code
stringlengths 2.5k
150k
| kind
stringclasses 1
value |
|---|---|
# Create TensorFlow Deep Neural Network Model
**Learning Objective**
- Create a DNN model using the high-level Estimator API
## Introduction
We'll begin by modeling our data using a Deep Neural Network. To achieve this we will use the high-level Estimator API in Tensorflow. Have a look at the various models available through the Estimator API in [the documentation here](https://www.tensorflow.org/api_docs/python/tf/estimator).
Start by setting the environment variables related to your project.
```
PROJECT = "cloud-training-demos" # Replace with your PROJECT
BUCKET = "cloud-training-bucket" # Replace with your BUCKET
REGION = "us-central1" # Choose an available region for Cloud MLE
TFVERSION = "1.14" # TF version for CMLE to use
import os
os.environ["BUCKET"] = BUCKET
os.environ["PROJECT"] = PROJECT
os.environ["REGION"] = REGION
os.environ["TFVERSION"] = TFVERSION
%%bash
if ! gsutil ls | grep -q gs://${BUCKET}/; then
gsutil mb -l ${REGION} gs://${BUCKET}
fi
%%bash
ls *.csv
```
## Create TensorFlow model using TensorFlow's Estimator API ##
We'll begin by writing an input function to read the data and define the csv column names and label column. We'll also set the default csv column values and set the number of training steps.
```
import shutil
import numpy as np
import tensorflow as tf
print(tf.__version__)
CSV_COLUMNS = "weight_pounds,is_male,mother_age,plurality,gestation_weeks".split(',')
LABEL_COLUMN = "weight_pounds"
# Set default values for each CSV column
DEFAULTS = [[0.0], ["null"], [0.0], ["null"], [0.0]]
TRAIN_STEPS = 1000
```
### Create the input function
Now we are ready to create an input function using the Dataset API.
```
def read_dataset(filename_pattern, mode, batch_size = 512):
def _input_fn():
def decode_csv(value_column):
columns = tf.decode_csv(records = value_column, record_defaults = DEFAULTS)
features = dict(zip(CSV_COLUMNS, columns))
label = features.pop(LABEL_COLUMN)
return features, label
# Create list of files that match pattern
file_list = tf.gfile.Glob(filename = filename_pattern)
# Create dataset from file list
dataset = (tf.data.TextLineDataset(filenames = file_list) # Read text file
.map(map_func = decode_csv)) # Transform each elem by applying decode_csv fn
if mode == tf.estimator.ModeKeys.TRAIN:
num_epochs = None # indefinitely
dataset = dataset.shuffle(buffer_size = 10 * batch_size)
else:
num_epochs = 1 # end-of-input after this
dataset = dataset.repeat(count = num_epochs).batch(batch_size = batch_size)
return dataset
return _input_fn
```
### Create the feature columns
Next, we define the feature columns
```
def get_categorical(name, values):
return tf.feature_column.indicator_column(
categorical_column = tf.feature_column.categorical_column_with_vocabulary_list(key = name, vocabulary_list = values))
def get_cols():
# Define column types
return [\
get_categorical("is_male", ["True", "False", "Unknown"]),
tf.feature_column.numeric_column(key = "mother_age"),
get_categorical("plurality",
["Single(1)", "Twins(2)", "Triplets(3)",
"Quadruplets(4)", "Quintuplets(5)","Multiple(2+)"]),
tf.feature_column.numeric_column(key = "gestation_weeks")
]
```
### Create the Serving Input function
To predict with the TensorFlow model, we also need a serving input function. This will allow us to serve prediction later using the predetermined inputs. We will want all the inputs from our user.
```
def serving_input_fn():
feature_placeholders = {
"is_male": tf.placeholder(dtype = tf.string, shape = [None]),
"mother_age": tf.placeholder(dtype = tf.float32, shape = [None]),
"plurality": tf.placeholder(dtype = tf.string, shape = [None]),
"gestation_weeks": tf.placeholder(dtype = tf.float32, shape = [None])
}
features = {
key: tf.expand_dims(input = tensor, axis = -1)
for key, tensor in feature_placeholders.items()
}
return tf.estimator.export.ServingInputReceiver(features = features, receiver_tensors = feature_placeholders)
```
### Create the model and run training and evaluation
Lastly, we'll create the estimator to train and evaluate. In the cell below, we'll set up a `DNNRegressor` estimator and the train and evaluation operations.
```
def train_and_evaluate(output_dir):
EVAL_INTERVAL = 300
run_config = tf.estimator.RunConfig(
save_checkpoints_secs = EVAL_INTERVAL,
keep_checkpoint_max = 3)
estimator = tf.estimator.DNNRegressor(
model_dir = output_dir,
feature_columns = get_cols(),
hidden_units = [64, 32],
config = run_config)
train_spec = tf.estimator.TrainSpec(
input_fn = read_dataset("train.csv", mode = tf.estimator.ModeKeys.TRAIN),
max_steps = TRAIN_STEPS)
exporter = tf.estimator.LatestExporter(name = "exporter", serving_input_receiver_fn = serving_input_fn)
eval_spec = tf.estimator.EvalSpec(
input_fn = read_dataset("eval.csv", mode = tf.estimator.ModeKeys.EVAL),
steps = None,
start_delay_secs = 60, # start evaluating after N seconds
throttle_secs = EVAL_INTERVAL, # evaluate every N seconds
exporters = exporter)
tf.estimator.train_and_evaluate(estimator = estimator, train_spec = train_spec, eval_spec = eval_spec)
```
Finally, we train the model!
```
# Run the model
shutil.rmtree(path = "babyweight_trained_dnn", ignore_errors = True) # start fresh each time
train_and_evaluate("babyweight_trained_dnn")
```
When I ran it, the final RMSE (the average_loss) is about **1.16**. You can explore the contents of the `exporter` directory to see the contains final model.
Copyright 2017-2018 Google Inc. Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License
| github_jupyter |
# Compare different DEMs for individual glaciers
For most glaciers in the world there are several digital elevation models (DEM) which cover the respective glacier. In OGGM we have currently implemented 10 different open access DEMs to choose from. Some are regional and only available in certain areas (e.g. Greenland or Antarctica) and some cover almost the entire globe. For more information, visit the [rgitools documentation about DEMs](https://rgitools.readthedocs.io/en/latest/dems.html).
This notebook allows to see which of the DEMs are available for a selected glacier and how they compare to each other. That way it is easy to spot systematic differences and also invalid points in the DEMs.
## Input parameters
This notebook can be run as a script with parameters using [papermill](https://github.com/nteract/papermill), but it is not necessary. The following cell contains the parameters you can choose from:
```
# The RGI Id of the glaciers you want to look for
# Use the original shapefiles or the GLIMS viewer to check for the ID: https://www.glims.org/maps/glims
rgi_id = 'RGI60-11.00897'
# The default is to test for all sources available for this glacier
# Set to a list of source names to override this
sources = None
# Where to write the plots. Default is in the current working directory
plot_dir = ''
# The RGI version to use
# V62 is an unofficial modification of V6 with only minor, backwards compatible modifications
prepro_rgi_version = 62
# Size of the map around the glacier. Currently only 10 and 40 are available
prepro_border = 10
# Degree of processing level. Currently only 1 is available.
from_prepro_level = 1
```
## Check input and set up
```
# The sources can be given as parameters
if sources is not None and isinstance(sources, str):
sources = sources.split(',')
# Plotting directory as well
if not plot_dir:
plot_dir = './' + rgi_id
import os
plot_dir = os.path.abspath(plot_dir)
import pandas as pd
import numpy as np
from oggm import cfg, utils, workflow, tasks, graphics, GlacierDirectory
import xarray as xr
import geopandas as gpd
import salem
import matplotlib.pyplot as plt
from mpl_toolkits.axes_grid1 import AxesGrid
import itertools
from oggm.utils import DEM_SOURCES
from oggm.workflow import init_glacier_directories
# Make sure the plot directory exists
utils.mkdir(plot_dir);
# Use OGGM to download the data
cfg.initialize()
cfg.PATHS['working_dir'] = utils.gettempdir(dirname='OGGM-DEMS', reset=True)
cfg.PARAMS['use_intersects'] = False
```
## Download the data using OGGM utility functions
Note that you could reach the same goal by downloading the data manually from https://cluster.klima.uni-bremen.de/~oggm/gdirs/oggm_v1.4/rgitopo/
```
# URL of the preprocessed GDirs
gdir_url = 'https://cluster.klima.uni-bremen.de/~oggm/gdirs/oggm_v1.4/rgitopo/'
# We use OGGM to download the data
gdir = init_glacier_directories([rgi_id], from_prepro_level=1, prepro_border=10,
prepro_rgi_version='62', prepro_base_url=gdir_url)[0]
```
## Read the DEMs and store them all in a dataset
```
if sources is None:
sources = [src for src in os.listdir(gdir.dir) if src in utils.DEM_SOURCES]
print('RGI ID:', rgi_id)
print('Available DEM sources:', sources)
print('Plotting directory:', plot_dir)
# We use xarray to store the data
ods = xr.Dataset()
for src in sources:
demfile = os.path.join(gdir.dir, src) + '/dem.tif'
with xr.open_rasterio(demfile) as ds:
data = ds.sel(band=1).load() * 1.
ods[src] = data.where(data > -100, np.NaN)
sy, sx = np.gradient(ods[src], gdir.grid.dx, gdir.grid.dx)
ods[src + '_slope'] = ('y', 'x'), np.arctan(np.sqrt(sy**2 + sx**2))
with xr.open_rasterio(gdir.get_filepath('glacier_mask')) as ds:
ods['mask'] = ds.sel(band=1).load()
# Decide on the number of plots and figure size
ns = len(sources)
x_size = 12
n_cols = 3
n_rows = -(-ns // n_cols)
y_size = x_size / n_cols * n_rows
```
## Raw topography data
```
smap = salem.graphics.Map(gdir.grid, countries=False)
smap.set_shapefile(gdir.read_shapefile('outlines'))
smap.set_plot_params(cmap='topo')
smap.set_lonlat_contours(add_tick_labels=False)
smap.set_plot_params(vmin=np.nanquantile([ods[s].min() for s in sources], 0.25),
vmax=np.nanquantile([ods[s].max() for s in sources], 0.75))
fig = plt.figure(figsize=(x_size, y_size))
grid = AxesGrid(fig, 111,
nrows_ncols=(n_rows, n_cols),
axes_pad=0.7,
cbar_mode='each',
cbar_location='right',
cbar_pad=0.1
)
for i, s in enumerate(sources):
data = ods[s]
smap.set_data(data)
ax = grid[i]
smap.visualize(ax=ax, addcbar=False, title=s)
if np.isnan(data).all():
grid[i].cax.remove()
continue
cax = grid.cbar_axes[i]
smap.colorbarbase(cax)
# take care of uneven grids
if ax != grid[-1]:
grid[-1].remove()
grid[-1].cax.remove()
plt.savefig(os.path.join(plot_dir, 'dem_topo_color.png'), dpi=150, bbox_inches='tight')
```
## Shaded relief
```
fig = plt.figure(figsize=(x_size, y_size))
grid = AxesGrid(fig, 111,
nrows_ncols=(n_rows, n_cols),
axes_pad=0.7,
cbar_mode='none',
cbar_location='right',
cbar_pad=0.1
)
smap.set_plot_params(cmap='Blues')
smap.set_shapefile()
for i, s in enumerate(sources):
data = ods[s].copy().where(np.isfinite(ods[s]), 0)
smap.set_data(data * 0)
ax = grid[i]
smap.set_topography(data)
smap.visualize(ax=ax, addcbar=False, title=s)
# take care of uneven grids
if ax != grid[-1]:
grid[-1].remove()
grid[-1].cax.remove()
plt.savefig(os.path.join(plot_dir, 'dem_topo_shade.png'), dpi=150, bbox_inches='tight')
```
## Slope
```
fig = plt.figure(figsize=(x_size, y_size))
grid = AxesGrid(fig, 111,
nrows_ncols=(n_rows, n_cols),
axes_pad=0.7,
cbar_mode='each',
cbar_location='right',
cbar_pad=0.1
)
smap.set_topography();
smap.set_plot_params(vmin=0, vmax=0.7, cmap='Blues')
for i, s in enumerate(sources):
data = ods[s + '_slope']
smap.set_data(data)
ax = grid[i]
smap.visualize(ax=ax, addcbar=False, title=s + ' (slope)')
cax = grid.cbar_axes[i]
smap.colorbarbase(cax)
# take care of uneven grids
if ax != grid[-1]:
grid[-1].remove()
grid[-1].cax.remove()
plt.savefig(os.path.join(plot_dir, 'dem_slope.png'), dpi=150, bbox_inches='tight')
```
## Some simple statistics about the DEMs
```
df = pd.DataFrame()
for s in sources:
df[s] = ods[s].data.flatten()[ods.mask.data.flatten() == 1]
dfs = pd.DataFrame()
for s in sources:
dfs[s] = ods[s + '_slope'].data.flatten()[ods.mask.data.flatten() == 1]
df.describe()
```
## Comparison matrix plot
```
# Table of differences between DEMS
df_diff = pd.DataFrame()
done = []
for s1, s2 in itertools.product(sources, sources):
if s1 == s2:
continue
if (s2, s1) in done:
continue
df_diff[s1 + '-' + s2] = df[s1] - df[s2]
done.append((s1, s2))
# Decide on plot levels
max_diff = df_diff.quantile(0.99).max()
base_levels = np.array([-8, -5, -3, -1.5, -1, -0.5, -0.2, -0.1, 0, 0.1, 0.2, 0.5, 1, 1.5, 3, 5, 8])
if max_diff < 10:
levels = base_levels
elif max_diff < 100:
levels = base_levels * 10
elif max_diff < 1000:
levels = base_levels * 100
else:
levels = base_levels * 1000
levels = [l for l in levels if abs(l) < max_diff]
if max_diff > 10:
levels = [int(l) for l in levels]
levels
smap.set_plot_params(levels=levels, cmap='PuOr', extend='both')
smap.set_shapefile(gdir.read_shapefile('outlines'))
fig = plt.figure(figsize=(14, 14))
grid = AxesGrid(fig, 111,
nrows_ncols=(ns - 1, ns - 1),
axes_pad=0.3,
cbar_mode='single',
cbar_location='right',
cbar_pad=0.1
)
done = []
for ax in grid:
ax.set_axis_off()
for s1, s2 in itertools.product(sources, sources):
if s1 == s2:
continue
if (s2, s1) in done:
continue
data = ods[s1] - ods[s2]
ax = grid[sources.index(s1) * (ns - 1) + sources[1:].index(s2)]
ax.set_axis_on()
smap.set_data(data)
smap.visualize(ax=ax, addcbar=False)
done.append((s1, s2))
ax.set_title(s1 + '-' + s2, fontsize=8)
cax = grid.cbar_axes[0]
smap.colorbarbase(cax);
plt.savefig(os.path.join(plot_dir, 'dem_diffs.png'), dpi=150, bbox_inches='tight')
```
## Comparison scatter plot
```
import seaborn as sns
sns.set(style="ticks")
l1, l2 = (utils.nicenumber(df.min().min(), binsize=50, lower=True),
utils.nicenumber(df.max().max(), binsize=50, lower=False))
def plot_unity(xdata, ydata, **kwargs):
points = np.linspace(l1, l2, 100)
plt.gca().plot(points, points, color='k', marker=None,
linestyle=':', linewidth=3.0)
g = sns.pairplot(df.dropna(how='all', axis=1).dropna(), plot_kws=dict(s=50, edgecolor="C0", linewidth=1));
g.map_offdiag(plot_unity)
for asx in g.axes:
for ax in asx:
ax.set_xlim((l1, l2))
ax.set_ylim((l1, l2))
plt.savefig(os.path.join(plot_dir, 'dem_scatter.png'), dpi=150, bbox_inches='tight')
```
## Table statistics
```
df.describe()
df.corr()
df_diff.describe()
df_diff.abs().describe()
```
## What's next?
- return to the [OGGM documentation](https://docs.oggm.org)
- back to the [table of contents](welcome.ipynb)
| github_jupyter |
Created from https://github.com/awslabs/amazon-sagemaker-examples/blob/master/introduction_to_amazon_algorithms/random_cut_forest/random_cut_forest.ipynb
```
import boto3
import botocore
import sagemaker
import sys
bucket = 'tdk-awsml-sagemaker-data.io-dev' # <--- specify a bucket you have access to
prefix = ''
execution_role = sagemaker.get_execution_role()
# check if the bucket exists
try:
boto3.Session().client('s3').head_bucket(Bucket=bucket)
except botocore.exceptions.ParamValidationError as e:
print('Hey! You either forgot to specify your S3 bucket'
' or you gave your bucket an invalid name!')
except botocore.exceptions.ClientError as e:
if e.response['Error']['Code'] == '403':
print("Hey! You don't have permission to access the bucket, {}.".format(bucket))
elif e.response['Error']['Code'] == '404':
print("Hey! Your bucket, {}, doesn't exist!".format(bucket))
else:
raise
else:
print('Training input/output will be stored in: s3://{}/{}'.format(bucket, prefix))
%%time
import pandas as pd
import urllib.request
data_filename = 'nyc_taxi.csv'
data_source = 'https://raw.githubusercontent.com/numenta/NAB/master/data/realKnownCause/nyc_taxi.csv'
urllib.request.urlretrieve(data_source, data_filename)
taxi_data = pd.read_csv(data_filename, delimiter=',')
from sagemaker import RandomCutForest
session = sagemaker.Session()
# specify general training job information
rcf = RandomCutForest(role=execution_role,
train_instance_count=1,
train_instance_type='ml.m5.large',
data_location='s3://{}/{}/'.format(bucket, prefix),
output_path='s3://{}/{}/output'.format(bucket, prefix),
num_samples_per_tree=512,
num_trees=50)
# automatically upload the training data to S3 and run the training job
# TK - had to modify this line to use to_numpy() instead of as_matrix()
rcf.fit(rcf.record_set(taxi_data.value.to_numpy().reshape(-1,1)))
rcf_inference = rcf.deploy(
initial_instance_count=1,
instance_type='ml.m5.large',
)
print('Endpoint name: {}'.format(rcf_inference.endpoint))
from sagemaker.predictor import csv_serializer, json_deserializer
rcf_inference.content_type = 'text/csv'
rcf_inference.serializer = csv_serializer
rcf_inference.accept = 'application/json'
rcf_inference.deserializer = json_deserializer
# TK - had to modify this line to use to_numpy() instead of as_matrix()
taxi_data_numpy = taxi_data.value.to_numpy().reshape(-1,1)
print(taxi_data_numpy[:6])
results = rcf_inference.predict(taxi_data_numpy[:6])
sagemaker.Session().delete_endpoint(rcf_inference.endpoint)
```
| github_jupyter |
<br>
# Analysis of Big Earth Data with Jupyter Notebooks
<img src='./img/opengeohub_logo.png' alt='OpenGeoHub Logo' align='right' width='25%'></img>
Lecture given for OpenGeoHub summer school 2020<br>
Tuesday, 18. August 2020 | 11:00-13:00 CEST
#### Lecturer
* [Julia Wagemann](https://jwagemann.com) | Independent consultant and Phd student at University of Marburg
#### Access to tutorial material
Notebooks are available on [GitHub](https://github.com/jwagemann/2020_analysis_of_big_earth_data_with_jupyter).
<hr>
### Access to the JupyterHub
You can access the lecture material on a JupyterHub instance, a pre-defined environment that gives you direct access to the data and Python packages required for following the lecture.
<div class="alert alert-block alert-success" align="left">
1. Web address: <a href='https://opengeohub.adamplatform.eu'>https://opengeohub.adamplatform.eu</a><br>
2. Create an account: <a href='https://meeoauth.adamplatform.eu'>https://meeoauth.adamplatform.eu</a><br>
3. Log into the <b>JupyterHub</b> with your account created.
</div>
<hr>
## What is this lecture about?
Growing volumes of `Big Earth Data` force us to change the way how we access and process large volumes of geospatial data. New (cloud-based) data systems are being developed, each offering different functionalities for users.
This lecture is split in two parts:
* **(Cloud-based) data access systems**<br>
This part will highlight five data access systems that allow you to access, download or process large volumes of Copernicus data related to climate and atmosphere. For each data system, an example is given how data can be retrieved.
Data access systems that will be covered:
* [Copernicus Climate Data Store (CDS)](https://cds.climate.copernicus.eu/) / [Copernicus Atmosphere Data Store (ADS)](https://ads.atmosphere.copernicus.eu/)
* [WEkEO - Copernicus Data and Information Access System](http://wekeo.eu/)
* [Open Data Registry on Amazon Web Services](http://registry.opendata.aws)
* [Google Earth Engine](https://code.earthengine.google.com/)
* **Case study: Analysis of Covid-19 with Sentinel-5P data**<br>
This example showcases a case study analysing daily Sentinel-5P data from 2019 and 2020 with Jupyter notebooks and the Python library [xarray](http://xarray.pydata.org/en/stable/) in order to analyse possible Covid-19 impacts in 2020.
## Lecture outline
This lecture has the following outline:
* [01 - Introduction to Project Jupyter (optional)](01_Intro_to_Python_and_Jupyter.ipynb)
* [02 - Copernicus Climate Data Store / Copernicus Atmosphere Data Store](02_copernicus_climate_atmosphere_data_store.ipynb)
* [03 - WEkEO - Copernicus Data and Information Access Service (DIAS)](03_WEkEO_dias_service.ipynb)
* [04 - Amazon Web Services Open Data Registry](04_aws_open_data_registry.ipynb)
* [05 - Google Earth Engine](05_google_earth_engine.ipynb)
* [11 - Covid-19 case study - Sentinel-5P anomaly map](11_covid19_case_study_s5p_anomaly_map.ipynb)
* [12 - Covid-19 case study - Sentinel-5P time-series analysis](12_covid19_case_study_s5p_time_series_analysis.ipynb)
<br>
<hr>
© 2020 | Julia Wagemann
<a rel="license" href="http://creativecommons.org/licenses/by/4.0/"><img style="float: right" alt="Creative Commons Lizenzvertrag" style="border-width:0" src="https://i.creativecommons.org/l/by/4.0/88x31.png" /></a>
| github_jupyter |
<a href="https://colab.research.google.com/github/sima97/unihobby/blob/master/test.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
from google.colab import drive
drive.mount('/content/drive')
pip install nilearn
pip install tables
pip install git+https://www.github.com/farizrahman4u/keras-contrib.git
pip install SimpleITK
#pip install tensorflow==1.4
import tensorflow as tf
from tensorflow.python.framework import ops
import tensorflow.compat.v1 as tf
tf.disable_v2_behavior()
def cross_entropy_loss_v1(y_true, y_pred, sample_weight=None, eps=1e-6):
"""
:param y_pred: output 5D tensor, [batch size, dim0, dim1, dim2, class]
:param y_true: 4D GT tensor, [batch size, dim0, dim1, dim2]
:param eps: avoid log0
:return: cross entropy loss
"""
log_y = tf.log(y_pred + eps)
num_samples = tf.cast(tf.reduce_prod(tf.shape(y_true)), "float32")
label_one_hot = tf.one_hot(indices=y_true, depth=y_pred.shape[-1], axis=-1, dtype=tf.float32)
if sample_weight is not None:
# ce = mean(- weight * y_true * log(y_pred)).
label_one_hot = label_one_hot * sample_weight
cross_entropy = - tf.reduce_sum(label_one_hot * log_y) / num_samples
return cross_entropy
def cross_entropy_loss(y_true, y_pred, sample_weight=None):
# may not use one_hot when use tf.keras.losses.CategoricalCrossentropy
y_true = tf.one_hot(indices=y_true, depth=y_pred.shape[-1], axis=-1, dtype=tf.float32)
if sample_weight is not None:
# ce = mean(weight * y_true * log(y_pred)).
y_true = y_true * sample_weight
return tf.keras.losses.BinaryCrossentropy()(y_true, y_pred)
def cross_entropy_loss_with_weight(y_true, y_pred, sample_weight_per_c=None, eps=1e-6):
# for simple calculate this batch.
# if possible, get weight per epoch before training.
num_dims, num_classes = [len(y_true.shape), y_pred.shape.as_list()[-1]]
if sample_weight_per_c is None:
print('use batch to calculate weight')
num_lbls_in_ygt = tf.cast(tf.reduce_prod(tf.shape(y_true)), dtype="float32")
num_lbls_in_ygt_per_c = tf.bincount(arr=tf.cast(y_true, tf.int32), minlength=num_classes, maxlength=num_classes,
dtype="float32") # without the min/max, length of vector can change.
sample_weight_per_c = (1. / (num_lbls_in_ygt_per_c + eps)) * (num_lbls_in_ygt / num_classes)
sample_weight_per_c = tf.reshape(sample_weight_per_c, [1] * num_dims + [num_classes])
# use cross_entropy_loss get negative value, while cross_entropy_loss and cross_entropy_loss_v1 get the same
# when no weight. I guess may some error when batch distribution is huge different from epoch distribution.
return cross_entropy_loss_v1(y_true, y_pred, sample_weight=sample_weight_per_c)
def dice_coef(y_true, y_pred, eps=1e-6):
# problem: when gt class-0 >> class-1, the pred p(class-0) >> p(class-1)
# eg. gt = [0, 0, 0, 0, 1] pred = [[1, 0], [1, 0], [1, 0], [1, 0], [1, 0]]. 2 * 4 / (5 + 5) = 0.8
# in fact, change every pred, 4/5 -> 0.6, 1/5 ->1, so the model just pred all 0. imbalance class problem.
# only calculate gt == 1 can fix my problem, but for multi-class task, weight needed like ce loss above.
y_true = tf.one_hot(indices=y_true, depth=y_pred.shape[-1], axis=-1, dtype=tf.float32)
abs_x_and_y = 2 * tf.reduce_sum(y_true * y_pred)
abs_x_plus_abs_y = tf.reduce_sum(y_true) + tf.reduce_sum(y_pred)
return (abs_x_and_y + eps) / (abs_x_plus_abs_y + eps)
def dice_coef_loss(y_true, y_pred):
return 1. - dice_coef(y_true, y_pred)
import numpy as np
from keras import backend as K
from keras.engine import Input, Model
from keras.layers import Conv3D, MaxPooling3D, UpSampling3D, Activation, BatchNormalization, PReLU#, Deconvolution3D
from keras.optimizers import Adam
#from unet3d.metrics import dice_coefficient_loss, get_label_dice_coefficient_function, dice_coefficient
K.set_image_data_format("channels_first")
try:
from keras.engine import merge
except ImportError:
from keras.layers.merge import concatenate
def unet_model_3d(input_shape, pool_size=(2, 2, 2), n_labels=1, initial_learning_rate=0.00001, deconvolution=False,
depth=4, n_base_filters=32, include_label_wise_dice_coefficients=False, metrics=dice_coef,
batch_normalization=False, activation_name="sigmoid"):
"""
Builds the 3D UNet Keras model.f
:param metrics: List metrics to be calculated during model training (default is dice coefficient).
:param include_label_wise_dice_coefficients: If True and n_labels is greater than 1, model will report the dice
coefficient for each label as metric.
:param n_base_filters: The number of filters that the first layer in the convolution network will have. Following
layers will contain a multiple of this number. Lowering this number will likely reduce the amount of memory required
to train the model.
:param depth: indicates the depth of the U-shape for the model. The greater the depth, the more max pooling
layers will be added to the model. Lowering the depth may reduce the amount of memory required for training.
:param input_shape: Shape of the input data (n_chanels, x_size, y_size, z_size). The x, y, and z sizes must be
divisible by the pool size to the power of the depth of the UNet, that is pool_size^depth.
:param pool_size: Pool size for the max pooling operations.
:param n_labels: Number of binary labels that the model is learning.
:param initial_learning_rate: Initial learning rate for the model. This will be decayed during training.
:param deconvolution: If set to True, will use transpose convolution(deconvolution) instead of up-sampling. This
increases the amount memory required during training.
:return: Untrained 3D UNet Model
"""
inputs = Input(input_shape)
current_layer = inputs
levels = list()
# add levels with max pooling
for layer_depth in range(depth):
layer1 = create_convolution_block(input_layer=current_layer, n_filters=n_base_filters*(2**layer_depth),
batch_normalization=batch_normalization)
layer2 = create_convolution_block(input_layer=layer1, n_filters=n_base_filters*(2**layer_depth)*2,
batch_normalization=batch_normalization)
if layer_depth < depth - 1:
current_layer = MaxPooling3D(pool_size=pool_size)(layer2)
levels.append([layer1, layer2, current_layer])
else:
current_layer = layer2
levels.append([layer1, layer2])
# add levels with up-convolution or up-sampling
for layer_depth in range(depth-2, -1, -1):
up_convolution = get_up_convolution(pool_size=pool_size, deconvolution=deconvolution,
n_filters=current_layer._keras_shape[1])(current_layer)
concat = concatenate([up_convolution, levels[layer_depth][1]], axis=1)
current_layer = create_convolution_block(n_filters=levels[layer_depth][1]._keras_shape[1],
input_layer=concat, batch_normalization=batch_normalization)
current_layer = create_convolution_block(n_filters=levels[layer_depth][1]._keras_shape[1],
input_layer=current_layer,
batch_normalization=batch_normalization)
final_convolution = Conv3D(n_labels, (1, 1, 1))(current_layer)
act = Activation(activation_name)(final_convolution)
model = Model(inputs=inputs, outputs=act)
if not isinstance(metrics, list):
metrics = [metrics]
if include_label_wise_dice_coefficients and n_labels > 1:
label_wise_dice_metrics = [get_label_dice_coefficient_function(index) for index in range(n_labels)]
if metrics:
metrics = metrics + label_wise_dice_metrics
else:
metrics = label_wise_dice_metrics
model.compile(optimizer=Adam(lr=initial_learning_rate), loss=dice_coefficient_loss, metrics=metrics)
return model
def create_convolution_block(input_layer, n_filters, batch_normalization=False, kernel=(3, 3, 3), activation=None,
padding='same', strides=(1, 1, 1), instance_normalization=False):
"""
:param strides:
:param input_layer:
:param n_filters:
:param batch_normalization:
:param kernel:
:param activation: Keras activation layer to use. (default is 'relu')
:param padding:
:return:
"""
layer = Conv3D(n_filters, kernel, padding=padding, strides=strides)(input_layer)
if batch_normalization:
layer = BatchNormalization(axis=1)(layer)
elif instance_normalization:
try:
from keras_contrib.layers.normalization.instancenormalization import InstanceNormalization
except ImportError:
raise ImportError("Install keras_contrib in order to use instance normalization."
"\nTry: pip install git+https://www.github.com/farizrahman4u/keras-contrib.git")
layer = InstanceNormalization(axis=1)(layer)
if activation is None:
return Activation('relu')(layer)
else:
return activation()(layer)
def compute_level_output_shape(n_filters, depth, pool_size, image_shape):
"""
Each level has a particular output shape based on the number of filters used in that level and the depth or number
of max pooling operations that have been done on the data at that point.
:param image_shape: shape of the 3d image.
:param pool_size: the pool_size parameter used in the max pooling operation.
:param n_filters: Number of filters used by the last node in a given level.
:param depth: The number of levels down in the U-shaped model a given node is.
:return: 5D vector of the shape of the output node
"""
output_image_shape = np.asarray(np.divide(image_shape, np.power(pool_size, depth)), dtype=np.int32).tolist()
return tuple([None, n_filters] + output_image_shape)
def get_up_convolution(n_filters, pool_size, kernel_size=(2, 2, 2), strides=(2, 2, 2),
deconvolution=False):
if deconvolution:
return Deconvolution3D(filters=n_filters, kernel_size=kernel_size,
strides=strides)
else:
return UpSampling3D(size=pool_size)
import os
import glob
#from unet3d.data import write_data_to_file, open_data_file
#from unet3d.generator import get_training_and_validation_generators
#from unet3d.model import unet_model_3d
#from unet3d.training import load_old_model, train_model
config = dict()
config["pool_size"] = (2, 2, 2) # pool size for the max pooling operations
config["image_shape"] = (144, 144, 144) # This determines what shape the images will be cropped/resampled to.
config["patch_shape"] = (64, 64, 64) # switch to None to train on the whole image
config["labels"] = (1, 2, 4) # the label numbers on the input image
config["n_labels"] = len(config["labels"])
config["all_modalities"] = ["t1", "t1ce", "flair", "t2"]
config["training_modalities"] = config["all_modalities"] # change this if you want to only use some of the modalities
config["nb_channels"] = len(config["training_modalities"])
if "patch_shape" in config and config["patch_shape"] is not None:
config["input_shape"] = tuple([config["nb_channels"]] + list(config["patch_shape"]))
else:
config["input_shape"] = tuple([config["nb_channels"]] + list(config["image_shape"]))
config["truth_channel"] = config["nb_channels"]
config["deconvolution"] = True # if False, will use upsampling instead of deconvolution
config["batch_size"] = 6
config["validation_batch_size"] = 12
config["n_epochs"] = 500 # cutoff the training after this many epochs
config["patience"] = 10 # learning rate will be reduced after this many epochs if the validation loss is not improving
config["early_stop"] = 50 # training will be stopped after this many epochs without the validation loss improving
config["initial_learning_rate"] = 0.00001
config["learning_rate_drop"] = 0.5 # factor by which the learning rate will be reduced
config["validation_split"] = 0.8 # portion of the data that will be used for training
config["flip"] = False # augments the data by randomly flipping an axis during
config["permute"] = True # data shape must be a cube. Augments the data by permuting in various directions
config["distort"] = None # switch to None if you want no distortion
config["augment"] = config["flip"] or config["distort"]
config["validation_patch_overlap"] = 0 # if > 0, during training, validation patches will be overlapping
config["training_patch_start_offset"] = (16, 16, 16) # randomly offset the first patch index by up to this offset
config["skip_blank"] = True # if True, then patches without any target will be skipped
config["data_file"] = os.path.abspath("/content/drive/My Drive/Brats2019/data.h5")
config["model_file"] = os.path.abspath("/content/drive/My Drive/Brats2019/tumor_segmentation_model.h5")
config["training_file"] = os.path.abspath("/content/drive/My Drive/Brats2019/pkl/training_ids.pkl")
config["validation_file"] = os.path.abspath("/content/drive/My Drive/Brats2019/pkl/validation_ids.pkl")
config["overwrite"] = False # If True, will previous files. If False, will use previously written files.
def fetch_training_data_files():
training_data_files = list()
for subject_dir in glob.glob(os.path.join(os.path.dirname(__file__), "data", "preprocessed", "*", "*")):
subject_files = list()
for modality in config["training_modalities"] + ["truth"]:
subject_files.append(os.path.join(subject_dir, modality + ".nii.gz"))
training_data_files.append(tuple(subject_files))
return training_data_files
def main(overwrite=False):
# convert input images into an hdf5 file
if overwrite or not os.path.exists(config["data_file"]):
training_files = fetch_training_data_files()
write_data_to_file(training_files, config["data_file"], image_shape=config["image_shape"])
data_file_opened = open_data_file(config["data_file"])
if not overwrite and os.path.exists(config["model_file"]):
model = load_old_model(config["model_file"])
else:
# instantiate new model
model = unet_model_3d(input_shape=config["input_shape"],
pool_size=config["pool_size"],
n_labels=config["n_labels"],
initial_learning_rate=config["initial_learning_rate"],
deconvolution=config["deconvolution"])
# get training and testing generators
train_generator, validation_generator, n_train_steps, n_validation_steps = get_training_and_validation_generators(
data_file_opened,
batch_size=config["batch_size"],
data_split=config["validation_split"],
overwrite=overwrite,
validation_keys_file=config["validation_file"],
training_keys_file=config["training_file"],
n_labels=config["n_labels"],
labels=config["labels"],
patch_shape=config["patch_shape"],
validation_batch_size=config["validation_batch_size"],
validation_patch_overlap=config["validation_patch_overlap"],
training_patch_start_offset=config["training_patch_start_offset"],
permute=config["permute"],
augment=config["augment"],
skip_blank=config["skip_blank"],
augment_flip=config["flip"],
augment_distortion_factor=config["distort"])
# run training
train_model(model=model,
model_file=config["model_file"],
training_generator=train_generator,
validation_generator=validation_generator,
steps_per_epoch=n_train_steps,
validation_steps=n_validation_steps,
initial_learning_rate=config["initial_learning_rate"],
learning_rate_drop=config["learning_rate_drop"],
learning_rate_patience=config["patience"],
early_stopping_patience=config["early_stop"],
n_epochs=config["n_epochs"])
data_file_opened.close()
if __name__ == "__main__":
main(overwrite=config["overwrite"])
```
| github_jupyter |
```
import pandas as pd
import numpy as np
from tools import acc_score
df_train = pd.read_csv("../data/train.csv", index_col=0)
df_test = pd.read_csv("../data/test.csv", index_col=0)
train_bins = seq_to_num(df_train.Sequence, target_split=True, pad=True, pad_adaptive=True,
pad_maxlen=100, dtype=np.float32, drop_na_inf=True,
nbins=5, bins_by='terms')
test_bins = seq_to_num(df_test.Sequence, target_split=True, pad_adaptive=True,
dtype=np.float32, drop_na_inf=True, nbins=5, bins_by='terms')
train_X, train_y, _ = train_bins[4]
test_X, test_y, test_idx = test_bins[4]
from sklearn.tree import DecisionTreeRegressor, ExtraTreeRegressor
dt = DecisionTreeRegressor(random_state=42)
dt.fit(train_X, train_y)
acc_score(dt.predict(test_X), test_y)
train_X2, train_y2, _ = train_bins[1]
test_X2, test_y2, _ = test_bins[1]
etr = ExtraTreeRegressor(max_depth=100, random_state=42)
etr.fit(train_X2, train_y2)
acc_score(etr.predict(test_X2), test_y2)
# too long sequence?
train_X2, train_y2 = train_bins[2]
test_X2, test_y2 = test_bins[2]
etr = DecisionTreeRegressor(max_depth=5, random_state=42)
etr.fit(train_X2, train_y2)
acc_score(etr.predict(test_X2), test_y2)
from sklearn.neural_network import MLPRegressor
# NNet still doesn't work
mlp = MLPRegressor(hidden_layer_sizes=(10, 1))
mlp.fit(train_X, train_y)
acc_score(mlp.predict(test_X), test_y)
# Try to combine predictions for bin 3 and 4 (by terms), while
# fallback to mode on bin 0, 1, 2
def mmode(arr):
modes = []
for row in arr:
counts = {i: row.tolist().count(i) for i in row}
if len(counts) > 0:
modes.append(max(counts.items(), key=lambda x:x[1])[0])
else:
modes.append(0)
return modes
kg_train = pd.read_csv('../data/kaggle_train.csv', index_col=0)
kg_test = pd.read_csv('../data/kaggle_test.csv', index_col=0)
train_bins = seq_to_num(kg_train.Sequence, target_split=True,
pad_adaptive=True, dtype=np.float32, drop_na_inf=True,
nbins=5, bins_by='terms')
test_bins = seq_to_num(kg_test.Sequence, target_split=False, pad_adaptive=True,
dtype=np.float32, drop_na_inf=True, nbins=5, bins_by='terms')
bin3_X, bin3_y, _ = train_bins[3]
bin4_X, bin4_y, _ = train_bins[4]
dt_bin3 = DecisionTreeRegressor(random_state=42)
dt_bin4 = DecisionTreeRegressor(random_state=42)
dt_bin3.fit(bin3_X, bin3_y)
dt_bin4.fit(bin4_X, bin4_y)
pred_bin3 = dt_bin3.predict(test_bins[3][0])
pred_bin4 = dt_bin4.predict(test_bins[4][0])
test_bins[3][1].shape, pred_bin3.shape
pred_bin0 = mmode(test_bins[0])
pred_bin1 = mmode(test_bins[1])
pred_bin2 = mmode(test_bins[2])
pred3 = pd.Series(pred_bin3, index=test_bins[3][1], dtype=object).map(lambda x: int(x))
pred4 = pd.Series(pred_bin4, index=test_bins[4][1], dtype=object).map(lambda x: int(x))
pred_total = pd.Series(np.zeros(kg_test.shape[0]), index=kg_test.index, dtype=np.int64)
pred_total[test_bins[3][1]] = pred_bin3
pred_total[test_bins[4][1]] = pred_bin4
prep_submit(pred_total)
```
| github_jupyter |
# 📃 Solution of Exercise M6.01
The aim of this notebook is to investigate if we can tune the hyperparameters
of a bagging regressor and evaluate the gain obtained.
We will load the California housing dataset and split it into a training and
a testing set.
```
from sklearn.datasets import fetch_california_housing
from sklearn.model_selection import train_test_split
data, target = fetch_california_housing(as_frame=True, return_X_y=True)
target *= 100 # rescale the target in k$
data_train, data_test, target_train, target_test = train_test_split(
data, target, random_state=0, test_size=0.5)
```
<div class="admonition note alert alert-info">
<p class="first admonition-title" style="font-weight: bold;">Note</p>
<p class="last">If you want a deeper overview regarding this dataset, you can refer to the
Appendix - Datasets description section at the end of this MOOC.</p>
</div>
Create a `BaggingRegressor` and provide a `DecisionTreeRegressor`
to its parameter `base_estimator`. Train the regressor and evaluate its
statistical performance on the testing set using the mean absolute error.
```
from sklearn.metrics import mean_absolute_error
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import BaggingRegressor
tree = DecisionTreeRegressor()
bagging = BaggingRegressor(base_estimator=tree, n_jobs=-1)
bagging.fit(data_train, target_train)
target_predicted = bagging.predict(data_test)
print(f"Basic mean absolute error of the bagging regressor:\n"
f"{mean_absolute_error(target_test, target_predicted):.2f} k$")
abs(target_test - target_predicted).mean()
```
Now, create a `RandomizedSearchCV` instance using the previous model and
tune the important parameters of the bagging regressor. Find the best
parameters and check if you are able to find a set of parameters that
improve the default regressor still using the mean absolute error as a
metric.
<div class="admonition tip alert alert-warning">
<p class="first admonition-title" style="font-weight: bold;">Tip</p>
<p class="last">You can list the bagging regressor's parameters using the <tt class="docutils literal">get_params</tt>
method.</p>
</div>
```
for param in bagging.get_params().keys():
print(param)
from scipy.stats import randint
from sklearn.model_selection import RandomizedSearchCV
param_grid = {
"n_estimators": randint(10, 30),
"max_samples": [0.5, 0.8, 1.0],
"max_features": [0.5, 0.8, 1.0],
"base_estimator__max_depth": randint(3, 10),
}
search = RandomizedSearchCV(
bagging, param_grid, n_iter=20, scoring="neg_mean_absolute_error"
)
_ = search.fit(data_train, target_train)
import pandas as pd
columns = [f"param_{name}" for name in param_grid.keys()]
columns += ["mean_test_score", "std_test_score", "rank_test_score"]
cv_results = pd.DataFrame(search.cv_results_)
cv_results = cv_results[columns].sort_values(by="rank_test_score")
cv_results["mean_test_score"] = -cv_results["mean_test_score"]
cv_results
target_predicted = search.predict(data_test)
print(f"Mean absolute error after tuning of the bagging regressor:\n"
f"{mean_absolute_error(target_test, target_predicted):.2f} k$")
```
We see that the predictor provided by the bagging regressor does not need
much hyperparameter tuning compared to a single decision tree. We see that
the bagging regressor provides a predictor for which tuning the
hyperparameters is not as important as in the case of fitting a single
decision tree.
| github_jupyter |
## Recommendations with MovieTweetings: Collaborative Filtering
One of the most popular methods for making recommendations is **collaborative filtering**. In collaborative filtering, you are using the collaboration of user-item recommendations to assist in making new recommendations.
There are two main methods of performing collaborative filtering:
1. **Neighborhood-Based Collaborative Filtering**, which is based on the idea that we can either correlate items that are similar to provide recommendations or we can correlate users to one another to provide recommendations.
2. **Model Based Collaborative Filtering**, which is based on the idea that we can use machine learning and other mathematical models to understand the relationships that exist amongst items and users to predict ratings and provide ratings.
In this notebook, you will be working on performing **neighborhood-based collaborative filtering**. There are two main methods for performing collaborative filtering:
1. **User-based collaborative filtering:** In this type of recommendation, users related to the user you would like to make recommendations for are used to create a recommendation.
2. **Item-based collaborative filtering:** In this type of recommendation, first you need to find the items that are most related to each other item (based on similar ratings). Then you can use the ratings of an individual on those similar items to understand if a user will like the new item.
In this notebook you will be implementing **user-based collaborative filtering**. However, it is easy to extend this approach to make recommendations using **item-based collaborative filtering**. First, let's read in our data and necessary libraries.
**NOTE**: Because of the size of the datasets, some of your code cells here will take a while to execute, so be patient!
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import tests as t
from scipy.sparse import csr_matrix
from IPython.display import HTML
%matplotlib inline
# Read in the datasets
movies = pd.read_csv('movies_clean.csv')
reviews = pd.read_csv('reviews_clean.csv')
del movies['Unnamed: 0']
del reviews['Unnamed: 0']
print(reviews.head())
```
### Measures of Similarity
When using **neighborhood** based collaborative filtering, it is important to understand how to measure the similarity of users or items to one another.
There are a number of ways in which we might measure the similarity between two vectors (which might be two users or two items). In this notebook, we will look specifically at two measures used to compare vectors:
* **Pearson's correlation coefficient**
Pearson's correlation coefficient is a measure of the strength and direction of a linear relationship. The value for this coefficient is a value between -1 and 1 where -1 indicates a strong, negative linear relationship and 1 indicates a strong, positive linear relationship.
If we have two vectors x and y, we can define the correlation between the vectors as:
$$CORR(x, y) = \frac{\text{COV}(x, y)}{\text{STDEV}(x)\text{ }\text{STDEV}(y)}$$
where
$$\text{STDEV}(x) = \sqrt{\frac{1}{n-1}\sum_{i=1}^{n}(x_i - \bar{x})^2}$$
and
$$\text{COV}(x, y) = \frac{1}{n-1}\sum_{i=1}^{n}(x_i - \bar{x})(y_i - \bar{y})$$
where n is the length of the vector, which must be the same for both x and y and $\bar{x}$ is the mean of the observations in the vector.
We can use the correlation coefficient to indicate how alike two vectors are to one another, where the closer to 1 the coefficient, the more alike the vectors are to one another. There are some potential downsides to using this metric as a measure of similarity. You will see some of these throughout this workbook.
* **Euclidean distance**
Euclidean distance is a measure of the straightline distance from one vector to another. Because this is a measure of distance, larger values are an indication that two vectors are different from one another (which is different than Pearson's correlation coefficient).
Specifically, the euclidean distance between two vectors x and y is measured as:
$$ \text{EUCL}(x, y) = \sqrt{\sum_{i=1}^{n}(x_i - y_i)^2}$$
Different from the correlation coefficient, no scaling is performed in the denominator. Therefore, you need to make sure all of your data are on the same scale when using this metric.
**Note:** Because measuring similarity is often based on looking at the distance between vectors, it is important in these cases to scale your data or to have all data be in the same scale. In this case, we will not need to scale data because they are all on a 10 point scale, but it is always something to keep in mind!
------------
### User-Item Matrix
In order to calculate the similarities, it is common to put values in a matrix. In this matrix, users are identified by each row, and items are represented by columns.

In the above matrix, you can see that **User 1** and **User 2** both used **Item 1**, and **User 2**, **User 3**, and **User 4** all used **Item 2**. However, there are also a large number of missing values in the matrix for users who haven't used a particular item. A matrix with many missing values (like the one above) is considered **sparse**.
Our first goal for this notebook is to create the above matrix with the **reviews** dataset. However, instead of 1 values in each cell, you should have the actual rating.
The users will indicate the rows, and the movies will exist across the columns. To create the user-item matrix, we only need the first three columns of the **reviews** dataframe, which you can see by running the cell below.
```
user_items = reviews[['user_id', 'movie_id', 'rating']]
user_items.head()
```
### Creating the User-Item Matrix
In order to create the user-items matrix (like the one above), I personally started by using a [pivot table](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.pivot_table.html).
However, I quickly ran into a memory error (a common theme throughout this notebook). I will help you navigate around many of the errors I had, and achieve useful collaborative filtering results!
_____
`1.` Create a matrix where the users are the rows, the movies are the columns, and the ratings exist in each cell, or a NaN exists in cells where a user hasn't rated a particular movie. If you get a memory error (like I did), [this link here](https://stackoverflow.com/questions/39648991/pandas-dataframe-pivot-memory-error) might help you!
```
# Create user-by-item matrix
user_by_movie = user_items.groupby(['user_id', 'movie_id'])['rating'].max().unstack()
```
Check your results below to make sure your matrix is ready for the upcoming sections.
```
assert movies.shape[0] == user_by_movie.shape[1], "Oh no! Your matrix should have {} columns, and yours has {}!".format(movies.shape[0], user_by_movie.shape[1])
assert reviews.user_id.nunique() == user_by_movie.shape[0], "Oh no! Your matrix should have {} rows, and yours has {}!".format(reviews.user_id.nunique(), user_by_movie.shape[0])
print("Looks like you are all set! Proceed!")
HTML('<img src="images/greatjob.webp">')
```
`2.` Now that you have a matrix of users by movies, use this matrix to create a dictionary where the key is each user and the value is an array of the movies each user has rated.
```
# Create a dictionary with users and corresponding movies seen
def movies_watched(user_id):
'''
INPUT:
user_id - the user_id of an individual as int
OUTPUT:
movies - an array of movies the user has watched
'''
movies = user_by_movie.loc[user_id][user_by_movie.loc[user_id].isnull() == False].index.values
return movies
def create_user_movie_dict():
'''
INPUT: None
OUTPUT: movies_seen - a dictionary where each key is a user_id and the value is an array of movie_ids
Creates the movies_seen dictionary
'''
n_users = user_by_movie.shape[0]
movies_seen = dict()
for user1 in range(1, n_users+1):
# assign list of movies to each user key
movies_seen[user1] = movies_watched(user1)
return movies_seen
movies_seen = create_user_movie_dict()
```
`3.` If a user hasn't rated more than 2 movies, we consider these users "too new". Create a new dictionary that only contains users who have rated more than 2 movies. This dictionary will be used for all the final steps of this workbook.
```
# Remove individuals who have watched 2 or fewer movies - don't have enough data to make recs
def create_movies_to_analyze(movies_seen, lower_bound=2):
'''
INPUT:
movies_seen - a dictionary where each key is a user_id and the value is an array of movie_ids
lower_bound - (an int) a user must have more movies seen than the lower bound to be added to the movies_to_analyze dictionary
OUTPUT:
movies_to_analyze - a dictionary where each key is a user_id and the value is an array of movie_ids
The movies_seen and movies_to_analyze dictionaries should be the same except that the output dictionary has removed
'''
movies_to_analyze = dict()
for user, movies in movies_seen.items():
if len(movies) > lower_bound:
movies_to_analyze[user] = movies
return movies_to_analyze
movies_to_analyze = create_movies_to_analyze(movies_seen)
# Run the tests below to check that your movies_to_analyze matches the solution
assert len(movies_to_analyze) == 23512, "Oops! It doesn't look like your dictionary has the right number of individuals."
assert len(movies_to_analyze[2]) == 23, "Oops! User 2 didn't match the number of movies we thought they would have."
assert len(movies_to_analyze[7]) == 3, "Oops! User 7 didn't match the number of movies we thought they would have."
print("If this is all you see, you are good to go!")
```
### Calculating User Similarities
Now that you have set up the **movies_to_analyze** dictionary, it is time to take a closer look at the similarities between users. Below is the pseudocode for how I thought about determining the similarity between users:
```
for user1 in movies_to_analyze
for user2 in movies_to_analyze
see how many movies match between the two users
if more than two movies in common
pull the overlapping movies
compute the distance/similarity metric between ratings on the same movies for the two users
store the users and the distance metric
```
However, this took a very long time to run, and other methods of performing these operations did not fit on the workspace memory!
Therefore, rather than creating a dataframe with all possible pairings of users in our data, your task for this question is to look at a few specific examples of the correlation between ratings given by two users. For this question consider you want to compute the [correlation](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.corr.html) between users.
`4.` Using the **movies_to_analyze** dictionary and **user_by_movie** dataframe, create a function that computes the correlation between the ratings of similar movies for two users. Then use your function to compare your results to ours using the tests below.
```
def compute_correlation(user1, user2):
'''
INPUT
user1 - int user_id
user2 - int user_id
OUTPUT
the correlation between the matching ratings between the two users
'''
# Pull movies for each user
movies1 = movies_to_analyze[user1]
movies2 = movies_to_analyze[user2]
# Find Similar Movies
sim_movs = np.intersect1d(movies1, movies2, assume_unique=True)
# Calculate correlation between the users
df = user_by_movie.loc[(user1, user2), sim_movs]
corr = df.transpose().corr().iloc[0,1]
return corr #return the correlation
# Test your function against the solution
assert compute_correlation(2,2) == 1.0, "Oops! The correlation between a user and itself should be 1.0."
assert round(compute_correlation(2,66), 2) == 0.76, "Oops! The correlation between user 2 and 66 should be about 0.76."
assert np.isnan(compute_correlation(2,104)), "Oops! The correlation between user 2 and 104 should be a NaN."
print("If this is all you see, then it looks like your function passed all of our tests!")
```
### Why the NaN's?
If the function you wrote passed all of the tests, then you have correctly set up your function to calculate the correlation between any two users.
`5.` But one question is, why are we still obtaining **NaN** values? As you can see in the code cell above, users 2 and 104 have a correlation of **NaN**. Why?
Think and write your ideas here about why these NaNs exist, and use the cells below to do some coding to validate your thoughts. You can check other pairs of users and see that there are actually many NaNs in our data - 2,526,710 of them in fact. These NaN's ultimately make the correlation coefficient a less than optimal measure of similarity between two users.
```
In the denominator of the correlation coefficient, we calculate the standard deviation for each user's ratings. The ratings for user 2 are all the same rating on the movies that match with user 104. Therefore, the standard deviation is 0. Because a 0 is in the denominator of the correlation coefficient, we end up with a **NaN** correlation coefficient. Therefore, a different approach is likely better for this particular situation.
```
```
# Which movies did both user 2 and user 104 see?
set_2 = set(movies_to_analyze[2])
set_104 = set(movies_to_analyze[104])
set_2.intersection(set_104)
# What were the ratings for each user on those movies?
print(user_by_movie.loc[2, set_2.intersection(set_104)])
print(user_by_movie.loc[104, set_2.intersection(set_104)])
```
`6.` Because the correlation coefficient proved to be less than optimal for relating user ratings to one another, we could instead calculate the euclidean distance between the ratings. I found [this post](https://stackoverflow.com/questions/1401712/how-can-the-euclidean-distance-be-calculated-with-numpy) particularly helpful when I was setting up my function. This function should be very similar to your previous function. When you feel confident with your function, test it against our results.
```
def compute_euclidean_dist(user1, user2):
'''
INPUT
user1 - int user_id
user2 - int user_id
OUTPUT
the euclidean distance between user1 and user2
'''
# Pull movies for each user
movies1 = movies_to_analyze[user1]
movies2 = movies_to_analyze[user2]
# Find Similar Movies
sim_movs = np.intersect1d(movies1, movies2, assume_unique=True)
# Calculate euclidean distance between the users
df = user_by_movie.loc[(user1, user2), sim_movs]
dist = np.linalg.norm(df.loc[user1] - df.loc[user2])
return dist #return the euclidean distance
# Read in solution euclidean distances"
import pickle
df_dists = pd.read_pickle("data/Term2/recommendations/lesson1/data/dists.p")
# Test your function against the solution
assert compute_euclidean_dist(2,2) == df_dists.query("user1 == 2 and user2 == 2")['eucl_dist'][0], "Oops! The distance between a user and itself should be 0.0."
assert round(compute_euclidean_dist(2,66), 2) == round(df_dists.query("user1 == 2 and user2 == 66")['eucl_dist'][1], 2), "Oops! The distance between user 2 and 66 should be about 2.24."
assert np.isnan(compute_euclidean_dist(2,104)) == np.isnan(df_dists.query("user1 == 2 and user2 == 104")['eucl_dist'][4]), "Oops! The distance between user 2 and 104 should be 2."
print("If this is all you see, then it looks like your function passed all of our tests!")
```
### Using the Nearest Neighbors to Make Recommendations
In the previous question, you read in **df_dists**. Therefore, you have a measure of distance between each user and every other user. This dataframe holds every possible pairing of users, as well as the corresponding euclidean distance.
Because of the **NaN** values that exist within the correlations of the matching ratings for many pairs of users, as we discussed above, we will proceed using **df_dists**. You will want to find the users that are 'nearest' each user. Then you will want to find the movies the closest neighbors have liked to recommend to each user.
I made use of the following objects:
* df_dists (to obtain the neighbors)
* user_items (to obtain the movies the neighbors and users have rated)
* movies (to obtain the names of the movies)
`7.` Complete the functions below, which allow you to find the recommendations for any user. There are five functions which you will need:
* **find_closest_neighbors** - this returns a list of user_ids from closest neighbor to farthest neighbor using euclidean distance
* **movies_liked** - returns an array of movie_ids
* **movie_names** - takes the output of movies_liked and returns a list of movie names associated with the movie_ids
* **make_recommendations** - takes a user id and goes through closest neighbors to return a list of movie names as recommendations
* **all_recommendations** = loops through every user and returns a dictionary of with the key as a user_id and the value as a list of movie recommendations
```
def find_closest_neighbors(user):
'''
INPUT:
user - (int) the user_id of the individual you want to find the closest users
OUTPUT:
closest_neighbors - an array of the id's of the users sorted from closest to farthest away
'''
# I treated ties as arbitrary and just kept whichever was easiest to keep using the head method
# You might choose to do something less hand wavy
closest_users = df_dists[df_dists['user1']==user].sort_values(by='eucl_dist').iloc[1:]['user2']
closest_neighbors = np.array(closest_users)
return closest_neighbors
def movies_liked(user_id, min_rating=7):
'''
INPUT:
user_id - the user_id of an individual as int
min_rating - the minimum rating considered while still a movie is still a "like" and not a "dislike"
OUTPUT:
movies_liked - an array of movies the user has watched and liked
'''
movies_liked = np.array(user_items.query('user_id == @user_id and rating > (@min_rating -1)')['movie_id'])
return movies_liked
def movie_names(movie_ids):
'''
INPUT
movie_ids - a list of movie_ids
OUTPUT
movies - a list of movie names associated with the movie_ids
'''
movie_lst = list(movies[movies['movie_id'].isin(movie_ids)]['movie'])
return movie_lst
def make_recommendations(user, num_recs=10):
'''
INPUT:
user - (int) a user_id of the individual you want to make recommendations for
num_recs - (int) number of movies to return
OUTPUT:
recommendations - a list of movies - if there are "num_recs" recommendations return this many
otherwise return the total number of recommendations available for the "user"
which may just be an empty list
'''
# I wanted to make recommendations by pulling different movies than the user has already seen
# Go in order from closest to farthest to find movies you would recommend
# I also only considered movies where the closest user rated the movie as a 9 or 10
# movies_seen by user (we don't want to recommend these)
movies_seen = movies_watched(user)
closest_neighbors = find_closest_neighbors(user)
# Keep the recommended movies here
recs = np.array([])
# Go through the neighbors and identify movies they like the user hasn't seen
for neighbor in closest_neighbors:
neighbs_likes = movies_liked(neighbor)
#Obtain recommendations for each neighbor
new_recs = np.setdiff1d(neighbs_likes, movies_seen, assume_unique=True)
# Update recs with new recs
recs = np.unique(np.concatenate([new_recs, recs], axis=0))
# If we have enough recommendations exit the loop
if len(recs) > num_recs-1:
break
# Pull movie titles using movie ids
recommendations = movie_names(recs)
return recommendations
def all_recommendations(num_recs=10):
'''
INPUT
num_recs (int) the (max) number of recommendations for each user
OUTPUT
all_recs - a dictionary where each key is a user_id and the value is an array of recommended movie titles
'''
# All the users we need to make recommendations for
users = np.unique(df_dists['user1'])
n_users = len(users)
#Store all recommendations in this dictionary
all_recs = dict()
# Make the recommendations for each user
for user in users:
all_recs[user] = make_recommendations(user, num_recs)
return all_recs
all_recs = all_recommendations(10)
# This loads our solution dictionary so you can compare results - FULL PATH IS "data/Term2/recommendations/lesson1/data/all_recs.p"
all_recs_sol = pd.read_pickle("data/Term2/recommendations/lesson1/data/all_recs.p")
assert all_recs[2] == make_recommendations(2), "Oops! Your recommendations for user 2 didn't match ours."
assert all_recs[26] == make_recommendations(26), "Oops! It actually wasn't possible to make any recommendations for user 26."
assert all_recs[1503] == make_recommendations(1503), "Oops! Looks like your solution for user 1503 didn't match ours."
print("If you made it here, you now have recommendations for many users using collaborative filtering!")
HTML('<img src="images/greatjob.webp">')
```
### Now What?
If you made it this far, you have successfully implemented a solution to making recommendations using collaborative filtering.
`8.` Let's do a quick recap of the steps taken to obtain recommendations using collaborative filtering.
```
# Check your understanding of the results by correctly filling in the dictionary below
a = "pearson's correlation and spearman's correlation"
b = 'item based collaborative filtering'
c = "there were too many ratings to get a stable metric"
d = 'user based collaborative filtering'
e = "euclidean distance and pearson's correlation coefficient"
f = "manhattan distance and euclidean distance"
g = "spearman's correlation and euclidean distance"
h = "the spread in some ratings was zero"
i = 'content based recommendation'
sol_dict = {
'The type of recommendation system implemented here was a ...': d,
'The two methods used to estimate user similarity were: ': e,
'There was an issue with using the correlation coefficient. What was it?': h
}
t.test_recs(sol_dict)
```
Additionally, let's take a closer look at some of the results. There are two solution files that you read in to check your results, and you created these objects
* **df_dists** - a dataframe of user1, user2, euclidean distance between the two users
* **all_recs_sol** - a dictionary of all recommendations (key = user, value = list of recommendations)
`9.` Use these two objects along with the cells below to correctly fill in the dictionary below and complete this notebook!
```
a = 567
b = 1503
c = 1319
d = 1325
e = 2526710
f = 0
g = 'Use another method to make recommendations - content based, knowledge based, or model based collaborative filtering'
sol_dict2 = {
'For how many pairs of users were we not able to obtain a measure of similarity using correlation?': e,
'For how many pairs of users were we not able to obtain a measure of similarity using euclidean distance?': f,
'For how many users were we unable to make any recommendations for using collaborative filtering?': c,
'For how many users were we unable to make 10 recommendations for using collaborative filtering?': d,
'What might be a way for us to get 10 recommendations for every user?': g
}
t.test_recs2(sol_dict2)
# Use the cells below for any work you need to do!
# Users without recs
users_without_recs = []
for user, movie_recs in all_recs.items():
if len(movie_recs) == 0:
users_without_recs.append(user)
len(users_without_recs)
# NaN euclidean distance values
df_dists['eucl_dist'].isnull().sum()
# Users with fewer than 10 recs
users_with_less_than_10recs = []
for user, movie_recs in all_recs.items():
if len(movie_recs) < 10:
users_with_less_than_10recs.append(user)
len(users_with_less_than_10recs)
```
| github_jupyter |
### Feature Engineering notebook
This is a demo notebook to play with feature engineering toolkit. In this notebook we will see some capabilities of the toolkit like filling missing values, PCA, Random Projections, Normalizing values, and etc.
```
%load_ext autoreload
%autoreload 1
%matplotlib inline
from Pipeline import Pipeline
from Compare import Compare
from StructuredData.LoadCSV import LoadCSV
from StructuredData.MissingValues import MissingValues
from StructuredData.Normalize import Normalize
from StructuredData.Factorize import Factorize
from StructuredData.PCAFeatures import PCAFeatures
from StructuredData.RandomProjection import RandomProjection
csv_path = './DemoData/synthetic_classification.csv'
df = LoadCSV(csv_path)()
df.head(5)
```
### Filling missing values
By default, median of the values of the column is applied for filling out the missing values
```
pipelineObj = Pipeline([MissingValues()])
new_df = pipelineObj(df, '0')
new_df.head(5)
```
However, the imputation type is a configurable parameter to customize it as per needs.
```
pipelineObj = Pipeline([MissingValues(imputation_type = 'mean')])
new_df = pipelineObj(df, '0')
new_df.head(5)
```
### Normalize data
By default, Min max normalization is applied. Please note that assertion has been set such that normlization cant be applied if there rae missing values in that column. This is part of validation phase
```
pipelineObj = Pipeline([MissingValues(), Normalize(['1','2', '3'])])
new_df = pipelineObj(df, '0')
df.head(5)
```
### Factorize data
Encode the object as an enumerated type or categorical variable for column 4 and 8, but we must remove missing values before Factorizing
```
pipelineObj = Pipeline([MissingValues(), Factorize(['4','8'])])
new_df = pipelineObj(df, '0')
new_df.head(5)
```
### Principal Component Analysis
Use n_components to play around with how many dimensions you want to keep. Please note that assertions will validate if a data frame has any missing values before applying PCA. In the below example, the pipeline first removed missing values before applying PCA.
```
pipelineObj = Pipeline([MissingValues(imputation_type = 'mean'), PCAFeatures(n_components = 5)])
pca_df = pipelineObj(df, '0')
pca_df.head(5)
```
### Random Projections
Use n_components to play around with how many dimensions you want to keep. Please note that assertions will validate if a data frame has any missing values before applying Random Projections. Type of projections can be specified as an argument, by default GaussianRandomProjection is applied. In the below example, the pipeline first removed missing values before applying Sparse Random Projection. As of now, 'auto' deduction of number of dimensions which are sufficient to represent the features with minimal loss of information has not been implemeted, hence default value for ouput columns is 2 (Use n_components to specify custom value)
```
pipelineObj = Pipeline([MissingValues(imputation_type = 'mean'), RandomProjection(n_components = 6, proj_type = 'Sparse')])
new_df = pipelineObj(df, '0')
new_df.head()
```
### Download the modified CSV
At any point, the new tranformed features can be downloaded using below command
```
csv_path = './DemoData/synthetic_classification_transformed.csv'
new_df.to_csv(csv_path)
```
| github_jupyter |
# Figure 4: NIRCam Grism + Filter Sensitivities ($1^{st}$ order)
***
### Table of Contents
1. [Information](#Information)
2. [Imports](#Imports)
3. [Data](#Data)
4. [Generate the First Order Grism + Filter Sensitivity Plot](#Generate-the-First-Order-Grism-+-Filter-Sensitivity-Plot)
5. [Issues](#Issues)
6. [About this Notebook](#About-this-Notebook)
***
## Information
#### JDox links:
* [NIRCam Grisms](https://jwst-docs.stsci.edu/display/JTI/NIRCam+Grisms#NIRCamGrisms-Sensitivity)
* Figure 4. NIRCam grism + filter sensitivities ($1^{st}$ order)
## Imports
```
import os
import pylab
import numpy as np
from astropy.io import ascii, fits
from astropy.table import Table
from scipy.optimize import fmin
from scipy.interpolate import interp1d
import requests
import matplotlib.pyplot as plt
%matplotlib inline
```
## Data
#### Data Location:
The data is stored in a NIRCam JDox Box folder here:
[ST-INS-NIRCAM -> JDox -> nircam_grisms](https://stsci.box.com/s/wu9mo54vi957x50rdirlcg9zkkr3xiaw)
```
files = [('https://stsci.box.com/shared/static/i0a9dkp02nnuw6w0xcfd7b42ctxfb8es.fits', 'NIRCam.F250M.R.A.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/vfnyk9veote92dz1edpbu83un5n20rsw.fits', 'NIRCam.F250M.R.A.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/ssvltwzt7f4y5lfvch2o1prdk5hb2gz2.fits', 'NIRCam.F250M.R.B.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/56wjvzx1jf2i5yg7l1gg77vtvi01ec5p.fits', 'NIRCam.F250M.R.B.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/v1621dcm44be21n381mbgd2hzxxqrb2e.fits', 'NIRCam.F277W.R.A.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/8slec91wj6ety6d8qvest09msklpypi8.fits', 'NIRCam.F277W.R.A.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/r42hdv64x6skqqszv24qkxohiijitqcf.fits', 'NIRCam.F277W.R.B.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/3vye6ni05i3kdqyd5vs1jk2q59yyms2e.fits', 'NIRCam.F277W.R.B.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/twcxbe6lxrjckqph980viiijv8fpmm8b.fits', 'NIRCam.F300M.R.A.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/bpvluysg3zsl3q4b4l5rj5nue84ydjem.fits', 'NIRCam.F300M.R.A.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/15x7rbwngsxiubbexy7zcezxqm3ndq54.fits', 'NIRCam.F300M.R.B.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/a7tqdp0feqcttw3d9vaioy7syzfsftz6.fits', 'NIRCam.F300M.R.B.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/i76sb53pthieh4kn62fpxhcxn8lreffj.fits', 'NIRCam.F322W2.R.A.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/wgbyfi3ofs7i19b7zsf2iceupzkbkokq.fits', 'NIRCam.F322W2.R.A.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/jhk3deym5wbc68djtcahy3otk2xfjdb5.fits', 'NIRCam.F322W2.R.B.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/zu3xqnicbyfjn54yb4kgzvnglanf13ak.fits', 'NIRCam.F322W2.R.B.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/e2srtf52wnh6vvxsy2aiknbcr8kx2xr5.fits', 'NIRCam.F335M.R.A.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/bav3tswdd7lemsyd53bnpj4b6yke5bgd.fits', 'NIRCam.F335M.R.A.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/81wm768mjemzj84w1ogzqddgmrk3exvt.fits', 'NIRCam.F335M.R.B.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/fhopmyongqifibdtwt3qr682lwdjaf7a.fits', 'NIRCam.F335M.R.B.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/j9gd8bclethgex40o7qi1e79hgj2hsyt.fits', 'NIRCam.F356W.R.A.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/s23novi3p6qwm9f9hj9wutgju08be776.fits', 'NIRCam.F356W.R.A.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/41fnmswn1ttnwts6jj5fu73m4hs6icxd.fits', 'NIRCam.F356W.R.B.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/wx3rvjt0mvf0hnhv4wvqcmxu61gamwmm.fits', 'NIRCam.F356W.R.B.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/e0p6vkiow4jlp49deqkji9kekzdt4oon.fits', 'NIRCam.F360M.R.A.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/xbh0rjjvxn0x22k9ktiyikol7c4ep6ka.fits', 'NIRCam.F360M.R.A.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/e7artuotyv8l9wfoa3rk1k00o5mv8so8.fits', 'NIRCam.F360M.R.B.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/9r5bmick13ti22l6hcsw0uod75vqartw.fits', 'NIRCam.F360M.R.B.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/tqd1uqsf8nj12he5qa3hna0zodnlzfea.fits', 'NIRCam.F410M.R.A.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/4szffesvswh0h8fjym5m5ht37sj0jzrl.fits', 'NIRCam.F410M.R.A.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/iur0tpbts23lc5rn5n0tplzndlkoudel.fits', 'NIRCam.F410M.R.B.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/rvz8iznsnl0bsjrqiw7rv74jj24b0otb.fits', 'NIRCam.F410M.R.B.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/sv3g82qbb4u2umksgu5zdl7rp569sdi7.fits', 'NIRCam.F430M.R.A.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/mmqv1pkuzpj6abtufxxfo960z2v1oygc.fits', 'NIRCam.F430M.R.A.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/84q83haic2h6eq5c6p2frkybz551hp8d.fits', 'NIRCam.F430M.R.B.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/3osceplhq6kmvmm2a72jsgrg6z1ggw1p.fits', 'NIRCam.F430M.R.B.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/kitx7gdo5kool6jus2g19vdy7q7hmxck.fits', 'NIRCam.F444W.R.A.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/ug7y93v0en9c84hfp6d3vtjogmjou9u3.fits', 'NIRCam.F444W.R.A.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/0p9h9ofayq8q6dbfsccf3tn5lvxxod9i.fits', 'NIRCam.F444W.R.B.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/34hbqzibt5h72hm0rj9wylttj7m9wd19.fits', 'NIRCam.F444W.R.B.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/vj0rkyebg0afny1khdyiho4mktmtsi1q.fits', 'NIRCam.F460M.R.A.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/ky1z1dpewsjqab1o9hstihrec7h52oq4.fits', 'NIRCam.F460M.R.A.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/s93cwpcvnxfjwqbulnkh9ts9ln0fu9cz.fits', 'NIRCam.F460M.R.B.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/1178in8zg462es1fkl0mgcbpgp6kgb6t.fits', 'NIRCam.F460M.R.B.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/b855uj293klac8hnoqhrnv8ei0rcvudj.fits', 'NIRCam.F480M.R.A.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/werzjlp3ybxk2ovg6u689zsfpts2t8w3.fits', 'NIRCam.F480M.R.A.2nd.sensitivity.fits'),
('https://stsci.box.com/shared/static/yrh5mylru1upbo5rifbz77acn8k1ud6i.fits', 'NIRCam.F480M.R.B.1st.sensitivity.fits'),
('https://stsci.box.com/shared/static/oxu6jsg9cn9yqkh3nh646fx0flhw8rej.fits', 'NIRCam.F480M.R.B.2nd.sensitivity.fits')]
def download_file(url, file_name, output_directory='./', overwrite=False):
"""Download a file from Box given the direct URL
Parameters
----------
url : str
URL to the file to be downloaded
file_name : str
The name of the file being downloaded
output_directory : str
Directory to download file_name into
overwrite : str
If False and the file to download already exists, the download
will be skipped. If True, the file will be downloaded regardless
of whether it already exists in output_directory
Returns
-------
download_filename : str
Name of the downloaded file
"""
download_filename = os.path.join(output_directory, file_name)
if not os.path.isfile(download_filename) or overwrite is True:
print("Downloading {}".format(file_name))
with requests.get(url, stream=True) as response:
if response.status_code != 200:
raise RuntimeError("Wrong URL - {}".format(url))
with open(download_filename, 'wb') as f:
for chunk in response.iter_content(chunk_size=2048):
if chunk:
f.write(chunk)
else:
print("{} already exists. Skipping download.".format(download_filename))
return download_filename
```
#### Load the data
(The next cell assumes you downloaded the data into your ```Users/$(logname)/``` home directory)
```
if os.environ.get('LOGNAME') is None:
raise ValueError("WARNING: LOGNAME environment variable not set!")
box_directory = os.path.join("/Users/", os.environ['LOGNAME'], "box_data")
box_directory
if not os.path.isdir(box_directory):
try:
os.mkdir(box_directory)
except:
raise OSError("Unable to create {}".format(box_directory))
for file_info in files:
file_url, filename = file_info
outfile = download_file(file_url, filename, output_directory=box_directory)
grism = "R"
mod = "A"
filters = ["F250M","F277W","F300M","F322W2","F335M","F356W","F360M","F410M","F430M","F444W","F460M","F480M"]
filenames = []
for fil in filters:
filenames.append(os.path.join(box_directory, "NIRCam.%s.%s.%s.1st.sensitivity.fits" % (fil,grism,mod)))
filenames
```
## Generate the First Order Grism + Filter Sensitivity Plot
### Define some convenience functions
```
def find_nearest(array,value):
idx = (np.abs(array-value)).argmin()
return array[idx]
def find_nearest(array,value):
idx = (np.abs(array-value)).argmin()
return array[idx]
def find_mid(w,s,w0,thr=0.05):
fct = interp1d(w,s,bounds_error=None,fill_value='extrapolate')
def func(x):
#print "x:",x
return np.abs(fct(x)-thr)
res = fmin(func,w0)
return res[0]
```
### Create the plots
```
f, ax1 = plt.subplots(1, figsize=(15, 10))
NUM_COLORS = len(filters)
cm = pylab.get_cmap('tab10')
grism = "R"
mod = "A"
for i,fname in zip(range(NUM_COLORS),filenames):
color = cm(1.*i/NUM_COLORS)
d = fits.open(fname)
w = d[1].data["WAVELENGTH"]
s = d[1].data["SENSITIVITY"]/(1e17)
ax1.plot(w,s,label=fil,lw=4,color=color)
ax1.legend(fontsize=16)
miny,maxy = ax1.get_ylim()
minx,maxx = ax1.get_xlim()
ax1.set_ylim(miny,2.15)
ax1.set_xlim(2.1,maxx)
ax1.tick_params(labelsize=18)
f.text(0.5, 0.04, 'Wavelength ($\mu m$)', ha='center', fontsize=22)
f.text(0.03, 0.5, 'Sensitivity ('+r'$1 \times 10^{17}\ \frac{e^{-} s^{-1}}{erg s^{-1} cm^{-2} A^{-1}}$'+')', va='center', rotation='vertical', fontsize=22)
```
### Figure option 2: filter name positions
```
f, ax1 = plt.subplots(1, figsize=(15, 10))
thr = 0.05 # 5% of peak boundaries
NUM_COLORS = len(filters)
cm = pylab.get_cmap('tab10')
for i,fil,fname in zip(range(NUM_COLORS),filters,filenames):
color = cm(1.*i/NUM_COLORS)
d = fits.open(fname)
w = d[1].data["WAVELENGTH"]
s = d[1].data["SENSITIVITY"]/(1e17)
wmin,wmax = np.min(w),np.max(w)
vg = w<(wmax+wmin)/2.
w1 = find_mid(w[vg],s[vg],wmin,thr)
vg = w>(wmax+wmin)/2.
w2 = find_mid(w[vg],s[vg],wmax,thr)
if fil == 'F356W':
ax1.text((w2+w1)/2 -0.04, s[np.where(w == find_nearest(w, (w2+w1)/2))]+0.25, fil, ha='center',color=color,fontsize=16,weight='bold')
elif fil == 'F335M':
ax1.text((w2+w1)/2 -0.03, s[np.where(w == find_nearest(w, (w2+w1)/2))]+0.22, fil, ha='center',color=color,fontsize=16,weight='bold')
elif fil == 'F460M':
ax1.text((w2+w1)/2+0.15, s[np.where(w == find_nearest(w, (w2+w1)/2))]+0.12, fil, ha='center',color=color,fontsize=16,weight='bold')
elif fil == 'F480M':
ax1.text((w2+w1)/2+0.15, s[np.where(w == find_nearest(w, (w2+w1)/2))]+0.1, fil, ha='center',color=color,fontsize=16,weight='bold')
else:
ax1.text((w2+w1)/2 -0.04, s[np.where(w == find_nearest(w, (w2+w1)/2))]+0.2, fil, ha='center',color=color,fontsize=16,weight='bold')
ax1.plot(w,s,label=fil,lw=4,color=color)
miny,maxy = ax1.get_ylim()
minx,maxx = ax1.get_xlim()
ax1.set_ylim(miny,2.15)
ax1.set_xlim(2.1,maxx)
ax1.tick_params(labelsize=18)
f.text(0.5, 0.04, 'Wavelength ($\mu m$)', ha='center', fontsize=22)
f.text(0.03, 0.5, 'Sensitivity ('+r'$1 \times 10^{17}\ \frac{e^{-} s^{-1}}{erg\ s^{-1} cm^{-2} A^{-1}}$'+')', va='center', rotation='vertical', fontsize=22)
```
## Issues
* None
## About this Notebook
**Authors:**
Nor Pirzkal & Alicia Canipe
**Updated On:**
April 10, 2019
| github_jupyter |
**Version 2**: disable unfreezing for speed
## setup for pytorch/xla on TPU
```
import os
import collections
from datetime import datetime, timedelta
os.environ["XRT_TPU_CONFIG"] = "tpu_worker;0;10.0.0.2:8470"
_VersionConfig = collections.namedtuple('_VersionConfig', 'wheels,server')
VERSION = "torch_xla==nightly"
CONFIG = {
'torch_xla==nightly': _VersionConfig('nightly', 'XRT-dev{}'.format(
(datetime.today() - timedelta(1)).strftime('%Y%m%d')))}[VERSION]
DIST_BUCKET = 'gs://tpu-pytorch/wheels'
TORCH_WHEEL = 'torch-{}-cp36-cp36m-linux_x86_64.whl'.format(CONFIG.wheels)
TORCH_XLA_WHEEL = 'torch_xla-{}-cp36-cp36m-linux_x86_64.whl'.format(CONFIG.wheels)
TORCHVISION_WHEEL = 'torchvision-{}-cp36-cp36m-linux_x86_64.whl'.format(CONFIG.wheels)
!export LD_LIBRARY_PATH=/usr/local/lib:$LD_LIBRARY_PATH
!apt-get install libomp5 -y
!apt-get install libopenblas-dev -y
!pip uninstall -y torch torchvision
!gsutil cp "$DIST_BUCKET/$TORCH_WHEEL" .
!gsutil cp "$DIST_BUCKET/$TORCH_XLA_WHEEL" .
!gsutil cp "$DIST_BUCKET/$TORCHVISION_WHEEL" .
!pip install "$TORCH_WHEEL"
!pip install "$TORCH_XLA_WHEEL"
!pip install "$TORCHVISION_WHEEL"
```
## Imports
```
import os
import re
import cv2
import time
import tensorflow
import collections
import numpy as np
import pandas as pd
from tqdm import tqdm
from glob import glob
from PIL import Image
import requests, threading
import matplotlib.pyplot as plt
from datetime import datetime, timedelta
import torch
import torchvision
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
from torchvision import datasets
from torchvision import transforms
from torch.autograd import Variable
from torch.utils.data import Dataset, DataLoader
from torch.optim.lr_scheduler import OneCycleLR
import torch_xla
import torch_xla.utils.utils as xu
import torch_xla.core.xla_model as xm
import torch_xla.debug.metrics as met
import torch_xla.distributed.data_parallel as dp
import torch_xla.distributed.parallel_loader as pl
import torch_xla.distributed.xla_multiprocessing as xmp
import warnings
warnings.filterwarnings("ignore")
torch.manual_seed(42)
torch.set_default_tensor_type('torch.FloatTensor')
# do not uncomment see https://github.com/pytorch/xla/issues/1587
# xm.get_xla_supported_devices()
# xm.xrt_world_size() # 1
```
## Dataset
```
DATASET_DIR = '/kaggle/input/104-flowers-garden-of-eden/jpeg-512x512'
TRAIN_DIR = DATASET_DIR + '/train'
VAL_DIR = DATASET_DIR + '/val'
TEST_DIR = DATASET_DIR + '/test'
BATCH_SIZE = 16 # per core
NUM_EPOCH = 25
normalize = transforms.Normalize(mean=[0.485, 0.456, 0.406],std=[0.229, 0.224, 0.225])
train_transform = transforms.Compose([transforms.RandomResizedCrop(224),
transforms.RandomHorizontalFlip(0.5),
transforms.ToTensor(),
normalize])
valid_transform = transforms.Compose([transforms.Resize((224,224)),
transforms.ToTensor(),
normalize])
train = datasets.ImageFolder(TRAIN_DIR, transform=train_transform)
valid = datasets.ImageFolder(VAL_DIR, transform=train_transform)
train = torch.utils.data.ConcatDataset([train, valid])
# print out some data stats
print('Num training images: ', len(train))
print('Num test images: ', len(valid))
```
## Model
```
class MyModel(nn.Module):
def __init__(self):
super(MyModel, self).__init__()
self.base_model = torchvision.models.densenet201(pretrained=True)
self.base_model.classifier = nn.Identity()
self.fc = torch.nn.Sequential(
torch.nn.Linear(1920, 1024, bias = True),
torch.nn.BatchNorm1d(1024),
torch.nn.ReLU(inplace=True),
torch.nn.Dropout(0.3),
torch.nn.Linear(1024, 512, bias = True),
torch.nn.BatchNorm1d(512),
torch.nn.ReLU(inplace=True),
torch.nn.Dropout(0.3),
torch.nn.Linear(512, 104))
def forward(self, inputs):
x = self.base_model(inputs)
return self.fc(x)
model = MyModel()
print(model)
del model
```
## Training
```
def train_model():
train = datasets.ImageFolder(TRAIN_DIR, transform=train_transform)
valid = datasets.ImageFolder(VAL_DIR, transform=train_transform)
train = torch.utils.data.ConcatDataset([train, valid])
torch.manual_seed(42)
train_sampler = torch.utils.data.distributed.DistributedSampler(
train,
num_replicas=xm.xrt_world_size(),
rank=xm.get_ordinal(),
shuffle=True)
train_loader = torch.utils.data.DataLoader(
train,
batch_size=BATCH_SIZE,
sampler=train_sampler,
num_workers=0,
drop_last=True) # print(len(train_loader))
xm.master_print(f"Train for {len(train_loader)} steps per epoch")
# Scale learning rate to num cores
learning_rate = 0.0001 * xm.xrt_world_size()
# Get loss function, optimizer, and model
device = xm.xla_device()
model = MyModel()
for param in model.base_model.parameters(): # freeze some layers
param.requires_grad = False
model = model.to(device)
loss_fn = nn.CrossEntropyLoss()
optimizer = optim.Adam(model.parameters(), lr=learning_rate, weight_decay=5e-4)
scheduler = OneCycleLR(optimizer,
learning_rate,
div_factor=10.0,
final_div_factor=50.0,
epochs=NUM_EPOCH,
steps_per_epoch=len(train_loader))
def train_loop_fn(loader):
tracker = xm.RateTracker()
model.train()
total_samples, correct = 0, 0
for x, (data, target) in enumerate(loader):
optimizer.zero_grad()
output = model(data)
loss = loss_fn(output, target)
loss.backward()
xm.optimizer_step(optimizer)
tracker.add(data.shape[0])
pred = output.max(1, keepdim=True)[1]
correct += pred.eq(target.view_as(pred)).sum().item()
total_samples += data.size()[0]
scheduler.step()
if x % 40 == 0:
print('[xla:{}]({})\tLoss={:.3f}\tRate={:.2f}\tGlobalRate={:.2f}'.format(
xm.get_ordinal(), x, loss.item(), tracker.rate(),
tracker.global_rate()), flush=True)
accuracy = 100.0 * correct / total_samples
print('[xla:{}] Accuracy={:.2f}%'.format(xm.get_ordinal(), accuracy), flush=True)
return accuracy
# Train loops
accuracy = []
for epoch in range(1, NUM_EPOCH + 1):
start = time.time()
para_loader = pl.ParallelLoader(train_loader, [device])
accuracy.append(train_loop_fn(para_loader.per_device_loader(device)))
xm.master_print("Finished training epoch {} train-acc {:.2f} in {:.2f} sec"\
.format(epoch, accuracy[-1], time.time() - start))
xm.save(model.state_dict(), "./model.pt")
# if epoch == 15: #unfreeze
# for param in model.base_model.parameters():
# param.requires_grad = True
return accuracy
# Start training processes
def _mp_fn(rank, flags):
global acc_list
torch.set_default_tensor_type('torch.FloatTensor')
a = train_model()
FLAGS={}
xmp.spawn(_mp_fn, args=(FLAGS,), nprocs=8, start_method='fork')
```
| github_jupyter |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sb
import gc
prop_data = pd.read_csv("properties_2017.csv")
# prop_data
train_data = pd.read_csv("train_2017.csv")
train_data
# missing_val = prop_data.isnull().sum().reset_index()
# missing_val.columns = ['column_name', 'missing_count']
# missing_val = missing_val.loc[missing_val['missing_count']>0]
# missing_val = missing_val.sort_values(by='missing_count')
# missing_val['missing_ratio'] = missing_val["missing_count"]/prop_data.shape[0]
# missing_val = missing_val.loc[missing_val["missing_ratio"]>0.6]
# missing_val
# ind = np.arange(missing_val.shape[0])
# width = 0.9
# fig, ax = plt.subplots(figsize=(12,18))
# rects = ax.barh(ind, missing_val.missing_ratio.values, color='blue')
# ax.set_yticks(ind)
# ax.set_yticklabels(missing_val.column_name.values, rotation='horizontal')
# ax.set_xlabel("Count of missing values")
# ax.set_title("Number of missing values in each column")
# plt.show()
# del ind
# prop_data.drop(missing_val.column_name.values, axis=1, inplace=True)
prop_data
# prop_data_temp = prop_data.fillna(prop_data.mean(), )
plt.plot(prop_data.groupby("regionidcounty")["taxvaluedollarcnt"].mean())
plt.show()
original = prop_data.copy()
prop_data = original.copy()
# prop_data['actual_area'] = prop_data[['finishedfloor1squarefeet','calculatedfinishedsquarefeet','finishedsquarefeet12', 'finishedsquarefeet13',
# 'finishedsquarefeet15', 'finishedsquarefeet50', 'finishedsquarefeet6']].max(axis=1)
prop_data['actual_area'] = prop_data['calculatedfinishedsquarefeet']#.value_counts(dropna = False)
prop_data['calculatedbathnbr'].fillna(prop_data['calculatedbathnbr'].median(),inplace = True)
prop_data['bedroomcnt'].fillna(prop_data['bedroomcnt'].median(), inplace = True)
prop_data['taxvaluedollarcnt'].fillna(prop_data["taxvaluedollarcnt"].mean(), inplace=True)
prop_data['actual_area'].replace(to_replace=1.0,value=np.nan,inplace=True)
prop_data['actual_area'].fillna(prop_data['actual_area'].median(),inplace=True)
prop_data['unitcnt'].fillna(1, inplace = True)
prop_data['latitude'].fillna(prop_data['latitude'].median(),inplace = True)
prop_data['longitude'].fillna(prop_data['longitude'].median(),inplace = True)
prop_data['lotsizesquarefeet'].fillna(prop_data['lotsizesquarefeet'].median(), inplace = True)
prop_data["poolcnt"].fillna(0, inplace=True)
prop_data["fireplacecnt"].fillna(0, inplace=True)
prop_data["hashottuborspa"].fillna(0, inplace=True)
prop_data['hashottuborspa'] = pd.to_numeric(prop_data['hashottuborspa'])
prop_data["taxdelinquencyflag"].fillna(-1, inplace=True)
prop_data["taxdelinquencyflag"] = prop_data["taxdelinquencyflag"].map({'Y':1, -1:-1})
prop_data.loc[(prop_data["heatingorsystemtypeid"]==2.0) & (pd.isnull(prop_data["airconditioningtypeid"])), "airconditioningtypeid"] = 1.0
prop_data["airconditioningtypeid"].fillna(-1, inplace=True)
prop_data["buildingqualitytypeid"].fillna(7, inplace=True)
prop_data["yearbuilt"].fillna(prop_data["yearbuilt"].mean(), inplace=True)
prop_data["age"] = 2017 - prop_data["yearbuilt"]
#imputing garagecarcnt on basis of propertylandusetypeid
#All the residential places have 1 or 2 garagecarcnt, hence using random filling for those values.
prop_data.loc[(prop_data["propertylandusetypeid"]==261) & (pd.isnull(prop_data["garagecarcnt"])), "garagecarcnt"] = np.random.randint(1,3)
prop_data.loc[(prop_data["propertylandusetypeid"]==266) & (pd.isnull(prop_data["garagecarcnt"])), "garagecarcnt"] = np.random.randint(1,3)
prop_data["garagecarcnt"].fillna(0, inplace=True)
prop_data["taxamount"].fillna(prop_data.taxamount.mean(), inplace=True)
prop_data['longitude'] = prop_data['longitude'].abs()
prop_data['calculatedfinishedsquarefeet'].describe()
```
### Normalizing the data
```
colsList = ["actual_area",
"poolcnt",
"latitude",
"longitude",
"unitcnt",
"lotsizesquarefeet",
"bedroomcnt",
"calculatedbathnbr",
"hashottuborspa",
"fireplacecnt",
"taxvaluedollarcnt",
"buildingqualitytypeid",
"garagecarcnt",
"age",
"taxamount"]
prop_data_ahp = prop_data[colsList]
# prop_data_ahp
for col in prop_data_ahp.columns:
prop_data_ahp[col] = (prop_data_ahp[col] - prop_data_ahp[col].mean())/prop_data_ahp[col].std(ddof=0)
# prop_data_ahp.isnull().sum()
for cols in prop_data_ahp.columns.values:
print prop_data_ahp[cols].value_counts(dropna=False)
```
## Analytical Hierarchical Processing
```
rel_imp_matrix = pd.read_csv("rel_imp_matrix.csv", index_col=0)
# rel_imp_matrix
import fractions
for col in rel_imp_matrix.columns.values:
temp_list = rel_imp_matrix[col].tolist()
rel_imp_matrix[col] = [float(fractions.Fraction(x)) for x in temp_list]
# data = [float(fractions.Fraction(x)) for x in data]
# rel_imp_matrix
for col in rel_imp_matrix.columns.values:
rel_imp_matrix[col] /= rel_imp_matrix[col].sum()
# rel_imp_matrix
rel_imp_matrix["row_sum"] = rel_imp_matrix.sum(axis=1)
rel_imp_matrix["score"] = rel_imp_matrix["row_sum"]/rel_imp_matrix.shape[0]
rel_imp_matrix.to_csv("final_score_matrix.csv", index=False)
# rel_imp_matrix
ahp_column_score = rel_imp_matrix["score"]
ahp_column_score
prop_data_ahp.info()
prop_data_ahp.drop('sum', axis=1,inplace=True)
prop_data_ahp.keys()
```
# SAW
```
sum_series = pd.Series(0, index=prop_data_ahp.index,dtype='float32')
for col in prop_data_ahp.columns:
sum_series = sum_series+ prop_data_ahp[col] * ahp_column_score[col]
prop_data_ahp["sum"] = sum_series.astype('float32')
prop_data_ahp["sum"]
# prop_data_ahp["sum"] = prop_data_ahp.sum(axis=1)
prop_data_ahp["sum"].describe()
prop_data_ahp.sort_values(by='sum', inplace=True)
prop_data_ahp.head(n=10)
prop_data_ahp.tail(n=10)
print prop_data[colsList].iloc[1252741],"\n\n"
print prop_data[colsList].iloc[342941]
# #imputing airconditioningtypeid, making some NaN to 1.0 where heatingorsystemtypeid == 2
# prop_data.loc[(prop_data["heatingorsystemtypeid"]==2.0) & (pd.isnull(prop_data["airconditioningtypeid"])), "airconditioningtypeid"] = 1.0
# prop_data["airconditioningtypeid"].fillna(-1, inplace=True)
# print prop_data["airconditioningtypeid"].value_counts()
# prop_data[["airconditioningtypeid", "heatingorsystemtypeid"]].head()
# duplicate_or_not_useful_cols = pd.Series(['calculatedbathnbr', 'assessmentyear', 'fullbathcnt',
# 'regionidneighborhood', 'propertyzoningdesc', 'censustractandblock'])#,'finishedsquarefeet12'])
# prop_data.drop(duplicate_or_not_useful_cols, axis=1, inplace=True)
# prop_data["buildingqualitytypeid"].fillna(prop_data["buildingqualitytypeid"].mean(), inplace=True)
# prop_data["calculatedfinishedsquarefeet"].interpolate(inplace=True)
# prop_data["heatingorsystemtypeid"].fillna(-1, inplace=True)
# prop_data["lotsizesquarefeet"].fillna(prop_data["lotsizesquarefeet"].median(), inplace=True)
# prop_data.drop(["numberofstories"], axis=1, inplace=True)
# #removing propertycountylandusecode because it is not in interpretable format
# prop_data.drop(["propertycountylandusecode"], axis=1, inplace=True)
# prop_data["regionidcity"].interpolate(inplace=True)
# prop_data["regionidzip"].interpolate(inplace=True)
# prop_data["yearbuilt"].fillna(prop_data["yearbuilt"].mean(), inplace=True)
# #impute structuretaxvaluedollarcnt, taxvaluedollarcnt, landtaxvaluedollarcnt, taxamount by interpolation
# cols_to_interpolate = ["structuretaxvaluedollarcnt", "taxvaluedollarcnt", "landtaxvaluedollarcnt", "taxamount"]
# for c in cols_to_interpolate:
# prop_data[c].interpolate(inplace=True)
# #imputing garagecarcnt on basis of propertylandusetypeid
# #All the residential places have 1 or 2 garagecarcnt, hence using random filling for those values.
# prop_data.loc[(prop_data["propertylandusetypeid"]==261) & (pd.isnull(prop_data["garagecarcnt"])), "garagecarcnt"] = np.random.randint(1,3)
# prop_data.loc[(prop_data["propertylandusetypeid"]==266) & (pd.isnull(prop_data["garagecarcnt"])), "garagecarcnt"] = np.random.randint(1,3)
# prop_data["garagecarcnt"].fillna(-1, inplace=True)
# prop_data["garagecarcnt"].value_counts(dropna=False)
# #imputing garagetotalsqft using the garagecarcnt
# prop_data.loc[(prop_data["garagecarcnt"]==-1) & (pd.isnull(prop_data["garagetotalsqft"]) | (prop_data["garagetotalsqft"] == 0)), "garagetotalsqft"] = -1
# prop_data.loc[(prop_data["garagecarcnt"]==1) & (pd.isnull(prop_data["garagetotalsqft"]) | (prop_data["garagetotalsqft"] == 0)), "garagetotalsqft"] = np.random.randint(180, 400)
# prop_data.loc[(prop_data["garagecarcnt"]==2) & (pd.isnull(prop_data["garagetotalsqft"]) | (prop_data["garagetotalsqft"] == 0)), "garagetotalsqft"] = np.random.randint(400, 720)
# prop_data.loc[(prop_data["garagecarcnt"]==3) & (pd.isnull(prop_data["garagetotalsqft"]) | (prop_data["garagetotalsqft"] == 0)), "garagetotalsqft"] = np.random.randint(720, 880)
# prop_data.loc[(prop_data["garagecarcnt"]==4) & (pd.isnull(prop_data["garagetotalsqft"]) | (prop_data["garagetotalsqft"] == 0)), "garagetotalsqft"] = np.random.randint(880, 1200)
# #interpolate the remaining missing values
# prop_data["garagetotalsqft"].interpolate(inplace=True)
# prop_data["garagetotalsqft"].value_counts(dropna=False)
# #imputing unitcnt using propertylandusetypeid
# prop_data.loc[(prop_data["propertylandusetypeid"]==261) & pd.isnull(prop_data["unitcnt"]), "unitcnt"] = 1
# prop_data.loc[(prop_data["propertylandusetypeid"]==266) & pd.isnull(prop_data["unitcnt"]), "unitcnt"] = 1
# prop_data.loc[(prop_data["propertylandusetypeid"]==269) & pd.isnull(prop_data["unitcnt"]), "unitcnt"] = 1
# prop_data.loc[(prop_data["propertylandusetypeid"]==246) & pd.isnull(prop_data["unitcnt"]), "unitcnt"] = 2
# prop_data.loc[(prop_data["propertylandusetypeid"]==247) & pd.isnull(prop_data["unitcnt"]), "unitcnt"] = 3
# prop_data.loc[(prop_data["propertylandusetypeid"]==248) & pd.isnull(prop_data["unitcnt"]), "unitcnt"] = 4
# prop_data["unitcnt"].value_counts(dropna=False)
```
## Distance Metric
We will be using weighted Manhattan distance as a distance metric
```
dist_imp_matrix = pd.read_csv("./dist_metric.csv", index_col=0)
dist_imp_matrix
import fractions
for col in dist_imp_matrix.columns.values:
temp_list = dist_imp_matrix[col].tolist()
dist_imp_matrix[col] = [float(fractions.Fraction(x)) for x in temp_list]
# dist_imp_matrix
for col in dist_imp_matrix.columns.values:
dist_imp_matrix[col] /= dist_imp_matrix[col].sum()
dist_imp_matrix["row_sum"] = dist_imp_matrix.sum(axis=1)
dist_imp_matrix["score"] = dist_imp_matrix["row_sum"]/dist_imp_matrix.shape[0]
dist_imp_matrix.to_csv("final_score_matrix_Q2.csv")
```
| github_jupyter |
```
from IPython.core.display import HTML
with open('../style.css', 'r') as file:
css = file.read()
HTML(css)
```
# A Crypto-Arithmetic Puzzle
In this exercise we will solve the crypto-arithmetic puzzle shown in the picture below:
<img src="send-more-money.png">
The idea is that the letters
"$\texttt{S}$", "$\texttt{E}$", "$\texttt{N}$", "$\texttt{D}$", "$\texttt{M}$", "$\texttt{O}$", "$\texttt{R}$", "$\texttt{Y}$" occurring in this puzzle
are interpreted as variables ranging over the set of decimal digits, i.e. these variables can take values in
the set $\{0,1,2,3,4,5,6,7,8,9\}$. Then, the string "$\texttt{SEND}$" is interpreted as a decimal number,
i.e. it is interpreted as the number
$$\texttt{S} \cdot 10^3 + \texttt{E} \cdot 10^2 + \texttt{N} \cdot 10^1 + \texttt{D} \cdot 10^0.$$
The strings "$\texttt{MORE}$ and "$\texttt{MONEY}$" are interpreted similarly. To make the problem
interesting, the assumption is that different variables have different values. Furthermore, the
digits at the beginning of a number should be different from $0$. Then, we have to find values for the variables
"$\texttt{S}$", "$\texttt{E}$", "$\texttt{N}$", "$\texttt{D}$", "$\texttt{M}$", "$\texttt{O}$", "$\texttt{R}$", "$\texttt{Y}$" such that the formula
$$ (\texttt{S} \cdot 10^3 + \texttt{E} \cdot 10^2 + \texttt{N} \cdot 10 + \texttt{D})
+ (\texttt{M} \cdot 10^3 + \texttt{O} \cdot 10^2 + \texttt{R} \cdot 10 + \texttt{E})
= \texttt{M} \cdot 10^4 + \texttt{O} \cdot 10^3 + \texttt{N} \cdot 10^2 + \texttt{E} \cdot 10 + \texttt{Y}
$$
is true. The problem with this constraint is that it involves far too many variables. As this constraint can only be
checked when all the variables have values assigned to them, the backtracking search would essentially
boil down to a mere brute force search. We would have 8 variables and hence we would have to test $8^{10}$
possible assignments. In order to do better, we have to perform the addition in the figure shown above
column by column, just as it is taught in elementary school. To be able to do this, we have to introduce <a href="https://en.wikipedia.org/wiki/Carry_(arithmetic)">carry digits</a> "$\texttt{C1}$", "$\texttt{C2}$", "$\texttt{C3}$" where $\texttt{C1}$ is the carry produced by adding
$\texttt{D}$ and $\texttt{E}$, $\texttt{C2}$ is the carry produced by adding
$\texttt{N}$, $\texttt{R}$ and $\texttt{C1}$, and $\texttt{C3}$ is the carry produced by adding
$\texttt{E}$, $\texttt{O}$ and $\texttt{C2}$.
```
import cspSolver
```
For a set $V$ of variables, the function $\texttt{allDifferent}(V)$ generates a set of formulas that express that all the variables of $V$ are different.
```
def allDifferent(Variables):
return { f'{x} != {y}' for x in Variables
for y in Variables
if x < y
}
allDifferent({ 'a', 'b', 'c' })
```
# Pause bis 14:23
```
def createCSP():
Variables = "your code here"
Values = "your code here"
Constraints = "much more code here"
return [Variables, Values, Constraints];
puzzle = createCSP()
puzzle
%%time
solution = cspSolver.solve(puzzle)
print(f'Time needed: {round((stop-start) * 1000)} milliseconds.')
solution
def printSolution(A):
if A == None:
print("no solution found")
return
for v in { "S", "E", "N", "D", "M", "O", "R", "Y" }:
print(f"{v} = {A[v]}")
print("\nThe solution of\n")
print(" S E N D")
print(" + M O R E")
print(" ---------")
print(" M O N E Y")
print("\nis as follows\n")
print(f" {A['S']} {A['E']} {A['N']} {A['D']}")
print(f" + {A['M']} {A['O']} {A['R']} {A['E']}")
print(f" ==========")
print(f" {A['M']} {A['O']} {A['N']} {A['E']} {A['Y']}")
printSolution(solution)
```
| github_jupyter |
# Solution based on Multiple Models
```
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all"
```
# Tokenize and Numerize - Make it ready
```
training_size = 20000
training_sentences = sentences[0:training_size]
testing_sentences = sentences[training_size:]
training_labels = labels[0:training_size]
testing_labels = labels[training_size:]
vocab_size = 1000
max_length = 120
embedding_dim = 16
trunc_type='post'
padding_type='post'
oov_tok = "<OOV>"
tokenizer = Tokenizer(num_words=vocab_size, oov_token=oov_tok)
tokenizer.fit_on_texts(training_sentences)
word_index = tokenizer.word_index
training_sequences = tokenizer.texts_to_sequences(training_sentences)
training_padded = pad_sequences(training_sequences,
maxlen=max_length,
padding=padding_type,
truncating=trunc_type)
testing_sequences = tokenizer.texts_to_sequences(testing_sentences)
testing_padded = pad_sequences(testing_sequences,
maxlen=max_length,
padding=padding_type,
truncating=trunc_type)
```
# Plot
```
def plot_graphs(history, string):
plt.plot(history.history[string])
plt.plot(history.history['val_'+string])
plt.xlabel("Epochs")
plt.ylabel(string)
plt.legend([string, 'val_'+string])
plt.show()
plot_graphs(history, "accuracy")
plot_graphs(history, "loss")
```
## Function to train and show
```
def fit_model_and_show_results (model, reviews):
model.summary()
history = model.fit(training_padded,
training_labels_final,
epochs=num_epochs,
validation_data=(validation_padded, validation_labels_final))
plot_graphs(history, "accuracy")
plot_graphs(history, "loss")
predict_review(model, reviews)
```
# ANN Embedding
```
model = tf.keras.Sequential([
tf.keras.layers.Embedding(vocab_size, embedding_dim, input_length=max_length),
tf.keras.layers.GlobalAveragePooling1D(),
tf.keras.layers.Dense(1, activation='sigmoid')
])
model.compile(loss='binary_crossentropy',optimizer='adam',metrics=['accuracy'])
model.summary()
num_epochs = 20
history = model.fit(training_padded, training_labels_final, epochs=num_epochs,
validation_data=(validation_padded, validation_labels_final))
plot_graphs(history, "accuracy")
plot_graphs(history, "loss")
```
# CNN
```
num_epochs = 30
model_cnn = tf.keras.Sequential([
tf.keras.layers.Embedding(vocab_size, embedding_dim, input_length=max_length),
tf.keras.layers.Conv1D(16, 5, activation='relu'),
tf.keras.layers.GlobalMaxPooling1D(),
tf.keras.layers.Dense(1, activation='sigmoid')
])
# Default learning rate for the Adam optimizer is 0.001
# Let's slow down the learning rate by 10.
learning_rate = 0.0001
model_cnn.compile(loss='binary_crossentropy',
optimizer=tf.keras.optimizers.Adam(learning_rate),
metrics=['accuracy'])
fit_model_and_show_results(model_cnn, new_reviews)
```
# GRU
```
num_epochs = 30
model_gru = tf.keras.Sequential([
tf.keras.layers.Embedding(vocab_size, embedding_dim, input_length=max_length),
tf.keras.layers.Bidirectional(tf.keras.layers.GRU(32)),
tf.keras.layers.Dense(1, activation='sigmoid')
])
learning_rate = 0.00003 # slower than the default learning rate
model_gru.compile(loss='binary_crossentropy',
optimizer=tf.keras.optimizers.Adam(learning_rate),
metrics=['accuracy'])
fit_model_and_show_results(model_gru, new_reviews)
```
# Bidirectional LSTM
```
num_epochs = 30
model_bidi_lstm = tf.keras.Sequential([
tf.keras.layers.Embedding(vocab_size, embedding_dim, input_length=max_length),
tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(embedding_dim)),
tf.keras.layers.Dense(1, activation='sigmoid')
])
learning_rate = 0.00003
model_bidi_lstm.compile(loss='binary_crossentropy',
optimizer=tf.keras.optimizers.Adam(learning_rate),
metrics=['accuracy'])
fit_model_and_show_results(model_bidi_lstm, new_reviews)
```
# Multiple bidirectional LSTMs
```
num_epochs = 30
model_multiple_bidi_lstm = tf.keras.Sequential([
tf.keras.layers.Embedding(vocab_size, embedding_dim, input_length=max_length),
tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(embedding_dim,
return_sequences=True)),
tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(embedding_dim)),
tf.keras.layers.Dense(1, activation='sigmoid')
])
learning_rate = 0.0003
model_multiple_bidi_lstm.compile(loss='binary_crossentropy',
optimizer=tf.keras.optimizers.Adam(learning_rate),
metrics=['accuracy'])
fit_model_and_show_results(model_multiple_bidi_lstm, new_reviews)
```
# Prediction
Define a function to prepare the new reviews for use with a model
and then use the model to predict the sentiment of the new reviews
```
def predict_review(model, reviews):
# Create the sequences
padding_type='post'
sample_sequences = tokenizer.texts_to_sequences(reviews)
reviews_padded = pad_sequences(sample_sequences,
padding=padding_type,
maxlen=max_length)
classes = model.predict(reviews_padded)
for x in range(len(reviews_padded)):
print(reviews[x])
print(classes[x])
print('\n')
```
## How to use examples
more_reviews = [review1, review2, review3, review4, review5, review6, review7,
review8, review9, review10]
predict_review(model, new_reviews)
```
print("============================\n","Embeddings only:\n", "============================")
predict_review(model, more_reviews)
print("============================\n","With CNN\n", "============================")
predict_review(model_cnn, more_reviews)
print("===========================\n","With bidirectional GRU\n", "============================")
predict_review(model_gru, more_reviews)
print("===========================\n", "With a single bidirectional LSTM:\n", "===========================")
predict_review(model_bidi_lstm, more_reviews)
print("===========================\n", "With multiple bidirectional LSTM:\n", "==========================")
predict_review(model_multiple_bidi_lstm, more_reviews)
```
| github_jupyter |
#1. Install Dependencies
First install the libraries needed to execute recipes, this only needs to be done once, then click play.
```
!pip install git+https://github.com/google/starthinker
```
#2. Get Cloud Project ID
To run this recipe [requires a Google Cloud Project](https://github.com/google/starthinker/blob/master/tutorials/cloud_project.md), this only needs to be done once, then click play.
```
CLOUD_PROJECT = 'PASTE PROJECT ID HERE'
print("Cloud Project Set To: %s" % CLOUD_PROJECT)
```
#3. Get Client Credentials
To read and write to various endpoints requires [downloading client credentials](https://github.com/google/starthinker/blob/master/tutorials/cloud_client_installed.md), this only needs to be done once, then click play.
```
CLIENT_CREDENTIALS = 'PASTE CLIENT CREDENTIALS HERE'
print("Client Credentials Set To: %s" % CLIENT_CREDENTIALS)
```
#4. Enter DV360 Data Warehouse Parameters
Deploy a BigQuery dataset mirroring DV360 account structure. Foundation for solutions on top.
1. Wait for <b>BigQuery->->->*</b> to be created.
1. Every table mimics the <a href='https://developers.google.com/display-video/api/reference/rest' target='_blank'>DV360 API Endpoints</a>.
Modify the values below for your use case, can be done multiple times, then click play.
```
FIELDS = {
'auth_bigquery': 'service', # Credentials used for writing data.
'auth_dv': 'service', # Credentials used for reading data.
'auth_cm': 'service', # Credentials used for reading data.
'recipe_slug': '', # Name of Google BigQuery dataset to create.
'partners': [], # List of account ids to pull.
}
print("Parameters Set To: %s" % FIELDS)
```
#5. Execute DV360 Data Warehouse
This does NOT need to be modified unless you are changing the recipe, click play.
```
from starthinker.util.configuration import Configuration
from starthinker.util.configuration import execute
from starthinker.util.recipe import json_set_fields
USER_CREDENTIALS = '/content/user.json'
TASKS = [
{
'dataset': {
'description': 'Create a dataset for bigquery tables.',
'hour': [
4
],
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','description': 'Place where tables will be created in BigQuery.'}}
}
},
{
'google_api': {
'auth': 'user',
'api': 'displayvideo',
'version': 'v1',
'function': 'partners.get',
'kwargs_remote': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','description': 'Place where tables will be created in BigQuery.'}},
'legacy': False,
'query': 'SELECT CAST(partnerId AS STRING) partnerId FROM (SELECT DISTINCT * FROM UNNEST({partners}) AS partnerId)',
'parameters': {
'partners': {'field': {'name': 'partners','kind': 'integer_list','order': 4,'default': [],'description': 'List of account ids to pull.'}}
}
}
},
'iterate': False,
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV360_Partners'
}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'displayvideo',
'version': 'v1',
'function': 'advertisers.list',
'kwargs_remote': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 0,'default': '','description': 'Google BigQuery dataset to create tables in.'}},
'query': 'SELECT DISTINCT CAST(partnerId AS STRING) partnerId FROM `DV360_Partners`',
'legacy': False
}
},
'iterate': True,
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV360_Advertisers'
}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'displayvideo',
'version': 'v1',
'function': 'advertisers.insertionOrders.list',
'kwargs_remote': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 0,'default': '','description': 'Google BigQuery dataset to create tables in.'}},
'query': 'SELECT DISTINCT CAST(advertiserId AS STRING) AS advertiserId FROM `DV360_Advertisers`',
'legacy': False
}
},
'iterate': True,
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV360_InsertionOrders'
}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'displayvideo',
'version': 'v1',
'function': 'advertisers.lineItems.list',
'kwargs_remote': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 0,'default': '','description': 'Google BigQuery dataset to create tables in.'}},
'query': 'SELECT DISTINCT CAST(advertiserId AS STRING) AS advertiserId FROM `DV360_Advertisers`',
'legacy': False
}
},
'iterate': True,
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV360_LineItems'
}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'displayvideo',
'version': 'v1',
'function': 'advertisers.campaigns.list',
'kwargs_remote': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 0,'default': '','description': 'Google BigQuery dataset to create tables in.'}},
'query': 'SELECT DISTINCT CAST(advertiserId AS STRING) AS advertiserId FROM `DV360_Advertisers`',
'legacy': False
}
},
'iterate': True,
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV360_Campaigns'
}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'displayvideo',
'version': 'v1',
'function': 'advertisers.channels.list',
'kwargs_remote': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 0,'default': '','description': 'Google BigQuery dataset to create tables in.'}},
'query': 'SELECT DISTINCT CAST(advertiserId AS STRING) AS advertiserId FROM `DV360_Advertisers`',
'legacy': False
}
},
'iterate': True,
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV360_Channels'
}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'displayvideo',
'version': 'v1',
'function': 'advertisers.creatives.list',
'kwargs_remote': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 0,'default': '','description': 'Google BigQuery dataset to create tables in.'}},
'query': 'SELECT DISTINCT CAST(advertiserId AS STRING) AS advertiserId FROM `DV360_Advertisers`',
'legacy': False
}
},
'iterate': True,
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV360_Creatives'
}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'displayvideo',
'version': 'v1',
'function': 'inventorySources.list',
'kwargs_remote': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 0,'default': '','description': 'Google BigQuery dataset to create tables in.'}},
'query': 'SELECT DISTINCT CAST(advertiserId AS STRING) AS advertiserId FROM `DV360_Advertisers`',
'legacy': False
}
},
'iterate': True,
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV360_Inventory_Sources'
}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'displayvideo',
'version': 'v1',
'function': 'googleAudiences.list',
'kwargs_remote': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 0,'default': '','description': 'Google BigQuery dataset to create tables in.'}},
'query': 'SELECT DISTINCT CAST(advertiserId AS STRING) AS advertiserId FROM `DV360_Advertisers`',
'legacy': False
}
},
'iterate': True,
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV360_Google_Audiences'
}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'displayvideo',
'version': 'v1',
'function': 'combinedAudiences.list',
'kwargs_remote': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 0,'default': '','description': 'Google BigQuery dataset to create tables in.'}},
'query': 'SELECT DISTINCT CAST(advertiserId AS STRING) AS advertiserId FROM `DV360_Advertisers`',
'legacy': False
}
},
'iterate': True,
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV360_Combined_Audiences'
}
}
}
}
]
json_set_fields(TASKS, FIELDS)
execute(Configuration(project=CLOUD_PROJECT, client=CLIENT_CREDENTIALS, user=USER_CREDENTIALS, verbose=True), TASKS, force=True)
```
| github_jupyter |
# Tutorial on Python for scientific computing
Marcos Duarte
This tutorial is a short introduction to programming and a demonstration of the basic features of Python for scientific computing. To use Python for scientific computing we need the Python program itself with its main modules and specific packages for scientific computing. [See this notebook on how to install Python for scientific computing](http://nbviewer.ipython.org/github/demotu/BMC/blob/master/notebooks/PythonInstallation.ipynb).
Once you get Python and the necessary packages for scientific computing ready to work, there are different ways to run Python, the main ones are:
- open a terminal window in your computer and type `python` or `ipython` that the Python interpreter will start
- run the IPython notebook and start working with Python in a browser
- run Spyder, an interactive development environment (IDE)
- run the IPython qtconsole, a more featured terminal
- run IPython completely in the cloud with for example, [https://cloud.sagemath.com](https://cloud.sagemath.com) or [https://www.wakari.io](https://www.wakari.io)
- run Python online in a website such as [https://www.pythonanywhere.com/](https://www.pythonanywhere.com/)
- run Python using any other Python editor or IDE
We will use the IPython Notebook for this tutorial but you can run almost all the things we will see here using the other forms listed above.
## Python as a calculator
Once in the IPython notebook, if you type a simple mathematical expression and press `Shift+Enter` it will give the result of the expression:
```
1 + 2 - 30
4/5
```
If you are using Python version 2.x instead of Python 3.x, you should have got 0 as the result of 4 divided by 5, which is wrong! The problem is that for Python versions up to 2.x, the operator '/' performs division with integers and the result will also be an integer (this behavior was changed in version 3.x).
If you want the normal behavior for division, in Python 2.x you have two options: tell Python that at least one of the numbers is not an integer or import the new division operator (which is inoffensive if you are already using Python 3), let's see these two options:
```
4/5.
from __future__ import division
4/5
```
I prefer to use the import division option (from future!); if we put this statement in the beginning of a file or IPython notebook, it will work for all subsequent commands.
Another command that changed its behavior from Python 2.x to 3.x is the `print` command.
In Python 2.x, the print command could be used as a statement:
```
print 4/5
```
With Python 3.x, the print command bahaves as a true function and has to be called with parentheses. Let's also import this future command to Python 2.x and use it from now on:
```
from __future__ import print_function
print(4/5)
```
With the `print` function, let's explore the mathematical operations available in Python:
```
print('1+2 = ', 1+2, '\n', '4*5 = ', 4*5, '\n', '6/7 = ', 6/7, '\n', '8**2 = ', 8**2, sep='')
```
And if we want the square-root of a number:
```
sqrt(9)
```
We get an error message saying that the `sqrt` function if not defined. This is because `sqrt` and other mathematical functions are available with the `math` module:
```
import math
math.sqrt(9)
from math import sqrt
sqrt(9)
```
## The import function
We used the command '`import`' to be able to call certain functions. In Python functions are organized in modules and packages and they have to be imported in order to be used.
A module is a file containing Python definitions (e.g., functions) and statements. Packages are a way of structuring Python’s module namespace by using “dotted module names”. For example, the module name A.B designates a submodule named B in a package named A. To be used, modules and packages have to be imported in Python with the import function.
Namespace is a container for a set of identifiers (names), and allows the disambiguation of homonym identifiers residing in different namespaces. For example, with the command import math, we will have all the functions and statements defined in this module in the namespace '`math.`', for example, '`math.pi`' is the π constant and '`math.cos()`', the cosine function.
By the way, to know which Python version you are running, we can use one of the following modules:
```
import sys
sys.version
```
And if you are in an IPython session:
```
from IPython import sys_info
print(sys_info())
```
The first option gives information about the Python version; the latter also includes the IPython version, operating system, etc.
## Object-oriented programming
Python is designed as an object-oriented programming (OOP) language. OOP is a paradigm that represents concepts as "objects" that have data fields (attributes that describe the object) and associated procedures known as methods.
This means that all elements in Python are objects and they have attributes which can be acessed with the dot (.) operator after the name of the object. We already experimented with that when we imported the module `sys`, it became an object, and we acessed one of its attribute: `sys.version`.
OOP as a paradigm is much more than defining objects, attributes, and methods, but for now this is enough to get going with Python.
## Python and IPython help
To get help about any Python command, use `help()`:
```
help(math.degrees)
```
Or if you are in the IPython environment, simply add '?' to the function that a window will open at the bottom of your browser with the same help content:
```
math.degrees?
```
And if you add a second '?' to the statement you get access to the original script file of the function (an advantage of an open source language), unless that function is a built-in function that does not have a script file, which is the case of the standard modules in Python (but you can access the Python source code if you want; it just does not come with the standard program for installation).
So, let's see this feature with another function:
```
import scipy.fftpack
scipy.fftpack.fft??
```
To know all the attributes of an object, for example all the functions available in `math`, we can use the function `dir`:
```
print(dir(math))
```
### Tab completion in IPython
IPython has tab completion: start typing the name of the command (object) and press `tab` to see the names of objects available with these initials letters. When the name of the object is typed followed by a dot (`math.`), pressing `tab` will show all available attribites, scroll down to the desired attribute and press `Enter` to select it.
### The four most helpful commands in IPython
These are the most helpful commands in IPython (from [IPython tutorial](http://ipython.org/ipython-doc/dev/interactive/tutorial.html)):
- `?` : Introduction and overview of IPython’s features.
- `%quickref` : Quick reference.
- `help` : Python’s own help system.
- `object?` : Details about ‘object’, use ‘object??’ for extra details.
[See these IPython Notebooks for more on IPython and the Notebook capabilities](http://nbviewer.ipython.org/github/ipython/ipython/tree/master/examples/Notebook/).
### Comments
Comments in Python start with the hash character, #, and extend to the end of the physical line:
```
# Import the math library to access more math stuff
import math
math.pi # this is the pi constant; a useless comment since this is obvious
```
To insert comments spanning more than one line, use a multi-line string with a pair of matching triple-quotes: `"""` or `'''` (we will see the string data type later). A typical use of a multi-line comment is as documentation strings and are meant for anyone reading the code:
```
"""Documentation strings are typically written like that.
A docstring is a string literal that occurs as the first statement
in a module, function, class, or method definition.
"""
```
A docstring like above is useless and its output as a standalone statement looks uggly in IPython Notebook, but you will see its real importance when reading and writting codes.
Commenting a programming code is an important step to make the code more readable, which Python cares a lot.
There is a style guide for writting Python code ([PEP 8](http://www.python.org/dev/peps/pep-0008/)) with a session about [how to write comments](http://www.python.org/dev/peps/pep-0008/#comments).
### Magic functions
IPython has a set of predefined ‘magic functions’ that you can call with a command line style syntax.
There are two kinds of magics, line-oriented and cell-oriented.
Line magics are prefixed with the % character and work much like OS command-line calls: they get as an argument the rest of the line, where arguments are passed without parentheses or quotes.
Cell magics are prefixed with a double %%, and they are functions that get as an argument not only the rest of the line, but also the lines below it in a separate argument.
## Assignment and expressions
The equal sign ('=') is used to assign a value to a variable. Afterwards, no result is displayed before the next interactive prompt:
```
x = 1
```
Spaces between the statements are optional but it helps for readability.
To see the value of the variable, call it again or use the print function:
```
x
print(x)
```
Of course, the last assignment is that holds:
```
x = 2
x = 3
x
```
In mathematics '=' is the symbol for identity, but in computer programming '=' is used for assignment, it means that the right part of the expresssion is assigned to its left part.
For example, 'x=x+1' does not make sense in mathematics but it does in computer programming:
```
x = 1
print(x)
x = x + 1
print(x)
```
A value can be assigned to several variables simultaneously:
```
x = y = 4
print(x)
print(y)
```
Several values can be assigned to several variables at once:
```
x, y = 5, 6
print(x)
print(y)
```
And with that, you can do (!):
```
x, y = y, x
print(x)
print(y)
```
Variables must be “defined” (assigned a value) before they can be used, or an error will occur:
```
x = z
```
## Variables and types
There are different types of built-in objects in Python (and remember that everything in Python is an object):
```
import types
print(dir(types))
```
Let's see some of them now.
### Numbers: int, float, complex
Numbers can an integer (int), float, and complex (with imaginary part).
Let's use the function `type` to show the type of number (and later for any other object):
```
type(6)
```
A float is a non-integer number:
```
math.pi
type(math.pi)
```
Python (IPython) is showing `math.pi` with only 15 decimal cases, but internally a float is represented with higher precision.
Floating point numbers in Python are implemented using a double (eight bytes) word; the precison and internal representation of floating point numbers are machine specific and are available in:
```
sys.float_info
```
Be aware that floating-point numbers can be trick in computers:
```
0.1 + 0.2
0.1 + 0.2 - 0.3
```
These results are not correct (and the problem is not due to Python). The error arises from the fact that floating-point numbers are represented in computer hardware as base 2 (binary) fractions and most decimal fractions cannot be represented exactly as binary fractions. As consequence, decimal floating-point numbers are only approximated by the binary floating-point numbers actually stored in the machine. [See here for more on this issue](http://docs.python.org/2/tutorial/floatingpoint.html).
A complex number has real and imaginary parts:
```
1+2j
print(type(1+2j))
```
Each part of a complex number is represented as a floating-point number. We can see them using the attributes `.real` and `.imag`:
```
print((1+2j).real)
print((1+2j).imag)
```
### Strings
Strings can be enclosed in single quotes or double quotes:
```
s = 'string (str) is a built-in type in Python'
s
type(s)
```
String enclosed with single and double quotes are equal, but it may be easier to use one instead of the other:
```
'string (str) is a Python's built-in type'
"string (str) is a Python's built-in type"
```
But you could have done that using the Python escape character '\':
```
'string (str) is a Python\'s built-in type'
```
Strings can be concatenated (glued together) with the + operator, and repeated with *:
```
s = 'P' + 'y' + 't' + 'h' + 'o' + 'n'
print(s)
print(s*5)
```
Strings can be subscripted (indexed); like in C, the first character of a string has subscript (index) 0:
```
print('s[0] = ', s[0], ' (s[index], start at 0)')
print('s[5] = ', s[5])
print('s[-1] = ', s[-1], ' (last element)')
print('s[:] = ', s[:], ' (all elements)')
print('s[1:] = ', s[1:], ' (from this index (inclusive) till the last (inclusive))')
print('s[2:4] = ', s[2:4], ' (from first index (inclusive) till second index (exclusive))')
print('s[:2] = ', s[:2], ' (till this index, exclusive)')
print('s[:10] = ', s[:10], ' (Python handles the index if it is larger than the string length)')
print('s[-10:] = ', s[-10:])
print('s[0:5:2] = ', s[0:5:2], ' (s[ini:end:step])')
print('s[::2] = ', s[::2], ' (s[::step], initial and final indexes can be omitted)')
print('s[0:5:-1] = ', s[::-1], ' (s[::-step] reverses the string)')
print('s[:2] + s[2:] = ', s[:2] + s[2:], ' (because of Python indexing, this sounds natural)')
```
### len()
Python has a built-in functon to get the number of itens of a sequence:
```
help(len)
s = 'Python'
len(s)
```
The function len() helps to understand how the backward indexing works in Python.
The index s[-i] should be understood as s[len(s) - i] rather than accessing directly the i-th element from back to front. This is why the last element of a string is s[-1]:
```
print('s = ', s)
print('len(s) = ', len(s))
print('len(s)-1 = ',len(s) - 1)
print('s[-1] = ', s[-1])
print('s[len(s) - 1] = ', s[len(s) - 1])
```
Or, strings can be surrounded in a pair of matching triple-quotes: """ or '''. End of lines do not need to be escaped when using triple-quotes, but they will be included in the string. This is how we created a multi-line comment earlier:
```
"""Strings can be surrounded in a pair of matching triple-quotes: \""" or '''.
End of lines do not need to be escaped when using triple-quotes,
but they will be included in the string.
"""
```
### Lists
Values can be grouped together using different types, one of them is list, which can be written as a list of comma-separated values between square brackets. List items need not all have the same type:
```
x = ['spam', 'eggs', 100, 1234]
x
```
Lists can be indexed and the same indexing rules we saw for strings are applied:
```
x[0]
```
The function len() works for lists:
```
len(x)
```
### Tuples
A tuple consists of a number of values separated by commas, for instance:
```
t = ('spam', 'eggs', 100, 1234)
t
```
The type tuple is why multiple assignments in a single line works; elements separated by commas (with or without surrounding parentheses) are a tuple and in an expression with an '=', the right-side tuple is attributed to the left-side tuple:
```
a, b = 1, 2
print('a = ', a, '\nb = ', b)
```
Is the same as:
```
(a, b) = (1, 2)
print('a = ', a, '\nb = ', b)
```
### Sets
Python also includes a data type for sets. A set is an unordered collection with no duplicate elements.
```
basket = ['apple', 'orange', 'apple', 'pear', 'orange', 'banana']
fruit = set(basket) # create a set without duplicates
fruit
```
As set is an unordered collection, it can not be indexed as lists and tuples.
```
set(['orange', 'pear', 'apple', 'banana'])
'orange' in fruit # fast membership testing
```
### Dictionaries
Dictionary is a collection of elements organized keys and values. Unlike lists and tuples, which are indexed by a range of numbers, dictionaries are indexed by their keys:
```
tel = {'jack': 4098, 'sape': 4139}
tel
tel['guido'] = 4127
tel
tel['jack']
del tel['sape']
tel['irv'] = 4127
tel
tel.keys()
'guido' in tel
```
The dict() constructor builds dictionaries directly from sequences of key-value pairs:
```
tel = dict([('sape', 4139), ('guido', 4127), ('jack', 4098)])
tel
```
## Built-in Constants
- **False** : false value of the bool type
- **True** : true value of the bool type
- **None** : sole value of types.NoneType. None is frequently used to represent the absence of a value.
In computer science, the Boolean or logical data type is composed by two values, true and false, intended to represent the values of logic and Boolean algebra. In Python, 1 and 0 can also be used in most situations as equivalent to the Boolean values.
## Logical (Boolean) operators
### and, or, not
- **and** : logical AND operator. If both the operands are true then condition becomes true. (a and b) is true.
- **or** : logical OR Operator. If any of the two operands are non zero then condition becomes true. (a or b) is true.
- **not** : logical NOT Operator. Reverses the logical state of its operand. If a condition is true then logical NOT operator will make false.
### Comparisons
The following comparison operations are supported by objects in Python:
- **==** : equal
- **!=** : not equal
- **<** : strictly less than
- **<=** : less than or equal
- **\>** : strictly greater than
- **\>=** : greater than or equal
- **is** : object identity
- **is not** : negated object identity
```
True == False
not True == False
1 < 2 > 1
True != (False or True)
True != False or True
```
## Indentation and whitespace
In Python, statement grouping is done by indentation (this is mandatory), which are done by inserting whitespaces, not tabs. Indentation is also recommended for alignment of function calling that span more than one line for better clarity.
We will see examples of indentation in the next session.
## Control of flow
### `if`...`elif`...`else`
Conditional statements (to peform something if another thing is True or False) can be implemmented using the `if` statement:
```
if expression:
statement
elif:
statement
else:
statement
```
`elif` (one or more) and `else` are optionals.
The indentation is obligatory.
For example:
```
if True:
pass
```
Which does nothing useful.
Let's use the `if`...`elif`...`else` statements to categorize the [body mass index](http://en.wikipedia.org/wiki/Body_mass_index) of a person:
```
# body mass index
weight = 100 # kg
height = 1.70 # m
bmi = weight / height**2
if bmi < 15:
c = 'very severely underweight'
elif 15 <= bmi < 16:
c = 'severely underweight'
elif 16 <= bmi < 18.5:
c = 'underweight'
elif 18.5 <= bmi < 25:
c = 'normal'
elif 25 <= bmi < 30:
c = 'overweight'
elif 30 <= bmi < 35:
c = 'moderately obese'
elif 35 <= bmi < 40:
c = 'severely obese'
else:
c = 'very severely obese'
print('For a weight of {0:.1f} kg and a height of {1:.2f} m,\n\
the body mass index (bmi) is {2:.1f} kg/m2,\nwhich is considered {3:s}.'\
.format(weight, height, bmi, c))
```
### for
The `for` statement iterates over a sequence to perform operations (a loop event).
```
for iterating_var in sequence:
statements
```
```
for i in [3, 2, 1, 'go!']:
print(i),
for letter in 'Python':
print(letter),
```
#### The `range()` function
The built-in function range() is useful if we need to create a sequence of numbers, for example, to iterate over this list. It generates lists containing arithmetic progressions:
```
help(range)
range(10)
range(1, 10, 2)
for i in range(10):
n2 = i**2
print(n2),
```
### while
The `while` statement is used for repeating sections of code in a loop until a condition is met (this different than the `for` statement which executes n times):
```
while expression:
statement
```
Let's generate the Fibonacci series using a `while` loop:
```
# Fibonacci series: the sum of two elements defines the next
a, b = 0, 1
while b < 1000:
print(b, end=' ')
a, b = b, a+b
```
## Function definition
A function in a programming language is a piece of code that performs a specific task. Functions are used to reduce duplication of code making easier to reuse it and to decompose complex problems into simpler parts. The use of functions contribute to the clarity of the code.
A function is created with the `def` keyword and the statements in the block of the function must be indented:
```
def function():
pass
```
As per construction, this function does nothing when called:
```
function()
```
The general syntax of a function definition is:
```
def function_name( parameters ):
"""Function docstring.
The help for the function
"""
function body
return variables
```
A more useful function:
```
def fibo(N):
"""Fibonacci series: the sum of two elements defines the next.
The series is calculated till the input parameter N and
returned as an ouput variable.
"""
a, b, c = 0, 1, []
while b < N:
c.append(b)
a, b = b, a + b
return c
fibo(100)
if 3 > 2:
print('teste')
```
Let's implemment the body mass index calculus and categorization as a function:
```
def bmi(weight, height):
"""Body mass index calculus and categorization.
Enter the weight in kg and the height in m.
See http://en.wikipedia.org/wiki/Body_mass_index
"""
bmi = weight / height**2
if bmi < 15:
c = 'very severely underweight'
elif 15 <= bmi < 16:
c = 'severely underweight'
elif 16 <= bmi < 18.5:
c = 'underweight'
elif 18.5 <= bmi < 25:
c = 'normal'
elif 25 <= bmi < 30:
c = 'overweight'
elif 30 <= bmi < 35:
c = 'moderately obese'
elif 35 <= bmi < 40:
c = 'severely obese'
else:
c = 'very severely obese'
s = 'For a weight of {0:.1f} kg and a height of {1:.2f} m,\
the body mass index (bmi) is {2:.1f} kg/m2,\
which is considered {3:s}.'\
.format(weight, height, bmi, c)
print(s)
bmi(73, 1.70)
```
## Numeric data manipulation with Numpy
Numpy is the fundamental package for scientific computing in Python and has a N-dimensional array package convenient to work with numerical data. With Numpy it's much easier and faster to work with numbers grouped as 1-D arrays (a vector), 2-D arrays (like a table or matrix), or higher dimensions. Let's create 1-D and 2-D arrays in Numpy:
```
import numpy as np
x1d = np.array([1, 2, 3, 4, 5, 6])
print(type(x1d))
x1d
x2d = np.array([[1, 2, 3], [4, 5, 6]])
x2d
```
len() and the Numpy functions size() and shape() give information aboout the number of elements and the structure of the Numpy array:
```
print('1-d array:')
print(x1d)
print('len(x1d) = ', len(x1d))
print('np.size(x1d) = ', np.size(x1d))
print('np.shape(x1d) = ', np.shape(x1d))
print('np.ndim(x1d) = ', np.ndim(x1d))
print('\n2-d array:')
print(x2d)
print('len(x2d) = ', len(x2d))
print('np.size(x2d) = ', np.size(x2d))
print('np.shape(x2d) = ', np.shape(x2d))
print('np.ndim(x2d) = ', np.ndim(x2d))
```
Create random data
```
x = np.random.randn(4,3)
x
```
Joining (stacking together) arrays
```
x = np.random.randint(0, 5, size=(2, 3))
print(x)
y = np.random.randint(5, 10, size=(2, 3))
print(y)
np.vstack((x,y))
np.hstack((x,y))
```
Create equally spaced data
```
np.arange(start = 1, stop = 10, step = 2)
np.linspace(start = 0, stop = 1, num = 11)
```
### Interpolation
Consider the following data:
```
y = [5, 4, 10, 8, 1, 10, 2, 7, 1, 3]
```
Suppose we want to create data in between the given data points (interpolation); for instance, let's try to double the resolution of the data by generating twice as many data:
```
t = np.linspace(0, len(y), len(y)) # time vector for the original data
tn = np.linspace(0, len(y), 2 * len(y)) # new time vector for the new time-normalized data
yn = np.interp(tn, t, y) # new time-normalized data
yn
```
The key is the Numpy `interp` function, from its help:
interp(x, xp, fp, left=None, right=None)
One-dimensional linear interpolation.
Returns the one-dimensional piecewise linear interpolant to a function with given values at discrete data-points.
A plot of the data will show what we have done:
```
%matplotlib inline
import matplotlib.pyplot as plt
plt.figure(figsize=(10,5))
plt.plot(t, y, 'bo-', lw=2, label='original data')
plt.plot(tn, yn, '.-', color=[1, 0, 0, .5], lw=2, label='interpolated')
plt.legend(loc='best', framealpha=.5)
plt.show()
```
For more about Numpy, see [http://wiki.scipy.org/Tentative_NumPy_Tutorial](http://wiki.scipy.org/Tentative_NumPy_Tutorial).
## Read and save files
There are two kinds of computer files: text files and binary files:
> Text file: computer file where the content is structured as a sequence of lines of electronic text. Text files can contain plain text (letters, numbers, and symbols) but they are not limited to such. The type of content in the text file is defined by the Unicode encoding (a computing industry standard for the consistent encoding, representation and handling of text expressed in most of the world's writing systems).
>
> Binary file: computer file where the content is encoded in binary form, a sequence of integers representing byte values.
Let's see how to save and read numeric data stored in a text file:
**Using plain Python**
```
f = open("newfile.txt", "w") # open file for writing
f.write("This is a test\n") # save to file
f.write("And here is another line\n") # save to file
f.close()
f = open('newfile.txt', 'r') # open file for reading
f = f.read() # read from file
print(f)
help(open)
```
**Using Numpy**
```
import numpy as np
data = np.random.randn(3,3)
np.savetxt('myfile.txt', data, fmt="%12.6G") # save to file
data = np.genfromtxt('myfile.txt', unpack=True) # read from file
data
```
## Ploting with matplotlib
Matplotlib is the most-widely used packge for plotting data in Python. Let's see some examples of it.
```
import matplotlib.pyplot as plt
```
Use the IPython magic `%matplotlib inline` to plot a figure inline in the notebook with the rest of the text:
```
%matplotlib notebook
import numpy as np
t = np.linspace(0, 0.99, 100)
x = np.sin(2 * np.pi * 2 * t)
n = np.random.randn(100) / 5
plt.Figure(figsize=(12,8))
plt.plot(t, x, label='sine', linewidth=2)
plt.plot(t, x + n, label='noisy sine', linewidth=2)
plt.annotate(s='$sin(4 \pi t)$', xy=(.2, 1), fontsize=20, color=[0, 0, 1])
plt.legend(loc='best', framealpha=.5)
plt.xlabel('Time [s]')
plt.ylabel('Amplitude')
plt.title('Data plotting using matplotlib')
plt.show()
```
Use the IPython magic `%matplotlib qt` to plot a figure in a separate window (from where you will be able to change some of the figure proprerties):
```
%matplotlib qt
mu, sigma = 10, 2
x = mu + sigma * np.random.randn(1000)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 4))
ax1.plot(x, 'ro')
ax1.set_title('Data')
ax1.grid()
n, bins, patches = ax2.hist(x, 25, normed=True, facecolor='r') # histogram
ax2.set_xlabel('Bins')
ax2.set_ylabel('Probability')
ax2.set_title('Histogram')
fig.suptitle('Another example using matplotlib', fontsize=18, y=1)
ax2.grid()
plt.tight_layout()
plt.show()
```
And a window with the following figure should appear:
```
from IPython.display import Image
Image(url="./../images/plot.png")
```
You can switch back and forth between inline and separate figure using the `%matplotlib` magic commands used above. There are plenty more examples with the source code in the [matplotlib gallery](http://matplotlib.org/gallery.html).
```
# get back the inline plot
%matplotlib inline
```
## Signal processing with Scipy
The Scipy package has a lot of functions for signal processing, among them: Integration (scipy.integrate), Optimization (scipy.optimize), Interpolation (scipy.interpolate), Fourier Transforms (scipy.fftpack), Signal Processing (scipy.signal), Linear Algebra (scipy.linalg), and Statistics (scipy.stats). As an example, let's see how to use a low-pass Butterworth filter to attenuate high-frequency noise and how the differentiation process of a signal affects the signal-to-noise content. We will also calculate the Fourier transform of these data to look at their frequencies content.
```
from scipy.signal import butter, filtfilt
import scipy.fftpack
freq = 100.
t = np.arange(0,1,.01);
w = 2*np.pi*1 # 1 Hz
y = np.sin(w*t)+0.1*np.sin(10*w*t)
# Butterworth filter
b, a = butter(4, (5/(freq/2)), btype = 'low')
y2 = filtfilt(b, a, y)
# 2nd derivative of the data
ydd = np.diff(y,2)*freq*freq # raw data
y2dd = np.diff(y2,2)*freq*freq # filtered data
# frequency content
yfft = np.abs(scipy.fftpack.fft(y))/(y.size/2); # raw data
y2fft = np.abs(scipy.fftpack.fft(y2))/(y.size/2); # filtered data
freqs = scipy.fftpack.fftfreq(y.size, 1./freq)
yddfft = np.abs(scipy.fftpack.fft(ydd))/(ydd.size/2);
y2ddfft = np.abs(scipy.fftpack.fft(y2dd))/(ydd.size/2);
freqs2 = scipy.fftpack.fftfreq(ydd.size, 1./freq)
```
And the plots:
```
fig, ((ax1,ax2),(ax3,ax4)) = plt.subplots(2, 2, figsize=(10, 4))
ax1.set_title('Temporal domain', fontsize=14)
ax1.plot(t, y, 'r', linewidth=2, label = 'raw data')
ax1.plot(t, y2, 'b', linewidth=2, label = 'filtered @ 5 Hz')
ax1.set_ylabel('f')
ax1.legend(frameon=False, fontsize=12)
ax2.set_title('Frequency domain', fontsize=14)
ax2.plot(freqs[:yfft.size/4], yfft[:yfft.size/4],'r', lw=2,label='raw data')
ax2.plot(freqs[:yfft.size/4],y2fft[:yfft.size/4],'b--',lw=2,label='filtered @ 5 Hz')
ax2.set_ylabel('FFT(f)')
ax2.legend(frameon=False, fontsize=12)
ax3.plot(t[:-2], ydd, 'r', linewidth=2, label = 'raw')
ax3.plot(t[:-2], y2dd, 'b', linewidth=2, label = 'filtered @ 5 Hz')
ax3.set_xlabel('Time [s]'); ax3.set_ylabel("f ''")
ax4.plot(freqs[:yddfft.size/4], yddfft[:yddfft.size/4], 'r', lw=2, label = 'raw')
ax4.plot(freqs[:yddfft.size/4],y2ddfft[:yddfft.size/4],'b--',lw=2, label='filtered @ 5 Hz')
ax4.set_xlabel('Frequency [Hz]'); ax4.set_ylabel("FFT(f '')")
plt.show()
```
For more about Scipy, see [http://docs.scipy.org/doc/scipy/reference/tutorial/](http://docs.scipy.org/doc/scipy/reference/tutorial/).
## Symbolic mathematics with Sympy
Sympy is a package to perform symbolic mathematics in Python. Let's see some of its features:
```
from IPython.display import display
import sympy as sym
from sympy.interactive import printing
printing.init_printing()
```
Define some symbols and the create a second-order polynomial function (a.k.a., parabola):
```
x, y = sym.symbols('x y')
y = x**2 - 2*x - 3
y
```
Plot the parabola at some given range:
```
from sympy.plotting import plot
%matplotlib inline
plot(y, (x, -3, 5));
```
And the roots of the parabola are given by:
```
sym.solve(y, x)
```
We can also do symbolic differentiation and integration:
```
dy = sym.diff(y, x)
dy
sym.integrate(dy, x)
```
For example, let's use Sympy to represent three-dimensional rotations. Consider the problem of a coordinate system xyz rotated in relation to other coordinate system XYZ. The single rotations around each axis are illustrated by:
```
from IPython.display import Image
Image(url="./../images/rotations.png")
```
The single 3D rotation matrices around Z, Y, and X axes can be expressed in Sympy:
```
from IPython.core.display import Math
from sympy import symbols, cos, sin, Matrix, latex
a, b, g = symbols('alpha beta gamma')
RX = Matrix([[1, 0, 0], [0, cos(a), -sin(a)], [0, sin(a), cos(a)]])
display(Math(latex('\\mathbf{R_{X}}=') + latex(RX, mat_str = 'matrix')))
RY = Matrix([[cos(b), 0, sin(b)], [0, 1, 0], [-sin(b), 0, cos(b)]])
display(Math(latex('\\mathbf{R_{Y}}=') + latex(RY, mat_str = 'matrix')))
RZ = Matrix([[cos(g), -sin(g), 0], [sin(g), cos(g), 0], [0, 0, 1]])
display(Math(latex('\\mathbf{R_{Z}}=') + latex(RZ, mat_str = 'matrix')))
```
And using Sympy, a sequence of elementary rotations around X, Y, Z axes is given by:
```
RXYZ = RZ*RY*RX
display(Math(latex('\\mathbf{R_{XYZ}}=') + latex(RXYZ, mat_str = 'matrix')))
```
Suppose there is a rotation only around X ($\alpha$) by $\pi/2$; we can get the numerical value of the rotation matrix by substituing the angle values:
```
r = RXYZ.subs({a: np.pi/2, b: 0, g: 0})
r
```
And we can prettify this result:
```
display(Math(latex(r'\mathbf{R_{(\alpha=\pi/2)}}=') +
latex(r.n(chop=True, prec=3), mat_str = 'matrix')))
```
For more about Sympy, see [http://docs.sympy.org/latest/tutorial/](http://docs.sympy.org/latest/tutorial/).
## Data analysis with pandas
> "[pandas](http://pandas.pydata.org/) is a Python package providing fast, flexible, and expressive data structures designed to make working with “relational” or “labeled” data both easy and intuitive. It aims to be the fundamental high-level building block for doing practical, real world data analysis in Python."
To work with labellled data, pandas has a type called DataFrame (basically, a matrix where columns and rows have may names and may be of different types) and it is also the main type of the software [R](http://www.r-project.org/). Fo ezample:
```
import pandas as pd
x = 5*['A'] + 5*['B']
x
df = pd.DataFrame(np.random.rand(10,2), columns=['Level 1', 'Level 2'] )
df['Group'] = pd.Series(['A']*5 + ['B']*5)
plot = df.boxplot(by='Group')
from pandas.tools.plotting import scatter_matrix
df = pd.DataFrame(np.random.randn(100, 3), columns=['A', 'B', 'C'])
plot = scatter_matrix(df, alpha=0.5, figsize=(8, 6), diagonal='kde')
```
pandas is aware the data is structured and give you basic statistics considerint that and nicely formatted:
```
df.describe()
```
For more on pandas, see this tutorial: [http://pandas.pydata.org/pandas-docs/stable/10min.html](http://pandas.pydata.org/pandas-docs/stable/10min.html).
## To learn more about Python
There is a lot of good material in the internet about Python for scientific computing, here is a small list of interesting stuff:
- [How To Think Like A Computer Scientist](http://www.openbookproject.net/thinkcs/python/english2e/) or [the interactive edition](http://interactivepython.org/courselib/static/thinkcspy/index.html) (book)
- [Python Scientific Lecture Notes](http://scipy-lectures.github.io/) (lecture notes)
- [Lectures on scientific computing with Python](https://github.com/jrjohansson/scientific-python-lectures#lectures-on-scientific-computing-with-python) (lecture notes)
- [IPython in depth: high-productivity interactive and parallel python](http://youtu.be/bP8ydKBCZiY) (video lectures)
| github_jupyter |
```
import numpy as np
import pandas as pd
import json as json
from scipy import stats
from statsmodels.formula.api import ols
import matplotlib.pyplot as plt
from scipy.signal import savgol_filter
from o_plot import opl # a small local package dedicated to this project
# Prepare the data
# loading the data
file_name = 'Up_to_Belem_TE4AL2_data_new.json'
f = open(file_name)
All_data = json.load(f)
print(len(All_data))
```
## Note for the interpretation of the curves and definition of the statistical variables
The quantum state classifier (QSC) error rates $\widehat{r}_i$ in function of the number of experimental shots $n$ were determined for each highly entangled quantum state $\omega_i$ in the $\Omega$ set, with $i=1...m$.
The curves seen on the figures represents the mean of the QSC error rate $\widehat{r}_{mean}$ over the $m$ quantum states at each $n$ value.
This Monte Carlo simulation allowed to determine a safe shot number $n_s$ such that $\forall i\; \widehat{r}_i\le \epsilon_s$. The value of $\epsilon_s$ was set at 0.001.
$\widehat{r}_{max}$ is the maximal value observed among all the $\widehat{r}_i$ values for the determined number of shots $n_s$.
Similarly, from the error curves stored in the data file, was computed the safe shot number $n_t$ such that $\widehat{r}_{mean}\le \epsilon_t$. The value of $\epsilon_t$ was set at 0.0005 after verifying that all $\widehat{r}_{mean}$ at $n_s$ were $\le \epsilon_s$ in the different experimental settings.
Correspondance between variables names in the text and in the data base:
- $\widehat{r}_{mean}$: error_curve
- $n_s$: shots
- max ($\widehat{r}_i$) at $n_s$: shot_rate
- $\widehat{r}_{mean}$ at $n_s$: mns_rate
- $n_t$: m_shots
- $\widehat{r}_{mean}$ at $n_t$: m_shot_rate
```
# Calculate shot number 'm_shots' for mean error rate 'm_shot_rates' <= epsilon_t
len_data = len(All_data)
epsilon_t = 0.0005
window = 11
for i in range(len_data):
curve = np.array(All_data[i]['error_curve'])
# filter the curve only for real devices:
if All_data[i]['device']!="ideal_device":
curve = savgol_filter(curve,window,2)
# find the safe shot number:
len_c = len(curve)
n_a = np.argmin(np.flip(curve)<=epsilon_t)+1
if n_a == 1:
n_a = np.nan
m_r = np.nan
else:
m_r = curve[len_c-n_a+1]
All_data[i]['min_r_shots'] = len_c-n_a
All_data[i]['min_r'] = m_r
# find mean error rate at n_s
for i in range(len_data):
i_shot = All_data[i]["shots"]
if not np.isnan(i_shot):
j = int(i_shot)-1
All_data[i]['mns_rate'] = All_data[i]['error_curve'][j]
else:
All_data[i]['mns_rate'] = np.nan
#defining the pandas data frame for statistics excluding from here ibmqx2 data
df_All= pd.DataFrame(All_data,columns=['shot_rates','shots', 'device', 'fidelity',
'mitigation','model','id_gates',
'QV', 'metric','error_curve',
'mns_rate','min_r_shots',
'min_r']).query("device != 'ibmqx2'")
# any shot number >= 488 indicates that the curve calculation
# was ended after reaching n = 500, hence this data correction:
df_All.loc[df_All.shots>=488,"shots"]=np.nan
# add the variable neperian log of safe shot number:
df_All['log_shots'] = np.log(df_All['shots'])
df_All['log_min_r_shots'] = np.log(df_All['min_r_shots'])
```
### Error rates in function of chosen $\epsilon_s$ and $\epsilon_t$
```
print("max mean error rate at n_s over all experiments =", round(max(df_All.mns_rate[:-2]),6))
print("min mean error rate at n_t over all experiments =", round(min(df_All.min_r[:-2]),6))
print("max mean error rate at n_t over all experiments =", round(max(df_All.min_r[:-2]),6))
df_All.mns_rate[:-2].plot.hist(alpha=0.5, legend = True)
df_All.min_r[:-2].plot.hist(alpha=0.5, legend = True)
```
# Statistical overview
For this section, an ordinary linear least square estimation is performed.
The dependent variables tested are $ln\;n_s$ (log_shots) and $ln\;n_t$ (log_min_r_shots)
```
stat_model = ols("log_shots ~ metric",
df_All.query("device != 'ideal_device'")).fit()
print(stat_model.summary())
stat_model = ols("log_min_r_shots ~ metric",
df_All.query("device != 'ideal_device'")).fit()
print(stat_model.summary())
stat_model = ols("log_shots ~ model+mitigation+id_gates+fidelity+QV",
df_All.query("device != 'ideal_device' & metric == 'sqeuclidean'")).fit()
print(stat_model.summary())
stat_model = ols("log_min_r_shots ~ model+mitigation+id_gates+fidelity+QV",
df_All.query("device != 'ideal_device'& metric == 'sqeuclidean'")).fit()
print(stat_model.summary())
```
#### Comments:
For the QSC, two different metrics were compared and at the end they gave the same output. For further analysis, the results obtained using the squared euclidean distance between distribution will be illustrated in this notebook, as it is more classical and strictly equivalent to the other classical Hellinger and Bhattacharyya distances. The Jensen-Shannon metric has however the theoretical advantage of being bayesian in nature and is therefore presented as an option for the result analysis.
Curves obtained for counts corrected by measurement error mitigation (MEM) are used in this presentation. MEM significantly reduces $n_s$ and $n_t$. However, using counts distribution before MEM is presented as an option because they anticipate how the method could perform in devices with more qubits where obtaining the mitigation filter is a problem.
Introducing a delay time $\delta t$ of 256 identity gates between state creation and measurement significantly increased $ln\;n_s$ and $ln\;n_t$ .
# Detailed statistical analysis
### Determine the options
Running sequentially these cells will end up with the main streaming options
```
# this for Jensen-Shannon metric
s_metric = 'jensenshannon'
sm = np.array([96+16+16+16]) # added Quito and Lima and Belem
SAD=0
# ! will be unselected by running the next cell
# mainstream option for metric: squared euclidean distance
# skip this cell if you don't want this option
s_metric = 'sqeuclidean'
sm = np.array([97+16+16+16]) # added Quito and Lima and Belem
SAD=2
# this for no mitigation
mit = 'no'
MIT=-4
# ! will be unselected by running the next cell
# mainstream option: this for measurement mitigation
# skip this cell if you don't want this option
mit = 'yes'
MIT=0
```
## 1. Compare distribution models
```
# select data according to the options
df_mod = df_All[df_All.mitigation == mit][df_All.metric == s_metric]
```
### A look at $n_s$ and $n_t$
```
print("mitigation:",mit," metric:",s_metric )
df_mod.groupby('device')[['shots','min_r_shots']].describe(percentiles=[0.5])
```
### Ideal vs empirical model: no state creation - measurements delay
```
ADD=0+SAD+MIT
#opl.plot_curves(All_data, np.append(sm,ADD+np.array([4,5,12,13,20,21,28,29,36,37,44,45])),
opl.plot_curves(All_data, np.append(sm,ADD+np.array([4,5,12,13,20,21,28,29,36,37,52,53,60,61,68,69])),
"Monte Carlo Simulation: Theoretical PDM vs Empirical PDM - no $\delta_t0$",
["metric","mitigation"],
["device","model"], right_xlimit = 90)
```
#### Paired t-test and Wilcoxon test
```
for depvar in ['log_shots', 'log_min_r_shots']:
#for depvar in ['shots', 'min_r_shots']:
print("mitigation:",mit," metric:",s_metric, "variable:", depvar)
df_dep = df_mod.query("id_gates == 0.0").groupby(['model'])[depvar]
print(df_dep.describe(percentiles=[0.5]),"\n")
# no error rate curve obtained for ibmqx2 with the ideal model, hence this exclusion:
df_emp=df_mod.query("model == 'empirical' & id_gates == 0.0")
df_ide=df_mod.query("model == 'ideal_sim' & id_gates == 0.0") #.reindex_like(df_emp,'nearest')
# back to numpy arrays from pandas:
print("paired data")
print(np.asarray(df_emp[depvar]))
print(np.asarray(df_ide[depvar]),"\n")
print(stats.ttest_rel(np.asarray(df_emp[depvar]),np.asarray(df_ide[depvar])))
print(stats.wilcoxon(np.asarray(df_emp[depvar]),np.asarray(df_ide[depvar])),"\n")
print("mitigation:",mit," metric:",s_metric, "id_gates == 0.0 ")
stat_model = ols("log_shots ~ model + device + fidelity + QV" ,
df_mod.query("id_gates == 0.0 ")).fit()
print(stat_model.summary())
print("mitigation:",mit," metric:",s_metric, "id_gates == 0.0 " )
stat_model = ols("log_min_r_shots ~ model + device + fidelity+QV",
df_mod.query("id_gates == 0.0 ")).fit()
print(stat_model.summary())
```
### Ideal vs empirical model: with state creation - measurements delay of 256 id gates
```
ADD=72+SAD+MIT
opl.plot_curves(All_data, np.append(sm,ADD+np.array([4,5,12,13,20,21,28,29,36,37,52,53,60,61,68,69])),
"No noise simulator vs empirical model - $\epsilon=0.001$ - with delay",
["metric","mitigation"],
["device","model"], right_xlimit = 90)
```
#### Paired t-test and Wilcoxon test
```
for depvar in ['log_shots', 'log_min_r_shots']:
print("mitigation:",mit," metric:",s_metric, "variable:", depvar)
df_dep = df_mod.query("id_gates == 256.0 ").groupby(['model'])[depvar]
print(df_dep.describe(percentiles=[0.5]),"\n")
# no error rate curve obtained for ibmqx2 with the ideal model, hence their exclusion:
df_emp=df_mod.query("model == 'empirical' & id_gates == 256.0 ")
df_ide=df_mod.query("model == 'ideal_sim' & id_gates == 256.0") #.reindex_like(df_emp,'nearest')
# back to numpy arrays from pandas:
print("paired data")
print(np.asarray(df_emp[depvar]))
print(np.asarray(df_ide[depvar]),"\n")
print(stats.ttest_rel(np.asarray(df_emp[depvar]),np.asarray(df_ide[depvar])))
print(stats.wilcoxon(np.asarray(df_emp[depvar]),np.asarray(df_ide[depvar])),"\n")
print("mitigation:",mit," metric:",s_metric , "id_gates == 256.0 ")
stat_model = ols("log_shots ~ model + device + fidelity + QV" ,
df_mod.query("id_gates == 256.0 ")).fit()
print(stat_model.summary())
print("mitigation:",mit," metric:",s_metric, "id_gates == 256.0 " )
stat_model = ols("log_min_r_shots ~ model + device +fidelity+QV",
df_mod.query("id_gates == 256.0 ")).fit()
print(stat_model.summary())
```
### Pooling results obtained in circuit sets with and without creation-measurement delay
#### Paired t-test and Wilcoxon test
```
#for depvar in ['log_shots', 'log_min_r_shots']:
for depvar in ['log_shots', 'log_min_r_shots']:
print("mitigation:",mit," metric:",s_metric, "variable:", depvar)
df_dep = df_mod.groupby(['model'])[depvar]
print(df_dep.describe(percentiles=[0.5]),"\n")
# no error rate curve obtained for ibmqx2 with the ideal model, hence this exclusion:
df_emp=df_mod.query("model == 'empirical'")
df_ide=df_mod.query("model == 'ideal_sim'") #.reindex_like(df_emp,'nearest')
# back to numpy arrays from pandas:
print("paired data")
print(np.asarray(df_emp[depvar]))
print(np.asarray(df_ide[depvar]),"\n")
print(stats.ttest_rel(np.asarray(df_emp[depvar]),np.asarray(df_ide[depvar])))
print(stats.wilcoxon(np.asarray(df_emp[depvar]),np.asarray(df_ide[depvar])),"\n")
```
#### Statsmodel Ordinary Least Square (OLS) Analysis
```
print("mitigation:",mit," metric:",s_metric )
stat_model = ols("log_shots ~ model + id_gates + device + fidelity + QV" ,
df_mod).fit()
print(stat_model.summary())
print("mitigation:",mit," metric:",s_metric )
stat_model = ols("log_min_r_shots ~ model + id_gates + device + fidelity+QV ",
df_mod).fit()
print(stat_model.summary())
```
| github_jupyter |
# Continuous Control
---
## 1. Import the Necessary Packages
```
from unityagents import UnityEnvironment
import random
import torch
import numpy as np
from collections import deque
import matplotlib.pyplot as plt
%matplotlib inline
from ddpg_agent import Agent
```
## 2. Instantiate the Environment and 20 Agents
```
# initialize the environment
env = UnityEnvironment(file_name='./Reacher_20.app')
# get the default brain
brain_name = env.brain_names[0]
brain = env.brains[brain_name]
# reset the environment
env_info = env.reset(train_mode=True)[brain_name]
# number of agents
num_agents = len(env_info.agents)
print('Number of agents:', num_agents)
# size of each action
action_size = brain.vector_action_space_size
print('Size of each action:', action_size)
# examine the state space
states = env_info.vector_observations
state_size = states.shape[1]
print('There are {} agents. Each observes a state with length: {}'.format(states.shape[0], state_size))
print('The state for the first agent looks like:', states[0])
# initialize agents
agent = Agent(state_size=33,
action_size=4,
random_seed=2,
num_agents=20)
```
## 3. Train the 20 Agents with DDPG
To amend the `ddpg` code to work for 20 agents instead of 1, here are the modifications I did in `ddpg_agent.py`:
- With each step, each agent adds its experience to a replay buffer shared by all agents (line 61-61).
- At first, the (local) actor and critic networks are updated 20 times in a row (one for each agent), using 20 different samples from the replay buffer as below:
```
def step(self, states, actions, rewards, next_states, dones):
...
# Learn (with each agent), if enough samples are available in memory
if len(self.memory) > BATCH_SIZE:
for i in range(self.num_agents):
experiences = self.memory.sample()
self.learn(experiences, GAMMA)
```
Then in order to get less aggressive with the number of updates per time step, instead of updating the actor and critic networks __20 times__ at __every timestep__, we amended the code to update the networks __10 times__ after every __20 timesteps__ (line )
```
def ddpg(n_episodes=1000, max_t=300, print_every=100,
num_agents=1):
"""
Params
======
n_episodes (int): maximum number of training episodes
max_t (int): maximum number of timesteps per episode
print_every (int): episodes interval to print training scores
num_agents (int): the number of agents
"""
scores_deque = deque(maxlen=print_every)
scores = []
for i_episode in range(1, n_episodes+1):
# reset the environment
env_info = env.reset(train_mode=True)[brain_name]
# get the current state (for each agent)
states = env_info.vector_observations
# initialize the scores (for each agent) of the current episode
scores_i = np.zeros(num_agents)
for t in range(max_t):
# select an action (for each agent)
actions = agent.act(states)
# send action to the environment
env_info = env.step(actions)[brain_name]
# get the next_state, reward, done (for each agent)
next_states = env_info.vector_observations
rewards = env_info.rewards
dones = env_info.local_done
# store experience and train the agent
agent.step(states, actions, rewards, next_states, dones,
update_every=20, update_times=10)
# roll over state to next time step
states = next_states
# update the score
scores_i += rewards
# exit loop if episode finished
if np.any(dones):
break
# save average of the most recent scores
scores_deque.append(scores_i.mean())
scores.append(scores_i.mean())
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_deque)), end="")
torch.save(agent.actor_local.state_dict(), 'd')
torch.save(agent.critic_local.state_dict(), 'checkpoint_critic.pth')
if i_episode % print_every == 0:
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_deque)))
return scores
scores = ddpg(n_episodes=200, max_t=1000, print_every=20, num_agents=20)
fig = plt.figure()
ax = fig.add_subplot(111)
plt.plot(np.arange(1, len(scores)+1), scores)
plt.ylabel('Score')
plt.xlabel('Episode #')
plt.show()
plt.savefig('ddpg_20_agents.png')
#env.close()
# load Actor-Critic policy
agent.actor_local.state_dict() = torch.load('checkpoint_actor.pth')
agent.critic_local.state_dict() = torch.load('checkpoint_critic.pth')
scores = ddpg(n_episodes=100, max_t=300, print_every=10, num_agents=20)
fig = plt.figure()
ax = fig.add_subplot(111)
plt.plot(np.arange(1, len(scores)+1), scores)
plt.ylabel('Score')
plt.xlabel('Episode #')
plt.show()
plt.savefig('ddpg_20_agents_101to200.png')
```
| github_jupyter |
# **OPTICS Algorithm**
Ordering Points to Identify the Clustering Structure (OPTICS) is a Clustering Algorithm which locates region of high density that are seperated from one another by regions of low density.
For using this library in Python this comes under Scikit Learn Library.
## Parameters:
**Reachability Distance** -It is defined with respect to another data point q(Let). The Reachability distance between a point p and q is the maximum of the Core Distance of p and the Euclidean Distance(or some other distance metric) between p and q. Note that The Reachability Distance is not defined if q is not a Core point.<br><br>
**Core Distance** – It is the minimum value of radius required to classify a given point as a core point. If the given point is not a Core point, then it’s Core Distance is undefined.
## OPTICS Pointers
<ol>
<li>Produces a special order of the database with respect to its density-based clustering structure.This cluster-ordering contains info equivalent to the density-based clustering corresponding to a broad range of parameter settings.</li>
<li>Good for both automatic and interactive cluster analysis, including finding intrinsic clustering structure</li>
<li>Can be represented graphically or using visualization technique</li>
</ol>
In this file , we will showcase how a basic OPTICS Algorithm works in Python , on a randomly created Dataset.
## Importing Libraries
```
import matplotlib.pyplot as plt #Used for plotting graphs
from sklearn.datasets import make_blobs #Used for creating random dataset
from sklearn.cluster import OPTICS #OPTICS is provided under Scikit-Learn Extra
from sklearn.metrics import silhouette_score #silhouette score for checking accuracy
import numpy as np
import pandas as pd
```
## Generating Data
```
data, clusters = make_blobs(
n_samples=800, centers=4, cluster_std=0.3, random_state=0
)
# Originally created plot with data
plt.scatter(data[:,0], data[:,1])
plt.show()
```
## Model Creation
```
# Creating OPTICS Model
optics_model = OPTICS(min_samples=50, xi=.05, min_cluster_size=.05)
#min_samples : The number of samples in a neighborhood for a point to be considered as a core point.
#xi : Determines the minimum steepness on the reachability plot that constitutes a cluster boundary
#min_cluster_size : Minimum number of samples in an OPTICS cluster, expressed as an absolute number or a fraction of the number of samples
pred =optics_model.fit(data) #Fitting the data
optics_labels = optics_model.labels_ #storing labels predicted by our model
no_clusters = len(np.unique(optics_labels) ) #determining the no. of unique clusters and noise our model predicted
no_noise = np.sum(np.array(optics_labels) == -1, axis=0)
```
## Plotting our observations
```
print('Estimated no. of clusters: %d' % no_clusters)
print('Estimated no. of noise points: %d' % no_noise)
colors = list(map(lambda x: '#aa2211' if x == 1 else '#120416', optics_labels))
plt.scatter(data[:,0], data[:,1], c=colors, marker="o", picker=True)
plt.title(f'OPTICS clustering')
plt.xlabel('Axis X[0]')
plt.ylabel('Axis X[1]')
plt.show()
# Generate reachability plot , this helps understand the working of our Model in OPTICS
reachability = optics_model.reachability_[optics_model.ordering_]
plt.plot(reachability)
plt.title('Reachability plot')
plt.show()
```
## Accuracy of OPTICS Clustering
```
OPTICS_score = silhouette_score(data, optics_labels)
OPTICS_score
```
On this randomly created dataset we got an accuracy of 84.04 %
### Hence , we can see the implementation of OPTICS Clustering Algorithm on a randomly created Dataset .As we can observe from our result . the score which we got is around 84% , which is really good for a unsupervised learning algorithm.However , this accuracy definitely comes with the additonal cost of higher computational power
## Thanks a lot!
| github_jupyter |
```
from datascience import *
path_data = '../data/'
import numpy as np
import matplotlib.pyplot as plots
plots.style.use('fivethirtyeight')
%matplotlib inline
```
# Finding Probabilities
Over the centuries, there has been considerable philosophical debate about what probabilities are. Some people think that probabilities are relative frequencies; others think they are long run relative frequencies; still others think that probabilities are a subjective measure of their own personal degree of uncertainty.
In this course, most probabilities will be relative frequencies, though many will have subjective interpretations. Regardless, the ways in which probabilities are calculated and combined are consistent across the different interpretations.
By convention, probabilities are numbers between 0 and 1, or, equivalently, 0% and 100%. Impossible events have probability 0. Events that are certain have probability 1.
Math is the main tool for finding probabilities exactly, though computers are useful for this purpose too. Simulation can provide excellent approximations, with high probability. In this section, we will informally develop a few simple rules that govern the calculation of probabilities. In subsequent sections we will return to simulations to approximate probabilities of complex events.
We will use the standard notation $P(\mbox{event})$ to denote the probability that "event" happens, and we will use the words "chance" and "probability" interchangeably.
## When an Event Doesn't Happen
If the chance that event happens is 40%, then the chance that it doesn't happen is 60%. This natural calculation can be described in general as follows:
$$
P(\mbox{an event doesn't happen}) ~=~ 1 - P(\mbox{the event happens})
$$
## When All Outcomes are Equally Likely
If you are rolling an ordinary die, a natural assumption is that all six faces are equally likely. Under this assumption, the probabilities of how one roll comes out can be easily calculated as a ratio. For example, the chance that the die shows an even number is
$$
\frac{\mbox{number of even faces}}{\mbox{number of all faces}}
~=~ \frac{\#\{2, 4, 6\}}{\#\{1, 2, 3, 4, 5, 6\}}
~=~ \frac{3}{6}
$$
Similarly,
$$
P(\mbox{die shows a multiple of 3}) ~=~
\frac{\#\{3, 6\}}{\#\{1, 2, 3, 4, 5, 6\}}
~=~ \frac{2}{6}
$$
In general, **if all outcomes are equally likely**,
$$
P(\mbox{an event happens}) ~=~
\frac{\#\{\mbox{outcomes that make the event happen}\}}
{\#\{\mbox{all outcomes}\}}
$$
Not all random phenomena are as simple as one roll of a die. The two main rules of probability, developed below, allow mathematicians to find probabilities even in complex situations.
## When Two Events Must Both Happen
Suppose you have a box that contains three tickets: one red, one blue, and one green. Suppose you draw two tickets at random without replacement; that is, you shuffle the three tickets, draw one, shuffle the remaining two, and draw another from those two. What is the chance you get the green ticket first, followed by the red one?
There are six possible pairs of colors: RB, BR, RG, GR, BG, GB (we've abbreviated the names of each color to just its first letter). All of these are equally likely by the sampling scheme, and only one of them (GR) makes the event happen. So
$$
P(\mbox{green first, then red}) ~=~
\frac{\#\{\mbox{GR}\}}{\#\{\mbox{RB, BR, RG, GR, BG, GB}\}} ~=~ \frac{1}{6}
$$
But there is another way of arriving at the answer, by thinking about the event in two stages. First, the green ticket has to be drawn. That has chance $1/3$, which means that the green ticket is drawn first in about $1/3$ of all repetitions of the experiment. But that doesn't complete the event. *Among the 1/3 of repetitions when green is drawn first*, the red ticket has to be drawn next. That happens in about $1/2$ of those repetitions, and so:
$$
P(\mbox{green first, then red}) ~=~ \frac{1}{2} ~\mbox{of}~ \frac{1}{3}
~=~ \frac{1}{6}
$$
This calculation is usually written "in chronological order," as follows.
$$
P(\mbox{green first, then red}) ~=~ \frac{1}{3} ~\times~ \frac{1}{2}
~=~ \frac{1}{6}
$$
The factor of $1/2$ is called " the conditional chance that the red ticket appears second, given that the green ticket appeared first."
In general, we have the **multiplication rule**:
$$
P(\mbox{two events both happen})
~=~ P(\mbox{one event happens}) \times
P(\mbox{the other event happens, given that the first one happened})
$$
Thus, when there are two conditions – one event must happen, as well as another – the chance is *a fraction of a fraction*, which is smaller than either of the two component fractions. The more conditions that have to be satisfied, the less likely they are to all be satisfied.
## When an Event Can Happen in Two Different Ways
Suppose instead we want the chance that one of the two tickets is green and the other red. This event doesn't specify the order in which the colors must appear. So they can appear in either order.
A good way to tackle problems like this is to *partition* the event so that it can happen in exactly one of several different ways. The natural partition of "one green and one red" is: GR, RG.
Each of GR and RG has chance $1/6$ by the calculation above. So you can calculate the chance of "one green and one red" by adding them up.
$$
P(\mbox{one green and one red}) ~=~ P(\mbox{GR}) + P(\mbox{RG}) ~=~
\frac{1}{6} + \frac{1}{6} ~=~ \frac{2}{6}
$$
In general, we have the **addition rule**:
$$
P(\mbox{an event happens}) ~=~
P(\mbox{first way it can happen}) + P(\mbox{second way it can happen}) ~~~
\mbox{}
$$
provided the event happens in exactly one of the two ways.
Thus, when an event can happen in one of two different ways, the chance that it happens is a sum of chances, and hence bigger than the chance of either of the individual ways.
The multiplication rule has a natural extension to more than two events, as we will see below. So also the addition rule has a natural extension to events that can happen in one of several different ways.
We end the section with examples that use combinations of all these rules.
## At Least One Success
Data scientists often work with random samples from populations. A question that sometimes arises is about the likelihood that a particular individual in the population is selected to be in the sample. To work out the chance, that individual is called a "success," and the problem is to find the chance that the sample contains a success.
To see how such chances might be calculated, we start with a simpler setting: tossing a coin two times.
If you toss a coin twice, there are four equally likely outcomes: HH, HT, TH, and TT. We have abbreviated "Heads" to H and "Tails" to T. The chance of getting at least one head in two tosses is therefore 3/4.
Another way of coming up with this answer is to work out what happens if you *don't* get at least one head. That is when both the tosses land tails. So
$$
P(\mbox{at least one head in two tosses}) ~=~ 1 - P(\mbox{both tails}) ~=~ 1 - \frac{1}{4}
~=~ \frac{3}{4}
$$
Notice also that
$$
P(\mbox{both tails}) ~=~ \frac{1}{4} ~=~ \frac{1}{2} \cdot \frac{1}{2} ~=~ \left(\frac{1}{2}\right)^2
$$
by the multiplication rule.
These two observations allow us to find the chance of at least one head in any given number of tosses. For example,
$$
P(\mbox{at least one head in 17 tosses}) ~=~ 1 - P(\mbox{all 17 are tails})
~=~ 1 - \left(\frac{1}{2}\right)^{17}
$$
And now we are in a position to find the chance that the face with six spots comes up at least once in rolls of a die.
For example,
$$
P(\mbox{a single roll is not 6}) ~=~ 1 - P(6)
~=~ \frac{5}{6}
$$
Therefore,
$$
P(\mbox{at least one 6 in two rolls}) ~=~ 1 - P(\mbox{both rolls are not 6})
~=~ 1 - \left(\frac{5}{6}\right)^2
$$
and
$$
P(\mbox{at least one 6 in 17 rolls})
~=~ 1 - \left(\frac{5}{6}\right)^{17}
$$
The table below shows these probabilities as the number of rolls increases from 1 to 50.
```
rolls = np.arange(1, 51, 1)
results = Table().with_columns(
'Rolls', rolls,
'Chance of at least one 6', 1 - (5/6)**rolls
)
results
```
The chance that a 6 appears at least once rises rapidly as the number of rolls increases.
```
results.scatter('Rolls')
```
In 50 rolls, you are almost certain to get at least one 6.
```
results.where('Rolls', are.equal_to(50))
```
Calculations like these can be used to find the chance that a particular individual is selected in a random sample. The exact calculation will depend on the sampling scheme. But what we have observed above can usually be generalized: increasing the size of the random sample increases the chance that an individual is selected.
| github_jupyter |
#
# <p style="color:red">Chapter 7</p>
### 1. What makes dictionaries different from sequence type containers like lists and tuples is the way the data are stored and accessed.
### 2.Sequence types use numeric keys only (numbered sequentially as indexed offsets from the beginning of the sequence). Mapping types may use most other object types as keys; strings are the most common.
### 3.Hash tabel: They store each piece of data, called a value, based on an associated data item, called a key.
Hash tables generally provide good performance because lookups occur fairly quickly once you have a key.
### 4. Dictionary is an unordered collection of data.
The only kind of ordering you can obtain is by taking either a dictionary’s set of keys or values. The keys() or values() method returns lists, which are sortable. You can also call items() to get a list of keys and values as tuple pairs and sort that. Dictionaries themselves have no implicit ordering because they are hashes.
### 5.create dictionary
The syntax of a dictionary entry is key:value. Also, dictionary entries are enclosed in braces ( { } ).
#### a.dictionary can be created by using {} with K,V pairs
```
adict={}
bdict={"k":"v"}
bdict
```
#### b. another way to create dictionary is using dict() method (factory fucntion)
```
fdict = dict((['x', 1], ['y', 2]))
cdict=dict([("k1",2),("k2",3)])
```
#### c.dictionaries may also be created using a very convenient built-in method for creating a “default” dictionary whose elements all have the same value (defaulting to None if not given), fromkeys():
```
ddict = {}.fromkeys(('x', 'y'), -1)
ddict
ddict={}.fromkeys(('x','y'),(2,3))
ddict
```
### 6.How to Access Values in Dictionaries
To traverse a dictionary (normally by key), you only need to cycle through its keys, like this:
```
dict2 = {'name': 'earth', 'port': 80}
for key in dict2.keys():
print("key=%s,value=%s" %(key,dict2[key]))
```
### a.Beginning with Python 2.2, you no longer need to use the keys() method to extract a list of keys to loop over. Iterators were created to simplify access- ing of sequence-like objects such as dictionaries and files. Using just the dictionary name itself will cause an iterator over that dictionary to be used in a for loop:
```
dict2 = {'name': 'earth', 'port': 80}
for key in dict2:
print("key=%s,value=%s" %(key,dict2[key]))
```
#### b.To access individual dictionary elements, you use the familiar square brackets along with the key to obtain its value:
```
dict2['name']
```
#### b. If we attempt to access a data item with a key that is not part of the dictio- nary, we get an error:
```
dict['service']
```
## c. The best way to check if a dic- tionary has a specific key is to use the in or not in operators
```
'service' in dict2
```
#### d. number can be the keys for dictionary
```
dict3 = {3.2: 'xyz', 1: 'abc', '1': 3.14159}
```
#### e. Not allowing keys to change during execution makes sense keys must be hashable, so numbers and strings are fine, but lists and other dictionaries are not.
### 7. update and add new dictionary
```
dict2['port'] = 6969 # update existing entry or add new entry
```
#### a. the string format operator (%) is specific for dictionary
```
print('host %(name)s is running on port %(port)d' % dict2)
dict2
```
### b. You may also add the contents of an entire dictionary to another dictionary by using the update() built-in method.3
### 8. remove dictionary elements:
#### a. use the del statement to delete an entire dictionary
```
del dict2['name'] # remove entry with key 'name'
dict2.pop('port') # remove and return entry with key
adict.clear() # remove all entries in adict
del bdict # delete entire dictionary
```
### Note: dict() is now a type and factory function, overriding it may cause you headaches and potential bugsDo NOT create variables with built-in names like: dict, list, file, bool, str, input, or len!
### 9.Dictionaries will work with all of the standard type operators but do not support operations such as concatenation and repetition.
#### a.Dictionary Key-Lookup Operator ( [ ] ). The key-lookup operator is used for both assigning values to and retrieving values from a dictionary
```
adict={"k":2}
adict["k"] = 3 # set value in dictionary. Dictionary Key-Lookup Operator ( [ ] )
cdict = {'fruits':1}
ddict = {'fruits':1}
```
## 10. dict() function
### a. The dict() factory function is used for creating dictionaries. If no argument is provided, then an empty dictionary is created. The fun happens when a container object is passed in as an argument to dict().
#### dict()
* dict() -> new empty dictionary
* dict(mapping) -> new dictionary initialized from a mapping object's (key, value) pairs
* dict(iterable) -> new dictionary initialized as if via:
d = {}
for k, v in iterable:
d[k] = v
* dict(**kwargs) -> new dictionary initialized with the name=value pairs in the keyword argument list. For example: dict(one=1, two=2)
```
dict()
dict({'k':1,'k2':2})
dict([(1,2),(2,3)])
dict(((1,2),(2,3)))
dict(([2,3],[3,4]))
dict(zip(('x','y'),(1,2)))
```
### 11. If it is a(nother) mapping object, i.e., a dictionary, then dict() will just create a new dictionary and copy the contents of the existing one. The new dictionary is actually a shallow copy of the original one and the same results can be accomplished by using a dictionary’s copy() built-in method. Because creating a new dictionary from an existing one using dict() is measurably slower than using copy(), we recommend using the latter.
### 12.it is possible to call dict() with an existing dic- tionary or keyword argument dictionary ( function operator)
```
dict7=dict(x=1, y=2)
dict8 = dict(x=1, y=2)
dict9 = dict(**dict8) # not a realistic example, better use copy()
dict10=dict(dict7)
dict9
dict10
dict9 = dict8.copy() # better than dict9 = dict(**dict8)
```
### 13.The len() BIF is flexible. It works with sequences, mapping types, and sets
```
dict2 = {'name': 'earth', 'port': 80}
len(dict2)
```
### 14. We can see that above, when referencing dict2, the items are listed in reverse order from which they were entered into the dictionary. ??????
```
dict2 = {'name': 'earth', 'port': 80}
dict2
```
### 15.The hash() BIF is not really meant to be used for dictionaries per se, but it can be used to determine whether an object is fit to be a dictionary key (or not).
* Given an object as its argument, hash() returns the hash value of that object.
* Numeric val- ues that are equal hash to the same value.
* A TypeError will occur if an unhashable type is given as the argument to hash()
```
hash([])
dict2={}
dict2[{}]="foo"
```
### 16. Mapping Type Built-in Methods
* has_key() and its replacements in and not in
* keys(), which returns a list of the dictionary’s keys,
* values(), which returns a list of the dictionary’s values, and
* items(), which returns a list of (key, value) tuple pairs.
```
dict2={"k1":1,"k2":2,"k3":3}
for eachKey in dict2.keys():
print(eachKey,dict2[eachKey])
```
#### * dict.fromkeysc (seq, val=None): create dict where all the key in seq have teh same value val
```
{}.fromkeys(("k1","k2","3"),None) # fromkeysc(seq, val=None)
```
#### * get(key,default=None): return the value corresponding to key, otherwise return None if key is not in dictionary
#### * dict.setdefault (key, default=None): Similar to get(), but sets dict[key]=default if key is not already in dict
#### * dict.setdefault(key, default=None): Add the key-value pairs of dict2 to dict
#### * keys() method to get the list of its keys, then call that list’s sort() method to get a sorted list to iterate over. sorted(), made especially for iterators, exists, which returns a sorted iterator:
```
for eachKey in sorted(dict2):
print(eachKey,dict2[eachKey])
```
#### * The update() method can be used to add the contents of one dictionary to another. Any existing entries with duplicate keys will be overridden by the new incoming entries. Nonexistent ones will be added. All entries in a dictio- nary can be removed with the clear() method.
```
dict2
dict3={"k1":"ka","kb":"kb"}
dict2.update(dict3)
dict2
dict3.clear()
dict3
del dict3
dict3
```
#### * The copy() method simply returns a copy of a dictionary.
#### * the get() method is similar to using the key-lookup operator ( [ ] ), but allows you to provide a default value returned if a key does not exist.
```
dict2
dict4=dict2.copy()
dict4
dict4.get('xgag')
type(dict4.get("agasg"))
type(dict4.get("agasg", "no such key"))
dict2
```
#### * f the dictionary does not have the key you are seeking, you want to set a default value and then return it. That is precisely what setdefault() does
```
dict2.setdefault('kk','k1')
dict2
```
#### * Currently,thekeys(),items(),andvalues()methodsreturnlists. This can be unwieldy if such data collections are large, and the main reason why iteritems(), iterkeys(), and itervalues() were added to Python
#### * In python3, iter*() names are no longer supported. The new keys(), values(), and items() all return views
#### * When key collisions are detected (meaning duplicate keys encountered during assignment), the last (most recent) assignment wins.
```
dict1 = {' foo':789, 'foo': 'xyz'}
dict1
dict1['foo']
```
#### * most Python objects can serve as keys; however they have to be hashable objects—mutable types such as lists and dictionaries are disallowed because they cannot be hashed
* All immutable types are hashable
* Numbers of the same value represent the same key. In other words, the integer 1 and the float 1.0 hash to the same value, meaning that they are identical as keys.
* there are some mutable objects that are (barely) hashable, so they are eligible as keys, but there are very few of them. One example would be a class that has implemented the __hash__() special method. In the end, an immutable value is used anyway as __hash__() must return an integer.
* Why must keys be hashable? The hash function used by the interpreter to calculate where to store your data is based on the value of your key. If the key was a mutable object, its value could be changed. If a key changes, the hash function will map to a different place to store the data. If that was the case, then the hash function could never reliably store or retrieve the associated value
* : Tuples are valid keys only if they only contain immutable arguments like numbers and strings.
### 19.A set object is an unordered collection of distinct values that are hashable.
#### a.Like other container types, sets support membership testing via in and not in operators, cardinality using the len()BIF, and iteration over the set membership using for loops. However, since sets are unordered, you do not index into or slice them, and there are no keys used to access a value.
#### b.There are two different types of sets available, mutable (set) and immuta- ble (frozenset).
#### c.Note that mutable sets are not hashable and thus cannot be used as either a dictionary key or as an element of another set.
#### d.sets module and accessed via the ImmutableSet and Set classes.
#### d. sets can be created, using their factory functions set() and frozenset():
```
s1=set('asgag')
s2=frozenset('sbag')
len(s1)
type(s1)
type(s2)
len(s2)
```
#### f. iterate over set or check if an item is a member of a set
```
'k' in s1
```
#### g. update set
```
s1.add('z')
s1
s1.update('abc')
s1
s1.remove('a')
s1
```
#### h.As mentioned before, only mutable sets can be updated. Any attempt at such operations on immutable sets is met with an exception
```
s2.add('c')
```
#### i. mixed set type operation
```
type(s1|s2) # set | frozenset, mix operation
s3=frozenset('agg') #frozenset
type(s2|s3)
```
#### j. update mutable set: (Union) Update ( |= )
```
s=set('abc')
s1=set("123")
s |=s1
s
```
### k.The retention (or intersection update) operation keeps only the existing set members that are also elements of the other set. The method equivalent is intersection_update()
```
s=set('abc')
s1=set('ab')
s &=s1
s
```
### l.The difference update operation returns a set whose elements are members of the original set after removing elements that are (also) members of the other set. The method equivalent is difference_update().
```
s = set('cheeseshop')
s
u = frozenset(s)
s -= set('shop')
s
```
#### m.The symmetric difference update operation returns a set whose members are either elements of the original or other set but not both. The method equiva- lent is symmetric_difference_update()
```
s=set('cheeseshop')
s
u=set('bookshop')
u
s ^=u
s
vari='abc'
set(vari) # vari must be iterable
```
### n.The new methods here are
* add(),
* remove(),
* discard(),
* pop(), and
* clear().
* s.copy(), Copy operation: return (shallow) copy of s
For the methods that take an object, the argument must be hashable.
| github_jupyter |
# Chapter 7. 텍스트 문서의 범주화 - (4) IMDB 전체 데이터로 전이학습
- 앞선 전이학습 실습과는 달리, IMDB 영화리뷰 데이터셋 전체를 사용하며 문장 수는 10개 -> 20개로 조정한다
- IMDB 영화 리뷰 데이터를 다운로드 받아 data 디렉토리에 압축 해제한다
- 다운로드 : http://ai.stanford.edu/~amaas/data/sentiment/
- 저장경로 : data/aclImdb
```
import os
import config
from dataloader.loader import Loader
from preprocessing.utils import Preprocess, remove_empty_docs
from dataloader.embeddings import GloVe
from model.cnn_document_model import DocumentModel, TrainingParameters
from keras.callbacks import ModelCheckpoint, EarlyStopping
import numpy as np
```
## 학습 파라미터 설정
```
# 학습된 모델을 저장할 디렉토리 생성
if not os.path.exists(os.path.join(config.MODEL_DIR, 'imdb')):
os.makedirs(os.path.join(config.MODEL_DIR, 'imdb'))
# 학습 파라미터 설정
train_params = TrainingParameters('imdb_transfer_tanh_activation',
model_file_path = config.MODEL_DIR+ '/imdb/full_model_10.hdf5',
model_hyper_parameters = config.MODEL_DIR+ '/imdb/full_model_10.json',
model_train_parameters = config.MODEL_DIR+ '/imdb/full_model_10_meta.json',
num_epochs=30,
batch_size=128)
```
## IMDB 데이터셋 로드
```
# 다운받은 IMDB 데이터 로드: 학습셋 전체 사용
train_df = Loader.load_imdb_data(directory = 'train')
# train_df = train_df.sample(frac=0.05, random_state = train_params.seed)
print(f'train_df.shape : {train_df.shape}')
test_df = Loader.load_imdb_data(directory = 'test')
print(f'test_df.shape : {test_df.shape}')
# 텍스트 데이터, 레이블 추출
corpus = train_df['review'].tolist()
target = train_df['sentiment'].tolist()
corpus, target = remove_empty_docs(corpus, target)
print(f'corpus size : {len(corpus)}')
print(f'target size : {len(target)}')
```
## 인덱스 시퀀스 생성
```
# 앞선 전이학습 실습과 달리, 문장 개수를 10개 -> 20개로 상향
Preprocess.NUM_SENTENCES = 20
# 학습셋을 인덱스 시퀀스로 변환
preprocessor = Preprocess(corpus=corpus)
corpus_to_seq = preprocessor.fit()
print(f'corpus_to_seq size : {len(corpus_to_seq)}')
print(f'corpus_to_seq[0] size : {len(corpus_to_seq[0])}')
# 테스트셋을 인덱스 시퀀스로 변환
test_corpus = test_df['review'].tolist()
test_target = test_df['sentiment'].tolist()
test_corpus, test_target = remove_empty_docs(test_corpus, test_target)
test_corpus_to_seq = preprocessor.transform(test_corpus)
print(f'test_corpus_to_seq size : {len(test_corpus_to_seq)}')
print(f'test_corpus_to_seq[0] size : {len(test_corpus_to_seq[0])}')
# 학습셋, 테스트셋 준비
x_train = np.array(corpus_to_seq)
x_test = np.array(test_corpus_to_seq)
y_train = np.array(target)
y_test = np.array(test_target)
print(f'x_train.shape : {x_train.shape}')
print(f'y_train.shape : {y_train.shape}')
print(f'x_test.shape : {x_test.shape}')
print(f'y_test.shape : {y_test.shape}')
```
## GloVe 임베딩 초기화
```
# GloVe 임베딩 초기화 - glove.6B.50d.txt pretrained 벡터 사용
glove = GloVe(50)
initial_embeddings = glove.get_embedding(preprocessor.word_index)
print(f'initial_embeddings.shape : {initial_embeddings.shape}')
```
## 훈련된 모델 로드
- HandsOn03에서 아마존 리뷰 데이터로 학습한 CNN 모델을 로드한다.
- DocumentModel 클래스의 load_model로 모델을 로드하고, load_model_weights로 학습된 가중치를 가져온다.
- 그 후, GloVe.update_embeddings 함수로 GloVe 초기화 임베딩을 업데이트한다
```
# 모델 하이퍼파라미터 로드
model_json_path = os.path.join(config.MODEL_DIR, 'amazonreviews/model_06.json')
amazon_review_model = DocumentModel.load_model(model_json_path)
# 모델 가중치 로드
model_hdf5_path = os.path.join(config.MODEL_DIR, 'amazonreviews/model_06.hdf5')
amazon_review_model.load_model_weights(model_hdf5_path)
# 모델 임베딩 레이어 추출
learned_embeddings = amazon_review_model.get_classification_model().get_layer('imdb_embedding').get_weights()[0]
print(f'learned_embeddings size : {len(learned_embeddings)}')
# 기존 GloVe 모델을 학습된 임베딩 행렬로 업데이트한다
glove.update_embeddings(preprocessor.word_index,
np.array(learned_embeddings),
amazon_review_model.word_index)
# 업데이트된 임베딩을 얻는다
initial_embeddings = glove.get_embedding(preprocessor.word_index)
```
## IMDB 전이학습 모델 생성
```
# 분류 모델 생성 : IMDB 리뷰 데이터를 입력받아 이진분류를 수행하는 모델 생성
imdb_model = DocumentModel(vocab_size=preprocessor.get_vocab_size(),
word_index = preprocessor.word_index,
num_sentences=Preprocess.NUM_SENTENCES,
embedding_weights=initial_embeddings,
embedding_regularizer_l2 = 0.0,
conv_activation = 'tanh',
train_embedding = True, # 임베딩 레이어의 가중치 학습함
learn_word_conv = False, # 단어 수준 conv 레이어의 가중치 학습 안 함
learn_sent_conv = False, # 문장 수준 conv 레이어의 가중치 학습 안 함
hidden_dims=64,
input_dropout=0.1,
hidden_layer_kernel_regularizer=0.01,
final_layer_kernel_regularizer=0.01)
# 가중치 업데이트 : 생성한 imdb_model 모델에서 다음의 각 레이어들의 가중치를 위에서 로드한 가중치로 갱신한다
for l_name in ['word_conv','sentence_conv','hidden_0', 'final']:
new_weights = amazon_review_model.get_classification_model().get_layer(l_name).get_weights()
imdb_model.get_classification_model().get_layer(l_name).set_weights(weights=new_weights)
```
## 모델 학습 및 평가
```
# 모델 컴파일
imdb_model.get_classification_model().compile(loss="binary_crossentropy",
optimizer='rmsprop',
metrics=["accuracy"])
# callback (1) - 체크포인트
checkpointer = ModelCheckpoint(filepath=train_params.model_file_path,
verbose=1,
save_best_only=True,
save_weights_only=True)
# callback (2) - 조기종료
early_stop = EarlyStopping(patience=2)
# 학습 시작
imdb_model.get_classification_model().fit(x_train,
y_train,
batch_size=train_params.batch_size,
epochs=train_params.num_epochs,
verbose=2,
validation_split=0.01,
callbacks=[checkpointer])
# 모델 저장
imdb_model._save_model(train_params.model_hyper_parameters)
train_params.save()
# 모델 평가
imdb_model.get_classification_model().evaluate(x_test,
y_test,
batch_size=train_params.batch_size*10,
verbose=2)
```
| github_jupyter |
```
import pandas as pd
from bs4 import BeautifulSoup as soup
from splinter import Browser
import requests
import time
from webdriver_manager.chrome import ChromeDriverManager
from selenium import webdriver
!pip install chromedriver
driver = webdriver.Chrome(ChromeDriverManager().install())
#driver = webdriver.chrome(executable_path='C:/path/to/chromedriver.exe')
#pointing to the directory where chromedriver exists
executable_path = {"executable_path":"C:\\Users\\alex2\\OneDrive\Desktop\chromedriver"}
browser = Browser('chrome', **executable_path, headless=False)
#url = "https://mars.nasa.gov/news/"
# Mars news url
Marsnews_url = 'https://mars.nasa.gov/news/'
browser.visit(Marsnews_url)
# create beautiful soup object
html = browser.html
MarsNews_soup = soup(html, 'html.parser')
# First news Title
News_title = MarsNews_soup.body.find("div", class_="content_title").text
NewsParagraph = MarsNews_soup.body.find("div", class_="article_teaser_body").text
print(f"The title is: \n{News_title}")
print()
print(f"The descriptive paragraph is: \n{NewsParagraph}")
# JPL Mars Space Images
## define the url and visit it with browser
mars_image_url = "https://www.jpl.nasa.gov/spaceimages/?search=&category=Mars"
base_url = 'https://www.jpl.nasa.gov'
browser.visit(mars_image_url)
#browser.visit(url)
#html = browser.html
#MarsNews_soup = soup(html, 'html.parser')
#image_url = soup.find('article')['style'].replace('background-image: url(','').replace(');', '')[1:-1]
# Website Url
#main_url = "https://www.jpl.nasa.gov"
# Concatenate website url with scrapped route
#image_url = main_url + image_url
# Display full link to featured image
#image_url
# HTML object
html=browser.html
# Parse HTML
MarsNews_soup = soup(html,"html.parser")
# Retrieve image url
image_url=soup.find_all('article')
image_url=soup.find("a", class_ = "button fancybox")["data-fancybox-href"]
featured_image_url = base_url + image_url
print(featured_image_url)
# Mars Facts
url = 'https://space-facts.com/mars/'
browser.visit(url)
# Use Pandas to "read_html" to parse the URL
tables = pd.read_html(url)
# Mars Facts DataFrame
facts_df = tables[0]
facts_df.columns = ['Fact', 'Value']
facts_df['Fact'] = facts_df['Fact'].str.replace(':', '')
facts_df
# Show as html table string
facts_df = tables[0]
facts_df.columns = ['Fact', 'Value']
facts_df['Fact'] = facts_df['Fact'].str.replace(':', '')
facts_df
facts_html = facts_df.to_html()
print(facts_html)
# Mars Hemispheres
hemispheres_url = "https://astrogeology.usgs.gov/search/results?q=hemisphere+enhanced&k1=target&v1=Mars"
browser.visit(hemispheres_url)
# HTML Object
html_hemispheres = browser.html
# Parse HTML with Beautiful Soup
MarsNews_soup = soup(html_hemispheres, 'html.parser')
# Retreive all items that contain mars hemispheres information
items = soup.find_all('div', class_='item')
# Create empty list for hemisphere urls
hemisphere_image_urls = []
# Store the main_ul
hemispheres_main_url = 'https://astrogeology.usgs.gov'
# Loop through the items previously stored
for i in items:
# Store title
title = i.find('h3').text
# Store link that leads to full image website
partial_img_url = i.find('a', class_='itemLink product-item')['href']
# Visit the link that contains the full image website
browser.visit(hemispheres_main_url + partial_img_url)
# HTML Object of individual hemisphere information website
partial_img_html = browser.html
# Parse HTML with Beautiful Soup for every individual hemisphere information website
MarsNews_soup = soup( partial_img_html, 'html.parser')
# Retrieve full image source
image_url = hemispheres_main_url + soup.find('img',class_='wide-image')['src']
# Append the retreived information into a list of dictionaries
hemisphere_image_urls.append({"title" : title, "img_url" : img_url})
# Display hemisphere_image_urls
hemisphere_image_urls
```
| github_jupyter |
### Lgbm and Optuna
* changed with cross validation
```
import pandas as pd
import numpy as np
# the GBM used
mport xgboost as xgb
import catboost as cat
import lightgbm as lgb
from sklearn.model_selection import cross_validate
from sklearn.metrics import make_scorer
# to encode categoricals
from sklearn.preprocessing import LabelEncoder
# see utils.py
from utils import add_features, rmsle, train_encoders, apply_encoders
import warnings
warnings.filterwarnings('ignore')
import optuna
# globals and load train dataset
FILE_TRAIN = "train.csv"
# load train dataset
data_orig = pd.read_csv(FILE_TRAIN)
#
# Data preparation, feature engineering
#
# add features (hour, year) extracted form timestamp
data_extended = add_features(data_orig)
# ok, we will treat as categorical: holiday, hour, season, weather, workingday, year
all_columns = data_extended.columns
# cols to be ignored
# atemp and temp are strongly correlated (0.98) we're taking only one
del_columns = ['datetime', 'casual', 'registered', 'temp']
TARGET = "count"
cat_cols = ['season', 'holiday','workingday', 'weather', 'hour', 'year']
num_cols = list(set(all_columns) - set([TARGET]) - set(del_columns) - set(cat_cols))
features = sorted(cat_cols + num_cols)
# drop ignored columns
data_used = data_extended.drop(del_columns, axis=1)
# Code categorical columns (only season, weather, year)
le_list = train_encoders(data_used)
# coding
data_used = apply_encoders(data_used, le_list)
# define indexes for cat_cols
# cat boost want indexes
cat_columns_idxs = [i for i, col in enumerate(features) if col in cat_cols]
# finally we have the train dataset
X = data_used[features].values
y = data_used[TARGET].values
# general
FOLDS = 5
SEED = 4321
N_TRIALS = 5
STUDY_NAME = "gbm3"
#
# Here we define what we do using Optuna
#
def objective(trial):
# tuning on max_depth, n_estimators for the example
dict_params = {
"num_iterations": trial.suggest_categorical("num_iterations", [3000, 4000, 5000]),
"learning_rate": trial.suggest_loguniform("learning_rate", low=1e-4, high=1e-2),
"metrics" : ["rmse"],
"verbose" : -1,
}
max_depth = trial.suggest_int("max_depth", 4, 10)
num_leaves = trial.suggest_int("num_leaves", 2**(max_depth), 2**(max_depth))
dict_params['max_depth'] = max_depth
dict_params['num_leaves'] = num_leaves
regr = lgb.LGBMRegressor(**dict_params)
# using rmsle for scoring
scorer = make_scorer(rmsle, greater_is_better=False)
scores = cross_validate(regr, X, y, cv=FOLDS, scoring=scorer)
avg_test_score = round(np.mean(scores['test_score']), 4)
return avg_test_score
# launch Optuna Study
study = optuna.create_study(study_name=STUDY_NAME, direction="maximize")
study.optimize(objective, n_trials=N_TRIALS)
study.best_params
# visualize trials as an ordered Pandas df
df = study.trials_dataframe()
result_df = df[df['state'] == 'COMPLETE'].sort_values(by=['value'], ascending=False)
# best on top
result_df.head()
```
### train the model on entire train set and save
```
%%time
# maybe I shoud add save best model (see nu_iteration in cell below)
model = lgb.LGBMRegressor(**study.best_params)
model.fit(X, y)
model_file = "lgboost.txt"
model.booster_.save_model(model_file, num_iteration=study.best_params['num_iterations'])
```
| github_jupyter |
# "Tuesday Wonderland and PLOT Fidel Huancas"
> "In this blog post we head back to the fine folks at PLOT coffee roasting this time looking at a Peruvian competition lot. We pair this with the Esbjörn Svennson Trio classic 'Tuesday Wonderland' from 2006"
- toc: false
- author: Lewis Cole (2020)
- branch: master
- badges: false
- comments: false
- categories: [Jazz, Coffee, EST, 2000s, Plot, Peru, Washed-Process, Caturra]
- hide: false
- search_exclude: false
- image: https://github.com/jazzcoffeestuff/blog/raw/master/images/013-Tuesday-Wonderland/Tuesday-Wonderland.jpg
> youtube: https://youtu.be/9VF8kxFEEsA
This week we are heading back to a relatively new roaster for me: PLOT coffee roasters in Woolwich. Readers may remember a trio of their coffees featured on the page a couple of months ago which were heavily favoured. This is part of my second order from them. I'm hesitant to judge a roaster based on a single order so we will see if this one lives up to the previous coffees from them.
Again I am very impressed with the packaging, the pink and black branded packing boxes through to the "sound wave" style logos on the bags it is all very well done. The packaging is something I always look at when judging a roaster/coffee, it is my opinion that you make judgements about a coffee from the instant you see it and having attractive packaging adds to this. You "first drink with your eyes" is the expression I believe! Further good packaging to me suggests that a roaster is leaving no stone unturned, they are concerned with how their coffee is perceived at every step of the coffee drinking journey - I think this is a good indicator of attention to detail and passion, both of which are crucially important to find in a roaster.
The coffee today is called: "Fidel Huancas - Chirinos Competition Lot" - which is a bit of a mouthful. Fidel Huancas is the man behind the coffee, the producer who is responsible for ensuring the quality of the on site processing, growing, etc. Chirinos is the district of the province San Ignacio where the farm is located. This is in the north west corner of the country, not too far from the border with Ecuador. The term "competition lot" has sprung up more and more in recent years, it does not seem to be a protected term but is meant to be an indicator that the coffee is of a quality that it could be used in barita or brewing competitions.
A piece of information you often find on the label of a coffee or on the roasters website is "altitude" or "elevation" - this is measured in meters above sea level (MASL) and as one would expect represents how "high" the coffee bushes are grown. When I was first getting into specialty coffee many many moons ago people were obsessing over altitude, people often judging whether to buy a coffee based on this metric alone. These days it appears as though the metric is not given the weight it once was, which in my opinion is a good thing. Generally speaking a coffee at a higher altitude will be favoured as being sweeter and more complex, and so some aim for higher altitude coffees as a result. Unfortunately it is not quite that simple. Higher does not necessarily mean better, what really matters (or so it seems) is the speed at which the coffee plants grow which is related to temperature. At a higher altitude generally the temperature will be lower which means the plants will grow more slowly and develop the complex flavours and sugars we are after. However we can equally find lower altitude locations that have lower temperatures and slower growing cherries which are equally delicious!
Unfortunately temperature is not something that is easy to quantify, each day the temperature will fluctuate and over the course of a growing season there may be large swings. Even the "average temperature" nor the temperature range tell us very much at all. As such altitude has become the proxy metric used. Personally I never look at altitude at all when deciding on whether to buy or try a coffee, I have not found it a particularly useful indicator for any particular characteristic or quality. Everybody is different though, so if you find that there is a strong correlation between coffees you enjoy and the altitude they're grown at stick with it!
Moving onto the tasting of this coffee. The first thing that strikes me with this coffee is the dry aroma of the beans (both whole and ground) - the smell is very intense. I'd describe it as a dried fruit sweet sort of smell laced with a delicate floral note. I usually find that the very best tasting coffees have a very intense dry aroma, it is not universally true and a great dry aroma does not always translate to a good cup but this coffee is onto a good start! As usual starting with my usual filter set up. The first thing I get hit with is apricot, this coffee has it in spades! Those that know me know that, while I'm not big on desserts, an apricot tart is one of my favourite things. This coffee reminds me a bit of that. If apricot is the head note of the coffee the heart is sweet caramel which gives some body to the coffee, perhaps apricot compote is a good descriptor given this syrupy body. As the coffee cools floral notes begin to appear and these linger in the aftertaste which is long lived and clean. Moving onto the espresso the flavour profile is much the same. I found that this coffee likes to be pulled long, about a 2.7:1 works for me. Too short and the flavours get a bit muddled and it's hard to discern individual notes. As a longer pull the flavours are defined and totally delicious! At this level the syrupy mouthfeel has all but gone but I feel this is a fair compromise. Pulling out floral notes in an espresso can be difficult, they are often dominated by the "heavier" notes but they manage to punch through in the aftertaste here.
This is another stand out coffee for me from the folks at PLOT. If this one is not in my shortlist for "coffees of the year" I have some outstanding coffees to look forward to in the last 6 months of the year!
This week's jazz is the 2006 seminal album "Tuesday Wonderland" by Esbjörn Svennson Trio (EST) on the ACT record label:

I remember the lead up to this album being released very vividly, at the time I was a regular reader of Jazzwise magazine and it felt like every issue for the year leading up to the release (and subsequently) had an article about this album. By this point EST were already big names in the jazz world. In the late 1990s to early 2000s jazz was in a bit of a lull, by this point the whole "fusion" thing had lost its way and there was a movement back towards more "traditional" styles, however what was lacking was a young act who won international acclaim, this is where EST came in. They almost singlehandedly kept jazz alive during this period and spawned many imitators who are still playing with the "EST sound" today. Unusually for an album surrounded in such hype and expectation "Tuesday Wonderland" not only lived up to it but possibly exceeded it, which is not something you can say often.
The EST style is hard to put down in words but unmistakable once you hear it. There is a clear influence of modern European classical composers (they list Bartok in particular as an influence regularly). Esbjörn's piano style also takes inspiration from Keith Jarrett to my ear. While each member plays (primarily) on acoustic instruments they layer these with multi-track recording and run electronic effects to keep things fresh. Magnus Öström is on drum duty for the band and plays with both manic urgency as well as tender subtlty, feeling most at home playing drum and bass and techno grooves. Dan Berglund on bass brings with him the thunderous weight through the rock and heavy metal influence to preceedings. EST represents "fusion" in its truest sense being a melting pot of ideas. In doing this they managed to create a sound all to themselves that is not like anything that came before it.
One of my favourite pieces from the entire EST back catalogue is "The Goldhearted Miner" (embedded above). It is a beautiful subtle piece with a very strong melody. It is the sort of tune I could imagine being a standard if it had been released some 60 years earlier. It does not have quite the "broad influence" that some of the other pieces on the album have however. Perhaps their biggest hit is "Goldwrap":
> youtube: https://youtu.be/xCuGTuDsVoY
This even had a "proper" music video associated with it! Back in 2006 this was essentially unheard of, I can't think of another record from the era that had that sort of reach. It shows how big the band had become, I seem to remember even seeing this appear on the music TV channels on satelite television at the time. Compared to "The Goldhearted Miner" this one features more prominently some of the electronic effects on the instruments and some of the EST-isms that made the band so popular.
Live the band were also a force to be reckoned with. They managed to attract a young "hip" crowd that comprised of jazz fans, electronic music fans and rock fans alike. The crowd hanging on every single the note, and the band acting like a conductor whipping the audience up into a frenzy. There was also a running "bit" that Esbjörn did; the band would play 3 or 4 tunes on the bounce and then Esbjörn would try to announce what was just played to the crowd but "forgot" the names and the more eager members of the audience would shout out to remind him. I'm not sure if it was a genuine slip of the mind from being so deep into the moment or whether it was just a shtick, either way it certainly added a little humour to the gigs.
Unfortunately Esbjörn died tragically young, at just the age of 44, in a freak scuba diving accident in 2008 only 2 years after the release of "Tuesday Wonderland". This was a devestating event in recent jazz history and he will be sorely missed. I was forunate enough to catch, what turned out to be, one of the last UK shows. It was truly fantastic and a totally memorable experience. Afterwards I even got to meet the band as they hung around for autographs and the like. Unfortunately camera phones of the day were still fairly rudimentary (at least the one I had) so I do not have a high-def picture but you can see me as a fresh-faced wee nipper below:

To end this post I'll leave one of the more experimental cuts from the album called: "Brewery of Beggars". I feel this really captures the "melting pot" ethos of the band and highlights the use of electronic effects and rockier sounds the band were known for.
> youtube: https://youtu.be/3umiaQLwR38
| github_jupyter |
```
import pandas as pd
import numpy as np
data = np.array([1,2,3,4,5,6])
name = np.array(['' for x in range(6)])
besio = np.array(['' for x in range(6)])
entity = besio
columns = ['name/doi', 'data', 'BESIO', 'entity']
df = pd.DataFrame(np.array([name, data, besio, entity]).transpose(), columns=columns)
df.iloc[1,0] = 'doi'
hey = np.random.shuffle(data)
for piece in np.random.shuffle(data):
print(piece)
df
filename = 'carbon_ner_labels.xlsx'
append_df_to_excel(filename, df, startcol=0)
append_df_to_excel(filename, df, startcol=6)
def append_df_to_excel(filename, df, sheet_name='Sheet1', startrow=0, startcol=None,
truncate_sheet=False,
**to_excel_kwargs):
"""
Append a DataFrame [df] to existing Excel file [filename]
into [sheet_name] Sheet.
If [filename] doesn't exist, then this function will create it.
Parameters:
filename : File path or existing ExcelWriter
(Example: '/path/to/file.xlsx')
df : dataframe to save to workbook
sheet_name : Name of sheet which will contain DataFrame.
(default: 'Sheet1')
startrow : upper left cell row to dump data frame.
Per default (startrow=None) calculate the last row
in the existing DF and write to the next row...
startcol : upper left cell column to dump data frame.
truncate_sheet : truncate (remove and recreate) [sheet_name]
before writing DataFrame to Excel file
to_excel_kwargs : arguments which will be passed to `DataFrame.to_excel()`
[can be dictionary]
Returns: None
"""
from openpyxl import load_workbook
import pandas as pd
# ignore [engine] parameter if it was passed
if 'engine' in to_excel_kwargs:
to_excel_kwargs.pop('engine')
writer = pd.ExcelWriter(filename, engine='openpyxl')
# Python 2.x: define [FileNotFoundError] exception if it doesn't exist
try:
FileNotFoundError
except NameError:
FileNotFoundError = IOError
try:
# try to open an existing workbook
writer.book = load_workbook(filename)
if startcol is None and sheet_name in writer.book.sheetnames:
startcol = writer.book[sheet_name].max_col
# truncate sheet
if truncate_sheet and sheet_name in writer.book.sheetnames:
# index of [sheet_name] sheet
idx = writer.book.sheetnames.index(sheet_name)
# remove [sheet_name]
writer.book.remove(writer.book.worksheets[idx])
# create an empty sheet [sheet_name] using old index
writer.book.create_sheet(sheet_name, idx)
# copy existing sheets
writer.sheets = {ws.title:ws for ws in writer.book.worksheets}
except FileNotFoundError:
# file does not exist yet, we will create it
pass
if startcol is None:
startcol = 0
# write out the new sheet
df.to_excel(writer, sheet_name, startrow=startrow, startcol=startcol, **to_excel_kwargs)
# save the workbook
writer.save()
```
| github_jupyter |
# TensorFlow Tutorial #01
# Simple Linear Model
by [Magnus Erik Hvass Pedersen](http://www.hvass-labs.org/)
/ [GitHub](https://github.com/Hvass-Labs/TensorFlow-Tutorials) / [Videos on YouTube](https://www.youtube.com/playlist?list=PL9Hr9sNUjfsmEu1ZniY0XpHSzl5uihcXZ)
## Introduction
This tutorial demonstrates the basic workflow of using TensorFlow with a simple linear model. After loading the so-called MNIST data-set with images of hand-written digits, we define and optimize a simple mathematical model in TensorFlow. The results are then plotted and discussed.
You should be familiar with basic linear algebra, Python and the Jupyter Notebook editor. It also helps if you have a basic understanding of Machine Learning and classification.
## Imports
```
%matplotlib inline
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
import tensorflow as tf
import numpy as np
from sklearn.metrics import confusion_matrix
```
This was developed using Python 3.6 (Anaconda) and TensorFlow version:
```
tf.__version__
```
## Load Data
The MNIST data-set is about 12 MB and will be downloaded automatically if it is not located in the given path.
```
from mnist import MNIST
data = MNIST(data_dir="data/MNIST/")
```
The MNIST data-set has now been loaded and consists of 70.000 images and class-numbers for the images. The data-set is split into 3 mutually exclusive sub-sets. We will only use the training and test-sets in this tutorial.
```
print("Size of:")
print("- Training-set:\t\t{}".format(data.num_train))
print("- Validation-set:\t{}".format(data.num_val))
print("- Test-set:\t\t{}".format(data.num_test))
```
Copy some of the data-dimensions for convenience.
```
# The images are stored in one-dimensional arrays of this length.
img_size_flat = data.img_size_flat
# Tuple with height and width of images used to reshape arrays.
img_shape = data.img_shape
# Number of classes, one class for each of 10 digits.
num_classes = data.num_classes
```
### One-Hot Encoding
The output-data is loaded as both integer class-numbers and so-called One-Hot encoded arrays. This means the class-numbers have been converted from a single integer to a vector whose length equals the number of possible classes. All elements of the vector are zero except for the $i$'th element which is 1 and means the class is $i$. For example, the One-Hot encoded labels for the first 5 images in the test-set are:
```
data.y_test[0:5, :]
```
We also need the classes as integers for various comparisons and performance measures. These can be found from the One-Hot encoded arrays by taking the index of the highest element using the `np.argmax()` function. But this has already been done for us when the data-set was loaded, so we can see the class-number for the first five images in the test-set. Compare these to the One-Hot encoded arrays above.
```
data.y_test_cls[0:5]
```
### Helper-function for plotting images
Function used to plot 9 images in a 3x3 grid, and writing the true and predicted classes below each image.
```
def plot_images(images, cls_true, cls_pred=None):
assert len(images) == len(cls_true) == 9
# Create figure with 3x3 sub-plots.
fig, axes = plt.subplots(3, 3)
fig.subplots_adjust(hspace=0.3, wspace=0.3)
for i, ax in enumerate(axes.flat):
# Plot image.
ax.imshow(images[i].reshape(img_shape), cmap='binary')
# Show true and predicted classes.
if cls_pred is None:
xlabel = "True: {0}".format(cls_true[i])
else:
xlabel = "True: {0}, Pred: {1}".format(cls_true[i], cls_pred[i])
ax.set_xlabel(xlabel)
# Remove ticks from the plot.
ax.set_xticks([])
ax.set_yticks([])
# Ensure the plot is shown correctly with multiple plots
# in a single Notebook cell.
plt.show()
```
### Plot a few images to see if data is correct
```
# Get the first images from the test-set.
images = data.x_test[0:9]
# Get the true classes for those images.
cls_true = data.y_test_cls[0:9]
# Plot the images and labels using our helper-function above.
plot_images(images=images, cls_true=cls_true)
```
## TensorFlow Graph
The entire purpose of TensorFlow is to have a so-called computational graph that can be executed much more efficiently than if the same calculations were to be performed directly in Python. TensorFlow can be more efficient than NumPy because TensorFlow knows the entire computation graph that must be executed, while NumPy only knows the computation of a single mathematical operation at a time.
TensorFlow can also automatically calculate the gradients that are needed to optimize the variables of the graph so as to make the model perform better. This is because the graph is a combination of simple mathematical expressions so the gradient of the entire graph can be calculated using the chain-rule for derivatives.
TensorFlow can also take advantage of multi-core CPUs as well as GPUs - and Google has even built special chips just for TensorFlow which are called TPUs (Tensor Processing Units) that are even faster than GPUs.
A TensorFlow graph consists of the following parts which will be detailed below:
* Placeholder variables used to feed input into the graph.
* Model variables that are going to be optimized so as to make the model perform better.
* The model which is essentially just a mathematical function that calculates some output given the input in the placeholder variables and the model variables.
* A cost measure that can be used to guide the optimization of the variables.
* An optimization method which updates the variables of the model.
In addition, the TensorFlow graph may also contain various debugging statements e.g. for logging data to be displayed using TensorBoard, which is not covered in this tutorial.
### Placeholder variables
Placeholder variables serve as the input to the graph that we may change each time we execute the graph. We call this feeding the placeholder variables and it is demonstrated further below.
First we define the placeholder variable for the input images. This allows us to change the images that are input to the TensorFlow graph. This is a so-called tensor, which just means that it is a multi-dimensional vector or matrix. The data-type is set to `float32` and the shape is set to `[None, img_size_flat]`, where `None` means that the tensor may hold an arbitrary number of images with each image being a vector of length `img_size_flat`.
```
x = tf.placeholder(tf.float32, [None, img_size_flat])
```
Next we have the placeholder variable for the true labels associated with the images that were input in the placeholder variable `x`. The shape of this placeholder variable is `[None, num_classes]` which means it may hold an arbitrary number of labels and each label is a vector of length `num_classes` which is 10 in this case.
```
y_true = tf.placeholder(tf.float32, [None, num_classes])
```
Finally we have the placeholder variable for the true class of each image in the placeholder variable `x`. These are integers and the dimensionality of this placeholder variable is set to `[None]` which means the placeholder variable is a one-dimensional vector of arbitrary length.
```
y_true_cls = tf.placeholder(tf.int64, [None])
```
### Variables to be optimized
Apart from the placeholder variables that were defined above and which serve as feeding input data into the model, there are also some model variables that must be changed by TensorFlow so as to make the model perform better on the training data.
The first variable that must be optimized is called `weights` and is defined here as a TensorFlow variable that must be initialized with zeros and whose shape is `[img_size_flat, num_classes]`, so it is a 2-dimensional tensor (or matrix) with `img_size_flat` rows and `num_classes` columns.
```
weights = tf.Variable(tf.zeros([img_size_flat, num_classes]))
```
The second variable that must be optimized is called `biases` and is defined as a 1-dimensional tensor (or vector) of length `num_classes`.
```
biases = tf.Variable(tf.zeros([num_classes]))
```
### Model
This simple mathematical model multiplies the images in the placeholder variable `x` with the `weights` and then adds the `biases`.
The result is a matrix of shape `[num_images, num_classes]` because `x` has shape `[num_images, img_size_flat]` and `weights` has shape `[img_size_flat, num_classes]`, so the multiplication of those two matrices is a matrix with shape `[num_images, num_classes]` and then the `biases` vector is added to each row of that matrix.
Note that the name `logits` is typical TensorFlow terminology, but other people may call the variable something else.
```
logits = tf.matmul(x, weights) + biases
```
Now `logits` is a matrix with `num_images` rows and `num_classes` columns, where the element of the $i$'th row and $j$'th column is an estimate of how likely the $i$'th input image is to be of the $j$'th class.
However, these estimates are a bit rough and difficult to interpret because the numbers may be very small or large, so we want to normalize them so that each row of the `logits` matrix sums to one, and each element is limited between zero and one. This is calculated using the so-called softmax function and the result is stored in `y_pred`.
```
y_pred = tf.nn.softmax(logits)
```
The predicted class can be calculated from the `y_pred` matrix by taking the index of the largest element in each row.
```
y_pred_cls = tf.argmax(y_pred, axis=1)
```
### Cost-function to be optimized
To make the model better at classifying the input images, we must somehow change the variables for `weights` and `biases`. To do this we first need to know how well the model currently performs by comparing the predicted output of the model `y_pred` to the desired output `y_true`.
The cross-entropy is a performance measure used in classification. The cross-entropy is a continuous function that is always positive and if the predicted output of the model exactly matches the desired output then the cross-entropy equals zero. The goal of optimization is therefore to minimize the cross-entropy so it gets as close to zero as possible by changing the `weights` and `biases` of the model.
TensorFlow has a built-in function for calculating the cross-entropy. Note that it uses the values of the `logits` because it also calculates the softmax internally.
```
cross_entropy = tf.nn.softmax_cross_entropy_with_logits_v2(logits=logits,
labels=y_true)
```
We have now calculated the cross-entropy for each of the image classifications so we have a measure of how well the model performs on each image individually. But in order to use the cross-entropy to guide the optimization of the model's variables we need a single scalar value, so we simply take the average of the cross-entropy for all the image classifications.
```
cost = tf.reduce_mean(cross_entropy)
```
### Optimization method
Now that we have a cost measure that must be minimized, we can then create an optimizer. In this case it is the basic form of Gradient Descent where the step-size is set to 0.5.
Note that optimization is not performed at this point. In fact, nothing is calculated at all, we just add the optimizer-object to the TensorFlow graph for later execution.
```
optimizer = tf.train.GradientDescentOptimizer(learning_rate=0.5).minimize(cost)
```
### Performance measures
We need a few more performance measures to display the progress to the user.
This is a vector of booleans whether the predicted class equals the true class of each image.
```
correct_prediction = tf.equal(y_pred_cls, y_true_cls)
```
This calculates the classification accuracy by first type-casting the vector of booleans to floats, so that False becomes 0 and True becomes 1, and then calculating the average of these numbers.
```
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
```
## TensorFlow Run
### Create TensorFlow session
Once the TensorFlow graph has been created, we have to create a TensorFlow session which is used to execute the graph.
```
session = tf.Session()
```
### Initialize variables
The variables for `weights` and `biases` must be initialized before we start optimizing them.
```
session.run(tf.global_variables_initializer())
```
### Helper-function to perform optimization iterations
There are 55.000 images in the training-set. It takes a long time to calculate the gradient of the model using all these images. We therefore use Stochastic Gradient Descent which only uses a small batch of images in each iteration of the optimizer.
```
batch_size = 100
```
Function for performing a number of optimization iterations so as to gradually improve the `weights` and `biases` of the model. In each iteration, a new batch of data is selected from the training-set and then TensorFlow executes the optimizer using those training samples.
```
def optimize(num_iterations):
for i in range(num_iterations):
# Get a batch of training examples.
# x_batch now holds a batch of images and
# y_true_batch are the true labels for those images.
x_batch, y_true_batch, _ = data.random_batch(batch_size=batch_size)
# Put the batch into a dict with the proper names
# for placeholder variables in the TensorFlow graph.
# Note that the placeholder for y_true_cls is not set
# because it is not used during training.
feed_dict_train = {x: x_batch,
y_true: y_true_batch}
# Run the optimizer using this batch of training data.
# TensorFlow assigns the variables in feed_dict_train
# to the placeholder variables and then runs the optimizer.
session.run(optimizer, feed_dict=feed_dict_train)
```
### Helper-functions to show performance
Dict with the test-set data to be used as input to the TensorFlow graph. Note that we must use the correct names for the placeholder variables in the TensorFlow graph.
```
feed_dict_test = {x: data.x_test,
y_true: data.y_test,
y_true_cls: data.y_test_cls}
```
Function for printing the classification accuracy on the test-set.
```
def print_accuracy():
# Use TensorFlow to compute the accuracy.
acc = session.run(accuracy, feed_dict=feed_dict_test)
# Print the accuracy.
print("Accuracy on test-set: {0:.1%}".format(acc))
```
Function for printing and plotting the confusion matrix using scikit-learn.
```
def print_confusion_matrix():
# Get the true classifications for the test-set.
cls_true = data.y_test_cls
# Get the predicted classifications for the test-set.
cls_pred = session.run(y_pred_cls, feed_dict=feed_dict_test)
# Get the confusion matrix using sklearn.
cm = confusion_matrix(y_true=cls_true,
y_pred=cls_pred)
# Print the confusion matrix as text.
print(cm)
# Plot the confusion matrix as an image.
plt.imshow(cm, interpolation='nearest', cmap=plt.cm.Blues, norm=LogNorm())
# Make various adjustments to the plot.
plt.tight_layout()
plt.colorbar()
tick_marks = np.arange(num_classes)
plt.xticks(tick_marks, range(num_classes))
plt.yticks(tick_marks, range(num_classes))
plt.xlabel('Predicted')
plt.ylabel('True')
# Ensure the plot is shown correctly with multiple plots
# in a single Notebook cell.
plt.show()
```
Function for plotting examples of images from the test-set that have been mis-classified.
```
def plot_example_errors():
# Use TensorFlow to get a list of boolean values
# whether each test-image has been correctly classified,
# and a list for the predicted class of each image.
correct, cls_pred = session.run([correct_prediction, y_pred_cls],
feed_dict=feed_dict_test)
# Negate the boolean array.
incorrect = (correct == False)
# Get the images from the test-set that have been
# incorrectly classified.
images = data.x_test[incorrect]
# Get the predicted classes for those images.
cls_pred = cls_pred[incorrect]
# Get the true classes for those images.
cls_true = data.y_test_cls[incorrect]
# Plot the first 9 images.
plot_images(images=images[0:9],
cls_true=cls_true[0:9],
cls_pred=cls_pred[0:9])
```
### Helper-function to plot the model weights
Function for plotting the `weights` of the model. 10 images are plotted, one for each digit that the model is trained to recognize.
```
def plot_weights():
# Get the values for the weights from the TensorFlow variable.
w = session.run(weights)
# Get the lowest and highest values for the weights.
# This is used to correct the colour intensity across
# the images so they can be compared with each other.
w_min = np.min(w)
w_max = np.max(w)
# Create figure with 3x4 sub-plots,
# where the last 2 sub-plots are unused.
fig, axes = plt.subplots(3, 4)
fig.subplots_adjust(hspace=0.3, wspace=0.3)
for i, ax in enumerate(axes.flat):
# Only use the weights for the first 10 sub-plots.
if i<10:
# Get the weights for the i'th digit and reshape it.
# Note that w.shape == (img_size_flat, 10)
image = w[:, i].reshape(img_shape)
# Set the label for the sub-plot.
ax.set_xlabel("Weights: {0}".format(i))
# Plot the image.
ax.imshow(image, vmin=w_min, vmax=w_max, cmap='seismic')
# Remove ticks from each sub-plot.
ax.set_xticks([])
ax.set_yticks([])
# Ensure the plot is shown correctly with multiple plots
# in a single Notebook cell.
plt.show()
```
## Performance before any optimization
The accuracy on the test-set is 9.8%. This is because the model has only been initialized and not optimized at all, so it always predicts that the image shows a zero digit, as demonstrated in the plot below, and it turns out that 9.8% of the images in the test-set happens to be zero digits.
```
print_accuracy()
plot_example_errors()
```
## Performance after 1 optimization iteration
Already after a single optimization iteration, the model has increased its accuracy on the test-set significantly.
```
optimize(num_iterations=1)
print_accuracy()
plot_example_errors()
```
The weights can also be plotted as shown below. Positive weights are red and negative weights are blue. These weights can be intuitively understood as image-filters.
For example, the weights used to determine if an image shows a zero-digit have a positive reaction (red) to an image of a circle, and have a negative reaction (blue) to images with content in the centre of the circle.
Similarly, the weights used to determine if an image shows a one-digit react positively (red) to a vertical line in the centre of the image, and react negatively (blue) to images with content surrounding that line.
Note that the weights mostly look like the digits they're supposed to recognize. This is because only one optimization iteration has been performed so the weights are only trained on 100 images. After training on several thousand images, the weights become more difficult to interpret because they have to recognize many variations of how digits can be written.
```
plot_weights()
```
## Performance after 10 optimization iterations
```
# We have already performed 1 iteration.
optimize(num_iterations=9)
print_accuracy()
plot_example_errors()
plot_weights()
```
## Performance after 1000 optimization iterations
After 1000 optimization iterations, the model only mis-classifies about one in ten images. As demonstrated below, some of the mis-classifications are justified because the images are very hard to determine with certainty even for humans, while others are quite obvious and should have been classified correctly by a good model. But this simple model cannot reach much better performance and more complex models are therefore needed.
```
# We have already performed 10 iterations.
optimize(num_iterations=990)
print_accuracy()
plot_example_errors()
```
The model has now been trained for 1000 optimization iterations, with each iteration using 100 images from the training-set. Because of the great variety of the images, the weights have now become difficult to interpret and we may doubt whether the model truly understands how digits are composed from lines, or whether the model has just memorized many different variations of pixels.
```
plot_weights()
```
We can also print and plot the so-called confusion matrix which lets us see more details about the mis-classifications. For example, it shows that images actually depicting a 5 have sometimes been mis-classified as all other possible digits, but mostly as 6 or 8.
```
print_confusion_matrix()
```
We are now done using TensorFlow, so we close the session to release its resources.
```
# This has been commented out in case you want to modify and experiment
# with the Notebook without having to restart it.
# session.close()
```
## Exercises
These are a few suggestions for exercises that may help improve your skills with TensorFlow. It is important to get hands-on experience with TensorFlow in order to learn how to use it properly.
You may want to backup this Notebook before making any changes.
* Change the learning-rate for the optimizer.
* Change the optimizer to e.g. `AdagradOptimizer` or `AdamOptimizer`.
* Change the batch-size to e.g. 1 or 1000.
* How do these changes affect the performance?
* Do you think these changes will have the same effect (if any) on other classification problems and mathematical models?
* Do you get the exact same results if you run the Notebook multiple times without changing any parameters? Why or why not?
* Change the function `plot_example_errors()` so it also prints the `logits` and `y_pred` values for the mis-classified examples.
* Use `sparse_softmax_cross_entropy_with_logits` instead of `softmax_cross_entropy_with_logits`. This may require several changes to multiple places in the source-code. Discuss the advantages and disadvantages of using the two methods.
* Remake the program yourself without looking too much at this source-code.
* Explain to a friend how the program works.
## License (MIT)
Copyright (c) 2016 by [Magnus Erik Hvass Pedersen](http://www.hvass-labs.org/)
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
| github_jupyter |
# Modelling trend life cycles in scientific research
**Authors:** E. Tattershall, G. Nenadic, and R.D. Stevens
**Abstract:** Scientific topics vary in popularity over time. In this paper, we model the life-cycles of 200 topics by fitting the Logistic and Gompertz models to their frequency over time in published abstracts. Unlike other work, the topics we use are algorithmically extracted from large datasets of abstracts covering computer science, particle physics, cancer research, and mental health. We find that the Gompertz model produces lower median error, leading us to conclude that it is the more appropriate model. Since the Gompertz model is asymmetric, with a steep rise followed a long tail, this implies that scientific topics follow a similar trajectory. We also explore the case of double-peaking curves and find that in some cases, topics will peak multiple times as interest resurges. Finally, when looking at the different scientific disciplines, we find that the lifespan of topics is longer in some disciplines (e.g. cancer research and mental health) than it is others, which may indicate differences in research process and culture between these disciplines.
**Requirements**
- Data. Data ingress is excluded from this notebook, but we alraedy have four large datasets of abstracts. The documents in these datasets have been cleaned (described in sections below) and separated by year. Anecdotally, this method works best when there are >100,000 documents in the dataset (and more is even better).
- The other utility files in this directory, including burst_detection.py, my_stopwords.py, etc...
**In this notebook**
- Vectorisation
- Burst detection
- Clustering
- Model fitting
- Comparing the error of the two models
- Calculating trend duration
- Double peaked curves
- Trends and fitted models in full
```
import os
import csv
import pandas as pd
from collections import defaultdict
import matplotlib.pyplot as plt
from matplotlib.ticker import FormatStrFormatter
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_extraction.text import TfidfTransformer
import numpy as np
import scipy
from scipy.spatial.distance import squareform
from scipy.cluster import hierarchy
import pickle
import burst_detection
import my_stopwords
import cleaning
import tools
import logletlab
import scipy.optimize as opt
from sklearn.metrics import mean_squared_error
stop = my_stopwords.get_stopwords()
burstiness_threshold = 0.004
cluster_distance_threshold = 7
# Burst detection internal parameters
# These are the same as in our earlier paper [Tattershall 2020]
parameters = {
"min_yearly_df": 5,
"significance_threshold": 0.0015,
"years_above_significance": 3,
"long_ma_length": 8,
"short_ma_length": 4,
"signal_line_ma": 3,
"significance_ma_length": 3
}
# Number of bursty terms to extract for each dataset. This will later be filtered down to 50 for each dataset after clustering.
max_bursts = 300
dataset_names = ['pubmed_mh', 'arxiv_hep', 'pubmed_cancer', 'dblp_cs']
dataset_titles = ['Computer science (dblp)', 'Particle physics (arXiv)', 'Mental health (PubMed)', 'Cancer (PubMed)']
datasets = {}
def reverse_cumsum(ls):
reverse = np.zeros_like(ls)
for i in range(len(ls)):
if i == 0:
reverse[i] = ls[i]
else:
reverse[i] = ls[i]-ls[i-1]
if reverse[0]>reverse[1]:
reverse[0]=reverse[1]
return reverse
def detransform_fit(ypc, F, dataset_name):
'''
The Gompertz and Logistic curves actually model *cumulative* frequency over time, not raw frequency.
However, raw frequency is more intuitive for graphs, so we use this function to change a cumulative
time series into a non-cumulative one. Additionally, the models were originally fitted to scaled curves
(such that the minumum frequency was zero and the maximum was one). This was done to make it possible to
directly compare the error between different time series without a much more frequent term dwarfing the calculation.
We now transform back.
'''
s = document_count_per_year[dataset_name]
yf = reverse_cumsum(F*(max(ypc)-min(ypc)) + min(ypc))
return yf
# Location where the cleaned data is stored
data_root = 'cleaned_data/'
# Location where we will store the results of this notebook
root = 'results/'
os.mkdir(root+'clusters')
os.mkdir(root+'images')
os.mkdir(root+'fitted_curves')
os.mkdir(root+'vectors')
for dataset_name in dataset_names:
os.mkdir(root+'vectors/'+dataset_name)
os.mkdir(root+'fitted_curves/'+dataset_name)
```
## The data
We have four datasets:
- **Computer Science (dblp_cs):** This dataset contains 2.6 million abstracts downloaded from Semantic Scholar. We select all abstracts with the dblp tag.
- **Particle Physics (arxiv_hep):** This dataset of 0.2 million abstracts was downloaded from arXiv's public API. We extracted particle physics-reladed documents by selecting everything under the categroies hep-ex, hep-lat, hep-ph and hep-th.
- **Mental Health (pubmed_mh):** 0.7 million abstracts downloaded from PubMed. This dataset was created by filtering on the MeSH keyword "Mental Health" and all its subterms.
- **Cancer (pubmed_cancer):** 1.9 million abstracts downloaded from PubMed. This dataset was created by filtering on the MeSH keyword "Neoplasms" and all its subterms.
The data in each dataset has already been cleaned. We removed punctuation, set all characters to lowercase and lemmatised each word using WordNetLemmatizer. The cleaned data is stored in pickled pandas dataframes in files named 1988.p, 1989.p, 1990.p. Each dataframe has a column "cleaned" which contains the cleaned and lemmatized text for each document in that dataset in the given year.
### How many documents are in each dataset in each year?
```
document_count_per_year = {}
for dataset_name in dataset_names:
# For each dataset, we want a list of document counts for each year
document_count_per_year[dataset_name] = []
# The files in the directory are named 1988.p, 1989.p, 1990.p....
files = os.listdir(data_root+dataset_name)
min_year = np.min([int(file[0:4]) for file in files])
max_year = np.max([int(file[0:4]) for file in files])
for year in range(min_year, max_year+1):
df = pickle.load(open(data_root+dataset_name+'/'+str(year)+'.p', "rb"))
document_count_per_year[dataset_name].append(len(df))
pickle.dump(document_count_per_year, open(root + 'document_count_per_year.p', "wb"))
plt.figure(figsize=(6,3.7))
ax1=plt.subplot(111)
plt.subplots_adjust(left=0.2, right=0.9)
ax1.set_title('Documents per year in each dataset', fontsize=11)
ax1.plot(np.arange(1988, 2018), document_count_per_year['dblp_cs'], 'k', label='dblp')
ax1.plot(np.arange(1994, 2018), document_count_per_year['arxiv_hep'], 'k', linestyle= '-.', label='arXiv')
ax1.plot(np.arange(1975, 2018), document_count_per_year['pubmed_mh'], 'k', linestyle= '--', label='PubMed (Mental Health)')
ax1.plot(np.arange(1975, 2018), document_count_per_year['pubmed_cancer'], 'k', linestyle= ':', label='PubMed (Cancer)')
ax1.grid()
ax1.set_xlim([1975, 2018])
ax1.set_ylabel('Documents', fontsize=10)
ax1.set_xlabel('Year', fontsize=10)
ax1.set_ylim([0,200000])
ax1.legend(fontsize=10)
plt.savefig(root+'images/documents_per_year.eps', format='eps', dpi=1200)
```
### Create a vocabulary for each dataset
- For each dataset, we find all **1-5 word terms** (after stopwords are removed). This allows us to use relatively complex phrases.
- Since the set of all 1-5 word terms is very large and contains much noise, we filter out terms that fail to meet a **minimum threshold of "significance"**. For significance we require that they occur at least six times in at least one year. We find that this also gets rid of spelling erros and cuts down the size of the data.
```
for dataset_name in dataset_names:
vocabulary = set()
files = os.listdir(data_root+dataset_name)
min_year = np.min([int(file[0:4]) for file in files])
max_year = np.max([int(file[0:4]) for file in files])
for year in range(min_year, max_year+1):
df = pickle.load(open(data_root+dataset_name+"/"+str(year)+".p", "rb"))
# Create an initial vocabulary based on the list of text files
vectorizer = CountVectorizer(strip_accents='ascii',
ngram_range=(1,5),
stop_words=stop,
min_df=6
)
# Vectorise the data in order to get the vocabulary
vector = vectorizer.fit_transform(df['cleaned'])
# Add the harvested vocabulary to the set. This removes duplicates of terms that occur in multiple years
vocabulary = vocabulary.union(set(vectorizer.vocabulary_))
# To conserve memory, delete the vector here
del vector
print('Overall vocabulary created for ', dataset_name)
# We now vectorise the dataset again based on the unifying vocabulary
vocabulary = list(vocabulary)
vectors = []
vectorizer = CountVectorizer(strip_accents='ascii',
ngram_range=(1,5),
stop_words=stop,
vocabulary=vocabulary)
for year in range(min_year, max_year+1):
df = pickle.load(open(data_root+dataset_name+"/"+str(year)+".p", "rb"))
vector = vectorizer.fit_transform(df['cleaned'])
# Set all elements of the vector that are greater than 1 to 1. This is because we only care about
# the overall document frequency of each term. If a word is used multiple times in a single document
# it only contributes 1 to the document frequency.
vector[vector>1] = 1
# Sum the vector along its columns in order to get the total document frequency of each term in a year
summed = np.squeeze(np.asarray(np.sum(vector, axis=0)))
vectors.append(summed)
# Turn the vector into a pandas dataframe
df = pd.DataFrame(vectors, columns=vocabulary)
# THE PART BELOW IS OPTIONAL
# We found that the process works better if very similar terms are removed from the vocabulary
# Therefore, for each 2-5 ngram, we identify all possible subterms, then attempt to calculate whether
# the subterms are legitimate terms in their own right (i.e. they appear in documents without their
# superterm parent). For example, the term "long short-term memory" is made up of the subterms
# ["long short", "short term", "term memory", "long short term", "short term memory"]
# However, when we calculate the document frequency of each subterm divided by the document frequency of
# "long short term memory", we find:
#
# long short 1.4
# short term 6.1
# term memory 2.2
# long short term 1.1
# short term memory 1.4
#
# Since the term "long short term" occurs only very rarely outside the phrase "long short term memory", we
# omit this term by setting an arbitrary threshold of 1.1. This preserves most of the subterms while removing the rarest.
removed = []
# for each term in the vocabulary
for i, term in enumerate(list(df.columns)):
# If the term is a 2-5 ngram (i.e. not a single word)
if ' ' in term:
# Find the overall term document frequency over the entire dataset
term_total_document_frequency = df[term].sum()
# Find all possible subterms of the term.
subterms = tools.all_subterms(term)
for subterm in subterms:
try:
# If the subterm is in the vocabulary, check whether it often occurs on its own
# without the superterm being present
subterm_total_document_frequency = df[subterm].sum()
if subterm_total_document_frequency < term_total_document_frequency*1.1:
removed.append([subterm, term])
except:
pass
# Remove the removed terms from the dataframe
df = df.drop(list(set([r[0] for r in removed])), axis=1)
# END OPTIONAL PART
# Store the stacked vectors for later use
pickle.dump(df, open(root+'vectors/'+dataset_name+"/stacked_vector.p", "wb"))
pickle.dump(list(df.columns), open(root+'vectors/'+dataset_name+"/vocabulary.p", "wb"))
```
### Detect bursty terms
Now that we have vectors representing the document frequency of each term over time, we can use our MACD-based burst detection, as described in our earlier paper [Tattershall 2020].
```
bursts = dict()
for dataset_name in dataset_names:
files = os.listdir(data_root+dataset_name)
min_year = np.min([int(file[0:4]) for file in files])
max_year = np.max([int(file[0:4]) for file in files])
# Create a dataset object for the burst detection algorithm
bd_dataset = burst_detection.Dataset(
name = dataset_name,
years = (min_year, max_year),
# We divide the term-document frequency for each year by the number of documents in that year
stacked_vectors = pickle.load(open(root+dataset_name+"/stacked_vector.p", "rb")).divide(document_count_per_year[dataset_name],axis=0)
)
# We apply the significance threshold from the burst detection methodology. This cuts the size of the dataset by
# removing terms that occur only in one year
bd_dataset.get_sig_stacked_vectors(parameters["significance_threshold"], parameters["years_above_significance"])
bd_dataset.get_burstiness(parameters["short_ma_length"], parameters["long_ma_length"], parameters["significance_ma_length"], parameters["signal_line_ma"])
datasets[dataset_name] = bd_dataset
bursts[dataset_name] = tools.get_top_n_bursts(datasets[dataset_name].burstiness, max_bursts)
pickle.dump(bursts, open(root+'vectors/'+'bursts.p', "wb"))
```
### Calculate burst co-occurence
We now have 300 bursts per dataset. Some of these describe very similar concepts, such as "internet of things" and "iot". The purpose of this section is the merge similar terms into clusters to prevent redundancy within the dataset. We calculate the relatedness of terms using term co-occurrence within the same document (terms that appear together are grouped together).
```
for dataset_name in dataset_names:
vectors = []
vectorizer = CountVectorizer(strip_accents='ascii',
ngram_range=(1,5),
stop_words=stop,
vocabulary=bursts[dataset_name])
for year in range(min_year, max_year+1):
df = pickle.load(open(data_root+dataset_name+"/"+str(year)+".p", "rb"))
vector = vectorizer.fit_transform(df['cleaned'])
# Set all elements of the vector that are greater than 1 to 1. This is because we only care about
# the overall document frequency of each term. If a word is used multiple times in a single document
# it only contributes 1 to the document frequency.
vector[vector>1] = 1
vectors.append(vector)
# Calculate the cooccurrence matrix
v = vectors[0]
c = v.T*v
c.setdiag(0)
c = c.todense()
cooccurrence = c
for v in vectors[1:]:
c = v.T*v
c.setdiag(0)
c = c.toarray()
cooccurrence += c
pickle.dump(cooccurrence, open(root+'vectors/'+dataset_name+"/cooccurrence_matrix.p", "wb"))
```
### Use burst co-occurrence to cluster terms
We use a hierarchichal clustering method to group terms together. This is highly customisable due to threshold setting, allowing us to group more or less conservatively if required.
```
# Reload bursts if required by uncommenting this line
#bursts = pickle.load(open(root+'bursts.p', "rb"))
dataset_clusters = dict()
for dataset_name in dataset_names:
#cooccurrence = pickle.load(open('Data/stacked_vectors/'+dataset_name+"/cooccurrence_matrix.p", "rb"))
# Translate co-occurence into a distance
dists = np.log(cooccurrence+1).max()- np.log(cooccurrence+1)
# Remove the diagonal (squareform requires diagonals be zero)
dists -= np.diag(np.diagonal(dists))
# Put the distance matrix into the format required by hierachy.linkage
flat_dists = squareform(dists)
# Get the linkage matrix
linkage_matrix = hierarchy.linkage(flat_dists, "ward")
assignments = hierarchy.fcluster(linkage_matrix, t=cluster_distance_threshold, criterion='distance')
clusters = defaultdict(list)
for term, assign, co in zip(bursts[dataset_name], assignments, cooccurrence):
clusters[assign].append(term)
dataset_clusters[dataset_name] = list(clusters.values())
dataset_clusters['arxiv_hep']
```
### Manual choice of clusters
We now sort the clusters in order of burstiness (using the burstiness of the most bursty term in the cluster) and manually exclude clusters that include publishing artefacts such as "elsevier science bv right reserved". From the remainder, we select the top fifty. We do this for all four datasets, giving 200 clusters. The selected clusters are stored in the file "200clusters.csv".
### For each cluster, create a time series of mentions in abstracts over time
We now need to search for the clusters to pull out the frequency of appearance in abstracts over time. For the cluster ["Internet of things", "IoT"], all abstracts that mention **either** term are included (i.e. an abstract that uses "Internet of things" without the abbreviation "IoT" still counts towards the total for that year). We take document frequency, not term frequency, so the number of times the terms are mentioned in each document do not matter, so long as they are mentioned once.
```
raw_clusters = pd.read_csv('200clusters.csv')
cluster_dict = defaultdict(list)
for dataset_name in dataset_names:
for raw_cluster in raw_clusters[dataset_name]:
cluster_dict[dataset_name].append(raw_cluster.split(','))
for dataset_name in dataset_names:
# List all the cluster terms. This will be more than the total number of clusters.
all_cluster_terms = sum(cluster_dict[dataset_name], [])
# Get the cluster titles. This is the list of terms in each cluster
all_cluster_titles = [','.join(cluster) for cluster in cluster_dict[dataset_name]]
# Get a list of files from the directory
files = os.listdir(data_root + dataset_name)
# This is where we will store the data. The columns correspond to clusters, the rows to years
prevalence_array = np.zeros([len(files),len(cluster_dict[dataset_name])])
# Open each year file in turn
for i, file in enumerate(files):
print(file)
year_data = pickle.load(open(data_root + dataset_name + '/' + file, 'rb'))
# Vectorise the data for that year
vectorizer = CountVectorizer(strip_accents='ascii',
ngram_range=(1,5),
stop_words=stop,
vocabulary=all_cluster_terms
)
vector = vectorizer.fit_transform(year_data['cleaned'])
# Get the index of each cluster term. This will allows us to map the full vocabulary
# e.g. (60 items) back onto the original clusters (e.g. 50 items)
for j, cluster in enumerate(cluster_dict[dataset_name]):
indices = []
for term in cluster:
indices.append(all_cluster_terms.index(term))
# If there are multiple terms in a cluster, sum the cluster columns together
summed_column = np.squeeze(np.asarray(vector[:,indices].sum(axis=1).flatten()))
# Set any element greater than one to one--we're only counting documents here, not
# total occurrences
summed_column[summed_column!=0] = 1
# This is the total number of occurrences of the cluster per year
prevalence_array[i, j] = np.sum(summed_column)
# Save the data
df = pd.DataFrame(data=prevalence_array, index=[f[0:4] for f in files], columns=all_cluster_titles)
pickle.dump(df, open(root+'clusters/'+dataset_name+'.p', 'wb'))
```
### Curve fitting
The below is a pythonic version of the Loglet Lab 4 code found at https://github.com/pheguest/logletlab4. Loglet Lab also has a web interface at https://logletlab.com/ which allows you to create amazing graphs. However, the issue with the web interface is that it is not designed for processing hundreds of time series, and in order to do this, each time series must be laboriously copy-pasted into the input box, the parameters set, and then the results saved individually. With 200 time series and multiple parameter sets, this process is quite slow! Therefore, we have adapted the code from the github repository, but the original should be seen at https://github.com/pheguest/logletlab4/blob/master/javascript/src/psmlogfunc3.js.
```
curve_header_1 = ['', 'd', 'k', 'a', 'b', 'RMS']
curve_header_2 = ['', 'd', 'k1', 'a1', 'b1', 'k2', 'a2', 'b2', 'RMS']
dataset_names = ['arxiv_hep', 'pubmed_mh', 'pubmed_cancer', 'dblp_cs']
for dataset_name in dataset_names:
print('-'*50)
print(dataset_name.upper())
for curve_type in ['logistic', 'gompertz']:
for number_of_peaks in [1, 2]:
with open('our_loglet_lab/'+dataset_name+'/'+curve_type+str(number_of_peaks)+'.csv', 'w', newline='') as f:
writer = csv.writer(f)
if number_of_peaks == 1:
writer.writerow(curve_header_1)
elif number_of_peaks == 2:
writer.writerow(curve_header_2)
df = pickle.load(open(root+'clusters/'+dataset_name+'.p', 'rb'))
document_count_per_year = pickle.load(open(root+"/document_count_per_year.p", 'rb'))[dataset_name]
df = df.divide(document_count_per_year, axis=0)
for term in df.keys():
y = tools.normalise_time_series(df[term].cumsum())
x = np.array([int(i) for i in y.index])
y = y.values
if number_of_peaks == 1:
logobj = logletlab.LogObj(x, y, 1)
constraints = logletlab.estimate_constraints(x, y, 1)
if curve_type == 'logistic':
logobj = logletlab.loglet_MC_anneal_regression(logobj, constraints=constraints, number_of_loglets=1,
curve_type='logistic', anneal_iterations=20,
mc_iterations=1000, anneal_sample_size=100)
else:
logobj = logletlab.loglet_MC_anneal_regression(logobj, constraints=constraints, number_of_loglets=1,
curve_type='gompertz', anneal_iterations=20,
mc_iterations=1000, anneal_sample_size=100)
line = [term, logobj.parameters['d'], logobj.parameters['k'][0], logobj.parameters['a'][0], logobj.parameters['b'][0], logobj.energy_best]
print(curve_type, number_of_peaks, term, 'RMSE='+str(np.round(logobj.energy_best,3)))
with open(root+'fitted_curves/'+dataset_name+'/'+curve_type+'_single.csv', 'a', newline='') as f:
writer = csv.writer(f)
writer.writerow(line)
elif number_of_peaks == 2:
logobj = logletlab.LogObj(x, y, 2)
constraints = logletlab.estimate_constraints(x, y, 2)
if curve_type == 'logistic':
logobj = logletlab.loglet_MC_anneal_regression(logobj, constraints=constraints, number_of_loglets=2,
curve_type='logistic', anneal_iterations=30,
mc_iterations=1000, anneal_sample_size=100)
else:
logobj = logletlab.loglet_MC_anneal_regression(logobj, constraints=constraints, number_of_loglets=2,
curve_type='gompertz', anneal_iterations=30,
mc_iterations=1000, anneal_sample_size=100)
line = [term, logobj.parameters['d'],
logobj.parameters['k'][0],
logobj.parameters['a'][0],
logobj.parameters['b'][0],
logobj.parameters['k'][1],
logobj.parameters['a'][1],
logobj.parameters['b'][1],
logobj.energy_best]
print(curve_type, number_of_peaks, term, 'RMSE='+str(np.round(logobj.energy_best,3)))
with open(root+'fitted_curves/'+dataset_name+'/'+curve_type+'_double.csv', 'a', newline='') as f:
writer = csv.writer(f)
writer.writerow(line)
```
## Reload the data
The preceding step is very long, and may take many hours to complete. Therefore, since we did it in chunks, we now reload the results from memory.
```
# Load the data back up (since the steps above store the results in files, not local memory)
document_count_per_year = pickle.load(open(root+'document_count_per_year.p', "rb"))
datasets = {}
for dataset_name in dataset_names:
datasets[dataset_name] = {}
for curve_type in ['logistic', 'gompertz']:
datasets[dataset_name][curve_type] = {}
for peaks in ['single', 'double']:
df = pd.read_csv(root+'fitted_curves/'+dataset_name+'/'+curve_type+'_'+peaks+'.csv', index_col=0)
datasets[dataset_name][curve_type][peaks] = df
```
### Graph: Example single-peaked fit for XML
```
x = range(1988,2018)
term = 'xml'
# Load the original time series for xml
df = pickle.load(open(root+'clusters/dblp_cs.p', 'rb'))
# Divide the data for each year by the document count in each year
y_proportional = df[term].divide(document_count_per_year['dblp_cs'])
# Calculate Logistic and Gompertz curves from the parameters estimated earlier
y_logistic = logletlab.calculate_series(x,
datasets['dblp_cs']['logistic']['single']['a'][term],
datasets['dblp_cs']['logistic']['single']['k'][term],
datasets['dblp_cs']['logistic']['single']['b'][term],
'logistic'
)
# Since the fitting was done with a normalised version of the curve, we detransform it back into the original scale
y_logistic = detransform_fit(y_proportional.cumsum(), y_logistic, 'dblp_cs')
y_gompertz = logletlab.calculate_series(x,
datasets['dblp_cs']['gompertz']['single']['a'][term],
datasets['dblp_cs']['gompertz']['single']['k'][term],
datasets['dblp_cs']['gompertz']['single']['b'][term],
'gompertz'
)
y_gompertz = detransform_fit(y_proportional.cumsum(), y_gompertz, 'dblp_cs')
plt.figure(figsize=(6,3.7))
# Multiply by 100 so that values will be percentages
plt.plot(x, 100*y_proportional, label='Data', color='k')
plt.plot(x, 100*y_logistic, label='Logistic', color='k', linestyle=':')
plt.plot(x, 100*y_gompertz, label='Gompertz', color='k', linestyle='--')
plt.legend()
plt.grid()
plt.title("Logistic and Gompertz models fitted to the data for 'XML'", fontsize=12)
plt.xlim([1988,2017])
plt.ylim(0,2)
plt.ylabel("Documents containing term (%)", fontsize=11)
plt.xlabel("Year", fontsize=11)
plt.savefig(root+'images/xmlexamplefit.eps', format='eps', dpi=1200)
```
### Table of results for Logistic vs Gompertz
Compare the error of the Logistic and Gompertz models across the entire dataset of 200 trends.
```
def statistics(df):
mean = df.mean()
ci = 1.96*logistic_error.std()/np.sqrt(len(logistic_error))
median = df.median()
std = df.std()
return [mean, mean-ci, mean+ci, median, std]
logistic_error = pd.concat([datasets['arxiv_hep']['logistic']['single']['RMS'],
datasets['dblp_cs']['logistic']['single']['RMS'],
datasets['pubmed_mh']['logistic']['single']['RMS'],
datasets['pubmed_cancer']['logistic']['single']['RMS']])
gompertz_error = pd.concat([datasets['arxiv_hep']['gompertz']['single']['RMS'],
datasets['dblp_cs']['gompertz']['single']['RMS'],
datasets['pubmed_mh']['gompertz']['single']['RMS'],
datasets['pubmed_cancer']['gompertz']['single']['RMS']])
print('Logistic')
mean = logistic_error.mean()
ci = 1.96*logistic_error.std()/np.sqrt(len(logistic_error))
print('Mean =', np.round(mean,3))
print('95% CI = [', np.round(mean-ci, 3), ',', np.round(mean+ci, 3), ']')
print('Median =', np.round(logistic_error.median(), 3))
print('STDEV =', np.round(logistic_error.std(), 3))
print('')
print('Gompertz')
mean = gompertz_error.mean()
ci = 1.96*gompertz_error.std()/np.sqrt(len(logistic_error))
print('Mean =', np.round(mean,3))
print('95% CI = [', np.round(mean-ci, 3), ',', np.round(mean+ci, 3), ']')
print('Median =', np.round(gompertz_error.median(), 3))
print('STDEV =', np.round(gompertz_error.std(), 3))
```
### Is the difference between the means significant?
Here we use an independent t-test to investigate significance.
```
scipy.stats.ttest_ind(logistic_error, gompertz_error, axis=0, equal_var=True, nan_policy='propagate')
```
Yes, it is significant! However, since the data is slightly skewed, we can also test the signficance of the difference between medians using Mood's median test:
```
stat, p, med, tbl = scipy.stats.median_test(logistic_error, gompertz_error)
print(p)
```
So either way, the p-value is very low, causing us to reject the null hypothesis. This leads us to the conclusion that the **Gompertz model** is more appropriate for the task of modelling publishing activity over time.
### Box and whisker plots of Logistic and Gompertz model error
```
axs = pd.DataFrame({
'CS Logistic': datasets['dblp_cs']['logistic']['single']['RMS'],
'CS Gompertz': datasets['dblp_cs']['gompertz']['single']['RMS'],
'Physics Logistic': datasets['arxiv_hep']['logistic']['single']['RMS'],
'Physics Gompertz': datasets['arxiv_hep']['gompertz']['single']['RMS'],
'MH Logistic': datasets['pubmed_mh']['logistic']['single']['RMS'],
'MH Gompertz': datasets['pubmed_mh']['gompertz']['single']['RMS'],
'Cancer Logistic': datasets['pubmed_cancer']['logistic']['single']['RMS'],
'Cancer Gompertz': datasets['pubmed_cancer']['gompertz']['single']['RMS'],
}).boxplot(figsize=(13,4), return_type='dict')
[item.set_color('k') for item in axs['boxes']]
[item.set_color('k') for item in axs['whiskers']]
[item.set_color('k') for item in axs['medians']]
plt.suptitle("")
p = plt.gca()
p.set_ylabel('RMSE error')
p.set_title('Distribution of RMSE error of models fitted to the four datasets', fontsize=12)
p.set_ylim([0,0.12])
```
There is some variation across the datasets, although the Gompertz model is consistent in producing a lower median error than the Logistic model. It's worth noting also that the Particle Physics and Mental Health datasets are smaller than the Cancer and Computer Science ones. They also have higher error.
### Calculation of trend duration
The Loglet Lab documentation (https://logletlab.com/loglet/documentation/index) contains a formula for the time taken for a Gompertz curve to go from 10% to 90% of its eventual maximum cumulative frequency ($\Delta t$). Their calculation is that
$\Delta t = -\frac{\ln(\ln(81))}{r}$
However, our observation was that this did not remotely describe the observed span of the fitted curves. We have therefore done the derivation ourselves and found that the correct parameterisation is:
$\Delta t = \frac{1}{\ln(-(\ln(0.9))-\ln(-\ln(0.1))}$
Unfortunately, the LogletLab initial parameter guesses are tailored to this incorrect parameterisation so it is much simpler to use it when fitting the curve (and irrelevant, except when it comes to calculating curve span). Therefore we use it, then convert to the correct value using the conversion factor below:
```
conversion_factor = -((np.log(-np.log(0.9))-np.log(-np.log(0.1)))/np.log(np.log(81)))
spans = pd.DataFrame({
'Computer Science': datasets['dblp_cs']['gompertz']['single']['a']*conversion_factor,
'Particle Physics': datasets['arxiv_hep']['gompertz']['single']['a']*conversion_factor,
'Mental Health': datasets['pubmed_mh']['gompertz']['single']['a']*conversion_factor,
'Cancer': datasets['pubmed_cancer']['gompertz']['single']['a']*conversion_factor
})
axs = spans.boxplot(figsize=(7.5,3.7), return_type='dict', fontsize=11)
[item.set_color('k') for item in axs['boxes']]
[item.set_color('k') for item in axs['whiskers']]
[item.set_color('k') for item in axs['medians']]
#plt.figure(figsize=(6,3.7))
plt.suptitle("")
p = plt.gca()
p.set_ylabel('Peak width (years)', fontsize=11)
p.set_title('Distribution of peak widths by dataset (Gomperz model)', fontsize=12)
p.set_ylim([0,100])
plt.savefig(root+'images/curvespans.eps', format='eps', dpi=1200)
```
The data is quite skewed here...something to bear in mind when testing for significance later.
### Median trend durations in different disciplines
```
for i , dataset_name in enumerate(dataset_names):
print(dataset_titles[i], '| Median trend duration =', np.round(np.median(datasets[dataset_name]['gompertz']['single']['a']*conversion_factor),1), 'years')
```
### Testing for significance between disciplines
There are substantial differences between the median trend durations, with Computer Science and Particle Physics having shorter durations and the two PubMed datasets having longer ones. But are these significant? Since the data is somewhat skewed, we use Mood's median test to find p-values for the differences (Mood's median test does not require normal data).
```
for i in range(4):
for j in range(i,4):
if i == j:
pass
else:
spans1 = datasets[dataset_names[i]]['gompertz']['single']['a']*conversion_factor
spans2 = datasets[dataset_names[j]]['gompertz']['single']['a']*conversion_factor
stat, p, med, tbl = scipy.stats.median_test(spans1, spans2)
print(dataset_titles[i], 'vs', dataset_titles[j], 'p-value =', np.round(p,3))
```
So the p value between Particle Physics and Computer Science is not acceptable, and neither is the p-value between Mental Health and Cancer. How about between these two groups?
```
dblp_spans = datasets['dblp_cs']['gompertz']['single']['a']*conversion_factor
cancer_spans = datasets['pubmed_cancer']['gompertz']['single']['a']*conversion_factor
arxiv_spans = datasets['arxiv_hep']['gompertz']['single']['a']*conversion_factor
mh_spans = datasets['pubmed_mh']['gompertz']['single']['a']*conversion_factor
stat, p, med, tbl = scipy.stats.median_test(pd.concat([arxiv_spans, dblp_spans]), pd.concat([cancer_spans, mh_spans]))
print(np.round(p,5))
```
This difference IS significant!
### Double-peaking curves
We now move to analyse the data for double-peaked curves. For each term, we have calculated the error when two peaks are fitted, and the error when a single peak is fitted. We can compare the error in each case like so:
```
print('Neural networks, single peak | error =', np.round(datasets['dblp_cs']['gompertz']['single']['RMS']['neural network'],3))
print('Neural networks, double peak| error =', np.round(datasets['dblp_cs']['gompertz']['double']['RMS']['neural network'],3))
```
Where do we see the largest reductions?
```
difference = datasets['dblp_cs']['gompertz']['single']['RMS']-datasets['dblp_cs']['gompertz']['double']['RMS']
for term in difference.index:
if difference[term] > 0.015:
print(term, np.round(difference[term], 3))
```
### Examples of double peaking curves
So in some cases there is an error reduction from moving from the single- to double-peaked model. What does this look like in practice?
```
x = range(1988,2018)
# Load the original data
df = pickle.load(open(root+'clusters/dblp_cs.p', 'rb'))
# Choose four example terms
terms = ['big data', 'cloud', 'internet', 'neural network']
titles = ['a) Big Data', 'b) Cloud', 'c) Internet', 'd) Neural network']
# We want to set an overall y-label. The solution(found at https://stackoverflow.com/a/27430940) is to
# create an overall plot first, give it a y-label, then hide it by removing plot borders.
fig, big_ax = plt.subplots(figsize=(9.0, 6.0) , nrows=1, ncols=1, sharex=True)
big_ax.tick_params(labelcolor=(1,1,1,0.0), top=False, bottom=False, left=False, right=False)
big_ax._frameon = False
big_ax.set_ylabel("Documents containing term (%)", fontsize=11)
axs = [0,0,0,0]
axs[0]=fig.add_subplot(2,2,1)
axs[1]=fig.add_subplot(2,2,2)
axs[2]=fig.add_subplot(2,2,3)
axs[3]=fig.add_subplot(2,2,4)
fig.subplots_adjust(wspace=0.25, hspace=0.5, right=0.9)
# Set y limits manually beforehand
limits = [2, 4, 6, 8]
for i, term in enumerate(terms):
# Get the proportional document frequency of the term over time
y_proportional = df[term].divide(document_count_per_year['dblp_cs'])
# Multiply by 100 when plotting so that it reads as a percentage
axs[i].plot(x, 100*y_proportional, color='k')
axs[i].grid(True)
axs[i].set_xlabel("Year", fontsize=11)
axs[i].yaxis.set_major_formatter(FormatStrFormatter('%.1f'))
# Now plot single and double peaked models
for j, curve_type in enumerate(['single', 'double']):
if curve_type == 'single':
y_overall = logletlab.calculate_series(x,
datasets['dblp_cs']['gompertz'][curve_type]['a'][term],
datasets['dblp_cs']['gompertz'][curve_type]['k'][term],
datasets['dblp_cs']['gompertz'][curve_type]['b'][term],
'gompertz')
y_overall = detransform_fit(y_proportional.cumsum(), y_overall, 'dblp_cs')
error = datasets['dblp_cs']['gompertz'][curve_type]['RMS'][term]
axs[i].plot(x, 100*y_overall, color='k', linestyle='--', label="single peak, error="+str(np.round(error,3)))
else:
y_overall, y_1, y_2 = logletlab.calculate_series_double(x,
datasets['dblp_cs']['gompertz'][curve_type]['a1'][term],
datasets['dblp_cs']['gompertz'][curve_type]['k1'][term],
datasets['dblp_cs']['gompertz'][curve_type]['b1'][term],
datasets['dblp_cs']['gompertz'][curve_type]['a2'][term],
datasets['dblp_cs']['gompertz'][curve_type]['k2'][term],
datasets['dblp_cs']['gompertz'][curve_type]['b2'][term],
'gompertz')
y_overall = detransform_fit(y_proportional.cumsum(), y_overall, 'dblp_cs')
error = datasets['dblp_cs']['gompertz'][curve_type]['RMS'][term]
axs[i].plot(x, 100*y_overall, color='k', linestyle=':', label="double peak, error="+str(np.round(error,3)))
axs[i].set_title(titles[i], fontsize=12)
axs[i].legend( fontsize=11)
axs[i].set_ylim([0, limits[i]])
# We want the same number of y ticks for each axis so that it reads more neatly
axs[2].set_yticks([0, 1.5, 3, 4.5, 6])
fig.savefig(root+'images/doublepeaked.eps', format='eps', dpi=1200)
```
### Graphs of all four datasets
In this section we try to show as many graphs of fitted models as can reasonably fit on a page. The two functions used to make the graphs [below] are very hacky! However they work for this specific purpose.
```
def choose_ylimit(prevalence):
'''
This function works to find the most appropriate upper y limit to make the plots look good
'''
if max(prevalence) < 0.5:
return 0.5
elif max(prevalence) > 0.5 and max(prevalence) < 0.8:
return 0.8
elif max(prevalence) > 10 and max(prevalence) < 12:
return 12
elif max(prevalence) > 12 and max(prevalence) < 15:
return 15
elif max(prevalence) > 15 and max(prevalence) < 20:
return 20
else:
return np.ceil(max(prevalence))
def prettyplot(df, dataset_name, gompertz_params, yplots, xplots, title, ylabel, xlabel, xlims, plot_titles):
'''
Plot a nicely formatted set of trends with their fitted models. This function is rather hacky and made
for this specific purpose!
'''
fig, axs = plt.subplots(yplots, xplots)
plt.subplots_adjust(right=1, hspace=0.5, wspace=0.25)
plt.suptitle(title, fontsize=14)
fig.subplots_adjust(top=0.95)
fig.set_figheight(15)
fig.set_figwidth(9)
x = [int(i) for i in list(df.index)]
for i, term in enumerate(df.columns[0:yplots*xplots]):
prevalence = df[term].divide(document_count_per_year[dataset_name], axis=0)
if plot_titles == None:
title = term.split(',')[0]
else:
title = titles[i]
# Now get the gompertz representation of it
if gompertz_params['single']['RMS'][term]-gompertz_params['double']['RMS'][term] < 0.005:
# Use the single peaked version
y_overall = logletlab.calculate_series(x,
gompertz_params['single']['a'][term],
gompertz_params['single']['k'][term],
gompertz_params['single']['b'][term],
'gompertz')
y_overall = detransform_fit(prevalence.cumsum(), y_overall, dataset_name)
else:
y_overall, y_1, y_2 = logletlab.calculate_series_double(x,
gompertz_params['double']['a1'][term],
gompertz_params['double']['k1'][term],
gompertz_params['double']['b1'][term],
gompertz_params['double']['a2'][term],
gompertz_params['double']['k2'][term],
gompertz_params['double']['b2'][term],
'gompertz')
y_overall = detransform_fit(prevalence.cumsum(), y_overall, dataset_name)
axs[int(np.floor((i/xplots)%yplots)), i%xplots].plot(x, 100*prevalence, color='k', ls='-', label=title)
axs[int(np.floor((i/xplots)%yplots)), i%xplots].plot(x, 100*y_overall, color='k', ls='--', label='gompertz')
axs[int(np.floor((i/xplots)%yplots)), i%xplots].grid()
axs[int(np.floor((i/xplots)%yplots)), i%xplots].set_xlim(xlims[0], xlims[1])
axs[int(np.floor((i/xplots)%yplots)), i%xplots].set_ylim(0,choose_ylimit(100*prevalence))
axs[int(np.floor((i/xplots)%yplots)), i%xplots].set_title(title, fontsize=12)
axs[int(np.floor((i/xplots)%yplots)), i%xplots].yaxis.set_major_formatter(FormatStrFormatter('%.1f'))
if i%yplots != yplots-1:
axs[i%yplots, int(np.floor((i/yplots)%xplots))].set_xticklabels([])
axs[5,0].set_ylabel(ylabel, fontsize=12)
dataset_name = 'arxiv_hep'
df = pickle.load(open(root+'clusters/'+dataset_name+'.p', 'rb'))
titles = ['125 GeV', 'Pentaquark', 'WMAP', 'LHC Run', 'PAMELA', 'Lattice Gauge',
'Tensor-to-Scalar Ratio', 'Brane', 'ATLAS', 'Horava-Lifshitz', 'LHC',
'Noncommutative', 'Black Hole', 'Anomalous Magnetic Moment', 'Unparticle',
'Superluminal', 'M2 Brane', '126 GeV', 'pp-Wave', 'Lambert', 'Tevatron', 'Higgs',
'Brane World', 'Extra Dimension', 'Entropic', 'KamLAND', 'Solar Neutrino',
'Neutrino Oscillation', 'Chern Simon', 'Forward-Backward Asymmetry', 'Dark Energy',
'Bulk', 'Holographic', 'International Linear Collider', 'ABJM', 'BaBar']
prettyplot(df, 'arxiv_hep', datasets[dataset_name]['gompertz'], 12, 3, "Gompertz model fitted to trends in particle physics (1994-2017)", "Documents containing term (%)", None, [1990,2020], titles)
plt.savefig(root+'images/arxiv_hep.eps', format='eps', dpi=1200, bbox_inches='tight')
dataset_name = 'dblp_cs'
df = pickle.load(open(root+'clusters/'+dataset_name+'.p', 'rb'))
titles = ['Deep Learning', 'Neural Network', 'Machine Learning', 'Convolutional Neural Network',
'Java', 'Web', 'XML', 'Internet', 'Web Service', 'Internet of Things', 'World Wide Web',
'Speech', '5G', 'Discrete Mathematics', 'Parallel', 'Agent', 'Recurrent', 'SUP', 'Cloud',
'Big Data', 'Peer-to-peer', 'Wireless', 'Sensor Network', 'Electronic Commerce', 'ATM', 'Gene',
'Packet', 'Multimedia', 'Smart Grid', 'Embeddings', 'Ontology', 'Ad-hoc Network', 'Service Oriented',
'Web Site', 'RAC', 'Distributed Memory']
prettyplot(df, 'dblp_cs', datasets[dataset_name]['gompertz'], 12, 3, 'Gompertz model fitted to trends in computer science (1988-2017)', "Documents containing term (%)", None, [1980,2020], titles)
plt.savefig(root+'images/dblp_cs.eps', format='eps', dpi=1200, bbox_inches='tight')
dataset_name = 'pubmed_mh'
df = pickle.load(open(root+'clusters/'+dataset_name+'.p', 'rb'))
titles = titles = ['Alcoholic', 'Abeta', 'Psycinfo', 'Dexamethasone', 'Human Immunodeficiency Virus',
'Database', 'Alzheimers Disease', 'Amitriptyline', 'Intravenous Drug', 'Bupropion',
'DSM iii', 'Depression', 'Drug User', 'Apolipoprotein', 'Epsilon4 Allele', 'Rett Syndrome',
'Cocaine', 'Heroin', 'Panic', 'Imipramine', 'Papaverine', 'Cortisol', 'Presenilin', 'Plasma',
'Tricyclic', 'Epsilon Allele', 'HTLV iii', 'Learning Disability', 'DSM IV', 'DSM',
'Retardation', 'Aldehyde', 'Protein Precursor', 'Bulimia', 'Narcoleptic', 'Acquired Immunodeficiency Syndrome']
prettyplot(df, 'pubmed_mh', datasets[dataset_name]['gompertz'], 12, 3, 'Gompertz model fitted to trends in mental health research (1975-2017)', 'Documents containing term (%)', None, [1970,2020], titles)
plt.savefig(root+'images/pubmed_mh.eps', format='eps', dpi=1200, bbox_inches='tight')
dataset_name = 'pubmed_cancer'
df = pickle.load(open(root+'clusters/'+dataset_name+'.p', 'rb'))
titles = ['Immunohistochemical', 'Monoclonal Antibody', 'NF KappaB', 'Polymerase Chain Reaction',
'Immune Checkpoint', 'Tumor Suppressor Gene', 'Beta Catenin', 'PD-L1', 'Interleukin',
'Oncogene', 'Microarray', '1Alpha', 'PC12 Cell', 'Magnetic Resonance',
'Proliferating Cell Nuclear Antigen', 'Human T-cell Leukemia', 'Adult T-cell Leukemia',
'lncRNA', 'Apoptosis', 'CD4', 'Recombinant', 'Acquired Immunodeficiency Syndrome',
'HR', 'Meta Analysis', 'IC50', 'Immunoperoxidase', 'Blot', 'Interfering RNA', '18F',
'(Estrogen) Receptor Alpha', 'OKT4', 'kDa', 'CA', 'OKT8', 'Imatinib', 'Helper (T-cells)']
prettyplot(df, 'pubmed_cancer', datasets[dataset_name]['gompertz'], 12, 3, 'Gompertz model fitted to trends in cancer research (1988-2017)', 'Documents containing term (%)', None, [1970,2020], titles)
plt.savefig(root+'images/pubmed_cancer.eps', format='eps', dpi=1200, bbox_inches='tight')
```
| github_jupyter |
# CTR预估(1)
资料&&代码整理by[@寒小阳](https://blog.csdn.net/han_xiaoyang)([email protected])
reference:
* [《广告点击率预估是怎么回事?》](https://zhuanlan.zhihu.com/p/23499698)
* [从ctr预估问题看看f(x)设计—DNN篇](https://zhuanlan.zhihu.com/p/28202287)
* [Atomu2014 product_nets](https://github.com/Atomu2014/product-nets)
关于CTR预估的背景推荐大家看欧阳辰老师在知乎的文章[《广告点击率预估是怎么回事?》](https://zhuanlan.zhihu.com/p/23499698),感谢欧阳辰老师并在这里做一点小小的摘抄。
>点击率预估是广告技术的核心算法之一,它是很多广告算法工程师喜爱的战场。一直想介绍一下点击率预估,但是涉及公式和模型理论太多,怕说不清楚,读者也不明白。所以,这段时间花了一些时间整理点击率预估的知识,希望在尽量不使用数据公式的情况下,把大道理讲清楚,给一些不愿意看公式的同学一个Cook Book。
> ### 点击率预测是什么?
> * 点击率预测是对每次广告的点击情况做出预测,可以判定这次为点击或不点击,也可以给出点击的概率,有时也称作pClick。
> ### 点击率预测和推荐算法的不同?
> * 广告中点击率预估需要给出精准的点击概率,A点击率0.3% , B点击率0.13%等,需要结合出价用于排序使用;推荐算法很多时候只需要得出一个最优的次序A>B>C即可;
> ### 搜索和非搜索广告点击率预测的区别
> * 搜索中有强搜索信号-“查询词(Query)”,查询词和广告内容的匹配程度很大程度影响了点击概率; 点击率也高,PC搜索能到达百分之几的点击率。
> * 非搜索广告(例如展示广告,信息流广告),点击率的计算很多来源于用户的兴趣和广告特征,上下文环境;移动信息流广告的屏幕比较大,用户关注度也比较集中,好位置也能到百分之几的点击率。对于很多文章底部的广告,点击率非常低,用户关注度也不高,常常是千分之几,甚至更低;
> ### 如何衡量点击率预测的准确性?
> AUC是常常被用于衡量点击率预估的准确性的方法;理解AUC之前,需要理解一下Precision/Recall;对于一个分类器,我们通常将结果分为:TP,TN,FP,FN。
> 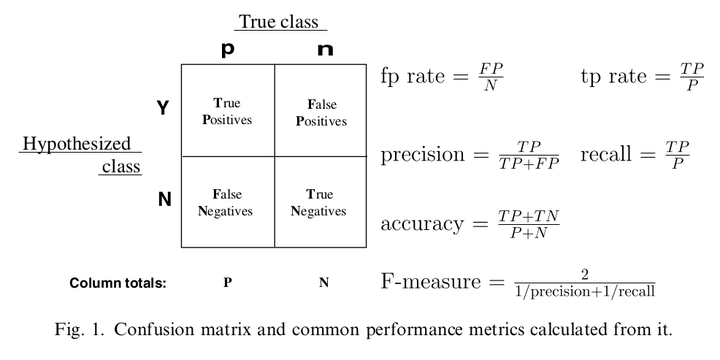
> 本来用Precision=TP/(TP+FP),Recall=TP/P,也可以用于评估点击率算法的好坏,毕竟这是一种监督学习,每一次预测都有正确答案。但是,这种方法对于测试数据样本的依赖性非常大,稍微不同的测试数据集合,结果差异非常大。那么,既然无法使用简单的单点Precision/Recall来描述,我们可以考虑使用一系列的点来描述准确性。做法如下:
> * 找到一系列的测试数据,点击率预估分别会对每个测试数据给出点击/不点击,和Confidence Score。
> * 按照给出的Score进行排序,那么考虑如果将Score作为一个Thresholds的话,考虑这个时候所有数据的 TP Rate 和 FP Rate; 当Thresholds分数非常高时,例如0.9,TP数很小,NP数很大,因此TP率不会太高;
> 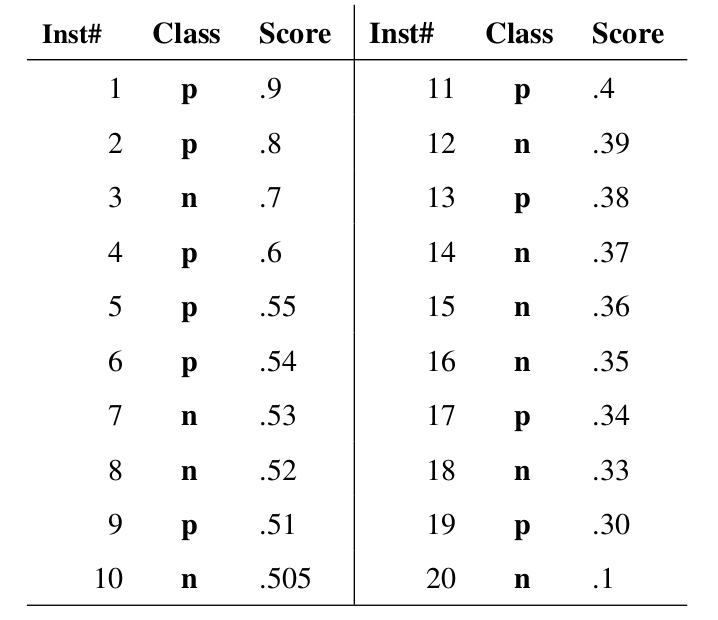
> 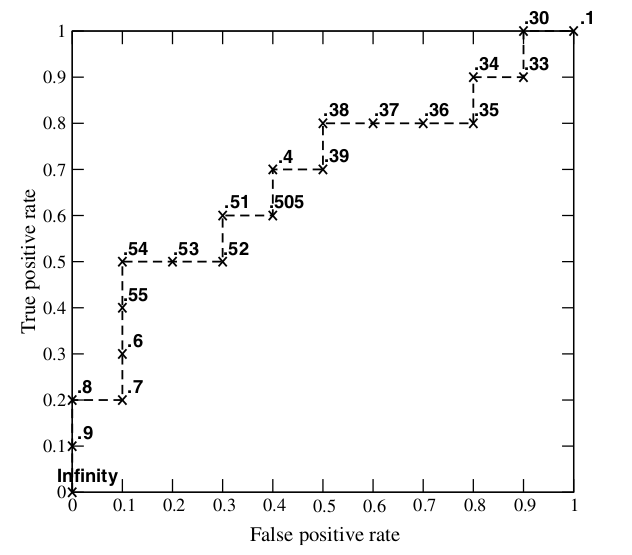
> 
> * 当选用不同Threshold时候,画出来的ROC曲线,以及下方AUC面积
> * 我们计算这个曲线下面的面积就是所谓的AUC值;AUC值越大,预测约准确。
> ### 为什么要使用AUC曲线
> 既然已经这么多评价标准,为什么还要使用ROC和AUC呢?因为ROC曲线有个很好的特性:当测试集中的正负样本的分布变化的时候,ROC曲线能够保持不变。在实际的数据集中经常会出现类不平衡(class imbalance)现象,即负样本比正样本多很多(或者相反),而且测试数据中的正负样本的分布也可能随着时间变化。AUC对样本的比例变化有一定的容忍性。AUC的值通常在0.6-0.85之间。
> ### 如何来进行点击率预测?
> 点击率预测可以考虑为一个黑盒,输入一堆信号,输出点击的概率。这些信号就包括如下信号
> * **广告**:历史点击率,文字,格式,图片等等
> * **环境**:手机型号,时间媒体,位置,尺寸,曝光时间,网络IP,上网方式,代理等
> * **用户**:基础属性(男女,年龄等),兴趣属性(游戏,旅游等),历史浏览,点击行为,电商行为
> * **信号的粒度**:
> `Low Level : 数据来自一些原始访问行为的记录,例如用户是否点击过Landing Page,流量IP等。这些特征可以用于粗选,模型简单,`
> `High Level: 特征来自一些可解释的数据,例如兴趣标签,性别等`
> * **特征编码Feature Encoding:**
> `特征离散化:把连续的数字,变成离散化,例如温度值可以办成多个温度区间。`
> `特征交叉: 把多个特征进行叫交叉的出的值,用于训练,这种值可以表示一些非线性的关系。例如,点击率预估中应用最多的就是广告跟用户的交叉特征、广告跟性别的交叉特征,广告跟年龄的交叉特征,广告跟手机平台的交叉特征,广告跟地域的交叉特征等等。`
> * **特征选取(Feature Selection):**
> `特征选择就是选择那些靠谱的Feature,去掉冗余的Feature,对于搜索广告Query和广告的匹配程度很关键;对于展示广告,广告本身的历史表现,往往是最重要的Feature。`
> * **独热编码(One-Hot encoding)**
```假设有三组特征,分别表示年龄,城市,设备;
["男", "女"]
["北京", "上海", "广州"]
["苹果", "小米", "华为", "微软"]
传统变化: 对每一组特征,使用枚举类型,从0开始;
["男“,”上海“,”小米“]=[ 0,1,1]
["女“,”北京“,”苹果“] =[1,0,0]
传统变化后的数据不是连续的,而是随机分配的,不容易应用在分类器中。
热独编码是一种经典编码,是使用N位状态寄存器来对N个状态进行编码,每个状态都由他独立的寄存器位,并且在任意时候,其中只有一位有效。
["男“,”上海“,”小米“]=[ 1,0,0,1,0,0,1,0,0]
["女“,”北京“,”苹果“] =[0,1,1,0,0,1,0,0,0]
经过热独编码,数据会变成稀疏的,方便分类器处理。
```
> ### 点击率预估整体过程:
> 三个基本过程:特征工程,模型训练,线上服务
> 
> * 特征工程:准备各种特征,编码,去掉冗余特征(用PCA等)
> * 模型训练:选定训练,测试等数据集,计算AUC,如果AUC有提升,通常可以在进一步在线上分流实验。
> * 线上服务:线上服务,需要实时计算CTR,实时计算相关特征和利用模型计算CTR,对于不同来源的CTR,可能需要一个Calibration的服务。
```
## 用tensorflow构建各种模型完成ctr预估
```
!head -5 ./data/train.txt
!head -10 ./data/featindex.txt
from __future__ import print_function
from __future__ import absolute_import
from __future__ import division
import cPickle as pkl
import numpy as np
import tensorflow as tf
from scipy.sparse import coo_matrix
# 读取数据,统计基本的信息,field等
DTYPE = tf.float32
FIELD_SIZES = [0] * 26
with open('./data/featindex.txt') as fin:
for line in fin:
line = line.strip().split(':')
if len(line) > 1:
f = int(line[0]) - 1
FIELD_SIZES[f] += 1
print('field sizes:', FIELD_SIZES)
FIELD_OFFSETS = [sum(FIELD_SIZES[:i]) for i in range(len(FIELD_SIZES))]
INPUT_DIM = sum(FIELD_SIZES)
OUTPUT_DIM = 1
STDDEV = 1e-3
MINVAL = -1e-3
MAXVAL = 1e-3
# 读取libsvm格式数据成稀疏矩阵形式
# 0 5:1 9:1 140858:1 445908:1 446177:1 446293:1 449140:1 490778:1 491626:1 491634:1 491641:1 491645:1 491648:1 491668:1 491700:1 491708:1
def read_data(file_name):
X = []
D = []
y = []
with open(file_name) as fin:
for line in fin:
fields = line.strip().split()
y_i = int(fields[0])
X_i = [int(x.split(':')[0]) for x in fields[1:]]
D_i = [int(x.split(':')[1]) for x in fields[1:]]
y.append(y_i)
X.append(X_i)
D.append(D_i)
y = np.reshape(np.array(y), [-1])
X = libsvm_2_coo(zip(X, D), (len(X), INPUT_DIM)).tocsr()
return X, y
# 数据乱序
def shuffle(data):
X, y = data
ind = np.arange(X.shape[0])
for i in range(7):
np.random.shuffle(ind)
return X[ind], y[ind]
# 工具函数,libsvm格式转成coo稀疏存储格式
def libsvm_2_coo(libsvm_data, shape):
coo_rows = []
coo_cols = []
coo_data = []
n = 0
for x, d in libsvm_data:
coo_rows.extend([n] * len(x))
coo_cols.extend(x)
coo_data.extend(d)
n += 1
coo_rows = np.array(coo_rows)
coo_cols = np.array(coo_cols)
coo_data = np.array(coo_data)
return coo_matrix((coo_data, (coo_rows, coo_cols)), shape=shape)
# csr转成输入格式
def csr_2_input(csr_mat):
if not isinstance(csr_mat, list):
coo_mat = csr_mat.tocoo()
indices = np.vstack((coo_mat.row, coo_mat.col)).transpose()
values = csr_mat.data
shape = csr_mat.shape
return indices, values, shape
else:
inputs = []
for csr_i in csr_mat:
inputs.append(csr_2_input(csr_i))
return inputs
# 数据切片
def slice(csr_data, start=0, size=-1):
if not isinstance(csr_data[0], list):
if size == -1 or start + size >= csr_data[0].shape[0]:
slc_data = csr_data[0][start:]
slc_labels = csr_data[1][start:]
else:
slc_data = csr_data[0][start:start + size]
slc_labels = csr_data[1][start:start + size]
else:
if size == -1 or start + size >= csr_data[0][0].shape[0]:
slc_data = []
for d_i in csr_data[0]:
slc_data.append(d_i[start:])
slc_labels = csr_data[1][start:]
else:
slc_data = []
for d_i in csr_data[0]:
slc_data.append(d_i[start:start + size])
slc_labels = csr_data[1][start:start + size]
return csr_2_input(slc_data), slc_labels
# 数据切分
def split_data(data, skip_empty=True):
fields = []
for i in range(len(FIELD_OFFSETS) - 1):
start_ind = FIELD_OFFSETS[i]
end_ind = FIELD_OFFSETS[i + 1]
if skip_empty and start_ind == end_ind:
continue
field_i = data[0][:, start_ind:end_ind]
fields.append(field_i)
fields.append(data[0][:, FIELD_OFFSETS[-1]:])
return fields, data[1]
# 在tensorflow中初始化各种参数变量
def init_var_map(init_vars, init_path=None):
if init_path is not None:
load_var_map = pkl.load(open(init_path, 'rb'))
print('load variable map from', init_path, load_var_map.keys())
var_map = {}
for var_name, var_shape, init_method, dtype in init_vars:
if init_method == 'zero':
var_map[var_name] = tf.Variable(tf.zeros(var_shape, dtype=dtype), name=var_name, dtype=dtype)
elif init_method == 'one':
var_map[var_name] = tf.Variable(tf.ones(var_shape, dtype=dtype), name=var_name, dtype=dtype)
elif init_method == 'normal':
var_map[var_name] = tf.Variable(tf.random_normal(var_shape, mean=0.0, stddev=STDDEV, dtype=dtype),
name=var_name, dtype=dtype)
elif init_method == 'tnormal':
var_map[var_name] = tf.Variable(tf.truncated_normal(var_shape, mean=0.0, stddev=STDDEV, dtype=dtype),
name=var_name, dtype=dtype)
elif init_method == 'uniform':
var_map[var_name] = tf.Variable(tf.random_uniform(var_shape, minval=MINVAL, maxval=MAXVAL, dtype=dtype),
name=var_name, dtype=dtype)
elif init_method == 'xavier':
maxval = np.sqrt(6. / np.sum(var_shape))
minval = -maxval
var_map[var_name] = tf.Variable(tf.random_uniform(var_shape, minval=minval, maxval=maxval, dtype=dtype),
name=var_name, dtype=dtype)
elif isinstance(init_method, int) or isinstance(init_method, float):
var_map[var_name] = tf.Variable(tf.ones(var_shape, dtype=dtype) * init_method, name=var_name, dtype=dtype)
elif init_method in load_var_map:
if load_var_map[init_method].shape == tuple(var_shape):
var_map[var_name] = tf.Variable(load_var_map[init_method], name=var_name, dtype=dtype)
else:
print('BadParam: init method', init_method, 'shape', var_shape, load_var_map[init_method].shape)
else:
print('BadParam: init method', init_method)
return var_map
# 不同的激活函数选择
def activate(weights, activation_function):
if activation_function == 'sigmoid':
return tf.nn.sigmoid(weights)
elif activation_function == 'softmax':
return tf.nn.softmax(weights)
elif activation_function == 'relu':
return tf.nn.relu(weights)
elif activation_function == 'tanh':
return tf.nn.tanh(weights)
elif activation_function == 'elu':
return tf.nn.elu(weights)
elif activation_function == 'none':
return weights
else:
return weights
# 不同的优化器选择
def get_optimizer(opt_algo, learning_rate, loss):
if opt_algo == 'adaldeta':
return tf.train.AdadeltaOptimizer(learning_rate).minimize(loss)
elif opt_algo == 'adagrad':
return tf.train.AdagradOptimizer(learning_rate).minimize(loss)
elif opt_algo == 'adam':
return tf.train.AdamOptimizer(learning_rate).minimize(loss)
elif opt_algo == 'ftrl':
return tf.train.FtrlOptimizer(learning_rate).minimize(loss)
elif opt_algo == 'gd':
return tf.train.GradientDescentOptimizer(learning_rate).minimize(loss)
elif opt_algo == 'padagrad':
return tf.train.ProximalAdagradOptimizer(learning_rate).minimize(loss)
elif opt_algo == 'pgd':
return tf.train.ProximalGradientDescentOptimizer(learning_rate).minimize(loss)
elif opt_algo == 'rmsprop':
return tf.train.RMSPropOptimizer(learning_rate).minimize(loss)
else:
return tf.train.GradientDescentOptimizer(learning_rate).minimize(loss)
# 工具函数
# 提示:tf.slice(input_, begin, size, name=None):按照指定的下标范围抽取连续区域的子集
# tf.gather(params, indices, validate_indices=None, name=None):按照指定的下标集合从axis=0中抽取子集,适合抽取不连续区域的子集
def gather_2d(params, indices):
shape = tf.shape(params)
flat = tf.reshape(params, [-1])
flat_idx = indices[:, 0] * shape[1] + indices[:, 1]
flat_idx = tf.reshape(flat_idx, [-1])
return tf.gather(flat, flat_idx)
def gather_3d(params, indices):
shape = tf.shape(params)
flat = tf.reshape(params, [-1])
flat_idx = indices[:, 0] * shape[1] * shape[2] + indices[:, 1] * shape[2] + indices[:, 2]
flat_idx = tf.reshape(flat_idx, [-1])
return tf.gather(flat, flat_idx)
def gather_4d(params, indices):
shape = tf.shape(params)
flat = tf.reshape(params, [-1])
flat_idx = indices[:, 0] * shape[1] * shape[2] * shape[3] + \
indices[:, 1] * shape[2] * shape[3] + indices[:, 2] * shape[3] + indices[:, 3]
flat_idx = tf.reshape(flat_idx, [-1])
return tf.gather(flat, flat_idx)
# 池化2d
def max_pool_2d(params, k):
_, indices = tf.nn.top_k(params, k, sorted=False)
shape = tf.shape(indices)
r1 = tf.reshape(tf.range(shape[0]), [-1, 1])
r1 = tf.tile(r1, [1, k])
r1 = tf.reshape(r1, [-1, 1])
indices = tf.concat([r1, tf.reshape(indices, [-1, 1])], 1)
return tf.reshape(gather_2d(params, indices), [-1, k])
# 池化3d
def max_pool_3d(params, k):
_, indices = tf.nn.top_k(params, k, sorted=False)
shape = tf.shape(indices)
r1 = tf.reshape(tf.range(shape[0]), [-1, 1])
r2 = tf.reshape(tf.range(shape[1]), [-1, 1])
r1 = tf.tile(r1, [1, k * shape[1]])
r2 = tf.tile(r2, [1, k])
r1 = tf.reshape(r1, [-1, 1])
r2 = tf.tile(tf.reshape(r2, [-1, 1]), [shape[0], 1])
indices = tf.concat([r1, r2, tf.reshape(indices, [-1, 1])], 1)
return tf.reshape(gather_3d(params, indices), [-1, shape[1], k])
# 池化4d
def max_pool_4d(params, k):
_, indices = tf.nn.top_k(params, k, sorted=False)
shape = tf.shape(indices)
r1 = tf.reshape(tf.range(shape[0]), [-1, 1])
r2 = tf.reshape(tf.range(shape[1]), [-1, 1])
r3 = tf.reshape(tf.range(shape[2]), [-1, 1])
r1 = tf.tile(r1, [1, shape[1] * shape[2] * k])
r2 = tf.tile(r2, [1, shape[2] * k])
r3 = tf.tile(r3, [1, k])
r1 = tf.reshape(r1, [-1, 1])
r2 = tf.tile(tf.reshape(r2, [-1, 1]), [shape[0], 1])
r3 = tf.tile(tf.reshape(r3, [-1, 1]), [shape[0] * shape[1], 1])
indices = tf.concat([r1, r2, r3, tf.reshape(indices, [-1, 1])], 1)
return tf.reshape(gather_4d(params, indices), [-1, shape[1], shape[2], k])
```
## 定义不同的模型
```
# 定义基类模型
dtype = DTYPE
class Model:
def __init__(self):
self.sess = None
self.X = None
self.y = None
self.layer_keeps = None
self.vars = None
self.keep_prob_train = None
self.keep_prob_test = None
# run model
def run(self, fetches, X=None, y=None, mode='train'):
# 通过feed_dict传入数据
feed_dict = {}
if type(self.X) is list:
for i in range(len(X)):
feed_dict[self.X[i]] = X[i]
else:
feed_dict[self.X] = X
if y is not None:
feed_dict[self.y] = y
if self.layer_keeps is not None:
if mode == 'train':
feed_dict[self.layer_keeps] = self.keep_prob_train
elif mode == 'test':
feed_dict[self.layer_keeps] = self.keep_prob_test
#通过session.run去执行op
return self.sess.run(fetches, feed_dict)
# 模型参数持久化
def dump(self, model_path):
var_map = {}
for name, var in self.vars.iteritems():
var_map[name] = self.run(var)
pkl.dump(var_map, open(model_path, 'wb'))
print('model dumped at', model_path)
```
### 1.LR逻辑回归
输入输出:{X,y}<br>
映射函数f(x):单层单节点的“DNN”, 宽而不深,sigmoid(wx+b)输出概率,需要大量的人工特征工程,非线性来源于特征处理<br>
损失函数:logloss/... + L1/L2/...<br>
优化方法:sgd/...<br>
评估:logloss/auc/...<br>
```
class LR(Model):
def __init__(self, input_dim=None, output_dim=1, init_path=None, opt_algo='gd', learning_rate=1e-2, l2_weight=0,
random_seed=None):
Model.__init__(self)
# 声明参数
init_vars = [('w', [input_dim, output_dim], 'xavier', dtype),
('b', [output_dim], 'zero', dtype)]
self.graph = tf.Graph()
with self.graph.as_default():
if random_seed is not None:
tf.set_random_seed(random_seed)
# 用稀疏的placeholder
self.X = tf.sparse_placeholder(dtype)
self.y = tf.placeholder(dtype)
# init参数
self.vars = init_var_map(init_vars, init_path)
w = self.vars['w']
b = self.vars['b']
# sigmoid(wx+b)
xw = tf.sparse_tensor_dense_matmul(self.X, w)
logits = tf.reshape(xw + b, [-1])
self.y_prob = tf.sigmoid(logits)
self.loss = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(labels=self.y, logits=logits)) + \
l2_weight * tf.nn.l2_loss(xw)
self.optimizer = get_optimizer(opt_algo, learning_rate, self.loss)
# GPU设定
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
self.sess = tf.Session(config=config)
# 初始化图里的参数
tf.global_variables_initializer().run(session=self.sess)
import numpy as np
from sklearn.metrics import roc_auc_score
import progressbar
train_file = './data/train.txt'
test_file = './data/test.txt'
input_dim = INPUT_DIM
# 读取数据
#train_data = read_data(train_file)
#test_data = read_data(test_file)
train_data = pkl.load(open('./data/train.pkl', 'rb'))
#train_data = shuffle(train_data)
test_data = pkl.load(open('./data/test.pkl', 'rb'))
# pkl.dump(train_data, open('./data/train.pkl', 'wb'))
# pkl.dump(test_data, open('./data/test.pkl', 'wb'))
# 输出数据信息维度
if train_data[1].ndim > 1:
print('label must be 1-dim')
exit(0)
print('read finish')
print('train data size:', train_data[0].shape)
print('test data size:', test_data[0].shape)
# 训练集与测试集
train_size = train_data[0].shape[0]
test_size = test_data[0].shape[0]
num_feas = len(FIELD_SIZES)
# 超参数设定
min_round = 1
num_round = 200
early_stop_round = 5
# train + val
batch_size = 1024
field_sizes = FIELD_SIZES
field_offsets = FIELD_OFFSETS
# 逻辑回归参数设定
lr_params = {
'input_dim': input_dim,
'opt_algo': 'gd',
'learning_rate': 0.1,
'l2_weight': 0,
'random_seed': 0
}
print(lr_params)
model = LR(**lr_params)
print("training LR...")
def train(model):
history_score = []
# 执行num_round轮
for i in range(num_round):
# 主要的2个op是优化器和损失
fetches = [model.optimizer, model.loss]
if batch_size > 0:
ls = []
# 进度条工具
bar = progressbar.ProgressBar()
print('[%d]\ttraining...' % i)
for j in bar(range(int(train_size / batch_size + 1))):
X_i, y_i = slice(train_data, j * batch_size, batch_size)
# 训练,run op
_, l = model.run(fetches, X_i, y_i)
ls.append(l)
elif batch_size == -1:
X_i, y_i = slice(train_data)
_, l = model.run(fetches, X_i, y_i)
ls = [l]
train_preds = []
print('[%d]\tevaluating...' % i)
bar = progressbar.ProgressBar()
for j in bar(range(int(train_size / 10000 + 1))):
X_i, _ = slice(train_data, j * 10000, 10000)
preds = model.run(model.y_prob, X_i, mode='test')
train_preds.extend(preds)
test_preds = []
bar = progressbar.ProgressBar()
for j in bar(range(int(test_size / 10000 + 1))):
X_i, _ = slice(test_data, j * 10000, 10000)
preds = model.run(model.y_prob, X_i, mode='test')
test_preds.extend(preds)
# 把预估的结果和真实结果拿出来计算auc
train_score = roc_auc_score(train_data[1], train_preds)
test_score = roc_auc_score(test_data[1], test_preds)
# 输出auc信息
print('[%d]\tloss (with l2 norm):%f\ttrain-auc: %f\teval-auc: %f' % (i, np.mean(ls), train_score, test_score))
history_score.append(test_score)
# early stopping
if i > min_round and i > early_stop_round:
if np.argmax(history_score) == i - early_stop_round and history_score[-1] - history_score[
-1 * early_stop_round] < 1e-5:
print('early stop\nbest iteration:\n[%d]\teval-auc: %f' % (
np.argmax(history_score), np.max(history_score)))
break
train(model)
```
### 2.FM
FM可以视作有二次交叉的LR,为了控制参数量和充分学习,提出了user vector和item vector的概念
```
class FM(Model):
def __init__(self, input_dim=None, output_dim=1, factor_order=10, init_path=None, opt_algo='gd', learning_rate=1e-2,
l2_w=0, l2_v=0, random_seed=None):
Model.__init__(self)
# 一次、二次交叉、偏置项
init_vars = [('w', [input_dim, output_dim], 'xavier', dtype),
('v', [input_dim, factor_order], 'xavier', dtype),
('b', [output_dim], 'zero', dtype)]
self.graph = tf.Graph()
with self.graph.as_default():
if random_seed is not None:
tf.set_random_seed(random_seed)
self.X = tf.sparse_placeholder(dtype)
self.y = tf.placeholder(dtype)
self.vars = init_var_map(init_vars, init_path)
w = self.vars['w']
v = self.vars['v']
b = self.vars['b']
# [(x1+x2+x3)^2 - (x1^2+x2^2+x3^2)]/2
# 先计算所有的交叉项,再减去平方项(自己和自己相乘)
X_square = tf.SparseTensor(self.X.indices, tf.square(self.X.values), tf.to_int64(tf.shape(self.X)))
xv = tf.square(tf.sparse_tensor_dense_matmul(self.X, v))
p = 0.5 * tf.reshape(
tf.reduce_sum(xv - tf.sparse_tensor_dense_matmul(X_square, tf.square(v)), 1),
[-1, output_dim])
xw = tf.sparse_tensor_dense_matmul(self.X, w)
logits = tf.reshape(xw + b + p, [-1])
self.y_prob = tf.sigmoid(logits)
self.loss = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=logits, labels=self.y)) + \
l2_w * tf.nn.l2_loss(xw) + \
l2_v * tf.nn.l2_loss(xv)
self.optimizer = get_optimizer(opt_algo, learning_rate, self.loss)
#GPU设定
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
self.sess = tf.Session(config=config)
# 图中所有variable初始化
tf.global_variables_initializer().run(session=self.sess)
import numpy as np
from sklearn.metrics import roc_auc_score
import progressbar
train_file = './data/train.txt'
test_file = './data/test.txt'
input_dim = INPUT_DIM
train_data = pkl.load(open('./data/train.pkl', 'rb'))
train_data = shuffle(train_data)
test_data = pkl.load(open('./data/test.pkl', 'rb'))
if train_data[1].ndim > 1:
print('label must be 1-dim')
exit(0)
print('read finish')
print('train data size:', train_data[0].shape)
print('test data size:', test_data[0].shape)
# 训练集与测试集
train_size = train_data[0].shape[0]
test_size = test_data[0].shape[0]
num_feas = len(FIELD_SIZES)
# 超参数设定
min_round = 1
num_round = 200
early_stop_round = 5
batch_size = 1024
field_sizes = FIELD_SIZES
field_offsets = FIELD_OFFSETS
# FM参数设定
fm_params = {
'input_dim': input_dim,
'factor_order': 10,
'opt_algo': 'gd',
'learning_rate': 0.1,
'l2_w': 0,
'l2_v': 0,
}
print(fm_params)
model = FM(**fm_params)
print("training FM...")
def train(model):
history_score = []
for i in range(num_round):
# 同样是优化器和损失两个op
fetches = [model.optimizer, model.loss]
if batch_size > 0:
ls = []
bar = progressbar.ProgressBar()
print('[%d]\ttraining...' % i)
for j in bar(range(int(train_size / batch_size + 1))):
X_i, y_i = slice(train_data, j * batch_size, batch_size)
# 训练
_, l = model.run(fetches, X_i, y_i)
ls.append(l)
elif batch_size == -1:
X_i, y_i = slice(train_data)
_, l = model.run(fetches, X_i, y_i)
ls = [l]
train_preds = []
print('[%d]\tevaluating...' % i)
bar = progressbar.ProgressBar()
for j in bar(range(int(train_size / 10000 + 1))):
X_i, _ = slice(train_data, j * 10000, 10000)
preds = model.run(model.y_prob, X_i, mode='test')
train_preds.extend(preds)
test_preds = []
bar = progressbar.ProgressBar()
for j in bar(range(int(test_size / 10000 + 1))):
X_i, _ = slice(test_data, j * 10000, 10000)
preds = model.run(model.y_prob, X_i, mode='test')
test_preds.extend(preds)
train_score = roc_auc_score(train_data[1], train_preds)
test_score = roc_auc_score(test_data[1], test_preds)
print('[%d]\tloss (with l2 norm):%f\ttrain-auc: %f\teval-auc: %f' % (i, np.mean(ls), train_score, test_score))
history_score.append(test_score)
if i > min_round and i > early_stop_round:
if np.argmax(history_score) == i - early_stop_round and history_score[-1] - history_score[
-1 * early_stop_round] < 1e-5:
print('early stop\nbest iteration:\n[%d]\teval-auc: %f' % (
np.argmax(history_score), np.max(history_score)))
break
train(model)
```
### FNN
FNN的考虑是模型的capacity可以进一步提升,以对更复杂的场景建模。<br>
FNN可以视作FM + MLP = LR + MF + MLP
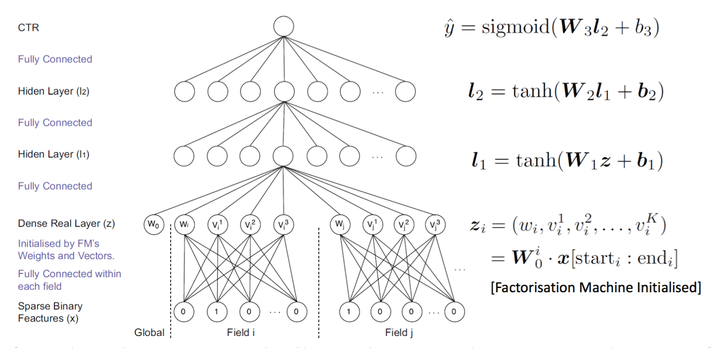
```
class FNN(Model):
def __init__(self, field_sizes=None, embed_size=10, layer_sizes=None, layer_acts=None, drop_out=None,
embed_l2=None, layer_l2=None, init_path=None, opt_algo='gd', learning_rate=1e-2, random_seed=None):
Model.__init__(self)
init_vars = []
num_inputs = len(field_sizes)
for i in range(num_inputs):
init_vars.append(('embed_%d' % i, [field_sizes[i], embed_size], 'xavier', dtype))
node_in = num_inputs * embed_size
for i in range(len(layer_sizes)):
init_vars.append(('w%d' % i, [node_in, layer_sizes[i]], 'xavier', dtype))
init_vars.append(('b%d' % i, [layer_sizes[i]], 'zero', dtype))
node_in = layer_sizes[i]
self.graph = tf.Graph()
with self.graph.as_default():
if random_seed is not None:
tf.set_random_seed(random_seed)
self.X = [tf.sparse_placeholder(dtype) for i in range(num_inputs)]
self.y = tf.placeholder(dtype)
self.keep_prob_train = 1 - np.array(drop_out)
self.keep_prob_test = np.ones_like(drop_out)
self.layer_keeps = tf.placeholder(dtype)
self.vars = init_var_map(init_vars, init_path)
w0 = [self.vars['embed_%d' % i] for i in range(num_inputs)]
xw = tf.concat([tf.sparse_tensor_dense_matmul(self.X[i], w0[i]) for i in range(num_inputs)], 1)
l = xw
for i in range(len(layer_sizes)):
wi = self.vars['w%d' % i]
bi = self.vars['b%d' % i]
print(l.shape, wi.shape, bi.shape)
l = tf.nn.dropout(
activate(
tf.matmul(l, wi) + bi,
layer_acts[i]),
self.layer_keeps[i])
l = tf.squeeze(l)
self.y_prob = tf.sigmoid(l)
self.loss = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=l, labels=self.y))
if layer_l2 is not None:
self.loss += embed_l2 * tf.nn.l2_loss(xw)
for i in range(len(layer_sizes)):
wi = self.vars['w%d' % i]
self.loss += layer_l2[i] * tf.nn.l2_loss(wi)
self.optimizer = get_optimizer(opt_algo, learning_rate, self.loss)
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
self.sess = tf.Session(config=config)
tf.global_variables_initializer().run(session=self.sess)
import numpy as np
from sklearn.metrics import roc_auc_score
import progressbar
train_file = './data/train.txt'
test_file = './data/test.txt'
input_dim = INPUT_DIM
train_data = pkl.load(open('./data/train.pkl', 'rb'))
train_data = shuffle(train_data)
test_data = pkl.load(open('./data/test.pkl', 'rb'))
if train_data[1].ndim > 1:
print('label must be 1-dim')
exit(0)
print('read finish')
print('train data size:', train_data[0].shape)
print('test data size:', test_data[0].shape)
train_size = train_data[0].shape[0]
test_size = test_data[0].shape[0]
num_feas = len(FIELD_SIZES)
min_round = 1
num_round = 200
early_stop_round = 5
batch_size = 1024
field_sizes = FIELD_SIZES
field_offsets = FIELD_OFFSETS
train_data = split_data(train_data)
test_data = split_data(test_data)
tmp = []
for x in field_sizes:
if x > 0:
tmp.append(x)
field_sizes = tmp
print('remove empty fields', field_sizes)
fnn_params = {
'field_sizes': field_sizes,
'embed_size': 10,
'layer_sizes': [500, 1],
'layer_acts': ['relu', None],
'drop_out': [0, 0],
'opt_algo': 'gd',
'learning_rate': 0.1,
'embed_l2': 0,
'layer_l2': [0, 0],
'random_seed': 0
}
print(fnn_params)
model = FNN(**fnn_params)
def train(model):
history_score = []
for i in range(num_round):
fetches = [model.optimizer, model.loss]
if batch_size > 0:
ls = []
bar = progressbar.ProgressBar()
print('[%d]\ttraining...' % i)
for j in bar(range(int(train_size / batch_size + 1))):
X_i, y_i = slice(train_data, j * batch_size, batch_size)
_, l = model.run(fetches, X_i, y_i)
ls.append(l)
elif batch_size == -1:
X_i, y_i = slice(train_data)
_, l = model.run(fetches, X_i, y_i)
ls = [l]
train_preds = []
print('[%d]\tevaluating...' % i)
bar = progressbar.ProgressBar()
for j in bar(range(int(train_size / 10000 + 1))):
X_i, _ = slice(train_data, j * 10000, 10000)
preds = model.run(model.y_prob, X_i, mode='test')
train_preds.extend(preds)
test_preds = []
bar = progressbar.ProgressBar()
for j in bar(range(int(test_size / 10000 + 1))):
X_i, _ = slice(test_data, j * 10000, 10000)
preds = model.run(model.y_prob, X_i, mode='test')
test_preds.extend(preds)
train_score = roc_auc_score(train_data[1], train_preds)
test_score = roc_auc_score(test_data[1], test_preds)
print('[%d]\tloss (with l2 norm):%f\ttrain-auc: %f\teval-auc: %f' % (i, np.mean(ls), train_score, test_score))
history_score.append(test_score)
if i > min_round and i > early_stop_round:
if np.argmax(history_score) == i - early_stop_round and history_score[-1] - history_score[
-1 * early_stop_round] < 1e-5:
print('early stop\nbest iteration:\n[%d]\teval-auc: %f' % (
np.argmax(history_score), np.max(history_score)))
break
train(model)
```
### CCPM
reference:[ctr模型汇总](https://zhuanlan.zhihu.com/p/32523455)
FM只能学习特征的二阶组合,但CNN能学习更高阶的组合,可学习的阶数和卷积的视野相关。
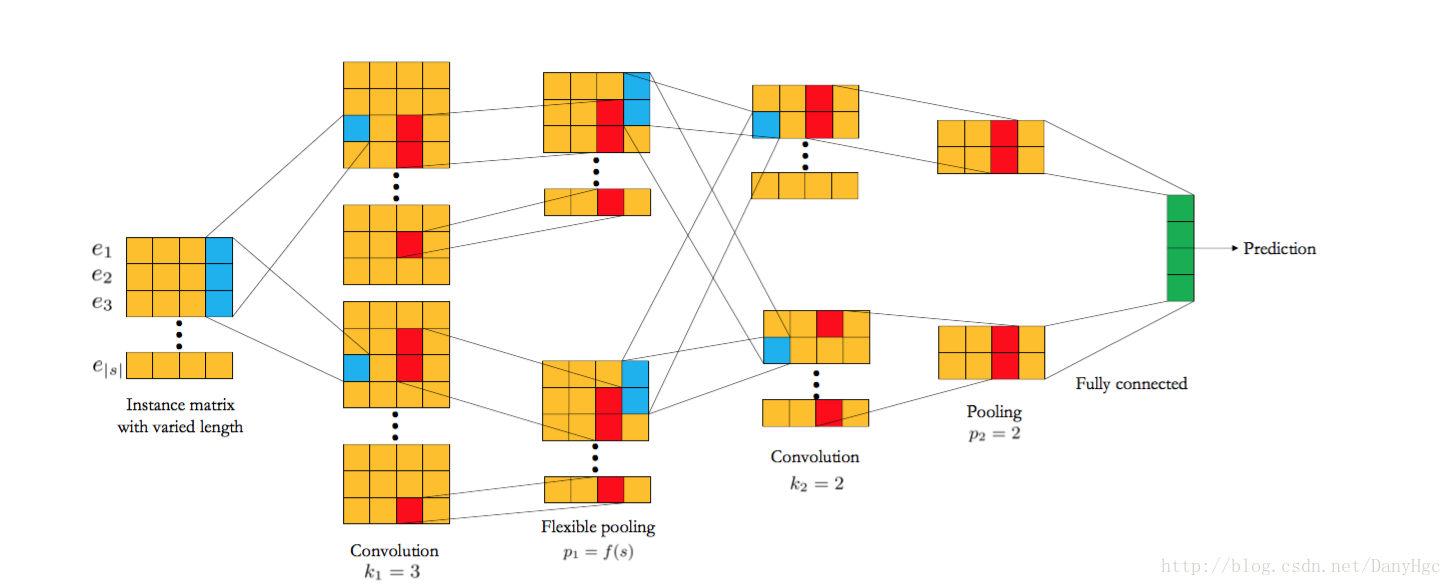
mbedding层:e1, e2…en是某特定用户被展示的一系列广告。如果在预测广告是否会点击时不考虑历史展示广告的点击情况,则n=1。同时embedding矩阵的具体值是随着模型训练学出来的。Embedding矩阵为S,向量维度为d。
卷积层:卷积参数W有d*w个,即对于矩阵S,上图每一列对应一个参数不共享的一维卷积,其视野为w,卷积共有d个,每个输出向量维度为(n+w-1),输出矩阵维度d*(n+w-1)。因为对于ctr预估而言,矩阵S每一列都对应特定的描述维度,所以需要分别处理,得到的输出矩阵的每一列就都是描述广告特定方面的特征。
Pooling层:flexible p-max pooling。
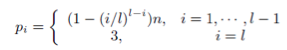
L是模型总卷积层数,n是输入序列长度,pi就是第i层的pooling参数。这样最后一层卷积层都是输出3个最大的元素,长度固定方便后面接全连接层。同时这个指数型的参数,一开始改变比较小,几乎都是n,后面就减少得比较快。这样可以防止在模型浅层的时候就损失太多信息,众所周知深度模型在前面几层最好不要做得太简单,容易损失很多信息。文章还提到p-max pooling输出的几个最大的元素是保序的,可输入时的顺序一致,这点对于保留序列信息是重要的。
激活层:tanh
最后,
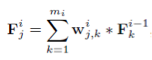
Fij是指低i层的第j个feature map。感觉是不同输入通道的卷积参数也不共享,对应输出是所有输入通道卷积的输出的求和。
```
class CCPM(Model):
def __init__(self, field_sizes=None, embed_size=10, filter_sizes=None, layer_acts=None, drop_out=None,
init_path=None, opt_algo='gd', learning_rate=1e-2, random_seed=None):
Model.__init__(self)
init_vars = []
num_inputs = len(field_sizes)
for i in range(num_inputs):
init_vars.append(('embed_%d' % i, [field_sizes[i], embed_size], 'xavier', dtype))
init_vars.append(('f1', [embed_size, filter_sizes[0], 1, 2], 'xavier', dtype))
init_vars.append(('f2', [embed_size, filter_sizes[1], 2, 2], 'xavier', dtype))
init_vars.append(('w1', [2 * 3 * embed_size, 1], 'xavier', dtype))
init_vars.append(('b1', [1], 'zero', dtype))
self.graph = tf.Graph()
with self.graph.as_default():
if random_seed is not None:
tf.set_random_seed(random_seed)
self.X = [tf.sparse_placeholder(dtype) for i in range(num_inputs)]
self.y = tf.placeholder(dtype)
self.keep_prob_train = 1 - np.array(drop_out)
self.keep_prob_test = np.ones_like(drop_out)
self.layer_keeps = tf.placeholder(dtype)
self.vars = init_var_map(init_vars, init_path)
w0 = [self.vars['embed_%d' % i] for i in range(num_inputs)]
xw = tf.concat([tf.sparse_tensor_dense_matmul(self.X[i], w0[i]) for i in range(num_inputs)], 1)
l = xw
l = tf.transpose(tf.reshape(l, [-1, num_inputs, embed_size, 1]), [0, 2, 1, 3])
f1 = self.vars['f1']
l = tf.nn.conv2d(l, f1, [1, 1, 1, 1], 'SAME')
l = tf.transpose(
max_pool_4d(
tf.transpose(l, [0, 1, 3, 2]),
int(num_inputs / 2)),
[0, 1, 3, 2])
f2 = self.vars['f2']
l = tf.nn.conv2d(l, f2, [1, 1, 1, 1], 'SAME')
l = tf.transpose(
max_pool_4d(
tf.transpose(l, [0, 1, 3, 2]), 3),
[0, 1, 3, 2])
l = tf.nn.dropout(
activate(
tf.reshape(l, [-1, embed_size * 3 * 2]),
layer_acts[0]),
self.layer_keeps[0])
w1 = self.vars['w1']
b1 = self.vars['b1']
l = tf.matmul(l, w1) + b1
l = tf.squeeze(l)
self.y_prob = tf.sigmoid(l)
self.loss = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=l, labels=self.y))
self.optimizer = get_optimizer(opt_algo, learning_rate, self.loss)
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
self.sess = tf.Session(config=config)
tf.global_variables_initializer().run(session=self.sess)
```
### PNN
reference:<br>
[深度学习在CTR预估中的应用](https://zhuanlan.zhihu.com/p/35484389)
可以视作FNN+product layer
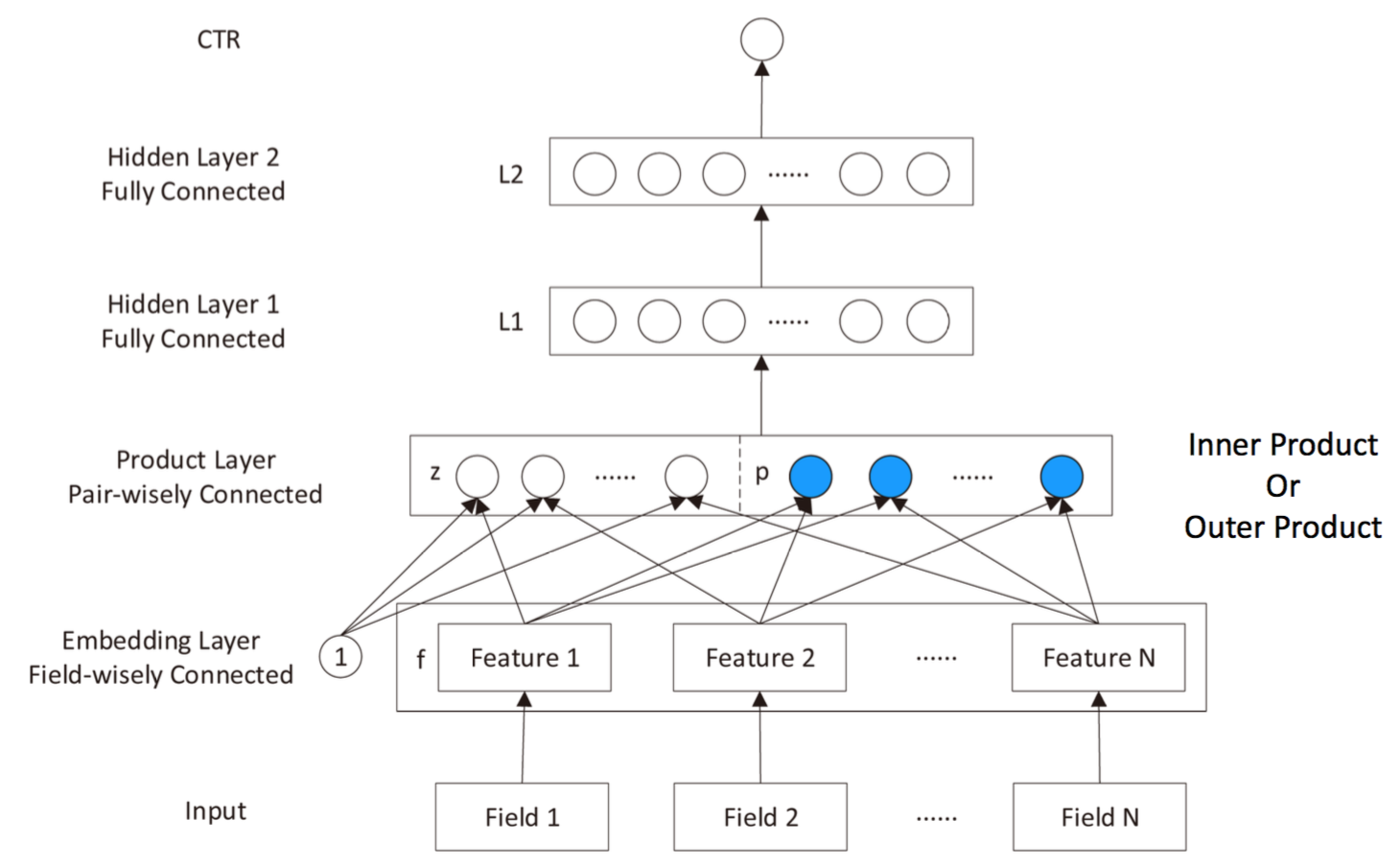
PNN和FNN的主要不同在于除了得到z向量,还增加了一个p向量,即Product向量。Product向量由每个category field的feature vector做inner product 或则 outer product 得到,作者认为这样做有助于特征交叉。另外PNN中Embeding层不再由FM生成,可以在整个网络中训练得到。
对比 FNN 网络,PNN的区别在于中间多了一层 Product Layer 层。Product Layer 层由两部分组成,左边z为 embedding 层的线性部分,右边为 embedding 层的特征交叉部分。
除了 Product Layer 不同,PNN 和 FNN 的 MLP 结构是一样的。这种 product 思想来源于,在 CTR 预估中,认为特征之间的关系更多是一种 and“且”的关系,而非 add"加”的关系。例如,性别为男且喜欢游戏的人群,比起性别男和喜欢游戏的人群,前者的组合比后者更能体现特征交叉的意义。
根据 product 的方式不同,可以分为 inner product (IPNN) 和 outer product (OPNN),如下图所示。
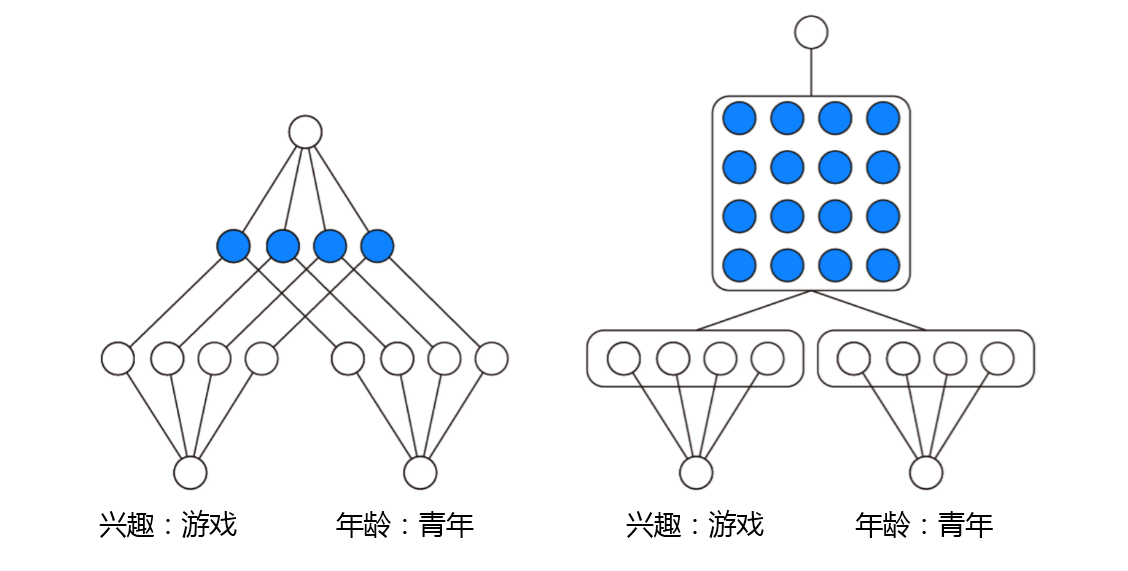
### PNN1
```
class PNN1(Model):
def __init__(self, field_sizes=None, embed_size=10, layer_sizes=None, layer_acts=None, drop_out=None,
embed_l2=None, layer_l2=None, init_path=None, opt_algo='gd', learning_rate=1e-2, random_seed=None):
Model.__init__(self)
init_vars = []
num_inputs = len(field_sizes)
for i in range(num_inputs):
init_vars.append(('embed_%d' % i, [field_sizes[i], embed_size], 'xavier', dtype))
num_pairs = int(num_inputs * (num_inputs - 1) / 2)
node_in = num_inputs * embed_size + num_pairs
# node_in = num_inputs * (embed_size + num_inputs)
for i in range(len(layer_sizes)):
init_vars.append(('w%d' % i, [node_in, layer_sizes[i]], 'xavier', dtype))
init_vars.append(('b%d' % i, [layer_sizes[i]], 'zero', dtype))
node_in = layer_sizes[i]
self.graph = tf.Graph()
with self.graph.as_default():
if random_seed is not None:
tf.set_random_seed(random_seed)
self.X = [tf.sparse_placeholder(dtype) for i in range(num_inputs)]
self.y = tf.placeholder(dtype)
self.keep_prob_train = 1 - np.array(drop_out)
self.keep_prob_test = np.ones_like(drop_out)
self.layer_keeps = tf.placeholder(dtype)
self.vars = init_var_map(init_vars, init_path)
w0 = [self.vars['embed_%d' % i] for i in range(num_inputs)]
xw = tf.concat([tf.sparse_tensor_dense_matmul(self.X[i], w0[i]) for i in range(num_inputs)], 1)
xw3d = tf.reshape(xw, [-1, num_inputs, embed_size])
row = []
col = []
for i in range(num_inputs-1):
for j in range(i+1, num_inputs):
row.append(i)
col.append(j)
# batch * pair * k
p = tf.transpose(
# pair * batch * k
tf.gather(
# num * batch * k
tf.transpose(
xw3d, [1, 0, 2]),
row),
[1, 0, 2])
# batch * pair * k
q = tf.transpose(
tf.gather(
tf.transpose(
xw3d, [1, 0, 2]),
col),
[1, 0, 2])
p = tf.reshape(p, [-1, num_pairs, embed_size])
q = tf.reshape(q, [-1, num_pairs, embed_size])
ip = tf.reshape(tf.reduce_sum(p * q, [-1]), [-1, num_pairs])
# simple but redundant
# batch * n * 1 * k, batch * 1 * n * k
# ip = tf.reshape(
# tf.reduce_sum(
# tf.expand_dims(xw3d, 2) *
# tf.expand_dims(xw3d, 1),
# 3),
# [-1, num_inputs**2])
l = tf.concat([xw, ip], 1)
for i in range(len(layer_sizes)):
wi = self.vars['w%d' % i]
bi = self.vars['b%d' % i]
l = tf.nn.dropout(
activate(
tf.matmul(l, wi) + bi,
layer_acts[i]),
self.layer_keeps[i])
l = tf.squeeze(l)
self.y_prob = tf.sigmoid(l)
self.loss = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=l, labels=self.y))
if layer_l2 is not None:
self.loss += embed_l2 * tf.nn.l2_loss(xw)
for i in range(len(layer_sizes)):
wi = self.vars['w%d' % i]
self.loss += layer_l2[i] * tf.nn.l2_loss(wi)
self.optimizer = get_optimizer(opt_algo, learning_rate, self.loss)
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
self.sess = tf.Session(config=config)
tf.global_variables_initializer().run(session=self.sess)
```
### PNN2
```
class PNN2(Model):
def __init__(self, field_sizes=None, embed_size=10, layer_sizes=None, layer_acts=None, drop_out=None,
embed_l2=None, layer_l2=None, init_path=None, opt_algo='gd', learning_rate=1e-2, random_seed=None,
layer_norm=True):
Model.__init__(self)
init_vars = []
num_inputs = len(field_sizes)
for i in range(num_inputs):
init_vars.append(('embed_%d' % i, [field_sizes[i], embed_size], 'xavier', dtype))
num_pairs = int(num_inputs * (num_inputs - 1) / 2)
node_in = num_inputs * embed_size + num_pairs
init_vars.append(('kernel', [embed_size, num_pairs, embed_size], 'xavier', dtype))
for i in range(len(layer_sizes)):
init_vars.append(('w%d' % i, [node_in, layer_sizes[i]], 'xavier', dtype))
init_vars.append(('b%d' % i, [layer_sizes[i]], 'zero', dtype))
node_in = layer_sizes[i]
self.graph = tf.Graph()
with self.graph.as_default():
if random_seed is not None:
tf.set_random_seed(random_seed)
self.X = [tf.sparse_placeholder(dtype) for i in range(num_inputs)]
self.y = tf.placeholder(dtype)
self.keep_prob_train = 1 - np.array(drop_out)
self.keep_prob_test = np.ones_like(drop_out)
self.layer_keeps = tf.placeholder(dtype)
self.vars = init_var_map(init_vars, init_path)
w0 = [self.vars['embed_%d' % i] for i in range(num_inputs)]
xw = tf.concat([tf.sparse_tensor_dense_matmul(self.X[i], w0[i]) for i in range(num_inputs)], 1)
xw3d = tf.reshape(xw, [-1, num_inputs, embed_size])
row = []
col = []
for i in range(num_inputs - 1):
for j in range(i + 1, num_inputs):
row.append(i)
col.append(j)
# batch * pair * k
p = tf.transpose(
# pair * batch * k
tf.gather(
# num * batch * k
tf.transpose(
xw3d, [1, 0, 2]),
row),
[1, 0, 2])
# batch * pair * k
q = tf.transpose(
tf.gather(
tf.transpose(
xw3d, [1, 0, 2]),
col),
[1, 0, 2])
# b * p * k
p = tf.reshape(p, [-1, num_pairs, embed_size])
# b * p * k
q = tf.reshape(q, [-1, num_pairs, embed_size])
# k * p * k
k = self.vars['kernel']
# batch * 1 * pair * k
p = tf.expand_dims(p, 1)
# batch * pair
kp = tf.reduce_sum(
# batch * pair * k
tf.multiply(
# batch * pair * k
tf.transpose(
# batch * k * pair
tf.reduce_sum(
# batch * k * pair * k
tf.multiply(
p, k),
-1),
[0, 2, 1]),
q),
-1)
#
# if layer_norm:
# # x_mean, x_var = tf.nn.moments(xw, [1], keep_dims=True)
# # xw = (xw - x_mean) / tf.sqrt(x_var)
# # x_g = tf.Variable(tf.ones([num_inputs * embed_size]), name='x_g')
# # x_b = tf.Variable(tf.zeros([num_inputs * embed_size]), name='x_b')
# # x_g = tf.Print(x_g, [x_g[:10], x_b])
# # xw = xw * x_g + x_b
# p_mean, p_var = tf.nn.moments(op, [1], keep_dims=True)
# op = (op - p_mean) / tf.sqrt(p_var)
# p_g = tf.Variable(tf.ones([embed_size**2]), name='p_g')
# p_b = tf.Variable(tf.zeros([embed_size**2]), name='p_b')
# # p_g = tf.Print(p_g, [p_g[:10], p_b])
# op = op * p_g + p_b
l = tf.concat([xw, kp], 1)
for i in range(len(layer_sizes)):
wi = self.vars['w%d' % i]
bi = self.vars['b%d' % i]
l = tf.nn.dropout(
activate(
tf.matmul(l, wi) + bi,
layer_acts[i]),
self.layer_keeps[i])
l = tf.squeeze(l)
self.y_prob = tf.sigmoid(l)
self.loss = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=l, labels=self.y))
if layer_l2 is not None:
self.loss += embed_l2 * tf.nn.l2_loss(xw)#tf.concat(w0, 0))
for i in range(len(layer_sizes)):
wi = self.vars['w%d' % i]
self.loss += layer_l2[i] * tf.nn.l2_loss(wi)
self.optimizer = get_optimizer(opt_algo, learning_rate, self.loss)
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
self.sess = tf.Session(config=config)
tf.global_variables_initializer().run(session=self.sess)
```
| github_jupyter |
```
import pandas as pd
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
from os import listdir
import seaborn as sns
sns.set_style("white")
from keras.preprocessing import sequence
import tensorflow as tf
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import LSTM
from keras.layers import Flatten
from keras.layers import Dropout
from keras.callbacks import EarlyStopping
from keras import optimizers
from keras.regularizers import l1,l2,l1_l2
from keras.optimizers import Adam
from keras.models import load_model
from keras.callbacks import ModelCheckpoint
from keras.models import model_from_json
df_test = pd.read_csv('df_test.csv')
df_test
df_list = []
for i in range(18):
df_split = df_test[df_test['target'] == i]
df_split.reset_index(drop=True, inplace=True)
df_list.append(df_split)
myorder = [0, 1, 2, 5, 4, 3, 6, 7, 8, 11, 10, 9, 12, 13, 14, 17, 16, 15]
test_list = [df_list[i] for i in myorder]
test_list[3]
df_test = pd.concat(test_list)
df_med = df_test.drop(df_test.iloc[:, :36], axis = 1)
df_med.drop(df_med.iloc[:, 72:108], inplace = True, axis = 1)
df_small = df_test.drop(df_test.iloc[:, :54], axis = 1)
df_small.drop(df_small.iloc[:, 36:90], inplace = True, axis = 1)
df_smaller = df_test.drop(df_test.iloc[:, :64], axis = 1)
df_smaller.drop(df_smaller.iloc[:, 18:80], inplace = True, axis = 1)
df_smooth = df_test.T
df_med = df_med.T
df_small = df_small.T
df_smaller = df_smaller.T
sequences_smooth = list()
for i in range(df_smooth.shape[1]):
values = df_smooth.iloc[:-1,i].values
sequences_smooth.append(values)
targets_smooth = df_smooth.iloc[-1, :].values
sequences_med = list()
for i in range(df_med.shape[1]):
values = df_med.iloc[:-1,i].values
sequences_med.append(values)
targets_med = df_med.iloc[-1, :].values
sequences_small = list()
for i in range(df_small.shape[1]):
values = df_small.iloc[:-1,i].values
sequences_small.append(values)
targets_small = df_small.iloc[-1, :].values
sequences_smaller = list()
for i in range(df_smaller.shape[1]):
values = df_smaller.iloc[:-1,i].values
sequences_smaller.append(values)
targets_smaller = df_smaller.iloc[-1, :].values
targets = targets_smooth
targets_smooth
from sklearn.preprocessing import LabelEncoder, OneHotEncoder
from keras.utils import np_utils
# encode class values as integers
encoder = LabelEncoder()
encoder.fit(targets)
encoded_y = encoder.transform(targets)
# convert integers to dummy variables (i.e. one hot encoded)
dummy_y = np_utils.to_categorical(encoded_y)
targets = dummy_y
X_test_smooth, X_test_med, X_test_small, X_test_smaller, y_test = sequences_smooth, sequences_med, sequences_small, sequences_smaller, targets
# Feature Scaling
from sklearn.preprocessing import StandardScaler, MinMaxScaler
from sklearn.externals.joblib import dump, load
sc1 = load('std_scaler_smooth.bin')
X_test_smooth = sc1.transform(X_test_smooth)
sc2 = load('std_scaler_med.bin')
X_test_med = sc2.transform(X_test_med)
sc3 = load('std_scaler_small.bin')
X_test_small = sc3.transform(X_test_small)
sc4 = load('std_scaler_smaller.bin')
X_test_smaller = sc4.transform(X_test_smaller)
X_test_smooth.shape, X_test_med.shape, X_test_small.shape, X_test_smaller.shape
X_test_smooth = np.reshape(X_test_smooth, (X_test_smooth.shape[0], X_test_smooth.shape[1], 1))
X_test_med = np.reshape(X_test_med, (X_test_med.shape[0], X_test_med.shape[1], 1))
X_test_small = np.reshape(X_test_small, (X_test_small.shape[0], X_test_small.shape[1], 1))
X_test_smaller = np.reshape(X_test_smaller, (X_test_smaller.shape[0], X_test_smaller.shape[1], 1))
y_test.argmax(axis=1)
class Model:
def __init__(self, path_model, path_weight):
self.model = self.loadmodel(path_model, path_weight)
self.graph = tf.get_default_graph()
@staticmethod
def loadmodel(path_model, path_weight):
json_file = open(path_model, 'r')
loaded_model_json = json_file.read()
json_file.close()
model = model_from_json(loaded_model_json)
model.load_weights(path_weight)
return model
def predict(self, X):
with self.graph.as_default():
return self.model.predict(X)
work_dir_model = '/home/hongyu/Documents/Spring2020/ECE_research/signal_analysis/data_18points/3_section_sliding5/MSLSTM_models/'
work_dir_weight = '/home/hongyu/Documents/Spring2020/ECE_research/signal_analysis/data_18points/3_section_sliding5/MSLSTM_weights/'
# work_dir_model = '/home/wuh007/Desktop/signal/signal_analysis/data_18points/3_section/models_mixed/'
# work_dir_weight = '/home/wuh007/Desktop/signal/signal_analysis/data_18points/3_section/weights_mixed/'
# model_2sec = Model(work_dir_model + 'MSLSTM_wholetotwo_model.json', work_dir_weight + 'model-015-0.976846-0.930156-0.194448-wtt.h5')
model_2sec = Model(work_dir_model + 'MSLSTM_18ptswithdense_model.json', work_dir_weight + 'model-012-0.994497-0.930156-0.256649-18ptswithdense.h5')
# model_TopToNine = Model(work_dir_model + 'MSLSTM_toptonine_model.json', work_dir_weight + 'MSLSTM_toptonine_weight.h5')
# model_MiddleToSix = Model(work_dir_model + 'LSTM_MiddleToSix_model.json', work_dir_weight + 'LSTM_MiddleToSix_weight.h5')
# model_ButtomToNine = Model(work_dir_model + 'MSLSTM_buttomtonine_model.json', work_dir_weight + 'MSLSTM_buttomtonine_weight.h5')
# model_TopTopToThree = Model(work_dir_model + 'LSTM_TopTopToThree_model.json', work_dir_weight + 'LSTM_TopTopToThree_weight.h5')
# model_TopButtomToThree = Model(work_dir_model + 'LSTM_TopButtomToThree_model.json', work_dir_weight + 'LSTM_TopButtomToThree_weight.h5')
# model_MiddleTopToThree = Model(work_dir_model + 'LSTM_MiddleTopToThree_model.json', work_dir_weight + 'LSTM_MiddleTopToThree_weight.h5')
# model_MiddleButtomToThree = Model(work_dir_model + 'LSTM_MiddleButtomToThree_model.json', work_dir_weight + 'LSTM_MiddleButtomToThree_weight.h5')
# model_ButtomTopToThree = Model(work_dir_model + 'LSTM_ButtomTopToThree_model.json', work_dir_weight + 'LSTM_ButtomTopToThree_weight.h5')
# model_ButtomButtomToThree = Model(work_dir_model + 'LSTM_ButtomButtomToThree_model.json', work_dir_weight + 'LSTM_ButtomButtomToThree_weight.h5')
y_pred_2sec = model_2sec.predict([X_test_small, X_test_med, X_test_smaller, X_test_smooth])
# y_pred_TopToNine = model_TopToNine.predict([X_test_small, X_test_med, X_test_smaller, X_test_smooth][(y_pred_2sec.max(axis=1) > 0.00) & (y_pred_2sec.argmax(axis=1) == 0)])
# y_pred_MiddleToSix = model_MiddleToSix.predict(X_test[(y_pred_3sec.max(axis=1) > 0.85) & (y_pred_3sec.argmax(axis=1) == 1)])
# y_pred_ButtomToNine = model_ButtomToNine.predict([X_test_small, X_test_med, X_test_smaller, X_test_smooth][(y_pred_2sec.max(axis=1) > 0.00) & (y_pred_2sec.argmax(axis=1) == 1)])
# y_pred_TopTopToThree = model_TopTopToThree.predict(X_test[(y_pred_3sec.max(axis=1) > 0.85) & (y_pred_3sec.argmax(axis=1) == 0)][(y_pred_TopToTwo.max(axis=1) > 0.85) & (y_pred_TopToTwo.argmax(axis=1) == 0)])
# y_pred_TopButtomToThree = model_TopButtomToThree.predict(X_test[(y_pred_3sec.max(axis=1) > 0.85) & (y_pred_3sec.argmax(axis=1) == 0)][(y_pred_TopToTwo.max(axis=1) > 0.85) & (y_pred_TopToTwo.argmax(axis=1) == 1)])
# y_pred_MiddleTopToThree = model_MiddleTopToThree.predict(X_test[(y_pred_3sec.max(axis=1) > 0.85) & (y_pred_3sec.argmax(axis=1) == 1)][(y_pred_MiddleToTwo.max(axis=1) > 0.85) & (y_pred_MiddleToTwo.argmax(axis=1) == 0)])
# y_pred_MiddleButtomToThree = model_MiddleButtomToThree.predict(X_test[(y_pred_3sec.max(axis=1) > 0.85) & (y_pred_3sec.argmax(axis=1) == 1)][(y_pred_MiddleToTwo.max(axis=1) > 0.85) & (y_pred_MiddleToTwo.argmax(axis=1) == 1)])
# y_pred_ButtomTopToThree = model_ButtomTopToThree.predict(X_test[(y_pred_3sec.max(axis=1) > 0.85) & (y_pred_3sec.argmax(axis=1) == 2)][(y_pred_ButtomToTwo.max(axis=1) > 0.85) & (y_pred_ButtomToTwo.argmax(axis=1) == 0)])
# y_pred_ButtomButtomToThree = model_ButtomButtomToThree.predict(X_test[(y_pred_3sec.max(axis=1) > 0.85) & (y_pred_3sec.argmax(axis=1) == 2)][(y_pred_ButtomToTwo.max(axis=1) > 0.85) & (y_pred_ButtomToTwo.argmax(axis=1) == 1)])
# len(y_pred_TopToNine), len(y_pred_ButtomToNine)
# len(y_pred_TopToNine) + len(y_pred_ButtomToNine)
len(y_pred_2sec), len(y_test)
# y_pred_TopToNine.argmax(axis=1)
# y_test[(y_pred_2sec.max(axis=1) > 0.00) & (y_pred_2sec.argmax(axis=1) == 0)].argmax(axis=1)
# y_pred_ButtomToNine.argmax(axis=1)
# y_test[(y_pred_2sec.max(axis=1) > 0.00) & (y_pred_2sec.argmax(axis=1) == 2)].argmax(axis=1)
# y_pred_BTN = y_pred_ButtomToNine.argmax(axis=1)
# y_pred_BTN[y_pred_BTN == 0] = 9
# y_pred_BTN[y_pred_BTN == 1] = 10
# y_pred_BTN[y_pred_BTN == 2] = 11
# y_pred_BTN[y_pred_BTN == 3] = 12
# y_pred_BTN[y_pred_BTN == 4] = 13
# y_pred_BTN[y_pred_BTN == 5] = 14
# y_pred_BTN[y_pred_BTN == 6] = 15
# y_pred_BTN[y_pred_BTN == 7] = 16
# y_pred_BTN[y_pred_BTN == 8] = 17
# y_pred_BTN
# y_pred_final = np.concatenate((y_pred_TopToNine.argmax(axis=1), y_pred_BTN), axis=0)
# y_pred_final, len(y_pred_final)
# y_test_final = np.concatenate((y_test[(y_pred_3sec.max(axis=1) > 0.00) & (y_pred_3sec.argmax(axis=1) == 0)].argmax(axis=1),
# y_test[(y_pred_3sec.max(axis=1) > 0.00) & (y_pred_3sec.argmax(axis=1) == 1)].argmax(axis=1),
# axis=0)
y_pred_2sec
y_pred_2sec.argmax(axis=1)
y_test.argmax(axis=1)
len(y_test.argmax(axis=1))
# len(set([1,1,1]))
column_map = dict()
row_map = dict()
y_test_new = list()
for j in range(len(y_test.argmax(axis=1)) - 2):
window = [y_test.argmax(axis=1)[j],
y_test.argmax(axis=1)[j+1],
y_test.argmax(axis=1)[j+2]]
# print(window)
# print(len(set(window)))
if len(set(window)) == 1 or 2:
for item in set(window):
if window.count(item) > 1:
y_test_new.append(item)
# print(y_test_new)
else:
print('windows', window)
y_test_new.append(18)
y_test_new = list()
for j in range(len(y_test[y_pred_2sec.max(axis=1) > 0.70].argmax(axis=1)) - 2):
window = [y_test[y_pred_2sec.max(axis=1) > 0.70].argmax(axis=1)[j],
y_test[y_pred_2sec.max(axis=1) > 0.70].argmax(axis=1)[j+1],
y_test[y_pred_2sec.max(axis=1) > 0.70].argmax(axis=1)[j+2]]
# print(window)
# print(len(set(window)))
if len(set(window)) == 1 or 2:
for item in set(window):
if window.count(item) > 1:
y_test_new.append(item)
# print(y_test_new)
else:
print('windows', window)
y_test_new.append(18)
# print(j)
len(y_test_new)
y_pred_new = list()
for j in range(len(y_pred_2sec.argmax(axis=1)) - 2):
window = [y_pred_2sec.argmax(axis=1)[j],
y_pred_2sec.argmax(axis=1)[j+1],
y_pred_2sec.argmax(axis=1)[j+2]]
print('windows', window)
# print('unique elements', len(set(window)))
if len(set(window)) == 1 or len(set(window)) == 2:
for item in set(window):
if window.count(item) > 1:
y_pred_new.append(item)
# print(y_pred_new)
else:
print('unique window', window)
y_pred
print(y_test_new[j])
y_pred_new.append(18)
# print('special', j)
# print(j)
y_pred_new = list()
for j in range(len(y_pred_2sec[y_pred_2sec.max(axis=1) > 0.70].argmax(axis=1)) - 2):
window = [y_pred_2sec[y_pred_2sec.max(axis=1) > 0.70].argmax(axis=1)[j],
y_pred_2sec[y_pred_2sec.max(axis=1) > 0.70].argmax(axis=1)[j+1],
y_pred_2sec[y_pred_2sec.max(axis=1) > 0.70].argmax(axis=1)[j+2]]
# print('windows', window)
# print('unique elements', len(set(window)))
if len(set(window)) == 1 or len(set(window)) == 2:
for item in set(window):
if window.count(item) > 1:
y_pred_new.append(item)
# print(y_pred_new)
else:
print('windows', window)
y_pred_new.append(18)
# print('special', j)
# print(j)
len(y_pred_new)
count = 0
for i in range(len(y_pred_new)):
if y_pred_new[i] == 18:
count += 1
count
count = 0
for i in range(len(y_pred_new)):
if y_pred_new[i] == 18:
count += 1
count
y_test_new = np.asarray(y_test_new)
y_pred_new = np.asarray(y_pred_new)
# Creating the Confusion Matrix
from sklearn.metrics import confusion_matrix
matrix = confusion_matrix(y_test_new, y_pred_new)
# matrix = confusion_matrix(y_test[y_pred_2sec.max(axis=1) > 0.80].argmax(axis=1), y_pred_2sec[y_pred_2sec.max(axis=1) > 0.80].argmax(axis=1))
matrix
plot_confusion_matrix(matrix, [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17])
plot_confusion_matrix(matrix, [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17])
# Creating the Confusion Matrix
from sklearn.metrics import confusion_matrix
# matrix = confusion_matrix(y_pred_2sec.argmax(axis=1), y_test.argmax(axis=1))
matrix = confusion_matrix(y_test[y_pred_2sec.max(axis=1) > 0.70].argmax(axis=1), y_pred_2sec[y_pred_2sec.max(axis=1) > 0.70].argmax(axis=1))
matrix
len(y_test), len(y_test[y_pred_2sec.max(axis=1) > 0.70])
def plot_confusion_matrix(cm,
target_names,
title='Confusion matrix',
cmap=None,
normalize=True):
"""
given a sklearn confusion matrix (cm), make a nice plot
Arguments
---------
cm: confusion matrix from sklearn.metrics.confusion_matrix
target_names: given classification classes such as [0, 1, 2]
the class names, for example: ['high', 'medium', 'low']
title: the text to display at the top of the matrix
cmap: the gradient of the values displayed from matplotlib.pyplot.cm
see http://matplotlib.org/examples/color/colormaps_reference.html
plt.get_cmap('jet') or plt.cm.Blues
normalize: If False, plot the raw numbers
If True, plot the proportions
Usage|
-----
plot_confusion_matrix(cm = cm, # confusion matrix created by
# sklearn.metrics.confusion_matrix
normalize = True, # show proportions
target_names = y_labels_vals, # list of names of the classes
title = best_estimator_name) # title of graph
Citiation
---------
http://scikit-learn.org/stable/auto_examples/model_selection/plot_confusion_matrix.html
"""
import matplotlib.pyplot as plt
import numpy as np
import itertools
accuracy = np.trace(cm) / np.sum(cm).astype('float')
misclass = 1 - accuracy
if cmap is None:
cmap = plt.get_cmap('Blues')
plt.figure(figsize=(16, 14))
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
if target_names is not None:
tick_marks = np.arange(len(target_names))
plt.xticks(tick_marks, target_names, rotation=45)
plt.yticks(tick_marks, target_names)
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
thresh = cm.max() / 1.5 if normalize else cm.max() / 2
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
if normalize:
plt.text(j, i, "{:0.4f}".format(cm[i, j]),
horizontalalignment="center",
color="red" if cm[i, j] > thresh else "black")
else:
plt.text(j, i, "{:,}".format(cm[i, j]),
horizontalalignment="center",
color="red" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label\naccuracy={:0.4f}; misclass={:0.4f}'.format(accuracy, misclass))
plt.show()
plot_confusion_matrix(matrix, [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17])
plot_confusion_matrix(matrix, [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17])
tl = np.sum(matrix[:9, :9])
tr = np.sum(matrix[:9, 9:])
bl = np.sum(matrix[9:, :9])
br = np.sum(matrix[9:, 9:])
(tl+br)/(tl+tr+bl+br)
matrix[:9, :9]
```
| github_jupyter |
```
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from keras import initializers
import keras.backend as K
import numpy as np
import pandas as pd
from tensorflow.keras.layers import *
from keras.regularizers import l2#正则化
# 12-0.2
# 13-2.4
# 18-12.14
import pandas as pd
import numpy as np
normal = np.loadtxt(r'E:\水泵代码调试\试验数据(包括压力脉动和振动)\2013.9.12-未发生缠绕前\2013-9.12振动\2013-9-12振动-1250rmin-mat\1250rnormalvibx.txt', delimiter=',')
chanrao = np.loadtxt(r'E:\水泵代码调试\试验数据(包括压力脉动和振动)\2013.9.17-发生缠绕后\振动\9-18上午振动1250rmin-mat\1250r_chanraovibx.txt', delimiter=',')
print(normal.shape,chanrao.shape,"***************************************************")
data_normal=normal[6:8] #提取前两行
data_chanrao=chanrao[6:8] #提取前两行
print(data_normal.shape,data_chanrao.shape)
print(data_normal,"\r\n",data_chanrao,"***************************************************")
data_normal=data_normal.reshape(1,-1)
data_chanrao=data_chanrao.reshape(1,-1)
print(data_normal.shape,data_chanrao.shape)
print(data_normal,"\r\n",data_chanrao,"***************************************************")
#水泵的两种故障类型信号normal正常,chanrao故障
data_normal=data_normal.reshape(-1, 512)#(65536,1)-(128, 515)
data_chanrao=data_chanrao.reshape(-1,512)
print(data_normal.shape,data_chanrao.shape)
import numpy as np
def yuchuli(data,label):#(4:1)(51:13)
#打乱数据顺序
np.random.shuffle(data)
train = data[0:102,:]
test = data[102:128,:]
label_train = np.array([label for i in range(0,102)])
label_test =np.array([label for i in range(0,26)])
return train,test ,label_train ,label_test
def stackkk(a,b,c,d,e,f,g,h):
aa = np.vstack((a, e))
bb = np.vstack((b, f))
cc = np.hstack((c, g))
dd = np.hstack((d, h))
return aa,bb,cc,dd
x_tra0,x_tes0,y_tra0,y_tes0 = yuchuli(data_normal,0)
x_tra1,x_tes1,y_tra1,y_tes1 = yuchuli(data_chanrao,1)
tr1,te1,yr1,ye1=stackkk(x_tra0,x_tes0,y_tra0,y_tes0 ,x_tra1,x_tes1,y_tra1,y_tes1)
x_train=tr1
x_test=te1
y_train = yr1
y_test = ye1
#打乱数据
state = np.random.get_state()
np.random.shuffle(x_train)
np.random.set_state(state)
np.random.shuffle(y_train)
state = np.random.get_state()
np.random.shuffle(x_test)
np.random.set_state(state)
np.random.shuffle(y_test)
#对训练集和测试集标准化
def ZscoreNormalization(x):
"""Z-score normaliaztion"""
x = (x - np.mean(x)) / np.std(x)
return x
x_train=ZscoreNormalization(x_train)
x_test=ZscoreNormalization(x_test)
# print(x_test[0])
#转化为一维序列
x_train = x_train.reshape(-1,512,1)
x_test = x_test.reshape(-1,512,1)
print(x_train.shape,x_test.shape)
def to_one_hot(labels,dimension=2):
results = np.zeros((len(labels),dimension))
for i,label in enumerate(labels):
results[i,label] = 1
return results
one_hot_train_labels = to_one_hot(y_train)
one_hot_test_labels = to_one_hot(y_test)
#定义挤压函数
def squash(vectors, axis=-1):
"""
对向量的非线性激活函数
## vectors: some vectors to be squashed, N-dim tensor
## axis: the axis to squash
:return: a Tensor with same shape as input vectors
"""
s_squared_norm = K.sum(K.square(vectors), axis, keepdims=True)
scale = s_squared_norm / (1 + s_squared_norm) / K.sqrt(s_squared_norm + K.epsilon())
return scale * vectors
class Length(layers.Layer):
"""
计算向量的长度。它用于计算与margin_loss中的y_true具有相同形状的张量
Compute the length of vectors. This is used to compute a Tensor that has the same shape with y_true in margin_loss
inputs: shape=[dim_1, ..., dim_{n-1}, dim_n]
output: shape=[dim_1, ..., dim_{n-1}]
"""
def call(self, inputs, **kwargs):
return K.sqrt(K.sum(K.square(inputs), -1))
def compute_output_shape(self, input_shape):
return input_shape[:-1]
def get_config(self):
config = super(Length, self).get_config()
return config
#定义预胶囊层
def PrimaryCap(inputs, dim_capsule, n_channels, kernel_size, strides, padding):
"""
进行普通二维卷积 `n_channels` 次, 然后将所有的胶囊重叠起来
:param inputs: 4D tensor, shape=[None, width, height, channels]
:param dim_capsule: the dim of the output vector of capsule
:param n_channels: the number of types of capsules
:return: output tensor, shape=[None, num_capsule, dim_capsule]
"""
output = layers.Conv2D(filters=dim_capsule*n_channels, kernel_size=kernel_size, strides=strides,
padding=padding,name='primarycap_conv2d')(inputs)
outputs = layers.Reshape(target_shape=[-1, dim_capsule], name='primarycap_reshape')(output)
return layers.Lambda(squash, name='primarycap_squash')(outputs)
class DenseCapsule(layers.Layer):
"""
胶囊层. 输入输出都为向量.
## num_capsule: 本层包含的胶囊数量
## dim_capsule: 输出的每一个胶囊向量的维度
## routings: routing 算法的迭代次数
"""
def __init__(self, num_capsule, dim_capsule, routings=3, kernel_initializer='glorot_uniform',**kwargs):
super(DenseCapsule, self).__init__(**kwargs)
self.num_capsule = num_capsule
self.dim_capsule = dim_capsule
self.routings = routings
self.kernel_initializer = kernel_initializer
def build(self, input_shape):
assert len(input_shape) >= 3, '输入的 Tensor 的形状[None, input_num_capsule, input_dim_capsule]'#(None,1152,8)
self.input_num_capsule = input_shape[1]
self.input_dim_capsule = input_shape[2]
#转换矩阵
self.W = self.add_weight(shape=[self.num_capsule, self.input_num_capsule,
self.dim_capsule, self.input_dim_capsule],
initializer=self.kernel_initializer,name='W')
self.built = True
def call(self, inputs, training=None):
# inputs.shape=[None, input_num_capsuie, input_dim_capsule]
# inputs_expand.shape=[None, 1, input_num_capsule, input_dim_capsule]
inputs_expand = K.expand_dims(inputs, 1)
# 运算优化:将inputs_expand重复num_capsule 次,用于快速和W相乘
# inputs_tiled.shape=[None, num_capsule, input_num_capsule, input_dim_capsule]
inputs_tiled = K.tile(inputs_expand, [1, self.num_capsule, 1, 1])
# 将inputs_tiled的batch中的每一条数据,计算inputs+W
# x.shape = [num_capsule, input_num_capsule, input_dim_capsule]
# W.shape = [num_capsule, input_num_capsule, dim_capsule, input_dim_capsule]
# 将x和W的前两个维度看作'batch'维度,向量和矩阵相乘:
# [input_dim_capsule] x [dim_capsule, input_dim_capsule]^T -> [dim_capsule].
# inputs_hat.shape = [None, num_capsule, input_num_capsule, dim_capsutel
inputs_hat = K.map_fn(lambda x: K.batch_dot(x, self.W, [2, 3]),elems=inputs_tiled)
# Begin: Routing算法
# 将系数b初始化为0.
# b.shape = [None, self.num_capsule, self, input_num_capsule].
b = tf.zeros(shape=[K.shape(inputs_hat)[0], self.num_capsule, self.input_num_capsule])
assert self.routings > 0, 'The routings should be > 0.'
for i in range(self.routings):
# c.shape=[None, num_capsule, input_num_capsule]
C = tf.nn.softmax(b ,axis=1)
# c.shape = [None, num_capsule, input_num_capsule]
# inputs_hat.shape = [None, num_capsule, input_num_capsule, dim_capsule]
# 将c与inputs_hat的前两个维度看作'batch'维度,向量和矩阵相乘:
# [input_num_capsule] x [input_num_capsule, dim_capsule] -> [dim_capsule],
# outputs.shape= [None, num_capsule, dim_capsule]
outputs = squash(K. batch_dot(C, inputs_hat, [2, 2])) # [None, 10, 16]
if i < self.routings - 1:
# outputs.shape = [None, num_capsule, dim_capsule]
# inputs_hat.shape = [None, num_capsule, input_num_capsule, dim_capsule]
# 将outputs和inρuts_hat的前两个维度看作‘batch’ 维度,向量和矩阵相乘:
# [dim_capsule] x [imput_num_capsule, dim_capsule]^T -> [input_num_capsule]
# b.shape = [batch_size. num_capsule, input_nom_capsule]
# b += K.batch_dot(outputs, inputs_hat, [2, 3]) to this b += tf.matmul(self.W, x)
b += K.batch_dot(outputs, inputs_hat, [2, 3])
# End: Routing 算法
return outputs
def compute_output_shape(self, input_shape):
return tuple([None, self.num_capsule, self.dim_capsule])
def get_config(self):
config = {
'num_capsule': self.num_capsule,
'dim_capsule': self.dim_capsule,
'routings': self.routings
}
base_config = super(DenseCapsule, self).get_config()
return dict(list(base_config.items()) + list(config.items()))
from tensorflow import keras
from keras.regularizers import l2#正则化
x = layers.Input(shape=[512,1, 1])
#普通卷积层
conv1 = layers.Conv2D(filters=16, kernel_size=(2, 1),activation='relu',padding='valid',name='conv1')(x)
#池化层
POOL1 = MaxPooling2D((2,1))(conv1)
#普通卷积层
conv2 = layers.Conv2D(filters=32, kernel_size=(2, 1),activation='relu',padding='valid',name='conv2')(POOL1)
#池化层
# POOL2 = MaxPooling2D((2,1))(conv2)
#Dropout层
Dropout=layers.Dropout(0.1)(conv2)
# Layer 3: 使用“squash”激活的Conv2D层, 然后重塑 [None, num_capsule, dim_vector]
primarycaps = PrimaryCap(Dropout, dim_capsule=8, n_channels=12, kernel_size=(4, 1), strides=2, padding='valid')
# Layer 4: 数字胶囊层,动态路由算法在这里工作。
digitcaps = DenseCapsule(num_capsule=2, dim_capsule=16, routings=3, name='digit_caps')(primarycaps)
# Layer 5:这是一个辅助层,用它的长度代替每个胶囊。只是为了符合标签的形状。
out_caps = Length(name='out_caps')(digitcaps)
model = keras.Model(x, out_caps)
model.summary()
#定义优化
model.compile(loss='categorical_crossentropy',
optimizer='adam',metrics=['accuracy'])
import time
time_begin = time.time()
history = model.fit(x_train,one_hot_train_labels,
validation_split=0.1,
epochs=50,batch_size=10,
shuffle=True)
time_end = time.time()
time = time_end - time_begin
print('time:', time)
import time
time_begin = time.time()
score = model.evaluate(x_test,one_hot_test_labels, verbose=0)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
time_end = time.time()
time = time_end - time_begin
print('time:', time)
#绘制acc-loss曲线
import matplotlib.pyplot as plt
plt.plot(history.history['loss'],color='r')
plt.plot(history.history['val_loss'],color='g')
plt.plot(history.history['accuracy'],color='b')
plt.plot(history.history['val_accuracy'],color='k')
plt.title('model loss and acc')
plt.ylabel('Accuracy')
plt.xlabel('epoch')
plt.legend(['train_loss', 'test_loss','train_acc', 'test_acc'], loc='center right')
# plt.legend(['train_loss','train_acc'], loc='upper left')
#plt.savefig('1.png')
plt.show()
import matplotlib.pyplot as plt
plt.plot(history.history['loss'],color='r')
plt.plot(history.history['accuracy'],color='b')
plt.title('model loss and sccuracy ')
plt.ylabel('loss/sccuracy')
plt.xlabel('epoch')
plt.legend(['train_loss', 'train_sccuracy'], loc='center right')
plt.show()
```
| github_jupyter |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(color_codes=True)
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import os
destdir = '/Users/argha/Dropbox/CS/DatSci/nyc-data'
files = [ f for f in os.listdir(destdir) if os.path.isfile(os.path.join(destdir,f)) ]
files
#df2014 = pd.read_csv('/Users/argha/Dropbox/CS/DatSci/nyc-data/Parking_Violations_Issued_-_Fiscal_Year_2014.csv')
#df2015 = pd.read_csv('/Users/argha/Dropbox/CS/DatSci/nyc-data/Parking_Violations_Issued_-_Fiscal_Year_2015.csv')
df2016 = pd.read_csv('/Users/argha/Dropbox/CS/DatSci/nyc-data/Parking_Violations_Issued_-_Fiscal_Year_2016.csv')
#df2017 = pd.read_csv('/Users/argha/Dropbox/CS/DatSci/nyc-data/Parking_Violations_Issued_-_Fiscal_Year_2017.csv')
#df2018 = pd.read_csv('/Users/argha/Dropbox/CS/DatSci/nyc-data/Parking_Violations_Issued_-_Fiscal_Year_2018.csv')
```
## Take a look into the 2016 data
```
df2016.head(n=2)
df2016.shape
```
So in the 2016 dataset there are about 10.6 million entries for parking ticket, and each entry has 51 columns.
Lets take a look at the number of unique values for each column name...
```
d = {'Unique Entry': df2016.nunique(axis = 0),
'Nan Entry': df2016.isnull().any()}
pd.DataFrame(data = d, index = df2016.columns.values)
```
As it turns out, the last 11 columns in this dataset has no entry. So we can ignore those columns, while carrying out any visualization operation in this dataframe.
Also if the entry does not have a **Plate ID** it is very hard to locate those cars. Therefore I am going to drop those rows as well.
```
drop_column = ['No Standing or Stopping Violation', 'Hydrant Violation',
'Double Parking Violation', 'Latitude', 'Longitude',
'Community Board', 'Community Council ', 'Census Tract', 'BIN',
'BBL', 'NTA',
'Street Code1', 'Street Code2', 'Street Code3','Meter Number', 'Violation Post Code',
'Law Section', 'Sub Division', 'House Number', 'Street Name']
df2016.drop(drop_column, axis = 1, inplace = True)
drop_row = ['Plate ID']
df2016.dropna(axis = 0, how = 'any', subset = drop_row, inplace = True)
```
Check if there is anymore rows left without a **Plate ID**.
```
df2016['Plate ID'].isnull().any()
df2016.shape
```
# Create a sample data for visualization
The cleaned dataframe has 10624735 rows and 40 columns.
But this is still a lot of data points. I does not make sense to use all of them to get an idea of distribution of the data points. So for visualization I will use only 0.1% of the whole data. Assmuing that the entries are not sorted I pick my 0.1% data points from the main dataframe at random.
```
mini2016 = df2016.sample(frac = 0.01, replace = False)
mini2016.shape
```
My sample dataset has about 10K data points, which I will use for data visualization. Using the whole dataset is unnecessary and time consuming.
## Barplot of 'Registration State'
```
x_ticks = mini2016['Registration State'].value_counts().index
heights = mini2016['Registration State'].value_counts()
y_pos = np.arange(len(x_ticks))
fig = plt.figure(figsize=(15,14))
# Create horizontal bars
plt.barh(y_pos, heights)
# Create names on the y-axis
plt.yticks(y_pos, x_ticks)
# Show graphic
plt.show()
pd.DataFrame(mini2016['Registration State'].value_counts()/len(mini2016)).nlargest(10, columns = ['Registration State'])
```
You can see from the barplot above: in our sample ~77.67% cars are registered in state : **NY**. After that 9.15% cars are registered in state : **NJ**, followed by **PA**, **CT**, and **FL**.
## How the number of tickets given changes with each month?
```
month = []
for time_stamp in pd.to_datetime(mini2016['Issue Date']):
month.append(time_stamp.month)
m_count = pd.Series(month).value_counts()
plt.figure(figsize=(12,8))
sns.barplot(y=m_count.values, x=m_count.index, alpha=0.6)
plt.title("Number of Parking Ticket Given Each Month", fontsize=16)
plt.xlabel("Month", fontsize=16)
plt.ylabel("No. of cars", fontsize=16)
plt.show();
```
So from the barplot above **March** and **October** has the highest number of tickets!
## How many parking tickets are given for each violation code?
```
violation_code = mini2016['Violation Code'].value_counts()
plt.figure(figsize=(16,8))
f = sns.barplot(y=violation_code.values, x=violation_code.index, alpha=0.6)
#plt.xticks(np.arange(0,101, 10.0))
f.set(xticks=np.arange(0,100, 5.0))
plt.title("Number of Parking Tickets Given for Each Violation Code", fontsize=16)
plt.xlabel("Violation Code [ X5 ]", fontsize=16)
plt.ylabel("No. of cars", fontsize=16)
plt.show();
```
## How many parking tickets are given for each body type?
```
x_ticks = mini2016['Vehicle Body Type'].value_counts().index
heights = mini2016['Vehicle Body Type'].value_counts().values
y_pos = np.arange(len(x_ticks))
fig = plt.figure(figsize=(15,4))
f = sns.barplot(y=heights, x=y_pos, orient = 'v', alpha=0.6);
# remove labels
plt.tick_params(labelbottom='off')
plt.ylabel('No. of cars', fontsize=16);
plt.xlabel('Car models [Label turned off due to crowding. Too many types.]', fontsize=16);
plt.title('Parking ticket given for different type of car body', fontsize=16);
df_bodytype = pd.DataFrame(mini2016['Vehicle Body Type'].value_counts() / len(mini2016)).nlargest(10, columns = ['Vehicle Body Type'])
```
Top 10 car body types that get the most parking tickets are listed below :
```
df_bodytype
df_bodytype.sum(axis = 0)/len(mini2016)
```
Top 10 vehicle body type includes 93.42% of my sample dataset.
## How many parking tickets are given for each vehicle make?
Just for the sake of changing the flavor of visualization this time I will make a logplot of car no. vs make. In that case we will be able to see much smaller values in the same graph with larger values.
```
vehicle_make = mini2016['Vehicle Make'].value_counts()
plt.figure(figsize=(16,8))
f = sns.barplot(y=np.log(vehicle_make.values), x=vehicle_make.index, alpha=0.6)
# remove labels
plt.tick_params(labelbottom='off')
plt.ylabel('log(No. of cars)', fontsize=16);
plt.xlabel('Car make [Label turned off due to crowding. Too many companies!]', fontsize=16);
plt.title('Parking ticket given for different type of car make', fontsize=16);
plt.show();
pd.DataFrame(mini2016['Vehicle Make'].value_counts() / len(mini2016)).nlargest(10, columns = ['Vehicle Make'])
```
## Insight on violation time
In the raw data the **Violaation Time** is in a format, which is non-interpretable using standard **to_datatime** function in pandas. We need to change it in a useful format so that we can use the data. After formatting we may replace the old **Violation Time ** column with the new one.
```
timestamp = []
for time in mini2016['Violation Time']:
if len(str(time)) == 5:
time = time[:2] + ':' + time[2:]
timestamp.append(pd.to_datetime(time, errors='coerce'))
else:
timestamp.append(pd.NaT)
mini2016 = mini2016.assign(Violation_Time2 = timestamp)
mini2016.drop(['Violation Time'], axis = 1, inplace = True)
mini2016.rename(index=str, columns={"Violation_Time2": "Violation Time"}, inplace = True)
```
So in the new **Violation Time** column the data is in **Timestamp** format.
```
hours = [lambda x: x.hour, mini2016['Violation Time']]
# Getting the histogram
mini2016.set_index('Violation Time', drop=False, inplace=True)
plt.figure(figsize=(16,8))
mini2016['Violation Time'].groupby(pd.TimeGrouper(freq='30Min')).count().plot(kind='bar');
plt.tick_params(labelbottom='on')
plt.ylabel('No. of cars', fontsize=16);
plt.xlabel('Day Time', fontsize=16);
plt.title('Parking ticket given at different time of the day', fontsize=16);
```
## Parking ticket vs county
```
violation_county = mini2016['Violation County'].value_counts()
plt.figure(figsize=(16,8))
f = sns.barplot(y=violation_county.values, x=violation_county.index, alpha=0.6)
# remove labels
plt.tick_params(labelbottom='on')
plt.ylabel('No. of cars', fontsize=16);
plt.xlabel('County', fontsize=16);
plt.title('Parking ticket given in different counties', fontsize=16);
```
## Unregistered Vehicle?
```
sns.countplot(x = 'Unregistered Vehicle?', data = mini2016)
mini2016['Unregistered Vehicle?'].unique()
```
## Vehicle Year
```
pd.DataFrame(mini2016['Vehicle Year'].value_counts()).nlargest(10, columns = ['Vehicle Year'])
plt.figure(figsize=(20,8))
sns.countplot(x = 'Vehicle Year', data = mini2016.loc[(mini2016['Vehicle Year']>1980) & (mini2016['Vehicle Year'] <= 2018)]);
```
## Violation In Front Of Or Opposite
```
plt.figure(figsize=(16,8))
sns.countplot(x = 'Violation In Front Of Or Opposite', data = mini2016);
# create data
names = mini2016['Violation In Front Of Or Opposite'].value_counts().index
size = mini2016['Violation In Front Of Or Opposite'].value_counts().values
# Create a circle for the center of the plot
my_circle=plt.Circle( (0,0), 0.7, color='white')
plt.figure(figsize=(8,8))
from palettable.colorbrewer.qualitative import Pastel1_7
plt.pie(size, labels=names, colors=Pastel1_7.hex_colors)
p=plt.gcf()
p.gca().add_artist(my_circle)
plt.show()
```
| github_jupyter |
```
import os
from tensorflow.keras import layers
from tensorflow.keras import Model
from tensorflow.keras.applications.inception_v3 import InceptionV3
#!wget --no-check-certificate \
# https://storage.googleapis.com/mledu-datasets/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5 \
# -O /tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5
root = r'D:\Users\Arkady\Verint\Coursera_2019_Tensorflow_Specialization\Course2_CNN_in_TF'
local_weights_file = root + '/tmp/inception_v3_weights_tf_dim_ordering_tf_kernels_notop.h5'
pre_trained_model = InceptionV3(input_shape = (150, 150, 3),
include_top = False,
weights = None)
pre_trained_model.load_weights(local_weights_file)
for layer in pre_trained_model.layers:
layer.trainable = False
# pre_trained_model.summary()
last_layer = pre_trained_model.get_layer('mixed7')
print('last layer output shape: ', last_layer.output_shape)
last_output = last_layer.output
from tensorflow.keras.optimizers import RMSprop
# Flatten the output layer to 1 dimension
x = layers.Flatten()(last_output)
# Add a fully connected layer with 1,024 hidden units and ReLU activation
x = layers.Dense(1024, activation='relu')(x)
# Add a dropout rate of 0.2
x = layers.Dropout(0.2)(x)
# Add a final sigmoid layer for classification
x = layers.Dense (1, activation='sigmoid')(x)
model = Model( pre_trained_model.input, x)
model.compile(optimizer = RMSprop(lr=0.0001),
loss = 'binary_crossentropy',
metrics = ['acc'])
#!wget --no-check-certificate \
# https://storage.googleapis.com/mledu-datasets/cats_and_dogs_filtered.zip \
# -O /tmp/cats_and_dogs_filtered.zip
from tensorflow.keras.preprocessing.image import ImageDataGenerator
import os
import zipfile
#local_zip = '//tmp/cats_and_dogs_filtered.zip'
#zip_ref = zipfile.ZipFile(local_zip, 'r')
#zip_ref.extractall('/tmp')
#zip_ref.close()
# Define our example directories and files
base_dir = root + '/tmp/cats_and_dogs_filtered'
train_dir = os.path.join( base_dir, 'train')
validation_dir = os.path.join( base_dir, 'validation')
train_cats_dir = os.path.join(train_dir, 'cats') # Directory with our training cat pictures
train_dogs_dir = os.path.join(train_dir, 'dogs') # Directory with our training dog pictures
validation_cats_dir = os.path.join(validation_dir, 'cats') # Directory with our validation cat pictures
validation_dogs_dir = os.path.join(validation_dir, 'dogs')# Directory with our validation dog pictures
train_cat_fnames = os.listdir(train_cats_dir)
train_dog_fnames = os.listdir(train_dogs_dir)
# Add our data-augmentation parameters to ImageDataGenerator
train_datagen = ImageDataGenerator(rescale = 1./255.,
rotation_range = 40,
width_shift_range = 0.2,
height_shift_range = 0.2,
shear_range = 0.2,
zoom_range = 0.2,
horizontal_flip = True)
# Note that the validation data should not be augmented!
test_datagen = ImageDataGenerator( rescale = 1.0/255. )
# Flow training images in batches of 20 using train_datagen generator
train_generator = train_datagen.flow_from_directory(train_dir,
batch_size = 20,
class_mode = 'binary',
target_size = (150, 150))
# Flow validation images in batches of 20 using test_datagen generator
validation_generator = test_datagen.flow_from_directory( validation_dir,
batch_size = 20,
class_mode = 'binary',
target_size = (150, 150))
history = model.fit_generator(
train_generator,
validation_data = validation_generator,
steps_per_epoch = 100,
epochs = 20,
validation_steps = 50,
verbose = 2)
import matplotlib.pyplot as plt
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'r', label='Training accuracy')
plt.plot(epochs, val_acc, 'b', label='Validation accuracy')
plt.title('Training and validation accuracy')
plt.legend(loc=0)
plt.figure()
plt.show()
```
| github_jupyter |
CER041 - Install signed Knox certificate
========================================
This notebook installs into the Big Data Cluster the certificate signed
using:
- [CER031 - Sign Knox certificate with generated
CA](../cert-management/cer031-sign-knox-generated-cert.ipynb)
Steps
-----
### Parameters
```
app_name = "gateway"
scaledset_name = "gateway/pods/gateway-0"
container_name = "knox"
prefix_keyfile_name = "knox"
common_name = "gateway-svc"
test_cert_store_root = "/var/opt/secrets/test-certificates"
```
### Common functions
Define helper functions used in this notebook.
```
# Define `run` function for transient fault handling, suggestions on error, and scrolling updates on Windows
import sys
import os
import re
import json
import platform
import shlex
import shutil
import datetime
from subprocess import Popen, PIPE
from IPython.display import Markdown
retry_hints = {}
error_hints = {}
install_hint = {}
first_run = True
rules = None
def run(cmd, return_output=False, no_output=False, retry_count=0):
"""
Run shell command, stream stdout, print stderr and optionally return output
"""
MAX_RETRIES = 5
output = ""
retry = False
global first_run
global rules
if first_run:
first_run = False
rules = load_rules()
# shlex.split is required on bash and for Windows paths with spaces
#
cmd_actual = shlex.split(cmd)
# Store this (i.e. kubectl, python etc.) to support binary context aware error_hints and retries
#
user_provided_exe_name = cmd_actual[0].lower()
# When running python, use the python in the ADS sandbox ({sys.executable})
#
if cmd.startswith("python "):
cmd_actual[0] = cmd_actual[0].replace("python", sys.executable)
# On Mac, when ADS is not launched from terminal, LC_ALL may not be set, which causes pip installs to fail
# with:
#
# UnicodeDecodeError: 'ascii' codec can't decode byte 0xc5 in position 4969: ordinal not in range(128)
#
# Setting it to a default value of "en_US.UTF-8" enables pip install to complete
#
if platform.system() == "Darwin" and "LC_ALL" not in os.environ:
os.environ["LC_ALL"] = "en_US.UTF-8"
# To aid supportabilty, determine which binary file will actually be executed on the machine
#
which_binary = None
# Special case for CURL on Windows. The version of CURL in Windows System32 does not work to
# get JWT tokens, it returns "(56) Failure when receiving data from the peer". If another instance
# of CURL exists on the machine use that one. (Unfortunately the curl.exe in System32 is almost
# always the first curl.exe in the path, and it can't be uninstalled from System32, so here we
# look for the 2nd installation of CURL in the path)
if platform.system() == "Windows" and cmd.startswith("curl "):
path = os.getenv('PATH')
for p in path.split(os.path.pathsep):
p = os.path.join(p, "curl.exe")
if os.path.exists(p) and os.access(p, os.X_OK):
if p.lower().find("system32") == -1:
cmd_actual[0] = p
which_binary = p
break
# Find the path based location (shutil.which) of the executable that will be run (and display it to aid supportability), this
# seems to be required for .msi installs of azdata.cmd/az.cmd. (otherwise Popen returns FileNotFound)
#
# NOTE: Bash needs cmd to be the list of the space separated values hence shlex.split.
#
if which_binary == None:
which_binary = shutil.which(cmd_actual[0])
if which_binary == None:
if user_provided_exe_name in install_hint and install_hint[user_provided_exe_name] is not None:
display(Markdown(f'HINT: Use [{install_hint[user_provided_exe_name][0]}]({install_hint[user_provided_exe_name][1]}) to resolve this issue.'))
raise FileNotFoundError(f"Executable '{cmd_actual[0]}' not found in path (where/which)")
else:
cmd_actual[0] = which_binary
start_time = datetime.datetime.now().replace(microsecond=0)
print(f"START: {cmd} @ {start_time} ({datetime.datetime.utcnow().replace(microsecond=0)} UTC)")
print(f" using: {which_binary} ({platform.system()} {platform.release()} on {platform.machine()})")
print(f" cwd: {os.getcwd()}")
# Command-line tools such as CURL and AZDATA HDFS commands output
# scrolling progress bars, which causes Jupyter to hang forever, to
# workaround this, use no_output=True
#
# Work around a infinite hang when a notebook generates a non-zero return code, break out, and do not wait
#
wait = True
try:
if no_output:
p = Popen(cmd_actual)
else:
p = Popen(cmd_actual, stdout=PIPE, stderr=PIPE, bufsize=1)
with p.stdout:
for line in iter(p.stdout.readline, b''):
line = line.decode()
if return_output:
output = output + line
else:
if cmd.startswith("azdata notebook run"): # Hyperlink the .ipynb file
regex = re.compile(' "(.*)"\: "(.*)"')
match = regex.match(line)
if match:
if match.group(1).find("HTML") != -1:
display(Markdown(f' - "{match.group(1)}": "{match.group(2)}"'))
else:
display(Markdown(f' - "{match.group(1)}": "[{match.group(2)}]({match.group(2)})"'))
wait = False
break # otherwise infinite hang, have not worked out why yet.
else:
print(line, end='')
if rules is not None:
apply_expert_rules(line)
if wait:
p.wait()
except FileNotFoundError as e:
if install_hint is not None:
display(Markdown(f'HINT: Use {install_hint} to resolve this issue.'))
raise FileNotFoundError(f"Executable '{cmd_actual[0]}' not found in path (where/which)") from e
exit_code_workaround = 0 # WORKAROUND: azdata hangs on exception from notebook on p.wait()
if not no_output:
for line in iter(p.stderr.readline, b''):
line_decoded = line.decode()
# azdata emits a single empty line to stderr when doing an hdfs cp, don't
# print this empty "ERR:" as it confuses.
#
if line_decoded == "":
continue
print(f"STDERR: {line_decoded}", end='')
if line_decoded.startswith("An exception has occurred") or line_decoded.startswith("ERROR: An error occurred while executing the following cell"):
exit_code_workaround = 1
if user_provided_exe_name in error_hints:
for error_hint in error_hints[user_provided_exe_name]:
if line_decoded.find(error_hint[0]) != -1:
display(Markdown(f'HINT: Use [{error_hint[1]}]({error_hint[2]}) to resolve this issue.'))
if rules is not None:
apply_expert_rules(line_decoded)
if user_provided_exe_name in retry_hints:
for retry_hint in retry_hints[user_provided_exe_name]:
if line_decoded.find(retry_hint) != -1:
if retry_count < MAX_RETRIES:
print(f"RETRY: {retry_count} (due to: {retry_hint})")
retry_count = retry_count + 1
output = run(cmd, return_output=return_output, retry_count=retry_count)
if return_output:
return output
else:
return
elapsed = datetime.datetime.now().replace(microsecond=0) - start_time
# WORKAROUND: We avoid infinite hang above in the `azdata notebook run` failure case, by inferring success (from stdout output), so
# don't wait here, if success known above
#
if wait:
if p.returncode != 0:
raise SystemExit(f'Shell command:\n\n\t{cmd} ({elapsed}s elapsed)\n\nreturned non-zero exit code: {str(p.returncode)}.\n')
else:
if exit_code_workaround !=0 :
raise SystemExit(f'Shell command:\n\n\t{cmd} ({elapsed}s elapsed)\n\nreturned non-zero exit code: {str(exit_code_workaround)}.\n')
print(f'\nSUCCESS: {elapsed}s elapsed.\n')
if return_output:
return output
def load_json(filename):
with open(filename, encoding="utf8") as json_file:
return json.load(json_file)
def load_rules():
try:
# Load this notebook as json to get access to the expert rules in the notebook metadata.
#
j = load_json("cer041-install-knox-cert.ipynb")
except:
pass # If the user has renamed the book, we can't load ourself. NOTE: Is there a way in Jupyter, to know your own filename?
else:
if "metadata" in j and \
"azdata" in j["metadata"] and \
"expert" in j["metadata"]["azdata"] and \
"rules" in j["metadata"]["azdata"]["expert"]:
rules = j["metadata"]["azdata"]["expert"]["rules"]
rules.sort() # Sort rules, so they run in priority order (the [0] element). Lowest value first.
# print (f"EXPERT: There are {len(rules)} rules to evaluate.")
return rules
def apply_expert_rules(line):
global rules
for rule in rules:
# rules that have 9 elements are the injected (output) rules (the ones we want). Rules
# with only 8 elements are the source (input) rules, which are not expanded (i.e. TSG029,
# not ../repair/tsg029-nb-name.ipynb)
if len(rule) == 9:
notebook = rule[1]
cell_type = rule[2]
output_type = rule[3] # i.e. stream or error
output_type_name = rule[4] # i.e. ename or name
output_type_value = rule[5] # i.e. SystemExit or stdout
details_name = rule[6] # i.e. evalue or text
expression = rule[7].replace("\\*", "*") # Something escaped *, and put a \ in front of it!
# print(f"EXPERT: If rule '{expression}' satisfied', run '{notebook}'.")
if re.match(expression, line, re.DOTALL):
# print("EXPERT: MATCH: name = value: '{0}' = '{1}' matched expression '{2}', therefore HINT '{4}'".format(output_type_name, output_type_value, expression, notebook))
match_found = True
display(Markdown(f'HINT: Use [{notebook}]({notebook}) to resolve this issue.'))
print('Common functions defined successfully.')
# Hints for binary (transient fault) retry, (known) error and install guide
#
retry_hints = {'kubectl': ['A connection attempt failed because the connected party did not properly respond after a period of time, or established connection failed because connected host has failed to respond']}
error_hints = {'kubectl': [['no such host', 'TSG010 - Get configuration contexts', '../monitor-k8s/tsg010-get-kubernetes-contexts.ipynb'], ['no such host', 'TSG011 - Restart sparkhistory server', '../repair/tsg011-restart-sparkhistory-server.ipynb'], ['No connection could be made because the target machine actively refused it', 'TSG056 - Kubectl fails with No connection could be made because the target machine actively refused it', '../repair/tsg056-kubectl-no-connection-could-be-made.ipynb']]}
install_hint = {'kubectl': ['SOP036 - Install kubectl command line interface', '../install/sop036-install-kubectl.ipynb']}
```
### Get the Kubernetes namespace for the big data cluster
Get the namespace of the big data cluster use the kubectl command line
interface .
NOTE: If there is more than one big data cluster in the target
Kubernetes cluster, then set \[0\] to the correct value for the big data
cluster.
```
# Place Kubernetes namespace name for BDC into 'namespace' variable
try:
namespace = run(f'kubectl get namespace --selector=MSSQL_CLUSTER -o jsonpath={{.items[0].metadata.name}}', return_output=True)
except:
from IPython.display import Markdown
print(f"ERROR: Unable to find a Kubernetes namespace with label 'MSSQL_CLUSTER'. SQL Server Big Data Cluster Kubernetes namespaces contain the label 'MSSQL_CLUSTER'.")
display(Markdown(f'HINT: Use [TSG081 - Get namespaces (Kubernetes)](../monitor-k8s/tsg081-get-kubernetes-namespaces.ipynb) to resolve this issue.'))
display(Markdown(f'HINT: Use [TSG010 - Get configuration contexts](../monitor-k8s/tsg010-get-kubernetes-contexts.ipynb) to resolve this issue.'))
display(Markdown(f'HINT: Use [SOP011 - Set kubernetes configuration context](../common/sop011-set-kubernetes-context.ipynb) to resolve this issue.'))
raise
else:
print(f'The SQL Server Big Data Cluster Kubernetes namespace is: {namespace}')
```
### Create a temporary directory to stage files
```
# Create a temporary directory to hold configuration files
import tempfile
temp_dir = tempfile.mkdtemp()
print(f"Temporary directory created: {temp_dir}")
```
### Helper function to save configuration files to disk
```
# Define helper function 'save_file' to save configuration files to the temporary directory created above
import os
import io
def save_file(filename, contents):
with io.open(os.path.join(temp_dir, filename), "w", encoding='utf8', newline='\n') as text_file:
text_file.write(contents)
print("File saved: " + os.path.join(temp_dir, filename))
```
### Get name of the ‘Running’ `controller` `pod`
```
# Place the name of the 'Running' controller pod in variable `controller`
controller = run(f'kubectl get pod --selector=app=controller -n {namespace} -o jsonpath={{.items[0].metadata.name}} --field-selector=status.phase=Running', return_output=True)
print(f"Controller pod name: {controller}")
```
### Pod name for gateway
```
pod = 'gateway-0'
```
### Copy certifcate files from `controller` to local machine
```
import os
cwd = os.getcwd()
os.chdir(temp_dir) # Use chdir to workaround kubectl bug on Windows, which incorrectly processes 'c:\' on kubectl cp cmd line
run(f'kubectl cp {controller}:{test_cert_store_root}/{app_name}/{prefix_keyfile_name}-certificate.pem {prefix_keyfile_name}-certificate.pem -c controller -n {namespace}')
run(f'kubectl cp {controller}:{test_cert_store_root}/{app_name}/{prefix_keyfile_name}-privatekey.pem {prefix_keyfile_name}-privatekey.pem -c controller -n {namespace}')
os.chdir(cwd)
```
### Copy certifcate files from local machine to `controldb`
```
import os
cwd = os.getcwd()
os.chdir(temp_dir) # Workaround kubectl bug on Windows, can't put c:\ on kubectl cp cmd line
run(f'kubectl cp {prefix_keyfile_name}-certificate.pem controldb-0:/var/opt/mssql/{prefix_keyfile_name}-certificate.pem -c mssql-server -n {namespace}')
run(f'kubectl cp {prefix_keyfile_name}-privatekey.pem controldb-0:/var/opt/mssql/{prefix_keyfile_name}-privatekey.pem -c mssql-server -n {namespace}')
os.chdir(cwd)
```
### Get the `controller-db-rw-secret` secret
Get the controller SQL symmetric key password for decryption.
```
import base64
controller_db_rw_secret = run(f'kubectl get secret/controller-db-rw-secret -n {namespace} -o jsonpath={{.data.encryptionPassword}}', return_output=True)
controller_db_rw_secret = base64.b64decode(controller_db_rw_secret).decode('utf-8')
print("controller_db_rw_secret retrieved")
```
### Update the files table with the certificates through opened SQL connection
```
import os
sql = f"""
OPEN SYMMETRIC KEY ControllerDbSymmetricKey DECRYPTION BY PASSWORD = '{controller_db_rw_secret}'
DECLARE @FileData VARBINARY(MAX), @Key uniqueidentifier;
SELECT @Key = KEY_GUID('ControllerDbSymmetricKey');
SELECT TOP 1 @FileData = doc.BulkColumn FROM OPENROWSET(BULK N'/var/opt/mssql/{prefix_keyfile_name}-certificate.pem', SINGLE_BLOB) AS doc;
EXEC [dbo].[sp_set_file_data_encrypted] @FilePath = '/config/scaledsets/{scaledset_name}/containers/{container_name}/files/{prefix_keyfile_name}-certificate.pem',
@Data = @FileData,
@KeyGuid = @Key,
@Version = '0',
@User = '',
@Group = '',
@Mode = '';
SELECT TOP 1 @FileData = doc.BulkColumn FROM OPENROWSET(BULK N'/var/opt/mssql/{prefix_keyfile_name}-privatekey.pem', SINGLE_BLOB) AS doc;
EXEC [dbo].[sp_set_file_data_encrypted] @FilePath = '/config/scaledsets/{scaledset_name}/containers/{container_name}/files/{prefix_keyfile_name}-privatekey.pem',
@Data = @FileData,
@KeyGuid = @Key,
@Version = '0',
@User = '',
@Group = '',
@Mode = '';
"""
save_file("insert_certificates.sql", sql)
cwd = os.getcwd()
os.chdir(temp_dir) # Workaround kubectl bug on Windows, can't put c:\ on kubectl cp cmd line
run(f'kubectl cp insert_certificates.sql controldb-0:/var/opt/mssql/insert_certificates.sql -c mssql-server -n {namespace}')
run(f"""kubectl exec controldb-0 -c mssql-server -n {namespace} -- bash -c "SQLCMDPASSWORD=`cat /var/run/secrets/credentials/mssql-sa-password/password` /opt/mssql-tools/bin/sqlcmd -b -U sa -d controller -i /var/opt/mssql/insert_certificates.sql" """)
# Clean up
run(f"""kubectl exec controldb-0 -c mssql-server -n {namespace} -- bash -c "rm /var/opt/mssql/insert_certificates.sql" """)
run(f"""kubectl exec controldb-0 -c mssql-server -n {namespace} -- bash -c "rm /var/opt/mssql/{prefix_keyfile_name}-certificate.pem" """)
run(f"""kubectl exec controldb-0 -c mssql-server -n {namespace} -- bash -c "rm /var/opt/mssql/{prefix_keyfile_name}-privatekey.pem" """)
os.chdir(cwd)
```
### Clear out the controller\_db\_rw\_secret variable
```
controller_db_rw_secret= ""
```
### Clean up certificate staging area
Remove the certificate files generated on disk (they have now been
placed in the controller database).
```
cmd = f"rm -r {test_cert_store_root}/{app_name}"
run(f'kubectl exec {controller} -c controller -n {namespace} -- bash -c "{cmd}"')
```
### Restart knox gateway service
```
run(f'kubectl delete pod {pod} -n {namespace}')
```
### Clean up temporary directory for staging configuration files
```
# Delete the temporary directory used to hold configuration files
import shutil
shutil.rmtree(temp_dir)
print(f'Temporary directory deleted: {temp_dir}')
print('Notebook execution complete.')
```
Related
-------
- [CER042 - Install signed App-Proxy
certificate](../cert-management/cer042-install-app-proxy-cert.ipynb)
- [CER031 - Sign Knox certificate with generated
CA](../cert-management/cer031-sign-knox-generated-cert.ipynb)
- [CER021 - Create Knox
certificate](../cert-management/cer021-create-knox-cert.ipynb)
| github_jupyter |
```
import requests
!pip3 install requests
response = requests.get("https://api.spotify.com/v1/search?q=Lil&type=artist&market=US&limit=50")
print(response.text)
data = response.json()
type(data)
data.keys()
data['artists'].keys()
artists=data['artists']
type(artists['items'])
artist_info = artists['items']
for artist in artist_info:
print(artist['name'], artist['popularity'])
print(artist_info[5])
artists=data['artists']
artist_info = artists['items']
separator = ", "
for artist in artist_info:
if len(artist['genres']) == 0:
print("No genres listed.")
else:
print(artist['name'], ":", separator.join(artist['genres']))
most_popular_name = ""
most_popular_score = 0
for artist in artist_info:
if artist['popularity'] > most_popular_score and artist['name'] != "Lil Wayne":
most_popular_name = artist['name']
most_popular_score = artist['popularity']
else:
pass
print(most_popular_name,most_popular_score)
```
print a list of Lil's that are more popular than Lil's Kim
```
for artist in artist_info:
print(artist['name'])
if artist['name']== "Lil' Kim":
print("Found Lil Kim")
print(artist['popularity'])
else:
pass #print
Lil_kim_popularity = 62
more_popular_than_Lil_kim = []
for artist in artist_info:
if artist['popularity'] > Lil_kim_popularity:
#If yes, let's add them to our list
print(artist['name'], "is more popular with a score of", artist['popularity'])
more_popular_than_Lil_kim.append(artist['name'])
else:
print(artist['name'], "is less popular with a score of", artist['popularity'])
for artist_name in more_popular_than_Lil_kim:
print(artist_name)
```
Pick two of your favorite Lils to fight it out, and use their IDs to print out their top tracks
```
for artist in artist_info:
print(artist['name'], artist['id'])
#I chose Lil Fate and Lil' Flip, first I want to figure out the top track of Lil Fate
response = requests.get("https://api.spotify.com/v1/artists/6JUnsP7jmvYmdhbg7lTMQj/top-tracks?country=US")
print(response.text)
data = response.json()
type(data)
data.keys()
type(data['tracks'])
print(data['tracks'])
data['tracks'][0]
for item in data['tracks']:
print(item['name'])
# now to figure out the top track of Lil' Flip #things within {} or ALL Caps means to replace them
response = requests.get("https://api.spotify.com/v1/artists/4Q5sPmM8j4SpMqL4UA1DtS/top-tracks?country=US")
print(response.text)
data = response.json()
type(data)
data.keys()
type(data['tracks'])
for item in data['tracks']:
#type(item): dict
#print(item.keys()), saw 'name'
print(item['name'])
```
##Will the world explode if a musicians swears? Get an average popularity for their explicit songs vs. their non-explicit songs. How many minutes of explicit songs do they have? Non-explicit?
```
#for Lil' fate's top tracks
explicit_count = 0
non_explicit_count = 0
popularity_explicit = 0
popularity_non_explicit = 0
minutes_explicit = 0
minutes_non_explicit = 0
for track in data['tracks']:
if track['explicit']== True:
explicit_count = explicit_count + 1
popularity_explicit = popularity_explicit + track['popularity']
minutes_explicit = minutes_explicit + track['duration_ms']
elif track['explicit']== False:
non_explicit_count = non_explicit_count + 1
popularity_non_explicit = popularity_non_explicit + track['popularity']
minutes_non_explicit = minutes_non_explicit + track['duration_ms']
print("Lil' Flip has", (minutes_explicit/1000)/60, "of explicit songs")
print("Lil' Flip has", (minutes_non_explicit/1000)/60, "of non-explicit songs")
print("The average popularity of Lil' Flip explicits songs is", popularity_explicit/explicit_count)
print("The average popularity of Lil' Flip non-explicits songs is", popularity_non_explicit/non_explicit_count)
```
Since we're talking about Lils, what about Biggies? How many total "Biggie" artists are there? How many total "Lil"s? If you made 1 request every 5 seconds, how long would it take to download information on all the Lils vs the Biggies?
```
response = requests.get('https://api.spotify.com/v1/search?q=Lil&type=artist&market=US')
all_lil = response.json()
print(response.text)
all_lil.keys()
all_lil['artists'].keys()
print(all_lil['artists']['total'])
response = requests.get('https://api.spotify.com/v1/search?q=Biggie&type=artist&market=US')
all_biggies = response.json()
print(all_biggies['artists']['total'])
```
## how to count the genres
```
all_genres = []
for artist in artist_info:
print("All genres we've heard of:", all_genres)
print("Current artist has:", artist['genres'])
all_genres = all_genres + artist['genres']
all_genres.count('dirty south rap')
## There is a library that comes with Python called Collections, inside of it is a thing called Counter
from collections import Counter
```
## How to automate getting all of the results
```
response=requests.get('https://api.spotify.com/v1/search?q=Lil&type=artist&market=US&limit50')
small_data = response.json()
data['artists']
print(len(data['artists']['items'])) #we only get 10 artists
print(data['artists']['total'])
#first page: artists 1-50, offset of 0
# https://
```
| github_jupyter |
<img src="http://developer.download.nvidia.com/compute/machine-learning/frameworks/nvidia_logo.png" style="width: 90px; float: right;">
# HugeCTR Continuous Training and Inference Demo (Part I)
## Overview
In HugeCTR version 3.3, we finished the whole pipeline of parameter server, including
1. The parameter dumping interface from training to kafka.
2. CPU cache(Redis Cluster / Hash Map / Parallel Hash Map).
3. RocksDB as a persistence storage.
4. Embedding cache update mechanism.
The purpose of this notebook is to show how to do continuous traning and inference using HugeCTR Hierarchical Parameter Server.
## Table of Contents
- Data Preparation
- Data Preprocessing using Pandas
- Wide&Deep Training Demo
- Wide&Deep Model Inference using Python API
- Wide&Deep Model continuous training
- Wide&Deep Model continuous inference
## 1. Data preparation
### 1.1 Make a folder to store our data and data processing scripts:
```
!mkdir criteo_data
!mkdir criteo_script
```
### 1.2 Download Criteo Dataset
```
!wget http://azuremlsampleexperiments.blob.core.windows.net/criteo/day_1.gz
```
**NOTE**: Replace `1` with a value from [0, 23] to use a different day.
During preprocessing, the amount of data, which is used to speed up the preprocessing, fill missing values, and remove the feature values that are considered rare, is further reduced.
### 1.3 Write the preprocessing the script.
```
%%writefile preprocess.sh
#!/bin/bash
if [[ $# -lt 3 ]]; then
echo "Usage: preprocess.sh [DATASET_NO.] [DST_DATA_DIR] [SCRIPT_TYPE] [SCRIPT_TYPE_SPECIFIC_ARGS...]"
exit 2
fi
DST_DATA_DIR=$2
echo "Warning: existing $DST_DATA_DIR is erased"
rm -rf $DST_DATA_DIR
if [[ $3 == "nvt" ]]; then
if [[ $# -ne 6 ]]; then
echo "Usage: preprocess.sh [DATASET_NO.] [DST_DATA_DIR] nvt [IS_PARQUET_FORMAT] [IS_CRITEO_MODE] [IS_FEATURE_CROSSED]"
exit 2
fi
echo "Preprocessing script: NVTabular"
elif [[ $3 == "perl" ]]; then
if [[ $# -ne 4 ]]; then
echo "Usage: preprocess.sh [DATASET_NO.] [DST_DATA_DIR] perl [NUM_SLOTS]"
exit 2
fi
echo "Preprocessing script: Perl"
elif [[ $3 == "pandas" ]]; then
if [[ $# -lt 5 ]]; then
echo "Usage: preprocess.sh [DATASET_NO.] [DST_DATA_DIR] pandas [IS_DENSE_NORMALIZED] [IS_FEATURE_CROSSED] (FILE_LIST_LENGTH)"
exit 2
fi
echo "Preprocessing script: Pandas"
else
echo "Error: $3 is an invalid script type. Pick one from {nvt, perl, pandas}."
exit 2
fi
SCRIPT_TYPE=$3
echo "Getting the first few examples from the uncompressed dataset..."
mkdir -p $DST_DATA_DIR/train && \
mkdir -p $DST_DATA_DIR/val && \
head -n 500000 day_$1 > $DST_DATA_DIR/day_$1_small
if [ $? -ne 0 ]; then
echo "Warning: fallback to find original compressed data day_$1.gz..."
echo "Decompressing day_$1.gz..."
gzip -d -c day_$1.gz > day_$1
if [ $? -ne 0 ]; then
echo "Error: failed to decompress the file."
exit 2
fi
head -n 500000 day_$1 > $DST_DATA_DIR/day_$1_small
if [ $? -ne 0 ]; then
echo "Error: day_$1 file"
exit 2
fi
fi
echo "Counting the number of samples in day_$1 dataset..."
total_count=$(wc -l $DST_DATA_DIR/day_$1_small)
total_count=(${total_count})
echo "The first $total_count examples will be used in day_$1 dataset."
echo "Shuffling dataset..."
shuf $DST_DATA_DIR/day_$1_small > $DST_DATA_DIR/day_$1_shuf
train_count=$(( total_count * 8 / 10))
valtest_count=$(( total_count - train_count ))
val_count=$(( valtest_count * 5 / 10 ))
test_count=$(( valtest_count - val_count ))
split_dataset()
{
echo "Splitting into $train_count-sample training, $val_count-sample val, and $test_count-sample test datasets..."
head -n $train_count $DST_DATA_DIR/$1 > $DST_DATA_DIR/train/train.txt && \
tail -n $valtest_count $DST_DATA_DIR/$1 > $DST_DATA_DIR/val/valtest.txt && \
head -n $val_count $DST_DATA_DIR/val/valtest.txt > $DST_DATA_DIR/val/val.txt && \
tail -n $test_count $DST_DATA_DIR/val/valtest.txt > $DST_DATA_DIR/val/test.txt
if [ $? -ne 0 ]; then
exit 2
fi
}
echo "Preprocessing..."
if [[ $SCRIPT_TYPE == "nvt" ]]; then
IS_PARQUET_FORMAT=$4
IS_CRITEO_MODE=$5
FEATURE_CROSS_LIST_OPTION=""
if [[ ( $IS_CRITEO_MODE -eq 0 ) && ( $6 -eq 1 ) ]]; then
FEATURE_CROSS_LIST_OPTION="--feature_cross_list C1_C2,C3_C4"
echo $FEATURE_CROSS_LIST_OPTION
fi
split_dataset day_$1_shuf
python3 criteo_script/preprocess_nvt.py \
--data_path $DST_DATA_DIR \
--out_path $DST_DATA_DIR \
--freq_limit 6 \
--device_limit_frac 0.5 \
--device_pool_frac 0.5 \
--out_files_per_proc 8 \
--devices "0" \
--num_io_threads 2 \
--parquet_format=$IS_PARQUET_FORMAT \
--criteo_mode=$IS_CRITEO_MODE \
$FEATURE_CROSS_LIST_OPTION
elif [[ $SCRIPT_TYPE == "perl" ]]; then
NUM_SLOT=$4
split_dataset day_$1_shuf
perl criteo_script_legacy/preprocess.pl $DST_DATA_DIR/train/train.txt $DST_DATA_DIR/val/val.txt $DST_DATA_DIR/val/test.txt && \
criteo2hugectr_legacy $NUM_SLOT $DST_DATA_DIR/train/train.txt.out $DST_DATA_DIR/train/sparse_embedding $DST_DATA_DIR/file_list.txt && \
criteo2hugectr_legacy $NUM_SLOT $DST_DATA_DIR/val/test.txt.out $DST_DATA_DIR/val/sparse_embedding $DST_DATA_DIR/file_list_test.txt
elif [[ $SCRIPT_TYPE == "pandas" ]]; then
python3 criteo_script/preprocess.py \
--src_csv_path=$DST_DATA_DIR/day_$1_shuf \
--dst_csv_path=$DST_DATA_DIR/day_$1_shuf.out \
--normalize_dense=$4 --feature_cross=$5 && \
split_dataset day_$1_shuf.out
NUM_WIDE_KEYS=""
if [[ $5 -ne 0 ]]; then
NUM_WIDE_KEYS=2
fi
FILE_LIST_LENGTH=""
if [[ $# -gt 5 ]]; then
FILE_LIST_LENGTH=$6
fi
criteo2hugectr $DST_DATA_DIR/train/train.txt $DST_DATA_DIR/train/sparse_embedding $DST_DATA_DIR/file_list.txt $NUM_WIDE_KEYS $FILE_LIST_LENGTH && \
criteo2hugectr $DST_DATA_DIR/val/test.txt $DST_DATA_DIR/val/sparse_embedding $DST_DATA_DIR/file_list_test.txt $NUM_WIDE_KEYS $FILE_LIST_LENGTH
fi
if [ $? -ne 0 ]; then
exit 2
fi
echo "All done!"
```
**NOTE**: Here we only read the first 500000 lines of the data to do the demo.
```
%%writefile criteo_script/preprocess.py
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
from __future__ import unicode_literals
import argparse
import sys
import tempfile
from six.moves import urllib
import urllib.request
import sys
import os
import math
import time
import logging
import concurrent.futures as cf
from traceback import print_exc
import numpy as np
import pandas as pd
import sklearn.preprocessing as skp
logging.basicConfig(format='%(asctime)s %(message)s')
logging.root.setLevel(logging.NOTSET)
NUM_INTEGER_COLUMNS = 13
NUM_CATEGORICAL_COLUMNS = 26
NUM_TOTAL_COLUMNS = 1 + NUM_INTEGER_COLUMNS + NUM_CATEGORICAL_COLUMNS
MAX_NUM_WORKERS = NUM_TOTAL_COLUMNS
INT_NAN_VALUE = np.iinfo(np.int32).min
CAT_NAN_VALUE = '80000000'
def idx2key(idx):
if idx == 0:
return 'label'
return 'I' + str(idx) if idx <= NUM_INTEGER_COLUMNS else 'C' + str(idx - NUM_INTEGER_COLUMNS)
def _fill_missing_features_and_split(chunk, series_list_dict):
for cid, col in enumerate(chunk.columns):
NAN_VALUE = INT_NAN_VALUE if cid <= NUM_INTEGER_COLUMNS else CAT_NAN_VALUE
result_series = chunk[col].fillna(NAN_VALUE)
series_list_dict[col].append(result_series)
def _merge_and_transform_series(src_series_list, col, dense_cols,
normalize_dense):
result_series = pd.concat(src_series_list)
if col != 'label':
unique_value_counts = result_series.value_counts()
unique_value_counts = unique_value_counts.loc[unique_value_counts >= 6]
unique_value_counts = set(unique_value_counts.index.values)
NAN_VALUE = INT_NAN_VALUE if col.startswith('I') else CAT_NAN_VALUE
result_series = result_series.apply(
lambda x: x if x in unique_value_counts else NAN_VALUE)
if col == 'label' or col in dense_cols:
result_series = result_series.astype(np.int64)
le = skp.LabelEncoder()
result_series = pd.DataFrame(le.fit_transform(result_series))
if col != 'label':
result_series = result_series + 1
else:
oe = skp.OrdinalEncoder(dtype=np.int64)
result_series = pd.DataFrame(oe.fit_transform(pd.DataFrame(result_series)))
result_series = result_series + 1
if normalize_dense != 0:
if col in dense_cols:
mms = skp.MinMaxScaler(feature_range=(0,1))
result_series = pd.DataFrame(mms.fit_transform(result_series))
result_series.columns = [col]
min_max = (np.int64(result_series[col].min()), np.int64(result_series[col].max()))
if col != 'label':
logging.info('column {} [{}, {}]'.format(col, str(min_max[0]),str(min_max[1])))
return [result_series, min_max]
def _convert_to_string(series):
return series.astype(str)
def _merge_columns_and_feature_cross(series_list, min_max, feature_pairs,
feature_cross):
name_to_series = dict()
for series in series_list:
name_to_series[series.columns[0]] = series.iloc[:,0]
df = pd.DataFrame(name_to_series)
cols = [idx2key(idx) for idx in range(0, NUM_TOTAL_COLUMNS)]
df = df.reindex(columns=cols)
offset = np.int64(0)
for col in cols:
if col != 'label' and col.startswith('I') == False:
df[col] += offset
logging.info('column {} offset {}'.format(col, str(offset)))
offset += min_max[col][1]
if feature_cross != 0:
for idx, pair in enumerate(feature_pairs):
col0 = pair[0]
col1 = pair[1]
col1_width = int(min_max[col1][1] - min_max[col1][0] + 1)
crossed_column_series = df[col0] * col1_width + df[col1]
oe = skp.OrdinalEncoder(dtype=np.int64)
crossed_column_series = pd.DataFrame(oe.fit_transform(pd.DataFrame(crossed_column_series)))
crossed_column_series = crossed_column_series + 1
crossed_column = col0 + '_' + col1
df.insert(NUM_INTEGER_COLUMNS + 1 + idx, crossed_column, crossed_column_series)
crossed_column_max_val = np.int64(df[crossed_column].max())
logging.info('column {} [{}, {}]'.format(
crossed_column,
str(df[crossed_column].min()),
str(crossed_column_max_val)))
df[crossed_column] += offset
logging.info('column {} offset {}'.format(crossed_column, str(offset)))
offset += crossed_column_max_val
return df
def _wait_futures_and_reset(futures):
for future in futures:
result = future.result()
if result:
print(result)
futures = list()
def _process_chunks(executor, chunks_to_process, op, *argv):
futures = list()
for chunk in chunks_to_process:
argv_list = list(argv)
argv_list.insert(0, chunk)
new_argv = tuple(argv_list)
future = executor.submit(op, *new_argv)
futures.append(future)
_wait_futures_and_reset(futures)
def preprocess(src_txt_name, dst_txt_name, normalize_dense, feature_cross):
cols = [idx2key(idx) for idx in range(0, NUM_TOTAL_COLUMNS)]
series_list_dict = dict()
with cf.ThreadPoolExecutor(max_workers=MAX_NUM_WORKERS) as executor:
logging.info('read a CSV file')
reader = pd.read_csv(src_txt_name, sep='\t',
names=cols,
chunksize=131072)
logging.info('_fill_missing_features_and_split')
for col in cols:
series_list_dict[col] = list()
_process_chunks(executor, reader, _fill_missing_features_and_split,
series_list_dict)
with cf.ProcessPoolExecutor(max_workers=MAX_NUM_WORKERS) as executor:
logging.info('_merge_and_transform_series')
futures = list()
dense_cols = [idx2key(idx+1) for idx in range(NUM_INTEGER_COLUMNS)]
dst_series_list = list()
min_max = dict()
for col, src_series_list in series_list_dict.items():
future = executor.submit(_merge_and_transform_series,
src_series_list, col, dense_cols,
normalize_dense)
futures.append(future)
for future in futures:
col = None
for idx, ret in enumerate(future.result()):
try:
if idx == 0:
col = ret.columns[0]
dst_series_list.append(ret)
else:
min_max[col] = ret
except:
print_exc()
futures = list()
logging.info('_merge_columns_and_feature_cross')
feature_pairs = [('C1', 'C2'), ('C3', 'C4')]
df = _merge_columns_and_feature_cross(dst_series_list, min_max, feature_pairs,
feature_cross)
logging.info('_convert_to_string')
futures = dict()
for col in cols:
future = executor.submit(_convert_to_string, df[col])
futures[col] = future
if feature_cross != 0:
for pair in feature_pairs:
col = pair[0] + '_' + pair[1]
future = executor.submit(_convert_to_string, df[col])
futures[col] = future
logging.info('_store_to_df')
for col, future in futures.items():
ret = future.result()
try:
df[col] = ret
except:
print_exc()
futures = dict()
logging.info('write to a CSV file')
df.to_csv(dst_txt_name, sep=' ', header=False, index=False)
logging.info('done!')
if __name__ == '__main__':
arg_parser = argparse.ArgumentParser(description='Preprocssing Criteo Dataset')
arg_parser.add_argument('--src_csv_path', type=str, required=True)
arg_parser.add_argument('--dst_csv_path', type=str, required=True)
arg_parser.add_argument('--normalize_dense', type=int, default=1)
arg_parser.add_argument('--feature_cross', type=int, default=1)
args = arg_parser.parse_args()
src_csv_path = args.src_csv_path
dst_csv_path = args.dst_csv_path
normalize_dense = args.normalize_dense
feature_cross = args.feature_cross
if os.path.exists(src_csv_path) == False:
sys.exit('ERROR: the file \'{}\' doesn\'t exist'.format(src_csv_path))
if os.path.exists(dst_csv_path) == True:
sys.exit('ERROR: the file \'{}\' exists'.format(dst_csv_path))
preprocess(src_csv_path, dst_csv_path, normalize_dense, feature_cross)
```
### 1.4 Run the preprocess script
```
!bash preprocess.sh 0 criteo_data pandas 1 1 1
```
**IMPORTANT NOTES**:
Arguments may vary depend on your setting:
- The first argument represents the dataset postfix. For instance, if `day_1` is used, the postfix is `1`.
- The second argument, `criteo_data`, is where the preprocessed data is stored.
### 1.5 Generate data sample for inference
```
import pandas as pd
import numpy as np
df = pd.read_table("criteo_data/train/train.txt", header = None, sep= ' ', \
names = ['label'] + ['I'+str(i) for i in range(1, 14)] + \
['C1_C2', 'C3_C4'] + ['C'+str(i) for i in range(1, 27)])[:5]
left = df.iloc[:,:14].astype(np.float32)
right = df.iloc[:, 14:].astype(np.int64)
merged = pd.concat([left, right], axis = 1)
merged.to_csv("infer_data.csv", index = False)
```
## 2. Start the Kafka Broker
**Please refer to the README to start the Kafka Broker properly.**
## 3. Wide&Deep Model Demo
```
!rm -r *model
%%writefile wdl_demo.py
import hugectr
from mpi4py import MPI
solver = hugectr.CreateSolver(model_name = "wdl",
max_eval_batches = 5000,
batchsize_eval = 1024,
batchsize = 1024,
lr = 0.001,
vvgpu = [[0]],
i64_input_key = False,
use_mixed_precision = False,
repeat_dataset = False,
use_cuda_graph = True,
kafka_brockers = "10.23.137.25:9093") #Make sure this is consistent with your Kafka broker.)
reader = hugectr.DataReaderParams(data_reader_type = hugectr.DataReaderType_t.Norm,
source = ["criteo_data/file_list."+str(i)+".txt" for i in range(2)],
keyset = ["criteo_data/file_list."+str(i)+".keyset" for i in range(2)],
eval_source = "criteo_data/file_list.2.txt",
check_type = hugectr.Check_t.Sum)
optimizer = hugectr.CreateOptimizer(optimizer_type = hugectr.Optimizer_t.Adam)
hc_config = hugectr.CreateHMemCache(2, 0.5, 0)
etc = hugectr.CreateETC(ps_types = [hugectr.TrainPSType_t.Staged, hugectr.TrainPSType_t.Cached],\
sparse_models = ["./wdl_0_sparse_model", "./wdl_1_sparse_model"],\
local_paths = ["./"], hmem_cache_configs = [hc_config])
model = hugectr.Model(solver, reader, optimizer, etc)
model.add(hugectr.Input(label_dim = 1, label_name = "label",
dense_dim = 13, dense_name = "dense",
data_reader_sparse_param_array =
[hugectr.DataReaderSparseParam("wide_data", 2, True, 1),
hugectr.DataReaderSparseParam("deep_data", 1, True, 26)]))
model.add(hugectr.SparseEmbedding(embedding_type = hugectr.Embedding_t.DistributedSlotSparseEmbeddingHash,
workspace_size_per_gpu_in_mb = 23,
embedding_vec_size = 1,
combiner = "sum",
sparse_embedding_name = "sparse_embedding0",
bottom_name = "wide_data",
optimizer = optimizer))
model.add(hugectr.SparseEmbedding(embedding_type = hugectr.Embedding_t.DistributedSlotSparseEmbeddingHash,
workspace_size_per_gpu_in_mb = 358,
embedding_vec_size = 16,
combiner = "sum",
sparse_embedding_name = "sparse_embedding1",
bottom_name = "deep_data",
optimizer = optimizer))
model.add(hugectr.DenseLayer(layer_type = hugectr.Layer_t.Reshape,
bottom_names = ["sparse_embedding1"],
top_names = ["reshape1"],
leading_dim=416))
model.add(hugectr.DenseLayer(layer_type = hugectr.Layer_t.Reshape,
bottom_names = ["sparse_embedding0"],
top_names = ["reshape2"],
leading_dim=1))
model.add(hugectr.DenseLayer(layer_type = hugectr.Layer_t.Concat,
bottom_names = ["reshape1", "dense"], top_names = ["concat1"]))
model.add(hugectr.DenseLayer(layer_type = hugectr.Layer_t.InnerProduct,
bottom_names = ["concat1"],
top_names = ["fc1"],
num_output=1024))
model.add(hugectr.DenseLayer(layer_type = hugectr.Layer_t.ReLU,
bottom_names = ["fc1"],
top_names = ["relu1"]))
model.add(hugectr.DenseLayer(layer_type = hugectr.Layer_t.Dropout,
bottom_names = ["relu1"],
top_names = ["dropout1"],
dropout_rate=0.5))
model.add(hugectr.DenseLayer(layer_type = hugectr.Layer_t.InnerProduct,
bottom_names = ["dropout1"],
top_names = ["fc2"],
num_output=1024))
model.add(hugectr.DenseLayer(layer_type = hugectr.Layer_t.ReLU,
bottom_names = ["fc2"],
top_names = ["relu2"]))
model.add(hugectr.DenseLayer(layer_type = hugectr.Layer_t.Dropout,
bottom_names = ["relu2"],
top_names = ["dropout2"],
dropout_rate=0.5))
model.add(hugectr.DenseLayer(layer_type = hugectr.Layer_t.InnerProduct,
bottom_names = ["dropout2"],
top_names = ["fc3"],
num_output=1))
model.add(hugectr.DenseLayer(layer_type = hugectr.Layer_t.Add,
bottom_names = ["fc3", "reshape2"],
top_names = ["add1"]))
model.add(hugectr.DenseLayer(layer_type = hugectr.Layer_t.BinaryCrossEntropyLoss,
bottom_names = ["add1", "label"],
top_names = ["loss"]))
model.compile()
model.summary()
model.graph_to_json(graph_config_file = "wdl.json")
#model.save_params_to_files("wdl")
model.fit(num_epochs = 1, display = 500, eval_interval = 1000)
model.set_source(source = ["criteo_data/file_list."+str(i)+".txt" for i in range(3, 5)], \
keyset = ["criteo_data/file_list."+str(i)+".keyset" for i in range(3, 5)], \
eval_source = "criteo_data/file_list.9.txt")
model.save_params_to_files("wdl")
!python wdl_demo.py
```
## 4. WDL Inference
### 4.1 Inference using HugeCTR python API
```
#Create a folder for RocksDB
!mkdir /wdl_infer
!mkdir /wdl_infer/rocksdb
```
**Please make sure you have started Redis cluster following the README before you start doing inference.**
```
%%writefile 'wdl_predict.py'
from hugectr.inference import InferenceParams, CreateInferenceSession
import hugectr
import pandas as pd
import numpy as np
import sys
from mpi4py import MPI
def wdl_inference(model_name='wdl', network_file='wdl.json', dense_file='wdl_dense_0.model', \
embedding_file_list=['wdl_0_sparse_model', 'wdl_1_sparse_model'], data_file='infer_data.csv',\
enable_cache=False, rocksdb_path=""):
CATEGORICAL_COLUMNS=["C1_C2","C3_C4"] + ["C" + str(x) for x in range(1, 27)]
CONTINUOUS_COLUMNS=["I" + str(x) for x in range(1, 14)]
LABEL_COLUMNS = ['label']
test_df=pd.read_csv(data_file,sep=',')
config_file = network_file
row_ptrs = list(range(0, 11, 2)) + list(range(0, 131))
dense_features = list(test_df[CONTINUOUS_COLUMNS].values.flatten())
test_df[CATEGORICAL_COLUMNS].astype(np.int64)
embedding_columns = list((test_df[CATEGORICAL_COLUMNS]).values.flatten())
redisdatabase = hugectr.inference.DistributedDatabaseParams(
hugectr.DatabaseType_t.redis_cluster,
address="127.0.0.1:7000,127.0.0.1:7001,127.0.0.1:7002",
initial_cache_rate=0.2)
rocksdbdatabase = hugectr.inference.PersistentDatabaseParams(
hugectr.DatabaseType_t.rocks_db,
path="/wdl_infer/rocksdb/")
# create parameter server, embedding cache and inference session
inference_params = InferenceParams(model_name = model_name,
max_batchsize = 64,
hit_rate_threshold = 0.5,
dense_model_file = dense_file,
sparse_model_files = embedding_file_list,
device_id = 0,
use_gpu_embedding_cache = enable_cache,
cache_size_percentage = 0.9,
i64_input_key = True,
use_mixed_precision = False,
volatile_db=redisdatabase,
persistent_db=rocksdbdatabase)
inference_session = CreateInferenceSession(config_file, inference_params)
output = inference_session.predict(dense_features, embedding_columns, row_ptrs)
print("WDL multi-embedding table inference result is {}".format(output))
wdl_inference()
!python wdl_predict.py
```
### 4.2 Inference using Triton
**Please refer to the [Triton_Inference.ipynb](./Triton_Inference.ipynb) notebook to start Triton and do the inference.**
## 5. Continue Training WDL Model
```
%%writefile wdl_continue.py
import hugectr
from mpi4py import MPI
solver = hugectr.CreateSolver(model_name = "wdl",
max_eval_batches = 5000,
batchsize_eval = 1024,
batchsize = 1024,
lr = 0.001,
vvgpu = [[0]],
i64_input_key = False,
use_mixed_precision = False,
repeat_dataset = False,
use_cuda_graph = True,
kafka_brockers = "10.23.137.25:9093")
reader = hugectr.DataReaderParams(data_reader_type = hugectr.DataReaderType_t.Norm,
source = ["criteo_data/file_list."+str(i)+".txt" for i in range(6, 9)],
keyset = ["criteo_data/file_list."+str(i)+".keyset" for i in range(6, 9)],
eval_source = "criteo_data/file_list.9.txt",
check_type = hugectr.Check_t.Sum)
optimizer = hugectr.CreateOptimizer(optimizer_type = hugectr.Optimizer_t.Adam)
hc_config = hugectr.CreateHMemCache(2, 0.5, 0)
etc = hugectr.CreateETC(ps_types = [hugectr.TrainPSType_t.Staged, hugectr.TrainPSType_t.Cached],\
sparse_models = ["./wdl_0_sparse_model", "./wdl_1_sparse_model"],\
local_paths = ["./"], hmem_cache_configs = [hc_config])
model = hugectr.Model(solver, reader, optimizer, etc)
model.construct_from_json(graph_config_file = "wdl.json", include_dense_network = True)
model.compile()
model.load_dense_weights("wdl_dense_0_model")
model.load_dense_optimizer_states("dcn_opt_dense_1000.model")
model.summary()
model.graph_to_json(graph_config_file = "wdl.json")
model.fit(num_epochs = 1, display = 500, eval_interval = 1000)
model.dump_incremental_model_2kafka()
model.save_params_to_files("wdl_new")
!python wdl_continue.py
```
## 6. Inference with new model
### 6.1 Continuous inference using Python API
```
!python wdl_predict.py
```
### 6.2 Continuous inference using Triton
**Please refer to the [Triton_Inference.ipynb](./Triton_Inference.ipynb) notebook to do the inference.**
| github_jupyter |
## Observations and Insights
```
# Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
import scipy.stats as st
# Study data files
mouse_metadata_path = "data/Mouse_metadata.csv"
study_results_path = "data/Study_results.csv"
# Read the mouse data and the study results
mouse_metadata = pd.read_csv(mouse_metadata_path)
study_results = pd.read_csv(study_results_path)
# Combine the data into a single dataset
merged_df = pd.merge(study_results, mouse_metadata, how="left", on="Mouse ID")
# Preview of the merged dataset
merged_df.head()
# Checking the number of mice in the DataFrame.
len(merged_df["Mouse ID"].value_counts())
# Getting the duplicate mice by ID number that shows up for Mouse ID and Timepoint.
duplicate_df = merged_df[merged_df.duplicated(subset=["Mouse ID", "Timepoint"], keep=False)]
duplicate_df[["Mouse ID", "Timepoint"]]
# Optional: Get all the data for the duplicate mouse ID.
duplicate_df = merged_df.loc[merged_df["Mouse ID"] == "g989"]
duplicate_df
# Create a clean DataFrame by dropping the duplicate mouse by its ID.
clean_df = merged_df.drop(duplicate_df.index)
clean_df.head()
# Checking the number of mice in the clean DataFrame.
len(clean_df["Mouse ID"].value_counts())
```
## Summary Statistics
```
# Generate a summary statistics table of mean, median, variance, standard deviation, and SEM of the tumor volume for each regimen
# This method is the most straightforward, creating multiple series and putting them all together at the end.
drug_df = clean_df.groupby("Drug Regimen")
# calculating the statistics
mean_arr = drug_df["Tumor Volume (mm3)"].mean()
median_arr = drug_df["Tumor Volume (mm3)"].median()
var_arr = drug_df["Tumor Volume (mm3)"].var()
std_arr = drug_df["Tumor Volume (mm3)"].std()
sem_arr = drug_df["Tumor Volume (mm3)"].sem()
# creating statistic summary dataframe
stats_df = pd.DataFrame({
"Mean Tumor Volume": mean_arr,
"Median Tumor Volume": median_arr,
"Tumor Volume Variance": var_arr,
"Tumor Volume Std. Dev.": std_arr,
"Tumor Volume Std. Err.": sem_arr
})
# show statistic summary
stats_df
# Generate a summary statistics table of mean, median, variance, standard deviation, and SEM of the tumor volume for each regimen
# This method produces everything in a single groupby function.
stats2_df = clean_df.groupby("Drug Regimen").agg({"Tumor Volume (mm3)": ["mean", "median", "var", "std", "sem"]})
stats2_df
```
## Bar Plots
```
# Generate a bar plot showing the number of mice per time point for each treatment throughout the course of the study using pandas.
#creating count dataframe
bar_df = clean_df.groupby("Drug Regimen").count()
#creating bar chart dataframe
bar_df = bar_df.sort_values("Timepoint", ascending=False)
bars_df = bar_df["Timepoint"]
# creating bar chart
graph = bars_df.plot(kind="bar")
graph.set_ylabel("Number of Data Points")
# Generate a bar plot showing the number of mice per time point for each treatment throughout the course of the study using pyplot.
#creating count dataframe
bar_df = clean_df.groupby("Drug Regimen").count()
#creating bar chart dataframe
bar_df = bar_df.sort_values("Timepoint", ascending=False)
bars_df = bar_df["Timepoint"]
plt.bar(bar_df.index, bar_df["Timepoint"])
plt.ylabel("Number of Data Points")
plt.xticks(rotation="vertical")
```
## Pie Plots
```
# Generate a pie plot showing the distribution of female versus male mice using pandas
pie_chart = mouse_metadata["Sex"].value_counts()
pie_chart.plot(kind='pie', subplots=True, autopct="%0.1f%%")
# Generate a pie plot showing the distribution of female versus male mice using pyplot
f_vs_m = mouse_metadata["Sex"].value_counts()
plt.pie(f_vs_m, autopct="%1.1f%%")
```
## Quartiles, Outliers and Boxplots
```
# Calculate the final tumor volume of each mouse across four of the most promising treatment regimens. Calculate the IQR and quantitatively determine if there are any potential outliers.
# returning the max timepoints for Capomulin
temp_df = clean_df.loc[clean_df["Drug Regimen"] == "Capomulin", :]
max_capo = temp_df.groupby("Mouse ID").max()
# returning the max timepoints for Ramicane
temp_df = clean_df.loc[clean_df["Drug Regimen"] == "Ramicane", :]
max_rami = temp_df.groupby("Mouse ID").max()
# returning the max timepoints for Infubinol
temp_df = clean_df.loc[clean_df["Drug Regimen"] == "Infubinol", :]
max_infu = temp_df.groupby("Mouse ID").max()
# returning the max timepoints for Ketapril
temp_df = clean_df.loc[clean_df["Drug Regimen"] == "Ceftamin", :]
max_ceft = temp_df.groupby("Mouse ID").max()
# calculating IQR's
quartiles = max_capo["Tumor Volume (mm3)"].quantile([.25,.5,.75])
capo_iqr = quartiles[0.75] - quartiles[0.25]
lower_bound = quartiles[0.25] - (1.5*capo_iqr)
upper_bound = quartiles[0.75] + (1.5*capo_iqr)
capo_outliers = max_capo["Tumor Volume (mm3)"].loc[(max_capo["Tumor Volume (mm3)"] < lower_bound) | (max_capo["Tumor Volume (mm3)"] > upper_bound)]
quartiles = max_rami["Tumor Volume (mm3)"].quantile([.25,.5,.75])
rami_iqr = quartiles[0.75] - quartiles[0.25]
lower_bound = quartiles[0.25] - (1.5*rami_iqr)
upper_bound = quartiles[0.75] + (1.5*rami_iqr)
rami_outliers = max_rami["Tumor Volume (mm3)"].loc[(max_rami["Tumor Volume (mm3)"] < lower_bound) | (max_rami["Tumor Volume (mm3)"] > upper_bound)]
infu_quartiles = max_infu["Tumor Volume (mm3)"].quantile([.25,.5,.75])
infu_iqr = quartiles[0.75] - infu_quartiles[0.25]
lower_bound_infu = infu_quartiles[0.25] - (1.5*infu_iqr)
upper_bound_infu = infu_quartiles[0.75] + (1.5*infu_iqr)
infu_outliers = max_infu["Tumor Volume (mm3)"].loc[(max_infu["Tumor Volume (mm3)"] <= lower_bound_infu) | (max_infu["Tumor Volume (mm3)"] >= upper_bound_infu)]
quartiles = max_ceft["Tumor Volume (mm3)"].quantile([.25,.5,.75])
ceft_iqr = quartiles[0.75] - quartiles[0.25]
lower_bound = quartiles[0.25] - (1.5*ceft_iqr)
upper_bound = quartiles[0.75] + (1.5*ceft_iqr)
ceft_outliers = max_ceft["Tumor Volume (mm3)"].loc[(max_ceft["Tumor Volume (mm3)"] < lower_bound) | (max_ceft["Tumor Volume (mm3)"] > upper_bound)]
len(infu_outliers)
# Generate a box plot of the final tumor volume of each mouse across four regimens of interest
```
## Line and Scatter Plots
```
# Generate a line plot of time point versus tumor volume for a mouse treated with Capomulin
# Generate a scatter plot of mouse weight versus average tumor volume for the Capomulin regimen
```
## Correlation and Regression
```
# Calculate the correlation coefficient and linear regression model
# for mouse weight and average tumor volume for the Capomulin regimen
```
| github_jupyter |
```
!pip install -q condacolab
import condacolab
condacolab.install()
!conda install -c chembl chembl_structure_pipeline
import chembl_structure_pipeline
from chembl_structure_pipeline import standardizer
from IPython.display import clear_output
# https://www.dgl.ai/pages/start.html
# !pip install dgl
!pip install dgl-cu111 -f https://data.dgl.ai/wheels/repo.html # FOR CUDA VERSION
!pip install dgllife
!pip install rdkit-pypi
!pip install --pre deepchem
!pip install ipython-autotime
!pip install gputil
!pip install psutil
!pip install humanize
%load_ext autotime
clear = clear_output()
import os
from os import path
import statistics
import warnings
import random
import time
import itertools
import psutil
import humanize
import GPUtil as GPU
import subprocess
from datetime import datetime, timedelta
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import tqdm
from tqdm import trange, tqdm_notebook, tnrange
import deepchem as dc
import rdkit
from rdkit import Chem
from rdkit.Chem.MolStandardize import rdMolStandardize
import dgl
from dgl.dataloading import GraphDataLoader
from dgl.nn import GraphConv, SumPooling, MaxPooling
import dgl.function as fn
import dgllife
from dgllife import utils
# embedding
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.autograd import Variable
from torch.optim.lr_scheduler import ReduceLROnPlateau
from torch.profiler import profile, record_function, ProfilerActivity
from torch.utils.tensorboard import SummaryWriter
import sklearn
from sklearn.metrics import (auc, roc_curve, roc_auc_score, average_precision_score,
accuracy_score, ConfusionMatrixDisplay, confusion_matrix, precision_recall_curve,
f1_score, PrecisionRecallDisplay)
from sklearn.ensemble import RandomForestClassifier
warnings.filterwarnings("ignore", message="DGLGraph.__len__")
DGLBACKEND = 'pytorch'
clear
def get_cmd_output(command):
return subprocess.check_output(command,
stderr=subprocess.STDOUT,
shell=True).decode('UTF-8')
```
### Create Dataset
```
def create_dataset(df, name, bonds):
print(f"Creating Dataset and Saving to {drive_path}/data/{name}.pkl")
data = df.sample(frac=1)
data = data.reset_index(drop=True)
data['mol'] = data['smiles'].apply(lambda x: create_dgl_features(x, bonds))
data.to_pickle(f"{drive_path}/data/{name}.pkl")
return data
def featurize_atoms(mol):
feats = []
atom_features = utils.ConcatFeaturizer([
utils.atom_type_one_hot,
utils.atomic_number_one_hot,
utils.atom_degree_one_hot,
utils.atom_explicit_valence_one_hot,
utils.atom_formal_charge_one_hot,
utils.atom_num_radical_electrons_one_hot,
utils.atom_hybridization_one_hot,
utils.atom_is_aromatic_one_hot
])
for atom in mol.GetAtoms():
feats.append(atom_features(atom))
return {'feats': torch.tensor(feats).float()}
def featurize_bonds(mol):
feats = []
bond_features = utils.ConcatFeaturizer([
utils.bond_type_one_hot,
utils.bond_is_conjugated_one_hot,
utils.bond_is_in_ring_one_hot,
utils.bond_stereo_one_hot,
utils.bond_direction_one_hot,
])
for bond in mol.GetBonds():
feats.append(bond_features(bond))
feats.append(bond_features(bond))
return {'edge_feats': torch.tensor(feats).float()}
def create_dgl_features(smiles, bonds):
mol = Chem.MolFromSmiles(smiles)
mol = standardizer.standardize_mol(mol)
if bonds:
dgl_graph = utils.mol_to_bigraph(mol=mol,
node_featurizer=featurize_atoms,
edge_featurizer=featurize_bonds,
canonical_atom_order=True)
else:
dgl_graph = utils.mol_to_bigraph(mol=mol,
node_featurizer=featurize_atoms,
canonical_atom_order=True)
dgl_graph = dgl.add_self_loop(dgl_graph)
return dgl_graph
def load_dataset(dataset, bonds=False, feat='graph', create_new=False):
"""
dataset values: muv, tox21, dude-gpcr
feat values: graph, ecfp
"""
dataset_test_tasks = {
'tox21': ['SR-HSE', 'SR-MMP', 'SR-p53'],
'muv': ['MUV-832', 'MUV-846', 'MUV-852', 'MUV-858', 'MUV-859'],
'dude-gpcr': ['adrb2', 'cxcr4']
}
dataset_original = dataset
if bonds:
dataset = dataset + "_with_bonds"
if path.exists(f"{drive_path}/data/{dataset}_dgl.pkl") and not create_new:
# Load Dataset
print("Reading Pickle")
if feat == 'graph':
data = pd.read_pickle(f"{drive_path}/data/{dataset}_dgl.pkl")
else:
data = pd.read_pickle(f"{drive_path}/data/{dataset}_ecfp.pkl")
else:
# Create Dataset
df = pd.read_csv(f"{drive_path}/data/raw/{dataset_original}.csv")
if feat == 'graph':
data = create_dataset(df, f"{dataset}_dgl", bonds)
else:
data = create_ecfp_dataset(df, f"{dataset}_ecfp")
test_tasks = dataset_test_tasks.get(dataset_original)
drop_cols = test_tasks.copy()
drop_cols.extend(['mol_id', 'smiles', 'mol'])
train_tasks = [x for x in list(data.columns) if x not in drop_cols]
train_dfs = dict.fromkeys(train_tasks)
for task in train_tasks:
df = data[[task, 'mol']].dropna()
df.columns = ['y', 'mol']
# FOR BOND INFORMATION
if with_bonds:
for index, r in df.iterrows():
if r.mol.edata['edge_feats'].shape[-1] < 17:
df.drop(index, inplace=True)
train_dfs[task] = df
for key in train_dfs:
print(key, len(train_dfs[key]))
if feat == 'graph':
feat_length = data.iloc[0].mol.ndata['feats'].shape[-1]
print("Feature Length", feat_length)
if with_bonds:
feat_length = data.iloc[0].mol.edata['edge_feats'].shape[-1]
print("Feature Length", feat_length)
else:
print("Edge Features: ", with_bonds)
test_dfs = dict.fromkeys(test_tasks)
for task in test_tasks:
df = data[[task, 'mol']].dropna()
df.columns = ['y', 'mol']
# FOR BOND INFORMATION
if with_bonds:
for index, r in df.iterrows():
if r.mol.edata['edge_feats'].shape[-1] < 17:
df.drop(index, inplace=True)
test_dfs[task] = df
for key in test_dfs:
print(key, len(test_dfs[key]))
# return data, train_tasks, test_tasks
return train_tasks, train_dfs, test_tasks, test_dfs
```
## Create Episode
```
def create_episode(n_support_pos, n_support_neg, n_query, data, test=False, train_balanced=True):
"""
n_query = per class data points
Xy = dataframe dataset in format [['y', 'mol']]
"""
support = []
query = []
n_query_pos = n_query
n_query_neg = n_query
support_neg = data[data['y'] == 0].sample(n_support_neg)
support_pos = data[data['y'] == 1].sample(n_support_pos)
# organise support by class in array dimensions
support.append(support_neg.to_numpy())
support.append(support_pos.to_numpy())
support = np.array(support, dtype=object)
support_X = [rec[1] for sup_class in support for rec in sup_class]
support_y = np.asarray([rec[0] for sup_class in support for rec in sup_class], dtype=np.float16).flatten()
data = data.drop(support_neg.index)
data = data.drop(support_pos.index)
if len(data[data['y'] == 1]) < n_query:
n_query_pos = len(data[data['y'] == 1])
if test:
# test uses all data remaining
query_neg = data[data['y'] == 0].to_numpy()
query_pos = data[data['y'] == 1].to_numpy()
elif (not test) and train_balanced:
# for balanced queries, same size as support
query_neg = data[data['y'] == 0].sample(n_query_neg).to_numpy()
query_pos = data[data['y'] == 1].sample(n_query_pos).to_numpy()
elif (not test) and (not train_balanced):
# print('test')
query_neg = data[data['y'] == 0].sample(1).to_numpy()
query_pos = data[data['y'] == 1].sample(1).to_numpy()
query_rem = data.sample(n_query*2 - 2)
query_neg_rem = query_rem[query_rem['y'] == 0].to_numpy()
query_pos_rem = query_rem[query_rem['y'] == 1].to_numpy()
query_neg = np.concatenate((query_neg, query_neg_rem))
query_pos = np.concatenate((query_pos, query_pos_rem), axis=0)
query_X = np.concatenate([query_neg[:, 1], query_pos[:, 1]])
query_y = np.concatenate([query_neg[:, 0], query_pos[:, 0]])
return support_X, support_y, query_X, query_y
# task = 'NR-AR'
# df = data[[task, 'mol']]
# df = df.dropna()
# df.columns = ['y', 'mol']
# support_X, support_y, query_X, query_y = create_episode(1, 1, 64, df)
# support_y
# testing
# support = []
# query = []
# support_neg = df[df['y'] == 0].sample(2)
# support_pos = df[df['y'] == 1].sample(2)
# # organise support by class in array dimensions
# support.append(support_neg.to_numpy())
# support.append(support_pos.to_numpy())
# support = np.array(support)
# support.shape
# support[:, :, 1]
```
## Graph Embedding
```
class GCN(nn.Module):
def __init__(self, in_channels, out_channels=128):
super(GCN, self).__init__()
self.conv1 = GraphConv(in_channels, 64)
self.conv2 = GraphConv(64, 128)
self.conv3 = GraphConv(128, 64)
self.sum_pool = SumPooling()
self.dense = nn.Linear(64, out_channels)
def forward(self, graph, in_feat):
h = self.conv1(graph, in_feat)
h = F.relu(h)
graph.ndata['h'] = h
graph.update_all(fn.copy_u('h', 'm'), fn.max('m', 'h'))
h = self.conv2(graph, graph.ndata['h'])
h = F.relu(h)
graph.ndata['h'] = h
graph.update_all(fn.copy_u('h', 'm'), fn.max('m', 'h'))
h = self.conv3(graph, graph.ndata['h'])
h = F.relu(h)
graph.ndata['h'] = h
graph.update_all(fn.copy_u('h', 'm'), fn.max('m', 'h'))
output = self.sum_pool(graph, graph.ndata['h'])
output = torch.tanh(output)
output = self.dense(output)
output = torch.tanh(output)
return output
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.fc1 = nn.Linear(2048, 1000)
self.fc2 = nn.Linear(1000, 500)
self.fc3 = nn.Linear(500, 128)
def forward(self, x):
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = torch.tanh(self.fc3(x))
return x
```
## Distance Function
```
def euclidean_dist(x, y):
# x: N x D
# y: M x D
n = x.size(0)
m = y.size(0)
d = x.size(1)
assert d == y.size(1)
x = x.unsqueeze(1).expand(n, m, d)
y = y.unsqueeze(0).expand(n, m, d)
return torch.pow(x - y, 2).sum(2)
```
### LSTM
```
def cos(x, y):
transpose_shape = tuple(list(range(len(y.shape)))[::-1])
x = x.float()
denom = (
torch.sqrt(torch.sum(torch.square(x)) *
torch.sum(torch.square(y))) + torch.finfo(torch.float32).eps)
return torch.matmul(x, torch.permute(y, transpose_shape)) / denom
class ResiLSTMEmbedding(nn.Module):
def __init__(self, n_support, n_feat=128, max_depth=3):
super(ResiLSTMEmbedding, self).__init__()
self.max_depth = max_depth
self.n_support = n_support
self.n_feat = n_feat
self.support_lstm = nn.LSTMCell(input_size=2*self.n_feat, hidden_size=self.n_feat)
self.q_init = torch.nn.Parameter(torch.zeros((self.n_support, self.n_feat), dtype=torch.float, device="cuda"))
self.support_states_init_h = torch.nn.Parameter(torch.zeros(self.n_support, self.n_feat))
self.support_states_init_c = torch.nn.Parameter(torch.zeros(self.n_support, self.n_feat))
self.query_lstm = nn.LSTMCell(input_size=2*self.n_feat, hidden_size=self.n_feat)
if torch.cuda.is_available():
self.support_lstm = self.support_lstm.cuda()
self.query_lstm = self.query_lstm.cuda()
self.q_init = self.q_init.cuda()
# self.p_init = self.p_init.cuda()
def forward(self, x_support, x_query):
self.p_init = torch.zeros((len(x_query), self.n_feat)).to(device)
self.query_states_init_h = torch.zeros(len(x_query), self.n_feat).to(device)
self.query_states_init_c = torch.zeros(len(x_query), self.n_feat).to(device)
x_support = x_support
x_query = x_query
z_support = x_support
q = self.q_init
p = self.p_init
support_states_h = self.support_states_init_h
support_states_c = self.support_states_init_c
query_states_h = self.query_states_init_h
query_states_c = self.query_states_init_c
for i in range(self.max_depth):
sup_e = cos(z_support + q, x_support)
sup_a = torch.nn.functional.softmax(sup_e, dim=-1)
sup_r = torch.matmul(sup_a, x_support).float()
query_e = cos(x_query + p, z_support)
query_a = torch.nn.functional.softmax(query_e, dim=-1)
query_r = torch.matmul(query_a, z_support).float()
sup_qr = torch.cat((q, sup_r), 1)
support_hidden, support_out = self.support_lstm(sup_qr, (support_states_h, support_states_c))
q = support_hidden
query_pr = torch.cat((p, query_r), 1)
query_hidden, query_out = self.query_lstm(query_pr, (query_states_h, query_states_c))
p = query_hidden
z_support = sup_r
return x_support + q, x_query + p
```
## Protonet
https://colab.research.google.com/drive/1QDYIwg2-iiUpVU8YyAh0lOgFgFPhVgvx#scrollTo=BnLOgECOKG_y
```
class ProtoNet(nn.Module):
def __init__(self, with_bonds=False):
"""
Prototypical Network
"""
super(ProtoNet, self).__init__()
def forward(self, X_support, X_query, n_support_pos, n_support_neg):
n_support = len(X_support)
# prototypes
z_dim = X_support.size(-1) # size of the embedding - 128
z_proto_0 = X_support[:n_support_neg].view(n_support_neg, z_dim).mean(0)
z_proto_1 = X_support[n_support_neg:n_support].view(n_support_pos, z_dim).mean(0)
z_proto = torch.stack((z_proto_0, z_proto_1))
# queries
z_query = X_query
# compute distance
dists = euclidean_dist(z_query, z_proto) # [128, 2]
# compute probabilities
log_p_y = nn.LogSoftmax(dim=1)(-dists) # [128, 2]
return log_p_y
```
## Training Loop
```
def train(train_tasks, train_dfs, balanced_queries, k_pos, k_neg, n_query, episodes, lr):
writer = SummaryWriter()
start_time = time.time()
node_feat_size = 177
embedding_size = 128
encoder = Net()
resi_lstm = ResiLSTMEmbedding(k_pos+k_neg)
proto_net = ProtoNet()
loss_fn = nn.NLLLoss()
if torch.cuda.is_available():
encoder = encoder.cuda()
resi_lstm = resi_lstm.cuda()
proto_net = proto_net.cuda()
loss_fn = loss_fn.cuda()
encoder_optimizer = torch.optim.Adam(encoder.parameters(), lr = lr)
lstm_optimizer = torch.optim.Adam(resi_lstm.parameters(), lr = lr)
# proto_optimizer = torch.optim.Adam(proto_net.parameters(), lr = lr)
# encoder_scheduler = torch.optim.lr_scheduler.StepLR(encoder_optimizer, step_size=1, gamma=0.8)
encoder_scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(encoder_optimizer, patience=300, verbose=False)
lstm_scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(lstm_optimizer, patience=300, verbose=False)
# rn_scheduler = torch.optim.lr_scheduler.StepLR(rn_optimizer, step_size=1, gamma=0.8)
# rn_scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(rn_optimizer, patience=500, verbose=False)
episode_num = 1
early_stop = False
losses = []
running_loss = 0.0
running_acc = 0.0
running_roc = 0.0
running_prc = 0.0
# for task in shuffled_train_tasks:
pbar = trange(episodes, desc=f"Training")
# while episode_num < episodes and not early_stop:
for episode in pbar:
episode_loss = 0.0
# SET TRAINING MODE
encoder.train()
resi_lstm.train()
proto_net.train()
# RANDOMISE ORDER OF TASKS PER EPISODE
shuffled_train_tasks = random.sample(train_tasks, len(train_tasks))
# LOOP OVER TASKS
for task in shuffled_train_tasks:
# CREATE EPISODE FOR TASK
X = train_dfs[task]
X_support, y_support, X_query, y_query = create_episode(k_pos, k_neg, n_query, X, False, balanced_queries)
# TOTAL NUMBER OF QUERIES
total_query = int((y_query == 0).sum() + (y_query == 1).sum())
# ONE HOT QUERY TARGETS
# query_targets = torch.from_numpy(y_query.astype('int'))
# targets = F.one_hot(query_targets, num_classes=2)
target_inds = torch.from_numpy(y_query.astype('float32')).float()
target_inds = target_inds.unsqueeze(1).type(torch.int64)
targets = Variable(target_inds, requires_grad=False).to(device)
if torch.cuda.is_available():
targets=targets.cuda()
n_support = k_pos + k_neg
# flat_support = list(np.concatenate(X_support).flat)
# X = flat_support + list(X_query)
X = X_support + list(X_query)
# CREATE EMBEDDINGS
dataloader = torch.utils.data.DataLoader(X, batch_size=(n_support + total_query), shuffle=False, pin_memory=True)
for graph in dataloader:
graph = graph.to(device)
embeddings = encoder.forward(graph)
# LSTM EMBEDDINGS
emb_support = embeddings[:n_support]
emb_query = embeddings[n_support:]
emb_support, emb_query = resi_lstm(emb_support, emb_query)
# PROTO NETS
logits = proto_net(emb_support, emb_query, k_pos, k_neg)
# loss = loss_fn(logits, torch.max(query_targets, 1)[1])
loss = loss_fn(logits, targets.squeeze())
encoder.zero_grad()
resi_lstm.zero_grad()
proto_net.zero_grad()
loss.backward()
encoder_optimizer.step()
lstm_optimizer.step()
_, y_hat = logits.max(1)
# class_indices = torch.max(query_targets, 1)[1]
targets = targets.squeeze().cpu()
y_hat = y_hat.squeeze().detach().cpu()
roc = roc_auc_score(targets, y_hat)
prc = average_precision_score(targets, y_hat)
acc = accuracy_score(targets, y_hat)
# proto_optimizer.step()
# EVALUATE TRAINING LOOP ON TASK
episode_loss += loss.item()
running_loss += loss.item()
running_acc += acc
running_roc += roc
running_prc += prc
pbar.set_description(f"Episode {episode_num} - Loss {loss.item():.6f} - Acc {acc:.4f} - LR {encoder_optimizer.param_groups[0]['lr']}")
pbar.refresh()
losses.append(episode_loss / len(train_tasks))
writer.add_scalar('Loss/train', episode_loss / len(train_tasks), episode_num)
if encoder_optimizer.param_groups[0]['lr'] < 0.000001:
break # EARLY STOP
elif episode_num < episodes:
episode_num += 1
encoder_scheduler.step(loss)
lstm_scheduler.step(loss)
epoch_loss = running_loss / (episode_num*len(train_tasks))
epoch_acc = running_acc / (episode_num*len(train_tasks))
epoch_roc = running_roc / (episode_num*len(train_tasks))
epoch_prc = running_prc / (episode_num*len(train_tasks))
print(f'Loss: {epoch_loss:.5f} Acc: {epoch_acc:.4f} ROC: {epoch_roc:.4f} PRC: {epoch_prc:.4f}')
end_time = time.time()
train_info = {
"losses": losses,
"duration": str(timedelta(seconds=(end_time - start_time))),
"episodes": episode_num,
"train_roc": epoch_roc,
"train_prc": epoch_prc
}
return encoder, resi_lstm, proto_net, train_info
```
## Testing Loop
```
def test(encoder, lstm, proto_net, test_tasks, test_dfs, k_pos, k_neg, rounds):
encoder.eval()
lstm.eval()
proto_net.eval()
test_info = {}
with torch.no_grad():
for task in test_tasks:
Xy = test_dfs[task]
running_loss = []
running_acc = []
running_roc = [0]
running_prc = [0]
running_preds = []
running_targets = []
running_actuals = []
for round in trange(rounds):
X_support, y_support, X_query, y_query = create_episode(k_pos, k_neg, n_query=0, data=Xy, test=True, train_balanced=False)
total_query = int((y_query == 0).sum() + (y_query == 1).sum())
n_support = k_pos + k_neg
# flat_support = list(np.concatenate(X_support).flat)
# X = flat_support + list(X_query)
X = X_support + list(X_query)
# CREATE EMBEDDINGS
dataloader = torch.utils.data.DataLoader(X, batch_size=(n_support + total_query), shuffle=False, pin_memory=True)
for graph in dataloader:
graph = graph.to(device)
embeddings = encoder.forward(graph)
# LSTM EMBEDDINGS
emb_support = embeddings[:n_support]
emb_query = embeddings[n_support:]
emb_support, emb_query = lstm(emb_support, emb_query)
# PROTO NETS
logits = proto_net(emb_support, emb_query, k_pos, k_neg)
# PRED
_, y_hat_actual = logits.max(1)
y_hat = logits[:, 1]
# targets = targets.squeeze().cpu()
target_inds = torch.from_numpy(y_query.astype('float32')).float()
target_inds = target_inds.unsqueeze(1).type(torch.int64)
targets = Variable(target_inds, requires_grad=False)
y_hat = y_hat.squeeze().detach().cpu()
roc = roc_auc_score(targets, y_hat)
prc = average_precision_score(targets, y_hat)
# acc = accuracy_score(targets, y_hat)
running_preds.append(y_hat)
running_actuals.append(y_hat_actual)
running_targets.append(targets)
# running_acc.append(acc)
running_roc.append(roc)
running_prc.append(prc)
median_index = running_roc.index(statistics.median(running_roc))
if median_index == rounds:
median_index = median_index - 1
chart_preds = running_preds[median_index]
chart_actuals = running_actuals[median_index].detach().cpu()
chart_targets = running_targets[median_index]
c_auc = roc_auc_score(chart_targets, chart_preds)
c_fpr, c_tpr, _ = roc_curve(chart_targets, chart_preds)
plt.plot(c_fpr, c_tpr, marker='.', label = 'AUC = %0.2f' % c_auc)
plt.plot([0, 1], [0, 1],'r--', label='No Skill')
# plt.plot([0, 0, 1], [0, 1, 1], 'g--', label='Perfect Classifier')
plt.title('Receiver Operating Characteristic')
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.legend(loc = 'best')
plt.savefig(f"{drive_path}/{method_dir}/graphs/roc_{dataset}_{task}_ecfp_pos{n_pos}_neg{n_neg}.png")
plt.figure().clear()
# prc_graph = PrecisionRecallDisplay.from_predictions(chart_targets, chart_preds)
c_precision, c_recall, _ = precision_recall_curve(chart_targets, chart_preds)
plt.title('Precision Recall Curve')
# plt.plot([0, 1], [0, 0], 'r--', label='No Skill')
no_skill = len(chart_targets[chart_targets==1]) / len(chart_targets)
plt.plot([0, 1], [no_skill, no_skill], linestyle='--', label='No Skill')
# plt.plot([0, 1, 1], [1, 1, 0], 'g--', label='Perfect Classifier')
plt.plot(c_recall, c_precision, marker='.', label = 'AUC = %0.2f' % auc(c_recall, c_precision))
plt.xlabel('Recall')
plt.ylabel('Precision')
plt.legend(loc = 'best')
plt.savefig(f"{drive_path}/{method_dir}/graphs/prc_{dataset}_{task}_ecfp_pos{n_pos}_neg{n_neg}.png")
plt.figure().clear()
cm = ConfusionMatrixDisplay.from_predictions(chart_targets, chart_actuals)
plt.title('Confusion Matrix')
plt.savefig(f"{drive_path}/{method_dir}/graphs/cm_{dataset}_{task}_ecfp_pos{n_pos}_neg{n_neg}.png")
plt.figure().clear()
running_roc.pop(0) # remove the added 0
running_prc.pop(0) # remove the added 0
# round_acc = f"{statistics.mean(running_acc):.3f} \u00B1 {statistics.stdev(running_acc):.3f}"
round_roc = f"{statistics.mean(running_roc):.3f} \u00B1 {statistics.stdev(running_roc):.3f}"
round_prc = f"{statistics.mean(running_prc):.3f} \u00B1 {statistics.stdev(running_prc):.3f}"
test_info[task] = {
# "acc": round_acc,
"roc": round_roc,
"prc": round_prc,
"roc_values": running_roc,
"prc_values": running_prc
}
print(f'Test {task}')
# print(f"Acc: {round_acc}")
print(f"ROC: {round_roc}")
print(f"PRC: {round_prc}")
return targets, y_hat, test_info
```
## Initiate Training and Testing
```
from google.colab import drive
drive.mount('/content/drive')
# PATHS
drive_path = "/content/drive/MyDrive/Colab Notebooks/MSC_21"
method_dir = "ProtoNets"
log_path = f"{drive_path}/{method_dir}/logs/"
# PARAMETERS
dataset = 'tox21'
with_bonds = False
test_rounds = 20
n_query = 64 # per class
episodes = 10000
lr = 0.001
balanced_queries = True
#FOR DETERMINISTIC REPRODUCABILITY
randomseed = 12
torch.manual_seed(randomseed)
np.random.seed(randomseed)
random.seed(randomseed)
torch.cuda.manual_seed(randomseed)
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
torch.backends.cudnn.is_available()
torch.backends.cudnn.benchmark = False # selects fastest conv algo
torch.backends.cudnn.deterministic = True
# LOAD DATASET
# data, train_tasks, test_tasks = load_dataset(dataset, bonds=with_bonds, create_new=False)
train_tasks, train_dfs, test_tasks, test_dfs = load_dataset(dataset, bonds=with_bonds, feat='ecfp', create_new=False)
combinations = [
[10, 10],
[5, 10],
[1, 10],
[1, 5],
[1, 1]
]
# worksheet = gc.open_by_url("https://docs.google.com/spreadsheets/d/1K15Rx4IZqiLgjUsmMq0blB-WB16MDY-ENR2j8z7S6Ss/edit#gid=0").sheet1
cols = [
'DATE', 'CPU', 'CPU COUNT', 'GPU', 'GPU RAM', 'RAM', 'CUDA',
'REF', 'DATASET', 'ARCHITECTURE',
'SPLIT', 'TARGET', 'ACCURACY', 'ROC', 'PRC',
'ROC_VALUES', 'PRC_VALUES',
'TRAIN ROC', 'TRAIN PRC', 'EPISODES', 'TRAINING TIME'
]
load_from_saved = False
for comb in combinations:
n_pos = comb[0]
n_neg = comb[1]
results = pd.DataFrame(columns=cols)
print(f"\nRUNNING {n_pos}+/{n_neg}-")
if load_from_saved:
encoder = GCN(177, 128)
lstm = ResiLSTMEmbedding(n_pos+n_neg)
proto_net = ProtoNet()
encoder.load_state_dict(torch.load(f"{drive_path}/{method_dir}/{dataset}_ecfp_encoder_pos{n_pos}_neg{n_neg}.pt"))
lstm.load_state_dict(torch.load(f"{drive_path}/{method_dir}/{dataset}_ecfp__lstm_pos{n_pos}_neg{n_neg}.pt"))
proto_net.load_state_dict(torch.load(f"{drive_path}/{method_dir}/{dataset}_ecfp__proto_pos{n_pos}_neg{n_neg}.pt"))
encoder.to(device)
lstm.to(device)
proto_net.to(device)
else:
encoder, lstm, proto_net, train_info = train(train_tasks, train_dfs, balanced_queries, n_pos, n_neg, n_query, episodes, lr)
if with_bonds:
torch.save(encoder.state_dict(), f"{drive_path}/{method_dir}/{dataset}_ecfp__encoder_pos{n_pos}_neg{n_neg}_bonds.pt")
torch.save(lstm.state_dict(), f"{drive_path}/{method_dir}/{dataset}_ecfp__lstm_pos{n_pos}_neg{n_neg}_bonds.pt")
torch.save(proto_net.state_dict(), f"{drive_path}/{method_dir}/{dataset}_ecfp__proto_pos{n_pos}_neg{n_neg}_bonds.pt")
else:
torch.save(encoder.state_dict(), f"{drive_path}/{method_dir}/{dataset}_ecfp__encoder_pos{n_pos}_neg{n_neg}.pt")
torch.save(lstm.state_dict(), f"{drive_path}/{method_dir}/{dataset}_ecfp__lstm_pos{n_pos}_neg{n_neg}.pt")
torch.save(proto_net.state_dict(), f"{drive_path}/{method_dir}/{dataset}_ecfp__proto_pos{n_pos}_neg{n_neg}.pt")
loss_plot = plt.plot(train_info['losses'])[0]
loss_plot.figure.savefig(f"{drive_path}/{method_dir}/loss_plots/{dataset}_ecfp__pos{n_pos}_neg{n_neg}.png")
plt.figure().clear()
targets, preds, test_info = test(encoder, lstm, proto_net, test_tasks, test_dfs, n_pos, n_neg, test_rounds)
dt_string = datetime.now().strftime("%d/%m/%Y %H:%M:%S")
cpu = get_cmd_output('cat /proc/cpuinfo | grep -E "model name"')
cpu = cpu.split('\n')[0].split('\t: ')[-1]
cpu_count = psutil.cpu_count()
cuda_version = get_cmd_output('nvcc --version | grep -E "Build"')
gpu = get_cmd_output("nvidia-smi -L")
general_ram_gb = humanize.naturalsize(psutil.virtual_memory().available)
gpu_ram_total_mb = GPU.getGPUs()[0].memoryTotal
for target in test_info:
if load_from_saved:
rec = pd.DataFrame([[dt_string, cpu, cpu_count, gpu, gpu_ram_total_mb, general_ram_gb, cuda_version, "MSC",
dataset, {method_dir}, f"{n_pos}+/{n_neg}-", target, 0, test_info[target]['roc'], test_info[target]['prc'],
test_info[target]['roc_values'], test_info[target]['prc_values'],
99, 99, 99, 102]], columns=cols)
results = pd.concat([results, rec])
else:
rec = pd.DataFrame([[dt_string, cpu, cpu_count, gpu, gpu_ram_total_mb, general_ram_gb, cuda_version, "MSC",
dataset, {method_dir}, f"{n_pos}+/{n_neg}-", target, 0, test_info[target]['roc'], test_info[target]['prc'],
test_info[target]['roc_values'], test_info[target]['prc_values'],
train_info["train_roc"], train_info["train_prc"], train_info["episodes"], train_info["duration"]
]], columns=cols)
results = pd.concat([results, rec])
if load_from_saved:
results.to_csv(f"{drive_path}/results/{dataset}_{method_dir}_ecfp_pos{n_pos}_neg{n_neg}_from_saved.csv", index=False)
else:
results.to_csv(f"{drive_path}/results/{dataset}_{method_dir}_ecfp_pos{n_pos}_neg{n_neg}.csv", index=False)
```
| github_jupyter |
# Normalize text
```
herod_fp = '/Users/kyle/cltk_data/greek/text/tlg/plaintext/TLG0016.txt'
with open(herod_fp) as fo:
herod_raw = fo.read()
print(herod_raw[2000:2500]) # What do we notice needs help?
from cltk.corpus.utils.formatter import tlg_plaintext_cleanup
herod_clean = tlg_plaintext_cleanup(herod_raw, rm_punctuation=True, rm_periods=False)
print(herod_clean[2000:2500])
```
# Tokenize sentences
```
from cltk.tokenize.sentence import TokenizeSentence
tokenizer = TokenizeSentence('greek')
herod_sents = tokenizer.tokenize_sentences(herod_clean)
print(herod_sents[:5])
for sent in herod_sents:
print(sent)
print()
input()
```
# Make word tokens
```
from cltk.tokenize.word import nltk_tokenize_words
for sent in herod_sents:
words = nltk_tokenize_words(sent)
print(words)
input()
```
### Tokenize Latin enclitics
```
from cltk.corpus.utils.formatter import phi5_plaintext_cleanup
from cltk.tokenize.word import WordTokenizer
# 'LAT0474': 'Marcus Tullius Cicero, Cicero, Tully',
cicero_fp = '/Users/kyle/cltk_data/latin/text/phi5/plaintext/LAT0474.TXT'
with open(cicero_fp) as fo:
cicero_raw = fo.read()
cicero_clean = phi5_plaintext_cleanup(cicero_raw, rm_punctuation=True, rm_periods=False) # ~5 sec
print(cicero_clean[400:600])
sent_tokenizer = TokenizeSentence('latin')
cicero_sents = tokenizer.tokenize_sentences(cicero_clean)
print(cicero_sents[:3])
word_tokenizer = WordTokenizer('latin') # Patrick's tokenizer
for sent in cicero_sents:
#words = nltk_tokenize_words(sent)
sub_words = word_tokenizer.tokenize(sent)
print(sub_words)
input()
```
# POS Tagging
```
from cltk.tag.pos import POSTag
tagger = POSTag('greek')
# Heordotus again
for sent in herod_sents:
tagged_text = tagger.tag_unigram(sent)
print(tagged_text)
input()
```
# NER
```
## Latin -- decent, but see M, P, etc
from cltk.tag import ner
# Heordotus again
for sent in cicero_sents:
ner_tags = ner.tag_ner('latin', input_text=sent, output_type=list)
print(ner_tags)
input()
# Greek -- not as good!
from cltk.tag import ner
# Heordotus again
for sent in herod_sents:
ner_tags = ner.tag_ner('greek', input_text=sent, output_type=list)
print(ner_tags)
input()
```
# Stopword filtering
```
from cltk.stop.greek.stops import STOPS_LIST
#p = PunktLanguageVars()
for sent in herod_sents:
words = nltk_tokenize_words(sent)
print('W/ STOPS', words)
words = [w for w in words if not w in STOPS_LIST]
print('W/O STOPS', words)
input()
```
# Concordance
```
from cltk.utils.philology import Philology
p = Philology()
herod_fp = '/Users/kyle/cltk_data/greek/text/tlg/plaintext/TLG0016.txt'
p.write_concordance_from_file(herod_fp, 'kyle_herod')
```
# Word count
```
from nltk.text import Text
words = nltk_tokenize_words(herod_clean)
print(words[:15])
t = Text(words)
vocabulary_count = t.vocab()
vocabulary_count['ἱστορίης']
vocabulary_count['μήτε']
vocabulary_count['ἀνθρώπων']
```
# Word frequency
```
from cltk.utils.frequency import Frequency
freq = Frequency()
herod_frequencies = freq.counter_from_str(herod_clean)
herod_frequencies.most_common()
```
# Lemmatizing
| github_jupyter |
```
import pandas as pd
import numpy as np
# set the column names
colnames=['price', 'year_model', 'mileage', 'fuel_type', 'mark', 'model', 'fiscal_power', 'sector', 'type', 'city']
# read the csv file as a dataframe
df = pd.read_csv("./data/output.csv", sep=",", names=colnames, header=None)
# let's get some simple vision on our dataset
df.head()
# remove thos rows doesn't contain the price value
df = df[df.price.str.contains("DH") == True]
# remove the 'DH' caracters from the price
df.price = df.price.map(lambda x: x.rstrip('DH'))
# remove the space on it
df.price = df.price.str.replace(" ","")
# change it to integer value
df.price = pd.to_numeric(df.price, errors = 'coerce', downcast= 'integer')
# remove thos rows doesn't contain the year_model value
df = df[df.year_model.str.contains("Année-Modèle") == True]
# remove the 'Année-Modèle:' from the year_model
df.year_model = df.year_model.map(lambda x: x.lstrip('Année-Modèle:').rstrip('ou plus ancien'))
# df.year_model = df.year_model.map(lambda x: x.lstrip('Plus de '))
# remove those lines having the year_model not set
df = df[df.year_model != ' -']
df = df[df.year_model != '']
# change it to integer value
df.year_model = pd.to_numeric(df.year_model, errors = 'coerce', downcast = 'integer')
# remove thos rows doesn't contain the year_model value
df = df[df.mileage.str.contains("Kilométrage") == True]
# remove the 'Kilométrage:' string from the mileage feature
df.mileage = df.mileage.map(lambda x: x.lstrip('Kilométrage:'))
df.mileage = df.mileage.map(lambda x: x.lstrip('Plus de '))
# remove those lines having the mileage values null or '-'
df = df[df.mileage != '-']
# we have only one value type that is equal to 500 000, all the other ones contain two values
if any(df.mileage != '500 000'):
# create two columns minim and maxim to calculate the mileage mean
df['minim'], df['maxim'] = df.mileage.str.split('-', 1).str
# remove spaces from the maxim & minim values
df['maxim'] = df.maxim.str.replace(" ","")
df['minim'] = df.minim.str.replace(" ","")
df['maxim'] = df['maxim'].replace(np.nan, 500000)
# calculate the mean of mileage
df.mileage = df.apply(lambda row: (int(row.minim) + int(row.maxim)) / 2, axis=1)
# now that the mileage is calculated so we do not need the minim and maxim values anymore
df = df.drop(columns=['minim', 'maxim'])
```
#### Fuel type
```
# remove the 'Type de carburant:' string from the carburant_type feature
df.fuel_type = df.fuel_type.map(lambda x: x.lstrip('Type de carburant:'))
```
#### Mark & Model
```
# remove the 'Marque:' string from the mark feature
df['mark'] = df['mark'].map(lambda x: x.replace('Marque:', ''))
df = df[df.mark != '-']
# remove the 'Modèle:' string from model feature
df['model'] = df['model'].map(lambda x: x.replace('Modèle:', ''))
```
#### fiscal power
For the fiscal power we can see that there is exactly 5728 rows not announced, so we will fill them by the mean of the other columns, since it is an important feature in cars price prediction so we can not drop it.
```
# remove the 'Puissance fiscale:' from the fiscal_power feature
df.fiscal_power = df.fiscal_power.map(lambda x: x.lstrip('Puissance fiscale:Plus de').rstrip(' CV'))
# replace the - with NaN values and convert them to integer values
df.fiscal_power = df.fiscal_power.str.replace("-","0")
# convert all fiscal_power values to numerical ones
df.fiscal_power = pd.to_numeric(df.fiscal_power, errors = 'coerce', downcast= 'integer')
# now we need to fill those 0 values with the mean of all fiscal_power columns
df.fiscal_power = df.fiscal_power.map( lambda x : df.fiscal_power.mean() if x == 0 else x )
```
#### fuel type
```
# remove those lines having the fuel_type not set
df = df[df.fuel_type != '-']
```
#### drop unwanted columns
```
df = df.drop(columns=['sector', 'type'])
df = df[['price', 'year_model', 'mileage', 'fiscal_power', 'fuel_type', 'mark']]
df.to_csv('data/car_dataset.csv')
df.head()
from car_price.wsgi import application
from api.models import Car
for x in df.values[5598:]:
car = Car(
price=x[0],
year_model=x[1],
mileage=x[2],
fiscal_power=x[3],
fuel_type=x[4],
mark=x[5]
)
car.save()
Car.objects.all().count()
df.shape
```
| github_jupyter |
# Credit Risk Classification
Credit risk poses a classification problem that’s inherently imbalanced. This is because healthy loans easily outnumber risky loans. In this Challenge, you’ll use various techniques to train and evaluate models with imbalanced classes. You’ll use a dataset of historical lending activity from a peer-to-peer lending services company to build a model that can identify the creditworthiness of borrowers.
## Instructions:
This challenge consists of the following subsections:
* Split the Data into Training and Testing Sets
* Create a Logistic Regression Model with the Original Data
* Predict a Logistic Regression Model with Resampled Training Data
### Split the Data into Training and Testing Sets
Open the starter code notebook and then use it to complete the following steps.
1. Read the `lending_data.csv` data from the `Resources` folder into a Pandas DataFrame.
2. Create the labels set (`y`) from the “loan_status” column, and then create the features (`X`) DataFrame from the remaining columns.
> **Note** A value of `0` in the “loan_status” column means that the loan is healthy. A value of `1` means that the loan has a high risk of defaulting.
3. Check the balance of the labels variable (`y`) by using the `value_counts` function.
4. Split the data into training and testing datasets by using `train_test_split`.
### Create a Logistic Regression Model with the Original Data
Employ your knowledge of logistic regression to complete the following steps:
1. Fit a logistic regression model by using the training data (`X_train` and `y_train`).
2. Save the predictions on the testing data labels by using the testing feature data (`X_test`) and the fitted model.
3. Evaluate the model’s performance by doing the following:
* Calculate the accuracy score of the model.
* Generate a confusion matrix.
* Print the classification report.
4. Answer the following question: How well does the logistic regression model predict both the `0` (healthy loan) and `1` (high-risk loan) labels?
### Predict a Logistic Regression Model with Resampled Training Data
Did you notice the small number of high-risk loan labels? Perhaps, a model that uses resampled data will perform better. You’ll thus resample the training data and then reevaluate the model. Specifically, you’ll use `RandomOverSampler`.
To do so, complete the following steps:
1. Use the `RandomOverSampler` module from the imbalanced-learn library to resample the data. Be sure to confirm that the labels have an equal number of data points.
2. Use the `LogisticRegression` classifier and the resampled data to fit the model and make predictions.
3. Evaluate the model’s performance by doing the following:
* Calculate the accuracy score of the model.
* Generate a confusion matrix.
* Print the classification report.
4. Answer the following question: How well does the logistic regression model, fit with oversampled data, predict both the `0` (healthy loan) and `1` (high-risk loan) labels?
### Write a Credit Risk Analysis Report
For this section, you’ll write a brief report that includes a summary and an analysis of the performance of both machine learning models that you used in this challenge. You should write this report as the `README.md` file included in your GitHub repository.
Structure your report by using the report template that `Starter_Code.zip` includes, and make sure that it contains the following:
1. An overview of the analysis: Explain the purpose of this analysis.
2. The results: Using bulleted lists, describe the balanced accuracy scores and the precision and recall scores of both machine learning models.
3. A summary: Summarize the results from the machine learning models. Compare the two versions of the dataset predictions. Include your recommendation for the model to use, if any, on the original vs. the resampled data. If you don’t recommend either model, justify your reasoning.
```
# Import the modules
import numpy as np
import pandas as pd
from pathlib import Path
from sklearn.metrics import balanced_accuracy_score
from sklearn.metrics import confusion_matrix
from imblearn.metrics import classification_report_imbalanced
import warnings
warnings.filterwarnings('ignore')
```
---
## Split the Data into Training and Testing Sets
### Step 1: Read the `lending_data.csv` data from the `Resources` folder into a Pandas DataFrame.
```
# Read the CSV file from the Resources folder into a Pandas DataFrame
lending_data_df = pd.read_csv(Path("../Starter_Code/Resources/lending_data.csv"))
# Review the DataFrame
display(lending_data_df.head())
```
### Step 2: Create the labels set (`y`) from the “loan_status” column, and then create the features (`X`) DataFrame from the remaining columns.
```
# Separate the data into labels and features
# Separate the y variable, the labels
y = lending_data_df["loan_status"]
# Separate the X variable, the features
X = lending_data_df.drop(columns=["loan_status"])
# Review the y variable Series
y.head()
# Review the X variable DataFrame
X.head()
```
### Step 3: Check the balance of the labels variable (`y`) by using the `value_counts` function.
```
# Check the balance of our target values
y.value_counts()
```
### Step 4: Split the data into training and testing datasets by using `train_test_split`.
```
# Import the train_test_learn module
from sklearn.model_selection import train_test_split
# Split the data using train_test_split
# Assign a random_state of 1 to the function
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1)
```
---
## Create a Logistic Regression Model with the Original Data
### Step 1: Fit a logistic regression model by using the training data (`X_train` and `y_train`).
```
# Import the LogisticRegression module from SKLearn
from sklearn.linear_model import LogisticRegression
# Instantiate the Logistic Regression model
# Assign a random_state parameter of 1 to the model
model = LogisticRegression(random_state=1)
# Fit the model using training data
model.fit(X_train, y_train)
```
### Step 2: Save the predictions on the testing data labels by using the testing feature data (`X_test`) and the fitted model.
```
# Make a prediction using the testing data
y_pred = model.predict(X_test)
y_pred
```
### Step 3: Evaluate the model’s performance by doing the following:
* Calculate the accuracy score of the model.
* Generate a confusion matrix.
* Print the classification report.
```
# Print the balanced_accuracy score of the model
BAS = balanced_accuracy_score(y_test, y_pred)
print(BAS)
# Generate a confusion matrix for the model
print(confusion_matrix(y_test, y_pred))
# Print the classification report for the model
print(classification_report_imbalanced(y_test, y_pred))
```
### Step 4: Answer the following question.
**Question:** How well does the logistic regression model predict both the `0` (healthy loan) and `1` (high-risk loan) labels?
**Answer:** The regression models predicts both healthy loans and high-risk loans, for the most part, accurately. We have an average 99% for our F1 score, the summary statistic for both the precision and recall of the data. Although, there is some room for improvement for healthy loans for our PPV (positive predictive value) and recall.
---
## Predict a Logistic Regression Model with Resampled Training Data
### Step 1: Use the `RandomOverSampler` module from the imbalanced-learn library to resample the data. Be sure to confirm that the labels have an equal number of data points.
```
# Import the RandomOverSampler module form imbalanced-learn
from imblearn.over_sampling import RandomOverSampler
# Instantiate the random oversampler model
# # Assign a random_state parameter of 1 to the model
random_oversampler = RandomOverSampler(random_state=1)
# Fit the original training data to the random_oversampler model
X_resampled, y_resampled = random_oversampler.fit_resample(X_train, y_train)
# Count the distinct values of the resampled labels data
y_resampled.value_counts()
```
### Step 2: Use the `LogisticRegression` classifier and the resampled data to fit the model and make predictions.
```
# Instantiate the Logistic Regression model
# Assign a random_state parameter of 1 to the model
resampled_model = LogisticRegression(random_state=1)
# Fit the model using the resampled training data
resampled_model.fit(X_resampled, y_resampled)
# Make a prediction using the testing data
y_pred = resampled_model.predict(X_test)
```
### Step 3: Evaluate the model’s performance by doing the following:
* Calculate the accuracy score of the model.
* Generate a confusion matrix.
* Print the classification report.
```
# Print the balanced_accuracy score of the model
print(balanced_accuracy_score(y_test, y_pred))
# Generate a confusion matrix for the model
confusion_matrix(y_test, y_pred)
# Print the classification report for the model
print(classification_report_imbalanced(y_test, y_pred))
```
### Step 4: Answer the following question
**Question:** How well does the logistic regression model, fit with oversampled data, predict both the `0` (healthy loan) and `1` (high-risk loan) labels?
**Answer:** The logistic regression model, fit with oversampled data, predicts both the healthy loans and high-risk loans pretty accurately. We have an F1 score of 99%, which summarizes the precision and recall. Again, there's room for improvement for the high-risk loan portion in terms of precision. But for the most part, the model predicts the labels of both loans accurately.
| github_jupyter |
```
import glob
import os
import random
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import cv2
import math
from tqdm.auto import tqdm
from sklearn import linear_model
import optuna
import seaborn as sns
FEAT_OOFS = [
{
'model' : 'feat_lasso',
'fn' : '../output/2021011_segmentation_feature_model_v4/feature_model_oofs_0.csv'
},
{
'model' : 'feat_linreg',
'fn' : '../output/2021011_segmentation_feature_model_v4/feature_model_oofs_1.csv'
},
{
'model' : 'feat_ridge',
'fn' : '../output/2021011_segmentation_feature_model_v4/feature_model_oofs_2.csv',
}
]
CNN_OOFS = [
{
'model' : 'resnet50_rocstar',
'fn' : '../output/resnet50_bs32_ep10_rocstar_lr0.0001_ps0.8_ranger_sz256/'
},
{
'model' : 'resnet50_bce',
'fn' : '../output/resnet50_bs32_ep10_bce_lr0.0001_ps0.8_ranger_sz256/'
},
{
'model' : 'densenet169_rocstar',
'fn' : '../output/densenet169_bs32_ep10_rocstar_lr0.0001_ps0.8_ranger_sz256/'
},
{
'model' : 'resnet101_rocstar',
'fn' : '../output/resnet101_bs32_ep20_rocstar_lr0.0001_ps0.8_ranger_sz256/'
},
{
'model' : 'efficientnetv2_l_rocstar',
'fn' : '../output/tf_efficientnetv2_l_bs32_ep10_rocstar_lr0.0001_ps0.8_ranger_sz256/'
},
]
df = pd.read_csv('../output/20210925_segmentation_feature_model_v3/feature_model_oofs_0.csv')[
['BraTS21ID','MGMT_value','fold']]
df.head()
def read_feat_oof(fn):
return pd.read_csv(fn).sort_values('BraTS21ID')['oof_pred'].values
def read_cnn_oof(dir_path):
oof_fns = [os.path.join(dir_path, f'fold-{i}', 'oof.csv') for i in range(5)]
dfs = []
for fn in oof_fns:
dfs.append(pd.read_csv(fn))
df = pd.concat(dfs)
return df.sort_values('BraTS21ID')['pred_mgmt_tta'].values
def normalize_pred_distribution(preds, min_percentile=10, max_percentile=90):
""" Clips min and max percentiles and Z-score normalizes """
min_range = np.percentile(preds, min_percentile)
max_range = np.percentile(preds, max_percentile)
norm_preds = np.clip(preds, min_range, max_range)
pred_std = np.std(norm_preds)
pred_mean = np.mean(norm_preds)
norm_preds = (norm_preds - pred_mean) / (pred_std + 1e-6)
return norm_preds
def rescale_pred_distribution(preds):
""" Rescales pred distribution to 0-1 range. Doesn't affect AUC """
return (preds - np.min(preds)) / (np.max(preds) - np.min(preds) + 1e-6)
for d in FEAT_OOFS:
df[d['model']] = read_feat_oof(d['fn'])
for d in CNN_OOFS:
df[d['model']] = read_cnn_oof(d['fn'])
df_norm = df.copy()
for feat in df.columns.to_list()[3:]:
df_norm[feat] = rescale_pred_distribution(
normalize_pred_distribution(df_norm[feat].values)
)
df_norm.head()
df_raw = df.copy()
all_feat_names = df_norm.columns.to_list()[3:]
corr = df_norm[['MGMT_value'] + all_feat_names].corr()
# Generate a mask for the upper triangle
mask = np.triu(np.ones_like(corr, dtype=bool))
plt.close('all')
f, ax = plt.subplots(figsize=(5, 5))
cmap = sns.diverging_palette(230, 20, as_cmap=True)
sns.heatmap(corr, mask=mask, cmap=cmap, vmax=.3, center=0,
square=True, linewidths=.5, cbar_kws={"shrink": .5})
plt.title('OOF pred correlations')
plt.show()
mgmt_corr_sorted = corr['MGMT_value'].sort_values()
mgmt_corr_sorted
```
## Average
```
from sklearn.metrics import accuracy_score, roc_auc_score
from sklearn.preprocessing import StandardScaler
oof_preds = np.mean(df_norm[all_feat_names].to_numpy(),1)
oof_gts = df_norm['MGMT_value']
cv_preds = [np.mean(df_norm[df_norm.fold==fold][all_feat_names].to_numpy(),1) for fold in range(5)]
cv_gts = [df_norm[df_norm.fold==fold]['MGMT_value'] for fold in range(5)]
oof_acc = accuracy_score((np.array(oof_gts) > 0.5).flatten(), (np.array(oof_preds) > 0.5).flatten())
oof_auc = roc_auc_score(np.array(oof_gts).flatten().astype(np.float32), np.array(oof_preds).flatten())
cv_accs = np.array([accuracy_score((np.array(cv_gt) > 0.5).flatten(), (np.array(cv_pred) > 0.5).flatten())
for cv_gt,cv_pred in zip(cv_gts, cv_preds)])
cv_aucs = np.array([roc_auc_score(np.array(cv_gt).flatten().astype(np.float32), np.array(cv_pred).flatten())
for cv_gt,cv_pred in zip(cv_gts, cv_preds)])
print(f'OOF acc {oof_acc}, OOF auc {oof_auc}, CV AUC {np.mean(cv_aucs)} (std {np.std(cv_aucs)})')
plt.close('all')
df_plot = pd.DataFrame({'Pred-MGMT': oof_preds, 'GT-MGMT': oof_gts})
sns.histplot(x='Pred-MGMT', hue='GT-MGMT', data=df_plot)
plt.title(f'Average of all models # CV AUC = {np.mean(cv_aucs):.3f} (std: {np.std(cv_aucs):.3f}), Acc. = {np.mean(cv_accs):.3f}')
plt.show()
selected_feats = [
'feat_lasso',
'feat_ridge',
'feat_linreg',
'efficientnetv2_l_rocstar',
'resnet101_rocstar',
'densenet169_rocstar',
]
oof_acc = accuracy_score((np.array(oof_gts) > 0.5).flatten(), (np.mean(df_norm[selected_feats].to_numpy(),1) > 0.5).flatten())
oof_auc = roc_auc_score(np.array(oof_gts).flatten().astype(np.float32), np.mean(df_norm[selected_feats].to_numpy(),1).flatten())
cv_preds = [np.mean(df_norm[df_norm.fold==fold][selected_feats].to_numpy(),1) for fold in range(5)]
cv_gts = [df_norm[df_norm.fold==fold]['MGMT_value'] for fold in range(5)]
cv_accs = np.array([accuracy_score((np.array(cv_gt) > 0.5).flatten(), (np.array(cv_pred) > 0.5).flatten())
for cv_gt,cv_pred in zip(cv_gts, cv_preds)])
cv_aucs = np.array([roc_auc_score(np.array(cv_gt).flatten().astype(np.float32), np.array(cv_pred).flatten())
for cv_gt,cv_pred in zip(cv_gts, cv_preds)])
print(f'OOF acc {oof_acc}, OOF auc {oof_auc}, CV AUC {np.mean(cv_aucs)} (std {np.std(cv_aucs)})')
plt.close('all')
df_plot = pd.DataFrame({'Pred-MGMT': oof_preds, 'GT-MGMT': oof_gts})
sns.histplot(x='Pred-MGMT', hue='GT-MGMT', data=df_plot)
plt.title(f'Average of all models # CV AUC = {np.mean(cv_aucs):.3f} (std: {np.std(cv_aucs):.3f}), Acc. = {np.mean(cv_accs):.3f}')
plt.show()
```
## 2nd level models
```
import xgboost as xgb
def get_data(fold, features):
df = df_norm.dropna(inplace=False)
scaler = StandardScaler()
df_train = df[df.fold != fold]
df_val = df[df.fold == fold]
if len(df_val) == 0:
df_val = df[df.fold == 0]
# shuffle train
df_train = df_train.sample(frac=1)
y_train = df_train.MGMT_value.to_numpy().reshape((-1,1)).astype(np.float32)
y_val = df_val.MGMT_value.to_numpy().reshape((-1,1)).astype(np.float32)
X_train = df_train[features].to_numpy().astype(np.float32)
X_val = df_val[features].to_numpy().astype(np.float32)
scaler.fit(X_train)
X_train = scaler.transform(X_train)
X_val = scaler.transform(X_val)
return X_train, y_train, X_val, y_val, scaler, (df_train.index.values).flatten(), (df_val.index.values).flatten()
def measure_cv_score(parameters, verbose=False, train_one_model=False, plot=False, return_oof_preds=False):
val_preds = []
val_gts = []
val_aucs = []
val_accs = []
val_index_values = []
for fold in range(5):
if train_one_model: fold = -1
X_train, y_train, X_val, y_val, scaler, train_index, val_index = get_data(fold, features=parameters['features'])
val_index_values = val_index_values + list(val_index)
if parameters['model_type'] == 'xgb':
model = xgb.XGBRegressor(
n_estimators=parameters['n_estimators'],
max_depth=parameters['max_depth'],
eta=parameters['eta'],
subsample=parameters['subsample'],
colsample_bytree=parameters['colsample_bytree'],
gamma=parameters['gamma']
)
elif parameters['model_type'] == 'linreg':
model = linear_model.LinearRegression()
elif parameters['model_type'] == 'ridge':
model = linear_model.Ridge(parameters['alpha'])
elif parameters['model_type'] == 'bayesian':
model = linear_model.BayesianRidge(
n_iter = parameters['n_iter'],
lambda_1 = parameters['lambda_1'],
lambda_2 = parameters['lambda_2'],
alpha_1 = parameters['alpha_1'],
alpha_2 = parameters['alpha_2'],
)
elif parameters['model_type'] == 'logreg':
model = linear_model.LogisticRegression()
elif parameters['model_type'] == 'lassolarsic':
model = linear_model.LassoLarsIC(
max_iter = parameters['max_iter'],
eps = parameters['eps']
)
elif parameters['model_type'] == 'perceptron':
model = linear_model.Perceptron(
)
else:
raise NotImplementedError
model.fit(X_train, y_train.ravel())
if train_one_model:
return model, scaler
val_pred = model.predict(X_val)
val_preds += list(val_pred)
val_gts += list(y_val)
val_aucs.append(roc_auc_score(np.array(y_val).flatten().astype(np.float32), np.array(val_pred).flatten()))
val_accs.append(accuracy_score((np.array(y_val) > 0.5).flatten(), (np.array(val_pred) > 0.5).flatten()))
if return_oof_preds:
return np.array(val_preds).flatten(), np.array(val_gts).flatten(), val_index_values
oof_acc = accuracy_score((np.array(val_gts) > 0.5).flatten(), (np.array(val_preds) > 0.5).flatten())
oof_auc = roc_auc_score(np.array(val_gts).flatten().astype(np.float32), np.array(val_preds).flatten())
auc_std = np.std(np.array(val_aucs))
if plot:
df_plot = pd.DataFrame({'Pred-MGMT': np.array(val_preds).flatten(), 'GT-MGMT': np.array(val_gts).flatten()})
sns.histplot(x='Pred-MGMT', hue='GT-MGMT', data=df_plot)
plt.title(f'{parameters["model_type"]} # CV AUC = {oof_auc:.3f} (std {auc_std:.3f}), Acc. = {oof_acc:.3f}')
plt.show()
if verbose:
print(f'CV AUC = {oof_auc} (std {auc_std}), Acc. = {oof_acc}, aucs: {val_aucs}, accs: {val_accs}')
# optimize lower limit of the (2x std range around mean)
# This way, we choose the model which ranks well and performs ~equally well on all folds
return float(oof_auc) - auc_std
default_parameters = {
'model_type': 'linreg',
'n_estimators': 100,
'max_depth' : 3,
'eta': 0.1,
'subsample': 0.7,
'colsample_bytree' : 0.8,
'gamma' : 1.0,
'alpha' : 1.0,
'n_iter':300,
'lambda_1': 1e-6, # bayesian
'lambda_2':1e-6, # bayesian
'alpha_1': 1e-6, # bayesian
'alpha_2': 1e-6, # bayesian
'max_iter': 3, #lasso
'eps': 1e-6, #lasso
'features' : all_feat_names
}
measure_cv_score(default_parameters, verbose=True)
def feat_selection_linreg_objective(trial):
kept_feats = []
for i in range(len(all_feat_names)):
var = trial.suggest_int(all_feat_names[i], 0,1)
if var == 1:
kept_feats.append(all_feat_names[i])
parameters = default_parameters.copy()
parameters['features'] = kept_feats
return 1 - measure_cv_score(parameters, verbose=False)
if 1:
study = optuna.create_study()
study.optimize(feat_selection_linreg_objective, n_trials=20, show_progress_bar=True)
print(study.best_value, study.best_params)
study.best_params
pruned_features = default_parameters.copy()
pruned_features['features'] = ['feat_lasso', 'feat_linreg', 'feat_ridge', 'efficientnetv2_l_rocstar']
measure_cv_score(pruned_features, verbose=True)
random.randint(0,1)
```
| github_jupyter |
## Introduction to \LaTeX Math Mode
Jupyter notebooks integrate the MathJax Javascript library in order to render mathematical formulas and symbols in the same way as one would in \LaTeX (often used to typeset textbooks, research papers, or other technical documents).
First, we will take a look at a couple of rendered expressions and the corresponding way to render these in your notebooks, then follow-up with a few exercises which will help you become more familiar with these tools and their corresponding documentation.
For example, a common expression used in neural networks is the _weighted sum_ rendered as so:
$y=\sum_{i=1}^{N}{w_i x_i + b}$
where the variable $y$ is calculating the sum of the elements for a vector, $x_i$, each multiplied by a corresponding weight, $w_i$. An additional scalar term, $b$, known as the _bias_ is added to the overall result as well. This expression is more commonly written as:
$y=\boldsymbol{w}\boldsymbol{x}+b$
where $\boldsymbol{w}$ and $\boldsymbol{x}$ are both vectors of length $N$. Note the subtle difference in the notation where __ _vectors_ __ are in bold italic, while _scalars_ are only in italic.
These kinds of expressions can be rendered in your notebook by creating _markdown_ cells and populating them with the proper expressions. Normally, a cell in a Jupyter notebook is for code that you would like to hand off to the interpreter, but there is a drop-down menu at the top of the current notebook which can change the mode of the current cell to either _code_, _markdown_, or _raw_. We will rarely use _raw_ cells, but the _code_ and _markdown_ types are both quite useful.
To render both of the two expressions above, you will need to create a markdown cell, and then enter the following code into the cell:
```
$y = \sum_{i=1}^{N}{w_i x_i + b}$
$y = \boldsymbol{w}\boldsymbol{x}+b$
```
You should notice first that each expression is surrounded by a set of \$ symbols. Any text that you type between two \$ symbols is rendered using the \LaTeX mathematics mode. \LaTeX is a complete document preparation system that we will learn more about later in the semester. For now, the important thing to understand is that it has a special mode and markup language used to render mathematical expressions, and this markup language is supported in _markdown_ cells in Jupyter notebooks.
Second, you can see that special mathematical symbols such as a summation ($\sum$) can be rendered using the "sum" escape sequence (\\sum) where \\ is the math mode escape character. There are numerous different escape sequences that can be used in math mode, each representing a common mathematical symbol or operation.
Third, you can see that symbols can be attached to other symbols for rendering as sub- or super-scripts by using the _ and ^ operators, respectively. You can also use curly-braces (liberally) to group symbols together into these sub- or super-scripts and the curly-braces, themselves, will not be rendered in the equation. These delimeters only help the math mode interpreter understand which symbols you would like grouped together, and won't be displayed unless escaped.
Finally, it is clear that many symbols are rendered in a way that makes intuitive sense. For example, the bias term, $b$, is simply provided with no markup. Any text __not__ escaped or otherwise marked up will be rendered as a standard scalar is rendered (italic). However, the `\text{}` sequence can be used to render standard text when required. For example:
`$a\ \text{plus}\ b$`
$a\ \text{plus}\ b$
Notice also how a backslash followed by a space will add a space between the words. Normally, when two scalars are presented, it is assumed they are being multiplied together, and are placed closely to represent this fact. However, since ext
Here are a few other examples:
`$\boldsymbol{A}=\boldsymbol{U}\boldsymbol{\Sigma}\boldsymbol{V}^\top$`
$\boldsymbol{A}=\boldsymbol{U}\boldsymbol{\Sigma}\boldsymbol{V}^\top$
`$\alpha \beta \Theta \Omega$`
$\alpha \beta \Theta \Omega$
`$\int_{-\pi}^{\pi} \sin{x}\ dx$`
$\int_{-\pi}^{\pi} \sin{x}\ dx$
`$\prod_{i=1}^{N}{(x_i+y_i)^2}$`
$\prod_{i=1}^{N}{(x_i+y_i)^2}$
`$f(x)=\frac{1}{x^2}$`
$f(x)=\frac{1}{x^2}$
`$\frac{d}{dx}f(x) = -\frac{2}{x^3}$`
$\frac{d}{dx}f(x) = -\frac{2}{x^3}$
Let's make a simple table, and then also show the markdown source for the table...
| One | Two | Three | Four |
| --- | --- | --- | --- |
| 10% | Something | Else | 40% |
| 90% | To | Do | 50% |
```
| One | Two | Three | Four |
| --- | --- | --- | --- |
| 10% | Something | Else | 40% |
| 90% | To | Do | 50% |
```
| github_jupyter |
```
#import necessary modules, set up the plotting
import numpy as np
%matplotlib inline
%config InlineBackend.figure_format = 'svg'
import matplotlib;matplotlib.rcParams['figure.figsize'] = (8,6)
from matplotlib import pyplot as plt
import GPy
```
# Interacting with models
### November 2014, by Max Zwiessele
#### with edits by James Hensman
The GPy model class has a set of features which are designed to make it simple to explore the parameter space of the model. By default, the scipy optimisers are used to fit GPy models (via model.optimize()), for which we provide mechanisms for ‘free’ optimisation: GPy can ensure that naturally positive parameters (such as variances) remain positive. But these mechanisms are much more powerful than simple reparameterisation, as we shall see.
Along this tutorial we’ll use a sparse GP regression model as example. This example can be in GPy.examples.regression. All of the examples included in GPy return an instance of a model class, and therefore they can be called in the following way:
```
m = GPy.examples.regression.sparse_GP_regression_1D(plot=False, optimize=False)
```
## Examining the model using print
To see the current state of the model parameters, and the model’s (marginal) likelihood just print the model
print m
The first thing displayed on the screen is the log-likelihood value of the model with its current parameters. Below the log-likelihood, a table with all the model’s parameters is shown. For each parameter, the table contains the name of the parameter, the current value, and in case there are defined: constraints, ties and prior distrbutions associated.
```
m
```
In this case the kernel parameters (`bf.variance`, `bf.lengthscale`) as well as the likelihood noise parameter (`Gaussian_noise.variance`), are constrained to be positive, while the inducing inputs have no constraints associated. Also there are no ties or prior defined.
You can also print all subparts of the model, by printing the subcomponents individually; this will print the details of this particular parameter handle:
```
m.rbf
```
When you want to get a closer look into multivalue parameters, print them directly:
```
m.inducing_inputs
m.inducing_inputs[0] = 1
```
## Interacting with Parameters:
The preferred way of interacting with parameters is to act on the parameter handle itself. Interacting with parameter handles is simple. The names, printed by print m are accessible interactively and programatically. For example try to set the kernel's `lengthscale` to 0.2 and print the result:
```
m.rbf.lengthscale = 0.2
print m
```
This will already have updated the model’s inner state: note how the log-likelihood has changed. YOu can immediately plot the model or see the changes in the posterior (`m.posterior`) of the model.
## Regular expressions
The model’s parameters can also be accessed through regular expressions, by ‘indexing’ the model with a regular expression, matching the parameter name. Through indexing by regular expression, you can only retrieve leafs of the hierarchy, and you can retrieve the values matched by calling `values()` on the returned object
```
print m['.*var']
#print "variances as a np.array:", m['.*var'].values()
#print "np.array of rbf matches: ", m['.*rbf'].values()
```
There is access to setting parameters by regular expression, as well. Here are a few examples of how to set parameters by regular expression. Note that each time the values are set, computations are done internally to compute the log likeliood of the model.
```
m['.*var'] = 2.
print m
m['.*var'] = [2., 3.]
print m
```
A handy trick for seeing all of the parameters of the model at once is to regular-expression match every variable:
```
print m['']
```
## Setting and fetching parameters parameter_array
Another way to interact with the model’s parameters is through the parameter_array. The Parameter array holds all the parameters of the model in one place and is editable. It can be accessed through indexing the model for example you can set all the parameters through this mechanism:
```
new_params = np.r_[[-4,-2,0,2,4], [.1,2], [.7]]
print new_params
m[:] = new_params
print m
```
Parameters themselves (leafs of the hierarchy) can be indexed and used the same way as numpy arrays. First let us set a slice of the inducing_inputs:
```
m.inducing_inputs[2:, 0] = [1,3,5]
print m.inducing_inputs
```
Or you use the parameters as normal numpy arrays for calculations:
```
precision = 1./m.Gaussian_noise.variance
print precision
```
## Getting the model parameter’s gradients
The gradients of a model can shed light on understanding the (possibly hard) optimization process. The gradients of each parameter handle can be accessed through their gradient field.:
```
print "all gradients of the model:\n", m.gradient
print "\n gradients of the rbf kernel:\n", m.rbf.gradient
```
If we optimize the model, the gradients (should be close to) zero
```
m.optimize()
print m.gradient
```
## Adjusting the model’s constraints
When we initially call the example, it was optimized and hence the log-likelihood gradients were close to zero. However, since we have been changing the parameters, the gradients are far from zero now. Next we are going to show how to optimize the model setting different restrictions on the parameters.
Once a constraint has been set on a parameter, it is possible to remove it with the command unconstrain(), which can be called on any parameter handle of the model. The methods constrain() and unconstrain() return the indices which were actually unconstrained, relative to the parameter handle the method was called on. This is particularly handy for reporting which parameters where reconstrained, when reconstraining a parameter, which was already constrained:
```
m.rbf.variance.unconstrain()
print m
m.unconstrain()
print m
```
If you want to unconstrain only a specific constraint, you can call the respective method, such as `unconstrain_fixed()` (or `unfix()`) to only unfix fixed parameters:
```
m.inducing_inputs[0].fix()
m.rbf.constrain_positive()
print m
m.unfix()
print m
```
## Tying Parameters
Not yet implemented for GPy version 0.8.0
## Optimizing the model
Once we have finished defining the constraints, we can now optimize the model with the function optimize.:
```
m.Gaussian_noise.constrain_positive()
m.rbf.constrain_positive()
m.optimize()
```
By deafult, GPy uses the lbfgsb optimizer.
Some optional parameters may be discussed here.
* `optimizer`: which optimizer to use, currently there are lbfgsb, fmin_tnc, scg, simplex or any unique identifier uniquely identifying an optimizer.
Thus, you can say m.optimize('bfgs') for using the `lbfgsb` optimizer
* `messages`: if the optimizer is verbose. Each optimizer has its own way of printing, so do not be confused by differing messages of different optimizers
* `max_iters`: Maximum number of iterations to take. Some optimizers see iterations as function calls, others as iterations of the algorithm. Please be advised to look into scipy.optimize for more instructions, if the number of iterations matter, so you can give the right parameters to optimize()
* `gtol`: only for some optimizers. Will determine the convergence criterion, as the tolerance of gradient to finish the optimization.
## Plotting
Many of GPys models have built-in plot functionality. we distringuish between plotting the posterior of the function (`m.plot_f`) and plotting the posterior over predicted data values (`m.plot`). This becomes especially important for non-Gaussian likleihoods. Here we'll plot the sparse GP model we've been working with. for more information of the meaning of the plot, please refer to the accompanying `basic_gp_regression` and `sparse_gp` noteooks.
```
fig = m.plot()
```
We can even change the backend for plotting and plot the model using a different backend.
```
GPy.plotting.change_plotting_library('plotly')
fig = m.plot(plot_density=True)
GPy.plotting.show(fig, filename='gpy_sparse_gp_example')
```
| github_jupyter |
```
%matplotlib inline
```
# Partial Dependence Plots
Sigurd Carlsen Feb 2019
Holger Nahrstaedt 2020
.. currentmodule:: skopt
Plot objective now supports optional use of partial dependence as well as
different methods of defining parameter values for dependency plots.
```
print(__doc__)
import sys
from skopt.plots import plot_objective
from skopt import forest_minimize
import numpy as np
np.random.seed(123)
import matplotlib.pyplot as plt
```
## Objective function
Plot objective now supports optional use of partial dependence as well as
different methods of defining parameter values for dependency plots
```
# Here we define a function that we evaluate.
def funny_func(x):
s = 0
for i in range(len(x)):
s += (x[i] * i) ** 2
return s
```
## Optimisation using decision trees
We run forest_minimize on the function
```
bounds = [(-1, 1.), ] * 3
n_calls = 150
result = forest_minimize(funny_func, bounds, n_calls=n_calls,
base_estimator="ET",
random_state=4)
```
## Partial dependence plot
Here we see an example of using partial dependence. Even when setting
n_points all the way down to 10 from the default of 40, this method is
still very slow. This is because partial dependence calculates 250 extra
predictions for each point on the plots.
```
_ = plot_objective(result, n_points=10)
```
It is possible to change the location of the red dot, which normally shows
the position of the found minimum. We can set it 'expected_minimum',
which is the minimum value of the surrogate function, obtained by a
minimum search method.
```
_ = plot_objective(result, n_points=10, minimum='expected_minimum')
```
## Plot without partial dependence
Here we plot without partial dependence. We see that it is a lot faster.
Also the values for the other parameters are set to the default "result"
which is the parameter set of the best observed value so far. In the case
of funny_func this is close to 0 for all parameters.
```
_ = plot_objective(result, sample_source='result', n_points=10)
```
## Modify the shown minimum
Here we try with setting the `minimum` parameters to something other than
"result". First we try with "expected_minimum" which is the set of
parameters that gives the miniumum value of the surrogate function,
using scipys minimum search method.
```
_ = plot_objective(result, n_points=10, sample_source='expected_minimum',
minimum='expected_minimum')
```
"expected_minimum_random" is a naive way of finding the minimum of the
surrogate by only using random sampling:
```
_ = plot_objective(result, n_points=10, sample_source='expected_minimum_random',
minimum='expected_minimum_random')
```
We can also specify how many initial samples are used for the two different
"expected_minimum" methods. We set it to a low value in the next examples
to showcase how it affects the minimum for the two methods.
```
_ = plot_objective(result, n_points=10, sample_source='expected_minimum_random',
minimum='expected_minimum_random',
n_minimum_search=10)
_ = plot_objective(result, n_points=10, sample_source="expected_minimum",
minimum='expected_minimum', n_minimum_search=2)
```
## Set a minimum location
Lastly we can also define these parameters ourself by parsing a list
as the minimum argument:
```
_ = plot_objective(result, n_points=10, sample_source=[1, -0.5, 0.5],
minimum=[1, -0.5, 0.5])
```
| github_jupyter |
# Training Models
The central goal of machine learning is to train predictive models that can be used by applications. In Azure Machine Learning, you can use scripts to train models leveraging common machine learning frameworks like Scikit-Learn, Tensorflow, PyTorch, SparkML, and others. You can run these training scripts as experiments in order to track metrics and outputs - in particular, the trained models.
## Before You Start
Before you start this lab, ensure that you have completed the *Create an Azure Machine Learning Workspace* and *Create a Compute Instance* tasks in [Lab 1: Getting Started with Azure Machine Learning](./labdocs/Lab01.md). Then open this notebook in Jupyter on your Compute Instance.
## Connect to Your Workspace
The first thing you need to do is to connect to your workspace using the Azure ML SDK.
> **Note**: If you do not have a current authenticated session with your Azure subscription, you'll be prompted to authenticate. Follow the instructions to authenticate using the code provided.
```
import azureml.core
from azureml.core import Workspace
# Load the workspace from the saved config file
ws = Workspace.from_config()
print('Ready to use Azure ML {} to work with {}'.format(azureml.core.VERSION, ws.name))
```
## Create a Training Script
You're going to use a Python script to train a machine learning model based on the diabates data, so let's start by creating a folder for the script and data files.
```
import os, shutil
# Create a folder for the experiment files
training_folder = 'diabetes-training'
os.makedirs(training_folder, exist_ok=True)
# Copy the data file into the experiment folder
shutil.copy('data/diabetes.csv', os.path.join(training_folder, "diabetes.csv"))
```
Now you're ready to create the training script and save it in the folder.
```
%%writefile $training_folder/diabetes_training.py
# Import libraries
from azureml.core import Run
import pandas as pd
import numpy as np
import joblib
import os
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import roc_auc_score
from sklearn.metrics import roc_curve
# Get the experiment run context
run = Run.get_context()
# load the diabetes dataset
print("Loading Data...")
diabetes = pd.read_csv('diabetes.csv')
# Separate features and labels
X, y = diabetes[['Pregnancies','PlasmaGlucose','DiastolicBloodPressure','TricepsThickness','SerumInsulin','BMI','DiabetesPedigree','Age']].values, diabetes['Diabetic'].values
# Split data into training set and test set
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=0)
# Set regularization hyperparameter
reg = 0.01
# Train a logistic regression model
print('Training a logistic regression model with regularization rate of', reg)
run.log('Regularization Rate', np.float(reg))
model = LogisticRegression(C=1/reg, solver="liblinear").fit(X_train, y_train)
# calculate accuracy
y_hat = model.predict(X_test)
acc = np.average(y_hat == y_test)
print('Accuracy:', acc)
run.log('Accuracy', np.float(acc))
# calculate AUC
y_scores = model.predict_proba(X_test)
auc = roc_auc_score(y_test,y_scores[:,1])
print('AUC: ' + str(auc))
run.log('AUC', np.float(auc))
# Save the trained model in the outputs folder
os.makedirs('outputs', exist_ok=True)
joblib.dump(value=model, filename='outputs/diabetes_model.pkl')
run.complete()
```
## Use an Estimator to Run the Script as an Experiment
You can run experiment scripts using a **RunConfiguration** and a **ScriptRunConfig**, or you can use an **Estimator**, which abstracts both of these configurations in a single object.
In this case, we'll use a generic **Estimator** object to run the training experiment. Note that the default environment for this estimator does not include the **scikit-learn** package, so you need to explicitly add that to the configuration. The conda environment is built on-demand the first time the estimator is used, and cached for future runs that use the same configuration; so the first run will take a little longer. On subsequent runs, the cached environment can be re-used so they'll complete more quickly.
```
from azureml.train.estimator import Estimator
from azureml.core import Experiment
# Create an estimator
estimator = Estimator(source_directory=training_folder,
entry_script='diabetes_training.py',
compute_target='local',
conda_packages=['scikit-learn']
)
# Create an experiment
experiment_name = 'diabetes-training'
experiment = Experiment(workspace = ws, name = experiment_name)
# Run the experiment based on the estimator
run = experiment.submit(config=estimator)
run.wait_for_completion(show_output=True)
```
As with any experiment run, you can use the **RunDetails** widget to view information about the run and get a link to it in Azure Machine Learning studio.
```
from azureml.widgets import RunDetails
RunDetails(run).show()
```
You can also retrieve the metrics and outputs from the **Run** object.
```
# Get logged metrics
metrics = run.get_metrics()
for key in metrics.keys():
print(key, metrics.get(key))
print('\n')
for file in run.get_file_names():
print(file)
```
## Register the Trained Model
Note that the outputs of the experiment include the trained model file (**diabetes_model.pkl**). You can register this model in your Azure Machine Learning workspace, making it possible to track model versions and retrieve them later.
```
from azureml.core import Model
# Register the model
run.register_model(model_path='outputs/diabetes_model.pkl', model_name='diabetes_model',
tags={'Training context':'Estimator'},
properties={'AUC': run.get_metrics()['AUC'], 'Accuracy': run.get_metrics()['Accuracy']})
# List registered models
for model in Model.list(ws):
print(model.name, 'version:', model.version)
for tag_name in model.tags:
tag = model.tags[tag_name]
print ('\t',tag_name, ':', tag)
for prop_name in model.properties:
prop = model.properties[prop_name]
print ('\t',prop_name, ':', prop)
print('\n')
```
## Create a Parameterized Training Script
You can increase the flexibility of your training experiment by adding parameters to your script, enabling you to repeat the same training experiment with different settings. In this case, you'll add a parameter for the regularization rate used by the Logistic Regression algorithm when training the model.
Again, lets start by creating a folder for the parameterized script and the training data.
```
import os, shutil
# Create a folder for the experiment files
training_folder = 'diabetes-training-params'
os.makedirs(training_folder, exist_ok=True)
# Copy the data file into the experiment folder
shutil.copy('data/diabetes.csv', os.path.join(training_folder, "diabetes.csv"))
```
Now let's create a script containing a parameter for the regularization rate hyperparameter.
```
%%writefile $training_folder/diabetes_training.py
# Import libraries
from azureml.core import Run
import pandas as pd
import numpy as np
import joblib
import os
import argparse
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import roc_auc_score
from sklearn.metrics import roc_curve
# Get the experiment run context
run = Run.get_context()
# Set regularization hyperparameter
parser = argparse.ArgumentParser()
parser.add_argument('--reg_rate', type=float, dest='reg', default=0.01)
args = parser.parse_args()
reg = args.reg
# load the diabetes dataset
print("Loading Data...")
# load the diabetes dataset
diabetes = pd.read_csv('diabetes.csv')
# Separate features and labels
X, y = diabetes[['Pregnancies','PlasmaGlucose','DiastolicBloodPressure','TricepsThickness','SerumInsulin','BMI','DiabetesPedigree','Age']].values, diabetes['Diabetic'].values
# Split data into training set and test set
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=0)
# Train a logistic regression model
print('Training a logistic regression model with regularization rate of', reg)
run.log('Regularization Rate', np.float(reg))
model = LogisticRegression(C=1/reg, solver="liblinear").fit(X_train, y_train)
# calculate accuracy
y_hat = model.predict(X_test)
acc = np.average(y_hat == y_test)
print('Accuracy:', acc)
run.log('Accuracy', np.float(acc))
# calculate AUC
y_scores = model.predict_proba(X_test)
auc = roc_auc_score(y_test,y_scores[:,1])
print('AUC: ' + str(auc))
run.log('AUC', np.float(auc))
os.makedirs('outputs', exist_ok=True)
joblib.dump(value=model, filename='outputs/diabetes_model.pkl')
run.complete()
```
## Use a Framework-Specific Estimator
You used a generic **Estimator** class to run the training script, but you can also take advantage of framework-specific estimators that include environment definitions for common machine learning frameworks. In this case, you're using Scikit-Learn, so you can use the **SKLearn** estimator. This means that you don't need to specify the **scikit-learn** package in the configuration.
> **Note**: Once again, the training experiment uses a new environment; which must be created the first time it is run.
```
from azureml.train.sklearn import SKLearn
from azureml.widgets import RunDetails
# Create an estimator
estimator = SKLearn(source_directory=training_folder,
entry_script='diabetes_training.py',
script_params = {'--reg_rate': 0.1},
compute_target='local'
)
# Create an experiment
experiment_name = 'diabetes-training'
experiment = Experiment(workspace = ws, name = experiment_name)
# Run the experiment
run = experiment.submit(config=estimator)
# Show the run details while running
RunDetails(run).show()
run.wait_for_completion()
```
Once again, you can get the metrics and outputs from the run.
```
# Get logged metrics
metrics = run.get_metrics()
for key in metrics.keys():
print(key, metrics.get(key))
print('\n')
for file in run.get_file_names():
print(file)
```
## Register A New Version of the Model
Now that you've trained a new model, you can register it as a new version in the workspace.
```
from azureml.core import Model
# Register the model
run.register_model(model_path='outputs/diabetes_model.pkl', model_name='diabetes_model',
tags={'Training context':'Parameterized SKLearn Estimator'},
properties={'AUC': run.get_metrics()['AUC'], 'Accuracy': run.get_metrics()['Accuracy']})
# List registered models
for model in Model.list(ws):
print(model.name, 'version:', model.version)
for tag_name in model.tags:
tag = model.tags[tag_name]
print ('\t',tag_name, ':', tag)
for prop_name in model.properties:
prop = model.properties[prop_name]
print ('\t',prop_name, ':', prop)
print('\n')
```
## Clean Up
If you've finished exploring, you can close this notebook and shut down your Compute Instance.
| github_jupyter |
```
%load_ext autoreload
%autoreload 2
import tensorflow as tf
import numpy as np
import pandas as pd
import altair as alt
import shap
from interaction_effects.marginal import MarginalExplainer
from interaction_effects import utils
n = 3000
d = 3
batch_size = 50
learning_rate = 0.02
X = np.random.randn(n, d)
y = (np.sum(X, axis=-1) > 0.0).astype(np.float32)
model = tf.keras.Sequential()
model.add(tf.keras.Input(shape=(3,), batch_size=batch_size))
model.add(tf.keras.layers.Dense(2, activation=None, use_bias=True))
optimizer = tf.keras.optimizers.SGD(learning_rate=learning_rate)
model.compile(optimizer=optimizer,
loss=tf.keras.losses.SparseCategoricalCrossentropy(),
metrics=[tf.keras.metrics.SparseCategoricalAccuracy()])
model.fit(X, y, epochs=20, verbose=2)
primal_explainer = MarginalExplainer(model, X[20:], nsamples=800, representation='mobius')
primal_effects = primal_explainer.explain(X[:20], verbose=True, index_outputs=True, labels=y[:20].astype(int))
dual_explainer = MarginalExplainer(model, X[20:], nsamples=800, representation='comobius')
dual_effects = dual_explainer.explain(X[:20], verbose=True, index_outputs=True, labels=y[:20].astype(int))
average_explainer = MarginalExplainer(model, X[20:], nsamples=800, representation='average')
average_effects = average_explainer.explain(X[:20], verbose=True, index_outputs=True, labels=y[:20].astype(int))
model_func = lambda x: model(x).numpy()
kernel_explainer = shap.SamplingExplainer(model_func, X)
kernel_shap = kernel_explainer.shap_values(X[:20])
kernel_shap = np.stack(kernel_shap, axis=0)
kernel_shap_true_class = kernel_shap[y[:20].astype(int), np.arange(20), :]
def unroll(x):
ret = []
for i in range(x.shape[-1]):
ret.append(x[:, i])
return np.concatenate(ret)
data_df = pd.DataFrame({
'Sampled Primal Effects': unroll(primal_effects),
'Sampled Dual Effects': unroll(dual_effects),
'Sampled Average Effects': unroll(average_effects),
'Kernel SHAP Values': unroll(kernel_shap_true_class),
'Feature Values': unroll(X[:20]),
'Feature': [int(i / 20) for i in range(20 * d)],
'Label': np.tile(y[:20], 3).astype(int)
})
alt.Chart(data_df).mark_point(filled=True).encode(
alt.X('Kernel SHAP Values:Q'),
alt.Y(alt.repeat("column"), type='quantitative')
).properties(width=300, height=300).repeat(column=['Sampled Primal Effects', 'Sampled Dual Effects', 'Sampled Average Effects'])
melted_df = pd.melt(data_df, id_vars=['Feature Values', 'Feature', 'Label'], var_name='Effect Type', value_name='Effect Value')
alt.Chart(melted_df).mark_point(filled=True).encode(
alt.X('Feature Values:Q'),
alt.Y('Effect Value:Q'),
alt.Color('Label:N')
).properties(width=200, height=200).facet(column='Effect Type', row='Feature')
```
| github_jupyter |
#CHALLENGE TASK
#Stats Challege notebook
#Fit multiple linear regression for the following data and check for the assumptions using python
#X1 22 22 25 26 24 28 29 27 24 33 39 42
#X2 15 14 18 13 12 11 11 10 5 9 7 3
#Y 55 56 55 59 66 65 69 70 75 75 78 79
```
import numpy as np
import pandas as pd
import statsmodels.api as sm
import matplotlib.pyplot as plt
""" Convert the data values into DataFrames"""
stats_chal={"X1":[22, 22, 25, 26, 24, 28, 29, 27, 24, 33, 39, 42],
"X2":[15, 14, 18, 13, 12, 11, 11, 10, 5, 9, 7, 3],
"Y":[55, 56, 55, 59, 66, 65, 69, 70, 75, 75, 78, 79]}
df = pd.DataFrame(stats_chal,columns=['X1','X2','Y'])
print (df)
"""Check for the linearity"""
plt.scatter(df['X1'], df['Y'], color='green')
plt.xlabel('X1 values', fontsize=14)
plt.ylabel('Y values', fontsize=14)
plt.grid(True)
plt.show()
"""its clear that indeed a linear relationship exists between the X1 values and the Y values.
Specifically, when X1 values go up, the Y values also goes up"""
"""Check for the linearity"""
plt.scatter(df['X2'], df['Y'], color='blue')
plt.xlabel('X2 values', fontsize=14)
plt.ylabel('Y values', fontsize=14)
plt.grid(True)
plt.show()
"""its clear that indeed a linear relationship exists between the X2 values and the Y values.
Specifically, when X2 values go up, the Y values also goes down but with a negative slope"""
"""Performing the Multiple Linear Regression"""
X = df[['X1','X2']] # here we have 2 variables for multiple regression.
Y = df['Y']
# with statsmodels
X = sm.add_constant(X) # adding a constant
mlr_model = sm.OLS(Y, X).fit()
predictions = mlr_model.predict(X)
print_model = mlr_model.summary()
print(print_model)
"""If you plug that X1=22, X2=15 data into the regression equation,
you’ll get the same predicted result of Y values """
y = (74.5958) + (0.3314)*(22)+(-1.6106)*(15)
y
y = (74.5958) + (0.3314)*(X1)+(-1.6106)*(X2)
predicted_values=(74.5958) + (0.3314)*(X1)+(-1.6106)*(X2)
predicted_values
X = df[['X1','X2']].values
X
sns.regplot(data=df,x="X",y="Y",color="green") #OLS
y=df["Y"]
y
X = df[['X1','X2']]
X
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42)
X_train.head()
len(X_train)
len(X_test)
from sklearn.linear_model import LinearRegression
model =LinearRegression()
model.fit(X_train,y_train)
test_model=model.predict(X_test)
test_model
from sklearn.metrics import mean_squared_error,mean_absolute_error
import seaborn as sns
sns.histplot(data=df,x="X1",bins=20)
import seaborn as sns
sns.histplot(data=df,x="X2",bins=20)
mean_absolute_error(y_test,test_model)
mean_squared_error(y_test,test_model)
np.sqrt(mean_squared_error(y_test,test_model))
sns.scatterplot(x="X",y="y",data=df)#scatter plot
plt.plot(potential_spend,predicted_sales,color="green")
# with sklearn
from sklearn import linear_model
ml_regr = linear_model.LinearRegression()
ml_regr.fit(X, Y)
print('Intercept: \n', ml_regr.intercept_)
print('Coefficients: \n', ml_regr.coef_)
```
#CHECKING FOR LINEAR REGRESSION ASSUMPTIONS
1.Linear Relationship
Aims at finding linear relationship between the independent and dependent variables
TEST
A simple visual way of determining this is through the use of scatter plot
2.Variables follow a normal Distribution
This assumption ensures that for each value of independent variable, the dependent variable is a random variable following a normal distribution and its mean lies on the regression line.
TEST
One of the ways to visually test for this assumption is through the use of the Quantile-Quantile plot(Q-Q_Plot)
```
#Multicollinearity test
corr =df.corr()
print(corr)
#Linearity and Normality Test
import seaborn as sns
sns.set(style="ticks", color_codes=True, font_scale=2)
g=sns.pairplot(df, height=3, diag_kind="hist",kind="reg")
g.fig.suptitle("Scatter Plot",y=1.08)
X_test = sm.add_constant(X_test)
X_test
y_pred=mlr_model.predict(X_test)
residual = y_test - y_pred
#No Multicolinearity
from statsmodels.stats.outliers_influence import variance_inflation_factor
vif = [variance_inflation_factor(X_train.values, i) for i in range(X_train.shape[1])]
pd.DataFrame({'vif': vif[0:]}, index=X_train.columns).T
"""Little or no multicollinearity
This assumption aims to test correlation between independent variables.
If multicollinearity exists between them (i.e independent variables are highly correlated), they are no longer independent.
TEST
Correlation Analysis (others are variance inflation factor (VIF)) and condition Index
If you find any values in which the absolute value of their correlation is >=0.8, the multicollinearity
Assumption is being broken.
"""
#Normailty of Residual
sns.distplot(residual)
import scipy as sp
fig, ax = plt.subplots(figsize=(6,2.5))
_, (__, ___, r) = sp.stats.probplot(residual, plot=ax, fit=True)
np.mean(residual)
#Normality of error / residue
import scipy.stats as stats
fig, ax=plt.subplots(figsize=(10,6))
stats.probplot(residual, dist="norm",plot=plt)
plt.show
#Homoscedasticity
fig, ax = plt.subplots(figsize=(6,2.5))
_ = ax.scatter(y_pred, residual)
plt.title("Homoscedasticity")
"""Data is homoscedastic
The linear regression analysis makes is homoscedasticity (i.e the error terms along the regression line are equal)
This analysis is also applied to the residuals of your linear regression model.
TEST
Homoscedasticity can be easily tested with a Scatterplot of the residuals.
"""
#No autocorrelation of residuals
import statsmodels.tsa.api as smt
acf = smt.graphics.plot_acf(residual, lags=3 , alpha=0.05)
acf.show()
"""Little or No Autocorrelation
This next assumption is much like our previous one, except it applies to the residuals of your linear regression model.
Linear regression analysis requires that there is little or no autocorrelation in the data.
TEST
You can test the liner regression model for autocorrelation with Durbin-Watson test(d),
while d can assume values between 0 and 4 ,values around 2 indicates no autocorrelation. As a rule of thumb values of 1.5
"""
```
#Conclusion
Here performd multiple linear regression in Python using both
sklearn and statsmodels in this both models the coefficient values are same.
we got consistent results by applying both sklearn and statsmodels.
#Multicollinearity test : If the VIF value of greater than 10 signifies that heavy Multicollinearity in the dataset,
a value less than 5 for a given feature specifies that there is a little relatioship that feature holds with
other feature.
In this case the VIF score are 4.085853 with independet having vey weak VIF score within them,
so the assumption of Multicollinearity holds True in our given schemes of things.
#Normailty of Residual
Residual is the differenec berween y_test & y_pred, if you check the plot for Normal distrubution ,
the plot is near Normal distrubution but its not entirely Normal distrubution and centered near to zero,
which one of the assumptions of Normal distrubution.
One more way to validate Normal distrubution is Q-Q Plot, Here I can see that the theoritical values fall on same line,
i.e most of the values near the line, this shows clear that the overall distribution is near Normal distrubution.
here we can observe the mean value 0.87 it inferes that genereaal this not enough good value, meanschnege cureve towards right.
i.e to ge normal distribution the value of mean should be zero or near to zero. so our assumption is True.
#Homoscedasticity or Constant variance
Here we need to observe the visuvalization, overall distribution is randomly sampled,
does it kind of inrecare with increase in residual values, here my predections on X-axis,
overall residuals on Y-axis,i find that there is no pattern based on the predection and residual
values, and move cenetred arond zero value.so our assumption is True.
#No autocorrelation of residuals
Here there shoulb be absolutely no correleation of the residual value is with any of its
lagged verstions which is called as autocorrelation, I observed in tha plot the residuals itself have a heavy correlation,
if you observe the most of auto correlated values non of the values corss the threashold of beying significant,
so the blue color signified here shows the significance level of the autocorrelation which it should cross
in orderd to be significant in termd of auto correlation , so here there no value cross the blue bounndary ,
so this linear regression model followign the assumptios of the linear regression.
| github_jupyter |
### Prepare stimuli in stereo with sync tone in the L channel
To syncrhonize the recording systems, each stimulus file goes in stereo, the L channel has the stimulus, and the R channel has a pure tone (500-5Khz).
This is done here, with the help of the rigmq.util.stimprep module
It uses (or creates) a dictionary of {stim_file: tone_freq} which is stored as a .json file for offline processing.
```
import socket
import os
import sys
import logging
import warnings
import numpy as np
import glob
from rigmq.util import stimprep as sp
# setup the logger
logger = logging.getLogger()
handler = logging.StreamHandler()
formatter = logging.Formatter(
'%(asctime)s %(name)-12s %(levelname)-8s %(message)s')
handler.setFormatter(formatter)
logger.addHandler(handler)
logger.setLevel(logging.INFO)
# Check wich computer to decide where the things are mounted
comp_name=socket.gethostname()
logger.info('Computer: ' + comp_name)
exp_folder = os.path.abspath('/Users/zeke/experiment/birds')
bird = 'g3v3'
sess = 'acute_0'
stim_sf = 48000 # sampling frequency of the stimulus system
stim_folder = os.path.join(exp_folder, bird, 'SongData', sess)
glob.glob(os.path.join(stim_folder, '*.wav'))
from scipy.io import wavfile
from scipy.signal import resample
a_file = glob.glob(os.path.join(stim_folder, '*.wav'))[0]
in_sf, data = wavfile.read(a_file)
%matplotlib inline
from matplotlib import pyplot as plt
plt.plot(data)
data.dtype
np.iinfo(data.dtype).min
def normalize(x: np.array, max_amp: np.float=0.9)-> np.array:
y = x.astype(np.float)
y = y - np.mean(y)
y = y / np.max(np.abs(y)) # if it is still of-centered, scale to avoid clipping in the widest varyng sign
return y * max_amp
data_float = normalize(data)
plt.plot(data_float)
def int_range(x: np.array, dtype: np.dtype):
min_int = np.iinfo(dtype).min
max_int = np.iinfo(dtype).max
if min_int==0: # for unsigned types shift everything
x = x + np.min(x)
y = x * max_int
return y.astype(dtype)
data_int = int_range(data_float, data.dtype)
plt.plot(data_int)
data_tagged = sp.make_stereo_stim(a_file, 48000, tag_freq=1000)
plt.plot(data_tagged[:480,1])
### Define stim_tags
There is a dictionary of {wav_file: tag_frequency} can be done by hand when there are few stimuli
stim_tags_dict = {'bos': 1000,
'bos-lo': 2000,
'bos-rev': 3000}
stims_list = list(stim_tags_dict.keys())
sp.create_sbc_stim(stims_list, stim_folder, stim_sf, stim_tag_dict=stim_tags_dict)
```
| github_jupyter |
# Scaling and Normalization
```
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler, MinMaxScaler, RobustScaler
from scipy.cluster.vq import whiten
```
Terminology (from [this post](https://towardsdatascience.com/scale-standardize-or-normalize-with-scikit-learn-6ccc7d176a02)):
* Scale generally means to change the range of the values. The shape of the distribution doesn’t change. Think about how a scale model of a building has the same proportions as the original, just smaller. That’s why we say it is drawn to scale. The range is often set at 0 to 1.
* Standardize generally means changing the values so that the distribution standard deviation from the mean equals one. It outputs something very close to a normal distribution. Scaling is often implied.
* Normalize can be used to mean either of the above things (and more!). I suggest you avoid the term normalize, because it has many definitions and is prone to creating confusion.
via [Machine Learning Mastery](https://machinelearningmastery.com/standardscaler-and-minmaxscaler-transforms-in-python/):
* If the distribution of the quantity is normal, then it should be standardized, otherwise, the data should be normalized.
```
house_prices = pd.read_csv("data/house-prices.csv")
house_prices["AgeWhenSold"] = house_prices["YrSold"] - house_prices["YearBuilt"]
house_prices.head()
```
## Unscaled Housing Prices Age When Sold
```
sns.displot(house_prices["AgeWhenSold"])
plt.xticks(rotation=90)
plt.show()
```
## StandardScaler
Note that DataFrame.var and DataFrame.std default to using 1 degree of freedom (ddof=1) but StandardScaler is using numpy's versions which default to ddof=0. That's why when printing the variance and standard deviation of the original data frame, we're specifying ddof=0. ddof=1 is known as Bessel's correction.
```
df = pd.DataFrame({
'col1': [1, 2, 3],
'col2': [10, 20, 30],
'col3': [0, 20, 22]
})
print("Original:\n")
print(df)
print("\nColumn means:\n")
print(df.mean())
print("\nOriginal variance:\n")
print(df.var(ddof=0))
print("\nOriginal standard deviations:\n")
print(df.std(ddof=0))
scaler = StandardScaler()
df1 = pd.DataFrame(scaler.fit_transform(df), columns=df.columns)
print("\nAfter scaling:\n")
print(df1)
print("\nColumn means:\n")
print(round(df1.mean(), 3))
print("\nVariance:\n")
print(df1.var(ddof=0))
print("\nStandard deviations:\n")
print(df1.std(ddof=0))
print("\nExample calculation for col2:")
print("z = (x - mean) / std")
print("z = (10 - 20) / 8.164966 = -1.224745")
```
### Standard Scaler with Age When Sold
```
scaler = StandardScaler()
age_when_sold_scaled = scaler.fit_transform(house_prices["AgeWhenSold"].values.reshape(-1, 1))
sns.displot(age_when_sold_scaled)
plt.xticks(rotation=90)
plt.show()
```
## Whiten
```
x_new = x / std(x)
```
```
data = [5, 1, 3, 3, 2, 3, 8, 1, 2, 2, 3, 5]
print("Original:", data)
print("\nStd Dev:", np.std(data))
scaled = whiten(data)
print("\nScaled with Whiten:", scaled)
scaled_manual = data / np.std(data)
print("\nScaled Manuallly:", scaled_manual)
```
## MinMax
Scales to a value between 0 and 1.
More suspectible to influence by outliers.
### Housing Prices Age When Sold
```
scaler = MinMaxScaler()
age_when_sold_scaled = scaler.fit_transform(house_prices["AgeWhenSold"].values.reshape(-1, 1))
sns.displot(age_when_sold_scaled)
plt.xticks(rotation=90)
plt.show()
```
## Robust Scaler
```
scaler = RobustScaler()
age_when_sold_scaled = scaler.fit_transform(house_prices["AgeWhenSold"].values.reshape(-1, 1))
sns.displot(age_when_sold_scaled)
plt.xticks(rotation=90)
plt.show()
```
| github_jupyter |
# Tutorial 6.3. Advanced Topics on Extreme Value Analysis
### Description: Some advanced topics on Extreme Value Analysis are presented.
#### Students are advised to complete the exercises.
Project: Structural Wind Engineering WS19-20
Chair of Structural Analysis @ TUM - R. Wüchner, M. Péntek
Author: [email protected], [email protected]
Created on: 24.12.2019
Last update: 08.01.2020
##### Contents:
1. Prediction of the extreme value of a time series - MaxMin Estimation
2. Lieblein's BLUE method
The worksheet is based on the knowledge base and scripts provided by [NIST](https://www.itl.nist.gov/div898/winds/overview.htm) as well as work available from [Christopher Howlett](https://github.com/chowlet5) from UWO.
```
# import
import matplotlib.pyplot as plt
import numpy as np
from scipy.stats import gumbel_r as gumbel
from ipywidgets import interactive
#external files
from peakpressure import maxminest
from blue4pressure import *
import custom_utilities as c_utils
```
## 1. Prediction of the extreme value of a time series - MaxMin Estimation
#### This method is based on [the procedure (and sample Matlab file](https://www.itl.nist.gov/div898/winds/peakest_files/peakest.htm) by Sadek, F. and Simiu, E. (2002). "Peak non-gaussian wind effects for database-assisted low-rise building design." Journal of Engineering Mechanics, 128(5), 530-539. Please find it [here](https://www.itl.nist.gov/div898/winds/pdf_files/b02030.pdf).
The method uses
* gamma distribution for estimating the peaks corresponding to the longer tail of time series
* normal distribution for estimating the peaks corresponding to the shorter tail of time series
The distribution of the peaks is then estimated by using the standard translation processes approach.
#### implementation details :
INPUT ARGUMENTS:
Each row of *record* is a time series.
The optional input argument *dur_ratio* allows peaks to be estimated for
a duration that differs from the duration of the record itself:
*dur_ratio* = [duration for peak estimation]/[duration of record]
(If unspecified, a value of 1 is used.)
OUTPUT ARGUMENTS:
* *max_est* gives the expected maximum values of each row of *record*
* *min_est* gives the expected minimum values of each row of *record*
* *max_std* gives the standard deviations of the maximum value for each row of *record*
* *min_std* gives the standard deviations of the minimum value for each row of *record*
#### Let us test the method for a given time series
```
# using as sample input some pre-generated generalized extreme value random series
given_series = np.loadtxt('test_data_gevrnd.dat', skiprows=0, usecols = (0,))
# print results
dur_ratio = 1
result = maxminest(given_series, dur_ratio)
maxv = result[0][0][0]
minv = result[1][0][0]
print('estimation of maximum value ', np.around(maxv,3))
print('estimation of minimum value ', np.around(minv,3))
plt.figure(num=1, figsize=(8, 6))
x_series = np.arange(0.0, len(given_series), 1.0)
plt.plot(x_series, given_series)
plt.ylabel('Amplitude')
plt.xlabel('Time [s]')
plt.hlines([maxv, minv], x_series[0], x_series[-1])
plt.title('Predicted extrema')
plt.grid(True)
plt.show()
```
#### Let us plot the pdf and cdf
```
[pdf_x, pdf_y] = c_utils.get_pdf(given_series)
ecdf_y = c_utils.get_ecdf(pdf_x, pdf_y)
plt.figure(num=2, figsize=(16, 6))
plt.subplot(1,2,1)
plt.plot(pdf_x, pdf_y)
plt.ylabel('PDF(Amplitude)')
plt.grid(True)
plt.subplot(1,2,2)
plt.plot(pdf_x, ecdf_y)
plt.vlines([maxv, minv], 0, 1)
plt.ylabel('CDF(Amplitude)')
plt.grid(True)
plt.show()
```
## 2. Lieblein's BLUE method
From a time series of pressure coefficients, *blue4pressure.py* estimates
extremes of positive and negative pressures based on Lieblein's BLUE
(Best Linear Unbiased Estimate) method applied to n epochs.
Extremes are estimated for 1 and dur epochs for probabilities of non-exceedance
P1 and P2 of the Gumbel distribution fitted to the epochal peaks.
*n* = integer, dur need not be an integer.
Written by Dat Duthinh 8_25_2015, 2_2_2016, 2_6_2017.
For further reference check out the material provided by [NIST](https://www.itl.nist.gov/div898/winds/gumbel_blue/gumbblue.htm).
Reference:
1) Julius Lieblein "Efficient Methods of Extreme-Value
Methodology" NBSIR 74-602 OCT 1974 for n = 4:16
2) Nicholas John Cook "The designer's guide to wind loading of
building structures" part 1, British Research Establishment 1985 Table C3
pp. 321-323 for n = 17:24. Extension to n=100 by Adam Pintar Feb 12 2016.
3) INTERNATIONAL STANDARD, ISO 4354 (2009-06-01), 2nd edition, “Wind
actions on structures,” Annex D (informative) “Aerodynamic pressure and
force coefficients,” Geneva, Switzerland, p. 22
#### implementation details :
INPUT ARGUMENTS
* *cp* = vector of time history of pressure coefficients
* *n* = number of epochs (integer)of cp data, 4 <= n <= 100
* *dur* = number of epochs for estimation of extremes. Default dur = n dur need not be an integer
* *P1, P2* = probabilities of non-exceedance of extremes in EV1 (Gumbel), P1 defaults to 0.80 (ISO)and P2 to 0.5704 (mean) for the Gumbel distribution .
OUTPUT ARGUMENTS
* *suffix max* for + peaks, min for - peaks of pressure coeff.
* *p1_max* (p1_min)= extreme value of positive (negative) peaks with probability of non-exceedance P1 for 1 epoch
* *p2_max* (p2_min)= extreme value of positive (negative) peaks with probability of exceedance P2 for 1 epoch
* *p1_rmax* (p1_rmin)= extreme value of positive (negative) peaks with probability of non-exceedance P1 for dur epochs
* *p2_rmax* (p2_rmin)= extreme value of positive (negative) peaks with probability of non-exceedance P2 for for dur epochs
* *cp_max* (cp_min)= vector of n positive (negative) epochal peaks
* *u_max, b_max* (u_min, b_min) = location and scale parameters of EV1 (Gumbel) for positive (negative) peaks
```
# n = number of epochs (integer)of cp data, 4 <= n <= 100
n=4
# P1, P2 = probabilities of non-exceedance of extremes in EV1 (Gumbel).
P1=0.80
P2=0.5704 # this corresponds to the mean of gumbel distribution
# dur = number of epochs for estimation of extremes. Default dur = n
# dur need not be an integer
dur=1
# Call function
result = blue4pressure(given_series, n, P1, P2, dur)
p1_max = result[0][0]
p2_max = result[1][0]
umax = result[4][0] # location parameters
b_max = result[5][0] # sclae parameters
p1_min = result[7][0]
p2_min = result[8][0]
umin = result[11][0] # location parameters
b_min = result[12][0] # scale parameters
# print results
## maximum
print('estimation of maximum value with probability of non excedence of p1', np.around(p1_max,3))
print('estimation of maximum value with probability of non excedence of p2', np.around(p2_max,3))
## minimum
print('estimation of minimum value with probability of non excedence of p1', np.around(p1_min,3))
print('estimation of minimum value with probability of non excedence of p2', np.around(p2_min,3))
```
#### Let us plot the pdf and cdf for the maximum values
```
max_pdf_x = np.linspace(1, 3, 100)
max_pdf_y = gumbel.pdf(max_pdf_x, umax, b_max)
max_ecdf_y = c_utils.get_ecdf(max_pdf_x, max_pdf_y)
plt.figure(num=3, figsize=(16, 6))
plt.subplot(1,2,1)
# PDF generated as a fitted curve using generalized extreme distribution
plt.plot(max_pdf_x, max_pdf_y, label = 'PDF from the fitted Gumbel')
plt.xlabel('Max values')
plt.ylabel('PDF(Amplitude)')
plt.title('PDF of Maxima')
plt.grid(True)
plt.legend()
plt.subplot(1,2,2)
plt.plot(max_pdf_x, max_ecdf_y)
plt.vlines([p1_max, p2_max], 0, 1)
plt.ylabel('CDF(Amplitude)')
plt.grid(True)
plt.show()
```
#### Try plotting these for the minimum values. Discuss among groups the advanced extreme value evaluation methods.
| github_jupyter |
# What's this TensorFlow business?
You've written a lot of code in this assignment to provide a whole host of neural network functionality. Dropout, Batch Norm, and 2D convolutions are some of the workhorses of deep learning in computer vision. You've also worked hard to make your code efficient and vectorized.
For the last part of this assignment, though, we're going to leave behind your beautiful codebase and instead migrate to one of two popular deep learning frameworks: in this instance, TensorFlow (or PyTorch, if you choose to work with that notebook).
#### What is it?
TensorFlow is a system for executing computational graphs over Tensor objects, with native support for performing backpropogation for its Variables. In it, we work with Tensors which are n-dimensional arrays analogous to the numpy ndarray.
#### Why?
* Our code will now run on GPUs! Much faster training. Writing your own modules to run on GPUs is beyond the scope of this class, unfortunately.
* We want you to be ready to use one of these frameworks for your project so you can experiment more efficiently than if you were writing every feature you want to use by hand.
* We want you to stand on the shoulders of giants! TensorFlow and PyTorch are both excellent frameworks that will make your lives a lot easier, and now that you understand their guts, you are free to use them :)
* We want you to be exposed to the sort of deep learning code you might run into in academia or industry.
## How will I learn TensorFlow?
TensorFlow has many excellent tutorials available, including those from [Google themselves](https://www.tensorflow.org/get_started/get_started).
Otherwise, this notebook will walk you through much of what you need to do to train models in TensorFlow. See the end of the notebook for some links to helpful tutorials if you want to learn more or need further clarification on topics that aren't fully explained here.
**NOTE: This notebook is meant to teach you the latest version of Tensorflow which is as of this homework version `2.2.0-rc3`. Most examples on the web today are still in 1.x, so be careful not to confuse the two when looking up documentation**.
## Install Tensorflow 2.0 (ONLY IF YOU ARE WORKING LOCALLY)
1. Have the latest version of Anaconda installed on your machine.
2. Create a new conda environment starting from Python 3.7. In this setup example, we'll call it `tf_20_env`.
3. Run the command: `source activate tf_20_env`
4. Then pip install TF 2.0 as described here: https://www.tensorflow.org/install
# Table of Contents
This notebook has 5 parts. We will walk through TensorFlow at **three different levels of abstraction**, which should help you better understand it and prepare you for working on your project.
1. Part I, Preparation: load the CIFAR-10 dataset.
2. Part II, Barebone TensorFlow: **Abstraction Level 1**, we will work directly with low-level TensorFlow graphs.
3. Part III, Keras Model API: **Abstraction Level 2**, we will use `tf.keras.Model` to define arbitrary neural network architecture.
4. Part IV, Keras Sequential + Functional API: **Abstraction Level 3**, we will use `tf.keras.Sequential` to define a linear feed-forward network very conveniently, and then explore the functional libraries for building unique and uncommon models that require more flexibility.
5. Part V, CIFAR-10 open-ended challenge: please implement your own network to get as high accuracy as possible on CIFAR-10. You can experiment with any layer, optimizer, hyperparameters or other advanced features.
We will discuss Keras in more detail later in the notebook.
Here is a table of comparison:
| API | Flexibility | Convenience |
|---------------|-------------|-------------|
| Barebone | High | Low |
| `tf.keras.Model` | High | Medium |
| `tf.keras.Sequential` | Low | High |
# Part I: Preparation
First, we load the CIFAR-10 dataset. This might take a few minutes to download the first time you run it, but after that the files should be cached on disk and loading should be faster.
In previous parts of the assignment we used CS231N-specific code to download and read the CIFAR-10 dataset; however the `tf.keras.datasets` package in TensorFlow provides prebuilt utility functions for loading many common datasets.
For the purposes of this assignment we will still write our own code to preprocess the data and iterate through it in minibatches. The `tf.data` package in TensorFlow provides tools for automating this process, but working with this package adds extra complication and is beyond the scope of this notebook. However using `tf.data` can be much more efficient than the simple approach used in this notebook, so you should consider using it for your project.
```
import os
import tensorflow as tf
import numpy as np
import math
import timeit
import matplotlib.pyplot as plt
%matplotlib inline
def load_cifar10(num_training=49000, num_validation=1000, num_test=10000):
"""
Fetch the CIFAR-10 dataset from the web and perform preprocessing to prepare
it for the two-layer neural net classifier. These are the same steps as
we used for the SVM, but condensed to a single function.
"""
# Load the raw CIFAR-10 dataset and use appropriate data types and shapes
cifar10 = tf.keras.datasets.cifar10.load_data()
(X_train, y_train), (X_test, y_test) = cifar10
X_train = np.asarray(X_train, dtype=np.float32)
y_train = np.asarray(y_train, dtype=np.int32).flatten()
X_test = np.asarray(X_test, dtype=np.float32)
y_test = np.asarray(y_test, dtype=np.int32).flatten()
# Subsample the data
mask = range(num_training, num_training + num_validation)
X_val = X_train[mask]
y_val = y_train[mask]
mask = range(num_training)
X_train = X_train[mask]
y_train = y_train[mask]
mask = range(num_test)
X_test = X_test[mask]
y_test = y_test[mask]
# Normalize the data: subtract the mean pixel and divide by std
mean_pixel = X_train.mean(axis=(0, 1, 2), keepdims=True)
std_pixel = X_train.std(axis=(0, 1, 2), keepdims=True)
X_train = (X_train - mean_pixel) / std_pixel
X_val = (X_val - mean_pixel) / std_pixel
X_test = (X_test - mean_pixel) / std_pixel
return X_train, y_train, X_val, y_val, X_test, y_test
# If there are errors with SSL downloading involving self-signed certificates,
# it may be that your Python version was recently installed on the current machine.
# See: https://github.com/tensorflow/tensorflow/issues/10779
# To fix, run the command: /Applications/Python\ 3.7/Install\ Certificates.command
# ...replacing paths as necessary.
# Invoke the above function to get our data.
NHW = (0, 1, 2)
X_train, y_train, X_val, y_val, X_test, y_test = load_cifar10()
print('Train data shape: ', X_train.shape)
print('Train labels shape: ', y_train.shape, y_train.dtype)
print('Validation data shape: ', X_val.shape)
print('Validation labels shape: ', y_val.shape)
print('Test data shape: ', X_test.shape)
print('Test labels shape: ', y_test.shape)
class Dataset(object):
def __init__(self, X, y, batch_size, shuffle=False):
"""
Construct a Dataset object to iterate over data X and labels y
Inputs:
- X: Numpy array of data, of any shape
- y: Numpy array of labels, of any shape but with y.shape[0] == X.shape[0]
- batch_size: Integer giving number of elements per minibatch
- shuffle: (optional) Boolean, whether to shuffle the data on each epoch
"""
assert X.shape[0] == y.shape[0], 'Got different numbers of data and labels'
self.X, self.y = X, y
self.batch_size, self.shuffle = batch_size, shuffle
def __iter__(self):
N, B = self.X.shape[0], self.batch_size
idxs = np.arange(N)
if self.shuffle:
np.random.shuffle(idxs)
return iter((self.X[i:i+B], self.y[i:i+B]) for i in range(0, N, B))
train_dset = Dataset(X_train, y_train, batch_size=64, shuffle=True)
val_dset = Dataset(X_val, y_val, batch_size=64, shuffle=False)
test_dset = Dataset(X_test, y_test, batch_size=64)
# We can iterate through a dataset like this:
for t, (x, y) in enumerate(train_dset):
print(t, x.shape, y.shape)
if t > 5: break
```
You can optionally **use GPU by setting the flag to True below**.
## Colab Users
If you are using Colab, you need to manually switch to a GPU device. You can do this by clicking `Runtime -> Change runtime type` and selecting `GPU` under `Hardware Accelerator`. Note that you have to rerun the cells from the top since the kernel gets restarted upon switching runtimes.
```
# Set up some global variables
USE_GPU = True
if USE_GPU:
device = '/device:GPU:0'
else:
device = '/cpu:0'
# Constant to control how often we print when training models
print_every = 100
print('Using device: ', device)
```
# Part II: Barebones TensorFlow
TensorFlow ships with various high-level APIs which make it very convenient to define and train neural networks; we will cover some of these constructs in Part III and Part IV of this notebook. In this section we will start by building a model with basic TensorFlow constructs to help you better understand what's going on under the hood of the higher-level APIs.
**"Barebones Tensorflow" is important to understanding the building blocks of TensorFlow, but much of it involves concepts from TensorFlow 1.x.** We will be working with legacy modules such as `tf.Variable`.
Therefore, please read and understand the differences between legacy (1.x) TF and the new (2.0) TF.
### Historical background on TensorFlow 1.x
TensorFlow 1.x is primarily a framework for working with **static computational graphs**. Nodes in the computational graph are Tensors which will hold n-dimensional arrays when the graph is run; edges in the graph represent functions that will operate on Tensors when the graph is run to actually perform useful computation.
Before Tensorflow 2.0, we had to configure the graph into two phases. There are plenty of tutorials online that explain this two-step process. The process generally looks like the following for TF 1.x:
1. **Build a computational graph that describes the computation that you want to perform**. This stage doesn't actually perform any computation; it just builds up a symbolic representation of your computation. This stage will typically define one or more `placeholder` objects that represent inputs to the computational graph.
2. **Run the computational graph many times.** Each time the graph is run (e.g. for one gradient descent step) you will specify which parts of the graph you want to compute, and pass a `feed_dict` dictionary that will give concrete values to any `placeholder`s in the graph.
### The new paradigm in Tensorflow 2.0
Now, with Tensorflow 2.0, we can simply adopt a functional form that is more Pythonic and similar in spirit to PyTorch and direct Numpy operation. Instead of the 2-step paradigm with computation graphs, making it (among other things) easier to debug TF code. You can read more details at https://www.tensorflow.org/guide/eager.
The main difference between the TF 1.x and 2.0 approach is that the 2.0 approach doesn't make use of `tf.Session`, `tf.run`, `placeholder`, `feed_dict`. To get more details of what's different between the two version and how to convert between the two, check out the official migration guide: https://www.tensorflow.org/alpha/guide/migration_guide
Later, in the rest of this notebook we'll focus on this new, simpler approach.
### TensorFlow warmup: Flatten Function
We can see this in action by defining a simple `flatten` function that will reshape image data for use in a fully-connected network.
In TensorFlow, data for convolutional feature maps is typically stored in a Tensor of shape N x H x W x C where:
- N is the number of datapoints (minibatch size)
- H is the height of the feature map
- W is the width of the feature map
- C is the number of channels in the feature map
This is the right way to represent the data when we are doing something like a 2D convolution, that needs spatial understanding of where the intermediate features are relative to each other. When we use fully connected affine layers to process the image, however, we want each datapoint to be represented by a single vector -- it's no longer useful to segregate the different channels, rows, and columns of the data. So, we use a "flatten" operation to collapse the `H x W x C` values per representation into a single long vector.
Notice the `tf.reshape` call has the target shape as `(N, -1)`, meaning it will reshape/keep the first dimension to be N, and then infer as necessary what the second dimension is in the output, so we can collapse the remaining dimensions from the input properly.
**NOTE**: TensorFlow and PyTorch differ on the default Tensor layout; TensorFlow uses N x H x W x C but PyTorch uses N x C x H x W.
```
def flatten(x):
"""
Input:
- TensorFlow Tensor of shape (N, D1, ..., DM)
Output:
- TensorFlow Tensor of shape (N, D1 * ... * DM)
"""
N = tf.shape(x)[0]
return tf.reshape(x, (N, -1))
def test_flatten():
# Construct concrete values of the input data x using numpy
x_np = np.arange(24).reshape((2, 3, 4))
print('x_np:\n', x_np, '\n')
# Compute a concrete output value.
x_flat_np = flatten(x_np)
print('x_flat_np:\n', x_flat_np, '\n')
test_flatten()
```
### Barebones TensorFlow: Define a Two-Layer Network
We will now implement our first neural network with TensorFlow: a fully-connected ReLU network with two hidden layers and no biases on the CIFAR10 dataset. For now we will use only low-level TensorFlow operators to define the network; later we will see how to use the higher-level abstractions provided by `tf.keras` to simplify the process.
We will define the forward pass of the network in the function `two_layer_fc`; this will accept TensorFlow Tensors for the inputs and weights of the network, and return a TensorFlow Tensor for the scores.
After defining the network architecture in the `two_layer_fc` function, we will test the implementation by checking the shape of the output.
**It's important that you read and understand this implementation.**
```
def two_layer_fc(x, params):
"""
A fully-connected neural network; the architecture is:
fully-connected layer -> ReLU -> fully connected layer.
Note that we only need to define the forward pass here; TensorFlow will take
care of computing the gradients for us.
The input to the network will be a minibatch of data, of shape
(N, d1, ..., dM) where d1 * ... * dM = D. The hidden layer will have H units,
and the output layer will produce scores for C classes.
Inputs:
- x: A TensorFlow Tensor of shape (N, d1, ..., dM) giving a minibatch of
input data.
- params: A list [w1, w2] of TensorFlow Tensors giving weights for the
network, where w1 has shape (D, H) and w2 has shape (H, C).
Returns:
- scores: A TensorFlow Tensor of shape (N, C) giving classification scores
for the input data x.
"""
w1, w2 = params # Unpack the parameters
x = flatten(x) # Flatten the input; now x has shape (N, D)
h = tf.nn.relu(tf.matmul(x, w1)) # Hidden layer: h has shape (N, H)
scores = tf.matmul(h, w2) # Compute scores of shape (N, C)
return scores
def two_layer_fc_test():
hidden_layer_size = 42
# Scoping our TF operations under a tf.device context manager
# lets us tell TensorFlow where we want these Tensors to be
# multiplied and/or operated on, e.g. on a CPU or a GPU.
with tf.device(device):
x = tf.zeros((64, 32, 32, 3))
w1 = tf.zeros((32 * 32 * 3, hidden_layer_size))
w2 = tf.zeros((hidden_layer_size, 10))
# Call our two_layer_fc function for the forward pass of the network.
scores = two_layer_fc(x, [w1, w2])
print(scores.shape)
two_layer_fc_test()
```
### Barebones TensorFlow: Three-Layer ConvNet
Here you will complete the implementation of the function `three_layer_convnet` which will perform the forward pass of a three-layer convolutional network. The network should have the following architecture:
1. A convolutional layer (with bias) with `channel_1` filters, each with shape `KW1 x KH1`, and zero-padding of two
2. ReLU nonlinearity
3. A convolutional layer (with bias) with `channel_2` filters, each with shape `KW2 x KH2`, and zero-padding of one
4. ReLU nonlinearity
5. Fully-connected layer with bias, producing scores for `C` classes.
**HINT**: For convolutions: https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/nn/conv2d; be careful with padding!
**HINT**: For biases: https://www.tensorflow.org/performance/xla/broadcasting
```
def three_layer_convnet(x, params):
"""
A three-layer convolutional network with the architecture described above.
Inputs:
- x: A TensorFlow Tensor of shape (N, H, W, 3) giving a minibatch of images
- params: A list of TensorFlow Tensors giving the weights and biases for the
network; should contain the following:
- conv_w1: TensorFlow Tensor of shape (KH1, KW1, 3, channel_1) giving
weights for the first convolutional layer.
- conv_b1: TensorFlow Tensor of shape (channel_1,) giving biases for the
first convolutional layer.
- conv_w2: TensorFlow Tensor of shape (KH2, KW2, channel_1, channel_2)
giving weights for the second convolutional layer
- conv_b2: TensorFlow Tensor of shape (channel_2,) giving biases for the
second convolutional layer.
- fc_w: TensorFlow Tensor giving weights for the fully-connected layer.
Can you figure out what the shape should be?
- fc_b: TensorFlow Tensor giving biases for the fully-connected layer.
Can you figure out what the shape should be?
"""
conv_w1, conv_b1, conv_w2, conv_b2, fc_w, fc_b = params
scores = None
############################################################################
# TODO: Implement the forward pass for the three-layer ConvNet. #
############################################################################
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
paddings = tf.constant([[0,0], [2,2], [2,2], [0,0]])
x = tf.pad(x, paddings, 'CONSTANT')
conv1 = tf.nn.conv2d(x, conv_w1, strides=[1,1,1,1], padding="VALID")+conv_b1
relu1 = tf.nn.relu(conv1)
paddings = tf.constant([[0,0], [1,1], [1,1], [0,0]])
conv1 = tf.pad(conv1, paddings, 'CONSTANT')
conv2 = tf.nn.conv2d(conv1, conv_w2, strides=[1,1,1,1], padding="VALID")+conv_b2
relu2 = tf.nn.relu(conv2)
relu2 = flatten(relu2)
scores = tf.matmul(relu2, fc_w) + fc_b
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
############################################################################
# END OF YOUR CODE #
############################################################################
return scores
```
After defing the forward pass of the three-layer ConvNet above, run the following cell to test your implementation. Like the two-layer network, we run the graph on a batch of zeros just to make sure the function doesn't crash, and produces outputs of the correct shape.
When you run this function, `scores_np` should have shape `(64, 10)`.
```
def three_layer_convnet_test():
with tf.device(device):
x = tf.zeros((64, 32, 32, 3))
conv_w1 = tf.zeros((5, 5, 3, 6))
conv_b1 = tf.zeros((6,))
conv_w2 = tf.zeros((3, 3, 6, 9))
conv_b2 = tf.zeros((9,))
fc_w = tf.zeros((32 * 32 * 9, 10))
fc_b = tf.zeros((10,))
params = [conv_w1, conv_b1, conv_w2, conv_b2, fc_w, fc_b]
scores = three_layer_convnet(x, params)
# Inputs to convolutional layers are 4-dimensional arrays with shape
# [batch_size, height, width, channels]
print('scores_np has shape: ', scores.shape)
three_layer_convnet_test()
```
### Barebones TensorFlow: Training Step
We now define the `training_step` function performs a single training step. This will take three basic steps:
1. Compute the loss
2. Compute the gradient of the loss with respect to all network weights
3. Make a weight update step using (stochastic) gradient descent.
We need to use a few new TensorFlow functions to do all of this:
- For computing the cross-entropy loss we'll use `tf.nn.sparse_softmax_cross_entropy_with_logits`: https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/nn/sparse_softmax_cross_entropy_with_logits
- For averaging the loss across a minibatch of data we'll use `tf.reduce_mean`:
https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/reduce_mean
- For computing gradients of the loss with respect to the weights we'll use `tf.GradientTape` (useful for Eager execution): https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/GradientTape
- We'll mutate the weight values stored in a TensorFlow Tensor using `tf.assign_sub` ("sub" is for subtraction): https://www.tensorflow.org/api_docs/python/tf/assign_sub
```
def training_step(model_fn, x, y, params, learning_rate):
with tf.GradientTape() as tape:
scores = model_fn(x, params) # Forward pass of the model
loss = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=scores)
total_loss = tf.reduce_mean(loss)
grad_params = tape.gradient(total_loss, params)
# Make a vanilla gradient descent step on all of the model parameters
# Manually update the weights using assign_sub()
for w, grad_w in zip(params, grad_params):
w.assign_sub(learning_rate * grad_w)
return total_loss
def train_part2(model_fn, init_fn, learning_rate):
"""
Train a model on CIFAR-10.
Inputs:
- model_fn: A Python function that performs the forward pass of the model
using TensorFlow; it should have the following signature:
scores = model_fn(x, params) where x is a TensorFlow Tensor giving a
minibatch of image data, params is a list of TensorFlow Tensors holding
the model weights, and scores is a TensorFlow Tensor of shape (N, C)
giving scores for all elements of x.
- init_fn: A Python function that initializes the parameters of the model.
It should have the signature params = init_fn() where params is a list
of TensorFlow Tensors holding the (randomly initialized) weights of the
model.
- learning_rate: Python float giving the learning rate to use for SGD.
"""
params = init_fn() # Initialize the model parameters
for t, (x_np, y_np) in enumerate(train_dset):
# Run the graph on a batch of training data.
loss = training_step(model_fn, x_np, y_np, params, learning_rate)
# Periodically print the loss and check accuracy on the val set.
if t % print_every == 0:
print('Iteration %d, loss = %.4f' % (t, loss))
check_accuracy(val_dset, x_np, model_fn, params)
def check_accuracy(dset, x, model_fn, params):
"""
Check accuracy on a classification model, e.g. for validation.
Inputs:
- dset: A Dataset object against which to check accuracy
- x: A TensorFlow placeholder Tensor where input images should be fed
- model_fn: the Model we will be calling to make predictions on x
- params: parameters for the model_fn to work with
Returns: Nothing, but prints the accuracy of the model
"""
num_correct, num_samples = 0, 0
for x_batch, y_batch in dset:
scores_np = model_fn(x_batch, params).numpy()
y_pred = scores_np.argmax(axis=1)
num_samples += x_batch.shape[0]
num_correct += (y_pred == y_batch).sum()
acc = float(num_correct) / num_samples
print('Got %d / %d correct (%.2f%%)' % (num_correct, num_samples, 100 * acc))
```
### Barebones TensorFlow: Initialization
We'll use the following utility method to initialize the weight matrices for our models using Kaiming's normalization method.
[1] He et al, *Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification
*, ICCV 2015, https://arxiv.org/abs/1502.01852
```
def create_matrix_with_kaiming_normal(shape):
if len(shape) == 2:
fan_in, fan_out = shape[0], shape[1]
elif len(shape) == 4:
fan_in, fan_out = np.prod(shape[:3]), shape[3]
return tf.keras.backend.random_normal(shape) * np.sqrt(2.0 / fan_in)
```
### Barebones TensorFlow: Train a Two-Layer Network
We are finally ready to use all of the pieces defined above to train a two-layer fully-connected network on CIFAR-10.
We just need to define a function to initialize the weights of the model, and call `train_part2`.
Defining the weights of the network introduces another important piece of TensorFlow API: `tf.Variable`. A TensorFlow Variable is a Tensor whose value is stored in the graph and persists across runs of the computational graph; however unlike constants defined with `tf.zeros` or `tf.random_normal`, the values of a Variable can be mutated as the graph runs; these mutations will persist across graph runs. Learnable parameters of the network are usually stored in Variables.
You don't need to tune any hyperparameters, but you should achieve validation accuracies above 40% after one epoch of training.
```
def two_layer_fc_init():
"""
Initialize the weights of a two-layer network, for use with the
two_layer_network function defined above.
You can use the `create_matrix_with_kaiming_normal` helper!
Inputs: None
Returns: A list of:
- w1: TensorFlow tf.Variable giving the weights for the first layer
- w2: TensorFlow tf.Variable giving the weights for the second layer
"""
hidden_layer_size = 4000
w1 = tf.Variable(create_matrix_with_kaiming_normal((3 * 32 * 32, 4000)))
w2 = tf.Variable(create_matrix_with_kaiming_normal((4000, 10)))
return [w1, w2]
learning_rate = 1e-2
train_part2(two_layer_fc, two_layer_fc_init, learning_rate)
```
### Barebones TensorFlow: Train a three-layer ConvNet
We will now use TensorFlow to train a three-layer ConvNet on CIFAR-10.
You need to implement the `three_layer_convnet_init` function. Recall that the architecture of the network is:
1. Convolutional layer (with bias) with 32 5x5 filters, with zero-padding 2
2. ReLU
3. Convolutional layer (with bias) with 16 3x3 filters, with zero-padding 1
4. ReLU
5. Fully-connected layer (with bias) to compute scores for 10 classes
You don't need to do any hyperparameter tuning, but you should see validation accuracies above 43% after one epoch of training.
```
def three_layer_convnet_init():
"""
Initialize the weights of a Three-Layer ConvNet, for use with the
three_layer_convnet function defined above.
You can use the `create_matrix_with_kaiming_normal` helper!
Inputs: None
Returns a list containing:
- conv_w1: TensorFlow tf.Variable giving weights for the first conv layer
- conv_b1: TensorFlow tf.Variable giving biases for the first conv layer
- conv_w2: TensorFlow tf.Variable giving weights for the second conv layer
- conv_b2: TensorFlow tf.Variable giving biases for the second conv layer
- fc_w: TensorFlow tf.Variable giving weights for the fully-connected layer
- fc_b: TensorFlow tf.Variable giving biases for the fully-connected layer
"""
params = None
############################################################################
# TODO: Initialize the parameters of the three-layer network. #
############################################################################
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
conv_w1 = tf.Variable(kaiming_normal([5, 5, 3, 32]))
conv_b1 = tf.Variable(np.zeros([32]), dtype=tf.float32)
conv_w2 = tf.Variable(kaiming_normal([3, 3, 32, 16]))
conv_b2 = tf.Variable(np.zeros([16]), dtype=tf.float32)
fc_w = tf.Variable(kaiming_normal([32*32*16,10]))
fc_b = tf.Variable(np.zeros([10]), dtype=tf.float32)
params = (conv_w1, conv_b1, conv_w2, conv_b2, fc_w, fc_b)
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
############################################################################
# END OF YOUR CODE #
############################################################################
return params
learning_rate = 3e-3
train_part2(three_layer_convnet, three_layer_convnet_init, learning_rate)
```
# Part III: Keras Model Subclassing API
Implementing a neural network using the low-level TensorFlow API is a good way to understand how TensorFlow works, but it's a little inconvenient - we had to manually keep track of all Tensors holding learnable parameters. This was fine for a small network, but could quickly become unweildy for a large complex model.
Fortunately TensorFlow 2.0 provides higher-level APIs such as `tf.keras` which make it easy to build models out of modular, object-oriented layers. Further, TensorFlow 2.0 uses eager execution that evaluates operations immediately, without explicitly constructing any computational graphs. This makes it easy to write and debug models, and reduces the boilerplate code.
In this part of the notebook we will define neural network models using the `tf.keras.Model` API. To implement your own model, you need to do the following:
1. Define a new class which subclasses `tf.keras.Model`. Give your class an intuitive name that describes it, like `TwoLayerFC` or `ThreeLayerConvNet`.
2. In the initializer `__init__()` for your new class, define all the layers you need as class attributes. The `tf.keras.layers` package provides many common neural-network layers, like `tf.keras.layers.Dense` for fully-connected layers and `tf.keras.layers.Conv2D` for convolutional layers. Under the hood, these layers will construct `Variable` Tensors for any learnable parameters. **Warning**: Don't forget to call `super(YourModelName, self).__init__()` as the first line in your initializer!
3. Implement the `call()` method for your class; this implements the forward pass of your model, and defines the *connectivity* of your network. Layers defined in `__init__()` implement `__call__()` so they can be used as function objects that transform input Tensors into output Tensors. Don't define any new layers in `call()`; any layers you want to use in the forward pass should be defined in `__init__()`.
After you define your `tf.keras.Model` subclass, you can instantiate it and use it like the model functions from Part II.
### Keras Model Subclassing API: Two-Layer Network
Here is a concrete example of using the `tf.keras.Model` API to define a two-layer network. There are a few new bits of API to be aware of here:
We use an `Initializer` object to set up the initial values of the learnable parameters of the layers; in particular `tf.initializers.VarianceScaling` gives behavior similar to the Kaiming initialization method we used in Part II. You can read more about it here: https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/initializers/VarianceScaling
We construct `tf.keras.layers.Dense` objects to represent the two fully-connected layers of the model. In addition to multiplying their input by a weight matrix and adding a bias vector, these layer can also apply a nonlinearity for you. For the first layer we specify a ReLU activation function by passing `activation='relu'` to the constructor; the second layer uses softmax activation function. Finally, we use `tf.keras.layers.Flatten` to flatten the output from the previous fully-connected layer.
```
class TwoLayerFC(tf.keras.Model):
def __init__(self, hidden_size, num_classes):
super(TwoLayerFC, self).__init__()
initializer = tf.initializers.VarianceScaling(scale=2.0)
self.fc1 = tf.keras.layers.Dense(hidden_size, activation='relu',
kernel_initializer=initializer)
self.fc2 = tf.keras.layers.Dense(num_classes, activation='softmax',
kernel_initializer=initializer)
self.flatten = tf.keras.layers.Flatten()
def call(self, x, training=False):
x = self.flatten(x)
x = self.fc1(x)
x = self.fc2(x)
return x
def test_TwoLayerFC():
""" A small unit test to exercise the TwoLayerFC model above. """
input_size, hidden_size, num_classes = 50, 42, 10
x = tf.zeros((64, input_size))
model = TwoLayerFC(hidden_size, num_classes)
with tf.device(device):
scores = model(x)
print(scores.shape)
test_TwoLayerFC()
```
### Keras Model Subclassing API: Three-Layer ConvNet
Now it's your turn to implement a three-layer ConvNet using the `tf.keras.Model` API. Your model should have the same architecture used in Part II:
1. Convolutional layer with 5 x 5 kernels, with zero-padding of 2
2. ReLU nonlinearity
3. Convolutional layer with 3 x 3 kernels, with zero-padding of 1
4. ReLU nonlinearity
5. Fully-connected layer to give class scores
6. Softmax nonlinearity
You should initialize the weights of your network using the same initialization method as was used in the two-layer network above.
**Hint**: Refer to the documentation for `tf.keras.layers.Conv2D` and `tf.keras.layers.Dense`:
https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/keras/layers/Conv2D
https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/keras/layers/Dense
```
class ThreeLayerConvNet(tf.keras.Model):
def __init__(self, channel_1, channel_2, num_classes):
super(ThreeLayerConvNet, self).__init__()
########################################################################
# TODO: Implement the __init__ method for a three-layer ConvNet. You #
# should instantiate layer objects to be used in the forward pass. #
########################################################################
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
initializer = tf.variance_scaling_initializer(scale=2.0)
self.conv1 = tf.layers.Conv2D(channel_1, [5,5], [1,1], padding='valid',
kernel_initializer=initializer,
activation=tf.nn.relu)
self.conv2 = tf.layers.Conv2D(channel_2, [3,3], [1,1], padding='valid',
kernel_initializer=initializer,
activation=tf.nn.relu)
self.fc = tf.layers.Dense(num_classes, kernel_initializer=initializer)
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
########################################################################
# END OF YOUR CODE #
########################################################################
def call(self, x, training=False):
scores = None
########################################################################
# TODO: Implement the forward pass for a three-layer ConvNet. You #
# should use the layer objects defined in the __init__ method. #
########################################################################
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
padding = tf.constant([[0,0],[2,2],[2,2],[0,0]])
x = tf.pad(x, padding, 'CONSTANT')
x = self.conv1(x)
padding = tf.constant([[0,0],[1,1],[1,1],[0,0]])
x = tf.pad(x, padding, 'CONSTANT')
x = self.conv2(x)
x = tf.layers.flatten(x)
scores = self.fc(x)
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
########################################################################
# END OF YOUR CODE #
########################################################################
return scores
```
Once you complete the implementation of the `ThreeLayerConvNet` above you can run the following to ensure that your implementation does not crash and produces outputs of the expected shape.
```
def test_ThreeLayerConvNet():
channel_1, channel_2, num_classes = 12, 8, 10
model = ThreeLayerConvNet(channel_1, channel_2, num_classes)
with tf.device(device):
x = tf.zeros((64, 3, 32, 32))
scores = model(x)
print(scores.shape)
test_ThreeLayerConvNet()
```
### Keras Model Subclassing API: Eager Training
While keras models have a builtin training loop (using the `model.fit`), sometimes you need more customization. Here's an example, of a training loop implemented with eager execution.
In particular, notice `tf.GradientTape`. Automatic differentiation is used in the backend for implementing backpropagation in frameworks like TensorFlow. During eager execution, `tf.GradientTape` is used to trace operations for computing gradients later. A particular `tf.GradientTape` can only compute one gradient; subsequent calls to tape will throw a runtime error.
TensorFlow 2.0 ships with easy-to-use built-in metrics under `tf.keras.metrics` module. Each metric is an object, and we can use `update_state()` to add observations and `reset_state()` to clear all observations. We can get the current result of a metric by calling `result()` on the metric object.
```
def train_part34(model_init_fn, optimizer_init_fn, num_epochs=1, is_training=False):
"""
Simple training loop for use with models defined using tf.keras. It trains
a model for one epoch on the CIFAR-10 training set and periodically checks
accuracy on the CIFAR-10 validation set.
Inputs:
- model_init_fn: A function that takes no parameters; when called it
constructs the model we want to train: model = model_init_fn()
- optimizer_init_fn: A function which takes no parameters; when called it
constructs the Optimizer object we will use to optimize the model:
optimizer = optimizer_init_fn()
- num_epochs: The number of epochs to train for
Returns: Nothing, but prints progress during trainingn
"""
with tf.device(device):
# Compute the loss like we did in Part II
loss_fn = tf.keras.losses.SparseCategoricalCrossentropy()
model = model_init_fn()
optimizer = optimizer_init_fn()
train_loss = tf.keras.metrics.Mean(name='train_loss')
train_accuracy = tf.keras.metrics.SparseCategoricalAccuracy(name='train_accuracy')
val_loss = tf.keras.metrics.Mean(name='val_loss')
val_accuracy = tf.keras.metrics.SparseCategoricalAccuracy(name='val_accuracy')
t = 0
for epoch in range(num_epochs):
# Reset the metrics - https://www.tensorflow.org/alpha/guide/migration_guide#new-style_metrics
train_loss.reset_states()
train_accuracy.reset_states()
for x_np, y_np in train_dset:
with tf.GradientTape() as tape:
# Use the model function to build the forward pass.
scores = model(x_np, training=is_training)
loss = loss_fn(y_np, scores)
gradients = tape.gradient(loss, model.trainable_variables)
optimizer.apply_gradients(zip(gradients, model.trainable_variables))
# Update the metrics
train_loss.update_state(loss)
train_accuracy.update_state(y_np, scores)
if t % print_every == 0:
val_loss.reset_states()
val_accuracy.reset_states()
for test_x, test_y in val_dset:
# During validation at end of epoch, training set to False
prediction = model(test_x, training=False)
t_loss = loss_fn(test_y, prediction)
val_loss.update_state(t_loss)
val_accuracy.update_state(test_y, prediction)
template = 'Iteration {}, Epoch {}, Loss: {}, Accuracy: {}, Val Loss: {}, Val Accuracy: {}'
print (template.format(t, epoch+1,
train_loss.result(),
train_accuracy.result()*100,
val_loss.result(),
val_accuracy.result()*100))
t += 1
```
### Keras Model Subclassing API: Train a Two-Layer Network
We can now use the tools defined above to train a two-layer network on CIFAR-10. We define the `model_init_fn` and `optimizer_init_fn` that construct the model and optimizer respectively when called. Here we want to train the model using stochastic gradient descent with no momentum, so we construct a `tf.keras.optimizers.SGD` function; you can [read about it here](https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/optimizers/SGD).
You don't need to tune any hyperparameters here, but you should achieve validation accuracies above 40% after one epoch of training.
```
hidden_size, num_classes = 4000, 10
learning_rate = 1e-2
def model_init_fn():
return TwoLayerFC(hidden_size, num_classes)
def optimizer_init_fn():
return tf.keras.optimizers.SGD(learning_rate=learning_rate)
train_part34(model_init_fn, optimizer_init_fn)
```
### Keras Model Subclassing API: Train a Three-Layer ConvNet
Here you should use the tools we've defined above to train a three-layer ConvNet on CIFAR-10. Your ConvNet should use 32 filters in the first convolutional layer and 16 filters in the second layer.
To train the model you should use gradient descent with Nesterov momentum 0.9.
**HINT**: https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/optimizers/SGD
You don't need to perform any hyperparameter tuning, but you should achieve validation accuracies above 50% after training for one epoch.
```
learning_rate = 3e-3
channel_1, channel_2, num_classes = 32, 16, 10
def model_init_fn():
model = None
############################################################################
# TODO: Complete the implementation of model_fn. #
############################################################################
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
pass
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
############################################################################
# END OF YOUR CODE #
############################################################################
return model
def optimizer_init_fn():
optimizer = None
############################################################################
# TODO: Complete the implementation of model_fn. #
############################################################################
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
pass
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
############################################################################
# END OF YOUR CODE #
############################################################################
return optimizer
train_part34(model_init_fn, optimizer_init_fn)
```
# Part IV: Keras Sequential API
In Part III we introduced the `tf.keras.Model` API, which allows you to define models with any number of learnable layers and with arbitrary connectivity between layers.
However for many models you don't need such flexibility - a lot of models can be expressed as a sequential stack of layers, with the output of each layer fed to the next layer as input. If your model fits this pattern, then there is an even easier way to define your model: using `tf.keras.Sequential`. You don't need to write any custom classes; you simply call the `tf.keras.Sequential` constructor with a list containing a sequence of layer objects.
One complication with `tf.keras.Sequential` is that you must define the shape of the input to the model by passing a value to the `input_shape` of the first layer in your model.
### Keras Sequential API: Two-Layer Network
In this subsection, we will rewrite the two-layer fully-connected network using `tf.keras.Sequential`, and train it using the training loop defined above.
You don't need to perform any hyperparameter tuning here, but you should see validation accuracies above 40% after training for one epoch.
```
learning_rate = 1e-2
def model_init_fn():
input_shape = (32, 32, 3)
hidden_layer_size, num_classes = 4000, 10
initializer = tf.initializers.VarianceScaling(scale=2.0)
layers = [
tf.keras.layers.Flatten(input_shape=input_shape),
tf.keras.layers.Dense(hidden_layer_size, activation='relu',
kernel_initializer=initializer),
tf.keras.layers.Dense(num_classes, activation='softmax',
kernel_initializer=initializer),
]
model = tf.keras.Sequential(layers)
return model
def optimizer_init_fn():
return tf.keras.optimizers.SGD(learning_rate=learning_rate)
train_part34(model_init_fn, optimizer_init_fn)
```
### Abstracting Away the Training Loop
In the previous examples, we used a customised training loop to train models (e.g. `train_part34`). Writing your own training loop is only required if you need more flexibility and control during training your model. Alternately, you can also use built-in APIs like `tf.keras.Model.fit()` and `tf.keras.Model.evaluate` to train and evaluate a model. Also remember to configure your model for training by calling `tf.keras.Model.compile.
You don't need to perform any hyperparameter tuning here, but you should see validation and test accuracies above 42% after training for one epoch.
```
model = model_init_fn()
model.compile(optimizer=tf.keras.optimizers.SGD(learning_rate=learning_rate),
loss='sparse_categorical_crossentropy',
metrics=[tf.keras.metrics.sparse_categorical_accuracy])
model.fit(X_train, y_train, batch_size=64, epochs=1, validation_data=(X_val, y_val))
model.evaluate(X_test, y_test)
```
### Keras Sequential API: Three-Layer ConvNet
Here you should use `tf.keras.Sequential` to reimplement the same three-layer ConvNet architecture used in Part II and Part III. As a reminder, your model should have the following architecture:
1. Convolutional layer with 32 5x5 kernels, using zero padding of 2
2. ReLU nonlinearity
3. Convolutional layer with 16 3x3 kernels, using zero padding of 1
4. ReLU nonlinearity
5. Fully-connected layer giving class scores
6. Softmax nonlinearity
You should initialize the weights of the model using a `tf.initializers.VarianceScaling` as above.
You should train the model using Nesterov momentum 0.9.
You don't need to perform any hyperparameter search, but you should achieve accuracy above 45% after training for one epoch.
```
def model_init_fn():
model = None
############################################################################
# TODO: Construct a three-layer ConvNet using tf.keras.Sequential. #
############################################################################
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
pass
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
############################################################################
# END OF YOUR CODE #
############################################################################
return model
learning_rate = 5e-4
def optimizer_init_fn():
optimizer = None
############################################################################
# TODO: Complete the implementation of model_fn. #
############################################################################
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
pass
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
############################################################################
# END OF YOUR CODE #
############################################################################
return optimizer
train_part34(model_init_fn, optimizer_init_fn)
```
We will also train this model with the built-in training loop APIs provided by TensorFlow.
```
model = model_init_fn()
model.compile(optimizer='sgd',
loss='sparse_categorical_crossentropy',
metrics=[tf.keras.metrics.sparse_categorical_accuracy])
model.fit(X_train, y_train, batch_size=64, epochs=1, validation_data=(X_val, y_val))
model.evaluate(X_test, y_test)
```
## Part IV: Functional API
### Demonstration with a Two-Layer Network
In the previous section, we saw how we can use `tf.keras.Sequential` to stack layers to quickly build simple models. But this comes at the cost of losing flexibility.
Often we will have to write complex models that have non-sequential data flows: a layer can have **multiple inputs and/or outputs**, such as stacking the output of 2 previous layers together to feed as input to a third! (Some examples are residual connections and dense blocks.)
In such cases, we can use Keras functional API to write models with complex topologies such as:
1. Multi-input models
2. Multi-output models
3. Models with shared layers (the same layer called several times)
4. Models with non-sequential data flows (e.g. residual connections)
Writing a model with Functional API requires us to create a `tf.keras.Model` instance and explicitly write input tensors and output tensors for this model.
```
def two_layer_fc_functional(input_shape, hidden_size, num_classes):
initializer = tf.initializers.VarianceScaling(scale=2.0)
inputs = tf.keras.Input(shape=input_shape)
flattened_inputs = tf.keras.layers.Flatten()(inputs)
fc1_output = tf.keras.layers.Dense(hidden_size, activation='relu',
kernel_initializer=initializer)(flattened_inputs)
scores = tf.keras.layers.Dense(num_classes, activation='softmax',
kernel_initializer=initializer)(fc1_output)
# Instantiate the model given inputs and outputs.
model = tf.keras.Model(inputs=inputs, outputs=scores)
return model
def test_two_layer_fc_functional():
""" A small unit test to exercise the TwoLayerFC model above. """
input_size, hidden_size, num_classes = 50, 42, 10
input_shape = (50,)
x = tf.zeros((64, input_size))
model = two_layer_fc_functional(input_shape, hidden_size, num_classes)
with tf.device(device):
scores = model(x)
print(scores.shape)
test_two_layer_fc_functional()
```
### Keras Functional API: Train a Two-Layer Network
You can now train this two-layer network constructed using the functional API.
You don't need to perform any hyperparameter tuning here, but you should see validation accuracies above 40% after training for one epoch.
```
input_shape = (32, 32, 3)
hidden_size, num_classes = 4000, 10
learning_rate = 1e-2
def model_init_fn():
return two_layer_fc_functional(input_shape, hidden_size, num_classes)
def optimizer_init_fn():
return tf.keras.optimizers.SGD(learning_rate=learning_rate)
train_part34(model_init_fn, optimizer_init_fn)
```
# Part V: CIFAR-10 open-ended challenge
In this section you can experiment with whatever ConvNet architecture you'd like on CIFAR-10.
You should experiment with architectures, hyperparameters, loss functions, regularization, or anything else you can think of to train a model that achieves **at least 70%** accuracy on the **validation** set within 10 epochs. You can use the built-in train function, the `train_part34` function from above, or implement your own training loop.
Describe what you did at the end of the notebook.
### Some things you can try:
- **Filter size**: Above we used 5x5 and 3x3; is this optimal?
- **Number of filters**: Above we used 16 and 32 filters. Would more or fewer do better?
- **Pooling**: We didn't use any pooling above. Would this improve the model?
- **Normalization**: Would your model be improved with batch normalization, layer normalization, group normalization, or some other normalization strategy?
- **Network architecture**: The ConvNet above has only three layers of trainable parameters. Would a deeper model do better?
- **Global average pooling**: Instead of flattening after the final convolutional layer, would global average pooling do better? This strategy is used for example in Google's Inception network and in Residual Networks.
- **Regularization**: Would some kind of regularization improve performance? Maybe weight decay or dropout?
### NOTE: Batch Normalization / Dropout
If you are using Batch Normalization and Dropout, remember to pass `is_training=True` if you use the `train_part34()` function. BatchNorm and Dropout layers have different behaviors at training and inference time. `training` is a specific keyword argument reserved for this purpose in any `tf.keras.Model`'s `call()` function. Read more about this here : https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/keras/layers/BatchNormalization#methods
https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/keras/layers/Dropout#methods
### Tips for training
For each network architecture that you try, you should tune the learning rate and other hyperparameters. When doing this there are a couple important things to keep in mind:
- If the parameters are working well, you should see improvement within a few hundred iterations
- Remember the coarse-to-fine approach for hyperparameter tuning: start by testing a large range of hyperparameters for just a few training iterations to find the combinations of parameters that are working at all.
- Once you have found some sets of parameters that seem to work, search more finely around these parameters. You may need to train for more epochs.
- You should use the validation set for hyperparameter search, and save your test set for evaluating your architecture on the best parameters as selected by the validation set.
### Going above and beyond
If you are feeling adventurous there are many other features you can implement to try and improve your performance. You are **not required** to implement any of these, but don't miss the fun if you have time!
- Alternative optimizers: you can try Adam, Adagrad, RMSprop, etc.
- Alternative activation functions such as leaky ReLU, parametric ReLU, ELU, or MaxOut.
- Model ensembles
- Data augmentation
- New Architectures
- [ResNets](https://arxiv.org/abs/1512.03385) where the input from the previous layer is added to the output.
- [DenseNets](https://arxiv.org/abs/1608.06993) where inputs into previous layers are concatenated together.
- [This blog has an in-depth overview](https://chatbotslife.com/resnets-highwaynets-and-densenets-oh-my-9bb15918ee32)
### Have fun and happy training!
```
class CustomConvNet(tf.keras.Model):
def __init__(self):
super(CustomConvNet, self).__init__()
############################################################################
# TODO: Construct a model that performs well on CIFAR-10 #
############################################################################
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
pass
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
############################################################################
# END OF YOUR CODE #
############################################################################
def call(self, input_tensor, training=False):
############################################################################
# TODO: Construct a model that performs well on CIFAR-10 #
############################################################################
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
pass
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
############################################################################
# END OF YOUR CODE #
############################################################################
return x
print_every = 700
num_epochs = 10
model = CustomConvNet()
def model_init_fn():
return CustomConvNet()
def optimizer_init_fn():
learning_rate = 1e-3
return tf.keras.optimizers.Adam(learning_rate)
train_part34(model_init_fn, optimizer_init_fn, num_epochs=num_epochs, is_training=True)
```
## Describe what you did
In the cell below you should write an explanation of what you did, any additional features that you implemented, and/or any graphs that you made in the process of training and evaluating your network.
TODO: Tell us what you did
| github_jupyter |
<h1>Lists</h1>
<li>Sequential, Ordered Collection
<h2>Creating lists</h2>
```
x = [4,2,6,3] #Create a list with values
y = list() # Create an empty list
y = [] #Create an empty list
print(x)
print(y)
```
<h3>Adding items to a list</h3>
```
x=list()
print(x)
x.append('One') #Adds 'One' to the back of the empty list
print(x)
x.append('Two') #Adds 'Two' to the back of the list ['One']
print(x)
x.insert(0,'Half') #Inserts 'Half' at location 0. Items will shift to make roomw
print(x)
x=list()
x.extend([1,2,3]) #Unpacks the list and adds each item to the back of the list
print(x)
```
<h3>Indexing and slicing</h3>
```
x=[1,7,2,5,3,5,67,32]
print(len(x))
print(x[3])
print(x[2:5])
print(x[-1])
print(x[::-1])
```
<h3>Removing items from a list</h3>
```
x=[1,7,2,5,3,5,67,32]
x.pop() #Removes the last element from a list
print(x)
x.pop(3) #Removes element at item 3 from a list
print(x)
x.remove(7) #Removes the first 7 from the list
print(x)
```
<h3>Anything you want to remove must be in the list or the location must be inside the list</h3>
```
x.remove(20)
```
<h2>Mutablility of lists</h2>
```
y=['a','b']
x = [1,y,3]
print(x)
print(y)
y[1] = 4
print(y)
print(x)
x="Hello"
print(x,id(x))
x+=" You!"
print(x,id(x)) #x is not the same object it was
y=["Hello"]
print(y,id(y))
y+=["You!"]
print(y,id(y)) #y is still the same object. Lists are mutable. Strings are immutable
def eggs(item,total=0):
total+=item
return total
def spam(elem,some_list=[]):
some_list.append(elem)
return some_list
print(eggs(1))
print(eggs(2))
print(spam(1))
print(spam(2))
```
<h1>Iteration</h1>
<h2>Range iteration</h2>
```
#The for loop creates a new variable (e.g., index below)
#range(len(x)) generates values from 0 to len(x)
x=[1,7,2,5,3,5,67,32]
for index in range(len(x)):
print(x[index])
list(range(len(x)))
```
<h3>List element iteration</h3>
```
x=[1,7,2,5,3,5,67,32]
#The for draws elements - sequentially - from the list x and uses the variable "element" to store values
for element in x:
print(element)
```
<h3>Practice problem</h3>
Write a function search_list that searches a list of tuple pairs and returns the value associated with the first element of the pair
```
def search_list(list_of_tuples,value):
#Write the function here
for t in prices:
if t[0] == value:
return t[1]
prices = [('AAPL',96.43),('IONS',39.28),('GS',159.53)]
ticker = 'IONS'
print(search_list(prices,ticker))
```
<h1>Dictionaries</h1>
```
mktcaps = {'AAPL':538.7,'GOOG':68.7,'IONS':4.6}
mktcaps['AAPL'] #Returns the value associated with the key "AAPL"
mktcaps['GS'] #Error because GS is not in mktcaps
mktcaps.get('GS') #Returns None because GS is not in mktcaps
mktcaps['GS'] = 88.65 #Adds GS to the dictionary
print(mktcaps)
del(mktcaps['GOOG']) #Removes GOOG from mktcaps
print(mktcaps)
mktcaps.keys() #Returns all the keys
mktcaps.values() #Returns all the values
list1 = [1, 2, 3, 4, 5, 6, 7]
list1[0]
list1[:2]
list1[:-2]
list1[3:5]
data = [[[1, 2], [3, 4]], [[5, 6], [7, 8]]]
print(data[1][0][0])
numbers = [1, 2, 3, 4]
numbers.append([5, 6, 7, 8])
print(len(numbers))
list1 = [1, 2, 3, 4, 5, 6, 7]
print(list1[0])
print(list1[:2])
print(list1[:-2])
print(list1[3:5])
dict1 = {"john":40, "peter":45}
dict2 = {"john":466, "peter":45}
dict1 > dict2
dict1 = {"a":1, "b":2}# to delete the entry for "a":1, use ________.
#d.delete("a":1)
#dict1.delete("a")
#del dict1("a":1)
del dict1["a"]
dict1
s = {1, 2, 4, 3}# which of the following will result in an exception (error)? Multiple options may be correct.
#print(s[3])
print(max(s))
print(len(s))
#s[3] = 45
```
| github_jupyter |
```
import shapefile
import numpy as np
import xarray as xr
from shapely.geometry import mapping as mappy
from shapely.geometry import Polygon
import cartopy.crs as ccrs
import cartopy
import os, sys
import pandas as pd
import richdem as rd
import skimage
from matplotlib import pyplot as plt
%matplotlib inline
from skimage import measure
from skimage import util
from skimage import morphology
import numpy as np
import scipy
from scipy.ndimage import gaussian_filter
from skimage import data
from skimage import img_as_float
from skimage.morphology import reconstruction
tiffname = '38_49_8m_dem.tif'
metadata = '38_49_8m_dem_meta.txt'
DEM = rd.rdarray(xr.open_rasterio(tiffname),no_data = -9999)
DEMflat = DEM.squeeze()
DEMfig = rd.rdShow(DEMflat[6249:-1,6249:-1], ignore_colours=[0], axes=False, cmap='jet', figsize=(8,5.5))
geoDEM = xr.open_rasterio(tiffname)
geoDEM
from pyproj import Proj, transform
inProj = Proj(init= geoDEM.crs)
outProj = Proj(init='epsg:4326') #Plate Carree
x2,y2 = transform(inProj,outProj,list(geoDEM.x),list(geoDEM.y))
lat, lon = np.meshgrid(y2,x2)
latmesh = np.array([[lat],]*len(lat))
lonmesh = np.array([[lon],]*len(lat)).transpose()
DEMflat_ss = DEMflat[6249:-1,6249:-1]
import time
start=time.time()
#DEM40 = skimage.transform.resize(DEMflat_ss, (DEMflat_ss.shape[0] / 5, DEMflat_ss.shape[1] / 5),
#anti_aliasing=True)
#DEM40 = rd.rdarray(DEM40,no_data = -9999 )
DEMf = rd.FillDepressions(DEMflat)
lakes = DEMf-DEMflat
end = time.time()
print(end-start)
fig_diff = rd.rdShow(lakes, ignore_colours=[0], axes=False, cmap='jet', figsize=(8,5.5))
```
## Julian's Code
```
start = time.time()
# 5m meter lakes
DEM40 = skimage.transform.resize(DEMflat, (DEMflat.shape[0] / 10, DEMflat.shape[1] / 10),
anti_aliasing=True)
DEM_inv = skimage.util.invert(DEM40)
DEMinv_gaus = gaussian_filter(DEM_inv,1)
marker = np.copy(DEMinv_gaus)
marker[1:-1, 1:-1] = -9999
Inan = np.argwhere(np.isnan(DEMinv_gaus))
Inan_mask = np.isnan(DEMinv_gaus)
Inan_mask_not = np.logical_not(Inan_mask)
if((np.array(Inan)).size>0):
I = skimage.morphology.binary_dilation(Inan_mask,np.ones(3)) & Inan_mask_not
marker[I] = DEMinv_gaus[I]
mask = DEMinv_gaus
demfs = reconstruction(marker, mask, method='dilation')
D = DEMinv_gaus-demfs
index = list(Inan_mask_not)
maxdepth = 40
while np.any(D[index]>maxdepth):
lakemask = D>0
label_lakemask = measure.label(lakemask)
STATS = measure.regionprops(label_lakemask,D)
for r in np.arange(0,len(STATS)):
if(STATS[r].max_intensity < maxdepth):
pass
else:
poly_x = STATS[r].coords[:,0]
poly_y = STATS[r].coords[:,1]
poly = D[poly_x, poly_y]
ix = poly.argmax()
#ix = ix[1]
marker[STATS[r].coords[ix][0],STATS[r].coords[ix][1]] = DEMinv_gaus[STATS[r].coords[ix][0],STATS[r].coords[ix][1]]
demfs = reconstruction(marker,DEMinv_gaus, method='dilation');
D = DEMinv_gaus-demfs;
demfs = skimage.util.invert(demfs)
demfs[Inan_mask] = np.nan
end = time.time()
print(end-start)
fig, ax = plt.subplots(ncols = 2, figsize=(10,5))
lakes2 = demfs-DEM40
jjscodefig = ax[0].imshow(lakes2, cmap='jet')
jjscodefig.set_clim(0,8)
plt.colorbar(jjscodefig)
basecodefig = ax[1].imshow(lakes, cmap='jet')
basecodefig.set_clim(0,8)
#difference between methods
lakediff = lakes-lakes2
fig, ax = plt.subplots( figsize=(10,10))
plt.imshow(lakediff)
plt.colorbar()
plt.clim(-5,5)
```
Lake Properties
```
lakemask = lakes>0
label_lakes = measure.label(lakemask)
LakeProps = measure.regionprops(label_lakes,lakes)
numLakes = len(LakeProps)
Area = np.zeros((numLakes,1))
Orientation= np.zeros((numLakes,1))
Volume = np.zeros((numLakes,1))
Max_Depth = np.zeros((numLakes,1))
Mean_Depth = np.zeros((numLakes,1))
Min_Depth = np.zeros((numLakes,1))
Perimeter = np.zeros((numLakes,1))
PPscore = np.zeros((numLakes,1))
DVscore = np.zeros((numLakes,1))
Centroid = np.zeros((numLakes,2))
for lake in np.arange(0,numLakes):
Area[lake] = LakeProps[lake].area*8**2
Orientation[lake] = LakeProps[lake].orientation
Volume[lake] = LakeProps[lake].intensity_image.sum()*8**2
Max_Depth[lake] = LakeProps[lake].max_intensity
Mean_Depth[lake] = LakeProps[lake].mean_intensity
Min_Depth[lake] = LakeProps[lake].min_intensity
Perimeter[lake] = LakeProps[lake].perimeter*8
PPscore[lake] = (4*3.14*Area[lake])/(Perimeter[lake]**2)
DVscore[lake] = 3*Mean_Depth[lake]/Max_Depth[lake]
Centroid[lake] = LakeProps[lake].centroid
plt.scatter(Area, Max_Depth)
plt.xlim(0,1e5)
plt.ylim(0,5)
```
## Elevation Data
```
ElevationProps = measure.regionprops(label_lakes,DEM40)
numLakes = len(LakeProps)
Max_Elev = np.zeros((numLakes,1))
Mean_Elev = np.zeros((numLakes,1))
Min_Elev = np.zeros((numLakes,1))
for lake in np.arange(0,numLakes):
Max_Elev[lake] =ElevationProps[lake].max_intensity
Mean_Elev[lake] = ElevationProps[lake].mean_intensity
Min_Elev[lake] = ElevationProps[lake].min_intensity
```
## Full Tiles
```
xlength = DEMflat.shape[0]
ylength = DEMflat.shape[1]
quarter_tile = np.empty([int(xlength/2),int(ylength/2),4])
inProj = Proj(init= geoDEM.crs)
outProj = Proj(init='epsg:4326') #Plate Carree
lon,lat = transform(inProj,outProj,list(geoDEM.x),list(geoDEM.y))
quarter_tile[:,:,0] = DEMflat[0:int(xlength/2), 0:int(ylength/2)]
quarter_tile[:,:,1] = DEMflat[int(xlength/2):xlength,0:int(ylength/2)]
quarter_tile[:,:,2] = DEMflat[0:int(xlength/2),int(ylength/2):ylength]
quarter_tile[:,:,3] = DEMflat[int(xlength/2):xlength,int(ylength/2):ylength]
#coordinates_lat = np.empty([int(xlength/2),4])
#coordinates_lon = np.empty([int(ylength/2),4])
#coordinates_lon[:,0] = lon[0:int(xlength/2)]
#coordinates_lon[:,1] = lon[int(xlength/2):xlength]
#coordinates_lon[:,2] = lon[0:int(xlength/2)]
#coordinates_lon[:,3] = lon[int(xlength/2):xlength]
#coordinates_lat[:,0] = lat[0:int(ylength/2)]
#coordinates_lat[:,1] = lat[0:int(ylength/2)]
#coordinates_lat[:,2] = lat[int(ylength/2):ylength]
#coordinates_lat[:,3] = lat[int(ylength/2):ylength]
numLakes_total = 0
Area_total = []
Orientation_total= []
Volume_total = []
Max_Depth_total = []
Mean_Depth_total = []
Min_Depth_total = []
Perimeter_total = []
PPscore_total = []
DVscore_total = []
Max_Elev_total = []
Mean_Elev_total = []
Min_Elev_total = []
Centroidlat_total = []
Centroidlon_total = []
for tile in np.arange(0,3):
DEM40 = skimage.transform.resize(quarter_tile[:,:,tile], (quarter_tile[:,:,tile].shape[0] / 5,
quarter_tile[:,:,tile].shape[1] / 5), anti_aliasing=True)
DEM40 = rd.rdarray(DEM40,no_data = -9999 )
DEMf = rd.FillDepressions(DEM40)
lakes = DEMf-DEM40
lakemask = lakes>0
label_lakes = measure.label(lakemask)
LakeProps = measure.regionprops(label_lakes,lakes)
ElevationProps = measure.regionprops(label_lakes,DEM40)
numLakes = len(LakeProps)
Area = np.zeros((numLakes,1))
Orientation= np.zeros((numLakes,1))
Volume = np.zeros((numLakes,1))
Max_Depth = np.zeros((numLakes,1))
Mean_Depth = np.zeros((numLakes,1))
Min_Depth = np.zeros((numLakes,1))
Perimeter = np.zeros((numLakes,1))
PPscore = np.zeros((numLakes,1))
DVscore = np.zeros((numLakes,1))
Centroidlon = np.zeros((numLakes,1))
Centroidlat = np.zeros((numLakes,1))
for lake in np.arange(0,numLakes):
Area[lake] = LakeProps[lake].area*8**2
Orientation[lake] = LakeProps[lake].orientation
Volume[lake] = LakeProps[lake].intensity_image.sum()*8**2
Max_Depth[lake] = LakeProps[lake].max_intensity
Mean_Depth[lake] = LakeProps[lake].mean_intensity
Min_Depth[lake] = LakeProps[lake].min_intensity
Perimeter[lake] = LakeProps[lake].perimeter*8
PPscore[lake] = (4*3.14*Area[lake])/(Perimeter[lake]**2)
DVscore[lake] = 3*Mean_Depth[lake]/Max_Depth[lake]
Max_Elev[lake] =ElevationProps[lake].max_intensity
Mean_Elev[lake] = ElevationProps[lake].mean_intensity
Min_Elev[lake] = ElevationProps[lake].min_intensity
Centroidlat[lake] = coordinates_lat[int(round(LakeProps[lake].centroid[0])),tile]
Centroidlon[lake] = coordinates_lon[int(round(LakeProps[lake].centroid[1])),tile]
numLakes_total = numLakes_total+numLakes
Area_total = np.append(Area_total,Area)
Orientation_total= np.append(Orientation_total,Orientation)
Volume_total = np.append(Volume_total,Volume)
Max_Depth_total =np.append(Max_Depth_total,Max_Depth)
Mean_Depth_total = np.append(Mean_Depth_total,Mean_Depth)
Min_Depth_total = np.append(Min_Depth_total,Min_Depth)
Perimeter_total = np.append(Perimeter_total,Perimeter)
PPscore_total = np.append(PPscore_total,PPscore)
DVscore_total = np.append(DVscore_total,DVscore)
Max_Elev_total = np.append(Max_Elev_total,Max_Elev)
Mean_Elev_total = np.append(Mean_Elev_total,Mean_Elev)
Min_Elev_total = np.append(Min_Elev_total,Min_Elev)
Centroidlat_total = np.append(Centroidlat_total,Centroidlat)
Centroidlon_total = np.append(Centroidlon_total, Centroidlon)
coordinates_lat[int(round(LakeProps[1].centroid[0])),1]
plt.scatter(Centroidlat_total,Centroidlon_total)
len(lon)
```
| github_jupyter |
# Authoring repeatable processes aka AzureML pipelines
```
from azureml.core import Workspace
ws = Workspace.from_config()
dataset = ws.datasets["diabetes-tabular"]
compute_target = ws.compute_targets["cpu-cluster"]
from azureml.core import RunConfiguration
# To simplify we are going to use a big demo environment instead
# of creating our own specialized environment. We will also use
# the same environment for all steps, but this is not needed.
runconfig = RunConfiguration()
runconfig.environment = ws.environments["AzureML-lightgbm-3.2-ubuntu18.04-py37-cpu"]
```
## Step 1 - Convert data into LightGBM dataset
```
from azureml.pipeline.core import PipelineData
step01_output = PipelineData(
"training_data", datastore=ws.get_default_datastore(), is_directory=True
)
from azureml.pipeline.core import PipelineParameter
from azureml.data.dataset_consumption_config import DatasetConsumptionConfig
ds_pipeline_param = PipelineParameter(name="dataset", default_value=dataset)
step01_input_dataset = DatasetConsumptionConfig("input_dataset", ds_pipeline_param)
from azureml.pipeline.steps import PythonScriptStep
step_01 = PythonScriptStep(
"step01_data_prep.py",
source_directory="040_scripts",
arguments=["--dataset-id", step01_input_dataset, "--output-path", step01_output],
name="Prepare data",
runconfig=runconfig,
compute_target=compute_target,
inputs=[step01_input_dataset],
outputs=[step01_output],
allow_reuse=True,
)
```
## Step 2 - Train the LightGBM model
```
from azureml.pipeline.core import PipelineParameter
learning_rate_param = PipelineParameter(name="learning_rate", default_value=0.05)
step02_output = PipelineData(
"model_output", datastore=ws.get_default_datastore(), is_directory=True
)
step_02 = PythonScriptStep(
"step02_train.py",
source_directory="040_scripts",
arguments=[
"--learning-rate",
learning_rate_param,
"--input-path",
step01_output,
"--output-path",
step02_output,
],
name="Train model",
runconfig=runconfig,
compute_target=compute_target,
inputs=[step01_output],
outputs=[step02_output],
)
```
## Step 3 - Register model
```
step_03 = PythonScriptStep(
"step03_register.py",
source_directory="040_scripts",
arguments=[
"--input-path",
step02_output,
"--dataset-id",
step01_input_dataset,
],
name="Register model",
runconfig=runconfig,
compute_target=compute_target,
inputs=[step01_input_dataset, step02_output],
)
```
## Create pipeline
```
from azureml.pipeline.core import Pipeline
pipeline = Pipeline(workspace=ws, steps=[step_01, step_02, step_03])
```
## Trigger pipeline through SDK
```
from azureml.core import Experiment
# Using the SDK
experiment = Experiment(ws, "pipeline-run")
pipeline_run = experiment.submit(pipeline, pipeline_parameters={"learning_rate": 0.5})
pipeline_run.wait_for_completion()
```
## Register pipeline to reuse
```
published_pipeline = pipeline.publish(
"Training pipeline", description="A pipeline to train a LightGBM model"
)
```
## Trigger published pipeline through REST
```
from azureml.core.authentication import InteractiveLoginAuthentication
auth = InteractiveLoginAuthentication()
aad_token = auth.get_authentication_header()
import requests
response = requests.post(
published_pipeline.endpoint,
headers=aad_token,
json={
"ExperimentName": "pipeline-run",
"ParameterAssignments": {"learning_rate": 0.02},
},
)
print(
f"Made a POST request to {published_pipeline.endpoint} and got {response.status_code}."
)
print(f"The portal url for the run is {response.json()['RunUrl']}")
```
## Scheduling a pipeline
```
from azureml.pipeline.core.schedule import ScheduleRecurrence, Schedule
from datetime import datetime
recurrence = ScheduleRecurrence(
frequency="Month", interval=1, start_time=datetime.now()
)
schedule = Schedule.create(
workspace=ws,
name="pipeline-schedule",
pipeline_id=published_pipeline.id,
experiment_name="pipeline-schedule-run",
recurrence=recurrence,
wait_for_provisioning=True,
description="Schedule to retrain model",
)
print("Created schedule with id: {}".format(schedule.id))
from azureml.pipeline.core.schedule import Schedule
# Disable schedule
schedules = Schedule.list(ws, active_only=True)
print("Your workspace has the following schedules set up:")
for schedule in schedules:
print(f"Disabling {schedule.id} (Published pipeline: {schedule.pipeline_id}")
schedule.disable(wait_for_provisioning=True)
```
| github_jupyter |
# Local Feature Matching
By the end of this exercise, you will be able to transform images of a flat (planar) object, or images taken from the same point into a common reference frame. This is at the core of applications such as panorama stitching.
A quick overview:
1. We will start with histogram representations for images (or image regions).
2. Then we will detect robust keypoints in images and use simple histogram descriptors to describe the neighborhood of each keypoint.
3. After this we will compare descriptors from different images using a distance function and establish matching points.
4. Using these matching points we will estimate the homography transformation between two images of a planar object (wall with graffiti) and use this to warp one image to look like the other.
```
%matplotlib notebook
import numpy as np
import matplotlib.pyplot as plt
import imageio
import cv2
import math
from scipy import ndimage
from attrdict import AttrDict
from mpl_toolkits.mplot3d import Axes3D
# Many useful functions
def plot_multiple(images, titles=None, colormap='gray',
max_columns=np.inf, imwidth=4, imheight=4, share_axes=False):
"""Plot multiple images as subplots on a grid."""
if titles is None:
titles = [''] *len(images)
assert len(images) == len(titles)
n_images = len(images)
n_cols = min(max_columns, n_images)
n_rows = int(np.ceil(n_images / n_cols))
fig, axes = plt.subplots(
n_rows, n_cols, figsize=(n_cols * imwidth, n_rows * imheight),
squeeze=False, sharex=share_axes, sharey=share_axes)
axes = axes.flat
# Hide subplots without content
for ax in axes[n_images:]:
ax.axis('off')
if not isinstance(colormap, (list,tuple)):
colormaps = [colormap]*n_images
else:
colormaps = colormap
for ax, image, title, cmap in zip(axes, images, titles, colormaps):
ax.imshow(image, cmap=cmap)
ax.set_title(title)
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
fig.tight_layout()
def load_image(f_name):
return imageio.imread(f_name, as_gray=True).astype(np.float32)/255
def convolve_with_two(image, kernel1, kernel2):
"""Apply two filters, one after the other."""
image = ndimage.convolve(image, kernel1)
image = ndimage.convolve(image, kernel2)
return image
def gauss(x, sigma):
return 1 / np.sqrt(2 * np.pi) / sigma * np.exp(- x**2 / 2 / sigma**2)
def gaussdx(x, sigma):
return (-1 / np.sqrt(2 * np.pi) / sigma**3 * x *
np.exp(- x**2 / 2 / sigma**2))
def gauss_derivs(image, sigma):
kernel_radius = np.ceil(3.0 * sigma)
x = np.arange(-kernel_radius, kernel_radius + 1)[np.newaxis]
G = gauss(x, sigma)
D = gaussdx(x, sigma)
image_dx = convolve_with_two(image, D, G.T)
image_dy = convolve_with_two(image, G, D.T)
return image_dx, image_dy
def gauss_filter(image, sigma):
kernel_radius = np.ceil(3.0 * sigma)
x = np.arange(-kernel_radius, kernel_radius + 1)[np.newaxis]
G = gauss(x, sigma)
return convolve_with_two(image, G, G.T)
def gauss_second_derivs(image, sigma):
kernel_radius = np.ceil(3.0 * sigma)
x = np.arange(-kernel_radius, kernel_radius + 1)[np.newaxis]
G = gauss(x, sigma)
D = gaussdx(x, sigma)
image_dx, image_dy = gauss_derivs(image, sigma)
image_dxx = convolve_with_two(image_dx, D, G.T)
image_dyy = convolve_with_two(image_dy, G, D.T)
image_dxy = convolve_with_two(image_dx, G, D.T)
return image_dxx, image_dxy, image_dyy
def map_range(x, start, end):
"""Maps values `x` that are within the range [start, end) to the range [0, 1)
Values smaller than `start` become 0, values larger than `end` become
slightly smaller than 1."""
return np.clip((x-start)/(end-start), 0, 1-1e-10)
def draw_keypoints(image, points):
image = cv2.cvtColor(image, cv2.COLOR_GRAY2RGB)
radius = image.shape[1]//100+1
for x, y in points:
cv2.circle(image, (int(x), int(y)), radius, (1, 0, 0), thickness=2)
return image
def draw_point_matches(im1, im2, point_matches):
result = np.concatenate([im1, im2], axis=1)
result = (result.astype(float)*0.6).astype(np.uint8)
im1_width = im1.shape[1]
for x1, y1, x2, y2 in point_matches:
cv2.line(result, (x1, y1), (im1_width+x2, y2),
color=(0,255,255), thickness=2, lineType=cv2.LINE_AA)
return result
%%html
<!-- This adds heading numbers to each section header -->
<style>
body {counter-reset: section;}
h2:before {counter-increment: section;
content: counter(section) " ";}
</style>
```
## Histograms in 1D
If we have a grayscale image, creating a histogram of the gray values tells us how frequently each gray value appears in the image, at a certain discretization level, which is controlled by the number of bins.
Implement `compute_1d_histogram(im, n_bins)`. Given an grayscale image `im` with shape `[height, width]` and the number of bins `n_bins`, return a `histogram` array that contains the number of values falling into each bin. Assume that the values (of the image) are in the range \[0,1), so the specified number of bins should cover the range from 0 to 1. Normalize the resulting histogram to sum to 1.
```
def compute_1d_histogram(im, n_bins):
histogram = np.zeros(n_bins)
# YOUR CODE HERE
raise NotImplementedError()
return histogram
fig, axes = plt.subplots(1,4, figsize=(10,2), constrained_layout=True)
bin_counts = [2, 25, 256]
gray_img = imageio.imread('terrain.png', as_gray=True
).astype(np.float32)/256
axes[0].set_title('Image')
axes[0].imshow(gray_img, cmap='gray')
for ax, n_bins in zip(axes[1:], bin_counts):
ax.set_title(f'1D histogram with {n_bins} bins')
bin_size = 1/n_bins
x_axis = np.linspace(0, 1, n_bins, endpoint=False)+bin_size/2
hist = compute_1d_histogram(gray_img, n_bins)
ax.bar(x_axis, hist, bin_size)
```
What is the effect of the different bin counts?
YOUR ANSWER HERE
## Histograms in 3D
If the pixel values are more than one-dimensional (e.g. three-dimensional RGB, for red, green and blue color channels), we can build a multi-dimensional histogram. In the R, G, B example this will tell us how frequently each *combination* of R, G, B values occurs. (Note that this contains more information than simply building 3 one-dimensional histograms, each for R, G and B, separately. Why?)
Implement a new function `compute_3d_histogram(im, n_bins)`, which takes as input an array of shape `[height, width, 3]` and returns a histogram of shape `[n_bins, n_bins, n_bins]`. Again, assume that the range of values is \[0,1) and normalize the histogram at the end.
Visualize the RGB histograms of the images `sunset.png` and `terrain.png` using the provided code and describe what you see. We cannot use a bar chart in 3D. Instead, in the position of each 3D bin ("voxel"), we have a sphere, whose volume is proportional to the histogram's value in that bin. The color of the sphere is simply the RGB color that the bin represents. Which number of bins gives the best impression of the color distribution?
```
def compute_3d_histogram(im, n_bins):
histogram = np.zeros([n_bins, n_bins, n_bins], dtype=np.float32)
# YOUR CODE HERE
raise NotImplementedError()
return histogram
def plot_3d_histogram(ax, data, axis_names='xyz'):
"""Plot a 3D histogram. We plot a sphere for each bin,
with volume proportional to the bin content."""
r,g,b = np.meshgrid(*[np.linspace(0,1, dim) for dim in data.shape], indexing='ij')
colors = np.stack([r,g,b], axis=-1).reshape(-1, 3)
marker_sizes = 300 * data**(1/3)
ax.scatter(r.flat, g.flat, b.flat, s=marker_sizes.flat, c=colors, alpha=0.5)
ax.set_xlabel(axis_names[0])
ax.set_ylabel(axis_names[1])
ax.set_zlabel(axis_names[2])
paths = ['sunset.png', 'terrain.png']
images = [imageio.imread(p) for p in paths]
plot_multiple(images, paths)
fig, axes = plt.subplots(1, 2, figsize=(8, 4), subplot_kw={'projection': '3d'})
for path, ax in zip(paths, axes):
im = imageio.imread(path).astype(np.float32)/256
hist = compute_3d_histogram(im, n_bins=16) # <--- FIDDLE WITH N_BINS HERE
plot_3d_histogram(ax, hist, 'RGB')
fig.tight_layout()
```
## Histograms in 2D
Now modify your code to work in 2D. This can be useful, for example, for a gradient image that stores two values for each pixel: the vertical and horizontal derivative. Again, assume the values are in the range \[0,1).
Since gradients can be negative, we need to pick a relevant range of values an map them linearly to the range of \[0,1) before applying `compute_2d_histogram`. This is implemented by the function `map_range` provided at the beginning of the notebook.
In 2D we can plot the histogram as an image. For better visibility of small values, we plot the logarithm of each bin value. Yellowish colors mean high values. The center is (0,0). Can you explain why each histogram looks the way it does for the test images?
```
def compute_2d_histogram(im, n_bins):
histogram = np.zeros([n_bins, n_bins], dtype=np.float32)
# YOUR CODE HERE
raise NotImplementedError()
return histogram
def compute_gradient_histogram(rgb_im, n_bins):
# Convert to grayscale
gray_im = cv2.cvtColor(im, cv2.COLOR_RGB2GRAY).astype(float)
# Compute Gaussian derivatives
dx, dy = gauss_derivs(gray_im, sigma=2.0)
# Map the derivatives between -10 and 10 to be between 0 and 1
dx = map_range(dx, start=-10, end=10)
dy = map_range(dy, start=-10, end=10)
# Stack the two derivative images along a new
# axis at the end (-1 means "last")
gradients = np.stack([dy, dx], axis=-1)
return dx, dy, compute_2d_histogram(gradients, n_bins=16)
paths = ['model/obj4__0.png', 'model/obj42__0.png']
images, titles = [], []
for path in paths:
im = imageio.imread(path)
dx, dy, hist = compute_gradient_histogram(im, n_bins=16)
images += [im, dx, dy, np.log(hist+1e-3)]
titles += [path, 'dx', 'dy', 'Histogram (log)']
plot_multiple(images, titles, max_columns=4, imwidth=2,
imheight=2, colormap='viridis')
```
Similar to the function `compute_gradient_histogram` above, we can build a "Mag/Lap" histogram from the gradient magnitudes and the Laplacians at each pixel. Refer back to the first exercise to refresh your knowledge of the Laplacian. Implement this in `compute_maglap_histogram`!
Make sure to map the relevant range of the gradient magnitude and Laplacian values to \[0,1) using `map_range()`. For the magnitude you can assume that the values will mostly lie in the range \[0, 15) and the Laplacian in the range \[-5, 5).
```
def compute_maglap_histogram(rgb_im, n_bins):
# Convert to grayscale
gray_im = cv2.cvtColor(rgb_im, cv2.COLOR_RGB2GRAY).astype(float)
# Compute Gaussian derivatives
sigma = 2
kernel_radius = np.ceil(3.0 * sigma)
x = np.arange(-kernel_radius, kernel_radius + 1)[np.newaxis]
G = gauss(x, sigma)
D = gaussdx(x, sigma)
dx = convolve_with_two(gray_im, D, G.T)
dy = convolve_with_two(gray_im, G, D.T)
# Compute second derivatives
dxx = convolve_with_two(dx, D, G.T)
dyy = convolve_with_two(dy, G, D.T)
# Compute gradient magnitude and Laplacian
# YOUR CODE HERE
raise NotImplementedError()
mag_lap = np.stack([mag, lap], axis=-1)
return mag, lap, compute_2d_histogram(mag_lap, n_bins=16)
paths = [f'model/obj{i}__0.png' for i in [20, 37, 36, 55]]
images, titles = [], []
for path in paths:
im = imageio.imread(path)
mag, lap, hist = compute_maglap_histogram(im, n_bins=16)
images += [im, mag, lap, np.log(hist+1e-3)]
titles += [path, 'Gradient magn.', 'Laplacian', 'Histogram (log)']
plot_multiple(images, titles, imwidth=2, imheight=2,
max_columns=4, colormap='viridis')
```
## Comparing Histograms
The above histograms looked different, but to quantify this objectively, we need a **distance measure**. The Euclidean distance is a common one.
Implement the function `euclidean_distance`, which takes two histograms $P$ and $Q$ as input and returns their Euclidean distance:
$$
\textit{dist}_{\textit{Euclidean}}(P, Q) = \sqrt{\sum_{i=1}^{D}{(P_i - Q_i)^2}}
$$
Another commonly used distance for histograms is the so-called chi-squared ($\chi^2$) distance, commonly defined as:
$$
\chi^2(P, Q) = \frac{1}{2} \sum_{i=1}^{D}\frac{(P_i - Q_i)^2}{P_i + Q_i + \epsilon}
$$
Where we can use a small value $\epsilon$ is used to avoid division by zero.
Implement it as `chi_square_distance`. The inputs `hist1` and `hist2` are histogram vectors containing the bin values. Remember to use numpy array functions (such as `np.sum()`) instead of looping over each element in Python (looping is slow).
```
def euclidean_distance(hist1, hist2):
# YOUR CODE HERE
raise NotImplementedError()
def chi_square_distance(hist1, hist2, eps=1e-3):
# YOUR CODE HERE
raise NotImplementedError()
```
Now let's take the image `obj1__0.png` as reference and let's compare it to `obj91__0.png` and `obj94__0.png`, using an RGB histogram, both with Euclidean and chi-square distance. Can you interpret the results?
You can also try other images from the "model" folder.
```
im1 = imageio.imread('model/obj1__0.png')
im2 = imageio.imread('model/obj91__0.png')
im3 = imageio.imread('model/obj94__0.png')
n_bins = 8
h1 = compute_3d_histogram(im1/256, n_bins)
h2 = compute_3d_histogram(im2/256, n_bins)
h3 = compute_3d_histogram(im3/256, n_bins)
eucl_dist1 = euclidean_distance(h1, h2)
chisq_dist1 = chi_square_distance(h1, h2)
eucl_dist2 = euclidean_distance(h1, h3)
chisq_dist2 = chi_square_distance(h1, h3)
titles = ['Reference image',
f'Eucl: {eucl_dist1:.3f}, ChiSq: {chisq_dist1:.3f}',
f'Eucl: {eucl_dist2:.3f}, ChiSq: {chisq_dist2:.3f}']
plot_multiple([im1, im2, im3], titles, imheight=3)
```
# Keypoint Detection
Now we turn to finding keypoints in images.
## Harris Detector
The Harris detector searches for points, around which the second-moment matrix $M$ of the gradient vector has two large eigenvalues (This $M$ is denoted by $C$ in the Grauman & Leibe script). This matrix $M$ can be written as:
$$
M(\sigma, \tilde{\sigma}) = G(\tilde{\sigma}) \star \left[\begin{matrix} I_x^2(\sigma) & I_x(\sigma) \cdot I_y(\sigma) \cr I_x(\sigma)\cdot I_y(\sigma) & I_y^2(\sigma) \end{matrix}\right]
$$
Note that the matrix $M$ is computed for each pixel (we omitted the $x, y$ dependency in this formula for clarity). In the above notation the 4 elements of the second-moment matrix are considered as full 2D "images" (signals) and each of these 4 "images" are convolved with the Gaussian $G(\tilde{\sigma})$ independently. We have two sigmas $\sigma$ and $\tilde{\sigma}$ here for two different uses of Gaussian blurring:
* first for computing the derivatives themselves (as derivatives-of-Gaussian) with $\sigma$, and
* then another Gaussian with $\tilde{\sigma}$ that operates on "images" containing the *products* of the derivatives (such as $I_x^2(\sigma)$) in order to collect summary statistics from a window around each point.
Instead of explicitly computing the eigenvalues $\lambda_1$ and $\lambda_2$ of $M$, the following equivalences are used:
$$
\det(M) = \lambda_1 \lambda_2 = (G(\tilde{\sigma}) \star I_x^2)\cdot (G(\tilde{\sigma}) \star I_y^2) - (G(\tilde{\sigma}) \star (I_x\cdot I_y))^2
$$
$$
\mathrm{trace}(M) = \lambda_1 + \lambda_2 = G(\tilde{\sigma}) \star I_x^2 + G(\tilde{\sigma}) \star I_y^2
$$
The Harris criterion is then:
$$
\det(M) - \alpha \cdot \mathrm{trace}^2(M) > t
$$
In practice, the parameters are usually set as $\tilde{\sigma} = 2 \sigma, \alpha=0.06$.
Read more in Section 3.2.1.2 of the Grauman & Leibe script (grauman-leibe-ch3-local-features.pdf in the Moodle).
----
Write a function `harris_score(im, opts)` which:
- computes the values of $M$ **for each pixel** of the grayscale image `im`
- calculates the trace and the determinant at each pixel
- combines them to the Harris response and returns the resulting image
To handle the large number of configurable parameters in this exercise, we will store them in an `opts` object. Use `opts.sigma1` for $\sigma$, `opts.sigma2` for $\tilde{\sigma}$ and `opts.alpha` for $\alpha$.
Furthermore, implement `nms(scores)` to perform non-maximum suppression of the response image.
Then look at `score_map_to_keypoints(scores, opts)`. It takes a score map and returns an array of shape `[number_of_corners, 2]`, with each row being the $(x,y)$ coordinates of a found keypoint. We use `opts.score_threshold` as the threshold for considering a point to be a keypoint. (This is quite similar to how we found detections from score maps in the sliding-window detection exercise.)
```
def harris_scores(im, opts):
dx, dy = gauss_derivs(im, opts.sigma1)
# YOUR CODE HERE
raise NotImplementedError()
return scores
def nms(scores):
"""Non-maximum suppression"""
# YOUR CODE HERE
raise NotImplementedError()
return scores_out
def score_map_to_keypoints(scores, opts):
corner_ys, corner_xs = (scores > opts.score_threshold).nonzero()
return np.stack([corner_xs, corner_ys], axis=1)
```
Now check the score maps and keypoints:
```
opts = AttrDict()
opts.sigma1=2
opts.sigma2=opts.sigma1*2
opts.alpha=0.06
opts.score_threshold=1e-8
paths = ['checkboard.jpg', 'graf.png', 'gantrycrane.png']
images = []
titles = []
for path in paths:
image = load_image(path)
score_map = harris_scores(image, opts)
score_map_nms = nms(score_map)
keypoints = score_map_to_keypoints(score_map_nms, opts)
keypoint_image = draw_keypoints(image, keypoints)
images += [score_map, keypoint_image]
titles += ['Harris response scores', 'Harris keypoints']
plot_multiple(images, titles, max_columns=2, colormap='viridis')
```
## Hessian Detector
The Hessian detector operates on the second-derivative matrix $H$ (called the “Hessian” matrix)
$$
H = \left[\begin{matrix}I_{xx}(\sigma) & I_{xy}(\sigma) \cr I_{xy}(\sigma) & I_{yy}(\sigma)\end{matrix}\right] \tag{6}
$$
Note that these are *second* derivatives, while the Harris detector computes *products* of *first* derivatives! The score is computed as follows:
$$
\sigma^4 \det(H) = \sigma^4 (I_{xx}I_{yy} - I^2_{xy}) > t \tag{7}
$$
You can read more in Section 3.2.1.1 of the Grauman & Leibe script (grauman-leibe-ch3-local-features.pdf in the Moodle).
-----
Write a function `hessian_scores(im, opts)`, which:
- computes the four entries of the $H$ matrix for each pixel of a given image,
- calculates the determinant of $H$ to get the response image
Use `opts.sigma1` for computing the Gaussian second derivatives.
```
def hessian_scores(im, opts):
height, width = im.shape
# YOUR CODE HERE
raise NotImplementedError()
return scores
opts = AttrDict()
opts.sigma1=3
opts.score_threshold=5e-4
paths = ['checkboard.jpg', 'graf.png', 'gantrycrane.png']
images = []
titles = []
for path in paths:
image = load_image(path)
score_map = hessian_scores(image, opts)
score_map_nms = nms(score_map)
keypoints = score_map_to_keypoints(score_map_nms, opts)
keypoint_image = draw_keypoints(image, keypoints)
images += [score_map, keypoint_image]
titles += ['Hessian scores', 'Hessian keypoints']
plot_multiple(images, titles, max_columns=2, colormap='viridis')
```
## Region Descriptor Matching
Now that we can detect robust keypoints, we can try to match them across different images of the same object. For this we need a way to compare the neighborhood of a keypoint found in one image with the neighborhood of a keypoint found in another. If the neighborhoods are similar, then the keypoints may represent the same physical point on the object.
To compare two neighborhoods, we compute a **descriptor** vector for the image window around each keypoint and then compare these descriptors using a **distance function**.
Inspect the following `compute_rgb_descriptors` function that takes a window around each point in `points` and computes a 3D RGB histogram and returns these as row vectors in a `descriptors` array.
Now write the function `compute_maglap_descriptors`, which works very similarly to `compute_rgb_descriptors`, but computes two-dimensional gradient-magnitude/Laplacian histograms. (Compute the gradient magnitude and the Laplacian for the full image first. See also the beginning of this exercise.) Pay attention to the scale of the gradient-magnitude values.
```
def compute_rgb_descriptors(rgb_im, points, opts):
"""For each (x,y) point in `points` calculate the 3D RGB histogram
descriptor and stack these into a matrix
of shape [num_points, descriptor_length]
"""
win_half = opts.descriptor_window_halfsize
descriptors = []
rgb_im_01 = rgb_im.astype(np.float32)/256
for (x, y) in points:
y_start = max(0, y-win_half)
y_end = y+win_half+1
x_start = max(0, x-win_half)
x_end = x+win_half+1
window = rgb_im_01[y_start:y_end, x_start:x_end]
histogram = compute_3d_histogram(window, opts.n_histogram_bins)
descriptors.append(histogram.reshape(-1))
return np.array(descriptors)
def compute_maglap_descriptors(rgb_im, points, opts):
"""For each (x,y) point in `points` calculate the magnitude-Laplacian
2D histogram descriptor and stack these into a matrix of
shape [num_points, descriptor_length]
"""
# Compute the gradient magnitude and Laplacian for each pixel first
gray_im = cv2.cvtColor(rgb_im, cv2.COLOR_RGB2GRAY).astype(float)
kernel_radius = np.ceil(3.0 * opts.sigma1)
x = np.arange(-kernel_radius, kernel_radius + 1)[np.newaxis]
G = gauss(x, opts.sigma1)
D = gaussdx(x, opts.sigma1)
dx = convolve_with_two(gray_im, D, G.T)
dy = convolve_with_two(gray_im, G, D.T)
dxx = convolve_with_two(dx, D, G.T)
dyy = convolve_with_two(dy, G, D.T)
# YOUR CODE HERE
raise NotImplementedError()
return np.array(descriptors)
```
Now let's implement the distance computation between descriptors. Look at `compute_euclidean_distances`. It takes descriptors that were computed for keypoints found in two different images and returns the pairwise distances between all point pairs.
Implement `compute_chi_square_distances` in a similar manner.
```
def compute_euclidean_distances(descriptors1, descriptors2):
distances = np.empty((len(descriptors1), len(descriptors2)))
for i, desc1 in enumerate(descriptors1):
distances[i] = np.linalg.norm(descriptors2-desc1, axis=-1)
return distances
def compute_chi_square_distances(descriptors1, descriptors2):
distances = np.empty((len(descriptors1), len(descriptors2)))
# YOUR CODE HERE
raise NotImplementedError()
return distances
```
Given the distances, a simple way to produce point matches is to take each descriptor extracted from a keypoint of the first image, and find the keypoint in the second image with the nearest descriptor. The full pipeline from images to point matches is implemented below in the function `find_point_matches(im1, im2, opts)`.
Experiment with different parameter settings. Which keypoint detector, region descriptor and distance function works best?
```
def find_point_matches(im1, im2, opts):
# Process first image
im1_gray = cv2.cvtColor(im1, cv2.COLOR_RGB2GRAY).astype(float)/255
score_map1 = nms(opts.score_func(im1_gray, opts))
points1 = score_map_to_keypoints(score_map1, opts)
descriptors1 = opts.descriptor_func(im1, points1, opts)
# Process second image independently of first
im2_gray = cv2.cvtColor(im2, cv2.COLOR_RGB2GRAY).astype(float)/255
score_map2 = nms(opts.score_func(im2_gray, opts))
points2 = score_map_to_keypoints(score_map2, opts)
descriptors2 = opts.descriptor_func(im2, points2, opts)
# Compute descriptor distances
distances = opts.distance_func(descriptors1, descriptors2)
# Find the nearest neighbor of each descriptor from the first image
# among descriptors of the second image
closest_ids = np.argmin(distances, axis=1)
closest_dists = np.min(distances, axis=1)
# Sort the point pairs in increasing order of distance
# (most similar ones first)
ids1 = np.argsort(closest_dists)
ids2 = closest_ids[ids1]
points1 = points1[ids1]
points2 = points2[ids2]
# Stack the point matches into rows of (x1, y1, x2, y2) values
point_matches = np.concatenate([points1, points2], axis=1)
return point_matches
# Try changing these values in different ways and see if you can explain
# why the result changes the way it does.
opts = AttrDict()
opts.sigma1=2
opts.sigma2=opts.sigma1*2
opts.alpha=0.06
opts.score_threshold=1e-8
opts.descriptor_window_halfsize = 20
opts.n_histogram_bins = 16
opts.score_func = harris_scores
opts.descriptor_func = compute_maglap_descriptors
opts.distance_func = compute_chi_square_distances
# Or try these:
#opts.sigma1=3
#opts.n_histogram_bins = 8
#opts.score_threshold=5e-4
#opts.score_func = hessian_scores
#opts.descriptor_func = compute_rgb_descriptors
#opts.distance_func = compute_euclidean_distances
im1 = imageio.imread('graff5/img1.jpg')
im2 = imageio.imread('graff5/img2.jpg')
point_matches = find_point_matches(im1, im2, opts)
match_image = draw_point_matches(im1, im2, point_matches[:50])
plot_multiple([match_image], imwidth=16, imheight=8)
```
## Homography Estimation
Now that we have these pairs of matching points (also called point correspondences), what can we do with them? In the above case, the wall is planar (flat) and the camera was moved towards the left to take the second image compared to the first image. Therefore, the way that points on the wall are transformed across these two images can be modeled as a **homography**. Homographies can model two distinct effects:
* transformation across images of **any scene** taken from the **exact same camera position** (center of projection)
* transformation across images of a **planar object** taken from **any camera position**.
We are dealing with the second case in these graffiti images. Therefore if our point matches are correct, there should be a homography that transforms image points in the first image to the corresponding points in the second image. Recap the algorithm from the lecture for finding this homography (it's called the **Direct Linear Transformation**, DLT). There is a 2 page description of it in the Grauman & Leibe script (grauman-leibe-ch5-geometric-verification.pdf in the Moodle) in Section 5.1.3.
----
Now let's actually put this into practice. Implement `estimate_homography(point_matches)`, which returns a 3x3 homography matrix that transforms points of the first image to points of the second image.
The steps are:
1. Build the matrix $A$ from the point matches according to Eq. 5.7 from the script.
2. Apply SVD using `np.linalg.svd(A)`. It returns $U,d,V^T$. Note that the last return value is not $V$ but $V^T$.
3. Compute $\mathbf{h}$ from $V$ according to Eq. 5.9 or 5.10
4. Reshape $\mathbf{h}$ to the 3x3 matrix $H$ and return it.
The input `point_matches` contains as many rows as there are point matches (correspondences) and each row has 4 elements: $x, y, x', y'$.
```
def estimate_homography(point_matches):
n_matches = len(point_matches)
A = np.empty((n_matches*2, 9))
for i, (x1, y1, x2, y2) in enumerate(point_matches):
# YOUR CODE HERE
raise NotImplementedError()
return H
```
The `point_matches` have already been sorted in the `find_point_matches` function according to the descriptor distances, so the more accurate pairs will be near the beginning. We can use the top $k$, e.g. $k=10$ pairs in the homography estimation and have a reasonably accurate estimate. What $k$ give the best result? What happens if you use too many? Why?
We can use `cv2.warpPerspective` to warp the first image to the reference frame of the second. Does the result look good?
Can you interpret the entries of the resulting $H$ matrix and are the numbers as you would expect them for these images?
You can also try other image from the `graff5` folder or the `NewYork` folder.
```
# See what happens if you change top_k below
top_k = 10
H = estimate_homography(point_matches[:top_k])
H_string = np.array_str(H, precision=5, suppress_small=True)
print('The estimated homography matrix H is\n', H_string)
im1_warped = cv2.warpPerspective(im1, H, (im2.shape[1], im2.shape[0]))
absdiff = np.abs(im2.astype(np.float32)-im1_warped.astype(np.float32))/255
plot_multiple([im1, im2, im1_warped, absdiff],
['First image', 'Second image',
'Warped first image', 'Absolute difference'],
max_columns=2, colormap='viridis')
```
| github_jupyter |
```
# Load necessary modules and libraries
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import Perceptron
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.model_selection import learning_curve
from sklearn.neural_network import MLPRegressor
from sklearn.linear_model import LinearRegression
from sklearn.gaussian_process import GaussianProcessRegressor
from sklearn.gaussian_process.kernels import RBF, RationalQuadratic, Matern, ExpSineSquared,DotProduct
import pickle
import seaborn as sns
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
# Load the data
Geometry1 = pd.read_csv('Surface_features.csv',header=0, usecols=(4,8,9,10,11,12,14))
Geometry = pd.read_csv('Surface_features.csv',header=0, usecols=(4,6,7,8,9,10,11,12)).values
Ra_ch = pd.read_csv('Surface_features.csv',header=0,usecols=(5,)).values
Ra_ch = Ra_ch[:,0]
ks = pd.read_csv('Surface_features.csv',header=0,usecols=(13,)).values
ks = ks[:,0]
Geometry1["ks"]= np.divide(ks,Ra_ch)
Geometry1["krms_ch"]= np.divide(Geometry1["krms_ch"],Ra_ch)
Geometry1.rename({'krms_ch': '$k_{rms}/R_a$',
'pro_ch': '$P_o$',
'ESx_ch': '$E_x$',
'ESz_ch': '$E_z$',
'sk_ch': '$S_k$',
'ku_ch': '$K_u$',
'ks': '$k_s/R_a$',
'label': 'Label',
}, axis='columns', errors="raise",inplace = True)
# Plot raw data
plt.rc('text', usetex=True)
sns.set(context='paper',
style='ticks',
palette='deep',
font='sans-serif',
font_scale=3, color_codes=True, rc=None)
g = sns.pairplot(Geometry1,diag_kind="kde", #palette="seismic",
hue='Label',
plot_kws=dict(s=70,facecolor="w", edgecolor="w", linewidth=1),
diag_kws=dict(linewidth=1.5))
g.map_upper(sns.kdeplot)
g.map_lower(sns.scatterplot, s=50,)
plt.savefig('pair.pdf', dpi=None, facecolor='w', edgecolor='w',
orientation='portrait', papertype=None, format=None,
transparent=False, bbox_inches=None, pad_inches=0.1,
frameon=None, metadata=None)
# Data reconfiguration, to be used in ML
X = Geometry
y = np.divide(ks,Ra_ch)
X[:,0] = np.divide(X[:,0],Ra_ch)
X[:,2] = np.abs(X[:,2])
# Generate secondary features and append them to the original dataset
n,m = X.shape
X0 = np.ones((n,1))
X1 = np.ones((n,1))
X2 = np.ones((n,1))
X3 = np.ones((n,1))
X4 = np.ones((n,1))
X5 = np.ones((n,1))
X6 = np.ones((n,1))
X7 = np.ones((n,1))
X8 = np.ones((n,1))
X9 = np.ones((n,1))
X1[:,0] = np.transpose(X[:,4]*X[:,5])
X2[:,0] = np.transpose(X[:,4]*X[:,6])
X3[:,0] = np.transpose(X[:,4]*X[:,7])
X4[:,0] = np.transpose(X[:,5]*X[:,6])
X5[:,0] = np.transpose(X[:,5]*X[:,7])
X6[:,0] = np.transpose(X[:,6]*X[:,7])
X7[:,0] = np.transpose(X[:,4]*X[:,4])
X8[:,0] = np.transpose(X[:,5]*X[:,5])
X9[:,0] = np.transpose(X[:,6]*X[:,6])
X = np.hstack((X,X1))
X = np.hstack((X,X2))
X = np.hstack((X,X3))
X = np.hstack((X,X4))
X = np.hstack((X,X5))
X = np.hstack((X,X6))
X = np.hstack((X,X7))
X = np.hstack((X,X8))
X = np.hstack((X,X9))
# Best linear estimation
reg = LinearRegression().fit(X, y)
reg.score(X, y)
yn=reg.predict(X)
print("Mean err: %f" % np.mean(100.*abs(yn-y)/(y)))
print("Max err: %f" % max(100.*abs(yn-y)/(y)))
# Define two files that store the best ML prediction based on either L1 or L_\infty norms
filename1 = 'GPR_Linf.sav'
filename2 = 'GPR_L1.sav'
# Perform ML training --- it may take some time.
# Adjust ranges for by4 for faster (but potentially less accurate) results.
miny1=100
miny2=100
by4=0.
while by4<10000.:
by4=by4+1
kernel1 = RBF(10, (1e-3, 1e2))
kernel2 = RBF(5, (1e-3, 1e2))
kernel3 = RationalQuadratic(length_scale=1.0, alpha=0.1)
kernel4 = Matern(length_scale=1.0, length_scale_bounds=(1e-05, 100000.0), nu=4.5)
kernel5 = ExpSineSquared(length_scale=2.0,
periodicity=3.0,
length_scale_bounds=(1e-05, 100000.0),
periodicity_bounds=(1e-05, 100000.0))
kernel6 = DotProduct()
gpr = GaussianProcessRegressor(kernel=kernel1, n_restarts_optimizer=1000)
gpr = GaussianProcessRegressor(kernel=kernel3, n_restarts_optimizer=1000,alpha=.1)
print("by4: %f" % by4)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3)
gpr.fit(X_train, y_train)
yn, sigma = gpr.predict(X, return_std=True)
#print("Max err: %f" % max(100.*abs(yn-y)/y))
#print("Mean err: %f" % np.mean(100.*abs(yn-y)/y))
if miny1>max(100.*abs(yn-y)/y):
pickle.dump(gpr, open(filename1, 'wb'))
miny1=max(100.*abs(yn-y)/y)
print("Miny1: %f" % miny1)
if miny2>np.mean(100.*abs(yn-y)/y):
pickle.dump(gpr, open(filename2, 'wb'))
miny2=np.mean(100.*abs(yn-y)/y)
print("Miny2: %f" % miny2)
print("by4: %f" % by4)
# Load either file1 or file2 to extract the results
loaded_model = pickle.load(open(filename2, 'rb'))
loaded_model.get_params()
yn, sigma = loaded_model.predict(X,return_std=True)
print("PREDICTED k_s/R_a= ")
print(yn)
print("Max err: %f" % max(100.*abs(yn-y)/(y)))
print("mean err: %f" % np.mean(100.*abs(yn-y)/(y)))
Error=pd.DataFrame()
Error["$k_s/Ra$"]= y
Error["$k_{sp}/Ra$"]= yn
Error["$error(\%)$"]= (100.*(yn-y)/(y))
Error["Label"]= Geometry1["Label"]
print(Error)
# Plot the results
plt.rc('text', usetex=True)
sns.set(context='paper',
style='ticks',
palette='deep',
font='sans-serif',
font_scale=2, color_codes=True, rc=None)
g = sns.pairplot(Error,diag_kind="kde", hue='Label',
aspect=1.,
plot_kws=dict(s=50,facecolor="w", edgecolor="w", linewidth=1.),
diag_kws=dict(linewidth=1.5,kernel='gau'))
g.map_upper(sns.kdeplot)
g.map_lower(sns.scatterplot, s=50,legend='full')
g.axes[-2,0].plot(range(15), range(15),'k--', linewidth= 1.7)
for i in range(0,3):
for ax in g.axes[:,i]:
ax.spines['top'].set_visible(True)
ax.spines['right'].set_visible(True)
plt.savefig('GPR_result.pdf', dpi=None, facecolor='w', edgecolor='w',
orientation='portrait', papertype=None, format=None,
transparent=False, bbox_inches=None, pad_inches=0.1,
frameon=None, metadata=None)
# Plot confidence interval
sns.set(context='notebook',
style='ticks',
palette='seismic',
font='sans-serif',
font_scale=5, color_codes=True, rc=None)
plt.rc('text', usetex=True)
fig = plt.figure(figsize=(50,55))
plt.subplot(411)
Xm=X[np.argsort(X[:,0])]
Xm=Xm[:,0]
ym=y[np.argsort(X[:,0])]
ymp=yn[np.argsort(X[:,0])]
sigmap=sigma[np.argsort(X[:,0])]
plt.plot(Xm, ym, 'r.', markersize=26)
plt.plot(Xm, ymp, 'b-',linewidth=6)
plt.fill(np.concatenate([Xm, Xm[::-1]]),
np.concatenate([ymp - 1.900 * sigmap,
(ymp + 1.900 * sigmap)[::-1]]),
alpha=.5, fc='b', ec='None')
plt.xlabel('$k_{rms}/R_a$')
plt.ylabel('$k_s/R_a$')
plt.grid(alpha=0.15)
#plt.legend(loc='best')
plt.subplot(412)
Xm=X[np.argsort(X[:,4])]
Xm=Xm[:,4]
ym=y[np.argsort(X[:,4])]
ymp=yn[np.argsort(X[:,4])]
sigmap=sigma[np.argsort(X[:,4])]
plt.plot(Xm, ym, 'r.', markersize=26)
plt.plot(Xm, ymp, 'b-',linewidth=6)
plt.fill(np.concatenate([Xm, Xm[::-1]]),
np.concatenate([ymp - 1.900 * sigmap,
(ymp + 1.900 * sigmap)[::-1]]),
alpha=.5, fc='b', ec='None')
plt.xlabel('$E_x$')
plt.ylabel('$k_s/R_a$')
plt.grid(alpha=0.15)
plt.subplot(413)
Xm=X[np.argsort(X[:,3])]
Xm=Xm[:,3]
ym=y[np.argsort(X[:,3])]
ymp=yn[np.argsort(X[:,3])]
sigmap=sigma[np.argsort(X[:,3])]
plt.plot(Xm, ym, 'r.', markersize=26)
plt.plot(Xm, ymp, 'b-',linewidth=6)
plt.fill(np.concatenate([Xm, Xm[::-1]]),
np.concatenate([ymp - 1.900 * sigmap,
(ymp + 1.900 * sigmap)[::-1]]),
alpha=.5, fc='b', ec='None')
plt.xlabel('$P_o$')
plt.ylabel('$k_s/R_a$')
plt.grid(alpha=0.15)
plt.subplot(414)
Xm=X[np.argsort(X[:,6])]
Xm=Xm[:,6]
ym=y[np.argsort(X[:,6])]
ymp=yn[np.argsort(X[:,6])]
sigmap=sigma[np.argsort(X[:,6])]
plt.plot(Xm, ym, 'r.', markersize=26, label='$k_s/R_a$')
plt.plot(Xm, ymp, 'b-', linewidth=6,label='$k_{sp}/R_a$')
plt.fill(np.concatenate([Xm, Xm[::-1]]),
np.concatenate([ymp - 1.900 * sigmap,
(ymp + 1.900 * sigmap)[::-1]]),
alpha=.5, fc='b', ec='None', label='$90\%$ $CI$')
plt.xlabel('$S_k$')
plt.ylabel('$k_s/R_a$')
plt.grid(alpha=0.15)
plt.legend(loc='best')
plt.savefig('GPR_CI.pdf', dpi=None, facecolor='w', edgecolor='w',
orientation='portrait', papertype=None, format=None,
transparent=False, bbox_inches=None, pad_inches=0.1,
frameon=None, metadata=None)
```
| github_jupyter |
# SiteAlign features
We read the SiteAlign features from the respective [paper](https://onlinelibrary.wiley.com/doi/full/10.1002/prot.21858) and [SI table](https://onlinelibrary.wiley.com/action/downloadSupplement?doi=10.1002%2Fprot.21858&file=prot21858-SupplementaryTable.pdf) to verify `kissim`'s implementation of the SiteAlign definitions:
```
from kissim.definitions import SITEALIGN_FEATURES
SITEALIGN_FEATURES
```
## Size
SiteAlign's size definitions:
> Natural amino acids have been classified into three groups according to the number of heavy atoms (<4 heavy atoms: Ala, Cys, Gly, Pro, Ser, Thr, Val; 4–6 heavy atoms: Asn, Asp, Gln, Glu, His, Ile, Leu, Lys, Met; >6 heavy atoms: Arg, Phe, Trp, Tyr) and three values (“1,” “2,” “3”) are outputted according to the group to which the current residues belong to (Table I)
https://onlinelibrary.wiley.com/doi/full/10.1002/prot.21858
### Parse text from SiteAlign paper
```
size = {
1.0: "Ala, Cys, Gly, Pro, Ser, Thr, Val".split(", "),
2.0: "Asn, Asp, Gln, Glu, His, Ile, Leu, Lys, Met".split(", "),
3.0: "Arg, Phe, Trp, Tyr".split(", "),
}
```
### `kissim` definitions correct?
```
import pandas as pd
from IPython.display import display, HTML
# Format SiteAlign data
size_list = []
for value, amino_acids in size.items():
values = [(amino_acid.upper(), value) for amino_acid in amino_acids]
size_list = size_list + values
size_series = (
pd.DataFrame(size_list, columns=["amino_acid", "size"])
.sort_values("amino_acid")
.set_index("amino_acid")
.squeeze()
)
# KiSSim implementation of SiteAlign features correct?
diff = size_series == SITEALIGN_FEATURES["size"]
if not diff.all():
raise ValueError(
f"KiSSim implementation of SiteAlign features is incorrect!!!\n"
f"{display(HTML(diff.to_html()))}"
)
else:
print("KiSSim implementation of SiteAlign features is correct :)")
```
## HBA, HBD, charge, aromatic, aliphatic
### Parse table from SiteAlign SI
```
sitealign_table = """
Ala 0 0 0 1 0
Arg 3 0 +1 0 0
Asn 1 1 0 0 0
Asp 0 2 -1 0 0
Cys 1 0 0 1 0
Gly 0 0 0 0 0
Gln 1 1 0 0 0
Glu 0 2 -1 0 0
His/Hid/Hie 1 1 0 0 1
Hip 2 0 1 0 0
Ile 0 0 0 1 0
Leu 0 0 0 1 0
Lys 1 0 +1 0 0
Met 0 0 0 1 0
Phe 0 0 0 0 1
Pro 0 0 0 1 0
Ser 1 1 0 0 0
Thr 1 1 0 1 0
Trp 1 0 0 0 1
Tyr 1 1 0 0 1
Val 0 0 0 1 0
"""
sitealign_table = [i.split() for i in sitealign_table.split("\n")[1:-1]]
sitealign_dict = {i[0]: i[1:] for i in sitealign_table}
sitealign_df = pd.DataFrame.from_dict(sitealign_dict).transpose()
sitealign_df.columns = ["hbd", "hba", "charge", "aliphatic", "aromatic"]
sitealign_df = sitealign_df[["hbd", "hba", "charge", "aromatic", "aliphatic"]]
sitealign_df = sitealign_df.rename(index={"His/Hid/Hie": "His"})
sitealign_df = sitealign_df.drop("Hip", axis=0)
sitealign_df = sitealign_df.astype("float")
sitealign_df.index = [i.upper() for i in sitealign_df.index]
sitealign_df = sitealign_df.sort_index()
sitealign_df
```
### `kissim` definitions correct?
```
from IPython.display import display, HTML
diff = sitealign_df == SITEALIGN_FEATURES.drop("size", axis=1).sort_index()
if not diff.all().all():
raise ValueError(
f"KiSSim implementation of SiteAlign features is incorrect!!!\n"
f"{display(HTML(diff.to_html()))}"
)
else:
print("KiSSim implementation of SiteAlign features is correct :)")
```
## Table style
```
from Bio.Data.IUPACData import protein_letters_3to1
for feature_name in SITEALIGN_FEATURES.columns:
print(feature_name)
for name, group in SITEALIGN_FEATURES.groupby(feature_name):
amino_acids = {protein_letters_3to1[i.capitalize()] for i in group.index}
amino_acids = sorted(amino_acids)
print(f"{name:<7}{' '.join(amino_acids)}")
print()
```
| github_jupyter |
# "Building Excel dashboard using NYSE data"
> "A project for my Udacity certificate in business analysis"
- toc: false
- branch: master
- badges: false
- hide_github_badge: true
- comments: true
- categories: [Excel, Dashboards]
- image: images/dashboard_icon.webp
- hide: false
- search_exclude: false
- metadata_key1: Excel
- metadata_key2: Dashboards
This project is from my Udacityies' "Business Analysis" nanodegree last year. Udacity has famously known for thair project-based courses. Meaning that candidates can only get thair certificate if they apply what they have learned in real-life projects. Candidates' projects get checked by experts to evaluate their work and make sure they have fulfilled Udacityies' requirements.
This project uses the NYSE dataset for over 450 companies. It includes companies' financials data for four years, period date, sector, and industry.
Here's a look at the dataset head:
<iframe width="900" height="281" frameborder="0" scrolling="no" src="https://onedrive.live.com/embed?resid=946CEB56A706EBDB%219613&authkey=%21AJt2pyGN0wVXNN4&em=2&wdAllowInteractivity=False&Item=dataset_head&wdInConfigurator=True"></iframe>
To pass this project I've been asked to attain two requirements:
- Find insights from the data and tell a story about it through a presentation.
- Build a dynamic financial dashboard in excel.
## Find insights in the data
The dataset includes financial data from 2012 to 2016. The first thing that came to my mind of that period is when the oil price hit 120$ in 2012 and then fell in 2015.
I was wondering how the airline industry did during that period. Why airline? because during that time I was studying in China and I've noticed that airline ticket prices were getting more and more expensive. So I wanted to see if there's a correlation between ticket prices and oil prices to confirm my hypothesis. In general, if oil prices go up or down it affects many aspects of the global economy, some sectors benefit from high prices but most of them benefit from lower prices. my question was **"how was airline companies' financial performance during that period?"**
My assumption was that high oil prices will increase the cost of airline operations, which therefore increases the price of tickets. High ticket prices lead to lower demand and therefore lower profits.
### Extracting EBIT from data
The main benchmarks I used to answer my question are **total revenue** and **EBIT** (earnings before interest and tax). There are other factors that could tell you about companies' performance, but these two are good for my question. We don't have EBIT in the dataset, But luckily we have the raw data to extract **EBIT**.
To do that, First I found the ``Gross Profit`` by subtracting ``Cost of Goods Sold`` from ``Total Revenue`` then we get **EBIT** by subtracting ``Sales, General and Admin`` from ``Gross Profit``.
Lastly, I used the wonderful pivot table tool, to get ***average, median,***, and ***standard deviation*** of the two benchmarks mentioned earlier. Using them all together will give us more accurate insight.
Here's the result on excel:
- Average EBIT & revenue
 
<iframe width="900" height="615" frameborder="0" scrolling="no" src="https://onedrive.live.com/embed?resid=946CEB56A706EBDB%219613&authkey=%21AJt2pyGN0wVXNN4&em=2&wdAllowInteractivity=False&Item=average_EBIT&wdInConfigurator=True"></iframe>
---
 
- Median EBIT
 
<iframe width="900" height="480" frameborder="0" scrolling="no" src="https://onedrive.live.com/embed?resid=946CEB56A706EBDB%219613&authkey=%21AJt2pyGN0wVXNN4&em=2&wdAllowInteractivity=False&Item=median_EBIT&wdInConfigurator=True"></iframe>
---
 
- EBIT standard deviation
 
<iframe width="900" height="462" frameborder="0" scrolling="no" src="https://onedrive.live.com/embed?resid=946CEB56A706EBDB%219613&authkey=%21AJt2pyGN0wVXNN4&em=2&wdAllowInteractivity=False&Item=STD_EBIT&wdInConfigurator=True"></iframe>
---
 
Here are my insights in clean slides:
{% include info.html text="Use the full-screen button in the lower right corner." %}
<iframe src="https://onedrive.live.com/embed?cid=946CEB56A706EBDB&resid=946CEB56A706EBDB%219681&authkey=AHt-YAA_ZHUa-YI&em=2" width="900" height="480" frameborder="0" scrolling="no"></iframe>
---
## Building dynamic dashboard in Excel
Udacity required me to build two dynamic dashboards:
+ P/L (Profit and loss) dashboard.
+ Forecast analysis dashboard with three case scenarios.
A dynamic dashboard means that the user can choose the company symbol and read P/L or predictions for any company individually. The prediction dashboard predicts how a company would perform in the next two years.
### P/L statment dashboard
This dashboard is simple, I just brought the data from the dataset sheet into each cell using `INDEX` and `MATCH` functions and used `Ctrl`+`Shift`+`Enter` to turn it into an array formula.
Try it yourself:
 
<iframe width="900" height="370" frameborder="0" scrolling="no" src="https://onedrive.live.com/embed?resid=946CEB56A706EBDB%219613&authkey=%21AJt2pyGN0wVXNN4&em=2&wdAllowInteractivity=False&Item=profit_loss_dashboard&wdInConfigurator=True"></iframe>
 
## Forecast dashboard
This dashboard is different. Here I'm required to build a dynamic dashboard that can show each company forecast with:
- Three scenarios: *week*, *base*, and *strong* scenario.
- Operating scenarios
First, I created the ratios table like **Gross margin** and **Revenue growth** percentages because assumptions will be extracted from past years' ratios. Then under that table, I created the *operating scenario* table (sensitivity analysis). I could've implemented this table in the final formula but this will not allow the users to read ratios when they need it.
Finally, I built the assumption table with past data as a final result.
In all tables, I used `INDEX`, `OFFSET`, and `MATCH` but in a boolean way. This is an example of a formula from one of the cells:
```
{=INDEX(total_revenue,MATCH(1,($F$5=symbols)*(G$8=years),0))}
```
 
This is the forecasting dashboard, give it a try.
 
<iframe width="900" height="813" frameborder="0" scrolling="no" src="https://onedrive.live.com/embed?resid=946CEB56A706EBDB%219613&authkey=%21AJt2pyGN0wVXNN4&em=2&wdAllowInteractivity=False&Item=forecast_dashboard&wdHideGridlines=True&wdInConfigurator=True"></iframe>
 
 
If you would like to play with the file yourself [Click here](https://1drv.ms/x/s!AtvrBqdW62yUyw1J0gD7Z5hDorfQ?e=mmCYL1) to open the full file on OneDrive.
If you have any question please contact me on my [LinkedIn](https://www.linkedin.com/in/saleh-alhodaif) or [Twitter](https://twitter.com/salehalhodaif2)
| github_jupyter |
# Fashion MNIST Generative Adversarial Network (GAN)
[Мой блог](https://tiendil.org)
[Пост об этом notebook](https://tiendil.org/generative-adversarial-network-implementation)
[Все публичные notebooks](https://github.com/Tiendil/public-jupyter-notebooks)
Учебная реализация [GAN](https://en.wikipedia.org/wiki/Generative_adversarial_network) на данных [Fashion MNIST](https://github.com/zalandoresearch/fashion-mnist).
На основе следующих материалов:
- https://machinelearningmastery.com/practical-guide-to-gan-failure-modes/
- https://www.tensorflow.org/tutorials/generative/dcgan
- https://keras.io/examples/generative/dcgan_overriding_train_step/
Сходу у меня не получилось нагуглись «красивое» решение. Поэтому тут будет композиция разных уроков. На мой взгляд, получилось более идиоматично.
Про GAN лучше почитать по ссылке выше. Краткая суть:
- Тренируются две сети: generator & discriminator.
- Генератор учится создавать картинки из шума.
- Дискриминатор учится отличать поддельные картинки от настоящих.
- Ошибка дискриминатора определяется качеством предсказания фейковости изображения.
- Ошибка генератора определяется качеством обмана дискриминатора.
Подробнее про ошибки будет далее.
Если правильно подобрать топологии сетей и параметры обучения, то в итоге генератор научается создавать картинки неотличимые от оригинальных. ??????. Profit.
## Подготовка
Notebook запускался в кастомизированном docker контейнере.
Подробнее про мои злоключения с настройкой tensorflow + CUDA можно почитать в блоге: [нельзя просто так взять и запустить DL](https://tiendil.org/you-cant-just-take-and-run-dl).
Официальная документация о [запуске tensorflow через docker](https://www.tensorflow.org/install/docker).
Dockerfile:
```
FROM tensorflow/tensorflow:2.5.0-gpu-jupyter
RUN apt-get update && apt-get install -y graphviz
RUN pip install --upgrade pip
COPY requirements.txt ./
RUN pip install -r ./requirements.txt
```
requirements.txt:
```
pandas==1.1.5
kaggle==1.5.12
pydot==1.4.2 # requeired by tensorflow to visualize models
livelossplot==0.5.4 # required to plot loss while training
albumentations==1.0.3 # augument image data
jupyter-beeper==1.0.3
```
## Инициализация
Уже без комментариев, подробнее рассказано в [предыдущих notebooks](https://github.com/Tiendil/public-jupyter-notebooks).
```
import os
import random
import logging
import datetime
import PIL
import PIL.Image
import jupyter_beeper
from IPython.display import display, Markdown, Image
import ipywidgets as ipw
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '1'
import numpy as np
import pandas as pd
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
import matplotlib.pyplot as plt
from livelossplot import PlotLossesKerasTF
import cv2
logging.getLogger().setLevel(logging.WARNING)
tf.get_logger().setLevel(logging.WARNING)
tf.autograph.set_verbosity(1)
old_settings = np.seterr('raise')
gpus = tf.config.list_physical_devices("GPU")
display(Markdown(f'Num GPUs Available: {len(gpus)}'))
if not gpus:
raise RuntimeError('No GPUs found, learning process will be too slow. In Google Colab set runtime type — GPU.')
display(Markdown(f'Eager mode: {tf.executing_eagerly()}'))
tf.config.experimental.set_memory_growth(gpus[0], True)
SEED = 1
random.seed(SEED)
np.random.seed(SEED)
tf.random.set_seed(SEED)
tf.keras.backend.clear_session()
RNG = np.random.default_rng()
```
## Вспомогательные функции
Можно пролистать. Смотрите при необходимости.
```
def split_dataset(data, *parts, cache):
data_size = data.cardinality()
assert data_size == sum(parts), \
f"dataset size must be equal to sum of parts: {data_size} != sum{parts}"
result = []
for part in parts:
data_part = data.take(part)
if cache:
data_part = data_part.cache()
result.append(data_part)
data = data.skip(part)
return result
def normalizer(minimum, maximum):
def normalize_dataset(x):
return (x - minimum) / (maximum - minimum)
return normalize_dataset
def display_model(model, name):
filename = f'/tmp/tmp_model_schema_{name}.png'
keras.utils.plot_model(model,
show_shapes=True,
show_layer_names=True,
show_dtype=True,
expand_nested=True,
to_file=filename)
display(Image(filename))
class LayersNameGenerator:
__slots__ = ('prefix', 'number')
_version = 0
def __init__(self, prefix):
self.prefix = prefix
self.number = 0
self.__class__._version += 1
def __call__(self, type_name, name=None):
self.number += 1
if name is None:
name = str(self.number)
return f'{self.prefix}.{self._version}-{type_name}.{name}'
def display_examples(examples_number=1,
data_number=1,
image_getter=None,
label_getter='',
figsize=(16, 16),
subplot=None,
cmap=plt.get_cmap('gray')):
if image_getter is None:
raise ValueError('image_getter must be an image or a collable')
if not callable(image_getter):
image_value = image_getter
image_getter = lambda j: image_value
if not callable(label_getter):
label_value = label_getter
label_getter = lambda j: label_value
examples_number = min(examples_number, data_number)
if subplot is None:
subplot = (1, examples_number)
plt.figure(figsize=figsize)
if examples_number < data_number:
choices = RNG.choice(data_number, examples_number, replace=False)
else:
choices = list(range(data_number))
for i, j in enumerate(choices):
plt.subplot(*subplot, i+1)
plt.imshow(image_getter(j), cmap=cmap)
plt.title(label_getter(j))
plt.show()
def display_memory_stats():
stats = tf.config.experimental.get_memory_info('GPU:0')
message = f'''
current: {stats["current"]/1024/1024}Mb
peak: {stats["peak"]/1024/1024}Mb
'''
display(Markdown(message))
def make_report(history, main, metrics):
groups = {'main': {}}
for key in history.history.keys():
if key in ('loss', 'val_loss', 'accuracy', 'val_accuracy'):
if key.startswith('val_'):
metric = key
else:
metric = f'train_{key}'
groups['main'][metric] = history.history[key][-1]
continue
if not any(key.endswith(f'_{metric}') for metric in metrics):
continue
group, metric = key.rsplit('_', 1)
validation = False
if group.startswith('val_'):
group = group[4:]
validation = True
if group not in groups:
groups[group] = {}
if validation:
metric = f'val_{metric}'
else:
metric = f'train_{metric}'
groups[group][metric] = history.history[key][-1]
lines = []
for group, group_metrics in groups.items():
lines.append(f'**{group}:**')
lines.append(f'```')
for name, value in sorted(group_metrics.items()):
if name in ('accuracy', 'val_accuracy', 'train_accuracy'):
lines.append(f' {name}: {value:.4%} ({value})')
else:
lines.append(f' {name}: {value}')
lines.append(f'```')
train_loss = groups[main]['train_loss']
val_loss = groups[main].get('val_loss')
val_accuracy = groups[main].get('val_accuracy')
history.history[key][-1]
if val_loss is None:
description = f'train_loss: {train_loss:.4};'
else:
description = f'train_loss: {train_loss:.4}; val_loss: {val_loss:.4}; val_acc: {val_accuracy:.4%}'
lines.append(f'**description:** {description}')
return '\n\n'.join(lines), description
def crope_layer(input, expected_shape, names):
raw_shape = input.get_shape()
if raw_shape == (None, *expected_shape):
outputs = input
else:
dy = raw_shape[1] - expected_shape[0]
dx = raw_shape[2] - expected_shape[1]
x1 = dx // 2
x2 = dx - x1
y1 = dy // 2
y2 = dy - y1
outputs = layers.Cropping2D(cropping=((y1, y2), (x1, x2)),
name=names('Cropping2D'))(input)
return outputs
def neurons_in_shape(shape):
input_n = 1
for n in shape:
if n is not None:
input_n *= n
return input_n
def form_images_map(h, w, images, channels, scale=1):
map_image = np.empty((SPRITE_SIZE*h, SPRITE_SIZE*w, channels), dtype=np.float32)
for i in range(h):
y_1 = i * SPRITE_SIZE
for j in range(w):
sprite = images[i*w+j]
x_1 = j * SPRITE_SIZE
map_image[y_1:y_1+SPRITE_SIZE, x_1:x_1+SPRITE_SIZE, :] = sprite
if channels == 1:
mode = 'L'
map_image = np.squeeze(map_image)
elif channels == 3:
mode = 'RGB'
else:
raise ValueError(f'Unexpected channels value {channels}')
if scale != 1:
width, height = w * SPRITE_SIZE, h * SPRITE_SIZE
map_image = cv2.resize(map_image, dsize=(width * scale, height * scale), interpolation=cv2.INTER_NEAREST)
image = PIL.Image.fromarray((map_image * 255).astype(np.int8), mode)
return image
```
## Получение данных
```
# получаем картинки одежды средствами TensorFlow
(TRAIN_IMAGES, TRAIN_LABELS), (TEST_IMAGES, TEST_LABELS) = tf.keras.datasets.fashion_mnist.load_data()
# константы, описывающие данные
CHANNELS = 1
SPRITE_SIZE = 28
SPRITE_SHAPE = (SPRITE_SIZE, SPRITE_SIZE, CHANNELS)
# Подготавливаем данные. Для GAN нам нужны только картинки.
def transform(images):
images = (images / 255.0).astype(np.float32)
images = np.expand_dims(images, axis=-1)
return images
def filter_by_class(images, labels, classes):
_images = tf.data.Dataset.from_tensor_slices(transform(images))
_labels = tf.data.Dataset.from_tensor_slices(labels)
d = tf.data.Dataset.zip((_images, _labels))
d = d.filter(lambda i, l: tf.reduce_any(tf.equal(classes, l)))
d = d.map(lambda i, l: i)
return d
# Обучаться будем только на изображениях обуви:
#
# - сеть будет учиться быстрее;
# - результат будет лучше;
# - будет проще, интереснее играться с работой обученной сети.
#
# Впрочем, эта реализация нормально учится и на всех изображениях.
_classes = tf.constant((5, 7, 9), tf.uint8)
_train = filter_by_class(TRAIN_IMAGES, TRAIN_LABELS, _classes)
_test = filter_by_class(TEST_IMAGES, TEST_LABELS, _classes)
DATA = _train.concatenate(_test).cache()
# В некоторых местах нам потребуется знать размер обучающей выборки.
# Получать его таким образом — плохое решение, но на таких объёмах данных оно работает.
DATA_NUMBER = len(list(DATA))
display(Markdown(f'full data shape: {DATA}'))
# Визуально проверяем, что отобрали нужные классы
data = [image for image in DATA.take(100).as_numpy_iterator()]
form_images_map(5, 20, data, scale=1, channels=CHANNELS)
```
## Конструируем модель
По-сути, GAN — это три сети:
- Generator.
- Discriminator.
- GAN — объединение двух предыдущих.
Сам GAN можно не оформлять отдельной сетью, достаточно правильно описать взаимодействие генератора и дискриминатора при обучения. Но, поскольку они учатся совместно, как одно целое, я вижу логичным работать с ними как с единой сетью.
Поэтому мы отдельно создадим генератор с дискриминатором, после чего опишем класс сети, объединяющий их в единое целое.
### Обучение GAN
Обучение генератора и дискриминатора, само собой, происходит на основе функций ошибок. Функции каждой сети оценивают качество бинарной классификации входных данных, на фейковые и реальные. Обычно для этого используют [Binary Crossentropy](https://www.tensorflow.org/api_docs/python/tf/keras/losses/BinaryCrossentropy).
Дискриминатор на вход получает часть реальных изображений и часть созданных генератором. Поскольку класс каждого изображения мы знаем, мы можем легко определить ошибку дискриминатора.
Ошибку же генератора посчитать немного сложнее — качество его работы определяется дискриминатором. Чем хуже результат выдаёт дискриминатор на картинках генератора, тем лучше работает генератор. Поэтому мы скармиливаем дискриминатору созданные изображения с отметкой того, что они реальные (относятся к классу реальных), ошибка дискриминатора на таких данных и будет ошибкой генератора.
### Синхронизация сетей
Если генератор и дискриминатор будут учиться с разной скоростью или иметь разный потенциал выучиваемости, то они не смогут обучаться синхронно. Либо генератор превзойдёт дискриминатор и будет душить его тривиальными фейками, либо дискриминатор найдёт элементарный способ отличать подделки, который генератор не сможет обойти.
Поэтому очень рекомендую при экспериментах с GAN сначала запускать что-нибудь очень простое, но работающее. И только после этого усложнять и экспериментировать. Не будьте как я :-)
Эти же соображения предполагают визуализацию результатов обучения сети. **Убедитесь, что она корректно работает перед экспериментами.** Иначе можете как я сутки отлаживать работающую сеть с неработающей визуализацией.
### Метрики
У нас есть две конкурирующие сети, которые учатся на результатах работы друг друга. Такое обучение, потенциально, может происходить бесконечно.
Поэтому сходу не ясно какой критерий остановки использовать и на какие метрики смотреть, чтобы анализировать ход обучения.
На сколько я понял, по крайней мере для простых случаев качество обучения GAN оценивают визуально: видит человек косяки на выходе генератора или не видит. Альтернативой может быть либо использование другой, предобученной сети, либо метаанализ метрик. Ни в ту ни в другую сторону я не смотрел.
Касательно анализа самих метрик, есть одна эвристика, которую можно применить сразу в двух местах.
Поскольку сети конкурируют, обучаются совместно и на одних данных, мы можем ожидать, что их ошибки будут стабильны. Утрируя, если генератор и дискриминатор обучаются с одинаковой скоростью из одного состояния, то их ошибки не должны изменяться, так как на любое улучшение генератора последует соответствующее улучшение дискриминатора и наоборот.
Отсюда можно вывести метаметрики, которые позволяют оценить стабильность обучения GAN:
- Отношение ошибки генератора к ошибке диксриминатора должно колебаться около единицы. Конечно, если их функции ошибок совпадают.
- Отношение ошибки дискриминатора на реальных данных к ошибке дискриминатора на фейковых данных должно колебаться около единицы.
Если любое из этих отношений сильно отклоняется от единицы, значит GAN обучается неравномерно и могут возникнуть проблемы. В то же время необходимо помнить, что нейронные сети — сложная штука, и отклонения могут быть. Иногда даже большие. Главное чтобы GAN восстанавливался после них.
```
def construct_discriminator():
names = LayersNameGenerator('discriminator')
inputs = keras.Input(shape=SPRITE_SHAPE, name=names('Input'))
branch = inputs
n = 64
branch = layers.Conv2D(n, 4, 2, padding='same', name=names('Conv2D'))(branch)
branch = layers.LeakyReLU(alpha=0.2, name=names('LeakyReLU'))(branch)
branch = layers.Conv2D(n, 4, 2, padding='same', name=names('Conv2D'))(branch)
branch = layers.LeakyReLU(alpha=0.2, name=names('LeakyReLU'))(branch)
branch = layers.Flatten(name=names('Flatten'))(branch)
branch = layers.Dense(1, activation="sigmoid", name=names('Dense'))(branch)
outputs = branch
return keras.Model(inputs=inputs, outputs=outputs, name='Discimiantor')
def construct_generator(code_n):
names = LayersNameGenerator('generator')
inputs = keras.Input(shape=(code_n,), name=names('Input'))
branch = inputs
n = 128
branch = layers.Dense(7 * 7 * n, activation='elu', name=names('Dense'))(branch)
branch = layers.Reshape((7, 7, n), name=names('Reshape'))(branch)
branch = layers.Conv2DTranspose(n, 4, 2, activation='relu', padding='same', name=names('Conv2DTranspose'))(branch)
branch = layers.Conv2DTranspose(n, 4, 2, activation='relu', padding='same', name=names('Conv2DTranspose'))(branch)
branch = layers.Conv2D(CHANNELS, 7, activation="sigmoid", padding='same', name=names('Conv2D'))(branch)
outputs = branch
return keras.Model(inputs=inputs, outputs=outputs, name='Generator')
# Вспомогательный класс для сбора метрик GAN.
# Кроме трёх базовых метрик:
# - ошибка дискриминатора на реальных данных;
# - ошибка дискриминатора на фейковых данных;
# - ошибка генератора;
# Поддерживает две производные метрики:
# - отношение ошибок дискриминатора на реальных и фейковых данных;
# - отношение ошибок дискриминатора на фековых данных и генератора.
class GANMetrics:
def __init__(self):
self._define('discriminator_real_loss')
self._define('discriminator_fake_loss')
self._define('generator_loss')
self._define('discriminator_real_vs_fake_loss')
self._define('discriminator_vs_generator_loss')
def _define(self, name):
setattr(self, name, keras.metrics.Mean(name=name))
def update_state(self, d_real_loss, d_fake_loss, g_loss):
self.discriminator_real_loss.update_state(d_real_loss)
self.discriminator_fake_loss.update_state(d_fake_loss)
self.generator_loss.update_state(g_loss)
self.discriminator_real_vs_fake_loss.update_state(tf.math.divide_no_nan(d_real_loss, d_fake_loss))
self.discriminator_vs_generator_loss.update_state(tf.math.divide_no_nan(d_fake_loss, g_loss))
def result(self):
return {"discriminator_real_loss": self.discriminator_real_loss.result(),
"discriminator_fake_loss": self.discriminator_fake_loss.result(),
"generator_loss": self.generator_loss.result(),
"discriminator_real_vs_fake_loss": self.discriminator_real_vs_fake_loss.result(),
"discriminator_vs_generator_loss": self.discriminator_vs_generator_loss.result()}
def list(self):
return [self.discriminator_real_loss,
self.discriminator_fake_loss,
self.generator_loss,
self.discriminator_real_vs_fake_loss,
self.discriminator_vs_generator_loss]
# Группы графиков для livelossplot
def plotlosses_groups(self):
return {'discriminator loss': ['discriminator_real_loss', 'discriminator_fake_loss'],
'generator loss': ['generator_loss'],
'relations': ['discriminator_real_vs_fake_loss', 'discriminator_vs_generator_loss']}
# Короткие имена для графиков livelossplot
def plotlosses_group_patterns(self):
return ((r'^(discriminator_real_loss)(.*)', 'real'),
(r'^(discriminator_fake_loss)(.*)', 'fake'),
(r'^(generator_loss)(.*)', 'loss'),
(r'^(discriminator_real_vs_fake_loss)(.*)', 'real / fake'),
(r'^(discriminator_vs_generator_loss)(.*)', 'disciminator / generator'),)
# Класс сети, объединяющей генератор и дискриминатор в GAN.
# Делаем отдельный класс, так как нам необходимо переопределить шаг обучения.
# Плюс, оформление в виде класса позволяет проще визуализировать сеть.
class GAN(keras.Model):
def __init__(self, discriminator, generator, latent_dim, **kwargs):
inputs = layers.Input(shape=latent_dim)
super().__init__(inputs=inputs,
outputs=discriminator(generator(inputs)),
**kwargs)
self.discriminator = discriminator
self.generator = generator
self.latent_dim = latent_dim
self.batch_size = None
self.real_labels = None
self.fake_labels = None
def compile(self, batch_size):
super().compile()
self.custom_metrics = GANMetrics()
self.batch_size = batch_size
self.real_labels = tf.ones((self.batch_size, 1))
self.fake_labels = tf.zeros((self.batch_size, 1))
@property
def metrics(self):
return self.custom_metrics.list()
def latent_vector(self, n):
return tf.random.normal(shape=(n, self.latent_dim))
# Самый интересный метод — шаг обучения GAN.
# В куче примеров генератор и дискриминатор учатся отдельно и даже на разных данных.
# Такой лобовой подход имеет право на жизнь, но он точно не оптимален.
# Он приводит к генерации большого количества лишних данных и просто к лишним операциям над памятью.
# Поэтому мы будем учить обе сети в один проход.
@tf.function
def train_step(self, real_images):
# Генерируем шум для генератора, количество примеров берём равным количеству входных данных.
random_latent_vectors = self.latent_vector(self.batch_size)
# Генератор и дискриминатор должны учиться на разных операциях.
# Поэтому самостоятельно записываем операции для расчёта градиентов.
# Указываем persistent=True. TensorFlow по-умолчанию чистит GradientTape после расчёта первого градиаента,
# а нам надо рассчитывать два — по градиенту на сеть.
try:
with tf.GradientTape(persistent=True) as tape:
# генерируем поддельные картинки
fake_images = self.generator(random_latent_vectors)
# оцениваем их дискриминатором
fake_predictions = self.discriminator(fake_images)
# рассчитываем ошибку генератора, предполагая что сгенерированные картинки реальны
g_loss = self.discriminator.compiled_loss(self.real_labels, fake_predictions)
# рассчитываем ошибку дискриминатора на фейковых картинках, зная, что они фейковые
d_f_loss = self.discriminator.compiled_loss(self.fake_labels, fake_predictions)
# получаем предсказания дискриминатора для реальных картинок
real_predictions = self.discriminator(real_images)
# рассчитываем ошибку дискриминатора для реальных картинок
d_r_loss = self.discriminator.compiled_loss(self.real_labels, real_predictions)
# считаем градиент генератора и делаем шаг оптимизации
grads = tape.gradient(g_loss, self.generator.trainable_weights)
self.generator.optimizer.apply_gradients(zip(grads, self.generator.trainable_weights))
# считаем градиент дискриминатора и делаем шаг оптимизации
grads = tape.gradient((d_r_loss, d_f_loss), self.discriminator.trainable_weights)
self.discriminator.optimizer.apply_gradients(zip(grads, self.discriminator.trainable_weights))
# обновляем метрики
self.custom_metrics.update_state(d_r_loss, d_f_loss, g_loss)
finally:
# Удаляем лог градиента
del tape
return self.custom_metrics.result()
# Количество входов шума для генератора.
# 10 — очень малое значение! Я взял его, чтобы после обучения сети было проще с ней экспериментировать.
# По-хорошему, это значение надо установить в 100 или больше.
# Само собой, при большом количестве шума, сложно будет целенаправлено манипулировать сетью.
# Обойти эту проблему можно с использованием дополнительной autoencoder сети,
# которая учится «сжимать» данные до множества признаков.
# Подход с autoencoder мне видится логичным и потому, что GAN использует входные данные всё-таки как шум,
# а не как признаки. В то же время autoencoder ориентирован на выделение признаков.
CODE_N = 10
# Создаём генератор, дискриминатор и объединяем их в GAN.
# Обратите внимание на кастомные параметры оптимизаторов.
# Стандартные параметры TensorFlow плохо подходят для обучения GAN.
discriminator = construct_discriminator()
discriminator.compile(optimizer=keras.optimizers.Adam(learning_rate=0.0002, beta_1=0.5),
loss=keras.losses.BinaryCrossentropy())
generator = construct_generator(CODE_N)
generator.compile(optimizer=keras.optimizers.Adam(learning_rate=0.0002, beta_1=0.5),
loss=keras.losses.BinaryCrossentropy())
gan = GAN(discriminator=discriminator, generator=generator, latent_dim=CODE_N, name='GAN')
display_model(gan, 'GAN')
# Проверяем, что модель в принципе что-то считает
check_input = tf.constant(RNG.random((1, CODE_N)), shape=(1, CODE_N))
generator_output = gan.generator(check_input)
display(Markdown(f'Generator output'))
display_examples(image_getter=generator_output[0],
figsize=(3, 3))
discriminator_output = gan.discriminator(generator_output)
display(Markdown(f'Discriminator output: {discriminator_output}'))
# Проверяем, что визуализатор работает на реальных данных
data = [image for image in DATA.take(9).as_numpy_iterator()]
form_images_map(3, 3, data, scale=1, channels=CHANNELS)
# Определяем собственный callback для model.fit, который будет:
# - отображать работу генератора каждую эпоху;
# - сохранять картинки на файловую систему.
class GANMonitor(keras.callbacks.Callback):
def __init__(self, w, h, images_directory, scale):
self.w = w
self.h = h
self.images_directory = images_directory
self.scale = scale
def on_epoch_end(self, epoch, logs=None):
n = self.w * self.h
random_latent_vectors = self.model.latent_vector(n)
generated_images = self.model.generator(random_latent_vectors).numpy()
pil_world = form_images_map(self.h, self.w, generated_images, channels=CHANNELS, scale=self.scale)
pil_world.save(f"{IMAGES_DIRECTORY}/generated_img_%04d.png" % (epoch,))
display(pil_world)
# Задаём параметры обучения
# Сколько раз цикл обучения пройдёт по всем обучающим данным.
# Установите на свой вкус, 100 должно хватить, чтобу увидеть результат
EPOCHS = 100
BATCH_SIZE = 128
display(Markdown(f'batch size: {BATCH_SIZE}'))
display(Markdown(f'epochs: {EPOCHS}'))
%%time
# каталог с результатами работы генератора
IMAGES_DIRECTORY = 'generated-images'
# создаём каталог с картинками и чистим его, если он заполнен
!mkdir -p $IMAGES_DIRECTORY
!rm $IMAGES_DIRECTORY/*
# Явно формируем dataset для скармливания сети во время обучения.
# Разбиваем на куски и говорим готовить их заранее.
data_for_train = DATA.shuffle(DATA_NUMBER).batch(BATCH_SIZE, drop_remainder=True).prefetch(buffer_size=10)
# Подготавливаем модель.
gan.compile(batch_size=BATCH_SIZE)
# Запускаем обучение.
# Для PlotLossesKerasTF указываем дополнительную конфигурацию графиков.
# Для GANMonitor указываем параметры визуализации.
history = gan.fit(data_for_train,
epochs=EPOCHS,
callbacks=[PlotLossesKerasTF(from_step=-50,
groups=gan.custom_metrics.plotlosses_groups(),
group_patterns=gan.custom_metrics.plotlosses_group_patterns(),
outputs=['MatplotlibPlot']),
GANMonitor(h=3,
w=10,
images_directory=IMAGES_DIRECTORY,
scale=1)])
# Гудим противным звуком, чтобы сообщить об окончании обучения
jupyter_beeper.Beeper().beep(frequency=330, secs=3, blocking=True)
# Поиграем с результатом
start_index = random.randint(0, DATA_NUMBER-1)
def zero_input():
return tf.zeros((CODE_N,))
start_vector = gan.latent_vector(1)[0]
interact_args = {f'v_{i}': ipw.FloatSlider(min=-3.0, max=3.0, step=0.01, value=start_vector[i])
for i in range(CODE_N)}
@ipw.interact(**interact_args)
def generate_sprite(**kwargs):
vector = zero_input().numpy()
for i in range(CODE_N):
vector[i] = kwargs[f'v_{i}']
vector = vector.reshape((1, CODE_N))
sprite = gan.generator(vector)[0].numpy()
scale = 1
sprite = cv2.resize(sprite, dsize=(SPRITE_SIZE*scale, SPRITE_SIZE*scale), interpolation=cv2.INTER_NEAREST)
return PIL.Image.fromarray((sprite * 255).astype(np.uint8))
```
| github_jupyter |
```
#@markdown ■■■■■■■■■■■■■■■■■■
#@markdown 初始化openpose
#@markdown ■■■■■■■■■■■■■■■■■■
#设置版本为1.x
%tensorflow_version 1.x
import tensorflow as tf
tf.__version__
! nvcc --version
! nvidia-smi
! pip install PyQt5
import time
init_start_time = time.time()
#安装 cmake
#https://drive.google.com/file/d/1lAXs5X7qMnKQE48I0JqSob4FX1t6-mED/view?usp=sharing
file_id = "1lAXs5X7qMnKQE48I0JqSob4FX1t6-mED"
file_name = "cmake-3.13.4.zip"
! cd ./ && curl -sc ./cookie "https://drive.google.com/uc?export=download&id=$file_id" > /dev/null
code = "$(awk '/_warning_/ {print $NF}' ./cookie)"
! cd ./ && curl -Lb ./cookie "https://drive.google.com/uc?export=download&confirm=$code&id=$file_id" -o "$file_name"
! cd ./ && unzip cmake-3.13.4.zip
! cd cmake-3.13.4 && ./configure && make && sudo make install
# 依赖库安装
! sudo apt install caffe-cuda
! sudo apt-get --assume-yes update
! sudo apt-get --assume-yes install build-essential
# OpenCV
! sudo apt-get --assume-yes install libopencv-dev
# General dependencies
! sudo apt-get --assume-yes install libatlas-base-dev libprotobuf-dev libleveldb-dev libsnappy-dev libhdf5-serial-dev protobuf-compiler
! sudo apt-get --assume-yes install --no-install-recommends libboost-all-dev
# Remaining dependencies, 14.04
! sudo apt-get --assume-yes install libgflags-dev libgoogle-glog-dev liblmdb-dev
# Python3 libs
! sudo apt-get --assume-yes install python3-setuptools python3-dev build-essential
! sudo apt-get --assume-yes install python3-pip
! sudo -H pip3 install --upgrade numpy protobuf opencv-python
# OpenCL Generic
! sudo apt-get --assume-yes install opencl-headers ocl-icd-opencl-dev
! sudo apt-get --assume-yes install libviennacl-dev
# Openpose安装
ver_openpose = "v1.6.0"
# Openpose の clone
! git clone --depth 1 -b "$ver_openpose" https://github.com/CMU-Perceptual-Computing-Lab/openpose.git
# ! git clone --depth 1 https://github.com/CMU-Perceptual-Computing-Lab/openpose.git
# Openpose の モデルデータDL
! cd openpose/models && ./getModels.sh
#编译Openpose
! cd openpose && rm -r build || true && mkdir build && cd build && cmake .. && make -j`nproc` # example demo usage
# 执行示例确认
! cd /content/openpose && ./build/examples/openpose/openpose.bin --video examples/media/video.avi --write_json ./output/ --display 0 --write_video ./output/openpose.avi
#@markdown ■■■■■■■■■■■■■■■■■■
#@markdown 其他软件初始化
#@markdown ■■■■■■■■■■■■■■■■■■
ver_tag = "ver1.02.01"
# FCRN-DepthPrediction-vmd clone
! git clone --depth 1 -b "$ver_tag" https://github.com/miu200521358/FCRN-DepthPrediction-vmd.git
# FCRN-DepthPrediction-vmd 识别深度模型下载
# 建立模型数据文件夹
! mkdir -p ./FCRN-DepthPrediction-vmd/tensorflow/data
# 下载模型数据并解压
! cd ./FCRN-DepthPrediction-vmd/tensorflow/data && wget -c "http://campar.in.tum.de/files/rupprecht/depthpred/NYU_FCRN-checkpoint.zip" && unzip NYU_FCRN-checkpoint.zip
# 3d-pose-baseline-vmd clone
! git clone --depth 1 -b "$ver_tag" https://github.com/miu200521358/3d-pose-baseline-vmd.git
# 3d-pose-baseline-vmd Human3.6M 模型数据DL
# 建立Human3.6M模型数据文件夹
! mkdir -p ./3d-pose-baseline-vmd/data/h36m
# 下载Human3.6M模型数据并解压
file_id = "1W5WoWpCcJvGm4CHoUhfIB0dgXBDCEHHq"
file_name = "h36m.zip"
! cd ./ && curl -sc ./cookie "https://drive.google.com/uc?export=download&id=$file_id" > /dev/null
code = "$(awk '/_warning_/ {print $NF}' ./cookie)"
! cd ./ && curl -Lb ./cookie "https://drive.google.com/uc?export=download&confirm=$code&id=$file_id" -o "$file_name"
! cd ./ && unzip h36m.zip
! mv ./h36m ./3d-pose-baseline-vmd/data/
# 3d-pose-baseline-vmd 训练数据
# 3d-pose-baseline学习数据文件夹
! mkdir -p ./3d-pose-baseline-vmd/experiments
# 下载3d-pose-baseline训练后的数据
file_id = "1v7ccpms3ZR8ExWWwVfcSpjMsGscDYH7_"
file_name = "experiments.zip"
! cd ./3d-pose-baseline-vmd && curl -sc ./cookie "https://drive.google.com/uc?export=download&id=$file_id" > /dev/null
code = "$(awk '/_warning_/ {print $NF}' ./cookie)"
! cd ./3d-pose-baseline-vmd && curl -Lb ./cookie "https://drive.google.com/uc?export=download&confirm=$code&id=$file_id" -o "$file_name"
! cd ./3d-pose-baseline-vmd && unzip experiments.zip
# VMD-3d-pose-baseline-multi clone
! git clone --depth 1 -b "$ver_tag" https://github.com/miu200521358/VMD-3d-pose-baseline-multi.git
# 安装VMD-3d-pose-baseline-multi 依赖库
! sudo apt-get install python3-pyqt5
! sudo apt-get install pyqt5-dev-tools
! sudo apt-get install qttools5-dev-tools
#安装编码器
! sudo apt-get install mkvtoolnix
init_elapsed_time = (time.time() - init_start_time) / 60
! echo "■■■■■■■■■■■■■■■■■■■■■■■■"
! echo "■■所有初始化均已完成"
! echo "■■"
! echo "■■处理时间:" "$init_elapsed_time" "分"
! echo "■■■■■■■■■■■■■■■■■■■■■■■■"
! echo "Openpose执行结果"
! ls -l /content/openpose/output
#@markdown ■■■■■■■■■■■■■■■■■■
#@markdown 执行函数初始化
#@markdown ■■■■■■■■■■■■■■■■■■
import os
import cv2
import datetime
import time
import datetime
import cv2
import shutil
import glob
from google.colab import files
static_number_people_max = 1
static_frame_first = 0
static_end_frame_no = -1
static_reverse_specific = ""
static_order_specific = ""
static_born_model_csv = "born/animasa_miku_born.csv"
static_is_ik = 1
static_heel_position = 0.0
static_center_z_scale = 1
static_smooth_times = 1
static_threshold_pos = 0.5
static_threshold_rot = 3
static_src_input_video = ""
static_input_video = ""
#执行文件夹
openpose_path = "/content/openpose"
#输出文件夹
base_path = "/content/output"
output_json = "/content/output/json"
output_openpose_avi = "/content/output/openpose.avi"
now_str = ""
depth_dir_path = ""
drive_dir_path = ""
def video_hander( input_video):
global base_path
print("视频名称: ", os.path.basename(input_video))
print("视频大小: ", os.path.getsize(input_video))
video = cv2.VideoCapture(input_video)
# 宽
W = video.get(cv2.CAP_PROP_FRAME_WIDTH)
# 高
H = video.get(cv2.CAP_PROP_FRAME_HEIGHT)
# 总帧数
count = video.get(cv2.CAP_PROP_FRAME_COUNT)
# fps
fps = video.get(cv2.CAP_PROP_FPS)
print("宽: {0}, 高: {1}, 总帧数: {2}, fps: {3}".format(W, H, count, fps))
width = 1280
height = 720
if W != 1280 or (fps != 30 and fps != 60):
print("重新编码,因为大小或fps不在范围: "+ input_video)
# 縮尺
scale = width / W
# 高さ
height = int(H * scale)
# 出力ファイルパス
out_name = 'recode_{0}.mp4'.format("{0:%Y%m%d_%H%M%S}".format(datetime.datetime.now()))
out_path = '{0}/{1}'.format(base_path, out_name)
# try:
# fourcc = cv2.VideoWriter_fourcc(*"MP4V")
# out = cv2.VideoWriter(out_path, fourcc, 30.0, (width, height), True)
# # 入力ファイル
# cap = cv2.VideoCapture(input_video)
# while(cap.isOpened()):
# # 動画から1枚キャプチャして読み込む
# flag, frame = cap.read() # Capture frame-by-frame
# # 動画が終わっていたら終了
# if flag == False:
# break
# # 縮小
# output_frame = cv2.resize(frame, (width, height))
# # 出力
# out.write(output_frame)
# # 終わったら開放
# out.release()
# except Exception as e:
# print("重新编码失败", e)
# cap.release()
# cv2.destroyAllWindows()
# ! mkvmerge --default-duration 0:30fps --fix-bitstream-timing-information 0 "$input_video" -o temp-video.mkv
# ! ffmpeg -i temp-video.mkv -c:v copy side_video.mkv
# ! ffmpeg -i side_video.mkv -vf scale=1280:720 "$out_path"
! ffmpeg -i "$input_video" -qscale 0 -r 30 -y -vf scale=1280:720 "$out_path"
print('MMD重新生成MP4文件成功', out_path)
input_video_name = out_name
# 入力動画ファイル再設定
input_video = base_path + "/"+ input_video_name
video = cv2.VideoCapture(input_video)
# 幅
W = video.get(cv2.CAP_PROP_FRAME_WIDTH)
# 高さ
H = video.get(cv2.CAP_PROP_FRAME_HEIGHT)
# 総フレーム数
count = video.get(cv2.CAP_PROP_FRAME_COUNT)
# fps
fps = video.get(cv2.CAP_PROP_FPS)
print("【重新生成】宽: {0}, 高: {1}, 总帧数: {2}, fps: {3}, 名字: {4}".format(W, H, count, fps,input_video_name))
return input_video
def run_openpose(input_video,number_people_max,frame_first):
#建立临时文件夹
! mkdir -p "$output_json"
#开始执行
! cd "$openpose_path" && ./build/examples/openpose/openpose.bin --video "$input_video" --display 0 --model_pose COCO --write_json "$output_json" --write_video "$output_openpose_avi" --frame_first "$frame_first" --number_people_max "$number_people_max"
def run_fcrn_depth(input_video,end_frame_no,reverse_specific,order_specific):
global now_str,depth_dir_path,drive_dir_path
now_str = "{0:%Y%m%d_%H%M%S}".format(datetime.datetime.now())
! cd FCRN-DepthPrediction-vmd && python tensorflow/predict_video.py --model_path tensorflow/data/NYU_FCRN.ckpt --video_path "$input_video" --json_path "$output_json" --interval 10 --reverse_specific "$reverse_specific" --order_specific "$order_specific" --verbose 1 --now "$now_str" --avi_output "yes" --number_people_max "$number_people_max" --end_frame_no "$end_frame_no"
# 深度結果コピー
depth_dir_path = output_json + "_" + now_str + "_depth"
drive_dir_path = base_path + "/" + now_str
! mkdir -p "$drive_dir_path"
if os.path.exists( depth_dir_path + "/error.txt"):
# 发生错误
! cp "$depth_dir_path"/error.txt "$drive_dir_path"
! echo "■■■■■■■■■■■■■■■■■■■■■■■■"
! echo "■■由于发生错误,处理被中断。"
! echo "■■"
! echo "■■■■■■■■■■■■■■■■■■■■■■■■"
! echo "$drive_dir_path" "请检查 error.txt 的内容。"
else:
! cp "$depth_dir_path"/*.avi "$drive_dir_path"
! cp "$depth_dir_path"/message.log "$drive_dir_path"
! cp "$depth_dir_path"/reverse_specific.txt "$drive_dir_path"
! cp "$depth_dir_path"/order_specific.txt "$drive_dir_path"
for i in range(1, number_people_max+1):
! echo ------------------------------------------
! echo 3d-pose-baseline-vmd ["$i"]
! echo ------------------------------------------
target_name = "_" + now_str + "_idx0" + str(i)
target_dir = output_json + target_name
!cd ./3d-pose-baseline-vmd && python src/openpose_3dpose_sandbox_vmd.py --camera_frame --residual --batch_norm --dropout 0.5 --max_norm --evaluateActionWise --use_sh --epochs 200 --load 4874200 --gif_fps 30 --verbose 1 --openpose "$target_dir" --person_idx 1
def run_3d_to_vmd(number_people_max,born_model_csv,is_ik,heel_position,center_z_scale,smooth_times,threshold_pos,threshold_rot):
global now_str,depth_dir_path,drive_dir_path
for i in range(1, number_people_max+1):
target_name = "_" + now_str + "_idx0" + str(i)
target_dir = output_json + target_name
for f in glob.glob(target_dir +"/*.vmd"):
! rm "$f"
! cd ./VMD-3d-pose-baseline-multi && python applications/pos2vmd_multi.py -v 2 -t "$target_dir" -b "$born_model_csv" -c 30 -z "$center_z_scale" -s "$smooth_times" -p "$threshold_pos" -r "$threshold_rot" -k "$is_ik" -e "$heel_position"
# INDEX別結果コピー
idx_dir_path = drive_dir_path + "/idx0" + str(i)
! mkdir -p "$idx_dir_path"
# 日本語対策でpythonコピー
for f in glob.glob(target_dir +"/*.vmd"):
shutil.copy(f, idx_dir_path)
print(f)
files.download(f)
! cp "$target_dir"/pos.txt "$idx_dir_path"
! cp "$target_dir"/start_frame.txt "$idx_dir_path"
def run_mmd(input_video,number_people_max,frame_first,end_frame_no,reverse_specific,order_specific,born_model_csv,is_ik,heel_position,center_z_scale,smooth_times,threshold_pos,threshold_rot):
global static_input_video,static_number_people_max ,static_frame_first ,static_end_frame_no,static_reverse_specific ,static_order_specific,static_born_model_csv
global static_is_ik,static_heel_position ,static_center_z_scale ,static_smooth_times ,static_threshold_pos ,static_threshold_rot
global base_path,static_src_input_video
start_time = time.time()
video_check= False
openpose_check = False
Fcrn_depth_check = False
pose_to_vmd_check = False
#源文件对比
if static_src_input_video != input_video:
video_check = True
openpose_check = True
Fcrn_depth_check = True
pose_to_vmd_check = True
if (static_number_people_max != number_people_max) or (static_frame_first != frame_first):
openpose_check = True
Fcrn_depth_check = True
pose_to_vmd_check = True
if (static_end_frame_no != end_frame_no) or (static_reverse_specific != reverse_specific) or (static_order_specific != order_specific):
Fcrn_depth_check = True
pose_to_vmd_check = True
if (static_born_model_csv != born_model_csv) or (static_is_ik != is_ik) or (static_heel_position != heel_position) or (static_center_z_scale != center_z_scale) or \
(static_smooth_times != smooth_times) or (static_threshold_pos != threshold_pos) or (static_threshold_rot != threshold_rot):
pose_to_vmd_check = True
#因为视频源文件重置,所以如果无修改需要重命名文件
if video_check:
! rm -rf "$base_path"
! mkdir -p "$base_path"
static_src_input_video = input_video
input_video = video_hander(input_video)
static_input_video = input_video
else:
input_video = static_input_video
if openpose_check:
run_openpose(input_video,number_people_max,frame_first)
static_number_people_max = number_people_max
static_frame_first = frame_first
if Fcrn_depth_check:
run_fcrn_depth(input_video,end_frame_no,reverse_specific,order_specific)
static_end_frame_no = end_frame_no
static_reverse_specific = reverse_specific
static_order_specific = order_specific
if pose_to_vmd_check:
run_3d_to_vmd(number_people_max,born_model_csv,is_ik,heel_position,center_z_scale,smooth_times,threshold_pos,threshold_rot)
static_born_model_csv = born_model_csv
static_is_ik = is_ik
static_heel_position = heel_position
static_center_z_scale = center_z_scale
static_smooth_times = smooth_times
static_threshold_pos = threshold_pos
static_threshold_rot = threshold_rot
elapsed_time = (time.time() - start_time) / 60
print( "■■■■■■■■■■■■■■■■■■■■■■■■")
print( "■■所有处理完成")
print( "■■")
print( "■■处理時間:" + str(elapsed_time)+ "分")
print( "■■■■■■■■■■■■■■■■■■■■■■■■")
print( "")
print( "MMD自动跟踪执行结果")
print( base_path)
! ls -l "$base_path"
#@markdown ■■■■■■■■■■■■■■■■■■
#@markdown GO GO GO GO 执行本单元格,上传视频
#@markdown ■■■■■■■■■■■■■■■■■■
from google.colab import files
#@markdown ---
#@markdown ### 输入视频名称
#@markdown 可以选择手动拖入视频到文件中(比较快),然后输入视频文件名,或者直接运行,不输入文件名直接本地上传
input_video = "" #@param {type: "string"}
if input_video == "":
uploaded = files.upload()
for fn in uploaded.keys():
print('User uploaded file "{name}" with length {length} bytes'.format(
name=fn, length=len(uploaded[fn])))
input_video = fn
input_video = "/content/" + input_video
print("本次执行的转化视频文件名为: "+input_video)
#@markdown 输入用于跟踪图像的参数并执行单元。
#@markdown ---
#@markdown ### 【O】视频中的最大人数
#@markdown 请输入您希望从视频中获得的人数。
#@markdown 请与视频中人数尽量保持一致
number_people_max = 1#@param {type: "number"}
#@markdown ---
#@markdown ### 【O】要从第几帧开始分析
#@markdown 输入帧号以开始分析。(从0开始)
#@markdown 请指定在视频中显示所有人的第一帧,默认为0即可,除非你需要跳过某些片段(例如片头)。
frame_first = 0 #@param {type: "number"}
#@markdown ---
#@markdown ### 【F】要从第几帧结束
#@markdown 请输入要从哪一帧结束
#@markdown (从0开始)在“FCRN-DepthPrediction-vmd”中调整反向或顺序时,可以完成过程并查看结果,默认为-1 表示执行到最后
end_frame_no = -1 #@param {type: "number"}
#@markdown ---
#@markdown ### 【F】反转数据表
#@markdown 指定由Openpose反转的帧号(从0开始),人员INDEX顺序和反转的内容。
#@markdown 按照Openpose在 0F 识别的顺序,将INDEX分配为0,1,...。
#@markdown 格式: [{帧号}: 用于指定反转的人INDEX, {反转内容}]
#@markdown {反转内容}: R: 整体身体反转, U:上半身反转, L: 下半身反转, N: 无反转
#@markdown 例如:[10:1,R] 整个人在第10帧中反转第一个人。在message.log中会记录以上述格式输出内容
#@markdown 因此请参考与[10:1,R][30:0,U],中一样,可以在括号中指定多个项目 ps(不要带有中文标点符号))
reverse_specific = "" #@param {type: "string"}
#@markdown ---
#@markdown ### 【F】输出颜色(仅参考,如果多人时,某个人序号跟别人交换或者错误,可以用此项修改)
#@markdown 请在多人轨迹中的交点之后指定人索引顺序。如果要跟踪一个人,可以将其留为空白。
#@markdown 按照Openpose在0F时识别的顺序分配0、1和INDEX。格式:[<帧号>:第几个人的索引,第几个人的索引,…]示例)[10:1,0]…第帧10是从左数第1人按第0个人的顺序对其进行排序。
#@markdown message.log包含以上述格式输出的顺序,因此请参考它。可以在括号中指定多个项目,例如[10:1,0] [30:0,1]。在output_XXX.avi中,按照估计顺序为人们分配了颜色。身体的右半部分为红色,左半部分为以下颜色。
#@markdown 0:绿色,1:蓝色,2:白色,3:黄色,4:桃红色,5:浅蓝色,6:深绿色,7:深蓝色,8:灰色,9:深黄色,10:深桃红色,11:深浅蓝色
order_specific = "" #@param {type: "string"}
#@markdown ---
#@markdown ### 【V】骨骼结构CSV文件
#@markdown 选择或输入跟踪目标模型的骨骼结构CSV文件的路径。请将csv文件上传到Google云端硬盘的“ autotrace”文件夹。
#@markdown 您可以选择 "Animasa-Miku" 和 "Animasa-Miku semi-standard", 也可以输入任何模型的骨骼结构CSV文件
#@markdown 如果要输入任何模型骨骼结构CSV文件, 请将csv文件上传到Google云端硬盘的 "autotrace" 文件夹下
#@markdown 然后请输入「/gdrive/My Drive/autotrace/[csv file name]」
born_model_csv = "born/\u3042\u306B\u307E\u3055\u5F0F\u30DF\u30AF\u6E96\u6A19\u6E96\u30DC\u30FC\u30F3.csv" #@param ["born/animasa_miku_born.csv", "born/animasa_miku_semi_standard_born.csv"] {allow-input: true}
#@markdown ---
#@markdown ### 【V】是否使用IK输出
#@markdown 选择以IK输出,yes或no
#@markdown 如果输入no,则以输出FK
ik_flag = "yes" #@param ['yes', 'no']
is_ik = 1 if ik_flag == "yes" else 0
#@markdown ---
#@markdown ### 【V】脚与地面位置校正
#@markdown 请输入数值的鞋跟的Y轴校正值(可以为小数)
#@markdown 输入负值会接近地面,输入正值会远离地面。
#@markdown 尽管会自动在某种程度上自动校正,但如果无法校正,请进行设置。
heel_position = 0.0 #@param {type: "number"}
#@markdown ---
#@markdown ### 【V】Z中心放大倍率
#@markdown 以将放大倍数应用到Z轴中心移动(可以是小数)
#@markdown 值越小,中心Z移动的宽度越小
#@markdown 输入0时,不进行Z轴中心移动。
center_z_scale = 2#@param {type: "number"}
#@markdown ---
#@markdown ### 【V】平滑频率
#@markdown 指定运动的平滑频率
#@markdown 请仅输入1或更大的整数
#@markdown 频率越大,频率越平滑。(行为幅度会变小)
smooth_times = 1#@param {type: "number"}
#@markdown ---
#@markdown ### 【V】移动稀疏量 (低于该阀值的运动宽度,不会进行输出,防抖动)
#@markdown 用数值(允许小数)指定用于稀疏移动(IK /中心)的移动量
#@markdown 如果在指定范围内有移动,则将稀疏。如果移动抽取量设置为0,则不执行抽取。
#@markdown 当移动稀疏量设置为0时,不进行稀疏。
threshold_pos = 0.3 #@param {type: "number"}
#@markdown ---
#@markdown ### 【V】旋转稀疏角 (低于该阀值的运动角度,则不会进行输出)
#@markdown 指定用于稀疏旋转键的角度(0到180度的十进制数)
#@markdown 如果在指定角度范围内有旋转,则稀疏旋转键。
threshold_rot = 3#@param {type: "number"}
print(" 【O】Maximum number of people in the video: "+str(number_people_max))
print(" 【O】Frame number to start analysis: "+str(frame_first))
print(" 【F】Frame number to finish analysis: "+str(end_frame_no))
print(" 【F】Reverse specification list: "+str(reverse_specific))
print(" 【F】Ordered list: "+str(order_specific))
print(" 【V】Bone structure CSV file: "+str(born_model_csv))
print(" 【V】Whether to output with IK: "+str(ik_flag))
print(" 【V】Heel position correction: "+str(heel_position))
print(" 【V】Center Z moving magnification: "+str(center_z_scale))
print(" 【V】Smoothing frequency: "+str(smooth_times))
print(" 【V】Movement key thinning amount: "+str(threshold_pos))
print(" 【V】Rotating Key Culling Angle: "+str(threshold_rot))
print("")
print("If the above is correct, please proceed to the next.")
#input_video = "/content/openpose/examples/media/video.avi"
run_mmd(input_video,number_people_max,frame_first,end_frame_no,reverse_specific,order_specific,born_model_csv,is_ik,heel_position,center_z_scale,smooth_times,threshold_pos,threshold_rot)
```
# License许可
发布和分发MMD自动跟踪的结果时,请确保检查许可证。Unity也是如此。
如果您能列出您的许可证,我将不胜感激。
[MMD运动跟踪自动化套件许可证](https://ch.nicovideo.jp/miu200521358/blomaga/ar1686913)
原作者:Twitter miu200521358
修改与优化:B站 妖风瑟瑟
| github_jupyter |
**Important note:** You should always work on a duplicate of the course notebook. On the page you used to open this, tick the box next to the name of the notebook and click duplicate to easily create a new version of this notebook.
You will get errors each time you try to update your course repository if you don't do this, and your changes will end up being erased by the original course version.
# Welcome to Jupyter Notebooks!
If you want to learn how to use this tool you've come to the right place. This article will teach you all you need to know to use Jupyter Notebooks effectively. You only need to go through Section 1 to learn the basics and you can go into Section 2 if you want to further increase your productivity.
You might be reading this tutorial in a web page (maybe Github or the course's webpage). We strongly suggest to read this tutorial in a (yes, you guessed it) Jupyter Notebook. This way you will be able to actually *try* the different commands we will introduce here.
## Section 1: Need to Know
### Introduction
Let's build up from the basics, what is a Jupyter Notebook? Well, you are reading one. It is a document made of cells. You can write like I am writing now (markdown cells) or you can perform calculations in Python (code cells) and run them like this:
```
1+1
```
Cool huh? This combination of prose and code makes Jupyter Notebook ideal for experimentation: we can see the rationale for each experiment, the code and the results in one comprehensive document. In fast.ai, each lesson is documented in a notebook and you can later use that notebook to experiment yourself.
Other renowned institutions in academy and industry use Jupyter Notebook: Google, Microsoft, IBM, Bloomberg, Berkeley and NASA among others. Even Nobel-winning economists [use Jupyter Notebooks](https://paulromer.net/jupyter-mathematica-and-the-future-of-the-research-paper/) for their experiments and some suggest that Jupyter Notebooks will be the [new format for research papers](https://www.theatlantic.com/science/archive/2018/04/the-scientific-paper-is-obsolete/556676/).
### Writing
A type of cell in which you can write like this is called _Markdown_. [_Markdown_](https://en.wikipedia.org/wiki/Markdown) is a very popular markup language. To specify that a cell is _Markdown_ you need to click in the drop-down menu in the toolbar and select _Markdown_.
Click on the the '+' button on the left and select _Markdown_ from the toolbar.
Now you can type your first _Markdown_ cell. Write 'My first markdown cell' and press run.

You should see something like this:
My first markdown cell
Now try making your first _Code_ cell: follow the same steps as before but don't change the cell type (when you add a cell its default type is _Code_). Type something like 3/2. You should see '1.5' as output.
```
3/2
```
### Modes
If you made a mistake in your *Markdown* cell and you have already ran it, you will notice that you cannot edit it just by clicking on it. This is because you are in **Command Mode**. Jupyter Notebooks have two distinct modes:
1. **Edit Mode**: Allows you to edit a cell's content.
2. **Command Mode**: Allows you to edit the notebook as a whole and use keyboard shortcuts but not edit a cell's content.
You can toggle between these two by either pressing <kbd>ESC</kbd> and <kbd>Enter</kbd> or clicking outside a cell or inside it (you need to double click if its a Markdown cell). You can always know which mode you're on since the current cell has a green border if in **Edit Mode** and a blue border in **Command Mode**. Try it!
### Other Important Considerations
1. Your notebook is autosaved every 120 seconds. If you want to manually save it you can just press the save button on the upper left corner or press <kbd>s</kbd> in **Command Mode**.

2. To know if your kernel is computing or not you can check the dot in your upper right corner. If the dot is full, it means that the kernel is working. If not, it is idle. You can place the mouse on it and see the state of the kernel be displayed.

3. There are a couple of shortcuts you must know about which we use **all** the time (always in **Command Mode**). These are:
<kbd>Shift</kbd>+<kbd>Enter</kbd>: Runs the code or markdown on a cell
<kbd>Up Arrow</kbd>+<kbd>Down Arrow</kbd>: Toggle across cells
<kbd>b</kbd>: Create new cell
<kbd>0</kbd>+<kbd>0</kbd>: Reset Kernel
You can find more shortcuts in the Shortcuts section below.
4. You may need to use a terminal in a Jupyter Notebook environment (for example to git pull on a repository). That is very easy to do, just press 'New' in your Home directory and 'Terminal'. Don't know how to use the Terminal? We made a tutorial for that as well. You can find it [here](https://course.fast.ai/terminal_tutorial.html).

That's it. This is all you need to know to use Jupyter Notebooks. That said, we have more tips and tricks below ↓↓↓
## Section 2: Going deeper
### Markdown formatting
#### Italics, Bold, Strikethrough, Inline, Blockquotes and Links
The five most important concepts to format your code appropriately when using markdown are:
1. *Italics*: Surround your text with '\_' or '\*'
2. **Bold**: Surround your text with '\__' or '\**'
3. `inline`: Surround your text with '\`'
4. > blockquote: Place '\>' before your text.
5. [Links](https://course.fast.ai/): Surround the text you want to link with '\[\]' and place the link adjacent to the text, surrounded with '()'
#### Headings
Notice that including a hashtag before the text in a markdown cell makes the text a heading. The number of hashtags you include will determine the priority of the header ('#' is level one, '##' is level two, '###' is level three and '####' is level four). We will add three new cells with the '+' button on the left to see how every level of heading looks.
Double click on some headings and find out what level they are!
#### Lists
There are three types of lists in markdown.
Ordered list:
1. Step 1
2. Step 1B
3. Step 3
Unordered list
* learning rate
* cycle length
* weight decay
Task list
- [x] Learn Jupyter Notebooks
- [x] Writing
- [x] Modes
- [x] Other Considerations
- [ ] Change the world
Double click on each to see how they are built!
### Code Capabilities
**Code** cells are different than **Markdown** cells in that they have an output cell. This means that we can _keep_ the results of our code within the notebook and share them. Let's say we want to show a graph that explains the result of an experiment. We can just run the necessary cells and save the notebook. The output will be there when we open it again! Try it out by running the next four cells.
```
# Import necessary libraries
from fastai.vision import *
import matplotlib.pyplot as plt
from PIL import Image
a = 1
b = a + 1
c = b + a + 1
d = c + b + a + 1
a, b, c ,d
plt.plot([a,b,c,d])
plt.show()
```
We can also print images while experimenting. I am watching you.
```
Image.open('images/notebook_tutorial/cat_example.jpg')
```
### Running the app locally
You may be running Jupyter Notebook from an interactive coding environment like Gradient, Sagemaker or Salamander. You can also run a Jupyter Notebook server from your local computer. What's more, if you have installed Anaconda you don't even need to install Jupyter (if not, just `pip install jupyter`).
You just need to run `jupyter notebook` in your terminal. Remember to run it from a folder that contains all the folders/files you will want to access. You will be able to open, view and edit files located within the directory in which you run this command but not files in parent directories.
If a browser tab does not open automatically once you run the command, you should CTRL+CLICK the link starting with 'https://localhost:' and this will open a new tab in your default browser.
### Creating a notebook
Click on 'New' in the upper left corner and 'Python 3' in the drop-down list (we are going to use a [Python kernel](https://github.com/ipython/ipython) for all our experiments).

Note: You will sometimes hear people talking about the Notebook 'kernel'. The 'kernel' is just the Python engine that performs the computations for you.
### Shortcuts and tricks
#### Command Mode Shortcuts
There are a couple of useful keyboard shortcuts in `Command Mode` that you can leverage to make Jupyter Notebook faster to use. Remember that to switch back and forth between `Command Mode` and `Edit Mode` with <kbd>Esc</kbd> and <kbd>Enter</kbd>.
<kbd>m</kbd>: Convert cell to Markdown
<kbd>y</kbd>: Convert cell to Code
<kbd>D</kbd>+<kbd>D</kbd>: Delete cell
<kbd>o</kbd>: Toggle between hide or show output
<kbd>Shift</kbd>+<kbd>Arrow up/Arrow down</kbd>: Selects multiple cells. Once you have selected them you can operate on them like a batch (run, copy, paste etc).
<kbd>Shift</kbd>+<kbd>M</kbd>: Merge selected cells.
<kbd>Shift</kbd>+<kbd>Tab</kbd>: [press once] Tells you which parameters to pass on a function
<kbd>Shift</kbd>+<kbd>Tab</kbd>: [press three times] Gives additional information on the method
#### Cell Tricks
```
from fastai import*
from fastai.vision import *
```
There are also some tricks that you can code into a cell.
`?function-name`: Shows the definition and docstring for that function
```
?ImageDataBunch
```
`??function-name`: Shows the source code for that function
```
??ImageDataBunch
```
`doc(function-name)`: Shows the definition, docstring **and links to the documentation** of the function
(only works with fastai library imported)
```
doc(ImageDataBunch)
```
#### Line Magics
Line magics are functions that you can run on cells and take as an argument the rest of the line from where they are called. You call them by placing a '%' sign before the command. The most useful ones are:
`%matplotlib inline`: This command ensures that all matplotlib plots will be plotted in the output cell within the notebook and will be kept in the notebook when saved.
`%reload_ext autoreload`, `%autoreload 2`: Reload all modules before executing a new line. If a module is edited, it is not necessary to rerun the import commands, the modules will be reloaded automatically.
These three commands are always called together at the beginning of every notebook.
```
%matplotlib inline
%reload_ext autoreload
%autoreload 2
```
`%timeit`: Runs a line a ten thousand times and displays the average time it took to run it.
```
%timeit [i+1 for i in range(1000)]
```
`%debug`: Allows to inspect a function which is showing an error using the [Python debugger](https://docs.python.org/3/library/pdb.html).
```
for i in range(1000):
a = i+1
b = 'string'
c = b+1
%debug
```
| github_jupyter |
# Self-Driving Car Engineer Nanodegree
## Project: **Finding Lane Lines on the Road**
***
In this project, you will use the tools you learned about in the lesson to identify lane lines on the road. You can develop your pipeline on a series of individual images, and later apply the result to a video stream (really just a series of images). Check out the video clip "raw-lines-example.mp4" (also contained in this repository) to see what the output should look like after using the helper functions below.
Once you have a result that looks roughly like "raw-lines-example.mp4", you'll need to get creative and try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video "P1_example.mp4". Ultimately, you would like to draw just one line for the left side of the lane, and one for the right.
In addition to implementing code, there is a brief writeup to complete. The writeup should be completed in a separate file, which can be either a markdown file or a pdf document. There is a [write up template](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) that can be used to guide the writing process. Completing both the code in the Ipython notebook and the writeup template will cover all of the [rubric points](https://review.udacity.com/#!/rubrics/322/view) for this project.
---
Let's have a look at our first image called 'test_images/solidWhiteRight.jpg'. Run the 2 cells below (hit Shift-Enter or the "play" button above) to display the image.
**Note: If, at any point, you encounter frozen display windows or other confounding issues, you can always start again with a clean slate by going to the "Kernel" menu above and selecting "Restart & Clear Output".**
---
**The tools you have are color selection, region of interest selection, grayscaling, Gaussian smoothing, Canny Edge Detection and Hough Tranform line detection. You are also free to explore and try other techniques that were not presented in the lesson. Your goal is piece together a pipeline to detect the line segments in the image, then average/extrapolate them and draw them onto the image for display (as below). Once you have a working pipeline, try it out on the video stream below.**
---
<figure>
<img src="examples/line-segments-example.jpg" width="380" alt="Combined Image" />
<figcaption>
<p></p>
<p style="text-align: center;"> Your output should look something like this (above) after detecting line segments using the helper functions below </p>
</figcaption>
</figure>
<p></p>
<figure>
<img src="examples/laneLines_thirdPass.jpg" width="380" alt="Combined Image" />
<figcaption>
<p></p>
<p style="text-align: center;"> Your goal is to connect/average/extrapolate line segments to get output like this</p>
</figcaption>
</figure>
**Run the cell below to import some packages. If you get an `import error` for a package you've already installed, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.**
## Import Packages
```
#importing some useful packages
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import numpy as np
import cv2
%matplotlib inline
```
## Read in an Image
```
#reading in an image
image = mpimg.imread('test_images/solidWhiteRight.jpg')
#printing out some stats and plotting
print('This image is:', type(image), 'with dimensions:', image.shape)
plt.imshow(image) # if you wanted to show a single color channel image called 'gray', for example, call as plt.imshow(gray, cmap='gray')
```
## Ideas for Lane Detection Pipeline
**Some OpenCV functions (beyond those introduced in the lesson) that might be useful for this project are:**
`cv2.inRange()` for color selection
`cv2.fillPoly()` for regions selection
`cv2.line()` to draw lines on an image given endpoints
`cv2.addWeighted()` to coadd / overlay two images
`cv2.cvtColor()` to grayscale or change color
`cv2.imwrite()` to output images to file
`cv2.bitwise_and()` to apply a mask to an image
**Check out the OpenCV documentation to learn about these and discover even more awesome functionality!**
## Helper Functions
Below are some helper functions to help get you started. They should look familiar from the lesson!
```
import math
def grayscale(img):
"""Applies the Grayscale transform
This will return an image with only one color channel
but NOTE: to see the returned image as grayscale
(assuming your grayscaled image is called 'gray')
you should call plt.imshow(gray, cmap='gray')"""
return cv2.cvtColor(img, cv2.COLOR_RGB2GRAY)
# Or use BGR2GRAY if you read an image with cv2.imread()
# return cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
def canny(img, low_threshold, high_threshold):
"""Applies the Canny transform"""
return cv2.Canny(img, low_threshold, high_threshold)
def gaussian_blur(img, kernel_size):
"""Applies a Gaussian Noise kernel"""
return cv2.GaussianBlur(img, (kernel_size, kernel_size), 0)
def region_of_interest(img, vertices):
"""
Applies an image mask.
Only keeps the region of the image defined by the polygon
formed from `vertices`. The rest of the image is set to black.
`vertices` should be a numpy array of integer points.
"""
#defining a blank mask to start with
mask = np.zeros_like(img)
#defining a 3 channel or 1 channel color to fill the mask with depending on the input image
if len(img.shape) > 2:
channel_count = img.shape[2] # i.e. 3 or 4 depending on your image
ignore_mask_color = (255,) * channel_count
else:
ignore_mask_color = 255
#filling pixels inside the polygon defined by "vertices" with the fill color
cv2.fillPoly(mask, vertices, ignore_mask_color)
#returning the image only where mask pixels are nonzero
masked_image = cv2.bitwise_and(img, mask)
return masked_image
def draw_lines(img, lines, color=[255, 0, 0], thickness=8):
"""
NOTE: this is the function you might want to use as a starting point once you want to
average/extrapolate the line segments you detect to map out the full
extent of the lane (going from the result shown in raw-lines-example.mp4
to that shown in P1_example.mp4).
Think about things like separating line segments by their
slope ((y2-y1)/(x2-x1)) to decide which segments are part of the left
line vs. the right line. Then, you can average the position of each of
the lines and extrapolate to the top and bottom of the lane.
This function draws `lines` with `color` and `thickness`.
Lines are drawn on the image inplace (mutates the image).
If you want to make the lines semi-transparent, think about combining
this function with the weighted_img() function below
"""
negative_slopes = []
positive_slopes = []
negetive_intercepts = []
positive_intercepts = []
left_line_x = []
left_line_y = []
right_line_x = []
right_line_y = []
y_max = img.shape[0]
y_min = img.shape[0]
#Drawing Lines
for line in lines:
for x1,y1,x2,y2 in line:
current_slope = (y2-y1)/(x2-x1)
if current_slope < 0.0 and current_slope > -math.inf:
negative_slopes.append(current_slope) # left line
left_line_x.append(x1)
left_line_x.append(x2)
left_line_y.append(y1)
left_line_y.append(y2)
negetive_intercepts.append(y1 -current_slope*x1)
if current_slope > 0.0 and current_slope < math.inf:
positive_slopes.append(current_slope) # right line
right_line_x.append(x1)
right_line_x.append(x2)
right_line_y.append(y1)
right_line_y.append(y2)
positive_intercepts.append(y1 - current_slope*x1)
y_min = min(y_min, y1, y2)
y_min += 20 # add small threshold
if len(positive_slopes) > 0 and len(right_line_x) > 0 and len(right_line_y) > 0:
ave_positive_slope = sum(positive_slopes) / len(positive_slopes)
ave_right_line_x = sum(right_line_x) / len(right_line_x)
ave_right_line_y = sum(right_line_y ) / len(right_line_y)
intercept = sum(positive_intercepts) / len(positive_intercepts)
x_min=int((y_min-intercept)/ave_positive_slope)
x_max = int((y_max - intercept)/ ave_positive_slope)
cv2.line(img, (x_min, y_min), (x_max, y_max), color, thickness)
if len(negative_slopes) > 0 and len(left_line_x) > 0 and len(left_line_y) > 0:
ave_negative_slope = sum(negative_slopes) / len(negative_slopes)
ave_left_line_x = sum(left_line_x) / len(left_line_x)
ave_left_line_y = sum(left_line_y ) / len(left_line_y)
intercept = sum(negetive_intercepts) / len(negetive_intercepts)
x_min = int((y_min-intercept)/ave_negative_slope)
x_max = int((y_max - intercept)/ ave_negative_slope)
cv2.line(img, (x_min, y_min), (x_max, y_max), color, thickness)
def hough_lines(img, rho, theta, threshold, min_line_len, max_line_gap):
"""
`img` should be the output of a Canny transform.
Returns an image with hough lines drawn.
"""
lines = cv2.HoughLinesP(img, rho, theta, threshold, np.array([]), minLineLength=min_line_len, maxLineGap=max_line_gap)
line_img = np.zeros((img.shape[0], img.shape[1], 3), dtype=np.uint8)
draw_lines(line_img, lines)
return line_img
# Python 3 has support for cool math symbols.
def weighted_img(img, initial_img, α=0.8, β=1., γ=0.):
"""
`img` is the output of the hough_lines(), An image with lines drawn on it.
Should be a blank image (all black) with lines drawn on it.
`initial_img` should be the image before any processing.
The result image is computed as follows:
initial_img * α + img * β + γ
NOTE: initial_img and img must be the same shape!
"""
return cv2.addWeighted(initial_img, α, img, β, γ)
```
## Test Images
Build your pipeline to work on the images in the directory "test_images"
**You should make sure your pipeline works well on these images before you try the videos.**
```
import os
os.listdir("test_images/")
```
## Build a Lane Finding Pipeline
Build the pipeline and run your solution on all test_images. Make copies into the `test_images_output` directory, and you can use the images in your writeup report.
Try tuning the various parameters, especially the low and high Canny thresholds as well as the Hough lines parameters.
```
# TODO: Build your pipeline that will draw lane lines on the test_images
# then save them to the test_images_output directory.
##1) We Have To read our Image in a grey scale fromat
Input_Image = mpimg.imread('test_images/solidWhiteCurve.jpg')
Input_Grey_Img = grayscale(Input_Image)
plt.imshow(Input_Grey_Img, cmap='gray')
plt.title('Image in Grey Scale Format')
##2) Apply Canny Detection with a low threshold 1 : 3 to high threshold
## we do further smoothing before applying canny algorithm
Kernel_size = 3 #always put an odd number (3, 5, 7, ..)
img_Smoothed = gaussian_blur(Input_Grey_Img, Kernel_size)
High_threshold = 150
Low_threshold = 75
imga_fter_Canny = canny(img_Smoothed, Low_threshold, High_threshold)
plt.imshow(imga_fter_Canny, cmap='gray')
plt.title('Image after Applying Canny')
##3) Determine Region of interest to detect Lane lines in Image
## Set Verticies Parameter to determine regoin of interest first
#Vertices : Left_bottom, Right_bottom, Apex (Area of interest)
vertices = np.array([[(0,image.shape[0]),(470, 320), (500, 320), (image.shape[1],image.shape[0])]], dtype=np.int32)
Masked_Image = region_of_interest(imga_fter_Canny, vertices)
plt.imshow(Masked_Image,cmap='gray')
plt.title('Massked Image')
##4)using hough transfrom to find lines
# Define the Hough transform parameters
# Make a blank the same size as our image to draw on
rho = 2
theta = np.pi/180
threshold = 15
min_line_length = 40
max_line_gap = 20
lines = hough_lines(Masked_Image, rho, theta, threshold, min_line_length, max_line_gap)
plt.imshow(lines,cmap='gray')
plt.title('lines Image')
##5) Draw Lines on the real Image
Final_out = weighted_img(lines, Input_Image, α=0.8, β=1., γ=0.)
plt.imshow(Final_out)
plt.title('Final Image with lane detected')
```
## Test on Videos
You know what's cooler than drawing lanes over images? Drawing lanes over video!
We can test our solution on two provided videos:
`solidWhiteRight.mp4`
`solidYellowLeft.mp4`
**Note: if you get an import error when you run the next cell, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.**
**If you get an error that looks like this:**
```
NeedDownloadError: Need ffmpeg exe.
You can download it by calling:
imageio.plugins.ffmpeg.download()
```
**Follow the instructions in the error message and check out [this forum post](https://discussions.udacity.com/t/project-error-of-test-on-videos/274082) for more troubleshooting tips across operating systems.**
```
# Import everything needed to edit/save/watch video clips
from moviepy.editor import VideoFileClip
from IPython.display import HTML
def process_image(image):
# NOTE: The output you return should be a color image (3 channel) for processing video below
# TODO: put your pipeline here,
# you should return the final output (image where lines are drawn on lanes)
##1) We Have To read our Image in a grey scale fromat
Input_Grey_Img = grayscale(image)
##2) Apply Canny Detection with a low threshold 1 : 3 to high threshold
## we do further smoothing before applying canny algorithm
Kernel_size = 3 #always put an odd number (3, 5, 7, ..)
img_Smoothed = gaussian_blur(Input_Grey_Img, Kernel_size)
High_threshold = 150
Low_threshold = 50
imga_fter_Canny = canny(img_Smoothed, Low_threshold, High_threshold)
##3) Determine Region of interest to detect Lane lines in Image
## Set Verticies Parameter to determine regoin of interest first
#Vertices : Left_bottom, Right_bottom, Apex (Area of interest)
vertices = np.array([[(0,image.shape[0]),
(470, 320),
(500, 320),
(image.shape[1],
image.shape[0])]],
dtype=np.int32)
Masked_Image = region_of_interest(imga_fter_Canny, vertices)
##4)using hough transfrom to find lines
# Define the Hough transform parameters
# Make a blank the same size as our image to draw on
rho = 2
theta = np.pi/180
threshold = 55
min_line_length = 100
max_line_gap = 150
lines = hough_lines(Masked_Image, rho, theta, threshold, min_line_length, max_line_gap)
##5)Draw Lines on the real Image
result = weighted_img(lines, image, α=0.8, β=1., γ=0.)
return result
```
Let's try the one with the solid white lane on the right first ...
```
white_output = 'test_videos_output/solidWhiteRight.mp4'
## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video
## To do so add .subclip(start_second,end_second) to the end of the line below
## Where start_second and end_second are integer values representing the start and end of the subclip
## You may also uncomment the following line for a subclip of the first 5 seconds
##clip1 = VideoFileClip("test_videos/solidWhiteRight.mp4").subclip(0,5)
clip1 = VideoFileClip("test_videos/solidWhiteRight.mp4")
white_clip = clip1.fl_image(process_image) #NOTE: this function expects color images!!
%time white_clip.write_videofile(white_output, audio=False)
```
Play the video inline, or if you prefer find the video in your filesystem (should be in the same directory) and play it in your video player of choice.
```
HTML("""
<video width="960" height="540" controls>
<source src="{0}">
</video>
""".format(white_output))
```
## Improve the draw_lines() function
**At this point, if you were successful with making the pipeline and tuning parameters, you probably have the Hough line segments drawn onto the road, but what about identifying the full extent of the lane and marking it clearly as in the example video (P1_example.mp4)? Think about defining a line to run the full length of the visible lane based on the line segments you identified with the Hough Transform. As mentioned previously, try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video "P1_example.mp4".**
**Go back and modify your draw_lines function accordingly and try re-running your pipeline. The new output should draw a single, solid line over the left lane line and a single, solid line over the right lane line. The lines should start from the bottom of the image and extend out to the top of the region of interest.**
Now for the one with the solid yellow lane on the left. This one's more tricky!
```
yellow_output = 'test_videos_output/solidYellowLeft.mp4'
## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video
## To do so add .subclip(start_second,end_second) to the end of the line below
## Where start_second and end_second are integer values representing the start and end of the subclip
## You may also uncomment the following line for a subclip of the first 5 seconds
##clip2 = VideoFileClip('test_videos/solidYellowLeft.mp4').subclip(0,5)
clip2 = VideoFileClip('test_videos/solidYellowLeft.mp4')
yellow_clip = clip2.fl_image(process_image)
%time yellow_clip.write_videofile(yellow_output, audio=False)
HTML("""
<video width="960" height="540" controls>
<source src="{0}">
</video>
""".format(yellow_output))
```
## Writeup and Submission
If you're satisfied with your video outputs, it's time to make the report writeup in a pdf or markdown file. Once you have this Ipython notebook ready along with the writeup, it's time to submit for review! Here is a [link](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) to the writeup template file.
## Optional Challenge
Try your lane finding pipeline on the video below. Does it still work? Can you figure out a way to make it more robust? If you're up for the challenge, modify your pipeline so it works with this video and submit it along with the rest of your project!
```
def process_image1(image):
# NOTE: The output you return should be a color image (3 channel) for processing video below
# TODO: put your pipeline here,
# you should return the final output (image where lines are drawn on lanes)
##1) We Have To read our Image in a grey scale fromat
Input_Grey_Img = grayscale(image)
##2) Apply Canny Detection with a low threshold 1 : 3 to high threshold
## we do further smoothing before applying canny algorithm
Kernel_size = 3 #always put an odd number (3, 5, 7, ..)
img_Smoothed = gaussian_blur(Input_Grey_Img, Kernel_size)
High_threshold = 150
Low_threshold = 50
imga_fter_Canny = canny(img_Smoothed, Low_threshold, High_threshold)
##3) Determine Region of interest to detect Lane lines in Image
## Set Verticies Parameter to determine regoin of interest first
vertices = np.array([[(226, 680),
(614,436),
(714,436),
(1093,634)]])
Masked_Image = region_of_interest(imga_fter_Canny, vertices)
##4)using hough transfrom to find lines
# Define the Hough transform parameters
# Make a blank the same size as our image to draw on
rho = 2
theta = np.pi/180
threshold = 55
min_line_length = 100
max_line_gap = 150
lines = hough_lines(Masked_Image, rho, theta, threshold, min_line_length, max_line_gap)
##5)Draw Lines on the real Image
result = weighted_img(lines, image, α=0.8, β=1., γ=0.)
return result
challenge_output = 'test_videos_output/challenge.mp4'
## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video
## To do so add .subclip(start_second,end_second) to the end of the line below
## Where start_second and end_second are integer values representing the start and end of the subclip
## You may also uncomment the following line for a subclip of the first 5 seconds
##clip3 = VideoFileClip('test_videos/challenge.mp4').subclip(0,5)
clip3 = VideoFileClip('test_videos/challenge.mp4')
challenge_clip = clip3.fl_image(process_image1)
%time challenge_clip.write_videofile(challenge_output, audio=False)
HTML("""
<video width="960" height="540" controls>
<source src="{0}">
</video>
""".format(challenge_output))
```
| github_jupyter |
```
## Advanced Course in Machine Learning
## Week 4
## Exercise 2 / Probabilistic PCA
import numpy as np
import scipy
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib.animation as animation
from numpy import linalg as LA
sns.set_style("darkgrid")
def build_dataset(N, D, K, sigma=1):
x = np.zeros((D, N))
z = np.random.normal(0.0, 1.0, size=(K, N))
# Create a w with random values
w = np.random.normal(0.0, sigma**2, size=(D, K))
mean = np.dot(w, z)
for d in range(D):
for n in range(N):
x[d, n] = np.random.normal(mean[d, n], sigma**2)
print("True principal axes:")
print(w)
return x, mean, w, z
N = 5000 # number of data points
D = 2 # data dimensionality
K = 1 # latent dimensionality
sigma = 1.0
x, mean, w, z = build_dataset(N, D, K, sigma)
print(z)
print(w)
plt.figure(num=None, figsize=(8, 6), dpi=100, facecolor='w', edgecolor='k')
sns.scatterplot(z[0, :], 0, alpha=0.5, label='z')
origin = [0], [0] # origin point
plt.xlabel('x')
plt.ylabel('y')
plt.legend(loc='lower right')
plt.title('Probabilistic PCA, generated z')
plt.show()
plt.figure(num=None, figsize=(8, 6), dpi=100, facecolor='w', edgecolor='k')
sns.scatterplot(z[0, :], 0, alpha=0.5, label='z')
sns.scatterplot(mean[0, :], mean[1, :], color='red', alpha=0.5, label='Wz')
origin = [0], [0] # origin point
#Plot the principal axis
plt.quiver(*origin, w[0,0], w[1,0], color=['g'], scale=1, label='W')
plt.xlabel('x')
plt.ylabel('y')
plt.legend(loc='upper right')
plt.title('Probabilistic PCA, generated z')
plt.show()
print(x)
plt.figure(num=None, figsize=(8, 6), dpi=100, facecolor='w', edgecolor='k')
sns.scatterplot(x[0, :], x[1, :], color='orange', alpha=0.5)
#plt.axis([-5, 5, -5, 5])
plt.xlabel('x')
plt.ylabel('y')
#Plot the principal axis
plt.quiver(*origin, w[0,0], w[1,0], color=['g'], scale=10, label='W')
#Plot probability density contours
sns.kdeplot(x[0, :], x[1, :], n_levels=3, color='purple')
plt.title('Probabilistic PCA, generated x')
plt.show()
plt.figure(num=None, figsize=(8, 6), dpi=100, facecolor='w', edgecolor='k')
sns.scatterplot(x[0, :], x[1, :], color='orange', alpha=0.5, label='X')
sns.scatterplot(z[0, :], 0, alpha=0.5, label='z')
sns.scatterplot(mean[0, :], mean[1, :], color='red', alpha=0.5, label='Wz')
origin = [0], [0] # origin point
#Plot the principal axis
plt.quiver(*origin, w[0,0], w[1,0], color=['g'], scale=10, label='W')
plt.xlabel('x')
plt.ylabel('y')
plt.legend(loc='lower right')
plt.title('Probabilistic PCA')
plt.show()
plt.figure(num=None, figsize=(8, 6), dpi=100, facecolor='w', edgecolor='k')
sns.scatterplot(x[0, :], x[1, :], color='orange', alpha=0.5, label='X')
sns.scatterplot(z[0, :], 0, alpha=0.5, label='z')
sns.scatterplot(mean[0, :], mean[1, :], color='red', alpha=0.5, label='Wz')
origin = [0], [0] # origin point
#Plot the principal axis
plt.quiver(*origin, w[0,0], w[1,0], color=['g'], scale=10, label='W')
#Plot probability density contours
sns.kdeplot(x[0, :], x[1, :], n_levels=6, color='purple')
plt.xlabel('x')
plt.ylabel('y')
plt.legend(loc='lower right')
plt.title('Probabilistic PCA')
plt.show()
```
def main():
fig = plt.figure()
scat = plt.scatter(mean[0, :], color='red', alpha=0.5, label='Wz')
ani = animation.FuncAnimation(fig, update_plot, frames=xrange(N),
fargs=(scat))
plt.show()
def update_plot(i, scat):
scat.set_array(data[i])
return scat,
main()
| github_jupyter |
# Bayes Classifier
```
import util
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import multivariate_normal as mvn
%matplotlib inline
def clamp_sample(x):
x = np.minimum(x, 1)
x = np.maximum(x, 0)
return x
class BayesClassifier:
def fit(self, X, Y):
# assume classes are numbered 0...K-1
self.K = len(set(Y))
self.gaussians = []
self.p_y = np.zeros(self.K)
for k in range(self.K):
Xk = X[Y == k]
self.p_y[k] = len(Xk)
mean = Xk.mean(axis=0) # describe gaussian
cov = np.cov(Xk.T) # describe gaussian
g = {'m': mean, 'c': cov}
self.gaussians.append(g)
# normalize p(y)
self.p_y /= self.p_y.sum()
def sample_given_y(self, y):
g = self.gaussians[y]
return clamp_sample( mvn.rvs(mean=g['m'], cov=g['c']) )
def sample(self):
y = np.random.choice(self.K, p=self.p_y)
return clamp_sample( self.sample_given_y(y) )
X, Y = util.get_mnist()
clf = BayesClassifier()
clf.fit(X, Y)
for k in range(clf.K):
# show one sample for each class
# also show the mean image learned from Gaussian Bayes Classifier
sample = clf.sample_given_y(k).reshape(28, 28)
mean = clf.gaussians[k]['m'].reshape(28, 28)
plt.subplot(1,2,1)
plt.imshow(sample, cmap='gray')
plt.title("Sample")
plt.subplot(1,2,2)
plt.imshow(mean, cmap='gray')
plt.title("Mean")
plt.show()
```
# Bayes Classifier with Gaussian Mixture Models
```
from sklearn.mixture import BayesianGaussianMixture
class BayesClassifier:
def fit(self, X, Y):
# assume classes are numbered 0...K-1
self.K = len(set(Y))
self.gaussians = []
self.p_y = np.zeros(self.K)
for k in range(self.K):
print("Fitting gmm", k)
Xk = X[Y == k]
self.p_y[k] = len(Xk)
gmm = BayesianGaussianMixture(10) # number of clusters
gmm.fit(Xk)
self.gaussians.append(gmm)
# normalize p(y)
self.p_y /= self.p_y.sum()
def sample_given_y(self, y):
gmm = self.gaussians[y]
sample = gmm.sample()
# note: sample returns a tuple containing 2 things:
# 1) the sample
# 2) which cluster it came from
# we'll use (2) to obtain the means so we can plot
# them like we did in the previous script
# we cheat by looking at "non-public" params in
# the sklearn source code
mean = gmm.means_[sample[1]]
return clamp_sample( sample[0].reshape(28, 28) ), mean.reshape(28, 28)
def sample(self):
y = np.random.choice(self.K, p=self.p_y)
return clamp_sample( self.sample_given_y(y) )
clf = BayesClassifier()
clf.fit(X, Y)
for k in range(clf.K):
# show one sample for each class
# also show the mean image learned
sample, mean = clf.sample_given_y(k)
plt.subplot(1,2,1)
plt.imshow(sample, cmap='gray')
plt.title("Sample")
plt.subplot(1,2,2)
plt.imshow(mean, cmap='gray')
plt.title("Mean")
plt.show()
# generate a random sample
sample, mean = clf.sample()
plt.subplot(1,2,1)
plt.imshow(sample, cmap='gray')
plt.title("Random Sample from Random Class")
plt.subplot(1,2,2)
plt.imshow(mean, cmap='gray')
plt.title("Corresponding Cluster Mean")
plt.show()
```
# Neural Network and Autoencoder
```
import tensorflow as tf
class Autoencoder:
def __init__(self, D, M):
# represents a batch of training data
self.X = tf.placeholder(tf.float32, shape=(None, D))
# input -> hidden
self.W = tf.Variable(tf.random_normal(shape=(D, M)) * np.sqrt(2.0 / M))
self.b = tf.Variable(np.zeros(M).astype(np.float32))
# hidden -> output
self.V = tf.Variable(tf.random_normal(shape=(M, D)) * np.sqrt(2.0 / D))
self.c = tf.Variable(np.zeros(D).astype(np.float32))
# construct the reconstruction
self.Z = tf.nn.relu(tf.matmul(self.X, self.W) + self.b)
logits = tf.matmul(self.Z, self.V) + self.c
self.X_hat = tf.nn.sigmoid(logits)
# compute the cost
self.cost = tf.reduce_sum(
tf.nn.sigmoid_cross_entropy_with_logits(
labels=self.X,
logits=logits
)
)
# make the trainer
self.train_op = tf.train.RMSPropOptimizer(learning_rate=0.001).minimize(self.cost)
# set up session and variables for later
self.init_op = tf.global_variables_initializer()
self.sess = tf.InteractiveSession()
self.sess.run(self.init_op)
def fit(self, X, epochs=30, batch_sz=64):
costs = []
n_batches = len(X) // batch_sz
print("n_batches:", n_batches)
for i in range(epochs):
if i % 5 == 0:
print("epoch:", i)
np.random.shuffle(X)
for j in range(n_batches):
batch = X[j*batch_sz:(j+1)*batch_sz]
_, c, = self.sess.run((self.train_op, self.cost), feed_dict={self.X: batch})
c /= batch_sz # just debugging
costs.append(c)
if (j % 100 == 0) and (i % 5 == 0):
print("iter: %d, cost: %.3f" % (j, c))
plt.plot(costs)
plt.show()
def predict(self, X):
return self.sess.run(self.X_hat, feed_dict={self.X: X})
model = Autoencoder(784, 300)
model.fit(X)
done = False
while not done:
i = np.random.choice(len(X))
x = X[i]
im = model.predict([x]).reshape(28, 28)
plt.subplot(1,2,1)
plt.imshow(x.reshape(28, 28), cmap='gray')
plt.title("Original")
plt.subplot(1,2,2)
plt.imshow(im, cmap='gray')
plt.title("Reconstruction")
plt.show()
ans = input("Generate another?")
if ans and ans[0] in ('n' or 'N'):
done = True
```
| github_jupyter |
```
%matplotlib inline
import pandas as pd
import cv2
import numpy as np
from matplotlib import pyplot as plt
df = pd.read_csv("data/22800_SELECT_t___FROM_data_data_t.csv",header=None,index_col=0)
df = df.rename(columns={0:"no", 1: "CAPTDATA", 2: "CAPTIMAGE",3: "timestamp"})
df.info()
df.sample(5)
def alpha_to_gray(img):
alpha_channel = img[:, :, 3]
_, mask = cv2.threshold(alpha_channel, 128, 255, cv2.THRESH_BINARY) # binarize mask
color = img[:, :, :3]
img = cv2.bitwise_not(cv2.bitwise_not(color, mask=mask))
return cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
def preprocess(data):
data = bytes.fromhex(data[2:])
img = cv2.imdecode( np.asarray(bytearray(data), dtype=np.uint8), cv2.IMREAD_UNCHANGED )
img = alpha_to_gray(img)
kernel = np.ones((3, 3), np.uint8)
img = cv2.dilate(img, kernel, iterations=1)
img = cv2.medianBlur(img, 3)
kernel = np.ones((4, 4), np.uint8)
img = cv2.erode(img, kernel, iterations=1)
# plt.imshow(img)
return img
df["IMAGE"] = df["CAPTIMAGE"].apply(preprocess)
def bounding(gray):
# data = bytes.fromhex(df["CAPTIMAGE"][1][2:])
# image = cv2.imdecode( np.asarray(bytearray(data), dtype=np.uint8), cv2.IMREAD_UNCHANGED )
# alpha_channel = image[:, :, 3]
# _, mask = cv2.threshold(alpha_channel, 128, 255, cv2.THRESH_BINARY) # binarize mask
# color = image[:, :, :3]
# src = cv2.bitwise_not(cv2.bitwise_not(color, mask=mask))
ret, binary = cv2.threshold(gray, 127, 255, cv2.THRESH_BINARY)
binary = cv2.bitwise_not(binary)
contours, hierachy = cv2.findContours(binary, cv2.RETR_EXTERNAL , cv2.CHAIN_APPROX_NONE)
ans = []
for h, tcnt in enumerate(contours):
x,y,w,h = cv2.boundingRect(tcnt)
if h < 25:
continue
if 40 < w < 100: # 2개가 붙어 있는 경우
ans.append([x,y,w//2,h])
ans.append([x+(w//2),y,w//2,h])
continue
if 100 <= w < 170:
ans.append([x,y,w//3,h])
ans.append([x+(w//3),y,w//3,h])
ans.append([x+(2*w//3),y,w//3,h])
# cv2.rectangle(src,(x,y),(x+w,y+h),(255,0,0),1)
ans.append([x,y,w,h])
return ans
# cv2.destroyAllWindows()
df["bounding"] = df["IMAGE"].apply(bounding)
def draw_bounding(idx):
CAPTIMAGE = df["CAPTIMAGE"][idx]
bounding = df["bounding"][idx]
data = bytes.fromhex(CAPTIMAGE[2:])
image = cv2.imdecode( np.asarray(bytearray(data), dtype=np.uint8), cv2.IMREAD_UNCHANGED )
alpha_channel = image[:, :, 3]
_, mask = cv2.threshold(alpha_channel, 128, 255, cv2.THRESH_BINARY) # binarize mask
color = image[:, :, :3]
src = cv2.bitwise_not(cv2.bitwise_not(color, mask=mask))
for x,y,w,h in bounding:
# print(x,y,w,h)
cv2.rectangle(src,(x,y),(x+w,y+h),(255,0,0),1)
return src
import random
nrows = 4
ncols = 4
fig, axes = plt.subplots(nrows=nrows, ncols=ncols)
fig.set_size_inches((16, 6))
for i in range(nrows):
for j in range(ncols):
idx = random.randrange(20,22800)
axes[i][j].set_title(str(idx))
axes[i][j].imshow(draw_bounding(idx))
fig.tight_layout()
plt.savefig('sample.png')
plt.show()
charImg = []
for idx in df.index:
IMAGE = df["IMAGE"][idx]
bounding = df["bounding"][idx]
for x,y,w,h in bounding:
newImg = IMAGE[y:y+h,x:x+w]
newImg = cv2.resize(newImg, dsize=(41, 38), interpolation=cv2.INTER_NEAREST)
charImg.append(newImg/255.0)
# cast to numpy arrays
trainingImages = np.asarray(charImg)
# reshape img array to vector
def reshape_image(img):
return np.reshape(img,len(img)*len(img[0]))
img_reshape = np.zeros((len(trainingImages),len(trainingImages[0])*len(trainingImages[0][0])))
for i in range(0,len(trainingImages)):
img_reshape[i] = reshape_image(trainingImages[i])
from sklearn.cluster import KMeans
import matplotlib.pyplot as plt
import seaborn as sns
# create model and prediction
model = KMeans(n_clusters=40,algorithm='auto')
model.fit(img_reshape)
predict = pd.DataFrame(model.predict(img_reshape))
predict.columns=['predict']
import pickle
pickle.dump(model, open("KMeans_40_22800.pkl", "wb"))
import pickle
model = pickle.load(open("KMeans_40_22800.pkl", "rb"))
predict = pd.DataFrame(model.predict(img_reshape))
predict.columns=['predict']
import random
from tqdm import tqdm
r = pd.concat([pd.DataFrame(img_reshape),predict],axis=1)
!rm -rf res_40
!mkdir res_40
nrows = 4
ncols = 10
fig, axes = plt.subplots(nrows=nrows, ncols=ncols)
fig.set_size_inches((16, 6))
for j in tqdm(range(40)):
i = 0
nSample = min(nrows * ncols,len(r[r["predict"] == j]))
for idx in r[r["predict"] == j].sample(nSample).index:
axes[i // ncols][i % ncols].set_title(str(idx))
axes[i // ncols][i % ncols].imshow(trainingImages[idx])
i+=1
fig.tight_layout()
plt.savefig('res_40/sample_' + str(j) + '.png')
```
98 95 92 222 255
| github_jupyter |
```
# Import and create a new SQLContext
from pyspark.sql import SQLContext
sqlContext = SQLContext(sc)
# Read the country CSV file into an RDD.
country_lines = sc.textFile('file:///home/ubuntu/work/notebooks/UCSD/big-data-3/final-project/country-list.csv')
country_lines.collect()
# Convert each line into a pair of words
country_lines.map(lambda a: a.split(",")).collect()
# Convert each pair of words into a tuple
country_tuples = country_lines.map(lambda a: (a.split(",")[0].lower(), a.split(",")[1]))
# Create the DataFrame, look at schema and contents
countryDF = sqlContext.createDataFrame(country_tuples, ["country", "code"])
countryDF.printSchema()
countryDF.take(3)
# Read tweets CSV file into RDD of lines
tweets = sc.textFile('file:///home/ubuntu/work/notebooks/UCSD/big-data-3/final-project/tweets.csv')
tweets.count()
# Clean the data: some tweets are empty. Remove the empty tweets using filter()
filtered_tweets = tweets.filter(lambda a: len(a) > 0)
filtered_tweets.count()
# Perform WordCount on the cleaned tweet texts. (note: this is several lines.)
word_counts = filtered_tweets.flatMap(lambda a: a.split(" ")) \
.map(lambda word: (word.lower(), 1)) \
.reduceByKey(lambda a, b: a + b)
from pyspark.sql import HiveContext
from pyspark.sql.types import *
# sc is an existing SparkContext.
sqlContext = HiveContext(sc)
schemaString = "word count"
fields = [StructField(field_name, StringType(), True) for field_name in schemaString.split()]
schema = StructType(fields)
# Create the DataFrame of tweet word counts
tweetsDF = sqlContext.createDataFrame(word_counts, schema)
tweetsDF.printSchema()
tweetsDF.count()
# Join the country and tweet DataFrames (on the appropriate column)
joined = countryDF.join(tweetsDF, countryDF.country == tweetsDF.word)
joined.take(5)
joined.show()
# Question 1: number of distinct countries mentioned
distinct_countries = joined.select("country").distinct()
distinct_countries.show(100)
# Question 2: number of countries mentioned in tweets.
from pyspark.sql.functions import sum
from pyspark.sql import SparkSession
from pyspark.sql import Row
countries_count = joined.groupBy("country")
joined.createOrReplaceTempView("records")
spark.sql("SELECT country, count(*) count1 FROM records group by country order by count1 desc, country asc").show(100)
# Table 1: top three countries and their counts.
from pyspark.sql.functions import desc
from pyspark.sql.functions import col
top_3 = joined.sort(col("count").desc())
top_3.show()
# Table 2: counts for Wales, Iceland, and Japan.
```
| github_jupyter |
# Our data exists as vectors in matrixes
Linear algeabra helps us manipulate data to eventually find the smallest sum squared errors of our data which will give us our beta value for our regression model
```
import numpy as np
# create array to be transformed into vectors
x1 = np.array([1,2,1])
x2 = np.array([4,1,5])
x3 = np.array([6,8,6])
print("Array 1:", x1, sep="\n")
print("Array 2:", x2, sep="\n")
print("Array 3:", x3, sep="\n")
```
Next, transform these arrays into row vectors using matrix().
```
x1 = np.matrix(x1)
x2 = np.matrix(x2)
x3 = np.matrix(x3)
```
use np.concatenate() to combine the rows
```
X = np.concatenate((x1, x2, x3), axis = 0)
X
```
X.getI method gets inverse of matrix
```
X_inverse = X.getI()
X_inverse = np.round(X_inverse, 2)
X_inverse
```
# Regression function - Pulling necessary data
We now know the necessary operations for inverting matrices and minimizing squared residuals. We can import real data and begin to analyze how variables influence one another.
To start, we will use the Fraser economic freedom data.
```
import pandas as pd
import statsmodels.api as sm
import numpy as np
data = pd.read_csv('fraserDataWithRGDPPC.csv',
index_col = [0,1],
parse_dates = True)
data
years = np.array(sorted(list(set(data.index.get_level_values("Year")))))
years = pd.date_range(years[0], years[-2], freq = "AS")
countries = sorted(list(set(data.index.get_level_values("ISO_Code"))))
index_names = list(data.index.names)
multi_index = pd.MultiIndex.from_product([countries, years[:-1]], names = data.index.names)
data = data.reindex(multi_index)
data["RGDP Per Capita Lag"] = data.groupby("ISO_Code")["RGDP Per Capita"].shift()
data
data.dropna(axis = 0).loc['GBR']
```
# Running Regression Model:
```
y_vars = ['RGDP Per Capita']
x_vars = [
'Size of Government', 'Legal System & Property Rights', 'Sound Money',
'Freedom to trade internationally', 'Regulation'
]
reg_vars = y_vars + x_vars
reg_data = data[reg_vars].dropna()
reg_data.corr().round(2)
reg_data.describe().round(2)
y = reg_data[y_vars]
x = reg_data[x_vars]
x['Constant'] = 1
results = sm.OLS(y, x).fit()
results.summary()
predictor = results.predict()
reg_data[y_vars[0] + " Predictor"] = predictor
reg_data.loc["GBR", [y_vars[0], y_vars[0] + " Predictor"]].plot()
```
# OLS Statistics
We have calculated beta values for each independent variable, meaning that we estimated the average effect of a change in each independent variable upon the dependent variable. While this is useful, we have not yet measured the statistical significance of these estimations; neither have we determined the explanatory power of our particular regression.
Our regression has estimated predicted values for our dependent variable given the values of the independent variables for each observation. Together, these estimations for an array of predicted values that we will refer to as $y ̂ $. We will refer to individual predicted values as ($y_i$) ̂. We will also refer to the mean value of observations of our dependent variable as $y ̅ $ and individual observed values of our dependent variable as $y_i$. These values will be use to estimate the sum of squares due to regression ($SSR$), sum of squared errors ($SSE$), and the total sum of squares ($SST$). By comparing the estimated $y$ values, the observed $y$ values, and the mean of $y$, we will estimate the standard error for each coefficient and other values that estimate convey the significance of the estimation.
We define these values as follows:
$SSR = \sum_{i=0}^{n} (y ̂ _{i} - y ̅ )^2$
$SSE = \sum_{i=0}^{n} (y_{i} - y ̂ _{i})^2$
$SST = \sum_{i=0}^{n} (y_{i} - y ̅ _{i})^2$
It happens that the sum of the squared distances between the estimated values and mean of observed values and the squared distances between the observed and estimated values add up to the sum of the squared distances between the observed values and the mean of observed values. We indicate this as:
$SST = SSR + SSE$
The script below will estimate these statistics. It calls the sum_square_stats method from the which is passed in the calculate_regression_stats method.
```
y_name = y_vars[0]
y_hat = reg_data[y_name + " Predictor"]
y_mean = reg_data[y_name].mean()
y = reg_data[y_name]
y_hat, y_mean, y
reg_data["Residuals"] = y_hat.sub(y_mean)
reg_data["Squared Residuals"] = reg_data["Residuals"].pow(2)
reg_data["Squared Errors"] = (y.sub(y_hat)) ** 2
reg_data["Squared Totals"] = (y.sub(y_mean)) ** 2
SSR = reg_data["Squared Residuals"].sum()
SSE = reg_data["Squared Errors"].sum()
SST = reg_data["Squared Totals"].sum()
SSR, SSE, SST
n = results.nobs
k = len(results.params)
estimator_variance = SSE / (n-k)
n, k, estimator_variance
cov_matrix = results.cov_params()
cov_matrix
```
## Calculate t-stats
```
parameters = {}
for x_var in cov_matrix.keys():
parameters[x_var] = {}
parameters[x_var]["Beta"] = results.params[x_var]
parameters[x_var]["Standard Error"] = cov_matrix.loc[x_var, x_var]**(1 / 2)
parameters[x_var]["t_stats"] = parameters[x_var]["Beta"] / parameters[
x_var]["Standard Error"]
pd.DataFrame(parameters).T
r2 = SSR / SST
r2
results.summary()
```
# Plot Residuals
```
import matplotlib.pyplot as plt
plt.rcParams.update({"font.size": 26})
fig, ax = plt.subplots(figsize=(12, 8))
reg_data[["Residuals"]].plot.hist(bins=100, ax=ax)
plt.xticks(rotation=60)
```
slightly skewed left. Need to log the data in order to normally distrbute it
# Regression using rates
```
reg_data = data
reg_data["RGDP Per Capita"] = data.groupby("ISO_Code")["RGDP Per Capita"].pct_change()
reg_data["RGDP Per Capita Lag"] = reg_data["RGDP Per Capita"].shift()
reg_data = reg_data.replace([np.inf, -np.inf], np.nan).dropna(axis = 0, how = "any")
reg_data.loc["USA"]
reg_data.corr().round(2)
y_var = ["RGDP Per Capita"]
x_vars = ["Size of Government",
"Legal System & Property Rights",
"Sound Money",
"Freedom to trade internationally",
"Regulation",
"RGDP Per Capita Lag"]
y = reg_data[y_var]
X = reg_data[x_vars]
x["Constant"] = 1
results = sm.OLS(y, X).fit()
reg_data["Predictor"] = results.predict()
results.summary()
reg_data["Residuals"] = results.resid
fig, ax = plt.subplots(figsize = (12,8))
reg_data[["Residuals"]].plot.hist(bins = 100, ax = ax)
betaEstimates = results.params
tStats = results.tvalues
pValues = results.pvalues
stdErrors = results.bse
resultsDict = {"Beta Estimates" : betaEstimates,
"t-stats":tStats,
"p-values":pValues,
"Standard Errors":stdErrors}
resultsDF = pd.DataFrame(resultsDict)
resultsDF.round(3)
fig, ax = plt.subplots(figsize = (14,10))
reg_data.plot.scatter(x = y_var[0],
y = "Predictor",
s = 30, ax = ax)
plt.xticks(rotation=90)
plt.show()
plt.close()
fig, ax = plt.subplots(figsize = (14,10))
reg_data.plot.scatter(x = y_var[0],
y = "Residuals",
s = 30, ax = ax)
ax.axhline(0, ls = "--", color = "k")
plt.xticks(rotation=90)
plt.show()
plt.close()
```
| github_jupyter |
# Datafaucet
Datafaucet is a productivity framework for ETL, ML application. Simplifying some of the common activities which are typical in Data pipeline such as project scaffolding, data ingesting, start schema generation, forecasting etc.
```
import datafaucet as dfc
```
## Loading and Saving Data
```
dfc.project.load()
query = """
SELECT
p.payment_date,
p.amount,
p.rental_id,
p.staff_id,
c.*
FROM payment p
INNER JOIN customer c
ON p.customer_id = c.customer_id;
"""
df = dfc.load(query, 'pagila')
```
#### Select cols
```
df.cols.find('id').columns
df.cols.find(by_type='string').columns
df.cols.find(by_func=lambda x: x.startswith('st')).columns
df.cols.find('^st').columns
```
#### Collect data, oriented by rows or cols
```
df.cols.find(by_type='numeric').rows.collect(3)
df.cols.find(by_type='string').collect(3)
df.cols.find('name', 'date').data.collect(3)
```
#### Get just one row or column
```
df.cols.find('active', 'amount', 'name').one()
df.cols.find('active', 'amount', 'name').rows.one()
```
#### Grid view
```
df.cols.find('amount', 'id', 'name').data.grid(5)
```
#### Data Exploration
```
df.cols.find('amount', 'id', 'name').data.facets()
```
#### Rename columns
```
df.cols.find(by_type='timestamp').rename('new_', '***').columns
# to do
# df.cols.rename(transform=['unidecode', 'alnum', 'alpha', 'num', 'lower', 'trim', 'squeeze', 'slice', tr("abc", "_", mode='')'])
# df.cols.rename(transform=['unidecode', 'alnum', 'lower', 'trim("_")', 'squeeze("_")'])
# as a dictionary
mapping = {
'staff_id': 'foo',
'first_name': 'bar',
'email': 'qux',
'active':'active'
}
# or as a list of 2-tuples
mapping = [
('staff_id','foo'),
('first_name','bar'),
'active'
]
dict(zip(df.columns, df.cols.rename('new_', '***', mapping).columns))
```
#### Drop multiple columns
```
df.cols.find('id').drop().rows.collect(3)
```
#### Apply to multiple columns
```
from pyspark.sql import functions as F
(df
.cols.find(by_type='string').lower()
.cols.get('email').split('@')
.cols.get('email').expand(2)
.cols.find('name', 'email')
.rows.collect(3)
)
```
### Aggregations
```
from datafaucet.spark import aggregations as A
df.cols.find('amount', '^st.*id', 'first_name').agg(A.all).cols.collect(10)
```
##### group by a set of columns
```
df.cols.find('amount').groupby('staff_id', 'store_id').agg(A.all).cols.collect(4)
```
#### Aggregate specific metrics
```
# by function
df.cols.get('amount', 'active').groupby('customer_id').agg({'count':F.count, 'sum': F.sum}).rows.collect(10)
# or by alias
df.cols.get('amount', 'active').groupby('customer_id').agg('count','sum').rows.collect(10)
# or a mix of the two
df.cols.get('amount', 'active').groupby('customer_id').agg('count',{'sum': F.sum}).rows.collect(10)
```
#### Featurize specific metrics in a single row
```
(df
.cols.get('amount', 'active')
.groupby('customer_id', 'store_id')
.featurize({'count':A.count, 'sum':A.sum, 'avg':A.avg})
.rows.collect(10)
)
# todo:
# different features per different column
```
#### Plot dataset statistics
```
df.data.summary()
from bokeh.io import output_notebook
output_notebook()
from bokeh.plotting import figure, show, output_file
p = figure(plot_width=400, plot_height=400)
p.hbar(y=[1, 2, 3], height=0.5, left=0,
right=[1.2, 2.5, 3.7], color="navy")
show(p)
import seaborn as sns
import matplotlib.pyplot as plt
sns.set(style="whitegrid")
# Initialize the matplotlib figure
f, ax = plt.subplots(figsize=(6, 6))
# Load the example car crash dataset
crashes = sns.load_dataset("car_crashes").sort_values("total", ascending=False)[:10]
# Plot the total crashes
sns.set_color_codes("pastel")
sns.barplot(x="total", y="abbrev", data=crashes,
label="Total", color="b")
# Plot the crashes where alcohol was involved
sns.set_color_codes("muted")
sns.barplot(x="alcohol", y="abbrev", data=crashes,
label="Alcohol-involved", color="b")
# Add a legend and informative axis label
ax.legend(ncol=2, loc="lower right", frameon=True)
ax.set(xlim=(0, 24), ylabel="",
xlabel="Automobile collisions per billion miles")
sns.despine(left=True, bottom=True)
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
sns.set(style="white", palette="muted", color_codes=True)
# Generate a random univariate dataset
rs = np.random.RandomState(10)
d = rs.normal(size=100)
# Plot a simple histogram with binsize determined automatically
sns.distplot(d, hist=True, kde=True, rug=True, color="b");
import seaborn as sns
sns.set(style="ticks")
df = sns.load_dataset("iris")
sns.pairplot(df, hue="species")
from IPython.display import HTML
HTML('''
<!-- Bootstrap CSS -->
<link rel="stylesheet" href="https://stackpath.bootstrapcdn.com/bootstrap/4.1.3/css/bootstrap.min.css" crossorigin="anonymous">
<div class="container-fluid">
<div class="jumbotron">
<h1 class="display-4">Hello, world!</h1>
<p class="lead">This is a simple hero unit, a simple jumbotron-style component for calling extra attention to featured content or information.</p>
<hr class="my-4">
<p>It uses utility classes for typography and spacing to space content out within the larger container.</p>
<a class="btn btn-primary btn-lg" href="#" role="button">Learn more</a>
</div>
<button type="button" class="btn btn-secondary" data-toggle="tooltip" data-placement="top" title="Tooltip on top">
Tooltip on top
</button>
<button type="button" class="btn btn-secondary" data-toggle="tooltip" data-placement="right" title="Tooltip on right">
Tooltip on right
</button>
<button type="button" class="btn btn-secondary" data-toggle="tooltip" data-placement="bottom" title="Tooltip on bottom">
Tooltip on bottom
</button>
<button type="button" class="btn btn-secondary" data-toggle="tooltip" data-placement="left" title="Tooltip on left">
Tooltip on left
</button>
<table class="table">
<thead>
<tr>
<th scope="col">#</th>
<th scope="col">First</th>
<th scope="col">Last</th>
<th scope="col">Handle</th>
</tr>
</thead>
<tbody>
<tr>
<th scope="row">1</th>
<td>Mark</td>
<td>Otto</td>
<td>@mdo</td>
</tr>
<tr>
<th scope="row">2</th>
<td>Jacob</td>
<td>Thornton</td>
<td>@fat</td>
</tr>
<tr>
<th scope="row">3</th>
<td>Larry</td>
<td>the Bird</td>
<td>@twitter</td>
</tr>
</tbody>
</table>
<span class="badge badge-primary">Primary</span>
<span class="badge badge-secondary">Secondary</span>
<span class="badge badge-success">Success</span>
<span class="badge badge-danger">Danger</span>
<span class="badge badge-warning">Warning</span>
<span class="badge badge-info">Info</span>
<span class="badge badge-light">Light</span>
<span class="badge badge-dark">Dark</span>
<table class="table table-sm" style="text-align:left">
<thead>
<tr>
<th scope="col">#</th>
<th scope="col">First</th>
<th scope="col">Last</th>
<th scope="col">Handle</th>
<th scope="col">bar</th>
</tr>
</thead>
<tbody>
<tr>
<th scope="row">1</th>
<td>Mark</td>
<td>Otto</td>
<td>@mdo</td>
<td class="text-left"><span class="badge badge-primary" style="width: 75%">Primary</span></td>
</tr>
<tr>
<th scope="row">2</th>
<td>Jacob</td>
<td>Thornton</td>
<td>@fat</td>
<td class="text-left"><span class="badge badge-secondary" style="width: 25%">Primary</span></td>
</tr>
<tr>
<th scope="row">3</th>
<td colspan="2">Larry the Bird</td>
<td>@twitter</td>
<td class="text-left"><span class="badge badge-warning" style="width: 55%">Primary</span></td>
</div>
</tr>
</tbody>
</table>
</div>''')
tbl = '''
<table class="table table-sm">
<thead>
<tr>
<th scope="col">#</th>
<th scope="col">First</th>
<th scope="col">Last</th>
<th scope="col">Handle</th>
<th scope="col">bar</th>
</tr>
</thead>
<tbody>
<tr>
<th scope="row">1</th>
<td>Mark</td>
<td>Otto</td>
<td>@mdo</td>
<td class="text-left"><span class="badge badge-primary" style="width: 75%">75%</span></td>
</tr>
<tr>
<th scope="row">2</th>
<td>Jacob</td>
<td>Thornton</td>
<td>@fat</td>
<td class="text-left"><span class="badge badge-secondary" style="width: 25%" title="Tooltip on top">25%</span></td>
</tr>
<tr>
<th scope="row">3</th>
<td colspan="2">Larry the Bird</td>
<td>@twitter</td>
<td class="text-left"><span class="badge badge-warning" style="width: 0%">0%</span></td>
</tr>
</tbody>
</table>
'''
drp = '''
<div class="dropdown">
<button class="btn btn-secondary dropdown-toggle" type="button" id="dropdownMenuButton" data-toggle="dropdown" aria-haspopup="true" aria-expanded="false">
Dropdown button
</button>
<div class="dropdown-menu" aria-labelledby="dropdownMenuButton">
<a class="dropdown-item" href="#">Action</a>
<a class="dropdown-item" href="#">Another action</a>
<a class="dropdown-item" href="#">Something else here</a>
</div>
</div>'''
tabs = f'''
<nav>
<div class="nav nav-tabs" id="nav-tab" role="tablist">
<a class="nav-item nav-link active" id="nav-home-tab" data-toggle="tab" href="#nav-home" role="tab" aria-controls="nav-home" aria-selected="true">Home</a>
<a class="nav-item nav-link" id="nav-profile-tab" data-toggle="tab" href="#nav-profile" role="tab" aria-controls="nav-profile" aria-selected="false">Profile</a>
<a class="nav-item nav-link" id="nav-contact-tab" data-toggle="tab" href="#nav-contact" role="tab" aria-controls="nav-contact" aria-selected="false">Contact</a>
</div>
</nav>
<div class="tab-content" id="nav-tabContent">
<div class="tab-pane fade show active" id="nav-home" role="tabpanel" aria-labelledby="nav-home-tab">..jjj.</div>
<div class="tab-pane fade" id="nav-profile" role="tabpanel" aria-labelledby="nav-profile-tab">..kkk.</div>
<div class="tab-pane fade" id="nav-contact" role="tabpanel" aria-labelledby="nav-contact-tab">{tbl}</div>
</div>
'''
from IPython.display import HTML
HTML(f'''
<!-- Bootstrap CSS -->
<link rel="stylesheet" href="https://stackpath.bootstrapcdn.com/bootstrap/4.1.3/css/bootstrap.min.css" crossorigin="anonymous">
<div class="container-fluid">
<div class="row">
<div class="col">
{drp}
</div>
<div class="col">
{tabs}
</div>
<div class="col">
{tbl}
</div>
</div>
</div>
<script src="https://stackpath.bootstrapcdn.com/bootstrap/4.1.3/js/bootstrap.bundle.min.js" crossorigin="anonymous" >
''')
from IPython.display import HTML
HTML(f'''
<!-- Bootstrap CSS -->
<link rel="stylesheet" href="https://stackpath.bootstrapcdn.com/bootstrap/4.1.3/css/bootstrap.min.css" crossorigin="anonymous">
<script src="https://stackpath.bootstrapcdn.com/bootstrap/4.1.3/js/bootstrap.bundle.min.js" crossorigin="anonymous" >
''')
d =df.cols.find('id', 'name').sample(10)
d.columns
tbl_head = '''
<thead>
<tr>
'''
tbl_head += '\n'.join([' <th scope="col">'+str(x)+'</th>' for x in d.columns])
tbl_head +='''
</tr>
</thead>
'''
print(tbl_head)
tbl_body = '''
<tbody>
<tr>
<th scope="row">1</th>
<td>Mark</td>
<td>Otto</td>
<td>@mdo</td>
<td class="text-left"><span class="badge badge-primary" style="width: 75%">75%</span></td>
</tr>
<tr>
<th scope="row">2</th>
<td>Jacob</td>
<td>Thornton</td>
<td>@fat</td>
<td class="text-left"><span class="badge badge-secondary" style="width: 25%" title="Tooltip on top">25%</span></td>
</tr>
<tr>
<th scope="row">3</th>
<td colspan="2">Larry the Bird</td>
<td>@twitter</td>
<td class="text-left"><span class="badge badge-warning" style="width: 0%">0%</span></td>
</tr>
</tbody>
</table>
'''
HTML(f'''
<!-- Bootstrap CSS -->
<div class="container-fluid">
<div class="row">
<div class="col">
<table class="table table-sm">
{tbl_head}
{tbl_body}
</table>
</div>
</div>
</div>
''')
# .rows.sample()
# .cols.select('name', 'id', 'amount')\
# .cols.apply(F.lower, 'name')\
# .cols.apply(F.floor, 'amount', output_prefix='_')\
# .cols.drop('^amount$')\
# .cols.rename()
# .cols.unicode()
.grid()
df = df.cols.select('name')
df = df.rows.overwrite([('Nhập mật', 'khẩu')])
df.columns
# .rows.overwrite(['Nhập mật', 'khẩu'])\
# .cols.apply(F.lower)\
# .grid()
# #withColumn('pippo', F.lower(F.col('first_name'))).grid()
import pandas as pd
df = pd.DataFrame({'lab':['A', 'B', 'C'], 'val':[10, 30, 20]})
df.plot.bar(x='lab', y='val', rot=0);
```
| github_jupyter |
## Deciding on a Model Using Manual Analysis with Gradio
This notebook documents some of the steps taken to choose the final model for deployment.
For this project, we played around with four different models to see which performed best for our dataset. Our initial literature search showcased four different models that are popular for transfer learning including:
1. Densenet
2. Resnet
3. Vgg16
4. Inception
After conducting extensive runs to choose the [best image transformations](https://github.com/UBC-MDS/capstone-gdrl-lipo/blob/master/notebooks/manual-albumentation.ipynb) and doing hyperparameter tuning on the individual [models](https://github.com/UBC-MDS/capstone-gdrl-lipo/tree/master/notebooks), we used these optimized models to do a manual analysis of images to compare the models. We build a [local internal decision making tool app using gradio](https://github.com/UBC-MDS/capstone-gdrl-lipo/tree/master/notebooks/gradio_demo.ipynb) to analyze specific test cases.
## Reviewing Specific Images
Below are some screenshots from the gradio app of some negative and positive images that the model has never seen. Six negative images and five positives images were chosen for a manual review in hopes to pick out ways to see how the model would do on examples that are visually hard for the human eye to identify and label correctly. All models were able to catch negative examples relatively well. Densenet stood out was able to capture 4 out of the 6 images well compared to the rest of the models with very high confidence.
### Negative Image Example
We chose a difficult negative image example that features a circular ball that to the eye appears to be lipohypertrophy but it is not. We can see that although all models predict negative, Densenet is the most confident in its prediction.

## Positive Image Example
Identifying positives was hard for all models and the below figure shows an example where all model struggled. It makes sense that all the models are struggling as we don't have a very large dataset (~300 total images with a 62:38 split for negative:positive) and it's hard to tell visually where the lipohypertrophy is present or not. However, we noticed that even when Densenet is wrong, it is less confident in its prediction. This is ideal as our capstone partner has identified that the model should be less confident in its prediction when its wrong.

## Conclusion and Next Steps
From this manual visualization excercise, we were able to narrow down our model choice to Densenet. According to the recall and accuracy, this model has the highest score, so even when it is wrong, it is not as confident in its prediction. Lastly, due to resource limitation on the deployment of this application, DenseNet is also the smallest app. So, the next steps were to optimize the Densenet model to further improve the scores. Two steps taken were:
1. Increase the pos_weight argument of the optimizer so that there is a greater loss on positive examples. See the exploration [here](https://github.com/UBC-MDS/capstone-gdrl-lipo/blob/master/notebooks/pos-weight-exploration.ipynb).
2. Play around with the dropout rate in the model architechture. See the exploration [here](https://github.com/UBC-MDS/capstone-gdrl-lipo/blob/master/notebooks/densemodels-ax-dropout-layers.ipynb).
| github_jupyter |
<a href="https://colab.research.google.com/github/taniokah/where-is-santa/blob/master/Indexer_for_Santa.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Indexer for Santa
Script score queryedit
https://www.elastic.co/guide/en/elasticsearch/reference/current/query-dsl-script-score-query.html#vector-functions
ELASTICSEARCHで分散表現を使った類似文書検索
https://yag-ays.github.io/project/elasticsearch-similarity-search/
Image Search for ICDAR WML 2019
https://github.com/taniokah/icdar-wml-2019/blob/master/Image%20Search%20for%20ICDAR%20WML%202019.ipynb
```
# Crawling Santa images.
!pip install icrawler
!rm -rf google_images/*
!rm -rf bing_images/*
!rm -rf baidu_images/*
from icrawler.builtin import BaiduImageCrawler, BingImageCrawler, GoogleImageCrawler
crawler = GoogleImageCrawler(storage={"root_dir": "google_images"}, downloader_threads=4)
crawler.crawl(keyword="Santa", offset=0, max_num=1000)
#bing_crawler = BingImageCrawler(storage={'root_dir': 'bing_images'}, downloader_threads=4)
#bing_crawler.crawl(keyword='Santa', filters=None, offset=0, max_num=1000)
#baidu_crawler = BaiduImageCrawler(storage={'root_dir': 'baidu_images'})
#baidu_crawler.crawl(keyword='Santa', offset=0, max_num=1000)
!wget https://artifacts.elastic.co/downloads/elasticsearch/elasticsearch-7.5.1-linux-x86_64.tar.gz -q
!tar -xzf elasticsearch-7.5.1-linux-x86_64.tar.gz
!chown -R daemon:daemon elasticsearch-7.5.1/
#!elasticsearch-7.5.1/bin/elasticsearch
import os
from subprocess import Popen, PIPE, STDOUT
es_server = Popen(['elasticsearch-7.5.1/bin/elasticsearch'],
stdout=PIPE, stderr=STDOUT,
preexec_fn=lambda: os.setuid(1) # as daemon
)
!ps aux | grep elastic
!sleep 30
!curl -X GET "localhost:9200/"
!pip install elasticsearch
from datetime import datetime
from elasticsearch import Elasticsearch
es = Elasticsearch(timeout=60)
doc = {
'author': 'Santa Claus',
'text': 'Where is Santa Claus?',
'timestamp': datetime.now(),
}
res = es.index(index="test-index", doc_type='tweet', id=1, body=doc)
print(res['result'])
res = es.get(index="test-index", doc_type='tweet', id=1)
print(res['_source'])
es.indices.refresh(index="test-index")
res = es.search(index="test-index", body={"query": {"match_all": {}}})
print("Got %d Hits:" % res['hits']['total']['value'])
for hit in res['hits']['hits']:
print("%(timestamp)s %(author)s: %(text)s" % hit["_source"])
# Load libraries
from keras.applications.vgg16 import VGG16, preprocess_input, decode_predictions
from keras.preprocessing import image
from PIL import Image
import matplotlib.pyplot as plt
import numpy as np
import sys
model = VGG16(weights='imagenet')
def predict(filename, featuresize, scale=1.0):
img = image.load_img(filename, target_size=(224, 224))
return predictimg(img, featuresize, scale=1.0)
def predictpart(filename, featuresize, scale=1.0, size=1):
im = Image.open(filename)
width, height = im.size
im = im.resize((width * size, height * size))
im_list = np.asarray(im)
# partition
out_img = []
if size > 1:
v_split = size
h_split = size
[out_img.extend(np.hsplit(h_img, h_split)) for h_img in np.vsplit(im_list, v_split)]
else:
out_img.append(im_list)
reslist = []
for offset in range(size * size):
img = Image.fromarray(out_img[offset])
reslist.append(predictimg(img, featuresize, scale))
return reslist
def predictimg(img, featuresize, scale=1.0):
width, height = img.size
img = img.resize((int(width * scale), int(height * scale)))
img = img.resize((224, 224))
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
preds = model.predict(preprocess_input(x))
results = decode_predictions(preds, top=featuresize)[0]
return results
def showimg(filename, title, i, scale=1.0, col=2, row=5):
im = Image.open(filename)
width, height = im.size
im = im.resize((int(width * scale), int(height * scale)))
im = im.resize((width, height))
im_list = np.asarray(im)
plt.subplot(col, row, i)
plt.title(title)
plt.axis("off")
plt.imshow(im_list)
def showpartimg(filename, title, i, size, scale=1.0, col=2, row=5):
im = Image.open(filename)
width, height = im.size
im = im.resize((int(width * scale), int(height * scale)))
#im = im.resize((width, height))
im = im.resize((width * size, height * size))
im_list = np.asarray(im)
# partition
out_img = []
if size > 1:
v_split = size
h_split = size
[out_img.extend(np.hsplit(h_img, h_split)) for h_img in np.vsplit(im_list, v_split)]
else:
out_img.append(im_list)
# draw image
for offset in range(size * size):
im_list = out_img[offset]
pos = i + offset
print(str(col) + ' ' + str(row) + ' ' + str(pos))
plt.subplot(col, row, pos)
plt.title(title)
plt.axis("off")
plt.imshow(im_list)
out_img[offset] = Image.fromarray(im_list)
return out_img
# Predict an image
scale = 1.0
filename = "google_images/000046.jpg"
plt.figure(figsize=(20, 10))
#showimg(filename, "query", i+1, scale)
imgs = showpartimg(filename, "query", 1, 1, scale)
plt.show()
for img in imgs:
reslist = predictpart(filename, 10, scale)
for results in reslist:
for result in results:
print(result)
print()
def createindex(indexname):
if es.indices.exists(index=indexname):
es.indices.delete(index=indexname)
es.indices.create(index=indexname, body={
"settings": {
"index.mapping.total_fields.limit": 10000,
}
})
mapping = {
"image": {
"properties": {
"f": {
"type": "text"
},
's': {
"type": "sparse_vector"
}
}
}
}
es.indices.put_mapping(index=indexname, doc_type='image', body=mapping, include_type_name=True)
wnidmap = {}
def loadimages(directory):
imagefiles = []
for file in os.listdir(directory):
if file.rfind('.jpg') < 0:
continue
filepath = os.path.join(directory, file)
imagefiles.append(filepath)
return imagefiles
def indexfiles(indexname, directory, featuresize=10, docsize=1000):
imagefiles = loadimages(directory)
for i in range(len(imagefiles)):
if i >= docsize:
return
filename = imagefiles[i]
indexfile(indexname, filename, i, featuresize)
sys.stdout.write("\r%d" % (i + 1))
sys.stdout.flush()
es.indices.refresh(index=indexname)
def indexfile(indexname, filename, i, featuresize):
global wnidmap
rounddown = 16
doc = {'f': filename, 's':{}}
results = predict(filename, featuresize)
#print(len(results))
synset = doc['s']
for result in results:
score = float(str(result[2]))
wnid = result[0]
id = 0
if wnid in wnidmap.keys():
id = wnidmap[wnid]
else:
id = len(wnidmap)
wnidmap[wnid] = id
synset[str(id)] = score
#print(doc)
#count = es.count(index=indexname, doc_type='image')['count']
count = i
res = es.index(index=indexname, doc_type='image', id=count, body=doc)
createindex("santa-search")
directory = "google_images/"
indexfiles("santa-search", directory, 100, 1000)
#directory = "bing_images/"
#indexfiles("santa-search", directory, 100, 1000)
#directory = "baidu_images/"
#indexfiles("santa-search", directory, 100, 1000)
res = es.search(index="santa-search", request_timeout=60, body={"query": {"match_all": {}}})
print("Got " + str(res['hits']['total']) + " Hits:" )
for hit in res['hits']['hits']:
print(hit["_source"])
#print("%(timestamp)s %(author)s: %(text)s" % hit["_source"])
def searchimg(indexname, filename, num=10, topk=10, scoretype='dot', scale=1.0, partition=1):
plt.figure(figsize=(20, 10))
imgs = showpartimg(filename, "query", 1, partition, scale)
plt.show()
reslist = []
for img in imgs:
results = predictimg(img, num, scale)
for result in results:
print(result)
print()
res = search(indexname, results, num, topk, scoretype)
reslist.append(res)
return reslist
def search(indexname, synsets, num, topk, scoretype='dot', disp=True):
if scoretype == 'vcos':
inline = {}
for synset in synsets:
score = synset[2]
if score <= 0.0:
continue
wnid = synset[0]
if wnid not in wnidmap.keys():
continue
id = wnidmap[wnid]
inline[str(id)] = float(score)
if inline == {}:
print("Got " + str(0) + " Hits:")
return
#print('wnidmap = ' + str(wnidmap))
#print('inline = ' + str(inline))
b = {
"size": topk,
"query": {
"script_score": {
"query": {"match_all": {}},
"script": {
"source": "cosineSimilaritySparse(params.s, doc['s']) + 0.01",
"params": {
's': {}
}
}
}
}}
b['query']['script_score']['script']['params']['s'] = inline
res = es.search(index=indexname, body=b)
#print(str(b))
if disp==True:
print("Got " + str(res['hits']['total']['value']) + " Hits:")
topres = res['hits']['hits'][0:topk]
for hit in topres:
print(str(hit["_id"]) + " " + str(hit["_source"]["f"]) + " " + str(hit["_score"]))
plt.figure(figsize=(20, 10))
for i in range(len(topres)):
hit = topres[i]
row = 5
col = int(topk / 5)
if i >= 25:
break
showimg(hit["_source"]["f"], hit["_id"], i+1, col, row)
plt.show()
return res
filename = "google_images/000001.jpg"
_ = searchimg('santa-search', filename, 10, 10, 'vcos', 1.0, 1)
```
| github_jupyter |
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
from astropy.visualization import astropy_mpl_style
plt.style.use(astropy_mpl_style)
import astropy.units as u
from astropy.time import Time
from astropy.coordinates import SkyCoord, EarthLocation, AltAz, ICRS
```
The observing period is the whole year of -2000 B.C.E. ~ 0 B.C.
To represent the epoch before the common era, I use the Julian date.
I calculate the altitude and azimuth of Sun and Canopus among 4:00~8:00 in autumnal equinox and 16:00~20:00 in vernal equinox for every year.
```
def observable_duration(obs_time):
"""
"""
# Assume we have an observer in Tai Mountain.
taishan = EarthLocation(lat=36.2*u.deg, lon=117.1*u.deg, height=1500*u.m)
utcoffset = +8 * u.hour # Daylight Time
midnight = obs_time - utcoffset
# Position of the Canopus with the proper motion correction at the beginning of the year.
# This effect is very small.
dt_jyear = obs_time.jyear - 2000.0
ra = 95.98787790 * u.deg + 19.93 * u.mas * dt_jyear
dec = -52.69571787 * u.deg + 23.24 * u.mas * dt_jyear
hip30438 = SkyCoord(ra=ra, dec=dec, frame="icrs")
delta_midnight = np.arange(0, 24, 1./30) * u.hour # Interval of 2 minutes
obser_time = midnight + delta_midnight
local_frame = AltAz(obstime=obser_time,
location=taishan)
hip30438altazs = hip30438.transform_to(local_frame)
# position of Sun
from astropy.coordinates import get_sun
sunaltazs = get_sun(obser_time).transform_to(local_frame)
mask = (sunaltazs.alt < -0*u.deg) & (hip30438altazs.alt > 0)
observable_time = delta_midnight[mask]
# observable_time
if len(observable_time):
beg_time = observable_time.min().to('hr').value
end_time = observable_time.max().to('hr').value
else:
beg_time, end_time = 0, 0
return beg_time, end_time
year_arr = np.arange(0, 2000, 1)
# Number of days for every year
date_nb = np.ones_like(year_arr)
date_nb = np.where(year_arr % 4 == 0, 366, 365)
date_nb = np.where((year_arr % 100 == 0) & (
year_arr % 400 != 0), 365, date_nb)
total_date_nb = np.zeros_like(year_arr)
for i in range(year_arr.size):
total_date_nb[i] = np.sum(date_nb[:i+1])
# Autumnal equinox of every year
obs_time_aut = Time("0000-09-23 00:00:00") - total_date_nb * u.day
# Calculate the observable time of everyday
beg_time = np.zeros_like(obs_time_aut)
end_time = np.zeros_like(obs_time_aut)
obs_dur = np.zeros_like(obs_time_aut) # Observable duration
for i, obs_timei in enumerate(obs_time_aut):
# we calculate the 30 days before and after the equinox
delta_date = np.arange(-5, 5, 1) * u.day
obs_time0 = obs_timei + delta_date
beg_time_aut = np.zeros_like(obs_time0)
end_time_aut = np.zeros_like(obs_time0)
for j, obs_time0j in enumerate(obs_time0):
# Vernal equninox
beg_time_aut[j], end_time_aut[j] = observable_duration(obs_time0j)
obs_dur_aut = end_time_aut - beg_time_aut
obs_dur[i] = np.max(obs_dur_aut)
beg_time[i] = beg_time_aut[obs_dur_aut == obs_dur[i]][0]
end_time[i] = end_time_aut[obs_dur_aut == obs_dur[i]][0]
```
I assume that the Canopus can be observed by the local observer only when the observable duration in one day is longer than 10 minitues.
With such an assumption, I determine the observable period of the Canopus.
```
# Save data
np.save("multi_epoch-max-duration-Autumnal-output", [obs_time_aut.jyear, obs_dur])
# For Autumnal equinox
# mask = (obs_dur >= 1./6)
mask = (obs_dur >= 1.0/60)
observable_date = obs_time_aut[mask]
fig, ax = plt.subplots(figsize=(12, 8))
ax.plot(observable_date.jyear, obs_dur[mask],
"r.", ms=3, label="Autumnal")
# ax.fill_between(obs_time.jyear, 0, 24,
# (obs_dur1 >= 1./6) & (obs_dur2 >= 1./6), color="0.8", zorder=0)
ax.set_xlabel("Year", fontsize=15)
ax.set_xlim([-2000, 0])
ax.set_xticks(np.arange(-2000, 1, 100))
ax.set_ylim([0, 2.0])
ax.set_ylabel("Time (hour)", fontsize=15)
ax.set_title("Observable duration of Canopus among $-2000$ B.C.E and 0")
ax.legend(fontsize=15)
fig.tight_layout()
plt.savefig("multi_epoch-max-duration-Autumnal.eps", dpi=100)
plt.savefig("multi_epoch-max-duration-Autumnal.png", dpi=100)
```
| github_jupyter |
```
import sys
import os
import time
import torch
import torch.backends.cudnn as cudnn
import argparse
import socket
import pandas as pd
import csv
import numpy as np
import pickle
import re
from model_util import MyAlexNetCMC
from contrast_util import NCEAverage,AverageMeter,NCESoftmaxLoss
from torch.utils.data.sampler import SubsetRandomSampler
from data_pre import load_BCRdata,aamapping,datasetMap_nt,ntmapping
from data_util import Dataset
from random import sample,seed
from sklearn.metrics import roc_curve,auc
class opttest(object):
def __init__(self,path1,path2,path3,num,num2):
self.input_data=path1
self.atchley_factors=path2
self.resume=path3
self.encode_dim=num
self.pad_length=num2
opt=opttest('/home2/s421955/projects/scBCR/data/cleaned_BCRmltrain/IEDB.csv',
'/home2/s421955/projects/scBCR/data/Atchley_factors.csv',
'/home2/s421955/projects/scBCR/data/model_BCRmltrain',
40,
130)
test=load_BCRdata(opt)
test['strlen']=test['cdr3_nt'].str.len()
test=test[test['strlen']<=130]
test.index=range(0,test.shape[0])
import os
filedir=opt.input_data
if filedir.find('.csv')>(-1):
datasets=[filedir]
else:
datasets=[os.path.join(dp, f) for dp, dn, fn in os.walk(os.path.expanduser(filedir)) for f in fn]
print(datasets)
for index,file in enumerate(datasets):
if index % 10==0:
print('Reading file:')
print(index)
f=pd.read_csv(file,header=0)
test.shape
test['cdr3_nt'][0]
aa_dict=dict()
with open(opt.atchley_factors,'r') as aa:
aa_reader=csv.reader(aa)
next(aa_reader, None)
for rows in aa_reader:
aa_name=rows[0]
aa_factor=rows[1:len(rows)]
aa_dict[aa_name]=np.asarray(aa_factor,dtype='float')
cdr_test,vdj_test,cdr3_seq_test=datasetMap_nt(test,aa_dict,opt.encode_dim,opt.pad_length)
#cdr = open('/home2/s421955/projects/scBCR/data/model_BCRmltrain/cdr_test.pkl',"wb")
#pickle.dump(cdr_test,cdr)
#cdr.close()
#vdj = open('/home2/s421955/projects/scBCR/data/model_BCRmltrain/nt_test.pkl',"wb")
#pickle.dump(vdj_testtest,vdj)
#vdj.close()
#After data prep
# cdr = open('/home2/s421955/projects/scBCR/data/model_BCRmltrain/cdr_10ktest.pkl', 'rb')
# cdr_test = pickle.load(cdr)
# cdr.close()
#cdr_test={ind:cdr_test[ind][0:40,:] for ind in list(cdr_test.keys())}
# vdj = open('/home2/s421955/projects/scBCR/data/model_BCRmltrain/nt_10ktest.pkl', 'rb')
# vdj_test = pickle.load(vdj)
# vdj.close()
#Load data
batch_size = 64
random_seed= 123
test_indices = list(set(vdj_test.keys()))
cdr_shape = cdr_test[list(cdr_test.keys())[0]].shape[0]
test_set = Dataset(test_indices,cdr_test,vdj_test,cdr3_seq_test)
test_loader = torch.utils.data.DataLoader(test_set, batch_size=batch_size,
shuffle=False, sampler=None,batch_sampler=None,num_workers=1)
#Load model
device = "cuda: 0"
epoch=59
feat_dim=20
in_feature=130
n_out_features=feat_dim
nce_k = 1
nce_t = 0.2
nce_m = 0.9
n_vdj = vdj_test[list(vdj_test.keys())[0]].size()[0]
n_data = len(test_indices)
lr = 0.001
momentum = 0.9
weight_decay = 0.0001
gradient_clip = 5
state=torch.load(opt.resume+"/trained_model.pt")
test_model=MyAlexNetCMC(in_feature=in_feature,feat_dim=feat_dim,freeze=True).cuda()
contrast = NCEAverage(n_out_features, n_data, nce_k, nce_t, nce_m).cuda()
criterion_cdr = NCESoftmaxLoss().cuda()
criterion_vdj = NCESoftmaxLoss().cuda()
optimizer = torch.optim.SGD(test_model.parameters(),
lr=lr,
momentum=momentum,
weight_decay=weight_decay)
test_model.load_state_dict(state['model'])
optimizer.load_state_dict(state['optimizer'])
def predict(test_loader, model, contrast,criterion_cdr,criterion_vdj):
acc=dict()
roc_score=dict()
model.eval()
contrast.eval()
with torch.no_grad():
for idx, (data, index) in enumerate(test_loader):
index = index.to(device)
for _ in list(data.keys())[0:2]:
data[_] = data[_].float().to(device)
feat_cdr,feat_vdj,cdr3_seq = model(data)
if idx==0:
feature_array=pd.DataFrame(feat_cdr.cpu().numpy())
feature_array['index']=cdr3_seq
else:
feature_array_tmp=pd.DataFrame(feat_cdr.cpu().numpy())
feature_array_tmp['index']=cdr3_seq
feature_array=feature_array.append(feature_array_tmp)
out_cdr, out_vdj = contrast(feat_cdr, feat_vdj, index)
loss_cdr=criterion_cdr(out_cdr)
loss_vdj=criterion_vdj(out_vdj)
loss=loss_cdr+loss_vdj
print('Batch {0}: test loss {1:.3f}'.format(idx,loss))
out_cdr=out_cdr.squeeze()
out_vdj=out_vdj.squeeze()
acc_cdr=torch.argmax(out_cdr,dim=1)
acc_vdj=torch.argmax(out_vdj,dim=1)
acc_vdj=acc_vdj.squeeze()
if idx==0:
acc['cdr']=acc_cdr
acc['vdj']=acc_vdj
roc_score['cdr']=out_cdr.flatten()
roc_score['vdj']=out_vdj.flatten()
else:
acc['cdr']=torch.cat((acc['cdr'],acc_cdr),0)
acc['vdj']=torch.cat((acc['vdj'],acc_vdj),0)
roc_score['cdr']=torch.cat((roc_score['cdr'],out_cdr.flatten()),0)
roc_score['vdj']=torch.cat((roc_score['vdj'],out_vdj.flatten()),0)
return feature_array,acc,roc_score,loss
feature_array,acc,roc_score,test_loss=predict(test_loader,test_model,contrast,criterion_cdr,criterion_vdj)
acc['cdr']=acc['cdr'].cpu().numpy()
acc['vdj']=acc['vdj'].cpu().numpy()
print('cdr accuracy:\n')
print(len(np.where(acc['cdr']==0)[0])/len(acc['cdr']))
print('nt accuracy:\n')
print(len(np.where(acc['vdj']==0)[0])/len(acc['vdj']))
feature_array.to_csv('/home2/s421955/projects/scBCR/data/test_BCRmltrain/testoutput.csv',sep=',')
```
| github_jupyter |
# Build a Pipeline
> A tutorial on building pipelines to orchestrate your ML workflow
A Kubeflow pipeline is a portable and scalable definition of a machine learning
(ML) workflow. Each step in your ML workflow, such as preparing data or
training a model, is an instance of a pipeline component. This document
provides an overview of pipeline concepts and best practices, and instructions
describing how to build an ML pipeline.
## Before you begin
1. Run the following command to install the Kubeflow Pipelines SDK. If you run this command in a Jupyter
notebook, restart the kernel after installing the SDK.
```
!pip install kfp --upgrade
```
2. Import the `kfp` and `kfp.components` packages.
```
import kfp
import kfp.components as comp
```
## Understanding pipelines
A Kubeflow pipeline is a portable and scalable definition of an ML workflow,
based on containers. A pipeline is composed of a set of input parameters and a
list of the steps in this workflow. Each step in a pipeline is an instance of a
component, which is represented as an instance of
[`ContainerOp`][container-op].
You can use pipelines to:
* Orchestrate repeatable ML workflows.
* Accelerate experimentation by running a workflow with different sets of
hyperparameters.
### Understanding pipeline components
A pipeline component is a containerized application that performs one step in a
pipeline's workflow. Pipeline components are defined in
[component specifications][component-spec], which define the following:
* The component's interface, its inputs and outputs.
* The component's implementation, the container image and the command to
execute.
* The component's metadata, such as the name and description of the
component.
You can build components by [defining a component specification for a
containerized application][component-dev], or you can [use the Kubeflow
Pipelines SDK to generate a component specification for a Python
function][python-function-component]. You can also [reuse prebuilt components
in your pipeline][prebuilt-components].
### Understanding the pipeline graph
Each step in your pipeline's workflow is an instance of a component. When
you define your pipeline, you specify the source of each step's inputs. Step
inputs can be set from the pipeline's input arguments, constants, or step
inputs can depend on the outputs of other steps in this pipeline. Kubeflow
Pipelines uses these dependencies to define your pipeline's workflow as
a graph.
For example, consider a pipeline with the following steps: ingest data,
generate statistics, preprocess data, and train a model. The following
describes the data dependencies between each step.
* **Ingest data**: This step loads data from an external source which is
specified using a pipeline argument, and it outputs a dataset. Since
this step does not depend on the output of any other steps, this step
can run first.
* **Generate statistics**: This step uses the ingested dataset to generate
and output a set of statistics. Since this step depends on the dataset
produced by the ingest data step, it must run after the ingest data step.
* **Preprocess data**: This step preprocesses the ingested dataset and
transforms the data into a preprocessed dataset. Since this step depends
on the dataset produced by the ingest data step, it must run after the
ingest data step.
* **Train a model**: This step trains a model using the preprocessed dataset,
the generated statistics, and pipeline parameters, such as the learning
rate. Since this step depends on the preprocessed data and the generated
statistics, it must run after both the preprocess data and generate
statistics steps are complete.
Since the generate statistics and preprocess data steps both depend on the
ingested data, the generate statistics and preprocess data steps can run in
parallel. All other steps are executed once their data dependencies are
available.
## Designing your pipeline
When designing your pipeline, think about how to split your ML workflow into
pipeline components. The process of splitting an ML workflow into pipeline
components is similar to the process of splitting a monolithic script into
testable functions. The following rules can help you define the components
that you need to build your pipeline.
* Components should have a single responsibility. Having a single
responsibility makes it easier to test and reuse a component. For example,
if you have a component that loads data you can reuse that for similar
tasks that load data. If you have a component that loads and transforms
a dataset, the component can be less useful since you can use it only when
you need to load and transform that dataset.
* Reuse components when possible. Kubeflow Pipelines provides [components for
common pipeline tasks and for access to cloud
services][prebuilt-components].
* Consider what you need to know to debug your pipeline and research the
lineage of the models that your pipeline produces. Kubeflow Pipelines
stores the inputs and outputs of each pipeline step. By interrogating the
artifacts produced by a pipeline run, you can better understand the
variations in model quality between runs or track down bugs in your
workflow.
In general, you should design your components with composability in mind.
Pipelines are composed of component instances, also called steps. Steps can
define their inputs as depending on the output of another step. The
dependencies between steps define the pipeline workflow graph.
### Building pipeline components
Kubeflow pipeline components are containerized applications that perform a
step in your ML workflow. Here are the ways that you can define pipeline
components:
* If you have a containerized application that you want to use as a
pipeline component, create a component specification to define this
container image as a pipeline component.
This option provides the flexibility to include code written in any
language in your pipeline, so long as you can package the application
as a container image. Learn more about [building pipeline
components][component-dev].
* If your component code can be expressed as a Python function, [evaluate if
your component can be built as a Python function-based
component][python-function-component]. The Kubeflow Pipelines SDK makes it
easier to build lightweight Python function-based components by saving you
the effort of creating a component specification.
Whenever possible, [reuse prebuilt components][prebuilt-components] to save
yourself the effort of building custom components.
The example in this guide demonstrates how to build a pipeline that uses a
Python function-based component and reuses a prebuilt component.
### Understanding how data is passed between components
When Kubeflow Pipelines runs a component, a container image is started in a
Kubernetes Pod and your component’s inputs are passed in as command-line
arguments. When your component has finished, the component's outputs are
returned as files.
In your component's specification, you define the components inputs and outputs
and how the inputs and output paths are passed to your program as command-line
arguments. You can pass small inputs, such as short strings or numbers, to your
component by value. Large inputs, such as datasets, must be passed to your
component as file paths. Outputs are written to the paths that Kubeflow
Pipelines provides.
Python function-based components make it easier to build pipeline components
by building the component specification for you. Python function-based
components also handle the complexity of passing inputs into your component
and passing your function’s outputs back to your pipeline.
Learn more about how [Python function-based components handle inputs and
outputs][python-function-component-data-passing].
## Getting started building a pipeline
The following sections demonstrate how to get started building a Kubeflow
pipeline by walking through the process of converting a Python script into
a pipeline.
### Design your pipeline
The following steps walk through some of the design decisions you may face
when designing a pipeline.
1. Evaluate the process. In the following example, a Python function downloads
a zipped tar file (`.tar.gz`) that contains several CSV files, from a
public website. The function extracts the CSV files and then merges them
into a single file.
[container-op]: https://kubeflow-pipelines.readthedocs.io/en/latest/source/kfp.dsl.html#kfp.dsl.ContainerOp
[component-spec]: https://www.kubeflow.org/docs/components/pipelines/reference/component-spec/
[python-function-component]: https://www.kubeflow.org/docs/components/pipelines/sdk/python-function-components/
[component-dev]: https://www.kubeflow.org/docs/components/pipelines/sdk/component-development/
[python-function-component-data-passing]: https://www.kubeflow.org/docs/components/pipelines/sdk/python-function-components/#understanding-how-data-is-passed-between-components
[prebuilt-components]: https://www.kubeflow.org/docs/examples/shared-resources/
```
import glob
import pandas as pd
import tarfile
import urllib.request
def download_and_merge_csv(url: str, output_csv: str):
with urllib.request.urlopen(url) as res:
tarfile.open(fileobj=res, mode="r|gz").extractall('data')
df = pd.concat(
[pd.read_csv(csv_file, header=None)
for csv_file in glob.glob('data/*.csv')])
df.to_csv(output_csv, index=False, header=False)
```
2. Run the following Python command to test the function.
```
download_and_merge_csv(
url='https://storage.googleapis.com/ml-pipeline-playground/iris-csv-files.tar.gz',
output_csv='merged_data.csv')
```
3. Run the following to print the first few rows of the
merged CSV file.
```
!head merged_data.csv
```
4. Design your pipeline. For example, consider the following pipeline designs.
* Implement the pipeline using a single step. In this case, the pipeline
contains one component that works similarly to the example function.
This is a straightforward function, and implementing a single-step
pipeline is a reasonable approach in this case.
The down side of this approach is that the zipped tar file would not be
an artifact of your pipeline runs. Not having this artifact available
could make it harder to debug this component in production.
* Implement this as a two-step pipeline. The first step downloads a file
from a website. The second step extracts the CSV files from a zipped
tar file and merges them into a single file.
This approach has a few benefits:
* You can reuse the [Web Download component][web-download-component]
to implement the first step.
* Each step has a single responsibility, which makes the components
easier to reuse.
* The zipped tar file is an artifact of the first pipeline step.
This means that you can examine this artifact when debugging
pipelines that use this component.
This example implements a two-step pipeline.
### Build your pipeline components
1. Build your pipeline components. This example modifies the initial script to
extract the contents of a zipped tar file, merge the CSV files that were
contained in the zipped tar file, and return the merged CSV file.
This example builds a Python function-based component. You can also package
your component's code as a Docker container image and define the component
using a ComponentSpec.
In this case, the following modifications were required to the original
function.
* The file download logic was removed. The path to the zipped tar file
is passed as an argument to this function.
* The import statements were moved inside of the function. Python
function-based components require standalone Python functions. This
means that any required import statements must be defined within the
function, and any helper functions must be defined within the function.
Learn more about [building Python function-based
components][python-function-components].
* The function's arguments are decorated with the
[`kfp.components.InputPath`][input-path] and the
[`kfp.components.OutputPath`][output-path] annotations. These
annotations let Kubeflow Pipelines know to provide the path to the
zipped tar file and to create a path where your function stores the
merged CSV file.
The following example shows the updated `merge_csv` function.
[web-download-component]: https://github.com/kubeflow/pipelines/blob/master/components/web/Download/component.yaml
[python-function-components]: https://www.kubeflow.org/docs/components/pipelines/sdk/python-function-components/
[input-path]: https://kubeflow-pipelines.readthedocs.io/en/latest/source/kfp.components.html?highlight=inputpath#kfp.components.InputPath
[output-path]: https://kubeflow-pipelines.readthedocs.io/en/latest/source/kfp.components.html?highlight=outputpath#kfp.components.OutputPath
```
def merge_csv(file_path: comp.InputPath('Tarball'),
output_csv: comp.OutputPath('CSV')):
import glob
import pandas as pd
import tarfile
tarfile.open(name=file_path, mode="r|gz").extractall('data')
df = pd.concat(
[pd.read_csv(csv_file, header=None)
for csv_file in glob.glob('data/*.csv')])
df.to_csv(output_csv, index=False, header=False)
```
2. Use [`kfp.components.create_component_from_func`][create_component_from_func]
to return a factory function that you can use to create pipeline steps.
This example also specifies the base container image to run this function
in, the path to save the component specification to, and a list of PyPI
packages that need to be installed in the container at runtime.
[create_component_from_func]: (https://kubeflow-pipelines.readthedocs.io/en/latest/source/kfp.components.html#kfp.components.create_component_from_func
[container-op]: https://kubeflow-pipelines.readthedocs.io/en/stable/source/kfp.dsl.html#kfp.dsl.ContainerOp
```
create_step_merge_csv = kfp.components.create_component_from_func(
func=merge_csv,
output_component_file='component.yaml', # This is optional. It saves the component spec for future use.
base_image='python:3.7',
packages_to_install=['pandas==1.1.4'])
```
### Build your pipeline
1. Use [`kfp.components.load_component_from_url`][load_component_from_url]
to load the component specification YAML for any components that you are
reusing in this pipeline.
[load_component_from_url]: https://kubeflow-pipelines.readthedocs.io/en/latest/source/kfp.components.html?highlight=load_component_from_url#kfp.components.load_component_from_url
```
web_downloader_op = kfp.components.load_component_from_url(
'https://raw.githubusercontent.com/kubeflow/pipelines/master/components/contrib/web/Download/component.yaml')
```
2. Define your pipeline as a Python function.
Your pipeline function's arguments define your pipeline's parameters. Use
pipeline parameters to experiment with different hyperparameters, such as
the learning rate used to train a model, or pass run-level inputs, such as
the path to an input file, into a pipeline run.
Use the factory functions created by
`kfp.components.create_component_from_func` and
`kfp.components.load_component_from_url` to create your pipeline's tasks.
The inputs to the component factory functions can be pipeline parameters,
the outputs of other tasks, or a constant value. In this case, the
`web_downloader_task` task uses the `url` pipeline parameter, and the
`merge_csv_task` uses the `data` output of the `web_downloader_task`.
```
# Define a pipeline and create a task from a component:
def my_pipeline(url):
web_downloader_task = web_downloader_op(url=url)
merge_csv_task = create_step_merge_csv(file=web_downloader_task.outputs['data'])
# The outputs of the merge_csv_task can be referenced using the
# merge_csv_task.outputs dictionary: merge_csv_task.outputs['output_csv']
```
### Compile and run your pipeline
After defining the pipeline in Python as described in the preceding section, use one of the following options to compile the pipeline and submit it to the Kubeflow Pipelines service.
#### Option 1: Compile and then upload in UI
1. Run the following to compile your pipeline and save it as `pipeline.yaml`.
```
kfp.compiler.Compiler().compile(
pipeline_func=my_pipeline,
package_path='pipeline.yaml')
```
2. Upload and run your `pipeline.yaml` using the Kubeflow Pipelines user interface.
See the guide to [getting started with the UI][quickstart].
[quickstart]: https://www.kubeflow.org/docs/components/pipelines/overview/quickstart
#### Option 2: run the pipeline using Kubeflow Pipelines SDK client
1. Create an instance of the [`kfp.Client` class][kfp-client] following steps in [connecting to Kubeflow Pipelines using the SDK client][connect-api].
[kfp-client]: https://kubeflow-pipelines.readthedocs.io/en/latest/source/kfp.client.html#kfp.Client
[connect-api]: https://www.kubeflow.org/docs/components/pipelines/sdk/connect-api
```
client = kfp.Client() # change arguments accordingly
```
2. Run the pipeline using the `kfp.Client` instance:
```
client.create_run_from_pipeline_func(
my_pipeline,
arguments={
'url': 'https://storage.googleapis.com/ml-pipeline-playground/iris-csv-files.tar.gz'
})
```
## Next steps
* Learn about advanced pipeline features, such as [authoring recursive
components][recursion] and [using conditional execution in a
pipeline][conditional].
* Learn how to [manipulate Kubernetes resources in a
pipeline][k8s-resources] (Experimental).
[conditional]: https://github.com/kubeflow/pipelines/blob/master/samples/tutorials/DSL%20-%20Control%20structures/DSL%20-%20Control%20structures.py
[recursion]: https://www.kubeflow.org/docs/components/pipelines/sdk/dsl-recursion/
[k8s-resources]: https://www.kubeflow.org/docs/components/pipelines/sdk/manipulate-resources/
| github_jupyter |

https://www.kaggle.com/danofer/sarcasm
<div class="markdown-converter__text--rendered"><h3>Context</h3>
<p>This dataset contains 1.3 million Sarcastic comments from the Internet commentary website Reddit. The dataset was generated by scraping comments from Reddit (not by me :)) containing the <code>\s</code> ( sarcasm) tag. This tag is often used by Redditors to indicate that their comment is in jest and not meant to be taken seriously, and is generally a reliable indicator of sarcastic comment content.</p>
<h3>Content</h3>
<p>Data has balanced and imbalanced (i.e true distribution) versions. (True ratio is about 1:100). The
corpus has 1.3 million sarcastic statements, along with what they responded to as well as many non-sarcastic comments from the same source.</p>
<p>Labelled comments are in the <code>train-balanced-sarcasm.csv</code> file.</p>
<h3>Acknowledgements</h3>
<p>The data was gathered by: Mikhail Khodak and Nikunj Saunshi and Kiran Vodrahalli for their article "<a href="https://arxiv.org/abs/1704.05579" rel="nofollow">A Large Self-Annotated Corpus for Sarcasm</a>". The data is hosted <a href="http://nlp.cs.princeton.edu/SARC/0.0/" rel="nofollow">here</a>.</p>
<p>Citation:</p>
<pre><code>@unpublished{SARC,
authors={Mikhail Khodak and Nikunj Saunshi and Kiran Vodrahalli},
title={A Large Self-Annotated Corpus for Sarcasm},
url={https://arxiv.org/abs/1704.05579},
year=2017
}
</code></pre>
<p><a href="http://nlp.cs.princeton.edu/SARC/0.0/readme.txt" rel="nofollow">Annotation of files in the original dataset: readme.txt</a>.</p>
<h3>Inspiration</h3>
<ul>
<li>Predicting sarcasm and relevant NLP features (e.g. subjective determinant, racism, conditionals, sentiment heavy words, "Internet Slang" and specific phrases). </li>
<li>Sarcasm vs Sentiment</li>
<li>Unusual linguistic features such as caps, italics, or elongated words. e.g., "Yeahhh, I'm sure THAT is the right answer".</li>
<li>Topics that people tend to react to sarcastically</li>
</ul></div>
```
import os
# Install java
! apt-get update -qq
! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null
os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64"
os.environ["PATH"] = os.environ["JAVA_HOME"] + "/bin:" + os.environ["PATH"]
! java -version
# Install pyspark
! pip install --ignore-installed pyspark==2.4.4
# Install Spark NLP
! pip install --ignore-installed spark-nlp
import sys
import time
import sparknlp
from pyspark.sql import SparkSession
packages = [
'JohnSnowLabs:spark-nlp: 2.5.5'
]
spark = SparkSession \
.builder \
.appName("ML SQL session") \
.config('spark.jars.packages', ','.join(packages)) \
.config('spark.executor.instances','2') \
.config("spark.executor.memory", "2g") \
.config("spark.driver.memory","16g") \
.getOrCreate()
print("Spark NLP version: ", sparknlp.version())
print("Apache Spark version: ", spark.version)
! wget -N https://s3.amazonaws.com/auxdata.johnsnowlabs.com/public/resources/en/sarcasm/train-balanced-sarcasm.csv -P /tmp
from pyspark.sql import SQLContext
sql = SQLContext(spark)
trainBalancedSarcasmDF = spark.read.option("header", True).option("inferSchema", True).csv("/tmp/train-balanced-sarcasm.csv")
trainBalancedSarcasmDF.printSchema()
# Let's create a temp view (table) for our SQL queries
trainBalancedSarcasmDF.createOrReplaceTempView('data')
sql.sql('SELECT COUNT(*) FROM data').collect()
sql.sql('select * from data limit 20').show()
sql.sql('select label,count(*) as cnt from data group by label order by cnt desc').show()
sql.sql('select count(*) from data where comment is null').collect()
df = sql.sql('select label,concat(parent_comment,"\n",comment) as comment from data where comment is not null and parent_comment is not null limit 100000')
print(type(df))
df.printSchema()
df.show()
from sparknlp.annotator import *
from sparknlp.common import *
from sparknlp.base import *
from pyspark.ml import Pipeline
document_assembler = DocumentAssembler() \
.setInputCol("comment") \
.setOutputCol("document")
sentence_detector = SentenceDetector() \
.setInputCols(["document"]) \
.setOutputCol("sentence") \
.setUseAbbreviations(True)
tokenizer = Tokenizer() \
.setInputCols(["sentence"]) \
.setOutputCol("token")
stemmer = Stemmer() \
.setInputCols(["token"]) \
.setOutputCol("stem")
normalizer = Normalizer() \
.setInputCols(["stem"]) \
.setOutputCol("normalized")
finisher = Finisher() \
.setInputCols(["normalized"]) \
.setOutputCols(["ntokens"]) \
.setOutputAsArray(True) \
.setCleanAnnotations(True)
nlp_pipeline = Pipeline(stages=[document_assembler, sentence_detector, tokenizer, stemmer, normalizer, finisher])
nlp_model = nlp_pipeline.fit(df)
processed = nlp_model.transform(df).persist()
processed.count()
processed.show()
train, test = processed.randomSplit(weights=[0.7, 0.3], seed=123)
print(train.count())
print(test.count())
from pyspark.ml import feature as spark_ft
stopWords = spark_ft.StopWordsRemover.loadDefaultStopWords('english')
sw_remover = spark_ft.StopWordsRemover(inputCol='ntokens', outputCol='clean_tokens', stopWords=stopWords)
tf = spark_ft.CountVectorizer(vocabSize=500, inputCol='clean_tokens', outputCol='tf')
idf = spark_ft.IDF(minDocFreq=5, inputCol='tf', outputCol='idf')
feature_pipeline = Pipeline(stages=[sw_remover, tf, idf])
feature_model = feature_pipeline.fit(train)
train_featurized = feature_model.transform(train).persist()
train_featurized.count()
train_featurized.show()
train_featurized.groupBy("label").count().show()
train_featurized.printSchema()
from pyspark.ml import classification as spark_cls
rf = spark_cls. RandomForestClassifier(labelCol="label", featuresCol="idf", numTrees=100)
model = rf.fit(train_featurized)
test_featurized = feature_model.transform(test)
preds = model.transform(test_featurized)
preds.show()
pred_df = preds.select('comment', 'label', 'prediction').toPandas()
pred_df.head()
import pandas as pd
from sklearn import metrics as skmetrics
pd.DataFrame(
data=skmetrics.confusion_matrix(pred_df['label'], pred_df['prediction']),
columns=['pred ' + l for l in ['0','1']],
index=['true ' + l for l in ['0','1']]
)
print(skmetrics.classification_report(pred_df['label'], pred_df['prediction'],
target_names=['0','1']))
spark.stop()
```
| github_jupyter |
<a href="https://colab.research.google.com/github/RamSaw/NLP/blob/master/HW_03_LSTM.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import re
from collections import defaultdict
from tqdm import tnrange, tqdm_notebook
import random
from tqdm.auto import tqdm
import os
from sklearn.model_selection import train_test_split
import numpy as np
from matplotlib import pyplot as plt
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torch.utils.data.dataset import Dataset
from torch.nn.utils.rnn import pad_sequence
from nltk.stem.snowball import SnowballStemmer
def make_reproducible(seed, make_cuda_reproducible):
random.seed(seed)
os.environ['PYTHONHASHSEED'] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
if make_cuda_reproducible:
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
SEED = 2341
make_reproducible(SEED, False)
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
print(device)
def indices_from_sentence(words):
cur_id = 0
result = []
for word in words:
result.append((cur_id, len(word)))
cur_id += len(word)
return result
print(indices_from_sentence(['word1', 'a', ',', 'word2']))
print(indices_from_sentence(re.split('(\W)', 'Барак Обама принимает в Белом доме своего французского коллегу Николя Саркози.')))
test_words = re.split('(\W)', 'Скотланд-Ярд{ORG} вызвал на допрос Руперта{PERSON} Мердока{PERSON}')
test_words_clean = re.split('(\W)', 'Скотланд-Ярд вызвал на допрос Руперта Мердока')
print(test_words)
print(indices_from_sentence(test_words_clean))
def extract_tags(words):
i = 0
res_tags = []
res_source = []
cur_id = 0
while i < len(words):
if words[i] == '{':
res_tags.append((cur_id - len(words[i - 1]), len(words[i - 1]), words[i + 1]))
i += 2
else:
res_source.append(words[i])
cur_id += len(words[i])
i += 1
return res_tags, res_source
extract_tags(test_words)
def combine_datasets():
with open('train_nes.txt', 'r') as train_nes, \
open('train_sentences.txt', 'r') as train_sentences, \
open('train_sentences_enhanced.txt', 'r') as train_sentences_enhanced, \
open('combined_sentences.txt', 'w') as combined_sentences, \
open('combined_nes.txt', 'w') as combined_nes:
combined_nes.write(train_nes.read())
combined_sentences.write(train_sentences.read())
for line in train_sentences_enhanced:
words = re.split('(\W)', line)
res_tags, res_source = extract_tags(words)
res_tags_flatten = []
for tag in res_tags:
res_tags_flatten.append(str(tag[0]))
res_tags_flatten.append(str(tag[1]))
res_tags_flatten.append(tag[2])
res_tags_flatten.append('EOL')
combined_nes.write(' '.join(res_tags_flatten) + '\n')
combined_sentences.write(''.join(res_source))
combine_datasets()
def read_training_data():
with open('train_nes.txt', 'r') as combined_nes, open('train_sentences.txt', 'r') as combined_sentences:
X, y = [], []
for line in combined_sentences:
X.append(re.split('(\W)', line))
for i, line in enumerate(combined_nes):
words = line.split()[:-1]
tags_in_line = []
i = 0
while i < len(words):
tags_in_line.append((int(words[i]), int(words[i + 1]), words[i + 2]))
i += 3
y.append(tags_in_line)
return X, y
X, y = read_training_data()
print(X[0])
print(y[0])
print(X[-1])
print(y[-1])
stemmer = SnowballStemmer("russian")
def preprocess(word):
return stemmer.stem(word.lower())
def build_vocab(data):
vocab = defaultdict(lambda: 0)
for sent in data:
for word in sent:
stemmed = preprocess(word)
if stemmed not in vocab:
vocab[stemmed] = len(vocab) + 1
return vocab
VOCAB = build_vocab(X)
PAD_VALUE = len(VOCAB) + 1
print(len(VOCAB))
def get_positions(sent):
pos = []
idx = 0
for word in sent:
cur_l = len(word)
pos.append((idx, cur_l))
idx += cur_l
return pos
def pad_dataset(dataset, vocab):
num_dataset = [torch.tensor([vocab[preprocess(word)] for word in sent]) for sent in dataset]
return pad_sequence(num_dataset, batch_first=True, padding_value=PAD_VALUE)
X_padded = pad_dataset(X, VOCAB)
def pos_dataset(dataset):
return [get_positions(sent) for sent in dataset]
X_pos = pos_dataset(X)
def pair_X_Y(X_padded, X_pos, Y):
dataset = []
tag_to_int = {
'NONE': 0,
'PERSON': 1,
'ORG': 2
}
for sent, pos, tags in zip(X_padded, X_pos, Y):
y = []
pos_i = 0
tag_i = 0
for word in sent:
if pos_i < len(pos) and tag_i < len(tags) and pos[pos_i][0] == tags[tag_i][0]:
y.append(tag_to_int[tags[tag_i][2]])
tag_i += 1
else:
y.append(tag_to_int['NONE'])
pos_i += 1
dataset.append([sent.numpy(), y])
return np.array(dataset)
pairs_dataset = pair_X_Y(X_padded, X_pos, y)
print(pairs_dataset.shape)
TRAIN_X_Y, VAL_X_Y = train_test_split(pairs_dataset, test_size=0.1, random_state=SEED)
class Model(nn.Module):
def __init__(self, embedding_dim, hidden_dim, vocab_size):
super(Model, self).__init__()
self.emb = nn.Embedding(vocab_size, embedding_dim)
self.lstm = nn.LSTM(embedding_dim, hidden_dim, num_layers=2, bidirectional=False, batch_first=True)
self.fc2 = nn.Linear(hidden_dim, 3)
def forward(self, batch):
emb = self.emb(batch)
out, _ = self.lstm(emb)
tag_hidden = self.fc2(out)
tag_probs = F.log_softmax(tag_hidden, dim=-1)
return tag_probs
def train(model, train, val, epoch_cnt, batch_size):
train_loader = torch.utils.data.DataLoader(train, batch_size=batch_size, shuffle=True)
val_loader = torch.utils.data.DataLoader(val, batch_size=batch_size)
loss_function = nn.NLLLoss()
optimizer = optim.Adam(model.parameters(), lr=5e-4)
train_loss_values = []
val_loss_values = []
for epoch in tnrange(epoch_cnt, desc='Epoch'):
for batch_data in train_loader:
x, y = batch_data[:, 0].to(device), batch_data[:, 1].to(device)
optimizer.zero_grad()
output = model(x.long())
output = output.view(-1, 3)
y = y.reshape(-1)
loss = loss_function(output, y.long())
train_loss_values.append(loss)
loss.backward()
nn.utils.clip_grad_norm_(model.parameters(), 5)
optimizer.step()
with torch.no_grad():
loss_values = []
for batch_data in val_loader:
x, y = batch_data[:, 0].to(device), batch_data[:, 1].to(device)
output = model(x.long())
output = output.view(-1, 3)
y = y.reshape(-1)
loss = loss_function(output, y.long())
loss_values.append(loss.item())
val_loss_values.append(np.mean(np.array(loss_values)))
return train_loss_values, val_loss_values
embed = 128
hidden_dim = 256
vocab_size = len(VOCAB) + 1
epoch_cnt = 290
batch_size = 512
model = Model(embed, hidden_dim, vocab_size)
model = model.float()
model = model.to(device)
train_loss_values, val_loss_values =\
train(model, TRAIN_X_Y, VAL_X_Y, epoch_cnt, batch_size)
plt.plot(train_loss_values, label='train')
plt.plot(np.arange(0, len(train_loss_values), len(train_loss_values) / epoch_cnt), val_loss_values, label='validation')
plt.legend()
plt.title("Loss values")
plt.show()
def read_test():
test_filename = "test.txt"
lines = []
with open(test_filename, 'r') as test_file:
for line in test_file:
lines.append(re.split('(\W)', line))
return lines
TEST = read_test()
print(TEST[0])
def produce_test_results():
test_padded = pad_dataset(TEST, VOCAB)
test_pos = pos_dataset(TEST)
with torch.no_grad():
test_loader = torch.utils.data.DataLoader(test_padded, batch_size=batch_size)
ans = None
for batch_data in test_loader:
x = batch_data.to(device)
output = model(x.long())
_, ansx = output.max(dim=-1)
ansx = ansx.cpu().numpy()
if ans is None:
ans = ansx
else:
ans = np.append(ans, ansx, axis=0)
out_filename = "out.txt"
int_to_tag = {1:"PERSON" , 2:"ORG"}
with open(out_filename, "w") as out:
for sent, pos, tags in zip(test_padded, test_pos, ans):
for i in range(len(pos)):
if tags[i] in int_to_tag:
out.write("%d %d %s " % (pos[i][0], pos[i][1], int_to_tag[tags[i]]))
out.write("EOL\n")
produce_test_results()
```
| github_jupyter |
```
# Import Splinter, BeautifulSoup, and Pandas
from splinter import Browser
from bs4 import BeautifulSoup as soup
import pandas as pd
from webdriver_manager.chrome import ChromeDriverManager
# Set up Splinter
executable_path = {'executable_path': ChromeDriverManager().install()}
browser = Browser('chrome', **executable_path, headless=False)
```
## Visit the NASA mars news site
```
# Visit the Mars news site
url = 'https://redplanetscience.com/'
browser.visit(url)
# Optional delay for loading the page
browser.is_element_present_by_css('div.list_text', wait_time=1)
# Convert the browser html to a soup object
html = browser.html
news_soup = soup(html, 'html.parser')
slide_elem = news_soup.select_one('div.list_text')
print(news_soup.prettify())
slide_elem = news_soup.body.find('div', class_="content_title")
#display the current title content
news_title = slide_elem.find('div', class_="content_title").get_text()
news_title
# Use the parent element to find the first a tag and save it as `news_title`
news_title
news_p = slide_elem.find('div', class_="article_teaser_body").get_text()
news_p
# Use the parent element to find the paragraph text
news_p
```
## JPL Space Images Featured Image
```
# Visit URL
url = 'https://spaceimages-mars.com'
browser.visit(url)
# Find and click the full image button
full_image_link = browser.find_by_tag('button')[1]
full_image_link.click()
# Parse the resulting html with soup
html = browser.html
img_soup = soup(html, 'html.parser')
print(img_soup.prettify())
img_url_rel = img_soup.find('img',class_='fancybox-image').get('src')
img_url_rel
# find the relative image url
img_url_rel
img_url = f'https://spaceimages-mars.com/{img_url_rel}'
img_url
# Use the base url to create an absolute url
img_url
```
## Mars Facts
```
url = 'https://galaxyfacts-mars.com'
browser.visit(url)
html = browser.html
facts_soup = soup(html, 'html.parser')
html = browser.html
facts_soup = soup(html, 'html.parser')
tables = pd.read_html(url)
tables
df = tables[0]
df.head()
# Use `pd.read_html` to pull the data from the Mars-Earth Comparison section
# hint use index 0 to find the table
df.head()
df.columns = ['Description','Mars','Earth']
df = df.iloc[1:]
df.set_index('Description',inplace=True)
df
df
df.to_html()
df.to_html()
```
## Hemispheres
```
url = 'https://marshemispheres.com/'
browser.visit(url)
html = browser.html
hems_soup = soup(html, 'html.parser')
print(hems_soup.prettify())
# Create a list to hold the images and titles.
hemisphere_image_urls = []
# Get a list of all of the hemispheres
links = browser.find_by_css('a.product-item img')
# Next, loop through those links, click the link, find the sample anchor, return the href
for i in range(len(links)):
hemisphereInfo = {}
# We have to find the elements on each loop to avoid a stale element exception
browser.find_by_css('a.product-item img')[i].click()
# Next, we find the Sample image anchor tag and extract the href
sample = browser.links.find_by_text('Sample').first
hemisphereInfo['img_url'] = sample['href']
# Get Hemisphere title
titleA = browser.find_by_css('h2.title').text
hemisphereInfo['title'] = titleA.rpartition(' Enhanced')[0]
# Append hemisphere object to list
hemisphere_image_urls.append(hemisphereInfo)
# Finally, we navigate backwards
browser.back()
hemisphere_image_urls
hemisphere_image_urls
browser.quit()
```
| github_jupyter |
```
# Copyright 2020 NVIDIA Corporation. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
```
<img src="http://developer.download.nvidia.com/compute/machine-learning/frameworks/nvidia_logo.png" style="width: 90px; float: right;">
# Object Detection with TRTorch (SSD)
---
## Overview
In PyTorch 1.0, TorchScript was introduced as a method to separate your PyTorch model from Python, make it portable and optimizable.
TRTorch is a compiler that uses TensorRT (NVIDIA's Deep Learning Optimization SDK and Runtime) to optimize TorchScript code. It compiles standard TorchScript modules into ones that internally run with TensorRT optimizations.
TensorRT can take models from any major framework and specifically tune them to perform better on specific target hardware in the NVIDIA family, and TRTorch enables us to continue to remain in the PyTorch ecosystem whilst doing so. This allows us to leverage the great features in PyTorch, including module composability, its flexible tensor implementation, data loaders and more. TRTorch is available to use with both PyTorch and LibTorch.
To get more background information on this, we suggest the **lenet-getting-started** notebook as a primer for getting started with TRTorch.
### Learning objectives
This notebook demonstrates the steps for compiling a TorchScript module with TRTorch on a pretrained SSD network, and running it to test the speedup obtained.
## Contents
1. [Requirements](#1)
2. [SSD Overview](#2)
3. [Creating TorchScript modules](#3)
4. [Compiling with TRTorch](#4)
5. [Running Inference](#5)
6. [Measuring Speedup](#6)
7. [Conclusion](#7)
---
<a id="1"></a>
## 1. Requirements
Follow the steps in `notebooks/README` to prepare a Docker container, within which you can run this demo notebook.
In addition to that, run the following cell to obtain additional libraries specific to this demo.
```
# Known working versions
!pip install numpy==1.21.2 scipy==1.5.2 Pillow==6.2.0 scikit-image==0.17.2 matplotlib==3.3.0
```
---
<a id="2"></a>
## 2. SSD
### Single Shot MultiBox Detector model for object detection
_ | _
- | -
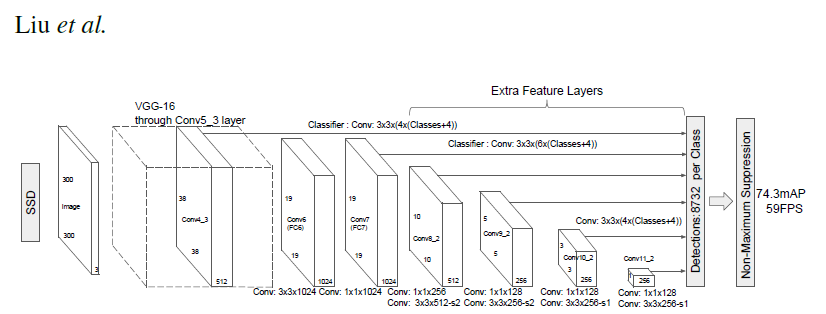 | 
PyTorch has a model repository called the PyTorch Hub, which is a source for high quality implementations of common models. We can get our SSD model pretrained on [COCO](https://cocodataset.org/#home) from there.
### Model Description
This SSD300 model is based on the
[SSD: Single Shot MultiBox Detector](https://arxiv.org/abs/1512.02325) paper, which
describes SSD as “a method for detecting objects in images using a single deep neural network".
The input size is fixed to 300x300.
The main difference between this model and the one described in the paper is in the backbone.
Specifically, the VGG model is obsolete and is replaced by the ResNet-50 model.
From the
[Speed/accuracy trade-offs for modern convolutional object detectors](https://arxiv.org/abs/1611.10012)
paper, the following enhancements were made to the backbone:
* The conv5_x, avgpool, fc and softmax layers were removed from the original classification model.
* All strides in conv4_x are set to 1x1.
The backbone is followed by 5 additional convolutional layers.
In addition to the convolutional layers, we attached 6 detection heads:
* The first detection head is attached to the last conv4_x layer.
* The other five detection heads are attached to the corresponding 5 additional layers.
Detector heads are similar to the ones referenced in the paper, however,
they are enhanced by additional BatchNorm layers after each convolution.
More information about this SSD model is available at Nvidia's "DeepLearningExamples" Github [here](https://github.com/NVIDIA/DeepLearningExamples/tree/master/PyTorch/Detection/SSD).
```
import torch
torch.hub._validate_not_a_forked_repo=lambda a,b,c: True
# List of available models in PyTorch Hub from Nvidia/DeepLearningExamples
torch.hub.list('NVIDIA/DeepLearningExamples:torchhub')
# load SSD model pretrained on COCO from Torch Hub
precision = 'fp32'
ssd300 = torch.hub.load('NVIDIA/DeepLearningExamples:torchhub', 'nvidia_ssd', model_math=precision);
```
Setting `precision="fp16"` will load a checkpoint trained with mixed precision
into architecture enabling execution on Tensor Cores. Handling mixed precision data requires the Apex library.
### Sample Inference
We can now run inference on the model. This is demonstrated below using sample images from the COCO 2017 Validation set.
```
# Sample images from the COCO validation set
uris = [
'http://images.cocodataset.org/val2017/000000397133.jpg',
'http://images.cocodataset.org/val2017/000000037777.jpg',
'http://images.cocodataset.org/val2017/000000252219.jpg'
]
# For convenient and comprehensive formatting of input and output of the model, load a set of utility methods.
utils = torch.hub.load('NVIDIA/DeepLearningExamples:torchhub', 'nvidia_ssd_processing_utils')
# Format images to comply with the network input
inputs = [utils.prepare_input(uri) for uri in uris]
tensor = utils.prepare_tensor(inputs, False)
# The model was trained on COCO dataset, which we need to access in order to
# translate class IDs into object names.
classes_to_labels = utils.get_coco_object_dictionary()
# Next, we run object detection
model = ssd300.eval().to("cuda")
detections_batch = model(tensor)
# By default, raw output from SSD network per input image contains 8732 boxes with
# localization and class probability distribution.
# Let’s filter this output to only get reasonable detections (confidence>40%) in a more comprehensive format.
results_per_input = utils.decode_results(detections_batch)
best_results_per_input = [utils.pick_best(results, 0.40) for results in results_per_input]
```
### Visualize results
```
from matplotlib import pyplot as plt
import matplotlib.patches as patches
# The utility plots the images and predicted bounding boxes (with confidence scores).
def plot_results(best_results):
for image_idx in range(len(best_results)):
fig, ax = plt.subplots(1)
# Show original, denormalized image...
image = inputs[image_idx] / 2 + 0.5
ax.imshow(image)
# ...with detections
bboxes, classes, confidences = best_results[image_idx]
for idx in range(len(bboxes)):
left, bot, right, top = bboxes[idx]
x, y, w, h = [val * 300 for val in [left, bot, right - left, top - bot]]
rect = patches.Rectangle((x, y), w, h, linewidth=1, edgecolor='r', facecolor='none')
ax.add_patch(rect)
ax.text(x, y, "{} {:.0f}%".format(classes_to_labels[classes[idx] - 1], confidences[idx]*100), bbox=dict(facecolor='white', alpha=0.5))
plt.show()
# Visualize results without TRTorch/TensorRT
plot_results(best_results_per_input)
```
### Benchmark utility
```
import time
import numpy as np
import torch.backends.cudnn as cudnn
cudnn.benchmark = True
# Helper function to benchmark the model
def benchmark(model, input_shape=(1024, 1, 32, 32), dtype='fp32', nwarmup=50, nruns=1000):
input_data = torch.randn(input_shape)
input_data = input_data.to("cuda")
if dtype=='fp16':
input_data = input_data.half()
print("Warm up ...")
with torch.no_grad():
for _ in range(nwarmup):
features = model(input_data)
torch.cuda.synchronize()
print("Start timing ...")
timings = []
with torch.no_grad():
for i in range(1, nruns+1):
start_time = time.time()
pred_loc, pred_label = model(input_data)
torch.cuda.synchronize()
end_time = time.time()
timings.append(end_time - start_time)
if i%10==0:
print('Iteration %d/%d, avg batch time %.2f ms'%(i, nruns, np.mean(timings)*1000))
print("Input shape:", input_data.size())
print("Output location prediction size:", pred_loc.size())
print("Output label prediction size:", pred_label.size())
print('Average batch time: %.2f ms'%(np.mean(timings)*1000))
```
We check how well the model performs **before** we use TRTorch/TensorRT
```
# Model benchmark without TRTorch/TensorRT
model = ssd300.eval().to("cuda")
benchmark(model, input_shape=(128, 3, 300, 300), nruns=100)
```
---
<a id="3"></a>
## 3. Creating TorchScript modules
To compile with TRTorch, the model must first be in **TorchScript**. TorchScript is a programming language included in PyTorch which removes the Python dependency normal PyTorch models have. This conversion is done via a JIT compiler which given a PyTorch Module will generate an equivalent TorchScript Module. There are two paths that can be used to generate TorchScript: **Tracing** and **Scripting**. <br>
- Tracing follows execution of PyTorch generating ops in TorchScript corresponding to what it sees. <br>
- Scripting does an analysis of the Python code and generates TorchScript, this allows the resulting graph to include control flow which tracing cannot do.
Tracing however due to its simplicity is more likely to compile successfully with TRTorch (though both systems are supported).
```
model = ssd300.eval().to("cuda")
traced_model = torch.jit.trace(model, [torch.randn((1,3,300,300)).to("cuda")])
```
If required, we can also save this model and use it independently of Python.
```
# This is just an example, and not required for the purposes of this demo
torch.jit.save(traced_model, "ssd_300_traced.jit.pt")
# Obtain the average time taken by a batch of input with Torchscript compiled modules
benchmark(traced_model, input_shape=(128, 3, 300, 300), nruns=100)
```
---
<a id="4"></a>
## 4. Compiling with TRTorch
TorchScript modules behave just like normal PyTorch modules and are intercompatible. From TorchScript we can now compile a TensorRT based module. This module will still be implemented in TorchScript but all the computation will be done in TensorRT.
```
import trtorch
# The compiled module will have precision as specified by "op_precision".
# Here, it will have FP16 precision.
trt_model = trtorch.compile(traced_model, {
"inputs": [trtorch.Input((3, 3, 300, 300))],
"enabled_precisions": {torch.float, torch.half}, # Run with FP16
"workspace_size": 1 << 20
})
```
---
<a id="5"></a>
## 5. Running Inference
Next, we run object detection
```
# using a TRTorch module is exactly the same as how we usually do inference in PyTorch i.e. model(inputs)
detections_batch = trt_model(tensor.to(torch.half)) # convert the input to half precision
# By default, raw output from SSD network per input image contains 8732 boxes with
# localization and class probability distribution.
# Let’s filter this output to only get reasonable detections (confidence>40%) in a more comprehensive format.
results_per_input = utils.decode_results(detections_batch)
best_results_per_input_trt = [utils.pick_best(results, 0.40) for results in results_per_input]
```
Now, let's visualize our predictions!
```
# Visualize results with TRTorch/TensorRT
plot_results(best_results_per_input_trt)
```
We get similar results as before!
---
## 6. Measuring Speedup
We can run the benchmark function again to see the speedup gained! Compare this result with the same batch-size of input in the case without TRTorch/TensorRT above.
```
batch_size = 128
# Recompiling with batch_size we use for evaluating performance
trt_model = trtorch.compile(traced_model, {
"inputs": [trtorch.Input((batch_size, 3, 300, 300))],
"enabled_precisions": {torch.float, torch.half}, # Run with FP16
"workspace_size": 1 << 20
})
benchmark(trt_model, input_shape=(batch_size, 3, 300, 300), nruns=100, dtype="fp16")
```
---
## 7. Conclusion
In this notebook, we have walked through the complete process of compiling a TorchScript SSD300 model with TRTorch, and tested the performance impact of the optimization. We find that using the TRTorch compiled model, we gain significant speedup in inference without any noticeable drop in performance!
### Details
For detailed information on model input and output,
training recipies, inference and performance visit:
[github](https://github.com/NVIDIA/DeepLearningExamples/tree/master/PyTorch/Detection/SSD)
and/or [NGC](https://ngc.nvidia.com/catalog/model-scripts/nvidia:ssd_for_pytorch)
### References
- [SSD: Single Shot MultiBox Detector](https://arxiv.org/abs/1512.02325) paper
- [Speed/accuracy trade-offs for modern convolutional object detectors](https://arxiv.org/abs/1611.10012) paper
- [SSD on NGC](https://ngc.nvidia.com/catalog/model-scripts/nvidia:ssd_for_pytorch)
- [SSD on github](https://github.com/NVIDIA/DeepLearningExamples/tree/master/PyTorch/Detection/SSD)
| github_jupyter |
# 3. Markov Models Example Problems
We will now look at a model that examines our state of healthiness vs. being sick. Keep in mind that this is very much like something you could do in real life. If you wanted to model a certain situation or environment, we could take some data that we have gathered, build a maximum likelihood model on it, and do things like study the properties that emerge from the model, or make predictions from the model, or generate the next most likely state.
Let's say we have 2 states: **sick** and **healthy**. We know that we spend most of our time in a healthy state, so the probability of transitioning from healthy to sick is very low:
$$p(sick \; | \; healthy) = 0.005$$
Hence, the probability of going from healthy to healthy is:
$$p(healthy \; | \; healthy) = 0.995$$
Now, on the other hand the probability of going from sick to sick is also very high. This is because if you just got sick yesterday then you are very likely to be sick tomorrow.
$$p(sick \; | \; sick) = 0.8$$
However, the probability of transitioning from sick to healthy should be higher than the reverse, because you probably won't stay sick for as long as you would stay healthy:
$$p(healthy \; | \; sick) = 0.02$$
We have now fully defined our state transition matrix, and we can now do some calculations.
## 1.1 Example Calculations
### 1.1.1
What is the probability of being healthy for 10 days in a row, given that we already start out as healthy? Well that is:
$$p(healthy \; 10 \; days \; in \; a \; row \; | \; healthy \; at \; t=0) = 0.995^9 = 95.6 \%$$
How about the probability of being healthy for 100 days in a row?
$$p(healthy \; 100 \; days \; in \; a \; row \; | \; healthy \; at \; t=0) = 0.995^{99} = 60.9 \%$$
## 2. Expected Number of Continuously Sick Days
We can now look at the expected number of days that you would remain in the same state (e.g. how many days would you expect to stay sick given the model?). This is a bit more difficult than the last problem, but completely doable, only involving the mathematics of <a href="https://en.wikipedia.org/wiki/Geometric_series">infinite sums</a>.
First, we can look at the probability of being in state $i$, and going to state $i$ in the next state. That is just $A(i,i)$:
$$p \big(s(t)=i \; | \; s(t-1)=i \big) = A(i, i)$$
Now, what is the probability distribution that we actually want to calculate? How about we calculate the probability that we stay in state $i$ for $n$ transitions, at which point we move to another state:
$$p \big(s(t) \;!=i \; | \; s(t-1)=i \big) = 1 - A(i, i)$$
So, the joint probability that we are trying to model is:
$$p\big(s(1)=i, s(2)=i,...,s(n)=i, s(n+1) \;!= i\big) = A(i,i)^{n-1}\big(1-A(i,i)\big)$$
In english this means that we are multiplying the transition probability of staying in the same state, $A(i,i)$, times the number of times we stayed in the same state, $n$, (note it is $n-1$ because we are given that we start in that state, hence there is no transition associated with it) times $1 - A(i,i)$, the probability of transitioning from that state. This leaves us with an expected value for $n$ of:
$$E(n) = \sum np(n) = \sum_{n=1..\infty} nA(i,i)^{n-1}(1-A(i,i))$$
Note, in the above equation $p(n)$ is the probability that we will see state $i$ $n-1$ times after starting from $i$ and then see a state that is not $i$. Also, we know that the expected value of $n$ should be the sum of all possible values of $n$ times $p(n)$.
### 2.1 Expected $n$
So, we can now expand this function and calculate the two sums separately.
$$E(n) = \sum_{n=1..\infty}nA(i,i)^{n-1}(1 - A(i,i)) = \sum nA(i, i)^{n-1} - \sum nA(i,i)^n$$
**First Sum**<br>
With our first sum, we can say that:
$$S = \sum na(i, i)^{n-1}$$
$$S = 1 + 2a + 3a^2 + 4a^3+ ...$$
And we can then multiply that sum, $S$, by $a$, to get:
$$aS = a + 2a^2 + 3a^3 + 4a^4+...$$
And then we can subtract $aS$ from $S$:
$$S - aS = S'= 1 + a + a^2 + a^3+...$$
This $S'$ is another infinite sum, but it is one that is much easier to solve!
$$S'= 1 + a + a^2 + a^3+...$$
And then $aS'$ is:
$$aS' = a + a^2 + a^3+ + a^4 + ...$$
Which, when we then do $S' - aS'$, we end up with:
$$S' - aS' = 1$$
$$S' = \frac{1}{1 - a}$$
And if we then substitute that value in for $S'$ above:
$$S - aS = S'= 1 + a + a^2 + a^3+... = \frac{1}{1 - a}$$
$$S - aS = \frac{1}{1 - a}$$
$$S = \frac{1}{(1 - a)^2}$$
**Second Sum**<br>
We can now look at our second sum:
$$S = \sum na(i,i)^n$$
$$S = 1a + 2a^2 + 3a^3 +...$$
$$Sa = 1a^2 + 2a^3 +...$$
$$S - aS = S' = a + a^2 + a^3 + ...$$
$$aS' = a^2 + a^3 + a^4 +...$$
$$S' - aS' = a$$
$$S' = \frac{a}{1 - a}$$
And we can plug back in $S'$ to get:
$$S - aS = \frac{a}{1 - a}$$
$$S = \frac{a}{(1 - a)^2}$$
**Combine** <br>
We can now combine these two sums as follows:
$$E(n) = \frac{1}{(1 - a)^2} - \frac{a}{(1-a)^2}$$
$$E(n) = \frac{1}{1-a}$$
**Calculate Number of Sick Days**<br>
So, how do we calculate the correct number of sick days? That is just:
$$\frac{1}{1 - 0.8} = 5$$
## 3. SEO and Bounce Rate Optimization
We are now going to look at SEO and Bounch Rate Optimization. This is a problem that every developer and website owner can relate to. You have a website and obviously you would like to increase traffic, increase conversions, and avoid a high bounce rate (which could lead to google assigning your page a low ranking). What would a good way of modeling this data be? Without even looking at any code we can look at some examples of things that we want to know, and how they relate to markov models.
### 3.1 Arrival
First and foremost, how do people arrive on your page? Is it your home page? Your landing page? Well, this is just the very first page of what is hopefully a sequence of pages. So, the markov analogy here is that this is just the initial state distribution or $\pi$. So, once we have our markov model, the $\pi$ vector will tell us which of our pages a user is most likely to start on.
### 3.2 Sequences of Pages
What about sequences of pages? Well, if you think people are getting to your landing page, hitting the buy button, checking out, and then closing the browser window, you can test the validity of that assumption by calculating the probability of that sequence. Of course, the probability of any sequence is probability going to be much less than 1. This is because for a longer sequence, we have more multiplication, and hence smaller final numbers. We do have two alternatives however:
> * 1) You can compare the probability of two different sequences. So, are people going through the entire checkout process? Or is it more probable that they are just bouncing?
* 2) Another option is to just find the transition probabilities themselves. These are conditional probabilities instead of joint probabilities. You want to know, once they have made it to the landing page, what is the probability of hitting buy. Then, once they have hit buy, what is the probability of them completing the checkout.
### 3.3 Bounce Rate
This is hard to measure, unless you are google and hence have analytics on nearly every page on the web. This is because once a user has left your site, you can no longer run code on their computer or track what they are doing. However, let's pretend that we can determine this information. Once we have done this, we can measure which page has the highest bounce rate. At this point we can manually analyze that page and ask our marketing people "what is different about this page that people don't find it useful/want to leave?" We can then address that problem, and the hopefully later analysis shows that the fixed page no longer has a high bounce right. In the markov model, we can just represents this as the null state.
### 3.4 Data
So, the data we are going to be working with has two columns: `last_page_id` and `next_page_id`. This can be interpreted as the current page and the next page. The site has 10 pages with the id's 0-9. We can represent start pages by making the current page -1, and the next page the actual page. We can represent the end of the page with two different codes, `B`(bounce) or `C` (close). In the case of bounce, the user saw the page and then immediately bounced. In the case of close, the user saw the page stayed and potentially saw some useful information, and then closed the window. So, you can imagine that our engineer may use time as a factor in determining if it is a bounce or a close.
```
import numpy as np
import pandas as pd
"""Goal here is to store start page and end page, and the count how many times that happens. After that
we are going to turn it into a probability distribution. We can divide all transitions that start with specific
start state, by row_sum"""
transitions = {} # getting all specific transitions from start pg to end pg, tallying up # of times each occurs
row_sums = {} # start date as key -> getting number of times each starting pg occurs
# Collect our counts
for line in open('../../../data/site/site_data.csv'):
s, e = line.rstrip().split(',') # get start and end page
transitions[(s, e)] = transitions.get((s, e), 0.) + 1
row_sums[s] = row_sums.get(s, 0.) + 1
# Normalize the counts so they become real probability distributions
for k, v in transitions.items():
s, e = k
transitions[k] = v / row_sums[s]
# Calculate initial state distribution
print('Initial state distribution')
for k, v in transitions.items():
s, e = k
if s == '-1': # this means it is the start of the sequence.
print (e, v)
# Which page has the highest bounce rate?
for k, v in transitions.items():
s, e = k
if e == 'B':
print(f'Bounce rate for {s}: {v}')
```
We can see that page with `id` 9 has the highest value in the initial state distribution, so we are most likely to start on that page. We can then see that the page with highest bounce rate is also at page `id` 9.
## 4. Build a 2nd-order language model and generate phrases
So, we are now going to work with non first order markov chains for a little bit. In this example we are going to try and create a language model. So we are going to first train a model on some data to determine the distribution of a word given the previous two words. We can then use this model to generate new phrases. Note that another step of this model would be to calculate the probability of a phrase.
So the data that we are going to look at is just a collection of Robert Frost Poems. It is just a text file with all of the poems concatenated together. So, the first thing we are going to want to do is tokenize each sentence, and remove punctuation. It will look similar to this:
```
def remove_punctuation(s):
return s.translate(None, string.punctuation)
tokens = [t for t in remove_puncuation(line.rstrip().lower()).split()]
```
Once we have tokenized each line, we want to perform various counts in addition to the second order model counts. We need to measure the initial distribution of words, or stated another way the distribution of the first word of a sentence. We also want to know the distribution of the second word of a sentence. Both of these do not have two previous words, so they are not second order. We could technically include them in the second order measurement by using `None` in place of the previous words, but we won't do that here. We also want to keep track of how to end the sentence (end of sentence distribution, will look similar to (w(t-2), w(t-1) -> END)), so we will include a special token for that too.
When we do this counting, what we first want to do is create an array of all possibilities. So, for example if we had two sentences:
```
I love dogs
I love cats
```
Then we could have a dictionary where the key was `(I, love)` and the value was an array `[dogs, cats]`. If "I love" was also a stand alone sentence, then the value would be `[dogs, cats, END]`. The function below can help us with this, since we first need to check if there is any value for the key, create an array if not, otherwise just append to the array.
```
def add2dict(d, k, v):
if k not in d:
d[k] = []
else:
d[k].append(v)
```
One we have collected all of these arrays of possible next words, we need to turn them into **probability distributions**. For example, the array `[cat, cat, dog]` would become the dictionary `{"cat": 2/3, "dog": 1/3}`. Here is a function that can do this:
```
def list2pdict(ts):
d = {}
n = len(ts)
for t in ts:
d[t] = d.get(t, 0.) + 1
for t, c in d.items():
d[t] = c / n
return d
```
Next, we will need a function that can sample from this dictionary. To do this we will need to generate a random number between 0 and 1, and then use the distribution of the words to sample a word given a random number. Here is a function that can do that:
```
def sample_word(d):
p0 = np.random.random()
cumulative = 0
for t, p in d.items():
cumulative += p
if p0 < cumulative:
return t
assert(False) # should never get here
```
Because all of our distributions are structured as dictionaries, we can use the same function for all of them.
```
import numpy as np
import string
"""3 dicts. 1st store pdist for the start of a phrase, then a second word dict which stores the distributions
for the 2nd word of a sentence, and then we are going to have a dict for all second order transitions"""
initial = {}
second_word = {}
transitions = {}
def remove_punctuation(s):
return s.translate(str.maketrans('', '', string.punctuation))
def add2dict(d, k, v):
"""Parameters: Dictionary, Key, Value"""
if k not in d:
d[k] = []
d[k].append(v)
# Loop through file of poems
for line in open('../../../data/poems/robert_frost.txt'):
tokens = remove_punctuation(line.rstrip().lower()).split() # Get all tokens for specific line we are looping over
T = len(tokens) # Length of sequence
for i in range(T): # Loop through every token in sequence
t = tokens[i]
if i == 0: # We are looking at first word
initial[t] = initial.get(t, 0.) + 1
else:
t_1 = tokens[i - 1]
if i == T - 1: # Looking at last word
add2dict(transitions, (t_1, t), 'END')
if i == 1: # second word of sentence, hence only 1 previous word
add2dict(second_word, t_1, t)
else:
t_2 = tokens[i - 2] # Get second previous word
add2dict(transitions, (t_2, t_1), t) # add previous and 2nd previous word as key, and current word as val
# Normalize the distributions
initial_total = sum(initial.values())
for t, c in initial.items():
initial[t] = c / initial_total
# Take our list and turn it into a dictionary of probabilities
def list2pdict(ts):
d = {}
n = len(ts) # get total number of values
for t in ts: # look at each token
d[t] = d.get(t, 0.) + 1
for t, c in d.items(): # go through dictionary, divide frequency by sum
d[t] = c / n
return d
for t_1, ts in second_word.items():
second_word[t_1] = list2pdict(ts)
for k, ts in transitions.items():
transitions[k] = list2pdict(ts)
def sample_word(d):
p0 = np.random.random() # Generate random number from 0 to 1
cumulative = 0 # cumulative count for all probabilities seen so far
for t, p in d.items():
cumulative += p
if p0 < cumulative:
return t
assert(False) # should never hit this
"""Function to generate a poem"""
def generate():
for i in range(4):
sentence = []
# initial word
w0 = sample_word(initial)
sentence.append(w0)
# sample second word
w1 = sample_word(second_word[w0])
sentence.append(w1)
# second-order transitions until END -> enter infinite loop
while True:
w2 = sample_word(transitions[(w0, w1)]) # sample next word given previous two words
if w2 == 'END':
break
sentence.append(w2)
w0 = w1
w1 = w2
print(' '.join(sentence))
generate()
```
## 5. Google's PageRank Algorithm
Markov models were even used in Google's PageRank algorithm. The basic problem we face is:
> * We have $M$ webpages that link to eachother, and we would like to assign importance scores $x(1),...,x(M)$
* All of these scores are greater than or equal to 0
* So, we want to assign a page rank to all of these pages
How can we go about doing this? Well, we can think of a webpage as a sequence, and the page you are on as the state. Where does the ranking come from? Well, the ranking actually comes from the limiting distribution. That is, in the long run, the proportion of visits that will be spent on this page. Now, if you think "great that is all I need to know", slow down. How can we actually do this in practice? How do we train the markov model, and what are the values we assign to the state transition matrix? And how can we ensure that the limiting distribution exists and is unique? The key insight was that **we can use the linked structure of the web to determine the ranking**.
The main idea is that a *link to a page* is like a *vote for its importance*. So, as a first attempt we could just use a frequency count to measure the votes. Of course, that wouldn't be a valid probability distribution, so we could just divide each row by its sum to make it sum to 1. So we set:
$$A(i, j) = \frac{1}{n(i)} \; if \; i \; links \; to \; j$$
$$A(i, j) = 0 \; otherwise$$
Here $n(i)$ stands for the total number of links on a page, and you can confirm that the sum of a row is $\frac{n(i)}{n(i)} = 1$, so this is a valid markov matrix. Now, we still aren't sure if the limiting distribution is unique.
### 5.1 This is already a good start
Let's keep in mind that the above solution already solves a few problems. For instance, let's say you are a spammer and you want to sell 1000 links on your webpage. Well, because the transition matrix must remain a valid probability matrix, the rows must sum to 1, which means that each of your links now only has a strength of $\frac{1}{1000}$. For example the frequency matrix would look like:
| |abc.com|amazon.com|facebook.com|github.com|
|--- |--- |--- | --- |--- |
|thespammer.com|1 |1 |1 |1 |
And then if we transformed that into a probability matrix it would just be each value divided by the total number of links, 4:
| |abc.com|amazon.com|facebook.com|github.com|
|--- |--- |--- | --- |--- |
|thespammer.com|0.25 |0.25 |0.25 |0.25 |
You may then think, I will just create 1000 pages and each of them will only have 1 link. Unfortunately, since nobody knows about those 1000 pages you just created nobody is going to link to them, which means they are impossible to get to. So, in the limiting distribution, those states will have 0 probability because you can't even get to them, so there outgoing links are worthless. Remember, the markov chains limiting distribution will model the long running proportion of visits to a state. So, if you never visit that state, its probability will be 0.
We still have not ensure that the limiting distribution exists and is unique.
### 5.2 Perron-Frobenius Theorem
How can we ensure that our model has a unique stationary distribution. In 1910, this was actually determined. It is known as the **Perron-Frobenius Theorem**, and it states that:
> *If our transition matrix is a markov matrix -meaning that all of the rows sum to 1, and all of the values are strictly positive, i.e. no values that are 0- then the stationary distribution exists and is unique*.
In fact, we can start in any initial state and as time approaches infinity we will always end up with the same stationary distribution, therefore this is also the limiting distribution.
So, how can we satisfy the PF criterion? Let's return to this idea of **smoothing**, which we first talked about when discussing how to train a markov model. The basic idea was that we can make things that were 0, non-zero, so there is still a small possibility that we can get to that state. This might be good news for the spammer. So, we can create a uniform probability distribution $U = \frac{1}{M}$, which is an $M x M$ matrix ($M$ is the number of states). PageRanks solution was to take the matrix we had before and multiply it by 0.85, and to take the uniform distribution and multiply it by 0.15, and add them together to get the final pagerank matrix.
$$G = 0.85A + 0.15U$$
Now all of the elements are strictly positive, and we can convince ourselves that G is still a valid markov matrix.
| github_jupyter |
# Quantization of Signals
*This jupyter notebook is part of a [collection of notebooks](../index.ipynb) on various topics of Digital Signal Processing. Please direct questions and suggestions to [[email protected]](mailto:[email protected]).*
## Spectral Shaping of the Quantization Noise
The quantized signal $x_Q[k]$ can be expressed by the continuous amplitude signal $x[k]$ and the quantization error $e[k]$ as
\begin{equation}
x_Q[k] = \mathcal{Q} \{ x[k] \} = x[k] + e[k]
\end{equation}
According to the [introduced model](linear_uniform_quantization_error.ipynb#Model-for-the-Quantization-Error), the quantization noise can be modeled as uniformly distributed white noise. Hence, the noise is distributed over the entire frequency range. The basic concept of [noise shaping](https://en.wikipedia.org/wiki/Noise_shaping) is a feedback of the quantization error to the input of the quantizer. This way the spectral characteristics of the quantization noise can be modified, i.e. spectrally shaped. Introducing a generic filter $h[k]$ into the feedback loop yields the following structure

The quantized signal can be deduced from the block diagram above as
\begin{equation}
x_Q[k] = \mathcal{Q} \{ x[k] - e[k] * h[k] \} = x[k] + e[k] - e[k] * h[k]
\end{equation}
where the additive noise model from above has been introduced and it has been assumed that the impulse response $h[k]$ is normalized such that the magnitude of $e[k] * h[k]$ is below the quantization step $Q$. The overall quantization error is then
\begin{equation}
e_H[k] = x_Q[k] - x[k] = e[k] * (\delta[k] - h[k])
\end{equation}
The power spectral density (PSD) of the quantization error with noise shaping is calculated to
\begin{equation}
\Phi_{e_H e_H}(\mathrm{e}^{\,\mathrm{j}\,\Omega}) = \Phi_{ee}(\mathrm{e}^{\,\mathrm{j}\,\Omega}) \cdot \left| 1 - H(\mathrm{e}^{\,\mathrm{j}\,\Omega}) \right|^2
\end{equation}
Hence the PSD $\Phi_{ee}(\mathrm{e}^{\,\mathrm{j}\,\Omega})$ of the quantizer without noise shaping is weighted by $| 1 - H(\mathrm{e}^{\,\mathrm{j}\,\Omega}) |^2$. Noise shaping allows a spectral modification of the quantization error. The desired shaping depends on the application scenario. For some applications, high-frequency noise is less disturbing as low-frequency noise.
### Example - First-Order Noise Shaping
If the feedback of the error signal is delayed by one sample we get with $h[k] = \delta[k-1]$
\begin{equation}
\Phi_{e_H e_H}(\mathrm{e}^{\,\mathrm{j}\,\Omega}) = \Phi_{ee}(\mathrm{e}^{\,\mathrm{j}\,\Omega}) \cdot \left| 1 - \mathrm{e}^{\,-\mathrm{j}\,\Omega} \right|^2
\end{equation}
For linear uniform quantization $\Phi_{ee}(\mathrm{e}^{\,\mathrm{j}\,\Omega}) = \sigma_e^2$ is constant. Hence, the spectral shaping constitutes a high-pass characteristic of first order. The following simulation evaluates the noise shaping quantizer of first order.
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import scipy.signal as sig
w = 8 # wordlength of the quantized signal
xmin = -1 # minimum of input signal
N = 32768 # number of samples
def uniform_midtread_quantizer_w_ns(x, Q):
# limiter
x = np.copy(x)
idx = np.where(x <= -1)
x[idx] = -1
idx = np.where(x > 1 - Q)
x[idx] = 1 - Q
# linear uniform quantization with noise shaping
xQ = Q * np.floor(x/Q + 1/2)
e = xQ - x
xQ = xQ - np.concatenate(([0], e[0:-1]))
return xQ[1:]
# quantization step
Q = 1/(2**(w-1))
# compute input signal
np.random.seed(5)
x = np.random.uniform(size=N, low=xmin, high=(-xmin-Q))
# quantize signal
xQ = uniform_midtread_quantizer_w_ns(x, Q)
e = xQ - x[1:]
# estimate PSD of error signal
nf, Pee = sig.welch(e, nperseg=64)
# estimate SNR
SNR = 10*np.log10((np.var(x)/np.var(e)))
print('SNR = {:2.1f} dB'.format(SNR))
plt.figure(figsize=(10,5))
Om = nf*2*np.pi
plt.plot(Om, Pee*6/Q**2, label='estimated PSD')
plt.plot(Om, np.abs(1 - np.exp(-1j*Om))**2, label='theoretic PSD')
plt.plot(Om, np.ones(Om.shape), label='PSD w/o noise shaping')
plt.title('PSD of quantization error')
plt.xlabel(r'$\Omega$')
plt.ylabel(r'$\hat{\Phi}_{e_H e_H}(e^{j \Omega}) / \sigma_e^2$')
plt.axis([0, np.pi, 0, 4.5]);
plt.legend(loc='upper left')
plt.grid()
```
**Exercise**
* The overall average SNR is lower than for the quantizer without noise shaping. Why?
Solution: The average power per frequency is lower that without noise shaping for frequencies below $\Omega \approx \pi$. However, this comes at the cost of a larger average power per frequency for frequencies above $\Omega \approx \pi$. The average power of the quantization noise is given as the integral over the PSD of the quantization noise. It is larger for noise shaping and the resulting SNR is consequently lower. Noise shaping is nevertheless beneficial in applications where a lower quantization error in a limited frequency region is desired.
**Copyright**
This notebook is provided as [Open Educational Resource](https://en.wikipedia.org/wiki/Open_educational_resources). Feel free to use the notebook for your own purposes. The text is licensed under [Creative Commons Attribution 4.0](https://creativecommons.org/licenses/by/4.0/), the code of the IPython examples under the [MIT license](https://opensource.org/licenses/MIT). Please attribute the work as follows: *Sascha Spors, Digital Signal Processing - Lecture notes featuring computational examples, 2016-2018*.
| github_jupyter |
# Neural networks with PyTorch
Deep learning networks tend to be massive with dozens or hundreds of layers, that's where the term "deep" comes from. You can build one of these deep networks using only weight matrices as we did in the previous notebook, but in general it's very cumbersome and difficult to implement. PyTorch has a nice module `nn` that provides a nice way to efficiently build large neural networks.
```
# Import necessary packages
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import numpy as np
import torch
import helper
import matplotlib.pyplot as plt
```
Now we're going to build a larger network that can solve a (formerly) difficult problem, identifying text in an image. Here we'll use the MNIST dataset which consists of greyscale handwritten digits. Each image is 28x28 pixels, you can see a sample below
<img src='assets/mnist.png'>
Our goal is to build a neural network that can take one of these images and predict the digit in the image.
First up, we need to get our dataset. This is provided through the `torchvision` package. The code below will download the MNIST dataset, then create training and test datasets for us. Don't worry too much about the details here, you'll learn more about this later.
```
### Run this cell
from torchvision import datasets, transforms
# Define a transform to normalize the data
transform = transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.5,), (0.5,)),
])
# Download and load the training data
trainset = datasets.MNIST('~/.pytorch/MNIST_data/', download=True, train=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)
```
We have the training data loaded into `trainloader` and we make that an iterator with `iter(trainloader)`. Later, we'll use this to loop through the dataset for training, like
```python
for image, label in trainloader:
## do things with images and labels
```
You'll notice I created the `trainloader` with a batch size of 64, and `shuffle=True`. The batch size is the number of images we get in one iteration from the data loader and pass through our network, often called a *batch*. And `shuffle=True` tells it to shuffle the dataset every time we start going through the data loader again. But here I'm just grabbing the first batch so we can check out the data. We can see below that `images` is just a tensor with size `(64, 1, 28, 28)`. So, 64 images per batch, 1 color channel, and 28x28 images.
```
dataiter = iter(trainloader)
images, labels = dataiter.next()
print(type(images))
print(images.shape)
print(labels.shape)
```
This is what one of the images looks like.
```
plt.imshow(images[1].numpy().squeeze(), cmap='Greys_r');
```
First, let's try to build a simple network for this dataset using weight matrices and matrix multiplications. Then, we'll see how to do it using PyTorch's `nn` module which provides a much more convenient and powerful method for defining network architectures.
The networks you've seen so far are called *fully-connected* or *dense* networks. Each unit in one layer is connected to each unit in the next layer. In fully-connected networks, the input to each layer must be a one-dimensional vector (which can be stacked into a 2D tensor as a batch of multiple examples). However, our images are 28x28 2D tensors, so we need to convert them into 1D vectors. Thinking about sizes, we need to convert the batch of images with shape `(64, 1, 28, 28)` to a have a shape of `(64, 784)`, 784 is 28 times 28. This is typically called *flattening*, we flattened the 2D images into 1D vectors.
Previously you built a network with one output unit. Here we need 10 output units, one for each digit. We want our network to predict the digit shown in an image, so what we'll do is calculate probabilities that the image is of any one digit or class. This ends up being a discrete probability distribution over the classes (digits) that tells us the most likely class for the image. That means we need 10 output units for the 10 classes (digits). We'll see how to convert the network output into a probability distribution next.
> **Exercise:** Flatten the batch of images `images`. Then build a multi-layer network with 784 input units, 256 hidden units, and 10 output units using random tensors for the weights and biases. For now, use a sigmoid activation for the hidden layer. Leave the output layer without an activation, we'll add one that gives us a probability distribution next.
```
## Your solution
images_flat = images.view(64, 784)
def act(x):
return 1/(1+torch.exp(-x))
torch.manual_seed(42)
n_input = 784
n_hidden = 256
n_output = 10
W1 = torch.randn((n_input, n_hidden))
W2 = torch.randn((n_hidden, n_output))
B1 = torch.randn((1, 1))
B2 = torch.randn((1, 1))
def network(features):
return act(torch.mm(act(torch.mm(features, W1) + B1) ,W2) + B2)
out = network(images_flat)
out.shape
#out = # output of your network, should have shape (64,10)
```
Now we have 10 outputs for our network. We want to pass in an image to our network and get out a probability distribution over the classes that tells us the likely class(es) the image belongs to. Something that looks like this:
<img src='assets/image_distribution.png' width=500px>
Here we see that the probability for each class is roughly the same. This is representing an untrained network, it hasn't seen any data yet so it just returns a uniform distribution with equal probabilities for each class.
To calculate this probability distribution, we often use the [**softmax** function](https://en.wikipedia.org/wiki/Softmax_function). Mathematically this looks like
$$
\Large \sigma(x_i) = \cfrac{e^{x_i}}{\sum_k^K{e^{x_k}}}
$$
What this does is squish each input $x_i$ between 0 and 1 and normalizes the values to give you a proper probability distribution where the probabilites sum up to one.
> **Exercise:** Implement a function `softmax` that performs the softmax calculation and returns probability distributions for each example in the batch. Note that you'll need to pay attention to the shapes when doing this. If you have a tensor `a` with shape `(64, 10)` and a tensor `b` with shape `(64,)`, doing `a/b` will give you an error because PyTorch will try to do the division across the columns (called broadcasting) but you'll get a size mismatch. The way to think about this is for each of the 64 examples, you only want to divide by one value, the sum in the denominator. So you need `b` to have a shape of `(64, 1)`. This way PyTorch will divide the 10 values in each row of `a` by the one value in each row of `b`. Pay attention to how you take the sum as well. You'll need to define the `dim` keyword in `torch.sum`. Setting `dim=0` takes the sum across the rows while `dim=1` takes the sum across the columns.
```
def softmax(x):
return torch.exp(x)/torch.sum(torch.exp(x), dim=1).reshape(64, 1)
## TODO: Implement the softmax function here
# Here, out should be the output of the network in the previous excercise with shape (64,10)
probabilities = softmax(out)
# Does it have the right shape? Should be (64, 10)
print(probabilities.shape)
# Does it sum to 1?
print(probabilities.sum(dim=1))
```
## Building networks with PyTorch
PyTorch provides a module `nn` that makes building networks much simpler. Here I'll show you how to build the same one as above with 784 inputs, 256 hidden units, 10 output units and a softmax output.
```
from torch import nn
class Network(nn.Module):
def __init__(self):
super().__init__()
# Inputs to hidden layer linear transformation
self.hidden = nn.Linear(784, 256)
# Output layer, 10 units - one for each digit
self.output = nn.Linear(256, 10)
# Define sigmoid activation and softmax output
self.sigmoid = nn.Sigmoid()
self.softmax = nn.Softmax(dim=1)
def forward(self, x):
# Pass the input tensor through each of our operations
x = self.hidden(x)
x = self.sigmoid(x)
x = self.output(x)
x = self.softmax(x)
return x
```
Let's go through this bit by bit.
```python
class Network(nn.Module):
```
Here we're inheriting from `nn.Module`. Combined with `super().__init__()` this creates a class that tracks the architecture and provides a lot of useful methods and attributes. It is mandatory to inherit from `nn.Module` when you're creating a class for your network. The name of the class itself can be anything.
```python
self.hidden = nn.Linear(784, 256)
```
This line creates a module for a linear transformation, $x\mathbf{W} + b$, with 784 inputs and 256 outputs and assigns it to `self.hidden`. The module automatically creates the weight and bias tensors which we'll use in the `forward` method. You can access the weight and bias tensors once the network (`net`) is created with `net.hidden.weight` and `net.hidden.bias`.
```python
self.output = nn.Linear(256, 10)
```
Similarly, this creates another linear transformation with 256 inputs and 10 outputs.
```python
self.sigmoid = nn.Sigmoid()
self.softmax = nn.Softmax(dim=1)
```
Here I defined operations for the sigmoid activation and softmax output. Setting `dim=1` in `nn.Softmax(dim=1)` calculates softmax across the columns.
```python
def forward(self, x):
```
PyTorch networks created with `nn.Module` must have a `forward` method defined. It takes in a tensor `x` and passes it through the operations you defined in the `__init__` method.
```python
x = self.hidden(x)
x = self.sigmoid(x)
x = self.output(x)
x = self.softmax(x)
```
Here the input tensor `x` is passed through each operation and reassigned to `x`. We can see that the input tensor goes through the hidden layer, then a sigmoid function, then the output layer, and finally the softmax function. It doesn't matter what you name the variables here, as long as the inputs and outputs of the operations match the network architecture you want to build. The order in which you define things in the `__init__` method doesn't matter, but you'll need to sequence the operations correctly in the `forward` method.
Now we can create a `Network` object.
```
# Create the network and look at it's text representation
model = Network()
model
```
You can define the network somewhat more concisely and clearly using the `torch.nn.functional` module. This is the most common way you'll see networks defined as many operations are simple element-wise functions. We normally import this module as `F`, `import torch.nn.functional as F`.
```
import torch.nn.functional as F
class Network(nn.Module):
def __init__(self):
super().__init__()
# Inputs to hidden layer linear transformation
self.hidden = nn.Linear(784, 256)
# Output layer, 10 units - one for each digit
self.output = nn.Linear(256, 10)
def forward(self, x):
# Hidden layer with sigmoid activation
x = F.sigmoid(self.hidden(x))
# Output layer with softmax activation
x = F.softmax(self.output(x), dim=1)
return x
```
### Activation functions
So far we've only been looking at the sigmoid activation function, but in general any function can be used as an activation function. The only requirement is that for a network to approximate a non-linear function, the activation functions must be non-linear. Here are a few more examples of common activation functions: Tanh (hyperbolic tangent), and ReLU (rectified linear unit).
<img src="assets/activation.png" width=700px>
In practice, the ReLU function is used almost exclusively as the activation function for hidden layers.
### Your Turn to Build a Network
<img src="assets/mlp_mnist.png" width=600px>
> **Exercise:** Create a network with 784 input units, a hidden layer with 128 units and a ReLU activation, then a hidden layer with 64 units and a ReLU activation, and finally an output layer with a softmax activation as shown above. You can use a ReLU activation with the `nn.ReLU` module or `F.relu` function.
It's good practice to name your layers by their type of network, for instance 'fc' to represent a fully-connected layer. As you code your solution, use `fc1`, `fc2`, and `fc3` as your layer names.
```
## Your solution here
class FirstNet(nn.Module):
def __init__(self):
super().__init__()
self.fc1 = nn.Linear(784, 128)
self.fc2 = nn.Linear(128, 64)
self.fc3 = nn.Linear(64, 10)
def forward(self, x):
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = F.softmax(self.fc3(x), dim=1)
return x
model = FirstNet()
```
### Initializing weights and biases
The weights and such are automatically initialized for you, but it's possible to customize how they are initialized. The weights and biases are tensors attached to the layer you defined, you can get them with `model.fc1.weight` for instance.
```
print(model.fc1.weight)
print(model.fc1.bias)
```
For custom initialization, we want to modify these tensors in place. These are actually autograd *Variables*, so we need to get back the actual tensors with `model.fc1.weight.data`. Once we have the tensors, we can fill them with zeros (for biases) or random normal values.
```
# Set biases to all zeros
model.fc1.bias.data.fill_(0)
# sample from random normal with standard dev = 0.01
model.fc1.weight.data.normal_(std=0.01)
```
### Forward pass
Now that we have a network, let's see what happens when we pass in an image.
```
# Grab some data
dataiter = iter(trainloader)
images, labels = dataiter.next()
# Resize images into a 1D vector, new shape is (batch size, color channels, image pixels)
images.resize_(64, 1, 784)
# or images.resize_(images.shape[0], 1, 784) to automatically get batch size
# Forward pass through the network
img_idx = 0
ps = model.forward(images[img_idx,:])
img = images[img_idx]
helper.view_classify(img.view(1, 28, 28), ps)
```
As you can see above, our network has basically no idea what this digit is. It's because we haven't trained it yet, all the weights are random!
### Using `nn.Sequential`
PyTorch provides a convenient way to build networks like this where a tensor is passed sequentially through operations, `nn.Sequential` ([documentation](https://pytorch.org/docs/master/nn.html#torch.nn.Sequential)). Using this to build the equivalent network:
```
# Hyperparameters for our network
input_size = 784
hidden_sizes = [128, 64]
output_size = 10
# Build a feed-forward network
model = nn.Sequential(nn.Linear(input_size, hidden_sizes[0]),
nn.ReLU(),
nn.Linear(hidden_sizes[0], hidden_sizes[1]),
nn.ReLU(),
nn.Linear(hidden_sizes[1], output_size),
nn.Softmax(dim=1))
print(model)
# Forward pass through the network and display output
images, labels = next(iter(trainloader))
images.resize_(images.shape[0], 1, 784)
ps = model.forward(images[0,:])
helper.view_classify(images[0].view(1, 28, 28), ps)
```
Here our model is the same as before: 784 input units, a hidden layer with 128 units, ReLU activation, 64 unit hidden layer, another ReLU, then the output layer with 10 units, and the softmax output.
The operations are available by passing in the appropriate index. For example, if you want to get first Linear operation and look at the weights, you'd use `model[0]`.
```
print(model[0])
model[0].weight
```
You can also pass in an `OrderedDict` to name the individual layers and operations, instead of using incremental integers. Note that dictionary keys must be unique, so _each operation must have a different name_.
```
from collections import OrderedDict
model = nn.Sequential(OrderedDict([
('fc1', nn.Linear(input_size, hidden_sizes[0])),
('relu1', nn.ReLU()),
('fc2', nn.Linear(hidden_sizes[0], hidden_sizes[1])),
('relu2', nn.ReLU()),
('output', nn.Linear(hidden_sizes[1], output_size)),
('softmax', nn.Softmax(dim=1))]))
model
```
Now you can access layers either by integer or the name
```
print(model[0])
print(model.fc1)
```
In the next notebook, we'll see how we can train a neural network to accuractly predict the numbers appearing in the MNIST images.
| github_jupyter |
# ------------ First A.I. activity ------------
## 1. IBOVESPA volume prediction
-> Importing libraries that are going to be used in the code
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
```
-> Importing the datasets
```
dataset = pd.read_csv("datasets/ibovespa.csv",delimiter = ";")
```
-> Converting time to datetime in order to make it easy to manipulate
```
dataset['Data/Hora'] = dataset['Data/Hora'].str.replace("/","-")
dataset['Data/Hora'] = pd.to_datetime(dataset['Data/Hora'])
```
-> Visualizing the data
```
dataset.head()
```
-> creating date dataframe and splitting its features
date = dataset.iloc[:,0:1]
date['day'] = date['Data/Hora'].dt.day
date['month'] = date['Data/Hora'].dt.month
date['year'] = date['Data/Hora'].dt.year
date = date.drop(columns = ['Data/Hora'])
-> removing useless columns
```
dataset = dataset.drop(columns = ['Data/Hora','Unnamed: 7','Unnamed: 8','Unnamed: 9'])
```
-> transforming atributes to the correct format
```
for key, value in dataset.head().iteritems():
dataset[key] = dataset[key].str.replace(".","").str.replace(",",".").astype(float)
"""
for key, value in date.head().iteritems():
dataset[key] = date[key]
"""
```
-> Means
```
dataset.mean()
```
-> plotting graphics
```
plt.boxplot(dataset['Volume'])
plt.title('boxplot')
plt.xlabel('volume')
plt.ylabel('valores')
plt.ticklabel_format(style='sci', axis='y', useMathText = True)
dataset['Maxima'].median()
dataset['Minima'].mean()
```
-> Média truncada
```
from scipy import stats
m = stats.trim_mean(dataset['Minima'], 0.1)
print(m)
```
-> variancia e standard deviation
```
v = dataset['Cotacao'].var()
print(v)
d = dataset['Cotacao'].std()
print(v)
m = dataset['Cotacao'].mean()
print(m)
```
-> covariancia dos atributos, mas antes fazer uma standard scaler pra facilitar a visão e depois transforma de volta pra dataframe pandas
#### correlation shows us the relationship between the two variables and how are they related while covariance shows us how the two variables vary from each other.
```
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
dataset_cov = sc.fit_transform(dataset)
dataset_cov = pd.DataFrame(dataset_cov)
dataset_cov.cov()
```
-> plotting the graph may be easier to observe the correlation
```
corr = dataset.corr()
corr.style.background_gradient(cmap = 'coolwarm')
pd.plotting.scatter_matrix(dataset, figsize=(6, 6))
plt.show()
plt.matshow(dataset.corr())
plt.xticks(range(len(dataset.columns)), dataset.columns)
plt.yticks(range(len(dataset.columns)), dataset.columns)
plt.colorbar()
plt.show()
```
| github_jupyter |
# Exercise 02 - Functions and Getting Help !
## 1. Complete Your Very First Function
Complete the body of the following function according to its docstring.
*HINT*: Python has a builtin function `round`
```
def round_to_two_places(num):
"""Return the given number rounded to two decimal places.
>>> round_to_two_places(3.14159)
3.14
"""
# Replace this body with your own code.
# ("pass" is a keyword that does literally nothing. We used it as a placeholder,
# so that it will not raise any errors,
# because after we begin a code block, Python requires at least one line of code)
pass
def round_to_two_places(num):
num = round(num,2)
print('The number after rounded to two decimal places is: ', num)
round_to_two_places(3.4455)
```
## 2. Explore the Built-in Function
The help for `round` says that `ndigits` (the second argument) may be negative.
What do you think will happen when it is? Try some examples in the following cell?
Can you think of a case where this would be useful?
```
print(round(122.3444,-3))
print(round(122.3456,-2))
print(round(122.5454,-1))
print(round(122.13432,0))
#round with ndigits <=0 - the rounding will begin from the decimal point to the left
```
## 3. More Function
Giving the problem of candy-sharing friends Alice, Bob and Carol tried to split candies evenly. For the sake of their friendship, any candies left over would be smashed. For example, if they collectively bring home 91 candies, they will take 30 each and smash 1.
Below is a simple function that will calculate the number of candies to smash for *any* number of total candies.
**Your task**:
- Modify it so that it optionally takes a second argument representing the number of friends the candies are being split between. If no second argument is provided, it should assume 3 friends, as before.
- Update the docstring to reflect this new behaviour.
```
def to_smash(total_candies,n = 3):
"""Return the number of leftover candies that must be smashed after distributing
the given number of candies evenly between 3 friends.
>>> to_smash(91)
1
"""
return total_candies % n
print('#no. of candies to smash = ', to_smash(31))
print('#no. of candies to smash = ', to_smash(32,5))
```
## 4. Taste some Errors
It may not be fun, but reading and understanding **error messages** will help you improve solving problem skills.
Each code cell below contains some commented-out buggy code. For each cell...
1. Read the code and predict what you think will happen when it's run.
2. Then uncomment the code and run it to see what happens. *(**Tips**: In the kernel editor, you can highlight several lines and press `ctrl`+`/` to toggle commenting.)*
3. Fix the code (so that it accomplishes its intended purpose without throwing an exception)
<!-- TODO: should this be autochecked? Delta is probably pretty small. -->
```
round_to_two_places(9.9999)
x = -10
y = 5
# Which of the two variables above has the smallest absolute value?
smallest_abs = min(abs(x),abs(y))
print(smallest_abs)
def f(x):
y = abs(x)
return y
print(f(5))
```
## 5. More and more Functions
For this question, we'll be using two functions imported from Python's `time` module.
### Time Function
The [time](https://docs.python.org/3/library/time.html#time.time) function returns the number of seconds that have passed since the Epoch (aka [Unix time](https://en.wikipedia.org/wiki/Unix_time)).
<!-- We've provided a function called `seconds_since_epoch` which returns the number of seconds that have passed since the Epoch (aka [Unix time](https://en.wikipedia.org/wiki/Unix_time)). -->
Try it out below. Each time you run it, you should get a slightly larger number.
```
# Importing the function 'time' from the module of the same name.
# (We'll discuss imports in more depth later)
from time import time
t = time()
print(t, "seconds since the Epoch")
```
### Sleep Function
We'll also be using a function called [sleep](https://docs.python.org/3/library/time.html#time.sleep), which makes us wait some number of seconds while it does nothing particular. (Sounds useful, right?)
You can see it in action by running the cell below:
```
from time import sleep
duration = 5
print("Getting sleepy. See you in", duration, "seconds")
sleep(duration)
print("I'm back. What did I miss?")
```
### Your Own Function
With the help of these functions, complete the function **`time_call`** below according to its docstring.
<!-- (The sleep function will be useful for testing here since we have a pretty good idea of what something like `time_call(sleep, 1)` should return.) -->
```
def time_call(fn, arg):
"""Return the amount of time the given function takes (in seconds) when called with the given argument.
"""
from time import time
start_time = time()
fn(arg)
end_time = time()
duration = end_time - start_time
return duration
```
How would you verify that `time_call` is working correctly? Think about it...
```
#solution? use sleep function?
```
## 6. 🌶️ Reuse your Function
*Note: this question depends on a working solution to the previous question.*
Complete the function below according to its docstring.
```
def slowest_call(fn, arg1, arg2, arg3):
"""Return the amount of time taken by the slowest of the following function
calls: fn(arg1), fn(arg2), fn(arg3)
"""
slowest = min(time_call(fn, arg1), time_call(fn, arg2), time_call(fn,arg3))
return slowest
print(slowest_call(sleep,1,2,3))
```
# Keep Going
| github_jupyter |
```
import matplotlib.pyplot as plt
import netCDF4 as nc
import numpy as np
from salishsea_tools import geo_tools
%matplotlib inline
bathyfile = '/home/sallen/MEOPAR/grid/bathymetry_201702.nc'
meshfile = '/home/sallen/MEOPAR/grid/mesh_mask201702.nc'
mesh = nc.Dataset(meshfile)
model_lats = nc.Dataset(bathyfile).variables['nav_lat'][:]
model_lons = nc.Dataset(bathyfile).variables['nav_lon'][:]
t_mask = mesh.variables['tmask'][0, 0]
windfile = './ubcSSaAtmosphereGridV1_0f03_6268_df4b.nc'
wind_lats = nc.Dataset(windfile).variables['latitude'][:]
wind_lons = nc.Dataset(windfile).variables['longitude'][:] -360
wavefile = '/results/SalishSea/wwatch3-forecast/SoG_ww3_fields_20170515_20170517.nc'
wave_lats = nc.Dataset(wavefile).variables['latitude'][:]
wave_lons = nc.Dataset(wavefile).variables['longitude'][:] -360.
wave_lons, wave_lats = np.meshgrid(wave_lons, wave_lats)
hs = nc.Dataset(wavefile).variables['hs'][0]
wave_mask = np.where(hs !=0, 1, 0)
def get_tidal_stations(lon, lat, model_lons, model_lats, wind_lons, wind_lats,
wave_lons, wave_lats, t_mask, wave_mask, size=20):
y, x = geo_tools.find_closest_model_point(lon, lat, model_lons, model_lats, grid='NEMO', land_mask=1-t_mask)
ywind, xwind = geo_tools.find_closest_model_point(lon, lat, wind_lons, wind_lats, grid='GEM2.5')
ywave, xwave = geo_tools.find_closest_model_point(lon, lat, wave_lons, wave_lats, grid='NEMO', land_mask=1-wave_mask)
fig, ax = plt.subplots(1, 1, figsize=(7, 7))
bigx = min(x+size, model_lons.shape[1]-1)
imin, imax = model_lats[y-size, x-size], model_lats[y+size, bigx]
jmin, jmax = model_lons[y+size, x-size], model_lons[y-size, bigx]
dlon = model_lons[y+1, x+1] - model_lons[y, x]
dlat = model_lats[y+1, x+1] - model_lats[y, x]
ax.pcolormesh(model_lons - dlon/2., model_lats-dlat/2., t_mask, cmap='Greys_r')
ax.set_xlim(jmin, jmax)
ax.set_ylim(imin, imax)
ax.plot(model_lons[y, x], model_lats[y, x], 'ro', label='NEMO')
ax.plot(wind_lons[ywind, xwind], wind_lats[ywind, xwind], 'ys', label='GEM2.5')
ax.plot(wave_lons[ywave, xwave], wave_lats[ywave, xwave], 'bo', label='WW3')
ax.legend()
return "NEMO y, x: {0}, Wind y, x: {1}, Wave y, x: {2}".format([y, x], [ywind, xwind], [ywave, xwave])
```
### Patricia Bay
7277 Patricia Bay 48.6536 123.4515
```
get_tidal_stations(-123.4515, 48.6536, model_lons, model_lats, wind_lons, wind_lats,
wave_lons, wave_lats, t_mask, wave_mask, size=10)
```
### Woodwards
7610 Woodwards's Landing 49.1251 123.0754
```
get_tidal_stations(-123.0754, 49.1251, model_lons, model_lats, wind_lons, wind_lats,
wave_lons, wave_lats, t_mask, wave_mask, size=10)
```
### New Westminster
7654 New Westminster 49.203683 122.90535
```
get_tidal_stations(-122.90535, 49.203683, model_lons, model_lats, wind_lons, wind_lats,
wave_lons, wave_lats, t_mask, wave_mask, size=10)
```
### Sandy Cove
7786 Sandy Cove 49.34 123.23
```
get_tidal_stations(-123.23, 49.34, model_lons, model_lats, wind_lons, wind_lats,
wave_lons, wave_lats, t_mask, wave_mask, size=10)
```
### Port Renfrew
check
8525 Port Renfrew 48.555 124.421
```
get_tidal_stations(-124.421, 48.555, model_lons, model_lats, wind_lons, wind_lats,
wave_lons, wave_lats, t_mask, wave_mask, size=10)
```
### Victoria
7120 Victoria 48.424666 123.3707
```
get_tidal_stations(-123.3707, 48.424666, model_lons, model_lats, wind_lons, wind_lats,
wave_lons, wave_lats, t_mask, wave_mask, size=10)
```
### Sand Heads
7594 Sand Heads 49.125 123.195
From Marlene's email
49º 06’ 21.1857’’, -123º 18’ 12.4789’’
we are using 426, 292
end of jetty is 429, 295
```
lat_sh = 49+6/60.+21.1857/3600.
lon_sh = -(123+18/60.+12.4789/3600.)
print(lon_sh, lat_sh)
get_tidal_stations(lon_sh, lat_sh, model_lons, model_lats, wind_lons, wind_lats,
wave_lons, wave_lats, t_mask, wave_mask, size=20)
```
### Nanaimo
7917 Nanaimo 49.17 123.93
```
get_tidal_stations(-123.93, 49.17, model_lons, model_lats, wind_lons, wind_lats,
wave_lons, wave_lats, t_mask, wave_mask, size=10)
```
In our code its at 484, 208 with lon,lat at -123.93 and 49.16: leave as is for now
### Boundary Bay
Guesstimated from Map
-122.925 49.0
```
get_tidal_stations(-122.925, 49.0, model_lons, model_lats, wind_lons, wind_lats,
wave_lons, wave_lats, t_mask, wave_mask, size=15)
```
### Squamish
49 41.675 N 123 09.299 W
```
print (49+41.675/60, -(123+9.299/60.))
print (model_lons.shape)
get_tidal_stations(-(123+9.299/60.), 49.+41.675/60., model_lons, model_lats, wind_lons, wind_lats,
wave_lons, wave_lats, t_mask, wave_mask, size=10)
```
### Half Moon Bay
49 30.687 N 123 54.726 W
```
print (49+30.687/60, -(123+54.726/60.))
get_tidal_stations(-(123+54.726/60.), 49.+30.687/60., model_lons, model_lats, wind_lons, wind_lats,
wave_lons, wave_lats, t_mask, wave_mask, size=10)
```
### Friday Harbour
-123.016667, 48.55
```
get_tidal_stations(-123.016667, 48.55, model_lons, model_lats, wind_lons, wind_lats,
wave_lons, wave_lats, t_mask, wave_mask, size=10)
```
### Neah Bay
-124.6, 48.4
```
get_tidal_stations(-124.6, 48.4, model_lons, model_lats, wind_lons, wind_lats,
wave_lons, wave_lats, t_mask, wave_mask, size=10)
from salishsea_tools import places
```
| github_jupyter |
# Plotting massive data sets
This notebook plots about half a million LIDAR points around Toronto from the KITTI data set. ([Source](http://www.cvlibs.net/datasets/kitti/raw_data.php)) The data is meant to be played over time. With pydeck, we can render these points and interact with them.
### Cleaning the data
First we need to import the data. Each row of data represents one x/y/z coordinate for a point in space at a point in time, with each frame representing about 115,000 points.
We also need to scale the points to plot closely on a map. These point coordinates are not given in latitude and longitude, so as a workaround we'll plot them very close to (0, 0) on the earth.
In future versions of pydeck other viewports, like a flat plane, will be supported out-of-the-box. For now, we'll make do with scaling the points.
```
import pandas as pd
all_lidar = pd.concat([
pd.read_csv('https://raw.githubusercontent.com/ajduberstein/kitti_subset/master/kitti_1.csv'),
pd.read_csv('https://raw.githubusercontent.com/ajduberstein/kitti_subset/master/kitti_2.csv'),
pd.read_csv('https://raw.githubusercontent.com/ajduberstein/kitti_subset/master/kitti_3.csv'),
pd.read_csv('https://raw.githubusercontent.com/ajduberstein/kitti_subset/master/kitti_4.csv'),
])
# Filter to one frame of data
lidar = all_lidar[all_lidar['source'] == 136]
lidar.loc[: , ['x', 'y']] = lidar[['x', 'y']] / 10000
```
### Plotting the data
We'll define a single `PointCloudLayer` and plot it.
Pydeck by default expects the input of `get_position` to be a string name indicating a single position value. For convenience, you can pass in a string indicating the X/Y/Z coordinate, here `get_position='[x, y, z]'`. You also have access to a small expression parser--in our `get_position` function here, we increase the size of the z coordinate times 10.
Using `pydeck.data_utils.compute_view`, we'll zoom to the approximate center of the data.
```
import pydeck as pdk
point_cloud = pdk.Layer(
'PointCloudLayer',
lidar[['x', 'y', 'z']],
get_position=['x', 'y', 'z * 10'],
get_normal=[0, 0, 1],
get_color=[255, 0, 100, 200],
pickable=True,
auto_highlight=True,
point_size=1)
view_state = pdk.data_utils.compute_view(lidar[['x', 'y']], 0.9)
view_state.max_pitch = 360
view_state.pitch = 80
view_state.bearing = 120
r = pdk.Deck(
point_cloud,
initial_view_state=view_state,
map_provider=None,
)
r.show()
import time
from collections import deque
# Choose a handful of frames to loop through
frame_buffer = deque([42, 56, 81, 95])
print('Press the stop icon to exit')
while True:
current_frame = frame_buffer[0]
lidar = all_lidar[all_lidar['source'] == current_frame]
r.layers[0].get_position = '@@=[x / 10000, y / 10000, z * 10]'
r.layers[0].data = lidar.to_dict(orient='records')
frame_buffer.rotate()
r.update()
time.sleep(0.5)
```
| github_jupyter |
# Seq2Seq with Attention for Korean-English Neural Machine Translation
- Network architecture based on this [paper](https://arxiv.org/abs/1409.0473)
- Fit to run on Google Colaboratory
```
import os
import io
import tarfile
import torch
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
import torchtext
from torchtext.data import Dataset
from torchtext.data import Example
from torchtext.data import Field
from torchtext.data import BucketIterator
```
# 1. Upload Data to Colab Workspace
로컬에 존재하는 다음 3개의 데이터를 가상 머신에 업로드. 파일의 원본은 [여기](https://github.com/jungyeul/korean-parallel-corpora/tree/master/korean-english-news-v1/)에서도 확인
- korean-english-park.train.tar.gz
- korean-english-park.dev.tar.gz
- korean.english-park.test.tar.gz
```
# 현재 작업경로를 확인 & 'data' 폴더 생성
!echo 'Current working directory:' ${PWD}
!mkdir -p data/
!ls -al
# 로컬의 데이터 업로드
from google.colab import files
uploaded = files.upload()
# 'data' 폴더 하위로 이동, 잘 옮겨졌는지 확인
!mv *.tar.gz data/
!ls -al data/
```
# 2. Check Packages
## KoNLPy (설치 필요)
```
# Java 1.8 & KoNLPy 설치
!apt-get update
!apt-get install g++ openjdk-8-jdk python-dev python3-dev
!pip3 install JPype1-py3
!pip3 install konlpy
from konlpy.tag import Okt
ko_tokens = Okt().pos('트위터 데이터로 학습한 형태소 분석기가 잘 실행이 되는지 확인해볼까요?') # list of (word, POS TAG) tuples
ko_tokens = [t[0] for t in ko_tokens] # Only get words
print(ko_tokens)
del ko_tokens # 필요 없으니까 삭제
```
## Spacy (이미 설치되어 있음)
```
# 설치가 되어있는지 확인
!pip show spacy
# 설치가 되어있는지 확인 (없다면 자동설치됨)
!python -m spacy download en_core_web_sm
import spacy
spacy_en = spacy.load('en_core_web_sm')
en_tokens = [t.text for t in spacy_en.tokenizer('Check that spacy tokenizer works.')]
print(en_tokens)
del en_tokens # 필요 없으니까 삭제
```
# 3. Define Tokenizing Functions
문장을 받아 그보다 작은 어절 혹은 형태소 단위의 리스트로 반환해주는 함수를 각 언어에 대해 작성
- Korean: konlpy.tag.Okt() <- Twitter()에서 명칭변경
- English: spacy.tokenizer
## Korean Tokenizer
```
#from konlpy.tag import Okt
class KoTokenizer(object):
"""For Korean."""
def __init__(self):
self.tokenizer = Okt()
def tokenize(self, text):
tokens = self.tokenizer.pos(text)
tokens = [t[0] for t in tokens]
return tokens
# Usage example
print(KoTokenizer().tokenize('전처리는 언제나 지겨워요.'))
```
## English Tokenizer
```
#import spacy
class EnTokenizer(object):
"""For English."""
def __init__(self):
self.spacy_en = spacy.load('en_core_web_sm')
def tokenize(self, text):
tokens = [t.text for t in self.spacy_en.tokenizer(text)]
return tokens
# Usage example
print(EnTokenizer().tokenize("What I cannot create, I don't understand."))
```
# 4. Data Preprocessing
## Load data
```
# Current working directory & list of files
!echo 'Current working directory:' ${PWD}
!ls -al
DATA_DIR = './data/'
print('Data directory exists:', os.path.isdir(DATA_DIR))
print('List of files:')
print(*os.listdir(DATA_DIR), sep='\n')
def get_data_from_tar_gz(filename):
"""
Retrieve contents from a `tar.gz` file without extraction.
Arguments:
filename: path to `tar.gz` file.
Returns:
dict, (name, content) pairs
"""
assert os.path.exists(filename)
out = {}
with tarfile.open(filename, 'r:gz') as tar:
for member in tar.getmembers():
lang = member.name.split('.')[-1] # ex) korean-english-park.train.ko -> ko
f = tar.extractfile(member)
if f is not None:
content = f.read().decode('utf-8')
content = content.splitlines()
out[lang] = content
assert isinstance(out, dict)
return out
# Each 'xxx_data' is a dictionary with keys; 'ko', 'en'
train_dict= get_data_from_tar_gz(os.path.join(DATA_DIR, 'korean-english-park.train.tar.gz')) # train
dev_dict = get_data_from_tar_gz(os.path.join(DATA_DIR, 'korean-english-park.dev.tar.gz')) # dev
test_dict = get_data_from_tar_gz(os.path.join(DATA_DIR, 'korean-english-park.test.tar.gz')) # test
# Some samples (ko)
train_dict['ko'][100:105]
# Some samples (en)
train_dict['en'][100:105]
```
## Define Datasets
```
#from torchtext.data import Dataset
#from torchtext.data import Example
class KoEnTranslationDataset(Dataset):
"""A dataset for Korean-English Neural Machine Translation."""
@staticmethod
def sort_key(ex):
return torchtext.data.interleave_keys(len(ex.src), len(ex.trg))
def __init__(self, data_dict, field_dict, source_lang='ko', max_samples=None, **kwargs):
"""
Only 'ko' and 'en' supported for `language`
Arguments:
data_dict: dict of (`language`, text) pairs.
field_dict: dict of (`language`, Field instance) pairs.
source_lang: str, default 'ko'.
Other kwargs are passed to the constructor of `torchtext.data.Dataset`.
"""
if not all(k in ['ko', 'en'] for k in data_dict.keys()):
raise KeyError("Check data keys.")
if not all(k in ['ko', 'en'] for k in field_dict.keys()):
raise KeyError("Check field keys.")
if source_lang == 'ko':
fields = [('src', field_dict['ko']), ('trg', field_dict['en'])]
src_data = data_dict['ko']
trg_data = data_dict['en']
elif source_lang == 'en':
fields = [('src', field_dict['en']), ('trg', field_dict['ko'])]
src_data = data_dict['en']
trg_data = data_dict['ko']
else:
raise NotImplementedError
if not len(src_data) == len(trg_data):
raise ValueError('Inconsistent number of instances between two languages.')
examples = []
for i, (src_line, trg_line) in enumerate(zip(src_data, trg_data)):
src_line = src_line.strip()
trg_line = trg_line.strip()
if src_line != '' and trg_line != '':
examples.append(
torchtext.data.Example.fromlist(
[src_line, trg_line], fields
)
)
i += 1
if max_samples is not None:
if i >= max_samples:
break
super(KoEnTranslationDataset, self).__init__(examples, fields, **kwargs)
```
## Define Fields
- Instantiate tokenizers; one for each language.
- The 'tokenize' argument of `Field` requires a tokenizing function.
```
#from torchtext.data import Field
ko_tokenizer = KoTokenizer() # korean tokenizer
en_tokenizer = EnTokenizer() # english tokenizer
# Field instance for korean
KOREAN = Field(
init_token='<sos>',
eos_token='<eos>',
tokenize=ko_tokenizer.tokenize,
batch_first=True,
lower=False
)
# Field instance for english
ENGLISH = Field(
init_token='<sos>',
eos_token='<eos>',
tokenize=en_tokenizer.tokenize,
batch_first=True,
lower=True
)
# Store Field instances in a dictionary
field_dict = {
'ko': KOREAN,
'en': ENGLISH,
}
```
## Instantiate datasets
- one for each set (train, dev, test)
```
# 학습시간 단축을 위해 학습 데이터 줄이기
MAX_TRAIN_SAMPLES = 10000
# Instantiate with data
train_set = KoEnTranslationDataset(train_dict, field_dict, max_samples=MAX_TRAIN_SAMPLES)
print('Train set ready.')
print('#. examples:', len(train_set.examples))
dev_set = KoEnTranslationDataset(dev_dict, field_dict)
print('Dev set ready...')
print('#. examples:', len(dev_set.examples))
test_set = KoEnTranslationDataset(test_dict, field_dict)
print('Test set ready...')
print('#. examples:', len(test_set.examples))
# Training example (KO, source language)
train_set.examples[50].src
# Training example (EN, target language)
train_set.examples[50].trg
```
## Build Vocabulary
- 각 언어별 생성: `Field`의 인스턴스를 활용
- 최소 빈도수(`MIN_FREQ`) 값을 작게 하면 vocabulary의 크기가 커짐.
- 최소 빈도수(`MIN_FREQ`) 값을 크게 하면 vocabulary의 크기가 작아짐.
```
MIN_FREQ = 2 # TODO: try different values
# Build vocab for Korean
KOREAN.build_vocab(train_set, dev_set, test_set, min_freq=MIN_FREQ) # ko
print('Size of source vocab (ko):', len(KOREAN.vocab))
# Check indices of some important tokens
tokens = ['<unk>', '<pad>', '<sos>', '<eos>']
for token in tokens:
print(f"{token} -> {KOREAN.vocab.stoi[token]}")
# Build vocab for English
ENGLISH.build_vocab(train_set, dev_set, test_set, min_freq=MIN_FREQ) # en
print('Size of target vocab (en):', len(ENGLISH.vocab))
# Check indices of some important tokens
tokens = ['<unk>', '<pad>', '<sos>', '<eos>']
for token in tokens:
print(f"{token} -> {KOREAN.vocab.stoi[token]}")
```
## Configure Device
- *'런타임' -> '런타임 유형변경'* 에서 하드웨어 가속기로 **GPU** 선택
```
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
print('Device to use:', device)
```
## Create Data Iterators
- 데이터를 미니배치(mini-batch) 단위로 반환해주는 역할
- `train_set`, `dev_set`, `test_set`에 대해 개별적으로 정의해야 함
- `BATCH_SIZE`를 정의해주어야 함
- `torchtext.data.BucketIterator`는 하나의 미니배치를 서로 비슷한 길이의 관측치들로 구성함
- [Bucketing](https://medium.com/@rashmi.margani/how-to-speed-up-the-training-of-the-sequence-model-using-bucketing-techniques-9e302b0fd976)의 효과: 하나의 미니배치 내 padding을 최소화하여 연산의 낭비를 줄여줌
```
BATCH_SIZE = 128
#from torchtext.data import BucketIterator
# Train iterator
train_iterator = BucketIterator(
train_set,
batch_size=BATCH_SIZE,
train=True,
shuffle=True,
device=device
)
print(f'Number of minibatches per epoch: {len(train_iterator)}')
#from torchtext.data import BucketIterator
# Dev iterator
dev_iterator = BucketIterator(
dev_set,
batch_size=100,
train=False,
shuffle=False,
device=device
)
print(f'Number of minibatches per epoch: {len(dev_iterator)}')
#from torchtext.data import BucketIterator
# Test iterator
test_iterator = BucketIterator(
test_set,
batch_size=200,
train=False,
shuffle=False,
device=device
)
print(f'Number of minibatches per epoch: {len(test_iterator)}')
train_batch = next(iter(train_iterator))
print('a batch of source examples has shape:', train_batch.src.size()) # (b, s)
print('a batch of target examples has shape:', train_batch.trg.size()) # (b, s)
# Checking first sample in mini-batch (KO, source lang)
ko_indices = train_batch.src[0]
ko_tokens = [KOREAN.vocab.itos[i] for i in ko_indices]
for t, i in zip(ko_tokens, ko_indices):
print(f"{t} ({i})")
del ko_indices, ko_tokens
# Checking first sample in mini-batch (EN, target lang)
en_indices = train_batch.trg[0]
en_tokens = [ENGLISH.vocab.itos[i] for i in en_indices]
for t, i in zip(en_tokens, en_indices):
print(f"{t} ({i})")
del en_indices, en_tokens
del train_batch # 더 이상 필요 없으니까 삭제
```
# 5. Building Seq2Seq Model
## Hyperparameters
```
# Hyperparameters
INPUT_DIM = len(KOREAN.vocab)
OUTPUT_DIM = len(ENGLISH.vocab)
ENC_EMB_DIM = DEC_EMB_DIM = 100
ENC_HID_DIM = DEC_HID_DIM = 60
USE_BIDIRECTIONAL = False
```
## Encoder
```
class Encoder(nn.Module):
"""
Learns an embedding for the source text.
Arguments:
input_dim: int, size of input language vocabulary.
emb_dim: int, size of embedding layer output.
enc_hid_dim: int, size of encoder hidden state.
dec_hid_dim: int, size of decoder hidden state.
bidirectional: uses bidirectional RNNs if True. default is False.
"""
def __init__(self, input_dim, emb_dim, enc_hid_dim, dec_hid_dim, bidirectional=False):
super(Encoder, self).__init__()
self.input_dim = input_dim
self.emb_dim = emb_dim
self.enc_hid_dim = enc_hid_dim
self.dec_hid_dim = dec_hid_dim
self.bidirectional = bidirectional
self.embedding = nn.Embedding(
num_embeddings=self.input_dim,
embedding_dim=self.emb_dim
)
self.rnn = nn.GRU(
input_size=self.emb_dim,
hidden_size=self.enc_hid_dim,
bidirectional=self.bidirectional,
batch_first=True
)
self.rnn_output_dim = self.enc_hid_dim
if self.bidirectional:
self.rnn_output_dim *= 2
self.fc = nn.Linear(self.rnn_output_dim, self.dec_hid_dim)
self.dropout = nn.Dropout(.2)
def forward(self, src):
"""
Arguments:
src: 2d tensor of shape (batch_size, input_seq_len)
Returns:
outputs: 3d tensor of shape (batch_size, input_seq_len, num_directions * enc_h)
hidden: 2d tensor of shape (b, dec_h). This tensor will be used as the initial
hidden state value of the decoder (h0 of decoder).
"""
assert len(src.size()) == 2, 'Input requires dimension (batch_size, seq_len).'
# Shape: (b, s, h)
embedded = self.embedding(src)
embedded = self.dropout(embedded)
outputs, hidden = self.rnn(embedded)
if self.bidirectional:
# (2, b, enc_h) -> (b, 2 * enc_h)
hidden = torch.cat((hidden[-2, :, :], hidden[-1, :, :]), dim=1)
else:
# (1, b, enc_h) -> (b, enc_h)
hidden = hidden.squeeze(0)
# (b, num_directions * enc_h) -> (b, dec_h)
hidden = self.fc(hidden)
hidden = torch.tanh(hidden)
return outputs, hidden
```
## Attention
```
class Attention(nn.Module):
def __init__(self, enc_hid_dim, dec_hid_dim, encoder_is_bidirectional=False):
super(Attention, self).__init__()
self.enc_hid_dim = enc_hid_dim
self.dec_hid_dim = dec_hid_dim
self.encoder_is_bidirectional = encoder_is_bidirectional
self.attention_input_dim = enc_hid_dim + dec_hid_dim
if self.encoder_is_bidirectional:
self.attention_input_dim += enc_hid_dim # 2 * h_enc + h_dec
self.linear = nn.Linear(self.attention_input_dim, dec_hid_dim)
self.v = nn.Parameter(torch.rand(dec_hid_dim))
def forward(self, hidden, encoder_outputs):
"""
Arguments:
hidden: 2d tensor with shape (batch_size, dec_hid_dim).
encoder_outputs: 3d tensor with shape (batch_size, input_seq_len, enc_hid_dim).
if encoder is bidirectional, expects (batch_size, input_seq_len, 2 * enc_hid_dim).
"""
# Shape check
assert hidden.dim() == 2
assert encoder_outputs.dim() == 3
batch_size, seq_len, _ = encoder_outputs.size()
# (b, dec_h) -> (b, s, dec_h)
hidden = hidden.unsqueeze(1).expand(-1, seq_len, -1)
# concat; shape results in (b, s, enc_h + dec_h).
# if encoder is bidirectional, (b, s, 2 * h_enc + h_dec).
concat = torch.cat((hidden, encoder_outputs), dim=2)
# concat; shape is (b, s, dec_h)
concat = self.linear(concat)
concat = torch.tanh(concat)
# tile v; (dec_h, ) -> (b, dec_h, 1)
v = self.v.repeat(batch_size, 1).unsqueeze(2)
# attn; (b, s, dec_h) @ (b, dec_h, 1) -> (b, s, 1) -> (b, s)
attn_scores = torch.bmm(concat, v).squeeze(-1)
assert attn_scores.dim() == 2 # Final shape check: (b, s)
return F.softmax(attn_scores, dim=1)
```
## Decoder
```
class Decoder(nn.Module):
"""
Unlike the encoder, a single forward pass of
a `Decoder` instance is defined for only a single timestep.
Arguments:
output_dim: int,
emb_dim: int,
enc_hid_dim: int,
dec_hid_dim: int,
attention_module: torch.nn.Module,
encoder_is_bidirectional: False
"""
def __init__(self, output_dim, emb_dim, enc_hid_dim, dec_hid_dim, attention_module, encoder_is_bidirectional=False):
super(Decoder, self).__init__()
self.emb_dim = emb_dim
self.enc_hid_dim = enc_hid_dim
self.dec_hid_dim = dec_hid_dim
self.output_dim = output_dim
self.encoder_is_bidirectional = encoder_is_bidirectional
if isinstance(attention_module, nn.Module):
self.attention_module = attention_module
else:
raise ValueError
self.rnn_input_dim = enc_hid_dim + emb_dim # enc_h + dec_emb_dim
if self.encoder_is_bidirectional:
self.rnn_input_dim += enc_hid_dim # 2 * enc_h + dec_emb_dim
self.embedding = nn.Embedding(output_dim, emb_dim)
self.rnn = nn.GRU(
input_size=self.rnn_input_dim,
hidden_size=dec_hid_dim,
bidirectional=False,
batch_first=True,
)
out_input_dim = 2 * dec_hid_dim + emb_dim # hidden + dec_hidden_dim + dec_emb_dim
self.out = nn.Linear(out_input_dim, output_dim)
self.dropout = nn.Dropout(.2)
def forward(self, inp, hidden, encoder_outputs):
"""
Arguments:
inp: 1d tensor with shape (batch_size, )
hidden: 2d tensor with shape (batch_size, dec_hid_dim).
This `hidden` tensor is the hidden state vector from the previous timestep.
encoder_outputs: 3d tensor with shape (batch_size, seq_len, enc_hid_dim).
If encoder_is_bidirectional is True, expects shape (batch_size, seq_len, 2 * enc_hid_dim).
"""
assert inp.dim() == 1
assert hidden.dim() == 2
assert encoder_outputs.dim() == 3
# (batch_size, ) -> (batch_size, 1)
inp = inp.unsqueeze(1)
# (batch_size, 1) -> (batch_size, 1, emb_dim)
embedded = self.embedding(inp)
embedded = self.dropout(embedded)
# attention probabilities; (batch_size, seq_len)
attn_probs = self.attention_module(hidden, encoder_outputs)
# (batch_size, 1, seq_len)
attn_probs = attn_probs.unsqueeze(1)
# (b, 1, s) @ (b, s, enc_hid_dim) -> (b, 1, enc_hid_dim)
weighted = torch.bmm(attn_probs, encoder_outputs)
# (batch_size, 1, emb_dim + enc_hid_dim)
rnn_input = torch.cat((embedded, weighted), dim=2)
# output; (batch_size, 1, dec_hid_dim)
# new_hidden; (1, batch_size, dec_hid_dim)
output, new_hidden = self.rnn(rnn_input, hidden.unsqueeze(0))
embedded = embedded.squeeze(1) # (b, 1, emb) -> (b, emb)
output = output.squeeze(1) # (b, 1, dec_h) -> (b, dec_h)
weighted = weighted.squeeze(1) # (b, 1, dec_h) -> (b, dec_h)
# output; (batch_size, emb + 2 * dec_h) -> (batch_size, output_dim)
output = self.out(torch.cat((output, weighted, embedded), dim=1))
return output, new_hidden.squeeze(0)
```
## Seq2Seq
```
class Seq2Seq(nn.Module):
def __init__(self, encoder, decoder, device):
super(Seq2Seq, self).__init__()
self.encoder = encoder
self.decoder = decoder
self.device = device
def forward(self, src, trg, teacher_forcing_ratio=.5):
batch_size, max_seq_len = trg.size()
trg_vocab_size = self.decoder.output_dim
# An empty tesnor to store decoder outputs (time index first for indexing)
outputs_shape = (max_seq_len, batch_size, trg_vocab_size)
outputs = torch.zeros(outputs_shape).to(self.device)
encoder_outputs, hidden = self.encoder(src)
# first input to the decoder is '<sos>'
# trg; shape (batch_size, seq_len)
initial_dec_input = output = trg[:, 0] # get first timestep token
for t in range(1, max_seq_len):
output, hidden = self.decoder(output, hidden, encoder_outputs)
outputs[t] = output # Save output for timestep t, for 1 <= t <= max_len
top1_val, top1_idx = output.max(dim=1)
teacher_force = torch.rand(1).item() >= teacher_forcing_ratio
output = trg[:, t] if teacher_force else top1_idx
# Switch batch and time dimensions for consistency (batch_first=True)
outputs = outputs.permute(1, 0, 2) # (s, b, trg_vocab) -> (b, s, trg_vocab)
return outputs
```
## Build Model
```
# Define encoder
enc = Encoder(
input_dim=INPUT_DIM,
emb_dim=ENC_EMB_DIM,
enc_hid_dim=ENC_HID_DIM,
dec_hid_dim=DEC_HID_DIM,
bidirectional=USE_BIDIRECTIONAL
)
print(enc)
# Define attention layer
attn = Attention(
enc_hid_dim=ENC_HID_DIM,
dec_hid_dim=DEC_HID_DIM,
encoder_is_bidirectional=USE_BIDIRECTIONAL
)
print(attn)
# Define decoder
dec = Decoder(
output_dim=OUTPUT_DIM,
emb_dim=DEC_EMB_DIM,
enc_hid_dim=ENC_HID_DIM,
dec_hid_dim=DEC_HID_DIM,
attention_module=attn,
encoder_is_bidirectional=USE_BIDIRECTIONAL
)
print(dec)
model = Seq2Seq(enc, dec, device).to(device)
print(model)
def count_parameters(model):
return sum(p.numel() for p in model.parameters() if p.requires_grad)
print(f'The model has {count_parameters(model):,} trainable parameters.')
```
# 6. Train
## Optimizer
- Use `optim.Adam` or `optim.RMSprop`.
```
optimizer = optim.Adam(model.parameters(), lr=0.001)
#optimizer = optim.RMSprop(model.parameters(), lr=0.01)
```
## Loss function
```
# Padding indices should not be considered when loss is calculated.
PAD_IDX = ENGLISH.vocab.stoi['<pad>']
criterion = nn.CrossEntropyLoss(ignore_index=PAD_IDX)
```
## Train function
```
def train(seq2seq_model, iterator, optimizer, criterion, grad_clip=1.0):
seq2seq_model.train()
epoch_loss = .0
for i, batch in enumerate(iterator):
print('.', end='')
src = batch.src
trg = batch.trg
optimizer.zero_grad()
decoder_outputs = seq2seq_model(src, trg, teacher_forcing_ratio=.5)
seq_len, batch_size, trg_vocab_size = decoder_outputs.size() # (b, s, trg_vocab)
# (b-1, s, trg_vocab)
decoder_outputs = decoder_outputs[:, 1:, :]
# ((b-1) * s, trg_vocab)
decoder_outputs = decoder_outputs.contiguous().view(-1, trg_vocab_size)
# ((b-1) * s, )
trg = trg[:, 1:].contiguous().view(-1)
loss = criterion(decoder_outputs, trg)
loss.backward()
# Gradient clipping; remedy for exploding gradients
torch.nn.utils.clip_grad_norm_(seq2seq_model.parameters(), grad_clip)
optimizer.step()
epoch_loss += loss.item()
return epoch_loss / len(iterator)
```
## Evaluate function
```
def evaluate(seq2seq_model, iterator, criterion):
seq2seq_model.eval()
epoch_loss = 0.
with torch.no_grad():
for i, batch in enumerate(iterator):
print('.', end='')
src = batch.src
trg = batch.trg
decoder_outputs = seq2seq_model(src, trg, teacher_forcing_ratio=0.)
seq_len, batch_size, trg_vocab_size = decoder_outputs.size() # (b, s, trg_vocab)
# (b-1, s, trg_vocab)
decoder_outputs = decoder_outputs[:, 1:, :]
# ((b-1) * s, trg_vocab)
decoder_outputs = decoder_outputs.contiguous().view(-1, trg_vocab_size)
# ((b-1) * s, )
trg = trg[:, 1:].contiguous().view(-1)
loss = criterion(decoder_outputs, trg)
epoch_loss += loss.item()
return epoch_loss / len(iterator)
```
## Epoch time measure function
```
def epoch_time(start_time, end_time):
"""Returns elapsed time in mins & secs."""
elapsed_time = end_time - start_time
elapsed_mins = int(elapsed_time / 60)
elapsed_secs = int(elapsed_time - (elapsed_mins * 60))
return elapsed_mins, elapsed_secs
```
## Train for multiple epochs
```
NUM_EPOCHS = 50
import time
import math
best_dev_loss = float('inf')
for epoch in range(NUM_EPOCHS):
start_time = time.time()
train_loss = train(model, train_iterator, optimizer, criterion)
dev_loss = evaluate(model, dev_iterator, criterion)
end_time = time.time()
epoch_mins, epoch_secs = epoch_time(start_time, end_time)
if dev_loss < best_dev_loss:
best_dev_loss = dev_loss
torch.save(model.state_dict(), './best_model.pt')
print("\n")
print(f"Epoch: {epoch + 1:>02d} | Time: {epoch_mins}m {epoch_secs}s")
print(f"Train Loss: {train_loss:>.4f} | Train Perplexity: {math.exp(train_loss):7.3f}")
print(f"Dev Loss: {dev_loss:>.4f} | Dev Perplexity: {math.exp(dev_loss):7.3f}")
```
## Save last model (overfitted)
```
torch.save(model.state_dict(), './last_model.pt')
```
# 7. Test
## Function to convert indices to original text strings
```
def indices_to_text(src_or_trg, lang_field):
assert src_or_trg.dim() == 1, f'{src_or_trg.dim()}' #(seq_len, )
assert isinstance(lang_field, torchtext.data.Field)
assert hasattr(lang_field, 'vocab')
return [lang_field.vocab.itos[t] for t in src_or_trg]
```
## Function to make predictions
- Returns a list of examples, where each example is a (src, trg, prediction) tuple.
```
def predict(seq2seq_model, iterator):
seq2seq_model.eval()
out = []
with torch.no_grad():
for i, batch in enumerate(iterator):
src = batch.src
trg = batch.trg
decoder_outputs = seq2seq_model(src, trg, teacher_forcing_ratio=0.)
seq_len, batch_size, trg_vocab_size = decoder_outputs.size() # (b, s, trg_vocab)
# Discard initial decoder input (index = 0)
#decoder_outputs = decoder_outputs[:, 1:, :]
decoder_predictions = decoder_outputs.argmax(dim=-1) # (b, s)
for i, pred in enumerate(decoder_predictions):
out.append((src[i], trg[i], pred))
return out
```
## Load best model
```
!ls -al
# Load model
model.load_state_dict(torch.load('./best_model.pt'))
```
## Make predictions
```
# Make prediction
test_predictions = predict(model, dev_iterator)
for i, prediction in enumerate(test_predictions):
src, trg, pred = prediction
src_text = indices_to_text(src, lang_field=KOREAN)
trg_text = indices_to_text(trg, lang_field=ENGLISH)
pred_text = indices_to_text(pred, lang_field=ENGLISH)
print('source:\n', src_text)
print('target:\n', trg_text)
print('prediction:\n', pred_text)
print('-' * 160)
if i > 5:
break
```
# 8. Download Model
```
!ls -al
from google.colab import files
print('Downloading models...') # Known bug; if using Firefox, a print statement in the same cell is necessary.
files.download('./best_model.pt')
files.download('./last_model.pt')
```
# 9. Discussions
```
```
| github_jupyter |
# Imports and Paths
```
import urllib3
http = urllib3.PoolManager()
from urllib import request
from bs4 import BeautifulSoup, Comment
import pandas as pd
from datetime import datetime
# from shutil import copyfile
# import time
import json
```
# Load in previous list of games
```
df_gms_lst = pd.read_csv('../data/bgg_top2000_2018-10-06.csv')
df_gms_lst.columns
metadata_dict = {"title": "BGG Top 2000",
"subtitle": "Board Game Geek top 2000 games rankings",
"description": "Board Game Geek top 2000 games rankings and other info",
"id": "mseinstein/bgg_top2000",
"licenses": [{"name": "CC-BY-SA-4.0"}],
"resources":[
{"path": "bgg_top2000_2018-10-06.csv",
"description": "Board Game Geek top 2000 games on 2018-10-06"
}
] }
with open('../data/kaggle/dataset-metadata.json', 'w') as fp:
json.dump(metadata_dict, fp)
```
# Get the id's of the top 2000 board games
```
pg_gm_rnks = 'https://boardgamegeek.com/browse/boardgame/page/'
def extract_gm_id(soup):
rows = soup.find('div', {'id': 'collection'}).find_all('tr')[1:]
id_list = []
for row in rows:
id_list.append(int(row.find_all('a')[1]['href'].split('/')[2]))
return id_list
def top_2k_gms(pg_gm_rnks):
gm_ids = []
for pg_num in range(1,21):
pg = request.urlopen(f'{pg_gm_rnks}{str(pg_num)}')
soup = BeautifulSoup(pg, 'html.parser')
gm_ids += extract_gm_id(soup)
return gm_ids
gm_ids = top_2k_gms(pg_gm_rnks)
len(gm_ids)
```
# Extract the info for each game in the top 2k using the extracted game id's
```
bs_pg = 'https://www.boardgamegeek.com/xmlapi2/'
bs_pg_gm = f'{bs_pg}thing?type=boardgame&stats=1&ratingcomments=1&page=1&pagesize=10&id='
def extract_game_item(item):
gm_dict = {}
field_int = ['yearpublished', 'minplayers', 'maxplayers', 'playingtime', 'minplaytime', 'maxplaytime', 'minage']
field_categ = ['boardgamecategory', 'boardgamemechanic', 'boardgamefamily','boardgamedesigner', 'boardgameartist', 'boardgamepublisher']
field_rank = [x['friendlyname'] for x in item.find_all('rank')]
field_stats = ['usersrated', 'average', 'bayesaverage', 'stddev', 'median', 'owned', 'trading', 'wanting', 'wishing', 'numcomments', 'numweights', 'averageweight']
gm_dict['name'] = item.find('name')['value']
gm_dict['id'] = item['id']
gm_dict['num_of_rankings'] = int(item.find('comments')['totalitems'])
for i in field_int:
field_val = item.find(i)
if field_val is None:
gm_dict[i] = -1
else:
gm_dict[i] = int(field_val['value'])
for i in field_categ:
gm_dict[i] = [x['value'] for x in item.find_all('link',{'type':i})]
for i in field_rank:
field_val = item.find('rank',{'friendlyname':i})
if field_val is None or field_val['value'] == 'Not Ranked':
gm_dict[i.replace(' ','')] = -1
else:
gm_dict[i.replace(' ','')] = int(field_val['value'])
for i in field_stats:
field_val = item.find(i)
if field_val is None:
gm_dict[i] = -1
else:
gm_dict[i] = float(field_val['value'])
return gm_dict
len(gm_ids)
gm_list = []
idx_split = 4
idx_size = int(len(gm_ids)/idx_split)
for i in range(idx_split):
idx = str(gm_ids[i*idx_size:(i+1)*idx_size]).replace(' ','')[1:-1]
pg = request.urlopen(f'{bs_pg_gm}{str(idx)}')
item_ct = 0
xsoup = BeautifulSoup(pg, 'xml')
# while item_ct < 500:
# xsoup = BeautifulSoup(pg, 'xml')
# item_ct = len(xsoup.find_all('item'))
gm_list += [extract_game_item(x) for x in xsoup.find_all('item')]
# break
df2 = pd.DataFrame(gm_list)
df2.shape
df2.head()
df2.loc[df2["Children'sGameRank"].notnull(),:].head().T
df2.isnull().sum()
gm_list = []
idx_split = 200
idx_size = int(len(gm_ids)/idx_split)
for i in range(idx_split):
idx = str(gm_ids[i*idx_size:(i+1)*idx_size]).replace(' ','')[1:-1]
break
# pg = request.urlopen(f'{bs_pg_gm}{str(idx)}')
# item_ct = 0
# xsoup = BeautifulSoup(pg, 'xml')
# # while item_ct < 500:
# # xsoup = BeautifulSoup(pg, 'xml')
# # item_ct = len(xsoup.find_all('item'))
# gm_list += [extract_game_item(x) for x in xsoup.find_all('item')]
# # break
# df2 = pd.DataFrame(gm_list)
# df2.shape
idx
def create_df_gm_ranks(gm_ids, bs_pg_gm):
gm_list = []
idx_split = 4
idx_size = int(len(gm_ids)/idx_split)
for i in range(idx_split):
idx = str(gm_ids[i*idx_size:(i+1)*idx_size]).replace(' ','')[1:-1]
pg = request.urlopen(f'{bs_pg_gm}{str(idx)}')
xsoup = BeautifulSoup(pg, 'xml')
gm_list += [extract_game_item(x) for x in xsoup.find_all('item')]
df = pd.DataFrame(gm_list)
return df
df = create_df_gm_ranks(gm_ids, bs_pg_gm)
df2.to_csv(f'../data/kaggle/{str(datetime.now().date())}_bgg_top{len(gm_ids)}.csv', index=False)
with open('../data/kaggle/dataset-metadata.json', 'rb') as f:
meta_dict = json.load(f)
meta_dict['resources'].append({
'path': f'{str(datetime.now().date())}_bgg_top{len(gm_ids)}.csv',
'description': f'Board Game Geek top 2000 games on {str(datetime.now().date())}'
})
meta_dict
meta_dict['title'] = 'Board Game Geek (BGG) Top 2000'
meta_dict['resources'][-1]['path'] = '2018-12-15_bgg_top2000.csv'
meta_dict['resources'][-1]['description']= 'Board Game Geek top 2000 games on 2018-12-15'
with open('../data/kaggle/dataset-metadata.json', 'w') as fp:
json.dump(meta_dict, fp)
```
Code for kaggle
kaggle datasets version -m "week of 2018-10-20" -p .\ -d
```
meta_dict
gm_list = []
idx_split = 4
idx_size = int(len(gm_ids)/idx_split)
for i in range(idx_split):
idx = str(gm_ids[i*idx_size:(i+1)*idx_size]).replace(' ','')[1:-1]
break
idx2 = '174430,161936,182028,167791,12333,187645,169786,220308,120677,193738,84876,173346,180263,115746,3076,102794,205637'
pg = request.urlopen(f'{bs_pg_gm}{str(idx)}')
xsoup = BeautifulSoup(pg, 'xml')
aa = xsoup.find_all('item')
len(aa)
http.urlopen()
r = http.request('GET', f'{bs_pg_gm}{str(idx)}')
xsoup2 = BeautifulSoup(r.data, 'xml')
bb = xsoup.find_all('item')
len(bb)
```
# XML2 API
Base URI: /xmlapi2/thing?parameters
- id=NNN
- Specifies the id of the thing(s) to retrieve. To request multiple things with a single query, NNN can specify a comma-delimited list of ids.
- type=THINGTYPE
- Specifies that, regardless of the type of thing asked for by id, the results are filtered by the THINGTYPE(s) specified. Multiple THINGTYPEs can be specified in a comma-delimited list.
- versions=1
- Returns version info for the item.
- videos = 1
- Returns videos for the item.
- stats=1
- Returns ranking and rating stats for the item.
- historical=1
- Returns historical data over time. See page parameter.
- marketplace=1
- Returns marketplace data.
- comments=1
- Returns all comments about the item. Also includes ratings when commented. See page parameter.
- ratingcomments=1
- Returns all ratings for the item. Also includes comments when rated. See page parameter. The ratingcomments and comments parameters cannot be used together, as the output always appears in the \<comments\> node of the XML; comments parameter takes precedence if both are specified. Ratings are sorted in descending rating value, based on the highest rating they have assigned to that item (each item in the collection can have a different rating).
- page=NNN
- Defaults to 1, controls the page of data to see for historical info, comments, and ratings data.
- pagesize=NNN
- Set the number of records to return in paging. Minimum is 10, maximum is 100.
- from=YYYY-MM-DD
- Not currently supported.
- to=YYYY-MM-DD
- Not currently supported.
| github_jupyter |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.ticker import FuncFormatter
import matplotlib as mpl
import matplotlib.dates as mdates
import datetime
# Set the matplotlib settings (eventually this will go at the top of the graph_util)
mpl.rcParams['axes.labelsize'] = 16
mpl.rcParams['axes.titlesize'] = 20
mpl.rcParams['legend.fontsize'] = 16
mpl.rcParams['font.size'] = 16.0
mpl.rcParams['figure.figsize'] = [15,10]
mpl.rcParams['xtick.labelsize'] = 16
mpl.rcParams['ytick.labelsize'] = 16
# Set the style for the graphs
plt.style.use('bmh')
# Additional matplotlib formatting settings
months = mdates.MonthLocator()
# This formats the months as three-letter abbreviations
months_format = mdates.DateFormatter('%b')
def area_cost_distribution(df, fiscal_year_col, utility_col_list, filename):
# Inputs include the dataframe, the column name for the fiscal year column, and the list of column names for the
# different utility bills. The dataframe should already include the summed bills for each fiscal year.
fig, ax = plt.subplots()
# Take costs for each utility type and convert to percent of total cost by fiscal year
df['total_costs'] = df[utility_col_list].sum(axis=1)
percent_columns = []
for col in utility_col_list:
percent_col = "Percent " + col
percent_columns.append(percent_col)
df[percent_col] = df[col] / df.total_costs
# Create stacked area plot
ax.stackplot(df[fiscal_year_col], df[percent_columns].T, labels=percent_columns)
# Format the y axis to be in percent
ax.yaxis.set_major_formatter(FuncFormatter('{0:.0%}'.format))
# Format the x-axis to include all fiscal years
plt.xticks(np.arange(df[fiscal_year_col].min(), df[fiscal_year_col].max()+1, 1.0))
# Add title and axis labels
plt.title('Annual Utility Cost Distribution')
plt.ylabel('Utility Cost Distribution')
plt.xlabel('Fiscal Year')
# Add legend
plt.legend()
# Make sure file goes in the proper directory
folder_and_filename = 'output/images/' + filename
# Save and show
plt.savefig(folder_and_filename)
plt.show()
def area_use_distribution(df, fiscal_year_col, utility_col_list, filename):
# Inputs include the dataframe, the column name for the fiscal year column, and the list of column names for the
# different utility bills. The dataframe should already include the summed bills for each fiscal year.
fig, ax = plt.subplots()
# Take usage for each utility type and convert to percent of total cost by fiscal year
df['total_use'] = df[utility_col_list].sum(axis=1)
percent_columns = []
for col in utility_col_list:
percent_col = "Percent " + col
percent_columns.append(percent_col)
df[percent_col] = df[col] / df.total_use
# Create stacked area plot
ax.stackplot(df[fiscal_year_col], df[percent_columns].T, labels=percent_columns)
# Format the y axis to be in percent
ax.yaxis.set_major_formatter(FuncFormatter('{0:.0%}'.format))
# Format the x-axis to include all fiscal years
plt.xticks(np.arange(df[fiscal_year_col].min(), df[fiscal_year_col].max()+1, 1.0))
# Add title and axis labels
plt.title('Annual Energy Usage Distribution')
plt.ylabel('Annual Energy Usage Distribution')
plt.xlabel('Fiscal Year')
# Add legend
plt.legend()
# Make sure file goes in the proper directory
folder_and_filename = 'output/images/' + filename
# Save and show
plt.savefig(folder_and_filename)
plt.show()
def create_stacked_bar(df, fiscal_year_col, column_name_list, filename):
# Parameters include the dataframe, the name of the column where the fiscal year is listed, a list of the column names
# with the correct data for the chart, and the filename where the output should be saved.
# Create the figure
plt.figure()
# Set the bar width
width = 0.50
# Create the stacked bars. The "bottom" is the sum of all previous bars to set the starting point for the next bar.
previous_col_name = 0
for col in column_name_list:
short_col_name = col.split(" Cost")[0]
short_col_name = plt.bar(df[fiscal_year_col], df[col], width, label=short_col_name, bottom=previous_col_name)
previous_col_name = previous_col_name + df[col]
# label axes
plt.ylabel('Utility Cost [$]')
plt.xlabel('Fiscal Year')
plt.title('Total Annual Utility Costs')
# Make one bar for each fiscal year
plt.xticks(np.arange(df[fiscal_year_col].min(), df[fiscal_year_col].max()+1, 1.0),
np.sort(list(df[fiscal_year_col].unique())))
# Set the yticks to go up to the total cost in increments of 100,000
df['total_cost'] = df[column_name_list].sum(axis=1)
plt.yticks(np.arange(0, df.total_cost.max(), 100000))
plt.legend()
# Make sure file goes in the proper directory
folder_and_filename = 'output/images/' + filename
# Save and show
plt.savefig(filename)
plt.show()
def energy_use_stacked_bar(df, fiscal_year_col, column_name_list, filename):
# Parameters include the dataframe, the name of the column where the fiscal year is listed, a list of the column names
# with the correct data for the chart, and the filename where the output should be saved.
# Create the figure
plt.figure()
# Set the bar width
width = 0.50
# Create the stacked bars. The "bottom" is the sum of all previous bars to set the starting point for the next bar.
previous_col_name = 0
for col in column_name_list:
short_col_name = col.split(" [MMBTU")[0]
short_col_name = plt.bar(df[fiscal_year_col], df[col], width, label=short_col_name, bottom=previous_col_name)
previous_col_name = previous_col_name + df[col]
# label axes
plt.ylabel('Annual Energy Usage [MMBTU]')
plt.xlabel('Fiscal Year')
plt.title('Total Annual Energy Usage')
# Make one bar for each fiscal year
plt.xticks(np.arange(df[fiscal_year_col].min(), df[fiscal_year_col].max()+1, 1.0),
np.sort(list(df[fiscal_year_col].unique())))
# Set the yticks to go up to the total usage in increments of 1,000
df['total_use'] = df[column_name_list].sum(axis=1)
plt.yticks(np.arange(0, df.total_use.max(), 1000))
plt.legend()
# Make sure file goes in the proper directory
folder_and_filename = 'output/images/' + filename
# Save and show
plt.savefig(folder_and_filename)
plt.show()
def usage_pie_charts(df, use_or_cost_cols, chart_type, filename):
# df: A dataframe with the fiscal_year as the index and needs to include the values for the passed in list of columns.
# use_or_cost_cols: a list of the energy usage or energy cost column names
# chart_type: 1 for an energy use pie chart, 2 for an energy cost pie chart
# Get the three most recent complete years of data
complete_years = df.query("month_count == 12.0")
sorted_completes = complete_years.sort_index(ascending=False)
most_recent_complete_years = sorted_completes[0:3]
years = list(most_recent_complete_years.index.values)
# Create percentages from usage
most_recent_complete_years = most_recent_complete_years[use_or_cost_cols]
most_recent_complete_years['Totals'] = most_recent_complete_years.sum(axis=1)
for col in use_or_cost_cols:
most_recent_complete_years[col] = most_recent_complete_years[col] / most_recent_complete_years.Totals
most_recent_complete_years = most_recent_complete_years.drop('Totals', axis=1)
for col in use_or_cost_cols:
if most_recent_complete_years[col].iloc[0] == 0:
most_recent_complete_years = most_recent_complete_years.drop(col, axis=1)
# Create a pie chart for each of 3 most recent complete years
for year in years:
year_df = most_recent_complete_years.query("fiscal_year == @year")
plt.figure()
fig, ax = plt.subplots()
ax.pie(list(year_df.iloc[0].values), labels=list(year_df.columns.values), autopct='%1.1f%%',
shadow=True, startangle=90)
# Create the title based on whether it is an energy use or energy cost pie chart.
if chart_type == 1:
title = "FY " + str(year) + " Energy Usage [MMBTU]"
else:
titel = "FY " + str(year) + " Energy Cost [$]"
plt.title(title)
ax.axis('equal') # Equal aspect ratio ensures that pie is drawn as a circle.
# Make sure file goes in the proper directory
folder_and_filename = 'output/images/' + filename + str(year)
# Save and show
plt.savefig(folder_and_filename)
plt.show()
def create_monthly_profile(df, graph_column_name, yaxis_name, color_choice, filename):
# Parameters:
# df: A dataframe with the fiscal_year, fiscal_mo, and appropriate graph column name ('kWh', 'kW', etc.)
# graph_column_name: The name of the column containing the data to be graphed on the y-axis
# yaxis_name: A string that will be displayed on the y-axis
# color_choice: 'blue', 'red', or 'green' depending on the desired color palette.
# Additional matplotlib formatting settings
months = mdates.MonthLocator()
# This formats the months as three-letter abbreviations
months_format = mdates.DateFormatter('%b')
# Get five most recent years
recent_years = (sorted(list(df.index.levels[0].values), reverse=True)[0:5])
# Reset the index of the dataframe for more straightforward queries
df_reset = df.reset_index()
def get_date(row):
# Converts the fiscal year and fiscal month columns to a datetime object for graphing
# Year is set to 2016-17 so that the charts overlap; otherwise they will be spread out by year.
# The "year trick" allows the graph to start from July so the seasonal energy changes are easier to identify
if row['fiscal_mo'] > 6:
year_trick = 2016
else:
year_trick = 2017
return datetime.date(year=year_trick, month=row['fiscal_mo'], day=1)
# This creates a new date column with data in the datetime format for graphing
df_reset['date'] = df_reset[['fiscal_year', 'fiscal_mo']].apply(get_date, axis=1)
# Create a color dictionary of progressively lighter colors of three different shades and convert to dataframe
color_dict = {'blue': ['#08519c', '#3182bd', '#6baed6', '#bdd7e7', '#eff3ff'],
'red': ['#a50f15', '#de2d26', '#fb6a4a', '#fcae91', '#fee5d9'],
'green': ['#006d2c', '#31a354', '#74c476', '#bae4b3', '#edf8e9']
}
color_df = pd.DataFrame.from_dict(color_dict)
# i is the counter for the different colors
i=0
# Create the plots
fig, ax = plt.subplots()
for year in recent_years:
# Create df for one year only so it's plotted as a single line
year_df = electric_pivot_monthly_reset.query("fiscal_year == @year")
year_df = year_df.sort_values(by='date')
# Plot the data
ax.plot_date(year_df['date'], year_df[graph_column_name], fmt='-', color=color_df.iloc[i][color_choice],
label=str(year_df.fiscal_year.iloc[0]))
# Increase counter by one to use the next color
i += 1
# Format the dates
ax.xaxis.set_major_locator(months)
ax.xaxis.set_major_formatter(months_format)
fig.autofmt_xdate()
# Add the labels
plt.xlabel('Month of Year')
plt.ylabel(yaxis_name)
plt.legend()
# Make sure file goes in the proper directory
folder_and_filename = 'output/images/' + filename
# Save and show
plt.savefig(folder_and_filename)
plt.show()
```
| github_jupyter |
# Artificial Intelligence Nanodegree
## Voice User Interfaces
## Project: Speech Recognition with Neural Networks
---
In this notebook, some template code has already been provided for you, and you will need to implement additional functionality to successfully complete this project. You will not need to modify the included code beyond what is requested. Sections that begin with **'(IMPLEMENTATION)'** in the header indicate that the following blocks of code will require additional functionality which you must provide. Please be sure to read the instructions carefully!
> **Note**: Once you have completed all of the code implementations, you need to finalize your work by exporting the Jupyter Notebook as an HTML document. Before exporting the notebook to html, all of the code cells need to have been run so that reviewers can see the final implementation and output. You can then export the notebook by using the menu above and navigating to \n",
"**File -> Download as -> HTML (.html)**. Include the finished document along with this notebook as your submission.
In addition to implementing code, there will be questions that you must answer which relate to the project and your implementation. Each section where you will answer a question is preceded by a **'Question X'** header. Carefully read each question and provide thorough answers in the following text boxes that begin with **'Answer:'**. Your project submission will be evaluated based on your answers to each of the questions and the implementation you provide.
>**Note:** Code and Markdown cells can be executed using the **Shift + Enter** keyboard shortcut. Markdown cells can be edited by double-clicking the cell to enter edit mode.
The rubric contains _optional_ "Stand Out Suggestions" for enhancing the project beyond the minimum requirements. If you decide to pursue the "Stand Out Suggestions", you should include the code in this Jupyter notebook.
---
## Introduction
In this notebook, you will build a deep neural network that functions as part of an end-to-end automatic speech recognition (ASR) pipeline! Your completed pipeline will accept raw audio as input and return a predicted transcription of the spoken language. The full pipeline is summarized in the figure below.
<img src="https://github.com/RomansWorks/AIND-VUI-Capstone/blob/master/images/pipeline.png?raw=1">
- **STEP 1** is a pre-processing step that converts raw audio to one of two feature representations that are commonly used for ASR.
- **STEP 2** is an acoustic model which accepts audio features as input and returns a probability distribution over all potential transcriptions. After learning about the basic types of neural networks that are often used for acoustic modeling, you will engage in your own investigations, to design your own acoustic model!
- **STEP 3** in the pipeline takes the output from the acoustic model and returns a predicted transcription.
Feel free to use the links below to navigate the notebook:
- [The Data](#thedata)
- [**STEP 1**](#step1): Acoustic Features for Speech Recognition
- [**STEP 2**](#step2): Deep Neural Networks for Acoustic Modeling
- [Model 0](#model0): RNN
- [Model 1](#model1): RNN + TimeDistributed Dense
- [Model 2](#model2): CNN + RNN + TimeDistributed Dense
- [Model 3](#model3): Deeper RNN + TimeDistributed Dense
- [Model 4](#model4): Bidirectional RNN + TimeDistributed Dense
- [Models 5+](#model5)
- [Compare the Models](#compare)
- [Final Model](#final)
- [**STEP 3**](#step3): Obtain Predictions
<a id='thedata'></a>
## The Data
We begin by investigating the dataset that will be used to train and evaluate your pipeline. [LibriSpeech](http://www.danielpovey.com/files/2015_icassp_librispeech.pdf) is a large corpus of English-read speech, designed for training and evaluating models for ASR. The dataset contains 1000 hours of speech derived from audiobooks. We will work with a small subset in this project, since larger-scale data would take a long while to train. However, after completing this project, if you are interested in exploring further, you are encouraged to work with more of the data that is provided [online](http://www.openslr.org/12/).
In the code cells below, you will use the `vis_train_features` module to visualize a training example. The supplied argument `index=0` tells the module to extract the first example in the training set. (You are welcome to change `index=0` to point to a different training example, if you like, but please **DO NOT** amend any other code in the cell.) The returned variables are:
- `vis_text` - transcribed text (label) for the training example.
- `vis_raw_audio` - raw audio waveform for the training example.
- `vis_mfcc_feature` - mel-frequency cepstral coefficients (MFCCs) for the training example.
- `vis_spectrogram_feature` - spectrogram for the training example.
- `vis_audio_path` - the file path to the training example.
```
%load_ext autoreload
%autoreload 1
%pip install python_speech_features
!rm -rf AIND-VUI-Capstone/
!git clone https://github.com/RomansWorks/AIND-VUI-Capstone
!cp -r ./AIND-VUI-Capstone/* .
!wget https://filebin.net/archive/s14yfd2p3q0sj1r2/zip
!unzip zip
!7z x capstone-ds.zip
!mv aind-vui-capstone-ds-processed/* .
!rm zip capstone-ds.*
from data_generator import vis_train_features
# extract label and audio features for a single training example
vis_text, vis_raw_audio, vis_mfcc_feature, vis_spectrogram_feature, vis_audio_path = vis_train_features()
```
The following code cell visualizes the audio waveform for your chosen example, along with the corresponding transcript. You also have the option to play the audio in the notebook!
```
from IPython.display import Markdown, display
from data_generator import vis_train_features, plot_raw_audio
from IPython.display import Audio
%matplotlib inline
# plot audio signal
plot_raw_audio(vis_raw_audio)
# print length of audio signal
display(Markdown('**Shape of Audio Signal** : ' + str(vis_raw_audio.shape)))
# print transcript corresponding to audio clip
display(Markdown('**Transcript** : ' + str(vis_text)))
# play the audio file
Audio(vis_audio_path)
```
<a id='step1'></a>
## STEP 1: Acoustic Features for Speech Recognition
For this project, you won't use the raw audio waveform as input to your model. Instead, we provide code that first performs a pre-processing step to convert the raw audio to a feature representation that has historically proven successful for ASR models. Your acoustic model will accept the feature representation as input.
In this project, you will explore two possible feature representations. _After completing the project_, if you'd like to read more about deep learning architectures that can accept raw audio input, you are encouraged to explore this [research paper](https://pdfs.semanticscholar.org/a566/cd4a8623d661a4931814d9dffc72ecbf63c4.pdf).
### Spectrograms
The first option for an audio feature representation is the [spectrogram](https://www.youtube.com/watch?v=_FatxGN3vAM). In order to complete this project, you will **not** need to dig deeply into the details of how a spectrogram is calculated; but, if you are curious, the code for calculating the spectrogram was borrowed from [this repository](https://github.com/baidu-research/ba-dls-deepspeech). The implementation appears in the `utils.py` file in your repository.
The code that we give you returns the spectrogram as a 2D tensor, where the first (_vertical_) dimension indexes time, and the second (_horizontal_) dimension indexes frequency. To speed the convergence of your algorithm, we have also normalized the spectrogram. (You can see this quickly in the visualization below by noting that the mean value hovers around zero, and most entries in the tensor assume values close to zero.)
```
from data_generator import plot_spectrogram_feature
# plot normalized spectrogram
plot_spectrogram_feature(vis_spectrogram_feature)
# print shape of spectrogram
display(Markdown('**Shape of Spectrogram** : ' + str(vis_spectrogram_feature.shape)))
```
### Mel-Frequency Cepstral Coefficients (MFCCs)
The second option for an audio feature representation is [MFCCs](https://en.wikipedia.org/wiki/Mel-frequency_cepstrum). You do **not** need to dig deeply into the details of how MFCCs are calculated, but if you would like more information, you are welcome to peruse the [documentation](https://github.com/jameslyons/python_speech_features) of the `python_speech_features` Python package. Just as with the spectrogram features, the MFCCs are normalized in the supplied code.
The main idea behind MFCC features is the same as spectrogram features: at each time window, the MFCC feature yields a feature vector that characterizes the sound within the window. Note that the MFCC feature is much lower-dimensional than the spectrogram feature, which could help an acoustic model to avoid overfitting to the training dataset.
```
from data_generator import plot_mfcc_feature
# plot normalized MFCC
plot_mfcc_feature(vis_mfcc_feature)
# print shape of MFCC
display(Markdown('**Shape of MFCC** : ' + str(vis_mfcc_feature.shape)))
```
When you construct your pipeline, you will be able to choose to use either spectrogram or MFCC features. If you would like to see different implementations that make use of MFCCs and/or spectrograms, please check out the links below:
- This [repository](https://github.com/baidu-research/ba-dls-deepspeech) uses spectrograms.
- This [repository](https://github.com/mozilla/DeepSpeech) uses MFCCs.
- This [repository](https://github.com/buriburisuri/speech-to-text-wavenet) also uses MFCCs.
- This [repository](https://github.com/pannous/tensorflow-speech-recognition/blob/master/speech_data.py) experiments with raw audio, spectrograms, and MFCCs as features.
<a id='step2'></a>
## STEP 2: Deep Neural Networks for Acoustic Modeling
In this section, you will experiment with various neural network architectures for acoustic modeling.
You will begin by training five relatively simple architectures. **Model 0** is provided for you. You will write code to implement **Models 1**, **2**, **3**, and **4**. If you would like to experiment further, you are welcome to create and train more models under the **Models 5+** heading.
All models will be specified in the `sample_models.py` file. After importing the `sample_models` module, you will train your architectures in the notebook.
After experimenting with the five simple architectures, you will have the opportunity to compare their performance. Based on your findings, you will construct a deeper architecture that is designed to outperform all of the shallow models.
For your convenience, we have designed the notebook so that each model can be specified and trained on separate occasions. That is, say you decide to take a break from the notebook after training **Model 1**. Then, you need not re-execute all prior code cells in the notebook before training **Model 2**. You need only re-execute the code cell below, that is marked with **`RUN THIS CODE CELL IF YOU ARE RESUMING THE NOTEBOOK AFTER A BREAK`**, before transitioning to the code cells corresponding to **Model 2**.
```
#####################################################################
# RUN THIS CODE CELL IF YOU ARE RESUMING THE NOTEBOOK AFTER A BREAK #
#####################################################################
# allocate 50% of GPU memory (if you like, feel free to change this)
# from keras.backend.tensorflow_backend import set_session
import tensorflow as tf
# config = tf.ConfigProto()
# config.gpu_options.per_process_gpu_memory_fraction = 0.5
# set_session(tf.Session(config=config))
from tensorflow.keras.optimizers import Adam, SGD
# watch for any changes in the sample_models module, and reload it automatically
%load_ext autoreload
%autoreload 2
# import NN architectures for speech recognition
from sample_models import *
# import function for training acoustic model
from train_utils import train_model
```
<a id='model0'></a>
### Model 0: RNN
Given their effectiveness in modeling sequential data, the first acoustic model you will use is an RNN. As shown in the figure below, the RNN we supply to you will take the time sequence of audio features as input.
<img src="https://github.com/RomansWorks/AIND-VUI-Capstone/blob/master/images/simple_rnn.png?raw=1" width="50%">
At each time step, the speaker pronounces one of 28 possible characters, including each of the 26 letters in the English alphabet, along with a space character (" "), and an apostrophe (').
The output of the RNN at each time step is a vector of probabilities with 29 entries, where the $i$-th entry encodes the probability that the $i$-th character is spoken in the time sequence. (The extra 29th character is an empty "character" used to pad training examples within batches containing uneven lengths.) If you would like to peek under the hood at how characters are mapped to indices in the probability vector, look at the `char_map.py` file in the repository. The figure below shows an equivalent, rolled depiction of the RNN that shows the output layer in greater detail.
<img src="https://github.com/RomansWorks/AIND-VUI-Capstone/blob/master/images/simple_rnn_unrolled.png?raw=1" width="60%">
The model has already been specified for you in Keras. To import it, you need only run the code cell below.
```
model_0 = simple_rnn_model(input_dim=161) # change to 13 if you would like to use MFCC features
```
As explored in the lesson, you will train the acoustic model with the [CTC loss](http://www.cs.toronto.edu/~graves/icml_2006.pdf) criterion. Custom loss functions take a bit of hacking in Keras, and so we have implemented the CTC loss function for you, so that you can focus on trying out as many deep learning architectures as possible :). If you'd like to peek at the implementation details, look at the `add_ctc_loss` function within the `train_utils.py` file in the repository.
To train your architecture, you will use the `train_model` function within the `train_utils` module; it has already been imported in one of the above code cells. The `train_model` function takes three **required** arguments:
- `input_to_softmax` - a Keras model instance.
- `pickle_path` - the name of the pickle file where the loss history will be saved.
- `save_model_path` - the name of the HDF5 file where the model will be saved.
If we have already supplied values for `input_to_softmax`, `pickle_path`, and `save_model_path`, please **DO NOT** modify these values.
There are several **optional** arguments that allow you to have more control over the training process. You are welcome to, but not required to, supply your own values for these arguments.
- `minibatch_size` - the size of the minibatches that are generated while training the model (default: `20`).
- `spectrogram` - Boolean value dictating whether spectrogram (`True`) or MFCC (`False`) features are used for training (default: `True`).
- `mfcc_dim` - the size of the feature dimension to use when generating MFCC features (default: `13`).
- `optimizer` - the Keras optimizer used to train the model (default: `SGD(lr=0.02, decay=1e-6, momentum=0.9, nesterov=True, clipnorm=5)`).
- `epochs` - the number of epochs to use to train the model (default: `20`). If you choose to modify this parameter, make sure that it is *at least* 20.
- `verbose` - controls the verbosity of the training output in the `model.fit_generator` method (default: `1`).
- `sort_by_duration` - Boolean value dictating whether the training and validation sets are sorted by (increasing) duration before the start of the first epoch (default: `False`).
The `train_model` function defaults to using spectrogram features; if you choose to use these features, note that the acoustic model in `simple_rnn_model` should have `input_dim=161`. Otherwise, if you choose to use MFCC features, the acoustic model should have `input_dim=13`.
We have chosen to use `GRU` units in the supplied RNN. If you would like to experiment with `LSTM` or `SimpleRNN` cells, feel free to do so here. If you change the `GRU` units to `SimpleRNN` cells in `simple_rnn_model`, you may notice that the loss quickly becomes undefined (`nan`) - you are strongly encouraged to check this for yourself! This is due to the [exploding gradients problem](http://www.wildml.com/2015/10/recurrent-neural-networks-tutorial-part-3-backpropagation-through-time-and-vanishing-gradients/). We have already implemented [gradient clipping](https://arxiv.org/pdf/1211.5063.pdf) in your optimizer to help you avoid this issue.
__IMPORTANT NOTE:__ If you notice that your gradient has exploded in any of the models below, feel free to explore more with gradient clipping (the `clipnorm` argument in your optimizer) or swap out any `SimpleRNN` cells for `LSTM` or `GRU` cells. You can also try restarting the kernel to restart the training process.
```
from tensorflow.keras.optimizers import Adam
train_model(input_to_softmax=model_0,
pickle_path='model_0.pickle',
save_model_path='model_0.h5',
minibatch_size=25,
optimizer=Adam(learning_rate=0.1, clipnorm=5), #SGD(lr=0.002, decay=1e-6, momentum=0.9, nesterov=True, clipnorm=5),
spectrogram=True) # change to False if you would like to use MFCC features
```
<a id='model1'></a>
### (IMPLEMENTATION) Model 1: RNN + TimeDistributed Dense
Read about the [TimeDistributed](https://keras.io/layers/wrappers/) wrapper and the [BatchNormalization](https://keras.io/layers/normalization/) layer in the Keras documentation. For your next architecture, you will add [batch normalization](https://arxiv.org/pdf/1510.01378.pdf) to the recurrent layer to reduce training times. The `TimeDistributed` layer will be used to find more complex patterns in the dataset. The unrolled snapshot of the architecture is depicted below.
<img src="https://github.com/RomansWorks/AIND-VUI-Capstone/blob/master/images/rnn_model.png?raw=1" width="60%">
The next figure shows an equivalent, rolled depiction of the RNN that shows the (`TimeDistrbuted`) dense and output layers in greater detail.
<img src="https://github.com/RomansWorks/AIND-VUI-Capstone/blob/master/images/rnn_model_unrolled.png?raw=1" width="60%">
Use your research to complete the `rnn_model` function within the `sample_models.py` file. The function should specify an architecture that satisfies the following requirements:
- The first layer of the neural network should be an RNN (`SimpleRNN`, `LSTM`, or `GRU`) that takes the time sequence of audio features as input. We have added `GRU` units for you, but feel free to change `GRU` to `SimpleRNN` or `LSTM`, if you like!
- Whereas the architecture in `simple_rnn_model` treated the RNN output as the final layer of the model, you will use the output of your RNN as a hidden layer. Use `TimeDistributed` to apply a `Dense` layer to each of the time steps in the RNN output. Ensure that each `Dense` layer has `output_dim` units.
Use the code cell below to load your model into the `model_1` variable. Use a value for `input_dim` that matches your chosen audio features, and feel free to change the values for `units` and `activation` to tweak the behavior of your recurrent layer.
```
model_1 = rnn_model(input_dim=161, # change to 13 if you would like to use MFCC features
units=200,
activation='relu')
```
Please execute the code cell below to train the neural network you specified in `input_to_softmax`. After the model has finished training, the model is [saved](https://keras.io/getting-started/faq/#how-can-i-save-a-keras-model) in the HDF5 file `model_1.h5`. The loss history is [saved](https://wiki.python.org/moin/UsingPickle) in `model_1.pickle`. You are welcome to tweak any of the optional parameters while calling the `train_model` function, but this is not required.
```
train_model(input_to_softmax=model_1,
pickle_path='model_1.pickle',
save_model_path='model_1.h5',
optimizer=Adam(clipvalue=0.5, clipnorm=1.0),
spectrogram=True) # change to False if you would like to use MFCC features
```
<a id='model2'></a>
### (IMPLEMENTATION) Model 2: CNN + RNN + TimeDistributed Dense
The architecture in `cnn_rnn_model` adds an additional level of complexity, by introducing a [1D convolution layer](https://keras.io/layers/convolutional/#conv1d).
<img src="https://github.com/RomansWorks/AIND-VUI-Capstone/blob/master/images/cnn_rnn_model.png?raw=1" width="100%">
This layer incorporates many arguments that can be (optionally) tuned when calling the `cnn_rnn_model` module. We provide sample starting parameters, which you might find useful if you choose to use spectrogram audio features.
If you instead want to use MFCC features, these arguments will have to be tuned. Note that the current architecture only supports values of `'same'` or `'valid'` for the `conv_border_mode` argument.
When tuning the parameters, be careful not to choose settings that make the convolutional layer overly small. If the temporal length of the CNN layer is shorter than the length of the transcribed text label, your code will throw an error.
Before running the code cell below, you must modify the `cnn_rnn_model` function in `sample_models.py`. Please add batch normalization to the recurrent layer, and provide the same `TimeDistributed` layer as before.
```
model_2 = cnn_rnn_model(input_dim=161, # change to 13 if you would like to use MFCC features
filters=200,
kernel_size=11,
conv_stride=2,
conv_border_mode='valid',
units=100)
```
Please execute the code cell below to train the neural network you specified in `input_to_softmax`. After the model has finished training, the model is [saved](https://keras.io/getting-started/faq/#how-can-i-save-a-keras-model) in the HDF5 file `model_2.h5`. The loss history is [saved](https://wiki.python.org/moin/UsingPickle) in `model_2.pickle`. You are welcome to tweak any of the optional parameters while calling the `train_model` function, but this is not required.
```
train_model(input_to_softmax=model_2,
pickle_path='model_2.pickle',
save_model_path='model_2.h5',
optimizer=Adam(clipvalue=0.5),
spectrogram=True) # change to False if you would like to use MFCC features
```
<a id='model3'></a>
### (IMPLEMENTATION) Model 3: Deeper RNN + TimeDistributed Dense
Review the code in `rnn_model`, which makes use of a single recurrent layer. Now, specify an architecture in `deep_rnn_model` that utilizes a variable number `recur_layers` of recurrent layers. The figure below shows the architecture that should be returned if `recur_layers=2`. In the figure, the output sequence of the first recurrent layer is used as input for the next recurrent layer.
<img src="https://github.com/RomansWorks/AIND-VUI-Capstone/blob/master/images/deep_rnn_model.png?raw=1" width="80%">
Feel free to change the supplied values of `units` to whatever you think performs best. You can change the value of `recur_layers`, as long as your final value is greater than 1. (As a quick check that you have implemented the additional functionality in `deep_rnn_model` correctly, make sure that the architecture that you specify here is identical to `rnn_model` if `recur_layers=1`.)
```
model_3 = deep_rnn_model(input_dim=161, # change to 13 if you would like to use MFCC features
units=100,
recur_layers=2)
```
Please execute the code cell below to train the neural network you specified in `input_to_softmax`. After the model has finished training, the model is [saved](https://keras.io/getting-started/faq/#how-can-i-save-a-keras-model) in the HDF5 file `model_3.h5`. The loss history is [saved](https://wiki.python.org/moin/UsingPickle) in `model_3.pickle`. You are welcome to tweak any of the optional parameters while calling the `train_model` function, but this is not required.
```
train_model(input_to_softmax=model_3,
pickle_path='model_3.pickle',
save_model_path='model_3.h5',
optimizer=Adam(clipvalue=0.5),
spectrogram=True) # change to False if you would like to use MFCC features
```
<a id='model4'></a>
### (IMPLEMENTATION) Model 4: Bidirectional RNN + TimeDistributed Dense
Read about the [Bidirectional](https://keras.io/layers/wrappers/) wrapper in the Keras documentation. For your next architecture, you will specify an architecture that uses a single bidirectional RNN layer, before a (`TimeDistributed`) dense layer. The added value of a bidirectional RNN is described well in [this paper](http://www.cs.toronto.edu/~hinton/absps/DRNN_speech.pdf).
> One shortcoming of conventional RNNs is that they are only able to make use of previous context. In speech recognition, where whole utterances are transcribed at once, there is no reason not to exploit future context as well. Bidirectional RNNs (BRNNs) do this by processing the data in both directions with two separate hidden layers which are then fed forwards to the same output layer.
<img src="https://github.com/RomansWorks/AIND-VUI-Capstone/blob/master/images/bidirectional_rnn_model.png?raw=1" width="80%">
Before running the code cell below, you must complete the `bidirectional_rnn_model` function in `sample_models.py`. Feel free to use `SimpleRNN`, `LSTM`, or `GRU` units. When specifying the `Bidirectional` wrapper, use `merge_mode='concat'`.
```
model_4 = bidirectional_rnn_model(input_dim=161, # change to 13 if you would like to use MFCC features
units=100)
```
Please execute the code cell below to train the neural network you specified in `input_to_softmax`. After the model has finished training, the model is [saved](https://keras.io/getting-started/faq/#how-can-i-save-a-keras-model) in the HDF5 file `model_4.h5`. The loss history is [saved](https://wiki.python.org/moin/UsingPickle) in `model_4.pickle`. You are welcome to tweak any of the optional parameters while calling the `train_model` function, but this is not required.
```
train_model(input_to_softmax=model_4,
pickle_path='model_4.pickle',
save_model_path='model_4.h5',
optimizer=Adam(clipvalue=0.5),
spectrogram=True) # change to False if you would like to use MFCC features
```
<a id='model5'></a>
### (OPTIONAL IMPLEMENTATION) Models 5+
If you would like to try out more architectures than the ones above, please use the code cell below. Please continue to follow the same convention for saving the models; for the $i$-th sample model, please save the loss at **`model_i.pickle`** and saving the trained model at **`model_i.h5`**.
```
## (Optional) TODO: Try out some more models!
### Feel free to use as many code cells as needed.
model_5 = dilated_double_cnn_rnn_model(input_dim=161,
filters=200,
kernel_size=6,
conv_border_mode='valid',
units=200,
dilation=2)
train_model(input_to_softmax=model_5,
pickle_path='model_5.pickle',
save_model_path='model_5.h5',
optimizer=Adam(clipvalue=0.5, amsgrad=True),
spectrogram=True)
```
<a id='compare'></a>
### Compare the Models
Execute the code cell below to evaluate the performance of the drafted deep learning models. The training and validation loss are plotted for each model.
```
from glob import glob
import numpy as np
import _pickle as pickle
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
sns.set_style(style='white')
# obtain the paths for the saved model history
all_pickles = sorted(glob("results/*.pickle"))
# extract the name of each model
model_names = [item[8:-7] for item in all_pickles]
# extract the loss history for each model
valid_loss = [pickle.load( open( i, "rb" ) )['val_loss'] for i in all_pickles]
train_loss = [pickle.load( open( i, "rb" ) )['loss'] for i in all_pickles]
# save the number of epochs used to train each model
num_epochs = [len(valid_loss[i]) for i in range(len(valid_loss))]
fig = plt.figure(figsize=(16,5))
# plot the training loss vs. epoch for each model
ax1 = fig.add_subplot(121)
for i in range(len(all_pickles)):
ax1.plot(np.linspace(1, num_epochs[i], num_epochs[i]),
train_loss[i], label=model_names[i])
# clean up the plot
ax1.legend()
ax1.set_xlim([1, max(num_epochs)])
plt.xlabel('Epoch')
plt.ylabel('Training Loss')
# plot the validation loss vs. epoch for each model
ax2 = fig.add_subplot(122)
for i in range(len(all_pickles)):
ax2.plot(np.linspace(1, num_epochs[i], num_epochs[i]),
valid_loss[i], label=model_names[i])
# clean up the plot
ax2.legend()
ax2.set_xlim([1, max(num_epochs)])
plt.xlabel('Epoch')
plt.ylabel('Validation Loss')
plt.show()
```
__Question 1:__ Use the plot above to analyze the performance of each of the attempted architectures. Which performs best? Provide an explanation regarding why you think some models perform better than others.
__Answer:__
<a id='final'></a>
### (IMPLEMENTATION) Final Model
Now that you've tried out many sample models, use what you've learned to draft your own architecture! While your final acoustic model should not be identical to any of the architectures explored above, you are welcome to merely combine the explored layers above into a deeper architecture. It is **NOT** necessary to include new layer types that were not explored in the notebook.
However, if you would like some ideas for even more layer types, check out these ideas for some additional, optional extensions to your model:
- If you notice your model is overfitting to the training dataset, consider adding **dropout**! To add dropout to [recurrent layers](https://faroit.github.io/keras-docs/1.0.2/layers/recurrent/), pay special attention to the `dropout_W` and `dropout_U` arguments. This [paper](http://arxiv.org/abs/1512.05287) may also provide some interesting theoretical background.
- If you choose to include a convolutional layer in your model, you may get better results by working with **dilated convolutions**. If you choose to use dilated convolutions, make sure that you are able to accurately calculate the length of the acoustic model's output in the `model.output_length` lambda function. You can read more about dilated convolutions in Google's [WaveNet paper](https://arxiv.org/abs/1609.03499). For an example of a speech-to-text system that makes use of dilated convolutions, check out this GitHub [repository](https://github.com/buriburisuri/speech-to-text-wavenet). You can work with dilated convolutions [in Keras](https://keras.io/layers/convolutional/) by paying special attention to the `padding` argument when you specify a convolutional layer.
- If your model makes use of convolutional layers, why not also experiment with adding **max pooling**? Check out [this paper](https://arxiv.org/pdf/1701.02720.pdf) for example architecture that makes use of max pooling in an acoustic model.
- So far, you have experimented with a single bidirectional RNN layer. Consider stacking the bidirectional layers, to produce a [deep bidirectional RNN](https://www.cs.toronto.edu/~graves/asru_2013.pdf)!
All models that you specify in this repository should have `output_length` defined as an attribute. This attribute is a lambda function that maps the (temporal) length of the input acoustic features to the (temporal) length of the output softmax layer. This function is used in the computation of CTC loss; to see this, look at the `add_ctc_loss` function in `train_utils.py`. To see where the `output_length` attribute is defined for the models in the code, take a look at the `sample_models.py` file. You will notice this line of code within most models:
```
model.output_length = lambda x: x
```
The acoustic model that incorporates a convolutional layer (`cnn_rnn_model`) has a line that is a bit different:
```
model.output_length = lambda x: cnn_output_length(
x, kernel_size, conv_border_mode, conv_stride)
```
In the case of models that use purely recurrent layers, the lambda function is the identity function, as the recurrent layers do not modify the (temporal) length of their input tensors. However, convolutional layers are more complicated and require a specialized function (`cnn_output_length` in `sample_models.py`) to determine the temporal length of their output.
You will have to add the `output_length` attribute to your final model before running the code cell below. Feel free to use the `cnn_output_length` function, if it suits your model.
```
# specify the model
model_end = final_model()
```
Please execute the code cell below to train the neural network you specified in `input_to_softmax`. After the model has finished training, the model is [saved](https://keras.io/getting-started/faq/#how-can-i-save-a-keras-model) in the HDF5 file `model_end.h5`. The loss history is [saved](https://wiki.python.org/moin/UsingPickle) in `model_end.pickle`. You are welcome to tweak any of the optional parameters while calling the `train_model` function, but this is not required.
```
train_model(input_to_softmax=model_end,
pickle_path='model_end.pickle',
save_model_path='model_end.h5',
optimizer=Adam(clipvalue=0.5, amsgrad=True),
spectrogram=True) # change to False if you would like to use MFCC features
```
__Question 2:__ Describe your final model architecture and your reasoning at each step.
__Answer:__
<a id='step3'></a>
## STEP 3: Obtain Predictions
We have written a function for you to decode the predictions of your acoustic model. To use the function, please execute the code cell below.
```
import numpy as np
from data_generator import AudioGenerator
from keras import backend as K
from utils import int_sequence_to_text
from IPython.display import Audio
def get_predictions(index, partition, input_to_softmax, model_path):
""" Print a model's decoded predictions
Params:
index (int): The example you would like to visualize
partition (str): One of 'train' or 'validation'
input_to_softmax (Model): The acoustic model
model_path (str): Path to saved acoustic model's weights
"""
# load the train and test data
data_gen = AudioGenerator()
data_gen.load_train_data()
data_gen.load_validation_data()
# obtain the true transcription and the audio features
if partition == 'validation':
transcr = data_gen.valid_texts[index]
audio_path = data_gen.valid_audio_paths[index]
data_point = data_gen.normalize(data_gen.featurize(audio_path))
elif partition == 'train':
transcr = data_gen.train_texts[index]
audio_path = data_gen.train_audio_paths[index]
data_point = data_gen.normalize(data_gen.featurize(audio_path))
else:
raise Exception('Invalid partition! Must be "train" or "validation"')
# obtain and decode the acoustic model's predictions
input_to_softmax.load_weights(model_path)
prediction = input_to_softmax.predict(np.expand_dims(data_point, axis=0))
output_length = [input_to_softmax.output_length(data_point.shape[0])]
pred_ints = (K.eval(K.ctc_decode(
prediction, output_length)[0][0])+1).flatten().tolist()
# play the audio file, and display the true and predicted transcriptions
print('-'*80)
Audio(audio_path)
print('True transcription:\n' + '\n' + transcr)
print('-'*80)
print('Predicted transcription:\n' + '\n' + ''.join(int_sequence_to_text(pred_ints)))
print('-'*80)
```
Use the code cell below to obtain the transcription predicted by your final model for the first example in the training dataset.
```
get_predictions(index=0,
partition='train',
input_to_softmax=final_model(),
model_path='model_end.h5')
```
Use the next code cell to visualize the model's prediction for the first example in the validation dataset.
```
get_predictions(index=0,
partition='validation',
input_to_softmax=final_model(),
model_path='model_end.h5')
```
One standard way to improve the results of the decoder is to incorporate a language model. We won't pursue this in the notebook, but you are welcome to do so as an _optional extension_.
If you are interested in creating models that provide improved transcriptions, you are encouraged to download [more data](http://www.openslr.org/12/) and train bigger, deeper models. But beware - the model will likely take a long while to train. For instance, training this [state-of-the-art](https://arxiv.org/pdf/1512.02595v1.pdf) model would take 3-6 weeks on a single GPU!
| github_jupyter |
<h1> Logistic Regression <h1>
<h2> ROMÂNĂ <h2>
<blockquote><p>În final, o să observăm dacă Google PlayStore a avut destule date pentru a putea prezice popularitatea unei aplicații de trading sau pentru topul jocurilor plătite.
Lucrul acesta se va face prin împărțirea descărcărilor în 2 variabile dummy. Cu mai mult de 1.000.000 pentru variabila 1 și cu mai puțin de 1.000.000 pentru variabila 0, pentru
aplicațiile de Trading și pentru jocurile plătite cu mai mult de 670.545 de descărcari pentru variabila 1 iar 0 corespunde celorlalte aplicații.</p></blockquote>
<h2>ENGLISH<h2>
<blockquote><p>Lastly, we shall see if Google PlayStore had enough data in order to predict the popularity of a trading app or for the top paid games of the store .
This will be done by dividing the downloads into 2 dummy variables. With more than 1,000,000 for variable 1 and less than 1,000,000 for variable 0, for Trading applications and for paid games with more than 670,545 downloads for variable 1 and 0 corresponding to the other applications.</p></blockquote>
<h3>Now we shall create a logistic regression model using a 80/20 ratio between the training sample and the testing sample<h3>
```
from sklearn.model_selection import cross_val_score
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report, confusion_matrix
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
def Log_reg(x,y):
model = LogisticRegression(solver='liblinear',C=10, random_state=0).fit(x,y)
print("Model accuracy",model.score(x,y))
cm = confusion_matrix(y, model.predict(x))
fig, ax = plt.subplots(figsize=(8, 8))
ax.imshow(cm)
ax.grid(False)
ax.xaxis.set(ticks=(0, 1), ticklabels=('Predicted 0s', 'Predicted 1s'))
ax.yaxis.set(ticks=(0, 1), ticklabels=('Actual 0s', 'Actual 1s'))
ax.set_ylim(1.5, -0.5)
for i in range(2):
for j in range(2):
ax.text(j, i, cm[i, j], ha='center', va='center', color='black')
plt.title('Confusion Matrix')
plt.show()
print(classification_report(y, model.predict(x)))
scores = cross_val_score(model, x,y, cv=10)
print('Cross-Validation Accuracy Scores', scores)
scores = pd.Series(scores)
print("Mean Accuracy: ",scores.mean())
import pandas as pd
import numpy as np
#path = "D:\Java\VS-CodPitonul\\GAME.xlsx"
#df = pd.read_excel (path, sheet_name='Sheet1')
path = "D:\Java\VS-CodPitonul\\Trading_Apps.xlsx"
df = pd.read_excel (path, sheet_name='Results')
'''
RO: Folosește dropna daca ai valori lipsă, altfel îți va da eroare
ENG: Use dropna only if you have missing values else you will recive an error message
'''
#df = df.dropna()
#Log_reg(df[['Score','Ratings','Reviews','Months_From_Release','Price']],df['Instalari_Bin']) #For GAME.xlsx
Log_reg(df[['Score','Ratings','Reviews','Months_From_Release']],df['Instalari_Bin']) #For Trading_Apps.xlsx
```
| github_jupyter |
# Import libraries and data
Dataset was obtained in the capstone project description (direct link [here](https://d3c33hcgiwev3.cloudfront.net/_429455574e396743d399f3093a3cc23b_capstone.zip?Expires=1530403200&Signature=FECzbTVo6TH7aRh7dXXmrASucl~Cy5mlO94P7o0UXygd13S~Afi38FqCD7g9BOLsNExNB0go0aGkYPtodekxCGblpc3I~R8TCtWRrys~2gciwuJLGiRp4CfNtfp08sFvY9NENaRb6WE2H4jFsAo2Z2IbXV~llOJelI3k-9Waj~M_&Key-Pair-Id=APKAJLTNE6QMUY6HBC5A)) and splited manually in separated csv files. They were stored at my personal github account (folder link [here](https://github.com/caiomiyashiro/RecommenderSystemsNotebooks/tree/master/data/capstone)) and you can download and paste inside your working directory in order for this notebook to run.
```
import pandas as pd
import numpy as np
```
## Preprocess data
Float data came with ',' in the csv and python works with '.', so it treated the number as text. In order to convert them to numbers, I first replaced all the commas by punct and then converted the columns to float.
```
items = pd.read_csv('data/capstone/Capstone Data - Office Products - Items.csv', index_col=0)
actual_ratings = pd.read_csv('data/capstone/Capstone Data - Office Products - Ratings.csv', index_col=0)
content_based = pd.read_csv('data/capstone/Capstone Data - Office Products - CBF.csv', index_col=0)
user_user = pd.read_csv('data/capstone/Capstone Data - Office Products - User-User.csv', index_col=0)
item_item = pd.read_csv('data/capstone/Capstone Data - Office Products - Item-Item.csv', index_col=0)
matrix_fact = pd.read_csv('data/capstone/Capstone Data - Office Products - MF.csv', index_col=0)
pers_bias = pd.read_csv('data/capstone/Capstone Data - Office Products - PersBias.csv', index_col=0)
items[['Availability','Price']] = items[['Availability','Price']].apply(lambda col: col.apply(lambda elem: str(elem).replace(',', '.'))).astype(float)
# preprocess
content_based = content_based.apply(lambda col: col.apply(lambda elem: str(elem).replace(',', '.'))).astype(float)
user_user = user_user.apply(lambda col: col.apply(lambda elem: str(elem).replace(',', '.'))).astype(float)
item_item = item_item.apply(lambda col: col.apply(lambda elem: str(elem).replace(',', '.'))).astype(float)
matrix_fact = matrix_fact.apply(lambda col: col.apply(lambda elem: str(elem).replace(',', '.'))).astype(float)
pers_bias = pers_bias.apply(lambda col: col.apply(lambda elem: str(elem).replace(',', '.'))).astype(float)
print('items.shape = ' + str(items.shape))
print('actual_ratings.shape = ' + str(actual_ratings.shape))
print('content_based.shape = ' + str(content_based.shape))
print('user_user.shape = ' + str(user_user.shape))
print('item_item.shape = ' + str(item_item.shape))
print('matrix_fact.shape = ' + str(matrix_fact.shape))
print('pers_bias.shape = ' + str(pers_bias.shape))
actual_ratings.head()
```
# Class RecommenderEvaluator
In order to become easier to evaluate the metrics, I created a class that receives all the original ratings and predicted ratings for every recommender system and defined functions to extract all the metrics established in section 1 of the capstone report. Lets take a look at a summary of the class before looking at the code:
- **Constructor (init)**: receive all recommendation algorithms, besides the actual rating list and the list of items. All data is contained in the data downloaded from Coursera. Besides storing all recommendation algorithms, the constructor also calculate the 20 most frequent items, which is used in the popularity metric calculation.
- **get_observed_ratings**: as the ratings matrix is sparse, this method only returns the items a user with id userId has purchased.
- **get_top_n**: by ordering all the predicted ratings for each recommendation algorithm, we can extract what would be their 'top' recommendation for a given user. Given a parameter $n$, we can then return all the top $n$ recommendations for all the recommendation algorithms.
- **rmse**: by comparing the observed ratings a given user has given to an item and the predicted rating an algorithm has defined for a user, we can have an idea of how much error the algorithm is predicting the user's ratings. Here we don't work with lists, as usually each user has rated only a few amount of items. So here we get all the items the user has rated, recover these items from the algorithms' recommendations and them calculate the error.
- **nDCG**: By looking at lists now, we can have an idea of how optimal the ranked lists are. By using the scoring factor defined in the report, we can calculate the overall DCG for the recommenders' lists and then normalise them using the concepts of the nDCG.
- **Price and avalaibility diversity**: Diversity metric which evaluate how the recommended items' prices vary, *i.e.*, how is the standard deviation of the price. The higher, the better in this case. The same is for the availability index, but here, with higher standard deviations, it means the models are recommending items which are present and not present in local stores.
- **Popularity**: A popular recommender tries to recommend items which has a high chance of being purchased. In the formulation of this metric, an item has a high chance of being purchased if lots of people have purchased them. In the class constructor, we take the observed ratings data and the item list and select which were the top $n$ (standard = 20) most purchased data. In a recommendation list, we return the ration of how many items were inside this list of top $n$ ones.
```
class RecommenderEvaluator:
def __init__(self, items, actual_ratings, content_based, user_user, item_item, matrix_fact, pers_bias):
self.items = items
self.actual_ratings = actual_ratings
# static data containing the average score given by each user
self.average_rating_per_userid = actual_ratings.apply(lambda row: np.average(row[~np.isnan(row)]))
self.content_based = content_based
self.user_user = user_user
self.item_item = item_item
self.matrix_fact = matrix_fact
self.pers_bias = pers_bias
# aggregate list. Makes for loops among all recommenders' predictions easier
self.recommenders_list = [self.content_based, self.user_user, self.item_item, self.matrix_fact,self.pers_bias]
self.recommenders_list_names = ['content_based', 'user_user', 'item_item', 'matrix_fact','pers_bias']
# Used for item popularity metric.
# Calculate the 20 most popular items (item which most of the customers bought)
N_LIM = 20
perc_users_bought_item = self.actual_ratings.apply(lambda item: np.sum(~np.isnan(item)), axis=0)/actual_ratings.shape[1]
sort_pop_items = np.argsort(perc_users_bought_item)[::-1]
self.pop_items = perc_users_bought_item.iloc[sort_pop_items][:N_LIM].index.values.astype(np.int)
def get_observed_ratings(self, userId):
"""
Returns all the items a given user evaluated and their ratings. Used mainly by all the metrics calculation
:parameter: userId - user id
:return: array of rated items. Index is the item id and value is the item rating
"""
userId = str(userId)
filtered_ratings = self.actual_ratings[userId]
rated_items = filtered_ratings[~np.isnan(filtered_ratings)]
return rated_items
def get_top_n(self, userId, n):
"""
Get the top n recommendations for every recommender in the list given a user id
:parameter: userId - user id
:parameter: n - max number of recommendations to return
:return: dictionary where the key is the recommender's name and the value is an array of size n for the top n recommnendations.
"""
userId = str(userId)
predicted_ratings = dict()
for recommender, recommender_name in zip(self.recommenders_list,self.recommenders_list_names):
item_ids = recommender[userId].argsort().sort_values()[:n].index.values
predicted_ratings[recommender_name] = item_ids
return predicted_ratings
def rmse(self, userId):
"""
Root Mean Square Error of the predicted and observed values between the recommender's prediction and the actual ratings
:parameter: userId - user id
:return: dataframe of containing the rmse from all recommenders given user id
"""
userId = str(userId)
observed_ratings = self.get_observed_ratings(userId)
rmse_list = {'rmse': []}
for recommender in self.recommenders_list:
predicted_ratings = recommender.loc[observed_ratings.index, userId]
rmse_list['rmse'].append(np.sqrt(np.average((predicted_ratings - observed_ratings)**2)))
rmse_list = pd.DataFrame(rmse_list, index = self.recommenders_list_names)
return rmse_list
def nDCG(self, userId, top_n = 5, individual_recommendation = None):
"""
Normalised Discounted Cumulative Gain for all recommenders given user id
:parameter: userId - user id
:return: dataframe of containing the nDCG from all recommenders given user id
"""
ri = self.get_observed_ratings(userId)
if(individual_recommendation is None):
topn = self.get_top_n(userId,top_n)
results_pandas_index = self.recommenders_list_names
else:
topn = individual_recommendation
results_pandas_index = list(individual_recommendation.keys())
# 1st step: Given recommendations, transform list into scores (see score transcriptions in the capstone report)
scores_all = []
for name, item_list in topn.items():
scores = np.empty_like(item_list) # initialise 'random' array
scores[:] = -10 ###########################
# check which items returned by the recommender
is_already_rated = np.isin(item_list, ri.index.values) # the user already rated. Items users didn't rate
scores[~is_already_rated] = 0 # receive score = 0
for index, score in enumerate(scores):
if(score != 0): # for each recommended items the user rated
if(ri[item_list[index]] < self.average_rating_per_userid[userId] - 1): # score accordingly the report
scores[index] = -1
elif((ri[item_list[index]] >= self.average_rating_per_userid[userId] - 1) &
(ri[item_list[index]] < self.average_rating_per_userid[userId] + 0.5)):
scores[index] = 1
else:
scores[index] = 2
scores_all.append(scores) # append all the transformed scores
scores_all
# 2nd step: Given scores, calculate the model's DCG, ideal DCG and then nDCG
nDCG_all = dict()
for index_model, scores_model in enumerate(scores_all): # for each model
model_DCG = 0 # calculate model's DCG
for index, score in enumerate(scores_model): #
index_ = index + 1 #
model_DCG = model_DCG + score/np.log2(index_ + 1) #
ideal_rank_items = np.sort(scores_model)[::-1] # calculate model's ideal DCG
ideal_rank_DCG = 0 #
for index, ideal_score in enumerate(ideal_rank_items): #
index_ = index + 1 #
ideal_rank_DCG = ideal_rank_DCG + ideal_score/np.log2(index_ + 1) #
if((ideal_rank_DCG == 0) | (np.abs(ideal_rank_DCG) < np.abs(model_DCG))): # if nDCG is 0 or only negative scores came up
nDCG = 0
else: # calculate final nDCG when ideal DCG is != 0
nDCG = model_DCG/ideal_rank_DCG
nDCG_all[results_pandas_index[index_model]] = nDCG # save each model's nDCG in a dict
# convert it to dataframe
result_final = pd.DataFrame(nDCG_all, index=range(1)).transpose()
result_final.columns = ['nDCG']
return result_final
def price_diversity(self,userId,top_n = 5,individual_recommendation = None):
"""
Mean and standard deviation of the price of the top n products recommended by each algorithm.
Intuition for a high price wise diversity recommender is to have a high price standard deviation
:parameter: userId - user id
:return: dataframe of containing the price's mean and standard deviation from all recommenders given user id
"""
if(individual_recommendation is None):
topn = self.get_top_n(userId,top_n)
else:
topn = individual_recommendation
stats = pd.DataFrame()
for key, value in topn.items():
data_filtered = self.items.loc[topn[key]][['Price']].agg(['mean','std']).transpose()
data_filtered.index = [key]
stats = stats.append(data_filtered)
return stats
def availability_diversity(self,userId,top_n = 5,individual_recommendation = None):
"""
Mean and standard deviation of the availabity index of the top n products recommended by each algorithm.
Intuition for a high availabity diversity is to have a small mean value in the availabity index
:parameter: userId - user id
:return: dataframe of containing the availabity index's mean and standard deviation from all recommenders given user id
"""
if(individual_recommendation is None):
topn = self.get_top_n(userId,top_n)
else:
topn = individual_recommendation
stats = pd.DataFrame()
for key, value in topn.items():
data_filtered = self.items.loc[topn[key]][['Availability']].agg(['mean','std']).transpose()
data_filtered.index = [key]
stats = stats.append(data_filtered)
return stats
def popularity(self, userId,top_n = 5,individual_recommendation = None):
"""
Return the ratio of how many items of the top n items are among the most popular purchased items. Default is
the 20 most purchased items.
:parameter: userId - user id
:return: dataframe of containing ratio of popular items in the recommended list from all recommenders given user id
"""
if(individual_recommendation is None):
topn = self.get_top_n(userId,top_n)
results_pandas_index = self.recommenders_list_names
else:
topn = individual_recommendation
results_pandas_index = list(individual_recommendation.keys())
results = {'popularity': []}
for recommender, recommendations in topn.items():
popularity = np.sum(np.isin(recommendations,self.pop_items))
results['popularity'].append(popularity)
return pd.DataFrame(results,index = results_pandas_index)
def precision_at_n(self, userId, top_n = 5, individual_recommendation = None):
if(individual_recommendation is None):
topn = self.get_top_n(userId,top_n)
results_pandas_index = self.recommenders_list_names
else:
topn = individual_recommendation
results_pandas_index = list(individual_recommendation.keys())
observed_ratings = self.get_observed_ratings(userId).index.values
precisions = {'precision_at_'+str(top_n): []}
for recommender, recommendations in topn.items():
precisions['precision_at_'+str(top_n)].append(np.sum(np.isin(recommendations, observed_ratings))/top_n)
return pd.DataFrame(precisions,index = results_pandas_index)
```
# Test methods:
Just to have an idea of the output of each method, lets call all them with a test user. At the next section we will calculate these metrics for all users.
```
userId = '64'
re = RecommenderEvaluator(items, actual_ratings, content_based, user_user, item_item, matrix_fact, pers_bias)
```
## Test RMSE
```
re.rmse(userId)
```
## Test nDCG
```
re.nDCG(userId)
```
## Test Diversity - Price and Availability
```
re.price_diversity(userId)
re.availability_diversity(userId)
```
## Test Popularity
```
re.popularity(userId)
```
## Test Precision@N
```
re.precision_at_n(userId)
```
# Average metrics by all users
Espefically for user 907, the recommendations from the user user came with all nulls (original dataset). This specifically impacted the RMSE calculation, as one Nan damaged the entire average calculation. So specifically for RMSE we did a separate calculation section. All the other metrics are going the be calculated in the next code block.
```
re = RecommenderEvaluator(items, actual_ratings, content_based, user_user, item_item, matrix_fact, pers_bias)
i = 0
count = np.array([0,0,0,0,0])
for userId in actual_ratings.columns:
if(userId == '907'):
rmse_recommenders = re.rmse(userId).fillna(0)
else:
rmse_recommenders = re.rmse(userId)
count = count + rmse_recommenders['rmse']
# as we didn't use user 907 for user user, divide it by the number of users - 1
denominator = [len(actual_ratings.columns)] * 5
denominator[1] = len(actual_ratings.columns) - 1
print('Average RMSE for all users')
count/ denominator
count_nDCG = np.array([0,0,0,0,0])
count_diversity_price = np.ndarray([5,2])
count_diversity_availability = np.ndarray([5,2])
count_popularity = np.array([0,0,0,0,0])
count_precision_at_5 = np.array([0,0,0,0,0])
for userId in actual_ratings.columns:
nDCG_recommenders = re.nDCG(userId)
count_nDCG = count_nDCG + nDCG_recommenders['nDCG']
diversity_price_recommenders = re.price_diversity(userId)
count_diversity_price = count_diversity_price + diversity_price_recommenders[['mean','std']]
diversity_availability_recommenders = re.availability_diversity(userId)
count_diversity_availability = count_diversity_availability + diversity_availability_recommenders[['mean','std']]
popularity_recommenders = re.popularity(userId)
count_popularity = count_popularity + popularity_recommenders['popularity']
precision_recommenders = re.precision_at_n(userId)
count_precision_at_5 = count_precision_at_5 + precision_recommenders['precision_at_5']
print('\n---')
print('Average nDCG')
print('---\n')
print(count_nDCG/len(actual_ratings.columns))
print('\n---')
print('Average Price - Diversity Measure')
print('---\n')
print(count_diversity_price/len(actual_ratings.columns))
print('\n---')
print('Average Availability - Diversity Measure')
print('---\n')
print(count_diversity_availability/len(actual_ratings.columns))
print('\n---')
print('Average Popularity')
print('---\n')
print(count_popularity/len(actual_ratings.columns))
print('---\n')
print('Average Precision@5')
print('---\n')
print(count_precision_at_5/len(actual_ratings.columns))
```
# Final Analysis
In terms of **RMSE**, the user-user collaborative filtering showed to be the most effective, despite it not being significantly better.
For nDCG rank score, again user user and now item item collaborative filtering were the best.
In terms of price diversity, the item item algorith was the most diverse, providing products varying ~32 dollars from the mean item price list. Matrix factorisation and user user follow right behind, with price standard deviation around 25 dollars. An interesting factor here was the *pers_bias* algorithm, as it recommended basically cheap products with a low standard deviation.
For the availabity index, all the algorithms besides the user user managed to recommend items not so present in the local stores **together** with items present in local stores, as we can see they also provided items with availability index high (high standard deviation).
In terms of popularity, no algorithm actually managed to obtain good scores in the way we defined. So, if the popularity is focused in the future, we can either change the popularity concept or improve mechanics in the recommender so it predict higher scores for the most popular items in the store.
After this evaluation, it seemed to us that the item-item recommender system had an overall better performance, highlighted in terms of its diversity scores. Unfortunately, the items that item item recommender has suggested are in overall pricy, and we can check if there is any mixture possibility with the pers_bias algorithm, as it really indicated cheap prices and a low price standard deviation. Matrix factorization performed good as well but it didn't outperform any of the other recommenders.
# Hibridization Techniques - Part III
We are trying four different types of hibridization here.
1. Linear ensemble
2. Non linear ensemble
3. Top 1 from each recommender
4. Recommender switching
The first two options approach the recommender's performance in terms of how good it predicts the users' ratings, so its only evaluation will be in terms of RMSE.
The third approach have the intuition that, if we get the top 1 recommendation from each algorithm, the resulting 5 item list will have a better performance in terms of identyfing 'good' items to users. In this case, we defined the good items if the recommender suggested an already bought item for an user. Therefore, the final measurement of this hibridization mechanism is through the precision@5, as we end up with a 5 item list.
The final mixing algorithm has the underlying theory of how collaborative filtering mechanisms perform with items that had not enough users/items in its calculations. As a well known weakness of these recommenders, the idea was to check how many items we would affect if we established a threshold of enough data in order for us to use a collaborative filtering. Otherwise, if the item doesn't have enough support in form of users' ratings we could have a support of a content based recommendation, or even, in last case, a non personalised one.
## Dataset Creation and User Sample Definition
### Dataset
For the first and second approach, we need another perspective on the data. The dataset contains all the existing ratings from all users and concatenates all the predictions made the 5 traditional recommenders. The idea is to use the observed rating as target variable and all recommenders' predictions as dependent variable, *i.e.* treat this as a regression problems.
```
obs_ratings_list = []
content_based_list = []
user_user_list = []
item_item_list = []
matrix_fact_list = []
pers_bias_list = []
re = RecommenderEvaluator(items, actual_ratings, content_based, user_user, item_item, matrix_fact, pers_bias)
for userId in actual_ratings.columns:
observed_ratings = re.get_observed_ratings(userId)
obs_ratings_list.extend(observed_ratings.values)
content_based_list.extend(content_based.loc[observed_ratings.index, userId].values)
user_user_list.extend(user_user.loc[observed_ratings.index, userId].values)
item_item_list.extend(item_item.loc[observed_ratings.index, userId].values)
matrix_fact_list.extend(matrix_fact.loc[observed_ratings.index, userId].values)
pers_bias_list.extend(pers_bias.loc[observed_ratings.index, userId].values)
dataset = pd.DataFrame({'rating': obs_ratings_list, 'content_based':content_based_list, 'user_user': user_user_list,
'item_item':item_item_list, 'matrix_fact':matrix_fact_list,'pers_bias':pers_bias_list})
dataset = dataset.dropna()
dataset.head()
```
### In order to have an idea of the results, let's choose 3 users randomly to show the predictions using the new hybrid models
```
np.random.seed(42)
sample_users = np.random.choice(actual_ratings.columns, 3).astype(str)
print('sample_users: ' + str(sample_users))
```
### Get recommenders' predictions for sample users in order to create input for ensemble models (hybridization I and II)
```
from collections import OrderedDict
df_sample = pd.DataFrame()
for user in sample_users:
content_based_ = re.content_based[user]
user_user_ = re.user_user[user]
item_item_ = re.item_item[user]
matrix_fact_ = re.matrix_fact[user]
pers_bias_ = re.pers_bias[user]
df_sample = df_sample.append(pd.DataFrame(OrderedDict({'user':user,'item':actual_ratings.index.values,'content_based':content_based_, 'user_user':user_user_, 'item_item':item_item_,
'matrix_fact':matrix_fact_,'pers_bias':pers_bias_})), ignore_index=True)
df_sample.head()
```
## Focus on Performance (RMSE) I - Linear Model
```
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import cross_val_score
linear = LinearRegression()
print('RMSE for linear ensemble of recommender systems:')
np.mean(cross_val_score(linear, dataset.drop('rating', axis=1), dataset['rating'], cv=5))
```
### Predictions for sample users: Creating top 5 recommendations for sample users
```
pred_cols = ['content_based','user_user','item_item','matrix_fact','pers_bias']
predictions = linear.fit(dataset.drop('rating', axis=1), dataset['rating']).predict(df_sample[pred_cols])
recommendations = pd.DataFrame(OrderedDict({'user':df_sample['user'], 'item':df_sample['item'], 'predictions':predictions}))
recommendations.groupby('user').apply(lambda df_user : df_user.loc[df_user['predictions'].sort_values(ascending=False)[:5].index.values])
```
## Focus on Performance (RMSE) II - Emsemble
```
from sklearn.ensemble import RandomForestRegressor
rf = RandomForestRegressor(random_state=42)
print('RMSE for non linear ensemble of recommender systems:')
np.mean(cross_val_score(rf, dataset.drop('rating', axis=1), dataset['rating'], cv=5))
```
### Predictions for sample users:
```
predictions = rf.fit(dataset.drop('rating', axis=1), dataset['rating']).predict(df_sample[pred_cols])
recommendations = pd.DataFrame(OrderedDict({'user':df_sample['user'], 'item':df_sample['item'], 'predictions':predictions}))
recommendations.groupby('user').apply(lambda df_user : df_user.loc[df_user['predictions'].sort_values(ascending=False)[:5].index.values])
```
## Focus on Recommendations - Top 1 from each Recommender
With the all top 1 recommender, we can evaluate its performance not just with RMSE, but all the list metrics we evaluated before. As a business constraint, we will also pay more attention to the *precision@5* metric, as a general information on how good is the recommender on providing suggestions that the user will buy, or already bought in this case.
The majority of metrics were in the same scale as the best metrics in the all models comparison. However, it's good to highlight the the top 1 all recommender had the best *precision@5* metric among all recommender, showing to be a **good suitable hibridization mechanism**.
```
count_nDCG = np.array([0])
count_diversity_price = np.ndarray([1,2])
count_diversity_availability = np.ndarray([1,2])
count_popularity = np.array([0])
count_precision = np.array([0])
for userId in actual_ratings.columns:
top_n_1 = re.get_top_n(userId,1)
user_items = {}
user_items['top_1_all'] = [a[0] for a in top_n_1.values()]
nDCG_recommenders = re.nDCG(userId, individual_recommendation = user_items)
count_nDCG = count_nDCG + nDCG_recommenders['nDCG']
diversity_price_recommenders = re.price_diversity(userId, individual_recommendation = user_items)
count_diversity_price = count_diversity_price + diversity_price_recommenders[['mean','std']]
diversity_availability_recommenders = re.availability_diversity(userId, individual_recommendation = user_items)
count_diversity_availability = count_diversity_availability + diversity_availability_recommenders[['mean','std']]
popularity_recommenders = re.popularity(userId, individual_recommendation = user_items)
count_popularity = count_popularity + popularity_recommenders['popularity']
precision_recommenders = re.precision_at_n(userId, individual_recommendation = user_items)
count_precision = count_precision + precision_recommenders['precision_at_5']
print('\n---')
print('Average nDCG')
print('---\n')
print(count_nDCG/len(actual_ratings.columns))
print('\n---')
print('Average Price - Diversity Measure')
print('---\n')
print(count_diversity_price/len(actual_ratings.columns))
print('\n---')
print('Average Availability - Diversity Measure')
print('---\n')
print(count_diversity_availability/len(actual_ratings.columns))
print('\n---')
print('Average Popularity')
print('---\n')
print(count_popularity/len(actual_ratings.columns))
print('\n---')
print('Average Precision@5')
print('---\n')
print(count_precision/len(actual_ratings.columns))
```
### Predictions for sample users:
```
results = {}
for user_sample in sample_users:
results[user_sample] = [a[0] for a in list(re.get_top_n(user_sample, 1).values())]
results
```
## Focus on Recommendations - Switching algorithm
### Can we use a Content Based Recommender for items with less evaluations?
We can see in the cumulative histogram that only around 20% of the rated items had 10 or more ratings. This signals us that maybe we can prioritize the use of a content based recommender or even a non personalised one for the majority of the items which don't have a sufficient amount of ratings in order to make the collaborative filtering algorithms to be stable.
```
import matplotlib.pyplot as plt
item_nbr_ratings = actual_ratings.apply(lambda col: np.sum(~np.isnan(col)), axis=1)
item_max_nbr_ratings = item_nbr_ratings.max()
range_item_max_nbr_ratings = range(item_max_nbr_ratings+1)
plt.figure(figsize=(15,3))
plt.subplot(121)
nbr_ratings_items = []
for i in range_item_max_nbr_ratings:
nbr_ratings_items.append(len(item_nbr_ratings[item_nbr_ratings == i]))
plt.plot(nbr_ratings_items)
plt.xlabel('Number of ratings')
plt.ylabel('Amount of items')
plt.title('Histogram of amount of ratings')
plt.subplot(122)
cum_nbr_ratings_items = []
for i in range(len(nbr_ratings_items)):
cum_nbr_ratings_items.append(np.sum(nbr_ratings_items[:i]))
cum_nbr_ratings_items = np.array(cum_nbr_ratings_items)
plt.plot(cum_nbr_ratings_items/actual_ratings.shape[0])
plt.xlabel('Number of ratings')
plt.ylabel('Cumulative distribution')
plt.title('Cumulative histogram of amount of ratings');
```
| github_jupyter |
```
import os
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
STATS_DIR = "/hg191/corpora/legaldata/data/stats/"
SEM_FEATS_FILE = os.path.join (STATS_DIR, "ops.temp.semfeat")
INDEG_FILE = os.path.join (STATS_DIR, "ops.ind")
ind = pd.read_csv (INDEG_FILE, sep=",", header=None, names=["opid", "indeg"])
semfeat = pd.read_csv (SEM_FEATS_FILE, sep=",", header=None, names=["opid", "semfeat"])
indegs = pd.Series([ind[ind["opid"] == opid]["indeg"].values[0] for opid in semfeat.opid.values])
semfeat["indeg"] = indegs
def labelPercentile (series):
labels = list ()
p50 = np.percentile (series, q=50)
p75 = np.percentile (series, q=75)
p90 = np.percentile (series, q=90)
for value in series:
if value <= p50:
labels.append ("<=50")
elif value <= p90:
labels.append (">50")
elif value > p90:
labels.append (">90")
return labels
semfeat["percentile"] = pd.Series (labelPercentile(semfeat["semfeat"].values))
df = semfeat[semfeat["indeg"] > 0]
df["log(indeg)"] = np.log(df["indeg"])
ax = sns.boxplot(x="percentile", y="log(indeg)", data=df, order=["<=50", ">50", ">90"])
vals = df[df["percentile"] == ">50"]["log(indeg)"].values
np.sort(vals)[int(len(vals)/2)]
print(len(df[df["percentile"] == ">50"]))
print(len(df[df["percentile"] == ">90"]))
print (df[df["percentile"] == "<=50"]["log(indeg)"].median())
print (df[df["percentile"] == ">50"]["log(indeg)"].median())
print (df[df["percentile"] == ">90"]["log(indeg)"].median())
#print (semfeat[semfeat["percentile"] == ">P99"]["logindeg"].mean())
print (semfeat[semfeat["percentile"] == "<=P50"]["logindeg"].mean())
print (semfeat[semfeat["percentile"] == ">P50"]["logindeg"].mean())
print (semfeat[semfeat["percentile"] == ">P90"]["logindeg"].mean())
print (semfeat[semfeat["percentile"] == "<=P50"]["indeg"].median())
print (semfeat[semfeat["percentile"] == ">P50"]["indeg"].median())
print (semfeat[semfeat["percentile"] == ">P90"]["indeg"].median())
print (semfeat[semfeat["percentile"] == "<=P50"]["indeg"].median())
print (semfeat[semfeat["percentile"] == ">P50"]["indeg"].median())
print (semfeat[semfeat["percentile"] == ">P90"]["indeg"].median())
np.percentile(semfeat["semfeat"].values, q=90)
[semfeat["percentile"] == ">P90"]["indeg"].mean()
semfeat[semfeat["percentile"] == ">P90"].tail(500)
sorted(semfeat["indeg"], reverse=True)[0:10]
semfeat[semfeat["indeg"].isin(sorted(semfeat["indeg"], reverse=True)[0:10])]
semfeat.loc[48004,]["semfeat"] = 1
semfeat[semfeat["indeg"].isin(sorted(semfeat["indeg"], reverse=True)[0:10])]
print(np.mean((semfeat[semfeat["percentile"] == "<=P50"]["indeg"] > 0).values))
print(np.mean((semfeat[semfeat["percentile"] == ">P50"]["indeg"] > 0).values))
print(np.mean((semfeat[semfeat["percentile"] == ">P90"]["indeg"] > 0).values))
print (len(semfeat[(semfeat["percentile"] == "<=P50") & (semfeat["indeg"] > 0)]))
print (len(semfeat[(semfeat["percentile"] == ">P50") & (semfeat["indeg"] > 0)]))
print (len(semfeat[(semfeat["percentile"] == ">P90") & (semfeat["indeg"] > 0)]))
print (semfeat[(semfeat["percentile"] == "<=P50") & (semfeat["indeg"] > 0)]["indeg"].mean())
print (semfeat[(semfeat["percentile"] == ">P50") & (semfeat["indeg"] > 0)]["indeg"].mean())
print (semfeat[(semfeat["percentile"] == ">P90") & (semfeat["indeg"] > 0)]["indeg"].mean())
print (semfeat[(semfeat["percentile"] == "<=P50") & (semfeat["indeg"] > 0)]["logindeg"].mean())
print (semfeat[(semfeat["percentile"] == ">P50") & (semfeat["indeg"] > 0)]["logindeg"].mean())
print (semfeat[(semfeat["percentile"] == ">P90") & (semfeat["indeg"] > 0)]["logindeg"].mean())
ax = sns.violinplot(x="percentile", y="logindeg", data=df, order=["<=P50", ">P50", ">P90"])
semfeat[semfeat["indeg"] == 1]
```
| github_jupyter |
# Geometric operations
## Overlay analysis
In this tutorial, the aim is to make an overlay analysis where we create a new layer based on geometries from a dataset that `intersect` with geometries of another layer. As our test case, we will select Polygon grid cells from `TravelTimes_to_5975375_RailwayStation_Helsinki.shp` that intersects with municipality borders of Helsinki found in `Helsinki_borders.shp`.
Typical overlay operations are (source: [QGIS docs](https://docs.qgis.org/2.8/en/docs/gentle_gis_introduction/vector_spatial_analysis_buffers.html#more-spatial-analysis-tools)):

## Download data
For this lesson, you should [download a data package](https://github.com/AutoGIS/data/raw/master/L4_data.zip) that includes 3 files:
1. Helsinki_borders.shp
2. Travel_times_to_5975375_RailwayStation.shp
3. Amazon_river.shp
```
$ cd /home/jovyan/notebooks/L4
$ wget https://github.com/AutoGIS/data/raw/master/L4_data.zip
$ unzip L4_data.zip
```
Let's first read the data and see how they look like.
- Import required packages and read in the input data:
```
import geopandas as gpd
import matplotlib.pyplot as plt
import shapely.speedups
%matplotlib inline
# File paths
border_fp = "data/Helsinki_borders.shp"
grid_fp = "data/TravelTimes_to_5975375_RailwayStation.shp"
# Read files
grid = gpd.read_file(grid_fp)
hel = gpd.read_file(border_fp)
```
- Visualize the layers:
```
# Plot the layers
ax = grid.plot(facecolor='gray')
hel.plot(ax=ax, facecolor='None', edgecolor='blue')
```
Here the grey area is the Travel Time Matrix grid (13231 grid squares) that covers the Helsinki region, and the blue area represents the municipality of Helsinki. Our goal is to conduct an overlay analysis and select the geometries from the grid polygon layer that intersect with the Helsinki municipality polygon.
When conducting overlay analysis, it is important to check that the CRS of the layers match!
- Check if Helsinki polygon and the grid polygon are in the same crs:
```
# Ensure that the CRS matches, if not raise an AssertionError
assert hel.crs == grid.crs, "CRS differs between layers!"
```
Indeed, they do. Hence, the pre-requisite to conduct spatial operations between the layers is fullfilled (also the map we plotted indicated this).
- Let's do an overlay analysis and create a new layer from polygons of the grid that `intersect` with our Helsinki layer. We can use a function called `overlay()` to conduct the overlay analysis that takes as an input 1) the GeoDataFrame where the selection is taken, 2) the GeoDataFrame used for making the selection, and 3) parameter `how` that can be used to control how the overlay analysis is conducted (possible values are `'intersection'`, `'union'`, `'symmetric_difference'`, `'difference'`, and `'identity'`):
```
intersection = gpd.overlay(grid, hel, how='intersection')
```
- Let's plot our data and see what we have:
```
intersection.plot(color="b")
```
As a result, we now have only those grid cells that intersect with the Helsinki borders. As we can see **the grid cells are clipped based on the boundary.**
- Whatabout the data attributes? Let's see what we have:
```
print(intersection.head())
```
As we can see, due to the overlay analysis, the dataset contains the attributes from both input layers.
- Let's save our result grid as a GeoJSON file that is commonly used file format nowadays for storing spatial data.
```
# Output filepath
outfp = "data/TravelTimes_to_5975375_RailwayStation_Helsinki.geojson"
# Use GeoJSON driver
intersection.to_file(outfp, driver="GeoJSON")
```
There are many more examples for different types of overlay analysis in [Geopandas documentation](http://geopandas.org/set_operations.html) where you can go and learn more.
## Aggregating data
Data aggregation refers to a process where we combine data into groups. When doing spatial data aggregation, we merge the geometries together into coarser units (based on some attribute), and can also calculate summary statistics for these combined geometries from the original, more detailed values. For example, suppose that we are interested in studying continents, but we only have country-level data like the country dataset. If we aggregate the data by continent, we would convert the country-level data into a continent-level dataset.
In this tutorial, we will aggregate our travel time data by car travel times (column `car_r_t`), i.e. the grid cells that have the same travel time to Railway Station will be merged together.
- For doing the aggregation we will use a function called `dissolve()` that takes as input the column that will be used for conducting the aggregation:
```
# Conduct the aggregation
dissolved = intersection.dissolve(by="car_r_t")
# What did we get
print(dissolved.head())
```
- Let's compare the number of cells in the layers before and after the aggregation:
```
print('Rows in original intersection GeoDataFrame:', len(intersection))
print('Rows in dissolved layer:', len(dissolved))
```
Indeed the number of rows in our data has decreased and the Polygons were merged together.
What actually happened here? Let's take a closer look.
- Let's see what columns we have now in our GeoDataFrame:
```
print(dissolved.columns)
```
As we can see, the column that we used for conducting the aggregation (`car_r_t`) can not be found from the columns list anymore. What happened to it?
- Let's take a look at the indices of our GeoDataFrame:
```
print(dissolved.index)
```
Aha! Well now we understand where our column went. It is now used as index in our `dissolved` GeoDataFrame.
- Now, we can for example select only such geometries from the layer that are for example exactly 15 minutes away from the Helsinki Railway Station:
```
# Select only geometries that are within 15 minutes away
dissolved.iloc[15]
# See the data type
print(type(dissolved.iloc[15]))
# See the data
print(dissolved.iloc[15].head())
```
As we can see, as a result, we have now a Pandas `Series` object containing basically one row from our original aggregated GeoDataFrame.
Let's also visualize those 15 minute grid cells.
- First, we need to convert the selected row back to a GeoDataFrame:
```
# Create a GeoDataFrame
selection = gpd.GeoDataFrame([dissolved.iloc[15]], crs=dissolved.crs)
```
- Plot the selection on top of the entire grid:
```
# Plot all the grid cells, and the grid cells that are 15 minutes a way from the Railway Station
ax = dissolved.plot(facecolor='gray')
selection.plot(ax=ax, facecolor='red')
```
## Simplifying geometries
Sometimes it might be useful to be able to simplify geometries. This could be something to consider for example when you have very detailed spatial features that cover the whole world. If you make a map that covers the whole world, it is unnecessary to have really detailed geometries because it is simply impossible to see those small details from your map. Furthermore, it takes a long time to actually render a large quantity of features into a map. Here, we will see how it is possible to simplify geometric features in Python.
As an example we will use data representing the Amazon river in South America, and simplify it's geometries.
- Let's first read the data and see how the river looks like:
```
import geopandas as gpd
# File path
fp = "data/Amazon_river.shp"
data = gpd.read_file(fp)
# Print crs
print(data.crs)
# Plot the river
data.plot();
```
The LineString that is presented here is quite detailed, so let's see how we can generalize them a bit. As we can see from the coordinate reference system, the data is projected in a metric system using [Mercator projection based on SIRGAS datum](http://spatialreference.org/ref/sr-org/7868/).
- Generalization can be done easily by using a Shapely function called `.simplify()`. The `tolerance` parameter can be used to adjusts how much geometries should be generalized. **The tolerance value is tied to the coordinate system of the geometries**. Hence, the value we pass here is 20 000 **meters** (20 kilometers).
```
# Generalize geometry
data2 = data.copy()
data2['geom_gen'] = data2.simplify(tolerance=20000)
# Set geometry to be our new simlified geometry
data2 = data2.set_geometry('geom_gen')
# Plot
data2.plot()
# plot them side-by-side
%matplotlib inline
import matplotlib.pyplot as plt
#basic config
fig, (ax1,ax2) = plt.subplots(nrows=1, ncols=2, figsize=(20, 16))
#ax1, ax2 = axes
#1st plot
ax1 = data.plot(ax=ax1, color='red', alpha=0.5)
ax1.set_title('Original')
#2nd plot
ax2 = data2.plot(ax=ax2, color='orange', alpha=0.5)
ax2.set_title('Generalize')
fig.tight_layout()
```
Nice! As a result, now we have simplified our LineString quite significantly as we can see from the map.
| github_jupyter |
# GNN Implementation
- Name: Abhishek Aditya BS
- SRN: PES1UG19CS019
- VI Semester 'A' Section
- Date: 27-04-2022
```
import sys
if 'google.colab' in sys.modules:
%pip install -q stellargraph[demos]==1.2.1
import pandas as pd
import os
import stellargraph as sg
from stellargraph.mapper import FullBatchNodeGenerator
from stellargraph.layer import GCN
from tensorflow.keras import layers, optimizers, losses, metrics, Model
from sklearn import preprocessing, model_selection
from IPython.display import display, HTML
import matplotlib.pyplot as plt
dataset=sg.datasets.Cora()
display(HTML(dataset.description))
G, node_subjects = dataset.load()
print(G.info())
node_subjects.value_counts().to_frame()
train_subjects, test_subjects = model_selection.train_test_split(node_subjects, train_size=140, test_size=None, stratify=node_subjects)
val_subjects, test_subjects = model_selection.train_test_split(test_subjects, train_size=500, test_size=None, stratify=test_subjects)
train_subjects.value_counts().to_frame()
target_encoding=preprocessing.LabelBinarizer()
train_targets=target_encoding.fit_transform(train_subjects)
val_targets=target_encoding.transform(val_subjects)
test_targets=target_encoding.transform(test_subjects)
from stellargraph.mapper.full_batch_generators import FullBatchGenerator
generator = FullBatchNodeGenerator(G, method="gcn")
train_gen=generator.flow(train_subjects.index, train_targets)
gcn=GCN(layer_sizes=[16,16], activations=['relu', 'relu'], generator=generator, dropout=0.5)
x_inp, x_out = gcn.in_out_tensors()
x_out
predictions=layers.Dense(units=train_targets.shape[1], activation="softmax")(x_out)
model=Model(inputs=x_inp, outputs=predictions)
model.compile(optimizer=optimizers.Adam(lr=0.01), loss=losses.categorical_crossentropy, metrics=["acc"])
val_gen = generator.flow(val_subjects.index, val_targets)
from tensorflow.keras.callbacks import EarlyStopping
os_callback = EarlyStopping(monitor="val_acc", patience=50, restore_best_weights=True)
history = model.fit(train_gen, epochs=200, validation_data=val_gen, verbose=2, shuffle=False, callbacks=[os_callback])
sg.utils.plot_history(history)
test_gen=generator.flow(test_subjects.index, test_targets)
all_nodes=node_subjects.index
all_gen=generator.flow(all_nodes)
all_predictions=model.predict(all_gen)
node_predictions=target_encoding.inverse_transform(all_predictions.squeeze())
df=pd.DataFrame({"Predicted":node_predictions, "True":node_subjects})
df.head(20)
embedding_model=Model(inputs=x_inp, outputs=x_out)
emb=embedding_model.predict(all_gen)
emb.shape
from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
X=emb.squeeze(0)
X.shape
transform = TSNE
trans=transform(n_components=2)
X_reduced=trans.fit_transform(X)
X_reduced.shape
fig, ax = plt.subplots(figsize=(7, 7))
ax.scatter(
X_reduced[:, 0],
X_reduced[:, 1],
c=node_subjects.astype("category").cat.codes,
cmap="jet",
alpha=0.7,
)
ax.set(
aspect="equal",
xlabel="$X_1$",
ylabel="$X_2$",
title=f"{transform.__name__} visualization of GCN embeddings for cora dataset",
)
```
| github_jupyter |
# Solution to puzzle number 5
```
import pandas as pd
import numpy as np
data = pd.read_csv('../inputs/puzzle5_input.csv')
data = [val for val in data.columns]
data[:10]
```
## Part 5.1
### After providing 1 to the only input instruction and passing all the tests, what diagnostic code does the program produce?
More Rules:
- Opcode 3 takes a single integer as input and saves it to the position given by its only parameter.
- Opcode 4 outputs the value of its only parameter.
Functions now need to support the parameter mode 1 (Immediate mode):
- Immediate mode
- In immediate mode, a parameter is interpreted as a value - if the parameter is 50, its value is 50.
```
user_ID = 1
numbers = 1002,4,3,4,33
def opcode_instructions(intcode):
"Function that breaks the opcode instructions into pieces"
str_intcode = str(intcode)
opcode = str_intcode[-2:]
int_opcode = int(opcode)
return int_opcode
def extract_p_modes(intcode):
"Function that extracts the p_modes"
str_p_modes = str(intcode)
p_modes_dic = {}
for n, val in enumerate(str_p_modes[:-2]):
p_modes_dic[f'p_mode_{n+1}'] = val
return p_modes_dic
def opcode_1(i, new_numbers, p_modes):
"Function that adds together numbers read from two positions and stores the result in a third position"
second_item = new_numbers[i+1]
third_item = new_numbers[i+2]
position_item = new_numbers[i+3]
if (p_modes[0] == 0) & (p_modes[1] == 0):
sum_of_second_and_third = new_numbers[second_item] + new_numbers[third_item]
elif (p_modes[0] == 1) & (p_modes[1] == 0):
sum_of_second_and_third = second_item + new_numbers[third_item]
elif (p_modes[0] == 0) & (p_modes[1] == 1):
sum_of_second_and_third = new_numbers[second_item] + third_item
else:
sum_of_second_and_third = second_item + third_item
new_numbers[position_item] = sum_of_second_and_third
return new_numbers
def opcode_2(i, new_numbers, p_modes):
"Function that multiplies together numbers read from two positions and stores the result in a third position"
second_item = new_numbers[i+1]
third_item = new_numbers[i+2]
position_item = new_numbers[i+3]
if (p_modes[0] == 0) & (p_modes[1] == 0):
m_of_second_and_third = new_numbers[second_item] * new_numbers[third_item]
elif (p_modes[0] == 1) & (p_modes[1] == 0):
m_of_second_and_third = second_item * new_numbers[third_item]
elif (p_modes[0] == 0) & (p_modes[1] == 1):
m_of_second_and_third = new_numbers[second_item] * third_item
else:
m_of_second_and_third = second_item * third_item
new_numbers[position_item] = m_of_second_and_third
return new_numbers
def opcode_3(i, new_numbers, inpt):
"Function takes a single integer as input and saves it to the position given by its only parameter"
val = input_value
second_item = new_numbers[i+1]
new_numbers[second_item] = val
return new_numbers
# from puzzle n2 copy the intcode function
def modifiedintcodefunction(numbers, input_value):
"Function that similates that of an Intcode program but takes into account extra information."
new_numbers = [num for num in numbers]
i = 0
output_values = []
while i < len(new_numbers):
opcode = opcode_instructions(new_numbers[i])
p_modes = extract_p_modes(new_numbers[i])
if new_numbers[i] == 1:
new_numbers = opcode_1(i, new_numbers, p_modes)
i = i + 4
elif new_numbers[i] == 2:
new_numbers = opcode_2(i, new_numbers, p_modes)
i = i + 4
elif new_numbers[i] == 3:
new_numbers = opcode_3(i, new_numbers, inpt)
i = i + 2
elif new_numbers[i] == 4:
output_values.append(new_numbers[i+1])
i = i + 2
elif new_numbers[i] == 99:
break
else:
continue
#Return the first item after the code has run.
first_item = new_numbers[0]
return first_item
```
| github_jupyter |
```
import nltk
from nltk.corpus import state_union
import pandas as pd
import os
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.decomposition import TruncatedSVD
from sklearn.decomposition import NMF
#from sklearn.metrics.pairwise import cosine_similarity
import matplotlib.pyplot as plt
%matplotlib inline
import pickle
from sklearn.feature_extraction.text import TfidfVectorizer
#path = 'state-of-the-union-corpus-1989-2017'
#path = 'c:\\Users\\gabri\\AppData\\Roaming\\nltk_data\\corpora\\state_union' # file location
path = 'c:\\Users\\gabri\\OneDrive\\Documents\\Metis_NLP_Kaggle\\Speeches\\sotu' # file location
dirs = os.listdir(path) # reads all the files in that directory
print (len(dirs)) #tell how many files
dirs[:] # file names
```
# Selecting the first speech to see what we need to clean.
```
filename = os.path.join(path, dirs[0]) # dirs is a list, and we are going to study the first element dirs[0]
text_file = open(filename, 'r') #open the first file dirs[0]
lines = text_file.read() # read the file
lines # print what is in the file
lines.replace('\n', ' ') # remove the \n symbols by replacing with an empty space
#print (lines)
sotu_data = [] #create an empty list
sotu_dict = {} # create an empty dictionary so that we can use file names to list the speeches by name
```
# Putting all the speeches into a list, after cleaning them
```
#The filter() function returns an iterator were the items are filtered
#through a function to test if the item is accepted or not.
# str.isalpha : checks if it is an alpha character.
# lower() : transform everything to lower case
# split() : Split a string into a list where each word is a list item
# loop over all the files:
for i in range(len(dirs)): # loop on all the speeches, dirs is the list of speeches
filename = os.path.join(path, dirs[i]) # location of the speeches
text_file = open(filename, 'r') # read the speeches
lines = text_file.read() #read the speeches
lines = lines.replace('\n', ' ') #replace \n by an empty string
# tranform the speeches in lower cases, split them into a list and then filter to accept only alpha characters
# finally it joins the words with an empty space
clean_lines = ' '.join(filter(str.isalpha, lines.lower().split()))
#print(clean_lines)
sotu_data.append(clean_lines) # append the clean speeches to the sotu_data list.
sotu_dict[filename] = clean_lines # store in dict so we can access clean_lines by filename.
sotu_data[10] #11th speech/element
speech_name = 'Wilson_1919.txt'
sotu_dict[path + '\\' + speech_name]
```
# Count Vectorize
```
#from notebook
#vectorizer = CountVectorizer(stop_words='english') #remove stop words: a, the, and, etc.
vectorizer = TfidfVectorizer(stop_words='english', max_df = 0.42, min_df = 0.01) #remove stop words: a, the, and, etc.
doc_word = vectorizer.fit_transform(sotu_data) #transform into sparse matrix (0, 1, 2, etc. for instance(s) in document)
pairwise_similarity = doc_word * doc_word.T
doc_word.shape # 228 = number of documents, 20932 = # of unique words)
#pairwise_similarity.toarray()
```
# Compare how similar speeches are to one another
```
df_similarity = pd.DataFrame(pairwise_similarity.toarray(), index = dirs, columns = dirs)
df_similarity.head() #similarity dataframe, compares each document to eachother
df_similarity.to_pickle("df_similarity.pkl") #pickle file
df_similarity['Speech_str'] = dirs #matrix comparing speech similarity
df_similarity['Year'] =df_similarity['Speech_str'].replace('[^0-9]', '', regex=True)
df_similarity.drop(['Speech_str'], axis=1)
df_similarity = df_similarity.sort_values(by=['Year'])
df_similarity.head()
plt.subplots(2, 2, figsize=(30, 15), sharex=True) #4 speeches similarity
#
plt.rcParams.update({'font.size': 20})
plt.subplot(2, 2, 1)
plt.plot(df_similarity['Year'], df_similarity['Adams_1797.txt'])
plt.title("Similarity for Adams 1797 speech")
plt.xlabel("Year")
plt.ylabel("Similarity")
plt.axhline(y=0.0, color='k', linestyle='-')
plt.xticks(['1800', '1850','1900','1950','2000']) # Set label locations.
plt.subplot(2, 2, 2)
plt.plot(df_similarity['Year'], df_similarity['Roosevelt_1945.txt'])
plt.title("Similarity for Roosevelt 1945 speech")
plt.xlabel("Year")
plt.ylabel("Similarity")
plt.axhline(y=0.0, color='k', linestyle='-')
plt.xticks(['1800', '1850','1900','1950','2000']) # Set label locations.
plt.subplot(2, 2, 3)
plt.plot(df_similarity['Year'], df_similarity['Obama_2014.txt'])
plt.title("Similarity for Obama 2014 speech")
plt.xlabel("Year")
plt.ylabel("Similarity")
plt.axhline(y=0.0, color='k', linestyle='-')
plt.xticks(['1800', '1850','1900','1950','2000']) # Set label locations.
plt.subplot(2, 2, 4)
plt.plot(df_similarity['Year'], df_similarity['Trump_2018.txt'])
plt.title("Similarity for Trump 2018 speech")
plt.xlabel("Year")
plt.ylabel("Similarity")
plt.axhline(y=0.0, color='k', linestyle='-')
plt.xticks(['1800', '1850','1900','1950','2000']) # Set label locations.
plt.subplots_adjust(top=0.90, bottom=0.02, wspace=0.30, hspace=0.3)
#sns.set()
plt.show()
#(sotu_dict.keys())
#for i in range(0,len(dirs)):
# print(dirs[i])
```
# Transforming the doc into a dataframe
```
# We have to convert `.toarray()` because the vectorizer returns a sparse matrix.
# For a big corpus, we would skip the dataframe and keep the output sparse.
#pd.DataFrame(doc_word.toarray(), index=sotu_data, columns=vectorizer.get_feature_names()).head(10) #doc_word.toarray() makes 7x19 table, otherwise it would be
#represented in 2 columns
#from notebook
pd.DataFrame(doc_word.toarray(), index=dirs, columns=vectorizer.get_feature_names()).head(95) #doc_word.toarray() makes 7x19 table, otherwise it would be
#represented in 2 columns
```
# Topic Modeling using nmf
```
n_topics = 8 # number of topics
nmf_model = NMF(n_topics) # create an object
doc_topic = nmf_model.fit_transform(doc_word) #break into 10 components like SVD
topic_word = pd.DataFrame(nmf_model.components_.round(3), #,"component_9","component_10","component_11","component_12"
index = ["component_1","component_2","component_3","component_4","component_5","component_6","component_7","component_8"],
columns = vectorizer.get_feature_names()) #8 components in final draft
topic_word
#https://stackoverflow.com/questions/16486252/is-it-possible-to-use-argsort-in-descending-order/16486299
#list the top words for each Component:
def print_top_words(model, feature_names, n_top_words):
for topic_idx, topic in enumerate(model.components_): # loop over the model components
print("Component_" + "%d:" % topic_idx ) # print the component
# join the top words by an empty space
# argsort : sorts the list in increasing order, meaning the top are the last words
# then select the top words
# -1 loops backwards
# reading from the tail to find the largest elements
print(" ".join([feature_names[i]
for i in topic.argsort()[:-n_top_words - 1:-1]]))
print()
```
# Top 15 words in each component
```
n_top_words = 15
feature_names = vectorizer.get_feature_names()
print_top_words(nmf_model, feature_names, n_top_words)
#Component x Speech
H = pd.DataFrame(doc_topic.round(5), index=dirs, #,"component_9","component_10"
columns = ["component_1","component_2", "component_3","component_4","component_5","component_6","component_7","component_8"])
H.head()
H.iloc[30:35]
H.iloc[60:70]
H.iloc[225:230]
```
# Use NMF to plot top 15 words for each of 8 components
def plot_top_words(model, feature_names, n_top_words, title):
fig, axes = plt.subplots(2, 4, figsize=(30, 15), sharex=True)
axes = axes.flatten()
for topic_idx, topic in enumerate(model.components_):
top_features_ind = topic.argsort()[:-n_top_words - 1:-1]
top_features = [feature_names[i] for i in top_features_ind]
weights = topic[top_features_ind]
ax = axes[topic_idx]
ax.barh(top_features, weights, height=0.7)
ax.set_title(f'Topic {topic_idx +1}',
fontdict={'fontsize': 30})
ax.invert_yaxis()
ax.tick_params(axis='both', which='major', labelsize=20)
for i in 'top right left'.split():
ax.spines[i].set_visible(False)
fig.suptitle(title, fontsize=40)
plt.subplots_adjust(top=0.90, bottom=0.05, wspace=0.90, hspace=0.3)
plt.show()
```
n_top_words = 12
feature_names = vectorizer.get_feature_names()
plot_top_words(nmf_model, feature_names, n_top_words,
'Topics in NMF model') #title
```
# Sort speeches Chronologically
```
H1 = H
H1['Speech_str'] = dirs
H1['Year'] = H1['Speech_str'].replace('[^0-9]', '', regex=True)
H1 = H1.sort_values(by = ['Year'])
H1.to_csv("Data_H1.csv", index = False) #Save chronologically sorted speeches in this csv
H1.head()
H1.to_pickle("H1.pkl") #pickle chronological csv file
```
# Plots of Components over Time (check Powerpoint/Readme for more insights)
```
plt.subplots(4, 2, figsize=(30, 15), sharex=True)
plt.rcParams.update({'font.size': 20})
plt.subplot(4, 2, 1)
plt.plot(H1['Year'], H1['component_1'] ) #Label axis and titles for all plots
plt.title("19th Century Economic Terms")
plt.xlabel("Year")
plt.ylabel("Component_1")
plt.axhline(y=0.0, color='k', linestyle='-')
plt.xticks(['1800', '1850','1900','1950','2000']) # Set label locations.
plt.subplot(4, 2, 2)
plt.plot(H1['Year'], H1['component_2'])
plt.title("Modern Economic Language")
plt.xlabel("Year")
plt.ylabel("Component_2")
plt.axhline(y=0.0, color='k', linestyle='-')
plt.xticks(['1800', '1850','1900','1950','2000']) # Set label locations.
plt.subplot(4, 2, 3)
plt.plot(H1['Year'], H1['component_3'])
plt.title("Growth of US Gov't & Programs")
plt.xlabel("Year")
plt.ylabel("Component_3")
plt.axhline(y=0.0, color='k', linestyle='-')
plt.xticks(['1800', '1850','1900','1950','2000']) # Set label locations.
plt.subplot(4, 2, 4)
plt.plot(H1['Year'], H1['component_4'])
plt.title("Early Foreign Policy & War")
plt.xlabel("Year")
plt.ylabel("Component_4")
plt.axhline(y=0.0, color='k', linestyle='-')
plt.xticks(['1800', '1850','1900','1950','2000']) # Set label locations.
plt.subplot(4, 2, 5)
plt.plot(H1['Year'], H1['component_5'])
plt.title("Progressive Era & Roaring 20s")
plt.xlabel("Year")
plt.ylabel("Component_5")
plt.axhline(y=0.0, color='k', linestyle='-')
plt.xticks(['1800', '1850','1900','1950','2000']) # Set label locations.
plt.subplot(4, 2, 6)
plt.plot(H1['Year'], H1['component_6'])
plt.title("Before, During, After the Civil War")
plt.xlabel("Year")
plt.ylabel("Component_6")
plt.axhline(y=0.0, color='k', linestyle='-')
plt.xticks(['1800', '1850','1900','1950','2000']) # Set label locations.
plt.subplot(4, 2, 7)
plt.plot(H1['Year'], H1['component_7'])
plt.title("World War & Cold War")
plt.xlabel("Year")
plt.ylabel("Component_7")
plt.axhline(y=0.0, color='k', linestyle='-')
plt.xticks(['1800', '1850','1900','1950','2000']) # Set label locations.
plt.subplot(4, 2, 8)
plt.plot(H1['Year'], H1['component_8'])
plt.title("Iraq War & Terrorism")
plt.xlabel("Year")
plt.ylabel("Component_8")
plt.axhline(y=0.0, color='k', linestyle='-')
plt.xticks(['1800', '1850','1900','1950','2000']) # Set label locations.
plt.subplots_adjust(top=0.90, bottom=0.02, wspace=0.30, hspace=0.4)
plt.show()
```
## Component 1: 19th Century Economics
```
H1.iloc[75:85] #Starts 1831. Peak starts 1868 (apex=1894), Nosedive in 1901 w/ Teddy. 4 Yr resurgence under Taft (1909-1912)
```
## Component 2: Modern Economic Language
```
H1.iloc[205:215] #1960s: Starts under JFK in 1961, peaks w/ Clinton, dips post 9/11 Bush, resurgence under Obama
```
## Component 3: Growth of US Government and Federal Programs
```
H1.iloc[155:165] #1921, 1929-1935. Big peak in 1946-1950 (1951 Cold War). 1954-1961 Eisenhower. Low after Reagan Revolution (1984)
```
## Component 4: Early Foreign Policy and War
```
H1.iloc[30:40] #Highest from 1790-1830, Washington to Jackson
```
## Component 5: Progressive Era, Roaring 20s
```
H1.iloc[115:125] #Peaks in 1900-1930.Especially Teddy Roosevelt. Dip around WW1
```
## Component 6: War Before, During, and After the Civil War
```
H1.iloc[70:80] #Starts w/ Jackson 1829, Peaks w/ Mexican-American War (1846-1848). Drops 60% w/ Lincoln. Peak ends w/ Johnson 1868. Remains pretty low after 1876 (Reconstruction ends)
```
## Component 7: World Wars and Korean War
```
H1.iloc[155:165] #Minor Peak around WW1. Masssive spike a response of Cold War, Korean War (1951). Eisenhower drops (except 1960 U2). Johnson Vietnam. Peaks again 1980 (Jimmy Carter foreign policy crises)
```
## Component 8: Iraq War and Terrorism
```
H1.iloc[210:220] #Minor peak w/ Bush 1990. BIG peak w/ Bush 2002. Ends w/ Obama 2009. Resurgence in 2016/18 (ISIS?)
```
# Word Cloud
```
from wordcloud import WordCloud, STOPWORDS, ImageColorGenerator
speech_name = 'Lincoln_1864.txt'
sotu_dict[path + '\\' + speech_name]
#example = sotu_data[0]
example = sotu_dict[path + '\\' + speech_name]
wordcloud = WordCloud(max_words=100).generate(example)
plt.title("WordCloud of " + speech_name)
plt.imshow(wordcloud, interpolation='bilinear')
plt.axis("off")
plt.show()
```
| github_jupyter |
# Average Monthly Temperatures, 1970-2004
**Date:** 2021-12-02
**Reference:**
```
library(TTR)
options(
jupyter.plot_mimetypes = "image/svg+xml",
repr.plot.width = 7,
repr.plot.height = 5
)
```
## Summary
The aim of this notebook was to show how to decompose seasonal time series data using **R** so the trend, seasonal and irregular components can be estimated.
Data on the average monthly temperatures in central England from January 1970 to December 2004 was plotted.
The series was decomposed using the `decompose` function from `R.stats` and the seasonal factors displayed as a `matrix`.
A seasonally adjusted series was calculated by subtracting the seasonal factors from the original series.
The seasonally adjusted series was used to plot an estimate of the trend component by taking a simple moving average.
The irregular component was estimated by subtracting the estimate of the trend and seasonal components from the original time series.
## Get the data
Data on the average monthly temperatures in central England January 1970 to December 2004 is shown below.
```
monthlytemps <- read.csv("..\\..\\data\\moderntemps.csv")
head(monthlytemps)
modtemps <- monthlytemps$temperature
```
## Plot the time series
```
ts_modtemps <- ts(modtemps, start = c(1970, 1), frequency = 12)
plot.ts(ts_modtemps, xlab = "year", ylab = "temperature")
```
The time series is highly seasonal with little evidence of a trend.
There appears to be a constant level of approximately 10$^{\circ}$C.
## Decompose the data
Use the `decompose` function from `R.stats` to return estimates of the trend, seasonal, and irregular components of the time series.
```
decomp_ts <- decompose(ts_modtemps)
```
## Seasonal factors
Calculate the seasonal factors of the decomposed time series.
Cast the `seasonal` time series object held in `decomp_ts` to a `vector`, slice the new vector to isolate a single period, and then cast the sliced vector to a named `matrix`.
```
sf <- as.vector(decomp_ts$seasonal)
(matrix(sf[1:12], dimnames = list(month.abb, c("factors"))))
```
_Add a comment_
## Plot the components
Plot the trend, seasonal, and irregular components in a single graphic.
```
plot(decomp_ts, xlab = "year")
```
Plot the individual components of the decomposition by accessing the variables held in the `tsdecomp`.
This will generally make the components easier to understand.
```
plot(decomp_ts$trend, xlab = "year", ylab = "temperature (Celsius)")
title(main = "Trend component")
plot(decomp_ts$seasonal, xlab = "year", ylab = "temperature (Celsius)")
title(main = "Seasonal component")
plot(decomp_ts$random, xlab = "year", ylab = "temperature (Celsius)")
title(main = "Irregular component")
```
_Add comment on trend, seasonal, and irregular components._
_Which component dominates the series?_
## Seasonal adjusted plot
Plot the seasonally adjusted series by subtracting the seasonal factors from the original series.
```
adjusted_ts <- ts_modtemps - decomp_ts$seasonal
plot(adjusted_ts, xlab = "year", ylab = "temperature (Celsius)")
title(main = "Seasonally adjusted series")
```
This new seasonally adjusted series only contains the trend and irregular components, so it can be treated as if it is non-seasonal data.
Estimate the trend component by taking the simple moving order of order 35.
```
sma35_adjusted_ts <- SMA(adjusted_ts, n = 35)
plot.ts(sma35_adjusted_ts, xlab = "year", ylab = "temperature (Celsius)")
title(main = "Trend component (ma35)")
```
Note that this is a different estimate of the trend component to what is contained in `decomp_ts`, as it uses a different order for the simple moving average.
| github_jupyter |
```
import pandas as pd
import numpy as np
import math
from pprint import pprint
import pandas as pd
import numpy as np
import nltk
import matplotlib.pyplot as plt
import seaborn as sns
nltk.download('vader_lexicon')
nltk.download('stopwords')
from nltk.corpus import stopwords
stop_words = stopwords.words('english')
from nltk.tokenize import word_tokenize, RegexpTokenizer
tokenizer = RegexpTokenizer(r'\w+')
import datetime as dt
from langdetect import detect
# detects the language of the comment
def language_detection(text):
try:
return detect(text)
except:
return None
raw_data = pd.read_csv("/IS5126airbnb_reviews_full.csv",low_memory = False)
raw_data.head()
raw_data_filtered= raw_data[['listing_id','id','comments']]
raw_data_filtered.dtypes
# group by hosts and count the number of unique listings --> cast it to a dataframe
reviews_per_listing = pd.DataFrame(raw_data.groupby('listing_id')['id'].nunique())
# sort unique values descending and show the Top20
reviews_per_listing.sort_values(by=['id'], ascending=False, inplace=True)
reviews_per_listing.head(20)
def language_detection(text):
try:
return detect(text)
except:
return None
raw_data_filtered['language'] = raw_data_filtered['comments'].apply(language_detection)
raw_data_filtered.language.value_counts().head(10)
# visualizing the comments' languages a) quick and dirty
ax = raw_data_filtered.language.value_counts(normalize=True).head(6).sort_values().plot(kind='barh', figsize=(9,5));
df_eng = raw_data_filtered[(raw_data_filtered['language']=='en')]
# import necessary libraries
from nltk.corpus import stopwords
from wordcloud import WordCloud
from collections import Counter
from PIL import Image
import re
import string
def plot_wordcloud(wordcloud, language):
plt.figure(figsize=(12, 10))
plt.imshow(wordcloud, interpolation = 'bilinear')
plt.axis("off")
plt.title('Word Cloud for Comments\n', fontsize=18, fontweight='bold')
plt.show()
wordcloud = WordCloud(max_font_size=None, max_words=200, background_color="lightgrey",
width=3000, height=2000,
stopwords=stopwords.words('english')).generate(str(raw_data_filtered.comments.values))
plot_wordcloud(wordcloud, 'English')
# load the SentimentIntensityAnalyser object in
from nltk.sentiment.vader import SentimentIntensityAnalyzer
# assign it to another name to make it easier to use
analyzer = SentimentIntensityAnalyzer()
# use the polarity_scores() method to get the sentiment metrics
def print_sentiment_scores(sentence):
snt = analyzer.polarity_scores(sentence)
print("{:-<40} {}".format(sentence, str(snt)))
# getting only the negative score
def negative_score(text):
negative_value = analyzer.polarity_scores(str(text))['neg']
return negative_value
# getting only the neutral score
def neutral_score(text):
neutral_value = analyzer.polarity_scores(str(text))['neu']
return neutral_value
# getting only the positive score
def positive_score(text):
positive_value = analyzer.polarity_scores(str(text))['pos']
return positive_value
# getting only the compound score
def compound_score(text):
compound_value = analyzer.polarity_scores(str(text))['compound']
return compound_value
raw_data_filtered['sentiment_neg'] = raw_data_filtered['comments'].apply(negative_score)
raw_data_filtered['sentiment_neu'] = raw_data_filtered['comments'].apply(neutral_score)
raw_data_filtered['sentiment_pos'] = raw_data_filtered['comments'].apply(positive_score)
raw_data_filtered['sentiment_compound'] = raw_data_filtered['comments'].apply(compound_score)
# all scores in 4 histograms
fig, axes = plt.subplots(2, 2, figsize=(10,8))
# plot all 4 histograms
df_eng.hist('sentiment_neg', bins=25, ax=axes[0,0], color='lightcoral', alpha=0.6)
axes[0,0].set_title('Negative Sentiment Score')
df_eng.hist('sentiment_neu', bins=25, ax=axes[0,1], color='lightsteelblue', alpha=0.6)
axes[0,1].set_title('Neutral Sentiment Score')
df_eng.hist('sentiment_pos', bins=25, ax=axes[1,0], color='chartreuse', alpha=0.6)
axes[1,0].set_title('Positive Sentiment Score')
df_eng.hist('sentiment_compound', bins=25, ax=axes[1,1], color='navajowhite', alpha=0.6)
axes[1,1].set_title('Compound')
# plot common x- and y-label
fig.text(0.5, 0.04, 'Sentiment Scores', fontweight='bold', ha='center')
fig.text(0.04, 0.5, 'Number of Reviews', fontweight='bold', va='center', rotation='vertical')
# plot title
plt.suptitle('Sentiment Analysis of Airbnb Reviews for Singapore\n\n', fontsize=12, fontweight='bold');
percentiles = df_eng.sentiment_compound.describe(percentiles=[.05, .1, .2, .3, .4, .5, .6, .7, .8, .9])
percentiles
# assign the data
neg = percentiles['10%']
mid = percentiles['30%']
pos = percentiles['max']
names = ['Negative Comments', 'Okayish Comments','Positive Comments']
size = [neg, mid, pos]
# call a pie chart
plt.pie(size, labels=names, colors=['lightcoral', 'lightsteelblue', 'chartreuse'],
autopct='%.5f%%', pctdistance=0.8,
wedgeprops={'linewidth':7, 'edgecolor':'white' })
# create circle for the center of the plot to make the pie look like a donut
my_circle = plt.Circle((0,0), 0.6, color='white')
# plot the donut chart
fig = plt.gcf()
fig.set_size_inches(7,7)
fig.gca().add_artist(my_circle)
plt.show()
df_eng.head()
pd.set_option("max_colwidth", 1000)
df_neu = df_eng.loc[df_eng.sentiment_compound <= 0.5]
# full dataframe with POSITIVE comments
df_pos = df_eng.loc[df_eng.sentiment_compound >= 0.95]
# only corpus of POSITIVE comments
pos_comments = df_pos['comments'].tolist()
# full dataframe with NEGATIVE comments
df_neg = df_eng.loc[df_eng.sentiment_compound < 0.0]
# only corpus of NEGATIVE comments
neg_comments = df_neg['comments'].tolist()
df_pos['text_length'] = df_pos['comments'].apply(len)
df_neg['text_length'] = df_neg['comments'].apply(len)
sns.set_style("whitegrid")
plt.figure(figsize=(8,5))
sns.distplot(df_pos['text_length'], kde=True, bins=50, color='chartreuse')
sns.distplot(df_neg['text_length'], kde=True, bins=50, color='lightcoral')
plt.title('\nDistribution Plot for Length of Comments\n')
plt.legend(['Positive Comments', 'Negative Comments'])
plt.xlabel('\nText Length')
plt.ylabel('Percentage of Comments\n');
df_eng.head()
merged_reviews = pd.merge(left=df_eng, right=raw_data, left_on='id', right_on='id')
merged_reviews.head()
merged_reviews.drop(['id', 'comments_x', 'language', 'listing_id_y','reviewer_id','reviewer_name','comments_y'], inplace=True, axis=1)
merged_reviews['month'] = pd.to_datetime(merged_reviews['date']).dt.month
merged_reviews['year'] = pd.to_datetime(merged_reviews['date']).dt.year
summary_reviews=merged_reviews.groupby(['year','month','listing_id_x'],as_index=False).mean()
summary_reviews['time_period']=summary_reviews['month'].astype(str)+'-'+summary_reviews['year'].astype(str)
summary_reviews.head()
summary_reviews.to_csv('/Users/sonakshimendiratta/Documents/NUS/YR 2 SEM 2/IS5126/Final Project/data/review_sentiments.csv')
```
| github_jupyter |
# PySchools without Thomas High School 9th graders
### Dependencies and data
```
# Dependencies
import os
import numpy as np
import pandas as pd
# School data
school_path = os.path.join('data', 'schools.csv') # school data path
school_df = pd.read_csv(school_path)
# Student data
student_path = os.path.join('data', 'students.csv') # student data path
student_df = pd.read_csv(student_path)
school_df.shape, student_df.shape
# Change Thomas High School 9th grade scores to NaN
student_df.loc[(student_df['school_name'].str.contains('Thomas')) & (student_df['grade'] == '9th'),
['reading_score', 'math_score']] = np.NaN
student_df.loc[(student_df['school_name'].str.contains('Thomas')) & (student_df['grade'] == '9th'),
['reading_score', 'math_score']].head(3)
```
### Clean student names
```
# Prefixes to remove: "Miss ", "Dr. ", "Mr. ", "Ms. ", "Mrs. "
# Suffixes to remove: " MD", " DDS", " DVM", " PhD"
fixes_to_remove = ['Miss ', '\w+\. ', ' [DMP]\w?[DMS]'] # regex for prefixes and suffixes
str_to_remove = r'|'.join(fixes_to_remove) # join into a single raw str
# Remove inappropriate prefixes and suffixes
student_df['student_name'] = student_df['student_name'].str.replace(str_to_remove, '', regex=True)
# Check prefixes and suffixes
student_names = [n.split() for n in student_df['student_name'].tolist() if len(n.split()) > 2]
pre = list(set([name[0] for name in student_names if len(name[0]) <= 4])) # prefixes
suf = list(set([name[-1] for name in student_names if len(name[-1]) <= 4])) # suffixes
print(pre, suf)
```
### Merge data
```
# Add binary vars for passing score
student_df['pass_read'] = (student_df.reading_score >= 70).astype(int) # passing reading score
student_df['pass_math'] = (student_df.math_score >= 70).astype(int) # passing math score
student_df['pass_both'] = np.min([student_df.pass_read, student_df.pass_math], axis=0) # passing both scores
student_df.head(3)
# Add budget per student var
school_df['budget_per_student'] = (school_df['budget'] / school_df['size']).round().astype(int)
# Bin budget per student
school_df['spending_lvl'] = pd.qcut(school_df['budget_per_student'], 4,
labels=range(1, 5))
# Bin school size
school_df['school_size'] = pd.qcut(school_df['size'], 3, labels=['Small', 'Medium', 'Large'])
school_df
# Merge data
df = pd.merge(student_df, school_df, on='school_name', how='left')
df.info()
```
### District summary
```
# District summary
district_summary = pd.DataFrame(school_df[['size', 'budget']].sum(), columns=['District']).T
district_summary['Total Schools'] = school_df.shape[0]
district_summary = district_summary[['Total Schools', 'size', 'budget']]
district_summary_cols = ['Total Schools', 'Total Students', 'Total Budget']
district_summary
# Score cols
score_cols = ['reading_score', 'math_score', 'pass_read', 'pass_math', 'pass_both']
score_cols_new = ['Average Reading Score', 'Average Math Score', '% Passing Reading', '% Passing Math', '% Passing Overall']
# Add scores to district summary
for col, val in df[score_cols].mean().items():
if 'pass' in col:
val *= 100
district_summary[col] = val
district_summary
# Rename cols
district_summary.columns = district_summary_cols + score_cols_new
district_summary
# Format columns
for col in district_summary.columns:
if 'Total' in col:
district_summary[col] = district_summary[col].apply('{:,}'.format)
if 'Average' in col:
district_summary[col] = district_summary[col].round(2)
if '%' in col:
district_summary[col] = district_summary[col].round().astype(int)
district_summary
```
### School summary
```
# School cols
school_cols = ['type', 'size', 'budget', 'budget_per_student',
'reading_score', 'math_score', 'pass_read', 'pass_math', 'pass_both']
school_cols_new = ['School Type', 'Total Students', 'Total Budget', 'Budget Per Student']
school_cols_new += score_cols_new
# School summary
school_summary = df.groupby('school_name')[school_cols].agg({
'type': 'max',
'size': 'max',
'budget': 'max',
'budget_per_student': 'max',
'reading_score': 'mean',
'math_score': 'mean',
'pass_read': 'mean',
'pass_math': 'mean',
'pass_both': 'mean'
})
school_summary.head(3)
# Rename cols
school_summary.index.name = None
school_summary.columns = school_cols_new
# Format values
for col in school_summary.columns:
if 'Total' in col:
school_summary[col] = school_summary[col].apply('{:,}'.format)
if 'Average' in col:
school_summary[col] = school_summary[col].round(2)
if '%' in col:
school_summary[col] = (school_summary[col] * 100).round().astype(int)
school_summary
```
### Scores by grade
```
# Reading scores by grade of each school
grade_read_scores = pd.pivot_table(df, index='school_name', columns='grade',
values='reading_score', aggfunc='mean').round(2)
grade_read_scores.index.name = None
grade_read_scores.columns.name = 'Reading scores'
grade_read_scores = grade_read_scores[['9th', '10th', '11th', '12th']]
grade_read_scores
# Math scores by grade of each school
grade_math_scores = pd.pivot_table(df, index='school_name', columns='grade',
values='math_score', aggfunc='mean').round(2)
grade_math_scores.index.name = None
grade_math_scores.columns.name = 'Math Scores'
grade_math_scores = grade_math_scores[['9th', '10th', '11th', '12th']]
grade_math_scores
```
### Scores by budget per student
```
# Scores by spending
spending_scores = df.groupby('spending_lvl')[score_cols].mean().round(2)
for col in spending_scores.columns:
if "pass" in col:
spending_scores[col] = (spending_scores[col] * 100).astype(int)
spending_scores
# Formatting
spending_scores.index.name = 'Spending Level'
spending_scores.columns = score_cols_new
spending_scores
```
### Scores by school size
```
# Scores by school size
size_scores = df.groupby('school_size')[score_cols].mean().round(2)
for col in size_scores.columns:
if "pass" in col:
size_scores[col] = (size_scores[col] * 100).astype(int)
size_scores
# Formatting
size_scores.index.name = 'School Size'
size_scores.columns = score_cols_new
size_scores
```
### Scores by school type
```
# Scores by school type
type_scores = df.groupby('type')[score_cols].mean().round(2)
for col in type_scores.columns:
if "pass" in col:
type_scores[col] = (type_scores[col] * 100).astype(int)
type_scores
# Formatting
type_scores.index.name = 'School Type'
type_scores.columns = score_cols_new
type_scores
```
| github_jupyter |
# **ANALYSIS OF FINANCIAL INCLUSION IN EAST AFRICA BETWEEN 2016 TO 2018**
##DEFINING QUESTION
The research problem is to figure out how we can predict which individuals are most likely to have or use a bank account.
### METRIC FOR SUCCESS
My solution procedure will be to help provide an indication of the state of financial inclusion in Kenya, Rwanda, Tanzania, and Uganda, while providing insights into some of the key demographic factors that might drive individuals financial outcomes.
###THE CONTEXT
Financial Inclusion remains one of the main obstacles to economic and human development in Africa. For example, across Kenya, Rwanda, Tanzania, and Uganda only 9.1 million adults (or 13.9% of the adult population) have access to or use a commercial bank account.
Traditionally, access to bank accounts has been regarded as an indicator of financial inclusion. Despite the proliferation of mobile money in Africa and the growth of innovative fintech solutions, banks still play a pivotal role in facilitating access to financial services. Access to bank accounts enables households to save and facilitate payments while also helping businesses build up their credit-worthiness and improve their access to other financial services. Therefore, access to bank accounts is an essential contributor to long-term economic growth.
### EXPERIMENTAL DESIGN TAKEN
The procedure taken is:
1. Definition of the question
2. Reading and checking of the data
3. External data source validation
4. Cleaning of the dataset
5. Exploratory analysis
### DATA RELEVANCE
Data to be used contains demographic information and what financial services are used by individuals in East Africa. The data is extracted from various Finsscope surveys and is ranging from 2016 to 2018. The data files include:
Variable Definitions: http://bit.ly/VariableDefinitions
Dataset: http://bit.ly/FinancialDataset
FinAccess Kenya 2018: https://fsdkenya.org/publication/finaccess2019/
Finscope Rwanda 2016: http://www.statistics.gov.rw/publication/finscope-rwanda-2016
Finscope Tanzania 2017: http://www.fsdt.or.tz/finscope/
Finscope Uganda 2018: http://fsduganda.or.ug/finscope-2018-survey-report/
This data is relevant in this project since it provides important insights that will help in solving the research question.
## LOADING LIBRARIES
```
# importing libraries
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
```
## READING AND CHECKING DATA
```
# loading and viewing variable definitions dataset
url = "http://bit.ly/VariableDefinitions"
vb_df = pd.read_csv(url)
vb_df
# loading and viewing financial dataset
url2 = "http://bit.ly/FinancialDataset"
fds = pd.read_csv(url2)
fds
fds.shape
fds.head()
fds.tail()
fds.dtypes
fds.columns
fds.info()
fds.describe()
fds.describe(include=object)
len(fds)
fds.nunique()
fds.count()
```
## EXTERNAL DATA SOURCE VALIDATION
FinAccess Kenya 2018: https://fsdkenya.org/publication/finaccess2019/
Finscope Rwanda 2016: http://www.statistics.gov.rw/publication/finscope-rwanda-2016
Finscope Tanzania 2017: http://www.fsdt.or.tz/finscope/
Finscope Uganda 2018: http://fsduganda.or.ug/finscope-2018-survey-report/
## CLEANING THE DATASET
```
fds.head(2)
# CHECKING FOR OUTLIERS IN YEAR COLUMN
sns.boxplot(x=fds['year'])
fds.shape
# dropping year column outliers
fds1= fds[fds['year']<2020]
fds1.shape
# CHECKING FOR OUTLIERS IN HOUSEHOLD SIZE COLUMN
sns.boxplot(x=fds1['household_size'])
# dropping household size outliers
fds2 =fds1[fds1['household_size']<10]
fds2.shape
# CHECKING FOR OUTLIERS IN AGE OF RESPONDENT
sns.boxplot(x=fds2['Respondent Age'])
# dropping age of respondent outliers
fds3 = fds2[fds2['Respondent Age']<82]
fds3.shape
# plotting the final boxplots
fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(2,2, figsize=(10, 7))
fig.suptitle('Boxplots')
sns.boxplot(fds3['Respondent Age'], ax=ax1)
sns.boxplot(fds3['year'], ax=ax2)
sns.boxplot(fds3['household_size'], ax=ax3)
plt.show()
# the outliers have finally been droppped
# CHECKING FOR NULLL OR MISSING DATA
fds3.isnull().sum()
# dropping nulls
fds4 = fds3.dropna()
fds4.shape
# dropping duplicates
fds4.drop_duplicates().head(2)
# changing column names and columns to lowercase
#for columns in fds.columns:
#fds1[columns] = fds[columns].astype(str).str.lower()
#fds1
#fds1.rename(columns=str.lower)
# renaming columns
fds5 = fds4.rename(columns={'Type of Location':'location_type', 'Has a Bank account' : 'bank account','Cell Phone Access':'cellphone_access', 'Respondent Age': 'age_of_respondent',
'The relathip with head': 'relationship_with_head', 'Level of Educuation' : 'education_level', 'Type of Job': 'job_type'})
fds5.head(2)
fds5.shape
fds5.size
fds5.nunique()
fds5['country'].unique()
fds5['year'].unique()
fds5['bank account'].unique()
fds5['location_type'].unique()
fds5['cellphone_access'].unique()
fds5['education_level'].unique()
fds5.drop(fds5.loc[fds5['education_level'] ==6].index, inplace=True)
fds5['education_level'].unique()
fds5['gender_of_respondent'].unique()
fds5['household_size'].unique()
fds5.drop(fds5.loc[fds5['household_size'] == 0].index, inplace=True)
fds5['household_size'].unique()
fds5['job_type'].unique()
fds5['relationship_with_head'].unique()
fds5['age_of_respondent'].unique()
```
## **EXPLORATORY ANALYSIS**
### 1.UNIVARIATE ANALYSIS
#### a. NUMERICAL VARIABLES
##### MODE
```
fds5['year'].mode()
fds5['household_size'].mode()
fds5['age_of_respondent'].mode()
```
##### MEAN
```
fds5['age_of_respondent'].mean()
fds5['household_size'].mean()
fds5.mean()
```
##### MEDIAN
```
fds5['age_of_respondent'].median()
fds5['household_size'].median()
fds5.median()
```
##### RANGE
```
a = fds5['age_of_respondent'].max()
b = fds5['age_of_respondent'].min()
c = a-b
print('The range of the age for the respondents is', c)
d = fds5['household_size'].max()
e = fds5['household_size'].min()
f = d-e
print('The range of the household_sizes is', f)
```
##### QUANTILE AND INTERQUANTILE
```
fds5.quantile([0.25,0.5,0.75])
# FINDING THE INTERQUANTILE RANGE = IQR
Q3 = fds5['age_of_respondent'].quantile(0.75)
Q2 = fds5['age_of_respondent'].quantile(0.25)
IQR= Q3-Q2
print('The IQR for the respondents age is', IQR)
q3 = fds5['household_size'].quantile(0.75)
q2 = fds5['household_size'].quantile(0.25)
iqr = q3-q2
print('The IQR for household sizes is', iqr)
```
##### STANDARD DEVIATION
```
fds5.std()
```
##### VARIANCE
```
fds5.var()
```
##### KURTOSIS
```
fds5.kurt()
```
##### SKEWNESS
```
fds5.skew()
```
#### b. CATEGORICAL
##### MODE
```
fds5.mode().head(1)
fds5['age_of_respondent'].plot(kind="hist")
plt.xlabel('ages of respondents')
plt.ylabel('frequency')
plt.title(' Frequency of the ages of the respondents')
country=fds5['country'].value_counts()
print(country)
# Plotting the pie chart
colors=['pink','white','cyan','yellow']
country.plot(kind='pie',colors=colors,autopct='%1.1f%%',shadow=True,startangle=90)
plt.title('Distribution of the respondents by country')
bank =fds5['bank account'].value_counts()
# Plotting the pie chart
colors=['plum', 'aqua']
bank.plot(kind='pie',colors=colors,autopct='%1.1f%%',shadow=True,startangle=90)
plt.title('availability of bank accounts')
location=fds5['location_type'].value_counts()
# Plotting the pie chart
colors=['aquamarine','beige']
location.plot(kind='pie',colors=colors,autopct='%1.3f%%',shadow=True,startangle=00)
plt.title('Distribution of the respondents according to location')
celly =fds5['cellphone_access'].value_counts()
# Plotting the pie chart
colors=['plum','lavender']
celly.plot(kind='pie',colors=colors,autopct='%1.1f%%',shadow=True,startangle=0)
plt.title('cellphone access for the respondents')
gen =fds5['gender_of_respondent'].value_counts()
# Plotting the pie chart
colors=['red','lavender']
gen.plot(kind='pie',colors=colors,autopct='%1.1f%%',shadow=True,startangle=0)
plt.title('gender distribution')
```
##### CONCLUSION AND RECOMMENDATION
Most of the data was collected in Rwanda.
Most of the data was collected in Rural areas.
Most of those who were interviewed were women.
Most of the population has mobile phones.
There were several outliers.
Since 75% of the population has phones, phones should be used as the main channel for information and awareness of bank accessories.
### 2. BIVARIATE ANALYSIS
```
fds5.head()
#@title Since i am predicting the likelihood of the respondents using the bank,I shall be comparing all variables against the bank account column.
```
##### NUMERICAL VS NUMERICAL
```
sns.pairplot(fds5)
plt.show()
# pearson correlation of numerical variables
sns.heatmap(fds5.corr(),annot=True)
plt.show()
# possible weak correlation
fds5.corr()
```
##### CATEGORICAL VS CATEGORICAL
```
# Grouping bank usage by country
country1 = fds5.groupby('country')['bank account'].value_counts(normalize=True).unstack()
colors= ['lightpink', 'skyblue']
country1.plot(kind='bar', figsize=(8, 6), color=colors, stacked=True)
plt.title('Bank usage by country', fontsize=15, y=1.015)
plt.xlabel('country', fontsize=14, labelpad=15)
plt.xticks(rotation = 360)
plt.ylabel('Bank usage by country', fontsize=14, labelpad=15)
plt.show()
# Bank usage by gender
gender1 = fds5.groupby('gender_of_respondent')['bank account'].value_counts(normalize=True).unstack()
colors= ['lightpink', 'skyblue']
gender1.plot(kind='bar', figsize=(8, 6), color=colors, stacked=True)
plt.title('Bank usage by gender', fontsize=17, y=1.015)
plt.xlabel('gender', fontsize=17, labelpad=17)
plt.xticks(rotation = 360)
plt.ylabel('Bank usage by gender', fontsize=17, labelpad=17)
plt.show()
# Bank usage depending on level of education
ed2 = fds5.groupby('education_level')['bank account'].value_counts(normalize=True).unstack()
colors= ['cyan', 'darkcyan']
ed2.plot(kind='barh', figsize=(8, 6), color=colors, stacked=True)
plt.title('Bank usage by level of education', fontsize=17, y=1.015)
plt.xlabel('frequency', fontsize=17, labelpad=17)
plt.xticks(rotation = 360)
plt.ylabel('level of education', fontsize=17, labelpad=17)
plt.show()
ms = fds5.groupby('marital_status')['bank account'].value_counts(normalize=True).unstack()
colors= ['coral', 'orange']
ms.plot(kind='barh', figsize=(8, 6), color=colors, stacked=True)
plt.title('Bank usage by marital status', fontsize=17, y=1.015)
plt.xlabel('frequency', fontsize=17, labelpad=17)
plt.xticks(rotation = 360)
plt.ylabel('marital status', fontsize=17, labelpad=17)
gj = fds5.groupby('gender_of_respondent')['job_type'].value_counts(normalize=True).unstack()
#colors= ['coral', 'orange']
gj.plot(kind='bar', figsize=(8, 6), stacked=True)
plt.title('job type by gender', fontsize=17, y=1.015)
plt.xlabel('gender_of_respondent', fontsize=17, labelpad=17)
plt.xticks(rotation = 360)
plt.ylabel('job type', fontsize=17, labelpad=17)
```
##### NUMERICAL VS CATEGORICAL
##### IMPLEMENTING AND CHALLENGING SOLUTION
```
```
Most of those interviewed do not have bank accounts of which 80% is the uneducated.
Most of the population that participated is married,followed by single/never married.
Most of the population has primary school education level.
Most of the population is involved in farming followed by self employment.
Bank usage has more males than females.
More channeling needs to be done in Kenya as it has the least bank users.
###3. MULTIVARIATE ANALYSIS
```
# Multivariate analysis - This is a statistical analysis that involves observation and analysis of more than one statistical outcome variable at a time
# LETS MAKE A COPY
fds_new = fds5.copy()
fds_new.columns
fds_new.dtypes
# IMPORTING THE LABEL ENCODER
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
# encoding categorial values
fds_new['country']=le.fit_transform(fds_new['country'].astype(str))
fds_new['location_type']=le.fit_transform(fds_new['location_type'].astype(str))
fds_new['cellphone_access']=le.fit_transform(fds_new['cellphone_access'].astype(str))
fds_new['gender_of_respondent']=le.fit_transform(fds_new['gender_of_respondent'].astype(str))
fds_new['relationship_with_head']=le.fit_transform(fds_new['relationship_with_head'].astype(str))
fds_new['marital_status']=le.fit_transform(fds_new['marital_status'].astype(str))
fds_new['education_level']=le.fit_transform(fds_new['education_level'].astype(str))
fds_new['job_type']=le.fit_transform(fds_new['job_type'].astype(str))
fds_new.sample(5)
# dropping unnecessary columns
fds_new.drop(['age_of_respondent','uniqueid','year'], axis=1).head(2)
```
##### FACTOR ANALYSIS
```
# Installing factor analyzer
!pip install factor_analyzer==0.2.3
from factor_analyzer.factor_analyzer import calculate_bartlett_sphericity
chi_square_value,p_value=calculate_bartlett_sphericity(fds_new)
chi_square_value, p_value
# In Bartlett ’s test, the p-value is 0. The test was statistically significant,
# indicating that the observed correlation matrix is not an identity matrix.
# Value of KMO less than 0.6 is considered inadequate.
#
#from factor_analyzer.factor_analyzer import calculate_kmo
#kmo_all,kmo_model=calculate_kmo(fds_new)
#calculate_kmo(fds_new)
# Choosing the Number of Factors
from factor_analyzer.factor_analyzer import FactorAnalyzer
# Creating factor analysis object and perform factor analysis
fa = FactorAnalyzer()
fa.analyze(fds_new, 10, rotation=None)
# Checking the Eigenvalues
ev, v = fa.get_eigenvalues()
ev
# We choose the factors that are > 1.
# so we choose 4 factors only
# PERFOMING FACTOR ANALYSIS FOR 4 FACTORS
fa = FactorAnalyzer()
fa.analyze(fds_new, 4, rotation="varimax")
fa.loadings
# GETTING VARIANCE FOR THE FACTORS
fa.get_factor_variance()
```
##### CONCLUSION
The reduction method used was factor analysis.
Four factors had an eigen value greater than 1.
##### CHALLENGING SOLUTION
There is room for modification because there are other methodologies that could be used for analysis.
There is also an assumption that this data is accurate to the real world.
| github_jupyter |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.