code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
```
__name__ = "k1lib.callbacks"
#export
from .callbacks import Callback, Callbacks, Cbs
import k1lib, time, torch, math, logging, numpy as np, torch.nn as nn
from functools import partial
import matplotlib.pyplot as plt
__all__ = ["Profiler"]
#export
import k1lib.callbacks.profilers as ps
ComputationProfiler = ps.computation.ComputationProfiler
IOProfiler = ps.io.IOProfiler
MemoryProfiler = ps.memory.MemoryProfiler
TimeProfiler = ps.time.TimeProfiler
#export
@k1lib.patch(Cbs)
class Profiler(Callback):
"""Profiles memory, time, and computational complexity of the network. See over
:mod:`k1lib.callbacks.profilers` for more details on each of these profilers"""
def __init__(self):
super().__init__(); self.clear(); self.dependsOn=["Recorder"]
def clear(self):
"""Clears every child profilers"""
self._mpCache=None; self._tpCache=None
self._cpCache=None; self._ioCache=None
def _memory(self): # do this to quickly debug, cause if not, Callback will just raise AttributeError on .memory
if self._mpCache != None: return self._mpCache
with self.cbs.context():
mp = MemoryProfiler(); self.cbs.add(mp)
mp._run(); self._mpCache = mp; return mp
@property
def memory(self) -> MemoryProfiler:
"""Gets :class:`~k1lib.callbacks.profilers.memory.MemoryProfiler`"""
return self._memory()
def _computation(self):
if self._cpCache != None: return self._cpCache
with self.cbs.context():
cp = ComputationProfiler(self); self.cbs.add(cp)
cp._run(); self._cpCache = cp; return cp
@property
def computation(self) -> ComputationProfiler:
"""Gets :class:`~k1lib.callbacks.profilers.computation.ComputationProfiler`"""
return self._computation()
def _time(self):
if self._tpCache != None: return self._tpCache
with self.cbs.context():
tp = TimeProfiler(); self.cbs.add(tp)
tp._run(); self._tpCache = tp; return tp
@property
def time(self) -> TimeProfiler:
"""Gets :class:`~k1lib.callbacks.profilers.time.TimeProfiler`"""
return self._time()
def _io(self):
if self._ioCache != None: return self._ioCache
with self.cbs.context():
io = IOProfiler(); self.cbs.add(io)
io._run(); self._ioCache = io; return io
@property
def io(self) -> IOProfiler:
"""Gets :class:`~k1lib.callbacks.profilers.io.IOProfiler`"""
return self._io()
def __repr__(self):
return f"""{self._reprHead}, can...
- p.memory: to profile module memory requirements
- p.time: to profile module execution times
- p.computation: to estimate module computation
- p.io: to get input and output shapes of
{self._reprCan}"""
!../../export.py callbacks/profiler
```
|
github_jupyter
|
__name__ = "k1lib.callbacks"
#export
from .callbacks import Callback, Callbacks, Cbs
import k1lib, time, torch, math, logging, numpy as np, torch.nn as nn
from functools import partial
import matplotlib.pyplot as plt
__all__ = ["Profiler"]
#export
import k1lib.callbacks.profilers as ps
ComputationProfiler = ps.computation.ComputationProfiler
IOProfiler = ps.io.IOProfiler
MemoryProfiler = ps.memory.MemoryProfiler
TimeProfiler = ps.time.TimeProfiler
#export
@k1lib.patch(Cbs)
class Profiler(Callback):
"""Profiles memory, time, and computational complexity of the network. See over
:mod:`k1lib.callbacks.profilers` for more details on each of these profilers"""
def __init__(self):
super().__init__(); self.clear(); self.dependsOn=["Recorder"]
def clear(self):
"""Clears every child profilers"""
self._mpCache=None; self._tpCache=None
self._cpCache=None; self._ioCache=None
def _memory(self): # do this to quickly debug, cause if not, Callback will just raise AttributeError on .memory
if self._mpCache != None: return self._mpCache
with self.cbs.context():
mp = MemoryProfiler(); self.cbs.add(mp)
mp._run(); self._mpCache = mp; return mp
@property
def memory(self) -> MemoryProfiler:
"""Gets :class:`~k1lib.callbacks.profilers.memory.MemoryProfiler`"""
return self._memory()
def _computation(self):
if self._cpCache != None: return self._cpCache
with self.cbs.context():
cp = ComputationProfiler(self); self.cbs.add(cp)
cp._run(); self._cpCache = cp; return cp
@property
def computation(self) -> ComputationProfiler:
"""Gets :class:`~k1lib.callbacks.profilers.computation.ComputationProfiler`"""
return self._computation()
def _time(self):
if self._tpCache != None: return self._tpCache
with self.cbs.context():
tp = TimeProfiler(); self.cbs.add(tp)
tp._run(); self._tpCache = tp; return tp
@property
def time(self) -> TimeProfiler:
"""Gets :class:`~k1lib.callbacks.profilers.time.TimeProfiler`"""
return self._time()
def _io(self):
if self._ioCache != None: return self._ioCache
with self.cbs.context():
io = IOProfiler(); self.cbs.add(io)
io._run(); self._ioCache = io; return io
@property
def io(self) -> IOProfiler:
"""Gets :class:`~k1lib.callbacks.profilers.io.IOProfiler`"""
return self._io()
def __repr__(self):
return f"""{self._reprHead}, can...
- p.memory: to profile module memory requirements
- p.time: to profile module execution times
- p.computation: to estimate module computation
- p.io: to get input and output shapes of
{self._reprCan}"""
!../../export.py callbacks/profiler
| 0.884021 | 0.121529 |
```
import pandas as pd
import numpy as np
from sklearn.svm import SVC
from sklearn.metrics import confusion_matrix, plot_roc_curve, balanced_accuracy_score
from sklearn.linear_model import LogisticRegression, LassoCV, LinearRegression
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
import seaborn as sns
```
## Task 2
```
df = pd.read_csv('sgn.csv')
def sgn(m, model):
plt.close()
sns.scatterplot(x=df['x'], y=df['y'], s=3)
global df1
df1 = df.copy()
for i in range(m):
df1[f'sin{i+1}']=np.sin(df['x']*i)
df1[f'cos{i+1}']=np.cos(df['x']*i)
X = df1.iloc[:,2:]
y = df1['y']
model.fit(X,y)
print(model.coef_)
print(model.intercept_)
y_pred = model.predict(X)
sns.lineplot(x=df['x'], y=y_pred, color = 'Red')
reg = LinearRegression()
lasso = LassoCV()
sgn(1, reg)
sgn(5, reg)
sgn(20, reg)
sgn(100, reg)
sgn(1000, reg)
```
## Task 3
```
sgn(1000, lasso)
```
Как видим, Lasso труднее переобучить, чем обычную линейную регрессию без регуляризации
## Task 4
```
df = pd.read_csv("../Bonus19/BRCA_pam50.tsv", sep="\t", index_col=0)
df = df.loc[df["Subtype"].isin(["Luminal A","Luminal B"])]
X = df.iloc[:, :-1].to_numpy()
y = df["Subtype"].to_numpy()
X_train, X_test, y_train, y_test = train_test_split(
X, y, stratify=y, random_state=17
)
svm = SVC(kernel="linear", C=0.01)
svm.fit(X_train, y_train); pass
y_pred = svm.predict(X_test)
print("Balanced accuracy score:", balanced_accuracy_score(y_pred, y_test))
M = confusion_matrix(y_test, y_pred)
print(M)
TPR = M[0, 0] / (M[0, 0] + M[0, 1])
TNR = M[1, 1] / (M[1, 0] + M[1, 1])
print("TPR:", round(TPR, 3), "TNR:", round(TNR, 3))
plot_roc_curve(svm, X_test, y_test)
plt.plot(1 - TPR, TNR, "x", c="red")
plt.title("SVC 50 genes")
plt.show()
coef = np.argsort(np.abs(svm.coef_[0]))[-2:]
X = df.iloc[:, coef].to_numpy()
X_train, X_test, y_train, y_test = train_test_split(
X, y, stratify=y, random_state=17
)
svm.fit(X_train, y_train); pass
y_pred = svm.predict(X_test)
print("Balanced accuracy score:", balanced_accuracy_score(y_pred, y_test))
M = confusion_matrix(y_test, y_pred)
print(M)
TPR = M[0, 0] / (M[0, 0] + M[0, 1])
TNR = M[1, 1] / (M[1, 0] + M[1, 1])
print("TPR:", round(TPR, 3), "TNR:", round(TNR, 3))
plot_roc_curve(svm, X_test, y_test)
plt.plot(1 - TPR, TNR, "x", c="red")
plt.title("SVC 2 genes")
plt.show()
X = df.iloc[:, :-1].to_numpy()
X_train, X_test, y_train, y_test = train_test_split(
X, y, stratify=y, random_state=17
)
log = LogisticRegression(class_weight = 'balanced', C=0.01, penalty='l1', solver='liblinear')
log.fit(X_train, y_train)
print(log.coef_)
y_pred = log.predict(X_test)
print("Balanced accuracy score:", balanced_accuracy_score(y_pred, y_test))
M = confusion_matrix(y_test, y_pred)
print(M)
TPR = M[0, 0] / (M[0, 0] + M[0, 1])
TNR = M[1, 1] / (M[1, 0] + M[1, 1])
print("TPR:", round(TPR, 3), "TNR:", round(TNR, 3))
plot_roc_curve(log, X_test, y_test)
plt.plot(1 - TPR, TNR, "x", c="red")
plt.title("Logistic regression")
plt.show()
```
|
github_jupyter
|
import pandas as pd
import numpy as np
from sklearn.svm import SVC
from sklearn.metrics import confusion_matrix, plot_roc_curve, balanced_accuracy_score
from sklearn.linear_model import LogisticRegression, LassoCV, LinearRegression
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
import seaborn as sns
df = pd.read_csv('sgn.csv')
def sgn(m, model):
plt.close()
sns.scatterplot(x=df['x'], y=df['y'], s=3)
global df1
df1 = df.copy()
for i in range(m):
df1[f'sin{i+1}']=np.sin(df['x']*i)
df1[f'cos{i+1}']=np.cos(df['x']*i)
X = df1.iloc[:,2:]
y = df1['y']
model.fit(X,y)
print(model.coef_)
print(model.intercept_)
y_pred = model.predict(X)
sns.lineplot(x=df['x'], y=y_pred, color = 'Red')
reg = LinearRegression()
lasso = LassoCV()
sgn(1, reg)
sgn(5, reg)
sgn(20, reg)
sgn(100, reg)
sgn(1000, reg)
sgn(1000, lasso)
df = pd.read_csv("../Bonus19/BRCA_pam50.tsv", sep="\t", index_col=0)
df = df.loc[df["Subtype"].isin(["Luminal A","Luminal B"])]
X = df.iloc[:, :-1].to_numpy()
y = df["Subtype"].to_numpy()
X_train, X_test, y_train, y_test = train_test_split(
X, y, stratify=y, random_state=17
)
svm = SVC(kernel="linear", C=0.01)
svm.fit(X_train, y_train); pass
y_pred = svm.predict(X_test)
print("Balanced accuracy score:", balanced_accuracy_score(y_pred, y_test))
M = confusion_matrix(y_test, y_pred)
print(M)
TPR = M[0, 0] / (M[0, 0] + M[0, 1])
TNR = M[1, 1] / (M[1, 0] + M[1, 1])
print("TPR:", round(TPR, 3), "TNR:", round(TNR, 3))
plot_roc_curve(svm, X_test, y_test)
plt.plot(1 - TPR, TNR, "x", c="red")
plt.title("SVC 50 genes")
plt.show()
coef = np.argsort(np.abs(svm.coef_[0]))[-2:]
X = df.iloc[:, coef].to_numpy()
X_train, X_test, y_train, y_test = train_test_split(
X, y, stratify=y, random_state=17
)
svm.fit(X_train, y_train); pass
y_pred = svm.predict(X_test)
print("Balanced accuracy score:", balanced_accuracy_score(y_pred, y_test))
M = confusion_matrix(y_test, y_pred)
print(M)
TPR = M[0, 0] / (M[0, 0] + M[0, 1])
TNR = M[1, 1] / (M[1, 0] + M[1, 1])
print("TPR:", round(TPR, 3), "TNR:", round(TNR, 3))
plot_roc_curve(svm, X_test, y_test)
plt.plot(1 - TPR, TNR, "x", c="red")
plt.title("SVC 2 genes")
plt.show()
X = df.iloc[:, :-1].to_numpy()
X_train, X_test, y_train, y_test = train_test_split(
X, y, stratify=y, random_state=17
)
log = LogisticRegression(class_weight = 'balanced', C=0.01, penalty='l1', solver='liblinear')
log.fit(X_train, y_train)
print(log.coef_)
y_pred = log.predict(X_test)
print("Balanced accuracy score:", balanced_accuracy_score(y_pred, y_test))
M = confusion_matrix(y_test, y_pred)
print(M)
TPR = M[0, 0] / (M[0, 0] + M[0, 1])
TNR = M[1, 1] / (M[1, 0] + M[1, 1])
print("TPR:", round(TPR, 3), "TNR:", round(TNR, 3))
plot_roc_curve(log, X_test, y_test)
plt.plot(1 - TPR, TNR, "x", c="red")
plt.title("Logistic regression")
plt.show()
| 0.506836 | 0.805288 |
```
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from keras import initializers
import keras.backend as K
import numpy as np
import pandas as pd
from tensorflow.keras.layers import *
from keras.regularizers import l2#正则化
import pandas as pd
import numpy as np
normal = np.loadtxt(r'F:\data_all\试验数据(包括压力脉动和振动)\2013.9.12-未发生缠绕前\2013-9.12振动\2013-9-12振动-1450rmin-mat\1450r_normalvib4.txt', delimiter=',')
chanrao = np.loadtxt(r'F:\data_all\试验数据(包括压力脉动和振动)\2013.9.17-发生缠绕后\振动\9-17下午振动1450rmin-mat\1450r_chanraovib4.txt', delimiter=',')
print(normal.shape,chanrao.shape,"***************************************************")
data_normal=normal[16:18] #提取前两行
data_chanrao=chanrao[16:18] #提取前两行
print(data_normal.shape,data_chanrao.shape)
print(data_normal,"\r\n",data_chanrao,"***************************************************")
data_normal=data_normal.reshape(1,-1)
data_chanrao=data_chanrao.reshape(1,-1)
print(data_normal.shape,data_chanrao.shape)
print(data_normal,"\r\n",data_chanrao,"***************************************************")
#水泵的两种故障类型信号normal正常,chanrao故障
data_normal=data_normal.reshape(-1, 512)#(65536,1)-(128, 515)
data_chanrao=data_chanrao.reshape(-1,512)
print(data_normal.shape,data_chanrao.shape)
import numpy as np
def yuchuli(data,label):#(4:1)(51:13)
#打乱数据顺序
np.random.shuffle(data)
train = data[0:102,:]
test = data[102:128,:]
label_train = np.array([label for i in range(0,102)])
label_test =np.array([label for i in range(0,26)])
return train,test ,label_train ,label_test
def stackkk(a,b,c,d,e,f,g,h):
aa = np.vstack((a, e))
bb = np.vstack((b, f))
cc = np.hstack((c, g))
dd = np.hstack((d, h))
return aa,bb,cc,dd
x_tra0,x_tes0,y_tra0,y_tes0 = yuchuli(data_normal,0)
x_tra1,x_tes1,y_tra1,y_tes1 = yuchuli(data_chanrao,1)
tr1,te1,yr1,ye1=stackkk(x_tra0,x_tes0,y_tra0,y_tes0 ,x_tra1,x_tes1,y_tra1,y_tes1)
x_train=tr1
x_test=te1
y_train = yr1
y_test = ye1
#打乱数据
state = np.random.get_state()
np.random.shuffle(x_train)
np.random.set_state(state)
np.random.shuffle(y_train)
state = np.random.get_state()
np.random.shuffle(x_test)
np.random.set_state(state)
np.random.shuffle(y_test)
#对训练集和测试集标准化
def ZscoreNormalization(x):
"""Z-score normaliaztion"""
x = (x - np.mean(x)) / np.std(x)
return x
x_train=ZscoreNormalization(x_train)
x_test=ZscoreNormalization(x_test)
# print(x_test[0])
#转化为一维序列
x_train = x_train.reshape(-1,512,1)
x_test = x_test.reshape(-1,512,1)
print(x_train.shape,x_test.shape)
def to_one_hot(labels,dimension=2):
results = np.zeros((len(labels),dimension))
for i,label in enumerate(labels):
results[i,label] = 1
return results
one_hot_train_labels = to_one_hot(y_train)
one_hot_test_labels = to_one_hot(y_test)
#定义挤压函数
def squash(vectors, axis=-1):
"""
对向量的非线性激活函数
## vectors: some vectors to be squashed, N-dim tensor
## axis: the axis to squash
:return: a Tensor with same shape as input vectors
"""
s_squared_norm = K.sum(K.square(vectors), axis, keepdims=True)
scale = s_squared_norm / (1 + s_squared_norm) / K.sqrt(s_squared_norm + K.epsilon())
return scale * vectors
class Length(layers.Layer):
"""
计算向量的长度。它用于计算与margin_loss中的y_true具有相同形状的张量
Compute the length of vectors. This is used to compute a Tensor that has the same shape with y_true in margin_loss
inputs: shape=[dim_1, ..., dim_{n-1}, dim_n]
output: shape=[dim_1, ..., dim_{n-1}]
"""
def call(self, inputs, **kwargs):
return K.sqrt(K.sum(K.square(inputs), -1))
def compute_output_shape(self, input_shape):
return input_shape[:-1]
def get_config(self):
config = super(Length, self).get_config()
return config
#定义预胶囊层
def PrimaryCap(inputs, dim_capsule, n_channels, kernel_size, strides, padding):
"""
进行普通二维卷积 `n_channels` 次, 然后将所有的胶囊重叠起来
:param inputs: 4D tensor, shape=[None, width, height, channels]
:param dim_capsule: the dim of the output vector of capsule
:param n_channels: the number of types of capsules
:return: output tensor, shape=[None, num_capsule, dim_capsule]
"""
output = layers.Conv2D(filters=dim_capsule*n_channels, kernel_size=kernel_size, strides=strides,
padding=padding,name='primarycap_conv2d')(inputs)
outputs = layers.Reshape(target_shape=[-1, dim_capsule], name='primarycap_reshape')(output)
return layers.Lambda(squash, name='primarycap_squash')(outputs)
class DenseCapsule(layers.Layer):
"""
胶囊层. 输入输出都为向量.
## num_capsule: 本层包含的胶囊数量
## dim_capsule: 输出的每一个胶囊向量的维度
## routings: routing 算法的迭代次数
"""
def __init__(self, num_capsule, dim_capsule, routings=3, kernel_initializer='glorot_uniform',**kwargs):
super(DenseCapsule, self).__init__(**kwargs)
self.num_capsule = num_capsule
self.dim_capsule = dim_capsule
self.routings = routings
self.kernel_initializer = kernel_initializer
def build(self, input_shape):
assert len(input_shape) >= 3, '输入的 Tensor 的形状[None, input_num_capsule, input_dim_capsule]'#(None,1152,8)
self.input_num_capsule = input_shape[1]
self.input_dim_capsule = input_shape[2]
#转换矩阵
self.W = self.add_weight(shape=[self.num_capsule, self.input_num_capsule,
self.dim_capsule, self.input_dim_capsule],
initializer=self.kernel_initializer,name='W')
self.built = True
def call(self, inputs, training=None):
# inputs.shape=[None, input_num_capsuie, input_dim_capsule]
# inputs_expand.shape=[None, 1, input_num_capsule, input_dim_capsule]
inputs_expand = K.expand_dims(inputs, 1)
# 运算优化:将inputs_expand重复num_capsule 次,用于快速和W相乘
# inputs_tiled.shape=[None, num_capsule, input_num_capsule, input_dim_capsule]
inputs_tiled = K.tile(inputs_expand, [1, self.num_capsule, 1, 1])
# 将inputs_tiled的batch中的每一条数据,计算inputs+W
# x.shape = [num_capsule, input_num_capsule, input_dim_capsule]
# W.shape = [num_capsule, input_num_capsule, dim_capsule, input_dim_capsule]
# 将x和W的前两个维度看作'batch'维度,向量和矩阵相乘:
# [input_dim_capsule] x [dim_capsule, input_dim_capsule]^T -> [dim_capsule].
# inputs_hat.shape = [None, num_capsule, input_num_capsule, dim_capsutel
inputs_hat = K.map_fn(lambda x: K.batch_dot(x, self.W, [2, 3]),elems=inputs_tiled)
# Begin: Routing算法
# 将系数b初始化为0.
# b.shape = [None, self.num_capsule, self, input_num_capsule].
b = tf.zeros(shape=[K.shape(inputs_hat)[0], self.num_capsule, self.input_num_capsule])
assert self.routings > 0, 'The routings should be > 0.'
for i in range(self.routings):
# c.shape=[None, num_capsule, input_num_capsule]
C = tf.nn.softmax(b ,axis=1)
# c.shape = [None, num_capsule, input_num_capsule]
# inputs_hat.shape = [None, num_capsule, input_num_capsule, dim_capsule]
# 将c与inputs_hat的前两个维度看作'batch'维度,向量和矩阵相乘:
# [input_num_capsule] x [input_num_capsule, dim_capsule] -> [dim_capsule],
# outputs.shape= [None, num_capsule, dim_capsule]
outputs = squash(K. batch_dot(C, inputs_hat, [2, 2])) # [None, 10, 16]
if i < self.routings - 1:
# outputs.shape = [None, num_capsule, dim_capsule]
# inputs_hat.shape = [None, num_capsule, input_num_capsule, dim_capsule]
# 将outputs和inρuts_hat的前两个维度看作‘batch’ 维度,向量和矩阵相乘:
# [dim_capsule] x [imput_num_capsule, dim_capsule]^T -> [input_num_capsule]
# b.shape = [batch_size. num_capsule, input_nom_capsule]
# b += K.batch_dot(outputs, inputs_hat, [2, 3]) to this b += tf.matmul(self.W, x)
b += K.batch_dot(outputs, inputs_hat, [2, 3])
# End: Routing 算法
return outputs
def compute_output_shape(self, input_shape):
return tuple([None, self.num_capsule, self.dim_capsule])
def get_config(self):
config = {
'num_capsule': self.num_capsule,
'dim_capsule': self.dim_capsule,
'routings': self.routings
}
base_config = super(DenseCapsule, self).get_config()
return dict(list(base_config.items()) + list(config.items()))
from tensorflow import keras
from keras.regularizers import l2#正则化
x = layers.Input(shape=[512,1, 1])
#普通卷积层
conv1 = layers.Conv2D(filters=16, kernel_size=(2, 1),activation='relu',padding='valid',name='conv1')(x)
#池化层
POOL1 = MaxPooling2D((2,1))(conv1)
#普通卷积层
conv2 = layers.Conv2D(filters=32, kernel_size=(2, 1),activation='relu',padding='valid',name='conv2')(POOL1)
#池化层
# POOL2 = MaxPooling2D((2,1))(conv2)
#Dropout层
Dropout=layers.Dropout(0.1)(conv2)
# Layer 3: 使用“squash”激活的Conv2D层, 然后重塑 [None, num_capsule, dim_vector]
primarycaps = PrimaryCap(Dropout, dim_capsule=8, n_channels=12, kernel_size=(4, 1), strides=2, padding='valid')
# Layer 4: 数字胶囊层,动态路由算法在这里工作。
digitcaps = DenseCapsule(num_capsule=2, dim_capsule=16, routings=3, name='digit_caps')(primarycaps)
# Layer 5:这是一个辅助层,用它的长度代替每个胶囊。只是为了符合标签的形状。
out_caps = Length(name='out_caps')(digitcaps)
model = keras.Model(x, out_caps)
model.summary()
#定义优化
model.compile(metrics=['accuracy'],
optimizer='adam',
loss=lambda y_true,y_pred: y_true*K.relu(0.9-y_pred)**2 + 0.25*(1-y_true)*K.relu(y_pred-0.1)**2
)
import time
time_begin = time.time()
history = model.fit(x_train,one_hot_train_labels,
validation_split=0.1,
epochs=50,batch_size=10,
shuffle=True)
time_end = time.time()
time = time_end - time_begin
print('time:', time)
score = model.evaluate(x_test,one_hot_test_labels, verbose=0)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
#绘制acc-loss曲线
import matplotlib.pyplot as plt
plt.plot(history.history['loss'],color='r')
plt.plot(history.history['val_loss'],color='g')
plt.plot(history.history['accuracy'],color='b')
plt.plot(history.history['val_accuracy'],color='k')
plt.title('model loss and acc')
plt.ylabel('Accuracy')
plt.xlabel('epoch')
plt.legend(['train_loss', 'test_loss','train_acc', 'test_acc'], loc='upper left')
# plt.legend(['train_loss','train_acc'], loc='upper left')
#plt.savefig('1.png')
plt.show()
import matplotlib.pyplot as plt
plt.plot(history.history['loss'],color='r')
plt.plot(history.history['accuracy'],color='b')
plt.title('model loss and sccuracy ')
plt.ylabel('loss/sccuracy')
plt.xlabel('epoch')
plt.legend(['train_loss', 'train_sccuracy'], loc='upper left')
plt.show()
```
|
github_jupyter
|
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from keras import initializers
import keras.backend as K
import numpy as np
import pandas as pd
from tensorflow.keras.layers import *
from keras.regularizers import l2#正则化
import pandas as pd
import numpy as np
normal = np.loadtxt(r'F:\data_all\试验数据(包括压力脉动和振动)\2013.9.12-未发生缠绕前\2013-9.12振动\2013-9-12振动-1450rmin-mat\1450r_normalvib4.txt', delimiter=',')
chanrao = np.loadtxt(r'F:\data_all\试验数据(包括压力脉动和振动)\2013.9.17-发生缠绕后\振动\9-17下午振动1450rmin-mat\1450r_chanraovib4.txt', delimiter=',')
print(normal.shape,chanrao.shape,"***************************************************")
data_normal=normal[16:18] #提取前两行
data_chanrao=chanrao[16:18] #提取前两行
print(data_normal.shape,data_chanrao.shape)
print(data_normal,"\r\n",data_chanrao,"***************************************************")
data_normal=data_normal.reshape(1,-1)
data_chanrao=data_chanrao.reshape(1,-1)
print(data_normal.shape,data_chanrao.shape)
print(data_normal,"\r\n",data_chanrao,"***************************************************")
#水泵的两种故障类型信号normal正常,chanrao故障
data_normal=data_normal.reshape(-1, 512)#(65536,1)-(128, 515)
data_chanrao=data_chanrao.reshape(-1,512)
print(data_normal.shape,data_chanrao.shape)
import numpy as np
def yuchuli(data,label):#(4:1)(51:13)
#打乱数据顺序
np.random.shuffle(data)
train = data[0:102,:]
test = data[102:128,:]
label_train = np.array([label for i in range(0,102)])
label_test =np.array([label for i in range(0,26)])
return train,test ,label_train ,label_test
def stackkk(a,b,c,d,e,f,g,h):
aa = np.vstack((a, e))
bb = np.vstack((b, f))
cc = np.hstack((c, g))
dd = np.hstack((d, h))
return aa,bb,cc,dd
x_tra0,x_tes0,y_tra0,y_tes0 = yuchuli(data_normal,0)
x_tra1,x_tes1,y_tra1,y_tes1 = yuchuli(data_chanrao,1)
tr1,te1,yr1,ye1=stackkk(x_tra0,x_tes0,y_tra0,y_tes0 ,x_tra1,x_tes1,y_tra1,y_tes1)
x_train=tr1
x_test=te1
y_train = yr1
y_test = ye1
#打乱数据
state = np.random.get_state()
np.random.shuffle(x_train)
np.random.set_state(state)
np.random.shuffle(y_train)
state = np.random.get_state()
np.random.shuffle(x_test)
np.random.set_state(state)
np.random.shuffle(y_test)
#对训练集和测试集标准化
def ZscoreNormalization(x):
"""Z-score normaliaztion"""
x = (x - np.mean(x)) / np.std(x)
return x
x_train=ZscoreNormalization(x_train)
x_test=ZscoreNormalization(x_test)
# print(x_test[0])
#转化为一维序列
x_train = x_train.reshape(-1,512,1)
x_test = x_test.reshape(-1,512,1)
print(x_train.shape,x_test.shape)
def to_one_hot(labels,dimension=2):
results = np.zeros((len(labels),dimension))
for i,label in enumerate(labels):
results[i,label] = 1
return results
one_hot_train_labels = to_one_hot(y_train)
one_hot_test_labels = to_one_hot(y_test)
#定义挤压函数
def squash(vectors, axis=-1):
"""
对向量的非线性激活函数
## vectors: some vectors to be squashed, N-dim tensor
## axis: the axis to squash
:return: a Tensor with same shape as input vectors
"""
s_squared_norm = K.sum(K.square(vectors), axis, keepdims=True)
scale = s_squared_norm / (1 + s_squared_norm) / K.sqrt(s_squared_norm + K.epsilon())
return scale * vectors
class Length(layers.Layer):
"""
计算向量的长度。它用于计算与margin_loss中的y_true具有相同形状的张量
Compute the length of vectors. This is used to compute a Tensor that has the same shape with y_true in margin_loss
inputs: shape=[dim_1, ..., dim_{n-1}, dim_n]
output: shape=[dim_1, ..., dim_{n-1}]
"""
def call(self, inputs, **kwargs):
return K.sqrt(K.sum(K.square(inputs), -1))
def compute_output_shape(self, input_shape):
return input_shape[:-1]
def get_config(self):
config = super(Length, self).get_config()
return config
#定义预胶囊层
def PrimaryCap(inputs, dim_capsule, n_channels, kernel_size, strides, padding):
"""
进行普通二维卷积 `n_channels` 次, 然后将所有的胶囊重叠起来
:param inputs: 4D tensor, shape=[None, width, height, channels]
:param dim_capsule: the dim of the output vector of capsule
:param n_channels: the number of types of capsules
:return: output tensor, shape=[None, num_capsule, dim_capsule]
"""
output = layers.Conv2D(filters=dim_capsule*n_channels, kernel_size=kernel_size, strides=strides,
padding=padding,name='primarycap_conv2d')(inputs)
outputs = layers.Reshape(target_shape=[-1, dim_capsule], name='primarycap_reshape')(output)
return layers.Lambda(squash, name='primarycap_squash')(outputs)
class DenseCapsule(layers.Layer):
"""
胶囊层. 输入输出都为向量.
## num_capsule: 本层包含的胶囊数量
## dim_capsule: 输出的每一个胶囊向量的维度
## routings: routing 算法的迭代次数
"""
def __init__(self, num_capsule, dim_capsule, routings=3, kernel_initializer='glorot_uniform',**kwargs):
super(DenseCapsule, self).__init__(**kwargs)
self.num_capsule = num_capsule
self.dim_capsule = dim_capsule
self.routings = routings
self.kernel_initializer = kernel_initializer
def build(self, input_shape):
assert len(input_shape) >= 3, '输入的 Tensor 的形状[None, input_num_capsule, input_dim_capsule]'#(None,1152,8)
self.input_num_capsule = input_shape[1]
self.input_dim_capsule = input_shape[2]
#转换矩阵
self.W = self.add_weight(shape=[self.num_capsule, self.input_num_capsule,
self.dim_capsule, self.input_dim_capsule],
initializer=self.kernel_initializer,name='W')
self.built = True
def call(self, inputs, training=None):
# inputs.shape=[None, input_num_capsuie, input_dim_capsule]
# inputs_expand.shape=[None, 1, input_num_capsule, input_dim_capsule]
inputs_expand = K.expand_dims(inputs, 1)
# 运算优化:将inputs_expand重复num_capsule 次,用于快速和W相乘
# inputs_tiled.shape=[None, num_capsule, input_num_capsule, input_dim_capsule]
inputs_tiled = K.tile(inputs_expand, [1, self.num_capsule, 1, 1])
# 将inputs_tiled的batch中的每一条数据,计算inputs+W
# x.shape = [num_capsule, input_num_capsule, input_dim_capsule]
# W.shape = [num_capsule, input_num_capsule, dim_capsule, input_dim_capsule]
# 将x和W的前两个维度看作'batch'维度,向量和矩阵相乘:
# [input_dim_capsule] x [dim_capsule, input_dim_capsule]^T -> [dim_capsule].
# inputs_hat.shape = [None, num_capsule, input_num_capsule, dim_capsutel
inputs_hat = K.map_fn(lambda x: K.batch_dot(x, self.W, [2, 3]),elems=inputs_tiled)
# Begin: Routing算法
# 将系数b初始化为0.
# b.shape = [None, self.num_capsule, self, input_num_capsule].
b = tf.zeros(shape=[K.shape(inputs_hat)[0], self.num_capsule, self.input_num_capsule])
assert self.routings > 0, 'The routings should be > 0.'
for i in range(self.routings):
# c.shape=[None, num_capsule, input_num_capsule]
C = tf.nn.softmax(b ,axis=1)
# c.shape = [None, num_capsule, input_num_capsule]
# inputs_hat.shape = [None, num_capsule, input_num_capsule, dim_capsule]
# 将c与inputs_hat的前两个维度看作'batch'维度,向量和矩阵相乘:
# [input_num_capsule] x [input_num_capsule, dim_capsule] -> [dim_capsule],
# outputs.shape= [None, num_capsule, dim_capsule]
outputs = squash(K. batch_dot(C, inputs_hat, [2, 2])) # [None, 10, 16]
if i < self.routings - 1:
# outputs.shape = [None, num_capsule, dim_capsule]
# inputs_hat.shape = [None, num_capsule, input_num_capsule, dim_capsule]
# 将outputs和inρuts_hat的前两个维度看作‘batch’ 维度,向量和矩阵相乘:
# [dim_capsule] x [imput_num_capsule, dim_capsule]^T -> [input_num_capsule]
# b.shape = [batch_size. num_capsule, input_nom_capsule]
# b += K.batch_dot(outputs, inputs_hat, [2, 3]) to this b += tf.matmul(self.W, x)
b += K.batch_dot(outputs, inputs_hat, [2, 3])
# End: Routing 算法
return outputs
def compute_output_shape(self, input_shape):
return tuple([None, self.num_capsule, self.dim_capsule])
def get_config(self):
config = {
'num_capsule': self.num_capsule,
'dim_capsule': self.dim_capsule,
'routings': self.routings
}
base_config = super(DenseCapsule, self).get_config()
return dict(list(base_config.items()) + list(config.items()))
from tensorflow import keras
from keras.regularizers import l2#正则化
x = layers.Input(shape=[512,1, 1])
#普通卷积层
conv1 = layers.Conv2D(filters=16, kernel_size=(2, 1),activation='relu',padding='valid',name='conv1')(x)
#池化层
POOL1 = MaxPooling2D((2,1))(conv1)
#普通卷积层
conv2 = layers.Conv2D(filters=32, kernel_size=(2, 1),activation='relu',padding='valid',name='conv2')(POOL1)
#池化层
# POOL2 = MaxPooling2D((2,1))(conv2)
#Dropout层
Dropout=layers.Dropout(0.1)(conv2)
# Layer 3: 使用“squash”激活的Conv2D层, 然后重塑 [None, num_capsule, dim_vector]
primarycaps = PrimaryCap(Dropout, dim_capsule=8, n_channels=12, kernel_size=(4, 1), strides=2, padding='valid')
# Layer 4: 数字胶囊层,动态路由算法在这里工作。
digitcaps = DenseCapsule(num_capsule=2, dim_capsule=16, routings=3, name='digit_caps')(primarycaps)
# Layer 5:这是一个辅助层,用它的长度代替每个胶囊。只是为了符合标签的形状。
out_caps = Length(name='out_caps')(digitcaps)
model = keras.Model(x, out_caps)
model.summary()
#定义优化
model.compile(metrics=['accuracy'],
optimizer='adam',
loss=lambda y_true,y_pred: y_true*K.relu(0.9-y_pred)**2 + 0.25*(1-y_true)*K.relu(y_pred-0.1)**2
)
import time
time_begin = time.time()
history = model.fit(x_train,one_hot_train_labels,
validation_split=0.1,
epochs=50,batch_size=10,
shuffle=True)
time_end = time.time()
time = time_end - time_begin
print('time:', time)
score = model.evaluate(x_test,one_hot_test_labels, verbose=0)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
#绘制acc-loss曲线
import matplotlib.pyplot as plt
plt.plot(history.history['loss'],color='r')
plt.plot(history.history['val_loss'],color='g')
plt.plot(history.history['accuracy'],color='b')
plt.plot(history.history['val_accuracy'],color='k')
plt.title('model loss and acc')
plt.ylabel('Accuracy')
plt.xlabel('epoch')
plt.legend(['train_loss', 'test_loss','train_acc', 'test_acc'], loc='upper left')
# plt.legend(['train_loss','train_acc'], loc='upper left')
#plt.savefig('1.png')
plt.show()
import matplotlib.pyplot as plt
plt.plot(history.history['loss'],color='r')
plt.plot(history.history['accuracy'],color='b')
plt.title('model loss and sccuracy ')
plt.ylabel('loss/sccuracy')
plt.xlabel('epoch')
plt.legend(['train_loss', 'train_sccuracy'], loc='upper left')
plt.show()
| 0.622459 | 0.396389 |
```
# Initialize logging.
import logging
logging.basicConfig(format='%(asctime)s : %(levelname)s : %(message)s')
import pandas as pd
import numpy as np
```
## Define preprocessor
```
# Import and download stopwords from NLTK.
from nltk.corpus import stopwords
from nltk import download
from nltk.tokenize import RegexpTokenizer
download('stopwords') # Download stopwords list.
# Remove stopwords.
stop_words = stopwords.words('english')
tokenizer = RegexpTokenizer(r'\w+')
def preprocess(text):
text = text.lower()
tokens = tokenizer.tokenize(text)
return [w for w in tokens if w not in stop_words]
```
## Load word2vec model
[GoogleNews pretrained model](https://drive.google.com/file/d/0B7XkCwpI5KDYNlNUTTlSS21pQmM/edit)
```
%%time
from gensim.models import Word2Vec
model = Word2Vec.load_word2vec_format('data/GoogleNews-vectors-negative300.bin.gz', binary=True)
%%time
# Normalizing word2vec vectors.
model.init_sims(replace=True) # Normalizes the vectors in the word2vec class.
```
#### Test WMDistance
```
candidate = "A young child is wearing blue goggles and sitting in a float in a pool."
ref0 = "A blond woman in a blue shirt appears to wait for a ride."
ref1 = "A blond woman is on the street hailing a taxi."
ref2 = "A woman is signaling is to traffic , as seen from behind."
ref3 = "A woman with blonde hair wearing a blue tube top is waving on the side of the street."
ref4 = "The woman in the blue dress is holding out her arm at oncoming traffic."
ref5 = "Sooners football player weas the number 28 and black armbands."
print(model.wmdistance(preprocess(ref0), preprocess(candidate)))
print(model.wmdistance(preprocess(candidate), preprocess(ref1)))
print(model.wmdistance(preprocess(candidate), preprocess(ref2)))
print(model.wmdistance(preprocess(candidate), preprocess(ref3)))
print(model.wmdistance(preprocess(candidate), preprocess(ref4)))
print(model.wmdistance(preprocess(candidate), preprocess(ref5)))
```
## Load candidates and refs
```
candiadates = []
with open('flickr8k/candidates') as f:
candiadates = [preprocess(text) for text in f.readlines()]
refs = {}
for i in range(0, 5):
with open('flickr8k/ref-' + str(i)) as f:
refs[i] = [preprocess(text) for text in f.readlines()]
```
## Calculate distances
```
distances = {}
for i in range(0, 5):
distances[i] = [model.wmdistance(candidate, refs[i][j]) for j, candidate in enumerate(candiadates)]
distances_df = pd.DataFrame(distances)
def normalize(distance):
return 1 / (1 + distance)
for i in range(0, 5):
distances_df[i] = distances_df[i].map(normalize)
```
#### Average by MEAN and MAX
```
def calculate_metrics(raw):
raw['mean'] = np.mean(raw[:5])
raw['max'] = np.max(raw[:5])
return raw
distances_df = distances_df.apply(calculate_metrics, axis=1)
distances_df['id'] = distances_df.index
distances_df['text'] = ''
```
#### Save to csv
```
distances_df.to_csv('scores.csv', columns=["id", "text", 0, 1, 2, 3, 4, "mean", "max",], index=False, sep='\t')
```
|
github_jupyter
|
# Initialize logging.
import logging
logging.basicConfig(format='%(asctime)s : %(levelname)s : %(message)s')
import pandas as pd
import numpy as np
# Import and download stopwords from NLTK.
from nltk.corpus import stopwords
from nltk import download
from nltk.tokenize import RegexpTokenizer
download('stopwords') # Download stopwords list.
# Remove stopwords.
stop_words = stopwords.words('english')
tokenizer = RegexpTokenizer(r'\w+')
def preprocess(text):
text = text.lower()
tokens = tokenizer.tokenize(text)
return [w for w in tokens if w not in stop_words]
%%time
from gensim.models import Word2Vec
model = Word2Vec.load_word2vec_format('data/GoogleNews-vectors-negative300.bin.gz', binary=True)
%%time
# Normalizing word2vec vectors.
model.init_sims(replace=True) # Normalizes the vectors in the word2vec class.
candidate = "A young child is wearing blue goggles and sitting in a float in a pool."
ref0 = "A blond woman in a blue shirt appears to wait for a ride."
ref1 = "A blond woman is on the street hailing a taxi."
ref2 = "A woman is signaling is to traffic , as seen from behind."
ref3 = "A woman with blonde hair wearing a blue tube top is waving on the side of the street."
ref4 = "The woman in the blue dress is holding out her arm at oncoming traffic."
ref5 = "Sooners football player weas the number 28 and black armbands."
print(model.wmdistance(preprocess(ref0), preprocess(candidate)))
print(model.wmdistance(preprocess(candidate), preprocess(ref1)))
print(model.wmdistance(preprocess(candidate), preprocess(ref2)))
print(model.wmdistance(preprocess(candidate), preprocess(ref3)))
print(model.wmdistance(preprocess(candidate), preprocess(ref4)))
print(model.wmdistance(preprocess(candidate), preprocess(ref5)))
candiadates = []
with open('flickr8k/candidates') as f:
candiadates = [preprocess(text) for text in f.readlines()]
refs = {}
for i in range(0, 5):
with open('flickr8k/ref-' + str(i)) as f:
refs[i] = [preprocess(text) for text in f.readlines()]
distances = {}
for i in range(0, 5):
distances[i] = [model.wmdistance(candidate, refs[i][j]) for j, candidate in enumerate(candiadates)]
distances_df = pd.DataFrame(distances)
def normalize(distance):
return 1 / (1 + distance)
for i in range(0, 5):
distances_df[i] = distances_df[i].map(normalize)
def calculate_metrics(raw):
raw['mean'] = np.mean(raw[:5])
raw['max'] = np.max(raw[:5])
return raw
distances_df = distances_df.apply(calculate_metrics, axis=1)
distances_df['id'] = distances_df.index
distances_df['text'] = ''
distances_df.to_csv('scores.csv', columns=["id", "text", 0, 1, 2, 3, 4, "mean", "max",], index=False, sep='\t')
| 0.46563 | 0.677197 |
### Update Test
```
from cvfw import CVFW_MODEL, CVFW_UPDATE
hand_model = CVFW_MODEL(dsize=(28, 28))
hand_model.add_directory(class_name="A", path="C:\\kimdonghwan\\python\\CVFW\\image\\train\\hand_sign\\A")
hand_model.add_directory(class_name="B", path="C:\\kimdonghwan\\python\\CVFW\\image\\train\\hand_sign\\B")
hand_model.add_directory(class_name="C", path="C:\\kimdonghwan\\python\\CVFW\\image\\train\\hand_sign\\C")
hand_model.train()
cvfw_update = CVFW_UPDATE(hand_model, feature_group_number = [5, 3000], feature_weight_number = [80, 120, 300])
cvfw_update.add_validation(class_name="A", path="C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\A")
cvfw_update.add_validation(class_name="B", path="C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\B")
cvfw_update.add_validation(class_name="C", path="C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\C")
cvfw_update.update()
cvfw_update.set(feature_group_number = [50, 100, 250], feature_weight_number = [0, 20, 40])
cvfw_update.update()
cvfw_update.set(feature_group_number = [10, 15], feature_weight_number = [18])
cvfw_update.update()
```
### 최적의 feature_group_number 와 feature_weight_number
- feature_group_number: 10
- feature_weight_number: 18
- accuracy: 92.91705498602051%
### Modeling Test
```
from time import time
start_time = time()
hand_model = CVFW_MODEL(dsize=(28, 28), feature_group_number = 10, feature_weight_number = 18)
hand_model.add_directory(class_name="A", path="C:\\kimdonghwan\\python\\CVFW\\image\\train\\hand_sign\\A")
hand_model.add_directory(class_name="B", path="C:\\kimdonghwan\\python\\CVFW\\image\\train\\hand_sign\\B")
hand_model.add_directory(class_name="C", path="C:\\kimdonghwan\\python\\CVFW\\image\\train\\hand_sign\\C")
hand_model.train()
print(time() - start_time)
import matplotlib.pyplot as plt
A_img = hand_model.modeling(class_name="A")
plt.imshow(A_img, cmap="gray")
B_img = hand_model.modeling(class_name='B')
plt.imshow(B_img, cmap="gray")
C_img = hand_model.modeling(class_name="C")
plt.imshow(C_img, cmap="gray")
```
### Modeling Predict Class 는 괜찮은 predict 방법일까?
- 각 클래스당 modeling 된 이미지로 유사도를 계산하여 predict를 하였을 때 86.1% 로 꽤 높은 정확도를 보여주었다.
- 원래 있었던 predict class 보다 좋은 방법인지 확인해보자.
### 결론
- 기존의 predict 방법이 더 좋다고 판단이 된다.
- 이유: modeling predict class 는 각 cost 들의 값이 너무 많이 비슷하여 불안정한 예측 방법이라고 판단되었기 때문이다.
```
from cvfw import Weight
import cv2
from os import listdir
import numpy as np
import matplotlib.pyplot as plt
A_Weight = Weight(28, 28, A_img.flatten().tolist())
B_Weight = Weight(28, 28, B_img.flatten().tolist())
C_Weight = Weight(28, 28, C_img.flatten().tolist())
count = 0
answer = 0
A_files = listdir("C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\A")
B_files = listdir("C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\B")
C_files = listdir("C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\C")
predicts = []
for file in A_files:
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\A\\{file}", 1), cv2.COLOR_BGR2GRAY).flatten().tolist()
img_weight = Weight(28, 28, img)
A_cost = sum(sum(abs(A_Weight - img_weight)))
B_cost = sum(sum(abs(B_Weight - img_weight)))
C_cost = sum(sum(abs(C_Weight - img_weight)))
sum_cost = sum([A_cost, B_cost, C_cost])
predict = [A_cost / sum_cost, B_cost / sum_cost, C_cost / sum_cost]
predicts.append(predict)
if min(predict) == predict[0]: answer += 1
count += 1
for file in B_files:
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\B\\{file}", 1), cv2.COLOR_BGR2GRAY).flatten().tolist()
img_weight = Weight(28, 28, img)
A_cost = sum(sum(abs(A_Weight - img_weight)))
B_cost = sum(sum(abs(B_Weight - img_weight)))
C_cost = sum(sum(abs(C_Weight - img_weight)))
sum_cost = sum([A_cost, B_cost, C_cost])
predict = [A_cost / sum_cost, B_cost / sum_cost, C_cost / sum_cost]
predicts.append(predict)
if min(predict) == predict[1]: answer += 1
count += 1
for file in C_files:
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\C\\{file}", 1), cv2.COLOR_BGR2GRAY).flatten().tolist()
img_weight = Weight(28, 28, img)
A_cost = sum(sum(abs(A_Weight - img_weight)))
B_cost = sum(sum(abs(B_Weight - img_weight)))
C_cost = sum(sum(abs(C_Weight - img_weight)))
sum_cost = sum([A_cost, B_cost, C_cost])
predict = [A_cost / sum_cost, B_cost / sum_cost, C_cost / sum_cost]
predicts.append(predict)
if min(predict) == predict[2]: answer += 1
count += 1
print(answer / count)
print("정확도가 약 95.7%로 꽤나 높은 수치임을 알 수 있다.")
print("하지만 cost 들의 값이 많이 비슷하기 때문에 불안정한 예측 방법으로 판단된다.")
print("but 속도는 기존의 predict_class 방법보다는 우월한 성능을 보여준다.")
print("하나의 predict 안에서의 cost 가 전체 30% 를 넘는 원소의 개수: ", (np.array(predicts) > 0.3).flatten().tolist().count(True))
print("하나의 predict 안에서의 cost 가 전체 20% 를 넘는 원소의 개수: ", (np.array(predicts) > 0.20).flatten().tolist().count(True))
x = [i for i in range(len(predicts))]
rock_y = [i[0] for i in predicts]
palm_y = [i[1] for i in predicts]
c_y = [i[2] for i in predicts]
plt.scatter(x, rock_y, color="red", label="rock", alpha=0.8)
plt.scatter(x, palm_y, color="blue", label="palm", alpha=0.8)
plt.scatter(x, c_y, color="green", label="c", alpha=0.8)
plt.ylim(0, 1.0)
plt.title("modeling predict class")
plt.legend()
count = 0
answer = 0
predicts = []
for file in A_files:
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\A\\{file}", 1), cv2.COLOR_BGR2GRAY).flatten().tolist()
predict = hand_model.predict_class(img)
predicts.append(predict)
if min(predict) == predict[0]: answer += 1
count += 1
for file in B_files:
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\B\\{file}", 1), cv2.COLOR_BGR2GRAY).flatten().tolist()
predict = hand_model.predict_class(img)
predicts.append(predict)
if min(predict) == predict[1]: answer += 1
count += 1
for file in C_files:
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\C\\{file}", 1), cv2.COLOR_BGR2GRAY).flatten().tolist()
predict = hand_model.predict_class(img)
predicts.append(predict)
if min(predict) == predict[2]: answer += 1
count += 1
print(answer / count)
x = [i for i in range(len(predicts))]
rock_y = [i[0] for i in predicts]
palm_y = [i[1] for i in predicts]
c_y = [i[2] for i in predicts]
plt.scatter(x, rock_y, color="red", label="rock", alpha=0.8)
plt.scatter(x, palm_y, color="blue", label="palm", alpha=0.8)
plt.scatter(x, c_y, color="green", label="c", alpha=0.8)
plt.ylim(0, 1.0)
plt.title("predict class")
plt.legend()
```
### Predict Test
```
import cv2
import matplotlib.pyplot as plt
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\A\\3_A.jpg", 1), cv2.COLOR_BGR2GRAY)
plt.imshow(img, cmap="gray")
img = cv2.resize(img, dsize=(28, 28)).flatten().tolist()
predict = hand_model.predict_class(img)
print(predict)
# CVIW 의 predict 는 cost 비율이기 때문에 predict 값이 가장 작은 것이 예측값이다.
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\B\\66_B.jpg", 1), cv2.COLOR_BGR2GRAY)
plt.imshow(img, cmap="gray")
img = cv2.resize(img, dsize=(28, 28)).flatten().tolist()
predict = hand_model.predict_class(img)
print(predict)
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\C\\20_c.jpg", 1), cv2.COLOR_BGR2GRAY)
plt.imshow(img, cmap="gray")
img = cv2.resize(img, dsize=(28, 28)).flatten().tolist()
predict = hand_model.predict_class(img)
print(predict)
```
|
github_jupyter
|
from cvfw import CVFW_MODEL, CVFW_UPDATE
hand_model = CVFW_MODEL(dsize=(28, 28))
hand_model.add_directory(class_name="A", path="C:\\kimdonghwan\\python\\CVFW\\image\\train\\hand_sign\\A")
hand_model.add_directory(class_name="B", path="C:\\kimdonghwan\\python\\CVFW\\image\\train\\hand_sign\\B")
hand_model.add_directory(class_name="C", path="C:\\kimdonghwan\\python\\CVFW\\image\\train\\hand_sign\\C")
hand_model.train()
cvfw_update = CVFW_UPDATE(hand_model, feature_group_number = [5, 3000], feature_weight_number = [80, 120, 300])
cvfw_update.add_validation(class_name="A", path="C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\A")
cvfw_update.add_validation(class_name="B", path="C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\B")
cvfw_update.add_validation(class_name="C", path="C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\C")
cvfw_update.update()
cvfw_update.set(feature_group_number = [50, 100, 250], feature_weight_number = [0, 20, 40])
cvfw_update.update()
cvfw_update.set(feature_group_number = [10, 15], feature_weight_number = [18])
cvfw_update.update()
from time import time
start_time = time()
hand_model = CVFW_MODEL(dsize=(28, 28), feature_group_number = 10, feature_weight_number = 18)
hand_model.add_directory(class_name="A", path="C:\\kimdonghwan\\python\\CVFW\\image\\train\\hand_sign\\A")
hand_model.add_directory(class_name="B", path="C:\\kimdonghwan\\python\\CVFW\\image\\train\\hand_sign\\B")
hand_model.add_directory(class_name="C", path="C:\\kimdonghwan\\python\\CVFW\\image\\train\\hand_sign\\C")
hand_model.train()
print(time() - start_time)
import matplotlib.pyplot as plt
A_img = hand_model.modeling(class_name="A")
plt.imshow(A_img, cmap="gray")
B_img = hand_model.modeling(class_name='B')
plt.imshow(B_img, cmap="gray")
C_img = hand_model.modeling(class_name="C")
plt.imshow(C_img, cmap="gray")
from cvfw import Weight
import cv2
from os import listdir
import numpy as np
import matplotlib.pyplot as plt
A_Weight = Weight(28, 28, A_img.flatten().tolist())
B_Weight = Weight(28, 28, B_img.flatten().tolist())
C_Weight = Weight(28, 28, C_img.flatten().tolist())
count = 0
answer = 0
A_files = listdir("C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\A")
B_files = listdir("C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\B")
C_files = listdir("C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\C")
predicts = []
for file in A_files:
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\A\\{file}", 1), cv2.COLOR_BGR2GRAY).flatten().tolist()
img_weight = Weight(28, 28, img)
A_cost = sum(sum(abs(A_Weight - img_weight)))
B_cost = sum(sum(abs(B_Weight - img_weight)))
C_cost = sum(sum(abs(C_Weight - img_weight)))
sum_cost = sum([A_cost, B_cost, C_cost])
predict = [A_cost / sum_cost, B_cost / sum_cost, C_cost / sum_cost]
predicts.append(predict)
if min(predict) == predict[0]: answer += 1
count += 1
for file in B_files:
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\B\\{file}", 1), cv2.COLOR_BGR2GRAY).flatten().tolist()
img_weight = Weight(28, 28, img)
A_cost = sum(sum(abs(A_Weight - img_weight)))
B_cost = sum(sum(abs(B_Weight - img_weight)))
C_cost = sum(sum(abs(C_Weight - img_weight)))
sum_cost = sum([A_cost, B_cost, C_cost])
predict = [A_cost / sum_cost, B_cost / sum_cost, C_cost / sum_cost]
predicts.append(predict)
if min(predict) == predict[1]: answer += 1
count += 1
for file in C_files:
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\C\\{file}", 1), cv2.COLOR_BGR2GRAY).flatten().tolist()
img_weight = Weight(28, 28, img)
A_cost = sum(sum(abs(A_Weight - img_weight)))
B_cost = sum(sum(abs(B_Weight - img_weight)))
C_cost = sum(sum(abs(C_Weight - img_weight)))
sum_cost = sum([A_cost, B_cost, C_cost])
predict = [A_cost / sum_cost, B_cost / sum_cost, C_cost / sum_cost]
predicts.append(predict)
if min(predict) == predict[2]: answer += 1
count += 1
print(answer / count)
print("정확도가 약 95.7%로 꽤나 높은 수치임을 알 수 있다.")
print("하지만 cost 들의 값이 많이 비슷하기 때문에 불안정한 예측 방법으로 판단된다.")
print("but 속도는 기존의 predict_class 방법보다는 우월한 성능을 보여준다.")
print("하나의 predict 안에서의 cost 가 전체 30% 를 넘는 원소의 개수: ", (np.array(predicts) > 0.3).flatten().tolist().count(True))
print("하나의 predict 안에서의 cost 가 전체 20% 를 넘는 원소의 개수: ", (np.array(predicts) > 0.20).flatten().tolist().count(True))
x = [i for i in range(len(predicts))]
rock_y = [i[0] for i in predicts]
palm_y = [i[1] for i in predicts]
c_y = [i[2] for i in predicts]
plt.scatter(x, rock_y, color="red", label="rock", alpha=0.8)
plt.scatter(x, palm_y, color="blue", label="palm", alpha=0.8)
plt.scatter(x, c_y, color="green", label="c", alpha=0.8)
plt.ylim(0, 1.0)
plt.title("modeling predict class")
plt.legend()
count = 0
answer = 0
predicts = []
for file in A_files:
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\A\\{file}", 1), cv2.COLOR_BGR2GRAY).flatten().tolist()
predict = hand_model.predict_class(img)
predicts.append(predict)
if min(predict) == predict[0]: answer += 1
count += 1
for file in B_files:
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\B\\{file}", 1), cv2.COLOR_BGR2GRAY).flatten().tolist()
predict = hand_model.predict_class(img)
predicts.append(predict)
if min(predict) == predict[1]: answer += 1
count += 1
for file in C_files:
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\C\\{file}", 1), cv2.COLOR_BGR2GRAY).flatten().tolist()
predict = hand_model.predict_class(img)
predicts.append(predict)
if min(predict) == predict[2]: answer += 1
count += 1
print(answer / count)
x = [i for i in range(len(predicts))]
rock_y = [i[0] for i in predicts]
palm_y = [i[1] for i in predicts]
c_y = [i[2] for i in predicts]
plt.scatter(x, rock_y, color="red", label="rock", alpha=0.8)
plt.scatter(x, palm_y, color="blue", label="palm", alpha=0.8)
plt.scatter(x, c_y, color="green", label="c", alpha=0.8)
plt.ylim(0, 1.0)
plt.title("predict class")
plt.legend()
import cv2
import matplotlib.pyplot as plt
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\A\\3_A.jpg", 1), cv2.COLOR_BGR2GRAY)
plt.imshow(img, cmap="gray")
img = cv2.resize(img, dsize=(28, 28)).flatten().tolist()
predict = hand_model.predict_class(img)
print(predict)
# CVIW 의 predict 는 cost 비율이기 때문에 predict 값이 가장 작은 것이 예측값이다.
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\B\\66_B.jpg", 1), cv2.COLOR_BGR2GRAY)
plt.imshow(img, cmap="gray")
img = cv2.resize(img, dsize=(28, 28)).flatten().tolist()
predict = hand_model.predict_class(img)
print(predict)
img = cv2.cvtColor(cv2.imread(f"C:\\kimdonghwan\\python\\CVFW\\image\\test\\hand_sign\\C\\20_c.jpg", 1), cv2.COLOR_BGR2GRAY)
plt.imshow(img, cmap="gray")
img = cv2.resize(img, dsize=(28, 28)).flatten().tolist()
predict = hand_model.predict_class(img)
print(predict)
| 0.369998 | 0.482551 |
```
import json
import spacy
from glob import glob
nlp = spacy.load('en_core_web_md')
files = glob('KPTimes.*.jsonl')
files
def case_of(text):
return (
str.upper
if text.isupper()
else str.lower
if text.islower()
else str.title
if text.istitle()
else str
)
def capitalize(string, keyword):
keywords = keyword.split(';')
doc = nlp(string)
entities = [entity.text for entity in doc.ents]
entities_short = [''.join([w[0] for w in word.split()]) for word in entities]
tainted = {}
actual_keywords = []
for key in keywords:
for no, entity in enumerate(entities_short):
if entity.lower().find(key) > 0:
result = []
index = 0
for i in range(entity.lower().index(key), len(entity)):
result.append(case_of(entity[i])(key[index]))
index += 1
actual_keywords.append(''.join(result))
actual_keywords.append(entities[no])
tainted[key] = True
string_lower = string.lower().split()
string = string.split()
for key in keywords:
if key in string_lower:
actual_keywords.append(string[string_lower.index(key)])
tainted[key] = True
for key in keywords:
if not tainted.get(key, False):
result = []
for token in nlp(key):
if len(token.ent_type_):
if token.ent_type_ in ['TIME', 'MONEY']:
t = token.text.upper()
else:
t = token.text.title()
else:
t = token.text
result.append(t)
actual_keywords.append(' '.join(result))
actual_keywords = list(set(actual_keywords))
return actual_keywords
from tqdm import tqdm
X, Y, titles = [], [], []
for file in files:
with open(file) as fopen:
data = list(filter(None, fopen.read().split('\n')))
print(file)
for i in tqdm(range(len(data))):
row = json.loads(data[i])
keywords = capitalize(row['abstract'], row['keyword'])
X.append(row['abstract'])
Y.append(keywords)
titles.append(row['title'])
file = 'KPTimes.test.jsonl'
with open(file) as fopen:
data = list(filter(None, fopen.read().split('\n')))
print(file)
for i in tqdm(range(165, len(data))):
try:
row = json.loads(data[i])
keywords = capitalize(row['abstract'], row['keyword'])
X.append(row['abstract'])
Y.append(keywords)
titles.append(row['title'])
except:
pass
titles[-1]
Y[-1]
with open('kptimes.json', 'w') as fopen:
json.dump({'X': X, 'Y': Y, 'titles': titles}, fopen)
import re
import json
def cleaning(string):
string = re.sub(r'[ ]+', ' ', string.replace('\n',' ')).strip()
return string
def limit(string, max_len = 3500):
string = string.split()
r = ''
for s in string:
if len(r + ' ' + s) > max_len:
break
r = r + ' ' + s
return cleaning(r)
limit(X[-1])
titles[-1]
combined = f"{titles[-1]} [[EENNDD]] {'; '.join(Y[-1])}"
combined
import re
from tqdm import tqdm
import json
data = []
for i in tqdm(range(len(X))):
string = X[i]
string = cleaning(string)
string = limit(string)
keywords = Y[i]
combined = f"{string} [[EENNDD]] {'; '.join(keywords)}"
data.append({'string': string, 'keywords': keywords, 'combined': combined})
combined = f"{titles[i]} [[EENNDD]] {'; '.join(keywords)}"
data.append({'string': titles[i], 'keywords': keywords, 'combined': combined})
len(data)
batch_size = 50000
for i in range(0, len(data), batch_size):
index = min(i + batch_size, len(data))
x = data[i: index]
with open(f'kptimes-{i}.json', 'w') as fopen:
json.dump(x, fopen)
```
|
github_jupyter
|
import json
import spacy
from glob import glob
nlp = spacy.load('en_core_web_md')
files = glob('KPTimes.*.jsonl')
files
def case_of(text):
return (
str.upper
if text.isupper()
else str.lower
if text.islower()
else str.title
if text.istitle()
else str
)
def capitalize(string, keyword):
keywords = keyword.split(';')
doc = nlp(string)
entities = [entity.text for entity in doc.ents]
entities_short = [''.join([w[0] for w in word.split()]) for word in entities]
tainted = {}
actual_keywords = []
for key in keywords:
for no, entity in enumerate(entities_short):
if entity.lower().find(key) > 0:
result = []
index = 0
for i in range(entity.lower().index(key), len(entity)):
result.append(case_of(entity[i])(key[index]))
index += 1
actual_keywords.append(''.join(result))
actual_keywords.append(entities[no])
tainted[key] = True
string_lower = string.lower().split()
string = string.split()
for key in keywords:
if key in string_lower:
actual_keywords.append(string[string_lower.index(key)])
tainted[key] = True
for key in keywords:
if not tainted.get(key, False):
result = []
for token in nlp(key):
if len(token.ent_type_):
if token.ent_type_ in ['TIME', 'MONEY']:
t = token.text.upper()
else:
t = token.text.title()
else:
t = token.text
result.append(t)
actual_keywords.append(' '.join(result))
actual_keywords = list(set(actual_keywords))
return actual_keywords
from tqdm import tqdm
X, Y, titles = [], [], []
for file in files:
with open(file) as fopen:
data = list(filter(None, fopen.read().split('\n')))
print(file)
for i in tqdm(range(len(data))):
row = json.loads(data[i])
keywords = capitalize(row['abstract'], row['keyword'])
X.append(row['abstract'])
Y.append(keywords)
titles.append(row['title'])
file = 'KPTimes.test.jsonl'
with open(file) as fopen:
data = list(filter(None, fopen.read().split('\n')))
print(file)
for i in tqdm(range(165, len(data))):
try:
row = json.loads(data[i])
keywords = capitalize(row['abstract'], row['keyword'])
X.append(row['abstract'])
Y.append(keywords)
titles.append(row['title'])
except:
pass
titles[-1]
Y[-1]
with open('kptimes.json', 'w') as fopen:
json.dump({'X': X, 'Y': Y, 'titles': titles}, fopen)
import re
import json
def cleaning(string):
string = re.sub(r'[ ]+', ' ', string.replace('\n',' ')).strip()
return string
def limit(string, max_len = 3500):
string = string.split()
r = ''
for s in string:
if len(r + ' ' + s) > max_len:
break
r = r + ' ' + s
return cleaning(r)
limit(X[-1])
titles[-1]
combined = f"{titles[-1]} [[EENNDD]] {'; '.join(Y[-1])}"
combined
import re
from tqdm import tqdm
import json
data = []
for i in tqdm(range(len(X))):
string = X[i]
string = cleaning(string)
string = limit(string)
keywords = Y[i]
combined = f"{string} [[EENNDD]] {'; '.join(keywords)}"
data.append({'string': string, 'keywords': keywords, 'combined': combined})
combined = f"{titles[i]} [[EENNDD]] {'; '.join(keywords)}"
data.append({'string': titles[i], 'keywords': keywords, 'combined': combined})
len(data)
batch_size = 50000
for i in range(0, len(data), batch_size):
index = min(i + batch_size, len(data))
x = data[i: index]
with open(f'kptimes-{i}.json', 'w') as fopen:
json.dump(x, fopen)
| 0.14013 | 0.169303 |
### Custom dataset.
In this notebook we are going to leran how we can load our own custom dataset from files. The dataset that i am using was found on [this site](http://www.statmt.org/europarl/).
First we will define the path where our files are located as the base_path. In my case i am using google drive.
```
base_path = '/content/drive/MyDrive/NLP Data/seq2seq/fr-eng'
```
### Imports
```
import os
import torch
from torchtext.legacy import data, datasets
import json
import pandas as pd
from sklearn.model_selection import train_test_split
```
We have two text files for the french and english sentences with the following file names:
```py
fr = "europarl-v7.fr-en.fr"
en = "europarl-v7.fr-en.en"
```
```
fr_path = "europarl-v7.fr-en.fr"
en_path = "europarl-v7.fr-en.en"
```
Now let's load the text into list of strings. We are going to use the new line as the surperator of each sentence.
```
eng_sentences = open(os.path.join(base_path, en_path), encoding='utf8').read().split('\n')
fr_sentences = open(os.path.join(base_path, fr_path), encoding='utf8').read().split('\n')
```
### Next we will check how many examples do we have for each language.
```
print("eng: ", len(eng_sentences))
print("fr: ", len(fr_sentences))
```
### Creating a pandas dataframe
Creatting the pd dataframe will help us to split the sets into train and test and the convert the splitted dataframes into either `.json` or `.csv` files which are the formats that are accepted by the `torchtext`. To make this very simple Im going to use only `500` sentence french to english pairs.
```
size = 500
raw_data ={
'eng': [sent for sent in eng_sentences[:size]],
'fr': [sent for sent in fr_sentences[:size]],
}
dataframe = pd.DataFrame(raw_data, columns=['eng', 'fr'])
```
### Checking our dataframe
```
dataframe.head(4)
```
### Spliting the datasets.
We are going to use `sklearn` `train_test_split` to split these two datasets for the train and validation sets.
```
train, val = train_test_split(dataframe, test_size=.2)
len(train), len(val)
```
### Creating json files.
We are going to create `json` files and save them to the `base_path` for these two sets. We will be using the `.to_json()` method to do this.
**Note** you can also use the `.to_csv()` to create `csv` files for example:
```py
train.to_csv("train.csv", index=False)
val.to_csv("val.csv", index=False)
```
**Note**: When you are using `.to_json()` we should pass the arg `orient="records"` so that these json files will be the files that can be accepted by the `torchtext`. Basically what this is doing is to add json files as records by removing the list `[]` brakets
```
train.to_json(os.path.join(base_path, 'train.json'), orient="records", lines=True)
val.to_json(os.path.join(base_path, 'val.json'), orient="records", lines=True)
```
Now each record has the following format:
```json
{"eng":"For us new members, it was the first time, and this was a very interesting process.","fr":"C' \u00e9tait pour nous, nouveaux d\u00e9put\u00e9s, la premi\u00e8re fois, et c' est un processus extr\u00eamement int\u00e9ressant."}
```
### Let's load the tokenizer models
```
import spacy
import spacy.cli
spacy.cli.download('fr_core_news_sm')
import fr_core_news_sm, en_core_web_sm
spacy_fr = spacy.load('fr_core_news_sm')
spacy_en = spacy.load('en_core_web_sm')
def tokenize_fr(sent):
sent = sent.lower()
return [tok for tok in spacy_fr.tokenizer(sent)]
def tokenize_en(sent):
sent = sent.lower()
return [tok for tok in spacy_en.tokenizer(sent)]
```
### Creating fields
```
SRC = data.Field(
tokenize = tokenize_fr,
init_token = "<sos>",
eos_token = "<eos>"
)
TRG = data.Field(
tokenize = tokenize_en,
init_token = "<sos>",
eos_token = "<eos>"
)
fields ={
"fr": ("src", SRC),
"eng": ("trg", TRG)
}
```
### We are now ready to create our dataset.
We are going to use the `TabularDataset.splits()` method to create the train and validation datasets.
```
train_data, val_data = data.TabularDataset.splits(
base_path,
format="json",
train="train.json",
validation= 'val.json',
fields=fields
)
print(vars(train_data.examples[0]))
```
### Building the vocabulary
Now we are ready to build the vocabulary.
**Note** In this simple example we will build the vocab on both sets. It is recomended that _when building the vocabulary we only need to build it on the train set_.
We will be building the vocab as follows without `min_freq=2` args since our dataset is small:
**Note**: The `min_freq=2` allows us to set the minimum frequency of each word meaning a word that appears less than two times will be converted to `<unk>` token.
```py
SRC.build_vocab(train_data, val_data, max_size=1000)
TRG.build_vocab(train_data, val_data, max_size=1000)
```
```
SRC.build_vocab(train_data, val_data, max_size=1000)
TRG.build_vocab(train_data, val_data, max_size=1000)
TRG.vocab.itos[11]
```
### Creating iterators
Now you can create iterators and then load the iterators to the models. Again we are going to use the `BucketIterator`.
```
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
BATCH_SIZE = 128
train_iter, val_iter = data.BucketIterator.splits(
(train_data, val_data),
batch_size=BATCH_SIZE,
device=device,
sort_key=lambda x: len(x.src)
)
```
### Checking the a single batch
```
batch = next(iter(train_iter))
batch.src
```
### Resources used.
1. [This Blog Post](https://towardsdatascience.com/how-to-use-torchtext-for-neural-machine-translation-plus-hack-to-make-it-5x-faster-77f3884d95)
2. [Datasets List](http://www.statmt.org/europarl/)
3. [Alen Nie](https://anie.me/On-Torchtext/)
### Extra resources
1 [Harvard](http://nlp.seas.harvard.edu/2018/04/03/attention.html)
```
```
|
github_jupyter
|
base_path = '/content/drive/MyDrive/NLP Data/seq2seq/fr-eng'
import os
import torch
from torchtext.legacy import data, datasets
import json
import pandas as pd
from sklearn.model_selection import train_test_split
fr = "europarl-v7.fr-en.fr"
en = "europarl-v7.fr-en.en"
fr_path = "europarl-v7.fr-en.fr"
en_path = "europarl-v7.fr-en.en"
eng_sentences = open(os.path.join(base_path, en_path), encoding='utf8').read().split('\n')
fr_sentences = open(os.path.join(base_path, fr_path), encoding='utf8').read().split('\n')
print("eng: ", len(eng_sentences))
print("fr: ", len(fr_sentences))
size = 500
raw_data ={
'eng': [sent for sent in eng_sentences[:size]],
'fr': [sent for sent in fr_sentences[:size]],
}
dataframe = pd.DataFrame(raw_data, columns=['eng', 'fr'])
dataframe.head(4)
train, val = train_test_split(dataframe, test_size=.2)
len(train), len(val)
train.to_csv("train.csv", index=False)
val.to_csv("val.csv", index=False)
train.to_json(os.path.join(base_path, 'train.json'), orient="records", lines=True)
val.to_json(os.path.join(base_path, 'val.json'), orient="records", lines=True)
{"eng":"For us new members, it was the first time, and this was a very interesting process.","fr":"C' \u00e9tait pour nous, nouveaux d\u00e9put\u00e9s, la premi\u00e8re fois, et c' est un processus extr\u00eamement int\u00e9ressant."}
import spacy
import spacy.cli
spacy.cli.download('fr_core_news_sm')
import fr_core_news_sm, en_core_web_sm
spacy_fr = spacy.load('fr_core_news_sm')
spacy_en = spacy.load('en_core_web_sm')
def tokenize_fr(sent):
sent = sent.lower()
return [tok for tok in spacy_fr.tokenizer(sent)]
def tokenize_en(sent):
sent = sent.lower()
return [tok for tok in spacy_en.tokenizer(sent)]
SRC = data.Field(
tokenize = tokenize_fr,
init_token = "<sos>",
eos_token = "<eos>"
)
TRG = data.Field(
tokenize = tokenize_en,
init_token = "<sos>",
eos_token = "<eos>"
)
fields ={
"fr": ("src", SRC),
"eng": ("trg", TRG)
}
train_data, val_data = data.TabularDataset.splits(
base_path,
format="json",
train="train.json",
validation= 'val.json',
fields=fields
)
print(vars(train_data.examples[0]))
SRC.build_vocab(train_data, val_data, max_size=1000)
TRG.build_vocab(train_data, val_data, max_size=1000)
SRC.build_vocab(train_data, val_data, max_size=1000)
TRG.build_vocab(train_data, val_data, max_size=1000)
TRG.vocab.itos[11]
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
BATCH_SIZE = 128
train_iter, val_iter = data.BucketIterator.splits(
(train_data, val_data),
batch_size=BATCH_SIZE,
device=device,
sort_key=lambda x: len(x.src)
)
batch = next(iter(train_iter))
batch.src
| 0.496338 | 0.938857 |
# NumPy Giriş
### Neden NumPy
```
# NumPy kullanılmadan yapılan bir işlem.
a = [1,2,3,4]
b = [2,3,4,5]
ab = list()
for i in range(0,len(a)):
ab.append(a[i]*b[i])
print(ab)
# Aynı işlmei Numpy da yaparsak.
import numpy as np
a = np.array([1,2,3,4])
b = np.array([2,3,4,5])
a*b
```
### C int VS Python int
```
x = 9
who
x # Python da x e atanana değer 4 farklı satır ile tanımlanır.
```
```
from IPython.display import Image
path = "/Users/ilgar/OneDrive\Programlama/Python A-Z - Veri Bilimi ve Machine Learning/Python Programlama/JupyterLab/Veri Manipulasyonu/img/"
Image(filename = path + "cint_vs_pyint.png", width=400, height=400)
```
### NumPy Array VS Python List
```
L = [1,2,"a",1.2]
L
# Python da liste oluşturmak çok maliyetli bir işlemdir.
# Liste yerine NumPy Array oluşturusak belli kısıtlamalar gelmesine rağmen daha performanslı çalışır.
[type(i) for i in L]
from IPython.display import Image
path = "/Users/ilgar/OneDrive/Programlama/Python A-Z - Veri Bilimi ve Machine Learning/Python Programlama/JupyterLab/Veri Manipulasyonu/img/"
Image(filename = path + "array_vs_list.png", width=500, height=500)
```
# Numpy Array'i Oluşturmak
### Listelerden Array Oluşturmak
```
import numpy as np # numpy'ı dahil ediyoruz
a = np.array([1,2,3,4,5]) # NumPy array oluşturduk
type(a) # tipini sorguladık
np.array([3, 4.5, 3, 12.5, 7]) # Fixed Type ==> Sabit Tip
np.array([3, 4.5, 3, 12.5, 7], dtype = "int") # Kendimiz veri tipi tanımlayabiliriz "dtype" ile
```
### Sıfırdan Array Oluşturmak
```
np.zeros(10, dtype = "int")
# Sıfırlardan oluşan int değerinde bir array.
np.zeros(10, dtype = "str")
# Sıfır değerlerinden oluşan string değerine sahip bir array
```
### Birlerden İki Boyutlu Bir Array Oluşturmak
```
np.ones(10) # 10 tane 1 den oluşur
```
```
np.ones((2,4)) # 2 tane 4'lük yapıdan oluşur. Bu bir Matrix dir.
```
### İstenilen Değerlerden İki Boyutlu Array Oluşturmak
```
np.full((2,3), 5)
```
### Doğrusal Bir Diziden Array Oluşturma
```
np.arange(0,10, 2)
```
### İki Değer Arasında İstenilen Kadar Array Oluşturma
```
np.linspace(0,1,30)
```
### Dağılımlara Göre Array Oluşturmak
```
np.random.normal(0,1, (3,4))
```
.
### İki Sayı Arasında İnt Değerlerden Oluşan Bir Matrix Oluşturma
```
np.random.randint(0, 10, (2,2))
```
### Köşegen Elemanları 1 Olan Bir Matrix Oluşturma
```
np.eye(4)
```
# NumPy Biçimlendirme
### Özellikleri
* **ndim:** Boyut Sayısı
* **shape:** Boyut bilgisi
* **size:** Toplam eleman sayısı
* **dtype:** Array veri tipi
```
import numpy as np
a = np.random.randint(1, 10, 10)
a
a.ndim # 1 Boyutlu,
a.shape # (10,) Boyutundan oluşan,
a.size # 10 elemanlı,
a.dtype # Integer tipinde bir array.
b = np.random.randint(0,12, (4,3))
b
b.ndim # 2 Boyutlu,
b.shape # (4,3) Boyutundan oluşan,
b.size # 12 elemanlı,
b.dtype # Integer tipinde olan bir array.
c = np.random.randint(1,20, size = (4,3,5))
c
c.ndim # 3 Boyutlu
c.shape # (4,3,5) boyutundan oluşan,
c.size # 60 elemanlı
c.dtype # Integer tipinde bir array
```
### Reshape (Yeniden Şekilldendirme) (Vektörler üzerinden matrix oluşturma)
```
np.arange(1,17).reshape((4,4))
# 1,2,3,4,5,...,14,15,16 elemanlarından oluşan 1 Boyutlu arrayi,
# (4,4) lük iki boyutlu bir arraya çevirdik
x = np.array([1,2,3])
x # 1 Boyutlu array.
y = x.reshape((1,3)) # 2 Boyutlu arraye çevirdik
y
y.ndim
x[np.newaxis, :] # Alternatif Yöntem # Satır vektörü
x[: ,np.newaxis] # Sütun Vektörü
```
### Array Birleştirme İşlemleri
##### Tek Boyutlu Birleştirme
```
x = np.array([1,2,3])
y = np.array([4,5,6])
np.concatenate([x,y])
my_list = [7,8,9]
np.concatenate([x,y,my_list])
# Liste kullanarak da arraye eleman eklenebilir
```
##### İki Boyutlu Birleştirme
```
a = np.array([[1,2,3],
[4,5,6]])
np.concatenate([a,a])
np.concatenate([a,a], axis = 1)
# Birinci ile birinci, ikinci ile ikinci birleşti.
```
##### Farklı Boyutlu Array Birleştirme
```
a = np.array([1,2,3])
b = np.array([[9,8,7],
[6,5,4]])
np.vstack([a,b]) # Dikey birleştirme
a = np.array([[99],
[99]])
np.hstack([a,b]) # Yatay birleştirme
```
### Array Ayırma İşlemleri (Splitting)
#### 1 Boyutlu Array Ayırma
```
x = [1,2,3,99,99,3,2,1]
x
np.split(x, [3,5]) # n indeks ise n+1 bölüme ayırma işlemi.
a, b, c = np.split(x, [3,5]) # Arrayleri değişik değişkenlere atar.
a
b
c
```
#### 2 Boyutlu Array Ayırma
##### Dikey Ayırma
```
m = np.arange(16).reshape((4,4))
m # 2.satırdan itibaren böl
np.vsplit(m,[2])
# Dikey bölme işlemi gerçekleştiriyoruz
ust, alt = np.vsplit(m,[2])
ust
alt
```
##### Yatay Ayırma
```
m
np.hsplit(m, [2])
# Yatay Bölme işlemi
sol, sağ = np.hsplit(m, [2])
sol
sağ
```
### Array Sıralama İşlemleri
```
v = np.array([2,1,4,3,5])
v
np.sort(v)
# Küçülten büyüğe sıralama arrayin orijinali bozulmaz
v
v.sort()
v # Arrayimizin ilk hali bozulur
v = np.array([2,1,4,3,5])
# v yi yeniden tanomladık
np.sort(v) # Yeniden sıraladık
i = np.argsort(v)
# Orijinal arrayde, sıralama sonrası değişiklikleri belirtir
i
v # arrayin orijinal hali
v[i]
```
.
.
.
.
.
# NumPy Eleman İşlemleri
### Index
##### 1 Boyutlu Array İşlemleri
```
import numpy as np
a = np.random.randint(0,10,10)
a
a[0]
a[-1]
a[-2]
a[0] = 1 # eleman değiştirme işlemi
a[0]
```
.
.
##### 2 Boyutlu Array İşlemleri
```
a = np.random.randint(10, size = (3,5))
a
```
```
a[1,3]
a[2,2]
a[2,2] = 2 # eleman değiştirme işlemi
a[2,2]
a[2,2] = 2.2
# Fixed Type özelliği nedeni ile tam sayı değerini alır
a[2,2]
```
.
.
### Slicing İle Array Alt Kümesine Erişmek
#### 1 Boyutlu Array
```
import numpy as np
a = np.arange(20,30)
a
a[0:3]
a[:3]
a[3:]
a[::2]
a[1::2]
```
.
.
#### 2 Boyutlu Array
```
a = np.random.randint(10, size = (5,5))
a
```
x[**satır_aralık_baslangici** : **satır_aralık_bitisi** , **sutun_aralık_baslangici** : **sutun_aralık_bitisi**]
##### Sütuna Erişmek
```
a[:,0] # 0.Sütun
a[:,1] # 1.Sütun
```
##### Satıra Erişmek
```
a[0,:] # 0.Satır
a[3] # 3.Satır
```
.
.
##### Satır ve Sütunlara Erişmek
x[**satır_aralık_baslangici** : **satır_aralık_bitisi** , **sutun_aralık_baslangici** : **sutun_aralık_bitisi**]
```
a
a[:2,:3]
a[0:2,0:3]
a[2:5,2:5]
a[:,:2]
a[1:3,:3]
```
#### Array Alt Kümelerini Bağımsızlaştırma
```
a = np.random.randint(10, size = (5,5))
a
alt_a = a[0:3,0:2]
alt_a # Arrayin alt kümesini "alt_a" değişkenine atadık
alt_a[0,0] = 9999 # Arrayin alt kümelerini değiştirdik
alt_a[1,1] = 9999
alt_a
a # Arrayin alt kümesini değiştirince orijinal hali de değişir
```
#### Copy Method
```
b = np.random.randint(10, size = (5,5))
b
alt_b = a[0:3,0:2].copy()
alt_b # .copy() methodu ile arrayin orijinal hali değişmez
alt_b[0,0] = 9999
alt_b[1,1] = 9999
alt_b # Arrayin orijinal hali değişmez
b
```
### Fancy Index ile Eleman İşlemleri
#### 1 Boyutlu Array
```
v = np.arange(0,30,3)
v
v[1]
v[3]
[v[1],v[3]]
al_getir = [1,3,5]
v[al_getir]
```
.
#### 2 Boyutlu Array
```
m = np.arange(9).reshape((3,3))
m
satir = np.array([0,1])
sutun = np.array([1,2])
m[satir, sutun]
m
m[0,[1,2]]
m[:, [1,2]]
```
**===================================================**
```
v = np.arange(10)
v
index = np.array([0,1,2])
v[index] = 99
v
v[[0,1,2]] = 54,76,32
v
```
### Koşullu Eleman İşlemleri
```
v = np.array([1,2,3,4,5])
v
v > 3
v <= 3
v == 3
v != 3
(2*v)
(v**2)
(v*2) == (v**2)
```
#### Ufunc
```
np.equal(3,v)
np.not_equal(3,v)
np.equal([0,1,3], np.arange(3))
v = np.random.randint(10, size = (3,3))
v
v > 5
np.sum(v > 5) # True sayısını buluruz.
(v > 3) & (v < 7)
np.sum((v > 3) & (v < 7)) # ve
np.sum((v > 3) | (v < 7)) # veya
np.sum((v > 4), axis = 1) # Satır bazında true sayısı
np.sum((v > 4), axis = 0) # Sütun bazında sayısı
```
#### all() & any()
```
v
np.all(v > 4)
# V'nin tüm elemanları 4'den büyük mü?
np.all(v > 4, axis = 1) # Satır bazında işlem.
np.all(v > 4, axis = 0) # Sütun bazında işlem.
np.all(v >= 0)
```
**======================================================================**
```
np.any(v > 4)
# Yanlızca 1 eleman True ise işlem için yeterlidir.
np.any(v > 4, axis = 1) # Satır bazında işlem.
np.any(v > 4, axis = 0) # Sütun bazında işlem.
```
#### Koşullar İle Elemanlara Erişmek
```
v = np.array([1,2,3,4,5])
v
v[0]
v > 3
v[v>3]
v[(v>1) & (v<5)]
```
# Matematiksel İşlemler
### Tek Boyutlu Arraylerde Matematiksel İşlemler
```
a = np.arange(5)
a
a - 1
a / 2
a * 5
a ** 2
a % 2
5*(a*9/2)
np.add(a,3) # Her bir elemana 3 ekler
np.subtract(a,2) # Her bir elemandan 2 çıkartır
np.multiply(a,2) # Her bir elemanı 2 ile çarpar
np.divide(a,2) # Her bir elemanı 2'ye böler
np.power(a,2) # Her bir elemanın karesini alır
```
==================================================================
```
a = np.arange(1,6)
a
np.add.reduce(a) # Elemanların hepsini toplar.
np.add.accumulate(a) # Elemanların hepsini aşamalı olarak toplar.
a = np.random.normal(0,1,30)
a
np.mean(a) # Ortalama getirir
np.std(a) # Standart sapma
np.var(a) # Varyans
np.median(a) # Median
np.min(a) # min değeri.
np.max(a) # max değeri.
```
### İki Boyutlu Arraylerde Matematiksel İşlemler
```
a = np.random.normal(0,1,(3,3))
a # YUKARIDAKİ İŞLEMLERİN AYNISI UYGULANABİLİR.
a.sum()
a.sum(axis = 1)
```
# Farklı Boyutlu Arrayler İle Birlikte Çalışmak (Broadcasting)
```
import numpy as np
a = np.array([1,2,3])
b = np.array([1,2,3])
a + b
m = np.ones((3,3))
m
a + m
```
=================================================
```
a = np.arange(3)
a
b = np.arange(3)[:,np.newaxis]
b
a + b
```
**Kural 1:** Eğer iki array boyut sayısı olarak birbirinden farklı ise boyutu az olanın boyutuna 1 eklenerek boyutu çoğaltılır.
**Kural 1:** Eğer eşleşmeyen boyut sayısı varsa 1 olan boyutu diğer arrayin boyutuna eşitlenir.
**Kural 3:** Halen uyuşmazlık varsa hata üretir.
```
m = np.ones((2,3))
m
a = np.arange(3)
a
```
**KURAL 1**
m: (2,3) **====>** m:(2,3)
a: (3,) **====>** a:(1,3)
**KURAL 2**
m: (2,3) **====>** m:(2,3)
a: (1,3) **====>** a:(2,3)
```
a + m
```
==============================================================
```
a = np.arange(3).reshape((3,1))
a
b = np.arange(3)
b
a.shape
b.shape
a + b
```
==============================================================
```
m = np.ones((3,2))
m
a = np.arange(3)
a
m.shape
a.shape
a + m # Kural 3: Hata
```
# Numpy Yapısal Array'ler
```
isim = ["ali","veli","isik"]
yas = [25,22,19]
boy = [168,159,172]
data = np.zeros(3, dtype = {"names" : ("isim","yas","boy"),
"formats" : ("U10","i4","f8")})
data
data["isim"] = isim
data["yas"] = yas
data["boy"] = boy
data
data["isim"]
data[0]
data[data["yas"] < 25]["isim"]
```
|
github_jupyter
|
# NumPy kullanılmadan yapılan bir işlem.
a = [1,2,3,4]
b = [2,3,4,5]
ab = list()
for i in range(0,len(a)):
ab.append(a[i]*b[i])
print(ab)
# Aynı işlmei Numpy da yaparsak.
import numpy as np
a = np.array([1,2,3,4])
b = np.array([2,3,4,5])
a*b
x = 9
who
x # Python da x e atanana değer 4 farklı satır ile tanımlanır.
from IPython.display import Image
path = "/Users/ilgar/OneDrive\Programlama/Python A-Z - Veri Bilimi ve Machine Learning/Python Programlama/JupyterLab/Veri Manipulasyonu/img/"
Image(filename = path + "cint_vs_pyint.png", width=400, height=400)
L = [1,2,"a",1.2]
L
# Python da liste oluşturmak çok maliyetli bir işlemdir.
# Liste yerine NumPy Array oluşturusak belli kısıtlamalar gelmesine rağmen daha performanslı çalışır.
[type(i) for i in L]
from IPython.display import Image
path = "/Users/ilgar/OneDrive/Programlama/Python A-Z - Veri Bilimi ve Machine Learning/Python Programlama/JupyterLab/Veri Manipulasyonu/img/"
Image(filename = path + "array_vs_list.png", width=500, height=500)
import numpy as np # numpy'ı dahil ediyoruz
a = np.array([1,2,3,4,5]) # NumPy array oluşturduk
type(a) # tipini sorguladık
np.array([3, 4.5, 3, 12.5, 7]) # Fixed Type ==> Sabit Tip
np.array([3, 4.5, 3, 12.5, 7], dtype = "int") # Kendimiz veri tipi tanımlayabiliriz "dtype" ile
np.zeros(10, dtype = "int")
# Sıfırlardan oluşan int değerinde bir array.
np.zeros(10, dtype = "str")
# Sıfır değerlerinden oluşan string değerine sahip bir array
np.ones(10) # 10 tane 1 den oluşur
np.ones((2,4)) # 2 tane 4'lük yapıdan oluşur. Bu bir Matrix dir.
np.full((2,3), 5)
np.arange(0,10, 2)
np.linspace(0,1,30)
np.random.normal(0,1, (3,4))
np.random.randint(0, 10, (2,2))
np.eye(4)
import numpy as np
a = np.random.randint(1, 10, 10)
a
a.ndim # 1 Boyutlu,
a.shape # (10,) Boyutundan oluşan,
a.size # 10 elemanlı,
a.dtype # Integer tipinde bir array.
b = np.random.randint(0,12, (4,3))
b
b.ndim # 2 Boyutlu,
b.shape # (4,3) Boyutundan oluşan,
b.size # 12 elemanlı,
b.dtype # Integer tipinde olan bir array.
c = np.random.randint(1,20, size = (4,3,5))
c
c.ndim # 3 Boyutlu
c.shape # (4,3,5) boyutundan oluşan,
c.size # 60 elemanlı
c.dtype # Integer tipinde bir array
np.arange(1,17).reshape((4,4))
# 1,2,3,4,5,...,14,15,16 elemanlarından oluşan 1 Boyutlu arrayi,
# (4,4) lük iki boyutlu bir arraya çevirdik
x = np.array([1,2,3])
x # 1 Boyutlu array.
y = x.reshape((1,3)) # 2 Boyutlu arraye çevirdik
y
y.ndim
x[np.newaxis, :] # Alternatif Yöntem # Satır vektörü
x[: ,np.newaxis] # Sütun Vektörü
x = np.array([1,2,3])
y = np.array([4,5,6])
np.concatenate([x,y])
my_list = [7,8,9]
np.concatenate([x,y,my_list])
# Liste kullanarak da arraye eleman eklenebilir
a = np.array([[1,2,3],
[4,5,6]])
np.concatenate([a,a])
np.concatenate([a,a], axis = 1)
# Birinci ile birinci, ikinci ile ikinci birleşti.
a = np.array([1,2,3])
b = np.array([[9,8,7],
[6,5,4]])
np.vstack([a,b]) # Dikey birleştirme
a = np.array([[99],
[99]])
np.hstack([a,b]) # Yatay birleştirme
x = [1,2,3,99,99,3,2,1]
x
np.split(x, [3,5]) # n indeks ise n+1 bölüme ayırma işlemi.
a, b, c = np.split(x, [3,5]) # Arrayleri değişik değişkenlere atar.
a
b
c
m = np.arange(16).reshape((4,4))
m # 2.satırdan itibaren böl
np.vsplit(m,[2])
# Dikey bölme işlemi gerçekleştiriyoruz
ust, alt = np.vsplit(m,[2])
ust
alt
m
np.hsplit(m, [2])
# Yatay Bölme işlemi
sol, sağ = np.hsplit(m, [2])
sol
sağ
v = np.array([2,1,4,3,5])
v
np.sort(v)
# Küçülten büyüğe sıralama arrayin orijinali bozulmaz
v
v.sort()
v # Arrayimizin ilk hali bozulur
v = np.array([2,1,4,3,5])
# v yi yeniden tanomladık
np.sort(v) # Yeniden sıraladık
i = np.argsort(v)
# Orijinal arrayde, sıralama sonrası değişiklikleri belirtir
i
v # arrayin orijinal hali
v[i]
import numpy as np
a = np.random.randint(0,10,10)
a
a[0]
a[-1]
a[-2]
a[0] = 1 # eleman değiştirme işlemi
a[0]
a = np.random.randint(10, size = (3,5))
a
a[1,3]
a[2,2]
a[2,2] = 2 # eleman değiştirme işlemi
a[2,2]
a[2,2] = 2.2
# Fixed Type özelliği nedeni ile tam sayı değerini alır
a[2,2]
import numpy as np
a = np.arange(20,30)
a
a[0:3]
a[:3]
a[3:]
a[::2]
a[1::2]
a = np.random.randint(10, size = (5,5))
a
a[:,0] # 0.Sütun
a[:,1] # 1.Sütun
a[0,:] # 0.Satır
a[3] # 3.Satır
a
a[:2,:3]
a[0:2,0:3]
a[2:5,2:5]
a[:,:2]
a[1:3,:3]
a = np.random.randint(10, size = (5,5))
a
alt_a = a[0:3,0:2]
alt_a # Arrayin alt kümesini "alt_a" değişkenine atadık
alt_a[0,0] = 9999 # Arrayin alt kümelerini değiştirdik
alt_a[1,1] = 9999
alt_a
a # Arrayin alt kümesini değiştirince orijinal hali de değişir
b = np.random.randint(10, size = (5,5))
b
alt_b = a[0:3,0:2].copy()
alt_b # .copy() methodu ile arrayin orijinal hali değişmez
alt_b[0,0] = 9999
alt_b[1,1] = 9999
alt_b # Arrayin orijinal hali değişmez
b
v = np.arange(0,30,3)
v
v[1]
v[3]
[v[1],v[3]]
al_getir = [1,3,5]
v[al_getir]
m = np.arange(9).reshape((3,3))
m
satir = np.array([0,1])
sutun = np.array([1,2])
m[satir, sutun]
m
m[0,[1,2]]
m[:, [1,2]]
v = np.arange(10)
v
index = np.array([0,1,2])
v[index] = 99
v
v[[0,1,2]] = 54,76,32
v
v = np.array([1,2,3,4,5])
v
v > 3
v <= 3
v == 3
v != 3
(2*v)
(v**2)
(v*2) == (v**2)
np.equal(3,v)
np.not_equal(3,v)
np.equal([0,1,3], np.arange(3))
v = np.random.randint(10, size = (3,3))
v
v > 5
np.sum(v > 5) # True sayısını buluruz.
(v > 3) & (v < 7)
np.sum((v > 3) & (v < 7)) # ve
np.sum((v > 3) | (v < 7)) # veya
np.sum((v > 4), axis = 1) # Satır bazında true sayısı
np.sum((v > 4), axis = 0) # Sütun bazında sayısı
v
np.all(v > 4)
# V'nin tüm elemanları 4'den büyük mü?
np.all(v > 4, axis = 1) # Satır bazında işlem.
np.all(v > 4, axis = 0) # Sütun bazında işlem.
np.all(v >= 0)
np.any(v > 4)
# Yanlızca 1 eleman True ise işlem için yeterlidir.
np.any(v > 4, axis = 1) # Satır bazında işlem.
np.any(v > 4, axis = 0) # Sütun bazında işlem.
v = np.array([1,2,3,4,5])
v
v[0]
v > 3
v[v>3]
v[(v>1) & (v<5)]
a = np.arange(5)
a
a - 1
a / 2
a * 5
a ** 2
a % 2
5*(a*9/2)
np.add(a,3) # Her bir elemana 3 ekler
np.subtract(a,2) # Her bir elemandan 2 çıkartır
np.multiply(a,2) # Her bir elemanı 2 ile çarpar
np.divide(a,2) # Her bir elemanı 2'ye böler
np.power(a,2) # Her bir elemanın karesini alır
a = np.arange(1,6)
a
np.add.reduce(a) # Elemanların hepsini toplar.
np.add.accumulate(a) # Elemanların hepsini aşamalı olarak toplar.
a = np.random.normal(0,1,30)
a
np.mean(a) # Ortalama getirir
np.std(a) # Standart sapma
np.var(a) # Varyans
np.median(a) # Median
np.min(a) # min değeri.
np.max(a) # max değeri.
a = np.random.normal(0,1,(3,3))
a # YUKARIDAKİ İŞLEMLERİN AYNISI UYGULANABİLİR.
a.sum()
a.sum(axis = 1)
import numpy as np
a = np.array([1,2,3])
b = np.array([1,2,3])
a + b
m = np.ones((3,3))
m
a + m
a = np.arange(3)
a
b = np.arange(3)[:,np.newaxis]
b
a + b
m = np.ones((2,3))
m
a = np.arange(3)
a
a + m
a = np.arange(3).reshape((3,1))
a
b = np.arange(3)
b
a.shape
b.shape
a + b
m = np.ones((3,2))
m
a = np.arange(3)
a
m.shape
a.shape
a + m # Kural 3: Hata
isim = ["ali","veli","isik"]
yas = [25,22,19]
boy = [168,159,172]
data = np.zeros(3, dtype = {"names" : ("isim","yas","boy"),
"formats" : ("U10","i4","f8")})
data
data["isim"] = isim
data["yas"] = yas
data["boy"] = boy
data
data["isim"]
data[0]
data[data["yas"] < 25]["isim"]
| 0.088539 | 0.93276 |
# Inference in a Propositional Knowledge Base
This Jupyter notebook demonstrates how to create a knowledge base for propositional logic and how to apply different inference algorithms to ask questions to the knowledge base.
Import modules:
```
from utils import *
from logic import *
from notebook import psource
```
## Wumpus World Knowledge Base
We first construct the knowledge base for the Wumpus World example from the lecture. The proposition symbols are:
<br/>
$P_{y, x}$ is true if there is a pit at position [y,x].<br/>
$B_{y, x}$ is true if the agent senses breeze at position [y,x].<br/>
We create all required proposition symbols with the `symbols` function:
```
(P11,P12,P13,P21,P22,P31,B11,B12,B21) = symbols('P11, P12, P13, P21, P22, P31, B11, B12, B21')
```
Next, we create an empty propositional knowledge base by calling the constructor of the class `PropKB`:
```
wumpus_kb = PropKB()
```
Now we can add all the knowledge we have about the Wumpus World environment to our knowledge base:
There is no pit in `[1,1]`.
```
wumpus_kb.tell(~P11)
```
A square is breezy if and only if there is a pit in a neighboring square. This has to be stated for each square but for now, we include just the relevant squares.
```
wumpus_kb.tell(B11 | '<=>' | ((P12 | P21)))
wumpus_kb.tell(B21 | '<=>' | ((P11 | P22 | P31)))
```
Finally, we include the breeze percepts for the first two squares:
```
wumpus_kb.tell(~B11)
wumpus_kb.tell(B21)
```
We can check the clauses stored in a knowledge base by accessing its `clauses` property:
```
wumpus_kb.clauses
```
We see that some equivalences automatically got converted into two implications which afterwards got converted to Conjunctive Normal Form (CNF).
### Inference by Enumeration
Given our knowledge base descibing the Wumpus World environment we now want to infere if the adjacent squares are safe. To solve this problem, we can use logical inference. The inference algorithm that is considered first is *Truth Table Enumeration*. To deside if a sentence $\alpha$ is entailed by the knowledge base, *Truth Table Enumeration* enumerates all models and checks if $\alpha$ is true for all models where the knowledge base is true. The algorithm is implemented in the function `tt_entails`:
```
psource(tt_entails)
```
Internally, the function `tt_entails` calls the function `tt_check_all` to check if the sentence $\alpha$ and the knowledge base are true for a certain model:
```
psource(tt_check_all)
```
Before we apply *Truth Table Enumeration* to our Wumpus World knowledge base we demonstrate the usage of the function `tt_entails` on some simple examples. Given two proposition symbols $P$ and $Q$, let us consider the knowledge base $KB = P \wedge Q$ and the sentence $\alpha = Q$. Since $P \wedge Q$ can only become true if $Q$ is true, the knowledge base obviously entails the sentence $\alpha$:
```
(P,Q) = symbols('P, Q')
tt_entails(P & Q, Q)
```
If we instead consider the knowledge base $KB = P \vee Q$, the sentence $\alpha$ is not entailed by the knowledge base:
```
tt_entails(P | Q, Q)
```
Now let's come back to our Wumpus World problem. The function `tt_entails()` takes an object of the class `Expr` which is a conjunction of clauses as the input instead of the `KB` itself. Instead of manually converting our knowledge base we can use the `ask_if_true` method of the `PropKB` class which does all the required conversions automatically. Let's check if there is a pit in the square [1,2]:
```
wumpus_kb.ask_if_true(~P12)
```
### Proof by Resolution
One alternative to the computational expensive *Truth Table Enumeration* is to proof the entailment using resolution. The algorithm that implements proof by resolution requires a knowledge base in Conjunctive Normal Form (CNF). We can apply the function `to_cnf` to convert logical sentences to CNF:
```
psource(to_cnf)
```
Here some examples that demsonstrate the automated conversion to CNF:
```
(A,B,C,D) = symbols('A, B, C, D')
to_cnf(A |'<=>'| B)
to_cnf((A |'<=>'| ~B) |'==>'| (C | ~D))
```
The resolution algorithm, which is implemented by the function `pl_resolution`, internally utilizes the function `to_cnf` to convert the knowledge base to CNF:
```
psource(pl_resolution)
```
Let's come back to our Wumpus World example. As done in the lecture slides, we first add some additional facts to our knowledge base:
There is no breeze in square [1,2]:
```
wumpus_kb.tell(~B12)
```
A square is breezy if and only if there is a pit in a neighboring square:
```
wumpus_kb.tell(B12 | '<=>' | ((P11 | P22 | P13)))
```
Now we apply the resolution algorithm to check if there is a pit in the square [3,1]:
```
pl_resolution(wumpus_kb, P31)
```
|
github_jupyter
|
from utils import *
from logic import *
from notebook import psource
(P11,P12,P13,P21,P22,P31,B11,B12,B21) = symbols('P11, P12, P13, P21, P22, P31, B11, B12, B21')
wumpus_kb = PropKB()
wumpus_kb.tell(~P11)
wumpus_kb.tell(B11 | '<=>' | ((P12 | P21)))
wumpus_kb.tell(B21 | '<=>' | ((P11 | P22 | P31)))
wumpus_kb.tell(~B11)
wumpus_kb.tell(B21)
wumpus_kb.clauses
psource(tt_entails)
psource(tt_check_all)
(P,Q) = symbols('P, Q')
tt_entails(P & Q, Q)
tt_entails(P | Q, Q)
wumpus_kb.ask_if_true(~P12)
psource(to_cnf)
(A,B,C,D) = symbols('A, B, C, D')
to_cnf(A |'<=>'| B)
to_cnf((A |'<=>'| ~B) |'==>'| (C | ~D))
psource(pl_resolution)
wumpus_kb.tell(~B12)
wumpus_kb.tell(B12 | '<=>' | ((P11 | P22 | P13)))
pl_resolution(wumpus_kb, P31)
| 0.291989 | 0.984381 |
```
%matplotlib inline
%config InlineBackend.figure_format='retina'
%load_ext autoreload
%autoreload 2
import numpy as np
import matplotlib.pyplot as plt
import h5py
import os
from sample_generators import CustomArgumentParser, TimeSeries
from sample_generation_tools import get_psd, get_waveforms_as_dataframe, apply_psd
```
## Read in the raw strain data
```
# -------------------------------------------------------------------------
# Read in the real strain data from the LIGO website
# -------------------------------------------------------------------------
event = 'GW150914'
data_path = '../data/'
# Names of the files containing the real strains, i.e. detector recordings
real_strain_file = {'H1': '{}_H1_STRAIN_4096.h5'.format(event),
'L1': '{}_L1_STRAIN_4096.h5'.format(event)}
# Read the HDF files into numpy arrays and store them in a dict
real_strains = dict()
for ifo in ['H1', 'L1']:
# Make the full path for the strain file
strain_path = os.path.join(data_path, 'strain', real_strain_file[ifo])
# Read the HDF file into a numpy array
with h5py.File(strain_path, 'r') as file:
real_strains[ifo] = np.array(file['strain/Strain'])
```
## Calculate the Power Spectral Densities
```
# -------------------------------------------------------------------------
# Pre-calculate the Power Spectral Density from the real strain data
# -------------------------------------------------------------------------
psds = dict()
psds['H1'] = get_psd(real_strains['H1'])
psds['L1'] = get_psd(real_strains['L1'])
```
## Calculate the Standard Deviation of the Whitened Strain
```
white_strain = dict()
white_strain['H1'] = apply_psd(real_strains['H1'], psds['H1'])
white_strain['L1'] = apply_psd(real_strains['L1'], psds['L1'])
white_strain_std = {'H1':np.std(white_strain['H1']), 'L1':np.std(white_strain['L1'])}
```
## Load Pre-Computed Waveforms
```
# -------------------------------------------------------------------------
# Load the pre-calculated waveforms from an HDF file into a DataFrame
# -------------------------------------------------------------------------
waveforms_file = 'waveforms_3s_0700_1200_training.h5'
waveforms_path = os.path.join(data_path, 'waveforms', waveforms_file)
waveforms = get_waveforms_as_dataframe(waveforms_path)
```
## Create a sample timeseries
```
np.random.seed(423)
sample = TimeSeries(sample_length=12,
sampling_rate=4096,
max_n_injections=2,
loudness=1.0,
noise_type='real',
pad=3.0,
waveforms=waveforms,
psds=psds,
real_strains=real_strains,
white_strain_std=white_strain_std,
max_delta_t=0.01,
event_position=2048)
timeseries_H1 = sample.get_timeseries()[0, :, 0]
timeseries_L1 = sample.get_timeseries()[0, :, 1]
signals_H1 = sample.signals['H1']
signals_L1 = sample.signals['L1']
labels = sample.get_label()
chirpmasses = sample.get_chirpmass()
distances = sample.get_distance()
snrs = sample.get_snr()
print(sample.delta_t)
print(snrs)
grid = np.linspace(0, 12, 12*2048)
# For poster
# fig, axes = plt.subplots(nrows=4, ncols=1, sharex='col',
# gridspec_kw={'height_ratios': [5, 5, 2, 2]},
# figsize=(17.52, 6.1))
# For paper:
fig, axes = plt.subplots(nrows=4, ncols=1, sharex='col',
gridspec_kw={'height_ratios': [4, 4, 2.25, 2.25]},
figsize=(4*4.50461, 4*1.235817933))
axes[0].plot(grid, timeseries_H1, color='C0')
axes[0].plot(grid, signals_H1, color='C1')
axes[0].plot(grid, [0 for _ in grid], color='Black', lw=0.75, ls=':')
axes[0].set_ylim(-6, 6)
axes[0].set_yticklabels(['', -4, -2, 0, 2, 4, ''])
axes[0].set_ylabel('Strain H1', fontsize=11)
axes[0].annotate("Coalescence", xy=(8.495, 3), xycoords='data', va='center', ha='center', xytext=(8.495, 5), textcoords='data', arrowprops=dict(arrowstyle='->'))
axes[0].annotate("Inspiral", xy=(8.385, -0.25), xycoords='data', va='center', ha='center', xytext=(8.085, -5), textcoords='data', arrowprops=dict(arrowstyle='->'))
axes[0].annotate("Ringdown", xy=(8.555, -0.25), xycoords='data', va='center', ha='center', xytext=(8.885, -5), textcoords='data', arrowprops=dict(arrowstyle='->'))
axes[1].plot(grid, timeseries_L1, color='C0')
axes[1].plot(grid, signals_L1, color='C1')
axes[1].set_ylim(-6, 6)
axes[1].plot(grid, [0 for _ in grid], color='Black', lw=0.75, ls=':')
axes[1].set_yticklabels(['', -4, -2, 0, 2, 4, ''])
axes[1].set_ylabel('Strain L1', fontsize=11)
# Calculate and plot the fuzzy-zones
THRESHOLD = 0.5
fuzzy_zones = -1 * np.ones(len(labels))
for j in range(len(labels)):
if 0.5 < labels[j] < 0.6:
fuzzy_zones[j] = 1
axes[2].fill_between(grid, -10, 10*fuzzy_zones, color='Gray', alpha=0.25, lw=0)
axes[2].plot(grid, labels, color='C2', lw=2)
axes[2].plot(grid, [THRESHOLD for _ in grid], color='Gray', lw=1, ls='--', )
axes[2].set_ylim(-0.3*np.max(labels), 1.3*np.max(labels))
axes[2].set_ylabel('Normed\nSignal\nEnvelope', fontsize=11)
axes[2].text(6, 0.65, 'Threshold at 0.5 to get FWHM interval', ha='center', va='center', color='Gray')
axes[3].plot(grid, [1 if _ > 0 else 0 for _ in labels], color='C3', lw=2, ls=':', alpha=0.5, label='Naive label')
axes[3].plot(grid, [1 if _ > THRESHOLD else 0 for _ in labels], color='C3', lw=2, label='FWHM label')
axes[3].fill_between(grid, -2, 2*fuzzy_zones, color='Gray', alpha=0.5, lw=0)
axes[3].set_ylim(-0.3, 1.3)
axes[3].annotate("Fuzzy Zone", xy=(8.485, 0.5), xycoords='data', va='center', ha='center', xytext=(7.65, 0.5), textcoords='data', color='Gray', arrowprops=dict(arrowstyle='->', color='Gray'))
axes[3].set_ylabel('Label', fontsize=11)
axes[3].legend(loc='upper right')
fig.subplots_adjust(hspace=0)
plt.setp([a.get_xticklabels() for a in fig.axes[:-1]], visible=False)
plt.xlim(0, 12)
plt.xticks(np.linspace(0, 12, 25))
plt.xlabel('Time (s)', fontsize=11)
plt.savefig('training_sample.png', dpi=600, bbox_inches='tight')
plt.show()
```
|
github_jupyter
|
%matplotlib inline
%config InlineBackend.figure_format='retina'
%load_ext autoreload
%autoreload 2
import numpy as np
import matplotlib.pyplot as plt
import h5py
import os
from sample_generators import CustomArgumentParser, TimeSeries
from sample_generation_tools import get_psd, get_waveforms_as_dataframe, apply_psd
# -------------------------------------------------------------------------
# Read in the real strain data from the LIGO website
# -------------------------------------------------------------------------
event = 'GW150914'
data_path = '../data/'
# Names of the files containing the real strains, i.e. detector recordings
real_strain_file = {'H1': '{}_H1_STRAIN_4096.h5'.format(event),
'L1': '{}_L1_STRAIN_4096.h5'.format(event)}
# Read the HDF files into numpy arrays and store them in a dict
real_strains = dict()
for ifo in ['H1', 'L1']:
# Make the full path for the strain file
strain_path = os.path.join(data_path, 'strain', real_strain_file[ifo])
# Read the HDF file into a numpy array
with h5py.File(strain_path, 'r') as file:
real_strains[ifo] = np.array(file['strain/Strain'])
# -------------------------------------------------------------------------
# Pre-calculate the Power Spectral Density from the real strain data
# -------------------------------------------------------------------------
psds = dict()
psds['H1'] = get_psd(real_strains['H1'])
psds['L1'] = get_psd(real_strains['L1'])
white_strain = dict()
white_strain['H1'] = apply_psd(real_strains['H1'], psds['H1'])
white_strain['L1'] = apply_psd(real_strains['L1'], psds['L1'])
white_strain_std = {'H1':np.std(white_strain['H1']), 'L1':np.std(white_strain['L1'])}
# -------------------------------------------------------------------------
# Load the pre-calculated waveforms from an HDF file into a DataFrame
# -------------------------------------------------------------------------
waveforms_file = 'waveforms_3s_0700_1200_training.h5'
waveforms_path = os.path.join(data_path, 'waveforms', waveforms_file)
waveforms = get_waveforms_as_dataframe(waveforms_path)
np.random.seed(423)
sample = TimeSeries(sample_length=12,
sampling_rate=4096,
max_n_injections=2,
loudness=1.0,
noise_type='real',
pad=3.0,
waveforms=waveforms,
psds=psds,
real_strains=real_strains,
white_strain_std=white_strain_std,
max_delta_t=0.01,
event_position=2048)
timeseries_H1 = sample.get_timeseries()[0, :, 0]
timeseries_L1 = sample.get_timeseries()[0, :, 1]
signals_H1 = sample.signals['H1']
signals_L1 = sample.signals['L1']
labels = sample.get_label()
chirpmasses = sample.get_chirpmass()
distances = sample.get_distance()
snrs = sample.get_snr()
print(sample.delta_t)
print(snrs)
grid = np.linspace(0, 12, 12*2048)
# For poster
# fig, axes = plt.subplots(nrows=4, ncols=1, sharex='col',
# gridspec_kw={'height_ratios': [5, 5, 2, 2]},
# figsize=(17.52, 6.1))
# For paper:
fig, axes = plt.subplots(nrows=4, ncols=1, sharex='col',
gridspec_kw={'height_ratios': [4, 4, 2.25, 2.25]},
figsize=(4*4.50461, 4*1.235817933))
axes[0].plot(grid, timeseries_H1, color='C0')
axes[0].plot(grid, signals_H1, color='C1')
axes[0].plot(grid, [0 for _ in grid], color='Black', lw=0.75, ls=':')
axes[0].set_ylim(-6, 6)
axes[0].set_yticklabels(['', -4, -2, 0, 2, 4, ''])
axes[0].set_ylabel('Strain H1', fontsize=11)
axes[0].annotate("Coalescence", xy=(8.495, 3), xycoords='data', va='center', ha='center', xytext=(8.495, 5), textcoords='data', arrowprops=dict(arrowstyle='->'))
axes[0].annotate("Inspiral", xy=(8.385, -0.25), xycoords='data', va='center', ha='center', xytext=(8.085, -5), textcoords='data', arrowprops=dict(arrowstyle='->'))
axes[0].annotate("Ringdown", xy=(8.555, -0.25), xycoords='data', va='center', ha='center', xytext=(8.885, -5), textcoords='data', arrowprops=dict(arrowstyle='->'))
axes[1].plot(grid, timeseries_L1, color='C0')
axes[1].plot(grid, signals_L1, color='C1')
axes[1].set_ylim(-6, 6)
axes[1].plot(grid, [0 for _ in grid], color='Black', lw=0.75, ls=':')
axes[1].set_yticklabels(['', -4, -2, 0, 2, 4, ''])
axes[1].set_ylabel('Strain L1', fontsize=11)
# Calculate and plot the fuzzy-zones
THRESHOLD = 0.5
fuzzy_zones = -1 * np.ones(len(labels))
for j in range(len(labels)):
if 0.5 < labels[j] < 0.6:
fuzzy_zones[j] = 1
axes[2].fill_between(grid, -10, 10*fuzzy_zones, color='Gray', alpha=0.25, lw=0)
axes[2].plot(grid, labels, color='C2', lw=2)
axes[2].plot(grid, [THRESHOLD for _ in grid], color='Gray', lw=1, ls='--', )
axes[2].set_ylim(-0.3*np.max(labels), 1.3*np.max(labels))
axes[2].set_ylabel('Normed\nSignal\nEnvelope', fontsize=11)
axes[2].text(6, 0.65, 'Threshold at 0.5 to get FWHM interval', ha='center', va='center', color='Gray')
axes[3].plot(grid, [1 if _ > 0 else 0 for _ in labels], color='C3', lw=2, ls=':', alpha=0.5, label='Naive label')
axes[3].plot(grid, [1 if _ > THRESHOLD else 0 for _ in labels], color='C3', lw=2, label='FWHM label')
axes[3].fill_between(grid, -2, 2*fuzzy_zones, color='Gray', alpha=0.5, lw=0)
axes[3].set_ylim(-0.3, 1.3)
axes[3].annotate("Fuzzy Zone", xy=(8.485, 0.5), xycoords='data', va='center', ha='center', xytext=(7.65, 0.5), textcoords='data', color='Gray', arrowprops=dict(arrowstyle='->', color='Gray'))
axes[3].set_ylabel('Label', fontsize=11)
axes[3].legend(loc='upper right')
fig.subplots_adjust(hspace=0)
plt.setp([a.get_xticklabels() for a in fig.axes[:-1]], visible=False)
plt.xlim(0, 12)
plt.xticks(np.linspace(0, 12, 25))
plt.xlabel('Time (s)', fontsize=11)
plt.savefig('training_sample.png', dpi=600, bbox_inches='tight')
plt.show()
| 0.679072 | 0.87674 |
# Exploratory Analysis of San Diego County TMDL Measurements
This dataset, which contains multiple TDML measurement series, was extracted from CEDEN, and is stored in the [Data Library data repository](https://data.sandiegodata.org/dataset/water-quality-project-example-data/resource/4d8d1b40-a70f-450b-b1a2-2f7571643cbb). More information about the dataset is avialble at the [program overview website.](https://www.waterboards.ca.gov/sandiego/water_issues/programs/tmdls/).
The largest number of records are for Rainbow Creek, which [has had a long term monitoring and remediation project.](http://missionrcd.org/residential/rainbow-creek-watershed/)
```
import pandas as pd
import missingno as msno
df = pd.read_csv('http://ds.civicknowledge.org.s3.amazonaws.com/ceden.waterboards.ca.gov/CEDEN%20TDML.csv',
skiprows=2, low_memory=False)
df['SampleDate'] = pd.to_datetime(df.SampleDate) # Dates are often not automatically convered
df['Result'] = pd.to_numeric(df['Result'],errors='coerce') # Column has some non-numerics, so is recognized as a string
df.head().T
df.Program.value_counts()
# Names of all of the parent projects
df.ParentProject.value_counts()
df.Project.value_counts()
# Identifiers for the various measurements
df.Analyte.value_counts()
```
# Records Per Stations
The largest number of records are for Rainbow Creek, which [has had a long term monitoring and remediation project.](http://missionrcd.org/residential/rainbow-creek-watershed/)
```
df.StationName.value_counts().head(20)
len(df[df.StationName.str.contains('Rainbow Creek')])
```
# Record Coverage
In these stacked event charts, it is easy to see the Rainbow Creek records, which cover a much longer time range than other locations, but which are not measured as frequently.
```
def event_time_plot(df,yaxis='StationName'):
from datetime import datetime
import matplotlib.pyplot as plt
import matplotlib.dates as dt
_ = df[(df.SampleDate > pd.datetime(1980,1,1))].copy()
_['secs_since_epoch'] = (_.SampleDate - pd.datetime(1970,1,1))
fig = plt.figure(figsize=(10,10))
ax = fig.add_subplot(111)
ax = plt.plot(_.SampleDate, _[yaxis], marker='|', markersize=2, linestyle='None')
```
Here is the time coverage per project
```
event_time_plot(df, 'Project')
```
And a more detailed coverage per station.
```
event_time_plot(df)
```
Here is what just the Beaches and Creeks Bacteria TDML records look like.
```
bac = df[df.ParentProject == 'Bacteria TMDL 20 Beaches and Creeks'].copy()
event_time_plot(bac)
msno.matrix(bac.sample(1000)[bac.columns[:40]])
msno.matrix(bac.sample(1000)[bac.columns[40:]])
bac.SampleDate.min(), bac.SampleDate.max()
bac.Analyte.value_counts()
bac['SampleDays'] = (bac.SampleDate - bac.SampleDate.min()).dt.days
import seaborn as sns
import matplotlib.pyplot as plt
g = sns.lmplot('SampleDays', 'Result', data=bac[bac.Analyte == 'Enterococcus'],
hue='StationName', fit_reg=False, size=10)
g.set( yscale="log")
ax = plt.gca()
ax.set_title("Enterococcus measurements by time for all stations in Bacteria TMDL project")
plt.show()
```
|
github_jupyter
|
import pandas as pd
import missingno as msno
df = pd.read_csv('http://ds.civicknowledge.org.s3.amazonaws.com/ceden.waterboards.ca.gov/CEDEN%20TDML.csv',
skiprows=2, low_memory=False)
df['SampleDate'] = pd.to_datetime(df.SampleDate) # Dates are often not automatically convered
df['Result'] = pd.to_numeric(df['Result'],errors='coerce') # Column has some non-numerics, so is recognized as a string
df.head().T
df.Program.value_counts()
# Names of all of the parent projects
df.ParentProject.value_counts()
df.Project.value_counts()
# Identifiers for the various measurements
df.Analyte.value_counts()
df.StationName.value_counts().head(20)
len(df[df.StationName.str.contains('Rainbow Creek')])
def event_time_plot(df,yaxis='StationName'):
from datetime import datetime
import matplotlib.pyplot as plt
import matplotlib.dates as dt
_ = df[(df.SampleDate > pd.datetime(1980,1,1))].copy()
_['secs_since_epoch'] = (_.SampleDate - pd.datetime(1970,1,1))
fig = plt.figure(figsize=(10,10))
ax = fig.add_subplot(111)
ax = plt.plot(_.SampleDate, _[yaxis], marker='|', markersize=2, linestyle='None')
event_time_plot(df, 'Project')
event_time_plot(df)
bac = df[df.ParentProject == 'Bacteria TMDL 20 Beaches and Creeks'].copy()
event_time_plot(bac)
msno.matrix(bac.sample(1000)[bac.columns[:40]])
msno.matrix(bac.sample(1000)[bac.columns[40:]])
bac.SampleDate.min(), bac.SampleDate.max()
bac.Analyte.value_counts()
bac['SampleDays'] = (bac.SampleDate - bac.SampleDate.min()).dt.days
import seaborn as sns
import matplotlib.pyplot as plt
g = sns.lmplot('SampleDays', 'Result', data=bac[bac.Analyte == 'Enterococcus'],
hue='StationName', fit_reg=False, size=10)
g.set( yscale="log")
ax = plt.gca()
ax.set_title("Enterococcus measurements by time for all stations in Bacteria TMDL project")
plt.show()
| 0.49292 | 0.966124 |
Copyright (c) Microsoft Corporation. All rights reserved.
Licensed under the MIT License.

# Automated Machine Learning
_**Multilabel Text Classification Using AutoML NLP**_
## Contents
1. [Introduction](#Introduction)
1. [Setup](#Setup)
1. [Data](#Data)
1. [Train](#Train)
1. [Inference](#Inference)
## Introduction
This notebook demonstrates multilabel classification with text data using AutoML NLP.
AutoML highlights here include using end to end deep learning for NLP tasks like multilabel text classification.
Make sure you have executed the [configuration](../../../configuration.ipynb) before running this notebook.
Notebook synopsis:
1. Creating an Experiment in an existing Workspace
2. Configuration and remote run of AutoML for a multilabel text classification dataset from [Kaggle](www.kaggle.com), [arXiv Paper Abstracts](https://www.kaggle.com/spsayakpaul/arxiv-paper-abstracts).
3. Evaluating the trained model on a test set
## Setup
```
import logging
import numpy as np
import pandas as pd
import azureml.core
from azureml.core import Dataset
from azureml.core.experiment import Experiment
from azureml.core.workspace import Workspace
from azureml.core.dataset import Dataset
from azureml.core.compute import AmlCompute
from azureml.core.compute import ComputeTarget
from azureml.core.run import Run
from azureml.train.automl import AutoMLConfig
```
This sample notebook may use features that are not available in previous versions of the Azure ML SDK.
```
print("This notebook was created using version 1.39.0 of the Azure ML SDK")
print("You are currently using version", azureml.core.VERSION, "of the Azure ML SDK")
```
As part of the setup you have already created a <b>Workspace</b>. To run AutoML, you also need to create an <b>Experiment</b>. An Experiment corresponds to a prediction problem you are trying to solve, while a Run corresponds to a specific approach to the problem.
```
ws = Workspace.from_config()
# Choose an experiment name.
experiment_name = "automl-nlp-text-multilabel"
experiment = Experiment(ws, experiment_name)
output = {}
output["Subscription ID"] = ws.subscription_id
output["Workspace Name"] = ws.name
output["Resource Group"] = ws.resource_group
output["Location"] = ws.location
output["Experiment Name"] = experiment.name
pd.set_option("display.max_colwidth", None)
outputDf = pd.DataFrame(data=output, index=[""])
outputDf.T
```
## Set up a compute cluster
This section uses a user-provided compute cluster (named "dist-compute" in this example). If a cluster with this name does not exist in the user's workspace, the below code will create a new cluster. You can choose the parameters of the cluster as mentioned in the comments.
```
from azureml.core.compute import ComputeTarget, AmlCompute
from azureml.core.compute_target import ComputeTargetException
number_of_vms = 2 # change this number to your vm size
# Choose a name for your cluster.
amlcompute_cluster_name = "parallel-{}".format(number_of_vms)
# Verify that cluster does not exist already
try:
compute_target = ComputeTarget(workspace=ws, name=amlcompute_cluster_name)
print("Found existing cluster, use it.")
except ComputeTargetException:
compute_config = AmlCompute.provisioning_configuration(
vm_size="STANDARD_NC6", max_nodes=number_of_vms # Use GPU Nodes
)
compute_target = ComputeTarget.create(ws, amlcompute_cluster_name, compute_config)
compute_target.wait_for_completion(show_output=True)
```
## Data
Since the original dataset is very large, we leverage a subsampled dataset to allow for faster training for the purposes of running this example notebook. To run the full dataset (50K+ samples and 1k+ labels) you might need a GPU instance with larger memory and it may take longer to finish training.
To run the code below, please first download `arxiv_data.csv` from [this link](https://www.kaggle.com/spsayakpaul/arxiv-paper-abstracts) and save it under the same directory as this notebook, and then run `preprocessing.py` to create a subset of the data for training and evaluation
After this preprocessing for labels, we split the dataset into three parts: train, valid, and test. Now we register train and valid for training purpose. We will register the test part later.
```
# Upload dataset to datastore
data_dir = "data" # Local directory to store data
blobstore_datadir = data_dir # Blob store directory to store data in
datastore = ws.get_default_datastore()
datastore.upload(src_dir=data_dir, target_path=blobstore_datadir, overwrite=True)
# Obtain training data as a Tabular dataset to pass into AutoMLConfig
full_dataset = Dataset.Tabular.from_delimited_files(
path=[(datastore, blobstore_datadir + "/arxiv_abstract.csv")]
)
train_split, valid_split = full_dataset.random_split(percentage=0.2, seed=101)
valid_split, test_split = valid_split.random_split(percentage=0.5, seed=47)
train_dataset = train_split.register(
workspace=ws,
name="arxiv_abstract_train",
description="Multilabel train dataset",
create_new_version=True,
)
valid_dataset = valid_split.register(
workspace=ws,
name="arxiv_abstract_valid",
description="Multilabel validation dataset",
create_new_version=True,
)
```
# Train
## Submit AutoML run
Now we can start the run with the prepared compute resource and datasets. On a `STANDARD_NC6` compute instance with one node, the training would take around 25 minutes and evaluation on valid dataset would take around 10 minutes. Here, to make training faster, we will use a `STANDARD_NC6` instance with 2 nodes and enable parallel training.
To use distributed training, we need to set `enable_distributed_dnn_training = True` and `max_concurrent_iterations` to be the number of vms available in your cluster.
Here we do not set `primary_metric` parameter as we only train one model and we do not need to rank trained models. The run will use default primary metrics, `accuracy`. But it is only for reporting purpose.
```
automl_settings = {
"max_concurrent_iterations": number_of_vms,
"enable_distributed_dnn_training": True,
"verbosity": logging.INFO,
}
target_column_name = "terms"
automl_config = AutoMLConfig(
task="text-classification-multilabel",
debug_log="automl_errors.log",
compute_target=compute_target,
training_data=train_dataset,
validation_data=valid_dataset,
label_column_name=target_column_name,
**automl_settings,
)
automl_run = experiment.submit(
automl_config, show_output=False
) # You might see a warning about "enable_distributed_dnn_training". Please simply ignore.
_ = automl_run.wait_for_completion(show_output=False)
```
## Download Metrics
These metrics logged with the training run are computed with the trained model on validation dataset
```
validation_metrics = automl_run.get_metrics()
pd.DataFrame(
{"metric_name": validation_metrics.keys(), "value": validation_metrics.values()}
)
```
You can also get the best run id and the best model with `get_output` method.
```
(
best_run,
best_model,
) = (
automl_run.get_output()
) # You might see a warning about "enable_distributed_dnn_training". Please simply ignore.
best_run
```
# Inference
Now you can use the trained model to do inference on unseen data. We use a `ScriptRun` to do this, with script that we provide. The following blocks will register the test dataset, download the inference script and trigger the inference run. Our inference run do not directly log the metrics. So we need to download the results and calculate the metrics offline
## Submit Inference Run
```
test_dataset = test_split.register(
workspace=ws,
name="arxiv_abstract_test",
description="Multilabel text dataset",
create_new_version=True,
)
# Load training script run corresponding to AutoML run above.
training_run_id = automl_run.id + "_HD_0"
training_run = Run(experiment, training_run_id)
# Inference script run arguments
arguments = [
"--run_id",
training_run_id,
"--experiment_name",
experiment.name,
"--input_dataset_id",
test_dataset.as_named_input("test_data"),
]
import os
import tempfile
from azureml.core.script_run_config import ScriptRunConfig
scoring_args = arguments
with tempfile.TemporaryDirectory() as tmpdir:
# Download required files from training run into temp folder.
entry_script_name = "score_script.py"
output_path = os.path.join(tmpdir, entry_script_name)
training_run.download_file(
"outputs/" + entry_script_name, os.path.join(tmpdir, entry_script_name)
)
script_run_config = ScriptRunConfig(
source_directory=tmpdir,
script=entry_script_name,
compute_target=compute_target,
environment=training_run.get_environment(),
arguments=scoring_args,
)
scoring_run = experiment.submit(script_run_config)
scoring_run
_ = scoring_run.wait_for_completion(show_output=False)
```
## Download Prediction
```
output_prediction_file = "./preds_multilabel.csv"
scoring_run.download_file(
"outputs/predictions.csv", output_file_path=output_prediction_file
)
test_data_df = test_dataset.to_pandas_dataframe()
test_set_predictions_df = pd.read_csv("preds_multilabel.csv")
test_set_predictions_df["label_confidence"] = test_set_predictions_df[
"label_confidence"
].apply(lambda x: [float(num) for num in x.split(",")])
# install this package to run the following block
# !pip install azureml-automl-dnn-nlp
from azureml.automl.dnn.nlp.classification.io.read.read_utils import load_model_wrapper
y_transformer = load_model_wrapper(training_run).y_transformer
```
## Offline Evaluation
We will use the evaluation module within AzureML to calculate the metrics.
```
import ast
test_y = y_transformer.transform(
test_data_df[target_column_name].apply(ast.literal_eval)
).toarray()
from azureml.automl.runtime.shared.score.scoring import score_classification
from azureml.automl.runtime.shared import metrics
test_pred_probs = []
for i in range(test_set_predictions_df.shape[0]):
test_pred_probs.append(test_set_predictions_df.loc[i, "label_confidence"])
test_pred_probs = np.array(test_pred_probs)
L = len(y_transformer.classes_)
test_metrics = score_classification(
test_y,
test_pred_probs,
list(validation_metrics.keys()),
np.arange(L),
np.arange(L),
y_transformer=y_transformer,
multilabel=True,
)
pd.DataFrame({"metric_name": test_metrics.keys(), "value": test_metrics.values()})
```
## Classification Report
We also provide the following function, which enables you to evaluate the trained model, for each class and average among classes, with any value of threshold you would like
```
from sklearn.metrics import classification_report
def classification_report_multilabel(
test_df, pred_df, label_col, y_transformer, threshold=0.5
):
message = (
"test_df and pred_df should have the same number of rows, but get {} and {}"
)
assert test_df.shape[0] == pred_df.shape[0], message.format(
test_df.shape[0], pred_df.shape[0]
)
label_set = y_transformer.classes_
n = len(label_set)
y_true = []
y_pred = []
for row in range(test_df.shape[0]):
true_labels = y_transformer.transform(
[ast.literal_eval(test_df.loc[row, label_col])]
).toarray()[0]
pred_labels = pred_df.loc[row, "label_confidence"]
for ind, (label, prob) in enumerate(zip(true_labels, pred_labels)):
predict_positive = prob >= threshold
if label or predict_positive:
y_true.append(label_set[ind] if label else "")
y_pred.append(label_set[ind] if predict_positive else "")
print(classification_report(y_true, y_pred, label_set))
classification_report_multilabel(
test_data_df,
test_set_predictions_df,
target_column_name,
y_transformer,
threshold=0.1,
)
classification_report_multilabel(
test_data_df,
test_set_predictions_df,
target_column_name,
y_transformer,
threshold=0.5,
)
classification_report_multilabel(
test_data_df,
test_set_predictions_df,
target_column_name,
y_transformer,
threshold=0.9,
)
```
|
github_jupyter
|
import logging
import numpy as np
import pandas as pd
import azureml.core
from azureml.core import Dataset
from azureml.core.experiment import Experiment
from azureml.core.workspace import Workspace
from azureml.core.dataset import Dataset
from azureml.core.compute import AmlCompute
from azureml.core.compute import ComputeTarget
from azureml.core.run import Run
from azureml.train.automl import AutoMLConfig
print("This notebook was created using version 1.39.0 of the Azure ML SDK")
print("You are currently using version", azureml.core.VERSION, "of the Azure ML SDK")
ws = Workspace.from_config()
# Choose an experiment name.
experiment_name = "automl-nlp-text-multilabel"
experiment = Experiment(ws, experiment_name)
output = {}
output["Subscription ID"] = ws.subscription_id
output["Workspace Name"] = ws.name
output["Resource Group"] = ws.resource_group
output["Location"] = ws.location
output["Experiment Name"] = experiment.name
pd.set_option("display.max_colwidth", None)
outputDf = pd.DataFrame(data=output, index=[""])
outputDf.T
from azureml.core.compute import ComputeTarget, AmlCompute
from azureml.core.compute_target import ComputeTargetException
number_of_vms = 2 # change this number to your vm size
# Choose a name for your cluster.
amlcompute_cluster_name = "parallel-{}".format(number_of_vms)
# Verify that cluster does not exist already
try:
compute_target = ComputeTarget(workspace=ws, name=amlcompute_cluster_name)
print("Found existing cluster, use it.")
except ComputeTargetException:
compute_config = AmlCompute.provisioning_configuration(
vm_size="STANDARD_NC6", max_nodes=number_of_vms # Use GPU Nodes
)
compute_target = ComputeTarget.create(ws, amlcompute_cluster_name, compute_config)
compute_target.wait_for_completion(show_output=True)
# Upload dataset to datastore
data_dir = "data" # Local directory to store data
blobstore_datadir = data_dir # Blob store directory to store data in
datastore = ws.get_default_datastore()
datastore.upload(src_dir=data_dir, target_path=blobstore_datadir, overwrite=True)
# Obtain training data as a Tabular dataset to pass into AutoMLConfig
full_dataset = Dataset.Tabular.from_delimited_files(
path=[(datastore, blobstore_datadir + "/arxiv_abstract.csv")]
)
train_split, valid_split = full_dataset.random_split(percentage=0.2, seed=101)
valid_split, test_split = valid_split.random_split(percentage=0.5, seed=47)
train_dataset = train_split.register(
workspace=ws,
name="arxiv_abstract_train",
description="Multilabel train dataset",
create_new_version=True,
)
valid_dataset = valid_split.register(
workspace=ws,
name="arxiv_abstract_valid",
description="Multilabel validation dataset",
create_new_version=True,
)
automl_settings = {
"max_concurrent_iterations": number_of_vms,
"enable_distributed_dnn_training": True,
"verbosity": logging.INFO,
}
target_column_name = "terms"
automl_config = AutoMLConfig(
task="text-classification-multilabel",
debug_log="automl_errors.log",
compute_target=compute_target,
training_data=train_dataset,
validation_data=valid_dataset,
label_column_name=target_column_name,
**automl_settings,
)
automl_run = experiment.submit(
automl_config, show_output=False
) # You might see a warning about "enable_distributed_dnn_training". Please simply ignore.
_ = automl_run.wait_for_completion(show_output=False)
validation_metrics = automl_run.get_metrics()
pd.DataFrame(
{"metric_name": validation_metrics.keys(), "value": validation_metrics.values()}
)
(
best_run,
best_model,
) = (
automl_run.get_output()
) # You might see a warning about "enable_distributed_dnn_training". Please simply ignore.
best_run
test_dataset = test_split.register(
workspace=ws,
name="arxiv_abstract_test",
description="Multilabel text dataset",
create_new_version=True,
)
# Load training script run corresponding to AutoML run above.
training_run_id = automl_run.id + "_HD_0"
training_run = Run(experiment, training_run_id)
# Inference script run arguments
arguments = [
"--run_id",
training_run_id,
"--experiment_name",
experiment.name,
"--input_dataset_id",
test_dataset.as_named_input("test_data"),
]
import os
import tempfile
from azureml.core.script_run_config import ScriptRunConfig
scoring_args = arguments
with tempfile.TemporaryDirectory() as tmpdir:
# Download required files from training run into temp folder.
entry_script_name = "score_script.py"
output_path = os.path.join(tmpdir, entry_script_name)
training_run.download_file(
"outputs/" + entry_script_name, os.path.join(tmpdir, entry_script_name)
)
script_run_config = ScriptRunConfig(
source_directory=tmpdir,
script=entry_script_name,
compute_target=compute_target,
environment=training_run.get_environment(),
arguments=scoring_args,
)
scoring_run = experiment.submit(script_run_config)
scoring_run
_ = scoring_run.wait_for_completion(show_output=False)
output_prediction_file = "./preds_multilabel.csv"
scoring_run.download_file(
"outputs/predictions.csv", output_file_path=output_prediction_file
)
test_data_df = test_dataset.to_pandas_dataframe()
test_set_predictions_df = pd.read_csv("preds_multilabel.csv")
test_set_predictions_df["label_confidence"] = test_set_predictions_df[
"label_confidence"
].apply(lambda x: [float(num) for num in x.split(",")])
# install this package to run the following block
# !pip install azureml-automl-dnn-nlp
from azureml.automl.dnn.nlp.classification.io.read.read_utils import load_model_wrapper
y_transformer = load_model_wrapper(training_run).y_transformer
import ast
test_y = y_transformer.transform(
test_data_df[target_column_name].apply(ast.literal_eval)
).toarray()
from azureml.automl.runtime.shared.score.scoring import score_classification
from azureml.automl.runtime.shared import metrics
test_pred_probs = []
for i in range(test_set_predictions_df.shape[0]):
test_pred_probs.append(test_set_predictions_df.loc[i, "label_confidence"])
test_pred_probs = np.array(test_pred_probs)
L = len(y_transformer.classes_)
test_metrics = score_classification(
test_y,
test_pred_probs,
list(validation_metrics.keys()),
np.arange(L),
np.arange(L),
y_transformer=y_transformer,
multilabel=True,
)
pd.DataFrame({"metric_name": test_metrics.keys(), "value": test_metrics.values()})
from sklearn.metrics import classification_report
def classification_report_multilabel(
test_df, pred_df, label_col, y_transformer, threshold=0.5
):
message = (
"test_df and pred_df should have the same number of rows, but get {} and {}"
)
assert test_df.shape[0] == pred_df.shape[0], message.format(
test_df.shape[0], pred_df.shape[0]
)
label_set = y_transformer.classes_
n = len(label_set)
y_true = []
y_pred = []
for row in range(test_df.shape[0]):
true_labels = y_transformer.transform(
[ast.literal_eval(test_df.loc[row, label_col])]
).toarray()[0]
pred_labels = pred_df.loc[row, "label_confidence"]
for ind, (label, prob) in enumerate(zip(true_labels, pred_labels)):
predict_positive = prob >= threshold
if label or predict_positive:
y_true.append(label_set[ind] if label else "")
y_pred.append(label_set[ind] if predict_positive else "")
print(classification_report(y_true, y_pred, label_set))
classification_report_multilabel(
test_data_df,
test_set_predictions_df,
target_column_name,
y_transformer,
threshold=0.1,
)
classification_report_multilabel(
test_data_df,
test_set_predictions_df,
target_column_name,
y_transformer,
threshold=0.5,
)
classification_report_multilabel(
test_data_df,
test_set_predictions_df,
target_column_name,
y_transformer,
threshold=0.9,
)
| 0.639286 | 0.923764 |
# Heart attack Prediction using Machine Learning
In this notebook we are going to perform Exploratory Data Analysis and use various Machine Learning Models to predict whether the patient has heart disease or not depending on the values of various features. We will be using Bokeh and a little bit of Seaborn to plot the graphs.
Firstly we will import all necessary libraries
```
import numpy as np
import pandas as pd
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn import svm
import warnings
warnings.filterwarnings('ignore')
```
# About the Datasets
This notebook contains 4 databases concerning heart disease diagnosis. All attributes are numeric-valued. The data was collected from the four following locations:
1. Cleveland Clinic Foundation (cleveland.csv)
2. Hungarian Institute of Cardiology, Budapest (hungarian.csv)
3. V.A. Medical Center, Long Beach, CA (long-beach-va.csv)
4. University Hospital, Zurich, Switzerland (switzerland.csv)
Each database has the same instance format. While the databases have 76 raw attributes, only 14 of them are actually used. This database contains 76 attributes, but all published experiments refer to using a subset of 14 of them. In particular, the Cleveland database is the only one that has been used by ML researchers to this date. The "goal" field refers to the presence of heart disease in the patient. It is integer valued from 0 (no presence) to 4.
Experiments with the Cleveland database have concentrated on simply attempting to distinguish presence (values 1,2,3,4) from absence (value 0).
Number of Instances:
Database: Number of instances:
Cleveland: 303
Hungarian: 294
Switzerland: 123
Long Beach VA: 200
Number of Attributes: 76 (including the predicted attribute)
The authors of the databases have requested that any publications resulting from the use of the data include the names of the principal investigator responsible for the data collection at each institution. They would be:
a. Hungarian Institute of Cardiology. Budapest: Andras Janosi, M.D.
b. University Hospital, Zurich, Switzerland: William Steinbrunn, M.D.
c. University Hospital, Basel, Switzerland: Matthias Pfisterer, M.D.
d. V.A. Medical Center, Long Beach and Cleveland Clinic Foundation:
Robert Detrano, M.D., Ph.D.
The dataset consists of 303 individual data. There are 14 columns in the dataset, which are described below:
1. Age: displays the age of the individual.
2. Sex: displays the gender of the individual using the following format:
1 = male
0 = female
3. Cp (Chest-pain type): displays the type of chest-pain experienced by the
individual using the following format:
1 = typical angina
2 = atypical angina
3 = non-anginal pain
4 = asymptotic
4. TrestBPS (Resting Blood Pressure): Displays the resting blood pressure value of
an individual in mmHg (unit). It can take continuous values from 94 to 200.
5. Chol (Serum Cholesterol): Displays the serum cholesterol in mg/dl (unit)
6. Fbs (Fasting Blood Sugar): Compares the fasting blood sugar value of an
individual with 120mg/dl:
1 (true) = Fasting blood sugar > 120mg/dl
0 (False) = Fasting blood sugar < 120mg/dl
7. RestECG (Resting ECG): displays resting electrocardiographic results
0 = normal
1 = having ST-T wave abnormality
2 = left ventricular hypertrophy
8. Thalach (Max heart rate achieved): displays the max heart rate achieved by an
individual. It can take continuous value from 71 to 202.
9. Exang (Exercise induced angina): Angina is a type of chest pain caused by reduced
blood flow to the heart.
1 = yes
0 = no
10. OldPeak (ST depression induced by exercise relative to rest): displays the value
which is an integer or float.
11. Slope (Peak exercise ST segment):
1 = upsloping
2 = flat
3 = down sloping
12. Ca (Number of major vessels (0–3) colored by fluoroscopy): displays the value as
integer or float.
13. Thal: displays the thalassemia:
3 = normal
6 = fixed defect
7 = reversible defect
14. Target (Diagnosis of heart disease): Displays whether the individual is
suffering from heart disease or not:
0 = absence
1, 2, 3, 4 = present.
```
df0 = pd.read_csv('cleveland.csv')
df1 = pd.read_csv('hungarian.csv')
df2 = pd.read_csv('switzerland.csv')
df3 = pd.read_csv('va.csv')
df0.head()
df1.head()
df2.head()
df3.head()
df0_missing = df0.isna()
df0_missing.head()
df1_missing = df1.isna()
df1_missing.head()
df2_missing = df2.isna()
df2_missing.head()
df3_missing = df3.isna()
df3_missing.head()
df0_num_missing = df0_missing.sum()
df0_num_missing
df1_num_missing = df1_missing.sum()
df1_num_missing
df2_num_missing = df2_missing.sum()
df2_num_missing
df3_num_missing = df3_missing.sum()
df3_num_missing
df0.isna().mean().round(4) * 100
df1.isna().mean().round(4) * 100
df2.isna().mean().round(4) * 100
df3.isna().mean().round(4) * 100
df0.drop(['slope', 'ca','thal'], axis = 1,inplace=True)
df0.dropna(inplace=True, thresh=3)
df0.info()
df0.shape
df1.drop(['slope', 'ca','thal'], axis = 1,inplace=True)
df1.dropna(inplace=True, thresh=3)
df1.info()
df1.shape
df2.drop(['slope', 'ca','thal'], axis = 1,inplace=True)
df2.dropna(inplace=True, thresh=3)
df2.info()
df2.shape
df3.drop(['slope', 'ca','thal'], axis = 1,inplace=True)
df3.dropna(inplace=True, thresh=3)
df3.info()
df3.shape
df0.describe()
df1.describe()
df2.describe()
df3.describe()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn import svm
import warnings
warnings.filterwarnings('ignore')
df0 = pd.read_csv('cleveland.csv')
df1 = pd.read_csv('hungarian.csv')
df2 = pd.read_csv('switzerland.csv')
df3 = pd.read_csv('va.csv')
df0.head()
df1.head()
df2.head()
df3.head()
df0_missing = df0.isna()
df0_missing.head()
df1_missing = df1.isna()
df1_missing.head()
df2_missing = df2.isna()
df2_missing.head()
df3_missing = df3.isna()
df3_missing.head()
df0_num_missing = df0_missing.sum()
df0_num_missing
df1_num_missing = df1_missing.sum()
df1_num_missing
df2_num_missing = df2_missing.sum()
df2_num_missing
df3_num_missing = df3_missing.sum()
df3_num_missing
df0.isna().mean().round(4) * 100
df1.isna().mean().round(4) * 100
df2.isna().mean().round(4) * 100
df3.isna().mean().round(4) * 100
df0.drop(['slope', 'ca','thal'], axis = 1,inplace=True)
df0.dropna(inplace=True, thresh=3)
df0.info()
df0.shape
df1.drop(['slope', 'ca','thal'], axis = 1,inplace=True)
df1.dropna(inplace=True, thresh=3)
df1.info()
df1.shape
df2.drop(['slope', 'ca','thal'], axis = 1,inplace=True)
df2.dropna(inplace=True, thresh=3)
df2.info()
df2.shape
df3.drop(['slope', 'ca','thal'], axis = 1,inplace=True)
df3.dropna(inplace=True, thresh=3)
df3.info()
df3.shape
df0.describe()
df1.describe()
df2.describe()
df3.describe()
| 0.38827 | 0.978115 |
<div style="text-align: right"><i>Peter Norvig<br>April 2020</i></div>
# The Stable Matching Problem
The **[stable matching problem](https://en.wikipedia.org/wiki/Stable_marriage_problem#Algorithmic_solution)** involves two equally-sized disjoint sets of actors that want to pair off in a way that maximizes happiness. It could be a set of women and a set of men that want to pair off in heterosexual marriage; or a a set of job-seekers and a set of employers. Every year, there is a large-scale application of this problem in which:
- Graduating medical students state which hospitals they would prefer to be residents at.
- Hospitals in turn state which students they prefer.
- An algorithm finds a stable matching.
Each actor has **preferences** for who they would prefer to be matched with. In the default way of stating the problem, preferences are expressed as an **ordering**: each actor rates the possible matches on the other side from most preferred to least preferred. But we will go beyond that, allowing each actor to say more: to express their preference for each possible match as a **utility**: a number between 0 and 1. For example actor $A$ on one side could say that they would like to be paired with actor β on the other side with utility 0.9 (meaning a very desireable match) and with actor γ on the other side with utility 0.1 (meaning an undesireable match). The algorithm we present actually pays attention only to the ordering of preferences, but we will use the utilities to analyze how well each side does, on average.
A matching is **stable** if it is **not** the case that there is an actor from one side and an actor from the other side who both have a higher preference for each other than they have for who they are currently matched with.
# Gale-Shapley Matching Algorithm
The **[Gale-Shapley Stable Matching Algorithm](https://en.wikipedia.org/wiki/Gale%E2%80%93Shapley_algorithm)** (*Note: David Gale was my father's [PhD advisor](https://www.genealogy.math.ndsu.nodak.edu/id.php?id=10282&fChrono=1).*) works as follows: one side is chosen to be the **proposers** and the other side the **acceptors**. Until everyone has been matched the algorithm repeats the following steps:
- An unmatched proposer, $p$, proposes a match to the highest-ranked acceptor, $a$, that $p$ has not yet proposed to.
- If $a$ is unmatched, then $a$ tentatively accepts the proposal to be a match.
- If $a$ is matched, but prefers $p$ to their previous match, then $a$ breaks the previous match and tentatively accepts $p$.
- If $a$ is matched and prefers their previous match to $p$, then $a$ rejects the proposal.
I will define the function `stable_matching(P, A)`, which is passed two preference arrays: $N \times N$ arrays of utility values such that `P[p][a]` is the utility that proposer `p` has for being matched with `a`, and `A[a][p]` is the utility that acceptor `a` has for being matched with `p`. The function returns a set of matches, `{(p, a), ...}`. To implement the algorithm sketched above, we keep track of the following variables:
- `ids`: If there are $N$ actors on each side, we number them $0$ to $N-1$; `ids` is the collection of these numbers.
- `unmatched`: the set of proposers that have not yet been matched to any acceptor.
- `matched`: A mapping from acceptors to their matched proposers: `matched[a] = p`.
- `proposals`: Keeps track of who each proposer should propose to next. `proposals[p]` is a list of acceptors sorted by increasing utility, which means that `proposals[p].pop()` returns (and removes) the best acceptor for $p$ to propose to next.
```
import matplotlib.pyplot as plt
from statistics import mean, stdev
from typing import *
import random
import itertools
flatten = itertools.chain.from_iterable
ID = int
Match = Tuple[ID, ID]
def stable_matching(P, A) -> Set[Match]:
"""Compute a stable match, a set of (p, a) pairs.
P and A are square preference arrays: P[p][a] is how much p likes a;
A[a][p] is how much a likes p. Stable means there is no (p, a)
such that both prefer each other over the partner they are matched with."""
ids = range(len(P)) # ID numbers of all actors on (either) side
unmatched = set(ids) # Members of P that are not yet matched to anyone
matched = {} # {a: p} mapping of who acceptors are matched with
proposals = [sorted(ids, key=lambda a: P[p][a])
for p in ids] # proposals[p] is an ordered list of who p should propose to
while unmatched:
p = next(iter(unmatched)) # p is an arbitrary unmatched Proposer
a = proposals[p].pop() # a is p's most preferred remaining acceptor
if a not in matched:
unmatched.remove(p)
matched[a] = p
elif A[a][p] > A[a][matched[a]]:
unmatched.add(matched[a])
unmatched.remove(p)
matched[a] = p
return {(p, a) for (a, p) in matched.items()}
```
The algorithm has the following properties:
- The algorithm will always terminate.
- The output of the algorithm will always be a stable matching.
- Out of all possible stable matchings, it will produce the one that is optimal for proposers: each proposer gets the best possible match they could get. That's true because the proposers propose in order of preference, so the acceptor that they most prefer who also prefers them will accept their proposal.
- The acceptors have no such luck; they might not get their best possible match, because a proposer who is a better match for them might not ever propose to them.
What I want to get a handle on is: *how bad is this for the acceptors?* What's the gap in expected utility between the proposers and the acceptors?
# Preference Arrays
Let's define some preference arrays. `I` is the identity matrix: it says that every proposer number $i$ likes acceptor $i$ best, and dislikes the others equally. `X` is the same as the identity matrix for indexes 0, 1, and 2, but it says that the actor with index 3 would be happy with any of 2, 3, or 4, and actor 4 prefers 3.
```
I = [[1, 0, 0, 0, 0],
[0, 1, 0, 0, 0],
[0, 0, 1, 0, 0],
[0, 0, 0, 1, 0],
[0, 0, 0, 0, 1]]
X = [[1, 0, 0, 0, 0],
[0, 1, 0, 0, 0],
[0, 0, 1, 0, 0],
[0, 0, 1, 0, 0],
[0, 0, 0, 1, 0]]
I = [[.9, .4, .3, .2, .1],
[.1, .9, .4, .3, .2],
[.2, .1, .9, .4, .3],
[.3, .2, .1, .9, .4],
[.4, .3, .2, .1, .9]]
M = [[.9, .4, .3, .2, .1],
[.1, .9, .4, .3, .2],
[.2, .1, .9, .4, .3],
[.1, .2, .3, .4, .9],
[.9, .4, .3, .2, .1]]
def mean_utilities(P, A):
"""The mean utility over all members of P, and the mean utility over all members of A,
for the matching given by stable_matching(P, A)."""
matching = stable_matching(P, A)
return (mean(P[p][a] for (p, a) in matching),
mean(A[a][p] for (p, a) in matching))
```
Let's see what happens when `I` is the proposing side, and when `X` is the proposing side:
```
stable_matching(I, X)
mean_utilities(I, X)
stable_matching(X, I)
mean_utilities(X, I)
```
When `I` is the proposing side, every actor in `I` gets their first-choice match.
Likewise, when `X` is the proposer, every actor in `X` gets a first-choice match.
We can measure the average utility to each side for any matching:
# Is it Fair?
We see that in both cases, the proposers get 100% of their maximum possible utility, and the acceptors gets only 60% (averaged over all five acceptors). Is this a problem? If the Gale-Shapley algorithm is used in high-stakes applications like matching medical residents to hospitals, does it make a big difference which side is the proposers? I want to address that question with some experiments.
# Preferences with Common and Private Values
I will create a bunch of randomized preference arrays and get a feeling for how they perform. But I don't want them to be completely random; I want them to reflect, in a very abstract way, some properties of the real world:
- Some choices have intrinsic properties that make them widely popular (or unpopular). For example, Massachusetts General Hospital is considered an excellent choice by many aspiring residents. The amount of utility that is commonly agreed upon is called the **common value** of a choice.
- Some choices have idiosyncratic properties that appeal only to specific choosers. For example, you might really want to be a resident at your hometown hospital, even if it is not highly-regarded by others. This is the **private value** of a choice.
- In real world situations there is usually a mix of common and private value.
The function call `preferences(N, 0.75)`, for example, creates an NxN array of preferences, where each preference is 75% common value and 25% individual value. I implement individual value as being proportional to the ID number (`a` in the code):
```
def preferences(N=25, c=0.75):
"""Create an NxN preference array, weighted: c × common + (1 - c) × random."""
return [[round(c * (a + 0.5) / N + (1 - c) * random.uniform(0, 1), 4)
for a in range(N)]
for p in range(N)]
random.seed(42)
```
Below is a 7x7 preference array that is half common, half private. You can see as you go across a row that the utilities tend to increase, but not always monotonically:
```
preferences(7, 0.5)
```
Here's a preference array with no common value; the utilities are completely random, uncorrelated to their position:
```
preferences(7, 0.0)
```
And here's a preference array with 100% common value: every row is identical, and the utilities monotonically increase across the row:
```
preferences(5, 1.0)
```
The `preferences` function has been designed so that the average utility value is close to 0.5, for all values of `c`:
```
mean(flatten(preferences(100)))
mean(flatten(preferences(100, c=0.25)))
```
Now for one more helpful function: `examples` returns a list of the form `[(P, A), ...]` where `P` and `A` are preference arrays.
```
def examples(N=25, c=0.5, repeat=10000):
"""A list of pairs of preference arrays, (P, A), of length `repeat`."""
return [(preferences(N, c), preferences(N, c)) for _ in range(repeat)]
examples(N=3, repeat=2)
```
# Histograms of Acceptor/Proposer Utility
Now we're readsy to answer the original question: how much worse is it to be an acceptor rather than a proposer? The function `show` displays two overlapping histograms of mean utilities: one for acceptors and one for proposers.
```
def show(N=25, c=0.5, repeat=10000, bins=50):
"""Show two histograms of mean utility values over examples, for proposers and acceptors."""
pr, ac = transpose(mean_utilities(P, A) for (P, A) in examples(N, c, repeat))
plt.hist(pr, bins=bins, alpha=0.5)
plt.hist(ac, bins=bins, alpha=0.5);
print(f'''{repeat:,d} examples with N = {N} actors, common value ratio c = {c}
Acceptors: {mean(ac):.3f} ± {stdev(ac):.3f}
Proposers: {mean(pr):.3f} ± {stdev(pr):.3f}''')
def transpose(matrix): return list(zip(*matrix))
```
We'll start with preferences that are completely private; no common value:
```
show(c=0.0)
```
The acceptors (the orange histogram) have a mean utility of 0.730 while the proposers (blue histogram) do much better with a mean of 0.870. Both sides do much better than the 0.5 average utility that they would average if we just used a random (non-stable) matching.
It is clear that proposers do much better than acceptors. That suggests that the `stable_matching` algorithm is very unfair. But before drawing that conclusion, let's consider preferences with a 50/50 mix of private/common value. We'll do that for two different population sizes, 25 and 50:
```
show(c=0.5, N=25)
show(c=0.5, N=50)
```
We see that the gap between proposer and acceptor has been greatly reduced (but not eliminated). With more actors, the variance is smaller (the histogram is not as wide).
What happens with 90% common value? How aboout 99%?
```
show(c=0.9)
show(c=0.99)
```
We see that there is very little difference between the two sides. So the conclusion is: when there is a lot of common value, the Gale-Shapley Matching Algorithm is fair. So it is probably okay to use it for matching medical residents, because there is a lot of common value in the perception of the quality of hospitals, and likewise for the quality of students.
But when there is mostly private value, the algorithm is unfair, favoring the proposers over the acceptors.
|
github_jupyter
|
import matplotlib.pyplot as plt
from statistics import mean, stdev
from typing import *
import random
import itertools
flatten = itertools.chain.from_iterable
ID = int
Match = Tuple[ID, ID]
def stable_matching(P, A) -> Set[Match]:
"""Compute a stable match, a set of (p, a) pairs.
P and A are square preference arrays: P[p][a] is how much p likes a;
A[a][p] is how much a likes p. Stable means there is no (p, a)
such that both prefer each other over the partner they are matched with."""
ids = range(len(P)) # ID numbers of all actors on (either) side
unmatched = set(ids) # Members of P that are not yet matched to anyone
matched = {} # {a: p} mapping of who acceptors are matched with
proposals = [sorted(ids, key=lambda a: P[p][a])
for p in ids] # proposals[p] is an ordered list of who p should propose to
while unmatched:
p = next(iter(unmatched)) # p is an arbitrary unmatched Proposer
a = proposals[p].pop() # a is p's most preferred remaining acceptor
if a not in matched:
unmatched.remove(p)
matched[a] = p
elif A[a][p] > A[a][matched[a]]:
unmatched.add(matched[a])
unmatched.remove(p)
matched[a] = p
return {(p, a) for (a, p) in matched.items()}
I = [[1, 0, 0, 0, 0],
[0, 1, 0, 0, 0],
[0, 0, 1, 0, 0],
[0, 0, 0, 1, 0],
[0, 0, 0, 0, 1]]
X = [[1, 0, 0, 0, 0],
[0, 1, 0, 0, 0],
[0, 0, 1, 0, 0],
[0, 0, 1, 0, 0],
[0, 0, 0, 1, 0]]
I = [[.9, .4, .3, .2, .1],
[.1, .9, .4, .3, .2],
[.2, .1, .9, .4, .3],
[.3, .2, .1, .9, .4],
[.4, .3, .2, .1, .9]]
M = [[.9, .4, .3, .2, .1],
[.1, .9, .4, .3, .2],
[.2, .1, .9, .4, .3],
[.1, .2, .3, .4, .9],
[.9, .4, .3, .2, .1]]
def mean_utilities(P, A):
"""The mean utility over all members of P, and the mean utility over all members of A,
for the matching given by stable_matching(P, A)."""
matching = stable_matching(P, A)
return (mean(P[p][a] for (p, a) in matching),
mean(A[a][p] for (p, a) in matching))
stable_matching(I, X)
mean_utilities(I, X)
stable_matching(X, I)
mean_utilities(X, I)
def preferences(N=25, c=0.75):
"""Create an NxN preference array, weighted: c × common + (1 - c) × random."""
return [[round(c * (a + 0.5) / N + (1 - c) * random.uniform(0, 1), 4)
for a in range(N)]
for p in range(N)]
random.seed(42)
preferences(7, 0.5)
preferences(7, 0.0)
preferences(5, 1.0)
mean(flatten(preferences(100)))
mean(flatten(preferences(100, c=0.25)))
def examples(N=25, c=0.5, repeat=10000):
"""A list of pairs of preference arrays, (P, A), of length `repeat`."""
return [(preferences(N, c), preferences(N, c)) for _ in range(repeat)]
examples(N=3, repeat=2)
def show(N=25, c=0.5, repeat=10000, bins=50):
"""Show two histograms of mean utility values over examples, for proposers and acceptors."""
pr, ac = transpose(mean_utilities(P, A) for (P, A) in examples(N, c, repeat))
plt.hist(pr, bins=bins, alpha=0.5)
plt.hist(ac, bins=bins, alpha=0.5);
print(f'''{repeat:,d} examples with N = {N} actors, common value ratio c = {c}
Acceptors: {mean(ac):.3f} ± {stdev(ac):.3f}
Proposers: {mean(pr):.3f} ± {stdev(pr):.3f}''')
def transpose(matrix): return list(zip(*matrix))
show(c=0.0)
show(c=0.5, N=25)
show(c=0.5, N=50)
show(c=0.9)
show(c=0.99)
| 0.84699 | 0.987412 |
# Introductory Tutorial
## Tutorial Description
[Mesa](https://github.com/projectmesa/mesa) is a Python framework for [agent-based modeling](https://en.wikipedia.org/wiki/Agent-based_model). Getting started with Mesa is easy. In this tutorial, we will walk through creating a simple model and progressively add functionality which will illustrate Mesa's core features.
**Note:** This tutorial is a work-in-progress. If you find any errors or bugs, or just find something unclear or confusing, [let us know](https://github.com/projectmesa/mesa/issues)!
The base for this tutorial is a very simple model of agents exchanging money. Next, we add *space* to allow agents to move. Then, we'll cover two of Mesa's analytic tools: the *data collector* and *batch runner*. After that, we'll add an *interactive visualization* which lets us watch the model as it runs. Finally, we go over how to write your own visualization module, for users who are comfortable with JavaScript.
You can also find all the code this tutorial describes in the **examples/boltzmann_wealth_model** directory of the Mesa repository.
## Sample Model Description
The tutorial model is a very simple simulated agent-based economy, drawn from econophysics and presenting a statistical mechanics approach to wealth distribution [Dragulescu2002]_. The rules of our tutorial model:
1. There are some number of agents.
2. All agents begin with 1 unit of money.
3. At every step of the model, an agent gives 1 unit of money (if they have it) to some other agent.
Despite its simplicity, this model yields results that are often unexpected to those not familiar with it. For our purposes, it also easily demonstrates Mesa's core features.
Let's get started.
### Installation
To start, install Mesa. We recommend doing this in a [virtual environment](https://virtualenvwrapper.readthedocs.org/en/stable/), but make sure your environment is set up with Python 3. Mesa requires Python3 and does not work in Python 2 environments.
To install Mesa, simply:
```bash
$ pip install mesa
```
When you do that, it will install Mesa itself, as well as any dependencies that aren't in your setup yet. Additional dependencies required by this tutorial can be found in the **examples/boltzmann_wealth_model/requirements.txt** file, which can be installed directly form the github repository by running:
```bash
$ pip install -r https://raw.githubusercontent.com/projectmesa/mesa/main/examples/boltzmann_wealth_model/requirements.txt
```
This will install the dependencies listed in the requirements.txt file which are:
- jupyter (Ipython interactive notebook)
- matplotlib (Python's visualization library)
- mesa (this ABM library -- if not installed)
- numpy (Python's numerical python library)
## Building a sample model
Once Mesa is installed, you can start building our model. You can write models in two different ways:
1. Write the code in its own file with your favorite text editor, or
2. Write the model interactively in [Jupyter Notebook](http://jupyter.org/) cells.
Either way, it's good practice to put your model in its own folder -- especially if the project will end up consisting of multiple files (for example, Python files for the model and the visualization, a Notebook for analysis, and a Readme with some documentation and discussion).
Begin by creating a folder, and either launch a Notebook or create a new Python source file. We will use the name `money_model.py` here.
### Setting up the model
To begin writing the model code, we start with two core classes: one for the overall model, the other for the agents. The model class holds the model-level attributes, manages the agents, and generally handles the global level of our model. Each instantiation of the model class will be a specific model run. Each model will contain multiple agents, all of which are instantiations of the agent class. Both the model and agent classes are child classes of Mesa's generic `Model` and `Agent` classes.
Each agent has only one variable: how much wealth it currently has. (Each agent will also have a unique identifier (i.e., a name), stored in the `unique_id` variable. Giving each agent a unique id is a good practice when doing agent-based modeling.)
There is only one model-level parameter: how many agents the model contains. When a new model is started, we want it to populate itself with the given number of agents.
The beginning of both classes looks like this:
```
from mesa import Agent, Model
class MoneyAgent(Agent):
"""An agent with fixed initial wealth."""
def __init__(self, unique_id, model):
super().__init__(unique_id, model)
self.wealth = 1
class MoneyModel(Model):
"""A model with some number of agents."""
def __init__(self, N):
self.num_agents = N
# Create agents
for i in range(self.num_agents):
a = MoneyAgent(i, self)
```
### Adding the scheduler
Time in most agent-based models moves in steps, sometimes also called **ticks**. At each step of the model, one or more of the agents -- usually all of them -- are activated and take their own step, changing internally and/or interacting with one another or the environment.
The **scheduler** is a special model component which controls the order in which agents are activated. For example, all the agents may activate in the same order every step; their order might be shuffled; we may try to simulate all the agents acting at the same time; and more. Mesa offers a few different built-in scheduler classes, with a common interface. That makes it easy to change the activation regime a given model uses, and see whether it changes the model behavior. This may not seem important, but scheduling patterns can have an impact on your results [Comer2014].
For now, let's use one of the simplest ones: `RandomActivation`, which activates all the agents once per step, in random order. Every agent is expected to have a ``step`` method. The step method is the action the agent takes when it is activated by the model schedule. We add an agent to the schedule using the `add` method; when we call the schedule's `step` method, the model shuffles the order of the agents, then activates and executes each agent's ```step``` method.
With that in mind, the model code with the scheduler added looks like this:
```
from mesa import Agent, Model
from mesa.time import RandomActivation
class MoneyAgent(Agent):
"""An agent with fixed initial wealth."""
def __init__(self, unique_id, model):
super().__init__(unique_id, model)
self.wealth = 1
def step(self):
# The agent's step will go here.
# For demonstration purposes we will print the agent's unique_id
print("Hi, I am agent " + str(self.unique_id) + ".")
class MoneyModel(Model):
"""A model with some number of agents."""
def __init__(self, N):
self.num_agents = N
self.schedule = RandomActivation(self)
# Create agents
for i in range(self.num_agents):
a = MoneyAgent(i, self)
self.schedule.add(a)
def step(self):
"""Advance the model by one step."""
self.schedule.step()
```
At this point, we have a model which runs -- it just doesn't do anything. You can see for yourself with a few easy lines. If you've been working in an interactive session, you can create a model object directly. Otherwise, you need to open an interactive session in the same directory as your source code file, and import the classes. For example, if your code is in `money_model.py`:
```python
from money_model import MoneyModel
```
Then create the model object, and run it for one step:
```
empty_model = MoneyModel(10)
empty_model.step()
```
#### Exercise
Try modifying the code above to have every agent print out its `wealth` when it is activated. Run a few steps of the model to see how the agent activation order is shuffled each step.
### Agent Step
Now we just need to have the agents do what we intend for them to do: check their wealth, and if they have the money, give one unit of it away to another random agent. To allow the agent to choose another agent at random, we use the ``model.random`` random-number generator. This works just like Python's ``random`` module, but with a fixed seed set when the model is instantiated, that can be used to replicate a specific model run later.
To pick an agent at random, we need a list of all agents. Notice that there isn't such a list explicitly in the model. The scheduler, however, does have an internal list of all the agents it is scheduled to activate.
With that in mind, we rewrite the agent `step` method, like this:
```
class MoneyAgent(Agent):
"""An agent with fixed initial wealth."""
def __init__(self, unique_id, model):
super().__init__(unique_id, model)
self.wealth = 1
def step(self):
if self.wealth == 0:
return
other_agent = self.random.choice(self.model.schedule.agents)
other_agent.wealth += 1
self.wealth -= 1
```
### Running your first model
With that last piece in hand, it's time for the first rudimentary run of the model.
If you've written the code in its own file (`money_model.py` or a different name), launch an interpreter in the same directory as the file (either the plain Python command-line interpreter, or the IPython interpreter), or launch a Jupyter Notebook there. Then import the classes you created. (If you wrote the code in a Notebook, obviously this step isn't necessary).
```python
from money_model import *
```
Now let's create a model with 10 agents, and run it for 10 steps.
```
model = MoneyModel(10)
for i in range(10):
model.step()
```
Next, we need to get some data out of the model. Specifically, we want to see the distribution of the agent's wealth. We can get the wealth values with list comprehension, and then use matplotlib (or another graphics library) to visualize the data in a histogram.
If you are running from a text editor or IDE, you'll also need to add this line, to make the graph appear.
```python
plt.show()
```
```
# For a jupyter notebook add the following line:
%matplotlib inline
# The below is needed for both notebooks and scripts
import matplotlib.pyplot as plt
agent_wealth = [a.wealth for a in model.schedule.agents]
plt.hist(agent_wealth)
```
You'll should see something like the distribution above. Yours will almost certainly look at least slightly different, since each run of the model is random, after all.
To get a better idea of how a model behaves, we can create multiple model runs and see the distribution that emerges from all of them. We can do this with a nested for loop:
```
all_wealth = []
# This runs the model 100 times, each model executing 10 steps.
for j in range(100):
# Run the model
model = MoneyModel(10)
for i in range(10):
model.step()
# Store the results
for agent in model.schedule.agents:
all_wealth.append(agent.wealth)
plt.hist(all_wealth, bins=range(max(all_wealth) + 1))
```
This runs 100 instantiations of the model, and runs each for 10 steps. (Notice that we set the histogram bins to be integers, since agents can only have whole numbers of wealth). This distribution looks a lot smoother. By running the model 100 times, we smooth out some of the 'noise' of randomness, and get to the model's overall expected behavior.
This outcome might be surprising. Despite the fact that all agents, on average, give and receive one unit of money every step, the model converges to a state where most agents have a small amount of money and a small number have a lot of money.
### Adding space
Many ABMs have a spatial element, with agents moving around and interacting with nearby neighbors. Mesa currently supports two overall kinds of spaces: grid, and continuous. Grids are divided into cells, and agents can only be on a particular cell, like pieces on a chess board. Continuous space, in contrast, allows agents to have any arbitrary position. Both grids and continuous spaces are frequently [toroidal](https://en.wikipedia.org/wiki/Toroidal_graph), meaning that the edges wrap around, with cells on the right edge connected to those on the left edge, and the top to the bottom. This prevents some cells having fewer neighbors than others, or agents being able to go off the edge of the environment.
Let's add a simple spatial element to our model by putting our agents on a grid and make them walk around at random. Instead of giving their unit of money to any random agent, they'll give it to an agent on the same cell.
Mesa has two main types of grids: `SingleGrid` and `MultiGrid`. `SingleGrid` enforces at most one agent per cell; `MultiGrid` allows multiple agents to be in the same cell. Since we want agents to be able to share a cell, we use `MultiGrid`.
```
from mesa.space import MultiGrid
```
We instantiate a grid with width and height parameters, and a boolean as to whether the grid is toroidal. Let's make width and height model parameters, in addition to the number of agents, and have the grid always be toroidal. We can place agents on a grid with the grid's `place_agent` method, which takes an agent and an (x, y) tuple of the coordinates to place the agent.
```
class MoneyModel(Model):
"""A model with some number of agents."""
def __init__(self, N, width, height):
self.num_agents = N
self.grid = MultiGrid(width, height, True)
self.schedule = RandomActivation(self)
# Create agents
for i in range(self.num_agents):
a = MoneyAgent(i, self)
self.schedule.add(a)
# Add the agent to a random grid cell
x = self.random.randrange(self.grid.width)
y = self.random.randrange(self.grid.height)
self.grid.place_agent(a, (x, y))
```
Under the hood, each agent's position is stored in two ways: the agent is contained in the grid in the cell it is currently in, and the agent has a `pos` variable with an (x, y) coordinate tuple. The `place_agent` method adds the coordinate to the agent automatically.
Now we need to add to the agents' behaviors, letting them move around and only give money to other agents in the same cell.
First let's handle movement, and have the agents move to a neighboring cell. The grid object provides a `move_agent` method, which like you'd imagine, moves an agent to a given cell. That still leaves us to get the possible neighboring cells to move to. There are a couple ways to do this. One is to use the current coordinates, and loop over all coordinates +/- 1 away from it. For example:
```python
neighbors = []
x, y = self.pos
for dx in [-1, 0, 1]:
for dy in [-1, 0, 1]:
neighbors.append((x+dx, y+dy))
```
But there's an even simpler way, using the grid's built-in `get_neighborhood` method, which returns all the neighbors of a given cell. This method can get two types of cell neighborhoods: [Moore](https://en.wikipedia.org/wiki/Moore_neighborhood) (includes all 8 surrounding squares), and [Von Neumann](https://en.wikipedia.org/wiki/Von_Neumann_neighborhood)(only up/down/left/right). It also needs an argument as to whether to include the center cell itself as one of the neighbors.
With that in mind, the agent's `move` method looks like this:
```python
class MoneyAgent(Agent):
#...
def move(self):
possible_steps = self.model.grid.get_neighborhood(
self.pos,
moore=True,
include_center=False)
new_position = self.random.choice(possible_steps)
self.model.grid.move_agent(self, new_position)
```
Next, we need to get all the other agents present in a cell, and give one of them some money. We can get the contents of one or more cells using the grid's `get_cell_list_contents` method, or by accessing a cell directly. The method accepts a list of cell coordinate tuples, or a single tuple if we only care about one cell.
```python
class MoneyAgent(Agent):
#...
def give_money(self):
cellmates = self.model.grid.get_cell_list_contents([self.pos])
if len(cellmates) > 1:
other = self.random.choice(cellmates)
other.wealth += 1
self.wealth -= 1
```
And with those two methods, the agent's ``step`` method becomes:
```python
class MoneyAgent(Agent):
# ...
def step(self):
self.move()
if self.wealth > 0:
self.give_money()
```
Now, putting that all together should look like this:
```
class MoneyAgent(Agent):
"""An agent with fixed initial wealth."""
def __init__(self, unique_id, model):
super().__init__(unique_id, model)
self.wealth = 1
def move(self):
possible_steps = self.model.grid.get_neighborhood(
self.pos, moore=True, include_center=False
)
new_position = self.random.choice(possible_steps)
self.model.grid.move_agent(self, new_position)
def give_money(self):
cellmates = self.model.grid.get_cell_list_contents([self.pos])
if len(cellmates) > 1:
other_agent = self.random.choice(cellmates)
other_agent.wealth += 1
self.wealth -= 1
def step(self):
self.move()
if self.wealth > 0:
self.give_money()
class MoneyModel(Model):
"""A model with some number of agents."""
def __init__(self, N, width, height):
self.num_agents = N
self.grid = MultiGrid(width, height, True)
self.schedule = RandomActivation(self)
# Create agents
for i in range(self.num_agents):
a = MoneyAgent(i, self)
self.schedule.add(a)
# Add the agent to a random grid cell
x = self.random.randrange(self.grid.width)
y = self.random.randrange(self.grid.height)
self.grid.place_agent(a, (x, y))
def step(self):
self.schedule.step()
```
Let's create a model with 50 agents on a 10x10 grid, and run it for 20 steps.
```
model = MoneyModel(50, 10, 10)
for i in range(20):
model.step()
```
Now let's use matplotlib and numpy to visualize the number of agents residing in each cell. To do that, we create a numpy array of the same size as the grid, filled with zeros. Then we use the grid object's `coord_iter()` feature, which lets us loop over every cell in the grid, giving us each cell's coordinates and contents in turn.
```
import numpy as np
agent_counts = np.zeros((model.grid.width, model.grid.height))
for cell in model.grid.coord_iter():
cell_content, x, y = cell
agent_count = len(cell_content)
agent_counts[x][y] = agent_count
plt.imshow(agent_counts, interpolation="nearest")
plt.colorbar()
# If running from a text editor or IDE, remember you'll need the following:
# plt.show()
```
### Collecting Data
So far, at the end of every model run, we've had to go and write our own code to get the data out of the model. This has two problems: it isn't very efficient, and it only gives us end results. If we wanted to know the wealth of each agent at each step, we'd have to add that to the loop of executing steps, and figure out some way to store the data.
Since one of the main goals of agent-based modeling is generating data for analysis, Mesa provides a class which can handle data collection and storage for us and make it easier to analyze.
The data collector stores three categories of data: model-level variables, agent-level variables, and tables (which are a catch-all for everything else). Model- and agent-level variables are added to the data collector along with a function for collecting them. Model-level collection functions take a model object as an input, while agent-level collection functions take an agent object as an input. Both then return a value computed from the model or each agent at their current state. When the data collector’s `collect` method is called, with a model object as its argument, it applies each model-level collection function to the model, and stores the results in a dictionary, associating the current value with the current step of the model. Similarly, the method applies each agent-level collection function to each agent currently in the schedule, associating the resulting value with the step of the model, and the agent’s `unique_id`.
Let's add a DataCollector to the model, and collect two variables. At the agent level, we want to collect every agent's wealth at every step. At the model level, let's measure the model's [Gini Coefficient](https://en.wikipedia.org/wiki/Gini_coefficient), a measure of wealth inequality.
```
from mesa.datacollection import DataCollector
def compute_gini(model):
agent_wealths = [agent.wealth for agent in model.schedule.agents]
x = sorted(agent_wealths)
N = model.num_agents
B = sum(xi * (N - i) for i, xi in enumerate(x)) / (N * sum(x))
return 1 + (1 / N) - 2 * B
class MoneyAgent(Agent):
"""An agent with fixed initial wealth."""
def __init__(self, unique_id, model):
super().__init__(unique_id, model)
self.wealth = 1
def move(self):
possible_steps = self.model.grid.get_neighborhood(
self.pos, moore=True, include_center=False
)
new_position = self.random.choice(possible_steps)
self.model.grid.move_agent(self, new_position)
def give_money(self):
cellmates = self.model.grid.get_cell_list_contents([self.pos])
if len(cellmates) > 1:
other = self.random.choice(cellmates)
other.wealth += 1
self.wealth -= 1
def step(self):
self.move()
if self.wealth > 0:
self.give_money()
class MoneyModel(Model):
"""A model with some number of agents."""
def __init__(self, N, width, height):
self.num_agents = N
self.grid = MultiGrid(width, height, True)
self.schedule = RandomActivation(self)
# Create agents
for i in range(self.num_agents):
a = MoneyAgent(i, self)
self.schedule.add(a)
# Add the agent to a random grid cell
x = self.random.randrange(self.grid.width)
y = self.random.randrange(self.grid.height)
self.grid.place_agent(a, (x, y))
self.datacollector = DataCollector(
model_reporters={"Gini": compute_gini}, agent_reporters={"Wealth": "wealth"}
)
def step(self):
self.datacollector.collect(self)
self.schedule.step()
```
At every step of the model, the datacollector will collect and store the model-level current Gini coefficient, as well as each agent's wealth, associating each with the current step.
We run the model just as we did above. Now is when an interactive session, especially via a Notebook, comes in handy: the DataCollector can export the data it's collected as a pandas DataFrame, for easy interactive analysis.
```
model = MoneyModel(50, 10, 10)
for i in range(100):
model.step()
```
To get the series of Gini coefficients as a pandas DataFrame:
```
gini = model.datacollector.get_model_vars_dataframe()
gini.plot()
```
Similarly, we can get the agent-wealth data:
```
agent_wealth = model.datacollector.get_agent_vars_dataframe()
agent_wealth.head()
```
You'll see that the DataFrame's index is pairings of model step and agent ID. You can analyze it the way you would any other DataFrame. For example, to get a histogram of agent wealth at the model's end:
```
end_wealth = agent_wealth.xs(99, level="Step")["Wealth"]
end_wealth.hist(bins=range(agent_wealth.Wealth.max() + 1))
```
Or to plot the wealth of a given agent (in this example, agent 14):
```
one_agent_wealth = agent_wealth.xs(14, level="AgentID")
one_agent_wealth.Wealth.plot()
```
### Batch Run
Like we mentioned above, you usually won't run a model only once, but multiple times, with fixed parameters to find the overall distributions the model generates, and with varying parameters to analyze how they drive the model's outputs and behaviors. Instead of needing to write nested for-loops for each model, Mesa provides a BatchRunner class which automates it for you.
The BatchRunner also requires an additional variable `self.running` for the MoneyModel class. This variable enables conditional shut off of the model once a condition is met. In this example it will be set as True indefinitely.
```
def compute_gini(model):
agent_wealths = [agent.wealth for agent in model.schedule.agents]
x = sorted(agent_wealths)
N = model.num_agents
B = sum(xi * (N - i) for i, xi in enumerate(x)) / (N * sum(x))
return 1 + (1 / N) - 2 * B
class MoneyModel(Model):
"""A model with some number of agents."""
def __init__(self, N, width, height):
self.num_agents = N
self.grid = MultiGrid(width, height, True)
self.schedule = RandomActivation(self)
self.running = True
# Create agents
for i in range(self.num_agents):
a = MoneyAgent(i, self)
self.schedule.add(a)
# Add the agent to a random grid cell
x = self.random.randrange(self.grid.width)
y = self.random.randrange(self.grid.height)
self.grid.place_agent(a, (x, y))
self.datacollector = DataCollector(
model_reporters={"Gini": compute_gini}, agent_reporters={"Wealth": "wealth"}
)
def step(self):
self.datacollector.collect(self)
self.schedule.step()
```
We instantiate a BatchRunner with a model class to run, and two dictionaries: one of the fixed parameters (mapping model arguments to values) and one of varying parameters (mapping each parameter name to a sequence of values for it to take). The BatchRunner also takes an argument for how many model instantiations to create and run at each combination of parameter values, and how many steps to run each instantiation for. Finally, like the DataCollector, it takes dictionaries of model- and agent-level reporters to collect. Unlike the DataCollector, it won't collect the data every step of the model, but only at the end of each run.
In the following example, we hold the height and width fixed, and vary the number of agents. We tell the BatchRunner to run 5 instantiations of the model with each number of agents, and to run each for 100 steps.*
We have it collect the final Gini coefficient value.
Now, we can set up and run the BatchRunner:
*The total number of runs is 245. That is 10 agents to 490 increasing by 10, making 49 agents populations. Each agent population is then run 5 times (49 * 5) for 245 iterations
```
from mesa.batchrunner import BatchRunner
fixed_params = {"width": 10, "height": 10}
variable_params = {"N": range(10, 500, 10)}
batch_run = BatchRunner(
MoneyModel,
variable_params,
fixed_params,
iterations=5,
max_steps=100,
model_reporters={"Gini": compute_gini},
)
batch_run.run_all()
```
BatchRunner has two ways to collect data.
First, one can pass model collection via BatchRunner as seen above with output below.
```
run_data = batch_run.get_model_vars_dataframe()
run_data.head()
plt.scatter(run_data.N, run_data.Gini)
```
Notice that each row is a model run, and gives us the parameter values associated with that run. We can use this data to view a scatter-plot comparing the number of agents to the final Gini.
Second, BatchRunner can call the datacollector from the model. The output is a dictionary, where each key is the parameters with the iteration number and then the datacollector dataframe. So in this model (<#number of agents>, <iteration#>).
```
# Get the Agent DataCollection
data_collector_agents = batch_run.get_collector_agents()
data_collector_agents[(10, 2)]
# Get the Model DataCollection.
data_collector_model = batch_run.get_collector_model()
data_collector_model[(10, 1)]
```
### Happy Modeling!
This document is a work in progress. If you see any errors, exclusions or have any problems please contact [us](https://github.com/projectmesa/mesa/issues).
`virtual environment`: http://docs.python-guide.org/en/latest/dev/virtualenvs/
[Comer2014] Comer, Kenneth W. “Who Goes First? An Examination of the Impact of Activation on Outcome Behavior in AgentBased Models.” George Mason University, 2014. http://mars.gmu.edu/bitstream/handle/1920/9070/Comer_gmu_0883E_10539.pdf
[Dragulescu2002] Drăgulescu, Adrian A., and Victor M. Yakovenko. “Statistical Mechanics of Money, Income, and Wealth: A Short Survey.” arXiv Preprint Cond-mat/0211175, 2002. http://arxiv.org/abs/cond-mat/0211175.
|
github_jupyter
|
$ pip install mesa
$ pip install -r https://raw.githubusercontent.com/projectmesa/mesa/main/examples/boltzmann_wealth_model/requirements.txt
from mesa import Agent, Model
class MoneyAgent(Agent):
"""An agent with fixed initial wealth."""
def __init__(self, unique_id, model):
super().__init__(unique_id, model)
self.wealth = 1
class MoneyModel(Model):
"""A model with some number of agents."""
def __init__(self, N):
self.num_agents = N
# Create agents
for i in range(self.num_agents):
a = MoneyAgent(i, self)
from mesa import Agent, Model
from mesa.time import RandomActivation
class MoneyAgent(Agent):
"""An agent with fixed initial wealth."""
def __init__(self, unique_id, model):
super().__init__(unique_id, model)
self.wealth = 1
def step(self):
# The agent's step will go here.
# For demonstration purposes we will print the agent's unique_id
print("Hi, I am agent " + str(self.unique_id) + ".")
class MoneyModel(Model):
"""A model with some number of agents."""
def __init__(self, N):
self.num_agents = N
self.schedule = RandomActivation(self)
# Create agents
for i in range(self.num_agents):
a = MoneyAgent(i, self)
self.schedule.add(a)
def step(self):
"""Advance the model by one step."""
self.schedule.step()
from money_model import MoneyModel
empty_model = MoneyModel(10)
empty_model.step()
class MoneyAgent(Agent):
"""An agent with fixed initial wealth."""
def __init__(self, unique_id, model):
super().__init__(unique_id, model)
self.wealth = 1
def step(self):
if self.wealth == 0:
return
other_agent = self.random.choice(self.model.schedule.agents)
other_agent.wealth += 1
self.wealth -= 1
from money_model import *
model = MoneyModel(10)
for i in range(10):
model.step()
plt.show()
# For a jupyter notebook add the following line:
%matplotlib inline
# The below is needed for both notebooks and scripts
import matplotlib.pyplot as plt
agent_wealth = [a.wealth for a in model.schedule.agents]
plt.hist(agent_wealth)
all_wealth = []
# This runs the model 100 times, each model executing 10 steps.
for j in range(100):
# Run the model
model = MoneyModel(10)
for i in range(10):
model.step()
# Store the results
for agent in model.schedule.agents:
all_wealth.append(agent.wealth)
plt.hist(all_wealth, bins=range(max(all_wealth) + 1))
from mesa.space import MultiGrid
class MoneyModel(Model):
"""A model with some number of agents."""
def __init__(self, N, width, height):
self.num_agents = N
self.grid = MultiGrid(width, height, True)
self.schedule = RandomActivation(self)
# Create agents
for i in range(self.num_agents):
a = MoneyAgent(i, self)
self.schedule.add(a)
# Add the agent to a random grid cell
x = self.random.randrange(self.grid.width)
y = self.random.randrange(self.grid.height)
self.grid.place_agent(a, (x, y))
neighbors = []
x, y = self.pos
for dx in [-1, 0, 1]:
for dy in [-1, 0, 1]:
neighbors.append((x+dx, y+dy))
class MoneyAgent(Agent):
#...
def move(self):
possible_steps = self.model.grid.get_neighborhood(
self.pos,
moore=True,
include_center=False)
new_position = self.random.choice(possible_steps)
self.model.grid.move_agent(self, new_position)
class MoneyAgent(Agent):
#...
def give_money(self):
cellmates = self.model.grid.get_cell_list_contents([self.pos])
if len(cellmates) > 1:
other = self.random.choice(cellmates)
other.wealth += 1
self.wealth -= 1
class MoneyAgent(Agent):
# ...
def step(self):
self.move()
if self.wealth > 0:
self.give_money()
class MoneyAgent(Agent):
"""An agent with fixed initial wealth."""
def __init__(self, unique_id, model):
super().__init__(unique_id, model)
self.wealth = 1
def move(self):
possible_steps = self.model.grid.get_neighborhood(
self.pos, moore=True, include_center=False
)
new_position = self.random.choice(possible_steps)
self.model.grid.move_agent(self, new_position)
def give_money(self):
cellmates = self.model.grid.get_cell_list_contents([self.pos])
if len(cellmates) > 1:
other_agent = self.random.choice(cellmates)
other_agent.wealth += 1
self.wealth -= 1
def step(self):
self.move()
if self.wealth > 0:
self.give_money()
class MoneyModel(Model):
"""A model with some number of agents."""
def __init__(self, N, width, height):
self.num_agents = N
self.grid = MultiGrid(width, height, True)
self.schedule = RandomActivation(self)
# Create agents
for i in range(self.num_agents):
a = MoneyAgent(i, self)
self.schedule.add(a)
# Add the agent to a random grid cell
x = self.random.randrange(self.grid.width)
y = self.random.randrange(self.grid.height)
self.grid.place_agent(a, (x, y))
def step(self):
self.schedule.step()
model = MoneyModel(50, 10, 10)
for i in range(20):
model.step()
import numpy as np
agent_counts = np.zeros((model.grid.width, model.grid.height))
for cell in model.grid.coord_iter():
cell_content, x, y = cell
agent_count = len(cell_content)
agent_counts[x][y] = agent_count
plt.imshow(agent_counts, interpolation="nearest")
plt.colorbar()
# If running from a text editor or IDE, remember you'll need the following:
# plt.show()
from mesa.datacollection import DataCollector
def compute_gini(model):
agent_wealths = [agent.wealth for agent in model.schedule.agents]
x = sorted(agent_wealths)
N = model.num_agents
B = sum(xi * (N - i) for i, xi in enumerate(x)) / (N * sum(x))
return 1 + (1 / N) - 2 * B
class MoneyAgent(Agent):
"""An agent with fixed initial wealth."""
def __init__(self, unique_id, model):
super().__init__(unique_id, model)
self.wealth = 1
def move(self):
possible_steps = self.model.grid.get_neighborhood(
self.pos, moore=True, include_center=False
)
new_position = self.random.choice(possible_steps)
self.model.grid.move_agent(self, new_position)
def give_money(self):
cellmates = self.model.grid.get_cell_list_contents([self.pos])
if len(cellmates) > 1:
other = self.random.choice(cellmates)
other.wealth += 1
self.wealth -= 1
def step(self):
self.move()
if self.wealth > 0:
self.give_money()
class MoneyModel(Model):
"""A model with some number of agents."""
def __init__(self, N, width, height):
self.num_agents = N
self.grid = MultiGrid(width, height, True)
self.schedule = RandomActivation(self)
# Create agents
for i in range(self.num_agents):
a = MoneyAgent(i, self)
self.schedule.add(a)
# Add the agent to a random grid cell
x = self.random.randrange(self.grid.width)
y = self.random.randrange(self.grid.height)
self.grid.place_agent(a, (x, y))
self.datacollector = DataCollector(
model_reporters={"Gini": compute_gini}, agent_reporters={"Wealth": "wealth"}
)
def step(self):
self.datacollector.collect(self)
self.schedule.step()
model = MoneyModel(50, 10, 10)
for i in range(100):
model.step()
gini = model.datacollector.get_model_vars_dataframe()
gini.plot()
agent_wealth = model.datacollector.get_agent_vars_dataframe()
agent_wealth.head()
end_wealth = agent_wealth.xs(99, level="Step")["Wealth"]
end_wealth.hist(bins=range(agent_wealth.Wealth.max() + 1))
one_agent_wealth = agent_wealth.xs(14, level="AgentID")
one_agent_wealth.Wealth.plot()
def compute_gini(model):
agent_wealths = [agent.wealth for agent in model.schedule.agents]
x = sorted(agent_wealths)
N = model.num_agents
B = sum(xi * (N - i) for i, xi in enumerate(x)) / (N * sum(x))
return 1 + (1 / N) - 2 * B
class MoneyModel(Model):
"""A model with some number of agents."""
def __init__(self, N, width, height):
self.num_agents = N
self.grid = MultiGrid(width, height, True)
self.schedule = RandomActivation(self)
self.running = True
# Create agents
for i in range(self.num_agents):
a = MoneyAgent(i, self)
self.schedule.add(a)
# Add the agent to a random grid cell
x = self.random.randrange(self.grid.width)
y = self.random.randrange(self.grid.height)
self.grid.place_agent(a, (x, y))
self.datacollector = DataCollector(
model_reporters={"Gini": compute_gini}, agent_reporters={"Wealth": "wealth"}
)
def step(self):
self.datacollector.collect(self)
self.schedule.step()
from mesa.batchrunner import BatchRunner
fixed_params = {"width": 10, "height": 10}
variable_params = {"N": range(10, 500, 10)}
batch_run = BatchRunner(
MoneyModel,
variable_params,
fixed_params,
iterations=5,
max_steps=100,
model_reporters={"Gini": compute_gini},
)
batch_run.run_all()
run_data = batch_run.get_model_vars_dataframe()
run_data.head()
plt.scatter(run_data.N, run_data.Gini)
# Get the Agent DataCollection
data_collector_agents = batch_run.get_collector_agents()
data_collector_agents[(10, 2)]
# Get the Model DataCollection.
data_collector_model = batch_run.get_collector_model()
data_collector_model[(10, 1)]
| 0.896266 | 0.991563 |
# Grouping your data
```
import warnings
warnings.simplefilter('ignore', FutureWarning)
import matplotlib
matplotlib.rcParams['axes.grid'] = True # show gridlines by default
%matplotlib inline
import pandas as pd
import pandas_datareader as pdr
if pd.__version__.startswith('0.23'):
# this solves an incompatibility between pandas 0.23 and datareader 0.6
# taken from https://stackoverflow.com/questions/50394873/
core.common.is_list_like = api.types.is_list_like
from pandas_datareader.wb import download
?download
YEAR = 2013
GDP_INDICATOR = 'NY.GDP.MKTP.CD'
gdp = download(indicator=GDP_INDICATOR, country=['GB','CN'],
start=YEAR-5, end=YEAR)
gdp = gdp.reset_index()
gdp
gdp.groupby('country')['NY.GDP.MKTP.CD'].aggregate(sum)
gdp.groupby('year')['NY.GDP.MKTP.CD'].aggregate(sum)
LOCATION='comtrade_milk_uk_monthly_14.csv'
# LOCATION = 'http://comtrade.un.org/api/get?max=5000&type=C&freq=M&px=HS&ps=2014&r=826&p=all&rg=1%2C2&cc=0401%2C0402&fmt=csv'
milk = pd.read_csv(LOCATION, dtype={'Commodity Code':str, 'Reporter Code':str})
milk.head(3)
COLUMNS = ['Year', 'Period','Trade Flow','Reporter', 'Partner', 'Commodity','Commodity Code','Trade Value (US$)']
milk = milk[COLUMNS]
milk_world = milk[milk['Partner'] == 'World']
milk_countries = milk[milk['Partner'] != 'World']
milk_countries.to_csv('countrymilk.csv', index=False)
load_test = pd.read_csv('countrymilk.csv', dtype={'Commodity Code':str, 'Reporter Code':str})
load_test.head(2)
milk_imports = milk[milk['Trade Flow'] == 'Imports']
milk_countries_imports = milk_countries[milk_countries['Trade Flow'] == 'Imports']
milk_world_imports=milk_world[milk_world['Trade Flow'] == 'Imports']
milkImportsInJanuary2014 = milk_countries_imports[milk_countries_imports['Period'] == 201401]
milkImportsInJanuary2014.sort_values('Trade Value (US$)',ascending=False).head(10)
```
# Make sure you run all the cell above!
## Grouping data
On many occasions, a dataframe may be organised as groups of rows where the group membership is identified based on cell values within one or more 'key' columns. **Grouping** refers to the process whereby rows associated with a particular group are collated so that you can work with just those rows as distinct subsets of the whole dataset.
The number of groups the dataframe will be split into is based on the number of unique values identified within a single key column, or the number of unique combinations of values for two or more key columns.
The `groupby()` method runs down each row in a data frame, splitting the rows into separate groups based on the unique values associated with the key column or columns.
The following is an example of the steps and code needed to split a dataframe.
### Grouping the data
Split the data into two different subsets of data (imports and exports), by grouping on trade flow.
```
groups = milk_countries.groupby('Trade Flow')
```
Inspect the first few rows associated with a particular group:
```
groups.get_group('Imports').head()
```
As well as grouping on a single term, you can create groups based on multiple columns by passing in several column names as a list. For example, generate groups based on commodity code *and* trade flow, and then preview the keys used to define the groups.
```
GROUPING_COMMFLOW = ['Commodity Code','Trade Flow']
groups = milk_countries.groupby(GROUPING_COMMFLOW)
groups.groups.keys()
```
Retrieve a group based on multiple group levels by passing in a tuple that specifies a value for each index column. For example, if a grouping is based on the `'Partner'` and `'Trade Flow'` columns, the argument of `get_group` has to be a partner/flow pair, like `('France', 'Import')` to get all rows associated with imports from France.
```
GROUPING_PARTNERFLOW = ['Partner','Trade Flow']
groups = milk_countries.groupby(GROUPING_PARTNERFLOW)
GROUP_PARTNERFLOW= ('France','Imports')
groups.get_group( GROUP_PARTNERFLOW )
```
To find the leading partner for a particular commodity, group by commodity, get the desired group, and then sort the result.
```
groups = milk_countries.groupby(['Commodity Code'])
groups.get_group('0402').sort_values("Trade Value (US$)", ascending=False).head()
```
### Task
Using your own data set from Exercise 1, try to group the data in a variety of ways, finding the most significant trade partner in each case:
- by commodity, or commodity code
- by trade flow, commodity and year.
```
LOCATION='comtrade_flowers_kenya_monthly_20.csv'
# LOCATION = 'http://comtrade.un.org//api/get?max=5000&type=C&freq=M&px=HS&ps=2020&r=404&p=all&rg=1%2C2&cc=0603%2C0402&fmt=csv'
flowers = pd.read_csv(LOCATION, dtype={'Commodity Code':str, 'Reporter Code':str})
flowers.head(3)
COLUMNS = ['Year', 'Period','Trade Flow','Reporter', 'Partner', 'Commodity','Commodity Code','Trade Value (US$)']
flowers = flowers[COLUMNS]
flowers_world = flowers[flowers['Partner'] == 'World']
flowers_countries = flowers[flowers['Partner'] != 'World']
flowers_countries.to_csv('countryflowers.csv', index=False)
load_test = pd.read_csv('countryflowers.csv', dtype={'Commodity Code':str, 'Reporter Code':str})
load_test.head(5)
load_test=pd.read_csv('countryflowers.csv', dtype={'Commodity Code':str}, encoding = "ISO-8859-1")
load_test.head()
flowers_imports = flowers[flowers['Trade Flow'] == 'Imports']
flowers_countries_imports = flowers_countries[flowers_countries['Trade Flow'] == 'Imports']
flowers_world_imports=flowers_world[flowers_world['Trade Flow'] == 'Imports']
flowersImportsInJanuary2020 = flowers_countries_imports[flowers_countries_imports['Period'] == 202001]
flowersImportsInJanuary2020.sort_values('Trade Value (US$)',ascending=False).head(10)
flowers_exports = flowers[flowers['Trade Flow'] == 'Exports']
flowers_countries_exports = flowers_countries[flowers_countries['Trade Flow'] == 'Exports']
flowers_world_exports=flowers_world[flowers_world['Trade Flow'] == 'Exports']
flowersExportsInFebruary2020 = flowers_countries_exports[flowers_countries_exports['Period'] == 202002]
flowersExportsInFebruary2020.sort_values('Trade Value (US$)',ascending=False).head(10)
groups = flowers_countries.groupby('Trade Flow')
groups.get_group('Exports').head()
GROUPING_COMMFLOW = ['Commodity Code','Trade Flow']
groups = flowers_countries.groupby(GROUPING_COMMFLOW)
groups.groups.keys()
GROUPING_PARTNERFLOW = ['Partner','Trade Flow']
groups = flowers_countries.groupby(GROUPING_PARTNERFLOW)
GROUP_PARTNERFLOW= ('United Kingdom','Exports')
groups.get_group( GROUP_PARTNERFLOW )
groups = flowers_countries.groupby(['Commodity Code'])
groups.get_group('0603').sort_values("Trade Value (US$)", ascending=False).head()
```
Netherlands is a significant trade partner as both an importer and also the sole exporter of flowers to Kenya
|
github_jupyter
|
import warnings
warnings.simplefilter('ignore', FutureWarning)
import matplotlib
matplotlib.rcParams['axes.grid'] = True # show gridlines by default
%matplotlib inline
import pandas as pd
import pandas_datareader as pdr
if pd.__version__.startswith('0.23'):
# this solves an incompatibility between pandas 0.23 and datareader 0.6
# taken from https://stackoverflow.com/questions/50394873/
core.common.is_list_like = api.types.is_list_like
from pandas_datareader.wb import download
?download
YEAR = 2013
GDP_INDICATOR = 'NY.GDP.MKTP.CD'
gdp = download(indicator=GDP_INDICATOR, country=['GB','CN'],
start=YEAR-5, end=YEAR)
gdp = gdp.reset_index()
gdp
gdp.groupby('country')['NY.GDP.MKTP.CD'].aggregate(sum)
gdp.groupby('year')['NY.GDP.MKTP.CD'].aggregate(sum)
LOCATION='comtrade_milk_uk_monthly_14.csv'
# LOCATION = 'http://comtrade.un.org/api/get?max=5000&type=C&freq=M&px=HS&ps=2014&r=826&p=all&rg=1%2C2&cc=0401%2C0402&fmt=csv'
milk = pd.read_csv(LOCATION, dtype={'Commodity Code':str, 'Reporter Code':str})
milk.head(3)
COLUMNS = ['Year', 'Period','Trade Flow','Reporter', 'Partner', 'Commodity','Commodity Code','Trade Value (US$)']
milk = milk[COLUMNS]
milk_world = milk[milk['Partner'] == 'World']
milk_countries = milk[milk['Partner'] != 'World']
milk_countries.to_csv('countrymilk.csv', index=False)
load_test = pd.read_csv('countrymilk.csv', dtype={'Commodity Code':str, 'Reporter Code':str})
load_test.head(2)
milk_imports = milk[milk['Trade Flow'] == 'Imports']
milk_countries_imports = milk_countries[milk_countries['Trade Flow'] == 'Imports']
milk_world_imports=milk_world[milk_world['Trade Flow'] == 'Imports']
milkImportsInJanuary2014 = milk_countries_imports[milk_countries_imports['Period'] == 201401]
milkImportsInJanuary2014.sort_values('Trade Value (US$)',ascending=False).head(10)
groups = milk_countries.groupby('Trade Flow')
groups.get_group('Imports').head()
GROUPING_COMMFLOW = ['Commodity Code','Trade Flow']
groups = milk_countries.groupby(GROUPING_COMMFLOW)
groups.groups.keys()
GROUPING_PARTNERFLOW = ['Partner','Trade Flow']
groups = milk_countries.groupby(GROUPING_PARTNERFLOW)
GROUP_PARTNERFLOW= ('France','Imports')
groups.get_group( GROUP_PARTNERFLOW )
groups = milk_countries.groupby(['Commodity Code'])
groups.get_group('0402').sort_values("Trade Value (US$)", ascending=False).head()
LOCATION='comtrade_flowers_kenya_monthly_20.csv'
# LOCATION = 'http://comtrade.un.org//api/get?max=5000&type=C&freq=M&px=HS&ps=2020&r=404&p=all&rg=1%2C2&cc=0603%2C0402&fmt=csv'
flowers = pd.read_csv(LOCATION, dtype={'Commodity Code':str, 'Reporter Code':str})
flowers.head(3)
COLUMNS = ['Year', 'Period','Trade Flow','Reporter', 'Partner', 'Commodity','Commodity Code','Trade Value (US$)']
flowers = flowers[COLUMNS]
flowers_world = flowers[flowers['Partner'] == 'World']
flowers_countries = flowers[flowers['Partner'] != 'World']
flowers_countries.to_csv('countryflowers.csv', index=False)
load_test = pd.read_csv('countryflowers.csv', dtype={'Commodity Code':str, 'Reporter Code':str})
load_test.head(5)
load_test=pd.read_csv('countryflowers.csv', dtype={'Commodity Code':str}, encoding = "ISO-8859-1")
load_test.head()
flowers_imports = flowers[flowers['Trade Flow'] == 'Imports']
flowers_countries_imports = flowers_countries[flowers_countries['Trade Flow'] == 'Imports']
flowers_world_imports=flowers_world[flowers_world['Trade Flow'] == 'Imports']
flowersImportsInJanuary2020 = flowers_countries_imports[flowers_countries_imports['Period'] == 202001]
flowersImportsInJanuary2020.sort_values('Trade Value (US$)',ascending=False).head(10)
flowers_exports = flowers[flowers['Trade Flow'] == 'Exports']
flowers_countries_exports = flowers_countries[flowers_countries['Trade Flow'] == 'Exports']
flowers_world_exports=flowers_world[flowers_world['Trade Flow'] == 'Exports']
flowersExportsInFebruary2020 = flowers_countries_exports[flowers_countries_exports['Period'] == 202002]
flowersExportsInFebruary2020.sort_values('Trade Value (US$)',ascending=False).head(10)
groups = flowers_countries.groupby('Trade Flow')
groups.get_group('Exports').head()
GROUPING_COMMFLOW = ['Commodity Code','Trade Flow']
groups = flowers_countries.groupby(GROUPING_COMMFLOW)
groups.groups.keys()
GROUPING_PARTNERFLOW = ['Partner','Trade Flow']
groups = flowers_countries.groupby(GROUPING_PARTNERFLOW)
GROUP_PARTNERFLOW= ('United Kingdom','Exports')
groups.get_group( GROUP_PARTNERFLOW )
groups = flowers_countries.groupby(['Commodity Code'])
groups.get_group('0603').sort_values("Trade Value (US$)", ascending=False).head()
| 0.384103 | 0.827584 |
# Mempersiapkan Data
### Deskripsi Data
SIM --> 0 : Tidak punya SIM 1 : Punya SIM<br>
Kode_Daerah --> Kode area tempat tinggal pelanggan<br>
Sudah_Asuransi --> 1 : Pelanggan sudah memiliki asuransi kendaraan, 0 : Pelanggan belum memiliki asuransi kendaraan<br>
Umur_Kendaraan --> Umur kendaraan<br>
Kendaraan_Rusak --> 1 : Kendaraan pernah rusak sebelumnya. 0 : Kendaraan belum pernah rusak.<br>
Premi --> Jumlah premi yang harus dibayarkan per tahun.<br>
Kanal_Penjualan --> Kode kanal untuk menghubungi pelanggan (email, telpon, dll)<br>
Lama_Berlangganan --> Sudah berapa lama pelanggan menjadi klien perusahaan<br>
Tertarik --> 1 : Pelanggan tertarik, 0 : Pelanggan tidak tertarik
<br>
### Import Library
```
!pip install category-encoders
import os
import random
import pandas as pd
import numpy as np
import missingno as msno
import seaborn as sns
import matplotlib.cm as cm
import matplotlib.pyplot as plt
from category_encoders import *
from sklearn.preprocessing import RobustScaler
from sklearn.model_selection import train_test_split
# Set seed untuk mendapatkan reproducible result
def set_seed(s):
os.environ['PYTHONHASHSEED']=str(s)
random.seed(s)
np.random.seed(s)
```
### Load Datasets
```
path = "/content/drive/MyDrive/Semester 5/Pembelajaran Mesin/Tugas II"
dftrain = pd.read_csv(path+"/kendaraan_train.csv")
dftest = pd.read_csv(path+"/kendaraan_test.csv")
```
## Exploratory Data Analysis
### Melihat Cuplikan Data
```
df = dftrain.copy()
df.head()
```
### Melihat Informasi pada Data
```
df.info()
```
### Deteksi Missing Value
#### Menggunakan missingno library untuk melihat persebaran data yang hilang (missing value)
```
msno.matrix(df)
```
Melihat secara presisi berapa ukuran dari data training
```
df.shape
```
Data yang kita dapat cukup banyak, lalu kita lihat seberapa banyak missing value pada setiap kolomnya
```
df.isna().sum()
df.nunique()
```
Persebaran kelas label begitu timpang, maka akan dilakukan pula Synthetic Minority Over-sampling Technique (SMOTE) untuk menangani data yang imbalance.
## Prapemrosesan
### Drop NaN value
```
df = df.dropna()
print("Jumlah data setelah di-drop sebanyak : "+str(df.shape[0]))
print("Baris yang di-drop sebanyak : "+ str(int(100*(285831-171068)/285831)) + " persen")
```
### Drop Kolom id
```
df = df.drop(['id'], axis=1)
```
### Persebaran Kelas Label
```
x=df.Tertarik.value_counts()
sns.barplot(x.index, x)
plt.gca().set_ylabel('sampel')
```
### Handling Ouliers
### Mengubah Semua data menjadi Numerik (Hanya untuk Keperluan Boxplotting)
```
def convert_category(dfraw):
cat = dfraw.unique()
i2c = {}
c2i = {}
for i, v in enumerate(cat):
i2c[i] = v
c2i[v] = i
return i2c, c2i
def apply_transform(dfraw, col):
cache = {}
for x in col:
i2c, c2i = convert_category(dfraw[x])
dfraw[x] = dfraw[x].apply(lambda x: float(c2i[x]))
cache[x] = (i2c, c2i)
return dfraw, cache
col = ["Jenis_Kelamin", "Umur_Kendaraan", "Kendaraan_Rusak"]
cache = {}
df, cache = apply_transform(df, col)
df.head()
cache
```
#### Boxplotting
```
kolom = df.columns[:-1]
f, axes = plt.subplots(2, 5, figsize=(20,7), gridspec_kw={'wspace': 0.4,'hspace': 0.4})
for i, v in enumerate(kolom):
if i < 5:
sns.boxplot(y=df[v], orient='v' , ax=axes[0][i])
else:
sns.boxplot(y=df[v], orient='v' , ax=axes[1][i%5])
```
#### Melihat deskripsi data
```
df.describe()
```
#### Analisis
Pada data yang kita olah, **tidak terdapat outlier**. Walaupun data premi pada boxplot menunjukkan banyak titik-titik di luar plot, namun kita pahami bersama bahwa premi merupakan jumlah premi yang harus dibayarkan per tahun. Tentu saja bisa terjadi banyak data yang distribusinya beda, karena memang bisa saja setiap orang memiliki banyak produk asuransi yang berbeda, bahkan perbedaannya bisa signifikan. Oleh karena itu, bisa dinyatakan bahwa karakteristik data premi itu sendiri di dunia nyata merupakan sesuatu yang bukan anomali/outlier. Alasan yang sama dengan kolom SIM, bukanlah outlier karena tidak memiliki SIM bukanlah suatu anomali.
# Data tanpa Proses Encoding
```
dfw_enc = dftrain.copy()
dfw_enc = dfw_enc.dropna()
dfw_enc = dfw_enc.drop("id", axis=1)
dfw_enc.head()
dftw_enc = dftest.copy()
dftw_enc.head()
dfw_enc_ = dfw_enc.drop("Tertarik", axis=1)
set_seed(42)
xt, xte, yt, yte = train_test_split(dfw_enc_, dfw_enc["Tertarik"], test_size=0.1, random_state=42, stratify=dfw_enc["Tertarik"])
xt["Tertarik"] = yt
xte["Tertarik"] = yte
xt.to_csv(path+"/without encoding/training.csv", index=False)
xte.to_csv(path+"/without encoding/validation.csv", index=False)
dftw_enc.to_csv(path+"/without encoding/testing.csv", index=False)
```
# Data dengan Proses Encoding
### Count Encoding
```
col = ["Jenis_Kelamin", "Umur_Kendaraan", "Kendaraan_Rusak"]
Xt_count = xt.drop("Tertarik", axis=1)
cache = {}
Xt_count, cache = apply_transform(Xt_count, col)
Xt_count["Tertarik"] = yt.copy()
Xt_count
col = ["Jenis_Kelamin", "Umur_Kendaraan", "Kendaraan_Rusak"]
Xte_count = xte.drop("Tertarik", axis=1)
cache = {}
Xte_count, cache = apply_transform(Xte_count, col)
Xte_count["Tertarik"] = yte.copy()
Xte_count
col = ["Jenis_Kelamin", "Umur_Kendaraan", "Kendaraan_Rusak"]
Xtest_count = dftw_enc.drop("Tertarik", axis=1)
cache = {}
Xtest_count, cache = apply_transform(Xtest_count, col)
Xtest_count["Tertarik"] = dftw_enc["Tertarik"]
Xtest_count
#Xt_count.to_csv(path+"/encoded/count encoding/training.csv", index=False)
#Xte_count.to_csv(path+"/encoded/count encoding/validation.csv", index=False)
#Xtest_count.to_csv(path+"/encoded/count encoding/testing.csv", index=False)
```
## Target Encoding
```
bunch = dftrain.copy()
bunch = bunch.dropna()
bunch = bunch.drop("id", axis=1)
y = bunch.Tertarik
X = bunch.drop("Tertarik", axis=1)
enc = TargetEncoder(cols=X.columns).fit(X, y)
numeric_dataset = enc.transform(X)
test = dftest.drop("Tertarik", axis=1)
testing_numeric_dataset = enc.transform(test)
set_seed(42)
qt, qte, wt, wte = train_test_split(numeric_dataset, y, test_size=0.1, random_state=42, stratify=y)
qt["Tertarik"] = wt
qte["Tertarik"] = wte
testing_numeric_dataset["Tertarik"] = dftest["Tertarik"]
qt.to_csv(path+"/encoded/training.csv", index=False)
qte.to_csv(path+"/encoded/validation.csv", index=False)
testing_numeric_dataset.to_csv(path+"/encoded/testing.csv", index=False)
```
# Synthetic Minority Over-sampling Technique (SMOTE) pada Data Training
```
from imblearn.over_sampling import SMOTE
sm = SMOTE(random_state = 42)
```
### Data with Count Encoding
```
X_train = xt.drop("Tertarik", axis=1)
y_train = yt.copy()
print("Before OverSampling, counts of label '1': {}".format(sum(y_train == 1)))
print("Before OverSampling, counts of label '0': {} \n".format(sum(y_train == 0)))
col = ["Jenis_Kelamin", "Umur_Kendaraan", "Kendaraan_Rusak"]
cache = {}
X_train, cache = apply_transform(X_train, col)
X_train_res, y_train_res = sm.fit_resample(X_train, y_train.ravel())
print('After OverSampling, the shape of train_X: {}'.format(X_train_res.shape))
print('After OverSampling, the shape of train_y: {} \n'.format(y_train_res.shape))
print("After OverSampling, counts of label '1': {}".format(sum(y_train_res == 1)))
print("After OverSampling, counts of label '0': {}".format(sum(y_train_res == 0)))
X_train_res
def make_int(dat):
if dat > 0.5:
return 1
else:
return 0
X_train_res.Jenis_Kelamin = X_train_res.Jenis_Kelamin.apply(make_int)
X_train_res.Sudah_Asuransi= X_train_res.Sudah_Asuransi.apply(make_int)
X_train_res.Umur_Kendaraan= X_train_res.Umur_Kendaraan.apply(make_int)
for i in X_train_res.columns:
X_train_res[i] = X_train_res[i].apply(lambda x: int(x))
X_train_res["Tertarik"] = y_train_res
```
### Data without Encoding
```
cache
col = ["Jenis_Kelamin", "Umur_Kendaraan", "Kendaraan_Rusak"]
def inverse_transform(dfraw, col, cache):
for x in col:
i2c, _ = cache[x]
dfraw[x] = dfraw[x].apply(lambda x: i2c[x])
return dfraw
Xw_train_res = inverse_transform(X_train_res, col, cache)
Xw_train_res
```
### Data with Target Encoding
```
bunch = X_train_res.copy()
bunch = bunch.dropna()
y = bunch.Tertarik
X = bunch.drop("Tertarik", axis=1)
enc = TargetEncoder(cols=X.columns).fit(X, y)
numeric_dataset = enc.transform(X)
dftrain = numeric_dataset.copy()
dftrain["Tertarik"] = y
dfvalid = pd.read_csv("/content/drive/MyDrive/Semester 5/Pembelajaran Mesin/Tugas II/encoded/count encoding/validation.csv")
valid = dfvalid.drop("Tertarik", axis=1)
x_val = enc.transform(valid)
x_val["Tertarik"] = dfvalid["Tertarik"]
x_val
# x_val.to_csv("/content/drive/MyDrive/Semester 5/Pembelajaran Mesin/Tugas II/encoded/validation.csv",index=False)
dftest = pd.read_csv("/content/drive/MyDrive/Semester 5/Pembelajaran Mesin/Tugas II/encoded/count encoding/testing.csv")
test = dfvalid.drop("Tertarik", axis=1)
x_test = enc.transform(test)
x_test["Tertarik"] = dftest["Tertarik"]
x_test
#x_test.to_csv("/content/drive/MyDrive/Semester 5/Pembelajaran Mesin/Tugas II/encoded/testing.csv",index=False)
```
# TomekLinks UnderSampling
```
from imblearn.under_sampling import TomekLinks
tl = TomekLinks(sampling_strategy='not minority')
col = ["Jenis_Kelamin", "Umur_Kendaraan", "Kendaraan_Rusak"]
cache = {}
X_train, cache = apply_transform(X_train, col)
print("Before UnderSampling, counts of label '1': {}".format(sum(y_train == 1)))
print("Before UnderSampling, counts of label '0': {} \n".format(sum(y_train == 0)))
X_tl, y_tl = tl.fit_resample(X_train, y_train.ravel())
print('After UnderSampling, the shape of train_X: {}'.format(X_tl.shape))
print('After UnderSampling, the shape of train_y: {} \n'.format(y_tl.shape))
print("After UnderSampling, counts of label '1': {}".format(sum(y_tl == 1)))
print("After UnderSampling, counts of label '0': {}".format(sum(y_tl == 0)))
X_tl["Tertarik"] = y_tl
#X_tl.to_csv("tomek_training.csv", index=False)
bunch = X_tl.copy()
y = bunch.Tertarik
X = bunch.drop("Tertarik", axis=1)
enc = TargetEncoder(cols=X.columns).fit(X, y)
Xtom_enc = enc.transform(X)
Xtom_enc["Tertarik"] = y
Xtom_enc.to_csv("targetencoding_tomek_training.csv", index=False)
```
|
github_jupyter
|
!pip install category-encoders
import os
import random
import pandas as pd
import numpy as np
import missingno as msno
import seaborn as sns
import matplotlib.cm as cm
import matplotlib.pyplot as plt
from category_encoders import *
from sklearn.preprocessing import RobustScaler
from sklearn.model_selection import train_test_split
# Set seed untuk mendapatkan reproducible result
def set_seed(s):
os.environ['PYTHONHASHSEED']=str(s)
random.seed(s)
np.random.seed(s)
path = "/content/drive/MyDrive/Semester 5/Pembelajaran Mesin/Tugas II"
dftrain = pd.read_csv(path+"/kendaraan_train.csv")
dftest = pd.read_csv(path+"/kendaraan_test.csv")
df = dftrain.copy()
df.head()
df.info()
msno.matrix(df)
df.shape
df.isna().sum()
df.nunique()
df = df.dropna()
print("Jumlah data setelah di-drop sebanyak : "+str(df.shape[0]))
print("Baris yang di-drop sebanyak : "+ str(int(100*(285831-171068)/285831)) + " persen")
df = df.drop(['id'], axis=1)
x=df.Tertarik.value_counts()
sns.barplot(x.index, x)
plt.gca().set_ylabel('sampel')
def convert_category(dfraw):
cat = dfraw.unique()
i2c = {}
c2i = {}
for i, v in enumerate(cat):
i2c[i] = v
c2i[v] = i
return i2c, c2i
def apply_transform(dfraw, col):
cache = {}
for x in col:
i2c, c2i = convert_category(dfraw[x])
dfraw[x] = dfraw[x].apply(lambda x: float(c2i[x]))
cache[x] = (i2c, c2i)
return dfraw, cache
col = ["Jenis_Kelamin", "Umur_Kendaraan", "Kendaraan_Rusak"]
cache = {}
df, cache = apply_transform(df, col)
df.head()
cache
kolom = df.columns[:-1]
f, axes = plt.subplots(2, 5, figsize=(20,7), gridspec_kw={'wspace': 0.4,'hspace': 0.4})
for i, v in enumerate(kolom):
if i < 5:
sns.boxplot(y=df[v], orient='v' , ax=axes[0][i])
else:
sns.boxplot(y=df[v], orient='v' , ax=axes[1][i%5])
df.describe()
dfw_enc = dftrain.copy()
dfw_enc = dfw_enc.dropna()
dfw_enc = dfw_enc.drop("id", axis=1)
dfw_enc.head()
dftw_enc = dftest.copy()
dftw_enc.head()
dfw_enc_ = dfw_enc.drop("Tertarik", axis=1)
set_seed(42)
xt, xte, yt, yte = train_test_split(dfw_enc_, dfw_enc["Tertarik"], test_size=0.1, random_state=42, stratify=dfw_enc["Tertarik"])
xt["Tertarik"] = yt
xte["Tertarik"] = yte
xt.to_csv(path+"/without encoding/training.csv", index=False)
xte.to_csv(path+"/without encoding/validation.csv", index=False)
dftw_enc.to_csv(path+"/without encoding/testing.csv", index=False)
col = ["Jenis_Kelamin", "Umur_Kendaraan", "Kendaraan_Rusak"]
Xt_count = xt.drop("Tertarik", axis=1)
cache = {}
Xt_count, cache = apply_transform(Xt_count, col)
Xt_count["Tertarik"] = yt.copy()
Xt_count
col = ["Jenis_Kelamin", "Umur_Kendaraan", "Kendaraan_Rusak"]
Xte_count = xte.drop("Tertarik", axis=1)
cache = {}
Xte_count, cache = apply_transform(Xte_count, col)
Xte_count["Tertarik"] = yte.copy()
Xte_count
col = ["Jenis_Kelamin", "Umur_Kendaraan", "Kendaraan_Rusak"]
Xtest_count = dftw_enc.drop("Tertarik", axis=1)
cache = {}
Xtest_count, cache = apply_transform(Xtest_count, col)
Xtest_count["Tertarik"] = dftw_enc["Tertarik"]
Xtest_count
#Xt_count.to_csv(path+"/encoded/count encoding/training.csv", index=False)
#Xte_count.to_csv(path+"/encoded/count encoding/validation.csv", index=False)
#Xtest_count.to_csv(path+"/encoded/count encoding/testing.csv", index=False)
bunch = dftrain.copy()
bunch = bunch.dropna()
bunch = bunch.drop("id", axis=1)
y = bunch.Tertarik
X = bunch.drop("Tertarik", axis=1)
enc = TargetEncoder(cols=X.columns).fit(X, y)
numeric_dataset = enc.transform(X)
test = dftest.drop("Tertarik", axis=1)
testing_numeric_dataset = enc.transform(test)
set_seed(42)
qt, qte, wt, wte = train_test_split(numeric_dataset, y, test_size=0.1, random_state=42, stratify=y)
qt["Tertarik"] = wt
qte["Tertarik"] = wte
testing_numeric_dataset["Tertarik"] = dftest["Tertarik"]
qt.to_csv(path+"/encoded/training.csv", index=False)
qte.to_csv(path+"/encoded/validation.csv", index=False)
testing_numeric_dataset.to_csv(path+"/encoded/testing.csv", index=False)
from imblearn.over_sampling import SMOTE
sm = SMOTE(random_state = 42)
X_train = xt.drop("Tertarik", axis=1)
y_train = yt.copy()
print("Before OverSampling, counts of label '1': {}".format(sum(y_train == 1)))
print("Before OverSampling, counts of label '0': {} \n".format(sum(y_train == 0)))
col = ["Jenis_Kelamin", "Umur_Kendaraan", "Kendaraan_Rusak"]
cache = {}
X_train, cache = apply_transform(X_train, col)
X_train_res, y_train_res = sm.fit_resample(X_train, y_train.ravel())
print('After OverSampling, the shape of train_X: {}'.format(X_train_res.shape))
print('After OverSampling, the shape of train_y: {} \n'.format(y_train_res.shape))
print("After OverSampling, counts of label '1': {}".format(sum(y_train_res == 1)))
print("After OverSampling, counts of label '0': {}".format(sum(y_train_res == 0)))
X_train_res
def make_int(dat):
if dat > 0.5:
return 1
else:
return 0
X_train_res.Jenis_Kelamin = X_train_res.Jenis_Kelamin.apply(make_int)
X_train_res.Sudah_Asuransi= X_train_res.Sudah_Asuransi.apply(make_int)
X_train_res.Umur_Kendaraan= X_train_res.Umur_Kendaraan.apply(make_int)
for i in X_train_res.columns:
X_train_res[i] = X_train_res[i].apply(lambda x: int(x))
X_train_res["Tertarik"] = y_train_res
cache
col = ["Jenis_Kelamin", "Umur_Kendaraan", "Kendaraan_Rusak"]
def inverse_transform(dfraw, col, cache):
for x in col:
i2c, _ = cache[x]
dfraw[x] = dfraw[x].apply(lambda x: i2c[x])
return dfraw
Xw_train_res = inverse_transform(X_train_res, col, cache)
Xw_train_res
bunch = X_train_res.copy()
bunch = bunch.dropna()
y = bunch.Tertarik
X = bunch.drop("Tertarik", axis=1)
enc = TargetEncoder(cols=X.columns).fit(X, y)
numeric_dataset = enc.transform(X)
dftrain = numeric_dataset.copy()
dftrain["Tertarik"] = y
dfvalid = pd.read_csv("/content/drive/MyDrive/Semester 5/Pembelajaran Mesin/Tugas II/encoded/count encoding/validation.csv")
valid = dfvalid.drop("Tertarik", axis=1)
x_val = enc.transform(valid)
x_val["Tertarik"] = dfvalid["Tertarik"]
x_val
# x_val.to_csv("/content/drive/MyDrive/Semester 5/Pembelajaran Mesin/Tugas II/encoded/validation.csv",index=False)
dftest = pd.read_csv("/content/drive/MyDrive/Semester 5/Pembelajaran Mesin/Tugas II/encoded/count encoding/testing.csv")
test = dfvalid.drop("Tertarik", axis=1)
x_test = enc.transform(test)
x_test["Tertarik"] = dftest["Tertarik"]
x_test
#x_test.to_csv("/content/drive/MyDrive/Semester 5/Pembelajaran Mesin/Tugas II/encoded/testing.csv",index=False)
from imblearn.under_sampling import TomekLinks
tl = TomekLinks(sampling_strategy='not minority')
col = ["Jenis_Kelamin", "Umur_Kendaraan", "Kendaraan_Rusak"]
cache = {}
X_train, cache = apply_transform(X_train, col)
print("Before UnderSampling, counts of label '1': {}".format(sum(y_train == 1)))
print("Before UnderSampling, counts of label '0': {} \n".format(sum(y_train == 0)))
X_tl, y_tl = tl.fit_resample(X_train, y_train.ravel())
print('After UnderSampling, the shape of train_X: {}'.format(X_tl.shape))
print('After UnderSampling, the shape of train_y: {} \n'.format(y_tl.shape))
print("After UnderSampling, counts of label '1': {}".format(sum(y_tl == 1)))
print("After UnderSampling, counts of label '0': {}".format(sum(y_tl == 0)))
X_tl["Tertarik"] = y_tl
#X_tl.to_csv("tomek_training.csv", index=False)
bunch = X_tl.copy()
y = bunch.Tertarik
X = bunch.drop("Tertarik", axis=1)
enc = TargetEncoder(cols=X.columns).fit(X, y)
Xtom_enc = enc.transform(X)
Xtom_enc["Tertarik"] = y
Xtom_enc.to_csv("targetencoding_tomek_training.csv", index=False)
| 0.261142 | 0.759671 |
**[PE1-01]**
Import modules.
```
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib
matplotlib.rcParams['font.size'] = 12
```
**[PE1-02]**
Define the Gridworld class.
```
class Gridworld:
def __init__(self, size=8, goals=[7], penalty=0):
self.size = size
self.goals = goals
self.penalty = penalty
self.states = range(size)
self.actions = [-1, 1]
self.policy = {}
self.value = {}
for s in self.states:
self.value[s] = 0
def move(self, s, a):
if s in self.goals:
return 0, s # Reward, Next state
s_new = s + a
if s_new not in self.states:
return self.penalty, s # Reward, Next state
if s_new in self.goals:
return 1, s_new # Reward, Next state
return self.penalty, s_new # Reward, Next state
```
**[PE1-03]**
Define a function to show state values.
```
def show_values(world, subplot=None, title='Values'):
if not subplot:
fig = plt.figure(figsize=(world.size*0.8, 1.7))
subplot = fig.add_subplot(1, 1, 1)
result = np.zeros([1, world.size])
for s in world.states:
result[0][s] = world.value[s]
sns.heatmap(result, square=True, cbar=False, yticklabels=[],
annot=True, fmt='3.1f', cmap='coolwarm',
ax=subplot).set_title(title)
```
**[PE1-04]**
Define a function to apply the policy evaluation algorithm.
```
def policy_eval(world, gamma=1.0, trace=False):
if trace:
fig = plt.figure(figsize=(world.size*0.8, len(world.states)*1.7))
for s in world.states:
v_new = 0
for a in world.actions:
r, s_new = world.move(s, a)
v_new += world.policy[(s, a)] * (r + gamma * world.value[s_new])
world.value[s] = v_new
if trace:
subplot = fig.add_subplot(world.size, 1, s+1)
show_values(world, subplot, title='Update on s={}'.format(s))
```
**[PE1-05]**
Create a gridworld instance and define a policy to keep moving right.
```
world = Gridworld(size=8, goals=[7])
for s in world.states:
world.policy[(s, 1)] = 1
world.policy[(s, -1)] = 0
show_values(world, title='Initial values')
```
**[PE1-06]**
Apply the policy evaluation algorithm for a single iteration.
```
policy_eval(world, trace=True)
```
**[PE1-07]**
Apply the policy evaluation algorithm for one more iteration.
```
policy_eval(world, trace=True)
```
**[PE1-08]**
Apply the policy evaluation untile it acieves the correct values.
```
for _ in range(5):
policy_eval(world)
show_values(world, title='Final values')
```
**[PE1-09]**
Create a gridworld instance and define a policy to move randomly.
```
world = Gridworld(size=8, goals=[7])
for s in world.states:
world.policy[(s, -1)] = 1/2
world.policy[(s, 1)] = 1/2
show_values(world, title='Initial values')
```
**[PE1-10]**
Apply the policy evaluation algorithm for three iterations.
```
policy_eval(world)
show_values(world, title='First iteration')
policy_eval(world)
show_values(world, title='Second iteration')
policy_eval(world)
show_values(world, title='Third iteration')
```
**[PE1-11]**
Apply the policy evaluation algorithm until the conversion.
```
for _ in range(100):
policy_eval(world)
show_values(world, title='Final values')
```
**[PE1-12]**
Create a gridworld instance with a penalty, and define a policy to to keep moving right.
```
world = Gridworld(size=8, goals=[7], penalty=-1)
for s in world.states:
world.policy[(s, 1)] = 1
world.policy[(s, -1)] = 0
show_values(world, title='Initial values')
```
**[PE1-13]**
Apply the policy evaluation algorithm for three iterations.
```
policy_eval(world)
show_values(world, title='First iteration')
policy_eval(world)
show_values(world, title='Second iteration')
policy_eval(world)
show_values(world, title='Third iteration')
```
**[PE1-14]**
Apply the policy evaluation algorithm until the conversion.
```
for _ in range(100):
policy_eval(world)
show_values(world, title='Final values')
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib
matplotlib.rcParams['font.size'] = 12
class Gridworld:
def __init__(self, size=8, goals=[7], penalty=0):
self.size = size
self.goals = goals
self.penalty = penalty
self.states = range(size)
self.actions = [-1, 1]
self.policy = {}
self.value = {}
for s in self.states:
self.value[s] = 0
def move(self, s, a):
if s in self.goals:
return 0, s # Reward, Next state
s_new = s + a
if s_new not in self.states:
return self.penalty, s # Reward, Next state
if s_new in self.goals:
return 1, s_new # Reward, Next state
return self.penalty, s_new # Reward, Next state
def show_values(world, subplot=None, title='Values'):
if not subplot:
fig = plt.figure(figsize=(world.size*0.8, 1.7))
subplot = fig.add_subplot(1, 1, 1)
result = np.zeros([1, world.size])
for s in world.states:
result[0][s] = world.value[s]
sns.heatmap(result, square=True, cbar=False, yticklabels=[],
annot=True, fmt='3.1f', cmap='coolwarm',
ax=subplot).set_title(title)
def policy_eval(world, gamma=1.0, trace=False):
if trace:
fig = plt.figure(figsize=(world.size*0.8, len(world.states)*1.7))
for s in world.states:
v_new = 0
for a in world.actions:
r, s_new = world.move(s, a)
v_new += world.policy[(s, a)] * (r + gamma * world.value[s_new])
world.value[s] = v_new
if trace:
subplot = fig.add_subplot(world.size, 1, s+1)
show_values(world, subplot, title='Update on s={}'.format(s))
world = Gridworld(size=8, goals=[7])
for s in world.states:
world.policy[(s, 1)] = 1
world.policy[(s, -1)] = 0
show_values(world, title='Initial values')
policy_eval(world, trace=True)
policy_eval(world, trace=True)
for _ in range(5):
policy_eval(world)
show_values(world, title='Final values')
world = Gridworld(size=8, goals=[7])
for s in world.states:
world.policy[(s, -1)] = 1/2
world.policy[(s, 1)] = 1/2
show_values(world, title='Initial values')
policy_eval(world)
show_values(world, title='First iteration')
policy_eval(world)
show_values(world, title='Second iteration')
policy_eval(world)
show_values(world, title='Third iteration')
for _ in range(100):
policy_eval(world)
show_values(world, title='Final values')
world = Gridworld(size=8, goals=[7], penalty=-1)
for s in world.states:
world.policy[(s, 1)] = 1
world.policy[(s, -1)] = 0
show_values(world, title='Initial values')
policy_eval(world)
show_values(world, title='First iteration')
policy_eval(world)
show_values(world, title='Second iteration')
policy_eval(world)
show_values(world, title='Third iteration')
for _ in range(100):
policy_eval(world)
show_values(world, title='Final values')
| 0.592902 | 0.893449 |
```
#Importing Libraries
import torch
import random
import numpy as np
import os
from os import path
import pandas as pd
import matplotlib.pyplot as plt
import num2words
import re, string, timeit
import random
import glob
import transformers
from sklearn.metrics import confusion_matrix, accuracy_score, classification_report
from itertools import combinations
%matplotlib inline
tokenizer = transformers.BertTokenizerFast.from_pretrained("dccuchile/bert-base-spanish-wwm-uncased",do_lower_case=True)
model = transformers.BertForPreTraining.from_pretrained("dccuchile/bert-base-spanish-wwm-uncased")
def generate_next_sentence(text):
sentence_a = []
sentence_b = []
label = []
n_lines = len(text['sentence'])
for i, line in enumerate(text['sentence']):
# 50/50 whether is IsNextSentence or NotNextSentence
if random.random() >= 0.5:
# this is IsNextSentence
sentence_a.append(line)
sentence_b.append(text['next_sentence'][i])
label.append(0)
else:
index = random.randint(0, n_lines-1)
# this is NotNextSentence
sentence_a.append(line)
sentence_b.append(text['next_sentence'][index])
label.append(1)
return sentence_a, sentence_b, label
def mask_for_mlm(inputs):
# create random array of floats with equal dimensions to input_ids tensor
rand = torch.rand(inputs.input_ids.shape)
# create mask array
mask_arr = (rand < 0.15) * (inputs.input_ids != 4) * \
(inputs.input_ids != 5) * (inputs.input_ids != 1)
selection = []
for i in range(inputs.input_ids.shape[0]):
selection.append(
torch.flatten(mask_arr[i].nonzero()).tolist()
)
for i in range(inputs.input_ids.shape[0]):
inputs.input_ids[i, selection[i]] = 0
return(inputs)
def generate_inputs_from_text(text):
sentence_a, sentence_b, label = generate_next_sentence(text)
inputs = tokenizer(sentence_a, sentence_b, return_tensors='pt',
max_length=512, truncation=True, padding='max_length')
inputs['next_sentence_label'] = torch.LongTensor([label]).T
inputs['labels'] = inputs.input_ids.detach().clone()
inputs = mask_for_mlm(inputs)
return(inputs)
class CorpusDataset(torch.utils.data.Dataset):
def __init__(self, encodings):
self.encodings = encodings
def __getitem__(self, idx):
return {key: torch.tensor(val[idx]) for key, val in self.encodings.items()}
def __len__(self):
return len(self.encodings.input_ids)
def train_BertForPreTrainig(model, text):
inputs = generate_inputs_from_text(text)
dataset = CorpusDataset(inputs)
training_args = transformers.TrainingArguments(
output_dir='./results', # output directory
num_train_epochs=3, # total # of training epochs
per_device_train_batch_size=16, # batch size per device during training
per_device_eval_batch_size=64, # batch size for evaluation
warmup_steps=500, # number of warmup steps for learning rate scheduler
weight_decay=0.01, # strength of weight decay
logging_dir='./logs') # directory for storing logs
trainer = transformers.Trainer(
model=model, # the instantiated Transformers model to be trained
args=training_args, # training arguments, defined above
train_dataset=dataset, # training dataset
eval_dataset=dataset) # evaluation dataset
return(model)
```
|
github_jupyter
|
#Importing Libraries
import torch
import random
import numpy as np
import os
from os import path
import pandas as pd
import matplotlib.pyplot as plt
import num2words
import re, string, timeit
import random
import glob
import transformers
from sklearn.metrics import confusion_matrix, accuracy_score, classification_report
from itertools import combinations
%matplotlib inline
tokenizer = transformers.BertTokenizerFast.from_pretrained("dccuchile/bert-base-spanish-wwm-uncased",do_lower_case=True)
model = transformers.BertForPreTraining.from_pretrained("dccuchile/bert-base-spanish-wwm-uncased")
def generate_next_sentence(text):
sentence_a = []
sentence_b = []
label = []
n_lines = len(text['sentence'])
for i, line in enumerate(text['sentence']):
# 50/50 whether is IsNextSentence or NotNextSentence
if random.random() >= 0.5:
# this is IsNextSentence
sentence_a.append(line)
sentence_b.append(text['next_sentence'][i])
label.append(0)
else:
index = random.randint(0, n_lines-1)
# this is NotNextSentence
sentence_a.append(line)
sentence_b.append(text['next_sentence'][index])
label.append(1)
return sentence_a, sentence_b, label
def mask_for_mlm(inputs):
# create random array of floats with equal dimensions to input_ids tensor
rand = torch.rand(inputs.input_ids.shape)
# create mask array
mask_arr = (rand < 0.15) * (inputs.input_ids != 4) * \
(inputs.input_ids != 5) * (inputs.input_ids != 1)
selection = []
for i in range(inputs.input_ids.shape[0]):
selection.append(
torch.flatten(mask_arr[i].nonzero()).tolist()
)
for i in range(inputs.input_ids.shape[0]):
inputs.input_ids[i, selection[i]] = 0
return(inputs)
def generate_inputs_from_text(text):
sentence_a, sentence_b, label = generate_next_sentence(text)
inputs = tokenizer(sentence_a, sentence_b, return_tensors='pt',
max_length=512, truncation=True, padding='max_length')
inputs['next_sentence_label'] = torch.LongTensor([label]).T
inputs['labels'] = inputs.input_ids.detach().clone()
inputs = mask_for_mlm(inputs)
return(inputs)
class CorpusDataset(torch.utils.data.Dataset):
def __init__(self, encodings):
self.encodings = encodings
def __getitem__(self, idx):
return {key: torch.tensor(val[idx]) for key, val in self.encodings.items()}
def __len__(self):
return len(self.encodings.input_ids)
def train_BertForPreTrainig(model, text):
inputs = generate_inputs_from_text(text)
dataset = CorpusDataset(inputs)
training_args = transformers.TrainingArguments(
output_dir='./results', # output directory
num_train_epochs=3, # total # of training epochs
per_device_train_batch_size=16, # batch size per device during training
per_device_eval_batch_size=64, # batch size for evaluation
warmup_steps=500, # number of warmup steps for learning rate scheduler
weight_decay=0.01, # strength of weight decay
logging_dir='./logs') # directory for storing logs
trainer = transformers.Trainer(
model=model, # the instantiated Transformers model to be trained
args=training_args, # training arguments, defined above
train_dataset=dataset, # training dataset
eval_dataset=dataset) # evaluation dataset
return(model)
| 0.550607 | 0.269518 |
```
import re
def normalize_line(line):
line = re.sub(r"[\"-]+", r" ", line)
line = re.sub(r"[ ]+", r" ", line)
return line
movie_lines = ""
with open("./movie_lines.tsv", "r") as file_in:
for movie_line in file_in:
movie_line = normalize_line(movie_line)
movie_lines += movie_line
movie_lines[:100]
import pandas as pd
from io import StringIO
frame = None
frame = pd.read_csv(
StringIO(movie_lines),
sep = "\t",
header = None,
parse_dates = False,
error_bad_lines = False,
warn_bad_lines = False)
frame
from nltk import sent_tokenize
import unidecode
preprocessed_sentences = []
def preprocess_sentence(sentence):
sentence = unidecode.unidecode(sentence)
sentence = re.sub(r"[-+]?(\d+([.,]\d*)?|[.,]\d+)([eE][-+]?\d+)?", r"#", sentence)
sentence = re.sub(r"<[/]?[a-z0-9]+>", r"", sentence)
sentence = re.sub(r""", r"", sentence)
sentence = re.sub(r"&", r"", sentence)
sentence = re.sub(r"<", r"", sentence)
sentence = re.sub(r">", r"", sentence)
sentence = re.sub(r"&emdash;", r"", sentence)
sentence = re.sub(r"`", r"'", sentence)
sentence = re.sub(r"[ ]+['][ ]+", r"'", sentence)
sentence = re.sub(r"[ ]+['](?=[a-zA-Z ])", r" ", sentence)
sentence = re.sub(r"(?<=[a-zA-Z])['][ ]+", r" ", sentence)
sentence = re.sub(r"[\.]{2,}", r" ", sentence)
sentence = re.sub(r"[$%&=|~<>/_\^\[\]{}():;,+*!?]+", r" ", sentence)
sentence = re.sub(r"[ ]+", r" ", sentence)
sentence = sentence.strip()
sentence = re.sub(r"(?<=[a-zA-Z])[']$", r"", sentence)
sentence = re.sub(r"^['](?=[a-zA-Z])", r"", sentence)
sentence = re.sub(r"[\.][']$", r"", sentence)
sentence = re.sub(r"['][\.]$", r"", sentence)
sentence = re.sub(r"^[ ]", r"", sentence)
sentence = re.sub(r"[ ]$", r"", sentence)
sentence = re.sub(r"[\.]$", r"", sentence)
sentence = sentence.strip()
return sentence
def preprocess_and_append_line(line):
sentences = sent_tokenize(line)
for sentence in sentences:
sentence = sentence.strip()
sentence = preprocess_sentence(sentence)
if (sentence != ""):
preprocessed_sentences.append(sentence)
frame[~pd.isnull(frame[4])][4].apply(preprocess_and_append_line)
chars = set()
for s in preprocessed_sentences:
for c in s:
chars.add(c)
chars
preprocessed_sentences[:500]
from sklearn.model_selection import train_test_split
indices = range(len(preprocessed_sentences))
seed = 2092093
train, test, train_indices, test_indices = train_test_split(
preprocessed_sentences,
indices,
train_size = 0.8,
test_size = 0.2,
random_state = seed)
test, valid, test_indices, valid_indices = train_test_split(
test,
test_indices,
train_size = 0.5,
test_size = 0.5,
random_state = seed)
len(preprocessed_sentences)
len(train_indices)
len(test_indices)
len(valid_indices)
len(train_indices) / len(preprocessed_sentences)
len(test_indices) / len(preprocessed_sentences)
len(valid_indices) / len(preprocessed_sentences)
def write_data(data, file_path):
with open(file_path, "w") as file_out:
for line in data:
file_out.write(line + "\n")
write_data(train, "train.txt")
write_data(test, "test.txt")
write_data(valid, "valid.txt")
```
|
github_jupyter
|
import re
def normalize_line(line):
line = re.sub(r"[\"-]+", r" ", line)
line = re.sub(r"[ ]+", r" ", line)
return line
movie_lines = ""
with open("./movie_lines.tsv", "r") as file_in:
for movie_line in file_in:
movie_line = normalize_line(movie_line)
movie_lines += movie_line
movie_lines[:100]
import pandas as pd
from io import StringIO
frame = None
frame = pd.read_csv(
StringIO(movie_lines),
sep = "\t",
header = None,
parse_dates = False,
error_bad_lines = False,
warn_bad_lines = False)
frame
from nltk import sent_tokenize
import unidecode
preprocessed_sentences = []
def preprocess_sentence(sentence):
sentence = unidecode.unidecode(sentence)
sentence = re.sub(r"[-+]?(\d+([.,]\d*)?|[.,]\d+)([eE][-+]?\d+)?", r"#", sentence)
sentence = re.sub(r"<[/]?[a-z0-9]+>", r"", sentence)
sentence = re.sub(r""", r"", sentence)
sentence = re.sub(r"&", r"", sentence)
sentence = re.sub(r"<", r"", sentence)
sentence = re.sub(r">", r"", sentence)
sentence = re.sub(r"&emdash;", r"", sentence)
sentence = re.sub(r"`", r"'", sentence)
sentence = re.sub(r"[ ]+['][ ]+", r"'", sentence)
sentence = re.sub(r"[ ]+['](?=[a-zA-Z ])", r" ", sentence)
sentence = re.sub(r"(?<=[a-zA-Z])['][ ]+", r" ", sentence)
sentence = re.sub(r"[\.]{2,}", r" ", sentence)
sentence = re.sub(r"[$%&=|~<>/_\^\[\]{}():;,+*!?]+", r" ", sentence)
sentence = re.sub(r"[ ]+", r" ", sentence)
sentence = sentence.strip()
sentence = re.sub(r"(?<=[a-zA-Z])[']$", r"", sentence)
sentence = re.sub(r"^['](?=[a-zA-Z])", r"", sentence)
sentence = re.sub(r"[\.][']$", r"", sentence)
sentence = re.sub(r"['][\.]$", r"", sentence)
sentence = re.sub(r"^[ ]", r"", sentence)
sentence = re.sub(r"[ ]$", r"", sentence)
sentence = re.sub(r"[\.]$", r"", sentence)
sentence = sentence.strip()
return sentence
def preprocess_and_append_line(line):
sentences = sent_tokenize(line)
for sentence in sentences:
sentence = sentence.strip()
sentence = preprocess_sentence(sentence)
if (sentence != ""):
preprocessed_sentences.append(sentence)
frame[~pd.isnull(frame[4])][4].apply(preprocess_and_append_line)
chars = set()
for s in preprocessed_sentences:
for c in s:
chars.add(c)
chars
preprocessed_sentences[:500]
from sklearn.model_selection import train_test_split
indices = range(len(preprocessed_sentences))
seed = 2092093
train, test, train_indices, test_indices = train_test_split(
preprocessed_sentences,
indices,
train_size = 0.8,
test_size = 0.2,
random_state = seed)
test, valid, test_indices, valid_indices = train_test_split(
test,
test_indices,
train_size = 0.5,
test_size = 0.5,
random_state = seed)
len(preprocessed_sentences)
len(train_indices)
len(test_indices)
len(valid_indices)
len(train_indices) / len(preprocessed_sentences)
len(test_indices) / len(preprocessed_sentences)
len(valid_indices) / len(preprocessed_sentences)
def write_data(data, file_path):
with open(file_path, "w") as file_out:
for line in data:
file_out.write(line + "\n")
write_data(train, "train.txt")
write_data(test, "test.txt")
write_data(valid, "valid.txt")
| 0.274351 | 0.344788 |
```
import ephem
import matplotlib.pyplot as plt
import matplotlib.patches as patches
import numpy as np
import matplotlib as mpl
#Lets define some lines and angles to be used in our design, based on what we computed.
x = np.linspace(-30,30,30)
y = [line(x,equinox_rise[1]),line(x,summer_solstice_set[1]),line(x,summer_solstice_rise[1])]
angle=(-summer_solstice_rise[1]+np.pi/4)/np.pi*180
anglerad=-summer_solstice_rise[1]+np.pi/4
rayshighx=np.array([x[4],10*(-np.cos(anglerad)-np.sin(anglerad))])
rayshighy=np.array([y[2][4],10*(np.cos(anglerad)-np.sin(anglerad))])
rayslowx=np.array([x[4],10*(np.cos(anglerad)+np.sin(anglerad))])
rayslowy=np.array([y[2][4],10*(-np.cos(anglerad)+np.sin(anglerad))])
def line(x,m):
return np.tan(-m+np.pi/2)*x
def draw_lines(x,y,text=None):
for i in range(3):
ax.plot(x,y[i])
ax.scatter(x[4],y[i][4],marker='o',color='black')
ax.scatter(x[25],y[i][25],marker='o',color='black')
if text!=None:
ax.text(x[4]-2,y[i][4]+1,text[i][0])
ax.text(x[25]-14,y[i][4]+1,text[i][1])
def draw_pyramid(ax):
for i in range (2,10):
j=12-i
if (j==3):rectangle=patches.Rectangle((-j,-j),2*j,2*j, color='black', alpha=0.5)
else: rectangle=patches.Rectangle((-j,-j),2*j,2*j, color='red', alpha=0.2)
t = mpl.transforms.Affine2D().rotate_deg_around(0,0,angle) + ax.transData
rectangle.set_transform(t)
ax.add_patch(rectangle)
#ax.add_patch(rectangle2)
for i in range (4):
a=[[-1.5,3],[-1.5,-11],[3,-1.5],[-11,-1.5]]
if i<=1:rectangle=patches.Rectangle((a[i]),3,8, color='black', alpha=0.5)
else:rectangle=patches.Rectangle((a[i]),8,3, color='black', alpha=0.5)
t = mpl.transforms.Affine2D().rotate_deg_around(0,0,angle) + ax.transData
rectangle.set_transform(t)
ax.add_patch(rectangle)
fig1,ax= plt.subplots(figsize=(7,6))
ax.set_xlim(-25,25)
ax.set_ylim(-25,25)
text=[["Equinoccio otoño/verano\nAmanecer \n%s"%equinox_rise[0].strftime("%d/%m/%Y %H:%M:%S"),"Equinoccio otoño/verano\nAtardecer \n%s" %equinox_set[0].strftime("%d/%m/%Y %H:%M:%S")],
["Solsticio de verano\nAtardecer - Junio 21\n%s"%summer_solstice_rise[0].strftime("%d/%m/%Y %H:%M:%S"),"Solsticio de verano\nAmanecer - Junio 21\n%s"%summer_solstice_set[0].strftime("%d/%m/%Y %H:%M:%S")],
["Solsticio de invierno\nAtardecer - Diciembre 21\n%s"%winter_solstice_set[0].strftime("%d/%m/%Y %H:%M:%S"),"Solsticio de invierno\nAmanecer - Diciembre 21\n%s" %winter_solstice_rise[0].strftime("%d/%m/%Y %H:%M:%S")]]
draw_lines(x,y,text=text)
circle=patches.Circle((0,0),radius=4, color='cyan', alpha=1)
ax.add_patch(circle)
fig1.savefig("images/orientation.pdf")
fig2,ax= plt.subplots(figsize=(7,6))
ax.set_xlim(-25,25)
ax.set_ylim(-25,25)
text=[["Equinoccios\n\nAmanecer","Equinoccios\n\nAtardecer"],
["Atardecer\nJunio 21","Amanecer\nJunio 21"],["Atardecer\nDiciembre 21","Amanecer\nDiciembre 21"]]
draw_lines(x,y,text=text)
circle=patches.Circle((0,0),radius=4, color='cyan', alpha=1)
ax.add_patch(circle)
draw_pyramid(ax=ax)
fig2.savefig("images/PyramidOrientation.pdf")
fig3,ax= plt.subplots(figsize=(7,6))
ax.set_xlim(-25,25)
ax.set_ylim(-25,25)
for i in range(3):
ax.plot(x,y[i])
ax.scatter(x[4],y[i][4],marker='o',color='black')
ax.scatter(x[25],y[i][25],marker='o',color='black')
circle=patches.Circle((0,0),radius=4, color='cyan', alpha=1)
ax.add_patch(circle)
draw_pyramid(ax=ax)
ax.fill(np.append(rayshighx, rayslowx[::-1]),np.append(rayshighy, rayslowy[::-1]), color='yellow', alpha=0.2)
ax.text(x[4],y[2][4]-7,"Atardecer - %s\nSombra completa en mitad de la pirámide" %winter_solstice_set[0].strftime("%d/%m/%Y %H:%M:%S"))
fig3.savefig("images/WinterShadow.pdf")
fig4,ax= plt.subplots(figsize=(7,6))
ax.set_xlim(-25,25)
ax.set_ylim(-25,25)
for i in range(3):
ax.plot(x,y[i])
ax.scatter(x[4],y[i][4],marker='o',color='black')
ax.scatter(x[25],y[i][25],marker='o',color='black')
circle=patches.Circle((0,0),radius=4, color='cyan', alpha=1)
ax.add_patch(circle)
draw_pyramid(ax=ax)
for i in range(6):
raystairsx=np.array([x[4],-1.5*np.cos(anglerad)-(11-i*1.5)*np.sin(anglerad)])
raystairsy=np.array([y[0][4],(11-i*1.5)*np.cos(anglerad)-1.5*np.sin(anglerad)])
ax.plot(raystairsx,raystairsy,'y-')
ax.text(x[4],y[1][4]+5,"Atardecer - %s\nSe iluminan escalones de la serpiente (los 7 triángulos de luz)" %summer_solstice_set[0].strftime("%d/%m/%Y %H:%M:%S"))
fig4.savefig("images/TrianglesOfLight.pdf")
fig5,ax= plt.subplots(figsize=(7,6))
img= plt.imread("images/kukulkan.png")
ax.imshow(img)
rectangle=patches.Rectangle((188,178),190,190, color='red', alpha=0.5)
t = mpl.transforms.Affine2D().rotate_deg_around(294,264,-angle+0*45) + ax.transData
rectangle.set_transform(t)
ax.add_patch(rectangle)
ax.text(120,70,"Chichen Itza: 20.6829703, -88.5692032\nTu locación: %s, %s"%(lat,lon), backgroundcolor='white')
fig4.savefig("images/YourPyramidVsChichenItza.pdf")
```
|
github_jupyter
|
import ephem
import matplotlib.pyplot as plt
import matplotlib.patches as patches
import numpy as np
import matplotlib as mpl
#Lets define some lines and angles to be used in our design, based on what we computed.
x = np.linspace(-30,30,30)
y = [line(x,equinox_rise[1]),line(x,summer_solstice_set[1]),line(x,summer_solstice_rise[1])]
angle=(-summer_solstice_rise[1]+np.pi/4)/np.pi*180
anglerad=-summer_solstice_rise[1]+np.pi/4
rayshighx=np.array([x[4],10*(-np.cos(anglerad)-np.sin(anglerad))])
rayshighy=np.array([y[2][4],10*(np.cos(anglerad)-np.sin(anglerad))])
rayslowx=np.array([x[4],10*(np.cos(anglerad)+np.sin(anglerad))])
rayslowy=np.array([y[2][4],10*(-np.cos(anglerad)+np.sin(anglerad))])
def line(x,m):
return np.tan(-m+np.pi/2)*x
def draw_lines(x,y,text=None):
for i in range(3):
ax.plot(x,y[i])
ax.scatter(x[4],y[i][4],marker='o',color='black')
ax.scatter(x[25],y[i][25],marker='o',color='black')
if text!=None:
ax.text(x[4]-2,y[i][4]+1,text[i][0])
ax.text(x[25]-14,y[i][4]+1,text[i][1])
def draw_pyramid(ax):
for i in range (2,10):
j=12-i
if (j==3):rectangle=patches.Rectangle((-j,-j),2*j,2*j, color='black', alpha=0.5)
else: rectangle=patches.Rectangle((-j,-j),2*j,2*j, color='red', alpha=0.2)
t = mpl.transforms.Affine2D().rotate_deg_around(0,0,angle) + ax.transData
rectangle.set_transform(t)
ax.add_patch(rectangle)
#ax.add_patch(rectangle2)
for i in range (4):
a=[[-1.5,3],[-1.5,-11],[3,-1.5],[-11,-1.5]]
if i<=1:rectangle=patches.Rectangle((a[i]),3,8, color='black', alpha=0.5)
else:rectangle=patches.Rectangle((a[i]),8,3, color='black', alpha=0.5)
t = mpl.transforms.Affine2D().rotate_deg_around(0,0,angle) + ax.transData
rectangle.set_transform(t)
ax.add_patch(rectangle)
fig1,ax= plt.subplots(figsize=(7,6))
ax.set_xlim(-25,25)
ax.set_ylim(-25,25)
text=[["Equinoccio otoño/verano\nAmanecer \n%s"%equinox_rise[0].strftime("%d/%m/%Y %H:%M:%S"),"Equinoccio otoño/verano\nAtardecer \n%s" %equinox_set[0].strftime("%d/%m/%Y %H:%M:%S")],
["Solsticio de verano\nAtardecer - Junio 21\n%s"%summer_solstice_rise[0].strftime("%d/%m/%Y %H:%M:%S"),"Solsticio de verano\nAmanecer - Junio 21\n%s"%summer_solstice_set[0].strftime("%d/%m/%Y %H:%M:%S")],
["Solsticio de invierno\nAtardecer - Diciembre 21\n%s"%winter_solstice_set[0].strftime("%d/%m/%Y %H:%M:%S"),"Solsticio de invierno\nAmanecer - Diciembre 21\n%s" %winter_solstice_rise[0].strftime("%d/%m/%Y %H:%M:%S")]]
draw_lines(x,y,text=text)
circle=patches.Circle((0,0),radius=4, color='cyan', alpha=1)
ax.add_patch(circle)
fig1.savefig("images/orientation.pdf")
fig2,ax= plt.subplots(figsize=(7,6))
ax.set_xlim(-25,25)
ax.set_ylim(-25,25)
text=[["Equinoccios\n\nAmanecer","Equinoccios\n\nAtardecer"],
["Atardecer\nJunio 21","Amanecer\nJunio 21"],["Atardecer\nDiciembre 21","Amanecer\nDiciembre 21"]]
draw_lines(x,y,text=text)
circle=patches.Circle((0,0),radius=4, color='cyan', alpha=1)
ax.add_patch(circle)
draw_pyramid(ax=ax)
fig2.savefig("images/PyramidOrientation.pdf")
fig3,ax= plt.subplots(figsize=(7,6))
ax.set_xlim(-25,25)
ax.set_ylim(-25,25)
for i in range(3):
ax.plot(x,y[i])
ax.scatter(x[4],y[i][4],marker='o',color='black')
ax.scatter(x[25],y[i][25],marker='o',color='black')
circle=patches.Circle((0,0),radius=4, color='cyan', alpha=1)
ax.add_patch(circle)
draw_pyramid(ax=ax)
ax.fill(np.append(rayshighx, rayslowx[::-1]),np.append(rayshighy, rayslowy[::-1]), color='yellow', alpha=0.2)
ax.text(x[4],y[2][4]-7,"Atardecer - %s\nSombra completa en mitad de la pirámide" %winter_solstice_set[0].strftime("%d/%m/%Y %H:%M:%S"))
fig3.savefig("images/WinterShadow.pdf")
fig4,ax= plt.subplots(figsize=(7,6))
ax.set_xlim(-25,25)
ax.set_ylim(-25,25)
for i in range(3):
ax.plot(x,y[i])
ax.scatter(x[4],y[i][4],marker='o',color='black')
ax.scatter(x[25],y[i][25],marker='o',color='black')
circle=patches.Circle((0,0),radius=4, color='cyan', alpha=1)
ax.add_patch(circle)
draw_pyramid(ax=ax)
for i in range(6):
raystairsx=np.array([x[4],-1.5*np.cos(anglerad)-(11-i*1.5)*np.sin(anglerad)])
raystairsy=np.array([y[0][4],(11-i*1.5)*np.cos(anglerad)-1.5*np.sin(anglerad)])
ax.plot(raystairsx,raystairsy,'y-')
ax.text(x[4],y[1][4]+5,"Atardecer - %s\nSe iluminan escalones de la serpiente (los 7 triángulos de luz)" %summer_solstice_set[0].strftime("%d/%m/%Y %H:%M:%S"))
fig4.savefig("images/TrianglesOfLight.pdf")
fig5,ax= plt.subplots(figsize=(7,6))
img= plt.imread("images/kukulkan.png")
ax.imshow(img)
rectangle=patches.Rectangle((188,178),190,190, color='red', alpha=0.5)
t = mpl.transforms.Affine2D().rotate_deg_around(294,264,-angle+0*45) + ax.transData
rectangle.set_transform(t)
ax.add_patch(rectangle)
ax.text(120,70,"Chichen Itza: 20.6829703, -88.5692032\nTu locación: %s, %s"%(lat,lon), backgroundcolor='white')
fig4.savefig("images/YourPyramidVsChichenItza.pdf")
| 0.176281 | 0.558327 |

# Train POS Tagger in French by Spark NLP
### Based on Universal Dependency `UD_French-GSD` version 2.3
```
import sys
import time
#Spark ML and SQL
from pyspark.ml import Pipeline, PipelineModel
from pyspark.sql.functions import array_contains
from pyspark.sql import SparkSession
from pyspark.sql.types import StructType, StructField, IntegerType, StringType
#Spark NLP
import sparknlp
from sparknlp.annotator import *
from sparknlp.common import RegexRule
from sparknlp.base import DocumentAssembler, Finisher
```
### Let's create a Spark Session for our app
```
spark = sparknlp.start()
print("Spark NLP version: ", sparknlp.version())
print("Apache Spark version: ", spark.version)
```
Let's prepare our training datasets containing `token_posTag` like `de_DET`. You can download this data set from Amazon S3:
```
wget -N https://s3.amazonaws.com/auxdata.johnsnowlabs.com/public/resources/fr/pos/UD_French/UD_French-GSD_2.3.txt -P /tmp
```
```
! wget -N https://s3.amazonaws.com/auxdata.johnsnowlabs.com/public/resources/fr/pos/UD_French/UD_French-GSD_2.3.txt -P /tmp
from sparknlp.training import POS
training_data = POS().readDataset(
spark=spark,
path="/tmp/UD_French-GSD_2.3.txt",
delimiter="_",
outputPosCol="tags",
outputDocumentCol="document",
outputTextCol="text"
)
training_data.show()
document_assembler = DocumentAssembler() \
.setInputCol("text")
sentence_detector = SentenceDetector() \
.setInputCols(["document"]) \
.setOutputCol("sentence")
tokenizer = Tokenizer() \
.setInputCols(["sentence"]) \
.setOutputCol("token")\
.setPrefixPattern("\\A([^\\s\\p{L}\\d\\$\\.#]*)")\
.setSuffixPattern("([^\\s\\p{L}\\d]?)([^\\s\\p{L}\\d]*)\\z")\
.setInfixPatterns([
"([\\p{L}\\w]+'{1})",
"([\\$#]?\\d+(?:[^\\s\\d]{1}\\d+)*)",
"((?:\\p{L}\\.)+)",
"((?:\\p{L}+[^\\s\\p{L}]{1})+\\p{L}+)",
"([\\p{L}\\w]+)"
])
posTagger = PerceptronApproach() \
.setNIterations(6) \
.setInputCols(["sentence", "token"]) \
.setOutputCol("pos") \
.setPosCol("tags")
pipeline = Pipeline(stages=[
document_assembler,
sentence_detector,
tokenizer,
posTagger
])
%%time
# Let's train our Pipeline by using our training dataset
model = pipeline.fit(training_data)
```
This is our testing DataFrame where we get some sentences in French. We are going to use our trained Pipeline to transform these sentence and predict each token's `Part Of Speech`.
```
dfTest = spark.createDataFrame([
"Je sens qu'entre ça et les films de médecins et scientifiques fous que nous avons déjà vus, nous pourrions emprunter un autre chemin pour l'origine.",
"On pourra toujours parler à propos d'Averroès de décentrement du Sujet."
], StringType()).toDF("text")
predict = model.transform(dfTest)
predict.select("token.result", "pos.result").show()
```
|
github_jupyter
|
import sys
import time
#Spark ML and SQL
from pyspark.ml import Pipeline, PipelineModel
from pyspark.sql.functions import array_contains
from pyspark.sql import SparkSession
from pyspark.sql.types import StructType, StructField, IntegerType, StringType
#Spark NLP
import sparknlp
from sparknlp.annotator import *
from sparknlp.common import RegexRule
from sparknlp.base import DocumentAssembler, Finisher
spark = sparknlp.start()
print("Spark NLP version: ", sparknlp.version())
print("Apache Spark version: ", spark.version)
wget -N https://s3.amazonaws.com/auxdata.johnsnowlabs.com/public/resources/fr/pos/UD_French/UD_French-GSD_2.3.txt -P /tmp
! wget -N https://s3.amazonaws.com/auxdata.johnsnowlabs.com/public/resources/fr/pos/UD_French/UD_French-GSD_2.3.txt -P /tmp
from sparknlp.training import POS
training_data = POS().readDataset(
spark=spark,
path="/tmp/UD_French-GSD_2.3.txt",
delimiter="_",
outputPosCol="tags",
outputDocumentCol="document",
outputTextCol="text"
)
training_data.show()
document_assembler = DocumentAssembler() \
.setInputCol("text")
sentence_detector = SentenceDetector() \
.setInputCols(["document"]) \
.setOutputCol("sentence")
tokenizer = Tokenizer() \
.setInputCols(["sentence"]) \
.setOutputCol("token")\
.setPrefixPattern("\\A([^\\s\\p{L}\\d\\$\\.#]*)")\
.setSuffixPattern("([^\\s\\p{L}\\d]?)([^\\s\\p{L}\\d]*)\\z")\
.setInfixPatterns([
"([\\p{L}\\w]+'{1})",
"([\\$#]?\\d+(?:[^\\s\\d]{1}\\d+)*)",
"((?:\\p{L}\\.)+)",
"((?:\\p{L}+[^\\s\\p{L}]{1})+\\p{L}+)",
"([\\p{L}\\w]+)"
])
posTagger = PerceptronApproach() \
.setNIterations(6) \
.setInputCols(["sentence", "token"]) \
.setOutputCol("pos") \
.setPosCol("tags")
pipeline = Pipeline(stages=[
document_assembler,
sentence_detector,
tokenizer,
posTagger
])
%%time
# Let's train our Pipeline by using our training dataset
model = pipeline.fit(training_data)
dfTest = spark.createDataFrame([
"Je sens qu'entre ça et les films de médecins et scientifiques fous que nous avons déjà vus, nous pourrions emprunter un autre chemin pour l'origine.",
"On pourra toujours parler à propos d'Averroès de décentrement du Sujet."
], StringType()).toDF("text")
predict = model.transform(dfTest)
predict.select("token.result", "pos.result").show()
| 0.314051 | 0.915809 |
## Análisis de Simulated Annealing para una fuerza de ventas de 5 nodos
Simulated Annhealing es un algoritmo que recibe los siguientes hiperárametros de entrada:
+ Tmax
+ Tmin
+ steps
+ updates
Los resultados obtenidos pueden verse afectados al variar los valores de tales hiperparámetros. Por otro lado, dependiendo del número de nodos del grafo, estos hiperparámetros también podrían afectar la ruta mínima encontrada por el algoritmo.
El presente notebook considera la implementación de simulated annhealing para un grafo con 5 nodos. Como primero objetivo se variarán los hiperparámetros y se identificarán aquellos que den mejores resultados. Entendiéndose como mejores resultados; obtener la ruta con menor distancia. Adicionalmente, una vez seleccionados los mejores hiperparámetros, se correrá el algoritmo 100 veces con el fin de realizar un análisis sobre las rutas obtenidas. Particularmente se tiene interés en revisar variaciones en las rutas obtenidas en cada corrida.
Dentro del conjunto de datos que se tienen disponibles, existen varias fuerzas de venta que deben recorrer 5 nodos. Se decidió elegir la fuerza de venta **80993** perteneciente al estado de Nuevo León para llevar a cabo estas pruebas.
```
!pip install dynaconf
!pip install psycopg2-binary
!pip install simanneal
# Librerías
import pandas as pd
import sys
sys.path.append('../')
%load_ext autoreload
%autoreload 2
from src import Utileria as ut
from src.models import particle_swarm as ps
from src.models import simulated_annealing as sa
```
### 1. Búsqueda de mejores hiperparámetros
**1.1 Definición de datos de entrada**
+ Grafo completo de los puntos que debe vistar la fuerza de ventas
+ Hiperámetros con los que correrá el algoritmo
```
# Se obtiene el dataframe que contiene el grafo de la fuerza de venta a evaluar:
str_Query = 'select id_origen, id_destino, distancia from trabajo.grafos where id_fza_ventas={};'
# En el query se especifica el id_fza_venta del cual se quiere obtener su grafo
df_Grafo = ut.get_data(str_Query.format(80993))
df_Grafo
# Se crea el diccionario de hiper-parámetros que se evaluarán
#Default parameters
#Tmax = 25000.0 # Max (starting) temperature
#Tmin = 2.5 # Min (ending) temperature
#steps = 50000 # Number of iterations
#updates = 100 # Number of updates (by default an update prints to stdout)
dict_Hiper_SA = {'Tmax': {1000,10000, 25000.0, 50000},
'Tmin': {1,2.5,5,10},
'steps': {500,5000,10000},
'updates': {10,50,100,200}
}
```
**1.2 Gridsearch**
Dentro de la clase Utileria fue definido un método llamado *GridSearch*, el cual recibe como parámetros el grafo de una fuerza de ventas fijo, un diccionario de parámetros, el algoritmo a evaluar y el número de iteraciones que se correrá por cada combinación de hiperámetros. Este método evalúa el algoritmo con todas las combinaciones que se pueden generar a partir del diccionario de parámetros. En este caso se considerarán 3 valores de *Iteraciones*, 3 valores del *Número de Partículas*, 2 valores de $\alpha$ y 2 de $\beta$; dando lugar a un total de 36 combinaciones. Cada combinación de hiperarámetros se correrá 100 veces y como resultado se obtendrá una tabla indicando los Hiperámetros utilizados, las distancias mínima y máxima obtenidas dentro de las 100 corridas; y el número de corridas en que se repitió tal distancia mínima.
```
%%time
# Se corre el GridSearch para el grafo y los hiperparámetros previamente definidos
df_Resultado = ut.GridSearch(df_Grafo, sa.SimulatedAnnealing, dict_Hiper_SA, 100)
# Se muestra el dataframe con los resultados obtenidos de la corrida del GridSearch
pd.options.display.max_colwidth = 100
df_Resultado
```
**1.3 Análisis y Resultados**
En primer lugar es importante mencionar las motivaciones de ciertos de los hiperparámetros elegidos.
+ Con respecto al número de iteraciones, $100$ iteraciones fueron elegidas con base en pruebas realizadas en la literatura [](), tomando este número como base, se determinó hacer pruebas considerando la mitad de iteraciones ($50$); y finalmente para tener un valor extremo contra el cuál comparar se eligió dismniur el número de iteraciones incial en un $90%$, dando lugar a $10$ iteraciones.
+ Sobre el número de partículas, la primera idea fue consideran tantas partículas como número de nodos, en este caso $5$; y tomar un número de partículas considerablemente mayor ($100$) y uno considerablemente menor ($2$). En este caso, al tener únicamente $5$ nodos no es posible tomar un número menor de partículas extremo; sin embargo los resultados obtenidos con $1$ partícula aportan resultados muy relevantes para el análisis.
+ Puesto que $\alpha$ y $\beta$ representan probabilidades, los valores que pueden tomar están entre $0$ y $1$. De manera análoga a los otros parámetros, se consideraron valores bajos($0$), medios($0.5$) y altos($1$).
Para poder interpretar los resultados mostrados en el dataframe anterior es conveniente recordar que se realizaron 36 pruebas, es decir, se tuvieron 36 combinaciones de hiperparámetros; y cada una de esas combinaciones se corrió 100 veces, dando un total de 3,600 corridas. En las 100 corridas de cada prueba se registró la distancia mínima obtenida, la distancia máxima y la frecuencia relativa de la distancia mínima. A continuación se enlistan observaciones importantes derivadas de estas pruebas:
+ La distancia mínima obtenida: $5.604 km$ es la misma para las 36 combinaciones; sin embargo la distancia máxima obtenida varía entre $5.604 km$ y $8.559 km$. Esto demuestra que apesar de utilizar los mismos hiperparámetros, el algoritmo puede dar resultados distintos en cada corrida.
+ Al considerar únicamente $1$ partícula y variar el resto de los hiperparámetros, se observa que la frecuencia relativa de la distancia mínima en cada una de las pruebas ($100$ corridas por prueba) es menor al $5\%$.
+ Al fijar el número de partículas en $100$ y variar el resto de los hiperparámetros, se obtienen frecuencias relativas de la distancia mínima mayores al $90\%$. Éstas corresponden a las frecuencias relativas más altas de las 36 pruebas realizadas.
+ En general, al fijar tanto el número de iteraciones como el número de partículas y variar los valores de $\alpha$ y $\beta$, se observa que bajo la siguiente condición $\alpha = \beta = 1$ se obtiene los valores más chicos de frecuencias relativas de la distancia mínima. Al considerar estos parámetros igual a $1$ se está considerando que ambos influyen "totalmente" en la actualización de la posición de cada una de las partículas; ya que tanto $\alpha$ como $\beta$ pueden tomar valores entre $[0,1]$, representando una probabilidad.
+ Con respecto al número de iteraciones, puede decirse que este parámetro depende en gran medida del número de partículas. Por un lado, al considerar $100$ iteraciones y $1$ ó $5$ partículas, las frecuencias relativas de la distancia mínima son bajas. Por otro lado, al disminuir el número de iteraciones a $10$ y aumentar el número de partículas a $100$, tales frecuencias relativas son las más altas.
### 2. Análisis de rutas
```
rutas = pd.DataFrame(index=range(3),columns=['km', 'Ruta'])
# Definición de hiperparámetros
dict_Hiper = {'Iteraciones': 10,
'Particulas': 100,
'Alfa': .5,
'Beta': .5
}
rutas
for corrida in range(3):
PS = ps.ParticleSwarm(df_Grafo,dict_Hiper)
PS.Ejecutar()
min_distancia = round(PS.nbr_MejorCosto,3)
mejor_ruta = PS.lst_MejorCamino
rutas.km[corrida] = min_distancia
rutas.Ruta[corrida] = mejor_ruta
rutas
```
### Conclusiones:
+ Los resultados del algoritmo pueden variar en cada corrida, aún cuando se mantengan fijos los hiperparámetros.
+ Correr el algoritmo una sola vez no garantiza que se obtenga la distancia mínima.
+ Al considerar un número de partículas alrededor de $100$ se tiene mayor probabilidad de encontrar la distancia mínima al correr una sola vez el algoritmo.
+
```
def convert(ruta):
s = [str(i) for i in ruta]
ruta_c = "-".join(s)
return(ruta_c)
```
|
github_jupyter
|
!pip install dynaconf
!pip install psycopg2-binary
!pip install simanneal
# Librerías
import pandas as pd
import sys
sys.path.append('../')
%load_ext autoreload
%autoreload 2
from src import Utileria as ut
from src.models import particle_swarm as ps
from src.models import simulated_annealing as sa
# Se obtiene el dataframe que contiene el grafo de la fuerza de venta a evaluar:
str_Query = 'select id_origen, id_destino, distancia from trabajo.grafos where id_fza_ventas={};'
# En el query se especifica el id_fza_venta del cual se quiere obtener su grafo
df_Grafo = ut.get_data(str_Query.format(80993))
df_Grafo
# Se crea el diccionario de hiper-parámetros que se evaluarán
#Default parameters
#Tmax = 25000.0 # Max (starting) temperature
#Tmin = 2.5 # Min (ending) temperature
#steps = 50000 # Number of iterations
#updates = 100 # Number of updates (by default an update prints to stdout)
dict_Hiper_SA = {'Tmax': {1000,10000, 25000.0, 50000},
'Tmin': {1,2.5,5,10},
'steps': {500,5000,10000},
'updates': {10,50,100,200}
}
%%time
# Se corre el GridSearch para el grafo y los hiperparámetros previamente definidos
df_Resultado = ut.GridSearch(df_Grafo, sa.SimulatedAnnealing, dict_Hiper_SA, 100)
# Se muestra el dataframe con los resultados obtenidos de la corrida del GridSearch
pd.options.display.max_colwidth = 100
df_Resultado
rutas = pd.DataFrame(index=range(3),columns=['km', 'Ruta'])
# Definición de hiperparámetros
dict_Hiper = {'Iteraciones': 10,
'Particulas': 100,
'Alfa': .5,
'Beta': .5
}
rutas
for corrida in range(3):
PS = ps.ParticleSwarm(df_Grafo,dict_Hiper)
PS.Ejecutar()
min_distancia = round(PS.nbr_MejorCosto,3)
mejor_ruta = PS.lst_MejorCamino
rutas.km[corrida] = min_distancia
rutas.Ruta[corrida] = mejor_ruta
rutas
def convert(ruta):
s = [str(i) for i in ruta]
ruta_c = "-".join(s)
return(ruta_c)
| 0.353317 | 0.91611 |
This home assignment consists of several parts.
You are supposed to make some transformations, train the model, estimate its quality and comment on your results.
This task foreshadows the forthcoming large task on ML pipeline and should help you get ready for it.
Several comments:
* Don't hesitate to ask questions, it's a good practice.
* No private/public sharing, please. The copied assignments will be graded with 0 points.
## Data preprocessing, model training and evaluation
### 1. Reading the data
Today we work with the [wine dataset](https://scikit-learn.org/stable/datasets/toy_dataset.html#wine-dataset), describing several wine types for multiclass ($k=3$) classification problem.
The data is available from [scikit-learn](https://scikit-learn.org/stable/modules/generated/sklearn.datasets.load_wine.html) library.
```
import pandas as pd
import numpy as np
from sklearn.datasets import load_wine
from sklearn.model_selection import train_test_split
random_state = 42
features = pd.DataFrame(load_wine(return_X_y=False)['data'],
columns=load_wine(return_X_y=False)['feature_names'])
target = load_wine(return_X_y=False)['target']
features.head(5)
target
```
Let's select *30%* of the data for testing set.
```
# make a train/test split using 30% test size
X_train, X_test, y_train, y_test = train_test_split(features, target,
test_size=0.30,
random_state=random_state)
X_train.shape, X_test.shape, y_train.shape, y_test.shape
```
### 2. Machine Learning Pipeline
Here you are supposed to perform the desired transformations. Please, explain your results briefly after each task.
#### 2.0. Data preprocessing
* Make some transformations of the dataset (if necessary). Briefly explain the transformations in several sentences: do you need the transformation at all and if so, what transform should you applly and why? In case you decide to apply specific pre-processing, remember the selected transformations to re-use them in the necessary Pipeline.
We need to apply normalization to our data because they have different dimensions. I used StandardScaler.
```
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train_std = sc.fit_transform(X_train)
X_test_std = sc.transform(X_test)
X_train_std.shape, X_test_std.shape
```
#### 2.1. Basic logistic regression
* Find optimal hyperparameters for logistic regression with cross-validation on the `train` data (small grid/random search is enough, no need to find the *best* parameters).
* Estimate the model quality with [`f1`](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.f1_score.html) (macro-averaged) and `accuracy` scores.
* Plot a ROC-curve for the trained model. For the multiclass case you might use `scikitplot` library (e.g. `scikitplot.metrics.plot_roc(test_labels, predicted_proba)`).
*Note: please, use the following hyperparameters for logistic regression: `multi_class='multinomial'`, `solver='saga'` `tol=1e-3` and ` max_iter=500`.*
```
# You might use this command to install scikit-plot.
# Warning, if you a running locally, don't call pip from within jupyter, call it from terminal in the corresponding
# virtual environment instead
#! pip install scikit-plot
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import accuracy_score, f1_score
import matplotlib.pyplot as plt
import scikitplot
param_grid_lr = {
'penalty' : ['l1', 'l2'],
'C' : np.linspace(0.1, 10, num=50)
}
grid_lr = GridSearchCV(LogisticRegression(multi_class='multinomial', solver='saga', tol=1e-3, max_iter=500), param_grid_lr, cv=5, verbose=1)
grid_lr.fit(X_train_std, y_train);
lr = grid_lr.best_estimator_
y_pred = lr.predict(X_test_std)
y_pred_proba = lr.predict_proba(X_test_std)
print(f'f1: {f1_score(y_test, y_pred, average="macro")}, accuracy: {accuracy_score(y_test, y_pred)}')
scikitplot.metrics.plot_roc(y_test, y_pred_proba)
plt.show()
```
#### 2.2. PCA: explained variance plot
* Apply the PCA to the train part of the data. Build the explaided variance plot.
```
from sklearn.decomposition import PCA
pca = PCA()
X_train_pca = pca.fit_transform(X_train_std)
exp_var_pca = pca.explained_variance_ratio_
cum_sum_eigenvalues = np.cumsum(exp_var_pca)
plt.bar(range(0,len(exp_var_pca)), exp_var_pca, alpha=0.5, align='center', label='Individual explained variance')
plt.step(range(0,len(cum_sum_eigenvalues)), cum_sum_eigenvalues, where='mid',label='Cumulative explained variance')
plt.ylabel('Explained variance ratio')
plt.xlabel('Principal component index')
plt.legend(loc='best')
plt.tight_layout()
plt.show()
```
* Visualize your data in 2D using another `PCA(num_components=2)` model and `.transform()` method
```
pca = PCA(n_components=2)
X_train_pca = pca.fit_transform(X_train_std)
exp_var_pca = pca.explained_variance_ratio_
cum_sum_eigenvalues = np.cumsum(exp_var_pca)
plt.bar(range(0,len(exp_var_pca)), exp_var_pca, alpha=0.5, align='center', label='Individual explained variance')
plt.step(range(0,len(cum_sum_eigenvalues)), cum_sum_eigenvalues, where='mid',label='Cumulative explained variance')
plt.ylabel('Explained variance ratio')
plt.xlabel('Principal component index')
plt.legend(loc='best')
plt.tight_layout()
plt.show()
```
#### 2.3. PCA trasformation
* Select the appropriate number of components. Briefly explain your choice. Should you normalize the data?
*Use `fit` and `transform` methods to transform the `train` and `test` parts.*
I use n_components=10 because it's enough for good explaining of the data.
```
pca = PCA(n_components=10)
X_train_pca = pca.fit_transform(X_train_std)
X_test_pca = pca.transform(X_test_std)
exp_var_pca = pca.explained_variance_ratio_
cum_sum_eigenvalues = np.cumsum(exp_var_pca)
plt.bar(range(0,len(exp_var_pca)), exp_var_pca, alpha=0.5, align='center', label='Individual explained variance')
plt.step(range(0,len(cum_sum_eigenvalues)), cum_sum_eigenvalues, where='mid',label='Cumulative explained variance')
plt.ylabel('Explained variance ratio')
plt.xlabel('Principal component index')
plt.legend(loc='best')
plt.tight_layout()
plt.show()
```
#### 2.4. Logistic regression on (PCA-)preprocessed data.
* Find optimal hyperparameters for logistic regression with cross-validation on the transformed by PCA `train` data.
* Estimate the model quality with `f1` (macro-averaged) and `accuracy` scores.
* Plot a ROC-curve for the trained model. For the multiclass case you might use `scikitplot` library (e.g. `scikitplot.metrics.plot_roc(test_labels, predicted_proba)`).
*Note: please, use the following hyperparameters for logistic regression: `multi_class='multinomial'`, `solver='saga'` and `tol=1e-3`*
```
param_grid_lr = {
'penalty' : ['l1', 'l2'],
'C' : np.linspace(0.1, 10, num=50)
}
grid_lr = GridSearchCV(LogisticRegression(multi_class='multinomial', solver='saga', tol=1e-3, max_iter=500), param_grid_lr, cv=5, verbose=1)
grid_lr.fit(X_train_pca, y_train);
lr = grid_lr.best_estimator_
y_pred = lr.predict(X_test_pca)
y_pred_proba = lr.predict_proba(X_test_pca)
print(f'f1: {f1_score(y_test, y_pred, average="macro")}, accuracy: {accuracy_score(y_test, y_pred)}')
scikitplot.metrics.plot_roc(y_test, y_pred_proba)
plt.show()
```
#### 2.5. Evaluate resulting model performance on the testing set
* Use the model with the optimal hyperparamteres, fitted to the whole `train` set.
* Estimate resulting `f1` (macro-averaged) and `accuracy` scores for the `test` set from block 1.
```
lr = LogisticRegression(multi_class='multinomial',
solver='saga',
tol=1e-3,
max_iter=500,
C=grid_lr.best_params_['C'],
penalty=grid_lr.best_params_['penalty'])
lr.fit(X_train_std, y_train)
y_pred = lr.predict(X_test_std)
y_pred_proba = lr.predict_proba(X_test_std)
print(f'f1: {f1_score(y_test, y_pred, average="macro")}, accuracy: {accuracy_score(y_test, y_pred)}')
scikitplot.metrics.plot_roc(y_test, y_pred_proba)
plt.show()
```
|
github_jupyter
|
import pandas as pd
import numpy as np
from sklearn.datasets import load_wine
from sklearn.model_selection import train_test_split
random_state = 42
features = pd.DataFrame(load_wine(return_X_y=False)['data'],
columns=load_wine(return_X_y=False)['feature_names'])
target = load_wine(return_X_y=False)['target']
features.head(5)
target
# make a train/test split using 30% test size
X_train, X_test, y_train, y_test = train_test_split(features, target,
test_size=0.30,
random_state=random_state)
X_train.shape, X_test.shape, y_train.shape, y_test.shape
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train_std = sc.fit_transform(X_train)
X_test_std = sc.transform(X_test)
X_train_std.shape, X_test_std.shape
# You might use this command to install scikit-plot.
# Warning, if you a running locally, don't call pip from within jupyter, call it from terminal in the corresponding
# virtual environment instead
#! pip install scikit-plot
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import accuracy_score, f1_score
import matplotlib.pyplot as plt
import scikitplot
param_grid_lr = {
'penalty' : ['l1', 'l2'],
'C' : np.linspace(0.1, 10, num=50)
}
grid_lr = GridSearchCV(LogisticRegression(multi_class='multinomial', solver='saga', tol=1e-3, max_iter=500), param_grid_lr, cv=5, verbose=1)
grid_lr.fit(X_train_std, y_train);
lr = grid_lr.best_estimator_
y_pred = lr.predict(X_test_std)
y_pred_proba = lr.predict_proba(X_test_std)
print(f'f1: {f1_score(y_test, y_pred, average="macro")}, accuracy: {accuracy_score(y_test, y_pred)}')
scikitplot.metrics.plot_roc(y_test, y_pred_proba)
plt.show()
from sklearn.decomposition import PCA
pca = PCA()
X_train_pca = pca.fit_transform(X_train_std)
exp_var_pca = pca.explained_variance_ratio_
cum_sum_eigenvalues = np.cumsum(exp_var_pca)
plt.bar(range(0,len(exp_var_pca)), exp_var_pca, alpha=0.5, align='center', label='Individual explained variance')
plt.step(range(0,len(cum_sum_eigenvalues)), cum_sum_eigenvalues, where='mid',label='Cumulative explained variance')
plt.ylabel('Explained variance ratio')
plt.xlabel('Principal component index')
plt.legend(loc='best')
plt.tight_layout()
plt.show()
pca = PCA(n_components=2)
X_train_pca = pca.fit_transform(X_train_std)
exp_var_pca = pca.explained_variance_ratio_
cum_sum_eigenvalues = np.cumsum(exp_var_pca)
plt.bar(range(0,len(exp_var_pca)), exp_var_pca, alpha=0.5, align='center', label='Individual explained variance')
plt.step(range(0,len(cum_sum_eigenvalues)), cum_sum_eigenvalues, where='mid',label='Cumulative explained variance')
plt.ylabel('Explained variance ratio')
plt.xlabel('Principal component index')
plt.legend(loc='best')
plt.tight_layout()
plt.show()
pca = PCA(n_components=10)
X_train_pca = pca.fit_transform(X_train_std)
X_test_pca = pca.transform(X_test_std)
exp_var_pca = pca.explained_variance_ratio_
cum_sum_eigenvalues = np.cumsum(exp_var_pca)
plt.bar(range(0,len(exp_var_pca)), exp_var_pca, alpha=0.5, align='center', label='Individual explained variance')
plt.step(range(0,len(cum_sum_eigenvalues)), cum_sum_eigenvalues, where='mid',label='Cumulative explained variance')
plt.ylabel('Explained variance ratio')
plt.xlabel('Principal component index')
plt.legend(loc='best')
plt.tight_layout()
plt.show()
param_grid_lr = {
'penalty' : ['l1', 'l2'],
'C' : np.linspace(0.1, 10, num=50)
}
grid_lr = GridSearchCV(LogisticRegression(multi_class='multinomial', solver='saga', tol=1e-3, max_iter=500), param_grid_lr, cv=5, verbose=1)
grid_lr.fit(X_train_pca, y_train);
lr = grid_lr.best_estimator_
y_pred = lr.predict(X_test_pca)
y_pred_proba = lr.predict_proba(X_test_pca)
print(f'f1: {f1_score(y_test, y_pred, average="macro")}, accuracy: {accuracy_score(y_test, y_pred)}')
scikitplot.metrics.plot_roc(y_test, y_pred_proba)
plt.show()
lr = LogisticRegression(multi_class='multinomial',
solver='saga',
tol=1e-3,
max_iter=500,
C=grid_lr.best_params_['C'],
penalty=grid_lr.best_params_['penalty'])
lr.fit(X_train_std, y_train)
y_pred = lr.predict(X_test_std)
y_pred_proba = lr.predict_proba(X_test_std)
print(f'f1: {f1_score(y_test, y_pred, average="macro")}, accuracy: {accuracy_score(y_test, y_pred)}')
scikitplot.metrics.plot_roc(y_test, y_pred_proba)
plt.show()
| 0.658857 | 0.986572 |
# Fine Tuning `BERT` for `Disaster Tweets` Classification
Text classification is a technique for putting text into different categories and has a wide range of applications: email providers use text classification to detect to spam emails, marketing agencies use it for sentiment analysis of customer reviews, and moderators of discussion forums use it to detect inappropriate comments.
In the past, data scientists used methods such as [tf-idf](https://en.wikipedia.org/wiki/Tf%E2%80%93idf), [word2vec](https://en.wikipedia.org/wiki/Word2vec), or [bag-of-words (BOW)](https://en.wikipedia.org/wiki/Bag-of-words_model) to generate features for training classification models. While these techniques have been very successful in many NLP tasks, they don't always capture the meanings of words accurately when they appear in different contexts. Recently, we see increasing interest in using [Bidirectional Encoder Representations from Transformers (BERT)](https://arxiv.org/abs/1810.04805) to achieve better results in text classification tasks, due to its ability more accurately encode the meaning of words in different contexts.
BERT was trained on BookCorpus and English Wikipedia data, which contain 800 million words and 2,500 million words, respectively. Training BERT from scratch would be prohibitively expensive. By taking advantage of transfer learning, one can quickly fine tune BERT for another use case with a relatively small amount of training data to achieve state-of-the-art results for common NLP tasks, such as text classification and question answering.
[Amazon SageMaker](https://docs.aws.amazon.com/sagemaker/index.html) is a fully managed service that provides developers and data scientists with the ability to build, train, and deploy machine learning (ML) models quickly. Amazon SageMaker removes the heavy lifting from each step of the machine learning process to make it easier to develop high-quality models. The [SageMaker Python SDK](https://sagemaker.readthedocs.io/en/stable/) provides open source APIs and containers that make it easy to train and deploy models in Amazon SageMaker with several different machine learning and deep learning frameworks.
In this example, we walk through our dataset, the training process, and finally model deployment.
# About the `Problem`
Twitter has become an important communication channel in times of emergency. The ubiquitousness of smartphones enables people to announce an emergency they’re observing in real-time. Because of this, more agencies are interested in programmatically monitoring Twitter (i.e. disaster relief organizations and news agencies). However, identifying such tweets has always been a difficult task because of the ambiguity in the linguistic structure of the tweets and hence it is not always clear whether an individual’s words are actually announcing a disaster.
More details [here](https://www.kaggle.com/c/nlp-getting-started/overview)
<img src = "img/disaster.png" >
# Installation
```
#!pip install transformers
```
# Setup
To start, we import some Python libraries and initialize a SageMaker session, S3 bucket and prefix, and IAM role.
```
import os
import numpy as np
import pandas as pd
import sagemaker
sagemaker_session = sagemaker.Session() # Provides a collection of methods for working with SageMaker resources
bucket = "aws-sagemaker-nlp-2021"
prefix = "sagemaker/DEMO-pytorch-bert"
role = sagemaker.get_execution_role() # Get the execution role for the notebook instance.
# This is the IAM role that we created for our notebook instance.
# We pass the role to the tuning job(later on).
```
# Prepare training data
Kaggle hosted a challenge named `Real` or `Not` whose aim was to use the Twitter data of disaster tweets, originally created by the company figure-eight, to classify Tweets talking about `real disaster` against the ones talking about it metaphorically. (https://www.kaggle.com/c/nlp-getting-started/overview)
### Get `sentences` and `labels`
Let us take a quick look at our data and for that we need to first read the training data.
The only two columns we interested are:
- the `sentence` (the tweet)
- the `label` (the label, this denotes whether a tweet is about a real disaster (1) or not (0))
```
df = pd.read_csv(
"dataset/raw/data.csv",
header=None,
usecols=[1, 3],
names=["label", "sentence"],
)
sentences = df.sentence.values
labels = df.label.values
df.tail()
```
Printing few tweets with its class label
```
list(zip(sentences[80:85], labels[80:85]))
```
### Cleaning Text
As we can see from the above output, there are few information which are not that important, like `URLs`, `Emojis`, `Tags`, etc. So, now lets try to clean the dataset before we actually pass this data for training.
```
import string
import re
# Helper functions to clean text by removing urls, emojis, html tags and punctuations.
def remove_URL(text):
url = re.compile(r'https?://\S+|www\.\S+')
return url.sub(r'', text)
def remove_emoji(text):
emoji_pattern = re.compile(
'['
u'\U0001F600-\U0001F64F' # emoticons
u'\U0001F300-\U0001F5FF' # symbols & pictographs
u'\U0001F680-\U0001F6FF' # transport & map symbols
u'\U0001F1E0-\U0001F1FF' # flags (iOS)
u'\U00002702-\U000027B0'
u'\U000024C2-\U0001F251'
']+',
flags=re.UNICODE)
return emoji_pattern.sub(r'', text)
def remove_html(text):
html = re.compile(r'<.*?>|&([a-z0-9]+|#[0-9]{1,6}|#x[0-9a-f]{1,6});')
return re.sub(html, '', text)
def remove_punct(text):
table = str.maketrans('', '', string.punctuation)
return text.translate(table)
df['sentence'] = df['sentence'].apply(lambda x: remove_URL(x))
df['sentence'] = df['sentence'].apply(lambda x: remove_emoji(x))
df['sentence'] = df['sentence'].apply(lambda x: remove_html(x))
df['sentence'] = df['sentence'].apply(lambda x: remove_punct(x))
df.head()
sentences = df.sentence.values
labels = df.label.values
list(zip(sentences[80:85], labels[80:85]))
```
# EDA
Let's spend couple of minutes to explore the dataset
## a) Data Distribution
```
import matplotlib.pyplot as plt
import seaborn as sns
fig, axes = plt.subplots(ncols=2, nrows=1, figsize=(18, 6), dpi=100)
sns.countplot(df['label'], ax=axes[0])
axes[1].pie(df['label'].value_counts(),
labels=['Non Disaster', 'Real Disaster'],
autopct='%1.2f%%',
shadow=True,
explode=(0.05, 0),
startangle=60)
fig.suptitle('Distribution of the Tweets', fontsize=24)
plt.show()
```
## b) Word Count
```
import util
df['Word Count'] = df['sentence'].apply(lambda x: len(str(x).split()))
util.plot_dist_char(df[df['label'] == 0], 'Word Count', 'Words Per "Non Disaster Tweet"')
util.plot_dist_char(df[df['label'] == 1], 'Word Count', 'Words Per "Real Disaster"')
```
# Split the dataset for `training` and `testing`
We then split the dataset for training and testing.
```
from sklearn.model_selection import train_test_split
train, test = train_test_split(df) # Default split ratio 75/25, we can modify using "test_size"
train.to_csv("dataset/train.csv", index=False)
test.to_csv("dataset/test.csv", index=False)
```
### Upload both to Amazon S3 for use later
The SageMaker Python SDK provides a helpful function for uploading to Amazon S3:
```
inputs_train = sagemaker_session.upload_data("dataset/train.csv", bucket=bucket, key_prefix=prefix)
inputs_test = sagemaker_session.upload_data("dataset/test.csv", bucket=bucket, key_prefix=prefix)
```
# Amazon SageMaker Training
## Amazon SageMaker
- When running a training job, SageMaker reads input data from Amazon S3, uses that data to train a model.
- Training data from S3 is made available to the Model Training instance container, which is pulled from Amazon Elastic Container Registry(`ECR`).
- The training job persists model artifacts back to the output S3 location designated in the training job configuration.
- When we are ready to deploy a model, SageMaker spins up new ML instances and pulls in these model artifacts to use for batch or real-time model inference.
<img src = "img/sm-training.png" >
## Training script
We use the [PyTorch-Transformers library](https://pytorch.org/hub/huggingface_pytorch-transformers), which contains PyTorch implementations and pre-trained model weights for many NLP models, including BERT.
Our training script should save model artifacts learned during training to a file path called `model_dir`, as stipulated by the SageMaker PyTorch image. Upon completion of training, model artifacts saved in `model_dir` will be uploaded to S3 by SageMaker and will become available in S3 for deployment.
We save this script in a file named `train_deploy.py`, and put the file in a directory named `code/`. The full training script can be viewed under `code/`.
```
!pygmentize code/train_deploy.py
```
# Train on Amazon SageMaker
We use Amazon SageMaker to train and deploy a model using our custom PyTorch code. The Amazon SageMaker Python SDK makes it easier to run a PyTorch script in Amazon SageMaker using its PyTorch estimator. After that, we can use the SageMaker Python SDK to deploy the trained model and run predictions. For more information on how to use this SDK with PyTorch, see [the SageMaker Python SDK documentation](https://sagemaker.readthedocs.io/en/stable/using_pytorch.html).
To start, we use the `PyTorch` estimator class to train our model. When creating our estimator, we make sure to specify a few things:
* `entry_point`: the name of our PyTorch script. It contains our training script, which loads data from the input channels, configures training with hyperparameters, trains a model, and saves a model. It also contains code to load and run the model during inference.
* `source_dir`: the location of our training scripts and requirements.txt file. "requirements.txt" lists packages you want to use with your script.
* `framework_version`: the PyTorch version we want to use
The PyTorch estimator supports multi-machine, distributed PyTorch training. To use this, we just set train_instance_count to be greater than one. Our training script supports distributed training for only GPU instances.
After creating the estimator, we then call fit(), which launches a training job. We use the Amazon S3 URIs where we uploaded the training data earlier.
<img src = "img/sm-estimator.png" >
```
from sagemaker.pytorch import PyTorch
# 1. Defining the estimator
estimator = PyTorch(entry_point="train_deploy.py",
source_dir="code",
role=role,
framework_version="1.5.0",
py_version="py3",
instance_count=2, # Distributed training for GPU instances.
instance_type="ml.p3.2xlarge", # Type of instance we want the training to happen
hyperparameters={"epochs": 2,
"num_labels": 2,
"backend": "gloo", # gloo and tcp for cpu instances - gloo and nccl for gpu instances
}
)
# 2. Start the Training
estimator.fit({"training": inputs_train, "testing": inputs_test})
```
# Host the model on an `Amazon SageMaker Endpoint`
After training our model, we host it on an Amazon SageMaker Endpoint. To make the endpoint load the model and serve predictions, we implement a few methods in `train_deploy.py`.
* `model_fn()`: function defined to load the saved model and return a model object that can be used for model serving. The SageMaker PyTorch model server loads our model by invoking model_fn.
* `input_fn()`: deserializes and prepares the prediction input. In this example, our request body is first serialized to JSON and then sent to model serving endpoint. Therefore, in `input_fn()`, we first deserialize the JSON-formatted request body and return the input as a `torch.tensor`, as required for BERT.
* `predict_fn()`: performs the prediction and returns the result.
To deploy our endpoint, we call `deploy()` on our PyTorch estimator object, passing in our desired number of instances and instance type:
```
predictor = estimator.deploy(initial_instance_count=1, instance_type="ml.m4.xlarge")
```
We then configure the predictor to use `application/json` for the content type when sending requests to our endpoint:
```
predictor.serializer = sagemaker.serializers.JSONSerializer()
predictor.deserializer = sagemaker.deserializers.JSONDeserializer()
```
# Prediction/Inferance
Finally, we use the returned predictor object to call the endpoint:
```
class_label = {1: "Real disaster",
0: "Not a disaster"}
test_sentences = ["I met my friend today by accident",
"Frank had a severe head injury after the car accident last month",
"Just happened a terrible car crash"
]
result = predictor.predict(test_sentences)
result = list(np.argmax(result, axis=1))
predicted_labels = [class_label[l] for l in result]
predicted_labels
for tweet, label in zip(test_sentences, predicted_labels):
print(f"{tweet} ---> {label}")
```
Before moving on, let's delete the Amazon SageMaker endpoint to avoid charges:
# Cleanup
Lastly, please remember to delete the Amazon SageMaker endpoint to avoid charges:
```
#predictor.delete_endpoint()
```
|
github_jupyter
|
#!pip install transformers
import os
import numpy as np
import pandas as pd
import sagemaker
sagemaker_session = sagemaker.Session() # Provides a collection of methods for working with SageMaker resources
bucket = "aws-sagemaker-nlp-2021"
prefix = "sagemaker/DEMO-pytorch-bert"
role = sagemaker.get_execution_role() # Get the execution role for the notebook instance.
# This is the IAM role that we created for our notebook instance.
# We pass the role to the tuning job(later on).
df = pd.read_csv(
"dataset/raw/data.csv",
header=None,
usecols=[1, 3],
names=["label", "sentence"],
)
sentences = df.sentence.values
labels = df.label.values
df.tail()
list(zip(sentences[80:85], labels[80:85]))
import string
import re
# Helper functions to clean text by removing urls, emojis, html tags and punctuations.
def remove_URL(text):
url = re.compile(r'https?://\S+|www\.\S+')
return url.sub(r'', text)
def remove_emoji(text):
emoji_pattern = re.compile(
'['
u'\U0001F600-\U0001F64F' # emoticons
u'\U0001F300-\U0001F5FF' # symbols & pictographs
u'\U0001F680-\U0001F6FF' # transport & map symbols
u'\U0001F1E0-\U0001F1FF' # flags (iOS)
u'\U00002702-\U000027B0'
u'\U000024C2-\U0001F251'
']+',
flags=re.UNICODE)
return emoji_pattern.sub(r'', text)
def remove_html(text):
html = re.compile(r'<.*?>|&([a-z0-9]+|#[0-9]{1,6}|#x[0-9a-f]{1,6});')
return re.sub(html, '', text)
def remove_punct(text):
table = str.maketrans('', '', string.punctuation)
return text.translate(table)
df['sentence'] = df['sentence'].apply(lambda x: remove_URL(x))
df['sentence'] = df['sentence'].apply(lambda x: remove_emoji(x))
df['sentence'] = df['sentence'].apply(lambda x: remove_html(x))
df['sentence'] = df['sentence'].apply(lambda x: remove_punct(x))
df.head()
sentences = df.sentence.values
labels = df.label.values
list(zip(sentences[80:85], labels[80:85]))
import matplotlib.pyplot as plt
import seaborn as sns
fig, axes = plt.subplots(ncols=2, nrows=1, figsize=(18, 6), dpi=100)
sns.countplot(df['label'], ax=axes[0])
axes[1].pie(df['label'].value_counts(),
labels=['Non Disaster', 'Real Disaster'],
autopct='%1.2f%%',
shadow=True,
explode=(0.05, 0),
startangle=60)
fig.suptitle('Distribution of the Tweets', fontsize=24)
plt.show()
import util
df['Word Count'] = df['sentence'].apply(lambda x: len(str(x).split()))
util.plot_dist_char(df[df['label'] == 0], 'Word Count', 'Words Per "Non Disaster Tweet"')
util.plot_dist_char(df[df['label'] == 1], 'Word Count', 'Words Per "Real Disaster"')
from sklearn.model_selection import train_test_split
train, test = train_test_split(df) # Default split ratio 75/25, we can modify using "test_size"
train.to_csv("dataset/train.csv", index=False)
test.to_csv("dataset/test.csv", index=False)
inputs_train = sagemaker_session.upload_data("dataset/train.csv", bucket=bucket, key_prefix=prefix)
inputs_test = sagemaker_session.upload_data("dataset/test.csv", bucket=bucket, key_prefix=prefix)
!pygmentize code/train_deploy.py
from sagemaker.pytorch import PyTorch
# 1. Defining the estimator
estimator = PyTorch(entry_point="train_deploy.py",
source_dir="code",
role=role,
framework_version="1.5.0",
py_version="py3",
instance_count=2, # Distributed training for GPU instances.
instance_type="ml.p3.2xlarge", # Type of instance we want the training to happen
hyperparameters={"epochs": 2,
"num_labels": 2,
"backend": "gloo", # gloo and tcp for cpu instances - gloo and nccl for gpu instances
}
)
# 2. Start the Training
estimator.fit({"training": inputs_train, "testing": inputs_test})
predictor = estimator.deploy(initial_instance_count=1, instance_type="ml.m4.xlarge")
predictor.serializer = sagemaker.serializers.JSONSerializer()
predictor.deserializer = sagemaker.deserializers.JSONDeserializer()
class_label = {1: "Real disaster",
0: "Not a disaster"}
test_sentences = ["I met my friend today by accident",
"Frank had a severe head injury after the car accident last month",
"Just happened a terrible car crash"
]
result = predictor.predict(test_sentences)
result = list(np.argmax(result, axis=1))
predicted_labels = [class_label[l] for l in result]
predicted_labels
for tweet, label in zip(test_sentences, predicted_labels):
print(f"{tweet} ---> {label}")
#predictor.delete_endpoint()
| 0.463201 | 0.992244 |
```
import sys
sys.path.append('../rai/rai/ry')
import numpy as np
import libry as ry
C = ry.Config()
C.addFile('../test/boxProblem.g')
komo = C.komo_path(4., 10, .2, False)
komo.addTimeOptimization()
obj = 'ballR'
S = [
[0., .5], ry.SY.magic, [obj],
[.7, 4.], ry.SY.dynamicTrans, [obj],
[1., 1.], ry.SY.bounce, ["boxBo", obj],
[2., 2.], ry.SY.bounce, ["boxBo", obj],
[3., 3.], ry.SY.bounce, ["boxBo", obj],
[4., 4.], ry.SY.touch, ["target", obj]
]
komo.addSkeleton(S)
komo.optimize(True)
komoView = komo.view()
# read out solution: the full frame path, the tau path (time optimization), list of interaction forces
obj_path = komo.getPathFrames([obj])
tau_path = komo.getPathTau()
forces = komo.getForceInteractions()
forces
D=0
komoView=0
komo=0
C=0
C = ry.Config()
C.addFile("../rai-robotModels/pr2/pr2.g")
C.addFile("../models/tables.g")
C.addFrame("obj0", "table1", "type:ssBox size:[.1 .1 .2 .02] color:[1. 0. 0.], contact, logical={ object }, joint:rigid, Q:<t(0 0 .15)>" )
C.addFrame("obj1", "table1", "type:ssBox size:[.1 .1 .2 .02] color:[1. 0. 0.], contact, logical={ object }, joint:rigid, Q:<t(0 .2 .15)>" )
C.addFrame("obj2", "table1", "type:ssBox size:[.1 .1 .2 .02] color:[1. 0. 0.], contact, logical={ object }, joint:rigid, Q:<t(0 .4 .15)>" )
C.addFrame("obj3", "table1", "type:ssBox size:[.1 .1 .2 .02] color:[1. 0. 0.], contact, logical={ object }, joint:rigid, Q:<t(0 .6.15)>" )
C.addFrame("tray", "table2", "type:ssBox size:[.15 .15 .04 .02] color:[0. 1. 0.], logical={ table }, Q:<t(0 0 .07)>" );
C.addFrame("", "tray", "type:ssBox size:[.27 .27 .04 .02] color:[0. 1. 0.]" )
S =[
[1., 1.], ry.SY.touch, ['pr2R', 'obj0'],
[1., 2.], ry.SY.stable, ['pr2R', 'obj0'],
[2.,2.], ry.SY.touch, ['pr2L', 'obj3'],
[2.,3.], ry.SY.stable, ['pr2L', 'obj3'],
[3.,3.], ry.SY.touch, ['pr2R', 'obj0'],
[3.,3.], ry.SY.above, ['obj0', 'tray'],
[3.,3.], ry.SY.stableOn, ['tray', 'obj0']
]
komo = C.komo_path(3., 10, 10., True)
komo.addSkeletonBound(S, ry.BT.seq, True)
komo.optimize(True)
komoView = komo.view()
komo = C.komo_path(3., 10, 10., True)
komo.addSkeletonBound(S, ry.BT.path, True)
komo.optimize(True)
komoView = komo.view()
D=0
komoView=0
komo=0
C=0
C = ry.Config()
C.addFile("../models/RSSproblem-01.g")
S =[
[1, 1], ry.SY.touch, ['baxterR', 'stick'],
[1, 4], ry.SY.stable, ['baxterR', 'stick'],
[1, 1], ry.SY.liftDownUp, ['baxterR'],
[2, 2], ry.SY.touch, ['baxterL', 'stick'],
[2, 4], ry.SY.stable, ['baxterL', 'stick'],
[3, 3], ry.SY.touch, ['stickTip', 'redBall'],
[3, 3], ry.SY.impulse, ['stickTip', 'redBall'],
[3, 3], ry.SY.dynamicOn, ['table1', 'redBall'],
[4, 4], ry.SY.touch, ['baxterR', 'redBall'],
#[4, 5], ry.SY.stable, ['baxterR', 'redBall'],
[4, 5], ry.SY.graspSlide, ['baxterR', 'redBall', 'table1']
]
komo = C.komo_path(4., 10, 10., False)
komo.addSkeletonBound(S, ry.BT.path, False)
komo.optimize(True)
komoView = komo.view()
```
|
github_jupyter
|
import sys
sys.path.append('../rai/rai/ry')
import numpy as np
import libry as ry
C = ry.Config()
C.addFile('../test/boxProblem.g')
komo = C.komo_path(4., 10, .2, False)
komo.addTimeOptimization()
obj = 'ballR'
S = [
[0., .5], ry.SY.magic, [obj],
[.7, 4.], ry.SY.dynamicTrans, [obj],
[1., 1.], ry.SY.bounce, ["boxBo", obj],
[2., 2.], ry.SY.bounce, ["boxBo", obj],
[3., 3.], ry.SY.bounce, ["boxBo", obj],
[4., 4.], ry.SY.touch, ["target", obj]
]
komo.addSkeleton(S)
komo.optimize(True)
komoView = komo.view()
# read out solution: the full frame path, the tau path (time optimization), list of interaction forces
obj_path = komo.getPathFrames([obj])
tau_path = komo.getPathTau()
forces = komo.getForceInteractions()
forces
D=0
komoView=0
komo=0
C=0
C = ry.Config()
C.addFile("../rai-robotModels/pr2/pr2.g")
C.addFile("../models/tables.g")
C.addFrame("obj0", "table1", "type:ssBox size:[.1 .1 .2 .02] color:[1. 0. 0.], contact, logical={ object }, joint:rigid, Q:<t(0 0 .15)>" )
C.addFrame("obj1", "table1", "type:ssBox size:[.1 .1 .2 .02] color:[1. 0. 0.], contact, logical={ object }, joint:rigid, Q:<t(0 .2 .15)>" )
C.addFrame("obj2", "table1", "type:ssBox size:[.1 .1 .2 .02] color:[1. 0. 0.], contact, logical={ object }, joint:rigid, Q:<t(0 .4 .15)>" )
C.addFrame("obj3", "table1", "type:ssBox size:[.1 .1 .2 .02] color:[1. 0. 0.], contact, logical={ object }, joint:rigid, Q:<t(0 .6.15)>" )
C.addFrame("tray", "table2", "type:ssBox size:[.15 .15 .04 .02] color:[0. 1. 0.], logical={ table }, Q:<t(0 0 .07)>" );
C.addFrame("", "tray", "type:ssBox size:[.27 .27 .04 .02] color:[0. 1. 0.]" )
S =[
[1., 1.], ry.SY.touch, ['pr2R', 'obj0'],
[1., 2.], ry.SY.stable, ['pr2R', 'obj0'],
[2.,2.], ry.SY.touch, ['pr2L', 'obj3'],
[2.,3.], ry.SY.stable, ['pr2L', 'obj3'],
[3.,3.], ry.SY.touch, ['pr2R', 'obj0'],
[3.,3.], ry.SY.above, ['obj0', 'tray'],
[3.,3.], ry.SY.stableOn, ['tray', 'obj0']
]
komo = C.komo_path(3., 10, 10., True)
komo.addSkeletonBound(S, ry.BT.seq, True)
komo.optimize(True)
komoView = komo.view()
komo = C.komo_path(3., 10, 10., True)
komo.addSkeletonBound(S, ry.BT.path, True)
komo.optimize(True)
komoView = komo.view()
D=0
komoView=0
komo=0
C=0
C = ry.Config()
C.addFile("../models/RSSproblem-01.g")
S =[
[1, 1], ry.SY.touch, ['baxterR', 'stick'],
[1, 4], ry.SY.stable, ['baxterR', 'stick'],
[1, 1], ry.SY.liftDownUp, ['baxterR'],
[2, 2], ry.SY.touch, ['baxterL', 'stick'],
[2, 4], ry.SY.stable, ['baxterL', 'stick'],
[3, 3], ry.SY.touch, ['stickTip', 'redBall'],
[3, 3], ry.SY.impulse, ['stickTip', 'redBall'],
[3, 3], ry.SY.dynamicOn, ['table1', 'redBall'],
[4, 4], ry.SY.touch, ['baxterR', 'redBall'],
#[4, 5], ry.SY.stable, ['baxterR', 'redBall'],
[4, 5], ry.SY.graspSlide, ['baxterR', 'redBall', 'table1']
]
komo = C.komo_path(4., 10, 10., False)
komo.addSkeletonBound(S, ry.BT.path, False)
komo.optimize(True)
komoView = komo.view()
| 0.248443 | 0.477859 |
```
import numpy as np
import matplotlib.pyplot as plt
import torch
%matplotlib inline
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
```
## Neural Processes for Images
This notebook contains examples of Neural Processes for images and how these can be used for various tasks like inpainting.
### Load a trained model
```
import json
from neural_process import NeuralProcessImg
# Load config file for celebA model
folder = 'trained_models/celeba'
config_file = folder + '/config.json'
model_file = folder + '/model.pt'
with open(config_file) as f:
config = json.load(f)
# Load trained model
model = NeuralProcessImg(config["img_size"],
config["r_dim"],
config["h_dim"],
config["z_dim"]).to(device)
model.load_state_dict(torch.load(model_file, map_location=lambda storage, loc: storage))
```
### Visualize some CelebA samples
```
import imageio
from torchvision.utils import make_grid
# Read images into torch.Tensor
all_imgs = torch.zeros(8, 3, 32, 32)
for i in range(8):
img = imageio.imread('imgs/celeba-examples/{}.png'.format(i + 1))
all_imgs[i] = torch.Tensor(img.transpose(2, 0, 1) / 255.)
# Visualize sample on a grid
img_grid = make_grid(all_imgs, nrow=4, pad_value=1.)
plt.imshow(img_grid.permute(1, 2, 0).numpy())
```
### Inpainting images with Neural Processes
Inpainting is the task of inferring missing pixels in a partially occluded image. Here we show examples of how Neural Processes can be used to solve this problem.
#### Occluding image
```
# Select one of the images to perform inpainting
img = all_imgs[0]
# Define a binary mask to occlude image. For Neural Processes,
# the context points will be defined as the visible pixels
context_mask = torch.zeros((32, 32)).byte()
context_mask[:16, :] = 1 # Top half of pixels are visible
# Show occluded image
occluded_img = img * context_mask.float()
plt.imshow(occluded_img.permute(1, 2, 0).numpy())
```
#### Generating inpaintings
```
from utils import inpaint
num_inpaintings = 8 # Number of inpaintings to sample from model
all_inpaintings = torch.zeros(num_inpaintings, 3, 32, 32)
# Sample several inpaintings
for i in range(num_inpaintings):
all_inpaintings[i] = inpaint(model, img, context_mask, device)
# Visualize inpainting results on a grid
inpainting_grid = make_grid(all_inpaintings, nrow=4, pad_value=1.)
plt.imshow(inpainting_grid.permute(1, 2, 0).numpy())
```
As can be seen, the inpaintings match the context pixels and are fairly diverse.
#### Different masks
We can use a variety of masks and image to test the model.
```
# Select one of the images to perform inpainting
img = all_imgs[1]
# Define a random mask
context_mask = torch.Tensor(32, 32).uniform_() > 0.9
# Visualize occluded image
occluded_img = img * context_mask.float()
plt.imshow(occluded_img.permute(1, 2, 0).numpy())
num_inpaintings = 8 # Number of inpaintings to sample from model
all_inpaintings = torch.zeros(num_inpaintings, 3, 32, 32)
# Sample several inpaintings
for i in range(num_inpaintings):
all_inpaintings[i] = inpaint(model, img, context_mask, device)
# Visualize inpainting results on a grid
inpainting_grid = make_grid(all_inpaintings, nrow=4, pad_value=1.)
grid_as_np = inpainting_grid.permute(1, 2, 0).numpy()
# If NP returns out of range values for pixels, clip values
plt.imshow(np.clip(grid_as_np, 0, 1))
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import torch
%matplotlib inline
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
import json
from neural_process import NeuralProcessImg
# Load config file for celebA model
folder = 'trained_models/celeba'
config_file = folder + '/config.json'
model_file = folder + '/model.pt'
with open(config_file) as f:
config = json.load(f)
# Load trained model
model = NeuralProcessImg(config["img_size"],
config["r_dim"],
config["h_dim"],
config["z_dim"]).to(device)
model.load_state_dict(torch.load(model_file, map_location=lambda storage, loc: storage))
import imageio
from torchvision.utils import make_grid
# Read images into torch.Tensor
all_imgs = torch.zeros(8, 3, 32, 32)
for i in range(8):
img = imageio.imread('imgs/celeba-examples/{}.png'.format(i + 1))
all_imgs[i] = torch.Tensor(img.transpose(2, 0, 1) / 255.)
# Visualize sample on a grid
img_grid = make_grid(all_imgs, nrow=4, pad_value=1.)
plt.imshow(img_grid.permute(1, 2, 0).numpy())
# Select one of the images to perform inpainting
img = all_imgs[0]
# Define a binary mask to occlude image. For Neural Processes,
# the context points will be defined as the visible pixels
context_mask = torch.zeros((32, 32)).byte()
context_mask[:16, :] = 1 # Top half of pixels are visible
# Show occluded image
occluded_img = img * context_mask.float()
plt.imshow(occluded_img.permute(1, 2, 0).numpy())
from utils import inpaint
num_inpaintings = 8 # Number of inpaintings to sample from model
all_inpaintings = torch.zeros(num_inpaintings, 3, 32, 32)
# Sample several inpaintings
for i in range(num_inpaintings):
all_inpaintings[i] = inpaint(model, img, context_mask, device)
# Visualize inpainting results on a grid
inpainting_grid = make_grid(all_inpaintings, nrow=4, pad_value=1.)
plt.imshow(inpainting_grid.permute(1, 2, 0).numpy())
# Select one of the images to perform inpainting
img = all_imgs[1]
# Define a random mask
context_mask = torch.Tensor(32, 32).uniform_() > 0.9
# Visualize occluded image
occluded_img = img * context_mask.float()
plt.imshow(occluded_img.permute(1, 2, 0).numpy())
num_inpaintings = 8 # Number of inpaintings to sample from model
all_inpaintings = torch.zeros(num_inpaintings, 3, 32, 32)
# Sample several inpaintings
for i in range(num_inpaintings):
all_inpaintings[i] = inpaint(model, img, context_mask, device)
# Visualize inpainting results on a grid
inpainting_grid = make_grid(all_inpaintings, nrow=4, pad_value=1.)
grid_as_np = inpainting_grid.permute(1, 2, 0).numpy()
# If NP returns out of range values for pixels, clip values
plt.imshow(np.clip(grid_as_np, 0, 1))
| 0.827724 | 0.924959 |
```
!pip install --upgrade tables
!pip install eli5
!pip install xgboost
!pip instal hyperopt
import pandas as pd
import numpy as np
import xgboost as xgb
from sklearn.metrics import mean_absolute_error as mae
from sklearn.model_selection import cross_val_score, KFold
from hyperopt import hp,fmin,tpe,STATUS_OK
import eli5
from eli5.sklearn import PermutationImportance
cd "/content/drive/My Drive/Colab Notebooks/dw_matrix/matrix_two/dw_matrix_car"
df=pd.read_hdf('data/car.h5')
df.shape
```
##Feature engineering
```
SUFFIX_CAT='__cat'
for feat in df.columns:
if isinstance(df[feat][0],list):continue
factorized_values=df[feat].factorize()[0]
if SUFFIX_CAT in feat:
df[feat]=factorized_values
else:
df[feat+SUFFIX_CAT]=factorized_values
df['param_rok-produkcji']=df['param_rok-produkcji'].map(lambda x: -1 if str(x)=='None' else int(x))
df['param_moc']= df['param_moc'].map(lambda x: -1 if str(x)=='None' else int(x.split(' ')[0]))
df['param_pojemność-skokowa']=df['param_pojemność-skokowa'].map(lambda x: -1 if str(x)=='None' else int(x.split('cm')[0].replace(' ','')))
def run_model(model,feats):
X=df[feats].values
y=df['price_value'].values
scores=cross_val_score(model,X,y,cv=3,scoring='neg_mean_absolute_error')
return np.mean(scores), np.std(scores)
feats=['param_napęd__cat','param_rok-produkcji','param_stan__cat','param_skrzynia-biegów__cat','param_faktura-vat__cat','param_moc','param_marka-pojazdu__cat','feature_kamera-cofania__cat','param_typ__cat','param_pojemność-skokowa','seller_name__cat','feature_wspomaganie-kierownicy__cat','param_model-pojazdu__cat','param_wersja__cat','param_kod-silnika__cat','feature_system-start-stop__cat','feature_asystent-pasa-ruchu__cat','feature_czujniki-parkowania-przednie__cat','feature_łopatki-zmiany-biegów__cat','feature_regulowane-zawieszenie__cat']
xgb_params={
'max_depth':5,
'n_estimators':50,
'learning_rate':0.1,
'seed':0
}
run_model(xgb.XGBRegressor(**xgb_params),feats)
```
##Hyperopt
```
def obj_func(params):
print("Training with params: ")
print(params)
mean_mae,score_std, =run_model(xgb.XGBRegressor(**params),feats)
return {'loss': np.abs(mean_mae), 'status': STATUS_OK}
#space
xgb_reg_params={
'learning_rate': hp.choice('learning_rate', np.arange(0.05,0.31,0.05)),
'max_depth': hp.choice('max_depth', np.arange(5,16,1, dtype=int)),
'subsample': hp.quniform('subsample', 0.5, 1, 0.05),
'colsample_bytree': hp.quniform('colsample_bytree', 0.5, 1, 0.05),
'objective': 'reg:squarederror',
'n_estimators':100,
'seed':0,
}
##run
best=fmin(obj_func,xgb_reg_params,algo=tpe.suggest,max_evals=25)
best
i
```
|
github_jupyter
|
!pip install --upgrade tables
!pip install eli5
!pip install xgboost
!pip instal hyperopt
import pandas as pd
import numpy as np
import xgboost as xgb
from sklearn.metrics import mean_absolute_error as mae
from sklearn.model_selection import cross_val_score, KFold
from hyperopt import hp,fmin,tpe,STATUS_OK
import eli5
from eli5.sklearn import PermutationImportance
cd "/content/drive/My Drive/Colab Notebooks/dw_matrix/matrix_two/dw_matrix_car"
df=pd.read_hdf('data/car.h5')
df.shape
SUFFIX_CAT='__cat'
for feat in df.columns:
if isinstance(df[feat][0],list):continue
factorized_values=df[feat].factorize()[0]
if SUFFIX_CAT in feat:
df[feat]=factorized_values
else:
df[feat+SUFFIX_CAT]=factorized_values
df['param_rok-produkcji']=df['param_rok-produkcji'].map(lambda x: -1 if str(x)=='None' else int(x))
df['param_moc']= df['param_moc'].map(lambda x: -1 if str(x)=='None' else int(x.split(' ')[0]))
df['param_pojemność-skokowa']=df['param_pojemność-skokowa'].map(lambda x: -1 if str(x)=='None' else int(x.split('cm')[0].replace(' ','')))
def run_model(model,feats):
X=df[feats].values
y=df['price_value'].values
scores=cross_val_score(model,X,y,cv=3,scoring='neg_mean_absolute_error')
return np.mean(scores), np.std(scores)
feats=['param_napęd__cat','param_rok-produkcji','param_stan__cat','param_skrzynia-biegów__cat','param_faktura-vat__cat','param_moc','param_marka-pojazdu__cat','feature_kamera-cofania__cat','param_typ__cat','param_pojemność-skokowa','seller_name__cat','feature_wspomaganie-kierownicy__cat','param_model-pojazdu__cat','param_wersja__cat','param_kod-silnika__cat','feature_system-start-stop__cat','feature_asystent-pasa-ruchu__cat','feature_czujniki-parkowania-przednie__cat','feature_łopatki-zmiany-biegów__cat','feature_regulowane-zawieszenie__cat']
xgb_params={
'max_depth':5,
'n_estimators':50,
'learning_rate':0.1,
'seed':0
}
run_model(xgb.XGBRegressor(**xgb_params),feats)
def obj_func(params):
print("Training with params: ")
print(params)
mean_mae,score_std, =run_model(xgb.XGBRegressor(**params),feats)
return {'loss': np.abs(mean_mae), 'status': STATUS_OK}
#space
xgb_reg_params={
'learning_rate': hp.choice('learning_rate', np.arange(0.05,0.31,0.05)),
'max_depth': hp.choice('max_depth', np.arange(5,16,1, dtype=int)),
'subsample': hp.quniform('subsample', 0.5, 1, 0.05),
'colsample_bytree': hp.quniform('colsample_bytree', 0.5, 1, 0.05),
'objective': 'reg:squarederror',
'n_estimators':100,
'seed':0,
}
##run
best=fmin(obj_func,xgb_reg_params,algo=tpe.suggest,max_evals=25)
best
i
| 0.389198 | 0.415551 |
```
import numpy as np
import matplotlib.pyplot as plt
import scipy.stats
plt.style.use('ggplot')
population = np.random.normal(10,3,30000)
sample = population[np.random.randint(0, 30000, 1000)]
plt.figure(figsize=(10,5))
plt.hist(sample,bins=35)
plt.title("Distribution of 1000 observations sampled from a population of 30,000 with $\mu$=10, $\sigma$=3")
mu_obs = sample.mean()
mu_obs
```
$\sum_i^n -nLog(\sigma_{new}\sqrt{2\pi})-\dfrac{(d_i-\mu_{obs})^2}{2\sigma_{new}^2} + Log(prior(\mu_{obs},\sigma_{new})) \quad > $
$ \sum_i^n -nLog(\sigma_{current}\sqrt{2\pi})-\dfrac{(d_i-\mu_{obs})^2}{2\sigma_{current}^2}+Log(prior(\mu_{obs},\sigma_{current})) $
```
def prior(x):
if x[1] <= 0:
return 1e-7
return 1
def log_gaussian(x, data):
return np.sum(np.log(scipy.stats.norm(x[0],x[1]).pdf(data)))
def acceptance(x, x_new):
if x_new > x:
return True
else:
accept = np.random.uniform(0, 1)
return accept < (np.exp(x_new - x))
def metropolis_hastings(param_init, iterations, data):
x = param_init
accepted = []
rejected = []
for i in range(iterations):
if (i + 1) % 2000 == 0:
print(i + 1)
x_new = [x[0],np.random.normal(x[1],0.5,(1,))]
x_likehood = log_gaussian(x,data)
x_new_likehood = log_gaussian(x_new,data)
x_likehood_prior = x_likehood + np.log(prior(x))
x_new_likehood_prior = x_new_likehood + np.log(prior(x_new))
if acceptance(x_likehood_prior, x_new_likehood_prior):
x = x_new
accepted.append(x)
else:
rejected.append(x_new)
return np.array(accepted), np.array(rejected)
accepted, rejected = metropolis_hastings([mu_obs,0.1], 50000, sample)
plt.figure(figsize=(10,10))
plt.subplot(2, 1, 1)
plt.plot(rejected[0:50,1], 'rx', label='Rejected',alpha=0.5)
plt.plot(accepted[0:50,1], 'b.', label='Accepted',alpha=0.5)
plt.xlabel("Iteration")
plt.ylabel("$\sigma$")
plt.title("MCMC sampling for $\sigma$ with Metropolis-Hastings. First 50 samples are shown.")
plt.legend()
plt.subplot(2, 1, 2)
plt.plot(rejected[-accepted.shape[0]:,1], 'rx', label='Rejected',alpha=0.5)
plt.plot(accepted[-accepted.shape[0]:,1], 'b.', label='Accepted',alpha=0.5)
plt.xlabel("Iteration")
plt.ylabel("$\sigma$")
plt.title("MCMC sampling for $\sigma$ with Metropolis-Hastings.")
plt.legend()
plt.show()
sigmas = accepted[:,1]
sigmas_accept = sigmas.mean() - 0.3
fig = plt.figure(figsize=(15,5))
ax = fig.add_subplot(1,2,1)
ax.plot(sigmas[sigmas > sigmas_accept])
ax.set_title("Trace for $\sigma$")
ax.set_ylabel("$\sigma$")
ax.set_xlabel("Iteration")
ax = fig.add_subplot(1,2,2)
ax.hist(sigmas[sigmas > sigmas_accept], bins=20,density=True)
ax.set_ylabel("Frequency (normed)")
ax.set_xlabel("$\sigma$")
ax.set_title("Histogram of $\sigma$")
plt.show()
mu=accepted[sigmas > sigmas_accept,0].mean()
sigma=accepted[sigmas > sigmas_accept,1].mean()
observation_gen = np.random.normal(mu,sigma,population.shape[0])
fig = plt.figure(figsize=(15,7))
ax = fig.add_subplot(1,1,1)
ax.hist(observation_gen,bins=70 ,label="Predicted distribution of 30,000 individuals")
ax.hist(population,bins=70 ,alpha=0.5, label="Original values of the 30,000 individuals")
ax.set_xlabel("Mean")
ax.set_ylabel("Frequency")
ax.set_title("Posterior distribution of predicitons")
ax.legend()
plt.show()
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import scipy.stats
plt.style.use('ggplot')
population = np.random.normal(10,3,30000)
sample = population[np.random.randint(0, 30000, 1000)]
plt.figure(figsize=(10,5))
plt.hist(sample,bins=35)
plt.title("Distribution of 1000 observations sampled from a population of 30,000 with $\mu$=10, $\sigma$=3")
mu_obs = sample.mean()
mu_obs
def prior(x):
if x[1] <= 0:
return 1e-7
return 1
def log_gaussian(x, data):
return np.sum(np.log(scipy.stats.norm(x[0],x[1]).pdf(data)))
def acceptance(x, x_new):
if x_new > x:
return True
else:
accept = np.random.uniform(0, 1)
return accept < (np.exp(x_new - x))
def metropolis_hastings(param_init, iterations, data):
x = param_init
accepted = []
rejected = []
for i in range(iterations):
if (i + 1) % 2000 == 0:
print(i + 1)
x_new = [x[0],np.random.normal(x[1],0.5,(1,))]
x_likehood = log_gaussian(x,data)
x_new_likehood = log_gaussian(x_new,data)
x_likehood_prior = x_likehood + np.log(prior(x))
x_new_likehood_prior = x_new_likehood + np.log(prior(x_new))
if acceptance(x_likehood_prior, x_new_likehood_prior):
x = x_new
accepted.append(x)
else:
rejected.append(x_new)
return np.array(accepted), np.array(rejected)
accepted, rejected = metropolis_hastings([mu_obs,0.1], 50000, sample)
plt.figure(figsize=(10,10))
plt.subplot(2, 1, 1)
plt.plot(rejected[0:50,1], 'rx', label='Rejected',alpha=0.5)
plt.plot(accepted[0:50,1], 'b.', label='Accepted',alpha=0.5)
plt.xlabel("Iteration")
plt.ylabel("$\sigma$")
plt.title("MCMC sampling for $\sigma$ with Metropolis-Hastings. First 50 samples are shown.")
plt.legend()
plt.subplot(2, 1, 2)
plt.plot(rejected[-accepted.shape[0]:,1], 'rx', label='Rejected',alpha=0.5)
plt.plot(accepted[-accepted.shape[0]:,1], 'b.', label='Accepted',alpha=0.5)
plt.xlabel("Iteration")
plt.ylabel("$\sigma$")
plt.title("MCMC sampling for $\sigma$ with Metropolis-Hastings.")
plt.legend()
plt.show()
sigmas = accepted[:,1]
sigmas_accept = sigmas.mean() - 0.3
fig = plt.figure(figsize=(15,5))
ax = fig.add_subplot(1,2,1)
ax.plot(sigmas[sigmas > sigmas_accept])
ax.set_title("Trace for $\sigma$")
ax.set_ylabel("$\sigma$")
ax.set_xlabel("Iteration")
ax = fig.add_subplot(1,2,2)
ax.hist(sigmas[sigmas > sigmas_accept], bins=20,density=True)
ax.set_ylabel("Frequency (normed)")
ax.set_xlabel("$\sigma$")
ax.set_title("Histogram of $\sigma$")
plt.show()
mu=accepted[sigmas > sigmas_accept,0].mean()
sigma=accepted[sigmas > sigmas_accept,1].mean()
observation_gen = np.random.normal(mu,sigma,population.shape[0])
fig = plt.figure(figsize=(15,7))
ax = fig.add_subplot(1,1,1)
ax.hist(observation_gen,bins=70 ,label="Predicted distribution of 30,000 individuals")
ax.hist(population,bins=70 ,alpha=0.5, label="Original values of the 30,000 individuals")
ax.set_xlabel("Mean")
ax.set_ylabel("Frequency")
ax.set_title("Posterior distribution of predicitons")
ax.legend()
plt.show()
| 0.682468 | 0.937326 |
# Upload & Manage Annotations
```
item = dl.items.get(item_id="")
annotation = item.annotations.get(annotation_id="")
annotation.metadata["user"] = True
annotation.update()
```
## Convert Annotations To COCO Format
```
import dtlpy as dl
dataset = project.datasets.get(dataset_name='dataset_name')
converter = dl.Converter()
converter.upload_local_dataset(
from_format=dl.AnnotationFormat.COCO,
dataset=dataset,
local_items_path=r'C:/path/to/items', # Please make sure the names of the items are the same as written in the COCO JSON file
local_annotations_path=r'C:/path/to/annotations/file/coco.json'
)
```
## Upload Entire Directory and their Corresponding Dataloop JSON Annotations
```
import dtlpy as dl
if dl.token_expired():
dl.login()
project = dl.projects.get(project_name='project_name')
dataset = project.datasets.get(dataset_name='dataset_name')
# Local path to the items folder
# If you wish to upload items with your directory tree use : r'C:/home/project/images_folder'
local_items_path = r'C:/home/project/images_folder/*'
# Local path to the corresponding annotations - make sure the file names fit
local_annotations_path = r'C:/home/project/annotations_folder'
dataset.items.upload(local_path=local_items_path,
local_annotations_path=local_annotations_path)
```
## Upload Annotations To Video Item
Uploading annotations to video items needs to consider spanning between frames, and toggling visibility (occlusion). In this example, we will use the following CSV file.
In this file there is a single 'person' box annotation that begins on frame number 20, disappears on frame number 41, reappears on frame number 51 and ends on frame number 90.
[Video_annotations_example.CSV](https://cdn.document360.io/53f32fe9-1937-4652-8526-90c1bc78d3f8/Images/Documentation/video_annotation_example.csv)
```
import dtlpy as dl
import pandas as pd
project = dl.projects.get(project_name='my_project')
dataset = project.datasets.get(dataset_id='my_dataset')
# Read CSV file
df = pd.read_csv(r'C:/file.csv')
# Get item
item = dataset.items.get(item_id='my_item_id')
builder = item.annotations.builder()
# Read line by line from the csv file
for i_row, row in df.iterrows():
# Create box annotation from csv rows and add it to a builder
builder.add(annotation_definition=dl.Box(top=row['top'],
left=row['left'],
bottom=row['bottom'],
right=row['right'],
label=row['label']),
object_visible=row['visible'], # Support hidden annotations on the visible row
object_id=row['annotation id'], # Numbering system that separates different annotations
frame_num=row['frame'])
# Upload all created annotations
item.annotations.upload(annotations=builder)
```
# Show Annotations Over Image
After uploading items and annotations with their metadata, you might want to see some of them and perform visual validation.
To see only the annotations, use the annotation type *show* option.
```
# Use the show function for all annotation types
box = dl.Box()
# Must provide all inputs
box.show(image='', thickness='', with_text='', height='', width='', annotation_format='', color='')
```
To see the item itself with all annotations, use the Annotations option.
```
# Must input an image or height and width
annotation.show(image='', height='', width='', annotation_format='dl.ViewAnnotationOptions.*', thickness='', with_text='')
```
# Download Data, Annotations & Metadata
The item ID for a specific file can be found in the platform UI - Click BROWSE for a dataset, click on the selected file, and the file information will be displayed in the right-side panel. The item ID is detailed, and can be copied in a single click.
## Download Items and Annotations
Download dataset items and annotations to your computer folder in two separate folders.
See all annotation options [here](https://dataloop.ai/docs/sdk-download#annotation-options).
```
import dtlpy as dl
if dl.token_expired():
dl.login()
project = dl.projects.get(project_name='project_name')
dataset = project.datasets.get(dataset_name='dataset_name')
dataset.download(local_path=r'C:/home/project/images', # The default value is ".dataloop" folder
annotation_options=dl.VIEW_ANNOTATION_OPTIONS_JSON)
```
## Multiple Annotation Options
See all annotation options [here](https://dataloop.ai/docs/sdk-download#annotation-options).
```
import dtlpy as dl
if dl.token_expired():
dl.login()
project = dl.projects.get(project_name='project_name')
dataset = project.datasets.get(dataset_name='dataset_name')
dataset.download(local_path=r'C:/home/project/images', # The default value is ".dataloop" folder
annotation_options=[dl.VIEW_ANNOTATION_OPTIONS_MASK, dl.VIEW_ANNOTATION_OPTIONS_JSON, dl.ViewAnnotationOptions.INSTANCE])
```
## Filter by Item and/or Annotation
* **Items filter** - download filtered items based on multiple parameters, like their directory.
You can also download items based on different filters. Learn all about item filters [here](https://dataloop.ai/docs/sdk-sort-filter).
* **Annotation filter** - download filtered annotations based on multiple parameters like their label.
You can also download items annotations based on different filters, learn all about annotation filters [here](https://dataloop.ai/docs/sdk-sort-filter-annotation).
This example will download items and JSONS from a dog folder of the label 'dog'.
```
import dtlpy as dl
if dl.token_expired():
dl.login()
project = dl.projects.get(project_name='project_name')
dataset = project.datasets.get(dataset_name='dataset_name')
# Filter items from "folder_name" directory
item_filters = dl.Filters(resource='items',field='dir', values='/dog_name')
# Filter items with dog annotations
annotation_filters = dl.Filters(resource='annotations', field='label', values='dog')
dataset.download( # The default value is ".dataloop" folder
local_path=r'C:/home/project/images',
filters = item_filters,
annotation_filters=annotation_filters,
annotation_options=dl.VIEW_ANNOTATION_OPTIONS_JSON)
```
## Filter by Annotations
* **Annotation filter** - download filtered annotations based on multiple parameters like their label. You can also download items annotations based on different filters, learn all about annotation filters [here](https://dataloop.ai/docs/sdk-sort-filter-annotation).
```
import dtlpy as dl
if dl.token_expired():
dl.login()
project = dl.projects.get(project_name='project_name')
dataset = project.datasets.get(dataset_name='dataset_name')
item = dataset.items.get(item_id="item_id") #Get item from dataset to be able to view the dataset colors on Mask
# Filter items with dog annotations
annotation_filters = dl.Filters(resource='annotations', field='label', values='dog')
item.download( # the default value is ".dataloop" folder
local_path=r'C:/home/project/images',
annotation_filters=annotation_filters,
annotation_options=dl.VIEW_ANNOTATION_OPTIONS_JSON)
```
## Download Annotations in COCO Format
* **Items filter** - download filtered items based on multiple parameters like their directory. You can also download items based on different filters, learn all about item filters [here](https://dataloop.ai/docs/sdk-sort-filter).
* **Annotation filter** - download filtered annotations based on multiple parameters like their label. You can also download items annotations based on different filters, learn all about annotation filters [here](https://dataloop.ai/docs/sdk-sort-filter-annotation).
This example will download COCO from a dog items folder of the label 'dog'.
```
import dtlpy as dl
if dl.token_expired():
dl.login()
project = dl.projects.get(project_name='project_name')
dataset = project.datasets.get(dataset_name='dataset_name')
# Filter items from "folder_name" directory
item_filters = dl.Filters(resource='items',field='dir', values='/dog_name')
# Filter items with dog annotations
annotation_filters = dl.Filters(resource='annotations', field='label', values='dog')
converter = dl.Converter()
converter.convert_dataset(dataset=dataset, to_format='coco',
local_path=r'C:/home/coco_annotations',
filters = item_filters,
annotation_filters=annotation_filters)
```
|
github_jupyter
|
item = dl.items.get(item_id="")
annotation = item.annotations.get(annotation_id="")
annotation.metadata["user"] = True
annotation.update()
import dtlpy as dl
dataset = project.datasets.get(dataset_name='dataset_name')
converter = dl.Converter()
converter.upload_local_dataset(
from_format=dl.AnnotationFormat.COCO,
dataset=dataset,
local_items_path=r'C:/path/to/items', # Please make sure the names of the items are the same as written in the COCO JSON file
local_annotations_path=r'C:/path/to/annotations/file/coco.json'
)
import dtlpy as dl
if dl.token_expired():
dl.login()
project = dl.projects.get(project_name='project_name')
dataset = project.datasets.get(dataset_name='dataset_name')
# Local path to the items folder
# If you wish to upload items with your directory tree use : r'C:/home/project/images_folder'
local_items_path = r'C:/home/project/images_folder/*'
# Local path to the corresponding annotations - make sure the file names fit
local_annotations_path = r'C:/home/project/annotations_folder'
dataset.items.upload(local_path=local_items_path,
local_annotations_path=local_annotations_path)
import dtlpy as dl
import pandas as pd
project = dl.projects.get(project_name='my_project')
dataset = project.datasets.get(dataset_id='my_dataset')
# Read CSV file
df = pd.read_csv(r'C:/file.csv')
# Get item
item = dataset.items.get(item_id='my_item_id')
builder = item.annotations.builder()
# Read line by line from the csv file
for i_row, row in df.iterrows():
# Create box annotation from csv rows and add it to a builder
builder.add(annotation_definition=dl.Box(top=row['top'],
left=row['left'],
bottom=row['bottom'],
right=row['right'],
label=row['label']),
object_visible=row['visible'], # Support hidden annotations on the visible row
object_id=row['annotation id'], # Numbering system that separates different annotations
frame_num=row['frame'])
# Upload all created annotations
item.annotations.upload(annotations=builder)
# Use the show function for all annotation types
box = dl.Box()
# Must provide all inputs
box.show(image='', thickness='', with_text='', height='', width='', annotation_format='', color='')
# Must input an image or height and width
annotation.show(image='', height='', width='', annotation_format='dl.ViewAnnotationOptions.*', thickness='', with_text='')
import dtlpy as dl
if dl.token_expired():
dl.login()
project = dl.projects.get(project_name='project_name')
dataset = project.datasets.get(dataset_name='dataset_name')
dataset.download(local_path=r'C:/home/project/images', # The default value is ".dataloop" folder
annotation_options=dl.VIEW_ANNOTATION_OPTIONS_JSON)
import dtlpy as dl
if dl.token_expired():
dl.login()
project = dl.projects.get(project_name='project_name')
dataset = project.datasets.get(dataset_name='dataset_name')
dataset.download(local_path=r'C:/home/project/images', # The default value is ".dataloop" folder
annotation_options=[dl.VIEW_ANNOTATION_OPTIONS_MASK, dl.VIEW_ANNOTATION_OPTIONS_JSON, dl.ViewAnnotationOptions.INSTANCE])
import dtlpy as dl
if dl.token_expired():
dl.login()
project = dl.projects.get(project_name='project_name')
dataset = project.datasets.get(dataset_name='dataset_name')
# Filter items from "folder_name" directory
item_filters = dl.Filters(resource='items',field='dir', values='/dog_name')
# Filter items with dog annotations
annotation_filters = dl.Filters(resource='annotations', field='label', values='dog')
dataset.download( # The default value is ".dataloop" folder
local_path=r'C:/home/project/images',
filters = item_filters,
annotation_filters=annotation_filters,
annotation_options=dl.VIEW_ANNOTATION_OPTIONS_JSON)
import dtlpy as dl
if dl.token_expired():
dl.login()
project = dl.projects.get(project_name='project_name')
dataset = project.datasets.get(dataset_name='dataset_name')
item = dataset.items.get(item_id="item_id") #Get item from dataset to be able to view the dataset colors on Mask
# Filter items with dog annotations
annotation_filters = dl.Filters(resource='annotations', field='label', values='dog')
item.download( # the default value is ".dataloop" folder
local_path=r'C:/home/project/images',
annotation_filters=annotation_filters,
annotation_options=dl.VIEW_ANNOTATION_OPTIONS_JSON)
import dtlpy as dl
if dl.token_expired():
dl.login()
project = dl.projects.get(project_name='project_name')
dataset = project.datasets.get(dataset_name='dataset_name')
# Filter items from "folder_name" directory
item_filters = dl.Filters(resource='items',field='dir', values='/dog_name')
# Filter items with dog annotations
annotation_filters = dl.Filters(resource='annotations', field='label', values='dog')
converter = dl.Converter()
converter.convert_dataset(dataset=dataset, to_format='coco',
local_path=r'C:/home/coco_annotations',
filters = item_filters,
annotation_filters=annotation_filters)
| 0.321673 | 0.757974 |
```
from Scweet.scweet import scrape
from Scweet.user import get_user_information, get_users_following, get_users_followers
```
# Scrape tweets of a specific account or words or hashtag
```
# scrape top tweets with the words 'covid','covid19' in proximity and without replies.
# the process is slower as the interval is smaller (choose an interval that can divide the period of time betwee, start and max date)
data = scrape(words=['bitcoin','etherium'], since="2015-04-01", until="2015-04-2", from_account = None,interval=1,
headless=True, display_type="Top", save_images=False, proxy = None, save_dir = 'outputs',
resume=False, filter_replies=True, proximity=False)
# scrape top tweets of with the hashtag #covid19, in proximity and without replies.
# the process is slower as the interval is smaller (choose an interval that can divide the period of time betwee, start and max date)
data = scrap(hashtag="covid19", start_date="2020-04-01", max_date="2020-04-15", from_account = None,interval=1,
headless=True, display_type="Top", save_images=False, proxy = None, save_dir = 'outputs',
resume=False, filter_replies=True, proximity=True)
data.head()
```
# Get the main information of a given list of users
```
# These users belongs to my following.
users = ['nagouzil', '@yassineaitjeddi', 'TahaAlamIdrissi',]
#'@Nabila_Gl', 'geceeekusuu', '@pabu232', '@av_ahmet', '@x_born_to_die_x']
# this function return a list that contains :
# ["nb of following","nb of followers", "join date", "birthdate", "location", "website", "description"]
users_info = get_user_information(users, headless=True)
import pandas as pd
users_df = pd.DataFrame(users_info, index = ["nb of following","nb of followers", "join date",
"birthdate", "location", "website", "description"]).T
users_df
```
# Get followers and following of a given list of users
Enter your username and password in .env file. I recommande you dont use your main account.
Increase wait argument to avoid banning your account and maximise the crawling process if the internet is slow. I used 1 and it's safe.
```
users[0]
env_path = ".env"
following = get_users_following(users=['nagouzil'], env=env_path, verbose=0, headless = True, wait=2, file_path='outputs/')
following['nagouzil'][:10]
followers = get_users_followers(users=['nagouzil'], env=env_path, verbose=0, headless = True, wait=2, file_path='outputs/')
followers['nagouzil'][:10]
```
|
github_jupyter
|
from Scweet.scweet import scrape
from Scweet.user import get_user_information, get_users_following, get_users_followers
# scrape top tweets with the words 'covid','covid19' in proximity and without replies.
# the process is slower as the interval is smaller (choose an interval that can divide the period of time betwee, start and max date)
data = scrape(words=['bitcoin','etherium'], since="2015-04-01", until="2015-04-2", from_account = None,interval=1,
headless=True, display_type="Top", save_images=False, proxy = None, save_dir = 'outputs',
resume=False, filter_replies=True, proximity=False)
# scrape top tweets of with the hashtag #covid19, in proximity and without replies.
# the process is slower as the interval is smaller (choose an interval that can divide the period of time betwee, start and max date)
data = scrap(hashtag="covid19", start_date="2020-04-01", max_date="2020-04-15", from_account = None,interval=1,
headless=True, display_type="Top", save_images=False, proxy = None, save_dir = 'outputs',
resume=False, filter_replies=True, proximity=True)
data.head()
# These users belongs to my following.
users = ['nagouzil', '@yassineaitjeddi', 'TahaAlamIdrissi',]
#'@Nabila_Gl', 'geceeekusuu', '@pabu232', '@av_ahmet', '@x_born_to_die_x']
# this function return a list that contains :
# ["nb of following","nb of followers", "join date", "birthdate", "location", "website", "description"]
users_info = get_user_information(users, headless=True)
import pandas as pd
users_df = pd.DataFrame(users_info, index = ["nb of following","nb of followers", "join date",
"birthdate", "location", "website", "description"]).T
users_df
users[0]
env_path = ".env"
following = get_users_following(users=['nagouzil'], env=env_path, verbose=0, headless = True, wait=2, file_path='outputs/')
following['nagouzil'][:10]
followers = get_users_followers(users=['nagouzil'], env=env_path, verbose=0, headless = True, wait=2, file_path='outputs/')
followers['nagouzil'][:10]
| 0.446736 | 0.607576 |
```
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from auxiliary.auxiliary_figures import (get_figure1, get_figure2, get_figure3)
from auxiliary.auxiliary_tables import (get_table1, get_table2, get_table3, get_table4)
from auxiliary.auxiliary_data import process_data
from auxiliary.auxiliary_visuals import (background_negative_green, p_value_star)
from auxiliary.auxiliary_extensions import (get_flexible_table4, get_figure1_extension1, get_figure2_extension1,
get_bias, get_figure1_extension2, get_figure2_extension2)
import warnings
warnings.filterwarnings('ignore')
plt.rcParams['figure.figsize'] = [12, 6]
```
The code below is needed to automatically enumerate the equations used in this notebook.
```
%%javascript
MathJax.Hub.Config({
TeX: { equationNumbers: { autoNumber: "AMS" } }
});
MathJax.Hub.Queue(
["resetEquationNumbers", MathJax.InputJax.TeX],
["PreProcess", MathJax.Hub],
["Reprocess", MathJax.Hub]
);
```
---
# Replication of Angrist (1990)
---
This notebook replicates the core results of the following paper:
> Angrist, Joshua. (1990). [Lifetime Earnings and the Vietnam Era Draft Lottery: Evidence from Social Security Administrative Records](https://www.jstor.org/stable/2006669?seq=1#metadata_info_tab_contents). *American Economic Review*. 80. 313-36.
In the following just a few notes on how to read the remainder:
- In this excerpt I replicate the Figures 1 to 3 and the Tables 1 to 4 (in some extended form) while I do not consider Table 5 to be a core result of the paper which is why it cannot be found in this notebook.
- I follow the example of Angrist keeping his structure throughout the replication part of this notebook.
- The naming and order of appearance of the figures does not follow the original paper but the published [correction](https://economics.mit.edu/files/7769).
- The replication material including the partially processed data as well as some replication do-files can be found [here](https://economics.mit.edu/faculty/angrist/data1/data/angrist90).
# 1. Introduction
---
For a soft introduction to the topic, let us have a look at the goal of Angrist's article. Already in the first few lines Angrist states a clear-cut aim for his paper by making the remark that "yet, academic research has not shown conclusively that Vietnam (or other) veterans are worse off economically than nonveterans". He further elaborates on why research had yet been so inconclusive. He traces it back to the flaw that previous research had solely tried to estimate the effect of veteran status on subsequent earnings by comparing the latter across individuals differing in veteran status. He argues that this naive estimate might likely be biased as it is easily imaginable that specific types of men choose to enlist in the army whose unobserved characteristics imply low civilian earnings (self-selcetion on unobservables).
Angrist avoids this pitfall by employing an instrumental variable strategy to obtain unbiased estimates of the effect of veteran status on earnings. For that he exploits the random nature of the Vietnam draft lottery. This lottery randomly groups people into those that are eligible to be forced to join the army and those that are not. The idea is that this randomly affects the veteran status without being linked to any unobserved characteristics that cause earnings. This allows Angrist to obtain an estimate of the treatment effect that does not suffer from the same shortcomings as the ones of previous studies.
He finds that Vietnam era veterans are worse off when it comes to long term annual real earnings as opposed to those that have not served in the army. In a secondary point he traces this back to the loss of working experience for veterans due to their service by estimating a simple structural model.
In the following sections I first walk you through the identification idea and empirical strategy. Secondly, I replicate and explain the core findings of the paper with a rather extensive elaboration on the different data sources used and some additional visualizations. Thirdly, I critically assess the paper followed by my own two extensions concluding with some overall remarks right after.
# 2. Identification and Empirical Approach
---
As already mentioned above the main goal of Angrist's paper is to determine the causal effect of veteran status on subsequent earnings. He believes for several reasons that conventional estimates that only compare earnings by veteran status are biased due to unobservables that affect both the probability of serving in the military as well as earnings over lifetime. This is conveniently shown in the causal graph below. Angrist names two potential reasons why this might be likely. First of all, he makes the point that probably people with few civilian opportunities (lower expected earnings) are more likely to register for the army. Without a measure for civilian opportunities at hand a naive estimate of the effect of military service on earnings would not be capable of capturing the causal effect. Hence, he believes that there is probably some self-selection into treatment on unobservables by individuals. In a second point, Angrist states that the selection criteria of the army might be correlated with unobserved characteristics of individuals that makes them more prone to receiving future earnings pointing into a certain direction.
Econometrically spoken, Angrist argues with the following linear regression equation representing a version of the right triangle in the causal graph:
\begin{align}
y_{cti} = \beta_c + \delta_t + s_i \alpha + u_{it}.
\end{align}
He argues that estimating the above model with the real earnings $y_{cti}$ for an individual $i$ in cohort $c$ at time $t$ being determined by cohort and time fixed effects ($\beta_c$ and $\delta_t$) as well an individual effect for veteran status is biased. This is for the above given reasons that the indicator for veteran status $s_i$ is likely to be correlated with the error term $u_{it}$.
Angrist's approach to avoid bias is now to employ an instrumental variable approach which is based on the accuracy of the causal graph below.
<div>
<img src="material/fig-angrist-1990-valid.png" width="600"/>
</div>
The validity of this causal graph rests on the crucial reasoning that there is no common cause of the instrument (Draft Lottery) and the unobserved variables (U). Angrist provides the main argument that the draft lottery was essentially random in nature and hence is not correlated with any personal characteristics and therefore not linked to any unobservables that might determine military service and earnings. As will be later explained in more detail, the Vietnam draft lottery determined randomly on the basis of the birth dates whether a person is eligible to be drafted by the army in the year following the lottery. The directed graph from Draft Lottery to Military Service is therefore warranted as the fact of having a lottery number rendering a person draft-eligible increases the probability of joining the military as opposed to a person that has an excluded lottery number.
This argumentation leads Angrist to use the probability of being a veteran conditional on being draft-eligible in the lottery as an instrument for the effect of veteran status on earnings. In essence this is the Wald estimate which is equal to the following formula:
\begin{align*}
\hat{\alpha}_{IV, WALD} = \frac{E[earnings \mid eligible = 1] - E[earnings \mid eligible = 0]}{E[veteran \mid eligible = 1] - E[veteran \mid eligible = 0]}
\end{align*}
The nominator equals to the estimated $\alpha$ from equation (1) while the denominator can be obtained by a first stage regression which regresses veteran status on draft-eligibility. It reduces to estimating the difference in conditional probabilities of being a veteran $prob(veteran \mid eligible = 1) - prob(veteran \mid eligible = 0)$. Estimates for this are obtained by Angrist through weighted least squares (WLS). This is done as Angrist does not have micro data but just grouped data (for more details see the data section in the replication). In order to obtain the estimates of the underlying micro level data it is necessary to adjust OLS by the size of the respective groups as weights. The above formula is also equivalent to a Two Stage Least Squares (2SLS) procedure in which earnings are regressed on the fitted values from a first stage regression of veteran status on eligibility.
In a last step, Angrist generalizes the Wald grouping method to more than just one group as instrument. There are 365 lottery numbers that were split up into two groups (eligible and non-eligible) for the previous Wald estimate. Those lottery numbers can also be split up even further into many more subgroups than just two, resulting in many more dummy variables as instruments. Angrist splits the lottery numbers into intervals of five which determine a group $j$. By cohort $c$ he estimates for each group $j$ the conditional probability of being a veteran $p_{cj}$. This first stage is again run by WLS. The resulting estimate $\hat p_{cj}$ is then used to conduct the second stage regression below.
\begin{align}
\bar y_{ctj} = \beta_c + \delta_t + \hat p_{cj} \alpha + \bar u_{ctj}
\end{align}
The details and estimation technique will be further explained when presenting the results in the replication section below.
# 3. Replication
---
## 3.1 Background and Data
### The Vietnam Era Draft Lottery
Before discussing how the data looks like it is worthwhile to understand how the Vietnam era draft lottery was working in order to determine to which extent it might actually serve as a valid instrument. During the Vietnam war there were several draft lotteries. They were held in the years from 1970 to 1975. The first one took place at the end of 1969 determining which men might be drafted in the following year. This procedure of determining the lottery numbers for the following year continued until 1975. The table below shows for which years there were lotteries drawn and which birth years were affected by them in the respective year. For more details have a look [here](https://www.sss.gov/history-and-records/vietnam-lotteries/).
| **Year** | **Cohorts** | **Draft-Eligibility Ceiling**|
|--------------|---------------|------------------------------|
| 1970 | 1944-50 | 195 |
| 1971 | 1951 | 125 |
| 1972 | 1952 | 95 |
| 1973 | 1953 | 95 |
| 1974 | 1954 | 95 |
| 1975 | 1955 | 95 |
| 1976 | 1956 | 95 |
The authority of drafting men for the army through the lottery expired on June 30, 1973 and already before no one was drafted anymore. The last draft call took place on December 7, 1972.
The general functioning of those seven lotteries was that every possible birthday (365 days) was randomly assigned a number between 1 and 365 without replacement. Taking the 1969 lottery this meant that the birthdate that had the number 1 assigned to, it caused every man born on that day in the years 1944 to 1950 to be drafted first if it came to a draft call in the year 1970. In practice, later in the same year of the draft lottery, the army announced draft-eligibility ceilings determining up to which draft lottery number was called in the following year. In 1970, this means that every man having a lottery number of below 195 was called to join the army. As from 1973 on nobody was called anymore, the numbers for the ceiling are imputed from the last observed one which was 95 in the year 1972. Men with lottery numbers below the ceiling for their respective year are from here on called "draft-eligible".
Being drafted did not mean that one actually had to serve in the army, though. Those drafted had to pass mental and physical tests which in the end decided who had to join. Further it should be mentioned that Angrist decides to only use data on those that turned 19 when being at risk of induction which includes men born between 1950 and 1953.
### The Data
#### Continuous Work History Sample (CWHS)
This administrative data set constitutes a random one percent sample draw of all possible social security numbers in the US. For the years from 1964 until 1984 it includes the **FICA** (social security) earnings history censored to the Social Security maximum taxable amount. It further includes FICA taxable earnings from self-employment. For the years from 1978 on it also has a series on total earnings (**Total W-2**) including for instance cash payments but excluding earnings from self-employment. This data set has some confidentiality restrictions which means that only group averages and variances were available. This means that Angrist cannot rely on micro data but has to work with sample moment which is a crucial factor for the exact implementation of the IV method. A group is made of by year of earnings, year of birth, ethnicity and five consecutive lottery numbers. The statistics collected for those also include the number of people in the group, the fraction of them having taxable earnings equal and above the taxable maximum and the fraction having zero earnings.
Regarding the actual data sets available for replication we have the data set `cwhsa` which consists of the above data for the years from 1964 to 1977 and then `cwhsb` which consists of the CWHS for the years after.
Above that Angrist provides the data set `cwhsc_new` which includes the **adjusted FICA** earnings. For those Angrist employed a strategy to approximate the underlying uncensored FICA earnings from the reported censored ones. All of those three different earnings variables are used repeatedly throughout the replication.
```
process_data("cwhsa")
```
The above earnings data only consists of FICA earnings. The lottery intervals from 1 to 73 are equivalent to intervals of five consecutive lottery numbers. Consequently, the variable lottery interval equals to one for the lottery numbers 1 to 5 and so on. The ethnicity variable is encoded as 1 for a white person and 2 for a nonwhite person.
```
process_data("cwhsb")
```
As stated above this data now consists of earnings from 1978 to 1984 for FICA (here encoded as "TAXAB") and Total W-2 (encoded as "TOTAL").
#### Survey of Income and Program Participation (SIPP) and the Defense Manpower Data Center (DMDC)
Throughout the paper it is necessary to have a measure of the fraction of people serving in the military. For this purpose the above two data sources are used.
The **SIPP** is a longitudinal survey of around 20,000 households in the year 1984 for which is determined whether the persons in the household are Vietnam war veterans. The survey also collected data on ethnicity and birth data which made it possible to match the data to lottery numbers. The **DMDC** on the other hand is an administrative record which shows the total number of new entries into the army by ethnicity, cohort and lottery number per year from mid 1970 until the end of 1973.
Those sources are needed for the results in Table 3 and 4. A combination of those two are matched to the earnings data of the CWHS which constitutes the data set `chwsc_new` below.
```
data_cwhsc_new = process_data("cwhsc_new")
data_cwhsc_new
```
This data set now also includes the adjusted FICA earnings which are marked by "ADJ" as well as the probability of serving in the military conditional on being in a group made up by ethnicity, birth cohort and lottery interval.
Below we have a short look at how the distribution of the different earnings measures look like. In the table you see the real earnings in 1978 dollar terms for the years from 1974 to 1984 for FICA and adjusted FICA as well as the years 1978 until 1984 for Total W-2.
```
for data in ["ADJ", "TAXAB", "TOTAL"]:
ax = sns.kdeplot(data_cwhsc_new.loc[data, "earnings"], color=np.random.choice(
np.array([sns.color_palette()]).flatten(), 4))
ax.set_xlim(xmax=20000)
ax.legend(["Adjusted FICA", "FICA", "TOTAL W-2"], loc="upper left")
ax.set_title("Kernel Density of the different Earning Measures")
```
For a more detailed description of the somewhat confusing original variable names in the data sets please refer to the appendix at the very bottom of the notebook.
## 3.2 Establishing the Validity of the Instrument
In order to convincingly pursue the identification strategy outlined above it is necessary to establish an effect of draft eligibility (the draft lottery) on veteran status and to argue that draft eligibility is exogenous to any unobserved factor affecting both veteran status and subsequent earnings. As argued before one could easily construct reasonable patterns of unobservables that both cause veteran status and earnings rendering a naive regression of earnings on veteran status as biased.
The first requirement for IV to be valid holds as it is clearly observable that draft-eligibility has an effect on veteran status. The instrument is hence **relevant**. For the second part Angrist argues that the draft lottery itself is random in nature and hence not correlated with any unobserved characteristics (**exogenous**) a man might have that causes him to enroll in the army while at the same time making his earnings likely to go into a certain direction irrespective of veteran status.
On the basis of this, Angrist now shows that subsequent earnings are affected by draft eligibility. This is the foundation to find a nonzero effect of veteran status on earnings. Going back to the causal diagram from before, Angrist argued so far that there is no directed graph from Draft Lottery to the unobservables U but only to Military Service. Now he further establishes the point that there is an effect of draft-eligibility (Draft Lottery) that propagates through Military Service onto earnings (Wages).
In order to see this clearly let us have a look at **Figure 1** of the paper below. For white and nonwhite men separately the history of average FICA earnings in 1978 dollar terms is plotted. This is done by year within cohort across those that were draft-eligible and those that were not. The highest two lines represent the 1950 cohort going down to the cohort of men born in 1953. There is a clearly observable pattern among white men in the cohorts from 1950 to 52 which shows persistently lower earnings for those draft-eligible starting the year in which they could be drafted. This cannot be seen for those born in 1953 which is likely due to the fact that nobody was actually drafted in 1973 which would have otherwise been "their" year. For nonwhite men the picture is less clear. It seems that for cohorts 50 to 52 there is slightly higher earnings for those ineligible but this does not seem to be persistent over time. The cohort 1953 again does not present a conclusive image. Observable in all lines, though, is that before the year of conscription risk there is no difference in earnings among the group which is due to the random nature of the draft lottery.
```
# read in the original data sets
data_cwhsa = pd.read_stata("data/cwhsa.dta")
data_cwhsb = pd.read_stata("data/cwhsb.dta")
data_cwhsc_new = pd.read_stata("data/cwhsc_new.dta")
data_dmdc = pd.read_stata("data/dmdcdat.dta")
data_sipp = pd.read_stata("data/sipp2.dta")
get_figure1(data_cwhsa, data_cwhsb)
```
A more condensed view of the results in Figure 1 is given in **Figure 2**. It depicts the differences in earnings between the red and the black line in Figure 1 by cohort and ethnicity. This is just included for completeness as it does not provide any further insight in comparison to Figure 1.
```
get_figure2(data_cwhsa, data_cwhsb)
```
A further continuation of this line of argument is resulting in **Table 1**. Angrist makes the observations from the figures before even further fine-grained and explicit. In Table 1 Angrist estimates the expected difference in average FICA and Total W-2 earnings by ethnicity within cohort and year of earnings. In the table below for white men we can observe that there is no significant difference to the five percent level for the years before the year in which they might be drafted. This changes for the cohorts from 1950 to 52 in the years 1970 to 72, respectively. There we can observe a significantly lower income for those eligible in comparison to those ineligible. This seems to be persistent for the cohorts 1950 and 52 while less so for those born in 1951 and 1953. It should further be noted that Angrist reports that the quality of the Total W-2 earnings data was low in the first years (it was launched in 1972) explaining the inconlusive estimations in the periods at the beginning.
To focus the attention on the crucial points I mark all the negative estimates in different shades of green with more negative ones being darker. This clearly emphasizes the verbal arguments brought up before.
```
table1 = get_table1(data_cwhsa, data_cwhsb)
table1["white"].style.applymap(background_negative_green)
```
For the nonwhite males there is no clear cut pattern. Only few cells show significant results which is why Angrist in the following focuses on white males when constructing IV estimates. For completeness I present Table 1 for nonwhite males below although it is somewhat less important for the remainder of the paper.
```
table1["nonwhite"].style.applymap(background_negative_green)
```
## 3.3 Measuring the Effect of Military Service on Earnings
### 3.3.1 Wald-estimates
As discussed in the identification section a simple OLS regression estimating the model in equation (1) might suffer from bias due to elements of $s_i$ that are correlated with the error term $u_{it}$. This problem can be to a certain extent circumvented by the grouping method proposed by Abraham Wald (1940). Grouping the data by the instrument which is draft eligibility status makes it possible to uncover the effect of veteran status on earnings.
An unbiased estimate of $\alpha$ can therefore be found by adjusting the difference in mean earnings across eligibility status by the difference in probability of becoming a veteran conditional on being either draft eligible or not. This verbal explanation is translated in the following formula:
\begin{equation}
\hat\alpha = \frac{\bar y^e - \bar y^n}{\hat{p}(V|e) - \hat{p}(V|n)}
\end{equation}
The variable $\bar y$ captures the mean earnings within a certain cohort and year further defined by the superscript $e$ or $n$ which indicates draft-eligibility status. The above formula poses the problem that the conditional probabilities of being a veteran cannot be obtained from the CWHS data set alone. Therefore in **Table 2** Angrist attempts to estimate them from two other sources. First from the SIPP which has the problem that it is a quite small sample. And secondly, he matches the CWHS data to the DMDC. Here it is problematic, though, that the amount of people entering the army in 1970 (which is the year when those born 1950 were drafted) is only collected for the second half of the year. This is the reason why Angrist has to go with the estimates from the SIPP for the cohort of 1950 while taking the bigger sample of the matched DMDC/CWHS for the birth years 1951 to 53. The crucial estimates needed for the denominator of equation (3) are presented in the last column of Table 2 below. It can already be seen that the differences in earnings by eligibility that we found in Table 1 will be scaled up quite a bit to obtain the estimates for $\hat{\alpha}$. We will come back to that in Table 3.
<div class="alert alert-block alert-success">
<b>Note:</b> The cohort 1950 for the DMDC/CWHS could not be replicated as the data for cohort 1950 from the DMDC set is missing in the replication data. Above that the standard errors for the estimates coming form SIPP differ slightly from the published results but are equal to the results from the replication code.
</div>
```
table2 = get_table2(data_cwhsa, data_dmdc, data_sipp)
table2["white"]
table2["nonwhite"]
```
In the next step Angrist brings together the insights gained so far from his analysis. **Table 3** presents again differences in mean earnings across eligibility status for different earnings measures and within cohort and year. The values in column 1 and 3 are directly taken from Table 1. In column 2 we now encounter the adjusted FICA measure for the first time. As a reminder, it consists of the scaled up FICA earnings as FICA earnings are only reported to a certain maximum amount. The true average earnings are likely to be higher and Angrist transformed the data to account for this. We can see that the difference in mean earnings is most often in between the one of pure FICA earnings and Total W-2 compensation. In column three there is again the probability difference from the last column of Table 2. As mentioned before the measure is taken from the SIPP sample for the cohort of 1950 and the DMDC/CWHS sample for the other cohorts. Angrist decides to exclude cohort 1953 and nonwhite males as for those draft eligibility does not seem to be an efficient instrument (see Table 1 and Figure 1 and 2). Although Angrist does not, in this replication I also present Table 3 for nonwhites to give the reader a broader picture. Further Angrist focuses his derivations only on the years 1981 to 1984 as those are the latest after the Vietnam war for which there was data avalaible. Effects in those years are most likely to represent long term effects.
Let us now look at the most crucial column of Table 3 which is the last one. It captures the Wald estimate for the effect of veteran status on adjusted FICA earnings in 1978 dollar terms per year and cohort from equation (3). So this is our $\hat{\alpha}$ per year and cohort.
For white males the point estimates indicate that the annual loss in real earnings due to serving in the military was around 2000 dollars. Looking at the high standard errors, though, only few of the estimates are actually statistically significant. In order to see this more clearly I added a star to those values in the last column that are statistically significant to the five percent level.
<div class="alert alert-block alert-success">
<b>Note:</b> In the last column I obtain slightly different standard errors than in the paper. The same is the case, though, in the replication code my replication is building up on.
</div>
```
table3 = get_table3(data_cwhsa, data_cwhsb, data_dmdc, data_sipp, data_cwhsc_new)
p_value_star(table3["white"], slice(None), ("", "Service Effect in 1978 $"))
```
Looking at nonwhite males now, we observe what we already expected. All of the Wald estimates are actually far away from being statistically significant.
```
p_value_star(table3["nonwhite"], slice(None), ("", "Service Effect in 1978 $"))
```
### 3.3.2 More complex IV estimates
In the next step Angrist uses a more generalized version of the Wald estimate for the given data. While in the previous analysis the mean earnings were compared solely on the basis of two groups (eligibles and ineligibles, which were determined by the lottery numbers), in the following this is extended to more complex subgroups. The grouping is now based on intervals of five consecutive lottery numbers. As explained in the section on idenficication this boils down to estimating the model described in equation (2).
\begin{equation*}
\bar y_{ctj} = \beta_c + \delta_t + \hat p_{cj} \alpha + \bar u_{ctj}
\end{equation*}
$\bar y_{ctj}$ captures the mean earnings by cohort $c$, in year $t$ for group $j$. $\hat p_{cj}$ depicts the estimated probability of being a veteran conditional on being in cohort $c$ and group $j$. We are now interested in obtaining an estimate of $\alpha$. In our current set up $\alpha$ corresponds to a linear combination of the many different possible Wald estimates when comparing each of the subgroups in pairs. With this view in mind Angrist restricts the treatment effect to be same (i.e. equal to $\alpha$) for each comparison of subgroups. The above equation is equivalent to the second stage of the 2SLS estimation. Angrist estimates the above model using the mean real earnings averaged over the years 1981 to 84 and the cohorts from 1950 to 53. In the first stage Angrist has to estimate $\hat p_{cj}$ which is done again by using a combination of the SIPP sample and the matched DWDC/CWHS data set. With this at hand Angrist shows how the equation (2) looks like if it was estimated by OLS. The following **Figure 3** is also called Visual Instrumental Variables (VIV). In order to arrive there he takes the residuals from an OLS regression of $\bar y_{ctj}$ and $\hat p_{cj}$ on cohort and time dummies, respectively. Then he performs another OLS regression of the earnings residuals on the probability residuals. This is depicted in Figure 3 below. The slope of the regression line corresponds to an IV estimate of $\alpha$. The slope amounts to an estimate of -2384 dollars which serves as a reference for the treatment effect measured by another, more efficient method described below the Figure.
```
get_figure3(data_cwhsc_new)
```
We now shortly turn back to a remark from before. Angrist is forced to only work with sample means due to confidentiality restrictions on the underlying micro data. For the Wald estimates it is somewhat easily imaginable that this does not pose any problem. For the above estimation of $\alpha$ using 2SLS this is less obvious. Angrist argues, though, that there is a Generalized Method of Moments (GMM) interpretation of the 2SLS approach which allows him to work with sample moments alone. Another important implication thereof is that he is not restricted to using only one sample to obtain the sample moments. In our concrete case here, it is therefore unproblematic that the earnings data is coming from another sample than the conditional probabilities of being a veteran as both of the samples are drawn from the same population. This is a characteristic of the GMM.
In the following, Angrist estimates equation (2) by using the more efficient approach of Generalized Least Squares (GLS) as opposed to OLS. The GLS is more efficient if there is correlation between the residuals in a regression model. Angrist argues that this is the case in the above model equation and that this correlation can be estimated. GLS works such that coming from the estimated covariance matrix $\hat\Omega$ of the residuals, the regressors as well as the dependent variable are transformed using the upper triangle of the Cholesky decomposition of $\hat\Omega^{-1}$. Those transformed variables are then used to run a regular OLS model with nonrobust standard errors. The resulting estimate $\hat\alpha$ then is the most efficient one (if it is true that there is correlation between the residuals).
Angrist states that the optimal weigthing matrix $\Omega$ resulting in the most efficient estimate of $\hat\alpha$ looks the following:
\begin{equation}
\Omega = V(\bar y_{ctj}) + \alpha^2 V(\hat p_{cj}).
\end{equation}
All of the three elements on the right hand side can be estimated from the data at hand.
Now we have all the ingredients to have a look at the results in **Table 4**. In practice, Angrist estimates two models in the above manner based on the general form of the above regression equation. Model 1 allows the treatment effect to vary by cohort while Model 2 collapses them into a scalar estimate of $\alpha$.
The results for white men in Model 1 show that for each of the three earnings measures as dependent variable only few are statistically significant to the five percent level (indicated by a star added by me again). A look at Model 2 reveals, though, that the combined treatment effect is significant and it amounts to a minus of 2000 dollar (we look again at real earnings in 1978 dollar terms) annualy for those having served in the army. For cohort 1953 we obtain insignificant estimates which was to be expected given that actually nobody was drafted in that year.
<div class="alert alert-block alert-success">
<b>Note:</b> The results are again a bit different to those in the paper. The same is the case, though, in the replication code my replication is building up on.
</div>
```
table4 = get_table4(data_cwhsc_new)
p_value_star(table4["white"], (slice(None), slice(None), ["Value", "Standard Error"]), (slice(None)))
```
Angrist also reports those estimates for nonwhite men which are not significant. This was already expected as the the instrument was not clearly correlated with the endogenous variable of veteran status.
```
p_value_star(table4["nonwhite"], (slice(None), slice(None), ["Value", "Standard Error"]), (slice(None)))
```
This table concludes the replication of the core results of the paper. Summing up, Angrist constructed a causal graph for which he employs a plausible estimation strategy. Using his approach he concludes with the main result of having found a negative effect of serving in the military during the Vietnam era on subsequent earnings for white male in the United States.
Angrist provides some interpretation of the found effect and some concerns that might arise when reading his paper. I will discuss some of his points in the following critical assessment.
# 4. Critical Assessment
---
Considering the time back then and the consequently different state of research, the paper was a major contribution to instrumental variable estimation of treatment effects. More broadly, the paper is very conclusive and well written. Angrist discusses caveats quite thoroughly which makes the whole argumentation at first glance very concise. Methodologically, the paper is quite complex as due to the kind of data available. Angrist is quite innovative in that regard as he comes up with the two sample IV method in this paper which allows him to pratically follow his identification strategy. The attempt to explain the mechanisms behind the negative treatment effect found by him makes the paper comprehensive and shows the great sense of detail Angrist put into this paper.
While keeping in mind the positive sides of his paper, in hindsight, Angrist is a bit too vocal about the relevance and accuracy of his findings. Given our knowledge about the local average treatment effect (**LATE**) we encountered in our lecture, Angrist only identifies the average treatment effect of the compliers (those that enroll for the army if they are draft-eligible but do not if they are not) if there is individual level treatment heterogeneity and if the causal graph from before is accurate. Hence, the interpretation of the results gives only limited policy implications. For the discussion of veteran compensation the group of those who were induced by the lottery to join the military are not crucial. As there is no draft lottery anymore, what we are interested in is how to compensate veterans for their service who "voluntarily" decided to serve in the military. This question cannot be answered by Angrist's approach given the realistic assumption that there is treatment effect heterogeneity (which also Angrist argues might be warranted).
A related difficulty of interpretation arises because in the second part, Angrist uses an overidentified model. As already discussed before this amounts to a linear combination of the average treatment effects of subgroups. This mixes the LATEs of several subgroups making the policy implications even more blurred as it is not clear what the individual contributions of the different subgroups are. In this example here this might not make a big difference but should be kept in mind when using entirely different instrumental variables to identify the LATE.
In a last step, there are several possible scenarios to argue why **the given causal graph might be violated**. Angrist himself delivers one of them. After the lottery numbers were drawn, there was some time in between the drawing and the announcement of the draft-eligibility ceiling. This provoked behavioral responses of some individuals with low numbers to volunteer for the army in order to get better terms of service as well as enrolling in university which rendered them ineligible for the army. In our data, it is unobservable to see the fraction of individuals in each group to join university. If there was actually some avoidance behavior for those with low lottery numbers, then the instrument would be questionable as there would be a path from the Draft Lottery to unobservables (University) which affects earnings. At the same time there is also clearly a relation between University and Military Service.
Rosenzweig and Wolpin (2000) provide a causal graph that draws the general interpretability of the results in Angrist (1990) further into question. Let us look at the causal graph below now imagining that there was no directed graph from Draft Lottery to Civilian Experience. Their argument is that Military Service reduces Schooling and Civilian Experience which lowers Wages while affecting Wages directly and increasing them indirectly by reducing Schooling and increasing work experience. Those subtle mechanism are all collapsed into one measure by Angrist which gives an only insufficiently shallow answer to potentially more complex policy questions. Building up on this causal graph, Heckman (1997) challenges the validity of the instrument in general by making the point that there might be a directed graph from Draft Lottery to Civilian Experience. The argument goes as follows: Employers, after learning about their employees' lottery numbers, decrease the training on the job for those with a high risk of being drafted. If this is actually warranted the instrument Draft Lottery cannot produce unbiased estimates anymore.
<img src="material/fig-10-2.png" width="600" />
Morgan and Winship (2014) add to this that the bias introduced by this is further affected by how strongly Draft Lottery affects Military Service. Given the factor that the lottery alone does not determine military service but that there are tests, might cause the instrument to be rather weak and therefore a potential bias to be rather strong.
# 5. Extensions
---
## 5.1 Treatment effect with different years of earning
In the calculation of the average treatment in Table 4 Angrist chooses to calculate it for earnings in the years from 1981 to 84. While he plausibly argues that this most likely constitutes a long term effect (as those are the last years for which he has data) in comparison to earlier years, it does not give a complete picture. Looking at Table 1 again we can see that for the earnings differences in the years 81 to 84 quite big estimates are calculated. Assuming that the difference in probability of serving given eligibility versus noneligibility stays somewhat stable across the years, we would expect some heterogeneity in average treatment effects depending on which years we use the earnings data of. Angrist, though, does not investigate this although he has the data for it at hand. For example from a policy perspective one could easily argue that a look at the average treatment effect for earlier years (close to the years in which treatment happens) might be more relevant than the one for years after. This is because given the long time between the actual service and the earnings data of 1981 to 84 it is likely that second round effects are driving some of the results. These might be initially caused by veteran status but for later years the effect of veteran status might mainly act by means of other variables. For instance veterans after the war might be forced to take simple jobs due to their lack of work experience and from then on their path is determined by the lower quality of the job that they had to take right after war. For policy makers it might be of interest to see what happens to veterans right after service to see what needs to be done in order to stop second round effects from happening in the first place.
To give a more wholesome image, I estimate the results for Table 4 for different years of earnings of white men. As mentioned before the quality of the Total W-2 data set is rather low and the adjusted FICA is more plausible than the FICA data. This is why I only use the adjusted FICA data in the following. For the adjusted FICA I have data for Table 4 for the years from 1974 to 1984. For each possible four year range within those ten years I estimate Model 1 and 2 from Table 4 again.
Below I plot the average treatment effects obtained. On the x-axis I present the starting year of the range of the adjusted FICA data used. For starting value 74 it means that the average treatment effect is calculated for earnings data of the years 1974 to 77. The results at the starting year 81 are equivalent to the ones found by Angrist in Table 4 for white men.
```
# get the average treatment effects of Model 1 and 2 with adjusted FICA earnings for several different ranges of four years
results_model1 = np.empty((8, 4))
results_model2 = np.array([])
for number, start_year in enumerate(np.arange(74, 82)):
years = np.arange(start_year, start_year + 4)
flex_table4 = get_flexible_table4(data_cwhsc_new, years, ["ADJ"], [50, 51, 52, 53])
results_model1[number, :] = flex_table4["white"].loc[("Model 1", slice(None), "Value") , :].values.flatten()
results_model2 = np.append(results_model2, flex_table4["white"].loc[("Model 2", slice(None), "Value") , :].values)
# Plot the effects for white men in Model 1 and 2 (colors apart from Cohort 1950 are random, execute again to change them)
get_figure1_extension1(results_model1, results_model2)
```
The pattern is more complex than what we can see in the glimpse of Table 4 in the paper. We can see that there is quite some heterogeneity in average treatment effects across cohorts when looking at the data for early years. This changes when using data of later years. Further the fact of being a veteran does seem to play a role for the cohort 1953 right after the war but the treatment effect becomes insignificant when looking at later years. This is interesting as the cohort of 1953 was the one for which no one was drafted (remember that in 1973 no one was drafted as the last call was in December 1972).
Another observation is linked to the fact that draft eligibility does not matter for those born in 1953. These people appear to have voluntarily joined the army as no one of them could have possibly been drafted. This cannot be said for the cohorts before. Employers can only observe whether a person is a veteran and when they are born (and not if they are compliers or not). A theory could be that employers act on the loss of experience for initial wage setting for every army veteran right after the war. The fact that the cohort of 1953 could only be volunteers but not draftees could give them a boost in social status to catch up again in the long run, though. This mechanism might explain to a certain extent why we observe the upward sloping line for the cohort of 1953 (but not for the other groups).
As discussed in the critical assessment, we actually only capture the local average treatment effect of the compliers. Those are the ones who join the army when they are draft-eligible but do not when they are not. The identifying assumption for the LATE requires that everyone is a complier. This is probably not warranted for the cohort of 1953. In that year it is easily imaginable that there are both defiers and compliers which means that we do not capture the LATE for cohort 1953 in Model 1 and for cohort 1950-53 in Model 2 but something else we do not really know how to interpret. This might be another reason why we observe this peculiar pattern for the cohort of 1953. Following up on this remark I estimate the Model 2 again excluding the cohort of 1953 to focus on the cohorts for which the assumptions for LATE are likely to hold.
```
results_model2_53 = np.array([])
for number, start_year in enumerate(np.arange(74, 82)):
years = np.arange(start_year, start_year + 4)
flex_table4 = get_flexible_table4(data_cwhsc_new, years, ["ADJ"], [50, 51, 52])
results_model2_53 = np.append(results_model2_53, flex_table4["white"].loc[("Model 2", slice(None), "Value") , :].values)
get_figure2_extension1(results_model2, results_model2_53)
```
We can see that for later years the treatment effect is a bit lower when excluding the cohort of 1953. It confirms the findings of Angrist with the advantage of making it possible to attach a clearer interpretation to it.
Following the above path, it would also be interesting to vary the amount of instruments used by more than just the two ways Angrist has shown. It would be interesting to break down the interval size of lottery numbers further. Unfortunately I could no find a way to do that with the already pre-processed data I have at hand.
## 5.2 Bias Quantification
In the critical assessment I argued that the simple Wald estimate might be biased because employers know their employees' birth date and hence their draft eligibility. The argument was that employers invest less into the human capital of those that might be drafted. This would cause the instrument of draft eligibility to not be valid and hence suffer from bias. This bias can be calculated in the following way for a binary instrument:
\begin{align}
\frac{E[Y|Z=1] - E[Y|Z=0]}{E[D|Z=1] - E[D|Z=0]} = \delta + \frac{E[\epsilon|Z=1] - E[\epsilon|Z=0]}{E[D|Z=1] - E[D|Z=0]}
\end{align}
What has been done in the last column of Table 3 (the Wald estimate) is that Angrist calculated the left hand side of this equation. This calculation yields an unbiased estimate of the treatment effect of $D$ (veteran status) on $Y$ (earnings) $\delta$ if there is no effect of the instrument $Z$ (draft eligibility) on $Y$ through means of unobservables $\epsilon$. In our argumentation this assumption does not hold which means that $E[\epsilon|Z=1] - E[\epsilon|Z=0]$ is not equal to zero as draft eligibility affects $Y$ by the behavioral change of employers to make investing into human capital dependent on draft eligibility. Therefore the left hand side calculation is not equal to the true treatment effect $\delta$ but has to be adjusted by the bias $\frac{E[\epsilon|Z=1] - E[\epsilon|Z=0]}{E[D|Z=1] - E[D|Z=0]}$.
In this section I run a thought experiment in which I quantify this bias. The argumentation here is rather heuristic because I lack the resources to really find a robust estimate of the bias but it gives a rough idea of whether the bias might matter economically. My idea is the following. In order to get a measure of $E[\epsilon|Z=1] - E[\epsilon|Z=0]$ I have a look at estimates for the effect of work experience on earnings. Remember that the expected difference in earnings due to a difference in draft eligibility is caused by a loss in human capital for those draft eligible because they might miss out on on-the-job-training. This loss in on-the-job-training could be approximated by a general loss in working experience. For an estimate of that effect I rely on Keane and Wolpin (1997) who work with a sample for young men between 14 and 21 years old from the year 1979. The effect of working experience on real earnings could be at least not far off of the possible effect in our sample of adjusted FICA real earnings of 19 year old men for the years 1981 to 1984. Remember that lottery participants find out about whether they are draft eligible or not at the end of the year before they might be drafted. I assume that draft dates are spread evenly over the draft year. One could then argue that on average a draft eligible person stays in his job for another half a year after having found out about the eligibility and before being drafted. Hence, for on average half a year an employer might invest less into the human capital of this draft eligible man. I assume now that employers show a quite moderate behavioral response. During the six months of time, the employees only receive a five month equivalent of human capital gain (or work experience gain) as opposed to the six months they stay in the company. This means they loose one month of work experience on average in comparison to those that are not draft eligible.
To quantify this one month loss of work experience I take estimates from Keane and Wolpin (1997). For blue collar workers they roughly estimate the gain in real earnings in percent from an increase in a year of blue collar work experience to be 4.6 percent (actually their found effect depends on the years of work experience but I simplify this for my rough calculations). For white collar workers the equivalent estimate amounts to roughly 2.7 percent. I now take those as upper and lower bounds, calculate their one month counterparts and quantify the bias in the Wald estimates of the last column of Table 3. The bias $\frac{E[\epsilon|Z=1] - E[\epsilon|Z=0]}{E[D|Z=1] - E[D|Z=0]}$ is then roughly equal to the loss in annual real earnings due to one month less of work experience divided by the difference in probability of being a veteran conditional on draft eligibility.
The first table below depicts how the bias changes by cohort across the different years of real earnings with increasing estimates of how a loss in experience affects real earnings. Clearly with increasing estimates of how strong work experience contributes to real earnings, the bias gets stronger. This is logical as it is equivalent to an absolute increase in the nominator. Above that the bias is stronger for later years of earnings as the real earnings increase by year. Further the slope is steeper for later cohorts as the denominator is smaller for later cohorts. Given the still moderate assumption of a loss of one month of work experience we can see that the bias does not seem to be negligible economically especially when taking the blue collar percentage estimate.
```
# Calculate the bias, the true delta and the orginal Wald estimate for a ceratain interval of working experience effect
interval = np.linspace(0.025, 0.05, 50)/12
bias, true_delta, wald = get_bias(data_cwhsa, data_cwhsb, data_dmdc, data_sipp, data_cwhsc_new, interval)
# plot the bias by cohort
get_figure1_extension2(bias, interval)
```
To get a sense of how the size of the bias relates to the size of the previously estimated Wald coefficients, let us have look at the figure below. It shows for each cell consisting of a cohort and year combination, the Wald estimate from Table 3 as the horizontal line and the true $\delta$ depending on the weight of the loss in work experience as the upward sloping line. Given that our initial estimates of the Wald coefficients are in a range of only a few thousands, an estimated bias of roughly between 200 and 500 dollars cannot be characterized as incosiderable. Further given Angrist's policy question concerning Veteran compensation, even an estimate that is higher by 200 dollars makes a big difference when it is about compensating thousands of veterans.
```
# plot the the true delta (accounted for the bias) compared to the original Wald estimate
get_figure2_extension2(true_delta, wald, interval)
```
# 6. Conclusion
---
Regarding the overall quality and structure of Angrist (1990), reading it is a real treat. The controversy after its publication and the fact that it is highly cited clearly show how important its contribution was and still is. It is a great piece of discussion when it comes to the interpretability and policy relevance of instrumental variable approaches. As already reiterated in the critical assessment, one has to acknowledge the care Angrist put into this work. Although his results do not seem to prove reliable, it opened a whole discussion on how to use instrumental variables to get the most out of them. Another contribution that should not go unnoticed is that Angrist shows that instruments can be used even though they might not come from the same sample as the dependent and the endogenous variable. Practically, this is very useful as it widens possible areas of application for instrumental variables.
Overall, it has to be stated that the paper has some shortcomings but the care put into this paper and the good readibility allowed other researchers (and Angrist himself) to swoop in giving helpful remarks that improved the understanding of instrumental variable approaches for treatment effect evaluation.
# References
**Angrist, J.** (1990). [Lifetime Earnings and the Vietnam Era Draft Lottery: Evidence from Social Security Administrative Records](https://www.jstor.org/stable/2006669?seq=1#metadata_info_tab_contents). *American Economic Review*. 80. 313-36.
**Angrist, J. D., & Pischke, J.-S.** (2009). Mostly harmless econometrics: An empiricist's companion.
**Heckman, J.** (1997). Instrumental Variables: A Study of Implicit Behavioral Assumptions Used in Making Program Evaluations. *The Journal of Human Resources*, 32(3), 441-462. doi:10.2307/146178
**Keane, M., & Wolpin, K.** (1997). The Career Decisions of Young Men. *Journal of Political Economy*, 105(3), 473-522. doi:10.1086/262080
**Morgan, S., and Winship, C.** (2014). Counterfactuals and Causal Inference: Methods and Principles for Social Research (Analytical Methods for Social Research). Cambridge: Cambridge University Press. doi:10.1017/CBO9781107587991
**Rosenzweig, M. R. and Wolpin, K. I.**. (2000). “Natural ‘Natural Experiments’ in Economics.” *Journal of Economic Literature* 38:827–74.
**Wald, A.** (1940). The Fitting of Straight Lines if Both Variables are Subject to Error. *Ann. Math. Statist.* 11 , no. 3, 284--300.
# Appendix
### Key Variables in the Data Sets
#### data_cwhsa
| **Name** | **Description** |
|-----------------|--------------------------------------------|
| **index** | |
| byr | birth year |
| race | ethnicity, 1 for white and 2 for nonwhite |
| interval | interval of draft lottery numbers, 73 intervals with the size of five consecutive numbers |
| year | year for which earnings are collected |
| **variables** | |
| vmn1 | nominal earnings |
| vfin1 | fraction of people with zero earnings |
| vnu1 | sample size |
| vsd1 | standard deviation of earnings |
#### data_cwhsb
| **Name** | **Description** |
|-----------------|--------------------------------------------|
| **index** | |
| byr | birth year |
| race | ethnicity, 1 for white and 2 for nonwhite |
| interval | interval of draft lottery numbers, 73 intervals with the size of five consecutive numbers |
| year | year for which earnings are collected |
| type | source of the earnings data, "TAXAB" for FICA and "TOTAL" for Total W-2 |
| **variables** | |
| vmn1 | nominal earnings |
| vfin1 | fraction of people with zero earnings |
| vnu1 | sample size |
| vsd1 | standard deviation of earnings |
#### data_cwhsc_new
| **Name** | **Description** |
|-----------------|--------------------------------------------|
| **index** | |
| byr | birth year |
| race | ethnicity, 1 for white and 2 for nonwhite |
| interval | interval of draft lottery numbers, 73 intervals with the size of five consecutive numbers |
| year | year for which earnings are collected |
| type | source of the earnings data, "ADJ" for adjusted FICA, "TAXAB" for FICA and "TOTAL" for Total W-2 |
| **variables** | |
| earnings | real earnings in 1978 dollars |
| nj | sample size |
| nj0 | number of persons in the sample with zero earnings |
| iweight_old | weight for weighted least squares |
| ps_r | fraction of people having served in the army |
| ern74 to ern84 | unweighted covariance matrix of the real earnings |
#### data_dmdc
| **Name** | **Description** |
|-----------------|--------------------------------------------|
| **index** | |
| byr | birth year |
| race | ethnicity, 1 for white and 2 for nonwhite |
| interval | interval of draft lottery numbers, 73 intervals with the size of five consecutive numbers |
| **variables** | |
| nsrvd | number of people having served |
| ps_r | fraction of people having served |
#### data_sipp (this is the only micro data set)
| **Name** | **Description** |
|-----------------|--------------------------------------------|
| **index** | |
| u_brthyr | birth year |
| nrace | ethnicity, 0 for white and 1 for nonwhite |
| **variables** | |
| nvstat | 0 if man is not a veteran, 1 if he is |
| fnlwgt_5 | fraction of people with this index among overall sample |
| rsncode | 1 if person was draft eligible, else if not |
|
github_jupyter
|
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from auxiliary.auxiliary_figures import (get_figure1, get_figure2, get_figure3)
from auxiliary.auxiliary_tables import (get_table1, get_table2, get_table3, get_table4)
from auxiliary.auxiliary_data import process_data
from auxiliary.auxiliary_visuals import (background_negative_green, p_value_star)
from auxiliary.auxiliary_extensions import (get_flexible_table4, get_figure1_extension1, get_figure2_extension1,
get_bias, get_figure1_extension2, get_figure2_extension2)
import warnings
warnings.filterwarnings('ignore')
plt.rcParams['figure.figsize'] = [12, 6]
%%javascript
MathJax.Hub.Config({
TeX: { equationNumbers: { autoNumber: "AMS" } }
});
MathJax.Hub.Queue(
["resetEquationNumbers", MathJax.InputJax.TeX],
["PreProcess", MathJax.Hub],
["Reprocess", MathJax.Hub]
);
process_data("cwhsa")
process_data("cwhsb")
data_cwhsc_new = process_data("cwhsc_new")
data_cwhsc_new
for data in ["ADJ", "TAXAB", "TOTAL"]:
ax = sns.kdeplot(data_cwhsc_new.loc[data, "earnings"], color=np.random.choice(
np.array([sns.color_palette()]).flatten(), 4))
ax.set_xlim(xmax=20000)
ax.legend(["Adjusted FICA", "FICA", "TOTAL W-2"], loc="upper left")
ax.set_title("Kernel Density of the different Earning Measures")
# read in the original data sets
data_cwhsa = pd.read_stata("data/cwhsa.dta")
data_cwhsb = pd.read_stata("data/cwhsb.dta")
data_cwhsc_new = pd.read_stata("data/cwhsc_new.dta")
data_dmdc = pd.read_stata("data/dmdcdat.dta")
data_sipp = pd.read_stata("data/sipp2.dta")
get_figure1(data_cwhsa, data_cwhsb)
get_figure2(data_cwhsa, data_cwhsb)
table1 = get_table1(data_cwhsa, data_cwhsb)
table1["white"].style.applymap(background_negative_green)
table1["nonwhite"].style.applymap(background_negative_green)
table2 = get_table2(data_cwhsa, data_dmdc, data_sipp)
table2["white"]
table2["nonwhite"]
table3 = get_table3(data_cwhsa, data_cwhsb, data_dmdc, data_sipp, data_cwhsc_new)
p_value_star(table3["white"], slice(None), ("", "Service Effect in 1978 $"))
p_value_star(table3["nonwhite"], slice(None), ("", "Service Effect in 1978 $"))
get_figure3(data_cwhsc_new)
table4 = get_table4(data_cwhsc_new)
p_value_star(table4["white"], (slice(None), slice(None), ["Value", "Standard Error"]), (slice(None)))
p_value_star(table4["nonwhite"], (slice(None), slice(None), ["Value", "Standard Error"]), (slice(None)))
# get the average treatment effects of Model 1 and 2 with adjusted FICA earnings for several different ranges of four years
results_model1 = np.empty((8, 4))
results_model2 = np.array([])
for number, start_year in enumerate(np.arange(74, 82)):
years = np.arange(start_year, start_year + 4)
flex_table4 = get_flexible_table4(data_cwhsc_new, years, ["ADJ"], [50, 51, 52, 53])
results_model1[number, :] = flex_table4["white"].loc[("Model 1", slice(None), "Value") , :].values.flatten()
results_model2 = np.append(results_model2, flex_table4["white"].loc[("Model 2", slice(None), "Value") , :].values)
# Plot the effects for white men in Model 1 and 2 (colors apart from Cohort 1950 are random, execute again to change them)
get_figure1_extension1(results_model1, results_model2)
results_model2_53 = np.array([])
for number, start_year in enumerate(np.arange(74, 82)):
years = np.arange(start_year, start_year + 4)
flex_table4 = get_flexible_table4(data_cwhsc_new, years, ["ADJ"], [50, 51, 52])
results_model2_53 = np.append(results_model2_53, flex_table4["white"].loc[("Model 2", slice(None), "Value") , :].values)
get_figure2_extension1(results_model2, results_model2_53)
# Calculate the bias, the true delta and the orginal Wald estimate for a ceratain interval of working experience effect
interval = np.linspace(0.025, 0.05, 50)/12
bias, true_delta, wald = get_bias(data_cwhsa, data_cwhsb, data_dmdc, data_sipp, data_cwhsc_new, interval)
# plot the bias by cohort
get_figure1_extension2(bias, interval)
# plot the the true delta (accounted for the bias) compared to the original Wald estimate
get_figure2_extension2(true_delta, wald, interval)
| 0.692538 | 0.874238 |
# Original Voce-Chaboche Model Fitting Example 1
An example of fitting the original Voce-Chaboche model to a set of test data is provided.
Documentation for all the functions used in this example can be found by either looking at docstrings for any of the functions.
```
import RESSPyLab as rpl
import numpy as np
```
## Run optimization with multiple test data set
This is a simple example for fitting the Voce-Chaboche model to a set of test data.
We only use two backstresses in this model, additional backstresses can be specified by adding pairs of `0.1`'s to the list of `x_0`.
E.g., three backstresses would be
```
x_0 = [200000., 355., 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1]
```
Likewise, one backstress can be specified by removing a pair of `0.1`'s from the list below.
The overall steps to calibrate the model parameters are as follows:
1. Load the set of test data
2. Choose a starting point
3. Set the location to save the analysis history
4. Run the analysis
```
# Specify the true stress-strain to be used in the calibration
data_files = ['example_1.csv', 'example_2.csv']
# Set initial parameters for the Voce-Chaboche model with two backstresses
# [E, \sigma_{y0}, Q_\infty, b, C_1, \gamma_1, C_2, \gamma_2]
x_0 = [200000., 355., 0.1, 0.1, 0.1, 0.1, 0.1, 0.1]
# Log files for the parameters at each step, and values of the objective function at each step
x_log = './output/x_log.txt'
fxn_log = './output/fxn_log.txt'
# Run the calibration
# Set filter_data=True if you have NOT already filtered/reduced the data
# We recommend that you filter/reduce the data beforehand (i.e., filter_data=False is recommended)
x_sol = rpl.vc_param_opt(x_0, data_files, x_log, fxn_log, filter_data=False)
```
## Plot results
After the analysis is finished we can plot the test data versus the fitted model.
Note that we add two dummy parameters to the list of final parameters because the plotting function was written for the updated Voce-Chaboche model that has two additional parameters.
Setting the first of these two additional parameters equal to zero neglects the effects of the updated model.
If we set `output_dir='./output/'`, for example, instead of `output_dir=''` the `uvc_data_plotter` function will save pdf's of all the plots instead of displaying them below.
The function `uvc_data_multi_plotter` is also provided to give more fine-grained control over the plotting process, and can compare multiple analyses.
```
data = rpl.load_data_set(data_files)
# Added parameters are necessary for plotting the Voce-Chaboche model
x_sol_2 = np.insert(x_sol, 4, [0., 1.])
rpl.uvc_data_plotter(x_sol_2, data, output_dir='', file_name='vc_example_plots', plot_label='Fitted')
```
|
github_jupyter
|
import RESSPyLab as rpl
import numpy as np
x_0 = [200000., 355., 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1]
# Specify the true stress-strain to be used in the calibration
data_files = ['example_1.csv', 'example_2.csv']
# Set initial parameters for the Voce-Chaboche model with two backstresses
# [E, \sigma_{y0}, Q_\infty, b, C_1, \gamma_1, C_2, \gamma_2]
x_0 = [200000., 355., 0.1, 0.1, 0.1, 0.1, 0.1, 0.1]
# Log files for the parameters at each step, and values of the objective function at each step
x_log = './output/x_log.txt'
fxn_log = './output/fxn_log.txt'
# Run the calibration
# Set filter_data=True if you have NOT already filtered/reduced the data
# We recommend that you filter/reduce the data beforehand (i.e., filter_data=False is recommended)
x_sol = rpl.vc_param_opt(x_0, data_files, x_log, fxn_log, filter_data=False)
data = rpl.load_data_set(data_files)
# Added parameters are necessary for plotting the Voce-Chaboche model
x_sol_2 = np.insert(x_sol, 4, [0., 1.])
rpl.uvc_data_plotter(x_sol_2, data, output_dir='', file_name='vc_example_plots', plot_label='Fitted')
| 0.486332 | 0.989889 |
## 신경망 학습
- `학습`이란 훈련 데이터로부터 가중치 매개변수의 최적값을 자동으로 획득하는 것을 의미한다. 이번 장에서는 `신경망이 학습할 수 있도록 해주는 지표인 손실함수`에 대해 알아볼 것이다. 손실 함수의 결과값을 최소로 만드는 가중치 매개변수를 찾는 것이 학습의 목표이다.
### 데이터에서 학습한다!
- `신경망의 특징은 데이터를 보고 학습할 수 있다는 점이다.` 데이터에서 학습한다는 것은 가중치 매개변수의 값을 `데이터를 보고 자동으로 결정한다는 의미이다.` 기계학습 방식에서는 feature를 사람이 설계하지만, 신경망은 `이미지에 포함된 중요한 특징까지도 기계가 스스로 학습할 것이다.`
- 이미지에서 feature를 추출하고 그 특징의 패턴을 기계학습 기술로 학습하는 방법이 있다. 여기서 말하는 feature는 입력 데이터(입력 이미지)에서 본질적인 데이터(중요한 데이터)를 정확하게 추출할 수 있도록 설계된 변환기를 가리킨다. 이미지의 feature는 보통 벡터로 기술하고, 컴퓨터 비전 분야에서는 SIFT, SURF, HOG 등의 feature를 많이 사용한다. 이런 feature를 사용하여 이미지 데이터를 벡터로 변환하고, 변환된 벡터를 가지고 지도 학습 방식의 대표 분류 기법인 SVM, KNN 등으로 학습할 수 있다. 하지만 `위의 방법들 조차도 feature를 만드는 것은 사람이므로 적합한 feature를 사용하지 않으면 좋은 결과를 얻을 수 없다.`
- 신경망의 이점은 `모든 문제를 같은 맥락에서 풀 수 있다는 점이다.` 예를 들어 '5'를 인식하는 문제든, '개'를 인식하는 문제든, 아니면 '사람의 얼굴'을 인식하는 문제든, 세부사항과 관계없이 `신경망은 주어진 데이터를 온전히 학습하고, 주어진 문제의 패턴을 발견하려 시도한다. 즉, 신경망은 모든 문제를 주어진 데이터 그대로를 입력 데이터로 활용해 'end-to-end'로 학습할 수 있다.`
### 손실함수(loss function)
- 손실함수를 기준으로 최적의 매개변수 값을 탐색한다. 손실 함수는 `신경망 성능의 나쁨을 나타내는 지표`로, 현재의 신경망이 훈련 데이터를 얼마나 잘 처리하지 못하느냐를 나타낸다.
#### MSE(Mean squared error, 평균 제곱 오차)
$$E = \frac{1}{2} \sum_{k}(y_{k} - t_{k})^{2}$$
```
def mean_squared_error(y, t):
return 0.5 * np.sum((y-t)**2)
```
#### Cross entropy error(CEE, 교차 엔트로피 오차)
- 정답일 때의 출력이 전체 값을 정하게 된다.
$$E = - \sum_{k}t_{k}\log{y_{k}}$$
```
def cross_entropy_error(y, t):
delta = 1e-7
return -np.sum(t * log(y + delta))
```
### 미니배치 학습
기계학습 문제는 훈련 데이터를 사용해 학습한다. 더 구체적으로 말하면 훈련 데이터에 대한 손실 함수의 값을 구하고, 그 값을 최대한 줄여주는 매개변수를 찾아낸다. 이렇게 하려면 모든 훈련 데이터를 대상으로 손실 함수 값을 구해야 한다. 즉, 훈련 데이터가 100개 있으면 그로부터 계산한 100개의 손실 함수 값들의 합을 지표로 삼는 것이다. 허나, 더 많은 데이터를 학습하려는 경우 일일이 손실 함수를 계산하는 것은 현실적이지 않을 것이다. 이런 경우 데이터 일부를 추려 전체의 "근사치"로 이용할 수 있다. `신경망 학습에서도 훈련 데이터로부터 일부만 골라 학습을 수행`한다. 이 일부를 `미니배치(mini-batch)`라고 한다.
- 예를 들어, 60,000장의 훈련 데이터 중에서 100장을 무작위로 뽑아 그 100장만을 사용하여 학습하는 것이다. 이러한 학습 방법을 `미니배치 학습`이라고 한다. np.random.choice() 함수를 사용하여 랜덤으로 뽑아낸다. 예를 들면 np.random.choice(60000, 10)은 0이상 60000미만의 수 중에서 무작위로 10개를 골라낸다.
```
import sys, os
import numpy as np
from dataset.mnist import load_mnist
sys.path.append(os.pardir)
(x_train, label_train),(x_test, label_test) = load_mnist(normalize=True, one_hot_label=True)
print(x_train.shape)
print(label_train.shape)
train_size = x_train.shape[0]
batch_size = 10
batch_mask = np.random.choice(train_size, batch_size)
x_batch = x_train[batch_mask]
label_batch = label_train[batch_mask]
```
### (배치용) 교차 엔트로피 오차 구현하기
```
def cross_entropy_error(y, t):
if y.dim == 1:
t = t.reshape(1, t.size)
y = y.reshape(1, y.size)
batch_size = y.shape[0]
delta = 1e-7
return -np.sum(t * np.log(y + delta)) / batch_size
```
### 정답 레이블이 one-hot encoding이 아니라 '2'나 '7' 등의 숫자 레이블로 주어졌을때의 오차 구현하기
- 이 구현에서는 `one-hot encoding일 때 t가 0인 원소는 교차 엔트로피 오차도 0이므로, 그 계산은 무시해도 된다는 것이 핵심`이다.
- 이 예에서는 y[np.arange(batch_size), t]는 만약 batch_size=5이고, t에 레이블이 [2, 7, 0, 9, 4]와 같이 저장되어 있다고 가정하면, [y[0, 2], y[1, 7], y[2, 0], y[3, 9], y[4, 4]]인 넘파이 배열을 생성한다.
```
def cross_entropy_error(y, t):
if y.dim == 1:
t = t.reshape(1, t.size)
y = y.reshape(1, y.size)
batch_size = y.shape[0]
delta = 1e-7
return -np.sum(np.log(y[np.arange(batch_size), t] + delta)) / batch_size
```
### 경사 하강법
- 일반적인 문제의 손실 함수는 매우 복잡하다. 매개변수 공간이 광대하여 어디가 최솟값이 되는 곳인지를 짐작할 수 없다. 이런 상황에서 기울기를 잘 이용햐 함수의 최솟값(또는 가능한 한 작은값)을 찾으려는 것이 경사 하강법이다.
- `기울기가 가리키는 쪽은 해당 지점에서의 함수의 출력 값을 가장 크게 줄이는 방향이다.` 그러나, 기울기가 가리키는 곳에 정말 함수의 최솟값이 있는지, 그쪽이 정말로 나아갈 방향인지는 보장할 수 없다. 실제로 복잡한 함수에서는 기울기가 가리키는 방향에 최솟값이 없는 경우가 대부분이다.
- 함수가 극솟값, 최솟값, 또 saddle point(안장점)이 되는 곳에서 기울기가 0이다. 극솟값은 국소적인 최소값, 즉 한정된 범위에서의 최솟값인 점이다. 안장점은 어느 방향에서 보면 극댓값이고 다른 방향에서 보면 극솟값이 되는 점이다. 경사하강법은 기울기가 0인 지점인 곳을 찾지만 그것이 optimal solution이라고는 할 수 없다.(극솟값이나 안장점일 가능성이 있다.) 또, 복잡하고 찌그러진 모양의 함수라면 (대부분) 평평한 곳으로 파고들면서 Plateau(고원)이라 하는, 학습이 진행되지 않는 정체기에 빠질 수 있다.
```
def numerical_gradient_1d(f, x):
h = 1e-4
grad = np.zeros_like(x) # x와 형상이 같은 배열을 생성하며 원소들이 모두 0인 배열
for idx in range(x.size):
tmp_val = x[idx]
#f(x+h)계산
x[idx] = tmp_val + h
fxh1 = f(x)
#f(x-h)계산
x[idx] = tmp_val - h
fxh2 = f(x)
grad[idx] = (fxh1 - fxh2) / (2*h)
x[idx] = tmp_val # 값 복원
return grad
```
함수의 기울기는 numerical_gradient(f,x)로 구하고, 그 기울기에 lr을 곱한 값으로 갱신하는 처리를 step_num이 반복한다.
- 학습률 같은 매개변수를 `하이퍼 파라미터(hyper parameter)`라고 합니다. 이는 가중치와 편향 같은 신경망의 매개변수와는 성질이 다른 매개변수이다. 신경망의 가중치 매개변수는 훈련 데이터와 학습 알고리즘에 의해서 '자동'으로 획득되는 매개변수인 반면, `학습률 같은 하이퍼 파라미터는 사람이 직접 설정해야하는 매개변수인 것이다.` 일반적으로는 이 하이퍼 파라미터들은 여러 후보 값 중에서 시험을 통해 가장 잘 학습하는 값을 찾는 과정을 거쳐야 한다.
```
def gradient_descent(f, init_x, lr=0.01, step_num=100):
"""
--------------------------------------
f : 최적화하려는 함수
init_x : 초기값
lr : learning rate
step_num : 경사법에 따른 반복 횟수
--------------------------------------
"""
x = init_x
for i in range(step_num):
grad = numerical_gradient_1d(f, x)
x -= lr * grad
return x
```
문제 : 경사하강법으로 $f(x_{0}, x_{1}) = x_{0}^2 + x_{1}^2$의 최솟값을 구하라.
```
def function_2(x):
return x[0]**2 + x[1]**2
gradient_descent(function_2, init_x=np.array([-3.0, 4.0]), lr=0.1, step_num=100)
import sys, os
sys.path.append(os.pardir)
import numpy as np
from common.functions import softmax, cross_entropy_error
from common.gradient import numerical_gradient
class simpleNet:
def __init__(self):
self.W = np.random.randn(2,3) # 정규분포로 초기화
def predict(self, x):
return np.dot(x, self.W)
def loss(self, x, t):
z = self.predict(x)
y = softmax(z)
loss = cross_entropy_error(y, t)
return loss
net = simpleNet()
print(net.W)
x = np.array([0.6, 0.9])
p = net.predict(x)
print(p)
print(np.argmax(p))
t = np.array([0, 0, 1])
net.loss(x, t)
f = lambda w: net.loss(x, t)
dW = numerical_gradient(f, net.W)
print(dW)
```
dW의 내용을 보면, 예를 들어 $\frac{\delta L}{\delta W}$의 $\frac{\delta L}{\delta W_{11}}$은 대략 0.1이다.
### 이는 $W_{11}$을 h만큼 늘리면 손실함수의 값은 0.2h만큼 증가한다는 의미이다.
마찬가지로 $\frac{\delta L}{\delta W_{13}}$은 대략 -0.4이므로 $\frac{\delta L}{\delta W_{13}}$을 h만큼 늘리면 손실 함수의 값은 0.5h만큼 감소하는 것이다. `그래서 손실 함수를 줄인다는 관점에서는` $W_{23}$ `은 양의 방향으로 갱신하고,` $W_{11}$ `은 음의 방향으로 갱신해야 함을 알 수 있다.`
### 2층 신경망 클래스 구현하기
신경망의 학습 절차는 다음과 같다. 아래는 `경사하강법으로 매개변수를 갱신하는 방법이며 미니배치로 데이터를 무작위로 선정하기 때문에 SGD(Stochastic Gradient Descent)라고 부른다. 확률적으로 무작위로 골라낸 데이터에 대해 수행하는 경사 하강법이라는 의미이다.`
#### 전제
- 신경망에는 적응 가능한 가중치와 편향이있고 이 가중치와 편향을 훈련 데이터에 적응하도록 조정하는 과정을 '학습'이라 한다. 신경망 학습은 다음과 같이 4단계로 수행한다.
#### 1단계 - 미니배치
- 훈련 데이터 중 일부를 무작위로 가져온다. 이렇게 선별한 데이터를 미니배치라 하며, 그 미니배치의 손실함수 값을 줄이는 것이 목표이다.
#### 2단계 - 기울기 산출
- 미니배치의 손실 함수 값을 줄이기 위해 각 가중치 매개변수의 기울기를 구한다. 기울기는 손실 함수의 값을 가장 작게 하는 방향을 제시한다.
#### 3단계 - 매개변수 갱신
- 가중치 매개변수를 기울기 방향으로 아주 조금 갱신한다.
#### 4단계 - 반복
- 1~3단계 반복한다.
```
import sys, os
sys.path.append(os.pardir)
from common.functions import *
from common.gradient import numerical_gradient
class TwoLayerNet:
def __init__(self, input_size, hidden_size, output_size, weight_init_std=0.01):
# 가중치 초기화
self.params = {}
self.params["W1"] = weight_init_std * np.random.randn(input_size, hidden_size)
self.params["b1"] = np.zeros(hidden_size)
self.params["W2"] = weight_init_std * np.random.randn(hidden_size, output_size)
self.params["b2"] = np.zeros(output_size)
def predict(self, x):
W1, W2 = self.params["W1"], self.params["W2"]
b1, b2 = self.params["b1"], self.params["b2"]
a1 = np.dot(x, W1) + b1
z1 = sigmoid(a1)
a2 = np.dot(z1, W2) + b2
y = softmax(a2)
return y
# x : 입력 데이터, label : 정답 레이블
def loss(self, x, label):
y = self.predict(x)
return cross_entropy_error(y, label)
def accuracy(self, x, label):
y = self.predict(x)
y = np.argmax(y, axis=1)
y = np.argmax(label, axis=1)
accuracy = np.sum(y == label) / float(x.shape[0])
return accuracy
def numerical_gradient(self, x, label):
loss_W = lambda x : self.loss(x, label)
grads = {}
grads['W1'] = numerical_gradient(loss_W, self.params['W1'])
grads['b1'] = numerical_gradient(loss_W, self.params['b1'])
grads['W2'] = numerical_gradient(loss_W, self.params['W2'])
grads['b2'] = numerical_gradient(loss_W, self.params['b2'])
return grads
```
### 미니배치 학습 구현하기
```
import numpy as np
from dataset.mnist import load_mnist
from two_layer_net import TwoLayerNet
(x_train, label_train), (x_test, label_test)= load_mnist(normalize=True, one_hot_label=True)
train_loss_list = []
train_acc_list = []
test_acc_list =[]
#hyper parameter
iters_num = 10000 # 반복횟수
train_size = x_train.shape[0]
batch_size = 100 # mini-batch size
learning_rate = 0.1
network = TwoLayerNet(input_size=784, hidden_size=50, output_size=10)
iter_per_epoch = max(train_size/ batch_size, 1)
for i in range(iters_num):
# mini-batch
batch_mask = np.random.choice(train_size, batch_size)
x_batch = x_train[batch_mask]
label_batch = label_train[batch_mask]
# 기울기 계산
# grad = network.numerical_gradient(x_batch, label_batch) # 이건 너무 느림 진짜!!!
grad = network.gradient(x_batch, label_batch) # 성능 개선판!
# 매개변수 갱신
for key in ["W1", "b1", "W2", "b2"]:
network.params[key] -= learning_rate *grad[key]
# 학습경과 기록
loss = network.loss(x_batch, label_batch)
train_loss_list.append(loss)
# 1 epoch당 정확도 계산
if i % iter_per_epoch == 0:
train_acc = network.accuracy(x_train, label_train)
test_acc = network.accuracy(x_test, label_test)
train_acc_list.append(train_acc)
test_acc_list.append(test_acc)
print("train acc, test acc | " + str(train_acc) + ", " + str(test_acc))
```
### 시험 데이터로 평가하기
- 훈련 데이터의 mini-batch에 대한 손실 함수의 값이 서서히 내려가는 것을 확인할 수 있다. 신경망이 잘 학습하고 있다는 것이다. 그러나 `overfitting을 일으키지 않는지 확인해야한다.` overfitting이 되었다는 것은, 예를 들어 훈련 데이터에 포함된 이미지만 제대로 구분하고, 그렇지 않은 이미지는 식별할 수 없다는 뜻이다. `우리의 원래 목표는 범용적인 능력을 익히는 것이다.`
- 아래에서는 1epoch(훈련 데이터를 모두 소진했을 때의 횟수에 해당.예를 들어 훈련 데이터 10,000개를 100개의 mini-batch로 학습할 경우, SGD를 100회 반복하면 모든 훈련 데이터를 소진하게 되는데 이 경우 100회가 1epoch이다.)별로 훈련 데이터와 시험 데이터에 대한 정확도를 기록한다. 그 이유는 매번 for문 안에서 계산하기에는 시간이 걸리며 자주 기록할 필요는 없기 때문이다. 추세를 알 수 있으면 충분!!
- 아래의 그래프로 보아 위의 학습에서는 overfitting이 일어나지 않았음을 알 수 있다.
```
# 그래프 그리기
markers = {'train': 'o', 'test': 's'}
x = np.arange(len(train_acc_list))
plt.plot(x, train_acc_list, label='train acc')
plt.plot(x, test_acc_list, label='test acc', linestyle='--')
plt.xlabel("epochs")
plt.ylabel("accuracy")
plt.ylim(0, 1.0)
plt.legend(loc='lower right')
plt.show()
```
### 오차역전파법(Backpropagation)
수치 미분은 단순하고 구현하기도 쉽지만 계산 시간이 오래 걸린다는 것이 단점이다. 이러한 단점을 보완하는 가중치 매개변수의 기울기를 효율적으로 계산하는 `오차역전파법(Backpropagation)`
- 계산 그래프의 특징은 `국소적 계산`을 전파함으로써 최종결과를 얻는다는 점이다. 즉, 각 노드는 자신과 관련한 계산 외에는 아무것도 신경 쓸 것이 없다. 또한 `중간 계산 결과를 모두 보관`할 수 있다. 이런 특징을 바탕으로 `실제 계산 그래프를 사용하는 가장 큰 이유는 역전파를 통해 미분을 효율적으로 계산할 수 있는 점에 있다.`
- 덧셈의 역전파에서는 상류의 값을 그대로 흘려보내서 순방향 입력 신호의 값은 필요하지 않는다. 다만, 곱세의 역전파는 순방향 입력 신호의 값이 필요하다. 서로 입력된 것들을 반대로 곱해 주어야 하기 때문이다. 그래서 곱셈 노드를 구현할 때는 순전파의 입력 신호를 변수에 저장해둔다.
### 단순한 계층 구현
#### 곱셈 계층
```
class MulLayer:
def __init__(self):
self.x = None
self.y = None
def forward(self, x, y):
self.x = x
self.y = y
out = x * y
return out
def backward(self, dout):
dx = dout * self.y
dy = dout * self.x
return dx, dy
```
#### 덧셈 계층
```
class AddLayer:
def __init__(self):
pass
def forward(self, x, y):
out = x + y
return out
def backward(self, dout):
dx = dout * 1
dy = dout * 1
return dx, dy
```
### 활성화 함수 계층 구현하기
#### ReLU 계층
순전파 때의 입력인 x가 0보다 크면 역전파는 상류의 값을 그대로 하류로 흘린다. 반면, 순전파 때 x가 0이하이면 역전파 때는 하류로 신호를 보내지 않는다.
$$y=\begin{cases}
x & x > 0 \\
0 & x \leq 0
\end{cases}$$
$$\frac{\delta y}{\delta x}=\begin{cases}
1 & x > 0 \\
0 & x \leq 0
\end{cases}$$
- ReLU 계층은 전기 회로의 '스위치'에 비유할 수 있다. 순전파 때 전류가 흐르고 있으면 스위치를 ON으로 하고, 흐르지 않으면 OFF로 한다. 역전파 때는 스위치가 ON이라면 전류가 그대로 흐르고, OFF면 더 이상 흐르지 않는다.
```
class Relu:
def __init__(self):
self.mask = None
def forward(self, x):
self.mask = x <= 0
out = x.copy()
out[self.mask] = 0
return out
def backward(self, dout):
dout[self.mask] = 0
dx = dout
return dx
x = np.array([[1.0, -0.5], [-2.0, 3.0]])
mask = x<=0
mask
```
#### Sigmoid 계층
$$y = \frac{1}{1 + \exp(-x)}$$
순전파의 출력을 인스턴스 변수 out에 보관했다가, 역전파 계산 때 그 값을 사용한다.
```
class Sigmoid:
def __init__(self):
self.out = None
def forward(self, x):
out = 1 / (1 + np.exp(-x))
self.out = out
return out
def backward(self, dout):
dx = dout * (1.0 - self.out) * self.out
return dx
```
### Affine / Softmax 계층 구현하기
#### Affine 계층
신경망의 순전파에서는 가중치 신호의 총합을 게산하기 때문에 행렬의 곱을 사용했다. 그러한 행렬의 곱을 기하학에서는 `어파인 변환(Affine transformation)`이라고 한다.
```
class Affine:
def __init__(self, W, b):
self.W = W
self.b = b
self.x = None
self.dW = None
self.db = None
def forward(self, x):
self.x = x
out = np.dot(x, self.W) + self.b
return out
def backward(self, dout):
dx = np.dot(dout, self.W.T)
self.dW = np.dot(self.x.T, dout)
self.db = np.sum(dout, axis=0)
```
#### Softmax-with-Loss 계층
신경망에서 수행하는 작업은 학습과 추론 두가지이다. 추론할 때는 일반적으로 Softmax 계층을 사용하지 않는다. 신경망은 추론할 때는 마지막 Affine 계층의 출력을 인식 결과로 이용한다. 또한 `신경망에서 정규화하지 않는 출력 결과를 점수(Score)라고 한다.`
- 즉, 신경망 추론에서 답을 하나만 내는 경우에는 가장 높은 점수만 알면 되니 Softmax 계층은 필요 없다는 것이다. 반면, 신경망을 학습할 때는 Softmax계층이 필요하다.
- 아래의 코드에서 `역전파 때에는 전파하는 값을 배치의 수(batch_size)로 나눠서 데이터 1개당 오차를 앞 계층으로 전파하는 점에 주의하자!`
```
class SoftmaxWithLoss:
def __init__(self):
self.loss = None
self.y = None
self.t = None
def forward(self, x, t):
self.t = t
self.y = softmax(x)
self.loss = cross_entropy_error(self.y, self.t)
return self.loss
def backward(self, dout=1):
batch_size = self.t.shape[0]
dx = (self.y - self.t) / batch_size
return dx
```
### Backpropagation을 적용한 신경망 구현
- 계층을 사용함으로써 인식결과를 얻는 처리(predict())와 기울기를 구하는 처리(gradient()) 계층의 전파만으로 동작이 이루어질 수 있다.
- OrderedDict를 사용하여 순전파 때는 추가한 순서대로 각 계층의 forward() 메서드를 호출하기만 하면 처리가 완료된다.
```
# coding: utf-8
import sys, os
sys.path.append(os.pardir) # 부모 디렉터리의 파일을 가져올 수 있도록 설정
import numpy as np
from common.layers import *
from common.gradient import numerical_gradient
from collections import OrderedDict
class TwoLayerNet:
def __init__(self, input_size, hidden_size, output_size, weight_init_std = 0.01):
# 가중치 초기화
self.params = {}
self.params['W1'] = weight_init_std * np.random.randn(input_size, hidden_size)
self.params['b1'] = np.zeros(hidden_size)
self.params['W2'] = weight_init_std * np.random.randn(hidden_size, output_size)
self.params['b2'] = np.zeros(output_size)
# 계층 생성
self.layers = OrderedDict()
self.layers['Affine1'] = Affine(self.params['W1'], self.params['b1'])
self.layers['Relu1'] = Relu()
self.layers['Affine2'] = Affine(self.params['W2'], self.params['b2'])
self.lastLayer = SoftmaxWithLoss()
def predict(self, x):
for layer in self.layers.values():
x = layer.forward(x)
return x
# x : 입력 데이터, t : 정답 레이블
def loss(self, x, t):
y = self.predict(x)
return self.lastLayer.forward(y, t)
def accuracy(self, x, t):
y = self.predict(x)
y = np.argmax(y, axis=1)
if t.ndim != 1 : t = np.argmax(t, axis=1)
accuracy = np.sum(y == t) / float(x.shape[0])
return accuracy
# x : 입력 데이터, t : 정답 레이블
def numerical_gradient(self, x, t):
loss_W = lambda W: self.loss(x, t)
grads = {}
grads['W1'] = numerical_gradient(loss_W, self.params['W1'])
grads['b1'] = numerical_gradient(loss_W, self.params['b1'])
grads['W2'] = numerical_gradient(loss_W, self.params['W2'])
grads['b2'] = numerical_gradient(loss_W, self.params['b2'])
return grads
def gradient(self, x, t):
# forward
self.loss(x, t)
# backward
dout = 1
dout = self.lastLayer.backward(dout)
layers = list(self.layers.values())
layers.reverse()
for layer in layers:
dout = layer.backward(dout)
# 결과 저장
grads = {}
grads['W1'], grads['b1'] = self.layers['Affine1'].dW, self.layers['Affine1'].db
grads['W2'], grads['b2'] = self.layers['Affine2'].dW, self.layers['Affine2'].db
return grads
```
|
github_jupyter
|
def mean_squared_error(y, t):
return 0.5 * np.sum((y-t)**2)
def cross_entropy_error(y, t):
delta = 1e-7
return -np.sum(t * log(y + delta))
import sys, os
import numpy as np
from dataset.mnist import load_mnist
sys.path.append(os.pardir)
(x_train, label_train),(x_test, label_test) = load_mnist(normalize=True, one_hot_label=True)
print(x_train.shape)
print(label_train.shape)
train_size = x_train.shape[0]
batch_size = 10
batch_mask = np.random.choice(train_size, batch_size)
x_batch = x_train[batch_mask]
label_batch = label_train[batch_mask]
def cross_entropy_error(y, t):
if y.dim == 1:
t = t.reshape(1, t.size)
y = y.reshape(1, y.size)
batch_size = y.shape[0]
delta = 1e-7
return -np.sum(t * np.log(y + delta)) / batch_size
def cross_entropy_error(y, t):
if y.dim == 1:
t = t.reshape(1, t.size)
y = y.reshape(1, y.size)
batch_size = y.shape[0]
delta = 1e-7
return -np.sum(np.log(y[np.arange(batch_size), t] + delta)) / batch_size
def numerical_gradient_1d(f, x):
h = 1e-4
grad = np.zeros_like(x) # x와 형상이 같은 배열을 생성하며 원소들이 모두 0인 배열
for idx in range(x.size):
tmp_val = x[idx]
#f(x+h)계산
x[idx] = tmp_val + h
fxh1 = f(x)
#f(x-h)계산
x[idx] = tmp_val - h
fxh2 = f(x)
grad[idx] = (fxh1 - fxh2) / (2*h)
x[idx] = tmp_val # 값 복원
return grad
def gradient_descent(f, init_x, lr=0.01, step_num=100):
"""
--------------------------------------
f : 최적화하려는 함수
init_x : 초기값
lr : learning rate
step_num : 경사법에 따른 반복 횟수
--------------------------------------
"""
x = init_x
for i in range(step_num):
grad = numerical_gradient_1d(f, x)
x -= lr * grad
return x
def function_2(x):
return x[0]**2 + x[1]**2
gradient_descent(function_2, init_x=np.array([-3.0, 4.0]), lr=0.1, step_num=100)
import sys, os
sys.path.append(os.pardir)
import numpy as np
from common.functions import softmax, cross_entropy_error
from common.gradient import numerical_gradient
class simpleNet:
def __init__(self):
self.W = np.random.randn(2,3) # 정규분포로 초기화
def predict(self, x):
return np.dot(x, self.W)
def loss(self, x, t):
z = self.predict(x)
y = softmax(z)
loss = cross_entropy_error(y, t)
return loss
net = simpleNet()
print(net.W)
x = np.array([0.6, 0.9])
p = net.predict(x)
print(p)
print(np.argmax(p))
t = np.array([0, 0, 1])
net.loss(x, t)
f = lambda w: net.loss(x, t)
dW = numerical_gradient(f, net.W)
print(dW)
import sys, os
sys.path.append(os.pardir)
from common.functions import *
from common.gradient import numerical_gradient
class TwoLayerNet:
def __init__(self, input_size, hidden_size, output_size, weight_init_std=0.01):
# 가중치 초기화
self.params = {}
self.params["W1"] = weight_init_std * np.random.randn(input_size, hidden_size)
self.params["b1"] = np.zeros(hidden_size)
self.params["W2"] = weight_init_std * np.random.randn(hidden_size, output_size)
self.params["b2"] = np.zeros(output_size)
def predict(self, x):
W1, W2 = self.params["W1"], self.params["W2"]
b1, b2 = self.params["b1"], self.params["b2"]
a1 = np.dot(x, W1) + b1
z1 = sigmoid(a1)
a2 = np.dot(z1, W2) + b2
y = softmax(a2)
return y
# x : 입력 데이터, label : 정답 레이블
def loss(self, x, label):
y = self.predict(x)
return cross_entropy_error(y, label)
def accuracy(self, x, label):
y = self.predict(x)
y = np.argmax(y, axis=1)
y = np.argmax(label, axis=1)
accuracy = np.sum(y == label) / float(x.shape[0])
return accuracy
def numerical_gradient(self, x, label):
loss_W = lambda x : self.loss(x, label)
grads = {}
grads['W1'] = numerical_gradient(loss_W, self.params['W1'])
grads['b1'] = numerical_gradient(loss_W, self.params['b1'])
grads['W2'] = numerical_gradient(loss_W, self.params['W2'])
grads['b2'] = numerical_gradient(loss_W, self.params['b2'])
return grads
import numpy as np
from dataset.mnist import load_mnist
from two_layer_net import TwoLayerNet
(x_train, label_train), (x_test, label_test)= load_mnist(normalize=True, one_hot_label=True)
train_loss_list = []
train_acc_list = []
test_acc_list =[]
#hyper parameter
iters_num = 10000 # 반복횟수
train_size = x_train.shape[0]
batch_size = 100 # mini-batch size
learning_rate = 0.1
network = TwoLayerNet(input_size=784, hidden_size=50, output_size=10)
iter_per_epoch = max(train_size/ batch_size, 1)
for i in range(iters_num):
# mini-batch
batch_mask = np.random.choice(train_size, batch_size)
x_batch = x_train[batch_mask]
label_batch = label_train[batch_mask]
# 기울기 계산
# grad = network.numerical_gradient(x_batch, label_batch) # 이건 너무 느림 진짜!!!
grad = network.gradient(x_batch, label_batch) # 성능 개선판!
# 매개변수 갱신
for key in ["W1", "b1", "W2", "b2"]:
network.params[key] -= learning_rate *grad[key]
# 학습경과 기록
loss = network.loss(x_batch, label_batch)
train_loss_list.append(loss)
# 1 epoch당 정확도 계산
if i % iter_per_epoch == 0:
train_acc = network.accuracy(x_train, label_train)
test_acc = network.accuracy(x_test, label_test)
train_acc_list.append(train_acc)
test_acc_list.append(test_acc)
print("train acc, test acc | " + str(train_acc) + ", " + str(test_acc))
# 그래프 그리기
markers = {'train': 'o', 'test': 's'}
x = np.arange(len(train_acc_list))
plt.plot(x, train_acc_list, label='train acc')
plt.plot(x, test_acc_list, label='test acc', linestyle='--')
plt.xlabel("epochs")
plt.ylabel("accuracy")
plt.ylim(0, 1.0)
plt.legend(loc='lower right')
plt.show()
class MulLayer:
def __init__(self):
self.x = None
self.y = None
def forward(self, x, y):
self.x = x
self.y = y
out = x * y
return out
def backward(self, dout):
dx = dout * self.y
dy = dout * self.x
return dx, dy
class AddLayer:
def __init__(self):
pass
def forward(self, x, y):
out = x + y
return out
def backward(self, dout):
dx = dout * 1
dy = dout * 1
return dx, dy
class Relu:
def __init__(self):
self.mask = None
def forward(self, x):
self.mask = x <= 0
out = x.copy()
out[self.mask] = 0
return out
def backward(self, dout):
dout[self.mask] = 0
dx = dout
return dx
x = np.array([[1.0, -0.5], [-2.0, 3.0]])
mask = x<=0
mask
class Sigmoid:
def __init__(self):
self.out = None
def forward(self, x):
out = 1 / (1 + np.exp(-x))
self.out = out
return out
def backward(self, dout):
dx = dout * (1.0 - self.out) * self.out
return dx
class Affine:
def __init__(self, W, b):
self.W = W
self.b = b
self.x = None
self.dW = None
self.db = None
def forward(self, x):
self.x = x
out = np.dot(x, self.W) + self.b
return out
def backward(self, dout):
dx = np.dot(dout, self.W.T)
self.dW = np.dot(self.x.T, dout)
self.db = np.sum(dout, axis=0)
class SoftmaxWithLoss:
def __init__(self):
self.loss = None
self.y = None
self.t = None
def forward(self, x, t):
self.t = t
self.y = softmax(x)
self.loss = cross_entropy_error(self.y, self.t)
return self.loss
def backward(self, dout=1):
batch_size = self.t.shape[0]
dx = (self.y - self.t) / batch_size
return dx
# coding: utf-8
import sys, os
sys.path.append(os.pardir) # 부모 디렉터리의 파일을 가져올 수 있도록 설정
import numpy as np
from common.layers import *
from common.gradient import numerical_gradient
from collections import OrderedDict
class TwoLayerNet:
def __init__(self, input_size, hidden_size, output_size, weight_init_std = 0.01):
# 가중치 초기화
self.params = {}
self.params['W1'] = weight_init_std * np.random.randn(input_size, hidden_size)
self.params['b1'] = np.zeros(hidden_size)
self.params['W2'] = weight_init_std * np.random.randn(hidden_size, output_size)
self.params['b2'] = np.zeros(output_size)
# 계층 생성
self.layers = OrderedDict()
self.layers['Affine1'] = Affine(self.params['W1'], self.params['b1'])
self.layers['Relu1'] = Relu()
self.layers['Affine2'] = Affine(self.params['W2'], self.params['b2'])
self.lastLayer = SoftmaxWithLoss()
def predict(self, x):
for layer in self.layers.values():
x = layer.forward(x)
return x
# x : 입력 데이터, t : 정답 레이블
def loss(self, x, t):
y = self.predict(x)
return self.lastLayer.forward(y, t)
def accuracy(self, x, t):
y = self.predict(x)
y = np.argmax(y, axis=1)
if t.ndim != 1 : t = np.argmax(t, axis=1)
accuracy = np.sum(y == t) / float(x.shape[0])
return accuracy
# x : 입력 데이터, t : 정답 레이블
def numerical_gradient(self, x, t):
loss_W = lambda W: self.loss(x, t)
grads = {}
grads['W1'] = numerical_gradient(loss_W, self.params['W1'])
grads['b1'] = numerical_gradient(loss_W, self.params['b1'])
grads['W2'] = numerical_gradient(loss_W, self.params['W2'])
grads['b2'] = numerical_gradient(loss_W, self.params['b2'])
return grads
def gradient(self, x, t):
# forward
self.loss(x, t)
# backward
dout = 1
dout = self.lastLayer.backward(dout)
layers = list(self.layers.values())
layers.reverse()
for layer in layers:
dout = layer.backward(dout)
# 결과 저장
grads = {}
grads['W1'], grads['b1'] = self.layers['Affine1'].dW, self.layers['Affine1'].db
grads['W2'], grads['b2'] = self.layers['Affine2'].dW, self.layers['Affine2'].db
return grads
| 0.47317 | 0.979036 |
## La bibliothèque `numpy` de Python
`numpy` est une extension du langage Python qui permet de manipuler des tableaux multi-dimensionnels et/ou des matrices.
Elle est souvent utilisée conjointement à l'extension `scipy` qui contient des outils relatifs:
- aux intégrateurs numériques (`scipy.integrate`);
- à l'algèbre linéaire (`scipy.linalg`);
- etc.
Des fonctionnalités simples illustrant le fonctionnement de `numpy` sont présentées ci-dessous.
En cas de souci, n'hésitez pas à vous référer à:
- la documentation de `numpy` [ici](https://docs.scipy.org/doc/numpy/reference/);
- la documentation de `scipy` [ici](https://docs.scipy.org/doc/scipy/reference/) pour ce que vous ne trouvez pas dans `numpy`.
```
import numpy as np
```
### Types des données embarquées
On peut créer un tableau `numpy` à partir d'une structure itérable (tableau, tuple, liste) Python.
La puissance de `numpy` vient du fait que tous les éléments du tableau sont forcés au même type (le moins disant).
```
# Définition d'un tableau à partir d'une liste
tableau = [2, 7.3, 4]
print('>>> Liste Python: type %s' % type(tableau))
for l in tableau:
print('{%s, %s}' % (l, type(l)), end=" ")
print()
print()
# Création d'un tableau numpy
tableau = np.array(tableau)
print('>>> Tableau numpy: type %s' % type(tableau))
for l in tableau:
print('{%s, %s}' % (l, type(l)), end=" ")
print()
print('On retrouve alors le type de chaque élément dans dtype: %s' % tableau.dtype)
```
### Performance
On reproche souvent à Python d'être lent à l'exécution. C'est dû à de nombreux paramètres, notamment la flexibilité du langage, les nombreuses vérifications faites à notre insu (Python ne présume de rien sur vos données), et surtout au **typage dynamique**.
Avec `numpy`, on connaît désormais une fois pour toute le type de chaque élément du tableau; de plus les opérations mathématiques sur ces tableaux sont alors codées en C (rapide!)
Observez plutôt:
```
tableau = [i for i in range(1, 10000000)]
array = np.array(tableau)
%timeit double = [x * 2 for x in tableau]
%timeit double = array * 2
```
### Vues sur des sous-ensembles du tableau
Il est possible avec `numpy` de travailler sur des vues d'un tableau à $n$ dimensions qu'on aura construit.
On emploie ici le mot **vue** parce qu'une modification des données dans la vue modifie les données dans le tableau d'origine.
Observons plutôt:
```
tableau = np.array([[i+2*j for i in range(5)] for j in range(4)])
print(tableau)
# On affiche les lignes d'indices 0 à 1 (< 2), colonnes d'indices 2 à 3 (< 4)
sub = tableau[0:2, 2:4]
print(sub)
# L'absence d'indice signifie "début" ou "fin"
sub = tableau[:3, 2:]
print(sub)
# On modifie sub
sub *= 0
print(sub)
```
<div class="alert alert-danger">
<b>Attention</b>: voici pourquoi on parlait de vue !
</div>
```
print(tableau)
```
### Opérations matricielles
`numpy` donne accès aux opérations matricielles de base.
```
a = np.array([[4,6,7,6]])
b = np.array([[i+j for i in range(5)] for j in range(4)])
print(a.shape, a, sep="\n")
print()
print(b.shape, b, sep="\n")
# Produit matriciel (ou vectoriel)
print(np.dot (a, b))
```
<div class="alert alert-danger">
<b>Attention</b>: Contrairement à Matlab, les opérateurs arithmétiques +, -, * sont des opérations terme à terme.
</div>
Pour bien comprendre la différence:
```
import numpy.linalg
a = np.array([[abs(i-j) for i in range(5)] for j in range(5)])
inv_a = numpy.linalg.inv(a) # L'inverse
print(a)
print(inv_a)
print(inv_a * a)
print("\nDiantre!!")
print(np.dot(inv_a, a))
print("\nC'est si facile de se faire avoir...")
```
<div class="alert alert-success">
<b>Note</b>: Depuis Python 3.5, l'opérateur @ est utilisable pour la multiplication de matrice.
</div>
```
print(inv_a @ a)
```
# La bibliothèque `matplotlib` de Python
`matplotlib` propose un ensemble de commandes qui permettent d'afficher des données de manière graphique, d'afficher des lignes, de remplir des zones avec des couleurs, d'ajouter du texte, etc.
L'instruction `%matplotlib inline` avant l'import permet de rediriger la sortie graphique vers le notebook.
```
%matplotlib inline
import matplotlib.pyplot as plt
```
L'instruction `plot` prend une série de points en abscisses et une série de points en ordonnées:
```
plt.plot([1, 2, 3, 4], [1, 4, 9, 16])
```
Il y a un style par défaut qui est choisi de manière automatique, mais il est possible de sélectionner:
- les couleurs;
- le style des points de données;
- la longueur des axes;
- etc.
```
plt.plot([1, 2, 3, 4], [1, 4, 9, 16], 'ro-')
plt.xlim(0, 6)
plt.ylim(0, 20)
plt.xlabel("Temps")
plt.ylabel("Argent")
```
Il est recommandé d'utiliser `matplotlib` avec des tableaux `numpy`.
```
# échantillon à 200ms d'intervalle
t = np.arange(0., 5., 0.2)
# red dashes, blue squares and green triangles
plt.plot(t, t, 'r--', t, t**2, 'bs', t, t**3, 'g^')
```
Enfin il est possible d'afficher plusieurs graphes côte à côte.
Notez que l'on peut également gérer la taille de la figure (bitmap) produite.
```
fig, ax = plt.subplots(ncols=2, nrows=2, figsize=(10, 10))
# Vous pouvez choisir des palettes de couleurs « jolies » avec des sites comme celui-ci :
# http://paletton.com/#uid=7000u0kllllaFw0g0qFqFg0w0aF
ax[0,0].plot(t, np.sin(t), '#aa3939')
ax[0,1].plot(t, np.cos(t), '#aa6c39')
ax[1,0].plot(t, np.tan(t), '#226666')
ax[1,1].plot(t, np.sqrt(t), '#2d882d')
```
Un bon réflexe semble être de commencer tous les plots par:
```python
fig, ax = plt.subplots()
```
<div class="alert alert-warning">
<b>Exercice:</b> Tracer le graphe de la fonction $t \mapsto e^{-t} \cdot \cos(2\,\pi\,t)$ pour $t\in[0,5]$
</div>
```
fig, ax = plt.subplots()
ax.plot(t, np.exp(-t)*np.cos(2*np.pi*t), '#aa3939')
# %load solutions/trace_cos.py
fig, ax = plt.subplots(figsize=(10,10))
t = np.arange(0., 5., .2)
ax.plot(t, np.exp(-t)*np.cos(2*np.pi*t))
```
<div class="alert alert-warning">
<b>Exercice:</b> À partir des coordonnées polaires, produire les coordonnées $(x,y)$ pour la fonction $r=\sin(5\,\theta)$, puis les tracer.
</div>
<b>Consigne</b> : n'utiliser que des tableaux et des fonctions `numpy` pour produire les données à tracer.
```
teta = np.arange(0.0, 2*np.pi, 0.1)
r = np.sin(5*teta)
x = r*np.cos(teta)
y = r*np.sin(teta)
fig, (ax1, ax2) = plt.subplots(1, 2)
ax1.plot(teta, r)
ax2.plot(x, y)
# %load solutions/trace_sin.py
theta = np.linspace(0.0, 2*np.pi, 100)
r = np.sin(5 * theta)
x, y = r * np.cos(theta), r * np.sin(theta)
fig, ax = plt.subplots(figsize=(10,10))
plt.plot(x, y)
```
|
github_jupyter
|
import numpy as np
# Définition d'un tableau à partir d'une liste
tableau = [2, 7.3, 4]
print('>>> Liste Python: type %s' % type(tableau))
for l in tableau:
print('{%s, %s}' % (l, type(l)), end=" ")
print()
print()
# Création d'un tableau numpy
tableau = np.array(tableau)
print('>>> Tableau numpy: type %s' % type(tableau))
for l in tableau:
print('{%s, %s}' % (l, type(l)), end=" ")
print()
print('On retrouve alors le type de chaque élément dans dtype: %s' % tableau.dtype)
tableau = [i for i in range(1, 10000000)]
array = np.array(tableau)
%timeit double = [x * 2 for x in tableau]
%timeit double = array * 2
tableau = np.array([[i+2*j for i in range(5)] for j in range(4)])
print(tableau)
# On affiche les lignes d'indices 0 à 1 (< 2), colonnes d'indices 2 à 3 (< 4)
sub = tableau[0:2, 2:4]
print(sub)
# L'absence d'indice signifie "début" ou "fin"
sub = tableau[:3, 2:]
print(sub)
# On modifie sub
sub *= 0
print(sub)
print(tableau)
a = np.array([[4,6,7,6]])
b = np.array([[i+j for i in range(5)] for j in range(4)])
print(a.shape, a, sep="\n")
print()
print(b.shape, b, sep="\n")
# Produit matriciel (ou vectoriel)
print(np.dot (a, b))
import numpy.linalg
a = np.array([[abs(i-j) for i in range(5)] for j in range(5)])
inv_a = numpy.linalg.inv(a) # L'inverse
print(a)
print(inv_a)
print(inv_a * a)
print("\nDiantre!!")
print(np.dot(inv_a, a))
print("\nC'est si facile de se faire avoir...")
print(inv_a @ a)
%matplotlib inline
import matplotlib.pyplot as plt
plt.plot([1, 2, 3, 4], [1, 4, 9, 16])
plt.plot([1, 2, 3, 4], [1, 4, 9, 16], 'ro-')
plt.xlim(0, 6)
plt.ylim(0, 20)
plt.xlabel("Temps")
plt.ylabel("Argent")
# échantillon à 200ms d'intervalle
t = np.arange(0., 5., 0.2)
# red dashes, blue squares and green triangles
plt.plot(t, t, 'r--', t, t**2, 'bs', t, t**3, 'g^')
fig, ax = plt.subplots(ncols=2, nrows=2, figsize=(10, 10))
# Vous pouvez choisir des palettes de couleurs « jolies » avec des sites comme celui-ci :
# http://paletton.com/#uid=7000u0kllllaFw0g0qFqFg0w0aF
ax[0,0].plot(t, np.sin(t), '#aa3939')
ax[0,1].plot(t, np.cos(t), '#aa6c39')
ax[1,0].plot(t, np.tan(t), '#226666')
ax[1,1].plot(t, np.sqrt(t), '#2d882d')
fig, ax = plt.subplots()
fig, ax = plt.subplots()
ax.plot(t, np.exp(-t)*np.cos(2*np.pi*t), '#aa3939')
# %load solutions/trace_cos.py
fig, ax = plt.subplots(figsize=(10,10))
t = np.arange(0., 5., .2)
ax.plot(t, np.exp(-t)*np.cos(2*np.pi*t))
teta = np.arange(0.0, 2*np.pi, 0.1)
r = np.sin(5*teta)
x = r*np.cos(teta)
y = r*np.sin(teta)
fig, (ax1, ax2) = plt.subplots(1, 2)
ax1.plot(teta, r)
ax2.plot(x, y)
# %load solutions/trace_sin.py
theta = np.linspace(0.0, 2*np.pi, 100)
r = np.sin(5 * theta)
x, y = r * np.cos(theta), r * np.sin(theta)
fig, ax = plt.subplots(figsize=(10,10))
plt.plot(x, y)
| 0.351756 | 0.938969 |
# PersistentDataset, CacheDataset, and simple Dataset Tutorial and Speed Test
This tutorial shows how to accelerate PyTorch medical DL program based on
how data is loaded and preprocessed using different MONAI `Dataset` managers.
`Dataset` provides the simplest model of data loading. Each time a dataset is needed, it is reloaded from the original datasources, and processed through the all non-random and random transforms to generate analyzable tensors. This mechanism has the smallest memory footprint, and the smallest temporary disk footprint.
`CacheDataset` provides a mechanism to pre-load all original data and apply non-random transforms into analyzable tensors loaded in memory prior to starting analysis. The `CacheDataset` requires all tensor representations of data requested to be loaded into memory at once. The subset of random transforms is applied to the cached components before use. This is the highest performance dataset if all data fit in core memory.
`PersistentDataset` processes original data sources through the non-random transforms on first use, and stores these intermediate tensor values to an on-disk persistence representation. The intermediate processed tensors are loaded from disk on each use for processing by the random-transforms for each analysis request. The `PersistentDataset` has a similar memory footprint to the simple `Dataset`, with performance characteristics close to the `CacheDataset` at the expense of disk storage. Additionally, the cost of first time processing of data is distributed across each first use.
It's modified from the [Spleen 3D segmentation tutorial notebook](https://github.com/Project-MONAI/tutorials/blob/master/3d_segmentation/spleen_segmentation_3d.ipynb).
[](https://colab.research.google.com/github/Project-MONAI/tutorials/blob/master/acceleration/dataset_type_performance.ipynb)
## Setup environment
```
!python -c "import monai" || pip install -q "monai-weekly[pillow, tqdm]"
!python -c "import matplotlib" || pip install -q matplotlib
%matplotlib inline
```
## Setup imports
```
# Copyright 2020 MONAI Consortium
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
# http://www.apache.org/licenses/LICENSE-2.0
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import glob
import os
import pathlib
import shutil
import tempfile
import time
import matplotlib.pyplot as plt
import torch
from monai.apps import download_and_extract
from monai.config import print_config
from monai.data import (
CacheDataset,
Dataset,
PersistentDataset,
list_data_collate,
)
from monai.inferers import sliding_window_inference
from monai.losses import DiceLoss
from monai.metrics import compute_meandice
from monai.networks.layers import Norm
from monai.networks.nets import UNet
from monai.transforms import (
AddChanneld,
AsDiscrete,
Compose,
CropForegroundd,
LoadImaged,
Orientationd,
RandCropByPosNegLabeld,
ScaleIntensityRanged,
Spacingd,
ToTensord,
)
from monai.utils import set_determinism
print_config()
```
## Setup data directory
You can specify a directory with the `MONAI_DATA_DIRECTORY` environment variable.
This allows you to save results and reuse downloads.
If not specified a temporary directory will be used.
```
directory = os.environ.get("MONAI_DATA_DIRECTORY")
root_dir = tempfile.mkdtemp() if directory is None else directory
print(root_dir)
```
## Define a typical PyTorch training process
```
def train_process(train_ds, val_ds):
# use batch_size=2 to load images and use RandCropByPosNegLabeld
# to generate 2 x 4 images for network training
train_loader = torch.utils.data.DataLoader(
train_ds,
batch_size=2,
shuffle=True,
num_workers=4,
collate_fn=list_data_collate,
)
val_loader = torch.utils.data.DataLoader(
val_ds, batch_size=1, num_workers=4
)
device = torch.device("cuda:0")
model = UNet(
dimensions=3,
in_channels=1,
out_channels=2,
channels=(16, 32, 64, 128, 256),
strides=(2, 2, 2, 2),
num_res_units=2,
norm=Norm.BATCH,
).to(device)
loss_function = DiceLoss(to_onehot_y=True, softmax=True)
optimizer = torch.optim.Adam(model.parameters(), 1e-4)
post_pred = AsDiscrete(argmax=True, to_onehot=True, n_classes=2)
post_label = AsDiscrete(to_onehot=True, n_classes=2)
max_epochs = 600
val_interval = 1 # do validation for every epoch
best_metric = -1
best_metric_epoch = -1
epoch_loss_values = []
metric_values = []
epoch_times = []
total_start = time.time()
for epoch in range(max_epochs):
epoch_start = time.time()
print("-" * 10)
print(f"epoch {epoch + 1}/{max_epochs}")
model.train()
epoch_loss = 0
step = 0
for batch_data in train_loader:
step_start = time.time()
step += 1
inputs, labels = (
batch_data["image"].to(device),
batch_data["label"].to(device),
)
optimizer.zero_grad()
outputs = model(inputs)
loss = loss_function(outputs, labels)
loss.backward()
optimizer.step()
epoch_loss += loss.item()
print(
f"{step}/{len(train_ds) // train_loader.batch_size},"
f" train_loss: {loss.item():.4f}"
f" step time: {(time.time() - step_start):.4f}"
)
epoch_loss /= step
epoch_loss_values.append(epoch_loss)
print(f"epoch {epoch + 1} average loss: {epoch_loss:.4f}")
if (epoch + 1) % val_interval == 0:
model.eval()
with torch.no_grad():
metric_sum = 0.0
metric_count = 0
for val_data in val_loader:
val_inputs, val_labels = (
val_data["image"].to(device),
val_data["label"].to(device),
)
roi_size = (160, 160, 160)
sw_batch_size = 4
val_outputs = sliding_window_inference(
val_inputs, roi_size, sw_batch_size, model
)
val_outputs = post_pred(val_outputs)
val_labels = post_label(val_labels)
value = compute_meandice(
y_pred=val_outputs,
y=val_labels,
include_background=False,
)
metric_count += len(value)
metric_sum += value.sum().item()
metric = metric_sum / metric_count
metric_values.append(metric)
if metric > best_metric:
best_metric = metric
best_metric_epoch = epoch + 1
torch.save(
model.state_dict(),
os.path.join(root_dir, "best_metric_model.pth"),
)
print("saved new best metric model")
print(
f"current epoch: {epoch + 1} current"
f" mean dice: {metric:.4f}"
f" best mean dice: {best_metric:.4f}"
f" at epoch: {best_metric_epoch}"
)
print(
f"time of epoch {epoch + 1} is: {(time.time() - epoch_start):.4f}"
)
epoch_times.append(time.time() - epoch_start)
print(
f"train completed, best_metric: {best_metric:.4f}"
f" at epoch: {best_metric_epoch}"
f" total time: {(time.time() - total_start):.4f}"
)
return (
max_epochs,
time.time() - total_start,
epoch_loss_values,
metric_values,
epoch_times,
)
```
# Start of speed testing
The `PersistenceDataset`, `CacheDataset`, and `Dataset` are compared for speed for running 600 epochs.
## Download dataset
Downloads and extracts the dataset.
The dataset comes from http://medicaldecathlon.com/.
```
resource = "https://msd-for-monai.s3-us-west-2.amazonaws.com/Task09_Spleen.tar"
md5 = "410d4a301da4e5b2f6f86ec3ddba524e"
compressed_file = os.path.join(root_dir, "Task09_Spleen.tar")
data_dir = os.path.join(root_dir, "Task09_Spleen")
if not os.path.exists(data_dir):
download_and_extract(resource, compressed_file, root_dir, md5)
```
## Set MSD Spleen dataset path
```
train_images = sorted(
glob.glob(os.path.join(data_dir, "imagesTr", "*.nii.gz"))
)
train_labels = sorted(
glob.glob(os.path.join(data_dir, "labelsTr", "*.nii.gz"))
)
data_dicts = [
{"image": image_name, "label": label_name}
for image_name, label_name in zip(train_images, train_labels)
]
train_files, val_files = data_dicts[:-9], data_dicts[-9:]
```
## Setup transforms for training and validation
Deterministic transforms during training:
* LoadImaged
* AddChanneld
* Spacingd
* Orientationd
* ScaleIntensityRanged
Non-deterministic transforms:
* RandCropByPosNegLabeld
* ToTensord
All the validation transforms are deterministic.
The results of all the deterministic transforms will be cached to accelerate training.
```
def transformations():
train_transforms = Compose(
[
LoadImaged(keys=["image", "label"]),
AddChanneld(keys=["image", "label"]),
Spacingd(
keys=["image", "label"],
pixdim=(1.5, 1.5, 2.0),
mode=("bilinear", "nearest"),
),
Orientationd(keys=["image", "label"], axcodes="RAS"),
ScaleIntensityRanged(
keys=["image"],
a_min=-57,
a_max=164,
b_min=0.0,
b_max=1.0,
clip=True,
),
CropForegroundd(keys=["image", "label"], source_key="image"),
# randomly crop out patch samples from big
# image based on pos / neg ratio
# the image centers of negative samples
# must be in valid image area
RandCropByPosNegLabeld(
keys=["image", "label"],
label_key="label",
spatial_size=(96, 96, 96),
pos=1,
neg=1,
num_samples=4,
image_key="image",
image_threshold=0,
),
ToTensord(keys=["image", "label"]),
]
)
# NOTE: No random cropping in the validation data,
# we will evaluate the entire image using a sliding window.
val_transforms = Compose(
[
LoadImaged(keys=["image", "label"]),
AddChanneld(keys=["image", "label"]),
Spacingd(
keys=["image", "label"],
pixdim=(1.5, 1.5, 2.0),
mode=("bilinear", "nearest"),
),
Orientationd(keys=["image", "label"], axcodes="RAS"),
ScaleIntensityRanged(
keys=["image"],
a_min=-57,
a_max=164,
b_min=0.0,
b_max=1.0,
clip=True,
),
CropForegroundd(keys=["image", "label"], source_key="image"),
ToTensord(keys=["image", "label"]),
]
)
return train_transforms, val_transforms
```
## Enable deterministic training and regular `Dataset`
Load each original dataset and transform each time it is needed.
```
set_determinism(seed=0)
train_trans, val_trans = transformations()
train_ds = Dataset(data=train_files, transform=train_trans)
val_ds = Dataset(data=val_files, transform=val_trans)
(
max_epochs,
total_time,
epoch_loss_values,
metric_values,
epoch_times,
) = train_process(train_ds, val_ds)
print(
f"total training time of {max_epochs} epochs"
f" with regular Dataset: {total_time:.4f}"
)
```
## Enable deterministic training and `PersistentDataset`
Use persistent storage of non-random transformed training and validation data computed once and stored in persistently across runs
```
persistent_cache = pathlib.Path(root_dir, "persistent_cache")
persistent_cache.mkdir(parents=True, exist_ok=True)
set_determinism(seed=0)
train_trans, val_trans = transformations()
train_persitence_ds = PersistentDataset(
data=train_files, transform=train_trans, cache_dir=persistent_cache
)
val_persitence_ds = PersistentDataset(
data=val_files, transform=val_trans, cache_dir=persistent_cache
)
(
persistence_epoch_num,
persistence_total_time,
persistence_epoch_loss_values,
persistence_metric_values,
persistence_epoch_times,
) = train_process(train_persitence_ds, val_persitence_ds)
print(
f"total training time of {persistence_epoch_num}"
f" epochs with persistent storage Dataset: {persistence_total_time:.4f}"
)
```
## Enable deterministic training and `CacheDataset`
Precompute all non-random transforms of original data and store in memory.
```
set_determinism(seed=0)
train_trans, val_trans = transformations()
cache_init_start = time.time()
cache_train_ds = CacheDataset(
data=train_files, transform=train_trans, cache_rate=1.0, num_workers=4
)
cache_val_ds = CacheDataset(
data=val_files, transform=val_trans, cache_rate=1.0, num_workers=4
)
cache_init_time = time.time() - cache_init_start
(
cache_epoch_num,
cache_total_time,
cache_epoch_loss_values,
cache_metric_values,
cache_epoch_times,
) = train_process(cache_train_ds, cache_val_ds)
print(
f"total training time of {cache_epoch_num}"
f" epochs with CacheDataset: {cache_total_time:.4f}"
)
```
## Plot training loss and validation metrics
```
plt.figure("train", (12, 18))
plt.subplot(3, 2, 1)
plt.title("Regular Epoch Average Loss")
x = [i + 1 for i in range(len(epoch_loss_values))]
y = epoch_loss_values
plt.xlabel("epoch")
plt.grid(alpha=0.4, linestyle=":")
plt.plot(x, y, color="red")
plt.subplot(3, 2, 2)
plt.title("Regular Val Mean Dice")
x = [i + 1 for i in range(len(metric_values))]
y = cache_metric_values
plt.xlabel("epoch")
plt.grid(alpha=0.4, linestyle=":")
plt.plot(x, y, color="red")
plt.subplot(3, 2, 3)
plt.title("PersistentDataset Epoch Average Loss")
x = [i + 1 for i in range(len(persistence_epoch_loss_values))]
y = persistence_epoch_loss_values
plt.xlabel("epoch")
plt.grid(alpha=0.4, linestyle=":")
plt.plot(x, y, color="blue")
plt.subplot(3, 2, 4)
plt.title("PersistentDataset Val Mean Dice")
x = [i + 1 for i in range(len(persistence_metric_values))]
y = persistence_metric_values
plt.xlabel("epoch")
plt.grid(alpha=0.4, linestyle=":")
plt.plot(x, y, color="blue")
plt.subplot(3, 2, 5)
plt.title("Cache Epoch Average Loss")
x = [i + 1 for i in range(len(cache_epoch_loss_values))]
y = cache_epoch_loss_values
plt.xlabel("epoch")
plt.grid(alpha=0.4, linestyle=":")
plt.plot(x, y, color="green")
plt.subplot(3, 2, 6)
plt.title("Cache Val Mean Dice")
x = [i + 1 for i in range(len(cache_metric_values))]
y = cache_metric_values
plt.xlabel("epoch")
plt.grid(alpha=0.4, linestyle=":")
plt.plot(x, y, color="green")
plt.show()
```
## Plot total time and every epoch time
```
plt.figure("train", (12, 6))
plt.subplot(1, 2, 1)
plt.title("Total Train Time(600 epochs)")
plt.bar("regular", total_time, 1, label="Regular Dataset", color="red")
plt.bar(
"persistent",
persistence_total_time,
1,
label="Persistent Dataset",
color="blue",
)
plt.bar(
"cache",
cache_init_time + cache_total_time,
1,
label="Cache Dataset",
color="green",
)
plt.bar("cache", cache_init_time, 1, label="Cache Init", color="orange")
plt.ylabel("secs")
plt.grid(alpha=0.4, linestyle=":")
plt.legend(loc="best")
plt.subplot(1, 2, 2)
plt.title("Epoch Time")
x = [i + 1 for i in range(len(epoch_times))]
plt.xlabel("epoch")
plt.ylabel("secs")
plt.plot(x, epoch_times, label="Regular Dataset", color="red")
plt.plot(x, persistence_epoch_times, label="Persistent Dataset", color="blue")
plt.plot(x, cache_epoch_times, label="Cache Dataset", color="green")
plt.grid(alpha=0.4, linestyle=":")
plt.legend(loc="best")
plt.show()
```
## Cleanup data directory
Remove directory if a temporary was used.
```
if directory is None:
shutil.rmtree(root_dir)
```
|
github_jupyter
|
!python -c "import monai" || pip install -q "monai-weekly[pillow, tqdm]"
!python -c "import matplotlib" || pip install -q matplotlib
%matplotlib inline
# Copyright 2020 MONAI Consortium
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
# http://www.apache.org/licenses/LICENSE-2.0
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import glob
import os
import pathlib
import shutil
import tempfile
import time
import matplotlib.pyplot as plt
import torch
from monai.apps import download_and_extract
from monai.config import print_config
from monai.data import (
CacheDataset,
Dataset,
PersistentDataset,
list_data_collate,
)
from monai.inferers import sliding_window_inference
from monai.losses import DiceLoss
from monai.metrics import compute_meandice
from monai.networks.layers import Norm
from monai.networks.nets import UNet
from monai.transforms import (
AddChanneld,
AsDiscrete,
Compose,
CropForegroundd,
LoadImaged,
Orientationd,
RandCropByPosNegLabeld,
ScaleIntensityRanged,
Spacingd,
ToTensord,
)
from monai.utils import set_determinism
print_config()
directory = os.environ.get("MONAI_DATA_DIRECTORY")
root_dir = tempfile.mkdtemp() if directory is None else directory
print(root_dir)
def train_process(train_ds, val_ds):
# use batch_size=2 to load images and use RandCropByPosNegLabeld
# to generate 2 x 4 images for network training
train_loader = torch.utils.data.DataLoader(
train_ds,
batch_size=2,
shuffle=True,
num_workers=4,
collate_fn=list_data_collate,
)
val_loader = torch.utils.data.DataLoader(
val_ds, batch_size=1, num_workers=4
)
device = torch.device("cuda:0")
model = UNet(
dimensions=3,
in_channels=1,
out_channels=2,
channels=(16, 32, 64, 128, 256),
strides=(2, 2, 2, 2),
num_res_units=2,
norm=Norm.BATCH,
).to(device)
loss_function = DiceLoss(to_onehot_y=True, softmax=True)
optimizer = torch.optim.Adam(model.parameters(), 1e-4)
post_pred = AsDiscrete(argmax=True, to_onehot=True, n_classes=2)
post_label = AsDiscrete(to_onehot=True, n_classes=2)
max_epochs = 600
val_interval = 1 # do validation for every epoch
best_metric = -1
best_metric_epoch = -1
epoch_loss_values = []
metric_values = []
epoch_times = []
total_start = time.time()
for epoch in range(max_epochs):
epoch_start = time.time()
print("-" * 10)
print(f"epoch {epoch + 1}/{max_epochs}")
model.train()
epoch_loss = 0
step = 0
for batch_data in train_loader:
step_start = time.time()
step += 1
inputs, labels = (
batch_data["image"].to(device),
batch_data["label"].to(device),
)
optimizer.zero_grad()
outputs = model(inputs)
loss = loss_function(outputs, labels)
loss.backward()
optimizer.step()
epoch_loss += loss.item()
print(
f"{step}/{len(train_ds) // train_loader.batch_size},"
f" train_loss: {loss.item():.4f}"
f" step time: {(time.time() - step_start):.4f}"
)
epoch_loss /= step
epoch_loss_values.append(epoch_loss)
print(f"epoch {epoch + 1} average loss: {epoch_loss:.4f}")
if (epoch + 1) % val_interval == 0:
model.eval()
with torch.no_grad():
metric_sum = 0.0
metric_count = 0
for val_data in val_loader:
val_inputs, val_labels = (
val_data["image"].to(device),
val_data["label"].to(device),
)
roi_size = (160, 160, 160)
sw_batch_size = 4
val_outputs = sliding_window_inference(
val_inputs, roi_size, sw_batch_size, model
)
val_outputs = post_pred(val_outputs)
val_labels = post_label(val_labels)
value = compute_meandice(
y_pred=val_outputs,
y=val_labels,
include_background=False,
)
metric_count += len(value)
metric_sum += value.sum().item()
metric = metric_sum / metric_count
metric_values.append(metric)
if metric > best_metric:
best_metric = metric
best_metric_epoch = epoch + 1
torch.save(
model.state_dict(),
os.path.join(root_dir, "best_metric_model.pth"),
)
print("saved new best metric model")
print(
f"current epoch: {epoch + 1} current"
f" mean dice: {metric:.4f}"
f" best mean dice: {best_metric:.4f}"
f" at epoch: {best_metric_epoch}"
)
print(
f"time of epoch {epoch + 1} is: {(time.time() - epoch_start):.4f}"
)
epoch_times.append(time.time() - epoch_start)
print(
f"train completed, best_metric: {best_metric:.4f}"
f" at epoch: {best_metric_epoch}"
f" total time: {(time.time() - total_start):.4f}"
)
return (
max_epochs,
time.time() - total_start,
epoch_loss_values,
metric_values,
epoch_times,
)
resource = "https://msd-for-monai.s3-us-west-2.amazonaws.com/Task09_Spleen.tar"
md5 = "410d4a301da4e5b2f6f86ec3ddba524e"
compressed_file = os.path.join(root_dir, "Task09_Spleen.tar")
data_dir = os.path.join(root_dir, "Task09_Spleen")
if not os.path.exists(data_dir):
download_and_extract(resource, compressed_file, root_dir, md5)
train_images = sorted(
glob.glob(os.path.join(data_dir, "imagesTr", "*.nii.gz"))
)
train_labels = sorted(
glob.glob(os.path.join(data_dir, "labelsTr", "*.nii.gz"))
)
data_dicts = [
{"image": image_name, "label": label_name}
for image_name, label_name in zip(train_images, train_labels)
]
train_files, val_files = data_dicts[:-9], data_dicts[-9:]
def transformations():
train_transforms = Compose(
[
LoadImaged(keys=["image", "label"]),
AddChanneld(keys=["image", "label"]),
Spacingd(
keys=["image", "label"],
pixdim=(1.5, 1.5, 2.0),
mode=("bilinear", "nearest"),
),
Orientationd(keys=["image", "label"], axcodes="RAS"),
ScaleIntensityRanged(
keys=["image"],
a_min=-57,
a_max=164,
b_min=0.0,
b_max=1.0,
clip=True,
),
CropForegroundd(keys=["image", "label"], source_key="image"),
# randomly crop out patch samples from big
# image based on pos / neg ratio
# the image centers of negative samples
# must be in valid image area
RandCropByPosNegLabeld(
keys=["image", "label"],
label_key="label",
spatial_size=(96, 96, 96),
pos=1,
neg=1,
num_samples=4,
image_key="image",
image_threshold=0,
),
ToTensord(keys=["image", "label"]),
]
)
# NOTE: No random cropping in the validation data,
# we will evaluate the entire image using a sliding window.
val_transforms = Compose(
[
LoadImaged(keys=["image", "label"]),
AddChanneld(keys=["image", "label"]),
Spacingd(
keys=["image", "label"],
pixdim=(1.5, 1.5, 2.0),
mode=("bilinear", "nearest"),
),
Orientationd(keys=["image", "label"], axcodes="RAS"),
ScaleIntensityRanged(
keys=["image"],
a_min=-57,
a_max=164,
b_min=0.0,
b_max=1.0,
clip=True,
),
CropForegroundd(keys=["image", "label"], source_key="image"),
ToTensord(keys=["image", "label"]),
]
)
return train_transforms, val_transforms
set_determinism(seed=0)
train_trans, val_trans = transformations()
train_ds = Dataset(data=train_files, transform=train_trans)
val_ds = Dataset(data=val_files, transform=val_trans)
(
max_epochs,
total_time,
epoch_loss_values,
metric_values,
epoch_times,
) = train_process(train_ds, val_ds)
print(
f"total training time of {max_epochs} epochs"
f" with regular Dataset: {total_time:.4f}"
)
persistent_cache = pathlib.Path(root_dir, "persistent_cache")
persistent_cache.mkdir(parents=True, exist_ok=True)
set_determinism(seed=0)
train_trans, val_trans = transformations()
train_persitence_ds = PersistentDataset(
data=train_files, transform=train_trans, cache_dir=persistent_cache
)
val_persitence_ds = PersistentDataset(
data=val_files, transform=val_trans, cache_dir=persistent_cache
)
(
persistence_epoch_num,
persistence_total_time,
persistence_epoch_loss_values,
persistence_metric_values,
persistence_epoch_times,
) = train_process(train_persitence_ds, val_persitence_ds)
print(
f"total training time of {persistence_epoch_num}"
f" epochs with persistent storage Dataset: {persistence_total_time:.4f}"
)
set_determinism(seed=0)
train_trans, val_trans = transformations()
cache_init_start = time.time()
cache_train_ds = CacheDataset(
data=train_files, transform=train_trans, cache_rate=1.0, num_workers=4
)
cache_val_ds = CacheDataset(
data=val_files, transform=val_trans, cache_rate=1.0, num_workers=4
)
cache_init_time = time.time() - cache_init_start
(
cache_epoch_num,
cache_total_time,
cache_epoch_loss_values,
cache_metric_values,
cache_epoch_times,
) = train_process(cache_train_ds, cache_val_ds)
print(
f"total training time of {cache_epoch_num}"
f" epochs with CacheDataset: {cache_total_time:.4f}"
)
plt.figure("train", (12, 18))
plt.subplot(3, 2, 1)
plt.title("Regular Epoch Average Loss")
x = [i + 1 for i in range(len(epoch_loss_values))]
y = epoch_loss_values
plt.xlabel("epoch")
plt.grid(alpha=0.4, linestyle=":")
plt.plot(x, y, color="red")
plt.subplot(3, 2, 2)
plt.title("Regular Val Mean Dice")
x = [i + 1 for i in range(len(metric_values))]
y = cache_metric_values
plt.xlabel("epoch")
plt.grid(alpha=0.4, linestyle=":")
plt.plot(x, y, color="red")
plt.subplot(3, 2, 3)
plt.title("PersistentDataset Epoch Average Loss")
x = [i + 1 for i in range(len(persistence_epoch_loss_values))]
y = persistence_epoch_loss_values
plt.xlabel("epoch")
plt.grid(alpha=0.4, linestyle=":")
plt.plot(x, y, color="blue")
plt.subplot(3, 2, 4)
plt.title("PersistentDataset Val Mean Dice")
x = [i + 1 for i in range(len(persistence_metric_values))]
y = persistence_metric_values
plt.xlabel("epoch")
plt.grid(alpha=0.4, linestyle=":")
plt.plot(x, y, color="blue")
plt.subplot(3, 2, 5)
plt.title("Cache Epoch Average Loss")
x = [i + 1 for i in range(len(cache_epoch_loss_values))]
y = cache_epoch_loss_values
plt.xlabel("epoch")
plt.grid(alpha=0.4, linestyle=":")
plt.plot(x, y, color="green")
plt.subplot(3, 2, 6)
plt.title("Cache Val Mean Dice")
x = [i + 1 for i in range(len(cache_metric_values))]
y = cache_metric_values
plt.xlabel("epoch")
plt.grid(alpha=0.4, linestyle=":")
plt.plot(x, y, color="green")
plt.show()
plt.figure("train", (12, 6))
plt.subplot(1, 2, 1)
plt.title("Total Train Time(600 epochs)")
plt.bar("regular", total_time, 1, label="Regular Dataset", color="red")
plt.bar(
"persistent",
persistence_total_time,
1,
label="Persistent Dataset",
color="blue",
)
plt.bar(
"cache",
cache_init_time + cache_total_time,
1,
label="Cache Dataset",
color="green",
)
plt.bar("cache", cache_init_time, 1, label="Cache Init", color="orange")
plt.ylabel("secs")
plt.grid(alpha=0.4, linestyle=":")
plt.legend(loc="best")
plt.subplot(1, 2, 2)
plt.title("Epoch Time")
x = [i + 1 for i in range(len(epoch_times))]
plt.xlabel("epoch")
plt.ylabel("secs")
plt.plot(x, epoch_times, label="Regular Dataset", color="red")
plt.plot(x, persistence_epoch_times, label="Persistent Dataset", color="blue")
plt.plot(x, cache_epoch_times, label="Cache Dataset", color="green")
plt.grid(alpha=0.4, linestyle=":")
plt.legend(loc="best")
plt.show()
if directory is None:
shutil.rmtree(root_dir)
| 0.837686 | 0.961786 |
# Probability Distributions
## Topics
* Probability
* Random variables
* Probability distributions
* Uniform
* Normal
* Binomial
* Poisson
* Fat Tailed
## Probability
* Probability is a measure of the likelihood of a random phenomenon or chance behavior. Probability describes the long-term proportion with which a certain outcome will occur in situations with short-term uncertainty.
* Probability is expressed in numbers between 0 and 1. Probability = 0 means the event never happens; probability = 1 means it always happens.
* The total probability of all possible event always sums to 1.
## Sample Space
* Coin Toss ={head,tail}
* Two coins S = {HH, HT, TH, TT}
* Inspecting a part ={good,bad}
* Rolling a die S ={1,2,3,4,5,6}
## Random Variables
In probability and statistics, a random variable, or stochastic variable is a variable whose value is subject to variations due to chance (i.e. it can take on a range of values)
* Coin Toss ={head,tail}
* Rolling a die S ={1,2,3,4,5,6}
Discrete Random Variables
* Random variables (RVs) which may take on only a countable number of distinct values
E.g. the total number of tails X you get if you flip 100 coins
* X is a RV with arity k if it can take on exactly one value out of {x1, …, xk}
E.g. the possible values that X can take on are 0, 1, 2, …, 100
Continuous Random Variables
* Probability density function (pdf) instead of probability mass function (pmf)
* A pdf is any function f(x) that describes the probability density in terms of the input variable x.
## Probability distributions
* We use probability distributions because they model data in real world.
* They allow us to calculate what to expect and therefore understand what is unusual.
* They also provide insight in to the process in which real world data may have been generated.
* Many machine learning algorithms have assumptions based on certain probability distributions.
_Cumulative distribution function_
A probability distribution Pr on the real line is determined by the probability of a scalar random variable X being in a half-open interval (-$\infty$, x], the probability distribution is completely characterized by its cumulative distribution function:
$$
F(x) = \Pr[X \leq x] \quad \forall \quad x \in R .
$$
## Uniform Distribution
$$
X \equiv U[a,b]
$$
$$
f(x) = \frac{1}{b-a} \quad for \quad a \lt x \lt b
$$
$$
f(x) = 0 \quad for \quad a \leq x \quad or \quad \geq b
$$
$$
F(x) = \frac{x-a}{b-a} \quad for \quad a \leq x \lt b
$$
$$
F(x) = 0 \quad for \quad x \lt a \quad
F(x) = 1 \quad for \quad x \geq b
$$

_Continuous Uniform Distribution_
In probability theory and statistics, the continuous uniform distribution or rectangular distribution is a family of symmetric probability distributions such that for each member of the family, all intervals of the same length on the distribution's support are equally probable.
- from [Uniform distribution (continuous Wikipedia)](https://en.wikipedia.org/wiki/Uniform_distribution_(continuous))


_Discrete Uniform Distribution_
In probability theory and statistics, the discrete uniform distribution is a symmetric probability distribution whereby a finite number of values are equally likely to be observed; every one of n values has equal probability 1/n. Another way of saying "discrete uniform distribution" would be "a known, finite number of outcomes equally likely to happen".
- from [Uniform distribution (discrete) Wikipedia)](https://en.wikipedia.org/wiki/Uniform_distribution_(discrete))


## Uniform Distribution in python
```
%matplotlib inline
# %matplotlib inline is a magic function in IPython that displays images in the notebook
# Line magics are prefixed with the % character and work much like OS command-line calls
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from scipy import stats
import seaborn as sns
import warnings
warnings.filterwarnings('ignore')
# Make plots larger
plt.rcParams['figure.figsize'] = (10, 6)
#------------------------------------------------------------
# Define the distribution parameters to be plotted
W_values = [1.0, 3.0, 5.0]
linestyles = ['-', '--', ':']
mu = 0
x = np.linspace(-4, 4, 1000)
#------------------------------------------------------------
# plot the distributions
fig, ax = plt.subplots(figsize=(10, 5))
for W, ls in zip(W_values, linestyles):
left = mu - 0.5 * W
dist = stats.uniform(left, W)
plt.plot(x, dist.pdf(x), ls=ls, c='black',
label=r'$\mu=%i,\ W=%i$' % (mu, W))
plt.xlim(-4, 4)
plt.ylim(0, 1.2)
plt.xlabel('$x$')
plt.ylabel(r'$p(x|\mu, W)$')
plt.title('Uniform Distribution')
plt.legend()
plt.show()
# Adapted from http://www.astroml.org/book_figures/chapter3/fig_uniform_distribution.html
```
## Quiz Distribution of two dice
See if you can generate a distribution that models the output that would be generated by the sum of two dice. Self-test homework.
## Normal Distribution
In probability theory, the normal (or Gaussian) distribution is a very common continuous probability distribution. The normal distribution is remarkably useful because of the central limit theorem. In its most general form, under mild conditions, it states that averages of random variables independently drawn from independent distributions are normally distributed. Physical quantities that are expected to be the sum of many independent processes (such as measurement errors) often have distributions that are nearly normal.
- from [Normal Distribution - Wikipedia)](https://en.wikipedia.org/wiki/Normal_distribution)
$$
X \sim \quad N(\mu, \sigma^2)
$$
$$
f(x) = \frac{1}{\sigma \sqrt {2\pi }} e^{-\frac{( x - \mu)^2}{2\sigma^2}} \quad
$$


Normal cumulative distribution function

_Properties of normal distribution_
- symmetrical, unimodal, and bell-shaped
- on average, the error component will equal zero, the error above and below the mean will cancel out
- Z-Score is a statistical measurement is (above/below) the mean of the data
- important characteristics about z scores:
1. mean of z scores is 0
2. standard deviation of a standardized variable is always 1
3. the linear transformation does not change the _form_ of the distribution
The normal (or Gaussian) distribution was discovered in 1733 by Abraham de Moivre as an approximation to the binomial distribution when the number of trails is large.

- from [Abraham de Moivre - Wikipedia)](https://en.wikipedia.org/wiki/Abraham_de_Moivre)
The Gaussian distribution was derived in 1809 by Carl Friedrich Gauss.

- from [Carl Friedrich Gauss - Wikipedia)](https://en.wikipedia.org/wiki/Carl_Friedrich_Gauss)
Importance lies in the Central Limit Theorem, which states that the sum of a large number of independent random variables (binomial, Poisson, etc.) will approximate a normal distribution
## Central Limit Theorem
In probability theory, the central limit theorem (CLT) states that, given certain conditions, the arithmetic mean of a sufficiently large number of iterates of independent random variables, each with a well-defined expected value and well-defined variance, will be approximately normally distributed, regardless of the underlying distribution. The central limit theorem has a number of variants. In its common form, the random variables must be identically distributed.
- from [Central Limit Theorem - Wikipedia)](https://en.wikipedia.org/wiki/Central_limit_theorem)
The Central Limit Theorem tells us that when the sample size is large the average $\bar{Y}$ of a random sample follows a normal distribution centered at the population average $\mu_Y$ and with standard deviation equal to the population standard deviation $\sigma_Y$, divided by the square root of the sample size $N$.
This means that if we subtract a constant from a random variable, the mean of the new random variable shifts by that constant. If $X$ is a random variable with mean $\mu$ and $a$ is a constant, the mean of $X - a$ is $\mu-a$.
This property also holds for the spread, if $X$ is a random variable with mean $\mu$ and SD $\sigma$, and $a$ is a constant, then the mean and SD of $aX$ are $a \mu$ and $\|a\| \sigma$ respectively.
This implies that if we take many samples of size $N$ then the quantity
$$
\frac{\bar{Y} - \mu}{\sigma_Y/\sqrt{N}}
$$
is approximated with a normal distribution centered at 0 and with standard deviation 1.
## The t-distribution
In probability and statistics, Student's t-distribution (or simply the t-distribution) is any member of a family of continuous probability distributions that arises when estimating the mean of a normally distributed population in situations where the sample size is small and population standard deviation is unknown. Whereas a normal distribution describes a full population, t-distributions describe samples drawn from a full population; accordingly, the t-distribution for each sample size is different, and the larger the sample, the more the distribution resembles a normal distribution.
The t-distribution plays a role in a number of widely used statistical analyses, including the Student's t-test for assessing the statistical significance of the difference between two sample means, the construction of confidence intervals for the difference between two population means, and in linear regression analysis. The Student's t-distribution also arises in the Bayesian analysis of data from a normal family.
- from [The t-distribution - Wikipedia)](https://en.wikipedia.org/wiki/Student%27s_t-distribution)
When the CLT does not apply (i.e. as the number of samples is large), there is another option that does not rely on large samples When a the original population from which a random variable, say $Y$, is sampled is normally distributed with mean 0 then we can calculate the distribution of
number of variants. In its common form, the random variables must be identically distributed.
$$
\sqrt{N} \frac{\bar{Y}}{s_Y}
$$

Normal cumulative distribution function

## Normal Distribution in python
```
# Plot two normal distributions
domain = np.arange(-22, 22, 0.1)
values = stats.norm(3.3, 5.5).pdf(domain)
plt.plot(domain, values, color='r', linewidth=2)
plt.fill_between(domain, 0, values, color='#ffb6c1', alpha=0.3)
values = stats.norm(4.4, 2.3).pdf(domain)
plt.plot(domain, values, color='b', linewidth=2)
plt.ylabel("Probability")
plt.title("Two Normal Distributions")
plt.show()
```
## Binomial Distribution
$$
X \quad \sim \quad B(n, p)
$$
$$
P(X=k) = \binom{n}{k} p^k (1-p)^{n-k} \quad k=1,2,...,n
$$
$$
\binom{n}{k} = \frac{n!}{k!(n-k)!}
$$
_Binomial Distribution_
In probability theory and statistics, the binomial distribution with parameters n and p is the discrete probability distribution of the number of successes in a sequence of n independent yes/no experiments, each of which yields success with probability p. A success/failure experiment is also called a Bernoulli experiment or Bernoulli trial; when n = 1, the binomial distribution is a Bernoulli distribution.
- from [Binomial Distribution - Wikipedia](https://en.wikipedia.org/wiki/Binomial_distribution)
Binomial Distribution

Binomial cumulative distribution function
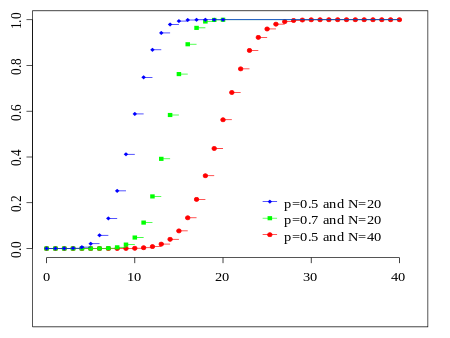
* The data arise from a sequence of n independent trials.
* At each trial there are only two possible outcomes, conventionally called success and failure.
* The probability of success, p, is the same in each trial.
* The random variable of interest is the number of successes, X, in the n trials.
* The assumptions of independence and constant p are important. If they are invalid, so is the binomial distribution
_Bernoulli Random Variables_
* Imagine a simple trial with only two possible outcomes
* Success (S) with probabilty p.
* Failure (F) with probabilty 1-p.
* Examples
* Toss of a coin (heads or tails)
* Gender of a newborn (male or female)
## Binomial Distribution in python
```
#------------------------------------------------------------
# Define the distribution parameters to be plotted
n_values = [20, 20, 40]
b_values = [0.2, 0.6, 0.6]
linestyles = ['-', '--', ':']
x = np.arange(-1, 200)
#------------------------------------------------------------
# plot the distributions
for (n, b, ls) in zip(n_values, b_values, linestyles):
# create a binomial distribution
dist = stats.binom(n, b)
plt.plot(x, dist.pmf(x), ls=ls, c='black',
label=r'$b=%.1f,\ n=%i$' % (b, n), linestyle='steps-mid')
plt.xlim(-0.5, 35)
plt.ylim(0, 0.25)
plt.xlabel('$x$')
plt.ylabel(r'$p(x|b, n)$')
plt.title('Binomial Distribution')
plt.legend()
plt.show()
# Adapted from http://www.astroml.org/book_figures/chapter3/fig_binomial_distribution.html
fair_coin_flips = stats.binom.rvs(n=33, # Number of flips per trial
p=0.4, # Success probability
size=1000) # Number of trials
pd.DataFrame(fair_coin_flips).hist(range=(-0.5,10.5), bins=11)
plt.fill_between(x=np.arange(-4,-1,0.01),
y1= stats.norm.pdf(np.arange(-4,-1,0.01)) ,
facecolor='red',
alpha=0.35)
plt.fill_between(x=np.arange(1,4,0.01),
y1= stats.norm.pdf(np.arange(1,4,0.01)) ,
facecolor='red',
alpha=0.35)
plt.fill_between(x=np.arange(-1,1,0.01),
y1= stats.norm.pdf(np.arange(-1,1,0.01)) ,
facecolor='blue',
alpha=0.35)
```
## Poisson Distribution
$X$ expresses the number of "rare" events
$$
X \quad \sim P( \lambda )\quad \lambda \gt 0
$$
$$
P(X = x) = \frac{ \mathrm{e}^{- \lambda } \lambda^x }{x!} \quad x=1,2,...,n
$$
_Poisson Distribution_
In probability theory and statistics, the Poisson distribution, named after French mathematician Siméon Denis Poisson, is a discrete probability distribution that expresses the probability of a given number of events occurring in a fixed interval of time and/or space if these events occur with a constant rate per time unit and independently of the time since the last event. The Poisson distribution can also be used for the number of events in other specified intervals such as distance, area or volume.
For instance, an individual keeping track of the amount of mail they receive each day may notice that they receive an average number of 4 letters per day. If receiving any particular piece of mail doesn't affect the arrival times of future pieces of mail, i.e., if pieces of mail from a wide range of sources arrive independently of one another, then a reasonable assumption is that the number of pieces of mail received per day obeys a Poisson distribution. Other examples that may follow a Poisson: the number of phone calls received by a call center per hour, the number of decay events per second from a radioactive source, or the number of taxis passing a particular street corner per hour.
The Poisson distribution gives us a probability mass for discrete natural numbers *k* given some mean value λ. Knowing that, on average, λ discrete events occur over some time period, the Poisson distribution gives us the probability of seeing exactly *k* events in that time period.
For example, if a call center gets, on average, 100 customers per day, the Poisson distribution can tell us the probability of getting exactly 150 customers today.
*k* ∈ **N** (i.e. is a natural number) because, on any particular day, you can't have a fraction of a phone call. The probability of any non-integer number of people calling in is zero. E.g., P(150.5) = 0.
λ ∈ **R** (i.e. is a real number) because, even though any *particular* day must have an integer number of people, the *mean* number of people taken over many days can be fractional (and usually is). It's why the "average" number of phone calls per day could be 3.5 even though half a phone call won't occur.
- from [Poisson Distribution - Wikipedia)](https://en.wikipedia.org/wiki/Poisson_distribution)
Poisson Distribution
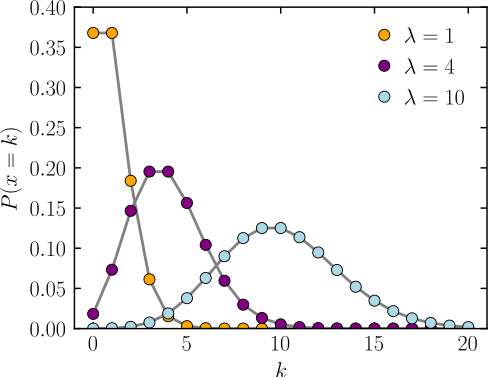
Poisson cumulative distribution function

_Properties of Poisson distribution_
* The mean number of successes from n trials is µ = np
* If we substitute µ/n for p, and let n tend to infinity, the binomial distribution becomes the Poisson distribution.
* Poisson distributions are often used to describe the number of occurrences of a ‘rare’ event. For example
* The number of storms in a season
* The number of occasions in a season when river levels exceed a certain value
* The main assumptions are that events occur
* at random (the occurrence of an event doesn’t change the probability of it happening again)
* at a constant rate
* Poisson distributions also arise as approximations to binomials when n is large and p is small.
* When there is a large number of trials, but a very small probability of success, binomial calculation becomes impractical
## Poisson Distribution in python
```
# Generate poisson counts
arrival_rate_1 = stats.poisson.rvs(size=10000, # Generate Poisson data
mu=1 ) # Average arrival time 1
# Plot histogram
pd.DataFrame(arrival_rate_1).hist(range=(-0.5,max(arrival_rate_1)+0.5)
, bins=max(arrival_rate_1)+1)
arrival_rate_10 = stats.poisson.rvs(size=10000, # Generate Poisson data
mu=10 ) # Average arrival time 10
# Plot histogram
pd.DataFrame(arrival_rate_10).hist(range=(-0.5,max(arrival_rate_10)+0.5)
, bins=max(arrival_rate_10)+1)
```
## Poisson and Binomial Distributions
The binomial distribution is usually shown with a fixed n, with different values of p that will affect the k successes from the fixed n trails. This supposes we know the number of trails beforehand. We can graph the binomial distribution as a set of curves with a fixed n, and varying probabilities of the probability of success, p, below.
## What if we knew the rate but not the probability, p, or the number of trails, n?
But what if we were to invert the problem? What if we knew only the number of heads we observed, but not the total number of flips? If we have a known expected number of heads but an unknown number of flips, then we don't really know the true probability for each individual head. Rather we know that, on average, p=mean(k)/n. However if we were to plot these all on the same graph in the vicinity of the same k, we can make them all have a convergent shape around mean(k) because, no matter how much we increase n, we decrease p proportionally so that, for all n, the peak stays at mean(k).
## Deriving the Poisson Distribution from the Binomial Distribution
Let’s make this a little more formal. The binomial distribution works when we have a fixed number of events n, each with a constant probability of success p. In the Poisson Distribution, we don't know the number of trials that will happen. Instead, we only know the average number of successes per time period, the rate $\lambda$. So we know the rate of successes per day, or per minute but not the number of trials n or the probability of success p that was used to estimate to that rate.
If n is the number of trails in our time period, then np is the success rate or $\lambda$, that is, $\lambda$ = np. Solving for p, we get:
$$
p=\frac{\lambda}{n} \quad(1)
$$
Since the Binomial distribution is defined as below
$$
P(X=k) = \binom{n}{k} p^k (1-p)^{n-k} \quad k=1,2,...,n
\quad (2)
$$
or equivelently
$$
P(X=k) = \frac{n!}{k!(n-k)!} p^k (1-p)^{n-k} \quad k=1,2,...,n
\quad (3)
$$
By substituting the above p from (1) into the binomial distribution (3)
$$
P(X=k) = \frac{n!}{k!(n-k)!} {\frac{\lambda!}{n}}^k (1-{\frac{\lambda!}{n} })^{n-k} \quad (4)
$$
For n large and p small:
$$
P(X = k) \equiv \frac{ \mathrm{e}^{- \lambda } \lambda^k }{k!} \quad k=1,2,...,n\quad (5)
$$
Which is the probability mass function for the Poisson distribution.
## Fat-Tailed Distribution
In probability theory, the Fat-Tailed (or Gaussian) distribution is a very common continuous probability distribution. The Fat-Tailed distribution is remarkably useful because of the central limit theorem. In its most general form, under mild conditions, it states that averages of random variables independently drawn from independent distributions are Fat-Tailedly distributed. Physical quantities that are expected to be the sum of many independent processes (such as measurement errors) often have distributions that are nearly Fat-Tailed.
- from [Fat-Tailed Distribution - Wikipedia)](https://en.wikipedia.org/wiki/Fat-Tailed_distribution)
_Properties of Fat-Tailed distribution_
* Power law distributions:
* for variables assuming integer values > 0
* Prob [X=k] ~ Ck-α
* typically 0 < alpha < 2; smaller a gives heavier tail
* For binomial, normal, and Poisson distributions the tail probabilities approach 0 exponentially fast
* What kind of phenomena does this distribution model?
* What kind of process would generate it?
## Cauchy Distribution
An example of a Fat-tailed distribution is the Cauchy distribution.
The Cauchy distribution, named after Augustin Cauchy, is a continuous probability distribution. It is also known, especially among physicists, as the Lorentz distribution (after Hendrik Lorentz), Cauchy–Lorentz distribution, Lorentz(ian) function, or Breit–Wigner distribution. The simplest Cauchy distribution is called the standard Cauchy distribution. It is the distribution of a random variable that is the ratio of two independent standard normal variables and has the probability density function
The Cauchy distribution is often used in statistics as the canonical example of a "pathological" distribution since both its mean and its variance are undefined. (But see the section Explanation of undefined moments below.) The Cauchy distribution does not have finite moments of order greater than or equal to one; only fractional absolute moments exist.[1] The Cauchy distribution has no moment generating function.
- from [Cauchy Distribution - Wikipedia)](https://en.wikipedia.org/wiki/Cauchy_distribution)
Cauchy Distribution

Cauchy cumulative distribution function

## Cauchy Distribution in python
```
# Define the distribution parameters to be plotted
gamma_values = [0.5, 1.0, 2.0]
linestyles = ['-', '--', ':']
mu = 0
x = np.linspace(-10, 10, 1000)
#------------------------------------------------------------
# plot the distributions
for gamma, ls in zip(gamma_values, linestyles):
dist = stats.cauchy(mu, gamma)
plt.plot(x, dist.pdf(x), ls=ls, color='black',
label=r'$\mu=%i,\ \gamma=%.1f$' % (mu, gamma))
plt.xlim(-4.5, 4.5)
plt.ylim(0, 0.65)
plt.xlabel('$x$')
plt.ylabel(r'$p(x|\mu,\gamma)$')
plt.title('Cauchy Distribution')
plt.legend()
plt.show()
# From http://www.astroml.org/book_figures/chapter3/fig_cauchy_distribution.html
n=50
def random_distributions(n=50):
mu, sigma, p = 5, 2*np.sqrt(2), 0.3# mean, standard deviation, probabilty of success
shape, scale = 2.5, 2. # mean=5, std=2*sqrt(2)
normal_dist = np.random.normal(mu, sigma, n)
lognormal_dist = np.random.lognormal(mu, sigma, n)
lognormal_dist = np.random.lognormal(np.log2(mu), np.log2(sigma), n)
pareto_dist = np.random.pareto(mu, n)
uniform_dist= np.random.uniform(np.amin(normal_dist),np.amax(normal_dist),n)
binomial_dist= np.random.binomial(n, p,n)
gamma_dist= np.random.gamma(shape, scale, n)
poisson_dist= np.random.poisson((n*0.05), n)
df = pd.DataFrame({'Normal' : normal_dist, 'Lognormal' : lognormal_dist, 'Pareto' : pareto_dist,'Gamma' : gamma_dist, 'Poisson' : poisson_dist, 'Binomial' : binomial_dist, 'Uniform' : uniform_dist})
return df
df=random_distributions(n=50)
df.head()
def show_distributions(df):
for col in list(df.columns.values):
sns.distplot(df[col])
sns.plt.show()
show_distributions(df)
def qqplot_stats(obs, c):
z = (obs-np.mean(obs))/np.std(obs)
stats.probplot(z, dist="norm", plot=plt)
plt.title("Normal Q-Q plot for " + c)
plt.show()
def qqplot_df(df):
for col in list(df.columns.values):
qqplot_stats(df[col], col)
qqplot_df(df)
```
## Statistical tests for normality (e.g. Shapiro-Wilk test, Anderson-Darling test, scipy.stats.normaltest, etc.)
```
def normality_stats(df):
s={}
for col in list(df.columns.values):
s[col]={}
for col in list(df.columns.values):
s[col].update({'shapiro':stats.shapiro(df[col])})
s[col].update({'anderson':stats.anderson(df[col], dist='norm')})
s[col].update({'normaltest':stats.normaltest(df[col])})
return s
```
## Shapiro-Wilk test
scipy.stats.shapiro [https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.shapiro.html](https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.shapiro.html)
scipy.stats.shapiro
scipy.stats.shapiro(x, a=None, reta=False)[source]
Perform the Shapiro-Wilk test for normality.
The Shapiro-Wilk test tests the null hypothesis that the data was drawn from a normal distribution.
Parameters:
x : array_like
Array of sample data.
a : array_like, optional
Array of internal parameters used in the calculation. If these are not given, they will be computed internally. If x has length n, then a must have length n/2.
reta : bool, optional
Whether or not to return the internally computed a values. The default is False.
Returns:
W : float
The test statistic.
p-value : float
The p-value for the hypothesis test.
a : array_like, optional
If reta is True, then these are the internally computed “a” values that may be passed into this function on future calls.
### Anderson-Darling test
scipy.stats.anderson [https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.anderson.html](https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.anderson.html)
scipy.stats.anderson(x, dist='norm')
Anderson-Darling test for data coming from a particular distribution
The Anderson-Darling test is a modification of the Kolmogorov- Smirnov test kstest for the null hypothesis that a sample is drawn from a population that follows a particular distribution. For the Anderson-Darling test, the critical values depend on which distribution is being tested against. This function works for normal, exponential, logistic, or Gumbel (Extreme Value Type I) distributions.
Parameters:
x : array_like
array of sample data
dist : {‘norm’,’expon’,’logistic’,’gumbel’,’gumbel_l’, gumbel_r’,
‘extreme1’}, optional the type of distribution to test against. The default is ‘norm’ and ‘extreme1’, ‘gumbel_l’ and ‘gumbel’ are synonyms.
Returns:
statistic : float
The Anderson-Darling test statistic
critical_values : list
The critical values for this distribution
significance_level : list
The significance levels for the corresponding critical values in percents. The function returns critical values for a differing set of significance levels depending on the distribution that is being tested against.
Note: The critical values are for a given significance level. When we want a smaller significance level, then we have to increase the critical values, assuming we are in the right, upper tail of the distribution.
### scipy.stats.normaltest
scipy.stats.normaltest [https://docs.scipy.org/doc/scipy-0.14.0/reference/generated/scipy.stats.normaltest.html](https://docs.scipy.org/doc/scipy-0.14.0/reference/generated/scipy.stats.normaltest.html)
scipy.stats.normaltest(a, axis=0)
Tests whether a sample differs from a normal distribution.
This function tests the null hypothesis that a sample comes from a normal distribution. It is based on D’Agostino and Pearson’s [R251], [R252] test that combines skew and kurtosis to produce an omnibus test of normality.
Parameters:
a : array_like
The array containing the data to be tested.
axis : int or None
If None, the array is treated as a single data set, regardless of its shape. Otherwise, each 1-d array along axis axis is tested.
Returns:
k2 : float or array
s^2 + k^2, where s is the z-score returned by skewtest and k is the z-score returned by kurtosistest.
p-value : float or array
A 2-sided chi squared probability for the hypothesis test.
```
norm_stats=normality_stats(df)
print norm_stats
df=random_distributions(n=500)
df.head()
show_distributions(df)
qqplot_df(df)
norm_stats=normality_stats(df)
print norm_stats
df=random_distributions(n=5000)
df.head()
show_distributions(df)
qqplot_df(df)
norm_stats=normality_stats(df)
print norm_stats
```
Last update September 5, 2017
|
github_jupyter
|
%matplotlib inline
# %matplotlib inline is a magic function in IPython that displays images in the notebook
# Line magics are prefixed with the % character and work much like OS command-line calls
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from scipy import stats
import seaborn as sns
import warnings
warnings.filterwarnings('ignore')
# Make plots larger
plt.rcParams['figure.figsize'] = (10, 6)
#------------------------------------------------------------
# Define the distribution parameters to be plotted
W_values = [1.0, 3.0, 5.0]
linestyles = ['-', '--', ':']
mu = 0
x = np.linspace(-4, 4, 1000)
#------------------------------------------------------------
# plot the distributions
fig, ax = plt.subplots(figsize=(10, 5))
for W, ls in zip(W_values, linestyles):
left = mu - 0.5 * W
dist = stats.uniform(left, W)
plt.plot(x, dist.pdf(x), ls=ls, c='black',
label=r'$\mu=%i,\ W=%i$' % (mu, W))
plt.xlim(-4, 4)
plt.ylim(0, 1.2)
plt.xlabel('$x$')
plt.ylabel(r'$p(x|\mu, W)$')
plt.title('Uniform Distribution')
plt.legend()
plt.show()
# Adapted from http://www.astroml.org/book_figures/chapter3/fig_uniform_distribution.html
# Plot two normal distributions
domain = np.arange(-22, 22, 0.1)
values = stats.norm(3.3, 5.5).pdf(domain)
plt.plot(domain, values, color='r', linewidth=2)
plt.fill_between(domain, 0, values, color='#ffb6c1', alpha=0.3)
values = stats.norm(4.4, 2.3).pdf(domain)
plt.plot(domain, values, color='b', linewidth=2)
plt.ylabel("Probability")
plt.title("Two Normal Distributions")
plt.show()
#------------------------------------------------------------
# Define the distribution parameters to be plotted
n_values = [20, 20, 40]
b_values = [0.2, 0.6, 0.6]
linestyles = ['-', '--', ':']
x = np.arange(-1, 200)
#------------------------------------------------------------
# plot the distributions
for (n, b, ls) in zip(n_values, b_values, linestyles):
# create a binomial distribution
dist = stats.binom(n, b)
plt.plot(x, dist.pmf(x), ls=ls, c='black',
label=r'$b=%.1f,\ n=%i$' % (b, n), linestyle='steps-mid')
plt.xlim(-0.5, 35)
plt.ylim(0, 0.25)
plt.xlabel('$x$')
plt.ylabel(r'$p(x|b, n)$')
plt.title('Binomial Distribution')
plt.legend()
plt.show()
# Adapted from http://www.astroml.org/book_figures/chapter3/fig_binomial_distribution.html
fair_coin_flips = stats.binom.rvs(n=33, # Number of flips per trial
p=0.4, # Success probability
size=1000) # Number of trials
pd.DataFrame(fair_coin_flips).hist(range=(-0.5,10.5), bins=11)
plt.fill_between(x=np.arange(-4,-1,0.01),
y1= stats.norm.pdf(np.arange(-4,-1,0.01)) ,
facecolor='red',
alpha=0.35)
plt.fill_between(x=np.arange(1,4,0.01),
y1= stats.norm.pdf(np.arange(1,4,0.01)) ,
facecolor='red',
alpha=0.35)
plt.fill_between(x=np.arange(-1,1,0.01),
y1= stats.norm.pdf(np.arange(-1,1,0.01)) ,
facecolor='blue',
alpha=0.35)
# Generate poisson counts
arrival_rate_1 = stats.poisson.rvs(size=10000, # Generate Poisson data
mu=1 ) # Average arrival time 1
# Plot histogram
pd.DataFrame(arrival_rate_1).hist(range=(-0.5,max(arrival_rate_1)+0.5)
, bins=max(arrival_rate_1)+1)
arrival_rate_10 = stats.poisson.rvs(size=10000, # Generate Poisson data
mu=10 ) # Average arrival time 10
# Plot histogram
pd.DataFrame(arrival_rate_10).hist(range=(-0.5,max(arrival_rate_10)+0.5)
, bins=max(arrival_rate_10)+1)
# Define the distribution parameters to be plotted
gamma_values = [0.5, 1.0, 2.0]
linestyles = ['-', '--', ':']
mu = 0
x = np.linspace(-10, 10, 1000)
#------------------------------------------------------------
# plot the distributions
for gamma, ls in zip(gamma_values, linestyles):
dist = stats.cauchy(mu, gamma)
plt.plot(x, dist.pdf(x), ls=ls, color='black',
label=r'$\mu=%i,\ \gamma=%.1f$' % (mu, gamma))
plt.xlim(-4.5, 4.5)
plt.ylim(0, 0.65)
plt.xlabel('$x$')
plt.ylabel(r'$p(x|\mu,\gamma)$')
plt.title('Cauchy Distribution')
plt.legend()
plt.show()
# From http://www.astroml.org/book_figures/chapter3/fig_cauchy_distribution.html
n=50
def random_distributions(n=50):
mu, sigma, p = 5, 2*np.sqrt(2), 0.3# mean, standard deviation, probabilty of success
shape, scale = 2.5, 2. # mean=5, std=2*sqrt(2)
normal_dist = np.random.normal(mu, sigma, n)
lognormal_dist = np.random.lognormal(mu, sigma, n)
lognormal_dist = np.random.lognormal(np.log2(mu), np.log2(sigma), n)
pareto_dist = np.random.pareto(mu, n)
uniform_dist= np.random.uniform(np.amin(normal_dist),np.amax(normal_dist),n)
binomial_dist= np.random.binomial(n, p,n)
gamma_dist= np.random.gamma(shape, scale, n)
poisson_dist= np.random.poisson((n*0.05), n)
df = pd.DataFrame({'Normal' : normal_dist, 'Lognormal' : lognormal_dist, 'Pareto' : pareto_dist,'Gamma' : gamma_dist, 'Poisson' : poisson_dist, 'Binomial' : binomial_dist, 'Uniform' : uniform_dist})
return df
df=random_distributions(n=50)
df.head()
def show_distributions(df):
for col in list(df.columns.values):
sns.distplot(df[col])
sns.plt.show()
show_distributions(df)
def qqplot_stats(obs, c):
z = (obs-np.mean(obs))/np.std(obs)
stats.probplot(z, dist="norm", plot=plt)
plt.title("Normal Q-Q plot for " + c)
plt.show()
def qqplot_df(df):
for col in list(df.columns.values):
qqplot_stats(df[col], col)
qqplot_df(df)
def normality_stats(df):
s={}
for col in list(df.columns.values):
s[col]={}
for col in list(df.columns.values):
s[col].update({'shapiro':stats.shapiro(df[col])})
s[col].update({'anderson':stats.anderson(df[col], dist='norm')})
s[col].update({'normaltest':stats.normaltest(df[col])})
return s
norm_stats=normality_stats(df)
print norm_stats
df=random_distributions(n=500)
df.head()
show_distributions(df)
qqplot_df(df)
norm_stats=normality_stats(df)
print norm_stats
df=random_distributions(n=5000)
df.head()
show_distributions(df)
qqplot_df(df)
norm_stats=normality_stats(df)
print norm_stats
| 0.811788 | 0.991505 |
```
import pymongo
import pandas as pd
import numpy as np
from tqdm import tqdm_notebook
import json
with open('../data/yelp_tset.json', 'r') as infile:
T = json.load(infile)
```
## Weighting schemes
- **tfidf**: tfidf weights
- **sentiwn**: average sentiwn
- **combo**: tfidf x average sentiwn
### tfidf
```
from sklearn.feature_extraction.text import CountVectorizer, TfidfTransformer
from sklearn.decomposition import TruncatedSVD
def tf_idf(data, components=200):
docs = [x for x in data.keys()]
texts = [data[k] for k in docs]
c = CountVectorizer()
tf_idf = TfidfTransformer(use_idf=True)
k = c.fit_transform(texts)
j = tf_idf.fit_transform(k)
return j, TruncatedSVD(n_components=components).fit_transform(j), docs, c, tf_idf
Tx = {}
for k, tset in T.items():
Tx[k + '_tfidf'] = tf_idf(tset, components=200)
Tx['raw_text_tfidf']
```
### sentiwn
```
from nltk.corpus import sentiwordnet as swn
from scipy.sparse import csr_matrix
def sentiwn(data, components=200):
docs = [x for x in data.keys()]
texts = tqdm_notebook([data[k].split() for k in docs])
indptr, indices, data, dictionary = [0], [], [], {}
for doc in texts:
for token in doc:
t_index = dictionary.setdefault(token, len(dictionary))
indices.append(t_index)
if token.startswith('NOT_'):
synsets = list(swn.senti_synsets(token.replace('NOT_', '')))
modifier = -1
else:
synsets = list(swn.senti_synsets(token))
modifier = 1
w = 0
for syn in synsets:
w += (syn.pos_score() - syn.neg_score()) * modifier
try:
data.append(w / len(synsets))
except ZeroDivisionError:
data.append(0)
indptr.append(len(indices))
csr = csr_matrix((data, indices, indptr), dtype=np.float64)
return csr, TruncatedSVD(n_components=components).fit_transform(csr), docs, dictionary
for k, tset in T.items():
Tx[k + '_sentiwn'] = sentiwn(tset)
```
### combo
```
def combo(case, Tx, T, components=200):
m1d, _, docs1, d1 = Tx['{}_sentiwn'.format(case)]
m2d, _, docs2, d2, _ = Tx['{}_tfidf'.format(case)]
m1 = m1d.toarray()
m2 = m2d.toarray()
M = np.zeros(m1.shape)
run = tqdm_notebook(list(enumerate(docs1)))
for i, doc in run:
tokens = T[case][doc].split()
d2_index = docs2.index(doc)
for t in tokens:
try:
t1_index = d1[t]
sw = m1[i,t1_index]
except KeyError:
t1_index = None
sw = 0
try:
t2_index = d2.vocabulary_[t]
tw = m2[d2_index,t2_index]
except KeyError:
tw = 0
if t1_index is not None:
M[i,t1_index] = sw * tw
out = csr_matrix(M, dtype=np.float64)
return out, TruncatedSVD(n_components=components).fit_transform(out), docs1, d1
for k, tset in T.items():
Tx[k + '_combo'] = combo(k, Tx, T)
```
### Save
```
import pickle
to_save = {}
for k, v in Tx.items():
to_save[k] = list(v)[1:]
with open('../data/yelp_training.pkl', 'wb') as out:
pickle.dump(to_save, out)
```
|
github_jupyter
|
import pymongo
import pandas as pd
import numpy as np
from tqdm import tqdm_notebook
import json
with open('../data/yelp_tset.json', 'r') as infile:
T = json.load(infile)
from sklearn.feature_extraction.text import CountVectorizer, TfidfTransformer
from sklearn.decomposition import TruncatedSVD
def tf_idf(data, components=200):
docs = [x for x in data.keys()]
texts = [data[k] for k in docs]
c = CountVectorizer()
tf_idf = TfidfTransformer(use_idf=True)
k = c.fit_transform(texts)
j = tf_idf.fit_transform(k)
return j, TruncatedSVD(n_components=components).fit_transform(j), docs, c, tf_idf
Tx = {}
for k, tset in T.items():
Tx[k + '_tfidf'] = tf_idf(tset, components=200)
Tx['raw_text_tfidf']
from nltk.corpus import sentiwordnet as swn
from scipy.sparse import csr_matrix
def sentiwn(data, components=200):
docs = [x for x in data.keys()]
texts = tqdm_notebook([data[k].split() for k in docs])
indptr, indices, data, dictionary = [0], [], [], {}
for doc in texts:
for token in doc:
t_index = dictionary.setdefault(token, len(dictionary))
indices.append(t_index)
if token.startswith('NOT_'):
synsets = list(swn.senti_synsets(token.replace('NOT_', '')))
modifier = -1
else:
synsets = list(swn.senti_synsets(token))
modifier = 1
w = 0
for syn in synsets:
w += (syn.pos_score() - syn.neg_score()) * modifier
try:
data.append(w / len(synsets))
except ZeroDivisionError:
data.append(0)
indptr.append(len(indices))
csr = csr_matrix((data, indices, indptr), dtype=np.float64)
return csr, TruncatedSVD(n_components=components).fit_transform(csr), docs, dictionary
for k, tset in T.items():
Tx[k + '_sentiwn'] = sentiwn(tset)
def combo(case, Tx, T, components=200):
m1d, _, docs1, d1 = Tx['{}_sentiwn'.format(case)]
m2d, _, docs2, d2, _ = Tx['{}_tfidf'.format(case)]
m1 = m1d.toarray()
m2 = m2d.toarray()
M = np.zeros(m1.shape)
run = tqdm_notebook(list(enumerate(docs1)))
for i, doc in run:
tokens = T[case][doc].split()
d2_index = docs2.index(doc)
for t in tokens:
try:
t1_index = d1[t]
sw = m1[i,t1_index]
except KeyError:
t1_index = None
sw = 0
try:
t2_index = d2.vocabulary_[t]
tw = m2[d2_index,t2_index]
except KeyError:
tw = 0
if t1_index is not None:
M[i,t1_index] = sw * tw
out = csr_matrix(M, dtype=np.float64)
return out, TruncatedSVD(n_components=components).fit_transform(out), docs1, d1
for k, tset in T.items():
Tx[k + '_combo'] = combo(k, Tx, T)
import pickle
to_save = {}
for k, v in Tx.items():
to_save[k] = list(v)[1:]
with open('../data/yelp_training.pkl', 'wb') as out:
pickle.dump(to_save, out)
| 0.27338 | 0.653445 |
<h1>Distances</h1>
<p>In this notebook, we will use sktime for time series distance computation</p>
<h3>Preliminaries</h3>
```
import matplotlib.pyplot as plt
import numpy as np
from sktime.datasets import load_macroeconomic
from sktime.distances import distance
```
<h2>Distances</h2>
The goal of a distance computation is to measure the similarity between the time series
'x' and 'y'. A distance function should take x and y as parameters and return a float
that is the computed distance between x and y. The value returned should be 0.0 when
the time series are the exact same, and a value greater than 0.0 that is a measure of
distance between them, when they are different.
Take the following two time series:
```
X = load_macroeconomic()
country_d, country_c, country_b, country_a = np.split(X["realgdp"].to_numpy()[3:], 4)
plt.plot(country_a, label="County D")
plt.plot(country_b, label="Country C")
plt.plot(country_c, label="Country B")
plt.plot(country_d, label="Country A")
plt.xlabel("Quarters from 1959")
plt.ylabel("Gdp")
plt.legend()
```
The above shows a made up scenario comparing the gdp growth of four countries (country
A, B, C and D) by quarter from 1959. If our task is to determine how different country
C is from our other countries one way to do this is to measure the distance between
each country.
<br>
How to use the distance module to perform tasks such as these, will now be outlined.
<h2>Distance module</h2>
To begin using the distance module we need at least two time series, x and y and they
must be numpy arrays. We've established the various time series we'll be using for this
example above as country_a, country_b, country_c and country_d. To compute the distance
between x and y we can use a euclidean distance as shown:
```
# Simple euclidean distance
distance(country_a, country_b, metric="euclidean")
```
Shown above taking the distance between country_a and country_b, returns a singular
float that represents their similarity (distance). We can do the same again but compare
country_d to country_a:
```
distance(country_a, country_d, metric="euclidean")
```
Now we can compare the result of the distance computation and we find that country_a is
closer to country_b than country_d (27014.7 < 58340.1).
We can further confirm this result by looking at the graph above and see the green line
(country_b) is closer to the red line (country_a) than the orange line (country d).
<br>
<h3>Different metric parameters</h3>
Above we used the metric "euclidean". While euclidean distance is appropriate for simple
example such as the one above, it has been shown to be inadequate when we have larger
and more complex timeseries (particularly multivariate). While the merits of each
different distance won't be described here (see documentation for descriptions of each),
a large number of specialised time series distances have been implement to get a better
accuracy in distance computation. These are:
<br><br>
'euclidean', 'squared', 'dtw', 'ddtw', 'wdtw', 'wddtw', 'lcss', 'edr', 'erp'
<br><br>
All of the above can be used as a metric parameter. This will now be demonstrated:
```
print("Euclidean distance: ", distance(country_a, country_d, metric="euclidean"))
print("Squared euclidean distance: ", distance(country_a, country_d, metric="squared"))
print("Dynamic time warping distance: ", distance(country_a, country_d, metric="dtw"))
print(
"Derivative dynamic time warping distance: ",
distance(country_a, country_d, metric="ddtw"),
)
print(
"Weighted dynamic time warping distance: ",
distance(country_a, country_d, metric="wdtw"),
)
print(
"Weighted derivative dynamic time warping distance: ",
distance(country_a, country_d, metric="wddtw"),
)
print(
"Longest common subsequence distance: ",
distance(country_a, country_d, metric="lcss"),
)
print(
"Edit distance for real sequences distance: ",
distance(country_a, country_d, metric="edr"),
)
print(
"Edit distance for real penalty distance: ",
distance(country_a, country_d, metric="erp"),
)
```
While many of the above use euclidean distance at their core, they change how it is
used to account for various problems we encounter with time series data such as:
alignment, phase, shape, dimensions etc. As mentioned for specific details on how to
best use each distance and what it does see the documentation for that distance.
<h3>Custom parameters for distances</h3>
In addition each distance has a different set of parameters. How these are passed to
the 'distance' function will now be outlined using the 'dtw' example. As stated for
specific parameters for each distance please refer to the documentation.
<br><br>
Dtw is a O(n^2) algorithm and as such a point of focus has been trying to optimise the
algorithm. A proposal to improve performance is to restrict the potential alignment
path by putting a 'bound' on values to consider when looking for an alignment. While
there have been many bounding algorithms proposed the two most popular are Sakoe-Chiba
bounding or Itakuras parallelogram bounding. How these two work will briefly be
outlined using the LowerBounding class:
```
from sktime.distances import LowerBounding
x = np.zeros((6, 6))
y = np.zeros((6, 6)) # Create dummy data to show the matrix
LowerBounding.NO_BOUNDING.create_bounding_matrix(x, y)
```
Above shows a matrix that maps each index in 'x' to each index in 'y'. Dtw without
bounding will consider all of these indexes (indexes in bound we define as finite
values (0.0)). However, we can change the indexes that are considered using
Sakoe-Chibas bounding like so:
```
LowerBounding.SAKOE_CHIBA.create_bounding_matrix(x, y, sakoe_chiba_window_radius=1)
```
The matrix that is produced follows the same concept as no bounding where each index
between x and y are assigned a value. If the value is finite (0.0) it is considered
inbound and infinite out of bounds. Using Sakoe-Chiba bounding matrix with a window
radius of 1 we can see we get a diagonal from 0,0 to 5,5 where values inside the
window are 0.0 and values outside are infinite. This reduces the compute time of
dtw as we are considering 12 less potential indexes (12 values are infinite).
<br><br>
As mentioned there are other bounding techniques that use different 'shapes' over the
matrix such a Itakuras parallelogram which as the name implies produces a parallelogram
shape over the matrix.
```
LowerBounding.ITAKURA_PARALLELOGRAM.create_bounding_matrix(x, y, itakura_max_slope=3.0)
```
With that base introductory to bounding algorithms and why we may want to use them
how do we use it in our distance computation. There are two ways:
```
# Create two random unaligned time series to better illustrate the difference
rng = np.random.RandomState(42)
n_timestamps, n_features = 10, 19
x = rng.randn(n_timestamps, n_features)
y = rng.randn(n_timestamps, n_features)
# First we can specify the bounding matrix to use either via enum or int (see
# documentation for potential values):
print(
"Dynamic time warping distance with Sakoe-Chiba: ",
distance(x, y, metric="dtw", lower_bounding=LowerBounding.SAKOE_CHIBA, window=1),
) # Sakoe chiba
print(
"Dynamic time warping distance with Itakura parallelogram: ",
distance(x, y, metric="dtw", lower_bounding=2, itakura_max_slope=1.0),
) # Itakura parallelogram using int to specify
print(
"Dynamic time warping distance with Sakoe-Chiba: ",
distance(x, y, metric="dtw", lower_bounding=LowerBounding.NO_BOUNDING),
) # No bounding
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import numpy as np
from sktime.datasets import load_macroeconomic
from sktime.distances import distance
X = load_macroeconomic()
country_d, country_c, country_b, country_a = np.split(X["realgdp"].to_numpy()[3:], 4)
plt.plot(country_a, label="County D")
plt.plot(country_b, label="Country C")
plt.plot(country_c, label="Country B")
plt.plot(country_d, label="Country A")
plt.xlabel("Quarters from 1959")
plt.ylabel("Gdp")
plt.legend()
# Simple euclidean distance
distance(country_a, country_b, metric="euclidean")
distance(country_a, country_d, metric="euclidean")
print("Euclidean distance: ", distance(country_a, country_d, metric="euclidean"))
print("Squared euclidean distance: ", distance(country_a, country_d, metric="squared"))
print("Dynamic time warping distance: ", distance(country_a, country_d, metric="dtw"))
print(
"Derivative dynamic time warping distance: ",
distance(country_a, country_d, metric="ddtw"),
)
print(
"Weighted dynamic time warping distance: ",
distance(country_a, country_d, metric="wdtw"),
)
print(
"Weighted derivative dynamic time warping distance: ",
distance(country_a, country_d, metric="wddtw"),
)
print(
"Longest common subsequence distance: ",
distance(country_a, country_d, metric="lcss"),
)
print(
"Edit distance for real sequences distance: ",
distance(country_a, country_d, metric="edr"),
)
print(
"Edit distance for real penalty distance: ",
distance(country_a, country_d, metric="erp"),
)
from sktime.distances import LowerBounding
x = np.zeros((6, 6))
y = np.zeros((6, 6)) # Create dummy data to show the matrix
LowerBounding.NO_BOUNDING.create_bounding_matrix(x, y)
LowerBounding.SAKOE_CHIBA.create_bounding_matrix(x, y, sakoe_chiba_window_radius=1)
LowerBounding.ITAKURA_PARALLELOGRAM.create_bounding_matrix(x, y, itakura_max_slope=3.0)
# Create two random unaligned time series to better illustrate the difference
rng = np.random.RandomState(42)
n_timestamps, n_features = 10, 19
x = rng.randn(n_timestamps, n_features)
y = rng.randn(n_timestamps, n_features)
# First we can specify the bounding matrix to use either via enum or int (see
# documentation for potential values):
print(
"Dynamic time warping distance with Sakoe-Chiba: ",
distance(x, y, metric="dtw", lower_bounding=LowerBounding.SAKOE_CHIBA, window=1),
) # Sakoe chiba
print(
"Dynamic time warping distance with Itakura parallelogram: ",
distance(x, y, metric="dtw", lower_bounding=2, itakura_max_slope=1.0),
) # Itakura parallelogram using int to specify
print(
"Dynamic time warping distance with Sakoe-Chiba: ",
distance(x, y, metric="dtw", lower_bounding=LowerBounding.NO_BOUNDING),
) # No bounding
| 0.714628 | 0.984826 |
```
import pandas as pd
import keras as ks
import sklearn
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
from keras.models import Sequential
from keras.layers import Dense, Activation, Embedding, LSTM,Dropout
from sklearn.preprocessing import StandardScaler
from keras.wrappers.scikit_learn import KerasClassifier
from sklearn.pipeline import Pipeline
from sklearn.model_selection import StratifiedKFold
from keras.models import Sequential
from keras.layers import Dense
from keras.wrappers.scikit_learn import KerasClassifier
from sklearn.model_selection import cross_val_score
x=['HR',
'O2Sat',
'Temp',
'SBP',
'MAP',
'DBP',
'Resp',
'BaseExcess',
'HCO3',
'FiO2',
'pH',
'PaCO2',
'AST',
'BUN',
'Calcium',
'Chloride',
'Glucose',
'Magnesium',
'Potassium',
'Hct',
'Hgb',
'WBC',
'Age',
'Gender',
'Unit1',
'Unit2',
'HospAdmTime',
'ICULOS']
def create_larger():
model = Sequential()
model.add(Dense(80,input_dim=28,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(1, activation='sigmoid'))
model.compile(loss='binary_crossentropy', optimizer='sgd', metrics=['accuracy'])
return model
model=create_larger()
x
data=pd.read_csv('./SetA/setAE.csv')
data=data.fillna(0)
X=data[x].copy()
y=data['SepsisLabel'].copy()
data
X=X.values
y=y.values
model.fit(X, y,epochs=200,batch_size=128)
y_pred=model.predict(x_train)
data_train=pd.read_csv('./SetA/setAA.csv')
data_train=data_train.fillna(0)
x_test=data_train[x].copy()
y_test=data_train['SepsisLabel'].copy()
y_pred=model.predict(x_test)
len(y_test[y_test==1])
y_t=y_pred
y_t[y_pred>0.5]=1
y_t[y_pred<0.5]=0
c=0
for i in range(0,len(y_test)):
if y_test[i]==1 and y_t[i]==1:
c=c+1
c
score = model.evaluate(x_test, y_test)
print(score)
c/len(y_test[y_test==1])
model.save('./sepsissihupdate.h5')
```
|
github_jupyter
|
import pandas as pd
import keras as ks
import sklearn
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
from keras.models import Sequential
from keras.layers import Dense, Activation, Embedding, LSTM,Dropout
from sklearn.preprocessing import StandardScaler
from keras.wrappers.scikit_learn import KerasClassifier
from sklearn.pipeline import Pipeline
from sklearn.model_selection import StratifiedKFold
from keras.models import Sequential
from keras.layers import Dense
from keras.wrappers.scikit_learn import KerasClassifier
from sklearn.model_selection import cross_val_score
x=['HR',
'O2Sat',
'Temp',
'SBP',
'MAP',
'DBP',
'Resp',
'BaseExcess',
'HCO3',
'FiO2',
'pH',
'PaCO2',
'AST',
'BUN',
'Calcium',
'Chloride',
'Glucose',
'Magnesium',
'Potassium',
'Hct',
'Hgb',
'WBC',
'Age',
'Gender',
'Unit1',
'Unit2',
'HospAdmTime',
'ICULOS']
def create_larger():
model = Sequential()
model.add(Dense(80,input_dim=28,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(120,activation='relu'))
model.add(Dense(1, activation='sigmoid'))
model.compile(loss='binary_crossentropy', optimizer='sgd', metrics=['accuracy'])
return model
model=create_larger()
x
data=pd.read_csv('./SetA/setAE.csv')
data=data.fillna(0)
X=data[x].copy()
y=data['SepsisLabel'].copy()
data
X=X.values
y=y.values
model.fit(X, y,epochs=200,batch_size=128)
y_pred=model.predict(x_train)
data_train=pd.read_csv('./SetA/setAA.csv')
data_train=data_train.fillna(0)
x_test=data_train[x].copy()
y_test=data_train['SepsisLabel'].copy()
y_pred=model.predict(x_test)
len(y_test[y_test==1])
y_t=y_pred
y_t[y_pred>0.5]=1
y_t[y_pred<0.5]=0
c=0
for i in range(0,len(y_test)):
if y_test[i]==1 and y_t[i]==1:
c=c+1
c
score = model.evaluate(x_test, y_test)
print(score)
c/len(y_test[y_test==1])
model.save('./sepsissihupdate.h5')
| 0.571288 | 0.256471 |
# <font color='blue'>Data Science Academy - Python Fundamentos - Capítulo 4</font>
## Download: http://github.com/dsacademybr
## Exercícios
```
# Exercício 1 - Crie uma lista de 3 elementos e calcule a terceira potência de cada elemento.
elementos = [1, 2, 3]
print(list(map(lambda x: x ** 3, elementos)))
# Exercício 2 - Reescreva o código abaixo, usando a função map(). O resultado final deve ser o mesmo!
palavras = 'A Data Science Academy oferce os melhores cursos de análise de dados do Brasil'.split()
resultado = [[w.upper(), w.lower(), len(w)] for w in palavras]
for i in resultado:
print (i)
# resposta exercicio 2 em map
palavras = 'A Data Science Academy oferce os melhores cursos de análise de dados do Brasil'.split()
resultdo = map(lambda x: [x.upper(), x.lower(), len(x)], palavras)
for i in resultado:
print(i)
# Exercício 3 - Calcule a matriz transposta da matriz abaixo.
# Caso não saiba o que é matriz transposta, visite este link: https://pt.wikipedia.org/wiki/Matriz_transposta
# Matriz transposta é um conceito fundamental na construção de redes neurais artificiais, base de sistemas de IA.
matrix = [[1, 2],[3,4],[5,6],[7,8]]
transponse = [[row[i] for row in matrix] for i in range(2)]
print(transponse)
# Exercício 4 - Crie duas funções, uma para elevar um número ao quadrado e outra para elevar ao cubo.
# Aplique as duas funções aos elementos da lista abaixo.
# Obs: as duas funções devem ser aplicadas simultaneamente.
lista = [0, 1, 2, 3, 4]
# quadrado
def quadrado(num): return (num**2)
#cubo
def cubo(num): return (num**3)
funcs = [quadrado, cubo]
for i in lista:
valor = map(lambda x: x(i), funcs)
print(list(valor))
# Exercício 5 - Abaixo você encontra duas listas. Faça com que cada elemento da listaA seja elevado
# ao elemento correspondente na listaB.
listaA = [2, 3, 4]
listaB = [10, 11, 12]
print(list(zip(listaA, listaB)))
# Exercício 6 - Considerando o range de valores abaixo, use a função filter() para retornar apenas os valores negativos.
range(-5, 5)
print(list(filter((lambda x: x < 0), range(-5, 5))))
# Exercício 7 - Usando a função filter(), encontre os valores que são comuns às duas listas abaixo.
a = [1,2,3,5,7,9]
b = [2,3,5,6,7,8]
print(list(filter(lambda x: x in a, b)))
# Exercício 8 - Considere o código abaixo. Obtenha o mesmo resultado usando o pacote time.
# Não conhece o pacote time? Pesquise!
import datetime
import time
print (datetime.datetime.now().strftime("%d/%m/%Y %H:%M"))
print(time.strftime('%d/%m/%Y %H:%M'))
# Exercício 9 - Considere os dois dicionários abaixo.
# Crie um terceiro dicionário com as chaves do dicionário 1 e os valores do dicionário 2.
dict1 = {'a':1,'b':2}
dict2 = {'c':4,'d':5}
def troca_valores(d1, d2):
'''
recebe os dois dicionários
une a chave do 1 com valores do 2
'''
troca = {}
for d1key, d2val in zip(d1, d2.values()):
troca[d1key] = d2val
return troca
dict3 = troca_valores(dict1, dict2)
print(dict3)
# Exercício 10 - Considere a lista abaixo e retorne apenas os elementos cujo índice for maior que 5.
lista = ['a', 'b', 'c', 'd', 'e', 'f', 'g', 'h']
for i, valor in enumerate(lista):
if i > 5:
```
# Fim
### Obrigado - Data Science Academy - <a href="http://facebook.com/dsacademybr">facebook.com/dsacademybr</a>
|
github_jupyter
|
# Exercício 1 - Crie uma lista de 3 elementos e calcule a terceira potência de cada elemento.
elementos = [1, 2, 3]
print(list(map(lambda x: x ** 3, elementos)))
# Exercício 2 - Reescreva o código abaixo, usando a função map(). O resultado final deve ser o mesmo!
palavras = 'A Data Science Academy oferce os melhores cursos de análise de dados do Brasil'.split()
resultado = [[w.upper(), w.lower(), len(w)] for w in palavras]
for i in resultado:
print (i)
# resposta exercicio 2 em map
palavras = 'A Data Science Academy oferce os melhores cursos de análise de dados do Brasil'.split()
resultdo = map(lambda x: [x.upper(), x.lower(), len(x)], palavras)
for i in resultado:
print(i)
# Exercício 3 - Calcule a matriz transposta da matriz abaixo.
# Caso não saiba o que é matriz transposta, visite este link: https://pt.wikipedia.org/wiki/Matriz_transposta
# Matriz transposta é um conceito fundamental na construção de redes neurais artificiais, base de sistemas de IA.
matrix = [[1, 2],[3,4],[5,6],[7,8]]
transponse = [[row[i] for row in matrix] for i in range(2)]
print(transponse)
# Exercício 4 - Crie duas funções, uma para elevar um número ao quadrado e outra para elevar ao cubo.
# Aplique as duas funções aos elementos da lista abaixo.
# Obs: as duas funções devem ser aplicadas simultaneamente.
lista = [0, 1, 2, 3, 4]
# quadrado
def quadrado(num): return (num**2)
#cubo
def cubo(num): return (num**3)
funcs = [quadrado, cubo]
for i in lista:
valor = map(lambda x: x(i), funcs)
print(list(valor))
# Exercício 5 - Abaixo você encontra duas listas. Faça com que cada elemento da listaA seja elevado
# ao elemento correspondente na listaB.
listaA = [2, 3, 4]
listaB = [10, 11, 12]
print(list(zip(listaA, listaB)))
# Exercício 6 - Considerando o range de valores abaixo, use a função filter() para retornar apenas os valores negativos.
range(-5, 5)
print(list(filter((lambda x: x < 0), range(-5, 5))))
# Exercício 7 - Usando a função filter(), encontre os valores que são comuns às duas listas abaixo.
a = [1,2,3,5,7,9]
b = [2,3,5,6,7,8]
print(list(filter(lambda x: x in a, b)))
# Exercício 8 - Considere o código abaixo. Obtenha o mesmo resultado usando o pacote time.
# Não conhece o pacote time? Pesquise!
import datetime
import time
print (datetime.datetime.now().strftime("%d/%m/%Y %H:%M"))
print(time.strftime('%d/%m/%Y %H:%M'))
# Exercício 9 - Considere os dois dicionários abaixo.
# Crie um terceiro dicionário com as chaves do dicionário 1 e os valores do dicionário 2.
dict1 = {'a':1,'b':2}
dict2 = {'c':4,'d':5}
def troca_valores(d1, d2):
'''
recebe os dois dicionários
une a chave do 1 com valores do 2
'''
troca = {}
for d1key, d2val in zip(d1, d2.values()):
troca[d1key] = d2val
return troca
dict3 = troca_valores(dict1, dict2)
print(dict3)
# Exercício 10 - Considere a lista abaixo e retorne apenas os elementos cujo índice for maior que 5.
lista = ['a', 'b', 'c', 'd', 'e', 'f', 'g', 'h']
for i, valor in enumerate(lista):
if i > 5:
| 0.329715 | 0.912163 |
```
import pandas as pd
import numpy as np
import seaborn as sns
import statsmodels
import linearmodels
import matplotlib as plt
import xlrd
import scipy
from scipy.stats import sem, t
from scipy import mean
import os
import statsmodels.api as sm
from pandas.plotting import autocorrelation_plot
import pandas_profiling
from linearmodels.panel import PanelOLS
import random
%matplotlib inline
# read data file, convert to csv
data = pd.read_excel('data/fpds_rates.xlsx')
data.head()
data.columns
data.shape
data['unique_name'] = data['department_name'] + '-' + data['contracting_agency_name']
data['unique_name'] = data['unique_name'].fillna('agency wide')
```
# Intro
Here we're trying to recreate a fixed effects panel regression written in the proprietary econometric software EViews. We only have the summary output of that model, which is specified as:
`EffCompRate ~ C + NotCompRate(-1) + OneBidRate + SdbRate`
These variables are defined as follows:
- **EffCompRate:** Total Competed Dollars with 2 or more bids / Total Competed Dollars
- **NotCompRate:** Total Not Competed Dollars / Total Obligations
- **OneBidRate:** Total Competed Dollars with only 1 bid / Total Competed Dollars
- **SdbRate:** Total Small Disadvantaged Business Dollars / Total Small Business Eligible Dollars
Some research into EViews leads us to believe that this model was fit using fixed entity and time effects:
$$
Y_{it} = \beta_0 + \beta_1 X_{it} + \gamma_2 D2_i + \cdots + \gamma_n DT_i + \delta_2 B2_t + \cdots + \delta_T BT_t + u_{it}
$$
Such a model allows us to eliminate bias from unobservables that change over time but are constant over entities (time fixed effects) and it controls for factors that differ across entities but are constant over time (entity fixed effects).
### First Thoughts
Inspecting the model specification, we notice that each variable is a ratio measurement, with a few common/similar terms in the numerators and denominators. For example, the OneBidRate should sum to one when added to the EffCompRate. Additionally, one should also be able to calculate the total competed dollars by summing the numerators in OneBidRate and EffCompRate. This knowledge will both help and hinder us, as interrealted ratio components like this bake a sort of statistical dependency into our data that can impact our coeffecient estimates as well as inferences. It also fortunately furnishes us with an opportunity to assess data quality, as all of these calculations should work out perfectly in the absence of data issues.
Moreover, we notice that there's a common denominator for both the IV (EffCompRate) and one of the DVs (OneBidRate). In this situation, we could re-write the regression equation by factoring out the common denominator as a separate right-hand term and then demonstrate that there is a correlation between the independent variable with the common component and the error term, which has now absorved the denominator. This essentially renders the relationship between that IV and the DV in the orginal model as definitionally spurious.
Finally, we suspect that there will be some multicolinearity issues with these variables. All of the variables are functions of total dollars obligated. If one moves a certain percentage then another will definitionally move in proportion. This lack of independence problematizes coefficient interpretation as we can no longer attribute a change in $y$ to a change in $x$ *holding all other things constant*.
In short, we'd advise against interpreting the results of this model specification. The negative consequences of doing so would be further compounded if data quality issues are discovered (spoiler alert: we find some).
## Purpose / Path Forward
We're building this model after-the-fact for documentation purposes. We do not intend to carry out a robust analysis. We're merely going to recreate it as originally specified. Even so, we'll run through several "what-if" scenarios using Monte Carlo simulations to demonstrate the impact of some of the methodological/data issues.
## Possible Sources of Error / Assumptions
Since we only have the model output, we're not certain how the data was cleansed prior to fitting. We're also not entirely sure whether or not time *and* cross-sectional fixed effects were modeled. We're assuming both were.
# Data Cleaning
```
data.head()
#drop agency-wide observations
agency_wide = False
if agency_wide:
clean_data = data
else:
print("Dropping agency wide observations")
clean_data = data[data['unique_name'] != 'agency wide']
# some records have not obligated dollars, so I'll drop those as it does not make sense to include agencies who
# did not participate in any contracting actions in a regression measuring the dollar impact of
#contracting actions. Also certain rates used in the regression use TotalDollars in the denominator
clean_data = clean_data[clean_data['TotalDollars']>0]
# remove negatives
clean_data= clean_data[clean_data['OneBidRate']>0]
clean_data = clean_data[clean_data['SdbRate']>0]
clean_data = clean_data[clean_data['NotCompRate']>0]
clean_data = clean_data[clean_data['EffCompRate']>0]
#ensure each unique_name contains 5 years of data
n_years_n_obs_map = {k:0 for k in range(1,6)}
for name in clean_data['unique_name'].unique():
if name is np.nan:
continue
n_years = clean_data[clean_data['unique_name'] == name].shape[0]
if n_years == 0:
print(name)
try:
n_years_n_obs_map[n_years] += 1
except KeyError:
n_years_n_obs_map[n_years] = 1
# this is the number of observations the OLS model should report
sum(n_years_n_obs_map.values())
```
# EDA
```
df = clean_data[['SdbRate', 'OneBidRate', 'NotCompRate', 'EffCompRate', 'FiscalYear', 'unique_name']]
pandas_profiling.ProfileReport(df)
```
## Data Validation
Above, we noted that the OneBidRate should sum to one when added to the EffCompRate. We'll check that below to identify data validity issues:
```
sum_to_one = df['OneBidRate'] + df['EffCompRate']
sns.distplot(sum_to_one)
```
It seems that not every value record sums to one, indicating some data quality issues. Let's inspect further by rounding each sum to the nearest tenth.
```
sum_to_one.round(decimals = 1).value_counts()
no_sum_to_one = sum_to_one.round(decimals = 1).value_counts().sum() - 662
n_records = sum_to_one.round(decimals = 1).value_counts().sum()
print(f"{no_sum_to_one} of {n_records} records do not sum to one.")
```
A lot of the records to do not sum to one as they should. We should either drop these observations from the analysis or fit models using probabilistic (e.g. bootstrap) methods.
Another data quality issue we could check is whether or not $OneBidDollars + EffCompDollars = CompetedDollars$
```
competed_dollars = data1['CompetedDollars']
one_bid_dollars = data1['OneBidDollars']
eff_comp_dollars = data1['EffCompDollars']
actual_competed_dollars = one_bid_dollars + eff_comp_dollars
dif = competed_dollars - actual_competed_dollars
dif.abs().round().describe()
```
The above finding is also alarming, suggesting that there are serious data quality issues at play here. Nevertheless, we'll proceed to fit a model for documentation purposes.
# Panel OLS
Despite the data quality issues identified above, here we'll fit a panel least sqaures model using fixed effects for both time and entities. This recreates the original analysis.
```
design_df = df.copy(deep = True)
design_df['NotCompRate_shift'] = design_df['NotCompRate'].shift(1)
design_df = design_df[design_df['FiscalYear'] != 2014]
year = pd.Categorical(design_df.FiscalYear)
design_df = design_df.set_index(['unique_name','FiscalYear'])
design_df['FiscalYear'] = year
exog_cols = ['SdbRate','OneBidRate','NotCompRate_shift']
exog = sm.add_constant(design_df[exog_cols])
model = PanelOLS(design_df.EffCompRate,
exog,
entity_effects = True,
time_effects = True).fit()
print(model)
```
## Interpretation
Above, we notice a few issues:
1. We have 161 entities whereas the original analysis had 165. We're not sure what those extra 4 observations are.
2. All of our coefficient estimates differ from those originally found. That could be due to either the 4 missing observations, but it could also be due to the fixed effects we've included.
# Data Validity Simulations
Here we'll randomly generate data such that the OneBidRate does sum to one when added to the EffCompRate. We'll do this a couple thousand times, generating the same model as specified above. For each fit, we'll save the coeffecient parameter estimates and then perform some summary statistics on them to get a measure of the vagaries introduced by our poor data.
```
exog_cols = ['SdbRate','OneBidRate','NotCompRate_shift']
sim_df = design_df.copy(deep = True)
sum_to_one = sim_df['OneBidRate'] + sim_df['EffCompRate']
sim_df['sums_to_one'] = sum_to_one.round(decimals = 1) >= 1
sim_df = sim_df.drop(labels = ['FiscalYear'], axis = 1)
#reset index due to duplciate contracting_agency and fiscal year combinations
#(bad for resetting individual cell values)
sim_df = sim_df.reset_index()
indices_to_adjust = sim_df.index[sim_df['sums_to_one'] == False].tolist()
params = []
for i in range(1000):
#create deep copy of sim_df so we're not reassigning values in place
temp_df = sim_df.copy(deep = True)
for ix in indices_to_adjust:
#get the values to adjust
one_bid_rate = sim_df.at[ix, 'OneBidRate']
eff_comp_rate = sim_df.at[ix, 'EffCompRate']
#get get the difference from one. This is the amount we'll randomly fill
diff = 1 - (one_bid_rate + eff_comp_rate)
#randomly choose a number between 0 and the diff
rand = random.uniform(0,diff)
diff_rand = diff - rand
#reassign these random filler values to the temp_df. This makes the two sum to one now
temp_df.at[ix, 'OneBidRate'] += rand
temp_df.at[ix, 'EffCompRate'] += diff_rand
#restructure temp_df so we can fit a model
year = pd.Categorical(temp_df.FiscalYear)
temp_df = temp_df.set_index(['unique_name','FiscalYear'])
temp_df['FiscalYear'] = year
temp_df['NotCompRate_shift'] = temp_df['NotCompRate'].shift(1)
temp_df = temp_df[temp_df['FiscalYear'] != 2014]
#fit the model
exog = sm.add_constant(temp_df[exog_cols])
temp_model = PanelOLS(temp_df.EffCompRate,
exog,
entity_effects = True,
time_effects = True).fit()
temp_params = temp_model.params
params.append(pd.DataFrame(temp_params).transpose())
params_df = pd.concat(params)
#get original model params
original_params = pd.DataFrame(model.params).transpose()
```
Now that we've got the estimated coefficients from 1,000 different simulations, we'll construct some 95% confidence intervals for each one and then see if the model's orginal parameteres fall within these ranges.
```
def get_ci(data, confidence = 0.95):
n = len(data)
m = mean(data)
std_err = sem(data)
h = std_err * t.ppf((1 + confidence) / 2, n - 1)
start = m - h
end = m + h
return start, end
print(original_params, end = "\n\n")
for col in params_df.columns:
col_values = params_df[col]
start, end = get_ci(col_values)
print(f"The 95% CI for {col} is {start:.4f} to {end:.4f}")
```
As we can see, the original estimates do not fall within the 95% CIs. Although the signs are the same, we could have surmised as much without ever fitting a model given the definitions of each variable.
# Summary
There are three main issues with this analysis:
1. Inability to replicate the original analysis<br>
>We have been unable to replicate the analysis in both the estimated coeffecients and the number of observations.
We've also been assuming that the model controlled for entity and time fixed effects. Theses issues need to be clarified.
1. Invalid Data<br>
>The simulations above demonstrated the bias introduced by the invalid OneBidRate and EffCompRate values. A recommended solution would be to drop records where the OneBidRate and EffCompRate do not sum to one. However, this might vastly reduce the number of observations and then undermine the analysis.
2. Model Specification<br>
>A common denominator for the IV (EffCompRate) and one of the DVs (OneBidRate) suggests that we'd be able to re-write the regression equation with the common denomonitor factored out as a separate term on the right-hand side of the equation. If we were to do this, we could likely demonstrate that there is a correlation between OneBidRate and the error term, as the denominator common to both this IV and the DV has been absorbed by the error term. This essentially renders the relationship between that IV and the DV as definitionally spurious.
|
github_jupyter
|
import pandas as pd
import numpy as np
import seaborn as sns
import statsmodels
import linearmodels
import matplotlib as plt
import xlrd
import scipy
from scipy.stats import sem, t
from scipy import mean
import os
import statsmodels.api as sm
from pandas.plotting import autocorrelation_plot
import pandas_profiling
from linearmodels.panel import PanelOLS
import random
%matplotlib inline
# read data file, convert to csv
data = pd.read_excel('data/fpds_rates.xlsx')
data.head()
data.columns
data.shape
data['unique_name'] = data['department_name'] + '-' + data['contracting_agency_name']
data['unique_name'] = data['unique_name'].fillna('agency wide')
data.head()
#drop agency-wide observations
agency_wide = False
if agency_wide:
clean_data = data
else:
print("Dropping agency wide observations")
clean_data = data[data['unique_name'] != 'agency wide']
# some records have not obligated dollars, so I'll drop those as it does not make sense to include agencies who
# did not participate in any contracting actions in a regression measuring the dollar impact of
#contracting actions. Also certain rates used in the regression use TotalDollars in the denominator
clean_data = clean_data[clean_data['TotalDollars']>0]
# remove negatives
clean_data= clean_data[clean_data['OneBidRate']>0]
clean_data = clean_data[clean_data['SdbRate']>0]
clean_data = clean_data[clean_data['NotCompRate']>0]
clean_data = clean_data[clean_data['EffCompRate']>0]
#ensure each unique_name contains 5 years of data
n_years_n_obs_map = {k:0 for k in range(1,6)}
for name in clean_data['unique_name'].unique():
if name is np.nan:
continue
n_years = clean_data[clean_data['unique_name'] == name].shape[0]
if n_years == 0:
print(name)
try:
n_years_n_obs_map[n_years] += 1
except KeyError:
n_years_n_obs_map[n_years] = 1
# this is the number of observations the OLS model should report
sum(n_years_n_obs_map.values())
df = clean_data[['SdbRate', 'OneBidRate', 'NotCompRate', 'EffCompRate', 'FiscalYear', 'unique_name']]
pandas_profiling.ProfileReport(df)
sum_to_one = df['OneBidRate'] + df['EffCompRate']
sns.distplot(sum_to_one)
sum_to_one.round(decimals = 1).value_counts()
no_sum_to_one = sum_to_one.round(decimals = 1).value_counts().sum() - 662
n_records = sum_to_one.round(decimals = 1).value_counts().sum()
print(f"{no_sum_to_one} of {n_records} records do not sum to one.")
competed_dollars = data1['CompetedDollars']
one_bid_dollars = data1['OneBidDollars']
eff_comp_dollars = data1['EffCompDollars']
actual_competed_dollars = one_bid_dollars + eff_comp_dollars
dif = competed_dollars - actual_competed_dollars
dif.abs().round().describe()
design_df = df.copy(deep = True)
design_df['NotCompRate_shift'] = design_df['NotCompRate'].shift(1)
design_df = design_df[design_df['FiscalYear'] != 2014]
year = pd.Categorical(design_df.FiscalYear)
design_df = design_df.set_index(['unique_name','FiscalYear'])
design_df['FiscalYear'] = year
exog_cols = ['SdbRate','OneBidRate','NotCompRate_shift']
exog = sm.add_constant(design_df[exog_cols])
model = PanelOLS(design_df.EffCompRate,
exog,
entity_effects = True,
time_effects = True).fit()
print(model)
exog_cols = ['SdbRate','OneBidRate','NotCompRate_shift']
sim_df = design_df.copy(deep = True)
sum_to_one = sim_df['OneBidRate'] + sim_df['EffCompRate']
sim_df['sums_to_one'] = sum_to_one.round(decimals = 1) >= 1
sim_df = sim_df.drop(labels = ['FiscalYear'], axis = 1)
#reset index due to duplciate contracting_agency and fiscal year combinations
#(bad for resetting individual cell values)
sim_df = sim_df.reset_index()
indices_to_adjust = sim_df.index[sim_df['sums_to_one'] == False].tolist()
params = []
for i in range(1000):
#create deep copy of sim_df so we're not reassigning values in place
temp_df = sim_df.copy(deep = True)
for ix in indices_to_adjust:
#get the values to adjust
one_bid_rate = sim_df.at[ix, 'OneBidRate']
eff_comp_rate = sim_df.at[ix, 'EffCompRate']
#get get the difference from one. This is the amount we'll randomly fill
diff = 1 - (one_bid_rate + eff_comp_rate)
#randomly choose a number between 0 and the diff
rand = random.uniform(0,diff)
diff_rand = diff - rand
#reassign these random filler values to the temp_df. This makes the two sum to one now
temp_df.at[ix, 'OneBidRate'] += rand
temp_df.at[ix, 'EffCompRate'] += diff_rand
#restructure temp_df so we can fit a model
year = pd.Categorical(temp_df.FiscalYear)
temp_df = temp_df.set_index(['unique_name','FiscalYear'])
temp_df['FiscalYear'] = year
temp_df['NotCompRate_shift'] = temp_df['NotCompRate'].shift(1)
temp_df = temp_df[temp_df['FiscalYear'] != 2014]
#fit the model
exog = sm.add_constant(temp_df[exog_cols])
temp_model = PanelOLS(temp_df.EffCompRate,
exog,
entity_effects = True,
time_effects = True).fit()
temp_params = temp_model.params
params.append(pd.DataFrame(temp_params).transpose())
params_df = pd.concat(params)
#get original model params
original_params = pd.DataFrame(model.params).transpose()
def get_ci(data, confidence = 0.95):
n = len(data)
m = mean(data)
std_err = sem(data)
h = std_err * t.ppf((1 + confidence) / 2, n - 1)
start = m - h
end = m + h
return start, end
print(original_params, end = "\n\n")
for col in params_df.columns:
col_values = params_df[col]
start, end = get_ci(col_values)
print(f"The 95% CI for {col} is {start:.4f} to {end:.4f}")
| 0.324128 | 0.816918 |
## Dependencies
```
import json, glob
from tweet_utility_scripts import *
from tweet_utility_preprocess_roberta_scripts_aux import *
from transformers import TFRobertaModel, RobertaConfig
from tokenizers import ByteLevelBPETokenizer
from tensorflow.keras import layers
from tensorflow.keras.models import Model
```
# Load data
```
test = pd.read_csv('/kaggle/input/tweet-sentiment-extraction/test.csv')
print('Test samples: %s' % len(test))
display(test.head())
```
# Model parameters
```
input_base_path = '/kaggle/input/260-robertabase/'
with open(input_base_path + 'config.json') as json_file:
config = json.load(json_file)
config
# vocab_path = input_base_path + 'vocab.json'
# merges_path = input_base_path + 'merges.txt'
base_path = '/kaggle/input/qa-transformers/roberta/'
vocab_path = base_path + 'roberta-base-vocab.json'
merges_path = base_path + 'roberta-base-merges.txt'
config['base_model_path'] = base_path + 'roberta-base-tf_model.h5'
config['config_path'] = base_path + 'roberta-base-config.json'
model_path_list = glob.glob(input_base_path + '*.h5')
model_path_list.sort()
print('Models to predict:')
print(*model_path_list, sep='\n')
```
# Tokenizer
```
tokenizer = ByteLevelBPETokenizer(vocab_file=vocab_path, merges_file=merges_path,
lowercase=True, add_prefix_space=True)
```
# Pre process
```
test['text'].fillna('', inplace=True)
test['text'] = test['text'].apply(lambda x: x.lower())
test['text'] = test['text'].apply(lambda x: x.strip())
x_test, x_test_aux, x_test_aux_2 = get_data_test(test, tokenizer, config['MAX_LEN'], preprocess_fn=preprocess_roberta_test)
```
# Model
```
module_config = RobertaConfig.from_pretrained(config['config_path'], output_hidden_states=True)
def model_fn(MAX_LEN):
input_ids = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='input_ids')
attention_mask = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='attention_mask')
base_model = TFRobertaModel.from_pretrained(config['base_model_path'], config=module_config, name='base_model')
_, _, hidden_states = base_model({'input_ids': input_ids, 'attention_mask': attention_mask})
h11 = hidden_states[-2]
logits = layers.Dense(3, use_bias=False, name='qa_outputs')(h11)
start_logits, end_logits, mask_logits = tf.split(logits, 3, axis=-1)
start_logits = tf.squeeze(start_logits, axis=-1, name='y_start')
end_logits = tf.squeeze(end_logits, axis=-1, name='y_end')
mask_logits = tf.squeeze(mask_logits, axis=-1, name='y_mask')
model = Model(inputs=[input_ids, attention_mask], outputs=[start_logits, end_logits, mask_logits])
return model
```
# Make predictions
```
NUM_TEST_IMAGES = len(test)
test_start_preds = np.zeros((NUM_TEST_IMAGES, config['MAX_LEN']))
test_end_preds = np.zeros((NUM_TEST_IMAGES, config['MAX_LEN']))
for model_path in model_path_list:
print(model_path)
model = model_fn(config['MAX_LEN'])
model.load_weights(model_path)
test_preds = model.predict(get_test_dataset(x_test, config['BATCH_SIZE']))
test_start_preds += test_preds[0]
test_end_preds += test_preds[1]
```
# Post process
```
test['start'] = test_start_preds.argmax(axis=-1)
test['end'] = test_end_preds.argmax(axis=-1)
test['selected_text'] = test.apply(lambda x: decode(x['start'], x['end'], x['text'], config['question_size'], tokenizer), axis=1)
# Post-process
test["selected_text"] = test.apply(lambda x: ' '.join([word for word in x['selected_text'].split() if word in x['text'].split()]), axis=1)
test['selected_text'] = test.apply(lambda x: x['text'] if (x['selected_text'] == '') else x['selected_text'], axis=1)
test['selected_text'].fillna(test['text'], inplace=True)
```
# Visualize predictions
```
test['text_len'] = test['text'].apply(lambda x : len(x))
test['label_len'] = test['selected_text'].apply(lambda x : len(x))
test['text_wordCnt'] = test['text'].apply(lambda x : len(x.split(' ')))
test['label_wordCnt'] = test['selected_text'].apply(lambda x : len(x.split(' ')))
test['text_tokenCnt'] = test['text'].apply(lambda x : len(tokenizer.encode(x).ids))
test['label_tokenCnt'] = test['selected_text'].apply(lambda x : len(tokenizer.encode(x).ids))
test['jaccard'] = test.apply(lambda x: jaccard(x['text'], x['selected_text']), axis=1)
display(test.head(10))
display(test.describe())
```
# Test set predictions
```
submission = pd.read_csv('/kaggle/input/tweet-sentiment-extraction/sample_submission.csv')
submission['selected_text'] = test['selected_text']
submission.to_csv('submission.csv', index=False)
submission.head(10)
```
|
github_jupyter
|
import json, glob
from tweet_utility_scripts import *
from tweet_utility_preprocess_roberta_scripts_aux import *
from transformers import TFRobertaModel, RobertaConfig
from tokenizers import ByteLevelBPETokenizer
from tensorflow.keras import layers
from tensorflow.keras.models import Model
test = pd.read_csv('/kaggle/input/tweet-sentiment-extraction/test.csv')
print('Test samples: %s' % len(test))
display(test.head())
input_base_path = '/kaggle/input/260-robertabase/'
with open(input_base_path + 'config.json') as json_file:
config = json.load(json_file)
config
# vocab_path = input_base_path + 'vocab.json'
# merges_path = input_base_path + 'merges.txt'
base_path = '/kaggle/input/qa-transformers/roberta/'
vocab_path = base_path + 'roberta-base-vocab.json'
merges_path = base_path + 'roberta-base-merges.txt'
config['base_model_path'] = base_path + 'roberta-base-tf_model.h5'
config['config_path'] = base_path + 'roberta-base-config.json'
model_path_list = glob.glob(input_base_path + '*.h5')
model_path_list.sort()
print('Models to predict:')
print(*model_path_list, sep='\n')
tokenizer = ByteLevelBPETokenizer(vocab_file=vocab_path, merges_file=merges_path,
lowercase=True, add_prefix_space=True)
test['text'].fillna('', inplace=True)
test['text'] = test['text'].apply(lambda x: x.lower())
test['text'] = test['text'].apply(lambda x: x.strip())
x_test, x_test_aux, x_test_aux_2 = get_data_test(test, tokenizer, config['MAX_LEN'], preprocess_fn=preprocess_roberta_test)
module_config = RobertaConfig.from_pretrained(config['config_path'], output_hidden_states=True)
def model_fn(MAX_LEN):
input_ids = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='input_ids')
attention_mask = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='attention_mask')
base_model = TFRobertaModel.from_pretrained(config['base_model_path'], config=module_config, name='base_model')
_, _, hidden_states = base_model({'input_ids': input_ids, 'attention_mask': attention_mask})
h11 = hidden_states[-2]
logits = layers.Dense(3, use_bias=False, name='qa_outputs')(h11)
start_logits, end_logits, mask_logits = tf.split(logits, 3, axis=-1)
start_logits = tf.squeeze(start_logits, axis=-1, name='y_start')
end_logits = tf.squeeze(end_logits, axis=-1, name='y_end')
mask_logits = tf.squeeze(mask_logits, axis=-1, name='y_mask')
model = Model(inputs=[input_ids, attention_mask], outputs=[start_logits, end_logits, mask_logits])
return model
NUM_TEST_IMAGES = len(test)
test_start_preds = np.zeros((NUM_TEST_IMAGES, config['MAX_LEN']))
test_end_preds = np.zeros((NUM_TEST_IMAGES, config['MAX_LEN']))
for model_path in model_path_list:
print(model_path)
model = model_fn(config['MAX_LEN'])
model.load_weights(model_path)
test_preds = model.predict(get_test_dataset(x_test, config['BATCH_SIZE']))
test_start_preds += test_preds[0]
test_end_preds += test_preds[1]
test['start'] = test_start_preds.argmax(axis=-1)
test['end'] = test_end_preds.argmax(axis=-1)
test['selected_text'] = test.apply(lambda x: decode(x['start'], x['end'], x['text'], config['question_size'], tokenizer), axis=1)
# Post-process
test["selected_text"] = test.apply(lambda x: ' '.join([word for word in x['selected_text'].split() if word in x['text'].split()]), axis=1)
test['selected_text'] = test.apply(lambda x: x['text'] if (x['selected_text'] == '') else x['selected_text'], axis=1)
test['selected_text'].fillna(test['text'], inplace=True)
test['text_len'] = test['text'].apply(lambda x : len(x))
test['label_len'] = test['selected_text'].apply(lambda x : len(x))
test['text_wordCnt'] = test['text'].apply(lambda x : len(x.split(' ')))
test['label_wordCnt'] = test['selected_text'].apply(lambda x : len(x.split(' ')))
test['text_tokenCnt'] = test['text'].apply(lambda x : len(tokenizer.encode(x).ids))
test['label_tokenCnt'] = test['selected_text'].apply(lambda x : len(tokenizer.encode(x).ids))
test['jaccard'] = test.apply(lambda x: jaccard(x['text'], x['selected_text']), axis=1)
display(test.head(10))
display(test.describe())
submission = pd.read_csv('/kaggle/input/tweet-sentiment-extraction/sample_submission.csv')
submission['selected_text'] = test['selected_text']
submission.to_csv('submission.csv', index=False)
submission.head(10)
| 0.394084 | 0.33689 |
# Parameter Estimation (and Hypothesis testing... which is just parameter estimation)
In our `MagicCoin` example `n` was an example of a parameter. It's some important value for our understanding of how things work that we don't know. By using probability and data we can come up with ways to estimate what these parameters might be.
In the rest of this tutorial we'll focus on an admittedly boring case of trying to estimate the rate that a product sells. The rate being the parameter we're trying to estimate.
We'll be using a simulated product and customer data...
```
from context import src
from src import customer as cust
from src import product as prod
from src import experiment as exp
import numpy as np
import pandas as pd
import seaborn as sns
from scipy import stats
```
Let's start by creating a new product. Our `Product` class is amazingly simple. It just takes a `name`, a `price` and a `quality`. Of course `quality` of a product is not so easy, but in this case we can just set it to whatever we want to simulate the average reviews are.
We'll start with a toothbrush:
<img src="./images/tooth_brush.jpg" alt="A toothbrush" style="width: 300px;"/>
```
toothbrush = prod.Product(name="alright brush",
price=4.99,
quality=3.9)
```
It's not the most amazing toothbrush, but let's try to sell it to somebody. Our `Customer` class can generate a random customer for us that has a certain price threshold and quality threshold that we don't know, and varies from customer to customer:
```
person_a = cust.Customer.get_random()
```
Next we can show our customer the product using the `will_purchase` method and we'll get a bool back telling us whether or not the decided to purchase that product:
```
person_a.will_purchase(toothbrush)
```
Like the `MagicCoin` we don't really know what's going on inside of our customer. This singular observation doesn't tell us very much about how our coin behaves. The best way to handle this is to run some sort of an `Experiment`!
We can create an experiment using our `Experiment` class which will help us collect data on customers that we've shown our `toothbrush` to!
```
toothbrush_test = exp.Experiment(toothbrush)
```
Now we can use our experiment to show this `toothbrush` to a bunch of customers and see how many will purchase it!
```
test_result = toothbrush_test.show_to_customers(20)
```
Now we can see how our test went...
```
test_result.purchased
```
With the `MagicCoin` we understood exactly how it worked so we didn't need a model to represent how we think the `ModelCoin` worked.
Even though the interaction between the `Customer` and the `Product` is more complicated that simply a probability that a `Customer` will purchase a `Product` from our view the best way to model this is to make the simplifying assumption that each `Product` sells on at a particular rate.
If you've worked in ecommerce or any similar conversion focused industry it might seem obvious that we want to estimate "the rate that a product sells", but it's good to remember that this is really an application of probability. If we knew everything about each customer and each product and how they interact we would know exactly how a product sells, there would be no rate involved at all!
So if you wanted to estimate a rate at what this product sells, what would be your first estimate? Most people intuitively might estimate:
```
sold = sum(test_result.purchased)
total = len(test_result.purchased)
rate_est = sold/total
print("{0} sold out of {1} total so our estimate is {2}".format(sold,total,rate_est))
```
Making our `rate_est` as `sold/total` seems like a pretty good idea. But how can we show this is the best estimate, and are there other good estimate?
For example what about `rate_est + 0.05` or `rate_est - 0.05`? Are these good estimate? certainly they explain the data pretty well. How would we compare these estimates?
One way we can do this is to use the Binomial distribution. The Binomial distribution will tell us exactly how likely data would be given this was the rate of a product selling. Let's look at the probability of the data for each of these alternate options:
```
bin_est = stats.binom(n=total,p=rate_est)
bin_est.pmf(sold)
bin_est_bit_smaller = stats.binom(n=total,p=rate_est-0.05)
bin_est_bit_smaller.pmf(sold)
bin_est_bit_bigger = stats.binom(n=total,p=rate_est+0.05)
bin_est_bit_bigger.pmf(sold)
```
In both cases we can see that the slightly different estimates are a bit more surprised by the data that we observed. This means that they are not quite a good of an explaination of the data.. but that doesn't mean they're *wrong*.
If you flipped a coin 3 times and got 1 head, you wouldn't assume the probability of heads is $\frac{1}{3}$ the coin being fair is still pretty likely and you know that most coins tend to be fair so the belief that the probability of heads is $\frac{1}{2}$
Rather than just worry about which estimate for the rate is the best, it might be a good idea to look at how strongly we believe in multi estimates. We could start by looking at estimates in intervals of every 0.05 and apply the same logic using the Binomial Distribution we did before. We can use `numpy` to quickly do this in a vectorized way:
```
est_dists = stats.binom(total,np.arange(0,1,0.1))
sns.lineplot(x=np.arange(0,1,0.1),
y=est_dists.pmf(sold))
```
Here we can see that if we look at each possibility between 0 and 1 incrementing by 0.05 we have some sort of distribution forming.
We can see that this continues to smooth as we shrink our increment size:
```
est_dists = stats.binom(total,np.arange(0,1,0.05))
sns.lineplot(x=np.arange(0,1,0.05),
y=est_dists.pmf(sold))
est_dists = stats.binom(total,np.arange(0,1,0.01))
sns.lineplot(x=np.arange(0,1,0.01),
y=est_dists.pmf(sold))
```
### The Beta Distribution
The distribution we're converging on is a very useful distribution call the *Beta distribution*. It differs from our plot above in two ways: First it is a continous distribution meaning it accounts for the infinitely many possible rates for what we've observed. The second is that it makes it so that if we sum up over all those possible points (technically integrate) the result is exactly 1. This let's us talk about probabilities for different values.
The Beta distribution takes two paramters $\alpha$ the number of successes or `True` values we oberved and $\beta$ the number failures or `False` values. Note that this is bit different than the Binomial where `n` = `alpha+beta`
Here's a plot of what this distribution looks like for our cases of `alpha = sold` and `beta = total - sold`
**note:** because the Beta distribution is continuous we'll use the `.pdf` method rather than the `.pmf` method.
```
alpha = sold
beta = total - sold
est_beta_dist = stats.beta(alpha,beta)
xs = np.arange(0,1,0.01)
sns.lineplot(x=xs,
y=est_beta_dist.pdf(xs)).set(xlabel='rate estimate',
ylabel='density')
```
Now we have a plot that shows the distribution of how strongly we believe in various possible rates for our `toothbrush` to sell to a custuomer.
The power of having a distribution like this is that we use it to ask questions about our beliefs.
For example: suppose the director of marketing came in an asked if you had a product she could feature on the company webiste. The catch is she wants a product that will have a rate of *at least* 0.4.
Looking at the distribution of our beliefs it's certainly possible that it could be at least 0.4, but how confindent are we?
We can answer this question a few ways. One way would be to use calculus to integrate between 0.4 and 1.0. But plenty of people are a bit nervous about doing calculus, and it turns out that integration gets tricky no matter what, so what we can also do is simply sample from this distribution:
```
n_samples = 10000
rate_est_samples = np.random.beta(a=sold,b=(total-sold),size=n_samples)
```
Here we have a vector of samples of our rate estimate and we can use them to answer questions like the directors:
```
sum(rate_est_samples > 0.4)/n_samples
```
Notice that if we compute the `mean` of these `rate_est_samples` get a result very similar to `sold/total`.
```
rate_est_samples.mean()
```
This is no coincidence. It turns out that the analytical *expectation* (or mean) of the Beta distribution is
$$E[Beta(\alpha,\beta)] = \frac{\alpha}{\alpha + \beta}$$
Which is the same as the successes over the total. If we sample more from out distribution the answers to the questions we ask of those samples are going to be closer and to the answer we would get if we perform the correct mathematical operations on our function. This is super important because it means we can use sampling as a subsitute for increasingly complicated integrals.
So if you aren't super comfortable with the math, definitely feel free to just use sample... and even if you *are* comfortable with the math, pretty soon you'll need to rely on sampling techniques anyway so you should start playing around with sampling in your analysis
## Hypothesis testing: comparing two products
Hypothesis testing is the essential parts of statistics. It's fancy way of saying you have some hypothesis about the data and you want to test out how likely that hypothesis is. In fact, we've already done hypthesis testing: we asked about the hypothesis of each possible rate that could explain the data, and also looked at the hypothesis that that rate is greater than 0.4
But typically when people think of hypothesis tests they think of comparing some things like:
- do patients that recieve the treatment get healthy faster
- is the new version of the website have more sign-ups than the old
- does saying "Florida" make people think of being old and then walk slower than people who don't hear it.
So to explore hypothesis testing we're going to have to have something to compare our `tooth brush` with, which is our `luxury_toothbrush`
<img src="./images/luxury_toothbrush.jpg" alt="A luxury toothbrush" style="width: 300px;"/>
```
luxury_toothbrush = prod.Product(name="luxury toothbrush",
price=7.99,
quality=4.8)
```
This is a much better toothbrush, but it also costs more, how well will it do? And more important, how well will it do compared to just our regular `toothbrush`?
To look at this let's set up a new experiment, this time one where we can compare both toothbrushes to each other:
```
toothbrush_ab_test = exp.Experiment(toothbrush,luxury_toothbrush)
```
We're calling this an AB test because it's similar in nature to an AB test for websites. We're going to show each variant, the original `toothbrush` and the `luxury_toothbrush` to different groups of people and see which one does better.
```
n_ab_test_samples = 30
ab_test_results = toothbrush_ab_test.show_to_customers(n_ab_test_samples)
```
We now showed each toothbrush to 30 different people here's the results for each tooth brush
For `toothbrush` we get:
```
ab_test_results[['a_purchased']]
```
And for `luxury_toothbrush`:
```
ab_test_results[['b_purchased']]
a_sold = sum(ab_test_results.a_purchased)
b_sold = sum(ab_test_results.b_purchased)
print("A sold {} (rate {:.2f}) and B sold {} (rate {:.2f})".format(a_sold,
float(a_sold/n_ab_test_samples),
b_sold,
float(b_sold/n_ab_test_samples)))
```
So which toothbrush is beter? And if you think you're sure rerun these cells a few times and you're likely to get different results!
To better understand what's happening here look at our parameter estimates for each tooth brush
```
alpha_a = sum(ab_test_results.a_purchased)
beta_a = n_ab_test_samples - alpha_a
a_beta_dist = stats.beta(alpha_a,beta_a)
alpha_b = sum(ab_test_results.b_purchased)
beta_b = n_ab_test_samples - alpha_b
b_beta_dist = stats.beta(alpha_b,beta_b)
rates = np.arange(0,0.5,0.005)
plot_df = pd.DataFrame({
'density':np.concatenate((a_beta_dist.pdf(rates),
b_beta_dist.pdf(rates))),
'rate': np.concatenate((rates,rates)),
'group':['regular']*len(rates) + ['luxury']*len(rates)
})
sns.lineplot(x='rate',
y='density',
hue='group',
data=plot_df)
```
We can se that we have two estimates now that might tell us very different things. If you look at your neighbors' plots (if you're in the live workshop) you might notice a very different plot based on which customers looked at the times.
this means that we don't have enough data to tell which distribution is different. What if we had more customers?
Let's try 100 customers for each brush and see what we learn:
```
n_ab_test_2_samples = 100
ab_test_2 = toothbrush_ab_test.show_to_customers(n_ab_test_2_samples)
#notice that we're over writing the variables here so be careful of what order you run these cells in!
alpha_a = sum(ab_test_2.a_purchased)
beta_a = n_ab_test_2_samples - alpha_a
a_beta_dist = stats.beta(alpha_a,beta_a)
alpha_b = sum(ab_test_2.b_purchased)
beta_b = n_ab_test_2_samples - alpha_b
b_beta_dist = stats.beta(alpha_b,beta_b)
rates = np.arange(0,0.5,0.001)
plot_df = pd.DataFrame({
'density':np.concatenate((a_beta_dist.pdf(rates),
b_beta_dist.pdf(rates))),
'rate': np.concatenate((rates,rates)),
'group':['regular']*len(rates) + ['luxury']*len(rates)
})
sns.lineplot(x='rate',
y='density',
hue='group',
data=plot_df)
```
Now we're getting much better results! There's still a lot of uncertainity around exactly what rate each toothbrush sells, but we can clearly see that our estimates for `luxury_toothbrush` are much lower than they are for regular. Even if you look at your neighbors' plots this time, they should look much more similar.
But we might want to quantify *exactly* how certain we are that the `toothbrush` is doing better than the `luxury_toothbrush`. The best way to do that is with sampling:
```
total_samples = 10000
regular_samples = np.random.beta(a=alpha_a,b=beta_a,size=total_samples)
luxury_samples = np.random.beta(a=alpha_b,b=beta_b,size=total_samples)
```
This will tell the probability that the regular toothbrushes sell better than luxury:
```
sum(regular_samples >luxury_samples)/total_samples
```
As we can see we are almost certain that this is the case, even though we have a lot different beliefs about how well the different toothbrushes convert.
Just to be clear, we just did a hypothesis test! The **hypothesis** was that variant A was better than variant B, and the result of that test was the probability we got from our simulation. The great thing about this test is we don't have a "p value" we have the actual probability that the `toothbrush` is superior based on our simulation.
## Homework
Here are some things you can experiment around with on your own to get a better sense of how hypothesis tests work!
We can see that the `luxury_toothbrush` sells at a lower rate than the `toothbrush` but there are still many questions we can ask. How *much* better do we think `toothbrush` is than `luxury_toothbrush` in terms of how many times better it converts?
In [this post on the Count Bayesie blog](https://www.countbayesie.com/blog/2015/4/25/bayesian-ab-testing) I go over ways that you can add a prior probability (and in [this post](https://www.countbayesie.com/blog/2015/2/18/hans-solo-and-bayesian-priors) talk a bit about what prior probabilities are). Experiment with using a reasonable prior probability for the different purchase rates and see how much sooner (or longer) it takes to conclude that `luxury_toothbrush` is inferior.
`luxury_toothbrush` sells at a lower rate, but it also is more expensive. When we take into account the price difference with the sales difference is the plain old `toothbrush` the better product? or would we be better off selling less of the better tooth brush?
|
github_jupyter
|
from context import src
from src import customer as cust
from src import product as prod
from src import experiment as exp
import numpy as np
import pandas as pd
import seaborn as sns
from scipy import stats
toothbrush = prod.Product(name="alright brush",
price=4.99,
quality=3.9)
person_a = cust.Customer.get_random()
person_a.will_purchase(toothbrush)
toothbrush_test = exp.Experiment(toothbrush)
test_result = toothbrush_test.show_to_customers(20)
test_result.purchased
sold = sum(test_result.purchased)
total = len(test_result.purchased)
rate_est = sold/total
print("{0} sold out of {1} total so our estimate is {2}".format(sold,total,rate_est))
bin_est = stats.binom(n=total,p=rate_est)
bin_est.pmf(sold)
bin_est_bit_smaller = stats.binom(n=total,p=rate_est-0.05)
bin_est_bit_smaller.pmf(sold)
bin_est_bit_bigger = stats.binom(n=total,p=rate_est+0.05)
bin_est_bit_bigger.pmf(sold)
est_dists = stats.binom(total,np.arange(0,1,0.1))
sns.lineplot(x=np.arange(0,1,0.1),
y=est_dists.pmf(sold))
est_dists = stats.binom(total,np.arange(0,1,0.05))
sns.lineplot(x=np.arange(0,1,0.05),
y=est_dists.pmf(sold))
est_dists = stats.binom(total,np.arange(0,1,0.01))
sns.lineplot(x=np.arange(0,1,0.01),
y=est_dists.pmf(sold))
alpha = sold
beta = total - sold
est_beta_dist = stats.beta(alpha,beta)
xs = np.arange(0,1,0.01)
sns.lineplot(x=xs,
y=est_beta_dist.pdf(xs)).set(xlabel='rate estimate',
ylabel='density')
n_samples = 10000
rate_est_samples = np.random.beta(a=sold,b=(total-sold),size=n_samples)
sum(rate_est_samples > 0.4)/n_samples
rate_est_samples.mean()
luxury_toothbrush = prod.Product(name="luxury toothbrush",
price=7.99,
quality=4.8)
toothbrush_ab_test = exp.Experiment(toothbrush,luxury_toothbrush)
n_ab_test_samples = 30
ab_test_results = toothbrush_ab_test.show_to_customers(n_ab_test_samples)
ab_test_results[['a_purchased']]
ab_test_results[['b_purchased']]
a_sold = sum(ab_test_results.a_purchased)
b_sold = sum(ab_test_results.b_purchased)
print("A sold {} (rate {:.2f}) and B sold {} (rate {:.2f})".format(a_sold,
float(a_sold/n_ab_test_samples),
b_sold,
float(b_sold/n_ab_test_samples)))
alpha_a = sum(ab_test_results.a_purchased)
beta_a = n_ab_test_samples - alpha_a
a_beta_dist = stats.beta(alpha_a,beta_a)
alpha_b = sum(ab_test_results.b_purchased)
beta_b = n_ab_test_samples - alpha_b
b_beta_dist = stats.beta(alpha_b,beta_b)
rates = np.arange(0,0.5,0.005)
plot_df = pd.DataFrame({
'density':np.concatenate((a_beta_dist.pdf(rates),
b_beta_dist.pdf(rates))),
'rate': np.concatenate((rates,rates)),
'group':['regular']*len(rates) + ['luxury']*len(rates)
})
sns.lineplot(x='rate',
y='density',
hue='group',
data=plot_df)
n_ab_test_2_samples = 100
ab_test_2 = toothbrush_ab_test.show_to_customers(n_ab_test_2_samples)
#notice that we're over writing the variables here so be careful of what order you run these cells in!
alpha_a = sum(ab_test_2.a_purchased)
beta_a = n_ab_test_2_samples - alpha_a
a_beta_dist = stats.beta(alpha_a,beta_a)
alpha_b = sum(ab_test_2.b_purchased)
beta_b = n_ab_test_2_samples - alpha_b
b_beta_dist = stats.beta(alpha_b,beta_b)
rates = np.arange(0,0.5,0.001)
plot_df = pd.DataFrame({
'density':np.concatenate((a_beta_dist.pdf(rates),
b_beta_dist.pdf(rates))),
'rate': np.concatenate((rates,rates)),
'group':['regular']*len(rates) + ['luxury']*len(rates)
})
sns.lineplot(x='rate',
y='density',
hue='group',
data=plot_df)
total_samples = 10000
regular_samples = np.random.beta(a=alpha_a,b=beta_a,size=total_samples)
luxury_samples = np.random.beta(a=alpha_b,b=beta_b,size=total_samples)
sum(regular_samples >luxury_samples)/total_samples
| 0.410402 | 0.993819 |
```
import base64
import io
import json
import os
import sys
import cv2 as cv
import jsonpickle
import numpy as np
import requests
from flask import Flask, Response, jsonify, request
from PIL import Image, ImageFont, ImageDraw
from titlecase import titlecase
def analyze_img(rdata):
#receive base64 encoded string and convert it to binary
decoded_data = base64.b64decode(rdata)
#convert binary image to numpy array
np_data = np.frombuffer(decoded_data,np.uint8)
#encode numpy array to jepg image
image = cv.imdecode(np_data,cv.IMREAD_UNCHANGED)
url = "https://microsoft-azure-microsoft-computer-vision-v1.p.rapidapi.com/analyze"
querystring = {"visualfeatures":"Categories,Tags,Color,Faces,Description"}
#sending the converted binary image
payload = decoded_data
headers = {
'x-rapidapi-host': "microsoft-azure-microsoft-computer-vision-v1.p.rapidapi.com",
'x-rapidapi-key': "f5cf55a5c2msh4f27fe5644b39bdp157623jsn37b8f0da0884",
'content-type': "application/octet-stream"
}
resp = requests.request("POST", url, data=payload, headers=headers, params=querystring)
loaded_json = json.loads(resp.text)
faces = loaded_json['faces']
num_face = len(faces)
desc = loaded_json['description']
cel = loaded_json['categories'][0]['detail']['celebrities']
num_cel = len(cel)
img_title = titlecase(desc['captions'][0]['text'])
tags = ', '.join([i for i in desc['tags'][0:8]])
x1=np.zeros(num_face)
y1=np.zeros(num_face)
x2=np.zeros(num_face)
y2=np.zeros(num_face)
age = np.zeros(num_face)
gender = list()
for i in range(0,num_face):
x1[i] = faces[i]['faceRectangle']['left']
y1[i] = faces[i]['faceRectangle']['top']
x2[i] = faces[i]['faceRectangle']['width'] + x1[i]
y2[i] = faces[i]['faceRectangle']['height'] + y1[i]
age[i] = faces[i]['age']
gender.append(faces[i]['gender'])
x_c = np.zeros(num_cel)
y_c = np.zeros(num_cel)
for i in range(0,num_cel):
x_c[i] = cel[i]['faceRectangle']['left']
y_c[i] = cel[i]['faceRectangle']['top']
if(x_c[i] in x1 and y_c[i] in y1):
if(np.where(x1==x_c[i])==np.where(y1==y_c[i])):
gender[i] = cel[i]['name']
#drawing and naming face rectangles
for i in range(0,num_face):
cv.rectangle(image,(int(x1[i]),int(y1[i])),(int(x2[i]),int(y2[i])),(255,255,255),2)
#cv.putText(image, (gender[i] +" "+ str(int(age[i]))) , (int(x1[i]), int(y1[i])-5), cv.FONT_HERSHEY_SIMPLEX, 0.5,(255,255,255), 1)
fontpath = "./Roboto-Medium.ttf"
font = ImageFont.truetype(fontpath, 16)
img_pil = Image.fromarray(image)
draw = ImageDraw.Draw(img_pil)
for i in range(0,num_face):
if (int(y1[i])-25)>0:
box_area = image[int(y1[i])-18:int(y1[i]),int(x1[i]):int(x2[i])]
textLoc = (int(x1[i]), int(y1[i])-18)
shadowLoc = (int(x1[i])+1, int(y1[i])-17)
elif (int(y1[i]))>0:
box_area = image[0:int(y1[i]),int(x1[i]):int(x2[i])]
textLoc = (int(x1[i]), int(y1[i])-18)
shadowLoc = (int(x1[i])+1, int(y1[i])-17)
elif (int(y1[i]))==0:
box_area = image[int(y2[i]):int(y2[i])+20,int(x1[i]):int(x2[i])]
textLoc = (int(x1[i]), int(y2[i])+4)
shadowLoc = (int(x1[i])+1, int(y2[i])+5)
box_area = cv.cvtColor(box_area,cv.COLOR_BGR2GRAY)
ret,thresh = cv.threshold(box_area,127,255,cv.THRESH_BINARY)
average = np.sum(thresh)/thresh.size
if average<127:
textColor = (255,255,255,0)
else:
textColor = (0,0,0,0)
draw.text(shadowLoc, gender[i] +" "+ str(int(age[i])), font = font, fill = (100,100,100,100))
draw.text(textLoc, gender[i] +" "+ str(int(age[i])), font = font, fill = textColor)
image = np.array(img_pil)
#converting jpeg to binary image
image = cv.imencode(".jpg",image)[1].tostring()
#converting binary to base64
img_base64 = base64.b64encode(image)
return img_base64, img_title, tags
# Initialize the Flask application
application = Flask(__name__)
# route http posts to this method
@application.route('/test',methods=['POST'])
def test():
r = request
#try:
img_base64,img_title,tags = analyze_img(r.data)
return jsonify({'Image':str(img_base64),'Image_Title':img_title,'Image_Tags':tags})
#except:
# return jsonify({'status':'An exception occurred'})
@application.route('/',methods=['GET','POST'])
def main():
return({'status':'Working'})
# start flask app
if __name__ == "__main__":
application.run()
```
|
github_jupyter
|
import base64
import io
import json
import os
import sys
import cv2 as cv
import jsonpickle
import numpy as np
import requests
from flask import Flask, Response, jsonify, request
from PIL import Image, ImageFont, ImageDraw
from titlecase import titlecase
def analyze_img(rdata):
#receive base64 encoded string and convert it to binary
decoded_data = base64.b64decode(rdata)
#convert binary image to numpy array
np_data = np.frombuffer(decoded_data,np.uint8)
#encode numpy array to jepg image
image = cv.imdecode(np_data,cv.IMREAD_UNCHANGED)
url = "https://microsoft-azure-microsoft-computer-vision-v1.p.rapidapi.com/analyze"
querystring = {"visualfeatures":"Categories,Tags,Color,Faces,Description"}
#sending the converted binary image
payload = decoded_data
headers = {
'x-rapidapi-host': "microsoft-azure-microsoft-computer-vision-v1.p.rapidapi.com",
'x-rapidapi-key': "f5cf55a5c2msh4f27fe5644b39bdp157623jsn37b8f0da0884",
'content-type': "application/octet-stream"
}
resp = requests.request("POST", url, data=payload, headers=headers, params=querystring)
loaded_json = json.loads(resp.text)
faces = loaded_json['faces']
num_face = len(faces)
desc = loaded_json['description']
cel = loaded_json['categories'][0]['detail']['celebrities']
num_cel = len(cel)
img_title = titlecase(desc['captions'][0]['text'])
tags = ', '.join([i for i in desc['tags'][0:8]])
x1=np.zeros(num_face)
y1=np.zeros(num_face)
x2=np.zeros(num_face)
y2=np.zeros(num_face)
age = np.zeros(num_face)
gender = list()
for i in range(0,num_face):
x1[i] = faces[i]['faceRectangle']['left']
y1[i] = faces[i]['faceRectangle']['top']
x2[i] = faces[i]['faceRectangle']['width'] + x1[i]
y2[i] = faces[i]['faceRectangle']['height'] + y1[i]
age[i] = faces[i]['age']
gender.append(faces[i]['gender'])
x_c = np.zeros(num_cel)
y_c = np.zeros(num_cel)
for i in range(0,num_cel):
x_c[i] = cel[i]['faceRectangle']['left']
y_c[i] = cel[i]['faceRectangle']['top']
if(x_c[i] in x1 and y_c[i] in y1):
if(np.where(x1==x_c[i])==np.where(y1==y_c[i])):
gender[i] = cel[i]['name']
#drawing and naming face rectangles
for i in range(0,num_face):
cv.rectangle(image,(int(x1[i]),int(y1[i])),(int(x2[i]),int(y2[i])),(255,255,255),2)
#cv.putText(image, (gender[i] +" "+ str(int(age[i]))) , (int(x1[i]), int(y1[i])-5), cv.FONT_HERSHEY_SIMPLEX, 0.5,(255,255,255), 1)
fontpath = "./Roboto-Medium.ttf"
font = ImageFont.truetype(fontpath, 16)
img_pil = Image.fromarray(image)
draw = ImageDraw.Draw(img_pil)
for i in range(0,num_face):
if (int(y1[i])-25)>0:
box_area = image[int(y1[i])-18:int(y1[i]),int(x1[i]):int(x2[i])]
textLoc = (int(x1[i]), int(y1[i])-18)
shadowLoc = (int(x1[i])+1, int(y1[i])-17)
elif (int(y1[i]))>0:
box_area = image[0:int(y1[i]),int(x1[i]):int(x2[i])]
textLoc = (int(x1[i]), int(y1[i])-18)
shadowLoc = (int(x1[i])+1, int(y1[i])-17)
elif (int(y1[i]))==0:
box_area = image[int(y2[i]):int(y2[i])+20,int(x1[i]):int(x2[i])]
textLoc = (int(x1[i]), int(y2[i])+4)
shadowLoc = (int(x1[i])+1, int(y2[i])+5)
box_area = cv.cvtColor(box_area,cv.COLOR_BGR2GRAY)
ret,thresh = cv.threshold(box_area,127,255,cv.THRESH_BINARY)
average = np.sum(thresh)/thresh.size
if average<127:
textColor = (255,255,255,0)
else:
textColor = (0,0,0,0)
draw.text(shadowLoc, gender[i] +" "+ str(int(age[i])), font = font, fill = (100,100,100,100))
draw.text(textLoc, gender[i] +" "+ str(int(age[i])), font = font, fill = textColor)
image = np.array(img_pil)
#converting jpeg to binary image
image = cv.imencode(".jpg",image)[1].tostring()
#converting binary to base64
img_base64 = base64.b64encode(image)
return img_base64, img_title, tags
# Initialize the Flask application
application = Flask(__name__)
# route http posts to this method
@application.route('/test',methods=['POST'])
def test():
r = request
#try:
img_base64,img_title,tags = analyze_img(r.data)
return jsonify({'Image':str(img_base64),'Image_Title':img_title,'Image_Tags':tags})
#except:
# return jsonify({'status':'An exception occurred'})
@application.route('/',methods=['GET','POST'])
def main():
return({'status':'Working'})
# start flask app
if __name__ == "__main__":
application.run()
| 0.138753 | 0.17259 |
# Multilayer Perceptrons
:label:`sec_mlp`
In :numref:`chap_linear`, we introduced
softmax regression (:numref:`sec_softmax`),
implementing the algorithm from scratch
(:numref:`sec_softmax_scratch`) and using high-level APIs
(:numref:`sec_softmax_concise`),
and training classifiers to recognize
10 categories of clothing from low-resolution images.
Along the way, we learned how to wrangle data,
coerce our outputs into a valid probability distribution,
apply an appropriate loss function,
and minimize it with respect to our model's parameters.
Now that we have mastered these mechanics
in the context of simple linear models,
we can launch our exploration of deep neural networks,
the comparatively rich class of models
with which this book is primarily concerned.
## Hidden Layers
We have described the affine transformation in
:numref:`subsec_linear_model`,
which is a linear transformation added by a bias.
To begin, recall the model architecture
corresponding to our softmax regression example,
illustrated in :numref:`fig_softmaxreg`.
This model mapped our inputs directly to our outputs
via a single affine transformation,
followed by a softmax operation.
If our labels truly were related
to our input data by an affine transformation,
then this approach would be sufficient.
But linearity in affine transformations is a *strong* assumption.
### Linear Models May Go Wrong
For example, linearity implies the *weaker*
assumption of *monotonicity*:
that any increase in our feature must
either always cause an increase in our model's output
(if the corresponding weight is positive),
or always cause a decrease in our model's output
(if the corresponding weight is negative).
Sometimes that makes sense.
For example, if we were trying to predict
whether an individual will repay a loan,
we might reasonably imagine that holding all else equal,
an applicant with a higher income
would always be more likely to repay
than one with a lower income.
While monotonic, this relationship likely
is not linearly associated with the probability of
repayment. An increase in income from 0 to 50 thousand
likely corresponds to a bigger increase
in likelihood of repayment
than an increase from 1 million to 1.05 million.
One way to handle this might be to preprocess
our data such that linearity becomes more plausible,
say, by using the logarithm of income as our feature.
Note that we can easily come up with examples
that violate monotonicity.
Say for example that we want to predict probability
of death based on body temperature.
For individuals with a body temperature
above 37°C (98.6°F),
higher temperatures indicate greater risk.
However, for individuals with body temperatures
below 37° C, higher temperatures indicate lower risk!
In this case too, we might resolve the problem
with some clever preprocessing.
Namely, we might use the distance from 37°C as our feature.
But what about classifying images of cats and dogs?
Should increasing the intensity
of the pixel at location (13, 17)
always increase (or always decrease)
the likelihood that the image depicts a dog?
Reliance on a linear model corresponds to the implicit
assumption that the only requirement
for differentiating cats vs. dogs is to assess
the brightness of individual pixels.
This approach is doomed to fail in a world
where inverting an image preserves the category.
And yet despite the apparent absurdity of linearity here,
as compared with our previous examples,
it is less obvious that we could address the problem
with a simple preprocessing fix.
That is because the significance of any pixel
depends in complex ways on its context
(the values of the surrounding pixels).
While there might exist a representation of our data
that would take into account
the relevant interactions among our features,
on top of which a linear model would be suitable,
we simply do not know how to calculate it by hand.
With deep neural networks, we used observational data
to jointly learn both a representation via hidden layers
and a linear predictor that acts upon that representation.
### Incorporating Hidden Layers
We can overcome these limitations of linear models
and handle a more general class of functions
by incorporating one or more hidden layers.
The easiest way to do this is to stack
many fully-connected layers on top of each other.
Each layer feeds into the layer above it,
until we generate outputs.
We can think of the first $L-1$ layers
as our representation and the final layer
as our linear predictor.
This architecture is commonly called
a *multilayer perceptron*,
often abbreviated as *MLP*.
Below, we depict an MLP diagrammatically (:numref:`fig_mlp`).

:label:`fig_mlp`
This MLP has 4 inputs, 3 outputs,
and its hidden layer contains 5 hidden units.
Since the input layer does not involve any calculations,
producing outputs with this network
requires implementing the computations
for both the hidden and output layers;
thus, the number of layers in this MLP is 2.
Note that these layers are both fully connected.
Every input influences every neuron in the hidden layer,
and each of these in turn influences
every neuron in the output layer.
However, as suggested by :numref:`subsec_parameterization-cost-fc-layers`,
the parameterization cost of MLPs
with fully-connected layers
can be prohibitively high,
which may motivate
tradeoff between parameter saving and model effectiveness even without changing the input or output size :cite:`Zhang.Tay.Zhang.ea.2021`.
### From Linear to Nonlinear
As before, by the matrix $\mathbf{X} \in \mathbb{R}^{n \times d}$,
we denote a minibatch of $n$ examples where each example has $d$ inputs (features).
For a one-hidden-layer MLP whose hidden layer has $h$ hidden units,
denote by $\mathbf{H} \in \mathbb{R}^{n \times h}$
the outputs of the hidden layer, which are
*hidden representations*.
In mathematics or code, $\mathbf{H}$ is also known as a *hidden-layer variable* or a *hidden variable*.
Since the hidden and output layers are both fully connected,
we have hidden-layer weights $\mathbf{W}^{(1)} \in \mathbb{R}^{d \times h}$ and biases $\mathbf{b}^{(1)} \in \mathbb{R}^{1 \times h}$
and output-layer weights $\mathbf{W}^{(2)} \in \mathbb{R}^{h \times q}$ and biases $\mathbf{b}^{(2)} \in \mathbb{R}^{1 \times q}$.
Formally, we calculate the outputs $\mathbf{O} \in \mathbb{R}^{n \times q}$
of the one-hidden-layer MLP as follows:
$$
\begin{aligned}
\mathbf{H} & = \mathbf{X} \mathbf{W}^{(1)} + \mathbf{b}^{(1)}, \\
\mathbf{O} & = \mathbf{H}\mathbf{W}^{(2)} + \mathbf{b}^{(2)}.
\end{aligned}
$$
Note that after adding the hidden layer,
our model now requires us to track and update
additional sets of parameters.
So what have we gained in exchange?
You might be surprised to find out
that---in the model defined above---*we
gain nothing for our troubles*!
The reason is plain.
The hidden units above are given by
an affine function of the inputs,
and the outputs (pre-softmax) are just
an affine function of the hidden units.
An affine function of an affine function
is itself an affine function.
Moreover, our linear model was already
capable of representing any affine function.
We can view the equivalence formally
by proving that for any values of the weights,
we can just collapse out the hidden layer,
yielding an equivalent single-layer model with parameters
$\mathbf{W} = \mathbf{W}^{(1)}\mathbf{W}^{(2)}$ and $\mathbf{b} = \mathbf{b}^{(1)} \mathbf{W}^{(2)} + \mathbf{b}^{(2)}$:
$$
\mathbf{O} = (\mathbf{X} \mathbf{W}^{(1)} + \mathbf{b}^{(1)})\mathbf{W}^{(2)} + \mathbf{b}^{(2)} = \mathbf{X} \mathbf{W}^{(1)}\mathbf{W}^{(2)} + \mathbf{b}^{(1)} \mathbf{W}^{(2)} + \mathbf{b}^{(2)} = \mathbf{X} \mathbf{W} + \mathbf{b}.
$$
In order to realize the potential of multilayer architectures,
we need one more key ingredient: a
nonlinear *activation function* $\sigma$
to be applied to each hidden unit
following the affine transformation.
The outputs of activation functions
(e.g., $\sigma(\cdot)$)
are called *activations*.
In general, with activation functions in place,
it is no longer possible to collapse our MLP into a linear model:
$$
\begin{aligned}
\mathbf{H} & = \sigma(\mathbf{X} \mathbf{W}^{(1)} + \mathbf{b}^{(1)}), \\
\mathbf{O} & = \mathbf{H}\mathbf{W}^{(2)} + \mathbf{b}^{(2)}.\\
\end{aligned}
$$
Since each row in $\mathbf{X}$ corresponds to an example in the minibatch,
with some abuse of notation, we define the nonlinearity
$\sigma$ to apply to its inputs in a rowwise fashion,
i.e., one example at a time.
Note that we used the notation for softmax
in the same way to denote a rowwise operation in :numref:`subsec_softmax_vectorization`.
Often, as in this section, the activation functions
that we apply to hidden layers are not merely rowwise,
but elementwise.
That means that after computing the linear portion of the layer,
we can calculate each activation
without looking at the values taken by the other hidden units.
This is true for most activation functions.
To build more general MLPs, we can continue stacking
such hidden layers,
e.g., $\mathbf{H}^{(1)} = \sigma_1(\mathbf{X} \mathbf{W}^{(1)} + \mathbf{b}^{(1)})$
and $\mathbf{H}^{(2)} = \sigma_2(\mathbf{H}^{(1)} \mathbf{W}^{(2)} + \mathbf{b}^{(2)})$,
one atop another, yielding ever more expressive models.
### Universal Approximators
MLPs can capture complex interactions
among our inputs via their hidden neurons,
which depend on the values of each of the inputs.
We can easily design hidden nodes
to perform arbitrary computation,
for instance, basic logic operations on a pair of inputs.
Moreover, for certain choices of the activation function,
it is widely known that MLPs are universal approximators.
Even with a single-hidden-layer network,
given enough nodes (possibly absurdly many),
and the right set of weights,
we can model any function,
though actually learning that function is the hard part.
You might think of your neural network
as being a bit like the C programming language.
The language, like any other modern language,
is capable of expressing any computable program.
But actually coming up with a program
that meets your specifications is the hard part.
Moreover, just because a single-hidden-layer network
*can* learn any function
does not mean that you should try
to solve all of your problems
with single-hidden-layer networks.
In fact, we can approximate many functions
much more compactly by using deeper (vs. wider) networks.
We will touch upon more rigorous arguments in subsequent chapters.
## Activation Functions
:label:`subsec_activation-functions`
Activation functions decide whether a neuron should be activated or not by
calculating the weighted sum and further adding bias with it.
They are differentiable operators to transform input signals to outputs,
while most of them add non-linearity.
Because activation functions are fundamental to deep learning,
(**let us briefly survey some common activation functions**).
```
%matplotlib inline
import tensorflow as tf
from d2l import tensorflow as d2l
```
### ReLU Function
The most popular choice,
due to both simplicity of implementation and
its good performance on a variety of predictive tasks,
is the *rectified linear unit* (*ReLU*).
[**ReLU provides a very simple nonlinear transformation**].
Given an element $x$, the function is defined
as the maximum of that element and $0$:
$$\operatorname{ReLU}(x) = \max(x, 0).$$
Informally, the ReLU function retains only positive
elements and discards all negative elements
by setting the corresponding activations to 0.
To gain some intuition, we can plot the function.
As you can see, the activation function is piecewise linear.
```
x = tf.Variable(tf.range(-8.0, 8.0, 0.1), dtype=tf.float32)
y = tf.nn.relu(x)
d2l.plot(x.numpy(), y.numpy(), 'x', 'relu(x)', figsize=(5, 2.5))
```
When the input is negative,
the derivative of the ReLU function is 0,
and when the input is positive,
the derivative of the ReLU function is 1.
Note that the ReLU function is not differentiable
when the input takes value precisely equal to 0.
In these cases, we default to the left-hand-side
derivative and say that the derivative is 0 when the input is 0.
We can get away with this because
the input may never actually be zero.
There is an old adage that if subtle boundary conditions matter,
we are probably doing (*real*) mathematics, not engineering.
That conventional wisdom may apply here.
We plot the derivative of the ReLU function plotted below.
```
with tf.GradientTape() as t:
y = tf.nn.relu(x)
d2l.plot(x.numpy(),
t.gradient(y, x).numpy(), 'x', 'grad of relu', figsize=(5, 2.5))
```
The reason for using ReLU is that
its derivatives are particularly well behaved:
either they vanish or they just let the argument through.
This makes optimization better behaved
and it mitigated the well-documented problem
of vanishing gradients that plagued
previous versions of neural networks (more on this later).
Note that there are many variants to the ReLU function,
including the *parameterized ReLU* (*pReLU*) function :cite:`He.Zhang.Ren.ea.2015`.
This variation adds a linear term to ReLU,
so some information still gets through,
even when the argument is negative:
$$\operatorname{pReLU}(x) = \max(0, x) + \alpha \min(0, x).$$
### Sigmoid Function
[**The *sigmoid function* transforms its inputs**],
for which values lie in the domain $\mathbb{R}$,
(**to outputs that lie on the interval (0, 1).**)
For that reason, the sigmoid is
often called a *squashing function*:
it squashes any input in the range (-inf, inf)
to some value in the range (0, 1):
$$\operatorname{sigmoid}(x) = \frac{1}{1 + \exp(-x)}.$$
In the earliest neural networks, scientists
were interested in modeling biological neurons
which either *fire* or *do not fire*.
Thus the pioneers of this field,
going all the way back to McCulloch and Pitts,
the inventors of the artificial neuron,
focused on thresholding units.
A thresholding activation takes value 0
when its input is below some threshold
and value 1 when the input exceeds the threshold.
When attention shifted to gradient based learning,
the sigmoid function was a natural choice
because it is a smooth, differentiable
approximation to a thresholding unit.
Sigmoids are still widely used as
activation functions on the output units,
when we want to interpret the outputs as probabilities
for binary classification problems
(you can think of the sigmoid as a special case of the softmax).
However, the sigmoid has mostly been replaced
by the simpler and more easily trainable ReLU
for most use in hidden layers.
In later chapters on recurrent neural networks,
we will describe architectures that leverage sigmoid units
to control the flow of information across time.
Below, we plot the sigmoid function.
Note that when the input is close to 0,
the sigmoid function approaches
a linear transformation.
```
y = tf.nn.sigmoid(x)
d2l.plot(x.numpy(), y.numpy(), 'x', 'sigmoid(x)', figsize=(5, 2.5))
```
The derivative of the sigmoid function is given by the following equation:
$$\frac{d}{dx} \operatorname{sigmoid}(x) = \frac{\exp(-x)}{(1 + \exp(-x))^2} = \operatorname{sigmoid}(x)\left(1-\operatorname{sigmoid}(x)\right).$$
The derivative of the sigmoid function is plotted below.
Note that when the input is 0,
the derivative of the sigmoid function
reaches a maximum of 0.25.
As the input diverges from 0 in either direction,
the derivative approaches 0.
```
with tf.GradientTape() as t:
y = tf.nn.sigmoid(x)
d2l.plot(x.numpy(),
t.gradient(y, x).numpy(), 'x', 'grad of sigmoid', figsize=(5, 2.5))
```
### Tanh Function
Like the sigmoid function, [**the tanh (hyperbolic tangent)
function also squashes its inputs**],
transforming them into elements on the interval (**between -1 and 1**):
$$\operatorname{tanh}(x) = \frac{1 - \exp(-2x)}{1 + \exp(-2x)}.$$
We plot the tanh function below.
Note that as the input nears 0, the tanh function approaches a linear transformation. Although the shape of the function is similar to that of the sigmoid function, the tanh function exhibits point symmetry about the origin of the coordinate system.
```
y = tf.nn.tanh(x)
d2l.plot(x.numpy(), y.numpy(), 'x', 'tanh(x)', figsize=(5, 2.5))
```
The derivative of the tanh function is:
$$\frac{d}{dx} \operatorname{tanh}(x) = 1 - \operatorname{tanh}^2(x).$$
The derivative of tanh function is plotted below.
As the input nears 0,
the derivative of the tanh function approaches a maximum of 1.
And as we saw with the sigmoid function,
as the input moves away from 0 in either direction,
the derivative of the tanh function approaches 0.
```
with tf.GradientTape() as t:
y = tf.nn.tanh(x)
d2l.plot(x.numpy(),
t.gradient(y, x).numpy(), 'x', 'grad of tanh', figsize=(5, 2.5))
```
In summary, we now know how to incorporate nonlinearities
to build expressive multilayer neural network architectures.
As a side note, your knowledge already
puts you in command of a similar toolkit
to a practitioner circa 1990.
In some ways, you have an advantage
over anyone working in the 1990s,
because you can leverage powerful
open-source deep learning frameworks
to build models rapidly, using only a few lines of code.
Previously, training these networks
required researchers to code up
thousands of lines of C and Fortran.
## Summary
* MLP adds one or multiple fully-connected hidden layers between the output and input layers and transforms the output of the hidden layer via an activation function.
* Commonly-used activation functions include the ReLU function, the sigmoid function, and the tanh function.
## Exercises
1. Compute the derivative of the pReLU activation function.
1. Show that an MLP using only ReLU (or pReLU) constructs a continuous piecewise linear function.
1. Show that $\operatorname{tanh}(x) + 1 = 2 \operatorname{sigmoid}(2x)$.
1. Assume that we have a nonlinearity that applies to one minibatch at a time. What kinds of problems do you expect this to cause?
[Discussions](https://discuss.d2l.ai/t/226)
|
github_jupyter
|
%matplotlib inline
import tensorflow as tf
from d2l import tensorflow as d2l
x = tf.Variable(tf.range(-8.0, 8.0, 0.1), dtype=tf.float32)
y = tf.nn.relu(x)
d2l.plot(x.numpy(), y.numpy(), 'x', 'relu(x)', figsize=(5, 2.5))
with tf.GradientTape() as t:
y = tf.nn.relu(x)
d2l.plot(x.numpy(),
t.gradient(y, x).numpy(), 'x', 'grad of relu', figsize=(5, 2.5))
y = tf.nn.sigmoid(x)
d2l.plot(x.numpy(), y.numpy(), 'x', 'sigmoid(x)', figsize=(5, 2.5))
with tf.GradientTape() as t:
y = tf.nn.sigmoid(x)
d2l.plot(x.numpy(),
t.gradient(y, x).numpy(), 'x', 'grad of sigmoid', figsize=(5, 2.5))
y = tf.nn.tanh(x)
d2l.plot(x.numpy(), y.numpy(), 'x', 'tanh(x)', figsize=(5, 2.5))
with tf.GradientTape() as t:
y = tf.nn.tanh(x)
d2l.plot(x.numpy(),
t.gradient(y, x).numpy(), 'x', 'grad of tanh', figsize=(5, 2.5))
| 0.679923 | 0.991398 |
## _*H2 ground state energy computation using Quantum Phase Estimation*_
This notebook demonstrates using QISKit ACQUA Chemistry to compute ground state energy of the Hydrogen (H2) molecule using QPE (Quantum Phase Estimation) algorithm. It is compared to the same energy as computed by the ExactEigensolver
This notebook populates a dictionary, that is a progammatic representation of an input file, in order to drive the qiskit_acqua_chemistry stack. Such a dictionary can be manipulated programmatically. An sibling notebook `h2_iqpe` is also provided, which showcases how the ground state energies over a range of inter-atomic distances can be computed and then plotted as well.
This notebook has been written to use the PYSCF chemistry driver. See the PYSCF chemistry driver readme if you need to install the external PySCF library that this driver requires.
```
from qiskit_acqua_chemistry import ACQUAChemistry
import time
distance = 0.735
molecule = 'H .0 .0 0; H .0 .0 {}'.format(distance)
# Input dictionary to configure QISKit ACQUA Chemistry for the chemistry problem.
acqua_chemistry_qpe_dict = {
'driver': {'name': 'PYSCF'},
'PYSCF': {
'atom': molecule,
'basis': 'sto3g'
},
'operator': {'name': 'hamiltonian', 'transformation': 'full', 'qubit_mapping': 'parity'},
'algorithm': {
'name': 'QPE',
'num_ancillae': 9,
'num_time_slices': 50,
'expansion_mode': 'suzuki',
'expansion_order': 2,
},
'initial_state': {'name': 'HartreeFock'},
'backend': {
'name': 'local_qasm_simulator',
'shots': 100,
}
}
acqua_chemistry_ees_dict = {
'driver': {'name': 'PYSCF'},
'PYSCF': {'atom': molecule, 'basis': 'sto3g'},
'operator': {'name': 'hamiltonian', 'transformation': 'full', 'qubit_mapping': 'parity'},
'algorithm': {
'name': 'ExactEigensolver',
},
}
```
With the two algorithms configured, we can then run them and check the results, as follows.
```
start_time = time.time()
result_qpe = ACQUAChemistry().run(acqua_chemistry_qpe_dict)
result_ees = ACQUAChemistry().run(acqua_chemistry_ees_dict)
print("--- computation completed in %s seconds ---" % (time.time() - start_time))
print('The groundtruth total ground state energy is {}.'.format(
result_ees['energy']
))
print('The total ground state energy as computed by QPE is {}.'.format(
result_qpe['energy']
))
print('In comparison, the Hartree-Fock ground state energy is {}.'.format(
result_ees['hf_energy']
))
```
|
github_jupyter
|
from qiskit_acqua_chemistry import ACQUAChemistry
import time
distance = 0.735
molecule = 'H .0 .0 0; H .0 .0 {}'.format(distance)
# Input dictionary to configure QISKit ACQUA Chemistry for the chemistry problem.
acqua_chemistry_qpe_dict = {
'driver': {'name': 'PYSCF'},
'PYSCF': {
'atom': molecule,
'basis': 'sto3g'
},
'operator': {'name': 'hamiltonian', 'transformation': 'full', 'qubit_mapping': 'parity'},
'algorithm': {
'name': 'QPE',
'num_ancillae': 9,
'num_time_slices': 50,
'expansion_mode': 'suzuki',
'expansion_order': 2,
},
'initial_state': {'name': 'HartreeFock'},
'backend': {
'name': 'local_qasm_simulator',
'shots': 100,
}
}
acqua_chemistry_ees_dict = {
'driver': {'name': 'PYSCF'},
'PYSCF': {'atom': molecule, 'basis': 'sto3g'},
'operator': {'name': 'hamiltonian', 'transformation': 'full', 'qubit_mapping': 'parity'},
'algorithm': {
'name': 'ExactEigensolver',
},
}
start_time = time.time()
result_qpe = ACQUAChemistry().run(acqua_chemistry_qpe_dict)
result_ees = ACQUAChemistry().run(acqua_chemistry_ees_dict)
print("--- computation completed in %s seconds ---" % (time.time() - start_time))
print('The groundtruth total ground state energy is {}.'.format(
result_ees['energy']
))
print('The total ground state energy as computed by QPE is {}.'.format(
result_qpe['energy']
))
print('In comparison, the Hartree-Fock ground state energy is {}.'.format(
result_ees['hf_energy']
))
| 0.560253 | 0.984913 |
As the name implies, a variable is something that can change. A variable is just a way of referring to a memory location used by a Python program. Based on the datatype of the variable, the Python interpreter allocates the memory and decides what can be stored in the reserved memory. This makes Python a dynamically-typed language
If you are familiar with other programming languages like C, C++ or Java it might be tempting to consider variable as just a container to store data. However in Python, you can at best, think of variables as pointers. This is why you can dynamically change the type of data that a variable is pointing at.
### Assigning values to variables
One of the main differences between Python and strongly typed languages like C++ or Java is the way it deals with the data types. In languages like C++ or Java, every variable must have a unique data type (i.e if a variable is of type string it cannot store integers or floats). Moreover, every variable has to be declared before it can be used, thus binding it to the data type that can be stored in it. Python variables do not need explicit declaration to reserve memory space. The declaration happens automatically when a value is assigned to a variable. This means that a variable that was used to store a string can now be used to store an integer. Try it out.
Do something like this
```
var = 'I am in NYC'
print(var)
```
**var** is a string in the above case.
Well.. don't take our word for it. Let's confirm. If we run the next line,
```
type(var)
```
As we saw in the grid in the 01.00, Type is a built-in function that returns any datatype If we point our variable var to, lets say, an integer, it will return int
```
var = 123
type(var)
```
**So what are the rules of naming a variable?**
Every language has some rules for naming the identifier of variables (aka the variable name). In Python, a valid identifier is a non-empty sequence of characters of any length with:
The start of the character can be an underscore _ or a capital or lowercase letter. However, it is generally recommended to use all uppercase for global variables and all lower case for local variables. The letters following the first letter can be a digit or a string. Python is a case-sensitive language. Therefore, **var** is not equal to **VAR** or **vAr**.
Apart from the above restrictions, Python keywords cannot be used as identifier names. These are:
||||||
|:-------|:--------|:---------|:--------|:----|
|and |del |from |not |while|
|as | elif |global |or |with |
|assert | else | if |pass |yield|
|break | except | import |print
|class | exec | in |raise
|continue| finally | is |return
|def | for | lambda |try
### Multiples
Python allows you to assign a single value to several variables simultaneously. For example:
```
x = y = z = a = 1
```
or even assign different values to different variables:
```
x, y, z, a = 'Hello', 'World', 1, 2
print (x,y,z,a)
```
Try printing the above variables. Hint: The key to printing lies above!
|
github_jupyter
|
var = 'I am in NYC'
print(var)
type(var)
var = 123
type(var)
x = y = z = a = 1
x, y, z, a = 'Hello', 'World', 1, 2
print (x,y,z,a)
| 0.065355 | 0.919534 |
```
# Import required packages
import pandas as pd
import gurobipy
import datetime
from typing import List, Dict
import altair as alt
import datapane as dp
def plot_load(planning: pd.DataFrame, need: pd.DataFrame, timeline: List[str]) -> None:
# Plot graph - Requirement
source = (
pd.Series(need).rename_axis(["Date", "Customer_Order"]).reset_index(name="Qty")
)
source = source.groupby(["Date"]).sum()
source["Date"] = source.index
# Plot graph - Optimized planning
source = planning.filter(like="Total hours", axis=0).copy()
source["Date"] = source.index
source = source.reset_index(drop=True)
source = source.rename(columns={"Solution": "Hours"}).reset_index()
source[["Date", "Line"]] = source["Date"].str.split(",", expand=True)
source["Date"] = source["Date"].str.split("[").str[1]
source["Line"] = source["Line"].str.split("]").str[0]
source["Min capacity"] = 7
source["Max capacity"] = 12
source = source.round({"Hours": 1})
source["Load%"] = pd.Series(
["{0:.0f}%".format(val / 8 * 100) for val in source["Hours"]],
index=source.index,
)
bars = (
alt.Chart(source)
.mark_bar()
.encode(
x="Line:N",
y="Hours:Q",
color="Line:N",
tooltip=["Date", "Line", "Hours", "Load%"],
)
.interactive()
.properties(width=550 / len(timeline) - 22, height=150)
)
line_min = alt.Chart(source).mark_rule(color="darkgrey").encode(y="Min capacity:Q")
line_max = (
alt.Chart(source)
.mark_rule(color="darkgrey")
.encode(y=alt.Y("Max capacity:Q", title="Load (hours)"))
)
chart = (
alt.layer(bars, line_min, line_max, data=source)
.facet(column="Date:N")
.properties(title="Daily working time")
)
chart.save("planning_load_model4.html")
dp.Report(
dp.Plot(chart, caption="Production schedule model 4 - Time")
).upload(name='Optimized production schedule - Time',
description="Optimized production schedule - Time", open=True, visibily="PUBLIC")
def plot_planning(
planning: pd.DataFrame, need: pd.DataFrame, timeline: List[str]
) -> None:
# Plot graph - Requirement
source = pd.Series(need).rename_axis(["Date", "Order"]).reset_index(name="Qty")
chart_need = (
alt.Chart(source)
.mark_bar()
.encode(
y=alt.Y("Qty", axis=alt.Axis(grid=False)),
column=alt.Column("Date:N"),
color="Order:N",
tooltip=["Order", "Qty"],
)
.interactive()
.properties(
width=800 / len(timeline) - 22,
height=100,
title="Customer's requirement",
)
)
df = (
planning.filter(like="plannedQty", axis=0)
.copy()
.rename(columns={"Solution": "Qty"})
.reset_index()
)
df[["Date", "Order", "Line"]] = df["index"].str.split(",", expand=True)
df["Date"] = df["Date"].str.split("[").str[1]
df["Line"] = df["Line"].str.split("]").str[0]
df = df[["Date", "Line", "Qty", "Order"]]
chart_planning = (
alt.Chart(df)
.mark_bar()
.encode(
y=alt.Y("Qty", axis=alt.Axis(grid=False)),
x="Line:N",
column=alt.Column("Date:N"),
color="Order:N",
tooltip=["Line", "Order", "Qty"],
)
.interactive()
.properties(
width=800 / len(timeline) - 22,
height=200,
title="Optimized Production Schedule",
)
)
chart = alt.vconcat(chart_planning, chart_need)
chart.save("planning_MO_model4.html")
dp.Report(
dp.Plot(chart, caption="Production schedule model 4 - Qty")
).upload(name='Optimized production schedule - Qty',
description="Optimized production schedule - Qty", open=True, visibily="PUBLIC")
def plot_inventory(
planning: pd.DataFrame, timeline: List[str], cust_orders,
) -> None:
# Plot inventory
df = (
planning.filter(like="early prod", axis=0)
.copy()
.rename(columns={"Solution": "Qty"})
.reset_index()
)
df[["Date", "Order"]] = df["index"].str.split(",", expand=True)
df["Date"] = df["Date"].str.split("[").str[1]
df["Order"] = df["Order"].str.split("]").str[0]
df = df[["Date", "Qty", "Order"]]
models_list = cust_orders[['Order', 'Product_Family']]
df = pd.merge(df, models_list, on='Order', how='inner')
df = df[["Date", "Qty", "Product_Family"]]
bars = (
alt.Chart(df)
.mark_bar()
.encode(
y="Qty:Q",
color="Product_Family:N",
tooltip=["Product_Family", "Qty"],
)
.interactive()
.properties(width=550 / len(timeline) - 22, height=60)
)
chart_inventory = (
alt.layer(bars, data=df)
.facet(column="Date:N")
.properties(title="Inventory")
)
# Plot shortage
df = (
planning.filter(like="late prod", axis=0)
.copy()
.rename(columns={"Solution": "Qty"})
.reset_index()
)
df[["Date", "Order"]] = df["index"].str.split(",", expand=True)
df["Date"] = df["Date"].str.split("[").str[1]
df["Order"] = df["Order"].str.split("]").str[0]
df = df[["Date", "Qty", "Order"]]
models_list = cust_orders[['Order', 'Product_Family']]
df = pd.merge(df, models_list, on='Order', how='inner')
df = df[["Date", "Qty", "Product_Family"]]
bars = (
alt.Chart(df)
.mark_bar()
.encode(
y="Qty:Q",
color="Product_Family:N",
tooltip=["Product_Family", "Qty"],
)
.interactive()
.properties(width=550 / len(timeline) - 22, height=60)
)
chart_shortage = (
alt.layer(bars, data=df)
.facet(column="Date:N")
.properties(title="Shortage")
)
chart = alt.vconcat(chart_inventory, chart_shortage)
chart.save("Inventory_Shortage.html")
dp.Report(
dp.Plot(chart, caption="Inventory_Shortage")
).upload(name='Inventory_Shortage',
description="Inventory_Shortage", open=True, visibily="PUBLIC")
def print_planning(planning: pd.DataFrame) -> None:
df = (
planning.filter(like="plannedQty", axis=0)
.copy()
.rename(columns={"Solution": "Qty"})
.reset_index()
)
df[["Date", "Customer_Order", "Line"]] = df["index"].str.split(",", expand=True)
df["Date"] = df["Date"].str.split("[").str[1]
df["Line"] = df["Line"].str.split("]").str[0]
df = df[["Date", "Line", "Qty", "Customer_Order"]]
df.to_csv(r"Planning_model4_list.csv", index=True)
print(df)
df.pivot_table(
values="Qty", index="Customer_Order", columns=["Date", "Line"]
).to_csv(r"Planning_model4v2.csv", index=True)
def optimize_planning(
timeline: List[str],
workcenters: List[str],
needs,
wc_cost_reg: Dict[str, int],
wc_cost_ot: Dict[str, int],
wc_cost_we: Dict[str, int],
inventory_carrying_cost: int,
customer_orders: List[str],
cycle_times,
delay_cost: int,
) -> pd.DataFrame:
# Split weekdays/weekends
weekdays = []
weekend = []
for date in timeline:
day = datetime.datetime.strptime(date, "%Y/%m/%d")
if day.weekday() < 5:
weekdays.append(date)
else:
weekend.append(date)
# Initiate optimization model
model = gurobipy.Model("Optimize production planning")
# DEFINE VARIABLES
# Quantity variable
x_qty = model.addVars(
timeline,
customer_orders,
workcenters,
lb=0,
vtype=gurobipy.GRB.INTEGER,
name="plannedQty",
)
# Time variable
x_time = model.addVars(
timeline,
customer_orders,
workcenters,
lb=0,
vtype=gurobipy.GRB.CONTINUOUS,
name="plannedTime",
)
# Set the value of x_time
model.addConstrs(
(
(
x_time[(date, mo, wc)] == x_qty[(date, mo, wc)] * cycle_times[(mo, wc)]
for date in timeline
for mo in customer_orders
for wc in workcenters
)
),
name="x_time_constr",
)
# Qty to display
quantity = model.addVars(
timeline, workcenters, lb=0, vtype=gurobipy.GRB.INTEGER, name="qty"
)
# Set the value of qty
model.addConstrs(
(
(
quantity[(date, wc)]
== gurobipy.quicksum(x_qty[(date, mo, wc)] for mo in customer_orders)
for date in timeline
for wc in workcenters
)
),
name="wty_time_constr",
)
# Variable status of the line ( 0 = closed, 1 = opened)
line_opening = model.addVars(
timeline, workcenters, vtype=gurobipy.GRB.BINARY, name="Open status"
)
# Load variables (hours) - regular and overtime
reg_hours = model.addVars(
timeline,
workcenters,
vtype=gurobipy.GRB.CONTINUOUS,
name="Regular hours",
)
ot_hours = model.addVars(
timeline,
workcenters,
vtype=gurobipy.GRB.CONTINUOUS,
name="Overtime hours",
)
reg_hours_bis = model.addVars(
timeline,
workcenters,
lb=7,
ub=8,
vtype=gurobipy.GRB.CONTINUOUS,
name="regHours",
)
ot_hours_bis = model.addVars(
timeline, workcenters, lb=0, ub=4, vtype=gurobipy.GRB.CONTINUOUS, name="OTHours"
)
# Set the value of reg and OT hours)
model.addConstrs(
(
reg_hours[(date, wc)]
== reg_hours_bis[(date, wc)] * line_opening[(date, wc)]
for date in timeline
for wc in workcenters
),
name="total_hours_constr",
)
model.addConstrs(
(
ot_hours[(date, wc)] == ot_hours_bis[(date, wc)] * line_opening[(date, wc)]
for date in timeline
for wc in workcenters
),
name="total_hours_constr",
)
# Variable total load (hours)
total_hours = model.addVars(
timeline,
workcenters,
vtype=gurobipy.GRB.CONTINUOUS,
name="Total hours",
)
# Set the value of total load (regular + overtime)
model.addConstrs(
(
total_hours[(date, wc)] == (reg_hours[(date, wc)] + ot_hours[(date, wc)])
for date in timeline
for wc in workcenters
),
name="Link total hours - reg/ot hours",
)
# Set total hours of production in link with the time variable
model.addConstrs(
(
(
total_hours[(date, wc)]
== gurobipy.quicksum(x_time[(date, mo, wc)] for mo in customer_orders)
for date in timeline
for wc in workcenters
)
),
name="total_hours_constr",
)
# Variable cost
labor_cost = model.addVars(
timeline, workcenters, lb=0, vtype=gurobipy.GRB.CONTINUOUS, name="Labor cost"
)
# Set the value of cost (hours * hourly cost)
model.addConstrs(
(
labor_cost[(date, wc)]
== reg_hours[(date, wc)] * wc_cost_reg[wc]
+ ot_hours[(date, wc)] * wc_cost_ot[wc]
for date in weekdays
for wc in workcenters
),
name="Link labor cost - working hours - wd",
)
model.addConstrs(
(
labor_cost[(date, wc)]
== total_hours[(date, wc)] * wc_cost_we[wc]
for date in weekend
for wc in workcenters
),
name="Link labor cost - working hours - we",
)
# Variable gap early/late production
gap_prod = model.addVars(
timeline,
customer_orders,
lb=-10000,
ub=10000,
vtype=gurobipy.GRB.CONTINUOUS,
name="gapProd",
)
abs_gap_prod = model.addVars(
timeline,
customer_orders,
vtype=gurobipy.GRB.CONTINUOUS,
name="absGapProd",
)
# Set the value of gap for early production
for l in range(len(timeline)):
model.addConstrs(
(
gap_prod[(timeline[l], mo)]
== gurobipy.quicksum(
x_qty[(date, mo, wc)]
for date in timeline[: l + 1]
for wc in workcenters
)
- (gurobipy.quicksum(needs[(date, mo)] for date in timeline[: l + 1]))
for mo in customer_orders
),
name="gap_prod",
)
# Set the value of ABS(gap for early production)
model.addConstrs(
(
(abs_gap_prod[(date, mo)] == gurobipy.abs_(gap_prod[(date, mo)]))
for date in timeline
for mo in customer_orders
),
name="abs gap prod",
)
# Create variable "early production" and "inventory costs"
early_prod = model.addVars(
timeline,
customer_orders,
vtype=gurobipy.GRB.CONTINUOUS,
name="early prod",
)
inventory_costs = model.addVars(
timeline,
customer_orders,
vtype=gurobipy.GRB.CONTINUOUS,
name="inventory costs",
)
# Set the value of early production
model.addConstrs(
(
early_prod[(date, m)] == (gap_prod[(date, m)] + abs_gap_prod[(date, m)]) / 2
for date in timeline
for m in customer_orders
),
name="early prod",
)
# Set the value of inventory costs
model.addConstrs(
(
(inventory_costs[(date, m)] == early_prod[(date, m)] * inventory_carrying_cost)
for date in timeline
for m in customer_orders
),
name="inventory costs",
)
# Create variable "late production" and "delay costs"
late_prod = model.addVars(
timeline,
customer_orders,
vtype=gurobipy.GRB.CONTINUOUS,
name="late prod",
)
delay_costs = model.addVars(
timeline,
customer_orders,
vtype=gurobipy.GRB.CONTINUOUS,
name="inventory costs",
)
# Set the value of late production
model.addConstrs(
(
late_prod[(date, m)] == (abs_gap_prod[(date, m)] - gap_prod[(date, m)]) / 2
for date in timeline
for m in customer_orders
),
name="late prod",
)
# Set the value of delay costs
model.addConstrs(
(
(delay_costs[(date, m)] == late_prod[(date, m)] * delay_cost)
for date in timeline
for m in customer_orders
),
name="delay costs",
)
# CONSTRAINT
# Constraint: Total hours of production = required production time
model.addConstr(
(
gurobipy.quicksum(
x_qty[(date, mo, wc)]
for date in timeline
for mo in customer_orders
for wc in workcenters
)
== (gurobipy.quicksum(needs[(date, mo)] for date in timeline for mo in customer_orders))
),
name="total_req",
)
# DEFINE MODEL
# Objective : minimize a function
model.ModelSense = gurobipy.GRB.MINIMIZE
# Function to minimize
objective = 0
objective += gurobipy.quicksum(
labor_cost[(date, wc)] for date in timeline for wc in workcenters
)
objective += gurobipy.quicksum(
inventory_costs[(date, mo)]
for date in timeline
for mo in customer_orders
)
objective += gurobipy.quicksum(
delay_costs[(date, mo)]
for date in timeline
for mo in customer_orders
)
# SOLVE MODEL
model.setObjective(objective)
model.optimize()
sol = pd.DataFrame(data={"Solution": model.X}, index=model.VarName)
print("Total cost = $" + str(model.ObjVal))
# model.write("Planning_optimization.lp")
# file = open("Planning_optimization.lp", 'r')
# print(file.read())
# file.close()
return sol
def check_duplicates(list_to_check):
if len(list_to_check) == len(set(list_to_check)):
return
else:
print("Duplicate order, please check the requirements file")
exit()
return
# Define hourly cost per line - regular, overtime and weekend
reg_costs_per_line = {"Line_1": 245, "Line_2": 315, "Line_3": 245}
ot_costs_per_line = {
k: 1.5 * reg_costs_per_line[k] for k, v in reg_costs_per_line.items()
}
we_costs_per_line = {
k: 2 * reg_costs_per_line[k] for k, w in reg_costs_per_line.items()
}
storage_cost = 5
late_prod_cost = 1000
lines: List[str] = list(reg_costs_per_line.keys())
# Get orders
customer_orders = pd.read_excel("customer_orders.xlsx")
# Get cycle times
capacity = pd.read_excel("line_capacity.xlsx", sheet_name="8h capacity").set_index("Line")
cycle_time = capacity.rdiv(8)
order_list = customer_orders["Order"].to_list()
check_duplicates(order_list)
# Create cycle times dictionnary
customer_orders = customer_orders.merge(
cycle_time, left_on="Product_Family", right_index=True
)
customer_orders["Delivery_Date"] = pd.to_datetime(
customer_orders["Delivery_Date"]
).dt.strftime("%Y/%m/%d")
customer_orders = customer_orders.sort_values(by=["Delivery_Date", "Order"])
cycle_times = {
(order, line): customer_orders[line][customer_orders.Order == order].item()
for order in order_list
for line in lines
}
# Define calendar
start_date = datetime.datetime.strptime(
customer_orders["Delivery_Date"].min(), "%Y/%m/%d"
)
end_date = datetime.datetime.strptime(
customer_orders["Delivery_Date"].max(), "%Y/%m/%d"
)
date_modified = start_date
calendar = [start_date.strftime("%Y/%m/%d")]
while date_modified < end_date:
date_modified += datetime.timedelta(days=1)
calendar.append(date_modified.strftime("%Y/%m/%d"))
# Create daily requirements dictionnary
daily_requirements = {}
for day in calendar:
for order in order_list:
try:
daily_requirements[(day, order)] = customer_orders[
(customer_orders.Order == order)
& (customer_orders.Delivery_Date == day)
]["Quantity"].item()
except ValueError:
daily_requirements[(day, order)] = 0
# Optimize planning
solution = optimize_planning(
calendar,
lines,
daily_requirements,
reg_costs_per_line,
ot_costs_per_line,
we_costs_per_line,
storage_cost,
order_list,
cycle_times,
late_prod_cost,
)
# Plot the new planning
plot_load(solution, daily_requirements, calendar)
print_planning(solution)
plot_planning(solution, daily_requirements, calendar)
plot_inventory(solution, calendar, customer_orders)
```
|
github_jupyter
|
# Import required packages
import pandas as pd
import gurobipy
import datetime
from typing import List, Dict
import altair as alt
import datapane as dp
def plot_load(planning: pd.DataFrame, need: pd.DataFrame, timeline: List[str]) -> None:
# Plot graph - Requirement
source = (
pd.Series(need).rename_axis(["Date", "Customer_Order"]).reset_index(name="Qty")
)
source = source.groupby(["Date"]).sum()
source["Date"] = source.index
# Plot graph - Optimized planning
source = planning.filter(like="Total hours", axis=0).copy()
source["Date"] = source.index
source = source.reset_index(drop=True)
source = source.rename(columns={"Solution": "Hours"}).reset_index()
source[["Date", "Line"]] = source["Date"].str.split(",", expand=True)
source["Date"] = source["Date"].str.split("[").str[1]
source["Line"] = source["Line"].str.split("]").str[0]
source["Min capacity"] = 7
source["Max capacity"] = 12
source = source.round({"Hours": 1})
source["Load%"] = pd.Series(
["{0:.0f}%".format(val / 8 * 100) for val in source["Hours"]],
index=source.index,
)
bars = (
alt.Chart(source)
.mark_bar()
.encode(
x="Line:N",
y="Hours:Q",
color="Line:N",
tooltip=["Date", "Line", "Hours", "Load%"],
)
.interactive()
.properties(width=550 / len(timeline) - 22, height=150)
)
line_min = alt.Chart(source).mark_rule(color="darkgrey").encode(y="Min capacity:Q")
line_max = (
alt.Chart(source)
.mark_rule(color="darkgrey")
.encode(y=alt.Y("Max capacity:Q", title="Load (hours)"))
)
chart = (
alt.layer(bars, line_min, line_max, data=source)
.facet(column="Date:N")
.properties(title="Daily working time")
)
chart.save("planning_load_model4.html")
dp.Report(
dp.Plot(chart, caption="Production schedule model 4 - Time")
).upload(name='Optimized production schedule - Time',
description="Optimized production schedule - Time", open=True, visibily="PUBLIC")
def plot_planning(
planning: pd.DataFrame, need: pd.DataFrame, timeline: List[str]
) -> None:
# Plot graph - Requirement
source = pd.Series(need).rename_axis(["Date", "Order"]).reset_index(name="Qty")
chart_need = (
alt.Chart(source)
.mark_bar()
.encode(
y=alt.Y("Qty", axis=alt.Axis(grid=False)),
column=alt.Column("Date:N"),
color="Order:N",
tooltip=["Order", "Qty"],
)
.interactive()
.properties(
width=800 / len(timeline) - 22,
height=100,
title="Customer's requirement",
)
)
df = (
planning.filter(like="plannedQty", axis=0)
.copy()
.rename(columns={"Solution": "Qty"})
.reset_index()
)
df[["Date", "Order", "Line"]] = df["index"].str.split(",", expand=True)
df["Date"] = df["Date"].str.split("[").str[1]
df["Line"] = df["Line"].str.split("]").str[0]
df = df[["Date", "Line", "Qty", "Order"]]
chart_planning = (
alt.Chart(df)
.mark_bar()
.encode(
y=alt.Y("Qty", axis=alt.Axis(grid=False)),
x="Line:N",
column=alt.Column("Date:N"),
color="Order:N",
tooltip=["Line", "Order", "Qty"],
)
.interactive()
.properties(
width=800 / len(timeline) - 22,
height=200,
title="Optimized Production Schedule",
)
)
chart = alt.vconcat(chart_planning, chart_need)
chart.save("planning_MO_model4.html")
dp.Report(
dp.Plot(chart, caption="Production schedule model 4 - Qty")
).upload(name='Optimized production schedule - Qty',
description="Optimized production schedule - Qty", open=True, visibily="PUBLIC")
def plot_inventory(
planning: pd.DataFrame, timeline: List[str], cust_orders,
) -> None:
# Plot inventory
df = (
planning.filter(like="early prod", axis=0)
.copy()
.rename(columns={"Solution": "Qty"})
.reset_index()
)
df[["Date", "Order"]] = df["index"].str.split(",", expand=True)
df["Date"] = df["Date"].str.split("[").str[1]
df["Order"] = df["Order"].str.split("]").str[0]
df = df[["Date", "Qty", "Order"]]
models_list = cust_orders[['Order', 'Product_Family']]
df = pd.merge(df, models_list, on='Order', how='inner')
df = df[["Date", "Qty", "Product_Family"]]
bars = (
alt.Chart(df)
.mark_bar()
.encode(
y="Qty:Q",
color="Product_Family:N",
tooltip=["Product_Family", "Qty"],
)
.interactive()
.properties(width=550 / len(timeline) - 22, height=60)
)
chart_inventory = (
alt.layer(bars, data=df)
.facet(column="Date:N")
.properties(title="Inventory")
)
# Plot shortage
df = (
planning.filter(like="late prod", axis=0)
.copy()
.rename(columns={"Solution": "Qty"})
.reset_index()
)
df[["Date", "Order"]] = df["index"].str.split(",", expand=True)
df["Date"] = df["Date"].str.split("[").str[1]
df["Order"] = df["Order"].str.split("]").str[0]
df = df[["Date", "Qty", "Order"]]
models_list = cust_orders[['Order', 'Product_Family']]
df = pd.merge(df, models_list, on='Order', how='inner')
df = df[["Date", "Qty", "Product_Family"]]
bars = (
alt.Chart(df)
.mark_bar()
.encode(
y="Qty:Q",
color="Product_Family:N",
tooltip=["Product_Family", "Qty"],
)
.interactive()
.properties(width=550 / len(timeline) - 22, height=60)
)
chart_shortage = (
alt.layer(bars, data=df)
.facet(column="Date:N")
.properties(title="Shortage")
)
chart = alt.vconcat(chart_inventory, chart_shortage)
chart.save("Inventory_Shortage.html")
dp.Report(
dp.Plot(chart, caption="Inventory_Shortage")
).upload(name='Inventory_Shortage',
description="Inventory_Shortage", open=True, visibily="PUBLIC")
def print_planning(planning: pd.DataFrame) -> None:
df = (
planning.filter(like="plannedQty", axis=0)
.copy()
.rename(columns={"Solution": "Qty"})
.reset_index()
)
df[["Date", "Customer_Order", "Line"]] = df["index"].str.split(",", expand=True)
df["Date"] = df["Date"].str.split("[").str[1]
df["Line"] = df["Line"].str.split("]").str[0]
df = df[["Date", "Line", "Qty", "Customer_Order"]]
df.to_csv(r"Planning_model4_list.csv", index=True)
print(df)
df.pivot_table(
values="Qty", index="Customer_Order", columns=["Date", "Line"]
).to_csv(r"Planning_model4v2.csv", index=True)
def optimize_planning(
timeline: List[str],
workcenters: List[str],
needs,
wc_cost_reg: Dict[str, int],
wc_cost_ot: Dict[str, int],
wc_cost_we: Dict[str, int],
inventory_carrying_cost: int,
customer_orders: List[str],
cycle_times,
delay_cost: int,
) -> pd.DataFrame:
# Split weekdays/weekends
weekdays = []
weekend = []
for date in timeline:
day = datetime.datetime.strptime(date, "%Y/%m/%d")
if day.weekday() < 5:
weekdays.append(date)
else:
weekend.append(date)
# Initiate optimization model
model = gurobipy.Model("Optimize production planning")
# DEFINE VARIABLES
# Quantity variable
x_qty = model.addVars(
timeline,
customer_orders,
workcenters,
lb=0,
vtype=gurobipy.GRB.INTEGER,
name="plannedQty",
)
# Time variable
x_time = model.addVars(
timeline,
customer_orders,
workcenters,
lb=0,
vtype=gurobipy.GRB.CONTINUOUS,
name="plannedTime",
)
# Set the value of x_time
model.addConstrs(
(
(
x_time[(date, mo, wc)] == x_qty[(date, mo, wc)] * cycle_times[(mo, wc)]
for date in timeline
for mo in customer_orders
for wc in workcenters
)
),
name="x_time_constr",
)
# Qty to display
quantity = model.addVars(
timeline, workcenters, lb=0, vtype=gurobipy.GRB.INTEGER, name="qty"
)
# Set the value of qty
model.addConstrs(
(
(
quantity[(date, wc)]
== gurobipy.quicksum(x_qty[(date, mo, wc)] for mo in customer_orders)
for date in timeline
for wc in workcenters
)
),
name="wty_time_constr",
)
# Variable status of the line ( 0 = closed, 1 = opened)
line_opening = model.addVars(
timeline, workcenters, vtype=gurobipy.GRB.BINARY, name="Open status"
)
# Load variables (hours) - regular and overtime
reg_hours = model.addVars(
timeline,
workcenters,
vtype=gurobipy.GRB.CONTINUOUS,
name="Regular hours",
)
ot_hours = model.addVars(
timeline,
workcenters,
vtype=gurobipy.GRB.CONTINUOUS,
name="Overtime hours",
)
reg_hours_bis = model.addVars(
timeline,
workcenters,
lb=7,
ub=8,
vtype=gurobipy.GRB.CONTINUOUS,
name="regHours",
)
ot_hours_bis = model.addVars(
timeline, workcenters, lb=0, ub=4, vtype=gurobipy.GRB.CONTINUOUS, name="OTHours"
)
# Set the value of reg and OT hours)
model.addConstrs(
(
reg_hours[(date, wc)]
== reg_hours_bis[(date, wc)] * line_opening[(date, wc)]
for date in timeline
for wc in workcenters
),
name="total_hours_constr",
)
model.addConstrs(
(
ot_hours[(date, wc)] == ot_hours_bis[(date, wc)] * line_opening[(date, wc)]
for date in timeline
for wc in workcenters
),
name="total_hours_constr",
)
# Variable total load (hours)
total_hours = model.addVars(
timeline,
workcenters,
vtype=gurobipy.GRB.CONTINUOUS,
name="Total hours",
)
# Set the value of total load (regular + overtime)
model.addConstrs(
(
total_hours[(date, wc)] == (reg_hours[(date, wc)] + ot_hours[(date, wc)])
for date in timeline
for wc in workcenters
),
name="Link total hours - reg/ot hours",
)
# Set total hours of production in link with the time variable
model.addConstrs(
(
(
total_hours[(date, wc)]
== gurobipy.quicksum(x_time[(date, mo, wc)] for mo in customer_orders)
for date in timeline
for wc in workcenters
)
),
name="total_hours_constr",
)
# Variable cost
labor_cost = model.addVars(
timeline, workcenters, lb=0, vtype=gurobipy.GRB.CONTINUOUS, name="Labor cost"
)
# Set the value of cost (hours * hourly cost)
model.addConstrs(
(
labor_cost[(date, wc)]
== reg_hours[(date, wc)] * wc_cost_reg[wc]
+ ot_hours[(date, wc)] * wc_cost_ot[wc]
for date in weekdays
for wc in workcenters
),
name="Link labor cost - working hours - wd",
)
model.addConstrs(
(
labor_cost[(date, wc)]
== total_hours[(date, wc)] * wc_cost_we[wc]
for date in weekend
for wc in workcenters
),
name="Link labor cost - working hours - we",
)
# Variable gap early/late production
gap_prod = model.addVars(
timeline,
customer_orders,
lb=-10000,
ub=10000,
vtype=gurobipy.GRB.CONTINUOUS,
name="gapProd",
)
abs_gap_prod = model.addVars(
timeline,
customer_orders,
vtype=gurobipy.GRB.CONTINUOUS,
name="absGapProd",
)
# Set the value of gap for early production
for l in range(len(timeline)):
model.addConstrs(
(
gap_prod[(timeline[l], mo)]
== gurobipy.quicksum(
x_qty[(date, mo, wc)]
for date in timeline[: l + 1]
for wc in workcenters
)
- (gurobipy.quicksum(needs[(date, mo)] for date in timeline[: l + 1]))
for mo in customer_orders
),
name="gap_prod",
)
# Set the value of ABS(gap for early production)
model.addConstrs(
(
(abs_gap_prod[(date, mo)] == gurobipy.abs_(gap_prod[(date, mo)]))
for date in timeline
for mo in customer_orders
),
name="abs gap prod",
)
# Create variable "early production" and "inventory costs"
early_prod = model.addVars(
timeline,
customer_orders,
vtype=gurobipy.GRB.CONTINUOUS,
name="early prod",
)
inventory_costs = model.addVars(
timeline,
customer_orders,
vtype=gurobipy.GRB.CONTINUOUS,
name="inventory costs",
)
# Set the value of early production
model.addConstrs(
(
early_prod[(date, m)] == (gap_prod[(date, m)] + abs_gap_prod[(date, m)]) / 2
for date in timeline
for m in customer_orders
),
name="early prod",
)
# Set the value of inventory costs
model.addConstrs(
(
(inventory_costs[(date, m)] == early_prod[(date, m)] * inventory_carrying_cost)
for date in timeline
for m in customer_orders
),
name="inventory costs",
)
# Create variable "late production" and "delay costs"
late_prod = model.addVars(
timeline,
customer_orders,
vtype=gurobipy.GRB.CONTINUOUS,
name="late prod",
)
delay_costs = model.addVars(
timeline,
customer_orders,
vtype=gurobipy.GRB.CONTINUOUS,
name="inventory costs",
)
# Set the value of late production
model.addConstrs(
(
late_prod[(date, m)] == (abs_gap_prod[(date, m)] - gap_prod[(date, m)]) / 2
for date in timeline
for m in customer_orders
),
name="late prod",
)
# Set the value of delay costs
model.addConstrs(
(
(delay_costs[(date, m)] == late_prod[(date, m)] * delay_cost)
for date in timeline
for m in customer_orders
),
name="delay costs",
)
# CONSTRAINT
# Constraint: Total hours of production = required production time
model.addConstr(
(
gurobipy.quicksum(
x_qty[(date, mo, wc)]
for date in timeline
for mo in customer_orders
for wc in workcenters
)
== (gurobipy.quicksum(needs[(date, mo)] for date in timeline for mo in customer_orders))
),
name="total_req",
)
# DEFINE MODEL
# Objective : minimize a function
model.ModelSense = gurobipy.GRB.MINIMIZE
# Function to minimize
objective = 0
objective += gurobipy.quicksum(
labor_cost[(date, wc)] for date in timeline for wc in workcenters
)
objective += gurobipy.quicksum(
inventory_costs[(date, mo)]
for date in timeline
for mo in customer_orders
)
objective += gurobipy.quicksum(
delay_costs[(date, mo)]
for date in timeline
for mo in customer_orders
)
# SOLVE MODEL
model.setObjective(objective)
model.optimize()
sol = pd.DataFrame(data={"Solution": model.X}, index=model.VarName)
print("Total cost = $" + str(model.ObjVal))
# model.write("Planning_optimization.lp")
# file = open("Planning_optimization.lp", 'r')
# print(file.read())
# file.close()
return sol
def check_duplicates(list_to_check):
if len(list_to_check) == len(set(list_to_check)):
return
else:
print("Duplicate order, please check the requirements file")
exit()
return
# Define hourly cost per line - regular, overtime and weekend
reg_costs_per_line = {"Line_1": 245, "Line_2": 315, "Line_3": 245}
ot_costs_per_line = {
k: 1.5 * reg_costs_per_line[k] for k, v in reg_costs_per_line.items()
}
we_costs_per_line = {
k: 2 * reg_costs_per_line[k] for k, w in reg_costs_per_line.items()
}
storage_cost = 5
late_prod_cost = 1000
lines: List[str] = list(reg_costs_per_line.keys())
# Get orders
customer_orders = pd.read_excel("customer_orders.xlsx")
# Get cycle times
capacity = pd.read_excel("line_capacity.xlsx", sheet_name="8h capacity").set_index("Line")
cycle_time = capacity.rdiv(8)
order_list = customer_orders["Order"].to_list()
check_duplicates(order_list)
# Create cycle times dictionnary
customer_orders = customer_orders.merge(
cycle_time, left_on="Product_Family", right_index=True
)
customer_orders["Delivery_Date"] = pd.to_datetime(
customer_orders["Delivery_Date"]
).dt.strftime("%Y/%m/%d")
customer_orders = customer_orders.sort_values(by=["Delivery_Date", "Order"])
cycle_times = {
(order, line): customer_orders[line][customer_orders.Order == order].item()
for order in order_list
for line in lines
}
# Define calendar
start_date = datetime.datetime.strptime(
customer_orders["Delivery_Date"].min(), "%Y/%m/%d"
)
end_date = datetime.datetime.strptime(
customer_orders["Delivery_Date"].max(), "%Y/%m/%d"
)
date_modified = start_date
calendar = [start_date.strftime("%Y/%m/%d")]
while date_modified < end_date:
date_modified += datetime.timedelta(days=1)
calendar.append(date_modified.strftime("%Y/%m/%d"))
# Create daily requirements dictionnary
daily_requirements = {}
for day in calendar:
for order in order_list:
try:
daily_requirements[(day, order)] = customer_orders[
(customer_orders.Order == order)
& (customer_orders.Delivery_Date == day)
]["Quantity"].item()
except ValueError:
daily_requirements[(day, order)] = 0
# Optimize planning
solution = optimize_planning(
calendar,
lines,
daily_requirements,
reg_costs_per_line,
ot_costs_per_line,
we_costs_per_line,
storage_cost,
order_list,
cycle_times,
late_prod_cost,
)
# Plot the new planning
plot_load(solution, daily_requirements, calendar)
print_planning(solution)
plot_planning(solution, daily_requirements, calendar)
plot_inventory(solution, calendar, customer_orders)
| 0.796372 | 0.382805 |
___
# Logistic Regression Project
In this project we will be working with a fake advertising data set, indicating whether or not a particular internet user clicked on an Advertisement. We will try to create a model that will predict whether or not they will click on an ad based off the features of that user.
This data set contains the following features:
* 'Daily Time Spent on Site': consumer time on site in minutes
* 'Age': cutomer age in years
* 'Area Income': Avg. Income of geographical area of consumer
* 'Daily Internet Usage': Avg. minutes a day consumer is on the internet
* 'Ad Topic Line': Headline of the advertisement
* 'City': City of consumer
* 'Male': Whether or not consumer was male
* 'Country': Country of consumer
* 'Timestamp': Time at which consumer clicked on Ad or closed window
* 'Clicked on Ad': 0 or 1 indicated clicking on Ad
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report
```
## Get the Data
**Read in the advertising.csv file and set it to a data frame called ad_data.**
```
adv_data = pd.read_csv('/home/abhishekranjan/Downloads/Py_DS_ML_Bootcamp-master/Refactored_Py_DS_ML_Bootcamp-master/13-Logistic-Regression/advertising.csv')
```
**Check the head of ad_data**
```
adv_data.head()
adv_data.info()
adv_data.describe()
```
## Exploratory Data Analysis
Let's use seaborn to explore the data!
Try recreating the plots shown below!
** Create a histogram of the Age**
```
sns.set_style('whitegrid')
sns.histplot(data = adv_data['Age'], bins = 30)
```
**Create a jointplot showing Area Income versus Age.**
```
sns.set_style('whitegrid')
sns.jointplot(x = adv_data['Age'], y = adv_data['Daily Time Spent on Site'], )
```
**Create a jointplot showing the kde distributions of Daily Time spent on site vs. Age.**
```
# sns.set_style('whitegrid')
sns.jointplot(x='Age',y='Daily Time Spent on Site',data=adv_data,color='red',kind='kde')
```
** Create a jointplot of 'Daily Time Spent on Site' vs. 'Daily Internet Usage'**
```
sns.jointplot(x= 'Daily Time Spent on Site', y = 'Daily Internet Usage', data = adv_data, color = 'green', kind= 'scatter')
```
** Finally, create a pairplot with the hue defined by the 'Clicked on Ad' column feature.**
```
sns.pairplot(data = adv_data, hue = 'Clicked on Ad')
```
# Logistic Regression
Now it's time to do a train test split, and train our model!
we have to predict whether the user will click on a particular ad or not. so our predicted column will be 'click on ad'.
** Split the data into training set and testing set using train_test_split**
```
X = adv_data[['Daily Time Spent on Site', 'Age', 'Area Income','Daily Internet Usage', 'Male']]
y = adv_data['Clicked on Ad']
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=101)
```
** Train and fit a logistic regression model on the training set.**
```
logisticPrediction = LogisticRegression(max_iter=500)
logisticPrediction.fit(X_train, y_train )
```
## Predictions and Evaluations
** Now predict values for the testing data.**
```
predictedVal = logisticPrediction.predict(X_test)
```
** Create a classification report for the model.**
```
#max iteration = 150
print(classification_report(y_test, predictedVal))
#max iteration = 500
print(classification_report(y_test, predictedVal))
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test, predictedVal)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report
adv_data = pd.read_csv('/home/abhishekranjan/Downloads/Py_DS_ML_Bootcamp-master/Refactored_Py_DS_ML_Bootcamp-master/13-Logistic-Regression/advertising.csv')
adv_data.head()
adv_data.info()
adv_data.describe()
sns.set_style('whitegrid')
sns.histplot(data = adv_data['Age'], bins = 30)
sns.set_style('whitegrid')
sns.jointplot(x = adv_data['Age'], y = adv_data['Daily Time Spent on Site'], )
# sns.set_style('whitegrid')
sns.jointplot(x='Age',y='Daily Time Spent on Site',data=adv_data,color='red',kind='kde')
sns.jointplot(x= 'Daily Time Spent on Site', y = 'Daily Internet Usage', data = adv_data, color = 'green', kind= 'scatter')
sns.pairplot(data = adv_data, hue = 'Clicked on Ad')
X = adv_data[['Daily Time Spent on Site', 'Age', 'Area Income','Daily Internet Usage', 'Male']]
y = adv_data['Clicked on Ad']
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=101)
logisticPrediction = LogisticRegression(max_iter=500)
logisticPrediction.fit(X_train, y_train )
predictedVal = logisticPrediction.predict(X_test)
#max iteration = 150
print(classification_report(y_test, predictedVal))
#max iteration = 500
print(classification_report(y_test, predictedVal))
from sklearn.metrics import confusion_matrix
confusion_matrix(y_test, predictedVal)
| 0.428951 | 0.956391 |
```
import numpy as np
import pandas as pd #Dataframe
from matplotlib import pyplot as plt #graph
import seaborn as sns #graph
sns.set(color_codes=True)
from sklearn.model_selection import train_test_split #Split Data into train-test
from sklearn.ensemble import RandomForestClassifier as Rf #RandomForestClassifer
from sklearn.preprocessing import LabelEncoder #label encoder to change value
from sklearn.metrics import confusion_matrix # consfusion matrix for true positive
from sklearn.metrics import accuracy_score # accuracy score
from sklearn.metrics import classification_report
df_train = pd.read_csv("train/train.csv")
df_train.head()
df_train.isnull().sum()
df_train.dtypes
df_train.columns
```
### Item Drop
- ID
- Year
- Year Type
- Organization Group Code
- department code
- union code
- job family code
- job code
- Employee Indentifier
- total benifits
```
df_train.shape
df_train["Class"].unique()
sns.countplot(df_train['Class'])
df_train.groupby('Department')['Class'].value_counts(normalize=True)
df_train.describe()
#Unique Value in Job Column
df_train['Job'].unique()
df_train.groupby('Job')['Union'].max()
# No. of job by post
df_train['Job'].value_counts()[:10]
plt.figure(figsize=(15,6))
plt.plot(df_train['Job'].value_counts()[:10])
plt.xticks(rotation=45)
df_train['Union'].value_counts()[:10]
plt.figure(figsize=(15,6))
plt.plot(df_train['Union'].value_counts()[:10])
plt.xticks(rotation=45)
df_train.groupby('Class')['Overtime'].max()
plt.figure(figsize=(12,6))
sns.countplot(df_train['Job'])
plt.xticks(rotation=45)
df_train.groupby("Class")['Job'].max()
df_youth=df_train.loc[df_train['Job'] == "Youth Comm Advisor"]
mean_class_wise = df_youth.groupby("Class")["Total Benefits"].mean()
mean_class_wise
df_train.groupby("Class")["Health/Dental"].mean()
df_train.drop(['ID' ,'Year' ,'Year Type','Organization Group Code','Department Code','Union Code','Job Family Code','Job Code','Employee Identifier','Total Benefits'],1,inplace=True)
df_train.columns
```
## Label Encoding
- Organization Group
- Department
- Union
- Job Family
```
le = LabelEncoder()
le.fit(df_train['Organization Group'])
df_train['Organization Group'] = le.transform(df_train['Organization Group'])
le.fit(df_train['Department'])
df_train['Department'] = le.transform(df_train['Department'])
le.fit(df_train['Union'])
df_train['Union'] = le.transform(df_train['Union'])
le.fit(df_train['Job Family'])
df_train['Job Family'] = le.transform(df_train['Job Family'])
le.fit(df_train['Job'])
df_train['Job'] = le.transform(df_train['Job'])
df_train.head()
X = df_train[['Organization Group', 'Department', 'Union', 'Job Family', 'Job',
'Overtime', 'Other Salaries', 'Retirement', 'Health/Dental',
'Other Benefits']]
y = df_train["Class"]
train_X,test_X,train_y, test_y =train_test_split(X, y , test_size = 0.2,random_state = 0)
rf = Rf(n_estimators =10)
rf.fit(train_X,train_y)
predict_y = rf.predict(test_X)
print(f'Confusion Matrix: \n {confusion_matrix(test_y,predict_y)}')
print(f"Train Accuracy Score : {accuracy_score(test_y,predict_y)}")
print(f'Classification Report : \n\n {classification_report(test_y,predict_y)}')
```
|
github_jupyter
|
import numpy as np
import pandas as pd #Dataframe
from matplotlib import pyplot as plt #graph
import seaborn as sns #graph
sns.set(color_codes=True)
from sklearn.model_selection import train_test_split #Split Data into train-test
from sklearn.ensemble import RandomForestClassifier as Rf #RandomForestClassifer
from sklearn.preprocessing import LabelEncoder #label encoder to change value
from sklearn.metrics import confusion_matrix # consfusion matrix for true positive
from sklearn.metrics import accuracy_score # accuracy score
from sklearn.metrics import classification_report
df_train = pd.read_csv("train/train.csv")
df_train.head()
df_train.isnull().sum()
df_train.dtypes
df_train.columns
df_train.shape
df_train["Class"].unique()
sns.countplot(df_train['Class'])
df_train.groupby('Department')['Class'].value_counts(normalize=True)
df_train.describe()
#Unique Value in Job Column
df_train['Job'].unique()
df_train.groupby('Job')['Union'].max()
# No. of job by post
df_train['Job'].value_counts()[:10]
plt.figure(figsize=(15,6))
plt.plot(df_train['Job'].value_counts()[:10])
plt.xticks(rotation=45)
df_train['Union'].value_counts()[:10]
plt.figure(figsize=(15,6))
plt.plot(df_train['Union'].value_counts()[:10])
plt.xticks(rotation=45)
df_train.groupby('Class')['Overtime'].max()
plt.figure(figsize=(12,6))
sns.countplot(df_train['Job'])
plt.xticks(rotation=45)
df_train.groupby("Class")['Job'].max()
df_youth=df_train.loc[df_train['Job'] == "Youth Comm Advisor"]
mean_class_wise = df_youth.groupby("Class")["Total Benefits"].mean()
mean_class_wise
df_train.groupby("Class")["Health/Dental"].mean()
df_train.drop(['ID' ,'Year' ,'Year Type','Organization Group Code','Department Code','Union Code','Job Family Code','Job Code','Employee Identifier','Total Benefits'],1,inplace=True)
df_train.columns
le = LabelEncoder()
le.fit(df_train['Organization Group'])
df_train['Organization Group'] = le.transform(df_train['Organization Group'])
le.fit(df_train['Department'])
df_train['Department'] = le.transform(df_train['Department'])
le.fit(df_train['Union'])
df_train['Union'] = le.transform(df_train['Union'])
le.fit(df_train['Job Family'])
df_train['Job Family'] = le.transform(df_train['Job Family'])
le.fit(df_train['Job'])
df_train['Job'] = le.transform(df_train['Job'])
df_train.head()
X = df_train[['Organization Group', 'Department', 'Union', 'Job Family', 'Job',
'Overtime', 'Other Salaries', 'Retirement', 'Health/Dental',
'Other Benefits']]
y = df_train["Class"]
train_X,test_X,train_y, test_y =train_test_split(X, y , test_size = 0.2,random_state = 0)
rf = Rf(n_estimators =10)
rf.fit(train_X,train_y)
predict_y = rf.predict(test_X)
print(f'Confusion Matrix: \n {confusion_matrix(test_y,predict_y)}')
print(f"Train Accuracy Score : {accuracy_score(test_y,predict_y)}")
print(f'Classification Report : \n\n {classification_report(test_y,predict_y)}')
| 0.558086 | 0.579787 |
# Collaboratibe Filtering (Item-Item) : Movie Recommendation System
We are creating a collaborative filtering based movie recommendatipon system where are trying to create user-movie rating matrix, where ratings given by users are playing features/ patterns role and based on nearest neighbours algorithm finding closest movies based on those patterns. Kind of finding similiar movies/ item-item similarity based recommendation system.
## References
https://www.geeksforgeeks.org/recommendation-system-in-python/
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
```
## Loading data
### Ratings
```
ratings = pd.read_csv("./datasets/ratings.csv")
ratings.head()
```
### Movies
```
movies = pd.read_csv("./datasets/movies.csv")
movies.head()
```
## General Shapes Information
```
print(f"""
ratings dataframe shape :{ratings.shape}
movies dataframe shape :{movies.shape}
total movies : {movies.movieId.unique().shape}
total rated movies : {ratings.movieId.unique().shape}
total users : {ratings.userId.unique().shape}
All kind of ratings given by users : {ratings.rating.unique()}""")
```
## Get unique ids
```
all_user_ids = ratings.userId.unique()
all_movie_ids = movies.movieId.unique()
```
## Creating Maps
### Map to convert user id to user mapping id
```
user_id_maps = { i[0]:i[1] for i in enumerate(all_user_ids) }
```
### Map to convert user mapping id to user id
```
user_id_inv_maps = { user_id_maps[i]:i for i in user_id_maps }
```
### Map to convert movie id to movie mapping id
```
movie_id_maps = { i[0]:i[1] for i in enumerate(all_movie_ids) }
```
### Map to convert movie mapping id to movie id
```
movie_id_inv_maps = { movie_id_maps[i]:i for i in movie_id_maps }
```
## user-movie average rating
```
user_movie_ratings_df = ratings[['userId','movieId','rating']]\
.groupby(by=['userId', 'movieId'],as_index=False)\
.mean()
user_movie_ratings_df.columns
user_movie_ratings_df.head()
```
## Converting existing ids to mapping id
```
user_movie_ratings_df['user_maps'] = user_movie_ratings_df['userId'].apply(lambda x: user_id_inv_maps[x])
user_movie_ratings_df['movie_maps'] = user_movie_ratings_df['movieId'].apply(lambda x: movie_id_inv_maps[x])
user_movie_ratings_df.head()
```
## Creating User-Movie Rating Matrix
```
from scipy.sparse import csr_matrix
user_maps = user_movie_ratings_df.user_maps.values
movie_maps = user_movie_ratings_df.movie_maps.values
rating_values = user_movie_ratings_df.rating.values
rating_matrix = csr_matrix((rating_values, (movie_maps, user_maps))).toarray()
rating_matrix
```
So How is this working actually
```
user_maps = [0 1 2 3 4 5 6 7 8 9 10]
movie_maps = [0 1 2 3 4 5 6 7 8 9 10]
ratings = [3 4 1 5 5 2 5 1 1 4]
Users maps
0 1 2 3 4 5 6 7 8 9 10
_ _ _ _ _ _ _ _ _ _ _
0 |3 0 0 0 0 0 0 0
1 |0 4 0 0 0 0 0
movies maps 2 |0 0 1 0 0 0 .
3 |0 0 0 5 0 0 .
4 |0 0 0 0 5 0 .
5 |0 0 0 0 0 2
6 |0 0 0 0 0 0
7 |. . . .
8 |. .
```
```
rating_matrix.shape
```
## Clustering Model
```
from sklearn.neighbors import NearestNeighbors
n_neighbors = 10
metric = 'cosine'
kNN = NearestNeighbors(n_neighbors= n_neighbors, algorithm='brute', metric=metric)
kNN.fit(rating_matrix)
```
## Testing Model
```
movie_id = 1
movies.query(f'movieId == {movie_id}')
input_movie_vector = rating_matrix[movie_id_inv_maps[movie_id]].reshape(1,-1)
print("Input Movie Vector:",input_movie_vector.shape)
distances, collected_neighbours = kNN.kneighbors(
X=input_movie_vector, n_neighbors=n_neighbors, return_distance=True)
iter_map = map(lambda x: movie_id_maps[x], collected_neighbours[0])
recoms = movies[movies.movieId.isin(iter_map)].reset_index()
recoms['distances'] = distances[0]
print("\nRecommendations :")
recoms.head(n_neighbors)
```
## Plotting distances
```
fig, ax = plt.subplots(1,1, figsize=(10,5))
sns.barplot(data=recoms, y='title', x='distances', orient='h')
plt.show()
```
## Recommendation System Modelling Class
```
from utils.engine import BasicMovieRecommedation
from utils.plot_utils import plot_recommendations
model = BasicMovieRecommedation(n_neighbors=20)
model.fit(rating_df=ratings[['userId','movieId','rating']], movie_df=movies[['movieId']])
movie_input = 1
print(movies[movies.movieId.isin([movie_input])])
movie_ids, distances = model.get_recommendations(movie_input, 5)
recommendations = movies[movies.movieId.isin(movie_ids)].reset_index()
recommendations['distances'] = distances
plot_recommendations(recommendations)
movie_input = 50
print(movies[movies.movieId.isin([movie_input])])
movie_ids, distances = model.get_recommendations(movie_input, 5)
recommendations = movies[movies.movieId.isin(movie_ids)].reset_index()
recommendations['distances'] = distances
plot_recommendations(recommendations)
movie_input = 605
print(movies[movies.movieId.isin([movie_input])])
movie_ids, distances = model.get_recommendations(movie_input, 5)
recommendations = movies[movies.movieId.isin(movie_ids)].reset_index()
recommendations['distances'] = distances
plot_recommendations(recommendations)
movie_input = 999
print(movies[movies.movieId.isin([movie_input])])
movie_ids, distances = model.get_recommendations(movie_input, 5)
recommendations = movies[movies.movieId.isin(movie_ids)].reset_index()
recommendations['distances'] = distances
plot_recommendations(recommendations)
movie_input = 189333
print(movies[movies.movieId.isin([movie_input])])
movie_ids, distances = model.get_recommendations(movie_input, 5)
recommendations = movies[movies.movieId.isin(movie_ids)].reset_index()
recommendations['distances'] = distances
plot_recommendations(recommendations)
movie_input = 187593
print(movies[movies.movieId.isin([movie_input])])
movie_ids, distances = model.get_recommendations(movie_input, 5)
recommendations = movies[movies.movieId.isin(movie_ids)].reset_index()
recommendations['distances'] = distances
plot_recommendations(recommendations)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
ratings = pd.read_csv("./datasets/ratings.csv")
ratings.head()
movies = pd.read_csv("./datasets/movies.csv")
movies.head()
print(f"""
ratings dataframe shape :{ratings.shape}
movies dataframe shape :{movies.shape}
total movies : {movies.movieId.unique().shape}
total rated movies : {ratings.movieId.unique().shape}
total users : {ratings.userId.unique().shape}
All kind of ratings given by users : {ratings.rating.unique()}""")
all_user_ids = ratings.userId.unique()
all_movie_ids = movies.movieId.unique()
user_id_maps = { i[0]:i[1] for i in enumerate(all_user_ids) }
user_id_inv_maps = { user_id_maps[i]:i for i in user_id_maps }
movie_id_maps = { i[0]:i[1] for i in enumerate(all_movie_ids) }
movie_id_inv_maps = { movie_id_maps[i]:i for i in movie_id_maps }
user_movie_ratings_df = ratings[['userId','movieId','rating']]\
.groupby(by=['userId', 'movieId'],as_index=False)\
.mean()
user_movie_ratings_df.columns
user_movie_ratings_df.head()
user_movie_ratings_df['user_maps'] = user_movie_ratings_df['userId'].apply(lambda x: user_id_inv_maps[x])
user_movie_ratings_df['movie_maps'] = user_movie_ratings_df['movieId'].apply(lambda x: movie_id_inv_maps[x])
user_movie_ratings_df.head()
from scipy.sparse import csr_matrix
user_maps = user_movie_ratings_df.user_maps.values
movie_maps = user_movie_ratings_df.movie_maps.values
rating_values = user_movie_ratings_df.rating.values
rating_matrix = csr_matrix((rating_values, (movie_maps, user_maps))).toarray()
rating_matrix
## Clustering Model
## Testing Model
## Plotting distances
## Recommendation System Modelling Class
| 0.297776 | 0.932515 |
# Data Cleaning and Preparation
```
import numpy as np
import pandas as pd
PREVIOUS_MAX_ROWS = pd.options.display.max_rows
pd.options.display.max_rows = 20
np.random.seed(12345)
import matplotlib.pyplot as plt
plt.rc('figure', figsize=(10, 6))
np.set_printoptions(precision=4, suppress=True)
```
## Handling Missing Data
```
string_data = pd.Series(['aardvark', 'artichoke', np.nan, 'avocado'])
string_data
string_data.isnull()
string_data[0] = None
string_data.isnull()
```
### Filtering Out Missing Data
```
from numpy import nan as NA
data = pd.Series([1, NA, 3.5, NA, 7])
data.dropna()
data[data.notnull()]
data = pd.DataFrame([[1., 6.5, 3.], [1., NA, NA],
[NA, NA, NA], [NA, 6.5, 3.]])
cleaned = data.dropna()
data
cleaned
data.dropna(how='all')
data[4] = NA
data
data.dropna(axis=1, how='all')
df = pd.DataFrame(np.random.randn(7, 3))
df.iloc[:4, 1] = NA
df.iloc[:2, 2] = NA
df
df.dropna()
df.dropna(thresh=2)
```
### Filling In Missing Data
```
df.fillna(0)
df.fillna({1: 0.5, 2: 0})
_ = df.fillna(0, inplace=True)
df
df = pd.DataFrame(np.random.randn(6, 3))
df.iloc[2:, 1] = NA
df.iloc[4:, 2] = NA
df
df.fillna(method='ffill')
df.fillna(method='ffill', limit=2)
data = pd.Series([1., NA, 3.5, NA, 7])
data.fillna(data.mean())
```
## Data Transformation
### Removing Duplicates
```
data = pd.DataFrame({'k1': ['one', 'two'] * 3 + ['two'],
'k2': [1, 1, 2, 3, 3, 4, 4]})
data
data.duplicated()
data.drop_duplicates()
data['v1'] = range(7)
data.drop_duplicates(['k1'])
data.drop_duplicates(['k1', 'k2'], keep='last')
```
### Transforming Data Using a Function or Mapping
```
data = pd.DataFrame({'food': ['bacon', 'pulled pork', 'bacon',
'Pastrami', 'corned beef', 'Bacon',
'pastrami', 'honey ham', 'nova lox'],
'ounces': [4, 3, 12, 6, 7.5, 8, 3, 5, 6]})
data
meat_to_animal = {
'bacon': 'pig',
'pulled pork': 'pig',
'pastrami': 'cow',
'corned beef': 'cow',
'honey ham': 'pig',
'nova lox': 'salmon'
}
lowercased = data['food'].str.lower()
lowercased
data['animal'] = lowercased.map(meat_to_animal)
data
data['food'].map(lambda x: meat_to_animal[x.lower()])
```
### Replacing Values
```
data = pd.Series([1., -999., 2., -999., -1000., 3.])
data
data.replace(-999, np.nan)
data.replace([-999, -1000], np.nan)
data.replace([-999, -1000], [np.nan, 0])
data.replace({-999: np.nan, -1000: 0})
```
### Renaming Axis Indexes
```
data = pd.DataFrame(np.arange(12).reshape((3, 4)),
index=['Ohio', 'Colorado', 'New York'],
columns=['one', 'two', 'three', 'four'])
transform = lambda x: x[:4].upper()
data.index.map(transform)
data.index = data.index.map(transform)
data
data.rename(index=str.title, columns=str.upper)
data.rename(index={'OHIO': 'INDIANA'},
columns={'three': 'peekaboo'})
data.rename(index={'OHIO': 'INDIANA'}, inplace=True)
data
```
### Discretization and Binning
```
ages = [20, 22, 25, 27, 21, 23, 37, 31, 61, 45, 41, 32]
bins = [18, 25, 35, 60, 100]
cats = pd.cut(ages, bins)
cats
cats.codes
cats.categories
pd.value_counts(cats)
pd.cut(ages, [18, 26, 36, 61, 100], right=False)
group_names = ['Youth', 'YoungAdult', 'MiddleAged', 'Senior']
pd.cut(ages, bins, labels=group_names)
data = np.random.rand(20)
pd.cut(data, 4, precision=2)
data = np.random.randn(1000) # Normally distributed
cats = pd.qcut(data, 4) # Cut into quartiles
cats
pd.value_counts(cats)
pd.qcut(data, [0, 0.1, 0.5, 0.9, 1.])
```
### Detecting and Filtering Outliers
```
data = pd.DataFrame(np.random.randn(1000, 4))
data.describe()
col = data[2]
col[np.abs(col) > 3]
data[(np.abs(data) > 3).any(1)]
data[np.abs(data) > 3] = np.sign(data) * 3
data.describe()
np.sign(data).head()
```
### Permutation and Random Sampling
```
df = pd.DataFrame(np.arange(5 * 4).reshape((5, 4)))
sampler = np.random.permutation(5)
sampler
df
df.take(sampler)
df.sample(n=3)
choices = pd.Series([5, 7, -1, 6, 4])
draws = choices.sample(n=10, replace=True)
draws
```
### Computing Indicator/Dummy Variables
```
df = pd.DataFrame({'key': ['b', 'b', 'a', 'c', 'a', 'b'],
'data1': range(6)})
pd.get_dummies(df['key'])
dummies = pd.get_dummies(df['key'], prefix='key')
df_with_dummy = df[['data1']].join(dummies)
df_with_dummy
mnames = ['movie_id', 'title', 'genres']
movies = pd.read_table('datasets/movielens/movies.dat', sep='::',
header=None, names=mnames)
movies[:10]
all_genres = []
for x in movies.genres:
all_genres.extend(x.split('|'))
genres = pd.unique(all_genres)
genres
zero_matrix = np.zeros((len(movies), len(genres)))
dummies = pd.DataFrame(zero_matrix, columns=genres)
gen = movies.genres[0]
gen.split('|')
dummies.columns.get_indexer(gen.split('|'))
for i, gen in enumerate(movies.genres):
indices = dummies.columns.get_indexer(gen.split('|'))
dummies.iloc[i, indices] = 1
movies_windic = movies.join(dummies.add_prefix('Genre_'))
movies_windic.iloc[0]
np.random.seed(12345)
values = np.random.rand(10)
values
bins = [0, 0.2, 0.4, 0.6, 0.8, 1]
pd.get_dummies(pd.cut(values, bins))
```
## String Manipulation
### String Object Methods
```
val = 'a,b, guido'
val.split(',')
pieces = [x.strip() for x in val.split(',')]
pieces
first, second, third = pieces
first + '::' + second + '::' + third
'::'.join(pieces)
'guido' in val
val.index(',')
val.find(':')
val.index(':')
val.count(',')
val.replace(',', '::')
val.replace(',', '')
```
### Regular Expressions
```
import re
text = "foo bar\t baz \tqux"
re.split('\s+', text)
regex = re.compile('\s+')
regex.split(text)
regex.findall(text)
text = """Dave [email protected]
Steve [email protected]
Rob [email protected]
Ryan [email protected]
"""
pattern = r'[A-Z0-9._%+-]+@[A-Z0-9.-]+\.[A-Z]{2,4}'
# re.IGNORECASE makes the regex case-insensitive
regex = re.compile(pattern, flags=re.IGNORECASE)
regex.findall(text)
m = regex.search(text)
m
text[m.start():m.end()]
print(regex.match(text))
print(regex.sub('REDACTED', text))
pattern = r'([A-Z0-9._%+-]+)@([A-Z0-9.-]+)\.([A-Z]{2,4})'
regex = re.compile(pattern, flags=re.IGNORECASE)
m = regex.match('[email protected]')
m.groups()
regex.findall(text)
print(regex.sub(r'Username: \1, Domain: \2, Suffix: \3', text))
```
### Vectorized String Functions in pandas
```
data = {'Dave': '[email protected]', 'Steve': '[email protected]',
'Rob': '[email protected]', 'Wes': np.nan}
data = pd.Series(data)
data
data.isnull()
data.str.contains('gmail')
pattern
data.str.findall(pattern, flags=re.IGNORECASE)
matches = data.str.match(pattern, flags=re.IGNORECASE)
matches
matches.str.get(1)
matches.str[0]
data.str[:5]
pd.options.display.max_rows = PREVIOUS_MAX_ROWS
```
## Conclusion
|
github_jupyter
|
import numpy as np
import pandas as pd
PREVIOUS_MAX_ROWS = pd.options.display.max_rows
pd.options.display.max_rows = 20
np.random.seed(12345)
import matplotlib.pyplot as plt
plt.rc('figure', figsize=(10, 6))
np.set_printoptions(precision=4, suppress=True)
string_data = pd.Series(['aardvark', 'artichoke', np.nan, 'avocado'])
string_data
string_data.isnull()
string_data[0] = None
string_data.isnull()
from numpy import nan as NA
data = pd.Series([1, NA, 3.5, NA, 7])
data.dropna()
data[data.notnull()]
data = pd.DataFrame([[1., 6.5, 3.], [1., NA, NA],
[NA, NA, NA], [NA, 6.5, 3.]])
cleaned = data.dropna()
data
cleaned
data.dropna(how='all')
data[4] = NA
data
data.dropna(axis=1, how='all')
df = pd.DataFrame(np.random.randn(7, 3))
df.iloc[:4, 1] = NA
df.iloc[:2, 2] = NA
df
df.dropna()
df.dropna(thresh=2)
df.fillna(0)
df.fillna({1: 0.5, 2: 0})
_ = df.fillna(0, inplace=True)
df
df = pd.DataFrame(np.random.randn(6, 3))
df.iloc[2:, 1] = NA
df.iloc[4:, 2] = NA
df
df.fillna(method='ffill')
df.fillna(method='ffill', limit=2)
data = pd.Series([1., NA, 3.5, NA, 7])
data.fillna(data.mean())
data = pd.DataFrame({'k1': ['one', 'two'] * 3 + ['two'],
'k2': [1, 1, 2, 3, 3, 4, 4]})
data
data.duplicated()
data.drop_duplicates()
data['v1'] = range(7)
data.drop_duplicates(['k1'])
data.drop_duplicates(['k1', 'k2'], keep='last')
data = pd.DataFrame({'food': ['bacon', 'pulled pork', 'bacon',
'Pastrami', 'corned beef', 'Bacon',
'pastrami', 'honey ham', 'nova lox'],
'ounces': [4, 3, 12, 6, 7.5, 8, 3, 5, 6]})
data
meat_to_animal = {
'bacon': 'pig',
'pulled pork': 'pig',
'pastrami': 'cow',
'corned beef': 'cow',
'honey ham': 'pig',
'nova lox': 'salmon'
}
lowercased = data['food'].str.lower()
lowercased
data['animal'] = lowercased.map(meat_to_animal)
data
data['food'].map(lambda x: meat_to_animal[x.lower()])
data = pd.Series([1., -999., 2., -999., -1000., 3.])
data
data.replace(-999, np.nan)
data.replace([-999, -1000], np.nan)
data.replace([-999, -1000], [np.nan, 0])
data.replace({-999: np.nan, -1000: 0})
data = pd.DataFrame(np.arange(12).reshape((3, 4)),
index=['Ohio', 'Colorado', 'New York'],
columns=['one', 'two', 'three', 'four'])
transform = lambda x: x[:4].upper()
data.index.map(transform)
data.index = data.index.map(transform)
data
data.rename(index=str.title, columns=str.upper)
data.rename(index={'OHIO': 'INDIANA'},
columns={'three': 'peekaboo'})
data.rename(index={'OHIO': 'INDIANA'}, inplace=True)
data
ages = [20, 22, 25, 27, 21, 23, 37, 31, 61, 45, 41, 32]
bins = [18, 25, 35, 60, 100]
cats = pd.cut(ages, bins)
cats
cats.codes
cats.categories
pd.value_counts(cats)
pd.cut(ages, [18, 26, 36, 61, 100], right=False)
group_names = ['Youth', 'YoungAdult', 'MiddleAged', 'Senior']
pd.cut(ages, bins, labels=group_names)
data = np.random.rand(20)
pd.cut(data, 4, precision=2)
data = np.random.randn(1000) # Normally distributed
cats = pd.qcut(data, 4) # Cut into quartiles
cats
pd.value_counts(cats)
pd.qcut(data, [0, 0.1, 0.5, 0.9, 1.])
data = pd.DataFrame(np.random.randn(1000, 4))
data.describe()
col = data[2]
col[np.abs(col) > 3]
data[(np.abs(data) > 3).any(1)]
data[np.abs(data) > 3] = np.sign(data) * 3
data.describe()
np.sign(data).head()
df = pd.DataFrame(np.arange(5 * 4).reshape((5, 4)))
sampler = np.random.permutation(5)
sampler
df
df.take(sampler)
df.sample(n=3)
choices = pd.Series([5, 7, -1, 6, 4])
draws = choices.sample(n=10, replace=True)
draws
df = pd.DataFrame({'key': ['b', 'b', 'a', 'c', 'a', 'b'],
'data1': range(6)})
pd.get_dummies(df['key'])
dummies = pd.get_dummies(df['key'], prefix='key')
df_with_dummy = df[['data1']].join(dummies)
df_with_dummy
mnames = ['movie_id', 'title', 'genres']
movies = pd.read_table('datasets/movielens/movies.dat', sep='::',
header=None, names=mnames)
movies[:10]
all_genres = []
for x in movies.genres:
all_genres.extend(x.split('|'))
genres = pd.unique(all_genres)
genres
zero_matrix = np.zeros((len(movies), len(genres)))
dummies = pd.DataFrame(zero_matrix, columns=genres)
gen = movies.genres[0]
gen.split('|')
dummies.columns.get_indexer(gen.split('|'))
for i, gen in enumerate(movies.genres):
indices = dummies.columns.get_indexer(gen.split('|'))
dummies.iloc[i, indices] = 1
movies_windic = movies.join(dummies.add_prefix('Genre_'))
movies_windic.iloc[0]
np.random.seed(12345)
values = np.random.rand(10)
values
bins = [0, 0.2, 0.4, 0.6, 0.8, 1]
pd.get_dummies(pd.cut(values, bins))
val = 'a,b, guido'
val.split(',')
pieces = [x.strip() for x in val.split(',')]
pieces
first, second, third = pieces
first + '::' + second + '::' + third
'::'.join(pieces)
'guido' in val
val.index(',')
val.find(':')
val.index(':')
val.count(',')
val.replace(',', '::')
val.replace(',', '')
import re
text = "foo bar\t baz \tqux"
re.split('\s+', text)
regex = re.compile('\s+')
regex.split(text)
regex.findall(text)
text = """Dave [email protected]
Steve [email protected]
Rob [email protected]
Ryan [email protected]
"""
pattern = r'[A-Z0-9._%+-]+@[A-Z0-9.-]+\.[A-Z]{2,4}'
# re.IGNORECASE makes the regex case-insensitive
regex = re.compile(pattern, flags=re.IGNORECASE)
regex.findall(text)
m = regex.search(text)
m
text[m.start():m.end()]
print(regex.match(text))
print(regex.sub('REDACTED', text))
pattern = r'([A-Z0-9._%+-]+)@([A-Z0-9.-]+)\.([A-Z]{2,4})'
regex = re.compile(pattern, flags=re.IGNORECASE)
m = regex.match('[email protected]')
m.groups()
regex.findall(text)
print(regex.sub(r'Username: \1, Domain: \2, Suffix: \3', text))
data = {'Dave': '[email protected]', 'Steve': '[email protected]',
'Rob': '[email protected]', 'Wes': np.nan}
data = pd.Series(data)
data
data.isnull()
data.str.contains('gmail')
pattern
data.str.findall(pattern, flags=re.IGNORECASE)
matches = data.str.match(pattern, flags=re.IGNORECASE)
matches
matches.str.get(1)
matches.str[0]
data.str[:5]
pd.options.display.max_rows = PREVIOUS_MAX_ROWS
| 0.221519 | 0.917266 |
# Use AutoAI with Watson Studio project `ibm-watson-machine-learning`
This notebook contains the steps and code to demonstrate support of AutoAI experiments in Watson Machine Learning service inside Watson Studio's projects. It introduces commands for data retrieval, training experiments and scoring.
Some familiarity with Python is helpful. This notebook uses Python 3.7.
## Learning goals
The learning goals of this notebook are:
- Work with Watson Machine Learning experiments to train AutoAI model using Watson Studio project.
- Online, Batch deployment and score the trained model trained.
## Contents
This notebook contains the following parts:
1. [Setup](#setup)
2. [Optimizer definition](#definition)
3. [Experiment Run](#run)
4. [Deploy and Score](#scoring)
5. [Clean up](#cleanup)
6. [Summary and next steps](#summary)
<a id="setup"></a>
## 1. Set up the environment
Before you use the sample code in this notebook, you must perform the following setup tasks:
- Contact with your Cloud Pack for Data administrator and ask him for your account credentials
### Connection to WML
Authenticate the Watson Machine Learning service on IBM Cloud Pack for Data. You need to provide platform `url`, your `username` and `api_key`.
```
username = 'PASTE YOUR USERNAME HERE'
api_key = 'PASTE YOUR API_KEY HERE'
url = 'PASTE THE PLATFORM URL HERE'
wml_credentials = {
"username": username,
"apikey": api_key,
"url": url,
"instance_id": 'openshift',
"version": '4.0'
}
```
Alternatively you can use `username` and `password` to authenticate WML services.
```
wml_credentials = {
"username": ***,
"password": ***,
"url": ***,
"instance_id": 'openshift',
"version": '4.0'
}
```
### Install and import the `ibm-watson-machine-learning` package
**Note:** `ibm-watson-machine-learning` documentation can be found <a href="http://ibm-wml-api-pyclient.mybluemix.net/" target="_blank" rel="noopener no referrer">here</a>.
```
!pip install -U ibm-watson-machine-learning | tail -n 1
from ibm_watson_machine_learning import APIClient
client = APIClient(wml_credentials)
```
### Working with spaces
First of all, you need to create a space that will be used for your work. If you do not have space already created, you can use `{PLATFORM_URL}/ml-runtime/spaces?context=icp4data` to create one.
- Click New Deployment Space
- Create an empty space
- Go to space `Settings` tab
- Copy `space_id` and paste it below
**Tip**: You can also use SDK to prepare the space for your work. More information can be found [here](https://github.com/IBM/watson-machine-learning-samples/blob/master/cpd4.0/notebooks/python_sdk/instance-management/Space%20management.ipynb).
**Action**: Assign space ID below
```
space_id = 'PASTE YOUR SPACE ID HERE'
```
You can use the `list` method to print all existing spaces.
```
client.spaces.list(limit=10)
```
### Working with projects
First of all, you need to create a project that will be used for your work. If you do not have space already created follow bellow steps.
- Open IBM Cloud Pak main page
- Click all projects
- Create an empty project
- Copy `project_id` from url and paste it below
**Action**: Assign project ID below
```
project_id = 'PASTE YOUR PROJECT ID HERE'
```
To be able to interact with all resources available in Watson Machine Learning, you need to set the **project** which you will be using.
```
client.set.default_project(project_id)
```
<a id="definition"></a>
## 2. Optimizer definition
### Training data connection
Define connection information to training data CSV file. This example uses the German Credit Risk dataset.
The dataset can be downloaded from [here](https://github.com/IBM/watson-machine-learning-samples/blob/master/data/credit_risk/german_credit_data_biased_training.csv).
```
filename = 'credit_risk_training.csv'
!wget -q https://raw.githubusercontent.com/IBM/watson-machine-learning-samples/master/cpd4.0/data/credit_risk/german_credit_data_biased_training.csv \
-O credit_risk_training.csv
asset_details = client.data_assets.create('german_credit_data_biased_training', filename)
asset_details
client.data_assets.get_id(asset_details)
from ibm_watson_machine_learning.helpers import DataConnection, AssetLocation
credit_risk_conn = DataConnection(data_asset_id=client.data_assets.get_id(asset_details))
training_data_reference=[credit_risk_conn]
```
### Optimizer configuration
Provide the input information for AutoAI optimizer:
- `name` - experiment name
- `prediction_type` - type of the problem
- `prediction_column` - target column name
- `scoring` - optimization metric
```
from ibm_watson_machine_learning.experiment import AutoAI
experiment = AutoAI(wml_credentials, project_id)
pipeline_optimizer = experiment.optimizer(
name='Credit Risk Prediction - AutoAI',
desc='Sample notebook',
prediction_type=AutoAI.PredictionType.BINARY,
prediction_column='Risk',
scoring=AutoAI.Metrics.ROC_AUC_SCORE,
)
```
Configuration parameters can be retrieved via `get_params()`.
```
pipeline_optimizer.get_params()
```
<a id="run"></a>
## 3. Experiment run
Call the `fit()` method to trigger the AutoAI experiment. You can either use interactive mode (synchronous job) or background mode (asychronous job) by specifying `background_model=True`.
```
run_details = pipeline_optimizer.fit(
training_data_reference=training_data_reference,
background_mode=False)
```
You can use the `get_run_status()` method to monitor AutoAI jobs in background mode.
```
pipeline_optimizer.get_run_status()
```
<a id="comparison"></a>
### 3.1 Pipelines comparison
You can list trained pipelines and evaluation metrics information in
the form of a Pandas DataFrame by calling the `summary()` method. You can
use the DataFrame to compare all discovered pipelines and select the one
you like for further testing.
```
summary = pipeline_optimizer.summary()
summary
```
You can visualize the scoring metric calculated on a holdout data set.
```
import pandas as pd
pd.options.plotting.backend = "plotly"
summary.holdout_roc_auc.plot()
```
<a id="scoring"></a>
## 4. Deploy and Score
In this section you will learn how to deploy and score trained model using project in a specified deployment space as a webservice and batch using WML instance.
### Webservice deployment creation
```
from ibm_watson_machine_learning.deployment import WebService
service = WebService(
source_wml_credentials=wml_credentials,
source_project_id=project_id,
target_wml_credentials=wml_credentials,
target_space_id=space_id
)
service.create(
experiment_run_id=run_details['metadata']['id'],
model='Pipeline_1',
deployment_name="Credit Risk Deployment AutoAI")
```
Deployment object could be printed to show basic information:
```
print(service)
```
To show all available information about the deployment use the `.get_params()` method:
```
service.get_params()
```
### Scoring of webservice
You can make scoring request by calling `score()` on deployed pipeline.
```
pipeline_optimizer.get_data_connections()[0]
train_df = pipeline_optimizer.get_data_connections()[0].read()
train_X = train_df.drop(['Risk'], axis=1)
train_y = train_df.Risk.values
predictions = service.score(payload=train_X.iloc[:10])
predictions
```
If you want to work with the web service in an external Python application you can retrieve the service object by:
- Initialize the service by `service = WebService(wml_credentials)`
- Get deployment_id by `service.list()` method
- Get webservice object by `service.get('deployment_id')` method
After that you can call `service.score()` method.
### Deleting deployment
You can delete the existing deployment by calling the `service.delete()` command.
To list the existing web services you can use `service.list()`.
### Batch deployment creation
A batch deployment processes input data from a inline data and return predictions in scoring details or processes from data asset and writes the output to a file.
```
batch_payload_df = train_df.drop(['Risk'], axis=1)[:5]
batch_payload_df
```
Create batch deployment for `Pipeline_2` created in AutoAI experiment with the `run_id`.
```
from ibm_watson_machine_learning.deployment import Batch
service_batch = Batch(
source_wml_credentials=wml_credentials,
source_project_id=project_id,
target_wml_credentials=wml_credentials,
target_space_id=space_id
)
service_batch.create(
experiment_run_id=run_details['metadata']['id'],
model="Pipeline_2",
deployment_name="Credit Risk Batch Deployment AutoAI")
```
### Score batch deployment with inline payload as pandas DataFrame.
```
scoring_params = service_batch.run_job(
payload=batch_payload_df,
background_mode=False)
scoring_params['entity']['scoring'].get('predictions')
```
### Deleting deployment
You can delete the existing deployment by calling the `service_batch.delete()` command.
To list the existing:
- batch services you can use `service_batch.list()`,
- scoring jobs you can use `service_batch.list_jobs()`.
<a id="cleanup"></a>
## 5. Clean up
If you want to clean up all created assets:
- experiments
- trainings
- pipelines
- model definitions
- models
- functions
- deployments
please follow up this sample [notebook](https://github.com/IBM/watson-machine-learning-samples/blob/master/cpd4.0/notebooks/python_sdk/instance-management/Machine%20Learning%20artifacts%20management.ipynb).
<a id="summary"></a>
## 6. Summary and next steps
You successfully completed this notebook!.
You learned how to use `ibm-watson-machine-learning` to run AutoAI experiments using project.
Check out our _[Online Documentation](https://www.ibm.com/cloud/watson-studio/autoai)_ for more samples, tutorials, documentation, how-tos, and blog posts.
### Authors
**Jan Sołtysik**, Intern in Watson Machine Learning
Copyright © 2020, 2021 IBM. This notebook and its source code are released under the terms of the MIT License.
|
github_jupyter
|
username = 'PASTE YOUR USERNAME HERE'
api_key = 'PASTE YOUR API_KEY HERE'
url = 'PASTE THE PLATFORM URL HERE'
wml_credentials = {
"username": username,
"apikey": api_key,
"url": url,
"instance_id": 'openshift',
"version": '4.0'
}
wml_credentials = {
"username": ***,
"password": ***,
"url": ***,
"instance_id": 'openshift',
"version": '4.0'
}
!pip install -U ibm-watson-machine-learning | tail -n 1
from ibm_watson_machine_learning import APIClient
client = APIClient(wml_credentials)
space_id = 'PASTE YOUR SPACE ID HERE'
client.spaces.list(limit=10)
project_id = 'PASTE YOUR PROJECT ID HERE'
client.set.default_project(project_id)
filename = 'credit_risk_training.csv'
!wget -q https://raw.githubusercontent.com/IBM/watson-machine-learning-samples/master/cpd4.0/data/credit_risk/german_credit_data_biased_training.csv \
-O credit_risk_training.csv
asset_details = client.data_assets.create('german_credit_data_biased_training', filename)
asset_details
client.data_assets.get_id(asset_details)
from ibm_watson_machine_learning.helpers import DataConnection, AssetLocation
credit_risk_conn = DataConnection(data_asset_id=client.data_assets.get_id(asset_details))
training_data_reference=[credit_risk_conn]
from ibm_watson_machine_learning.experiment import AutoAI
experiment = AutoAI(wml_credentials, project_id)
pipeline_optimizer = experiment.optimizer(
name='Credit Risk Prediction - AutoAI',
desc='Sample notebook',
prediction_type=AutoAI.PredictionType.BINARY,
prediction_column='Risk',
scoring=AutoAI.Metrics.ROC_AUC_SCORE,
)
pipeline_optimizer.get_params()
run_details = pipeline_optimizer.fit(
training_data_reference=training_data_reference,
background_mode=False)
pipeline_optimizer.get_run_status()
summary = pipeline_optimizer.summary()
summary
import pandas as pd
pd.options.plotting.backend = "plotly"
summary.holdout_roc_auc.plot()
from ibm_watson_machine_learning.deployment import WebService
service = WebService(
source_wml_credentials=wml_credentials,
source_project_id=project_id,
target_wml_credentials=wml_credentials,
target_space_id=space_id
)
service.create(
experiment_run_id=run_details['metadata']['id'],
model='Pipeline_1',
deployment_name="Credit Risk Deployment AutoAI")
print(service)
service.get_params()
pipeline_optimizer.get_data_connections()[0]
train_df = pipeline_optimizer.get_data_connections()[0].read()
train_X = train_df.drop(['Risk'], axis=1)
train_y = train_df.Risk.values
predictions = service.score(payload=train_X.iloc[:10])
predictions
batch_payload_df = train_df.drop(['Risk'], axis=1)[:5]
batch_payload_df
from ibm_watson_machine_learning.deployment import Batch
service_batch = Batch(
source_wml_credentials=wml_credentials,
source_project_id=project_id,
target_wml_credentials=wml_credentials,
target_space_id=space_id
)
service_batch.create(
experiment_run_id=run_details['metadata']['id'],
model="Pipeline_2",
deployment_name="Credit Risk Batch Deployment AutoAI")
scoring_params = service_batch.run_job(
payload=batch_payload_df,
background_mode=False)
scoring_params['entity']['scoring'].get('predictions')
| 0.489015 | 0.958886 |
### Deep Learning Homework 6
Taking inspiration from the last 2 pictures within the notebook (07-convnets.ipynb), implement a U-Net-style CNN with the following specs:
1. All convolutions must use a 3 x 3 kernel and leave the spatial dimensions (i.e. height, width) of the input untouched.
2. Downsampling in the contracting part is performed via maxpooling with a 2 x 2 kernel and stride of 2.
3. Upsampling is operated by a deconvolution with a 2 x 2 kernel and stride of 2. The PyTorch module that implements the deconvolution is `nn.ConvTranspose2d`
4. The final layer of the expanding part has only 1 channel
* between how many classes are we discriminating?
Create a network class with (at least) a `__init__` and a `forward` method. Please resort to additional structures (e.g., `nn.Module`s, private methods...) if you believe it helps readability of your code.
Test, at least with random data, that the network is doing the correct tensor operations and that the output has the correct shape (e.g., use `assert`s in your code to see if the byproduct is of the expected shape).
Note: the overall organization of your work can greatly improve readability and understanding of your code by others. Please consider preparing your notebook in an organized fashion so that we can better understand (and correct) your implementation.
```
import torch
from torch import nn
from torch.utils.data import Dataset, DataLoader
import pylab as pl
from IPython.display import clear_output
class VGG_block(nn.Module):
"""Implements a VGG layer with kernel size 3 and padding 1"""
def __init__(self, in_channels, out_channels, num_layers=2, maxpool=False, activation=nn.ReLU):
super().__init__()
layers = []
layers.append(nn.Conv2d(in_channels, out_channels, kernel_size=3, padding=1))
layers.append(activation())
for i in range(num_layers-1):
layers.append(nn.Conv2d(out_channels, out_channels, kernel_size=3, padding=1))
layers.append(activation())
if maxpool:
layers.append(nn.MaxPool2d(2))
self.layers = nn.Sequential(*layers)
def forward(self, X):
return self.layers(X)
class upsampling_block(nn.Module):
"""
Implements the upsampling block of the U-net, basically it's a VGG_block
with in_channels=in_channels and out_channels=mid_channels followed by a
deconvolution operation that doubles the size of the input image
"""
def __init__(self, in_channels, mid_channels, out_channels, num_mid_layers=2, activation=nn.ReLU):
super().__init__()
self.layers = nn.Sequential(
VGG_block(in_channels, mid_channels, num_layers=num_mid_layers, activation=activation),
nn.ConvTranspose2d(mid_channels, out_channels, kernel_size=2, stride=2),
activation()
)
def forward(self, X):
return self.layers(X)
class U_net(nn.Module):
"""
Implements a U-net, this architecture can be trained without changing anything
on non-square and non-power of two images, however the results can be worse
"""
def __init__(self, channels=3, depth=4, num_classes=10):
super().__init__()
# Downsampling layers
downsampling_layers = []
in_channels, out_channels = channels, 64
for i in range(depth):
downsampling_layers.append(VGG_block(in_channels, out_channels))
in_channels = out_channels
out_channels *= 2
self.downsampling_layers = nn.Sequential(*downsampling_layers)
# Ceil mode is required if I have uneven images, otherwise it's the same
self.maxpool = nn.MaxPool2d(2, ceil_mode=True)
# "Deepest" layer
mid_channels = out_channels
out_channels = in_channels
self.deep_layer = upsampling_block(in_channels, mid_channels, out_channels)
# Upsampling layers
upsampling_layers = []
in_channels, mid_channels, out_channels = in_channels*2, mid_channels//2, out_channels//2
for i in range(depth-1):
upsampling_layers.append(upsampling_block(in_channels, mid_channels, out_channels))
in_channels = in_channels//2
mid_channels = mid_channels//2
out_channels = out_channels//2
self.upsampling_layers = nn.Sequential(*upsampling_layers)
# Classifier or last layer
self.classifier = nn.Sequential(
VGG_block(in_channels, mid_channels),
nn.Conv2d(mid_channels, num_classes, kernel_size=1)
)
def center_crop(self, images, size_x, size_y):
"""Rectangle center crop of a set of images"""
# If the crop is bigger or equal to the images do nothing
if size_x>=images.size()[2] and size_y>=images.size()[3]:
return images
# Otherwise perform the cropping
center_x = images.size()[2] // 2
center_y = images.size()[3] // 2
bottom_left = [center_x - size_x//2, center_y - size_y//2]
top_right = [center_x + (size_x+1)//2, center_y + (size_y+1)//2]
return images[:, :, bottom_left[0]: top_right[0], bottom_left[1]: top_right[1]]
def forward(self, X):
skips = [] # Holds the skip connections
out = X
# Downsampling phase
for layer in self.downsampling_layers:
out = layer(out)
skips.append(out)
out = self.maxpool(out)
# Deepest layer
out = self.deep_layer(out)
# Upsampling phase
for i, layer in enumerate(self.upsampling_layers, start=1):
# The cropping is done only if the downsampling phase has uneven image sizes
# In that case in the upsampling the resulting image will be 1 pixel
# wider and I need to crop it, notice that this doesn't happen for
# power-of-two images and cropping does nothing
out = self.center_crop(out, *skips[-i].size()[2:])
out = torch.cat((skips[-i], out), dim=1) # Concatenate the previous output
out = layer(out)
# Classification phase
out = self.center_crop(out, *skips[0].size()[2:])
out = torch.cat((skips[0], out), dim=1)
return self.classifier(out)
from random import randint
# Test with images like the one in the U-net picture of the notebook, F for my pc
net = U_net(channels=3, depth=3, num_classes=2)
images = torch.randn((10, 3, 572, 572))
print("input size:", images.size(), "output size:", net(images).size(), "\n")
# Test with random images and random nets
for i in range(10):
channels = randint(1,10)
net = U_net(channels=channels, depth=randint(1,5), num_classes=randint(2, 20))
images = torch.randn((randint(1,10), channels, randint(1, 200), randint(1, 200)))
out = net(images)
assert out.size()[2:] == images.size()[2:]
print("input size:", images.size(), "output size:", out.size())
print("\nTest passed!")
# Summary of the model
from torchsummary import summary
summary(net)
net
```
|
github_jupyter
|
import torch
from torch import nn
from torch.utils.data import Dataset, DataLoader
import pylab as pl
from IPython.display import clear_output
class VGG_block(nn.Module):
"""Implements a VGG layer with kernel size 3 and padding 1"""
def __init__(self, in_channels, out_channels, num_layers=2, maxpool=False, activation=nn.ReLU):
super().__init__()
layers = []
layers.append(nn.Conv2d(in_channels, out_channels, kernel_size=3, padding=1))
layers.append(activation())
for i in range(num_layers-1):
layers.append(nn.Conv2d(out_channels, out_channels, kernel_size=3, padding=1))
layers.append(activation())
if maxpool:
layers.append(nn.MaxPool2d(2))
self.layers = nn.Sequential(*layers)
def forward(self, X):
return self.layers(X)
class upsampling_block(nn.Module):
"""
Implements the upsampling block of the U-net, basically it's a VGG_block
with in_channels=in_channels and out_channels=mid_channels followed by a
deconvolution operation that doubles the size of the input image
"""
def __init__(self, in_channels, mid_channels, out_channels, num_mid_layers=2, activation=nn.ReLU):
super().__init__()
self.layers = nn.Sequential(
VGG_block(in_channels, mid_channels, num_layers=num_mid_layers, activation=activation),
nn.ConvTranspose2d(mid_channels, out_channels, kernel_size=2, stride=2),
activation()
)
def forward(self, X):
return self.layers(X)
class U_net(nn.Module):
"""
Implements a U-net, this architecture can be trained without changing anything
on non-square and non-power of two images, however the results can be worse
"""
def __init__(self, channels=3, depth=4, num_classes=10):
super().__init__()
# Downsampling layers
downsampling_layers = []
in_channels, out_channels = channels, 64
for i in range(depth):
downsampling_layers.append(VGG_block(in_channels, out_channels))
in_channels = out_channels
out_channels *= 2
self.downsampling_layers = nn.Sequential(*downsampling_layers)
# Ceil mode is required if I have uneven images, otherwise it's the same
self.maxpool = nn.MaxPool2d(2, ceil_mode=True)
# "Deepest" layer
mid_channels = out_channels
out_channels = in_channels
self.deep_layer = upsampling_block(in_channels, mid_channels, out_channels)
# Upsampling layers
upsampling_layers = []
in_channels, mid_channels, out_channels = in_channels*2, mid_channels//2, out_channels//2
for i in range(depth-1):
upsampling_layers.append(upsampling_block(in_channels, mid_channels, out_channels))
in_channels = in_channels//2
mid_channels = mid_channels//2
out_channels = out_channels//2
self.upsampling_layers = nn.Sequential(*upsampling_layers)
# Classifier or last layer
self.classifier = nn.Sequential(
VGG_block(in_channels, mid_channels),
nn.Conv2d(mid_channels, num_classes, kernel_size=1)
)
def center_crop(self, images, size_x, size_y):
"""Rectangle center crop of a set of images"""
# If the crop is bigger or equal to the images do nothing
if size_x>=images.size()[2] and size_y>=images.size()[3]:
return images
# Otherwise perform the cropping
center_x = images.size()[2] // 2
center_y = images.size()[3] // 2
bottom_left = [center_x - size_x//2, center_y - size_y//2]
top_right = [center_x + (size_x+1)//2, center_y + (size_y+1)//2]
return images[:, :, bottom_left[0]: top_right[0], bottom_left[1]: top_right[1]]
def forward(self, X):
skips = [] # Holds the skip connections
out = X
# Downsampling phase
for layer in self.downsampling_layers:
out = layer(out)
skips.append(out)
out = self.maxpool(out)
# Deepest layer
out = self.deep_layer(out)
# Upsampling phase
for i, layer in enumerate(self.upsampling_layers, start=1):
# The cropping is done only if the downsampling phase has uneven image sizes
# In that case in the upsampling the resulting image will be 1 pixel
# wider and I need to crop it, notice that this doesn't happen for
# power-of-two images and cropping does nothing
out = self.center_crop(out, *skips[-i].size()[2:])
out = torch.cat((skips[-i], out), dim=1) # Concatenate the previous output
out = layer(out)
# Classification phase
out = self.center_crop(out, *skips[0].size()[2:])
out = torch.cat((skips[0], out), dim=1)
return self.classifier(out)
from random import randint
# Test with images like the one in the U-net picture of the notebook, F for my pc
net = U_net(channels=3, depth=3, num_classes=2)
images = torch.randn((10, 3, 572, 572))
print("input size:", images.size(), "output size:", net(images).size(), "\n")
# Test with random images and random nets
for i in range(10):
channels = randint(1,10)
net = U_net(channels=channels, depth=randint(1,5), num_classes=randint(2, 20))
images = torch.randn((randint(1,10), channels, randint(1, 200), randint(1, 200)))
out = net(images)
assert out.size()[2:] == images.size()[2:]
print("input size:", images.size(), "output size:", out.size())
print("\nTest passed!")
# Summary of the model
from torchsummary import summary
summary(net)
net
| 0.936095 | 0.930868 |
# Exercise 2 - R: Logistic Regression
<img src="http://www.saedsayad.com/images/LogReg_1.png">
## Part I: *Logistic regression without regularization*
Predicting if a student will be accepted into a university based off of two test scores
Beginning with package imports, data loading, and initial visualization
```
rm(list=ls()) # Clearing all environment variables
suppressPackageStartupMessages({
library(readr)
library(ggplot2)
library(dplyr)
})
theme_update(plot.title = element_text(hjust = 0.5)) # Centers ggplot2 titles
df <- read_csv("ex2/ex2data1.txt",
col_names = FALSE,
col_types = 'ddd')
colnames(df) <- c('Exam1Score', 'Exam2Score', 'Admitted')
# Inserting ones for the intercept
df <- cbind(Intercept=1, df)
X <- df %>% select(-Admitted)
y <- df$Admitted
# An array of 0s for starting values of theta to be used in many functions
initialTheta <- c(0, 0, 0)
ggplot(df) +
geom_point(aes(x=Exam1Score, y=Exam2Score, color=as.factor(Admitted))) +
labs(color='Admitted')
```
### Sigmoid Function
$g(z) = \frac{1}{1+e^{-z}}$
Converts $z$ into a value between 0 and 1
```
sigmoid <- function(z){
# Converts numerical input into a value between 0 and 1
return(1/(1+exp(-z)))
}
# Plotting to validate that the function is working
data.frame(X = -10:10) %>%
mutate(y = sigmoid(X)) %>%
ggplot() +
geom_line(aes(x=X, y=y))
```
### Logistic Regression Hypothesis
$h_\theta(x) = g(\theta^Tx)$
- Notation:
- $g$: Sigmoid function
- $\theta^T$: Transposed parameters
- E.x.: $\theta^T = \begin{bmatrix} \theta_1 \\ \theta_2 \\ \vdots \\ \theta_n \end{bmatrix}$
```
logistic_hypothesis <- function(theta, X){
# Calculates the hypothesis for X given values of
# theta for logistic regression
X <- as.matrix(X)
theta <- as.matrix(theta)
h <- X %*% theta
return(sigmoid(h))
}
logistic_hypothesis(initialTheta, head(X))
```
### Cost Function
$J(\theta) = \frac{1}{m} \sum_{i=1}^m[-y^{(i)}log(h_\theta(x^{(i)})) - (1-y^{(i)})log(1-h_\theta(x^{(i)}))]$
- Notation:
- $m$: Number of records
- $h_\theta$: Logistic hypothesis $(h)$ given specific values of $\theta$ for parameters
- $i$: Index of the record (e.x. if $i = 46$, then 46th row)
```
cost_function <- function(theta, X, y){
# Computes cost for logistic regression
m <- length(y)
h <- logistic_hypothesis(theta, X)
error <- sum(-y*log(h)-(1-y)*log(1-h))
J <- (1/m)*error
return(J)
}
cost_function(initialTheta, X, y)
```
### Gradient
$\frac{\partial J(\theta)}{\partial \theta_j} = \frac{1}{m} \sum_{i=1}^m (h_\theta(x^{(i)}) - y^{(i)})x_j^{(i)}$
- Notation:
- $\partial$: Partial derivative
- $J(\theta)$: Cost given $\theta$
- $m$: Number of records
- $h_\theta$: Logistic hypothesis $(h)$ given specific values of $\theta$ for parameters
- $i$: Index of the record (e.x. if $i = 46$, then 46th row)
```
gradient <- function(theta, X, y){
# Computes the gradient for logistic regression
X <- as.matrix(X)
y <- as.matrix(y)
theta <- as.matrix(theta)
m <- length(y)
h <- logistic_hypothesis(theta, X)
gradient <- (1/m) * (t(X) %*% (h-y))
return(gradient)
}
gradient(initialTheta, X, y)
```
Finding the optimal values of $\theta_j$ for the cost function using the base R optim function. This is similar to MATLAB's fminunc function.
```
# Find values of theta that minimize the cost function
optimalTheta <- optim(
# Specifying function parameters
par=initialTheta, # Initial guess
fn=cost_function, # Function to minimize
X=X,
y=y,
method="BFGS", # Optimization function to use
control = list(maxit = 400) # Maximum number of iterations
)$par # Specifying that we only want the obtained thetas
# Pretty printing the obtained values for theta
cat('Cost:', x = cost_function(optimalTheta, X, y), '\n')
cat('\nOptimal Thetas\n',
'Intercept:', optimalTheta[1], '\n',
'Theta 1:', optimalTheta[2], '\n',
'Theta 2:', optimalTheta[3])
```
Comparing the obtained parameters to what base R's glm function provides
```
glm(Admitted ~ Exam1Score+Exam2Score, data=df, family=binomial)
```
Not bad!
Calculating the class probability and generating predictions of acceptance using values of $\theta_j$ obtained from the optimization function
The outputs from logistic regression are just the class probability, or $P(y = 1 \mid x; \theta)$, so we are predicting the classes (accepted or not) as follows:
$Prediction(y \mid x; \theta) = \begin{cases} 1, \quad\mbox{ if } P(y = 1 \mid x; \theta) > 0.50 \\ 0, \quad\mbox{ if } P(y = 1 \mid x; \theta) \leq 0.50 \end{cases} $
```
# Predicting the class probability with the obtained thetas
df$ClassProbability <- logistic_hypothesis(optimalTheta, X)
# Assigning those with a class probability above 0.5 as admitted
df$Prediction <- ifelse(df$ClassProbability > 0.5, 1, 0)
head(df)
```
Plotting the decision boundary over the data
```
# Calculating and plotting the decision boundary
decisionSlope <- optimalTheta[2]/(-optimalTheta[3])
decisionIntercept <- optimalTheta[1]/(-optimalTheta[3])
ggplot(df) +
geom_point(aes(x=Exam1Score, y=Exam2Score, color=as.factor(Admitted))) +
labs(color='Admitted') +
geom_abline(intercept=decisionIntercept, slope=decisionSlope,
linetype='dashed')
```
---
## Part II: *Logistic regression with regularization*
Predicting if a microchip passes QA after two tests
```
df <- read_csv("ex2/ex2data2.txt",
col_names = FALSE,
col_types = 'ddd')
colnames(df) <- c('Test1', 'Test2', 'Accepted')
ggplot(df) +
geom_point(aes(x=Test1, y=Test2, color=as.factor(Accepted))) +
labs(color='Accepted') +
xlab('Microchip Test 1') +
ylab('Microchip Test 2')
```
### Feature Mapping
Maps the features into all polynomial terms of $x_1$ and $x_2$ up to the sixth power. This allows for a more complex and nonlinear decision boundary.
The feature space prior to feature mapping (3-dimensional vector):
$\hspace{1cm} Feature(x) = \begin{bmatrix} 1 \\ x_1 \\ x_2 \end{bmatrix}$
The feature space after feature mapping:
$\hspace{1cm} mapFeature(x) = \begin{bmatrix} 1 \\ x_1 \\ x_2 \\ x_1^2 \\ x_1x_2 \\ x_2^2 \\ x_1^3 \\ \vdots \\ x_1x_2^5 \\ x_2^6 \end{bmatrix}$
**Note:** I made a few adjustments on the Octave/MATLAB code provided for this assignment in order to maintain the names of the polynomials
Octave/MATLAB code:
```
degree = 6;
out = ones(size(X1(:,1)));
for i = 1:degree
for j = 0:i
out(:, end+1) = (X1.^(i-j)).*(X2.^j);
end
end
```
```
X <- df %>% select(Test1, Test2)
y <- df$Accepted
# Creating function for use in plotting decision boundaries later
mapFeatures <- function(X, degree) {
# Creates polynomial features of X up to the degree specified
for (i in 1:degree) {
for (j in 0:i) {
# Assigning names to the columns
if (j == 0) {
if (i == 1) {
colName <- 'x1'
} else {
colName <- paste0('x1_', as.character(i))
}
} else if (i-j == 0) {
if (j == 1) {
colName <- 'x2'
} else {
colName <- paste0('x2_', as.character(j))
}
} else {
colName <- paste0('x1_', as.character(i-j), ':', 'x2_', as.character(j))
}
# Calculating polynomial features
X[colName] <- (X[, 1]^(i-j)) * (X[, 2]^j)
}
}
X <- X[, 3:length(X)] # Removing original columns to keep naming conventions
X <- cbind(x0 = 1, X) # Inserting the intercept term
return(X)
}
X <- mapFeatures(X, 6)
# Creating a new list of initial thetas
initialTheta <- numeric(length(X))
print(dim(X))
head(X)
```
### Regularized Cost Function
$J(\theta) = \frac{1}{m} \sum_{i=1}^m[-y^{(i)}log(h_\theta(x^{(i)})) - (1-y^{(i)})log(1-h_\theta(x^{(i)}))] + \frac{\lambda}{2m} \sum_{j=1}^n \theta_j^2$
The only change from the other cost function we used earlier is the addition of the regularization parameter:
#### Regularization Parameter
$\frac{\lambda}{2m} \sum_{j=1}^n \theta_j^2$
- Notation:
- $\lambda$: The weight which the parameters are adjusted by. A lower $\lambda$ has little effect on the parameters, and a higher $\lambda$ (e.x. $\lambda = 1,000$) will adjust the parameters to be close to 0.
- $m$: Number of records
- $j$: The index for the parameter. E.x. $\theta_{j=1}$ is the score for Microchip Test #1
**Note:** $\theta_0$ should not be regularized as denoted by the summation in the regularization parameter
```
regularized_cost <- function(theta, X, y, C){
# Computes cost for logistic regression
m <- length(y)
h <- logistic_hypothesis(theta, X)
error <- sum(-y*log(h)-(1-y)*log(1-h))
# Calculating the regularization parameter
# Avoiding the regularization parameter for the first theta
regularizedTheta <- theta[-1] ** 2
regularization <- (C/(2*m)) * sum(regularizedTheta)
J <- (1/m)*error + regularization
return(J)
}
# Testing how cost differs with regularization
# Using thetas above 0 so the regularization parameter has an impact
regTestTheta <- rep(0.5, length(X))
cat('Cost at lambda=0:', regularized_cost(regTestTheta, X, y, C=0), '\n')
cat('Cost at lambda=1:', regularized_cost(regTestTheta, X, y, C=1), '\n')
cat('Cost at lambda=100:', regularized_cost(regTestTheta, X, y, C=100))
```
### Regularized Gradient
$\frac{\partial J(\theta)}{\partial \theta_j} = \begin{cases}
\hspace{0.25cm} \frac{1}{m} \sum_{i=1}^m (h_\theta(x^{(i)}) - y^{(i)})x_j^{(i)} & \text{for}\ j = 0 \\
\Big(\frac{1}{m} \sum_{i=1}^m (h_\theta(x^{(i)}) - y^{(i)})x_j^{(i)}\Big) + \frac{\lambda}{m}\theta_j & \text{for}\ j \geq 1
\end{cases}$
This is also the same as the last gradient with the exception of the regularization parameter
#### Regularization Parameter
$\frac{\lambda}{m}\theta_j \hspace{0.5cm}$for $j \geq 1$
- Notation:
- $\lambda$: The weight which the parameters are adjusted by. A lower $\lambda$ has little effect on the parameters, and a higher $\lambda$ (e.x. $\lambda = 1,000$) will adjust the parameters to be close to 0.
- $m$: Number of records
- $j$: The index for the parameter. E.x. $\theta_{j=1}$ is the score for Microchip Test #1
```
regularized_gradient <- function(theta, X, y, C){
# Computes the gradient for logistic regression
X <- as.matrix(X)
y <- as.matrix(y)
theta <- as.matrix(theta)
m <- length(y)
h <- logistic_hypothesis(theta, X)
# Calculating the regularization parameter for all thetas but the intercept
regParam <- (C/m) * theta[-1]
gradient <- (1/m) * (t(X) %*% (h-y))
gradient[-1] <- gradient[-1] + regParam
return(gradient)
}
t(regularized_gradient(initialTheta, X, y, C=1.0))
```
Finding the optimal values of $\theta$. This chunk is surprisingly much quicker than the similar function I used in Python.
```
# Find values of theta that minimize the cost function
optimalTheta <- optim(
# Specifying function parameters
par=initialTheta, # Initial guess
fn=regularized_cost, # Function to minimize
X=X,
y=y,
C=1.0,
method="BFGS", # Optimization function to use
control = list(maxit = 4000) # Maximum number of iterations
)$par # Specifying that we only want the obtained thetas
```
Checking against R's glm logistic regression. This is an unregularized approach, but since we used a blanket value of lambda, there shouldn't be a huge difference.
```
# Fitting the model
model <- glm(y ~ ., data=cbind(X[, -1], y), family='binomial')
modelCoef <- model$coefficients %>% data.frame(glm=.)
cbind(modelCoef, optimalTheta) %>% t()
```
R's glm produced much more extreme values than our implementation. We'll see how this affects the performance.
Lastly, comparing the accuracy between the two models. Classification accuracy is just the percentage of records correctly classified (precision, recall, f-1 score, etc. offer more nuanced information on performance), so we will have to calculate the class probabilities and assign predictions like we did for part one:
```
# Predicting the class probability with the obtained thetas
df$ClassProbability <- logistic_hypothesis(optimalTheta, X)
df$glmClassProbability <- predict(model, X)
# Assigning those with a class probability above 0.5 as admitted
df$Prediction <- ifelse(df$ClassProbability > 0.5, 1, 0)
df$glmPrediction <- ifelse(df$glmClassProbability > 0.5, 1, 0)
head(df)
ourAccuracy <- mean(ifelse(df$Accepted == df$Prediction, 1, 0))
glmAccuracy <- mean(ifelse(df$Accepted == df$glmPrediction, 1, 0))
cat('Our accuracy:', ourAccuracy, '\n')
cat('R\'s glm accuracy:', glmAccuracy)
```
Our model had a lower accuracy, but it still did relatively well.
Plotting the decision boundary using inspiration from [this post](http://www.onthelambda.com/2014/07/24/interactive-visualization-of-non-linear-logistic-regression-decision-boundaries-with-shiny/)
```
# Timing this cell
timeStart <- proc.time()
createBoundary <- function(X, y, C=0) {
# Returns a data frame used for plotting the decision boundary.
# Uses the optimization function to find the optimal values of
# theta, so it is not needed as a parameter.
# Find values of theta that minimize the cost function
initialTheta <- numeric(length(X))
optimalTheta <- optim(
# Specifying function parameters
par=initialTheta, # Initial guess
fn=regularized_cost, # Function to minimize
X=X,
y=y,
C=C,
method="BFGS", # Optimization function to use
control = list(maxit = 4000) # Maximum number of iterations
)$par # Specifying that we only want the obtained thetas
# Creating a mesh grid to generate class probabilities for
grid <- data.frame()
dx <- seq(min(df$Test1), max(df$Test1), by=0.05)
dy <- seq(min(df$Test2), max(df$Test2), length.out=length(dx))
# Filling the grid with values
for (i in 1:length(dx)) {
for (j in 1:length(dx)) {
v <- t(matrix(c(dx[i], dy[j])))
grid <- rbind(grid, v)
}
}
# Generating class probabilities on the polynomial features
grid <- mapFeatures(grid, 6) # Creating polynomial features
grid$z <- logistic_hypothesis(optimalTheta, grid) # Class probabilities
grid <- grid %>% select(x1, x2, z) # Removing polynomial features
return(grid)
}
# Generating the decision boundaries for various levels of lambda
lambda0 <- createBoundary(X, y)
lambda1 <- createBoundary(X, y, 1)
lambda10 <- createBoundary(X, y, 10)
lambda100 <- createBoundary(X, y, 100)
# Plotting the original data
ggplot() +
geom_point(data=df, aes(x=Test1, y=Test2,
# Using alpha instead of color to clean up legend
alpha=as.factor(df$Accepted))) +
xlab('Microchip Test 1') +
ylab('Microchip Test 2') +
# Plotting the decision boundaries
geom_contour(data=lambda0, aes(x=x1, y=x2, z=z, color='Lambda = 0'),
breaks=0.5, alpha=0.4) +
geom_contour(data=lambda1, aes(x=x1, y=x2, z=z, color='Lambda = 1'),
breaks=0.5, alpha=0.4) +
geom_contour(data=lambda10, aes(x=x1, y=x2, z=z, color='Lambda = 10'),
breaks=0.5, alpha=0.4) +
geom_contour(data=lambda100, aes(x=x1, y=x2, z=z, color='Lambda = 100'),
breaks=0.5, alpha=0.4) +
labs(alpha='Accepted', color='Decision Boundaries') +
theme(panel.background = element_blank()) # To aid in seeing rejected points
# Reporting time results
timeStop <- proc.time()
timeStop - timeStart
```
|
github_jupyter
|
rm(list=ls()) # Clearing all environment variables
suppressPackageStartupMessages({
library(readr)
library(ggplot2)
library(dplyr)
})
theme_update(plot.title = element_text(hjust = 0.5)) # Centers ggplot2 titles
df <- read_csv("ex2/ex2data1.txt",
col_names = FALSE,
col_types = 'ddd')
colnames(df) <- c('Exam1Score', 'Exam2Score', 'Admitted')
# Inserting ones for the intercept
df <- cbind(Intercept=1, df)
X <- df %>% select(-Admitted)
y <- df$Admitted
# An array of 0s for starting values of theta to be used in many functions
initialTheta <- c(0, 0, 0)
ggplot(df) +
geom_point(aes(x=Exam1Score, y=Exam2Score, color=as.factor(Admitted))) +
labs(color='Admitted')
sigmoid <- function(z){
# Converts numerical input into a value between 0 and 1
return(1/(1+exp(-z)))
}
# Plotting to validate that the function is working
data.frame(X = -10:10) %>%
mutate(y = sigmoid(X)) %>%
ggplot() +
geom_line(aes(x=X, y=y))
logistic_hypothesis <- function(theta, X){
# Calculates the hypothesis for X given values of
# theta for logistic regression
X <- as.matrix(X)
theta <- as.matrix(theta)
h <- X %*% theta
return(sigmoid(h))
}
logistic_hypothesis(initialTheta, head(X))
cost_function <- function(theta, X, y){
# Computes cost for logistic regression
m <- length(y)
h <- logistic_hypothesis(theta, X)
error <- sum(-y*log(h)-(1-y)*log(1-h))
J <- (1/m)*error
return(J)
}
cost_function(initialTheta, X, y)
gradient <- function(theta, X, y){
# Computes the gradient for logistic regression
X <- as.matrix(X)
y <- as.matrix(y)
theta <- as.matrix(theta)
m <- length(y)
h <- logistic_hypothesis(theta, X)
gradient <- (1/m) * (t(X) %*% (h-y))
return(gradient)
}
gradient(initialTheta, X, y)
# Find values of theta that minimize the cost function
optimalTheta <- optim(
# Specifying function parameters
par=initialTheta, # Initial guess
fn=cost_function, # Function to minimize
X=X,
y=y,
method="BFGS", # Optimization function to use
control = list(maxit = 400) # Maximum number of iterations
)$par # Specifying that we only want the obtained thetas
# Pretty printing the obtained values for theta
cat('Cost:', x = cost_function(optimalTheta, X, y), '\n')
cat('\nOptimal Thetas\n',
'Intercept:', optimalTheta[1], '\n',
'Theta 1:', optimalTheta[2], '\n',
'Theta 2:', optimalTheta[3])
glm(Admitted ~ Exam1Score+Exam2Score, data=df, family=binomial)
# Predicting the class probability with the obtained thetas
df$ClassProbability <- logistic_hypothesis(optimalTheta, X)
# Assigning those with a class probability above 0.5 as admitted
df$Prediction <- ifelse(df$ClassProbability > 0.5, 1, 0)
head(df)
# Calculating and plotting the decision boundary
decisionSlope <- optimalTheta[2]/(-optimalTheta[3])
decisionIntercept <- optimalTheta[1]/(-optimalTheta[3])
ggplot(df) +
geom_point(aes(x=Exam1Score, y=Exam2Score, color=as.factor(Admitted))) +
labs(color='Admitted') +
geom_abline(intercept=decisionIntercept, slope=decisionSlope,
linetype='dashed')
df <- read_csv("ex2/ex2data2.txt",
col_names = FALSE,
col_types = 'ddd')
colnames(df) <- c('Test1', 'Test2', 'Accepted')
ggplot(df) +
geom_point(aes(x=Test1, y=Test2, color=as.factor(Accepted))) +
labs(color='Accepted') +
xlab('Microchip Test 1') +
ylab('Microchip Test 2')
degree = 6;
out = ones(size(X1(:,1)));
for i = 1:degree
for j = 0:i
out(:, end+1) = (X1.^(i-j)).*(X2.^j);
end
end
X <- df %>% select(Test1, Test2)
y <- df$Accepted
# Creating function for use in plotting decision boundaries later
mapFeatures <- function(X, degree) {
# Creates polynomial features of X up to the degree specified
for (i in 1:degree) {
for (j in 0:i) {
# Assigning names to the columns
if (j == 0) {
if (i == 1) {
colName <- 'x1'
} else {
colName <- paste0('x1_', as.character(i))
}
} else if (i-j == 0) {
if (j == 1) {
colName <- 'x2'
} else {
colName <- paste0('x2_', as.character(j))
}
} else {
colName <- paste0('x1_', as.character(i-j), ':', 'x2_', as.character(j))
}
# Calculating polynomial features
X[colName] <- (X[, 1]^(i-j)) * (X[, 2]^j)
}
}
X <- X[, 3:length(X)] # Removing original columns to keep naming conventions
X <- cbind(x0 = 1, X) # Inserting the intercept term
return(X)
}
X <- mapFeatures(X, 6)
# Creating a new list of initial thetas
initialTheta <- numeric(length(X))
print(dim(X))
head(X)
regularized_cost <- function(theta, X, y, C){
# Computes cost for logistic regression
m <- length(y)
h <- logistic_hypothesis(theta, X)
error <- sum(-y*log(h)-(1-y)*log(1-h))
# Calculating the regularization parameter
# Avoiding the regularization parameter for the first theta
regularizedTheta <- theta[-1] ** 2
regularization <- (C/(2*m)) * sum(regularizedTheta)
J <- (1/m)*error + regularization
return(J)
}
# Testing how cost differs with regularization
# Using thetas above 0 so the regularization parameter has an impact
regTestTheta <- rep(0.5, length(X))
cat('Cost at lambda=0:', regularized_cost(regTestTheta, X, y, C=0), '\n')
cat('Cost at lambda=1:', regularized_cost(regTestTheta, X, y, C=1), '\n')
cat('Cost at lambda=100:', regularized_cost(regTestTheta, X, y, C=100))
regularized_gradient <- function(theta, X, y, C){
# Computes the gradient for logistic regression
X <- as.matrix(X)
y <- as.matrix(y)
theta <- as.matrix(theta)
m <- length(y)
h <- logistic_hypothesis(theta, X)
# Calculating the regularization parameter for all thetas but the intercept
regParam <- (C/m) * theta[-1]
gradient <- (1/m) * (t(X) %*% (h-y))
gradient[-1] <- gradient[-1] + regParam
return(gradient)
}
t(regularized_gradient(initialTheta, X, y, C=1.0))
# Find values of theta that minimize the cost function
optimalTheta <- optim(
# Specifying function parameters
par=initialTheta, # Initial guess
fn=regularized_cost, # Function to minimize
X=X,
y=y,
C=1.0,
method="BFGS", # Optimization function to use
control = list(maxit = 4000) # Maximum number of iterations
)$par # Specifying that we only want the obtained thetas
# Fitting the model
model <- glm(y ~ ., data=cbind(X[, -1], y), family='binomial')
modelCoef <- model$coefficients %>% data.frame(glm=.)
cbind(modelCoef, optimalTheta) %>% t()
# Predicting the class probability with the obtained thetas
df$ClassProbability <- logistic_hypothesis(optimalTheta, X)
df$glmClassProbability <- predict(model, X)
# Assigning those with a class probability above 0.5 as admitted
df$Prediction <- ifelse(df$ClassProbability > 0.5, 1, 0)
df$glmPrediction <- ifelse(df$glmClassProbability > 0.5, 1, 0)
head(df)
ourAccuracy <- mean(ifelse(df$Accepted == df$Prediction, 1, 0))
glmAccuracy <- mean(ifelse(df$Accepted == df$glmPrediction, 1, 0))
cat('Our accuracy:', ourAccuracy, '\n')
cat('R\'s glm accuracy:', glmAccuracy)
# Timing this cell
timeStart <- proc.time()
createBoundary <- function(X, y, C=0) {
# Returns a data frame used for plotting the decision boundary.
# Uses the optimization function to find the optimal values of
# theta, so it is not needed as a parameter.
# Find values of theta that minimize the cost function
initialTheta <- numeric(length(X))
optimalTheta <- optim(
# Specifying function parameters
par=initialTheta, # Initial guess
fn=regularized_cost, # Function to minimize
X=X,
y=y,
C=C,
method="BFGS", # Optimization function to use
control = list(maxit = 4000) # Maximum number of iterations
)$par # Specifying that we only want the obtained thetas
# Creating a mesh grid to generate class probabilities for
grid <- data.frame()
dx <- seq(min(df$Test1), max(df$Test1), by=0.05)
dy <- seq(min(df$Test2), max(df$Test2), length.out=length(dx))
# Filling the grid with values
for (i in 1:length(dx)) {
for (j in 1:length(dx)) {
v <- t(matrix(c(dx[i], dy[j])))
grid <- rbind(grid, v)
}
}
# Generating class probabilities on the polynomial features
grid <- mapFeatures(grid, 6) # Creating polynomial features
grid$z <- logistic_hypothesis(optimalTheta, grid) # Class probabilities
grid <- grid %>% select(x1, x2, z) # Removing polynomial features
return(grid)
}
# Generating the decision boundaries for various levels of lambda
lambda0 <- createBoundary(X, y)
lambda1 <- createBoundary(X, y, 1)
lambda10 <- createBoundary(X, y, 10)
lambda100 <- createBoundary(X, y, 100)
# Plotting the original data
ggplot() +
geom_point(data=df, aes(x=Test1, y=Test2,
# Using alpha instead of color to clean up legend
alpha=as.factor(df$Accepted))) +
xlab('Microchip Test 1') +
ylab('Microchip Test 2') +
# Plotting the decision boundaries
geom_contour(data=lambda0, aes(x=x1, y=x2, z=z, color='Lambda = 0'),
breaks=0.5, alpha=0.4) +
geom_contour(data=lambda1, aes(x=x1, y=x2, z=z, color='Lambda = 1'),
breaks=0.5, alpha=0.4) +
geom_contour(data=lambda10, aes(x=x1, y=x2, z=z, color='Lambda = 10'),
breaks=0.5, alpha=0.4) +
geom_contour(data=lambda100, aes(x=x1, y=x2, z=z, color='Lambda = 100'),
breaks=0.5, alpha=0.4) +
labs(alpha='Accepted', color='Decision Boundaries') +
theme(panel.background = element_blank()) # To aid in seeing rejected points
# Reporting time results
timeStop <- proc.time()
timeStop - timeStart
| 0.592902 | 0.98881 |
Copyright (c) 2020, salesforce.com, inc.
All rights reserved.
SPDX-License-Identifier: BSD-3-Clause
For full license text, see the LICENSE file in the repo root or https://opensource.org/licenses/BSD-3-Clause
### Colab
Try this notebook on Colab: [http://colab.research.google.com/github/salesforce/ai-economist/blob/master/tutorials/economic_simulation_basic.ipynb](http://colab.research.google.com/github/salesforce/ai-economist/blob/master/tutorials/economic_simulation_basic.ipynb)
# Welcome to Foundation!
Foundation is the name of the economic simulator built for the AI Economist ([paper here](https://arxiv.org/abs/2004.13332)). This is the first of several tutorials designed to explain how Foundation works and how it can be used to build simulation environments for studying economic problems.
Just to orient you a bit, Foundation is specially designed for modeling economies in spatial, 2D grid worlds. The AI Economist paper uses a scenario with 4 agents in a world with *Stone* and *Wood*, which can be *collected*, *traded*, and used to build *Houses*. Here's a (nicely rendered) example of what such an environment looks like:

This image just shows what you might see spatially. Behind the scenes, agents have inventories of Stone, Wood, and *Coin*, which they can exchange through a commodities marketplace. In addition, they periodically pay taxes on income earned through trading and building.
**We've open-sourced Foundation to foster transparency and to enable others to build on it!** With that goal in mind, this first tutorial should give you enough to see how to create the type of simulation environment described above and how to interact with it. If you're interested to learn how it all works and how to build on it, make sure to check out the advanced tutorial as well! If, after that, you want to understand more about designing the simulation around economic problems, check out our tutorial explaining how the AI Economist uses Foundation to study the optimal taxation problem!
# Introduction
In this **basic** tutorial, we will demonstrate the basics of how to create an instance of a simulation environment and how to interact with it.
We will cover the following:
1. Markov Decision Processes
2. Creating a Simulation Environment (a Scenario Instance)
3. Interacting with the Simulation
4. Sampling and Visualizing an Episode
## Dependencies:
You can install the ai-economist package using
- the pip package manager OR
- by cloning the ai-economist package and installing the requirements (we shall use this when running on Colab):
```
import sys
IN_COLAB = 'google.colab' in sys.modules
if IN_COLAB:
! git clone https://github.com/salesforce/ai-economist.git
% cd ai-economist
! pip install -e .
else:
! pip install ai-economist
# Import foundation
from ai_economist import foundation
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
from IPython import display
if IN_COLAB:
from tutorials.utils import plotting # plotting utilities for visualizing env state
else:
from utils import plotting
```
# 1. Markov Decision Process
Formally, our economic simulation is a key part of a Markov Decision Process (MDP).
MDPs describe episodes in which agents interact with a stateful environment in a continuous feedback loop. At each timestep, agents receive an observation and use a policy to choose actions. The environment then advances to a new state, using the old state and the chosen actions. The agents then receive new observations and rewards. This process repeats over $T$ timesteps (possibly infinite).
The goal of each agent is to maximize its expected sum of future (discounted) rewards, by finding its optimal policy. Intuitively, this means that an agent needs to understand which (sequence of) actions lead to high rewards (in expectation).
### References
For more information on reinforcement learning and MDPs, check out:
- Richard S. Sutton and Andrew G. Barto, Reinforcement Learning: An Introduction. [http://incompleteideas.net/book/bookdraft2017nov5.pdf](http://incompleteideas.net/book/bookdraft2017nov5.pdf)
# 2. Creating a Simulation Environment (a Scenario Instance)
The Scenario class implements an economic simulation with multiple agents and (optionally) a social planner.
Scenarios provide a high-level gym-style API that lets agents interact with it. The API supports multi-agent observations, actions, etc.
Each Scenario is stateful and implements two main methods:
- __step__, which advances the simulation to the next state, and
- __reset__, which puts the simulation back in an initial state.
Each Scenario is customizable: you can specify options in a dictionary. Here is an example for a scenario with 4 agents:
**Note: This config dictionary will likely seem fairly incomprehensible at this point in the tutorials. Don't worry. The advanced tutorial offers much more context. This is just to get things started and to provide a reference for how to create a "free market" economy from the AI Economist.**
```
# Define the configuration of the environment that will be built
env_config = {
# ===== SCENARIO CLASS =====
# Which Scenario class to use: the class's name in the Scenario Registry (foundation.scenarios).
# The environment object will be an instance of the Scenario class.
'scenario_name': 'layout_from_file/simple_wood_and_stone',
# ===== COMPONENTS =====
# Which components to use (specified as list of ("component_name", {component_kwargs}) tuples).
# "component_name" refers to the Component class's name in the Component Registry (foundation.components)
# {component_kwargs} is a dictionary of kwargs passed to the Component class
# The order in which components reset, step, and generate obs follows their listed order below.
'components': [
# (1) Building houses
('Build', {'skill_dist': "pareto", 'payment_max_skill_multiplier': 3}),
# (2) Trading collectible resources
('ContinuousDoubleAuction', {'max_num_orders': 5}),
# (3) Movement and resource collection
('Gather', {}),
# (4) Taxes
('PeriodicBracketTax', {})
],
# ===== SCENARIO CLASS ARGUMENTS =====
# (optional) kwargs that are added by the Scenario class (i.e. not defined in BaseEnvironment)
'env_layout_file': 'quadrant_25x25_20each_30clump.txt',
'starting_agent_coin': 10,
'fixed_four_skill_and_loc': True,
# ===== STANDARD ARGUMENTS ======
# kwargs that are used by every Scenario class (i.e. defined in BaseEnvironment)
'n_agents': 4, # Number of non-planner agents (must be > 1)
'world_size': [25, 25], # [Height, Width] of the env world
'episode_length': 1000, # Number of timesteps per episode
# In multi-action-mode, the policy selects an action for each action subspace (defined in component code).
# Otherwise, the policy selects only 1 action.
'multi_action_mode_agents': False,
'multi_action_mode_planner': True,
# When flattening observations, concatenate scalar & vector observations before output.
# Otherwise, return observations with minimal processing.
'flatten_observations': False,
# When Flattening masks, concatenate each action subspace mask into a single array.
# Note: flatten_masks = True is required for masking action logits in the code below.
'flatten_masks': True,
}
```
Create an environment instance using this configuration:
```
env = foundation.make_env_instance(**env_config)
```
# 3. Interacting with the Simulation
### Agents
The Agent class holds the state of agents in the simulation. Each Agent instance represents a _logical_ agent.
_Note that this might be separate from a Policy model that lives outside the Scenario and controls the Agent's behavior._
```
env.get_agent(0)
```
### A random policy
Now let's interact with the simulation.
Each Agent needs to choose which actions to execute using a __policy__.
Agents might not always be allowed to execute all actions. For instance, a mobile Agent cannot move beyond the boundary of the world. Hence, in position (0, 0), a mobile cannot move "Left" or "Down". This information is given by a mask, which is provided under ```obs[<agent_id_str>]["action_mask"]``` in the observation dictionary ```obs``` returned by the scenario.
Let's use a random policy to step through the simulation. The methods below implement a random policy.
```
# Note: The code for sampling actions (this cell), and playing an episode (below) are general.
# That is, it doesn't depend on the Scenario and Component classes used in the environment!
def sample_random_action(agent, mask):
"""Sample random UNMASKED action(s) for agent."""
# Return a list of actions: 1 for each action subspace
if agent.multi_action_mode:
split_masks = np.split(mask, agent.action_spaces.cumsum()[:-1])
print(len(split_masks))
print(split_masks[0]/split_masks[0].sum())
return [np.random.choice(np.arange(len(m_)), p=m_/m_.sum()) for m_ in split_masks]
# Return a single action
else:
return np.random.choice(np.arange(agent.action_spaces), p=mask/mask.sum())
def sample_random_actions(env, obs):
"""Samples random UNMASKED actions for each agent in obs."""
actions = {
a_idx: sample_random_action(env.get_agent(a_idx), a_obs['action_mask'])
for a_idx, a_obs in obs.items()
}
return actions
```
Now we're ready to interact with the simulation...
First, environments can be put in an initial state by using __reset__.
```
obs = env.reset()
```
Then, we call __step__ to advance the state and advance time by one tick.
```
actions = sample_random_actions(env, obs)
obs, rew, done, info = env.step(actions)
print(obs)
```
Internally, the __step__ method composes several Components (which act almost like modular sub-Environments) that implement various agent affordances and environment dynamics. For more detailed information on Components and how to implement custom Component classes, see the advanced tutorial. For this tutorial, we will continue to inspect the information that __step__ returns and run a full episode in the simulation.
### Observation
Each observation is a dictionary that contains information for the $N$ agents and (optionally) social planner (with id "p").
```
obs.keys()
```
For each agent, the agent-specific observation is a dictionary. Each Component can contribute information to the agent-specific observation. For instance, the Build Component contributes the
- Build-build_payment (float)
- Build-build_skill (int)
fields, which are defined in the ```generate_observations``` method in [foundation/components/build.py](https://github.com/salesforce/ai-economist/blob/master/ai_economist/foundation/components/build.py).
```
for key, val in obs['0'].items():
print("{:50} {}".format(key, type(val)))
```
### Reward
For each agent / planner, the reward dictionary contains a scalar reward:
```
for agent_idx, reward in rew.items():
print("{:2} {:.3f}".format(agent_idx, reward))
```
### Done
The __done__ object is a dictionary that by default records whether all agents have seen the end of the episode. The default criterion for each agent is to 'stop' their episode once $H$ steps have been executed. Once an agent is 'done', they do not change their state anymore. So, while it's not currently implemented, this could be used to indicate that the episode has ended *for a specific Agent*.
In general, this is useful for telling a Reinforcement Learning framework when to reset the environment and how to organize the trajectories of individual Agents.
```
done
```
### Info
The __info__ object can record any auxiliary information from the simulator, which can be useful, e.g., for visualization. By default, this is empty. To modify this behavior, modify the step() method in [foundation/base/base_env.py](https://github.com/salesforce/ai-economist/blob/master/ai_economist/foundation/base/base_env.py).
```
info
```
# 3. Sampling and Visualizing an Episode
Let's step multiple times with this random policy and visualize the result:
```
def do_plot(env, ax, fig):
"""Plots world state during episode sampling."""
plotting.plot_env_state(env, ax)
ax.set_aspect('equal')
display.display(fig)
display.clear_output(wait=True)
def play_random_episode(env, plot_every=100, do_dense_logging=False):
"""Plays an episode with randomly sampled actions.
Demonstrates gym-style API:
obs <-- env.reset(...) # Reset
obs, rew, done, info <-- env.step(actions, ...) # Interaction loop
"""
fig, ax = plt.subplots(1, 1, figsize=(10, 10))
# Reset
obs = env.reset(force_dense_logging=do_dense_logging)
# Interaction loop (w/ plotting)
for t in range(env.episode_length):
actions = sample_random_actions(env, obs)
obs, rew, done, info = env.step(actions)
if ((t+1) % plot_every) == 0:
do_plot(env, ax, fig)
if ((t+1) % plot_every) != 0:
do_plot(env, ax, fig)
play_random_episode(env, plot_every=100)
```
We see four agents (indicated by a circled __\*__) that move around in the 2-dimensional world. Light brown cells contain Stone, green cells contain Wood. Each agent can build Houses, indicated by corresponding colored cells. Water tiles (blue squares), which prevent movement, divide the map into four quadrants.
Note: this is showing the same information as the image at the top of the tutorial -- it just uses a much more simplistic rendering.
# Visualize using dense logging
Environments built with Foundation provide a couple tools for logging. Perhaps the most useful are **dense logs**. When you reset the environment, you can tell it to create a dense log for the new episode. This will store Agent states at each point in time along with any Component-specific dense log information (say, about builds, trades, etc.) that the Components provide. In addition, it will periodically store a snapshot of the world state.
We provide a few plotting tools that work well with the type of environment showcased here.
```
# Play another episode. This time, tell the environment to do dense logging
play_random_episode(env, plot_every=100, do_dense_logging=True)
# Grab the dense log from the env
dense_log = env.previous_episode_dense_log
# Show the evolution of the world state from t=0 to t=200
fig = plotting.vis_world_range(dense_log, t0=0, tN=200, N=5)
# Show the evolution of the world state over the full episode
fig = plotting.vis_world_range(dense_log, N=5)
# Use the "breakdown" tool to visualize the world state, agent-wise quantities, movement, and trading events
plotting.breakdown(dense_log);
```
# Next
Now that you've seen how to interact with the simulation and generate episodes, try the next tutorial ([economic_simulation_advanced.ipynb](https://github.com/salesforce/ai-economist/blob/master/tutorials/economic_simulation_advanced.ipynb)) that explains how the simulation is composed of low-level Components and Entities. This structure enables flexible extensions of the economic simulation.
|
github_jupyter
|
import sys
IN_COLAB = 'google.colab' in sys.modules
if IN_COLAB:
! git clone https://github.com/salesforce/ai-economist.git
% cd ai-economist
! pip install -e .
else:
! pip install ai-economist
# Import foundation
from ai_economist import foundation
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
from IPython import display
if IN_COLAB:
from tutorials.utils import plotting # plotting utilities for visualizing env state
else:
from utils import plotting
# Define the configuration of the environment that will be built
env_config = {
# ===== SCENARIO CLASS =====
# Which Scenario class to use: the class's name in the Scenario Registry (foundation.scenarios).
# The environment object will be an instance of the Scenario class.
'scenario_name': 'layout_from_file/simple_wood_and_stone',
# ===== COMPONENTS =====
# Which components to use (specified as list of ("component_name", {component_kwargs}) tuples).
# "component_name" refers to the Component class's name in the Component Registry (foundation.components)
# {component_kwargs} is a dictionary of kwargs passed to the Component class
# The order in which components reset, step, and generate obs follows their listed order below.
'components': [
# (1) Building houses
('Build', {'skill_dist': "pareto", 'payment_max_skill_multiplier': 3}),
# (2) Trading collectible resources
('ContinuousDoubleAuction', {'max_num_orders': 5}),
# (3) Movement and resource collection
('Gather', {}),
# (4) Taxes
('PeriodicBracketTax', {})
],
# ===== SCENARIO CLASS ARGUMENTS =====
# (optional) kwargs that are added by the Scenario class (i.e. not defined in BaseEnvironment)
'env_layout_file': 'quadrant_25x25_20each_30clump.txt',
'starting_agent_coin': 10,
'fixed_four_skill_and_loc': True,
# ===== STANDARD ARGUMENTS ======
# kwargs that are used by every Scenario class (i.e. defined in BaseEnvironment)
'n_agents': 4, # Number of non-planner agents (must be > 1)
'world_size': [25, 25], # [Height, Width] of the env world
'episode_length': 1000, # Number of timesteps per episode
# In multi-action-mode, the policy selects an action for each action subspace (defined in component code).
# Otherwise, the policy selects only 1 action.
'multi_action_mode_agents': False,
'multi_action_mode_planner': True,
# When flattening observations, concatenate scalar & vector observations before output.
# Otherwise, return observations with minimal processing.
'flatten_observations': False,
# When Flattening masks, concatenate each action subspace mask into a single array.
# Note: flatten_masks = True is required for masking action logits in the code below.
'flatten_masks': True,
}
env = foundation.make_env_instance(**env_config)
env.get_agent(0)
# Note: The code for sampling actions (this cell), and playing an episode (below) are general.
# That is, it doesn't depend on the Scenario and Component classes used in the environment!
def sample_random_action(agent, mask):
"""Sample random UNMASKED action(s) for agent."""
# Return a list of actions: 1 for each action subspace
if agent.multi_action_mode:
split_masks = np.split(mask, agent.action_spaces.cumsum()[:-1])
print(len(split_masks))
print(split_masks[0]/split_masks[0].sum())
return [np.random.choice(np.arange(len(m_)), p=m_/m_.sum()) for m_ in split_masks]
# Return a single action
else:
return np.random.choice(np.arange(agent.action_spaces), p=mask/mask.sum())
def sample_random_actions(env, obs):
"""Samples random UNMASKED actions for each agent in obs."""
actions = {
a_idx: sample_random_action(env.get_agent(a_idx), a_obs['action_mask'])
for a_idx, a_obs in obs.items()
}
return actions
obs = env.reset()
actions = sample_random_actions(env, obs)
obs, rew, done, info = env.step(actions)
print(obs)
obs.keys()
for key, val in obs['0'].items():
print("{:50} {}".format(key, type(val)))
for agent_idx, reward in rew.items():
print("{:2} {:.3f}".format(agent_idx, reward))
done
info
def do_plot(env, ax, fig):
"""Plots world state during episode sampling."""
plotting.plot_env_state(env, ax)
ax.set_aspect('equal')
display.display(fig)
display.clear_output(wait=True)
def play_random_episode(env, plot_every=100, do_dense_logging=False):
"""Plays an episode with randomly sampled actions.
Demonstrates gym-style API:
obs <-- env.reset(...) # Reset
obs, rew, done, info <-- env.step(actions, ...) # Interaction loop
"""
fig, ax = plt.subplots(1, 1, figsize=(10, 10))
# Reset
obs = env.reset(force_dense_logging=do_dense_logging)
# Interaction loop (w/ plotting)
for t in range(env.episode_length):
actions = sample_random_actions(env, obs)
obs, rew, done, info = env.step(actions)
if ((t+1) % plot_every) == 0:
do_plot(env, ax, fig)
if ((t+1) % plot_every) != 0:
do_plot(env, ax, fig)
play_random_episode(env, plot_every=100)
# Play another episode. This time, tell the environment to do dense logging
play_random_episode(env, plot_every=100, do_dense_logging=True)
# Grab the dense log from the env
dense_log = env.previous_episode_dense_log
# Show the evolution of the world state from t=0 to t=200
fig = plotting.vis_world_range(dense_log, t0=0, tN=200, N=5)
# Show the evolution of the world state over the full episode
fig = plotting.vis_world_range(dense_log, N=5)
# Use the "breakdown" tool to visualize the world state, agent-wise quantities, movement, and trading events
plotting.breakdown(dense_log);
| 0.533397 | 0.884639 |
# X To A Testis 2
```
import os
import sys
from pathlib import Path
from collections import defaultdict
from IPython.display import display, HTML, Markdown
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
# Project level imports
sys.path.insert(0, '../lib')
from larval_gonad.notebook import Nb
from larval_gonad.plotting import make_figs
from larval_gonad.config import memory
from larval_gonad.x_to_a import CHROMS_CHR, AUTOSOMES_CHR, commonly_expressed, multi_chrom_boxplot, get_gene_sets
# Setup notebook
nbconfig = Nb.setup_notebook('2018-03-27_x2a_testis2', seurat_dir='../output/testis2_scRNAseq')
clusters = nbconfig.seurat.get_clusters()
clus6 = clusters['res.0.6']
soma = [2, 9, 10]
early = [4, 8]
late = [0, 3, 5, 7]
```
## Gene Groups
Chromosomal distribution of gene lists.
```
gene_sets = get_gene_sets()
# Get list of genes in 1/3 of samples
norm = nbconfig.seurat.get_normalized_read_counts()
oneThird = np.array(commonly_expressed(norm))
oneThird.shape
_dat = norm.T.join(clus6).groupby('res.0.6').sum().T
melted = _dat.reset_index().melt(id_vars=['index'])
melted.columns = ['FBgn', 'cluster', 'value']
melted.
melted.
expressed = norm.index[norm.index.isin(housekeeping)]
norm.head()
expressed = norm[((norm > 0).sum(axis=1) > 1200)].index
data = norm.loc[expressed, :].T.join(clus6)
data.rename({'res.0.6': 'cluster'}, axis=1, inplace=True)
dat = data.groupby('cluster').median().T.reset_index()\
.melt(id_vars='index')\
.merge(nbconfig.fbgn2chrom, left_on='index', right_index=True)\
.set_index('index')
def _plot(dat):
num_cells = data.groupby('cluster').count().iloc[:, 0].to_dict()
g = sns.FacetGrid(dat, col='cluster', col_wrap=2, size=4)
g.map_dataframe(multi_chrom_boxplot, 'chrom', 'value', num_cells=num_cells, palette=nbconfig.color_chrom,
notch=True, flierprops=nbconfig.fliersprops)
#g.set(ylim=(0, 600))
_plot(dat)
Ydata = norm.join(nbconfig.fbgn2chrom).query('chrom == "chrY"').drop('chrom', axis=1)
Ydata = Ydata.T.loc[clus6.sort_values().index]
Ydata.columns = Ydata.columns.map(lambda x: nbconfig.fbgn2symbol[x])
levels = sorted(clus6.unique())
colors = sns.color_palette('tab20', n_colors=len(levels))
mapper = dict(zip(levels, colors))
cmap = clus6.sort_values().map(mapper)
g = sns.clustermap(Ydata, row_cluster=False, col_cluster=True, yticklabels=False, row_colors=cmap, figsize=(20, 10))
g.ax_col_dendrogram.set_visible(False)
for label in levels:
g.ax_row_dendrogram.bar(0, 0, color=mapper[label],
label=label, linewidth=0)
g.ax_row_dendrogram.legend(loc="center", ncol=2)
soma = clus12[clus12 == 13].index.tolist()
early = clus12[clus12 == 8].index.tolist()
late = clus12[clus12 == 6].index.tolist()
from larval_gonad.x_to_a import estimate_dcc, clean_pvalue
from scipy.stats import mannwhitneyu
def boxplot(data, expressed, mask, chrom, ax, name):
_data = data.loc[expressed, mask]
_data['median'] = _data.median(axis=1)
_data = _data.join(chrom, how='inner')
med_x, med_major, prop_dcc = estimate_dcc('chrom', 'median', _data)
_data['chrom'] = _data['chrom'].map(lambda x: x.replace('chr', ''))
ORDER = ['X', '2L', '2R', '3L', '3R', '4']
sns.boxplot(_data['chrom'], _data['median'], order=ORDER, notch=True, boxprops={"facecolor": 'w'}, ax=ax, flierprops={'alpha': .6})
ax.axhline(med_major, ls=':', lw=2, color=nbconfig.color_c1)
ax.set_title(name)
ax.set_xlabel('Chromosome')
ax.set_ylabel('Median Normalized Expression')
# Clean up the pvalue for plotting
pvalues = {}
iqr = 0
chromX = _data[_data.chrom == 'X']
for g, df in _data.groupby('chrom'):
_iqr = sns.utils.iqr(df['median'])
if _iqr > iqr:
iqr = _iqr
if g == 'X':
continue
if g == 'M':
continue
_, pval = mannwhitneyu(chromX['median'], df['median'], alternative='two-sided')
if pval <= 0.001:
pvalues[g] = '***'
multiplier = .35
xloc = ORDER.index('X')
for k, v in pvalues.items():
oloc = ORDER.index(k)
pval = v
y, h, col = iqr + iqr * multiplier, .1, 'k'
plt.plot([xloc, xloc, oloc, oloc], [y, y+h, y+h, y], lw=1, c=col)
plt.text((xloc+oloc)*.5, y+h+.01, f"{pval}", ha='center',
va='bottom', color=col)
multiplier += .2
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(8.5, 3.3), sharex=True, sharey=True)
chrom = nbconfig.fbgn2chrom
boxplot(norm, expressed, soma, chrom, ax1, 'Somatic Cells')
boxplot(norm, expressed, early, chrom, ax2, 'Early Germ Cells')
boxplot(norm, expressed, late, chrom, ax3, 'Late Germ Cells')
ax2.set_ylabel('')
ax3.set_ylabel('')
plt.savefig('../output/figures/2018-03-16_x2a_combined_forced_simple_boxplot.png', dpi=300)
```
|
github_jupyter
|
import os
import sys
from pathlib import Path
from collections import defaultdict
from IPython.display import display, HTML, Markdown
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
# Project level imports
sys.path.insert(0, '../lib')
from larval_gonad.notebook import Nb
from larval_gonad.plotting import make_figs
from larval_gonad.config import memory
from larval_gonad.x_to_a import CHROMS_CHR, AUTOSOMES_CHR, commonly_expressed, multi_chrom_boxplot, get_gene_sets
# Setup notebook
nbconfig = Nb.setup_notebook('2018-03-27_x2a_testis2', seurat_dir='../output/testis2_scRNAseq')
clusters = nbconfig.seurat.get_clusters()
clus6 = clusters['res.0.6']
soma = [2, 9, 10]
early = [4, 8]
late = [0, 3, 5, 7]
gene_sets = get_gene_sets()
# Get list of genes in 1/3 of samples
norm = nbconfig.seurat.get_normalized_read_counts()
oneThird = np.array(commonly_expressed(norm))
oneThird.shape
_dat = norm.T.join(clus6).groupby('res.0.6').sum().T
melted = _dat.reset_index().melt(id_vars=['index'])
melted.columns = ['FBgn', 'cluster', 'value']
melted.
melted.
expressed = norm.index[norm.index.isin(housekeeping)]
norm.head()
expressed = norm[((norm > 0).sum(axis=1) > 1200)].index
data = norm.loc[expressed, :].T.join(clus6)
data.rename({'res.0.6': 'cluster'}, axis=1, inplace=True)
dat = data.groupby('cluster').median().T.reset_index()\
.melt(id_vars='index')\
.merge(nbconfig.fbgn2chrom, left_on='index', right_index=True)\
.set_index('index')
def _plot(dat):
num_cells = data.groupby('cluster').count().iloc[:, 0].to_dict()
g = sns.FacetGrid(dat, col='cluster', col_wrap=2, size=4)
g.map_dataframe(multi_chrom_boxplot, 'chrom', 'value', num_cells=num_cells, palette=nbconfig.color_chrom,
notch=True, flierprops=nbconfig.fliersprops)
#g.set(ylim=(0, 600))
_plot(dat)
Ydata = norm.join(nbconfig.fbgn2chrom).query('chrom == "chrY"').drop('chrom', axis=1)
Ydata = Ydata.T.loc[clus6.sort_values().index]
Ydata.columns = Ydata.columns.map(lambda x: nbconfig.fbgn2symbol[x])
levels = sorted(clus6.unique())
colors = sns.color_palette('tab20', n_colors=len(levels))
mapper = dict(zip(levels, colors))
cmap = clus6.sort_values().map(mapper)
g = sns.clustermap(Ydata, row_cluster=False, col_cluster=True, yticklabels=False, row_colors=cmap, figsize=(20, 10))
g.ax_col_dendrogram.set_visible(False)
for label in levels:
g.ax_row_dendrogram.bar(0, 0, color=mapper[label],
label=label, linewidth=0)
g.ax_row_dendrogram.legend(loc="center", ncol=2)
soma = clus12[clus12 == 13].index.tolist()
early = clus12[clus12 == 8].index.tolist()
late = clus12[clus12 == 6].index.tolist()
from larval_gonad.x_to_a import estimate_dcc, clean_pvalue
from scipy.stats import mannwhitneyu
def boxplot(data, expressed, mask, chrom, ax, name):
_data = data.loc[expressed, mask]
_data['median'] = _data.median(axis=1)
_data = _data.join(chrom, how='inner')
med_x, med_major, prop_dcc = estimate_dcc('chrom', 'median', _data)
_data['chrom'] = _data['chrom'].map(lambda x: x.replace('chr', ''))
ORDER = ['X', '2L', '2R', '3L', '3R', '4']
sns.boxplot(_data['chrom'], _data['median'], order=ORDER, notch=True, boxprops={"facecolor": 'w'}, ax=ax, flierprops={'alpha': .6})
ax.axhline(med_major, ls=':', lw=2, color=nbconfig.color_c1)
ax.set_title(name)
ax.set_xlabel('Chromosome')
ax.set_ylabel('Median Normalized Expression')
# Clean up the pvalue for plotting
pvalues = {}
iqr = 0
chromX = _data[_data.chrom == 'X']
for g, df in _data.groupby('chrom'):
_iqr = sns.utils.iqr(df['median'])
if _iqr > iqr:
iqr = _iqr
if g == 'X':
continue
if g == 'M':
continue
_, pval = mannwhitneyu(chromX['median'], df['median'], alternative='two-sided')
if pval <= 0.001:
pvalues[g] = '***'
multiplier = .35
xloc = ORDER.index('X')
for k, v in pvalues.items():
oloc = ORDER.index(k)
pval = v
y, h, col = iqr + iqr * multiplier, .1, 'k'
plt.plot([xloc, xloc, oloc, oloc], [y, y+h, y+h, y], lw=1, c=col)
plt.text((xloc+oloc)*.5, y+h+.01, f"{pval}", ha='center',
va='bottom', color=col)
multiplier += .2
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(8.5, 3.3), sharex=True, sharey=True)
chrom = nbconfig.fbgn2chrom
boxplot(norm, expressed, soma, chrom, ax1, 'Somatic Cells')
boxplot(norm, expressed, early, chrom, ax2, 'Early Germ Cells')
boxplot(norm, expressed, late, chrom, ax3, 'Late Germ Cells')
ax2.set_ylabel('')
ax3.set_ylabel('')
plt.savefig('../output/figures/2018-03-16_x2a_combined_forced_simple_boxplot.png', dpi=300)
| 0.344664 | 0.761006 |
# Transpiling Quantum Circuits
In this chapter we will investigate how quantum circuits are transformed when run on quantum devices. That we need to modify the circuits at all is a consequence of the limitations of current quantum computing hardware. Namely, the limited connectivity inherent in most quantum hardware, restricted gate sets, as well as environmental noise and gate errors, all conspire to limit the effective computational power on today's quantum devices. Fortunately, quantum circuit rewriting tool chains have been developed that directly address these issues, and return heavily optimized circuits mapped to targeted quantum devices. Here we will explore the IBM Qiskit 'transpiler' circuit rewriting framework.
```
import numpy as np
from qiskit import *
from qiskit.tools.jupyter import *
from qiskit.providers.ibmq import least_busy
%matplotlib inline
IBMQ.load_account()
```
## Core Steps in Circuit Rewriting
As we will see, rewriting quantum circuits to match hardware constraints and optimize for performance can be far from trivial. The flow of logic in the rewriting tool chain need not be linear, and can often have iterative sub-loops, conditional branches, and other complex behaviors. That being said, the basic building blocks follow the structure given below.
<img src="images/transpiling_core_steps.png" style="width=auto;"/>
Our goal in this section is to see what each of these "passes" does at a high-level, and then begin exploring their usage on a set of common circuits.
### Unrolling to Basis Gates
When writing a quantum circuit you are free to use any quantum gate (unitary operator) that you like, along with a collection of non-gate operations such as qubit measurements and reset operations. However, when running a circuit on a real quantum device one no longer has this flexibility. Due to limitations in, for example, the physical interactions between qubits, difficulty in implementing multi-qubit gates, control electronics etc, a quantum computing device can only natively support a handful of quantum gates and non-gate operations. In the present case of IBM Q devices, the native gate set can be found by querying the devices themselves:
```
provider = IBMQ.get_provider(group='open')
provider.backends(simulator=False)
backend = least_busy(provider.backends(filters=lambda x: x.configuration().n_qubits >= 5 and not x.configuration().simulator and x.status().operational==True))
backend.configuration().basis_gates
```
We see that the our device supports five native gates: three single-qubit gates (`u1`, `u2`, `u3`, and `id`) and one two-qubit entangling gate `cx`. In addition, the device supports qubit measurements (otherwise we can not read out an answer!). Although we have queried only a single device, all IBM Q devices support this gate set.
The `u*` gates represent arbitrary single-qubit rotations of one, two, and three angles. The `u1` gates are single-parameter rotations that represent generalized phase gates of the form
$$
U_{1}(\lambda) = \begin{bmatrix}
1 & 0 \\
0 & e^{i\lambda}
\end{bmatrix}
$$
This set includes common gates such as $Z$, $T$, $T^{\dagger}$, $S$, and $S^{\dagger}$. It turns out that these gates do not actually need to be performed on hardware, but instead, can be implemented in software as "virtual gates". These virtual gates are called "frame changes" and take zero time, and have no associated error; they are free gates on hardware.
Two-angle rotations, $U_{2}(\phi,\lambda)$, are actually two frame changes with a single $X_{\pi/2}$ gate in between them, and can be used to synthesize gates like the Hadamard ($U_{2}(0,\pi)$) gate. As the only actual gate performed is the $X_{\pi/2}$ gate, the error and gate time associated with any $U_{2}$ gate is the same as an $X_{\pi/2}$ gate. Similarly, $U_{3}(\theta,\phi,\lambda)$ gates are formed from three frame changes with two $X_{\pi/2}$ gates in between them. The errors and gate times are twice those of a single $X_{\pi/2}$. The identity gate, $id$, is straightforward, and is a placeholder gate with a fixed time-interval.
The only entangling gate supported by the IBM Q devices is the CNOT gate (`cx`) that, in the computational basis, can be written as:
$$
\mathrm{CNOT}(0,1) = \begin{bmatrix}
1 & 0 & 0 & 0 \\
0 & 0 & 0 & 1 \\
0 & 0 & 1 & 0 \\
0 & 1 & 0 & 0
\end{bmatrix}
$$,
where we see that the matrix form follows from the specific bit-ordering convention used in Qiskit.
Every quantum circuit run on a IBM Q device must be expressed using only these basis gates. For example, suppose one wants to run a simple phase estimation circuit:
```
qr = QuantumRegister(2, 'q')
cr = ClassicalRegister(1, 'c')
qc = QuantumCircuit(qr, cr)
qc.h(qr[0])
qc.x(qr[1])
qc.cu1(np.pi/4, qr[0], qr[1])
qc.h(qr[0])
qc.measure(qr[0], cr[0])
qc.draw(output='mpl')
```
We have $H$, $X$, and controlled-$U_{1}$ gates, all of which are not in our devices basis gate set, and must be expanded. We will see that this expansion is taken care of for you, but for now let us just rewrite the circuit in the basis gate set:
```
qr = QuantumRegister(2, 'q')
cr = ClassicalRegister(1, 'c')
qc_basis = QuantumCircuit(qr, cr)
# Hadamard in U2 format
qc_basis.u2(0, np.pi, qr[0])
# X gate in U3 format
qc_basis.u3(np.pi, 0, np.pi, qr[1])
# Decomposition for controlled-U1 with lambda=pi/4
qc_basis.u1(np.pi/8, qr[0])
qc_basis.cx(qr[0], qr[1])
qc_basis.u1(-np.pi/8, qr[1])
qc_basis.cx(qr[0], qr[1])
qc_basis.u1(np.pi/8, qr[1])
# Hadamard in U2 format
qc_basis.u2(0, np.pi, qr[0])
qc_basis.measure(qr[0], cr[0])
qc_basis.draw(output='mpl')
```
A few things to highlight. One, the circuit has gotten longer with respect to the initial one. This can be verified by checking the depth of the circuits:
```
print(qc.depth(), ',', qc_basis.depth())
```
Second, although we had a single controlled gate, the fact that it was not in the basis set means that, when expanded, it requires more than a single `cx` gate to implement. All said, unrolling to the basis set of gates leads to an increase in the length of a quantum circuit and the number of gates. Both of these increases lead to more errors from the environment and gate errors, respectively, and further circuit rewriting steps must try to mitigate this effect through circuit optimizations.
Finally, we will look at the particularly important example of a Toffoli, or controlled-controlled-not gate:
```
qr = QuantumRegister(3, 'q')
qc = QuantumCircuit(qr)
qc.ccx(qr[0], qr[1], qr[2])
qc.draw(output='mpl')
```
As a three-qubit gate, it should already be clear that this is not in the basis set of our devices. We have already seen that controlled gates not in the basis set are typically decomposed into multiple CNOT gates. This is doubly true for controlled gates with more than two qubits, where multiple CNOT gates are needed to implement the entangling across the multiple qubits. In our basis set, the Toffoli gate can be written as:
```
qr = QuantumRegister(3, 'q')
qc_basis = QuantumCircuit(qr)
qc_basis.u2(0,np.pi, qr[2])
qc_basis.cx(qr[1], qr[2])
qc_basis.u1(-np.pi/4, qr[2])
qc_basis.cx(qr[0], qr[2])
qc_basis.u1(np.pi/4, qr[2])
qc_basis.cx(qr[1], qr[2])
qc_basis.u1(np.pi/4, qr[1])
qc_basis.u1(-np.pi/4, qr[2])
qc_basis.cx(qr[0], qr[2])
qc_basis.cx(qr[0], qr[1])
qc_basis.u1(np.pi/4, qr[2])
qc_basis.u1(np.pi/4, qr[0])
qc_basis.u1(-np.pi/4, qr[1])
qc_basis.u2(0,np.pi, qr[2])
qc_basis.cx(qr[0], qr[1])
qc_basis.draw(output='mpl')
```
Therefore, for every Toffoli gate in a quantum circuit, the IBM Q hardware must execute six CNOT gates, and a handful of single-qubit gates. From this example, it should be clear that any algorithm that makes use of multiple Toffoli gates will end up as a circuit with large depth and with therefore be appreciably affected by noise and gate errors.
### Initial Layout
```
qr = QuantumRegister(5, 'q')
cr = ClassicalRegister(5, 'c')
qc = QuantumCircuit(qr, cr)
qc.h(qr[0])
qc.cx(qr[0], qr[4])
qc.cx(qr[4], qr[3])
qc.cx(qr[3], qr[1])
qc.cx(qr[1], qr[2])
qc.draw(output='mpl')
from qiskit.visualization.gate_map import plot_gate_map
plot_gate_map(backend, plot_directed=True)
```
|
github_jupyter
|
import numpy as np
from qiskit import *
from qiskit.tools.jupyter import *
from qiskit.providers.ibmq import least_busy
%matplotlib inline
IBMQ.load_account()
provider = IBMQ.get_provider(group='open')
provider.backends(simulator=False)
backend = least_busy(provider.backends(filters=lambda x: x.configuration().n_qubits >= 5 and not x.configuration().simulator and x.status().operational==True))
backend.configuration().basis_gates
qr = QuantumRegister(2, 'q')
cr = ClassicalRegister(1, 'c')
qc = QuantumCircuit(qr, cr)
qc.h(qr[0])
qc.x(qr[1])
qc.cu1(np.pi/4, qr[0], qr[1])
qc.h(qr[0])
qc.measure(qr[0], cr[0])
qc.draw(output='mpl')
qr = QuantumRegister(2, 'q')
cr = ClassicalRegister(1, 'c')
qc_basis = QuantumCircuit(qr, cr)
# Hadamard in U2 format
qc_basis.u2(0, np.pi, qr[0])
# X gate in U3 format
qc_basis.u3(np.pi, 0, np.pi, qr[1])
# Decomposition for controlled-U1 with lambda=pi/4
qc_basis.u1(np.pi/8, qr[0])
qc_basis.cx(qr[0], qr[1])
qc_basis.u1(-np.pi/8, qr[1])
qc_basis.cx(qr[0], qr[1])
qc_basis.u1(np.pi/8, qr[1])
# Hadamard in U2 format
qc_basis.u2(0, np.pi, qr[0])
qc_basis.measure(qr[0], cr[0])
qc_basis.draw(output='mpl')
print(qc.depth(), ',', qc_basis.depth())
qr = QuantumRegister(3, 'q')
qc = QuantumCircuit(qr)
qc.ccx(qr[0], qr[1], qr[2])
qc.draw(output='mpl')
qr = QuantumRegister(3, 'q')
qc_basis = QuantumCircuit(qr)
qc_basis.u2(0,np.pi, qr[2])
qc_basis.cx(qr[1], qr[2])
qc_basis.u1(-np.pi/4, qr[2])
qc_basis.cx(qr[0], qr[2])
qc_basis.u1(np.pi/4, qr[2])
qc_basis.cx(qr[1], qr[2])
qc_basis.u1(np.pi/4, qr[1])
qc_basis.u1(-np.pi/4, qr[2])
qc_basis.cx(qr[0], qr[2])
qc_basis.cx(qr[0], qr[1])
qc_basis.u1(np.pi/4, qr[2])
qc_basis.u1(np.pi/4, qr[0])
qc_basis.u1(-np.pi/4, qr[1])
qc_basis.u2(0,np.pi, qr[2])
qc_basis.cx(qr[0], qr[1])
qc_basis.draw(output='mpl')
qr = QuantumRegister(5, 'q')
cr = ClassicalRegister(5, 'c')
qc = QuantumCircuit(qr, cr)
qc.h(qr[0])
qc.cx(qr[0], qr[4])
qc.cx(qr[4], qr[3])
qc.cx(qr[3], qr[1])
qc.cx(qr[1], qr[2])
qc.draw(output='mpl')
from qiskit.visualization.gate_map import plot_gate_map
plot_gate_map(backend, plot_directed=True)
| 0.464659 | 0.993448 |
<a href="https://colab.research.google.com/github/blkbamboo74/DS-Unit-1-Sprint-2-Data-Wrangling-and-Storytelling/blob/master/Dennis_Batiste_LS_DSPT3_123_Make_Explanatory_Visualizations.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
_Lambda School Data Science_
# Make Explanatory Visualizations
### Objectives
- identify misleading visualizations and how to fix them
- use Seaborn to visualize distributions and relationships with continuous and discrete variables
- add emphasis and annotations to transform visualizations from exploratory to explanatory
- remove clutter from visualizations
### Links
- [How to Spot Visualization Lies](https://flowingdata.com/2017/02/09/how-to-spot-visualization-lies/)
- [Visual Vocabulary - Vega Edition](http://ft.com/vocabulary)
- [Choosing a Python Visualization Tool flowchart](http://pbpython.com/python-vis-flowchart.html)
- [Searborn example gallery](http://seaborn.pydata.org/examples/index.html) & [tutorial](http://seaborn.pydata.org/tutorial.html)
- [Strong Titles Are The Biggest Bang for Your Buck](http://stephanieevergreen.com/strong-titles/)
- [Remove to improve (the data-ink ratio)](https://www.darkhorseanalytics.com/blog/data-looks-better-naked)
- [How to Generate FiveThirtyEight Graphs in Python](https://www.dataquest.io/blog/making-538-plots/)
# Avoid Misleading Visualizations
Did you find/discuss any interesting misleading visualizations in your Walkie Talkie?
## What makes a visualization misleading?
[5 Ways Writers Use Misleading Graphs To Manipulate You](https://venngage.com/blog/misleading-graphs/)
## Two y-axes
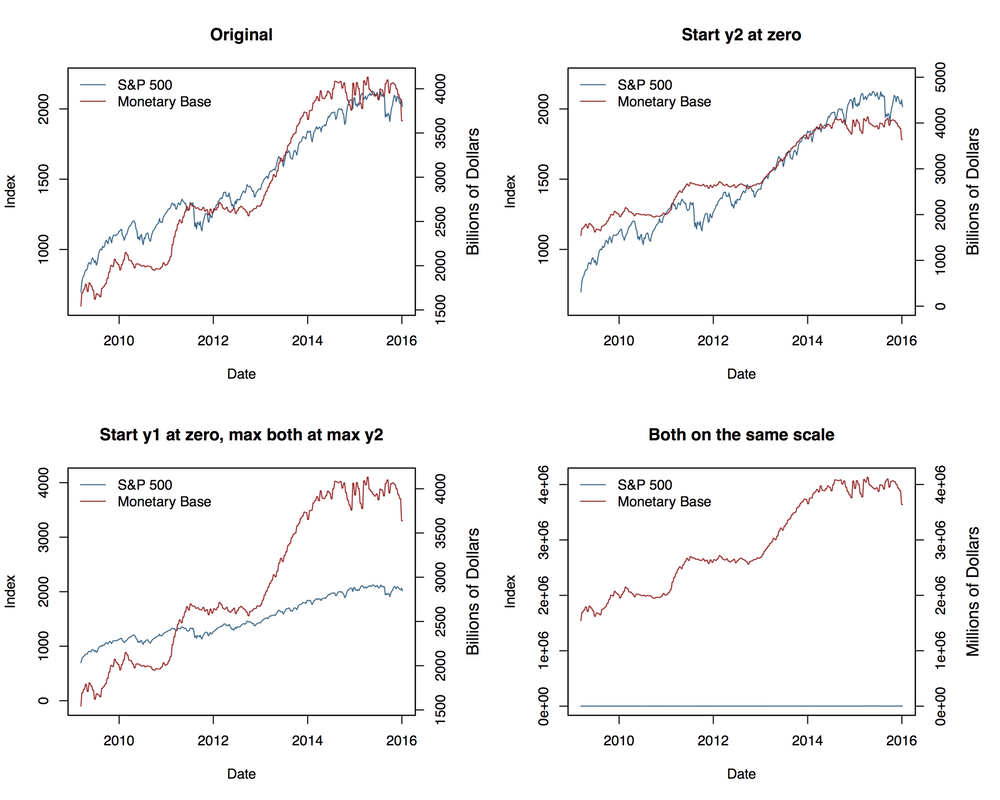
Other Examples:
- [Spurious Correlations](https://tylervigen.com/spurious-correlations)
- <https://blog.datawrapper.de/dualaxis/>
- <https://kieranhealy.org/blog/archives/2016/01/16/two-y-axes/>
- <http://www.storytellingwithdata.com/blog/2016/2/1/be-gone-dual-y-axis>
## Y-axis doesn't start at zero.
<img src="https://i.pinimg.com/originals/22/53/a9/2253a944f54bb61f1983bc076ff33cdd.jpg" width="600">
## Pie Charts are bad
<img src="https://i1.wp.com/flowingdata.com/wp-content/uploads/2009/11/Fox-News-pie-chart.png?fit=620%2C465&ssl=1" width="600">
## Pie charts that omit data are extra bad
- A guy makes a misleading chart that goes viral
What does this chart imply at first glance? You don't want your user to have to do a lot of work in order to be able to interpret you graph correctly. You want that first-glance conclusions to be the correct ones.
<img src="https://pbs.twimg.com/media/DiaiTLHWsAYAEEX?format=jpg&name=medium" width='600'>
<https://twitter.com/michaelbatnick/status/1019680856837849090?lang=en>
- It gets picked up by overworked journalists (assuming incompetency before malice)
<https://www.marketwatch.com/story/this-1-chart-puts-mega-techs-trillions-of-market-value-into-eye-popping-perspective-2018-07-18>
- Even after the chart's implications have been refuted, it's hard a bad (although compelling) visualization from being passed around.
<https://www.linkedin.com/pulse/good-bad-pie-charts-karthik-shashidhar/>
**["yea I understand a pie chart was probably not the best choice to present this data."](https://twitter.com/michaelbatnick/status/1037036440494985216)**
## Pie Charts that compare unrelated things are next-level extra bad
<img src="http://www.painting-with-numbers.com/download/document/186/170403+Legalizing+Marijuana+Graph.jpg" width="600">
## Be careful about how you use volume to represent quantities:
radius vs diameter vs volume
<img src="https://static1.squarespace.com/static/5bfc8dbab40b9d7dd9054f41/t/5c32d86e0ebbe80a25873249/1546836082961/5474039-25383714-thumbnail.jpg?format=1500w" width="600">
## Don't cherrypick timelines or specific subsets of your data:
<img src="https://wattsupwiththat.com/wp-content/uploads/2019/02/Figure-1-1.png" width="600">
Look how specifically the writer has selected what years to show in the legend on the right side.
<https://wattsupwiththat.com/2019/02/24/strong-arctic-sea-ice-growth-this-year/>
Try the tool that was used to make the graphic for yourself
<http://nsidc.org/arcticseaicenews/charctic-interactive-sea-ice-graph/>
## Use Relative units rather than Absolute Units
<img src="https://imgs.xkcd.com/comics/heatmap_2x.png" width="600">
## Avoid 3D graphs unless having the extra dimension is effective
Usually you can Split 3D graphs into multiple 2D graphs
3D graphs that are interactive can be very cool. (See Plotly and Bokeh)
<img src="https://thumbor.forbes.com/thumbor/1280x868/https%3A%2F%2Fblogs-images.forbes.com%2Fthumbnails%2Fblog_1855%2Fpt_1855_811_o.jpg%3Ft%3D1339592470" width="600">
## Don't go against typical conventions
<img src="http://www.callingbullshit.org/twittercards/tools_misleading_axes.png" width="600">
# Tips for choosing an appropriate visualization:
## Use Appropriate "Visual Vocabulary"
[Visual Vocabulary - Vega Edition](http://ft.com/vocabulary)
## What are the properties of your data?
- Is your primary variable of interest continuous or discrete?
- Is in wide or long (tidy) format?
- Does your visualization involve multiple variables?
- How many dimensions do you need to include on your plot?
Can you express the main idea of your visualization in a single sentence?
How hard does your visualization make the user work in order to draw the intended conclusion?
## Which Visualization tool is most appropriate?
[Choosing a Python Visualization Tool flowchart](http://pbpython.com/python-vis-flowchart.html)
## Anatomy of a Matplotlib Plot

```
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.ticker import AutoMinorLocator, MultipleLocator, FuncFormatter
np.random.seed(19680801)
X = np.linspace(0.5, 3.5, 100)
Y1 = 3+np.cos(X)
Y2 = 1+np.cos(1+X/0.75)/2
Y3 = np.random.uniform(Y1, Y2, len(X))
fig = plt.figure(figsize=(8, 8))
ax = fig.add_subplot(1, 1, 1, aspect=1)
def minor_tick(x, pos):
if not x % 1.0:
return ""
return "%.2f" % x
ax.xaxis.set_major_locator(MultipleLocator(1.000))
ax.xaxis.set_minor_locator(AutoMinorLocator(4))
ax.yaxis.set_major_locator(MultipleLocator(1.000))
ax.yaxis.set_minor_locator(AutoMinorLocator(4))
ax.xaxis.set_minor_formatter(FuncFormatter(minor_tick))
ax.set_xlim(0, 4)
ax.set_ylim(0, 4)
ax.tick_params(which='major', width=1.0)
ax.tick_params(which='major', length=10)
ax.tick_params(which='minor', width=1.0, labelsize=10)
ax.tick_params(which='minor', length=5, labelsize=10, labelcolor='0.25')
ax.grid(linestyle="--", linewidth=0.5, color='.25', zorder=-10)
ax.plot(X, Y1, c=(0.25, 0.25, 1.00), lw=2, label="Blue signal", zorder=10)
ax.plot(X, Y2, c=(1.00, 0.25, 0.25), lw=2, label="Red signal")
ax.plot(X, Y3, linewidth=0,
marker='o', markerfacecolor='w', markeredgecolor='k')
ax.set_title("Anatomy of a figure", fontsize=20, verticalalignment='bottom')
ax.set_xlabel("X axis label")
ax.set_ylabel("Y axis label")
ax.legend()
def circle(x, y, radius=0.15):
from matplotlib.patches import Circle
from matplotlib.patheffects import withStroke
circle = Circle((x, y), radius, clip_on=False, zorder=10, linewidth=1,
edgecolor='black', facecolor=(0, 0, 0, .0125),
path_effects=[withStroke(linewidth=5, foreground='w')])
ax.add_artist(circle)
def text(x, y, text):
ax.text(x, y, text, backgroundcolor="white",
ha='center', va='top', weight='bold', color='blue')
# Minor tick
circle(0.50, -0.10)
text(0.50, -0.32, "Minor tick label")
# Major tick
circle(-0.03, 4.00)
text(0.03, 3.80, "Major tick")
# Minor tick
circle(0.00, 3.50)
text(0.00, 3.30, "Minor tick")
# Major tick label
circle(-0.15, 3.00)
text(-0.15, 2.80, "Major tick label")
# X Label
circle(1.80, -0.27)
text(1.80, -0.45, "X axis label")
# Y Label
circle(-0.27, 1.80)
text(-0.27, 1.6, "Y axis label")
# Title
circle(1.60, 4.13)
text(1.60, 3.93, "Title")
# Blue plot
circle(1.75, 2.80)
text(1.75, 2.60, "Line\n(line plot)")
# Red plot
circle(1.20, 0.60)
text(1.20, 0.40, "Line\n(line plot)")
# Scatter plot
circle(3.20, 1.75)
text(3.20, 1.55, "Markers\n(scatter plot)")
# Grid
circle(3.00, 3.00)
text(3.00, 2.80, "Grid")
# Legend
circle(3.70, 3.80)
text(3.70, 3.60, "Legend")
# Axes
circle(0.5, 0.5)
text(0.5, 0.3, "Axes")
# Figure
circle(-0.3, 0.65)
text(-0.3, 0.45, "Figure")
color = 'blue'
ax.annotate('Spines', xy=(4.0, 0.35), xytext=(3.3, 0.5),
weight='bold', color=color,
arrowprops=dict(arrowstyle='->',
connectionstyle="arc3",
color=color))
ax.annotate('', xy=(3.15, 0.0), xytext=(3.45, 0.45),
weight='bold', color=color,
arrowprops=dict(arrowstyle='->',
connectionstyle="arc3",
color=color))
ax.text(4.0, -0.4, "Made with http://matplotlib.org",
fontsize=10, ha="right", color='.5')
plt.show()
```
# Making Explanatory Visualizations with Matplotlib
Today we will reproduce this [example by FiveThirtyEight:](https://fivethirtyeight.com/features/al-gores-new-movie-exposes-the-big-flaw-in-online-movie-ratings/)
```
from IPython.display import display, Image
url = 'https://fivethirtyeight.com/wp-content/uploads/2017/09/mehtahickey-inconvenient-0830-1.png'
example = Image(url=url, width=400)
display(example)
```
Using this data: https://github.com/fivethirtyeight/data/tree/master/inconvenient-sequel
Links
- [Strong Titles Are The Biggest Bang for Your Buck](http://stephanieevergreen.com/strong-titles/)
- [Remove to improve (the data-ink ratio)](https://www.darkhorseanalytics.com/blog/data-looks-better-naked)
- [How to Generate FiveThirtyEight Graphs in Python](https://www.dataquest.io/blog/making-538-plots/)
## Make prototypes
This helps us understand the problem
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
plt.style.use('fivethirtyeight')
fake = pd.Series([38, 3, 2, 1, 2, 4, 6, 5, 5, 33],
index=range(1,11))
fake.plot.bar(color='C1', width=0.9);
fake2 = pd.Series(
[1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
2, 2, 2,
3, 3, 3,
4, 4,
5, 5, 5,
6, 6, 6, 6,
7, 7, 7, 7, 7,
8, 8, 8, 8,
9, 9, 9, 9,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10])
fake2.value_counts().sort_index().plot.bar(color='C1', width=0.9);
```
## Annotate with text
```
counts = [38, 3, 2, 1, 2, 4, 6, 5, 5, 33]
data_list = []
for i, c in enumerate (counts,1):
data_list = data_list + [i]*c
fake2 = pd.Series(data_list)
plt.style.use('fivethirtyeight')
fake2.value_counts().sort_index().plot.bar(color='#ed713a',width=0.9, rot=0);
plt.text(x=-1,
y=50,
fontsize=16,
fontweight='bold',
s="'An Incovenient Sequel: Truth To Power' is divisive")
plt.text(x=-1,
y=46,
fontsize=16,
s="IMDb ratings for the film as of Aug.29")
plt.xlabel('Rating')
plt.ylabel('Percent of Total Votes')
plt.yticks(range(0, 50, 10));
```
## Reproduce with real data
```
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
df = pd.read_csv('https://raw.githubusercontent.com/fivethirtyeight/data/master/inconvenient-sequel/ratings.csv')
df.describe
df.head()
df.sample(1).T
df['timestamp'] = pd.to_datetime(df['timestamp'])
df.head()
df['timestamp'].describe()
df = df.set_index('timestamp')
df.head()
df['2017-07-17']
df['category'].value_counts()
df_imdb = df[df['category'] == 'IMDb users']
df_imdb.shape
lastday = df['2017-08-29']
lastday[lastday['category'] == 'IMDb users']['respondents'].plot()
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.ticker import AutoMinorLocator, MultipleLocator, FuncFormatter
np.random.seed(19680801)
X = np.linspace(0.5, 3.5, 100)
Y1 = 3+np.cos(X)
Y2 = 1+np.cos(1+X/0.75)/2
Y3 = np.random.uniform(Y1, Y2, len(X))
fig = plt.figure(figsize=(8, 8))
ax = fig.add_subplot(1, 1, 1, aspect=1)
def minor_tick(x, pos):
if not x % 1.0:
return ""
return "%.2f" % x
ax.xaxis.set_major_locator(MultipleLocator(1.000))
ax.xaxis.set_minor_locator(AutoMinorLocator(4))
ax.yaxis.set_major_locator(MultipleLocator(1.000))
ax.yaxis.set_minor_locator(AutoMinorLocator(4))
ax.xaxis.set_minor_formatter(FuncFormatter(minor_tick))
ax.set_xlim(0, 4)
ax.set_ylim(0, 4)
ax.tick_params(which='major', width=1.0)
ax.tick_params(which='major', length=10)
ax.tick_params(which='minor', width=1.0, labelsize=10)
ax.tick_params(which='minor', length=5, labelsize=10, labelcolor='0.25')
ax.grid(linestyle="--", linewidth=0.5, color='.25', zorder=-10)
ax.plot(X, Y1, c=(0.25, 0.25, 1.00), lw=2, label="Blue signal", zorder=10)
ax.plot(X, Y2, c=(1.00, 0.25, 0.25), lw=2, label="Red signal")
ax.plot(X, Y3, linewidth=0,
marker='o', markerfacecolor='w', markeredgecolor='k')
ax.set_title("Anatomy of a figure", fontsize=20, verticalalignment='bottom')
ax.set_xlabel("X axis label")
ax.set_ylabel("Y axis label")
ax.legend()
def circle(x, y, radius=0.15):
from matplotlib.patches import Circle
from matplotlib.patheffects import withStroke
circle = Circle((x, y), radius, clip_on=False, zorder=10, linewidth=1,
edgecolor='black', facecolor=(0, 0, 0, .0125),
path_effects=[withStroke(linewidth=5, foreground='w')])
ax.add_artist(circle)
def text(x, y, text):
ax.text(x, y, text, backgroundcolor="white",
ha='center', va='top', weight='bold', color='blue')
# Minor tick
circle(0.50, -0.10)
text(0.50, -0.32, "Minor tick label")
# Major tick
circle(-0.03, 4.00)
text(0.03, 3.80, "Major tick")
# Minor tick
circle(0.00, 3.50)
text(0.00, 3.30, "Minor tick")
# Major tick label
circle(-0.15, 3.00)
text(-0.15, 2.80, "Major tick label")
# X Label
circle(1.80, -0.27)
text(1.80, -0.45, "X axis label")
# Y Label
circle(-0.27, 1.80)
text(-0.27, 1.6, "Y axis label")
# Title
circle(1.60, 4.13)
text(1.60, 3.93, "Title")
# Blue plot
circle(1.75, 2.80)
text(1.75, 2.60, "Line\n(line plot)")
# Red plot
circle(1.20, 0.60)
text(1.20, 0.40, "Line\n(line plot)")
# Scatter plot
circle(3.20, 1.75)
text(3.20, 1.55, "Markers\n(scatter plot)")
# Grid
circle(3.00, 3.00)
text(3.00, 2.80, "Grid")
# Legend
circle(3.70, 3.80)
text(3.70, 3.60, "Legend")
# Axes
circle(0.5, 0.5)
text(0.5, 0.3, "Axes")
# Figure
circle(-0.3, 0.65)
text(-0.3, 0.45, "Figure")
color = 'blue'
ax.annotate('Spines', xy=(4.0, 0.35), xytext=(3.3, 0.5),
weight='bold', color=color,
arrowprops=dict(arrowstyle='->',
connectionstyle="arc3",
color=color))
ax.annotate('', xy=(3.15, 0.0), xytext=(3.45, 0.45),
weight='bold', color=color,
arrowprops=dict(arrowstyle='->',
connectionstyle="arc3",
color=color))
ax.text(4.0, -0.4, "Made with http://matplotlib.org",
fontsize=10, ha="right", color='.5')
plt.show()
from IPython.display import display, Image
url = 'https://fivethirtyeight.com/wp-content/uploads/2017/09/mehtahickey-inconvenient-0830-1.png'
example = Image(url=url, width=400)
display(example)
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
plt.style.use('fivethirtyeight')
fake = pd.Series([38, 3, 2, 1, 2, 4, 6, 5, 5, 33],
index=range(1,11))
fake.plot.bar(color='C1', width=0.9);
fake2 = pd.Series(
[1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
2, 2, 2,
3, 3, 3,
4, 4,
5, 5, 5,
6, 6, 6, 6,
7, 7, 7, 7, 7,
8, 8, 8, 8,
9, 9, 9, 9,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10])
fake2.value_counts().sort_index().plot.bar(color='C1', width=0.9);
counts = [38, 3, 2, 1, 2, 4, 6, 5, 5, 33]
data_list = []
for i, c in enumerate (counts,1):
data_list = data_list + [i]*c
fake2 = pd.Series(data_list)
plt.style.use('fivethirtyeight')
fake2.value_counts().sort_index().plot.bar(color='#ed713a',width=0.9, rot=0);
plt.text(x=-1,
y=50,
fontsize=16,
fontweight='bold',
s="'An Incovenient Sequel: Truth To Power' is divisive")
plt.text(x=-1,
y=46,
fontsize=16,
s="IMDb ratings for the film as of Aug.29")
plt.xlabel('Rating')
plt.ylabel('Percent of Total Votes')
plt.yticks(range(0, 50, 10));
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
df = pd.read_csv('https://raw.githubusercontent.com/fivethirtyeight/data/master/inconvenient-sequel/ratings.csv')
df.describe
df.head()
df.sample(1).T
df['timestamp'] = pd.to_datetime(df['timestamp'])
df.head()
df['timestamp'].describe()
df = df.set_index('timestamp')
df.head()
df['2017-07-17']
df['category'].value_counts()
df_imdb = df[df['category'] == 'IMDb users']
df_imdb.shape
lastday = df['2017-08-29']
lastday[lastday['category'] == 'IMDb users']['respondents'].plot()
| 0.61451 | 0.931836 |
```
#hide
%reload_ext autoreload
%autoreload 2
%matplotlib inline
```
# blurr
> A library that integrates huggingface transformers with version 2 of the fastai framework
Named after the **fast**est **transformer** (well, at least of the Autobots), ***blurr*** provides both a comprehensive and extensible framework for training and deploying *all* 🤗 [huggingface](https://huggingface.co/transformers/) transformer models with [fastai](http://docs.fast.ai/) v2.
Utilizing features like fastai's new `@typedispatch` and `@patch` decorators, and a simple class hiearchy, **blurr** provides fastai developers with the ability to train and deploy transformers for sequence classification, question answer, token classification, summarization, and language modeling tasks. Though much of this works out-of-the-box, users will be able to customize the tokenization strategies and model inputs based on task and/or architecture as needed.
**Supports**:
- Sequence Classification (multiclassification and multi-label classification)
- Token Classification
- Question Answering
- Summarization
*Support for language modeling, translation tasks and more forthcoming!!!*
## Install
You can now pip install blurr via `pip install ohmeow-blurr`
Or, even better as this library is under *very* active development, create an editable install like this:
```
git clone https://github.com/ohmeow/blurr.git
cd blurr
pip install -e ".[dev]"
```
## How to use
The initial release includes everything you need for sequence classification and question answering tasks. Support for token classification and summarization are incoming. Please check the documentation for more thorough examples of how to use this package.
The following two packages need to be installed for blurr to work:
1. fastai2 (see http://docs.fast.ai/ for installation instructions)
2. huggingface transformers (see https://huggingface.co/transformers/installation.html for details)
### Imports
```
import torch
from transformers import *
from fastai.text.all import *
from blurr.data.all import *
from blurr.modeling.all import *
```
### Get your data
```
path = untar_data(URLs.IMDB_SAMPLE)
model_path = Path('models')
imdb_df = pd.read_csv(path/'texts.csv')
```
### Get your 🤗 objects
```
task = HF_TASKS_AUTO.SequenceClassification
pretrained_model_name = "bert-base-uncased"
hf_arch, hf_config, hf_tokenizer, hf_model = BLURR_MODEL_HELPER.get_hf_objects(pretrained_model_name, task=task)
```
### Build your Data 🧱 and your DataLoaders
```
# single input
blocks = (HF_TextBlock(hf_arch=hf_arch, hf_tokenizer=hf_tokenizer), CategoryBlock)
dblock = DataBlock(blocks=blocks,
get_x=ColReader('text'), get_y=ColReader('label'),
splitter=ColSplitter(col='is_valid'))
dls = dblock.dataloaders(imdb_df, bs=4)
dls.show_batch(dataloaders=dls, max_n=2)
```
### ... and 🚂
```
#slow
model = HF_BaseModelWrapper(hf_model)
learn = Learner(dls,
model,
opt_func=partial(Adam, decouple_wd=True),
loss_func=CrossEntropyLossFlat(),
metrics=[accuracy],
cbs=[HF_BaseModelCallback],
splitter=hf_splitter)
learn.create_opt()
learn.freeze()
learn.fit_one_cycle(3, lr_max=1e-3)
#slow
learn.show_results(learner=learn, max_n=2)
```
## ❗ Updates
**11/12/2020**
* Updated documentation
* Updated model callbacks to support mixed precision training regardless of whether you are calculating the loss yourself or letting huggingface do it for you.
**11/10/2020**
* Major update just about everywhere to facilitate a breaking change in fastai's treatment of `before_batch` transforms.
* Reorganized code as I being to work on LM and other text2text tasks
* Misc. fixes
**10/08/2020**
* Updated all models to use [ModelOutput](https://huggingface.co/transformers/main_classes/output.html) classes instead of traditional tuples. `ModelOutput` attributes are assigned to the appropriate fastai bits like `Learner.pred` and `Learner.loss` and anything else you've requested the huggingface model to return is available via the `Learner.blurr_model_outputs` dictionary (see next two bullet items)
* Added ability to grab attentions and hidden state from `Learner`. You can get at them via `Learner.blurr_model_outputs` dictionary if you tell `HF_BaseModelWrapper` to provide them.
* Added `model_kwargs` to `HF_BaseModelWrapper` should you need to request a huggingface model to return something specific to it's type. These outputs will be available via the `Learner.blurr_model_outputs` dictionary as well.
**09/16/2020**
* Major overhaul to do *everything* at batch time (including tokenization/numericalization). If this backfires, I'll roll everything back but as of now, I think this approach not only meshes better with how huggingface tokenization works and reduce RAM utilization for big datasets, but also opens up opportunities for incorporating augmentation, building adversarial models, etc.... Thoughts?
* Added tests for summarization bits
* New change may require some small modifications (see docs or ask on issues thread if you have problems you can't fiture out). I'm NOT doing a release until pypi until folks have a chance to work with the latest.
**09/07/2020**
* Added tests for question/answer and summarization transformer models
* Updated summarization to support BART, T5, and Pegasus
**08/20/2020**
* Updated everything to work latest version of fastai (tested against 2.0.0)
* Added batch-time padding, so that by default now, `HF_TokenizerTransform` doesn't add any padding tokens and all huggingface inputs are padded simply to the max sequence length in each batch rather than to the max length (passed in and/or acceptable to the model). This should create efficiencies across the board, from memory consumption to GPU utilization. The old tried and true method of padding during tokenization requires you to pass in `padding='max_length` to `HF_TextBlock`.
* Removed code to remove fastai2 @patched summary methods which had previously conflicted with a couple of the huggingface transformers
**08/13/2020**
* Updated everything to work latest transformers and fastai
* Reorganized code to bring it more inline with how huggingface separates out their "tasks".
**07/06/2020**
* Updated everything to work huggingface>=3.02
* Changed a lot of the internals to make everything more efficient and performant along with the latest version of huggingface ... meaning, I have broken things for folks using previous versions of blurr :).
**06/27/2020**
* Simplified the `BLURR_MODEL_HELPER.get_hf_objects` method to support a wide range of options in terms of building the necessary huggingface objects (architecture, config, tokenizer, and model). Also added `cache_dir` for saving pre-trained objects in a custom directory.
* Misc. renaming and cleanup that may break existing code (please see the docs/source if things blow up)
* Added missing required libraries to requirements.txt (e.g., nlp)
**05/23/2020**
* Initial support for text generation (e.g., summarization, conversational agents) models now included. Only tested with BART so if you try it with other models before I do, lmk what works ... and what doesn't
**05/17/2020**
* Major code restructuring to make it easier to build out the library.
* `HF_TokenizerTransform` replaces `HF_Tokenizer`, handling the tokenization and numericalization in one place. DataBlock code has been dramatically simplified.
* Tokenization correctly handles huggingface tokenizers that require `add_prefix_space=True`.
* `HF_BaseModelCallback` and `HF_BaseModelCallback` are required and work together in order to allow developers to tie into any callback friendly event exposed by fastai2 and also pass in named arguments to the huggingface models.
* `show_batch` and `show_results` have been updated for Question/Answer and Token Classification models to represent the data and results in a more easily intepretable manner than the defaults.
**05/06/2020**
* Initial support for Token classification (e.g., NER) models now included
* Extended fastai's `Learner` object with a `predict_tokens` method used specifically in token classification
* `HF_BaseModelCallback` can be used (or extended) instead of the model wrapper to ensure your inputs into the huggingface model is correct (recommended). See docs for examples (and thanks to fastai's Sylvain for the suggestion!)
* `HF_Tokenizer` can work with strings or a string representation of a list (the later helpful for token classification tasks)
* `show_batch` and `show_results` methods have been updated to allow better control on how huggingface tokenized data is represented in those methods
## ⭐ Props
A word of gratitude to the following individuals, repos, and articles upon which much of this work is inspired from:
- The wonderful community that is the [fastai forum](https://forums.fast.ai/) and especially the tireless work of both Jeremy and Sylvain in building this amazing framework and place to learn deep learning.
- All the great tokenizers, transformers, docs and examples over at [huggingface](https://huggingface.co/)
- [FastHugs](https://github.com/morganmcg1/fasthugs)
- [Fastai with 🤗Transformers (BERT, RoBERTa, XLNet, XLM, DistilBERT)](https://towardsdatascience.com/fastai-with-transformers-bert-roberta-xlnet-xlm-distilbert-4f41ee18ecb2)
- [Fastai integration with BERT: Multi-label text classification identifying toxicity in texts](https://medium.com/@abhikjha/fastai-integration-with-bert-a0a66b1cecbe)
- [A Tutorial to Fine-Tuning BERT with Fast AI](https://mlexplained.com/2019/05/13/a-tutorial-to-fine-tuning-bert-with-fast-ai/)
|
github_jupyter
|
#hide
%reload_ext autoreload
%autoreload 2
%matplotlib inline
git clone https://github.com/ohmeow/blurr.git
cd blurr
pip install -e ".[dev]"
import torch
from transformers import *
from fastai.text.all import *
from blurr.data.all import *
from blurr.modeling.all import *
path = untar_data(URLs.IMDB_SAMPLE)
model_path = Path('models')
imdb_df = pd.read_csv(path/'texts.csv')
task = HF_TASKS_AUTO.SequenceClassification
pretrained_model_name = "bert-base-uncased"
hf_arch, hf_config, hf_tokenizer, hf_model = BLURR_MODEL_HELPER.get_hf_objects(pretrained_model_name, task=task)
# single input
blocks = (HF_TextBlock(hf_arch=hf_arch, hf_tokenizer=hf_tokenizer), CategoryBlock)
dblock = DataBlock(blocks=blocks,
get_x=ColReader('text'), get_y=ColReader('label'),
splitter=ColSplitter(col='is_valid'))
dls = dblock.dataloaders(imdb_df, bs=4)
dls.show_batch(dataloaders=dls, max_n=2)
#slow
model = HF_BaseModelWrapper(hf_model)
learn = Learner(dls,
model,
opt_func=partial(Adam, decouple_wd=True),
loss_func=CrossEntropyLossFlat(),
metrics=[accuracy],
cbs=[HF_BaseModelCallback],
splitter=hf_splitter)
learn.create_opt()
learn.freeze()
learn.fit_one_cycle(3, lr_max=1e-3)
#slow
learn.show_results(learner=learn, max_n=2)
| 0.536556 | 0.882225 |
## Deploy a Grafana Dashboard
To track the different stocks on a live dashboard we will use **Grafana**. <br>
We will use [Grafwiz](https://github.com/v3io/grafwiz) to define and deploy the dashboard directly from this notebook
```
# nuclio: ignore
import nuclio
# nuclio: start-code
from grafwiz import *
import os
import v3io_frames as v3f
def handler(context,streamview_url,readvector_url,rnn_serving_url,v3io_container,stocks_kv,stocks_tsdb,grafana_url):
context.logger.info("Initializing DataSources1")
context.logger.info(streamview_url)
context.logger.info(readvector_url)
DataSource(name='iguazio').deploy(grafana_url, use_auth=True)
DataSource(name='stream-viewer', frames_url=streamview_url).deploy(grafana_url, use_auth=False, overwrite=False)
DataSource(name='read-vector', frames_url=readvector_url).deploy(grafana_url, use_auth=False, overwrite=False)
DataSource(name='rnn-serving', frames_url=rnn_serving_url).deploy(grafana_url, use_auth=False, overwrite=False)
dash = Dashboard("stocks", start='now-7d', dataSource='iguazio')
# Add a symbol combo box (template) with data from the stocks table
dash.template(name="SYMBOL", label="Symbol", query="fields=symbol;table=" + os.getenv('V3IO_USERNAME') + "/stocks/stocks_kv;backend=kv;container=users")
# Create a table and log viewer in one row
tbl = Table('Current Stocks Value', span=12).source(table=stocks_kv,fields=['symbol','price', 'volume','last_updated'],container=v3io_container)
dash.row([tbl])
# Create 2 charts on the second row
metrics_row = [Graph(metric).series(table=stocks_tsdb, fields=[metric], filter='symbol=="$SYMBOL"',container=v3io_container) for metric in ['price','volume']]
dash.row(metrics_row)
log = Table('Articles Log', dataSource='stream-viewer', span=12)
dash.row([log])
log = Table('Vector Log', dataSource='read-vector', span=12)
dash.row([log])
log = Table('Predictions', dataSource='rnn-serving', span=12)
dash.row([log])
dash.deploy(grafana_url)
return "Done"
def init_context(context):
context.logger.info("init context")
# nuclio: end-code
# create a test event and invoke the function locally
init_context(context)
event = nuclio.Event(body='')
resp = handler(context,
"", # here you need to insert the nuclio function endpoint - streamview_url
"", # here you need to insert the nuclio function endpoint - readvector_url
"", # here you need to insert the nuclio function endpoint - rnn_serving_url
"users",
os.getenv('V3IO_USERNAME') + 'stocks/stocks_kv',
os.getenv('V3IO_USERNAME') + '/stocks/stocks_tsdb',
"http://grafana")
import mlrun
import os
fn = mlrun.code_to_function('grafana-tryout',
handler='handler', kind='job', image='mlrun/mlrun:0.6.5')
fn.apply(mlrun.platforms.v3io_cred())
fn.apply(mlrun.mount_v3io())
fn.spec.build.commands = ['pip install git+https://github.com/v3io/grafwiz --upgrade', 'pip install v3io_frames', 'pip install attrs==19.1.0']
fn.deploy()
fn.run(project = "stocks-" + os.getenv('V3IO_USERNAME'),
params = {"streamview_url": "",# here you need to insert the nuclio function endpoint - streamview_url
"readvector_url" : "",# here you need to insert the nuclio function endpoint - readvector_url
"rnn_serving_url" : "",# here you need to insert the nuclio function endpoint - rnn_serving_url
"v3io_container" : "users",
"stocks_kv" : os.getenv('V3IO_USERNAME') + 'stocks/stocks_kv',
"stocks_tsdb" : os.getenv('V3IO_USERNAME') + '/stocks/stocks_tsdb',
"grafana_url" : "http://grafana"})
```
|
github_jupyter
|
# nuclio: ignore
import nuclio
# nuclio: start-code
from grafwiz import *
import os
import v3io_frames as v3f
def handler(context,streamview_url,readvector_url,rnn_serving_url,v3io_container,stocks_kv,stocks_tsdb,grafana_url):
context.logger.info("Initializing DataSources1")
context.logger.info(streamview_url)
context.logger.info(readvector_url)
DataSource(name='iguazio').deploy(grafana_url, use_auth=True)
DataSource(name='stream-viewer', frames_url=streamview_url).deploy(grafana_url, use_auth=False, overwrite=False)
DataSource(name='read-vector', frames_url=readvector_url).deploy(grafana_url, use_auth=False, overwrite=False)
DataSource(name='rnn-serving', frames_url=rnn_serving_url).deploy(grafana_url, use_auth=False, overwrite=False)
dash = Dashboard("stocks", start='now-7d', dataSource='iguazio')
# Add a symbol combo box (template) with data from the stocks table
dash.template(name="SYMBOL", label="Symbol", query="fields=symbol;table=" + os.getenv('V3IO_USERNAME') + "/stocks/stocks_kv;backend=kv;container=users")
# Create a table and log viewer in one row
tbl = Table('Current Stocks Value', span=12).source(table=stocks_kv,fields=['symbol','price', 'volume','last_updated'],container=v3io_container)
dash.row([tbl])
# Create 2 charts on the second row
metrics_row = [Graph(metric).series(table=stocks_tsdb, fields=[metric], filter='symbol=="$SYMBOL"',container=v3io_container) for metric in ['price','volume']]
dash.row(metrics_row)
log = Table('Articles Log', dataSource='stream-viewer', span=12)
dash.row([log])
log = Table('Vector Log', dataSource='read-vector', span=12)
dash.row([log])
log = Table('Predictions', dataSource='rnn-serving', span=12)
dash.row([log])
dash.deploy(grafana_url)
return "Done"
def init_context(context):
context.logger.info("init context")
# nuclio: end-code
# create a test event and invoke the function locally
init_context(context)
event = nuclio.Event(body='')
resp = handler(context,
"", # here you need to insert the nuclio function endpoint - streamview_url
"", # here you need to insert the nuclio function endpoint - readvector_url
"", # here you need to insert the nuclio function endpoint - rnn_serving_url
"users",
os.getenv('V3IO_USERNAME') + 'stocks/stocks_kv',
os.getenv('V3IO_USERNAME') + '/stocks/stocks_tsdb',
"http://grafana")
import mlrun
import os
fn = mlrun.code_to_function('grafana-tryout',
handler='handler', kind='job', image='mlrun/mlrun:0.6.5')
fn.apply(mlrun.platforms.v3io_cred())
fn.apply(mlrun.mount_v3io())
fn.spec.build.commands = ['pip install git+https://github.com/v3io/grafwiz --upgrade', 'pip install v3io_frames', 'pip install attrs==19.1.0']
fn.deploy()
fn.run(project = "stocks-" + os.getenv('V3IO_USERNAME'),
params = {"streamview_url": "",# here you need to insert the nuclio function endpoint - streamview_url
"readvector_url" : "",# here you need to insert the nuclio function endpoint - readvector_url
"rnn_serving_url" : "",# here you need to insert the nuclio function endpoint - rnn_serving_url
"v3io_container" : "users",
"stocks_kv" : os.getenv('V3IO_USERNAME') + 'stocks/stocks_kv',
"stocks_tsdb" : os.getenv('V3IO_USERNAME') + '/stocks/stocks_tsdb',
"grafana_url" : "http://grafana"})
| 0.391988 | 0.72829 |
This is a companion notebook for the book [Deep Learning with Python, Second Edition](https://www.manning.com/books/deep-learning-with-python-second-edition?a_aid=ml-ninja&a_cid=11111111&chan=c2). For readability, it only contains runnable code blocks and section titles, and omits everything else in the book: text paragraphs, figures, and pseudocode.
**If you want to be able to follow what's going on, I recommend reading the notebook side by side with your copy of the book.**
This notebook was generated for TensorFlow 2.6.
## Generating images with variational autoencoders
### Sampling from latent spaces of images
### Concept vectors for image editing
### Variational autoencoders
### Implementing a VAE with Keras
**VAE encoder network**
```
from tensorflow import keras
from tensorflow.keras import layers
latent_dim = 2
encoder_inputs = keras.Input(shape=(28, 28, 1))
x = layers.Conv2D(32, 3, activation="relu", strides=2, padding="same")(encoder_inputs)
x = layers.Conv2D(64, 3, activation="relu", strides=2, padding="same")(x)
x = layers.Flatten()(x)
x = layers.Dense(16, activation="relu")(x)
z_mean = layers.Dense(latent_dim, name="z_mean")(x)
z_log_var = layers.Dense(latent_dim, name="z_log_var")(x)
encoder = keras.Model(encoder_inputs, [z_mean, z_log_var], name="encoder")
encoder.summary()
```
**Latent-space-sampling layer**
```
import tensorflow as tf
class Sampler(layers.Layer):
def call(self, z_mean, z_log_var):
batch_size = tf.shape(z_mean)[0]
z_size = tf.shape(z_mean)[1]
epsilon = tf.random.normal(shape=(batch_size, z_size))
return z_mean + tf.exp(0.5 * z_log_var) * epsilon
```
**VAE decoder network, mapping latent space points to images**
```
latent_inputs = keras.Input(shape=(latent_dim,))
x = layers.Dense(7 * 7 * 64, activation="relu")(latent_inputs)
x = layers.Reshape((7, 7, 64))(x)
x = layers.Conv2DTranspose(64, 3, activation="relu", strides=2, padding="same")(x)
x = layers.Conv2DTranspose(32, 3, activation="relu", strides=2, padding="same")(x)
decoder_outputs = layers.Conv2D(1, 3, activation="sigmoid", padding="same")(x)
decoder = keras.Model(latent_inputs, decoder_outputs, name="decoder")
decoder.summary()
```
**VAE model with custom `train_step()`**
```
class VAE(keras.Model):
def __init__(self, encoder, decoder, **kwargs):
super().__init__(**kwargs)
self.encoder = encoder
self.decoder = decoder
self.sampler = Sampler()
self.total_loss_tracker = keras.metrics.Mean(name="total_loss")
self.reconstruction_loss_tracker = keras.metrics.Mean(
name="reconstruction_loss")
self.kl_loss_tracker = keras.metrics.Mean(name="kl_loss")
@property
def metrics(self):
return [self.total_loss_tracker,
self.reconstruction_loss_tracker,
self.kl_loss_tracker]
def train_step(self, data):
with tf.GradientTape() as tape:
z_mean, z_log_var = self.encoder(data)
z = self.sampler(z_mean, z_log_var)
reconstruction = decoder(z)
reconstruction_loss = tf.reduce_mean(
tf.reduce_sum(
keras.losses.binary_crossentropy(data, reconstruction),
axis=(1, 2)
)
)
kl_loss = -0.5 * (1 + z_log_var - tf.square(z_mean) - tf.exp(z_log_var))
total_loss = reconstruction_loss + tf.reduce_mean(kl_loss)
grads = tape.gradient(total_loss, self.trainable_weights)
self.optimizer.apply_gradients(zip(grads, self.trainable_weights))
self.total_loss_tracker.update_state(total_loss)
self.reconstruction_loss_tracker.update_state(reconstruction_loss)
self.kl_loss_tracker.update_state(kl_loss)
return {
"total_loss": self.total_loss_tracker.result(),
"reconstruction_loss": self.reconstruction_loss_tracker.result(),
"kl_loss": self.kl_loss_tracker.result(),
}
```
**Training the VAE**
```
import numpy as np
(x_train, _), (x_test, _) = keras.datasets.mnist.load_data()
mnist_digits = np.concatenate([x_train, x_test], axis=0)
mnist_digits = np.expand_dims(mnist_digits, -1).astype("float32") / 255
vae = VAE(encoder, decoder)
vae.compile(optimizer=keras.optimizers.Adam(), run_eagerly=True)
vae.fit(mnist_digits, epochs=30, batch_size=128)
```
**Sampling a grid of images from the 2D latent space**
```
import matplotlib.pyplot as plt
n = 30
digit_size = 28
figure = np.zeros((digit_size * n, digit_size * n))
grid_x = np.linspace(-1, 1, n)
grid_y = np.linspace(-1, 1, n)[::-1]
for i, yi in enumerate(grid_y):
for j, xi in enumerate(grid_x):
z_sample = np.array([[xi, yi]])
x_decoded = vae.decoder.predict(z_sample)
digit = x_decoded[0].reshape(digit_size, digit_size)
figure[
i * digit_size : (i + 1) * digit_size,
j * digit_size : (j + 1) * digit_size,
] = digit
plt.figure(figsize=(15, 15))
start_range = digit_size // 2
end_range = n * digit_size + start_range
pixel_range = np.arange(start_range, end_range, digit_size)
sample_range_x = np.round(grid_x, 1)
sample_range_y = np.round(grid_y, 1)
plt.xticks(pixel_range, sample_range_x)
plt.yticks(pixel_range, sample_range_y)
plt.xlabel("z[0]")
plt.ylabel("z[1]")
plt.axis("off")
plt.imshow(figure, cmap="Greys_r")
```
### Wrapping up
|
github_jupyter
|
from tensorflow import keras
from tensorflow.keras import layers
latent_dim = 2
encoder_inputs = keras.Input(shape=(28, 28, 1))
x = layers.Conv2D(32, 3, activation="relu", strides=2, padding="same")(encoder_inputs)
x = layers.Conv2D(64, 3, activation="relu", strides=2, padding="same")(x)
x = layers.Flatten()(x)
x = layers.Dense(16, activation="relu")(x)
z_mean = layers.Dense(latent_dim, name="z_mean")(x)
z_log_var = layers.Dense(latent_dim, name="z_log_var")(x)
encoder = keras.Model(encoder_inputs, [z_mean, z_log_var], name="encoder")
encoder.summary()
import tensorflow as tf
class Sampler(layers.Layer):
def call(self, z_mean, z_log_var):
batch_size = tf.shape(z_mean)[0]
z_size = tf.shape(z_mean)[1]
epsilon = tf.random.normal(shape=(batch_size, z_size))
return z_mean + tf.exp(0.5 * z_log_var) * epsilon
latent_inputs = keras.Input(shape=(latent_dim,))
x = layers.Dense(7 * 7 * 64, activation="relu")(latent_inputs)
x = layers.Reshape((7, 7, 64))(x)
x = layers.Conv2DTranspose(64, 3, activation="relu", strides=2, padding="same")(x)
x = layers.Conv2DTranspose(32, 3, activation="relu", strides=2, padding="same")(x)
decoder_outputs = layers.Conv2D(1, 3, activation="sigmoid", padding="same")(x)
decoder = keras.Model(latent_inputs, decoder_outputs, name="decoder")
decoder.summary()
class VAE(keras.Model):
def __init__(self, encoder, decoder, **kwargs):
super().__init__(**kwargs)
self.encoder = encoder
self.decoder = decoder
self.sampler = Sampler()
self.total_loss_tracker = keras.metrics.Mean(name="total_loss")
self.reconstruction_loss_tracker = keras.metrics.Mean(
name="reconstruction_loss")
self.kl_loss_tracker = keras.metrics.Mean(name="kl_loss")
@property
def metrics(self):
return [self.total_loss_tracker,
self.reconstruction_loss_tracker,
self.kl_loss_tracker]
def train_step(self, data):
with tf.GradientTape() as tape:
z_mean, z_log_var = self.encoder(data)
z = self.sampler(z_mean, z_log_var)
reconstruction = decoder(z)
reconstruction_loss = tf.reduce_mean(
tf.reduce_sum(
keras.losses.binary_crossentropy(data, reconstruction),
axis=(1, 2)
)
)
kl_loss = -0.5 * (1 + z_log_var - tf.square(z_mean) - tf.exp(z_log_var))
total_loss = reconstruction_loss + tf.reduce_mean(kl_loss)
grads = tape.gradient(total_loss, self.trainable_weights)
self.optimizer.apply_gradients(zip(grads, self.trainable_weights))
self.total_loss_tracker.update_state(total_loss)
self.reconstruction_loss_tracker.update_state(reconstruction_loss)
self.kl_loss_tracker.update_state(kl_loss)
return {
"total_loss": self.total_loss_tracker.result(),
"reconstruction_loss": self.reconstruction_loss_tracker.result(),
"kl_loss": self.kl_loss_tracker.result(),
}
import numpy as np
(x_train, _), (x_test, _) = keras.datasets.mnist.load_data()
mnist_digits = np.concatenate([x_train, x_test], axis=0)
mnist_digits = np.expand_dims(mnist_digits, -1).astype("float32") / 255
vae = VAE(encoder, decoder)
vae.compile(optimizer=keras.optimizers.Adam(), run_eagerly=True)
vae.fit(mnist_digits, epochs=30, batch_size=128)
import matplotlib.pyplot as plt
n = 30
digit_size = 28
figure = np.zeros((digit_size * n, digit_size * n))
grid_x = np.linspace(-1, 1, n)
grid_y = np.linspace(-1, 1, n)[::-1]
for i, yi in enumerate(grid_y):
for j, xi in enumerate(grid_x):
z_sample = np.array([[xi, yi]])
x_decoded = vae.decoder.predict(z_sample)
digit = x_decoded[0].reshape(digit_size, digit_size)
figure[
i * digit_size : (i + 1) * digit_size,
j * digit_size : (j + 1) * digit_size,
] = digit
plt.figure(figsize=(15, 15))
start_range = digit_size // 2
end_range = n * digit_size + start_range
pixel_range = np.arange(start_range, end_range, digit_size)
sample_range_x = np.round(grid_x, 1)
sample_range_y = np.round(grid_y, 1)
plt.xticks(pixel_range, sample_range_x)
plt.yticks(pixel_range, sample_range_y)
plt.xlabel("z[0]")
plt.ylabel("z[1]")
plt.axis("off")
plt.imshow(figure, cmap="Greys_r")
| 0.966976 | 0.959193 |
### Imports
```
import torch
import warnings
from tqdm import tqdm
from torch.autograd import Variable
from sklearn.metrics import mean_absolute_error
```
### Auglichem imports
```
from auglichem.crystal import (PerturbStructureTransformation,
RotationTransformation,
SwapAxesTransformation,
TranslateSitesTransformation,
SupercellTransformation,
)
from auglichem.crystal.data import CrystalDatasetWrapper
from auglichem.crystal.models import CrystalGraphConvNet as CGCNN
```
### Set up dataset
```
# Create transformation
transforms = [
PerturbStructureTransformation(distance=0.1, min_distance=0.01),
RotationTransformation(axis=[0,0,1], angle=90),
SwapAxesTransformation(),
TranslateSitesTransformation(indices_to_move=[0], translation_vector=[1,0,0],
vector_in_frac_coords=True),
SupercellTransformation(scaling_matrix=[[1,0,0],[0,1,0],[0,0,1]]),
]
# Initialize dataset object
dataset = CrystalDatasetWrapper("lanthanides", batch_size=256,
valid_size=0.1, test_size=0.1, cgcnn=True)
# Get train/valid/test splits as loaders
train_loader, valid_loader, test_loader = dataset.get_data_loaders(transform=transforms)
```
### Initialize model with task from data
```
# Get model
structures, _, _ = dataset[0]
orig_atom_fea_len = structures[0].shape[-1]
nbr_fea_len = structures[1].shape[-1]
model = CGCNN(orig_atom_fea_len, nbr_fea_len)
#model.cuda()
```
### Initialize traning loop
```
criterion = torch.nn.MSELoss()
optimizer = torch.optim.Adam(model.parameters(), lr=1e-3, weight_decay=1e-5)
```
### Train the model
```
with warnings.catch_warnings():
warnings.simplefilter("ignore")
for epoch in range(1):
for bn, (data, target, _) in tqdm(enumerate(train_loader)):
optimizer.zero_grad()
input_var = (Variable(data[0]),
Variable(data[1]),
data[2],
data[3])
# data -> GPU
#input_var = (Variable(data[0].cuda()),
# Variable(data[1].cuda()),
# data[2].cuda(),
# data[3])
pred = model(*input_var)
loss = criterion(pred, target)
#loss = criterion(pred, target.cuda())
loss.backward()
optimizer.step()
```
### Test the model
```
def evaluate(model, test_loader, validation=False):
with torch.no_grad():
model.eval()
preds = torch.Tensor([])
targets = torch.Tensor([])
for data, target, _ in test_loader:
input_var = (Variable(data[0]),
Variable(data[1]),
data[2],
data[3])
# data -> GPU
#input_var = (Variable(data[0].cuda()),
# Variable(data[1].cuda()),
# data[2].cuda(),
# data[3])
pred = model(*input_var)
preds = torch.cat((preds, pred.cpu().detach()))
targets = torch.cat((targets, target))
mae = mean_absolute_error(preds, targets)
set_str = "VALIDATION" if(validation) else "TEST"
print("{0} MAE: {1:.3f}".format(set_str, mae))
evaluate(model, valid_loader, validation=True)
evaluate(model, test_loader)
```
### Model saving/loading example
```
# Save model
torch.save(model.state_dict(), "./saved_models/example_cgcnn")
# Instantiate new model and evaluate
structures, _, _ = dataset[0]
orig_atom_fea_len = structures[0].shape[-1]
nbr_fea_len = structures[1].shape[-1]
model = CGCNN(orig_atom_fea_len, nbr_fea_len)
evaluate(model, valid_loader, validation=True)
evaluate(model, test_loader)
# Load saved model and evaluate
model.load_state_dict(torch.load("./saved_models/example_cgcnn"))
evaluate(model, valid_loader, validation=True)
evaluate(model, test_loader)
```
|
github_jupyter
|
import torch
import warnings
from tqdm import tqdm
from torch.autograd import Variable
from sklearn.metrics import mean_absolute_error
from auglichem.crystal import (PerturbStructureTransformation,
RotationTransformation,
SwapAxesTransformation,
TranslateSitesTransformation,
SupercellTransformation,
)
from auglichem.crystal.data import CrystalDatasetWrapper
from auglichem.crystal.models import CrystalGraphConvNet as CGCNN
# Create transformation
transforms = [
PerturbStructureTransformation(distance=0.1, min_distance=0.01),
RotationTransformation(axis=[0,0,1], angle=90),
SwapAxesTransformation(),
TranslateSitesTransformation(indices_to_move=[0], translation_vector=[1,0,0],
vector_in_frac_coords=True),
SupercellTransformation(scaling_matrix=[[1,0,0],[0,1,0],[0,0,1]]),
]
# Initialize dataset object
dataset = CrystalDatasetWrapper("lanthanides", batch_size=256,
valid_size=0.1, test_size=0.1, cgcnn=True)
# Get train/valid/test splits as loaders
train_loader, valid_loader, test_loader = dataset.get_data_loaders(transform=transforms)
# Get model
structures, _, _ = dataset[0]
orig_atom_fea_len = structures[0].shape[-1]
nbr_fea_len = structures[1].shape[-1]
model = CGCNN(orig_atom_fea_len, nbr_fea_len)
#model.cuda()
criterion = torch.nn.MSELoss()
optimizer = torch.optim.Adam(model.parameters(), lr=1e-3, weight_decay=1e-5)
with warnings.catch_warnings():
warnings.simplefilter("ignore")
for epoch in range(1):
for bn, (data, target, _) in tqdm(enumerate(train_loader)):
optimizer.zero_grad()
input_var = (Variable(data[0]),
Variable(data[1]),
data[2],
data[3])
# data -> GPU
#input_var = (Variable(data[0].cuda()),
# Variable(data[1].cuda()),
# data[2].cuda(),
# data[3])
pred = model(*input_var)
loss = criterion(pred, target)
#loss = criterion(pred, target.cuda())
loss.backward()
optimizer.step()
def evaluate(model, test_loader, validation=False):
with torch.no_grad():
model.eval()
preds = torch.Tensor([])
targets = torch.Tensor([])
for data, target, _ in test_loader:
input_var = (Variable(data[0]),
Variable(data[1]),
data[2],
data[3])
# data -> GPU
#input_var = (Variable(data[0].cuda()),
# Variable(data[1].cuda()),
# data[2].cuda(),
# data[3])
pred = model(*input_var)
preds = torch.cat((preds, pred.cpu().detach()))
targets = torch.cat((targets, target))
mae = mean_absolute_error(preds, targets)
set_str = "VALIDATION" if(validation) else "TEST"
print("{0} MAE: {1:.3f}".format(set_str, mae))
evaluate(model, valid_loader, validation=True)
evaluate(model, test_loader)
# Save model
torch.save(model.state_dict(), "./saved_models/example_cgcnn")
# Instantiate new model and evaluate
structures, _, _ = dataset[0]
orig_atom_fea_len = structures[0].shape[-1]
nbr_fea_len = structures[1].shape[-1]
model = CGCNN(orig_atom_fea_len, nbr_fea_len)
evaluate(model, valid_loader, validation=True)
evaluate(model, test_loader)
# Load saved model and evaluate
model.load_state_dict(torch.load("./saved_models/example_cgcnn"))
evaluate(model, valid_loader, validation=True)
evaluate(model, test_loader)
| 0.847369 | 0.743052 |
# Lesson 1 - What's your pet
Welcome to lesson 1! For those of you who are using a Jupyter Notebook for the first time, you can learn about this useful tool in a tutorial we prepared specially for you; click `File`->`Open` now and click `00_notebook_tutorial.ipynb`.
In this lesson we will build our first image classifier from scratch, and see if we can achieve world-class results. Let's dive in!
Every notebook starts with the following three lines; they ensure that any edits to libraries you make are reloaded here automatically, and also that any charts or images displayed are shown in this notebook.
```
%reload_ext autoreload
%autoreload 2
%matplotlib inline
```
We import all the necessary packages. We are going to work with the [fastai V1 library](http://www.fast.ai/2018/10/02/fastai-ai/) which sits on top of [Pytorch 1.0](https://hackernoon.com/pytorch-1-0-468332ba5163). The fastai library provides many useful functions that enable us to quickly and easily build neural networks and train our models.
```
from fastai.vision import *
from fastai.metrics import error_rate
from fastai.datasets import *
import numpy as np
```
If you're using a computer with an unusually small GPU, you may get an out of memory error when running this notebook. If this happens, click Kernel->Restart, uncomment the 2nd line below to use a smaller *batch size* (you'll learn all about what this means during the course), and try again.
```
bs = 64
# bs = 16 # uncomment this line if you run out of memory even after clicking Kernel->Restart
```
## Looking at the data
We are going to use the [Oxford-IIIT Pet Dataset](http://www.robots.ox.ac.uk/~vgg/data/pets/) by [O. M. Parkhi et al., 2012](http://www.robots.ox.ac.uk/~vgg/publications/2012/parkhi12a/parkhi12a.pdf) which features 12 cat breeds and 25 dogs breeds. Our model will need to learn to differentiate between these 37 distinct categories. According to their paper, the best accuracy they could get in 2012 was 59.21%, using a complex model that was specific to pet detection, with separate "Image", "Head", and "Body" models for the pet photos. Let's see how accurate we can be using deep learning!
We are going to use the `untar_data` function to which we must pass a URL as an argument and which will download and extract the data.
```
help(untar_data)
path = untar_data(URLs.PETS); path
path.ls()
path_anno = path/'annotations'
path_img = path/'images'
print(path_anno)
print(path_img)
```
The first thing we do when we approach a problem is to take a look at the data. We _always_ need to understand very well what the problem is and what the data looks like before we can figure out how to solve it. Taking a look at the data means understanding how the data directories are structured, what the labels are and what some sample images look like.
The main difference between the handling of image classification datasets is the way labels are stored. In this particular dataset, labels are stored in the filenames themselves. We will need to extract them to be able to classify the images into the correct categories. Fortunately, the fastai library has a handy function made exactly for this, `ImageDataBunch.from_name_re` gets the labels from the filenames using a [regular expression](https://docs.python.org/3.6/library/re.html).
```
fnames = get_image_files(path_img)
fnames[:5]
np.random.seed(2)
pat = r'/([^/]+)_\d+.jpg$'
data = ImageDataBunch.from_name_re(path_img, fnames, pat, ds_tfms=get_transforms(), size=224, bs=bs
).normalize(imagenet_stats)
data.show_batch(rows=3, figsize=(7,6))
print(data.classes)
len(data.classes),data.c
```
## Training: resnet34
Now we will start training our model. We will use a [convolutional neural network](http://cs231n.github.io/convolutional-networks/) backbone and a fully connected head with a single hidden layer as a classifier. Don't know what these things mean? Not to worry, we will dive deeper in the coming lessons. For the moment you need to know that we are building a model which will take images as input and will output the predicted probability for each of the categories (in this case, it will have 37 outputs).
We will train for 4 epochs (4 cycles through all our data).
```
learn = cnn_learner(data, models.resnet34, metrics=error_rate)
learn.model
learn.fit_one_cycle(4)
learn.save('stage-1')
```
## Results
Let's see what results we have got.
We will first see which were the categories that the model most confused with one another. We will try to see if what the model predicted was reasonable or not. In this case the mistakes look reasonable (none of the mistakes seems obviously naive). This is an indicator that our classifier is working correctly.
Furthermore, when we plot the confusion matrix, we can see that the distribution is heavily skewed: the model makes the same mistakes over and over again but it rarely confuses other categories. This suggests that it just finds it difficult to distinguish some specific categories between each other; this is normal behaviour.
```
interp = ClassificationInterpretation.from_learner(learn)
losses,idxs = interp.top_losses()
len(data.valid_ds)==len(losses)==len(idxs)
interp.plot_top_losses(9, figsize=(15,11))
doc(interp.plot_top_losses)
interp.plot_confusion_matrix(figsize=(12,12), dpi=60)
interp.most_confused(min_val=2)
```
## Unfreezing, fine-tuning, and learning rates
Since our model is working as we expect it to, we will *unfreeze* our model and train some more.
```
learn.unfreeze()
learn.fit_one_cycle(1)
learn.load('stage-1');
learn.lr_find()
learn.recorder.plot()
learn.unfreeze()
learn.fit_one_cycle(2, max_lr=slice(1e-6,1e-4))
```
That's a pretty accurate model!
## Training: resnet50
Now we will train in the same way as before but with one caveat: instead of using resnet34 as our backbone we will use resnet50 (resnet34 is a 34 layer residual network while resnet50 has 50 layers. It will be explained later in the course and you can learn the details in the [resnet paper](https://arxiv.org/pdf/1512.03385.pdf)).
Basically, resnet50 usually performs better because it is a deeper network with more parameters. Let's see if we can achieve a higher performance here. To help it along, let's us use larger images too, since that way the network can see more detail. We reduce the batch size a bit since otherwise this larger network will require more GPU memory.
```
data = ImageDataBunch.from_name_re(path_img, fnames, pat, ds_tfms=get_transforms(),
size=299, bs=bs//2).normalize(imagenet_stats)
learn = cnn_learner(data, models.resnet50, metrics=error_rate)
learn.lr_find()
learn.recorder.plot()
learn.fit_one_cycle(8)
learn.save('stage-1-50')
```
It's astonishing that it's possible to recognize pet breeds so accurately! Let's see if full fine-tuning helps:
```
learn.unfreeze()
learn.fit_one_cycle(3, max_lr=slice(1e-6,1e-4))
```
If it doesn't, you can always go back to your previous model.
```
learn.load('stage-1-50');
interp = ClassificationInterpretation.from_learner(learn)
interp.most_confused(min_val=2)
```
## Other data formats
```
path = untar_data(URLs.MNIST_SAMPLE); path
tfms = get_transforms(do_flip=False)
data = ImageDataBunch.from_folder(path, ds_tfms=tfms, size=26)
data.show_batch(rows=3, figsize=(5,5))
learn = cnn_learner(data, models.resnet18, metrics=accuracy)
learn.fit(2)
df = pd.read_csv(path/'labels.csv')
df.head()
data = ImageDataBunch.from_csv(path, ds_tfms=tfms, size=28)
data.show_batch(rows=3, figsize=(5,5))
data.classes
data = ImageDataBunch.from_df(path, df, ds_tfms=tfms, size=24)
data.classes
fn_paths = [path/name for name in df['name']]; fn_paths[:2]
pat = r"/(\d)/\d+\.png$"
data = ImageDataBunch.from_name_re(path, fn_paths, pat=pat, ds_tfms=tfms, size=24)
data.classes
data = ImageDataBunch.from_name_func(path, fn_paths, ds_tfms=tfms, size=24,
label_func = lambda x: '3' if '/3/' in str(x) else '7')
data.classes
labels = [('3' if '/3/' in str(x) else '7') for x in fn_paths]
labels[:5]
data = ImageDataBunch.from_lists(path, fn_paths, labels=labels, ds_tfms=tfms, size=24)
data.classes
```
|
github_jupyter
|
%reload_ext autoreload
%autoreload 2
%matplotlib inline
from fastai.vision import *
from fastai.metrics import error_rate
from fastai.datasets import *
import numpy as np
bs = 64
# bs = 16 # uncomment this line if you run out of memory even after clicking Kernel->Restart
help(untar_data)
path = untar_data(URLs.PETS); path
path.ls()
path_anno = path/'annotations'
path_img = path/'images'
print(path_anno)
print(path_img)
fnames = get_image_files(path_img)
fnames[:5]
np.random.seed(2)
pat = r'/([^/]+)_\d+.jpg$'
data = ImageDataBunch.from_name_re(path_img, fnames, pat, ds_tfms=get_transforms(), size=224, bs=bs
).normalize(imagenet_stats)
data.show_batch(rows=3, figsize=(7,6))
print(data.classes)
len(data.classes),data.c
learn = cnn_learner(data, models.resnet34, metrics=error_rate)
learn.model
learn.fit_one_cycle(4)
learn.save('stage-1')
interp = ClassificationInterpretation.from_learner(learn)
losses,idxs = interp.top_losses()
len(data.valid_ds)==len(losses)==len(idxs)
interp.plot_top_losses(9, figsize=(15,11))
doc(interp.plot_top_losses)
interp.plot_confusion_matrix(figsize=(12,12), dpi=60)
interp.most_confused(min_val=2)
learn.unfreeze()
learn.fit_one_cycle(1)
learn.load('stage-1');
learn.lr_find()
learn.recorder.plot()
learn.unfreeze()
learn.fit_one_cycle(2, max_lr=slice(1e-6,1e-4))
data = ImageDataBunch.from_name_re(path_img, fnames, pat, ds_tfms=get_transforms(),
size=299, bs=bs//2).normalize(imagenet_stats)
learn = cnn_learner(data, models.resnet50, metrics=error_rate)
learn.lr_find()
learn.recorder.plot()
learn.fit_one_cycle(8)
learn.save('stage-1-50')
learn.unfreeze()
learn.fit_one_cycle(3, max_lr=slice(1e-6,1e-4))
learn.load('stage-1-50');
interp = ClassificationInterpretation.from_learner(learn)
interp.most_confused(min_val=2)
path = untar_data(URLs.MNIST_SAMPLE); path
tfms = get_transforms(do_flip=False)
data = ImageDataBunch.from_folder(path, ds_tfms=tfms, size=26)
data.show_batch(rows=3, figsize=(5,5))
learn = cnn_learner(data, models.resnet18, metrics=accuracy)
learn.fit(2)
df = pd.read_csv(path/'labels.csv')
df.head()
data = ImageDataBunch.from_csv(path, ds_tfms=tfms, size=28)
data.show_batch(rows=3, figsize=(5,5))
data.classes
data = ImageDataBunch.from_df(path, df, ds_tfms=tfms, size=24)
data.classes
fn_paths = [path/name for name in df['name']]; fn_paths[:2]
pat = r"/(\d)/\d+\.png$"
data = ImageDataBunch.from_name_re(path, fn_paths, pat=pat, ds_tfms=tfms, size=24)
data.classes
data = ImageDataBunch.from_name_func(path, fn_paths, ds_tfms=tfms, size=24,
label_func = lambda x: '3' if '/3/' in str(x) else '7')
data.classes
labels = [('3' if '/3/' in str(x) else '7') for x in fn_paths]
labels[:5]
data = ImageDataBunch.from_lists(path, fn_paths, labels=labels, ds_tfms=tfms, size=24)
data.classes
| 0.461745 | 0.986085 |
# Сглаживание как способ быстрого решения негладких задач
$$
\min_x f(x)
$$
Основано на статье [Smooth minimization of non-smooth functions](https://www.math.ucdavis.edu/~sqma/MAT258A_Files/Nesterov-2005.pdf) by Y. Nesterov
## Текущие достижения для выпуклых функций
- Функция $f$ негладкая
$$
\epsilon \sim O\left(\frac{1}{\sqrt{k}}\right), \quad k \sim O\left(\frac{1}{\epsilon^2}\right)
$$
- Функция негладкая, но имеет структуру $f = h + g$, где $h$ - гладкая выпуклая, $g$ - негладкая выпуклая
- Проксимальный градиентный метод
$$
\epsilon \sim O\left(\frac{L}{k}\right), \quad k \sim O\left(\frac{L}{\epsilon}\right)
$$
- Ускоренный проксимальный градиентный метод
$$
\epsilon \sim O\left(\frac{L}{k^2}\right), \quad k \sim O\left(\sqrt{\frac{L}{\epsilon}}\right)
$$
## Вдохновляющий вывод и мотивирующий вопрос
Знание структуры функции позволяет получить быстро сходящиеся методы по сравнению с методами, которые воспринимают функцию, как чёрный ящик.
**Вопрос**: какая ещё структура негладкой функции позволяет использовать методы для минимизации гладких функций?
## План получения гладких аппроксимаций негладких функций
- Применять быстрые методы для негладких функций, как они есть, нельзя
- Вместо этого можно (почему и как именно будет изложено далее) ввести их гладкую аппроксимацию $f_{\mu}$
- Заменить $f$ на $f_{\mu}$ и решить задачу
$$
\min f_{\mu}(x)
$$
ускоренными методами для **гладких** задач
## Баланс между точностью и скоростью
- Параметр $\mu$ регулирует гладкость $f_{\mu}$
- Сложность решения задачи
$$
\min f_{\mu}(x)
$$
зависит от $\frac{L_{\mu}}{\epsilon_{\mu}}$, где $L_{\mu}$ - константа Липшица градиента функции $f_{\mu}$.
- Большое значение $L_{\mu}$ даёт более высокую точность аппроксимации, но приводит к замедлению метода для гладкой задачи - уменьшение сглаживания
- Малое значение для $L_{\mu}$ приводит к быстрой сходимости метода, но $f_{\mu}$ менее точно аппроксимирует $f$
## Пример: функция Хьюбера (Huber function)
- $f(x) = |x|$
- Функция Хьюбера
$$
h_{\mu}(x) = \begin{cases} \dfrac{x^2}{2\mu} & |x| \leq \mu \\ |x| - \mu/2 & |x| \geq \mu \end{cases}
$$
```
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
plt.rc("text", usetex=True)
x = np.linspace(-3, 3, 10000)
f = np.abs(x)
def huber(x_range, mu):
h = np.zeros_like(x_range)
for i, x in enumerate(x_range):
if np.abs(x) <= mu:
h[i] = x**2 / (2 * mu)
else:
h[i] = np.abs(x) - mu / 2
return h
mu = 0.5
huber_f = huber(x, mu)
plt.figure(figsize=(12, 8))
plt.plot(x, f, label="$|x|$")
plt.plot(x, huber_f, label=r"Huber $(\mu = {})$".format(mu))
plt.grid(True)
plt.xticks(fontsize=28)
plt.yticks(fontsize=28)
plt.legend(fontsize=28)
```
- Точность приближения
$$
|x| - \mu / 2 \leq h_{\mu}(x) \leq |x|
$$
- Степень гладкости
$$
h''_{\mu}(x) \leq \frac{1}{\mu}
$$
- **Вывод**: при малом $\mu$ действительно наблюдается повышение точности аппроксимации и уменьшение гладкости
## Пример: сглаживание в $\mathbb{R}^n$
- Задача
$$
\min f(x) = \min \|Ax - b\|_1
$$
- Сглаженная версия
$$
\min \sum_{i=1}^m h_{\mu}(a_i^{\top}x - b_i)
$$
- Точность сглаживания аналогично одномерному случаю
$$
f(x) -\frac{m\mu}{2} \leq f_{\mu}(x) \leq f(x)
$$
откуда получаем оценку на сходимость по функции
$$
f(x) - f^* \leq f_{\mu}(x) - f_{\mu}^* + \frac{m\mu}{2}
$$
- Если хотим $f(x) - f^* \leq \epsilon$, то точность для решения сглаженной задачи должна быть $\epsilon_{\mu} = \epsilon - m \mu / 2$
- Константа Липшица градиента для $f_{\mu}$
$$
L_{\mu} = \frac{\|A\|_2^2}{\mu}
$$
аналогично одномерному случаю + правила дифференцирования сложной функции (выведите это выражение!)
- Сложность работы ускоренного градиентного метода зависит от
$$
\frac{L_{\mu}}{\epsilon_{\mu}} = \frac{\|A\|_2^2}{\mu(\epsilon - m \mu / 2)} = \left(\mu = \frac{\epsilon}{m}\right) = \frac{2m\|A\|_2^2}{\epsilon^2}
$$
тогда
$$
O\left(\sqrt{\frac{L_{\mu}}{\epsilon_{\mu}}}\right) = O\left(\frac{1}{\epsilon}\right)
$$
для ускоренного градиентного метода
- **Вывод**: использование сглаженной версии целевой **негладкой** функции привело к снижению скорости сходимости ускоренного градиентного метода с $O\left(\frac{1}{\sqrt{\epsilon}}\right)$ до $O\left(\frac{1}{\epsilon}\right)$. Но это всё ещё быстрее субградиентного метода!
## Эксперименты
```
import liboptpy.unconstr_solvers as methods
import liboptpy.step_size as ss
m, n = 500, 100
A = np.random.randn(m, n)
x_true = np.random.randn(n)
b = A.dot(x_true)
f = lambda x: np.linalg.norm(A.dot(x) - b, 1)
subgrad = lambda x: A.T.dot(np.sign(A.dot(x) - b))
alpha = 1e-3
s = 1e-1
sg_methods = {
"SM 1/k": methods.fo.SubgradientMethod(f, subgrad, ss.InvIterStepSize()),
"SM fixed={}".format(alpha): methods.fo.SubgradientMethod(f, subgrad, ss.ConstantStepSize(alpha)),
"SM scaled fix, s={}".format(s): methods.fo.SubgradientMethod(f, subgrad,
ss.ScaledConstantStepSize(s)),
}
x0 = np.random.randn(n)
max_iter = 50000
for m in sg_methods:
_ = sg_methods[m].solve(x0=x0, max_iter=max_iter)
plt.figure(figsize=(10, 8))
for m in sg_methods:
plt.semilogy([f(x) for x in sg_methods[m].get_convergence()], label=m)
plt.legend(fontsize=20)
plt.xlabel(r"Number of iterations, $k$", fontsize=26)
plt.ylabel(r"Objective, $f(x_k)$", fontsize=26)
plt.xticks(fontsize=20)
plt.yticks(fontsize=20)
plt.grid(True)
def huber(x, mu):
res = np.zeros_like(x)
res[np.abs(x) <= mu] = x[np.abs(x) <= mu]**2 / (2 * mu)
res[np.abs(x) >= mu] = np.abs(x[np.abs(x) >= mu]) - mu / 2
return res
def grad_huber(x, mu):
res = np.zeros_like(x)
res[np.abs(x) <= mu] = x[np.abs(x) <= mu] / mu
res[np.abs(x) >= mu] = np.sign(x[np.abs(x) >= mu])
return res
mu = 1e-2
print(mu)
fmu = lambda x: np.sum(huber(A.dot(x) - b, mu))
grad_fmu = lambda x: A.T @ grad_huber(A.dot(x) - b, mu)
grad_methods = {
"GD Armijo": methods.fo.GradientDescent(fmu, grad_fmu, ss.Backtracking("Armijo", beta=0.1, rho=0.5, init_alpha=1)),
"GD": methods.fo.GradientDescent(fmu, grad_fmu, ss.ConstantStepSize(mu/np.linalg.norm(A, 2)**2)),
"Fast GD": methods.fo.AcceleratedGD(fmu, grad_fmu, ss.ConstantStepSize(mu/np.linalg.norm(A, 2)**2))
}
max_iter = 10000
for m in grad_methods:
_ = grad_methods[m].solve(x0=x0, max_iter=max_iter)
plt.figure(figsize=(10, 8))
for m in grad_methods:
plt.semilogy([fmu(x) for x in grad_methods[m].get_convergence()], label=m)
plt.legend(fontsize=20)
plt.xlabel(r"Number of iterations, $k$", fontsize=26)
plt.ylabel(r"Objective, $f_{\mu}(x_k)$", fontsize=26)
plt.xticks(fontsize=20)
plt.yticks(fontsize=20)
plt.grid(True)
plt.figure(figsize=(10, 8))
for m in grad_methods:
plt.semilogy([np.linalg.norm(grad_fmu(x)) for x in grad_methods[m].get_convergence()], label=m)
plt.legend(fontsize=20)
plt.xlabel(r"Number of iterations, $k$", fontsize=26)
plt.ylabel(r"$\|f_{\mu}'(x_k)\|_2$", fontsize=26)
plt.xticks(fontsize=20)
plt.yticks(fontsize=20)
plt.grid(True)
plt.figure(figsize=(10, 8))
for m in grad_methods:
plt.semilogy([f(x) for x in grad_methods[m].get_convergence()], label=m)
for m in sg_methods:
plt.semilogy([f(x) for x in sg_methods[m].get_convergence()], label=m)
plt.legend(fontsize=20)
plt.xlabel(r"Number of iterations, $k$", fontsize=26)
plt.ylabel(r"Objective, $f(x_k)$", fontsize=26)
plt.xticks(fontsize=20)
plt.yticks(fontsize=20)
plt.grid(True)
for m in grad_methods:
x_m = grad_methods[m].get_convergence()[-1]
print(m, "\n\t", np.linalg.norm(x_true - x_m), f(x_m) - f(x_true))
for m in sg_methods:
x_m = sg_methods[m].get_convergence()[-1]
print(m, "\n\t", np.linalg.norm(x_true - x_m), f(x_m) - f(x_true))
```
## Откуда взялась функцию Хьюбера и ей подобные?
- Придётся вспомнить теорию про сильно выпуклые функции и сопряжённые функции из прошлого семестра!
- **Теорема 1.** Если $x^*$ точка минимума для сильно выпуклой функции $f$ с константой $m > 0$, то $x^*$ единственная такая точка и
$$
f(y) \geq f(x^*) + \frac{m}{2} \|x^* - y\|^2_2, \quad y \in \mathrm{dom}(f)
$$
- Сопряжённая функция к $f$
$$
f^*(y) = \sup_x \; (y^{\top}x - f(x))
$$
выпукла всегда
### Сопряжённая функция к сильновыпуклой функции
**Теорема.** Если $f$ - сильно выпуклая функция с константой $m$, сопряжённая к которой
$$
f^*(y) = \sup_{x} \; (y^{\top}x - f(x))
$$
тогда
- $f^*$ определена и дифференцируема для всех $y$ и при этом
$$
\nabla f^*(y) = \arg\max_{x} \; (y^{\top}x - f(x))
$$
- $\nabla f^*(y)$ удовлетворяет условию Липшица с константой $\frac{1}{m}$
#### Доказательство (часть 1)
_1._ Так как $f(x)$ - сильно выпуклая функция, то у функции $y^{\top} x - f(x)$ единственная точка максимума для каждого $y$. Обозначим её $x_y$
_2._ В силу условия оптимальности выполнено
$$
y = f'(x_y) \quad f^*(y) = \langle y, x_y \rangle - f(x_y)
$$
_3._ Тогда для произвольного $u$ выполнено
$$
f^*(u) = \sup_v \; (u^{\top}v - f(v)) \geq u^{\top}x_y - f(x_y) = x_y^{\top}(u - y) + x_y^{\top}y - f(x_y) = x_y^{\top}(u - y) + f^*(y)
$$
_4._ То есть по определению субдифференциала $x_y \in \partial f^*(y)$, но так как $x_y$ единственная точка максимума функции $y^{\top} x - f(x)$, то $\partial f^*(y) = \{x_y\}$ и функция $f^*$ дифференцируема
_5._ В итоге $\nabla f^*(y) = x_y$
### Доказательство (часть 2)
1. Рассмотрим две точки $u$ и $v$, в которых соответственно
$$
x_u = \nabla f^*(u) \quad x_v = \nabla f^* (v)
$$
2. Тогда по теореме 1, приведённой выше и применённой к сильно выпуклой функции $f(x) - y^{\top}x$
\begin{align*}
& f(x_u) + v^{\top}x_u \geq f(x_v) + v^{\top}x_v + \frac{m}{2}\| x_v - x_u \|^2_2\\
& f(x_v) + u^{\top}x_v \geq f(x_u) + u^{\top}x_u + \frac{m}{2} \| x_v - x_u \|^2_2
\end{align*}
3. Сложим оба неравенства и получим
$$
m \| x_v - x_u \|^2_2 \leq (x_u - x_v)^{\top}(u - v) \leq \|x_u - x_v\|_2 \|u - v\|_2
$$
или
$$
\| \nabla f^* (v) - \nabla f^*(u) \|_2 \leq \frac{1}{m}\| u - v\|_2
$$
4. Таким образом, $\nabla f^* (u)$ Липшицев с константой Липшица $\frac{1}{m}$
### Функция близости (proximity function)
- **Определение**. Функция $d$ называется функцией близости на выпуклом замкнутом множестве $C$ если
- она непрерывна и сильно выпукла на этом множестве
- $C \subseteq \mathrm{dom} f$
- Центром множества будем называть точку
$$
x_d = \arg\min_{x \in C} d(x)
$$
- Дополнительные предположения:
- $d$ 1-сильно выпукла
- $\inf_{x \in C} d(x) = 0$
- Тогда выполнено
$$
d(x) \geq \frac{1}{2}\|x - x_d\|_2^2, \quad x \in C
$$
### Примеры
- $d(x) = \frac{1}{2}\|x - u\|_2^2$ для $x_d = u \in C$
- $d(x) = \sum_{i=1}^n w_i (x_i - u_i)^2 / 2$ при $w_i > 1$ и $x_d = u \in C$
- $d(x) = \sum_{i=1}^n x_i\log x_i + \log n$ для $C = \{ x \mid x \geq 0, \; x_1 + \ldots + x_n = 1 \}$ и $x_d = \frac{1}{n}\mathbf{1}$
## Сглаживание через сопряжение
- Пусть негладкая выпуклая функция $f$ представима в виде
$$
f(x) = \sup_{y \in \mathrm{dom}(h)} \; ((Ax + b)^{\top}y - h(y)) = h^*(Ax+ b),
$$
где $h$ - выпукла, замкнута и имеет ограниченную область определения
- Тогда выберем функцию близости $d$ на $C = \mathrm{dom}(h)$ и построим функцию
$$
f_{\mu}(x) = \sup_{y \in \mathrm{dom}(h)} \; ((Ax + b)^{\top}y - h(y) - \mu d(y)) = (h + \mu d)^*(Ax + b)
$$
- Функция $f_{\mu}(x)$ будет гладкой, так как функция $h + \mu d$ - сильно выпукла с константой $\mu$
### Примеры для $f(x) = |x|$
- Представление через сопряжённую функцию
$$
f(x) = \sup_{-1\leq y \leq 1} xy = h^*(x), \quad h(y) = I_{[-1, 1]}
$$
- Функция близости $d(x) = \frac{x^2}{2}$ даёт функцию Хьюбера
$$
f_{\mu}(x) = \sup_{-1\leq y \leq 1} \left(xy - \mu \frac{y^2}{2}\right)
$$
- Функция близости $d(x) = 1 - \sqrt{1 - y^2}$ даёт другую аппроксимацию
$$
f_{\mu}(x) = \sup_{-1\leq y \leq 1} \left(xy - \mu \left(1 - \sqrt{1 - y^2}\right)\right) = \sqrt{x^2 + \mu^2} - \mu
$$
### Градиент гладкой аппроксимации
$$
f_{\mu}(x) = \sup_{y \in \mathrm{dom}(h)} \; ((Ax + b)^{\top}y - h(y) - \mu d(y)) = (h + \mu d)^*(Ax + b)
$$
- Градиент
$$
\nabla f_{\mu}(x) = A^{\top} \arg\max_{y \in \mathrm{dom}(h)} \; ((Ax + b)^{\top}y - h(y) - \mu d(y))
$$
- Его константа Липшица
$$
L_{\mu} = \frac{\|A\|_2^2}{\mu}
$$
### Точность аппроксимации и сложность решения сглаженной задачи
- Точность аппроксимации
$$
f(x) - \mu D \leq f_{\mu}(x) \leq f(x), \quad D = \sup_{y \in \mathrm{dom}(h)} d(y)
$$
- Оценка сверху
$$
f_{\mu}(x) \leq \sup_{y \in \mathrm{dom}(h)} \; ((Ax + b)^{\top}y - h(y)) = f(x)
$$
- Оценка снизу
\begin{align*}
f_{\mu}(x) & = \sup_{y \in \mathrm{dom}(h)} \; ((Ax + b)^{\top}y - h(y) - \mu d(y)) \\
& \geq \sup_{y \in \mathrm{dom}(h)} \; ((Ax + b)^{\top}y - h(y) - \mu D) = f(x) - \mu D
\end{align*}
- Сложность аналогична примеру с функцией Хьюбера будет $O\left( \frac{1}{\epsilon}\right)$ при решении гладкой задачи ускоренным градиентным методом
## Основные шаги
- Представить негладкую функцию в виде сопряжённой к некоторой другой выпуклой функции $h$ с ограниченной областью определения
- Найти подходящую функцию близости $d$ для множества $\mathrm{dom}(h)$
- Сопряжённая функция к функции $h + \mu d$ будет гладкой аппроксимацией исходной негладкой функции $f$
## Сглаживание через регуляризацию Moreau-Yosida
- Рассмотрим выпуклую и негладкую функцию $g$
- Для неё построим следующую функцию
$$
g_{\mu}(x) = \inf_{y} \left( g(y) + \frac{1}{2\mu}\|y - x\|_2^2 \right) = \left(g \square \frac{1}{2\mu}\| \cdot\|_2^2\right)(x),
$$
где $\square$ обозначает инфимальную конволюцию двух функций
- Из свойств инфимальной конволюции следует, что $g_{\mu}$ выпукла
### Почему будет сглаживание?
- Перепишем $g_{\mu}(x)$ в виде
\begin{align*}
g_{\mu}(x) & = \frac{1}{2\mu}\|x\|_2^2 + \inf_y \left(g(y) - \frac{1}{\mu} y^{\top}x + \frac{1}{2\mu}\|y\|_2^2 \right) \\
& = \frac{1}{2\mu}\|x\|_2^2 - \frac{1}{\mu}\sup_y \left( y^{\top}x - \left( \mu g(y) + \frac{1}{2}\| y \|_2^2 \right) \right) \\
& = \frac{1}{2\mu}\|x\|_2^2 -\frac{1}{\mu} \left( \mu g + \frac{1}{2}\| \cdot \|_2^2 \right)^*(x)
\end{align*}
- Второе слагаемое есть сопряжённая функция к сумме выпуклой и сильной выпуклой функции, а значит является гладкой по ранее доказанной теореме
### Вычислим градиент
\begin{align*}
g'_{\mu}(x) &= \frac{x}{\mu} - \frac{1}{\mu}\arg\max_y \left( y^{\top}x - \left( \mu g(y) + \frac{1}{2}\| y \|_2^2 \right) \right) \\
& = \frac{x}{\mu} - \frac{1}{\mu}\arg\min_y \left( \mu g(y) + \frac{1}{2}\|y - x\|_2^2 \right) \\
& = \frac{x}{\mu} - \frac{1}{\mu} prox_{\mu g}(x)
\end{align*}
Таким образом, получаем следующую интерпретацию проксимального метода
$$
x_{k+1} = prox_{\mu g}(x_k) = x_k - \mu g'_{\mu}(x_k)
$$
как градиентного спуска для сглаженной аппроксимации исходной функции.
### Совпадение точек минимума
**Теорема.** $x^*$ точка минимума функции $g$ iff $x^*$ точка минимума для $g_{\mu}$
**Доказательство**
_1._ $x^*$ точка минимума функции $g$ iff $x^* = prox_{g}(x^*)$
_2._ Если $x^*$ точка минимума $g$, тогда $x^* = prox_{g}(x^*) = x^* - \mu g'_{\mu}(x^*)$, то есть $g'_{\mu}(x^*) = 0$, следовательно в силу выпуклости $g_{\mu}(x)$ точка $x^*$ и для неё является точкой минимума.
_3._ Если $x^*$ точка минимума для функции $g_{\mu}(x)$ тогда $g'_{\mu}(x^*) = 0$, но тогда $prox_{\mu g}(x^*) = x^*$, то есть $x^*$ неподвижная точка проксимального оператора и тем самым показано, что $x^*$ точка минимума $g$
## Связь между двумя подходами к сглаживанию
- Первый подход связан с представлением функции в виде сопряжённой к некоторой другой функции
- Второй подход - это регуляризация Moreau-Yosida
- Запишем $g_{\mu}(x)$ в таком виде
$$
g_{\mu}(x) = \inf_y \left( g(y) + \frac{1}{2\mu} \|x - y\|_2^2 \right) = \inf_{y, z} \left( g(y) + \frac{1}{2\mu} \|z\|_2^2 \right) \quad \text{s.t. } x - y = z
$$
- Построим двойственную задачу к этой задаче минимизации
$$
L(y, z, \lambda) = g(y) + \frac{1}{2\mu} \|z\|_2^2 + \lambda^{\top}(x - y - z)
$$
- Сгруппируем слагаемые
$$
L(y, z, \lambda) = (g(y) - \lambda^{\top}y) + \left(\frac{1}{2\mu} \|z\|_2^2 - \lambda^{\top}z\right) + \lambda^{\top}x
$$
- Двойственная функция
$$
\inf_{y, z} L(y, z, \lambda) = \inf_{y} (g(y) - \lambda^{\top}y) - \frac{\mu}{2}\| \lambda \|_2^2 + \lambda^{\top}x = -g^*(\lambda) - \frac{\mu}{2}\| \lambda \|_2^2 + \lambda^{\top}x
$$
- В силу сильной двойственность
$$
g_{\mu}(x) = \max_{\lambda} \left( \lambda^{\top}x -g^*(\lambda) - \frac{\mu}{2}\| \lambda \|_2^2 \right) = \left( g^* + \frac{\mu}{2} \|\cdot\|_2^2 \right)^*(x)
$$
- Сравним с результатом для первым подходом
$$
f_{\mu}(x) = (h + \mu d)^*(x),
$$
для $h$ такой что $f(x) = h^*(Ax + b)$
## Итоги сравнения
- Первый подход является обобщением подхода регуляризации Moreau-Yosida на произвольные функции близости $d$
- Также первый подход позволяет произвольно выбирать $h$ лишь бы выполнялось представление функции $f$
- На самом деле множество функции, которые подлежат сглаживанию не ограничивается рассмотренными: ключевая операция - это инфимальная конволюция
- Подробности [тут](https://pdfs.semanticscholar.org/9df1/6ee1828f8d46cfc6c817dfef540f4c1af51e.pdf) в разделе 4.4.
## Всегда ли нужно применять сглаживание?
- Если задача имеет вид суммы двух негладких функций
$$
f(x) = g(x) + h(x),
$$
то как правильнее подходить к её решению?
- Напоминаю, что сглаживание даёт сходимость вида $O\left( \frac{1}{\epsilon}\right)$
- Поэтому делать задачу гладкой по обеим функциям $g$ и $h$ может быть нецелесообразно!
- Вместо этого стоит оставить как есть слагаемое, для которого легко вычитслить проксимальный оператор, сгладить другое слагаемое и применить **ускоренный проксимальный градиентный метод**, чтобы получить сходимость порядка $O\left( \frac{1}{\sqrt{\epsilon}}\right)$
## Резюме
- Изложенная техника сглаживания применима для довольно большого набора функций, которые представимы как сопряжённые к некоторой другой функции (conjugate-like functions)
- Выбор различных параметризаций и различных функций близости даёт раздличные гладкие аппроксимации
- Все они приводят к ускоренному получению решения исходной задачи по сравнению с субградиентным методом
- Однако добиться полного совпадения скоростей сходиморсти как в случае задачи композитной оптимизации не удаётся
|
github_jupyter
|
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
plt.rc("text", usetex=True)
x = np.linspace(-3, 3, 10000)
f = np.abs(x)
def huber(x_range, mu):
h = np.zeros_like(x_range)
for i, x in enumerate(x_range):
if np.abs(x) <= mu:
h[i] = x**2 / (2 * mu)
else:
h[i] = np.abs(x) - mu / 2
return h
mu = 0.5
huber_f = huber(x, mu)
plt.figure(figsize=(12, 8))
plt.plot(x, f, label="$|x|$")
plt.plot(x, huber_f, label=r"Huber $(\mu = {})$".format(mu))
plt.grid(True)
plt.xticks(fontsize=28)
plt.yticks(fontsize=28)
plt.legend(fontsize=28)
import liboptpy.unconstr_solvers as methods
import liboptpy.step_size as ss
m, n = 500, 100
A = np.random.randn(m, n)
x_true = np.random.randn(n)
b = A.dot(x_true)
f = lambda x: np.linalg.norm(A.dot(x) - b, 1)
subgrad = lambda x: A.T.dot(np.sign(A.dot(x) - b))
alpha = 1e-3
s = 1e-1
sg_methods = {
"SM 1/k": methods.fo.SubgradientMethod(f, subgrad, ss.InvIterStepSize()),
"SM fixed={}".format(alpha): methods.fo.SubgradientMethod(f, subgrad, ss.ConstantStepSize(alpha)),
"SM scaled fix, s={}".format(s): methods.fo.SubgradientMethod(f, subgrad,
ss.ScaledConstantStepSize(s)),
}
x0 = np.random.randn(n)
max_iter = 50000
for m in sg_methods:
_ = sg_methods[m].solve(x0=x0, max_iter=max_iter)
plt.figure(figsize=(10, 8))
for m in sg_methods:
plt.semilogy([f(x) for x in sg_methods[m].get_convergence()], label=m)
plt.legend(fontsize=20)
plt.xlabel(r"Number of iterations, $k$", fontsize=26)
plt.ylabel(r"Objective, $f(x_k)$", fontsize=26)
plt.xticks(fontsize=20)
plt.yticks(fontsize=20)
plt.grid(True)
def huber(x, mu):
res = np.zeros_like(x)
res[np.abs(x) <= mu] = x[np.abs(x) <= mu]**2 / (2 * mu)
res[np.abs(x) >= mu] = np.abs(x[np.abs(x) >= mu]) - mu / 2
return res
def grad_huber(x, mu):
res = np.zeros_like(x)
res[np.abs(x) <= mu] = x[np.abs(x) <= mu] / mu
res[np.abs(x) >= mu] = np.sign(x[np.abs(x) >= mu])
return res
mu = 1e-2
print(mu)
fmu = lambda x: np.sum(huber(A.dot(x) - b, mu))
grad_fmu = lambda x: A.T @ grad_huber(A.dot(x) - b, mu)
grad_methods = {
"GD Armijo": methods.fo.GradientDescent(fmu, grad_fmu, ss.Backtracking("Armijo", beta=0.1, rho=0.5, init_alpha=1)),
"GD": methods.fo.GradientDescent(fmu, grad_fmu, ss.ConstantStepSize(mu/np.linalg.norm(A, 2)**2)),
"Fast GD": methods.fo.AcceleratedGD(fmu, grad_fmu, ss.ConstantStepSize(mu/np.linalg.norm(A, 2)**2))
}
max_iter = 10000
for m in grad_methods:
_ = grad_methods[m].solve(x0=x0, max_iter=max_iter)
plt.figure(figsize=(10, 8))
for m in grad_methods:
plt.semilogy([fmu(x) for x in grad_methods[m].get_convergence()], label=m)
plt.legend(fontsize=20)
plt.xlabel(r"Number of iterations, $k$", fontsize=26)
plt.ylabel(r"Objective, $f_{\mu}(x_k)$", fontsize=26)
plt.xticks(fontsize=20)
plt.yticks(fontsize=20)
plt.grid(True)
plt.figure(figsize=(10, 8))
for m in grad_methods:
plt.semilogy([np.linalg.norm(grad_fmu(x)) for x in grad_methods[m].get_convergence()], label=m)
plt.legend(fontsize=20)
plt.xlabel(r"Number of iterations, $k$", fontsize=26)
plt.ylabel(r"$\|f_{\mu}'(x_k)\|_2$", fontsize=26)
plt.xticks(fontsize=20)
plt.yticks(fontsize=20)
plt.grid(True)
plt.figure(figsize=(10, 8))
for m in grad_methods:
plt.semilogy([f(x) for x in grad_methods[m].get_convergence()], label=m)
for m in sg_methods:
plt.semilogy([f(x) for x in sg_methods[m].get_convergence()], label=m)
plt.legend(fontsize=20)
plt.xlabel(r"Number of iterations, $k$", fontsize=26)
plt.ylabel(r"Objective, $f(x_k)$", fontsize=26)
plt.xticks(fontsize=20)
plt.yticks(fontsize=20)
plt.grid(True)
for m in grad_methods:
x_m = grad_methods[m].get_convergence()[-1]
print(m, "\n\t", np.linalg.norm(x_true - x_m), f(x_m) - f(x_true))
for m in sg_methods:
x_m = sg_methods[m].get_convergence()[-1]
print(m, "\n\t", np.linalg.norm(x_true - x_m), f(x_m) - f(x_true))
| 0.501465 | 0.99476 |
# Ejercicio 3 - Simulación de distribuciones condicionadas
### Julian Ferres - Nro.Padrón 101483
## Enunciado:
Sea $X$ \~ $N(0,1)$ truncada al intervalo $[-1,1]$
Imagine $m(x) = E[Y | X=x]$ como:
\begin{equation}
m(x) := \left\{
\begin{array}{ll}
\frac{(x + 2)^2}{2} & \mathrm{si\ } si -1\leq x<-0.5 \\
\frac{x}{2}+0.875 & \mathrm{si\ } -0.5 \leq x \leq 0\\
-5(x-0.2)^2 +1.075 & \mathrm{si\ } 0 < x \leq 0.5 \\
x + 0.125 & \mathrm{si\ } 0.5 \leq x < 1
\end{array}
\right.
\end{equation}
Dado un $x$, la distribución condicional de $Y - m(x)$ es $N(0, \sigma ^2(x))$,
con $\sigma(x)=0.2-0.1 * \cos(2x)$
- Se pide simular $200$ puntos $(X,Y)$, y graficarlos en un plano. Además, vamos a necesitar
Los $200$ pares ordenados en cuestión, para hacer análisis posteriores
- Reconstruir $m(x)$ con los $200$ puntos, para eso:
Realizar una partición de $[-1,1]$ en intervalos de longitud $h$ y en cada intervalo encontrar el polinomio $f$ de grado $M$ que minimice el error cuadratico medio $$ \frac{1}{n} \sum |f(X_i)-Y_i|^2$$
Usar:
1. $h = 0.5$ , $M=1$
2. $h = 0.1$ , $M=1$
3. $h = 0.25$ , $M=2$
4. $h = 0.5$ , $M=2$
## Solución:
#### Importo todas las librerias e inicializo funciones
```
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
from math import cos, pi
from scipy.stats import truncnorm
m1 = lambda x: (x+2)**2/2
m2 = lambda x: x/2 + 0.875
m3 = lambda x: -5*(x-0.2)**2 + 1.075
m4 = lambda x: x + 0.125
def m(x):
if -1 <= x < -0.5:
return m1(x)
if -0.5 <= x < 0:
return m2(x)
if 0 <= x < 0.5:
return m3(x)
if 0.5 <= x < 1:
return m4(x)
m = np.vectorize(m)
x_0 = np.linspace(-1,1,1000) #Me genero 1000 valores entre -1 y 1 para graficar m(x) 'suave'
y_0 = m(x_0)
```
#### Normal truncada
```
a , b = -1 , 1 #Limites de la normal truncada
x1 = np.linspace(truncnorm.ppf(0.01, a, b),
truncnorm.ppf(0.99, a, b), 200) #Genero 200 cuantiles de la normal truncada
plt.plot(x1, truncnorm.pdf(x1, a, b),
'r-', lw=3, alpha=0.75, label='Normal truncada')
plt.title("Density Plot de X",fontsize='15')
plt.legend(loc='best', frameon= True)
plt.grid()
x1 = truncnorm.rvs(a, b, size=200)
#Me genero la muestra de distribucion X
sigma = np.vectorize(lambda x : 0.2 - 0.1 * cos(2*pi*x))
normal = np.vectorize(np.random.normal)
y1 = normal( m(x1),sigma(x1))
fig, ax = plt.subplots(figsize=(11,7))
plt.plot(x_0, y_0, 'g-', linewidth = 5, label = 'Función m(x)=E[Y|X=x]')
plt.legend(loc='best', frameon= True)
plt.plot(x1, y1, 'ro' ,markersize= 5, alpha = 0.5 ,label = 'Dispersion (X,Y)')
plt.legend(loc='best', frameon= True)
plt.title("Scatter Plot de (X,Y) y Line plot de m(x)", fontsize='15')
plt.xlabel('X')
plt.ylabel('Y')
plt.show()
```
#### La muestra de los $200$ pares con distribución $(X,Y)$ se encuentra en la variable output
## Reconstruyo la regresión
#### Con h=0.5 y M=1
```
partition = [[],[],[],[]]
for i in range(200):
partition[int(2*(x1[i]+1))].append(i)
polinomio_a_trozos = []
cuadrado_de_los_errores1 = 0
for i in range(4):
x_aux , y_aux = [x1[j] for j in partition[i]],[y1[j] for j in partition[i]]
z = np.polyfit(x_aux,y_aux,1)
polinomio_a_trozos.append(np.poly1d(z))
#sumo los errores para cada trozo del polinomio
for j in range(len(x_aux)):
cuadrado_de_los_errores1 += (polinomio_a_trozos[i](x_aux[j])-y_aux[j])**2
xp=[]
xp.append(np.linspace(-1, -0.5, 200))
xp.append(np.linspace(-0.5,0, 200))
xp.append(np.linspace(0, 0.5, 200))
xp.append(np.linspace(0.5,1, 200))
fig, ax = plt.subplots(figsize=(11,7))
plt.plot(x1, y1, 'ro', linewidth = 5, alpha = 0.5 ,label = 'Dispersion X,Y')
plt.legend(loc='best', frameon= True)
for i in range(4):
plt.plot(xp[i], polinomio_a_trozos[i](xp[i]) ,'b-', linewidth = 5 )
plt.plot(x_0, y_0, 'g-', linewidth = 5, alpha = 0.75 ,label = 'Función m(x)=E[Y|X=x]')
plt.legend(loc='best', frameon= True)
plt.title("Estimación m(x) con h=0.5 y M=1", fontsize='15')
plt.xlabel('X')
plt.ylabel('Y')
plt.show()
```
La estimación parece ajustarse bien a la función de regresion, no obstante, el error cuadrático medio es alto ya que no esta Overfitteando
a la muestra.
#### Estimación del error cuadrático medio
```
(cuadrado_de_los_errores1 / 200)**0.5
```
#### Con h=0.1 y M=1
```
partition = [[] for i in range(20)]
for i in range(200):
partition[int(10*(x1[i]+1))].append(i)
polinomio_a_trozos = []
cuadrado_de_los_errores2 = 0
for i in range(20):
x_aux , y_aux = [x1[j] for j in partition[i]],[y1[j] for j in partition[i]]
z = np.polyfit(x_aux,y_aux,1)
polinomio_a_trozos.append(np.poly1d(z))
#sumo los errores para cada trozo del polinomio
for j in range(len(x_aux)):
cuadrado_de_los_errores2 += (polinomio_a_trozos[i](x_aux[j])-y_aux[j])**2
xp=[]
for i in range(20):
xp.append(np.linspace(-1+i*(1/10), -0.9+i*(1/10), 200))
fig, ax = plt.subplots(figsize=(11,7))
plt.plot(x1, y1, 'ro', linewidth = 5, alpha = 0.5 ,label = 'Dispersion X,Y')
plt.legend(loc='best', frameon= True)
for i in range(20):
plt.plot(xp[i], polinomio_a_trozos[i](xp[i]) ,'b-', linewidth = 5 )
plt.plot(x_0, y_0, 'g-', linewidth = 5, alpha = 0.75,label = 'Función m(x)=E[Y|X=x]')
plt.legend(loc='best', frameon= True)
plt.title("Estimación m(x) con h=0.1 y M=1", fontsize='15')
plt.xlabel('X')
plt.ylabel('Y')
plt.show()
```
Se puede observar un claro caso de Overfitting, donde el error cuadrático medio es medianamente bajo, pero no estima correctamente la regresión.
#### Estimación del error cuadrático medio
```
(cuadrado_de_los_errores2 / 200)**0.5
```
#### Con h=0.25 y M=2
```
partition = [[] for i in range(8)]
for i in range(200):
partition[int(4*(x1[i]+1))].append(i)
polinomio_a_trozos = []
cuadrado_de_los_errores3 = 0
for i in range(8):
x_aux , y_aux = [x1[j] for j in partition[i]],[y1[j] for j in partition[i]]
z = np.polyfit(x_aux,y_aux,2)
polinomio_a_trozos.append(np.poly1d(z))
#sumo los errores para cada trozo del polinomio
for j in range(len(x_aux)):
cuadrado_de_los_errores3 += (polinomio_a_trozos[i](x_aux[j])-y_aux[j])**2
xp=[]
for i in range(8):
xp.append(np.linspace(-1+i*(1/4), -1+(i+1)*(1/4), 200))
fig, ax = plt.subplots(figsize=(11,7))
plt.plot(x1, y1, 'ro', linewidth = 5,alpha = 0.5, label ='Dispersion X,Y')
plt.legend(loc='best', frameon= True)
for i in range(8):
plt.plot(xp[i], polinomio_a_trozos[i](xp[i]) ,'b-', linewidth = 5 )
plt.plot(x_0, y_0, 'g-', linewidth = 5,alpha = 0.75 ,label = 'Función m(x)=E[Y|X=x]')
plt.legend(loc='best', frameon= True)
plt.title("Estimación m(x) con h=0.25 y M=2", fontsize='15')
plt.xlabel('X')
plt.ylabel('Y')
plt.show()
```
Se puede observar un claro caso de Overfitting, donde el error cuadrático medio es medianamente bajo, pero no estima correctamente la regresión.
#### Estimación del error cuadrático medio
```
(cuadrado_de_los_errores3 / 200)**0.5
```
#### Con h=0.5 y M=2
```
partition = [[] for i in range(4)]
for i in range(200):
partition[int(2*(x1[i]+1))].append(i)
polinomio_a_trozos = []
cuadrado_de_los_errores4 = 0
for i in range(4):
x_aux , y_aux = [x1[j] for j in partition[i]],[y1[j] for j in partition[i]]
z = np.polyfit(x_aux,y_aux,2)
polinomio_a_trozos.append(np.poly1d(z))
#sumo los errores para cada trozo del polinomio
for j in range(len(x_aux)):
cuadrado_de_los_errores4 += (polinomio_a_trozos[i](x_aux[j])-y_aux[j])**2
xp=[]
for i in range(4):
xp.append(np.linspace(-1+i*(1/2), -1+(i+1)*(1/2), 200))
fig, ax = plt.subplots(figsize=(11,7))
plt.plot(x1, y1, 'ro', linewidth = 5,alpha = 0.5, label = 'Dispersion X,Y')
plt.legend(loc='best', frameon= True)
for i in range(4):
plt.plot(xp[i], polinomio_a_trozos[i](xp[i]) ,'b-', linewidth = 5)
plt.plot(x_0, y_0, 'g-', linewidth = 5,alpha = 0.75 ,label = 'Función m(x)=E[Y|X=x]')
plt.legend(loc='best', frameon= True)
plt.title("Estimación m(x) con h=0.5 y M=2", fontsize='15')
plt.xlabel('X')
plt.ylabel('Y')
plt.show()
```
Se ve que el ECM es ligeramente superior a los casos con Overfitting, se ve que predice la regresión de forma bastante acertada.
#### Estimación del error cuadrático medio
```
(cuadrado_de_los_errores4 / 200)**0.5
(cuadrado_de_los_errores1 / 200)**0.5 , (cuadrado_de_los_errores2 / 200)**0.5 , (cuadrado_de_los_errores3 / 200)**0.5 , (cuadrado_de_los_errores4 / 200)**0.5
```
Link al Repo de GitHub: https://github.com/julianferres/Aprendizaje-Estadistico.git
|
github_jupyter
|
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
from math import cos, pi
from scipy.stats import truncnorm
m1 = lambda x: (x+2)**2/2
m2 = lambda x: x/2 + 0.875
m3 = lambda x: -5*(x-0.2)**2 + 1.075
m4 = lambda x: x + 0.125
def m(x):
if -1 <= x < -0.5:
return m1(x)
if -0.5 <= x < 0:
return m2(x)
if 0 <= x < 0.5:
return m3(x)
if 0.5 <= x < 1:
return m4(x)
m = np.vectorize(m)
x_0 = np.linspace(-1,1,1000) #Me genero 1000 valores entre -1 y 1 para graficar m(x) 'suave'
y_0 = m(x_0)
a , b = -1 , 1 #Limites de la normal truncada
x1 = np.linspace(truncnorm.ppf(0.01, a, b),
truncnorm.ppf(0.99, a, b), 200) #Genero 200 cuantiles de la normal truncada
plt.plot(x1, truncnorm.pdf(x1, a, b),
'r-', lw=3, alpha=0.75, label='Normal truncada')
plt.title("Density Plot de X",fontsize='15')
plt.legend(loc='best', frameon= True)
plt.grid()
x1 = truncnorm.rvs(a, b, size=200)
#Me genero la muestra de distribucion X
sigma = np.vectorize(lambda x : 0.2 - 0.1 * cos(2*pi*x))
normal = np.vectorize(np.random.normal)
y1 = normal( m(x1),sigma(x1))
fig, ax = plt.subplots(figsize=(11,7))
plt.plot(x_0, y_0, 'g-', linewidth = 5, label = 'Función m(x)=E[Y|X=x]')
plt.legend(loc='best', frameon= True)
plt.plot(x1, y1, 'ro' ,markersize= 5, alpha = 0.5 ,label = 'Dispersion (X,Y)')
plt.legend(loc='best', frameon= True)
plt.title("Scatter Plot de (X,Y) y Line plot de m(x)", fontsize='15')
plt.xlabel('X')
plt.ylabel('Y')
plt.show()
partition = [[],[],[],[]]
for i in range(200):
partition[int(2*(x1[i]+1))].append(i)
polinomio_a_trozos = []
cuadrado_de_los_errores1 = 0
for i in range(4):
x_aux , y_aux = [x1[j] for j in partition[i]],[y1[j] for j in partition[i]]
z = np.polyfit(x_aux,y_aux,1)
polinomio_a_trozos.append(np.poly1d(z))
#sumo los errores para cada trozo del polinomio
for j in range(len(x_aux)):
cuadrado_de_los_errores1 += (polinomio_a_trozos[i](x_aux[j])-y_aux[j])**2
xp=[]
xp.append(np.linspace(-1, -0.5, 200))
xp.append(np.linspace(-0.5,0, 200))
xp.append(np.linspace(0, 0.5, 200))
xp.append(np.linspace(0.5,1, 200))
fig, ax = plt.subplots(figsize=(11,7))
plt.plot(x1, y1, 'ro', linewidth = 5, alpha = 0.5 ,label = 'Dispersion X,Y')
plt.legend(loc='best', frameon= True)
for i in range(4):
plt.plot(xp[i], polinomio_a_trozos[i](xp[i]) ,'b-', linewidth = 5 )
plt.plot(x_0, y_0, 'g-', linewidth = 5, alpha = 0.75 ,label = 'Función m(x)=E[Y|X=x]')
plt.legend(loc='best', frameon= True)
plt.title("Estimación m(x) con h=0.5 y M=1", fontsize='15')
plt.xlabel('X')
plt.ylabel('Y')
plt.show()
(cuadrado_de_los_errores1 / 200)**0.5
partition = [[] for i in range(20)]
for i in range(200):
partition[int(10*(x1[i]+1))].append(i)
polinomio_a_trozos = []
cuadrado_de_los_errores2 = 0
for i in range(20):
x_aux , y_aux = [x1[j] for j in partition[i]],[y1[j] for j in partition[i]]
z = np.polyfit(x_aux,y_aux,1)
polinomio_a_trozos.append(np.poly1d(z))
#sumo los errores para cada trozo del polinomio
for j in range(len(x_aux)):
cuadrado_de_los_errores2 += (polinomio_a_trozos[i](x_aux[j])-y_aux[j])**2
xp=[]
for i in range(20):
xp.append(np.linspace(-1+i*(1/10), -0.9+i*(1/10), 200))
fig, ax = plt.subplots(figsize=(11,7))
plt.plot(x1, y1, 'ro', linewidth = 5, alpha = 0.5 ,label = 'Dispersion X,Y')
plt.legend(loc='best', frameon= True)
for i in range(20):
plt.plot(xp[i], polinomio_a_trozos[i](xp[i]) ,'b-', linewidth = 5 )
plt.plot(x_0, y_0, 'g-', linewidth = 5, alpha = 0.75,label = 'Función m(x)=E[Y|X=x]')
plt.legend(loc='best', frameon= True)
plt.title("Estimación m(x) con h=0.1 y M=1", fontsize='15')
plt.xlabel('X')
plt.ylabel('Y')
plt.show()
(cuadrado_de_los_errores2 / 200)**0.5
partition = [[] for i in range(8)]
for i in range(200):
partition[int(4*(x1[i]+1))].append(i)
polinomio_a_trozos = []
cuadrado_de_los_errores3 = 0
for i in range(8):
x_aux , y_aux = [x1[j] for j in partition[i]],[y1[j] for j in partition[i]]
z = np.polyfit(x_aux,y_aux,2)
polinomio_a_trozos.append(np.poly1d(z))
#sumo los errores para cada trozo del polinomio
for j in range(len(x_aux)):
cuadrado_de_los_errores3 += (polinomio_a_trozos[i](x_aux[j])-y_aux[j])**2
xp=[]
for i in range(8):
xp.append(np.linspace(-1+i*(1/4), -1+(i+1)*(1/4), 200))
fig, ax = plt.subplots(figsize=(11,7))
plt.plot(x1, y1, 'ro', linewidth = 5,alpha = 0.5, label ='Dispersion X,Y')
plt.legend(loc='best', frameon= True)
for i in range(8):
plt.plot(xp[i], polinomio_a_trozos[i](xp[i]) ,'b-', linewidth = 5 )
plt.plot(x_0, y_0, 'g-', linewidth = 5,alpha = 0.75 ,label = 'Función m(x)=E[Y|X=x]')
plt.legend(loc='best', frameon= True)
plt.title("Estimación m(x) con h=0.25 y M=2", fontsize='15')
plt.xlabel('X')
plt.ylabel('Y')
plt.show()
(cuadrado_de_los_errores3 / 200)**0.5
partition = [[] for i in range(4)]
for i in range(200):
partition[int(2*(x1[i]+1))].append(i)
polinomio_a_trozos = []
cuadrado_de_los_errores4 = 0
for i in range(4):
x_aux , y_aux = [x1[j] for j in partition[i]],[y1[j] for j in partition[i]]
z = np.polyfit(x_aux,y_aux,2)
polinomio_a_trozos.append(np.poly1d(z))
#sumo los errores para cada trozo del polinomio
for j in range(len(x_aux)):
cuadrado_de_los_errores4 += (polinomio_a_trozos[i](x_aux[j])-y_aux[j])**2
xp=[]
for i in range(4):
xp.append(np.linspace(-1+i*(1/2), -1+(i+1)*(1/2), 200))
fig, ax = plt.subplots(figsize=(11,7))
plt.plot(x1, y1, 'ro', linewidth = 5,alpha = 0.5, label = 'Dispersion X,Y')
plt.legend(loc='best', frameon= True)
for i in range(4):
plt.plot(xp[i], polinomio_a_trozos[i](xp[i]) ,'b-', linewidth = 5)
plt.plot(x_0, y_0, 'g-', linewidth = 5,alpha = 0.75 ,label = 'Función m(x)=E[Y|X=x]')
plt.legend(loc='best', frameon= True)
plt.title("Estimación m(x) con h=0.5 y M=2", fontsize='15')
plt.xlabel('X')
plt.ylabel('Y')
plt.show()
(cuadrado_de_los_errores4 / 200)**0.5
(cuadrado_de_los_errores1 / 200)**0.5 , (cuadrado_de_los_errores2 / 200)**0.5 , (cuadrado_de_los_errores3 / 200)**0.5 , (cuadrado_de_los_errores4 / 200)**0.5
| 0.405449 | 0.924142 |
```
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
df = pd.read_csv('data/df_preprocessed.csv')
df.info()
df.describe()
# create a list of our conditions
conditions = [
(df['state'] == 0),
(df['state'] == 1)
]
# create a list of the values we want to assign for each condition
values = ['failed', 'successful']
# create a new column and use np.select to assign values to it using our lists as arguments
df['outcome'] = np.select(conditions, values)
# display updated DataFrame
df.head()
fig, ax = plt.subplots()
sns.countplot(x='outcome', data=df, palette=['sandybrown', 'steelblue'])
plt.xlabel("Outcome", fontsize = 20)
plt.ylabel("Count", fontsize = 20)
#ax.tick_params(axis='x', rotation=45)
ax.set_facecolor('white')
plt.yticks(fontsize=20)
plt.xticks(fontsize=20)
fig.set_size_inches(11.7, 8.27)
sns.despine()
# add a column to bin goal_usd
# 100-1000
# 1000-5000
# 5000-10000
# 10000-50000
# 50000-100000
# >100000
# create a list of our conditions
conditions = [
(df['goal_usd'] <= 1000),
(df['goal_usd'] > 1000) & (df['goal_usd'] <= 5000),
(df['goal_usd'] > 5000) & (df['goal_usd'] <= 10000),
(df['goal_usd'] > 10000) & (df['goal_usd'] <= 50000),
(df['goal_usd'] > 50000) & (df['goal_usd'] <= 100000),
(df['goal_usd'] > 100000)
]
# create a list of the values we want to assign for each condition
values = ['< 1,000', '1,000 - 5,000', '5,000 - 10,000', '10,000 - 50,000', '50,000 - 100,000', '> 100,000']
# create a new column and use np.select to assign values to it using our lists as arguments
df['goal_bin'] = np.select(conditions, values)
# display updated DataFrame
df.head()
fig, ax = plt.subplots()
sns.countplot(x='goal_bin', hue='state', data=df, order=['< 1,000', '1,000 - 5,000', '5,000 - 10,000', '10,000 - 50,000', '50,000 - 100,000', '> 100,000'])
plt.legend(loc='upper right', labels=['failed', 'successful'], fontsize=20)
plt.xlabel("Goal in USD", fontsize = 20)
plt.ylabel("Count", fontsize = 20)
ax.tick_params(axis='x', rotation=45)
ax.set_facecolor('white')
plt.yticks(fontsize=20)
plt.xticks(fontsize=20)
fig.set_size_inches(11.7, 8.27)
sns.despine()
#sns.histplot(x='goal_usd', hue='state', data=goal, bins=[100,1000,5000,10000,50000,100000], multiple='dodge')
#plt.ylim(0,2000)
fig, ax = plt.subplots()
sns.countplot(x='country', data=df, hue='state')
plt.legend(loc='upper right', labels=['failed', 'successful'], fontsize=20)
plt.xlabel("Country", fontsize = 20)
plt.ylabel("Count", fontsize = 20)
ax.tick_params(axis='x', rotation=45)
ax.set_facecolor('white')
plt.yticks(fontsize=20)
plt.xticks(fontsize=20)
fig.set_size_inches(11.7, 8.27)
sns.despine()
# unfortunately, craft and crafts are listed as two separate categories
# combine them into crafts
df.loc[(df['category'] == 'craft'), 'category'] = 'crafts'
fig, ax = plt.subplots()
sns.countplot(x='category', data=df, hue='state')
plt.legend(loc='upper right', labels=['failed', 'successful'], fontsize=16)
plt.xlabel("Category", fontsize = 20)
plt.ylabel("Count", fontsize = 20)
ax.tick_params(axis='x', rotation=45)
ax.set_facecolor('white')
plt.yticks(fontsize=20)
plt.xticks(fontsize=16)
fig.set_size_inches(11.7, 8.27)
sns.despine()
## subset df so that it only includes the categories to avoid
df_avoid = df.query('category == "journalism" or category == "technology" or category =="food"')
df_avoid.info()
fig, ax = plt.subplots()
sns.countplot(x='category', data=df_avoid, hue='state')
#ax.tick_params(axis='x', rotation=45)
plt.legend(loc='upper right', labels=['failed', 'successful'], fontsize=20)
plt.xlabel("Category", fontsize = 24)
plt.ylabel("Count", fontsize = 20)
ax.set_facecolor('white')
plt.yticks(fontsize=20)
plt.xticks(fontsize=20)
fig.set_size_inches(11.7, 8.27)
sns.despine()
df.info()
fig, ax = plt.subplots()
sns.countplot(x='launch_day', data=df, hue='state')
ax.tick_params(axis='x', rotation=45)
plt.legend(title='Outcome', loc='upper right', labels=['failed', 'successful'])
fig.set_size_inches(11.7, 8.27)
sns.despine()
fig, ax = plt.subplots()
sns.countplot(x='launch_hour', data=df, hue='state')
ax.tick_params(axis='x', rotation=45)
plt.legend(title='Outcome', loc='upper right', labels=['failed', 'successful'])
fig.set_size_inches(11.7, 8.27)
sns.despine()
fig, ax = plt.subplots()
sns.histplot(x='name_len', data=df, hue='state')
ax.tick_params(axis='x', rotation=90)
plt.legend(loc='upper right', labels=[ 'successful', 'failed'], fontsize=20)
plt.xlabel("Length of Name", fontsize = 20)
plt.ylabel("Count", fontsize = 20)
ax.tick_params(axis='x', rotation=45)
ax.set_facecolor('white')
plt.yticks(fontsize=20)
plt.xticks(fontsize=20)
fig.set_size_inches(11.7, 8.27)
sns.despine()
fig, ax = plt.subplots()
sns.histplot(x='blurb_len', data=df, hue='state')
ax.tick_params(axis='x', rotation=90)
plt.legend(title='Outcome', loc='upper right', labels=['failed', 'successful'])
fig.set_size_inches(11.7, 8.27)
sns.despine()
df.head()
# bin time online
# create a list of our conditions
conditions = [
(df['delta_dead_laun'] <= 168),
(df['delta_dead_laun'] > 168) & (df['delta_dead_laun'] <= 336),
(df['delta_dead_laun'] > 336) & (df['delta_dead_laun'] <= 504),
(df['delta_dead_laun'] > 504) & (df['delta_dead_laun'] <= 672),
(df['delta_dead_laun'] > 672) & (df['delta_dead_laun'] <= 840),
(df['delta_dead_laun'] > 840) & (df['delta_dead_laun'] <= 1008),
(df['delta_dead_laun'] > 1008) & (df['delta_dead_laun'] <= 1176),
(df['delta_dead_laun'] > 1176) & (df['delta_dead_laun'] <= 1344),
(df['delta_dead_laun'] > 1344)
]
# create a list of the values we want to assign for each condition
values = ['1 week', '2 weeks', '3 weeks', '4 weeks', '5 weeks', '6 weeks', '7 weeks', '8 weeks', 'more than 8 weeks']
# create a new column and use np.select to assign values to it using our lists as arguments
df['time_online'] = np.select(conditions, values)
# display updated DataFrame
df.head()
fig, ax = plt.subplots()
sns.countplot(x='time_online', hue='state', data=df, order=['1 week', '2 weeks', '3 weeks', '4 weeks', '5 weeks', '6 weeks', '7 weeks', '8 weeks', 'more than 8 weeks'])
ax.tick_params(axis='x', rotation=45)
plt.legend(loc='upper right', labels=['failed', 'successful'], fontsize=20)
plt.xlabel("time online", fontsize = 20)
plt.ylabel("Count", fontsize = 20)
ax.set_facecolor('white')
plt.yticks(fontsize=20)
plt.xticks(fontsize=20)
fig.set_size_inches(11.7, 8.27)
sns.despine()
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
df = pd.read_csv('data/df_preprocessed.csv')
df.info()
df.describe()
# create a list of our conditions
conditions = [
(df['state'] == 0),
(df['state'] == 1)
]
# create a list of the values we want to assign for each condition
values = ['failed', 'successful']
# create a new column and use np.select to assign values to it using our lists as arguments
df['outcome'] = np.select(conditions, values)
# display updated DataFrame
df.head()
fig, ax = plt.subplots()
sns.countplot(x='outcome', data=df, palette=['sandybrown', 'steelblue'])
plt.xlabel("Outcome", fontsize = 20)
plt.ylabel("Count", fontsize = 20)
#ax.tick_params(axis='x', rotation=45)
ax.set_facecolor('white')
plt.yticks(fontsize=20)
plt.xticks(fontsize=20)
fig.set_size_inches(11.7, 8.27)
sns.despine()
# add a column to bin goal_usd
# 100-1000
# 1000-5000
# 5000-10000
# 10000-50000
# 50000-100000
# >100000
# create a list of our conditions
conditions = [
(df['goal_usd'] <= 1000),
(df['goal_usd'] > 1000) & (df['goal_usd'] <= 5000),
(df['goal_usd'] > 5000) & (df['goal_usd'] <= 10000),
(df['goal_usd'] > 10000) & (df['goal_usd'] <= 50000),
(df['goal_usd'] > 50000) & (df['goal_usd'] <= 100000),
(df['goal_usd'] > 100000)
]
# create a list of the values we want to assign for each condition
values = ['< 1,000', '1,000 - 5,000', '5,000 - 10,000', '10,000 - 50,000', '50,000 - 100,000', '> 100,000']
# create a new column and use np.select to assign values to it using our lists as arguments
df['goal_bin'] = np.select(conditions, values)
# display updated DataFrame
df.head()
fig, ax = plt.subplots()
sns.countplot(x='goal_bin', hue='state', data=df, order=['< 1,000', '1,000 - 5,000', '5,000 - 10,000', '10,000 - 50,000', '50,000 - 100,000', '> 100,000'])
plt.legend(loc='upper right', labels=['failed', 'successful'], fontsize=20)
plt.xlabel("Goal in USD", fontsize = 20)
plt.ylabel("Count", fontsize = 20)
ax.tick_params(axis='x', rotation=45)
ax.set_facecolor('white')
plt.yticks(fontsize=20)
plt.xticks(fontsize=20)
fig.set_size_inches(11.7, 8.27)
sns.despine()
#sns.histplot(x='goal_usd', hue='state', data=goal, bins=[100,1000,5000,10000,50000,100000], multiple='dodge')
#plt.ylim(0,2000)
fig, ax = plt.subplots()
sns.countplot(x='country', data=df, hue='state')
plt.legend(loc='upper right', labels=['failed', 'successful'], fontsize=20)
plt.xlabel("Country", fontsize = 20)
plt.ylabel("Count", fontsize = 20)
ax.tick_params(axis='x', rotation=45)
ax.set_facecolor('white')
plt.yticks(fontsize=20)
plt.xticks(fontsize=20)
fig.set_size_inches(11.7, 8.27)
sns.despine()
# unfortunately, craft and crafts are listed as two separate categories
# combine them into crafts
df.loc[(df['category'] == 'craft'), 'category'] = 'crafts'
fig, ax = plt.subplots()
sns.countplot(x='category', data=df, hue='state')
plt.legend(loc='upper right', labels=['failed', 'successful'], fontsize=16)
plt.xlabel("Category", fontsize = 20)
plt.ylabel("Count", fontsize = 20)
ax.tick_params(axis='x', rotation=45)
ax.set_facecolor('white')
plt.yticks(fontsize=20)
plt.xticks(fontsize=16)
fig.set_size_inches(11.7, 8.27)
sns.despine()
## subset df so that it only includes the categories to avoid
df_avoid = df.query('category == "journalism" or category == "technology" or category =="food"')
df_avoid.info()
fig, ax = plt.subplots()
sns.countplot(x='category', data=df_avoid, hue='state')
#ax.tick_params(axis='x', rotation=45)
plt.legend(loc='upper right', labels=['failed', 'successful'], fontsize=20)
plt.xlabel("Category", fontsize = 24)
plt.ylabel("Count", fontsize = 20)
ax.set_facecolor('white')
plt.yticks(fontsize=20)
plt.xticks(fontsize=20)
fig.set_size_inches(11.7, 8.27)
sns.despine()
df.info()
fig, ax = plt.subplots()
sns.countplot(x='launch_day', data=df, hue='state')
ax.tick_params(axis='x', rotation=45)
plt.legend(title='Outcome', loc='upper right', labels=['failed', 'successful'])
fig.set_size_inches(11.7, 8.27)
sns.despine()
fig, ax = plt.subplots()
sns.countplot(x='launch_hour', data=df, hue='state')
ax.tick_params(axis='x', rotation=45)
plt.legend(title='Outcome', loc='upper right', labels=['failed', 'successful'])
fig.set_size_inches(11.7, 8.27)
sns.despine()
fig, ax = plt.subplots()
sns.histplot(x='name_len', data=df, hue='state')
ax.tick_params(axis='x', rotation=90)
plt.legend(loc='upper right', labels=[ 'successful', 'failed'], fontsize=20)
plt.xlabel("Length of Name", fontsize = 20)
plt.ylabel("Count", fontsize = 20)
ax.tick_params(axis='x', rotation=45)
ax.set_facecolor('white')
plt.yticks(fontsize=20)
plt.xticks(fontsize=20)
fig.set_size_inches(11.7, 8.27)
sns.despine()
fig, ax = plt.subplots()
sns.histplot(x='blurb_len', data=df, hue='state')
ax.tick_params(axis='x', rotation=90)
plt.legend(title='Outcome', loc='upper right', labels=['failed', 'successful'])
fig.set_size_inches(11.7, 8.27)
sns.despine()
df.head()
# bin time online
# create a list of our conditions
conditions = [
(df['delta_dead_laun'] <= 168),
(df['delta_dead_laun'] > 168) & (df['delta_dead_laun'] <= 336),
(df['delta_dead_laun'] > 336) & (df['delta_dead_laun'] <= 504),
(df['delta_dead_laun'] > 504) & (df['delta_dead_laun'] <= 672),
(df['delta_dead_laun'] > 672) & (df['delta_dead_laun'] <= 840),
(df['delta_dead_laun'] > 840) & (df['delta_dead_laun'] <= 1008),
(df['delta_dead_laun'] > 1008) & (df['delta_dead_laun'] <= 1176),
(df['delta_dead_laun'] > 1176) & (df['delta_dead_laun'] <= 1344),
(df['delta_dead_laun'] > 1344)
]
# create a list of the values we want to assign for each condition
values = ['1 week', '2 weeks', '3 weeks', '4 weeks', '5 weeks', '6 weeks', '7 weeks', '8 weeks', 'more than 8 weeks']
# create a new column and use np.select to assign values to it using our lists as arguments
df['time_online'] = np.select(conditions, values)
# display updated DataFrame
df.head()
fig, ax = plt.subplots()
sns.countplot(x='time_online', hue='state', data=df, order=['1 week', '2 weeks', '3 weeks', '4 weeks', '5 weeks', '6 weeks', '7 weeks', '8 weeks', 'more than 8 weeks'])
ax.tick_params(axis='x', rotation=45)
plt.legend(loc='upper right', labels=['failed', 'successful'], fontsize=20)
plt.xlabel("time online", fontsize = 20)
plt.ylabel("Count", fontsize = 20)
ax.set_facecolor('white')
plt.yticks(fontsize=20)
plt.xticks(fontsize=20)
fig.set_size_inches(11.7, 8.27)
sns.despine()
| 0.511961 | 0.611672 |
# Now You Code 4: Guess A Number
Write a program to play the classic "Guess a number" game.
In this game the computer selects a random number between 1 and 10.
It's your job to guess the number. Whenever you guess, the computer will
give you a hint of higher or lower. This process repeats until you guess
the number, after which the computer reports the number of guesses it took you.
```
For Example:
I'm thinking of a number between 1 and 10...
Your guess: 5
Too low. Guess higher.
Your guess: 7
Too high. Guess lower.
Your guess: 6
You guessed it in 3 tries.
```
Your loop should continue until your input guess equals the
computer generated random number.
### How do you make Python generate a random number?
```
# Sample code which demostrates how to generate a number between 1 and 10
import random
number = random.randint(1,10)
print(number)
```
Run the cell above a couple of times. Notice how each time you execute the code, it comes up with a different number.
Here's a breakdown of the code
```
line 1 imports the random module
line 2 randomly selects an integer between 1 and 10
line 3 prints the number
```
Now that you understand how to generate a random number, try to design then write code for the program. The first step in your program should be to generate the random number.
## Step 1: Problem Analysis
Inputs: my guess and the random guessn
Outputs: if i was close or correct
Algorithm (Steps in Program):
```
import random
number = random.randint(1,100)
t = 0
while True:
guess = int(input("guess a number between 1 and 100: "))
if guess > number:
print("too high try again")
t = t + 1
if guess < number:
print("too low try again")
t = t + 1
if guess == number:
print('you got it! and only tried', t,'times!')
break
```
## Step 3: Questions
1. Which loop did you use to solve the problem? Was it a definite or indefinite loop?
Answer: an indefinite loop
2. Modify this program to allow you to guess a number between 1 and 100. How much of your code did you need to change to make this work?
Answer: not much just changed it in the range that the random number was selected and in the question.
3. This program is a good example of a difficult problem to conceptualize which has a simple solution when you look at actual lines of code. I assume you did not write this in a single try, so explain where you got stuck and describe your approach to overcoming it.
Answer: i actually did write it in 1 try shockingly but the more difficult thing to figure out was how to compact the program
## Step 4: Reflection
Reflect upon your experience completing this assignment. This should be a personal narrative, in your own voice, and cite specifics relevant to the activity as to help the grader understand how you arrived at the code you submitted. Things to consider touching upon: Elaborate on the process itself. Did your original problem analysis work as designed? How many iterations did you go through before you arrived at the solution? Where did you struggle along the way and how did you overcome it? What did you learn from completing the assignment? What do you need to work on to get better? What was most valuable and least valuable about this exercise? Do you have any suggestions for improvements?
To make a good reflection, you should journal your thoughts, questions and comments while you complete the exercise.
Keep your response to between 100 and 250 words.
`--== Write Your Reflection Below Here ==--`
i actually found it easy to code and not too difficult to output what i wanted
|
github_jupyter
|
For Example:
I'm thinking of a number between 1 and 10...
Your guess: 5
Too low. Guess higher.
Your guess: 7
Too high. Guess lower.
Your guess: 6
You guessed it in 3 tries.
# Sample code which demostrates how to generate a number between 1 and 10
import random
number = random.randint(1,10)
print(number)
line 1 imports the random module
line 2 randomly selects an integer between 1 and 10
line 3 prints the number
import random
number = random.randint(1,100)
t = 0
while True:
guess = int(input("guess a number between 1 and 100: "))
if guess > number:
print("too high try again")
t = t + 1
if guess < number:
print("too low try again")
t = t + 1
if guess == number:
print('you got it! and only tried', t,'times!')
break
| 0.516108 | 0.962568 |
```
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.model_selection import train_test_split
from scipy.sparse import csr_matrix, hstack
from wordbatch.models import FTRL, FM_FTRL
from nltk.corpus import stopwords
import re
import wordbatch
import pandas as pd
import numpy as np
def rmsle(y, y0): #defining metric
assert len(y) == len(y0)
return np.sqrt(np.mean(np.power(np.log1p(y) - np.log1p(y0), 2)))
stopwords = {x: 1 for x in stopwords.words('english')}
non_alphanums = re.compile(u'[^A-Za-z0-9]+') #using only numbers + english alphabet
def normalize_text(text):
return u" ".join(
[x for x in [y for y in non_alphanums.sub(' ', text).lower().strip().split(" ")] \
if len(x) > 1 and x not in stopwords]) #removing stop words and using only numbers + english alphabet
def handle_missing_inplace(df): #filling all nans
df['category_name'].fillna(value='missing/missing/missing', inplace=True)
df['brand_name'].fillna(value='missing', inplace=True)
df['item_description'].fillna(value='missing', inplace=True)
return df
train = pd.read_csv('./train.tsv', sep = '\t') #loading train
train.head()
sample = train.sample(frac = 0.05, random_state = 42)#using 5% sample
sample = handle_missing_inplace(sample) #filling all nans
y = sample.pop('price')
#splitting categories into 3 sub categories
sample['cat1'] = sample['category_name'].apply(lambda x: x.split('/')[0])
sample['cat2'] = sample['category_name'].apply(lambda x: x.split('/')[1])
sample['cat3'] = sample['category_name'].apply(lambda x: x.split('/')[2])
sample.head()
tf = TfidfVectorizer(max_features=10000,
max_df = 0.95, min_df = 100) #using tf-idf preprocessing to convert text in numerical matrix
#Evaluating tf-idf (transformig text into matrix)
print('Working with name')
x_name = tf.fit_transform(sample['name'].values)
print(7*'-')
print('Working with item_description')
x_description = tf.fit_transform(sample['item_description'].values)
print(7*'-')
print('Working with cat1')
x_cat1 = tf.fit_transform(sample['cat1'].values)
print(7*'-')
print('Working with cat2')
x_cat2 = tf.fit_transform(sample['cat2'].values)
print(7*'-')
print('Working with cat3')
x_cat3 = tf.fit_transform(sample['cat3'].values)
print(7*'-')
sample_preprocessed = hstack((x_name, x_description, x_cat1, x_cat2, x_cat3)).tocsr() #concatenating together and
#using scipy sparse for low-memory
#allocation of matrix
mask = np.array(np.clip(sample_preprocessed.getnnz(axis=0) - 1, 0, 1), dtype=bool)
sample_preprocessed = sample_preprocessed[:, mask]
x_train, x_val, y_train, y_val = train_test_split(sample_preprocessed, y, test_size = 0.15) #splitting into test and train
model = FM_FTRL(alpha=0.01, beta=0.01, L1=0.00001, L2=0.1, D = sample_preprocessed.shape[1], alpha_fm=0.01, L2_fm=0.0, init_fm=0.01,
D_fm=200, e_noise=0.0001, iters=15, inv_link="identity", threads=16) #defining model
model.fit(x_train, y_train) #training algorithm
y_pred = model.predict(x_val)#evaluating algorithm
print('RMSLE score using FM_FTRL:', rmsle(y_val, y_pred))
```
|
github_jupyter
|
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.model_selection import train_test_split
from scipy.sparse import csr_matrix, hstack
from wordbatch.models import FTRL, FM_FTRL
from nltk.corpus import stopwords
import re
import wordbatch
import pandas as pd
import numpy as np
def rmsle(y, y0): #defining metric
assert len(y) == len(y0)
return np.sqrt(np.mean(np.power(np.log1p(y) - np.log1p(y0), 2)))
stopwords = {x: 1 for x in stopwords.words('english')}
non_alphanums = re.compile(u'[^A-Za-z0-9]+') #using only numbers + english alphabet
def normalize_text(text):
return u" ".join(
[x for x in [y for y in non_alphanums.sub(' ', text).lower().strip().split(" ")] \
if len(x) > 1 and x not in stopwords]) #removing stop words and using only numbers + english alphabet
def handle_missing_inplace(df): #filling all nans
df['category_name'].fillna(value='missing/missing/missing', inplace=True)
df['brand_name'].fillna(value='missing', inplace=True)
df['item_description'].fillna(value='missing', inplace=True)
return df
train = pd.read_csv('./train.tsv', sep = '\t') #loading train
train.head()
sample = train.sample(frac = 0.05, random_state = 42)#using 5% sample
sample = handle_missing_inplace(sample) #filling all nans
y = sample.pop('price')
#splitting categories into 3 sub categories
sample['cat1'] = sample['category_name'].apply(lambda x: x.split('/')[0])
sample['cat2'] = sample['category_name'].apply(lambda x: x.split('/')[1])
sample['cat3'] = sample['category_name'].apply(lambda x: x.split('/')[2])
sample.head()
tf = TfidfVectorizer(max_features=10000,
max_df = 0.95, min_df = 100) #using tf-idf preprocessing to convert text in numerical matrix
#Evaluating tf-idf (transformig text into matrix)
print('Working with name')
x_name = tf.fit_transform(sample['name'].values)
print(7*'-')
print('Working with item_description')
x_description = tf.fit_transform(sample['item_description'].values)
print(7*'-')
print('Working with cat1')
x_cat1 = tf.fit_transform(sample['cat1'].values)
print(7*'-')
print('Working with cat2')
x_cat2 = tf.fit_transform(sample['cat2'].values)
print(7*'-')
print('Working with cat3')
x_cat3 = tf.fit_transform(sample['cat3'].values)
print(7*'-')
sample_preprocessed = hstack((x_name, x_description, x_cat1, x_cat2, x_cat3)).tocsr() #concatenating together and
#using scipy sparse for low-memory
#allocation of matrix
mask = np.array(np.clip(sample_preprocessed.getnnz(axis=0) - 1, 0, 1), dtype=bool)
sample_preprocessed = sample_preprocessed[:, mask]
x_train, x_val, y_train, y_val = train_test_split(sample_preprocessed, y, test_size = 0.15) #splitting into test and train
model = FM_FTRL(alpha=0.01, beta=0.01, L1=0.00001, L2=0.1, D = sample_preprocessed.shape[1], alpha_fm=0.01, L2_fm=0.0, init_fm=0.01,
D_fm=200, e_noise=0.0001, iters=15, inv_link="identity", threads=16) #defining model
model.fit(x_train, y_train) #training algorithm
y_pred = model.predict(x_val)#evaluating algorithm
print('RMSLE score using FM_FTRL:', rmsle(y_val, y_pred))
| 0.621081 | 0.529081 |
```
import csv
import cv2
import numpy as np
import matplotlib.pyplot as plt
import time
def read_file(filename):
"""
reads the file using csv library and returns rows in the file
"""
lines = []
with open(filename) as csvfile:
data_rows = csv.reader(csvfile)
for row in data_rows:
lines.append(row)
return lines
def crop_images(X, y):
"""
This method calculates the top and bottom percentages and crops the image
Resulting shape is (72, 320, 3)
No. of Output Images = No. of Input Images
"""
images = []
steering_angles = []
top_percent = 0.4
bottom_percent = 0.15
for i in range(len(X)):
ind_img = X[i]
top = int(np.ceil(ind_img.shape[0] * top_percent))
bottom = ind_img.shape[0] - int(np.ceil(ind_img.shape[0] * bottom_percent))
cropped_img = ind_img[top:bottom, :]
images.append(cropped_img)
steering_angles.append(y[i])
return images, steering_angles
#Without resizing gave better results, hence don't use this
def resize_images(X, y):
"""
This method resizes the images to height=66, widht=200
No. of Output Images = No. of Input Images
"""
images = []
steering_angles = []
for i in range(len(X)):
resized = cv2.resize(X[i], (200, 66))
images.append(resized)
steering_angles.append(y[i])
return images, steering_angles
def apply_gamma(X, y):
"""
This method applies gamma filter to the input images
Observe the gamma images are added to the original data set
No. of Output Images = 2 * (No. of Input Images)
"""
images = []
steering_angles = []
for i in range(len(X)):
gamma = np.random.uniform(0.7, 1.7)
inv_gamma = 1 / gamma
map_table = np.array([((i/255.0)**inv_gamma)*255 for i in np.arange(0,256)])
transformed_img = cv2.LUT(X[i], map_table)
images.append(X[i])
steering_angles.append(y[i])
images.append(transformed_img)
steering_angles.append(y[i])
return images, steering_angles
def vary_brightness(X, y):
"""
This method alters the brightness of the image by a random value
uses HSV color space as V represents brightness
No. of Output Images = No. of Input Images
"""
images = []
steering_angles = []
for i in range(len(X)):
# HSV (Hue, Saturation, Value) - Value is brightness
hsv_img = cv2.cvtColor(X[i], cv2.COLOR_RGB2HSV)
random_value = 1.0 + 0.6 * (np.random.rand() - 0.5)
hsv_img[:,:,2] = hsv_img[:,:,2] * random_value
transformed_img = cv2.cvtColor(hsv_img, cv2.COLOR_HSV2RGB)
images.append(transformed_img)
steering_angles.append(y[i])
return images, steering_angles
def flip_images_and_add(X, y):
"""
This method flips the input images
Flips are done only for those images where steering angles are outside the range of (-0.1, +0,1)
This means straight or near straight steering angle images are not flipped as it doens't add any value
No. of Output Images > No. of Input Images
"""
#print('size before', len(X))
images = []
steering_angles = []
for i in range(len(X)):
#print('less or greater {}'.format(y[i]))
images.append(X[i])
steering_angles.append(y[i])
#Flip only those images where there are curves
if y[i] < -0.1 or y[i] > 0.1 :
images.append(cv2.flip(X[i], 1))
steering_angles.append(y[i] * -1.0)
return images, steering_angles
def translate(X, y, range_x, range_y):
"""
This method randomly translates the image in any direction
and calculates the corresponding change in the steering angle
"""
images = []
steering_angles = []
for i in range(len(X)):
trans_x = range_x * (np.random.rand() - 0.5)
trans_y = range_y * (np.random.rand() - 0.5)
transformed_angle = y[i] + trans_x * 0.002
trans_m = np.float32([[1, 0, trans_x], [0, 1, trans_y]])
height, width = X[i].shape[:2]
transformed_img = cv2.warpAffine(X[i], trans_m, (width, height))
images.append(X[i])
steering_angles.append(y[i])
images.append(transformed_img)
steering_angles.append(transformed_angle)
return images, steering_angles
from sklearn.model_selection import train_test_split
from sklearn.utils import shuffle
def data_generator(rows, validation_flag, batch_size):
"""
This is the Python Generator that reads values in chunks
and makes it possible to run in modest CPUs
"""
correction_factor = 0.20
path = 'trainingdata/IMG/'
len_rows = len(rows)
rows = shuffle(rows)
while 1:
for offset in range(0, len_rows, batch_size):
batch_rows = rows[offset:offset+batch_size]
images = []
steering_values = []
#print('rows in batch', len(batch_rows))
for line in batch_rows:
center_image_path = line[0]
left_image_path = line[1]
right_image_path = line[2]
center_image_name = center_image_path.split('/')[-1] #Last token [-1] is the image
left_image_name = left_image_path.split('/')[-1]
right_image_name = right_image_path.split('/')[-1]
center_image_bgr = cv2.imread(path+center_image_name)
left_image_bgr = cv2.imread(path+left_image_name)
right_image_bgr = cv2.imread(path+right_image_name)
#Converting from BGR to RGB space as simulator reads RGB space
center_image = cv2.cvtColor(center_image_bgr, cv2.COLOR_BGR2RGB)
left_image = cv2.cvtColor(left_image_bgr, cv2.COLOR_BGR2RGB)
right_image = cv2.cvtColor(right_image_bgr, cv2.COLOR_BGR2RGB)
steering_value = float(line[3])
left_steering_value = steering_value + correction_factor
right_steering_value = steering_value - correction_factor
images.append(cv2.GaussianBlur(center_image, (3, 3), 0))
# images.append(center_image)
steering_values.append(steering_value)
images.append(cv2.GaussianBlur(left_image, (3, 3), 0))
# images.append(left_image)
steering_values.append(left_steering_value)
images.append(cv2.GaussianBlur(right_image, (3, 3), 0))
# images.append(right_image)
steering_values.append(right_steering_value)
X_train, y_train = images, steering_values
X_train, y_train = shuffle(X_train, y_train)
#Augmenting & Pre-processing
#X_train, y_train = crop_images(X_train, y_train)
#X_train, y_train = resize_images(X_train, y_train)
X_train, y_train = translate(X_train, y_train, 100, 10)
X_train, y_train = flip_images_and_add(X_train, y_train)
X_train, y_train = vary_brightness(X_train, y_train)
X_train, y_train = shuffle(X_train, y_train)
X_train = np.array(X_train)
y_train = np.array(y_train)
yield X_train, y_train
from keras.layers import Flatten, Dense, Lambda, Cropping2D, Dropout, Reshape
from keras.models import Sequential
from keras.layers.convolutional import Convolution2D
#Architecture based on NVIDIA
def train_model(train_generator, valid_generator, len_train, len_valid):
"""
This method contains the definition of the model
It also calls methods to train and validate the data set
"""
print('Training started...')
model = Sequential()
#model.add(Lambda(lambda x: (x / 255) - 0.5, input_shape=(72, 320, 3)))
model.add(Lambda(lambda x: (x / 255) - 0.5, input_shape=(160, 320, 3)))
model.add(Cropping2D(cropping=((70, 25), (0, 0))))
#model.add(Reshape((55, 135)))
model.add(Convolution2D(24, 5, 5, activation='elu', subsample=(2, 2)))
model.add(Convolution2D(36, 5, 5, activation='elu', subsample=(2, 2)))
model.add(Convolution2D(48, 5, 5, activation='elu', subsample=(2, 2)))
model.add(Dropout(0.5))
model.add(Convolution2D(64, 3, 3, activation='elu'))
model.add(Convolution2D(64, 3, 3, activation='elu'))
model.add(Dropout(0.5))
model.add(Flatten())
model.add(Dense(512, activation='elu'))
model.add(Dense(64, activation='elu'))
model.add(Dropout(0.3))
model.add(Dense(10, activation='elu'))
model.add(Dense(1))
model.summary()
start_time = time.time()
model.compile(loss='mse', optimizer='adam')
model.fit_generator(train_generator, samples_per_epoch= len_train, validation_data=valid_generator, nb_val_samples=len_valid, nb_epoch=10)
print('Training complete!')
print('Total time for training {:.3f}'.format(time.time() - start_time))
model.save('model.h5')
def mainfn():
"""
This is the main function that kicks-off the process
"""
data_rows = read_file('./trainingdata/driving_log.csv')
print('Length of the csv file {}'.format(len(data_rows)))
rows_train, rows_valid = train_test_split(data_rows, test_size=0.2)
#print('splitting done {} {}'.format(len(rows_train), len(rows_valid)))
train_generator = data_generator(rows_train, False, batch_size = 32)
valid_generator = data_generator(rows_valid, True, batch_size = 32)
#print('generator invoked train {} valid {}'.format(train_generator, valid_generator))
train_model(train_generator, valid_generator, len(rows_train), len(rows_valid))
#Calling the mainfn() to kick-off the process
mainfn()
```
|
github_jupyter
|
import csv
import cv2
import numpy as np
import matplotlib.pyplot as plt
import time
def read_file(filename):
"""
reads the file using csv library and returns rows in the file
"""
lines = []
with open(filename) as csvfile:
data_rows = csv.reader(csvfile)
for row in data_rows:
lines.append(row)
return lines
def crop_images(X, y):
"""
This method calculates the top and bottom percentages and crops the image
Resulting shape is (72, 320, 3)
No. of Output Images = No. of Input Images
"""
images = []
steering_angles = []
top_percent = 0.4
bottom_percent = 0.15
for i in range(len(X)):
ind_img = X[i]
top = int(np.ceil(ind_img.shape[0] * top_percent))
bottom = ind_img.shape[0] - int(np.ceil(ind_img.shape[0] * bottom_percent))
cropped_img = ind_img[top:bottom, :]
images.append(cropped_img)
steering_angles.append(y[i])
return images, steering_angles
#Without resizing gave better results, hence don't use this
def resize_images(X, y):
"""
This method resizes the images to height=66, widht=200
No. of Output Images = No. of Input Images
"""
images = []
steering_angles = []
for i in range(len(X)):
resized = cv2.resize(X[i], (200, 66))
images.append(resized)
steering_angles.append(y[i])
return images, steering_angles
def apply_gamma(X, y):
"""
This method applies gamma filter to the input images
Observe the gamma images are added to the original data set
No. of Output Images = 2 * (No. of Input Images)
"""
images = []
steering_angles = []
for i in range(len(X)):
gamma = np.random.uniform(0.7, 1.7)
inv_gamma = 1 / gamma
map_table = np.array([((i/255.0)**inv_gamma)*255 for i in np.arange(0,256)])
transformed_img = cv2.LUT(X[i], map_table)
images.append(X[i])
steering_angles.append(y[i])
images.append(transformed_img)
steering_angles.append(y[i])
return images, steering_angles
def vary_brightness(X, y):
"""
This method alters the brightness of the image by a random value
uses HSV color space as V represents brightness
No. of Output Images = No. of Input Images
"""
images = []
steering_angles = []
for i in range(len(X)):
# HSV (Hue, Saturation, Value) - Value is brightness
hsv_img = cv2.cvtColor(X[i], cv2.COLOR_RGB2HSV)
random_value = 1.0 + 0.6 * (np.random.rand() - 0.5)
hsv_img[:,:,2] = hsv_img[:,:,2] * random_value
transformed_img = cv2.cvtColor(hsv_img, cv2.COLOR_HSV2RGB)
images.append(transformed_img)
steering_angles.append(y[i])
return images, steering_angles
def flip_images_and_add(X, y):
"""
This method flips the input images
Flips are done only for those images where steering angles are outside the range of (-0.1, +0,1)
This means straight or near straight steering angle images are not flipped as it doens't add any value
No. of Output Images > No. of Input Images
"""
#print('size before', len(X))
images = []
steering_angles = []
for i in range(len(X)):
#print('less or greater {}'.format(y[i]))
images.append(X[i])
steering_angles.append(y[i])
#Flip only those images where there are curves
if y[i] < -0.1 or y[i] > 0.1 :
images.append(cv2.flip(X[i], 1))
steering_angles.append(y[i] * -1.0)
return images, steering_angles
def translate(X, y, range_x, range_y):
"""
This method randomly translates the image in any direction
and calculates the corresponding change in the steering angle
"""
images = []
steering_angles = []
for i in range(len(X)):
trans_x = range_x * (np.random.rand() - 0.5)
trans_y = range_y * (np.random.rand() - 0.5)
transformed_angle = y[i] + trans_x * 0.002
trans_m = np.float32([[1, 0, trans_x], [0, 1, trans_y]])
height, width = X[i].shape[:2]
transformed_img = cv2.warpAffine(X[i], trans_m, (width, height))
images.append(X[i])
steering_angles.append(y[i])
images.append(transformed_img)
steering_angles.append(transformed_angle)
return images, steering_angles
from sklearn.model_selection import train_test_split
from sklearn.utils import shuffle
def data_generator(rows, validation_flag, batch_size):
"""
This is the Python Generator that reads values in chunks
and makes it possible to run in modest CPUs
"""
correction_factor = 0.20
path = 'trainingdata/IMG/'
len_rows = len(rows)
rows = shuffle(rows)
while 1:
for offset in range(0, len_rows, batch_size):
batch_rows = rows[offset:offset+batch_size]
images = []
steering_values = []
#print('rows in batch', len(batch_rows))
for line in batch_rows:
center_image_path = line[0]
left_image_path = line[1]
right_image_path = line[2]
center_image_name = center_image_path.split('/')[-1] #Last token [-1] is the image
left_image_name = left_image_path.split('/')[-1]
right_image_name = right_image_path.split('/')[-1]
center_image_bgr = cv2.imread(path+center_image_name)
left_image_bgr = cv2.imread(path+left_image_name)
right_image_bgr = cv2.imread(path+right_image_name)
#Converting from BGR to RGB space as simulator reads RGB space
center_image = cv2.cvtColor(center_image_bgr, cv2.COLOR_BGR2RGB)
left_image = cv2.cvtColor(left_image_bgr, cv2.COLOR_BGR2RGB)
right_image = cv2.cvtColor(right_image_bgr, cv2.COLOR_BGR2RGB)
steering_value = float(line[3])
left_steering_value = steering_value + correction_factor
right_steering_value = steering_value - correction_factor
images.append(cv2.GaussianBlur(center_image, (3, 3), 0))
# images.append(center_image)
steering_values.append(steering_value)
images.append(cv2.GaussianBlur(left_image, (3, 3), 0))
# images.append(left_image)
steering_values.append(left_steering_value)
images.append(cv2.GaussianBlur(right_image, (3, 3), 0))
# images.append(right_image)
steering_values.append(right_steering_value)
X_train, y_train = images, steering_values
X_train, y_train = shuffle(X_train, y_train)
#Augmenting & Pre-processing
#X_train, y_train = crop_images(X_train, y_train)
#X_train, y_train = resize_images(X_train, y_train)
X_train, y_train = translate(X_train, y_train, 100, 10)
X_train, y_train = flip_images_and_add(X_train, y_train)
X_train, y_train = vary_brightness(X_train, y_train)
X_train, y_train = shuffle(X_train, y_train)
X_train = np.array(X_train)
y_train = np.array(y_train)
yield X_train, y_train
from keras.layers import Flatten, Dense, Lambda, Cropping2D, Dropout, Reshape
from keras.models import Sequential
from keras.layers.convolutional import Convolution2D
#Architecture based on NVIDIA
def train_model(train_generator, valid_generator, len_train, len_valid):
"""
This method contains the definition of the model
It also calls methods to train and validate the data set
"""
print('Training started...')
model = Sequential()
#model.add(Lambda(lambda x: (x / 255) - 0.5, input_shape=(72, 320, 3)))
model.add(Lambda(lambda x: (x / 255) - 0.5, input_shape=(160, 320, 3)))
model.add(Cropping2D(cropping=((70, 25), (0, 0))))
#model.add(Reshape((55, 135)))
model.add(Convolution2D(24, 5, 5, activation='elu', subsample=(2, 2)))
model.add(Convolution2D(36, 5, 5, activation='elu', subsample=(2, 2)))
model.add(Convolution2D(48, 5, 5, activation='elu', subsample=(2, 2)))
model.add(Dropout(0.5))
model.add(Convolution2D(64, 3, 3, activation='elu'))
model.add(Convolution2D(64, 3, 3, activation='elu'))
model.add(Dropout(0.5))
model.add(Flatten())
model.add(Dense(512, activation='elu'))
model.add(Dense(64, activation='elu'))
model.add(Dropout(0.3))
model.add(Dense(10, activation='elu'))
model.add(Dense(1))
model.summary()
start_time = time.time()
model.compile(loss='mse', optimizer='adam')
model.fit_generator(train_generator, samples_per_epoch= len_train, validation_data=valid_generator, nb_val_samples=len_valid, nb_epoch=10)
print('Training complete!')
print('Total time for training {:.3f}'.format(time.time() - start_time))
model.save('model.h5')
def mainfn():
"""
This is the main function that kicks-off the process
"""
data_rows = read_file('./trainingdata/driving_log.csv')
print('Length of the csv file {}'.format(len(data_rows)))
rows_train, rows_valid = train_test_split(data_rows, test_size=0.2)
#print('splitting done {} {}'.format(len(rows_train), len(rows_valid)))
train_generator = data_generator(rows_train, False, batch_size = 32)
valid_generator = data_generator(rows_valid, True, batch_size = 32)
#print('generator invoked train {} valid {}'.format(train_generator, valid_generator))
train_model(train_generator, valid_generator, len(rows_train), len(rows_valid))
#Calling the mainfn() to kick-off the process
mainfn()
| 0.433262 | 0.548553 |
The [previous notebook](2-Pipeline.ipynb) showed all the steps required to get a datashader rendering of your dataset, yielding raster images displayed using [Jupyter](http://jupyter.org)'s "rich display" support. However, these bare images do not show the data ranges or axis labels, making them difficult to interpret. Moreover, they are only static images, and datasets often need to be explored at multiple scales, which is much easier to do in an interactive program.
To get axes and interactivity, the images generated by datashader need to be embedded into a plot using an external library like [Matplotlib](http://matplotlib.org) or [Bokeh](http://bokeh.pydata.org). As we illustrate below, the most convenient way to make datashader plots using these libraries is via the [HoloViews](http://holoviews.org) high-level data-science API. Datashader itself also provides some limited support for use with Bokeh natively, and native datashader support for Matplotlib has been [sketched](https://github.com/bokeh/datashader/pull/200) but is not yet released.
In this notebook, we will first look at datashader's native Bokeh support, because it uses the same API introduced in the previous examples. We'll start with the same example from the [previous notebook](2-Pipeline.ipynb):
```
import pandas as pd
import numpy as np
import datashader as ds
import datashader.transfer_functions as tf
from collections import OrderedDict as odict
num=100000
np.random.seed(1)
dists = {cat: pd.DataFrame(odict([('x',np.random.normal(x,s,num)),
('y',np.random.normal(y,s,num)),
('val',val),
('cat',cat)]))
for x, y, s, val, cat in
[( 2, 2, 0.03, 10, "d1"),
( 2, -2, 0.10, 20, "d2"),
( -2, -2, 0.50, 30, "d3"),
( -2, 2, 1.00, 40, "d4"),
( 0, 0, 3.00, 50, "d5")] }
df = pd.concat(dists,ignore_index=True)
df["cat"]=df["cat"].astype("category")
```
Bokeh provides interactive plotting in a web browser. To make an interactive datashader plot when working with Bokeh directly, we'll first need to write a "callback" that wraps up the plotting steps shown in the previous notebook. A callback is a function that will render an image of the dataframe above when given some parameters:
```
def image_callback(x_range, y_range, w, h, name=None):
cvs = ds.Canvas(plot_width=w, plot_height=h, x_range=x_range, y_range=y_range)
agg = cvs.points(df, 'x', 'y', ds.count_cat('cat'))
img = tf.shade(agg)
return tf.dynspread(img, threshold=0.50, name=name)
```
As you can see, this callback is a function that lets us generate a Datashader image covering any range of data space that we want to examine:
```
tf.Images(image_callback(None, None, 300, 300, name="Original"),
image_callback(( 0, 4 ), ( 0, 4 ), 300, 300, name="Zoom 1"),
image_callback((1.9, 2.1), (1.9, 2.1), 300, 300, name="Zoom 2"))
```
You can now see that the single apparent "red dot" from the original image is actually a large collection of overlapping points (100,000, to be exact). However, you can also see that it would be awkward to explore a dataset using static images in this way, having to guess at numerical ranges as in the code above. Instead, let's make an interactive Bokeh plot using a convenience utility from Datashader called ``InteractiveImage``:
```
from datashader.bokeh_ext import InteractiveImage
import bokeh.plotting as bp
bp.output_notebook()
p = bp.figure(tools='pan,wheel_zoom,reset', x_range=(-5,5), y_range=(-5,5), plot_width=500, plot_height=500)
InteractiveImage(p, image_callback)
```
``InteractiveImage`` accepts any Bokeh figure and a callback that returns an image when given the range and pixel size. Now we can see the full axes corresponding to this data, and we can also zoom in using a scroll wheel (as long as the "wheel zoom" tool is enabled on the right) or pan by clicking and dragging (as long as the "pan" tool is enabled on the right). Each time you zoom or pan, the callback will be given the new viewport that's now visible, and datashader will render a new image to update the display. The result makes it look as if all of the data is available in the web browser interactively, while only ever storing a single image at any one time. In this way, full interactivity can be provided even for data that is far too large to display in a web browser directly. (Most web browsers can handle tens of thousands or hundreds of thousands of data points, but not millions or billions!)
***Note that you'll only see an updated image on zooming in if there is a live Python process running.*** Bokeh works by taking a Python specification for a plot and generating a corresponding JavaScript-based visualization in the browser. Whatever data has been given to the browser can be viewed interactively, but in this case only a single image of the data is given at a time, and so you will not be able to see more detail when zooming in unless the Python (and thus Datashader) process is running. In a static HTML export of this notebook, such as those on a website, you'll only see the original pixels getting larger, not a zoomed-in rendering as in the callback plots above.
``InteractiveImage`` lets you explore any Datashader pipeline you like, but unfortunately it only works in a Jupyter notebook (not a deployed Bokeh server), and it is not typically possible to combine such a plot with other Bokeh figures. The [dashboard.py](https://github.com/bokeh/datashader/blob/cb2f49f9/examples/dashboard/dashboard.py) from datashader 0.6 gives an example of building Bokeh+Datashader visualizations from the ground up, but this approach is quite difficult and is not recommended for most users. For these reasons, we do not recommend using InteractiveImage in new projects. Luckily, a much more practical approach to embedding and interactivity is available using HoloViews, as shown in the rest of this guide.
# Embedding Datashader with HoloViews
[HoloViews](http://holoviews.org) (1.7 and later) is a high-level data analysis and visualization library that makes it simple to generate interactive [Datashader](https://github.com/bokeh/datashader)-based plots. Here's an illustration of how this all fits together when using HoloViews+[Bokeh](http://bokeh.pydata.org):

HoloViews offers a data-centered approach for analysis, where the same tool can be used with small data (anything that fits in a web browser's memory, which can be visualized with Bokeh directly), and large data (which is first sent through Datashader to make it tractable) and with several different plotting frontends. A developer willing to do more programming can do all the same things separately, using Bokeh, Matplotlib, and Datashader's APIs directly, but with HoloViews it is much simpler to explore and analyze data. Of course, the [previous notebook](1-Pipeline.ipynb) showed that you can also use datashader without either any plotting library at all (the light gray pathways above), but then you wouldn't have interactivity, axes, and so on.
Most of this notebook will focus on HoloViews+Bokeh to support full interactive plots in web browsers, but we will also briefly illustrate the non-interactive HoloViews+Matplotlib approach. Let's start by importing some parts of HoloViews and setting some defaults:
```
import holoviews as hv
import holoviews.operation.datashader as hd
hd.shade.cmap=["lightblue", "darkblue"]
hv.extension("bokeh", "matplotlib")
```
### HoloViews+Bokeh
Rather than starting out by specifying a figure or plot, in HoloViews you specify an [``Element``](http://holoviews.org/reference/index.html#elements) object to contain your data, such as `Points` for a collection of 2D x,y points. To start, let's define a Points object wrapping around a small dataframe with 10,000 random samples from the ``df`` above:
```
points = hv.Points(df.sample(10000))
points
```
As you can see, the ``points`` object visualizes itself as a Bokeh plot, where you can already see many of the [problems that motivate datashader](https://anaconda.org/jbednar/plotting_pitfalls) (overplotting of points, being unable to detect the closely spaced dense collections of points shown in red above, and so on). But this visualization is just the default representation of ``points``, using Jupyter's [rich display](https://anaconda.org/jbednar/rich_display) support; the actual ``points`` object itself is merely a data container:
```
points.data.head()
```
### HoloViews+Datashader+Matplotlib
The default visualizations in HoloViews work well for small datasets, but larger ones will have overplotting issues as are already visible above, and will eventually either overwhelm the web browser (for the Bokeh frontend) or take many minutes to plot (for the Matplotlib backend). Luckily, HoloViews provides support for using Datashader to handle both of these problems:
```
%%output backend="matplotlib"
agg = ds.Canvas().points(df,'x','y')
hd.datashade(points) + hd.shade(hv.Image(agg)) + hv.RGB(np.array(tf.shade(agg).to_pil()))
```
Here we asked HoloViews to plot ``df`` using Datashader+Matplotlib, in three different ways:
- **A**: HoloViews aggregates and shades an image directly from the ``points`` object using its own datashader support, then passes the image to Matplotlib to embed into an appropriate set of axes.
- **B**: HoloViews accepts a pre-computed datashader aggregate, reads out the metadata about the plot ranges that is stored in the aggregate array, and passes it to Matplotlib for colormapping and then embedding.
- **C**: HoloViews accepts a PIL image computed beforehand and passes it to Matplotlib for embedding.
As you can see, option A is the most convenient; you can simply wrap your HoloViews element with ``datashade`` and the rest will be taken care of. But if you want to have more control by computing the aggregate or the full RGB image yourself using the API from the [previous notebook](2-Pipeline.ipynb) you are welcome to do so while using HoloViews+Matplotlib (or HoloViews+Bokeh, below) to embed the result into labelled axes.
### HoloViews+Datashader+Bokeh
The Matplotlib interface only produces a static plot, i.e., a PNG or SVG image, but the [Bokeh](http://bokeh.pydata.org) interface of HoloViews adds the dynamic zooming and panning necessary to understand datasets across scales:
```
hd.datashade(points)
```
Here, ``hd.datashade`` is not just a function call; it is an "operation" that dynamically calls datashader every time a new plot is needed by Bokeh, without the need for any explicit callback functions. The above plot will automatically be interactive when using the Bokeh frontend to HoloViews, and datashader will be called on each zoom or pan event if you have a live Python process running.
The powerful feature of operations is that you can chain them to make expressions for complex interactive visualizations. For instance, here is a Bokeh plot that works like the one created by ``InteractiveImage`` at the start of this notebook:
```
datashaded = hd.datashade(points, aggregator=ds.count_cat('cat')).redim.range(x=(-5,5),y=(-5,5))
hd.dynspread(datashaded, threshold=0.50, how='over').opts(plot=dict(height=500,width=500))
```
Compared to using ``InteractiveImage``, the HoloViews approach is simpler for the most basic plots (e.g. ``hd.datashade(hv.Points(df))``) while allowing plots to be overlaid and laid out together very flexibly. You can read more about HoloViews support for Datashader at [holoviews.org](http://holoviews.org/user_guide/Large_Data.html).
### HoloViews+Datashader+Bokeh Legends
Because the underlying plotting library only ever sees an image when using Datashader, providing legends and keys has to be handled separately from any underlying support for those features in the plotting library. We are working to simplify this process, but for now you can show a categorical legend by adding a suitable collection of labeled dummy points:
```
from datashader.colors import Sets1to3
datashaded = hd.datashade(points, aggregator=ds.count_cat('cat'), color_key=Sets1to3)
gaussspread = hd.dynspread(datashaded, threshold=0.50, how='over').opts(plot=dict(height=400,width=400))
color_key = [(name,color) for name,color in zip(["d1","d2","d3","d4","d5"], Sets1to3)]
color_points = hv.NdOverlay({n: hv.Points([0,0], label=str(n)).opts(style=dict(color=c)) for n,c in color_key})
color_points * gaussspread
```
### HoloViews+Datashader+Bokeh Hover info
As you can see, converting the data to an image using Datashader makes it feasible to work with even very large datasets interactively. One unfortunate side effect is that the original datapoints and line segments can no longer be used to support "tooltips" or "hover" information directly; that data simply is not present at the browser level, and so the browser cannot unambiguously report information about any specific datapoint. Luckily, you can still provide hover information that reports properties of a subset of the data in a separate layer, or you can provide information for a spatial region of the plot rather than for specific datapoints. For instance, in some small rectangle you can provide statistics such as the mean, count, standard deviation, etc. E.g. here let's calculate the count for each small square region:
```
%%opts QuadMesh [tools=['hover']] (alpha=0 hover_alpha=0.2)
from holoviews.streams import RangeXY
pts = hd.datashade(points, width=400, height=400)
(pts * hv.QuadMesh(hd.aggregate(points, width=10, height=10, dynamic=False))).relabel("Fixed hover") + \
\
(pts * hv.util.Dynamic(hd.aggregate(points, width=10, height=10, streams=[RangeXY]),
operation=hv.QuadMesh)).relabel("Dynamic hover")
```
In the above examples, the plot on the left provides hover information at a fixed spatial scale, while the one on the right reports on an area that scales with the zoom level so that arbitrarily small regions of data space can be examined, which is generally more useful.
As you can see, HoloViews makes it just about as simple to work with Datashader-based plots as regular Bokeh plots (at least if you don't need hover or color keys!), letting you visualize data of any size interactively in a browser using just a few lines of code. Because Datashader-based HoloViews plots are just one or two extra steps added on to regular HoloViews plots, they support all of the same features as regular HoloViews objects, and can freely be laid out, overlaid, and nested together with them. See [holoviews.org](http://holoviews.org) for examples and documentation for how to control the appearance of these plots and how to work with them in general.
|
github_jupyter
|
import pandas as pd
import numpy as np
import datashader as ds
import datashader.transfer_functions as tf
from collections import OrderedDict as odict
num=100000
np.random.seed(1)
dists = {cat: pd.DataFrame(odict([('x',np.random.normal(x,s,num)),
('y',np.random.normal(y,s,num)),
('val',val),
('cat',cat)]))
for x, y, s, val, cat in
[( 2, 2, 0.03, 10, "d1"),
( 2, -2, 0.10, 20, "d2"),
( -2, -2, 0.50, 30, "d3"),
( -2, 2, 1.00, 40, "d4"),
( 0, 0, 3.00, 50, "d5")] }
df = pd.concat(dists,ignore_index=True)
df["cat"]=df["cat"].astype("category")
def image_callback(x_range, y_range, w, h, name=None):
cvs = ds.Canvas(plot_width=w, plot_height=h, x_range=x_range, y_range=y_range)
agg = cvs.points(df, 'x', 'y', ds.count_cat('cat'))
img = tf.shade(agg)
return tf.dynspread(img, threshold=0.50, name=name)
tf.Images(image_callback(None, None, 300, 300, name="Original"),
image_callback(( 0, 4 ), ( 0, 4 ), 300, 300, name="Zoom 1"),
image_callback((1.9, 2.1), (1.9, 2.1), 300, 300, name="Zoom 2"))
from datashader.bokeh_ext import InteractiveImage
import bokeh.plotting as bp
bp.output_notebook()
p = bp.figure(tools='pan,wheel_zoom,reset', x_range=(-5,5), y_range=(-5,5), plot_width=500, plot_height=500)
InteractiveImage(p, image_callback)
import holoviews as hv
import holoviews.operation.datashader as hd
hd.shade.cmap=["lightblue", "darkblue"]
hv.extension("bokeh", "matplotlib")
points = hv.Points(df.sample(10000))
points
points.data.head()
%%output backend="matplotlib"
agg = ds.Canvas().points(df,'x','y')
hd.datashade(points) + hd.shade(hv.Image(agg)) + hv.RGB(np.array(tf.shade(agg).to_pil()))
hd.datashade(points)
datashaded = hd.datashade(points, aggregator=ds.count_cat('cat')).redim.range(x=(-5,5),y=(-5,5))
hd.dynspread(datashaded, threshold=0.50, how='over').opts(plot=dict(height=500,width=500))
from datashader.colors import Sets1to3
datashaded = hd.datashade(points, aggregator=ds.count_cat('cat'), color_key=Sets1to3)
gaussspread = hd.dynspread(datashaded, threshold=0.50, how='over').opts(plot=dict(height=400,width=400))
color_key = [(name,color) for name,color in zip(["d1","d2","d3","d4","d5"], Sets1to3)]
color_points = hv.NdOverlay({n: hv.Points([0,0], label=str(n)).opts(style=dict(color=c)) for n,c in color_key})
color_points * gaussspread
%%opts QuadMesh [tools=['hover']] (alpha=0 hover_alpha=0.2)
from holoviews.streams import RangeXY
pts = hd.datashade(points, width=400, height=400)
(pts * hv.QuadMesh(hd.aggregate(points, width=10, height=10, dynamic=False))).relabel("Fixed hover") + \
\
(pts * hv.util.Dynamic(hd.aggregate(points, width=10, height=10, streams=[RangeXY]),
operation=hv.QuadMesh)).relabel("Dynamic hover")
| 0.614972 | 0.973494 |
```
import os
# 設定環境變數來控制 keras, theano
os.environ['KERAS_BACKEND']="tensorflow"
#os.environ['THEANO_FLAGS']="floatX=float32, device=cuda"
import keras
from keras.models import Sequential
from PIL import Image
import numpy as np
import keras.backend as K
# 設定 channels_first 或 channels_last
K.set_image_data_format('channels_last')
from keras.preprocessing.image import load_img
from IPython.display import display
img_H, img_W = 360, 480
def preprocess_image(filename):
img = np.array(load_img(filename, target_size=(img_H, img_W)))
img = img[None, ...].astype('float32')
img = keras.applications.vgg16.preprocess_input(img)
return img
def show_image(arr):
arr = arr.reshape(img_H, img_W,3)+[103.939, 116.779, 123.68]
arr = arr.clip(0,255).astype('uint8')[:,:, ::-1]
display(Image.fromarray(arr))
from keras import backend as K
from keras.engine.topology import Layer
import numpy as np
class ImageLayer(Layer):
def __init__(self, init_img=None, **kwargs):
if init_img is None:
self.init_img = np.random.uniform(-50,50,size=(1,img_H, img_W, 3)).astype('float32')
else:
self.init_img = init_img
super().__init__(**kwargs)
def initializer(self, size):
return self.init_img
def build(self, input_shape):
# Create a trainable weight variable for this layer.
self.img = self.add_weight(shape=(1, img_H, img_W, 3),
initializer=self.initializer,
trainable=True)
super().build(input_shape) # Be sure to call this somewhere!
def call(self, x):
return self.img
def compute_output_shape(self, input_shape):
return (1, img_H, img_W, 3)
# 結構的圖片
#base_image = preprocess_image("img/tubingen.jpg")
base_image = preprocess_image("img/tubingen.jpg")
show_image(base_image)
style_image = preprocess_image("img/starry_night.jpg")
show_image(style_image)
image_layer = ImageLayer( init_img=.9*base_image +.1*style_image,
name='image_layer')(keras.layers.Input(shape=(0,)))
# Hack
_load_weights = keras.models.Model.load_weights
def my_load_weights(self, fn):
return _load_weights(self, fn, by_name=True)
keras.models.Model.load_weights = my_load_weights
# 將以上三個圖片送入 vgg16
vgg16_model = keras.applications.vgg16.VGG16(weights='imagenet', input_tensor=image_layer,
include_top=False, input_shape=(img_H, img_W,3))
# unhack
keras.models.Model.load_weights = _load_weights
# 比較簡單的方式取得各層
outputs_dict = {layer.name :layer.output for layer in vgg16_model.layers }
outputs_dict
import tensorflow as tf
w = vgg16_model.get_layer('image_layer').weights[0]
style_feature_names = ['block1_conv1', 'block2_conv1',
'block3_conv1', 'block4_conv1',
'block5_conv1']
style_features = [outputs_dict[x] for x in style_feature_names]
content_feature = outputs_dict['block4_conv2']
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
target_content_feature = sess.run(content_feature, feed_dict={w: base_image})
target_style_features = sess.run(style_features, feed_dict={w: style_image})
# 各種 Norms 和 loss function
# 取自 https://github.com/fchollet/keras/blob/master/examples/neural_style_transfer.py
# compute the neural style loss
# first we need to define 4 util functions
# the gram matrix of an image tensor (feature-wise outer product)
def gram_matrix(x):
assert K.ndim(x) == 3
features = K.batch_flatten(K.permute_dimensions(x, (2, 0, 1)))
gram = K.dot(features, K.transpose(features))
return gram
# the "style loss" is designed to maintain
# the style of the reference image in the generated image.
# It is based on the gram matrices (which capture style) of
# feature maps from the style reference image
# and from the generated image
def style_loss(combination, target):
assert K.ndim(combination) == 3
assert np.ndim(target) ==3
S = gram_matrix(K.constant(target))
C = gram_matrix(combination)
size = target.size
return K.sum(K.square(S - C)) / (4. * (size ** 2))
# an auxiliary loss function
# designed to maintain the "content" of the
# base image in the generated image
def content_loss(combination, target):
assert np.ndim(target) ==3
assert K.ndim(combination) == 3
size = target.size
return K.sum(K.square(combination - K.constant(target)))/size
# the 3rd loss function, total variation loss,
# designed to keep the generated image locally coherent
def total_variation_loss(x):
assert K.ndim(x) == 4
a = K.square(x[:, :-1, :-1, :] - x[:, 1: , :-1, :])
b = K.square(x[:, :-1, :-1, :] - x[:, :-1, 1: , :])
size = img_H * img_W * 3
return K.sum(K.pow(a + b, 1.25))/size
content_weight = .5
style_weight = 1.0
total_variation_weight = 1e-6
#content_weight = 20
#style_weight = 1.0
#total_variation_weight = 5e-4
loss_c = content_loss(content_feature[0], target_content_feature[0])
loss_s = K.variable(0.)
for layer, target_layer in zip(style_features, target_style_features):
loss_s = 2*loss_s + style_loss(layer[0], target_layer[0])
loss_s /= len(style_features)
loss_t = total_variation_loss(outputs_dict['image_layer'])
loss = content_weight * loss_c + style_weight*loss_s + total_variation_weight * loss_t
#train_step = tf.train.AdamOptimizer(5e-2).minimize(loss, var_list=[w])
train_step = tf.train.AdamOptimizer(0.1).minimize(loss, var_list=[w])
with tf.Session() as sess:
tf.global_variables_initializer().run()
for i in range(50000):
if i%100==0:
if i%500==0:
show_image(w.eval())
print(i, sess.run([loss, loss_s, loss_c, loss_t]))
train_step.run()
```
### 參考結果
<img src="img/result.png" />
|
github_jupyter
|
import os
# 設定環境變數來控制 keras, theano
os.environ['KERAS_BACKEND']="tensorflow"
#os.environ['THEANO_FLAGS']="floatX=float32, device=cuda"
import keras
from keras.models import Sequential
from PIL import Image
import numpy as np
import keras.backend as K
# 設定 channels_first 或 channels_last
K.set_image_data_format('channels_last')
from keras.preprocessing.image import load_img
from IPython.display import display
img_H, img_W = 360, 480
def preprocess_image(filename):
img = np.array(load_img(filename, target_size=(img_H, img_W)))
img = img[None, ...].astype('float32')
img = keras.applications.vgg16.preprocess_input(img)
return img
def show_image(arr):
arr = arr.reshape(img_H, img_W,3)+[103.939, 116.779, 123.68]
arr = arr.clip(0,255).astype('uint8')[:,:, ::-1]
display(Image.fromarray(arr))
from keras import backend as K
from keras.engine.topology import Layer
import numpy as np
class ImageLayer(Layer):
def __init__(self, init_img=None, **kwargs):
if init_img is None:
self.init_img = np.random.uniform(-50,50,size=(1,img_H, img_W, 3)).astype('float32')
else:
self.init_img = init_img
super().__init__(**kwargs)
def initializer(self, size):
return self.init_img
def build(self, input_shape):
# Create a trainable weight variable for this layer.
self.img = self.add_weight(shape=(1, img_H, img_W, 3),
initializer=self.initializer,
trainable=True)
super().build(input_shape) # Be sure to call this somewhere!
def call(self, x):
return self.img
def compute_output_shape(self, input_shape):
return (1, img_H, img_W, 3)
# 結構的圖片
#base_image = preprocess_image("img/tubingen.jpg")
base_image = preprocess_image("img/tubingen.jpg")
show_image(base_image)
style_image = preprocess_image("img/starry_night.jpg")
show_image(style_image)
image_layer = ImageLayer( init_img=.9*base_image +.1*style_image,
name='image_layer')(keras.layers.Input(shape=(0,)))
# Hack
_load_weights = keras.models.Model.load_weights
def my_load_weights(self, fn):
return _load_weights(self, fn, by_name=True)
keras.models.Model.load_weights = my_load_weights
# 將以上三個圖片送入 vgg16
vgg16_model = keras.applications.vgg16.VGG16(weights='imagenet', input_tensor=image_layer,
include_top=False, input_shape=(img_H, img_W,3))
# unhack
keras.models.Model.load_weights = _load_weights
# 比較簡單的方式取得各層
outputs_dict = {layer.name :layer.output for layer in vgg16_model.layers }
outputs_dict
import tensorflow as tf
w = vgg16_model.get_layer('image_layer').weights[0]
style_feature_names = ['block1_conv1', 'block2_conv1',
'block3_conv1', 'block4_conv1',
'block5_conv1']
style_features = [outputs_dict[x] for x in style_feature_names]
content_feature = outputs_dict['block4_conv2']
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
target_content_feature = sess.run(content_feature, feed_dict={w: base_image})
target_style_features = sess.run(style_features, feed_dict={w: style_image})
# 各種 Norms 和 loss function
# 取自 https://github.com/fchollet/keras/blob/master/examples/neural_style_transfer.py
# compute the neural style loss
# first we need to define 4 util functions
# the gram matrix of an image tensor (feature-wise outer product)
def gram_matrix(x):
assert K.ndim(x) == 3
features = K.batch_flatten(K.permute_dimensions(x, (2, 0, 1)))
gram = K.dot(features, K.transpose(features))
return gram
# the "style loss" is designed to maintain
# the style of the reference image in the generated image.
# It is based on the gram matrices (which capture style) of
# feature maps from the style reference image
# and from the generated image
def style_loss(combination, target):
assert K.ndim(combination) == 3
assert np.ndim(target) ==3
S = gram_matrix(K.constant(target))
C = gram_matrix(combination)
size = target.size
return K.sum(K.square(S - C)) / (4. * (size ** 2))
# an auxiliary loss function
# designed to maintain the "content" of the
# base image in the generated image
def content_loss(combination, target):
assert np.ndim(target) ==3
assert K.ndim(combination) == 3
size = target.size
return K.sum(K.square(combination - K.constant(target)))/size
# the 3rd loss function, total variation loss,
# designed to keep the generated image locally coherent
def total_variation_loss(x):
assert K.ndim(x) == 4
a = K.square(x[:, :-1, :-1, :] - x[:, 1: , :-1, :])
b = K.square(x[:, :-1, :-1, :] - x[:, :-1, 1: , :])
size = img_H * img_W * 3
return K.sum(K.pow(a + b, 1.25))/size
content_weight = .5
style_weight = 1.0
total_variation_weight = 1e-6
#content_weight = 20
#style_weight = 1.0
#total_variation_weight = 5e-4
loss_c = content_loss(content_feature[0], target_content_feature[0])
loss_s = K.variable(0.)
for layer, target_layer in zip(style_features, target_style_features):
loss_s = 2*loss_s + style_loss(layer[0], target_layer[0])
loss_s /= len(style_features)
loss_t = total_variation_loss(outputs_dict['image_layer'])
loss = content_weight * loss_c + style_weight*loss_s + total_variation_weight * loss_t
#train_step = tf.train.AdamOptimizer(5e-2).minimize(loss, var_list=[w])
train_step = tf.train.AdamOptimizer(0.1).minimize(loss, var_list=[w])
with tf.Session() as sess:
tf.global_variables_initializer().run()
for i in range(50000):
if i%100==0:
if i%500==0:
show_image(w.eval())
print(i, sess.run([loss, loss_s, loss_c, loss_t]))
train_step.run()
| 0.549641 | 0.363901 |
# <font color=#770000>ICPE 639 Introduction to Machine Learning </font>
## ------ With Energy Applications
<p> © 2021: Xiaoning Qian </p>
[Homepage](http://xqian37.github.io/)
**<font color=blue>[Note]</font>** This is currently a work in progress, will be updated as the material is tested in the class room.
All material open source under a Creative Commons license and free for use in non-commercial applications.
Source material used under the Creative Commons Attribution-NonCommercial 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc/3.0/ or send a letter to Creative Commons, PO Box 1866, Mountain View, CA 94042, USA.
# Reinforcement Learning
This section will provide a **very basic** introduction to Reinforcement Learning (RL):
- [1 Markov Decision Process](#1-Markov-Decision-Process)
- [2 Q Learning](#2-Q-Learning)
- [3 Deep RL](#3-Deep-RL)
- [4 Hands-on Exercise](#4-Exercise)
- [Reference](#Reference)
**<font color=blue>[Note]</font>**: Most of the materials here were based on the Microsoft free course: https://github.com/microsoft/ML-For-Beginners/blob/main/8-Reinforcement/1-QLearning/README.md and https://towardsdatascience.com/understanding-the-markov-decision-process-mdp-8f838510f150
## 1 Markov Decision Process
RL is to make the *optimal* decision, with model or model-free settings considering the underlying system, to achieve the maximum **reward**.
### Basics
RL is to make the *optimal* decision, with model or in a model-free setting for the underlying system, to achieve the maximum **reward**. For model-based decision making (control theory), Markov Decision Process (MDP) can serve as the foundation. The underlying dynamic model of MDP can be related to the state-space model that we just introduced.
MDP often involves the following critical components:
1. **State** in a state space $\mathcal{X}$;
2. **Action** in the action space $\mathcal{A}$;
3. **Transition** $\mathbf{P}$ governing the state dynamics (state-space models): $\mathbf{P}: \mathcal{X} \times \mathcal{A} \rightarrow \mathcal{X}$ (deterministic) or $\mathbf{P}: \mathcal{X} \times \mathcal{A} \times \mathcal{X} \rightarrow [0, 1]$ (probabilistic);
4. **Reward** $\mathbf{R}$ modeling the expected reward to take a certain action to reach different states: $\mathbf{R}: \mathcal{X} \times \mathcal{A} \times \mathcal{X} \rightarrow \mathbb{R}$.
With these four elements, we can model how an **agent** may interact with the **environment** based on $\mathbf{P}$. The goal of RL is to derive a good **policy** to achieve the best reward based on $\mathbf{R}$. If we have the model $\mathbf{P}$ and $\mathbf{R}$, we are with model-based RL. Otherwise, we are doing model-free RL, a famous one of which is **Q-learning**.
<img src="https://miro.medium.com/max/6000/1*p4JW6ibYcdmaeXVB_klivA.png" alt="MDP1">
<center>A Schematic for an artificial MDP</center>
<img src="https://miro.medium.com/max/1750/1*QLeo3MikUeGvNyVjl5mqoA.png" alt="MDP2">
<center>A Schematic for a Robot MDP</center>
### Markov Property
MDP can be considered as a **Markov Chain** with actions and assigned rewards. The introductory materials of MDP often starts with the finite state space and action space. Given the current state, the future is often assumed to be **conditionally independent** with the past, known as **Markov property**:
$$\mathbf{P}(x_{t+1} | x_t, a_t; \mbox{ the past state-action history }) = \mathbf{P}(x_{t+1} | x_{t}, a_t).$$
### MDP problem formulations
The goal is to derive a **control policy** to specific which action to take given the state at specific time:
$$\pi(a|x; t) = P(A_t = a | X_t = x). $$
The control policy can be either **deterministic** or **probabilistic**. If the policy is not dependent on time, $\pi(a|x; t) =\pi(a |x)$, then it is **stationary**, which is often the case in many applications.
### Value Iteration
#### Policy Value:
The **value** of a given policy $\pi(a|x)$ at $x$ is the **expected cumulative rewards** (often discounted) with the **discount factor** $\gamma \in (0, 1)$:
$$V(x) = \mathbb{E}_P[\mathbf{R}(x_0=x, a_0, x_1) + \gamma \mathbf{R}(x_1, a_1, x_2) + \ldots + \gamma^t \mathbf{R}(x_t, a_t, x_{t+1}) + \ldots|\pi]$$
By divide and conquer (an important trick for **dynamic programming**):
$$V(x) = \mathbb{E}_{x_1}[\mathbb{E}_P[\mathbf{R}(x_0=x, a_0, x_1) + \gamma V(x_1)]]$$
#### Optimal value and optimal action:
The logic of dynamic programming: To achieve the optimality of the whole sequence, each sub-sequence shall be also optimal. This leads to the famous **Bellman Optimality Equation**:
$$V_{opt}(x) = \max_{a\in\mathcal{A}}{\sum_{x_1} \mathbf{P}(x_1|x_0=x, a_0=a)(\mathbf{R}(x_0=x, a_0, x_1) + \gamma V_{opt}(x_1) )}$$
Assume that the iteration converges to a **stationary** policy (discounting). This immediate gives the **value iteration algorithm**:
$$V_{k+1} = \max_{a\in\mathcal{A}}{\sum_{x'} \mathbf{P}(x'|x, a)(\mathbf{R}(x, a, x') + \gamma V_{k}(x') )}.$$
$V_{k} \rightarrow V_{opt}$ for stationary control policies.
### State-Action (Q) Value
The Q value of a given policy $\pi(a|x)$ at $x$ with action $a$ is the expected cumulative rewards with the discount factor $\gamma$:
$$Q(x, a) = \mathbb{E}_{x_1}[\mathbb{E}_P[\mathbf{R}(x_0=x, a_0=a, x_1) + \gamma \max_{a_1} Q(x_1, a_1)]].$$
It has the same recursive structure.
### Q value iteration algorithm:
We can now derive the **optimal policy**:
$$\pi_{opt}(a|x) = \arg\max_a Q_{opt}(x,a),$$
where $Q_{opt}(x,a) = \mathbf{R}'(x_0=x, a_0=a) + \gamma \sum_{x_1} \mathbf{P}(x_1|x,a) V_{opt}(x_1)$ and $V_{opt}(x_1)$ can be solved:
$$V_{opt}(x_1) = \max_{a} Q(x_1, a)$$
as done by value iteration.
And
$$Q_{k+1}(x,a) = \sum_{x'} \mathbf{P}(x'|x, a)(\mathbf{R}(x, a, x') + \gamma \max_{a_1} Q_k(x_1, a_1))$$
(Q-value iteraction).
### Policy Iteration
We can also derive the **policy iteration** algorithm with the same dynamic programming trick:
$$\pi_k(a|x) = \arg\max_a{\sum_{x'} \mathbf{P}(x'|x, a)(\mathbf{R}(x, a, x') + \gamma V_{\pi_k}(x') )},$$
where $V_{\pi_k}(x')$ is evaluated value based on the previous policy.
## 2 Q Learning
Q Learning is a model-free RL method when we do not have the model for transition $\mathbf{P}$ or reward $\mathbf{R}$. Note that to be exact, RL often referrs to the model-free situations where you have to derive the policy based on the interaction between the agent and environment through experience/observations, *in an end-to-end fashion*. The policy $\pi(a|x)$ is learned only driven by the observed reward $r(x,a)$, for example as "relayed experience".
$$ \mathcal{D} = \{(x, a, r, x')_t\}_{t=0}^{T} \rightarrow Q_{opt}(x, a) \rightarrow \pi_{opt}(a|x) $$
### Q Learning Algorithm
1. Initialize Q-Table **$Q$** with equal numbers for all states and actions
2. Set **learning rate** $\alpha\leftarrow 1$
3. Repeat simulation many times
A. Start at random position
B. Repeat
1. Select an action $a$ at state $x$
2. Exectute action by moving to a new state $x'$
3. If we encounter end-of-game condition, or total reward is too small - exit simulation
4. Compute reward $r$ at the new state
5. Update Q-Function according to Bellman equation: $$Q(x,a)\leftarrow (1-\alpha)Q(x,a)+\alpha(r+\gamma\max_{a'}Q(x',a'))$$
6. $x\leftarrow x'$
7. Update total reward and decrease $\alpha$.
Check the above updating equation:
$$Q(x,a) \leftarrow Q(x,a) + \alpha(r+\gamma\max_{a'}Q(x',a') - Q(x,a))$$.
Similarity to what we have seen for other gradient descent updates.
### Exploration vs. Exploitation
At the step **3.B.2**, if random policy is adopted, it is simply **exploring** the state and action space randomly, which can be time-consuming to converge to the optimal policy.
**$\epsilon$-greedy policy**: With $1-\epsilon$ probabilty to take the action that gives the best Q value (**exploitation**).
### Temporal Difference Learning
Similarly, from the value iteration algorithm, we can derive the corresponding Temporal Difference Learning (TD Learning):
$$V(x)\leftarrow (1-\alpha)V(x)+\alpha(r+\gamma V(x'))$$
This is simply updating the running average (**incremental learning**).
Please check the following links for related examples:
https://github.com/ageron/handson-ml/blob/master/16_reinforcement_learning.ipynb
https://github.com/microsoft/ML-For-Beginners/blob/main/8-Reinforcement/1-QLearning/solution/notebook.ipynb
### Q Learning with Function Approximation
Instead of estimating Q value based on running average as above, it may be beneficial to have a model or function to **approximate** the Q value so that we may further improve the convergence properties if we can quickly learn a reasonably good function approximation:
$$Q(x, a) \leftarrow \tilde{Q}(x, a; \theta)$$
In the above algorithm, instead of directly estimating $Q$, we can update the model parameter $\theta$ (related to **SGD**):
$$\theta \leftarrow \theta + \alpha_t (r_t +\gamma \max_{a'}\tilde{Q}(x',a'; \theta) - \tilde{Q}(x,a; \theta) )\nabla_\theta \tilde{Q}(x, a; \theta) $$
Clearly, different function approximation can be adopted here as in other machine learning problems. This leads to the recent development of **Deep RL**.
### Policy Gradient
Another branch is to directly approximate the policy and learn it end-to-end:
$$\pi(a|x) \leftarrow \tilde{\pi}(a|x; \theta)$$
We will not discuss here this in detail. One of important challenges to address is to derive good gradient estimate of $\nabla_\theta \tilde{\pi}(a|x; \theta)$. One of famous estimation method is **REINFORCE**. There have been many new methods to improve policy gradient by either better modeling $\tilde{\pi}(a|x; \theta)$ or better gradient estimates.
## 3 Deep Q Learning
<img src="https://pylessons.com/static/images/Reinforcement-learning/03_CartPole-reinforcement-learning/Dueling-Q-architecture.jpg" alt="DQL" size=500>
<center>A Schematic for deep Q learning</center>
<img src="https://developer.ibm.com/developer/default/articles/machine-learning-and-gaming/images/Figure5.png" alt="DQL" size=500>
<center>A Schematic for deep Q learning for policy gradient in Gaming </center>
Note that once the approximating neural network, MLP, CNN, RNN, or GNN based on the state and action space, is specified, the learning is the same as all the other machine learning problems.
Here, also note that we are not restricted ourselves to finite state or action space any more!
There have been also many challenges to theoretically analyze DQN performances and empirically further improve DQN. Two main issues with vanilla DQN is the **bias** problem of overestimating Q values in noisy environment and the **moving target** problem due to using the same DQN to evaluate and choose actions. There have been recent efforts to address these problems, including **Prioritized experience replay** and **dueling/double DQN**, etc.
More tutorials can be found at:
https://github.com/pythonlessons/Reinforcement_Learning
## 4 Hands-on Exercise: Lunar Lander V2
We use an OpenAI Gym's example [LunarLander-v2](https://gym.openai.com/envs/LunarLander-v2/) as the environments for the DQN algorithm. [Shiva Verma's code](https://github.com/shivaverma/OpenAIGym/blob/master/lunar-lander/discrete/lunar_lander.py) is used for the following implementation.
<img src="https://miro.medium.com/max/1346/1*i7lxpgt2K3Q8lgEPJu3_xA.png" alt="DQL" size=500>
<center>A Schematic for LunarLander-v2</center>
LunarLander-v2 describes a qeustion that we need to control a lunar lander to land on a specific landing position. The specific settings for the problem are:
The action space consists of four actions: 0-do nothing; 1-Fire left orientation engine, 2-Fire down main engine, and 3-Fire right orientation engine.
Landing pad is always at coordinates (0,0). Coordinates are the first two numbers in state vector.
Reward for moving from the top of the screen to landing pad and zero speed is about 100..140 points. If lander moves away from landing pad it loses reward back.
Episode finishes if the lander crashes or comes to rest, receiving additional -100 or +100 points. Each leg ground contact is +10. Firing main engine is -0.3 points each frame. Solved is 200 points. Landing outside landing pad is possible. Fuel is infinite, so an agent can learn to fly and then land on its first attempt.
```
# Install the environments
!apt update
!apt install xvfb
!pip install pyvirtualdisplay
!pip install ribs[all] gym~=0.17.0 Box2D~=2.3.10 tqdm
# Start Xvfb and set "DISPLAY" environment properly.
import pyvirtualdisplay
d = pyvirtualdisplay.Display()
d.start()
import gym
import random
import numpy as np
from tensorflow.keras import Sequential
from collections import deque
from tensorflow.keras.layers import Dense
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.activations import relu, linear
import base64
import io
from gym.wrappers import Monitor
from IPython import display
from IPython.display import Image
from pyvirtualdisplay import Display
import matplotlib.pyplot as plt
env = gym.make("LunarLander-v2")
env = Monitor(env, 'videos', force=True)
np.random.seed(0)
d = Display()
d.start()
# Construct DQN model
class DQN:
""" Implementation of deep q learning algorithm """
def __init__(self, action_space, state_space):
self.action_space = action_space
self.state_space = state_space
self.epsilon = 1.0
self.gamma = .99
self.batch_size = 64
self.epsilon_min = .01
self.lr = 0.001
self.epsilon_decay = .996
self.memory = deque(maxlen=1000000)
self.model = self.build_model()
def build_model(self):
model = Sequential()
model.add(Dense(150, input_dim=self.state_space, activation=relu))
model.add(Dense(120, activation=relu))
model.add(Dense(self.action_space, activation=linear))
model.compile(loss='mse', optimizer=Adam(lr=self.lr))
return model
def remember(self, state, action, reward, next_state, done):
self.memory.append((state, action, reward, next_state, done))
def act(self, state):
if np.random.rand() <= self.epsilon:
return random.randrange(self.action_space)
act_values = self.model.predict(state)
return np.argmax(act_values[0])
def replay(self):
if len(self.memory) < self.batch_size:
return
minibatch = random.sample(self.memory, self.batch_size)
states = np.array([i[0] for i in minibatch])
actions = np.array([i[1] for i in minibatch])
rewards = np.array([i[2] for i in minibatch])
next_states = np.array([i[3] for i in minibatch])
dones = np.array([i[4] for i in minibatch])
states = np.squeeze(states)
next_states = np.squeeze(next_states)
targets = rewards + self.gamma*(np.amax(self.model.predict_on_batch(next_states), axis=1))*(1-dones)
targets_full = self.model.predict_on_batch(states)
ind = np.array([i for i in range(self.batch_size)])
targets_full[[ind], [actions]] = targets
self.model.fit(states, targets_full, epochs=1, verbose=0)
if self.epsilon > self.epsilon_min:
self.epsilon *= self.epsilon_decay
# Display the lunar lander actions for each episode
def render(env):
if len(env.videos) > 0:
f = env.videos[-1]
video = io.open(f[0], 'r+b').read()
encoded = base64.b64encode(video)
display.display(display.HTML(data="""
<video alt="test" controls>
<source src="data:video/mp4;base64,{0}" type="video/mp4" />
</video>
""".format(encoded.decode('ascii'))))
# train the DQN model
def train_dqn(episode):
loss = []
agent = DQN(env.action_space.n, env.observation_space.shape[0])
for e in range(episode):
state = env.reset()
state = np.reshape(state, (1, 8))
score = 0
max_steps = 3000
for i in range(max_steps):
action = agent.act(state)
next_state, reward, done, _ = env.step(action)
score += reward
next_state = np.reshape(next_state, (1, 8))
agent.remember(state, action, reward, next_state, done)
state = next_state
agent.replay()
if done:
print("episode: {}/{}, score: {}".format(e, episode, score))
break
loss.append(score)
render(env)
# Average score of last 100 episode
is_solved = np.mean(loss[-100:])
if is_solved > 200:
print('\n Task Completed! \n')
break
print("Average over last 100 episode: {0:.2f} \n".format(is_solved))
return loss
print(env.observation_space)
print(env.action_space)
episodes = 400
loss = train_dqn(episodes)
plt.plot([i+1 for i in range(0, len(loss), 2)], loss[::2])
plt.show()
```
## Reference
*Some materials in this section are adapted from several resources listed below:*
- https://towardsdatascience.com/
- An Introduction to Statistical Learning : with Applications in R by Gareth James, Daniela Witten, Trevor Hastie, Robert Tibshirani. New York: Springer, 2013.
- Open Machine Learning Course mlcourse.ai.
- Reinforcement Learning: An Introduction (textbook): http://incompleteideas.net/book/the-book-2nd.html
# Questions?
```
Image(url= "https://mirrors.creativecommons.org/presskit/buttons/88x31/png/by-nc-sa.png", width=100)
```
|
github_jupyter
|
# Install the environments
!apt update
!apt install xvfb
!pip install pyvirtualdisplay
!pip install ribs[all] gym~=0.17.0 Box2D~=2.3.10 tqdm
# Start Xvfb and set "DISPLAY" environment properly.
import pyvirtualdisplay
d = pyvirtualdisplay.Display()
d.start()
import gym
import random
import numpy as np
from tensorflow.keras import Sequential
from collections import deque
from tensorflow.keras.layers import Dense
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.activations import relu, linear
import base64
import io
from gym.wrappers import Monitor
from IPython import display
from IPython.display import Image
from pyvirtualdisplay import Display
import matplotlib.pyplot as plt
env = gym.make("LunarLander-v2")
env = Monitor(env, 'videos', force=True)
np.random.seed(0)
d = Display()
d.start()
# Construct DQN model
class DQN:
""" Implementation of deep q learning algorithm """
def __init__(self, action_space, state_space):
self.action_space = action_space
self.state_space = state_space
self.epsilon = 1.0
self.gamma = .99
self.batch_size = 64
self.epsilon_min = .01
self.lr = 0.001
self.epsilon_decay = .996
self.memory = deque(maxlen=1000000)
self.model = self.build_model()
def build_model(self):
model = Sequential()
model.add(Dense(150, input_dim=self.state_space, activation=relu))
model.add(Dense(120, activation=relu))
model.add(Dense(self.action_space, activation=linear))
model.compile(loss='mse', optimizer=Adam(lr=self.lr))
return model
def remember(self, state, action, reward, next_state, done):
self.memory.append((state, action, reward, next_state, done))
def act(self, state):
if np.random.rand() <= self.epsilon:
return random.randrange(self.action_space)
act_values = self.model.predict(state)
return np.argmax(act_values[0])
def replay(self):
if len(self.memory) < self.batch_size:
return
minibatch = random.sample(self.memory, self.batch_size)
states = np.array([i[0] for i in minibatch])
actions = np.array([i[1] for i in minibatch])
rewards = np.array([i[2] for i in minibatch])
next_states = np.array([i[3] for i in minibatch])
dones = np.array([i[4] for i in minibatch])
states = np.squeeze(states)
next_states = np.squeeze(next_states)
targets = rewards + self.gamma*(np.amax(self.model.predict_on_batch(next_states), axis=1))*(1-dones)
targets_full = self.model.predict_on_batch(states)
ind = np.array([i for i in range(self.batch_size)])
targets_full[[ind], [actions]] = targets
self.model.fit(states, targets_full, epochs=1, verbose=0)
if self.epsilon > self.epsilon_min:
self.epsilon *= self.epsilon_decay
# Display the lunar lander actions for each episode
def render(env):
if len(env.videos) > 0:
f = env.videos[-1]
video = io.open(f[0], 'r+b').read()
encoded = base64.b64encode(video)
display.display(display.HTML(data="""
<video alt="test" controls>
<source src="data:video/mp4;base64,{0}" type="video/mp4" />
</video>
""".format(encoded.decode('ascii'))))
# train the DQN model
def train_dqn(episode):
loss = []
agent = DQN(env.action_space.n, env.observation_space.shape[0])
for e in range(episode):
state = env.reset()
state = np.reshape(state, (1, 8))
score = 0
max_steps = 3000
for i in range(max_steps):
action = agent.act(state)
next_state, reward, done, _ = env.step(action)
score += reward
next_state = np.reshape(next_state, (1, 8))
agent.remember(state, action, reward, next_state, done)
state = next_state
agent.replay()
if done:
print("episode: {}/{}, score: {}".format(e, episode, score))
break
loss.append(score)
render(env)
# Average score of last 100 episode
is_solved = np.mean(loss[-100:])
if is_solved > 200:
print('\n Task Completed! \n')
break
print("Average over last 100 episode: {0:.2f} \n".format(is_solved))
return loss
print(env.observation_space)
print(env.action_space)
episodes = 400
loss = train_dqn(episodes)
plt.plot([i+1 for i in range(0, len(loss), 2)], loss[::2])
plt.show()
Image(url= "https://mirrors.creativecommons.org/presskit/buttons/88x31/png/by-nc-sa.png", width=100)
| 0.869798 | 0.972701 |
```
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
#Model that will later be evaluated must be first trained, we can't just train all the model at one time and then evaluate it all
#this might lead to model colapse
# Parameters
learning_rate = 0.001
training_epochs = 130
batch_size = 128 # the batch size can not exceed the size of the data.
#load the data matrics
matrix_1st=np.loadtxt('matrix81.txt')
print('1 state scenario matrics has the shape:',matrix_1st.shape)
matrix_2st=np.loadtxt('matrix82.txt')
print('2 state scenario matrics has the shape:',matrix_2st.shape)
matrix_3st=np.loadtxt('matrix83.txt')
print('3 state scenario matrics has the shape:',matrix_3st.shape)
matrix_4st=np.loadtxt('matrix84.txt')
print('4 state scenario matrics has the shape:',matrix_4st.shape)
test1 = np.unique(matrix_1st,axis =0)
print(test1.shape)
test2 = np.unique(matrix_2st,axis =0)
print(test2.shape)
test3 = np.unique(matrix_3st,axis =0)
print(test3.shape)
test4 = np.unique(matrix_4st,axis =0)
print(test4.shape)
from sklearn.preprocessing import MinMaxScaler
scaler1 = MinMaxScaler()
scaler1.fit(test1)
test1 = scaler1.transform(test1)
scaler2 = MinMaxScaler()
scaler2.fit(test2)
test2 = scaler2.transform(test2)
scaler3 = MinMaxScaler()
scaler3.fit(test3)
test3 = scaler3.transform(test3)
scaler4 = MinMaxScaler()
scaler4.fit(test4)
test4 = scaler4.transform(test4)
test1
```
## Evaluation für die test data using straight driving data

## the evaluation criterion
The testing set has both Known and unknown transactions in it. The Autoencoder will learn to identify the pattern of the input data. If an anomalous test point does not match the learned pattern, the autoencoder will likely have a high error rate in reconstructing this data, indicating anomalous data. So that we can identify the anomalies of the data. To calculate the error, it uses Mean Squared Error(MSE)
# 1 state scenario
```
tf.reset_default_graph()
with tf.Session() as sess1:
model_1st = tf.train.import_meta_graph('../Masterarbeit/model_1st/model_1st.meta')
model_1st.restore(sess1, '../Masterarbeit/model_1st/model_1st')
y_pred = tf.get_collection('pred_network')[0]
X = tf.get_collection('AE_input')[0]
encoder_decoder1st = sess1.run(y_pred, feed_dict={X: test1})
error1st = sess1.run(tf.reduce_mean(tf.pow(test1 - encoder_decoder1st,2)))
error_mse_1st = sess1.run(tf.pow(test1 - encoder_decoder1st,2))
summary_writer = tf.summary.FileWriter('./log/', sess1.graph)
```
## Visualization and compare for the 20 random data
```
f, a = plt.subplots(2,2, figsize=(20, 24))
for i in range(2):
a[0][i].matshow(test1[i:i+10])
a[1][i].matshow(encoder_decoder1st[i:i+10])
f.show()
import pandas as pd
plt.figure(figsize=(14, 5))
mse_df = pd.DataFrame(error_mse_1st)
temp = mse_df.mean(1)
plt.scatter(temp.index, temp, alpha = 0.7, marker = '^', c= 'black')
plt.title('Reconstruction MSE of 1-state sceanrios using TestRun2 data')
plt.ylabel('Reconstruction MSE'); plt.xlabel('Sample Index')
plt.ylim(0,0.25)
plt.show()
temp.sort_values().iloc()[189]
print('the average Mean Square Error for 1 state Scenario test data is:',error1st)
```
# 2 state scenario
```
tf.reset_default_graph()
with tf.Session() as sess2:
model_2st = tf.train.import_meta_graph('../Masterarbeit/model_2st/model_2st.meta')
model_2st.restore(sess2, '../Masterarbeit/model_2st/model_2st')
y_pred = tf.get_collection('pred_network')[0]
X = tf.get_collection('AE_input')[0]
encoder_decoder2st = sess2.run(y_pred, feed_dict={X: test2})
error2st = sess2.run(tf.reduce_mean(tf.pow(test2 - encoder_decoder2st,2)))
error_mse_2st = sess2.run((tf.pow(test2 - encoder_decoder2st,2)))
sess2.close()
summary_writer = tf.summary.FileWriter('./log/', sess2.graph)
```
## Visualization and compare for the 20 random data
```
f, a = plt.subplots(2,2, figsize=(14, 10))
for i in range(2):
print(error2st)
a[0][i].matshow(test2[i:i+10])
a[1][i].matshow(encoder_decoder2st[i:i+10])
f.show()
import pandas as pd
plt.figure(figsize=(14, 5))
mse_df = pd.DataFrame(error_mse_2st)
temp = mse_df.mean(1)
plt.scatter(temp.index, temp, alpha = 0.7, marker = '^', c= 'black')
plt.title('Reconstruction MSE of 2-state sceanrios using TestRun2 data')
plt.ylabel('Reconstruction MSE'); plt.xlabel('Index')
plt.ylim(0,0.15)
plt.show()
temp.sort_values().iloc()[679]
print('the average Mean Square Error for 2 state Scenario test data is:',error2st)
```
# 3 state scenario
```
tf.reset_default_graph()
with tf.Session() as sess3:
model_3st = tf.train.import_meta_graph('../Masterarbeit/model_3st/model_3st.meta')
model_3st.restore(sess3, '../Masterarbeit/model_3st/model_3st')
y_pred = tf.get_collection('pred_network')[0]
X = tf.get_collection('AE_input')[0]
encoder_decoder3st = sess3.run(y_pred, feed_dict={X: test3})
error3st = sess3.run(tf.reduce_mean(tf.pow(test3 - encoder_decoder3st,2)))
error_mse_3st = sess3.run(tf.pow(test3 - encoder_decoder3st,3))
summary_writer = tf.summary.FileWriter('./log/', sess3.graph)
```
## Visualization and compare for the 20 random data
```
f, a = plt.subplots(2,2, figsize=(16, 8))
for i in range(2):
a[0][i].matshow(test3[i:i+10])
a[1][i].matshow(encoder_decoder3st[i:i+10])
f.show()
import pandas as pd
plt.figure(figsize=(14, 5))
mse_df = pd.DataFrame(error_mse_3st)
temp = mse_df.mean(1)
plt.scatter(temp.index, temp, alpha = 0.7, marker = '^', c= 'black')
plt.title('Reconstruction MSE of 3-state sceanrios using TestRun2 data')
plt.ylabel('Reconstruction MSE'); plt.xlabel('Index')
plt.ylim(0,0.05)
plt.show()
temp.sort_values().iloc()[955]
print('the average Mean Square Error for 3 state Scenario test data is:',error3st)
```
# 4 state scenario
```
tf.reset_default_graph()
with tf.Session() as sess4:
model_1st = tf.train.import_meta_graph('../Masterarbeit/model_4st/model_4st.meta')
model_1st.restore(sess4, '../Masterarbeit/model_4st/model_4st')
y_pred = tf.get_collection('pred_network')[0]
X = tf.get_collection('AE_input')[0]
encoder_decoder4st = sess4.run(y_pred, feed_dict={X: test4})
error4st = sess4.run(tf.reduce_mean(tf.pow(test4 - encoder_decoder4st,2)))
error_mse_4st = sess4.run(tf.pow(test4 - encoder_decoder4st,4))
```
## Visualization and compare for the 20 random data
```
f, a = plt.subplots(2,2, figsize=(16, 8))
for i in range(2):
a[0][i].matshow(test4[i:i+10])
a[1][i].matshow(encoder_decoder4st[i:i+10])
f.show()
import pandas as pd
plt.figure(figsize=(14, 5))
mse_df = pd.DataFrame(error_mse_4st)
temp = mse_df.mean(1)
plt.scatter(temp.index, temp, alpha = 0.7, marker = '^', c= 'black')
plt.title('Reconstruction MSE of 4-state sceanrios using TestRun2 data')
plt.ylabel('Reconstruction MSE'); plt.xlabel('Index')
plt.ylim(0,0.04)
plt.show()
temp.sort_values().iloc()[1350]
print('the average Mean Square Error for 4 state Scenario test data is:',error4st)
```
|
github_jupyter
|
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
#Model that will later be evaluated must be first trained, we can't just train all the model at one time and then evaluate it all
#this might lead to model colapse
# Parameters
learning_rate = 0.001
training_epochs = 130
batch_size = 128 # the batch size can not exceed the size of the data.
#load the data matrics
matrix_1st=np.loadtxt('matrix81.txt')
print('1 state scenario matrics has the shape:',matrix_1st.shape)
matrix_2st=np.loadtxt('matrix82.txt')
print('2 state scenario matrics has the shape:',matrix_2st.shape)
matrix_3st=np.loadtxt('matrix83.txt')
print('3 state scenario matrics has the shape:',matrix_3st.shape)
matrix_4st=np.loadtxt('matrix84.txt')
print('4 state scenario matrics has the shape:',matrix_4st.shape)
test1 = np.unique(matrix_1st,axis =0)
print(test1.shape)
test2 = np.unique(matrix_2st,axis =0)
print(test2.shape)
test3 = np.unique(matrix_3st,axis =0)
print(test3.shape)
test4 = np.unique(matrix_4st,axis =0)
print(test4.shape)
from sklearn.preprocessing import MinMaxScaler
scaler1 = MinMaxScaler()
scaler1.fit(test1)
test1 = scaler1.transform(test1)
scaler2 = MinMaxScaler()
scaler2.fit(test2)
test2 = scaler2.transform(test2)
scaler3 = MinMaxScaler()
scaler3.fit(test3)
test3 = scaler3.transform(test3)
scaler4 = MinMaxScaler()
scaler4.fit(test4)
test4 = scaler4.transform(test4)
test1
tf.reset_default_graph()
with tf.Session() as sess1:
model_1st = tf.train.import_meta_graph('../Masterarbeit/model_1st/model_1st.meta')
model_1st.restore(sess1, '../Masterarbeit/model_1st/model_1st')
y_pred = tf.get_collection('pred_network')[0]
X = tf.get_collection('AE_input')[0]
encoder_decoder1st = sess1.run(y_pred, feed_dict={X: test1})
error1st = sess1.run(tf.reduce_mean(tf.pow(test1 - encoder_decoder1st,2)))
error_mse_1st = sess1.run(tf.pow(test1 - encoder_decoder1st,2))
summary_writer = tf.summary.FileWriter('./log/', sess1.graph)
f, a = plt.subplots(2,2, figsize=(20, 24))
for i in range(2):
a[0][i].matshow(test1[i:i+10])
a[1][i].matshow(encoder_decoder1st[i:i+10])
f.show()
import pandas as pd
plt.figure(figsize=(14, 5))
mse_df = pd.DataFrame(error_mse_1st)
temp = mse_df.mean(1)
plt.scatter(temp.index, temp, alpha = 0.7, marker = '^', c= 'black')
plt.title('Reconstruction MSE of 1-state sceanrios using TestRun2 data')
plt.ylabel('Reconstruction MSE'); plt.xlabel('Sample Index')
plt.ylim(0,0.25)
plt.show()
temp.sort_values().iloc()[189]
print('the average Mean Square Error for 1 state Scenario test data is:',error1st)
tf.reset_default_graph()
with tf.Session() as sess2:
model_2st = tf.train.import_meta_graph('../Masterarbeit/model_2st/model_2st.meta')
model_2st.restore(sess2, '../Masterarbeit/model_2st/model_2st')
y_pred = tf.get_collection('pred_network')[0]
X = tf.get_collection('AE_input')[0]
encoder_decoder2st = sess2.run(y_pred, feed_dict={X: test2})
error2st = sess2.run(tf.reduce_mean(tf.pow(test2 - encoder_decoder2st,2)))
error_mse_2st = sess2.run((tf.pow(test2 - encoder_decoder2st,2)))
sess2.close()
summary_writer = tf.summary.FileWriter('./log/', sess2.graph)
f, a = plt.subplots(2,2, figsize=(14, 10))
for i in range(2):
print(error2st)
a[0][i].matshow(test2[i:i+10])
a[1][i].matshow(encoder_decoder2st[i:i+10])
f.show()
import pandas as pd
plt.figure(figsize=(14, 5))
mse_df = pd.DataFrame(error_mse_2st)
temp = mse_df.mean(1)
plt.scatter(temp.index, temp, alpha = 0.7, marker = '^', c= 'black')
plt.title('Reconstruction MSE of 2-state sceanrios using TestRun2 data')
plt.ylabel('Reconstruction MSE'); plt.xlabel('Index')
plt.ylim(0,0.15)
plt.show()
temp.sort_values().iloc()[679]
print('the average Mean Square Error for 2 state Scenario test data is:',error2st)
tf.reset_default_graph()
with tf.Session() as sess3:
model_3st = tf.train.import_meta_graph('../Masterarbeit/model_3st/model_3st.meta')
model_3st.restore(sess3, '../Masterarbeit/model_3st/model_3st')
y_pred = tf.get_collection('pred_network')[0]
X = tf.get_collection('AE_input')[0]
encoder_decoder3st = sess3.run(y_pred, feed_dict={X: test3})
error3st = sess3.run(tf.reduce_mean(tf.pow(test3 - encoder_decoder3st,2)))
error_mse_3st = sess3.run(tf.pow(test3 - encoder_decoder3st,3))
summary_writer = tf.summary.FileWriter('./log/', sess3.graph)
f, a = plt.subplots(2,2, figsize=(16, 8))
for i in range(2):
a[0][i].matshow(test3[i:i+10])
a[1][i].matshow(encoder_decoder3st[i:i+10])
f.show()
import pandas as pd
plt.figure(figsize=(14, 5))
mse_df = pd.DataFrame(error_mse_3st)
temp = mse_df.mean(1)
plt.scatter(temp.index, temp, alpha = 0.7, marker = '^', c= 'black')
plt.title('Reconstruction MSE of 3-state sceanrios using TestRun2 data')
plt.ylabel('Reconstruction MSE'); plt.xlabel('Index')
plt.ylim(0,0.05)
plt.show()
temp.sort_values().iloc()[955]
print('the average Mean Square Error for 3 state Scenario test data is:',error3st)
tf.reset_default_graph()
with tf.Session() as sess4:
model_1st = tf.train.import_meta_graph('../Masterarbeit/model_4st/model_4st.meta')
model_1st.restore(sess4, '../Masterarbeit/model_4st/model_4st')
y_pred = tf.get_collection('pred_network')[0]
X = tf.get_collection('AE_input')[0]
encoder_decoder4st = sess4.run(y_pred, feed_dict={X: test4})
error4st = sess4.run(tf.reduce_mean(tf.pow(test4 - encoder_decoder4st,2)))
error_mse_4st = sess4.run(tf.pow(test4 - encoder_decoder4st,4))
f, a = plt.subplots(2,2, figsize=(16, 8))
for i in range(2):
a[0][i].matshow(test4[i:i+10])
a[1][i].matshow(encoder_decoder4st[i:i+10])
f.show()
import pandas as pd
plt.figure(figsize=(14, 5))
mse_df = pd.DataFrame(error_mse_4st)
temp = mse_df.mean(1)
plt.scatter(temp.index, temp, alpha = 0.7, marker = '^', c= 'black')
plt.title('Reconstruction MSE of 4-state sceanrios using TestRun2 data')
plt.ylabel('Reconstruction MSE'); plt.xlabel('Index')
plt.ylim(0,0.04)
plt.show()
temp.sort_values().iloc()[1350]
print('the average Mean Square Error for 4 state Scenario test data is:',error4st)
| 0.450601 | 0.913792 |
```
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
import os.path as op
import pickle
A1=np.empty((0,5),dtype='float32')
U1=np.empty((0,7),dtype='float32')
node=['150','149','147','144','142','140','136','61']
mon=['Apr','Mar','Aug','Jun','Jul','Sep','May','Oct']
for j in node:
for i in mon:
inp= pd.read_csv('data_gkv/AT510_Node_'+str(j)+'_'+str(i)+'19_OutputFile.csv',usecols=[1,2,3,15,16],low_memory=False)
out= pd.read_csv('data_gkv/AT510_Node_'+str(j)+'_'+str(i)+'19_OutputFile.csv',usecols=[5,6,7,8,17,18,19],low_memory=False)
inp=np.array(inp,dtype='float32')
out=np.array(out,dtype='float32')
A1=np.append(A1, inp, axis=0)
U1=np.append(U1, out, axis=0)
print(A1)
print(U1)
from sklearn.preprocessing import MinMaxScaler
import warnings
scaler_obj=MinMaxScaler()
X1=scaler_obj.fit_transform(A1)
Y1=scaler_obj.fit_transform(U1)
warnings.filterwarnings(action='ignore', category=UserWarning)
from sklearn.model_selection import train_test_split
x_train,x_test,y_train,y_test=train_test_split(X1,Y1,test_size=0.25,random_state=0)
import pickle
loaded_model_fit7 = pickle.load(open("Models_File/lasso.sav", 'rb'))
y_test_pred=loaded_model_fit7.predict(x_test)
y_test_pred
from numpy import savetxt
savetxt('ARRAY_DATA/lasso_y_test_pred.csv', y_test_pred[:1001], delimiter=',')
from numpy import loadtxt
data = loadtxt('ARRAY_DATA/linearregression_y_test.csv', delimiter=',')
data
import tensorflow as tf
from tensorflow import keras
from keras.models import Model,Sequential,load_model
from keras.layers import Input, Embedding
from keras.layers import Dense, Bidirectional
from keras.layers.recurrent import LSTM
import keras.metrics as metrics
import itertools
from tensorflow.python.keras.utils.data_utils import Sequence
from decimal import Decimal
from keras import backend as K
from keras.layers import Conv1D,MaxPooling1D,Flatten,Dense
def rmse(y_true, y_pred):
return K.sqrt(K.mean(K.square(y_pred - y_true), axis=-1))
def coeff_determination(y_true, y_pred):
SS_res = K.sum(K.square( y_true-y_pred ))
SS_tot = K.sum(K.square( y_true - K.mean(y_true) ) )
return ( 1 - SS_res/(SS_tot + K.epsilon()) )
from sklearn.preprocessing import MinMaxScaler
import warnings
scaler_obj=MinMaxScaler()
X1=scaler_obj.fit_transform(A1)
Y1=scaler_obj.fit_transform(U1)
warnings.filterwarnings(action='ignore', category=UserWarning)
X1=X1[:,np.newaxis,:]
Y1=Y1[:,np.newaxis,:]
from sklearn.model_selection import train_test_split
x_train,x_test,y_train,y_test=train_test_split(X1,Y1,test_size=0.25,random_state=0)
from keras.models import model_from_json
json_file = open('lstm.json', 'r')
loaded_model_json = json_file.read()
json_file.close()
loaded_model = model_from_json(loaded_model_json)
# load weights into new model
loaded_model.load_weights("lstm.h5")
print("Loaded model from disk")
loaded_model.compile(optimizer=keras.optimizers.Adam(learning_rate=0.001), loss='mse',metrics=['accuracy','mse','mae',rmse,coeff_determination])
print(loaded_model.evaluate(x_train, y_train, verbose=0))
y_test_pred=loaded_model.predict(x_test)
y_test_pred
y_test
y_test=y_test[:,0]
from numpy import savetxt
savetxt('ARRAY_DATA/lstm_y_test.csv', y_test[:1001], delimiter=',')
t=['Y_Actual']*10
t
l1=list()
l1.append(['Y_Actual']*10)
l1.append(np.round(y_test[:10,0],9))
temp1=np.array(l1).transpose()
x1=list(range(1,11))
chart_data1 = pd.DataFrame(temp1, x1,columns=['Data','y'])
l2=list()
l2.append(['Y_Predicted']*10)
l2.append(np.round(y_test_pred[:10,0],9))
temp2=np.array(l2).transpose()
x2=list(range(11,21))
chart_data2 = pd.DataFrame(temp2, x2,columns=['Data','y'])
frames=[chart_data1,chart_data2]
results=pd.concat(frames)
results['x']=np.tile(np.arange(1,11,1),2)
results
source = results.reset_index().melt('x', var_name='category', value_name='y')
# Create a selection that chooses the nearest point & selects based on x-value
nearest = alt.selection(type='single', nearest=True, on='mouseover',
fields=['x'], empty='none')
# The basic line
line = alt.Chart(source).mark_line(interpolate='basis').encode(
x='x',
y='y',
color='category:N'
)
# Transparent selectors across the chart. This is what tells us
# the x-value of the cursor
selectors = alt.Chart(source).mark_point().encode(
x='x',
opacity=alt.value(0),
).add_selection(
nearest
)
# Draw points on the line, and highlight based on selection
points = line.mark_point().encode(
opacity=alt.condition(nearest, alt.value(1), alt.value(0))
)
# Draw text labels near the points, and highlight based on selection
text = line.mark_text(align='left', dx=5, dy=-5).encode(
text=alt.condition(nearest, 'y', alt.value(' '))
)
# Draw a rule at the location of the selection
rules = alt.Chart(source).mark_rule(color='gray').encode(
x='x',
).transform_filter(
nearest
)
# Put the five layers into a chart and bind the data
alt.layer(
line, selectors, points, rules, text
).properties(
width=600, height=300
)
np.arange(11,21,1)
import altair as alt
from vega_datasets import data
nearest = alt.selection(type='single', nearest=True, on='mouseover',fields=[''], empty='none')
chart=alt.Chart(results).mark_line().encode(
x='y',
y='x',
color='Data',
strokeDash='Data',
).add_selection(
nearest
)
chart
from vega_datasets import data
data.stocks().head(100)
import pandas as pd
xgboost_list = [0.9310061554157045, 0.9315455074103038]
models=list()
models.append(["Xgboost",2,"Test"])
models.append(["Xgboost",3,"Train"])
models.append(["KNN",1,"Test"])
models.append(["KNN",2,"Train"])
x=list(range(1,5))
chart_data = pd.DataFrame(models, x,columns=['Models','R2 Score','Data Type'])
chart_data
chart=alt.Chart(chart_data).mark_bar().encode(
x='Data Type:O',
y='R2 Score:Q',
color='Data Type:N',
column='Models:N'
)
chart.configure_view(
continuousHeight=400,
continuousWidth=800,
)
chart
import altair as alt
from vega_datasets import data
source = data.barley()
alt.Chart(source).mark_bar().encode(
x='year:O',
y='sum(yield):Q',
color='year:N',
column='site:N'
)
source
```
|
github_jupyter
|
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
import os.path as op
import pickle
A1=np.empty((0,5),dtype='float32')
U1=np.empty((0,7),dtype='float32')
node=['150','149','147','144','142','140','136','61']
mon=['Apr','Mar','Aug','Jun','Jul','Sep','May','Oct']
for j in node:
for i in mon:
inp= pd.read_csv('data_gkv/AT510_Node_'+str(j)+'_'+str(i)+'19_OutputFile.csv',usecols=[1,2,3,15,16],low_memory=False)
out= pd.read_csv('data_gkv/AT510_Node_'+str(j)+'_'+str(i)+'19_OutputFile.csv',usecols=[5,6,7,8,17,18,19],low_memory=False)
inp=np.array(inp,dtype='float32')
out=np.array(out,dtype='float32')
A1=np.append(A1, inp, axis=0)
U1=np.append(U1, out, axis=0)
print(A1)
print(U1)
from sklearn.preprocessing import MinMaxScaler
import warnings
scaler_obj=MinMaxScaler()
X1=scaler_obj.fit_transform(A1)
Y1=scaler_obj.fit_transform(U1)
warnings.filterwarnings(action='ignore', category=UserWarning)
from sklearn.model_selection import train_test_split
x_train,x_test,y_train,y_test=train_test_split(X1,Y1,test_size=0.25,random_state=0)
import pickle
loaded_model_fit7 = pickle.load(open("Models_File/lasso.sav", 'rb'))
y_test_pred=loaded_model_fit7.predict(x_test)
y_test_pred
from numpy import savetxt
savetxt('ARRAY_DATA/lasso_y_test_pred.csv', y_test_pred[:1001], delimiter=',')
from numpy import loadtxt
data = loadtxt('ARRAY_DATA/linearregression_y_test.csv', delimiter=',')
data
import tensorflow as tf
from tensorflow import keras
from keras.models import Model,Sequential,load_model
from keras.layers import Input, Embedding
from keras.layers import Dense, Bidirectional
from keras.layers.recurrent import LSTM
import keras.metrics as metrics
import itertools
from tensorflow.python.keras.utils.data_utils import Sequence
from decimal import Decimal
from keras import backend as K
from keras.layers import Conv1D,MaxPooling1D,Flatten,Dense
def rmse(y_true, y_pred):
return K.sqrt(K.mean(K.square(y_pred - y_true), axis=-1))
def coeff_determination(y_true, y_pred):
SS_res = K.sum(K.square( y_true-y_pred ))
SS_tot = K.sum(K.square( y_true - K.mean(y_true) ) )
return ( 1 - SS_res/(SS_tot + K.epsilon()) )
from sklearn.preprocessing import MinMaxScaler
import warnings
scaler_obj=MinMaxScaler()
X1=scaler_obj.fit_transform(A1)
Y1=scaler_obj.fit_transform(U1)
warnings.filterwarnings(action='ignore', category=UserWarning)
X1=X1[:,np.newaxis,:]
Y1=Y1[:,np.newaxis,:]
from sklearn.model_selection import train_test_split
x_train,x_test,y_train,y_test=train_test_split(X1,Y1,test_size=0.25,random_state=0)
from keras.models import model_from_json
json_file = open('lstm.json', 'r')
loaded_model_json = json_file.read()
json_file.close()
loaded_model = model_from_json(loaded_model_json)
# load weights into new model
loaded_model.load_weights("lstm.h5")
print("Loaded model from disk")
loaded_model.compile(optimizer=keras.optimizers.Adam(learning_rate=0.001), loss='mse',metrics=['accuracy','mse','mae',rmse,coeff_determination])
print(loaded_model.evaluate(x_train, y_train, verbose=0))
y_test_pred=loaded_model.predict(x_test)
y_test_pred
y_test
y_test=y_test[:,0]
from numpy import savetxt
savetxt('ARRAY_DATA/lstm_y_test.csv', y_test[:1001], delimiter=',')
t=['Y_Actual']*10
t
l1=list()
l1.append(['Y_Actual']*10)
l1.append(np.round(y_test[:10,0],9))
temp1=np.array(l1).transpose()
x1=list(range(1,11))
chart_data1 = pd.DataFrame(temp1, x1,columns=['Data','y'])
l2=list()
l2.append(['Y_Predicted']*10)
l2.append(np.round(y_test_pred[:10,0],9))
temp2=np.array(l2).transpose()
x2=list(range(11,21))
chart_data2 = pd.DataFrame(temp2, x2,columns=['Data','y'])
frames=[chart_data1,chart_data2]
results=pd.concat(frames)
results['x']=np.tile(np.arange(1,11,1),2)
results
source = results.reset_index().melt('x', var_name='category', value_name='y')
# Create a selection that chooses the nearest point & selects based on x-value
nearest = alt.selection(type='single', nearest=True, on='mouseover',
fields=['x'], empty='none')
# The basic line
line = alt.Chart(source).mark_line(interpolate='basis').encode(
x='x',
y='y',
color='category:N'
)
# Transparent selectors across the chart. This is what tells us
# the x-value of the cursor
selectors = alt.Chart(source).mark_point().encode(
x='x',
opacity=alt.value(0),
).add_selection(
nearest
)
# Draw points on the line, and highlight based on selection
points = line.mark_point().encode(
opacity=alt.condition(nearest, alt.value(1), alt.value(0))
)
# Draw text labels near the points, and highlight based on selection
text = line.mark_text(align='left', dx=5, dy=-5).encode(
text=alt.condition(nearest, 'y', alt.value(' '))
)
# Draw a rule at the location of the selection
rules = alt.Chart(source).mark_rule(color='gray').encode(
x='x',
).transform_filter(
nearest
)
# Put the five layers into a chart and bind the data
alt.layer(
line, selectors, points, rules, text
).properties(
width=600, height=300
)
np.arange(11,21,1)
import altair as alt
from vega_datasets import data
nearest = alt.selection(type='single', nearest=True, on='mouseover',fields=[''], empty='none')
chart=alt.Chart(results).mark_line().encode(
x='y',
y='x',
color='Data',
strokeDash='Data',
).add_selection(
nearest
)
chart
from vega_datasets import data
data.stocks().head(100)
import pandas as pd
xgboost_list = [0.9310061554157045, 0.9315455074103038]
models=list()
models.append(["Xgboost",2,"Test"])
models.append(["Xgboost",3,"Train"])
models.append(["KNN",1,"Test"])
models.append(["KNN",2,"Train"])
x=list(range(1,5))
chart_data = pd.DataFrame(models, x,columns=['Models','R2 Score','Data Type'])
chart_data
chart=alt.Chart(chart_data).mark_bar().encode(
x='Data Type:O',
y='R2 Score:Q',
color='Data Type:N',
column='Models:N'
)
chart.configure_view(
continuousHeight=400,
continuousWidth=800,
)
chart
import altair as alt
from vega_datasets import data
source = data.barley()
alt.Chart(source).mark_bar().encode(
x='year:O',
y='sum(yield):Q',
color='year:N',
column='site:N'
)
source
| 0.510252 | 0.29324 |
# RadarCOVID-Report
## Data Extraction
```
import datetime
import json
import logging
import os
import shutil
import tempfile
import textwrap
import uuid
import matplotlib.pyplot as plt
import matplotlib.ticker
import numpy as np
import pandas as pd
import retry
import seaborn as sns
%matplotlib inline
current_working_directory = os.environ.get("PWD")
if current_working_directory:
os.chdir(current_working_directory)
sns.set()
matplotlib.rcParams["figure.figsize"] = (15, 6)
extraction_datetime = datetime.datetime.utcnow()
extraction_date = extraction_datetime.strftime("%Y-%m-%d")
extraction_previous_datetime = extraction_datetime - datetime.timedelta(days=1)
extraction_previous_date = extraction_previous_datetime.strftime("%Y-%m-%d")
extraction_date_with_hour = datetime.datetime.utcnow().strftime("%Y-%m-%d@%H")
current_hour = datetime.datetime.utcnow().hour
are_today_results_partial = current_hour != 23
```
### Constants
```
from Modules.ExposureNotification import exposure_notification_io
spain_region_country_code = "ES"
germany_region_country_code = "DE"
default_backend_identifier = spain_region_country_code
backend_generation_days = 7 * 2
daily_summary_days = 7 * 4 * 3
daily_plot_days = 7 * 4
tek_dumps_load_limit = daily_summary_days + 1
default_shared_diagnosis_generation_to_upload_days = 1
```
### Parameters
```
environment_backend_identifier = os.environ.get("RADARCOVID_REPORT__BACKEND_IDENTIFIER")
if environment_backend_identifier:
report_backend_identifier = environment_backend_identifier
else:
report_backend_identifier = default_backend_identifier
report_backend_identifier
efgs_supported_countries_backend_identifier = germany_region_country_code
efgs_supported_countries_backend_client = \
exposure_notification_io.get_backend_client_with_identifier(
backend_identifier=efgs_supported_countries_backend_identifier)
efgs_source_regions = efgs_supported_countries_backend_client.get_supported_countries()
environment_source_regions = os.environ.get("RADARCOVID_REPORT__EFGS_EXTRA_SOURCE_REGIONS")
if environment_source_regions:
efgs_source_regions = list(set(efgs_source_regions).union(environment_source_regions.split(",")))
efgs_source_regions = list(map(lambda x: x.upper(), efgs_source_regions))
if report_backend_identifier in efgs_source_regions:
default_source_regions = "EFGS"
else:
default_source_regions = report_backend_identifier.split("-")[0].split("@")[0]
environment_source_regions = os.environ.get("RADARCOVID_REPORT__SOURCE_REGIONS")
if environment_source_regions:
report_source_regions = environment_source_regions
else:
report_source_regions = default_source_regions
if report_source_regions == "EFGS":
if report_backend_identifier in efgs_source_regions:
efgs_source_regions = \
[report_backend_identifier] + \
sorted(list(set(efgs_source_regions).difference([report_backend_identifier])))
report_source_regions = efgs_source_regions
else:
report_source_regions = report_source_regions.split(",")
report_source_regions
environment_download_only_from_report_backend = \
os.environ.get("RADARCOVID_REPORT__DOWNLOAD_ONLY_FROM_REPORT_BACKEND")
if environment_download_only_from_report_backend:
report_backend_identifiers = [report_backend_identifier]
else:
report_backend_identifiers = None
report_backend_identifiers
environment_shared_diagnosis_generation_to_upload_days = \
os.environ.get("RADARCOVID_REPORT__SHARED_DIAGNOSIS_GENERATION_TO_UPLOAD_DAYS")
if environment_shared_diagnosis_generation_to_upload_days:
shared_diagnosis_generation_to_upload_days = \
int(environment_shared_diagnosis_generation_to_upload_days)
else:
shared_diagnosis_generation_to_upload_days = \
default_shared_diagnosis_generation_to_upload_days
shared_diagnosis_generation_to_upload_days
```
### COVID-19 Cases
```
@retry.retry(tries=10, delay=10, backoff=1.1, jitter=(0, 10))
def download_cases_dataframe_from_ecdc():
return pd.read_csv(
"https://opendata.ecdc.europa.eu/covid19/casedistribution/csv/data.csv")
confirmed_df = download_cases_dataframe_from_ecdc()
radar_covid_countries = set(report_source_regions)
confirmed_df = confirmed_df[["dateRep", "cases", "geoId"]]
confirmed_df.rename(
columns={
"dateRep":"sample_date",
"cases": "new_cases",
"geoId": "country_code",
},
inplace=True)
confirmed_df = confirmed_df[confirmed_df.country_code.isin(radar_covid_countries)]
confirmed_df["sample_date"] = pd.to_datetime(confirmed_df.sample_date, dayfirst=True)
confirmed_df = confirmed_df.groupby("sample_date").new_cases.sum().reset_index()
confirmed_df.sort_values("sample_date", inplace=True)
confirmed_df.tail()
confirmed_days = pd.date_range(
start=confirmed_df.iloc[0].sample_date,
end=extraction_datetime)
confirmed_days_df = pd.DataFrame(data=confirmed_days, columns=["sample_date"])
confirmed_df = confirmed_days_df.merge(confirmed_df, how="left")
confirmed_df["sample_date"] = confirmed_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_df.sort_values("sample_date", inplace=True)
confirmed_df.tail()
confirmed_df.columns = ["sample_date_string", "new_cases"]
confirmed_df.sort_values("sample_date_string", inplace=True)
confirmed_df["covid_cases"] = confirmed_df.new_cases.rolling(7).mean().round()
confirmed_df.fillna(method="ffill", inplace=True)
confirmed_df.tail()
confirmed_df[["new_cases", "covid_cases"]].plot()
```
### Extract API TEKs
```
raw_zip_path_prefix = "Data/TEKs/Raw/"
fail_on_error_backend_identifiers = [report_backend_identifier]
multi_backend_exposure_keys_df = \
exposure_notification_io.download_exposure_keys_from_backends(
backend_identifiers=report_backend_identifiers,
generation_days=backend_generation_days,
fail_on_error_backend_identifiers=fail_on_error_backend_identifiers,
save_raw_zip_path_prefix=raw_zip_path_prefix)
multi_backend_exposure_keys_df["region"] = multi_backend_exposure_keys_df["backend_identifier"]
multi_backend_exposure_keys_df.rename(
columns={
"generation_datetime": "sample_datetime",
"generation_date_string": "sample_date_string",
},
inplace=True)
multi_backend_exposure_keys_df.head()
early_teks_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.rolling_period < 144].copy()
early_teks_df["rolling_period_in_hours"] = early_teks_df.rolling_period / 6
early_teks_df[early_teks_df.sample_date_string != extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
early_teks_df[early_teks_df.sample_date_string == extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
multi_backend_exposure_keys_df = multi_backend_exposure_keys_df[[
"sample_date_string", "region", "key_data"]]
multi_backend_exposure_keys_df.head()
active_regions = \
multi_backend_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
active_regions
multi_backend_summary_df = multi_backend_exposure_keys_df.groupby(
["sample_date_string", "region"]).key_data.nunique().reset_index() \
.pivot(index="sample_date_string", columns="region") \
.sort_index(ascending=False)
multi_backend_summary_df.rename(
columns={"key_data": "shared_teks_by_generation_date"},
inplace=True)
multi_backend_summary_df.rename_axis("sample_date", inplace=True)
multi_backend_summary_df = multi_backend_summary_df.fillna(0).astype(int)
multi_backend_summary_df = multi_backend_summary_df.head(backend_generation_days)
multi_backend_summary_df.head()
def compute_keys_cross_sharing(x):
teks_x = x.key_data_x.item()
common_teks = set(teks_x).intersection(x.key_data_y.item())
common_teks_fraction = len(common_teks) / len(teks_x)
return pd.Series(dict(
common_teks=common_teks,
common_teks_fraction=common_teks_fraction,
))
multi_backend_exposure_keys_by_region_df = \
multi_backend_exposure_keys_df.groupby("region").key_data.unique().reset_index()
multi_backend_exposure_keys_by_region_df["_merge"] = True
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_df.merge(
multi_backend_exposure_keys_by_region_df, on="_merge")
multi_backend_exposure_keys_by_region_combination_df.drop(
columns=["_merge"], inplace=True)
if multi_backend_exposure_keys_by_region_combination_df.region_x.nunique() > 1:
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_combination_df[
multi_backend_exposure_keys_by_region_combination_df.region_x !=
multi_backend_exposure_keys_by_region_combination_df.region_y]
multi_backend_exposure_keys_cross_sharing_df = \
multi_backend_exposure_keys_by_region_combination_df \
.groupby(["region_x", "region_y"]) \
.apply(compute_keys_cross_sharing) \
.reset_index()
multi_backend_cross_sharing_summary_df = \
multi_backend_exposure_keys_cross_sharing_df.pivot_table(
values=["common_teks_fraction"],
columns="region_x",
index="region_y",
aggfunc=lambda x: x.item())
multi_backend_cross_sharing_summary_df
multi_backend_without_active_region_exposure_keys_df = \
multi_backend_exposure_keys_df[multi_backend_exposure_keys_df.region != report_backend_identifier]
multi_backend_without_active_region = \
multi_backend_without_active_region_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
multi_backend_without_active_region
exposure_keys_summary_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.region == report_backend_identifier]
exposure_keys_summary_df.drop(columns=["region"], inplace=True)
exposure_keys_summary_df = \
exposure_keys_summary_df.groupby(["sample_date_string"]).key_data.nunique().to_frame()
exposure_keys_summary_df = \
exposure_keys_summary_df.reset_index().set_index("sample_date_string")
exposure_keys_summary_df.sort_index(ascending=False, inplace=True)
exposure_keys_summary_df.rename(columns={"key_data": "shared_teks_by_generation_date"}, inplace=True)
exposure_keys_summary_df.head()
```
### Dump API TEKs
```
tek_list_df = multi_backend_exposure_keys_df[
["sample_date_string", "region", "key_data"]].copy()
tek_list_df["key_data"] = tek_list_df["key_data"].apply(str)
tek_list_df.rename(columns={
"sample_date_string": "sample_date",
"key_data": "tek_list"}, inplace=True)
tek_list_df = tek_list_df.groupby(
["sample_date", "region"]).tek_list.unique().reset_index()
tek_list_df["extraction_date"] = extraction_date
tek_list_df["extraction_date_with_hour"] = extraction_date_with_hour
tek_list_path_prefix = "Data/TEKs/"
tek_list_current_path = tek_list_path_prefix + f"/Current/RadarCOVID-TEKs.json"
tek_list_daily_path = tek_list_path_prefix + f"Daily/RadarCOVID-TEKs-{extraction_date}.json"
tek_list_hourly_path = tek_list_path_prefix + f"Hourly/RadarCOVID-TEKs-{extraction_date_with_hour}.json"
for path in [tek_list_current_path, tek_list_daily_path, tek_list_hourly_path]:
os.makedirs(os.path.dirname(path), exist_ok=True)
tek_list_df.drop(columns=["extraction_date", "extraction_date_with_hour"]).to_json(
tek_list_current_path,
lines=True, orient="records")
tek_list_df.drop(columns=["extraction_date_with_hour"]).to_json(
tek_list_daily_path,
lines=True, orient="records")
tek_list_df.to_json(
tek_list_hourly_path,
lines=True, orient="records")
tek_list_df.head()
```
### Load TEK Dumps
```
import glob
def load_extracted_teks(mode, region=None, limit=None) -> pd.DataFrame:
extracted_teks_df = pd.DataFrame(columns=["region"])
file_paths = list(reversed(sorted(glob.glob(tek_list_path_prefix + mode + "/RadarCOVID-TEKs-*.json"))))
if limit:
file_paths = file_paths[:limit]
for file_path in file_paths:
logging.info(f"Loading TEKs from '{file_path}'...")
iteration_extracted_teks_df = pd.read_json(file_path, lines=True)
extracted_teks_df = extracted_teks_df.append(
iteration_extracted_teks_df, sort=False)
extracted_teks_df["region"] = \
extracted_teks_df.region.fillna(spain_region_country_code).copy()
if region:
extracted_teks_df = \
extracted_teks_df[extracted_teks_df.region == region]
return extracted_teks_df
daily_extracted_teks_df = load_extracted_teks(
mode="Daily",
region=report_backend_identifier,
limit=tek_dumps_load_limit)
daily_extracted_teks_df.head()
exposure_keys_summary_df_ = daily_extracted_teks_df \
.sort_values("extraction_date", ascending=False) \
.groupby("sample_date").tek_list.first() \
.to_frame()
exposure_keys_summary_df_.index.name = "sample_date_string"
exposure_keys_summary_df_["tek_list"] = \
exposure_keys_summary_df_.tek_list.apply(len)
exposure_keys_summary_df_ = exposure_keys_summary_df_ \
.rename(columns={"tek_list": "shared_teks_by_generation_date"}) \
.sort_index(ascending=False)
exposure_keys_summary_df = exposure_keys_summary_df_
exposure_keys_summary_df.head()
```
### Daily New TEKs
```
tek_list_df = daily_extracted_teks_df.groupby("extraction_date").tek_list.apply(
lambda x: set(sum(x, []))).reset_index()
tek_list_df = tek_list_df.set_index("extraction_date").sort_index(ascending=True)
tek_list_df.head()
def compute_teks_by_generation_and_upload_date(date):
day_new_teks_set_df = tek_list_df.copy().diff()
try:
day_new_teks_set = day_new_teks_set_df[
day_new_teks_set_df.index == date].tek_list.item()
except ValueError:
day_new_teks_set = None
if pd.isna(day_new_teks_set):
day_new_teks_set = set()
day_new_teks_df = daily_extracted_teks_df[
daily_extracted_teks_df.extraction_date == date].copy()
day_new_teks_df["shared_teks"] = \
day_new_teks_df.tek_list.apply(lambda x: set(x).intersection(day_new_teks_set))
day_new_teks_df["shared_teks"] = \
day_new_teks_df.shared_teks.apply(len)
day_new_teks_df["upload_date"] = date
day_new_teks_df.rename(columns={"sample_date": "generation_date"}, inplace=True)
day_new_teks_df = day_new_teks_df[
["upload_date", "generation_date", "shared_teks"]]
day_new_teks_df["generation_to_upload_days"] = \
(pd.to_datetime(day_new_teks_df.upload_date) -
pd.to_datetime(day_new_teks_df.generation_date)).dt.days
day_new_teks_df = day_new_teks_df[day_new_teks_df.shared_teks > 0]
return day_new_teks_df
shared_teks_generation_to_upload_df = pd.DataFrame()
for upload_date in daily_extracted_teks_df.extraction_date.unique():
shared_teks_generation_to_upload_df = \
shared_teks_generation_to_upload_df.append(
compute_teks_by_generation_and_upload_date(date=upload_date))
shared_teks_generation_to_upload_df \
.sort_values(["upload_date", "generation_date"], ascending=False, inplace=True)
shared_teks_generation_to_upload_df.tail()
today_new_teks_df = \
shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.upload_date == extraction_date].copy()
today_new_teks_df.tail()
if not today_new_teks_df.empty:
today_new_teks_df.set_index("generation_to_upload_days") \
.sort_index().shared_teks.plot.bar()
generation_to_upload_period_pivot_df = \
shared_teks_generation_to_upload_df[
["upload_date", "generation_to_upload_days", "shared_teks"]] \
.pivot(index="upload_date", columns="generation_to_upload_days") \
.sort_index(ascending=False).fillna(0).astype(int) \
.droplevel(level=0, axis=1)
generation_to_upload_period_pivot_df.head()
new_tek_df = tek_list_df.diff().tek_list.apply(
lambda x: len(x) if not pd.isna(x) else None).to_frame().reset_index()
new_tek_df.rename(columns={
"tek_list": "shared_teks_by_upload_date",
"extraction_date": "sample_date_string",}, inplace=True)
new_tek_df.tail()
shared_teks_uploaded_on_generation_date_df = shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.generation_to_upload_days == 0] \
[["upload_date", "shared_teks"]].rename(
columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_teks_uploaded_on_generation_date",
})
shared_teks_uploaded_on_generation_date_df.head()
estimated_shared_diagnoses_df = shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.generation_to_upload_days == shared_diagnosis_generation_to_upload_days] \
[["upload_date", "shared_teks"]].rename(
columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_diagnoses",
})
estimated_shared_diagnoses_df.head()
```
### Hourly New TEKs
```
hourly_extracted_teks_df = load_extracted_teks(
mode="Hourly", region=report_backend_identifier, limit=25)
hourly_extracted_teks_df.head()
hourly_new_tek_count_df = hourly_extracted_teks_df \
.groupby("extraction_date_with_hour").tek_list. \
apply(lambda x: set(sum(x, []))).reset_index().copy()
hourly_new_tek_count_df = hourly_new_tek_count_df.set_index("extraction_date_with_hour") \
.sort_index(ascending=True)
hourly_new_tek_count_df["new_tek_list"] = hourly_new_tek_count_df.tek_list.diff()
hourly_new_tek_count_df["new_tek_count"] = hourly_new_tek_count_df.new_tek_list.apply(
lambda x: len(x) if not pd.isna(x) else 0)
hourly_new_tek_count_df.rename(columns={
"new_tek_count": "shared_teks_by_upload_date"}, inplace=True)
hourly_new_tek_count_df = hourly_new_tek_count_df.reset_index()[[
"extraction_date_with_hour", "shared_teks_by_upload_date"]]
hourly_new_tek_count_df.head()
hourly_summary_df = hourly_new_tek_count_df.copy()
hourly_summary_df.set_index("extraction_date_with_hour", inplace=True)
hourly_summary_df = hourly_summary_df.fillna(0).astype(int).reset_index()
hourly_summary_df["datetime_utc"] = pd.to_datetime(
hourly_summary_df.extraction_date_with_hour, format="%Y-%m-%d@%H")
hourly_summary_df.set_index("datetime_utc", inplace=True)
hourly_summary_df = hourly_summary_df.tail(-1)
hourly_summary_df.head()
```
### Data Merge
```
result_summary_df = exposure_keys_summary_df.merge(
new_tek_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
shared_teks_uploaded_on_generation_date_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
estimated_shared_diagnoses_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = confirmed_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df["sample_date"] = pd.to_datetime(result_summary_df.sample_date_string)
result_summary_df.set_index("sample_date", inplace=True)
result_summary_df.drop(columns=["sample_date_string"], inplace=True)
result_summary_df.sort_index(ascending=False, inplace=True)
result_summary_df.head()
with pd.option_context("mode.use_inf_as_na", True):
result_summary_df = result_summary_df.fillna(0).astype(int)
result_summary_df["teks_per_shared_diagnosis"] = \
(result_summary_df.shared_teks_by_upload_date / result_summary_df.shared_diagnoses).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case"] = \
(result_summary_df.shared_diagnoses / result_summary_df.covid_cases).fillna(0)
result_summary_df.head(daily_plot_days)
weekly_result_summary_df = result_summary_df \
.sort_index(ascending=True).fillna(0).rolling(7).agg({
"covid_cases": "sum",
"shared_teks_by_generation_date": "sum",
"shared_teks_by_upload_date": "sum",
"shared_diagnoses": "sum"
}).sort_index(ascending=False)
with pd.option_context("mode.use_inf_as_na", True):
weekly_result_summary_df = weekly_result_summary_df.fillna(0).astype(int)
weekly_result_summary_df["teks_per_shared_diagnosis"] = \
(weekly_result_summary_df.shared_teks_by_upload_date / weekly_result_summary_df.shared_diagnoses).fillna(0)
weekly_result_summary_df["shared_diagnoses_per_covid_case"] = \
(weekly_result_summary_df.shared_diagnoses / weekly_result_summary_df.covid_cases).fillna(0)
weekly_result_summary_df.head()
last_7_days_summary = weekly_result_summary_df.to_dict(orient="records")[1]
last_7_days_summary
```
## Report Results
```
display_column_name_mapping = {
"sample_date": "Sample\u00A0Date\u00A0(UTC)",
"datetime_utc": "Timestamp (UTC)",
"upload_date": "Upload Date (UTC)",
"generation_to_upload_days": "Generation to Upload Period in Days",
"region": "Backend",
"region_x": "Backend\u00A0(A)",
"region_y": "Backend\u00A0(B)",
"common_teks": "Common TEKs Shared Between Backends",
"common_teks_fraction": "Fraction of TEKs in Backend (A) Available in Backend (B)",
"covid_cases": "COVID-19 Cases in Source Countries (7-day Rolling Average)",
"shared_teks_by_generation_date": "Shared TEKs by Generation Date",
"shared_teks_by_upload_date": "Shared TEKs by Upload Date",
"shared_diagnoses": "Shared Diagnoses (Estimation)",
"teks_per_shared_diagnosis": "TEKs Uploaded per Shared Diagnosis",
"shared_diagnoses_per_covid_case": "Usage Ratio (Fraction of Cases in Source Countries Which Shared Diagnosis)",
"shared_teks_uploaded_on_generation_date": "Shared TEKs Uploaded on Generation Date",
}
summary_columns = [
"covid_cases",
"shared_teks_by_generation_date",
"shared_teks_by_upload_date",
"shared_teks_uploaded_on_generation_date",
"shared_diagnoses",
"teks_per_shared_diagnosis",
"shared_diagnoses_per_covid_case",
]
```
### Daily Summary Table
```
result_summary_df_ = result_summary_df.copy()
result_summary_df = result_summary_df[summary_columns]
result_summary_with_display_names_df = result_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
result_summary_with_display_names_df
```
### Daily Summary Plots
```
result_plot_summary_df = result_summary_df.head(daily_plot_days)[summary_columns] \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
summary_ax_list = result_plot_summary_df.sort_index(ascending=True).plot.bar(
title=f"Daily Summary",
rot=45, subplots=True, figsize=(15, 22), legend=False)
ax_ = summary_ax_list[-1]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.95)
ax_.yaxis.set_major_formatter(matplotlib.ticker.PercentFormatter(1.0))
_ = ax_.set_xticklabels(sorted(result_plot_summary_df.index.strftime("%Y-%m-%d").tolist()))
```
### Daily Generation to Upload Period Table
```
display_generation_to_upload_period_pivot_df = \
generation_to_upload_period_pivot_df \
.head(backend_generation_days)
display_generation_to_upload_period_pivot_df \
.head(backend_generation_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping)
fig, generation_to_upload_period_pivot_table_ax = plt.subplots(
figsize=(10, 1 + 0.5 * len(display_generation_to_upload_period_pivot_df)))
generation_to_upload_period_pivot_table_ax.set_title(
"Shared TEKs Generation to Upload Period Table")
sns.heatmap(
data=display_generation_to_upload_period_pivot_df
.rename_axis(columns=display_column_name_mapping)
.rename_axis(index=display_column_name_mapping),
fmt=".0f",
annot=True,
ax=generation_to_upload_period_pivot_table_ax)
generation_to_upload_period_pivot_table_ax.get_figure().tight_layout()
```
### Hourly Summary Plots
```
hourly_summary_ax_list = hourly_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.plot.bar(
title=f"Last 24h Summary",
rot=45, subplots=True, legend=False)
ax_ = hourly_summary_ax_list[-1]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.9)
_ = ax_.set_xticklabels(sorted(hourly_summary_df.index.strftime("%Y-%m-%d@%H").tolist()))
```
### Publish Results
```
def get_temporary_image_path() -> str:
return os.path.join(tempfile.gettempdir(), str(uuid.uuid4()) + ".png")
def save_temporary_plot_image(ax):
if isinstance(ax, np.ndarray):
ax = ax[0]
media_path = get_temporary_image_path()
ax.get_figure().savefig(media_path)
return media_path
def save_temporary_dataframe_image(df):
import dataframe_image as dfi
media_path = get_temporary_image_path()
dfi.export(df, media_path)
return media_path
github_repository = os.environ.get("GITHUB_REPOSITORY")
if github_repository is None:
github_repository = "pvieito/Radar-STATS"
github_project_base_url = "https://github.com/" + github_repository
display_formatters = {
display_column_name_mapping["teks_per_shared_diagnosis"]: lambda x: f"{x:.2f}",
display_column_name_mapping["shared_diagnoses_per_covid_case"]: lambda x: f"{x:.2%}",
}
daily_summary_table_html = result_summary_with_display_names_df \
.head(daily_plot_days) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.to_html(formatters=display_formatters)
multi_backend_summary_table_html = multi_backend_summary_df \
.head(daily_plot_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(formatters=display_formatters)
def format_multi_backend_cross_sharing_fraction(x):
if pd.isna(x):
return "-"
elif round(x * 100, 1) == 0:
return ""
else:
return f"{x:.1%}"
multi_backend_cross_sharing_summary_table_html = multi_backend_cross_sharing_summary_df \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(
classes="table-center",
formatters=display_formatters,
float_format=format_multi_backend_cross_sharing_fraction)
multi_backend_cross_sharing_summary_table_html = \
multi_backend_cross_sharing_summary_table_html \
.replace("<tr>","<tr style=\"text-align: center;\">")
extraction_date_result_summary_df = \
result_summary_df[result_summary_df.index == extraction_date]
extraction_date_result_hourly_summary_df = \
hourly_summary_df[hourly_summary_df.extraction_date_with_hour == extraction_date_with_hour]
covid_cases = \
extraction_date_result_summary_df.covid_cases.sum()
shared_teks_by_generation_date = \
extraction_date_result_summary_df.shared_teks_by_generation_date.sum()
shared_teks_by_upload_date = \
extraction_date_result_summary_df.shared_teks_by_upload_date.sum()
shared_diagnoses = \
extraction_date_result_summary_df.shared_diagnoses.sum()
teks_per_shared_diagnosis = \
extraction_date_result_summary_df.teks_per_shared_diagnosis.sum()
shared_diagnoses_per_covid_case = \
extraction_date_result_summary_df.shared_diagnoses_per_covid_case.sum()
shared_teks_by_upload_date_last_hour = \
extraction_date_result_hourly_summary_df.shared_teks_by_upload_date.sum().astype(int)
display_source_regions = ", ".join(report_source_regions)
display_brief_source_regions_limit = 2
if len(report_source_regions) <= display_brief_source_regions_limit:
display_brief_source_regions = display_source_regions
else:
prefix_countries = ", ".join(report_source_regions[:display_brief_source_regions_limit])
display_brief_source_regions = f"{len(report_source_regions)} ({prefix_countries}…)"
if len(report_source_regions) == 1:
display_brief_source_regions_warning_prefix_message = ""
else:
display_brief_source_regions_warning_prefix_message = "⚠️ "
summary_plots_image_path = save_temporary_plot_image(
ax=summary_ax_list)
summary_table_image_path = save_temporary_dataframe_image(
df=result_summary_with_display_names_df)
hourly_summary_plots_image_path = save_temporary_plot_image(
ax=hourly_summary_ax_list)
multi_backend_summary_table_image_path = save_temporary_dataframe_image(
df=multi_backend_summary_df)
generation_to_upload_period_pivot_table_image_path = save_temporary_plot_image(
ax=generation_to_upload_period_pivot_table_ax)
```
### Save Results
```
report_resources_path_prefix = "Data/Resources/Current/RadarCOVID-Report-"
result_summary_df.to_csv(
report_resources_path_prefix + "Summary-Table.csv")
result_summary_df.to_html(
report_resources_path_prefix + "Summary-Table.html")
hourly_summary_df.to_csv(
report_resources_path_prefix + "Hourly-Summary-Table.csv")
multi_backend_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Summary-Table.csv")
multi_backend_cross_sharing_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Cross-Sharing-Summary-Table.csv")
generation_to_upload_period_pivot_df.to_csv(
report_resources_path_prefix + "Generation-Upload-Period-Table.csv")
_ = shutil.copyfile(
summary_plots_image_path,
report_resources_path_prefix + "Summary-Plots.png")
_ = shutil.copyfile(
summary_table_image_path,
report_resources_path_prefix + "Summary-Table.png")
_ = shutil.copyfile(
hourly_summary_plots_image_path,
report_resources_path_prefix + "Hourly-Summary-Plots.png")
_ = shutil.copyfile(
multi_backend_summary_table_image_path,
report_resources_path_prefix + "Multi-Backend-Summary-Table.png")
_ = shutil.copyfile(
generation_to_upload_period_pivot_table_image_path,
report_resources_path_prefix + "Generation-Upload-Period-Table.png")
```
### Publish Results as JSON
```
summary_results_api_df = result_summary_df.reset_index()
summary_results_api_df["sample_date_string"] = \
summary_results_api_df["sample_date"].dt.strftime("%Y-%m-%d")
summary_results = dict(
source_regions=report_source_regions,
extraction_datetime=extraction_datetime,
extraction_date=extraction_date,
extraction_date_with_hour=extraction_date_with_hour,
last_hour=dict(
shared_teks_by_upload_date=shared_teks_by_upload_date_last_hour,
shared_diagnoses=0,
),
today=dict(
covid_cases=covid_cases,
shared_teks_by_generation_date=shared_teks_by_generation_date,
shared_teks_by_upload_date=shared_teks_by_upload_date,
shared_diagnoses=shared_diagnoses,
teks_per_shared_diagnosis=teks_per_shared_diagnosis,
shared_diagnoses_per_covid_case=shared_diagnoses_per_covid_case,
),
last_7_days=last_7_days_summary,
daily_results=summary_results_api_df.to_dict(orient="records"))
summary_results = \
json.loads(pd.Series([summary_results]).to_json(orient="records"))[0]
with open(report_resources_path_prefix + "Summary-Results.json", "w") as f:
json.dump(summary_results, f, indent=4)
```
### Publish on README
```
with open("Data/Templates/README.md", "r") as f:
readme_contents = f.read()
readme_contents = readme_contents.format(
extraction_date_with_hour=extraction_date_with_hour,
github_project_base_url=github_project_base_url,
daily_summary_table_html=daily_summary_table_html,
multi_backend_summary_table_html=multi_backend_summary_table_html,
multi_backend_cross_sharing_summary_table_html=multi_backend_cross_sharing_summary_table_html,
display_source_regions=display_source_regions)
with open("README.md", "w") as f:
f.write(readme_contents)
```
### Publish on Twitter
```
enable_share_to_twitter = os.environ.get("RADARCOVID_REPORT__ENABLE_PUBLISH_ON_TWITTER")
github_event_name = os.environ.get("GITHUB_EVENT_NAME")
if enable_share_to_twitter and github_event_name == "schedule" and \
(shared_teks_by_upload_date_last_hour or not are_today_results_partial):
import tweepy
twitter_api_auth_keys = os.environ["RADARCOVID_REPORT__TWITTER_API_AUTH_KEYS"]
twitter_api_auth_keys = twitter_api_auth_keys.split(":")
auth = tweepy.OAuthHandler(twitter_api_auth_keys[0], twitter_api_auth_keys[1])
auth.set_access_token(twitter_api_auth_keys[2], twitter_api_auth_keys[3])
api = tweepy.API(auth)
summary_plots_media = api.media_upload(summary_plots_image_path)
summary_table_media = api.media_upload(summary_table_image_path)
generation_to_upload_period_pivot_table_image_media = api.media_upload(generation_to_upload_period_pivot_table_image_path)
media_ids = [
summary_plots_media.media_id,
summary_table_media.media_id,
generation_to_upload_period_pivot_table_image_media.media_id,
]
if are_today_results_partial:
today_addendum = " (Partial)"
else:
today_addendum = ""
status = textwrap.dedent(f"""
#RadarCOVID – {extraction_date_with_hour}
{display_brief_source_regions_warning_prefix_message}Countries: {display_brief_source_regions}
Today{today_addendum}:
- Uploaded TEKs: {shared_teks_by_upload_date:.0f} ({shared_teks_by_upload_date_last_hour:+d} last hour)
- Shared Diagnoses: ≤{shared_diagnoses:.0f}
- Usage Ratio: ≤{shared_diagnoses_per_covid_case:.2%}
Last 7 Days:
- Shared Diagnoses: ≤{last_7_days_summary["shared_diagnoses"]:.0f}
- Usage Ratio: ≤{last_7_days_summary["shared_diagnoses_per_covid_case"]:.2%}
Info: {github_project_base_url}#documentation
""")
status = status.encode(encoding="utf-8")
api.update_status(status=status, media_ids=media_ids)
```
|
github_jupyter
|
import datetime
import json
import logging
import os
import shutil
import tempfile
import textwrap
import uuid
import matplotlib.pyplot as plt
import matplotlib.ticker
import numpy as np
import pandas as pd
import retry
import seaborn as sns
%matplotlib inline
current_working_directory = os.environ.get("PWD")
if current_working_directory:
os.chdir(current_working_directory)
sns.set()
matplotlib.rcParams["figure.figsize"] = (15, 6)
extraction_datetime = datetime.datetime.utcnow()
extraction_date = extraction_datetime.strftime("%Y-%m-%d")
extraction_previous_datetime = extraction_datetime - datetime.timedelta(days=1)
extraction_previous_date = extraction_previous_datetime.strftime("%Y-%m-%d")
extraction_date_with_hour = datetime.datetime.utcnow().strftime("%Y-%m-%d@%H")
current_hour = datetime.datetime.utcnow().hour
are_today_results_partial = current_hour != 23
from Modules.ExposureNotification import exposure_notification_io
spain_region_country_code = "ES"
germany_region_country_code = "DE"
default_backend_identifier = spain_region_country_code
backend_generation_days = 7 * 2
daily_summary_days = 7 * 4 * 3
daily_plot_days = 7 * 4
tek_dumps_load_limit = daily_summary_days + 1
default_shared_diagnosis_generation_to_upload_days = 1
environment_backend_identifier = os.environ.get("RADARCOVID_REPORT__BACKEND_IDENTIFIER")
if environment_backend_identifier:
report_backend_identifier = environment_backend_identifier
else:
report_backend_identifier = default_backend_identifier
report_backend_identifier
efgs_supported_countries_backend_identifier = germany_region_country_code
efgs_supported_countries_backend_client = \
exposure_notification_io.get_backend_client_with_identifier(
backend_identifier=efgs_supported_countries_backend_identifier)
efgs_source_regions = efgs_supported_countries_backend_client.get_supported_countries()
environment_source_regions = os.environ.get("RADARCOVID_REPORT__EFGS_EXTRA_SOURCE_REGIONS")
if environment_source_regions:
efgs_source_regions = list(set(efgs_source_regions).union(environment_source_regions.split(",")))
efgs_source_regions = list(map(lambda x: x.upper(), efgs_source_regions))
if report_backend_identifier in efgs_source_regions:
default_source_regions = "EFGS"
else:
default_source_regions = report_backend_identifier.split("-")[0].split("@")[0]
environment_source_regions = os.environ.get("RADARCOVID_REPORT__SOURCE_REGIONS")
if environment_source_regions:
report_source_regions = environment_source_regions
else:
report_source_regions = default_source_regions
if report_source_regions == "EFGS":
if report_backend_identifier in efgs_source_regions:
efgs_source_regions = \
[report_backend_identifier] + \
sorted(list(set(efgs_source_regions).difference([report_backend_identifier])))
report_source_regions = efgs_source_regions
else:
report_source_regions = report_source_regions.split(",")
report_source_regions
environment_download_only_from_report_backend = \
os.environ.get("RADARCOVID_REPORT__DOWNLOAD_ONLY_FROM_REPORT_BACKEND")
if environment_download_only_from_report_backend:
report_backend_identifiers = [report_backend_identifier]
else:
report_backend_identifiers = None
report_backend_identifiers
environment_shared_diagnosis_generation_to_upload_days = \
os.environ.get("RADARCOVID_REPORT__SHARED_DIAGNOSIS_GENERATION_TO_UPLOAD_DAYS")
if environment_shared_diagnosis_generation_to_upload_days:
shared_diagnosis_generation_to_upload_days = \
int(environment_shared_diagnosis_generation_to_upload_days)
else:
shared_diagnosis_generation_to_upload_days = \
default_shared_diagnosis_generation_to_upload_days
shared_diagnosis_generation_to_upload_days
@retry.retry(tries=10, delay=10, backoff=1.1, jitter=(0, 10))
def download_cases_dataframe_from_ecdc():
return pd.read_csv(
"https://opendata.ecdc.europa.eu/covid19/casedistribution/csv/data.csv")
confirmed_df = download_cases_dataframe_from_ecdc()
radar_covid_countries = set(report_source_regions)
confirmed_df = confirmed_df[["dateRep", "cases", "geoId"]]
confirmed_df.rename(
columns={
"dateRep":"sample_date",
"cases": "new_cases",
"geoId": "country_code",
},
inplace=True)
confirmed_df = confirmed_df[confirmed_df.country_code.isin(radar_covid_countries)]
confirmed_df["sample_date"] = pd.to_datetime(confirmed_df.sample_date, dayfirst=True)
confirmed_df = confirmed_df.groupby("sample_date").new_cases.sum().reset_index()
confirmed_df.sort_values("sample_date", inplace=True)
confirmed_df.tail()
confirmed_days = pd.date_range(
start=confirmed_df.iloc[0].sample_date,
end=extraction_datetime)
confirmed_days_df = pd.DataFrame(data=confirmed_days, columns=["sample_date"])
confirmed_df = confirmed_days_df.merge(confirmed_df, how="left")
confirmed_df["sample_date"] = confirmed_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_df.sort_values("sample_date", inplace=True)
confirmed_df.tail()
confirmed_df.columns = ["sample_date_string", "new_cases"]
confirmed_df.sort_values("sample_date_string", inplace=True)
confirmed_df["covid_cases"] = confirmed_df.new_cases.rolling(7).mean().round()
confirmed_df.fillna(method="ffill", inplace=True)
confirmed_df.tail()
confirmed_df[["new_cases", "covid_cases"]].plot()
raw_zip_path_prefix = "Data/TEKs/Raw/"
fail_on_error_backend_identifiers = [report_backend_identifier]
multi_backend_exposure_keys_df = \
exposure_notification_io.download_exposure_keys_from_backends(
backend_identifiers=report_backend_identifiers,
generation_days=backend_generation_days,
fail_on_error_backend_identifiers=fail_on_error_backend_identifiers,
save_raw_zip_path_prefix=raw_zip_path_prefix)
multi_backend_exposure_keys_df["region"] = multi_backend_exposure_keys_df["backend_identifier"]
multi_backend_exposure_keys_df.rename(
columns={
"generation_datetime": "sample_datetime",
"generation_date_string": "sample_date_string",
},
inplace=True)
multi_backend_exposure_keys_df.head()
early_teks_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.rolling_period < 144].copy()
early_teks_df["rolling_period_in_hours"] = early_teks_df.rolling_period / 6
early_teks_df[early_teks_df.sample_date_string != extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
early_teks_df[early_teks_df.sample_date_string == extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
multi_backend_exposure_keys_df = multi_backend_exposure_keys_df[[
"sample_date_string", "region", "key_data"]]
multi_backend_exposure_keys_df.head()
active_regions = \
multi_backend_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
active_regions
multi_backend_summary_df = multi_backend_exposure_keys_df.groupby(
["sample_date_string", "region"]).key_data.nunique().reset_index() \
.pivot(index="sample_date_string", columns="region") \
.sort_index(ascending=False)
multi_backend_summary_df.rename(
columns={"key_data": "shared_teks_by_generation_date"},
inplace=True)
multi_backend_summary_df.rename_axis("sample_date", inplace=True)
multi_backend_summary_df = multi_backend_summary_df.fillna(0).astype(int)
multi_backend_summary_df = multi_backend_summary_df.head(backend_generation_days)
multi_backend_summary_df.head()
def compute_keys_cross_sharing(x):
teks_x = x.key_data_x.item()
common_teks = set(teks_x).intersection(x.key_data_y.item())
common_teks_fraction = len(common_teks) / len(teks_x)
return pd.Series(dict(
common_teks=common_teks,
common_teks_fraction=common_teks_fraction,
))
multi_backend_exposure_keys_by_region_df = \
multi_backend_exposure_keys_df.groupby("region").key_data.unique().reset_index()
multi_backend_exposure_keys_by_region_df["_merge"] = True
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_df.merge(
multi_backend_exposure_keys_by_region_df, on="_merge")
multi_backend_exposure_keys_by_region_combination_df.drop(
columns=["_merge"], inplace=True)
if multi_backend_exposure_keys_by_region_combination_df.region_x.nunique() > 1:
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_combination_df[
multi_backend_exposure_keys_by_region_combination_df.region_x !=
multi_backend_exposure_keys_by_region_combination_df.region_y]
multi_backend_exposure_keys_cross_sharing_df = \
multi_backend_exposure_keys_by_region_combination_df \
.groupby(["region_x", "region_y"]) \
.apply(compute_keys_cross_sharing) \
.reset_index()
multi_backend_cross_sharing_summary_df = \
multi_backend_exposure_keys_cross_sharing_df.pivot_table(
values=["common_teks_fraction"],
columns="region_x",
index="region_y",
aggfunc=lambda x: x.item())
multi_backend_cross_sharing_summary_df
multi_backend_without_active_region_exposure_keys_df = \
multi_backend_exposure_keys_df[multi_backend_exposure_keys_df.region != report_backend_identifier]
multi_backend_without_active_region = \
multi_backend_without_active_region_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
multi_backend_without_active_region
exposure_keys_summary_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.region == report_backend_identifier]
exposure_keys_summary_df.drop(columns=["region"], inplace=True)
exposure_keys_summary_df = \
exposure_keys_summary_df.groupby(["sample_date_string"]).key_data.nunique().to_frame()
exposure_keys_summary_df = \
exposure_keys_summary_df.reset_index().set_index("sample_date_string")
exposure_keys_summary_df.sort_index(ascending=False, inplace=True)
exposure_keys_summary_df.rename(columns={"key_data": "shared_teks_by_generation_date"}, inplace=True)
exposure_keys_summary_df.head()
tek_list_df = multi_backend_exposure_keys_df[
["sample_date_string", "region", "key_data"]].copy()
tek_list_df["key_data"] = tek_list_df["key_data"].apply(str)
tek_list_df.rename(columns={
"sample_date_string": "sample_date",
"key_data": "tek_list"}, inplace=True)
tek_list_df = tek_list_df.groupby(
["sample_date", "region"]).tek_list.unique().reset_index()
tek_list_df["extraction_date"] = extraction_date
tek_list_df["extraction_date_with_hour"] = extraction_date_with_hour
tek_list_path_prefix = "Data/TEKs/"
tek_list_current_path = tek_list_path_prefix + f"/Current/RadarCOVID-TEKs.json"
tek_list_daily_path = tek_list_path_prefix + f"Daily/RadarCOVID-TEKs-{extraction_date}.json"
tek_list_hourly_path = tek_list_path_prefix + f"Hourly/RadarCOVID-TEKs-{extraction_date_with_hour}.json"
for path in [tek_list_current_path, tek_list_daily_path, tek_list_hourly_path]:
os.makedirs(os.path.dirname(path), exist_ok=True)
tek_list_df.drop(columns=["extraction_date", "extraction_date_with_hour"]).to_json(
tek_list_current_path,
lines=True, orient="records")
tek_list_df.drop(columns=["extraction_date_with_hour"]).to_json(
tek_list_daily_path,
lines=True, orient="records")
tek_list_df.to_json(
tek_list_hourly_path,
lines=True, orient="records")
tek_list_df.head()
import glob
def load_extracted_teks(mode, region=None, limit=None) -> pd.DataFrame:
extracted_teks_df = pd.DataFrame(columns=["region"])
file_paths = list(reversed(sorted(glob.glob(tek_list_path_prefix + mode + "/RadarCOVID-TEKs-*.json"))))
if limit:
file_paths = file_paths[:limit]
for file_path in file_paths:
logging.info(f"Loading TEKs from '{file_path}'...")
iteration_extracted_teks_df = pd.read_json(file_path, lines=True)
extracted_teks_df = extracted_teks_df.append(
iteration_extracted_teks_df, sort=False)
extracted_teks_df["region"] = \
extracted_teks_df.region.fillna(spain_region_country_code).copy()
if region:
extracted_teks_df = \
extracted_teks_df[extracted_teks_df.region == region]
return extracted_teks_df
daily_extracted_teks_df = load_extracted_teks(
mode="Daily",
region=report_backend_identifier,
limit=tek_dumps_load_limit)
daily_extracted_teks_df.head()
exposure_keys_summary_df_ = daily_extracted_teks_df \
.sort_values("extraction_date", ascending=False) \
.groupby("sample_date").tek_list.first() \
.to_frame()
exposure_keys_summary_df_.index.name = "sample_date_string"
exposure_keys_summary_df_["tek_list"] = \
exposure_keys_summary_df_.tek_list.apply(len)
exposure_keys_summary_df_ = exposure_keys_summary_df_ \
.rename(columns={"tek_list": "shared_teks_by_generation_date"}) \
.sort_index(ascending=False)
exposure_keys_summary_df = exposure_keys_summary_df_
exposure_keys_summary_df.head()
tek_list_df = daily_extracted_teks_df.groupby("extraction_date").tek_list.apply(
lambda x: set(sum(x, []))).reset_index()
tek_list_df = tek_list_df.set_index("extraction_date").sort_index(ascending=True)
tek_list_df.head()
def compute_teks_by_generation_and_upload_date(date):
day_new_teks_set_df = tek_list_df.copy().diff()
try:
day_new_teks_set = day_new_teks_set_df[
day_new_teks_set_df.index == date].tek_list.item()
except ValueError:
day_new_teks_set = None
if pd.isna(day_new_teks_set):
day_new_teks_set = set()
day_new_teks_df = daily_extracted_teks_df[
daily_extracted_teks_df.extraction_date == date].copy()
day_new_teks_df["shared_teks"] = \
day_new_teks_df.tek_list.apply(lambda x: set(x).intersection(day_new_teks_set))
day_new_teks_df["shared_teks"] = \
day_new_teks_df.shared_teks.apply(len)
day_new_teks_df["upload_date"] = date
day_new_teks_df.rename(columns={"sample_date": "generation_date"}, inplace=True)
day_new_teks_df = day_new_teks_df[
["upload_date", "generation_date", "shared_teks"]]
day_new_teks_df["generation_to_upload_days"] = \
(pd.to_datetime(day_new_teks_df.upload_date) -
pd.to_datetime(day_new_teks_df.generation_date)).dt.days
day_new_teks_df = day_new_teks_df[day_new_teks_df.shared_teks > 0]
return day_new_teks_df
shared_teks_generation_to_upload_df = pd.DataFrame()
for upload_date in daily_extracted_teks_df.extraction_date.unique():
shared_teks_generation_to_upload_df = \
shared_teks_generation_to_upload_df.append(
compute_teks_by_generation_and_upload_date(date=upload_date))
shared_teks_generation_to_upload_df \
.sort_values(["upload_date", "generation_date"], ascending=False, inplace=True)
shared_teks_generation_to_upload_df.tail()
today_new_teks_df = \
shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.upload_date == extraction_date].copy()
today_new_teks_df.tail()
if not today_new_teks_df.empty:
today_new_teks_df.set_index("generation_to_upload_days") \
.sort_index().shared_teks.plot.bar()
generation_to_upload_period_pivot_df = \
shared_teks_generation_to_upload_df[
["upload_date", "generation_to_upload_days", "shared_teks"]] \
.pivot(index="upload_date", columns="generation_to_upload_days") \
.sort_index(ascending=False).fillna(0).astype(int) \
.droplevel(level=0, axis=1)
generation_to_upload_period_pivot_df.head()
new_tek_df = tek_list_df.diff().tek_list.apply(
lambda x: len(x) if not pd.isna(x) else None).to_frame().reset_index()
new_tek_df.rename(columns={
"tek_list": "shared_teks_by_upload_date",
"extraction_date": "sample_date_string",}, inplace=True)
new_tek_df.tail()
shared_teks_uploaded_on_generation_date_df = shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.generation_to_upload_days == 0] \
[["upload_date", "shared_teks"]].rename(
columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_teks_uploaded_on_generation_date",
})
shared_teks_uploaded_on_generation_date_df.head()
estimated_shared_diagnoses_df = shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.generation_to_upload_days == shared_diagnosis_generation_to_upload_days] \
[["upload_date", "shared_teks"]].rename(
columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_diagnoses",
})
estimated_shared_diagnoses_df.head()
hourly_extracted_teks_df = load_extracted_teks(
mode="Hourly", region=report_backend_identifier, limit=25)
hourly_extracted_teks_df.head()
hourly_new_tek_count_df = hourly_extracted_teks_df \
.groupby("extraction_date_with_hour").tek_list. \
apply(lambda x: set(sum(x, []))).reset_index().copy()
hourly_new_tek_count_df = hourly_new_tek_count_df.set_index("extraction_date_with_hour") \
.sort_index(ascending=True)
hourly_new_tek_count_df["new_tek_list"] = hourly_new_tek_count_df.tek_list.diff()
hourly_new_tek_count_df["new_tek_count"] = hourly_new_tek_count_df.new_tek_list.apply(
lambda x: len(x) if not pd.isna(x) else 0)
hourly_new_tek_count_df.rename(columns={
"new_tek_count": "shared_teks_by_upload_date"}, inplace=True)
hourly_new_tek_count_df = hourly_new_tek_count_df.reset_index()[[
"extraction_date_with_hour", "shared_teks_by_upload_date"]]
hourly_new_tek_count_df.head()
hourly_summary_df = hourly_new_tek_count_df.copy()
hourly_summary_df.set_index("extraction_date_with_hour", inplace=True)
hourly_summary_df = hourly_summary_df.fillna(0).astype(int).reset_index()
hourly_summary_df["datetime_utc"] = pd.to_datetime(
hourly_summary_df.extraction_date_with_hour, format="%Y-%m-%d@%H")
hourly_summary_df.set_index("datetime_utc", inplace=True)
hourly_summary_df = hourly_summary_df.tail(-1)
hourly_summary_df.head()
result_summary_df = exposure_keys_summary_df.merge(
new_tek_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
shared_teks_uploaded_on_generation_date_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
estimated_shared_diagnoses_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = confirmed_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df["sample_date"] = pd.to_datetime(result_summary_df.sample_date_string)
result_summary_df.set_index("sample_date", inplace=True)
result_summary_df.drop(columns=["sample_date_string"], inplace=True)
result_summary_df.sort_index(ascending=False, inplace=True)
result_summary_df.head()
with pd.option_context("mode.use_inf_as_na", True):
result_summary_df = result_summary_df.fillna(0).astype(int)
result_summary_df["teks_per_shared_diagnosis"] = \
(result_summary_df.shared_teks_by_upload_date / result_summary_df.shared_diagnoses).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case"] = \
(result_summary_df.shared_diagnoses / result_summary_df.covid_cases).fillna(0)
result_summary_df.head(daily_plot_days)
weekly_result_summary_df = result_summary_df \
.sort_index(ascending=True).fillna(0).rolling(7).agg({
"covid_cases": "sum",
"shared_teks_by_generation_date": "sum",
"shared_teks_by_upload_date": "sum",
"shared_diagnoses": "sum"
}).sort_index(ascending=False)
with pd.option_context("mode.use_inf_as_na", True):
weekly_result_summary_df = weekly_result_summary_df.fillna(0).astype(int)
weekly_result_summary_df["teks_per_shared_diagnosis"] = \
(weekly_result_summary_df.shared_teks_by_upload_date / weekly_result_summary_df.shared_diagnoses).fillna(0)
weekly_result_summary_df["shared_diagnoses_per_covid_case"] = \
(weekly_result_summary_df.shared_diagnoses / weekly_result_summary_df.covid_cases).fillna(0)
weekly_result_summary_df.head()
last_7_days_summary = weekly_result_summary_df.to_dict(orient="records")[1]
last_7_days_summary
display_column_name_mapping = {
"sample_date": "Sample\u00A0Date\u00A0(UTC)",
"datetime_utc": "Timestamp (UTC)",
"upload_date": "Upload Date (UTC)",
"generation_to_upload_days": "Generation to Upload Period in Days",
"region": "Backend",
"region_x": "Backend\u00A0(A)",
"region_y": "Backend\u00A0(B)",
"common_teks": "Common TEKs Shared Between Backends",
"common_teks_fraction": "Fraction of TEKs in Backend (A) Available in Backend (B)",
"covid_cases": "COVID-19 Cases in Source Countries (7-day Rolling Average)",
"shared_teks_by_generation_date": "Shared TEKs by Generation Date",
"shared_teks_by_upload_date": "Shared TEKs by Upload Date",
"shared_diagnoses": "Shared Diagnoses (Estimation)",
"teks_per_shared_diagnosis": "TEKs Uploaded per Shared Diagnosis",
"shared_diagnoses_per_covid_case": "Usage Ratio (Fraction of Cases in Source Countries Which Shared Diagnosis)",
"shared_teks_uploaded_on_generation_date": "Shared TEKs Uploaded on Generation Date",
}
summary_columns = [
"covid_cases",
"shared_teks_by_generation_date",
"shared_teks_by_upload_date",
"shared_teks_uploaded_on_generation_date",
"shared_diagnoses",
"teks_per_shared_diagnosis",
"shared_diagnoses_per_covid_case",
]
result_summary_df_ = result_summary_df.copy()
result_summary_df = result_summary_df[summary_columns]
result_summary_with_display_names_df = result_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
result_summary_with_display_names_df
result_plot_summary_df = result_summary_df.head(daily_plot_days)[summary_columns] \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
summary_ax_list = result_plot_summary_df.sort_index(ascending=True).plot.bar(
title=f"Daily Summary",
rot=45, subplots=True, figsize=(15, 22), legend=False)
ax_ = summary_ax_list[-1]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.95)
ax_.yaxis.set_major_formatter(matplotlib.ticker.PercentFormatter(1.0))
_ = ax_.set_xticklabels(sorted(result_plot_summary_df.index.strftime("%Y-%m-%d").tolist()))
display_generation_to_upload_period_pivot_df = \
generation_to_upload_period_pivot_df \
.head(backend_generation_days)
display_generation_to_upload_period_pivot_df \
.head(backend_generation_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping)
fig, generation_to_upload_period_pivot_table_ax = plt.subplots(
figsize=(10, 1 + 0.5 * len(display_generation_to_upload_period_pivot_df)))
generation_to_upload_period_pivot_table_ax.set_title(
"Shared TEKs Generation to Upload Period Table")
sns.heatmap(
data=display_generation_to_upload_period_pivot_df
.rename_axis(columns=display_column_name_mapping)
.rename_axis(index=display_column_name_mapping),
fmt=".0f",
annot=True,
ax=generation_to_upload_period_pivot_table_ax)
generation_to_upload_period_pivot_table_ax.get_figure().tight_layout()
hourly_summary_ax_list = hourly_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.plot.bar(
title=f"Last 24h Summary",
rot=45, subplots=True, legend=False)
ax_ = hourly_summary_ax_list[-1]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.9)
_ = ax_.set_xticklabels(sorted(hourly_summary_df.index.strftime("%Y-%m-%d@%H").tolist()))
def get_temporary_image_path() -> str:
return os.path.join(tempfile.gettempdir(), str(uuid.uuid4()) + ".png")
def save_temporary_plot_image(ax):
if isinstance(ax, np.ndarray):
ax = ax[0]
media_path = get_temporary_image_path()
ax.get_figure().savefig(media_path)
return media_path
def save_temporary_dataframe_image(df):
import dataframe_image as dfi
media_path = get_temporary_image_path()
dfi.export(df, media_path)
return media_path
github_repository = os.environ.get("GITHUB_REPOSITORY")
if github_repository is None:
github_repository = "pvieito/Radar-STATS"
github_project_base_url = "https://github.com/" + github_repository
display_formatters = {
display_column_name_mapping["teks_per_shared_diagnosis"]: lambda x: f"{x:.2f}",
display_column_name_mapping["shared_diagnoses_per_covid_case"]: lambda x: f"{x:.2%}",
}
daily_summary_table_html = result_summary_with_display_names_df \
.head(daily_plot_days) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.to_html(formatters=display_formatters)
multi_backend_summary_table_html = multi_backend_summary_df \
.head(daily_plot_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(formatters=display_formatters)
def format_multi_backend_cross_sharing_fraction(x):
if pd.isna(x):
return "-"
elif round(x * 100, 1) == 0:
return ""
else:
return f"{x:.1%}"
multi_backend_cross_sharing_summary_table_html = multi_backend_cross_sharing_summary_df \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(
classes="table-center",
formatters=display_formatters,
float_format=format_multi_backend_cross_sharing_fraction)
multi_backend_cross_sharing_summary_table_html = \
multi_backend_cross_sharing_summary_table_html \
.replace("<tr>","<tr style=\"text-align: center;\">")
extraction_date_result_summary_df = \
result_summary_df[result_summary_df.index == extraction_date]
extraction_date_result_hourly_summary_df = \
hourly_summary_df[hourly_summary_df.extraction_date_with_hour == extraction_date_with_hour]
covid_cases = \
extraction_date_result_summary_df.covid_cases.sum()
shared_teks_by_generation_date = \
extraction_date_result_summary_df.shared_teks_by_generation_date.sum()
shared_teks_by_upload_date = \
extraction_date_result_summary_df.shared_teks_by_upload_date.sum()
shared_diagnoses = \
extraction_date_result_summary_df.shared_diagnoses.sum()
teks_per_shared_diagnosis = \
extraction_date_result_summary_df.teks_per_shared_diagnosis.sum()
shared_diagnoses_per_covid_case = \
extraction_date_result_summary_df.shared_diagnoses_per_covid_case.sum()
shared_teks_by_upload_date_last_hour = \
extraction_date_result_hourly_summary_df.shared_teks_by_upload_date.sum().astype(int)
display_source_regions = ", ".join(report_source_regions)
display_brief_source_regions_limit = 2
if len(report_source_regions) <= display_brief_source_regions_limit:
display_brief_source_regions = display_source_regions
else:
prefix_countries = ", ".join(report_source_regions[:display_brief_source_regions_limit])
display_brief_source_regions = f"{len(report_source_regions)} ({prefix_countries}…)"
if len(report_source_regions) == 1:
display_brief_source_regions_warning_prefix_message = ""
else:
display_brief_source_regions_warning_prefix_message = "⚠️ "
summary_plots_image_path = save_temporary_plot_image(
ax=summary_ax_list)
summary_table_image_path = save_temporary_dataframe_image(
df=result_summary_with_display_names_df)
hourly_summary_plots_image_path = save_temporary_plot_image(
ax=hourly_summary_ax_list)
multi_backend_summary_table_image_path = save_temporary_dataframe_image(
df=multi_backend_summary_df)
generation_to_upload_period_pivot_table_image_path = save_temporary_plot_image(
ax=generation_to_upload_period_pivot_table_ax)
report_resources_path_prefix = "Data/Resources/Current/RadarCOVID-Report-"
result_summary_df.to_csv(
report_resources_path_prefix + "Summary-Table.csv")
result_summary_df.to_html(
report_resources_path_prefix + "Summary-Table.html")
hourly_summary_df.to_csv(
report_resources_path_prefix + "Hourly-Summary-Table.csv")
multi_backend_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Summary-Table.csv")
multi_backend_cross_sharing_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Cross-Sharing-Summary-Table.csv")
generation_to_upload_period_pivot_df.to_csv(
report_resources_path_prefix + "Generation-Upload-Period-Table.csv")
_ = shutil.copyfile(
summary_plots_image_path,
report_resources_path_prefix + "Summary-Plots.png")
_ = shutil.copyfile(
summary_table_image_path,
report_resources_path_prefix + "Summary-Table.png")
_ = shutil.copyfile(
hourly_summary_plots_image_path,
report_resources_path_prefix + "Hourly-Summary-Plots.png")
_ = shutil.copyfile(
multi_backend_summary_table_image_path,
report_resources_path_prefix + "Multi-Backend-Summary-Table.png")
_ = shutil.copyfile(
generation_to_upload_period_pivot_table_image_path,
report_resources_path_prefix + "Generation-Upload-Period-Table.png")
summary_results_api_df = result_summary_df.reset_index()
summary_results_api_df["sample_date_string"] = \
summary_results_api_df["sample_date"].dt.strftime("%Y-%m-%d")
summary_results = dict(
source_regions=report_source_regions,
extraction_datetime=extraction_datetime,
extraction_date=extraction_date,
extraction_date_with_hour=extraction_date_with_hour,
last_hour=dict(
shared_teks_by_upload_date=shared_teks_by_upload_date_last_hour,
shared_diagnoses=0,
),
today=dict(
covid_cases=covid_cases,
shared_teks_by_generation_date=shared_teks_by_generation_date,
shared_teks_by_upload_date=shared_teks_by_upload_date,
shared_diagnoses=shared_diagnoses,
teks_per_shared_diagnosis=teks_per_shared_diagnosis,
shared_diagnoses_per_covid_case=shared_diagnoses_per_covid_case,
),
last_7_days=last_7_days_summary,
daily_results=summary_results_api_df.to_dict(orient="records"))
summary_results = \
json.loads(pd.Series([summary_results]).to_json(orient="records"))[0]
with open(report_resources_path_prefix + "Summary-Results.json", "w") as f:
json.dump(summary_results, f, indent=4)
with open("Data/Templates/README.md", "r") as f:
readme_contents = f.read()
readme_contents = readme_contents.format(
extraction_date_with_hour=extraction_date_with_hour,
github_project_base_url=github_project_base_url,
daily_summary_table_html=daily_summary_table_html,
multi_backend_summary_table_html=multi_backend_summary_table_html,
multi_backend_cross_sharing_summary_table_html=multi_backend_cross_sharing_summary_table_html,
display_source_regions=display_source_regions)
with open("README.md", "w") as f:
f.write(readme_contents)
enable_share_to_twitter = os.environ.get("RADARCOVID_REPORT__ENABLE_PUBLISH_ON_TWITTER")
github_event_name = os.environ.get("GITHUB_EVENT_NAME")
if enable_share_to_twitter and github_event_name == "schedule" and \
(shared_teks_by_upload_date_last_hour or not are_today_results_partial):
import tweepy
twitter_api_auth_keys = os.environ["RADARCOVID_REPORT__TWITTER_API_AUTH_KEYS"]
twitter_api_auth_keys = twitter_api_auth_keys.split(":")
auth = tweepy.OAuthHandler(twitter_api_auth_keys[0], twitter_api_auth_keys[1])
auth.set_access_token(twitter_api_auth_keys[2], twitter_api_auth_keys[3])
api = tweepy.API(auth)
summary_plots_media = api.media_upload(summary_plots_image_path)
summary_table_media = api.media_upload(summary_table_image_path)
generation_to_upload_period_pivot_table_image_media = api.media_upload(generation_to_upload_period_pivot_table_image_path)
media_ids = [
summary_plots_media.media_id,
summary_table_media.media_id,
generation_to_upload_period_pivot_table_image_media.media_id,
]
if are_today_results_partial:
today_addendum = " (Partial)"
else:
today_addendum = ""
status = textwrap.dedent(f"""
#RadarCOVID – {extraction_date_with_hour}
{display_brief_source_regions_warning_prefix_message}Countries: {display_brief_source_regions}
Today{today_addendum}:
- Uploaded TEKs: {shared_teks_by_upload_date:.0f} ({shared_teks_by_upload_date_last_hour:+d} last hour)
- Shared Diagnoses: ≤{shared_diagnoses:.0f}
- Usage Ratio: ≤{shared_diagnoses_per_covid_case:.2%}
Last 7 Days:
- Shared Diagnoses: ≤{last_7_days_summary["shared_diagnoses"]:.0f}
- Usage Ratio: ≤{last_7_days_summary["shared_diagnoses_per_covid_case"]:.2%}
Info: {github_project_base_url}#documentation
""")
status = status.encode(encoding="utf-8")
api.update_status(status=status, media_ids=media_ids)
| 0.304042 | 0.216643 |
```
import keras
from keras.models import Sequential, Model, load_model
from keras.layers import Dense, Dropout, Activation, Flatten, Input, Lambda
from keras.layers import Conv2D, MaxPooling2D, AveragePooling2D, Conv1D, MaxPooling1D, LSTM, ConvLSTM2D, GRU, CuDNNLSTM, CuDNNGRU, BatchNormalization, LocallyConnected2D, Permute, TimeDistributed, Bidirectional
from keras.layers import Concatenate, Reshape, Softmax, Conv2DTranspose, Embedding, Multiply
from keras.callbacks import ModelCheckpoint, EarlyStopping, Callback
from keras import regularizers
from keras import backend as K
from keras.utils.generic_utils import Progbar
from keras.layers.merge import _Merge
import keras.losses
from keras.datasets import mnist
from functools import partial
from collections import defaultdict
import tensorflow as tf
from tensorflow.python.framework import ops
import isolearn.keras as iso
import numpy as np
import tensorflow as tf
import logging
logging.getLogger('tensorflow').setLevel(logging.ERROR)
import os
import pickle
import numpy as np
import scipy.sparse as sp
import scipy.io as spio
import matplotlib.pyplot as plt
from keras.backend.tensorflow_backend import set_session
def contain_tf_gpu_mem_usage() :
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
sess = tf.Session(config=config)
set_session(sess)
contain_tf_gpu_mem_usage()
class EpochVariableCallback(Callback) :
def __init__(self, my_variable, my_func) :
self.my_variable = my_variable
self.my_func = my_func
def on_epoch_begin(self, epoch, logs={}) :
K.set_value(self.my_variable, self.my_func(K.get_value(self.my_variable), epoch))
#Load MNIST data
dataset_name = "mnist_3_vs_5"
img_rows, img_cols = 28, 28
num_classes = 10
batch_size = 32
included_classes = { 3, 5 }
(x_train, y_train), (x_test, y_test) = mnist.load_data()
keep_index_train = []
for i in range(y_train.shape[0]) :
if y_train[i] in included_classes :
keep_index_train.append(i)
keep_index_test = []
for i in range(y_test.shape[0]) :
if y_test[i] in included_classes :
keep_index_test.append(i)
x_train = x_train[keep_index_train]
x_test = x_test[keep_index_test]
y_train = y_train[keep_index_train]
y_test = y_test[keep_index_test]
n_train = int((x_train.shape[0] // batch_size) * batch_size)
n_test = int((x_test.shape[0] // batch_size) * batch_size)
x_train = x_train[:n_train]
x_test = x_test[:n_test]
y_train = y_train[:n_train]
y_test = y_test[:n_test]
x_train = x_train.reshape(x_train.shape[0], img_rows, img_cols, 1)
x_test = x_test.reshape(x_test.shape[0], img_rows, img_cols, 1)
input_shape = (img_rows, img_cols, 1)
x_train = x_train.astype('float32')
x_test = x_test.astype('float32')
x_train /= 255
x_test /= 255
print("x_train.shape = " + str(x_train.shape))
print("n train samples = " + str(x_train.shape[0]))
print("n test samples = " + str(x_test.shape[0]))
y_train = keras.utils.to_categorical(y_train, num_classes)
y_test = keras.utils.to_categorical(y_test, num_classes)
#Binarize images
def _binarize_images(x, val_thresh=0.5) :
x_bin = np.zeros(x.shape)
x_bin[x >= val_thresh] = 1.
return x_bin
x_train = _binarize_images(x_train, val_thresh=0.5)
x_test = _binarize_images(x_test, val_thresh=0.5)
from tensorflow.python.framework import ops
#Stochastic Binarized Neuron helper functions (Tensorflow)
#ST Estimator code adopted from https://r2rt.com/binary-stochastic-neurons-in-tensorflow.html
#See Github https://github.com/spitis/
def bernoulli_sample(x):
g = tf.get_default_graph()
with ops.name_scope("BernoulliSample") as name:
with g.gradient_override_map({"Ceil": "Identity","Sub": "BernoulliSample_ST"}):
return tf.ceil(x - tf.random_uniform(tf.shape(x)), name=name)
@ops.RegisterGradient("BernoulliSample_ST")
def bernoulliSample_ST(op, grad):
return [grad, tf.zeros(tf.shape(op.inputs[1]))]
#Scrambler network definition
#Gumbel Distribution Sampler
def gumbel_softmax(logits, temperature=0.5) :
gumbel_dist = tf.contrib.distributions.RelaxedOneHotCategorical(temperature, logits=logits)
batch_dim = logits.get_shape().as_list()[0]
onehot_dim = logits.get_shape().as_list()[1]
return gumbel_dist.sample()
def sample_mask_gumbel(pwm_logits) :
n_sequences = K.shape(pwm_logits)[0]
x_len = K.shape(pwm_logits)[1]
y_len = K.shape(pwm_logits)[2]
flat_pwm = K.reshape(pwm_logits, (n_sequences * x_len * y_len, 2))
sampled_pwm = gumbel_softmax(flat_pwm, temperature=0.5)
return K.reshape(sampled_pwm, (n_sequences, x_len, y_len, 2))[..., :1]
def make_resblock(n_channels=64, window_size=8, dilation_rate=1, group_ix=0, layer_ix=0, drop_rate=0.0, batchnorm_trainmode=True) :
#Initialize res block layers
batch_norm_0 = BatchNormalization(name='scrambler_resblock_' + str(group_ix) + '_' + str(layer_ix) + '_batch_norm_0')
relu_0 = Lambda(lambda x: K.relu(x, alpha=0.0))
conv_0 = Conv2D(n_channels, (window_size, window_size), dilation_rate=dilation_rate, strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_resblock_' + str(group_ix) + '_' + str(layer_ix) + '_conv_0')
batch_norm_1 = BatchNormalization(name='scrambler_resblock_' + str(group_ix) + '_' + str(layer_ix) + '_batch_norm_1')
relu_1 = Lambda(lambda x: K.relu(x, alpha=0.0))
conv_1 = Conv2D(n_channels, (window_size, window_size), dilation_rate=dilation_rate, strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_resblock_' + str(group_ix) + '_' + str(layer_ix) + '_conv_1')
skip_1 = Lambda(lambda x: x[0] + x[1], name='scrambler_resblock_' + str(group_ix) + '_' + str(layer_ix) + '_skip_1')
drop_1 = None
if drop_rate > 0.0 :
drop_1 = Dropout(drop_rate)
#Execute res block
def _resblock_func(input_tensor, training=batchnorm_trainmode) :
batch_norm_0_out = batch_norm_0(input_tensor, training=training)
relu_0_out = relu_0(batch_norm_0_out)
conv_0_out = conv_0(relu_0_out)
batch_norm_1_out = batch_norm_1(conv_0_out, training=training)
relu_1_out = relu_1(batch_norm_1_out)
if drop_rate > 0.0 :
conv_1_out = drop_1(conv_1(relu_1_out))
else :
conv_1_out = conv_1(relu_1_out)
skip_1_out = skip_1([conv_1_out, input_tensor])
return skip_1_out
return _resblock_func
def load_scrambler_network(n_groups=1, n_resblocks_per_group=4, n_channels=32, window_size=8, dilation_rates=[1], drop_rate=0.0, batchnorm_trainmode=True) :
#Discriminator network definition
conv_0 = Conv2D(n_channels, (1, 1), strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_conv_0')
skip_convs = []
resblock_groups = []
for group_ix in range(n_groups) :
skip_convs.append(Conv2D(n_channels, (1, 1), strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_skip_conv_' + str(group_ix)))
resblocks = []
for layer_ix in range(n_resblocks_per_group) :
resblocks.append(make_resblock(n_channels=n_channels, window_size=window_size, dilation_rate=dilation_rates[group_ix], group_ix=group_ix, layer_ix=layer_ix, drop_rate=drop_rate, batchnorm_trainmode=batchnorm_trainmode))
resblock_groups.append(resblocks)
last_block_conv = Conv2D(n_channels, (1, 1), strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_last_block_conv')
skip_add = Lambda(lambda x: x[0] + x[1], name='scrambler_skip_add')
final_conv_2_channels = Conv2D(2, (1, 1), strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_final_conv')
final_conv_sigm = Lambda(lambda x: K.softmax(x, axis=-1)[..., :1])
final_conv_gumbel = Lambda(lambda x: sample_mask_gumbel(x))
scale_inputs = Lambda(lambda x: x[1] * K.tile(x[0], (1, 1, 1, 1)), name='scrambler_input_scale')
def _scrambler_func(image_input) :
conv_0_out = conv_0(image_input)
#Connect group of res blocks
output_tensor = conv_0_out
#Res block group execution
skip_conv_outs = []
for group_ix in range(n_groups) :
skip_conv_out = skip_convs[group_ix](output_tensor)
skip_conv_outs.append(skip_conv_out)
for layer_ix in range(n_resblocks_per_group) :
output_tensor = resblock_groups[group_ix][layer_ix](output_tensor)
#Last res block extr conv
last_block_conv_out = last_block_conv(output_tensor)
skip_add_out = last_block_conv_out
for group_ix in range(n_groups) :
skip_add_out = skip_add([skip_add_out, skip_conv_outs[group_ix]])
#Final conv out
final_conv_2_channels_out = final_conv_2_channels(skip_add_out)
final_conv_sigm_out = final_conv_sigm(final_conv_2_channels_out)
final_conv_gumbel_out = final_conv_gumbel(final_conv_2_channels_out)
#Scale inputs by importance scores
scaled_inputs = scale_inputs([final_conv_gumbel_out, image_input])
return scaled_inputs, final_conv_sigm_out, final_conv_2_channels_out
return _scrambler_func
#Keras loss functions
def get_softmax_kl_divergence() :
def _softmax_kl_divergence(y_true, y_pred) :
y_true = K.clip(y_true, K.epsilon(), 1.0 - K.epsilon())
y_pred = K.clip(y_pred, K.epsilon(), 1.0 - K.epsilon())
return K.mean(K.sum(y_true * K.log(y_true / y_pred), axis=-1), axis=-1)
return _softmax_kl_divergence
def get_margin_lum_ame_masked(max_lum=0.1) :
def _margin_lum_ame(importance_scores) :
p_ons = importance_scores[..., 0]
mean_p_on = K.mean(p_ons, axis=(1, 2))
margin_p_on = K.switch(mean_p_on > max_lum, mean_p_on - max_lum, K.zeros_like(mean_p_on))
return margin_p_on
return _margin_lum_ame
def get_target_lum_sme_masked(target_lum=0.1) :
def _target_lum_sme(importance_scores) :
p_ons = importance_scores[..., 0]
mean_p_on = K.mean(p_ons, axis=(1, 2))
return (mean_p_on - target_lum)**2
return _target_lum_sme
def get_weighted_loss(loss_coeff=1.) :
def _min_pred(y_true, y_pred) :
return loss_coeff * y_pred
return _min_pred
#Initialize Encoder and Decoder networks
batch_size = 32
n_rows = 28
n_cols = 28
#Resnet parameters
resnet_n_groups = 5
resnet_n_resblocks_per_group = 4
resnet_n_channels = 32
resnet_window_size = 3
resnet_dilation_rates = [1, 2, 4, 2, 1]
resnet_drop_rate = 0.0#0.25
resnet_batchnorm_trainmode = True
#Load scrambler
scrambler = load_scrambler_network(
n_groups=resnet_n_groups,
n_resblocks_per_group=resnet_n_resblocks_per_group,
n_channels=resnet_n_channels, window_size=resnet_window_size,
dilation_rates=resnet_dilation_rates,
drop_rate=resnet_drop_rate,
batchnorm_trainmode=resnet_batchnorm_trainmode
)
#Load Predictor
predictor_path = 'saved_models/mnist_binarized_cnn_10_digits.h5'
predictor = load_model(predictor_path)
predictor.trainable = False
predictor.compile(optimizer=keras.optimizers.SGD(lr=0.1), loss='mean_squared_error')
#Build scrambler model
scrambler_input = Input(shape=(n_rows, n_cols, 1), name='scrambler_input')
image, importance_scores, importance_scores_log = scrambler(scrambler_input)
scrambler_model = Model([scrambler_input], [image, importance_scores, importance_scores_log])
scrambler_model.compile(
optimizer=keras.optimizers.Adam(lr=0.001, beta_1=0.9, beta_2=0.999),
loss='mean_squared_error'
)
#Build Auto-scrambler pipeline
#Define model inputs
ae_scrambler_input = Input(shape=(n_rows, n_cols, 1), name='ae_scrambler_input')
#Run encoder and decoder
scrambled_sample, importance_scores, importance_scores_log = scrambler(ae_scrambler_input)
#Make reference prediction on non-scrambled input sequence
y_pred_non_scrambled = predictor([ae_scrambler_input])
#Make prediction on scrambled sequence samples
y_pred_scrambled = predictor([scrambled_sample])
#Cost function parameters
initial_target_lum = 0.25
target_lum = 0.05
#NLL cost
nll_loss_func = get_softmax_kl_divergence()
#Conservation cost
conservation_loss_func = get_target_lum_sme_masked(target_lum=initial_target_lum)
#Entropy cost
entropy_loss_func = get_target_lum_sme_masked(target_lum=target_lum)
#entropy_loss_func = get_margin_lum_ame_masked(max_lum=target_lum)
#Define annealing coefficient
anneal_coeff = K.variable(1.0)
#Execute NLL cost
nll_loss = Lambda(lambda x: nll_loss_func(x[0], x[1]), name='nll')([y_pred_non_scrambled, y_pred_scrambled])
#Execute conservation cost
conservation_loss = Lambda(lambda x: anneal_coeff * conservation_loss_func(x), name='conservation')(importance_scores)
#Execute entropy cost
entropy_loss = Lambda(lambda x: (1. - anneal_coeff) * entropy_loss_func(x), name='entropy')(importance_scores)
loss_model = Model(
[ae_scrambler_input],
[nll_loss, conservation_loss, entropy_loss]
)
loss_model.compile(
optimizer=keras.optimizers.Adam(lr=0.0001, beta_1=0.5, beta_2=0.9),
loss={
'nll' : get_weighted_loss(loss_coeff=1.0),
'conservation' : get_weighted_loss(loss_coeff=1.0),
'entropy' : get_weighted_loss(loss_coeff=500.0)
}
)
scrambler_model.summary()
loss_model.summary()
#Training configuration
#Define number of training epochs
n_epochs = 50
#Define experiment suffix (optional)
experiment_suffix = "_kl_divergence_higher_entropy_penalty_gumbel_no_bg_lum"
#Define anneal function
def _anneal_func(val, epoch, n_epochs=n_epochs) :
if epoch in [0] :
return 1.0
return 0.0
architecture_str = "resnet_" + str(resnet_n_groups) + "_" + str(resnet_n_resblocks_per_group) + "_" + str(resnet_n_channels) + "_" + str(resnet_window_size) + "_" + str(resnet_drop_rate).replace(".", "")
model_name = "autoscrambler_dataset_" + dataset_name + "_" + architecture_str + "_n_epochs_" + str(n_epochs) + "_target_lum_" + str(target_lum).replace(".", "") + experiment_suffix
print("Model save name = " + model_name)
#Execute training procedure
callbacks =[
#ModelCheckpoint("model_checkpoints/" + model_name + "_epoch_{epoch:02d}.hdf5", monitor='val_loss', mode='min', period=10, save_weights_only=True),
EpochVariableCallback(anneal_coeff, _anneal_func)
]
s_train = np.zeros((x_train.shape[0], 1))
s_test = np.zeros((x_test.shape[0], 1))
# train the autoencoder
train_history = loss_model.fit(
[x_train],
[s_train, s_train, s_train],
shuffle=True,
epochs=n_epochs,
batch_size=batch_size,
validation_data=(
[x_test],
[s_test, s_test, s_test]
),
callbacks=callbacks
)
save_figs = False
f, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(3 * 4, 3))
n_epochs_actual = len(train_history.history['nll_loss'])
ax1.plot(np.arange(1, n_epochs_actual + 1), train_history.history['nll_loss'], linewidth=3, color='green')
ax1.plot(np.arange(1, n_epochs_actual + 1), train_history.history['val_nll_loss'], linewidth=3, color='orange')
plt.sca(ax1)
plt.xlabel("Epochs", fontsize=14)
plt.ylabel("NLL", fontsize=14)
plt.xlim(1, n_epochs_actual)
plt.xticks([1, n_epochs_actual], [1, n_epochs_actual], fontsize=12)
plt.yticks(fontsize=12)
ax2.plot(np.arange(1, n_epochs_actual + 1), train_history.history['entropy_loss'], linewidth=3, color='green')
ax2.plot(np.arange(1, n_epochs_actual + 1), train_history.history['val_entropy_loss'], linewidth=3, color='orange')
plt.sca(ax2)
plt.xlabel("Epochs", fontsize=14)
plt.ylabel("Entropy Loss", fontsize=14)
plt.xlim(1, n_epochs_actual)
plt.xticks([1, n_epochs_actual], [1, n_epochs_actual], fontsize=12)
plt.yticks(fontsize=12)
ax3.plot(np.arange(1, n_epochs_actual + 1), train_history.history['conservation_loss'], linewidth=3, color='green')
ax3.plot(np.arange(1, n_epochs_actual + 1), train_history.history['val_conservation_loss'], linewidth=3, color='orange')
plt.sca(ax3)
plt.xlabel("Epochs", fontsize=14)
plt.ylabel("Conservation Loss", fontsize=14)
plt.xlim(1, n_epochs_actual)
plt.xticks([1, n_epochs_actual], [1, n_epochs_actual], fontsize=12)
plt.yticks(fontsize=12)
plt.tight_layout()
if save_figs :
plt.savefig(model_name + "_loss.png", transparent=True, dpi=300)
plt.savefig(model_name + "_loss.eps")
plt.show()
# Save model and weights
save_dir = 'saved_models'
if not os.path.isdir(save_dir):
os.makedirs(save_dir)
model_path = os.path.join(save_dir, model_name + '.h5')
scrambler_model.save(model_path)
print('Saved scrambler model at %s ' % (model_path))
#Load models
save_dir = 'saved_models'
if not os.path.isdir(save_dir):
os.makedirs(save_dir)
model_path = os.path.join(save_dir, model_name + '.h5')
scrambler_model = load_model(model_path, custom_objects={
'bernoulli_sample' : bernoulli_sample,
'tf' : tf,
'sample_mask_gumbel' : sample_mask_gumbel
})
print('Loaded scrambler model %s ' % (model_path))
#Visualize a few reconstructed images
from numpy.ma import masked_array
digit_test = np.argmax(y_test, axis=1)
s_test = np.zeros((x_test.shape[0], 1))
sample_test, importance_scores_test, importance_scores_log_test = scrambler_model.predict_on_batch(x=[x_test[:32]])
importance_scores_test = (importance_scores_log_test[..., 0] - np.mean(importance_scores_log_test, axis=-1))[..., None]
save_images = [3, 4]
for plot_i in range(0, 20) :
print("Test image " + str(plot_i) + ":")
y_test_hat_ref = predictor.predict(x=[np.expand_dims(x_test[plot_i], axis=0)], batch_size=1)[0, digit_test[plot_i]]
y_test_hat = predictor.predict(x=[sample_test[plot_i:plot_i+1]], batch_size=1)[:, digit_test[plot_i]].tolist()
print(" - Prediction (original) = " + str(round(y_test_hat_ref, 2))[:4])
print(" - Predictions (scrambled) = " + str([float(str(round(y_test_hat[i], 2))[:4]) for i in range(len(y_test_hat))]))
f, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, figsize=(3 * 4, 3))
ax1.imshow(x_test[plot_i, :, :, 0], cmap="Greys", vmin=0.0, vmax=1.0, aspect='equal')
plt.sca(ax1)
plt.xticks([], [])
plt.yticks([], [])
ax2.imshow(x_test[plot_i, :, :, 0], cmap="Greys", vmin=0.0, vmax=1.0, aspect='equal')
plt.sca(ax2)
plt.xticks([], [])
plt.yticks([], [])
ax3.imshow(importance_scores_test[plot_i, :, :, 0], cmap="hot", vmin=0.0, vmax=np.max(importance_scores_test[plot_i, :, :, 0]), aspect='equal')
plt.sca(ax3)
plt.xticks([], [])
plt.yticks([], [])
ax4.imshow(x_test[plot_i, :, :, 0], cmap="Greys", vmin=0.0, vmax=1.0, aspect='equal')
plt.sca(ax4)
plt.xticks([], [])
plt.yticks([], [])
ax4.imshow(importance_scores_test[plot_i, :, :, 0], alpha=0.75, cmap="hot", vmin=0.0, vmax=np.max(importance_scores_test[plot_i, :, :, 0]), aspect='equal')
plt.sca(ax4)
plt.xticks([], [])
plt.yticks([], [])
plt.tight_layout()
if save_images is not None and plot_i in save_images :
plt.savefig(model_name + "_test_example_" + str(plot_i) + ".png", transparent=True, dpi=300)
plt.savefig(model_name + "_test_example_" + str(plot_i) + ".eps")
plt.show()
#Visualize a few reconstructed images
digit_test = np.argmax(y_test, axis=1)
s_test = np.zeros((x_test.shape[0], 1))
sample_test, importance_scores_test, importance_scores_log_test = scrambler_model.predict(x=[x_test], batch_size=32, verbose=True)
importance_scores_test = (importance_scores_log_test[..., 0] - np.mean(importance_scores_log_test, axis=-1))[..., None]
#Save predicted importance scores
np.save(model_name + "_importance_scores_test", importance_scores_test)
```
|
github_jupyter
|
import keras
from keras.models import Sequential, Model, load_model
from keras.layers import Dense, Dropout, Activation, Flatten, Input, Lambda
from keras.layers import Conv2D, MaxPooling2D, AveragePooling2D, Conv1D, MaxPooling1D, LSTM, ConvLSTM2D, GRU, CuDNNLSTM, CuDNNGRU, BatchNormalization, LocallyConnected2D, Permute, TimeDistributed, Bidirectional
from keras.layers import Concatenate, Reshape, Softmax, Conv2DTranspose, Embedding, Multiply
from keras.callbacks import ModelCheckpoint, EarlyStopping, Callback
from keras import regularizers
from keras import backend as K
from keras.utils.generic_utils import Progbar
from keras.layers.merge import _Merge
import keras.losses
from keras.datasets import mnist
from functools import partial
from collections import defaultdict
import tensorflow as tf
from tensorflow.python.framework import ops
import isolearn.keras as iso
import numpy as np
import tensorflow as tf
import logging
logging.getLogger('tensorflow').setLevel(logging.ERROR)
import os
import pickle
import numpy as np
import scipy.sparse as sp
import scipy.io as spio
import matplotlib.pyplot as plt
from keras.backend.tensorflow_backend import set_session
def contain_tf_gpu_mem_usage() :
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
sess = tf.Session(config=config)
set_session(sess)
contain_tf_gpu_mem_usage()
class EpochVariableCallback(Callback) :
def __init__(self, my_variable, my_func) :
self.my_variable = my_variable
self.my_func = my_func
def on_epoch_begin(self, epoch, logs={}) :
K.set_value(self.my_variable, self.my_func(K.get_value(self.my_variable), epoch))
#Load MNIST data
dataset_name = "mnist_3_vs_5"
img_rows, img_cols = 28, 28
num_classes = 10
batch_size = 32
included_classes = { 3, 5 }
(x_train, y_train), (x_test, y_test) = mnist.load_data()
keep_index_train = []
for i in range(y_train.shape[0]) :
if y_train[i] in included_classes :
keep_index_train.append(i)
keep_index_test = []
for i in range(y_test.shape[0]) :
if y_test[i] in included_classes :
keep_index_test.append(i)
x_train = x_train[keep_index_train]
x_test = x_test[keep_index_test]
y_train = y_train[keep_index_train]
y_test = y_test[keep_index_test]
n_train = int((x_train.shape[0] // batch_size) * batch_size)
n_test = int((x_test.shape[0] // batch_size) * batch_size)
x_train = x_train[:n_train]
x_test = x_test[:n_test]
y_train = y_train[:n_train]
y_test = y_test[:n_test]
x_train = x_train.reshape(x_train.shape[0], img_rows, img_cols, 1)
x_test = x_test.reshape(x_test.shape[0], img_rows, img_cols, 1)
input_shape = (img_rows, img_cols, 1)
x_train = x_train.astype('float32')
x_test = x_test.astype('float32')
x_train /= 255
x_test /= 255
print("x_train.shape = " + str(x_train.shape))
print("n train samples = " + str(x_train.shape[0]))
print("n test samples = " + str(x_test.shape[0]))
y_train = keras.utils.to_categorical(y_train, num_classes)
y_test = keras.utils.to_categorical(y_test, num_classes)
#Binarize images
def _binarize_images(x, val_thresh=0.5) :
x_bin = np.zeros(x.shape)
x_bin[x >= val_thresh] = 1.
return x_bin
x_train = _binarize_images(x_train, val_thresh=0.5)
x_test = _binarize_images(x_test, val_thresh=0.5)
from tensorflow.python.framework import ops
#Stochastic Binarized Neuron helper functions (Tensorflow)
#ST Estimator code adopted from https://r2rt.com/binary-stochastic-neurons-in-tensorflow.html
#See Github https://github.com/spitis/
def bernoulli_sample(x):
g = tf.get_default_graph()
with ops.name_scope("BernoulliSample") as name:
with g.gradient_override_map({"Ceil": "Identity","Sub": "BernoulliSample_ST"}):
return tf.ceil(x - tf.random_uniform(tf.shape(x)), name=name)
@ops.RegisterGradient("BernoulliSample_ST")
def bernoulliSample_ST(op, grad):
return [grad, tf.zeros(tf.shape(op.inputs[1]))]
#Scrambler network definition
#Gumbel Distribution Sampler
def gumbel_softmax(logits, temperature=0.5) :
gumbel_dist = tf.contrib.distributions.RelaxedOneHotCategorical(temperature, logits=logits)
batch_dim = logits.get_shape().as_list()[0]
onehot_dim = logits.get_shape().as_list()[1]
return gumbel_dist.sample()
def sample_mask_gumbel(pwm_logits) :
n_sequences = K.shape(pwm_logits)[0]
x_len = K.shape(pwm_logits)[1]
y_len = K.shape(pwm_logits)[2]
flat_pwm = K.reshape(pwm_logits, (n_sequences * x_len * y_len, 2))
sampled_pwm = gumbel_softmax(flat_pwm, temperature=0.5)
return K.reshape(sampled_pwm, (n_sequences, x_len, y_len, 2))[..., :1]
def make_resblock(n_channels=64, window_size=8, dilation_rate=1, group_ix=0, layer_ix=0, drop_rate=0.0, batchnorm_trainmode=True) :
#Initialize res block layers
batch_norm_0 = BatchNormalization(name='scrambler_resblock_' + str(group_ix) + '_' + str(layer_ix) + '_batch_norm_0')
relu_0 = Lambda(lambda x: K.relu(x, alpha=0.0))
conv_0 = Conv2D(n_channels, (window_size, window_size), dilation_rate=dilation_rate, strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_resblock_' + str(group_ix) + '_' + str(layer_ix) + '_conv_0')
batch_norm_1 = BatchNormalization(name='scrambler_resblock_' + str(group_ix) + '_' + str(layer_ix) + '_batch_norm_1')
relu_1 = Lambda(lambda x: K.relu(x, alpha=0.0))
conv_1 = Conv2D(n_channels, (window_size, window_size), dilation_rate=dilation_rate, strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_resblock_' + str(group_ix) + '_' + str(layer_ix) + '_conv_1')
skip_1 = Lambda(lambda x: x[0] + x[1], name='scrambler_resblock_' + str(group_ix) + '_' + str(layer_ix) + '_skip_1')
drop_1 = None
if drop_rate > 0.0 :
drop_1 = Dropout(drop_rate)
#Execute res block
def _resblock_func(input_tensor, training=batchnorm_trainmode) :
batch_norm_0_out = batch_norm_0(input_tensor, training=training)
relu_0_out = relu_0(batch_norm_0_out)
conv_0_out = conv_0(relu_0_out)
batch_norm_1_out = batch_norm_1(conv_0_out, training=training)
relu_1_out = relu_1(batch_norm_1_out)
if drop_rate > 0.0 :
conv_1_out = drop_1(conv_1(relu_1_out))
else :
conv_1_out = conv_1(relu_1_out)
skip_1_out = skip_1([conv_1_out, input_tensor])
return skip_1_out
return _resblock_func
def load_scrambler_network(n_groups=1, n_resblocks_per_group=4, n_channels=32, window_size=8, dilation_rates=[1], drop_rate=0.0, batchnorm_trainmode=True) :
#Discriminator network definition
conv_0 = Conv2D(n_channels, (1, 1), strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_conv_0')
skip_convs = []
resblock_groups = []
for group_ix in range(n_groups) :
skip_convs.append(Conv2D(n_channels, (1, 1), strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_skip_conv_' + str(group_ix)))
resblocks = []
for layer_ix in range(n_resblocks_per_group) :
resblocks.append(make_resblock(n_channels=n_channels, window_size=window_size, dilation_rate=dilation_rates[group_ix], group_ix=group_ix, layer_ix=layer_ix, drop_rate=drop_rate, batchnorm_trainmode=batchnorm_trainmode))
resblock_groups.append(resblocks)
last_block_conv = Conv2D(n_channels, (1, 1), strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_last_block_conv')
skip_add = Lambda(lambda x: x[0] + x[1], name='scrambler_skip_add')
final_conv_2_channels = Conv2D(2, (1, 1), strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_final_conv')
final_conv_sigm = Lambda(lambda x: K.softmax(x, axis=-1)[..., :1])
final_conv_gumbel = Lambda(lambda x: sample_mask_gumbel(x))
scale_inputs = Lambda(lambda x: x[1] * K.tile(x[0], (1, 1, 1, 1)), name='scrambler_input_scale')
def _scrambler_func(image_input) :
conv_0_out = conv_0(image_input)
#Connect group of res blocks
output_tensor = conv_0_out
#Res block group execution
skip_conv_outs = []
for group_ix in range(n_groups) :
skip_conv_out = skip_convs[group_ix](output_tensor)
skip_conv_outs.append(skip_conv_out)
for layer_ix in range(n_resblocks_per_group) :
output_tensor = resblock_groups[group_ix][layer_ix](output_tensor)
#Last res block extr conv
last_block_conv_out = last_block_conv(output_tensor)
skip_add_out = last_block_conv_out
for group_ix in range(n_groups) :
skip_add_out = skip_add([skip_add_out, skip_conv_outs[group_ix]])
#Final conv out
final_conv_2_channels_out = final_conv_2_channels(skip_add_out)
final_conv_sigm_out = final_conv_sigm(final_conv_2_channels_out)
final_conv_gumbel_out = final_conv_gumbel(final_conv_2_channels_out)
#Scale inputs by importance scores
scaled_inputs = scale_inputs([final_conv_gumbel_out, image_input])
return scaled_inputs, final_conv_sigm_out, final_conv_2_channels_out
return _scrambler_func
#Keras loss functions
def get_softmax_kl_divergence() :
def _softmax_kl_divergence(y_true, y_pred) :
y_true = K.clip(y_true, K.epsilon(), 1.0 - K.epsilon())
y_pred = K.clip(y_pred, K.epsilon(), 1.0 - K.epsilon())
return K.mean(K.sum(y_true * K.log(y_true / y_pred), axis=-1), axis=-1)
return _softmax_kl_divergence
def get_margin_lum_ame_masked(max_lum=0.1) :
def _margin_lum_ame(importance_scores) :
p_ons = importance_scores[..., 0]
mean_p_on = K.mean(p_ons, axis=(1, 2))
margin_p_on = K.switch(mean_p_on > max_lum, mean_p_on - max_lum, K.zeros_like(mean_p_on))
return margin_p_on
return _margin_lum_ame
def get_target_lum_sme_masked(target_lum=0.1) :
def _target_lum_sme(importance_scores) :
p_ons = importance_scores[..., 0]
mean_p_on = K.mean(p_ons, axis=(1, 2))
return (mean_p_on - target_lum)**2
return _target_lum_sme
def get_weighted_loss(loss_coeff=1.) :
def _min_pred(y_true, y_pred) :
return loss_coeff * y_pred
return _min_pred
#Initialize Encoder and Decoder networks
batch_size = 32
n_rows = 28
n_cols = 28
#Resnet parameters
resnet_n_groups = 5
resnet_n_resblocks_per_group = 4
resnet_n_channels = 32
resnet_window_size = 3
resnet_dilation_rates = [1, 2, 4, 2, 1]
resnet_drop_rate = 0.0#0.25
resnet_batchnorm_trainmode = True
#Load scrambler
scrambler = load_scrambler_network(
n_groups=resnet_n_groups,
n_resblocks_per_group=resnet_n_resblocks_per_group,
n_channels=resnet_n_channels, window_size=resnet_window_size,
dilation_rates=resnet_dilation_rates,
drop_rate=resnet_drop_rate,
batchnorm_trainmode=resnet_batchnorm_trainmode
)
#Load Predictor
predictor_path = 'saved_models/mnist_binarized_cnn_10_digits.h5'
predictor = load_model(predictor_path)
predictor.trainable = False
predictor.compile(optimizer=keras.optimizers.SGD(lr=0.1), loss='mean_squared_error')
#Build scrambler model
scrambler_input = Input(shape=(n_rows, n_cols, 1), name='scrambler_input')
image, importance_scores, importance_scores_log = scrambler(scrambler_input)
scrambler_model = Model([scrambler_input], [image, importance_scores, importance_scores_log])
scrambler_model.compile(
optimizer=keras.optimizers.Adam(lr=0.001, beta_1=0.9, beta_2=0.999),
loss='mean_squared_error'
)
#Build Auto-scrambler pipeline
#Define model inputs
ae_scrambler_input = Input(shape=(n_rows, n_cols, 1), name='ae_scrambler_input')
#Run encoder and decoder
scrambled_sample, importance_scores, importance_scores_log = scrambler(ae_scrambler_input)
#Make reference prediction on non-scrambled input sequence
y_pred_non_scrambled = predictor([ae_scrambler_input])
#Make prediction on scrambled sequence samples
y_pred_scrambled = predictor([scrambled_sample])
#Cost function parameters
initial_target_lum = 0.25
target_lum = 0.05
#NLL cost
nll_loss_func = get_softmax_kl_divergence()
#Conservation cost
conservation_loss_func = get_target_lum_sme_masked(target_lum=initial_target_lum)
#Entropy cost
entropy_loss_func = get_target_lum_sme_masked(target_lum=target_lum)
#entropy_loss_func = get_margin_lum_ame_masked(max_lum=target_lum)
#Define annealing coefficient
anneal_coeff = K.variable(1.0)
#Execute NLL cost
nll_loss = Lambda(lambda x: nll_loss_func(x[0], x[1]), name='nll')([y_pred_non_scrambled, y_pred_scrambled])
#Execute conservation cost
conservation_loss = Lambda(lambda x: anneal_coeff * conservation_loss_func(x), name='conservation')(importance_scores)
#Execute entropy cost
entropy_loss = Lambda(lambda x: (1. - anneal_coeff) * entropy_loss_func(x), name='entropy')(importance_scores)
loss_model = Model(
[ae_scrambler_input],
[nll_loss, conservation_loss, entropy_loss]
)
loss_model.compile(
optimizer=keras.optimizers.Adam(lr=0.0001, beta_1=0.5, beta_2=0.9),
loss={
'nll' : get_weighted_loss(loss_coeff=1.0),
'conservation' : get_weighted_loss(loss_coeff=1.0),
'entropy' : get_weighted_loss(loss_coeff=500.0)
}
)
scrambler_model.summary()
loss_model.summary()
#Training configuration
#Define number of training epochs
n_epochs = 50
#Define experiment suffix (optional)
experiment_suffix = "_kl_divergence_higher_entropy_penalty_gumbel_no_bg_lum"
#Define anneal function
def _anneal_func(val, epoch, n_epochs=n_epochs) :
if epoch in [0] :
return 1.0
return 0.0
architecture_str = "resnet_" + str(resnet_n_groups) + "_" + str(resnet_n_resblocks_per_group) + "_" + str(resnet_n_channels) + "_" + str(resnet_window_size) + "_" + str(resnet_drop_rate).replace(".", "")
model_name = "autoscrambler_dataset_" + dataset_name + "_" + architecture_str + "_n_epochs_" + str(n_epochs) + "_target_lum_" + str(target_lum).replace(".", "") + experiment_suffix
print("Model save name = " + model_name)
#Execute training procedure
callbacks =[
#ModelCheckpoint("model_checkpoints/" + model_name + "_epoch_{epoch:02d}.hdf5", monitor='val_loss', mode='min', period=10, save_weights_only=True),
EpochVariableCallback(anneal_coeff, _anneal_func)
]
s_train = np.zeros((x_train.shape[0], 1))
s_test = np.zeros((x_test.shape[0], 1))
# train the autoencoder
train_history = loss_model.fit(
[x_train],
[s_train, s_train, s_train],
shuffle=True,
epochs=n_epochs,
batch_size=batch_size,
validation_data=(
[x_test],
[s_test, s_test, s_test]
),
callbacks=callbacks
)
save_figs = False
f, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(3 * 4, 3))
n_epochs_actual = len(train_history.history['nll_loss'])
ax1.plot(np.arange(1, n_epochs_actual + 1), train_history.history['nll_loss'], linewidth=3, color='green')
ax1.plot(np.arange(1, n_epochs_actual + 1), train_history.history['val_nll_loss'], linewidth=3, color='orange')
plt.sca(ax1)
plt.xlabel("Epochs", fontsize=14)
plt.ylabel("NLL", fontsize=14)
plt.xlim(1, n_epochs_actual)
plt.xticks([1, n_epochs_actual], [1, n_epochs_actual], fontsize=12)
plt.yticks(fontsize=12)
ax2.plot(np.arange(1, n_epochs_actual + 1), train_history.history['entropy_loss'], linewidth=3, color='green')
ax2.plot(np.arange(1, n_epochs_actual + 1), train_history.history['val_entropy_loss'], linewidth=3, color='orange')
plt.sca(ax2)
plt.xlabel("Epochs", fontsize=14)
plt.ylabel("Entropy Loss", fontsize=14)
plt.xlim(1, n_epochs_actual)
plt.xticks([1, n_epochs_actual], [1, n_epochs_actual], fontsize=12)
plt.yticks(fontsize=12)
ax3.plot(np.arange(1, n_epochs_actual + 1), train_history.history['conservation_loss'], linewidth=3, color='green')
ax3.plot(np.arange(1, n_epochs_actual + 1), train_history.history['val_conservation_loss'], linewidth=3, color='orange')
plt.sca(ax3)
plt.xlabel("Epochs", fontsize=14)
plt.ylabel("Conservation Loss", fontsize=14)
plt.xlim(1, n_epochs_actual)
plt.xticks([1, n_epochs_actual], [1, n_epochs_actual], fontsize=12)
plt.yticks(fontsize=12)
plt.tight_layout()
if save_figs :
plt.savefig(model_name + "_loss.png", transparent=True, dpi=300)
plt.savefig(model_name + "_loss.eps")
plt.show()
# Save model and weights
save_dir = 'saved_models'
if not os.path.isdir(save_dir):
os.makedirs(save_dir)
model_path = os.path.join(save_dir, model_name + '.h5')
scrambler_model.save(model_path)
print('Saved scrambler model at %s ' % (model_path))
#Load models
save_dir = 'saved_models'
if not os.path.isdir(save_dir):
os.makedirs(save_dir)
model_path = os.path.join(save_dir, model_name + '.h5')
scrambler_model = load_model(model_path, custom_objects={
'bernoulli_sample' : bernoulli_sample,
'tf' : tf,
'sample_mask_gumbel' : sample_mask_gumbel
})
print('Loaded scrambler model %s ' % (model_path))
#Visualize a few reconstructed images
from numpy.ma import masked_array
digit_test = np.argmax(y_test, axis=1)
s_test = np.zeros((x_test.shape[0], 1))
sample_test, importance_scores_test, importance_scores_log_test = scrambler_model.predict_on_batch(x=[x_test[:32]])
importance_scores_test = (importance_scores_log_test[..., 0] - np.mean(importance_scores_log_test, axis=-1))[..., None]
save_images = [3, 4]
for plot_i in range(0, 20) :
print("Test image " + str(plot_i) + ":")
y_test_hat_ref = predictor.predict(x=[np.expand_dims(x_test[plot_i], axis=0)], batch_size=1)[0, digit_test[plot_i]]
y_test_hat = predictor.predict(x=[sample_test[plot_i:plot_i+1]], batch_size=1)[:, digit_test[plot_i]].tolist()
print(" - Prediction (original) = " + str(round(y_test_hat_ref, 2))[:4])
print(" - Predictions (scrambled) = " + str([float(str(round(y_test_hat[i], 2))[:4]) for i in range(len(y_test_hat))]))
f, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, figsize=(3 * 4, 3))
ax1.imshow(x_test[plot_i, :, :, 0], cmap="Greys", vmin=0.0, vmax=1.0, aspect='equal')
plt.sca(ax1)
plt.xticks([], [])
plt.yticks([], [])
ax2.imshow(x_test[plot_i, :, :, 0], cmap="Greys", vmin=0.0, vmax=1.0, aspect='equal')
plt.sca(ax2)
plt.xticks([], [])
plt.yticks([], [])
ax3.imshow(importance_scores_test[plot_i, :, :, 0], cmap="hot", vmin=0.0, vmax=np.max(importance_scores_test[plot_i, :, :, 0]), aspect='equal')
plt.sca(ax3)
plt.xticks([], [])
plt.yticks([], [])
ax4.imshow(x_test[plot_i, :, :, 0], cmap="Greys", vmin=0.0, vmax=1.0, aspect='equal')
plt.sca(ax4)
plt.xticks([], [])
plt.yticks([], [])
ax4.imshow(importance_scores_test[plot_i, :, :, 0], alpha=0.75, cmap="hot", vmin=0.0, vmax=np.max(importance_scores_test[plot_i, :, :, 0]), aspect='equal')
plt.sca(ax4)
plt.xticks([], [])
plt.yticks([], [])
plt.tight_layout()
if save_images is not None and plot_i in save_images :
plt.savefig(model_name + "_test_example_" + str(plot_i) + ".png", transparent=True, dpi=300)
plt.savefig(model_name + "_test_example_" + str(plot_i) + ".eps")
plt.show()
#Visualize a few reconstructed images
digit_test = np.argmax(y_test, axis=1)
s_test = np.zeros((x_test.shape[0], 1))
sample_test, importance_scores_test, importance_scores_log_test = scrambler_model.predict(x=[x_test], batch_size=32, verbose=True)
importance_scores_test = (importance_scores_log_test[..., 0] - np.mean(importance_scores_log_test, axis=-1))[..., None]
#Save predicted importance scores
np.save(model_name + "_importance_scores_test", importance_scores_test)
| 0.882098 | 0.377598 |
# MongoDB playing with Tags in python
```
%matplotlib inline
import pymongo
from pymongo import MongoClient
from pymongo.errors import ConnectionFailure
from bson import json_util, ObjectId
import pandas as pd
from pandas import DataFrame
from pandas.io.json import json_normalize
import numpy as np
import requests
import json, os
import configparser
import matplotlib.pyplot as plt
import numpy as np
from scipy import stats
import seaborn as sns
import warnings
import random
import pprint
from datetime import datetime
random.seed(datetime.now())
warnings.filterwarnings('ignore')
# Make plots larger
plt.rcParams['figure.figsize'] = (10, 6)
client = MongoClient('localhost', 27017)
db=client.tweets
db.collection_names(include_system_collections=False)
try:
result = db.tweets.drop()
print ("analytics tweets dropped")
except:
pass
def load_json(j):
p=os.path.join("data/", j)
print (p)
with open(p, 'rU') as f:
data = [json.loads(row) for row in f]
return data
tweets_j=load_json('db_tweets.json')
tweets_j[0]
print(tweets_j[0]['text'])
def tweet_json(tid,text,created,favorite_count,retweet_count,urls,tags):
j={
"tweet_id" : tid,
"text" : text,
"favorite_count" : favorite_count,
"retweet_count" : retweet_count,
"urls" : urls,
"tags" : tags,
"created_at" : created}
return j
# Twitter dates are of the form Sun Mar 15 21:41:54 +0000 2015
datestrings=['Sun Mar 15 21:41:54 +0000 2015','Tue Mar 29 08:11:25 +0000 2011']
from datetime import timedelta
from email.utils import parsedate_tz
from dateutil.parser import parse
def to_datetime(datestring):
time_tuple = parsedate_tz(datestring.strip())
dt = datetime(*time_tuple[:6])
return dt - timedelta(seconds=time_tuple[-1])
ts=to_datetime(datestrings[0])
print (ts.strftime("%Y-%m-%d %H:%M"))
ts=to_datetime(datestrings[1])
print (ts.strftime("%Y-%m-%d %H:%M"))
hashtags={}
starter_tags=['Bigdata','big data','algorithm','big data','AI','MongoDB','SQL','artificial intelligence','machine learning']
for tag in starter_tags:
hashtags[tag]=0
urls={}
tags={}
print(hashtags)
sample_tweet_text="RT @TheHesterView Tutorials on big data, big data, AI, MongoDB, SQL, artificial intelligence, machine learning. hackathons, crowdsourcing, #bigdata http://t.co/6HWjCv3BL5 Lets join "
print (sample_tweet_text)
import re
def tokenize(txt):
txt=re.sub(r'\n', ' ',txt)
txt=re.compile(r'[\.][ ]+').sub(' ',txt)
txt=re.compile(r'[\,][ ]+').sub(' ',txt)
txt=re.compile(r'[_+;=!@$%^&\*\"\?]').sub(' ',txt)
splitter=re.compile(r'[ ]+')
# Split the words by non-alpha characters
words=splitter.split(txt)
return words
print (tokenize(sample_tweet_text))
s='http://t.co/6HWjCv3BL5'
print (s[0:4].lower())
from nltk.corpus import stopwords
stop_words_list = list(stopwords.words('english'))
stop_words={}
for tag in stop_words_list:
stop_words[tag]=0
print (stop_words.keys())
def update_urls_tags(url_list,urls,hashtag_list,hashtags,tag_list,tags):
for url in url_list:
if url in urls:
urls[url]=urls[url]+1
else:
urls[url]=1
for tag in tag_list:
if tag in tags:
tags[tag]=tags[tag]+1
else:
tags[tag]=1
for hashtag in hashtag_list:
if hashtag in hashtags:
hashtags[hashtag]=hashtags[hashtag]+1
else:
hashtags[hashtag]=1
return urls,hashtags,tags
def extract_tags_urls(dct,words,stop):
i=0
tags={}
tokens={}
urls={}
size=len(words)
while i < size:
ngram = words[i]
i=i+1
if len(ngram) < 1: continue
if len(ngram) > 4:
if ngram[0:4].lower()=='http':
if ngram in urls:
urls[ngram]=urls[ngram]+1
else:
urls[ngram]=1
if ngram[0]=='#':
# ngram=re.sub(r'\#', '',ngram) if you want to remove the #
tags[ngram]=1
if ngram.lower() not in stop:
tokens[ngram]=1
if ngram in dct:
tags[ngram]=1
if i < (size-1):
ngram = words[i] + ' ' + words[i+1]
if words[i].lower() not in stop:
tokens[ngram]=1
if ngram in dct:
tags[ngram]=1
if i < (size-2):
ngram = words[i] + ' ' + words[i+1] + ' ' + words[i+2]
if ngram in dct:
tags[ngram]=1
return list(tags.keys()),list(urls.keys()),list(tokens.keys())
print (extract_tags_urls(hashtags,(tokenize(sample_tweet_text)),stop_words))
cnt=0
for tweet in tweets_j:
ts=datetime.now()
try:
ts=to_datetime(tweet['created_at'])
except:
continue
favorite_count=0
try:
favorite_count=int(tweet['favorite_count'])
except:
pass
retweet_count=0
try:
retweet_count=int(tweet['retweet_count'])
except:
pass
tweet_tags,tweet_urls,tweet_ngrams=extract_tags_urls(hashtags,(tokenize(tweet['text'])),stop_words)
urls,hashtags,tags=update_urls_tags(tweet_urls,urls,tweet_tags,hashtags,tweet_ngrams,tags)
try:
j=tweet_json(tweet['id'],tweet['text'],ts,favorite_count,retweet_count,tweet_urls,tweet_tags)
result = db.tweets.insert_one(j)
cnt+=1
except:
pass
print ("%d tweets inserted."%cnt)
for tweet in db.tweets.find().limit(9):
pprint.pprint(tweet)
for key, value in hashtags.items():
print ("%s count %d"%(key, value))
def trim_dct(dct,thresh):
tmp={}
keys=dct.keys()
for key in keys:
if dct[key] > thresh:
tmp[key]=dct[key]
return tmp
'''
for key, value in tags.items():
print ("%s count %d"%(key, value))
'''
tags=trim_dct(tags,3)
for key, value in tags.items():
print ("%s count %d"%(key, value))
for key, value in urls.items():
print ("%s count %d"%(key, value))
```
Updated October 3, 2017
|
github_jupyter
|
%matplotlib inline
import pymongo
from pymongo import MongoClient
from pymongo.errors import ConnectionFailure
from bson import json_util, ObjectId
import pandas as pd
from pandas import DataFrame
from pandas.io.json import json_normalize
import numpy as np
import requests
import json, os
import configparser
import matplotlib.pyplot as plt
import numpy as np
from scipy import stats
import seaborn as sns
import warnings
import random
import pprint
from datetime import datetime
random.seed(datetime.now())
warnings.filterwarnings('ignore')
# Make plots larger
plt.rcParams['figure.figsize'] = (10, 6)
client = MongoClient('localhost', 27017)
db=client.tweets
db.collection_names(include_system_collections=False)
try:
result = db.tweets.drop()
print ("analytics tweets dropped")
except:
pass
def load_json(j):
p=os.path.join("data/", j)
print (p)
with open(p, 'rU') as f:
data = [json.loads(row) for row in f]
return data
tweets_j=load_json('db_tweets.json')
tweets_j[0]
print(tweets_j[0]['text'])
def tweet_json(tid,text,created,favorite_count,retweet_count,urls,tags):
j={
"tweet_id" : tid,
"text" : text,
"favorite_count" : favorite_count,
"retweet_count" : retweet_count,
"urls" : urls,
"tags" : tags,
"created_at" : created}
return j
# Twitter dates are of the form Sun Mar 15 21:41:54 +0000 2015
datestrings=['Sun Mar 15 21:41:54 +0000 2015','Tue Mar 29 08:11:25 +0000 2011']
from datetime import timedelta
from email.utils import parsedate_tz
from dateutil.parser import parse
def to_datetime(datestring):
time_tuple = parsedate_tz(datestring.strip())
dt = datetime(*time_tuple[:6])
return dt - timedelta(seconds=time_tuple[-1])
ts=to_datetime(datestrings[0])
print (ts.strftime("%Y-%m-%d %H:%M"))
ts=to_datetime(datestrings[1])
print (ts.strftime("%Y-%m-%d %H:%M"))
hashtags={}
starter_tags=['Bigdata','big data','algorithm','big data','AI','MongoDB','SQL','artificial intelligence','machine learning']
for tag in starter_tags:
hashtags[tag]=0
urls={}
tags={}
print(hashtags)
sample_tweet_text="RT @TheHesterView Tutorials on big data, big data, AI, MongoDB, SQL, artificial intelligence, machine learning. hackathons, crowdsourcing, #bigdata http://t.co/6HWjCv3BL5 Lets join "
print (sample_tweet_text)
import re
def tokenize(txt):
txt=re.sub(r'\n', ' ',txt)
txt=re.compile(r'[\.][ ]+').sub(' ',txt)
txt=re.compile(r'[\,][ ]+').sub(' ',txt)
txt=re.compile(r'[_+;=!@$%^&\*\"\?]').sub(' ',txt)
splitter=re.compile(r'[ ]+')
# Split the words by non-alpha characters
words=splitter.split(txt)
return words
print (tokenize(sample_tweet_text))
s='http://t.co/6HWjCv3BL5'
print (s[0:4].lower())
from nltk.corpus import stopwords
stop_words_list = list(stopwords.words('english'))
stop_words={}
for tag in stop_words_list:
stop_words[tag]=0
print (stop_words.keys())
def update_urls_tags(url_list,urls,hashtag_list,hashtags,tag_list,tags):
for url in url_list:
if url in urls:
urls[url]=urls[url]+1
else:
urls[url]=1
for tag in tag_list:
if tag in tags:
tags[tag]=tags[tag]+1
else:
tags[tag]=1
for hashtag in hashtag_list:
if hashtag in hashtags:
hashtags[hashtag]=hashtags[hashtag]+1
else:
hashtags[hashtag]=1
return urls,hashtags,tags
def extract_tags_urls(dct,words,stop):
i=0
tags={}
tokens={}
urls={}
size=len(words)
while i < size:
ngram = words[i]
i=i+1
if len(ngram) < 1: continue
if len(ngram) > 4:
if ngram[0:4].lower()=='http':
if ngram in urls:
urls[ngram]=urls[ngram]+1
else:
urls[ngram]=1
if ngram[0]=='#':
# ngram=re.sub(r'\#', '',ngram) if you want to remove the #
tags[ngram]=1
if ngram.lower() not in stop:
tokens[ngram]=1
if ngram in dct:
tags[ngram]=1
if i < (size-1):
ngram = words[i] + ' ' + words[i+1]
if words[i].lower() not in stop:
tokens[ngram]=1
if ngram in dct:
tags[ngram]=1
if i < (size-2):
ngram = words[i] + ' ' + words[i+1] + ' ' + words[i+2]
if ngram in dct:
tags[ngram]=1
return list(tags.keys()),list(urls.keys()),list(tokens.keys())
print (extract_tags_urls(hashtags,(tokenize(sample_tweet_text)),stop_words))
cnt=0
for tweet in tweets_j:
ts=datetime.now()
try:
ts=to_datetime(tweet['created_at'])
except:
continue
favorite_count=0
try:
favorite_count=int(tweet['favorite_count'])
except:
pass
retweet_count=0
try:
retweet_count=int(tweet['retweet_count'])
except:
pass
tweet_tags,tweet_urls,tweet_ngrams=extract_tags_urls(hashtags,(tokenize(tweet['text'])),stop_words)
urls,hashtags,tags=update_urls_tags(tweet_urls,urls,tweet_tags,hashtags,tweet_ngrams,tags)
try:
j=tweet_json(tweet['id'],tweet['text'],ts,favorite_count,retweet_count,tweet_urls,tweet_tags)
result = db.tweets.insert_one(j)
cnt+=1
except:
pass
print ("%d tweets inserted."%cnt)
for tweet in db.tweets.find().limit(9):
pprint.pprint(tweet)
for key, value in hashtags.items():
print ("%s count %d"%(key, value))
def trim_dct(dct,thresh):
tmp={}
keys=dct.keys()
for key in keys:
if dct[key] > thresh:
tmp[key]=dct[key]
return tmp
'''
for key, value in tags.items():
print ("%s count %d"%(key, value))
'''
tags=trim_dct(tags,3)
for key, value in tags.items():
print ("%s count %d"%(key, value))
for key, value in urls.items():
print ("%s count %d"%(key, value))
| 0.187728 | 0.404449 |
# EPA-Moves: Network Analysis
Len Strnad <[email protected]>
Let's take a look to see how dropping the off-network instances changes the speed distribution. It might be wise to drop the off-network in order to get more accurate on-network data for analysis. In the plots, the blue is what would be missing if we filtered out the off-network data.
```
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib import pyplot
font = {'size' : 20}
plt.rc('font', **font)
plt.rcParams["figure.figsize"] = (20,10)
import seaborn as sns
sns.set(rc={'figure.figsize':(20,10)})
```
## CombShortHaul
Summary Statistics and plots.
```
final = pd.read_csv('./maps_plots_data/CombShortHaul/CombShortHaulAggData.csv')
gpd = pd.read_csv('./maps_plots_data/CombShortHaul/CombShortHaulGPDnCOUNT.csv')
print "One Network is about ", 42.5/54.0
final.groupby("network").speed.count()
# plot the difference
plt.hist(final[(final.speed!=0) & (final.network == True)].speed, 100, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final[(final.speed!=0)].speed, 100, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution")
plt.show()
# plot the difference
plt.hist(final[(final.network == True)].speed, 100, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final.speed, 100, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution Including Zero Speeds")
plt.show()
```
## CombLongHaul
Summary Statistics and plots.
```
final = pd.read_csv('./maps_plots_data/CombLongHaul/CombLongHaulAggData.csv')
gpd = pd.read_csv('./maps_plots_data/CombLongHaul/CombLongHaulGPDnCOUNT.csv')
print "One Network is about ", 60/71.0
final.groupby("network").speed.count()
# plot the difference
plt.hist(final[(final.speed!=0) & (final.network == True)].speed, 100, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final[(final.speed!=0)].speed, 100, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution")
plt.show()
# plot the difference
plt.hist(final[(final.network == True)].speed, 100, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final.speed, 100, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution Including Zero Speeds")
plt.show()
```
## School Buses
Summary Statistics and plots.
```
final = pd.read_csv('./maps_plots_data/Schoolbus/SchoolBusAggData.csv')
gpd = pd.read_csv('./maps_plots_data/Schoolbus/SchoolBusGPDnCOUNT.csv')
print "One Network is about ", 36/44.0
final.groupby("network").speed.count()
# plot the difference
plt.hist(final[(final.speed!=0) & (final.network == True)].speed, 50, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final[(final.speed!=0)].speed, 50, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution")
plt.show()
# plot the difference
plt.hist(final[(final.network == True)].speed, 100, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final.speed, 100, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution Including Zero Speeds")
plt.show()
```
## Refuse Trucks
Summary Statistics and plots.
```
final = pd.read_csv('./maps_plots_data/RefuseTrucks/RefuseTrucksAggData.csv')
gpd = pd.read_csv('./maps_plots_data/RefuseTrucks/RefuseTrucksGPDnCOUNT.csv')
print "One Network is about ", 11.5/12.0
final.groupby("network").speed.count()
# plot the difference
plt.hist(final[(final.speed!=0) & (final.network == True)].speed, 50, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final[(final.speed!=0)].speed, 50, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution")
plt.show()
# plot the difference
plt.hist(final[(final.network == True)].speed, 100, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final.speed, 100, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution Including Zero Speeds")
plt.show()
```
## Transit Buses
Summary Statistics and plots.
```
final = pd.read_csv('./maps_plots_data/Transitbus/TransitbusAggData.csv')
gpd = pd.read_csv('./maps_plots_data/Transitbus/TransitbusGPDnCOUNT.csv')
print "One Network is about ", 15/18.0
final.groupby("network").speed.count()
# plot the difference
plt.hist(final[(final.speed!=0) & (final.network == True)].speed, 50, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final[(final.speed!=0)].speed, 50, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution")
plt.show()
# plot the difference
plt.hist(final[(final.network == True)].speed, 100, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final.speed, 100, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution Including Zero Speeds")
plt.show()
```
## Single Short Haul
Summary Statistics and plots.
```
final = pd.read_csv('./maps_plots_data/SingleShortHaul/SingleShortAggData.csv')
gpd = pd.read_csv('./maps_plots_data/SingleShortHaul/SingleShortGPDnCOUNT.csv')
print "One Network is about ", 28.9/30.1
final.groupby("network").speed.count()
# plot the difference
plt.hist(final[(final.speed!=0) & (final.network == True)].speed, 50, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final[(final.speed!=0)].speed, 50, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution")
plt.show()
# plot the difference
plt.hist(final[(final.network == True)].speed, 100, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final.speed, 100, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution Including Zero Speeds")
plt.show()
```
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib import pyplot
font = {'size' : 20}
plt.rc('font', **font)
plt.rcParams["figure.figsize"] = (20,10)
import seaborn as sns
sns.set(rc={'figure.figsize':(20,10)})
final = pd.read_csv('./maps_plots_data/CombShortHaul/CombShortHaulAggData.csv')
gpd = pd.read_csv('./maps_plots_data/CombShortHaul/CombShortHaulGPDnCOUNT.csv')
print "One Network is about ", 42.5/54.0
final.groupby("network").speed.count()
# plot the difference
plt.hist(final[(final.speed!=0) & (final.network == True)].speed, 100, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final[(final.speed!=0)].speed, 100, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution")
plt.show()
# plot the difference
plt.hist(final[(final.network == True)].speed, 100, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final.speed, 100, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution Including Zero Speeds")
plt.show()
final = pd.read_csv('./maps_plots_data/CombLongHaul/CombLongHaulAggData.csv')
gpd = pd.read_csv('./maps_plots_data/CombLongHaul/CombLongHaulGPDnCOUNT.csv')
print "One Network is about ", 60/71.0
final.groupby("network").speed.count()
# plot the difference
plt.hist(final[(final.speed!=0) & (final.network == True)].speed, 100, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final[(final.speed!=0)].speed, 100, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution")
plt.show()
# plot the difference
plt.hist(final[(final.network == True)].speed, 100, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final.speed, 100, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution Including Zero Speeds")
plt.show()
final = pd.read_csv('./maps_plots_data/Schoolbus/SchoolBusAggData.csv')
gpd = pd.read_csv('./maps_plots_data/Schoolbus/SchoolBusGPDnCOUNT.csv')
print "One Network is about ", 36/44.0
final.groupby("network").speed.count()
# plot the difference
plt.hist(final[(final.speed!=0) & (final.network == True)].speed, 50, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final[(final.speed!=0)].speed, 50, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution")
plt.show()
# plot the difference
plt.hist(final[(final.network == True)].speed, 100, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final.speed, 100, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution Including Zero Speeds")
plt.show()
final = pd.read_csv('./maps_plots_data/RefuseTrucks/RefuseTrucksAggData.csv')
gpd = pd.read_csv('./maps_plots_data/RefuseTrucks/RefuseTrucksGPDnCOUNT.csv')
print "One Network is about ", 11.5/12.0
final.groupby("network").speed.count()
# plot the difference
plt.hist(final[(final.speed!=0) & (final.network == True)].speed, 50, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final[(final.speed!=0)].speed, 50, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution")
plt.show()
# plot the difference
plt.hist(final[(final.network == True)].speed, 100, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final.speed, 100, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution Including Zero Speeds")
plt.show()
final = pd.read_csv('./maps_plots_data/Transitbus/TransitbusAggData.csv')
gpd = pd.read_csv('./maps_plots_data/Transitbus/TransitbusGPDnCOUNT.csv')
print "One Network is about ", 15/18.0
final.groupby("network").speed.count()
# plot the difference
plt.hist(final[(final.speed!=0) & (final.network == True)].speed, 50, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final[(final.speed!=0)].speed, 50, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution")
plt.show()
# plot the difference
plt.hist(final[(final.network == True)].speed, 100, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final.speed, 100, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution Including Zero Speeds")
plt.show()
final = pd.read_csv('./maps_plots_data/SingleShortHaul/SingleShortAggData.csv')
gpd = pd.read_csv('./maps_plots_data/SingleShortHaul/SingleShortGPDnCOUNT.csv')
print "One Network is about ", 28.9/30.1
final.groupby("network").speed.count()
# plot the difference
plt.hist(final[(final.speed!=0) & (final.network == True)].speed, 50, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final[(final.speed!=0)].speed, 50, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution")
plt.show()
# plot the difference
plt.hist(final[(final.network == True)].speed, 100, alpha=.3, label='on-network',color='r',normed=True)
plt.hist(final.speed, 100, alpha=0.4, label='all', color='blue', normed=True)
plt.xlim(0,80)
plt.legend(loc='upper right')
plt.title("Comparison between On-Network vs All-Network Speed Distribution Including Zero Speeds")
plt.show()
| 0.461259 | 0.923696 |
# Explaining text sentiment analysis using SageMaker Clarify
1. [Overview](#Overview)
1. [Prerequisites and Data](#Prerequisites-and-Data)
1. [Initialize SageMaker](#Initialize-SageMaker)
1. [Loading the data: Women's Ecommerce clothing reviews Dataset](#Loading-the-data:-Women's-ecommerce-clothing-reviews-dataset)
1. [Data preparation for model training](#Data-preparation-for-model-training)
1. [Train and Deploy Hugging Face Model](#Train-and-Deploy-Hugging-Face-Model)
1. [Train model with Hugging Face estimator](#Train-model-with-Hugging-Face-estimator)
1. [Deploy Model to Endpoint](#Deploy-Model)
1. [Model Explainability with SageMaker Clarify for text features](#Model-Explainability-with-SageMaker-Clarify-for-text-features)
1. [Explaining Predictions](#Explaining-Predictions)
1. [Visualize local explanations](#Visualize-local-explanations)
1. [Clean Up](#Clean-Up)
## Overview
Amazon SageMaker Clarify helps improve your machine learning models by detecting potential bias and helping explain how these models make predictions. The fairness and explainability functionality provided by SageMaker Clarify takes a step towards enabling AWS customers to build trustworthy and understandable machine learning models. The product comes with the tools to help you with the following tasks.
* Measure biases that can occur during each stage of the ML lifecycle (data collection, model training and tuning, and monitoring of ML models deployed for inference).
* Generate model governance reports targeting risk and compliance teams and external regulators.
* Provide explanations of the data, models, and monitoring used to assess predictions for input containing data of various modalities like numerical data, categorical data, text, and images.
Learn more about SageMaker Clarify [here](https://aws.amazon.com/sagemaker/clarify/). This sample notebook walks you through:
1. Key terms and concepts needed to understand SageMaker Clarify
1. The incremental updates required to explain text features, along with other tabular features.
1. Explaining the importance of the various new input features on the model's decision
In doing so, the notebook will first train a [Hugging Face model](https://huggingface.co/models) using the [Hugging Face Estimator](https://sagemaker.readthedocs.io/en/stable/frameworks/huggingface/sagemaker.huggingface.html) in the SageMaker Python SDK using training dataset, then use SageMaker Clarify to analyze a testing dataset in CSV format, and then visualize the results.
## Prerequisites and Data
We require the following AWS resources to be able to successfully run this notebook.
1. Kernel: Python 3 (Data Science) kernel on SageMaker Studio or `conda_python3` kernel on notebook instances
2. Instance type: Any GPU instance. Here, we use `ml.g4dn.xlarge`
3. [SageMaker Python SDK](https://pypi.org/project/sagemaker/) version 2.70.0 or greater
4. [Transformers](https://pypi.org/project/transformers/) > 4.6.1
5. [Datasets](https://pypi.org/project/datasets/) > 1.6.2
```
!pip --quiet install "transformers==4.6.1" "datasets[s3]==1.6.2" "captum" --upgrade
```
Let's start by installing preview wheels of the Python SDK, boto and aws cli
```
! pip install sagemaker botocore boto3 awscli --upgrade
```
#### Initialize SageMaker
```
# Import libraries for data loading and pre-processing
import os
import numpy as np
import pandas as pd
import json
import botocore
import sagemaker
import tarfile
from sagemaker.huggingface import HuggingFace
from sagemaker.pytorch import PyTorchModel
from sagemaker import get_execution_role, clarify
from captum.attr import visualization
from sklearn.model_selection import train_test_split
from datasets import Dataset
from datasets.filesystems import S3FileSystem
# SageMaker session bucket is used to upload the dataset, model and model training logs
sess = sagemaker.Session()
sess = sagemaker.Session(default_bucket=sess.default_bucket())
region = sess.boto_region_name
bucket = sess.default_bucket()
prefix = "sagemaker/DEMO-sagemaker-clarify-text"
# Define the IAM role
role = sagemaker.get_execution_role()
# SageMaker Clarify model directory name
model_path = "model/"
```
If you change the value of `model_path` variable above, please be sure to update the `model_path` in [`code/inference.py`](./code/inference.py) script as well.
### Loading the data: Women's ecommerce clothing reviews dataset
#### Download the dataset
Data Source: `https://www.kaggle.com/nicapotato/womens-ecommerce-clothing-reviews/`
The Women’s E-Commerce Clothing Reviews dataset has been made available under a Creative Commons Public Domain license. A copy of the dataset has been saved in a sample data Amazon S3 bucket. In the first section of the notebook, we’ll walk through how to download the data and get started with building the ML workflow as a SageMaker pipeline
```
! curl https://sagemaker-sample-files.s3.amazonaws.com/datasets/tabular/womens_clothing_ecommerce/Womens_Clothing_E-Commerce_Reviews.csv > womens_clothing_reviews_dataset.csv
```
#### Load the dataset
```
df = pd.read_csv("womens_clothing_reviews_dataset.csv", index_col=[0])
df.head()
```
**Context**
The Women’s Clothing E-Commerce dataset contains reviews written by customers. Because the dataset contains real commercial data, it has been anonymized, and any references to the company in the review text and body have been replaced with “retailer”.
**Content**
The dataset contains 23486 rows and 10 columns. Each row corresponds to a customer review.
The columns include:
* Clothing ID: Integer Categorical variable that refers to the specific piece being reviewed.
* Age: Positive Integer variable of the reviewer's age.
* Title: String variable for the title of the review.
* Review Text: String variable for the review body.
* Rating: Positive Ordinal Integer variable for the product score granted by the customer from 1 Worst, to 5 Best.
* Recommended IND: Binary variable stating where the customer recommends the product where 1 is recommended, 0 is not recommended.
* Positive Feedback Count: Positive Integer documenting the number of other customers who found this review positive.
* Division Name: Categorical name of the product high level division.
* Department Name: Categorical name of the product department name.
* Class Name: Categorical name of the product class name.
**Goal**
To predict the sentiment of a review based on the text, and then explain the predictions using SageMaker Clarify.
### Data preparation for model training
#### Target Variable Creation
Since the dataset does not contain a column that indicates the sentiment of the customer reviews, lets create one. To do this, let's assume that reviews with a `Rating` of 4 or higher indicate positive sentiment and reviews with a `Rating` of 2 or lower indicate negative sentiment. Let's also assume that a `Rating` of 3 indicates neutral sentiment and exclude these rows from the dataset. Additionally, to predict the sentiment of a review, we are going to use the `Review Text` column; therefore let's remove rows that are empty in the `Review Text` column of the dataset
```
def create_target_column(df, min_positive_score, max_negative_score):
neutral_values = [i for i in range(max_negative_score + 1, min_positive_score)]
for neutral_value in neutral_values:
df = df[df["Rating"] != neutral_value]
df["Sentiment"] = df["Rating"] >= min_positive_score
replace_dict = {True: 1, False: 0}
df["Sentiment"] = df["Sentiment"].map(replace_dict)
return df
df = create_target_column(df, 4, 2)
df = df[~df["Review Text"].isna()]
```
#### Train-Validation-Test splits
The most common approach for model evaluation is using the train/validation/test split. Although this approach can be very effective in general, it can result in misleading results and potentially fail when used on classification problems with a severe class imbalance. Instead, the technique must be modified to stratify the sampling by the class label as below. Stratification ensures that all classes are well represented across the train, validation and test datasets.
```
target = "Sentiment"
cols = "Review Text"
X = df[cols]
y = df[target]
# Data split: 11%(val) of the 90% (train and test) of the dataset ~ 10%; resulting in 80:10:10split
test_dataset_size = 0.10
val_dataset_size = 0.11
RANDOM_STATE = 42
# Stratified train-val-test split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=test_dataset_size, stratify=y, random_state=RANDOM_STATE
)
X_train, X_val, y_train, y_val = train_test_split(
X_train, y_train, test_size=val_dataset_size, stratify=y_train, random_state=RANDOM_STATE
)
print(
"Dataset: train ",
X_train.shape,
y_train.shape,
y_train.value_counts(dropna=False, normalize=True).to_dict(),
)
print(
"Dataset: validation ",
X_val.shape,
y_val.shape,
y_val.value_counts(dropna=False, normalize=True).to_dict(),
)
print(
"Dataset: test ",
X_test.shape,
y_test.shape,
y_test.value_counts(dropna=False, normalize=True).to_dict(),
)
# Combine the independent columns with the label
df_train = pd.concat([X_train, y_train], axis=1).reset_index(drop=True)
df_test = pd.concat([X_test, y_test], axis=1).reset_index(drop=True)
df_val = pd.concat([X_val, y_val], axis=1).reset_index(drop=True)
```
We have split the dataset into train, test, and validation datasets. We use the train and validation datasets during training process, and run Clarify on the test dataset.
In the cell below, we convert the Pandas DataFrames into Hugging Face Datasets for downstream modeling
```
train_dataset = Dataset.from_pandas(df_train)
test_dataset = Dataset.from_pandas(df_val)
```
#### Upload prepared dataset to the S3
Here, we upload the prepared datasets to S3 buckets so that we can train the model with the Hugging Face Estimator.
```
# S3 key prefix for the datasets
s3_prefix = "samples/datasets/womens_clothing_ecommerce_reviews"
s3 = S3FileSystem()
# save train_dataset to s3
training_input_path = f"s3://{sess.default_bucket()}/{s3_prefix}/train"
train_dataset.save_to_disk(training_input_path, fs=s3)
# save test_dataset to s3
test_input_path = f"s3://{sess.default_bucket()}/{s3_prefix}/test"
test_dataset.save_to_disk(test_input_path, fs=s3)
```
## Train and Deploy Hugging Face Model
In this step of the workflow, we use the [Hugging Face Estimator](https://sagemaker.readthedocs.io/en/stable/frameworks/huggingface/sagemaker.huggingface.html) to load the pre-trained `distilbert-base-uncased` model and fine-tune the model on our dataset.
### Train model with Hugging Face estimator
The hyperparameters defined below are parameters that are passed to the custom PyTorch code in [`scripts/train.py`](./scripts/train.py). The only required parameter is `model_name`. The other parameters like `epoch`, `train_batch_size` all have default values which can be overriden by setting their values here.
```
# Hyperparameters passed into the training job
hyperparameters = {"epochs": 1, "model_name": "distilbert-base-uncased"}
huggingface_estimator = HuggingFace(
entry_point="train.py",
source_dir="scripts",
instance_type="ml.g4dn.xlarge",
instance_count=1,
transformers_version="4.6.1",
pytorch_version="1.7.1",
py_version="py36",
role=role,
hyperparameters=hyperparameters,
)
# starting the train job with our uploaded datasets as input
huggingface_estimator.fit({"train": training_input_path, "test": test_input_path})
```
### Download the trained model files for model inference
```
! aws s3 cp {huggingface_estimator.model_data} model.tar.gz
! mkdir -p {model_path}
! tar -xvf model.tar.gz -C {model_path}/
```
### Deploy Model
We are going to use the trained model files along with the PyTorch Inference container to deploy the model to a SageMaker endpoint.
```
with tarfile.open("hf_model.tar.gz", mode="w:gz") as archive:
archive.add(model_path, recursive=True)
archive.add("code/")
prefix = s3_prefix.split("/")[-1]
zipped_model_path = sess.upload_data(path="hf_model.tar.gz", key_prefix=prefix + "/hf-model-sm")
model_name = "womens-ecommerce-reviews-model"
endpoint_name = "womens-ecommerce-reviews-endpoint"
model = PyTorchModel(
entry_point="inference.py",
name=model_name,
model_data=zipped_model_path,
role=get_execution_role(),
framework_version="1.7.1",
py_version="py3",
)
predictor = model.deploy(
initial_instance_count=1, instance_type="ml.g4dn.xlarge", endpoint_name=endpoint_name
)
```
#### Test the model endpoint
Lets test the model endpoint to ensure that deployment was successful.
```
test_sentence1 = "A very versatile and cozy top. would look great dressed up or down for a casual comfy fall day. what a fun piece for my wardrobe!"
test_sentence2 = "Love the color! very soft. unique look. can't wait to wear it this fall"
test_sentence3 = (
"These leggings are loose fitting and the quality is just not there.. i am returning the item."
)
test_sentence4 = "Very disappointed the back of this blouse is plain, not as displayed."
predictor = sagemaker.predictor.Predictor(endpoint_name, sess)
predictor.serializer = sagemaker.serializers.CSVSerializer()
predictor.deserializer = sagemaker.deserializers.CSVDeserializer()
predictor.predict([[test_sentence1], [test_sentence2], [test_sentence3], [test_sentence4]])
```
## Model Explainability with SageMaker Clarify for text features
Now that the model is ready, and we are able to get predictions, we are ready to get explanations for text data from Clarify processing job. For a detailed example that showcases how to use the Clarify processing job, please refer to [this example](https://github.com/aws/amazon-sagemaker-examples/blob/master/sagemaker_processing/fairness_and_explainability/fairness_and_explainability.ipynb). This example shows how to get explanations for text data from Clarify.
In the cell below, we create the CSV file to pass on to the Clarify dataset. We are using 10 samples here to make it fast, but we can use entire dataset at a time. We are also filtering out any reviews with less than 500 characters as long reviews provide better visualization with `sentence` level granularity (When granularity is `sentence`, each sentence is a feature, and we need a few sentences per review for good visualization).
```
file_path = "clarify_data.csv"
num_examples = 10
df_test["len"] = df_test["Review Text"].apply(lambda ele: len(ele))
df_test_clarify = pd.DataFrame(
df_test[df_test["len"] > 500].sample(n=num_examples, random_state=RANDOM_STATE),
columns=["Review Text"],
)
df_test_clarify.to_csv(file_path, header=True, index=False)
df_test_clarify
```
### Explaining Predictions
There are expanding business needs and legislative regulations that require explanations of _why_ a model made the decision it did. SageMaker Clarify uses SHAP to explain the contribution that each input feature makes to the final decision.
How does the Kernel SHAP algorithm work? Kernel SHAP algorithm is a local explanation method. That is, it explains each instance or row of the dataset at a time. To explain each instance, it perturbs the features values - that is, it changes the values of some features to a baseline (or non-informative) value, and then get predictions from the model for the perturbed samples. It does this for a number of times per instance (determined by the optional parameter `num_samples` in `SHAPConfig`), and computes the importance of each feature based on how the model prediction changed.
We are now extending this functionality to text data. In order to be able to explain text, we need the `TextConfig`. The `TextConfig` is an optional parameter of `SHAPConfig`, which you need to provide if you need explanations for the text features in your dataset. `TextConfig` in turn requires three parameters:
1. `granularity` (required): To explain text features, Clarify further breaks down text into smaller text units, and considers each such text unit as a feature. The parameter `granularity` informs the level to which Clarify will break down the text: `token`, `sentence`, or `paragraph` are the allowed values for `granularity`.
2. `language` (required): the language of the text features. This is required to tokenize the text to break them down to their granular form.
3. `max_top_tokens` (optional): the number of top token attributions that will be shown in the output (we need this because the size of vocabulary can be very big). This is an optional parameter, and defaults to 50.
Kernel SHAP algorithm requires a baseline (also known as background dataset). In case of tabular features, the baseline value/s for a feature is ideally a non-informative or least informative value for that feature. However, for text feature, the baseline values must be the value you want to replace the individual text feature (token, sentence or paragraph) with. For instance, in the example below, we have chosen the baseline values for `review_text` as `<UNK>`, and `granularity` is `sentence`. Every time a sentence has to replaced in the perturbed inputs, we will replace it with `<UNK>`.
If baseline is not provided, a baseline is calculated automatically by SageMaker Clarify using K-means or K-prototypes in the input dataset for tabular features. For text features, if baseline is not provided, the default replacement value will be the string `<PAD>`.
```
clarify_processor = clarify.SageMakerClarifyProcessor(
role=role, instance_count=1, instance_type="ml.m5.xlarge", sagemaker_session=sess
)
model_config = clarify.ModelConfig(
model_name=model_name,
instance_type="ml.m5.xlarge",
instance_count=1,
accept_type="text/csv",
content_type="text/csv",
)
explainability_output_path = "s3://{}/{}/clarify-text-explainability".format(bucket, prefix)
explainability_data_config = clarify.DataConfig(
s3_data_input_path=file_path,
s3_output_path=explainability_output_path,
headers=["Review Text"],
dataset_type="text/csv",
)
shap_config = clarify.SHAPConfig(
baseline=[["<UNK>"]],
num_samples=1000,
agg_method="mean_abs",
save_local_shap_values=True,
text_config=clarify.TextConfig(granularity="sentence", language="english"),
)
# Running the clarify explainability job involves spinning up a processing job and a model endpoint which may take a few minutes.
# After this you will see a progress bar for the SHAP computation.
# The size of the dataset (num_examples) and the num_samples for shap will effect the running time.
clarify_processor.run_explainability(
data_config=explainability_data_config,
model_config=model_config,
explainability_config=shap_config,
)
```
### Visualize local explanations
We use Captum to visualize the feature importances computed by Clarify.
First, lets load the local explanations. Local text explanations can be found in the analysis results folder in a file named `out.jsonl` in the `explanations_shap` directory.
```
local_feature_attributions_file = "out.jsonl"
analysis_results = []
analysis_result = sagemaker.s3.S3Downloader.download(
explainability_output_path + "/explanations_shap/" + local_feature_attributions_file,
local_path="./",
)
shap_out = []
file = sagemaker.s3.S3Downloader.read_file(
explainability_output_path + "/explanations_shap/" + local_feature_attributions_file
)
for line in file.split("\n"):
if line:
shap_out.append(json.loads(line))
```
The local explanations file is a JSON Lines file, that contains the explanation of one instance per row. Let's examine the output format of the explanations.
```
print(json.dumps(shap_out[0], indent=2))
```
At the highest level of this JSON Line, there are two keys: `explanations`, `join_source_value` (Not present here as we have not included a `joinsource` column in the input dataset). `explanations` contains a list of attributions for each feature in the dataset. In this case, we have a single element, because the input dataset also had a single feature. It also contains details like `feature_name`, `data_type` of the features (indicating whether Clarify inferred the column as numerical, categorical or text). Each token attribution also contains a `description` field that contains the token itself, and the starting index of the token in original input. This allows you to reconstruct the original sentence from the output as well.
In the following block, we create a list of attributions and a list of tokens for use in visualizations.
```
attributions_dataset = [
np.array([attr["attribution"][0] for attr in expl["explanations"][0]["attributions"]])
for expl in shap_out
]
tokens_dataset = [
np.array(
[attr["description"]["partial_text"] for attr in expl["explanations"][0]["attributions"]]
)
for expl in shap_out
]
```
We obtain predictions as well so that they can be displayed alongside the feature attributions.
```
preds = predictor.predict([t for t in df_test_clarify.values])
# This method is a wrapper around the captum that helps produce visualizations for local explanations. It will
# visualize the attributions for the tokens with red or green colors for negative and positive attributions.
def visualization_record(
attributions, # list of attributions for the tokens
text, # list of tokens
pred, # the prediction value obtained from the endpoint
delta,
true_label, # the true label from the dataset
normalize=True, # normalizes the attributions so that the max absolute value is 1. Yields stronger colors.
max_frac_to_show=0.05, # what fraction of tokens to highlight, set to 1 for all.
match_to_pred=False, # whether to limit highlights to red for negative predictions and green for positive ones.
# By enabling `match_to_pred` you show what tokens contribute to a high/low prediction not those that oppose it.
):
if normalize:
attributions = attributions / max(max(attributions), max(-attributions))
if max_frac_to_show is not None and max_frac_to_show < 1:
num_show = int(max_frac_to_show * attributions.shape[0])
sal = attributions
if pred < 0.5:
sal = -sal
if not match_to_pred:
sal = np.abs(sal)
top_idxs = np.argsort(-sal)[:num_show]
mask = np.zeros_like(attributions)
mask[top_idxs] = 1
attributions = attributions * mask
return visualization.VisualizationDataRecord(
attributions,
pred,
int(pred > 0.5),
true_label,
attributions.sum() > 0,
attributions.sum(),
text,
delta,
)
# You can customize the following display settings
normalize = True
max_frac_to_show = 1
match_to_pred = False
labels = test_dataset["Sentiment"][:num_examples]
vis = []
for attr, token, pred, label in zip(attributions_dataset, tokens_dataset, preds, labels):
vis.append(
visualization_record(
attr, token, float(pred[0]), 0.0, label, normalize, max_frac_to_show, match_to_pred
)
)
```
Now that we compiled the record we are finally ready to render the visualization.
We see a row per review in the selected dataset. For each row we have the prediction, the label, and the highlighted text. Additionally, we show the total sum of attributions (as attribution score) and its label (as attribution label), which indicates whether it is greater than zero.
```
_ = visualization.visualize_text(vis)
```
# Cleanup
Finally, please remember to delete the Amazon SageMaker endpoint to avoid charges:
```
predictor.delete_endpoint()
```
|
github_jupyter
|
!pip --quiet install "transformers==4.6.1" "datasets[s3]==1.6.2" "captum" --upgrade
! pip install sagemaker botocore boto3 awscli --upgrade
# Import libraries for data loading and pre-processing
import os
import numpy as np
import pandas as pd
import json
import botocore
import sagemaker
import tarfile
from sagemaker.huggingface import HuggingFace
from sagemaker.pytorch import PyTorchModel
from sagemaker import get_execution_role, clarify
from captum.attr import visualization
from sklearn.model_selection import train_test_split
from datasets import Dataset
from datasets.filesystems import S3FileSystem
# SageMaker session bucket is used to upload the dataset, model and model training logs
sess = sagemaker.Session()
sess = sagemaker.Session(default_bucket=sess.default_bucket())
region = sess.boto_region_name
bucket = sess.default_bucket()
prefix = "sagemaker/DEMO-sagemaker-clarify-text"
# Define the IAM role
role = sagemaker.get_execution_role()
# SageMaker Clarify model directory name
model_path = "model/"
! curl https://sagemaker-sample-files.s3.amazonaws.com/datasets/tabular/womens_clothing_ecommerce/Womens_Clothing_E-Commerce_Reviews.csv > womens_clothing_reviews_dataset.csv
df = pd.read_csv("womens_clothing_reviews_dataset.csv", index_col=[0])
df.head()
def create_target_column(df, min_positive_score, max_negative_score):
neutral_values = [i for i in range(max_negative_score + 1, min_positive_score)]
for neutral_value in neutral_values:
df = df[df["Rating"] != neutral_value]
df["Sentiment"] = df["Rating"] >= min_positive_score
replace_dict = {True: 1, False: 0}
df["Sentiment"] = df["Sentiment"].map(replace_dict)
return df
df = create_target_column(df, 4, 2)
df = df[~df["Review Text"].isna()]
target = "Sentiment"
cols = "Review Text"
X = df[cols]
y = df[target]
# Data split: 11%(val) of the 90% (train and test) of the dataset ~ 10%; resulting in 80:10:10split
test_dataset_size = 0.10
val_dataset_size = 0.11
RANDOM_STATE = 42
# Stratified train-val-test split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=test_dataset_size, stratify=y, random_state=RANDOM_STATE
)
X_train, X_val, y_train, y_val = train_test_split(
X_train, y_train, test_size=val_dataset_size, stratify=y_train, random_state=RANDOM_STATE
)
print(
"Dataset: train ",
X_train.shape,
y_train.shape,
y_train.value_counts(dropna=False, normalize=True).to_dict(),
)
print(
"Dataset: validation ",
X_val.shape,
y_val.shape,
y_val.value_counts(dropna=False, normalize=True).to_dict(),
)
print(
"Dataset: test ",
X_test.shape,
y_test.shape,
y_test.value_counts(dropna=False, normalize=True).to_dict(),
)
# Combine the independent columns with the label
df_train = pd.concat([X_train, y_train], axis=1).reset_index(drop=True)
df_test = pd.concat([X_test, y_test], axis=1).reset_index(drop=True)
df_val = pd.concat([X_val, y_val], axis=1).reset_index(drop=True)
train_dataset = Dataset.from_pandas(df_train)
test_dataset = Dataset.from_pandas(df_val)
# S3 key prefix for the datasets
s3_prefix = "samples/datasets/womens_clothing_ecommerce_reviews"
s3 = S3FileSystem()
# save train_dataset to s3
training_input_path = f"s3://{sess.default_bucket()}/{s3_prefix}/train"
train_dataset.save_to_disk(training_input_path, fs=s3)
# save test_dataset to s3
test_input_path = f"s3://{sess.default_bucket()}/{s3_prefix}/test"
test_dataset.save_to_disk(test_input_path, fs=s3)
# Hyperparameters passed into the training job
hyperparameters = {"epochs": 1, "model_name": "distilbert-base-uncased"}
huggingface_estimator = HuggingFace(
entry_point="train.py",
source_dir="scripts",
instance_type="ml.g4dn.xlarge",
instance_count=1,
transformers_version="4.6.1",
pytorch_version="1.7.1",
py_version="py36",
role=role,
hyperparameters=hyperparameters,
)
# starting the train job with our uploaded datasets as input
huggingface_estimator.fit({"train": training_input_path, "test": test_input_path})
! aws s3 cp {huggingface_estimator.model_data} model.tar.gz
! mkdir -p {model_path}
! tar -xvf model.tar.gz -C {model_path}/
with tarfile.open("hf_model.tar.gz", mode="w:gz") as archive:
archive.add(model_path, recursive=True)
archive.add("code/")
prefix = s3_prefix.split("/")[-1]
zipped_model_path = sess.upload_data(path="hf_model.tar.gz", key_prefix=prefix + "/hf-model-sm")
model_name = "womens-ecommerce-reviews-model"
endpoint_name = "womens-ecommerce-reviews-endpoint"
model = PyTorchModel(
entry_point="inference.py",
name=model_name,
model_data=zipped_model_path,
role=get_execution_role(),
framework_version="1.7.1",
py_version="py3",
)
predictor = model.deploy(
initial_instance_count=1, instance_type="ml.g4dn.xlarge", endpoint_name=endpoint_name
)
test_sentence1 = "A very versatile and cozy top. would look great dressed up or down for a casual comfy fall day. what a fun piece for my wardrobe!"
test_sentence2 = "Love the color! very soft. unique look. can't wait to wear it this fall"
test_sentence3 = (
"These leggings are loose fitting and the quality is just not there.. i am returning the item."
)
test_sentence4 = "Very disappointed the back of this blouse is plain, not as displayed."
predictor = sagemaker.predictor.Predictor(endpoint_name, sess)
predictor.serializer = sagemaker.serializers.CSVSerializer()
predictor.deserializer = sagemaker.deserializers.CSVDeserializer()
predictor.predict([[test_sentence1], [test_sentence2], [test_sentence3], [test_sentence4]])
file_path = "clarify_data.csv"
num_examples = 10
df_test["len"] = df_test["Review Text"].apply(lambda ele: len(ele))
df_test_clarify = pd.DataFrame(
df_test[df_test["len"] > 500].sample(n=num_examples, random_state=RANDOM_STATE),
columns=["Review Text"],
)
df_test_clarify.to_csv(file_path, header=True, index=False)
df_test_clarify
clarify_processor = clarify.SageMakerClarifyProcessor(
role=role, instance_count=1, instance_type="ml.m5.xlarge", sagemaker_session=sess
)
model_config = clarify.ModelConfig(
model_name=model_name,
instance_type="ml.m5.xlarge",
instance_count=1,
accept_type="text/csv",
content_type="text/csv",
)
explainability_output_path = "s3://{}/{}/clarify-text-explainability".format(bucket, prefix)
explainability_data_config = clarify.DataConfig(
s3_data_input_path=file_path,
s3_output_path=explainability_output_path,
headers=["Review Text"],
dataset_type="text/csv",
)
shap_config = clarify.SHAPConfig(
baseline=[["<UNK>"]],
num_samples=1000,
agg_method="mean_abs",
save_local_shap_values=True,
text_config=clarify.TextConfig(granularity="sentence", language="english"),
)
# Running the clarify explainability job involves spinning up a processing job and a model endpoint which may take a few minutes.
# After this you will see a progress bar for the SHAP computation.
# The size of the dataset (num_examples) and the num_samples for shap will effect the running time.
clarify_processor.run_explainability(
data_config=explainability_data_config,
model_config=model_config,
explainability_config=shap_config,
)
local_feature_attributions_file = "out.jsonl"
analysis_results = []
analysis_result = sagemaker.s3.S3Downloader.download(
explainability_output_path + "/explanations_shap/" + local_feature_attributions_file,
local_path="./",
)
shap_out = []
file = sagemaker.s3.S3Downloader.read_file(
explainability_output_path + "/explanations_shap/" + local_feature_attributions_file
)
for line in file.split("\n"):
if line:
shap_out.append(json.loads(line))
print(json.dumps(shap_out[0], indent=2))
attributions_dataset = [
np.array([attr["attribution"][0] for attr in expl["explanations"][0]["attributions"]])
for expl in shap_out
]
tokens_dataset = [
np.array(
[attr["description"]["partial_text"] for attr in expl["explanations"][0]["attributions"]]
)
for expl in shap_out
]
preds = predictor.predict([t for t in df_test_clarify.values])
# This method is a wrapper around the captum that helps produce visualizations for local explanations. It will
# visualize the attributions for the tokens with red or green colors for negative and positive attributions.
def visualization_record(
attributions, # list of attributions for the tokens
text, # list of tokens
pred, # the prediction value obtained from the endpoint
delta,
true_label, # the true label from the dataset
normalize=True, # normalizes the attributions so that the max absolute value is 1. Yields stronger colors.
max_frac_to_show=0.05, # what fraction of tokens to highlight, set to 1 for all.
match_to_pred=False, # whether to limit highlights to red for negative predictions and green for positive ones.
# By enabling `match_to_pred` you show what tokens contribute to a high/low prediction not those that oppose it.
):
if normalize:
attributions = attributions / max(max(attributions), max(-attributions))
if max_frac_to_show is not None and max_frac_to_show < 1:
num_show = int(max_frac_to_show * attributions.shape[0])
sal = attributions
if pred < 0.5:
sal = -sal
if not match_to_pred:
sal = np.abs(sal)
top_idxs = np.argsort(-sal)[:num_show]
mask = np.zeros_like(attributions)
mask[top_idxs] = 1
attributions = attributions * mask
return visualization.VisualizationDataRecord(
attributions,
pred,
int(pred > 0.5),
true_label,
attributions.sum() > 0,
attributions.sum(),
text,
delta,
)
# You can customize the following display settings
normalize = True
max_frac_to_show = 1
match_to_pred = False
labels = test_dataset["Sentiment"][:num_examples]
vis = []
for attr, token, pred, label in zip(attributions_dataset, tokens_dataset, preds, labels):
vis.append(
visualization_record(
attr, token, float(pred[0]), 0.0, label, normalize, max_frac_to_show, match_to_pred
)
)
_ = visualization.visualize_text(vis)
predictor.delete_endpoint()
| 0.459076 | 0.99056 |
[View in Colaboratory](https://colab.research.google.com/github/mariosky/databook/blob/master/DataLoad.ipynb)
#Colab and Jupyter Notebooks Quick Tour
##Welcome
First we are going to visit a notebbok from [Colab](https://colab.research.google.com/notebooks/welcome.ipynb#scrollTo=9J7p406abzgl).
## Loading and saving data
There are several options for Google external sources:
* Local files
* Google Drive
* Google Sheets
* Google Cloud Storage
###Upload local files
With this [snippet](https://colab.research.google.com/notebooks/io.ipynb) we can choose a file to upload from our local system:
```
from google.colab import files
uploaded = files.upload()
for fn in uploaded.keys():
print('User uploaded file "{name}" with length {length} bytes'.format(
name=fn, length=len(uploaded[fn])))
```
We can check to see if it was uploaded correctly by using a system call:
```
!ls
```
### Upload a file from Google Drive using [PyDrive](https://googledrive.github.io/PyDrive/docs/build/html/index.html)
We must install the PyDrive library first:
```
!pip install -U -q PyDrive
```
We need to import the libraries we need
```
from pydrive.auth import GoogleAuth
from pydrive.drive import GoogleDrive
from google.colab import auth
from oauth2client.client import GoogleCredentials
```
We must first authenticate with google and create the `GoogleDrive` client.
```
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
```
Now we can download the file and print its contents.
If an output is to long we can delete it with the corresponding option in the *code cell* menu.
```
data = drive.CreateFile({'id': '1pi40wcRtqCdqwbBx9ISrAZVLHF-MMQox'})
print('Downloaded content "{}"'.format(data.GetContentString()))
```
###Upload a file from the web
Another option is to download the file from the [UCI ML Repository](https://archive.ics.uci.edu/ml/index.php). We can use the [requests](http://docs.python-requests.org/en/master/) library.
```
import requests
r = requests.get('https://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data')
print(r.text)
```
## Python + Numpy + Matplotlib
We are going to use **python** to read the file. We need to split the file into lines first:
```
iris_rows = r.text.split('\n')
print (iris_rows[:5])
```
We use **slicing** to get fragments from the list.
```
iris_rows[-4:]
```
We use ***list comprehensions*** to get only what we need:
```
[row.split(',')[:-1] for row in iris_rows[:-2]]
```
Finally by using the function map we can get the array we need.
```
[map(float, row.split(',')[:-1]) for row in iris_rows[:-2]]
```
In Python 3, **map** is a generator object, but we can extract a list.
```
[list(map(float, row.split(',')[:-1])) for row in iris_rows[:-2]]
```
Let's create a NumPy array:
```
import numpy
iris_data = [list(map(float, row.split(',')[:-1])) for row in iris_rows[:-2]]
iris = numpy.array(iris_data)
iris[:5]
```
We can explore the data with **matplotlib**:
```
import matplotlib.pyplot as plt
x = iris[:50,0]
y = iris[:50,1]
plt.plot(x, y, 'r.')
plt.show()
```
We can plot two types of flowers:
```
plt.plot( iris[:50,0], iris[:50,1], 'r.')
plt.plot( iris[51:100,0], iris[51:100,1], 'b.')
```
Let's do three
```
plt.plot( iris[:50,0], iris[:50,1], 'r.')
plt.plot( iris[51:100,0], iris[51:100,1], 'b.')
plt.plot( iris[101:,0], iris[101:,1], 'g.')
```
##DEAP notebook
Here we have an example [DEAP notebook](https://github.com/DEAP/notebooks/blob/master/OneMax.ipynb) from Github.
We can visit or we can invite.
If we invite we need to **!pip install deap **
|
github_jupyter
|
from google.colab import files
uploaded = files.upload()
for fn in uploaded.keys():
print('User uploaded file "{name}" with length {length} bytes'.format(
name=fn, length=len(uploaded[fn])))
!ls
!pip install -U -q PyDrive
from pydrive.auth import GoogleAuth
from pydrive.drive import GoogleDrive
from google.colab import auth
from oauth2client.client import GoogleCredentials
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
data = drive.CreateFile({'id': '1pi40wcRtqCdqwbBx9ISrAZVLHF-MMQox'})
print('Downloaded content "{}"'.format(data.GetContentString()))
import requests
r = requests.get('https://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data')
print(r.text)
iris_rows = r.text.split('\n')
print (iris_rows[:5])
iris_rows[-4:]
[row.split(',')[:-1] for row in iris_rows[:-2]]
[map(float, row.split(',')[:-1]) for row in iris_rows[:-2]]
[list(map(float, row.split(',')[:-1])) for row in iris_rows[:-2]]
import numpy
iris_data = [list(map(float, row.split(',')[:-1])) for row in iris_rows[:-2]]
iris = numpy.array(iris_data)
iris[:5]
import matplotlib.pyplot as plt
x = iris[:50,0]
y = iris[:50,1]
plt.plot(x, y, 'r.')
plt.show()
plt.plot( iris[:50,0], iris[:50,1], 'r.')
plt.plot( iris[51:100,0], iris[51:100,1], 'b.')
plt.plot( iris[:50,0], iris[:50,1], 'r.')
plt.plot( iris[51:100,0], iris[51:100,1], 'b.')
plt.plot( iris[101:,0], iris[101:,1], 'g.')
| 0.293607 | 0.9455 |
# [모듈 2.2] SageMaker 내장 알고리즘의 모델 훈련
Download | Structure | Preprocessing (Built-in) | **Train Model (Built-in)** (4단계 중의 4/4)
```
```
### [알림] <font coler="red"> conda_mxnet_latest_p37 커널 </font> 과 함께 사용해야 합니다.
* 이 노트북은 `1.1.download_data`, `1.2.structuring_data` 및 `2.1.builtin_preprocessing`으로 시작하는 일련의 노트북의 일부입니다.
<pre>
</pre>
# 노트북 요약
---
이 노트북에서는 SageMaker SDK를 사용하여 SageMaker의 내장 이미지 분류 알고리즘에 대한 Estimator를 생성하고 원격 EC2 인스턴스에서 훈련합니다.
# 1. 환경 설정
- 카테고리 레이블, 버킷 이름, RecordIO 훈련 및 검증 파일 경로를 로딩 합니다.
```
import boto3
import shutil
import urllib
import pickle
import pathlib
import tarfile
import subprocess
import sagemaker
%store -r bucket_name
%store -r train_builtin_s3_uri
%store -r val_builtin_s3_uri
with open("pickled_data/category_labels.pickle", "rb") as f:
category_labels = pickle.load(f)
```
# 2. 내장 SageMaker 이미지 분류 알고리즘 실행하기
___
## 훈련 및 검증 데이터의 입력 쳐널 정의
- 훈련 및 검증의 경로 및 RecordIO 포맷 , S3 다운로드를 'Pipe" 로 설정 합니다.
```
train_data = sagemaker.inputs.TrainingInput(
# s3_data=f"s3://{bucket_name}/data/train",
s3_data= train_builtin_s3_uri,
content_type="application/x-recordio",
s3_data_type="S3Prefix",
input_mode="Pipe",
)
val_data = sagemaker.inputs.TrainingInput(
# s3_data=f"s3://{bucket_name}/data/val",
s3_data=val_builtin_s3_uri,
content_type="application/x-recordio",
s3_data_type="S3Prefix",
input_mode="Pipe",
)
data_channels = {"train": train_data, "validation": val_data}
```
### 알고리즘의 하이퍼파라미터 구성
- 자세한 내역은 아래를 참조 하세요.
- https://docs.aws.amazon.com/sagemaker/latest/dg/IC-Hyperparameter.html
* **num_layers** - 내장된 이미지 분류 알고리즘은 ResNet 아키텍처를 기반으로 합니다. 이 아키텍처에는 사용하는 레이어 수에 따라 다양한 버전이 있습니다. 이 가이드에서는 훈련 속도를 높이기 위해 가장 작은 것을 사용합니다. 알고리즘의 정확도가 정체되어 더 나은 정확도가 필요한 경우 레이어 수를 늘리는 것이 도움이 될 수 있습니다.
* **use_pretrained_model** - 이것은 전이 학습을 위해 사전 훈련된 모델에서 가중치를 초기화합니다. 그렇지 않으면 가중치가 무작위로 초기화됩니다.
* **augmentation_type** - 훈련 세트에 증강을 추가하여 모델을 더 잘 일반화할 수 있습니다. 작은 데이터 세트의 경우 증강은 훈련을 크게 향상시킬 수 있습니다.
* **image_shape** - 모든 이미지의 채널, 높이, 너비
* **num_classes** - 데이터세트의 클래스 수
* **num_training_samples** - 훈련 세트의 총 이미지 수(진행률 계산에 사용)
* **mini_batch_size** - 훈련 중에 사용할 배치 크기입니다.
* **에포크** - 에포크는 훈련 세트의 한 주기를 나타내며 훈련할 에포크가 많다는 것은 정확도를 향상시킬 기회가 더 많다는 것을 의미합니다. 적절한 값은 시간과 예산 제약에 따라 5~25 Epoch 범위입니다. 이상적으로는 검증 정확도가 안정되기 직전에 올바른 Epoch 수가 있습니다.
* **learning_rate**: 훈련의 각 배치 후에 우리는 해당 배치에 대해 가능한 최상의 결과를 제공하기 위해 모델의 가중치를 업데이트합니다. 학습률은 가중치를 업데이트해야 하는 정도를 제어합니다. 모범 사례는 0.2에서 .001 사이의 값을 지정하며 일반적으로 1보다 높지 않습니다. 학습률이 높을수록 훈련이 최적의 가중치로 더 빨리 수렴되지만 너무 빠르면 목표를 초과할 수 있습니다. 이 예에서는 사전 훈련된 모델의 가중치를 사용하므로 가중치가 이미 최적화되어 있고 가중치에서 너무 멀리 이동하고 싶지 않기 때문에 더 낮은 학습률로 시작하려고 합니다.
* **precision_dtype** - 모델의 가중치에 대해 32비트 부동 데이터 유형을 사용할지 16비트를 사용할지 여부. 메모리 관리 문제가 있는 경우 16비트를 사용할 수 있습니다. 그러나 가중치는 빠르게 증가하거나 줄어들 수 있으므로 32비트 가중치를 사용하면 이러한 문제에 대한 훈련이 더욱 강력해지며 일반적으로 대부분의 프레임워크에서 기본값입니다.
```
num_classes = len(category_labels)
num_training_samples = len(set(pathlib.Path("data_structured/train").rglob("*.jpg")))
hyperparameters = {
"num_layers": 18,
"use_pretrained_model": 1, # Pretrained Model (ResNet) 의 가중치 가져오기
"augmentation_type": "crop_color_transform",
"image_shape": "3,224,224",
"num_classes": num_classes,
"num_training_samples": num_training_samples,
"mini_batch_size": 64,
"epochs": 5,
"learning_rate": 0.001,
"precision_dtype": "float32",
}
```
## 내장 알고리즘의 Docker 경로 및 컨피그 설정
```
training_image = sagemaker.image_uris.retrieve(
"image-classification", sagemaker.Session().boto_region_name
)
algo_config = {
"hyperparameters": hyperparameters,
"image_uri": training_image,
"role": sagemaker.get_execution_role(),
"instance_count": 1,
"instance_type": "ml.p3.2xlarge",
"volume_size": 100,
"max_run": 360000,
"output_path": f"s3://{bucket_name}/data/output",
}
```
## 알고리즘 Estimator 를 생성하고 훈련 시작
```
algorithm = sagemaker.estimator.Estimator(**algo_config)
algorithm.fit(inputs=data_channels, logs=True)
```
# 3. 훈련 결과 이해하기
___
```
[09/14/2020 05:37:38 INFO 139869866030912] Epoch[0] Batch [20]#011Speed: 111.811 samples/sec#011accuracy=0.452381
[09/14/2020 05:37:54 INFO 139869866030912] Epoch[0] Batch [40]#011Speed: 131.393 samples/sec#011accuracy=0.570503
[09/14/2020 05:38:10 INFO 139869866030912] Epoch[0] Batch [60]#011Speed: 139.540 samples/sec#011accuracy=0.617700
[09/14/2020 05:38:27 INFO 139869866030912] Epoch[0] Batch [80]#011Speed: 144.003 samples/sec#011accuracy=0.644483
[09/14/2020 05:38:43 INFO 139869866030912] Epoch[0] Batch [100]#011Speed: 146.600 samples/sec#011accuracy=0.664991
```
훈련이 시작되었습니다:
* Epoch[0]: 하나의 Epoch는 모든 데이터를 통한 하나의 훈련 주기에 해당합니다. SGD 및 Adam과 같은 확률적 최적화 프로그램은 여러 에포크를 실행하여 정확도를 개선합니다.
- 훈련 알고리즘이 랜덤 데이터 증강으로 인해 수정된 이미지 대해서 매 에포크마다 훈련을 수행 합니다.
- 즉 에포크마다 데이터 증강으로 인한 새로운 이미지가를 훈련한다는 의미 입니다.
* 배치: 훈련 알고리즘에 의해 처리된 배치의 수. 우리는 `mini_batch_size` 하이퍼파라미터에서 하나의 배치를 64개의 이미지로 지정했습니다. SGD와 같은 알고리즘의 경우 모델은 배치마다 자체적으로 업데이트할 기회를 얻습니다.
* 속도: 초당 훈련 알고리즘으로 전송되는 이미지의 수입니다. 이 정보는 데이터 세트의 변경 사항이 훈련 속도에 미치는 영향을 결정하는 데 중요합니다.
* 정확도: 각 간격(이 경우 20개 배치)에서 달성한 훈련 정확도.
```
[09/14/2020 05:38:58 INFO 139869866030912] Epoch[0] Train-accuracy=0.677083
[09/14/2020 05:38:58 INFO 139869866030912] Epoch[0] Time cost=102.745
[09/14/2020 05:39:02 INFO 139869866030912] Epoch[0] Validation-accuracy=0.729492
[09/14/2020 05:39:02 INFO 139869866030912] Storing the best model with validation accuracy: 0.729492
[09/14/2020 05:39:02 INFO 139869866030912] Saved checkpoint to "/opt/ml/model/image-classification-0001.params"
```
- 훈련의 첫 번째 에포크가 종료 되었습니다. (이 예에서는 한 에포크에 대해서만 훈련합니다).
- 최종 훈련 정확도와 검증 세트의 정확도가 보고됩니다. 이 두 숫자를 비교하는 것은 모델이 과적합인지 과소적합인지와 Bias/Varience 트레이드오프를 결정하는 데 중요합니다. 저장할 모델은 검증 데이터 세트의 정확도가 가장 큰 가중치를 사용합니다.
```
2020-09-14 05:39:03 Uploading - Uploading generated training model
2020-09-14 05:39:15 Completed - Training job completed
Training seconds: 235
Billable seconds: 235
```
최종 모델 가중치는 'algo_config'의 'output_path' 의 지정된 S3의 '.tar.gz'로 저장됩니다. EC2 인스턴스가 데이터에 대해 훈련하는 시간에 대해서만 비용이 청구되기 때문에 총 청구 가능 시간(초)도 보고되어 훈련 비용을 계산하는 데 도움이 됩니다.
## 다음 과정
이것으로 SageMaker의 내장 알고리즘에 대한 이미지 데이터 준비 가이드를 마칩니다. 모델을 배포하고 테스트 데이터에 대한 예측을 얻으려면 여기에서 찾을 수 있습니다.
- [추론을 위한 모델 배포](https://docs.aws.amazon.com/sagemaker/latest/dg/deploy-model.html)
|
github_jupyter
|
import boto3
import shutil
import urllib
import pickle
import pathlib
import tarfile
import subprocess
import sagemaker
%store -r bucket_name
%store -r train_builtin_s3_uri
%store -r val_builtin_s3_uri
with open("pickled_data/category_labels.pickle", "rb") as f:
category_labels = pickle.load(f)
train_data = sagemaker.inputs.TrainingInput(
# s3_data=f"s3://{bucket_name}/data/train",
s3_data= train_builtin_s3_uri,
content_type="application/x-recordio",
s3_data_type="S3Prefix",
input_mode="Pipe",
)
val_data = sagemaker.inputs.TrainingInput(
# s3_data=f"s3://{bucket_name}/data/val",
s3_data=val_builtin_s3_uri,
content_type="application/x-recordio",
s3_data_type="S3Prefix",
input_mode="Pipe",
)
data_channels = {"train": train_data, "validation": val_data}
num_classes = len(category_labels)
num_training_samples = len(set(pathlib.Path("data_structured/train").rglob("*.jpg")))
hyperparameters = {
"num_layers": 18,
"use_pretrained_model": 1, # Pretrained Model (ResNet) 의 가중치 가져오기
"augmentation_type": "crop_color_transform",
"image_shape": "3,224,224",
"num_classes": num_classes,
"num_training_samples": num_training_samples,
"mini_batch_size": 64,
"epochs": 5,
"learning_rate": 0.001,
"precision_dtype": "float32",
}
training_image = sagemaker.image_uris.retrieve(
"image-classification", sagemaker.Session().boto_region_name
)
algo_config = {
"hyperparameters": hyperparameters,
"image_uri": training_image,
"role": sagemaker.get_execution_role(),
"instance_count": 1,
"instance_type": "ml.p3.2xlarge",
"volume_size": 100,
"max_run": 360000,
"output_path": f"s3://{bucket_name}/data/output",
}
algorithm = sagemaker.estimator.Estimator(**algo_config)
algorithm.fit(inputs=data_channels, logs=True)
[09/14/2020 05:37:38 INFO 139869866030912] Epoch[0] Batch [20]#011Speed: 111.811 samples/sec#011accuracy=0.452381
[09/14/2020 05:37:54 INFO 139869866030912] Epoch[0] Batch [40]#011Speed: 131.393 samples/sec#011accuracy=0.570503
[09/14/2020 05:38:10 INFO 139869866030912] Epoch[0] Batch [60]#011Speed: 139.540 samples/sec#011accuracy=0.617700
[09/14/2020 05:38:27 INFO 139869866030912] Epoch[0] Batch [80]#011Speed: 144.003 samples/sec#011accuracy=0.644483
[09/14/2020 05:38:43 INFO 139869866030912] Epoch[0] Batch [100]#011Speed: 146.600 samples/sec#011accuracy=0.664991
[09/14/2020 05:38:58 INFO 139869866030912] Epoch[0] Train-accuracy=0.677083
[09/14/2020 05:38:58 INFO 139869866030912] Epoch[0] Time cost=102.745
[09/14/2020 05:39:02 INFO 139869866030912] Epoch[0] Validation-accuracy=0.729492
[09/14/2020 05:39:02 INFO 139869866030912] Storing the best model with validation accuracy: 0.729492
[09/14/2020 05:39:02 INFO 139869866030912] Saved checkpoint to "/opt/ml/model/image-classification-0001.params"
2020-09-14 05:39:03 Uploading - Uploading generated training model
2020-09-14 05:39:15 Completed - Training job completed
Training seconds: 235
Billable seconds: 235
| 0.440469 | 0.980015 |
# Model Layers
This module contains many layer classes that we might be interested in using in our models. These layers complement the default [Pytorch layers](https://pytorch.org/docs/stable/nn.html) which we can also use as predefined layers.
```
from fastai import *
from fastai.vision import *
from fastai.gen_doc.nbdoc import *
show_doc(AdaptiveConcatPool2d, doc_string=False)
from fastai.gen_doc.nbdoc import *
from fastai.layers import *
```
Layer that concats `AdaptiveAvgPool2d` and `AdaptiveMaxPool2d`. Output will be `2*sz` or 2 if `sz` is None.
The [`AdaptiveConcatPool2d`](/layers.html#AdaptiveConcatPool2d) object uses adaptive average pooling and adaptive max pooling and concatenates them both. We use this because it provides the model with the information of both methods and improves performance. This technique is called `adaptive` because it allows us to decide on what output dimensions we want, instead of choosing the input's dimensions to fit a desired output size.
Let's try training with Adaptive Average Pooling first, then with Adaptive Max Pooling and finally with the concatenation of them both to see how they fare in performance.
We will first define a [`simple_cnn`](/layers.html#simple_cnn) using [Adapative Max Pooling](https://pytorch.org/docs/stable/nn.html#torch.nn.AdaptiveMaxPool2d) by changing the source code a bit.
```
path = untar_data(URLs.MNIST_SAMPLE)
data = ImageDataBunch.from_folder(path)
def simple_cnn_max(actns:Collection[int], kernel_szs:Collection[int]=None,
strides:Collection[int]=None) -> nn.Sequential:
"CNN with `conv2d_relu` layers defined by `actns`, `kernel_szs` and `strides`"
nl = len(actns)-1
kernel_szs = ifnone(kernel_szs, [3]*nl)
strides = ifnone(strides , [2]*nl)
layers = [conv2d_relu(actns[i], actns[i+1], kernel_szs[i], stride=strides[i])
for i in range(len(strides))]
layers.append(nn.Sequential(nn.AdaptiveMaxPool2d(1), Flatten()))
return nn.Sequential(*layers)
model = simple_cnn_max((3,16,16,2))
learner = Learner(data, model, metrics=[accuracy])
learner.fit(1)
```
Now let's try with [Adapative Average Pooling](https://pytorch.org/docs/stable/nn.html#torch.nn.AdaptiveAvgPool2d) now.
```
def simple_cnn_avg(actns:Collection[int], kernel_szs:Collection[int]=None,
strides:Collection[int]=None) -> nn.Sequential:
"CNN with `conv2d_relu` layers defined by `actns`, `kernel_szs` and `strides`"
nl = len(actns)-1
kernel_szs = ifnone(kernel_szs, [3]*nl)
strides = ifnone(strides , [2]*nl)
layers = [conv2d_relu(actns[i], actns[i+1], kernel_szs[i], stride=strides[i])
for i in range(len(strides))]
layers.append(nn.Sequential(nn.AdaptiveAvgPool2d(1), Flatten()))
return nn.Sequential(*layers)
model = simple_cnn_avg((3,16,16,2))
learner = Learner(data, model, metrics=[accuracy])
learner.fit(1)
```
Finally we will try with the concatenation of them both [`AdaptiveConcatPool2d`](/layers.html#AdaptiveConcatPool2d). We will see that, in fact, it increases our accuracy and decreases our loss considerably!
```
def simple_cnn(actns:Collection[int], kernel_szs:Collection[int]=None,
strides:Collection[int]=None) -> nn.Sequential:
"CNN with `conv2d_relu` layers defined by `actns`, `kernel_szs` and `strides`"
nl = len(actns)-1
kernel_szs = ifnone(kernel_szs, [3]*nl)
strides = ifnone(strides , [2]*nl)
layers = [conv2d_relu(actns[i], actns[i+1], kernel_szs[i], stride=strides[i])
for i in range(len(strides))]
layers.append(nn.Sequential(AdaptiveConcatPool2d(1), Flatten()))
return nn.Sequential(*layers)
model = simple_cnn((3,16,16,2))
learner = Learner(data, model, metrics=[accuracy])
learner.fit(1)
show_doc(Lambda, doc_string=False)
```
Lambda allows us to define functions and use them as layers in our networks inside a [Sequential](https://pytorch.org/docs/stable/nn.html#torch.nn.Sequential) object.
So, for example, say we want to apply a [log_softmax loss](https://pytorch.org/docs/stable/nn.html#torch.nn.functional.log_softmax) and we need to change the shape of our output batches to be able to use this loss. We can add a layer that applies the necessary change in shape by calling:
`Lambda(lambda x: x.view(x.size(0),-1))`
Let's see an example of how the shape of our output can change when we add this layer.
```
model = nn.Sequential(
nn.Conv2d(3, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 10, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.AdaptiveAvgPool2d(1),
)
model.cuda()
for xb, yb in data.train_dl:
out = (model(*[xb]))
print(out.size())
break
model = nn.Sequential(
nn.Conv2d(3, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 10, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.AdaptiveAvgPool2d(1),
Lambda(lambda x: x.view(x.size(0),-1))
)
model.cuda()
for xb, yb in data.train_dl:
out = (model(*[xb]))
print(out.size())
break
show_doc(Flatten)
```
The function we build above is actually implemented in our library as [`Flatten`](/layers.html#Flatten). We can see that it returns the same size when we run it.
```
model = nn.Sequential(
nn.Conv2d(3, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 10, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.AdaptiveAvgPool2d(1),
Flatten(),
)
model.cuda()
for xb, yb in data.train_dl:
out = (model(*[xb]))
print(out.size())
break
show_doc(PoolFlatten)
```
We can combine these two final layers ([AdaptiveAvgPool2d](https://pytorch.org/docs/stable/nn.html#torch.nn.AdaptiveAvgPool2d) and [`Flatten`](/layers.html#Flatten)) by using [`PoolFlatten`](/layers.html#PoolFlatten).
```
model = nn.Sequential(
nn.Conv2d(3, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 10, kernel_size=3, stride=2, padding=1), nn.ReLU(),
PoolFlatten()
)
model.cuda()
for xb, yb in data.train_dl:
out = (model(*[xb]))
print(out.size())
break
show_doc(ResizeBatch)
```
Another use we give to the Lambda function is to resize batches with [`ResizeBatch`](/layers.html#ResizeBatch) when we have a layer that expects a different input than what comes from the previous one. Let's see an example:
```
a = torch.tensor([[1., -1.], [1., -1.]])
print(a)
out = ResizeBatch(4)
print(out(a))
show_doc(StdUpsample, doc_string=False)
```
Increases the dimensionality of our data from `n_in` to `n_out` by applying a transposed convolution layer to the input and with batchnorm and a RELU activation.
```
show_doc(CrossEntropyFlat, doc_string=False)
```
Same as [nn.CrossEntropyLoss](https://pytorch.org/docs/stable/nn.html#torch.nn.CrossEntropyLoss), but flattens input and target. Is used to calculate cross entropy on arrays (which Pytorch will not let us do with their [nn.CrossEntropyLoss](https://pytorch.org/docs/stable/nn.html#torch.nn.CrossEntropyLoss) function). An example of a use case is image segmentation models where the output in an image (or an array of pixels).
The parameters are the same as [nn.CrossEntropyLoss](https://pytorch.org/docs/stable/nn.html#torch.nn.CrossEntropyLoss): `weight` to rescale each class, `size_average` whether we want to sum the losses across elements in a batch or we want to add them up, `ignore_index` what targets do we want to ignore, `reduce` on whether we want to return a loss per batch element and `reduction` specifies which type of reduction (if any) we want to apply to our input.
```
show_doc(MSELossFlat)
show_doc(Debugger)
```
The debugger module allows us to peek inside a network while its training and see in detail what is going on. We can see inputs, ouputs and sizes at any point in the network.
For instance, if you run the following:
``` python
model = nn.Sequential(
nn.Conv2d(3, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
Debugger(),
nn.Conv2d(16, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 10, kernel_size=3, stride=2, padding=1), nn.ReLU(),
)
model.cuda()
learner = Learner(data, model, metrics=[accuracy])
learner.fit(5)
```
... you'll see something like this:
```
/home/ubuntu/fastai/fastai/layers.py(74)forward()
72 def forward(self,x:Tensor) -> Tensor:
73 set_trace()
---> 74 return x
75
76 class StdUpsample(nn.Module):
ipdb>
```
```
show_doc(bn_drop_lin, doc_string=False)
```
The [`bn_drop_lin`](/layers.html#bn_drop_lin) function returns a sequence of [batch normalization](https://arxiv.org/abs/1502.03167), [dropout](https://www.cs.toronto.edu/~hinton/absps/JMLRdropout.pdf) and a linear layer. This custom layer is usually used at the end of a model.
`n_in` represents the number of size of the input `n_out` the size of the output, `bn` whether we want batch norm or not, `p` is how much dropout and `actn` is an optional parameter to add an activation function at the end.
```
show_doc(conv2d)
show_doc(conv2d_relu, doc_string=False)
```
Create a [`conv2d`](/layers.html#conv2d) layer with [`nn.ReLU`](https://pytorch.org/docs/stable/nn.html#torch.nn.ReLU) activation and optional(`bn`) [`nn.BatchNorm2d`](https://pytorch.org/docs/stable/nn.html#torch.nn.BatchNorm2d): `ni` input, `nf` out filters, `ks` kernel, `stride`:stride, `padding`:padding, `bn`: batch normalization.
```
show_doc(conv2d_trans)
show_doc(conv_layer, doc_string=False)
```
The [`conv_layer`](/layers.html#conv_layer) function returns a sequence of [nn.Conv2D](https://pytorch.org/docs/stable/nn.html#torch.nn.Conv2d), [BatchNorm2d](https://arxiv.org/abs/1502.03167) and a [leaky RELU](https://ai.stanford.edu/~amaas/papers/relu_hybrid_icml2013_final.pdf) activation function.
`n_in` represents the number of size of the input `n_out` the size of the output, `ks` kernel size, `stride` the stride with which we want to apply the convolutions.
```
show_doc(get_embedding, doc_string=False)
```
Create an [embedding layer](https://arxiv.org/abs/1711.09160) with input size `ni` and output size `nf`.
```
show_doc(simple_cnn)
show_doc(std_upsample_head, doc_string=False)
```
Create a sequence of upsample layers with a RELU at the beggining and a [nn.ConvTranspose2d](https://pytorch.org/docs/stable/nn.html#torch.nn.ConvTranspose2d).
`nfs` is a list with the input and output sizes of each upsample layer and `c` is the output size of the final 2D Transpose Convolutional layer.
```
show_doc(trunc_normal_)
```
## Undocumented Methods - Methods moved below this line will intentionally be hidden
## New Methods - Please document or move to the undocumented section
```
show_doc(Debugger.forward)
show_doc(StdUpsample.forward)
show_doc(MSELossFlat.forward)
show_doc(CrossEntropyFlat.forward)
show_doc(Lambda.forward)
show_doc(AdaptiveConcatPool2d.forward)
```
|
github_jupyter
|
from fastai import *
from fastai.vision import *
from fastai.gen_doc.nbdoc import *
show_doc(AdaptiveConcatPool2d, doc_string=False)
from fastai.gen_doc.nbdoc import *
from fastai.layers import *
path = untar_data(URLs.MNIST_SAMPLE)
data = ImageDataBunch.from_folder(path)
def simple_cnn_max(actns:Collection[int], kernel_szs:Collection[int]=None,
strides:Collection[int]=None) -> nn.Sequential:
"CNN with `conv2d_relu` layers defined by `actns`, `kernel_szs` and `strides`"
nl = len(actns)-1
kernel_szs = ifnone(kernel_szs, [3]*nl)
strides = ifnone(strides , [2]*nl)
layers = [conv2d_relu(actns[i], actns[i+1], kernel_szs[i], stride=strides[i])
for i in range(len(strides))]
layers.append(nn.Sequential(nn.AdaptiveMaxPool2d(1), Flatten()))
return nn.Sequential(*layers)
model = simple_cnn_max((3,16,16,2))
learner = Learner(data, model, metrics=[accuracy])
learner.fit(1)
def simple_cnn_avg(actns:Collection[int], kernel_szs:Collection[int]=None,
strides:Collection[int]=None) -> nn.Sequential:
"CNN with `conv2d_relu` layers defined by `actns`, `kernel_szs` and `strides`"
nl = len(actns)-1
kernel_szs = ifnone(kernel_szs, [3]*nl)
strides = ifnone(strides , [2]*nl)
layers = [conv2d_relu(actns[i], actns[i+1], kernel_szs[i], stride=strides[i])
for i in range(len(strides))]
layers.append(nn.Sequential(nn.AdaptiveAvgPool2d(1), Flatten()))
return nn.Sequential(*layers)
model = simple_cnn_avg((3,16,16,2))
learner = Learner(data, model, metrics=[accuracy])
learner.fit(1)
def simple_cnn(actns:Collection[int], kernel_szs:Collection[int]=None,
strides:Collection[int]=None) -> nn.Sequential:
"CNN with `conv2d_relu` layers defined by `actns`, `kernel_szs` and `strides`"
nl = len(actns)-1
kernel_szs = ifnone(kernel_szs, [3]*nl)
strides = ifnone(strides , [2]*nl)
layers = [conv2d_relu(actns[i], actns[i+1], kernel_szs[i], stride=strides[i])
for i in range(len(strides))]
layers.append(nn.Sequential(AdaptiveConcatPool2d(1), Flatten()))
return nn.Sequential(*layers)
model = simple_cnn((3,16,16,2))
learner = Learner(data, model, metrics=[accuracy])
learner.fit(1)
show_doc(Lambda, doc_string=False)
model = nn.Sequential(
nn.Conv2d(3, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 10, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.AdaptiveAvgPool2d(1),
)
model.cuda()
for xb, yb in data.train_dl:
out = (model(*[xb]))
print(out.size())
break
model = nn.Sequential(
nn.Conv2d(3, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 10, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.AdaptiveAvgPool2d(1),
Lambda(lambda x: x.view(x.size(0),-1))
)
model.cuda()
for xb, yb in data.train_dl:
out = (model(*[xb]))
print(out.size())
break
show_doc(Flatten)
model = nn.Sequential(
nn.Conv2d(3, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 10, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.AdaptiveAvgPool2d(1),
Flatten(),
)
model.cuda()
for xb, yb in data.train_dl:
out = (model(*[xb]))
print(out.size())
break
show_doc(PoolFlatten)
model = nn.Sequential(
nn.Conv2d(3, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 16, kernel_size=3, stride=2, padding=1), nn.ReLU(),
nn.Conv2d(16, 10, kernel_size=3, stride=2, padding=1), nn.ReLU(),
PoolFlatten()
)
model.cuda()
for xb, yb in data.train_dl:
out = (model(*[xb]))
print(out.size())
break
show_doc(ResizeBatch)
a = torch.tensor([[1., -1.], [1., -1.]])
print(a)
out = ResizeBatch(4)
print(out(a))
show_doc(StdUpsample, doc_string=False)
show_doc(CrossEntropyFlat, doc_string=False)
show_doc(MSELossFlat)
show_doc(Debugger)
... you'll see something like this:
The [`bn_drop_lin`](/layers.html#bn_drop_lin) function returns a sequence of [batch normalization](https://arxiv.org/abs/1502.03167), [dropout](https://www.cs.toronto.edu/~hinton/absps/JMLRdropout.pdf) and a linear layer. This custom layer is usually used at the end of a model.
`n_in` represents the number of size of the input `n_out` the size of the output, `bn` whether we want batch norm or not, `p` is how much dropout and `actn` is an optional parameter to add an activation function at the end.
Create a [`conv2d`](/layers.html#conv2d) layer with [`nn.ReLU`](https://pytorch.org/docs/stable/nn.html#torch.nn.ReLU) activation and optional(`bn`) [`nn.BatchNorm2d`](https://pytorch.org/docs/stable/nn.html#torch.nn.BatchNorm2d): `ni` input, `nf` out filters, `ks` kernel, `stride`:stride, `padding`:padding, `bn`: batch normalization.
The [`conv_layer`](/layers.html#conv_layer) function returns a sequence of [nn.Conv2D](https://pytorch.org/docs/stable/nn.html#torch.nn.Conv2d), [BatchNorm2d](https://arxiv.org/abs/1502.03167) and a [leaky RELU](https://ai.stanford.edu/~amaas/papers/relu_hybrid_icml2013_final.pdf) activation function.
`n_in` represents the number of size of the input `n_out` the size of the output, `ks` kernel size, `stride` the stride with which we want to apply the convolutions.
Create an [embedding layer](https://arxiv.org/abs/1711.09160) with input size `ni` and output size `nf`.
Create a sequence of upsample layers with a RELU at the beggining and a [nn.ConvTranspose2d](https://pytorch.org/docs/stable/nn.html#torch.nn.ConvTranspose2d).
`nfs` is a list with the input and output sizes of each upsample layer and `c` is the output size of the final 2D Transpose Convolutional layer.
## Undocumented Methods - Methods moved below this line will intentionally be hidden
## New Methods - Please document or move to the undocumented section
| 0.757705 | 0.987448 |
<div align="center">
<img src='./img/header.png'/>
</div>
## [Global Ice Velocities](https://its-live.jpl.nasa.gov/)
The Inter-mission Time Series of Land Ice Velocity and Elevation (ITS_LIVE) project facilitates ice sheet, ice shelf and glacier research by providing a globally comprehensive and temporally dense multi-sensor record of land ice velocity and elevation with low latency.
Scene-pair velocities generated from satellite optical and radar imagery.
* Coverage: All land ice
* Date range: 1985-present
* Resolution: 240m
* Scene-pair separation: 6 to 546 days
---
* If you want to query our API directly using your own software here is the OpenApi endpoint https://staging.nsidc.org/apps/itslive-search/docs
* For questions about this notebook and the dataset please contact users services at [email protected]
```
#1: Now let's render our UI and pick up an hemisphere, if you update the hemisphere you need to execute the cell again.
import warnings
warnings.filterwarnings('ignore')
from itslive import itslive_ui
ui = itslive_ui('north')
ui.render()
#2: We build the parameters to query the ITS_LIVE Search API, we get the time coverage for our selected area
params = ui.build_params()
print(f'current parameters: {params}')
timeline = None
if params is not None:
timeline = ui.update_coverages()
total = sum(item['count'] for item in timeline)
print(f'Total data granules: {total:,}')
timeline
#3: Now we are going to get the velocity pair urls, this does not download the files yet just their location
urls = []
params = ui.build_params()
if params is not None:
urls = ui.get_granule_urls(params)
# Print the first 10 granule URLs
for url in urls[0:10]:
print(url)
```
## Filtering granules by year and month
```
#4: This will query AWS(where the granules are stored) so we know the total size of our first N granules
# This may take some time, try reducing the selected area or constraining the other parameters to download a reasonable number of granules.
url_list = [url['url'] for url in urls]
# urls
filtered_urls = ui.filter_urls(url_list, max_files_per_year=5, months=[3,4,5,6,7])
# max_granules = 100
# sizes = ui.calculate_file_sizes(filtered_urls, max_granules)
# total_zise = round(sum(sizes)/1024,2)
# print(f'Approx size to download for the first {max_granules:,} granules: {total_zise} MB')
print(len(filtered_urls))
filtered_urls
```
## Downloading the data
**Now that we have our list of data granules we can download them from AWS.**
If this notebook is running inside AWS we could load the granules into a Dask cluster and reduce our processing times and costs.
Let's get some coffee, some data requests are in the Gigabytes realm and may take a little while to be processed.
Once that your status URL says is completed we can grab the HDF5 data file using the URL on the same response!
```
#5 This will download the first 50 velocity pairs
files = ui.download_velocity_pairs(filtered_urls, start=0, end=50)
files
```
## Working with the data
```
import os
import glob
import xarray as xr
import pyproj
import warnings
import pandas as pd
import numpy as np
from datetime import datetime
from shapely.geometry import Polygon
warnings.filterwarnings('ignore')
centroid = ui.dc.last_draw['geometry']['coordinates']
# coord = [-49.59321, 69.210579]
#loads an array of xarray datasets from the nc files
velocity_pairs = ui.load_velocity_pairs('data')
centroid
velocities = []
mean_offset_meters = 1200
for ds in velocity_pairs:
proj = str(int(ds.UTM_Projection.spatial_epsg))
selected_coord = ui.transform_coord('4326',proj, centroid[0], centroid[1])
projected_lon = round(selected_coord[0])
projected_lat = round(selected_coord[1])
mid_date = datetime.strptime(ds.img_pair_info.date_center,'%Y%m%d')
# We are going to calculate the mean value of the neighboring pixels(each is 240m) 10 x 10 window
mask_lon = (ds.x >= projected_lon - mean_offset_meters) & (ds.x <= projected_lon + mean_offset_meters)
mask_lat = (ds.y >= projected_lat - mean_offset_meters) & (ds.y <= projected_lat + mean_offset_meters)
v = ds.where(mask_lon & mask_lat , drop=True).v.mean(skipna=True)
# If we have a valid value we add it to the velocities array.
if not np.isnan(v):
velocities.append({'date': mid_date, 'mean_velocity': v.values.ravel()[0]})
velocities
import pandas as pd
# order by date
df = pd.DataFrame(velocities)
df = df.sort_values(by='date').reset_index(drop=True)
df
df.plot(x ='date', y='mean_velocity', kind = 'bar')
velocity_pairs[0].plot.scatter(x='x', y='y', hue='v')
```
|
github_jupyter
|
#1: Now let's render our UI and pick up an hemisphere, if you update the hemisphere you need to execute the cell again.
import warnings
warnings.filterwarnings('ignore')
from itslive import itslive_ui
ui = itslive_ui('north')
ui.render()
#2: We build the parameters to query the ITS_LIVE Search API, we get the time coverage for our selected area
params = ui.build_params()
print(f'current parameters: {params}')
timeline = None
if params is not None:
timeline = ui.update_coverages()
total = sum(item['count'] for item in timeline)
print(f'Total data granules: {total:,}')
timeline
#3: Now we are going to get the velocity pair urls, this does not download the files yet just their location
urls = []
params = ui.build_params()
if params is not None:
urls = ui.get_granule_urls(params)
# Print the first 10 granule URLs
for url in urls[0:10]:
print(url)
#4: This will query AWS(where the granules are stored) so we know the total size of our first N granules
# This may take some time, try reducing the selected area or constraining the other parameters to download a reasonable number of granules.
url_list = [url['url'] for url in urls]
# urls
filtered_urls = ui.filter_urls(url_list, max_files_per_year=5, months=[3,4,5,6,7])
# max_granules = 100
# sizes = ui.calculate_file_sizes(filtered_urls, max_granules)
# total_zise = round(sum(sizes)/1024,2)
# print(f'Approx size to download for the first {max_granules:,} granules: {total_zise} MB')
print(len(filtered_urls))
filtered_urls
#5 This will download the first 50 velocity pairs
files = ui.download_velocity_pairs(filtered_urls, start=0, end=50)
files
import os
import glob
import xarray as xr
import pyproj
import warnings
import pandas as pd
import numpy as np
from datetime import datetime
from shapely.geometry import Polygon
warnings.filterwarnings('ignore')
centroid = ui.dc.last_draw['geometry']['coordinates']
# coord = [-49.59321, 69.210579]
#loads an array of xarray datasets from the nc files
velocity_pairs = ui.load_velocity_pairs('data')
centroid
velocities = []
mean_offset_meters = 1200
for ds in velocity_pairs:
proj = str(int(ds.UTM_Projection.spatial_epsg))
selected_coord = ui.transform_coord('4326',proj, centroid[0], centroid[1])
projected_lon = round(selected_coord[0])
projected_lat = round(selected_coord[1])
mid_date = datetime.strptime(ds.img_pair_info.date_center,'%Y%m%d')
# We are going to calculate the mean value of the neighboring pixels(each is 240m) 10 x 10 window
mask_lon = (ds.x >= projected_lon - mean_offset_meters) & (ds.x <= projected_lon + mean_offset_meters)
mask_lat = (ds.y >= projected_lat - mean_offset_meters) & (ds.y <= projected_lat + mean_offset_meters)
v = ds.where(mask_lon & mask_lat , drop=True).v.mean(skipna=True)
# If we have a valid value we add it to the velocities array.
if not np.isnan(v):
velocities.append({'date': mid_date, 'mean_velocity': v.values.ravel()[0]})
velocities
import pandas as pd
# order by date
df = pd.DataFrame(velocities)
df = df.sort_values(by='date').reset_index(drop=True)
df
df.plot(x ='date', y='mean_velocity', kind = 'bar')
velocity_pairs[0].plot.scatter(x='x', y='y', hue='v')
| 0.412767 | 0.934991 |
# Fastpages Notebook Blog Post
> A tutorial of fastpages for Jupyter notebooks.
- toc: true
- badges: true
- comments: true
- categories: [jupyter]
- image: images/chart-preview.png
# About
This notebook is a demonstration of some of capabilities of [fastpages](https://github.com/fastai/fastpages) with notebooks.
With `fastpages` you can save your jupyter notebooks into the `_notebooks` folder at the root of your repository, and they will be automatically be converted to Jekyll compliant blog posts!
## Front Matter
The first cell in your Jupyter Notebook or markdown blog post contains front matter. Front matter is metadata that can turn on/off options in your Notebook. It is formatted like this:
```
# "My Title"
> "Awesome summary"
- toc: true- branch: master- badges: true
- comments: true
- author: Hamel Husain & Jeremy Howard
- categories: [fastpages, jupyter]
```
- Setting `toc: true` will automatically generate a table of contents
- Setting `badges: true` will automatically include GitHub and Google Colab links to your notebook.
- Setting `comments: true` will enable commenting on your blog post, powered by [utterances](https://github.com/utterance/utterances).
The title and description need to be enclosed in double quotes only if they include special characters such as a colon. More details and options for front matter can be viewed on the [front matter section](https://github.com/fastai/fastpages#front-matter-related-options) of the README.
## Markdown Shortcuts
A `#hide` comment at the top of any code cell will hide **both the input and output** of that cell in your blog post.
A `#hide_input` comment at the top of any code cell will **only hide the input** of that cell.
```
#hide_input
print('The comment #hide_input was used to hide the code that produced this.')
```
put a `#collapse-hide` flag at the top of any cell if you want to **hide** that cell by default, but give the reader the option to show it:
```
#collapse-hide
import pandas as pd
import altair as alt
```
put a `#collapse-show` flag at the top of any cell if you want to **show** that cell by default, but give the reader the option to hide it:
```
#collapse-show
cars = 'https://vega.github.io/vega-datasets/data/cars.json'
movies = 'https://vega.github.io/vega-datasets/data/movies.json'
sp500 = 'https://vega.github.io/vega-datasets/data/sp500.csv'
stocks = 'https://vega.github.io/vega-datasets/data/stocks.csv'
flights = 'https://vega.github.io/vega-datasets/data/flights-5k.json'
```
## Interactive Charts With Altair
Charts made with Altair remain interactive. Example charts taken from [this repo](https://github.com/uwdata/visualization-curriculum), specifically [this notebook](https://github.com/uwdata/visualization-curriculum/blob/master/altair_interaction.ipynb).
```
# hide
df = pd.read_json(movies) # load movies data
genres = df['Major_Genre'].unique() # get unique field values
genres = list(filter(lambda d: d is not None, genres)) # filter out None values
genres.sort() # sort alphabetically
#hide
mpaa = ['G', 'PG', 'PG-13', 'R', 'NC-17', 'Not Rated']
```
### Example 1: DropDown
```
# single-value selection over [Major_Genre, MPAA_Rating] pairs
# use specific hard-wired values as the initial selected values
selection = alt.selection_single(
name='Select',
fields=['Major_Genre', 'MPAA_Rating'],
init={'Major_Genre': 'Drama', 'MPAA_Rating': 'R'},
bind={'Major_Genre': alt.binding_select(options=genres), 'MPAA_Rating': alt.binding_radio(options=mpaa)}
)
# scatter plot, modify opacity based on selection
alt.Chart(movies).mark_circle().add_selection(
selection
).encode(
x='Rotten_Tomatoes_Rating:Q',
y='IMDB_Rating:Q',
tooltip='Title:N',
opacity=alt.condition(selection, alt.value(0.75), alt.value(0.05))
)
```
### Example 2: Tooltips
```
alt.Chart(movies).mark_circle().add_selection(
alt.selection_interval(bind='scales', encodings=['x'])
).encode(
x='Rotten_Tomatoes_Rating:Q',
y=alt.Y('IMDB_Rating:Q', axis=alt.Axis(minExtent=30)), # use min extent to stabilize axis title placement
tooltip=['Title:N', 'Release_Date:N', 'IMDB_Rating:Q', 'Rotten_Tomatoes_Rating:Q']
).properties(
width=600,
height=400
)
```
### Example 3: More Tooltips
```
# select a point for which to provide details-on-demand
label = alt.selection_single(
encodings=['x'], # limit selection to x-axis value
on='mouseover', # select on mouseover events
nearest=True, # select data point nearest the cursor
empty='none' # empty selection includes no data points
)
# define our base line chart of stock prices
base = alt.Chart().mark_line().encode(
alt.X('date:T'),
alt.Y('price:Q', scale=alt.Scale(type='log')),
alt.Color('symbol:N')
)
alt.layer(
base, # base line chart
# add a rule mark to serve as a guide line
alt.Chart().mark_rule(color='#aaa').encode(
x='date:T'
).transform_filter(label),
# add circle marks for selected time points, hide unselected points
base.mark_circle().encode(
opacity=alt.condition(label, alt.value(1), alt.value(0))
).add_selection(label),
# add white stroked text to provide a legible background for labels
base.mark_text(align='left', dx=5, dy=-5, stroke='white', strokeWidth=2).encode(
text='price:Q'
).transform_filter(label),
# add text labels for stock prices
base.mark_text(align='left', dx=5, dy=-5).encode(
text='price:Q'
).transform_filter(label),
data=stocks
).properties(
width=700,
height=400
)
```
## Data Tables
You can display tables per the usual way in your blog:
```
movies = 'https://vega.github.io/vega-datasets/data/movies.json'
df = pd.read_json(movies)
# display table with pandas
df[['Title', 'Worldwide_Gross',
'Production_Budget', 'Distributor', 'MPAA_Rating', 'IMDB_Rating', 'Rotten_Tomatoes_Rating']].head()
```
## Images
### Local Images
You can reference local images and they will be copied and rendered on your blog automatically. You can include these with the following markdown syntax:
``

### Remote Images
Remote images can be included with the following markdown syntax:
``

### Animated Gifs
Animated Gifs work, too!
``

### Captions
You can include captions with markdown images like this:
```

```

# Other Elements
## GitHub Flavored Emojis
Typing `I give this post two :+1:!` will render this:
I give this post two :+1:!
## Tweetcards
Typing `> twitter: https://twitter.com/jakevdp/status/1204765621767901185?s=20` will render this:
> twitter: https://twitter.com/jakevdp/status/1204765621767901185?s=20
## Youtube Videos
Typing `> youtube: https://youtu.be/XfoYk_Z5AkI` will render this:
> youtube: https://youtu.be/XfoYk_Z5AkI
## Boxes / Callouts
Typing `> Warning: There will be no second warning!` will render this:
> Warning: There will be no second warning!
Typing `> Important: Pay attention! It's important.` will render this:
> Important: Pay attention! It's important.
Typing `> Tip: This is my tip.` will render this:
> Tip: This is my tip.
Typing `> Note: Take note of this.` will render this:
> Note: Take note of this.
Typing `> Note: A doc link to [an example website: fast.ai](https://www.fast.ai/) should also work fine.` will render in the docs:
> Note: A doc link to [an example website: fast.ai](https://www.fast.ai/) should also work fine.
## Footnotes
You can have footnotes in notebooks, however the syntax is different compared to markdown documents. [This guide provides more detail about this syntax](https://github.com/fastai/fastpages/blob/master/_fastpages_docs/NOTEBOOK_FOOTNOTES.md), which looks like this:
```
{% raw %}For example, here is a footnote {% fn 1 %}.
And another {% fn 2 %}
{{ 'This is the footnote.' | fndetail: 1 }}
{{ 'This is the other footnote. You can even have a [link](www.github.com)!' | fndetail: 2 }}{% endraw %}
```
For example, here is a footnote {% fn 1 %}.
And another {% fn 2 %}
{{ 'This is the footnote.' | fndetail: 1 }}
{{ 'This is the other footnote. You can even have a [link](www.github.com)!' | fndetail: 2 }}
|
github_jupyter
|
# "My Title"
> "Awesome summary"
- toc: true- branch: master- badges: true
- comments: true
- author: Hamel Husain & Jeremy Howard
- categories: [fastpages, jupyter]
#hide_input
print('The comment #hide_input was used to hide the code that produced this.')
#collapse-hide
import pandas as pd
import altair as alt
#collapse-show
cars = 'https://vega.github.io/vega-datasets/data/cars.json'
movies = 'https://vega.github.io/vega-datasets/data/movies.json'
sp500 = 'https://vega.github.io/vega-datasets/data/sp500.csv'
stocks = 'https://vega.github.io/vega-datasets/data/stocks.csv'
flights = 'https://vega.github.io/vega-datasets/data/flights-5k.json'
# hide
df = pd.read_json(movies) # load movies data
genres = df['Major_Genre'].unique() # get unique field values
genres = list(filter(lambda d: d is not None, genres)) # filter out None values
genres.sort() # sort alphabetically
#hide
mpaa = ['G', 'PG', 'PG-13', 'R', 'NC-17', 'Not Rated']
# single-value selection over [Major_Genre, MPAA_Rating] pairs
# use specific hard-wired values as the initial selected values
selection = alt.selection_single(
name='Select',
fields=['Major_Genre', 'MPAA_Rating'],
init={'Major_Genre': 'Drama', 'MPAA_Rating': 'R'},
bind={'Major_Genre': alt.binding_select(options=genres), 'MPAA_Rating': alt.binding_radio(options=mpaa)}
)
# scatter plot, modify opacity based on selection
alt.Chart(movies).mark_circle().add_selection(
selection
).encode(
x='Rotten_Tomatoes_Rating:Q',
y='IMDB_Rating:Q',
tooltip='Title:N',
opacity=alt.condition(selection, alt.value(0.75), alt.value(0.05))
)
alt.Chart(movies).mark_circle().add_selection(
alt.selection_interval(bind='scales', encodings=['x'])
).encode(
x='Rotten_Tomatoes_Rating:Q',
y=alt.Y('IMDB_Rating:Q', axis=alt.Axis(minExtent=30)), # use min extent to stabilize axis title placement
tooltip=['Title:N', 'Release_Date:N', 'IMDB_Rating:Q', 'Rotten_Tomatoes_Rating:Q']
).properties(
width=600,
height=400
)
# select a point for which to provide details-on-demand
label = alt.selection_single(
encodings=['x'], # limit selection to x-axis value
on='mouseover', # select on mouseover events
nearest=True, # select data point nearest the cursor
empty='none' # empty selection includes no data points
)
# define our base line chart of stock prices
base = alt.Chart().mark_line().encode(
alt.X('date:T'),
alt.Y('price:Q', scale=alt.Scale(type='log')),
alt.Color('symbol:N')
)
alt.layer(
base, # base line chart
# add a rule mark to serve as a guide line
alt.Chart().mark_rule(color='#aaa').encode(
x='date:T'
).transform_filter(label),
# add circle marks for selected time points, hide unselected points
base.mark_circle().encode(
opacity=alt.condition(label, alt.value(1), alt.value(0))
).add_selection(label),
# add white stroked text to provide a legible background for labels
base.mark_text(align='left', dx=5, dy=-5, stroke='white', strokeWidth=2).encode(
text='price:Q'
).transform_filter(label),
# add text labels for stock prices
base.mark_text(align='left', dx=5, dy=-5).encode(
text='price:Q'
).transform_filter(label),
data=stocks
).properties(
width=700,
height=400
)
movies = 'https://vega.github.io/vega-datasets/data/movies.json'
df = pd.read_json(movies)
# display table with pandas
df[['Title', 'Worldwide_Gross',
'Production_Budget', 'Distributor', 'MPAA_Rating', 'IMDB_Rating', 'Rotten_Tomatoes_Rating']].head()

{% raw %}For example, here is a footnote {% fn 1 %}.
And another {% fn 2 %}
{{ 'This is the footnote.' | fndetail: 1 }}
{{ 'This is the other footnote. You can even have a [link](www.github.com)!' | fndetail: 2 }}{% endraw %}
| 0.591133 | 0.878471 |
```
from __future__ import print_function, division
import time
import datetime
from dateutil import parser
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
%matplotlib inline
import re
import string
import pprint
from textblob import TextBlob
import os
import subprocess
from nltk.corpus import stopwords, wordnet
from nltk.tokenize import word_tokenize
from nltk.stem import SnowballStemmer
from pymongo import MongoClient
client = MongoClient(port=12345)# Tunnel to my mongoDB client
db = client.tech_news_data
arts = db.articles
arts.count()
```
# Random pull
```
#pull a random document from database
import random
def get_random_doc():
c = arts.find()
count = c.count()
return c[random.randrange(count)]
doc = get_random_doc()['title']
doc
```
# Extraction and processing
```
class Articles(object):
"""A memory-friendly way to load large corpora"""
def __init__(self, connection):
self.connection = connection
def __iter__(self):
# iterate through all file names in our directory
for item in self.connection.find():
yield item
def preprocess_series_text(data):
"""Perform complete preprocessing on a Pandas series
including removal of alpha numerical words, normalization,
punctuation removal, tokenization, stop word removal,
and lemmatization."""
# remove alpha numerical words and make lowercase
alphanum_re = re.compile(r"""\w*\d\w*""")
alphanum_lambda = lambda x: alphanum_re.sub('', x.strip().lower())
data = data.map(alphanum_lambda)
#some characters don't show up in standard punctuation
punct = string.punctuation + '‘’“”£€–…'
# remove punctuation
punc_re = re.compile('[%s]' % re.escape(punct))
punc_lambda = lambda x: punc_re.sub(' ', x)
data = data.map(punc_lambda)
# tokenize words
data = data.map(word_tokenize)
# remove stop words
sw = stopwords.words('english')
sw_lambda = lambda x: list(filter(lambda y: y not in sw, x))
data = data.map(sw_lambda)
# stem words
stemmer = SnowballStemmer('english')
stemmer_fun = lambda x: list(map(stemmer.stem, x))
data = data.map(stemmer_fun)
return data
from nltk.sentiment import SentimentAnalyzer
from nltk.sentiment.util import *
from nltk.sentiment.vader import SentimentIntensityAnalyzer
sent = SentimentIntensityAnalyzer()
#testing the sentiment analyzer
sent.polarity_scores('the quick brown fox jumps over the lazy dog')['compound']
#Load mongodb data into pandas and process
articles = Articles(arts)
art_df = pd.DataFrame.from_dict(list(articles))
art_df.drop('_id', axis=1, inplace=True)
art_df['tags'] = art_df['tags'].apply(lambda x: list(set([i.lower() for i in x])))
art_df['polarity'] = art_df['body'].apply(lambda x: sent.polarity_scores(x)['compound'])
art_df['polarity'].fillna(0)
#Wipe empty rows
#art_df['title2'] = preprocess_series_text(art_df['title'])
#mask = art_df['title2'].map(len) > 0
#art_df['title2'] = art_df['title2'].loc[mask].reset_index(drop=True)
#art_df['title2'] = art_df['title2'].apply(lambda x: ' '.join(x))
art_df['body'] = preprocess_series_text(art_df['body'])
mask = art_df['body'].map(len) > 0
art_df['body'] = art_df['body'].loc[mask].reset_index(drop=True)
art_df['body'] = art_df['body'].apply(lambda x: ' '.join(x))
date = datetime.datetime.today() - datetime.timedelta(days=7)
art_df = art_df[art_df.date >= date]
art_df.shape
art_df.groupby(['date'])['body'].count()
plt.hist(art_df['polarity'], bins=50);
plt.title('Distribution of sentiment scores across tech articles')
```
# WordCloud
```
from sklearn.feature_extraction.text import CountVectorizer
v = TfidfVectorizer(stop_words='english', strip_accents= 'ascii', min_df=0.005)
a = v.fit_transform(art_df[art_df['body'].apply(lambda x: 'hamze' not in x)]['body'])
df2 = pd.DataFrame(a.toarray(), columns=v.get_feature_names())
df2.columns = [i[0].upper() + i[1:] for i in df2.columns]
wCloud = df2.sum().sort_values(ascending=False).head(200)
wCloud = wCloud/wCloud.iloc[0]*100
wCloud = wCloud.apply(lambda x: int(x))
#dump words to a js file to build a D3 wordcloud (DONT STEM)
with open('wordcloud/static/article_dump.js', 'w') as f:
f.write('article_dump = [')
for i in range(wCloud.shape[0]):
f.write('{text: \'' + wCloud.index[i] + '\', size: ' + str(wCloud.iloc[i]) + '},\n')
f.write('];')
```
# TFIDF, LSA Vectorization
```
from sklearn.pipeline import Pipeline
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.decomposition import TruncatedSVD
from sklearn.preprocessing import Normalizer
dims = 250
pipe = Pipeline([('tfidf', TfidfVectorizer(stop_words='english', tokenizer = word_tokenize, \
min_df=0.005, strip_accents= 'ascii', ngram_range=(1,3))),
('lsa', TruncatedSVD(dims, algorithm = 'arpack')),
('normalizer', Normalizer())])
#pipe.fit_transform(art_df['body'])
df2 = pd.DataFrame(pipe.fit_transform(art_df['body']))
pipe.steps[1][1].explained_variance_ratio_.sum()
dims2 = range(1,dims+1)
plt.plot(dims2, np.cumsum(pipe.steps[1][1].explained_variance_ratio_))
plt.title('Total input variance across dimensions')
plt.xlabel('number of LSA dimensions')
plt.ylabel('Explained fraction of input variance')
```
# LSA Topic modeling
```
def print_topic_top_words(pipe, n_topics=15, n_words=6):
cv = pipe.steps[0][1]
model = pipe.steps[1][1]
feature_names = cv.get_feature_names()
for topic_num in range(n_topics):
topic_mat = model.components_[topic_num]
print('Topic {}:'.format(topic_num + 1).center(80))
topic_values = sorted(zip(topic_mat, feature_names),
reverse=True)[:n_words]
print(' '.join([y for x,y in topic_values]))
print('-'*80)
print_topic_top_words(pipe)
```
# K-means
```
from sklearn import metrics
from sklearn.metrics import pairwise_distances
from sklearn.metrics import silhouette_score
from sklearn.preprocessing import scale
from sklearn.datasets import fetch_mldata
from sklearn.cluster import KMeans
from sklearn.utils import shuffle
n_clus=20
km = KMeans(n_clusters=n_clus)
km.fit(df2)
art_df['labels'] = km.labels_
art_df.groupby('labels')['body'].count()
art_df[art_df['labels']==19].title
#extract keywords from clusters
def get_keywords(cluster):
tf = TfidfVectorizer(stop_words='english', tokenizer = word_tokenize, \
min_df=0.005, strip_accents= 'ascii', ngram_range=(1,1))
X = tf.fit_transform(cluster).toarray()
return ' '.join(pd.DataFrame(X, columns=tf.get_feature_names()).mean().sort_values(ascending=False).index[:3])
keywords = art_df.groupby('labels')['body'].agg(get_keywords)
results = art_df.groupby('labels').agg({'polarity': ['median'], 'title':'first'})
results['keywords'] = keywords
results
```
# tSNE; Further dimensionality reduction
```
#run tsne so we can get a 2D visualization of clusters
from sklearn.manifold import TSNE
tsne = TSNE()
df3 = pd.DataFrame(tsne.fit_transform(df2))
df3['labels'] = art_df['labels'].values
colors = ['red','blue', 'green', 'orange', 'purple', \
'yellow', 'black', 'brown', 'gray', 'darkgoldenrod',\
'fuchsia', 'lime', 'orchid','teal', 'chartreuse', \
'coral', 'gold', 'sandybrown', 'powderblue', 'seagreen']
for i in range(n_clus):
if 'tito' not in keywords.values[i]:
plt.scatter(df3[df3['labels']==i].iloc[:,0].values, df3[df3['labels']==i].iloc[:,1].values, c= colors[i % len(colors)], label=keywords.values[i])
plt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)
plt.title('Topic clusters: 08/11/2017-8/17/2017')
art_df.groupby('date').count().sum()
```
# Getting the elbow curve
```
SSEs = []
Sil_coefs = []
Inertia = []
k_range = range(2,30)
for k in k_range:
print('Cluster numbers: ' + str(k))
km = KMeans(n_clusters=k, random_state=1)
km.fit(df2)
labels = km.labels_
Sil_coefs.append(metrics.silhouette_score(df2, labels, metric='euclidean'))
# SSEs.append(get_SSE(X_digits, labels)) # The SSE is just inertia, we
SSEs.append(km.inertia_) # could have just said km.inertia_
fig, (ax1, ax2) = plt.subplots(1,2, figsize=(15,5), sharex=True)
k_clusters = k_range
# the silhouete coefficient is (b - a) / max(a, b), where b(i) is the average distance between a point i and every other point from the closest cluster;
# and a(i) is the average distance between a point i and every other point within its own cluster. It has a value from -1,1. 1 means it belongs to the cluster.
ax1.plot(k_clusters, Sil_coefs)
ax1.set_xlabel('number of clusters')
ax1.set_ylabel('silhouette coefficient')
ax1.set_title('silhouette plot')
# plot here on ax2
ax2.plot(k_clusters, SSEs)
ax2.set_xlabel('number of clusters')
ax2.set_ylabel('Inertia');
ax2.set_title('Inertia Curve')
```
# Other Clustering
```
from sklearn.cluster import DBSCAN,AgglomerativeClustering, SpectralClustering
from sklearn.metrics.pairwise import cosine_distances
#metric = 'euclidean'
metric = 'cosine'
clus = DBSCAN(eps=0.4, min_samples=3, metric=metric, algorithm='brute')
#clus = SpectralClustering(n_clusters=50, affinity='cosine', n_init=15, n_neighbors=5)
clus.fit(df2)
art_df['labels'] = clus.labels_
art_df.groupby('labels')['body'].count()
#What articles seem to be related?
art_df[art_df['labels'] == 12]['title'].values
```
# LDA Topic modeling
```
from sklearn.decomposition import LatentDirichletAllocation as LDA
pipe2 = [
('tfidf', TfidfVectorizer(stop_words='english', tokenizer = word_tokenize, \
min_df=0.001, strip_accents= 'ascii', ngram_range=(1,3))),
('lda', LDA(n_topics=10, learning_method='batch'))]
pipe2 = Pipeline(pipe2)
pipe2.fit_transform(art_df['body'])
def topics(pipe, n_words=5):
features = pipe.steps[0][1].get_feature_names()
for i, topic in enumerate(pipe.steps[1][1].components_):
print('Topic ID:', i+1)
print(' '.join([features[i] for i in topic.argsort()[::-1][:n_words]]))
topics(pipe2, 6)
```
|
github_jupyter
|
from __future__ import print_function, division
import time
import datetime
from dateutil import parser
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
%matplotlib inline
import re
import string
import pprint
from textblob import TextBlob
import os
import subprocess
from nltk.corpus import stopwords, wordnet
from nltk.tokenize import word_tokenize
from nltk.stem import SnowballStemmer
from pymongo import MongoClient
client = MongoClient(port=12345)# Tunnel to my mongoDB client
db = client.tech_news_data
arts = db.articles
arts.count()
#pull a random document from database
import random
def get_random_doc():
c = arts.find()
count = c.count()
return c[random.randrange(count)]
doc = get_random_doc()['title']
doc
class Articles(object):
"""A memory-friendly way to load large corpora"""
def __init__(self, connection):
self.connection = connection
def __iter__(self):
# iterate through all file names in our directory
for item in self.connection.find():
yield item
def preprocess_series_text(data):
"""Perform complete preprocessing on a Pandas series
including removal of alpha numerical words, normalization,
punctuation removal, tokenization, stop word removal,
and lemmatization."""
# remove alpha numerical words and make lowercase
alphanum_re = re.compile(r"""\w*\d\w*""")
alphanum_lambda = lambda x: alphanum_re.sub('', x.strip().lower())
data = data.map(alphanum_lambda)
#some characters don't show up in standard punctuation
punct = string.punctuation + '‘’“”£€–…'
# remove punctuation
punc_re = re.compile('[%s]' % re.escape(punct))
punc_lambda = lambda x: punc_re.sub(' ', x)
data = data.map(punc_lambda)
# tokenize words
data = data.map(word_tokenize)
# remove stop words
sw = stopwords.words('english')
sw_lambda = lambda x: list(filter(lambda y: y not in sw, x))
data = data.map(sw_lambda)
# stem words
stemmer = SnowballStemmer('english')
stemmer_fun = lambda x: list(map(stemmer.stem, x))
data = data.map(stemmer_fun)
return data
from nltk.sentiment import SentimentAnalyzer
from nltk.sentiment.util import *
from nltk.sentiment.vader import SentimentIntensityAnalyzer
sent = SentimentIntensityAnalyzer()
#testing the sentiment analyzer
sent.polarity_scores('the quick brown fox jumps over the lazy dog')['compound']
#Load mongodb data into pandas and process
articles = Articles(arts)
art_df = pd.DataFrame.from_dict(list(articles))
art_df.drop('_id', axis=1, inplace=True)
art_df['tags'] = art_df['tags'].apply(lambda x: list(set([i.lower() for i in x])))
art_df['polarity'] = art_df['body'].apply(lambda x: sent.polarity_scores(x)['compound'])
art_df['polarity'].fillna(0)
#Wipe empty rows
#art_df['title2'] = preprocess_series_text(art_df['title'])
#mask = art_df['title2'].map(len) > 0
#art_df['title2'] = art_df['title2'].loc[mask].reset_index(drop=True)
#art_df['title2'] = art_df['title2'].apply(lambda x: ' '.join(x))
art_df['body'] = preprocess_series_text(art_df['body'])
mask = art_df['body'].map(len) > 0
art_df['body'] = art_df['body'].loc[mask].reset_index(drop=True)
art_df['body'] = art_df['body'].apply(lambda x: ' '.join(x))
date = datetime.datetime.today() - datetime.timedelta(days=7)
art_df = art_df[art_df.date >= date]
art_df.shape
art_df.groupby(['date'])['body'].count()
plt.hist(art_df['polarity'], bins=50);
plt.title('Distribution of sentiment scores across tech articles')
from sklearn.feature_extraction.text import CountVectorizer
v = TfidfVectorizer(stop_words='english', strip_accents= 'ascii', min_df=0.005)
a = v.fit_transform(art_df[art_df['body'].apply(lambda x: 'hamze' not in x)]['body'])
df2 = pd.DataFrame(a.toarray(), columns=v.get_feature_names())
df2.columns = [i[0].upper() + i[1:] for i in df2.columns]
wCloud = df2.sum().sort_values(ascending=False).head(200)
wCloud = wCloud/wCloud.iloc[0]*100
wCloud = wCloud.apply(lambda x: int(x))
#dump words to a js file to build a D3 wordcloud (DONT STEM)
with open('wordcloud/static/article_dump.js', 'w') as f:
f.write('article_dump = [')
for i in range(wCloud.shape[0]):
f.write('{text: \'' + wCloud.index[i] + '\', size: ' + str(wCloud.iloc[i]) + '},\n')
f.write('];')
from sklearn.pipeline import Pipeline
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.decomposition import TruncatedSVD
from sklearn.preprocessing import Normalizer
dims = 250
pipe = Pipeline([('tfidf', TfidfVectorizer(stop_words='english', tokenizer = word_tokenize, \
min_df=0.005, strip_accents= 'ascii', ngram_range=(1,3))),
('lsa', TruncatedSVD(dims, algorithm = 'arpack')),
('normalizer', Normalizer())])
#pipe.fit_transform(art_df['body'])
df2 = pd.DataFrame(pipe.fit_transform(art_df['body']))
pipe.steps[1][1].explained_variance_ratio_.sum()
dims2 = range(1,dims+1)
plt.plot(dims2, np.cumsum(pipe.steps[1][1].explained_variance_ratio_))
plt.title('Total input variance across dimensions')
plt.xlabel('number of LSA dimensions')
plt.ylabel('Explained fraction of input variance')
def print_topic_top_words(pipe, n_topics=15, n_words=6):
cv = pipe.steps[0][1]
model = pipe.steps[1][1]
feature_names = cv.get_feature_names()
for topic_num in range(n_topics):
topic_mat = model.components_[topic_num]
print('Topic {}:'.format(topic_num + 1).center(80))
topic_values = sorted(zip(topic_mat, feature_names),
reverse=True)[:n_words]
print(' '.join([y for x,y in topic_values]))
print('-'*80)
print_topic_top_words(pipe)
from sklearn import metrics
from sklearn.metrics import pairwise_distances
from sklearn.metrics import silhouette_score
from sklearn.preprocessing import scale
from sklearn.datasets import fetch_mldata
from sklearn.cluster import KMeans
from sklearn.utils import shuffle
n_clus=20
km = KMeans(n_clusters=n_clus)
km.fit(df2)
art_df['labels'] = km.labels_
art_df.groupby('labels')['body'].count()
art_df[art_df['labels']==19].title
#extract keywords from clusters
def get_keywords(cluster):
tf = TfidfVectorizer(stop_words='english', tokenizer = word_tokenize, \
min_df=0.005, strip_accents= 'ascii', ngram_range=(1,1))
X = tf.fit_transform(cluster).toarray()
return ' '.join(pd.DataFrame(X, columns=tf.get_feature_names()).mean().sort_values(ascending=False).index[:3])
keywords = art_df.groupby('labels')['body'].agg(get_keywords)
results = art_df.groupby('labels').agg({'polarity': ['median'], 'title':'first'})
results['keywords'] = keywords
results
#run tsne so we can get a 2D visualization of clusters
from sklearn.manifold import TSNE
tsne = TSNE()
df3 = pd.DataFrame(tsne.fit_transform(df2))
df3['labels'] = art_df['labels'].values
colors = ['red','blue', 'green', 'orange', 'purple', \
'yellow', 'black', 'brown', 'gray', 'darkgoldenrod',\
'fuchsia', 'lime', 'orchid','teal', 'chartreuse', \
'coral', 'gold', 'sandybrown', 'powderblue', 'seagreen']
for i in range(n_clus):
if 'tito' not in keywords.values[i]:
plt.scatter(df3[df3['labels']==i].iloc[:,0].values, df3[df3['labels']==i].iloc[:,1].values, c= colors[i % len(colors)], label=keywords.values[i])
plt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)
plt.title('Topic clusters: 08/11/2017-8/17/2017')
art_df.groupby('date').count().sum()
SSEs = []
Sil_coefs = []
Inertia = []
k_range = range(2,30)
for k in k_range:
print('Cluster numbers: ' + str(k))
km = KMeans(n_clusters=k, random_state=1)
km.fit(df2)
labels = km.labels_
Sil_coefs.append(metrics.silhouette_score(df2, labels, metric='euclidean'))
# SSEs.append(get_SSE(X_digits, labels)) # The SSE is just inertia, we
SSEs.append(km.inertia_) # could have just said km.inertia_
fig, (ax1, ax2) = plt.subplots(1,2, figsize=(15,5), sharex=True)
k_clusters = k_range
# the silhouete coefficient is (b - a) / max(a, b), where b(i) is the average distance between a point i and every other point from the closest cluster;
# and a(i) is the average distance between a point i and every other point within its own cluster. It has a value from -1,1. 1 means it belongs to the cluster.
ax1.plot(k_clusters, Sil_coefs)
ax1.set_xlabel('number of clusters')
ax1.set_ylabel('silhouette coefficient')
ax1.set_title('silhouette plot')
# plot here on ax2
ax2.plot(k_clusters, SSEs)
ax2.set_xlabel('number of clusters')
ax2.set_ylabel('Inertia');
ax2.set_title('Inertia Curve')
from sklearn.cluster import DBSCAN,AgglomerativeClustering, SpectralClustering
from sklearn.metrics.pairwise import cosine_distances
#metric = 'euclidean'
metric = 'cosine'
clus = DBSCAN(eps=0.4, min_samples=3, metric=metric, algorithm='brute')
#clus = SpectralClustering(n_clusters=50, affinity='cosine', n_init=15, n_neighbors=5)
clus.fit(df2)
art_df['labels'] = clus.labels_
art_df.groupby('labels')['body'].count()
#What articles seem to be related?
art_df[art_df['labels'] == 12]['title'].values
from sklearn.decomposition import LatentDirichletAllocation as LDA
pipe2 = [
('tfidf', TfidfVectorizer(stop_words='english', tokenizer = word_tokenize, \
min_df=0.001, strip_accents= 'ascii', ngram_range=(1,3))),
('lda', LDA(n_topics=10, learning_method='batch'))]
pipe2 = Pipeline(pipe2)
pipe2.fit_transform(art_df['body'])
def topics(pipe, n_words=5):
features = pipe.steps[0][1].get_feature_names()
for i, topic in enumerate(pipe.steps[1][1].components_):
print('Topic ID:', i+1)
print(' '.join([features[i] for i in topic.argsort()[::-1][:n_words]]))
topics(pipe2, 6)
| 0.596198 | 0.573858 |
# Runge-Kutta Implementation
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
```
# Define a function to integrate
```
def dfdx(x,f):
return x**2 + x
```
# Define the integral of the function
```
def f_int(x,C):
return (x**3)/3. + 0.5*x**2 + C
```
# Define the 2nd order Runge-Kutta method
```
def rk2_core(x_i,f_i,h,g):
# advance f by a step h
# half step
x_ipoh = x_i + 0.5*h
f_ipoh = f_i + 0.5*h*g(x_i,f_i)
# full step
f_ipo = f_i + h*g(x_ipoh, f_ipoh)
return f_ipo
```
# Define a wrapper function for 2nd order Runge-Kutta
```
def rk2(dfdx,a,b,f_a,N):
# dfdx is the derivate wrt x
# a is lower bound
# b is upper bound
# f_a is the boundary condition at 'a'
# N is the number of steps to take
# define the steps
x = np.linspace(a,b,N)
# set the single step size
h = x[1]-x[0]
# create array to hold f
f = np.zeros(N,dtype=float)
f[0] = f_a # the value of f at a
# evolve function f along x
for i in range(1,N):
f[i] = rk2_core(x[i-1],f[i-1],h,dfdx)
return x,f
```
# Define the 4th order Runge-Kutta method
```
def rk4_core(x_i,f_i,h,g):
# Define x at 1/2 step
x_ipoh = x_i + 0.5*h
# Define x at 1 step
x_ipo = x_i + h
# Advance f by a step h
k_1 = h*g(x_i,f_i)
k_2 = h*g(x_ipoh, f_i + 0.5*k_1)
k_3 = h*g(x_ipoh, f_i + 0.5*k_2)
k_4 = h*g(x_ipo, f_i + k_3)
f_ipo = f_i + (k_1 + 2*k_2 + 2*k_3 + k_4)/6.
return f_ipo
```
# Define a wrapper function for 4th order Runge-Kutta
```
def rk4(dfdx,a,b,f_a,N):
# dfdx is the derivate wrt x
# a is lower bound
# b is upper bound
# f_a is the boundary condition at 'a'
# N is the number of steps to take
# define the steps
x = np.linspace(a,b,N)
# set the single step size
h = x[1]-x[0]
# create array to hold f
f = np.zeros(N,dtype=float)
f[0] = f_a # the value of f at a
# evolve function f along x
for i in range(1,N):
f[i] = rk4_core(x[i-1],f[i-1],h,dfdx)
return x,f
```
# Perform the integration
```
a = 0.0
b = 1.0
f_a = 0.0
N = 10
x_2, f_2 = rk2(dfdx,a,b,f_a,N)
x_4, f_4 = rk4(dfdx,a,b,f_a,N)
x = x_2.copy()
plt.plot(x_2,f_2,label='RK2')
plt.plot(x_4,f_4,label='RK4')
plt.plot(x,f_int(x,f_a),'o',label='Analytic')
plt.legend(frameon=False)
```
# Plot the error
```
plt.plot(x_2,f_2 - f_int(x,f_a),label='RK2')
plt.plot(x_4,f_4 - f_int(x,f_a),label='RK4')
plt.legend()
```
|
github_jupyter
|
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
def dfdx(x,f):
return x**2 + x
def f_int(x,C):
return (x**3)/3. + 0.5*x**2 + C
def rk2_core(x_i,f_i,h,g):
# advance f by a step h
# half step
x_ipoh = x_i + 0.5*h
f_ipoh = f_i + 0.5*h*g(x_i,f_i)
# full step
f_ipo = f_i + h*g(x_ipoh, f_ipoh)
return f_ipo
def rk2(dfdx,a,b,f_a,N):
# dfdx is the derivate wrt x
# a is lower bound
# b is upper bound
# f_a is the boundary condition at 'a'
# N is the number of steps to take
# define the steps
x = np.linspace(a,b,N)
# set the single step size
h = x[1]-x[0]
# create array to hold f
f = np.zeros(N,dtype=float)
f[0] = f_a # the value of f at a
# evolve function f along x
for i in range(1,N):
f[i] = rk2_core(x[i-1],f[i-1],h,dfdx)
return x,f
def rk4_core(x_i,f_i,h,g):
# Define x at 1/2 step
x_ipoh = x_i + 0.5*h
# Define x at 1 step
x_ipo = x_i + h
# Advance f by a step h
k_1 = h*g(x_i,f_i)
k_2 = h*g(x_ipoh, f_i + 0.5*k_1)
k_3 = h*g(x_ipoh, f_i + 0.5*k_2)
k_4 = h*g(x_ipo, f_i + k_3)
f_ipo = f_i + (k_1 + 2*k_2 + 2*k_3 + k_4)/6.
return f_ipo
def rk4(dfdx,a,b,f_a,N):
# dfdx is the derivate wrt x
# a is lower bound
# b is upper bound
# f_a is the boundary condition at 'a'
# N is the number of steps to take
# define the steps
x = np.linspace(a,b,N)
# set the single step size
h = x[1]-x[0]
# create array to hold f
f = np.zeros(N,dtype=float)
f[0] = f_a # the value of f at a
# evolve function f along x
for i in range(1,N):
f[i] = rk4_core(x[i-1],f[i-1],h,dfdx)
return x,f
a = 0.0
b = 1.0
f_a = 0.0
N = 10
x_2, f_2 = rk2(dfdx,a,b,f_a,N)
x_4, f_4 = rk4(dfdx,a,b,f_a,N)
x = x_2.copy()
plt.plot(x_2,f_2,label='RK2')
plt.plot(x_4,f_4,label='RK4')
plt.plot(x,f_int(x,f_a),'o',label='Analytic')
plt.legend(frameon=False)
plt.plot(x_2,f_2 - f_int(x,f_a),label='RK2')
plt.plot(x_4,f_4 - f_int(x,f_a),label='RK4')
plt.legend()
| 0.4917 | 0.938632 |
<a href="https://colab.research.google.com/github/jeffheaton/t81_558_deep_learning/blob/master/t81_558_class_05_2_kfold.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# T81-558: Applications of Deep Neural Networks
**Module 5: Regularization and Dropout**
* Instructor: [Jeff Heaton](https://sites.wustl.edu/jeffheaton/), McKelvey School of Engineering, [Washington University in St. Louis](https://engineering.wustl.edu/Programs/Pages/default.aspx)
* For more information visit the [class website](https://sites.wustl.edu/jeffheaton/t81-558/).
# Module 5 Material
* Part 5.1: Part 5.1: Introduction to Regularization: Ridge and Lasso [[Video]](https://www.youtube.com/watch?v=jfgRtCYjoBs&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_05_1_reg_ridge_lasso.ipynb)
* **Part 5.2: Using K-Fold Cross Validation with Keras** [[Video]](https://www.youtube.com/watch?v=maiQf8ray_s&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_05_2_kfold.ipynb)
* Part 5.3: Using L1 and L2 Regularization with Keras to Decrease Overfitting [[Video]](https://www.youtube.com/watch?v=JEWzWv1fBFQ&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_05_3_keras_l1_l2.ipynb)
* Part 5.4: Drop Out for Keras to Decrease Overfitting [[Video]](https://www.youtube.com/watch?v=bRyOi0L6Rs8&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_05_4_dropout.ipynb)
* Part 5.5: Benchmarking Keras Deep Learning Regularization Techniques [[Video]](https://www.youtube.com/watch?v=1NLBwPumUAs&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_05_5_bootstrap.ipynb)
# Google CoLab Instructions
The following code ensures that Google CoLab is running the correct version of TensorFlow.
```
try:
%tensorflow_version 2.x
COLAB = True
print("Note: using Google CoLab")
except:
print("Note: not using Google CoLab")
COLAB = False
```
# Part 5.2: Using K-Fold Cross-validation with Keras
Cross-validation can be used for a variety of purposes in predictive modeling. These include:
* Generating out-of-sample predictions from a neural network
* Estimate a good number of epochs to train a neural network for (early stopping)
* Evaluate the effectiveness of certain hyperparameters, such as activation functions, neuron counts, and layer counts
Cross-validation uses a number of folds, and multiple models, to provide each segment of data a chance to serve as both the validation and training set.

It is important to note that there will be one model (neural network) for each fold. To generate predictions for new data, which is data not present in the training set, predictions from the fold models can be handled in several ways:
* Choose the model that had the highest validation score as the final model.
* Preset new data to the 5 models (one for each fold) and average the result (this is an [ensemble](https://en.wikipedia.org/wiki/Ensemble_learning)).
* Retrain a new model (using the same settings as the cross-validation) on the entire dataset. Train for as many epochs, and with the same hidden layer structure.
Generally, I prefer the last approach and will retrain a model on the entire data set once I have selected hyper-parameters. Of course, I will always set aside a final holdout set for model validation that I do not use in any aspect of the training process.
### Regression vs Classification K-Fold Cross-Validation
Regression and classification are handled somewhat differently with regards to cross-validation. Regression is the simpler case where you can simply break up the data set into K folds with little regard for where each item lands. For regression it is best that the data items fall into the folds as randomly as possible. It is also important to remember that not every fold will necessarily have exactly the same number of data items. It is not always possible for the data set to be evenly divided into K folds. For regression cross-validation we will use the Scikit-Learn class **KFold**.
Cross validation for classification could also use the **KFold** object; however, this technique would not ensure that the class balance remains the same in each fold as it was in the original. It is very important that the balance of classes that a model was trained on remains the same (or similar) to the training set. A drift in this distribution is one of the most important things to monitor after a trained model has been placed into actual use. Because of this, we want to make sure that the cross-validation itself does not introduce an unintended shift. This is referred to as stratified sampling and is accomplished by using the Scikit-Learn object **StratifiedKFold** in place of **KFold** whenever you are using classification. In summary, the following two objects in Scikit-Learn should be used:
* **KFold** When dealing with a regression problem.
* **StratifiedKFold** When dealing with a classification problem.
The following two sections demonstrate cross-validation with classification and regression.
### Out-of-Sample Regression Predictions with K-Fold Cross-Validation
The following code trains the simple dataset using a 5-fold cross-validation. The expected performance of a neural network, of the type trained here, would be the score for the generated out-of-sample predictions. We begin by preparing a feature vector using the jh-simple-dataset to predict age. This is a regression problem.
```
import pandas as pd
from scipy.stats import zscore
from sklearn.model_selection import train_test_split
# Read the data set
df = pd.read_csv(
"https://data.heatonresearch.com/data/t81-558/jh-simple-dataset.csv",
na_values=['NA','?'])
# Generate dummies for job
df = pd.concat([df,pd.get_dummies(df['job'],prefix="job")],axis=1)
df.drop('job', axis=1, inplace=True)
# Generate dummies for area
df = pd.concat([df,pd.get_dummies(df['area'],prefix="area")],axis=1)
df.drop('area', axis=1, inplace=True)
# Generate dummies for product
df = pd.concat([df,pd.get_dummies(df['product'],prefix="product")],axis=1)
df.drop('product', axis=1, inplace=True)
# Missing values for income
med = df['income'].median()
df['income'] = df['income'].fillna(med)
# Standardize ranges
df['income'] = zscore(df['income'])
df['aspect'] = zscore(df['aspect'])
df['save_rate'] = zscore(df['save_rate'])
df['subscriptions'] = zscore(df['subscriptions'])
# Convert to numpy - Classification
x_columns = df.columns.drop('age').drop('id')
x = df[x_columns].values
y = df['age'].values
```
Now that the feature vector is created a 5-fold cross-validation can be performed to generate out of sample predictions. We will assume 500 epochs, and not use early stopping. Later we will see how we can estimate a more optimal epoch count.
```
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from scipy.stats import zscore
from sklearn.model_selection import KFold
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation
# Cross-Validate
kf = KFold(5, shuffle=True, random_state=42) # Use for KFold classification
oos_y = []
oos_pred = []
fold = 0
for train, test in kf.split(x):
fold+=1
print(f"Fold #{fold}")
x_train = x[train]
y_train = y[train]
x_test = x[test]
y_test = y[test]
model = Sequential()
model.add(Dense(20, input_dim=x.shape[1], activation='relu'))
model.add(Dense(10, activation='relu'))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
model.fit(x_train,y_train,validation_data=(x_test,y_test),verbose=0,epochs=500)
pred = model.predict(x_test)
oos_y.append(y_test)
oos_pred.append(pred)
# Measure this fold's RMSE
score = np.sqrt(metrics.mean_squared_error(pred,y_test))
print(f"Fold score (RMSE): {score}")
# Build the oos prediction list and calculate the error.
oos_y = np.concatenate(oos_y)
oos_pred = np.concatenate(oos_pred)
score = np.sqrt(metrics.mean_squared_error(oos_pred,oos_y))
print(f"Final, out of sample score (RMSE): {score}")
# Write the cross-validated prediction
oos_y = pd.DataFrame(oos_y)
oos_pred = pd.DataFrame(oos_pred)
oosDF = pd.concat( [df, oos_y, oos_pred],axis=1 )
#oosDF.to_csv(filename_write,index=False)
```
As you can see, the above code also reports the average number of epochs needed. A common technique is to then train on the entire dataset for the average number of epochs needed.
### Classification with Stratified K-Fold Cross-Validation
The following code trains and fits the jh-simple-dataset dataset with cross-validation to generate out-of-sample . It also writes out the out of sample (predictions on the test set) results.
It is good to perform a stratified k-fold cross validation with classification data. This ensures that the percentages of each class remains the same across all folds. To do this, make use of the **StratifiedKFold** object, instead of the **KFold** object used in regression.
```
import pandas as pd
from scipy.stats import zscore
# Read the data set
df = pd.read_csv(
"https://data.heatonresearch.com/data/t81-558/jh-simple-dataset.csv",
na_values=['NA','?'])
# Generate dummies for job
df = pd.concat([df,pd.get_dummies(df['job'],prefix="job")],axis=1)
df.drop('job', axis=1, inplace=True)
# Generate dummies for area
df = pd.concat([df,pd.get_dummies(df['area'],prefix="area")],axis=1)
df.drop('area', axis=1, inplace=True)
# Missing values for income
med = df['income'].median()
df['income'] = df['income'].fillna(med)
# Standardize ranges
df['income'] = zscore(df['income'])
df['aspect'] = zscore(df['aspect'])
df['save_rate'] = zscore(df['save_rate'])
df['age'] = zscore(df['age'])
df['subscriptions'] = zscore(df['subscriptions'])
# Convert to numpy - Classification
x_columns = df.columns.drop('product').drop('id')
x = df[x_columns].values
dummies = pd.get_dummies(df['product']) # Classification
products = dummies.columns
y = dummies.values
```
We will assume 500 epochs, and not use early stopping. Later we will see how we can estimate a more optimal epoch count.
```
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from sklearn.model_selection import StratifiedKFold
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation
# np.argmax(pred,axis=1)
# Cross-validate
kf = StratifiedKFold(5, shuffle=True, random_state=42) # Use for StratifiedKFold classification
oos_y = []
oos_pred = []
fold = 0
for train, test in kf.split(x,df['product']): # Must specify y StratifiedKFold for
fold+=1
print(f"Fold #{fold}")
x_train = x[train]
y_train = y[train]
x_test = x[test]
y_test = y[test]
model = Sequential()
model.add(Dense(50, input_dim=x.shape[1], activation='relu')) # Hidden 1
model.add(Dense(25, activation='relu')) # Hidden 2
model.add(Dense(y.shape[1],activation='softmax')) # Output
model.compile(loss='categorical_crossentropy', optimizer='adam')
model.fit(x_train,y_train,validation_data=(x_test,y_test),verbose=0,epochs=500)
pred = model.predict(x_test)
oos_y.append(y_test)
pred = np.argmax(pred,axis=1) # raw probabilities to chosen class (highest probability)
oos_pred.append(pred)
# Measure this fold's accuracy
y_compare = np.argmax(y_test,axis=1) # For accuracy calculation
score = metrics.accuracy_score(y_compare, pred)
print(f"Fold score (accuracy): {score}")
# Build the oos prediction list and calculate the error.
oos_y = np.concatenate(oos_y)
oos_pred = np.concatenate(oos_pred)
oos_y_compare = np.argmax(oos_y,axis=1) # For accuracy calculation
score = metrics.accuracy_score(oos_y_compare, oos_pred)
print(f"Final score (accuracy): {score}")
# Write the cross-validated prediction
oos_y = pd.DataFrame(oos_y)
oos_pred = pd.DataFrame(oos_pred)
oosDF = pd.concat( [df, oos_y, oos_pred],axis=1 )
#oosDF.to_csv(filename_write,index=False)
```
### Training with both a Cross-Validation and a Holdout Set
If you have a considerable amount of data, it is always valuable to set aside a holdout set before you cross-validate. This hold out set will be the final evaluation before you make use of your model for its real-world use.

The following program makes use of a holdout set, and then still cross-validates.
```
import pandas as pd
from scipy.stats import zscore
from sklearn.model_selection import train_test_split
# Read the data set
df = pd.read_csv(
"https://data.heatonresearch.com/data/t81-558/jh-simple-dataset.csv",
na_values=['NA','?'])
# Generate dummies for job
df = pd.concat([df,pd.get_dummies(df['job'],prefix="job")],axis=1)
df.drop('job', axis=1, inplace=True)
# Generate dummies for area
df = pd.concat([df,pd.get_dummies(df['area'],prefix="area")],axis=1)
df.drop('area', axis=1, inplace=True)
# Generate dummies for product
df = pd.concat([df,pd.get_dummies(df['product'],prefix="product")],axis=1)
df.drop('product', axis=1, inplace=True)
# Missing values for income
med = df['income'].median()
df['income'] = df['income'].fillna(med)
# Standardize ranges
df['income'] = zscore(df['income'])
df['aspect'] = zscore(df['aspect'])
df['save_rate'] = zscore(df['save_rate'])
df['subscriptions'] = zscore(df['subscriptions'])
# Convert to numpy - Classification
x_columns = df.columns.drop('age').drop('id')
x = df[x_columns].values
y = df['age'].values
from sklearn.model_selection import train_test_split
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from scipy.stats import zscore
from sklearn.model_selection import KFold
# Keep a 10% holdout
x_main, x_holdout, y_main, y_holdout = train_test_split(
x, y, test_size=0.10)
# Cross-validate
kf = KFold(5)
oos_y = []
oos_pred = []
fold = 0
for train, test in kf.split(x_main):
fold+=1
print(f"Fold #{fold}")
x_train = x_main[train]
y_train = y_main[train]
x_test = x_main[test]
y_test = y_main[test]
model = Sequential()
model.add(Dense(20, input_dim=x.shape[1], activation='relu'))
model.add(Dense(5, activation='relu'))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
model.fit(x_train,y_train,validation_data=(x_test,y_test),verbose=0,epochs=500)
pred = model.predict(x_test)
oos_y.append(y_test)
oos_pred.append(pred)
# Measure accuracy
score = np.sqrt(metrics.mean_squared_error(pred,y_test))
print(f"Fold score (RMSE): {score}")
# Build the oos prediction list and calculate the error.
oos_y = np.concatenate(oos_y)
oos_pred = np.concatenate(oos_pred)
score = np.sqrt(metrics.mean_squared_error(oos_pred,oos_y))
print()
print(f"Cross-validated score (RMSE): {score}")
# Write the cross-validated prediction (from the last neural network)
holdout_pred = model.predict(x_holdout)
score = np.sqrt(metrics.mean_squared_error(holdout_pred,y_holdout))
print(f"Holdout score (RMSE): {score}")
```
|
github_jupyter
|
try:
%tensorflow_version 2.x
COLAB = True
print("Note: using Google CoLab")
except:
print("Note: not using Google CoLab")
COLAB = False
import pandas as pd
from scipy.stats import zscore
from sklearn.model_selection import train_test_split
# Read the data set
df = pd.read_csv(
"https://data.heatonresearch.com/data/t81-558/jh-simple-dataset.csv",
na_values=['NA','?'])
# Generate dummies for job
df = pd.concat([df,pd.get_dummies(df['job'],prefix="job")],axis=1)
df.drop('job', axis=1, inplace=True)
# Generate dummies for area
df = pd.concat([df,pd.get_dummies(df['area'],prefix="area")],axis=1)
df.drop('area', axis=1, inplace=True)
# Generate dummies for product
df = pd.concat([df,pd.get_dummies(df['product'],prefix="product")],axis=1)
df.drop('product', axis=1, inplace=True)
# Missing values for income
med = df['income'].median()
df['income'] = df['income'].fillna(med)
# Standardize ranges
df['income'] = zscore(df['income'])
df['aspect'] = zscore(df['aspect'])
df['save_rate'] = zscore(df['save_rate'])
df['subscriptions'] = zscore(df['subscriptions'])
# Convert to numpy - Classification
x_columns = df.columns.drop('age').drop('id')
x = df[x_columns].values
y = df['age'].values
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from scipy.stats import zscore
from sklearn.model_selection import KFold
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation
# Cross-Validate
kf = KFold(5, shuffle=True, random_state=42) # Use for KFold classification
oos_y = []
oos_pred = []
fold = 0
for train, test in kf.split(x):
fold+=1
print(f"Fold #{fold}")
x_train = x[train]
y_train = y[train]
x_test = x[test]
y_test = y[test]
model = Sequential()
model.add(Dense(20, input_dim=x.shape[1], activation='relu'))
model.add(Dense(10, activation='relu'))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
model.fit(x_train,y_train,validation_data=(x_test,y_test),verbose=0,epochs=500)
pred = model.predict(x_test)
oos_y.append(y_test)
oos_pred.append(pred)
# Measure this fold's RMSE
score = np.sqrt(metrics.mean_squared_error(pred,y_test))
print(f"Fold score (RMSE): {score}")
# Build the oos prediction list and calculate the error.
oos_y = np.concatenate(oos_y)
oos_pred = np.concatenate(oos_pred)
score = np.sqrt(metrics.mean_squared_error(oos_pred,oos_y))
print(f"Final, out of sample score (RMSE): {score}")
# Write the cross-validated prediction
oos_y = pd.DataFrame(oos_y)
oos_pred = pd.DataFrame(oos_pred)
oosDF = pd.concat( [df, oos_y, oos_pred],axis=1 )
#oosDF.to_csv(filename_write,index=False)
import pandas as pd
from scipy.stats import zscore
# Read the data set
df = pd.read_csv(
"https://data.heatonresearch.com/data/t81-558/jh-simple-dataset.csv",
na_values=['NA','?'])
# Generate dummies for job
df = pd.concat([df,pd.get_dummies(df['job'],prefix="job")],axis=1)
df.drop('job', axis=1, inplace=True)
# Generate dummies for area
df = pd.concat([df,pd.get_dummies(df['area'],prefix="area")],axis=1)
df.drop('area', axis=1, inplace=True)
# Missing values for income
med = df['income'].median()
df['income'] = df['income'].fillna(med)
# Standardize ranges
df['income'] = zscore(df['income'])
df['aspect'] = zscore(df['aspect'])
df['save_rate'] = zscore(df['save_rate'])
df['age'] = zscore(df['age'])
df['subscriptions'] = zscore(df['subscriptions'])
# Convert to numpy - Classification
x_columns = df.columns.drop('product').drop('id')
x = df[x_columns].values
dummies = pd.get_dummies(df['product']) # Classification
products = dummies.columns
y = dummies.values
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from sklearn.model_selection import StratifiedKFold
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation
# np.argmax(pred,axis=1)
# Cross-validate
kf = StratifiedKFold(5, shuffle=True, random_state=42) # Use for StratifiedKFold classification
oos_y = []
oos_pred = []
fold = 0
for train, test in kf.split(x,df['product']): # Must specify y StratifiedKFold for
fold+=1
print(f"Fold #{fold}")
x_train = x[train]
y_train = y[train]
x_test = x[test]
y_test = y[test]
model = Sequential()
model.add(Dense(50, input_dim=x.shape[1], activation='relu')) # Hidden 1
model.add(Dense(25, activation='relu')) # Hidden 2
model.add(Dense(y.shape[1],activation='softmax')) # Output
model.compile(loss='categorical_crossentropy', optimizer='adam')
model.fit(x_train,y_train,validation_data=(x_test,y_test),verbose=0,epochs=500)
pred = model.predict(x_test)
oos_y.append(y_test)
pred = np.argmax(pred,axis=1) # raw probabilities to chosen class (highest probability)
oos_pred.append(pred)
# Measure this fold's accuracy
y_compare = np.argmax(y_test,axis=1) # For accuracy calculation
score = metrics.accuracy_score(y_compare, pred)
print(f"Fold score (accuracy): {score}")
# Build the oos prediction list and calculate the error.
oos_y = np.concatenate(oos_y)
oos_pred = np.concatenate(oos_pred)
oos_y_compare = np.argmax(oos_y,axis=1) # For accuracy calculation
score = metrics.accuracy_score(oos_y_compare, oos_pred)
print(f"Final score (accuracy): {score}")
# Write the cross-validated prediction
oos_y = pd.DataFrame(oos_y)
oos_pred = pd.DataFrame(oos_pred)
oosDF = pd.concat( [df, oos_y, oos_pred],axis=1 )
#oosDF.to_csv(filename_write,index=False)
import pandas as pd
from scipy.stats import zscore
from sklearn.model_selection import train_test_split
# Read the data set
df = pd.read_csv(
"https://data.heatonresearch.com/data/t81-558/jh-simple-dataset.csv",
na_values=['NA','?'])
# Generate dummies for job
df = pd.concat([df,pd.get_dummies(df['job'],prefix="job")],axis=1)
df.drop('job', axis=1, inplace=True)
# Generate dummies for area
df = pd.concat([df,pd.get_dummies(df['area'],prefix="area")],axis=1)
df.drop('area', axis=1, inplace=True)
# Generate dummies for product
df = pd.concat([df,pd.get_dummies(df['product'],prefix="product")],axis=1)
df.drop('product', axis=1, inplace=True)
# Missing values for income
med = df['income'].median()
df['income'] = df['income'].fillna(med)
# Standardize ranges
df['income'] = zscore(df['income'])
df['aspect'] = zscore(df['aspect'])
df['save_rate'] = zscore(df['save_rate'])
df['subscriptions'] = zscore(df['subscriptions'])
# Convert to numpy - Classification
x_columns = df.columns.drop('age').drop('id')
x = df[x_columns].values
y = df['age'].values
from sklearn.model_selection import train_test_split
import pandas as pd
import os
import numpy as np
from sklearn import metrics
from scipy.stats import zscore
from sklearn.model_selection import KFold
# Keep a 10% holdout
x_main, x_holdout, y_main, y_holdout = train_test_split(
x, y, test_size=0.10)
# Cross-validate
kf = KFold(5)
oos_y = []
oos_pred = []
fold = 0
for train, test in kf.split(x_main):
fold+=1
print(f"Fold #{fold}")
x_train = x_main[train]
y_train = y_main[train]
x_test = x_main[test]
y_test = y_main[test]
model = Sequential()
model.add(Dense(20, input_dim=x.shape[1], activation='relu'))
model.add(Dense(5, activation='relu'))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
model.fit(x_train,y_train,validation_data=(x_test,y_test),verbose=0,epochs=500)
pred = model.predict(x_test)
oos_y.append(y_test)
oos_pred.append(pred)
# Measure accuracy
score = np.sqrt(metrics.mean_squared_error(pred,y_test))
print(f"Fold score (RMSE): {score}")
# Build the oos prediction list and calculate the error.
oos_y = np.concatenate(oos_y)
oos_pred = np.concatenate(oos_pred)
score = np.sqrt(metrics.mean_squared_error(oos_pred,oos_y))
print()
print(f"Cross-validated score (RMSE): {score}")
# Write the cross-validated prediction (from the last neural network)
holdout_pred = model.predict(x_holdout)
score = np.sqrt(metrics.mean_squared_error(holdout_pred,y_holdout))
print(f"Holdout score (RMSE): {score}")
| 0.484868 | 0.991804 |
# Text-network Visualization and Analysis
This work is based upon:
(https://gitlab.com/mattiasostmar/discoursebias/blob/master/README.md)
And in the reference article cited in the same README.
Its only a draft, but it's intended to give birth to articles about the content public available of text with the same topics such Buddhism (my aim).
Thus any subsequential working related to these will have it's own place/repository.
Hope you enjoy this for awhile.
```
import tnread as tr
import nltk
import plotly.graph_objects as go
%matplotlib inline
#nltk.download('stopwords')
#nltk.download('rslp')
```
# Run on text from file
First we load a file and read it's contents
```
# Blazing Resplendor
file = open("texts/pt/tulkutest.txt")
name = file.name
text = file.read()
# Otavio conto um
file = open("texts/pt/um_otavio.txt")
name = file.name
text = file.read()
# Otavio conto dois
file = open("texts/pt/dois_otavio.txt")
name = file.name
text = file.read()
# random text
file = open("magic_happiness.txt")
name = file.name
text = file.read()
# Tantra Part 1.0
text = " Identificando o Criador de Todos os Fenômenos como Sendo a Mente Examine o corpo, a fala e a mente e reconheça qual é o primordial como o monarca que tudo cria. Aqui está a maneira de examinar o agente, ou o monarca, que cria todos os fenômenos como sendo a mente, que é primordial entre o corpo, a fala e a mente. Durante o dia, a noite e o período intermediário, devido à autofixação da mente, o corpo e a fala surgem para a mente. Ao longo da vida é a mente que experimenta alegria e tristeza. Ao final, quando o corpo e a mente se separam, o corpo é deixado para trás como um cadáver. Quando a fala desaparece sem deixar vestígios, a mente segue o carma e é o agente que vagueia no saṃsāra. De uma perspectiva, por essas três razões, reconheça aquilo que é primordial como sendo a mente. De outra perspectiva, nenhum desses três é outra coisa além da mente, portanto, ao determiná-los como sendo apenas a mente, reconheça aquilo que é primordial como sendo a mente. A primeira perspectiva é determinada de acordo com o modo convencional das aparências, enquanto a última perspectiva é determinada de acordo com o modo convencional da existência. Para explicar um pouco mais o significado dessa última perspectiva, o tratado do Mahāpaṇḍita Nāropā, A Síntese da Visão, afirma: O Tantra do Vajra Cortante – parte 1 Lama Alan Wallace – Retiro \n Todos os fenômenos que surgem e passam a existir não têm existência separada da mente autoconsciente, pois esta faz com que surjam e fiquem claros, assim como a experiência da própria lucidez. Se eles não fossem a mente, não teriam relação com a mente e nem mesmo surgiriam. Determine a realidade enganosa13 dessa maneira.14 Para a mente deludida, o corpo e a mente surgem como se fossem diferentes, mas em termos de seu modo de existência, eles se apresentam diretamente à consciência não conceitual e são claramente experienciados. Isso indica que eles existem não como algo material, mas simplesmente como a iluminação natural da própria consciência, como a consciência que experiencia alegria e tristeza. Se eles não fossem a mente, mas fossem matéria, como luz e escuridão, nunca poderiam interagir com a mente e, portanto, nunca poderiam surgir. Por essas razões, o corpo e a fala – na verdade todos os fenômenos aparentes – são estabelecidos como sendo a mente."
name = "Tantra Vajra Cortante"
```
Then create a discoursebias Text object, sending in the text and the name of the file as identifier for later use.
```
experiment = tr.TextNetwork(text, name, stemmer="porter", stem=False, max_nodes=100, window = 10, plot=False, lang="pt")
data = experiment.run()
```
Now we can look at the data in the returned DataFrame.
Communities plotting
```
vis=tr.TextnetVis(experiment)
vis.plot_textnet()
def maximal(attrdict,num):
attrdict = {k:v for k,v in sorted(attrdict.items(),key=lambda x: x[1], reverse=True)}
keys = list(attrdict.keys())[0:num]
vals = list(attrdict.values())[0:num]
attrdict = {k:v for k,v in zip(keys,vals)}
return attrdict
def func(rede, source, target=hubs, filtr=maximal,num=2):
'''
Extract connections from source nodes to target nodes, filtering by its weights.
kwargs:
target: target node names (list of str), to see for connections
filtr: callable, a filter function that receives a dict with 2-tuple of
strings as key and a float as value (the weight of the edge).
num: in the case of filtering for maximal weight, the number of top maximal
edges to get.
'''
edges=dict()
for node1 in source:
for node2 in target:
try:
if (node2,node1) not in edges:
edges[(node1,node2)]=rede[node1][node2]['weight']
except:
pass
edges=filtr(edges,num)
return edges
print(experiment.plottable.junctions)
print(experiment.plottable.hubs)
print(experiment.plottable.others)
print(experiment.plottable.info)
edges=dict()
for i in range(len(junc)):
for k,v in func(experiment.finalGraph,[junc[i]],target=hubs,filtr=maximal,num=1).items():
if (k[0],k[1]) not in edges:
edges[k]=v
```
# Plotly Vis (dev)
```
edge_x = []
edge_y = []
for edge in vis.templatedgraph.edges():
x0, y0 = vis.pos[edge[0]]
x1, y1 = vis.pos[edge[1]]
edge_x.append(x0)
edge_x.append(x1)
edge_x.append(None)
edge_y.append(y0)
edge_y.append(y1)
edge_y.append(None)
edge_trace = go.Scatter(
x=edge_x, y=edge_y,
line=dict(width=0.5, color='#888'),
hoverinfo='none',
mode='lines')
node_x = []
node_y = []
for node in vis.templatedgraph.nodes():
x, y = vis.pos[node]
node_x.append(x)
node_y.append(y)
node_trace = go.Scatter(
x=node_x, y=node_y,
mode='markers',
hoverinfo='text',
marker=dict(
showscale=True,
# colorscale options
#'Greys' | 'YlGnBu' | 'Greens' | 'YlOrRd' | 'Bluered' | 'RdBu' |
#'Reds' | 'Blues' | 'Picnic' | 'Rainbow' | 'Portland' | 'Jet' |
#'Hot' | 'Blackbody' | 'Earth' | 'Electric' | 'Viridis' |
colorscale='YlGnBu',
reversescale=True,
color=vis.node_color,
size=vis.node_size,
colorbar=dict(
thickness=15,
title='Node Connections',
xanchor='left',
titleside='right'
),
line_width=2))
vis.node_size=[a*0.05 for a in vis.node_size]
node_adjacencies = []
node_text = []
names=list(vis.templatedgraph.nodes())
for node, adjacencies in enumerate(vis.templatedgraph.adjacency()):
node_adjacencies.append(len(adjacencies[1]))
nodename=names[node]
node_text.append(nodename+'\n'+'# of connections: '+str(len(adjacencies[1])))
node_trace.text = node_text
fig = go.Figure(data=[edge_trace, node_trace],
layout=go.Layout(
title=experiment.textname,
titlefont_size=16,
showlegend=False,
hovermode='closest',
margin=dict(b=20,l=5,r=5,t=40),
annotations=[ dict(
text="",
showarrow=False,
xref="paper", yref="paper",
x=0.005, y=-0.002 ) ],
xaxis=dict(showgrid=False, zeroline=False, showticklabels=False),
yaxis=dict(showgrid=False, zeroline=False, showticklabels=False))
)
fig.show()
transposed = data.T # Flip X to Y
results = transposed[["modularity","entropy","nodesInTopCom","nodesInGiantCom"]]
results
```
# Run on a sequence of texts for comparison
We can run the process of reading several files and adding them into a single Pandas DataFrame for analysis.
```
import glob
results_list = []
for pathname in glob.glob('texts/sv/*'): # glob allows regex to be run in the filesystem
text = open(pathname).read()
filename = pathname.rpartition("/")[2]
exp = did.TextNetwork(text, filename, stemmer="porter")
data = exp.run()
print("----- new experiment ------")
# Now do a little data munging and add the result data for this file
transp_results = data.T[["modularity","entropyTopFirst","nodesInTopCom","nodesInGiantCom"]]
results_list.append(transp_results)
```
|
github_jupyter
|
import tnread as tr
import nltk
import plotly.graph_objects as go
%matplotlib inline
#nltk.download('stopwords')
#nltk.download('rslp')
# Blazing Resplendor
file = open("texts/pt/tulkutest.txt")
name = file.name
text = file.read()
# Otavio conto um
file = open("texts/pt/um_otavio.txt")
name = file.name
text = file.read()
# Otavio conto dois
file = open("texts/pt/dois_otavio.txt")
name = file.name
text = file.read()
# random text
file = open("magic_happiness.txt")
name = file.name
text = file.read()
# Tantra Part 1.0
text = " Identificando o Criador de Todos os Fenômenos como Sendo a Mente Examine o corpo, a fala e a mente e reconheça qual é o primordial como o monarca que tudo cria. Aqui está a maneira de examinar o agente, ou o monarca, que cria todos os fenômenos como sendo a mente, que é primordial entre o corpo, a fala e a mente. Durante o dia, a noite e o período intermediário, devido à autofixação da mente, o corpo e a fala surgem para a mente. Ao longo da vida é a mente que experimenta alegria e tristeza. Ao final, quando o corpo e a mente se separam, o corpo é deixado para trás como um cadáver. Quando a fala desaparece sem deixar vestígios, a mente segue o carma e é o agente que vagueia no saṃsāra. De uma perspectiva, por essas três razões, reconheça aquilo que é primordial como sendo a mente. De outra perspectiva, nenhum desses três é outra coisa além da mente, portanto, ao determiná-los como sendo apenas a mente, reconheça aquilo que é primordial como sendo a mente. A primeira perspectiva é determinada de acordo com o modo convencional das aparências, enquanto a última perspectiva é determinada de acordo com o modo convencional da existência. Para explicar um pouco mais o significado dessa última perspectiva, o tratado do Mahāpaṇḍita Nāropā, A Síntese da Visão, afirma: O Tantra do Vajra Cortante – parte 1 Lama Alan Wallace – Retiro \n Todos os fenômenos que surgem e passam a existir não têm existência separada da mente autoconsciente, pois esta faz com que surjam e fiquem claros, assim como a experiência da própria lucidez. Se eles não fossem a mente, não teriam relação com a mente e nem mesmo surgiriam. Determine a realidade enganosa13 dessa maneira.14 Para a mente deludida, o corpo e a mente surgem como se fossem diferentes, mas em termos de seu modo de existência, eles se apresentam diretamente à consciência não conceitual e são claramente experienciados. Isso indica que eles existem não como algo material, mas simplesmente como a iluminação natural da própria consciência, como a consciência que experiencia alegria e tristeza. Se eles não fossem a mente, mas fossem matéria, como luz e escuridão, nunca poderiam interagir com a mente e, portanto, nunca poderiam surgir. Por essas razões, o corpo e a fala – na verdade todos os fenômenos aparentes – são estabelecidos como sendo a mente."
name = "Tantra Vajra Cortante"
experiment = tr.TextNetwork(text, name, stemmer="porter", stem=False, max_nodes=100, window = 10, plot=False, lang="pt")
data = experiment.run()
vis=tr.TextnetVis(experiment)
vis.plot_textnet()
def maximal(attrdict,num):
attrdict = {k:v for k,v in sorted(attrdict.items(),key=lambda x: x[1], reverse=True)}
keys = list(attrdict.keys())[0:num]
vals = list(attrdict.values())[0:num]
attrdict = {k:v for k,v in zip(keys,vals)}
return attrdict
def func(rede, source, target=hubs, filtr=maximal,num=2):
'''
Extract connections from source nodes to target nodes, filtering by its weights.
kwargs:
target: target node names (list of str), to see for connections
filtr: callable, a filter function that receives a dict with 2-tuple of
strings as key and a float as value (the weight of the edge).
num: in the case of filtering for maximal weight, the number of top maximal
edges to get.
'''
edges=dict()
for node1 in source:
for node2 in target:
try:
if (node2,node1) not in edges:
edges[(node1,node2)]=rede[node1][node2]['weight']
except:
pass
edges=filtr(edges,num)
return edges
print(experiment.plottable.junctions)
print(experiment.plottable.hubs)
print(experiment.plottable.others)
print(experiment.plottable.info)
edges=dict()
for i in range(len(junc)):
for k,v in func(experiment.finalGraph,[junc[i]],target=hubs,filtr=maximal,num=1).items():
if (k[0],k[1]) not in edges:
edges[k]=v
edge_x = []
edge_y = []
for edge in vis.templatedgraph.edges():
x0, y0 = vis.pos[edge[0]]
x1, y1 = vis.pos[edge[1]]
edge_x.append(x0)
edge_x.append(x1)
edge_x.append(None)
edge_y.append(y0)
edge_y.append(y1)
edge_y.append(None)
edge_trace = go.Scatter(
x=edge_x, y=edge_y,
line=dict(width=0.5, color='#888'),
hoverinfo='none',
mode='lines')
node_x = []
node_y = []
for node in vis.templatedgraph.nodes():
x, y = vis.pos[node]
node_x.append(x)
node_y.append(y)
node_trace = go.Scatter(
x=node_x, y=node_y,
mode='markers',
hoverinfo='text',
marker=dict(
showscale=True,
# colorscale options
#'Greys' | 'YlGnBu' | 'Greens' | 'YlOrRd' | 'Bluered' | 'RdBu' |
#'Reds' | 'Blues' | 'Picnic' | 'Rainbow' | 'Portland' | 'Jet' |
#'Hot' | 'Blackbody' | 'Earth' | 'Electric' | 'Viridis' |
colorscale='YlGnBu',
reversescale=True,
color=vis.node_color,
size=vis.node_size,
colorbar=dict(
thickness=15,
title='Node Connections',
xanchor='left',
titleside='right'
),
line_width=2))
vis.node_size=[a*0.05 for a in vis.node_size]
node_adjacencies = []
node_text = []
names=list(vis.templatedgraph.nodes())
for node, adjacencies in enumerate(vis.templatedgraph.adjacency()):
node_adjacencies.append(len(adjacencies[1]))
nodename=names[node]
node_text.append(nodename+'\n'+'# of connections: '+str(len(adjacencies[1])))
node_trace.text = node_text
fig = go.Figure(data=[edge_trace, node_trace],
layout=go.Layout(
title=experiment.textname,
titlefont_size=16,
showlegend=False,
hovermode='closest',
margin=dict(b=20,l=5,r=5,t=40),
annotations=[ dict(
text="",
showarrow=False,
xref="paper", yref="paper",
x=0.005, y=-0.002 ) ],
xaxis=dict(showgrid=False, zeroline=False, showticklabels=False),
yaxis=dict(showgrid=False, zeroline=False, showticklabels=False))
)
fig.show()
transposed = data.T # Flip X to Y
results = transposed[["modularity","entropy","nodesInTopCom","nodesInGiantCom"]]
results
import glob
results_list = []
for pathname in glob.glob('texts/sv/*'): # glob allows regex to be run in the filesystem
text = open(pathname).read()
filename = pathname.rpartition("/")[2]
exp = did.TextNetwork(text, filename, stemmer="porter")
data = exp.run()
print("----- new experiment ------")
# Now do a little data munging and add the result data for this file
transp_results = data.T[["modularity","entropyTopFirst","nodesInTopCom","nodesInGiantCom"]]
results_list.append(transp_results)
| 0.326808 | 0.911022 |
# Tutorial: Herramientas básicas de aprendizaje profundo privado
Bienvenido al tutorial de introducción de PySyft para aprendizaje profundo, privado y descentralizado. Esta serie de archivos es una guía paso a paso para conocer las nuevas herramientas y técnicas requeridas para hacer aprendizaje profundo con modelos/datos secretos/privados sin centralizarlos bajo una autoridad.
**Alcance:** Nosotros no hablaremos solamente acerca de como encriptar datos de forma decentralizada, también sobre como PySyft puede ayudar a decentralizar un ecosistema completo alrededor de datos, incluyendo las bases de datos donde estos son presentados y guardados, y los modelos neuronales que son usados para extraer información de los datos. Cuando nuevas extensiones de PySyft sean creadas, estos archivos serán extendidos con nuevos tutoriales para explicar las nuevas funciones.
Autores:
- Andrew Trask - Twitter: [@iamtrask](https://twitter.com/iamtrask)
Traductores:
- Arturo Márquez Flores - Twitter: [@arturomf94](https://twitter.com/arturomf94)
- Ricardo Pretelt - Twitter: [@ricardopretelt](https://twitter.com/ricardopretelt)
## Descripción general:
- Parte 1: Herramientas básicas de aprendizaje profundo privado
## ¿Por qué hacer este tutorial?
**1) Una ventaja competitiva en tu carrera** - En los últimos 20 años, la revolución digital ha hecho que los datos sean más y más accesibles en grandes cantidades mientras que los procesos análogos se convierten en digitales. Sin embargo, con nuevas regulaciones como [GDPR](https://eugdpr.org/), las empresas están bajo presión para tener menos libertad sobre como usan y analizan información personal. **Para concluir:** Los científicos de datos no van a tener mucho acceso a datos de la misma forma que antes, pero aprendiendo las herramientas de aprendizaje profundo privado, puedes estar arriba de esta curva y ganar una ventaja competitiva en tu carrera.
**2) Oportunidades empresariales** - Hay muchos problemas en la sociedad que pueden ser resueltos con aprendizaje profundo pero muchos de estos no han sido explorados porque requieren acceso a información increíblemente sensible para las personas. (Considera por ejemplo usar aprendizaje profundo para ayudar a personas con problemas mentales o de relaciones). Por lo tanto, el aprendizaje profundo privado abre muchas oportunidades de emprendimiento para tí que antes no estaban disponibles a otros sin estas herramientas.
**3) Bien social** - El aprendizaje profundo puede ser usado para resolver una amplia variedad de problemas en el mundo real, pero aprendizaje profundo con *información personal* es aprendizaje profundo sobre personas, *para personas*.
Aprender como hacer aprendizaje profundo con datos que no te pertencen representan más que una oportunidad de emprendimiento para tu carrera, es la oportunidad de ayudar a resolver uno de los problemas más importantes y personales en la vida de las personas - y hacerlo a escala.
## ¿Cómo consigo créditos extra?
- Dale una estrella a PySyft en GitHub! - [https://github.com/OpenMined/PySyft](https://github.com/OpenMined/PySyft)
- Haz un video en Youtube enseñando este tutorial!
... ok ... ¡Hagámoslo!
# Parte -1: Prerequisitos
- Conocer PyTorch - Si no, entonces toma el curso de http://fast.ai y regresa después
- Lee el paper de la plataforma de PySyft https://arxiv.org/pdf/1811.04017.pdf! Esto te dará un conocimiento completo sobre como PySyft está construido y eso hará que las cosas tengan más sentido.
# Parte 0: Preparación
Para empezar, necesitarás estar seguro de que tienes lo necesario correctamente instalado. Para esto, dirígete al readme de PySyft y sigue las intruscciones de instalación. Para algunos, muy largo para leer.
- Instalar Python 3.5 o en adelante
- Instalar PyTorch 1.1
- Clona PySyft (git clone https://github.com/OpenMined/PySyft.git)
- cd PySyft
- pip install -r pip-dep/requirements.txt
- pip install -r pip-dep/requirements_udacity.txt
- python setup.py install udacity
- python setup.py test
Si alguna parte de esto no funciona (o alguna de las pruebas falla) - primero chequea el [README](https://github.com/OpenMined/PySyft.git) para ayuda de instalación o abre un Issue de Github o escribe en el canal #beginner en ¡nuestro slack! [slack.openmined.org](http://slack.openmined.org/)
```
# Corre esta celda para ver si todo funciona
import sys
import torch
from torch.nn import Parameter
import torch.nn as nn
import torch.nn.functional as F
import syft as sy
hook = sy.TorchHook(torch)
torch.tensor([1,2,3,4,5])
```
Si esta celda es ejecutada exitósamente, entonces ¡Estás listo para comenzar! ¡Hagámoslo!
# Parte 1: Las herramientas básicas de ciencia de datos privada y descentralizada.
Entonces - la primera pregunta que te debes estar haciendo es - ¿Cómo es posible entrenar un modelo con datos a los que no tenemos acceso?
Bien, la respuesta es sorprendentemente simple. Si estas acostumbrado a trabajar con Pytorch, entonces estás acostumbrado a trabajar con objetos torch.Tensor como estos!
```
x = torch.tensor([1,2,3,4,5])
y = x + x
print(y)
```
Obviamente, usar estos elegantes (y poderosos!) tensores es importante, pero también requiere que tengas los datos en tu máquina local. Aquí es donde nuestro viaje comienza.
# Sección 1.1 - Enviar tensores a la máquina de Bob.
Mientras normalmente haríamos ciencia de datos / aprendizaje profundo en la máquina que tiene los datos, ahora queremos realizar esta computación en **otra** máquina. Más específicamente, nosotros ya no podemos asumir que los datos estarán en nuestra máquina local.
Por lo tanto, en vez de usar tensores de Torch, vamos a trabajar con **punteros** a los tensores. Déjame mostrarte a lo que me refiero. Primero crearemos una "aparente" máquina, que pertenece a una "aparente" persona. La llamaremos Bob.
```
bob = sy.VirtualWorker(hook, id="bob")
```
Digamos que la máquina de Bob está en otro planeta - ¡quizás en Marte! Pero, en el momento la máquina está vacía. Vamos a crear algunos datos para enviarlos a Bob y aprender sobre punteros!
```
x = torch.tensor([1,2,3,4,5])
y = torch.tensor([1,1,1,1,1])
```
Y ahora - Enviemos nuestros tensores a Bob!!
```
x_ptr = x.send(bob)
y_ptr = y.send(bob)
x_ptr
```
BOOM! Ahora Bob tiene ¡dos tensores! ¿No me crees? ¡Mira por tí mismo!
```
bob._objects
z = x_ptr + x_ptr
z
bob._objects
```
Ahora mira algo. Cuando llamamos `x.send(bob)` se devolvió un nuevo objeto llamado `x_ptr`. Este es nuestro primer *puntero* a un tensor. Los punteros a tensores en realidad no tienen ningún dato por sí mismos. Sólo contienen metadatos sobre los tensores (con datos) guardados en otra máquina. El propósito de estos tensores es dar una API intuitiva para decirle a la otra máquina que compute funciones usando este tensor. Miremos la metadata que contienen estos punteros.
```
x_ptr
```
Mira la metadata!
Hay dos atributos principales específicamente para punteros:
- `x_ptr.location : bob`, la ubicación, una referencia al lugar que el puntero está apuntando.
- `x_ptr.id_at_location : <random integer>`, el id donde el tensor está guardado en la ubicación.
Están impresas en el formato `<id_at_location>@<location>`
Hay también otros atributos más genéricos:
- `x_ptr.id : <random integer>`, el id de nuestro tensor puntero, fue ubicado aleatoriamente.
- `x_ptr.owner : "me"`, el trabajador al que le pertenece el tensor puntero, aquí es el trabajador local, llamado "me"
```
x_ptr.location
bob
bob == x_ptr.location
x_ptr.id_at_location
x_ptr.owner
```
Tú te puedes preguntar ¿Por qué el trabajador local, el cuál también tiene un puntero, es también un VirtualWorker aunque nosotros no lo creamos?. De hecho, así como tenemos un objeto VirtualWorker para Bob, (por predeterminado) siempre tendremos uno para nosotros también. Este trabajador es automáticamente creado cuando llamamos `hook = sy.TorchHook()` entonces tú no siempre tendrás que crearlo por tí mismo.
```
me = sy.local_worker
me
me == x_ptr.owner
```
Y finalmente, así como podemos llamar .send() en un tensor, podemos llamar .get() en un puntero a un tensor para tenerlo de vuelta.
```
x_ptr
x_ptr.get()
y_ptr
y_ptr.get()
z.get()
bob._objects
```
Y así como puedes ver... ¡Bob ya no más tiene los tensores! ¡Se han movido devuelta a nuestra máquina!
# Sección 1.2 - Usar punteros de tensores
Entonces, enviar y recibir tensores de Bob es genial, pero ¡difícilmente esto es deep learning! Queremos poder hacer _operaciones_ de tensores en tensores remotos. Afortunadamente los punteros de tensores ¡lo hacen muy fácil! Puedes usar punteros ¡así como normalmente usas tensores!
```
x = torch.tensor([1,2,3,4,5]).send(bob)
y = torch.tensor([1,1,1,1,1]).send(bob)
z = x + y
z
```
Y voilà!
Detrás de escenas, algo muy poderoso sucedió. En vez de que _x_ y _y_ se sumaran localmente, un comando fue serializado y enviado a Bob, quién realizó la operación. creó un tensor z, y luego regresó el puntero a z ¡de vuelta a nosotros!
Si llamamos .get() en el puntero, ¡recibiremos el resultado de vuelta a nuestra máquina!
```
z.get()
```
### Funciones de Torch
Esta API ha sido extendida a ¡todas las operaciones de Torch!
```
x
y
z = torch.add(x,y)
z
z.get()
```
### Variables (Incluyendo propagación hacia atrás)
```
x = torch.tensor([1,2,3,4,5.], requires_grad=True).send(bob)
y = torch.tensor([1,1,1,1,1.], requires_grad=True).send(bob)
z = (x + y).sum()
z.backward()
x = x.get()
x
x.grad
```
Así como puedes ver, la API es muy flexible y capaz de realizar casi todas las operaciones que normalmente harías en Torch con datos remotos. Esto pone las bases para protocolos más avanzados para preservar privacidad como aprendizaje federado,
computación segura multipartita, y ¡privacidad diferencial!
# !Felicitaciones! - !Es hora de unirte a la comunidad!
¡Felicitaciones por completar esta parte del tutorial! Si te gustó y quieres unirte al movimiento para preservar la privacidad, propiedad descentralizada de IA y la cadena de suministro de IA (datos), puedes hacerlo de las ¡siguientes formas!
### Dale una estrella a PySyft en GitHub
La forma más fácil de ayudar a nuestra comunidad es por darle estrellas a ¡los repositorios de Github! Esto ayuda a crear consciencia de las interesantes herramientas que estamos construyendo.
- [Star PySyft](https://github.com/OpenMined/PySyft)
### ¡Únete a nuestro Slack!
La mejor manera de mantenerte actualizado con los últimos avances es ¡unirte a la comunidad! Tú lo puedes hacer llenando el formulario en [http://slack.openmined.org](http://slack.openmined.org)
### ¡Únete a un proyecto de código!
La mejor manera de contribuir a nuestra comunidad es convertirte en un ¡contribuidor de código! En cualquier momento puedes ir al _Github Issues_ de PySyft y filtrar por "Proyectos". Esto mostrará todos los tiquetes de nivel superior dando un resumen de los proyectos a los que ¡te puedes unir! Si no te quieres unir a un proyecto, pero quieres hacer un poco de código, también puedes mirar más mini-proyectos "de una persona" buscando por Github Issues con la etiqueta "good first issue".
- [PySyft Projects](https://github.com/OpenMined/PySyft/issues?q=is%3Aopen+is%3Aissue+label%3AProject)
- [Good First Issue Tickets](https://github.com/OpenMined/PySyft/issues?q=is%3Aopen+is%3Aissue+label%3A%22good+first+issue%22)
### Donar
Si no tienes tiempo para contribuir a nuestra base de código, pero quieres ofrecer tu ayuda, también puedes aportar a nuestro *Open Collective"*. Todas las donaciones van a nuestro *web hosting* y otros gastos de nuestra comunidad como ¡hackathons y meetups!
[OpenMined's Open Collective Page](https://opencollective.com/openmined)
|
github_jupyter
|
# Corre esta celda para ver si todo funciona
import sys
import torch
from torch.nn import Parameter
import torch.nn as nn
import torch.nn.functional as F
import syft as sy
hook = sy.TorchHook(torch)
torch.tensor([1,2,3,4,5])
x = torch.tensor([1,2,3,4,5])
y = x + x
print(y)
bob = sy.VirtualWorker(hook, id="bob")
x = torch.tensor([1,2,3,4,5])
y = torch.tensor([1,1,1,1,1])
x_ptr = x.send(bob)
y_ptr = y.send(bob)
x_ptr
bob._objects
z = x_ptr + x_ptr
z
bob._objects
x_ptr
x_ptr.location
bob
bob == x_ptr.location
x_ptr.id_at_location
x_ptr.owner
me = sy.local_worker
me
me == x_ptr.owner
x_ptr
x_ptr.get()
y_ptr
y_ptr.get()
z.get()
bob._objects
x = torch.tensor([1,2,3,4,5]).send(bob)
y = torch.tensor([1,1,1,1,1]).send(bob)
z = x + y
z
z.get()
x
y
z = torch.add(x,y)
z
z.get()
x = torch.tensor([1,2,3,4,5.], requires_grad=True).send(bob)
y = torch.tensor([1,1,1,1,1.], requires_grad=True).send(bob)
z = (x + y).sum()
z.backward()
x = x.get()
x
x.grad
| 0.288268 | 0.959231 |
<style type="text/css">div.cell {border-width: 0px; padding: 0px;} html body {background-color: #1b2a49;}</style>
<div style="background-image: linear-gradient(135deg, #008aff, #86d472); padding: 15px;">
<img src="https://d0.awsstatic.com/logos/powered-by-aws.png" alt="Powered by AWS Cloud Computing" style="float:right;">
<h1 style="color: white">FAST-TABULOUS HOMESITE QUOTE SUCCESS APP</h1>
<p>Fastai Deep Learning Part 1 2021. Source code available at <a href="https://github.com/timcu/fast-tabulous-app">https://github.com/timcu/fast-tabulous-app</a></p>
</div>
```
import logging
import random
import threading
import time
import ipywidgets as widgets
import numpy as np
import pandas as pd
import pymongo
from fastai.tabular.all import *
from IPython.display import display
from IPython.utils import io # using io.capture_output
from sklearn.metrics import roc_auc_score
# Now specify the folder which contains the original kaggle data (train.csv and test.csv)
# and the trained TabularModel (learn_model_cpu_0708.pkl) and DataLoaders (dls_cpu_0708.pkl)
pd.options.mode.chained_assignment = None # default='warn'
path = Path('../homesite-quote')
logger = logging.getLogger("load_pickled_model")
logging.basicConfig(level=logging.INFO)
# Heavyweight = "model_cpu_dls_cpu", Lightweight = "export_load_learner"
learner_source = "export_load_learner"
# Heavyweight solution - disabled for now
# On GPU instance run the following commands
# to = TabularPandas(df=df_train, procs=procs, cat_names=cat_names, cont_names=cont_names, y_names=y_names,splits=splits, y_block=y_block)dls = to.dataloaders(bs=bs, val_bs=val_bs, layers=layers, embed_ps=emb_dropout, ps=dropout)
# dls = to.dataloaders(bs=bs, val_bs=val_bs, layers=layers, embed_ps=emb_dropout, ps=dropout)
# learn = tabular_learner(dls, metrics=roc_auc_binary)
# save_pickle("to_0708.pkl", to)
# learn_model_cpu = learn.model.to('cpu')
# save_pickle("learn_model_cpu_0708.pkl", learn_model_cpu)
# dls.to('cpu')
# save_pickle("dls_cpu_0708.pkl", dls)
if learner_source == "model_cpu_dls_cpu":
learn_model_cpu = load_pickle(path/"learn_model_cpu_0708.pkl")
dls_cpu = load_pickle(path/"dls_cpu_0708.pkl")
to = load_pickle(path/"to_0708.pkl") # optional for now. needed for xgboost
learn=TabularLearner(dls=dls_cpu, model=learn_model_cpu)
preds, targs = learn.get_preds()
logger.warning(f"Trained deep learning model has a roc_auc_score of {roc_auc_score(to_np(targs), to_np(preds[:,1]))}")
# Lightweight solution - enabled for now
# On GPU instance run the following command
# learn.export(fname="learn_empty_dls_0708.pkl")
if learner_source == "export_load_learner":
learn = load_learner(path/"learn_empty_dls_0708.pkl")
def lst_ind_value(df, field):
"""Return the list of independent values to be tested for specified field"""
num_unique = df[field].nunique()
# If number of unique values is under 30 then try every value (or for objects try every value)
if num_unique < 30 or df.dtypes[field] == 'O':
return df[field].dropna().unique().tolist()
else:
if df.dtypes[field] == "int64":
vmin = df[field].min()
vmax = df[field].max()
return [int(vmin + (vmax - vmin) * i // 10) for i in range(11)]
elif df.dtypes[field] == "float64":
vmin = df[field].min()
vmax = df[field].max()
return [float(vmin + (vmax - vmin) * i / 10) for i in range(11)]
else:
logger.warning(f"Unknown type {field} {num_unique} {df.dtypes[field]!r}")
return []
mongo = pymongo.MongoClient("mongodb://localhost:27017/")
db = mongo['db_homesite']
clct_quote = db['clct_quote']
clct_conv = db['clct_conv']
clct_vals_by_col = db['clct_vals_by_col']
# If database exists we don't have to create it again
if clct_quote.count_documents({"QuoteNumber": 1}) == 0:
df_train = pd.read_csv(path/'train.csv', low_memory=False, parse_dates=['Original_Quote_Date'], index_col="QuoteNumber")
df_test = pd.read_csv(path/'test.csv', low_memory=False, parse_dates=['Original_Quote_Date'], index_col="QuoteNumber")
sr_conv = df_train['QuoteConversion_Flag']
df_train.drop('QuoteConversion_Flag', inplace=True, axis=1)
df = pd.concat([df_train, df_test])
df = add_datepart(df, 'Original_Quote_Date')
logger.debug(f"{df.shape} {df_train.shape} {df_test.shape} {sr_conv.shape}")
df_train = None
df_test = None
# Save to database
clct_quote.drop()
clct_quote.insert_many(df.reset_index().to_dict("records"))
clct_quote.create_index("QuoteNumber", name="idx_qn", unique=True)
clct_conv.drop()
clct_conv.insert_many(sr_conv.to_frame().reset_index().to_dict("records"))
clct_conv.create_index("QuoteNumber", name="idx_qn", unique=True)
clct_vals_by_col.drop()
clct_vals_by_col.create_index("vc_column", name="idx_vc_column", unique=True)
for f in df.columns:
num_unique = df[f].nunique()
tf_nan = int(df[f].isnull().sum()) > 0
lst_value = sorted(list(lst_ind_value(df, f)))
clct_vals_by_col.insert_one({"vc_column": f, "lst_value": lst_value, "num_unique": num_unique, "tf_nan": tf_nan, "dtype": str(df[f].dtype)})
cursor = clct_quote.aggregate([{"$group": {"_id": "QuoteNumber", "first": {"$min": "$QuoteNumber"}, "last": {"$max": "$QuoteNumber"}}}])
quote_range = list(cursor)[0]
qn_min = quote_range['first']
qn_max = quote_range['last']
qn = random.randint(qn_min, qn_max) # pick an initial quote at random
def tf_equal_or_nan(a, b):
"""same as normal equals except np.nan == np.nan which is not normally True"""
if a == b:
return True
try:
if np.isnan(a) and np.isnan(b):
return True
except TypeError:
pass
return False
def nan_if_nan(n):
"""Can't include np.nan in dropdowns as np.nan != np.nan. Instead use a str"""
try:
if np.isnan(n):
return "nan"
except TypeError as te:
pass
return n
def df_for_field(df_ind_original, f, lst_v):
"""predicts quote success after changing field f from v_original to each value in lst_v.
If prediction changes then quote is sensitive to the value of this field and True is returned
Keyword arguments
df_ind_original: all independent values from original quote (Pandas DataFrame with index = quote number, column names = independent value field names)
f: field name
lst_v: list of alternative values of independent value in field f
Returns
dataframe of alternative values in field f and all other fields staying the same and a column called fieldname
"""
# Create a DataFrame which has every row identical except for field in question
# Field f iterates through every value provided
ind_other = df_ind_original.drop(f, axis=1) # fields other than f
ind_f = pd.DataFrame(data={f: lst_v, "fieldname": [f] * len(lst_v)}, index=[df_ind_original.index[0]] * len(lst_v))
# Merge these two DataFrames to create one with all rows identical except field f
return pd.merge(ind_other, ind_f, right_index=True, left_index=True)
def sensitivity_analysis(ind_original):
"""Using data from Series of independent variables do a sensitivity analysis on all independent variables
ind_original is a Pandas Series of independent variables with their original values"""
time_start = datetime.now()
# Original prediction before changes
prd = learn.predict(ind_original)
logger.debug(f"After one predict time = {(datetime.now() - time_start).total_seconds()} seconds")
# Predicted quote conversion flag
qcf_original = prd[1].item()
# Probability that quote conversion flag is as predicted
prb_original = prd[2][qcf_original].item()
lst_df_for_field = []
# Loop through all fields. Check different values of each field to see if result is sensitive to it.
cursor = clct_vals_by_col.find({})
dct_types = {d['vc_column']: d['dtype'] for d in cursor}
df_ind_original = ind_original.to_frame().T.astype(dct_types)
for field in df_ind_original.columns:
val_original = ind_original[field]
dct_vals_by_col = clct_vals_by_col.find_one({"vc_column": field})
lst_val = dct_vals_by_col['lst_value']
if dct_vals_by_col['tf_nan']:
if len(lst_val) > 0 and isinstance(lst_val[0], str):
lst_val.append('nan')
else:
lst_val.append(np.nan)
lst_df_for_field.append(df_for_field(df_ind_original, field, lst_val))
logger.info(f"Build lst_df_for_field time = {(datetime.now() - time_start).total_seconds()} seconds")
df_sensitivity = pd.concat(lst_df_for_field, ignore_index=True)
# logger.info(f"{df_sensitivity['Field7'].unique()=}")
logger.info(f"Concat time = {(datetime.now() - time_start).total_seconds()} seconds {df_sensitivity.shape=}")
sr_fieldname = df_sensitivity['fieldname']
df_sensitivity.drop('fieldname', inplace=True, axis=1)
dl = learn.dls.test_dl(df_sensitivity)
logger.info(f"Dataloader time = {(datetime.now() - time_start).total_seconds()} seconds")
dl.dataset.conts = dl.dataset.conts.astype(np.float32)
# stop learn.get_preds() printing blank lines
with io.capture_output() as captured:
# using get_preds() rather than predict() because get_preds can do multiple rows at once
inp,preds,_,dec_preds = learn.get_preds(dl=dl, with_input=True, with_decoded=True)
logger.info(f"Time taken = {(datetime.now() - time_start).total_seconds()} seconds")
df_results=pd.DataFrame({'fieldname': sr_fieldname, 'prob_success': preds[:,1]})
df_results.sort_values(by='prob_success', ascending=False, inplace=True)
return df_results, df_sensitivity
def quote_as_series(quote_number):
"""Find quote and return as a Series"""
# Find quote and exclude fields _id and QuoteNumber
dct_quote = clct_quote.find_one({"QuoteNumber": quote_number}, projection={"_id": False, "QuoteNumber": False})
return pd.Series(dct_quote, name=quote_number)
def sensitivity_analysis_for_quote_number(quote_number):
return sensitivity_analysis(quote_as_series(quote_number))
# Widget event handlers
def configure_inputs():
"""Dynamically create inputs (dropdowns and radio buttons) for trialling combinations of values to improve quote success"""
qn = wdg_quote_number_slider.value
sr_quote = quote_as_series(qn)
# Get the top 10 fields most likely to make quote more successful, and their values which work the best
i = 0
dct_fields = defaultdict(list)
while len(dct_fields.keys()) < 10:
f = df_results.iloc[i, 0] # fieldname column
idx = df_results.index[i] # index into df_sensitivity
# independent variable value which has a good result
ind_val = df_sensitivity.loc[idx, f]
# create a list of all values which have a good result for this field
dct_fields[f].append(ind_val)
i += 1
priority = 0
# delete all elements of lst_input and lst_hbox without deleting references
del lst_input[:]
del lst_vbox[:]
for f, lst_recommend in dct_fields.items():
priority += 1
dct_vals_by_col = clct_vals_by_col.find_one({"vc_column": f})
num_unique = dct_vals_by_col['num_unique'] # df[f].nunique()
lst_unique = dct_vals_by_col['lst_value'] # df[f].unique()
if dct_vals_by_col['tf_nan']:
lst_unique.append('nan')
v = nan_if_nan(sr_quote[f])
if v not in lst_unique:
lst_unique.append(v)
tip = f"Priority {priority}. Initially {v}. Recommend {lst_recommend}"
lbl = widgets.HTML(value=f"{tip}")
if num_unique < 5:
wdg = widgets.RadioButtons(options=lst_unique,
description=f,
description_tooltip=tip,
style=style_input,
value=v)
else:
wdg = widgets.Dropdown(options=lst_unique,
description=f,
description_tooltip=tip,
style=style_input,
value=v)
wdg.observe(handle_input_change, names='value')
lst_vbox.append(widgets.HBox(children=[wdg, lbl]))
lst_input.append(wdg)
wdg_inputs.children=lst_vbox
return i # Number of field-value pairs required to show 10 best fields
def do_progress_bar(progress):
total = 100
for i in range(total):
time.sleep(0.2)
progress.value = float(i + 1) / total
def do_sensitivity_analysis(btn=None):
"""Do a fresh sensitivity analysis for selected quote number"""
global df_results, df_sensitivity
thread = threading.Thread(target=do_progress_bar, args=(wdg_progress,))
wdg_progress.layout.visibility = 'visible'
thread.start()
qn = wdg_quote_number_slider.value
wdg_logging_out.clear_output()
with wdg_logging_out:
df_results, df_sensitivity = sensitivity_analysis_for_quote_number(qn)
num_field_value_pairs = configure_inputs()
wdg_prob_success_out.clear_output()
with wdg_prob_success_out:
print(df_results.head(num_field_value_pairs))
handle_input_change(0)
wdg_progress.layout.visibility = 'hidden'
def handle_input_change(change):
qn = wdg_quote_number_slider.value
ind = quote_as_series(qn)
for w in lst_input:
if w.value == "nan":
v = np.nan
else:
v = w.value
ind[w.description] = v
with io.capture_output() as captured:
prd = learn.predict(ind)
qcf = prd[1].item()
prb = prd[2][qcf].item()
lst_conv = list(clct_conv.find({"QuoteNumber": qn}))
act = dct_success_label[lst_conv[0]["QuoteConversion_Flag"]] if len(lst_conv) > 0 else "unknown"
wdg_status.value = f"<h3>Quote {qn} actual: {act}, predicted: {prb:.2%} {dct_success_label[qcf]}</h3>"
def calc_quote_success(quote_number):
"""Calculate success of quote number and show result"""
with io.capture_output() as captured:
prd = learn.predict(quote_as_series(quote_number))
qcf = prd[1].item()
prb = prd[2][qcf].item()
lst_conv = list(clct_conv.find({"QuoteNumber": quote_number}))
act = dct_success_label[lst_conv[0]["QuoteConversion_Flag"]] if len(lst_conv) > 0 else "unknown"
wdg_quote_success.value = f"Quote {quote_number} actual: {act}, predicted {prb:.2%} {dct_success_label[qcf]}"
def handle_quote_number_change(change):
calc_quote_success(change.new)
lst_input = []
lst_vbox = []
# define all standard widgets
wdg_quote_success = widgets.Label(value="")
dct_success_label = {0: "unsuccessful", 1: "successful"}
style_qn = {'description_width': 'initial', 'width': '300px'}
style_input = {'description_width': 'initial'}
wdg_quote_number_text = widgets.BoundedIntText(
description="Quote number", min=qn_min, max=qn_max, value=qn, style=style_qn)
wdg_quote_number_slider = widgets.IntSlider(
description="", min=qn_min, max=qn_max, value=qn, style=style_qn, layout={'width': '600px'})
# link slider and textfield together
qn_link = widgets.jslink((wdg_quote_number_text, 'value'), (wdg_quote_number_slider, 'value'))
wdg_quote_number_slider.observe(handle_quote_number_change, names='value')
wdg_quote_number_row = widgets.HBox(children=[wdg_quote_number_text, wdg_quote_number_slider])
wdg_sensitivity_analysis_button = widgets.Button(
description='Sensitivity Analysis',
tooltip='Do a fresh sensitivity analysis for selected quote number and display top 10 inputs ',
layout_style="background-image: linear-gradient(135deg, #008aff, #86d472);"
)
wdg_sensitivity_analysis_button.on_click(do_sensitivity_analysis)
wdg_status = widgets.HTML(value=f"<h3>Please click on button 'Sensitivity Analysis' and wait 20 seconds</h3>")
wdg_logging_out = widgets.Output(layout={'border': '1px solid green'})
wdg_prob_success_out = widgets.Output()
wdg_inputs = widgets.VBox(children=lst_vbox)
wdg_progress = widgets.FloatProgress(value=0.0, min=0.0, max=1.0)
wdg_progress.layout.visibility = 'hidden'
calc_quote_success(qn)
display(wdg_quote_number_row)
display(wdg_quote_success)
display(wdg_sensitivity_analysis_button)
display(wdg_progress)
display(wdg_status)
display(wdg_inputs)
display(wdg_prob_success_out)
```
|
github_jupyter
|
import logging
import random
import threading
import time
import ipywidgets as widgets
import numpy as np
import pandas as pd
import pymongo
from fastai.tabular.all import *
from IPython.display import display
from IPython.utils import io # using io.capture_output
from sklearn.metrics import roc_auc_score
# Now specify the folder which contains the original kaggle data (train.csv and test.csv)
# and the trained TabularModel (learn_model_cpu_0708.pkl) and DataLoaders (dls_cpu_0708.pkl)
pd.options.mode.chained_assignment = None # default='warn'
path = Path('../homesite-quote')
logger = logging.getLogger("load_pickled_model")
logging.basicConfig(level=logging.INFO)
# Heavyweight = "model_cpu_dls_cpu", Lightweight = "export_load_learner"
learner_source = "export_load_learner"
# Heavyweight solution - disabled for now
# On GPU instance run the following commands
# to = TabularPandas(df=df_train, procs=procs, cat_names=cat_names, cont_names=cont_names, y_names=y_names,splits=splits, y_block=y_block)dls = to.dataloaders(bs=bs, val_bs=val_bs, layers=layers, embed_ps=emb_dropout, ps=dropout)
# dls = to.dataloaders(bs=bs, val_bs=val_bs, layers=layers, embed_ps=emb_dropout, ps=dropout)
# learn = tabular_learner(dls, metrics=roc_auc_binary)
# save_pickle("to_0708.pkl", to)
# learn_model_cpu = learn.model.to('cpu')
# save_pickle("learn_model_cpu_0708.pkl", learn_model_cpu)
# dls.to('cpu')
# save_pickle("dls_cpu_0708.pkl", dls)
if learner_source == "model_cpu_dls_cpu":
learn_model_cpu = load_pickle(path/"learn_model_cpu_0708.pkl")
dls_cpu = load_pickle(path/"dls_cpu_0708.pkl")
to = load_pickle(path/"to_0708.pkl") # optional for now. needed for xgboost
learn=TabularLearner(dls=dls_cpu, model=learn_model_cpu)
preds, targs = learn.get_preds()
logger.warning(f"Trained deep learning model has a roc_auc_score of {roc_auc_score(to_np(targs), to_np(preds[:,1]))}")
# Lightweight solution - enabled for now
# On GPU instance run the following command
# learn.export(fname="learn_empty_dls_0708.pkl")
if learner_source == "export_load_learner":
learn = load_learner(path/"learn_empty_dls_0708.pkl")
def lst_ind_value(df, field):
"""Return the list of independent values to be tested for specified field"""
num_unique = df[field].nunique()
# If number of unique values is under 30 then try every value (or for objects try every value)
if num_unique < 30 or df.dtypes[field] == 'O':
return df[field].dropna().unique().tolist()
else:
if df.dtypes[field] == "int64":
vmin = df[field].min()
vmax = df[field].max()
return [int(vmin + (vmax - vmin) * i // 10) for i in range(11)]
elif df.dtypes[field] == "float64":
vmin = df[field].min()
vmax = df[field].max()
return [float(vmin + (vmax - vmin) * i / 10) for i in range(11)]
else:
logger.warning(f"Unknown type {field} {num_unique} {df.dtypes[field]!r}")
return []
mongo = pymongo.MongoClient("mongodb://localhost:27017/")
db = mongo['db_homesite']
clct_quote = db['clct_quote']
clct_conv = db['clct_conv']
clct_vals_by_col = db['clct_vals_by_col']
# If database exists we don't have to create it again
if clct_quote.count_documents({"QuoteNumber": 1}) == 0:
df_train = pd.read_csv(path/'train.csv', low_memory=False, parse_dates=['Original_Quote_Date'], index_col="QuoteNumber")
df_test = pd.read_csv(path/'test.csv', low_memory=False, parse_dates=['Original_Quote_Date'], index_col="QuoteNumber")
sr_conv = df_train['QuoteConversion_Flag']
df_train.drop('QuoteConversion_Flag', inplace=True, axis=1)
df = pd.concat([df_train, df_test])
df = add_datepart(df, 'Original_Quote_Date')
logger.debug(f"{df.shape} {df_train.shape} {df_test.shape} {sr_conv.shape}")
df_train = None
df_test = None
# Save to database
clct_quote.drop()
clct_quote.insert_many(df.reset_index().to_dict("records"))
clct_quote.create_index("QuoteNumber", name="idx_qn", unique=True)
clct_conv.drop()
clct_conv.insert_many(sr_conv.to_frame().reset_index().to_dict("records"))
clct_conv.create_index("QuoteNumber", name="idx_qn", unique=True)
clct_vals_by_col.drop()
clct_vals_by_col.create_index("vc_column", name="idx_vc_column", unique=True)
for f in df.columns:
num_unique = df[f].nunique()
tf_nan = int(df[f].isnull().sum()) > 0
lst_value = sorted(list(lst_ind_value(df, f)))
clct_vals_by_col.insert_one({"vc_column": f, "lst_value": lst_value, "num_unique": num_unique, "tf_nan": tf_nan, "dtype": str(df[f].dtype)})
cursor = clct_quote.aggregate([{"$group": {"_id": "QuoteNumber", "first": {"$min": "$QuoteNumber"}, "last": {"$max": "$QuoteNumber"}}}])
quote_range = list(cursor)[0]
qn_min = quote_range['first']
qn_max = quote_range['last']
qn = random.randint(qn_min, qn_max) # pick an initial quote at random
def tf_equal_or_nan(a, b):
"""same as normal equals except np.nan == np.nan which is not normally True"""
if a == b:
return True
try:
if np.isnan(a) and np.isnan(b):
return True
except TypeError:
pass
return False
def nan_if_nan(n):
"""Can't include np.nan in dropdowns as np.nan != np.nan. Instead use a str"""
try:
if np.isnan(n):
return "nan"
except TypeError as te:
pass
return n
def df_for_field(df_ind_original, f, lst_v):
"""predicts quote success after changing field f from v_original to each value in lst_v.
If prediction changes then quote is sensitive to the value of this field and True is returned
Keyword arguments
df_ind_original: all independent values from original quote (Pandas DataFrame with index = quote number, column names = independent value field names)
f: field name
lst_v: list of alternative values of independent value in field f
Returns
dataframe of alternative values in field f and all other fields staying the same and a column called fieldname
"""
# Create a DataFrame which has every row identical except for field in question
# Field f iterates through every value provided
ind_other = df_ind_original.drop(f, axis=1) # fields other than f
ind_f = pd.DataFrame(data={f: lst_v, "fieldname": [f] * len(lst_v)}, index=[df_ind_original.index[0]] * len(lst_v))
# Merge these two DataFrames to create one with all rows identical except field f
return pd.merge(ind_other, ind_f, right_index=True, left_index=True)
def sensitivity_analysis(ind_original):
"""Using data from Series of independent variables do a sensitivity analysis on all independent variables
ind_original is a Pandas Series of independent variables with their original values"""
time_start = datetime.now()
# Original prediction before changes
prd = learn.predict(ind_original)
logger.debug(f"After one predict time = {(datetime.now() - time_start).total_seconds()} seconds")
# Predicted quote conversion flag
qcf_original = prd[1].item()
# Probability that quote conversion flag is as predicted
prb_original = prd[2][qcf_original].item()
lst_df_for_field = []
# Loop through all fields. Check different values of each field to see if result is sensitive to it.
cursor = clct_vals_by_col.find({})
dct_types = {d['vc_column']: d['dtype'] for d in cursor}
df_ind_original = ind_original.to_frame().T.astype(dct_types)
for field in df_ind_original.columns:
val_original = ind_original[field]
dct_vals_by_col = clct_vals_by_col.find_one({"vc_column": field})
lst_val = dct_vals_by_col['lst_value']
if dct_vals_by_col['tf_nan']:
if len(lst_val) > 0 and isinstance(lst_val[0], str):
lst_val.append('nan')
else:
lst_val.append(np.nan)
lst_df_for_field.append(df_for_field(df_ind_original, field, lst_val))
logger.info(f"Build lst_df_for_field time = {(datetime.now() - time_start).total_seconds()} seconds")
df_sensitivity = pd.concat(lst_df_for_field, ignore_index=True)
# logger.info(f"{df_sensitivity['Field7'].unique()=}")
logger.info(f"Concat time = {(datetime.now() - time_start).total_seconds()} seconds {df_sensitivity.shape=}")
sr_fieldname = df_sensitivity['fieldname']
df_sensitivity.drop('fieldname', inplace=True, axis=1)
dl = learn.dls.test_dl(df_sensitivity)
logger.info(f"Dataloader time = {(datetime.now() - time_start).total_seconds()} seconds")
dl.dataset.conts = dl.dataset.conts.astype(np.float32)
# stop learn.get_preds() printing blank lines
with io.capture_output() as captured:
# using get_preds() rather than predict() because get_preds can do multiple rows at once
inp,preds,_,dec_preds = learn.get_preds(dl=dl, with_input=True, with_decoded=True)
logger.info(f"Time taken = {(datetime.now() - time_start).total_seconds()} seconds")
df_results=pd.DataFrame({'fieldname': sr_fieldname, 'prob_success': preds[:,1]})
df_results.sort_values(by='prob_success', ascending=False, inplace=True)
return df_results, df_sensitivity
def quote_as_series(quote_number):
"""Find quote and return as a Series"""
# Find quote and exclude fields _id and QuoteNumber
dct_quote = clct_quote.find_one({"QuoteNumber": quote_number}, projection={"_id": False, "QuoteNumber": False})
return pd.Series(dct_quote, name=quote_number)
def sensitivity_analysis_for_quote_number(quote_number):
return sensitivity_analysis(quote_as_series(quote_number))
# Widget event handlers
def configure_inputs():
"""Dynamically create inputs (dropdowns and radio buttons) for trialling combinations of values to improve quote success"""
qn = wdg_quote_number_slider.value
sr_quote = quote_as_series(qn)
# Get the top 10 fields most likely to make quote more successful, and their values which work the best
i = 0
dct_fields = defaultdict(list)
while len(dct_fields.keys()) < 10:
f = df_results.iloc[i, 0] # fieldname column
idx = df_results.index[i] # index into df_sensitivity
# independent variable value which has a good result
ind_val = df_sensitivity.loc[idx, f]
# create a list of all values which have a good result for this field
dct_fields[f].append(ind_val)
i += 1
priority = 0
# delete all elements of lst_input and lst_hbox without deleting references
del lst_input[:]
del lst_vbox[:]
for f, lst_recommend in dct_fields.items():
priority += 1
dct_vals_by_col = clct_vals_by_col.find_one({"vc_column": f})
num_unique = dct_vals_by_col['num_unique'] # df[f].nunique()
lst_unique = dct_vals_by_col['lst_value'] # df[f].unique()
if dct_vals_by_col['tf_nan']:
lst_unique.append('nan')
v = nan_if_nan(sr_quote[f])
if v not in lst_unique:
lst_unique.append(v)
tip = f"Priority {priority}. Initially {v}. Recommend {lst_recommend}"
lbl = widgets.HTML(value=f"{tip}")
if num_unique < 5:
wdg = widgets.RadioButtons(options=lst_unique,
description=f,
description_tooltip=tip,
style=style_input,
value=v)
else:
wdg = widgets.Dropdown(options=lst_unique,
description=f,
description_tooltip=tip,
style=style_input,
value=v)
wdg.observe(handle_input_change, names='value')
lst_vbox.append(widgets.HBox(children=[wdg, lbl]))
lst_input.append(wdg)
wdg_inputs.children=lst_vbox
return i # Number of field-value pairs required to show 10 best fields
def do_progress_bar(progress):
total = 100
for i in range(total):
time.sleep(0.2)
progress.value = float(i + 1) / total
def do_sensitivity_analysis(btn=None):
"""Do a fresh sensitivity analysis for selected quote number"""
global df_results, df_sensitivity
thread = threading.Thread(target=do_progress_bar, args=(wdg_progress,))
wdg_progress.layout.visibility = 'visible'
thread.start()
qn = wdg_quote_number_slider.value
wdg_logging_out.clear_output()
with wdg_logging_out:
df_results, df_sensitivity = sensitivity_analysis_for_quote_number(qn)
num_field_value_pairs = configure_inputs()
wdg_prob_success_out.clear_output()
with wdg_prob_success_out:
print(df_results.head(num_field_value_pairs))
handle_input_change(0)
wdg_progress.layout.visibility = 'hidden'
def handle_input_change(change):
qn = wdg_quote_number_slider.value
ind = quote_as_series(qn)
for w in lst_input:
if w.value == "nan":
v = np.nan
else:
v = w.value
ind[w.description] = v
with io.capture_output() as captured:
prd = learn.predict(ind)
qcf = prd[1].item()
prb = prd[2][qcf].item()
lst_conv = list(clct_conv.find({"QuoteNumber": qn}))
act = dct_success_label[lst_conv[0]["QuoteConversion_Flag"]] if len(lst_conv) > 0 else "unknown"
wdg_status.value = f"<h3>Quote {qn} actual: {act}, predicted: {prb:.2%} {dct_success_label[qcf]}</h3>"
def calc_quote_success(quote_number):
"""Calculate success of quote number and show result"""
with io.capture_output() as captured:
prd = learn.predict(quote_as_series(quote_number))
qcf = prd[1].item()
prb = prd[2][qcf].item()
lst_conv = list(clct_conv.find({"QuoteNumber": quote_number}))
act = dct_success_label[lst_conv[0]["QuoteConversion_Flag"]] if len(lst_conv) > 0 else "unknown"
wdg_quote_success.value = f"Quote {quote_number} actual: {act}, predicted {prb:.2%} {dct_success_label[qcf]}"
def handle_quote_number_change(change):
calc_quote_success(change.new)
lst_input = []
lst_vbox = []
# define all standard widgets
wdg_quote_success = widgets.Label(value="")
dct_success_label = {0: "unsuccessful", 1: "successful"}
style_qn = {'description_width': 'initial', 'width': '300px'}
style_input = {'description_width': 'initial'}
wdg_quote_number_text = widgets.BoundedIntText(
description="Quote number", min=qn_min, max=qn_max, value=qn, style=style_qn)
wdg_quote_number_slider = widgets.IntSlider(
description="", min=qn_min, max=qn_max, value=qn, style=style_qn, layout={'width': '600px'})
# link slider and textfield together
qn_link = widgets.jslink((wdg_quote_number_text, 'value'), (wdg_quote_number_slider, 'value'))
wdg_quote_number_slider.observe(handle_quote_number_change, names='value')
wdg_quote_number_row = widgets.HBox(children=[wdg_quote_number_text, wdg_quote_number_slider])
wdg_sensitivity_analysis_button = widgets.Button(
description='Sensitivity Analysis',
tooltip='Do a fresh sensitivity analysis for selected quote number and display top 10 inputs ',
layout_style="background-image: linear-gradient(135deg, #008aff, #86d472);"
)
wdg_sensitivity_analysis_button.on_click(do_sensitivity_analysis)
wdg_status = widgets.HTML(value=f"<h3>Please click on button 'Sensitivity Analysis' and wait 20 seconds</h3>")
wdg_logging_out = widgets.Output(layout={'border': '1px solid green'})
wdg_prob_success_out = widgets.Output()
wdg_inputs = widgets.VBox(children=lst_vbox)
wdg_progress = widgets.FloatProgress(value=0.0, min=0.0, max=1.0)
wdg_progress.layout.visibility = 'hidden'
calc_quote_success(qn)
display(wdg_quote_number_row)
display(wdg_quote_success)
display(wdg_sensitivity_analysis_button)
display(wdg_progress)
display(wdg_status)
display(wdg_inputs)
display(wdg_prob_success_out)
| 0.437103 | 0.626096 |
# Markdown 與 LaTeX簡介
每個範例內容將範例語法與輸出分為不同的 Cell 放置,雙擊 Cell 也可以查看原始碼,進行進一步的研究與實驗。
## 1. Markdown 主要語法
### 1.1 段落和斷行
_範例語法:_
```
前軍至夏口,周瑜問:「荊州有人在前面接否?」人報:「劉皇叔使糜竺來見都督。」瑜喚至,問勞軍如何。糜竺曰:「主公皆準備安排下了。」瑜曰:「皇叔何在?」竺曰:「在荊州城門相等,與都督把盞。」瑜曰:「今為汝家之事,出兵遠征;勞軍之禮,休得輕易。」糜竺領了言語先回。<br />戰船密密排在江上,依次而進。
```
_輸出:(可以看到在"戰船"前面有換行)_
前軍至夏口,周瑜問:「荊州有人在前面接否?」人報:「劉皇叔使糜竺來見都督。」瑜喚至,問勞軍如何。糜竺曰:「主公皆準備安排下了。」瑜曰:「皇叔何在?」竺曰:「在荊州城門相等,與都督把盞。」瑜曰:「今為汝家之事,出兵遠征;勞軍之禮,休得輕易。」糜竺領了言語先回。<br />戰船密密排在江上,依次而進。
### 1.2 標題 (Heading)
_範例語法:_
```
# 主標題 (等同於 HTML `<h1></h1>`)
## 次標題 (等同於 `<h2></h12`)
### 第三階 (等同於 `<h3></h3>`)
#### 第四階 (等同於 `<h4></h4>`)
##### 第五階 (等同於 `<h5></h5>`)
###### 第六階 (等同於 `<h6></h6>`)
```
_輸出:_
# 主標題 (等同於 HTML `<h1></h1>`)
## 次標題 (等同於 `<h2></h12`)
### 第三階 (等同於 `<h3></h3>`)
#### 第四階 (等同於 `<h4></h4>`)
##### 第五階 (等同於 `<h5></h5>`)
###### 第六階 (等同於 `<h6></h6>`)
### 1.3 無序號列表 (Bullet list)
_範例語法:_
```
六都清單:
- 台北市
- 大安區
- 大同區
- 新北市
* 桃園市
* 台中市
+ 台南市
+ 高雄市
```
_輸出:_
六都清單:
- 台北市
- 大安區
- 大同區
- 新北市
* 桃園市
* 台中市
+ 台南市
+ 高雄市
### 1.4 序號列表 (Numbered list)
_範例語法:_
```
1. 第一項
2. 第二項
3. 第三項
```
_輸出:_
1. 第一項
2. 第二項
3. 第三項
### 1.5 區塊引言 (blockquoting)
_範例語法:_
```
> 卻說魯肅回見周瑜,說玄德,孔明歡喜不疑,準備出城勞軍。周瑜大笑曰:「原來今番也中了吾計!」便教魯肅稟報吳侯,並遣程普引兵接應。周瑜此時箭瘡已漸平愈,身軀無事,使甘寧為先鋒,自與徐盛,丁奉為第二;淩統,呂蒙為後隊。水陸大兵五百萬,望荊州而來。
周瑜在船中,時復歡笑,以為孔明中計。
> 前軍至夏口,周瑜問:「荊州有人在前面接否?」人報:「劉皇叔使糜竺來見都督。」瑜喚至,問勞軍如何。
```
_輸出:_
> 卻說魯肅回見周瑜,說玄德,孔明歡喜不疑,準備出城勞軍。周瑜大笑曰:「原來今番也中了吾計!」便教魯肅稟報吳侯,並遣程普引兵接應。周瑜此時箭瘡已漸平愈,身軀無事,使甘寧為先鋒,自與徐盛,丁奉為第二;淩統,呂蒙為後隊。水陸大兵五百萬,望荊州而來。
周瑜在船中,時復歡笑,以為孔明中計。
> 前軍至夏口,周瑜問:「荊州有人在前面接否?」人報:「劉皇叔使糜竺來見都督。」瑜喚至,問勞軍如何。
### 1.6 程式碼區塊
_範例語法:_
\`\`\`julia
println("Hello Julia")
\`\`\`
_輸出:_
```julia
println("Hello Julia")
```
### 1.7 分隔線
_範例語法:_
```
卻說魯肅回見周瑜,說玄德,孔明歡喜不疑,準備出城勞軍。周瑜大笑曰:「原來今番也中了吾計!」便教魯肅稟報吳侯,並遣程普引兵接應。周瑜此時箭瘡已漸平愈,身軀無事,使甘寧為先鋒,自與徐盛,丁奉為第二;淩統,呂蒙為後隊。水陸大兵五百萬,望荊州而來。周瑜在船中,時復歡笑,以為孔明中計。
---
前軍至夏口,周瑜問:「荊州有人在前面接否?」人報:「劉皇叔使糜竺來見都督。」瑜喚至,問勞軍如何。糜竺曰:「主公皆準備安排下了。」瑜曰:「皇叔何在?」竺曰:「在荊州城門相等,與都督把盞。」瑜曰:「今為汝家之事,出兵遠征;勞軍之禮,休得輕易。」糜竺領了言語先回。<br />戰船密密排在江上,依次而進。
```
_輸出:_
卻說魯肅回見周瑜,說玄德,孔明歡喜不疑,準備出城勞軍。周瑜大笑曰:「原來今番也中了吾計!」便教魯肅稟報吳侯,並遣程普引兵接應。周瑜此時箭瘡已漸平愈,身軀無事,使甘寧為先鋒,自與徐盛,丁奉為第二;淩統,呂蒙為後隊。水陸大兵五百萬,望荊州而來。周瑜在船中,時復歡笑,以為孔明中計。
---
前軍至夏口,周瑜問:「荊州有人在前面接否?」人報:「劉皇叔使糜竺來見都督。」瑜喚至,問勞軍如何。糜竺曰:「主公皆準備安排下了。」瑜曰:「皇叔何在?」竺曰:「在荊州城門相等,與都督把盞。」瑜曰:「今為汝家之事,出兵遠征;勞軍之禮,休得輕易。」糜竺領了言語先回。<br />戰船密密排在江上,依次而進。
### 1.8 超連結
_範例語法:_
```
[Cupoy - 為你探索世界的新知](https://www.cupoy.com)
```
_輸出:_
[Cupoy - 為你探索世界的新知](https://www.cupoy.com)
### 1.9 嵌入圖片
_範例語法:_
```

```
_輸出:_

### 1.10 表格
_範例語法:_
```
|姓名|國家|地址|年齡|
|---|---|---|---|
|John Doe|中華民國台灣|台北市大安區敦化街1號|25|
```
_輸出:_
|姓名|國家|地址|年齡|
|---|---|---|---|
|John Doe|中華民國台灣|台北市大安區敦化街1號|25|
## 2. 用 LaTeX 寫數學公式
### 2.1 Inline 模式
_範例語法:_
```
$\LARGE f(\displaystyle\sum_i w_i x_i + b)$
```
_輸出:_
$\LARGE f(\displaystyle\sum_i w_i x_i + b)$
### 2.2 Block 模式
_範例語法:_
```
$$\LARGE f(\displaystyle\sum_i w_i x_i + b)$$
```
_輸出:_
$$\LARGE f(\displaystyle\sum_i w_i x_i + b)$$
_範例語法:_
```
\begin{equation}
\LARGE f(\displaystyle\sum_i w_i x_i + b)
\end{equation}
```
_輸出:_
\begin{equation}
\LARGE f(\displaystyle\sum_i w_i x_i + b)
\end{equation}
_範例語法:_
```
\begin{align}
\LARGE f(\displaystyle\sum_i w_i x_i + b)
\end{align}
```
_輸出:_
\begin{align}
\LARGE f(\displaystyle\sum_i w_i x_i + b)
\end{align}
## 3. 結合 Markdown 和 LaTeX 數學公式
_範例語法:_
```
公式 $\LARGE f(\displaystyle\sum_i w_i x_i + b)$ 是 Deep Learning 課程中常會見到的基本公式
結合 Markdown 和 LaTeX 數學公式,我們可以撰寫出漂亮的文件筆記,讓學習更有效率。
```
_輸出:_
公式 $\LARGE f(\displaystyle\sum_i w_i x_i + b)$ 是 Deep Learning 課程中常會見到的基本公式

結合 Markdown 和 LaTeX 數學公式,我們可以撰寫出漂亮的文件筆記,讓學習更有效率。
```
```
|
github_jupyter
|
前軍至夏口,周瑜問:「荊州有人在前面接否?」人報:「劉皇叔使糜竺來見都督。」瑜喚至,問勞軍如何。糜竺曰:「主公皆準備安排下了。」瑜曰:「皇叔何在?」竺曰:「在荊州城門相等,與都督把盞。」瑜曰:「今為汝家之事,出兵遠征;勞軍之禮,休得輕易。」糜竺領了言語先回。<br />戰船密密排在江上,依次而進。
# 主標題 (等同於 HTML `<h1></h1>`)
## 次標題 (等同於 `<h2></h12`)
### 第三階 (等同於 `<h3></h3>`)
#### 第四階 (等同於 `<h4></h4>`)
##### 第五階 (等同於 `<h5></h5>`)
###### 第六階 (等同於 `<h6></h6>`)
六都清單:
- 台北市
- 大安區
- 大同區
- 新北市
* 桃園市
* 台中市
+ 台南市
+ 高雄市
1. 第一項
2. 第二項
3. 第三項
> 卻說魯肅回見周瑜,說玄德,孔明歡喜不疑,準備出城勞軍。周瑜大笑曰:「原來今番也中了吾計!」便教魯肅稟報吳侯,並遣程普引兵接應。周瑜此時箭瘡已漸平愈,身軀無事,使甘寧為先鋒,自與徐盛,丁奉為第二;淩統,呂蒙為後隊。水陸大兵五百萬,望荊州而來。
周瑜在船中,時復歡笑,以為孔明中計。
> 前軍至夏口,周瑜問:「荊州有人在前面接否?」人報:「劉皇叔使糜竺來見都督。」瑜喚至,問勞軍如何。
println("Hello Julia")
卻說魯肅回見周瑜,說玄德,孔明歡喜不疑,準備出城勞軍。周瑜大笑曰:「原來今番也中了吾計!」便教魯肅稟報吳侯,並遣程普引兵接應。周瑜此時箭瘡已漸平愈,身軀無事,使甘寧為先鋒,自與徐盛,丁奉為第二;淩統,呂蒙為後隊。水陸大兵五百萬,望荊州而來。周瑜在船中,時復歡笑,以為孔明中計。
---
前軍至夏口,周瑜問:「荊州有人在前面接否?」人報:「劉皇叔使糜竺來見都督。」瑜喚至,問勞軍如何。糜竺曰:「主公皆準備安排下了。」瑜曰:「皇叔何在?」竺曰:「在荊州城門相等,與都督把盞。」瑜曰:「今為汝家之事,出兵遠征;勞軍之禮,休得輕易。」糜竺領了言語先回。<br />戰船密密排在江上,依次而進。
[Cupoy - 為你探索世界的新知](https://www.cupoy.com)

|姓名|國家|地址|年齡|
|---|---|---|---|
|John Doe|中華民國台灣|台北市大安區敦化街1號|25|
$\LARGE f(\displaystyle\sum_i w_i x_i + b)$
$$\LARGE f(\displaystyle\sum_i w_i x_i + b)$$
\begin{equation}
\LARGE f(\displaystyle\sum_i w_i x_i + b)
\end{equation}
\begin{align}
\LARGE f(\displaystyle\sum_i w_i x_i + b)
\end{align}
公式 $\LARGE f(\displaystyle\sum_i w_i x_i + b)$ 是 Deep Learning 課程中常會見到的基本公式

結合 Markdown 和 LaTeX 數學公式,我們可以撰寫出漂亮的文件筆記,讓學習更有效率。
| 0.395368 | 0.870101 |
# Load
In this exercise, you'll load data into different formats: a csv file, a json file, and a SQLite database.
You'll work with the GDP, population, and projects data. Run the code cell below to read in and clean the World Bank population and gdp data. This code creates a dataframe called df_indicator with both the gdp and population data.
```
# run this code cell
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# read in the projects data set and do basic wrangling
gdp = pd.read_csv('../data/gdp_data.csv', skiprows=4)
gdp.drop(['Unnamed: 62', 'Indicator Name', 'Indicator Code'], inplace=True, axis=1)
population = pd.read_csv('../data/population_data.csv', skiprows=4)
population.drop(['Unnamed: 62', 'Indicator Name', 'Indicator Code'], inplace=True, axis=1)
# Reshape the data sets so that they are in long format
gdp_melt = gdp.melt(id_vars=['Country Name', 'Country Code'],
var_name='year',
value_name='gdp')
# Use back fill and forward fill to fill in missing gdp values
gdp_melt['gdp'] = gdp_melt.sort_values('year').groupby(['Country Name', 'Country Code'])['gdp'].fillna(method='ffill').fillna(method='bfill')
population_melt = population.melt(id_vars=['Country Name', 'Country Code'],
var_name='year',
value_name='population')
# Use back fill and forward fill to fill in missing population values
population_melt['population'] = population_melt.sort_values('year').groupby('Country Name')['population'].fillna(method='ffill').fillna(method='bfill')
# merge the population and gdp data together into one data frame
df_indicator = gdp_melt.merge(population_melt, on=('Country Name', 'Country Code', 'year'))
# filter out values that are not countries
non_countries = ['World',
'High income',
'OECD members',
'Post-demographic dividend',
'IDA & IBRD total',
'Low & middle income',
'Middle income',
'IBRD only',
'East Asia & Pacific',
'Europe & Central Asia',
'North America',
'Upper middle income',
'Late-demographic dividend',
'European Union',
'East Asia & Pacific (excluding high income)',
'East Asia & Pacific (IDA & IBRD countries)',
'Euro area',
'Early-demographic dividend',
'Lower middle income',
'Latin America & Caribbean',
'Latin America & the Caribbean (IDA & IBRD countries)',
'Latin America & Caribbean (excluding high income)',
'Europe & Central Asia (IDA & IBRD countries)',
'Middle East & North Africa',
'Europe & Central Asia (excluding high income)',
'South Asia (IDA & IBRD)',
'South Asia',
'Arab World',
'IDA total',
'Sub-Saharan Africa',
'Sub-Saharan Africa (IDA & IBRD countries)',
'Sub-Saharan Africa (excluding high income)',
'Middle East & North Africa (excluding high income)',
'Middle East & North Africa (IDA & IBRD countries)',
'Central Europe and the Baltics',
'Pre-demographic dividend',
'IDA only',
'Least developed countries: UN classification',
'IDA blend',
'Fragile and conflict affected situations',
'Heavily indebted poor countries (HIPC)',
'Low income',
'Small states',
'Other small states',
'Not classified',
'Caribbean small states',
'Pacific island small states']
# remove non countries from the data
df_indicator = df_indicator[~df_indicator['Country Name'].isin(non_countries)]
df_indicator.reset_index(inplace=True, drop=True)
df_indicator.columns = ['countryname', 'countrycode', 'year', 'gdp', 'population']
# output the first few rows of the data frame
df_indicator.head()
```
Run this code cell to read in the countries data set. This will create a data frame called df_projects containing the World Bank projects data. The data frame only has the 'id', 'countryname', 'countrycode', 'totalamt', and 'year' columns.
```
!pip install pycountry
from pycountry import countries
# read in the projects data set with all columns type string
df_projects = pd.read_csv('../data/projects_data.csv', dtype=str)
df_projects.drop(['Unnamed: 56'], axis=1, inplace=True)
df_projects['countryname'] = df_projects['countryname'].str.split(';').str.get(0)
# set up the libraries and variables
from collections import defaultdict
country_not_found = [] # stores countries not found in the pycountry library
project_country_abbrev_dict = defaultdict(str) # set up an empty dictionary of string values
# TODO: iterate through the country names in df_projects.
# Create a dictionary mapping the country name to the alpha_3 ISO code
for country in df_projects['countryname'].drop_duplicates().sort_values():
try:
# TODO: look up the country name in the pycountry library
# store the country name as the dictionary key and the ISO-3 code as the value
project_country_abbrev_dict[country] = countries.lookup(country).alpha_3
except:
# If the country name is not in the pycountry library, then print out the country name
# And store the results in the country_not_found list
country_not_found.append(country)
# run this code cell to load the dictionary
country_not_found_mapping = {'Co-operative Republic of Guyana': 'GUY',
'Commonwealth of Australia':'AUS',
'Democratic Republic of Sao Tome and Prin':'STP',
'Democratic Republic of the Congo':'COD',
'Democratic Socialist Republic of Sri Lan':'LKA',
'East Asia and Pacific':'EAS',
'Europe and Central Asia': 'ECS',
'Islamic Republic of Afghanistan':'AFG',
'Latin America':'LCN',
'Caribbean':'LCN',
'Macedonia':'MKD',
'Middle East and North Africa':'MEA',
'Oriental Republic of Uruguay':'URY',
'Republic of Congo':'COG',
"Republic of Cote d'Ivoire":'CIV',
'Republic of Korea':'KOR',
'Republic of Niger':'NER',
'Republic of Kosovo':'XKX',
'Republic of Rwanda':'RWA',
'Republic of The Gambia':'GMB',
'Republic of Togo':'TGO',
'Republic of the Union of Myanmar':'MMR',
'Republica Bolivariana de Venezuela':'VEN',
'Sint Maarten':'SXM',
"Socialist People's Libyan Arab Jamahiriy":'LBY',
'Socialist Republic of Vietnam':'VNM',
'Somali Democratic Republic':'SOM',
'South Asia':'SAS',
'St. Kitts and Nevis':'KNA',
'St. Lucia':'LCA',
'St. Vincent and the Grenadines':'VCT',
'State of Eritrea':'ERI',
'The Independent State of Papua New Guine':'PNG',
'West Bank and Gaza':'PSE',
'World':'WLD'}
project_country_abbrev_dict.update(country_not_found_mapping)
df_projects['countrycode'] = df_projects['countryname'].apply(lambda x: project_country_abbrev_dict[x])
df_projects['boardapprovaldate'] = pd.to_datetime(df_projects['boardapprovaldate'])
df_projects['year'] = df_projects['boardapprovaldate'].dt.year.astype(str).str.slice(stop=4)
df_projects['totalamt'] = pd.to_numeric(df_projects['totalamt'].str.replace(',',""))
df_projects = df_projects[['id', 'countryname', 'countrycode', 'totalamt', 'year']]
df_projects.head()
```
# Exercise Part 1
The first few cells in this workbook loaded and cleaned the World Bank Data. You now have two data frames:
* df_projects, which contain data from the projects data set
* df_indicator, which contain population and gdp data for various years
They both have country code variables. Note, however, that there could be countries represented in the projects data set that are not in the indicator data set and vice versus.
In this first exercise, merge the two data sets together using country code and year as common keys. When joining the data sets, keep all of the data in the df_projects dataframe even if there is no indicator data for that country code.
```
# TODO: merge the projects and indicator data frames together using countrycode and year as common keys
# Use a left join so that all projects are returned even if the country/year combination does not have
# indicator data
df_merged = df_projects.merge(df_indicator, how='left', on=['countrycode', 'year'])
```
If you look at the first few rows of the merged data set, you'll see NaN's for the indicator data. That is because the indicator data does not contain values for 2018. If you run the code cell below, you should get the following results:
||id | countryname_x | countrycode |totalamt |year |countryname_y |gdp | population|
|-|-|-|-|-|-|-|
|256| P161982| Hashemite Kingdom of Jordan| JOR| 0| 2017| Jordan |4.006831e+10| 9702353.0|
|301| P162407| Hashemite Kingdom of Jordan| JOR| 147700000| 2017| Jordan| 4.006831e+10| 9702353.0|
|318| P160103| Hashemite Kingdom of Jordan| JOR| 0| 2017| Jordan |4.006831e+10 |9702353.0|
|464| P161905| Hashemite Kingdom of Jordan| JOR| 50000000| 2017 |Jordan |4.006831e+10 |9702353.0|
|495| P163387| Hashemite Kingdom of Jordan| JOR| 36100000| 2017 |Jordan |4.006831e+10 |9702353.0|
|515| P163241| Hashemite Kingdom of Jordan| JOR| 0| 2017| Jordan| 4.006831e+10| 9702353.0|
```
# Run this code to check your work
df_merged[(df_merged['year'] == '2017') & (df_merged['countryname_y'] == 'Jordan')]
```
# Exercise Part 2
Output the df_merged dataframe as a json file. You can use the pandas [to_json() method](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.to_json.html).
```
# TODO: Output the df_merged data frame as a json file
# HINT: Pandas has a to_json() method
# HINT: use orient='records' to get one of the more common json formats
# HINT: be sure to specify the name of the json file you want to create as the first input into to_json
df_merged.to_json('countrydata.json', orient='records')
```
If you go to File->Open and then open the 17_load_exercise folder, you can see the json file you created.
The records should look like this (note that the order of the records might be different, but the format should look similar):
```
[{"id":"P162228","countryname_x":"World","countrycode":"WLD","totalamt":0,"year":"2018","countryname_y":null,"gdp":null,"population":null},{"id":"P163962","countryname_x":"Democratic Republic of the Congo","countrycode":"COD","totalamt":200000000,"year":"2018","countryname_y":null,"gdp":null,"population":null},{"id":"P167672","countryname_x":"People's Republic of Bangladesh","countrycode":"BGD","totalamt":58330000,"year":"2018","countryname_y":null,"gdp":null,"population":null},{"id":"P158768","countryname_x":"Islamic Republic of Afghanistan","countrycode":"AFG","totalamt":20000000,"year":"2018","countryname_y":null,"gdp":null,"population":null},{"id":"P161364","countryname_x":"Federal Republic of Nigeria","countrycode":"NGA","totalamt":100000000,"year":"2018","countryname_y":null,"gdp":null,"population":null},{"id":"P161483","countryname_x":"Republic of Tunisia","countrycode":"TUN","totalamt":500000000,"year":"2018","countryname_y":null,"gdp":null,"population":null}
```
# Exercise Part 3
Output the df_merged dataframe as a csv file. You can use the pandas [to_csv() method](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.to_csv.html).
```
# TODO: Output the df_merged data frame as a csv file
# HINT: The to_csv() method is similar to the to_json() method.
# HINT: If you do not want the data frame indices in your result, use index=False
df_merged.to_csv('countrydata.csv', index=False)
```
If you go to File->Open and then open the 17_load_exercise folder, you can see the csv file you created.
The records should look something like this:
```
id,countryname_x,countrycode,totalamt,year,countryname_y,gdp,population
P162228,World,WLD,0,2018,,,
P163962,Democratic Republic of the Congo,COD,200000000,2018,,,
P167672,People's Republic of Bangladesh,BGD,58330000,2018,,,
P158768,Islamic Republic of Afghanistan,AFG,20000000,2018,,,
P161364,Federal Republic of Nigeria,NGA,100000000,2018,,,
P161483,Republic of Tunisia,TUN,500000000,2018,,,
P161885,Federal Republic of Nigeria,NGA,350000000,2018,,,
P162069,Federal Republic of Nigeria,NGA,225000000,2018,,,
P163540,Federal Republic of Nigeria,NGA,125000000,2018,,,
P163576,Lebanese Republic,LBN,329900000,2018,,,
P163721,Democratic Socialist Republic of Sri Lan,LKA,200000000,2018,,,
P164082,Federal Republic of Nigeria,NGA,400000000,2018,,,
P164783,Nepal,NPL,0,2018,,,
```
# Exercise Part 4
Output the df_merged dataframe as a sqlite database file. For this exercise, you can put all of the data as one table. In the next exercise, you'll create a database with multiple tables.
```
import sqlite3
# connect to the database
# the database file will be worldbank.db
# note that sqlite3 will create this database file if it does not exist already
conn = sqlite3.connect('worldbank.db')
# TODO: output the df_merged dataframe to a SQL table called 'merged'.
# HINT: Use the to_sql() method
# HINT: Use the conn variable for the connection parameter
# HINT: You can use the if_exists parameter like if_exists='replace' to replace a table if it already exists
df_merged.to_sql('merged', con = conn, if_exists='replace', index=False)
```
Run the code cell below to make sure that the worldbank.db file was created and the merged table loaded. You should get an output that is formatted like this:
|id|countryname_x|countrycode|totalamt|year|countryname_y|gdp|population
|-|-|-|-|-|-|-|-|
|P162033|'Federative Republic of Brazil'|BRA|125000000|2017|Brazil|2.055506e+12|209288278.0|
```
pd.read_sql('SELECT * FROM merged WHERE year = "2017" AND countrycode = "BRA"', con = conn).head()
```
# Exercise Part 5
Output the data to a SQL database like in the previous exercise; however, this time, put the df_indicator data in one table and the df_projects data in another table. Call the df_indicator table 'indicator' and the df_projects table 'projects'.
```
import sqlite3
# connect to the database
# the database file will be worldbank.db
# note that sqlite3 will create this database file if it does not exist already
conn = sqlite3.connect('worldbank.db')
# TODO: output the df_merged dataframe to a SQL table called 'merged'.
# HINT: Use the to_sql() method
# HINT: Use the conn variable for the connection parameter
# HINT: You can use the if_exists parameter like if_exists='replace' to replace a table if it already exists
df_indicator.to_sql('indicator', con = conn, if_exists='replace', index=False)
df_projects.to_sql('projects', con = conn, if_exists='replace', index=False)
```
Run the code cell below to see if your code is working properly. The code cell below runs a SQL query against the worldbank.db database joining the indicator table with the projects table.
```
pd.read_sql('SELECT * FROM projects LEFT JOIN indicator ON \
projects.countrycode = indicator.countrycode AND \
projects.year = indicator.year WHERE \
projects.year = "2017" AND projects.countrycode = "BRA"', con = conn).head()
# commit any changes to the database and close the database
conn.commit()
conn.close()
```
# Exercise Part 6 (Challenge)
SQLite, as its name would suggest, is somewhat limited in its functionality. For example, the Alter Table command only allows you to change a table name or to add a new column to a table. You can't, for example, add a primary key to a table once the table is already created.
If you want more control over a sqlite3 database, it's better to use the sqlite3 library directly. Here is an example of how to use the sqlite3 library to create a table in the database, insert a value, and then run a SQL query on the database. Run the code cells below to see the example.
### Demo
```
# connect to the data base
conn = sqlite3.connect('worldbank.db')
# get a cursor
cur = conn.cursor()
# drop the test table in case it already exists
cur.execute("DROP TABLE IF EXISTS test")
# create the test table including project_id as a primary key
cur.execute("CREATE TABLE test (project_id TEXT PRIMARY KEY, countryname TEXT, countrycode TEXT, totalamt REAL, year INTEGER);")
# insert a value into the test table
cur.execute("INSERT INTO test (project_id, countryname, countrycode, totalamt, year) VALUES ('a', 'Brazil', 'BRA', '100,000', 1970);")
# commit changes made to the database
conn.commit()
# select all from the test table
cur.execute("SELECT * FROM test")
cur.fetchall()
# commit any changes and close the data base
conn.close()
```
### Exercise
Now, it's your turn. Use the sqlite3 library to connect to the worldbank.db database. Then:
* Create a table, called projects, for the projects data where the primary key is the id of each project.
* Create another table, called gdp, that contains the gdp data.
* And create another table, called population, that contains the population data.
Here is the schema for each table.
##### projects
* project_id text
* countryname text
* countrycode text
* totalamt real
* year integer
project_id is the primary key
##### gdp
* countryname text
* countrycode text
* year integer
* gdp real
(countrycode, year) is the primary key
##### population
* countryname text
* countrycode text
* year integer
* population integer
(countrycode, year) is the primary key
After setting up the tables, write code that inserts the data into each table. (Note that this database is not normalized. For example, countryname and countrycode are in all three tables. You could make another table with countrycode and countryname and then create a foreign key constraint in the projects, gdp, and population tables. If you'd like an extra challenge, create a country table with countryname and countrycode. Then create the other tables with foreign key constraints).
Follow the TODOs in the next few code cells to finish the exercise.
```
# connect to the data base
conn = sqlite3.connect('worldbank.db')
# get a cursor
cur = conn.cursor()
# drop tables created previously to start fresh
cur.execute("DROP TABLE IF EXISTS test")
cur.execute("DROP TABLE IF EXISTS indicator")
cur.execute("DROP TABLE IF EXISTS projects")
cur.execute("DROP TABLE IF EXISTS gdp")
cur.execute("DROP TABLE IF EXISTS population")
# TODO create the projects table including project_id as a primary key
# HINT: Use cur.execute("SQL Query")
cur.execute("CREATE TABLE projects (project_id TEXT PRIMARY KEY, countryname TEXT, countrycode TEXT, totalamt REAL, year INTEGER);")
# TODO: create the gdp table including (countrycode, year) as primary key
# HINT: To create a primary key on multiple columns, you can do this:
# CREATE TABLE tablename (columna datatype, columnb datatype, columnc dataype, PRIMARY KEY (columna, columnb));
cur.execute("CREATE TABLE gdp (countryname TEXT, countrycode TEXT, year INTEGER, gdp REAL, PRIMARY KEY (countrycode, year));")
# TODO: create the gdp table including (countrycode, year) as primary key
cur.execute("CREATE TABLE population (countryname TEXT, countrycode TEXT, year INTEGER, population REAL, PRIMARY KEY (countrycode, year));")
# commit changes to the database. Do this whenever modifying a database
conn.commit()
# TODO:insert project values into the projects table
# HINT: Use a for loop with the pandas iterrows() method
# HINT: The iterrows() method returns two values: an index for each row and a tuple of values
# HINT: Some of the values for totalamt and year are NaN. Because you've defined
# year and totalamt as numbers, you cannot insert NaN as a value into those columns.
# When totaamt or year equal NaN, you'll need to change the value to something numeric
# like, for example, zero
for index, values in df_projects.iterrows():
project_id, countryname, countrycode, totalamt, year = values
if totalamt == 'nan':
totalamt = 0
if year == 'nan':
year = 0
sql_string = 'INSERT INTO projects (project_id, countryname, countrycode, totalamt, year) VALUES ("{}", "{}", "{}", {}, {});'.format(project_id, countryname, countrycode, totalamt, year)
cur.execute(sql_string)
conn.commit()
# TODO: insert gdp values into the gdp table
for index, values in df_indicator[['countryname', 'countrycode', 'year', 'gdp']].iterrows():
countryname, countrycode, year, gdp = values
sql_string = 'INSERT INTO gdp (countryname, countrycode, year, gdp) VALUES ("{}", "{}", {}, {});'.format(countryname, countrycode, year, gdp)
cur.execute(sql_string)
conn.commit()
# TODO: insert population values into the population table
for index, values in df_indicator[['countryname', 'countrycode', 'year', 'population']].iterrows():
countryname, countrycode, year, population = values
sql_string = 'INSERT INTO population (countryname, countrycode, year, population) VALUES ("{}", "{}", {}, {});'.format(countryname, countrycode, year, population)
cur.execute(sql_string)
conn.commit()
# run this command to see if your tables were loaded as expected
sqlquery = "SELECT * FROM projects JOIN gdp JOIN population ON projects.year = gdp.year AND projects.countrycode = gdp.countrycode AND projects.countrycode = population.countrycode AND projects.year=population.year;"
result = pd.read_sql(sqlquery, con=conn)
result.shape
```
If everything went well, the code above should output (15558, 13) as the shape of the resulting data frame.
```
# commit any changes and close the database
conn.commit()
conn.close()
```
# Conclusion
Once you have extracted data and transformed it, the final step is to load the data into storage. In this exercise, you stored results into a csv file, a json file, and a SQLite database.
|
github_jupyter
|
# run this code cell
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# read in the projects data set and do basic wrangling
gdp = pd.read_csv('../data/gdp_data.csv', skiprows=4)
gdp.drop(['Unnamed: 62', 'Indicator Name', 'Indicator Code'], inplace=True, axis=1)
population = pd.read_csv('../data/population_data.csv', skiprows=4)
population.drop(['Unnamed: 62', 'Indicator Name', 'Indicator Code'], inplace=True, axis=1)
# Reshape the data sets so that they are in long format
gdp_melt = gdp.melt(id_vars=['Country Name', 'Country Code'],
var_name='year',
value_name='gdp')
# Use back fill and forward fill to fill in missing gdp values
gdp_melt['gdp'] = gdp_melt.sort_values('year').groupby(['Country Name', 'Country Code'])['gdp'].fillna(method='ffill').fillna(method='bfill')
population_melt = population.melt(id_vars=['Country Name', 'Country Code'],
var_name='year',
value_name='population')
# Use back fill and forward fill to fill in missing population values
population_melt['population'] = population_melt.sort_values('year').groupby('Country Name')['population'].fillna(method='ffill').fillna(method='bfill')
# merge the population and gdp data together into one data frame
df_indicator = gdp_melt.merge(population_melt, on=('Country Name', 'Country Code', 'year'))
# filter out values that are not countries
non_countries = ['World',
'High income',
'OECD members',
'Post-demographic dividend',
'IDA & IBRD total',
'Low & middle income',
'Middle income',
'IBRD only',
'East Asia & Pacific',
'Europe & Central Asia',
'North America',
'Upper middle income',
'Late-demographic dividend',
'European Union',
'East Asia & Pacific (excluding high income)',
'East Asia & Pacific (IDA & IBRD countries)',
'Euro area',
'Early-demographic dividend',
'Lower middle income',
'Latin America & Caribbean',
'Latin America & the Caribbean (IDA & IBRD countries)',
'Latin America & Caribbean (excluding high income)',
'Europe & Central Asia (IDA & IBRD countries)',
'Middle East & North Africa',
'Europe & Central Asia (excluding high income)',
'South Asia (IDA & IBRD)',
'South Asia',
'Arab World',
'IDA total',
'Sub-Saharan Africa',
'Sub-Saharan Africa (IDA & IBRD countries)',
'Sub-Saharan Africa (excluding high income)',
'Middle East & North Africa (excluding high income)',
'Middle East & North Africa (IDA & IBRD countries)',
'Central Europe and the Baltics',
'Pre-demographic dividend',
'IDA only',
'Least developed countries: UN classification',
'IDA blend',
'Fragile and conflict affected situations',
'Heavily indebted poor countries (HIPC)',
'Low income',
'Small states',
'Other small states',
'Not classified',
'Caribbean small states',
'Pacific island small states']
# remove non countries from the data
df_indicator = df_indicator[~df_indicator['Country Name'].isin(non_countries)]
df_indicator.reset_index(inplace=True, drop=True)
df_indicator.columns = ['countryname', 'countrycode', 'year', 'gdp', 'population']
# output the first few rows of the data frame
df_indicator.head()
!pip install pycountry
from pycountry import countries
# read in the projects data set with all columns type string
df_projects = pd.read_csv('../data/projects_data.csv', dtype=str)
df_projects.drop(['Unnamed: 56'], axis=1, inplace=True)
df_projects['countryname'] = df_projects['countryname'].str.split(';').str.get(0)
# set up the libraries and variables
from collections import defaultdict
country_not_found = [] # stores countries not found in the pycountry library
project_country_abbrev_dict = defaultdict(str) # set up an empty dictionary of string values
# TODO: iterate through the country names in df_projects.
# Create a dictionary mapping the country name to the alpha_3 ISO code
for country in df_projects['countryname'].drop_duplicates().sort_values():
try:
# TODO: look up the country name in the pycountry library
# store the country name as the dictionary key and the ISO-3 code as the value
project_country_abbrev_dict[country] = countries.lookup(country).alpha_3
except:
# If the country name is not in the pycountry library, then print out the country name
# And store the results in the country_not_found list
country_not_found.append(country)
# run this code cell to load the dictionary
country_not_found_mapping = {'Co-operative Republic of Guyana': 'GUY',
'Commonwealth of Australia':'AUS',
'Democratic Republic of Sao Tome and Prin':'STP',
'Democratic Republic of the Congo':'COD',
'Democratic Socialist Republic of Sri Lan':'LKA',
'East Asia and Pacific':'EAS',
'Europe and Central Asia': 'ECS',
'Islamic Republic of Afghanistan':'AFG',
'Latin America':'LCN',
'Caribbean':'LCN',
'Macedonia':'MKD',
'Middle East and North Africa':'MEA',
'Oriental Republic of Uruguay':'URY',
'Republic of Congo':'COG',
"Republic of Cote d'Ivoire":'CIV',
'Republic of Korea':'KOR',
'Republic of Niger':'NER',
'Republic of Kosovo':'XKX',
'Republic of Rwanda':'RWA',
'Republic of The Gambia':'GMB',
'Republic of Togo':'TGO',
'Republic of the Union of Myanmar':'MMR',
'Republica Bolivariana de Venezuela':'VEN',
'Sint Maarten':'SXM',
"Socialist People's Libyan Arab Jamahiriy":'LBY',
'Socialist Republic of Vietnam':'VNM',
'Somali Democratic Republic':'SOM',
'South Asia':'SAS',
'St. Kitts and Nevis':'KNA',
'St. Lucia':'LCA',
'St. Vincent and the Grenadines':'VCT',
'State of Eritrea':'ERI',
'The Independent State of Papua New Guine':'PNG',
'West Bank and Gaza':'PSE',
'World':'WLD'}
project_country_abbrev_dict.update(country_not_found_mapping)
df_projects['countrycode'] = df_projects['countryname'].apply(lambda x: project_country_abbrev_dict[x])
df_projects['boardapprovaldate'] = pd.to_datetime(df_projects['boardapprovaldate'])
df_projects['year'] = df_projects['boardapprovaldate'].dt.year.astype(str).str.slice(stop=4)
df_projects['totalamt'] = pd.to_numeric(df_projects['totalamt'].str.replace(',',""))
df_projects = df_projects[['id', 'countryname', 'countrycode', 'totalamt', 'year']]
df_projects.head()
# TODO: merge the projects and indicator data frames together using countrycode and year as common keys
# Use a left join so that all projects are returned even if the country/year combination does not have
# indicator data
df_merged = df_projects.merge(df_indicator, how='left', on=['countrycode', 'year'])
# Run this code to check your work
df_merged[(df_merged['year'] == '2017') & (df_merged['countryname_y'] == 'Jordan')]
# TODO: Output the df_merged data frame as a json file
# HINT: Pandas has a to_json() method
# HINT: use orient='records' to get one of the more common json formats
# HINT: be sure to specify the name of the json file you want to create as the first input into to_json
df_merged.to_json('countrydata.json', orient='records')
[{"id":"P162228","countryname_x":"World","countrycode":"WLD","totalamt":0,"year":"2018","countryname_y":null,"gdp":null,"population":null},{"id":"P163962","countryname_x":"Democratic Republic of the Congo","countrycode":"COD","totalamt":200000000,"year":"2018","countryname_y":null,"gdp":null,"population":null},{"id":"P167672","countryname_x":"People's Republic of Bangladesh","countrycode":"BGD","totalamt":58330000,"year":"2018","countryname_y":null,"gdp":null,"population":null},{"id":"P158768","countryname_x":"Islamic Republic of Afghanistan","countrycode":"AFG","totalamt":20000000,"year":"2018","countryname_y":null,"gdp":null,"population":null},{"id":"P161364","countryname_x":"Federal Republic of Nigeria","countrycode":"NGA","totalamt":100000000,"year":"2018","countryname_y":null,"gdp":null,"population":null},{"id":"P161483","countryname_x":"Republic of Tunisia","countrycode":"TUN","totalamt":500000000,"year":"2018","countryname_y":null,"gdp":null,"population":null}
# TODO: Output the df_merged data frame as a csv file
# HINT: The to_csv() method is similar to the to_json() method.
# HINT: If you do not want the data frame indices in your result, use index=False
df_merged.to_csv('countrydata.csv', index=False)
id,countryname_x,countrycode,totalamt,year,countryname_y,gdp,population
P162228,World,WLD,0,2018,,,
P163962,Democratic Republic of the Congo,COD,200000000,2018,,,
P167672,People's Republic of Bangladesh,BGD,58330000,2018,,,
P158768,Islamic Republic of Afghanistan,AFG,20000000,2018,,,
P161364,Federal Republic of Nigeria,NGA,100000000,2018,,,
P161483,Republic of Tunisia,TUN,500000000,2018,,,
P161885,Federal Republic of Nigeria,NGA,350000000,2018,,,
P162069,Federal Republic of Nigeria,NGA,225000000,2018,,,
P163540,Federal Republic of Nigeria,NGA,125000000,2018,,,
P163576,Lebanese Republic,LBN,329900000,2018,,,
P163721,Democratic Socialist Republic of Sri Lan,LKA,200000000,2018,,,
P164082,Federal Republic of Nigeria,NGA,400000000,2018,,,
P164783,Nepal,NPL,0,2018,,,
import sqlite3
# connect to the database
# the database file will be worldbank.db
# note that sqlite3 will create this database file if it does not exist already
conn = sqlite3.connect('worldbank.db')
# TODO: output the df_merged dataframe to a SQL table called 'merged'.
# HINT: Use the to_sql() method
# HINT: Use the conn variable for the connection parameter
# HINT: You can use the if_exists parameter like if_exists='replace' to replace a table if it already exists
df_merged.to_sql('merged', con = conn, if_exists='replace', index=False)
pd.read_sql('SELECT * FROM merged WHERE year = "2017" AND countrycode = "BRA"', con = conn).head()
import sqlite3
# connect to the database
# the database file will be worldbank.db
# note that sqlite3 will create this database file if it does not exist already
conn = sqlite3.connect('worldbank.db')
# TODO: output the df_merged dataframe to a SQL table called 'merged'.
# HINT: Use the to_sql() method
# HINT: Use the conn variable for the connection parameter
# HINT: You can use the if_exists parameter like if_exists='replace' to replace a table if it already exists
df_indicator.to_sql('indicator', con = conn, if_exists='replace', index=False)
df_projects.to_sql('projects', con = conn, if_exists='replace', index=False)
pd.read_sql('SELECT * FROM projects LEFT JOIN indicator ON \
projects.countrycode = indicator.countrycode AND \
projects.year = indicator.year WHERE \
projects.year = "2017" AND projects.countrycode = "BRA"', con = conn).head()
# commit any changes to the database and close the database
conn.commit()
conn.close()
# connect to the data base
conn = sqlite3.connect('worldbank.db')
# get a cursor
cur = conn.cursor()
# drop the test table in case it already exists
cur.execute("DROP TABLE IF EXISTS test")
# create the test table including project_id as a primary key
cur.execute("CREATE TABLE test (project_id TEXT PRIMARY KEY, countryname TEXT, countrycode TEXT, totalamt REAL, year INTEGER);")
# insert a value into the test table
cur.execute("INSERT INTO test (project_id, countryname, countrycode, totalamt, year) VALUES ('a', 'Brazil', 'BRA', '100,000', 1970);")
# commit changes made to the database
conn.commit()
# select all from the test table
cur.execute("SELECT * FROM test")
cur.fetchall()
# commit any changes and close the data base
conn.close()
# connect to the data base
conn = sqlite3.connect('worldbank.db')
# get a cursor
cur = conn.cursor()
# drop tables created previously to start fresh
cur.execute("DROP TABLE IF EXISTS test")
cur.execute("DROP TABLE IF EXISTS indicator")
cur.execute("DROP TABLE IF EXISTS projects")
cur.execute("DROP TABLE IF EXISTS gdp")
cur.execute("DROP TABLE IF EXISTS population")
# TODO create the projects table including project_id as a primary key
# HINT: Use cur.execute("SQL Query")
cur.execute("CREATE TABLE projects (project_id TEXT PRIMARY KEY, countryname TEXT, countrycode TEXT, totalamt REAL, year INTEGER);")
# TODO: create the gdp table including (countrycode, year) as primary key
# HINT: To create a primary key on multiple columns, you can do this:
# CREATE TABLE tablename (columna datatype, columnb datatype, columnc dataype, PRIMARY KEY (columna, columnb));
cur.execute("CREATE TABLE gdp (countryname TEXT, countrycode TEXT, year INTEGER, gdp REAL, PRIMARY KEY (countrycode, year));")
# TODO: create the gdp table including (countrycode, year) as primary key
cur.execute("CREATE TABLE population (countryname TEXT, countrycode TEXT, year INTEGER, population REAL, PRIMARY KEY (countrycode, year));")
# commit changes to the database. Do this whenever modifying a database
conn.commit()
# TODO:insert project values into the projects table
# HINT: Use a for loop with the pandas iterrows() method
# HINT: The iterrows() method returns two values: an index for each row and a tuple of values
# HINT: Some of the values for totalamt and year are NaN. Because you've defined
# year and totalamt as numbers, you cannot insert NaN as a value into those columns.
# When totaamt or year equal NaN, you'll need to change the value to something numeric
# like, for example, zero
for index, values in df_projects.iterrows():
project_id, countryname, countrycode, totalamt, year = values
if totalamt == 'nan':
totalamt = 0
if year == 'nan':
year = 0
sql_string = 'INSERT INTO projects (project_id, countryname, countrycode, totalamt, year) VALUES ("{}", "{}", "{}", {}, {});'.format(project_id, countryname, countrycode, totalamt, year)
cur.execute(sql_string)
conn.commit()
# TODO: insert gdp values into the gdp table
for index, values in df_indicator[['countryname', 'countrycode', 'year', 'gdp']].iterrows():
countryname, countrycode, year, gdp = values
sql_string = 'INSERT INTO gdp (countryname, countrycode, year, gdp) VALUES ("{}", "{}", {}, {});'.format(countryname, countrycode, year, gdp)
cur.execute(sql_string)
conn.commit()
# TODO: insert population values into the population table
for index, values in df_indicator[['countryname', 'countrycode', 'year', 'population']].iterrows():
countryname, countrycode, year, population = values
sql_string = 'INSERT INTO population (countryname, countrycode, year, population) VALUES ("{}", "{}", {}, {});'.format(countryname, countrycode, year, population)
cur.execute(sql_string)
conn.commit()
# run this command to see if your tables were loaded as expected
sqlquery = "SELECT * FROM projects JOIN gdp JOIN population ON projects.year = gdp.year AND projects.countrycode = gdp.countrycode AND projects.countrycode = population.countrycode AND projects.year=population.year;"
result = pd.read_sql(sqlquery, con=conn)
result.shape
# commit any changes and close the database
conn.commit()
conn.close()
| 0.382257 | 0.915847 |
This notebook is developed using the `Python 3 (TensorFlow 2.3 Python 3.7 CPU Optimized)` kernel on an `ml.t3.medium` instance.
```
import sagemaker
import json
import boto3
role = sagemaker.get_execution_role()
sess = sagemaker.Session()
region = sess.boto_region_name
bucket = sess.default_bucket()
prefix = 'sagemaker-studio-book/chapter07'
import numpy as np
import os
from tensorflow.python.keras.datasets import imdb
csv_test_dir_prefix = 'imdb_data/test'
csv_test_filename = 'test.csv'
x_test = np.loadtxt(f'{csv_test_dir_prefix}/{csv_test_filename}',
delimiter=',', dtype='int')
print('x_test shape:', x_test.shape)
max_features = 20000
maxlen = 400
_ , (_, y_test) = imdb.load_data(num_words=max_features)
from sagemaker.tensorflow import TensorFlow
# insert the training job name from chapter05/02-tensorflow_sentiment_analysis.ipynb
training_job_name='imdb-tf-2022-02-18-05-31-26'
estimator = TensorFlow.attach(training_job_name)
predictor = estimator.deploy(initial_instance_count=1,
instance_type='ml.c5.xlarge')
predictor.endpoint_name
data_index=199
prediction=predictor.predict(x_test[data_index])
print(prediction)
def get_sentiment(score):
return 'positive' if score > 0.5 else 'negative'
import re
regex = re.compile(r'^[\?\s]+')
word_index = imdb.get_word_index()
reverse_word_index = dict([(value, key) for (key, value) in word_index.items()])
first_decoded_review = ' '.join([reverse_word_index.get(i - 3, '?')
for i in x_test[data_index]])
regex.sub('', first_decoded_review)
print(f'Labeled sentiment for this review is {get_sentiment(y_test[data_index])}')
print(f'Predicted sentiment is {get_sentiment(prediction["predictions"][0][0])}')
predictor.predict(x_test[:5000]) # this would throw an error due to large volume
sagemaker_client = sess.boto_session.client('sagemaker')
autoscaling_client = sess.boto_session.client('application-autoscaling')
endpoint_name = predictor.endpoint_name
response = sagemaker_client.describe_endpoint(EndpointName=endpoint_name)
print(response)
# application autoscaling references the endpoint using string below
resource_id=f'endpoint/{endpoint_name}/variant/AllTraffic'
response = autoscaling_client.register_scalable_target(
ServiceNamespace='sagemaker',
ResourceId=resource_id,
ScalableDimension='sagemaker:variant:DesiredInstanceCount',
MinCapacity=1,
MaxCapacity=4
)
response = autoscaling_client.put_scaling_policy(
PolicyName='Invocations-ScalingPolicy',
ServiceNamespace='sagemaker', # The namespace of the AWS service that provides the resource.
ResourceId=resource_id, # Endpoint name
ScalableDimension='sagemaker:variant:DesiredInstanceCount',
PolicyType='TargetTrackingScaling', # Other options are 'StepScaling'|'TargetTrackingScaling'
TargetTrackingScalingPolicyConfiguration={
'TargetValue': 4000.0, # The target value for the metric below.
'PredefinedMetricSpecification': {
'PredefinedMetricType': 'SageMakerVariantInvocationsPerInstance',
},
'ScaleInCooldown': 600,
'ScaleOutCooldown': 300,
'DisableScaleIn': False # If true, scale-in is disabled.
}
)
response = autoscaling_client.describe_scaling_policies(
ServiceNamespace='sagemaker'
)
for i in response['ScalingPolicies']:
print('')
print(i['PolicyName'])
print('')
if('TargetTrackingScalingPolicyConfiguration' in i):
print(i['TargetTrackingScalingPolicyConfiguration'])
else:
print(i['StepScalingPolicyConfiguration'])
print('')
```
You reach the end of this example. Typically it's recommended to delete the endpoint to stop incurring cost. However, the endpoint deployed in this notebook will be used again in [04-load_testing.ipynb](./04-load_testing.ipynb), so deletion is optional when you will soon advance to [04-load_testing.ipynb](./04-load_testing.ipynb).
If you decide to use the endpoint for [04-load_testing.ipynb](./04-load_testing.ipynb), you need the endpoint name later.
```
print(endpoint_name)
# predictor.delete_endpoint()
```
|
github_jupyter
|
import sagemaker
import json
import boto3
role = sagemaker.get_execution_role()
sess = sagemaker.Session()
region = sess.boto_region_name
bucket = sess.default_bucket()
prefix = 'sagemaker-studio-book/chapter07'
import numpy as np
import os
from tensorflow.python.keras.datasets import imdb
csv_test_dir_prefix = 'imdb_data/test'
csv_test_filename = 'test.csv'
x_test = np.loadtxt(f'{csv_test_dir_prefix}/{csv_test_filename}',
delimiter=',', dtype='int')
print('x_test shape:', x_test.shape)
max_features = 20000
maxlen = 400
_ , (_, y_test) = imdb.load_data(num_words=max_features)
from sagemaker.tensorflow import TensorFlow
# insert the training job name from chapter05/02-tensorflow_sentiment_analysis.ipynb
training_job_name='imdb-tf-2022-02-18-05-31-26'
estimator = TensorFlow.attach(training_job_name)
predictor = estimator.deploy(initial_instance_count=1,
instance_type='ml.c5.xlarge')
predictor.endpoint_name
data_index=199
prediction=predictor.predict(x_test[data_index])
print(prediction)
def get_sentiment(score):
return 'positive' if score > 0.5 else 'negative'
import re
regex = re.compile(r'^[\?\s]+')
word_index = imdb.get_word_index()
reverse_word_index = dict([(value, key) for (key, value) in word_index.items()])
first_decoded_review = ' '.join([reverse_word_index.get(i - 3, '?')
for i in x_test[data_index]])
regex.sub('', first_decoded_review)
print(f'Labeled sentiment for this review is {get_sentiment(y_test[data_index])}')
print(f'Predicted sentiment is {get_sentiment(prediction["predictions"][0][0])}')
predictor.predict(x_test[:5000]) # this would throw an error due to large volume
sagemaker_client = sess.boto_session.client('sagemaker')
autoscaling_client = sess.boto_session.client('application-autoscaling')
endpoint_name = predictor.endpoint_name
response = sagemaker_client.describe_endpoint(EndpointName=endpoint_name)
print(response)
# application autoscaling references the endpoint using string below
resource_id=f'endpoint/{endpoint_name}/variant/AllTraffic'
response = autoscaling_client.register_scalable_target(
ServiceNamespace='sagemaker',
ResourceId=resource_id,
ScalableDimension='sagemaker:variant:DesiredInstanceCount',
MinCapacity=1,
MaxCapacity=4
)
response = autoscaling_client.put_scaling_policy(
PolicyName='Invocations-ScalingPolicy',
ServiceNamespace='sagemaker', # The namespace of the AWS service that provides the resource.
ResourceId=resource_id, # Endpoint name
ScalableDimension='sagemaker:variant:DesiredInstanceCount',
PolicyType='TargetTrackingScaling', # Other options are 'StepScaling'|'TargetTrackingScaling'
TargetTrackingScalingPolicyConfiguration={
'TargetValue': 4000.0, # The target value for the metric below.
'PredefinedMetricSpecification': {
'PredefinedMetricType': 'SageMakerVariantInvocationsPerInstance',
},
'ScaleInCooldown': 600,
'ScaleOutCooldown': 300,
'DisableScaleIn': False # If true, scale-in is disabled.
}
)
response = autoscaling_client.describe_scaling_policies(
ServiceNamespace='sagemaker'
)
for i in response['ScalingPolicies']:
print('')
print(i['PolicyName'])
print('')
if('TargetTrackingScalingPolicyConfiguration' in i):
print(i['TargetTrackingScalingPolicyConfiguration'])
else:
print(i['StepScalingPolicyConfiguration'])
print('')
print(endpoint_name)
# predictor.delete_endpoint()
| 0.336222 | 0.770335 |
<img width="10%" alt="Naas" src="https://landen.imgix.net/jtci2pxwjczr/assets/5ice39g4.png?w=160"/>
# EM-DAT - Natural disasters
<a href="https://app.naas.ai/user-redirect/naas/downloader?url=https://raw.githubusercontent.com/jupyter-naas/awesome-notebooks/master/EM-DAT/EM-DAT_natural_disasters.ipynb" target="_parent"><img src="https://naasai-public.s3.eu-west-3.amazonaws.com/open_in_naas.svg"/></a>
**Tags:** #emdat #opendata #analytics
**Author:** [MyDigitalSchool](https://www.mydigitalschool.com/)
In 1988, the Centre for Research on the Epidemiology of Disasters (CRED) launched the Emergency Events Database (EM-DAT). [EM-DAT](https://www.emdat.be/) was created with the initial support of the World Health Organisation (WHO) and the Belgian Government.
## Input
In order to use this script, you need to download the dataset on this link :
https://public.emdat.be/
- Create an account and connect
- Go to "Data" tab
- Enter your filters criteria
- Press "Download"
### Import librairies
```
import pandas as pd
import plotly.express as px
```
### Variable
```
PATH_CSV = 'path_to_your_file.csv'
```
## Model
### Read the CSV
```
df = pd.read_csv(PATH_CSV)
```
### Configure the plot
```
# Types
types_df = df[['Year', 'Disaster Type']]
total_line = types_df[['Year']].value_counts().reset_index(name="value")
total_line['Disaster Type'] = "All"
types_df = types_df.groupby(['Year', 'Disaster Type']).size().reset_index(name="value")
types_df = types_df.append(total_line).sort_values(by=["Year"])
# Countries
count_by_countries = df[['Year', 'ISO', 'Country']].groupby(['Year', 'ISO', 'Country']).size().reset_index(name='counts')
```
## Output
```
fig = px.choropleth(
count_by_countries, locations="ISO",
color="counts",
hover_name="Country",
animation_frame="Year",
title = "Number of natural disasters per country",
color_continuous_scale=px.colors.sequential.OrRd,
range_color=[0, count_by_countries['counts'].max()]
)
fig.update_layout(
width=850,
height=600,
autosize=False,
template="plotly_white",
title_x=0.5
)
fig.show()
common_kwargs = {'x': "Year", 'y': "value", 'title': "Number of natural disasters per year"}
line_fig = px.line(types_df[types_df['Disaster Type'] == "All"], **common_kwargs)
lineplt_all = px.line(types_df[types_df['Disaster Type'] == "All"], **common_kwargs)
lineplt_filtered = {
disaster_type: px.line(types_df[types_df['Disaster Type'] == disaster_type], **common_kwargs)
for disaster_type in types_df['Disaster Type'].unique() if disaster_type != "All"
}
# Add dropdown
line_fig.update_layout(
updatemenus=[
dict(
buttons=list(
[
dict(
label="All disasters",
method="restyle",
args=[{
"y": [data.y for data in lineplt_all.data]
}]
)
] + [
dict(
label=disaster_type,
method="restyle",
args=[
{
"y": [data.y for data in lineplt.data],
}
]
)
for disaster_type, lineplt in lineplt_filtered.items()
]
),
),
],
title_x=0.5,
plot_bgcolor='rgba(0,0,0,0)',
)
line_fig.update_xaxes(gridcolor="grey")
line_fig.update_yaxes(gridcolor="grey")
line_fig.show()
```
**Idea of improvements :**
- Put all the curves of natural disasters in a logarithmic graph
|
github_jupyter
|
import pandas as pd
import plotly.express as px
PATH_CSV = 'path_to_your_file.csv'
df = pd.read_csv(PATH_CSV)
# Types
types_df = df[['Year', 'Disaster Type']]
total_line = types_df[['Year']].value_counts().reset_index(name="value")
total_line['Disaster Type'] = "All"
types_df = types_df.groupby(['Year', 'Disaster Type']).size().reset_index(name="value")
types_df = types_df.append(total_line).sort_values(by=["Year"])
# Countries
count_by_countries = df[['Year', 'ISO', 'Country']].groupby(['Year', 'ISO', 'Country']).size().reset_index(name='counts')
fig = px.choropleth(
count_by_countries, locations="ISO",
color="counts",
hover_name="Country",
animation_frame="Year",
title = "Number of natural disasters per country",
color_continuous_scale=px.colors.sequential.OrRd,
range_color=[0, count_by_countries['counts'].max()]
)
fig.update_layout(
width=850,
height=600,
autosize=False,
template="plotly_white",
title_x=0.5
)
fig.show()
common_kwargs = {'x': "Year", 'y': "value", 'title': "Number of natural disasters per year"}
line_fig = px.line(types_df[types_df['Disaster Type'] == "All"], **common_kwargs)
lineplt_all = px.line(types_df[types_df['Disaster Type'] == "All"], **common_kwargs)
lineplt_filtered = {
disaster_type: px.line(types_df[types_df['Disaster Type'] == disaster_type], **common_kwargs)
for disaster_type in types_df['Disaster Type'].unique() if disaster_type != "All"
}
# Add dropdown
line_fig.update_layout(
updatemenus=[
dict(
buttons=list(
[
dict(
label="All disasters",
method="restyle",
args=[{
"y": [data.y for data in lineplt_all.data]
}]
)
] + [
dict(
label=disaster_type,
method="restyle",
args=[
{
"y": [data.y for data in lineplt.data],
}
]
)
for disaster_type, lineplt in lineplt_filtered.items()
]
),
),
],
title_x=0.5,
plot_bgcolor='rgba(0,0,0,0)',
)
line_fig.update_xaxes(gridcolor="grey")
line_fig.update_yaxes(gridcolor="grey")
line_fig.show()
| 0.558568 | 0.906818 |
# Simple Location Based Sevice
This program returns nearby places of interest given the location of a vehicle.
```
import json #to import json module
from math import radians, cos, sin, asin, sqrt
def haversine(lon1, lat1, lon2, lat2):
"""
Calculate the distance between two points on the earth in Kms
"""
# convert decimal degrees to radians
lon1, lat1, lon2, lat2 = map(radians, [lon1, lat1, lon2, lat2])
# haversine formula
dlon = lon2 - lon1
dlat = lat2 - lat1
a = sin(dlat/2)**2 + cos(lat1) * cos(lat2) * sin(dlon/2)**2
c = 2 * asin(sqrt(a))
r = 6371 # Radius of earth in kilometers. Use 3956 for miles
return c * r
```
Data about restaurants, parks, fire stations, police stations and bus stops are stored in json format.
```
with open('restaurants.json') as res_data:#reading json data of restaurants
restaurantsData = json.load(res_data)
with open('fireStations.json') as res_data:#reading json data of fire stations
fireStationsData = json.load(res_data)
with open('parks.json') as res_data:#reading json data of parks
parksData = json.load(res_data)
with open('policeStation.json') as res_data:#reading json data of police stations
policeStationData = json.load(res_data)
with open('busStops.json') as res_data:#reading json data of bus Stops
busStopsData = json.load(res_data)
```
Latitude and Longitudes of different points in Bangalore.
```
#lat = 12.961009 #domlur
#lon = 77.637938
#lat = 12.952240 #marathalli
#lon = 77.700233
#lat = 12.937037 #Sony world signal
#lon = 77.626488
lat = 12.876535
lon = 77.566612
```
Menu driven program to find services.
```
def returnFacilities(pref, dist):
if(pref == 1):#searching for restaurants
listOfFeasibleRestaurants = []
for eachRes in restaurantsData:#iterating over each result in restaurants data list
try:
restData = []
resLatitude = float(eachRes['latitude'])
resLongitude = float(eachRes['longitude'])
distFromYourLocation = haversine(lon, lat, resLongitude, resLatitude)
if(distFromYourLocation < dist):
#storing the restaurant's name, latitude and longitude
resName = eachRes['name']
restData.append(resName)
restData.append(resLatitude)
restData.append(resLongitude)
listOfFeasibleRestaurants.append(restData)
except Exception,e:
#print eachRes['id']
#print repr(e)
continue
if(len(listOfFeasibleRestaurants) != 0):
for eachRest in listOfFeasibleRestaurants:
print "name : ", eachRest[0], " latitude : ", eachRest[1], " longitude : ", eachRest[2]
else:
print "Please increase the radius of search distance!"
elif(pref == 2):#searching for bus stops
listOfFeasibleBusStops = []
for eachRes in busStopsData:#iterating over each result in busStop data list
try:
busStop = []
resLatitude = float(eachRes['latitude'])
resLongitude = float(eachRes['longitude'])
distFromYourLocation = haversine(lon, lat, resLongitude, resLatitude)
if(distFromYourLocation < dist):
#storing the bus stop's name, latitude and longitude
busStopName = eachRes['Bus Stops']
busStop.append(busStopName)
busStop.append(resLatitude)
busStop.append(resLongitude)
listOfFeasibleBusStops.append(busStop)
except Exception,e:
#print eachRes['id']
#print repr(e)
continue
if(len(listOfFeasibleBusStops) != 0):
for eachBusStop in listOfFeasibleBusStops:
print "name : ", eachBusStop[0], " latitude : ", eachBusStop[1], " longitude : ", eachBusStop[2]
else:
print "Please increase the radius of search distance!"
elif(pref == 3):#searching for Parks
listOfFeasibleParks = []
for eachRes in parksData:#iterating over each result in parks data list
try:
parks = []
resLatitude = float(eachRes['latitude'])
resLongitude = float(eachRes['longitude'])
distFromYourLocation = haversine(lon, lat, resLongitude, resLatitude)
if(distFromYourLocation < dist):
#storing the parks' address, latitude and longitude
parksAddress = eachRes['Address of the Park']
parks.append(parksAddress)
parks.append(resLatitude)
parks.append(resLongitude)
listOfFeasibleParks.append(parks)
except Exception,e:
#print eachRes['id']
#print repr(e)
continue
if(len(listOfFeasibleParks) != 0):
for eachPark in listOfFeasibleParks:
print "Address : ", eachPark[0], " latitude : ", eachPark[1], " longitude : ", eachPark[2]
else:
print "Please increase the radius of search distance!"
elif(pref == 4):#searching for Police stations
listOfFeasiblePoliceStations = []
for eachRes in policeStationData:#iterating over each result in police stations data list
try:
policeData = []
resLatitude = float(eachRes['latitude'])
resLongitude = float(eachRes['longitude'])
distFromYourLocation = haversine(lon, lat, resLongitude, resLatitude)
if(distFromYourLocation < dist):
#storing the police station's name, latitude and longitude
policeStationName = eachRes['Police Station Name']
policeData.append(policeStationName)
policeData.append(resLatitude)
policeData.append(resLongitude)
listOfFeasiblePoliceStations.append(policeData)
except Exception,e:
#print eachRes['id']
#print repr(e)
continue
if(len(listOfFeasiblePoliceStations) != 0):
for eachPoliceStation in listOfFeasiblePoliceStations:
print "name : ", eachPoliceStation[0], " latitude : ", eachPoliceStation[1], " longitude : ", eachPoliceStation[2]
else:
print "Please increase the radius of search distance!"
elif(pref == 5):#searching for Fire stations
listOfFeasibleFireStations = []
for eachRes in fireStationsData:#iterating over each result in fire stations data list
try:
fireStation = []
resLatitude = float(eachRes['latitude'])
resLongitude = float(eachRes['longitude'])
distFromYourLocation = haversine(lon, lat, resLongitude, resLatitude)
if(distFromYourLocation < dist):
#storing the fire stations's address, latitude and longitude
fireStationAddress = eachRes['Fire Station Address']
fireStation.append(fireStationAddress)
fireStation.append(resLatitude)
fireStation.append(resLongitude)
listOfFeasibleFireStations.append(fireStation)
except Exception,e:
#print eachRes['id']
#print repr(e)
continue
if(len(listOfFeasibleFireStations) != 0):
for eachFireStation in listOfFeasibleFireStations:
print "Address : ", eachFireStation[0], " latitude : ", eachFireStation[1], " longitude : ", eachFireStation[2]
else:
print "Please increase the radius of search distance!"
else:
print "invalid input. Please choose a valid option."
#askUser()
def askUser(lat, lon):
print "What services would you like to search for?"
print "1. Restaurants"
print "2. Bus Stops"
print "3. Parks"
print "4. Police Station"
print "5. Fire Stations"
pref = int(raw_input("Please choose an option : "))
dist = float(raw_input("Please input the nearby distance : "))
returnFacilities(pref, dist)
askUser(lat, lon)
```
|
github_jupyter
|
import json #to import json module
from math import radians, cos, sin, asin, sqrt
def haversine(lon1, lat1, lon2, lat2):
"""
Calculate the distance between two points on the earth in Kms
"""
# convert decimal degrees to radians
lon1, lat1, lon2, lat2 = map(radians, [lon1, lat1, lon2, lat2])
# haversine formula
dlon = lon2 - lon1
dlat = lat2 - lat1
a = sin(dlat/2)**2 + cos(lat1) * cos(lat2) * sin(dlon/2)**2
c = 2 * asin(sqrt(a))
r = 6371 # Radius of earth in kilometers. Use 3956 for miles
return c * r
with open('restaurants.json') as res_data:#reading json data of restaurants
restaurantsData = json.load(res_data)
with open('fireStations.json') as res_data:#reading json data of fire stations
fireStationsData = json.load(res_data)
with open('parks.json') as res_data:#reading json data of parks
parksData = json.load(res_data)
with open('policeStation.json') as res_data:#reading json data of police stations
policeStationData = json.load(res_data)
with open('busStops.json') as res_data:#reading json data of bus Stops
busStopsData = json.load(res_data)
#lat = 12.961009 #domlur
#lon = 77.637938
#lat = 12.952240 #marathalli
#lon = 77.700233
#lat = 12.937037 #Sony world signal
#lon = 77.626488
lat = 12.876535
lon = 77.566612
def returnFacilities(pref, dist):
if(pref == 1):#searching for restaurants
listOfFeasibleRestaurants = []
for eachRes in restaurantsData:#iterating over each result in restaurants data list
try:
restData = []
resLatitude = float(eachRes['latitude'])
resLongitude = float(eachRes['longitude'])
distFromYourLocation = haversine(lon, lat, resLongitude, resLatitude)
if(distFromYourLocation < dist):
#storing the restaurant's name, latitude and longitude
resName = eachRes['name']
restData.append(resName)
restData.append(resLatitude)
restData.append(resLongitude)
listOfFeasibleRestaurants.append(restData)
except Exception,e:
#print eachRes['id']
#print repr(e)
continue
if(len(listOfFeasibleRestaurants) != 0):
for eachRest in listOfFeasibleRestaurants:
print "name : ", eachRest[0], " latitude : ", eachRest[1], " longitude : ", eachRest[2]
else:
print "Please increase the radius of search distance!"
elif(pref == 2):#searching for bus stops
listOfFeasibleBusStops = []
for eachRes in busStopsData:#iterating over each result in busStop data list
try:
busStop = []
resLatitude = float(eachRes['latitude'])
resLongitude = float(eachRes['longitude'])
distFromYourLocation = haversine(lon, lat, resLongitude, resLatitude)
if(distFromYourLocation < dist):
#storing the bus stop's name, latitude and longitude
busStopName = eachRes['Bus Stops']
busStop.append(busStopName)
busStop.append(resLatitude)
busStop.append(resLongitude)
listOfFeasibleBusStops.append(busStop)
except Exception,e:
#print eachRes['id']
#print repr(e)
continue
if(len(listOfFeasibleBusStops) != 0):
for eachBusStop in listOfFeasibleBusStops:
print "name : ", eachBusStop[0], " latitude : ", eachBusStop[1], " longitude : ", eachBusStop[2]
else:
print "Please increase the radius of search distance!"
elif(pref == 3):#searching for Parks
listOfFeasibleParks = []
for eachRes in parksData:#iterating over each result in parks data list
try:
parks = []
resLatitude = float(eachRes['latitude'])
resLongitude = float(eachRes['longitude'])
distFromYourLocation = haversine(lon, lat, resLongitude, resLatitude)
if(distFromYourLocation < dist):
#storing the parks' address, latitude and longitude
parksAddress = eachRes['Address of the Park']
parks.append(parksAddress)
parks.append(resLatitude)
parks.append(resLongitude)
listOfFeasibleParks.append(parks)
except Exception,e:
#print eachRes['id']
#print repr(e)
continue
if(len(listOfFeasibleParks) != 0):
for eachPark in listOfFeasibleParks:
print "Address : ", eachPark[0], " latitude : ", eachPark[1], " longitude : ", eachPark[2]
else:
print "Please increase the radius of search distance!"
elif(pref == 4):#searching for Police stations
listOfFeasiblePoliceStations = []
for eachRes in policeStationData:#iterating over each result in police stations data list
try:
policeData = []
resLatitude = float(eachRes['latitude'])
resLongitude = float(eachRes['longitude'])
distFromYourLocation = haversine(lon, lat, resLongitude, resLatitude)
if(distFromYourLocation < dist):
#storing the police station's name, latitude and longitude
policeStationName = eachRes['Police Station Name']
policeData.append(policeStationName)
policeData.append(resLatitude)
policeData.append(resLongitude)
listOfFeasiblePoliceStations.append(policeData)
except Exception,e:
#print eachRes['id']
#print repr(e)
continue
if(len(listOfFeasiblePoliceStations) != 0):
for eachPoliceStation in listOfFeasiblePoliceStations:
print "name : ", eachPoliceStation[0], " latitude : ", eachPoliceStation[1], " longitude : ", eachPoliceStation[2]
else:
print "Please increase the radius of search distance!"
elif(pref == 5):#searching for Fire stations
listOfFeasibleFireStations = []
for eachRes in fireStationsData:#iterating over each result in fire stations data list
try:
fireStation = []
resLatitude = float(eachRes['latitude'])
resLongitude = float(eachRes['longitude'])
distFromYourLocation = haversine(lon, lat, resLongitude, resLatitude)
if(distFromYourLocation < dist):
#storing the fire stations's address, latitude and longitude
fireStationAddress = eachRes['Fire Station Address']
fireStation.append(fireStationAddress)
fireStation.append(resLatitude)
fireStation.append(resLongitude)
listOfFeasibleFireStations.append(fireStation)
except Exception,e:
#print eachRes['id']
#print repr(e)
continue
if(len(listOfFeasibleFireStations) != 0):
for eachFireStation in listOfFeasibleFireStations:
print "Address : ", eachFireStation[0], " latitude : ", eachFireStation[1], " longitude : ", eachFireStation[2]
else:
print "Please increase the radius of search distance!"
else:
print "invalid input. Please choose a valid option."
#askUser()
def askUser(lat, lon):
print "What services would you like to search for?"
print "1. Restaurants"
print "2. Bus Stops"
print "3. Parks"
print "4. Police Station"
print "5. Fire Stations"
pref = int(raw_input("Please choose an option : "))
dist = float(raw_input("Please input the nearby distance : "))
returnFacilities(pref, dist)
askUser(lat, lon)
| 0.220342 | 0.806358 |
# Create a 3D Point Cloud Labeling Job with Amazon SageMaker Ground Truth
This notebook will demonstrate how you can pre-process your 3D point cloud input data to create an [object tracking labeling job](https://docs.aws.amazon.com/sagemaker/latest/dg/sms-point-cloud-object-tracking.html) and include sensor and camera data for sensor fusion.
In object tracking, you are tracking the movement of an object (e.g., a pedestrian on the side walk) while your point of reference (e.g., the autonomous vehicle) is moving. When performing object tracking, your data must be in a global reference coordinate system such as [world coordinate system](https://docs.aws.amazon.com/sagemaker/latest/dg/sms-point-cloud-sensor-fusion-details.html#sms-point-cloud-world-coordinate-system) because the ego vehicle itself is moving in the world. You can transform point cloud data in local coordinates to the world coordinate system by multiplying each of the points in a 3D frame with the extrinsic matrix for the LiDAR sensor.
In this notebook, you will transform 3D frames from a local coordinate system to a world coordinate system using extrinsic matrices. You will use the KITTI dataset, an open source autonomous driving dataset. The KITTI dataset provides an extrinsic matrix for each 3D point cloud frame. You will use [pykitti](https://github.com/utiasSTARS/pykitti) and the [numpy matrix multiplication function](https://numpy.org/doc/1.18/reference/generated/numpy.matmul.html) to multiple this matrix with each point in the frame to translate that point to the world coordinate system used by the KITTI dataset.
You include camera image data and provide workers with more visual information about the scene they are labeling. Through sensor fusion, workers will be able to adjust labels in the 3D scene and in 2D images, and label adjustments will be mirrored in the other view.
Ground Truth computes your sensor and camera extrinsic matrices for sensor fusion using sensor and camera **pose data** - position and heading. The KITTI raw dataset includes rotation matrix and translations vectors for extrinsic transformations for each frame. This notebook will demonstrate how you can extract **position** and **heading** from KITTI rotation matrices and translations vectors using [pykitti](https://github.com/utiasSTARS/pykitti).
In summary, you will:
* Convert a dataset to a world coordinate system.
* Learn how you can extract pose data from your LiDAR and camera extrinsict matrices for sensor fusion.
* Create a sequence input manifest file for an object tracking labeling job.
* Create an object tracking labeling job.
* Preview the worker UI and tools provided by Ground Truth.
## Prerequisites
To run this notebook, you can simply execute each cell in order. To understand what's happening, you'll need:
* An S3 bucket you can write to -- please provide its name in `BUCKET`. The bucket must be in the same region as this SageMaker Notebook instance. You can also change the `EXP_NAME` to any valid S3 prefix. All the files related to this experiment will be stored in that prefix of your bucket. **Important: you must attach the CORS policy to this bucket. See the next section for more information**.
* Familiarity with the [Ground Truth 3D Point Cloud Labeling Job](https://docs.aws.amazon.com/sagemaker/latest/dg/sms-point-cloud.html).
* Familiarity with Python and [numpy](http://www.numpy.org/).
* Basic familiarity with [AWS S3](https://docs.aws.amazon.com/s3/index.html).
* Basic understanding of [AWS Sagemaker](https://aws.amazon.com/sagemaker/).
* Basic familiarity with [AWS Command Line Interface (CLI)](https://aws.amazon.com/cli/) -- ideally, you should have it set up with credentials to access the AWS account you're running this notebook from.
This notebook has only been tested on a SageMaker notebook instance. The runtimes given are approximate. We used an `ml.t2.medium` instance in our tests. However, you can likely run it on a local instance by first executing the cell below on SageMaker and then copying the `role` string to your local copy of the notebook.
### IMPORTANT: Attach CORS policy to your bucket
You must attach the following CORS policy to your S3 bucket for the labeling task to render. To learn how to add a CORS policy to your S3 bucket, follow the instructions in [How do I add cross-domain resource sharing with CORS?](https://docs.aws.amazon.com/AmazonS3/latest/user-guide/add-cors-configuration.html). Paste the following policy in the CORS configuration editor:
```
<?xml version="1.0" encoding="UTF-8"?>
<CORSConfiguration xmlns="http://s3.amazonaws.com/doc/2006-03-01/">
<CORSRule>
<AllowedOrigin>*</AllowedOrigin>
<AllowedMethod>GET</AllowedMethod>
<AllowedMethod>HEAD</AllowedMethod>
<AllowedMethod>PUT</AllowedMethod>
<MaxAgeSeconds>3000</MaxAgeSeconds>
<ExposeHeader>Access-Control-Allow-Origin</ExposeHeader>
<AllowedHeader>*</AllowedHeader>
</CORSRule>
<CORSRule>
<AllowedOrigin>*</AllowedOrigin>
<AllowedMethod>GET</AllowedMethod>
</CORSRule>
</CORSConfiguration>
```
```
import boto3
import time
import pprint
import json
import sagemaker
from sagemaker import get_execution_role
from datetime import datetime, timezone
pp = pprint.PrettyPrinter(indent=4)
sagemaker_client = boto3.client('sagemaker')
BUCKET = ''
EXP_NAME = '' # Any valid S3 prefix.
# Make sure the bucket is in the same region as this notebook.
sess = sagemaker.session.Session()
role = sagemaker.get_execution_role()
region = boto3.session.Session().region_name
s3 = boto3.client('s3')
bucket_region = s3.head_bucket(Bucket=BUCKET)['ResponseMetadata']['HTTPHeaders']['x-amz-bucket-region']
assert bucket_region == region, "Your S3 bucket {} and this notebook need to be in the same region.".format(BUCKET)
```
## The Dataset and Input Manifest Files
The dataset and resources used in this notebook are located in the following Amazon S3 bucket: https://aws-ml-blog.s3.amazonaws.com/artifacts/gt-point-cloud-demos/.
This bucket contains a single scene from the [KITTI datasets](http://www.cvlibs.net/datasets/kitti/raw_data.php). KITTI created datasets for computer vision and machine learning research, including for 2D and 3D object detection and object tracking. The datasets are captured by driving around the mid-size city of Karlsruhe, in rural areas and on highways.
## Download and unzip data
```
rm -rf sample_data*
!wget https://aws-ml-blog.s3.amazonaws.com/artifacts/gt-point-cloud-demos/sample_data.zip
!unzip -o sample_data
```
Let's take a look at the sample_data folder. You'll see that we have images which can be used for sensor fusion, and point cloud data in ASCII format (.txt files). We will use a script to convert this point cloud data from the LiDAR sensor's local coordinates to a world coordinate system.
```
!ls sample_data/2011_09_26/2011_09_26_drive_0005_sync/
!ls sample_data/2011_09_26/2011_09_26_drive_0005_sync/oxts/data
```
## Use the Kitti2GT script to convert the raw data to Ground Truth format
You can use this script to do the following:
* Transform the KITTI dataset with respect to the LIDAR sensor's orgin in the first frame as the world cooridinate system ( global frame of reference ), so that it can be consumed by SageMaker Ground Truth.
* Extract pose data in world coordinate system using the camera and LiDAR extrinsic matrices. You will supply this pose data in your sequence file to enable sensor fusion.
First, the script uses [pykitti](https://github.com/utiasSTARS/pykitti) python module to load the KITTI raw data and calibrations. Let's look at the two main data-transformation functions of the script:
### Data Transformation to a World Coordinate System
In general, multiplying a point in a LIDAR frame with a LIDAR extrinsic matrix transforms it into world coordinates.
Using pykitti `dataset.oxts[i].T_w_imu` gives the lidar extrinsic transform for the `i`th frame. This matrix can be multiplied with the points of the frame to convert it to a world frame using the numpy matrix multiplication, function, [matmul](https://numpy.org/doc/1.18/reference/generated/numpy.matmul.html): `matmul(lidar_transform_matrix, points)`. Let's look at the function that performs this transformation:
```
# transform points from lidar to global frame using lidar_extrinsic_matrix
def generate_transformed_pcd_from_point_cloud(points, lidar_extrinsic_matrix):
tps = []
for point in points:
transformed_points = np.matmul(lidar_extrinsic_matrix, np.array([point[0], point[1], point[2], 1], dtype=np.float32).reshape(4,1)).tolist()
if len(point) > 3 and point[3] is not None:
tps.append([transformed_points[0][0], transformed_points[1][0], transformed_points[2][0], point[3]])
return tps
```
If your point cloud data includes more than four elements for each point, for example, (x,y,z) and r,g,b, modify the `if` statement in the function above to ensure your r, g, b values are copied.
### Extracting Pose Data from LiDAR and Camera Extrinsic for Sensor Fusion
For sensor fusion, you provide your extrinsic matrix in the form of sensor-pose in terms of origin position (for translation) and heading in quaternion (for rotation of the 3 axis). The following is an example of the pose JSON you use in the sequence file.
```
{
"position": {
"y": -152.77584902657554,
"x": 311.21505956090624,
"z": -10.854137529636024
},
"heading": {
"qy": -0.7046155108831117,
"qx": 0.034278837280808494,
"qz": 0.7070617895701465,
"qw": -0.04904659893885366
}
}
```
All of the positional coordinates (x, y, z) are in meters. All the pose headings (qx, qy, qz, qw) are measured in Spatial Orientation in Quaternion. Separately for each camera, you provide pose data extracted from the extrinsic of that camera.
Both LIDAR sensors and and cameras have their own extrinsic matrices, and they are used by SageMaker Ground Truth to enable the sensor fusion feature. In order to project a label from 3D point cloud to camera image plane Ground Truth needs to transform 3D points from LIDAR’s own coordinate system to the camera’s coordinate system. This is typically done by first transforming 3D points from LIDAR’s own coordinate to a world coordinate system using the LIDAR extrinsic matrix. Then we use the camera inverse extrinsic (world to camera) to transform the 3D points from the world coordinate system we obtained in previous step into camera image plane. If your 3D data is already transformed into world coordinate system then the first transformation doesn’t have any impact and label translation depends only on the camera extrinsic.
If you have a rotation matrix (made up of the axis rotations) and translation vector (or origin) in world coordinate system instead of a single 4x4 rigid transformation matrix, then you can directly use rotation and translation to compute pose. For example:
```
import numpy as np
rotation = [[ 9.96714314e-01, -8.09890350e-02, 1.16333982e-03],
[ 8.09967396e-02, 9.96661051e-01, -1.03090934e-02],
[-3.24531964e-04, 1.03694477e-02, 9.99946183e-01]]
origin= [1.71104606e+00,
5.80000039e-01,
9.43144935e-01]
from scipy.spatial.transform import Rotation as R
# position is the origin
position = origin
r = R.from_matrix(np.asarray(rotation))
# heading in WCS using scipy
heading = r.as_quat()
print(f"position:{position}\nheading: {heading}")
```
If you indeed have a 4x4 extrinsic transformation matrix then the transformation matrix is just in the form of ```[R T; 0 0 0 1]``` where R is the rotation matrix and T is the origin translation vector. That means you can extract rotation matrix and translation vector from the transformation matrix as follows
```
import numpy as np
transformation = [[ 9.96714314e-01, -8.09890350e-02, 1.16333982e-03, 1.71104606e+00],
[ 8.09967396e-02, 9.96661051e-01, -1.03090934e-02, 5.80000039e-01],
[-3.24531964e-04, 1.03694477e-02, 9.99946183e-01, 9.43144935e-01],
[0, 0, 0, 1]]
transformation = np.array(transformation)
rotation = transformation[0:3, 0:3]
origin= transformation[0:3, 3]
from scipy.spatial.transform import Rotation as R
# position is the origin
position = origin
r = R.from_matrix(np.asarray(rotation))
# heading in WCS using scipy
heading = r.as_quat()
print(f"position:{position}\nheading: {heading}")
```
For convenience, in this blog you will use [pykitti](https://github.com/utiasSTARS/pykitti) development kit to load the raw data and calibrations. With pykitti you will extract sensor pose in the world coordinate system from KITTI extrinsic which is provided as a rotation matrix and translation vector in the raw calibrations data. We will then format this pose data using the JSON format required for the 3D point cloud sequence input manifest.
With pykitti the ```dataset.oxts[i].T_w_imu``` gives the LiDAR extrinsic matrix ( lidar_extrinsic_transform ) for the i'th frame. Similarly, with pykitti the camera extrinsic matrix ( camera_extrinsic_transform ) for cam0 in i'th frame can be calculated by ```inv(matmul(dataset.calib.T_cam0_velo, inv(dataset.oxts[i].T_w_imu)))``` and this can be converted into heading and position for cam0.
In the script, the following functions are used to extract this pose data from the LiDAR extrinsict and camera inverse extrinsic matrices.
```
# utility to convert extrinsic matrix to pose heading quaternion and position
def convert_extrinsic_matrix_to_trans_quaternion_mat(lidar_extrinsic_transform):
position = lidar_extrinsic_transform[0:3, 3]
rot = np.linalg.inv(lidar_extrinsic_transform[0:3, 0:3])
quaternion= R.from_matrix(np.asarray(rot)).as_quat()
trans_quaternions = {
"translation": {
"x": position[0],
"y": position[1],
"z": position[2]
},
"rotation": {
"qx": quaternion[0],
"qy": quaternion[1],
"qz": quaternion[2],
"qw": quaternion[3]
}
}
return trans_quaternions
def convert_camera_inv_extrinsic_matrix_to_trans_quaternion_mat(camera_extrinsic_transform):
position = camera_extrinsic_transform[0:3, 3]
rot = np.linalg.inv(camera_extrinsic_transform[0:3, 0:3])
quaternion= R.from_matrix(np.asarray(rot)).as_quat()
trans_quaternions = {
"translation": {
"x": position[0],
"y": position[1],
"z": position[2]
},
"rotation": {
"qx": quaternion[0],
"qy": quaternion[1],
"qz": quaternion[2],
"qw": -quaternion[3]
}
}
return trans_quaternions
```
### Generate a Sequence File
After you've converted your data to a world coordinate system and extracted sensor and camera pose data for sensor fusion, you can create a sequence file. This is accomplished with the function `convert_to_gt` in the python script.
A **sequence** specifies a temporal series of point cloud frames. When a task is created using a sequence file, all point cloud frames in the sequence are sent to a worker to label. Your input manifest file will contain a single sequence per line. To learn more about the sequence input manifest format, see [Create a Point Cloud Frame Sequence Input Manifest](https://docs.aws.amazon.com/sagemaker/latest/dg/sms-point-cloud-multi-frame-input-data.html).
If you want to use this script to create a frame input manifest file, which is required for 3D point cloud object tracking and semantic segmentation labeling jobs, you can modify the for-loop in the function `convert_to_gt`
to produce the required content for `source-ref-metadata`. To learn more about the frame input manifest format, see [Create a Point Cloud Frame Input Manifest File](https://docs.aws.amazon.com/sagemaker/latest/dg/sms-point-cloud-single-frame-input-data.html).
Now, let's download the script and run it on the KITTI dataset to process the data you'll use for your labeling job.
```
!wget https://aws-ml-blog.s3.amazonaws.com/artifacts/gt-point-cloud-demos/kitti2gt.py
!pygmentize kitti2gt.py
```
### Install pykitti
```
!pip install pykitti
!pip install --upgrade scipy
from kitti2gt import *
if(EXP_NAME == ''):
s3loc = f's3://{BUCKET}/frames/'
else:
s3loc = f's3://{BUCKET}/{EXP_NAME}/frames/'
convert_to_gt(basedir='sample_data',
date='2011_09_26',
drive='0005',
output_base='sample_data_out',
s3prefix = s3loc)
```
The following folders that will contain the data you'll use for the labeling job.
```
!ls sample_data_out/
!ls sample_data_out/frames
```
Now, you'll upload the data to your bucket in S3.
```
if(EXP_NAME == ''):
!aws s3 cp sample_data_out/kitti-gt-seq.json s3://{BUCKET}/
else:
!aws s3 cp sample_data_out/kitti-gt-seq.json s3://{BUCKET}/{EXP_NAME}/
if(EXP_NAME == ''):
!aws s3 sync sample_data_out/frames/ s3://{BUCKET}/frames/
else:
!aws s3 sync sample_data_out/frames s3://{BUCKET}/{EXP_NAME}/frames/
if(EXP_NAME == ''):
!aws s3 sync sample_data_out/images/ s3://{BUCKET}/frames/images/
else:
!aws s3 sync sample_data_out/images s3://{BUCKET}/{EXP_NAME}/frames/images/
```
### Write and Upload Multi-Frame Input Manifest File
Now, let's create a **sequence input manifest file**. Each line in the input manifest (in this demo, there is only one) will point to a sequence file in your S3 bucket, `BUCKET/EXP_NAME`.
```
with open('manifest.json','w') as f:
if(EXP_NAME == ''):
json.dump({"source-ref": "s3://{}/kitti-gt-seq.json".format(BUCKET)},f)
else:
json.dump({"source-ref": "s3://{}/{}/kitti-gt-seq.json".format(BUCKET,EXP_NAME)},f)
```
Our manifest file is one line long, and identifies a single sequence file in your S3 bucket.
```
!cat manifest.json
if(EXP_NAME == ''):
!aws s3 cp manifest.json s3://{BUCKET}/
input_manifest_s3uri = f's3://{BUCKET}/manifest.json'
else:
!aws s3 cp manifest.json s3://{BUCKET}/{EXP_NAME}/
input_manifest_s3uri = f's3://{BUCKET}/{EXP_NAME}/manifest.json'
input_manifest_s3uri
```
## Create a Labeling Job
In the following cell, we specify object tracking as our [3D Point Cloud Task Type](https://docs.aws.amazon.com/sagemaker/latest/dg/sms-point-cloud-task-types.html).
```
task_type = "3DPointCloudObjectTracking"
```
## Identify Resources for Labeling Job
### Specify Human Task UI ARN
The following will be used to identify the HumanTaskUiArn. When you create a 3D point cloud labeling job, Ground Truth provides a worker UI that is specific to your task type. You can learn more about this UI and the assistive labeling tools that Ground Truth provides for Object Tracking on the [Object Tracking task type page](https://docs.aws.amazon.com/sagemaker/latest/dg/sms-point-cloud-object-tracking.html).
```
## Set up human_task_ui_arn map, to be used in case you chose UI_CONFIG_USE_TASK_UI_ARN
## Supported for GA
## Set up human_task_ui_arn map, to be used in case you chose UI_CONFIG_USE_TASK_UI_ARN
human_task_ui_arn = f'arn:aws:sagemaker:{region}:394669845002:human-task-ui/PointCloudObjectTracking'
human_task_ui_arn
```
### Label Category Configuration File
Your label category configuration file is used to specify labels, or classes, for your labeling job.
When you use the object detection or object tracking task types, you can also include **label attributes** in your [label category configuration file](https://docs.aws.amazon.com/sagemaker/latest/dg/sms-point-cloud-label-category-config.html). Workers can assign one or more attributes you provide to annotations to give more information about that object. For example, you may want to use the attribute *occluded* to have workers identify when an object is partially obstructed.
Let's look at an example of the label category configuration file for an object detection or object tracking labeling job.
```
!wget https://aws-ml-blog.s3.amazonaws.com/artifacts/gt-point-cloud-demos/label-category-config/label-category.json
with open('label-category.json', 'r') as j:
json_data = json.load(j)
print("\nA label category configuration file: \n\n",json.dumps(json_data, indent=4, sort_keys=True))
if(EXP_NAME == ''):
!aws s3 cp label-category.json s3://{BUCKET}/label-category.json
label_category_config_s3uri = f's3://{BUCKET}/label-category.json'
else:
!aws s3 cp label-category.json s3://{BUCKET}/{EXP_NAME}/label-category.json
label_category_config_s3uri = f's3://{BUCKET}/{EXP_NAME}/label-category.json'
label_category_config_s3uri
```
To learn more about the label category configuration file, see [Create a Label Category Configuration File](https://docs.aws.amazon.com/sagemaker/latest/dg/sms-point-cloud-label-category-config.html)
Run the following cell to identify the labeling category configuration file.
### Set up a private work team
If you want to preview the worker task UI, create a private work team and add yourself as a worker.
If you have already created a private workforce, follow the instructions in [Add or Remove Workers](https://docs.aws.amazon.com/sagemaker/latest/dg/sms-workforce-management-private-console.html#add-remove-workers-sm) to add yourself to the work team you use to create a lableing job.
#### Create a private workforce and add yourself as a worker
To create and manage your private workforce, you can use the **Labeling workforces** page in the Amazon SageMaker console. When following the instructions below, you will have the option to create a private workforce by entering worker emails or importing a pre-existing workforce from an Amazon Cognito user pool. To import a workforce, see [Create a Private Workforce (Amazon Cognito Console)](https://docs.aws.amazon.com/sagemaker/latest/dg/sms-workforce-create-private-cognito.html).
To create a private workforce using worker emails:
* Open the Amazon SageMaker console at https://console.aws.amazon.com/sagemaker/.
* In the navigation pane, choose **Labeling workforces**.
* Choose Private, then choose **Create private team**.
* Choose **Invite new workers by email**.
* Paste or type a list of up to 50 email addresses, separated by commas, into the email addresses box.
* Enter an organization name and contact email.
* Optionally choose an SNS topic to subscribe the team to so workers are notified by email when new Ground Truth labeling jobs become available.
* Click the **Create private team** button.
After you import your private workforce, refresh the page. On the Private workforce summary page, you'll see your work team ARN. Enter this ARN in the following cell.
```
##Use Beta Private Team till GA
workteam_arn = ''
```
#### Task Time Limits
3D point cloud annotation jobs can take workers hours. Workers will be able to save their work as they go, and complete the task in multiple sittings. Ground Truth will also automatically save workers' annotations periodically as they work.
When you configure your task, you can set the total amount of time that workers can work on each task when you create a labeling job using `TaskTimeLimitInSeconds`. The maximum time you can set for workers to work on tasks is 7 days. The default value is 3 days. It is recommended that you create labeling tasks that can be completed within 12 hours.
If you set `TaskTimeLimitInSeconds` to be greater than 8 hours, you must set `MaxSessionDuration` for your IAM execution to at least 8 hours. To update your execution role's `MaxSessionDuration`, use [UpdateRole](https://docs.aws.amazon.com/IAM/latest/APIReference/API_UpdateRole.html) or use the [IAM console](https://docs.aws.amazon.com/IAM/latest/UserGuide/roles-managingrole-editing-console.html#roles-modify_max-session-duration). You an identify the name of your role at the end of your role ARN.
```
#See your execution role ARN. The role name is located at the end of the ARN.
role
ac_arn_map = {'us-west-2': '081040173940',
'us-east-1': '432418664414',
'us-east-2': '266458841044',
'eu-west-1': '568282634449',
'ap-northeast-1': '477331159723'}
prehuman_arn = 'arn:aws:lambda:{}:{}:function:PRE-{}'.format(region, ac_arn_map[region],task_type)
acs_arn = 'arn:aws:lambda:{}:{}:function:ACS-{}'.format(region, ac_arn_map[region],task_type)
```
## Set Up HumanTaskConfig
`HumanTaskConfig` is used to specify your work team, and configure your labeling job tasks. Modify the following cell to identify a `task_description`, `task_keywords`, `task_title`, and `job_name`.
```
from datetime import datetime
## Set up Human Task Config
## Modify the following
task_description = 'add a task description here'
#example keywords
task_keywords = ['lidar', 'pointcloud']
#add a task title
task_title = 'Add a Task Title Here - This is Displayed to Workers'
#add a job name to identify your labeling job
job_name = 'add-job-name'
human_task_config = {
"AnnotationConsolidationConfig": {
"AnnotationConsolidationLambdaArn": acs_arn,
},
"UiConfig": {
"HumanTaskUiArn": human_task_ui_arn,
},
"WorkteamArn": workteam_arn,
"PreHumanTaskLambdaArn": prehuman_arn,
"MaxConcurrentTaskCount": 200, # 200 images will be sent at a time to the workteam.
"NumberOfHumanWorkersPerDataObject": 1, # One worker will work on each task
"TaskAvailabilityLifetimeInSeconds": 18000, # Your workteam has 5 hours to complete all pending tasks.
"TaskDescription": task_description,
"TaskKeywords": task_keywords,
"TaskTimeLimitInSeconds": 3600, # Each seq must be labeled within 1 hour.
"TaskTitle": task_title
}
print(json.dumps(human_task_config, indent=4, sort_keys=True))
```
## Set up Create Labeling Request
The following formats your labeling job request. For Object Tracking task types, the `LabelAttributeName` must end in `-ref`.
```
if(EXP_NAME == ''):
s3_output_path = f's3://{BUCKET}'
else:
s3_output_path = f's3://{BUCKET}/{EXP_NAME}'
s3_output_path
## Set up Create Labeling Request
labelAttributeName = job_name + "-ref"
if task_type == "3DPointCloudObjectDetection" or task_type == "Adjustment3DPointCloudObjectDetection":
labelAttributeName = job_name
ground_truth_request = {
"InputConfig" : {
"DataSource": {
"S3DataSource": {
"ManifestS3Uri": input_manifest_s3uri,
}
},
"DataAttributes": {
"ContentClassifiers": [
"FreeOfPersonallyIdentifiableInformation",
"FreeOfAdultContent"
]
},
},
"OutputConfig" : {
"S3OutputPath": s3_output_path,
},
"HumanTaskConfig" : human_task_config,
"LabelingJobName": job_name,
"RoleArn": role,
"LabelAttributeName": labelAttributeName,
"LabelCategoryConfigS3Uri": label_category_config_s3uri
}
print(json.dumps(ground_truth_request, indent=4, sort_keys=True))
```
## Call CreateLabelingJob
```
sagemaker_client.create_labeling_job(**ground_truth_request)
print(f'Labeling Job Name: {job_name}')
```
## Check Status of Labeling Job
```
## call describeLabelingJob
describeLabelingJob = sagemaker_client.describe_labeling_job(
LabelingJobName=job_name
)
print(describeLabelingJob)
```
## Start Working on tasks
When you add yourself to a private work team, you recieve an email invitation to access the worker portal that looks similar to this [image](https://d2908q01vomqb2.cloudfront.net/f1f836cb4ea6efb2a0b1b99f41ad8b103eff4b59/2020/04/16/a2i-critical-documents-26.gif). Use this invitation to sign in to the protal and view your 3D point cloud annotation tasks. Tasks may take up to 10 minutes to show up the worker portal.
Once you are done working on the tasks, click **Submit**.
### View Output Data
Once you have completed all of the tasks, you can view your output data in the S3 location you specified in `OutputConfig`.
To read more about Ground Truth output data format for your task type, see [Output Data](https://docs.aws.amazon.com/sagemaker/latest/dg/sms-data-output.html#sms-output-point-cloud-object-tracking).
|
github_jupyter
|
<?xml version="1.0" encoding="UTF-8"?>
<CORSConfiguration xmlns="http://s3.amazonaws.com/doc/2006-03-01/">
<CORSRule>
<AllowedOrigin>*</AllowedOrigin>
<AllowedMethod>GET</AllowedMethod>
<AllowedMethod>HEAD</AllowedMethod>
<AllowedMethod>PUT</AllowedMethod>
<MaxAgeSeconds>3000</MaxAgeSeconds>
<ExposeHeader>Access-Control-Allow-Origin</ExposeHeader>
<AllowedHeader>*</AllowedHeader>
</CORSRule>
<CORSRule>
<AllowedOrigin>*</AllowedOrigin>
<AllowedMethod>GET</AllowedMethod>
</CORSRule>
</CORSConfiguration>
import boto3
import time
import pprint
import json
import sagemaker
from sagemaker import get_execution_role
from datetime import datetime, timezone
pp = pprint.PrettyPrinter(indent=4)
sagemaker_client = boto3.client('sagemaker')
BUCKET = ''
EXP_NAME = '' # Any valid S3 prefix.
# Make sure the bucket is in the same region as this notebook.
sess = sagemaker.session.Session()
role = sagemaker.get_execution_role()
region = boto3.session.Session().region_name
s3 = boto3.client('s3')
bucket_region = s3.head_bucket(Bucket=BUCKET)['ResponseMetadata']['HTTPHeaders']['x-amz-bucket-region']
assert bucket_region == region, "Your S3 bucket {} and this notebook need to be in the same region.".format(BUCKET)
rm -rf sample_data*
!wget https://aws-ml-blog.s3.amazonaws.com/artifacts/gt-point-cloud-demos/sample_data.zip
!unzip -o sample_data
!ls sample_data/2011_09_26/2011_09_26_drive_0005_sync/
!ls sample_data/2011_09_26/2011_09_26_drive_0005_sync/oxts/data
# transform points from lidar to global frame using lidar_extrinsic_matrix
def generate_transformed_pcd_from_point_cloud(points, lidar_extrinsic_matrix):
tps = []
for point in points:
transformed_points = np.matmul(lidar_extrinsic_matrix, np.array([point[0], point[1], point[2], 1], dtype=np.float32).reshape(4,1)).tolist()
if len(point) > 3 and point[3] is not None:
tps.append([transformed_points[0][0], transformed_points[1][0], transformed_points[2][0], point[3]])
return tps
{
"position": {
"y": -152.77584902657554,
"x": 311.21505956090624,
"z": -10.854137529636024
},
"heading": {
"qy": -0.7046155108831117,
"qx": 0.034278837280808494,
"qz": 0.7070617895701465,
"qw": -0.04904659893885366
}
}
import numpy as np
rotation = [[ 9.96714314e-01, -8.09890350e-02, 1.16333982e-03],
[ 8.09967396e-02, 9.96661051e-01, -1.03090934e-02],
[-3.24531964e-04, 1.03694477e-02, 9.99946183e-01]]
origin= [1.71104606e+00,
5.80000039e-01,
9.43144935e-01]
from scipy.spatial.transform import Rotation as R
# position is the origin
position = origin
r = R.from_matrix(np.asarray(rotation))
# heading in WCS using scipy
heading = r.as_quat()
print(f"position:{position}\nheading: {heading}")
import numpy as np
transformation = [[ 9.96714314e-01, -8.09890350e-02, 1.16333982e-03, 1.71104606e+00],
[ 8.09967396e-02, 9.96661051e-01, -1.03090934e-02, 5.80000039e-01],
[-3.24531964e-04, 1.03694477e-02, 9.99946183e-01, 9.43144935e-01],
[0, 0, 0, 1]]
transformation = np.array(transformation)
rotation = transformation[0:3, 0:3]
origin= transformation[0:3, 3]
from scipy.spatial.transform import Rotation as R
# position is the origin
position = origin
r = R.from_matrix(np.asarray(rotation))
# heading in WCS using scipy
heading = r.as_quat()
print(f"position:{position}\nheading: {heading}")
# utility to convert extrinsic matrix to pose heading quaternion and position
def convert_extrinsic_matrix_to_trans_quaternion_mat(lidar_extrinsic_transform):
position = lidar_extrinsic_transform[0:3, 3]
rot = np.linalg.inv(lidar_extrinsic_transform[0:3, 0:3])
quaternion= R.from_matrix(np.asarray(rot)).as_quat()
trans_quaternions = {
"translation": {
"x": position[0],
"y": position[1],
"z": position[2]
},
"rotation": {
"qx": quaternion[0],
"qy": quaternion[1],
"qz": quaternion[2],
"qw": quaternion[3]
}
}
return trans_quaternions
def convert_camera_inv_extrinsic_matrix_to_trans_quaternion_mat(camera_extrinsic_transform):
position = camera_extrinsic_transform[0:3, 3]
rot = np.linalg.inv(camera_extrinsic_transform[0:3, 0:3])
quaternion= R.from_matrix(np.asarray(rot)).as_quat()
trans_quaternions = {
"translation": {
"x": position[0],
"y": position[1],
"z": position[2]
},
"rotation": {
"qx": quaternion[0],
"qy": quaternion[1],
"qz": quaternion[2],
"qw": -quaternion[3]
}
}
return trans_quaternions
!wget https://aws-ml-blog.s3.amazonaws.com/artifacts/gt-point-cloud-demos/kitti2gt.py
!pygmentize kitti2gt.py
!pip install pykitti
!pip install --upgrade scipy
from kitti2gt import *
if(EXP_NAME == ''):
s3loc = f's3://{BUCKET}/frames/'
else:
s3loc = f's3://{BUCKET}/{EXP_NAME}/frames/'
convert_to_gt(basedir='sample_data',
date='2011_09_26',
drive='0005',
output_base='sample_data_out',
s3prefix = s3loc)
!ls sample_data_out/
!ls sample_data_out/frames
if(EXP_NAME == ''):
!aws s3 cp sample_data_out/kitti-gt-seq.json s3://{BUCKET}/
else:
!aws s3 cp sample_data_out/kitti-gt-seq.json s3://{BUCKET}/{EXP_NAME}/
if(EXP_NAME == ''):
!aws s3 sync sample_data_out/frames/ s3://{BUCKET}/frames/
else:
!aws s3 sync sample_data_out/frames s3://{BUCKET}/{EXP_NAME}/frames/
if(EXP_NAME == ''):
!aws s3 sync sample_data_out/images/ s3://{BUCKET}/frames/images/
else:
!aws s3 sync sample_data_out/images s3://{BUCKET}/{EXP_NAME}/frames/images/
with open('manifest.json','w') as f:
if(EXP_NAME == ''):
json.dump({"source-ref": "s3://{}/kitti-gt-seq.json".format(BUCKET)},f)
else:
json.dump({"source-ref": "s3://{}/{}/kitti-gt-seq.json".format(BUCKET,EXP_NAME)},f)
!cat manifest.json
if(EXP_NAME == ''):
!aws s3 cp manifest.json s3://{BUCKET}/
input_manifest_s3uri = f's3://{BUCKET}/manifest.json'
else:
!aws s3 cp manifest.json s3://{BUCKET}/{EXP_NAME}/
input_manifest_s3uri = f's3://{BUCKET}/{EXP_NAME}/manifest.json'
input_manifest_s3uri
task_type = "3DPointCloudObjectTracking"
## Set up human_task_ui_arn map, to be used in case you chose UI_CONFIG_USE_TASK_UI_ARN
## Supported for GA
## Set up human_task_ui_arn map, to be used in case you chose UI_CONFIG_USE_TASK_UI_ARN
human_task_ui_arn = f'arn:aws:sagemaker:{region}:394669845002:human-task-ui/PointCloudObjectTracking'
human_task_ui_arn
!wget https://aws-ml-blog.s3.amazonaws.com/artifacts/gt-point-cloud-demos/label-category-config/label-category.json
with open('label-category.json', 'r') as j:
json_data = json.load(j)
print("\nA label category configuration file: \n\n",json.dumps(json_data, indent=4, sort_keys=True))
if(EXP_NAME == ''):
!aws s3 cp label-category.json s3://{BUCKET}/label-category.json
label_category_config_s3uri = f's3://{BUCKET}/label-category.json'
else:
!aws s3 cp label-category.json s3://{BUCKET}/{EXP_NAME}/label-category.json
label_category_config_s3uri = f's3://{BUCKET}/{EXP_NAME}/label-category.json'
label_category_config_s3uri
##Use Beta Private Team till GA
workteam_arn = ''
#See your execution role ARN. The role name is located at the end of the ARN.
role
ac_arn_map = {'us-west-2': '081040173940',
'us-east-1': '432418664414',
'us-east-2': '266458841044',
'eu-west-1': '568282634449',
'ap-northeast-1': '477331159723'}
prehuman_arn = 'arn:aws:lambda:{}:{}:function:PRE-{}'.format(region, ac_arn_map[region],task_type)
acs_arn = 'arn:aws:lambda:{}:{}:function:ACS-{}'.format(region, ac_arn_map[region],task_type)
from datetime import datetime
## Set up Human Task Config
## Modify the following
task_description = 'add a task description here'
#example keywords
task_keywords = ['lidar', 'pointcloud']
#add a task title
task_title = 'Add a Task Title Here - This is Displayed to Workers'
#add a job name to identify your labeling job
job_name = 'add-job-name'
human_task_config = {
"AnnotationConsolidationConfig": {
"AnnotationConsolidationLambdaArn": acs_arn,
},
"UiConfig": {
"HumanTaskUiArn": human_task_ui_arn,
},
"WorkteamArn": workteam_arn,
"PreHumanTaskLambdaArn": prehuman_arn,
"MaxConcurrentTaskCount": 200, # 200 images will be sent at a time to the workteam.
"NumberOfHumanWorkersPerDataObject": 1, # One worker will work on each task
"TaskAvailabilityLifetimeInSeconds": 18000, # Your workteam has 5 hours to complete all pending tasks.
"TaskDescription": task_description,
"TaskKeywords": task_keywords,
"TaskTimeLimitInSeconds": 3600, # Each seq must be labeled within 1 hour.
"TaskTitle": task_title
}
print(json.dumps(human_task_config, indent=4, sort_keys=True))
if(EXP_NAME == ''):
s3_output_path = f's3://{BUCKET}'
else:
s3_output_path = f's3://{BUCKET}/{EXP_NAME}'
s3_output_path
## Set up Create Labeling Request
labelAttributeName = job_name + "-ref"
if task_type == "3DPointCloudObjectDetection" or task_type == "Adjustment3DPointCloudObjectDetection":
labelAttributeName = job_name
ground_truth_request = {
"InputConfig" : {
"DataSource": {
"S3DataSource": {
"ManifestS3Uri": input_manifest_s3uri,
}
},
"DataAttributes": {
"ContentClassifiers": [
"FreeOfPersonallyIdentifiableInformation",
"FreeOfAdultContent"
]
},
},
"OutputConfig" : {
"S3OutputPath": s3_output_path,
},
"HumanTaskConfig" : human_task_config,
"LabelingJobName": job_name,
"RoleArn": role,
"LabelAttributeName": labelAttributeName,
"LabelCategoryConfigS3Uri": label_category_config_s3uri
}
print(json.dumps(ground_truth_request, indent=4, sort_keys=True))
sagemaker_client.create_labeling_job(**ground_truth_request)
print(f'Labeling Job Name: {job_name}')
## call describeLabelingJob
describeLabelingJob = sagemaker_client.describe_labeling_job(
LabelingJobName=job_name
)
print(describeLabelingJob)
| 0.541409 | 0.982971 |
```
import json
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
base = '/Users/chuamelia/Google Drive/Spring 2020/Machine Learning/fake-review-detection-project/data/processed/dev/'
def readJsonFile(fname, base=base):
with open(base + fname + '.json', 'r') as f:
data = json.load(f)
return data
fname = 'sgd_attempts_ac4119_202005117'
all_attempts = readJsonFile(fname)
fname = 'sgd_attempts_ac4119_202005119b'
all_attempts_new = readJsonFile(fname)
eval_metrics_df
eval_metrics_df = pd.json_normalize(all_attempts)
eval_metrics_df_new = pd.json_normalize(all_attempts_new)
vs_dev_set = eval_metrics_df[eval_metrics_df['dev_set'] == 'ac4119_dev_w_tokens']
eval_metrics_df = pd.concat([vs_dev_set, eval_metrics_df_new])
```
Best Performing Overall
```
best_ap_score = max(eval_metrics_df['metrics.test_ap_score'])
best_auc_score = max(eval_metrics_df['metrics.test_auc_score'])
eval_metrics_df[eval_metrics_df['metrics.test_auc_score'] == best_auc_score]
eval_metrics_df[eval_metrics_df['metrics.test_ap_score'] == best_ap_score]
vs_dev_set = eval_metrics_df[eval_metrics_df['dev_set'] == 'ac4119_dev_w_tokens']
best_ap_score = max(vs_dev_set['metrics.test_ap_score'])
best_auc_score = max(vs_dev_set['metrics.test_auc_score'])
vs_dev_set[vs_dev_set['metrics.test_auc_score'] == best_auc_score]
vs_dev_set[vs_dev_set['metrics.test_ap_score'] == best_ap_score]
Hey guys, lmk if you guys think you are done with you implementations. One thing I don't think any of us have tried yet, it to incorporate the
vs_dev_set.sort_values(by=['metrics.test_auc_score'], ascending=False)
fname = 'l1_attempts_ac4119_20200516'
l1_attempts = readJsonFile(fname)
eval_metrics_df = pd.json_normalize(l1_attempts)
vs_dev_set = eval_metrics_df[eval_metrics_df['dev_set'] == 'ac4119_dev_w_tokens']
best_ap_score = max(vs_dev_set['metrics.test_ap_score'])
best_auc_score = max(vs_dev_set['metrics.test_auc_score'])
vs_dev_set[vs_dev_set['metrics.test_auc_score'] == best_auc_score]
fname = 'sgd_attempts_ac4119_202005117'
l1_attempts = readJsonFile(fname)
eval_metrics_df = pd.json_normalize(l1_attempts)
vs_dev_set = eval_metrics_df[eval_metrics_df['dev_set'] == 'ac4119_dev_w_tokens']
best_ap_score = max(vs_dev_set['metrics.test_ap_score'])
best_auc_score = max(vs_dev_set['metrics.test_auc_score'])
vs_dev_set[vs_dev_set['metrics.test_auc_score'] == best_auc_score]
best_auc_score = max(eval_metrics_df['metrics.test_auc_score'])
eval_metrics_df[eval_metrics_df['metrics.test_auc_score'] == best_auc_score]
vs_dev_set.sort_values(by=['metrics.test_auc_score'], ascending=False).head(100)
l1_attempts[1147]
all_attempts = [{"params": {"n_neighbors": 10}, "metrics": {"train_accuracy": 0.873087685451661, "test_accuracy": 0.8415279247174119, "test_auc_pred": 0.511729185694714, "test_auc_score": 0.5310443356900929, "test_ap_pred": 0.10429276470132186, "test_ap_score": 0.11094622963614453}}, {"params": {"n_neighbors": 11}, "metrics": {"train_accuracy": 0.8520093752242162, "test_accuracy": 0.8107355643409989, "test_auc_pred": 0.5110040896710323, "test_auc_score": 0.5303059115386078, "test_ap_pred": 0.10391791154049448, "test_ap_score": 0.11035856176038553}}, {"params": {"n_neighbors": 13}, "metrics": {"train_accuracy": 0.8543810837312754, "test_accuracy": 0.8203964586001448, "test_auc_pred": 0.5106669449518589, "test_auc_score": 0.5321886561237693, "test_ap_pred": 0.10387427935448956, "test_ap_score": 0.11096606083238872}}, {"params": {"n_neighbors": 14}, "metrics": {"train_accuracy": 0.8718958521010547, "test_accuracy": 0.8494626649590734, "test_auc_pred": 0.509094157913765, "test_auc_score": 0.5305561303611523, "test_ap_pred": 0.10363763504514066, "test_ap_score": 0.11034996607735038}}, {"params": {"n_neighbors": 15}, "metrics": {"train_accuracy": 0.8562545341486164, "test_accuracy": 0.8282476752603152, "test_auc_pred": 0.5098089414153606, "test_auc_score": 0.5308391938020214, "test_ap_pred": 0.10369562558104857, "test_ap_score": 0.11053444304133936}}, {"params": {"n_neighbors": 20}, "metrics": {"train_accuracy": 0.8732630722992418, "test_accuracy": 0.858037752658834, "test_auc_pred": 0.505478259776339, "test_auc_score": 0.5333016482366436, "test_ap_pred": 0.10274942473584053, "test_ap_score": 0.11007725822596078}}, {"params": {"n_neighbors": 5}, "metrics": {"train_accuracy": 0.8522286087836922, "test_accuracy": 0.7633776936355031, "test_auc_pred": 0.5149185582584443, "test_auc_score": 0.5241492296698905, "test_ap_pred": 0.10470122635315257, "test_ap_score": 0.10815341284068077}}]
eval_metrics_df = pd.json_normalize(all_attempts)
eval_metrics_df
```
|
github_jupyter
|
import json
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
base = '/Users/chuamelia/Google Drive/Spring 2020/Machine Learning/fake-review-detection-project/data/processed/dev/'
def readJsonFile(fname, base=base):
with open(base + fname + '.json', 'r') as f:
data = json.load(f)
return data
fname = 'sgd_attempts_ac4119_202005117'
all_attempts = readJsonFile(fname)
fname = 'sgd_attempts_ac4119_202005119b'
all_attempts_new = readJsonFile(fname)
eval_metrics_df
eval_metrics_df = pd.json_normalize(all_attempts)
eval_metrics_df_new = pd.json_normalize(all_attempts_new)
vs_dev_set = eval_metrics_df[eval_metrics_df['dev_set'] == 'ac4119_dev_w_tokens']
eval_metrics_df = pd.concat([vs_dev_set, eval_metrics_df_new])
best_ap_score = max(eval_metrics_df['metrics.test_ap_score'])
best_auc_score = max(eval_metrics_df['metrics.test_auc_score'])
eval_metrics_df[eval_metrics_df['metrics.test_auc_score'] == best_auc_score]
eval_metrics_df[eval_metrics_df['metrics.test_ap_score'] == best_ap_score]
vs_dev_set = eval_metrics_df[eval_metrics_df['dev_set'] == 'ac4119_dev_w_tokens']
best_ap_score = max(vs_dev_set['metrics.test_ap_score'])
best_auc_score = max(vs_dev_set['metrics.test_auc_score'])
vs_dev_set[vs_dev_set['metrics.test_auc_score'] == best_auc_score]
vs_dev_set[vs_dev_set['metrics.test_ap_score'] == best_ap_score]
Hey guys, lmk if you guys think you are done with you implementations. One thing I don't think any of us have tried yet, it to incorporate the
vs_dev_set.sort_values(by=['metrics.test_auc_score'], ascending=False)
fname = 'l1_attempts_ac4119_20200516'
l1_attempts = readJsonFile(fname)
eval_metrics_df = pd.json_normalize(l1_attempts)
vs_dev_set = eval_metrics_df[eval_metrics_df['dev_set'] == 'ac4119_dev_w_tokens']
best_ap_score = max(vs_dev_set['metrics.test_ap_score'])
best_auc_score = max(vs_dev_set['metrics.test_auc_score'])
vs_dev_set[vs_dev_set['metrics.test_auc_score'] == best_auc_score]
fname = 'sgd_attempts_ac4119_202005117'
l1_attempts = readJsonFile(fname)
eval_metrics_df = pd.json_normalize(l1_attempts)
vs_dev_set = eval_metrics_df[eval_metrics_df['dev_set'] == 'ac4119_dev_w_tokens']
best_ap_score = max(vs_dev_set['metrics.test_ap_score'])
best_auc_score = max(vs_dev_set['metrics.test_auc_score'])
vs_dev_set[vs_dev_set['metrics.test_auc_score'] == best_auc_score]
best_auc_score = max(eval_metrics_df['metrics.test_auc_score'])
eval_metrics_df[eval_metrics_df['metrics.test_auc_score'] == best_auc_score]
vs_dev_set.sort_values(by=['metrics.test_auc_score'], ascending=False).head(100)
l1_attempts[1147]
all_attempts = [{"params": {"n_neighbors": 10}, "metrics": {"train_accuracy": 0.873087685451661, "test_accuracy": 0.8415279247174119, "test_auc_pred": 0.511729185694714, "test_auc_score": 0.5310443356900929, "test_ap_pred": 0.10429276470132186, "test_ap_score": 0.11094622963614453}}, {"params": {"n_neighbors": 11}, "metrics": {"train_accuracy": 0.8520093752242162, "test_accuracy": 0.8107355643409989, "test_auc_pred": 0.5110040896710323, "test_auc_score": 0.5303059115386078, "test_ap_pred": 0.10391791154049448, "test_ap_score": 0.11035856176038553}}, {"params": {"n_neighbors": 13}, "metrics": {"train_accuracy": 0.8543810837312754, "test_accuracy": 0.8203964586001448, "test_auc_pred": 0.5106669449518589, "test_auc_score": 0.5321886561237693, "test_ap_pred": 0.10387427935448956, "test_ap_score": 0.11096606083238872}}, {"params": {"n_neighbors": 14}, "metrics": {"train_accuracy": 0.8718958521010547, "test_accuracy": 0.8494626649590734, "test_auc_pred": 0.509094157913765, "test_auc_score": 0.5305561303611523, "test_ap_pred": 0.10363763504514066, "test_ap_score": 0.11034996607735038}}, {"params": {"n_neighbors": 15}, "metrics": {"train_accuracy": 0.8562545341486164, "test_accuracy": 0.8282476752603152, "test_auc_pred": 0.5098089414153606, "test_auc_score": 0.5308391938020214, "test_ap_pred": 0.10369562558104857, "test_ap_score": 0.11053444304133936}}, {"params": {"n_neighbors": 20}, "metrics": {"train_accuracy": 0.8732630722992418, "test_accuracy": 0.858037752658834, "test_auc_pred": 0.505478259776339, "test_auc_score": 0.5333016482366436, "test_ap_pred": 0.10274942473584053, "test_ap_score": 0.11007725822596078}}, {"params": {"n_neighbors": 5}, "metrics": {"train_accuracy": 0.8522286087836922, "test_accuracy": 0.7633776936355031, "test_auc_pred": 0.5149185582584443, "test_auc_score": 0.5241492296698905, "test_ap_pred": 0.10470122635315257, "test_ap_score": 0.10815341284068077}}]
eval_metrics_df = pd.json_normalize(all_attempts)
eval_metrics_df
| 0.25303 | 0.443359 |
# Hepstat_Tutorial_Genhists
Generate some histograms
* Signal samples with different mass points, assuming Gaussian shapes
* Background samples: nominal and up/down variations, a simple ploynomial
**Author:** Lailin XU
<i><small>This notebook tutorial was automatically generated with <a href= "https://github.com/root-project/root/blob/master/documentation/doxygen/converttonotebook.py">ROOTBOOK-izer</a> from the macro found in the ROOT repository on Monday, April 05, 2021 at 10:55 PM.</small></i>
```
import os
```
Import the ROOT libraries
```
import ROOT as R
from math import pow, sqrt
R.gROOT.SetStyle("ATLAS")
odir = "data"
if not os.path.isdir(odir): os.makedirs(odir)
tfout = R.TFile("data/h4l_toy_hists.root", "RECREATE")
```
Signal samples
----------------
Number of toy events to obtain the signal/bkg distributions (only to get the shape)
```
nbins = 25
nevents = 100000
```
Number of toy events for the background
```
nbkg_exp = 100
mHs = [120., 125., 130., 135., 140., 145., 150.]
```
Gaussian
```
for mH in mHs:
x = R.RooRealVar("x", "x", 110, 160)
mean = R.RooRealVar("mean", "mean of gaussians", mH)
wH = mH*0.01
sigma = R.RooRealVar("sigma", "width of gaussians", wH)
sig = R.RooGaussian("sig", "Signal", x, mean, sigma)
# Generate pseudo data via sampling
data = sig.generate(R.RooArgSet(x), nevents)
x.setBins(nbins)
hname = "sig_{:d}".format(int(mH))
dh = R.RooDataHist(hname, hname, R.RooArgSet(x), data).createHistogram(hname, x)
dh.Scale(1./(dh.Integral()))
dh.SetName(hname)
tfout.cd()
dh.Write()
```
Nominal bkg
----------------
```
x = R.RooRealVar("x", "x", 110, 160)
p0 = 480.
p1 = -2.5
a0 = R.RooRealVar("a0", "a0", p0)
a1 = R.RooRealVar("a1", "a1", p1)
bkg = R.RooPolynomial("bkg", "Background", x, R.RooArgList(a0, a1))
```
Generate pseudo data via sampling
```
data = bkg.generate(R.RooArgSet(x), nevents)
x.setBins(nbins)
hname = "bkg"
dh = R.RooDataHist(hname, hname, R.RooArgSet(x), data).createHistogram(hname, x)
nint = dh.Integral()
dh.Scale(nbkg_exp/nint)
dh.SetName(hname)
```
Toy observed data
----------------
```
data_obs = bkg.generate(R.RooArgSet(x), nbkg_exp)
x.setBins(nbins)
hname = "obsData"
dh_obs = R.RooDataHist(hname, hname, R.RooArgSet(x), data_obs).createHistogram(hname, x)
dh_obs.SetName(hname)
```
Background variations
----------------
Variation up
```
a0 = R.RooRealVar("a0", "a0", p0*1.02)
a1 = R.RooRealVar("a1", "a1", p1*0.99)
bkg = R.RooPolynomial("bkg_up", "Background", x, R.RooArgList(a0, a1))
```
Generate pseudo data via sampling
```
data = bkg.generate(R.RooArgSet(x), nevents)
x.setBins(nbins)
hname = "bkg_up"
dh_up = R.RooDataHist(hname, hname, R.RooArgSet(x), data).createHistogram(hname, x)
dh_up.Scale(nbkg_exp/nint)
dh_up.SetName(hname)
```
Variation up
```
a0 = R.RooRealVar("a0", "a0", p0*0.98)
a1 = R.RooRealVar("a1", "a1", p1*1.01)
bkg = R.RooPolynomial("bkg_dn", "Background", x, R.RooArgList(a0, a1))
```
Generate pseudo data via sampling
```
data = bkg.generate(R.RooArgSet(x), nevents)
x.setBins(nbins)
hname = "bkg_dn"
dh_dn = R.RooDataHist(hname, hname, R.RooArgSet(x), data).createHistogram(hname, x)
dh_dn.Scale(nbkg_exp/nint)
dh_dn.SetName(hname)
tfout.cd()
dh_obs.Write()
dh.Write()
dh_up.Write()
dh_dn.Write()
tfout.Close()
```
Draw all canvases
```
from ROOT import gROOT
gROOT.GetListOfCanvases().Draw()
```
|
github_jupyter
|
import os
import ROOT as R
from math import pow, sqrt
R.gROOT.SetStyle("ATLAS")
odir = "data"
if not os.path.isdir(odir): os.makedirs(odir)
tfout = R.TFile("data/h4l_toy_hists.root", "RECREATE")
nbins = 25
nevents = 100000
nbkg_exp = 100
mHs = [120., 125., 130., 135., 140., 145., 150.]
for mH in mHs:
x = R.RooRealVar("x", "x", 110, 160)
mean = R.RooRealVar("mean", "mean of gaussians", mH)
wH = mH*0.01
sigma = R.RooRealVar("sigma", "width of gaussians", wH)
sig = R.RooGaussian("sig", "Signal", x, mean, sigma)
# Generate pseudo data via sampling
data = sig.generate(R.RooArgSet(x), nevents)
x.setBins(nbins)
hname = "sig_{:d}".format(int(mH))
dh = R.RooDataHist(hname, hname, R.RooArgSet(x), data).createHistogram(hname, x)
dh.Scale(1./(dh.Integral()))
dh.SetName(hname)
tfout.cd()
dh.Write()
x = R.RooRealVar("x", "x", 110, 160)
p0 = 480.
p1 = -2.5
a0 = R.RooRealVar("a0", "a0", p0)
a1 = R.RooRealVar("a1", "a1", p1)
bkg = R.RooPolynomial("bkg", "Background", x, R.RooArgList(a0, a1))
data = bkg.generate(R.RooArgSet(x), nevents)
x.setBins(nbins)
hname = "bkg"
dh = R.RooDataHist(hname, hname, R.RooArgSet(x), data).createHistogram(hname, x)
nint = dh.Integral()
dh.Scale(nbkg_exp/nint)
dh.SetName(hname)
data_obs = bkg.generate(R.RooArgSet(x), nbkg_exp)
x.setBins(nbins)
hname = "obsData"
dh_obs = R.RooDataHist(hname, hname, R.RooArgSet(x), data_obs).createHistogram(hname, x)
dh_obs.SetName(hname)
a0 = R.RooRealVar("a0", "a0", p0*1.02)
a1 = R.RooRealVar("a1", "a1", p1*0.99)
bkg = R.RooPolynomial("bkg_up", "Background", x, R.RooArgList(a0, a1))
data = bkg.generate(R.RooArgSet(x), nevents)
x.setBins(nbins)
hname = "bkg_up"
dh_up = R.RooDataHist(hname, hname, R.RooArgSet(x), data).createHistogram(hname, x)
dh_up.Scale(nbkg_exp/nint)
dh_up.SetName(hname)
a0 = R.RooRealVar("a0", "a0", p0*0.98)
a1 = R.RooRealVar("a1", "a1", p1*1.01)
bkg = R.RooPolynomial("bkg_dn", "Background", x, R.RooArgList(a0, a1))
data = bkg.generate(R.RooArgSet(x), nevents)
x.setBins(nbins)
hname = "bkg_dn"
dh_dn = R.RooDataHist(hname, hname, R.RooArgSet(x), data).createHistogram(hname, x)
dh_dn.Scale(nbkg_exp/nint)
dh_dn.SetName(hname)
tfout.cd()
dh_obs.Write()
dh.Write()
dh_up.Write()
dh_dn.Write()
tfout.Close()
from ROOT import gROOT
gROOT.GetListOfCanvases().Draw()
| 0.325413 | 0.937954 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.