset: IterableDataset, seed, epoch):
buffer_size = 10
shuffled_dataset = dataset.shuffle(seed, buffer_size=buffer_size)
base_generator = shuffled_dataset._shuffling.generator
if epoch is not None:
shuffled_dataset.set_epoch(epoch)
effective_generator = shuffled_dataset._effective_generator()
assert effective_generator is not None
if epoch is None or epoch == 0:
assert is_rng_equal(base_generator, shuffled_dataset._effective_generator())
else:
assert not is_rng_equal(base_generator, shuffled_dataset._effective_generator())
effective_seed = deepcopy(base_generator).integers(0, 1 << 63) - epoch
assert is_rng_equal(np.random.default_rng(effective_seed), shuffled_dataset._effective_generator())
def test_iterable_dataset_map(
dataset: IterableDataset,
):
func = lambda x: {"id+1": x["id"] + 1} # noqa: E731
mapped_dataset = dataset.map(func)
assert isinstance(mapped_dataset._ex_iterable, MappedExamplesIterable)
assert mapped_dataset._ex_iterable.function is func
assert mapped_dataset._ex_iterable.batched is False
assert next(iter(mapped_dataset)) == {**next(iter(dataset)), **func(next(iter(generate_examples_fn()))[1])}
def test_iterable_dataset_map_batched(
dataset: IterableDataset,
):
func = lambda x: {"id+1": [i + 1 for i in x["id"]]} # noqa: E731
batch_size = 3
dataset = dataset.map(func, batched=True, batch_size=batch_size)
assert isinstance(dataset._ex_iterable, MappedExamplesIterable)
assert dataset._ex_iterable.function is func
assert dataset._ex_iterable.batch_size == batch_size
assert next(iter(dataset)) == {"id": 0, "id+1": 1}
def test_iterable_dataset_map_complex_features(
dataset: IterableDataset,
):
# https://github.com/huggingface/datasets/issues/3505
ex_iterable = ExamplesIterable(generate_examples_fn, {"label": "positive"})
features = Features(
{
"id": Value("int64"),
"label": Value("string"),
}
)
dataset = IterableDataset(ex_iterable, info=DatasetInfo(features=features))
dataset = dataset.cast_column("label", ClassLabel(names=["negative", "positive"]))
dataset = dataset.map(lambda x: {"id+1": x["id"] + 1, **x})
assert isinstance(dataset._ex_iterable, MappedExamplesIterable)
features["label"] = ClassLabel(names=["negative", "positive"])
assert [{k: v for k, v in ex.items() if k != "id+1"} for ex in dataset] == [
features.encode_example(ex) for _, ex in ex_iterable
]
def test_iterable_dataset_map_with_features(dataset: IterableDataset) -> None:
# https://github.com/huggingface/datasets/issues/3888
ex_iterable = ExamplesIterable(generate_examples_fn, {"label": "positive"})
features_before_map = Features(
{
"id": Value("int64"),
"label": Value("string"),
}
)
dataset = IterableDataset(ex_iterable, info=DatasetInfo(features=features_before_map))
assert dataset.info.features is not None
assert dataset.info.features == features_before_map
features_after_map = Features(
{
"id": Value("int64"),
"label": Value("string"),
"target": Value("string"),
}
)
dataset = dataset.map(lambda x: {"target": x["label"]}, features=features_after_map)
assert dataset.info.features is not None
assert dataset.info.features == features_after_map
def test_iterable_dataset_map_with_fn_kwargs(dataset: IterableDataset) -> None:
fn_kwargs = {"y": 1}
mapped_dataset = dataset.map(lambda x, y: {"id+y": x["id"] + y}, fn_kwargs=fn_kwargs)
assert mapped_dataset._ex_iterable.batched is False
assert next(iter(mapped_dataset)) == {"id": 0, "id+y": 1}
batch_size = 3
mapped_dataset = dataset.map(
lambda x, y: {"id+y": [i + y for i in x["id"]]}, batched=True, batch_size=batch_size, fn_kwargs=fn_kwargs
)
assert isinstance(mapped_dataset._ex_iterable, MappedExamplesIterable)
assert mapped_dataset._ex_iterable.batch_size == batch_size
assert next(iter(mapped_dataset)) == {"id": 0, "id+y": 1}
def test_iterable_dataset_filter(dataset: IterableDataset) -> None:
fn_kwargs = {"y": 1}
filtered_dataset = dataset.filter(lambda x, y: x["id"] == y, fn_kwargs=fn_kwargs)
assert filtered_dataset._ex_iterable.batched is False
assert next(iter(filtered_dataset)) == {"id": 1}
@pytest.mark.parametrize("seed", [42, 1337, 101010, 123456])
@pytest.mark.parametrize("epoch", [None, 0, 1])
def test_iterable_dataset_shuffle(dataset: IterableDataset, seed, epoch):
buffer_size = 3
dataset = deepcopy(dataset)
dataset._ex_iterable.kwargs["filepaths"] = ["0.txt", "1.txt"]
dataset = dataset.shuffle(seed, buffer_size=buffer_size)
assert isinstance(dataset._shuffling, ShufflingConfig)
assert isinstance(dataset._shuffling.generator, np.random.Generator)
assert is_rng_equal(dataset._shuffling.generator, np.random.default_rng(seed))
# Effective seed is sum of seed and epoch
if epoch is None or epoch == 0:
effective_seed = seed
else:
dataset.set_epoch(epoch)
effective_seed = np.random.default_rng(seed).integers(0, 1 << 63) - epoch
# Shuffling adds a shuffle buffer
expected_first_example_index = next(
iter(BufferShuffledExamplesIterable._iter_random_indices(np.random.default_rng(effective_seed), buffer_size))
)
assert isinstance(dataset._ex_iterable, BufferShuffledExamplesIterable)
# It also shuffles the underlying examples iterable
expected_ex_iterable = ExamplesIterable(
generate_examples_fn, {"filepaths": ["0.txt", "1.txt"]}
).shuffle_data_sources(np.random.default_rng(effective_seed))
assert isinstance(dataset._ex_iterable.ex_iterable, ExamplesIterable)
assert next(iter(dataset)) == list(islice(expected_ex_iterable, expected_first_example_index + 1))[-1][1]
@pytest.mark.parametrize(
"features",
[
None,
Features(
{
"id": Value("int64"),
"label": Value("int64"),
}
),
Features(
{
"id": Value("int64"),
"label": ClassLabel(names=["negative", "positive"]),
}
),
],
)
def test_iterable_dataset_features(features):
ex_iterable = ExamplesIterable(generate_examples_fn, {"label": 0})
dataset = IterableDataset(ex_iterable, info=DatasetInfo(features=features))
if features:
expected = [features.encode_example(x) for _, x in ex_iterable]
else:
expected = [x for _, x in ex_iterable]
assert list(dataset) == expected
def test_iterable_dataset_features_cast_to_python():
ex_iterable = ExamplesIterable(
generate_examples_fn, {"timestamp": pd.Timestamp(2020, 1, 1), "array": np.ones(5), "n": 1}
)
features = Features(
{
"id": Value("int64"),
"timestamp": Value("timestamp[us]"),
"array": [Value("int64")],
}
)
dataset = IterableDataset(ex_iterable, info=DatasetInfo(features=features))
assert list(dataset) == [{"timestamp": pd.Timestamp(2020, 1, 1).to_pydatetime(), "array": [1] * 5, "id": 0}]
@pytest.mark.parametrize("format_type", [None, "torch", "python", "tf", "tensorflow", "np", "numpy", "jax"])
def test_iterable_dataset_with_format(dataset: IterableDataset, format_type):
formatted_dataset = dataset.with_format(format_type)
assert formatted_dataset._formatting.format_type == get_format_type_from_alias(format_type)
@require_torch
def test_iterable_dataset_is_torch_iterable_dataset(dataset: IterableDataset):
from torch.utils.data import DataLoader, _DatasetKind
dataloader = DataLoader(dataset)
assert dataloader._dataset_kind == _DatasetKind.Iterable
out = list(dataloader)
assert len(out) == DEFAULT_N_EXAMPLES
@pytest.mark.parametrize("n", [0, 2, int(1e10)])
def test_iterable_dataset_skip(dataset: IterableDataset, n):
skip_dataset = dataset.skip(n)
assert isinstance(skip_dataset._ex_iterable, SkipExamplesIterable)
assert skip_dataset._ex_iterable.n == n
assert list(skip_dataset) == list(dataset)[n:]
@pytest.mark.parametrize("n", [0, 2, int(1e10)])
def test_iterable_dataset_take(dataset: IterableDataset, n):
take_dataset = dataset.take(n)
assert isinstance(take_dataset._ex_iterable, TakeExamplesIterable)
assert take_dataset._ex_iterable.n == n
assert list(take_dataset) == list(dataset)[:n]
@pytest.mark.parametrize("method", ["skip", "take"])
def test_iterable_dataset_shuffle_after_skip_or_take(method):
seed = 42
n, n_shards = 3, 10
count = 7
ex_iterable = ExamplesIterable(generate_examples_fn, {"n": n, "filepaths": [f"{i}.txt" for i in range(n_shards)]})
dataset = IterableDataset(ex_iterable)
dataset = dataset.skip(n) if method == "skip" else dataset.take(count)
shuffled_dataset = dataset.shuffle(seed, buffer_size=DEFAULT_N_EXAMPLES)
# shuffling a skip/take dataset should keep the same examples and don't shuffle the shards
key = lambda x: f"{x['filepath']}_{x['id']}" # noqa: E731
assert sorted(dataset, key=key) == sorted(shuffled_dataset, key=key)
def test_iterable_dataset_add_column(dataset_with_several_columns):
new_column = list(range(DEFAULT_N_EXAMPLES))
new_dataset = dataset_with_several_columns.add_column("new_column", new_column)
assert list(new_dataset) == [
{**example, "new_column": idx} for idx, example in enumerate(dataset_with_several_columns)
]
new_dataset = new_dataset._resolve_features()
assert "new_column" in new_dataset.column_names
def test_iterable_dataset_rename_column(dataset_with_several_columns):
new_dataset = dataset_with_several_columns.rename_column("id", "new_id")
assert list(new_dataset) == [
{("new_id" if k == "id" else k): v for k, v in example.items()} for example in dataset_with_several_columns
]
assert new_dataset.features is None
assert new_dataset.column_names is None
# rename the column if ds.features was not None
new_dataset = dataset_with_several_columns._resolve_features().rename_column("id", "new_id")
assert new_dataset.features is not None
assert new_dataset.column_names is not None
assert "id" not in new_dataset.column_names
assert "new_id" in new_dataset.column_names
def test_iterable_dataset_rename_columns(dataset_with_several_columns):
column_mapping = {"id": "new_id", "filepath": "filename"}
new_dataset = dataset_with_several_columns.rename_columns(column_mapping)
assert list(new_dataset) == [
{column_mapping.get(k, k): v for k, v in example.items()} for example in dataset_with_several_columns
]
assert new_dataset.features is None
assert new_dataset.column_names is None
# rename the columns if ds.features was not None
new_dataset = dataset_with_several_columns._resolve_features().rename_columns(column_mapping)
assert new_dataset.features is not None
assert new_dataset.column_names is not None
assert all(c not in new_dataset.column_names for c in ["id", "filepath"])
assert all(c in new_dataset.column_names for c in ["new_id", "filename"])
def test_iterable_dataset_remove_columns(dataset_with_several_columns):
new_dataset = dataset_with_several_columns.remove_columns("id")
assert list(new_dataset) == [
{k: v for k, v in example.items() if k != "id"} for example in dataset_with_several_columns
]
assert new_dataset.features is None
new_dataset = dataset_with_several_columns.remove_columns(["id", "filepath"])
assert list(new_dataset) == [
{k: v for k, v in example.items() if k != "id" and k != "filepath"} for example in dataset_with_several_columns
]
assert new_dataset.features is None
assert new_dataset.column_names is None
# remove the columns if ds.features was not None
new_dataset = dataset_with_several_columns._resolve_features().remove_columns(["id", "filepath"])
assert new_dataset.features is not None
assert new_dataset.column_names is not None
assert all(c not in new_dataset.features for c in ["id", "filepath"])
assert all(c not in new_dataset.column_names for c in ["id", "filepath"])
def test_iterable_dataset_select_columns(dataset_with_several_columns):
new_dataset = dataset_with_several_columns.select_columns("id")
assert list(new_dataset) == [
{k: v for k, v in example.items() if k == "id"} for example in dataset_with_several_columns
]
assert new_dataset.features is None
new_dataset = dataset_with_several_columns.select_columns(["id", "filepath"])
assert list(new_dataset) == [
{k: v for k, v in example.items() if k in ("id", "filepath")} for example in dataset_with_several_columns
]
assert new_dataset.features is None
# select the columns if ds.features was not None
new_dataset = dataset_with_several_columns._resolve_features().select_columns(["id", "filepath"])
assert new_dataset.features is not None
assert new_dataset.column_names is not None
assert all(c in new_dataset.features for c in ["id", "filepath"])
assert all(c in new_dataset.column_names for c in ["id", "filepath"])
def test_iterable_dataset_cast_column():
ex_iterable = ExamplesIterable(generate_examples_fn, {"label": 10})
features = Features({"id": Value("int64"), "label": Value("int64")})
dataset = IterableDataset(ex_iterable, info=DatasetInfo(features=features))
casted_dataset = dataset.cast_column("label", Value("bool"))
casted_features = features.copy()
casted_features["label"] = Value("bool")
assert list(casted_dataset) == [casted_features.encode_example(ex) for _, ex in ex_iterable]
def test_iterable_dataset_cast():
ex_iterable = ExamplesIterable(generate_examples_fn, {"label": 10})
features = Features({"id": Value("int64"), "label": Value("int64")})
dataset = IterableDataset(ex_iterable, info=DatasetInfo(features=features))
new_features = Features({"id": Value("int64"), "label": Value("bool")})
casted_dataset = dataset.cast(new_features)
assert list(casted_dataset) == [new_features.encode_example(ex) for _, ex in ex_iterable]
def test_iterable_dataset_resolve_features():
ex_iterable = ExamplesIterable(generate_examples_fn, {})
dataset = IterableDataset(ex_iterable)
assert dataset.features is None
assert dataset.column_names is None
dataset = dataset._resolve_features()
assert dataset.features == Features(
{
"id": Value("int64"),
}
)
assert dataset.column_names == ["id"]
def test_iterable_dataset_resolve_features_keep_order():
def gen():
yield from zip(range(3), [{"a": 1}, {"c": 1}, {"b": 1}])
ex_iterable = ExamplesIterable(gen, {})
dataset = IterableDataset(ex_iterable)._resolve_features()
# columns appear in order of appearance in the dataset
assert list(dataset.features) == ["a", "c", "b"]
assert dataset.column_names == ["a", "c", "b"]
def test_iterable_dataset_with_features_fill_with_none():
def gen():
yield from zip(range(2), [{"a": 1}, {"b": 1}])
ex_iterable = ExamplesIterable(gen, {})
info = DatasetInfo(features=Features({"a": Value("int32"), "b": Value("int32")}))
dataset = IterableDataset(ex_iterable, info=info)
assert list(dataset) == [{"a": 1, "b": None}, {"b": 1, "a": None}]
def test_concatenate_datasets():
ex_iterable1 = ExamplesIterable(generate_examples_fn, {"label": 10})
dataset1 = IterableDataset(ex_iterable1)
ex_iterable2 = ExamplesIterable(generate_examples_fn, {"label": 5})
dataset2 = IterableDataset(ex_iterable2)
concatenated_dataset = concatenate_datasets([dataset1, dataset2])
assert list(concatenated_dataset) == list(dataset1) + list(dataset2)
def test_concatenate_datasets_resolves_features():
ex_iterable1 = ExamplesIterable(generate_examples_fn, {"label": 10})
dataset1 = IterableDataset(ex_iterable1)
ex_iterable2 = ExamplesIterable(generate_examples_fn, {"label": 5})
dataset2 = IterableDataset(ex_iterable2)
concatenated_dataset = concatenate_datasets([dataset1, dataset2])
assert concatenated_dataset.features is not None
assert sorted(concatenated_dataset.features) == ["id", "label"]
def test_concatenate_datasets_with_different_columns():
ex_iterable1 = ExamplesIterable(generate_examples_fn, {"label": 10})
dataset1 = IterableDataset(ex_iterable1)
ex_iterable2 = ExamplesIterable(generate_examples_fn, {})
dataset2 = IterableDataset(ex_iterable2)
# missing column "label" -> it should be replaced with nulls
extended_dataset2_list = [{"label": None, **x} for x in dataset2]
concatenated_dataset = concatenate_datasets([dataset1, dataset2])
assert list(concatenated_dataset) == list(dataset1) + extended_dataset2_list
# change order
concatenated_dataset = concatenate_datasets([dataset2, dataset1])
assert list(concatenated_dataset) == extended_dataset2_list + list(dataset1)
def test_concatenate_datasets_axis_1():
ex_iterable1 = ExamplesIterable(generate_examples_fn, {"label1": 10})
dataset1 = IterableDataset(ex_iterable1)
ex_iterable2 = ExamplesIterable(generate_examples_fn, {"label2": 5})
dataset2 = IterableDataset(ex_iterable2)
with pytest.raises(ValueError): # column "id" is duplicated -> raise an error
concatenate_datasets([dataset1, dataset2], axis=1)
concatenated_dataset = concatenate_datasets([dataset1, dataset2.remove_columns("id")], axis=1)
assert list(concatenated_dataset) == [{**x, **y} for x, y in zip(dataset1, dataset2)]
def test_concatenate_datasets_axis_1_resolves_features():
ex_iterable1 = ExamplesIterable(generate_examples_fn, {"label1": 10})
dataset1 = IterableDataset(ex_iterable1)
ex_iterable2 = ExamplesIterable(generate_examples_fn, {"label2": 5})
dataset2 = IterableDataset(ex_iterable2).remove_columns("id")
concatenated_dataset = concatenate_datasets([dataset1, dataset2], axis=1)
assert concatenated_dataset.features is not None
assert sorted(concatenated_dataset.features) == ["id", "label1", "label2"]
def test_concatenate_datasets_axis_1_with_different_lengths():
n1 = 10
ex_iterable1 = ExamplesIterable(generate_examples_fn, {"label1": 10, "n": n1})
dataset1 = IterableDataset(ex_iterable1)
n2 = 5
ex_iterable2 = ExamplesIterable(generate_examples_fn, {"label2": 5, "n": n2})
dataset2 = IterableDataset(ex_iterable2).remove_columns("id")
# missing rows -> they should be replaced with nulls
extended_dataset2_list = list(dataset2) + [{"label2": None}] * (n1 - n2)
concatenated_dataset = concatenate_datasets([dataset1, dataset2], axis=1)
assert list(concatenated_dataset) == [{**x, **y} for x, y in zip(dataset1, extended_dataset2_list)]
# change order
concatenated_dataset = concatenate_datasets([dataset2, dataset1], axis=1)
assert list(concatenated_dataset) == [{**x, **y} for x, y in zip(extended_dataset2_list, dataset1)]
@pytest.mark.parametrize(
"probas, seed, expected_length, stopping_strategy",
[
(None, None, 3 * (DEFAULT_N_EXAMPLES - 1) + 1, "first_exhausted"),
([1, 0, 0], None, DEFAULT_N_EXAMPLES, "first_exhausted"),
([0, 1, 0], None, DEFAULT_N_EXAMPLES, "first_exhausted"),
([0.2, 0.5, 0.3], 42, None, "first_exhausted"),
([0.1, 0.1, 0.8], 1337, None, "first_exhausted"),
([0.5, 0.2, 0.3], 101010, None, "first_exhausted"),
(None, None, 3 * DEFAULT_N_EXAMPLES, "all_exhausted"),
([0.2, 0.5, 0.3], 42, None, "all_exhausted"),
([0.1, 0.1, 0.8], 1337, None, "all_exhausted"),
([0.5, 0.2, 0.3], 101010, None, "all_exhausted"),
],
)
def test_interleave_datasets(dataset: IterableDataset, probas, seed, expected_length, stopping_strategy):
d1 = dataset
d2 = dataset.map(lambda x: {"id+1": x["id"] + 1, **x})
d3 = dataset.with_format("python")
datasets = [d1, d2, d3]
merged_dataset = interleave_datasets(
datasets, probabilities=probas, seed=seed, stopping_strategy=stopping_strategy
)
def fill_default(example):
return {"id": None, "id+1": None, **example}
# Check the examples iterable
assert isinstance(
merged_dataset._ex_iterable, (CyclingMultiSourcesExamplesIterable, RandomlyCyclingMultiSourcesExamplesIterable)
)
# Check that it is deterministic
if seed is not None:
merged_dataset2 = interleave_datasets(
[d1, d2, d3], probabilities=probas, seed=seed, stopping_strategy=stopping_strategy
)
assert list(merged_dataset) == list(merged_dataset2)
# Check features
assert merged_dataset.features == Features({"id": Value("int64"), "id+1": Value("int64")})
# Check first example
if seed is not None:
rng = np.random.default_rng(seed)
i = next(iter(RandomlyCyclingMultiSourcesExamplesIterable._iter_random_indices(rng, len(datasets), p=probas)))
assert next(iter(merged_dataset)) == fill_default(next(iter(datasets[i])))
else:
assert any(next(iter(merged_dataset)) == fill_default(next(iter(dataset))) for dataset in datasets)
# Compute length it case it's random
if expected_length is None:
expected_length = 0
counts = np.array([len(list(d)) for d in datasets])
bool_strategy_func = np.all if stopping_strategy == "all_exhausted" else np.any
rng = np.random.default_rng(seed)
for i in RandomlyCyclingMultiSourcesExamplesIterable._iter_random_indices(rng, len(datasets), p=probas):
counts[i] -= 1
expected_length += 1
if bool_strategy_func(counts <= 0):
break
# Check length
assert len(list(merged_dataset)) == expected_length
def test_interleave_datasets_with_features(
dataset: IterableDataset,
):
features = Features(
{
"id": Value("int64"),
"label": ClassLabel(names=["negative", "positive"]),
}
)
ex_iterable = ExamplesIterable(generate_examples_fn, {"label": 0})
dataset_with_features = IterableDataset(ex_iterable, info=DatasetInfo(features=features))
merged_dataset = interleave_datasets([dataset, dataset_with_features])
assert merged_dataset.features == features
def test_interleave_datasets_with_oversampling():
# Test hardcoded results
d1 = IterableDataset(ExamplesIterable((lambda: (yield from [(i, {"a": i}) for i in [0, 1, 2]])), {}))
d2 = IterableDataset(ExamplesIterable((lambda: (yield from [(i, {"a": i}) for i in [10, 11, 12, 13]])), {}))
d3 = IterableDataset(ExamplesIterable((lambda: (yield from [(i, {"a": i}) for i in [20, 21, 22, 23, 24]])), {}))
expected_values = [0, 10, 20, 1, 11, 21, 2, 12, 22, 0, 13, 23, 1, 10, 24]
# Check oversampling strategy without probabilities
assert [x["a"] for x in interleave_datasets([d1, d2, d3], stopping_strategy="all_exhausted")] == expected_values
# Check oversampling strategy with probabilities
expected_values = [20, 0, 21, 10, 1, 22, 23, 24, 2, 0, 1, 20, 11, 21, 2, 0, 12, 1, 22, 13]
values = [
x["a"]
for x in interleave_datasets(
[d1, d2, d3], probabilities=[0.5, 0.2, 0.3], seed=42, stopping_strategy="all_exhausted"
)
]
assert values == expected_values
@require_torch
def test_with_format_torch(dataset_with_several_columns: IterableDataset):
import torch
dset = dataset_with_several_columns.with_format(type="torch")
example = next(iter(dset))
batch = next(iter(dset.iter(batch_size=3)))
assert len(example) == 3
assert isinstance(example["id"], torch.Tensor)
assert list(example["id"].shape) == []
assert example["id"].item() == 0
assert isinstance(batch["id"], torch.Tensor)
assert isinstance(example["filepath"], list)
assert isinstance(example["filepath"][0], str)
assert example["filepath"][0] == "data0.txt"
assert isinstance(batch["filepath"], list)
assert isinstance(example["metadata"], dict)
assert isinstance(example["metadata"]["sources"], list)
assert isinstance(example["metadata"]["sources"][0], str)
assert isinstance(batch["metadata"], list)
@require_tf
def test_with_format_tf(dataset_with_several_columns: IterableDataset):
import tensorflow as tf
dset = dataset_with_several_columns.with_format(type="tensorflow")
example = next(iter(dset))
batch = next(iter(dset.iter(batch_size=3)))
assert isinstance(example["id"], tf.Tensor)
assert list(example["id"].shape) == []
assert example["id"].numpy().item() == 0
assert isinstance(batch["id"], tf.Tensor)
assert isinstance(example["filepath"], tf.Tensor)
assert example["filepath"][0] == b"data0.txt"
assert isinstance(batch["filepath"], tf.Tensor)
assert isinstance(example["metadata"], dict)
assert isinstance(example["metadata"]["sources"], tf.Tensor)
assert isinstance(batch["metadata"], list)
def test_map_array_are_not_converted_back_to_lists(dataset: IterableDataset):
def func(example):
return {"array": np.array([1, 2, 3])}
dset_test = dataset.map(func)
example = next(iter(dset_test))
# not aligned with Dataset.map because we don't convert back to lists after map()
assert isinstance(example["array"], np.ndarray)
def test_formatted_map(dataset: IterableDataset):
dataset = dataset.with_format("np")
assert isinstance(next(dataset.iter(batch_size=3))["id"], np.ndarray)
dataset = dataset.with_format(None)
assert isinstance(next(dataset.iter(batch_size=3))["id"], list)
def add_one_numpy(example):
assert isinstance(example["id"], np.ndarray)
return {"id": example["id"] + 1}
dataset = dataset.with_format("np")
dataset = dataset.map(add_one_numpy, batched=True)
assert isinstance(next(dataset.iter(batch_size=3))["id"], np.ndarray)
dataset = dataset.with_format(None)
assert isinstance(next(dataset.iter(batch_size=3))["id"], list)
@pytest.mark.parametrize("n_shards1, nshards2, num_workers", [(2, 1, 1), (2, 2, 2), (1, 3, 1), (4, 3, 3)])
def test_interleave_dataset_with_sharding(n_shards1, nshards2, num_workers):
from torch.utils.data import DataLoader
ex_iterable1 = ExamplesIterable(generate_examples_fn, {"filepaths": [f"{i}-1.txt" for i in range(n_shards1)]})
dataset1 = IterableDataset(ex_iterable1).with_format("torch")
ex_iterable2 = ExamplesIterable(generate_examples_fn, {"filepaths": [f"{i}-2.txt" for i in range(nshards2)]})
dataset2 = IterableDataset(ex_iterable2).with_format("torch")
dataset_merged = interleave_datasets([dataset1, dataset2], stopping_strategy="first_exhausted")
assert dataset_merged.n_shards == min(n_shards1, nshards2)
dataloader = DataLoader(dataset_merged, batch_size=None, num_workers=num_workers)
result = list(dataloader)
expected_length = 2 * min(
len([example for _, example in ex_iterable1]), len([example for _, example in ex_iterable2])
)
# some samples may be missing because the stopping strategy is applied per process
assert expected_length - num_workers <= len(result) <= expected_length
assert len(result) == len({str(x) for x in result})
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_arrow_dataset.py | import contextlib
import copy
import itertools
import json
import os
import pickle
import re
import sys
import tempfile
from functools import partial
from pathlib import Path
from unittest import TestCase
from unittest.mock import MagicMock, patch
import numpy as np
import numpy.testing as npt
import pandas as pd
import pyarrow as pa
import pytest
from absl.testing import parameterized
from packaging import version
import datasets.arrow_dataset
from datasets import concatenate_datasets, interleave_datasets, load_from_disk
from datasets.arrow_dataset import Dataset, transmit_format, update_metadata_with_features
from datasets.dataset_dict import DatasetDict
from datasets.features import (
Array2D,
Array3D,
Audio,
ClassLabel,
Features,
Image,
Sequence,
Translation,
TranslationVariableLanguages,
Value,
)
from datasets.filesystems import extract_path_from_uri
from datasets.info import DatasetInfo
from datasets.iterable_dataset import IterableDataset
from datasets.splits import NamedSplit
from datasets.table import ConcatenationTable, InMemoryTable, MemoryMappedTable
from datasets.tasks import (
AutomaticSpeechRecognition,
LanguageModeling,
QuestionAnsweringExtractive,
Summarization,
TextClassification,
)
from datasets.utils.logging import INFO, get_logger
from datasets.utils.py_utils import temp_seed
from .utils import (
assert_arrow_memory_doesnt_increase,
assert_arrow_memory_increases,
require_dill_gt_0_3_2,
require_jax,
require_not_windows,
require_pil,
require_pyspark,
require_sqlalchemy,
require_tf,
require_torch,
require_transformers,
set_current_working_directory_to_temp_dir,
)
class PickableMagicMock(MagicMock):
def __reduce__(self):
return MagicMock, ()
class Unpicklable:
def __getstate__(self):
raise pickle.PicklingError()
def picklable_map_function(x):
return {"id": int(x["filename"].split("_")[-1])}
def picklable_map_function_with_indices(x, i):
return {"id": i}
def picklable_map_function_with_rank(x, r):
return {"rank": r}
def picklable_map_function_with_indices_and_rank(x, i, r):
return {"id": i, "rank": r}
def picklable_filter_function(x):
return int(x["filename"].split("_")[-1]) < 10
def assert_arrow_metadata_are_synced_with_dataset_features(dataset: Dataset):
assert dataset.data.schema.metadata is not None
assert b"huggingface" in dataset.data.schema.metadata
metadata = json.loads(dataset.data.schema.metadata[b"huggingface"].decode())
assert "info" in metadata
features = DatasetInfo.from_dict(metadata["info"]).features
assert features is not None
assert features == dataset.features
assert features == Features.from_arrow_schema(dataset.data.schema)
assert list(features) == dataset.data.column_names
assert list(features) == list(dataset.features)
IN_MEMORY_PARAMETERS = [
{"testcase_name": name, "in_memory": im} for im, name in [(True, "in_memory"), (False, "on_disk")]
]
@parameterized.named_parameters(IN_MEMORY_PARAMETERS)
class BaseDatasetTest(TestCase):
@pytest.fixture(autouse=True)
def inject_fixtures(self, caplog, set_sqlalchemy_silence_uber_warning):
self._caplog = caplog
def _create_dummy_dataset(
self, in_memory: bool, tmp_dir: str, multiple_columns=False, array_features=False, nested_features=False
) -> Dataset:
assert int(multiple_columns) + int(array_features) + int(nested_features) < 2
if multiple_columns:
data = {"col_1": [3, 2, 1, 0], "col_2": ["a", "b", "c", "d"], "col_3": [False, True, False, True]}
dset = Dataset.from_dict(data)
elif array_features:
data = {
"col_1": [[[True, False], [False, True]]] * 4, # 2D
"col_2": [[[["a", "b"], ["c", "d"]], [["e", "f"], ["g", "h"]]]] * 4, # 3D array
"col_3": [[3, 2, 1, 0]] * 4, # Sequence
}
features = Features(
{
"col_1": Array2D(shape=(2, 2), dtype="bool"),
"col_2": Array3D(shape=(2, 2, 2), dtype="string"),
"col_3": Sequence(feature=Value("int64")),
}
)
dset = Dataset.from_dict(data, features=features)
elif nested_features:
data = {"nested": [{"a": i, "x": i * 10, "c": i * 100} for i in range(1, 11)]}
features = Features({"nested": {"a": Value("int64"), "x": Value("int64"), "c": Value("int64")}})
dset = Dataset.from_dict(data, features=features)
else:
dset = Dataset.from_dict({"filename": ["my_name-train" + "_" + str(x) for x in np.arange(30).tolist()]})
if not in_memory:
dset = self._to(in_memory, tmp_dir, dset)
return dset
def _to(self, in_memory, tmp_dir, *datasets):
if in_memory:
datasets = [dataset.map(keep_in_memory=True) for dataset in datasets]
else:
start = 0
while os.path.isfile(os.path.join(tmp_dir, f"dataset{start}.arrow")):
start += 1
datasets = [
dataset.map(cache_file_name=os.path.join(tmp_dir, f"dataset{start + i}.arrow"))
for i, dataset in enumerate(datasets)
]
return datasets if len(datasets) > 1 else datasets[0]
def test_dummy_dataset(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertEqual(dset[0]["filename"], "my_name-train_0")
self.assertEqual(dset["filename"][0], "my_name-train_0")
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
self.assertDictEqual(
dset.features,
Features({"col_1": Value("int64"), "col_2": Value("string"), "col_3": Value("bool")}),
)
self.assertEqual(dset[0]["col_1"], 3)
self.assertEqual(dset["col_1"][0], 3)
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, array_features=True) as dset:
self.assertDictEqual(
dset.features,
Features(
{
"col_1": Array2D(shape=(2, 2), dtype="bool"),
"col_2": Array3D(shape=(2, 2, 2), dtype="string"),
"col_3": Sequence(feature=Value("int64")),
}
),
)
self.assertEqual(dset[0]["col_2"], [[["a", "b"], ["c", "d"]], [["e", "f"], ["g", "h"]]])
self.assertEqual(dset["col_2"][0], [[["a", "b"], ["c", "d"]], [["e", "f"], ["g", "h"]]])
def test_dataset_getitem(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
self.assertEqual(dset[0]["filename"], "my_name-train_0")
self.assertEqual(dset["filename"][0], "my_name-train_0")
self.assertEqual(dset[-1]["filename"], "my_name-train_29")
self.assertEqual(dset["filename"][-1], "my_name-train_29")
self.assertListEqual(dset[:2]["filename"], ["my_name-train_0", "my_name-train_1"])
self.assertListEqual(dset["filename"][:2], ["my_name-train_0", "my_name-train_1"])
self.assertEqual(dset[:-1]["filename"][-1], "my_name-train_28")
self.assertEqual(dset["filename"][:-1][-1], "my_name-train_28")
self.assertListEqual(dset[[0, -1]]["filename"], ["my_name-train_0", "my_name-train_29"])
self.assertListEqual(dset[range(0, -2, -1)]["filename"], ["my_name-train_0", "my_name-train_29"])
self.assertListEqual(dset[np.array([0, -1])]["filename"], ["my_name-train_0", "my_name-train_29"])
self.assertListEqual(dset[pd.Series([0, -1])]["filename"], ["my_name-train_0", "my_name-train_29"])
with dset.select(range(2)) as dset_subset:
self.assertListEqual(dset_subset[-1:]["filename"], ["my_name-train_1"])
self.assertListEqual(dset_subset["filename"][-1:], ["my_name-train_1"])
def test_dummy_dataset_deepcopy(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir).select(range(10)) as dset:
with assert_arrow_memory_doesnt_increase():
dset2 = copy.deepcopy(dset)
# don't copy the underlying arrow data using memory
self.assertEqual(len(dset2), 10)
self.assertDictEqual(dset2.features, Features({"filename": Value("string")}))
self.assertEqual(dset2[0]["filename"], "my_name-train_0")
self.assertEqual(dset2["filename"][0], "my_name-train_0")
del dset2
def test_dummy_dataset_pickle(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
tmp_file = os.path.join(tmp_dir, "dset.pt")
with self._create_dummy_dataset(in_memory, tmp_dir).select(range(0, 10, 2)) as dset:
with open(tmp_file, "wb") as f:
pickle.dump(dset, f)
with open(tmp_file, "rb") as f:
with pickle.load(f) as dset:
self.assertEqual(len(dset), 5)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertEqual(dset[0]["filename"], "my_name-train_0")
self.assertEqual(dset["filename"][0], "my_name-train_0")
with self._create_dummy_dataset(in_memory, tmp_dir).select(
range(0, 10, 2), indices_cache_file_name=os.path.join(tmp_dir, "ind.arrow")
) as dset:
if not in_memory:
dset._data.table = Unpicklable()
dset._indices.table = Unpicklable()
with open(tmp_file, "wb") as f:
pickle.dump(dset, f)
with open(tmp_file, "rb") as f:
with pickle.load(f) as dset:
self.assertEqual(len(dset), 5)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertEqual(dset[0]["filename"], "my_name-train_0")
self.assertEqual(dset["filename"][0], "my_name-train_0")
def test_dummy_dataset_serialize(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with set_current_working_directory_to_temp_dir():
with self._create_dummy_dataset(in_memory, tmp_dir).select(range(10)) as dset:
dataset_path = "my_dataset" # rel path
dset.save_to_disk(dataset_path)
with Dataset.load_from_disk(dataset_path) as dset:
self.assertEqual(len(dset), 10)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertEqual(dset[0]["filename"], "my_name-train_0")
self.assertEqual(dset["filename"][0], "my_name-train_0")
expected = dset.to_dict()
with self._create_dummy_dataset(in_memory, tmp_dir).select(range(10)) as dset:
dataset_path = os.path.join(tmp_dir, "my_dataset") # abs path
dset.save_to_disk(dataset_path)
with Dataset.load_from_disk(dataset_path) as dset:
self.assertEqual(len(dset), 10)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertEqual(dset[0]["filename"], "my_name-train_0")
self.assertEqual(dset["filename"][0], "my_name-train_0")
with self._create_dummy_dataset(in_memory, tmp_dir).select(
range(10), indices_cache_file_name=os.path.join(tmp_dir, "ind.arrow")
) as dset:
with assert_arrow_memory_doesnt_increase():
dset.save_to_disk(dataset_path)
with Dataset.load_from_disk(dataset_path) as dset:
self.assertEqual(len(dset), 10)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertEqual(dset[0]["filename"], "my_name-train_0")
self.assertEqual(dset["filename"][0], "my_name-train_0")
with self._create_dummy_dataset(in_memory, tmp_dir, nested_features=True) as dset:
with assert_arrow_memory_doesnt_increase():
dset.save_to_disk(dataset_path)
with Dataset.load_from_disk(dataset_path) as dset:
self.assertEqual(len(dset), 10)
self.assertDictEqual(
dset.features,
Features({"nested": {"a": Value("int64"), "x": Value("int64"), "c": Value("int64")}}),
)
self.assertDictEqual(dset[0]["nested"], {"a": 1, "c": 100, "x": 10})
self.assertDictEqual(dset["nested"][0], {"a": 1, "c": 100, "x": 10})
with self._create_dummy_dataset(in_memory, tmp_dir).select(range(10)) as dset:
with assert_arrow_memory_doesnt_increase():
dset.save_to_disk(dataset_path, num_shards=4)
with Dataset.load_from_disk(dataset_path) as dset:
self.assertEqual(len(dset), 10)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset.to_dict(), expected)
self.assertEqual(len(dset.cache_files), 4)
with self._create_dummy_dataset(in_memory, tmp_dir).select(range(10)) as dset:
with assert_arrow_memory_doesnt_increase():
dset.save_to_disk(dataset_path, num_proc=2)
with Dataset.load_from_disk(dataset_path) as dset:
self.assertEqual(len(dset), 10)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset.to_dict(), expected)
self.assertEqual(len(dset.cache_files), 2)
with self._create_dummy_dataset(in_memory, tmp_dir).select(range(10)) as dset:
with assert_arrow_memory_doesnt_increase():
dset.save_to_disk(dataset_path, num_shards=7, num_proc=2)
with Dataset.load_from_disk(dataset_path) as dset:
self.assertEqual(len(dset), 10)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset.to_dict(), expected)
self.assertEqual(len(dset.cache_files), 7)
with self._create_dummy_dataset(in_memory, tmp_dir).select(range(10)) as dset:
with assert_arrow_memory_doesnt_increase():
max_shard_size = dset._estimate_nbytes() // 2 + 1
dset.save_to_disk(dataset_path, max_shard_size=max_shard_size)
with Dataset.load_from_disk(dataset_path) as dset:
self.assertEqual(len(dset), 10)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset.to_dict(), expected)
self.assertEqual(len(dset.cache_files), 2)
def test_dummy_dataset_load_from_disk(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir).select(range(10)) as dset:
dataset_path = os.path.join(tmp_dir, "my_dataset")
dset.save_to_disk(dataset_path)
with load_from_disk(dataset_path) as dset:
self.assertEqual(len(dset), 10)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertEqual(dset[0]["filename"], "my_name-train_0")
self.assertEqual(dset["filename"][0], "my_name-train_0")
def test_restore_saved_format(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
dset.set_format(type="numpy", columns=["col_1"], output_all_columns=True)
dataset_path = os.path.join(tmp_dir, "my_dataset")
dset.save_to_disk(dataset_path)
with load_from_disk(dataset_path) as loaded_dset:
self.assertEqual(dset.format, loaded_dset.format)
def test_set_format_numpy_multiple_columns(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
fingerprint = dset._fingerprint
dset.set_format(type="numpy", columns=["col_1"])
self.assertEqual(len(dset[0]), 1)
self.assertIsInstance(dset[0]["col_1"], np.int64)
self.assertEqual(dset[0]["col_1"].item(), 3)
self.assertIsInstance(dset["col_1"], np.ndarray)
self.assertListEqual(list(dset["col_1"].shape), [4])
np.testing.assert_array_equal(dset["col_1"], np.array([3, 2, 1, 0]))
self.assertNotEqual(dset._fingerprint, fingerprint)
dset.reset_format()
with dset.formatted_as(type="numpy", columns=["col_1"]):
self.assertEqual(len(dset[0]), 1)
self.assertIsInstance(dset[0]["col_1"], np.int64)
self.assertEqual(dset[0]["col_1"].item(), 3)
self.assertIsInstance(dset["col_1"], np.ndarray)
self.assertListEqual(list(dset["col_1"].shape), [4])
np.testing.assert_array_equal(dset["col_1"], np.array([3, 2, 1, 0]))
self.assertEqual(dset.format["type"], None)
self.assertEqual(dset.format["format_kwargs"], {})
self.assertEqual(dset.format["columns"], dset.column_names)
self.assertEqual(dset.format["output_all_columns"], False)
dset.set_format(type="numpy", columns=["col_1"], output_all_columns=True)
self.assertEqual(len(dset[0]), 3)
self.assertIsInstance(dset[0]["col_2"], str)
self.assertEqual(dset[0]["col_2"], "a")
dset.set_format(type="numpy", columns=["col_1", "col_2"])
self.assertEqual(len(dset[0]), 2)
self.assertIsInstance(dset[0]["col_2"], np.str_)
self.assertEqual(dset[0]["col_2"].item(), "a")
@require_torch
def test_set_format_torch(self, in_memory):
import torch
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
dset.set_format(type="torch", columns=["col_1"])
self.assertEqual(len(dset[0]), 1)
self.assertIsInstance(dset[0]["col_1"], torch.Tensor)
self.assertIsInstance(dset["col_1"], torch.Tensor)
self.assertListEqual(list(dset[0]["col_1"].shape), [])
self.assertEqual(dset[0]["col_1"].item(), 3)
dset.set_format(type="torch", columns=["col_1"], output_all_columns=True)
self.assertEqual(len(dset[0]), 3)
self.assertIsInstance(dset[0]["col_2"], str)
self.assertEqual(dset[0]["col_2"], "a")
dset.set_format(type="torch")
self.assertEqual(len(dset[0]), 3)
self.assertIsInstance(dset[0]["col_1"], torch.Tensor)
self.assertIsInstance(dset["col_1"], torch.Tensor)
self.assertListEqual(list(dset[0]["col_1"].shape), [])
self.assertEqual(dset[0]["col_1"].item(), 3)
self.assertIsInstance(dset[0]["col_2"], str)
self.assertEqual(dset[0]["col_2"], "a")
self.assertIsInstance(dset[0]["col_3"], torch.Tensor)
self.assertIsInstance(dset["col_3"], torch.Tensor)
self.assertListEqual(list(dset[0]["col_3"].shape), [])
@require_tf
def test_set_format_tf(self, in_memory):
import tensorflow as tf
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
dset.set_format(type="tensorflow", columns=["col_1"])
self.assertEqual(len(dset[0]), 1)
self.assertIsInstance(dset[0]["col_1"], tf.Tensor)
self.assertListEqual(list(dset[0]["col_1"].shape), [])
self.assertEqual(dset[0]["col_1"].numpy().item(), 3)
dset.set_format(type="tensorflow", columns=["col_1"], output_all_columns=True)
self.assertEqual(len(dset[0]), 3)
self.assertIsInstance(dset[0]["col_2"], str)
self.assertEqual(dset[0]["col_2"], "a")
dset.set_format(type="tensorflow", columns=["col_1", "col_2"])
self.assertEqual(len(dset[0]), 2)
self.assertEqual(dset[0]["col_2"].numpy().decode("utf-8"), "a")
def test_set_format_pandas(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
dset.set_format(type="pandas", columns=["col_1"])
self.assertEqual(len(dset[0].columns), 1)
self.assertIsInstance(dset[0], pd.DataFrame)
self.assertListEqual(list(dset[0].shape), [1, 1])
self.assertEqual(dset[0]["col_1"].item(), 3)
dset.set_format(type="pandas", columns=["col_1", "col_2"])
self.assertEqual(len(dset[0].columns), 2)
self.assertEqual(dset[0]["col_2"].item(), "a")
def test_set_transform(self, in_memory):
def transform(batch):
return {k: [str(i).upper() for i in v] for k, v in batch.items()}
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
dset.set_transform(transform=transform, columns=["col_1"])
self.assertEqual(dset.format["type"], "custom")
self.assertEqual(len(dset[0].keys()), 1)
self.assertEqual(dset[0]["col_1"], "3")
self.assertEqual(dset[:2]["col_1"], ["3", "2"])
self.assertEqual(dset["col_1"][:2], ["3", "2"])
prev_format = dset.format
dset.set_format(**dset.format)
self.assertEqual(prev_format, dset.format)
dset.set_transform(transform=transform, columns=["col_1", "col_2"])
self.assertEqual(len(dset[0].keys()), 2)
self.assertEqual(dset[0]["col_2"], "A")
def test_transmit_format(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
transform = datasets.arrow_dataset.transmit_format(lambda x: x)
# make sure identity transform doesn't apply unnecessary format
self.assertEqual(dset._fingerprint, transform(dset)._fingerprint)
dset.set_format(**dset.format)
self.assertEqual(dset._fingerprint, transform(dset)._fingerprint)
# check lists comparisons
dset.set_format(columns=["col_1"])
self.assertEqual(dset._fingerprint, transform(dset)._fingerprint)
dset.set_format(columns=["col_1", "col_2"])
self.assertEqual(dset._fingerprint, transform(dset)._fingerprint)
dset.set_format("numpy", columns=["col_1", "col_2"])
self.assertEqual(dset._fingerprint, transform(dset)._fingerprint)
def test_cast(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
features = dset.features
features["col_1"] = Value("float64")
features = Features({k: features[k] for k in list(features)[::-1]})
fingerprint = dset._fingerprint
# TODO: with assert_arrow_memory_increases() if in_memory else assert_arrow_memory_doesnt_increase():
with dset.cast(features) as casted_dset:
self.assertEqual(casted_dset.num_columns, 3)
self.assertEqual(casted_dset.features["col_1"], Value("float64"))
self.assertIsInstance(casted_dset[0]["col_1"], float)
self.assertNotEqual(casted_dset._fingerprint, fingerprint)
self.assertNotEqual(casted_dset, dset)
assert_arrow_metadata_are_synced_with_dataset_features(casted_dset)
def test_class_encode_column(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
with self.assertRaises(ValueError):
dset.class_encode_column(column="does not exist")
with dset.class_encode_column("col_1") as casted_dset:
self.assertIsInstance(casted_dset.features["col_1"], ClassLabel)
self.assertListEqual(casted_dset.features["col_1"].names, ["0", "1", "2", "3"])
self.assertListEqual(casted_dset["col_1"], [3, 2, 1, 0])
self.assertNotEqual(casted_dset._fingerprint, dset._fingerprint)
self.assertNotEqual(casted_dset, dset)
assert_arrow_metadata_are_synced_with_dataset_features(casted_dset)
with dset.class_encode_column("col_2") as casted_dset:
self.assertIsInstance(casted_dset.features["col_2"], ClassLabel)
self.assertListEqual(casted_dset.features["col_2"].names, ["a", "b", "c", "d"])
self.assertListEqual(casted_dset["col_2"], [0, 1, 2, 3])
self.assertNotEqual(casted_dset._fingerprint, dset._fingerprint)
self.assertNotEqual(casted_dset, dset)
assert_arrow_metadata_are_synced_with_dataset_features(casted_dset)
with dset.class_encode_column("col_3") as casted_dset:
self.assertIsInstance(casted_dset.features["col_3"], ClassLabel)
self.assertListEqual(casted_dset.features["col_3"].names, ["False", "True"])
self.assertListEqual(casted_dset["col_3"], [0, 1, 0, 1])
self.assertNotEqual(casted_dset._fingerprint, dset._fingerprint)
self.assertNotEqual(casted_dset, dset)
assert_arrow_metadata_are_synced_with_dataset_features(casted_dset)
# Test raises if feature is an array / sequence
with self._create_dummy_dataset(in_memory, tmp_dir, array_features=True) as dset:
for column in dset.column_names:
with self.assertRaises(ValueError):
dset.class_encode_column(column)
def test_remove_columns(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
fingerprint = dset._fingerprint
with dset.remove_columns(column_names="col_1") as new_dset:
self.assertEqual(new_dset.num_columns, 2)
self.assertListEqual(list(new_dset.column_names), ["col_2", "col_3"])
self.assertNotEqual(new_dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(new_dset)
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
with dset.remove_columns(column_names=["col_1", "col_2", "col_3"]) as new_dset:
self.assertEqual(new_dset.num_columns, 0)
self.assertNotEqual(new_dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(new_dset)
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
dset._format_columns = ["col_1", "col_2", "col_3"]
with dset.remove_columns(column_names=["col_1"]) as new_dset:
self.assertListEqual(new_dset._format_columns, ["col_2", "col_3"])
self.assertEqual(new_dset.num_columns, 2)
self.assertListEqual(list(new_dset.column_names), ["col_2", "col_3"])
self.assertNotEqual(new_dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(new_dset)
def test_rename_column(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
fingerprint = dset._fingerprint
with dset.rename_column(original_column_name="col_1", new_column_name="new_name") as new_dset:
self.assertEqual(new_dset.num_columns, 3)
self.assertListEqual(list(new_dset.column_names), ["new_name", "col_2", "col_3"])
self.assertListEqual(list(dset.column_names), ["col_1", "col_2", "col_3"])
self.assertNotEqual(new_dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(new_dset)
def test_rename_columns(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
fingerprint = dset._fingerprint
with dset.rename_columns({"col_1": "new_name"}) as new_dset:
self.assertEqual(new_dset.num_columns, 3)
self.assertListEqual(list(new_dset.column_names), ["new_name", "col_2", "col_3"])
self.assertListEqual(list(dset.column_names), ["col_1", "col_2", "col_3"])
self.assertNotEqual(new_dset._fingerprint, fingerprint)
with dset.rename_columns({"col_1": "new_name", "col_2": "new_name2"}) as new_dset:
self.assertEqual(new_dset.num_columns, 3)
self.assertListEqual(list(new_dset.column_names), ["new_name", "new_name2", "col_3"])
self.assertListEqual(list(dset.column_names), ["col_1", "col_2", "col_3"])
self.assertNotEqual(new_dset._fingerprint, fingerprint)
# Original column not in dataset
with self.assertRaises(ValueError):
dset.rename_columns({"not_there": "new_name"})
# Empty new name
with self.assertRaises(ValueError):
dset.rename_columns({"col_1": ""})
# Duplicates
with self.assertRaises(ValueError):
dset.rename_columns({"col_1": "new_name", "col_2": "new_name"})
def test_select_columns(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
fingerprint = dset._fingerprint
with dset.select_columns(column_names=[]) as new_dset:
self.assertEqual(new_dset.num_columns, 0)
self.assertListEqual(list(new_dset.column_names), [])
self.assertNotEqual(new_dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(new_dset)
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
fingerprint = dset._fingerprint
with dset.select_columns(column_names="col_1") as new_dset:
self.assertEqual(new_dset.num_columns, 1)
self.assertListEqual(list(new_dset.column_names), ["col_1"])
self.assertNotEqual(new_dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(new_dset)
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
with dset.select_columns(column_names=["col_1", "col_2", "col_3"]) as new_dset:
self.assertEqual(new_dset.num_columns, 3)
self.assertListEqual(list(new_dset.column_names), ["col_1", "col_2", "col_3"])
self.assertNotEqual(new_dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(new_dset)
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
with dset.select_columns(column_names=["col_3", "col_2", "col_1"]) as new_dset:
self.assertEqual(new_dset.num_columns, 3)
self.assertListEqual(list(new_dset.column_names), ["col_3", "col_2", "col_1"])
self.assertNotEqual(new_dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(new_dset)
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
dset._format_columns = ["col_1", "col_2", "col_3"]
with dset.select_columns(column_names=["col_1"]) as new_dset:
self.assertListEqual(new_dset._format_columns, ["col_1"])
self.assertEqual(new_dset.num_columns, 1)
self.assertListEqual(list(new_dset.column_names), ["col_1"])
self.assertNotEqual(new_dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(new_dset)
def test_concatenate(self, in_memory):
data1, data2, data3 = {"id": [0, 1, 2]}, {"id": [3, 4, 5]}, {"id": [6, 7]}
info1 = DatasetInfo(description="Dataset1")
info2 = DatasetInfo(description="Dataset2")
with tempfile.TemporaryDirectory() as tmp_dir:
dset1, dset2, dset3 = (
Dataset.from_dict(data1, info=info1),
Dataset.from_dict(data2, info=info2),
Dataset.from_dict(data3),
)
dset1, dset2, dset3 = self._to(in_memory, tmp_dir, dset1, dset2, dset3)
with concatenate_datasets([dset1, dset2, dset3]) as dset_concat:
self.assertTupleEqual((len(dset1), len(dset2), len(dset3)), (3, 3, 2))
self.assertEqual(len(dset_concat), len(dset1) + len(dset2) + len(dset3))
self.assertListEqual(dset_concat["id"], [0, 1, 2, 3, 4, 5, 6, 7])
self.assertEqual(len(dset_concat.cache_files), 0 if in_memory else 3)
self.assertEqual(dset_concat.info.description, "Dataset1\n\nDataset2")
del dset1, dset2, dset3
def test_concatenate_formatted(self, in_memory):
data1, data2, data3 = {"id": [0, 1, 2]}, {"id": [3, 4, 5]}, {"id": [6, 7]}
info1 = DatasetInfo(description="Dataset1")
info2 = DatasetInfo(description="Dataset2")
with tempfile.TemporaryDirectory() as tmp_dir:
dset1, dset2, dset3 = (
Dataset.from_dict(data1, info=info1),
Dataset.from_dict(data2, info=info2),
Dataset.from_dict(data3),
)
dset1, dset2, dset3 = self._to(in_memory, tmp_dir, dset1, dset2, dset3)
dset1.set_format("numpy")
with concatenate_datasets([dset1, dset2, dset3]) as dset_concat:
self.assertEqual(dset_concat.format["type"], None)
dset2.set_format("numpy")
dset3.set_format("numpy")
with concatenate_datasets([dset1, dset2, dset3]) as dset_concat:
self.assertEqual(dset_concat.format["type"], "numpy")
del dset1, dset2, dset3
def test_concatenate_with_indices(self, in_memory):
data1, data2, data3 = {"id": [0, 1, 2] * 2}, {"id": [3, 4, 5] * 2}, {"id": [6, 7, 8]}
info1 = DatasetInfo(description="Dataset1")
info2 = DatasetInfo(description="Dataset2")
with tempfile.TemporaryDirectory() as tmp_dir:
dset1, dset2, dset3 = (
Dataset.from_dict(data1, info=info1),
Dataset.from_dict(data2, info=info2),
Dataset.from_dict(data3),
)
dset1, dset2, dset3 = self._to(in_memory, tmp_dir, dset1, dset2, dset3)
dset1, dset2, dset3 = dset1.select([2, 1, 0]), dset2.select([2, 1, 0]), dset3
with concatenate_datasets([dset3, dset2, dset1]) as dset_concat:
self.assertTupleEqual((len(dset1), len(dset2), len(dset3)), (3, 3, 3))
self.assertEqual(len(dset_concat), len(dset1) + len(dset2) + len(dset3))
self.assertListEqual(dset_concat["id"], [6, 7, 8, 5, 4, 3, 2, 1, 0])
# in_memory = False:
# 3 cache files for the dset_concat._data table
# no cache file for the indices because it's in memory
# in_memory = True:
# no cache files since both dset_concat._data and dset_concat._indices are in memory
self.assertEqual(len(dset_concat.cache_files), 0 if in_memory else 3)
self.assertEqual(dset_concat.info.description, "Dataset2\n\nDataset1")
dset1 = dset1.rename_columns({"id": "id1"})
dset2 = dset2.rename_columns({"id": "id2"})
dset3 = dset3.rename_columns({"id": "id3"})
with concatenate_datasets([dset1, dset2, dset3], axis=1) as dset_concat:
self.assertTupleEqual((len(dset1), len(dset2), len(dset3)), (3, 3, 3))
self.assertEqual(len(dset_concat), len(dset1))
self.assertListEqual(dset_concat["id1"], [2, 1, 0])
self.assertListEqual(dset_concat["id2"], [5, 4, 3])
self.assertListEqual(dset_concat["id3"], [6, 7, 8])
# in_memory = False:
# 3 cache files for the dset_concat._data table
# no cache file for the indices because it's None
# in_memory = True:
# no cache files since dset_concat._data is in memory and dset_concat._indices is None
self.assertEqual(len(dset_concat.cache_files), 0 if in_memory else 3)
self.assertIsNone(dset_concat._indices)
self.assertEqual(dset_concat.info.description, "Dataset1\n\nDataset2")
with concatenate_datasets([dset1], axis=1) as dset_concat:
self.assertEqual(len(dset_concat), len(dset1))
self.assertListEqual(dset_concat["id1"], [2, 1, 0])
# in_memory = False:
# 1 cache file for the dset_concat._data table
# no cache file for the indices because it's in memory
# in_memory = True:
# no cache files since both dset_concat._data and dset_concat._indices are in memory
self.assertEqual(len(dset_concat.cache_files), 0 if in_memory else 1)
self.assertTrue(dset_concat._indices == dset1._indices)
self.assertEqual(dset_concat.info.description, "Dataset1")
del dset1, dset2, dset3
def test_concatenate_with_indices_from_disk(self, in_memory):
data1, data2, data3 = {"id": [0, 1, 2] * 2}, {"id": [3, 4, 5] * 2}, {"id": [6, 7]}
info1 = DatasetInfo(description="Dataset1")
info2 = DatasetInfo(description="Dataset2")
with tempfile.TemporaryDirectory() as tmp_dir:
dset1, dset2, dset3 = (
Dataset.from_dict(data1, info=info1),
Dataset.from_dict(data2, info=info2),
Dataset.from_dict(data3),
)
dset1, dset2, dset3 = self._to(in_memory, tmp_dir, dset1, dset2, dset3)
dset1, dset2, dset3 = (
dset1.select([2, 1, 0], indices_cache_file_name=os.path.join(tmp_dir, "i1.arrow")),
dset2.select([2, 1, 0], indices_cache_file_name=os.path.join(tmp_dir, "i2.arrow")),
dset3.select([1, 0], indices_cache_file_name=os.path.join(tmp_dir, "i3.arrow")),
)
with concatenate_datasets([dset3, dset2, dset1]) as dset_concat:
self.assertTupleEqual((len(dset1), len(dset2), len(dset3)), (3, 3, 2))
self.assertEqual(len(dset_concat), len(dset1) + len(dset2) + len(dset3))
self.assertListEqual(dset_concat["id"], [7, 6, 5, 4, 3, 2, 1, 0])
# in_memory = False:
# 3 cache files for the dset_concat._data table, and 1 for the dset_concat._indices_table
# There is only 1 for the indices tables (i1.arrow)
# Indeed, the others are brought to memory since an offset is applied to them.
# in_memory = True:
# 1 cache file for i1.arrow since both dset_concat._data and dset_concat._indices are in memory
self.assertEqual(len(dset_concat.cache_files), 1 if in_memory else 3 + 1)
self.assertEqual(dset_concat.info.description, "Dataset2\n\nDataset1")
del dset1, dset2, dset3
def test_concatenate_pickle(self, in_memory):
data1, data2, data3 = {"id": [0, 1, 2] * 2}, {"id": [3, 4, 5] * 2}, {"id": [6, 7], "foo": ["bar", "bar"]}
info1 = DatasetInfo(description="Dataset1")
info2 = DatasetInfo(description="Dataset2")
with tempfile.TemporaryDirectory() as tmp_dir:
dset1, dset2, dset3 = (
Dataset.from_dict(data1, info=info1),
Dataset.from_dict(data2, info=info2),
Dataset.from_dict(data3),
)
# mix from in-memory and on-disk datasets
dset1, dset2 = self._to(in_memory, tmp_dir, dset1, dset2)
dset3 = self._to(not in_memory, tmp_dir, dset3)
dset1, dset2, dset3 = (
dset1.select(
[2, 1, 0],
keep_in_memory=in_memory,
indices_cache_file_name=os.path.join(tmp_dir, "i1.arrow") if not in_memory else None,
),
dset2.select(
[2, 1, 0],
keep_in_memory=in_memory,
indices_cache_file_name=os.path.join(tmp_dir, "i2.arrow") if not in_memory else None,
),
dset3.select(
[1, 0],
keep_in_memory=in_memory,
indices_cache_file_name=os.path.join(tmp_dir, "i3.arrow") if not in_memory else None,
),
)
dset3 = dset3.rename_column("foo", "new_foo")
dset3 = dset3.remove_columns("new_foo")
if in_memory:
dset3._data.table = Unpicklable()
else:
dset1._data.table, dset2._data.table = Unpicklable(), Unpicklable()
dset1, dset2, dset3 = (pickle.loads(pickle.dumps(d)) for d in (dset1, dset2, dset3))
with concatenate_datasets([dset3, dset2, dset1]) as dset_concat:
if not in_memory:
dset_concat._data.table = Unpicklable()
with pickle.loads(pickle.dumps(dset_concat)) as dset_concat:
self.assertTupleEqual((len(dset1), len(dset2), len(dset3)), (3, 3, 2))
self.assertEqual(len(dset_concat), len(dset1) + len(dset2) + len(dset3))
self.assertListEqual(dset_concat["id"], [7, 6, 5, 4, 3, 2, 1, 0])
# in_memory = True: 1 cache file for dset3
# in_memory = False: 2 caches files for dset1 and dset2, and 1 cache file for i1.arrow
self.assertEqual(len(dset_concat.cache_files), 1 if in_memory else 2 + 1)
self.assertEqual(dset_concat.info.description, "Dataset2\n\nDataset1")
del dset1, dset2, dset3
def test_flatten(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict(
{"a": [{"b": {"c": ["text"]}}] * 10, "foo": [1] * 10},
features=Features({"a": {"b": Sequence({"c": Value("string")})}, "foo": Value("int64")}),
) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
fingerprint = dset._fingerprint
with dset.flatten() as dset:
self.assertListEqual(sorted(dset.column_names), ["a.b.c", "foo"])
self.assertListEqual(sorted(dset.features.keys()), ["a.b.c", "foo"])
self.assertDictEqual(
dset.features, Features({"a.b.c": Sequence(Value("string")), "foo": Value("int64")})
)
self.assertNotEqual(dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset)
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict(
{"a": [{"en": "Thank you", "fr": "Merci"}] * 10, "foo": [1] * 10},
features=Features({"a": Translation(languages=["en", "fr"]), "foo": Value("int64")}),
) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
fingerprint = dset._fingerprint
with dset.flatten() as dset:
self.assertListEqual(sorted(dset.column_names), ["a.en", "a.fr", "foo"])
self.assertListEqual(sorted(dset.features.keys()), ["a.en", "a.fr", "foo"])
self.assertDictEqual(
dset.features,
Features({"a.en": Value("string"), "a.fr": Value("string"), "foo": Value("int64")}),
)
self.assertNotEqual(dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset)
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict(
{"a": [{"en": "the cat", "fr": ["le chat", "la chatte"], "de": "die katze"}] * 10, "foo": [1] * 10},
features=Features(
{"a": TranslationVariableLanguages(languages=["en", "fr", "de"]), "foo": Value("int64")}
),
) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
fingerprint = dset._fingerprint
with dset.flatten() as dset:
self.assertListEqual(sorted(dset.column_names), ["a.language", "a.translation", "foo"])
self.assertListEqual(sorted(dset.features.keys()), ["a.language", "a.translation", "foo"])
self.assertDictEqual(
dset.features,
Features(
{
"a.language": Sequence(Value("string")),
"a.translation": Sequence(Value("string")),
"foo": Value("int64"),
}
),
)
self.assertNotEqual(dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset)
@require_pil
def test_flatten_complex_image(self, in_memory):
# decoding turned on
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict(
{"a": [np.arange(4 * 4 * 3, dtype=np.uint8).reshape(4, 4, 3)] * 10, "foo": [1] * 10},
features=Features({"a": Image(), "foo": Value("int64")}),
) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
fingerprint = dset._fingerprint
with dset.flatten() as dset:
self.assertListEqual(sorted(dset.column_names), ["a", "foo"])
self.assertListEqual(sorted(dset.features.keys()), ["a", "foo"])
self.assertDictEqual(dset.features, Features({"a": Image(), "foo": Value("int64")}))
self.assertNotEqual(dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset)
# decoding turned on + nesting
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict(
{"a": [{"b": np.arange(4 * 4 * 3, dtype=np.uint8).reshape(4, 4, 3)}] * 10, "foo": [1] * 10},
features=Features({"a": {"b": Image()}, "foo": Value("int64")}),
) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
fingerprint = dset._fingerprint
with dset.flatten() as dset:
self.assertListEqual(sorted(dset.column_names), ["a.b", "foo"])
self.assertListEqual(sorted(dset.features.keys()), ["a.b", "foo"])
self.assertDictEqual(dset.features, Features({"a.b": Image(), "foo": Value("int64")}))
self.assertNotEqual(dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset)
# decoding turned off
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict(
{"a": [np.arange(4 * 4 * 3, dtype=np.uint8).reshape(4, 4, 3)] * 10, "foo": [1] * 10},
features=Features({"a": Image(decode=False), "foo": Value("int64")}),
) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
fingerprint = dset._fingerprint
with dset.flatten() as dset:
self.assertListEqual(sorted(dset.column_names), ["a.bytes", "a.path", "foo"])
self.assertListEqual(sorted(dset.features.keys()), ["a.bytes", "a.path", "foo"])
self.assertDictEqual(
dset.features,
Features({"a.bytes": Value("binary"), "a.path": Value("string"), "foo": Value("int64")}),
)
self.assertNotEqual(dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset)
# decoding turned off + nesting
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict(
{"a": [{"b": np.arange(4 * 4 * 3, dtype=np.uint8).reshape(4, 4, 3)}] * 10, "foo": [1] * 10},
features=Features({"a": {"b": Image(decode=False)}, "foo": Value("int64")}),
) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
fingerprint = dset._fingerprint
with dset.flatten() as dset:
self.assertListEqual(sorted(dset.column_names), ["a.b.bytes", "a.b.path", "foo"])
self.assertListEqual(sorted(dset.features.keys()), ["a.b.bytes", "a.b.path", "foo"])
self.assertDictEqual(
dset.features,
Features(
{"a.b.bytes": Value("binary"), "a.b.path": Value("string"), "foo": Value("int64")}
),
)
self.assertNotEqual(dset._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset)
def test_map(self, in_memory):
# standard
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
fingerprint = dset._fingerprint
with dset.map(
lambda x: {"name": x["filename"][:-2], "id": int(x["filename"].split("_")[-1])}
) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(
dset_test.features,
Features({"filename": Value("string"), "name": Value("string"), "id": Value("int64")}),
)
self.assertListEqual(dset_test["id"], list(range(30)))
self.assertNotEqual(dset_test._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset_test)
# no transform
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
fingerprint = dset._fingerprint
with dset.map(lambda x: None) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertEqual(dset_test._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset_test)
# with indices
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(
lambda x, i: {"name": x["filename"][:-2], "id": i}, with_indices=True
) as dset_test_with_indices:
self.assertEqual(len(dset_test_with_indices), 30)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(
dset_test_with_indices.features,
Features({"filename": Value("string"), "name": Value("string"), "id": Value("int64")}),
)
self.assertListEqual(dset_test_with_indices["id"], list(range(30)))
assert_arrow_metadata_are_synced_with_dataset_features(dset_test_with_indices)
# interrupted
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
def func(x, i):
if i == 4:
raise KeyboardInterrupt()
return {"name": x["filename"][:-2], "id": i}
tmp_file = os.path.join(tmp_dir, "test.arrow")
self.assertRaises(
KeyboardInterrupt,
dset.map,
function=func,
with_indices=True,
cache_file_name=tmp_file,
writer_batch_size=2,
)
self.assertFalse(os.path.exists(tmp_file))
with dset.map(
lambda x, i: {"name": x["filename"][:-2], "id": i},
with_indices=True,
cache_file_name=tmp_file,
writer_batch_size=2,
) as dset_test_with_indices:
self.assertTrue(os.path.exists(tmp_file))
self.assertEqual(len(dset_test_with_indices), 30)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(
dset_test_with_indices.features,
Features({"filename": Value("string"), "name": Value("string"), "id": Value("int64")}),
)
self.assertListEqual(dset_test_with_indices["id"], list(range(30)))
assert_arrow_metadata_are_synced_with_dataset_features(dset_test_with_indices)
# formatted
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
dset.set_format("numpy", columns=["col_1"])
with dset.map(lambda x: {"col_1_plus_one": x["col_1"] + 1}) as dset_test:
self.assertEqual(len(dset_test), 4)
self.assertEqual(dset_test.format["type"], "numpy")
self.assertIsInstance(dset_test["col_1"], np.ndarray)
self.assertIsInstance(dset_test["col_1_plus_one"], np.ndarray)
self.assertListEqual(sorted(dset_test[0].keys()), ["col_1", "col_1_plus_one"])
self.assertListEqual(sorted(dset_test.column_names), ["col_1", "col_1_plus_one", "col_2", "col_3"])
assert_arrow_metadata_are_synced_with_dataset_features(dset_test)
def test_map_multiprocessing(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir: # standard
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
fingerprint = dset._fingerprint
with dset.map(picklable_map_function, num_proc=2) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(
dset_test.features,
Features({"filename": Value("string"), "id": Value("int64")}),
)
self.assertEqual(len(dset_test.cache_files), 0 if in_memory else 2)
if not in_memory:
self.assertIn("_of_00002.arrow", dset_test.cache_files[0]["filename"])
self.assertListEqual(dset_test["id"], list(range(30)))
self.assertNotEqual(dset_test._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset_test)
with tempfile.TemporaryDirectory() as tmp_dir: # num_proc > num rows
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
fingerprint = dset._fingerprint
with dset.select([0, 1], keep_in_memory=True).map(picklable_map_function, num_proc=10) as dset_test:
self.assertEqual(len(dset_test), 2)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(
dset_test.features,
Features({"filename": Value("string"), "id": Value("int64")}),
)
self.assertEqual(len(dset_test.cache_files), 0 if in_memory else 2)
self.assertListEqual(dset_test["id"], list(range(2)))
self.assertNotEqual(dset_test._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset_test)
with tempfile.TemporaryDirectory() as tmp_dir: # with_indices
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
fingerprint = dset._fingerprint
with dset.map(picklable_map_function_with_indices, num_proc=3, with_indices=True) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(
dset_test.features,
Features({"filename": Value("string"), "id": Value("int64")}),
)
self.assertEqual(len(dset_test.cache_files), 0 if in_memory else 3)
self.assertListEqual(dset_test["id"], list(range(30)))
self.assertNotEqual(dset_test._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset_test)
with tempfile.TemporaryDirectory() as tmp_dir: # with_rank
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
fingerprint = dset._fingerprint
with dset.map(picklable_map_function_with_rank, num_proc=3, with_rank=True) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(
dset_test.features,
Features({"filename": Value("string"), "rank": Value("int64")}),
)
self.assertEqual(len(dset_test.cache_files), 0 if in_memory else 3)
self.assertListEqual(dset_test["rank"], [0] * 10 + [1] * 10 + [2] * 10)
self.assertNotEqual(dset_test._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset_test)
with tempfile.TemporaryDirectory() as tmp_dir: # with_indices AND with_rank
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
fingerprint = dset._fingerprint
with dset.map(
picklable_map_function_with_indices_and_rank, num_proc=3, with_indices=True, with_rank=True
) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(
dset_test.features,
Features({"filename": Value("string"), "id": Value("int64"), "rank": Value("int64")}),
)
self.assertEqual(len(dset_test.cache_files), 0 if in_memory else 3)
self.assertListEqual(dset_test["id"], list(range(30)))
self.assertListEqual(dset_test["rank"], [0] * 10 + [1] * 10 + [2] * 10)
self.assertNotEqual(dset_test._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset_test)
with tempfile.TemporaryDirectory() as tmp_dir: # new_fingerprint
new_fingerprint = "foobar"
invalid_new_fingerprint = "foobar/hey"
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
fingerprint = dset._fingerprint
self.assertRaises(
ValueError, dset.map, picklable_map_function, num_proc=2, new_fingerprint=invalid_new_fingerprint
)
with dset.map(picklable_map_function, num_proc=2, new_fingerprint=new_fingerprint) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(
dset_test.features,
Features({"filename": Value("string"), "id": Value("int64")}),
)
self.assertEqual(len(dset_test.cache_files), 0 if in_memory else 2)
self.assertListEqual(dset_test["id"], list(range(30)))
self.assertNotEqual(dset_test._fingerprint, fingerprint)
self.assertEqual(dset_test._fingerprint, new_fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset_test)
file_names = sorted(Path(cache_file["filename"]).name for cache_file in dset_test.cache_files)
for i, file_name in enumerate(file_names):
self.assertIn(new_fingerprint + f"_{i:05d}", file_name)
with tempfile.TemporaryDirectory() as tmp_dir: # lambda (requires multiprocess from pathos)
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
fingerprint = dset._fingerprint
with dset.map(lambda x: {"id": int(x["filename"].split("_")[-1])}, num_proc=2) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(
dset_test.features,
Features({"filename": Value("string"), "id": Value("int64")}),
)
self.assertEqual(len(dset_test.cache_files), 0 if in_memory else 2)
self.assertListEqual(dset_test["id"], list(range(30)))
self.assertNotEqual(dset_test._fingerprint, fingerprint)
assert_arrow_metadata_are_synced_with_dataset_features(dset_test)
def test_map_new_features(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
features = Features({"filename": Value("string"), "label": ClassLabel(names=["positive", "negative"])})
with dset.map(
lambda x, i: {"label": i % 2}, with_indices=True, features=features
) as dset_test_with_indices:
self.assertEqual(len(dset_test_with_indices), 30)
self.assertDictEqual(
dset_test_with_indices.features,
features,
)
assert_arrow_metadata_are_synced_with_dataset_features(dset_test_with_indices)
def test_map_batched(self, in_memory):
def map_batched(example):
return {"filename_new": [x + "_extension" for x in example["filename"]]}
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(map_batched, batched=True) as dset_test_batched:
self.assertEqual(len(dset_test_batched), 30)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(
dset_test_batched.features,
Features({"filename": Value("string"), "filename_new": Value("string")}),
)
assert_arrow_metadata_are_synced_with_dataset_features(dset_test_batched)
# change batch size and drop the last batch
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
batch_size = 4
with dset.map(
map_batched, batched=True, batch_size=batch_size, drop_last_batch=True
) as dset_test_batched:
self.assertEqual(len(dset_test_batched), 30 // batch_size * batch_size)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(
dset_test_batched.features,
Features({"filename": Value("string"), "filename_new": Value("string")}),
)
assert_arrow_metadata_are_synced_with_dataset_features(dset_test_batched)
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.formatted_as("numpy", columns=["filename"]):
with dset.map(map_batched, batched=True) as dset_test_batched:
self.assertEqual(len(dset_test_batched), 30)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(
dset_test_batched.features,
Features({"filename": Value("string"), "filename_new": Value("string")}),
)
assert_arrow_metadata_are_synced_with_dataset_features(dset_test_batched)
def map_batched_with_indices(example, idx):
return {"filename_new": [x + "_extension_" + str(idx) for x in example["filename"]]}
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(
map_batched_with_indices, batched=True, with_indices=True
) as dset_test_with_indices_batched:
self.assertEqual(len(dset_test_with_indices_batched), 30)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(
dset_test_with_indices_batched.features,
Features({"filename": Value("string"), "filename_new": Value("string")}),
)
assert_arrow_metadata_are_synced_with_dataset_features(dset_test_with_indices_batched)
# check remove columns for even if the function modifies input in-place
def map_batched_modifying_inputs_inplace(example):
result = {"filename_new": [x + "_extension" for x in example["filename"]]}
del example["filename"]
return result
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(
map_batched_modifying_inputs_inplace, batched=True, remove_columns="filename"
) as dset_test_modifying_inputs_inplace:
self.assertEqual(len(dset_test_modifying_inputs_inplace), 30)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(
dset_test_modifying_inputs_inplace.features,
Features({"filename_new": Value("string")}),
)
assert_arrow_metadata_are_synced_with_dataset_features(dset_test_modifying_inputs_inplace)
def test_map_nested(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict({"field": ["a", "b"]}) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
with dset.map(lambda example: {"otherfield": {"capital": example["field"].capitalize()}}) as dset:
with dset.map(lambda example: {"otherfield": {"append_x": example["field"] + "x"}}) as dset:
self.assertEqual(dset[0], {"field": "a", "otherfield": {"append_x": "ax"}})
def test_map_return_example_as_dict_value(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict({"en": ["aa", "bb"], "fr": ["cc", "dd"]}) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
with dset.map(lambda example: {"translation": example}) as dset:
self.assertEqual(dset[0], {"en": "aa", "fr": "cc", "translation": {"en": "aa", "fr": "cc"}})
def test_map_fn_kwargs(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict({"id": range(10)}) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
fn_kwargs = {"offset": 3}
with dset.map(
lambda example, offset: {"id+offset": example["id"] + offset}, fn_kwargs=fn_kwargs
) as mapped_dset:
assert mapped_dset["id+offset"] == list(range(3, 13))
with dset.map(
lambda id, offset: {"id+offset": id + offset}, fn_kwargs=fn_kwargs, input_columns="id"
) as mapped_dset:
assert mapped_dset["id+offset"] == list(range(3, 13))
with dset.map(
lambda id, i, offset: {"id+offset": i + offset},
fn_kwargs=fn_kwargs,
input_columns="id",
with_indices=True,
) as mapped_dset:
assert mapped_dset["id+offset"] == list(range(3, 13))
def test_map_caching(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
self._caplog.clear()
with self._caplog.at_level(INFO, logger=get_logger().name):
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with patch(
"datasets.arrow_dataset.Dataset._map_single",
autospec=Dataset._map_single,
side_effect=Dataset._map_single,
) as mock_map_single:
with dset.map(lambda x: {"foo": "bar"}) as dset_test1:
dset_test1_data_files = list(dset_test1.cache_files)
self.assertEqual(mock_map_single.call_count, 1)
with dset.map(lambda x: {"foo": "bar"}) as dset_test2:
self.assertEqual(dset_test1_data_files, dset_test2.cache_files)
self.assertEqual(len(dset_test2.cache_files), 1 - int(in_memory))
self.assertTrue(("Loading cached processed dataset" in self._caplog.text) ^ in_memory)
self.assertEqual(mock_map_single.call_count, 2 if in_memory else 1)
with tempfile.TemporaryDirectory() as tmp_dir:
self._caplog.clear()
with self._caplog.at_level(INFO, logger=get_logger().name):
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(lambda x: {"foo": "bar"}) as dset_test1:
dset_test1_data_files = list(dset_test1.cache_files)
with dset.map(lambda x: {"foo": "bar"}, load_from_cache_file=False) as dset_test2:
self.assertEqual(dset_test1_data_files, dset_test2.cache_files)
self.assertEqual(len(dset_test2.cache_files), 1 - int(in_memory))
self.assertNotIn("Loading cached processed dataset", self._caplog.text)
with tempfile.TemporaryDirectory() as tmp_dir:
self._caplog.clear()
with self._caplog.at_level(INFO, logger=get_logger().name):
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with patch(
"datasets.arrow_dataset.Pool",
new_callable=PickableMagicMock,
side_effect=datasets.arrow_dataset.Pool,
) as mock_pool:
with dset.map(lambda x: {"foo": "bar"}, num_proc=2) as dset_test1:
dset_test1_data_files = list(dset_test1.cache_files)
self.assertEqual(mock_pool.call_count, 1)
with dset.map(lambda x: {"foo": "bar"}, num_proc=2) as dset_test2:
self.assertEqual(dset_test1_data_files, dset_test2.cache_files)
self.assertTrue(
(len(re.findall("Loading cached processed dataset", self._caplog.text)) == 1)
^ in_memory
)
self.assertEqual(mock_pool.call_count, 2 if in_memory else 1)
with tempfile.TemporaryDirectory() as tmp_dir:
self._caplog.clear()
with self._caplog.at_level(INFO, logger=get_logger().name):
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(lambda x: {"foo": "bar"}, num_proc=2) as dset_test1:
dset_test1_data_files = list(dset_test1.cache_files)
with dset.map(lambda x: {"foo": "bar"}, num_proc=2, load_from_cache_file=False) as dset_test2:
self.assertEqual(dset_test1_data_files, dset_test2.cache_files)
self.assertEqual(len(dset_test2.cache_files), (1 - int(in_memory)) * 2)
self.assertNotIn("Loading cached processed dataset", self._caplog.text)
if not in_memory:
try:
self._caplog.clear()
with tempfile.TemporaryDirectory() as tmp_dir:
with self._caplog.at_level(INFO, logger=get_logger().name):
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
datasets.disable_caching()
with dset.map(lambda x: {"foo": "bar"}) as dset_test1:
with dset.map(lambda x: {"foo": "bar"}) as dset_test2:
self.assertNotEqual(dset_test1.cache_files, dset_test2.cache_files)
self.assertEqual(len(dset_test1.cache_files), 1)
self.assertEqual(len(dset_test2.cache_files), 1)
self.assertNotIn("Loading cached processed dataset", self._caplog.text)
# make sure the arrow files are going to be removed
self.assertIn("tmp", dset_test1.cache_files[0]["filename"])
self.assertIn("tmp", dset_test2.cache_files[0]["filename"])
finally:
datasets.enable_caching()
def test_map_return_pa_table(self, in_memory):
def func_return_single_row_pa_table(x):
return pa.table({"id": [0], "text": ["a"]})
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(func_return_single_row_pa_table) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertDictEqual(
dset_test.features,
Features({"id": Value("int64"), "text": Value("string")}),
)
self.assertEqual(dset_test[0]["id"], 0)
self.assertEqual(dset_test[0]["text"], "a")
# Batched
def func_return_single_row_pa_table_batched(x):
batch_size = len(x[next(iter(x))])
return pa.table({"id": [0] * batch_size, "text": ["a"] * batch_size})
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(func_return_single_row_pa_table_batched, batched=True) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertDictEqual(
dset_test.features,
Features({"id": Value("int64"), "text": Value("string")}),
)
self.assertEqual(dset_test[0]["id"], 0)
self.assertEqual(dset_test[0]["text"], "a")
# Error when returning a table with more than one row in the non-batched mode
def func_return_multi_row_pa_table(x):
return pa.table({"id": [0, 1], "text": ["a", "b"]})
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
self.assertRaises(ValueError, dset.map, func_return_multi_row_pa_table)
def test_map_return_pd_dataframe(self, in_memory):
def func_return_single_row_pd_dataframe(x):
return pd.DataFrame({"id": [0], "text": ["a"]})
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(func_return_single_row_pd_dataframe) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertDictEqual(
dset_test.features,
Features({"id": Value("int64"), "text": Value("string")}),
)
self.assertEqual(dset_test[0]["id"], 0)
self.assertEqual(dset_test[0]["text"], "a")
# Batched
def func_return_single_row_pd_dataframe_batched(x):
batch_size = len(x[next(iter(x))])
return pd.DataFrame({"id": [0] * batch_size, "text": ["a"] * batch_size})
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(func_return_single_row_pd_dataframe_batched, batched=True) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertDictEqual(
dset_test.features,
Features({"id": Value("int64"), "text": Value("string")}),
)
self.assertEqual(dset_test[0]["id"], 0)
self.assertEqual(dset_test[0]["text"], "a")
# Error when returning a table with more than one row in the non-batched mode
def func_return_multi_row_pd_dataframe(x):
return pd.DataFrame({"id": [0, 1], "text": ["a", "b"]})
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
self.assertRaises(ValueError, dset.map, func_return_multi_row_pd_dataframe)
@require_torch
def test_map_torch(self, in_memory):
import torch
def func(example):
return {"tensor": torch.tensor([1.0, 2, 3])}
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(func) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertDictEqual(
dset_test.features,
Features({"filename": Value("string"), "tensor": Sequence(Value("float32"))}),
)
self.assertListEqual(dset_test[0]["tensor"], [1, 2, 3])
@require_tf
def test_map_tf(self, in_memory):
import tensorflow as tf
def func(example):
return {"tensor": tf.constant([1.0, 2, 3])}
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(func) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertDictEqual(
dset_test.features,
Features({"filename": Value("string"), "tensor": Sequence(Value("float32"))}),
)
self.assertListEqual(dset_test[0]["tensor"], [1, 2, 3])
@require_jax
def test_map_jax(self, in_memory):
import jax.numpy as jnp
def func(example):
return {"tensor": jnp.asarray([1.0, 2, 3])}
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(func) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertDictEqual(
dset_test.features,
Features({"filename": Value("string"), "tensor": Sequence(Value("float32"))}),
)
self.assertListEqual(dset_test[0]["tensor"], [1, 2, 3])
def test_map_numpy(self, in_memory):
def func(example):
return {"tensor": np.array([1.0, 2, 3])}
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(func) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertDictEqual(
dset_test.features,
Features({"filename": Value("string"), "tensor": Sequence(Value("float64"))}),
)
self.assertListEqual(dset_test[0]["tensor"], [1, 2, 3])
@require_torch
def test_map_tensor_batched(self, in_memory):
import torch
def func(batch):
return {"tensor": torch.tensor([[1.0, 2, 3]] * len(batch["filename"]))}
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(func, batched=True) as dset_test:
self.assertEqual(len(dset_test), 30)
self.assertDictEqual(
dset_test.features,
Features({"filename": Value("string"), "tensor": Sequence(Value("float32"))}),
)
self.assertListEqual(dset_test[0]["tensor"], [1, 2, 3])
def test_map_input_columns(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
with dset.map(lambda col_1: {"label": col_1 % 2}, input_columns="col_1") as mapped_dset:
self.assertEqual(mapped_dset[0].keys(), {"col_1", "col_2", "col_3", "label"})
self.assertEqual(
mapped_dset.features,
Features(
{
"col_1": Value("int64"),
"col_2": Value("string"),
"col_3": Value("bool"),
"label": Value("int64"),
}
),
)
def test_map_remove_columns(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.map(lambda x, i: {"name": x["filename"][:-2], "id": i}, with_indices=True) as dset:
self.assertTrue("id" in dset[0])
self.assertDictEqual(
dset.features,
Features({"filename": Value("string"), "name": Value("string"), "id": Value("int64")}),
)
assert_arrow_metadata_are_synced_with_dataset_features(dset)
with dset.map(lambda x: x, remove_columns=["id"]) as mapped_dset:
self.assertTrue("id" not in mapped_dset[0])
self.assertDictEqual(
mapped_dset.features, Features({"filename": Value("string"), "name": Value("string")})
)
assert_arrow_metadata_are_synced_with_dataset_features(mapped_dset)
with mapped_dset.with_format("numpy", columns=mapped_dset.column_names) as mapped_dset:
with mapped_dset.map(
lambda x: {"name": 1}, remove_columns=mapped_dset.column_names
) as mapped_dset:
self.assertTrue("filename" not in mapped_dset[0])
self.assertTrue("name" in mapped_dset[0])
self.assertDictEqual(mapped_dset.features, Features({"name": Value(dtype="int64")}))
assert_arrow_metadata_are_synced_with_dataset_features(mapped_dset)
# empty dataset
columns_names = dset.column_names
with dset.select([]) as empty_dset:
self.assertEqual(len(empty_dset), 0)
with empty_dset.map(lambda x: {}, remove_columns=columns_names[0]) as mapped_dset:
self.assertListEqual(columns_names[1:], mapped_dset.column_names)
assert_arrow_metadata_are_synced_with_dataset_features(mapped_dset)
def test_map_stateful_callable(self, in_memory):
# be sure that the state of the map callable is unaffected
# before processing the dataset examples
class ExampleCounter:
def __init__(self, batched=False):
self.batched = batched
# state
self.cnt = 0
def __call__(self, example):
if self.batched:
self.cnt += len(example)
else:
self.cnt += 1
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
ex_cnt = ExampleCounter()
dset.map(ex_cnt)
self.assertEqual(ex_cnt.cnt, len(dset))
ex_cnt = ExampleCounter(batched=True)
dset.map(ex_cnt)
self.assertEqual(ex_cnt.cnt, len(dset))
@require_not_windows
def test_map_crash_subprocess(self, in_memory):
# be sure that a crash in one of the subprocess will not
# hang dataset.map() call forever
def do_crash(row):
import os
os.kill(os.getpid(), 9)
return row
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with pytest.raises(RuntimeError) as excinfo:
dset.map(do_crash, num_proc=2)
assert str(excinfo.value) == (
"One of the subprocesses has abruptly died during map operation."
"To debug the error, disable multiprocessing."
)
def test_filter(self, in_memory):
# keep only first five examples
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
fingerprint = dset._fingerprint
with dset.filter(lambda x, i: i < 5, with_indices=True) as dset_filter_first_five:
self.assertEqual(len(dset_filter_first_five), 5)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_filter_first_five.features, Features({"filename": Value("string")}))
self.assertNotEqual(dset_filter_first_five._fingerprint, fingerprint)
# filter filenames with even id at the end + formatted
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
dset.set_format("numpy")
fingerprint = dset._fingerprint
with dset.filter(lambda x: (int(x["filename"][-1]) % 2 == 0)) as dset_filter_even_num:
self.assertEqual(len(dset_filter_even_num), 15)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_filter_even_num.features, Features({"filename": Value("string")}))
self.assertNotEqual(dset_filter_even_num._fingerprint, fingerprint)
self.assertEqual(dset_filter_even_num.format["type"], "numpy")
def test_filter_with_indices_mapping(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
dset = Dataset.from_dict({"col": [0, 1, 2]})
with self._to(in_memory, tmp_dir, dset) as dset:
with dset.filter(lambda x: x["col"] > 0) as dset:
self.assertListEqual(dset["col"], [1, 2])
with dset.filter(lambda x: x["col"] < 2) as dset:
self.assertListEqual(dset["col"], [1])
def test_filter_empty(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
self.assertIsNone(dset._indices, None)
tmp_file = os.path.join(tmp_dir, "test.arrow")
with dset.filter(lambda _: False, cache_file_name=tmp_file) as dset:
self.assertEqual(len(dset), 0)
self.assertIsNotNone(dset._indices, None)
tmp_file_2 = os.path.join(tmp_dir, "test_2.arrow")
with dset.filter(lambda _: False, cache_file_name=tmp_file_2) as dset2:
self.assertEqual(len(dset2), 0)
self.assertEqual(dset._indices, dset2._indices)
def test_filter_batched(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
dset = Dataset.from_dict({"col": [0, 1, 2]})
with self._to(in_memory, tmp_dir, dset) as dset:
with dset.filter(lambda x: [i > 0 for i in x["col"]], batched=True) as dset:
self.assertListEqual(dset["col"], [1, 2])
with dset.filter(lambda x: [i < 2 for i in x["col"]], batched=True) as dset:
self.assertListEqual(dset["col"], [1])
def test_filter_input_columns(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
dset = Dataset.from_dict({"col_1": [0, 1, 2], "col_2": ["a", "b", "c"]})
with self._to(in_memory, tmp_dir, dset) as dset:
with dset.filter(lambda x: x > 0, input_columns=["col_1"]) as filtered_dset:
self.assertListEqual(filtered_dset.column_names, dset.column_names)
self.assertListEqual(filtered_dset["col_1"], [1, 2])
self.assertListEqual(filtered_dset["col_2"], ["b", "c"])
def test_filter_fn_kwargs(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict({"id": range(10)}) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
fn_kwargs = {"max_offset": 3}
with dset.filter(
lambda example, max_offset: example["id"] < max_offset, fn_kwargs=fn_kwargs
) as filtered_dset:
assert len(filtered_dset) == 3
with dset.filter(
lambda id, max_offset: id < max_offset, fn_kwargs=fn_kwargs, input_columns="id"
) as filtered_dset:
assert len(filtered_dset) == 3
with dset.filter(
lambda id, i, max_offset: i < max_offset,
fn_kwargs=fn_kwargs,
input_columns="id",
with_indices=True,
) as filtered_dset:
assert len(filtered_dset) == 3
def test_filter_multiprocessing(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
fingerprint = dset._fingerprint
with dset.filter(picklable_filter_function, num_proc=2) as dset_filter_first_ten:
self.assertEqual(len(dset_filter_first_ten), 10)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_filter_first_ten.features, Features({"filename": Value("string")}))
self.assertEqual(len(dset_filter_first_ten.cache_files), 0 if in_memory else 2)
self.assertNotEqual(dset_filter_first_ten._fingerprint, fingerprint)
def test_filter_caching(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
self._caplog.clear()
with self._caplog.at_level(INFO, logger=get_logger().name):
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.filter(lambda x, i: i < 5, with_indices=True) as dset_filter_first_five1:
dset_test1_data_files = list(dset_filter_first_five1.cache_files)
with dset.filter(lambda x, i: i < 5, with_indices=True) as dset_filter_first_five2:
self.assertEqual(dset_test1_data_files, dset_filter_first_five2.cache_files)
self.assertEqual(len(dset_filter_first_five2.cache_files), 0 if in_memory else 2)
self.assertTrue(("Loading cached processed dataset" in self._caplog.text) ^ in_memory)
def test_keep_features_after_transform_specified(self, in_memory):
features = Features(
{"tokens": Sequence(Value("string")), "labels": Sequence(ClassLabel(names=["negative", "positive"]))}
)
def invert_labels(x):
return {"labels": [(1 - label) for label in x["labels"]]}
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict(
{"tokens": [["foo"] * 5] * 10, "labels": [[1] * 5] * 10}, features=features
) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
with dset.map(invert_labels, features=features) as inverted_dset:
self.assertEqual(inverted_dset.features.type, features.type)
self.assertDictEqual(inverted_dset.features, features)
assert_arrow_metadata_are_synced_with_dataset_features(inverted_dset)
def test_keep_features_after_transform_unspecified(self, in_memory):
features = Features(
{"tokens": Sequence(Value("string")), "labels": Sequence(ClassLabel(names=["negative", "positive"]))}
)
def invert_labels(x):
return {"labels": [(1 - label) for label in x["labels"]]}
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict(
{"tokens": [["foo"] * 5] * 10, "labels": [[1] * 5] * 10}, features=features
) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
with dset.map(invert_labels) as inverted_dset:
self.assertEqual(inverted_dset.features.type, features.type)
self.assertDictEqual(inverted_dset.features, features)
assert_arrow_metadata_are_synced_with_dataset_features(inverted_dset)
def test_keep_features_after_transform_to_file(self, in_memory):
features = Features(
{"tokens": Sequence(Value("string")), "labels": Sequence(ClassLabel(names=["negative", "positive"]))}
)
def invert_labels(x):
return {"labels": [(1 - label) for label in x["labels"]]}
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict(
{"tokens": [["foo"] * 5] * 10, "labels": [[1] * 5] * 10}, features=features
) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
tmp_file = os.path.join(tmp_dir, "test.arrow")
dset.map(invert_labels, cache_file_name=tmp_file)
with Dataset.from_file(tmp_file) as inverted_dset:
self.assertEqual(inverted_dset.features.type, features.type)
self.assertDictEqual(inverted_dset.features, features)
def test_keep_features_after_transform_to_memory(self, in_memory):
features = Features(
{"tokens": Sequence(Value("string")), "labels": Sequence(ClassLabel(names=["negative", "positive"]))}
)
def invert_labels(x):
return {"labels": [(1 - label) for label in x["labels"]]}
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict(
{"tokens": [["foo"] * 5] * 10, "labels": [[1] * 5] * 10}, features=features
) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
with dset.map(invert_labels, keep_in_memory=True) as inverted_dset:
self.assertEqual(inverted_dset.features.type, features.type)
self.assertDictEqual(inverted_dset.features, features)
def test_keep_features_after_loading_from_cache(self, in_memory):
features = Features(
{"tokens": Sequence(Value("string")), "labels": Sequence(ClassLabel(names=["negative", "positive"]))}
)
def invert_labels(x):
return {"labels": [(1 - label) for label in x["labels"]]}
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict(
{"tokens": [["foo"] * 5] * 10, "labels": [[1] * 5] * 10}, features=features
) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
tmp_file1 = os.path.join(tmp_dir, "test1.arrow")
tmp_file2 = os.path.join(tmp_dir, "test2.arrow")
# TODO: Why mapped twice?
inverted_dset = dset.map(invert_labels, cache_file_name=tmp_file1)
inverted_dset = dset.map(invert_labels, cache_file_name=tmp_file2)
self.assertGreater(len(inverted_dset.cache_files), 0)
self.assertEqual(inverted_dset.features.type, features.type)
self.assertDictEqual(inverted_dset.features, features)
del inverted_dset
def test_keep_features_with_new_features(self, in_memory):
features = Features(
{"tokens": Sequence(Value("string")), "labels": Sequence(ClassLabel(names=["negative", "positive"]))}
)
def invert_labels(x):
return {"labels": [(1 - label) for label in x["labels"]], "labels2": x["labels"]}
expected_features = Features(
{
"tokens": Sequence(Value("string")),
"labels": Sequence(ClassLabel(names=["negative", "positive"])),
"labels2": Sequence(Value("int64")),
}
)
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict(
{"tokens": [["foo"] * 5] * 10, "labels": [[1] * 5] * 10}, features=features
) as dset:
with self._to(in_memory, tmp_dir, dset) as dset:
with dset.map(invert_labels) as inverted_dset:
self.assertEqual(inverted_dset.features.type, expected_features.type)
self.assertDictEqual(inverted_dset.features, expected_features)
assert_arrow_metadata_are_synced_with_dataset_features(inverted_dset)
def test_select(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
# select every two example
indices = list(range(0, len(dset), 2))
tmp_file = os.path.join(tmp_dir, "test.arrow")
fingerprint = dset._fingerprint
with dset.select(indices, indices_cache_file_name=tmp_file) as dset_select_even:
self.assertIsNotNone(dset_select_even._indices) # an indices mapping is created
self.assertTrue(os.path.exists(tmp_file))
self.assertEqual(len(dset_select_even), 15)
for row in dset_select_even:
self.assertEqual(int(row["filename"][-1]) % 2, 0)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_select_even.features, Features({"filename": Value("string")}))
self.assertNotEqual(dset_select_even._fingerprint, fingerprint)
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
indices = list(range(0, len(dset)))
with dset.select(indices) as dset_select_all:
# no indices mapping, since the indices are contiguous
# (in this case the arrow table is simply sliced, which is more efficient)
self.assertIsNone(dset_select_all._indices)
self.assertEqual(len(dset_select_all), len(dset))
self.assertListEqual(list(dset_select_all), list(dset))
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_select_all.features, Features({"filename": Value("string")}))
self.assertNotEqual(dset_select_all._fingerprint, fingerprint)
indices = range(0, len(dset))
with dset.select(indices) as dset_select_all:
# same but with range
self.assertIsNone(dset_select_all._indices)
self.assertEqual(len(dset_select_all), len(dset))
self.assertListEqual(list(dset_select_all), list(dset))
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_select_all.features, Features({"filename": Value("string")}))
self.assertNotEqual(dset_select_all._fingerprint, fingerprint)
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
bad_indices = list(range(5))
bad_indices[-1] = len(dset) + 10 # out of bounds
tmp_file = os.path.join(tmp_dir, "test.arrow")
self.assertRaises(
Exception,
dset.select,
indices=bad_indices,
indices_cache_file_name=tmp_file,
writer_batch_size=2,
)
self.assertFalse(os.path.exists(tmp_file))
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
indices = iter(range(len(dset))) # iterator of contiguous indices
with dset.select(indices) as dset_select_all:
# no indices mapping, since the indices are contiguous
self.assertIsNone(dset_select_all._indices)
self.assertEqual(len(dset_select_all), len(dset))
indices = reversed(range(len(dset))) # iterator of not contiguous indices
tmp_file = os.path.join(tmp_dir, "test.arrow")
with dset.select(indices, indices_cache_file_name=tmp_file) as dset_select_all:
# new indices mapping, since the indices are not contiguous
self.assertIsNotNone(dset_select_all._indices)
self.assertEqual(len(dset_select_all), len(dset))
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
bad_indices = list(range(5))
bad_indices[3] = "foo" # wrong type
tmp_file = os.path.join(tmp_dir, "test.arrow")
self.assertRaises(
Exception,
dset.select,
indices=bad_indices,
indices_cache_file_name=tmp_file,
writer_batch_size=2,
)
self.assertFalse(os.path.exists(tmp_file))
dset.set_format("numpy")
with dset.select(
range(5),
indices_cache_file_name=tmp_file,
writer_batch_size=2,
) as dset_select_five:
self.assertIsNone(dset_select_five._indices)
self.assertEqual(len(dset_select_five), 5)
self.assertEqual(dset_select_five.format["type"], "numpy")
for i, row in enumerate(dset_select_five):
self.assertEqual(int(row["filename"][-1]), i)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_select_five.features, Features({"filename": Value("string")}))
def test_select_then_map(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.select([0]) as d1:
with d1.map(lambda x: {"id": int(x["filename"].split("_")[-1])}) as d1:
self.assertEqual(d1[0]["id"], 0)
with dset.select([1]) as d2:
with d2.map(lambda x: {"id": int(x["filename"].split("_")[-1])}) as d2:
self.assertEqual(d2[0]["id"], 1)
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
with dset.select([0], indices_cache_file_name=os.path.join(tmp_dir, "i1.arrow")) as d1:
with d1.map(lambda x: {"id": int(x["filename"].split("_")[-1])}) as d1:
self.assertEqual(d1[0]["id"], 0)
with dset.select([1], indices_cache_file_name=os.path.join(tmp_dir, "i2.arrow")) as d2:
with d2.map(lambda x: {"id": int(x["filename"].split("_")[-1])}) as d2:
self.assertEqual(d2[0]["id"], 1)
def test_pickle_after_many_transforms_on_disk(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
self.assertEqual(len(dset.cache_files), 0 if in_memory else 1)
with dset.rename_column("filename", "file") as dset:
self.assertListEqual(dset.column_names, ["file"])
with dset.select(range(5)) as dset:
self.assertEqual(len(dset), 5)
with dset.map(lambda x: {"id": int(x["file"][-1])}) as dset:
self.assertListEqual(sorted(dset.column_names), ["file", "id"])
with dset.rename_column("id", "number") as dset:
self.assertListEqual(sorted(dset.column_names), ["file", "number"])
with dset.select([1, 0]) as dset:
self.assertEqual(dset[0]["file"], "my_name-train_1")
self.assertEqual(dset[0]["number"], 1)
self.assertEqual(dset._indices["indices"].to_pylist(), [1, 0])
if not in_memory:
self.assertIn(
("rename_columns", (["file", "number"],), {}),
dset._data.replays,
)
if not in_memory:
dset._data.table = Unpicklable() # check that we don't pickle the entire table
pickled = pickle.dumps(dset)
with pickle.loads(pickled) as loaded:
self.assertEqual(loaded[0]["file"], "my_name-train_1")
self.assertEqual(loaded[0]["number"], 1)
def test_shuffle(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
tmp_file = os.path.join(tmp_dir, "test.arrow")
fingerprint = dset._fingerprint
with dset.shuffle(seed=1234, keep_in_memory=True) as dset_shuffled:
self.assertEqual(len(dset_shuffled), 30)
self.assertEqual(dset_shuffled[0]["filename"], "my_name-train_28")
self.assertEqual(dset_shuffled[2]["filename"], "my_name-train_10")
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_shuffled.features, Features({"filename": Value("string")}))
self.assertNotEqual(dset_shuffled._fingerprint, fingerprint)
with dset.shuffle(seed=1234, indices_cache_file_name=tmp_file) as dset_shuffled:
self.assertEqual(len(dset_shuffled), 30)
self.assertEqual(dset_shuffled[0]["filename"], "my_name-train_28")
self.assertEqual(dset_shuffled[2]["filename"], "my_name-train_10")
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_shuffled.features, Features({"filename": Value("string")}))
self.assertNotEqual(dset_shuffled._fingerprint, fingerprint)
# Reproducibility
tmp_file = os.path.join(tmp_dir, "test_2.arrow")
with dset.shuffle(seed=1234, indices_cache_file_name=tmp_file) as dset_shuffled_2:
self.assertListEqual(dset_shuffled["filename"], dset_shuffled_2["filename"])
# Compatible with temp_seed
with temp_seed(42), dset.shuffle() as d1:
with temp_seed(42), dset.shuffle() as d2, dset.shuffle() as d3:
self.assertListEqual(d1["filename"], d2["filename"])
self.assertEqual(d1._fingerprint, d2._fingerprint)
self.assertNotEqual(d3["filename"], d2["filename"])
self.assertNotEqual(d3._fingerprint, d2._fingerprint)
def test_sort(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
# Sort on a single key
with self._create_dummy_dataset(in_memory=in_memory, tmp_dir=tmp_dir) as dset:
# Keep only 10 examples
tmp_file = os.path.join(tmp_dir, "test.arrow")
with dset.select(range(10), indices_cache_file_name=tmp_file) as dset:
tmp_file = os.path.join(tmp_dir, "test_2.arrow")
with dset.shuffle(seed=1234, indices_cache_file_name=tmp_file) as dset:
self.assertEqual(len(dset), 10)
self.assertEqual(dset[0]["filename"], "my_name-train_8")
self.assertEqual(dset[1]["filename"], "my_name-train_9")
# Sort
tmp_file = os.path.join(tmp_dir, "test_3.arrow")
fingerprint = dset._fingerprint
with dset.sort("filename", indices_cache_file_name=tmp_file) as dset_sorted:
for i, row in enumerate(dset_sorted):
self.assertEqual(int(row["filename"][-1]), i)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_sorted.features, Features({"filename": Value("string")}))
self.assertNotEqual(dset_sorted._fingerprint, fingerprint)
# Sort reversed
tmp_file = os.path.join(tmp_dir, "test_4.arrow")
fingerprint = dset._fingerprint
with dset.sort("filename", indices_cache_file_name=tmp_file, reverse=True) as dset_sorted:
for i, row in enumerate(dset_sorted):
self.assertEqual(int(row["filename"][-1]), len(dset_sorted) - 1 - i)
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_sorted.features, Features({"filename": Value("string")}))
self.assertNotEqual(dset_sorted._fingerprint, fingerprint)
# formatted
dset.set_format("numpy")
with dset.sort("filename") as dset_sorted_formatted:
self.assertEqual(dset_sorted_formatted.format["type"], "numpy")
# Sort on multiple keys
with self._create_dummy_dataset(in_memory=in_memory, tmp_dir=tmp_dir, multiple_columns=True) as dset:
tmp_file = os.path.join(tmp_dir, "test_5.arrow")
fingerprint = dset._fingerprint
# Throw error when reverse is a list of bools that does not match the length of column_names
with pytest.raises(ValueError):
dset.sort(["col_1", "col_2", "col_3"], reverse=[False])
with dset.shuffle(seed=1234, indices_cache_file_name=tmp_file) as dset:
# Sort
with dset.sort(["col_1", "col_2", "col_3"], reverse=[False, True, False]) as dset_sorted:
for i, row in enumerate(dset_sorted):
self.assertEqual(row["col_1"], i)
self.assertDictEqual(
dset.features,
Features(
{
"col_1": Value("int64"),
"col_2": Value("string"),
"col_3": Value("bool"),
}
),
)
self.assertDictEqual(
dset_sorted.features,
Features(
{
"col_1": Value("int64"),
"col_2": Value("string"),
"col_3": Value("bool"),
}
),
)
self.assertNotEqual(dset_sorted._fingerprint, fingerprint)
# Sort reversed
with dset.sort(["col_1", "col_2", "col_3"], reverse=[True, False, True]) as dset_sorted:
for i, row in enumerate(dset_sorted):
self.assertEqual(row["col_1"], len(dset_sorted) - 1 - i)
self.assertDictEqual(
dset.features,
Features(
{
"col_1": Value("int64"),
"col_2": Value("string"),
"col_3": Value("bool"),
}
),
)
self.assertDictEqual(
dset_sorted.features,
Features(
{
"col_1": Value("int64"),
"col_2": Value("string"),
"col_3": Value("bool"),
}
),
)
self.assertNotEqual(dset_sorted._fingerprint, fingerprint)
# formatted
dset.set_format("numpy")
with dset.sort(
["col_1", "col_2", "col_3"], reverse=[False, True, False]
) as dset_sorted_formatted:
self.assertEqual(dset_sorted_formatted.format["type"], "numpy")
@require_tf
def test_export(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
# Export the data
tfrecord_path = os.path.join(tmp_dir, "test.tfrecord")
with dset.map(
lambda ex, i: {
"id": i,
"question": f"Question {i}",
"answers": {"text": [f"Answer {i}-0", f"Answer {i}-1"], "answer_start": [0, 1]},
},
with_indices=True,
remove_columns=["filename"],
) as formatted_dset:
with formatted_dset.flatten() as formatted_dset:
formatted_dset.set_format("numpy")
formatted_dset.export(filename=tfrecord_path, format="tfrecord")
# Import the data
import tensorflow as tf
tf_dset = tf.data.TFRecordDataset([tfrecord_path])
feature_description = {
"id": tf.io.FixedLenFeature([], tf.int64),
"question": tf.io.FixedLenFeature([], tf.string),
"answers.text": tf.io.VarLenFeature(tf.string),
"answers.answer_start": tf.io.VarLenFeature(tf.int64),
}
tf_parsed_dset = tf_dset.map(
lambda example_proto: tf.io.parse_single_example(example_proto, feature_description)
)
# Test that keys match original dataset
for i, ex in enumerate(tf_parsed_dset):
self.assertEqual(ex.keys(), formatted_dset[i].keys())
# Test for equal number of elements
self.assertEqual(i, len(formatted_dset) - 1)
def test_to_csv(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
# File path argument
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
file_path = os.path.join(tmp_dir, "test_path.csv")
bytes_written = dset.to_csv(path_or_buf=file_path)
self.assertTrue(os.path.isfile(file_path))
self.assertEqual(bytes_written, os.path.getsize(file_path))
csv_dset = pd.read_csv(file_path)
self.assertEqual(csv_dset.shape, dset.shape)
self.assertListEqual(list(csv_dset.columns), list(dset.column_names))
# File buffer argument
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
file_path = os.path.join(tmp_dir, "test_buffer.csv")
with open(file_path, "wb+") as buffer:
bytes_written = dset.to_csv(path_or_buf=buffer)
self.assertTrue(os.path.isfile(file_path))
self.assertEqual(bytes_written, os.path.getsize(file_path))
csv_dset = pd.read_csv(file_path)
self.assertEqual(csv_dset.shape, dset.shape)
self.assertListEqual(list(csv_dset.columns), list(dset.column_names))
# After a select/shuffle transform
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
dset = dset.select(range(0, len(dset), 2)).shuffle()
file_path = os.path.join(tmp_dir, "test_path.csv")
bytes_written = dset.to_csv(path_or_buf=file_path)
self.assertTrue(os.path.isfile(file_path))
self.assertEqual(bytes_written, os.path.getsize(file_path))
csv_dset = pd.read_csv(file_path)
self.assertEqual(csv_dset.shape, dset.shape)
self.assertListEqual(list(csv_dset.columns), list(dset.column_names))
# With array features
with self._create_dummy_dataset(in_memory, tmp_dir, array_features=True) as dset:
file_path = os.path.join(tmp_dir, "test_path.csv")
bytes_written = dset.to_csv(path_or_buf=file_path)
self.assertTrue(os.path.isfile(file_path))
self.assertEqual(bytes_written, os.path.getsize(file_path))
csv_dset = pd.read_csv(file_path)
self.assertEqual(csv_dset.shape, dset.shape)
self.assertListEqual(list(csv_dset.columns), list(dset.column_names))
def test_to_dict(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
# Full
dset_to_dict = dset.to_dict()
self.assertIsInstance(dset_to_dict, dict)
self.assertListEqual(sorted(dset_to_dict.keys()), sorted(dset.column_names))
for col_name in dset.column_names:
self.assertLessEqual(len(dset_to_dict[col_name]), len(dset))
# With index mapping
with dset.select([1, 0, 3]) as dset:
dset_to_dict = dset.to_dict()
self.assertIsInstance(dset_to_dict, dict)
self.assertEqual(len(dset_to_dict), 3)
self.assertListEqual(sorted(dset_to_dict.keys()), sorted(dset.column_names))
for col_name in dset.column_names:
self.assertIsInstance(dset_to_dict[col_name], list)
self.assertEqual(len(dset_to_dict[col_name]), len(dset))
def test_to_list(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
dset_to_list = dset.to_list()
self.assertIsInstance(dset_to_list, list)
for row in dset_to_list:
self.assertIsInstance(row, dict)
self.assertListEqual(sorted(row.keys()), sorted(dset.column_names))
# With index mapping
with dset.select([1, 0, 3]) as dset:
dset_to_list = dset.to_list()
self.assertIsInstance(dset_to_list, list)
self.assertEqual(len(dset_to_list), 3)
for row in dset_to_list:
self.assertIsInstance(row, dict)
self.assertListEqual(sorted(row.keys()), sorted(dset.column_names))
def test_to_pandas(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
# Batched
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
batch_size = dset.num_rows - 1
to_pandas_generator = dset.to_pandas(batched=True, batch_size=batch_size)
for batch in to_pandas_generator:
self.assertIsInstance(batch, pd.DataFrame)
self.assertListEqual(sorted(batch.columns), sorted(dset.column_names))
for col_name in dset.column_names:
self.assertLessEqual(len(batch[col_name]), batch_size)
# Full
dset_to_pandas = dset.to_pandas()
self.assertIsInstance(dset_to_pandas, pd.DataFrame)
self.assertListEqual(sorted(dset_to_pandas.columns), sorted(dset.column_names))
for col_name in dset.column_names:
self.assertEqual(len(dset_to_pandas[col_name]), len(dset))
# With index mapping
with dset.select([1, 0, 3]) as dset:
dset_to_pandas = dset.to_pandas()
self.assertIsInstance(dset_to_pandas, pd.DataFrame)
self.assertEqual(len(dset_to_pandas), 3)
self.assertListEqual(sorted(dset_to_pandas.columns), sorted(dset.column_names))
for col_name in dset.column_names:
self.assertEqual(len(dset_to_pandas[col_name]), dset.num_rows)
def test_to_parquet(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
# File path argument
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
file_path = os.path.join(tmp_dir, "test_path.parquet")
dset.to_parquet(path_or_buf=file_path)
self.assertTrue(os.path.isfile(file_path))
# self.assertEqual(bytes_written, os.path.getsize(file_path)) # because of compression, the number of bytes doesn't match
parquet_dset = pd.read_parquet(file_path)
self.assertEqual(parquet_dset.shape, dset.shape)
self.assertListEqual(list(parquet_dset.columns), list(dset.column_names))
# File buffer argument
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
file_path = os.path.join(tmp_dir, "test_buffer.parquet")
with open(file_path, "wb+") as buffer:
dset.to_parquet(path_or_buf=buffer)
self.assertTrue(os.path.isfile(file_path))
# self.assertEqual(bytes_written, os.path.getsize(file_path)) # because of compression, the number of bytes doesn't match
parquet_dset = pd.read_parquet(file_path)
self.assertEqual(parquet_dset.shape, dset.shape)
self.assertListEqual(list(parquet_dset.columns), list(dset.column_names))
# After a select/shuffle transform
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
dset = dset.select(range(0, len(dset), 2)).shuffle()
file_path = os.path.join(tmp_dir, "test_path.parquet")
dset.to_parquet(path_or_buf=file_path)
self.assertTrue(os.path.isfile(file_path))
# self.assertEqual(bytes_written, os.path.getsize(file_path)) # because of compression, the number of bytes doesn't match
parquet_dset = pd.read_parquet(file_path)
self.assertEqual(parquet_dset.shape, dset.shape)
self.assertListEqual(list(parquet_dset.columns), list(dset.column_names))
# With array features
with self._create_dummy_dataset(in_memory, tmp_dir, array_features=True) as dset:
file_path = os.path.join(tmp_dir, "test_path.parquet")
dset.to_parquet(path_or_buf=file_path)
self.assertTrue(os.path.isfile(file_path))
# self.assertEqual(bytes_written, os.path.getsize(file_path)) # because of compression, the number of bytes doesn't match
parquet_dset = pd.read_parquet(file_path)
self.assertEqual(parquet_dset.shape, dset.shape)
self.assertListEqual(list(parquet_dset.columns), list(dset.column_names))
@require_sqlalchemy
def test_to_sql(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
# Destionation specified as database URI string
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
file_path = os.path.join(tmp_dir, "test_path.sqlite")
_ = dset.to_sql("data", "sqlite:///" + file_path)
self.assertTrue(os.path.isfile(file_path))
sql_dset = pd.read_sql("data", "sqlite:///" + file_path)
self.assertEqual(sql_dset.shape, dset.shape)
self.assertListEqual(list(sql_dset.columns), list(dset.column_names))
# Destionation specified as sqlite3 connection
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
import sqlite3
file_path = os.path.join(tmp_dir, "test_path.sqlite")
with contextlib.closing(sqlite3.connect(file_path)) as con:
_ = dset.to_sql("data", con, if_exists="replace")
self.assertTrue(os.path.isfile(file_path))
sql_dset = pd.read_sql("data", "sqlite:///" + file_path)
self.assertEqual(sql_dset.shape, dset.shape)
self.assertListEqual(list(sql_dset.columns), list(dset.column_names))
# Test writing to a database in chunks
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
file_path = os.path.join(tmp_dir, "test_path.sqlite")
_ = dset.to_sql("data", "sqlite:///" + file_path, batch_size=1, if_exists="replace")
self.assertTrue(os.path.isfile(file_path))
sql_dset = pd.read_sql("data", "sqlite:///" + file_path)
self.assertEqual(sql_dset.shape, dset.shape)
self.assertListEqual(list(sql_dset.columns), list(dset.column_names))
# After a select/shuffle transform
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
dset = dset.select(range(0, len(dset), 2)).shuffle()
file_path = os.path.join(tmp_dir, "test_path.sqlite")
_ = dset.to_sql("data", "sqlite:///" + file_path, if_exists="replace")
self.assertTrue(os.path.isfile(file_path))
sql_dset = pd.read_sql("data", "sqlite:///" + file_path)
self.assertEqual(sql_dset.shape, dset.shape)
self.assertListEqual(list(sql_dset.columns), list(dset.column_names))
# With array features
with self._create_dummy_dataset(in_memory, tmp_dir, array_features=True) as dset:
file_path = os.path.join(tmp_dir, "test_path.sqlite")
_ = dset.to_sql("data", "sqlite:///" + file_path, if_exists="replace")
self.assertTrue(os.path.isfile(file_path))
sql_dset = pd.read_sql("data", "sqlite:///" + file_path)
self.assertEqual(sql_dset.shape, dset.shape)
self.assertListEqual(list(sql_dset.columns), list(dset.column_names))
def test_train_test_split(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
fingerprint = dset._fingerprint
dset_dict = dset.train_test_split(test_size=10, shuffle=False)
self.assertListEqual(list(dset_dict.keys()), ["train", "test"])
dset_train = dset_dict["train"]
dset_test = dset_dict["test"]
self.assertEqual(len(dset_train), 20)
self.assertEqual(len(dset_test), 10)
self.assertEqual(dset_train[0]["filename"], "my_name-train_0")
self.assertEqual(dset_train[-1]["filename"], "my_name-train_19")
self.assertEqual(dset_test[0]["filename"], "my_name-train_20")
self.assertEqual(dset_test[-1]["filename"], "my_name-train_29")
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_train.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_test.features, Features({"filename": Value("string")}))
self.assertNotEqual(dset_train._fingerprint, fingerprint)
self.assertNotEqual(dset_test._fingerprint, fingerprint)
self.assertNotEqual(dset_train._fingerprint, dset_test._fingerprint)
dset_dict = dset.train_test_split(test_size=0.5, shuffle=False)
self.assertListEqual(list(dset_dict.keys()), ["train", "test"])
dset_train = dset_dict["train"]
dset_test = dset_dict["test"]
self.assertEqual(len(dset_train), 15)
self.assertEqual(len(dset_test), 15)
self.assertEqual(dset_train[0]["filename"], "my_name-train_0")
self.assertEqual(dset_train[-1]["filename"], "my_name-train_14")
self.assertEqual(dset_test[0]["filename"], "my_name-train_15")
self.assertEqual(dset_test[-1]["filename"], "my_name-train_29")
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_train.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_test.features, Features({"filename": Value("string")}))
dset_dict = dset.train_test_split(train_size=10, shuffle=False)
self.assertListEqual(list(dset_dict.keys()), ["train", "test"])
dset_train = dset_dict["train"]
dset_test = dset_dict["test"]
self.assertEqual(len(dset_train), 10)
self.assertEqual(len(dset_test), 20)
self.assertEqual(dset_train[0]["filename"], "my_name-train_0")
self.assertEqual(dset_train[-1]["filename"], "my_name-train_9")
self.assertEqual(dset_test[0]["filename"], "my_name-train_10")
self.assertEqual(dset_test[-1]["filename"], "my_name-train_29")
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_train.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_test.features, Features({"filename": Value("string")}))
dset.set_format("numpy")
dset_dict = dset.train_test_split(train_size=10, seed=42)
self.assertListEqual(list(dset_dict.keys()), ["train", "test"])
dset_train = dset_dict["train"]
dset_test = dset_dict["test"]
self.assertEqual(len(dset_train), 10)
self.assertEqual(len(dset_test), 20)
self.assertEqual(dset_train.format["type"], "numpy")
self.assertEqual(dset_test.format["type"], "numpy")
self.assertNotEqual(dset_train[0]["filename"].item(), "my_name-train_0")
self.assertNotEqual(dset_train[-1]["filename"].item(), "my_name-train_9")
self.assertNotEqual(dset_test[0]["filename"].item(), "my_name-train_10")
self.assertNotEqual(dset_test[-1]["filename"].item(), "my_name-train_29")
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_train.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_test.features, Features({"filename": Value("string")}))
del dset_test, dset_train, dset_dict # DatasetDict
def test_shard(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir, self._create_dummy_dataset(in_memory, tmp_dir) as dset:
tmp_file = os.path.join(tmp_dir, "test.arrow")
with dset.select(range(10), indices_cache_file_name=tmp_file) as dset:
self.assertEqual(len(dset), 10)
# Shard
tmp_file_1 = os.path.join(tmp_dir, "test_1.arrow")
fingerprint = dset._fingerprint
with dset.shard(num_shards=8, index=1, indices_cache_file_name=tmp_file_1) as dset_sharded:
self.assertEqual(2, len(dset_sharded))
self.assertEqual(["my_name-train_1", "my_name-train_9"], dset_sharded["filename"])
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_sharded.features, Features({"filename": Value("string")}))
self.assertNotEqual(dset_sharded._fingerprint, fingerprint)
# Shard contiguous
tmp_file_2 = os.path.join(tmp_dir, "test_2.arrow")
with dset.shard(
num_shards=3, index=0, contiguous=True, indices_cache_file_name=tmp_file_2
) as dset_sharded_contiguous:
self.assertEqual([f"my_name-train_{i}" for i in (0, 1, 2, 3)], dset_sharded_contiguous["filename"])
self.assertDictEqual(dset.features, Features({"filename": Value("string")}))
self.assertDictEqual(dset_sharded_contiguous.features, Features({"filename": Value("string")}))
# Test lengths of sharded contiguous
self.assertEqual(
[4, 3, 3],
[
len(dset.shard(3, index=i, contiguous=True, indices_cache_file_name=tmp_file_2 + str(i)))
for i in range(3)
],
)
# formatted
dset.set_format("numpy")
with dset.shard(num_shards=3, index=0) as dset_sharded_formatted:
self.assertEqual(dset_sharded_formatted.format["type"], "numpy")
def test_flatten_indices(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
self.assertIsNone(dset._indices)
tmp_file = os.path.join(tmp_dir, "test.arrow")
with dset.select(range(0, 10, 2), indices_cache_file_name=tmp_file) as dset:
self.assertEqual(len(dset), 5)
self.assertIsNotNone(dset._indices)
tmp_file_2 = os.path.join(tmp_dir, "test_2.arrow")
fingerprint = dset._fingerprint
dset.set_format("numpy")
with dset.flatten_indices(cache_file_name=tmp_file_2) as dset:
self.assertEqual(len(dset), 5)
self.assertEqual(len(dset.data), len(dset))
self.assertIsNone(dset._indices)
self.assertNotEqual(dset._fingerprint, fingerprint)
self.assertEqual(dset.format["type"], "numpy")
# Test unique works
dset.unique(dset.column_names[0])
assert_arrow_metadata_are_synced_with_dataset_features(dset)
# Empty indices mapping
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir) as dset:
self.assertIsNone(dset._indices, None)
tmp_file = os.path.join(tmp_dir, "test.arrow")
with dset.filter(lambda _: False, cache_file_name=tmp_file) as dset:
self.assertEqual(len(dset), 0)
self.assertIsNotNone(dset._indices, None)
tmp_file_2 = os.path.join(tmp_dir, "test_2.arrow")
fingerprint = dset._fingerprint
dset.set_format("numpy")
with dset.flatten_indices(cache_file_name=tmp_file_2) as dset:
self.assertEqual(len(dset), 0)
self.assertEqual(len(dset.data), len(dset))
self.assertIsNone(dset._indices, None)
self.assertNotEqual(dset._fingerprint, fingerprint)
self.assertEqual(dset.format["type"], "numpy")
# Test unique works
dset.unique(dset.column_names[0])
assert_arrow_metadata_are_synced_with_dataset_features(dset)
@require_tf
@require_torch
def test_format_vectors(self, in_memory):
import numpy as np
import tensorflow as tf
import torch
with tempfile.TemporaryDirectory() as tmp_dir, self._create_dummy_dataset(
in_memory, tmp_dir
) as dset, dset.map(lambda ex, i: {"vec": np.ones(3) * i}, with_indices=True) as dset:
columns = dset.column_names
self.assertIsNotNone(dset[0])
self.assertIsNotNone(dset[:2])
for col in columns:
self.assertIsInstance(dset[0][col], (str, list))
self.assertIsInstance(dset[:2][col], list)
self.assertDictEqual(
dset.features, Features({"filename": Value("string"), "vec": Sequence(Value("float64"))})
)
dset.set_format("tensorflow")
self.assertIsNotNone(dset[0])
self.assertIsNotNone(dset[:2])
for col in columns:
self.assertIsInstance(dset[0][col], (tf.Tensor, tf.RaggedTensor))
self.assertIsInstance(dset[:2][col], (tf.Tensor, tf.RaggedTensor))
self.assertIsInstance(dset[col], (tf.Tensor, tf.RaggedTensor))
self.assertTupleEqual(tuple(dset[:2]["vec"].shape), (2, 3))
self.assertTupleEqual(tuple(dset["vec"][:2].shape), (2, 3))
dset.set_format("numpy")
self.assertIsNotNone(dset[0])
self.assertIsNotNone(dset[:2])
self.assertIsInstance(dset[0]["filename"], np.str_)
self.assertIsInstance(dset[:2]["filename"], np.ndarray)
self.assertIsInstance(dset["filename"], np.ndarray)
self.assertIsInstance(dset[0]["vec"], np.ndarray)
self.assertIsInstance(dset[:2]["vec"], np.ndarray)
self.assertIsInstance(dset["vec"], np.ndarray)
self.assertTupleEqual(dset[:2]["vec"].shape, (2, 3))
self.assertTupleEqual(dset["vec"][:2].shape, (2, 3))
dset.set_format("torch", columns=["vec"])
self.assertIsNotNone(dset[0])
self.assertIsNotNone(dset[:2])
# torch.Tensor is only for numerical columns
self.assertIsInstance(dset[0]["vec"], torch.Tensor)
self.assertIsInstance(dset[:2]["vec"], torch.Tensor)
self.assertIsInstance(dset["vec"][:2], torch.Tensor)
self.assertTupleEqual(dset[:2]["vec"].shape, (2, 3))
self.assertTupleEqual(dset["vec"][:2].shape, (2, 3))
@require_tf
@require_torch
def test_format_ragged_vectors(self, in_memory):
import numpy as np
import tensorflow as tf
import torch
with tempfile.TemporaryDirectory() as tmp_dir, self._create_dummy_dataset(
in_memory, tmp_dir
) as dset, dset.map(lambda ex, i: {"vec": np.ones(3 + i) * i}, with_indices=True) as dset:
columns = dset.column_names
self.assertIsNotNone(dset[0])
self.assertIsNotNone(dset[:2])
for col in columns:
self.assertIsInstance(dset[0][col], (str, list))
self.assertIsInstance(dset[:2][col], list)
self.assertDictEqual(
dset.features, Features({"filename": Value("string"), "vec": Sequence(Value("float64"))})
)
dset.set_format("tensorflow")
self.assertIsNotNone(dset[0])
self.assertIsNotNone(dset[:2])
for col in columns:
self.assertIsInstance(dset[0][col], tf.Tensor)
self.assertIsInstance(dset[:2][col], tf.RaggedTensor if col == "vec" else tf.Tensor)
self.assertIsInstance(dset[col], tf.RaggedTensor if col == "vec" else tf.Tensor)
# dim is None for ragged vectors in tensorflow
self.assertListEqual(dset[:2]["vec"].shape.as_list(), [2, None])
self.assertListEqual(dset["vec"][:2].shape.as_list(), [2, None])
dset.set_format("numpy")
self.assertIsNotNone(dset[0])
self.assertIsNotNone(dset[:2])
self.assertIsInstance(dset[0]["filename"], np.str_)
self.assertIsInstance(dset[:2]["filename"], np.ndarray)
self.assertIsInstance(dset["filename"], np.ndarray)
self.assertIsInstance(dset[0]["vec"], np.ndarray)
self.assertIsInstance(dset[:2]["vec"], np.ndarray)
self.assertIsInstance(dset["vec"], np.ndarray)
# array is flat for ragged vectors in numpy
self.assertTupleEqual(dset[:2]["vec"].shape, (2,))
self.assertTupleEqual(dset["vec"][:2].shape, (2,))
dset.set_format("torch")
self.assertIsNotNone(dset[0])
self.assertIsNotNone(dset[:2])
self.assertIsInstance(dset[0]["filename"], str)
self.assertIsInstance(dset[:2]["filename"], list)
self.assertIsInstance(dset["filename"], list)
self.assertIsInstance(dset[0]["vec"], torch.Tensor)
self.assertIsInstance(dset[:2]["vec"][0], torch.Tensor)
self.assertIsInstance(dset["vec"][0], torch.Tensor)
# pytorch doesn't support ragged tensors, so we should have lists
self.assertIsInstance(dset[:2]["vec"], list)
self.assertIsInstance(dset[:2]["vec"][0], torch.Tensor)
self.assertIsInstance(dset["vec"][:2], list)
self.assertIsInstance(dset["vec"][0], torch.Tensor)
@require_tf
@require_torch
def test_format_nested(self, in_memory):
import numpy as np
import tensorflow as tf
import torch
with tempfile.TemporaryDirectory() as tmp_dir, self._create_dummy_dataset(
in_memory, tmp_dir
) as dset, dset.map(lambda ex: {"nested": [{"foo": np.ones(3)}] * len(ex["filename"])}, batched=True) as dset:
self.assertDictEqual(
dset.features, Features({"filename": Value("string"), "nested": {"foo": Sequence(Value("float64"))}})
)
dset.set_format("tensorflow")
self.assertIsNotNone(dset[0])
self.assertIsInstance(dset[0]["nested"]["foo"], (tf.Tensor, tf.RaggedTensor))
self.assertIsNotNone(dset[:2])
self.assertIsInstance(dset[:2]["nested"][0]["foo"], (tf.Tensor, tf.RaggedTensor))
self.assertIsInstance(dset["nested"][0]["foo"], (tf.Tensor, tf.RaggedTensor))
dset.set_format("numpy")
self.assertIsNotNone(dset[0])
self.assertIsInstance(dset[0]["nested"]["foo"], np.ndarray)
self.assertIsNotNone(dset[:2])
self.assertIsInstance(dset[:2]["nested"][0]["foo"], np.ndarray)
self.assertIsInstance(dset["nested"][0]["foo"], np.ndarray)
dset.set_format("torch", columns="nested")
self.assertIsNotNone(dset[0])
self.assertIsInstance(dset[0]["nested"]["foo"], torch.Tensor)
self.assertIsNotNone(dset[:2])
self.assertIsInstance(dset[:2]["nested"][0]["foo"], torch.Tensor)
self.assertIsInstance(dset["nested"][0]["foo"], torch.Tensor)
def test_format_pandas(self, in_memory):
import pandas as pd
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
dset.set_format("pandas")
self.assertIsInstance(dset[0], pd.DataFrame)
self.assertIsInstance(dset[:2], pd.DataFrame)
self.assertIsInstance(dset["col_1"], pd.Series)
def test_transmit_format_single(self, in_memory):
@transmit_format
def my_single_transform(self, return_factory, *args, **kwargs):
return return_factory()
with tempfile.TemporaryDirectory() as tmp_dir:
return_factory = partial(
self._create_dummy_dataset, in_memory=in_memory, tmp_dir=tmp_dir, multiple_columns=True
)
with return_factory() as dset:
dset.set_format("numpy", columns=["col_1"])
prev_format = dset.format
with my_single_transform(dset, return_factory) as transformed_dset:
self.assertDictEqual(transformed_dset.format, prev_format)
def test_transmit_format_dict(self, in_memory):
@transmit_format
def my_split_transform(self, return_factory, *args, **kwargs):
return DatasetDict({"train": return_factory()})
with tempfile.TemporaryDirectory() as tmp_dir:
return_factory = partial(
self._create_dummy_dataset, in_memory=in_memory, tmp_dir=tmp_dir, multiple_columns=True
)
with return_factory() as dset:
dset.set_format("numpy", columns=["col_1"])
prev_format = dset.format
transformed_dset = my_split_transform(dset, return_factory)["train"]
self.assertDictEqual(transformed_dset.format, prev_format)
del transformed_dset # DatasetDict
def test_with_format(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
with dset.with_format("numpy", columns=["col_1"]) as dset2:
dset.set_format("numpy", columns=["col_1"])
self.assertDictEqual(dset.format, dset2.format)
self.assertEqual(dset._fingerprint, dset2._fingerprint)
# dset.reset_format()
# self.assertNotEqual(dset.format, dset2.format)
# self.assertNotEqual(dset._fingerprint, dset2._fingerprint)
def test_with_transform(self, in_memory):
with tempfile.TemporaryDirectory() as tmp_dir:
with self._create_dummy_dataset(in_memory, tmp_dir, multiple_columns=True) as dset:
transform = lambda x: {"foo": x["col_1"]} # noqa: E731
with dset.with_transform(transform, columns=["col_1"]) as dset2:
dset.set_transform(transform, columns=["col_1"])
self.assertDictEqual(dset.format, dset2.format)
self.assertEqual(dset._fingerprint, dset2._fingerprint)
dset.reset_format()
self.assertNotEqual(dset.format, dset2.format)
self.assertNotEqual(dset._fingerprint, dset2._fingerprint)
@require_tf
def test_tf_dataset_conversion(self, in_memory):
tmp_dir = tempfile.TemporaryDirectory()
for num_workers in [0, 1, 2]:
if num_workers > 0 and sys.platform == "win32" and not in_memory:
continue # This test hangs on the Py3.10 test worker, but it runs fine locally on my Windows machine
with self._create_dummy_dataset(in_memory, tmp_dir.name, array_features=True) as dset:
tf_dataset = dset.to_tf_dataset(columns="col_3", batch_size=2, num_workers=num_workers)
batch = next(iter(tf_dataset))
self.assertEqual(batch.shape.as_list(), [2, 4])
self.assertEqual(batch.dtype.name, "int64")
with self._create_dummy_dataset(in_memory, tmp_dir.name, multiple_columns=True) as dset:
tf_dataset = dset.to_tf_dataset(columns="col_1", batch_size=2, num_workers=num_workers)
batch = next(iter(tf_dataset))
self.assertEqual(batch.shape.as_list(), [2])
self.assertEqual(batch.dtype.name, "int64")
with self._create_dummy_dataset(in_memory, tmp_dir.name, multiple_columns=True) as dset:
# Check that it works with all default options (except batch_size because the dummy dataset only has 4)
tf_dataset = dset.to_tf_dataset(batch_size=2, num_workers=num_workers)
batch = next(iter(tf_dataset))
self.assertEqual(batch["col_1"].shape.as_list(), [2])
self.assertEqual(batch["col_2"].shape.as_list(), [2])
self.assertEqual(batch["col_1"].dtype.name, "int64")
self.assertEqual(batch["col_2"].dtype.name, "string") # Assert that we're converting strings properly
with self._create_dummy_dataset(in_memory, tmp_dir.name, multiple_columns=True) as dset:
# Check that when we use a transform that creates a new column from existing column values
# but don't load the old columns that the new column depends on in the final dataset,
# that they're still kept around long enough to be used in the transform
transform_dset = dset.with_transform(
lambda x: {"new_col": [val * 2 for val in x["col_1"]], "col_1": x["col_1"]}
)
tf_dataset = transform_dset.to_tf_dataset(columns="new_col", batch_size=2, num_workers=num_workers)
batch = next(iter(tf_dataset))
self.assertEqual(batch.shape.as_list(), [2])
self.assertEqual(batch.dtype.name, "int64")
del transform_dset
del tf_dataset # For correct cleanup
@require_tf
def test_tf_index_reshuffling(self, in_memory):
# This test checks that when we do two epochs over a tf.data.Dataset from to_tf_dataset
# that we get a different shuffle order each time
# It also checks that when we aren't shuffling, that the dataset order is fully preserved
# even when loading is split across multiple workers
data = {"col_1": list(range(20))}
for num_workers in [0, 1, 2, 3]:
with Dataset.from_dict(data) as dset:
tf_dataset = dset.to_tf_dataset(batch_size=10, shuffle=True, num_workers=num_workers)
indices = []
for batch in tf_dataset:
indices.append(batch["col_1"])
indices = np.concatenate([arr.numpy() for arr in indices])
second_indices = []
for batch in tf_dataset:
second_indices.append(batch["col_1"])
second_indices = np.concatenate([arr.numpy() for arr in second_indices])
self.assertFalse(np.array_equal(indices, second_indices))
self.assertEqual(len(indices), len(np.unique(indices)))
self.assertEqual(len(second_indices), len(np.unique(second_indices)))
tf_dataset = dset.to_tf_dataset(batch_size=1, shuffle=False, num_workers=num_workers)
for i, batch in enumerate(tf_dataset):
# Assert that the unshuffled order is fully preserved even when multiprocessing
self.assertEqual(i, batch["col_1"].numpy())
@require_tf
def test_tf_label_renaming(self, in_memory):
# Protect TF-specific imports in here
import tensorflow as tf
from datasets.utils.tf_utils import minimal_tf_collate_fn_with_renaming
tmp_dir = tempfile.TemporaryDirectory()
with self._create_dummy_dataset(in_memory, tmp_dir.name, multiple_columns=True) as dset:
with dset.rename_columns({"col_1": "features", "col_2": "label"}) as new_dset:
tf_dataset = new_dset.to_tf_dataset(collate_fn=minimal_tf_collate_fn_with_renaming, batch_size=4)
batch = next(iter(tf_dataset))
self.assertTrue("labels" in batch and "features" in batch)
tf_dataset = new_dset.to_tf_dataset(
columns=["features", "labels"], collate_fn=minimal_tf_collate_fn_with_renaming, batch_size=4
)
batch = next(iter(tf_dataset))
self.assertTrue("labels" in batch and "features" in batch)
tf_dataset = new_dset.to_tf_dataset(
columns=["features", "label"], collate_fn=minimal_tf_collate_fn_with_renaming, batch_size=4
)
batch = next(iter(tf_dataset))
self.assertTrue("labels" in batch and "features" in batch) # Assert renaming was handled correctly
tf_dataset = new_dset.to_tf_dataset(
columns=["features"],
label_cols=["labels"],
collate_fn=minimal_tf_collate_fn_with_renaming,
batch_size=4,
)
batch = next(iter(tf_dataset))
self.assertEqual(len(batch), 2)
# Assert that we don't have any empty entries here
self.assertTrue(isinstance(batch[0], tf.Tensor) and isinstance(batch[1], tf.Tensor))
tf_dataset = new_dset.to_tf_dataset(
columns=["features"],
label_cols=["label"],
collate_fn=minimal_tf_collate_fn_with_renaming,
batch_size=4,
)
batch = next(iter(tf_dataset))
self.assertEqual(len(batch), 2)
# Assert that we don't have any empty entries here
self.assertTrue(isinstance(batch[0], tf.Tensor) and isinstance(batch[1], tf.Tensor))
tf_dataset = new_dset.to_tf_dataset(
columns=["features"],
collate_fn=minimal_tf_collate_fn_with_renaming,
batch_size=4,
)
batch = next(iter(tf_dataset))
# Assert that labels didn't creep in when we don't ask for them
# just because the collate_fn added them
self.assertTrue(isinstance(batch, tf.Tensor))
del tf_dataset # For correct cleanup
@require_tf
def test_tf_dataset_options(self, in_memory):
tmp_dir = tempfile.TemporaryDirectory()
# Test that batch_size option works as expected
with self._create_dummy_dataset(in_memory, tmp_dir.name, array_features=True) as dset:
tf_dataset = dset.to_tf_dataset(columns="col_3", batch_size=2)
batch = next(iter(tf_dataset))
self.assertEqual(batch.shape.as_list(), [2, 4])
self.assertEqual(batch.dtype.name, "int64")
# Test that batch_size=None (optional) works as expected
with self._create_dummy_dataset(in_memory, tmp_dir.name, multiple_columns=True) as dset:
tf_dataset = dset.to_tf_dataset(columns="col_3", batch_size=None)
single_example = next(iter(tf_dataset))
self.assertEqual(single_example.shape.as_list(), [])
self.assertEqual(single_example.dtype.name, "int64")
# Assert that we can batch it with `tf.data.Dataset.batch` method
batched_dataset = tf_dataset.batch(batch_size=2)
batch = next(iter(batched_dataset))
self.assertEqual(batch.shape.as_list(), [2])
self.assertEqual(batch.dtype.name, "int64")
# Test that batching a batch_size=None dataset produces the same results as using batch_size arg
with self._create_dummy_dataset(in_memory, tmp_dir.name, multiple_columns=True) as dset:
batch_size = 2
tf_dataset_no_batch = dset.to_tf_dataset(columns="col_3")
tf_dataset_batch = dset.to_tf_dataset(columns="col_3", batch_size=batch_size)
self.assertEqual(tf_dataset_no_batch.element_spec, tf_dataset_batch.unbatch().element_spec)
self.assertEqual(tf_dataset_no_batch.cardinality(), tf_dataset_batch.cardinality() * batch_size)
for batch_1, batch_2 in zip(tf_dataset_no_batch.batch(batch_size=batch_size), tf_dataset_batch):
self.assertEqual(batch_1.shape, batch_2.shape)
self.assertEqual(batch_1.dtype, batch_2.dtype)
self.assertListEqual(batch_1.numpy().tolist(), batch_2.numpy().tolist())
# Test that requesting label_cols works as expected
with self._create_dummy_dataset(in_memory, tmp_dir.name, multiple_columns=True) as dset:
tf_dataset = dset.to_tf_dataset(columns="col_1", label_cols=["col_2", "col_3"], batch_size=4)
batch = next(iter(tf_dataset))
self.assertEqual(len(batch), 2)
self.assertEqual(set(batch[1].keys()), {"col_2", "col_3"})
self.assertEqual(batch[0].dtype.name, "int64")
# Assert data comes out as expected and isn't shuffled
self.assertEqual(batch[0].numpy().tolist(), [3, 2, 1, 0])
self.assertEqual(batch[1]["col_2"].numpy().tolist(), [b"a", b"b", b"c", b"d"])
self.assertEqual(batch[1]["col_3"].numpy().tolist(), [0, 1, 0, 1])
# Check that incomplete batches are dropped if requested
with self._create_dummy_dataset(in_memory, tmp_dir.name, multiple_columns=True) as dset:
tf_dataset = dset.to_tf_dataset(columns="col_1", batch_size=3)
tf_dataset_with_drop = dset.to_tf_dataset(columns="col_1", batch_size=3, drop_remainder=True)
self.assertEqual(len(tf_dataset), 2) # One batch of 3 and one batch of 1
self.assertEqual(len(tf_dataset_with_drop), 1) # Incomplete batch of 1 is dropped
# Test that `NotImplementedError` is raised `batch_size` is None and `num_workers` is > 0
if sys.version_info >= (3, 8):
with self._create_dummy_dataset(in_memory, tmp_dir.name, multiple_columns=True) as dset:
with self.assertRaisesRegex(
NotImplementedError, "`batch_size` must be specified when using multiple workers"
):
dset.to_tf_dataset(columns="col_1", batch_size=None, num_workers=2)
del tf_dataset # For correct cleanup
del tf_dataset_with_drop
class MiscellaneousDatasetTest(TestCase):
def test_from_pandas(self):
data = {"col_1": [3, 2, 1, 0], "col_2": ["a", "b", "c", "d"]}
df = pd.DataFrame.from_dict(data)
with Dataset.from_pandas(df) as dset:
self.assertListEqual(dset["col_1"], data["col_1"])
self.assertListEqual(dset["col_2"], data["col_2"])
self.assertListEqual(list(dset.features.keys()), ["col_1", "col_2"])
self.assertDictEqual(dset.features, Features({"col_1": Value("int64"), "col_2": Value("string")}))
features = Features({"col_1": Value("int64"), "col_2": Value("string")})
with Dataset.from_pandas(df, features=features) as dset:
self.assertListEqual(dset["col_1"], data["col_1"])
self.assertListEqual(dset["col_2"], data["col_2"])
self.assertListEqual(list(dset.features.keys()), ["col_1", "col_2"])
self.assertDictEqual(dset.features, Features({"col_1": Value("int64"), "col_2": Value("string")}))
features = Features({"col_1": Value("int64"), "col_2": Value("string")})
with Dataset.from_pandas(df, features=features, info=DatasetInfo(features=features)) as dset:
self.assertListEqual(dset["col_1"], data["col_1"])
self.assertListEqual(dset["col_2"], data["col_2"])
self.assertListEqual(list(dset.features.keys()), ["col_1", "col_2"])
self.assertDictEqual(dset.features, Features({"col_1": Value("int64"), "col_2": Value("string")}))
features = Features({"col_1": Sequence(Value("string")), "col_2": Value("string")})
self.assertRaises(TypeError, Dataset.from_pandas, df, features=features)
def test_from_dict(self):
data = {"col_1": [3, 2, 1, 0], "col_2": ["a", "b", "c", "d"], "col_3": pa.array([True, False, True, False])}
with Dataset.from_dict(data) as dset:
self.assertListEqual(dset["col_1"], data["col_1"])
self.assertListEqual(dset["col_2"], data["col_2"])
self.assertListEqual(dset["col_3"], data["col_3"].to_pylist())
self.assertListEqual(list(dset.features.keys()), ["col_1", "col_2", "col_3"])
self.assertDictEqual(
dset.features, Features({"col_1": Value("int64"), "col_2": Value("string"), "col_3": Value("bool")})
)
features = Features({"col_1": Value("int64"), "col_2": Value("string"), "col_3": Value("bool")})
with Dataset.from_dict(data, features=features) as dset:
self.assertListEqual(dset["col_1"], data["col_1"])
self.assertListEqual(dset["col_2"], data["col_2"])
self.assertListEqual(dset["col_3"], data["col_3"].to_pylist())
self.assertListEqual(list(dset.features.keys()), ["col_1", "col_2", "col_3"])
self.assertDictEqual(
dset.features, Features({"col_1": Value("int64"), "col_2": Value("string"), "col_3": Value("bool")})
)
features = Features({"col_1": Value("int64"), "col_2": Value("string"), "col_3": Value("bool")})
with Dataset.from_dict(data, features=features, info=DatasetInfo(features=features)) as dset:
self.assertListEqual(dset["col_1"], data["col_1"])
self.assertListEqual(dset["col_2"], data["col_2"])
self.assertListEqual(dset["col_3"], data["col_3"].to_pylist())
self.assertListEqual(list(dset.features.keys()), ["col_1", "col_2", "col_3"])
self.assertDictEqual(
dset.features, Features({"col_1": Value("int64"), "col_2": Value("string"), "col_3": Value("bool")})
)
features = Features({"col_1": Value("string"), "col_2": Value("string"), "col_3": Value("int32")})
with Dataset.from_dict(data, features=features) as dset:
# the integers are converted to strings
self.assertListEqual(dset["col_1"], [str(x) for x in data["col_1"]])
self.assertListEqual(dset["col_2"], data["col_2"])
self.assertListEqual(dset["col_3"], [int(x) for x in data["col_3"].to_pylist()])
self.assertListEqual(list(dset.features.keys()), ["col_1", "col_2", "col_3"])
self.assertDictEqual(
dset.features, Features({"col_1": Value("string"), "col_2": Value("string"), "col_3": Value("int32")})
)
features = Features({"col_1": Value("int64"), "col_2": Value("int64"), "col_3": Value("bool")})
self.assertRaises(ValueError, Dataset.from_dict, data, features=features)
def test_concatenate_mixed_memory_and_disk(self):
data1, data2, data3 = {"id": [0, 1, 2]}, {"id": [3, 4, 5]}, {"id": [6, 7]}
info1 = DatasetInfo(description="Dataset1")
info2 = DatasetInfo(description="Dataset2")
with tempfile.TemporaryDirectory() as tmp_dir:
with Dataset.from_dict(data1, info=info1).map(
cache_file_name=os.path.join(tmp_dir, "d1.arrow")
) as dset1, Dataset.from_dict(data2, info=info2).map(
cache_file_name=os.path.join(tmp_dir, "d2.arrow")
) as dset2, Dataset.from_dict(
data3
) as dset3:
with concatenate_datasets([dset1, dset2, dset3]) as concatenated_dset:
self.assertEqual(len(concatenated_dset), len(dset1) + len(dset2) + len(dset3))
self.assertListEqual(concatenated_dset["id"], dset1["id"] + dset2["id"] + dset3["id"])
@require_transformers
@pytest.mark.integration
def test_set_format_encode(self):
from transformers import BertTokenizer
tokenizer = BertTokenizer.from_pretrained("bert-base-uncased")
def encode(batch):
return tokenizer(batch["text"], padding="longest", return_tensors="np")
with Dataset.from_dict({"text": ["hello there", "foo"]}) as dset:
dset.set_transform(transform=encode)
self.assertEqual(str(dset[:2]), str(encode({"text": ["hello there", "foo"]})))
@require_tf
def test_tf_string_encoding(self):
data = {"col_1": ["á", "é", "í", "ó", "ú"], "col_2": ["à", "è", "ì", "ò", "ù"]}
with Dataset.from_dict(data) as dset:
tf_dset_wo_batch = dset.to_tf_dataset(columns=["col_1", "col_2"])
for tf_row, row in zip(tf_dset_wo_batch, dset):
self.assertEqual(tf_row["col_1"].numpy().decode("utf-8"), row["col_1"])
self.assertEqual(tf_row["col_2"].numpy().decode("utf-8"), row["col_2"])
tf_dset_w_batch = dset.to_tf_dataset(columns=["col_1", "col_2"], batch_size=2)
for tf_row, row in zip(tf_dset_w_batch.unbatch(), dset):
self.assertEqual(tf_row["col_1"].numpy().decode("utf-8"), row["col_1"])
self.assertEqual(tf_row["col_2"].numpy().decode("utf-8"), row["col_2"])
self.assertEqual(tf_dset_w_batch.unbatch().element_spec, tf_dset_wo_batch.element_spec)
self.assertEqual(tf_dset_w_batch.element_spec, tf_dset_wo_batch.batch(2).element_spec)
def test_cast_with_sliced_list():
old_features = Features({"foo": Sequence(Value("int64"))})
new_features = Features({"foo": Sequence(Value("int32"))})
dataset = Dataset.from_dict({"foo": [[i] * (i % 3) for i in range(20)]}, features=old_features)
casted_dataset = dataset.cast(new_features, batch_size=2) # small batch size to slice the ListArray
assert dataset["foo"] == casted_dataset["foo"]
assert casted_dataset.features == new_features
@pytest.mark.parametrize("include_nulls", [False, True])
def test_class_encode_column_with_none(include_nulls):
dataset = Dataset.from_dict({"col_1": ["a", "b", "c", None, "d", None]})
dataset = dataset.class_encode_column("col_1", include_nulls=include_nulls)
class_names = ["a", "b", "c", "d"]
if include_nulls:
class_names += ["None"]
assert isinstance(dataset.features["col_1"], ClassLabel)
assert set(dataset.features["col_1"].names) == set(class_names)
assert (None in dataset.unique("col_1")) == (not include_nulls)
@pytest.mark.parametrize("null_placement", ["first", "last"])
def test_sort_with_none(null_placement):
dataset = Dataset.from_dict({"col_1": ["item_2", "item_3", "item_1", None, "item_4", None]})
dataset = dataset.sort("col_1", null_placement=null_placement)
if null_placement == "first":
assert dataset["col_1"] == [None, None, "item_1", "item_2", "item_3", "item_4"]
else:
assert dataset["col_1"] == ["item_1", "item_2", "item_3", "item_4", None, None]
def test_update_metadata_with_features(dataset_dict):
table1 = pa.Table.from_pydict(dataset_dict)
features1 = Features.from_arrow_schema(table1.schema)
features2 = features1.copy()
features2["col_2"] = ClassLabel(num_classes=len(table1))
assert features1 != features2
table2 = update_metadata_with_features(table1, features2)
metadata = json.loads(table2.schema.metadata[b"huggingface"].decode())
assert features2 == Features.from_dict(metadata["info"]["features"])
with Dataset(table1) as dset1, Dataset(table2) as dset2:
assert dset1.features == features1
assert dset2.features == features2
@pytest.mark.parametrize("dataset_type", ["in_memory", "memory_mapped", "mixed"])
@pytest.mark.parametrize("axis, expected_shape", [(0, (4, 3)), (1, (2, 6))])
def test_concatenate_datasets(dataset_type, axis, expected_shape, dataset_dict, arrow_path):
table = {
"in_memory": InMemoryTable.from_pydict(dataset_dict),
"memory_mapped": MemoryMappedTable.from_file(arrow_path),
}
tables = [
table[dataset_type if dataset_type != "mixed" else "memory_mapped"].slice(0, 2), # shape = (2, 3)
table[dataset_type if dataset_type != "mixed" else "in_memory"].slice(2, 4), # shape = (2, 3)
]
if axis == 1: # don't duplicate columns
tables[1] = tables[1].rename_columns([col + "_bis" for col in tables[1].column_names])
datasets = [Dataset(table) for table in tables]
dataset = concatenate_datasets(datasets, axis=axis)
assert dataset.shape == expected_shape
assert_arrow_metadata_are_synced_with_dataset_features(dataset)
def test_concatenate_datasets_new_columns():
dataset1 = Dataset.from_dict({"col_1": ["a", "b", "c"]})
dataset2 = Dataset.from_dict({"col_1": ["d", "e", "f"], "col_2": [True, False, True]})
dataset = concatenate_datasets([dataset1, dataset2])
assert dataset.data.shape == (6, 2)
assert dataset.features == Features({"col_1": Value("string"), "col_2": Value("bool")})
assert dataset[:] == {"col_1": ["a", "b", "c", "d", "e", "f"], "col_2": [None, None, None, True, False, True]}
dataset3 = Dataset.from_dict({"col_3": ["a_1"]})
dataset = concatenate_datasets([dataset, dataset3])
assert dataset.data.shape == (7, 3)
assert dataset.features == Features({"col_1": Value("string"), "col_2": Value("bool"), "col_3": Value("string")})
assert dataset[:] == {
"col_1": ["a", "b", "c", "d", "e", "f", None],
"col_2": [None, None, None, True, False, True, None],
"col_3": [None, None, None, None, None, None, "a_1"],
}
@pytest.mark.parametrize("axis", [0, 1])
def test_concatenate_datasets_complex_features(axis):
n = 5
dataset1 = Dataset.from_dict(
{"col_1": [0] * n, "col_2": list(range(n))},
features=Features({"col_1": Value("int32"), "col_2": ClassLabel(num_classes=n)}),
)
if axis == 1:
dataset2 = dataset1.rename_columns({col: col + "_" for col in dataset1.column_names})
expected_features = Features({**dataset1.features, **dataset2.features})
else:
dataset2 = dataset1
expected_features = dataset1.features
assert concatenate_datasets([dataset1, dataset2], axis=axis).features == expected_features
@pytest.mark.parametrize("other_dataset_type", ["in_memory", "memory_mapped", "concatenation"])
@pytest.mark.parametrize("axis, expected_shape", [(0, (8, 3)), (1, (4, 6))])
def test_concatenate_datasets_with_concatenation_tables(
axis, expected_shape, other_dataset_type, dataset_dict, arrow_path
):
def _create_concatenation_table(axis):
if axis == 0: # shape: (4, 3) = (4, 1) + (4, 2)
concatenation_table = ConcatenationTable.from_blocks(
[
[
InMemoryTable.from_pydict({"col_1": dataset_dict["col_1"]}),
MemoryMappedTable.from_file(arrow_path).remove_column(0),
]
]
)
elif axis == 1: # shape: (4, 3) = (1, 3) + (3, 3)
concatenation_table = ConcatenationTable.from_blocks(
[
[InMemoryTable.from_pydict(dataset_dict).slice(0, 1)],
[MemoryMappedTable.from_file(arrow_path).slice(1, 4)],
]
)
return concatenation_table
concatenation_table = _create_concatenation_table(axis)
assert concatenation_table.shape == (4, 3)
if other_dataset_type == "in_memory":
other_table = InMemoryTable.from_pydict(dataset_dict)
elif other_dataset_type == "memory_mapped":
other_table = MemoryMappedTable.from_file(arrow_path)
elif other_dataset_type == "concatenation":
other_table = _create_concatenation_table(axis)
assert other_table.shape == (4, 3)
tables = [concatenation_table, other_table]
if axis == 1: # don't duplicate columns
tables[1] = tables[1].rename_columns([col + "_bis" for col in tables[1].column_names])
for tables in [tables, reversed(tables)]:
datasets = [Dataset(table) for table in tables]
dataset = concatenate_datasets(datasets, axis=axis)
assert dataset.shape == expected_shape
def test_concatenate_datasets_duplicate_columns(dataset):
with pytest.raises(ValueError) as excinfo:
concatenate_datasets([dataset, dataset], axis=1)
assert "duplicated" in str(excinfo.value)
def test_interleave_datasets():
d1 = Dataset.from_dict({"a": [0, 1, 2]})
d2 = Dataset.from_dict({"a": [10, 11, 12, 13]})
d3 = Dataset.from_dict({"a": [22, 21, 20]}).select([2, 1, 0])
dataset = interleave_datasets([d1, d2, d3])
expected_length = 3 * min(len(d1), len(d2), len(d3))
expected_values = [x["a"] for x in itertools.chain(*zip(d1, d2, d3))]
assert isinstance(dataset, Dataset)
assert len(dataset) == expected_length
assert dataset["a"] == expected_values
assert dataset._fingerprint == interleave_datasets([d1, d2, d3])._fingerprint
def test_interleave_datasets_probabilities():
seed = 42
probabilities = [0.3, 0.5, 0.2]
d1 = Dataset.from_dict({"a": [0, 1, 2]})
d2 = Dataset.from_dict({"a": [10, 11, 12, 13]})
d3 = Dataset.from_dict({"a": [22, 21, 20]}).select([2, 1, 0])
dataset = interleave_datasets([d1, d2, d3], probabilities=probabilities, seed=seed)
expected_length = 7 # hardcoded
expected_values = [10, 11, 20, 12, 0, 21, 13] # hardcoded
assert isinstance(dataset, Dataset)
assert len(dataset) == expected_length
assert dataset["a"] == expected_values
assert (
dataset._fingerprint == interleave_datasets([d1, d2, d3], probabilities=probabilities, seed=seed)._fingerprint
)
def test_interleave_datasets_oversampling_strategy():
d1 = Dataset.from_dict({"a": [0, 1, 2]})
d2 = Dataset.from_dict({"a": [10, 11, 12, 13]})
d3 = Dataset.from_dict({"a": [22, 21, 20]}).select([2, 1, 0])
dataset = interleave_datasets([d1, d2, d3], stopping_strategy="all_exhausted")
expected_length = 3 * max(len(d1), len(d2), len(d3))
expected_values = [0, 10, 20, 1, 11, 21, 2, 12, 22, 0, 13, 20] # hardcoded
assert isinstance(dataset, Dataset)
assert len(dataset) == expected_length
assert dataset["a"] == expected_values
assert dataset._fingerprint == interleave_datasets([d1, d2, d3], stopping_strategy="all_exhausted")._fingerprint
def test_interleave_datasets_probabilities_oversampling_strategy():
seed = 42
probabilities = [0.3, 0.5, 0.2]
d1 = Dataset.from_dict({"a": [0, 1, 2]})
d2 = Dataset.from_dict({"a": [10, 11, 12, 13]})
d3 = Dataset.from_dict({"a": [22, 21, 20]}).select([2, 1, 0])
dataset = interleave_datasets(
[d1, d2, d3], stopping_strategy="all_exhausted", probabilities=probabilities, seed=seed
)
expected_length = 16 # hardcoded
expected_values = [10, 11, 20, 12, 0, 21, 13, 10, 1, 11, 12, 22, 13, 20, 10, 2] # hardcoded
assert isinstance(dataset, Dataset)
assert len(dataset) == expected_length
assert dataset["a"] == expected_values
assert (
dataset._fingerprint
== interleave_datasets(
[d1, d2, d3], stopping_strategy="all_exhausted", probabilities=probabilities, seed=seed
)._fingerprint
)
@pytest.mark.parametrize("batch_size", [4, 5])
@pytest.mark.parametrize("drop_last_batch", [False, True])
def test_dataset_iter_batch(batch_size, drop_last_batch):
n = 25
dset = Dataset.from_dict({"i": list(range(n))})
all_col_values = list(range(n))
batches = []
for i, batch in enumerate(dset.iter(batch_size, drop_last_batch=drop_last_batch)):
assert batch == {"i": all_col_values[i * batch_size : (i + 1) * batch_size]}
batches.append(batch)
if drop_last_batch:
assert all(len(batch["i"]) == batch_size for batch in batches)
else:
assert all(len(batch["i"]) == batch_size for batch in batches[:-1])
assert len(batches[-1]["i"]) <= batch_size
@pytest.mark.parametrize(
"column, expected_dtype",
[(["a", "b", "c", "d"], "string"), ([1, 2, 3, 4], "int64"), ([1.0, 2.0, 3.0, 4.0], "float64")],
)
@pytest.mark.parametrize("in_memory", [False, True])
@pytest.mark.parametrize(
"transform",
[
None,
("shuffle", (42,), {}),
("with_format", ("pandas",), {}),
("class_encode_column", ("col_2",), {}),
("select", (range(3),), {}),
],
)
def test_dataset_add_column(column, expected_dtype, in_memory, transform, dataset_dict, arrow_path):
column_name = "col_4"
original_dataset = (
Dataset(InMemoryTable.from_pydict(dataset_dict))
if in_memory
else Dataset(MemoryMappedTable.from_file(arrow_path))
)
if transform is not None:
transform_name, args, kwargs = transform
original_dataset: Dataset = getattr(original_dataset, transform_name)(*args, **kwargs)
column = column[:3] if transform is not None and transform_name == "select" else column
dataset = original_dataset.add_column(column_name, column)
assert dataset.data.shape == (3, 4) if transform is not None and transform_name == "select" else (4, 4)
expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
# Sort expected features as in the original dataset
expected_features = {feature: expected_features[feature] for feature in original_dataset.features}
# Add new column feature
expected_features[column_name] = expected_dtype
assert dataset.data.column_names == list(expected_features.keys())
for feature, expected_dtype in expected_features.items():
assert dataset.features[feature].dtype == expected_dtype
assert len(dataset.data.blocks) == 1 if in_memory else 2 # multiple InMemoryTables are consolidated as one
assert dataset.format["type"] == original_dataset.format["type"]
assert dataset._fingerprint != original_dataset._fingerprint
dataset.reset_format()
original_dataset.reset_format()
assert all(dataset[col] == original_dataset[col] for col in original_dataset.column_names)
assert set(dataset["col_4"]) == set(column)
if dataset._indices is not None:
dataset_indices = dataset._indices["indices"].to_pylist()
expected_dataset_indices = original_dataset._indices["indices"].to_pylist()
assert dataset_indices == expected_dataset_indices
assert_arrow_metadata_are_synced_with_dataset_features(dataset)
@pytest.mark.parametrize(
"transform",
[None, ("shuffle", (42,), {}), ("with_format", ("pandas",), {}), ("class_encode_column", ("col_2",), {})],
)
@pytest.mark.parametrize("in_memory", [False, True])
@pytest.mark.parametrize(
"item",
[
{"col_1": "2", "col_2": 2, "col_3": 2.0},
{"col_1": "2", "col_2": "2", "col_3": "2"},
{"col_1": 2, "col_2": 2, "col_3": 2},
{"col_1": 2.0, "col_2": 2.0, "col_3": 2.0},
],
)
def test_dataset_add_item(item, in_memory, dataset_dict, arrow_path, transform):
dataset_to_test = (
Dataset(InMemoryTable.from_pydict(dataset_dict))
if in_memory
else Dataset(MemoryMappedTable.from_file(arrow_path))
)
if transform is not None:
transform_name, args, kwargs = transform
dataset_to_test: Dataset = getattr(dataset_to_test, transform_name)(*args, **kwargs)
dataset = dataset_to_test.add_item(item)
assert dataset.data.shape == (5, 3)
expected_features = dataset_to_test.features
assert sorted(dataset.data.column_names) == sorted(expected_features.keys())
for feature, expected_dtype in expected_features.items():
assert dataset.features[feature] == expected_dtype
assert len(dataset.data.blocks) == 1 if in_memory else 2 # multiple InMemoryTables are consolidated as one
assert dataset.format["type"] == dataset_to_test.format["type"]
assert dataset._fingerprint != dataset_to_test._fingerprint
dataset.reset_format()
dataset_to_test.reset_format()
assert dataset[:-1] == dataset_to_test[:]
assert {k: int(v) for k, v in dataset[-1].items()} == {k: int(v) for k, v in item.items()}
if dataset._indices is not None:
dataset_indices = dataset._indices["indices"].to_pylist()
dataset_to_test_indices = dataset_to_test._indices["indices"].to_pylist()
assert dataset_indices == dataset_to_test_indices + [len(dataset_to_test._data)]
def test_dataset_add_item_new_columns():
dataset = Dataset.from_dict({"col_1": [0, 1, 2]}, features=Features({"col_1": Value("uint8")}))
dataset = dataset.add_item({"col_1": 3, "col_2": "a"})
assert dataset.data.shape == (4, 2)
assert dataset.features == Features({"col_1": Value("uint8"), "col_2": Value("string")})
assert dataset[:] == {"col_1": [0, 1, 2, 3], "col_2": [None, None, None, "a"]}
dataset = dataset.add_item({"col_3": True})
assert dataset.data.shape == (5, 3)
assert dataset.features == Features({"col_1": Value("uint8"), "col_2": Value("string"), "col_3": Value("bool")})
assert dataset[:] == {
"col_1": [0, 1, 2, 3, None],
"col_2": [None, None, None, "a", None],
"col_3": [None, None, None, None, True],
}
def test_dataset_add_item_introduce_feature_type():
dataset = Dataset.from_dict({"col_1": [None, None, None]})
dataset = dataset.add_item({"col_1": "a"})
assert dataset.data.shape == (4, 1)
assert dataset.features == Features({"col_1": Value("string")})
assert dataset[:] == {"col_1": [None, None, None, "a"]}
def test_dataset_filter_batched_indices():
ds = Dataset.from_dict({"num": [0, 1, 2, 3]})
ds = ds.filter(lambda num: num % 2 == 0, input_columns="num", batch_size=2)
assert all(item["num"] % 2 == 0 for item in ds)
@pytest.mark.parametrize("in_memory", [False, True])
def test_dataset_from_file(in_memory, dataset, arrow_file):
filename = arrow_file
with assert_arrow_memory_increases() if in_memory else assert_arrow_memory_doesnt_increase():
dataset_from_file = Dataset.from_file(filename, in_memory=in_memory)
assert dataset_from_file.features.type == dataset.features.type
assert dataset_from_file.features == dataset.features
assert dataset_from_file.cache_files == ([{"filename": filename}] if not in_memory else [])
def _check_csv_dataset(dataset, expected_features):
assert isinstance(dataset, Dataset)
assert dataset.num_rows == 4
assert dataset.num_columns == 3
assert dataset.column_names == ["col_1", "col_2", "col_3"]
for feature, expected_dtype in expected_features.items():
assert dataset.features[feature].dtype == expected_dtype
@pytest.mark.parametrize("keep_in_memory", [False, True])
def test_dataset_from_csv_keep_in_memory(keep_in_memory, csv_path, tmp_path):
cache_dir = tmp_path / "cache"
expected_features = {"col_1": "int64", "col_2": "int64", "col_3": "float64"}
with assert_arrow_memory_increases() if keep_in_memory else assert_arrow_memory_doesnt_increase():
dataset = Dataset.from_csv(csv_path, cache_dir=cache_dir, keep_in_memory=keep_in_memory)
_check_csv_dataset(dataset, expected_features)
@pytest.mark.parametrize(
"features",
[
None,
{"col_1": "string", "col_2": "int64", "col_3": "float64"},
{"col_1": "string", "col_2": "string", "col_3": "string"},
{"col_1": "int32", "col_2": "int32", "col_3": "int32"},
{"col_1": "float32", "col_2": "float32", "col_3": "float32"},
],
)
def test_dataset_from_csv_features(features, csv_path, tmp_path):
cache_dir = tmp_path / "cache"
# CSV file loses col_1 string dtype information: default now is "int64" instead of "string"
default_expected_features = {"col_1": "int64", "col_2": "int64", "col_3": "float64"}
expected_features = features.copy() if features else default_expected_features
features = (
Features({feature: Value(dtype) for feature, dtype in features.items()}) if features is not None else None
)
dataset = Dataset.from_csv(csv_path, features=features, cache_dir=cache_dir)
_check_csv_dataset(dataset, expected_features)
@pytest.mark.parametrize("split", [None, NamedSplit("train"), "train", "test"])
def test_dataset_from_csv_split(split, csv_path, tmp_path):
cache_dir = tmp_path / "cache"
expected_features = {"col_1": "int64", "col_2": "int64", "col_3": "float64"}
dataset = Dataset.from_csv(csv_path, cache_dir=cache_dir, split=split)
_check_csv_dataset(dataset, expected_features)
assert dataset.split == split if split else "train"
@pytest.mark.parametrize("path_type", [str, list])
def test_dataset_from_csv_path_type(path_type, csv_path, tmp_path):
if issubclass(path_type, str):
path = csv_path
elif issubclass(path_type, list):
path = [csv_path]
cache_dir = tmp_path / "cache"
expected_features = {"col_1": "int64", "col_2": "int64", "col_3": "float64"}
dataset = Dataset.from_csv(path, cache_dir=cache_dir)
_check_csv_dataset(dataset, expected_features)
def _check_json_dataset(dataset, expected_features):
assert isinstance(dataset, Dataset)
assert dataset.num_rows == 4
assert dataset.num_columns == 3
assert dataset.column_names == ["col_1", "col_2", "col_3"]
for feature, expected_dtype in expected_features.items():
assert dataset.features[feature].dtype == expected_dtype
@pytest.mark.parametrize("keep_in_memory", [False, True])
def test_dataset_from_json_keep_in_memory(keep_in_memory, jsonl_path, tmp_path):
cache_dir = tmp_path / "cache"
expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
with assert_arrow_memory_increases() if keep_in_memory else assert_arrow_memory_doesnt_increase():
dataset = Dataset.from_json(jsonl_path, cache_dir=cache_dir, keep_in_memory=keep_in_memory)
_check_json_dataset(dataset, expected_features)
@pytest.mark.parametrize(
"features",
[
None,
{"col_1": "string", "col_2": "int64", "col_3": "float64"},
{"col_1": "string", "col_2": "string", "col_3": "string"},
{"col_1": "int32", "col_2": "int32", "col_3": "int32"},
{"col_1": "float32", "col_2": "float32", "col_3": "float32"},
],
)
def test_dataset_from_json_features(features, jsonl_path, tmp_path):
cache_dir = tmp_path / "cache"
default_expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
expected_features = features.copy() if features else default_expected_features
features = (
Features({feature: Value(dtype) for feature, dtype in features.items()}) if features is not None else None
)
dataset = Dataset.from_json(jsonl_path, features=features, cache_dir=cache_dir)
_check_json_dataset(dataset, expected_features)
def test_dataset_from_json_with_class_label_feature(jsonl_str_path, tmp_path):
features = Features(
{"col_1": ClassLabel(names=["s0", "s1", "s2", "s3"]), "col_2": Value("int64"), "col_3": Value("float64")}
)
cache_dir = tmp_path / "cache"
dataset = Dataset.from_json(jsonl_str_path, features=features, cache_dir=cache_dir)
assert dataset.features["col_1"].dtype == "int64"
@pytest.mark.parametrize("split", [None, NamedSplit("train"), "train", "test"])
def test_dataset_from_json_split(split, jsonl_path, tmp_path):
cache_dir = tmp_path / "cache"
expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
dataset = Dataset.from_json(jsonl_path, cache_dir=cache_dir, split=split)
_check_json_dataset(dataset, expected_features)
assert dataset.split == split if split else "train"
@pytest.mark.parametrize("path_type", [str, list])
def test_dataset_from_json_path_type(path_type, jsonl_path, tmp_path):
if issubclass(path_type, str):
path = jsonl_path
elif issubclass(path_type, list):
path = [jsonl_path]
cache_dir = tmp_path / "cache"
expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
dataset = Dataset.from_json(path, cache_dir=cache_dir)
_check_json_dataset(dataset, expected_features)
def _check_parquet_dataset(dataset, expected_features):
assert isinstance(dataset, Dataset)
assert dataset.num_rows == 4
assert dataset.num_columns == 3
assert dataset.column_names == ["col_1", "col_2", "col_3"]
for feature, expected_dtype in expected_features.items():
assert dataset.features[feature].dtype == expected_dtype
@pytest.mark.parametrize("keep_in_memory", [False, True])
def test_dataset_from_parquet_keep_in_memory(keep_in_memory, parquet_path, tmp_path):
cache_dir = tmp_path / "cache"
expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
with assert_arrow_memory_increases() if keep_in_memory else assert_arrow_memory_doesnt_increase():
dataset = Dataset.from_parquet(parquet_path, cache_dir=cache_dir, keep_in_memory=keep_in_memory)
_check_parquet_dataset(dataset, expected_features)
@pytest.mark.parametrize(
"features",
[
None,
{"col_1": "string", "col_2": "int64", "col_3": "float64"},
{"col_1": "string", "col_2": "string", "col_3": "string"},
{"col_1": "int32", "col_2": "int32", "col_3": "int32"},
{"col_1": "float32", "col_2": "float32", "col_3": "float32"},
],
)
def test_dataset_from_parquet_features(features, parquet_path, tmp_path):
cache_dir = tmp_path / "cache"
default_expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
expected_features = features.copy() if features else default_expected_features
features = (
Features({feature: Value(dtype) for feature, dtype in features.items()}) if features is not None else None
)
dataset = Dataset.from_parquet(parquet_path, features=features, cache_dir=cache_dir)
_check_parquet_dataset(dataset, expected_features)
@pytest.mark.parametrize("split", [None, NamedSplit("train"), "train", "test"])
def test_dataset_from_parquet_split(split, parquet_path, tmp_path):
cache_dir = tmp_path / "cache"
expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
dataset = Dataset.from_parquet(parquet_path, cache_dir=cache_dir, split=split)
_check_parquet_dataset(dataset, expected_features)
assert dataset.split == split if split else "train"
@pytest.mark.parametrize("path_type", [str, list])
def test_dataset_from_parquet_path_type(path_type, parquet_path, tmp_path):
if issubclass(path_type, str):
path = parquet_path
elif issubclass(path_type, list):
path = [parquet_path]
cache_dir = tmp_path / "cache"
expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
dataset = Dataset.from_parquet(path, cache_dir=cache_dir)
_check_parquet_dataset(dataset, expected_features)
def _check_text_dataset(dataset, expected_features):
assert isinstance(dataset, Dataset)
assert dataset.num_rows == 4
assert dataset.num_columns == 1
assert dataset.column_names == ["text"]
for feature, expected_dtype in expected_features.items():
assert dataset.features[feature].dtype == expected_dtype
@pytest.mark.parametrize("keep_in_memory", [False, True])
def test_dataset_from_text_keep_in_memory(keep_in_memory, text_path, tmp_path):
cache_dir = tmp_path / "cache"
expected_features = {"text": "string"}
with assert_arrow_memory_increases() if keep_in_memory else assert_arrow_memory_doesnt_increase():
dataset = Dataset.from_text(text_path, cache_dir=cache_dir, keep_in_memory=keep_in_memory)
_check_text_dataset(dataset, expected_features)
@pytest.mark.parametrize(
"features",
[
None,
{"text": "string"},
{"text": "int32"},
{"text": "float32"},
],
)
def test_dataset_from_text_features(features, text_path, tmp_path):
cache_dir = tmp_path / "cache"
default_expected_features = {"text": "string"}
expected_features = features.copy() if features else default_expected_features
features = (
Features({feature: Value(dtype) for feature, dtype in features.items()}) if features is not None else None
)
dataset = Dataset.from_text(text_path, features=features, cache_dir=cache_dir)
_check_text_dataset(dataset, expected_features)
@pytest.mark.parametrize("split", [None, NamedSplit("train"), "train", "test"])
def test_dataset_from_text_split(split, text_path, tmp_path):
cache_dir = tmp_path / "cache"
expected_features = {"text": "string"}
dataset = Dataset.from_text(text_path, cache_dir=cache_dir, split=split)
_check_text_dataset(dataset, expected_features)
assert dataset.split == split if split else "train"
@pytest.mark.parametrize("path_type", [str, list])
def test_dataset_from_text_path_type(path_type, text_path, tmp_path):
if issubclass(path_type, str):
path = text_path
elif issubclass(path_type, list):
path = [text_path]
cache_dir = tmp_path / "cache"
expected_features = {"text": "string"}
dataset = Dataset.from_text(path, cache_dir=cache_dir)
_check_text_dataset(dataset, expected_features)
@pytest.fixture
def data_generator():
def _gen():
data = [
{"col_1": "0", "col_2": 0, "col_3": 0.0},
{"col_1": "1", "col_2": 1, "col_3": 1.0},
{"col_1": "2", "col_2": 2, "col_3": 2.0},
{"col_1": "3", "col_2": 3, "col_3": 3.0},
]
for item in data:
yield item
return _gen
def _check_generator_dataset(dataset, expected_features):
assert isinstance(dataset, Dataset)
assert dataset.num_rows == 4
assert dataset.num_columns == 3
assert dataset.column_names == ["col_1", "col_2", "col_3"]
for feature, expected_dtype in expected_features.items():
assert dataset.features[feature].dtype == expected_dtype
@pytest.mark.parametrize("keep_in_memory", [False, True])
def test_dataset_from_generator_keep_in_memory(keep_in_memory, data_generator, tmp_path):
cache_dir = tmp_path / "cache"
expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
with assert_arrow_memory_increases() if keep_in_memory else assert_arrow_memory_doesnt_increase():
dataset = Dataset.from_generator(data_generator, cache_dir=cache_dir, keep_in_memory=keep_in_memory)
_check_generator_dataset(dataset, expected_features)
@pytest.mark.parametrize(
"features",
[
None,
{"col_1": "string", "col_2": "int64", "col_3": "float64"},
{"col_1": "string", "col_2": "string", "col_3": "string"},
{"col_1": "int32", "col_2": "int32", "col_3": "int32"},
{"col_1": "float32", "col_2": "float32", "col_3": "float32"},
],
)
def test_dataset_from_generator_features(features, data_generator, tmp_path):
cache_dir = tmp_path / "cache"
default_expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
expected_features = features.copy() if features else default_expected_features
features = (
Features({feature: Value(dtype) for feature, dtype in features.items()}) if features is not None else None
)
dataset = Dataset.from_generator(data_generator, features=features, cache_dir=cache_dir)
_check_generator_dataset(dataset, expected_features)
@require_not_windows
@require_dill_gt_0_3_2
@require_pyspark
def test_from_spark():
import pyspark
spark = pyspark.sql.SparkSession.builder.master("local[*]").appName("pyspark").getOrCreate()
data = [
("0", 0, 0.0),
("1", 1, 1.0),
("2", 2, 2.0),
("3", 3, 3.0),
]
df = spark.createDataFrame(data, "col_1: string, col_2: int, col_3: float")
dataset = Dataset.from_spark(df)
assert isinstance(dataset, Dataset)
assert dataset.num_rows == 4
assert dataset.num_columns == 3
assert dataset.column_names == ["col_1", "col_2", "col_3"]
@require_not_windows
@require_dill_gt_0_3_2
@require_pyspark
def test_from_spark_features():
import PIL.Image
import pyspark
spark = pyspark.sql.SparkSession.builder.master("local[*]").appName("pyspark").getOrCreate()
data = [(0, np.arange(4 * 4 * 3).reshape(4, 4, 3).tolist())]
df = spark.createDataFrame(data, "idx: int, image: array<array<array<int>>>")
features = Features({"idx": Value("int64"), "image": Image()})
dataset = Dataset.from_spark(
df,
features=features,
)
assert isinstance(dataset, Dataset)
assert dataset.num_rows == 1
assert dataset.num_columns == 2
assert dataset.column_names == ["idx", "image"]
assert isinstance(dataset[0]["image"], PIL.Image.Image)
assert dataset.features == features
assert_arrow_metadata_are_synced_with_dataset_features(dataset)
@require_not_windows
@require_dill_gt_0_3_2
@require_pyspark
def test_from_spark_different_cache():
import pyspark
spark = pyspark.sql.SparkSession.builder.master("local[*]").appName("pyspark").getOrCreate()
df = spark.createDataFrame([("0", 0)], "col_1: string, col_2: int")
dataset = Dataset.from_spark(df)
assert isinstance(dataset, Dataset)
different_df = spark.createDataFrame([("1", 1)], "col_1: string, col_2: int")
different_dataset = Dataset.from_spark(different_df)
assert isinstance(different_dataset, Dataset)
assert dataset[0]["col_1"] == "0"
# Check to make sure that the second dataset wasn't read from the cache.
assert different_dataset[0]["col_1"] == "1"
def _check_sql_dataset(dataset, expected_features):
assert isinstance(dataset, Dataset)
assert dataset.num_rows == 4
assert dataset.num_columns == 3
assert dataset.column_names == ["col_1", "col_2", "col_3"]
for feature, expected_dtype in expected_features.items():
assert dataset.features[feature].dtype == expected_dtype
@require_sqlalchemy
@pytest.mark.parametrize("con_type", ["string", "engine"])
def test_dataset_from_sql_con_type(con_type, sqlite_path, tmp_path, set_sqlalchemy_silence_uber_warning):
cache_dir = tmp_path / "cache"
expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
if con_type == "string":
con = "sqlite:///" + sqlite_path
elif con_type == "engine":
import sqlalchemy
con = sqlalchemy.create_engine("sqlite:///" + sqlite_path)
# # https://github.com/huggingface/datasets/issues/2832 needs to be fixed first for this to work
# with caplog.at_level(INFO):
# dataset = Dataset.from_sql(
# "dataset",
# con,
# cache_dir=cache_dir,
# )
# if con_type == "string":
# assert "couldn't be hashed properly" not in caplog.text
# elif con_type == "engine":
# assert "couldn't be hashed properly" in caplog.text
dataset = Dataset.from_sql(
"dataset",
con,
cache_dir=cache_dir,
)
_check_sql_dataset(dataset, expected_features)
@require_sqlalchemy
@pytest.mark.parametrize(
"features",
[
None,
{"col_1": "string", "col_2": "int64", "col_3": "float64"},
{"col_1": "string", "col_2": "string", "col_3": "string"},
{"col_1": "int32", "col_2": "int32", "col_3": "int32"},
{"col_1": "float32", "col_2": "float32", "col_3": "float32"},
],
)
def test_dataset_from_sql_features(features, sqlite_path, tmp_path, set_sqlalchemy_silence_uber_warning):
cache_dir = tmp_path / "cache"
default_expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
expected_features = features.copy() if features else default_expected_features
features = (
Features({feature: Value(dtype) for feature, dtype in features.items()}) if features is not None else None
)
dataset = Dataset.from_sql("dataset", "sqlite:///" + sqlite_path, features=features, cache_dir=cache_dir)
_check_sql_dataset(dataset, expected_features)
@require_sqlalchemy
@pytest.mark.parametrize("keep_in_memory", [False, True])
def test_dataset_from_sql_keep_in_memory(keep_in_memory, sqlite_path, tmp_path, set_sqlalchemy_silence_uber_warning):
cache_dir = tmp_path / "cache"
expected_features = {"col_1": "string", "col_2": "int64", "col_3": "float64"}
with assert_arrow_memory_increases() if keep_in_memory else assert_arrow_memory_doesnt_increase():
dataset = Dataset.from_sql(
"dataset", "sqlite:///" + sqlite_path, cache_dir=cache_dir, keep_in_memory=keep_in_memory
)
_check_sql_dataset(dataset, expected_features)
def test_dataset_to_json(dataset, tmp_path):
file_path = tmp_path / "test_path.jsonl"
bytes_written = dataset.to_json(path_or_buf=file_path)
assert file_path.is_file()
assert bytes_written == file_path.stat().st_size
df = pd.read_json(file_path, orient="records", lines=True)
assert df.shape == dataset.shape
assert list(df.columns) == list(dataset.column_names)
@pytest.mark.parametrize("in_memory", [False, True])
@pytest.mark.parametrize(
"method_and_params",
[
("rename_column", (), {"original_column_name": "labels", "new_column_name": "label"}),
("remove_columns", (), {"column_names": "labels"}),
(
"cast",
(),
{
"features": Features(
{
"tokens": Sequence(Value("string")),
"labels": Sequence(Value("int16")),
"answers": Sequence(
{
"text": Value("string"),
"answer_start": Value("int32"),
}
),
"id": Value("int32"),
}
)
},
),
("flatten", (), {}),
],
)
def test_pickle_dataset_after_transforming_the_table(in_memory, method_and_params, arrow_file):
method, args, kwargs = method_and_params
with Dataset.from_file(arrow_file, in_memory=in_memory) as dataset, Dataset.from_file(
arrow_file, in_memory=in_memory
) as reference_dataset:
out = getattr(dataset, method)(*args, **kwargs)
dataset = out if out is not None else dataset
pickled_dataset = pickle.dumps(dataset)
reloaded_dataset = pickle.loads(pickled_dataset)
assert dataset._data != reference_dataset._data
assert dataset._data.table == reloaded_dataset._data.table
def test_dummy_dataset_serialize_fs(dataset, mockfs):
dataset_path = "mock://my_dataset"
dataset.save_to_disk(dataset_path, storage_options=mockfs.storage_options)
assert mockfs.isdir(dataset_path)
assert mockfs.glob(dataset_path + "/*")
reloaded = dataset.load_from_disk(dataset_path, storage_options=mockfs.storage_options)
assert len(reloaded) == len(dataset)
assert reloaded.features == dataset.features
assert reloaded.to_dict() == dataset.to_dict()
@pytest.mark.parametrize(
"uri_or_path",
[
"relative/path",
"/absolute/path",
"s3://bucket/relative/path",
"hdfs://relative/path",
"hdfs:///absolute/path",
],
)
def test_build_local_temp_path(uri_or_path):
extracted_path = extract_path_from_uri(uri_or_path)
local_temp_path = Dataset._build_local_temp_path(extracted_path)
path_relative_to_tmp_dir = local_temp_path.as_posix().split("tmp")[-1].split("/", 1)[1]
assert (
"tmp" in local_temp_path.as_posix()
and "hdfs" not in path_relative_to_tmp_dir
and "s3" not in path_relative_to_tmp_dir
and not local_temp_path.as_posix().startswith(extracted_path)
and local_temp_path.as_posix().endswith(extracted_path)
), f"Local temp path: {local_temp_path.as_posix()}"
class TaskTemplatesTest(TestCase):
def test_task_text_classification(self):
labels = sorted(["pos", "neg"])
features_before_cast = Features(
{
"input_text": Value("string"),
"input_labels": ClassLabel(names=labels),
}
)
# Labels are cast to tuple during `TextClassification.__post_init_`, so we do the same here
features_after_cast = Features(
{
"text": Value("string"),
"labels": ClassLabel(names=labels),
}
)
# Label names are added in `DatasetInfo.__post_init__` so not needed here
task_without_labels = TextClassification(text_column="input_text", label_column="input_labels")
info1 = DatasetInfo(
features=features_before_cast,
task_templates=task_without_labels,
)
# Label names are required when passing a TextClassification template directly to `Dataset.prepare_for_task`
# However they also can be used to define `DatasetInfo` so we include a test for this too
task_with_labels = TextClassification(text_column="input_text", label_column="input_labels")
info2 = DatasetInfo(
features=features_before_cast,
task_templates=task_with_labels,
)
data = {"input_text": ["i love transformers!"], "input_labels": [1]}
# Test we can load from task name when label names not included in template (default behaviour)
with Dataset.from_dict(data, info=info1) as dset:
self.assertSetEqual({"input_text", "input_labels"}, set(dset.column_names))
self.assertDictEqual(features_before_cast, dset.features)
with dset.prepare_for_task(task="text-classification") as dset:
self.assertSetEqual({"labels", "text"}, set(dset.column_names))
self.assertDictEqual(features_after_cast, dset.features)
# Test we can load from task name when label names included in template
with Dataset.from_dict(data, info=info2) as dset:
self.assertSetEqual({"input_text", "input_labels"}, set(dset.column_names))
self.assertDictEqual(features_before_cast, dset.features)
with dset.prepare_for_task(task="text-classification") as dset:
self.assertSetEqual({"labels", "text"}, set(dset.column_names))
self.assertDictEqual(features_after_cast, dset.features)
# Test we can load from TextClassification template
info1.task_templates = None
with Dataset.from_dict(data, info=info1) as dset:
with dset.prepare_for_task(task=task_with_labels) as dset:
self.assertSetEqual({"labels", "text"}, set(dset.column_names))
self.assertDictEqual(features_after_cast, dset.features)
def test_task_question_answering(self):
features_before_cast = Features(
{
"input_context": Value("string"),
"input_question": Value("string"),
"input_answers": Sequence(
{
"text": Value("string"),
"answer_start": Value("int32"),
}
),
}
)
features_after_cast = Features(
{
"context": Value("string"),
"question": Value("string"),
"answers": Sequence(
{
"text": Value("string"),
"answer_start": Value("int32"),
}
),
}
)
task = QuestionAnsweringExtractive(
context_column="input_context", question_column="input_question", answers_column="input_answers"
)
info = DatasetInfo(features=features_before_cast, task_templates=task)
data = {
"input_context": ["huggingface is going to the moon!"],
"input_question": ["where is huggingface going?"],
"input_answers": [{"text": ["to the moon!"], "answer_start": [2]}],
}
# Test we can load from task name
with Dataset.from_dict(data, info=info) as dset:
self.assertSetEqual(
{"input_context", "input_question", "input_answers.text", "input_answers.answer_start"},
set(dset.flatten().column_names),
)
self.assertDictEqual(features_before_cast, dset.features)
with dset.prepare_for_task(task="question-answering-extractive") as dset:
self.assertSetEqual(
{"context", "question", "answers.text", "answers.answer_start"},
set(dset.flatten().column_names),
)
self.assertDictEqual(features_after_cast, dset.features)
# Test we can load from QuestionAnsweringExtractive template
info.task_templates = None
with Dataset.from_dict(data, info=info) as dset:
with dset.prepare_for_task(task=task) as dset:
self.assertSetEqual(
{"context", "question", "answers.text", "answers.answer_start"},
set(dset.flatten().column_names),
)
self.assertDictEqual(features_after_cast, dset.features)
def test_task_summarization(self):
# Include a dummy extra column `dummy` to test we drop it correctly
features_before_cast = Features(
{"input_text": Value("string"), "input_summary": Value("string"), "dummy": Value("string")}
)
features_after_cast = Features({"text": Value("string"), "summary": Value("string")})
task = Summarization(text_column="input_text", summary_column="input_summary")
info = DatasetInfo(features=features_before_cast, task_templates=task)
data = {
"input_text": ["jack and jill took a taxi to attend a super duper party in the city."],
"input_summary": ["jack and jill attend party"],
"dummy": ["123456"],
}
# Test we can load from task name
with Dataset.from_dict(data, info=info) as dset:
with dset.prepare_for_task(task="summarization") as dset:
self.assertSetEqual(
{"text", "summary"},
set(dset.column_names),
)
self.assertDictEqual(features_after_cast, dset.features)
# Test we can load from Summarization template
info.task_templates = None
with Dataset.from_dict(data, info=info) as dset:
with dset.prepare_for_task(task=task) as dset:
self.assertSetEqual(
{"text", "summary"},
set(dset.column_names),
)
self.assertDictEqual(features_after_cast, dset.features)
def test_task_automatic_speech_recognition(self):
# Include a dummy extra column `dummy` to test we drop it correctly
features_before_cast = Features(
{
"input_audio": Audio(sampling_rate=16_000),
"input_transcription": Value("string"),
"dummy": Value("string"),
}
)
features_after_cast = Features({"audio": Audio(sampling_rate=16_000), "transcription": Value("string")})
task = AutomaticSpeechRecognition(audio_column="input_audio", transcription_column="input_transcription")
info = DatasetInfo(features=features_before_cast, task_templates=task)
data = {
"input_audio": [{"bytes": None, "path": "path/to/some/audio/file.wav"}],
"input_transcription": ["hello, my name is bob!"],
"dummy": ["123456"],
}
# Test we can load from task name
with Dataset.from_dict(data, info=info) as dset:
with dset.prepare_for_task(task="automatic-speech-recognition") as dset:
self.assertSetEqual(
{"audio", "transcription"},
set(dset.column_names),
)
self.assertDictEqual(features_after_cast, dset.features)
# Test we can load from Summarization template
info.task_templates = None
with Dataset.from_dict(data, info=info) as dset:
with dset.prepare_for_task(task=task) as dset:
self.assertSetEqual(
{"audio", "transcription"},
set(dset.column_names),
)
self.assertDictEqual(features_after_cast, dset.features)
def test_task_with_no_template(self):
data = {"input_text": ["i love transformers!"], "input_labels": [1]}
with Dataset.from_dict(data) as dset:
with self.assertRaises(ValueError):
dset.prepare_for_task("text-classification")
def test_task_with_incompatible_templates(self):
labels = sorted(["pos", "neg"])
features = Features(
{
"input_text": Value("string"),
"input_labels": ClassLabel(names=labels),
}
)
task = TextClassification(text_column="input_text", label_column="input_labels")
info = DatasetInfo(
features=features,
task_templates=task,
)
data = {"input_text": ["i love transformers!"], "input_labels": [1]}
with Dataset.from_dict(data, info=info) as dset:
# Invalid task name
self.assertRaises(ValueError, dset.prepare_for_task, "this-task-does-not-exist")
# Invalid task type
self.assertRaises(ValueError, dset.prepare_for_task, 1)
def test_task_with_multiple_compatible_task_templates(self):
features = Features(
{
"text1": Value("string"),
"text2": Value("string"),
}
)
task1 = LanguageModeling(text_column="text1")
task2 = LanguageModeling(text_column="text2")
info = DatasetInfo(
features=features,
task_templates=[task1, task2],
)
data = {"text1": ["i love transformers!"], "text2": ["i love datasets!"]}
with Dataset.from_dict(data, info=info) as dset:
self.assertRaises(ValueError, dset.prepare_for_task, "language-modeling", id=3)
with dset.prepare_for_task("language-modeling") as dset1:
self.assertEqual(dset1[0]["text"], "i love transformers!")
with dset.prepare_for_task("language-modeling", id=1) as dset2:
self.assertEqual(dset2[0]["text"], "i love datasets!")
def test_task_templates_empty_after_preparation(self):
features = Features(
{
"input_text": Value("string"),
"input_labels": ClassLabel(names=["pos", "neg"]),
}
)
task = TextClassification(text_column="input_text", label_column="input_labels")
info = DatasetInfo(
features=features,
task_templates=task,
)
data = {"input_text": ["i love transformers!"], "input_labels": [1]}
with Dataset.from_dict(data, info=info) as dset:
with dset.prepare_for_task(task="text-classification") as dset:
self.assertIsNone(dset.info.task_templates)
def test_align_labels_with_mapping_classification(self):
features = Features(
{
"input_text": Value("string"),
"input_labels": ClassLabel(num_classes=3, names=["entailment", "neutral", "contradiction"]),
}
)
data = {"input_text": ["a", "a", "b", "b", "c", "c"], "input_labels": [0, 0, 1, 1, 2, 2]}
label2id = {"CONTRADICTION": 0, "ENTAILMENT": 2, "NEUTRAL": 1}
id2label = {v: k for k, v in label2id.items()}
expected_labels = [2, 2, 1, 1, 0, 0]
expected_label_names = [id2label[idx] for idx in expected_labels]
with Dataset.from_dict(data, features=features) as dset:
with dset.align_labels_with_mapping(label2id, "input_labels") as dset:
self.assertListEqual(expected_labels, dset["input_labels"])
aligned_label_names = [dset.features["input_labels"].int2str(idx) for idx in dset["input_labels"]]
self.assertListEqual(expected_label_names, aligned_label_names)
def test_align_labels_with_mapping_ner(self):
features = Features(
{
"input_text": Value("string"),
"input_labels": Sequence(
ClassLabel(
names=[
"b-per",
"i-per",
"o",
]
)
),
}
)
data = {"input_text": [["Optimus", "Prime", "is", "a", "Transformer"]], "input_labels": [[0, 1, 2, 2, 2]]}
label2id = {"B-PER": 2, "I-PER": 1, "O": 0}
id2label = {v: k for k, v in label2id.items()}
expected_labels = [[2, 1, 0, 0, 0]]
expected_label_names = [[id2label[idx] for idx in seq] for seq in expected_labels]
with Dataset.from_dict(data, features=features) as dset:
with dset.align_labels_with_mapping(label2id, "input_labels") as dset:
self.assertListEqual(expected_labels, dset["input_labels"])
aligned_label_names = [
dset.features["input_labels"].feature.int2str(idx) for idx in dset["input_labels"]
]
self.assertListEqual(expected_label_names, aligned_label_names)
def test_concatenate_with_no_task_templates(self):
info = DatasetInfo(task_templates=None)
data = {"text": ["i love transformers!"], "labels": [1]}
with Dataset.from_dict(data, info=info) as dset1, Dataset.from_dict(
data, info=info
) as dset2, Dataset.from_dict(data, info=info) as dset3:
with concatenate_datasets([dset1, dset2, dset3]) as dset_concat:
self.assertEqual(dset_concat.info.task_templates, None)
def test_concatenate_with_equal_task_templates(self):
labels = ["neg", "pos"]
task_template = TextClassification(text_column="text", label_column="labels")
info = DatasetInfo(
features=Features({"text": Value("string"), "labels": ClassLabel(names=labels)}),
# Label names are added in `DatasetInfo.__post_init__` so not included here
task_templates=TextClassification(text_column="text", label_column="labels"),
)
data = {"text": ["i love transformers!"], "labels": [1]}
with Dataset.from_dict(data, info=info) as dset1, Dataset.from_dict(
data, info=info
) as dset2, Dataset.from_dict(data, info=info) as dset3:
with concatenate_datasets([dset1, dset2, dset3]) as dset_concat:
self.assertListEqual(dset_concat.info.task_templates, [task_template])
def test_concatenate_with_mixed_task_templates_in_common(self):
tc_template = TextClassification(text_column="text", label_column="labels")
qa_template = QuestionAnsweringExtractive(
question_column="question", context_column="context", answers_column="answers"
)
info1 = DatasetInfo(
task_templates=[qa_template],
features=Features(
{
"text": Value("string"),
"labels": ClassLabel(names=["pos", "neg"]),
"context": Value("string"),
"question": Value("string"),
"answers": Sequence(
{
"text": Value("string"),
"answer_start": Value("int32"),
}
),
}
),
)
info2 = DatasetInfo(
task_templates=[qa_template, tc_template],
features=Features(
{
"text": Value("string"),
"labels": ClassLabel(names=["pos", "neg"]),
"context": Value("string"),
"question": Value("string"),
"answers": Sequence(
{
"text": Value("string"),
"answer_start": Value("int32"),
}
),
}
),
)
data = {
"text": ["i love transformers!"],
"labels": [1],
"context": ["huggingface is going to the moon!"],
"question": ["where is huggingface going?"],
"answers": [{"text": ["to the moon!"], "answer_start": [2]}],
}
with Dataset.from_dict(data, info=info1) as dset1, Dataset.from_dict(
data, info=info2
) as dset2, Dataset.from_dict(data, info=info2) as dset3:
with concatenate_datasets([dset1, dset2, dset3]) as dset_concat:
self.assertListEqual(dset_concat.info.task_templates, [qa_template])
def test_concatenate_with_no_mixed_task_templates_in_common(self):
tc_template1 = TextClassification(text_column="text", label_column="labels")
tc_template2 = TextClassification(text_column="text", label_column="sentiment")
qa_template = QuestionAnsweringExtractive(
question_column="question", context_column="context", answers_column="answers"
)
info1 = DatasetInfo(
features=Features(
{
"text": Value("string"),
"labels": ClassLabel(names=["pos", "neg"]),
"sentiment": ClassLabel(names=["pos", "neg", "neutral"]),
"context": Value("string"),
"question": Value("string"),
"answers": Sequence(
{
"text": Value("string"),
"answer_start": Value("int32"),
}
),
}
),
task_templates=[tc_template1],
)
info2 = DatasetInfo(
features=Features(
{
"text": Value("string"),
"labels": ClassLabel(names=["pos", "neg"]),
"sentiment": ClassLabel(names=["pos", "neg", "neutral"]),
"context": Value("string"),
"question": Value("string"),
"answers": Sequence(
{
"text": Value("string"),
"answer_start": Value("int32"),
}
),
}
),
task_templates=[tc_template2],
)
info3 = DatasetInfo(
features=Features(
{
"text": Value("string"),
"labels": ClassLabel(names=["pos", "neg"]),
"sentiment": ClassLabel(names=["pos", "neg", "neutral"]),
"context": Value("string"),
"question": Value("string"),
"answers": Sequence(
{
"text": Value("string"),
"answer_start": Value("int32"),
}
),
}
),
task_templates=[qa_template],
)
data = {
"text": ["i love transformers!"],
"labels": [1],
"sentiment": [0],
"context": ["huggingface is going to the moon!"],
"question": ["where is huggingface going?"],
"answers": [{"text": ["to the moon!"], "answer_start": [2]}],
}
with Dataset.from_dict(data, info=info1) as dset1, Dataset.from_dict(
data, info=info2
) as dset2, Dataset.from_dict(data, info=info3) as dset3:
with concatenate_datasets([dset1, dset2, dset3]) as dset_concat:
self.assertEqual(dset_concat.info.task_templates, None)
def test_task_text_classification_when_columns_removed(self):
labels = sorted(["pos", "neg"])
features_before_map = Features(
{
"input_text": Value("string"),
"input_labels": ClassLabel(names=labels),
}
)
features_after_map = Features({"new_column": Value("int64")})
# Label names are added in `DatasetInfo.__post_init__` so not needed here
task = TextClassification(text_column="input_text", label_column="input_labels")
info = DatasetInfo(
features=features_before_map,
task_templates=task,
)
data = {"input_text": ["i love transformers!"], "input_labels": [1]}
with Dataset.from_dict(data, info=info) as dset:
with dset.map(lambda x: {"new_column": 0}, remove_columns=dset.column_names) as dset:
self.assertDictEqual(dset.features, features_after_map)
class StratifiedTest(TestCase):
def test_errors_train_test_split_stratify(self):
ys = [
np.array([0, 0, 0, 0, 1, 1, 1, 2, 2, 2, 2, 2]),
np.array([0, 1, 1, 1, 2, 2, 2, 3, 3, 3]),
np.array([0, 1, 2, 3, 0, 1, 2, 3, 0, 1, 2, 3, 0, 1, 2] * 2),
np.array([0, 0, 1, 1, 2, 2, 3, 3, 4, 4, 5, 5]),
np.array([0, 0, 1, 1, 2, 2, 3, 3, 4, 4, 5, 5]),
]
for i in range(len(ys)):
features = Features({"text": Value("int64"), "label": ClassLabel(len(np.unique(ys[i])))})
data = {"text": np.ones(len(ys[i])), "label": ys[i]}
d1 = Dataset.from_dict(data, features=features)
# For checking stratify_by_column exist as key in self.features.keys()
if i == 0:
self.assertRaises(ValueError, d1.train_test_split, 0.33, stratify_by_column="labl")
# For checking minimum class count error
elif i == 1:
self.assertRaises(ValueError, d1.train_test_split, 0.33, stratify_by_column="label")
# For check typeof label as ClassLabel type
elif i == 2:
d1 = Dataset.from_dict(data)
self.assertRaises(ValueError, d1.train_test_split, 0.33, stratify_by_column="label")
# For checking test_size should be greater than or equal to number of classes
elif i == 3:
self.assertRaises(ValueError, d1.train_test_split, 0.30, stratify_by_column="label")
# For checking train_size should be greater than or equal to number of classes
elif i == 4:
self.assertRaises(ValueError, d1.train_test_split, 0.60, stratify_by_column="label")
def test_train_test_split_startify(self):
ys = [
np.array([0, 0, 0, 0, 1, 1, 1, 2, 2, 2, 2, 2]),
np.array([0, 0, 0, 1, 1, 1, 2, 2, 2, 3, 3, 3]),
np.array([0, 1, 2, 3, 0, 1, 2, 3, 0, 1, 2, 3, 0, 1, 2] * 2),
np.array([0, 0, 1, 1, 1, 2, 2, 2, 3, 3, 3, 3, 3, 3, 3, 3]),
np.array([0] * 800 + [1] * 50),
]
for y in ys:
features = Features({"text": Value("int64"), "label": ClassLabel(len(np.unique(y)))})
data = {"text": np.ones(len(y)), "label": y}
d1 = Dataset.from_dict(data, features=features)
d1 = d1.train_test_split(test_size=0.33, stratify_by_column="label")
y = np.asanyarray(y) # To make it indexable for y[train]
test_size = np.ceil(0.33 * len(y))
train_size = len(y) - test_size
npt.assert_array_equal(np.unique(d1["train"]["label"]), np.unique(d1["test"]["label"]))
# checking classes proportion
p_train = np.bincount(np.unique(d1["train"]["label"], return_inverse=True)[1]) / float(
len(d1["train"]["label"])
)
p_test = np.bincount(np.unique(d1["test"]["label"], return_inverse=True)[1]) / float(
len(d1["test"]["label"])
)
npt.assert_array_almost_equal(p_train, p_test, 1)
assert len(d1["train"]["text"]) + len(d1["test"]["text"]) == y.size
assert len(d1["train"]["text"]) == train_size
assert len(d1["test"]["text"]) == test_size
def test_dataset_estimate_nbytes():
ds = Dataset.from_dict({"a": ["0" * 100] * 100})
assert 0.9 * ds._estimate_nbytes() < 100 * 100, "must be smaller than full dataset size"
ds = Dataset.from_dict({"a": ["0" * 100] * 100}).select([0])
assert 0.9 * ds._estimate_nbytes() < 100 * 100, "must be smaller than one chunk"
ds = Dataset.from_dict({"a": ["0" * 100] * 100})
ds = concatenate_datasets([ds] * 100)
assert 0.9 * ds._estimate_nbytes() < 100 * 100 * 100, "must be smaller than full dataset size"
assert 1.1 * ds._estimate_nbytes() > 100 * 100 * 100, "must be bigger than full dataset size"
ds = Dataset.from_dict({"a": ["0" * 100] * 100})
ds = concatenate_datasets([ds] * 100).select([0])
assert 0.9 * ds._estimate_nbytes() < 100 * 100, "must be smaller than one chunk"
def test_dataset_to_iterable_dataset(dataset: Dataset):
iterable_dataset = dataset.to_iterable_dataset()
assert isinstance(iterable_dataset, IterableDataset)
assert list(iterable_dataset) == list(dataset)
assert iterable_dataset.features == dataset.features
iterable_dataset = dataset.to_iterable_dataset(num_shards=3)
assert isinstance(iterable_dataset, IterableDataset)
assert list(iterable_dataset) == list(dataset)
assert iterable_dataset.features == dataset.features
assert iterable_dataset.n_shards == 3
with pytest.raises(ValueError):
dataset.to_iterable_dataset(num_shards=len(dataset) + 1)
with pytest.raises(NotImplementedError):
dataset.with_format("torch").to_iterable_dataset()
@pytest.mark.parametrize("batch_size", [1, 4])
@require_torch
def test_dataset_with_torch_dataloader(dataset, batch_size):
from torch.utils.data import DataLoader
from datasets import config
dataloader = DataLoader(dataset, batch_size=batch_size)
with patch.object(dataset, "_getitem", wraps=dataset._getitem) as mock_getitem:
out = list(dataloader)
getitem_call_count = mock_getitem.call_count
assert len(out) == len(dataset) // batch_size + int(len(dataset) % batch_size > 0)
# calling dataset[list_of_indices] is much more efficient than [dataset[idx] for idx in list of indices]
if config.TORCH_VERSION >= version.parse("1.13.0"):
assert getitem_call_count == len(dataset) // batch_size + int(len(dataset) % batch_size > 0)
@pytest.mark.parametrize("return_lazy_dict", [True, False, "mix"])
def test_map_cases(return_lazy_dict):
def f(x):
"""May return a mix of LazyDict and regular Dict"""
if x["a"] < 2:
x["a"] = -1
return dict(x) if return_lazy_dict is False else x
else:
return x if return_lazy_dict is True else {}
ds = Dataset.from_dict({"a": [0, 1, 2, 3]})
ds = ds.map(f)
outputs = ds[:]
assert outputs == {"a": [-1, -1, 2, 3]}
def f(x):
"""May return a mix of LazyDict and regular Dict, but sometimes with None values"""
if x["a"] < 2:
x["a"] = None
return dict(x) if return_lazy_dict is False else x
else:
return x if return_lazy_dict is True else {}
ds = Dataset.from_dict({"a": [0, 1, 2, 3]})
ds = ds.map(f)
outputs = ds[:]
assert outputs == {"a": [None, None, 2, 3]}
def f(x):
"""Return a LazyDict, but we remove a lazy column and add a new one"""
if x["a"] < 2:
x["b"] = -1
return x
else:
x["b"] = x["a"]
return x
ds = Dataset.from_dict({"a": [0, 1, 2, 3]})
ds = ds.map(f, remove_columns=["a"])
outputs = ds[:]
assert outputs == {"b": [-1, -1, 2, 3]}
# The formatted dataset version removes the lazy column from a different dictionary, hence it should be preserved in the output
ds = Dataset.from_dict({"a": [0, 1, 2, 3]})
ds = ds.with_format("numpy")
ds = ds.map(f, remove_columns=["a"])
ds = ds.with_format(None)
outputs = ds[:]
assert outputs == {"a": [0, 1, 2, 3], "b": [-1, -1, 2, 3]}
def f(x):
"""May return a mix of LazyDict and regular Dict, but we replace a lazy column"""
if x["a"] < 2:
x["a"] = -1
return dict(x) if return_lazy_dict is False else x
else:
x["a"] = x["a"]
return x if return_lazy_dict is True else {"a": x["a"]}
ds = Dataset.from_dict({"a": [0, 1, 2, 3]})
ds = ds.map(f, remove_columns=["a"])
outputs = ds[:]
assert outputs == ({"a": [-1, -1, 2, 3]} if return_lazy_dict is False else {})
def f(x):
"""May return a mix of LazyDict and regular Dict, but we modify a nested lazy column in-place"""
if x["a"]["b"] < 2:
x["a"]["c"] = -1
return dict(x) if return_lazy_dict is False else x
else:
x["a"]["c"] = x["a"]["b"]
return x if return_lazy_dict is True else {}
ds = Dataset.from_dict({"a": [{"b": 0}, {"b": 1}, {"b": 2}, {"b": 3}]})
ds = ds.map(f)
outputs = ds[:]
assert outputs == {"a": [{"b": 0, "c": -1}, {"b": 1, "c": -1}, {"b": 2, "c": 2}, {"b": 3, "c": 3}]}
def f(x):
"""May return a mix of LazyDict and regular Dict, but using an extension type"""
if x["a"][0][0] < 2:
x["a"] = [[-1]]
return dict(x) if return_lazy_dict is False else x
else:
return x if return_lazy_dict is True else {}
features = Features({"a": Array2D(shape=(1, 1), dtype="int32")})
ds = Dataset.from_dict({"a": [[[i]] for i in [0, 1, 2, 3]]}, features=features)
ds = ds.map(f)
outputs = ds[:]
assert outputs == {"a": [[[i]] for i in [-1, -1, 2, 3]]}
def f(x):
"""May return a mix of LazyDict and regular Dict, but using a nested extension type"""
if x["a"]["nested"][0][0] < 2:
x["a"] = {"nested": [[-1]]}
return dict(x) if return_lazy_dict is False else x
else:
return x if return_lazy_dict is True else {}
features = Features({"a": {"nested": Array2D(shape=(1, 1), dtype="int64")}})
ds = Dataset.from_dict({"a": [{"nested": [[i]]} for i in [0, 1, 2, 3]]}, features=features)
ds = ds.map(f)
outputs = ds[:]
assert outputs == {"a": [{"nested": [[i]]} for i in [-1, -1, 2, 3]]}
def test_dataset_getitem_raises():
ds = Dataset.from_dict({"a": [0, 1, 2, 3]})
with pytest.raises(TypeError):
ds[False]
with pytest.raises(TypeError):
ds._getitem(True)
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_beam.py | import os
import tempfile
from functools import partial
from unittest import TestCase
from unittest.mock import patch
import datasets
import datasets.config
from .utils import require_beam
class DummyBeamDataset(datasets.BeamBasedBuilder):
"""Dummy beam dataset."""
def _info(self):
return datasets.DatasetInfo(
features=datasets.Features({"content": datasets.Value("string")}),
# No default supervised_keys.
supervised_keys=None,
)
def _split_generators(self, dl_manager, pipeline):
return [datasets.SplitGenerator(name=datasets.Split.TRAIN, gen_kwargs={"examples": get_test_dummy_examples()})]
def _build_pcollection(self, pipeline, examples):
import apache_beam as beam
return pipeline | "Load Examples" >> beam.Create(examples)
class NestedBeamDataset(datasets.BeamBasedBuilder):
"""Dummy beam dataset."""
def _info(self):
return datasets.DatasetInfo(
features=datasets.Features({"a": datasets.Sequence({"b": datasets.Value("string")})}),
# No default supervised_keys.
supervised_keys=None,
)
def _split_generators(self, dl_manager, pipeline):
return [
datasets.SplitGenerator(name=datasets.Split.TRAIN, gen_kwargs={"examples": get_test_nested_examples()})
]
def _build_pcollection(self, pipeline, examples):
import apache_beam as beam
return pipeline | "Load Examples" >> beam.Create(examples)
def get_test_dummy_examples():
return [(i, {"content": content}) for i, content in enumerate(["foo", "bar", "foobar"])]
def get_test_nested_examples():
return [(i, {"a": {"b": [content]}}) for i, content in enumerate(["foo", "bar", "foobar"])]
class BeamBuilderTest(TestCase):
@require_beam
def test_download_and_prepare(self):
expected_num_examples = len(get_test_dummy_examples())
with tempfile.TemporaryDirectory() as tmp_cache_dir:
builder = DummyBeamDataset(cache_dir=tmp_cache_dir, beam_runner="DirectRunner")
builder.download_and_prepare()
self.assertTrue(
os.path.exists(
os.path.join(tmp_cache_dir, builder.name, "default", "0.0.0", f"{builder.name}-train.arrow")
)
)
self.assertDictEqual(builder.info.features, datasets.Features({"content": datasets.Value("string")}))
dset = builder.as_dataset()
self.assertEqual(dset["train"].num_rows, expected_num_examples)
self.assertEqual(dset["train"].info.splits["train"].num_examples, expected_num_examples)
self.assertDictEqual(dset["train"][0], get_test_dummy_examples()[0][1])
self.assertDictEqual(
dset["train"][expected_num_examples - 1], get_test_dummy_examples()[expected_num_examples - 1][1]
)
self.assertTrue(
os.path.exists(os.path.join(tmp_cache_dir, builder.name, "default", "0.0.0", "dataset_info.json"))
)
del dset
@require_beam
def test_download_and_prepare_sharded(self):
import apache_beam as beam
original_write_parquet = beam.io.parquetio.WriteToParquet
expected_num_examples = len(get_test_dummy_examples())
with tempfile.TemporaryDirectory() as tmp_cache_dir:
builder = DummyBeamDataset(cache_dir=tmp_cache_dir, beam_runner="DirectRunner")
with patch("apache_beam.io.parquetio.WriteToParquet") as write_parquet_mock:
write_parquet_mock.side_effect = partial(original_write_parquet, num_shards=2)
builder.download_and_prepare()
self.assertTrue(
os.path.exists(
os.path.join(
tmp_cache_dir, builder.name, "default", "0.0.0", f"{builder.name}-train-00000-of-00002.arrow"
)
)
)
self.assertTrue(
os.path.exists(
os.path.join(
tmp_cache_dir, builder.name, "default", "0.0.0", f"{builder.name}-train-00000-of-00002.arrow"
)
)
)
self.assertDictEqual(builder.info.features, datasets.Features({"content": datasets.Value("string")}))
dset = builder.as_dataset()
self.assertEqual(dset["train"].num_rows, expected_num_examples)
self.assertEqual(dset["train"].info.splits["train"].num_examples, expected_num_examples)
# Order is not preserved when sharding, so we just check that all the elements are there
self.assertListEqual(sorted(dset["train"]["content"]), sorted(["foo", "bar", "foobar"]))
self.assertTrue(
os.path.exists(os.path.join(tmp_cache_dir, builder.name, "default", "0.0.0", "dataset_info.json"))
)
del dset
@require_beam
def test_no_beam_options(self):
with tempfile.TemporaryDirectory() as tmp_cache_dir:
builder = DummyBeamDataset(cache_dir=tmp_cache_dir)
self.assertRaises(datasets.builder.MissingBeamOptions, builder.download_and_prepare)
@require_beam
def test_nested_features(self):
expected_num_examples = len(get_test_nested_examples())
with tempfile.TemporaryDirectory() as tmp_cache_dir:
builder = NestedBeamDataset(cache_dir=tmp_cache_dir, beam_runner="DirectRunner")
builder.download_and_prepare()
self.assertTrue(
os.path.exists(
os.path.join(tmp_cache_dir, builder.name, "default", "0.0.0", f"{builder.name}-train.arrow")
)
)
self.assertDictEqual(
builder.info.features, datasets.Features({"a": datasets.Sequence({"b": datasets.Value("string")})})
)
dset = builder.as_dataset()
self.assertEqual(dset["train"].num_rows, expected_num_examples)
self.assertEqual(dset["train"].info.splits["train"].num_examples, expected_num_examples)
self.assertDictEqual(dset["train"][0], get_test_nested_examples()[0][1])
self.assertDictEqual(
dset["train"][expected_num_examples - 1], get_test_nested_examples()[expected_num_examples - 1][1]
)
self.assertTrue(
os.path.exists(os.path.join(tmp_cache_dir, builder.name, "default", "0.0.0", "dataset_info.json"))
)
del dset
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_builder.py | import importlib
import os
import tempfile
import types
from contextlib import nullcontext as does_not_raise
from multiprocessing import Process
from pathlib import Path
from unittest import TestCase
from unittest.mock import patch
import numpy as np
import pyarrow as pa
import pyarrow.parquet as pq
import pytest
from multiprocess.pool import Pool
from datasets.arrow_dataset import Dataset
from datasets.arrow_reader import DatasetNotOnHfGcsError
from datasets.arrow_writer import ArrowWriter
from datasets.builder import ArrowBasedBuilder, BeamBasedBuilder, BuilderConfig, DatasetBuilder, GeneratorBasedBuilder
from datasets.dataset_dict import DatasetDict, IterableDatasetDict
from datasets.download.download_manager import DownloadMode
from datasets.features import Features, Value
from datasets.info import DatasetInfo, PostProcessedInfo
from datasets.iterable_dataset import IterableDataset
from datasets.splits import Split, SplitDict, SplitGenerator, SplitInfo
from datasets.streaming import xjoin
from datasets.utils.file_utils import is_local_path
from datasets.utils.info_utils import VerificationMode
from datasets.utils.logging import INFO, get_logger
from .utils import (
assert_arrow_memory_doesnt_increase,
assert_arrow_memory_increases,
require_beam,
require_faiss,
set_current_working_directory_to_temp_dir,
)
class DummyBuilder(DatasetBuilder):
def _info(self):
return DatasetInfo(features=Features({"text": Value("string")}))
def _split_generators(self, dl_manager):
return [SplitGenerator(name=Split.TRAIN)]
def _prepare_split(self, split_generator, **kwargs):
fname = f"{self.dataset_name}-{split_generator.name}.arrow"
with ArrowWriter(features=self.info.features, path=os.path.join(self._output_dir, fname)) as writer:
writer.write_batch({"text": ["foo"] * 100})
num_examples, num_bytes = writer.finalize()
split_generator.split_info.num_examples = num_examples
split_generator.split_info.num_bytes = num_bytes
class DummyGeneratorBasedBuilder(GeneratorBasedBuilder):
def _info(self):
return DatasetInfo(features=Features({"text": Value("string")}))
def _split_generators(self, dl_manager):
return [SplitGenerator(name=Split.TRAIN)]
def _generate_examples(self):
for i in range(100):
yield i, {"text": "foo"}
class DummyArrowBasedBuilder(ArrowBasedBuilder):
def _info(self):
return DatasetInfo(features=Features({"text": Value("string")}))
def _split_generators(self, dl_manager):
return [SplitGenerator(name=Split.TRAIN)]
def _generate_tables(self):
for i in range(10):
yield i, pa.table({"text": ["foo"] * 10})
class DummyBeamBasedBuilder(BeamBasedBuilder):
def _info(self):
return DatasetInfo(features=Features({"text": Value("string")}))
def _split_generators(self, dl_manager):
return [SplitGenerator(name=Split.TRAIN)]
def _build_pcollection(self, pipeline):
import apache_beam as beam
def _process(item):
for i in range(10):
yield f"{i}_{item}", {"text": "foo"}
return pipeline | "Initialize" >> beam.Create(range(10)) | "Extract content" >> beam.FlatMap(_process)
class DummyGeneratorBasedBuilderWithIntegers(GeneratorBasedBuilder):
def _info(self):
return DatasetInfo(features=Features({"id": Value("int8")}))
def _split_generators(self, dl_manager):
return [SplitGenerator(name=Split.TRAIN)]
def _generate_examples(self):
for i in range(100):
yield i, {"id": i}
class DummyGeneratorBasedBuilderConfig(BuilderConfig):
def __init__(self, content="foo", times=2, *args, **kwargs):
super().__init__(*args, **kwargs)
self.content = content
self.times = times
class DummyGeneratorBasedBuilderWithConfig(GeneratorBasedBuilder):
BUILDER_CONFIG_CLASS = DummyGeneratorBasedBuilderConfig
def _info(self):
return DatasetInfo(features=Features({"text": Value("string")}))
def _split_generators(self, dl_manager):
return [SplitGenerator(name=Split.TRAIN)]
def _generate_examples(self):
for i in range(100):
yield i, {"text": self.config.content * self.config.times}
class DummyBuilderWithMultipleConfigs(DummyBuilder):
BUILDER_CONFIGS = [
DummyGeneratorBasedBuilderConfig(name="a"),
DummyGeneratorBasedBuilderConfig(name="b"),
]
class DummyBuilderWithDefaultConfig(DummyBuilderWithMultipleConfigs):
DEFAULT_CONFIG_NAME = "a"
class DummyBuilderWithDownload(DummyBuilder):
def __init__(self, *args, rel_path=None, abs_path=None, **kwargs):
super().__init__(*args, **kwargs)
self._rel_path = rel_path
self._abs_path = abs_path
def _split_generators(self, dl_manager):
if self._rel_path is not None:
assert os.path.exists(dl_manager.download(self._rel_path)), "dl_manager must support relative paths"
if self._abs_path is not None:
assert os.path.exists(dl_manager.download(self._abs_path)), "dl_manager must support absolute paths"
return [SplitGenerator(name=Split.TRAIN)]
class DummyBuilderWithManualDownload(DummyBuilderWithMultipleConfigs):
@property
def manual_download_instructions(self):
return "To use the dataset you have to download some stuff manually and pass the data path to data_dir"
def _split_generators(self, dl_manager):
if not os.path.exists(self.config.data_dir):
raise FileNotFoundError(f"data_dir {self.config.data_dir} doesn't exist.")
return [SplitGenerator(name=Split.TRAIN)]
class DummyArrowBasedBuilderWithShards(ArrowBasedBuilder):
def _info(self):
return DatasetInfo(features=Features({"id": Value("int8"), "filepath": Value("string")}))
def _split_generators(self, dl_manager):
return [SplitGenerator(name=Split.TRAIN, gen_kwargs={"filepaths": [f"data{i}.txt" for i in range(4)]})]
def _generate_tables(self, filepaths):
idx = 0
for filepath in filepaths:
for i in range(10):
yield idx, pa.table({"id": range(10 * i, 10 * (i + 1)), "filepath": [filepath] * 10})
idx += 1
class DummyGeneratorBasedBuilderWithShards(GeneratorBasedBuilder):
def _info(self):
return DatasetInfo(features=Features({"id": Value("int8"), "filepath": Value("string")}))
def _split_generators(self, dl_manager):
return [SplitGenerator(name=Split.TRAIN, gen_kwargs={"filepaths": [f"data{i}.txt" for i in range(4)]})]
def _generate_examples(self, filepaths):
idx = 0
for filepath in filepaths:
for i in range(100):
yield idx, {"id": i, "filepath": filepath}
idx += 1
class DummyArrowBasedBuilderWithAmbiguousShards(ArrowBasedBuilder):
def _info(self):
return DatasetInfo(features=Features({"id": Value("int8"), "filepath": Value("string")}))
def _split_generators(self, dl_manager):
return [
SplitGenerator(
name=Split.TRAIN,
gen_kwargs={
"filepaths": [f"data{i}.txt" for i in range(4)],
"dummy_kwarg_with_different_length": [f"dummy_data{i}.txt" for i in range(3)],
},
)
]
def _generate_tables(self, filepaths, dummy_kwarg_with_different_length):
idx = 0
for filepath in filepaths:
for i in range(10):
yield idx, pa.table({"id": range(10 * i, 10 * (i + 1)), "filepath": [filepath] * 10})
idx += 1
class DummyGeneratorBasedBuilderWithAmbiguousShards(GeneratorBasedBuilder):
def _info(self):
return DatasetInfo(features=Features({"id": Value("int8"), "filepath": Value("string")}))
def _split_generators(self, dl_manager):
return [
SplitGenerator(
name=Split.TRAIN,
gen_kwargs={
"filepaths": [f"data{i}.txt" for i in range(4)],
"dummy_kwarg_with_different_length": [f"dummy_data{i}.txt" for i in range(3)],
},
)
]
def _generate_examples(self, filepaths, dummy_kwarg_with_different_length):
idx = 0
for filepath in filepaths:
for i in range(100):
yield idx, {"id": i, "filepath": filepath}
idx += 1
def _run_concurrent_download_and_prepare(tmp_dir):
builder = DummyBuilder(cache_dir=tmp_dir)
builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.REUSE_DATASET_IF_EXISTS)
return builder
def check_streaming(builder):
builders_module = importlib.import_module(builder.__module__)
assert builders_module._patched_for_streaming
assert builders_module.os.path.join is xjoin
class BuilderTest(TestCase):
def test_download_and_prepare(self):
with tempfile.TemporaryDirectory() as tmp_dir:
builder = DummyBuilder(cache_dir=tmp_dir)
builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
self.assertTrue(
os.path.exists(
os.path.join(
tmp_dir, builder.dataset_name, "default", "0.0.0", f"{builder.dataset_name}-train.arrow"
)
)
)
self.assertDictEqual(builder.info.features, Features({"text": Value("string")}))
self.assertEqual(builder.info.splits["train"].num_examples, 100)
self.assertTrue(
os.path.exists(os.path.join(tmp_dir, builder.dataset_name, "default", "0.0.0", "dataset_info.json"))
)
def test_download_and_prepare_checksum_computation(self):
with tempfile.TemporaryDirectory() as tmp_dir:
builder_no_verification = DummyBuilder(cache_dir=tmp_dir)
builder_no_verification.download_and_prepare(
try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD
)
self.assertTrue(
all(v["checksum"] is not None for _, v in builder_no_verification.info.download_checksums.items())
)
builder_with_verification = DummyBuilder(cache_dir=tmp_dir)
builder_with_verification.download_and_prepare(
try_from_hf_gcs=False,
download_mode=DownloadMode.FORCE_REDOWNLOAD,
verification_mode=VerificationMode.ALL_CHECKS,
)
self.assertTrue(
all(v["checksum"] is None for _, v in builder_with_verification.info.download_checksums.items())
)
def test_concurrent_download_and_prepare(self):
with tempfile.TemporaryDirectory() as tmp_dir:
processes = 2
with Pool(processes=processes) as pool:
jobs = [
pool.apply_async(_run_concurrent_download_and_prepare, kwds={"tmp_dir": tmp_dir})
for _ in range(processes)
]
builders = [job.get() for job in jobs]
for builder in builders:
self.assertTrue(
os.path.exists(
os.path.join(
tmp_dir,
builder.dataset_name,
"default",
"0.0.0",
f"{builder.dataset_name}-train.arrow",
)
)
)
self.assertDictEqual(builder.info.features, Features({"text": Value("string")}))
self.assertEqual(builder.info.splits["train"].num_examples, 100)
self.assertTrue(
os.path.exists(
os.path.join(tmp_dir, builder.dataset_name, "default", "0.0.0", "dataset_info.json")
)
)
def test_download_and_prepare_with_base_path(self):
with tempfile.TemporaryDirectory() as tmp_dir:
rel_path = "dummy1.data"
abs_path = os.path.join(tmp_dir, "dummy2.data")
# test relative path is missing
builder = DummyBuilderWithDownload(cache_dir=tmp_dir, rel_path=rel_path)
with self.assertRaises(FileNotFoundError):
builder.download_and_prepare(
try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD, base_path=tmp_dir
)
# test absolute path is missing
builder = DummyBuilderWithDownload(cache_dir=tmp_dir, abs_path=abs_path)
with self.assertRaises(FileNotFoundError):
builder.download_and_prepare(
try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD, base_path=tmp_dir
)
# test that they are both properly loaded when they exist
open(os.path.join(tmp_dir, rel_path), "w")
open(abs_path, "w")
builder = DummyBuilderWithDownload(cache_dir=tmp_dir, rel_path=rel_path, abs_path=abs_path)
builder.download_and_prepare(
try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD, base_path=tmp_dir
)
self.assertTrue(
os.path.exists(
os.path.join(
tmp_dir,
builder.dataset_name,
"default",
"0.0.0",
f"{builder.dataset_name}-train.arrow",
)
)
)
def test_as_dataset_with_post_process(self):
def _post_process(self, dataset, resources_paths):
def char_tokenize(example):
return {"tokens": list(example["text"])}
return dataset.map(char_tokenize, cache_file_name=resources_paths["tokenized_dataset"])
def _post_processing_resources(self, split):
return {"tokenized_dataset": f"tokenized_dataset-{split}.arrow"}
with tempfile.TemporaryDirectory() as tmp_dir:
builder = DummyBuilder(cache_dir=tmp_dir)
builder.info.post_processed = PostProcessedInfo(
features=Features({"text": Value("string"), "tokens": [Value("string")]})
)
builder._post_process = types.MethodType(_post_process, builder)
builder._post_processing_resources = types.MethodType(_post_processing_resources, builder)
os.makedirs(builder.cache_dir)
builder.info.splits = SplitDict()
builder.info.splits.add(SplitInfo("train", num_examples=10))
builder.info.splits.add(SplitInfo("test", num_examples=10))
for split in builder.info.splits:
with ArrowWriter(
path=os.path.join(builder.cache_dir, f"{builder.dataset_name}-{split}.arrow"),
features=Features({"text": Value("string")}),
) as writer:
writer.write_batch({"text": ["foo"] * 10})
writer.finalize()
with ArrowWriter(
path=os.path.join(builder.cache_dir, f"tokenized_dataset-{split}.arrow"),
features=Features({"text": Value("string"), "tokens": [Value("string")]}),
) as writer:
writer.write_batch({"text": ["foo"] * 10, "tokens": [list("foo")] * 10})
writer.finalize()
dsets = builder.as_dataset()
self.assertIsInstance(dsets, DatasetDict)
self.assertListEqual(list(dsets.keys()), ["train", "test"])
self.assertEqual(len(dsets["train"]), 10)
self.assertEqual(len(dsets["test"]), 10)
self.assertDictEqual(
dsets["train"].features, Features({"text": Value("string"), "tokens": [Value("string")]})
)
self.assertDictEqual(
dsets["test"].features, Features({"text": Value("string"), "tokens": [Value("string")]})
)
self.assertListEqual(dsets["train"].column_names, ["text", "tokens"])
self.assertListEqual(dsets["test"].column_names, ["text", "tokens"])
del dsets
dset = builder.as_dataset("train")
self.assertIsInstance(dset, Dataset)
self.assertEqual(dset.split, "train")
self.assertEqual(len(dset), 10)
self.assertDictEqual(dset.features, Features({"text": Value("string"), "tokens": [Value("string")]}))
self.assertListEqual(dset.column_names, ["text", "tokens"])
self.assertGreater(builder.info.post_processing_size, 0)
self.assertGreater(
builder.info.post_processed.resources_checksums["train"]["tokenized_dataset"]["num_bytes"], 0
)
del dset
dset = builder.as_dataset("train+test[:30%]")
self.assertIsInstance(dset, Dataset)
self.assertEqual(dset.split, "train+test[:30%]")
self.assertEqual(len(dset), 13)
self.assertDictEqual(dset.features, Features({"text": Value("string"), "tokens": [Value("string")]}))
self.assertListEqual(dset.column_names, ["text", "tokens"])
del dset
dset = builder.as_dataset("all")
self.assertIsInstance(dset, Dataset)
self.assertEqual(dset.split, "train+test")
self.assertEqual(len(dset), 20)
self.assertDictEqual(dset.features, Features({"text": Value("string"), "tokens": [Value("string")]}))
self.assertListEqual(dset.column_names, ["text", "tokens"])
del dset
def _post_process(self, dataset, resources_paths):
return dataset.select([0, 1], keep_in_memory=True)
with tempfile.TemporaryDirectory() as tmp_dir:
builder = DummyBuilder(cache_dir=tmp_dir)
builder._post_process = types.MethodType(_post_process, builder)
os.makedirs(builder.cache_dir)
builder.info.splits = SplitDict()
builder.info.splits.add(SplitInfo("train", num_examples=10))
builder.info.splits.add(SplitInfo("test", num_examples=10))
for split in builder.info.splits:
with ArrowWriter(
path=os.path.join(builder.cache_dir, f"{builder.dataset_name}-{split}.arrow"),
features=Features({"text": Value("string")}),
) as writer:
writer.write_batch({"text": ["foo"] * 10})
writer.finalize()
with ArrowWriter(
path=os.path.join(builder.cache_dir, f"small_dataset-{split}.arrow"),
features=Features({"text": Value("string")}),
) as writer:
writer.write_batch({"text": ["foo"] * 2})
writer.finalize()
dsets = builder.as_dataset()
self.assertIsInstance(dsets, DatasetDict)
self.assertListEqual(list(dsets.keys()), ["train", "test"])
self.assertEqual(len(dsets["train"]), 2)
self.assertEqual(len(dsets["test"]), 2)
self.assertDictEqual(dsets["train"].features, Features({"text": Value("string")}))
self.assertDictEqual(dsets["test"].features, Features({"text": Value("string")}))
self.assertListEqual(dsets["train"].column_names, ["text"])
self.assertListEqual(dsets["test"].column_names, ["text"])
del dsets
dset = builder.as_dataset("train")
self.assertIsInstance(dset, Dataset)
self.assertEqual(dset.split, "train")
self.assertEqual(len(dset), 2)
self.assertDictEqual(dset.features, Features({"text": Value("string")}))
self.assertListEqual(dset.column_names, ["text"])
del dset
dset = builder.as_dataset("train+test[:30%]")
self.assertIsInstance(dset, Dataset)
self.assertEqual(dset.split, "train+test[:30%]")
self.assertEqual(len(dset), 2)
self.assertDictEqual(dset.features, Features({"text": Value("string")}))
self.assertListEqual(dset.column_names, ["text"])
del dset
@require_faiss
def test_as_dataset_with_post_process_with_index(self):
def _post_process(self, dataset, resources_paths):
if os.path.exists(resources_paths["index"]):
dataset.load_faiss_index("my_index", resources_paths["index"])
return dataset
else:
dataset.add_faiss_index_from_external_arrays(
external_arrays=np.ones((len(dataset), 8)), string_factory="Flat", index_name="my_index"
)
dataset.save_faiss_index("my_index", resources_paths["index"])
return dataset
def _post_processing_resources(self, split):
return {"index": f"Flat-{split}.faiss"}
with tempfile.TemporaryDirectory() as tmp_dir:
builder = DummyBuilder(cache_dir=tmp_dir)
builder._post_process = types.MethodType(_post_process, builder)
builder._post_processing_resources = types.MethodType(_post_processing_resources, builder)
os.makedirs(builder.cache_dir)
builder.info.splits = SplitDict()
builder.info.splits.add(SplitInfo("train", num_examples=10))
builder.info.splits.add(SplitInfo("test", num_examples=10))
for split in builder.info.splits:
with ArrowWriter(
path=os.path.join(builder.cache_dir, f"{builder.dataset_name}-{split}.arrow"),
features=Features({"text": Value("string")}),
) as writer:
writer.write_batch({"text": ["foo"] * 10})
writer.finalize()
with ArrowWriter(
path=os.path.join(builder.cache_dir, f"small_dataset-{split}.arrow"),
features=Features({"text": Value("string")}),
) as writer:
writer.write_batch({"text": ["foo"] * 2})
writer.finalize()
dsets = builder.as_dataset()
self.assertIsInstance(dsets, DatasetDict)
self.assertListEqual(list(dsets.keys()), ["train", "test"])
self.assertEqual(len(dsets["train"]), 10)
self.assertEqual(len(dsets["test"]), 10)
self.assertDictEqual(dsets["train"].features, Features({"text": Value("string")}))
self.assertDictEqual(dsets["test"].features, Features({"text": Value("string")}))
self.assertListEqual(dsets["train"].column_names, ["text"])
self.assertListEqual(dsets["test"].column_names, ["text"])
self.assertListEqual(dsets["train"].list_indexes(), ["my_index"])
self.assertListEqual(dsets["test"].list_indexes(), ["my_index"])
self.assertGreater(builder.info.post_processing_size, 0)
self.assertGreater(builder.info.post_processed.resources_checksums["train"]["index"]["num_bytes"], 0)
del dsets
dset = builder.as_dataset("train")
self.assertIsInstance(dset, Dataset)
self.assertEqual(dset.split, "train")
self.assertEqual(len(dset), 10)
self.assertDictEqual(dset.features, Features({"text": Value("string")}))
self.assertListEqual(dset.column_names, ["text"])
self.assertListEqual(dset.list_indexes(), ["my_index"])
del dset
dset = builder.as_dataset("train+test[:30%]")
self.assertIsInstance(dset, Dataset)
self.assertEqual(dset.split, "train+test[:30%]")
self.assertEqual(len(dset), 13)
self.assertDictEqual(dset.features, Features({"text": Value("string")}))
self.assertListEqual(dset.column_names, ["text"])
self.assertListEqual(dset.list_indexes(), ["my_index"])
del dset
def test_download_and_prepare_with_post_process(self):
def _post_process(self, dataset, resources_paths):
def char_tokenize(example):
return {"tokens": list(example["text"])}
return dataset.map(char_tokenize, cache_file_name=resources_paths["tokenized_dataset"])
def _post_processing_resources(self, split):
return {"tokenized_dataset": f"tokenized_dataset-{split}.arrow"}
with tempfile.TemporaryDirectory() as tmp_dir:
builder = DummyBuilder(cache_dir=tmp_dir)
builder.info.post_processed = PostProcessedInfo(
features=Features({"text": Value("string"), "tokens": [Value("string")]})
)
builder._post_process = types.MethodType(_post_process, builder)
builder._post_processing_resources = types.MethodType(_post_processing_resources, builder)
builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
self.assertTrue(
os.path.exists(
os.path.join(
tmp_dir, builder.dataset_name, "default", "0.0.0", f"{builder.dataset_name}-train.arrow"
)
)
)
self.assertDictEqual(builder.info.features, Features({"text": Value("string")}))
self.assertDictEqual(
builder.info.post_processed.features,
Features({"text": Value("string"), "tokens": [Value("string")]}),
)
self.assertEqual(builder.info.splits["train"].num_examples, 100)
self.assertTrue(
os.path.exists(os.path.join(tmp_dir, builder.dataset_name, "default", "0.0.0", "dataset_info.json"))
)
def _post_process(self, dataset, resources_paths):
return dataset.select([0, 1], keep_in_memory=True)
with tempfile.TemporaryDirectory() as tmp_dir:
builder = DummyBuilder(cache_dir=tmp_dir)
builder._post_process = types.MethodType(_post_process, builder)
builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
self.assertTrue(
os.path.exists(
os.path.join(
tmp_dir, builder.dataset_name, "default", "0.0.0", f"{builder.dataset_name}-train.arrow"
)
)
)
self.assertDictEqual(builder.info.features, Features({"text": Value("string")}))
self.assertIsNone(builder.info.post_processed)
self.assertEqual(builder.info.splits["train"].num_examples, 100)
self.assertTrue(
os.path.exists(os.path.join(tmp_dir, builder.dataset_name, "default", "0.0.0", "dataset_info.json"))
)
def _post_process(self, dataset, resources_paths):
if os.path.exists(resources_paths["index"]):
dataset.load_faiss_index("my_index", resources_paths["index"])
return dataset
else:
dataset = dataset.add_faiss_index_from_external_arrays(
external_arrays=np.ones((len(dataset), 8)), string_factory="Flat", index_name="my_index"
)
dataset.save_faiss_index("my_index", resources_paths["index"])
return dataset
def _post_processing_resources(self, split):
return {"index": f"Flat-{split}.faiss"}
with tempfile.TemporaryDirectory() as tmp_dir:
builder = DummyBuilder(cache_dir=tmp_dir)
builder._post_process = types.MethodType(_post_process, builder)
builder._post_processing_resources = types.MethodType(_post_processing_resources, builder)
builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
self.assertTrue(
os.path.exists(
os.path.join(
tmp_dir, builder.dataset_name, "default", "0.0.0", f"{builder.dataset_name}-train.arrow"
)
)
)
self.assertDictEqual(builder.info.features, Features({"text": Value("string")}))
self.assertIsNone(builder.info.post_processed)
self.assertEqual(builder.info.splits["train"].num_examples, 100)
self.assertTrue(
os.path.exists(os.path.join(tmp_dir, builder.dataset_name, "default", "0.0.0", "dataset_info.json"))
)
def test_error_download_and_prepare(self):
def _prepare_split(self, split_generator, **kwargs):
raise ValueError()
with tempfile.TemporaryDirectory() as tmp_dir:
builder = DummyBuilder(cache_dir=tmp_dir)
builder._prepare_split = types.MethodType(_prepare_split, builder)
self.assertRaises(
ValueError,
builder.download_and_prepare,
try_from_hf_gcs=False,
download_mode=DownloadMode.FORCE_REDOWNLOAD,
)
self.assertRaises(FileNotFoundError, builder.as_dataset)
def test_generator_based_download_and_prepare(self):
with tempfile.TemporaryDirectory() as tmp_dir:
builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir)
builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
self.assertTrue(
os.path.exists(
os.path.join(
tmp_dir,
builder.dataset_name,
"default",
"0.0.0",
f"{builder.dataset_name}-train.arrow",
)
)
)
self.assertDictEqual(builder.info.features, Features({"text": Value("string")}))
self.assertEqual(builder.info.splits["train"].num_examples, 100)
self.assertTrue(
os.path.exists(os.path.join(tmp_dir, builder.dataset_name, "default", "0.0.0", "dataset_info.json"))
)
# Test that duplicated keys are ignored if verification_mode is "no_checks"
with tempfile.TemporaryDirectory() as tmp_dir:
builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir)
with patch("datasets.builder.ArrowWriter", side_effect=ArrowWriter) as mock_arrow_writer:
builder.download_and_prepare(
download_mode=DownloadMode.FORCE_REDOWNLOAD, verification_mode=VerificationMode.NO_CHECKS
)
mock_arrow_writer.assert_called_once()
args, kwargs = mock_arrow_writer.call_args_list[0]
self.assertFalse(kwargs["check_duplicates"])
mock_arrow_writer.reset_mock()
builder.download_and_prepare(
download_mode=DownloadMode.FORCE_REDOWNLOAD, verification_mode=VerificationMode.BASIC_CHECKS
)
mock_arrow_writer.assert_called_once()
args, kwargs = mock_arrow_writer.call_args_list[0]
self.assertTrue(kwargs["check_duplicates"])
def test_cache_dir_no_args(self):
with tempfile.TemporaryDirectory() as tmp_dir:
builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, data_dir=None, data_files=None)
relative_cache_dir_parts = Path(builder._relative_data_dir()).parts
self.assertTupleEqual(relative_cache_dir_parts, (builder.dataset_name, "default", "0.0.0"))
def test_cache_dir_for_data_files(self):
with tempfile.TemporaryDirectory() as tmp_dir:
dummy_data1 = os.path.join(tmp_dir, "dummy_data1.txt")
with open(dummy_data1, "w", encoding="utf-8") as f:
f.writelines("foo bar")
dummy_data2 = os.path.join(tmp_dir, "dummy_data2.txt")
with open(dummy_data2, "w", encoding="utf-8") as f:
f.writelines("foo bar\n")
builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, data_files=dummy_data1)
other_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, data_files=dummy_data1)
self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, data_files=[dummy_data1])
self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, data_files={"train": dummy_data1})
self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, data_files={Split.TRAIN: dummy_data1})
self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, data_files={"train": [dummy_data1]})
self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, data_files={"test": dummy_data1})
self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, data_files=dummy_data2)
self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, data_files=[dummy_data2])
self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, data_files=[dummy_data1, dummy_data2])
self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, data_files=[dummy_data1, dummy_data2])
other_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, data_files=[dummy_data1, dummy_data2])
self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(cache_dir=tmp_dir, data_files=[dummy_data2, dummy_data1])
self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, data_files={"train": dummy_data1, "test": dummy_data2}
)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, data_files={"train": dummy_data1, "test": dummy_data2}
)
self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, data_files={"train": [dummy_data1], "test": dummy_data2}
)
self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, data_files={"test": dummy_data2, "train": dummy_data1}
)
self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir, data_files={"train": dummy_data1, "validation": dummy_data2}
)
self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilder(
cache_dir=tmp_dir,
data_files={"train": [dummy_data1, dummy_data2], "test": dummy_data2},
)
self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
def test_cache_dir_for_features(self):
with tempfile.TemporaryDirectory() as tmp_dir:
f1 = Features({"id": Value("int8")})
f2 = Features({"id": Value("int32")})
builder = DummyGeneratorBasedBuilderWithIntegers(cache_dir=tmp_dir, features=f1)
other_builder = DummyGeneratorBasedBuilderWithIntegers(cache_dir=tmp_dir, features=f1)
self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilderWithIntegers(cache_dir=tmp_dir, features=f2)
self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
def test_cache_dir_for_config_kwargs(self):
with tempfile.TemporaryDirectory() as tmp_dir:
# create config on the fly
builder = DummyGeneratorBasedBuilderWithConfig(cache_dir=tmp_dir, content="foo", times=2)
other_builder = DummyGeneratorBasedBuilderWithConfig(cache_dir=tmp_dir, times=2, content="foo")
self.assertEqual(builder.cache_dir, other_builder.cache_dir)
self.assertIn("content=foo", builder.cache_dir)
self.assertIn("times=2", builder.cache_dir)
other_builder = DummyGeneratorBasedBuilderWithConfig(cache_dir=tmp_dir, content="bar", times=2)
self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyGeneratorBasedBuilderWithConfig(cache_dir=tmp_dir, content="foo")
self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
with tempfile.TemporaryDirectory() as tmp_dir:
# overwrite an existing config
builder = DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, config_name="a", content="foo", times=2)
other_builder = DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, config_name="a", times=2, content="foo")
self.assertEqual(builder.cache_dir, other_builder.cache_dir)
self.assertIn("content=foo", builder.cache_dir)
self.assertIn("times=2", builder.cache_dir)
other_builder = DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, config_name="a", content="bar", times=2)
self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, config_name="a", content="foo")
self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
def test_config_names(self):
with tempfile.TemporaryDirectory() as tmp_dir:
with self.assertRaises(ValueError) as error_context:
DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, data_files=None, data_dir=None)
self.assertIn("Please pick one among the available configs", str(error_context.exception))
builder = DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, config_name="a")
self.assertEqual(builder.config.name, "a")
builder = DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir, config_name="b")
self.assertEqual(builder.config.name, "b")
with self.assertRaises(ValueError):
DummyBuilderWithMultipleConfigs(cache_dir=tmp_dir)
builder = DummyBuilderWithDefaultConfig(cache_dir=tmp_dir)
self.assertEqual(builder.config.name, "a")
def test_cache_dir_for_data_dir(self):
with tempfile.TemporaryDirectory() as tmp_dir, tempfile.TemporaryDirectory() as data_dir:
builder = DummyBuilderWithManualDownload(cache_dir=tmp_dir, config_name="a", data_dir=data_dir)
other_builder = DummyBuilderWithManualDownload(cache_dir=tmp_dir, config_name="a", data_dir=data_dir)
self.assertEqual(builder.cache_dir, other_builder.cache_dir)
other_builder = DummyBuilderWithManualDownload(cache_dir=tmp_dir, config_name="a", data_dir=tmp_dir)
self.assertNotEqual(builder.cache_dir, other_builder.cache_dir)
def test_arrow_based_download_and_prepare(tmp_path):
builder = DummyArrowBasedBuilder(cache_dir=tmp_path)
builder.download_and_prepare()
assert os.path.exists(
os.path.join(
tmp_path,
builder.dataset_name,
"default",
"0.0.0",
f"{builder.dataset_name}-train.arrow",
)
)
assert builder.info.features, Features({"text": Value("string")})
assert builder.info.splits["train"].num_examples == 100
assert os.path.exists(os.path.join(tmp_path, builder.dataset_name, "default", "0.0.0", "dataset_info.json"))
@require_beam
def test_beam_based_download_and_prepare(tmp_path):
builder = DummyBeamBasedBuilder(cache_dir=tmp_path, beam_runner="DirectRunner")
builder.download_and_prepare()
assert os.path.exists(
os.path.join(
tmp_path,
builder.dataset_name,
"default",
"0.0.0",
f"{builder.dataset_name}-train.arrow",
)
)
assert builder.info.features, Features({"text": Value("string")})
assert builder.info.splits["train"].num_examples == 100
assert os.path.exists(os.path.join(tmp_path, builder.dataset_name, "default", "0.0.0", "dataset_info.json"))
@require_beam
def test_beam_based_as_dataset(tmp_path):
builder = DummyBeamBasedBuilder(cache_dir=tmp_path, beam_runner="DirectRunner")
builder.download_and_prepare()
dataset = builder.as_dataset()
assert dataset
assert isinstance(dataset["train"], Dataset)
assert len(dataset["train"]) > 0
@pytest.mark.parametrize(
"split, expected_dataset_class, expected_dataset_length",
[
(None, DatasetDict, 10),
("train", Dataset, 10),
("train+test[:30%]", Dataset, 13),
],
)
@pytest.mark.parametrize("in_memory", [False, True])
def test_builder_as_dataset(split, expected_dataset_class, expected_dataset_length, in_memory, tmp_path):
cache_dir = str(tmp_path)
builder = DummyBuilder(cache_dir=cache_dir)
os.makedirs(builder.cache_dir)
builder.info.splits = SplitDict()
builder.info.splits.add(SplitInfo("train", num_examples=10))
builder.info.splits.add(SplitInfo("test", num_examples=10))
for info_split in builder.info.splits:
with ArrowWriter(
path=os.path.join(builder.cache_dir, f"{builder.dataset_name}-{info_split}.arrow"),
features=Features({"text": Value("string")}),
) as writer:
writer.write_batch({"text": ["foo"] * 10})
writer.finalize()
with assert_arrow_memory_increases() if in_memory else assert_arrow_memory_doesnt_increase():
dataset = builder.as_dataset(split=split, in_memory=in_memory)
assert isinstance(dataset, expected_dataset_class)
if isinstance(dataset, DatasetDict):
assert list(dataset.keys()) == ["train", "test"]
datasets = dataset.values()
expected_splits = ["train", "test"]
elif isinstance(dataset, Dataset):
datasets = [dataset]
expected_splits = [split]
for dataset, expected_split in zip(datasets, expected_splits):
assert dataset.split == expected_split
assert len(dataset) == expected_dataset_length
assert dataset.features == Features({"text": Value("string")})
dataset.column_names == ["text"]
@pytest.mark.parametrize("in_memory", [False, True])
def test_generator_based_builder_as_dataset(in_memory, tmp_path):
cache_dir = tmp_path / "data"
cache_dir.mkdir()
cache_dir = str(cache_dir)
builder = DummyGeneratorBasedBuilder(cache_dir=cache_dir)
builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
with assert_arrow_memory_increases() if in_memory else assert_arrow_memory_doesnt_increase():
dataset = builder.as_dataset("train", in_memory=in_memory)
assert dataset.data.to_pydict() == {"text": ["foo"] * 100}
@pytest.mark.parametrize(
"writer_batch_size, default_writer_batch_size, expected_chunks", [(None, None, 1), (None, 5, 20), (10, None, 10)]
)
def test_custom_writer_batch_size(tmp_path, writer_batch_size, default_writer_batch_size, expected_chunks):
cache_dir = str(tmp_path)
if default_writer_batch_size:
DummyGeneratorBasedBuilder.DEFAULT_WRITER_BATCH_SIZE = default_writer_batch_size
builder = DummyGeneratorBasedBuilder(cache_dir=cache_dir, writer_batch_size=writer_batch_size)
assert builder._writer_batch_size == (writer_batch_size or default_writer_batch_size)
builder.download_and_prepare(try_from_hf_gcs=False, download_mode=DownloadMode.FORCE_REDOWNLOAD)
dataset = builder.as_dataset("train")
assert len(dataset.data[0].chunks) == expected_chunks
def test_builder_as_streaming_dataset(tmp_path):
dummy_builder = DummyGeneratorBasedBuilder(cache_dir=str(tmp_path))
check_streaming(dummy_builder)
dsets = dummy_builder.as_streaming_dataset()
assert isinstance(dsets, IterableDatasetDict)
assert isinstance(dsets["train"], IterableDataset)
assert len(list(dsets["train"])) == 100
dset = dummy_builder.as_streaming_dataset(split="train")
assert isinstance(dset, IterableDataset)
assert len(list(dset)) == 100
@require_beam
def test_beam_based_builder_as_streaming_dataset(tmp_path):
builder = DummyBeamBasedBuilder(cache_dir=tmp_path)
check_streaming(builder)
with pytest.raises(DatasetNotOnHfGcsError):
builder.as_streaming_dataset()
def _run_test_builder_streaming_works_in_subprocesses(builder):
check_streaming(builder)
dset = builder.as_streaming_dataset(split="train")
assert isinstance(dset, IterableDataset)
assert len(list(dset)) == 100
def test_builder_streaming_works_in_subprocess(tmp_path):
dummy_builder = DummyGeneratorBasedBuilder(cache_dir=str(tmp_path))
p = Process(target=_run_test_builder_streaming_works_in_subprocesses, args=(dummy_builder,))
p.start()
p.join()
class DummyBuilderWithVersion(GeneratorBasedBuilder):
VERSION = "2.0.0"
def _info(self):
return DatasetInfo(features=Features({"text": Value("string")}))
def _split_generators(self, dl_manager):
pass
def _generate_examples(self):
pass
class DummyBuilderWithBuilderConfigs(GeneratorBasedBuilder):
BUILDER_CONFIGS = [BuilderConfig(name="custom", version="2.0.0")]
def _info(self):
return DatasetInfo(features=Features({"text": Value("string")}))
def _split_generators(self, dl_manager):
pass
def _generate_examples(self):
pass
class CustomBuilderConfig(BuilderConfig):
def __init__(self, date=None, language=None, version="2.0.0", **kwargs):
name = f"{date}.{language}"
super().__init__(name=name, version=version, **kwargs)
self.date = date
self.language = language
class DummyBuilderWithCustomBuilderConfigs(GeneratorBasedBuilder):
BUILDER_CONFIGS = [CustomBuilderConfig(date="20220501", language="en")]
BUILDER_CONFIG_CLASS = CustomBuilderConfig
def _info(self):
return DatasetInfo(features=Features({"text": Value("string")}))
def _split_generators(self, dl_manager):
pass
def _generate_examples(self):
pass
@pytest.mark.parametrize(
"builder_class, kwargs",
[
(DummyBuilderWithVersion, {}),
(DummyBuilderWithBuilderConfigs, {"config_name": "custom"}),
(DummyBuilderWithCustomBuilderConfigs, {"config_name": "20220501.en"}),
(DummyBuilderWithCustomBuilderConfigs, {"date": "20220501", "language": "ca"}),
],
)
def test_builder_config_version(builder_class, kwargs, tmp_path):
cache_dir = str(tmp_path)
builder = builder_class(cache_dir=cache_dir, **kwargs)
assert builder.config.version == "2.0.0"
def test_builder_download_and_prepare_with_absolute_output_dir(tmp_path):
builder = DummyGeneratorBasedBuilder()
output_dir = str(tmp_path)
builder.download_and_prepare(output_dir)
assert builder._output_dir.startswith(tmp_path.resolve().as_posix())
assert os.path.exists(os.path.join(output_dir, "dataset_info.json"))
assert os.path.exists(os.path.join(output_dir, f"{builder.dataset_name}-train.arrow"))
assert not os.path.exists(os.path.join(output_dir + ".incomplete"))
def test_builder_download_and_prepare_with_relative_output_dir():
with set_current_working_directory_to_temp_dir():
builder = DummyGeneratorBasedBuilder()
output_dir = "test-out"
builder.download_and_prepare(output_dir)
assert Path(builder._output_dir).resolve().as_posix().startswith(Path(output_dir).resolve().as_posix())
assert os.path.exists(os.path.join(output_dir, "dataset_info.json"))
assert os.path.exists(os.path.join(output_dir, f"{builder.dataset_name}-train.arrow"))
assert not os.path.exists(os.path.join(output_dir + ".incomplete"))
def test_builder_with_filesystem_download_and_prepare(tmp_path, mockfs):
builder = DummyGeneratorBasedBuilder(cache_dir=tmp_path)
builder.download_and_prepare("mock://my_dataset", storage_options=mockfs.storage_options)
assert builder._output_dir.startswith("mock://my_dataset")
assert is_local_path(builder._cache_downloaded_dir)
assert isinstance(builder._fs, type(mockfs))
assert builder._fs.storage_options == mockfs.storage_options
assert mockfs.exists("my_dataset/dataset_info.json")
assert mockfs.exists(f"my_dataset/{builder.dataset_name}-train.arrow")
assert not mockfs.exists("my_dataset.incomplete")
def test_builder_with_filesystem_download_and_prepare_reload(tmp_path, mockfs, caplog):
builder = DummyGeneratorBasedBuilder(cache_dir=tmp_path)
mockfs.makedirs("my_dataset")
DatasetInfo().write_to_directory("mock://my_dataset", storage_options=mockfs.storage_options)
mockfs.touch(f"my_dataset/{builder.dataset_name}-train.arrow")
caplog.clear()
with caplog.at_level(INFO, logger=get_logger().name):
builder.download_and_prepare("mock://my_dataset", storage_options=mockfs.storage_options)
assert "Found cached dataset" in caplog.text
def test_generator_based_builder_download_and_prepare_as_parquet(tmp_path):
builder = DummyGeneratorBasedBuilder(cache_dir=tmp_path)
builder.download_and_prepare(file_format="parquet")
assert builder.info.splits["train"].num_examples == 100
parquet_path = os.path.join(
tmp_path, builder.dataset_name, "default", "0.0.0", f"{builder.dataset_name}-train.parquet"
)
assert os.path.exists(parquet_path)
assert pq.ParquetFile(parquet_path) is not None
def test_generator_based_builder_download_and_prepare_sharded(tmp_path):
writer_batch_size = 25
builder = DummyGeneratorBasedBuilder(cache_dir=tmp_path, writer_batch_size=writer_batch_size)
with patch("datasets.config.MAX_SHARD_SIZE", 1): # one batch per shard
builder.download_and_prepare(file_format="parquet")
expected_num_shards = 100 // writer_batch_size
assert builder.info.splits["train"].num_examples == 100
parquet_path = os.path.join(
tmp_path,
builder.dataset_name,
"default",
"0.0.0",
f"{builder.dataset_name}-train-00000-of-{expected_num_shards:05d}.parquet",
)
assert os.path.exists(parquet_path)
parquet_files = [
pq.ParquetFile(parquet_path)
for parquet_path in Path(tmp_path).rglob(
f"{builder.dataset_name}-train-*-of-{expected_num_shards:05d}.parquet"
)
]
assert len(parquet_files) == expected_num_shards
assert sum(parquet_file.metadata.num_rows for parquet_file in parquet_files) == 100
def test_generator_based_builder_download_and_prepare_with_max_shard_size(tmp_path):
writer_batch_size = 25
builder = DummyGeneratorBasedBuilder(cache_dir=tmp_path, writer_batch_size=writer_batch_size)
builder.download_and_prepare(file_format="parquet", max_shard_size=1) # one batch per shard
expected_num_shards = 100 // writer_batch_size
assert builder.info.splits["train"].num_examples == 100
parquet_path = os.path.join(
tmp_path,
builder.dataset_name,
"default",
"0.0.0",
f"{builder.dataset_name}-train-00000-of-{expected_num_shards:05d}.parquet",
)
assert os.path.exists(parquet_path)
parquet_files = [
pq.ParquetFile(parquet_path)
for parquet_path in Path(tmp_path).rglob(
f"{builder.dataset_name}-train-*-of-{expected_num_shards:05d}.parquet"
)
]
assert len(parquet_files) == expected_num_shards
assert sum(parquet_file.metadata.num_rows for parquet_file in parquet_files) == 100
def test_generator_based_builder_download_and_prepare_with_num_proc(tmp_path):
builder = DummyGeneratorBasedBuilderWithShards(cache_dir=tmp_path)
builder.download_and_prepare(num_proc=2)
expected_num_shards = 2
assert builder.info.splits["train"].num_examples == 400
assert builder.info.splits["train"].shard_lengths == [200, 200]
arrow_path = os.path.join(
tmp_path,
builder.dataset_name,
"default",
"0.0.0",
f"{builder.dataset_name}-train-00000-of-{expected_num_shards:05d}.arrow",
)
assert os.path.exists(arrow_path)
ds = builder.as_dataset("train")
assert len(ds) == 400
assert ds.to_dict() == {
"id": [i for _ in range(4) for i in range(100)],
"filepath": [f"data{i}.txt" for i in range(4) for _ in range(100)],
}
@pytest.mark.parametrize(
"num_proc, expectation", [(None, does_not_raise()), (1, does_not_raise()), (2, pytest.raises(RuntimeError))]
)
def test_generator_based_builder_download_and_prepare_with_ambiguous_shards(num_proc, expectation, tmp_path):
builder = DummyGeneratorBasedBuilderWithAmbiguousShards(cache_dir=tmp_path)
with expectation:
builder.download_and_prepare(num_proc=num_proc)
def test_arrow_based_builder_download_and_prepare_as_parquet(tmp_path):
builder = DummyArrowBasedBuilder(cache_dir=tmp_path)
builder.download_and_prepare(file_format="parquet")
assert builder.info.splits["train"].num_examples == 100
parquet_path = os.path.join(
tmp_path, builder.dataset_name, "default", "0.0.0", f"{builder.dataset_name}-train.parquet"
)
assert os.path.exists(parquet_path)
assert pq.ParquetFile(parquet_path) is not None
def test_arrow_based_builder_download_and_prepare_sharded(tmp_path):
builder = DummyArrowBasedBuilder(cache_dir=tmp_path)
with patch("datasets.config.MAX_SHARD_SIZE", 1): # one batch per shard
builder.download_and_prepare(file_format="parquet")
expected_num_shards = 10
assert builder.info.splits["train"].num_examples == 100
parquet_path = os.path.join(
tmp_path,
builder.dataset_name,
"default",
"0.0.0",
f"{builder.dataset_name}-train-00000-of-{expected_num_shards:05d}.parquet",
)
assert os.path.exists(parquet_path)
parquet_files = [
pq.ParquetFile(parquet_path)
for parquet_path in Path(tmp_path).rglob(
f"{builder.dataset_name}-train-*-of-{expected_num_shards:05d}.parquet"
)
]
assert len(parquet_files) == expected_num_shards
assert sum(parquet_file.metadata.num_rows for parquet_file in parquet_files) == 100
def test_arrow_based_builder_download_and_prepare_with_max_shard_size(tmp_path):
builder = DummyArrowBasedBuilder(cache_dir=tmp_path)
builder.download_and_prepare(file_format="parquet", max_shard_size=1) # one table per shard
expected_num_shards = 10
assert builder.info.splits["train"].num_examples == 100
parquet_path = os.path.join(
tmp_path,
builder.dataset_name,
"default",
"0.0.0",
f"{builder.dataset_name}-train-00000-of-{expected_num_shards:05d}.parquet",
)
assert os.path.exists(parquet_path)
parquet_files = [
pq.ParquetFile(parquet_path)
for parquet_path in Path(tmp_path).rglob(
f"{builder.dataset_name}-train-*-of-{expected_num_shards:05d}.parquet"
)
]
assert len(parquet_files) == expected_num_shards
assert sum(parquet_file.metadata.num_rows for parquet_file in parquet_files) == 100
def test_arrow_based_builder_download_and_prepare_with_num_proc(tmp_path):
builder = DummyArrowBasedBuilderWithShards(cache_dir=tmp_path)
builder.download_and_prepare(num_proc=2)
expected_num_shards = 2
assert builder.info.splits["train"].num_examples == 400
assert builder.info.splits["train"].shard_lengths == [200, 200]
arrow_path = os.path.join(
tmp_path,
builder.dataset_name,
"default",
"0.0.0",
f"{builder.dataset_name}-train-00000-of-{expected_num_shards:05d}.arrow",
)
assert os.path.exists(arrow_path)
ds = builder.as_dataset("train")
assert len(ds) == 400
assert ds.to_dict() == {
"id": [i for _ in range(4) for i in range(100)],
"filepath": [f"data{i}.txt" for i in range(4) for _ in range(100)],
}
@pytest.mark.parametrize(
"num_proc, expectation", [(None, does_not_raise()), (1, does_not_raise()), (2, pytest.raises(RuntimeError))]
)
def test_arrow_based_builder_download_and_prepare_with_ambiguous_shards(num_proc, expectation, tmp_path):
builder = DummyArrowBasedBuilderWithAmbiguousShards(cache_dir=tmp_path)
with expectation:
builder.download_and_prepare(num_proc=num_proc)
@require_beam
def test_beam_based_builder_download_and_prepare_as_parquet(tmp_path):
builder = DummyBeamBasedBuilder(cache_dir=tmp_path, beam_runner="DirectRunner")
builder.download_and_prepare(file_format="parquet")
assert builder.info.splits["train"].num_examples == 100
parquet_path = os.path.join(
tmp_path, builder.dataset_name, "default", "0.0.0", f"{builder.dataset_name}-train.parquet"
)
assert os.path.exists(parquet_path)
assert pq.ParquetFile(parquet_path) is not None
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_download_manager.py | import json
import os
from pathlib import Path
import pytest
from datasets.download.download_config import DownloadConfig
from datasets.download.download_manager import DownloadManager
from datasets.utils.file_utils import hash_url_to_filename
URL = "http://www.mocksite.com/file1.txt"
CONTENT = '"text": ["foo", "foo"]'
HASH = "6d8ce9aa78a471c7477201efbeabd3bb01ac2e7d100a6dc024ba1608361f90a8"
class MockResponse:
status_code = 200
headers = {"Content-Length": "100"}
cookies = {}
def iter_content(self, **kwargs):
return [bytes(CONTENT, "utf-8")]
def mock_request(*args, **kwargs):
return MockResponse()
@pytest.mark.parametrize("urls_type", [str, list, dict])
def test_download_manager_download(urls_type, tmp_path, monkeypatch):
import requests
monkeypatch.setattr(requests, "request", mock_request)
url = URL
if issubclass(urls_type, str):
urls = url
elif issubclass(urls_type, list):
urls = [url]
elif issubclass(urls_type, dict):
urls = {"train": url}
dataset_name = "dummy"
cache_subdir = "downloads"
cache_dir_root = tmp_path
download_config = DownloadConfig(
cache_dir=os.path.join(cache_dir_root, cache_subdir),
use_etag=False,
)
dl_manager = DownloadManager(dataset_name=dataset_name, download_config=download_config)
downloaded_paths = dl_manager.download(urls)
input_urls = urls
for downloaded_paths in [downloaded_paths]:
if isinstance(urls, str):
downloaded_paths = [downloaded_paths]
input_urls = [urls]
elif isinstance(urls, dict):
assert "train" in downloaded_paths.keys()
downloaded_paths = downloaded_paths.values()
input_urls = urls.values()
assert downloaded_paths
for downloaded_path, input_url in zip(downloaded_paths, input_urls):
assert downloaded_path == dl_manager.downloaded_paths[input_url]
downloaded_path = Path(downloaded_path)
parts = downloaded_path.parts
assert parts[-1] == HASH
assert parts[-2] == cache_subdir
assert downloaded_path.exists()
content = downloaded_path.read_text()
assert content == CONTENT
metadata_downloaded_path = downloaded_path.with_suffix(".json")
assert metadata_downloaded_path.exists()
metadata_content = json.loads(metadata_downloaded_path.read_text())
assert metadata_content == {"url": URL, "etag": None}
@pytest.mark.parametrize("paths_type", [str, list, dict])
def test_download_manager_extract(paths_type, xz_file, text_file):
filename = str(xz_file)
if issubclass(paths_type, str):
paths = filename
elif issubclass(paths_type, list):
paths = [filename]
elif issubclass(paths_type, dict):
paths = {"train": filename}
dataset_name = "dummy"
cache_dir = xz_file.parent
extracted_subdir = "extracted"
download_config = DownloadConfig(
cache_dir=cache_dir,
use_etag=False,
)
dl_manager = DownloadManager(dataset_name=dataset_name, download_config=download_config)
extracted_paths = dl_manager.extract(paths)
input_paths = paths
for extracted_paths in [extracted_paths]:
if isinstance(paths, str):
extracted_paths = [extracted_paths]
input_paths = [paths]
elif isinstance(paths, dict):
assert "train" in extracted_paths.keys()
extracted_paths = extracted_paths.values()
input_paths = paths.values()
assert extracted_paths
for extracted_path, input_path in zip(extracted_paths, input_paths):
assert extracted_path == dl_manager.extracted_paths[input_path]
extracted_path = Path(extracted_path)
parts = extracted_path.parts
assert parts[-1] == hash_url_to_filename(input_path, etag=None)
assert parts[-2] == extracted_subdir
assert extracted_path.exists()
extracted_file_content = extracted_path.read_text()
expected_file_content = text_file.read_text()
assert extracted_file_content == expected_file_content
def _test_jsonl(path, file):
assert path.endswith(".jsonl")
for num_items, line in enumerate(file, start=1):
item = json.loads(line.decode("utf-8"))
assert item.keys() == {"col_1", "col_2", "col_3"}
assert num_items == 4
@pytest.mark.parametrize("archive_jsonl", ["tar_jsonl_path", "zip_jsonl_path"])
def test_iter_archive_path(archive_jsonl, request):
archive_jsonl_path = request.getfixturevalue(archive_jsonl)
dl_manager = DownloadManager()
for num_jsonl, (path, file) in enumerate(dl_manager.iter_archive(archive_jsonl_path), start=1):
_test_jsonl(path, file)
assert num_jsonl == 2
@pytest.mark.parametrize("archive_nested_jsonl", ["tar_nested_jsonl_path", "zip_nested_jsonl_path"])
def test_iter_archive_file(archive_nested_jsonl, request):
archive_nested_jsonl_path = request.getfixturevalue(archive_nested_jsonl)
dl_manager = DownloadManager()
for num_tar, (path, file) in enumerate(dl_manager.iter_archive(archive_nested_jsonl_path), start=1):
for num_jsonl, (subpath, subfile) in enumerate(dl_manager.iter_archive(file), start=1):
_test_jsonl(subpath, subfile)
assert num_tar == 1
assert num_jsonl == 2
def test_iter_files(data_dir_with_hidden_files):
dl_manager = DownloadManager()
for num_file, file in enumerate(dl_manager.iter_files(data_dir_with_hidden_files), start=1):
assert os.path.basename(file) == ("test.txt" if num_file == 1 else "train.txt")
assert num_file == 2
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_upstream_hub.py | import fnmatch
import os
import tempfile
import time
import unittest
from io import BytesIO
from pathlib import Path
from unittest.mock import patch
import numpy as np
import pytest
from huggingface_hub import DatasetCard, HfApi
from datasets import (
Audio,
ClassLabel,
Dataset,
DatasetDict,
Features,
Image,
Value,
load_dataset,
load_dataset_builder,
)
from datasets.config import METADATA_CONFIGS_FIELD
from datasets.utils.file_utils import cached_path
from datasets.utils.hub import hf_hub_url
from tests.fixtures.hub import CI_HUB_ENDPOINT, CI_HUB_USER, CI_HUB_USER_TOKEN
from tests.utils import for_all_test_methods, require_pil, require_sndfile, xfail_if_500_502_http_error
pytestmark = pytest.mark.integration
@for_all_test_methods(xfail_if_500_502_http_error)
@pytest.mark.usefixtures("set_ci_hub_access_token", "ci_hfh_hf_hub_url")
class TestPushToHub:
_api = HfApi(endpoint=CI_HUB_ENDPOINT)
_token = CI_HUB_USER_TOKEN
def test_push_dataset_dict_to_hub_no_token(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
local_ds = DatasetDict({"train": ds})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
local_ds.push_to_hub(ds_name)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there is a single file on the repository that has the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset"))
assert all(
fnmatch.fnmatch(file, expected_file)
for file, expected_file in zip(
files, [".gitattributes", "README.md", "data/train-00000-of-00001-*.parquet"]
)
)
def test_push_dataset_dict_to_hub_name_without_namespace(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
local_ds = DatasetDict({"train": ds})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
local_ds.push_to_hub(ds_name.split("/")[-1], token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there is a single file on the repository that has the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset"))
assert all(
fnmatch.fnmatch(file, expected_file)
for file, expected_file in zip(
files, [".gitattributes", "README.md", "data/train-00000-of-00001-*.parquet"]
)
)
def test_push_dataset_dict_to_hub_datasets_with_different_features(self, cleanup_repo):
ds_train = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_test = Dataset.from_dict({"x": [True, False, True], "y": ["a", "b", "c"]})
local_ds = DatasetDict({"train": ds_train, "test": ds_test})
ds_name = f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}"
try:
with pytest.raises(ValueError):
local_ds.push_to_hub(ds_name.split("/")[-1], token=self._token)
except AssertionError:
cleanup_repo(ds_name)
raise
def test_push_dataset_dict_to_hub_private(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
local_ds = DatasetDict({"train": ds})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
local_ds.push_to_hub(ds_name, token=self._token, private=True)
hub_ds = load_dataset(ds_name, download_mode="force_redownload", token=self._token)
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there is a single file on the repository that has the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset", token=self._token))
assert all(
fnmatch.fnmatch(file, expected_file)
for file, expected_file in zip(
files, [".gitattributes", "README.md", "data/train-00000-of-00001-*.parquet"]
)
)
def test_push_dataset_dict_to_hub(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
local_ds = DatasetDict({"train": ds})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
local_ds.push_to_hub(ds_name, token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there is a single file on the repository that has the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset", token=self._token))
assert all(
fnmatch.fnmatch(file, expected_file)
for file, expected_file in zip(
files, [".gitattributes", "README.md", "data/train-00000-of-00001-*.parquet"]
)
)
def test_push_dataset_dict_to_hub_multiple_files(self, temporary_repo):
ds = Dataset.from_dict({"x": list(range(1000)), "y": list(range(1000))})
local_ds = DatasetDict({"train": ds})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
with patch("datasets.config.MAX_SHARD_SIZE", "16KB"):
local_ds.push_to_hub(ds_name, token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there are two files on the repository that have the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset", token=self._token))
assert all(
fnmatch.fnmatch(file, expected_file)
for file, expected_file in zip(
files,
[
".gitattributes",
"README.md",
"data/train-00000-of-00002-*.parquet",
"data/train-00001-of-00002-*.parquet",
],
)
)
def test_push_dataset_dict_to_hub_multiple_files_with_max_shard_size(self, temporary_repo):
ds = Dataset.from_dict({"x": list(range(1000)), "y": list(range(1000))})
local_ds = DatasetDict({"train": ds})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
local_ds.push_to_hub(ds_name, token=self._token, max_shard_size="16KB")
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there are two files on the repository that have the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset", token=self._token))
assert all(
fnmatch.fnmatch(file, expected_file)
for file, expected_file in zip(
files,
[
".gitattributes",
"README.md",
"data/train-00000-of-00002-*.parquet",
"data/train-00001-of-00002-*.parquet",
],
)
)
def test_push_dataset_dict_to_hub_multiple_files_with_num_shards(self, temporary_repo):
ds = Dataset.from_dict({"x": list(range(1000)), "y": list(range(1000))})
local_ds = DatasetDict({"train": ds})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
local_ds.push_to_hub(ds_name, token=self._token, num_shards={"train": 2})
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there are two files on the repository that have the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset", token=self._token))
assert all(
fnmatch.fnmatch(file, expected_file)
for file, expected_file in zip(
files,
[
".gitattributes",
"README.md",
"data/train-00000-of-00002-*.parquet",
"data/train-00001-of-00002-*.parquet",
],
)
)
def test_push_dataset_dict_to_hub_overwrite_files(self, temporary_repo):
ds = Dataset.from_dict({"x": list(range(1000)), "y": list(range(1000))})
ds2 = Dataset.from_dict({"x": list(range(100)), "y": list(range(100))})
local_ds = DatasetDict({"train": ds, "random": ds2})
ds_name = f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}"
# Push to hub two times, but the second time with a larger amount of files.
# Verify that the new files contain the correct dataset.
with temporary_repo(ds_name) as ds_name:
local_ds.push_to_hub(ds_name, token=self._token)
with tempfile.TemporaryDirectory() as tmp:
# Add a file starting with "data" to ensure it doesn't get deleted.
path = Path(tmp) / "datafile.txt"
with open(path, "w") as f:
f.write("Bogus file")
self._api.upload_file(
path_or_fileobj=str(path),
path_in_repo="datafile.txt",
repo_id=ds_name,
repo_type="dataset",
token=self._token,
)
local_ds.push_to_hub(ds_name, token=self._token, max_shard_size=500 << 5)
# Ensure that there are two files on the repository that have the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset", token=self._token))
assert all(
fnmatch.fnmatch(file, expected_file)
for file, expected_file in zip(
files,
[
".gitattributes",
"README.md",
"data/random-00000-of-00001-*.parquet",
"data/train-00000-of-00002-*.parquet",
"data/train-00001-of-00002-*.parquet",
"datafile.txt",
],
)
)
self._api.delete_file("datafile.txt", repo_id=ds_name, repo_type="dataset", token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
del hub_ds
# Push to hub two times, but the second time with fewer files.
# Verify that the new files contain the correct dataset and that non-necessary files have been deleted.
with temporary_repo(ds_name) as ds_name:
local_ds.push_to_hub(ds_name, token=self._token, max_shard_size=500 << 5)
with tempfile.TemporaryDirectory() as tmp:
# Add a file starting with "data" to ensure it doesn't get deleted.
path = Path(tmp) / "datafile.txt"
with open(path, "w") as f:
f.write("Bogus file")
self._api.upload_file(
path_or_fileobj=str(path),
path_in_repo="datafile.txt",
repo_id=ds_name,
repo_type="dataset",
token=self._token,
)
local_ds.push_to_hub(ds_name, token=self._token)
# Ensure that there are two files on the repository that have the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset", token=self._token))
assert all(
fnmatch.fnmatch(file, expected_file)
for file, expected_file in zip(
files,
[
".gitattributes",
"README.md",
"data/random-00000-of-00001-*.parquet",
"data/train-00000-of-00001-*.parquet",
"datafile.txt",
],
)
)
# Keeping the "datafile.txt" breaks the load_dataset to think it's a text-based dataset
self._api.delete_file("datafile.txt", repo_id=ds_name, repo_type="dataset", token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
def test_push_dataset_to_hub(self, temporary_repo):
local_ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
local_ds.push_to_hub(ds_name, split="train", token=self._token)
local_ds_dict = {"train": local_ds}
hub_ds_dict = load_dataset(ds_name, download_mode="force_redownload")
assert list(local_ds_dict.keys()) == list(hub_ds_dict.keys())
for ds_split_name in local_ds_dict.keys():
local_ds = local_ds_dict[ds_split_name]
hub_ds = hub_ds_dict[ds_split_name]
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds.features.keys()) == list(hub_ds.features.keys())
assert local_ds.features == hub_ds.features
def test_push_dataset_to_hub_custom_features(self, temporary_repo):
features = Features({"x": Value("int64"), "y": ClassLabel(names=["neg", "pos"])})
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [0, 0, 1]}, features=features)
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
ds.push_to_hub(ds_name, token=self._token)
hub_ds = load_dataset(ds_name, split="train", download_mode="force_redownload")
assert ds.column_names == hub_ds.column_names
assert list(ds.features.keys()) == list(hub_ds.features.keys())
assert ds.features == hub_ds.features
assert ds[:] == hub_ds[:]
@require_sndfile
def test_push_dataset_to_hub_custom_features_audio(self, temporary_repo):
audio_path = os.path.join(os.path.dirname(__file__), "features", "data", "test_audio_44100.wav")
data = {"x": [audio_path, None], "y": [0, -1]}
features = Features({"x": Audio(), "y": Value("int32")})
ds = Dataset.from_dict(data, features=features)
for embed_external_files in [True, False]:
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
ds.push_to_hub(ds_name, embed_external_files=embed_external_files, token=self._token)
hub_ds = load_dataset(ds_name, split="train", download_mode="force_redownload")
assert ds.column_names == hub_ds.column_names
assert list(ds.features.keys()) == list(hub_ds.features.keys())
assert ds.features == hub_ds.features
np.testing.assert_equal(ds[0]["x"]["array"], hub_ds[0]["x"]["array"])
assert ds[1] == hub_ds[1] # don't test hub_ds[0] since audio decoding might be slightly different
hub_ds = hub_ds.cast_column("x", Audio(decode=False))
elem = hub_ds[0]["x"]
path, bytes_ = elem["path"], elem["bytes"]
assert isinstance(path, str)
assert os.path.basename(path) == "test_audio_44100.wav"
assert bool(bytes_) == embed_external_files
@require_pil
def test_push_dataset_to_hub_custom_features_image(self, temporary_repo):
image_path = os.path.join(os.path.dirname(__file__), "features", "data", "test_image_rgb.jpg")
data = {"x": [image_path, None], "y": [0, -1]}
features = Features({"x": Image(), "y": Value("int32")})
ds = Dataset.from_dict(data, features=features)
for embed_external_files in [True, False]:
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
ds.push_to_hub(ds_name, embed_external_files=embed_external_files, token=self._token)
hub_ds = load_dataset(ds_name, split="train", download_mode="force_redownload")
assert ds.column_names == hub_ds.column_names
assert list(ds.features.keys()) == list(hub_ds.features.keys())
assert ds.features == hub_ds.features
assert ds[:] == hub_ds[:]
hub_ds = hub_ds.cast_column("x", Image(decode=False))
elem = hub_ds[0]["x"]
path, bytes_ = elem["path"], elem["bytes"]
assert isinstance(path, str)
assert bool(bytes_) == embed_external_files
@require_pil
def test_push_dataset_to_hub_custom_features_image_list(self, temporary_repo):
image_path = os.path.join(os.path.dirname(__file__), "features", "data", "test_image_rgb.jpg")
data = {"x": [[image_path], [image_path, image_path]], "y": [0, -1]}
features = Features({"x": [Image()], "y": Value("int32")})
ds = Dataset.from_dict(data, features=features)
for embed_external_files in [True, False]:
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
ds.push_to_hub(ds_name, embed_external_files=embed_external_files, token=self._token)
hub_ds = load_dataset(ds_name, split="train", download_mode="force_redownload")
assert ds.column_names == hub_ds.column_names
assert list(ds.features.keys()) == list(hub_ds.features.keys())
assert ds.features == hub_ds.features
assert ds[:] == hub_ds[:]
hub_ds = hub_ds.cast_column("x", [Image(decode=False)])
elem = hub_ds[0]["x"][0]
path, bytes_ = elem["path"], elem["bytes"]
assert isinstance(path, str)
assert bool(bytes_) == embed_external_files
def test_push_dataset_dict_to_hub_custom_features(self, temporary_repo):
features = Features({"x": Value("int64"), "y": ClassLabel(names=["neg", "pos"])})
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [0, 0, 1]}, features=features)
local_ds = DatasetDict({"test": ds})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
local_ds.push_to_hub(ds_name, token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["test"].features.keys()) == list(hub_ds["test"].features.keys())
assert local_ds["test"].features == hub_ds["test"].features
def test_push_dataset_to_hub_custom_splits(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
ds.push_to_hub(ds_name, split="random", token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert ds.column_names == hub_ds["random"].column_names
assert list(ds.features.keys()) == list(hub_ds["random"].features.keys())
assert ds.features == hub_ds["random"].features
def test_push_dataset_to_hub_skip_identical_files(self, temporary_repo):
ds = Dataset.from_dict({"x": list(range(1000)), "y": list(range(1000))})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
with patch("datasets.arrow_dataset.HfApi.upload_file", side_effect=self._api.upload_file) as mock_hf_api:
# Initial push
ds.push_to_hub(ds_name, token=self._token, max_shard_size="1KB")
call_count_old = mock_hf_api.call_count
mock_hf_api.reset_mock()
# Remove a data file
files = self._api.list_repo_files(ds_name, repo_type="dataset", token=self._token)
data_files = [f for f in files if f.startswith("data/")]
assert len(data_files) > 1
self._api.delete_file(data_files[0], repo_id=ds_name, repo_type="dataset", token=self._token)
# "Resume" push - push missing files
ds.push_to_hub(ds_name, token=self._token, max_shard_size="1KB")
call_count_new = mock_hf_api.call_count
assert call_count_old > call_count_new
hub_ds = load_dataset(ds_name, split="train", download_mode="force_redownload")
assert ds.column_names == hub_ds.column_names
assert list(ds.features.keys()) == list(hub_ds.features.keys())
assert ds.features == hub_ds.features
def test_push_dataset_to_hub_multiple_splits_one_by_one(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
ds.push_to_hub(ds_name, split="train", token=self._token)
ds.push_to_hub(ds_name, split="test", token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert sorted(hub_ds) == ["test", "train"]
assert ds.column_names == hub_ds["train"].column_names
assert list(ds.features.keys()) == list(hub_ds["train"].features.keys())
assert ds.features == hub_ds["train"].features
def test_push_dataset_dict_to_hub_custom_splits(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
local_ds = DatasetDict({"random": ds})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
local_ds.push_to_hub(ds_name, token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["random"].features.keys()) == list(hub_ds["random"].features.keys())
assert local_ds["random"].features == hub_ds["random"].features
@unittest.skip("This test cannot pass until iterable datasets have push to hub")
def test_push_streaming_dataset_dict_to_hub(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
local_ds = DatasetDict({"train": ds})
with tempfile.TemporaryDirectory() as tmp:
local_ds.save_to_disk(tmp)
local_ds = load_dataset(tmp, streaming=True)
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
local_ds.push_to_hub(ds_name, token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
def test_push_multiple_dataset_configs_to_hub_load_dataset_builder(self, temporary_repo):
ds_default = Dataset.from_dict({"a": [0], "b": [1]})
ds_config1 = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_config2 = Dataset.from_dict({"foo": [1, 2], "bar": [4, 5]})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
ds_default.push_to_hub(ds_name, token=self._token)
ds_config1.push_to_hub(ds_name, "config1", token=self._token)
ds_config2.push_to_hub(ds_name, "config2", token=self._token)
ds_builder_default = load_dataset_builder(ds_name, download_mode="force_redownload") # default config
assert len(ds_builder_default.BUILDER_CONFIGS) == 3
assert len(ds_builder_default.config.data_files["train"]) == 1
assert fnmatch.fnmatch(
ds_builder_default.config.data_files["train"][0],
"*/data/train-*",
)
ds_builder_config1 = load_dataset_builder(ds_name, "config1", download_mode="force_redownload")
assert len(ds_builder_config1.BUILDER_CONFIGS) == 3
assert len(ds_builder_config1.config.data_files["train"]) == 1
assert fnmatch.fnmatch(
ds_builder_config1.config.data_files["train"][0],
"*/config1/train-*",
)
ds_builder_config2 = load_dataset_builder(ds_name, "config2", download_mode="force_redownload")
assert len(ds_builder_config2.BUILDER_CONFIGS) == 3
assert len(ds_builder_config2.config.data_files["train"]) == 1
assert fnmatch.fnmatch(
ds_builder_config2.config.data_files["train"][0],
"*/config2/train-*",
)
with pytest.raises(ValueError): # no config 'config3'
load_dataset_builder(ds_name, "config3", download_mode="force_redownload")
def test_push_multiple_dataset_configs_to_hub_load_dataset(self, temporary_repo):
ds_default = Dataset.from_dict({"a": [0], "b": [1]})
ds_config1 = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_config2 = Dataset.from_dict({"foo": [1, 2], "bar": [4, 5]})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
ds_default.push_to_hub(ds_name, token=self._token)
ds_config1.push_to_hub(ds_name, "config1", token=self._token)
ds_config2.push_to_hub(ds_name, "config2", token=self._token)
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset"))
expected_files = sorted(
[
".gitattributes",
"README.md",
"config1/train-00000-of-00001-*.parquet",
"config2/train-00000-of-00001-*.parquet",
"data/train-00000-of-00001-*.parquet",
]
)
assert all(fnmatch.fnmatch(file, expected_file) for file, expected_file in zip(files, expected_files))
hub_ds_default = load_dataset(ds_name, download_mode="force_redownload")
hub_ds_config1 = load_dataset(ds_name, "config1", download_mode="force_redownload")
hub_ds_config2 = load_dataset(ds_name, "config2", download_mode="force_redownload")
# only "train" split
assert len(hub_ds_default) == len(hub_ds_config1) == len(hub_ds_config2) == 1
assert ds_default.column_names == hub_ds_default["train"].column_names == ["a", "b"]
assert ds_config1.column_names == hub_ds_config1["train"].column_names == ["x", "y"]
assert ds_config2.column_names == hub_ds_config2["train"].column_names == ["foo", "bar"]
assert ds_default.features == hub_ds_default["train"].features
assert ds_config1.features == hub_ds_config1["train"].features
assert ds_config2.features == hub_ds_config2["train"].features
assert ds_default.num_rows == hub_ds_default["train"].num_rows == 1
assert ds_config1.num_rows == hub_ds_config1["train"].num_rows == 3
assert ds_config2.num_rows == hub_ds_config2["train"].num_rows == 2
with pytest.raises(ValueError): # no config 'config3'
load_dataset(ds_name, "config3", download_mode="force_redownload")
def test_push_multiple_dataset_configs_to_hub_readme_metadata_content(self, temporary_repo):
ds_default = Dataset.from_dict({"a": [0], "b": [2]})
ds_config1 = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_config2 = Dataset.from_dict({"foo": [1, 2], "bar": [4, 5]})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
ds_default.push_to_hub(ds_name, token=self._token)
ds_config1.push_to_hub(ds_name, "config1", token=self._token)
ds_config2.push_to_hub(ds_name, "config2", token=self._token)
# check that configs args was correctly pushed to README.md
ds_readme_path = cached_path(hf_hub_url(ds_name, "README.md"))
dataset_card_data = DatasetCard.load(ds_readme_path).data
assert METADATA_CONFIGS_FIELD in dataset_card_data
assert isinstance(dataset_card_data[METADATA_CONFIGS_FIELD], list)
assert sorted(dataset_card_data[METADATA_CONFIGS_FIELD], key=lambda x: x["config_name"]) == [
{
"config_name": "config1",
"data_files": [
{"split": "train", "path": "config1/train-*"},
],
},
{
"config_name": "config2",
"data_files": [
{"split": "train", "path": "config2/train-*"},
],
},
{
"config_name": "default",
"data_files": [
{"split": "train", "path": "data/train-*"},
],
},
]
def test_push_multiple_dataset_dict_configs_to_hub_load_dataset_builder(self, temporary_repo):
ds_default = Dataset.from_dict({"a": [0], "b": [1]})
ds_config1 = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_config2 = Dataset.from_dict({"foo": [1, 2], "bar": [4, 5]})
ds_default = DatasetDict({"random": ds_default})
ds_config1 = DatasetDict({"random": ds_config1})
ds_config2 = DatasetDict({"random": ds_config2})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
ds_default.push_to_hub(ds_name, token=self._token)
ds_config1.push_to_hub(ds_name, "config1", token=self._token)
ds_config2.push_to_hub(ds_name, "config2", token=self._token)
ds_builder_default = load_dataset_builder(ds_name, download_mode="force_redownload") # default config
assert len(ds_builder_default.BUILDER_CONFIGS) == 3
assert len(ds_builder_default.config.data_files["random"]) == 1
assert fnmatch.fnmatch(
ds_builder_default.config.data_files["random"][0],
"*/data/random-*",
)
ds_builder_config1 = load_dataset_builder(ds_name, "config1", download_mode="force_redownload")
assert len(ds_builder_config1.BUILDER_CONFIGS) == 3
assert len(ds_builder_config1.config.data_files["random"]) == 1
assert fnmatch.fnmatch(
ds_builder_config1.config.data_files["random"][0],
"*/config1/random-*",
)
ds_builder_config2 = load_dataset_builder(ds_name, "config2", download_mode="force_redownload")
assert len(ds_builder_config2.BUILDER_CONFIGS) == 3
assert len(ds_builder_config2.config.data_files["random"]) == 1
assert fnmatch.fnmatch(
ds_builder_config2.config.data_files["random"][0],
"*/config2/random-*",
)
with pytest.raises(ValueError): # no config named 'config3'
load_dataset_builder(ds_name, "config3", download_mode="force_redownload")
def test_push_multiple_dataset_dict_configs_to_hub_load_dataset(self, temporary_repo):
ds_default = Dataset.from_dict({"a": [0], "b": [1]})
ds_config1 = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_config2 = Dataset.from_dict({"foo": [1, 2], "bar": [4, 5]})
ds_default = DatasetDict({"train": ds_default, "random": ds_default})
ds_config1 = DatasetDict({"train": ds_config1, "random": ds_config1})
ds_config2 = DatasetDict({"train": ds_config2, "random": ds_config2})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
ds_default.push_to_hub(ds_name, token=self._token)
ds_config1.push_to_hub(ds_name, "config1", token=self._token)
ds_config2.push_to_hub(ds_name, "config2", token=self._token)
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset"))
expected_files = sorted(
[
".gitattributes",
"README.md",
"config1/random-00000-of-00001-*.parquet",
"config1/train-00000-of-00001-*.parquet",
"config2/random-00000-of-00001-*.parquet",
"config2/train-00000-of-00001-*.parquet",
"data/random-00000-of-00001-*.parquet",
"data/train-00000-of-00001-*.parquet",
]
)
assert all(fnmatch.fnmatch(file, expected_file) for file, expected_file in zip(files, expected_files))
hub_ds_default = load_dataset(ds_name, download_mode="force_redownload")
hub_ds_config1 = load_dataset(ds_name, "config1", download_mode="force_redownload")
hub_ds_config2 = load_dataset(ds_name, "config2", download_mode="force_redownload")
# two splits
expected_splits = ["random", "train"]
assert len(hub_ds_default) == len(hub_ds_config1) == len(hub_ds_config2) == 2
assert sorted(hub_ds_default) == sorted(hub_ds_config1) == sorted(hub_ds_config2) == expected_splits
for split in expected_splits:
assert ds_default[split].column_names == hub_ds_default[split].column_names == ["a", "b"]
assert ds_config1[split].column_names == hub_ds_config1[split].column_names == ["x", "y"]
assert ds_config2[split].column_names == hub_ds_config2[split].column_names == ["foo", "bar"]
assert ds_default[split].features == hub_ds_default[split].features
assert ds_config1[split].features == hub_ds_config1[split].features
assert ds_config2[split].features == hub_ds_config2["train"].features
assert ds_default[split].num_rows == hub_ds_default[split].num_rows == 1
assert ds_config1[split].num_rows == hub_ds_config1[split].num_rows == 3
assert ds_config2[split].num_rows == hub_ds_config2[split].num_rows == 2
with pytest.raises(ValueError): # no config 'config3'
load_dataset(ds_name, "config3", download_mode="force_redownload")
def test_push_multiple_dataset_dict_configs_to_hub_readme_metadata_content(self, temporary_repo):
ds_default = Dataset.from_dict({"a": [0], "b": [1]})
ds_config1 = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_config2 = Dataset.from_dict({"foo": [1, 2], "bar": [4, 5]})
ds_default = DatasetDict({"train": ds_default, "random": ds_default})
ds_config1 = DatasetDict({"train": ds_config1, "random": ds_config1})
ds_config2 = DatasetDict({"train": ds_config2, "random": ds_config2})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
ds_default.push_to_hub(ds_name, token=self._token)
ds_config1.push_to_hub(ds_name, "config1", token=self._token)
ds_config2.push_to_hub(ds_name, "config2", token=self._token)
# check that configs args was correctly pushed to README.md
ds_readme_path = cached_path(hf_hub_url(ds_name, "README.md"))
dataset_card_data = DatasetCard.load(ds_readme_path).data
assert METADATA_CONFIGS_FIELD in dataset_card_data
assert isinstance(dataset_card_data[METADATA_CONFIGS_FIELD], list)
assert sorted(dataset_card_data[METADATA_CONFIGS_FIELD], key=lambda x: x["config_name"]) == [
{
"config_name": "config1",
"data_files": [
{"split": "train", "path": "config1/train-*"},
{"split": "random", "path": "config1/random-*"},
],
},
{
"config_name": "config2",
"data_files": [
{"split": "train", "path": "config2/train-*"},
{"split": "random", "path": "config2/random-*"},
],
},
{
"config_name": "default",
"data_files": [
{"split": "train", "path": "data/train-*"},
{"split": "random", "path": "data/random-*"},
],
},
]
def test_push_dataset_to_hub_with_config_no_metadata_configs(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_another_config = Dataset.from_dict({"foo": [1, 2], "bar": [4, 5]})
parquet_buf = BytesIO()
ds.to_parquet(parquet_buf)
parquet_content = parquet_buf.getvalue()
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
self._api.create_repo(ds_name, token=self._token, repo_type="dataset")
# old push_to_hub was uploading the parquet files only - without metadata configs
self._api.upload_file(
path_or_fileobj=parquet_content,
path_in_repo="data/train-00000-of-00001.parquet",
repo_id=ds_name,
repo_type="dataset",
)
ds_another_config.push_to_hub(ds_name, "another_config", token=self._token)
ds_builder = load_dataset_builder(ds_name, download_mode="force_redownload")
assert len(ds_builder.config.data_files) == 1
assert len(ds_builder.config.data_files["train"]) == 1
assert fnmatch.fnmatch(ds_builder.config.data_files["train"][0], "*/data/train-00000-of-00001.parquet")
ds_another_config_builder = load_dataset_builder(
ds_name, "another_config", download_mode="force_redownload"
)
assert len(ds_another_config_builder.config.data_files) == 1
assert len(ds_another_config_builder.config.data_files["train"]) == 1
assert fnmatch.fnmatch(
ds_another_config_builder.config.data_files["train"][0],
"*/another_config/train-00000-of-00001-*.parquet",
)
def test_push_dataset_dict_to_hub_with_config_no_metadata_configs(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_another_config = Dataset.from_dict({"foo": [1, 2], "bar": [4, 5]})
parquet_buf = BytesIO()
ds.to_parquet(parquet_buf)
parquet_content = parquet_buf.getvalue()
local_ds_another_config = DatasetDict({"random": ds_another_config})
with temporary_repo(f"{CI_HUB_USER}/test-{int(time.time() * 10e3)}") as ds_name:
self._api.create_repo(ds_name, token=self._token, repo_type="dataset")
# old push_to_hub was uploading the parquet files only - without metadata configs
self._api.upload_file(
path_or_fileobj=parquet_content,
path_in_repo="data/random-00000-of-00001.parquet",
repo_id=ds_name,
repo_type="dataset",
)
local_ds_another_config.push_to_hub(ds_name, "another_config", token=self._token)
ds_builder = load_dataset_builder(ds_name, download_mode="force_redownload")
assert len(ds_builder.config.data_files) == 1
assert len(ds_builder.config.data_files["random"]) == 1
assert fnmatch.fnmatch(ds_builder.config.data_files["random"][0], "*/data/random-00000-of-00001.parquet")
ds_another_config_builder = load_dataset_builder(
ds_name, "another_config", download_mode="force_redownload"
)
assert len(ds_another_config_builder.config.data_files) == 1
assert len(ds_another_config_builder.config.data_files["random"]) == 1
assert fnmatch.fnmatch(
ds_another_config_builder.config.data_files["random"][0],
"*/another_config/random-00000-of-00001-*.parquet",
)
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_arrow_writer.py | import copy
import os
import tempfile
from unittest import TestCase
from unittest.mock import patch
import numpy as np
import pyarrow as pa
import pyarrow.parquet as pq
import pytest
from datasets.arrow_writer import ArrowWriter, OptimizedTypedSequence, ParquetWriter, TypedSequence
from datasets.features import Array2D, ClassLabel, Features, Image, Value
from datasets.features.features import Array2DExtensionType, cast_to_python_objects
from datasets.keyhash import DuplicatedKeysError, InvalidKeyError
from .utils import require_pil
class TypedSequenceTest(TestCase):
def test_no_type(self):
arr = pa.array(TypedSequence([1, 2, 3]))
self.assertEqual(arr.type, pa.int64())
def test_array_type_forbidden(self):
with self.assertRaises(ValueError):
_ = pa.array(TypedSequence([1, 2, 3]), type=pa.int64())
def test_try_type_and_type_forbidden(self):
with self.assertRaises(ValueError):
_ = pa.array(TypedSequence([1, 2, 3], try_type=Value("bool"), type=Value("int64")))
def test_compatible_type(self):
arr = pa.array(TypedSequence([1, 2, 3], type=Value("int32")))
self.assertEqual(arr.type, pa.int32())
def test_incompatible_type(self):
with self.assertRaises((TypeError, pa.lib.ArrowInvalid)):
_ = pa.array(TypedSequence(["foo", "bar"], type=Value("int64")))
def test_try_compatible_type(self):
arr = pa.array(TypedSequence([1, 2, 3], try_type=Value("int32")))
self.assertEqual(arr.type, pa.int32())
def test_try_incompatible_type(self):
arr = pa.array(TypedSequence(["foo", "bar"], try_type=Value("int64")))
self.assertEqual(arr.type, pa.string())
def test_compatible_extension_type(self):
arr = pa.array(TypedSequence([[[1, 2, 3]]], type=Array2D((1, 3), "int64")))
self.assertEqual(arr.type, Array2DExtensionType((1, 3), "int64"))
def test_incompatible_extension_type(self):
with self.assertRaises((TypeError, pa.lib.ArrowInvalid)):
_ = pa.array(TypedSequence(["foo", "bar"], type=Array2D((1, 3), "int64")))
def test_try_compatible_extension_type(self):
arr = pa.array(TypedSequence([[[1, 2, 3]]], try_type=Array2D((1, 3), "int64")))
self.assertEqual(arr.type, Array2DExtensionType((1, 3), "int64"))
def test_try_incompatible_extension_type(self):
arr = pa.array(TypedSequence(["foo", "bar"], try_type=Array2D((1, 3), "int64")))
self.assertEqual(arr.type, pa.string())
@require_pil
def test_exhaustive_cast(self):
import PIL.Image
pil_image = PIL.Image.fromarray(np.arange(10, dtype=np.uint8).reshape(2, 5))
with patch(
"datasets.arrow_writer.cast_to_python_objects", side_effect=cast_to_python_objects
) as mock_cast_to_python_objects:
_ = pa.array(TypedSequence([{"path": None, "bytes": b"image_bytes"}, pil_image], type=Image()))
args, kwargs = mock_cast_to_python_objects.call_args_list[-1]
self.assertIn("optimize_list_casting", kwargs)
self.assertFalse(kwargs["optimize_list_casting"])
def _check_output(output, expected_num_chunks: int):
stream = pa.BufferReader(output) if isinstance(output, pa.Buffer) else pa.memory_map(output)
f = pa.ipc.open_stream(stream)
pa_table: pa.Table = f.read_all()
assert len(pa_table.to_batches()) == expected_num_chunks
assert pa_table.to_pydict() == {"col_1": ["foo", "bar"], "col_2": [1, 2]}
del pa_table
@pytest.mark.parametrize("writer_batch_size", [None, 1, 10])
@pytest.mark.parametrize(
"fields", [None, {"col_1": pa.string(), "col_2": pa.int64()}, {"col_1": pa.string(), "col_2": pa.int32()}]
)
def test_write(fields, writer_batch_size):
output = pa.BufferOutputStream()
schema = pa.schema(fields) if fields else None
with ArrowWriter(stream=output, schema=schema, writer_batch_size=writer_batch_size) as writer:
writer.write({"col_1": "foo", "col_2": 1})
writer.write({"col_1": "bar", "col_2": 2})
num_examples, num_bytes = writer.finalize()
assert num_examples == 2
assert num_bytes > 0
if not fields:
fields = {"col_1": pa.string(), "col_2": pa.int64()}
assert writer._schema == pa.schema(fields, metadata=writer._schema.metadata)
_check_output(output.getvalue(), expected_num_chunks=num_examples if writer_batch_size == 1 else 1)
def test_write_with_features():
output = pa.BufferOutputStream()
features = Features({"labels": ClassLabel(names=["neg", "pos"])})
with ArrowWriter(stream=output, features=features) as writer:
writer.write({"labels": 0})
writer.write({"labels": 1})
num_examples, num_bytes = writer.finalize()
assert num_examples == 2
assert num_bytes > 0
assert writer._schema == features.arrow_schema
assert writer._schema.metadata == features.arrow_schema.metadata
stream = pa.BufferReader(output.getvalue())
f = pa.ipc.open_stream(stream)
pa_table: pa.Table = f.read_all()
schema = pa_table.schema
assert pa_table.num_rows == 2
assert schema == features.arrow_schema
assert schema.metadata == features.arrow_schema.metadata
assert features == Features.from_arrow_schema(schema)
@pytest.mark.parametrize("writer_batch_size", [None, 1, 10])
def test_key_datatype(writer_batch_size):
output = pa.BufferOutputStream()
with ArrowWriter(
stream=output,
writer_batch_size=writer_batch_size,
hash_salt="split_name",
check_duplicates=True,
) as writer:
with pytest.raises(InvalidKeyError):
writer.write({"col_1": "foo", "col_2": 1}, key=[1, 2])
num_examples, num_bytes = writer.finalize()
@pytest.mark.parametrize("writer_batch_size", [None, 2, 10])
def test_duplicate_keys(writer_batch_size):
output = pa.BufferOutputStream()
with ArrowWriter(
stream=output,
writer_batch_size=writer_batch_size,
hash_salt="split_name",
check_duplicates=True,
) as writer:
with pytest.raises(DuplicatedKeysError):
writer.write({"col_1": "foo", "col_2": 1}, key=10)
writer.write({"col_1": "bar", "col_2": 2}, key=10)
num_examples, num_bytes = writer.finalize()
@pytest.mark.parametrize("writer_batch_size", [None, 2, 10])
def test_write_with_keys(writer_batch_size):
output = pa.BufferOutputStream()
with ArrowWriter(
stream=output,
writer_batch_size=writer_batch_size,
hash_salt="split_name",
check_duplicates=True,
) as writer:
writer.write({"col_1": "foo", "col_2": 1}, key=1)
writer.write({"col_1": "bar", "col_2": 2}, key=2)
num_examples, num_bytes = writer.finalize()
assert num_examples == 2
assert num_bytes > 0
_check_output(output.getvalue(), expected_num_chunks=num_examples if writer_batch_size == 1 else 1)
@pytest.mark.parametrize("writer_batch_size", [None, 1, 10])
@pytest.mark.parametrize(
"fields", [None, {"col_1": pa.string(), "col_2": pa.int64()}, {"col_1": pa.string(), "col_2": pa.int32()}]
)
def test_write_batch(fields, writer_batch_size):
output = pa.BufferOutputStream()
schema = pa.schema(fields) if fields else None
with ArrowWriter(stream=output, schema=schema, writer_batch_size=writer_batch_size) as writer:
writer.write_batch({"col_1": ["foo", "bar"], "col_2": [1, 2]})
writer.write_batch({"col_1": [], "col_2": []})
num_examples, num_bytes = writer.finalize()
assert num_examples == 2
assert num_bytes > 0
if not fields:
fields = {"col_1": pa.string(), "col_2": pa.int64()}
assert writer._schema == pa.schema(fields, metadata=writer._schema.metadata)
_check_output(output.getvalue(), expected_num_chunks=num_examples if writer_batch_size == 1 else 1)
@pytest.mark.parametrize("writer_batch_size", [None, 1, 10])
@pytest.mark.parametrize(
"fields", [None, {"col_1": pa.string(), "col_2": pa.int64()}, {"col_1": pa.string(), "col_2": pa.int32()}]
)
def test_write_table(fields, writer_batch_size):
output = pa.BufferOutputStream()
schema = pa.schema(fields) if fields else None
with ArrowWriter(stream=output, schema=schema, writer_batch_size=writer_batch_size) as writer:
writer.write_table(pa.Table.from_pydict({"col_1": ["foo", "bar"], "col_2": [1, 2]}))
num_examples, num_bytes = writer.finalize()
assert num_examples == 2
assert num_bytes > 0
if not fields:
fields = {"col_1": pa.string(), "col_2": pa.int64()}
assert writer._schema == pa.schema(fields, metadata=writer._schema.metadata)
_check_output(output.getvalue(), expected_num_chunks=num_examples if writer_batch_size == 1 else 1)
@pytest.mark.parametrize("writer_batch_size", [None, 1, 10])
@pytest.mark.parametrize(
"fields", [None, {"col_1": pa.string(), "col_2": pa.int64()}, {"col_1": pa.string(), "col_2": pa.int32()}]
)
def test_write_row(fields, writer_batch_size):
output = pa.BufferOutputStream()
schema = pa.schema(fields) if fields else None
with ArrowWriter(stream=output, schema=schema, writer_batch_size=writer_batch_size) as writer:
writer.write_row(pa.Table.from_pydict({"col_1": ["foo"], "col_2": [1]}))
writer.write_row(pa.Table.from_pydict({"col_1": ["bar"], "col_2": [2]}))
num_examples, num_bytes = writer.finalize()
assert num_examples == 2
assert num_bytes > 0
if not fields:
fields = {"col_1": pa.string(), "col_2": pa.int64()}
assert writer._schema == pa.schema(fields, metadata=writer._schema.metadata)
_check_output(output.getvalue(), expected_num_chunks=num_examples if writer_batch_size == 1 else 1)
def test_write_file():
with tempfile.TemporaryDirectory() as tmp_dir:
fields = {"col_1": pa.string(), "col_2": pa.int64()}
output = os.path.join(tmp_dir, "test.arrow")
with ArrowWriter(path=output, schema=pa.schema(fields)) as writer:
writer.write_batch({"col_1": ["foo", "bar"], "col_2": [1, 2]})
num_examples, num_bytes = writer.finalize()
assert num_examples == 2
assert num_bytes > 0
assert writer._schema == pa.schema(fields, metadata=writer._schema.metadata)
_check_output(output, 1)
def get_base_dtype(arr_type):
if pa.types.is_list(arr_type):
return get_base_dtype(arr_type.value_type)
else:
return arr_type
def change_first_primitive_element_in_list(lst, value):
if isinstance(lst[0], list):
change_first_primitive_element_in_list(lst[0], value)
else:
lst[0] = value
@pytest.mark.parametrize("optimized_int_type, expected_dtype", [(None, pa.int64()), (Value("int32"), pa.int32())])
@pytest.mark.parametrize("sequence", [[1, 2, 3], [[1, 2, 3]], [[[1, 2, 3]]]])
def test_optimized_int_type_for_typed_sequence(sequence, optimized_int_type, expected_dtype):
arr = pa.array(TypedSequence(sequence, optimized_int_type=optimized_int_type))
assert get_base_dtype(arr.type) == expected_dtype
@pytest.mark.parametrize(
"col, expected_dtype",
[
("attention_mask", pa.int8()),
("special_tokens_mask", pa.int8()),
("token_type_ids", pa.int8()),
("input_ids", pa.int32()),
("other", pa.int64()),
],
)
@pytest.mark.parametrize("sequence", [[1, 2, 3], [[1, 2, 3]], [[[1, 2, 3]]]])
def test_optimized_typed_sequence(sequence, col, expected_dtype):
# in range
arr = pa.array(OptimizedTypedSequence(sequence, col=col))
assert get_base_dtype(arr.type) == expected_dtype
# not in range
if col != "other":
# avoids errors due to in-place modifications
sequence = copy.deepcopy(sequence)
value = np.iinfo(expected_dtype.to_pandas_dtype()).max + 1
change_first_primitive_element_in_list(sequence, value)
arr = pa.array(OptimizedTypedSequence(sequence, col=col))
assert get_base_dtype(arr.type) == pa.int64()
@pytest.mark.parametrize("raise_exception", [False, True])
def test_arrow_writer_closes_stream(raise_exception, tmp_path):
path = str(tmp_path / "dataset-train.arrow")
try:
with ArrowWriter(path=path) as writer:
if raise_exception:
raise pa.lib.ArrowInvalid()
else:
writer.stream.close()
except pa.lib.ArrowInvalid:
pass
finally:
assert writer.stream.closed
def test_arrow_writer_with_filesystem(mockfs):
path = "mock://dataset-train.arrow"
with ArrowWriter(path=path, storage_options=mockfs.storage_options) as writer:
assert isinstance(writer._fs, type(mockfs))
assert writer._fs.storage_options == mockfs.storage_options
writer.write({"col_1": "foo", "col_2": 1})
writer.write({"col_1": "bar", "col_2": 2})
num_examples, num_bytes = writer.finalize()
assert num_examples == 2
assert num_bytes > 0
assert mockfs.exists(path)
def test_parquet_writer_write():
output = pa.BufferOutputStream()
with ParquetWriter(stream=output) as writer:
writer.write({"col_1": "foo", "col_2": 1})
writer.write({"col_1": "bar", "col_2": 2})
num_examples, num_bytes = writer.finalize()
assert num_examples == 2
assert num_bytes > 0
stream = pa.BufferReader(output.getvalue())
pa_table: pa.Table = pq.read_table(stream)
assert pa_table.to_pydict() == {"col_1": ["foo", "bar"], "col_2": [1, 2]}
@require_pil
@pytest.mark.parametrize("embed_local_files", [False, True])
def test_writer_embed_local_files(tmp_path, embed_local_files):
import PIL.Image
image_path = str(tmp_path / "test_image_rgb.jpg")
PIL.Image.fromarray(np.zeros((5, 5), dtype=np.uint8)).save(image_path, format="png")
output = pa.BufferOutputStream()
with ParquetWriter(
stream=output, features=Features({"image": Image()}), embed_local_files=embed_local_files
) as writer:
writer.write({"image": image_path})
writer.finalize()
stream = pa.BufferReader(output.getvalue())
pa_table: pa.Table = pq.read_table(stream)
out = pa_table.to_pydict()
if embed_local_files:
assert isinstance(out["image"][0]["path"], str)
with open(image_path, "rb") as f:
assert out["image"][0]["bytes"] == f.read()
else:
assert out["image"][0]["path"] == image_path
assert out["image"][0]["bytes"] is None
def test_always_nullable():
non_nullable_schema = pa.schema([pa.field("col_1", pa.string(), nullable=False)])
output = pa.BufferOutputStream()
with ArrowWriter(stream=output) as writer:
writer._build_writer(inferred_schema=non_nullable_schema)
assert writer._schema == pa.schema([pa.field("col_1", pa.string())])
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_inspect.py | import os
import pytest
from datasets import (
get_dataset_config_info,
get_dataset_config_names,
get_dataset_infos,
get_dataset_split_names,
inspect_dataset,
inspect_metric,
)
pytestmark = pytest.mark.integration
@pytest.mark.parametrize("path", ["paws", "csv"])
def test_inspect_dataset(path, tmp_path):
inspect_dataset(path, tmp_path)
script_name = path + ".py"
assert script_name in os.listdir(tmp_path)
assert "__pycache__" not in os.listdir(tmp_path)
@pytest.mark.filterwarnings("ignore:inspect_metric is deprecated:FutureWarning")
@pytest.mark.filterwarnings("ignore:metric_module_factory is deprecated:FutureWarning")
@pytest.mark.parametrize("path", ["accuracy"])
def test_inspect_metric(path, tmp_path):
inspect_metric(path, tmp_path)
script_name = path + ".py"
assert script_name in os.listdir(tmp_path)
assert "__pycache__" not in os.listdir(tmp_path)
@pytest.mark.parametrize(
"path, config_name, expected_splits",
[
("squad", "plain_text", ["train", "validation"]),
("dalle-mini/wit", "default", ["train"]),
("paws", "labeled_final", ["train", "test", "validation"]),
],
)
def test_get_dataset_config_info(path, config_name, expected_splits):
info = get_dataset_config_info(path, config_name=config_name)
assert info.config_name == config_name
assert list(info.splits.keys()) == expected_splits
@pytest.mark.parametrize(
"path, config_name, expected_exception",
[
("paws", None, ValueError),
],
)
def test_get_dataset_config_info_error(path, config_name, expected_exception):
with pytest.raises(expected_exception):
get_dataset_config_info(path, config_name=config_name)
@pytest.mark.parametrize(
"path, expected",
[
("squad", ["plain_text"]),
("acronym_identification", ["default"]),
("lhoestq/squad", ["plain_text"]),
("lhoestq/test", ["default"]),
("lhoestq/demo1", ["default"]),
("dalle-mini/wit", ["default"]),
("datasets-maintainers/audiofolder_no_configs_in_metadata", ["default"]),
("datasets-maintainers/audiofolder_single_config_in_metadata", ["custom"]),
("datasets-maintainers/audiofolder_two_configs_in_metadata", ["v1", "v2"]),
],
)
def test_get_dataset_config_names(path, expected):
config_names = get_dataset_config_names(path)
assert config_names == expected
@pytest.mark.parametrize(
"path, expected_configs, expected_splits_in_first_config",
[
("squad", ["plain_text"], ["train", "validation"]),
("dalle-mini/wit", ["default"], ["train"]),
("paws", ["labeled_final", "labeled_swap", "unlabeled_final"], ["train", "test", "validation"]),
],
)
def test_get_dataset_info(path, expected_configs, expected_splits_in_first_config):
infos = get_dataset_infos(path)
assert list(infos.keys()) == expected_configs
expected_config = expected_configs[0]
assert expected_config in infos
info = infos[expected_config]
assert info.config_name == expected_config
assert list(info.splits.keys()) == expected_splits_in_first_config
@pytest.mark.parametrize(
"path, expected_config, expected_splits",
[
("squad", "plain_text", ["train", "validation"]),
("dalle-mini/wit", "default", ["train"]),
("paws", "labeled_final", ["train", "test", "validation"]),
],
)
def test_get_dataset_split_names(path, expected_config, expected_splits):
infos = get_dataset_infos(path)
assert expected_config in infos
info = infos[expected_config]
assert info.config_name == expected_config
assert list(info.splits.keys()) == expected_splits
@pytest.mark.parametrize(
"path, config_name, expected_exception",
[
("paws", None, ValueError),
],
)
def test_get_dataset_split_names_error(path, config_name, expected_exception):
with pytest.raises(expected_exception):
get_dataset_split_names(path, config_name=config_name)
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_version.py | import pytest
from datasets.utils.version import Version
@pytest.mark.parametrize(
"other, expected_equality",
[
(Version("1.0.0"), True),
("1.0.0", True),
(Version("2.0.0"), False),
("2.0.0", False),
("1", False),
("a", False),
(1, False),
(None, False),
],
)
def test_version_equality_and_hash(other, expected_equality):
version = Version("1.0.0")
assert (version == other) is expected_equality
assert (version != other) is not expected_equality
assert (hash(version) == hash(other)) is expected_equality
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/conftest.py | import pytest
import datasets
# Import fixture modules as plugins
pytest_plugins = ["tests.fixtures.files", "tests.fixtures.hub", "tests.fixtures.fsspec"]
def pytest_collection_modifyitems(config, items):
# Mark tests as "unit" by default if not marked as "integration" (or already marked as "unit")
for item in items:
if any(marker in item.keywords for marker in ["integration", "unit"]):
continue
item.add_marker(pytest.mark.unit)
def pytest_configure(config):
config.addinivalue_line("markers", "torchaudio_latest: mark test to run with torchaudio>=0.12")
@pytest.fixture(autouse=True)
def set_test_cache_config(tmp_path_factory, monkeypatch):
# test_hf_cache_home = tmp_path_factory.mktemp("cache") # TODO: why a cache dir per test function does not work?
test_hf_cache_home = tmp_path_factory.getbasetemp() / "cache"
test_hf_datasets_cache = test_hf_cache_home / "datasets"
test_hf_metrics_cache = test_hf_cache_home / "metrics"
test_hf_modules_cache = test_hf_cache_home / "modules"
monkeypatch.setattr("datasets.config.HF_DATASETS_CACHE", str(test_hf_datasets_cache))
monkeypatch.setattr("datasets.config.HF_METRICS_CACHE", str(test_hf_metrics_cache))
monkeypatch.setattr("datasets.config.HF_MODULES_CACHE", str(test_hf_modules_cache))
test_downloaded_datasets_path = test_hf_datasets_cache / "downloads"
monkeypatch.setattr("datasets.config.DOWNLOADED_DATASETS_PATH", str(test_downloaded_datasets_path))
test_extracted_datasets_path = test_hf_datasets_cache / "downloads" / "extracted"
monkeypatch.setattr("datasets.config.EXTRACTED_DATASETS_PATH", str(test_extracted_datasets_path))
@pytest.fixture(autouse=True, scope="session")
def disable_tqdm_output():
datasets.disable_progress_bar()
@pytest.fixture(autouse=True)
def set_update_download_counts_to_false(monkeypatch):
# don't take tests into account when counting downloads
monkeypatch.setattr("datasets.config.HF_UPDATE_DOWNLOAD_COUNTS", False)
@pytest.fixture
def set_sqlalchemy_silence_uber_warning(monkeypatch):
# Required to suppress RemovedIn20Warning when feature(s) are not compatible with SQLAlchemy 2.0
# To be removed once SQLAlchemy 2.0 supported
monkeypatch.setattr("sqlalchemy.util.deprecations.SILENCE_UBER_WARNING", True)
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_tasks.py | from copy import deepcopy
from unittest.case import TestCase
import pytest
from datasets.arrow_dataset import Dataset
from datasets.features import Audio, ClassLabel, Features, Image, Sequence, Value
from datasets.info import DatasetInfo
from datasets.tasks import (
AudioClassification,
AutomaticSpeechRecognition,
ImageClassification,
LanguageModeling,
QuestionAnsweringExtractive,
Summarization,
TextClassification,
task_template_from_dict,
)
from datasets.utils.py_utils import asdict
SAMPLE_QUESTION_ANSWERING_EXTRACTIVE = {
"id": "5733be284776f41900661182",
"title": "University_of_Notre_Dame",
"context": 'Architecturally, the school has a Catholic character. Atop the Main Building\'s gold dome is a golden statue of the Virgin Mary. Immediately in front of the Main Building and facing it, is a copper statue of Christ with arms upraised with the legend "Venite Ad Me Omnes". Next to the Main Building is the Basilica of the Sacred Heart. Immediately behind the basilica is the Grotto, a Marian place of prayer and reflection. It is a replica of the grotto at Lourdes, France where the Virgin Mary reputedly appeared to Saint Bernadette Soubirous in 1858. At the end of the main drive (and in a direct line that connects through 3 statues and the Gold Dome), is a simple, modern stone statue of Mary.',
"question": "To whom did the Virgin Mary allegedly appear in 1858 in Lourdes France?",
"answers": {"text": ["Saint Bernadette Soubirous"], "answer_start": [515]},
}
@pytest.mark.parametrize(
"task_cls",
[
AudioClassification,
AutomaticSpeechRecognition,
ImageClassification,
LanguageModeling,
QuestionAnsweringExtractive,
Summarization,
TextClassification,
],
)
def test_reload_task_from_dict(task_cls):
task = task_cls()
task_dict = asdict(task)
reloaded = task_template_from_dict(task_dict)
assert task == reloaded
class TestLanguageModeling:
def test_column_mapping(self):
task = LanguageModeling(text_column="input_text")
assert {"input_text": "text"} == task.column_mapping
def test_from_dict(self):
input_schema = Features({"text": Value("string")})
template_dict = {"text_column": "input_text"}
task = LanguageModeling.from_dict(template_dict)
assert "language-modeling" == task.task
assert input_schema == task.input_schema
class TextClassificationTest(TestCase):
def setUp(self):
self.labels = sorted(["pos", "neg"])
def test_column_mapping(self):
task = TextClassification(text_column="input_text", label_column="input_label")
self.assertDictEqual({"input_text": "text", "input_label": "labels"}, task.column_mapping)
def test_from_dict(self):
input_schema = Features({"text": Value("string")})
# Labels are cast to tuple during `TextClassification.__post_init__`, so we do the same here
label_schema = Features({"labels": ClassLabel})
template_dict = {"text_column": "input_text", "label_column": "input_labels"}
task = TextClassification.from_dict(template_dict)
self.assertEqual("text-classification", task.task)
self.assertEqual(input_schema, task.input_schema)
self.assertEqual(label_schema, task.label_schema)
def test_align_with_features(self):
task = TextClassification(text_column="input_text", label_column="input_label")
self.assertEqual(task.label_schema["labels"], ClassLabel)
task = task.align_with_features(Features({"input_label": ClassLabel(names=self.labels)}))
self.assertEqual(task.label_schema["labels"], ClassLabel(names=self.labels))
class QuestionAnsweringTest(TestCase):
def test_column_mapping(self):
task = QuestionAnsweringExtractive(
context_column="input_context", question_column="input_question", answers_column="input_answers"
)
self.assertDictEqual(
{"input_context": "context", "input_question": "question", "input_answers": "answers"}, task.column_mapping
)
def test_from_dict(self):
input_schema = Features({"question": Value("string"), "context": Value("string")})
label_schema = Features(
{
"answers": Sequence(
{
"text": Value("string"),
"answer_start": Value("int32"),
}
)
}
)
template_dict = {
"context_column": "input_input_context",
"question_column": "input_question",
"answers_column": "input_answers",
}
task = QuestionAnsweringExtractive.from_dict(template_dict)
self.assertEqual("question-answering-extractive", task.task)
self.assertEqual(input_schema, task.input_schema)
self.assertEqual(label_schema, task.label_schema)
class SummarizationTest(TestCase):
def test_column_mapping(self):
task = Summarization(text_column="input_text", summary_column="input_summary")
self.assertDictEqual({"input_text": "text", "input_summary": "summary"}, task.column_mapping)
def test_from_dict(self):
input_schema = Features({"text": Value("string")})
label_schema = Features({"summary": Value("string")})
template_dict = {"text_column": "input_text", "summary_column": "input_summary"}
task = Summarization.from_dict(template_dict)
self.assertEqual("summarization", task.task)
self.assertEqual(input_schema, task.input_schema)
self.assertEqual(label_schema, task.label_schema)
class AutomaticSpeechRecognitionTest(TestCase):
def test_column_mapping(self):
task = AutomaticSpeechRecognition(audio_column="input_audio", transcription_column="input_transcription")
self.assertDictEqual({"input_audio": "audio", "input_transcription": "transcription"}, task.column_mapping)
def test_from_dict(self):
input_schema = Features({"audio": Audio()})
label_schema = Features({"transcription": Value("string")})
template_dict = {
"audio_column": "input_audio",
"transcription_column": "input_transcription",
}
task = AutomaticSpeechRecognition.from_dict(template_dict)
self.assertEqual("automatic-speech-recognition", task.task)
self.assertEqual(input_schema, task.input_schema)
self.assertEqual(label_schema, task.label_schema)
class AudioClassificationTest(TestCase):
def setUp(self):
self.labels = sorted(["pos", "neg"])
def test_column_mapping(self):
task = AudioClassification(audio_column="input_audio", label_column="input_label")
self.assertDictEqual({"input_audio": "audio", "input_label": "labels"}, task.column_mapping)
def test_from_dict(self):
input_schema = Features({"audio": Audio()})
label_schema = Features({"labels": ClassLabel})
template_dict = {
"audio_column": "input_image",
"label_column": "input_label",
}
task = AudioClassification.from_dict(template_dict)
self.assertEqual("audio-classification", task.task)
self.assertEqual(input_schema, task.input_schema)
self.assertEqual(label_schema, task.label_schema)
def test_align_with_features(self):
task = AudioClassification(audio_column="input_audio", label_column="input_label")
self.assertEqual(task.label_schema["labels"], ClassLabel)
task = task.align_with_features(Features({"input_label": ClassLabel(names=self.labels)}))
self.assertEqual(task.label_schema["labels"], ClassLabel(names=self.labels))
class ImageClassificationTest(TestCase):
def setUp(self):
self.labels = sorted(["pos", "neg"])
def test_column_mapping(self):
task = ImageClassification(image_column="input_image", label_column="input_label")
self.assertDictEqual({"input_image": "image", "input_label": "labels"}, task.column_mapping)
def test_from_dict(self):
input_schema = Features({"image": Image()})
label_schema = Features({"labels": ClassLabel})
template_dict = {
"image_column": "input_image",
"label_column": "input_label",
}
task = ImageClassification.from_dict(template_dict)
self.assertEqual("image-classification", task.task)
self.assertEqual(input_schema, task.input_schema)
self.assertEqual(label_schema, task.label_schema)
def test_align_with_features(self):
task = ImageClassification(image_column="input_image", label_column="input_label")
self.assertEqual(task.label_schema["labels"], ClassLabel)
task = task.align_with_features(Features({"input_label": ClassLabel(names=self.labels)}))
self.assertEqual(task.label_schema["labels"], ClassLabel(names=self.labels))
class DatasetWithTaskProcessingTest(TestCase):
def test_map_on_task_template(self):
info = DatasetInfo(task_templates=QuestionAnsweringExtractive())
dataset = Dataset.from_dict({k: [v] for k, v in SAMPLE_QUESTION_ANSWERING_EXTRACTIVE.items()}, info=info)
assert isinstance(dataset.info.task_templates, list)
assert len(dataset.info.task_templates) == 1
def keep_task(x):
return x
def dont_keep_task(x):
out = deepcopy(SAMPLE_QUESTION_ANSWERING_EXTRACTIVE)
out["answers"]["foobar"] = 0
return out
mapped_dataset = dataset.map(keep_task)
assert mapped_dataset.info.task_templates == dataset.info.task_templates
# reload from cache
mapped_dataset = dataset.map(keep_task)
assert mapped_dataset.info.task_templates == dataset.info.task_templates
mapped_dataset = dataset.map(dont_keep_task)
assert mapped_dataset.info.task_templates == []
# reload from cache
mapped_dataset = dataset.map(dont_keep_task)
assert mapped_dataset.info.task_templates == []
def test_remove_and_map_on_task_template(self):
features = Features({"text": Value("string"), "label": ClassLabel(names=("pos", "neg"))})
task_templates = TextClassification(text_column="text", label_column="label")
info = DatasetInfo(features=features, task_templates=task_templates)
dataset = Dataset.from_dict({"text": ["A sentence."], "label": ["pos"]}, info=info)
def process(example):
return example
modified_dataset = dataset.remove_columns("label")
mapped_dataset = modified_dataset.map(process)
assert mapped_dataset.info.task_templates == []
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_formatting.py | import datetime
from pathlib import Path
from unittest import TestCase
import numpy as np
import pandas as pd
import pyarrow as pa
import pytest
from datasets import Audio, Features, Image, IterableDataset
from datasets.formatting import NumpyFormatter, PandasFormatter, PythonFormatter, query_table
from datasets.formatting.formatting import (
LazyBatch,
LazyRow,
NumpyArrowExtractor,
PandasArrowExtractor,
PythonArrowExtractor,
)
from datasets.table import InMemoryTable
from .utils import require_jax, require_pil, require_sndfile, require_tf, require_torch
class AnyArray:
def __init__(self, data) -> None:
self.data = data
def __array__(self) -> np.ndarray:
return np.asarray(self.data)
def _gen_any_arrays():
for _ in range(10):
yield {"array": AnyArray(list(range(10)))}
@pytest.fixture
def any_arrays_dataset():
return IterableDataset.from_generator(_gen_any_arrays)
_COL_A = [0, 1, 2]
_COL_B = ["foo", "bar", "foobar"]
_COL_C = [[[1.0, 0.0, 0.0]] * 2, [[0.0, 1.0, 0.0]] * 2, [[0.0, 0.0, 1.0]] * 2]
_COL_D = [datetime.datetime(2023, 1, 1, 0, 0, tzinfo=datetime.timezone.utc)] * 3
_INDICES = [1, 0]
IMAGE_PATH_1 = Path(__file__).parent / "features" / "data" / "test_image_rgb.jpg"
IMAGE_PATH_2 = Path(__file__).parent / "features" / "data" / "test_image_rgba.png"
AUDIO_PATH_1 = Path(__file__).parent / "features" / "data" / "test_audio_44100.wav"
class ArrowExtractorTest(TestCase):
def _create_dummy_table(self):
return pa.Table.from_pydict({"a": _COL_A, "b": _COL_B, "c": _COL_C, "d": _COL_D})
def test_python_extractor(self):
pa_table = self._create_dummy_table()
extractor = PythonArrowExtractor()
row = extractor.extract_row(pa_table)
self.assertEqual(row, {"a": _COL_A[0], "b": _COL_B[0], "c": _COL_C[0], "d": _COL_D[0]})
col = extractor.extract_column(pa_table)
self.assertEqual(col, _COL_A)
batch = extractor.extract_batch(pa_table)
self.assertEqual(batch, {"a": _COL_A, "b": _COL_B, "c": _COL_C, "d": _COL_D})
def test_numpy_extractor(self):
pa_table = self._create_dummy_table().drop(["c", "d"])
extractor = NumpyArrowExtractor()
row = extractor.extract_row(pa_table)
np.testing.assert_equal(row, {"a": _COL_A[0], "b": _COL_B[0]})
col = extractor.extract_column(pa_table)
np.testing.assert_equal(col, np.array(_COL_A))
batch = extractor.extract_batch(pa_table)
np.testing.assert_equal(batch, {"a": np.array(_COL_A), "b": np.array(_COL_B)})
def test_numpy_extractor_nested(self):
pa_table = self._create_dummy_table().drop(["a", "b"])
extractor = NumpyArrowExtractor()
row = extractor.extract_row(pa_table)
self.assertEqual(row["c"][0].dtype, np.float64)
self.assertEqual(row["c"].dtype, object)
col = extractor.extract_column(pa_table)
self.assertEqual(col[0][0].dtype, np.float64)
self.assertEqual(col[0].dtype, object)
self.assertEqual(col.dtype, object)
batch = extractor.extract_batch(pa_table)
self.assertEqual(batch["c"][0][0].dtype, np.float64)
self.assertEqual(batch["c"][0].dtype, object)
self.assertEqual(batch["c"].dtype, object)
def test_numpy_extractor_temporal(self):
pa_table = self._create_dummy_table().drop(["a", "b", "c"])
extractor = NumpyArrowExtractor()
row = extractor.extract_row(pa_table)
self.assertTrue(np.issubdtype(row["d"].dtype, np.datetime64))
col = extractor.extract_column(pa_table)
self.assertTrue(np.issubdtype(col[0].dtype, np.datetime64))
self.assertTrue(np.issubdtype(col.dtype, np.datetime64))
batch = extractor.extract_batch(pa_table)
self.assertTrue(np.issubdtype(batch["d"][0].dtype, np.datetime64))
self.assertTrue(np.issubdtype(batch["d"].dtype, np.datetime64))
def test_pandas_extractor(self):
pa_table = self._create_dummy_table()
extractor = PandasArrowExtractor()
row = extractor.extract_row(pa_table)
self.assertIsInstance(row, pd.DataFrame)
pd.testing.assert_series_equal(row["a"], pd.Series(_COL_A, name="a")[:1])
pd.testing.assert_series_equal(row["b"], pd.Series(_COL_B, name="b")[:1])
pd.testing.assert_series_equal(row["d"], pd.Series(_COL_D, name="d")[:1])
col = extractor.extract_column(pa_table)
pd.testing.assert_series_equal(col, pd.Series(_COL_A, name="a"))
batch = extractor.extract_batch(pa_table)
self.assertIsInstance(batch, pd.DataFrame)
pd.testing.assert_series_equal(batch["a"], pd.Series(_COL_A, name="a"))
pd.testing.assert_series_equal(batch["b"], pd.Series(_COL_B, name="b"))
pd.testing.assert_series_equal(batch["d"], pd.Series(_COL_D, name="d"))
class LazyDictTest(TestCase):
def _create_dummy_table(self):
return pa.Table.from_pydict({"a": _COL_A, "b": _COL_B, "c": _COL_C})
def _create_dummy_formatter(self):
return PythonFormatter(lazy=True)
def test_lazy_dict_copy(self):
pa_table = self._create_dummy_table()
formatter = self._create_dummy_formatter()
lazy_batch = formatter.format_batch(pa_table)
lazy_batch_copy = lazy_batch.copy()
self.assertEqual(type(lazy_batch), type(lazy_batch_copy))
self.assertEqual(lazy_batch.items(), lazy_batch_copy.items())
lazy_batch["d"] = [1, 2, 3]
self.assertNotEqual(lazy_batch.items(), lazy_batch_copy.items())
class FormatterTest(TestCase):
def _create_dummy_table(self):
return pa.Table.from_pydict({"a": _COL_A, "b": _COL_B, "c": _COL_C})
def test_python_formatter(self):
pa_table = self._create_dummy_table()
formatter = PythonFormatter()
row = formatter.format_row(pa_table)
self.assertEqual(row, {"a": _COL_A[0], "b": _COL_B[0], "c": _COL_C[0]})
col = formatter.format_column(pa_table)
self.assertEqual(col, _COL_A)
batch = formatter.format_batch(pa_table)
self.assertEqual(batch, {"a": _COL_A, "b": _COL_B, "c": _COL_C})
def test_python_formatter_lazy(self):
pa_table = self._create_dummy_table()
formatter = PythonFormatter(lazy=True)
row = formatter.format_row(pa_table)
self.assertIsInstance(row, LazyRow)
self.assertEqual(row["a"], _COL_A[0])
self.assertEqual(row["b"], _COL_B[0])
self.assertEqual(row["c"], _COL_C[0])
batch = formatter.format_batch(pa_table)
self.assertIsInstance(batch, LazyBatch)
self.assertEqual(batch["a"], _COL_A)
self.assertEqual(batch["b"], _COL_B)
self.assertEqual(batch["c"], _COL_C)
def test_numpy_formatter(self):
pa_table = self._create_dummy_table()
formatter = NumpyFormatter()
row = formatter.format_row(pa_table)
np.testing.assert_equal(row, {"a": _COL_A[0], "b": _COL_B[0], "c": np.array(_COL_C[0])})
col = formatter.format_column(pa_table)
np.testing.assert_equal(col, np.array(_COL_A))
batch = formatter.format_batch(pa_table)
np.testing.assert_equal(batch, {"a": np.array(_COL_A), "b": np.array(_COL_B), "c": np.array(_COL_C)})
assert batch["c"].shape == np.array(_COL_C).shape
def test_numpy_formatter_np_array_kwargs(self):
pa_table = self._create_dummy_table().drop(["b"])
formatter = NumpyFormatter(dtype=np.float16)
row = formatter.format_row(pa_table)
self.assertEqual(row["c"].dtype, np.dtype(np.float16))
col = formatter.format_column(pa_table)
self.assertEqual(col.dtype, np.float16)
batch = formatter.format_batch(pa_table)
self.assertEqual(batch["a"].dtype, np.dtype(np.float16))
self.assertEqual(batch["c"].dtype, np.dtype(np.float16))
@require_pil
def test_numpy_formatter_image(self):
# same dimensions
pa_table = pa.table({"image": [{"bytes": None, "path": str(IMAGE_PATH_1)}] * 2})
formatter = NumpyFormatter(features=Features({"image": Image()}))
row = formatter.format_row(pa_table)
self.assertEqual(row["image"].dtype, np.uint8)
self.assertEqual(row["image"].shape, (480, 640, 3))
col = formatter.format_column(pa_table)
self.assertEqual(col.dtype, np.uint8)
self.assertEqual(col.shape, (2, 480, 640, 3))
batch = formatter.format_batch(pa_table)
self.assertEqual(batch["image"].dtype, np.uint8)
self.assertEqual(batch["image"].shape, (2, 480, 640, 3))
# different dimensions
pa_table = pa.table(
{"image": [{"bytes": None, "path": str(IMAGE_PATH_1)}, {"bytes": None, "path": str(IMAGE_PATH_2)}]}
)
formatter = NumpyFormatter(features=Features({"image": Image()}))
row = formatter.format_row(pa_table)
self.assertEqual(row["image"].dtype, np.uint8)
self.assertEqual(row["image"].shape, (480, 640, 3))
col = formatter.format_column(pa_table)
self.assertIsInstance(col, np.ndarray)
self.assertEqual(col.dtype, object)
self.assertEqual(col[0].dtype, np.uint8)
self.assertEqual(col[0].shape, (480, 640, 3))
batch = formatter.format_batch(pa_table)
self.assertIsInstance(batch["image"], np.ndarray)
self.assertEqual(batch["image"].dtype, object)
self.assertEqual(batch["image"][0].dtype, np.uint8)
self.assertEqual(batch["image"][0].shape, (480, 640, 3))
@require_sndfile
def test_numpy_formatter_audio(self):
pa_table = pa.table({"audio": [{"bytes": None, "path": str(AUDIO_PATH_1)}]})
formatter = NumpyFormatter(features=Features({"audio": Audio()}))
row = formatter.format_row(pa_table)
self.assertEqual(row["audio"]["array"].dtype, np.dtype(np.float32))
col = formatter.format_column(pa_table)
self.assertEqual(col[0]["array"].dtype, np.float32)
batch = formatter.format_batch(pa_table)
self.assertEqual(batch["audio"][0]["array"].dtype, np.dtype(np.float32))
def test_pandas_formatter(self):
pa_table = self._create_dummy_table()
formatter = PandasFormatter()
row = formatter.format_row(pa_table)
self.assertIsInstance(row, pd.DataFrame)
pd.testing.assert_series_equal(row["a"], pd.Series(_COL_A, name="a")[:1])
pd.testing.assert_series_equal(row["b"], pd.Series(_COL_B, name="b")[:1])
col = formatter.format_column(pa_table)
pd.testing.assert_series_equal(col, pd.Series(_COL_A, name="a"))
batch = formatter.format_batch(pa_table)
self.assertIsInstance(batch, pd.DataFrame)
pd.testing.assert_series_equal(batch["a"], pd.Series(_COL_A, name="a"))
pd.testing.assert_series_equal(batch["b"], pd.Series(_COL_B, name="b"))
@require_torch
def test_torch_formatter(self):
import torch
from datasets.formatting import TorchFormatter
pa_table = self._create_dummy_table()
formatter = TorchFormatter()
row = formatter.format_row(pa_table)
torch.testing.assert_close(row["a"], torch.tensor(_COL_A, dtype=torch.int64)[0])
assert row["b"] == _COL_B[0]
torch.testing.assert_close(row["c"], torch.tensor(_COL_C, dtype=torch.float32)[0])
col = formatter.format_column(pa_table)
torch.testing.assert_close(col, torch.tensor(_COL_A, dtype=torch.int64))
batch = formatter.format_batch(pa_table)
torch.testing.assert_close(batch["a"], torch.tensor(_COL_A, dtype=torch.int64))
assert batch["b"] == _COL_B
torch.testing.assert_close(batch["c"], torch.tensor(_COL_C, dtype=torch.float32))
assert batch["c"].shape == np.array(_COL_C).shape
@require_torch
def test_torch_formatter_torch_tensor_kwargs(self):
import torch
from datasets.formatting import TorchFormatter
pa_table = self._create_dummy_table().drop(["b"])
formatter = TorchFormatter(dtype=torch.float16)
row = formatter.format_row(pa_table)
self.assertEqual(row["c"].dtype, torch.float16)
col = formatter.format_column(pa_table)
self.assertEqual(col.dtype, torch.float16)
batch = formatter.format_batch(pa_table)
self.assertEqual(batch["a"].dtype, torch.float16)
self.assertEqual(batch["c"].dtype, torch.float16)
@require_torch
@require_pil
def test_torch_formatter_image(self):
import torch
from datasets.formatting import TorchFormatter
# same dimensions
pa_table = pa.table({"image": [{"bytes": None, "path": str(IMAGE_PATH_1)}] * 2})
formatter = TorchFormatter(features=Features({"image": Image()}))
row = formatter.format_row(pa_table)
self.assertEqual(row["image"].dtype, torch.uint8)
self.assertEqual(row["image"].shape, (480, 640, 3))
col = formatter.format_column(pa_table)
self.assertEqual(col.dtype, torch.uint8)
self.assertEqual(col.shape, (2, 480, 640, 3))
batch = formatter.format_batch(pa_table)
self.assertEqual(batch["image"].dtype, torch.uint8)
self.assertEqual(batch["image"].shape, (2, 480, 640, 3))
# different dimensions
pa_table = pa.table(
{"image": [{"bytes": None, "path": str(IMAGE_PATH_1)}, {"bytes": None, "path": str(IMAGE_PATH_2)}]}
)
formatter = TorchFormatter(features=Features({"image": Image()}))
row = formatter.format_row(pa_table)
self.assertEqual(row["image"].dtype, torch.uint8)
self.assertEqual(row["image"].shape, (480, 640, 3))
col = formatter.format_column(pa_table)
self.assertIsInstance(col, list)
self.assertEqual(col[0].dtype, torch.uint8)
self.assertEqual(col[0].shape, (480, 640, 3))
batch = formatter.format_batch(pa_table)
self.assertIsInstance(batch["image"], list)
self.assertEqual(batch["image"][0].dtype, torch.uint8)
self.assertEqual(batch["image"][0].shape, (480, 640, 3))
@require_torch
@require_sndfile
def test_torch_formatter_audio(self):
import torch
from datasets.formatting import TorchFormatter
pa_table = pa.table({"audio": [{"bytes": None, "path": str(AUDIO_PATH_1)}]})
formatter = TorchFormatter(features=Features({"audio": Audio()}))
row = formatter.format_row(pa_table)
self.assertEqual(row["audio"]["array"].dtype, torch.float32)
col = formatter.format_column(pa_table)
self.assertEqual(col[0]["array"].dtype, torch.float32)
batch = formatter.format_batch(pa_table)
self.assertEqual(batch["audio"][0]["array"].dtype, torch.float32)
@require_tf
def test_tf_formatter(self):
import tensorflow as tf
from datasets.formatting import TFFormatter
pa_table = self._create_dummy_table()
formatter = TFFormatter()
row = formatter.format_row(pa_table)
tf.debugging.assert_equal(row["a"], tf.convert_to_tensor(_COL_A, dtype=tf.int64)[0])
tf.debugging.assert_equal(row["b"], tf.convert_to_tensor(_COL_B, dtype=tf.string)[0])
tf.debugging.assert_equal(row["c"], tf.convert_to_tensor(_COL_C, dtype=tf.float32)[0])
col = formatter.format_column(pa_table)
tf.debugging.assert_equal(col, tf.ragged.constant(_COL_A, dtype=tf.int64))
batch = formatter.format_batch(pa_table)
tf.debugging.assert_equal(batch["a"], tf.convert_to_tensor(_COL_A, dtype=tf.int64))
tf.debugging.assert_equal(batch["b"], tf.convert_to_tensor(_COL_B, dtype=tf.string))
self.assertIsInstance(batch["c"], tf.Tensor)
self.assertEqual(batch["c"].dtype, tf.float32)
tf.debugging.assert_equal(
batch["c"].shape.as_list(), tf.convert_to_tensor(_COL_C, dtype=tf.float32).shape.as_list()
)
tf.debugging.assert_equal(tf.convert_to_tensor(batch["c"]), tf.convert_to_tensor(_COL_C, dtype=tf.float32))
@require_tf
def test_tf_formatter_tf_tensor_kwargs(self):
import tensorflow as tf
from datasets.formatting import TFFormatter
pa_table = self._create_dummy_table().drop(["b"])
formatter = TFFormatter(dtype=tf.float16)
row = formatter.format_row(pa_table)
self.assertEqual(row["c"].dtype, tf.float16)
col = formatter.format_column(pa_table)
self.assertEqual(col.dtype, tf.float16)
batch = formatter.format_batch(pa_table)
self.assertEqual(batch["a"].dtype, tf.float16)
self.assertEqual(batch["c"].dtype, tf.float16)
@require_tf
@require_pil
def test_tf_formatter_image(self):
import tensorflow as tf
from datasets.formatting import TFFormatter
# same dimensions
pa_table = pa.table({"image": [{"bytes": None, "path": str(IMAGE_PATH_1)}] * 2})
formatter = TFFormatter(features=Features({"image": Image()}))
row = formatter.format_row(pa_table)
self.assertEqual(row["image"].dtype, tf.uint8)
self.assertEqual(row["image"].shape, (480, 640, 3))
col = formatter.format_column(pa_table)
self.assertEqual(col.dtype, tf.uint8)
self.assertEqual(col.shape, (2, 480, 640, 3))
batch = formatter.format_batch(pa_table)
self.assertEqual(batch["image"][0].dtype, tf.uint8)
self.assertEqual(batch["image"].shape, (2, 480, 640, 3))
# different dimensions
pa_table = pa.table(
{"image": [{"bytes": None, "path": str(IMAGE_PATH_1)}, {"bytes": None, "path": str(IMAGE_PATH_2)}]}
)
formatter = TFFormatter(features=Features({"image": Image()}))
row = formatter.format_row(pa_table)
self.assertEqual(row["image"].dtype, tf.uint8)
self.assertEqual(row["image"].shape, (480, 640, 3))
col = formatter.format_column(pa_table)
self.assertIsInstance(col, list)
self.assertEqual(col[0].dtype, tf.uint8)
self.assertEqual(col[0].shape, (480, 640, 3))
batch = formatter.format_batch(pa_table)
self.assertIsInstance(batch["image"], list)
self.assertEqual(batch["image"][0].dtype, tf.uint8)
self.assertEqual(batch["image"][0].shape, (480, 640, 3))
@require_tf
@require_sndfile
def test_tf_formatter_audio(self):
import tensorflow as tf
from datasets.formatting import TFFormatter
pa_table = pa.table({"audio": [{"bytes": None, "path": str(AUDIO_PATH_1)}]})
formatter = TFFormatter(features=Features({"audio": Audio()}))
row = formatter.format_row(pa_table)
self.assertEqual(row["audio"]["array"].dtype, tf.float32)
col = formatter.format_column(pa_table)
self.assertEqual(col[0]["array"].dtype, tf.float32)
batch = formatter.format_batch(pa_table)
self.assertEqual(batch["audio"][0]["array"].dtype, tf.float32)
@require_jax
def test_jax_formatter(self):
import jax
import jax.numpy as jnp
from datasets.formatting import JaxFormatter
pa_table = self._create_dummy_table()
formatter = JaxFormatter()
row = formatter.format_row(pa_table)
jnp.allclose(row["a"], jnp.array(_COL_A, dtype=jnp.int64 if jax.config.jax_enable_x64 else jnp.int32)[0])
assert row["b"] == _COL_B[0]
jnp.allclose(row["c"], jnp.array(_COL_C, dtype=jnp.float32)[0])
col = formatter.format_column(pa_table)
jnp.allclose(col, jnp.array(_COL_A, dtype=jnp.int64 if jax.config.jax_enable_x64 else jnp.int32))
batch = formatter.format_batch(pa_table)
jnp.allclose(batch["a"], jnp.array(_COL_A, dtype=jnp.int64 if jax.config.jax_enable_x64 else jnp.int32))
assert batch["b"] == _COL_B
jnp.allclose(batch["c"], jnp.array(_COL_C, dtype=jnp.float32))
assert batch["c"].shape == np.array(_COL_C).shape
@require_jax
def test_jax_formatter_jnp_array_kwargs(self):
import jax.numpy as jnp
from datasets.formatting import JaxFormatter
pa_table = self._create_dummy_table().drop(["b"])
formatter = JaxFormatter(dtype=jnp.float16)
row = formatter.format_row(pa_table)
self.assertEqual(row["c"].dtype, jnp.float16)
col = formatter.format_column(pa_table)
self.assertEqual(col.dtype, jnp.float16)
batch = formatter.format_batch(pa_table)
self.assertEqual(batch["a"].dtype, jnp.float16)
self.assertEqual(batch["c"].dtype, jnp.float16)
@require_jax
@require_pil
def test_jax_formatter_image(self):
import jax.numpy as jnp
from datasets.formatting import JaxFormatter
# same dimensions
pa_table = pa.table({"image": [{"bytes": None, "path": str(IMAGE_PATH_1)}] * 2})
formatter = JaxFormatter(features=Features({"image": Image()}))
row = formatter.format_row(pa_table)
self.assertEqual(row["image"].dtype, jnp.uint8)
self.assertEqual(row["image"].shape, (480, 640, 3))
col = formatter.format_column(pa_table)
self.assertEqual(col.dtype, jnp.uint8)
self.assertEqual(col.shape, (2, 480, 640, 3))
batch = formatter.format_batch(pa_table)
self.assertEqual(batch["image"].dtype, jnp.uint8)
self.assertEqual(batch["image"].shape, (2, 480, 640, 3))
# different dimensions
pa_table = pa.table(
{"image": [{"bytes": None, "path": str(IMAGE_PATH_1)}, {"bytes": None, "path": str(IMAGE_PATH_2)}]}
)
formatter = JaxFormatter(features=Features({"image": Image()}))
row = formatter.format_row(pa_table)
self.assertEqual(row["image"].dtype, jnp.uint8)
self.assertEqual(row["image"].shape, (480, 640, 3))
col = formatter.format_column(pa_table)
self.assertIsInstance(col, list)
self.assertEqual(col[0].dtype, jnp.uint8)
self.assertEqual(col[0].shape, (480, 640, 3))
batch = formatter.format_batch(pa_table)
self.assertIsInstance(batch["image"], list)
self.assertEqual(batch["image"][0].dtype, jnp.uint8)
self.assertEqual(batch["image"][0].shape, (480, 640, 3))
@require_jax
@require_sndfile
def test_jax_formatter_audio(self):
import jax.numpy as jnp
from datasets.formatting import JaxFormatter
pa_table = pa.table({"audio": [{"bytes": None, "path": str(AUDIO_PATH_1)}]})
formatter = JaxFormatter(features=Features({"audio": Audio()}))
row = formatter.format_row(pa_table)
self.assertEqual(row["audio"]["array"].dtype, jnp.float32)
col = formatter.format_column(pa_table)
self.assertEqual(col[0]["array"].dtype, jnp.float32)
batch = formatter.format_batch(pa_table)
self.assertEqual(batch["audio"][0]["array"].dtype, jnp.float32)
@require_jax
def test_jax_formatter_device(self):
import jax
from datasets.formatting import JaxFormatter
pa_table = self._create_dummy_table()
device = jax.devices()[0]
formatter = JaxFormatter(device=str(device))
row = formatter.format_row(pa_table)
assert row["a"].device() == device
assert row["c"].device() == device
col = formatter.format_column(pa_table)
assert col.device() == device
batch = formatter.format_batch(pa_table)
assert batch["a"].device() == device
assert batch["c"].device() == device
class QueryTest(TestCase):
def _create_dummy_table(self):
return pa.Table.from_pydict({"a": _COL_A, "b": _COL_B, "c": _COL_C})
def _create_dummy_arrow_indices(self):
return pa.Table.from_arrays([pa.array(_INDICES, type=pa.uint64())], names=["indices"])
def assertTableEqual(self, first: pa.Table, second: pa.Table):
self.assertEqual(first.schema, second.schema)
for first_array, second_array in zip(first, second):
self.assertEqual(first_array, second_array)
self.assertEqual(first, second)
def test_query_table_int(self):
pa_table = self._create_dummy_table()
table = InMemoryTable(pa_table)
n = pa_table.num_rows
# classical usage
subtable = query_table(table, 0)
self.assertTableEqual(subtable, pa.Table.from_pydict({"a": _COL_A[:1], "b": _COL_B[:1], "c": _COL_C[:1]}))
subtable = query_table(table, 1)
self.assertTableEqual(subtable, pa.Table.from_pydict({"a": _COL_A[1:2], "b": _COL_B[1:2], "c": _COL_C[1:2]}))
subtable = query_table(table, -1)
self.assertTableEqual(subtable, pa.Table.from_pydict({"a": _COL_A[-1:], "b": _COL_B[-1:], "c": _COL_C[-1:]}))
# raise an IndexError
with self.assertRaises(IndexError):
query_table(table, n)
with self.assertRaises(IndexError):
query_table(table, -(n + 1))
# with indices
indices = InMemoryTable(self._create_dummy_arrow_indices())
subtable = query_table(table, 0, indices=indices)
self.assertTableEqual(
subtable,
pa.Table.from_pydict({"a": [_COL_A[_INDICES[0]]], "b": [_COL_B[_INDICES[0]]], "c": [_COL_C[_INDICES[0]]]}),
)
with self.assertRaises(IndexError):
assert len(indices) < n
query_table(table, len(indices), indices=indices)
def test_query_table_slice(self):
pa_table = self._create_dummy_table()
table = InMemoryTable(pa_table)
n = pa_table.num_rows
# classical usage
subtable = query_table(table, slice(0, 1))
self.assertTableEqual(subtable, pa.Table.from_pydict({"a": _COL_A[:1], "b": _COL_B[:1], "c": _COL_C[:1]}))
subtable = query_table(table, slice(1, 2))
self.assertTableEqual(subtable, pa.Table.from_pydict({"a": _COL_A[1:2], "b": _COL_B[1:2], "c": _COL_C[1:2]}))
subtable = query_table(table, slice(-2, -1))
self.assertTableEqual(
subtable, pa.Table.from_pydict({"a": _COL_A[-2:-1], "b": _COL_B[-2:-1], "c": _COL_C[-2:-1]})
)
# usage with None
subtable = query_table(table, slice(-1, None))
self.assertTableEqual(subtable, pa.Table.from_pydict({"a": _COL_A[-1:], "b": _COL_B[-1:], "c": _COL_C[-1:]}))
subtable = query_table(table, slice(None, n + 1))
self.assertTableEqual(
subtable, pa.Table.from_pydict({"a": _COL_A[: n + 1], "b": _COL_B[: n + 1], "c": _COL_C[: n + 1]})
)
self.assertTableEqual(subtable, pa.Table.from_pydict({"a": _COL_A, "b": _COL_B, "c": _COL_C}))
subtable = query_table(table, slice(-(n + 1), None))
self.assertTableEqual(
subtable, pa.Table.from_pydict({"a": _COL_A[-(n + 1) :], "b": _COL_B[-(n + 1) :], "c": _COL_C[-(n + 1) :]})
)
self.assertTableEqual(subtable, pa.Table.from_pydict({"a": _COL_A, "b": _COL_B, "c": _COL_C}))
# usage with step
subtable = query_table(table, slice(None, None, 2))
self.assertTableEqual(subtable, pa.Table.from_pydict({"a": _COL_A[::2], "b": _COL_B[::2], "c": _COL_C[::2]}))
# empty ouput but no errors
subtable = query_table(table, slice(-1, 0)) # usage with both negative and positive idx
assert len(_COL_A[-1:0]) == 0
self.assertTableEqual(subtable, pa_table.slice(0, 0))
subtable = query_table(table, slice(2, 1))
assert len(_COL_A[2:1]) == 0
self.assertTableEqual(subtable, pa_table.slice(0, 0))
subtable = query_table(table, slice(n, n))
assert len(_COL_A[n:n]) == 0
self.assertTableEqual(subtable, pa_table.slice(0, 0))
subtable = query_table(table, slice(n, n + 1))
assert len(_COL_A[n : n + 1]) == 0
self.assertTableEqual(subtable, pa_table.slice(0, 0))
# it's not possible to get an error with a slice
# with indices
indices = InMemoryTable(self._create_dummy_arrow_indices())
subtable = query_table(table, slice(0, 1), indices=indices)
self.assertTableEqual(
subtable,
pa.Table.from_pydict({"a": [_COL_A[_INDICES[0]]], "b": [_COL_B[_INDICES[0]]], "c": [_COL_C[_INDICES[0]]]}),
)
subtable = query_table(table, slice(n - 1, n), indices=indices)
assert len(indices.column(0).to_pylist()[n - 1 : n]) == 0
self.assertTableEqual(subtable, pa_table.slice(0, 0))
def test_query_table_range(self):
pa_table = self._create_dummy_table()
table = InMemoryTable(pa_table)
n = pa_table.num_rows
np_A, np_B, np_C = np.array(_COL_A, dtype=np.int64), np.array(_COL_B), np.array(_COL_C)
# classical usage
subtable = query_table(table, range(0, 1))
self.assertTableEqual(
subtable,
pa.Table.from_pydict({"a": np_A[range(0, 1)], "b": np_B[range(0, 1)], "c": np_C[range(0, 1)].tolist()}),
)
subtable = query_table(table, range(1, 2))
self.assertTableEqual(
subtable,
pa.Table.from_pydict({"a": np_A[range(1, 2)], "b": np_B[range(1, 2)], "c": np_C[range(1, 2)].tolist()}),
)
subtable = query_table(table, range(-2, -1))
self.assertTableEqual(
subtable,
pa.Table.from_pydict(
{"a": np_A[range(-2, -1)], "b": np_B[range(-2, -1)], "c": np_C[range(-2, -1)].tolist()}
),
)
# usage with both negative and positive idx
subtable = query_table(table, range(-1, 0))
self.assertTableEqual(
subtable,
pa.Table.from_pydict({"a": np_A[range(-1, 0)], "b": np_B[range(-1, 0)], "c": np_C[range(-1, 0)].tolist()}),
)
subtable = query_table(table, range(-1, n))
self.assertTableEqual(
subtable,
pa.Table.from_pydict({"a": np_A[range(-1, n)], "b": np_B[range(-1, n)], "c": np_C[range(-1, n)].tolist()}),
)
# usage with step
subtable = query_table(table, range(0, n, 2))
self.assertTableEqual(
subtable,
pa.Table.from_pydict(
{"a": np_A[range(0, n, 2)], "b": np_B[range(0, n, 2)], "c": np_C[range(0, n, 2)].tolist()}
),
)
subtable = query_table(table, range(0, n + 1, 2 * n))
self.assertTableEqual(
subtable,
pa.Table.from_pydict(
{
"a": np_A[range(0, n + 1, 2 * n)],
"b": np_B[range(0, n + 1, 2 * n)],
"c": np_C[range(0, n + 1, 2 * n)].tolist(),
}
),
)
# empty ouput but no errors
subtable = query_table(table, range(2, 1))
assert len(np_A[range(2, 1)]) == 0
self.assertTableEqual(subtable, pa.Table.from_batches([], schema=pa_table.schema))
subtable = query_table(table, range(n, n))
assert len(np_A[range(n, n)]) == 0
self.assertTableEqual(subtable, pa.Table.from_batches([], schema=pa_table.schema))
# raise an IndexError
with self.assertRaises(IndexError):
with self.assertRaises(IndexError):
np_A[range(0, n + 1)]
query_table(table, range(0, n + 1))
with self.assertRaises(IndexError):
with self.assertRaises(IndexError):
np_A[range(-(n + 1), -1)]
query_table(table, range(-(n + 1), -1))
with self.assertRaises(IndexError):
with self.assertRaises(IndexError):
np_A[range(n, n + 1)]
query_table(table, range(n, n + 1))
# with indices
indices = InMemoryTable(self._create_dummy_arrow_indices())
subtable = query_table(table, range(0, 1), indices=indices)
self.assertTableEqual(
subtable,
pa.Table.from_pydict({"a": [_COL_A[_INDICES[0]]], "b": [_COL_B[_INDICES[0]]], "c": [_COL_C[_INDICES[0]]]}),
)
with self.assertRaises(IndexError):
assert len(indices) < n
query_table(table, range(len(indices), len(indices) + 1), indices=indices)
def test_query_table_str(self):
pa_table = self._create_dummy_table()
table = InMemoryTable(pa_table)
subtable = query_table(table, "a")
self.assertTableEqual(subtable, pa.Table.from_pydict({"a": _COL_A}))
with self.assertRaises(KeyError):
query_table(table, "z")
indices = InMemoryTable(self._create_dummy_arrow_indices())
subtable = query_table(table, "a", indices=indices)
self.assertTableEqual(subtable, pa.Table.from_pydict({"a": [_COL_A[i] for i in _INDICES]}))
def test_query_table_iterable(self):
pa_table = self._create_dummy_table()
table = InMemoryTable(pa_table)
n = pa_table.num_rows
np_A, np_B, np_C = np.array(_COL_A, dtype=np.int64), np.array(_COL_B), np.array(_COL_C)
# classical usage
subtable = query_table(table, [0])
self.assertTableEqual(
subtable, pa.Table.from_pydict({"a": np_A[[0]], "b": np_B[[0]], "c": np_C[[0]].tolist()})
)
subtable = query_table(table, [1])
self.assertTableEqual(
subtable, pa.Table.from_pydict({"a": np_A[[1]], "b": np_B[[1]], "c": np_C[[1]].tolist()})
)
subtable = query_table(table, [-1])
self.assertTableEqual(
subtable, pa.Table.from_pydict({"a": np_A[[-1]], "b": np_B[[-1]], "c": np_C[[-1]].tolist()})
)
subtable = query_table(table, [0, -1, 1])
self.assertTableEqual(
subtable,
pa.Table.from_pydict({"a": np_A[[0, -1, 1]], "b": np_B[[0, -1, 1]], "c": np_C[[0, -1, 1]].tolist()}),
)
# numpy iterable
subtable = query_table(table, np.array([0, -1, 1]))
self.assertTableEqual(
subtable,
pa.Table.from_pydict({"a": np_A[[0, -1, 1]], "b": np_B[[0, -1, 1]], "c": np_C[[0, -1, 1]].tolist()}),
)
# empty ouput but no errors
subtable = query_table(table, [])
assert len(np_A[[]]) == 0
self.assertTableEqual(subtable, pa.Table.from_batches([], schema=pa_table.schema))
# raise an IndexError
with self.assertRaises(IndexError):
with self.assertRaises(IndexError):
np_A[[n]]
query_table(table, [n])
with self.assertRaises(IndexError):
with self.assertRaises(IndexError):
np_A[[-(n + 1)]]
query_table(table, [-(n + 1)])
# with indices
indices = InMemoryTable(self._create_dummy_arrow_indices())
subtable = query_table(table, [0], indices=indices)
self.assertTableEqual(
subtable,
pa.Table.from_pydict({"a": [_COL_A[_INDICES[0]]], "b": [_COL_B[_INDICES[0]]], "c": [_COL_C[_INDICES[0]]]}),
)
with self.assertRaises(IndexError):
assert len(indices) < n
query_table(table, [len(indices)], indices=indices)
def test_query_table_invalid_key_type(self):
pa_table = self._create_dummy_table()
table = InMemoryTable(pa_table)
with self.assertRaises(TypeError):
query_table(table, 0.0)
with self.assertRaises(TypeError):
query_table(table, [0, "a"])
with self.assertRaises(TypeError):
query_table(table, int)
with self.assertRaises(TypeError):
def iter_to_inf(start=0):
while True:
yield start
start += 1
query_table(table, iter_to_inf())
@pytest.fixture(scope="session")
def arrow_table():
return pa.Table.from_pydict({"col_int": [0, 1, 2], "col_float": [0.0, 1.0, 2.0]})
@require_tf
@pytest.mark.parametrize(
"cast_schema",
[
None,
[("col_int", pa.int64()), ("col_float", pa.float64())],
[("col_int", pa.int32()), ("col_float", pa.float64())],
[("col_int", pa.int64()), ("col_float", pa.float32())],
],
)
def test_tf_formatter_sets_default_dtypes(cast_schema, arrow_table):
import tensorflow as tf
from datasets.formatting import TFFormatter
if cast_schema:
arrow_table = arrow_table.cast(pa.schema(cast_schema))
arrow_table_dict = arrow_table.to_pydict()
list_int = arrow_table_dict["col_int"]
list_float = arrow_table_dict["col_float"]
formatter = TFFormatter()
row = formatter.format_row(arrow_table)
tf.debugging.assert_equal(row["col_int"], tf.ragged.constant(list_int, dtype=tf.int64)[0])
tf.debugging.assert_equal(row["col_float"], tf.ragged.constant(list_float, dtype=tf.float32)[0])
col = formatter.format_column(arrow_table)
tf.debugging.assert_equal(col, tf.ragged.constant(list_int, dtype=tf.int64))
batch = formatter.format_batch(arrow_table)
tf.debugging.assert_equal(batch["col_int"], tf.ragged.constant(list_int, dtype=tf.int64))
tf.debugging.assert_equal(batch["col_float"], tf.ragged.constant(list_float, dtype=tf.float32))
@require_torch
@pytest.mark.parametrize(
"cast_schema",
[
None,
[("col_int", pa.int64()), ("col_float", pa.float64())],
[("col_int", pa.int32()), ("col_float", pa.float64())],
[("col_int", pa.int64()), ("col_float", pa.float32())],
],
)
def test_torch_formatter_sets_default_dtypes(cast_schema, arrow_table):
import torch
from datasets.formatting import TorchFormatter
if cast_schema:
arrow_table = arrow_table.cast(pa.schema(cast_schema))
arrow_table_dict = arrow_table.to_pydict()
list_int = arrow_table_dict["col_int"]
list_float = arrow_table_dict["col_float"]
formatter = TorchFormatter()
row = formatter.format_row(arrow_table)
torch.testing.assert_close(row["col_int"], torch.tensor(list_int, dtype=torch.int64)[0])
torch.testing.assert_close(row["col_float"], torch.tensor(list_float, dtype=torch.float32)[0])
col = formatter.format_column(arrow_table)
torch.testing.assert_close(col, torch.tensor(list_int, dtype=torch.int64))
batch = formatter.format_batch(arrow_table)
torch.testing.assert_close(batch["col_int"], torch.tensor(list_int, dtype=torch.int64))
torch.testing.assert_close(batch["col_float"], torch.tensor(list_float, dtype=torch.float32))
def test_iterable_dataset_of_arrays_format_to_arrow(any_arrays_dataset: IterableDataset):
formatted = any_arrays_dataset.with_format("arrow")
assert all(isinstance(example, pa.Table) for example in formatted)
def test_iterable_dataset_of_arrays_format_to_numpy(any_arrays_dataset: IterableDataset):
formatted = any_arrays_dataset.with_format("np")
assert all(isinstance(example["array"], np.ndarray) for example in formatted)
@require_torch
def test_iterable_dataset_of_arrays_format_to_torch(any_arrays_dataset: IterableDataset):
import torch
formatted = any_arrays_dataset.with_format("torch")
assert all(isinstance(example["array"], torch.Tensor) for example in formatted)
@require_tf
def test_iterable_dataset_of_arrays_format_to_tf(any_arrays_dataset: IterableDataset):
import tensorflow as tf
formatted = any_arrays_dataset.with_format("tf")
assert all(isinstance(example["array"], tf.Tensor) for example in formatted)
@require_jax
def test_iterable_dataset_of_arrays_format_to_jax(any_arrays_dataset: IterableDataset):
import jax.numpy as jnp
formatted = any_arrays_dataset.with_format("jax")
assert all(isinstance(example["array"], jnp.ndarray) for example in formatted)
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_info_utils.py | import pytest
import datasets.config
from datasets.utils.info_utils import is_small_dataset
@pytest.mark.parametrize("dataset_size", [None, 400 * 2**20, 600 * 2**20])
@pytest.mark.parametrize("input_in_memory_max_size", ["default", 0, 100 * 2**20, 900 * 2**20])
def test_is_small_dataset(dataset_size, input_in_memory_max_size, monkeypatch):
if input_in_memory_max_size != "default":
monkeypatch.setattr(datasets.config, "IN_MEMORY_MAX_SIZE", input_in_memory_max_size)
in_memory_max_size = datasets.config.IN_MEMORY_MAX_SIZE
if input_in_memory_max_size == "default":
assert in_memory_max_size == 0
else:
assert in_memory_max_size == input_in_memory_max_size
if dataset_size and in_memory_max_size:
expected = dataset_size < in_memory_max_size
else:
expected = False
result = is_small_dataset(dataset_size)
assert result == expected
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_distributed.py | import os
import sys
from pathlib import Path
import pytest
from datasets import Dataset, IterableDataset
from datasets.distributed import split_dataset_by_node
from .utils import execute_subprocess_async, get_torch_dist_unique_port, require_torch
def test_split_dataset_by_node_map_style():
full_ds = Dataset.from_dict({"i": range(17)})
full_size = len(full_ds)
world_size = 3
datasets_per_rank = [
split_dataset_by_node(full_ds, rank=rank, world_size=world_size) for rank in range(world_size)
]
assert sum(len(ds) for ds in datasets_per_rank) == full_size
assert len({tuple(x.values()) for ds in datasets_per_rank for x in ds}) == full_size
def test_split_dataset_by_node_iterable():
def gen():
return ({"i": i} for i in range(17))
world_size = 3
full_ds = IterableDataset.from_generator(gen)
full_size = len(list(full_ds))
datasets_per_rank = [
split_dataset_by_node(full_ds, rank=rank, world_size=world_size) for rank in range(world_size)
]
assert sum(len(list(ds)) for ds in datasets_per_rank) == full_size
assert len({tuple(x.values()) for ds in datasets_per_rank for x in ds}) == full_size
@pytest.mark.parametrize("shards_per_node", [1, 2, 3])
def test_split_dataset_by_node_iterable_sharded(shards_per_node):
def gen(shards):
for shard in shards:
yield from ({"i": i, "shard": shard} for i in range(17))
world_size = 3
num_shards = shards_per_node * world_size
gen_kwargs = {"shards": [f"shard_{shard_idx}.txt" for shard_idx in range(num_shards)]}
full_ds = IterableDataset.from_generator(gen, gen_kwargs=gen_kwargs)
full_size = len(list(full_ds))
assert full_ds.n_shards == world_size * shards_per_node
datasets_per_rank = [
split_dataset_by_node(full_ds, rank=rank, world_size=world_size) for rank in range(world_size)
]
assert [ds.n_shards for ds in datasets_per_rank] == [shards_per_node] * world_size
assert sum(len(list(ds)) for ds in datasets_per_rank) == full_size
assert len({tuple(x.values()) for ds in datasets_per_rank for x in ds}) == full_size
def test_distributed_shuffle_iterable():
def gen():
return ({"i": i} for i in range(17))
world_size = 2
full_ds = IterableDataset.from_generator(gen)
full_size = len(list(full_ds))
ds_rank0 = split_dataset_by_node(full_ds, rank=0, world_size=world_size).shuffle(seed=42)
assert len(list(ds_rank0)) == 1 + full_size // world_size
with pytest.raises(RuntimeError):
split_dataset_by_node(full_ds, rank=0, world_size=world_size).shuffle()
ds_rank0 = split_dataset_by_node(full_ds.shuffle(seed=42), rank=0, world_size=world_size)
assert len(list(ds_rank0)) == 1 + full_size // world_size
with pytest.raises(RuntimeError):
split_dataset_by_node(full_ds.shuffle(), rank=0, world_size=world_size)
@pytest.mark.parametrize("streaming", [False, True])
@require_torch
@pytest.mark.skipif(os.name == "nt", reason="execute_subprocess_async doesn't support windows")
@pytest.mark.integration
def test_torch_distributed_run(streaming):
nproc_per_node = 2
master_port = get_torch_dist_unique_port()
test_script = Path(__file__).resolve().parent / "distributed_scripts" / "run_torch_distributed.py"
distributed_args = f"""
-m torch.distributed.run
--nproc_per_node={nproc_per_node}
--master_port={master_port}
{test_script}
""".split()
args = f"""
--streaming={streaming}
""".split()
cmd = [sys.executable] + distributed_args + args
execute_subprocess_async(cmd, env=os.environ.copy())
@pytest.mark.parametrize(
"nproc_per_node, num_workers",
[
(2, 2), # each node has 2 shards and each worker has 1 shards
(3, 2), # each node uses all the shards but skips examples, and each worker has 2 shards
],
)
@require_torch
@pytest.mark.skipif(os.name == "nt", reason="execute_subprocess_async doesn't support windows")
@pytest.mark.integration
def test_torch_distributed_run_streaming_with_num_workers(nproc_per_node, num_workers):
streaming = True
master_port = get_torch_dist_unique_port()
test_script = Path(__file__).resolve().parent / "distributed_scripts" / "run_torch_distributed.py"
distributed_args = f"""
-m torch.distributed.run
--nproc_per_node={nproc_per_node}
--master_port={master_port}
{test_script}
""".split()
args = f"""
--streaming={streaming}
--num_workers={num_workers}
""".split()
cmd = [sys.executable] + distributed_args + args
execute_subprocess_async(cmd, env=os.environ.copy())
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_splits.py | import pytest
from datasets.splits import SplitDict, SplitInfo
from datasets.utils.py_utils import asdict
@pytest.mark.parametrize(
"split_dict",
[
SplitDict(),
SplitDict({"train": SplitInfo(name="train", num_bytes=1337, num_examples=42, dataset_name="my_dataset")}),
SplitDict({"train": SplitInfo(name="train", num_bytes=1337, num_examples=42)}),
SplitDict({"train": SplitInfo()}),
],
)
def test_split_dict_to_yaml_list(split_dict: SplitDict):
split_dict_yaml_list = split_dict._to_yaml_list()
assert len(split_dict_yaml_list) == len(split_dict)
reloaded = SplitDict._from_yaml_list(split_dict_yaml_list)
for split_name, split_info in split_dict.items():
# dataset_name field is deprecated, and is therefore not part of the YAML dump
split_info.dataset_name = None
# the split name of split_dict takes over the name of the split info object
split_info.name = split_name
assert split_dict == reloaded
@pytest.mark.parametrize(
"split_info", [SplitInfo(), SplitInfo(dataset_name=None), SplitInfo(dataset_name="my_dataset")]
)
def test_split_dict_asdict_has_dataset_name(split_info):
# For backward compatibility, we need asdict(split_dict) to return split info dictrionaries with the "dataset_name"
# field even if it's deprecated. This way old versionso of `datasets` can still reload dataset_infos.json files
split_dict_asdict = asdict(SplitDict({"train": split_info}))
assert "dataset_name" in split_dict_asdict["train"]
assert split_dict_asdict["train"]["dataset_name"] == split_info.dataset_name
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_table.py | import copy
import pickle
import warnings
from typing import List, Union
import numpy as np
import pyarrow as pa
import pytest
import datasets
from datasets import Sequence, Value
from datasets.features.features import Array2DExtensionType, ClassLabel, Features, Image
from datasets.table import (
ConcatenationTable,
InMemoryTable,
MemoryMappedTable,
Table,
TableBlock,
_in_memory_arrow_table_from_buffer,
_in_memory_arrow_table_from_file,
_interpolation_search,
_is_extension_type,
_memory_mapped_arrow_table_from_file,
array_concat,
cast_array_to_feature,
concat_tables,
embed_array_storage,
embed_table_storage,
inject_arrow_table_documentation,
table_cast,
table_iter,
)
from .utils import assert_arrow_memory_doesnt_increase, assert_arrow_memory_increases, slow
@pytest.fixture(scope="session")
def in_memory_pa_table(arrow_file) -> pa.Table:
return pa.ipc.open_stream(arrow_file).read_all()
def _to_testing_blocks(table: TableBlock) -> List[List[TableBlock]]:
assert len(table) > 2
blocks = [
[table.slice(0, 2)],
[table.slice(2).drop([c for c in table.column_names if c != "tokens"]), table.slice(2).drop(["tokens"])],
]
return blocks
@pytest.fixture(scope="session")
def in_memory_blocks(in_memory_pa_table):
table = InMemoryTable(in_memory_pa_table)
return _to_testing_blocks(table)
@pytest.fixture(scope="session")
def memory_mapped_blocks(arrow_file):
table = MemoryMappedTable.from_file(arrow_file)
return _to_testing_blocks(table)
@pytest.fixture(scope="session")
def mixed_in_memory_and_memory_mapped_blocks(in_memory_blocks, memory_mapped_blocks):
return in_memory_blocks[:1] + memory_mapped_blocks[1:]
def assert_deepcopy_without_bringing_data_in_memory(table: MemoryMappedTable):
with assert_arrow_memory_doesnt_increase():
copied_table = copy.deepcopy(table)
assert isinstance(copied_table, MemoryMappedTable)
assert copied_table.table == table.table
def assert_deepcopy_does_bring_data_in_memory(table: MemoryMappedTable):
with assert_arrow_memory_increases():
copied_table = copy.deepcopy(table)
assert isinstance(copied_table, MemoryMappedTable)
assert copied_table.table == table.table
def assert_pickle_without_bringing_data_in_memory(table: MemoryMappedTable):
with assert_arrow_memory_doesnt_increase():
pickled_table = pickle.dumps(table)
unpickled_table = pickle.loads(pickled_table)
assert isinstance(unpickled_table, MemoryMappedTable)
assert unpickled_table.table == table.table
def assert_pickle_does_bring_data_in_memory(table: MemoryMappedTable):
with assert_arrow_memory_increases():
pickled_table = pickle.dumps(table)
unpickled_table = pickle.loads(pickled_table)
assert isinstance(unpickled_table, MemoryMappedTable)
assert unpickled_table.table == table.table
def assert_index_attributes_equal(table: Table, other: Table):
assert table._batches == other._batches
np.testing.assert_array_equal(table._offsets, other._offsets)
assert table._schema == other._schema
def add_suffix_to_column_names(table, suffix):
return table.rename_columns([f"{name}{suffix}" for name in table.column_names])
def test_inject_arrow_table_documentation(in_memory_pa_table):
method = pa.Table.slice
def function_to_wrap(*args):
return method(*args)
args = (0, 1)
wrapped_method = inject_arrow_table_documentation(method)(function_to_wrap)
assert method(in_memory_pa_table, *args) == wrapped_method(in_memory_pa_table, *args)
assert "pyarrow.Table" not in wrapped_method.__doc__
assert "Table" in wrapped_method.__doc__
def test_in_memory_arrow_table_from_file(arrow_file, in_memory_pa_table):
with assert_arrow_memory_increases():
pa_table = _in_memory_arrow_table_from_file(arrow_file)
assert in_memory_pa_table == pa_table
def test_in_memory_arrow_table_from_buffer(in_memory_pa_table):
with assert_arrow_memory_increases():
buf_writer = pa.BufferOutputStream()
writer = pa.RecordBatchStreamWriter(buf_writer, schema=in_memory_pa_table.schema)
writer.write_table(in_memory_pa_table)
writer.close()
buf_writer.close()
pa_table = _in_memory_arrow_table_from_buffer(buf_writer.getvalue())
assert in_memory_pa_table == pa_table
def test_memory_mapped_arrow_table_from_file(arrow_file, in_memory_pa_table):
with assert_arrow_memory_doesnt_increase():
pa_table = _memory_mapped_arrow_table_from_file(arrow_file)
assert in_memory_pa_table == pa_table
def test_table_init(in_memory_pa_table):
table = Table(in_memory_pa_table)
assert table.table == in_memory_pa_table
def test_table_validate(in_memory_pa_table):
table = Table(in_memory_pa_table)
assert table.validate() == in_memory_pa_table.validate()
def test_table_equals(in_memory_pa_table):
table = Table(in_memory_pa_table)
assert table.equals(in_memory_pa_table)
def test_table_to_batches(in_memory_pa_table):
table = Table(in_memory_pa_table)
assert table.to_batches() == in_memory_pa_table.to_batches()
def test_table_to_pydict(in_memory_pa_table):
table = Table(in_memory_pa_table)
assert table.to_pydict() == in_memory_pa_table.to_pydict()
def test_table_to_string(in_memory_pa_table):
table = Table(in_memory_pa_table)
assert table.to_string() == in_memory_pa_table.to_string()
def test_table_field(in_memory_pa_table):
assert "tokens" in in_memory_pa_table.column_names
table = Table(in_memory_pa_table)
assert table.field("tokens") == in_memory_pa_table.field("tokens")
def test_table_column(in_memory_pa_table):
assert "tokens" in in_memory_pa_table.column_names
table = Table(in_memory_pa_table)
assert table.column("tokens") == in_memory_pa_table.column("tokens")
def test_table_itercolumns(in_memory_pa_table):
table = Table(in_memory_pa_table)
assert isinstance(table.itercolumns(), type(in_memory_pa_table.itercolumns()))
assert list(table.itercolumns()) == list(in_memory_pa_table.itercolumns())
def test_table_getitem(in_memory_pa_table):
table = Table(in_memory_pa_table)
assert table[0] == in_memory_pa_table[0]
def test_table_len(in_memory_pa_table):
table = Table(in_memory_pa_table)
assert len(table) == len(in_memory_pa_table)
def test_table_str(in_memory_pa_table):
table = Table(in_memory_pa_table)
assert str(table) == str(in_memory_pa_table).replace("pyarrow.Table", "Table")
assert repr(table) == repr(in_memory_pa_table).replace("pyarrow.Table", "Table")
@pytest.mark.parametrize(
"attribute", ["schema", "columns", "num_columns", "num_rows", "shape", "nbytes", "column_names"]
)
def test_table_attributes(in_memory_pa_table, attribute):
table = Table(in_memory_pa_table)
assert getattr(table, attribute) == getattr(in_memory_pa_table, attribute)
def test_in_memory_table_from_file(arrow_file, in_memory_pa_table):
with assert_arrow_memory_increases():
table = InMemoryTable.from_file(arrow_file)
assert table.table == in_memory_pa_table
assert isinstance(table, InMemoryTable)
def test_in_memory_table_from_buffer(in_memory_pa_table):
with assert_arrow_memory_increases():
buf_writer = pa.BufferOutputStream()
writer = pa.RecordBatchStreamWriter(buf_writer, schema=in_memory_pa_table.schema)
writer.write_table(in_memory_pa_table)
writer.close()
buf_writer.close()
table = InMemoryTable.from_buffer(buf_writer.getvalue())
assert table.table == in_memory_pa_table
assert isinstance(table, InMemoryTable)
def test_in_memory_table_from_pandas(in_memory_pa_table):
df = in_memory_pa_table.to_pandas()
with assert_arrow_memory_increases():
# with no schema it might infer another order of the fields in the schema
table = InMemoryTable.from_pandas(df)
assert isinstance(table, InMemoryTable)
# by specifying schema we get the same order of features, and so the exact same table
table = InMemoryTable.from_pandas(df, schema=in_memory_pa_table.schema)
assert table.table == in_memory_pa_table
assert isinstance(table, InMemoryTable)
def test_in_memory_table_from_arrays(in_memory_pa_table):
arrays = list(in_memory_pa_table.columns)
names = list(in_memory_pa_table.column_names)
table = InMemoryTable.from_arrays(arrays, names=names)
assert table.table == in_memory_pa_table
assert isinstance(table, InMemoryTable)
def test_in_memory_table_from_pydict(in_memory_pa_table):
pydict = in_memory_pa_table.to_pydict()
with assert_arrow_memory_increases():
table = InMemoryTable.from_pydict(pydict)
assert isinstance(table, InMemoryTable)
assert table.table == pa.Table.from_pydict(pydict)
def test_in_memory_table_from_pylist(in_memory_pa_table):
pylist = InMemoryTable(in_memory_pa_table).to_pylist()
table = InMemoryTable.from_pylist(pylist)
assert isinstance(table, InMemoryTable)
assert pylist == table.to_pylist()
def test_in_memory_table_from_batches(in_memory_pa_table):
batches = list(in_memory_pa_table.to_batches())
table = InMemoryTable.from_batches(batches)
assert table.table == in_memory_pa_table
assert isinstance(table, InMemoryTable)
def test_in_memory_table_deepcopy(in_memory_pa_table):
table = InMemoryTable(in_memory_pa_table)
copied_table = copy.deepcopy(table)
assert table.table == copied_table.table
assert_index_attributes_equal(table, copied_table)
# deepcopy must return the exact same arrow objects since they are immutable
assert table.table is copied_table.table
assert all(batch1 is batch2 for batch1, batch2 in zip(table._batches, copied_table._batches))
def test_in_memory_table_pickle(in_memory_pa_table):
table = InMemoryTable(in_memory_pa_table)
pickled_table = pickle.dumps(table)
unpickled_table = pickle.loads(pickled_table)
assert unpickled_table.table == table.table
assert_index_attributes_equal(table, unpickled_table)
@slow
def test_in_memory_table_pickle_big_table():
big_table_4GB = InMemoryTable.from_pydict({"col": [0] * ((4 * 8 << 30) // 64)})
length = len(big_table_4GB)
big_table_4GB = pickle.dumps(big_table_4GB)
big_table_4GB = pickle.loads(big_table_4GB)
assert len(big_table_4GB) == length
def test_in_memory_table_slice(in_memory_pa_table):
table = InMemoryTable(in_memory_pa_table).slice(1, 2)
assert table.table == in_memory_pa_table.slice(1, 2)
assert isinstance(table, InMemoryTable)
def test_in_memory_table_filter(in_memory_pa_table):
mask = pa.array([i % 2 == 0 for i in range(len(in_memory_pa_table))])
table = InMemoryTable(in_memory_pa_table).filter(mask)
assert table.table == in_memory_pa_table.filter(mask)
assert isinstance(table, InMemoryTable)
def test_in_memory_table_flatten(in_memory_pa_table):
table = InMemoryTable(in_memory_pa_table).flatten()
assert table.table == in_memory_pa_table.flatten()
assert isinstance(table, InMemoryTable)
def test_in_memory_table_combine_chunks(in_memory_pa_table):
table = InMemoryTable(in_memory_pa_table).combine_chunks()
assert table.table == in_memory_pa_table.combine_chunks()
assert isinstance(table, InMemoryTable)
def test_in_memory_table_cast(in_memory_pa_table):
assert pa.list_(pa.int64()) in in_memory_pa_table.schema.types
schema = pa.schema(
{
k: v if v != pa.list_(pa.int64()) else pa.list_(pa.int32())
for k, v in zip(in_memory_pa_table.schema.names, in_memory_pa_table.schema.types)
}
)
table = InMemoryTable(in_memory_pa_table).cast(schema)
assert table.table == in_memory_pa_table.cast(schema)
assert isinstance(table, InMemoryTable)
def test_in_memory_table_cast_reorder_struct():
table = InMemoryTable(
pa.Table.from_pydict(
{
"top": [
{
"foo": "a",
"bar": "b",
}
]
}
)
)
schema = pa.schema({"top": pa.struct({"bar": pa.string(), "foo": pa.string()})})
assert table.cast(schema).schema == schema
def test_in_memory_table_cast_with_hf_features():
table = InMemoryTable(pa.Table.from_pydict({"labels": [0, 1]}))
features = Features({"labels": ClassLabel(names=["neg", "pos"])})
schema = features.arrow_schema
assert table.cast(schema).schema == schema
assert Features.from_arrow_schema(table.cast(schema).schema) == features
def test_in_memory_table_replace_schema_metadata(in_memory_pa_table):
metadata = {"huggingface": "{}"}
table = InMemoryTable(in_memory_pa_table).replace_schema_metadata(metadata)
assert table.table.schema.metadata == in_memory_pa_table.replace_schema_metadata(metadata).schema.metadata
assert isinstance(table, InMemoryTable)
def test_in_memory_table_add_column(in_memory_pa_table):
i = len(in_memory_pa_table.column_names)
field_ = "new_field"
column = pa.array(list(range(len(in_memory_pa_table))))
table = InMemoryTable(in_memory_pa_table).add_column(i, field_, column)
assert table.table == in_memory_pa_table.add_column(i, field_, column)
assert isinstance(table, InMemoryTable)
def test_in_memory_table_append_column(in_memory_pa_table):
field_ = "new_field"
column = pa.array(list(range(len(in_memory_pa_table))))
table = InMemoryTable(in_memory_pa_table).append_column(field_, column)
assert table.table == in_memory_pa_table.append_column(field_, column)
assert isinstance(table, InMemoryTable)
def test_in_memory_table_remove_column(in_memory_pa_table):
table = InMemoryTable(in_memory_pa_table).remove_column(0)
assert table.table == in_memory_pa_table.remove_column(0)
assert isinstance(table, InMemoryTable)
def test_in_memory_table_set_column(in_memory_pa_table):
i = len(in_memory_pa_table.column_names)
field_ = "new_field"
column = pa.array(list(range(len(in_memory_pa_table))))
table = InMemoryTable(in_memory_pa_table).set_column(i, field_, column)
assert table.table == in_memory_pa_table.set_column(i, field_, column)
assert isinstance(table, InMemoryTable)
def test_in_memory_table_rename_columns(in_memory_pa_table):
assert "tokens" in in_memory_pa_table.column_names
names = [name if name != "tokens" else "new_tokens" for name in in_memory_pa_table.column_names]
table = InMemoryTable(in_memory_pa_table).rename_columns(names)
assert table.table == in_memory_pa_table.rename_columns(names)
assert isinstance(table, InMemoryTable)
def test_in_memory_table_drop(in_memory_pa_table):
names = [in_memory_pa_table.column_names[0]]
table = InMemoryTable(in_memory_pa_table).drop(names)
assert table.table == in_memory_pa_table.drop(names)
assert isinstance(table, InMemoryTable)
def test_memory_mapped_table_init(arrow_file, in_memory_pa_table):
table = MemoryMappedTable(_memory_mapped_arrow_table_from_file(arrow_file), arrow_file)
assert table.table == in_memory_pa_table
assert isinstance(table, MemoryMappedTable)
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
def test_memory_mapped_table_from_file(arrow_file, in_memory_pa_table):
with assert_arrow_memory_doesnt_increase():
table = MemoryMappedTable.from_file(arrow_file)
assert table.table == in_memory_pa_table
assert isinstance(table, MemoryMappedTable)
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
def test_memory_mapped_table_from_file_with_replay(arrow_file, in_memory_pa_table):
replays = [("slice", (0, 1), {}), ("flatten", (), {})]
with assert_arrow_memory_doesnt_increase():
table = MemoryMappedTable.from_file(arrow_file, replays=replays)
assert len(table) == 1
for method, args, kwargs in replays:
in_memory_pa_table = getattr(in_memory_pa_table, method)(*args, **kwargs)
assert table.table == in_memory_pa_table
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
def test_memory_mapped_table_deepcopy(arrow_file):
table = MemoryMappedTable.from_file(arrow_file)
copied_table = copy.deepcopy(table)
assert table.table == copied_table.table
assert table.path == copied_table.path
assert_index_attributes_equal(table, copied_table)
# deepcopy must return the exact same arrow objects since they are immutable
assert table.table is copied_table.table
assert all(batch1 is batch2 for batch1, batch2 in zip(table._batches, copied_table._batches))
def test_memory_mapped_table_pickle(arrow_file):
table = MemoryMappedTable.from_file(arrow_file)
pickled_table = pickle.dumps(table)
unpickled_table = pickle.loads(pickled_table)
assert unpickled_table.table == table.table
assert unpickled_table.path == table.path
assert_index_attributes_equal(table, unpickled_table)
def test_memory_mapped_table_pickle_doesnt_fill_memory(arrow_file):
with assert_arrow_memory_doesnt_increase():
table = MemoryMappedTable.from_file(arrow_file)
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
def test_memory_mapped_table_pickle_applies_replay(arrow_file):
replays = [("slice", (0, 1), {}), ("flatten", (), {})]
with assert_arrow_memory_doesnt_increase():
table = MemoryMappedTable.from_file(arrow_file, replays=replays)
assert isinstance(table, MemoryMappedTable)
assert table.replays == replays
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
def test_memory_mapped_table_slice(arrow_file, in_memory_pa_table):
table = MemoryMappedTable.from_file(arrow_file).slice(1, 2)
assert table.table == in_memory_pa_table.slice(1, 2)
assert isinstance(table, MemoryMappedTable)
assert table.replays == [("slice", (1, 2), {})]
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
def test_memory_mapped_table_filter(arrow_file, in_memory_pa_table):
mask = pa.array([i % 2 == 0 for i in range(len(in_memory_pa_table))])
table = MemoryMappedTable.from_file(arrow_file).filter(mask)
assert table.table == in_memory_pa_table.filter(mask)
assert isinstance(table, MemoryMappedTable)
assert table.replays == [("filter", (mask,), {})]
assert_deepcopy_without_bringing_data_in_memory(table)
# filter DOES increase memory
# assert_pickle_without_bringing_data_in_memory(table)
assert_pickle_does_bring_data_in_memory(table)
def test_memory_mapped_table_flatten(arrow_file, in_memory_pa_table):
table = MemoryMappedTable.from_file(arrow_file).flatten()
assert table.table == in_memory_pa_table.flatten()
assert isinstance(table, MemoryMappedTable)
assert table.replays == [("flatten", (), {})]
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
def test_memory_mapped_table_combine_chunks(arrow_file, in_memory_pa_table):
table = MemoryMappedTable.from_file(arrow_file).combine_chunks()
assert table.table == in_memory_pa_table.combine_chunks()
assert isinstance(table, MemoryMappedTable)
assert table.replays == [("combine_chunks", (), {})]
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
def test_memory_mapped_table_cast(arrow_file, in_memory_pa_table):
assert pa.list_(pa.int64()) in in_memory_pa_table.schema.types
schema = pa.schema(
{
k: v if v != pa.list_(pa.int64()) else pa.list_(pa.int32())
for k, v in zip(in_memory_pa_table.schema.names, in_memory_pa_table.schema.types)
}
)
table = MemoryMappedTable.from_file(arrow_file).cast(schema)
assert table.table == in_memory_pa_table.cast(schema)
assert isinstance(table, MemoryMappedTable)
assert table.replays == [("cast", (schema,), {})]
assert_deepcopy_without_bringing_data_in_memory(table)
# cast DOES increase memory when converting integers precision for example
# assert_pickle_without_bringing_data_in_memory(table)
assert_pickle_does_bring_data_in_memory(table)
def test_memory_mapped_table_replace_schema_metadata(arrow_file, in_memory_pa_table):
metadata = {"huggingface": "{}"}
table = MemoryMappedTable.from_file(arrow_file).replace_schema_metadata(metadata)
assert table.table.schema.metadata == in_memory_pa_table.replace_schema_metadata(metadata).schema.metadata
assert isinstance(table, MemoryMappedTable)
assert table.replays == [("replace_schema_metadata", (metadata,), {})]
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
def test_memory_mapped_table_add_column(arrow_file, in_memory_pa_table):
i = len(in_memory_pa_table.column_names)
field_ = "new_field"
column = pa.array(list(range(len(in_memory_pa_table))))
table = MemoryMappedTable.from_file(arrow_file).add_column(i, field_, column)
assert table.table == in_memory_pa_table.add_column(i, field_, column)
assert isinstance(table, MemoryMappedTable)
assert table.replays == [("add_column", (i, field_, column), {})]
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
def test_memory_mapped_table_append_column(arrow_file, in_memory_pa_table):
field_ = "new_field"
column = pa.array(list(range(len(in_memory_pa_table))))
table = MemoryMappedTable.from_file(arrow_file).append_column(field_, column)
assert table.table == in_memory_pa_table.append_column(field_, column)
assert isinstance(table, MemoryMappedTable)
assert table.replays == [("append_column", (field_, column), {})]
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
def test_memory_mapped_table_remove_column(arrow_file, in_memory_pa_table):
table = MemoryMappedTable.from_file(arrow_file).remove_column(0)
assert table.table == in_memory_pa_table.remove_column(0)
assert isinstance(table, MemoryMappedTable)
assert table.replays == [("remove_column", (0,), {})]
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
def test_memory_mapped_table_set_column(arrow_file, in_memory_pa_table):
i = len(in_memory_pa_table.column_names)
field_ = "new_field"
column = pa.array(list(range(len(in_memory_pa_table))))
table = MemoryMappedTable.from_file(arrow_file).set_column(i, field_, column)
assert table.table == in_memory_pa_table.set_column(i, field_, column)
assert isinstance(table, MemoryMappedTable)
assert table.replays == [("set_column", (i, field_, column), {})]
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
def test_memory_mapped_table_rename_columns(arrow_file, in_memory_pa_table):
assert "tokens" in in_memory_pa_table.column_names
names = [name if name != "tokens" else "new_tokens" for name in in_memory_pa_table.column_names]
table = MemoryMappedTable.from_file(arrow_file).rename_columns(names)
assert table.table == in_memory_pa_table.rename_columns(names)
assert isinstance(table, MemoryMappedTable)
assert table.replays == [("rename_columns", (names,), {})]
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
def test_memory_mapped_table_drop(arrow_file, in_memory_pa_table):
names = [in_memory_pa_table.column_names[0]]
table = MemoryMappedTable.from_file(arrow_file).drop(names)
assert table.table == in_memory_pa_table.drop(names)
assert isinstance(table, MemoryMappedTable)
assert table.replays == [("drop", (names,), {})]
assert_deepcopy_without_bringing_data_in_memory(table)
assert_pickle_without_bringing_data_in_memory(table)
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concatenation_table_init(
blocks_type, in_memory_pa_table, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = (
in_memory_blocks
if blocks_type == "in_memory"
else memory_mapped_blocks
if blocks_type == "memory_mapped"
else mixed_in_memory_and_memory_mapped_blocks
)
table = ConcatenationTable(in_memory_pa_table, blocks)
assert table.table == in_memory_pa_table
assert table.blocks == blocks
def test_concatenation_table_from_blocks(in_memory_pa_table, in_memory_blocks):
assert len(in_memory_pa_table) > 2
in_memory_table = InMemoryTable(in_memory_pa_table)
t1, t2 = in_memory_table.slice(0, 2), in_memory_table.slice(2)
table = ConcatenationTable.from_blocks(in_memory_table)
assert isinstance(table, ConcatenationTable)
assert table.table == in_memory_pa_table
assert table.blocks == [[in_memory_table]]
table = ConcatenationTable.from_blocks([t1, t2])
assert isinstance(table, ConcatenationTable)
assert table.table == in_memory_pa_table
assert table.blocks == [[in_memory_table]]
table = ConcatenationTable.from_blocks([[t1], [t2]])
assert isinstance(table, ConcatenationTable)
assert table.table == in_memory_pa_table
assert table.blocks == [[in_memory_table]]
table = ConcatenationTable.from_blocks(in_memory_blocks)
assert isinstance(table, ConcatenationTable)
assert table.table == in_memory_pa_table
assert table.blocks == [[in_memory_table]]
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concatenation_table_from_blocks_doesnt_increase_memory(
blocks_type, in_memory_pa_table, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = {
"in_memory": in_memory_blocks,
"memory_mapped": memory_mapped_blocks,
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
with assert_arrow_memory_doesnt_increase():
table = ConcatenationTable.from_blocks(blocks)
assert isinstance(table, ConcatenationTable)
assert table.table == in_memory_pa_table
if blocks_type == "in_memory":
assert table.blocks == [[InMemoryTable(in_memory_pa_table)]]
else:
assert table.blocks == blocks
@pytest.mark.parametrize("axis", [0, 1])
def test_concatenation_table_from_tables(axis, in_memory_pa_table, arrow_file):
in_memory_table = InMemoryTable(in_memory_pa_table)
concatenation_table = ConcatenationTable.from_blocks(in_memory_table)
memory_mapped_table = MemoryMappedTable.from_file(arrow_file)
tables = [in_memory_pa_table, in_memory_table, concatenation_table, memory_mapped_table]
if axis == 0:
expected_table = pa.concat_tables([in_memory_pa_table] * len(tables))
else:
# avoids error due to duplicate column names
tables[1:] = [add_suffix_to_column_names(table, i) for i, table in enumerate(tables[1:], 1)]
expected_table = in_memory_pa_table
for table in tables[1:]:
for name, col in zip(table.column_names, table.columns):
expected_table = expected_table.append_column(name, col)
with assert_arrow_memory_doesnt_increase():
table = ConcatenationTable.from_tables(tables, axis=axis)
assert isinstance(table, ConcatenationTable)
assert table.table == expected_table
# because of consolidation, we end up with 1 InMemoryTable and 1 MemoryMappedTable
assert len(table.blocks) == 1 if axis == 1 else 2
assert len(table.blocks[0]) == 1 if axis == 0 else 2
assert axis == 1 or len(table.blocks[1]) == 1
assert isinstance(table.blocks[0][0], InMemoryTable)
assert isinstance(table.blocks[1][0] if axis == 0 else table.blocks[0][1], MemoryMappedTable)
def test_concatenation_table_from_tables_axis1_misaligned_blocks(arrow_file):
table = MemoryMappedTable.from_file(arrow_file)
t1 = table.slice(0, 2)
t2 = table.slice(0, 3).rename_columns([col + "_1" for col in table.column_names])
concatenated = ConcatenationTable.from_tables(
[
ConcatenationTable.from_blocks([[t1], [t1], [t1]]),
ConcatenationTable.from_blocks([[t2], [t2]]),
],
axis=1,
)
assert len(concatenated) == 6
assert [len(row_blocks[0]) for row_blocks in concatenated.blocks] == [2, 1, 1, 2]
concatenated = ConcatenationTable.from_tables(
[
ConcatenationTable.from_blocks([[t2], [t2]]),
ConcatenationTable.from_blocks([[t1], [t1], [t1]]),
],
axis=1,
)
assert len(concatenated) == 6
assert [len(row_blocks[0]) for row_blocks in concatenated.blocks] == [2, 1, 1, 2]
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concatenation_table_deepcopy(
blocks_type, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = {
"in_memory": in_memory_blocks,
"memory_mapped": memory_mapped_blocks,
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
table = ConcatenationTable.from_blocks(blocks)
copied_table = copy.deepcopy(table)
assert table.table == copied_table.table
assert table.blocks == copied_table.blocks
assert_index_attributes_equal(table, copied_table)
# deepcopy must return the exact same arrow objects since they are immutable
assert table.table is copied_table.table
assert all(batch1 is batch2 for batch1, batch2 in zip(table._batches, copied_table._batches))
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concatenation_table_pickle(
blocks_type, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = {
"in_memory": in_memory_blocks,
"memory_mapped": memory_mapped_blocks,
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
table = ConcatenationTable.from_blocks(blocks)
pickled_table = pickle.dumps(table)
unpickled_table = pickle.loads(pickled_table)
assert unpickled_table.table == table.table
assert unpickled_table.blocks == table.blocks
assert_index_attributes_equal(table, unpickled_table)
def test_concat_tables_with_features_metadata(arrow_file, in_memory_pa_table):
input_features = Features.from_arrow_schema(in_memory_pa_table.schema)
input_features["id"] = Value("int64", id="my_id")
intput_schema = input_features.arrow_schema
t0 = in_memory_pa_table.replace_schema_metadata(intput_schema.metadata)
t1 = MemoryMappedTable.from_file(arrow_file)
tables = [t0, t1]
concatenated_table = concat_tables(tables, axis=0)
output_schema = concatenated_table.schema
output_features = Features.from_arrow_schema(output_schema)
assert output_schema == intput_schema
assert output_schema.metadata == intput_schema.metadata
assert output_features == input_features
assert output_features["id"].id == "my_id"
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concatenation_table_slice(
blocks_type, in_memory_pa_table, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = {
"in_memory": in_memory_blocks,
"memory_mapped": memory_mapped_blocks,
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
table = ConcatenationTable.from_blocks(blocks).slice(1, 2)
assert table.table == in_memory_pa_table.slice(1, 2)
assert isinstance(table, ConcatenationTable)
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concatenation_table_filter(
blocks_type, in_memory_pa_table, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = {
"in_memory": in_memory_blocks,
"memory_mapped": memory_mapped_blocks,
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
mask = pa.array([i % 2 == 0 for i in range(len(in_memory_pa_table))])
table = ConcatenationTable.from_blocks(blocks).filter(mask)
assert table.table == in_memory_pa_table.filter(mask)
assert isinstance(table, ConcatenationTable)
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concatenation_table_flatten(
blocks_type, in_memory_pa_table, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = {
"in_memory": in_memory_blocks,
"memory_mapped": memory_mapped_blocks,
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
table = ConcatenationTable.from_blocks(blocks).flatten()
assert table.table == in_memory_pa_table.flatten()
assert isinstance(table, ConcatenationTable)
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concatenation_table_combine_chunks(
blocks_type, in_memory_pa_table, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = {
"in_memory": in_memory_blocks,
"memory_mapped": memory_mapped_blocks,
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
table = ConcatenationTable.from_blocks(blocks).combine_chunks()
assert table.table == in_memory_pa_table.combine_chunks()
assert isinstance(table, ConcatenationTable)
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concatenation_table_cast(
blocks_type, in_memory_pa_table, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = {
"in_memory": in_memory_blocks,
"memory_mapped": memory_mapped_blocks,
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
assert pa.list_(pa.int64()) in in_memory_pa_table.schema.types
assert pa.int64() in in_memory_pa_table.schema.types
schema = pa.schema(
{
k: v if v != pa.list_(pa.int64()) else pa.list_(pa.int32())
for k, v in zip(in_memory_pa_table.schema.names, in_memory_pa_table.schema.types)
}
)
table = ConcatenationTable.from_blocks(blocks).cast(schema)
assert table.table == in_memory_pa_table.cast(schema)
assert isinstance(table, ConcatenationTable)
schema = pa.schema(
{
k: v if v != pa.int64() else pa.int32()
for k, v in zip(in_memory_pa_table.schema.names, in_memory_pa_table.schema.types)
}
)
table = ConcatenationTable.from_blocks(blocks).cast(schema)
assert table.table == in_memory_pa_table.cast(schema)
assert isinstance(table, ConcatenationTable)
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concat_tables_cast_with_features_metadata(
blocks_type, in_memory_pa_table, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = {
"in_memory": in_memory_blocks,
"memory_mapped": memory_mapped_blocks,
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
input_features = Features.from_arrow_schema(in_memory_pa_table.schema)
input_features["id"] = Value("int64", id="my_id")
intput_schema = input_features.arrow_schema
concatenated_table = ConcatenationTable.from_blocks(blocks).cast(intput_schema)
output_schema = concatenated_table.schema
output_features = Features.from_arrow_schema(output_schema)
assert output_schema == intput_schema
assert output_schema.metadata == intput_schema.metadata
assert output_features == input_features
assert output_features["id"].id == "my_id"
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concatenation_table_replace_schema_metadata(
blocks_type, in_memory_pa_table, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = {
"in_memory": in_memory_blocks,
"memory_mapped": memory_mapped_blocks,
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
metadata = {"huggingface": "{}"}
table = ConcatenationTable.from_blocks(blocks).replace_schema_metadata(metadata)
assert table.table.schema.metadata == in_memory_pa_table.replace_schema_metadata(metadata).schema.metadata
assert isinstance(table, ConcatenationTable)
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concatenation_table_add_column(
blocks_type, in_memory_pa_table, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = {
"in_memory": in_memory_blocks,
"memory_mapped": memory_mapped_blocks,
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
i = len(in_memory_pa_table.column_names)
field_ = "new_field"
column = pa.array(list(range(len(in_memory_pa_table))))
with pytest.raises(NotImplementedError):
ConcatenationTable.from_blocks(blocks).add_column(i, field_, column)
# assert table.table == in_memory_pa_table.add_column(i, field_, column)
# unpickled_table = pickle.loads(pickle.dumps(table))
# assert unpickled_table.table == in_memory_pa_table.add_column(i, field_, column)
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concatenation_table_append_column(
blocks_type, in_memory_pa_table, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = {
"in_memory": in_memory_blocks,
"memory_mapped": memory_mapped_blocks,
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
field_ = "new_field"
column = pa.array(list(range(len(in_memory_pa_table))))
with pytest.raises(NotImplementedError):
ConcatenationTable.from_blocks(blocks).append_column(field_, column)
# assert table.table == in_memory_pa_table.append_column(field_, column)
# unpickled_table = pickle.loads(pickle.dumps(table))
# assert unpickled_table.table == in_memory_pa_table.append_column(field_, column)
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concatenation_table_remove_column(
blocks_type, in_memory_pa_table, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = {
"in_memory": in_memory_blocks,
"memory_mapped": memory_mapped_blocks,
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
table = ConcatenationTable.from_blocks(blocks).remove_column(0)
assert table.table == in_memory_pa_table.remove_column(0)
assert isinstance(table, ConcatenationTable)
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concatenation_table_set_column(
blocks_type, in_memory_pa_table, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = {
"in_memory": in_memory_blocks,
"memory_mapped": memory_mapped_blocks,
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
i = len(in_memory_pa_table.column_names)
field_ = "new_field"
column = pa.array(list(range(len(in_memory_pa_table))))
with pytest.raises(NotImplementedError):
ConcatenationTable.from_blocks(blocks).set_column(i, field_, column)
# assert table.table == in_memory_pa_table.set_column(i, field_, column)
# unpickled_table = pickle.loads(pickle.dumps(table))
# assert unpickled_table.table == in_memory_pa_table.set_column(i, field_, column)
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concatenation_table_rename_columns(
blocks_type, in_memory_pa_table, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = {
"in_memory": in_memory_blocks,
"memory_mapped": memory_mapped_blocks,
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
assert "tokens" in in_memory_pa_table.column_names
names = [name if name != "tokens" else "new_tokens" for name in in_memory_pa_table.column_names]
table = ConcatenationTable.from_blocks(blocks).rename_columns(names)
assert isinstance(table, ConcatenationTable)
assert table.table == in_memory_pa_table.rename_columns(names)
@pytest.mark.parametrize("blocks_type", ["in_memory", "memory_mapped", "mixed"])
def test_concatenation_table_drop(
blocks_type, in_memory_pa_table, in_memory_blocks, memory_mapped_blocks, mixed_in_memory_and_memory_mapped_blocks
):
blocks = {
"in_memory": in_memory_blocks,
"memory_mapped": memory_mapped_blocks,
"mixed": mixed_in_memory_and_memory_mapped_blocks,
}[blocks_type]
names = [in_memory_pa_table.column_names[0]]
table = ConcatenationTable.from_blocks(blocks).drop(names)
assert table.table == in_memory_pa_table.drop(names)
assert isinstance(table, ConcatenationTable)
def test_concat_tables(arrow_file, in_memory_pa_table):
t0 = in_memory_pa_table
t1 = InMemoryTable(t0)
t2 = MemoryMappedTable.from_file(arrow_file)
t3 = ConcatenationTable.from_blocks(t1)
tables = [t0, t1, t2, t3]
concatenated_table = concat_tables(tables, axis=0)
assert concatenated_table.table == pa.concat_tables([t0] * 4)
assert concatenated_table.table.shape == (40, 4)
assert isinstance(concatenated_table, ConcatenationTable)
assert len(concatenated_table.blocks) == 3 # t0 and t1 are consolidated as a single InMemoryTable
assert isinstance(concatenated_table.blocks[0][0], InMemoryTable)
assert isinstance(concatenated_table.blocks[1][0], MemoryMappedTable)
assert isinstance(concatenated_table.blocks[2][0], InMemoryTable)
# add suffix to avoid error due to duplicate column names
concatenated_table = concat_tables(
[add_suffix_to_column_names(table, i) for i, table in enumerate(tables)], axis=1
)
assert concatenated_table.table.shape == (10, 16)
assert len(concatenated_table.blocks[0]) == 3 # t0 and t1 are consolidated as a single InMemoryTable
assert isinstance(concatenated_table.blocks[0][0], InMemoryTable)
assert isinstance(concatenated_table.blocks[0][1], MemoryMappedTable)
assert isinstance(concatenated_table.blocks[0][2], InMemoryTable)
def _interpolation_search_ground_truth(arr: List[int], x: int) -> Union[int, IndexError]:
for i in range(len(arr) - 1):
if arr[i] <= x < arr[i + 1]:
return i
return IndexError
class _ListWithGetitemCounter(list):
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs)
self.unique_getitem_calls = set()
def __getitem__(self, i):
out = super().__getitem__(i)
self.unique_getitem_calls.add(i)
return out
@property
def getitem_unique_count(self):
return len(self.unique_getitem_calls)
@pytest.mark.parametrize(
"arr, x",
[(np.arange(0, 14, 3), x) for x in range(-1, 22)]
+ [(list(np.arange(-5, 5)), x) for x in range(-6, 6)]
+ [([0, 1_000, 1_001, 1_003], x) for x in [-1, 0, 2, 100, 999, 1_000, 1_001, 1_002, 1_003, 1_004]]
+ [(list(range(1_000)), x) for x in [-1, 0, 1, 10, 666, 999, 1_000, 1_0001]],
)
def test_interpolation_search(arr, x):
ground_truth = _interpolation_search_ground_truth(arr, x)
if isinstance(ground_truth, int):
arr = _ListWithGetitemCounter(arr)
output = _interpolation_search(arr, x)
assert ground_truth == output
# 4 maximum unique getitem calls is expected for the cases of this test
# but it can be bigger for large and messy arrays.
assert arr.getitem_unique_count <= 4
else:
with pytest.raises(ground_truth):
_interpolation_search(arr, x)
def test_indexed_table_mixin():
n_rows_per_chunk = 10
n_chunks = 4
pa_table = pa.Table.from_pydict({"col": [0] * n_rows_per_chunk})
pa_table = pa.concat_tables([pa_table] * n_chunks)
table = Table(pa_table)
assert all(table._offsets.tolist() == np.cumsum([0] + [n_rows_per_chunk] * n_chunks))
assert table.fast_slice(5) == pa_table.slice(5)
assert table.fast_slice(2, 13) == pa_table.slice(2, 13)
@pytest.mark.parametrize(
"arrays",
[
[pa.array([[1, 2, 3, 4]]), pa.array([[10, 2]])],
[
pa.array([[[1, 2], [3]]], pa.list_(pa.list_(pa.int32()), 2)),
pa.array([[[10, 2, 3], [2]]], pa.list_(pa.list_(pa.int32()), 2)),
],
[pa.array([[[1, 2, 3]], [[2, 3], [20, 21]], [[4]]]).slice(1), pa.array([[[1, 2, 3]]])],
],
)
def test_concat_arrays(arrays):
assert array_concat(arrays) == pa.concat_arrays(arrays)
def test_concat_arrays_nested_with_nulls():
arrays = [pa.array([{"a": 21, "b": [[1, 2], [3]]}]), pa.array([{"a": 100, "b": [[1], None]}])]
concatenated_arrays = array_concat(arrays)
assert concatenated_arrays == pa.array([{"a": 21, "b": [[1, 2], [3]]}, {"a": 100, "b": [[1], None]}])
def test_concat_extension_arrays():
arrays = [pa.array([[[1, 2], [3, 4]]]), pa.array([[[10, 2], [3, 4]]])]
extension_type = Array2DExtensionType((2, 2), "int64")
assert array_concat([extension_type.wrap_array(array) for array in arrays]) == extension_type.wrap_array(
pa.concat_arrays(arrays)
)
def test_cast_array_to_features():
arr = pa.array([[0, 1]])
assert cast_array_to_feature(arr, Sequence(Value("string"))).type == pa.list_(pa.string())
with pytest.raises(TypeError):
cast_array_to_feature(arr, Sequence(Value("string")), allow_number_to_str=False)
def test_cast_array_to_features_nested():
arr = pa.array([[{"foo": [0]}]])
assert cast_array_to_feature(arr, [{"foo": Sequence(Value("string"))}]).type == pa.list_(
pa.struct({"foo": pa.list_(pa.string())})
)
def test_cast_array_to_features_to_nested_with_no_fields():
arr = pa.array([{}])
assert cast_array_to_feature(arr, {}).type == pa.struct({})
assert cast_array_to_feature(arr, {}).to_pylist() == arr.to_pylist()
def test_cast_array_to_features_nested_with_null_values():
# same type
arr = pa.array([{"foo": [None, [0]]}], pa.struct({"foo": pa.list_(pa.list_(pa.int64()))}))
casted_array = cast_array_to_feature(arr, {"foo": [[Value("int64")]]})
assert casted_array.type == pa.struct({"foo": pa.list_(pa.list_(pa.int64()))})
assert casted_array.to_pylist() == arr.to_pylist()
# different type
arr = pa.array([{"foo": [None, [0]]}], pa.struct({"foo": pa.list_(pa.list_(pa.int64()))}))
if datasets.config.PYARROW_VERSION.major < 10:
with pytest.warns(UserWarning, match="None values are converted to empty lists.+"):
casted_array = cast_array_to_feature(arr, {"foo": [[Value("int32")]]})
assert casted_array.type == pa.struct({"foo": pa.list_(pa.list_(pa.int32()))})
assert casted_array.to_pylist() == [
{"foo": [[], [0]]}
] # empty list because of https://github.com/huggingface/datasets/issues/3676
else:
with warnings.catch_warnings():
warnings.simplefilter("error")
casted_array = cast_array_to_feature(arr, {"foo": [[Value("int32")]]})
assert casted_array.type == pa.struct({"foo": pa.list_(pa.list_(pa.int32()))})
assert casted_array.to_pylist() == [{"foo": [None, [0]]}]
def test_cast_array_to_features_to_null_type():
# same type
arr = pa.array([[None, None]])
assert cast_array_to_feature(arr, Sequence(Value("null"))).type == pa.list_(pa.null())
# different type
arr = pa.array([[None, 1]])
with pytest.raises(TypeError):
cast_array_to_feature(arr, Sequence(Value("null")))
def test_cast_array_to_features_sequence_classlabel():
arr = pa.array([[], [1], [0, 1]], pa.list_(pa.int64()))
assert cast_array_to_feature(arr, Sequence(ClassLabel(names=["foo", "bar"]))).type == pa.list_(pa.int64())
arr = pa.array([[], ["bar"], ["foo", "bar"]], pa.list_(pa.string()))
assert cast_array_to_feature(arr, Sequence(ClassLabel(names=["foo", "bar"]))).type == pa.list_(pa.int64())
# Test empty arrays
arr = pa.array([[], []], pa.list_(pa.int64()))
assert cast_array_to_feature(arr, Sequence(ClassLabel(names=["foo", "bar"]))).type == pa.list_(pa.int64())
arr = pa.array([[], []], pa.list_(pa.string()))
assert cast_array_to_feature(arr, Sequence(ClassLabel(names=["foo", "bar"]))).type == pa.list_(pa.int64())
# Test invalid class labels
arr = pa.array([[2]], pa.list_(pa.int64()))
with pytest.raises(ValueError):
assert cast_array_to_feature(arr, Sequence(ClassLabel(names=["foo", "bar"])))
arr = pa.array([["baz"]], pa.list_(pa.string()))
with pytest.raises(ValueError):
assert cast_array_to_feature(arr, Sequence(ClassLabel(names=["foo", "bar"])))
def test_cast_sliced_fixed_size_array_to_features():
arr = pa.array([[0, 1, 2], [3, 4, 5], [6, 7, 8]], pa.list_(pa.int32(), 3))
casted_array = cast_array_to_feature(arr[1:], Sequence(Value("int64"), length=3))
assert casted_array.type == pa.list_(pa.int64(), 3)
assert casted_array.to_pylist() == arr[1:].to_pylist()
def test_embed_array_storage(image_file):
array = pa.array([{"bytes": None, "path": image_file}], type=Image.pa_type)
embedded_images_array = embed_array_storage(array, Image())
assert isinstance(embedded_images_array.to_pylist()[0]["path"], str)
assert embedded_images_array.to_pylist()[0]["path"] == "test_image_rgb.jpg"
assert isinstance(embedded_images_array.to_pylist()[0]["bytes"], bytes)
def test_embed_array_storage_nested(image_file):
array = pa.array([[{"bytes": None, "path": image_file}]], type=pa.list_(Image.pa_type))
embedded_images_array = embed_array_storage(array, [Image()])
assert isinstance(embedded_images_array.to_pylist()[0][0]["path"], str)
assert isinstance(embedded_images_array.to_pylist()[0][0]["bytes"], bytes)
array = pa.array([{"foo": {"bytes": None, "path": image_file}}], type=pa.struct({"foo": Image.pa_type}))
embedded_images_array = embed_array_storage(array, {"foo": Image()})
assert isinstance(embedded_images_array.to_pylist()[0]["foo"]["path"], str)
assert isinstance(embedded_images_array.to_pylist()[0]["foo"]["bytes"], bytes)
def test_embed_table_storage(image_file):
features = Features({"image": Image()})
table = table_cast(pa.table({"image": [image_file]}), features.arrow_schema)
embedded_images_table = embed_table_storage(table)
assert isinstance(embedded_images_table.to_pydict()["image"][0]["path"], str)
assert isinstance(embedded_images_table.to_pydict()["image"][0]["bytes"], bytes)
@pytest.mark.parametrize(
"table",
[
InMemoryTable(pa.table({"foo": range(10)})),
InMemoryTable(pa.concat_tables([pa.table({"foo": range(0, 5)}), pa.table({"foo": range(5, 10)})])),
InMemoryTable(pa.concat_tables([pa.table({"foo": [i]}) for i in range(10)])),
],
)
@pytest.mark.parametrize("batch_size", [1, 2, 3, 9, 10, 11, 20])
@pytest.mark.parametrize("drop_last_batch", [False, True])
def test_table_iter(table, batch_size, drop_last_batch):
num_rows = len(table) if not drop_last_batch else len(table) // batch_size * batch_size
num_batches = (num_rows // batch_size) + 1 if num_rows % batch_size else num_rows // batch_size
subtables = list(table_iter(table, batch_size=batch_size, drop_last_batch=drop_last_batch))
assert len(subtables) == num_batches
if drop_last_batch:
assert all(len(subtable) == batch_size for subtable in subtables)
else:
assert all(len(subtable) == batch_size for subtable in subtables[:-1])
assert len(subtables[-1]) <= batch_size
if num_rows > 0:
reloaded = pa.concat_tables(subtables)
assert table.slice(0, num_rows).to_pydict() == reloaded.to_pydict()
@pytest.mark.parametrize(
"pa_type, expected",
[
(pa.int8(), False),
(pa.struct({"col1": pa.int8(), "col2": pa.int64()}), False),
(pa.struct({"col1": pa.list_(pa.int8()), "col2": Array2DExtensionType((1, 3), "int64")}), True),
(pa.list_(pa.int8()), False),
(pa.list_(Array2DExtensionType((1, 3), "int64"), 4), True),
],
)
def test_is_extension_type(pa_type, expected):
assert _is_extension_type(pa_type) == expected
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_parallel.py | import pytest
from datasets.parallel import ParallelBackendConfig, parallel_backend
from datasets.utils.py_utils import map_nested
from .utils import require_dill_gt_0_3_2, require_joblibspark, require_not_windows
def add_one(i): # picklable for multiprocessing
return i + 1
@require_dill_gt_0_3_2
@require_joblibspark
@require_not_windows
def test_parallel_backend_input():
with parallel_backend("spark"):
assert ParallelBackendConfig.backend_name == "spark"
lst = [1, 2, 3]
with pytest.raises(ValueError):
with parallel_backend("unsupported backend"):
map_nested(add_one, lst, num_proc=2)
with pytest.raises(ValueError):
with parallel_backend("unsupported backend"):
map_nested(add_one, lst, num_proc=-1)
@require_dill_gt_0_3_2
@require_joblibspark
@require_not_windows
@pytest.mark.parametrize("num_proc", [2, -1])
def test_parallel_backend_map_nested(num_proc):
s1 = [1, 2]
s2 = {"a": 1, "b": 2}
s3 = {"a": [1, 2], "b": [3, 4]}
s4 = {"a": {"1": 1}, "b": 2}
s5 = {"a": 1, "b": 2, "c": 3, "d": 4}
expected_map_nested_s1 = [2, 3]
expected_map_nested_s2 = {"a": 2, "b": 3}
expected_map_nested_s3 = {"a": [2, 3], "b": [4, 5]}
expected_map_nested_s4 = {"a": {"1": 2}, "b": 3}
expected_map_nested_s5 = {"a": 2, "b": 3, "c": 4, "d": 5}
with parallel_backend("spark"):
assert map_nested(add_one, s1, num_proc=num_proc) == expected_map_nested_s1
assert map_nested(add_one, s2, num_proc=num_proc) == expected_map_nested_s2
assert map_nested(add_one, s3, num_proc=num_proc) == expected_map_nested_s3
assert map_nested(add_one, s4, num_proc=num_proc) == expected_map_nested_s4
assert map_nested(add_one, s5, num_proc=num_proc) == expected_map_nested_s5
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_dataset_list.py | from unittest import TestCase
from datasets import Sequence, Value
from datasets.arrow_dataset import Dataset
class DatasetListTest(TestCase):
def _create_example_records(self):
return [
{"col_1": 3, "col_2": "a"},
{"col_1": 2, "col_2": "b"},
{"col_1": 1, "col_2": "c"},
{"col_1": 0, "col_2": "d"},
]
def _create_example_dict(self):
data = {"col_1": [3, 2, 1, 0], "col_2": ["a", "b", "c", "d"]}
return Dataset.from_dict(data)
def test_create(self):
example_records = self._create_example_records()
dset = Dataset.from_list(example_records)
self.assertListEqual(dset.column_names, ["col_1", "col_2"])
for i, r in enumerate(dset):
self.assertDictEqual(r, example_records[i])
def test_list_dict_equivalent(self):
example_records = self._create_example_records()
dset = Dataset.from_list(example_records)
dset_from_dict = Dataset.from_dict({k: [r[k] for r in example_records] for k in example_records[0]})
self.assertEqual(dset.info, dset_from_dict.info)
def test_uneven_records(self): # checks what happens with missing columns
uneven_records = [{"col_1": 1}, {"col_2": "x"}]
dset = Dataset.from_list(uneven_records)
self.assertDictEqual(dset[0], {"col_1": 1})
self.assertDictEqual(dset[1], {"col_1": None}) # NB: first record is used for columns
def test_variable_list_records(self): # checks if the type can be inferred from the second record
list_records = [{"col_1": []}, {"col_1": [1, 2]}]
dset = Dataset.from_list(list_records)
self.assertEqual(dset.info.features["col_1"], Sequence(Value("int64")))
def test_create_empty(self):
dset = Dataset.from_list([])
self.assertEqual(len(dset), 0)
self.assertListEqual(dset.column_names, [])
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_fingerprint.py | import json
import os
import pickle
import subprocess
from hashlib import md5
from pathlib import Path
from tempfile import gettempdir
from textwrap import dedent
from types import FunctionType
from unittest import TestCase
from unittest.mock import patch
import pytest
from multiprocess import Pool
import datasets
from datasets.fingerprint import Hasher, fingerprint_transform
from datasets.table import InMemoryTable
from .utils import (
require_regex,
require_spacy,
require_spacy_model,
require_tiktoken,
require_torch,
require_transformers,
)
class Foo:
def __init__(self, foo):
self.foo = foo
def __call__(self):
return self.foo
class DatasetChild(datasets.Dataset):
@fingerprint_transform(inplace=False)
def func1(self, new_fingerprint, *args, **kwargs):
return DatasetChild(self.data, fingerprint=new_fingerprint)
@fingerprint_transform(inplace=False)
def func2(self, new_fingerprint, *args, **kwargs):
return DatasetChild(self.data, fingerprint=new_fingerprint)
class UnpicklableCallable:
def __init__(self, callable):
self.callable = callable
def __call__(self, *args, **kwargs):
if self.callable is not None:
return self.callable(*args, **kwargs)
def __getstate__(self):
raise pickle.PicklingError()
class TokenizersDumpTest(TestCase):
@require_transformers
@pytest.mark.integration
def test_hash_tokenizer(self):
from transformers import AutoTokenizer
def encode(x):
return tokenizer(x)
# TODO: add hash consistency tests across sessions
tokenizer = AutoTokenizer.from_pretrained("bert-base-uncased")
hash1 = md5(datasets.utils.py_utils.dumps(tokenizer)).hexdigest()
hash1_lambda = md5(datasets.utils.py_utils.dumps(lambda x: tokenizer(x))).hexdigest()
hash1_encode = md5(datasets.utils.py_utils.dumps(encode)).hexdigest()
tokenizer = AutoTokenizer.from_pretrained("bert-base-cased")
hash2 = md5(datasets.utils.py_utils.dumps(tokenizer)).hexdigest()
hash2_lambda = md5(datasets.utils.py_utils.dumps(lambda x: tokenizer(x))).hexdigest()
hash2_encode = md5(datasets.utils.py_utils.dumps(encode)).hexdigest()
tokenizer = AutoTokenizer.from_pretrained("bert-base-uncased")
hash3 = md5(datasets.utils.py_utils.dumps(tokenizer)).hexdigest()
hash3_lambda = md5(datasets.utils.py_utils.dumps(lambda x: tokenizer(x))).hexdigest()
hash3_encode = md5(datasets.utils.py_utils.dumps(encode)).hexdigest()
self.assertEqual(hash1, hash3)
self.assertNotEqual(hash1, hash2)
self.assertEqual(hash1_lambda, hash3_lambda)
self.assertNotEqual(hash1_lambda, hash2_lambda)
self.assertEqual(hash1_encode, hash3_encode)
self.assertNotEqual(hash1_encode, hash2_encode)
@require_transformers
@pytest.mark.integration
def test_hash_tokenizer_with_cache(self):
from transformers import AutoTokenizer
tokenizer = AutoTokenizer.from_pretrained("gpt2")
hash1 = md5(datasets.utils.py_utils.dumps(tokenizer)).hexdigest()
tokenizer("Hello world !") # call once to change the tokenizer's cache
hash2 = md5(datasets.utils.py_utils.dumps(tokenizer)).hexdigest()
self.assertEqual(hash1, hash2)
@require_regex
def test_hash_regex(self):
import regex
pat = regex.Regex("foo")
hash1 = md5(datasets.utils.py_utils.dumps(pat)).hexdigest()
pat = regex.Regex("bar")
hash2 = md5(datasets.utils.py_utils.dumps(pat)).hexdigest()
pat = regex.Regex("foo")
hash3 = md5(datasets.utils.py_utils.dumps(pat)).hexdigest()
self.assertEqual(hash1, hash3)
self.assertNotEqual(hash1, hash2)
class RecurseDumpTest(TestCase):
def test_recurse_dump_for_function(self):
def func():
return foo
foo = [0]
hash1 = md5(datasets.utils.py_utils.dumps(func)).hexdigest()
foo = [1]
hash2 = md5(datasets.utils.py_utils.dumps(func)).hexdigest()
foo = [0]
hash3 = md5(datasets.utils.py_utils.dumps(func)).hexdigest()
self.assertEqual(hash1, hash3)
self.assertNotEqual(hash1, hash2)
def test_dump_ignores_line_definition_of_function(self):
def func():
pass
hash1 = md5(datasets.utils.py_utils.dumps(func)).hexdigest()
def func():
pass
hash2 = md5(datasets.utils.py_utils.dumps(func)).hexdigest()
self.assertEqual(hash1, hash2)
def test_recurse_dump_for_class(self):
hash1 = md5(datasets.utils.py_utils.dumps(Foo([0]))).hexdigest()
hash2 = md5(datasets.utils.py_utils.dumps(Foo([1]))).hexdigest()
hash3 = md5(datasets.utils.py_utils.dumps(Foo([0]))).hexdigest()
self.assertEqual(hash1, hash3)
self.assertNotEqual(hash1, hash2)
def test_recurse_dump_for_method(self):
hash1 = md5(datasets.utils.py_utils.dumps(Foo([0]).__call__)).hexdigest()
hash2 = md5(datasets.utils.py_utils.dumps(Foo([1]).__call__)).hexdigest()
hash3 = md5(datasets.utils.py_utils.dumps(Foo([0]).__call__)).hexdigest()
self.assertEqual(hash1, hash3)
self.assertNotEqual(hash1, hash2)
def test_dump_ipython_function(self):
def create_ipython_func(co_filename, returned_obj):
def func():
return returned_obj
code = func.__code__
# Use _create_code from dill in order to make it work for different python versions
code = code.replace(co_filename=co_filename)
return FunctionType(code, func.__globals__, func.__name__, func.__defaults__, func.__closure__)
co_filename, returned_obj = "<ipython-input-2-e0383a102aae>", [0]
hash1 = md5(datasets.utils.py_utils.dumps(create_ipython_func(co_filename, returned_obj))).hexdigest()
co_filename, returned_obj = "<ipython-input-2-e0383a102aae>", [1]
hash2 = md5(datasets.utils.py_utils.dumps(create_ipython_func(co_filename, returned_obj))).hexdigest()
co_filename, returned_obj = "<ipython-input-5-713f6613acf3>", [0]
hash3 = md5(datasets.utils.py_utils.dumps(create_ipython_func(co_filename, returned_obj))).hexdigest()
self.assertEqual(hash1, hash3)
self.assertNotEqual(hash1, hash2)
co_filename, returned_obj = os.path.join(gettempdir(), "ipykernel_12345", "321456789.py"), [0]
hash4 = md5(datasets.utils.py_utils.dumps(create_ipython_func(co_filename, returned_obj))).hexdigest()
co_filename, returned_obj = os.path.join(gettempdir(), "ipykernel_12345", "321456789.py"), [1]
hash5 = md5(datasets.utils.py_utils.dumps(create_ipython_func(co_filename, returned_obj))).hexdigest()
co_filename, returned_obj = os.path.join(gettempdir(), "ipykernel_12345", "654123987.py"), [0]
hash6 = md5(datasets.utils.py_utils.dumps(create_ipython_func(co_filename, returned_obj))).hexdigest()
self.assertEqual(hash4, hash6)
self.assertNotEqual(hash4, hash5)
def test_recurse_dump_for_function_with_shuffled_globals(self):
foo, bar = [0], [1]
def func():
return foo, bar
func.__module__ = "__main__"
def globalvars_mock1_side_effect(func, *args, **kwargs):
return {"foo": foo, "bar": bar}
def globalvars_mock2_side_effect(func, *args, **kwargs):
return {"bar": bar, "foo": foo}
with patch("dill.detect.globalvars", side_effect=globalvars_mock1_side_effect) as globalvars_mock1:
hash1 = md5(datasets.utils.py_utils.dumps(func)).hexdigest()
self.assertGreater(globalvars_mock1.call_count, 0)
with patch("dill.detect.globalvars", side_effect=globalvars_mock2_side_effect) as globalvars_mock2:
hash2 = md5(datasets.utils.py_utils.dumps(func)).hexdigest()
self.assertGreater(globalvars_mock2.call_count, 0)
self.assertEqual(hash1, hash2)
class HashingTest(TestCase):
def test_hash_simple(self):
hash1 = Hasher.hash("hello")
hash2 = Hasher.hash("hello")
hash3 = Hasher.hash("there")
self.assertEqual(hash1, hash2)
self.assertNotEqual(hash1, hash3)
def test_hash_class_instance(self):
hash1 = Hasher.hash(Foo("hello"))
hash2 = Hasher.hash(Foo("hello"))
hash3 = Hasher.hash(Foo("there"))
self.assertEqual(hash1, hash2)
self.assertNotEqual(hash1, hash3)
def test_hash_update(self):
hasher = Hasher()
for x in ["hello", Foo("hello")]:
hasher.update(x)
hash1 = hasher.hexdigest()
hasher = Hasher()
for x in ["hello", Foo("hello")]:
hasher.update(x)
hash2 = hasher.hexdigest()
hasher = Hasher()
for x in ["there", Foo("there")]:
hasher.update(x)
hash3 = hasher.hexdigest()
self.assertEqual(hash1, hash2)
self.assertNotEqual(hash1, hash3)
def test_hash_unpicklable(self):
with self.assertRaises(pickle.PicklingError):
Hasher.hash(UnpicklableCallable(Foo("hello")))
def test_hash_same_strings(self):
string = "abc"
obj1 = [string, string] # two strings have the same ids
obj2 = [string, string]
obj3 = json.loads(f'["{string}", "{string}"]') # two strings have different ids
self.assertIs(obj1[0], string)
self.assertIs(obj1[0], obj1[1])
self.assertIs(obj2[0], string)
self.assertIs(obj2[0], obj2[1])
self.assertIsNot(obj3[0], string)
self.assertIsNot(obj3[0], obj3[1])
hash1 = Hasher.hash(obj1)
hash2 = Hasher.hash(obj2)
hash3 = Hasher.hash(obj3)
self.assertEqual(hash1, hash2)
self.assertEqual(hash1, hash3)
@require_tiktoken
def test_hash_tiktoken_encoding(self):
import tiktoken
enc = tiktoken.get_encoding("gpt2")
hash1 = md5(datasets.utils.py_utils.dumps(enc)).hexdigest()
enc = tiktoken.get_encoding("r50k_base")
hash2 = md5(datasets.utils.py_utils.dumps(enc)).hexdigest()
enc = tiktoken.get_encoding("gpt2")
hash3 = md5(datasets.utils.py_utils.dumps(enc)).hexdigest()
self.assertEqual(hash1, hash3)
self.assertNotEqual(hash1, hash2)
@require_torch
def test_hash_torch_tensor(self):
import torch
t = torch.tensor([1.0])
hash1 = md5(datasets.utils.py_utils.dumps(t)).hexdigest()
t = torch.tensor([2.0])
hash2 = md5(datasets.utils.py_utils.dumps(t)).hexdigest()
t = torch.tensor([1.0])
hash3 = md5(datasets.utils.py_utils.dumps(t)).hexdigest()
self.assertEqual(hash1, hash3)
self.assertNotEqual(hash1, hash2)
@require_spacy
@require_spacy_model("en_core_web_sm")
@require_spacy_model("fr_core_news_sm")
@pytest.mark.integration
def test_hash_spacy_model(self):
import spacy
nlp = spacy.load("en_core_web_sm")
hash1 = md5(datasets.utils.py_utils.dumps(nlp)).hexdigest()
nlp = spacy.load("fr_core_news_sm")
hash2 = md5(datasets.utils.py_utils.dumps(nlp)).hexdigest()
nlp = spacy.load("en_core_web_sm")
hash3 = md5(datasets.utils.py_utils.dumps(nlp)).hexdigest()
self.assertEqual(hash1, hash3)
self.assertNotEqual(hash1, hash2)
@pytest.mark.integration
def test_move_script_doesnt_change_hash(tmp_path: Path):
dir1 = tmp_path / "dir1"
dir2 = tmp_path / "dir2"
dir1.mkdir()
dir2.mkdir()
script_filename = "script.py"
code = dedent(
"""
from datasets.fingerprint import Hasher
def foo():
pass
print(Hasher.hash(foo))
"""
)
script_path1 = dir1 / script_filename
script_path2 = dir2 / script_filename
with script_path1.open("w") as f:
f.write(code)
with script_path2.open("w") as f:
f.write(code)
fingerprint1 = subprocess.check_output(["python", str(script_path1)])
fingerprint2 = subprocess.check_output(["python", str(script_path2)])
assert fingerprint1 == fingerprint2
def test_fingerprint_in_multiprocessing():
data = {"a": [0, 1, 2]}
dataset = DatasetChild(InMemoryTable.from_pydict(data))
expected_fingerprint = dataset.func1()._fingerprint
assert expected_fingerprint == dataset.func1()._fingerprint
assert expected_fingerprint != dataset.func2()._fingerprint
with Pool(2) as p:
assert expected_fingerprint == p.apply_async(dataset.func1).get()._fingerprint
assert expected_fingerprint != p.apply_async(dataset.func2).get()._fingerprint
def test_fingerprint_when_transform_version_changes():
data = {"a": [0, 1, 2]}
class DummyDatasetChild(datasets.Dataset):
@fingerprint_transform(inplace=False)
def func(self, new_fingerprint):
return DummyDatasetChild(self.data, fingerprint=new_fingerprint)
fingeprint_no_version = DummyDatasetChild(InMemoryTable.from_pydict(data)).func()
class DummyDatasetChild(datasets.Dataset):
@fingerprint_transform(inplace=False, version="1.0.0")
def func(self, new_fingerprint):
return DummyDatasetChild(self.data, fingerprint=new_fingerprint)
fingeprint_1 = DummyDatasetChild(InMemoryTable.from_pydict(data)).func()
class DummyDatasetChild(datasets.Dataset):
@fingerprint_transform(inplace=False, version="2.0.0")
def func(self, new_fingerprint):
return DummyDatasetChild(self.data, fingerprint=new_fingerprint)
fingeprint_2 = DummyDatasetChild(InMemoryTable.from_pydict(data)).func()
assert len({fingeprint_no_version, fingeprint_1, fingeprint_2}) == 3
def test_dependency_on_dill():
# AttributeError: module 'dill._dill' has no attribute 'stack'
hasher = Hasher()
hasher.update(lambda x: x)
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_extract.py | import os
import zipfile
import pytest
from datasets.utils.extract import (
Bzip2Extractor,
Extractor,
GzipExtractor,
Lz4Extractor,
SevenZipExtractor,
TarExtractor,
XzExtractor,
ZipExtractor,
ZstdExtractor,
)
from .utils import require_lz4, require_py7zr, require_zstandard
@pytest.mark.parametrize(
"compression_format, is_archive",
[
("7z", True),
("bz2", False),
("gzip", False),
("lz4", False),
("tar", True),
("xz", False),
("zip", True),
("zstd", False),
],
)
def test_base_extractors(
compression_format,
is_archive,
bz2_file,
gz_file,
lz4_file,
seven_zip_file,
tar_file,
xz_file,
zip_file,
zstd_file,
tmp_path,
text_file,
):
input_paths_and_base_extractors = {
"7z": (seven_zip_file, SevenZipExtractor),
"bz2": (bz2_file, Bzip2Extractor),
"gzip": (gz_file, GzipExtractor),
"lz4": (lz4_file, Lz4Extractor),
"tar": (tar_file, TarExtractor),
"xz": (xz_file, XzExtractor),
"zip": (zip_file, ZipExtractor),
"zstd": (zstd_file, ZstdExtractor),
}
input_path, base_extractor = input_paths_and_base_extractors[compression_format]
if input_path is None:
reason = f"for '{compression_format}' compression_format, "
if compression_format == "7z":
reason += require_py7zr.kwargs["reason"]
elif compression_format == "lz4":
reason += require_lz4.kwargs["reason"]
elif compression_format == "zstd":
reason += require_zstandard.kwargs["reason"]
pytest.skip(reason)
assert base_extractor.is_extractable(input_path)
output_path = tmp_path / ("extracted" if is_archive else "extracted.txt")
base_extractor.extract(input_path, output_path)
if is_archive:
assert output_path.is_dir()
for file_path in output_path.iterdir():
assert file_path.name == text_file.name
extracted_file_content = file_path.read_text(encoding="utf-8")
else:
extracted_file_content = output_path.read_text(encoding="utf-8")
expected_file_content = text_file.read_text(encoding="utf-8")
assert extracted_file_content == expected_file_content
@pytest.mark.parametrize(
"compression_format, is_archive",
[
("7z", True),
("bz2", False),
("gzip", False),
("lz4", False),
("tar", True),
("xz", False),
("zip", True),
("zstd", False),
],
)
def test_extractor(
compression_format,
is_archive,
bz2_file,
gz_file,
lz4_file,
seven_zip_file,
tar_file,
xz_file,
zip_file,
zstd_file,
tmp_path,
text_file,
):
input_paths = {
"7z": seven_zip_file,
"bz2": bz2_file,
"gzip": gz_file,
"lz4": lz4_file,
"tar": tar_file,
"xz": xz_file,
"zip": zip_file,
"zstd": zstd_file,
}
input_path = input_paths[compression_format]
if input_path is None:
reason = f"for '{compression_format}' compression_format, "
if compression_format == "7z":
reason += require_py7zr.kwargs["reason"]
elif compression_format == "lz4":
reason += require_lz4.kwargs["reason"]
elif compression_format == "zstd":
reason += require_zstandard.kwargs["reason"]
pytest.skip(reason)
extractor_format = Extractor.infer_extractor_format(input_path)
assert extractor_format is not None
output_path = tmp_path / ("extracted" if is_archive else "extracted.txt")
Extractor.extract(input_path, output_path, extractor_format)
if is_archive:
assert output_path.is_dir()
for file_path in output_path.iterdir():
assert file_path.name == text_file.name
extracted_file_content = file_path.read_text(encoding="utf-8")
else:
extracted_file_content = output_path.read_text(encoding="utf-8")
expected_file_content = text_file.read_text(encoding="utf-8")
assert extracted_file_content == expected_file_content
@pytest.fixture
def tar_file_with_dot_dot(tmp_path, text_file):
import tarfile
directory = tmp_path / "data_dot_dot"
directory.mkdir()
path = directory / "tar_file_with_dot_dot.tar"
with tarfile.TarFile(path, "w") as f:
f.add(text_file, arcname=os.path.join("..", text_file.name))
return path
@pytest.fixture
def tar_file_with_sym_link(tmp_path):
import tarfile
directory = tmp_path / "data_sym_link"
directory.mkdir()
path = directory / "tar_file_with_sym_link.tar"
os.symlink("..", directory / "subdir", target_is_directory=True)
with tarfile.TarFile(path, "w") as f:
f.add(str(directory / "subdir"), arcname="subdir") # str required by os.readlink on Windows and Python < 3.8
return path
@pytest.mark.parametrize(
"insecure_tar_file, error_log",
[("tar_file_with_dot_dot", "illegal path"), ("tar_file_with_sym_link", "Symlink")],
)
def test_tar_extract_insecure_files(
insecure_tar_file, error_log, tar_file_with_dot_dot, tar_file_with_sym_link, tmp_path, caplog
):
insecure_tar_files = {
"tar_file_with_dot_dot": tar_file_with_dot_dot,
"tar_file_with_sym_link": tar_file_with_sym_link,
}
input_path = insecure_tar_files[insecure_tar_file]
output_path = tmp_path / "extracted"
TarExtractor.extract(input_path, output_path)
assert caplog.text
for record in caplog.records:
assert record.levelname == "ERROR"
assert error_log in record.msg
def test_is_zipfile_false_positive(tmpdir):
# We should have less false positives than zipfile.is_zipfile
# We do that by checking only the magic number
not_a_zip_file = tmpdir / "not_a_zip_file"
# From: https://github.com/python/cpython/pull/5053
data = (
b"\x89PNG\r\n\x1a\n\x00\x00\x00\rIHDR\x00\x00\x00\x01\x00\x00"
b"\x00\x02\x08\x06\x00\x00\x00\x99\x81\xb6'\x00\x00\x00\x15I"
b"DATx\x01\x01\n\x00\xf5\xff\x00PK\x05\x06\x00PK\x06\x06\x07"
b"\xac\x01N\xc6|a\r\x00\x00\x00\x00IEND\xaeB`\x82"
)
with not_a_zip_file.open("wb") as f:
f.write(data)
assert zipfile.is_zipfile(str(not_a_zip_file)) # is a false positive for `zipfile`
assert not ZipExtractor.is_extractable(not_a_zip_file) # but we're right
| 0 |
hf_public_repos/datasets | hf_public_repos/datasets/tests/test_load.py | import importlib
import os
import pickle
import shutil
import tempfile
import time
from hashlib import sha256
from multiprocessing import Pool
from pathlib import Path
from unittest import TestCase
from unittest.mock import patch
import dill
import pyarrow as pa
import pytest
import requests
import datasets
from datasets import config, load_dataset, load_from_disk
from datasets.arrow_dataset import Dataset
from datasets.arrow_writer import ArrowWriter
from datasets.builder import DatasetBuilder
from datasets.config import METADATA_CONFIGS_FIELD
from datasets.data_files import DataFilesDict
from datasets.dataset_dict import DatasetDict, IterableDatasetDict
from datasets.download.download_config import DownloadConfig
from datasets.features import Features, Value
from datasets.iterable_dataset import IterableDataset
from datasets.load import (
CachedDatasetModuleFactory,
CachedMetricModuleFactory,
GithubMetricModuleFactory,
HubDatasetModuleFactoryWithoutScript,
HubDatasetModuleFactoryWithScript,
LocalDatasetModuleFactoryWithoutScript,
LocalDatasetModuleFactoryWithScript,
LocalMetricModuleFactory,
PackagedDatasetModuleFactory,
infer_module_for_data_files_list,
infer_module_for_data_files_list_in_archives,
)
from datasets.packaged_modules.audiofolder.audiofolder import AudioFolder, AudioFolderConfig
from datasets.packaged_modules.imagefolder.imagefolder import ImageFolder, ImageFolderConfig
from datasets.utils.logging import INFO, get_logger
from .utils import (
OfflineSimulationMode,
assert_arrow_memory_doesnt_increase,
assert_arrow_memory_increases,
offline,
require_pil,
require_sndfile,
set_current_working_directory_to_temp_dir,
)
DATASET_LOADING_SCRIPT_NAME = "__dummy_dataset1__"
DATASET_LOADING_SCRIPT_CODE = """
import os
import datasets
from datasets import DatasetInfo, Features, Split, SplitGenerator, Value
class __DummyDataset1__(datasets.GeneratorBasedBuilder):
def _info(self) -> DatasetInfo:
return DatasetInfo(features=Features({"text": Value("string")}))
def _split_generators(self, dl_manager):
return [
SplitGenerator(Split.TRAIN, gen_kwargs={"filepath": os.path.join(dl_manager.manual_dir, "train.txt")}),
SplitGenerator(Split.TEST, gen_kwargs={"filepath": os.path.join(dl_manager.manual_dir, "test.txt")}),
]
def _generate_examples(self, filepath, **kwargs):
with open(filepath, "r", encoding="utf-8") as f:
for i, line in enumerate(f):
yield i, {"text": line.strip()}
"""
SAMPLE_DATASET_IDENTIFIER = "lhoestq/test" # has dataset script
SAMPLE_DATASET_IDENTIFIER2 = "lhoestq/test2" # only has data files
SAMPLE_DATASET_IDENTIFIER3 = "mariosasko/test_multi_dir_dataset" # has multiple data directories
SAMPLE_DATASET_IDENTIFIER4 = "mariosasko/test_imagefolder_with_metadata" # imagefolder with a metadata file outside of the train/test directories
SAMPLE_NOT_EXISTING_DATASET_IDENTIFIER = "lhoestq/_dummy"
SAMPLE_DATASET_NAME_THAT_DOESNT_EXIST = "_dummy"
SAMPLE_DATASET_NO_CONFIGS_IN_METADATA = "datasets-maintainers/audiofolder_no_configs_in_metadata"
SAMPLE_DATASET_SINGLE_CONFIG_IN_METADATA = "datasets-maintainers/audiofolder_single_config_in_metadata"
SAMPLE_DATASET_TWO_CONFIG_IN_METADATA = "datasets-maintainers/audiofolder_two_configs_in_metadata"
SAMPLE_DATASET_TWO_CONFIG_IN_METADATA_WITH_DEFAULT = (
"datasets-maintainers/audiofolder_two_configs_in_metadata_with_default"
)
METRIC_LOADING_SCRIPT_NAME = "__dummy_metric1__"
METRIC_LOADING_SCRIPT_CODE = """
import datasets
from datasets import MetricInfo, Features, Value
class __DummyMetric1__(datasets.Metric):
def _info(self):
return MetricInfo(features=Features({"predictions": Value("int"), "references": Value("int")}))
def _compute(self, predictions, references):
return {"__dummy_metric1__": sum(int(p == r) for p, r in zip(predictions, references))}
"""
@pytest.fixture
def data_dir(tmp_path):
data_dir = tmp_path / "data_dir"
data_dir.mkdir()
with open(data_dir / "train.txt", "w") as f:
f.write("foo\n" * 10)
with open(data_dir / "test.txt", "w") as f:
f.write("bar\n" * 10)
return str(data_dir)
@pytest.fixture
def data_dir_with_arrow(tmp_path):
data_dir = tmp_path / "data_dir"
data_dir.mkdir()
output_train = os.path.join(data_dir, "train.arrow")
with ArrowWriter(path=output_train) as writer:
writer.write_table(pa.Table.from_pydict({"col_1": ["foo"] * 10}))
num_examples, num_bytes = writer.finalize()
assert num_examples == 10
assert num_bytes > 0
output_test = os.path.join(data_dir, "test.arrow")
with ArrowWriter(path=output_test) as writer:
writer.write_table(pa.Table.from_pydict({"col_1": ["bar"] * 10}))
num_examples, num_bytes = writer.finalize()
assert num_examples == 10
assert num_bytes > 0
return str(data_dir)
@pytest.fixture
def data_dir_with_metadata(tmp_path):
data_dir = tmp_path / "data_dir_with_metadata"
data_dir.mkdir()
with open(data_dir / "train.jpg", "wb") as f:
f.write(b"train_image_bytes")
with open(data_dir / "test.jpg", "wb") as f:
f.write(b"test_image_bytes")
with open(data_dir / "metadata.jsonl", "w") as f:
f.write(
"""\
{"file_name": "train.jpg", "caption": "Cool tran image"}
{"file_name": "test.jpg", "caption": "Cool test image"}
"""
)
return str(data_dir)
@pytest.fixture
def data_dir_with_single_config_in_metadata(tmp_path):
data_dir = tmp_path / "data_dir_with_one_default_config_in_metadata"
cats_data_dir = data_dir / "cats"
cats_data_dir.mkdir(parents=True)
dogs_data_dir = data_dir / "dogs"
dogs_data_dir.mkdir(parents=True)
with open(cats_data_dir / "cat.jpg", "wb") as f:
f.write(b"this_is_a_cat_image_bytes")
with open(dogs_data_dir / "dog.jpg", "wb") as f:
f.write(b"this_is_a_dog_image_bytes")
with open(data_dir / "README.md", "w") as f:
f.write(
f"""\
---
{METADATA_CONFIGS_FIELD}:
- config_name: custom
drop_labels: true
---
"""
)
return str(data_dir)
@pytest.fixture
def data_dir_with_two_config_in_metadata(tmp_path):
data_dir = tmp_path / "data_dir_with_two_configs_in_metadata"
cats_data_dir = data_dir / "cats"
cats_data_dir.mkdir(parents=True)
dogs_data_dir = data_dir / "dogs"
dogs_data_dir.mkdir(parents=True)
with open(cats_data_dir / "cat.jpg", "wb") as f:
f.write(b"this_is_a_cat_image_bytes")
with open(dogs_data_dir / "dog.jpg", "wb") as f:
f.write(b"this_is_a_dog_image_bytes")
with open(data_dir / "README.md", "w") as f:
f.write(
f"""\
---
{METADATA_CONFIGS_FIELD}:
- config_name: "v1"
drop_labels: true
default: true
- config_name: "v2"
drop_labels: false
---
"""
)
return str(data_dir)
@pytest.fixture
def data_dir_with_data_dir_configs_in_metadata(tmp_path):
data_dir = tmp_path / "data_dir_with_two_configs_in_metadata"
cats_data_dir = data_dir / "cats"
cats_data_dir.mkdir(parents=True)
dogs_data_dir = data_dir / "dogs"
dogs_data_dir.mkdir(parents=True)
with open(cats_data_dir / "cat.jpg", "wb") as f:
f.write(b"this_is_a_cat_image_bytes")
with open(dogs_data_dir / "dog.jpg", "wb") as f:
f.write(b"this_is_a_dog_image_bytes")
@pytest.fixture
def sub_data_dirs(tmp_path):
data_dir2 = tmp_path / "data_dir2"
relative_subdir1 = "subdir1"
sub_data_dir1 = data_dir2 / relative_subdir1
sub_data_dir1.mkdir(parents=True)
with open(sub_data_dir1 / "train.txt", "w") as f:
f.write("foo\n" * 10)
with open(sub_data_dir1 / "test.txt", "w") as f:
f.write("bar\n" * 10)
relative_subdir2 = "subdir2"
sub_data_dir2 = tmp_path / data_dir2 / relative_subdir2
sub_data_dir2.mkdir(parents=True)
with open(sub_data_dir2 / "train.txt", "w") as f:
f.write("foo\n" * 10)
with open(sub_data_dir2 / "test.txt", "w") as f:
f.write("bar\n" * 10)
return str(data_dir2), relative_subdir1
@pytest.fixture
def complex_data_dir(tmp_path):
data_dir = tmp_path / "complex_data_dir"
data_dir.mkdir()
(data_dir / "data").mkdir()
with open(data_dir / "data" / "train.txt", "w") as f:
f.write("foo\n" * 10)
with open(data_dir / "data" / "test.txt", "w") as f:
f.write("bar\n" * 10)
with open(data_dir / "README.md", "w") as f:
f.write("This is a readme")
with open(data_dir / ".dummy", "w") as f:
f.write("this is a dummy file that is not a data file")
return str(data_dir)
@pytest.fixture
def dataset_loading_script_dir(tmp_path):
script_name = DATASET_LOADING_SCRIPT_NAME
script_dir = tmp_path / script_name
script_dir.mkdir()
script_path = script_dir / f"{script_name}.py"
with open(script_path, "w") as f:
f.write(DATASET_LOADING_SCRIPT_CODE)
return str(script_dir)
@pytest.fixture
def dataset_loading_script_dir_readonly(tmp_path):
script_name = DATASET_LOADING_SCRIPT_NAME
script_dir = tmp_path / "readonly" / script_name
script_dir.mkdir(parents=True)
script_path = script_dir / f"{script_name}.py"
with open(script_path, "w") as f:
f.write(DATASET_LOADING_SCRIPT_CODE)
dataset_loading_script_dir = str(script_dir)
# Make this directory readonly
os.chmod(dataset_loading_script_dir, 0o555)
os.chmod(os.path.join(dataset_loading_script_dir, f"{script_name}.py"), 0o555)
return dataset_loading_script_dir
@pytest.fixture
def metric_loading_script_dir(tmp_path):
script_name = METRIC_LOADING_SCRIPT_NAME
script_dir = tmp_path / script_name
script_dir.mkdir()
script_path = script_dir / f"{script_name}.py"
with open(script_path, "w") as f:
f.write(METRIC_LOADING_SCRIPT_CODE)
return str(script_dir)
@pytest.mark.parametrize(
"data_files, expected_module, expected_builder_kwargs",
[
(["train.csv"], "csv", {}),
(["train.tsv"], "csv", {"sep": "\t"}),
(["train.json"], "json", {}),
(["train.jsonl"], "json", {}),
(["train.parquet"], "parquet", {}),
(["train.arrow"], "arrow", {}),
(["train.txt"], "text", {}),
(["uppercase.TXT"], "text", {}),
(["unsupported.ext"], None, {}),
([""], None, {}),
],
)
def test_infer_module_for_data_files(data_files, expected_module, expected_builder_kwargs):
module, builder_kwargs = infer_module_for_data_files_list(data_files)
assert module == expected_module
assert builder_kwargs == expected_builder_kwargs
@pytest.mark.parametrize(
"data_file, expected_module",
[
("zip_csv_path", "csv"),
("zip_csv_with_dir_path", "csv"),
("zip_uppercase_csv_path", "csv"),
("zip_unsupported_ext_path", None),
],
)
def test_infer_module_for_data_files_in_archives(
data_file, expected_module, zip_csv_path, zip_csv_with_dir_path, zip_uppercase_csv_path, zip_unsupported_ext_path
):
data_file_paths = {
"zip_csv_path": zip_csv_path,
"zip_csv_with_dir_path": zip_csv_with_dir_path,
"zip_uppercase_csv_path": zip_uppercase_csv_path,
"zip_unsupported_ext_path": zip_unsupported_ext_path,
}
data_files = [str(data_file_paths[data_file])]
inferred_module, _ = infer_module_for_data_files_list_in_archives(data_files)
assert inferred_module == expected_module
class ModuleFactoryTest(TestCase):
@pytest.fixture(autouse=True)
def inject_fixtures(
self,
jsonl_path,
data_dir,
data_dir_with_metadata,
data_dir_with_single_config_in_metadata,
data_dir_with_two_config_in_metadata,
sub_data_dirs,
dataset_loading_script_dir,
metric_loading_script_dir,
):
self._jsonl_path = jsonl_path
self._data_dir = data_dir
self._data_dir_with_metadata = data_dir_with_metadata
self._data_dir_with_single_config_in_metadata = data_dir_with_single_config_in_metadata
self._data_dir_with_two_config_in_metadata = data_dir_with_two_config_in_metadata
self._data_dir2 = sub_data_dirs[0]
self._sub_data_dir = sub_data_dirs[1]
self._dataset_loading_script_dir = dataset_loading_script_dir
self._metric_loading_script_dir = metric_loading_script_dir
def setUp(self):
self.hf_modules_cache = tempfile.mkdtemp()
self.cache_dir = tempfile.mkdtemp()
self.download_config = DownloadConfig(cache_dir=self.cache_dir)
self.dynamic_modules_path = datasets.load.init_dynamic_modules(
name="test_datasets_modules_" + os.path.basename(self.hf_modules_cache),
hf_modules_cache=self.hf_modules_cache,
)
def test_HubDatasetModuleFactoryWithScript_with_github_dataset(self):
# "wmt_t2t" has additional imports (internal)
factory = HubDatasetModuleFactoryWithScript(
"wmt_t2t", download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert module_factory_result.builder_kwargs["base_path"].startswith(config.HF_ENDPOINT)
def test_GithubMetricModuleFactory_with_internal_import(self):
# "squad_v2" requires additional imports (internal)
factory = GithubMetricModuleFactory(
"squad_v2", download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
@pytest.mark.filterwarnings("ignore:GithubMetricModuleFactory is deprecated:FutureWarning")
def test_GithubMetricModuleFactory_with_external_import(self):
# "bleu" requires additional imports (external from github)
factory = GithubMetricModuleFactory(
"bleu", download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
def test_LocalMetricModuleFactory(self):
path = os.path.join(self._metric_loading_script_dir, f"{METRIC_LOADING_SCRIPT_NAME}.py")
factory = LocalMetricModuleFactory(
path, download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
def test_LocalDatasetModuleFactoryWithScript(self):
path = os.path.join(self._dataset_loading_script_dir, f"{DATASET_LOADING_SCRIPT_NAME}.py")
factory = LocalDatasetModuleFactoryWithScript(
path, download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert os.path.isdir(module_factory_result.builder_kwargs["base_path"])
def test_LocalDatasetModuleFactoryWithoutScript(self):
factory = LocalDatasetModuleFactoryWithoutScript(self._data_dir)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert os.path.isdir(module_factory_result.builder_kwargs["base_path"])
def test_LocalDatasetModuleFactoryWithoutScript_with_data_dir(self):
factory = LocalDatasetModuleFactoryWithoutScript(self._data_dir2, data_dir=self._sub_data_dir)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert (
module_factory_result.builder_kwargs["data_files"] is not None
and len(module_factory_result.builder_kwargs["data_files"]["train"]) == 1
and len(module_factory_result.builder_kwargs["data_files"]["test"]) == 1
)
assert all(
self._sub_data_dir in Path(data_file).parts
for data_file in module_factory_result.builder_kwargs["data_files"]["train"]
+ module_factory_result.builder_kwargs["data_files"]["test"]
)
def test_LocalDatasetModuleFactoryWithoutScript_with_metadata(self):
factory = LocalDatasetModuleFactoryWithoutScript(self._data_dir_with_metadata)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert (
module_factory_result.builder_kwargs["data_files"] is not None
and len(module_factory_result.builder_kwargs["data_files"]["train"]) > 0
and len(module_factory_result.builder_kwargs["data_files"]["test"]) > 0
)
assert any(
Path(data_file).name == "metadata.jsonl"
for data_file in module_factory_result.builder_kwargs["data_files"]["train"]
)
assert any(
Path(data_file).name == "metadata.jsonl"
for data_file in module_factory_result.builder_kwargs["data_files"]["test"]
)
def test_LocalDatasetModuleFactoryWithoutScript_with_single_config_in_metadata(self):
factory = LocalDatasetModuleFactoryWithoutScript(
self._data_dir_with_single_config_in_metadata,
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
module_metadata_configs = module_factory_result.builder_configs_parameters.metadata_configs
assert module_metadata_configs is not None
assert len(module_metadata_configs) == 1
assert next(iter(module_metadata_configs)) == "custom"
assert "drop_labels" in next(iter(module_metadata_configs.values()))
assert next(iter(module_metadata_configs.values()))["drop_labels"] is True
module_builder_configs = module_factory_result.builder_configs_parameters.builder_configs
assert module_builder_configs is not None
assert len(module_builder_configs) == 1
assert isinstance(module_builder_configs[0], ImageFolderConfig)
assert module_builder_configs[0].name == "custom"
assert module_builder_configs[0].data_files is not None
assert isinstance(module_builder_configs[0].data_files, DataFilesDict)
assert len(module_builder_configs[0].data_files) == 1 # one train split
assert len(module_builder_configs[0].data_files["train"]) == 2 # two files
assert module_builder_configs[0].drop_labels is True # parameter is passed from metadata
# config named "default" is automatically considered to be a default config
assert module_factory_result.builder_configs_parameters.default_config_name is None
# we don't pass config params to builder in builder_kwargs, they are stored in builder_configs directly
assert "drop_labels" not in module_factory_result.builder_kwargs
def test_LocalDatasetModuleFactoryWithoutScript_with_two_configs_in_metadata(self):
factory = LocalDatasetModuleFactoryWithoutScript(
self._data_dir_with_two_config_in_metadata,
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
module_metadata_configs = module_factory_result.builder_configs_parameters.metadata_configs
assert module_metadata_configs is not None
assert len(module_metadata_configs) == 2
assert list(module_metadata_configs) == ["v1", "v2"]
assert "drop_labels" in module_metadata_configs["v1"]
assert module_metadata_configs["v1"]["drop_labels"] is True
assert "drop_labels" in module_metadata_configs["v2"]
assert module_metadata_configs["v2"]["drop_labels"] is False
module_builder_configs = module_factory_result.builder_configs_parameters.builder_configs
assert module_builder_configs is not None
assert len(module_builder_configs) == 2
module_builder_config_v1, module_builder_config_v2 = module_builder_configs
assert module_builder_config_v1.name == "v1"
assert module_builder_config_v2.name == "v2"
assert isinstance(module_builder_config_v1, ImageFolderConfig)
assert isinstance(module_builder_config_v2, ImageFolderConfig)
assert isinstance(module_builder_config_v1.data_files, DataFilesDict)
assert isinstance(module_builder_config_v2.data_files, DataFilesDict)
assert sorted(module_builder_config_v1.data_files) == ["train"]
assert len(module_builder_config_v1.data_files["train"]) == 2
assert sorted(module_builder_config_v2.data_files) == ["train"]
assert len(module_builder_config_v2.data_files["train"]) == 2
assert module_builder_config_v1.drop_labels is True # parameter is passed from metadata
assert module_builder_config_v2.drop_labels is False # parameter is passed from metadata
assert (
module_factory_result.builder_configs_parameters.default_config_name == "v1"
) # it's marked as a default one in yaml
# we don't pass config params to builder in builder_kwargs, they are stored in builder_configs directly
assert "drop_labels" not in module_factory_result.builder_kwargs
def test_PackagedDatasetModuleFactory(self):
factory = PackagedDatasetModuleFactory(
"json", data_files=self._jsonl_path, download_config=self.download_config
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
def test_PackagedDatasetModuleFactory_with_data_dir(self):
factory = PackagedDatasetModuleFactory("json", data_dir=self._data_dir, download_config=self.download_config)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert (
module_factory_result.builder_kwargs["data_files"] is not None
and len(module_factory_result.builder_kwargs["data_files"]["train"]) > 0
and len(module_factory_result.builder_kwargs["data_files"]["test"]) > 0
)
assert Path(module_factory_result.builder_kwargs["data_files"]["train"][0]).parent.samefile(self._data_dir)
assert Path(module_factory_result.builder_kwargs["data_files"]["test"][0]).parent.samefile(self._data_dir)
def test_PackagedDatasetModuleFactory_with_data_dir_and_metadata(self):
factory = PackagedDatasetModuleFactory(
"imagefolder", data_dir=self._data_dir_with_metadata, download_config=self.download_config
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert (
module_factory_result.builder_kwargs["data_files"] is not None
and len(module_factory_result.builder_kwargs["data_files"]["train"]) > 0
and len(module_factory_result.builder_kwargs["data_files"]["test"]) > 0
)
assert Path(module_factory_result.builder_kwargs["data_files"]["train"][0]).parent.samefile(
self._data_dir_with_metadata
)
assert Path(module_factory_result.builder_kwargs["data_files"]["test"][0]).parent.samefile(
self._data_dir_with_metadata
)
assert any(
Path(data_file).name == "metadata.jsonl"
for data_file in module_factory_result.builder_kwargs["data_files"]["train"]
)
assert any(
Path(data_file).name == "metadata.jsonl"
for data_file in module_factory_result.builder_kwargs["data_files"]["test"]
)
@pytest.mark.integration
def test_HubDatasetModuleFactoryWithoutScript(self):
factory = HubDatasetModuleFactoryWithoutScript(
SAMPLE_DATASET_IDENTIFIER2, download_config=self.download_config
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert module_factory_result.builder_kwargs["base_path"].startswith(config.HF_ENDPOINT)
@pytest.mark.integration
def test_HubDatasetModuleFactoryWithoutScript_with_data_dir(self):
data_dir = "data2"
factory = HubDatasetModuleFactoryWithoutScript(
SAMPLE_DATASET_IDENTIFIER3, data_dir=data_dir, download_config=self.download_config
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert module_factory_result.builder_kwargs["base_path"].startswith(config.HF_ENDPOINT)
assert (
module_factory_result.builder_kwargs["data_files"] is not None
and len(module_factory_result.builder_kwargs["data_files"]["train"]) == 1
and len(module_factory_result.builder_kwargs["data_files"]["test"]) == 1
)
assert all(
data_dir in Path(data_file).parts
for data_file in module_factory_result.builder_kwargs["data_files"]["train"]
+ module_factory_result.builder_kwargs["data_files"]["test"]
)
@pytest.mark.integration
def test_HubDatasetModuleFactoryWithoutScript_with_metadata(self):
factory = HubDatasetModuleFactoryWithoutScript(
SAMPLE_DATASET_IDENTIFIER4, download_config=self.download_config
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert module_factory_result.builder_kwargs["base_path"].startswith(config.HF_ENDPOINT)
assert (
module_factory_result.builder_kwargs["data_files"] is not None
and len(module_factory_result.builder_kwargs["data_files"]["train"]) > 0
and len(module_factory_result.builder_kwargs["data_files"]["test"]) > 0
)
assert any(
Path(data_file).name == "metadata.jsonl"
for data_file in module_factory_result.builder_kwargs["data_files"]["train"]
)
assert any(
Path(data_file).name == "metadata.jsonl"
for data_file in module_factory_result.builder_kwargs["data_files"]["test"]
)
@pytest.mark.integration
def test_HubDatasetModuleFactoryWithoutScript_with_one_default_config_in_metadata(self):
factory = HubDatasetModuleFactoryWithoutScript(
SAMPLE_DATASET_SINGLE_CONFIG_IN_METADATA,
download_config=self.download_config,
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert module_factory_result.builder_kwargs["base_path"].startswith(config.HF_ENDPOINT)
module_metadata_configs = module_factory_result.builder_configs_parameters.metadata_configs
assert module_metadata_configs is not None
assert len(module_metadata_configs) == 1
assert next(iter(module_metadata_configs)) == "custom"
assert "drop_labels" in next(iter(module_metadata_configs.values()))
assert next(iter(module_metadata_configs.values()))["drop_labels"] is True
module_builder_configs = module_factory_result.builder_configs_parameters.builder_configs
assert module_builder_configs is not None
assert len(module_builder_configs) == 1
assert isinstance(module_builder_configs[0], AudioFolderConfig)
assert module_builder_configs[0].name == "custom"
assert module_builder_configs[0].data_files is not None
assert isinstance(module_builder_configs[0].data_files, DataFilesDict)
assert sorted(module_builder_configs[0].data_files) == ["test", "train"]
assert len(module_builder_configs[0].data_files["train"]) == 3
assert len(module_builder_configs[0].data_files["test"]) == 3
assert module_builder_configs[0].drop_labels is True # parameter is passed from metadata
# config named "default" is automatically considered to be a default config
assert module_factory_result.builder_configs_parameters.default_config_name is None
# we don't pass config params to builder in builder_kwargs, they are stored in builder_configs directly
assert "drop_labels" not in module_factory_result.builder_kwargs
@pytest.mark.integration
def test_HubDatasetModuleFactoryWithoutScript_with_two_configs_in_metadata(self):
datasets_names = [SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, SAMPLE_DATASET_TWO_CONFIG_IN_METADATA_WITH_DEFAULT]
for dataset_name in datasets_names:
factory = HubDatasetModuleFactoryWithoutScript(dataset_name, download_config=self.download_config)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
module_metadata_configs = module_factory_result.builder_configs_parameters.metadata_configs
assert module_metadata_configs is not None
assert len(module_metadata_configs) == 2
assert list(module_metadata_configs) == ["v1", "v2"]
assert "drop_labels" in module_metadata_configs["v1"]
assert module_metadata_configs["v1"]["drop_labels"] is True
assert "drop_labels" in module_metadata_configs["v2"]
assert module_metadata_configs["v2"]["drop_labels"] is False
module_builder_configs = module_factory_result.builder_configs_parameters.builder_configs
assert module_builder_configs is not None
assert len(module_builder_configs) == 2
module_builder_config_v1, module_builder_config_v2 = module_builder_configs
assert module_builder_config_v1.name == "v1"
assert module_builder_config_v2.name == "v2"
assert isinstance(module_builder_config_v1, AudioFolderConfig)
assert isinstance(module_builder_config_v2, AudioFolderConfig)
assert isinstance(module_builder_config_v1.data_files, DataFilesDict)
assert isinstance(module_builder_config_v2.data_files, DataFilesDict)
assert sorted(module_builder_config_v1.data_files) == ["test", "train"]
assert len(module_builder_config_v1.data_files["train"]) == 3
assert len(module_builder_config_v1.data_files["test"]) == 3
assert sorted(module_builder_config_v2.data_files) == ["test", "train"]
assert len(module_builder_config_v2.data_files["train"]) == 2
assert len(module_builder_config_v2.data_files["test"]) == 1
assert module_builder_config_v1.drop_labels is True # parameter is passed from metadata
assert module_builder_config_v2.drop_labels is False # parameter is passed from metadata
# we don't pass config params to builder in builder_kwargs, they are stored in builder_configs directly
assert "drop_labels" not in module_factory_result.builder_kwargs
if dataset_name == SAMPLE_DATASET_TWO_CONFIG_IN_METADATA_WITH_DEFAULT:
assert module_factory_result.builder_configs_parameters.default_config_name == "v1"
else:
assert module_factory_result.builder_configs_parameters.default_config_name is None
@pytest.mark.integration
def test_HubDatasetModuleFactoryWithScript(self):
factory = HubDatasetModuleFactoryWithScript(
SAMPLE_DATASET_IDENTIFIER,
download_config=self.download_config,
dynamic_modules_path=self.dynamic_modules_path,
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert module_factory_result.builder_kwargs["base_path"].startswith(config.HF_ENDPOINT)
def test_CachedDatasetModuleFactory(self):
path = os.path.join(self._dataset_loading_script_dir, f"{DATASET_LOADING_SCRIPT_NAME}.py")
factory = LocalDatasetModuleFactoryWithScript(
path, download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
module_factory_result = factory.get_module()
for offline_mode in OfflineSimulationMode:
with offline(offline_mode):
factory = CachedDatasetModuleFactory(
DATASET_LOADING_SCRIPT_NAME,
dynamic_modules_path=self.dynamic_modules_path,
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
@pytest.mark.filterwarnings("ignore:LocalMetricModuleFactory is deprecated:FutureWarning")
@pytest.mark.filterwarnings("ignore:CachedMetricModuleFactory is deprecated:FutureWarning")
def test_CachedMetricModuleFactory(self):
path = os.path.join(self._metric_loading_script_dir, f"{METRIC_LOADING_SCRIPT_NAME}.py")
factory = LocalMetricModuleFactory(
path, download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
module_factory_result = factory.get_module()
for offline_mode in OfflineSimulationMode:
with offline(offline_mode):
factory = CachedMetricModuleFactory(
METRIC_LOADING_SCRIPT_NAME,
dynamic_modules_path=self.dynamic_modules_path,
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
@pytest.mark.parametrize(
"factory_class",
[
CachedDatasetModuleFactory,
CachedMetricModuleFactory,
GithubMetricModuleFactory,
HubDatasetModuleFactoryWithoutScript,
HubDatasetModuleFactoryWithScript,
LocalDatasetModuleFactoryWithoutScript,
LocalDatasetModuleFactoryWithScript,
LocalMetricModuleFactory,
PackagedDatasetModuleFactory,
],
)
def test_module_factories(factory_class):
name = "dummy_name"
factory = factory_class(name)
assert factory.name == name
@pytest.mark.integration
class LoadTest(TestCase):
@pytest.fixture(autouse=True)
def inject_fixtures(self, caplog):
self._caplog = caplog
def setUp(self):
self.hf_modules_cache = tempfile.mkdtemp()
self.dynamic_modules_path = datasets.load.init_dynamic_modules(
name="test_datasets_modules2", hf_modules_cache=self.hf_modules_cache
)
def tearDown(self):
shutil.rmtree(self.hf_modules_cache)
def _dummy_module_dir(self, modules_dir, dummy_module_name, dummy_code):
assert dummy_module_name.startswith("__")
module_dir = os.path.join(modules_dir, dummy_module_name)
os.makedirs(module_dir, exist_ok=True)
module_path = os.path.join(module_dir, dummy_module_name + ".py")
with open(module_path, "w") as f:
f.write(dummy_code)
return module_dir
def test_dataset_module_factory(self):
with tempfile.TemporaryDirectory() as tmp_dir:
# prepare module from directory path
dummy_code = "MY_DUMMY_VARIABLE = 'hello there'"
module_dir = self._dummy_module_dir(tmp_dir, "__dummy_module_name1__", dummy_code)
dataset_module = datasets.load.dataset_module_factory(
module_dir, dynamic_modules_path=self.dynamic_modules_path
)
dummy_module = importlib.import_module(dataset_module.module_path)
self.assertEqual(dummy_module.MY_DUMMY_VARIABLE, "hello there")
self.assertEqual(dataset_module.hash, sha256(dummy_code.encode("utf-8")).hexdigest())
# prepare module from file path + check resolved_file_path
dummy_code = "MY_DUMMY_VARIABLE = 'general kenobi'"
module_dir = self._dummy_module_dir(tmp_dir, "__dummy_module_name1__", dummy_code)
module_path = os.path.join(module_dir, "__dummy_module_name1__.py")
dataset_module = datasets.load.dataset_module_factory(
module_path, dynamic_modules_path=self.dynamic_modules_path
)
dummy_module = importlib.import_module(dataset_module.module_path)
self.assertEqual(dummy_module.MY_DUMMY_VARIABLE, "general kenobi")
self.assertEqual(dataset_module.hash, sha256(dummy_code.encode("utf-8")).hexdigest())
# missing module
for offline_simulation_mode in list(OfflineSimulationMode):
with offline(offline_simulation_mode):
with self.assertRaises((FileNotFoundError, ConnectionError, requests.exceptions.ConnectionError)):
datasets.load.dataset_module_factory(
"__missing_dummy_module_name__", dynamic_modules_path=self.dynamic_modules_path
)
def test_offline_dataset_module_factory(self):
with tempfile.TemporaryDirectory() as tmp_dir:
dummy_code = "MY_DUMMY_VARIABLE = 'hello there'"
module_dir = self._dummy_module_dir(tmp_dir, "__dummy_module_name2__", dummy_code)
dataset_module_1 = datasets.load.dataset_module_factory(
module_dir, dynamic_modules_path=self.dynamic_modules_path
)
time.sleep(0.1) # make sure there's a difference in the OS update time of the python file
dummy_code = "MY_DUMMY_VARIABLE = 'general kenobi'"
module_dir = self._dummy_module_dir(tmp_dir, "__dummy_module_name2__", dummy_code)
dataset_module_2 = datasets.load.dataset_module_factory(
module_dir, dynamic_modules_path=self.dynamic_modules_path
)
for offline_simulation_mode in list(OfflineSimulationMode):
with offline(offline_simulation_mode):
self._caplog.clear()
# allow provide the module name without an explicit path to remote or local actual file
dataset_module_3 = datasets.load.dataset_module_factory(
"__dummy_module_name2__", dynamic_modules_path=self.dynamic_modules_path
)
# it loads the most recent version of the module
self.assertEqual(dataset_module_2.module_path, dataset_module_3.module_path)
self.assertNotEqual(dataset_module_1.module_path, dataset_module_3.module_path)
self.assertIn("Using the latest cached version of the module", self._caplog.text)
def test_load_dataset_from_hub(self):
with self.assertRaises(FileNotFoundError) as context:
datasets.load_dataset("_dummy")
self.assertIn(
"Dataset '_dummy' doesn't exist on the Hub",
str(context.exception),
)
with self.assertRaises(FileNotFoundError) as context:
datasets.load_dataset("_dummy", revision="0.0.0")
self.assertIn(
"Dataset '_dummy' doesn't exist on the Hub",
str(context.exception),
)
self.assertIn(
"at revision '0.0.0'",
str(context.exception),
)
for offline_simulation_mode in list(OfflineSimulationMode):
with offline(offline_simulation_mode):
with self.assertRaises(ConnectionError) as context:
datasets.load_dataset("_dummy")
if offline_simulation_mode != OfflineSimulationMode.HF_DATASETS_OFFLINE_SET_TO_1:
self.assertIn(
"Couldn't reach '_dummy' on the Hub",
str(context.exception),
)
def test_load_dataset_users(self):
with self.assertRaises(FileNotFoundError) as context:
datasets.load_dataset("lhoestq/_dummy")
self.assertIn(
"lhoestq/_dummy",
str(context.exception),
)
for offline_simulation_mode in list(OfflineSimulationMode):
with offline(offline_simulation_mode):
with self.assertRaises(ConnectionError) as context:
datasets.load_dataset("lhoestq/_dummy")
self.assertIn("lhoestq/_dummy", str(context.exception), msg=offline_simulation_mode)
@pytest.mark.integration
def test_load_dataset_builder_with_metadata():
builder = datasets.load_dataset_builder(SAMPLE_DATASET_IDENTIFIER4)
assert isinstance(builder, ImageFolder)
assert builder.config.name == "default"
assert builder.config.data_files is not None
assert builder.config.drop_metadata is None
builder = datasets.load_dataset_builder(SAMPLE_DATASET_IDENTIFIER4, "non-existing-config")
assert isinstance(builder, ImageFolder)
assert builder.config.name == "non-existing-config"
@pytest.mark.integration
def test_load_dataset_builder_config_kwargs_passed_as_arguments():
builder_default = datasets.load_dataset_builder(SAMPLE_DATASET_IDENTIFIER4)
builder_custom = datasets.load_dataset_builder(SAMPLE_DATASET_IDENTIFIER4, drop_metadata=True)
assert builder_custom.config.drop_metadata != builder_default.config.drop_metadata
assert builder_custom.config.drop_metadata is True
@pytest.mark.integration
def test_load_dataset_builder_with_two_configs_in_metadata():
builder = datasets.load_dataset_builder(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, "v1")
assert isinstance(builder, AudioFolder)
assert builder.config.name == "v1"
assert builder.config.data_files is not None
with pytest.raises(ValueError):
datasets.load_dataset_builder(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA)
with pytest.raises(ValueError):
datasets.load_dataset_builder(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, "non-existing-config")
@pytest.mark.parametrize("serializer", [pickle, dill])
def test_load_dataset_builder_with_metadata_configs_pickable(serializer):
builder = datasets.load_dataset_builder(SAMPLE_DATASET_SINGLE_CONFIG_IN_METADATA)
builder_unpickled = serializer.loads(serializer.dumps(builder))
assert builder.BUILDER_CONFIGS == builder_unpickled.BUILDER_CONFIGS
assert list(builder_unpickled.builder_configs) == ["custom"]
assert isinstance(builder_unpickled.builder_configs["custom"], AudioFolderConfig)
builder2 = datasets.load_dataset_builder(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, "v1")
builder2_unpickled = serializer.loads(serializer.dumps(builder2))
assert builder2.BUILDER_CONFIGS == builder2_unpickled.BUILDER_CONFIGS != builder_unpickled.BUILDER_CONFIGS
assert list(builder2_unpickled.builder_configs) == ["v1", "v2"]
assert isinstance(builder2_unpickled.builder_configs["v1"], AudioFolderConfig)
assert isinstance(builder2_unpickled.builder_configs["v2"], AudioFolderConfig)
def test_load_dataset_builder_for_absolute_script_dir(dataset_loading_script_dir, data_dir):
builder = datasets.load_dataset_builder(dataset_loading_script_dir, data_dir=data_dir)
assert isinstance(builder, DatasetBuilder)
assert builder.name == DATASET_LOADING_SCRIPT_NAME
assert builder.dataset_name == DATASET_LOADING_SCRIPT_NAME
assert builder.info.features == Features({"text": Value("string")})
def test_load_dataset_builder_for_relative_script_dir(dataset_loading_script_dir, data_dir):
with set_current_working_directory_to_temp_dir():
relative_script_dir = DATASET_LOADING_SCRIPT_NAME
shutil.copytree(dataset_loading_script_dir, relative_script_dir)
builder = datasets.load_dataset_builder(relative_script_dir, data_dir=data_dir)
assert isinstance(builder, DatasetBuilder)
assert builder.name == DATASET_LOADING_SCRIPT_NAME
assert builder.dataset_name == DATASET_LOADING_SCRIPT_NAME
assert builder.info.features == Features({"text": Value("string")})
def test_load_dataset_builder_for_script_path(dataset_loading_script_dir, data_dir):
builder = datasets.load_dataset_builder(
os.path.join(dataset_loading_script_dir, DATASET_LOADING_SCRIPT_NAME + ".py"), data_dir=data_dir
)
assert isinstance(builder, DatasetBuilder)
assert builder.name == DATASET_LOADING_SCRIPT_NAME
assert builder.dataset_name == DATASET_LOADING_SCRIPT_NAME
assert builder.info.features == Features({"text": Value("string")})
def test_load_dataset_builder_for_absolute_data_dir(complex_data_dir):
builder = datasets.load_dataset_builder(complex_data_dir)
assert isinstance(builder, DatasetBuilder)
assert builder.name == "text"
assert builder.dataset_name == Path(complex_data_dir).name
assert builder.config.name == "default"
assert isinstance(builder.config.data_files, DataFilesDict)
assert len(builder.config.data_files["train"]) > 0
assert len(builder.config.data_files["test"]) > 0
def test_load_dataset_builder_for_relative_data_dir(complex_data_dir):
with set_current_working_directory_to_temp_dir():
relative_data_dir = "relative_data_dir"
shutil.copytree(complex_data_dir, relative_data_dir)
builder = datasets.load_dataset_builder(relative_data_dir)
assert isinstance(builder, DatasetBuilder)
assert builder.name == "text"
assert builder.dataset_name == relative_data_dir
assert builder.config.name == "default"
assert isinstance(builder.config.data_files, DataFilesDict)
assert len(builder.config.data_files["train"]) > 0
assert len(builder.config.data_files["test"]) > 0
@pytest.mark.integration
def test_load_dataset_builder_for_community_dataset_with_script():
builder = datasets.load_dataset_builder(SAMPLE_DATASET_IDENTIFIER)
assert isinstance(builder, DatasetBuilder)
assert builder.name == SAMPLE_DATASET_IDENTIFIER.split("/")[-1]
assert builder.dataset_name == SAMPLE_DATASET_IDENTIFIER.split("/")[-1]
assert builder.config.name == "default"
assert builder.info.features == Features({"text": Value("string")})
namespace = SAMPLE_DATASET_IDENTIFIER[: SAMPLE_DATASET_IDENTIFIER.index("/")]
assert builder._relative_data_dir().startswith(namespace)
assert SAMPLE_DATASET_IDENTIFIER.replace("/", "--") in builder.__module__
@pytest.mark.integration
def test_load_dataset_builder_for_community_dataset_without_script():
builder = datasets.load_dataset_builder(SAMPLE_DATASET_IDENTIFIER2)
assert isinstance(builder, DatasetBuilder)
assert builder.name == "text"
assert builder.dataset_name == SAMPLE_DATASET_IDENTIFIER2.split("/")[-1]
assert builder.config.name == "default"
assert isinstance(builder.config.data_files, DataFilesDict)
assert len(builder.config.data_files["train"]) > 0
assert len(builder.config.data_files["test"]) > 0
def test_load_dataset_builder_fail():
with pytest.raises(FileNotFoundError):
datasets.load_dataset_builder("blabla")
@pytest.mark.parametrize("keep_in_memory", [False, True])
def test_load_dataset_local(dataset_loading_script_dir, data_dir, keep_in_memory, caplog):
with assert_arrow_memory_increases() if keep_in_memory else assert_arrow_memory_doesnt_increase():
dataset = load_dataset(dataset_loading_script_dir, data_dir=data_dir, keep_in_memory=keep_in_memory)
assert isinstance(dataset, DatasetDict)
assert all(isinstance(d, Dataset) for d in dataset.values())
assert len(dataset) == 2
assert isinstance(next(iter(dataset["train"])), dict)
for offline_simulation_mode in list(OfflineSimulationMode):
with offline(offline_simulation_mode):
caplog.clear()
# Load dataset from cache
dataset = datasets.load_dataset(DATASET_LOADING_SCRIPT_NAME, data_dir=data_dir)
assert len(dataset) == 2
assert "Using the latest cached version of the module" in caplog.text
with pytest.raises(FileNotFoundError) as exc_info:
datasets.load_dataset(SAMPLE_DATASET_NAME_THAT_DOESNT_EXIST)
assert f"Dataset '{SAMPLE_DATASET_NAME_THAT_DOESNT_EXIST}' doesn't exist on the Hub" in str(exc_info.value)
assert os.path.abspath(SAMPLE_DATASET_NAME_THAT_DOESNT_EXIST) in str(exc_info.value)
def test_load_dataset_streaming(dataset_loading_script_dir, data_dir):
dataset = load_dataset(dataset_loading_script_dir, streaming=True, data_dir=data_dir)
assert isinstance(dataset, IterableDatasetDict)
assert all(isinstance(d, IterableDataset) for d in dataset.values())
assert len(dataset) == 2
assert isinstance(next(iter(dataset["train"])), dict)
def test_load_dataset_streaming_gz_json(jsonl_gz_path):
data_files = jsonl_gz_path
ds = load_dataset("json", split="train", data_files=data_files, streaming=True)
assert isinstance(ds, IterableDataset)
ds_item = next(iter(ds))
assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
@pytest.mark.integration
@pytest.mark.parametrize(
"path", ["sample.jsonl", "sample.jsonl.gz", "sample.tar", "sample.jsonl.xz", "sample.zip", "sample.jsonl.zst"]
)
def test_load_dataset_streaming_compressed_files(path):
repo_id = "albertvillanova/datasets-tests-compression"
data_files = f"https://huggingface.co/datasets/{repo_id}/resolve/main/{path}"
if data_files[-3:] in ("zip", "tar"): # we need to glob "*" inside archives
data_files = data_files[-3:] + "://*::" + data_files
return # TODO(QL, albert): support re-add support for ZIP and TAR archives streaming
ds = load_dataset("json", split="train", data_files=data_files, streaming=True)
assert isinstance(ds, IterableDataset)
ds_item = next(iter(ds))
assert ds_item == {
"tokens": ["Ministeri", "de", "Justícia", "d'Espanya"],
"ner_tags": [1, 2, 2, 2],
"langs": ["ca", "ca", "ca", "ca"],
"spans": ["PER: Ministeri de Justícia d'Espanya"],
}
@pytest.mark.parametrize("path_extension", ["csv", "csv.bz2"])
@pytest.mark.parametrize("streaming", [False, True])
def test_load_dataset_streaming_csv(path_extension, streaming, csv_path, bz2_csv_path):
paths = {"csv": csv_path, "csv.bz2": bz2_csv_path}
data_files = str(paths[path_extension])
features = Features({"col_1": Value("string"), "col_2": Value("int32"), "col_3": Value("float32")})
ds = load_dataset("csv", split="train", data_files=data_files, features=features, streaming=streaming)
assert isinstance(ds, IterableDataset if streaming else Dataset)
ds_item = next(iter(ds))
assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
@pytest.mark.parametrize("streaming", [False, True])
@pytest.mark.parametrize("data_file", ["zip_csv_path", "zip_csv_with_dir_path", "csv_path"])
def test_load_dataset_zip_csv(data_file, streaming, zip_csv_path, zip_csv_with_dir_path, csv_path):
data_file_paths = {
"zip_csv_path": zip_csv_path,
"zip_csv_with_dir_path": zip_csv_with_dir_path,
"csv_path": csv_path,
}
data_files = str(data_file_paths[data_file])
expected_size = 8 if data_file.startswith("zip") else 4
features = Features({"col_1": Value("string"), "col_2": Value("int32"), "col_3": Value("float32")})
ds = load_dataset("csv", split="train", data_files=data_files, features=features, streaming=streaming)
if streaming:
ds_item_counter = 0
for ds_item in ds:
if ds_item_counter == 0:
assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
ds_item_counter += 1
assert ds_item_counter == expected_size
else:
assert ds.shape[0] == expected_size
ds_item = next(iter(ds))
assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
@pytest.mark.parametrize("streaming", [False, True])
@pytest.mark.parametrize("data_file", ["zip_jsonl_path", "zip_jsonl_with_dir_path", "jsonl_path"])
def test_load_dataset_zip_jsonl(data_file, streaming, zip_jsonl_path, zip_jsonl_with_dir_path, jsonl_path):
data_file_paths = {
"zip_jsonl_path": zip_jsonl_path,
"zip_jsonl_with_dir_path": zip_jsonl_with_dir_path,
"jsonl_path": jsonl_path,
}
data_files = str(data_file_paths[data_file])
expected_size = 8 if data_file.startswith("zip") else 4
features = Features({"col_1": Value("string"), "col_2": Value("int32"), "col_3": Value("float32")})
ds = load_dataset("json", split="train", data_files=data_files, features=features, streaming=streaming)
if streaming:
ds_item_counter = 0
for ds_item in ds:
if ds_item_counter == 0:
assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
ds_item_counter += 1
assert ds_item_counter == expected_size
else:
assert ds.shape[0] == expected_size
ds_item = next(iter(ds))
assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
@pytest.mark.parametrize("streaming", [False, True])
@pytest.mark.parametrize("data_file", ["zip_text_path", "zip_text_with_dir_path", "text_path"])
def test_load_dataset_zip_text(data_file, streaming, zip_text_path, zip_text_with_dir_path, text_path):
data_file_paths = {
"zip_text_path": zip_text_path,
"zip_text_with_dir_path": zip_text_with_dir_path,
"text_path": text_path,
}
data_files = str(data_file_paths[data_file])
expected_size = 8 if data_file.startswith("zip") else 4
ds = load_dataset("text", split="train", data_files=data_files, streaming=streaming)
if streaming:
ds_item_counter = 0
for ds_item in ds:
if ds_item_counter == 0:
assert ds_item == {"text": "0"}
ds_item_counter += 1
assert ds_item_counter == expected_size
else:
assert ds.shape[0] == expected_size
ds_item = next(iter(ds))
assert ds_item == {"text": "0"}
@pytest.mark.parametrize("streaming", [False, True])
def test_load_dataset_arrow(streaming, data_dir_with_arrow):
ds = load_dataset("arrow", split="train", data_dir=data_dir_with_arrow, streaming=streaming)
expected_size = 10
if streaming:
ds_item_counter = 0
for ds_item in ds:
if ds_item_counter == 0:
assert ds_item == {"col_1": "foo"}
ds_item_counter += 1
assert ds_item_counter == 10
else:
assert ds.num_rows == 10
assert ds.shape[0] == expected_size
ds_item = next(iter(ds))
assert ds_item == {"col_1": "foo"}
def test_load_dataset_text_with_unicode_new_lines(text_path_with_unicode_new_lines):
data_files = str(text_path_with_unicode_new_lines)
ds = load_dataset("text", split="train", data_files=data_files)
assert ds.num_rows == 3
def test_load_dataset_with_unsupported_extensions(text_dir_with_unsupported_extension):
data_files = str(text_dir_with_unsupported_extension)
ds = load_dataset("text", split="train", data_files=data_files)
assert ds.num_rows == 4
@pytest.mark.integration
def test_loading_from_the_datasets_hub():
with tempfile.TemporaryDirectory() as tmp_dir:
dataset = load_dataset(SAMPLE_DATASET_IDENTIFIER, cache_dir=tmp_dir)
assert len(dataset["train"]) == 2
assert len(dataset["validation"]) == 3
del dataset
@pytest.mark.integration
def test_loading_from_the_datasets_hub_with_token():
true_request = requests.Session().request
def assert_auth(method, url, *args, headers, **kwargs):
assert headers["authorization"] == "Bearer foo"
return true_request(method, url, *args, headers=headers, **kwargs)
with patch("requests.Session.request") as mock_request:
mock_request.side_effect = assert_auth
with tempfile.TemporaryDirectory() as tmp_dir:
with offline():
with pytest.raises((ConnectionError, requests.exceptions.ConnectionError)):
load_dataset(SAMPLE_NOT_EXISTING_DATASET_IDENTIFIER, cache_dir=tmp_dir, token="foo")
mock_request.assert_called()
@pytest.mark.integration
def test_load_streaming_private_dataset(hf_token, hf_private_dataset_repo_txt_data):
ds = load_dataset(hf_private_dataset_repo_txt_data, streaming=True)
assert next(iter(ds)) is not None
@pytest.mark.integration
def test_load_streaming_private_dataset_with_zipped_data(hf_token, hf_private_dataset_repo_zipped_txt_data):
ds = load_dataset(hf_private_dataset_repo_zipped_txt_data, streaming=True)
assert next(iter(ds)) is not None
@pytest.mark.integration
def test_load_dataset_config_kwargs_passed_as_arguments():
ds_default = load_dataset(SAMPLE_DATASET_IDENTIFIER4)
ds_custom = load_dataset(SAMPLE_DATASET_IDENTIFIER4, drop_metadata=True)
assert list(ds_default["train"].features) == ["image", "caption"]
assert list(ds_custom["train"].features) == ["image"]
@require_sndfile
@pytest.mark.integration
def test_load_hub_dataset_without_script_with_single_config_in_metadata():
# load the same dataset but with no configurations (=with default parameters)
ds = load_dataset(SAMPLE_DATASET_NO_CONFIGS_IN_METADATA)
assert list(ds["train"].features) == ["audio", "label"] # assert label feature is here as expected by default
assert len(ds["train"]) == 5 and len(ds["test"]) == 4
ds2 = load_dataset(SAMPLE_DATASET_SINGLE_CONFIG_IN_METADATA) # single config -> no need to specify it
assert list(ds2["train"].features) == ["audio"] # assert param `drop_labels=True` from metadata is passed
assert len(ds2["train"]) == 3 and len(ds2["test"]) == 3
ds3 = load_dataset(SAMPLE_DATASET_SINGLE_CONFIG_IN_METADATA, "custom")
assert list(ds3["train"].features) == ["audio"] # assert param `drop_labels=True` from metadata is passed
assert len(ds3["train"]) == 3 and len(ds3["test"]) == 3
with pytest.raises(ValueError):
# no config named "default"
_ = load_dataset(SAMPLE_DATASET_SINGLE_CONFIG_IN_METADATA, "default")
@require_sndfile
@pytest.mark.integration
def test_load_hub_dataset_without_script_with_two_config_in_metadata():
ds = load_dataset(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, "v1")
assert list(ds["train"].features) == ["audio"] # assert param `drop_labels=True` from metadata is passed
assert len(ds["train"]) == 3 and len(ds["test"]) == 3
ds2 = load_dataset(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, "v2")
assert list(ds2["train"].features) == [
"audio",
"label",
] # assert param `drop_labels=False` from metadata is passed
assert len(ds2["train"]) == 2 and len(ds2["test"]) == 1
with pytest.raises(ValueError):
# config is required but not specified
_ = load_dataset(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA)
with pytest.raises(ValueError):
# no config named "default"
_ = load_dataset(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, "default")
ds_with_default = load_dataset(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA_WITH_DEFAULT)
# it's a dataset with the same data but "v1" config is marked as a default one
assert list(ds_with_default["train"].features) == list(ds["train"].features)
assert len(ds_with_default["train"]) == len(ds["train"]) and len(ds_with_default["test"]) == len(ds["test"])
@require_sndfile
@pytest.mark.integration
def test_load_hub_dataset_without_script_with_metadata_config_in_parallel():
# assert it doesn't fail (pickling of dynamically created class works)
ds = load_dataset(SAMPLE_DATASET_SINGLE_CONFIG_IN_METADATA, num_proc=2)
assert "label" not in ds["train"].features # assert param `drop_labels=True` from metadata is passed
assert len(ds["train"]) == 3 and len(ds["test"]) == 3
ds = load_dataset(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, "v1", num_proc=2)
assert "label" not in ds["train"].features # assert param `drop_labels=True` from metadata is passed
assert len(ds["train"]) == 3 and len(ds["test"]) == 3
ds = load_dataset(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, "v2", num_proc=2)
assert "label" in ds["train"].features
assert len(ds["train"]) == 2 and len(ds["test"]) == 1
@require_pil
@pytest.mark.integration
@pytest.mark.parametrize("implicit_token", [True])
@pytest.mark.parametrize("streaming", [True])
def test_load_dataset_private_zipped_images(
hf_private_dataset_repo_zipped_img_data, hf_token, streaming, implicit_token
):
token = None if implicit_token else hf_token
ds = load_dataset(hf_private_dataset_repo_zipped_img_data, split="train", streaming=streaming, token=token)
assert isinstance(ds, IterableDataset if streaming else Dataset)
ds_items = list(ds)
assert len(ds_items) == 2
def test_load_dataset_then_move_then_reload(dataset_loading_script_dir, data_dir, tmp_path, caplog):
cache_dir1 = tmp_path / "cache1"
cache_dir2 = tmp_path / "cache2"
dataset = load_dataset(dataset_loading_script_dir, data_dir=data_dir, split="train", cache_dir=cache_dir1)
fingerprint1 = dataset._fingerprint
del dataset
os.rename(cache_dir1, cache_dir2)
caplog.clear()
with caplog.at_level(INFO, logger=get_logger().name):
dataset = load_dataset(dataset_loading_script_dir, data_dir=data_dir, split="train", cache_dir=cache_dir2)
assert "Found cached dataset" in caplog.text
assert dataset._fingerprint == fingerprint1, "for the caching mechanism to work, fingerprint should stay the same"
dataset = load_dataset(dataset_loading_script_dir, data_dir=data_dir, split="test", cache_dir=cache_dir2)
assert dataset._fingerprint != fingerprint1
def test_load_dataset_readonly(dataset_loading_script_dir, dataset_loading_script_dir_readonly, data_dir, tmp_path):
cache_dir1 = tmp_path / "cache1"
cache_dir2 = tmp_path / "cache2"
dataset = load_dataset(dataset_loading_script_dir, data_dir=data_dir, split="train", cache_dir=cache_dir1)
fingerprint1 = dataset._fingerprint
del dataset
# Load readonly dataset and check that the fingerprint is the same.
dataset = load_dataset(dataset_loading_script_dir_readonly, data_dir=data_dir, split="train", cache_dir=cache_dir2)
assert dataset._fingerprint == fingerprint1, "Cannot load a dataset in a readonly folder."
@pytest.mark.parametrize("max_in_memory_dataset_size", ["default", 0, 50, 500])
def test_load_dataset_local_with_default_in_memory(
max_in_memory_dataset_size, dataset_loading_script_dir, data_dir, monkeypatch
):
current_dataset_size = 148
if max_in_memory_dataset_size == "default":
max_in_memory_dataset_size = 0 # default
else:
monkeypatch.setattr(datasets.config, "IN_MEMORY_MAX_SIZE", max_in_memory_dataset_size)
if max_in_memory_dataset_size:
expected_in_memory = current_dataset_size < max_in_memory_dataset_size
else:
expected_in_memory = False
with assert_arrow_memory_increases() if expected_in_memory else assert_arrow_memory_doesnt_increase():
dataset = load_dataset(dataset_loading_script_dir, data_dir=data_dir)
assert (dataset["train"].dataset_size < max_in_memory_dataset_size) is expected_in_memory
@pytest.mark.parametrize("max_in_memory_dataset_size", ["default", 0, 100, 1000])
def test_load_from_disk_with_default_in_memory(
max_in_memory_dataset_size, dataset_loading_script_dir, data_dir, tmp_path, monkeypatch
):
current_dataset_size = 512 # arrow file size = 512, in-memory dataset size = 148
if max_in_memory_dataset_size == "default":
max_in_memory_dataset_size = 0 # default
else:
monkeypatch.setattr(datasets.config, "IN_MEMORY_MAX_SIZE", max_in_memory_dataset_size)
if max_in_memory_dataset_size:
expected_in_memory = current_dataset_size < max_in_memory_dataset_size
else:
expected_in_memory = False
dset = load_dataset(dataset_loading_script_dir, data_dir=data_dir, keep_in_memory=True)
dataset_path = os.path.join(tmp_path, "saved_dataset")
dset.save_to_disk(dataset_path)
with assert_arrow_memory_increases() if expected_in_memory else assert_arrow_memory_doesnt_increase():
_ = load_from_disk(dataset_path)
@pytest.mark.integration
def test_remote_data_files():
repo_id = "albertvillanova/tests-raw-jsonl"
filename = "wikiann-bn-validation.jsonl"
data_files = f"https://huggingface.co/datasets/{repo_id}/resolve/main/{filename}"
ds = load_dataset("json", split="train", data_files=data_files, streaming=True)
assert isinstance(ds, IterableDataset)
ds_item = next(iter(ds))
assert ds_item.keys() == {"langs", "ner_tags", "spans", "tokens"}
@pytest.mark.parametrize("deleted", [False, True])
def test_load_dataset_deletes_extracted_files(deleted, jsonl_gz_path, tmp_path):
data_files = jsonl_gz_path
cache_dir = tmp_path / "cache"
if deleted:
download_config = DownloadConfig(delete_extracted=True, cache_dir=cache_dir / "downloads")
ds = load_dataset(
"json", split="train", data_files=data_files, cache_dir=cache_dir, download_config=download_config
)
else: # default
ds = load_dataset("json", split="train", data_files=data_files, cache_dir=cache_dir)
assert ds[0] == {"col_1": "0", "col_2": 0, "col_3": 0.0}
assert (
[path for path in (cache_dir / "downloads" / "extracted").iterdir() if path.suffix != ".lock"] == []
) is deleted
def distributed_load_dataset(args):
data_name, tmp_dir, datafiles = args
dataset = load_dataset(data_name, cache_dir=tmp_dir, data_files=datafiles)
return dataset
def test_load_dataset_distributed(tmp_path, csv_path):
num_workers = 5
args = "csv", str(tmp_path), csv_path
with Pool(processes=num_workers) as pool: # start num_workers processes
datasets = pool.map(distributed_load_dataset, [args] * num_workers)
assert len(datasets) == num_workers
assert all(len(dataset) == len(datasets[0]) > 0 for dataset in datasets)
assert len(datasets[0].cache_files) > 0
assert all(dataset.cache_files == datasets[0].cache_files for dataset in datasets)
| 0 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.