code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
# The framework and why do we need it
In the previous notebooks, we introduce some concepts regarding the
evaluation of predictive models. While this section could be slightly
redundant, we intend to go into details into the cross-validation framework.
Before we dive in, let's linger on the reasons for always having training and
testing sets. Let's first look at the limitation of using a dataset without
keeping any samples out.
To illustrate the different concepts, we will use the California housing
dataset.
```
from sklearn.datasets import fetch_california_housing
housing = fetch_california_housing(as_frame=True)
data, target = housing.data, housing.target
```
<div class="admonition caution alert alert-warning">
<p class="first admonition-title" style="font-weight: bold;">Caution!</p>
<p class="last">Here and later, we use the name <tt class="docutils literal">data</tt> and <tt class="docutils literal">target</tt> to be explicit. In
scikit-learn documentation, <tt class="docutils literal">data</tt> is commonly named <tt class="docutils literal">X</tt> and <tt class="docutils literal">target</tt> is
commonly called <tt class="docutils literal">y</tt>.</p>
</div>
In this dataset, the aim is to predict the median value of houses in an area
in California. The features collected are based on general real-estate and
geographical information.
Therefore, the task to solve is different from the one shown in the previous
notebook. The target to be predicted is a continuous variable and not anymore
discrete. This task is called regression.
Therefore, we will use predictive model specific to regression and not to
classification.
```
print(housing.DESCR)
data.head()
```
To simplify future visualization, let's transform the prices from the
dollar (\\$) range to the thousand dollars (k\\$) range.
```
target *= 100
target.head()
```
<div class="admonition note alert alert-info">
<p class="first admonition-title" style="font-weight: bold;">Note</p>
<p class="last">If you want a deeper overview regarding this dataset, you can refer to the
Appendix - Datasets description section at the end of this MOOC.</p>
</div>
## Training error vs testing error
To solve this regression task, we will use a decision tree regressor.
```
from sklearn.tree import DecisionTreeRegressor
regressor = DecisionTreeRegressor(random_state=0)
regressor.fit(data, target)
```
After training the regressor, we would like to know its potential statistical
performance once deployed in production. For this purpose, we use the mean
absolute error, which gives us an error in the native unit, i.e. k\\$.
```
from sklearn.metrics import mean_absolute_error
target_predicted = regressor.predict(data)
score = mean_absolute_error(target, target_predicted)
print(f"On average, our regressor makes an error of {score:.2f} k$")
```
We get perfect prediction with no error. It is too optimistic and almost
always revealing a methodological problem when doing machine learning.
Indeed, we trained and predicted on the same dataset. Since our decision tree
was fully grown, every sample in the dataset is stored in a leaf node.
Therefore, our decision tree fully memorized the dataset given during `fit`
and therefore made no error when predicting.
This error computed above is called the **empirical error** or **training
error**.
<div class="admonition note alert alert-info">
<p class="first admonition-title" style="font-weight: bold;">Note</p>
<p class="last">In this MOOC, we will consistently use the term "training error".</p>
</div>
We trained a predictive model to minimize the training error but our aim is
to minimize the error on data that has not been seen during training.
This error is also called the **generalization error** or the "true"
**testing error**.
<div class="admonition note alert alert-info">
<p class="first admonition-title" style="font-weight: bold;">Note</p>
<p class="last">In this MOOC, we will consistently use the term "testing error".</p>
</div>
Thus, the most basic evaluation involves:
* splitting our dataset into two subsets: a training set and a testing set;
* fitting the model on the training set;
* estimating the training error on the training set;
* estimating the testing error on the testing set.
So let's split our dataset.
```
from sklearn.model_selection import train_test_split
data_train, data_test, target_train, target_test = train_test_split(
data, target, random_state=0)
```
Then, let's train our model.
```
regressor.fit(data_train, target_train)
```
Finally, we estimate the different types of errors. Let's start by computing
the training error.
```
target_predicted = regressor.predict(data_train)
score = mean_absolute_error(target_train, target_predicted)
print(f"The training error of our model is {score:.2f} k$")
```
We observe the same phenomena as in the previous experiment: our model
memorized the training set. However, we now compute the testing error.
```
target_predicted = regressor.predict(data_test)
score = mean_absolute_error(target_test, target_predicted)
print(f"The testing error of our model is {score:.2f} k$")
```
This testing error is actually about what we would expect from our model if
it was used in a production environment.
## Stability of the cross-validation estimates
When doing a single train-test split we don't give any indication regarding
the robustness of the evaluation of our predictive model: in particular, if
the test set is small, this estimate of the testing error will be
unstable and wouldn't reflect the "true error rate" we would have observed
with the same model on an unlimited amount of test data.
For instance, we could have been lucky when we did our random split of our
limited dataset and isolated some of the easiest cases to predict in the
testing set just by chance: the estimation of the testing error would be
overly optimistic, in this case.
**Cross-validation** allows estimating the robustness of a predictive model
by repeating the splitting procedure. It will give several training and
testing errors and thus some **estimate of the variability of the
model statistical performance**.
There are different cross-validation strategies, for now we are going to
focus on one called "shuffle-split". At each iteration of this strategy we:
- randomly shuffle the order of the samples of a copy of the full dataset;
- split the shuffled dataset into a train and a test set;
- train a new model on the train set;
- evaluate the testing error on the test set.
We repeat this procedure `n_splits` times. Using `n_splits=40` means that we
will train 40 models in total and all of them will be discarded: we just
record their statistical performance on each variant of the test set.
To evaluate the statistical performance of our regressor, we can use
`cross_validate` with a `ShuffleSplit` object:
```
from sklearn.model_selection import cross_validate
from sklearn.model_selection import ShuffleSplit
cv = ShuffleSplit(n_splits=40, test_size=0.3, random_state=0)
cv_results = cross_validate(
regressor, data, target, cv=cv, scoring="neg_mean_absolute_error")
```
The results `cv_results` are stored into a Python dictionary. We will convert
it into a pandas dataframe to ease visualization and manipulation.
```
import pandas as pd
cv_results = pd.DataFrame(cv_results)
cv_results.head()
```
<div class="admonition tip alert alert-warning">
<p class="first admonition-title" style="font-weight: bold;">Tip</p>
<p>A score is a metric for which higher values mean better results. On the
contrary, an error is a metric for which lower values mean better results.
The parameter <tt class="docutils literal">scoring</tt> in <tt class="docutils literal">cross_validate</tt> always expect a function that is
a score.</p>
<p class="last">To make it easy, all error metrics in scikit-learn, like
<tt class="docutils literal">mean_absolute_error</tt>, can be transformed into a score to be used in
<tt class="docutils literal">cross_validate</tt>. To do so, you need to pass a string of the error metric
with an additional <tt class="docutils literal">neg_</tt> string at the front to the parameter <tt class="docutils literal">scoring</tt>;
for instance <tt class="docutils literal"><span class="pre">scoring="neg_mean_absolute_error"</span></tt>. In this case, the negative
of the mean absolute error will be computed which would be equivalent to a
score.</p>
</div>
Let us revert the negation to get the actual error:
```
cv_results["test_error"] = -cv_results["test_score"]
```
Let's check the results reported by the cross-validation.
```
cv_results.head(10)
```
We get timing information to fit and predict at each round of
cross-validation. Also, we get the test score, which corresponds to the
testing error on each of the split.
```
len(cv_results)
```
We get 40 entries in our resulting dataframe because we performed 40
splits. Therefore, we can show the testing error distribution and thus, have
an estimate of its variability.
```
import matplotlib.pyplot as plt
cv_results["test_error"].plot.hist(bins=10, edgecolor="black", density=True)
plt.xlabel("Mean absolute error (k$)")
_ = plt.title("Test error distribution")
```
We observe that the testing error is clustered around 47 k\\$ and
ranges from 43 k\\$ to 50 k\\$.
```
print(f"The mean cross-validated testing error is: "
f"{cv_results['test_error'].mean():.2f} k$")
print(f"The standard deviation of the testing error is: "
f"{cv_results['test_error'].std():.2f} k$")
```
Note that the standard deviation is much smaller than the mean: we could
summarize that our cross-validation estimate of the testing error is
46.36 +/- 1.17 k\\$.
If we were to train a single model on the full dataset (without
cross-validation) and then had later access to an unlimited amount of test
data, we would expect its true testing error to fall close to that
region.
While this information is interesting in itself, it should be contrasted to
the scale of the natural variability of the vector `target` in our dataset.
Let us plot the distribution of the target variable:
```
target.plot.hist(bins=20, edgecolor="black", density=True)
plt.xlabel("Median House Value (k$)")
_ = plt.title("Target distribution")
print(f"The standard deviation of the target is: {target.std():.2f} k$")
```
The target variable ranges from close to 0 k\\$ up to 500 k\\$ and, with a
standard deviation around 115 k\\$.
We notice that the mean estimate of the testing error obtained by
cross-validation is a bit smaller than the natural scale of variation of the
target variable. Furthermore the standard deviation of the cross validation
estimate of the testing error is even smaller.
This is a good start, but not necessarily enough to decide whether the
generalization performance is good enough to make our prediction useful in
practice.
We recall that our model makes, on average, an error around 47 k\\$. With this
information and looking at the target distribution, such an error might be
acceptable when predicting houses with a 500 k\\$. However, it would be an
issue with a house with a value of 50 k\\$. Thus, this indicates that our
metric (Mean Absolute Error) is not ideal.
We might instead choose a metric relative to the target value to predict: the
mean absolute percentage error would have been a much better choice.
But in all cases, an error of 47 k\\$ might be too large to automatically use
our model to tag house value without expert supervision.
## More detail regarding `cross_validate`
During cross-validation, many models are trained and evaluated. Indeed, the
number of elements in each array of the output of `cross_validate` is a
result from one of this `fit`/`score`. To make it explicit, it is possible
to retrieve theses fitted models for each of the fold by passing the option
`return_estimator=True` in `cross_validate`.
```
cv_results = cross_validate(regressor, data, target, return_estimator=True)
cv_results
cv_results["estimator"]
```
The five decision tree regressors corresponds to the five fitted decision
trees on the different folds. Having access to these regressors is handy
because it allows to inspect the internal fitted parameters of these
regressors.
In the case where you are interested only about the test score, scikit-learn
provide a `cross_val_score` function. It is identical to calling the
`cross_validate` function and to select the `test_score` only (as we
extensively did in the previous notebooks).
```
from sklearn.model_selection import cross_val_score
scores = cross_val_score(regressor, data, target)
scores
```
## Summary
In this notebook, we saw:
* the necessity of splitting the data into a train and test set;
* the meaning of the training and testing errors;
* the overall cross-validation framework with the possibility to study
statistical performance variations;
|
github_jupyter
|
from sklearn.datasets import fetch_california_housing
housing = fetch_california_housing(as_frame=True)
data, target = housing.data, housing.target
print(housing.DESCR)
data.head()
target *= 100
target.head()
from sklearn.tree import DecisionTreeRegressor
regressor = DecisionTreeRegressor(random_state=0)
regressor.fit(data, target)
from sklearn.metrics import mean_absolute_error
target_predicted = regressor.predict(data)
score = mean_absolute_error(target, target_predicted)
print(f"On average, our regressor makes an error of {score:.2f} k$")
from sklearn.model_selection import train_test_split
data_train, data_test, target_train, target_test = train_test_split(
data, target, random_state=0)
regressor.fit(data_train, target_train)
target_predicted = regressor.predict(data_train)
score = mean_absolute_error(target_train, target_predicted)
print(f"The training error of our model is {score:.2f} k$")
target_predicted = regressor.predict(data_test)
score = mean_absolute_error(target_test, target_predicted)
print(f"The testing error of our model is {score:.2f} k$")
from sklearn.model_selection import cross_validate
from sklearn.model_selection import ShuffleSplit
cv = ShuffleSplit(n_splits=40, test_size=0.3, random_state=0)
cv_results = cross_validate(
regressor, data, target, cv=cv, scoring="neg_mean_absolute_error")
import pandas as pd
cv_results = pd.DataFrame(cv_results)
cv_results.head()
cv_results["test_error"] = -cv_results["test_score"]
cv_results.head(10)
len(cv_results)
import matplotlib.pyplot as plt
cv_results["test_error"].plot.hist(bins=10, edgecolor="black", density=True)
plt.xlabel("Mean absolute error (k$)")
_ = plt.title("Test error distribution")
print(f"The mean cross-validated testing error is: "
f"{cv_results['test_error'].mean():.2f} k$")
print(f"The standard deviation of the testing error is: "
f"{cv_results['test_error'].std():.2f} k$")
target.plot.hist(bins=20, edgecolor="black", density=True)
plt.xlabel("Median House Value (k$)")
_ = plt.title("Target distribution")
print(f"The standard deviation of the target is: {target.std():.2f} k$")
cv_results = cross_validate(regressor, data, target, return_estimator=True)
cv_results
cv_results["estimator"]
from sklearn.model_selection import cross_val_score
scores = cross_val_score(regressor, data, target)
scores
| 0.879639 | 0.987794 |
```
import numpy as np
```
# Exceptions
An exception is an event, which occurs during the execution of a program, that disrupts the normal flow of the program's instructions.
You've already seen some exceptions in the **Debugging** lesson.
*
Many programs want to know about exceptions when they occur. For example, if the input to a program is a file path. If the user inputs an invalid or non-existent path, the program generates an exception. It may be desired to provide a response to the user in this case.
It may also be that programs will *generate* exceptions. This is a way of indicating that there is an error in the inputs provided. In general, this is the preferred style for dealing with invalid inputs or states inside a python function rather than having an error return.
## Catching Exceptions
Python provides a way to detect when an exception occurs. This is done by the use of a block of code surrounded by a "try" and "except" statement.
```
def divide(numerator, denominator):
result = numerator/denominator
print("result = %f" % result)
divide(1.0, 0)
def divide1(numerator, denominator):
try:
result = numerator/denominator
print("result = %f" % result)
except:
print("You can't divide by 0!")
divide1(1.0, 'a')
divide1(1.0, 2)
divide1("x", 2)
def divide2(numerator, denominator):
try:
result = numerator / denominator
print("result = %f" % result)
except (ZeroDivisionError, TypeError) as err:
print("Got an exception: %s" % err)
divide2(1, "X")
divide2("x, 2)
```
#### Why didn't we catch this `SyntaxError`?
```
# Handle division by 0 by using a small number
SMALL_NUMBER = 1e-3
def divide3(numerator, denominator):
try:
result = numerator/denominator
except ZeroDivisionError:
result = numerator/SMALL_NUMBER
print("result = %f" % result)
except Exception as err:
print("Different error than division by zero:", err)
divide3(1,0)
divide3("1",0)
```
#### What do you do when you get an exception?
First, you can feel relieved that you caught a problematic element of your software! Yes, relieved. Silent fails are much worse. (Again, another plug for testing.)
## Generating Exceptions
#### Why *generate* exceptions? (Don't I have enough unintentional errors?)
```
import pandas as pd
def validateDF(df):
""""
:param pd.DataFrame df: should have a column named "hours"
"""
if not "hours" in df.columns:
raise ValueError("DataFrame should have a column named 'hours'.")
df = pd.DataFrame({'hours': range(10) })
validateDF(df)
df = pd.DataFrame({'years': range(10) })
validateDF(df)
```
## Class exercise
Choose one of the functions from the last exercise. Create two new functions:
- The first function throws an exception if there is a negative argument.
- The second function catches an exception if the modulo operator (`%`) throws an exception and attempts to correct it by coercing the argument to a positive integer.
|
github_jupyter
|
import numpy as np
def divide(numerator, denominator):
result = numerator/denominator
print("result = %f" % result)
divide(1.0, 0)
def divide1(numerator, denominator):
try:
result = numerator/denominator
print("result = %f" % result)
except:
print("You can't divide by 0!")
divide1(1.0, 'a')
divide1(1.0, 2)
divide1("x", 2)
def divide2(numerator, denominator):
try:
result = numerator / denominator
print("result = %f" % result)
except (ZeroDivisionError, TypeError) as err:
print("Got an exception: %s" % err)
divide2(1, "X")
divide2("x, 2)
# Handle division by 0 by using a small number
SMALL_NUMBER = 1e-3
def divide3(numerator, denominator):
try:
result = numerator/denominator
except ZeroDivisionError:
result = numerator/SMALL_NUMBER
print("result = %f" % result)
except Exception as err:
print("Different error than division by zero:", err)
divide3(1,0)
divide3("1",0)
import pandas as pd
def validateDF(df):
""""
:param pd.DataFrame df: should have a column named "hours"
"""
if not "hours" in df.columns:
raise ValueError("DataFrame should have a column named 'hours'.")
df = pd.DataFrame({'hours': range(10) })
validateDF(df)
df = pd.DataFrame({'years': range(10) })
validateDF(df)
| 0.357231 | 0.840193 |
# Use Case 1: Kögur
In this example we will subsample a dataset stored on SciServer using methods resembling field-work procedures.
Specifically, we will estimate volume fluxes through the [Kögur section](http://kogur.whoi.edu) using (i) mooring arrays, and (ii) ship surveys.
```
# Import oceanspy
import oceanspy as ospy
# Import additional packages used in this notebook
import numpy as np
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
```
The following cell starts a dask client (see the [Dask Client section in the tutorial](Tutorial.ipynb#Dask-Client)).
```
# Start client
from dask.distributed import Client
client = Client()
client
```
This command opens one of the datasets avaiable on SciServer.
```
# Open dataset stored on SciServer.
od = ospy.open_oceandataset.from_catalog("EGshelfIIseas2km_ASR_full")
```
The following cell changes the default parameters used by the plotting functions.
```
import matplotlib as mpl
%matplotlib inline
mpl.rcParams["figure.figsize"] = [10.0, 5.0]
```
## Mooring array
The following diagram shows the instrumentation deployed by observational oceanographers to monitor the Kögur section (source: http://kogur.whoi.edu/img/array_boxes.png).
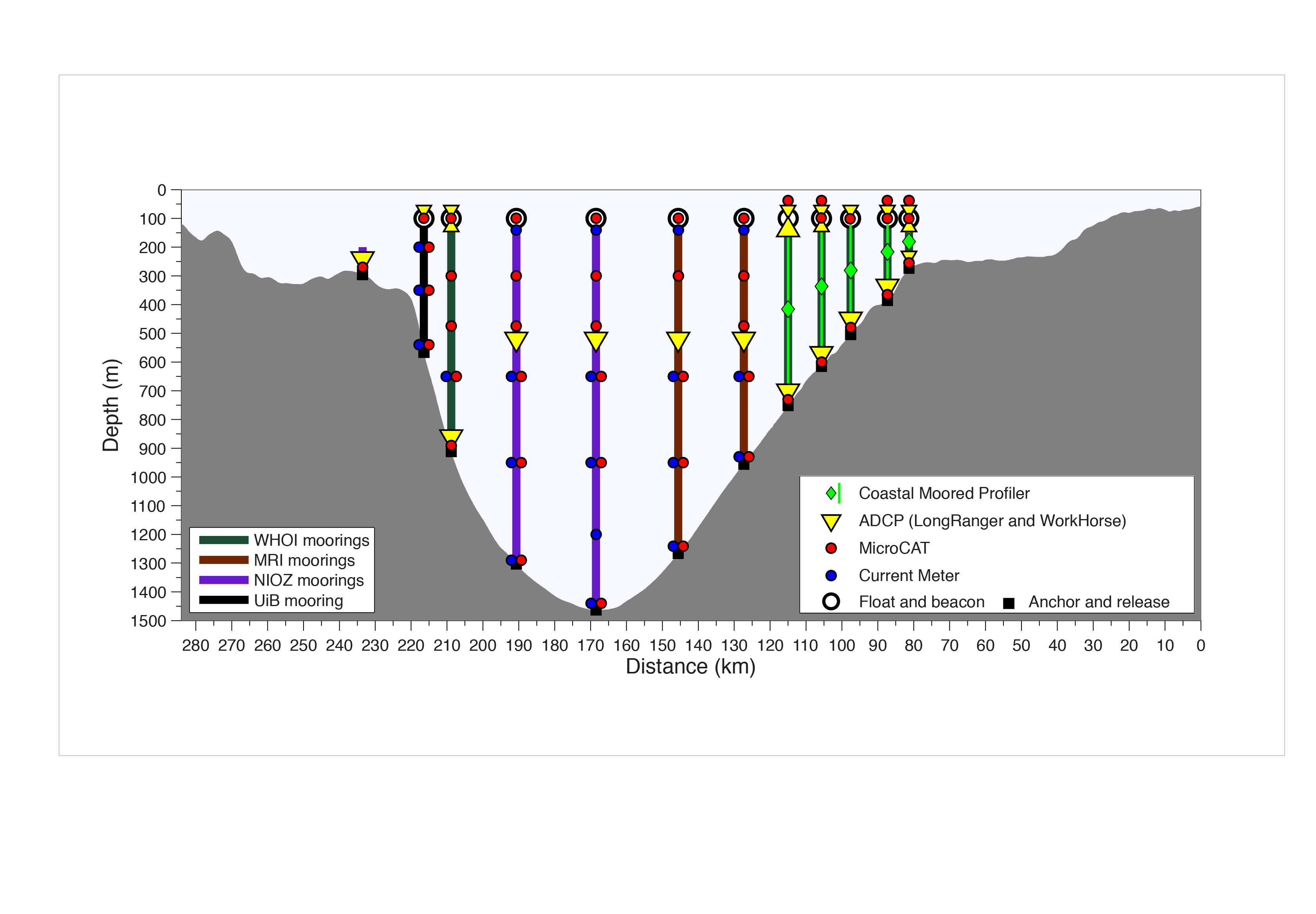
The analogous OceanSpy function (`compute.mooring_array`) extracts vertical sections from the model using two criteria:
* Vertical sections follow great circle paths (unless cartesian coordinates are used).
* Vertical sections follow the grid of the model (extracted moorings are adjacent to each other, and the native grid of the model is preserved).
```
# Kögur information
lats_Kogur = [68.68, 67.52, 66.49]
lons_Kogur = [-26.28, -23.77, -22.99]
depth_Kogur = [0, -1750]
# Select time range:
# September 2007, extracting one snapshot every 3 days
timeRange = ["2007-09-01", "2007-09-30T18"]
timeFreq = "3D"
# Extract mooring array and fields used by this notebook
od_moor = od.subsample.mooring_array(
Xmoor=lons_Kogur,
Ymoor=lats_Kogur,
ZRange=depth_Kogur,
timeRange=timeRange,
timeFreq=timeFreq,
varList=["Temp", "S", "U", "V", "dyG", "dxG", "drF", "HFacS", "HFacW"],
)
```
The following cell shows how to store the mooring array in a NetCDF file. In this use case, we only use this feature to create a checkpoint. Another option could be to move the file to other servers or computers. If the NetCDF is re-opened using OceanSpy (as shown below), all OceanSpy functions are enabled and can be applied to the `oceandataset`.
```
# Store the new mooring dataset
filename = "Kogur_mooring.nc"
od_moor.to_netcdf(filename)
# The NetCDF can now be re-opened with oceanspy at any time,
# and on any computer
od_moor = ospy.open_oceandataset.from_netcdf(filename)
# Print size
print("Size:")
print(" * Original dataset: {0:.1f} TB".format(od.dataset.nbytes * 1.0e-12))
print(" * Mooring dataset: {0:.1f} MB".format(od_moor.dataset.nbytes * 1.0e-6))
print()
```
The following map shows the location of the moorings forming the Kögur section.
```
# Plot map and mooring locations
fig = plt.figure(figsize=(5, 5))
ax = od.plot.horizontal_section(varName="Depth")
XC = od_moor.dataset["XC"].squeeze()
YC = od_moor.dataset["YC"].squeeze()
line = ax.plot(XC, YC, "r.", transform=ccrs.PlateCarree())
```
The following figure shows the grid structure of the mooring array. The original grid structure of the model is unchaged, and each mooring is associated with one C-gridpoint (e.g., hydrography), two U-gridpoint and two V-gridpoint (e.g., velocities), and four G-gripoint (e.g., vertical component of relative vorticity).
```
# Print grid
print(od_moor.grid)
print()
print(od_moor.dataset.coords)
print()
# Plot 10 moorings and their grid points
fig, ax = plt.subplots(1, 1)
n_moorings = 10
# Markers:
for _, (pos, mark, col) in enumerate(
zip(["C", "G", "U", "V"], ["o", "x", ">", "^"], ["k", "m", "r", "b"])
):
X = od_moor.dataset["X" + pos].values[:n_moorings].flatten()
Y = od_moor.dataset["Y" + pos].values[:n_moorings].flatten()
ax.plot(X, Y, col + mark, markersize=20, label=pos)
if pos == "C":
for i in range(n_moorings):
ax.annotate(
str(i),
(X[i], Y[i]),
size=15,
weight="bold",
color="w",
ha="center",
va="center",
)
ax.set_xticks(X, minor=False)
ax.set_yticks(Y, minor=False)
elif pos == "G":
ax.set_xticks(X, minor=True)
ax.set_yticks(Y, minor=True)
ax.legend(prop={"size": 20})
ax.grid(which="major", linestyle="-")
ax.grid(which="minor", linestyle="--")
```
## Plots
### Vertical sections
We can now use OceanSpy to plot vertical sections. Here we plot isopycnal contours on top of the mean meridional velocities (`V`). Although there are two V-points associated with each mooring, the plot can be displayed because OceanSpy automatically performs a linear interpolation using the grid object.
```
# Plot time mean
ax = od_moor.plot.vertical_section(
varName="V",
contourName="Sigma0",
meanAxes="time",
robust=True,
cmap="coolwarm",
)
```
It is possible to visualize all the snapshots by omitting the `meanAxes='time'` argument:
```
# Plot all snapshots
ax = od_moor.plot.vertical_section(
varName="V", contourName="Sigma0", robust=True, cmap="coolwarm", col_wrap=5
)
# Alternatively, use the following command to produce a movie:
# anim = od_moor.animate.vertical_section(varName='V', contourName='Sigma0', ...)
```
### TS-diagrams
Here we use OceanSpy to plot a Temperature-Salinity diagram.
```
ax = od_moor.plot.TS_diagram()
# Alternatively, use the following command
# to explore how the water masses change with time:
# anim = od_moor.animate.TS_diagram()
```
We can also color each TS point using any field in the original dataset, or any field computed by OceanSpy. Fields that are not on the same grid of temperature and salinity are automatically regridded by OceanSpy.
```
ax = od_moor.plot.TS_diagram(
colorName="V",
meanAxes="time",
cmap_kwargs={"robust": True, "cmap": "coolwarm"},
)
```
## Volume flux
OceanSpy can be used to compute accurate volume fluxes through vertical sections.
The function `compute.mooring_volume_transport` calculates the inflow/outflow through all grid faces of the vertical section.
This function creates a new dimension named `path` because transports can be computed using two paths (see the plot below).
```
# Show volume flux variables
ds_Vflux = ospy.compute.mooring_volume_transport(od_moor)
od_moor = od_moor.merge_into_oceandataset(ds_Vflux)
print(ds_Vflux)
# Plot 10 moorings and volume flux directions.
fig, ax = plt.subplots(1, 1)
ms = 10
s = 100
ds = od_moor.dataset
_ = ax.step(
ds["XU"].isel(Xp1=0).squeeze().values,
ds["YV"].isel(Yp1=0).squeeze().values,
"C0.-",
ms=ms,
label="path0",
)
_ = ax.step(
ds["XU"].isel(Xp1=1).squeeze().values,
ds["YV"].isel(Yp1=1).squeeze().values,
"C1.-",
ms=ms,
label="path1",
)
_ = ax.plot(
ds["XC"].squeeze(), ds["YC"].squeeze(), "k.", ms=ms, label="mooring"
)
_ = ax.scatter(
ds["X_Vtransport"].where(ds["dir_Vtransport"] == 1),
ds["Y_Vtransport"].where(ds["dir_Vtransport"] == 1),
s=s,
c="k",
marker="^",
label="meridional direction",
)
_ = ax.scatter(
ds["X_Utransport"].where(ds["dir_Utransport"] == 1),
ds["Y_Utransport"].where(ds["dir_Utransport"] == 1),
s=s,
c="k",
marker=">",
label="zonal direction",
)
_ = ax.scatter(
ds["X_Vtransport"].where(ds["dir_Vtransport"] == -1),
ds["Y_Vtransport"].where(ds["dir_Vtransport"] == -1),
s=s,
c="k",
marker="v",
label="meridional direction",
)
_ = ax.scatter(
ds["X_Utransport"].where(ds["dir_Utransport"] == -1),
ds["Y_Utransport"].where(ds["dir_Utransport"] == -1),
s=s,
c="k",
marker="<",
label="zonal direction",
)
# Only show a few moorings
m_start = 50
m_end = 70
xlim = ax.set_xlim(
sorted(
[
ds["XC"].isel(mooring=m_start).values,
ds["XC"].isel(mooring=m_end).values,
]
)
)
ylim = ax.set_ylim(
sorted(
[
ds["YC"].isel(mooring=m_start).values,
ds["YC"].isel(mooring=m_end).values,
]
)
)
ax.legend()
```
Here we compute and plot the cumulative mean transport through the Kögur mooring array.
```
# Compute cumulative transport
tran_moor = od_moor.dataset["transport"]
cum_tran_moor = tran_moor.sum("Z").mean("time").cumsum("mooring")
cum_tran_moor.attrs = tran_moor.attrs
fig, ax = plt.subplots(1, 1)
lines = cum_tran_moor.squeeze().plot.line(hue="path", linewidth=3)
tot_mean_tran_moor = cum_tran_moor.isel(mooring=-1).mean("path")
title = ax.set_title(
"TOTAL MEAN TRANSPORT: {0:.1f} Sv" "".format(tot_mean_tran_moor.values)
)
```
Here we compute the transport of the overflow, defined as water with density greater than 27.8 kg m$^{-3}$.
```
# Mask transport using density
od_moor = od_moor.compute.potential_density_anomaly()
density = od_moor.dataset["Sigma0"].squeeze()
oflow_moor = tran_moor.where(density > 27.8)
# Compute cumulative transport as before
cum_oflow_moor = oflow_moor.sum("Z").mean("time").cumsum("mooring")
cum_oflow_moor.attrs = oflow_moor.attrs
fig, ax = plt.subplots(1, 1)
lines = cum_oflow_moor.squeeze().plot.line(hue="path", linewidth=3)
tot_mean_oflow_moor = cum_oflow_moor.isel(mooring=-1).mean("path")
title = ax.set_title(
"TOTAL MEAN OVERFLOW TRANSPORT: {0:.1f} Sv"
"".format(tot_mean_oflow_moor.values)
)
```
## Ship survey
The following picture shows the NATO Research Vessel Alliance, a ship designed to carry out research at sea (source: http://www.marina.difesa.it/noi-siamo-la-marina/mezzi/forze-navali/PublishingImages/_alliance.jpg).

The OceanSpy function analogous to a ship survey (`compute.survey_stations`) extracts vertical sections from the model using two criteria:
* Vertical sections follow great circle paths (unless cartesian coordinates are used) with constant horizontal spacing between stations.
* Interpolation is performed and all fields are returned at the same locations (the native grid of the model is NOT preserved).
```
# Spacing between interpolated stations
delta_Kogur = 2 # km
# Extract survey stations
# Reduce dataset to speed things up:
od_surv = od.subsample.survey_stations(
Xsurv=lons_Kogur,
Ysurv=lats_Kogur,
delta=delta_Kogur,
ZRange=depth_Kogur,
timeRange=timeRange,
timeFreq=timeFreq,
varList=["Temp", "S", "U", "V", "drC", "drF", "HFacC", "HFacW", "HFacS"],
)
# Plot map and survey stations
fig = plt.figure(figsize=(5, 5))
ax = od.plot.horizontal_section(varName="Depth")
XC = od_surv.dataset["XC"].squeeze()
YC = od_surv.dataset["YC"].squeeze()
line = ax.plot(XC, YC, "r.", transform=ccrs.PlateCarree())
```
## Orthogonal velocities
We can use OceanSpy to compute the velocity components orthogonal and tangential to the Kögur section.
```
od_surv = od_surv.compute.survey_aligned_velocities()
```
The following animation shows isopycnal contours on top of the velocity component orthogonal to the Kögur section.
```
anim = od_surv.animate.vertical_section(
varName="ort_Vel",
contourName="Sigma0",
robust=True,
cmap="coolwarm",
display=False,
)
# The following code is necessary to display the animation in the documentation.
# When the notebook is executed, remove the code below and set
# display=True in the command above to show the animation.
import matplotlib.pyplot as plt
dirName = "_static"
import os
try:
os.mkdir(dirName)
except FileExistsError:
pass
anim.save("{}/Kogur.mp4".format(dirName))
plt.close()
!ffmpeg -loglevel panic -y -i _static/Kogur.mp4 -filter_complex "[0:v] fps=12,scale=480:-1,split [a][b];[a] palettegen [p];[b][p] paletteuse" _static/Kogur.gif
!rm -f _static/Kogur.mp4
```

Finally, we can infer the volume flux by integrating the orthogonal velocities.
```
# Integrate along Z
od_surv = od_surv.compute.integral(varNameList="ort_Vel", axesList=["Z"])
# Compute transport using weights
od_surv = od_surv.compute.weighted_mean(
varNameList="I(ort_Vel)dZ", axesList=["station"]
)
transport_surv = (
od_surv.dataset["I(ort_Vel)dZ"] * od_surv.dataset["weight_I(ort_Vel)dZ"]
)
# Convert in Sverdrup
transport_surv = transport_surv * 1.0e-6
# Compute cumulative transport
cum_transport_surv = transport_surv.cumsum("station").rename(
"Horizontal volume transport"
)
cum_transport_surv.attrs["units"] = "Sv"
```
Here we plot the cumulative transport for each snapshot.
```
# Plot
fig, ax = plt.subplots(figsize=(13, 5))
lines = cum_transport_surv.squeeze().plot.line(hue="time", linewidth=3)
tot_mean_transport = cum_transport_surv.isel(station=-1).mean("time")
title = ax.set_title(
"TOTAL MEAN TRANSPORT: {0:.1f} Sv".format(tot_mean_transport.values)
)
```
|
github_jupyter
|
# Import oceanspy
import oceanspy as ospy
# Import additional packages used in this notebook
import numpy as np
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
# Start client
from dask.distributed import Client
client = Client()
client
# Open dataset stored on SciServer.
od = ospy.open_oceandataset.from_catalog("EGshelfIIseas2km_ASR_full")
import matplotlib as mpl
%matplotlib inline
mpl.rcParams["figure.figsize"] = [10.0, 5.0]
# Kögur information
lats_Kogur = [68.68, 67.52, 66.49]
lons_Kogur = [-26.28, -23.77, -22.99]
depth_Kogur = [0, -1750]
# Select time range:
# September 2007, extracting one snapshot every 3 days
timeRange = ["2007-09-01", "2007-09-30T18"]
timeFreq = "3D"
# Extract mooring array and fields used by this notebook
od_moor = od.subsample.mooring_array(
Xmoor=lons_Kogur,
Ymoor=lats_Kogur,
ZRange=depth_Kogur,
timeRange=timeRange,
timeFreq=timeFreq,
varList=["Temp", "S", "U", "V", "dyG", "dxG", "drF", "HFacS", "HFacW"],
)
# Store the new mooring dataset
filename = "Kogur_mooring.nc"
od_moor.to_netcdf(filename)
# The NetCDF can now be re-opened with oceanspy at any time,
# and on any computer
od_moor = ospy.open_oceandataset.from_netcdf(filename)
# Print size
print("Size:")
print(" * Original dataset: {0:.1f} TB".format(od.dataset.nbytes * 1.0e-12))
print(" * Mooring dataset: {0:.1f} MB".format(od_moor.dataset.nbytes * 1.0e-6))
print()
# Plot map and mooring locations
fig = plt.figure(figsize=(5, 5))
ax = od.plot.horizontal_section(varName="Depth")
XC = od_moor.dataset["XC"].squeeze()
YC = od_moor.dataset["YC"].squeeze()
line = ax.plot(XC, YC, "r.", transform=ccrs.PlateCarree())
# Print grid
print(od_moor.grid)
print()
print(od_moor.dataset.coords)
print()
# Plot 10 moorings and their grid points
fig, ax = plt.subplots(1, 1)
n_moorings = 10
# Markers:
for _, (pos, mark, col) in enumerate(
zip(["C", "G", "U", "V"], ["o", "x", ">", "^"], ["k", "m", "r", "b"])
):
X = od_moor.dataset["X" + pos].values[:n_moorings].flatten()
Y = od_moor.dataset["Y" + pos].values[:n_moorings].flatten()
ax.plot(X, Y, col + mark, markersize=20, label=pos)
if pos == "C":
for i in range(n_moorings):
ax.annotate(
str(i),
(X[i], Y[i]),
size=15,
weight="bold",
color="w",
ha="center",
va="center",
)
ax.set_xticks(X, minor=False)
ax.set_yticks(Y, minor=False)
elif pos == "G":
ax.set_xticks(X, minor=True)
ax.set_yticks(Y, minor=True)
ax.legend(prop={"size": 20})
ax.grid(which="major", linestyle="-")
ax.grid(which="minor", linestyle="--")
# Plot time mean
ax = od_moor.plot.vertical_section(
varName="V",
contourName="Sigma0",
meanAxes="time",
robust=True,
cmap="coolwarm",
)
# Plot all snapshots
ax = od_moor.plot.vertical_section(
varName="V", contourName="Sigma0", robust=True, cmap="coolwarm", col_wrap=5
)
# Alternatively, use the following command to produce a movie:
# anim = od_moor.animate.vertical_section(varName='V', contourName='Sigma0', ...)
ax = od_moor.plot.TS_diagram()
# Alternatively, use the following command
# to explore how the water masses change with time:
# anim = od_moor.animate.TS_diagram()
ax = od_moor.plot.TS_diagram(
colorName="V",
meanAxes="time",
cmap_kwargs={"robust": True, "cmap": "coolwarm"},
)
# Show volume flux variables
ds_Vflux = ospy.compute.mooring_volume_transport(od_moor)
od_moor = od_moor.merge_into_oceandataset(ds_Vflux)
print(ds_Vflux)
# Plot 10 moorings and volume flux directions.
fig, ax = plt.subplots(1, 1)
ms = 10
s = 100
ds = od_moor.dataset
_ = ax.step(
ds["XU"].isel(Xp1=0).squeeze().values,
ds["YV"].isel(Yp1=0).squeeze().values,
"C0.-",
ms=ms,
label="path0",
)
_ = ax.step(
ds["XU"].isel(Xp1=1).squeeze().values,
ds["YV"].isel(Yp1=1).squeeze().values,
"C1.-",
ms=ms,
label="path1",
)
_ = ax.plot(
ds["XC"].squeeze(), ds["YC"].squeeze(), "k.", ms=ms, label="mooring"
)
_ = ax.scatter(
ds["X_Vtransport"].where(ds["dir_Vtransport"] == 1),
ds["Y_Vtransport"].where(ds["dir_Vtransport"] == 1),
s=s,
c="k",
marker="^",
label="meridional direction",
)
_ = ax.scatter(
ds["X_Utransport"].where(ds["dir_Utransport"] == 1),
ds["Y_Utransport"].where(ds["dir_Utransport"] == 1),
s=s,
c="k",
marker=">",
label="zonal direction",
)
_ = ax.scatter(
ds["X_Vtransport"].where(ds["dir_Vtransport"] == -1),
ds["Y_Vtransport"].where(ds["dir_Vtransport"] == -1),
s=s,
c="k",
marker="v",
label="meridional direction",
)
_ = ax.scatter(
ds["X_Utransport"].where(ds["dir_Utransport"] == -1),
ds["Y_Utransport"].where(ds["dir_Utransport"] == -1),
s=s,
c="k",
marker="<",
label="zonal direction",
)
# Only show a few moorings
m_start = 50
m_end = 70
xlim = ax.set_xlim(
sorted(
[
ds["XC"].isel(mooring=m_start).values,
ds["XC"].isel(mooring=m_end).values,
]
)
)
ylim = ax.set_ylim(
sorted(
[
ds["YC"].isel(mooring=m_start).values,
ds["YC"].isel(mooring=m_end).values,
]
)
)
ax.legend()
# Compute cumulative transport
tran_moor = od_moor.dataset["transport"]
cum_tran_moor = tran_moor.sum("Z").mean("time").cumsum("mooring")
cum_tran_moor.attrs = tran_moor.attrs
fig, ax = plt.subplots(1, 1)
lines = cum_tran_moor.squeeze().plot.line(hue="path", linewidth=3)
tot_mean_tran_moor = cum_tran_moor.isel(mooring=-1).mean("path")
title = ax.set_title(
"TOTAL MEAN TRANSPORT: {0:.1f} Sv" "".format(tot_mean_tran_moor.values)
)
# Mask transport using density
od_moor = od_moor.compute.potential_density_anomaly()
density = od_moor.dataset["Sigma0"].squeeze()
oflow_moor = tran_moor.where(density > 27.8)
# Compute cumulative transport as before
cum_oflow_moor = oflow_moor.sum("Z").mean("time").cumsum("mooring")
cum_oflow_moor.attrs = oflow_moor.attrs
fig, ax = plt.subplots(1, 1)
lines = cum_oflow_moor.squeeze().plot.line(hue="path", linewidth=3)
tot_mean_oflow_moor = cum_oflow_moor.isel(mooring=-1).mean("path")
title = ax.set_title(
"TOTAL MEAN OVERFLOW TRANSPORT: {0:.1f} Sv"
"".format(tot_mean_oflow_moor.values)
)
# Spacing between interpolated stations
delta_Kogur = 2 # km
# Extract survey stations
# Reduce dataset to speed things up:
od_surv = od.subsample.survey_stations(
Xsurv=lons_Kogur,
Ysurv=lats_Kogur,
delta=delta_Kogur,
ZRange=depth_Kogur,
timeRange=timeRange,
timeFreq=timeFreq,
varList=["Temp", "S", "U", "V", "drC", "drF", "HFacC", "HFacW", "HFacS"],
)
# Plot map and survey stations
fig = plt.figure(figsize=(5, 5))
ax = od.plot.horizontal_section(varName="Depth")
XC = od_surv.dataset["XC"].squeeze()
YC = od_surv.dataset["YC"].squeeze()
line = ax.plot(XC, YC, "r.", transform=ccrs.PlateCarree())
od_surv = od_surv.compute.survey_aligned_velocities()
anim = od_surv.animate.vertical_section(
varName="ort_Vel",
contourName="Sigma0",
robust=True,
cmap="coolwarm",
display=False,
)
# The following code is necessary to display the animation in the documentation.
# When the notebook is executed, remove the code below and set
# display=True in the command above to show the animation.
import matplotlib.pyplot as plt
dirName = "_static"
import os
try:
os.mkdir(dirName)
except FileExistsError:
pass
anim.save("{}/Kogur.mp4".format(dirName))
plt.close()
!ffmpeg -loglevel panic -y -i _static/Kogur.mp4 -filter_complex "[0:v] fps=12,scale=480:-1,split [a][b];[a] palettegen [p];[b][p] paletteuse" _static/Kogur.gif
!rm -f _static/Kogur.mp4
# Integrate along Z
od_surv = od_surv.compute.integral(varNameList="ort_Vel", axesList=["Z"])
# Compute transport using weights
od_surv = od_surv.compute.weighted_mean(
varNameList="I(ort_Vel)dZ", axesList=["station"]
)
transport_surv = (
od_surv.dataset["I(ort_Vel)dZ"] * od_surv.dataset["weight_I(ort_Vel)dZ"]
)
# Convert in Sverdrup
transport_surv = transport_surv * 1.0e-6
# Compute cumulative transport
cum_transport_surv = transport_surv.cumsum("station").rename(
"Horizontal volume transport"
)
cum_transport_surv.attrs["units"] = "Sv"
# Plot
fig, ax = plt.subplots(figsize=(13, 5))
lines = cum_transport_surv.squeeze().plot.line(hue="time", linewidth=3)
tot_mean_transport = cum_transport_surv.isel(station=-1).mean("time")
title = ax.set_title(
"TOTAL MEAN TRANSPORT: {0:.1f} Sv".format(tot_mean_transport.values)
)
| 0.670932 | 0.991067 |
# Grove Temperature Sensor 1.2
This example shows how to use the [Grove Temperature Sensor v1.2](http://wiki.seeedstudio.com/Grove-Temperature_Sensor_V1.2/). You will also see how to plot a graph using matplotlib. The Grove Temperature sensor produces an analog signal, and requires an ADC.
A Grove Temperature sensor and Pynq Grove Adapter, or Pynq Shield is required. The Grove Temperature Sensor, Pynq Grove Adapter, and Grove I2C ADC are used for this example.
You can read a single value of temperature or read multiple values at regular intervals for a desired duration.
At the end of this notebook, a Python only solution with single-sample read functionality is provided.
### 1. Load overlay
```
from pynq.overlays.base import BaseOverlay
base = BaseOverlay("base.bit")
```
### 2. Read single temperature
This example shows on how to get a single temperature sample from the Grove TMP sensor.
The Grove ADC is assumed to be attached to the GR4 connector of the StickIt. The StickIt module is assumed to be plugged in the 1st PMOD labeled JB. The Grove TMP sensor is connected to the other connector of the Grove ADC.
Grove ADC provides a raw sample which is converted into resistance first and then converted into temperature.
```
import math
from pynq.lib.pmod import Grove_TMP
from pynq.lib.pmod import PMOD_GROVE_G4
tmp = Grove_TMP(base.PMODB,PMOD_GROVE_G4)
temperature = tmp.read()
print(float("{0:.2f}".format(temperature)),'degree Celsius')
```
### 3. Start logging once every 100ms for 10 seconds
Executing the next cell will start logging the temperature sensor values every 100ms, and will run for 10s. You can try touch/hold the temperature sensor to vary the measured temperature.
You can vary the logging interval and the duration by changing the values 100 and 10 in the cellbelow. The raw samples are stored in the internal memory, and converted into temperature values.
```
import time
%matplotlib inline
import matplotlib.pyplot as plt
tmp.set_log_interval_ms(100)
tmp.start_log()
# Change input during this time
time.sleep(10)
tmp_log = tmp.get_log()
plt.plot(range(len(tmp_log)), tmp_log, 'ro')
plt.title('Grove Temperature Plot')
min_tmp_log = min(tmp_log)
max_tmp_log = max(tmp_log)
plt.axis([0, len(tmp_log), min_tmp_log, max_tmp_log])
plt.show()
```
### 4. A Pure Python class to exercise the AXI IIC Controller inheriting from PMOD_IIC
This class is ported from http://wiki.seeedstudio.com/Grove-Temperature_Sensor/
```
from time import sleep
from math import log
from pynq.lib.pmod import PMOD_GROVE_G3
from pynq.lib.pmod import PMOD_GROVE_G4
from pynq.lib import Pmod_IIC
class Python_Grove_TMP(Pmod_IIC):
"""This class controls the grove temperature sensor.
This class inherits from the PMODIIC class.
Attributes
----------
iop : _IOP
The _IOP object returned from the DevMode.
scl_pin : int
The SCL pin number.
sda_pin : int
The SDA pin number.
iic_addr : int
The IIC device address.
"""
def __init__(self, pmod_id, gr_pins, model = 'v1.2'):
"""Return a new instance of a grove OLED object.
Parameters
----------
pmod_id : int
The PMOD ID (1, 2) corresponding to (PMODA, PMODB).
gr_pins: list
The group pins on Grove Adapter. G3 or G4 is valid.
model : string
Temperature sensor model (can be found on the device).
"""
if gr_pins in [PMOD_GROVE_G3, PMOD_GROVE_G4]:
[scl_pin,sda_pin] = gr_pins
else:
raise ValueError("Valid group numbers are G3 and G4.")
# Each revision has its own B value
if model == 'v1.2':
# v1.2 uses thermistor NCP18WF104F03RC
self.bValue = 4250
elif model == 'v1.1':
# v1.1 uses thermistor NCP18WF104F03RC
self.bValue = 4250
else:
# v1.0 uses thermistor TTC3A103*39H
self.bValue = 3975
super().__init__(pmod_id, scl_pin, sda_pin, 0x50)
# Initialize the Grove ADC
self.send([0x2,0x20]);
def read(self):
"""Read temperature in Celsius from grove temperature sensor.
Parameters
----------
None
Returns
-------
float
Temperature reading in Celsius.
"""
val = self._read_grove_adc()
R = 4095.0/val - 1.0
temp = 1.0/(log(R)/self.bValue + 1/298.15)-273.15
return temp
def _read_grove_adc(self):
self.send([0])
bytes = self.receive(2)
return 2*(((bytes[0] & 0x0f) << 8) | bytes[1])
from pynq import PL
# Flush IOP state
PL.reset()
py_tmp = Python_Grove_TMP(base.PMODB, PMOD_GROVE_G4)
temperature = py_tmp.read()
print(float("{0:.2f}".format(temperature)),'degree Celsius')
```
Copyright (C) 2020 Xilinx, Inc
|
github_jupyter
|
from pynq.overlays.base import BaseOverlay
base = BaseOverlay("base.bit")
import math
from pynq.lib.pmod import Grove_TMP
from pynq.lib.pmod import PMOD_GROVE_G4
tmp = Grove_TMP(base.PMODB,PMOD_GROVE_G4)
temperature = tmp.read()
print(float("{0:.2f}".format(temperature)),'degree Celsius')
import time
%matplotlib inline
import matplotlib.pyplot as plt
tmp.set_log_interval_ms(100)
tmp.start_log()
# Change input during this time
time.sleep(10)
tmp_log = tmp.get_log()
plt.plot(range(len(tmp_log)), tmp_log, 'ro')
plt.title('Grove Temperature Plot')
min_tmp_log = min(tmp_log)
max_tmp_log = max(tmp_log)
plt.axis([0, len(tmp_log), min_tmp_log, max_tmp_log])
plt.show()
from time import sleep
from math import log
from pynq.lib.pmod import PMOD_GROVE_G3
from pynq.lib.pmod import PMOD_GROVE_G4
from pynq.lib import Pmod_IIC
class Python_Grove_TMP(Pmod_IIC):
"""This class controls the grove temperature sensor.
This class inherits from the PMODIIC class.
Attributes
----------
iop : _IOP
The _IOP object returned from the DevMode.
scl_pin : int
The SCL pin number.
sda_pin : int
The SDA pin number.
iic_addr : int
The IIC device address.
"""
def __init__(self, pmod_id, gr_pins, model = 'v1.2'):
"""Return a new instance of a grove OLED object.
Parameters
----------
pmod_id : int
The PMOD ID (1, 2) corresponding to (PMODA, PMODB).
gr_pins: list
The group pins on Grove Adapter. G3 or G4 is valid.
model : string
Temperature sensor model (can be found on the device).
"""
if gr_pins in [PMOD_GROVE_G3, PMOD_GROVE_G4]:
[scl_pin,sda_pin] = gr_pins
else:
raise ValueError("Valid group numbers are G3 and G4.")
# Each revision has its own B value
if model == 'v1.2':
# v1.2 uses thermistor NCP18WF104F03RC
self.bValue = 4250
elif model == 'v1.1':
# v1.1 uses thermistor NCP18WF104F03RC
self.bValue = 4250
else:
# v1.0 uses thermistor TTC3A103*39H
self.bValue = 3975
super().__init__(pmod_id, scl_pin, sda_pin, 0x50)
# Initialize the Grove ADC
self.send([0x2,0x20]);
def read(self):
"""Read temperature in Celsius from grove temperature sensor.
Parameters
----------
None
Returns
-------
float
Temperature reading in Celsius.
"""
val = self._read_grove_adc()
R = 4095.0/val - 1.0
temp = 1.0/(log(R)/self.bValue + 1/298.15)-273.15
return temp
def _read_grove_adc(self):
self.send([0])
bytes = self.receive(2)
return 2*(((bytes[0] & 0x0f) << 8) | bytes[1])
from pynq import PL
# Flush IOP state
PL.reset()
py_tmp = Python_Grove_TMP(base.PMODB, PMOD_GROVE_G4)
temperature = py_tmp.read()
print(float("{0:.2f}".format(temperature)),'degree Celsius')
| 0.664105 | 0.971483 |
# The *new* **Network** Class
## Intro
This notebook demonstrates the new design of skrf's **Network** Class. The new class utilizes a more object-oriented approach which is cleaner and more scalable. The draw-back is that it breaks backward compatibility.
Creating a *new style* **Network** from an old
```
import skrf as rf
%matplotlib inline
from pylab import *
rf.stylely()
from skrf import network2
a = network2.Network.from_ntwkv1(rf.data.ring_slot)
```
The new **Network** class employs nested objects, which makes for a cleaner and more logical namespace. The basic structure is:
* **Network**
* **Frequency ** (same as before)
* **Parameter** (s,z,y,etc)
* **Projection** ( db10, db20, deg, mag,etc)
## Network Parameters
Accessing a Network's parameters like `s`,`z`,or `y` returns a **Parameters** object,
```
type(a.s)
```
You can get at the array by accessing the property `val`.
```
a.s.val[:3]
```
You can also slice the parameter directly. This can be used as an alternative way to access the values.
```
a.s[:2]
```
This nested-object desgin allows for more concise function calls. For example, plot functions become members of the parameters, which behave like you expect
```
a.s.plot()
a.z.plot()
axis('equal')
```
## Projections
Each parameter has members for various scalar projections.
```
type(a.s.db)
type(a.s.deg)
```
Their numerical values may be accessed through `val` attribute or by direct slicing, just like a **Parameter**
```
a.s.db[:2]
```
Projections also `plot()` as you expect
```
a.s.db.plot();
a.s.deg.plot(1,0);
```
## Ipython Notebook display system
One interesting advantage of using an object-oriented model for parameters and projections is that we can create [custom display logic](http://nbviewer.ipython.org/github/ipython/ipython/blob/master/examples/notebooks/Custom%20Display%20Logic.ipynb) for the ipython notebook. This allows us to define graphical representations for an object, removing the need to call any plot method.
```
a.s.db
a.z.im
```
## Accessing numpy array properties
Numpy ndarray properties are accessable on both **Parameters** and **Projections **. These are implemented using python `__getattr__()` operator so they wont tab out, but you can still use em.
```
a.s.db.plot()
axhline(a.s.db.min(),color='k')
axhline(a.s.db.max(),color='k')
```
## Frequency Band Selection
Networks can sliced by an index on the frequency axis or by a human readable frequency selections,
```
a[40:100] #slice by frequency index
a['82-92ghz'] # slice by a human readable string
a['82-92ghz'].s.db
```
## Subnetworks
Individual s-parameters can be accessed by calling a **Network** with the desired port indecies (*index starting from 0*)
```
s11 = a(0,0) # s11
s22 = a(1,1) # s22
s12 = a(0,1) # s12
s21 = a(1,1) # s21
s11
```
## Time domain
Time domain transform is implemented as a Parameter named `s_time`. Note that accessing this parameter implicitly windows the s-parameters before taking the FFT. For finer control over the transform, use the functions `s2time` and `windowed`.
```
b = network2.Network.from_ntwkv1(rf.data.ring_slot_meas)
b.s_time.db
```
|
github_jupyter
|
import skrf as rf
%matplotlib inline
from pylab import *
rf.stylely()
from skrf import network2
a = network2.Network.from_ntwkv1(rf.data.ring_slot)
type(a.s)
a.s.val[:3]
a.s[:2]
a.s.plot()
a.z.plot()
axis('equal')
type(a.s.db)
type(a.s.deg)
a.s.db[:2]
a.s.db.plot();
a.s.deg.plot(1,0);
a.s.db
a.z.im
a.s.db.plot()
axhline(a.s.db.min(),color='k')
axhline(a.s.db.max(),color='k')
a[40:100] #slice by frequency index
a['82-92ghz'] # slice by a human readable string
a['82-92ghz'].s.db
s11 = a(0,0) # s11
s22 = a(1,1) # s22
s12 = a(0,1) # s12
s21 = a(1,1) # s21
s11
b = network2.Network.from_ntwkv1(rf.data.ring_slot_meas)
b.s_time.db
| 0.371365 | 0.921287 |
## Ghana Climate Data
### Exploratory Data Analysis
```
# Climate data source from https://www.ncdc.noaa.gov/cdo-web
# Period of record from 1973
# Contains data records of 17 stations of varying lengths
# Original units of variables - imperial units
%matplotlib inline
# Import libraries
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
# Seaborn for additional plotting and styling
import seaborn as sns
# Define file path to connect data source:
fp = 'C:/Users/Narteh/earth_analytics/Gh_Climate_EDA/data_weatherGH/1619559410439.csv'
# Read in data from the csv file
# Low memory set to false to remove memory errors
data = pd.read_csv(fp, parse_dates=['DATE'], dayfirst=True, low_memory=False)
# Print data types
data.dtypes
#Print column names
data.columns.values
# Check DATE type format
type(data['DATE'])
# Check dataframe shape (number of rows, number of columns)
data.shape
# Confirm output of dataframe
data.head()
# Check sum of no-data, not a number (NaN) values in the data set
data[['PRCP', 'TAVG', 'TMAX', 'TMIN']].isna().sum()
# Confirm output of parse date
data.loc[0, 'DATE'].day_name()
# Drop column Unnamed: 0, which contains station unique code
# Drop PRCP column, work only with temperature data
data.drop(columns=['Unnamed: 0', 'PRCP'], inplace=True)
# Check drop of Unnamed: 0 and PRCP column
data.tail()
# Rename column heading of Unnamed: 1 to TOWN and TAVG to TEMP for familiarity
data = data.rename(columns={'Unnamed: 1': 'TOWN', 'TAVG': 'TEMP'})
# Confirm rename of columns
data.head()
# Replace inadvertantly named double named towns falling under 2 columns to its original double word names & KIA for Kotoka International Airport
data.replace(to_replace=['AKIM', 'KOTOKA', 'KETE', 'SEFWI'],
value= ['AKIM ODA', 'KIA', 'KETE KRACHI', 'SEFWI WIASO'],
inplace=True)
# Check confirmation of output
data.head()
# Drop STATION column due to renamed town heading
data.drop(columns=['STATION'], inplace=True)
# Check drop of STATION column
data.head()
# The date of first observation
first_obs = data.at[0, "DATE"]
print('The first record of observation was on', data.at[0, "DATE"])
# Convert to string to change capital case to lower case for TOWN names
data['TOWN'] = data['TOWN'].astype(str)
data['TOWN'] = data['TOWN'].str.title()
# Check conversion string change
data.head()
# Convert imperial units to metric through def function
def fahr_to_celsius(temp_fahrenheit):
"""Function to convert Fahrenheit temperature into Celsius.
Parameters
----------
temp_fahrenheit: int | float
Input temperature in Fahrenheit (should be a number)
Returns
-------
Temperature in Celsius (float)
"""
# Convert the Fahrenheit into Celsius
converted_temp = (temp_fahrenheit - 32) / 1.8
return converted_temp # Use apply to convert each row of value
data[['TEMP_C', 'TMAX_C', 'TMIN_C']] = round(data[['TEMP', 'TMAX', 'TMIN']].apply(fahr_to_celsius), 2)
# Check conversion to metric units
data.head()
# Drop columns with fahrenheit units since they are no longer needed
data.drop(columns=['TEMP', 'TMAX', 'TMIN'], inplace=True)
# print head to evalaute
data.head()
# For later analysis
data['DayofWeek'] = data['DATE'].dt.day_name()
data.head()
# Take mean of mean temperatures of the data set
temp_mean = round(data['TEMP_C'].mean(), 2)
temp_mean
# Set DATE as index
data.set_index('DATE', inplace=True)
data.head()
# Mean daily temperature across all the weather stations
daily_temp = round(data['TEMP_C'].resample('D').mean(), 2)
daily_temp.head()
# Indicate the mean minimum and maximum daily temperature
print(daily_temp.min(),'°C')
print('')
print(daily_temp.max(),'°C')
# Show which dates the minimum and maximum daily mean temperature were recorded
min_daily = daily_temp.loc[daily_temp == 16.67]
print(min_daily)
print(' ')
max_daily = daily_temp.loc[daily_temp == 35.0]
print(max_daily)
# Mean monthly temperature across all the weather stations
monthly_temp = round(data['TEMP_C'].resample('M').mean(), 2)
monthly_temp.head()
# Indicate the mean minimum and maximum monthly temperature
min_monthly_temp = monthly_temp.min()
print(min_monthly_temp)
print('')
max_monthly_temp = monthly_temp.max()
print(max_monthly_temp)
# Show the date the mean minimum and maximum monthly temperature were recorded
print(monthly_temp.loc[monthly_temp == min_monthly_temp])
print(' ')
print(monthly_temp.loc[monthly_temp == max_monthly_temp])
# Mean yearly temperature across all the weather stations
yearly_temp = round(data[['TEMP_C', 'TMAX_C', 'TMIN_C']].resample('Y').mean(), 2)
# Check output
yearly_temp.head()
# Indicate the mean minimum and maximum yearly temperature
min_yearly_temp = yearly_temp['TEMP_C'].min()
print(min_yearly_temp)
print(' ')
max_yearly_temp = yearly_temp['TEMP_C'].max()
print(max_yearly_temp)
# Show the year the minimum and maximum yearly temperature were recorded
print(yearly_temp['TEMP_C'].loc[yearly_temp['TEMP_C'] == min_yearly_temp])
print(' ')
print(yearly_temp['TEMP_C'].loc[yearly_temp['TEMP_C'] == max_yearly_temp])
# Reset DATE index to column
yearly_temp = yearly_temp.reset_index()
yearly_temp.head()
# Convert DATE to year using dt.year
yearly_temp['YEAR']= yearly_temp['DATE'].dt.year
yearly_temp.head()
# Plot mean yearly temperatures
import matplotlib.dates as mdates
import matplotlib.ticker as ticker
# Plot style use Seaborn
plt.style.use('seaborn-whitegrid')
fig, ax = plt.subplots(1, figsize=(10, 6))
# Data values for X and Y
x = yearly_temp.YEAR
y = yearly_temp.TEMP_C
# Plot x, y values & parameters
plt.plot(x, y, color = 'red', linestyle='-', marker='o', mfc='blue')
# Highlight coalescencing temps around 28 °C
plt.plot(x[-6:], y[-6:], 'y*', ms='12')
fig.autofmt_xdate()
# Plot title & attributes
plt.title('Yearly Mean Temperatures, Ghana', fontdict={'fontname': 'comic sans ms',
'fontsize': 15, 'weight': 'bold'})
# Add label to the plot
#plt.text(1983, 30.190, ' Severe Drought & Bushfires', fontsize='large')
# Annotate peak temp with description & arrow
ax.annotate('Severe drought & bushfires', fontsize='large', weight='semibold',
xy=(1983, 30.190), xytext=(1986, 29.5),
arrowprops=dict(arrowstyle='->',
connectionstyle='arc'),
xycoords='data',)
# Labels for axes
plt.xlabel('Year', fontdict={'fontsize': 14})
plt.ylabel('Temperature °C', fontdict={'fontsize': 14})
# legend
plt.legend(['Mean Temperature'], frameon=True, fontsize='x-large')
# major ticks locators
plt.xticks(np.arange(min(x), max(x), 4))
def setup(ax):
ax.xaxis.set_ticks_position('bottom')
ax.tick_params(which='minor', width=0.75, length=2.5)
setup(ax)
ax.xaxis.set_minor_locator(ticker.FixedLocator((x)))
plt.tight_layout()
# To save graph
plt.savefig('C:/Users/Narteh/earth_analytics/Gh_Climate_EDA/Graphs/yearly_mean_gh.jpg')
import pandas_bokeh
pandas_bokeh.output_notebook()
pd.set_option('plotting.backend', 'pandas_bokeh')
# Plot interactive map with Bokeh
from bokeh.models import HoverTool
ax = yearly_temp.plot(x = 'YEAR', y = 'TEMP_C', title='Yearly Mean Temperatures, Ghana',
xlabel = 'Year', ylabel = 'Temperature °C', xticks = (yearly_temp.YEAR[::4]),
legend = False, plot_data_points = True, hovertool_string= r'''<h5> @{YEAR} </h5>
<h5> @{TEMP_C} °C </h5>''') # Change hoovertool stringe text to values in dataframe only
# set output to static HTML file
from bokeh.plotting import figure, output_file, show
output_file(filename='Ghana_Climate_Interactive.html', title='Ghana Climate HTML file')
#show(ax)
# Group by towns
sta_town_grp = data.groupby(['TOWN'])
# Check groupy operation with one example
sta_town_grp.get_group('Akuse').head()
# Filter out the maximum temps ever recorded in each town from TMAX_C
sta_town_max = sta_town_grp['TMAX_C'].max()
sta_town_max = sta_town_max.sort_values(ascending=False)
sta_town_max
# Filter out row values of the date of occurence of highest maximum temp for first few towns
data.loc[(data.TMAX_C == 48.89) | (data.TMAX_C == 43.89)]
# Sort DATE at index to allow slicing, done chronologically
data.sort_index(inplace=True)
# Calculate decadel mean temperatures
avg_temp_1970s = data['1970': '1979']['TEMP_C'].mean()
avg_temp_1980s = data['1980': '1989']['TEMP_C'].mean()
avg_temp_1990s = data['1990': '1999']['TEMP_C'].mean()
avg_temp_2000s = data['2000': '2009']['TEMP_C'].mean()
avg_temp_2010s = data['2010': '2019']['TEMP_C'].mean()
# Print to screen the mean decadal temperatures
print(round(avg_temp_1970s, 2))
print(' ')
print(round(avg_temp_1980s, 2))
print(' ')
print(round(avg_temp_1990s, 2))
print(' ')
print(round(avg_temp_2000s, 2))
print(' ')
print(round(avg_temp_2010s, 2))
# Take monthly mean of all daily records
monthly_data = round(data.resample('M').mean(), 2)
monthly_data.head()
# Return to column heading
monthly_data.reset_index(inplace=True)
# Convert to date month
monthly_data['MONTH'] = monthly_data['DATE'].dt.month_name().str.slice(stop=3)
monthly_data.head()
# Take mean of all indivdual months
monthly_mean = round(monthly_data.groupby(['MONTH']).mean(), 2)
monthly_mean.head()
# Create new order for actual calender months
new_order = ['Jan', 'Feb', 'Mar', 'Apr', 'May', 'Jun', 'Jul',
'Aug', 'Sep', 'Oct', 'Nov', 'Dec']
monthly_mean = monthly_mean.reindex(new_order, axis=0)
# check output of reindex
monthly_mean.head()
monthly_mean = monthly_mean.reset_index()
monthly_mean.head()
# Plot mean monthly temperatures for the period of the record
# Plot style use Seaborn
plt.style.use('seaborn')
fig, ax1 = plt.subplots(1, figsize=(10, 6))
x = monthly_mean['MONTH']
y = monthly_mean['TEMP_C']
plt.ylim(23, 31)
plt.plot(x, y, color = 'tomato', linestyle='-', marker='o', mfc='orange', linewidth = 3, markersize = 8)
plt.grid(axis = 'x')
plt.title('Monthly Mean Temperatures, Ghana', fontdict={'fontname': 'comic sans ms',
'fontsize': 15, 'weight': 'bold'})
# Labels for axes
plt.xlabel('Month', fontdict={'fontsize': 14})
plt.ylabel('Temperature °C', fontdict={'fontsize': 14})
# To save graph
plt.savefig('C:/Users/Narteh/earth_analytics/Gh_Climate_EDA/Graphs/monthly_mean_temp.jpg')
# Use bokeh for interactivity
ax1 = monthly_mean.plot.line(x = monthly_mean['MONTH'],
y = 'TEMP_C', title='Mean Monthly Temperatures, Ghana',
xlabel ='Month', ylabel='Temperature °C', ylim=(21, 33),
marker='o', plot_data_points_size = 8, line_width = 3,
legend = False, color = 'tomato', plot_data_points=True,
hovertool_string= r'''<h5> @{MONTH} </h5>
<h5> @{TEMP_C} °C </h5>''') # Change hoovertool stringe text to values in dataframe only
# set output to static HTML file
# Remove x grid line for different fig style
ax1.xgrid.grid_line_color = None
output_file(filename='mean_monthly_gh.html', title='Monthly Mean Temperatures, Ghana')
show(ax1)
# Create new column of Climatic zones in Ghana according "Climatic Regions of Ghana (Abass, 2009)
# To help evaluate changes since the period of the record
data['CLIMATE_ZONES'] = data['TOWN'].map({'Tamale': 'Zone_4', 'Wenchi': 'Zone_4', 'Navrongo': 'Zone_4',
'Wa': 'Zone_4', 'Bole': 'Zone_4', 'Kete Krachi': 'Zone_4',
'Akim Oda': 'Zone_3', 'Sefwi Wiaso': 'Zone_3', 'Koforidua': 'Zone_3',
'Kumasi': 'Zone_3', 'Sunyani': 'Zone_3', 'Ho': 'Zone_3','Kia': 'Zone_2',
'Akuse': 'Zone_2', 'Ada': 'Zone_2', 'Axim': 'Zone_1', 'Takoradi': 'Zone_1'})
data.tail()
# Find mean yearly temp per climate zone
climate_zones_yearly = data.groupby('CLIMATE_ZONES').resample('Y').mean()
climate_zones_yearly.head()
# Return to column heading without column index
climate_zones_yearly.reset_index(inplace=True)
climate_zones_yearly.head()
# Pivot table to get temps for each climate zone per year in column format
climate_zones_yearly = climate_zones_yearly.pivot_table(values = 'TEMP_C', index = 'DATE', columns = 'CLIMATE_ZONES')
climate_zones_yearly.head()
# Check for missing values
climate_zones_yearly.isnull().sum()
# Drop NA values due to grouping operation
climate_zones_yearly = climate_zones_yearly.dropna()
climate_zones_yearly.head()
# Plot mean monthly temperatures for the 4 climate zones in Ghana
# Plot style use Seaborn
plt.style.use('seaborn')
#sns.set_style('whitegrid')
fig, ax1 = plt.subplots(1, figsize=(10, 6))
x = climate_zones_yearly.reset_index()['DATE']
y_1 = climate_zones_yearly['Zone_1']
y_2 = climate_zones_yearly['Zone_2']
y_3 = climate_zones_yearly['Zone_3']
y_4 = climate_zones_yearly['Zone_4']
# x, y limits of axes
plt.ylim(23, 33)
# Plot each lines on same axis
plt.plot(x, y_1, '.-g', label = 'Zone 1', linewidth = 2)
plt.plot(x, y_2, '.--c', label = 'Zone 2', linewidth = 2)
plt.plot(x, y_3, '.-.b', label = 'Zone 3', linewidth = 2)
plt.plot(x, y_4, '.:r', label = 'Zone 4', linewidth = 2)
leg = plt.legend()
# get the lines and texts inside legend box
leg_lines = leg.get_lines()
leg_texts = leg.get_texts()
# bulk-set the properties of all lines and texts
plt.setp(leg_lines, linewidth=4)
plt.setp(leg_texts, fontsize='x-large')
plt.title('Mean Yearly Temperature per Climate Zone, Ghana',
fontdict={'fontname': 'comic sans ms',
'fontsize': 15, 'weight': 'bold'})
# Labels for axes
plt.xlabel('Year', fontdict={'fontsize': 14})
plt.ylabel('Temperature °C', fontdict={'fontsize': 14})
# To save graph
plt.savefig('C:/Users/Narteh/earth_analytics/Gh_Climate_EDA/Graphs/zone_yearly_mean.png')
# Find mean monthly temp per climate zone
climate_zones_monthly = data.groupby('CLIMATE_ZONES').resample('M').mean()
climate_zones_monthly.head()
# Return to column heading without column index
climate_zones_monthly.reset_index(inplace=True)
# Convert to date month in short format for temps months evaluation
climate_zones_monthly['MONTH'] = climate_zones_monthly['DATE'].dt.month_name().str.slice(stop=3)
climate_zones_monthly.tail()
# Take mean of each mean monthly temp for each zone
monthly_mean_zone = climate_zones_monthly.groupby(['CLIMATE_ZONES', 'MONTH'], as_index=False).mean()
monthly_mean_zone.head()
# Re order months by custom sorting per calendar months
months_categories = ('Jan', 'Feb', 'Mar', 'Apr', 'May', 'Jun', 'Jul', 'Aug', 'Sep', 'Oct', 'Nov', 'Dec')
monthly_mean_zone['MONTH'] = pd.Categorical(monthly_mean_zone['MONTH'], categories = months_categories)
monthly_mean_zone.head()
# Sort values under MONTH by calender months
monthly_mean_zone = monthly_mean_zone.sort_values(by = 'MONTH')
monthly_mean_zone.head()
# Pivot table to get temps for each climate zone per month in column format
monthly_mean_zone = monthly_mean_zone.pivot_table(values = 'TEMP_C', index = 'MONTH', columns = 'CLIMATE_ZONES')
monthly_mean_zone.head()
# Reset MONTH index to column
monthly_mean_zone = monthly_mean_zone.reset_index()
monthly_mean_zone
# Plot mean monthly temperatures for the 4 climate zones in Ghana
# Plot style use Seaborn
plt.style.use('seaborn')
#sns.set_style('whitegrid')
fig, ax11 = plt.subplots(1, figsize=(10, 6))
x = monthly_mean_zone['MONTH']
y_1 = monthly_mean_zone['Zone_1']
y_2 = monthly_mean_zone['Zone_2']
y_3 = monthly_mean_zone['Zone_3']
y_4 = monthly_mean_zone['Zone_4']
# x, y limits of axes
plt.ylim(23, 33)
# Plot each lines on same axis
plt.plot(x, y_1, '.-g', label = 'Zone 1', linewidth = 2)
plt.plot(x, y_2, '.--c', label = 'Zone 2', linewidth = 2)
plt.plot(x, y_3, '.-.b', label = 'Zone 3', linewidth = 2)
plt.plot(x, y_4, '.:r', label = 'Zone 4', linewidth = 2)
leg = plt.legend()
# get the lines and texts inside legend box
leg_lines = leg.get_lines()
leg_texts = leg.get_texts()
# bulk-set the properties of all lines and texts
plt.setp(leg_lines, linewidth=4)
plt.setp(leg_texts, fontsize='x-large')
plt.title('Mean Monthly Climatic Zone Temperatures, Ghana', fontdict={'fontname': 'comic sans ms',
'fontsize': 15, 'weight': 'bold'})
# Labels for axes
plt.xlabel('Month', fontdict={'fontsize': 14})
plt.ylabel('Temperature °C', fontdict={'fontsize': 14})
# To save graph
plt.savefig('C:/Users/Narteh/earth_analytics/Gh_Climate_EDA/Graphs/zone_monthly_mean.png')
# Plot mean monthly temperatures for the 4 climate zones in Ghana in comparison to the national average
# Plot style use Seaborn
plt.style.use('seaborn')
#sns.set_style('whitegrid')
fig, ax11 = plt.subplots(1, figsize=(10, 6))
x = monthly_mean_zone['MONTH']
y_1 = monthly_mean_zone['Zone_1']
y_2 = monthly_mean_zone['Zone_2']
y_3 = monthly_mean_zone['Zone_3']
y_4 = monthly_mean_zone['Zone_4']
y_5 = monthly_mean['TEMP_C']
# x, y limits of axes
plt.ylim(23, 33)
# Plot each lines on same axis
plt.plot(x, y_1, '.-g', label = 'Zone 1', linewidth = 2)
plt.plot(x, y_2, '.--c', label = 'Zone 2', linewidth = 2)
plt.plot(x, y_3, '.-.b', label = 'Zone 3', linewidth = 2)
plt.plot(x, y_4, '.:r', label = 'Zone 4', linewidth = 2)
plt.plot(x, y_5, '-m', label = 'Average', linewidth = 2)
leg = plt.legend()
# get the lines and texts inside legend box
leg_lines = leg.get_lines()
leg_texts = leg.get_texts()
# bulk-set the properties of all lines and texts
plt.setp(leg_lines, linewidth=4)
plt.setp(leg_texts, fontsize='x-large')
plt.title('Mean Monthly National & Zonal Climatic Temperatures, Ghana', fontdict={'fontname': 'comic sans ms',
'fontsize': 15, 'weight': 'bold'})
# Labels for axes
plt.xlabel('Month', fontdict={'fontsize': 14})
plt.ylabel('Temperature °C', fontdict={'fontsize': 14})
# To save graph
plt.savefig('C:/Users/Narteh/earth_analytics/Gh_Climate_EDA/Graphs/zone_nat_monthly_mean.png')
```
|
github_jupyter
|
# Climate data source from https://www.ncdc.noaa.gov/cdo-web
# Period of record from 1973
# Contains data records of 17 stations of varying lengths
# Original units of variables - imperial units
%matplotlib inline
# Import libraries
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
# Seaborn for additional plotting and styling
import seaborn as sns
# Define file path to connect data source:
fp = 'C:/Users/Narteh/earth_analytics/Gh_Climate_EDA/data_weatherGH/1619559410439.csv'
# Read in data from the csv file
# Low memory set to false to remove memory errors
data = pd.read_csv(fp, parse_dates=['DATE'], dayfirst=True, low_memory=False)
# Print data types
data.dtypes
#Print column names
data.columns.values
# Check DATE type format
type(data['DATE'])
# Check dataframe shape (number of rows, number of columns)
data.shape
# Confirm output of dataframe
data.head()
# Check sum of no-data, not a number (NaN) values in the data set
data[['PRCP', 'TAVG', 'TMAX', 'TMIN']].isna().sum()
# Confirm output of parse date
data.loc[0, 'DATE'].day_name()
# Drop column Unnamed: 0, which contains station unique code
# Drop PRCP column, work only with temperature data
data.drop(columns=['Unnamed: 0', 'PRCP'], inplace=True)
# Check drop of Unnamed: 0 and PRCP column
data.tail()
# Rename column heading of Unnamed: 1 to TOWN and TAVG to TEMP for familiarity
data = data.rename(columns={'Unnamed: 1': 'TOWN', 'TAVG': 'TEMP'})
# Confirm rename of columns
data.head()
# Replace inadvertantly named double named towns falling under 2 columns to its original double word names & KIA for Kotoka International Airport
data.replace(to_replace=['AKIM', 'KOTOKA', 'KETE', 'SEFWI'],
value= ['AKIM ODA', 'KIA', 'KETE KRACHI', 'SEFWI WIASO'],
inplace=True)
# Check confirmation of output
data.head()
# Drop STATION column due to renamed town heading
data.drop(columns=['STATION'], inplace=True)
# Check drop of STATION column
data.head()
# The date of first observation
first_obs = data.at[0, "DATE"]
print('The first record of observation was on', data.at[0, "DATE"])
# Convert to string to change capital case to lower case for TOWN names
data['TOWN'] = data['TOWN'].astype(str)
data['TOWN'] = data['TOWN'].str.title()
# Check conversion string change
data.head()
# Convert imperial units to metric through def function
def fahr_to_celsius(temp_fahrenheit):
"""Function to convert Fahrenheit temperature into Celsius.
Parameters
----------
temp_fahrenheit: int | float
Input temperature in Fahrenheit (should be a number)
Returns
-------
Temperature in Celsius (float)
"""
# Convert the Fahrenheit into Celsius
converted_temp = (temp_fahrenheit - 32) / 1.8
return converted_temp # Use apply to convert each row of value
data[['TEMP_C', 'TMAX_C', 'TMIN_C']] = round(data[['TEMP', 'TMAX', 'TMIN']].apply(fahr_to_celsius), 2)
# Check conversion to metric units
data.head()
# Drop columns with fahrenheit units since they are no longer needed
data.drop(columns=['TEMP', 'TMAX', 'TMIN'], inplace=True)
# print head to evalaute
data.head()
# For later analysis
data['DayofWeek'] = data['DATE'].dt.day_name()
data.head()
# Take mean of mean temperatures of the data set
temp_mean = round(data['TEMP_C'].mean(), 2)
temp_mean
# Set DATE as index
data.set_index('DATE', inplace=True)
data.head()
# Mean daily temperature across all the weather stations
daily_temp = round(data['TEMP_C'].resample('D').mean(), 2)
daily_temp.head()
# Indicate the mean minimum and maximum daily temperature
print(daily_temp.min(),'°C')
print('')
print(daily_temp.max(),'°C')
# Show which dates the minimum and maximum daily mean temperature were recorded
min_daily = daily_temp.loc[daily_temp == 16.67]
print(min_daily)
print(' ')
max_daily = daily_temp.loc[daily_temp == 35.0]
print(max_daily)
# Mean monthly temperature across all the weather stations
monthly_temp = round(data['TEMP_C'].resample('M').mean(), 2)
monthly_temp.head()
# Indicate the mean minimum and maximum monthly temperature
min_monthly_temp = monthly_temp.min()
print(min_monthly_temp)
print('')
max_monthly_temp = monthly_temp.max()
print(max_monthly_temp)
# Show the date the mean minimum and maximum monthly temperature were recorded
print(monthly_temp.loc[monthly_temp == min_monthly_temp])
print(' ')
print(monthly_temp.loc[monthly_temp == max_monthly_temp])
# Mean yearly temperature across all the weather stations
yearly_temp = round(data[['TEMP_C', 'TMAX_C', 'TMIN_C']].resample('Y').mean(), 2)
# Check output
yearly_temp.head()
# Indicate the mean minimum and maximum yearly temperature
min_yearly_temp = yearly_temp['TEMP_C'].min()
print(min_yearly_temp)
print(' ')
max_yearly_temp = yearly_temp['TEMP_C'].max()
print(max_yearly_temp)
# Show the year the minimum and maximum yearly temperature were recorded
print(yearly_temp['TEMP_C'].loc[yearly_temp['TEMP_C'] == min_yearly_temp])
print(' ')
print(yearly_temp['TEMP_C'].loc[yearly_temp['TEMP_C'] == max_yearly_temp])
# Reset DATE index to column
yearly_temp = yearly_temp.reset_index()
yearly_temp.head()
# Convert DATE to year using dt.year
yearly_temp['YEAR']= yearly_temp['DATE'].dt.year
yearly_temp.head()
# Plot mean yearly temperatures
import matplotlib.dates as mdates
import matplotlib.ticker as ticker
# Plot style use Seaborn
plt.style.use('seaborn-whitegrid')
fig, ax = plt.subplots(1, figsize=(10, 6))
# Data values for X and Y
x = yearly_temp.YEAR
y = yearly_temp.TEMP_C
# Plot x, y values & parameters
plt.plot(x, y, color = 'red', linestyle='-', marker='o', mfc='blue')
# Highlight coalescencing temps around 28 °C
plt.plot(x[-6:], y[-6:], 'y*', ms='12')
fig.autofmt_xdate()
# Plot title & attributes
plt.title('Yearly Mean Temperatures, Ghana', fontdict={'fontname': 'comic sans ms',
'fontsize': 15, 'weight': 'bold'})
# Add label to the plot
#plt.text(1983, 30.190, ' Severe Drought & Bushfires', fontsize='large')
# Annotate peak temp with description & arrow
ax.annotate('Severe drought & bushfires', fontsize='large', weight='semibold',
xy=(1983, 30.190), xytext=(1986, 29.5),
arrowprops=dict(arrowstyle='->',
connectionstyle='arc'),
xycoords='data',)
# Labels for axes
plt.xlabel('Year', fontdict={'fontsize': 14})
plt.ylabel('Temperature °C', fontdict={'fontsize': 14})
# legend
plt.legend(['Mean Temperature'], frameon=True, fontsize='x-large')
# major ticks locators
plt.xticks(np.arange(min(x), max(x), 4))
def setup(ax):
ax.xaxis.set_ticks_position('bottom')
ax.tick_params(which='minor', width=0.75, length=2.5)
setup(ax)
ax.xaxis.set_minor_locator(ticker.FixedLocator((x)))
plt.tight_layout()
# To save graph
plt.savefig('C:/Users/Narteh/earth_analytics/Gh_Climate_EDA/Graphs/yearly_mean_gh.jpg')
import pandas_bokeh
pandas_bokeh.output_notebook()
pd.set_option('plotting.backend', 'pandas_bokeh')
# Plot interactive map with Bokeh
from bokeh.models import HoverTool
ax = yearly_temp.plot(x = 'YEAR', y = 'TEMP_C', title='Yearly Mean Temperatures, Ghana',
xlabel = 'Year', ylabel = 'Temperature °C', xticks = (yearly_temp.YEAR[::4]),
legend = False, plot_data_points = True, hovertool_string= r'''<h5> @{YEAR} </h5>
<h5> @{TEMP_C} °C </h5>''') # Change hoovertool stringe text to values in dataframe only
# set output to static HTML file
from bokeh.plotting import figure, output_file, show
output_file(filename='Ghana_Climate_Interactive.html', title='Ghana Climate HTML file')
#show(ax)
# Group by towns
sta_town_grp = data.groupby(['TOWN'])
# Check groupy operation with one example
sta_town_grp.get_group('Akuse').head()
# Filter out the maximum temps ever recorded in each town from TMAX_C
sta_town_max = sta_town_grp['TMAX_C'].max()
sta_town_max = sta_town_max.sort_values(ascending=False)
sta_town_max
# Filter out row values of the date of occurence of highest maximum temp for first few towns
data.loc[(data.TMAX_C == 48.89) | (data.TMAX_C == 43.89)]
# Sort DATE at index to allow slicing, done chronologically
data.sort_index(inplace=True)
# Calculate decadel mean temperatures
avg_temp_1970s = data['1970': '1979']['TEMP_C'].mean()
avg_temp_1980s = data['1980': '1989']['TEMP_C'].mean()
avg_temp_1990s = data['1990': '1999']['TEMP_C'].mean()
avg_temp_2000s = data['2000': '2009']['TEMP_C'].mean()
avg_temp_2010s = data['2010': '2019']['TEMP_C'].mean()
# Print to screen the mean decadal temperatures
print(round(avg_temp_1970s, 2))
print(' ')
print(round(avg_temp_1980s, 2))
print(' ')
print(round(avg_temp_1990s, 2))
print(' ')
print(round(avg_temp_2000s, 2))
print(' ')
print(round(avg_temp_2010s, 2))
# Take monthly mean of all daily records
monthly_data = round(data.resample('M').mean(), 2)
monthly_data.head()
# Return to column heading
monthly_data.reset_index(inplace=True)
# Convert to date month
monthly_data['MONTH'] = monthly_data['DATE'].dt.month_name().str.slice(stop=3)
monthly_data.head()
# Take mean of all indivdual months
monthly_mean = round(monthly_data.groupby(['MONTH']).mean(), 2)
monthly_mean.head()
# Create new order for actual calender months
new_order = ['Jan', 'Feb', 'Mar', 'Apr', 'May', 'Jun', 'Jul',
'Aug', 'Sep', 'Oct', 'Nov', 'Dec']
monthly_mean = monthly_mean.reindex(new_order, axis=0)
# check output of reindex
monthly_mean.head()
monthly_mean = monthly_mean.reset_index()
monthly_mean.head()
# Plot mean monthly temperatures for the period of the record
# Plot style use Seaborn
plt.style.use('seaborn')
fig, ax1 = plt.subplots(1, figsize=(10, 6))
x = monthly_mean['MONTH']
y = monthly_mean['TEMP_C']
plt.ylim(23, 31)
plt.plot(x, y, color = 'tomato', linestyle='-', marker='o', mfc='orange', linewidth = 3, markersize = 8)
plt.grid(axis = 'x')
plt.title('Monthly Mean Temperatures, Ghana', fontdict={'fontname': 'comic sans ms',
'fontsize': 15, 'weight': 'bold'})
# Labels for axes
plt.xlabel('Month', fontdict={'fontsize': 14})
plt.ylabel('Temperature °C', fontdict={'fontsize': 14})
# To save graph
plt.savefig('C:/Users/Narteh/earth_analytics/Gh_Climate_EDA/Graphs/monthly_mean_temp.jpg')
# Use bokeh for interactivity
ax1 = monthly_mean.plot.line(x = monthly_mean['MONTH'],
y = 'TEMP_C', title='Mean Monthly Temperatures, Ghana',
xlabel ='Month', ylabel='Temperature °C', ylim=(21, 33),
marker='o', plot_data_points_size = 8, line_width = 3,
legend = False, color = 'tomato', plot_data_points=True,
hovertool_string= r'''<h5> @{MONTH} </h5>
<h5> @{TEMP_C} °C </h5>''') # Change hoovertool stringe text to values in dataframe only
# set output to static HTML file
# Remove x grid line for different fig style
ax1.xgrid.grid_line_color = None
output_file(filename='mean_monthly_gh.html', title='Monthly Mean Temperatures, Ghana')
show(ax1)
# Create new column of Climatic zones in Ghana according "Climatic Regions of Ghana (Abass, 2009)
# To help evaluate changes since the period of the record
data['CLIMATE_ZONES'] = data['TOWN'].map({'Tamale': 'Zone_4', 'Wenchi': 'Zone_4', 'Navrongo': 'Zone_4',
'Wa': 'Zone_4', 'Bole': 'Zone_4', 'Kete Krachi': 'Zone_4',
'Akim Oda': 'Zone_3', 'Sefwi Wiaso': 'Zone_3', 'Koforidua': 'Zone_3',
'Kumasi': 'Zone_3', 'Sunyani': 'Zone_3', 'Ho': 'Zone_3','Kia': 'Zone_2',
'Akuse': 'Zone_2', 'Ada': 'Zone_2', 'Axim': 'Zone_1', 'Takoradi': 'Zone_1'})
data.tail()
# Find mean yearly temp per climate zone
climate_zones_yearly = data.groupby('CLIMATE_ZONES').resample('Y').mean()
climate_zones_yearly.head()
# Return to column heading without column index
climate_zones_yearly.reset_index(inplace=True)
climate_zones_yearly.head()
# Pivot table to get temps for each climate zone per year in column format
climate_zones_yearly = climate_zones_yearly.pivot_table(values = 'TEMP_C', index = 'DATE', columns = 'CLIMATE_ZONES')
climate_zones_yearly.head()
# Check for missing values
climate_zones_yearly.isnull().sum()
# Drop NA values due to grouping operation
climate_zones_yearly = climate_zones_yearly.dropna()
climate_zones_yearly.head()
# Plot mean monthly temperatures for the 4 climate zones in Ghana
# Plot style use Seaborn
plt.style.use('seaborn')
#sns.set_style('whitegrid')
fig, ax1 = plt.subplots(1, figsize=(10, 6))
x = climate_zones_yearly.reset_index()['DATE']
y_1 = climate_zones_yearly['Zone_1']
y_2 = climate_zones_yearly['Zone_2']
y_3 = climate_zones_yearly['Zone_3']
y_4 = climate_zones_yearly['Zone_4']
# x, y limits of axes
plt.ylim(23, 33)
# Plot each lines on same axis
plt.plot(x, y_1, '.-g', label = 'Zone 1', linewidth = 2)
plt.plot(x, y_2, '.--c', label = 'Zone 2', linewidth = 2)
plt.plot(x, y_3, '.-.b', label = 'Zone 3', linewidth = 2)
plt.plot(x, y_4, '.:r', label = 'Zone 4', linewidth = 2)
leg = plt.legend()
# get the lines and texts inside legend box
leg_lines = leg.get_lines()
leg_texts = leg.get_texts()
# bulk-set the properties of all lines and texts
plt.setp(leg_lines, linewidth=4)
plt.setp(leg_texts, fontsize='x-large')
plt.title('Mean Yearly Temperature per Climate Zone, Ghana',
fontdict={'fontname': 'comic sans ms',
'fontsize': 15, 'weight': 'bold'})
# Labels for axes
plt.xlabel('Year', fontdict={'fontsize': 14})
plt.ylabel('Temperature °C', fontdict={'fontsize': 14})
# To save graph
plt.savefig('C:/Users/Narteh/earth_analytics/Gh_Climate_EDA/Graphs/zone_yearly_mean.png')
# Find mean monthly temp per climate zone
climate_zones_monthly = data.groupby('CLIMATE_ZONES').resample('M').mean()
climate_zones_monthly.head()
# Return to column heading without column index
climate_zones_monthly.reset_index(inplace=True)
# Convert to date month in short format for temps months evaluation
climate_zones_monthly['MONTH'] = climate_zones_monthly['DATE'].dt.month_name().str.slice(stop=3)
climate_zones_monthly.tail()
# Take mean of each mean monthly temp for each zone
monthly_mean_zone = climate_zones_monthly.groupby(['CLIMATE_ZONES', 'MONTH'], as_index=False).mean()
monthly_mean_zone.head()
# Re order months by custom sorting per calendar months
months_categories = ('Jan', 'Feb', 'Mar', 'Apr', 'May', 'Jun', 'Jul', 'Aug', 'Sep', 'Oct', 'Nov', 'Dec')
monthly_mean_zone['MONTH'] = pd.Categorical(monthly_mean_zone['MONTH'], categories = months_categories)
monthly_mean_zone.head()
# Sort values under MONTH by calender months
monthly_mean_zone = monthly_mean_zone.sort_values(by = 'MONTH')
monthly_mean_zone.head()
# Pivot table to get temps for each climate zone per month in column format
monthly_mean_zone = monthly_mean_zone.pivot_table(values = 'TEMP_C', index = 'MONTH', columns = 'CLIMATE_ZONES')
monthly_mean_zone.head()
# Reset MONTH index to column
monthly_mean_zone = monthly_mean_zone.reset_index()
monthly_mean_zone
# Plot mean monthly temperatures for the 4 climate zones in Ghana
# Plot style use Seaborn
plt.style.use('seaborn')
#sns.set_style('whitegrid')
fig, ax11 = plt.subplots(1, figsize=(10, 6))
x = monthly_mean_zone['MONTH']
y_1 = monthly_mean_zone['Zone_1']
y_2 = monthly_mean_zone['Zone_2']
y_3 = monthly_mean_zone['Zone_3']
y_4 = monthly_mean_zone['Zone_4']
# x, y limits of axes
plt.ylim(23, 33)
# Plot each lines on same axis
plt.plot(x, y_1, '.-g', label = 'Zone 1', linewidth = 2)
plt.plot(x, y_2, '.--c', label = 'Zone 2', linewidth = 2)
plt.plot(x, y_3, '.-.b', label = 'Zone 3', linewidth = 2)
plt.plot(x, y_4, '.:r', label = 'Zone 4', linewidth = 2)
leg = plt.legend()
# get the lines and texts inside legend box
leg_lines = leg.get_lines()
leg_texts = leg.get_texts()
# bulk-set the properties of all lines and texts
plt.setp(leg_lines, linewidth=4)
plt.setp(leg_texts, fontsize='x-large')
plt.title('Mean Monthly Climatic Zone Temperatures, Ghana', fontdict={'fontname': 'comic sans ms',
'fontsize': 15, 'weight': 'bold'})
# Labels for axes
plt.xlabel('Month', fontdict={'fontsize': 14})
plt.ylabel('Temperature °C', fontdict={'fontsize': 14})
# To save graph
plt.savefig('C:/Users/Narteh/earth_analytics/Gh_Climate_EDA/Graphs/zone_monthly_mean.png')
# Plot mean monthly temperatures for the 4 climate zones in Ghana in comparison to the national average
# Plot style use Seaborn
plt.style.use('seaborn')
#sns.set_style('whitegrid')
fig, ax11 = plt.subplots(1, figsize=(10, 6))
x = monthly_mean_zone['MONTH']
y_1 = monthly_mean_zone['Zone_1']
y_2 = monthly_mean_zone['Zone_2']
y_3 = monthly_mean_zone['Zone_3']
y_4 = monthly_mean_zone['Zone_4']
y_5 = monthly_mean['TEMP_C']
# x, y limits of axes
plt.ylim(23, 33)
# Plot each lines on same axis
plt.plot(x, y_1, '.-g', label = 'Zone 1', linewidth = 2)
plt.plot(x, y_2, '.--c', label = 'Zone 2', linewidth = 2)
plt.plot(x, y_3, '.-.b', label = 'Zone 3', linewidth = 2)
plt.plot(x, y_4, '.:r', label = 'Zone 4', linewidth = 2)
plt.plot(x, y_5, '-m', label = 'Average', linewidth = 2)
leg = plt.legend()
# get the lines and texts inside legend box
leg_lines = leg.get_lines()
leg_texts = leg.get_texts()
# bulk-set the properties of all lines and texts
plt.setp(leg_lines, linewidth=4)
plt.setp(leg_texts, fontsize='x-large')
plt.title('Mean Monthly National & Zonal Climatic Temperatures, Ghana', fontdict={'fontname': 'comic sans ms',
'fontsize': 15, 'weight': 'bold'})
# Labels for axes
plt.xlabel('Month', fontdict={'fontsize': 14})
plt.ylabel('Temperature °C', fontdict={'fontsize': 14})
# To save graph
plt.savefig('C:/Users/Narteh/earth_analytics/Gh_Climate_EDA/Graphs/zone_nat_monthly_mean.png')
| 0.778818 | 0.848031 |
## Interpretability - Text Explainers
In this example, we use LIME and Kernel SHAP explainers to explain a text classification model.
First we import the packages and define some UDFs and a plotting function we will need later.
```
from pyspark.sql.functions import *
from pyspark.sql.types import *
from pyspark.ml.feature import StopWordsRemover, HashingTF, IDF, Tokenizer
from pyspark.ml import Pipeline
from pyspark.ml.classification import LogisticRegression
from synapse.ml.explainers import *
from synapse.ml.featurize.text import TextFeaturizer
import os
if os.environ.get("AZURE_SERVICE", None) == "Microsoft.ProjectArcadia":
from pyspark.sql import SparkSession
spark = SparkSession.builder.getOrCreate()
from notebookutils.visualization import display
vec2array = udf(lambda vec: vec.toArray().tolist(), ArrayType(FloatType()))
vec_access = udf(lambda v, i: float(v[i]), FloatType())
```
Load training data, and convert rating to binary label.
```
data = (
spark.read.parquet("wasbs://[email protected]/BookReviewsFromAmazon10K.parquet")
.withColumn("label", (col("rating") > 3).cast(LongType()))
.select("label", "text")
.cache()
)
display(data)
```
We train a text classification model, and randomly sample 10 rows to explain.
```
train, test = data.randomSplit([0.60, 0.40])
pipeline = Pipeline(
stages=[
TextFeaturizer(
inputCol="text",
outputCol="features",
useStopWordsRemover=True,
useIDF=True,
minDocFreq=20,
numFeatures=1 << 16,
),
LogisticRegression(maxIter=100, regParam=0.005, labelCol="label", featuresCol="features"),
]
)
model = pipeline.fit(train)
prediction = model.transform(test)
explain_instances = prediction.orderBy(rand()).limit(10)
def plotConfusionMatrix(df, label, prediction, classLabels):
from synapse.ml.plot import confusionMatrix
import matplotlib.pyplot as plt
fig = plt.figure(figsize=(4.5, 4.5))
confusionMatrix(df, label, prediction, classLabels)
if os.environ.get("AZURE_SERVICE", None) == "Microsoft.ProjectArcadia":
plt.show()
else:
display(fig)
plotConfusionMatrix(model.transform(test), "label", "prediction", [0, 1])
```
First we use the LIME text explainer to explain the model's predicted probability for a given observation.
```
lime = TextLIME(
model=model,
outputCol="weights",
inputCol="text",
targetCol="probability",
targetClasses=[1],
tokensCol="tokens",
samplingFraction=0.7,
numSamples=2000,
)
lime_results = (
lime.transform(explain_instances)
.select("tokens", "weights", "r2", "probability", "text")
.withColumn("probability", vec_access("probability", lit(1)))
.withColumn("weights", vec2array(col("weights").getItem(0)))
.withColumn("r2", vec_access("r2", lit(0)))
.withColumn("tokens_weights", arrays_zip("tokens", "weights"))
)
display(lime_results.select("probability", "r2", "tokens_weights", "text").orderBy(col("probability").desc()))
```
Then we use the Kernel SHAP text explainer to explain the model's predicted probability for a given observation.
> Notice that we drop the base value from the SHAP output before displaying the SHAP values. The base value is the model output for an empty string.
```
shap = TextSHAP(
model=model,
outputCol="shaps",
inputCol="text",
targetCol="probability",
targetClasses=[1],
tokensCol="tokens",
numSamples=5000,
)
shap_results = (
shap.transform(explain_instances)
.select("tokens", "shaps", "r2", "probability", "text")
.withColumn("probability", vec_access("probability", lit(1)))
.withColumn("shaps", vec2array(col("shaps").getItem(0)))
.withColumn("shaps", slice(col("shaps"), lit(2), size(col("shaps"))))
.withColumn("r2", vec_access("r2", lit(0)))
.withColumn("tokens_shaps", arrays_zip("tokens", "shaps"))
)
display(shap_results.select("probability", "r2", "tokens_shaps", "text").orderBy(col("probability").desc()))
```
|
github_jupyter
|
from pyspark.sql.functions import *
from pyspark.sql.types import *
from pyspark.ml.feature import StopWordsRemover, HashingTF, IDF, Tokenizer
from pyspark.ml import Pipeline
from pyspark.ml.classification import LogisticRegression
from synapse.ml.explainers import *
from synapse.ml.featurize.text import TextFeaturizer
import os
if os.environ.get("AZURE_SERVICE", None) == "Microsoft.ProjectArcadia":
from pyspark.sql import SparkSession
spark = SparkSession.builder.getOrCreate()
from notebookutils.visualization import display
vec2array = udf(lambda vec: vec.toArray().tolist(), ArrayType(FloatType()))
vec_access = udf(lambda v, i: float(v[i]), FloatType())
data = (
spark.read.parquet("wasbs://[email protected]/BookReviewsFromAmazon10K.parquet")
.withColumn("label", (col("rating") > 3).cast(LongType()))
.select("label", "text")
.cache()
)
display(data)
train, test = data.randomSplit([0.60, 0.40])
pipeline = Pipeline(
stages=[
TextFeaturizer(
inputCol="text",
outputCol="features",
useStopWordsRemover=True,
useIDF=True,
minDocFreq=20,
numFeatures=1 << 16,
),
LogisticRegression(maxIter=100, regParam=0.005, labelCol="label", featuresCol="features"),
]
)
model = pipeline.fit(train)
prediction = model.transform(test)
explain_instances = prediction.orderBy(rand()).limit(10)
def plotConfusionMatrix(df, label, prediction, classLabels):
from synapse.ml.plot import confusionMatrix
import matplotlib.pyplot as plt
fig = plt.figure(figsize=(4.5, 4.5))
confusionMatrix(df, label, prediction, classLabels)
if os.environ.get("AZURE_SERVICE", None) == "Microsoft.ProjectArcadia":
plt.show()
else:
display(fig)
plotConfusionMatrix(model.transform(test), "label", "prediction", [0, 1])
lime = TextLIME(
model=model,
outputCol="weights",
inputCol="text",
targetCol="probability",
targetClasses=[1],
tokensCol="tokens",
samplingFraction=0.7,
numSamples=2000,
)
lime_results = (
lime.transform(explain_instances)
.select("tokens", "weights", "r2", "probability", "text")
.withColumn("probability", vec_access("probability", lit(1)))
.withColumn("weights", vec2array(col("weights").getItem(0)))
.withColumn("r2", vec_access("r2", lit(0)))
.withColumn("tokens_weights", arrays_zip("tokens", "weights"))
)
display(lime_results.select("probability", "r2", "tokens_weights", "text").orderBy(col("probability").desc()))
shap = TextSHAP(
model=model,
outputCol="shaps",
inputCol="text",
targetCol="probability",
targetClasses=[1],
tokensCol="tokens",
numSamples=5000,
)
shap_results = (
shap.transform(explain_instances)
.select("tokens", "shaps", "r2", "probability", "text")
.withColumn("probability", vec_access("probability", lit(1)))
.withColumn("shaps", vec2array(col("shaps").getItem(0)))
.withColumn("shaps", slice(col("shaps"), lit(2), size(col("shaps"))))
.withColumn("r2", vec_access("r2", lit(0)))
.withColumn("tokens_shaps", arrays_zip("tokens", "shaps"))
)
display(shap_results.select("probability", "r2", "tokens_shaps", "text").orderBy(col("probability").desc()))
| 0.771069 | 0.954647 |
```
import os
import glob
from collections import defaultdict
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import watchcbb.utils as utils
dfs = []
for fname in glob.glob("../data/game_data/*.csv"):
dfs.append(pd.read_csv(fname))
df = pd.concat(dfs)
df.Date = pd.to_datetime(df.Date)
df = df.sort_values("Date").reset_index()
# df = df.query("Season==2019").reset_index()
df["poss"] = 0.5*(df["WFGA"] + 0.44*df["WFTA"] - df["WOR"] + df["WTO"] + df["LFGA"] + 0.44*df["LFTA"] - df["LOR"] + df["LTO"])
print("Shape:",df.shape)
df.head(10)
first, second = utils.partition_games(df, frac=0.7)
print(df.iloc[first].shape[0], df.iloc[second].shape[0])
season_stats_dict = utils.compute_season_stats(df.iloc[first])
season_stats_df = utils.stats_dict_to_df(season_stats_dict)
utils.add_advanced_stats(season_stats_df)
season_stats_dict = utils.stats_df_to_dict(season_stats_df)
print(season_stats_df.shape)
season_stats_df.head()
season_stats_df.query('year==2019').sort_values('rawpace').tail()
year = 2019
tid1, tid2 = 'north-carolina', 'virginia'
poss1 = df.query(f'Season=={year} & (WTeamID=="{tid1}" | LTeamID=="tid1")').poss.values
poss2 = df.query(f'Season=={year} & (WTeamID=="{tid2}" | LTeamID=="tid2")').poss.values
plt.figure(figsize=(9,7))
plt.hist(poss1, bins=np.linspace(50,100,26), histtype='step', lw=2, label=str(year)+' '+tid1, color='c')
plt.hist(poss2, bins=np.linspace(50,100,26), histtype='step', lw=2, label=str(year)+' '+tid2, color='r')
plt.legend(fontsize='xx-large')
plt.xlabel('Pace', fontsize='x-large')
mean_poss = {year:season_stats_df.query(f'year=={year}').rawpace.mean() for year in season_stats_df.year.unique()}
for i,row in season_stats_df.iterrows():
season_stats_dict[row.year][row.team_id]["rawpace"] = row.rawpace
def predict_poss(row):
p1 = season_stats_dict[row.Season][row.WTeamID]["rawpace"]
p2 = season_stats_dict[row.Season][row.WTeamID]["rawpace"]
return p1*p2/mean_poss[row.Season]
pred_poss = df.iloc[second].apply(predict_poss, axis=1).values
from sklearn.linear_model import LinearRegression
plt.figure(figsize=(9,7))
xs = pred_poss.reshape(-1,1)
ys = df.iloc[second].poss.values.reshape(-1,1)
plt.scatter(xs, ys, s=20, alpha=0.05)
linreg = LinearRegression()
linreg.fit(xs, ys)
linreg.score(xs,ys)
xl = np.array([50,100]).reshape(-1,1)
yl = linreg.predict(xl)
plt.plot(xl, yl, 'r-')
plt.figure(figsize=(9,8))
xs = season_stats_df.query('year>=2016').Teff
ys = season_stats_df.query('year>=2016').Oeff
plt.scatter(xs, ys, s=20, alpha=0.3)
plt.xlabel("Offensive efficiency", fontsize='large')
plt.ylabel("Defensive efficiency", fontsize='large')
season_stats_df.info()
```
|
github_jupyter
|
import os
import glob
from collections import defaultdict
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import watchcbb.utils as utils
dfs = []
for fname in glob.glob("../data/game_data/*.csv"):
dfs.append(pd.read_csv(fname))
df = pd.concat(dfs)
df.Date = pd.to_datetime(df.Date)
df = df.sort_values("Date").reset_index()
# df = df.query("Season==2019").reset_index()
df["poss"] = 0.5*(df["WFGA"] + 0.44*df["WFTA"] - df["WOR"] + df["WTO"] + df["LFGA"] + 0.44*df["LFTA"] - df["LOR"] + df["LTO"])
print("Shape:",df.shape)
df.head(10)
first, second = utils.partition_games(df, frac=0.7)
print(df.iloc[first].shape[0], df.iloc[second].shape[0])
season_stats_dict = utils.compute_season_stats(df.iloc[first])
season_stats_df = utils.stats_dict_to_df(season_stats_dict)
utils.add_advanced_stats(season_stats_df)
season_stats_dict = utils.stats_df_to_dict(season_stats_df)
print(season_stats_df.shape)
season_stats_df.head()
season_stats_df.query('year==2019').sort_values('rawpace').tail()
year = 2019
tid1, tid2 = 'north-carolina', 'virginia'
poss1 = df.query(f'Season=={year} & (WTeamID=="{tid1}" | LTeamID=="tid1")').poss.values
poss2 = df.query(f'Season=={year} & (WTeamID=="{tid2}" | LTeamID=="tid2")').poss.values
plt.figure(figsize=(9,7))
plt.hist(poss1, bins=np.linspace(50,100,26), histtype='step', lw=2, label=str(year)+' '+tid1, color='c')
plt.hist(poss2, bins=np.linspace(50,100,26), histtype='step', lw=2, label=str(year)+' '+tid2, color='r')
plt.legend(fontsize='xx-large')
plt.xlabel('Pace', fontsize='x-large')
mean_poss = {year:season_stats_df.query(f'year=={year}').rawpace.mean() for year in season_stats_df.year.unique()}
for i,row in season_stats_df.iterrows():
season_stats_dict[row.year][row.team_id]["rawpace"] = row.rawpace
def predict_poss(row):
p1 = season_stats_dict[row.Season][row.WTeamID]["rawpace"]
p2 = season_stats_dict[row.Season][row.WTeamID]["rawpace"]
return p1*p2/mean_poss[row.Season]
pred_poss = df.iloc[second].apply(predict_poss, axis=1).values
from sklearn.linear_model import LinearRegression
plt.figure(figsize=(9,7))
xs = pred_poss.reshape(-1,1)
ys = df.iloc[second].poss.values.reshape(-1,1)
plt.scatter(xs, ys, s=20, alpha=0.05)
linreg = LinearRegression()
linreg.fit(xs, ys)
linreg.score(xs,ys)
xl = np.array([50,100]).reshape(-1,1)
yl = linreg.predict(xl)
plt.plot(xl, yl, 'r-')
plt.figure(figsize=(9,8))
xs = season_stats_df.query('year>=2016').Teff
ys = season_stats_df.query('year>=2016').Oeff
plt.scatter(xs, ys, s=20, alpha=0.3)
plt.xlabel("Offensive efficiency", fontsize='large')
plt.ylabel("Defensive efficiency", fontsize='large')
season_stats_df.info()
| 0.348756 | 0.323981 |
# DEMO: Survey module functionalities
This notebook provides a demo of how to utilise the survey module.
```
import sys
sys.path.append('../')
import numpy as np
import pandas as pd
import niimpy
from niimpy.survey import *
from niimpy.EDA import EDA_categorical
```
## Load data
We will load a mock survey data file.
```
# Load a mock dataframe
df = niimpy.read_csv('mock-survey.csv')
df.head()
```
## Preprocessing
The dataframe's columns are raw questions from a survey. Some questions belong to a specific category, so we will annotate them with ids. The id is constructed from a prefix (the questionnaire category: GAD, PHQ, PSQI etc.), followed by the question number (1,2,3). Similarly, we will also the answers to meaningful numerical values.
**Note:** It's important that the dataframe follows the below schema before passing into niimpy.
```
# Convert column name to id, based on provided mappers from niimpy
col_id = {**PHQ2_MAP, **PSQI_MAP, **PSS10_MAP, **PANAS_MAP, **GAD2_MAP}
selected_cols = [col for col in df.columns if col in col_id.keys()]
# Convert from wide to long format
m_df = pd.melt(df, id_vars=['user', 'age', 'gender'], value_vars=selected_cols, var_name='question', value_name='raw_answer')
# Assign questions to codes
m_df['id'] = m_df['question'].replace(col_id)
m_df.head()
# Transform raw answers to numerical values
m_df['answer'] = niimpy.survey.convert_to_numerical_answer(m_df, answer_col = 'raw_answer',
question_id = 'id', id_map=ID_MAP_PREFIX, use_prefix=True)
m_df.head()
```
We can also make a summary of the questionaire's score. This function can describe aggregated score over the whole population, or specific subgroups.
```
d = niimpy.survey.print_statistic(m_df, group='gender')
pd.DataFrame(d)
```
## Visualization
We can now make some plots for the preprocessed data frame. First, we can display the summary for a specific question.
```
fig = niimpy.EDA.EDA_categorical.questionnaire_summary(m_df, question = 'PHQ2_1', column = 'answer',
title='PHQ2_1', xlabel='value', ylabel='count',
width=900, height=400)
fig.show()
```
We can also display the summary for each subgroup.
```
fig = niimpy.EDA.EDA_categorical.questionnaire_grouped_summary(m_df, question='PSS10_9', group='gender',
title='PSS10_9',
xlabel='score', ylabel='count',
width=800, height=600)
fig.show()
```
With some quick preprocessing, we can display the score distribution of each questionaire.
```
pss_sum_df = m_df[m_df['id'].str.startswith('PSS')] \
.groupby(['user', 'gender']) \
.agg({'answer':sum}) \
.reset_index()
pss_sum_df['id'] = 'PSS'
fig = niimpy.EDA.EDA_categorical.questionnaire_grouped_summary(pss_sum_df, question='PSS', group='gender',
title='PSS10',
xlabel='score', ylabel='count',
width=800, height=600)
fig.show()
```
|
github_jupyter
|
import sys
sys.path.append('../')
import numpy as np
import pandas as pd
import niimpy
from niimpy.survey import *
from niimpy.EDA import EDA_categorical
# Load a mock dataframe
df = niimpy.read_csv('mock-survey.csv')
df.head()
# Convert column name to id, based on provided mappers from niimpy
col_id = {**PHQ2_MAP, **PSQI_MAP, **PSS10_MAP, **PANAS_MAP, **GAD2_MAP}
selected_cols = [col for col in df.columns if col in col_id.keys()]
# Convert from wide to long format
m_df = pd.melt(df, id_vars=['user', 'age', 'gender'], value_vars=selected_cols, var_name='question', value_name='raw_answer')
# Assign questions to codes
m_df['id'] = m_df['question'].replace(col_id)
m_df.head()
# Transform raw answers to numerical values
m_df['answer'] = niimpy.survey.convert_to_numerical_answer(m_df, answer_col = 'raw_answer',
question_id = 'id', id_map=ID_MAP_PREFIX, use_prefix=True)
m_df.head()
d = niimpy.survey.print_statistic(m_df, group='gender')
pd.DataFrame(d)
fig = niimpy.EDA.EDA_categorical.questionnaire_summary(m_df, question = 'PHQ2_1', column = 'answer',
title='PHQ2_1', xlabel='value', ylabel='count',
width=900, height=400)
fig.show()
fig = niimpy.EDA.EDA_categorical.questionnaire_grouped_summary(m_df, question='PSS10_9', group='gender',
title='PSS10_9',
xlabel='score', ylabel='count',
width=800, height=600)
fig.show()
pss_sum_df = m_df[m_df['id'].str.startswith('PSS')] \
.groupby(['user', 'gender']) \
.agg({'answer':sum}) \
.reset_index()
pss_sum_df['id'] = 'PSS'
fig = niimpy.EDA.EDA_categorical.questionnaire_grouped_summary(pss_sum_df, question='PSS', group='gender',
title='PSS10',
xlabel='score', ylabel='count',
width=800, height=600)
fig.show()
| 0.430626 | 0.938237 |
# Testing with [pytest](https://docs.pytest.org/en/latest/) - part 2
```
# Let's make sure pytest and ipytest packages are installed
# ipytest is required for running pytest inside Jupyter notebooks
import sys
!{sys.executable} -m pip install pytest
!{sys.executable} -m pip install ipytest
import ipytest
ipytest.autoconfig()
import pytest
__file__ = 'testing2.ipynb'
```
## [`@pytest.fixture`](https://docs.pytest.org/en/latest/fixture.html#pytest-fixtures-explicit-modular-scalable)
Let's consider we have an implemention of `Person` class which we want to test.
```
# This would be e.g. in person.py
class Person:
def __init__(self, first_name, last_name, age):
self.first_name = first_name
self.last_name = last_name
self.age = age
@property
def full_name(self):
return '{} {}'.format(self.first_name, self.last_name)
@property
def as_dict(self):
return {'name': self.full_name, 'age': self.age}
def increase_age(self, years):
if years < 0:
raise ValueError('Can not make people younger :(')
self.age += years
```
You can easily create resusable testing code by using pytest fixtures. If you introduce your fixtures inside [_conftest.py_](https://docs.pytest.org/en/latest/fixture.html#conftest-py-sharing-fixture-functions), the fixtures are available for all your test cases. In general, the location of _conftest.py_ is at the root of your _tests_ directory.
```
# This would be in either conftest.py or test_person.py
@pytest.fixture()
def default_person():
person = Person(first_name='John', last_name='Doe', age=82)
return person
```
Then you can utilize `default_person` fixture in the actual test cases.
```
%%run_pytest[clean]
# These would be in test_person.py
def test_full_name(default_person): # Note: we use fixture as an argument of the test case
result = default_person.full_name
assert result == 'John Doe'
def test_as_dict(default_person):
expected = {'name': 'John Doe', 'age': 82}
result = default_person.as_dict
assert result == expected
def test_increase_age(default_person):
default_person.increase_age(1)
assert default_person.age == 83
default_person.increase_age(10)
assert default_person.age == 93
def test_increase_age_with_negative_number(default_person):
with pytest.raises(ValueError):
default_person.increase_age(-1)
```
By using a fixture, we could use the same `default_person` for all our four test cases!
In the `test_increase_age_with_negative_number` we used [`pytest.raises`](https://docs.pytest.org/en/latest/assert.html#assertions-about-expected-exceptions) to verify that an exception is raised.
## [`@pytest.mark.parametrize`](https://docs.pytest.org/en/latest/parametrize.html#pytest-mark-parametrize-parametrizing-test-functions)
Sometimes you want to test the same functionality with multiple different inputs. `pytest.mark.parametrize` is your solution for defining multiple different inputs with expected outputs. Let's consider the following implementation of `replace_names` function.
```
# This would be e.g. in string_manipulate.py
def replace_names(original_str, new_name):
"""Replaces names (uppercase words) of original_str by new_name"""
words = original_str.split()
manipulated_words = [new_name if w.istitle() else w for w in words]
return ' '.join(manipulated_words)
```
We can test the `replace_names` function with multiple inputs by using `pytest.mark.parametrize`.
```
%%run_pytest[clean]
# This would be in your test module
@pytest.mark.parametrize("original,new_name,expected", [
('this is Lisa', 'John Doe', 'this is John Doe'),
('how about Frank and Amy', 'John', 'how about John and John'),
('no names here', 'John Doe', 'no names here'),
])
def test_replace_names(original, new_name, expected):
result = replace_names(original, new_name)
assert result == expected
```
|
github_jupyter
|
# Let's make sure pytest and ipytest packages are installed
# ipytest is required for running pytest inside Jupyter notebooks
import sys
!{sys.executable} -m pip install pytest
!{sys.executable} -m pip install ipytest
import ipytest
ipytest.autoconfig()
import pytest
__file__ = 'testing2.ipynb'
# This would be e.g. in person.py
class Person:
def __init__(self, first_name, last_name, age):
self.first_name = first_name
self.last_name = last_name
self.age = age
@property
def full_name(self):
return '{} {}'.format(self.first_name, self.last_name)
@property
def as_dict(self):
return {'name': self.full_name, 'age': self.age}
def increase_age(self, years):
if years < 0:
raise ValueError('Can not make people younger :(')
self.age += years
# This would be in either conftest.py or test_person.py
@pytest.fixture()
def default_person():
person = Person(first_name='John', last_name='Doe', age=82)
return person
%%run_pytest[clean]
# These would be in test_person.py
def test_full_name(default_person): # Note: we use fixture as an argument of the test case
result = default_person.full_name
assert result == 'John Doe'
def test_as_dict(default_person):
expected = {'name': 'John Doe', 'age': 82}
result = default_person.as_dict
assert result == expected
def test_increase_age(default_person):
default_person.increase_age(1)
assert default_person.age == 83
default_person.increase_age(10)
assert default_person.age == 93
def test_increase_age_with_negative_number(default_person):
with pytest.raises(ValueError):
default_person.increase_age(-1)
# This would be e.g. in string_manipulate.py
def replace_names(original_str, new_name):
"""Replaces names (uppercase words) of original_str by new_name"""
words = original_str.split()
manipulated_words = [new_name if w.istitle() else w for w in words]
return ' '.join(manipulated_words)
%%run_pytest[clean]
# This would be in your test module
@pytest.mark.parametrize("original,new_name,expected", [
('this is Lisa', 'John Doe', 'this is John Doe'),
('how about Frank and Amy', 'John', 'how about John and John'),
('no names here', 'John Doe', 'no names here'),
])
def test_replace_names(original, new_name, expected):
result = replace_names(original, new_name)
assert result == expected
| 0.367384 | 0.983925 |
# <font color='blue'>Data Science Academy - Python Fundamentos - Capítulo 6</font>
## Download: http://github.com/dsacademybr
## Stream de Dados do Twitter com MongoDB, Pandas e Scikit Learn
## Preparando a Conexão com o Twitter
```
# Instala o pacote tweepy
!pip install tweepy
# Importando os módulos Tweepy, Datetime e Json
from tweepy.streaming import StreamListener
from tweepy import OAuthHandler
from tweepy import Stream
from datetime import datetime
import json
```
Veja no manual em pdf como criar sua API no Twitter e configure as suas chaves abaixo.
```
# Adicione aqui sua Consumer Key
consumer_key = "Chave"
# Adicione aqui sua Consumer Secret
consumer_secret = "Chave"
# Adicione aqui seu Access Token
access_token = "Chave"
# Adicione aqui seu Access Token Secret
access_token_secret = "Chave"
# Criando as chaves de autenticação
auth = OAuthHandler(consumer_key, consumer_secret)
auth.set_access_token(access_token, access_token_secret)
# Criando uma classe para capturar os stream de dados do Twitter e
# armazenar no MongoDB
class MyListener(StreamListener):
def on_data(self, dados):
tweet = json.loads(dados)
created_at = tweet["created_at"]
id_str = tweet["id_str"]
text = tweet["text"]
obj = {"created_at":created_at,"id_str":id_str,"text":text,}
tweetind = col.insert_one(obj).inserted_id
print (obj)
return True
# Criando o objeto mylistener
mylistener = MyListener()
# Criando o objeto mystream
mystream = Stream(auth, listener = mylistener)
```
## Preparando a Conexão com o MongoDB
```
# Importando do PyMongo o módulo MongoClient
from pymongo import MongoClient
# Criando a conexão ao MongoDB
client = MongoClient('localhost', 27017)
# Criando o banco de dados twitterdb
db = client.twitterdb
# Criando a collection "col"
col = db.tweets
# Criando uma lista de palavras chave para buscar nos Tweets
keywords = ['Big Data', 'Python', 'Data Mining', 'Data Science']
# Iniciando o filtro e gravando os tweets no MongoDB
mystream.filter(track=keywords)
mystream.disconnect()
```
## Coletando os Tweets
## --> Pressione o botão Stop na barra de ferramentas para encerrar a captura dos Tweets
## Consultando os Dados no MongoDB
```
mystream.disconnect()
# Verificando um documento no collection
col.find_one()
```
## Análise de Dados com Pandas e Scikit-Learn
```
# criando um dataset com dados retornados do MongoDB
dataset = [{"created_at": item["created_at"], "text": item["text"],} for item in col.find()]
# Importando o módulo Pandas para trabalhar com datasets em Python
import pandas as pd
# Criando um dataframe a partir do dataset
df = pd.DataFrame(dataset)
# Imprimindo o dataframe
df
# Importando o módulo Scikit Learn
from sklearn.feature_extraction.text import CountVectorizer
# Usando o método CountVectorizer para criar uma matriz de documentos
cv = CountVectorizer()
count_matrix = cv.fit_transform(df.text)
# Contando o número de ocorrências das principais palavras em nosso dataset
word_count = pd.DataFrame(cv.get_feature_names(), columns=["word"])
word_count["count"] = count_matrix.sum(axis=0).tolist()[0]
word_count = word_count.sort_values("count", ascending=False).reset_index(drop=True)
word_count[:50]
```
# Fim
### Obrigado - Data Science Academy - <a href="http://facebook.com/dsacademybr">facebook.com/dsacademybr</a>
|
github_jupyter
|
# Instala o pacote tweepy
!pip install tweepy
# Importando os módulos Tweepy, Datetime e Json
from tweepy.streaming import StreamListener
from tweepy import OAuthHandler
from tweepy import Stream
from datetime import datetime
import json
# Adicione aqui sua Consumer Key
consumer_key = "Chave"
# Adicione aqui sua Consumer Secret
consumer_secret = "Chave"
# Adicione aqui seu Access Token
access_token = "Chave"
# Adicione aqui seu Access Token Secret
access_token_secret = "Chave"
# Criando as chaves de autenticação
auth = OAuthHandler(consumer_key, consumer_secret)
auth.set_access_token(access_token, access_token_secret)
# Criando uma classe para capturar os stream de dados do Twitter e
# armazenar no MongoDB
class MyListener(StreamListener):
def on_data(self, dados):
tweet = json.loads(dados)
created_at = tweet["created_at"]
id_str = tweet["id_str"]
text = tweet["text"]
obj = {"created_at":created_at,"id_str":id_str,"text":text,}
tweetind = col.insert_one(obj).inserted_id
print (obj)
return True
# Criando o objeto mylistener
mylistener = MyListener()
# Criando o objeto mystream
mystream = Stream(auth, listener = mylistener)
# Importando do PyMongo o módulo MongoClient
from pymongo import MongoClient
# Criando a conexão ao MongoDB
client = MongoClient('localhost', 27017)
# Criando o banco de dados twitterdb
db = client.twitterdb
# Criando a collection "col"
col = db.tweets
# Criando uma lista de palavras chave para buscar nos Tweets
keywords = ['Big Data', 'Python', 'Data Mining', 'Data Science']
# Iniciando o filtro e gravando os tweets no MongoDB
mystream.filter(track=keywords)
mystream.disconnect()
mystream.disconnect()
# Verificando um documento no collection
col.find_one()
# criando um dataset com dados retornados do MongoDB
dataset = [{"created_at": item["created_at"], "text": item["text"],} for item in col.find()]
# Importando o módulo Pandas para trabalhar com datasets em Python
import pandas as pd
# Criando um dataframe a partir do dataset
df = pd.DataFrame(dataset)
# Imprimindo o dataframe
df
# Importando o módulo Scikit Learn
from sklearn.feature_extraction.text import CountVectorizer
# Usando o método CountVectorizer para criar uma matriz de documentos
cv = CountVectorizer()
count_matrix = cv.fit_transform(df.text)
# Contando o número de ocorrências das principais palavras em nosso dataset
word_count = pd.DataFrame(cv.get_feature_names(), columns=["word"])
word_count["count"] = count_matrix.sum(axis=0).tolist()[0]
word_count = word_count.sort_values("count", ascending=False).reset_index(drop=True)
word_count[:50]
| 0.447943 | 0.59564 |
### Imports
```
from sys import argv
import numpy as np
import pandas as pd
import scipy as sp
from scipy import ndimage
import matplotlib.pyplot as plt
from matplotlib.colors import Normalize
import shapefile as sf
from scipy.interpolate import RegularGridInterpolator
from gnam.model.gridmod3d import gridmod3d as gm
from gnam.model.bbox import bbox as bb
from shapely.geometry import Point, Polygon
```
### Unpickle Smooth Subsampled Model
```
#this is a pickled dictionary with 4D ndarray, and 1D meta data arrays
#ifilename = './subsamp_smooth_z10.0m_nam_model_vp_vs_rho_Q_props.npz'
ifilename = './subsamp_smooth_z200m_nam_model_vp_vs_rho_Q_props.npz'
#Unpickle
data = np.load(ifilename)
props = data['props'] #4D ndarray
#meta data arrays
xdata = data['xd']
ydata = data['yd']
zdata = data['zd']
print('xd:\n',xdata)
print('yd:\n',ydata)
print('zd:\n',zdata)
# Setup Coordinate related vars
xmin = xdata[0]
dx = xdata[1]
nx = int(xdata[2])
xmax = xmin + (nx-1)*dx
ymin = ydata[0]
dy = ydata[1]
ny = int(ydata[2])
ymax = ymin + (ny-1)*dy
zmin = zdata[0]
dz = zdata[1]
nz = int(zdata[2])
zmax = (-zmin) + (nz-1)*dz
nsub_props = props.shape[0]
axes_order = {'X':0,'Y':1,'Z':2} #this dict keeps track of axes order
gm3d = gm(props,nsub_props,axes_order,(nx,ny,nz),(dx,dy,dz),(xmin,ymin,zmin))
print('gm3d.shape:',gm3d.shape)
#free up some memory
del props
```
### Confirm axes order
```
gm3d.changeAxOrder({'X':2,'Y':1,'Z':0})
print(gm3d.shape)
gm3d.changeAxOrder({'X':0,'Y':1,'Z':2})
print(gm3d.shape)
gm3d.changeAxOrder({'X':1,'Y':2,'Z':0})
print(gm3d.shape)
gm3d.changeAxOrder({'X':0,'Y':1,'Z':2})
print(gm3d.shape)
```
### Setup all coordinates (also get bbox, etc...)
```
mysf = sf.Reader('FieldShapeFile/Groningen_field')
print('mysf:',mysf)
print('mysf.shapes():',mysf.shapes())
s = mysf.shape(0)
sub_dxyz = 200
mybbox = s.bbox #this will be used for slicing (look further down)
print('mybbox:',mybbox)
#shrink and create y coordinates for slicing box
vl = np.array([0,0.87*(mybbox[3]-mybbox[1])])
dvl = ((0.87*(mybbox[3]-mybbox[1]))**2)**0.5
nvl = dvl//sub_dxyz + 1
y = np.arange(nvl)*sub_dxyz
print('nvl:',nvl)
#shrink and create x coordinates for slicing box
vb = np.array([0.85*(mybbox[2]-mybbox[0]),0])
dvb = ((0.85*(mybbox[2]-mybbox[0]))**2)**0.5
nvb = dvb//sub_dxyz + 1
x = np.arange(nvb)*sub_dxyz
print('nvb:',nvb)
#create set of xy coordinates for slicing box
xy = np.transpose([np.tile(x, len(y)), np.repeat(y, len(x))])
print('xy.shape:',xy.shape)
#setup rotation matrices
degree = 30
theta = degree*np.pi/180
rm = np.array([[np.cos(theta),-np.sin(theta)],[np.sin(theta),np.cos(theta)]])
#rotate coordinates
for i in range(len(xy[:,0])):
xy[i,:] = rm.dot(xy[i,:])
#get translated coordinates
xshift = 12600
yshift = -2600
rxy = np.copy(xy)
rxy[:,0] += mybbox[0] + xshift
rxy[:,1] += mybbox[1] + yshift
print('rxy.shape:',rxy.shape)
```
### Slice Volume
```
import time
#get sliced subsurface volume
start = time.time()
slice_props = gm3d.sliceVolumeValsFromCoordsXY(x,y,rxy,local=False)
end = time.time()
print('runtime:', end - start)
```
### Pickle the interpolated model
```
import numpy as np
orrssslfqn = './rect_rot_subsamp_smooth_z' + str(dz) + 'm_nam_model_vp_vs_rho_Q_props.npz'
print(orrssslfqn)
np.savez_compressed(orrssslfqn,props=slice_props,xc=x,yc=y,rxyc=rxy)
```
### Unpickle the sliced volume if need be
```
ifilename = './rect_rot_subsamp_smooth_z200.0m_nam_model_vp_vs_rho_Q_props.npz'
#Unpickle
data = np.load(ifilename)
slice_props = data['props'] #4D ndarray
xc=data['xc']
yc=data['yc']
rxy=data['rxyc']
print('slice_props.shape',slice_props.shape)
sprops = np.copy(slice_props.reshape((4,31, 193, 146)),order='C')
print('sprops.shape:',sprops.shape)
print('gm3d.shape:',gm3d.shape)
rdep_surf = sprops[0,10,:,:].copy()
print(rdep_surf.shape)
print('nxy:',rdep_surf.shape[0]*rdep_surf.shape[1])
print('nrxy:', rxy.shape)
# get new min max to normalize surface
vp_min = np.min(rdep_surf)
vp_max = np.max(rdep_surf)
surf_norm = Normalize(vp_min,vp_max)
xy = np.transpose([np.tile(xc, len(yc)), np.repeat(yc, len(xc))])
print('xy.shape:',xy.shape)
print('xy:',xy)
fig, ax = plt.subplots(1,figsize=(6,6))
ax.scatter(rxy[:,0],rxy[:,1],s=1,c=rdep_surf.flatten(),cmap=plt.cm.jet,norm=surf_norm,zorder=0)
plt.show()
sub_surf = gm3d[0,10,:,:].copy()
print(sub_surf.shape)
print('nxy:',sub_surf.shape[0]*sub_surf.shape[1])
print('nrxy:', rxy.shape)
# get new min max to normalize surface
vp_min = np.min(rdep_surf)
vp_max = np.max(rdep_surf)
surf_norm = Normalize(vp_min,vp_max)
sxc =
xy = np.transpose([np.tile(xc, len(yc)), np.repeat(yc, len(xc))])
print('xy.shape:',xy.shape)
fig, ax = plt.subplots(1,figsize=(6,6))
ax.scatter(xy[:,0],xy[:,1],s=1,c=sub_surf.flatten(),cmap=plt.cm.jet,norm=surf_norm,zorder=0)
plt.show()
sprops = sprops.transpose(0,3,2,1).copy()
print('sprops.shape:',sprops.shape)
print(np.isfortran(sprops))
import
write_vtk_gridded_model_3d(fqpname,props,xdata,ydata,zdata):
```
|
github_jupyter
|
from sys import argv
import numpy as np
import pandas as pd
import scipy as sp
from scipy import ndimage
import matplotlib.pyplot as plt
from matplotlib.colors import Normalize
import shapefile as sf
from scipy.interpolate import RegularGridInterpolator
from gnam.model.gridmod3d import gridmod3d as gm
from gnam.model.bbox import bbox as bb
from shapely.geometry import Point, Polygon
#this is a pickled dictionary with 4D ndarray, and 1D meta data arrays
#ifilename = './subsamp_smooth_z10.0m_nam_model_vp_vs_rho_Q_props.npz'
ifilename = './subsamp_smooth_z200m_nam_model_vp_vs_rho_Q_props.npz'
#Unpickle
data = np.load(ifilename)
props = data['props'] #4D ndarray
#meta data arrays
xdata = data['xd']
ydata = data['yd']
zdata = data['zd']
print('xd:\n',xdata)
print('yd:\n',ydata)
print('zd:\n',zdata)
# Setup Coordinate related vars
xmin = xdata[0]
dx = xdata[1]
nx = int(xdata[2])
xmax = xmin + (nx-1)*dx
ymin = ydata[0]
dy = ydata[1]
ny = int(ydata[2])
ymax = ymin + (ny-1)*dy
zmin = zdata[0]
dz = zdata[1]
nz = int(zdata[2])
zmax = (-zmin) + (nz-1)*dz
nsub_props = props.shape[0]
axes_order = {'X':0,'Y':1,'Z':2} #this dict keeps track of axes order
gm3d = gm(props,nsub_props,axes_order,(nx,ny,nz),(dx,dy,dz),(xmin,ymin,zmin))
print('gm3d.shape:',gm3d.shape)
#free up some memory
del props
gm3d.changeAxOrder({'X':2,'Y':1,'Z':0})
print(gm3d.shape)
gm3d.changeAxOrder({'X':0,'Y':1,'Z':2})
print(gm3d.shape)
gm3d.changeAxOrder({'X':1,'Y':2,'Z':0})
print(gm3d.shape)
gm3d.changeAxOrder({'X':0,'Y':1,'Z':2})
print(gm3d.shape)
mysf = sf.Reader('FieldShapeFile/Groningen_field')
print('mysf:',mysf)
print('mysf.shapes():',mysf.shapes())
s = mysf.shape(0)
sub_dxyz = 200
mybbox = s.bbox #this will be used for slicing (look further down)
print('mybbox:',mybbox)
#shrink and create y coordinates for slicing box
vl = np.array([0,0.87*(mybbox[3]-mybbox[1])])
dvl = ((0.87*(mybbox[3]-mybbox[1]))**2)**0.5
nvl = dvl//sub_dxyz + 1
y = np.arange(nvl)*sub_dxyz
print('nvl:',nvl)
#shrink and create x coordinates for slicing box
vb = np.array([0.85*(mybbox[2]-mybbox[0]),0])
dvb = ((0.85*(mybbox[2]-mybbox[0]))**2)**0.5
nvb = dvb//sub_dxyz + 1
x = np.arange(nvb)*sub_dxyz
print('nvb:',nvb)
#create set of xy coordinates for slicing box
xy = np.transpose([np.tile(x, len(y)), np.repeat(y, len(x))])
print('xy.shape:',xy.shape)
#setup rotation matrices
degree = 30
theta = degree*np.pi/180
rm = np.array([[np.cos(theta),-np.sin(theta)],[np.sin(theta),np.cos(theta)]])
#rotate coordinates
for i in range(len(xy[:,0])):
xy[i,:] = rm.dot(xy[i,:])
#get translated coordinates
xshift = 12600
yshift = -2600
rxy = np.copy(xy)
rxy[:,0] += mybbox[0] + xshift
rxy[:,1] += mybbox[1] + yshift
print('rxy.shape:',rxy.shape)
import time
#get sliced subsurface volume
start = time.time()
slice_props = gm3d.sliceVolumeValsFromCoordsXY(x,y,rxy,local=False)
end = time.time()
print('runtime:', end - start)
import numpy as np
orrssslfqn = './rect_rot_subsamp_smooth_z' + str(dz) + 'm_nam_model_vp_vs_rho_Q_props.npz'
print(orrssslfqn)
np.savez_compressed(orrssslfqn,props=slice_props,xc=x,yc=y,rxyc=rxy)
ifilename = './rect_rot_subsamp_smooth_z200.0m_nam_model_vp_vs_rho_Q_props.npz'
#Unpickle
data = np.load(ifilename)
slice_props = data['props'] #4D ndarray
xc=data['xc']
yc=data['yc']
rxy=data['rxyc']
print('slice_props.shape',slice_props.shape)
sprops = np.copy(slice_props.reshape((4,31, 193, 146)),order='C')
print('sprops.shape:',sprops.shape)
print('gm3d.shape:',gm3d.shape)
rdep_surf = sprops[0,10,:,:].copy()
print(rdep_surf.shape)
print('nxy:',rdep_surf.shape[0]*rdep_surf.shape[1])
print('nrxy:', rxy.shape)
# get new min max to normalize surface
vp_min = np.min(rdep_surf)
vp_max = np.max(rdep_surf)
surf_norm = Normalize(vp_min,vp_max)
xy = np.transpose([np.tile(xc, len(yc)), np.repeat(yc, len(xc))])
print('xy.shape:',xy.shape)
print('xy:',xy)
fig, ax = plt.subplots(1,figsize=(6,6))
ax.scatter(rxy[:,0],rxy[:,1],s=1,c=rdep_surf.flatten(),cmap=plt.cm.jet,norm=surf_norm,zorder=0)
plt.show()
sub_surf = gm3d[0,10,:,:].copy()
print(sub_surf.shape)
print('nxy:',sub_surf.shape[0]*sub_surf.shape[1])
print('nrxy:', rxy.shape)
# get new min max to normalize surface
vp_min = np.min(rdep_surf)
vp_max = np.max(rdep_surf)
surf_norm = Normalize(vp_min,vp_max)
sxc =
xy = np.transpose([np.tile(xc, len(yc)), np.repeat(yc, len(xc))])
print('xy.shape:',xy.shape)
fig, ax = plt.subplots(1,figsize=(6,6))
ax.scatter(xy[:,0],xy[:,1],s=1,c=sub_surf.flatten(),cmap=plt.cm.jet,norm=surf_norm,zorder=0)
plt.show()
sprops = sprops.transpose(0,3,2,1).copy()
print('sprops.shape:',sprops.shape)
print(np.isfortran(sprops))
import
write_vtk_gridded_model_3d(fqpname,props,xdata,ydata,zdata):
| 0.205854 | 0.659631 |
# LSTM Training
```
import sys
import numpy as np
from math import sqrt
from numpy import concatenate
from matplotlib import pyplot
from pandas import read_csv
from pandas import DataFrame
from pandas import concat
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import LabelEncoder
from sklearn.metrics import mean_squared_error
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import r2_score
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import LSTM
import os
# load dataset
dataset = read_csv('../datasets/bss/dublin/reorg/station_2.csv')
dataset = dataset.drop('TIME', axis=1)
values = dataset.values
# ensure all data is float
values = values.astype('float32')
# normalize data
scaler = MinMaxScaler(feature_range=(0, 1))
scaled = scaler.fit_transform(values)
print(scaled)
# split into train and test sets
# Manually setting train and test sets
train_start = 0
train_end = 8760
test_start = 99144
test_end = 108071
n_train_hours = 365 * 24
train = scaled[train_start:train_end, :]
test = scaled[test_start:test_end, :]
# split into input and outputs
train_X, train_y = train[:, 1:], train[:, 0]
test_X, test_y = test[:, 1:], test[:, 0]
# reshape input to be 3D [samples, timesteps, features]
train_X = train_X.reshape((train_X.shape[0], 1, train_X.shape[1]))
test_X = test_X.reshape((test_X.shape[0], 1, test_X.shape[1]))
print(train_X.shape, train_y.shape, test_X.shape, test_y.shape)
# design network
model = Sequential()
model.add(LSTM(40, input_shape=(train_X.shape[1], train_X.shape[2])))
model.add(Dense(1))
model.compile(loss='mean_squared_error')
# fit network
history = model.fit(train_X, train_y,
epochs=150,
batch_size=64,
validation_data=(test_X, test_y),
verbose=2,
shuffle=False)
# plot history
pyplot.plot(history.history['loss'], label='train')
pyplot.plot(history.history['val_loss'], label='test')
pyplot.legend()
pyplot.show()
model.summary()
# make a bunch of predictions
yhat = model.predict(test_X)
test_X = test_X.reshape((test_X.shape[0], test_X.shape[2]))
# invert scaling for forecast
inv_yhat = concatenate((yhat, test_X), axis=1)
inv_yhat = scaler.inverse_transform(inv_yhat)
inv_yhat = inv_yhat[:, 0]
# invert scaling for actual
test_y = test_y.reshape((len(test_y), 1))
inv_y = concatenate((test_y, test_X), axis=1)
inv_y = scaler.inverse_transform(inv_y)
inv_y = inv_y[:, 0]
rmse = sqrt(mean_squared_error(inv_y, inv_yhat))
mae = mean_absolute_error(inv_y, inv_yhat)
mse = mean_squared_error(inv_y, inv_yhat)
r2 = r2_score(inv_y, inv_yhat)
print('Test MAE: %.3f' % mae)
print('Test MSE: %.3f' % mse)
print('Test RMSE: %.3f' % rmse)
print('Test R2: %.30f' % r2)
```
|
github_jupyter
|
import sys
import numpy as np
from math import sqrt
from numpy import concatenate
from matplotlib import pyplot
from pandas import read_csv
from pandas import DataFrame
from pandas import concat
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import LabelEncoder
from sklearn.metrics import mean_squared_error
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import r2_score
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import LSTM
import os
# load dataset
dataset = read_csv('../datasets/bss/dublin/reorg/station_2.csv')
dataset = dataset.drop('TIME', axis=1)
values = dataset.values
# ensure all data is float
values = values.astype('float32')
# normalize data
scaler = MinMaxScaler(feature_range=(0, 1))
scaled = scaler.fit_transform(values)
print(scaled)
# split into train and test sets
# Manually setting train and test sets
train_start = 0
train_end = 8760
test_start = 99144
test_end = 108071
n_train_hours = 365 * 24
train = scaled[train_start:train_end, :]
test = scaled[test_start:test_end, :]
# split into input and outputs
train_X, train_y = train[:, 1:], train[:, 0]
test_X, test_y = test[:, 1:], test[:, 0]
# reshape input to be 3D [samples, timesteps, features]
train_X = train_X.reshape((train_X.shape[0], 1, train_X.shape[1]))
test_X = test_X.reshape((test_X.shape[0], 1, test_X.shape[1]))
print(train_X.shape, train_y.shape, test_X.shape, test_y.shape)
# design network
model = Sequential()
model.add(LSTM(40, input_shape=(train_X.shape[1], train_X.shape[2])))
model.add(Dense(1))
model.compile(loss='mean_squared_error')
# fit network
history = model.fit(train_X, train_y,
epochs=150,
batch_size=64,
validation_data=(test_X, test_y),
verbose=2,
shuffle=False)
# plot history
pyplot.plot(history.history['loss'], label='train')
pyplot.plot(history.history['val_loss'], label='test')
pyplot.legend()
pyplot.show()
model.summary()
# make a bunch of predictions
yhat = model.predict(test_X)
test_X = test_X.reshape((test_X.shape[0], test_X.shape[2]))
# invert scaling for forecast
inv_yhat = concatenate((yhat, test_X), axis=1)
inv_yhat = scaler.inverse_transform(inv_yhat)
inv_yhat = inv_yhat[:, 0]
# invert scaling for actual
test_y = test_y.reshape((len(test_y), 1))
inv_y = concatenate((test_y, test_X), axis=1)
inv_y = scaler.inverse_transform(inv_y)
inv_y = inv_y[:, 0]
rmse = sqrt(mean_squared_error(inv_y, inv_yhat))
mae = mean_absolute_error(inv_y, inv_yhat)
mse = mean_squared_error(inv_y, inv_yhat)
r2 = r2_score(inv_y, inv_yhat)
print('Test MAE: %.3f' % mae)
print('Test MSE: %.3f' % mse)
print('Test RMSE: %.3f' % rmse)
print('Test R2: %.30f' % r2)
| 0.601828 | 0.77343 |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
```
## Load the Yelp data
```
df = pd.read_csv(r"D:\LIUZHICHENG\Udemy\Machine Learning\8 Real World Projects\Natural Language Processing - Yelp Reviews\yelp.csv")
df = df.drop(columns=["business_id", "date", "review_id", "type", "user_id"])
df.head()
df["length"] = df["text"].apply(func=len)
df
```
## EDA
```
df["length"].describe()
df.nlargest(n=1, columns="length")
df["length"].idxmax()
df.iloc[df["length"].idxmax()]
df.nsmallest(n=1, columns="length")
df["length"].idxmin()
df.iloc[df["length"].idxmin()]
sns.countplot(data=df, x="stars")
g = sns.FacetGrid(data=df, col="stars", col_wrap=3)
g.map(plt.hist, "length", bins=20, color="g");
tmp = df[df["stars"].isin(values=[1, 5])]
tmp["stars"].value_counts() / len(tmp) * 100
sns.countplot(data=tmp, x="stars");
text1 = 'Hello Mr. Future, I am so happy to be learning AI now!!'
import string
string.punctuation
from nltk.corpus import stopwords
stopwords.words("english")
def message_cleaning(text):
# char
text_punc_removed = "".join([char for char in text if char not in string.punctuation])
# word
text_stopwords_removed = " ".join([word for word in text_punc_removed.split() if word.lower()
not in stopwords.words("english")])
# text_stopwords_removed = [word for word in text_punc_removed.split() if word.lower() not in stopwords.words("english")]
return text_stopwords_removed
message_cleaning(text1)
%%time
df["text"].apply(func=message_cleaning)
df = pd.read_csv(r"D:\LIUZHICHENG\Udemy\Machine Learning\8 Real World Projects\Natural Language Processing - Yelp Reviews\yelp.csv")
df = df.drop(columns=["business_id", "date", "review_id", "type", "user_id"])
df.head()
# df = df[df["stars"].isin(values=[1, 5])]
```
## Text Preprocessing
```
%%time
import string
import re
import nltk
from nltk.corpus import stopwords
def remove_punctuation(input_text):
""" Remove punctuations like '!"#$%&\'()*+,-./:;<=>?@[\\]^_`{|}~' """
#print("in remove_punctuation\n",input_text)
# Make translation table
input_text = str(input_text) # avoid thedange of being a series object
punct = string.punctuation
trantab = str.maketrans(punct, len(punct)*' ') # Every punctuation symbol will be replaced by a space
return input_text.translate(trantab).encode('ascii', 'ignore').decode('utf8') # -> Final kick to clean up :)
def remove_digits(input_text):
""" Remove numerical digits ranging from 0-9 """
#print("in remove_digits\n",input_text)
return re.sub('\d+', '', input_text)
def to_lower(input_text):
""" String handling, returns the lowercased strings from the given string """
#print("in to_lower\n",input_text)
return input_text.lower()
def remove_stopwords(input_text):
""" Remove the low-level information from our text in order to give more focus to the important information """
#print("in remove_stopwords\n",input_text)
stopwords_list = stopwords.words('english')
newStopWords = ['citi']
stopwords_list.extend(newStopWords)
# Some words which might indicate a certain sentiment are kept via a whitelist
#whitelist = ["n't", "not", "no"]
whitelist = ["n't"]
words = input_text.split()
clean_words = [word for word in words if (word not in stopwords_list or word in whitelist) and len(word) > 2]
return " ".join(clean_words) # list -> string
def expandShortsForms(input_text):
#print("in expandShortsForms\n",input_text)
return input_text.replace("can't", "can not").replace("won't", "will not")
def lemmatize(input_text):
""" Return the base or dictionary form of a word, lemma """
#print("in lemmatize\n",input_text)
outtext= ""
# Lemmatize
from nltk.stem import WordNetLemmatizer
from nltk import pos_tag, word_tokenize, wordnet
from nltk.corpus.reader.wordnet import WordNetError
lemmatizer = WordNetLemmatizer()
input_text = input_text.split()
for word in input_text:
# Get the single character pos constant from pos_tag like this:
pos_label = (pos_tag(word_tokenize(word))[0][1][0]).lower()
# pos_refs = {'n': ['NN', 'NNS', 'NNP', 'NNPS'],
# 'v': ['VB', 'VBD', 'VBG', 'VBN', 'VBP', 'VBZ'],
# 'r': ['RB', 'RBR', 'RBS'],
# 'a': ['JJ', 'JJR', 'JJS']}
if pos_label == 'j': pos_label = 'a' # 'j' <--> 'a' reassignment
if pos_label in ['r']: # For adverbs it's a bit different
try:
if len(wordnet.wordnet.synset(word+'.r.1').lemmas()[0].pertainyms()) > 0:
outtext = outtext + ' ' + (wordnet.wordnet.synset(word+'.r.1').lemmas()[0].pertainyms()[0].name())
except WordNetError:
pass
outtext = outtext + ' ' + word # To keep the word in the list
elif pos_label in ['a', 's', 'v']: # For adjectives and verbs
outtext = outtext +' ' + (lemmatizer.lemmatize(word, pos=pos_label))
else: # For nouns and everything else as it is the default kwarg
outtext = outtext +' ' + (lemmatizer.lemmatize(word))
return outtext
def execute_funcs(input_text, *args):
funcs = list(args)
for func in funcs:
input_text = func(input_text)
return input_text
def apply_funcs(input_text):
clean_X = execute_funcs(input_text, to_lower, remove_punctuation, remove_digits,
remove_stopwords,
expandShortsForms,
# lemmatize
)
return clean_X
```
### Pipeline & ColumnTransformer
```
%%time
import warnings
warnings.filterwarnings('ignore')
df["clean_text"] = df["text"].apply(func=apply_funcs)
df["length"] = df["clean_text"].apply(func=len)
feature_columns = ['cool', 'useful', 'funny', 'clean_text', 'length']
X = df[feature_columns]
y = df["stars"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0, stratify=y)
from sklearn.preprocessing import LabelEncoder
enc = LabelEncoder()
y_train = enc.fit_transform(y=y_train)
y_test = enc.transform(y=y_test)
from sklearn.compose import ColumnTransformer
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.preprocessing import OneHotEncoder
from sklearn.preprocessing import StandardScaler, RobustScaler, Normalizer
# Whenever the transformer expects a 1D array as input, the columns were specified as a string ("xxx").
# For the transformers which expects 2D data, we need to specify the column as a list of strings (["xxx"]).
ct = ColumnTransformer(transformers=[
("TfidfVectorizer", TfidfVectorizer(), ("clean_text")),
("OneHotEncoder", OneHotEncoder(handle_unknown='ignore'), (["cool", "useful", "funny"])),
('Normalizer', Normalizer(), (["length"])),
],
n_jobs=-1)
from sklearn.pipeline import Pipeline
from sklearn.naive_bayes import MultinomialNB
clf = Pipeline(steps=[
("ColumnTransformer", ct),
("MultinomialNB", MultinomialNB())
])
clf.fit(X_train, y_train)
from sklearn.metrics import confusion_matrix, accuracy_score, classification_report
predictions = clf.predict(X_test)
print(classification_report(y_test, predictions))
print(accuracy_score(y_test, predictions))
print(confusion_matrix(y_test, predictions))
from sklearn import set_config
set_config(display="diagram")
clf
```
### make_pipeline & make_column_transformer
```
%%time
import warnings
warnings.filterwarnings('ignore')
df["clean_text"] = df["text"].apply(func=apply_funcs)
df["length"] = df["clean_text"].apply(func=len)
feature_columns = ['cool', 'useful', 'funny', 'clean_text', 'length']
X = df[feature_columns]
y = df["stars"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0, stratify=y)
from sklearn.preprocessing import LabelEncoder
enc = LabelEncoder()
y_train = enc.fit_transform(y=y_train)
y_test = enc.transform(y=y_test)
from sklearn.compose import make_column_transformer
from sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer
from sklearn.preprocessing import OneHotEncoder
from sklearn.preprocessing import StandardScaler, RobustScaler, Normalizer
# This is a shorthand for the ColumnTransformer constructor; it does not require, and does not permit, naming the
# transformers.
ct = make_column_transformer(
(CountVectorizer(), ("clean_text")),
(OneHotEncoder(handle_unknown='ignore'), (["cool", "useful", "funny"])),
(StandardScaler(), (["length"])),
n_jobs=-1)
from sklearn.pipeline import make_pipeline
from lightgbm import LGBMClassifier
from sklearn.naive_bayes import MultinomialNB
# This is a shorthand for the Pipeline constructor; it does not require, and does not permit, naming the estimators.
# Instead, their names will be set to the lowercase of their types automatically.
clf = make_pipeline(ct, LGBMClassifier())
clf.fit(X_train, y_train)
from sklearn.metrics import confusion_matrix, accuracy_score, classification_report
predictions = clf.predict(X_test)
print(classification_report(y_test, predictions))
print(accuracy_score(y_test, predictions))
print(confusion_matrix(y_test, predictions))
from sklearn import set_config
set_config(display="diagram")
clf
from sklearn.metrics import plot_confusion_matrix
display = plot_confusion_matrix(estimator=clf, X=X_test, y_true=y_test, cmap="Blues", values_format='.3g')
display.confusion_matrix
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
df = pd.read_csv(r"D:\LIUZHICHENG\Udemy\Machine Learning\8 Real World Projects\Natural Language Processing - Yelp Reviews\yelp.csv")
df = df.drop(columns=["business_id", "date", "review_id", "type", "user_id"])
df.head()
df["length"] = df["text"].apply(func=len)
df
df["length"].describe()
df.nlargest(n=1, columns="length")
df["length"].idxmax()
df.iloc[df["length"].idxmax()]
df.nsmallest(n=1, columns="length")
df["length"].idxmin()
df.iloc[df["length"].idxmin()]
sns.countplot(data=df, x="stars")
g = sns.FacetGrid(data=df, col="stars", col_wrap=3)
g.map(plt.hist, "length", bins=20, color="g");
tmp = df[df["stars"].isin(values=[1, 5])]
tmp["stars"].value_counts() / len(tmp) * 100
sns.countplot(data=tmp, x="stars");
text1 = 'Hello Mr. Future, I am so happy to be learning AI now!!'
import string
string.punctuation
from nltk.corpus import stopwords
stopwords.words("english")
def message_cleaning(text):
# char
text_punc_removed = "".join([char for char in text if char not in string.punctuation])
# word
text_stopwords_removed = " ".join([word for word in text_punc_removed.split() if word.lower()
not in stopwords.words("english")])
# text_stopwords_removed = [word for word in text_punc_removed.split() if word.lower() not in stopwords.words("english")]
return text_stopwords_removed
message_cleaning(text1)
%%time
df["text"].apply(func=message_cleaning)
df = pd.read_csv(r"D:\LIUZHICHENG\Udemy\Machine Learning\8 Real World Projects\Natural Language Processing - Yelp Reviews\yelp.csv")
df = df.drop(columns=["business_id", "date", "review_id", "type", "user_id"])
df.head()
# df = df[df["stars"].isin(values=[1, 5])]
%%time
import string
import re
import nltk
from nltk.corpus import stopwords
def remove_punctuation(input_text):
""" Remove punctuations like '!"#$%&\'()*+,-./:;<=>?@[\\]^_`{|}~' """
#print("in remove_punctuation\n",input_text)
# Make translation table
input_text = str(input_text) # avoid thedange of being a series object
punct = string.punctuation
trantab = str.maketrans(punct, len(punct)*' ') # Every punctuation symbol will be replaced by a space
return input_text.translate(trantab).encode('ascii', 'ignore').decode('utf8') # -> Final kick to clean up :)
def remove_digits(input_text):
""" Remove numerical digits ranging from 0-9 """
#print("in remove_digits\n",input_text)
return re.sub('\d+', '', input_text)
def to_lower(input_text):
""" String handling, returns the lowercased strings from the given string """
#print("in to_lower\n",input_text)
return input_text.lower()
def remove_stopwords(input_text):
""" Remove the low-level information from our text in order to give more focus to the important information """
#print("in remove_stopwords\n",input_text)
stopwords_list = stopwords.words('english')
newStopWords = ['citi']
stopwords_list.extend(newStopWords)
# Some words which might indicate a certain sentiment are kept via a whitelist
#whitelist = ["n't", "not", "no"]
whitelist = ["n't"]
words = input_text.split()
clean_words = [word for word in words if (word not in stopwords_list or word in whitelist) and len(word) > 2]
return " ".join(clean_words) # list -> string
def expandShortsForms(input_text):
#print("in expandShortsForms\n",input_text)
return input_text.replace("can't", "can not").replace("won't", "will not")
def lemmatize(input_text):
""" Return the base or dictionary form of a word, lemma """
#print("in lemmatize\n",input_text)
outtext= ""
# Lemmatize
from nltk.stem import WordNetLemmatizer
from nltk import pos_tag, word_tokenize, wordnet
from nltk.corpus.reader.wordnet import WordNetError
lemmatizer = WordNetLemmatizer()
input_text = input_text.split()
for word in input_text:
# Get the single character pos constant from pos_tag like this:
pos_label = (pos_tag(word_tokenize(word))[0][1][0]).lower()
# pos_refs = {'n': ['NN', 'NNS', 'NNP', 'NNPS'],
# 'v': ['VB', 'VBD', 'VBG', 'VBN', 'VBP', 'VBZ'],
# 'r': ['RB', 'RBR', 'RBS'],
# 'a': ['JJ', 'JJR', 'JJS']}
if pos_label == 'j': pos_label = 'a' # 'j' <--> 'a' reassignment
if pos_label in ['r']: # For adverbs it's a bit different
try:
if len(wordnet.wordnet.synset(word+'.r.1').lemmas()[0].pertainyms()) > 0:
outtext = outtext + ' ' + (wordnet.wordnet.synset(word+'.r.1').lemmas()[0].pertainyms()[0].name())
except WordNetError:
pass
outtext = outtext + ' ' + word # To keep the word in the list
elif pos_label in ['a', 's', 'v']: # For adjectives and verbs
outtext = outtext +' ' + (lemmatizer.lemmatize(word, pos=pos_label))
else: # For nouns and everything else as it is the default kwarg
outtext = outtext +' ' + (lemmatizer.lemmatize(word))
return outtext
def execute_funcs(input_text, *args):
funcs = list(args)
for func in funcs:
input_text = func(input_text)
return input_text
def apply_funcs(input_text):
clean_X = execute_funcs(input_text, to_lower, remove_punctuation, remove_digits,
remove_stopwords,
expandShortsForms,
# lemmatize
)
return clean_X
%%time
import warnings
warnings.filterwarnings('ignore')
df["clean_text"] = df["text"].apply(func=apply_funcs)
df["length"] = df["clean_text"].apply(func=len)
feature_columns = ['cool', 'useful', 'funny', 'clean_text', 'length']
X = df[feature_columns]
y = df["stars"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0, stratify=y)
from sklearn.preprocessing import LabelEncoder
enc = LabelEncoder()
y_train = enc.fit_transform(y=y_train)
y_test = enc.transform(y=y_test)
from sklearn.compose import ColumnTransformer
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.preprocessing import OneHotEncoder
from sklearn.preprocessing import StandardScaler, RobustScaler, Normalizer
# Whenever the transformer expects a 1D array as input, the columns were specified as a string ("xxx").
# For the transformers which expects 2D data, we need to specify the column as a list of strings (["xxx"]).
ct = ColumnTransformer(transformers=[
("TfidfVectorizer", TfidfVectorizer(), ("clean_text")),
("OneHotEncoder", OneHotEncoder(handle_unknown='ignore'), (["cool", "useful", "funny"])),
('Normalizer', Normalizer(), (["length"])),
],
n_jobs=-1)
from sklearn.pipeline import Pipeline
from sklearn.naive_bayes import MultinomialNB
clf = Pipeline(steps=[
("ColumnTransformer", ct),
("MultinomialNB", MultinomialNB())
])
clf.fit(X_train, y_train)
from sklearn.metrics import confusion_matrix, accuracy_score, classification_report
predictions = clf.predict(X_test)
print(classification_report(y_test, predictions))
print(accuracy_score(y_test, predictions))
print(confusion_matrix(y_test, predictions))
from sklearn import set_config
set_config(display="diagram")
clf
%%time
import warnings
warnings.filterwarnings('ignore')
df["clean_text"] = df["text"].apply(func=apply_funcs)
df["length"] = df["clean_text"].apply(func=len)
feature_columns = ['cool', 'useful', 'funny', 'clean_text', 'length']
X = df[feature_columns]
y = df["stars"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0, stratify=y)
from sklearn.preprocessing import LabelEncoder
enc = LabelEncoder()
y_train = enc.fit_transform(y=y_train)
y_test = enc.transform(y=y_test)
from sklearn.compose import make_column_transformer
from sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer
from sklearn.preprocessing import OneHotEncoder
from sklearn.preprocessing import StandardScaler, RobustScaler, Normalizer
# This is a shorthand for the ColumnTransformer constructor; it does not require, and does not permit, naming the
# transformers.
ct = make_column_transformer(
(CountVectorizer(), ("clean_text")),
(OneHotEncoder(handle_unknown='ignore'), (["cool", "useful", "funny"])),
(StandardScaler(), (["length"])),
n_jobs=-1)
from sklearn.pipeline import make_pipeline
from lightgbm import LGBMClassifier
from sklearn.naive_bayes import MultinomialNB
# This is a shorthand for the Pipeline constructor; it does not require, and does not permit, naming the estimators.
# Instead, their names will be set to the lowercase of their types automatically.
clf = make_pipeline(ct, LGBMClassifier())
clf.fit(X_train, y_train)
from sklearn.metrics import confusion_matrix, accuracy_score, classification_report
predictions = clf.predict(X_test)
print(classification_report(y_test, predictions))
print(accuracy_score(y_test, predictions))
print(confusion_matrix(y_test, predictions))
from sklearn import set_config
set_config(display="diagram")
clf
from sklearn.metrics import plot_confusion_matrix
display = plot_confusion_matrix(estimator=clf, X=X_test, y_true=y_test, cmap="Blues", values_format='.3g')
display.confusion_matrix
| 0.246715 | 0.594257 |
This notebook handles the processing of PA docket data that has been downloaded in JSON format and converted into a CSV with the following columns:
* docket_no: Court docket number
* status: Status of this docket
* gender: Offender's gender
* race: Offender's race
* county: County of the court managing this docket
* offender_id: Hashed value for the Offender
* offense_age: Age computed from DOB
* seq_no: Sequential numbering of charges
* statute: Statute code in violation
* grade: Grade of the crime
* statute_description: Statute description
* offense_date: Date of the offense
* description: Most likely the same as statute description
* offense_tracking_no: Tracking number for the offense for multiple offenders involved
* disposition: Disposition of the charge
* sentence_date: Sentencing date (if any)
* sentence_start: Start of the sentence to be served (if any)
* sentence_type: Type of the sentence meted (if any)
* sentence_min_pd: Minimum sentence (if any)
* sentence_max_pd: Maximum sentence (if any)
```
import json
import os
import pandas as pd
import hashlib
from dateutil.relativedelta import relativedelta
from tqdm import tqdm_notebook
def get_bio(json_data):
"""
Retrieves the biographical information
"""
return dict(
docket_no = json_data["docketNumber"],
status = json_data["statusName"],
gender = json_data["caseParticipants"][0]["gender"],
dob = json_data["caseParticipants"][0]["primaryDateOfBirth"],
race = json_data["caseParticipants"][0]["race"],
first_name = json_data["caseParticipants"][0]["participantName"]["firstName"],
middle_name = json_data["caseParticipants"][0]["participantName"]["middleName"],
last_name = json_data["caseParticipants"][0]["participantName"]["lastName"],
county = json_data["county"]["name"]
)
def get_offenses(json_data):
"""
Retrieves the list of offenses
"""
offenses = map(
lambda x: (
x["sequenceNumber"],
x["statuteName"],
x["grade"],
x["statuteDescription"],
x["offenseDate"],
x["description"],
x["otn"]),
json_data["offenses"])
return pd.DataFrame(
offenses,
columns=['seq_no', 'statute', 'grade', 'statute_description', 'offense_date',
'description', 'offense_tracking_no'])
def get_dispositions(json_data):
"""Retrieves the disposition (if applicable) of the offenses"""
def process_sentencing(sentence_section):
"""Extracts sentencing as part of the disposition"""
if len(sentence_section) == 0:
return (None, None, None, None, None)
else:
latest_sentence = sentence_section[-1]
return (latest_sentence["eventDate"],
latest_sentence["sentenceTypes"][0]["startDateTime"],
latest_sentence["sentenceTypes"][0]["sentenceType"],
latest_sentence["sentenceTypes"][0]["minPeriod"],
latest_sentence["sentenceTypes"][0]["maxPeriod"])
if len(json_data["dispositionEvents"]) > 0:
disposition_section = json_data["dispositionEvents"][-1]["offenseDispositions"]
dispositions = map(lambda x: (
x["sequenceNumber"],
x["disposition"]) +
process_sentencing(x["sentences"]), disposition_section)
else:
dispositions = None
return pd.DataFrame(
dispositions,
columns=['seq_no', 'disposition', 'sentence_date', 'sentence_start',
'sentence_type', 'sentence_min_pd', 'sentence_max_pd']
)
def offense_age(row):
"""Computes the age of the offender at the time of the offense"""
if row["offense_date"] is pd.NaT or row["dob"] is pd.NaT:
# If the date is not valid return None
return None
else:
# Else get the number of years between offense date and DOB
return relativedelta(row["offense_date"].date(), row["dob"].date()).years
def get_records(json_data):
"""Pieces together all relevant pieces from the docket"""
# Retrieve components of the data
bio = get_bio(json_data) # Biographical information
off = get_offenses(json_data) # Charges
disps = get_dispositions(json_data) # Disposition of the charges
# Merge the data together
merged = off.merge(disps, on="seq_no", how='left')
# Federate out the biographical data so this is de-normalized
for k, v in get_bio(json_data).items():
merged[k] = v
# Convert date fields into datetime
merged["dob"] = pd.to_datetime(merged["dob"], errors = 'coerce')
merged["offense_date"] = pd.to_datetime(merged["offense_date"], errors = 'coerce')
merged["sentence_date"] = pd.to_datetime(merged["sentence_date"], errors = 'coerce')
# Construct a unique ID by hashing the names and DOB
uid_str = "".join(filter(None, (bio["first_name"], bio["middle_name"], bio["last_name"], bio["dob"])))
merged["offender_id"] = hashlib.sha256(uid_str.encode("utf-8")).hexdigest()[:12]
# Compute age at time of each offense
merged["offense_age"] = merged.apply(offense_age, axis=1)
# Drop sensitive columns
merged = merged.drop(columns=["first_name", "middle_name", "last_name", "dob"])
# Re-order columns
cols = merged.columns.tolist()
cols = cols[len(cols)-7:] + cols[0:-7]
return merged[cols]
input_path = "data/pa_json/"
output_path = "data/output/"
appended_data = []
def process_file(json_file):
with open(json_file) as f:
try:
data = json.load(f)
appended_data.append(get_records(data))
except:
print(json_file)
raise
for i, input_file in enumerate(tqdm_notebook(os.listdir(input_path))):
if input_file.endswith(".json"):
process_file(path + input_file)
if i > 0 and i % 10000 == 0:
df = pd.concat(appended_data)
df.to_csv(f"data/output/pa_data_{i}.csv")
appended_data = []
df = pd.concat(appended_data)
df.to_csv(f"{output_path}pa_data_{i}.csv")
appended_data = []
pa_data = pd.concat([pd.read_csv(f"{output_path}{x}", low_memory=False) for x in os.listdir(output_path)], axis=0)
# Create mapping for salted/hashed docket id
import os
salt = os.urandom(32)
hashed_docket_id = pa_data.apply(lambda row: hashlib.sha256(f"{salt}{row['docket_no']}".encode("utf-8")).hexdigest()[:12], axis=1)
docket_map = pd.concat([hashed_docket_id, pa_data["docket_no"]], axis=1)
docket_map.columns = ["hash_docket_no","real_docket_no"]
docket_map.drop_duplicates().to_csv(f"{output_path}docket_mapping.csv")
# Replace docket number
pa_data["docket_no"] = hashed_docket_id
pa_data = pa_data.drop(["Unnamed: 0"], axis=1)
pa_data.to_csv(f"{output_path}pa_data_all.csv.gz", compression='gzip')
len(pa_data)
```
|
github_jupyter
|
import json
import os
import pandas as pd
import hashlib
from dateutil.relativedelta import relativedelta
from tqdm import tqdm_notebook
def get_bio(json_data):
"""
Retrieves the biographical information
"""
return dict(
docket_no = json_data["docketNumber"],
status = json_data["statusName"],
gender = json_data["caseParticipants"][0]["gender"],
dob = json_data["caseParticipants"][0]["primaryDateOfBirth"],
race = json_data["caseParticipants"][0]["race"],
first_name = json_data["caseParticipants"][0]["participantName"]["firstName"],
middle_name = json_data["caseParticipants"][0]["participantName"]["middleName"],
last_name = json_data["caseParticipants"][0]["participantName"]["lastName"],
county = json_data["county"]["name"]
)
def get_offenses(json_data):
"""
Retrieves the list of offenses
"""
offenses = map(
lambda x: (
x["sequenceNumber"],
x["statuteName"],
x["grade"],
x["statuteDescription"],
x["offenseDate"],
x["description"],
x["otn"]),
json_data["offenses"])
return pd.DataFrame(
offenses,
columns=['seq_no', 'statute', 'grade', 'statute_description', 'offense_date',
'description', 'offense_tracking_no'])
def get_dispositions(json_data):
"""Retrieves the disposition (if applicable) of the offenses"""
def process_sentencing(sentence_section):
"""Extracts sentencing as part of the disposition"""
if len(sentence_section) == 0:
return (None, None, None, None, None)
else:
latest_sentence = sentence_section[-1]
return (latest_sentence["eventDate"],
latest_sentence["sentenceTypes"][0]["startDateTime"],
latest_sentence["sentenceTypes"][0]["sentenceType"],
latest_sentence["sentenceTypes"][0]["minPeriod"],
latest_sentence["sentenceTypes"][0]["maxPeriod"])
if len(json_data["dispositionEvents"]) > 0:
disposition_section = json_data["dispositionEvents"][-1]["offenseDispositions"]
dispositions = map(lambda x: (
x["sequenceNumber"],
x["disposition"]) +
process_sentencing(x["sentences"]), disposition_section)
else:
dispositions = None
return pd.DataFrame(
dispositions,
columns=['seq_no', 'disposition', 'sentence_date', 'sentence_start',
'sentence_type', 'sentence_min_pd', 'sentence_max_pd']
)
def offense_age(row):
"""Computes the age of the offender at the time of the offense"""
if row["offense_date"] is pd.NaT or row["dob"] is pd.NaT:
# If the date is not valid return None
return None
else:
# Else get the number of years between offense date and DOB
return relativedelta(row["offense_date"].date(), row["dob"].date()).years
def get_records(json_data):
"""Pieces together all relevant pieces from the docket"""
# Retrieve components of the data
bio = get_bio(json_data) # Biographical information
off = get_offenses(json_data) # Charges
disps = get_dispositions(json_data) # Disposition of the charges
# Merge the data together
merged = off.merge(disps, on="seq_no", how='left')
# Federate out the biographical data so this is de-normalized
for k, v in get_bio(json_data).items():
merged[k] = v
# Convert date fields into datetime
merged["dob"] = pd.to_datetime(merged["dob"], errors = 'coerce')
merged["offense_date"] = pd.to_datetime(merged["offense_date"], errors = 'coerce')
merged["sentence_date"] = pd.to_datetime(merged["sentence_date"], errors = 'coerce')
# Construct a unique ID by hashing the names and DOB
uid_str = "".join(filter(None, (bio["first_name"], bio["middle_name"], bio["last_name"], bio["dob"])))
merged["offender_id"] = hashlib.sha256(uid_str.encode("utf-8")).hexdigest()[:12]
# Compute age at time of each offense
merged["offense_age"] = merged.apply(offense_age, axis=1)
# Drop sensitive columns
merged = merged.drop(columns=["first_name", "middle_name", "last_name", "dob"])
# Re-order columns
cols = merged.columns.tolist()
cols = cols[len(cols)-7:] + cols[0:-7]
return merged[cols]
input_path = "data/pa_json/"
output_path = "data/output/"
appended_data = []
def process_file(json_file):
with open(json_file) as f:
try:
data = json.load(f)
appended_data.append(get_records(data))
except:
print(json_file)
raise
for i, input_file in enumerate(tqdm_notebook(os.listdir(input_path))):
if input_file.endswith(".json"):
process_file(path + input_file)
if i > 0 and i % 10000 == 0:
df = pd.concat(appended_data)
df.to_csv(f"data/output/pa_data_{i}.csv")
appended_data = []
df = pd.concat(appended_data)
df.to_csv(f"{output_path}pa_data_{i}.csv")
appended_data = []
pa_data = pd.concat([pd.read_csv(f"{output_path}{x}", low_memory=False) for x in os.listdir(output_path)], axis=0)
# Create mapping for salted/hashed docket id
import os
salt = os.urandom(32)
hashed_docket_id = pa_data.apply(lambda row: hashlib.sha256(f"{salt}{row['docket_no']}".encode("utf-8")).hexdigest()[:12], axis=1)
docket_map = pd.concat([hashed_docket_id, pa_data["docket_no"]], axis=1)
docket_map.columns = ["hash_docket_no","real_docket_no"]
docket_map.drop_duplicates().to_csv(f"{output_path}docket_mapping.csv")
# Replace docket number
pa_data["docket_no"] = hashed_docket_id
pa_data = pa_data.drop(["Unnamed: 0"], axis=1)
pa_data.to_csv(f"{output_path}pa_data_all.csv.gz", compression='gzip')
len(pa_data)
| 0.390127 | 0.736045 |
# SAS Viya, CAS & Python Integration Workshop
### Notebook Summary
1. [Set Up](#1)
2. [Exploring CAS Action Sets and the CASResults Object](#2)
3. [Working with a SASDataFrame](#3)
4. [Exploring the CAS File Structure](#4)
5. [Loading Data Into CAS](#5)
6. [Exploring Table Details](#6)
7. [Data Exploration](#7)
8. [Filtering Data](#8)
9. [Data Preparation](#9)
10. [SQL](#10)
11. [Analyzing Data](#11)
12. [Promote the Table to use in SAS Visual Analytics](#12)
# SAS Viya
### What is SAS Viya
SAS Viya extends the SAS Platform, operates in the cloud (as well as in hybrid and on-prem solutions) and is open source-friendly. For better performance while manipulating data and running analytical procedures, SAS Viya can run your code in Cloud Analytic Services (CAS). CAS operates on in-memory data, removing the read/write transfer overhead. Further, it enables everyone in an organization to collaborate and work with data by providing a variety of [products and solutions](https://www.sas.com/en_us/software/viya.html) running in CAS.

### Cloud Analytic Services (CAS)
SAS Viya processes data and performs analytics using *SAS Cloud Analytic Services*, or *CAS* for short. CAS provides a powerful distributed computing environment designed to store large data sets in memory for fast and efficient processing. It uses scalable, high-performance, multi-threaded algorithms to rapidly perform analytical processing on in-memory data of any size.

#### For more information about Cloud Analytic Services, visit the documentation: [SAS® Cloud Analytic Services 3.5: Fundamentals](https://go.documentation.sas.com/?docsetId=casfun&docsetTarget=titlepage.htm&docsetVersion=3.5&locale=en)
### SAS Viya is Open
SAS Viya is open. Business analysts and data scientists can explore, prepare and manage data to provide insights, create visualizations or analytical models using the SAS programming language or a variety of open source languages like Python, R, Lua, or Java. Because of this, programmers can easily process data in CAS, using a language of their choice.

## <a id='1'>1. Set Up
### a. Import Packages
Visit the documentation for the SWAT [(SAS Scripting Wrapper for Analytics Transfer)](https://sassoftware.github.io/python-swat/index.html) package.
```
## Data Management
import swat
import pandas as pd
## Data Visualization
from matplotlib import pyplot as plt
import seaborn as sns
%matplotlib inline
## Global Options
swat.options.cas.trace_actions = False # Enabling tracing of actions (Default is False. Will change to true later)
swat.options.cas.trace_ui_actions = False # Display the actions behind “UI” methods (Default is False. Will change to true later)
pd.set_option('display.max_columns', 500) # Modify DataFrame max columns shown
pd.set_option('display.max_colwidth', 1000) # Modify DataFrame max column width
```
### b. Make a Connection to CAS</a>
##### To connect to the CAS server you will need:
1. the host name,
2. the portnumber,
3. your user name, and your password.
Visit the documentation [Getting Started with SAS® Viya® 3.5 for Python](https://go.documentation.sas.com/api/docsets/caspg3/3.5/content/caspg3.pdf) for more information about connecting to CAS.
**Be aware that connecting to the CAS server can be implemented in various ways, so you might need to see your system administrator about how to make a connection. Please follow company policy regarding authentication.**
```
conn = swat.CAS("server", 8777, "student", "Metadata0", protocol="http")
conn
```
### c. Obtain Data for the Demo
```
conn.fileinfo()
## Download the data from github and load to the CAS server
conn.read_csv(r"https://raw.githubusercontent.com/sassoftware/sas-viya-programming/master/data/cars.csv",
casout={"name":"cars", "caslib":"casuser", "replace":True})
## Save the in-memory table as a physical file
conn.save(table="cars", name="cars.sashdat",
caslib="casuser",
replace=True)
## Drop the in-memory table
conn.droptable(name='cars', caslib="casuser")
```
## <a id='2'>2. Exploring CAS Action Sets and the CASResults Object</a>
- Think of **action sets** as a *package*, and all the **actions** inside an action set as a *method*.
- CAS actions interact with the CAS server and return a **CASResults** object.
- A **CASResults** object is simply an ordered **Python dictionary** with a few extra methods and attributes added.
- You can also use the SWAT package API to interact with the CAS server. The SWAT package contains *many* of the methods defined by **Pandas DataFrames**. Using methods from the SWAT API will typically return a CASTable, CASColumn, pandas.DataFrame, or pandas.Series object.
**Documentation**:
- To view all CAS action sets and actions visit the documentation: [SAS® Viya® 3.5 Actions and Action Sets by Name and Product](https://go.documentation.sas.com/?docsetId=allprodsactions&docsetTarget=titlepage.htm&docsetVersion=3.5&locale=en)
- To view the SWAT API Reference visit: [API Reference](https://sassoftware.github.io/python-swat/api.html)
### a. View All the CAS Action Sets that are Loaded in CAS.
- From the **Builtins** action set, use the **actionSetInfo** action, to view all *loaded* action sets.
- CAS action sets and actions are case insensitive.
- CAS actions return a CASResults object.
```
conn.builtins.actionSetInfo()
```
View the available CAS actions in the **builtins** action set using the **help** function.
```
conn.help(actionSet="builtins")
```
You do not need to specify the CAS action set prior to the CAS action. Moving forward, all actions will not include the CAS action set.
```
conn.actionSetInfo()
```
All CAS actions return a **CASResults** object.
```
type(conn.actionSetInfo())
```
### b. CASResults Object
- A **CASResults** object is an ordered Python dictionary with *keys* and *values*.
- A **CASResults** object is local data returned by the CAS server.
- While all **CAS actions** return a **CASResults** object, there are no rules about how many keys are contained in the object, or what values are returned.
View the *keys* in the **CASResults** object. This specific **CASResults** object contains a *single key*, and a *single value*.
```
conn.actionSetInfo().keys()
```
Call the **setinfo** key to return the *value*.
```
conn.actionSetInfo()['setinfo']
```
The **setinfo** key holds a **SASDataFrame** object.
```
type(conn.actionSetInfo()['setinfo'])
```
## <a id='3'>3. Working with a SASDataFrame
- A **SASDataFrame** object contains local data.
- A **SASDataFrame** object is a subclass of a **Pandas DataFrame**. You can work with them as you normally do a **Pandas DataFrame**.
**NOTE: When bringing data from CAS locally, remember that CAS can hold larger data than your local computer can handle.**
### a. Create a **SASDataFrame** Object Named *df*.
```
df = conn.actionSetInfo()['setinfo']
type(df)
```
A SASDataFrame is **local** data. Work with it as you would a Pandas DataFrame.
### b. Use Pandas Methods on a SASDataFrame.
View the first *5* rows of the **SASDataFrame** using the pandas **head** method.
```
df.head()
```
Find all rows where the value in the **actionset** column equals *simple* using the pandas **loc** method.
```
df.loc[df['actionset']=='simple',['actionset','label']]
```
View counts of unique values using the pandas **value_counts** method and plot a bar chart.
```
df['product_name'].value_counts().plot(kind="bar")
```
## <a id='4'> 4. Exploring the CAS File Structure</a>
### **Caslib Overview**:
1. A **caslib** has two parts:
1. **Data Source** - Connection information about the **data source** gives access to a resource that contains data. These can be files that are located in a file system, a database, streaming data from an ESP (Event Stream Processing) server, or other data sources that SAS can access.
2. **In-Memory Space** - The **in-memory** portion of a caslib that contains data that is uploaded into memory and ready for processing.

2. Think of your active **caslib** as the *current working directory* of your CAS session, and it's only possible to have one active caslib.
3. When you want to work with data from your **data source**, you must load the data into the **in-memory portion** for processing. This loaded table is known as a **CAS Table**.
### **Types of Caslibs**:
1. **Personal Caslib** - By default, all users are given access to their own caslib, named CASUSER, within a CAS session. This is a personal caslib and is only accessible to the user who owns the CAS session.
2. **Pre-defined Caslib** - These are defined by an administrator and are available to all CAS sessions (dependent on access controls). Think of these as different folders for different units of a business. You can have an HR caslib with HR data, Marketing caslib with Marketing data, etc.
3. **Manually added Caslib** - These can be added at any point to perform ad-hoc analysis within CAS.
### **Caslib Scope**
1. **Session Caslib** - When a caslib is defined without including the GLOBAL option, the caslib is a session-scoped caslib. When a table is loaded to the CAS server with session-scoped caslib, the table is available to that specific CAS user session only. Think of session scope as *local* to that specific session only.
2. **Global Caslib** -These are available to anyone who has access to the CAS Server (dependent on access controls). The name of these caslibs must be unique across all CAS sessions on the server.
**For additional information about caslibs**:
- [Watch SAS® Viya™ CAS Libraries (Caslibs) Simplified](https://video.sas.com/detail/video/5343952274001/sas%C2%AE-viya%E2%84%A2-cas-libraries-caslibs-simplified)
- [SAS® Cloud Analytic Services 3.5: Fundamentals - Caslibs](https://go.documentation.sas.com/?docsetId=casfun&docsetTarget=n1i11h5hggxv65n1m5i4nw9s5cli.htm&docsetVersion=3.5&locale=en)
### a. View all Available Caslibs
- Depending on your CAS server setup, you might already have one or more caslibs configured and ready to use.
- If you do not have **ReadInfo** permissions on a caslib, then you will *not see* the caslib.
View all available caslibs using the **casLibInfo** action.
```
conn.caslibInfo()
```
### b. View Available Files in the *casuser* Caslib
```
conn.fileInfo(caslib="casuser")
```
### c. View All Available In-Memory Tables in the *casuser* Caslib
NOTE: Tables need to be **in-memory** to be processed by CAS.
```
conn.tableInfo(caslib="casuser")
```
## <a id='5'>5. Loading Data Into CAS
There are various ways of loading data into CAS:
1. server-side data
2. client-side parsed
3. client-side files uploaded and parsed on the server
They follow these naming conventions:
- **load***: Loads server-side data
- **read_***: Uses client-side parsers and then uploads the result into CAS
- **upload***: Uploads client-side files as is, which are parsed on the server
For more information about loading client side files to CAS: [Two Simple Ways to Import Local Files with Python in CAS (Viya 3.5)](www.google.com)
### a. Loading Server-Side Data into Memory.
View the available files in the casuser caslib.
```
conn.fileInfo(caslib="casuser")
```
There are two methods that can be used to load server-side data into CAS:
- **loadtable** - Loads a table into CAS and returns a CASResults object.
- **load_path** - Convenience method. Similar to loadtable, load_path loads a table into CAS and returns a reference to that CAS table in one step.
**loadtable**
```
# 1. Load the table into CAS. Will return a CASResults object.
conn.loadtable(path="cars.sashdat", caslib="casuser",
casout={"caslib":"casuser","name":"cars", "replace":True})
conn.tableInfo(caslib="casuser")
# 2. Create a reference to the in-memory table
castbl = conn.CASTable("cars",caslib="casuser")
```
**load_path**
```
# Load the table into CAS and create a reference to that table in one step.
##castbl = conn.load_path(path="cars.sashdat", caslib="casuser",
## casout={"caslib":"casuser","name":"cars", "replace":True})
```
## b. Local vs CAS Data
A CASTable object is a reference to data **in the CAS server**. Actions or methods run on a CASTable object are processed in CAS.
```
type(castbl)
print(castbl)
```
View the first 5 rows of the in-memory table using the **head** method. The head method is not a CAS action, so it will not return a CASResults object. The head method is using the API to CAS. The API to CAS contains many of the pandas methods you are familiar with. These methods **process the data in CAS** and can return a variety of **different objects locally**.
[SWAT API Reference](https://sassoftware.github.io/python-swat/api.html)

```
castbl.head()
```
The results of using the head method returns a SASDataFrame. SASDataFrames are located on locally.
```
type(castbl.head())
```
You can use the **fetch** CAS action to return similar results. The processing of the fetch CAS action occurs in CAS and returns a CASResults object to your local machine. When using a CAS action a CASResults object is always returned.
```
castbl.fetch(to=5)
```
CASResults objects are local.
```
type(castbl.fetch(to=5))
```
SASDataFrame objects can be contained in the CASResults object.
```
type(castbl.fetch(to=5)['Fetch'])
```
Turn on tracing.
```
swat.options.cas.trace_actions = True
swat.options.cas.trace_ui_actions = True
```
## <a id='6'>6. Exploring Table Details
### a. View the Number of Rows and Columns in the In-Memory Table.
Use **shape** to return a tuple of the CAS data.
```
castbl.shape
```
Use the **numRows** CAS action to shows the number of rows in a CAS table.
```
castbl.numRows()
```
Use the **tableInfo** CAS action to show information about a CAS table.
```
castbl.tableInfo()
```
Create a function to return the in-memory table name, number of rows and columns.
```
def details(tbl):
sasdf = tbl.tableInfo()["TableInfo"].set_index("Name").loc[:,["Rows","Columns"]]
return sasdf
details(castbl)
```
### b. View the Column Information
```
castbl.columnInfo()
castbl.dtypes
```
## <a id='7'>7. Data Exploration
### a. Summary Statistics
Using the **summary** CAS action to generate descriptive statistics of numeric variables.
```
castbl.summary()
```
Using the **describe** method.
```
castbl.describe()
```
Turn off tracing.
```
swat.options.cas.trace_actions = False
swat.options.cas.trace_ui_actions = False
```
### b. Distinct Values
Use the **distinct** CAS action to calculate the number of distinct values in the cars table.
```
castbl.distinct()
```
Plot the number of missing values for each column.
```
castbl.distinct()['Distinct'] \
.set_index("Column") \
.loc[:,['NMiss']] \
.plot(kind='bar')
```
Use the **distinct** CAS action to calculate the number of distinct values in the Origin, Type and Make columns using the distinct CAS action.
```
castbl.distinct(inputs=["Origin","Type","Make"])
```
Create a new CAS table named **castblDistinct** with the number of distinct values for the specified inputs.
```
castbl.distinct(inputs=["Origin","Type","Make"],
casout={"caslib":"casuser", ## Create a new CAS table in casuser
"name":"castblDistinct", ## Name the table castblDistinct
"replace":True}) ## Replace if exists
```
View the available in-memory tables.
```
conn.tableInfo()
```
Using Pandas methods.
```
castbl.Cylinders.nunique()
castbl.Cylinders.isnull().sum()
```
### c. Frequency
View the frequency of the Origin column using the **freq** CAS action.
```
castbl.freq(inputs=["Origin"])
```
Plot the resuls of the **freq** CAS action in a bar chart.
```
## Perform the processing in CAS and store the summary in the originFreq object.
originFreq = castbl.freq(inputs=["Origin"])['Frequency']
## Graph the summarized local data.
originFreq.loc[:,["CharVar","Frequency"]] \
.sort_values(by="Frequency", ascending=False) \
.set_index("CharVar") \
.plot(kind="bar")
```
Use the **value_counts** method. The value_counts method will process in CAS and return the summary locally. The plot method will create the graph locally.
```
castbl['Origin'].value_counts().plot(kind='bar')
```
Perform a frequency on mulitple columns. The final CASResults object will contain a SASDataFrame with a frequency of each of the specified columns in one table.
```
castbl.freq(inputs=["Origin","Make","Type","DriveTrain"])
```
### D. Create a Frequency Table of all Columns with Less Than 20 Distinct Values.
Use the distinct CAS action to find the number of distinct values for each column and filter for all columns with less than 20 distinct values.
```
distinctCars = castbl.distinct()['Distinct']
distinctCars.loc[distinctCars["NDistinct"]<=20,:]
```
Create a variable named **distinctCars** that holds the SASDataFrame from the results above.
```
distinctCars = distinctCars.loc[distinctCars["NDistinct"]<=20,:]
```
Create a list of column names that have less than 20 distinct values named **listCars**.
```
listCars = distinctCars.Column.unique().tolist()
print(listCars)
```
Use the list from above to create a frequency table of columns with less than 20 distinct values.
```
castbl.freq(inputs=listCars)
```
## <a id='8'>8. Filtering Data
### a. Subset Using Pandas Indexing Expressions.
```
castbl[castbl["Make"]=="Toyota"].head()
castbl[(castbl["Make"]=="Toyota") & (castbl["Type"]=="Hybrid")].head()
```
### b. Subset Using the Query Method.
```
castbl.query("Make='Toyota'").head()
castbl.query("Make='Toyota' and Type='Hybrid'").head()
```
## <a id='9'>9. Data Preparation
Create a new column that calculates the average of MPG_City and MPG_Highway. Processing done in CAS.
```
castbl["avgMPG"] = (castbl["MPG_City"] + castbl["MPG_Highway"])/2
castbl
castbl.head()
```
Remove the Model and MSRP columns.
```
cols = ['Make', 'Type', 'Origin', 'DriveTrain','Invoice',
'EngineSize', 'Cylinders', 'Horsepower', 'MPG_City',
'MPG_Highway', 'Weight', 'Wheelbase', 'Length', 'avgMPG']
castbl = castbl[cols]
castbl
castbl.head()
```
## <a id='10'>10. SQL
### a. Load the fedSQL CAS Action Set
View all **available** (not just loaded) CAS action sets by using the all=True parameter.
```
conn.actionSetInfo(all=True)['setinfo']
```
Search the actionset column for any CAS action set that contains the string *sql*.
```
actionSets = conn.actionSetInfo(all=True)['setinfo']
actionSets.loc[actionSets['actionset'].str.upper().str.contains("SQL")]
```
Load the **fedSQL** action set using the **loadActionSet** action.
```
conn.loadActionSet(actionSet="fedSQL")
conn.actionSetInfo()
conn.help(actionSet="fedSQL")
```
### b. Run SQL Queries in CAS
Run a query to view the first 10 rows of the cars table.
```
conn.execdirect("""select *
from cars
limit 10""")
```
Find the average MSRP of each car make.
```
conn.execdirect("""select Make, round(avg(MSRP)) as avgMSRP
from cars
group by Make""")
```
Create a table named **make_avg** that contains the average MSRP of each car make.
```
conn.execdirect("""create table make_avg as
select Make, round(avg(MSRP)) as test
from cars
group by Make""")
conn.tableInfo(caslib="casuser")
```
## <a id='11'>11. Analyzing Data
Preview the table.
```
castbl.head()
```
### a. Correlation with a Heat Map
Use the **correlation** action and remove the simple statistics. Processing will be done in CAS and the summary table will be returned locally.
```
castbl.correlation(inputs=["MSRP","EngineSize","HorsePower","MPG_City"], simple=False)
```
Store the SASDataFrame object in the **dfCorr** variable. A SASDataFrame object is local.
```
dfCorr = castbl.correlation(inputs=["MSRP","EngineSize","HorsePower","MPG_City"], simple=False)['Correlation']
dfCorr
```
Replace the default index with the Variable column
```
dfCorr.set_index("Variable", inplace=True)
dfCorr
```
Use seaborn to produce a heatmap.
```
fig, ax = plt.subplots(figsize=(12,8))
ax = sns.heatmap(dfCorr, cmap="YlGnBu", annot=True)
ax.set_ylim(len(dfCorr),-.05) ## Truncation with defaults. Need to adjust limits. Fixed in newer verison of matplotlib.
```
### b. Histogram
Run the **histogram** action to return a summary of the midpoints and percents. Processing occurs in CAS.
```
castbl.histogram(inputs=["avgMPG"])
```
Store the BinDetails in the variable **mpgHist**.
```
mpgHist = castbl.histogram(inputs="avgMPG")['BinDetails']
```
Round the columns Percent and MidPoint.
```
mpgHist['Percent'] = mpgHist['Percent'].round(1)
mpgHist['MidPoint'] = mpgHist['MidPoint'].round(1)
mpgHist[["MidPoint","Percent"]].head()
```
Plot the histogram.
```
fig, ax = plt.subplots(figsize=(12,8))
ax = sns.barplot(x="MidPoint", y="Percent", data=mpgHist)
ax.set_title("Histogram of MPG")
```
Specify multiple columns in the **histogram** action.
```
castbl.histogram(inputs=["avgMPG", "HorsePower"])
```
Store the results from the histogram CAS action in the **carsHist** variable.
```
carsHist = castbl.histogram(inputs=["avgMPG", "HorsePower"])['BinDetails']
```
Find the unique values in the carsHist SASDataFrame.
```
list(carsHist.Variable.unique())
```
Run a loop through the list of unique values and plot a histogram for each.
```
for i in list(carsHist.Variable.unique()):
carsHist['Percent'] = carsHist['Percent'].round(1)
carsHist['MidPoint'] = carsHist['MidPoint'].round(1)
df = carsHist[carsHist["Variable"]==i]
df.plot.bar(x='MidPoint', y='Percent')
```
## <a id='12'>12. Promote the Table to use in SAS Visual Analytics
```
castbl.head()
castbl
```
### Two Options:
- Save the castbl object as a physical file
- Create a new in-memory table from the castbl object.
### a. Save the castbl Object as a Physical File.
Use the **save** CAS action to save the castbl object as a physical file. Here we will save it as a sashdat file.
```
castbl.save(name="updatedCars.sashdat", caslib="casuser")
```
View the available files in the casuser caslib. Notice the updatedCars.sashdat file is available.
```
conn.fileInfo(caslib="casuser")
```
### b. Create a New In-Memory Table From the castbl Object.
The partition CAS action has a variety of options, but if we leave the defaults we can take the castbl object (reference to the cars table with a few columns dropped and the new avgMPG column) and create a new in-memory table without saving a physical file.
Here a new in-memory table will be created called **cars_update** in the casuser caslib from the **castbl** object.
```
castbl.partition(casout={"caslib":"casuser","name":"cars_update"})
```
View the new in-memory table **cars_update**.
```
conn.tableInfo(caslib="casuser")
```
View the files in the casuser caslib. Notice no new files were created.
```
conn.fileInfo(caslib="casuser")
```
### c. Promote a Table to Global Scope.
View all the tables in the casuser caslib. Focus on the specified columns. Notice no table is global scope.
```
conn.tableInfo(caslib="casuser")['TableInfo'][['Name','Rows','Columns','Global']]
```
Use the **promote** CAS action to promote a table to global scope. Global scope allows other users and software like SAS Visual Analtyics to use the in-memory table. Currently, all the in-memory tables are session scope. That is, only this account on this connection to CAS can see the in-memory tables.
In this example, the **cars_update** table is promoted to global scope in the casuser caslib. This only allows the current account (student) to access this table since it is promoted in the casuser caslib. If a table is promoted to global scope in a shared caslib, other users can see that table.
***DEMO: Go to SAS Visual Analyics and see cars_update does not exist outside of this session.***
Promote the **cars_update** in-memory table to global scope
```
conn.promote(name="cars_update", caslib="casuser")
```
Notice only the **cars_update** table is global.
```
conn.tableInfo(caslib="casuser")['TableInfo'][['Name','Rows','Columns','Global']]
```
***DEMO: Go to SAS Visual Analtyics and view the cars_update table outside of this session now that the in-memory table is global.***
|
github_jupyter
|
## Data Management
import swat
import pandas as pd
## Data Visualization
from matplotlib import pyplot as plt
import seaborn as sns
%matplotlib inline
## Global Options
swat.options.cas.trace_actions = False # Enabling tracing of actions (Default is False. Will change to true later)
swat.options.cas.trace_ui_actions = False # Display the actions behind “UI” methods (Default is False. Will change to true later)
pd.set_option('display.max_columns', 500) # Modify DataFrame max columns shown
pd.set_option('display.max_colwidth', 1000) # Modify DataFrame max column width
conn = swat.CAS("server", 8777, "student", "Metadata0", protocol="http")
conn
conn.fileinfo()
## Download the data from github and load to the CAS server
conn.read_csv(r"https://raw.githubusercontent.com/sassoftware/sas-viya-programming/master/data/cars.csv",
casout={"name":"cars", "caslib":"casuser", "replace":True})
## Save the in-memory table as a physical file
conn.save(table="cars", name="cars.sashdat",
caslib="casuser",
replace=True)
## Drop the in-memory table
conn.droptable(name='cars', caslib="casuser")
conn.builtins.actionSetInfo()
conn.help(actionSet="builtins")
conn.actionSetInfo()
type(conn.actionSetInfo())
conn.actionSetInfo().keys()
conn.actionSetInfo()['setinfo']
type(conn.actionSetInfo()['setinfo'])
df = conn.actionSetInfo()['setinfo']
type(df)
df.head()
df.loc[df['actionset']=='simple',['actionset','label']]
df['product_name'].value_counts().plot(kind="bar")
conn.caslibInfo()
conn.fileInfo(caslib="casuser")
conn.tableInfo(caslib="casuser")
conn.fileInfo(caslib="casuser")
# 1. Load the table into CAS. Will return a CASResults object.
conn.loadtable(path="cars.sashdat", caslib="casuser",
casout={"caslib":"casuser","name":"cars", "replace":True})
conn.tableInfo(caslib="casuser")
# 2. Create a reference to the in-memory table
castbl = conn.CASTable("cars",caslib="casuser")
# Load the table into CAS and create a reference to that table in one step.
##castbl = conn.load_path(path="cars.sashdat", caslib="casuser",
## casout={"caslib":"casuser","name":"cars", "replace":True})
type(castbl)
print(castbl)
castbl.head()
type(castbl.head())
castbl.fetch(to=5)
type(castbl.fetch(to=5))
type(castbl.fetch(to=5)['Fetch'])
swat.options.cas.trace_actions = True
swat.options.cas.trace_ui_actions = True
castbl.shape
castbl.numRows()
castbl.tableInfo()
def details(tbl):
sasdf = tbl.tableInfo()["TableInfo"].set_index("Name").loc[:,["Rows","Columns"]]
return sasdf
details(castbl)
castbl.columnInfo()
castbl.dtypes
castbl.summary()
castbl.describe()
swat.options.cas.trace_actions = False
swat.options.cas.trace_ui_actions = False
castbl.distinct()
castbl.distinct()['Distinct'] \
.set_index("Column") \
.loc[:,['NMiss']] \
.plot(kind='bar')
castbl.distinct(inputs=["Origin","Type","Make"])
castbl.distinct(inputs=["Origin","Type","Make"],
casout={"caslib":"casuser", ## Create a new CAS table in casuser
"name":"castblDistinct", ## Name the table castblDistinct
"replace":True}) ## Replace if exists
conn.tableInfo()
castbl.Cylinders.nunique()
castbl.Cylinders.isnull().sum()
castbl.freq(inputs=["Origin"])
## Perform the processing in CAS and store the summary in the originFreq object.
originFreq = castbl.freq(inputs=["Origin"])['Frequency']
## Graph the summarized local data.
originFreq.loc[:,["CharVar","Frequency"]] \
.sort_values(by="Frequency", ascending=False) \
.set_index("CharVar") \
.plot(kind="bar")
castbl['Origin'].value_counts().plot(kind='bar')
castbl.freq(inputs=["Origin","Make","Type","DriveTrain"])
distinctCars = castbl.distinct()['Distinct']
distinctCars.loc[distinctCars["NDistinct"]<=20,:]
distinctCars = distinctCars.loc[distinctCars["NDistinct"]<=20,:]
listCars = distinctCars.Column.unique().tolist()
print(listCars)
castbl.freq(inputs=listCars)
castbl[castbl["Make"]=="Toyota"].head()
castbl[(castbl["Make"]=="Toyota") & (castbl["Type"]=="Hybrid")].head()
castbl.query("Make='Toyota'").head()
castbl.query("Make='Toyota' and Type='Hybrid'").head()
castbl["avgMPG"] = (castbl["MPG_City"] + castbl["MPG_Highway"])/2
castbl
castbl.head()
cols = ['Make', 'Type', 'Origin', 'DriveTrain','Invoice',
'EngineSize', 'Cylinders', 'Horsepower', 'MPG_City',
'MPG_Highway', 'Weight', 'Wheelbase', 'Length', 'avgMPG']
castbl = castbl[cols]
castbl
castbl.head()
conn.actionSetInfo(all=True)['setinfo']
actionSets = conn.actionSetInfo(all=True)['setinfo']
actionSets.loc[actionSets['actionset'].str.upper().str.contains("SQL")]
conn.loadActionSet(actionSet="fedSQL")
conn.actionSetInfo()
conn.help(actionSet="fedSQL")
conn.execdirect("""select *
from cars
limit 10""")
conn.execdirect("""select Make, round(avg(MSRP)) as avgMSRP
from cars
group by Make""")
conn.execdirect("""create table make_avg as
select Make, round(avg(MSRP)) as test
from cars
group by Make""")
conn.tableInfo(caslib="casuser")
castbl.head()
castbl.correlation(inputs=["MSRP","EngineSize","HorsePower","MPG_City"], simple=False)
dfCorr = castbl.correlation(inputs=["MSRP","EngineSize","HorsePower","MPG_City"], simple=False)['Correlation']
dfCorr
dfCorr.set_index("Variable", inplace=True)
dfCorr
fig, ax = plt.subplots(figsize=(12,8))
ax = sns.heatmap(dfCorr, cmap="YlGnBu", annot=True)
ax.set_ylim(len(dfCorr),-.05) ## Truncation with defaults. Need to adjust limits. Fixed in newer verison of matplotlib.
castbl.histogram(inputs=["avgMPG"])
mpgHist = castbl.histogram(inputs="avgMPG")['BinDetails']
mpgHist['Percent'] = mpgHist['Percent'].round(1)
mpgHist['MidPoint'] = mpgHist['MidPoint'].round(1)
mpgHist[["MidPoint","Percent"]].head()
fig, ax = plt.subplots(figsize=(12,8))
ax = sns.barplot(x="MidPoint", y="Percent", data=mpgHist)
ax.set_title("Histogram of MPG")
castbl.histogram(inputs=["avgMPG", "HorsePower"])
carsHist = castbl.histogram(inputs=["avgMPG", "HorsePower"])['BinDetails']
list(carsHist.Variable.unique())
for i in list(carsHist.Variable.unique()):
carsHist['Percent'] = carsHist['Percent'].round(1)
carsHist['MidPoint'] = carsHist['MidPoint'].round(1)
df = carsHist[carsHist["Variable"]==i]
df.plot.bar(x='MidPoint', y='Percent')
castbl.head()
castbl
castbl.save(name="updatedCars.sashdat", caslib="casuser")
conn.fileInfo(caslib="casuser")
castbl.partition(casout={"caslib":"casuser","name":"cars_update"})
conn.tableInfo(caslib="casuser")
conn.fileInfo(caslib="casuser")
conn.tableInfo(caslib="casuser")['TableInfo'][['Name','Rows','Columns','Global']]
conn.promote(name="cars_update", caslib="casuser")
conn.tableInfo(caslib="casuser")['TableInfo'][['Name','Rows','Columns','Global']]
| 0.526343 | 0.989811 |
## Deliverable 2. Create a Customer Travel Destinations Map.
```
# Dependencies and Setup
import pandas as pd
import requests
import gmaps
# Import API key
from config import g_key
# Configure gmaps API key
gmaps.configure(api_key=g_key)
# 1. Import the WeatherPy_database.csv file.
city_data_df = pd.read_csv("../Weather_Database/WeatherPy_database.csv")
city_data_df.head()
# 2. Prompt the user to enter minimum and maximum temperature criteria
min_temp = float(input("What is the minimum temperature you would like for your trip? "))
max_temp = float(input("What is the maximum temperature you would like for your trip? "))
# 3. Filter the city_data_df DataFrame using the input statements to create a new DataFrame using the loc method.
preferred_cities_df = city_data_df.loc[(city_data_df["Max Temp"] <= max_temp) &
(city_data_df["Max Temp"] >= min_temp)]
preferred_cities_df.head(10)
# 4a. Determine if there are any empty rows.
preferred_cities_df.count()
# 4b. Drop any empty rows and create a new DataFrame that doesn’t have empty rows.
preferred_cities_df = preferred_cities_df.dropna()
preferred_cities_df.count()
# 5a. Create DataFrame called hotel_df to store hotel names along with city, country, max temp, and coordinates.
hotel_df = preferred_cities_df[["City", "Country", "Max Temp", "Current Description", "Lat", "Lng"]].copy()
# 5b. Create a new column "Hotel Name"
hotel_df["Hotel Name"] = ""
hotel_df.head(10)
# 6a. Set parameters to search for hotels with 5000 meters.
params = {"radius": 5000,
"type": "lodging",
"key": g_key}
# 6b. Iterate through the hotel DataFrame.
for index, row in hotel_df.iterrows():
# 6c. Get latitude and longitude from DataFrame
lat = row["Lat"]
lng = row["Lng"]
params["location"] = f'{lat},{lng}'
# 6d. Set up the base URL for the Google Directions API to get JSON data.
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
# 6e. Make request and retrieve the JSON data from the search.
hotels = requests.get(base_url, params=params).json()
# 6f. Get the first hotel from the results and store the name, if a hotel isn't found skip the city.
try:
hotel_df.loc[index, "Hotel Name"] = hotels["results"][0]["name"]
except (IndexError):
print("Hotel not found... skipping.")
hotel_df
# 7. Drop the rows where there is no Hotel Name.
import numpy as np
hotel_df["Hotel Name"].replace("", np.nan, inplace=True)
hotel_df
hotel_df = hotel_df.dropna()
hotel_df.count()
hotel_df
# 8a. Create the output File (CSV)
output_data_file = "./WeatherPy_vacation.csv"
# 8b. Export the City_Data into a csv
hotel_df.to_csv(output_data_file, index_label="City_ID")
# 9. Using the template add city name, the country code, the weather description and maximum temperature for the city.
info_box_template = """
<dl>
<dt>Hotel Name</dt><dd>{Hotel Name}</dd>
<dt>City</dt><dd>{City}</dd>
<dt>Country</dt><dd>{Country}</dd>
<dt>Weather Description</dt><dd>{Current Description}</dd>
<dt>Max Temp</dt><dd>{Max Temp} °F</dd>
</dl>
"""
# 10a. Get the data from each row and add it to the formatting template and store the data in a list.
hotel_info = [info_box_template.format(**row) for index, row in hotel_df.iterrows()]
# 10b. Get the latitude and longitude from each row and store in a new DataFrame.
locations = hotel_df[["Lat", "Lng"]]
# 11a. Add a marker layer for each city to the map.
fig = gmaps.figure(center=(30.0, 31.0), zoom_level=1.5)
marker_layer = gmaps.marker_layer(locations, info_box_content=hotel_info)
fig.add_layer(marker_layer)
# 11b. Display the figure
fig
```
|
github_jupyter
|
# Dependencies and Setup
import pandas as pd
import requests
import gmaps
# Import API key
from config import g_key
# Configure gmaps API key
gmaps.configure(api_key=g_key)
# 1. Import the WeatherPy_database.csv file.
city_data_df = pd.read_csv("../Weather_Database/WeatherPy_database.csv")
city_data_df.head()
# 2. Prompt the user to enter minimum and maximum temperature criteria
min_temp = float(input("What is the minimum temperature you would like for your trip? "))
max_temp = float(input("What is the maximum temperature you would like for your trip? "))
# 3. Filter the city_data_df DataFrame using the input statements to create a new DataFrame using the loc method.
preferred_cities_df = city_data_df.loc[(city_data_df["Max Temp"] <= max_temp) &
(city_data_df["Max Temp"] >= min_temp)]
preferred_cities_df.head(10)
# 4a. Determine if there are any empty rows.
preferred_cities_df.count()
# 4b. Drop any empty rows and create a new DataFrame that doesn’t have empty rows.
preferred_cities_df = preferred_cities_df.dropna()
preferred_cities_df.count()
# 5a. Create DataFrame called hotel_df to store hotel names along with city, country, max temp, and coordinates.
hotel_df = preferred_cities_df[["City", "Country", "Max Temp", "Current Description", "Lat", "Lng"]].copy()
# 5b. Create a new column "Hotel Name"
hotel_df["Hotel Name"] = ""
hotel_df.head(10)
# 6a. Set parameters to search for hotels with 5000 meters.
params = {"radius": 5000,
"type": "lodging",
"key": g_key}
# 6b. Iterate through the hotel DataFrame.
for index, row in hotel_df.iterrows():
# 6c. Get latitude and longitude from DataFrame
lat = row["Lat"]
lng = row["Lng"]
params["location"] = f'{lat},{lng}'
# 6d. Set up the base URL for the Google Directions API to get JSON data.
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
# 6e. Make request and retrieve the JSON data from the search.
hotels = requests.get(base_url, params=params).json()
# 6f. Get the first hotel from the results and store the name, if a hotel isn't found skip the city.
try:
hotel_df.loc[index, "Hotel Name"] = hotels["results"][0]["name"]
except (IndexError):
print("Hotel not found... skipping.")
hotel_df
# 7. Drop the rows where there is no Hotel Name.
import numpy as np
hotel_df["Hotel Name"].replace("", np.nan, inplace=True)
hotel_df
hotel_df = hotel_df.dropna()
hotel_df.count()
hotel_df
# 8a. Create the output File (CSV)
output_data_file = "./WeatherPy_vacation.csv"
# 8b. Export the City_Data into a csv
hotel_df.to_csv(output_data_file, index_label="City_ID")
# 9. Using the template add city name, the country code, the weather description and maximum temperature for the city.
info_box_template = """
<dl>
<dt>Hotel Name</dt><dd>{Hotel Name}</dd>
<dt>City</dt><dd>{City}</dd>
<dt>Country</dt><dd>{Country}</dd>
<dt>Weather Description</dt><dd>{Current Description}</dd>
<dt>Max Temp</dt><dd>{Max Temp} °F</dd>
</dl>
"""
# 10a. Get the data from each row and add it to the formatting template and store the data in a list.
hotel_info = [info_box_template.format(**row) for index, row in hotel_df.iterrows()]
# 10b. Get the latitude and longitude from each row and store in a new DataFrame.
locations = hotel_df[["Lat", "Lng"]]
# 11a. Add a marker layer for each city to the map.
fig = gmaps.figure(center=(30.0, 31.0), zoom_level=1.5)
marker_layer = gmaps.marker_layer(locations, info_box_content=hotel_info)
fig.add_layer(marker_layer)
# 11b. Display the figure
fig
| 0.506591 | 0.70458 |
# Iris Flower Classification using Supervised Machine Learning
## Introduction
The [Iris flower data set](https://en.wikipedia.org/wiki/Iris_flower_data_set) is widely used as a beginner's dataset for machine learning purposes.
The data set consists of 50 samples from each of three species of Iris (Iris setosa, Iris virginica and Iris versicolor). Four features were measured from each sample: the length and the width of the sepals and petals, in centimeters.
We are going to build a machine learning model to distinguish the species from each other based on the combination of these four features.
## Importing the Libraries
```
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
sns.set(font_scale=1.1)
%matplotlib inline
```
## Importing the Dataset
Iris flower data is available in file './data/iris.csv' and the columns are separated by ',' (comma). Let us use [pandas.read_csv](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.read_csv.html) method to read the file into a DataFrame.
```
field_names = ['sepal-length', 'sepal-width', 'petal-length', 'petal-width', 'class']
dataset = pd.read_csv('./data/iris.csv', names=field_names)
X = dataset.iloc[:, :-1].values
y = dataset.iloc[:, -1].values
```
## Exploratory Data Analysis (EDA)
Let us understand the data first and try to gather as many insights from it.
### DataFrame summary
To start with, let us print a concise summary of the DataFrame using the [info()](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.info.html) method.
```
dataset.info()
```
From the above output we can understand that:
- There are 150 rows and 5 columns.
- Data has only float and object values.
- Data looks very clean and there are no missing values.
### A closer look at the Data
Let us check the first ten rows of the dataset.
```
print(dataset.head(10))
```
By looking at the above output we can undertand that first 4 columns are input variables and the last coumn (class) is the output variable. We will be using various classification techniques to predict the value of the class column.
### Statistical Summary
Let us take a look at a summary of each attribute.
This includes the count, mean, the min and max values as well as some percentiles.
```
print(dataset.describe())
```
We can see that all of the numerical values have the same scale (centimeters) and similar ranges between 0 and 8 centimeters.
### Distribution of the class variable
Let us have a look at the distribution of the class variable we are trying to predict.
```
print(dataset.groupby('class').size())
```
From the above results we can understand that each class has 50 observations. This indicates that the dataset is balanced.
### Data Visualization
Let us visualize the data to get more insights about the data.
#### Box and Whisker Plots
Let us start with box and whisker plots of the input variables. These plots will help us to get a clear idea of the distribution of input variables.
```
sns.boxplot(data=dataset)
plt.tight_layout()
plt.show()
```
#### Histogram
A histogram can also give us an idea about the distribution of input variables.
```
dataset.hist()
plt.tight_layout()
plt.show()
```
For both petal_length and petal_width, it looks like there are a group of data points that have smaller values than others.
#### Plot Pairwise Relationships
Let us see the pairplot of all pairs of attributes. This will help us to get a much better understanding of the relationships between the variables.
```
sns.pairplot(dataset, hue='class', diag_kind="hist")
plt.show()
```
From the above plot it looks like some variables are highly correlated (Example:- petal-length & petal-width). Another observation is that petal measurements of Iris-setosa species are smaller than other species.
#### Correlation matrix
Let us make a correlation matrix to quantitatively examine the relationship between variables.
```
corrmat = dataset.corr()
plt.figure(figsize=(5,5))
sns.heatmap(corrmat, annot = True, square = True, linewidths=.5, cbar=False)
plt.yticks(rotation=0)
plt.show()
```
From above plot we can understand that petal-length & petal-width have high positive correlation and sepal-length & sepal-width are uncorrelated.
### Insights after EDA
- There are 150 rows and 5 columns.
- Data has only float and object values.
- Data looks very clean and there are no missing values.
- Each class has 50 observations. This indicates that the dataset is balanced.
- Variables petal-length & petal-width have high positive correlation and sepal-length & sepal-width are uncorrelated.
## ML Modeling
Let us split the dataset into the training and test sets and then apply different classification algorithms to see which algorithm gives better accuracy.
### Splitting the dataset into Training set and Test set
```
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state = 1)
```
### Evaluate Models
Let us build and evaluate models using the training set.
```
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import StratifiedKFold
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier
# List of models
models = []
models.append(('LR', LogisticRegression(solver='liblinear', multi_class='ovr')))
models.append(('LDA', LinearDiscriminantAnalysis()))
models.append(('KNN', KNeighborsClassifier()))
models.append(('NB', GaussianNB()))
models.append(('SVM', SVC(gamma='auto')))
models.append(('DTC', DecisionTreeClassifier()))
models.append(('RFC', RandomForestClassifier(n_estimators=50)))
# Evaluate each model
results = []
names = []
for name, model in models:
kfold = StratifiedKFold(n_splits=10, random_state=1, shuffle=True)
cv_results = cross_val_score(model, X_train, y_train, cv=kfold, scoring='accuracy')
results.append(cv_results)
names.append(name)
print('%s: %f (%f)' % (name, cv_results.mean(), cv_results.std()))
```
### Model Evaluation
From the above results we can see that Support Vector Machine (SVM) model has the highest accuracy (0.983333) and hence we will choose this as the final model.
Let us check the performance of this SVM model using the test set. We will check the classification report, confusion matrix and accuracy score.
#### Classification Report
```
from sklearn.metrics import accuracy_score
from sklearn.metrics import confusion_matrix
from sklearn.metrics import classification_report
model = SVC(gamma='auto')
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
print(classification_report(y_test, y_pred))
```
#### Confusion Matrix & Accuracy Score
```
cm = confusion_matrix(y_test, y_pred)
cm_df = pd.DataFrame(cm,
index = dataset['class'].unique(),
columns = dataset['class'].unique())
plt.figure(figsize=(5,5))
sns.heatmap(cm_df, annot=True, linewidths=.5, cbar=False)
plt.yticks(rotation=0)
plt.title('Support Vector Machine\nAccuracy: {0:.4f}'.format(accuracy_score(y_test, y_pred)))
plt.ylabel('True Label')
plt.xlabel('Predicted Label')
plt.show()
```
From the above results we can understand that accuracy of the SVM model on test set is 0.9667. Confusion matrix and classification matrix also looks excellent.
## Conclusion
We were able to predict the classes of the Iris dataset with very high accuracy.
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
sns.set(font_scale=1.1)
%matplotlib inline
field_names = ['sepal-length', 'sepal-width', 'petal-length', 'petal-width', 'class']
dataset = pd.read_csv('./data/iris.csv', names=field_names)
X = dataset.iloc[:, :-1].values
y = dataset.iloc[:, -1].values
dataset.info()
print(dataset.head(10))
print(dataset.describe())
print(dataset.groupby('class').size())
sns.boxplot(data=dataset)
plt.tight_layout()
plt.show()
dataset.hist()
plt.tight_layout()
plt.show()
sns.pairplot(dataset, hue='class', diag_kind="hist")
plt.show()
corrmat = dataset.corr()
plt.figure(figsize=(5,5))
sns.heatmap(corrmat, annot = True, square = True, linewidths=.5, cbar=False)
plt.yticks(rotation=0)
plt.show()
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state = 1)
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import StratifiedKFold
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier
# List of models
models = []
models.append(('LR', LogisticRegression(solver='liblinear', multi_class='ovr')))
models.append(('LDA', LinearDiscriminantAnalysis()))
models.append(('KNN', KNeighborsClassifier()))
models.append(('NB', GaussianNB()))
models.append(('SVM', SVC(gamma='auto')))
models.append(('DTC', DecisionTreeClassifier()))
models.append(('RFC', RandomForestClassifier(n_estimators=50)))
# Evaluate each model
results = []
names = []
for name, model in models:
kfold = StratifiedKFold(n_splits=10, random_state=1, shuffle=True)
cv_results = cross_val_score(model, X_train, y_train, cv=kfold, scoring='accuracy')
results.append(cv_results)
names.append(name)
print('%s: %f (%f)' % (name, cv_results.mean(), cv_results.std()))
from sklearn.metrics import accuracy_score
from sklearn.metrics import confusion_matrix
from sklearn.metrics import classification_report
model = SVC(gamma='auto')
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
print(classification_report(y_test, y_pred))
cm = confusion_matrix(y_test, y_pred)
cm_df = pd.DataFrame(cm,
index = dataset['class'].unique(),
columns = dataset['class'].unique())
plt.figure(figsize=(5,5))
sns.heatmap(cm_df, annot=True, linewidths=.5, cbar=False)
plt.yticks(rotation=0)
plt.title('Support Vector Machine\nAccuracy: {0:.4f}'.format(accuracy_score(y_test, y_pred)))
plt.ylabel('True Label')
plt.xlabel('Predicted Label')
plt.show()
| 0.627152 | 0.990615 |
## Reading the midi File and finding the chords
```
from Tonnetz_Select import fromMidiToPCS
from structural_functions import testInput, getKeyByValue
from os.path import isdir
from Data_and_Dicts import dictOfTonnetze
```
Will ask you the directory and the name of the file and returns the Modified list of chords and the appropriate Tonnetz based on vertical compactness
```
print("Enter the directory of the MIDI file")
directory = testInput(isdir)
file = input("Enter the name of the MIDI file(without the extension)")
complete_name = directory + '/' + file + '.mid'
chordListConnectNoDoubles, Tonnetz, connectivity = fromMidiToPCS(complete_name)
print(file, "is complete with Tonnetz", Tonnetz)
```
## Trajectory Calculations
### Computing Trajectory for every Tonnetz
```
from TrajectoryCalculationsWithClass import *
trajectory345 = NewTrajectory(chordListConnectNoDoubles, [3,4,5])
trajectory147 = NewTrajectory(chordListConnectNoDoubles, [1,4,7])
trajectory138 = NewTrajectory(chordListConnectNoDoubles, [1,3,8])
trajectory237 = NewTrajectory(chordListConnectNoDoubles, [2,3,7])
trajectory129 = NewTrajectory(chordListConnectNoDoubles, [1,2,9])
```
### Let's find the edges
```
import itertools as itt
def TrajectoryNoteEdges(TrajectoryPoints):
TotalEdges = []
dist = [-1, 0, 1]
for dicts in TrajectoryPoints:
chordEdges = []
l = list(itt.product(dicts.values(), dicts.values()))
for couple in l:
(x1, y1), (x2, y2) = couple
if (x1 - x2) in dist and (y1 - y2) in dist:
if not (((x1 - x2) == 1 and (y1 - y2) == -1) or ((x1 - x2) == -1 and (y1 - y2) == 1)) :
chordEdges.append(couple)
TotalEdges.append(chordEdges)
return TotalEdges
TrajectoryEdges345 = TrajectoryNoteEdges(trajectory345.chordPositions)
TrajectoryEdges147 = TrajectoryNoteEdges(trajectory147.chordPositions)
TrajectoryEdges237 = TrajectoryNoteEdges(trajectory237.chordPositions)
TrajectoryEdges129 = TrajectoryNoteEdges(trajectory129.chordPositions)
TrajectoryEdges138 = TrajectoryNoteEdges(trajectory138.chordPositions)
```
### Let's plot that!
We plot all five trajectories and compare
```
%matplotlib notebook
import numpy as np
import pylab as plt
from matplotlib import collections as mc
def plot_trajectory(TrajectoryEdges, Tonnetz):
fig, ax = plt.subplots()
for el in TrajectoryEdges:
line = []
line = mc.LineCollection(el, linewidths=0.3)
ax.add_collection(line)
ax.autoscale()
ax.margins(0.1)
plt.title(Tonnetz)
plt.grid()
plt.axis('equal')
plt.show()
plot_trajectory(TrajectoryEdges345, "T345")
plot_trajectory(TrajectoryEdges147, "T147")
plot_trajectory(TrajectoryEdges237, "T237")
plot_trajectory(TrajectoryEdges129, "T129")
plot_trajectory(TrajectoryEdges138, "T138")
```
### Measuring horizontal Compactness
Let's try graph libraries
```
import numpy as np
from scipy.ndimage.measurements import label
def createList(r1, r2):
"""Create a list from a range."""
return list(range(r1, r2 + 1))
def addCouples(v, u):
x, y = v
z, r = u
return x+z, y+r
def squarematrixcreate(maxWidth, minWidth, maxHeight, minHeight, points):
"""Create a square matrix of zeros."""
width = maxWidth - minWidth + 1
height = maxHeight - minHeight + 1
matrix = np.zeros((width, height))
nlist = list(map(lambda x: addCouples(x, (abs(minWidth), abs(minHeight))), points))
for el in nlist:
x, y = el
matrix[x, y] = 1
return matrix
def ccl(matrix):
structure = np.array([[1, 1, 0], [1, 1, 1], [0, 1, 1]])
labeled, ncomponents = label(matrix, structure)
return ncomponents
def dimensionsOfTrajectory(TrajectoryPoints):
totalPoints = []
for dicts in TrajectoryPoints:
totalPoints = totalPoints + list(dicts.values())
totalPoints = list(set(totalPoints))
x, y = zip(*totalPoints)
maxW = max(x)
minW = min(x)
maxH = max(y)
minH = min(y)
numberOfComponents = ccl(squarematrixcreate(maxW, minW, maxH, minH, totalPoints))
width = maxW - minW
height = maxH - minH
return numberOfComponents, width*height
D345 = dimensionsOfTrajectory(trajectory345.chordPositions)
D147 = dimensionsOfTrajectory(trajectory147.chordPositions)
D237 = dimensionsOfTrajectory(trajectory237.chordPositions)
D129 = dimensionsOfTrajectory(trajectory129.chordPositions)
D138 = dimensionsOfTrajectory(trajectory138.chordPositions)
D345 = [sorted(trajectory345.Tonnetz), D345[0], D345[1], 0]
D147 = [sorted(trajectory147.Tonnetz), D147[0], D147[1], 0]
D237 = [sorted(trajectory237.Tonnetz), D237[0], D237[1], 0]
D129 = [sorted(trajectory129.Tonnetz), D129[0], D129[1], 0]
D138 = [sorted(trajectory138.Tonnetz), D138[0], D138[1], 0]
TonnetzList = [D345, D147, D237, D129, D138]
print(TonnetzList)
def addConnectivity(TonnetzList):
for el in TonnetzList:
el[3] = connectivity[getKeyByValue(dictOfTonnetze, el[0])]
return TonnetzList
TonnetzList = addConnectivity(TonnetzList)
print(TonnetzList)
def applyingCoefficients(maxChords, maxComponents, maxDimensions, TonnetzDetails):
coef1 = 1 - TonnetzDetails[3]/maxChords
coef2 = TonnetzDetails[1]/maxComponents
coef3 = TonnetzDetails[2]/maxDimensions
coefGen = (coef1*2 + coef2 + coef3)/4
return coefGen
def finalCompliance(TonnetzList):
Tonnetze, components, dimensions, chords = zip(*TonnetzList)
maxChords = max(chords)
maxComponents = max(components)
maxDimensions = max(dimensions)
newlist = []
for el in TonnetzList:
coefGen = applyingCoefficients(maxChords, maxComponents, maxDimensions, el)
newlist.append((el[0], coefGen))
sortedList = sorted(newlist, key = lambda x: x[1])
return sortedList[0][0], sortedList[1][0]
finalCompliance(TonnetzList)
```
|
github_jupyter
|
from Tonnetz_Select import fromMidiToPCS
from structural_functions import testInput, getKeyByValue
from os.path import isdir
from Data_and_Dicts import dictOfTonnetze
print("Enter the directory of the MIDI file")
directory = testInput(isdir)
file = input("Enter the name of the MIDI file(without the extension)")
complete_name = directory + '/' + file + '.mid'
chordListConnectNoDoubles, Tonnetz, connectivity = fromMidiToPCS(complete_name)
print(file, "is complete with Tonnetz", Tonnetz)
from TrajectoryCalculationsWithClass import *
trajectory345 = NewTrajectory(chordListConnectNoDoubles, [3,4,5])
trajectory147 = NewTrajectory(chordListConnectNoDoubles, [1,4,7])
trajectory138 = NewTrajectory(chordListConnectNoDoubles, [1,3,8])
trajectory237 = NewTrajectory(chordListConnectNoDoubles, [2,3,7])
trajectory129 = NewTrajectory(chordListConnectNoDoubles, [1,2,9])
import itertools as itt
def TrajectoryNoteEdges(TrajectoryPoints):
TotalEdges = []
dist = [-1, 0, 1]
for dicts in TrajectoryPoints:
chordEdges = []
l = list(itt.product(dicts.values(), dicts.values()))
for couple in l:
(x1, y1), (x2, y2) = couple
if (x1 - x2) in dist and (y1 - y2) in dist:
if not (((x1 - x2) == 1 and (y1 - y2) == -1) or ((x1 - x2) == -1 and (y1 - y2) == 1)) :
chordEdges.append(couple)
TotalEdges.append(chordEdges)
return TotalEdges
TrajectoryEdges345 = TrajectoryNoteEdges(trajectory345.chordPositions)
TrajectoryEdges147 = TrajectoryNoteEdges(trajectory147.chordPositions)
TrajectoryEdges237 = TrajectoryNoteEdges(trajectory237.chordPositions)
TrajectoryEdges129 = TrajectoryNoteEdges(trajectory129.chordPositions)
TrajectoryEdges138 = TrajectoryNoteEdges(trajectory138.chordPositions)
%matplotlib notebook
import numpy as np
import pylab as plt
from matplotlib import collections as mc
def plot_trajectory(TrajectoryEdges, Tonnetz):
fig, ax = plt.subplots()
for el in TrajectoryEdges:
line = []
line = mc.LineCollection(el, linewidths=0.3)
ax.add_collection(line)
ax.autoscale()
ax.margins(0.1)
plt.title(Tonnetz)
plt.grid()
plt.axis('equal')
plt.show()
plot_trajectory(TrajectoryEdges345, "T345")
plot_trajectory(TrajectoryEdges147, "T147")
plot_trajectory(TrajectoryEdges237, "T237")
plot_trajectory(TrajectoryEdges129, "T129")
plot_trajectory(TrajectoryEdges138, "T138")
import numpy as np
from scipy.ndimage.measurements import label
def createList(r1, r2):
"""Create a list from a range."""
return list(range(r1, r2 + 1))
def addCouples(v, u):
x, y = v
z, r = u
return x+z, y+r
def squarematrixcreate(maxWidth, minWidth, maxHeight, minHeight, points):
"""Create a square matrix of zeros."""
width = maxWidth - minWidth + 1
height = maxHeight - minHeight + 1
matrix = np.zeros((width, height))
nlist = list(map(lambda x: addCouples(x, (abs(minWidth), abs(minHeight))), points))
for el in nlist:
x, y = el
matrix[x, y] = 1
return matrix
def ccl(matrix):
structure = np.array([[1, 1, 0], [1, 1, 1], [0, 1, 1]])
labeled, ncomponents = label(matrix, structure)
return ncomponents
def dimensionsOfTrajectory(TrajectoryPoints):
totalPoints = []
for dicts in TrajectoryPoints:
totalPoints = totalPoints + list(dicts.values())
totalPoints = list(set(totalPoints))
x, y = zip(*totalPoints)
maxW = max(x)
minW = min(x)
maxH = max(y)
minH = min(y)
numberOfComponents = ccl(squarematrixcreate(maxW, minW, maxH, minH, totalPoints))
width = maxW - minW
height = maxH - minH
return numberOfComponents, width*height
D345 = dimensionsOfTrajectory(trajectory345.chordPositions)
D147 = dimensionsOfTrajectory(trajectory147.chordPositions)
D237 = dimensionsOfTrajectory(trajectory237.chordPositions)
D129 = dimensionsOfTrajectory(trajectory129.chordPositions)
D138 = dimensionsOfTrajectory(trajectory138.chordPositions)
D345 = [sorted(trajectory345.Tonnetz), D345[0], D345[1], 0]
D147 = [sorted(trajectory147.Tonnetz), D147[0], D147[1], 0]
D237 = [sorted(trajectory237.Tonnetz), D237[0], D237[1], 0]
D129 = [sorted(trajectory129.Tonnetz), D129[0], D129[1], 0]
D138 = [sorted(trajectory138.Tonnetz), D138[0], D138[1], 0]
TonnetzList = [D345, D147, D237, D129, D138]
print(TonnetzList)
def addConnectivity(TonnetzList):
for el in TonnetzList:
el[3] = connectivity[getKeyByValue(dictOfTonnetze, el[0])]
return TonnetzList
TonnetzList = addConnectivity(TonnetzList)
print(TonnetzList)
def applyingCoefficients(maxChords, maxComponents, maxDimensions, TonnetzDetails):
coef1 = 1 - TonnetzDetails[3]/maxChords
coef2 = TonnetzDetails[1]/maxComponents
coef3 = TonnetzDetails[2]/maxDimensions
coefGen = (coef1*2 + coef2 + coef3)/4
return coefGen
def finalCompliance(TonnetzList):
Tonnetze, components, dimensions, chords = zip(*TonnetzList)
maxChords = max(chords)
maxComponents = max(components)
maxDimensions = max(dimensions)
newlist = []
for el in TonnetzList:
coefGen = applyingCoefficients(maxChords, maxComponents, maxDimensions, el)
newlist.append((el[0], coefGen))
sortedList = sorted(newlist, key = lambda x: x[1])
return sortedList[0][0], sortedList[1][0]
finalCompliance(TonnetzList)
| 0.412175 | 0.830525 |
```
import pandas as pd
import json
"hola"[0]
def add_type(codigo_comision):
if(codigo_comision[0] != 'M' and codigo_comision[0] != 'T'):
codigo_comision = 'M' + codigo_comision # Lamentablemente no tengo forma de saber si es M o T...
return codigo_comision
def get_data():
GSPREADHSEET_DOWNLOAD_URL = (
"https://docs.google.com/spreadsheets/d/{gid}/export?format=csv&id={gid}".format
)
GID = '10ztDqFAi2HvbZNtOkGXK6dy-M22lXJkc6KHvpxVyOYQ'
df = pd.read_csv(GSPREADHSEET_DOWNLOAD_URL(gid=GID))
del df["Turno"]
del df["Cupo"]
df['Comisión'] = df["Comisión"].apply(add_type)
return df.dropna()
def normalize_day(s):
replacements = (
("á", "a"),
("é", "e"),
("í", "i"),
("ó", "o"),
("ú", "u"),
)
s = s.lower()
for a, b in replacements:
s = s.replace(a, b)
return s
def nday(s):
if s is None:
return 0
nday_dict = {'lunes':1, 'martes':2, 'miercoles':3, 'jueves':4, 'viernes':5, 'sabado':6}
return nday_dict[normalize_day(s)]
df = get_data()
def get_horario():
horarios = df["Dia"].str.split(' ', expand=True).drop([3,8], axis=1)
horarios.set_axis(['tipo_1', 'dia_1', 'base_1', 'tope_1',
'tipo_2', 'dia_2', 'base_2', 'tope_2'],
axis='columns', inplace=True)
horarios['dia_1'] = horarios['dia_1'].apply(nday)
horarios['dia_2'] = horarios['dia_2'].apply(nday)
return horarios
horarios = get_horario()
del df['Dia']
df = df.join(horarios)
def format_clases(clases, row):
clases.append({"dia": row["dia_1"],
"inicio": row["base_1"],
"fin": row["tope_1"]})
if(row["dia_2"] != 0):
clases.append({"dia": row["dia_2"],
"inicio": row["base_2"],
"fin": row["tope_2"]})
return clases
def format_materia(cursos, row):
cursos.append({ "codigo": row["Comisión"],
"materia": row["Comisión"].split("-")[0],
"docentes": row["Docente"]+' - '+row["Comisión"].split("-")[2],
"clases": format_clases([], row)})
return cursos
DATA = {
"cuatrimestre": "2C2022",
"timestamp": 0,
}
DATA["materias"] = []
DATA["cursos"] = []
for index, row in df.iterrows():
format_materia(DATA["cursos"], row)
for materia in df["Actividad / Materia"].unique():
cursos = list(df[df["Actividad / Materia"] == materia]["Comisión"].values)
cod = cursos[0].split("-")[0]
DATA["materias"].append({"codigo": cod,
"cursos": cursos,
"nombre": materia})
dump = json.dumps(DATA, indent=2, ensure_ascii=False, sort_keys=True)
with open('src/data/horarios.js', 'w') as fw:
fw.write("export const data = ")
fw.write("\n") # Do not remove me. Make me easy to parse.
fw.write(dump)
```
|
github_jupyter
|
import pandas as pd
import json
"hola"[0]
def add_type(codigo_comision):
if(codigo_comision[0] != 'M' and codigo_comision[0] != 'T'):
codigo_comision = 'M' + codigo_comision # Lamentablemente no tengo forma de saber si es M o T...
return codigo_comision
def get_data():
GSPREADHSEET_DOWNLOAD_URL = (
"https://docs.google.com/spreadsheets/d/{gid}/export?format=csv&id={gid}".format
)
GID = '10ztDqFAi2HvbZNtOkGXK6dy-M22lXJkc6KHvpxVyOYQ'
df = pd.read_csv(GSPREADHSEET_DOWNLOAD_URL(gid=GID))
del df["Turno"]
del df["Cupo"]
df['Comisión'] = df["Comisión"].apply(add_type)
return df.dropna()
def normalize_day(s):
replacements = (
("á", "a"),
("é", "e"),
("í", "i"),
("ó", "o"),
("ú", "u"),
)
s = s.lower()
for a, b in replacements:
s = s.replace(a, b)
return s
def nday(s):
if s is None:
return 0
nday_dict = {'lunes':1, 'martes':2, 'miercoles':3, 'jueves':4, 'viernes':5, 'sabado':6}
return nday_dict[normalize_day(s)]
df = get_data()
def get_horario():
horarios = df["Dia"].str.split(' ', expand=True).drop([3,8], axis=1)
horarios.set_axis(['tipo_1', 'dia_1', 'base_1', 'tope_1',
'tipo_2', 'dia_2', 'base_2', 'tope_2'],
axis='columns', inplace=True)
horarios['dia_1'] = horarios['dia_1'].apply(nday)
horarios['dia_2'] = horarios['dia_2'].apply(nday)
return horarios
horarios = get_horario()
del df['Dia']
df = df.join(horarios)
def format_clases(clases, row):
clases.append({"dia": row["dia_1"],
"inicio": row["base_1"],
"fin": row["tope_1"]})
if(row["dia_2"] != 0):
clases.append({"dia": row["dia_2"],
"inicio": row["base_2"],
"fin": row["tope_2"]})
return clases
def format_materia(cursos, row):
cursos.append({ "codigo": row["Comisión"],
"materia": row["Comisión"].split("-")[0],
"docentes": row["Docente"]+' - '+row["Comisión"].split("-")[2],
"clases": format_clases([], row)})
return cursos
DATA = {
"cuatrimestre": "2C2022",
"timestamp": 0,
}
DATA["materias"] = []
DATA["cursos"] = []
for index, row in df.iterrows():
format_materia(DATA["cursos"], row)
for materia in df["Actividad / Materia"].unique():
cursos = list(df[df["Actividad / Materia"] == materia]["Comisión"].values)
cod = cursos[0].split("-")[0]
DATA["materias"].append({"codigo": cod,
"cursos": cursos,
"nombre": materia})
dump = json.dumps(DATA, indent=2, ensure_ascii=False, sort_keys=True)
with open('src/data/horarios.js', 'w') as fw:
fw.write("export const data = ")
fw.write("\n") # Do not remove me. Make me easy to parse.
fw.write(dump)
| 0.257392 | 0.288485 |
# Part 8 bis - Introduction to Protocols
### Context
Now that we've been through Plans, we'll introduce a new object called the Protocol. A Protocol coordinates a sequence of Plans, deploys them on distant workers
and run them in a single pass.
It's a high level object which contains the logic of a complex computation
distributed across several workers. The main feature of Protocol is the
ability to be sent / searched / fetched back between workers, and finally
deployed to identified workers. So a user can design a protocol, upload it
to a cloud worker, and any other workers will be able to search for it,
download it, and apply the computation program it contains on the workers
that it is connected to.
Let's see how to use it!
Authors:
- Théo Ryffel - Twitter [@theoryffel](https://twitter.com/theoryffel) - GitHub: [@LaRiffle](https://github.com/LaRiffle)
### 1. Create and deploy
Protocol are created by providing a list of pairs `(worker, plan)`. `worker` can be either a real
worker or a worker id or a string to represent a fictive worker. This
last case can be used at creation to specify that two plans should be
owned (or not owned) by the same worker at deployment. `plan` can
either be a Plan or a PointerPlan.
```
import torch as th
import syft as sy
hook = sy.TorchHook(th)
# IMPORTANT: Local worker should not be a client worker
hook.local_worker.is_client_worker = False
```
Let's define 3 plans and feed them to a protocol. They all perform an increment operation.
```
@sy.func2plan(args_shape=[(1,)])
def inc1(x):
return x + 1
@sy.func2plan(args_shape=[(1,)])
def inc2(x):
return x + 1
@sy.func2plan(args_shape=[(1,)])
def inc3(x):
return x + 1
protocol = sy.Protocol([("worker1", inc1), ("worker2", inc2), ("worker3", inc3)])
```
Now we need to bind the Protocol to workers, which is done by calling `.deploy(*workers)`. Let's create some workers.
```
bob = sy.VirtualWorker(hook, id="bob")
alice = sy.VirtualWorker(hook, id="alice")
charlie = sy.VirtualWorker(hook, id="charlie")
workers = alice, bob, charlie
protocol.deploy(*workers)
```
You can see that the plans have already been sent to the appropriate workers: it has been deployed!
This has been done in 2 phases: first, we map the fictive workers provided at creation
(named by strings) to the provided workers, and second, we send the corresponding
plans to each of them.
### 2. Run a protocol
Running a protocol means executing all the plans sequentially. Do do so, you provide some input data which is sent to the first plan location. This first plan is
run and its output is moved to the second plan location, and so on. The final
result is returned after all plans have run, and it is composed of pointers to
the last plan location.
```
x = th.tensor([1.0])
ptr = protocol.run(x)
ptr
ptr.get()
```
The input 1.0 has been through the 3 plans and so has been incremented 3 times, that's why it now equals 4.0!
Actually, you can also **run a protocol remotely** on some pointers to data:
```
james = sy.VirtualWorker(hook, id="james")
protocol.send(james)
x = th.tensor([1.0]).send(james)
ptr = protocol.run(x)
ptr
```
As you see the result is a pointer to james
```
ptr = ptr.get()
ptr
ptr = ptr.get()
ptr
```
### 3. Search for a protocol
In real settings you might want to download a remote protocol, to deploy it on your workers and to run it with you data:
Let's initialize a protocol **which is not deployed**, and put it on a remote worker
```
protocol = sy.Protocol([("worker1", inc1), ("worker2", inc2), ("worker3", inc3)])
protocol.tag('my_protocol')
protocol.send(james)
me = sy.hook.local_worker # get access to me as a local worker
```
Now we launch a search to find the protocol
```
responses = me.request_search(['my_protocol'], location=james)
responses
```
You have access to a pointer to a Protocol
```
ptr_protocol = responses[0]
```
Like usual pointer you can get it back:
```
protocol_back = ptr_protocol.get()
protocol_back
```
And we can do like we did in parts 1. & 2.
```
protocol_back.deploy(alice, bob, charlie)
x = th.tensor([1.0])
ptr = protocol_back.run(x)
ptr.get()
```
More real world examples will come with Protocols, but you can already see all the possibilities opened by this new object!
### Star PySyft on GitHub
The easiest way to help our community is just by starring the repositories! This helps raise awareness of the cool tools we're building.
- [Star PySyft](https://github.com/OpenMined/PySyft)
### Pick our tutorials on GitHub!
We made really nice tutorials to get a better understanding of what Federated and Privacy-Preserving Learning should look like and how we are building the bricks for this to happen.
- [Checkout the PySyft tutorials](https://github.com/OpenMined/PySyft/tree/master/examples/tutorials)
### Join our Slack!
The best way to keep up to date on the latest advancements is to join our community!
- [Join slack.openmined.org](http://slack.openmined.org)
### Join a Code Project!
The best way to contribute to our community is to become a code contributor! If you want to start "one off" mini-projects, you can go to PySyft GitHub Issues page and search for issues marked `Good First Issue`.
- [Good First Issue Tickets](https://github.com/OpenMined/PySyft/issues?q=is%3Aopen+is%3Aissue+label%3A%22good+first+issue%22)
### Donate
If you don't have time to contribute to our codebase, but would still like to lend support, you can also become a Backer on our Open Collective. All donations go toward our web hosting and other community expenses such as hackathons and meetups!
- [Donate through OpenMined's Open Collective Page](https://opencollective.com/openmined)
|
github_jupyter
|
import torch as th
import syft as sy
hook = sy.TorchHook(th)
# IMPORTANT: Local worker should not be a client worker
hook.local_worker.is_client_worker = False
@sy.func2plan(args_shape=[(1,)])
def inc1(x):
return x + 1
@sy.func2plan(args_shape=[(1,)])
def inc2(x):
return x + 1
@sy.func2plan(args_shape=[(1,)])
def inc3(x):
return x + 1
protocol = sy.Protocol([("worker1", inc1), ("worker2", inc2), ("worker3", inc3)])
bob = sy.VirtualWorker(hook, id="bob")
alice = sy.VirtualWorker(hook, id="alice")
charlie = sy.VirtualWorker(hook, id="charlie")
workers = alice, bob, charlie
protocol.deploy(*workers)
x = th.tensor([1.0])
ptr = protocol.run(x)
ptr
ptr.get()
james = sy.VirtualWorker(hook, id="james")
protocol.send(james)
x = th.tensor([1.0]).send(james)
ptr = protocol.run(x)
ptr
ptr = ptr.get()
ptr
ptr = ptr.get()
ptr
protocol = sy.Protocol([("worker1", inc1), ("worker2", inc2), ("worker3", inc3)])
protocol.tag('my_protocol')
protocol.send(james)
me = sy.hook.local_worker # get access to me as a local worker
responses = me.request_search(['my_protocol'], location=james)
responses
ptr_protocol = responses[0]
protocol_back = ptr_protocol.get()
protocol_back
protocol_back.deploy(alice, bob, charlie)
x = th.tensor([1.0])
ptr = protocol_back.run(x)
ptr.get()
| 0.458834 | 0.959724 |
## Eno-gastronomic Heritage Collection
Data integration and Data Profile Project - 02/2022
Rachel Fanti Coelho Lima
## Summary
* 1. [Libraries](#libraries)
* 2. [Setup](#setup)
* 3. [Visualization](#visualization)
* 3.1 [Producers - location map](#producers_map)
* 3.2 [Producers by grape variety - location map](#producers_by_grape_map)
* 3.3 [Number of producers by Location](#producers_by_loc)
* 3.4 [Percentage of wines by colour](#wines_by_colour)
* 3.5 [Number of wines by certification](#wines_by_certification)
* 3.6 [Number of wines by producer and locality](#wine_by_producer_locality)
## 1. Libraries <a class="anchor" id="libraries"></a>
```
import numpy as np
import pandas as pd
import plotly.express as px
from jupyter_dash import JupyterDash
import dash
from dash import dcc
from dash import html
from dash.dependencies import Input, Output
from SPARQLWrapper import SPARQLWrapper, JSON
import sparql_dataframe
import warnings
warnings.filterwarnings("ignore")
#pip install sparqlwrapper
#pip install plotly
#pip install "jupyterlab>=3" "ipywidgets>=7.6"
#pip install jupyter-dash
```
## 2. Setup <a class="anchor" id="setup"></a>
```
# Set up the endpoint and the URL (http://localhost:8080/sparql).
# SPARQLWrapper library https://rdflib.github.io/sparqlwrapper/ is used to send SPARQL queries and get results.
# The following code gets the result as JSON documents and convert it to a Python dict object.
sparql = SPARQLWrapper("http://localhost:8080/sparql")
q1 = """
PREFIX : <http://www.semanticweb.org/rachel/ontologies/2022/0/untitled-ontology-30#>
SELECT ?cod ?wn ?sour ?col ?alc ?org_desc ?p ?lat ?long ?lau ?reg ?prov
WHERE {?w :isProducedBy ?ac;
:wWineCode ?cod;
:wName ?wn;
:hasDescription ?d.
?d :wdColour ?col.
?d:hasOrganolepticDescription ?od.
?ac a :Actor;
:hasMainAddress ?ad;
:acName ?p.
?ad :hasGeolocalization ?g.
?g :gLatitude ?lat;
:gLongitude ?long.
?ad :hasMunicipality ?m.
?m :mLauNameNational ?lau;
:mNUTSLevel2 ?reg;
:mNUTSLevel3 ?prov.
OPTIONAL {?w:wSource ?sour.}
OPTIONAL {?d :wdAlcoholContent ?alc.}
OPTIONAL {?od :wodOrganolepticDescription ?org_desc.}
}
"""
#sparql.setQuery(q1)
#sparql.setReturnFormat(JSON)
#results = sparql.query().convert()
#print(results)
# The SPARQL results are converted to a pandas DataFrame for data analysis.
# The library sparql-dataframe https://github.com/lawlesst/sparql-dataframe is handy for this.
endpoint = "http://localhost:8080/sparql"
#dataset of wine
df_w = sparql_dataframe.get(endpoint, q1)
df_w.head()
q2 = """
PREFIX : <http://www.semanticweb.org/rachel/ontologies/2022/0/untitled-ontology-30#>
SELECT ?cod ?wn ?sour ?col ?alc ?org_desc ?perc ?grap ?p ?lat ?long ?lau ?reg ?prov
WHERE {?w :isProducedBy ?ac;
:wWineCode ?cod;
:wName ?wn;
:hasDescription ?d.
?d :wdColour ?col.
?d:hasOrganolepticDescription ?od.
?ac a :Actor;
:hasMainAddress ?ad;
:acName ?p.
?ad :hasGeolocalization ?g.
?g :gLatitude ?lat;
:gLongitude ?long.
?ad :hasMunicipality ?m.
?m :mLauNameNational ?lau;
:mNUTSLevel2 ?reg;
:mNUTSLevel3 ?prov.
OPTIONAL {?w:wSource ?sour.}
OPTIONAL {?d :wdAlcoholContent ?alc.}
OPTIONAL {?od :wodOrganolepticDescription ?org_desc.}
?w :hasGrapeComposition ?gc.
?gc :hasGrape ?gv.
?gv :gvName ?grap. optional {?gc :wgcPercentageOfGrape ?perc.}
}
"""
#dataset of grape_composition
#(only data for the wines that has grape_composition, more than one line per wine, since they can have more than one grape) )
df_g = sparql_dataframe.get(endpoint, q2)
df_g.head()
q3 = """
PREFIX : <http://www.semanticweb.org/rachel/ontologies/2022/0/untitled-ontology-30#>
SELECT ?w ?c ?cert ?ext_cert
WHERE {?w a :Wine;
:hasCertification ?c.
?c :cName ?cert;
:cExtendedCode ?ext_cert.
}
"""
# dataset of certificates
#(only data for the wines that has certfication, it can have more than one line per wine)
df_c = sparql_dataframe.get(endpoint, q3)
df_c.head()
q4 = """
PREFIX : <http://www.semanticweb.org/rachel/ontologies/2022/0/untitled-ontology-30#>
SELECT ?p ?r ?lat ?long ?lau ?reg ?prov
WHERE {?ac :hasMainAddress ?d;
:acName ?p.
?d :hasGeolocalization ?g.
?g :gLatitude ?lat;
:gLongitude ?long.
?d :hasMunicipality ?m.
?m :mLauNameNational ?lau;
:mNUTSLevel2 ?reg;
:mNUTSLevel3 ?prov.
}
"""
# dataset for producer
df_p = sparql_dataframe.get(endpoint, q4)
df_p.head()
```
## 3. Visualization <a class="anchor" id="visualization"></a>
### 3.1 Producers - location map <a class="anchor" id="producers_map"></a>
```
fig = px.scatter_mapbox(df_w, lat="lat", lon="long", color = "prov", hover_name="p", hover_data=["lau", "prov"], zoom=6.5)
fig.update_layout(mapbox_style="open-street-map") #carto-positron, open-street-map
fig.update_layout(margin={"r":0,"t":30,"l":0,"b":0})
fig.update_geos(fitbounds = 'locations')
fig.show()
```
### 3.2 Producers by grape variety - location map <a class="anchor" id="producers_by_grape_map"></a>
```
grapes = sorted(df_g['grap'].unique())
app = JupyterDash(__name__)
server = app.server
app.layout = html.Div([
dcc.Dropdown(
id="dropdown_grape",
options=[{"label": x, "value": x} for x in grapes],
value=grapes[0],
clearable=False,
),
dcc.Graph(id="map_producers"),
dcc.Textarea(
#placeholder="Enter a value",
value="The map shows only producers who have provided information about grape variety.",
style={'width': '86.5%'}
)
])
@app.callback(
Output("map_producers", "figure"),
[Input("dropdown_grape", "value")])
def display_map_producers(dropdown_grape):
df1 = df_g[df_g['grap'] == dropdown_grape]
fig = px.scatter_mapbox(df1, lat="lat", lon="long", color = "prov", hover_name="p", hover_data=["lau", "prov"], zoom=6.5)
fig.update_layout(mapbox_style="open-street-map")
fig.update_layout(margin={"r":0,"t":30,"l":0,"b":0})
fig.update_geos(fitbounds = 'locations')
return fig
#app.run_server(port=8051)
app.run_server('inline')
```
### 3.3 Number of producers by Location <a class="anchor" id="producers_by_loc"></a>
```
list_local = ['lau', 'prov', 'reg']
app = JupyterDash(__name__)
server = app.server
app.layout = html.Div([
dcc.Dropdown(
id="dropdown_locality",
options=[{"label": x, "value": x} for x in list_local],
value=list_local[0],
clearable=False,
),
dcc.Graph(id="bar-chart"),
])
@app.callback(
Output("bar-chart", "figure"),
[Input("dropdown_locality", "value")])
def update_bar_chart(local):
fig = px.histogram(df_p, x=local).update_xaxes(categoryorder="total descending")
return fig
app.run_server('inline')
```
### 3.4 Percentage of wines by colour <a class="anchor" id="wines_by_colour"></a>
```
df_col = df_w['col'].value_counts()
df_col = df_col.to_frame()
fig = px.pie(df_col, values='col', names=df_col.index, title='Percentage of wines by colour')
fig.update_traces(textposition='inside', textinfo='percent+value+label')
fig.show()
```
### 3.5 Number of wines by certification <a class="anchor" id="wines_by_certificate"></a>
```
fig = px.histogram(df_c, x="cert").update_xaxes(categoryorder="total descending", title='Number of wines by certification')
fig.show()
```
### 3.6 Number of wines by producer and locality <a class="anchor" id="wines_by_producer_locality"></a>
```
list_local = ['lau', 'prov', 'reg']
list_values = sorted(df_g['prov'].unique())
app = JupyterDash(__name__)
server = app.server
app.layout = html.Div([
dcc.Dropdown(
id="dropdown_locality",
options=[{"label": x, "value": x} for x in list_local],
value=list_local[1],
clearable=False,
),
dcc.Dropdown(
id="dropdown_chosen_locality",
value=list_values[0],
clearable=False,
),
dcc.Graph(id="bar-chart"),
])
@app.callback(
Output("dropdown_chosen_locality", 'options'),
Input("dropdown_locality", "value"))
def update_dropdown (locality):
if locality=='lau':
list_values = sorted(df_g['lau'].unique())
elif locality=='prov':
list_values = sorted(df_g['prov'].unique())
else:
list_values = sorted(df_g['reg'].unique())
return [{'label': i, 'value': i} for i in list_values]
@app.callback(
Output("bar-chart", "figure"),
[Input("dropdown_locality", "value"),
Input("dropdown_chosen_locality", "value")])
def update_bar_chart(locality, chosen_locality):
df2 = df_w[df_w[locality]==chosen_locality]
fig = px.histogram(df2, x='p').update_xaxes(categoryorder="total descending")
return fig
app.run_server('inline')
#app.run_server(port=8050)
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import plotly.express as px
from jupyter_dash import JupyterDash
import dash
from dash import dcc
from dash import html
from dash.dependencies import Input, Output
from SPARQLWrapper import SPARQLWrapper, JSON
import sparql_dataframe
import warnings
warnings.filterwarnings("ignore")
#pip install sparqlwrapper
#pip install plotly
#pip install "jupyterlab>=3" "ipywidgets>=7.6"
#pip install jupyter-dash
# Set up the endpoint and the URL (http://localhost:8080/sparql).
# SPARQLWrapper library https://rdflib.github.io/sparqlwrapper/ is used to send SPARQL queries and get results.
# The following code gets the result as JSON documents and convert it to a Python dict object.
sparql = SPARQLWrapper("http://localhost:8080/sparql")
q1 = """
PREFIX : <http://www.semanticweb.org/rachel/ontologies/2022/0/untitled-ontology-30#>
SELECT ?cod ?wn ?sour ?col ?alc ?org_desc ?p ?lat ?long ?lau ?reg ?prov
WHERE {?w :isProducedBy ?ac;
:wWineCode ?cod;
:wName ?wn;
:hasDescription ?d.
?d :wdColour ?col.
?d:hasOrganolepticDescription ?od.
?ac a :Actor;
:hasMainAddress ?ad;
:acName ?p.
?ad :hasGeolocalization ?g.
?g :gLatitude ?lat;
:gLongitude ?long.
?ad :hasMunicipality ?m.
?m :mLauNameNational ?lau;
:mNUTSLevel2 ?reg;
:mNUTSLevel3 ?prov.
OPTIONAL {?w:wSource ?sour.}
OPTIONAL {?d :wdAlcoholContent ?alc.}
OPTIONAL {?od :wodOrganolepticDescription ?org_desc.}
}
"""
#sparql.setQuery(q1)
#sparql.setReturnFormat(JSON)
#results = sparql.query().convert()
#print(results)
# The SPARQL results are converted to a pandas DataFrame for data analysis.
# The library sparql-dataframe https://github.com/lawlesst/sparql-dataframe is handy for this.
endpoint = "http://localhost:8080/sparql"
#dataset of wine
df_w = sparql_dataframe.get(endpoint, q1)
df_w.head()
q2 = """
PREFIX : <http://www.semanticweb.org/rachel/ontologies/2022/0/untitled-ontology-30#>
SELECT ?cod ?wn ?sour ?col ?alc ?org_desc ?perc ?grap ?p ?lat ?long ?lau ?reg ?prov
WHERE {?w :isProducedBy ?ac;
:wWineCode ?cod;
:wName ?wn;
:hasDescription ?d.
?d :wdColour ?col.
?d:hasOrganolepticDescription ?od.
?ac a :Actor;
:hasMainAddress ?ad;
:acName ?p.
?ad :hasGeolocalization ?g.
?g :gLatitude ?lat;
:gLongitude ?long.
?ad :hasMunicipality ?m.
?m :mLauNameNational ?lau;
:mNUTSLevel2 ?reg;
:mNUTSLevel3 ?prov.
OPTIONAL {?w:wSource ?sour.}
OPTIONAL {?d :wdAlcoholContent ?alc.}
OPTIONAL {?od :wodOrganolepticDescription ?org_desc.}
?w :hasGrapeComposition ?gc.
?gc :hasGrape ?gv.
?gv :gvName ?grap. optional {?gc :wgcPercentageOfGrape ?perc.}
}
"""
#dataset of grape_composition
#(only data for the wines that has grape_composition, more than one line per wine, since they can have more than one grape) )
df_g = sparql_dataframe.get(endpoint, q2)
df_g.head()
q3 = """
PREFIX : <http://www.semanticweb.org/rachel/ontologies/2022/0/untitled-ontology-30#>
SELECT ?w ?c ?cert ?ext_cert
WHERE {?w a :Wine;
:hasCertification ?c.
?c :cName ?cert;
:cExtendedCode ?ext_cert.
}
"""
# dataset of certificates
#(only data for the wines that has certfication, it can have more than one line per wine)
df_c = sparql_dataframe.get(endpoint, q3)
df_c.head()
q4 = """
PREFIX : <http://www.semanticweb.org/rachel/ontologies/2022/0/untitled-ontology-30#>
SELECT ?p ?r ?lat ?long ?lau ?reg ?prov
WHERE {?ac :hasMainAddress ?d;
:acName ?p.
?d :hasGeolocalization ?g.
?g :gLatitude ?lat;
:gLongitude ?long.
?d :hasMunicipality ?m.
?m :mLauNameNational ?lau;
:mNUTSLevel2 ?reg;
:mNUTSLevel3 ?prov.
}
"""
# dataset for producer
df_p = sparql_dataframe.get(endpoint, q4)
df_p.head()
fig = px.scatter_mapbox(df_w, lat="lat", lon="long", color = "prov", hover_name="p", hover_data=["lau", "prov"], zoom=6.5)
fig.update_layout(mapbox_style="open-street-map") #carto-positron, open-street-map
fig.update_layout(margin={"r":0,"t":30,"l":0,"b":0})
fig.update_geos(fitbounds = 'locations')
fig.show()
grapes = sorted(df_g['grap'].unique())
app = JupyterDash(__name__)
server = app.server
app.layout = html.Div([
dcc.Dropdown(
id="dropdown_grape",
options=[{"label": x, "value": x} for x in grapes],
value=grapes[0],
clearable=False,
),
dcc.Graph(id="map_producers"),
dcc.Textarea(
#placeholder="Enter a value",
value="The map shows only producers who have provided information about grape variety.",
style={'width': '86.5%'}
)
])
@app.callback(
Output("map_producers", "figure"),
[Input("dropdown_grape", "value")])
def display_map_producers(dropdown_grape):
df1 = df_g[df_g['grap'] == dropdown_grape]
fig = px.scatter_mapbox(df1, lat="lat", lon="long", color = "prov", hover_name="p", hover_data=["lau", "prov"], zoom=6.5)
fig.update_layout(mapbox_style="open-street-map")
fig.update_layout(margin={"r":0,"t":30,"l":0,"b":0})
fig.update_geos(fitbounds = 'locations')
return fig
#app.run_server(port=8051)
app.run_server('inline')
list_local = ['lau', 'prov', 'reg']
app = JupyterDash(__name__)
server = app.server
app.layout = html.Div([
dcc.Dropdown(
id="dropdown_locality",
options=[{"label": x, "value": x} for x in list_local],
value=list_local[0],
clearable=False,
),
dcc.Graph(id="bar-chart"),
])
@app.callback(
Output("bar-chart", "figure"),
[Input("dropdown_locality", "value")])
def update_bar_chart(local):
fig = px.histogram(df_p, x=local).update_xaxes(categoryorder="total descending")
return fig
app.run_server('inline')
df_col = df_w['col'].value_counts()
df_col = df_col.to_frame()
fig = px.pie(df_col, values='col', names=df_col.index, title='Percentage of wines by colour')
fig.update_traces(textposition='inside', textinfo='percent+value+label')
fig.show()
fig = px.histogram(df_c, x="cert").update_xaxes(categoryorder="total descending", title='Number of wines by certification')
fig.show()
list_local = ['lau', 'prov', 'reg']
list_values = sorted(df_g['prov'].unique())
app = JupyterDash(__name__)
server = app.server
app.layout = html.Div([
dcc.Dropdown(
id="dropdown_locality",
options=[{"label": x, "value": x} for x in list_local],
value=list_local[1],
clearable=False,
),
dcc.Dropdown(
id="dropdown_chosen_locality",
value=list_values[0],
clearable=False,
),
dcc.Graph(id="bar-chart"),
])
@app.callback(
Output("dropdown_chosen_locality", 'options'),
Input("dropdown_locality", "value"))
def update_dropdown (locality):
if locality=='lau':
list_values = sorted(df_g['lau'].unique())
elif locality=='prov':
list_values = sorted(df_g['prov'].unique())
else:
list_values = sorted(df_g['reg'].unique())
return [{'label': i, 'value': i} for i in list_values]
@app.callback(
Output("bar-chart", "figure"),
[Input("dropdown_locality", "value"),
Input("dropdown_chosen_locality", "value")])
def update_bar_chart(locality, chosen_locality):
df2 = df_w[df_w[locality]==chosen_locality]
fig = px.histogram(df2, x='p').update_xaxes(categoryorder="total descending")
return fig
app.run_server('inline')
#app.run_server(port=8050)
| 0.421314 | 0.773601 |
# **Deep learning for image analysis with Python**
#### Fernando Cervantes, Systems Analyst I, Imaging Solutions, Research IT
#### [email protected] (slack) @fernando.cervantes
## 6 Monitoring and logging the training process
It is important to track the training process. By doing that, we can detect interesting behavior of our network, possible failures, and even *overfitting*.<br>
This also helps to save the results of different experiments performed using distinct configurations.
### 6.1 _Logging the network performance_
```
from torchvision.datasets import CIFAR100
from torch.utils.data import DataLoader
from torchvision.transforms import ToTensor
cifar_data = CIFAR100(root=r'/home/cervaf/data', # '/mnt/data'
download=False,
train=True,
transform=ToTensor()
)
cifar_loader = DataLoader(cifar_data,
batch_size=128,
shuffle=True,
pin_memory=True
)
import torch
import torch.nn as nn
class LeNet(nn.Module):
def __init__(self, in_channels=1, num_classes=10):
"""
Always call the initialization function from the nn.Module parent class.
This way all parameters from the operations defined as members of *this* class are tracked for their optimization.
"""
super(LeNet, self).__init__()
self.conv_1 = nn.Conv2d(in_channels=in_channels, out_channels=6, kernel_size=5)
self.sub_1 = nn.MaxPool2d(kernel_size=2, stride=2)
self.conv_2 = nn.Conv2d(in_channels=6, out_channels=16, kernel_size=5)
self.sub_2 = nn.MaxPool2d(kernel_size=2, stride=2)
self.fc_1 = nn.Linear(in_features=5*5*16, out_features=120)
self.fc_2 = nn.Linear(in_features=120, out_features=84)
self.fc_3 = nn.Linear(in_features=84, out_features=num_classes)
self.act_fn = nn.ReLU()
def forward(self, x):
# Apply convolution layers to extract feature maps with image context
fx = self.act_fn(self.conv_1(x))
fx = self.sub_1(fx)
fx = self.act_fn(self.conv_2(fx))
fx = self.sub_2(fx)
# Flatten the feature maps to perform linear operations
fx = fx.view(-1, 16*5*5)
fx = self.act_fn(self.fc_1(fx))
fx = self.act_fn(self.fc_2(fx))
y = self.fc_3(fx)
return y
net = LeNet(in_channels=3, num_classes=100)
criterion = nn.CrossEntropyLoss()
net.cuda()
criterion.cuda()
import torch.optim as optim
optimizer = optim.Adam(
params=net.parameters(),
lr=1e-3
)
```
***
Now that we have set up our experiment, lets create a summary writer for our training stage
```
from torch.utils.tensorboard import SummaryWriter
```
Create a summary writter using TensorBoard
```
writer = SummaryWriter('runs/LR_0_001_BATCH_128')
net.train()
for e in range(10):
avg_loss = 0
avg_acc = 0
for i, (x, t) in enumerate(cifar_loader):
optimizer.zero_grad()
x = x.cuda()
t = t.cuda()
y = net(x)
loss = criterion(y, t)
loss.backward()
curr_acc = torch.sum(y.argmax(dim=1) == t)
avg_loss += loss.item()
avg_acc += curr_acc
optimizer.step()
writer.add_scalar('training loss', loss.item(), e * len(cifar_loader) + i)
writer.add_scalar('training acc', curr_acc / x.size(0), e * len(cifar_loader) + i)
avg_loss = avg_loss / len(cifar_loader)
avg_acc = avg_acc / len(cifar_data)
writer.add_scalar('training loss', loss.item(), e)
writer.add_scalar('training loss', loss.item(), e)
torch.save(net.state_dict(), 'lenet_700epochs_20220519.pth')
```
|
github_jupyter
|
from torchvision.datasets import CIFAR100
from torch.utils.data import DataLoader
from torchvision.transforms import ToTensor
cifar_data = CIFAR100(root=r'/home/cervaf/data', # '/mnt/data'
download=False,
train=True,
transform=ToTensor()
)
cifar_loader = DataLoader(cifar_data,
batch_size=128,
shuffle=True,
pin_memory=True
)
import torch
import torch.nn as nn
class LeNet(nn.Module):
def __init__(self, in_channels=1, num_classes=10):
"""
Always call the initialization function from the nn.Module parent class.
This way all parameters from the operations defined as members of *this* class are tracked for their optimization.
"""
super(LeNet, self).__init__()
self.conv_1 = nn.Conv2d(in_channels=in_channels, out_channels=6, kernel_size=5)
self.sub_1 = nn.MaxPool2d(kernel_size=2, stride=2)
self.conv_2 = nn.Conv2d(in_channels=6, out_channels=16, kernel_size=5)
self.sub_2 = nn.MaxPool2d(kernel_size=2, stride=2)
self.fc_1 = nn.Linear(in_features=5*5*16, out_features=120)
self.fc_2 = nn.Linear(in_features=120, out_features=84)
self.fc_3 = nn.Linear(in_features=84, out_features=num_classes)
self.act_fn = nn.ReLU()
def forward(self, x):
# Apply convolution layers to extract feature maps with image context
fx = self.act_fn(self.conv_1(x))
fx = self.sub_1(fx)
fx = self.act_fn(self.conv_2(fx))
fx = self.sub_2(fx)
# Flatten the feature maps to perform linear operations
fx = fx.view(-1, 16*5*5)
fx = self.act_fn(self.fc_1(fx))
fx = self.act_fn(self.fc_2(fx))
y = self.fc_3(fx)
return y
net = LeNet(in_channels=3, num_classes=100)
criterion = nn.CrossEntropyLoss()
net.cuda()
criterion.cuda()
import torch.optim as optim
optimizer = optim.Adam(
params=net.parameters(),
lr=1e-3
)
from torch.utils.tensorboard import SummaryWriter
writer = SummaryWriter('runs/LR_0_001_BATCH_128')
net.train()
for e in range(10):
avg_loss = 0
avg_acc = 0
for i, (x, t) in enumerate(cifar_loader):
optimizer.zero_grad()
x = x.cuda()
t = t.cuda()
y = net(x)
loss = criterion(y, t)
loss.backward()
curr_acc = torch.sum(y.argmax(dim=1) == t)
avg_loss += loss.item()
avg_acc += curr_acc
optimizer.step()
writer.add_scalar('training loss', loss.item(), e * len(cifar_loader) + i)
writer.add_scalar('training acc', curr_acc / x.size(0), e * len(cifar_loader) + i)
avg_loss = avg_loss / len(cifar_loader)
avg_acc = avg_acc / len(cifar_data)
writer.add_scalar('training loss', loss.item(), e)
writer.add_scalar('training loss', loss.item(), e)
torch.save(net.state_dict(), 'lenet_700epochs_20220519.pth')
| 0.945311 | 0.944689 |
# Fictional Army - Filtering and Sorting
### Introduction:
This exercise was inspired by this [page](http://chrisalbon.com/python/)
Special thanks to: https://github.com/chrisalbon for sharing the dataset and materials.
### Step 1. Import the necessary libraries
```
import pandas as pd
```
### Step 2. This is the data given as a dictionary
```
# Create an example dataframe about a fictional army
raw_data = {'regiment': ['Nighthawks', 'Nighthawks', 'Nighthawks', 'Nighthawks', 'Dragoons', 'Dragoons', 'Dragoons', 'Dragoons', 'Scouts', 'Scouts', 'Scouts', 'Scouts'],
'company': ['1st', '1st', '2nd', '2nd', '1st', '1st', '2nd', '2nd','1st', '1st', '2nd', '2nd'],
'deaths': [523, 52, 25, 616, 43, 234, 523, 62, 62, 73, 37, 35],
'battles': [5, 42, 2, 2, 4, 7, 8, 3, 4, 7, 8, 9],
'size': [1045, 957, 1099, 1400, 1592, 1006, 987, 849, 973, 1005, 1099, 1523],
'veterans': [1, 5, 62, 26, 73, 37, 949, 48, 48, 435, 63, 345],
'readiness': [1, 2, 3, 3, 2, 1, 2, 3, 2, 1, 2, 3],
'armored': [1, 0, 1, 1, 0, 1, 0, 1, 0, 0, 1, 1],
'deserters': [4, 24, 31, 2, 3, 4, 24, 31, 2, 3, 2, 3],
'origin': ['Arizona', 'California', 'Texas', 'Florida', 'Maine', 'Iowa', 'Alaska', 'Washington', 'Oregon', 'Wyoming', 'Louisana', 'Georgia']}
```
### Step 3. Create a dataframe and assign it to a variable called army.
#### Don't forget to include the columns names in the order presented in the dictionary ('regiment', 'company', 'deaths'...) so that the column index order is consistent with the solutions. If omitted, pandas will order the columns alphabetically.
```
army = pd.DataFrame(raw_data)
army
```
### Step 4. Set the 'origin' colum as the index of the dataframe
```
army = army.set_index('origin')
```
### Step 5. Print only the column veterans
```
army.veterans
```
### Step 6. Print the columns 'veterans' and 'deaths'
```
army[['veterans', 'deaths']]
```
### Step 7. Print the name of all the columns.
```
army.columns
```
### Step 8. Select the 'deaths', 'size' and 'deserters' columns from Maine and Alaska
```
army.loc[['Maine','Alaska'] , ["deaths","size","deserters"]]
```
### Step 9. Select the rows 3 to 7 and the columns 3 to 6
```
army.iloc[3:7, 3:6]
```
### Step 10. Select every row after the fourth row
```
army.iloc[4:]
```
### Step 11. Select every row up to the 4th row
```
army.iloc[:4]
```
### Step 12. Select the 3rd column up to the 7th column
```
army.iloc[:, 3:7]
```
### Step 13. Select rows where df.deaths is greater than 50
```
army[army.deaths > 50]
```
### Step 14. Select rows where df.deaths is greater than 500 or less than 50
```
army[(army.deaths < 50) | (army.deaths > 500)]
```
### Step 15. Select all the regiments not named "Dragoons"
```
army[army.regiment != 'Dragoons']
```
### Step 16. Select the rows called Texas and Arizona
```
army.loc[['Texas', 'Arizona']]
```
### Step 17. Select the third cell in the row named Arizona
```
army.iloc[[0], army.columns.get_loc('deaths')]
```
### Step 18. Select the third cell down in the column named deaths
```
army.iloc[[2], army.columns.get_loc('deaths')]
```
|
github_jupyter
|
import pandas as pd
# Create an example dataframe about a fictional army
raw_data = {'regiment': ['Nighthawks', 'Nighthawks', 'Nighthawks', 'Nighthawks', 'Dragoons', 'Dragoons', 'Dragoons', 'Dragoons', 'Scouts', 'Scouts', 'Scouts', 'Scouts'],
'company': ['1st', '1st', '2nd', '2nd', '1st', '1st', '2nd', '2nd','1st', '1st', '2nd', '2nd'],
'deaths': [523, 52, 25, 616, 43, 234, 523, 62, 62, 73, 37, 35],
'battles': [5, 42, 2, 2, 4, 7, 8, 3, 4, 7, 8, 9],
'size': [1045, 957, 1099, 1400, 1592, 1006, 987, 849, 973, 1005, 1099, 1523],
'veterans': [1, 5, 62, 26, 73, 37, 949, 48, 48, 435, 63, 345],
'readiness': [1, 2, 3, 3, 2, 1, 2, 3, 2, 1, 2, 3],
'armored': [1, 0, 1, 1, 0, 1, 0, 1, 0, 0, 1, 1],
'deserters': [4, 24, 31, 2, 3, 4, 24, 31, 2, 3, 2, 3],
'origin': ['Arizona', 'California', 'Texas', 'Florida', 'Maine', 'Iowa', 'Alaska', 'Washington', 'Oregon', 'Wyoming', 'Louisana', 'Georgia']}
army = pd.DataFrame(raw_data)
army
army = army.set_index('origin')
army.veterans
army[['veterans', 'deaths']]
army.columns
army.loc[['Maine','Alaska'] , ["deaths","size","deserters"]]
army.iloc[3:7, 3:6]
army.iloc[4:]
army.iloc[:4]
army.iloc[:, 3:7]
army[army.deaths > 50]
army[(army.deaths < 50) | (army.deaths > 500)]
army[army.regiment != 'Dragoons']
army.loc[['Texas', 'Arizona']]
army.iloc[[0], army.columns.get_loc('deaths')]
army.iloc[[2], army.columns.get_loc('deaths')]
| 0.342242 | 0.981275 |
<a href="https://colab.research.google.com/github/kapil9236/Crop-Production/blob/master/Kapil_Crop_Production.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Import the Libraries and read the data
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
df=pd.read_csv('/content/drive/MyDrive/Crop_recommendation.csv')
df.head(10)
```
* N - ratio of Nitrogen content in soil
* P - ratio of Phosphorous content in soil
* K - ratio of Potassium content in soil
* temperature - temperature in degree Celsius
* humidity - relative humidity in %
* ph - ph value of the soil
* rainfall - rainfall in mm
# Data Wrangling
```
df.info()
df.columns
```
**Since all input variables contributes to our predicton, we don't make any changes in the given data set.**
```
df.describe()
df.label.unique()
```
# Missing Values Treatment
**When values are missing in some colums of a dataset then we deal with it as follows**
1. No values missing then skip this step
1. If more than 50% values are missing in a column then drop that particular column
2. Else for continuos values always replace with Median(robost)
1. For descrete values always replace with Mode of that particular column
```
df.isnull().sum()
sns.heatmap(df.isnull())
```
# Exploratory Data Analysis
* It is a way of visualizing, summarizing and interpreting the information that is hidden in given dataset.
1. Variable Identification
2. Univariate analysis - histogram and Box Plot
3. Bivariate Analysis - three types
4. Feature Selection
5. Outlier detection
```
# Variable Identification
df.describe()
# Continuous Variables = [N,P,K, temperature,humidity,ph,rainfall]
# Categorical Variables =[label]
# Univariate Analysis
f= plt.figure(figsize=(16,4))
ax=ax=f.add_subplot(121)
sns.distplot(df['N'], color='purple', ax=ax)
ax=f.add_subplot(122)
sns.distplot(df['P'], color='blue', ax=ax)
sns.displot(df['K'], color='green', binwidth=12)
sns.set_style("whitegrid")
sns.boxplot('ph', palette='Spectral', data=df)
plt.figure(figsize=(6,4))
sns.boxplot('rainfall', data=df)
# Bi variate analysis
df.head()
df.corr()
sns.heatmap(df.corr())
sns.relplot(x="P", y="K", hue="label", data=df);
sns.relplot(x="humidity", y="rainfall", hue="label", data=df);
f= plt.figure(figsize=(12,4))
sns.countplot(df['label'] , palette = 'Spectral')
plt.xticks(rotation=90)
plt.show()
# We can clearly see that for every label we have 100 training examples
# We don't need to remove any label
sns.set_style("whitegrid")
plt.figure(figsize=(24,6))
sns.boxplot(x = 'label', y = 'P', width=0.8, data = df)
sns.set_style("whitegrid")
plt.figure(figsize=(24,6))
sns.boxplot(x = 'label', y = 'rainfall', width=0.8, data = df)
df.groupby('label').rainfall.mean()
```
# Convert the data in Numeric Form
1. One Hot Encoding
2. Label Encoding
```
df.label.unique()
```
**Label** **Encoding**
```
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
df['label'] = le.fit_transform(df['label'])
df.head()
```
# Separating Features and Target Label
```
X=df.drop('label', axis=1)
X.head()
y=df['label']
y.head()
```
**Train Test Split**
```
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X,y,random_state=0,test_size=0.3)
```
# Machine Learning Model
Since it is a classification problem of supervised machine learning, we can use different classification problems as given below:
1. Logistic Regression
2. Neural Network or MLP
3. SVM
4. Decision Tree
5. Random Forest
6. Naive Bayes
7. KNN
```
accuracy=[]
model=[]
```
# Logistic Regression
```
from sklearn.linear_model import LogisticRegression
lg=LogisticRegression()
lg.fit(X_train,y_train)
a=lg.score(X_test,y_test)
accuracy.append(a)
model.append('Logistic Regression')
a
```
# Neural Network or MLP
```
from sklearn.neural_network import MLPClassifier
mlp=MLPClassifier(hidden_layer_sizes=[100,100],activation='relu', alpha=0.001).fit(X_train,y_train)
b=mlp.score(X_test,y_test)
accuracy.append(b)
model.append('Neural Network')
b
```
# SVM
```
from sklearn.svm import SVC
svm=SVC(C= 2, kernel='rbf').fit(X_train, y_train)
c=svm.score(X_test,y_test)
accuracy.append(c)
model.append('SVM')
c
```
# Decision Tree
```
from sklearn.tree import DecisionTreeClassifier
tree= DecisionTreeClassifier().fit(X_train, y_train)
d=tree.score(X_test, y_test)
accuracy.append(d)
model.append('Decision Tree')
d
```
# **Random Forest**
```
from sklearn.ensemble import RandomForestClassifier
rm= RandomForestClassifier( max_depth=12).fit(X_train, y_train)
e=rm.score(X_test,y_test)
accuracy.append(e)
model.append('Random Forest')
e
```
#Naive Bayes
```
from sklearn.naive_bayes import GaussianNB
nb= GaussianNB().fit(X_train, y_train)
f=nb.score(X_test,y_test)
accuracy.append(f)
model.append('Naive Bayes')
f
```
#KNN
```
from sklearn.neighbors import KNeighborsClassifier
knn=KNeighborsClassifier(n_neighbors= 31).fit(X_train,y_train)
g=knn.score(X_test,y_test)
accuracy.append(g)
model.append('KNN')
g
z= pd.DataFrame({'Logistic Regression':[a], 'Neural Network':[b],'SVM':[c],'Decision Tree':[d],'Random Forest':[e],'Naive Bayes':[f], 'KNN':[g]}).astype(float)
```
# **Model Accuracy**
```
z.T
plt.figure(figsize=(11,6))
sns.barplot(x = model , y = accuracy ,palette ='Spectral')
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
df=pd.read_csv('/content/drive/MyDrive/Crop_recommendation.csv')
df.head(10)
df.info()
df.columns
df.describe()
df.label.unique()
df.isnull().sum()
sns.heatmap(df.isnull())
# Variable Identification
df.describe()
# Continuous Variables = [N,P,K, temperature,humidity,ph,rainfall]
# Categorical Variables =[label]
# Univariate Analysis
f= plt.figure(figsize=(16,4))
ax=ax=f.add_subplot(121)
sns.distplot(df['N'], color='purple', ax=ax)
ax=f.add_subplot(122)
sns.distplot(df['P'], color='blue', ax=ax)
sns.displot(df['K'], color='green', binwidth=12)
sns.set_style("whitegrid")
sns.boxplot('ph', palette='Spectral', data=df)
plt.figure(figsize=(6,4))
sns.boxplot('rainfall', data=df)
# Bi variate analysis
df.head()
df.corr()
sns.heatmap(df.corr())
sns.relplot(x="P", y="K", hue="label", data=df);
sns.relplot(x="humidity", y="rainfall", hue="label", data=df);
f= plt.figure(figsize=(12,4))
sns.countplot(df['label'] , palette = 'Spectral')
plt.xticks(rotation=90)
plt.show()
# We can clearly see that for every label we have 100 training examples
# We don't need to remove any label
sns.set_style("whitegrid")
plt.figure(figsize=(24,6))
sns.boxplot(x = 'label', y = 'P', width=0.8, data = df)
sns.set_style("whitegrid")
plt.figure(figsize=(24,6))
sns.boxplot(x = 'label', y = 'rainfall', width=0.8, data = df)
df.groupby('label').rainfall.mean()
df.label.unique()
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
df['label'] = le.fit_transform(df['label'])
df.head()
X=df.drop('label', axis=1)
X.head()
y=df['label']
y.head()
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X,y,random_state=0,test_size=0.3)
accuracy=[]
model=[]
from sklearn.linear_model import LogisticRegression
lg=LogisticRegression()
lg.fit(X_train,y_train)
a=lg.score(X_test,y_test)
accuracy.append(a)
model.append('Logistic Regression')
a
from sklearn.neural_network import MLPClassifier
mlp=MLPClassifier(hidden_layer_sizes=[100,100],activation='relu', alpha=0.001).fit(X_train,y_train)
b=mlp.score(X_test,y_test)
accuracy.append(b)
model.append('Neural Network')
b
from sklearn.svm import SVC
svm=SVC(C= 2, kernel='rbf').fit(X_train, y_train)
c=svm.score(X_test,y_test)
accuracy.append(c)
model.append('SVM')
c
from sklearn.tree import DecisionTreeClassifier
tree= DecisionTreeClassifier().fit(X_train, y_train)
d=tree.score(X_test, y_test)
accuracy.append(d)
model.append('Decision Tree')
d
from sklearn.ensemble import RandomForestClassifier
rm= RandomForestClassifier( max_depth=12).fit(X_train, y_train)
e=rm.score(X_test,y_test)
accuracy.append(e)
model.append('Random Forest')
e
from sklearn.naive_bayes import GaussianNB
nb= GaussianNB().fit(X_train, y_train)
f=nb.score(X_test,y_test)
accuracy.append(f)
model.append('Naive Bayes')
f
from sklearn.neighbors import KNeighborsClassifier
knn=KNeighborsClassifier(n_neighbors= 31).fit(X_train,y_train)
g=knn.score(X_test,y_test)
accuracy.append(g)
model.append('KNN')
g
z= pd.DataFrame({'Logistic Regression':[a], 'Neural Network':[b],'SVM':[c],'Decision Tree':[d],'Random Forest':[e],'Naive Bayes':[f], 'KNN':[g]}).astype(float)
z.T
plt.figure(figsize=(11,6))
sns.barplot(x = model , y = accuracy ,palette ='Spectral')
| 0.524151 | 0.967194 |
# Deep Convolutional GANs
In this notebook, you'll build a GAN using convolutional layers in the generator and discriminator. This is called a Deep Convolutional GAN, or DCGAN for short. The DCGAN architecture was first explored last year and has seen impressive results in generating new images, you can read the [original paper here](https://arxiv.org/pdf/1511.06434.pdf).
You'll be training DCGAN on the [Street View House Numbers](http://ufldl.stanford.edu/housenumbers/) (SVHN) dataset. These are color images of house numbers collected from Google street view. SVHN images are in color and much more variable than MNIST.

So, we'll need a deeper and more powerful network. This is accomplished through using convolutional layers in the discriminator and generator. It's also necessary to use batch normalization to get the convolutional networks to train. The only real changes compared to what [you saw previously](https://github.com/udacity/deep-learning/tree/master/gan_mnist) are in the generator and discriminator, otherwise the rest of the implementation is the same.
```
%matplotlib inline
import pickle as pkl
import matplotlib.pyplot as plt
import numpy as np
from scipy.io import loadmat
import tensorflow as tf
!mkdir data
```
## Getting the data
Here you can download the SVHN dataset. Run the cell above and it'll download to your machine.
```
from urllib.request import urlretrieve
from os.path import isfile, isdir
from tqdm import tqdm
data_dir = 'data/'
if not isdir(data_dir):
raise Exception("Data directory doesn't exist!")
class DLProgress(tqdm):
last_block = 0
def hook(self, block_num=1, block_size=1, total_size=None):
self.total = total_size
self.update((block_num - self.last_block) * block_size)
self.last_block = block_num
if not isfile(data_dir + "train_32x32.mat"):
with DLProgress(unit='B', unit_scale=True, miniters=1, desc='SVHN Training Set') as pbar:
urlretrieve(
'http://ufldl.stanford.edu/housenumbers/train_32x32.mat',
data_dir + 'train_32x32.mat',
pbar.hook)
if not isfile(data_dir + "test_32x32.mat"):
with DLProgress(unit='B', unit_scale=True, miniters=1, desc='SVHN Testing Set') as pbar:
urlretrieve(
'http://ufldl.stanford.edu/housenumbers/test_32x32.mat',
data_dir + 'test_32x32.mat',
pbar.hook)
```
These SVHN files are `.mat` files typically used with Matlab. However, we can load them in with `scipy.io.loadmat` which we imported above.
```
trainset = loadmat(data_dir + 'train_32x32.mat')
testset = loadmat(data_dir + 'test_32x32.mat')
```
Here I'm showing a small sample of the images. Each of these is 32x32 with 3 color channels (RGB). These are the real images we'll pass to the discriminator and what the generator will eventually fake.
```
idx = np.random.randint(0, trainset['X'].shape[3], size=36)
fig, axes = plt.subplots(6, 6, sharex=True, sharey=True, figsize=(5,5),)
for ii, ax in zip(idx, axes.flatten()):
ax.imshow(trainset['X'][:,:,:,ii], aspect='equal')
ax.xaxis.set_visible(False)
ax.yaxis.set_visible(False)
plt.subplots_adjust(wspace=0, hspace=0)
```
Here we need to do a bit of preprocessing and getting the images into a form where we can pass batches to the network. First off, we need to rescale the images to a range of -1 to 1, since the output of our generator is also in that range. We also have a set of test and validation images which could be used if we're trying to identify the numbers in the images.
```
def scale(x, feature_range=(-1, 1)):
# scale to (0, 1)
x = ((x - x.min())/(255 - x.min()))
# scale to feature_range
min, max = feature_range
x = x * (max - min) + min
return x
class Dataset:
def __init__(self, train, test, val_frac=0.5, shuffle=False, scale_func=None):
split_idx = int(len(test['y'])*(1 - val_frac))
self.test_x, self.valid_x = test['X'][:,:,:,:split_idx], test['X'][:,:,:,split_idx:]
self.test_y, self.valid_y = test['y'][:split_idx], test['y'][split_idx:]
self.train_x, self.train_y = train['X'], train['y']
self.train_x = np.rollaxis(self.train_x, 3)
self.valid_x = np.rollaxis(self.valid_x, 3)
self.test_x = np.rollaxis(self.test_x, 3)
if scale_func is None:
self.scaler = scale
else:
self.scaler = scale_func
self.shuffle = shuffle
def batches(self, batch_size):
if self.shuffle:
idx = np.arange(len(dataset.train_x))
np.random.shuffle(idx)
self.train_x = self.train_x[idx]
self.train_y = self.train_y[idx]
n_batches = len(self.train_y)//batch_size
for ii in range(0, len(self.train_y), batch_size):
x = self.train_x[ii:ii+batch_size]
y = self.train_y[ii:ii+batch_size]
yield self.scaler(x), y
```
## Network Inputs
Here, just creating some placeholders like normal.
```
def model_inputs(real_dim, z_dim):
inputs_real = tf.placeholder(tf.float32, (None, *real_dim), name='input_real')
inputs_z = tf.placeholder(tf.float32, (None, z_dim), name='input_z')
return inputs_real, inputs_z
```
## Generator
Here you'll build the generator network. The input will be our noise vector `z` as before. Also as before, the output will be a $tanh$ output, but this time with size 32x32 which is the size of our SVHN images.
What's new here is we'll use convolutional layers to create our new images. The first layer is a fully connected layer which is reshaped into a deep and narrow layer, something like 4x4x1024 as in the original DCGAN paper. Then we use batch normalization and a leaky ReLU activation. Next is a transposed convolution where typically you'd halve the depth and double the width and height of the previous layer. Again, we use batch normalization and leaky ReLU. For each of these layers, the general scheme is convolution > batch norm > leaky ReLU.
You keep stacking layers up like this until you get the final transposed convolution layer with shape 32x32x3. Below is the archicture used in the original DCGAN paper:

Note that the final layer here is 64x64x3, while for our SVHN dataset, we only want it to be 32x32x3.
>**Exercise:** Build the transposed convolutional network for the generator in the function below. Be sure to use leaky ReLUs on all the layers except for the last tanh layer, as well as batch normalization on all the transposed convolutional layers except the last one.
```
def generator(z, output_dim, reuse=False, alpha=0.2, training=True):
with tf.variable_scope('generator', reuse=reuse):
# First fully connected layer
x = tf.layers.Dense(z, 512)
x = tf.nn.conv2d(x, 256, 2)
x = tf.layers.
x = tf.nn.conv2d(x, 128, 2)
x = tf.nn.conv2d(x, 64, 2)
x = tf.nn.conv2d(x, 32, 2)
# Output layer, 32x32x3
logits =
out = tf.tanh(logits)
return out
```
## Discriminator
Here you'll build the discriminator. This is basically just a convolutional classifier like you've built before. The input to the discriminator are 32x32x3 tensors/images. You'll want a few convolutional layers, then a fully connected layer for the output. As before, we want a sigmoid output, and you'll need to return the logits as well. For the depths of the convolutional layers I suggest starting with 16, 32, 64 filters in the first layer, then double the depth as you add layers. Note that in the DCGAN paper, they did all the downsampling using only strided convolutional layers with no maxpool layers.
You'll also want to use batch normalization with `tf.layers.batch_normalization` on each layer except the first convolutional and output layers. Again, each layer should look something like convolution > batch norm > leaky ReLU.
Note: in this project, your batch normalization layers will always use batch statistics. (That is, always set `training` to `True`.) That's because we are only interested in using the discriminator to help train the generator. However, if you wanted to use the discriminator for inference later, then you would need to set the `training` parameter appropriately.
>**Exercise:** Build the convolutional network for the discriminator. The input is a 32x32x3 images, the output is a sigmoid plus the logits. Again, use Leaky ReLU activations and batch normalization on all the layers except the first.
```
def discriminator(x, reuse=False, alpha=0.2):
with tf.variable_scope('discriminator', reuse=reuse):
# Input layer is 32x32x3
x =
logits =
out =
return out, logits
```
## Model Loss
Calculating the loss like before, nothing new here.
```
def model_loss(input_real, input_z, output_dim, alpha=0.2):
"""
Get the loss for the discriminator and generator
:param input_real: Images from the real dataset
:param input_z: Z input
:param out_channel_dim: The number of channels in the output image
:return: A tuple of (discriminator loss, generator loss)
"""
g_model = generator(input_z, output_dim, alpha=alpha)
d_model_real, d_logits_real = discriminator(input_real, alpha=alpha)
d_model_fake, d_logits_fake = discriminator(g_model, reuse=True, alpha=alpha)
d_loss_real = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=d_logits_real, labels=tf.ones_like(d_model_real)))
d_loss_fake = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=d_logits_fake, labels=tf.zeros_like(d_model_fake)))
g_loss = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=d_logits_fake, labels=tf.ones_like(d_model_fake)))
d_loss = d_loss_real + d_loss_fake
return d_loss, g_loss
```
## Optimizers
Not much new here, but notice how the train operations are wrapped in a `with tf.control_dependencies` block so the batch normalization layers can update their population statistics.
```
def model_opt(d_loss, g_loss, learning_rate, beta1):
"""
Get optimization operations
:param d_loss: Discriminator loss Tensor
:param g_loss: Generator loss Tensor
:param learning_rate: Learning Rate Placeholder
:param beta1: The exponential decay rate for the 1st moment in the optimizer
:return: A tuple of (discriminator training operation, generator training operation)
"""
# Get weights and bias to update
t_vars = tf.trainable_variables()
d_vars = [var for var in t_vars if var.name.startswith('discriminator')]
g_vars = [var for var in t_vars if var.name.startswith('generator')]
# Optimize
with tf.control_dependencies(tf.get_collection(tf.GraphKeys.UPDATE_OPS)):
d_train_opt = tf.train.AdamOptimizer(learning_rate, beta1=beta1).minimize(d_loss, var_list=d_vars)
g_train_opt = tf.train.AdamOptimizer(learning_rate, beta1=beta1).minimize(g_loss, var_list=g_vars)
return d_train_opt, g_train_opt
```
## Building the model
Here we can use the functions we defined about to build the model as a class. This will make it easier to move the network around in our code since the nodes and operations in the graph are packaged in one object.
```
class GAN:
def __init__(self, real_size, z_size, learning_rate, alpha=0.2, beta1=0.5):
tf.reset_default_graph()
self.input_real, self.input_z = model_inputs(real_size, z_size)
self.d_loss, self.g_loss = model_loss(self.input_real, self.input_z,
real_size[2], alpha=alpha)
self.d_opt, self.g_opt = model_opt(self.d_loss, self.g_loss, learning_rate, beta1)
```
Here is a function for displaying generated images.
```
def view_samples(epoch, samples, nrows, ncols, figsize=(5,5)):
fig, axes = plt.subplots(figsize=figsize, nrows=nrows, ncols=ncols,
sharey=True, sharex=True)
for ax, img in zip(axes.flatten(), samples[epoch]):
ax.axis('off')
img = ((img - img.min())*255 / (img.max() - img.min())).astype(np.uint8)
ax.set_adjustable('box-forced')
im = ax.imshow(img, aspect='equal')
plt.subplots_adjust(wspace=0, hspace=0)
return fig, axes
```
And another function we can use to train our network. Notice when we call `generator` to create the samples to display, we set `training` to `False`. That's so the batch normalization layers will use the population statistics rather than the batch statistics. Also notice that we set the `net.input_real` placeholder when we run the generator's optimizer. The generator doesn't actually use it, but we'd get an error without it because of the `tf.control_dependencies` block we created in `model_opt`.
```
def train(net, dataset, epochs, batch_size, print_every=10, show_every=100, figsize=(5,5)):
saver = tf.train.Saver()
sample_z = np.random.uniform(-1, 1, size=(72, z_size))
samples, losses = [], []
steps = 0
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
for e in range(epochs):
for x, y in dataset.batches(batch_size):
steps += 1
# Sample random noise for G
batch_z = np.random.uniform(-1, 1, size=(batch_size, z_size))
# Run optimizers
_ = sess.run(net.d_opt, feed_dict={net.input_real: x, net.input_z: batch_z})
_ = sess.run(net.g_opt, feed_dict={net.input_z: batch_z, net.input_real: x})
if steps % print_every == 0:
# At the end of each epoch, get the losses and print them out
train_loss_d = net.d_loss.eval({net.input_z: batch_z, net.input_real: x})
train_loss_g = net.g_loss.eval({net.input_z: batch_z})
print("Epoch {}/{}...".format(e+1, epochs),
"Discriminator Loss: {:.4f}...".format(train_loss_d),
"Generator Loss: {:.4f}".format(train_loss_g))
# Save losses to view after training
losses.append((train_loss_d, train_loss_g))
if steps % show_every == 0:
gen_samples = sess.run(
generator(net.input_z, 3, reuse=True, training=False),
feed_dict={net.input_z: sample_z})
samples.append(gen_samples)
_ = view_samples(-1, samples, 6, 12, figsize=figsize)
plt.show()
saver.save(sess, './checkpoints/generator.ckpt')
with open('samples.pkl', 'wb') as f:
pkl.dump(samples, f)
return losses, samples
```
## Hyperparameters
GANs are very sensitive to hyperparameters. A lot of experimentation goes into finding the best hyperparameters such that the generator and discriminator don't overpower each other. Try out your own hyperparameters or read [the DCGAN paper](https://arxiv.org/pdf/1511.06434.pdf) to see what worked for them.
>**Exercise:** Find hyperparameters to train this GAN. The values found in the DCGAN paper work well, or you can experiment on your own. In general, you want the discriminator loss to be around 0.3, this means it is correctly classifying images as fake or real about 50% of the time.
```
real_size = (32,32,3)
z_size = 100
learning_rate = 0.001
batch_size = 64
epochs = 1
alpha = 0.01
beta1 = 0.9
# Create the network
net = GAN(real_size, z_size, learning_rate, alpha=alpha, beta1=beta1)
# Load the data and train the network here
dataset = Dataset(trainset, testset)
losses, samples = train(net, dataset, epochs, batch_size, figsize=(10,5))
fig, ax = plt.subplots()
losses = np.array(losses)
plt.plot(losses.T[0], label='Discriminator', alpha=0.5)
plt.plot(losses.T[1], label='Generator', alpha=0.5)
plt.title("Training Losses")
plt.legend()
_ = view_samples(-1, samples, 6, 12, figsize=(10,5))
```
|
github_jupyter
|
%matplotlib inline
import pickle as pkl
import matplotlib.pyplot as plt
import numpy as np
from scipy.io import loadmat
import tensorflow as tf
!mkdir data
from urllib.request import urlretrieve
from os.path import isfile, isdir
from tqdm import tqdm
data_dir = 'data/'
if not isdir(data_dir):
raise Exception("Data directory doesn't exist!")
class DLProgress(tqdm):
last_block = 0
def hook(self, block_num=1, block_size=1, total_size=None):
self.total = total_size
self.update((block_num - self.last_block) * block_size)
self.last_block = block_num
if not isfile(data_dir + "train_32x32.mat"):
with DLProgress(unit='B', unit_scale=True, miniters=1, desc='SVHN Training Set') as pbar:
urlretrieve(
'http://ufldl.stanford.edu/housenumbers/train_32x32.mat',
data_dir + 'train_32x32.mat',
pbar.hook)
if not isfile(data_dir + "test_32x32.mat"):
with DLProgress(unit='B', unit_scale=True, miniters=1, desc='SVHN Testing Set') as pbar:
urlretrieve(
'http://ufldl.stanford.edu/housenumbers/test_32x32.mat',
data_dir + 'test_32x32.mat',
pbar.hook)
trainset = loadmat(data_dir + 'train_32x32.mat')
testset = loadmat(data_dir + 'test_32x32.mat')
idx = np.random.randint(0, trainset['X'].shape[3], size=36)
fig, axes = plt.subplots(6, 6, sharex=True, sharey=True, figsize=(5,5),)
for ii, ax in zip(idx, axes.flatten()):
ax.imshow(trainset['X'][:,:,:,ii], aspect='equal')
ax.xaxis.set_visible(False)
ax.yaxis.set_visible(False)
plt.subplots_adjust(wspace=0, hspace=0)
def scale(x, feature_range=(-1, 1)):
# scale to (0, 1)
x = ((x - x.min())/(255 - x.min()))
# scale to feature_range
min, max = feature_range
x = x * (max - min) + min
return x
class Dataset:
def __init__(self, train, test, val_frac=0.5, shuffle=False, scale_func=None):
split_idx = int(len(test['y'])*(1 - val_frac))
self.test_x, self.valid_x = test['X'][:,:,:,:split_idx], test['X'][:,:,:,split_idx:]
self.test_y, self.valid_y = test['y'][:split_idx], test['y'][split_idx:]
self.train_x, self.train_y = train['X'], train['y']
self.train_x = np.rollaxis(self.train_x, 3)
self.valid_x = np.rollaxis(self.valid_x, 3)
self.test_x = np.rollaxis(self.test_x, 3)
if scale_func is None:
self.scaler = scale
else:
self.scaler = scale_func
self.shuffle = shuffle
def batches(self, batch_size):
if self.shuffle:
idx = np.arange(len(dataset.train_x))
np.random.shuffle(idx)
self.train_x = self.train_x[idx]
self.train_y = self.train_y[idx]
n_batches = len(self.train_y)//batch_size
for ii in range(0, len(self.train_y), batch_size):
x = self.train_x[ii:ii+batch_size]
y = self.train_y[ii:ii+batch_size]
yield self.scaler(x), y
def model_inputs(real_dim, z_dim):
inputs_real = tf.placeholder(tf.float32, (None, *real_dim), name='input_real')
inputs_z = tf.placeholder(tf.float32, (None, z_dim), name='input_z')
return inputs_real, inputs_z
def generator(z, output_dim, reuse=False, alpha=0.2, training=True):
with tf.variable_scope('generator', reuse=reuse):
# First fully connected layer
x = tf.layers.Dense(z, 512)
x = tf.nn.conv2d(x, 256, 2)
x = tf.layers.
x = tf.nn.conv2d(x, 128, 2)
x = tf.nn.conv2d(x, 64, 2)
x = tf.nn.conv2d(x, 32, 2)
# Output layer, 32x32x3
logits =
out = tf.tanh(logits)
return out
def discriminator(x, reuse=False, alpha=0.2):
with tf.variable_scope('discriminator', reuse=reuse):
# Input layer is 32x32x3
x =
logits =
out =
return out, logits
def model_loss(input_real, input_z, output_dim, alpha=0.2):
"""
Get the loss for the discriminator and generator
:param input_real: Images from the real dataset
:param input_z: Z input
:param out_channel_dim: The number of channels in the output image
:return: A tuple of (discriminator loss, generator loss)
"""
g_model = generator(input_z, output_dim, alpha=alpha)
d_model_real, d_logits_real = discriminator(input_real, alpha=alpha)
d_model_fake, d_logits_fake = discriminator(g_model, reuse=True, alpha=alpha)
d_loss_real = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=d_logits_real, labels=tf.ones_like(d_model_real)))
d_loss_fake = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=d_logits_fake, labels=tf.zeros_like(d_model_fake)))
g_loss = tf.reduce_mean(
tf.nn.sigmoid_cross_entropy_with_logits(logits=d_logits_fake, labels=tf.ones_like(d_model_fake)))
d_loss = d_loss_real + d_loss_fake
return d_loss, g_loss
def model_opt(d_loss, g_loss, learning_rate, beta1):
"""
Get optimization operations
:param d_loss: Discriminator loss Tensor
:param g_loss: Generator loss Tensor
:param learning_rate: Learning Rate Placeholder
:param beta1: The exponential decay rate for the 1st moment in the optimizer
:return: A tuple of (discriminator training operation, generator training operation)
"""
# Get weights and bias to update
t_vars = tf.trainable_variables()
d_vars = [var for var in t_vars if var.name.startswith('discriminator')]
g_vars = [var for var in t_vars if var.name.startswith('generator')]
# Optimize
with tf.control_dependencies(tf.get_collection(tf.GraphKeys.UPDATE_OPS)):
d_train_opt = tf.train.AdamOptimizer(learning_rate, beta1=beta1).minimize(d_loss, var_list=d_vars)
g_train_opt = tf.train.AdamOptimizer(learning_rate, beta1=beta1).minimize(g_loss, var_list=g_vars)
return d_train_opt, g_train_opt
class GAN:
def __init__(self, real_size, z_size, learning_rate, alpha=0.2, beta1=0.5):
tf.reset_default_graph()
self.input_real, self.input_z = model_inputs(real_size, z_size)
self.d_loss, self.g_loss = model_loss(self.input_real, self.input_z,
real_size[2], alpha=alpha)
self.d_opt, self.g_opt = model_opt(self.d_loss, self.g_loss, learning_rate, beta1)
def view_samples(epoch, samples, nrows, ncols, figsize=(5,5)):
fig, axes = plt.subplots(figsize=figsize, nrows=nrows, ncols=ncols,
sharey=True, sharex=True)
for ax, img in zip(axes.flatten(), samples[epoch]):
ax.axis('off')
img = ((img - img.min())*255 / (img.max() - img.min())).astype(np.uint8)
ax.set_adjustable('box-forced')
im = ax.imshow(img, aspect='equal')
plt.subplots_adjust(wspace=0, hspace=0)
return fig, axes
def train(net, dataset, epochs, batch_size, print_every=10, show_every=100, figsize=(5,5)):
saver = tf.train.Saver()
sample_z = np.random.uniform(-1, 1, size=(72, z_size))
samples, losses = [], []
steps = 0
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
for e in range(epochs):
for x, y in dataset.batches(batch_size):
steps += 1
# Sample random noise for G
batch_z = np.random.uniform(-1, 1, size=(batch_size, z_size))
# Run optimizers
_ = sess.run(net.d_opt, feed_dict={net.input_real: x, net.input_z: batch_z})
_ = sess.run(net.g_opt, feed_dict={net.input_z: batch_z, net.input_real: x})
if steps % print_every == 0:
# At the end of each epoch, get the losses and print them out
train_loss_d = net.d_loss.eval({net.input_z: batch_z, net.input_real: x})
train_loss_g = net.g_loss.eval({net.input_z: batch_z})
print("Epoch {}/{}...".format(e+1, epochs),
"Discriminator Loss: {:.4f}...".format(train_loss_d),
"Generator Loss: {:.4f}".format(train_loss_g))
# Save losses to view after training
losses.append((train_loss_d, train_loss_g))
if steps % show_every == 0:
gen_samples = sess.run(
generator(net.input_z, 3, reuse=True, training=False),
feed_dict={net.input_z: sample_z})
samples.append(gen_samples)
_ = view_samples(-1, samples, 6, 12, figsize=figsize)
plt.show()
saver.save(sess, './checkpoints/generator.ckpt')
with open('samples.pkl', 'wb') as f:
pkl.dump(samples, f)
return losses, samples
real_size = (32,32,3)
z_size = 100
learning_rate = 0.001
batch_size = 64
epochs = 1
alpha = 0.01
beta1 = 0.9
# Create the network
net = GAN(real_size, z_size, learning_rate, alpha=alpha, beta1=beta1)
# Load the data and train the network here
dataset = Dataset(trainset, testset)
losses, samples = train(net, dataset, epochs, batch_size, figsize=(10,5))
fig, ax = plt.subplots()
losses = np.array(losses)
plt.plot(losses.T[0], label='Discriminator', alpha=0.5)
plt.plot(losses.T[1], label='Generator', alpha=0.5)
plt.title("Training Losses")
plt.legend()
_ = view_samples(-1, samples, 6, 12, figsize=(10,5))
| 0.668015 | 0.98366 |
```
import xml.etree.ElementTree as ET
# libraries
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
```
# Execution times analysis
## Parsing and aggregation
We first parse the execution times for Micro and TPC-H queries in the XML report.
```
tree = ET.parse('TEST-ch.epfl.dias.cs422.QueryTest.xml')
print(tree)
root = tree.getroot()[1:-2]
for child in root[:10]:
print(child.tag, child.attrib)
tests = ["volcano (row store)", "operator-at-a-time (row store)","block-at-a-time (row store)",
"late-operator-at-a-time (row store)","volcano (column store)",
"operator-at-a-time (column store)",
"block-at-a-time (column store)",
"late-operator-at-a-time (column store)",
"volcano (pax store)", "operator-at-a-time (pax store)",
"block-at-a-time (pax store)",
"late-operator-at-a-time (pax store)" ]
groups = zip(*[iter(root)]*3)
data = {}
for idx,test in enumerate(tests):
data[test] = root[45*idx: (idx + 1)*45]
```
We associate the list of tests with the correct execution times.
```
print(data[tests[0]][0].attrib['time'])
bars = {}
limit = 12
count = 0
for k,v in data.items():
if count < limit:
tmp = []
for stat in v:
tmp.append(float(stat.attrib['time']))
bars[k] = tmp
count += 1
block = "block-at-a-time"
volcano = "volcano"
operator = "operator-at-a-time"
late ="late"
models = [volcano, operator, block, late]
```
We group the execution times by execution models and type of queries
```
def byModel(bars, start=0, end=45):
groupedByModel = {}
for m in models:
groupedByModel[m] = {}
for name, times in bars.items():
for model in models:
if name.startswith(model):
groupedByModel[model][name] = times[start:end]
return groupedByModel
tcph = byModel(bars, 35, 45)
micro = byModel(bars,1,35)
tcph
```
# Visualization
We plot bar charts of execution times for all execution models under different data layout
```
def plot(bars, n, title):
ind = np.arange(n) # the x locations for the groups
width = 0.35 # the width of the bars: can also be len(x) sequence
barWidth = 0.25
#p1 = plt.bar(ind, menMeans, width, yerr=menStd)
#p2 = plt.bar(ind, womenMeans, width,
#bottom=menMeans, yerr=womenStd)
ps = []
r=ind
plt.figure(figsize=(10,5))
for name, times in bars.items():
ps.append(plt.bar(r, times, barWidth))
r = [x + barWidth for x in r]
plt.ylabel('Execution Time (s)')
plt.title(title)
#plt.xticks(ind, tuple([str(i) for i in range(n)]))
plt.xticks([y + barWidth for y in range(n)], tuple([str(i) for i in range(n)]))
plt.xlabel('query #')
#plt.yticks(np.linspace(0, 0.1 , 10))
plt.legend(tuple(ps), tuple(bars.keys()))
plt.show()
models
for model in models:
plot(micro[model], 34, "Execution time of Micro queries by data layout")
for model in models:
plot(tcph[model], 10, "Execution time of TPC-H queries by data layout")
```
We select the best data layout in each execution model and compare the execution times
```
def bestLayouts(results, qtype):
bests = {}
for model, layouts in results.items():
sum = float('inf')
#print(layouts)
for layout, times in layouts.items():
if np.sum(times) < sum:
sum = np.sum(times)
bests[model] ={layout: times, 'time': sum}
cleanUp = {}
if qtype == 'micro':
qnames = ['Micro q' + str(i) for i in range(35)]
elif qtype == 'tpch':
qnames = ['TPC-H q01', 'TPC-H q02', 'TPC-H q03','TPC-H q04','TPC-H q05', 'TPC-H q06','TPC-H q07','TPC-H q09', 'TPC-H q17','TPC-H q18','TPC-H q19']
print(bests)
for name,v in bests.items():
zippedTimes = zip(qnames, list(v.values())[0])
model_layout = list(v.keys())[0]
cleanUp[model_layout] = {}
for (qname, time) in zippedTimes:
cleanUp[model_layout][qname] = time
cleanUp[model_layout]['Total time (s)'] = list(v.values())[1]
cleanUp[model_layout]['Average time (s)'] = np.mean(list(v.values())[0] )
cleanUp[model_layout]['Standard deviation (s)'] = np.std(list(v.values())[0] )
return cleanUp
bestsMicro = bestLayouts(micro, 'micro')
bestsTPCH = bestLayouts(tcph, 'tpch')
pd.DataFrame(bestsMicro)
pd.DataFrame(bestsTPCH)
```
|
github_jupyter
|
import xml.etree.ElementTree as ET
# libraries
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
tree = ET.parse('TEST-ch.epfl.dias.cs422.QueryTest.xml')
print(tree)
root = tree.getroot()[1:-2]
for child in root[:10]:
print(child.tag, child.attrib)
tests = ["volcano (row store)", "operator-at-a-time (row store)","block-at-a-time (row store)",
"late-operator-at-a-time (row store)","volcano (column store)",
"operator-at-a-time (column store)",
"block-at-a-time (column store)",
"late-operator-at-a-time (column store)",
"volcano (pax store)", "operator-at-a-time (pax store)",
"block-at-a-time (pax store)",
"late-operator-at-a-time (pax store)" ]
groups = zip(*[iter(root)]*3)
data = {}
for idx,test in enumerate(tests):
data[test] = root[45*idx: (idx + 1)*45]
print(data[tests[0]][0].attrib['time'])
bars = {}
limit = 12
count = 0
for k,v in data.items():
if count < limit:
tmp = []
for stat in v:
tmp.append(float(stat.attrib['time']))
bars[k] = tmp
count += 1
block = "block-at-a-time"
volcano = "volcano"
operator = "operator-at-a-time"
late ="late"
models = [volcano, operator, block, late]
def byModel(bars, start=0, end=45):
groupedByModel = {}
for m in models:
groupedByModel[m] = {}
for name, times in bars.items():
for model in models:
if name.startswith(model):
groupedByModel[model][name] = times[start:end]
return groupedByModel
tcph = byModel(bars, 35, 45)
micro = byModel(bars,1,35)
tcph
def plot(bars, n, title):
ind = np.arange(n) # the x locations for the groups
width = 0.35 # the width of the bars: can also be len(x) sequence
barWidth = 0.25
#p1 = plt.bar(ind, menMeans, width, yerr=menStd)
#p2 = plt.bar(ind, womenMeans, width,
#bottom=menMeans, yerr=womenStd)
ps = []
r=ind
plt.figure(figsize=(10,5))
for name, times in bars.items():
ps.append(plt.bar(r, times, barWidth))
r = [x + barWidth for x in r]
plt.ylabel('Execution Time (s)')
plt.title(title)
#plt.xticks(ind, tuple([str(i) for i in range(n)]))
plt.xticks([y + barWidth for y in range(n)], tuple([str(i) for i in range(n)]))
plt.xlabel('query #')
#plt.yticks(np.linspace(0, 0.1 , 10))
plt.legend(tuple(ps), tuple(bars.keys()))
plt.show()
models
for model in models:
plot(micro[model], 34, "Execution time of Micro queries by data layout")
for model in models:
plot(tcph[model], 10, "Execution time of TPC-H queries by data layout")
def bestLayouts(results, qtype):
bests = {}
for model, layouts in results.items():
sum = float('inf')
#print(layouts)
for layout, times in layouts.items():
if np.sum(times) < sum:
sum = np.sum(times)
bests[model] ={layout: times, 'time': sum}
cleanUp = {}
if qtype == 'micro':
qnames = ['Micro q' + str(i) for i in range(35)]
elif qtype == 'tpch':
qnames = ['TPC-H q01', 'TPC-H q02', 'TPC-H q03','TPC-H q04','TPC-H q05', 'TPC-H q06','TPC-H q07','TPC-H q09', 'TPC-H q17','TPC-H q18','TPC-H q19']
print(bests)
for name,v in bests.items():
zippedTimes = zip(qnames, list(v.values())[0])
model_layout = list(v.keys())[0]
cleanUp[model_layout] = {}
for (qname, time) in zippedTimes:
cleanUp[model_layout][qname] = time
cleanUp[model_layout]['Total time (s)'] = list(v.values())[1]
cleanUp[model_layout]['Average time (s)'] = np.mean(list(v.values())[0] )
cleanUp[model_layout]['Standard deviation (s)'] = np.std(list(v.values())[0] )
return cleanUp
bestsMicro = bestLayouts(micro, 'micro')
bestsTPCH = bestLayouts(tcph, 'tpch')
pd.DataFrame(bestsMicro)
pd.DataFrame(bestsTPCH)
| 0.17037 | 0.79538 |
```
import nltk
import gensim
import gensim.corpora as corpora
from gensim.utils import simple_preprocess
from gensim.models import CoherenceModel
import pyLDAvis
from nltk.corpus import stopwords
import pandas as pd
import warnings
import pyLDAvis.gensim_models
import spacy
import warnings
warnings.filterwarnings("ignore",category=DeprecationWarning)
from nltk.corpus import stopwords
stop = stopwords.words('indonesian')
import nltk
from nltk.corpus import stopwords
model = spacy.load("xx_ent_wiki_sm", disable=['parser', 'ner'])
df = pd.read_excel("data training.xlsx")
df = df.replace({"POSITIF":3,"NEUTRAL":2,"NEGATIF":1})
df = df.replace({3:2,2:1,1:0})
df_neg = df[df['sentiment'] == 0]
df_net = df[df['sentiment'] == 1]
df_pos = df[df['sentiment'] == 2]
print(f"negatif shape {df_neg.shape}")
print(f"netral shape {df_net.shape}")
print(f"postif shape {df_pos.shape}")
print(f"total shape {df.shape}")
import re
df['sentiment'] = df['sentiment'].astype(int)
df['berita'] = df['berita'].replace({'"':' ',
'\d+':' ',
':':' ',
';':' ',
'#':' ',
'@':' ',
'_':' ',
',': ' ',
"'": ' ',
}, regex=True)
df['berita'] = df['berita'].str.replace(r'[https]+[?://]+[^\s<>"]+|www\.[^\s<>"]+[?()]+[(??)]+[)*]+[(\xa0]+[->...]', " ",regex=True)
df['berita'] = df['berita'].replace('\n',' ', regex=True)
df['berita'] = df['berita'].replace({'\.':' ','(/)':' ','\(':' ','\)':' ','\-':' ','\“':' ','\”':' ','\*':' ','\?':' '},regex=True)
df['berita'] = df['berita'].replace('[\.:"]',' ',regex =True)
df['berita'] = df['berita'].replace('[\–"]',' ',regex =True)
df['berita'].astype(str)
letters_only = re.sub("[^a-zA-Z]", # Search for all non-letters
" ", # Replace all non-letters with spaces
str(df['berita']))
df['berita'] = df['berita'].str.strip()
df['berita'] = df['berita'].str.lower()
df['berita'] = df['berita'].replace('\s+', ' ', regex=True)
for text in df['berita'].iteritems():
text = [text]
print(text)
def lemmatization(texts, allowed_postags=["NOUN", "ADJ", "VERB", "ADV"]):
nlp = spacy.load("xx_ent_wiki_sm", disable=["parser", "ner"])
texts_out = []
for text in texts.iteritems():
doc = nlp(text)
new_text = []
for token in doc:
if token.pos_ in allowed_postags:
new_text.append(token.lemma_)
final = " ".join(new_text)
texts_out.append(final)
return (texts_out)
lemmatized_texts = lemmatization(df['berita'])
lemmatized_texts
bigrams_phrases = gensim.models.Phrases(data_words,min_count=6,threshold=50)
trigrams_phrases = gensim.models.Phrases(bigrams_phrases[data_words],min_count=6,threshold=50)
bigram = gensim.models.phrases.Phraser(bigrams_phrases)
trigram = gensim.models.phrases.Phraser(trigrams_phrases)
def make_bigrams(texts):
re(bigram[doc] for dic in texts)
id2word = corpora.Dictionary(lemma)
corpus = []
for text in lemma:
new = id2word.doc2bow(text)
corpus.append(new)
print(corpus[0])
print(lemma[0])
lda_model = gensim.models.ldamodel.LdaModel(corpus=corpus,
id2word=id2word,
num_topics=30,
random_state=50,
update_every=5,
chunksize=100,
passes=10,
alpha='auto')
py = pyLDAvis.gensim_models.prepare(lda_model,corpus,id2word,mds='mmds',R=30)
'''
#extract = pyLDAvis.save_html(py,"after.html")
w_tokenizer = nltk.tokenize.WhitespaceTokenizer()
lemmatizer = nltk.stem.WordNetLemmatizer()
def lemmatize_text(text):
return [lemmatizer.lemmatize(w) for w in w_tokenizer.tokenize(text)]
df['berita'] = df['berita'].apply(lemmatize_text)
data_words = df['berita']'''
```
|
github_jupyter
|
import nltk
import gensim
import gensim.corpora as corpora
from gensim.utils import simple_preprocess
from gensim.models import CoherenceModel
import pyLDAvis
from nltk.corpus import stopwords
import pandas as pd
import warnings
import pyLDAvis.gensim_models
import spacy
import warnings
warnings.filterwarnings("ignore",category=DeprecationWarning)
from nltk.corpus import stopwords
stop = stopwords.words('indonesian')
import nltk
from nltk.corpus import stopwords
model = spacy.load("xx_ent_wiki_sm", disable=['parser', 'ner'])
df = pd.read_excel("data training.xlsx")
df = df.replace({"POSITIF":3,"NEUTRAL":2,"NEGATIF":1})
df = df.replace({3:2,2:1,1:0})
df_neg = df[df['sentiment'] == 0]
df_net = df[df['sentiment'] == 1]
df_pos = df[df['sentiment'] == 2]
print(f"negatif shape {df_neg.shape}")
print(f"netral shape {df_net.shape}")
print(f"postif shape {df_pos.shape}")
print(f"total shape {df.shape}")
import re
df['sentiment'] = df['sentiment'].astype(int)
df['berita'] = df['berita'].replace({'"':' ',
'\d+':' ',
':':' ',
';':' ',
'#':' ',
'@':' ',
'_':' ',
',': ' ',
"'": ' ',
}, regex=True)
df['berita'] = df['berita'].str.replace(r'[https]+[?://]+[^\s<>"]+|www\.[^\s<>"]+[?()]+[(??)]+[)*]+[(\xa0]+[->...]', " ",regex=True)
df['berita'] = df['berita'].replace('\n',' ', regex=True)
df['berita'] = df['berita'].replace({'\.':' ','(/)':' ','\(':' ','\)':' ','\-':' ','\“':' ','\”':' ','\*':' ','\?':' '},regex=True)
df['berita'] = df['berita'].replace('[\.:"]',' ',regex =True)
df['berita'] = df['berita'].replace('[\–"]',' ',regex =True)
df['berita'].astype(str)
letters_only = re.sub("[^a-zA-Z]", # Search for all non-letters
" ", # Replace all non-letters with spaces
str(df['berita']))
df['berita'] = df['berita'].str.strip()
df['berita'] = df['berita'].str.lower()
df['berita'] = df['berita'].replace('\s+', ' ', regex=True)
for text in df['berita'].iteritems():
text = [text]
print(text)
def lemmatization(texts, allowed_postags=["NOUN", "ADJ", "VERB", "ADV"]):
nlp = spacy.load("xx_ent_wiki_sm", disable=["parser", "ner"])
texts_out = []
for text in texts.iteritems():
doc = nlp(text)
new_text = []
for token in doc:
if token.pos_ in allowed_postags:
new_text.append(token.lemma_)
final = " ".join(new_text)
texts_out.append(final)
return (texts_out)
lemmatized_texts = lemmatization(df['berita'])
lemmatized_texts
bigrams_phrases = gensim.models.Phrases(data_words,min_count=6,threshold=50)
trigrams_phrases = gensim.models.Phrases(bigrams_phrases[data_words],min_count=6,threshold=50)
bigram = gensim.models.phrases.Phraser(bigrams_phrases)
trigram = gensim.models.phrases.Phraser(trigrams_phrases)
def make_bigrams(texts):
re(bigram[doc] for dic in texts)
id2word = corpora.Dictionary(lemma)
corpus = []
for text in lemma:
new = id2word.doc2bow(text)
corpus.append(new)
print(corpus[0])
print(lemma[0])
lda_model = gensim.models.ldamodel.LdaModel(corpus=corpus,
id2word=id2word,
num_topics=30,
random_state=50,
update_every=5,
chunksize=100,
passes=10,
alpha='auto')
py = pyLDAvis.gensim_models.prepare(lda_model,corpus,id2word,mds='mmds',R=30)
'''
#extract = pyLDAvis.save_html(py,"after.html")
w_tokenizer = nltk.tokenize.WhitespaceTokenizer()
lemmatizer = nltk.stem.WordNetLemmatizer()
def lemmatize_text(text):
return [lemmatizer.lemmatize(w) for w in w_tokenizer.tokenize(text)]
df['berita'] = df['berita'].apply(lemmatize_text)
data_words = df['berita']'''
| 0.16654 | 0.164315 |
# 16 - Adding Formation Data to a Well Log Plot
**Created by: Andy McDonald**
Link to article: https://andymcdonaldgeo.medium.com/adding-formation-data-to-a-well-log-plot-3897b96a3967
Well log plots are a common visualization tool within geoscience and petrophysics. They allow easy visualization of data (for example, Gamma Ray, Neutron Porosity, Bulk Density, etc) that has been acquired along the length (depth) of a wellbore.
I have previously covered different aspects of making these plots in the following articles:
- [Loading Multiple Well Log LAS Files Using Python
](https://towardsdatascience.com/loading-multiple-well-log-las-files-using-python-39ac35de99dd)
- [Displaying Logging While Drilling (LWD) Image Logs in Python](https://towardsdatascience.com/displaying-logging-while-drilling-lwd-image-logs-in-python-4babb6e577ba)
- [Enhancing Visualization of Well Logs With Plot Fills](https://towardsdatascience.com/enhancing-visualization-of-well-logs-with-plot-fills-72d9dcd10c1b)
- [Loading and Displaying Well Log Data](https://andymcdonaldgeo.medium.com/loading-and-displaying-well-log-data-b9568efd1d8)
In this article, I will show how to combine these different methods into a single plot function allow you to easily reuse the code with similar data.
For the examples below you can find my Jupyter Notebook and dataset on my GitHub repository at the following link.
https://github.com/andymcdgeo/Petrophysics-Python-Series
## Creating a Reusable Log Plot and Showing Formation Data
### Setting up the Libraries
For this article and notebook we will be using a number of different libraries.
We will be using five libraries: pandas, matplotlib, csv, collecitons, and lasio. Pandas and lasio, will be used to load and store the log data, collections allow us to use a defaultdict to load in our formation tops to a dictionary. Finally, matplotlib will let us plot out well log data.
```
import pandas as pd
import matplotlib.pyplot as plt
import lasio as las
import csv
from collections import defaultdict
import numpy as np
```
### Loading LAS Data
The first set of data we will load will be the LAS file from the Volve datset. To do this we call upon the las.read() function and pass in the file. Once the file has been read, we can convert it quickly to a pandas dataframe using .df(). This will make it easier for us to work with.
To see how to load multiple las files, check out my previous article [here](https://towardsdatascience.com/loading-multiple-well-log-las-files-using-python-39ac35de99dd).
```
data = las.read('Data/15-9-19_SR_COMP.las')
well = data.df()
```
When lasio converts the las file to a dataframe it assigns the depth curve as the dataframe index. This can be converted to a column as seen below. This is especially important when we are working with multiple las files and we do not wish to cause clashes with similar depth values.
```
well['DEPTH'] = well.index
```
We can then print the header of the dataframe to verify our data has loaded correctly.
```
well.head()
```
### Loading Formation Tops
For this article, three formation tops are stored within a simple csv file. For each formation, the top and bottom depth are stored. To load this file in, we can use the code snippet below.
```
formations_dict= {}
with open('Data/Formations/15_9_19_SR_Formations.csv', 'r') as file:
next(file) #skip header row
for row in csv.DictReader(file, fieldnames=['Formation', 'Top', 'Bottom']):
formations_dict[row['Formation']]=[float(row['Top']), float(row['Bottom'])]
```
When we call formations_dict we can preview what our dictionary contains.
```
formations_dict
formations_dict['Hugin Fm.'][0]
```
In order for the tops to be plotted in the correct place on a log plot we need to calculate the midpoint between the formation top and bottom depths. As the depth values are in list form, they can be called using the index number, with 0 being the top depth and 1 being the bottom depth.
```
formation_midpoints = []
for key, value in formations_dict.items():
formation_midpoints.append(value[0] + (value[1]-value[0])/2)
formation_midpoints
```
Finally, we can assign some colors to our formations. in this case I have selected red, blue, and green.
```
# Select the same number of colors as there are formations
zone_colors = ["red", "blue", "green"]
```
### Setting up the Log Plot
In my previous articles, I created the plots on the fly and the code was only usable for a particular dataset. Generalizing a well log plot or petrophysics plot can be difficult due to the variety of curve mnemonics and log plot setups. In this example, I have placed my plotting section within a function.
Using functions is a great way to increase the reusability of code and reduces the amount of duplication that can occur.
Below the following code snippet, I have explained some of the key sections that make up our well log plot function.
```
def makeplot(depth, gamma, res, neut, dens, dtc, formations, topdepth, bottomdepth,
colors):
fig, ax = plt.subplots(figsize=(15,10))
#Set up the plot axes
ax1 = plt.subplot2grid((1,10), (0,0), rowspan=1, colspan = 3)
ax2 = plt.subplot2grid((1,10), (0,3), rowspan=1, colspan = 3, sharey = ax1)
ax3 = plt.subplot2grid((1,10), (0,6), rowspan=1, colspan = 3, sharey = ax1)
ax4 = ax3.twiny()
ax5 = plt.subplot2grid((1,10), (0,9), rowspan=1, colspan = 1, sharey = ax1)
# As our curve scales will be detached from the top of the track,
# this code adds the top border back in without dealing with splines
ax10 = ax1.twiny()
ax10.xaxis.set_visible(False)
ax11 = ax2.twiny()
ax11.xaxis.set_visible(False)
ax12 = ax3.twiny()
ax12.xaxis.set_visible(False)
# Gamma Ray track
## Setting up the track and curve
ax1.plot(gamma, depth, color = "green", linewidth = 0.5)
ax1.set_xlabel("Gamma")
ax1.xaxis.label.set_color("green")
ax1.set_xlim(0, 150)
ax1.set_ylabel("Depth (m)")
ax1.tick_params(axis='x', colors="green")
ax1.spines["top"].set_edgecolor("green")
ax1.title.set_color('green')
ax1.set_xticks([0, 50, 100, 150])
ax1.text(0.05, 1.04, 0, color='green',
horizontalalignment='left', transform=ax1.transAxes)
ax1.text(0.95, 1.04, 150, color='green',
horizontalalignment='right', transform=ax1.transAxes)
ax1.set_xticklabels([])
## Setting Up Shading for GR
left_col_value = 0
right_col_value = 150
span = abs(left_col_value - right_col_value)
cmap = plt.get_cmap('hot_r')
color_index = np.arange(left_col_value, right_col_value, span / 100)
#loop through each value in the color_index
for index in sorted(color_index):
index_value = (index - left_col_value)/span
color = cmap(index_value) #obtain color for color index value
ax1.fill_betweenx(depth, gamma , right_col_value, where = gamma >= index, color = color)
# Resistivity track
ax2.plot(res, depth, color = "red", linewidth = 0.5)
ax2.set_xlabel("Resistivity")
ax2.set_xlim(0.2, 2000)
ax2.xaxis.label.set_color("red")
ax2.tick_params(axis='x', colors="red")
ax2.spines["top"].set_edgecolor("red")
ax2.set_xticks([0.1, 1, 10, 100, 1000])
ax2.semilogx()
ax2.text(0.05, 1.04, 0.1, color='red',
horizontalalignment='left', transform=ax2.transAxes)
ax2.text(0.95, 1.04, 1000, color='red',
horizontalalignment='right', transform=ax2.transAxes)
ax2.set_xticklabels([])
# Density track
ax3.plot(dens, depth, color = "red", linewidth = 0.5)
ax3.set_xlabel("Density")
ax3.set_xlim(1.95, 2.95)
ax3.xaxis.label.set_color("red")
ax3.tick_params(axis='x', colors="red")
ax3.spines["top"].set_edgecolor("red")
ax3.set_xticks([1.95, 2.45, 2.95])
ax3.text(0.05, 1.04, 1.95, color='red',
horizontalalignment='left', transform=ax3.transAxes)
ax3.text(0.95, 1.04, 2.95, color='red',
horizontalalignment='right', transform=ax3.transAxes)
ax3.set_xticklabels([])
# Neutron track placed ontop of density track
ax4.plot(neut, depth, color = "blue", linewidth = 0.5)
ax4.set_xlabel('Neutron')
ax4.xaxis.label.set_color("blue")
ax4.set_xlim(45, -15)
ax4.tick_params(axis='x', colors="blue")
ax4.spines["top"].set_position(("axes", 1.08))
ax4.spines["top"].set_visible(True)
ax4.spines["top"].set_edgecolor("blue")
ax4.set_xticks([45, 15, -15])
ax4.text(0.05, 1.1, 45, color='blue',
horizontalalignment='left', transform=ax4.transAxes)
ax4.text(0.95, 1.1, -15, color='blue',
horizontalalignment='right', transform=ax4.transAxes)
ax4.set_xticklabels([])
ax5.set_xticklabels([])
ax5.text(0.5, 1.1, 'Formations', fontweight='bold',
horizontalalignment='center', transform=ax5.transAxes)
# Adding in neutron density shading
x1=dens
x2=neut
x = np.array(ax3.get_xlim())
z = np.array(ax4.get_xlim())
nz=((x2-np.max(z))/(np.min(z)-np.max(z)))*(np.max(x)-np.min(x))+np.min(x)
ax3.fill_betweenx(depth, x1, nz, where=x1>=nz, interpolate=True, color='green')
ax3.fill_betweenx(depth, x1, nz, where=x1<=nz, interpolate=True, color='yellow')
# Common functions for setting up the plot can be extracted into
# a for loop. This saves repeating code.
for ax in [ax1, ax2, ax3]:
ax.set_ylim(bottomdepth, topdepth)
ax.grid(which='major', color='lightgrey', linestyle='-')
ax.xaxis.set_ticks_position("top")
ax.xaxis.set_label_position("top")
ax.spines["top"].set_position(("axes", 1.02))
for ax in [ax1, ax2, ax3, ax5]:
# loop through the formations dictionary and zone colors
for depth, color in zip(formations.values(), colors):
# use the depths and colors to shade across the subplots
ax.axhspan(depth[0], depth[1], color=color, alpha=0.1)
for ax in [ax2, ax3, ax4, ax5]:
plt.setp(ax.get_yticklabels(), visible = False)
for label, formation_mid in zip(formations_dict.keys(),
formation_midpoints):
ax5.text(0.5, formation_mid, label, rotation=90,
verticalalignment='center', fontweight='bold',
fontsize='large')
plt.tight_layout()
fig.subplots_adjust(wspace = 0)
```
**Lines 3–10** sets up the log tracks. Here, I am using subplot2grid to control the number of tracks. suplot2grid((1,10), (0,0), rowspan=1, colspan=3) translates to creating a plot that is 10 columns wide, 1 row high and the first few axes spans 3 columns each. This allows us to control the width of each track.
The last track (ax5) will be used to plot our formation tops information.
```python
fig, ax = plt.subplots(figsize=(15,10))
#Set up the plot axes
ax1 = plt.subplot2grid((1,10), (0,0), rowspan=1, colspan = 3)
ax2 = plt.subplot2grid((1,10), (0,3), rowspan=1, colspan = 3, sharey = ax1)
ax3 = plt.subplot2grid((1,10), (0,6), rowspan=1, colspan = 3, sharey = ax1)
ax4 = ax3.twiny()
ax5 = plt.subplot2grid((1,10), (0,9), rowspan=1, colspan = 1, sharey = ax1)
```
**Lines 14–19** adds a second set of the axis on top of the existing one. This allows maintaining a border around each track when we come to detach the scale.
```python
ax10 = ax1.twiny()
ax10.xaxis.set_visible(False)
ax11 = ax2.twiny()
ax11.xaxis.set_visible(False)
ax12 = ax3.twiny()
ax12.xaxis.set_visible(False)
```
**Lines 21–49** set up the gamma ray track. First, we use ax1.plot to set up the data, line width, and color. Next, we set up the x-axis with a label, an axis color, and a set of limits.
As ax1 is going to be the first track on the plot, we can assign the y axis label of Depth (m).
After defining the curve setup, we can add some colored fill between the curve and the right-hand side of the track. See Enhancing Visualization of Well Logs with Plot Fills for details on how this was setup.
```python
# Gamma Ray track
## Setting up the track and curve
ax1.plot(gamma, depth, color = "green", linewidth = 0.5)
ax1.set_xlabel("Gamma")
ax1.xaxis.label.set_color("green")
ax1.set_xlim(0, 150)
ax1.set_ylabel("Depth (m)")
ax1.tick_params(axis='x', colors="green")
ax1.spines["top"].set_edgecolor("green")
ax1.title.set_color('green')
ax1.set_xticks([0, 50, 100, 150])
ax1.text(0.05, 1.04, 0, color='green',
horizontalalignment='left', transform=ax1.transAxes)
ax1.text(0.95, 1.04, 150, color='green',
horizontalalignment='right', transform=ax1.transAxes)
ax1.set_xticklabels([])
## Setting Up Shading for GR
left_col_value = 0
right_col_value = 150
span = abs(left_col_value - right_col_value)
cmap = plt.get_cmap('hot_r')
color_index = np.arange(left_col_value, right_col_value, span / 100)
#loop through each value in the color_index
for index in sorted(color_index):
index_value = (index - left_col_value)/span
color = cmap(index_value) #obtain color for color index value
ax1.fill_betweenx(depth, gamma , right_col_value, where = gamma >= index, color = color)
```
In my previous articles with log plots, there have been gaps between each of the tracks. When this gap was reduced the scale information for each track became muddled up. A better solution for this is to turn of the axis labels using ax1.set_xticklabels([]) and use a text label like below.
```python
ax1.text(0.05, 1.04, 0, color='green',
horizontalalignment='left', transform=ax1.transAxes)
ax1.text(0.95, 1.04, 150, color='green',
horizontalalignment='right', transform=ax1.transAxes)
```
The ax.text function can take in a number of arguments, but the basics are ax.text(xposition, yposition, textstring). In our example, we also pass in the horizontal alignment and a transform argument.
We then repeat this for each axis, until ax5, where all we need to do is add a track header and hide the x ticks.
Note that ax4 is twinned with ax3 and sits on top of it. This allows easy plotting of the neutron porosity data.
**Lines 103–113** contain the code for setting up the fill between the neutron porosity and density logs. See Enhancing Visualization of Well Logs with Plot Fills for details on this method.
```python
ax5.set_xticklabels([])
ax5.text(0.5, 1.1, 'Formations', fontweight='bold',
horizontalalignment='center', transform=ax5.transAxes)
```
In **Lines 118–123** we can save some lines of code by bundling common functions we want to apply to each track into a single for loop and allows us to set parameters for the axes in one go.
In this loop we are:
- setting the y axis limits to the bottom and top depth we supply to the function
- setting the grid line style
- setting the position of the x axis label and tick marks
- offsetting the top part (spine) of the track so it floats above the track
```python
for ax in [ax1, ax2, ax3]:
ax.set_ylim(bottomdepth, topdepth)
ax.grid(which='major', color='lightgrey', linestyle='-')
ax.xaxis.set_ticks_position("top")
ax.xaxis.set_label_position("top")
ax.spines["top"].set_position(("axes", 1.02))
```
**Lines 125–129** contains the next for loop that applies to ax1, ax2, ax3, and ax5 and lets us add formation shading across all tracks. We can loop through a zipped object of our formation depth values in our formations_dict and the zone_colours list. We will use these values to create a horizontal span object using ax.axhspan. Basically, it adds a rectangle on top of our axes between two Y-values (depths).
```python
for ax in [ax1, ax2, ax3, ax5]:
# loop through the formations dictionary and zone colors
for depth, color in zip(formations.values(), colors):
# use the depths and colors to shade across subplots
ax.axhspan(depth[0], depth[1], color=color, alpha=0.1)
```
**Lines 132–133** hides the yticklabels (depth labels) on each of the tracks (subplots).
```python
for ax in [ax2, ax3, ax4, ax5]:
plt.setp(ax.get_yticklabels(), visible = False)
```
**Lines 135–139** is our final and key for loop for adding our formation labels directly onto ax5. Here we are using the ax.text function and passing in an x position (0.5 = middle of the track) and a y position, which is our calculated formation midpoint depths. We then want to align the text vertically from the center so that the center of the text string sits in the middle of the formation.
```python
for label, formation_mid in zip(formations_dict.keys(),
formation_midpoints):
ax5.text(0.5, formation_mid, label, rotation=90,
verticalalignment='center', fontweight='bold',
fontsize='large')
```
## Creating the Plot With Our Data
Now that our function is setup the way want it, we can now pass in the columns from the well dataframe. The power of the function comes into its own when we use other wells to make plots that are set up the same.
```
makeplot(well['DEPTH'], well['GR'], well['RDEP'], well['NEU'],
well['DEN'], well['AC'], formations_dict, 4300, 4650,
zone_colors)
```
### Summary
In this article, we have covered how to load a las file and formation data from a csv file. This data was then plotted on our log plot. Also, we have seen how to turn our log plot code into a function, which allows us to reuse the code with other wells. This makes future plotting much simpler and quicker.
***Thanks for reading!***
*If you have found this article useful, please feel free to check out my other articles looking at various aspects of Python and well log data. You can also find my code used in this article and others at GitHub.*
*If you want to get in touch you can find me on LinkedIn or at my website.*
*Interested in learning more about python and well log data or petrophysics? Follow me on [Medium](https://medium.com/@andymcdonaldgeo).*
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
import lasio as las
import csv
from collections import defaultdict
import numpy as np
data = las.read('Data/15-9-19_SR_COMP.las')
well = data.df()
well['DEPTH'] = well.index
well.head()
formations_dict= {}
with open('Data/Formations/15_9_19_SR_Formations.csv', 'r') as file:
next(file) #skip header row
for row in csv.DictReader(file, fieldnames=['Formation', 'Top', 'Bottom']):
formations_dict[row['Formation']]=[float(row['Top']), float(row['Bottom'])]
formations_dict
formations_dict['Hugin Fm.'][0]
formation_midpoints = []
for key, value in formations_dict.items():
formation_midpoints.append(value[0] + (value[1]-value[0])/2)
formation_midpoints
# Select the same number of colors as there are formations
zone_colors = ["red", "blue", "green"]
def makeplot(depth, gamma, res, neut, dens, dtc, formations, topdepth, bottomdepth,
colors):
fig, ax = plt.subplots(figsize=(15,10))
#Set up the plot axes
ax1 = plt.subplot2grid((1,10), (0,0), rowspan=1, colspan = 3)
ax2 = plt.subplot2grid((1,10), (0,3), rowspan=1, colspan = 3, sharey = ax1)
ax3 = plt.subplot2grid((1,10), (0,6), rowspan=1, colspan = 3, sharey = ax1)
ax4 = ax3.twiny()
ax5 = plt.subplot2grid((1,10), (0,9), rowspan=1, colspan = 1, sharey = ax1)
# As our curve scales will be detached from the top of the track,
# this code adds the top border back in without dealing with splines
ax10 = ax1.twiny()
ax10.xaxis.set_visible(False)
ax11 = ax2.twiny()
ax11.xaxis.set_visible(False)
ax12 = ax3.twiny()
ax12.xaxis.set_visible(False)
# Gamma Ray track
## Setting up the track and curve
ax1.plot(gamma, depth, color = "green", linewidth = 0.5)
ax1.set_xlabel("Gamma")
ax1.xaxis.label.set_color("green")
ax1.set_xlim(0, 150)
ax1.set_ylabel("Depth (m)")
ax1.tick_params(axis='x', colors="green")
ax1.spines["top"].set_edgecolor("green")
ax1.title.set_color('green')
ax1.set_xticks([0, 50, 100, 150])
ax1.text(0.05, 1.04, 0, color='green',
horizontalalignment='left', transform=ax1.transAxes)
ax1.text(0.95, 1.04, 150, color='green',
horizontalalignment='right', transform=ax1.transAxes)
ax1.set_xticklabels([])
## Setting Up Shading for GR
left_col_value = 0
right_col_value = 150
span = abs(left_col_value - right_col_value)
cmap = plt.get_cmap('hot_r')
color_index = np.arange(left_col_value, right_col_value, span / 100)
#loop through each value in the color_index
for index in sorted(color_index):
index_value = (index - left_col_value)/span
color = cmap(index_value) #obtain color for color index value
ax1.fill_betweenx(depth, gamma , right_col_value, where = gamma >= index, color = color)
# Resistivity track
ax2.plot(res, depth, color = "red", linewidth = 0.5)
ax2.set_xlabel("Resistivity")
ax2.set_xlim(0.2, 2000)
ax2.xaxis.label.set_color("red")
ax2.tick_params(axis='x', colors="red")
ax2.spines["top"].set_edgecolor("red")
ax2.set_xticks([0.1, 1, 10, 100, 1000])
ax2.semilogx()
ax2.text(0.05, 1.04, 0.1, color='red',
horizontalalignment='left', transform=ax2.transAxes)
ax2.text(0.95, 1.04, 1000, color='red',
horizontalalignment='right', transform=ax2.transAxes)
ax2.set_xticklabels([])
# Density track
ax3.plot(dens, depth, color = "red", linewidth = 0.5)
ax3.set_xlabel("Density")
ax3.set_xlim(1.95, 2.95)
ax3.xaxis.label.set_color("red")
ax3.tick_params(axis='x', colors="red")
ax3.spines["top"].set_edgecolor("red")
ax3.set_xticks([1.95, 2.45, 2.95])
ax3.text(0.05, 1.04, 1.95, color='red',
horizontalalignment='left', transform=ax3.transAxes)
ax3.text(0.95, 1.04, 2.95, color='red',
horizontalalignment='right', transform=ax3.transAxes)
ax3.set_xticklabels([])
# Neutron track placed ontop of density track
ax4.plot(neut, depth, color = "blue", linewidth = 0.5)
ax4.set_xlabel('Neutron')
ax4.xaxis.label.set_color("blue")
ax4.set_xlim(45, -15)
ax4.tick_params(axis='x', colors="blue")
ax4.spines["top"].set_position(("axes", 1.08))
ax4.spines["top"].set_visible(True)
ax4.spines["top"].set_edgecolor("blue")
ax4.set_xticks([45, 15, -15])
ax4.text(0.05, 1.1, 45, color='blue',
horizontalalignment='left', transform=ax4.transAxes)
ax4.text(0.95, 1.1, -15, color='blue',
horizontalalignment='right', transform=ax4.transAxes)
ax4.set_xticklabels([])
ax5.set_xticklabels([])
ax5.text(0.5, 1.1, 'Formations', fontweight='bold',
horizontalalignment='center', transform=ax5.transAxes)
# Adding in neutron density shading
x1=dens
x2=neut
x = np.array(ax3.get_xlim())
z = np.array(ax4.get_xlim())
nz=((x2-np.max(z))/(np.min(z)-np.max(z)))*(np.max(x)-np.min(x))+np.min(x)
ax3.fill_betweenx(depth, x1, nz, where=x1>=nz, interpolate=True, color='green')
ax3.fill_betweenx(depth, x1, nz, where=x1<=nz, interpolate=True, color='yellow')
# Common functions for setting up the plot can be extracted into
# a for loop. This saves repeating code.
for ax in [ax1, ax2, ax3]:
ax.set_ylim(bottomdepth, topdepth)
ax.grid(which='major', color='lightgrey', linestyle='-')
ax.xaxis.set_ticks_position("top")
ax.xaxis.set_label_position("top")
ax.spines["top"].set_position(("axes", 1.02))
for ax in [ax1, ax2, ax3, ax5]:
# loop through the formations dictionary and zone colors
for depth, color in zip(formations.values(), colors):
# use the depths and colors to shade across the subplots
ax.axhspan(depth[0], depth[1], color=color, alpha=0.1)
for ax in [ax2, ax3, ax4, ax5]:
plt.setp(ax.get_yticklabels(), visible = False)
for label, formation_mid in zip(formations_dict.keys(),
formation_midpoints):
ax5.text(0.5, formation_mid, label, rotation=90,
verticalalignment='center', fontweight='bold',
fontsize='large')
plt.tight_layout()
fig.subplots_adjust(wspace = 0)
fig, ax = plt.subplots(figsize=(15,10))
#Set up the plot axes
ax1 = plt.subplot2grid((1,10), (0,0), rowspan=1, colspan = 3)
ax2 = plt.subplot2grid((1,10), (0,3), rowspan=1, colspan = 3, sharey = ax1)
ax3 = plt.subplot2grid((1,10), (0,6), rowspan=1, colspan = 3, sharey = ax1)
ax4 = ax3.twiny()
ax5 = plt.subplot2grid((1,10), (0,9), rowspan=1, colspan = 1, sharey = ax1)
ax10 = ax1.twiny()
ax10.xaxis.set_visible(False)
ax11 = ax2.twiny()
ax11.xaxis.set_visible(False)
ax12 = ax3.twiny()
ax12.xaxis.set_visible(False)
# Gamma Ray track
## Setting up the track and curve
ax1.plot(gamma, depth, color = "green", linewidth = 0.5)
ax1.set_xlabel("Gamma")
ax1.xaxis.label.set_color("green")
ax1.set_xlim(0, 150)
ax1.set_ylabel("Depth (m)")
ax1.tick_params(axis='x', colors="green")
ax1.spines["top"].set_edgecolor("green")
ax1.title.set_color('green')
ax1.set_xticks([0, 50, 100, 150])
ax1.text(0.05, 1.04, 0, color='green',
horizontalalignment='left', transform=ax1.transAxes)
ax1.text(0.95, 1.04, 150, color='green',
horizontalalignment='right', transform=ax1.transAxes)
ax1.set_xticklabels([])
## Setting Up Shading for GR
left_col_value = 0
right_col_value = 150
span = abs(left_col_value - right_col_value)
cmap = plt.get_cmap('hot_r')
color_index = np.arange(left_col_value, right_col_value, span / 100)
#loop through each value in the color_index
for index in sorted(color_index):
index_value = (index - left_col_value)/span
color = cmap(index_value) #obtain color for color index value
ax1.fill_betweenx(depth, gamma , right_col_value, where = gamma >= index, color = color)
ax1.text(0.05, 1.04, 0, color='green',
horizontalalignment='left', transform=ax1.transAxes)
ax1.text(0.95, 1.04, 150, color='green',
horizontalalignment='right', transform=ax1.transAxes)
ax5.set_xticklabels([])
ax5.text(0.5, 1.1, 'Formations', fontweight='bold',
horizontalalignment='center', transform=ax5.transAxes)
for ax in [ax1, ax2, ax3]:
ax.set_ylim(bottomdepth, topdepth)
ax.grid(which='major', color='lightgrey', linestyle='-')
ax.xaxis.set_ticks_position("top")
ax.xaxis.set_label_position("top")
ax.spines["top"].set_position(("axes", 1.02))
```
**Lines 125–129** contains the next for loop that applies to ax1, ax2, ax3, and ax5 and lets us add formation shading across all tracks. We can loop through a zipped object of our formation depth values in our formations_dict and the zone_colours list. We will use these values to create a horizontal span object using ax.axhspan. Basically, it adds a rectangle on top of our axes between two Y-values (depths).
**Lines 132–133** hides the yticklabels (depth labels) on each of the tracks (subplots).
**Lines 135–139** is our final and key for loop for adding our formation labels directly onto ax5. Here we are using the ax.text function and passing in an x position (0.5 = middle of the track) and a y position, which is our calculated formation midpoint depths. We then want to align the text vertically from the center so that the center of the text string sits in the middle of the formation.
## Creating the Plot With Our Data
Now that our function is setup the way want it, we can now pass in the columns from the well dataframe. The power of the function comes into its own when we use other wells to make plots that are set up the same.
| 0.481941 | 0.96862 |
```
import torch
import torch.nn as nn
from torch.autograd import Variable
def conv3x3(in_, out):
return nn.Conv2d(in_, out, 3, padding=1)
class ConvRelu(nn.Module):
def __init__(self, in_, out):
super().__init__()
self.conv = conv3x3(in_, out)
self.activation = nn.ReLU(inplace=True)
def forward(self, x):
x = self.conv(x)
x = self.activation(x)
return x
class NoOperation(nn.Module):
def forward(self, x):
return x
class DecoderBlock(nn.Module):
def __init__(self, in_channels, middle_channels, out_channels):
super().__init__()
self.block = nn.Sequential(
ConvRelu(in_channels, middle_channels),
nn.ConvTranspose2d(middle_channels, out_channels, kernel_size=3, stride=2, padding=1, output_padding=1),
nn.ReLU(inplace=True)
)
def forward(self, x):
return self.block(x)
class DecoderBlockV2(nn.Module):
def __init__(self, in_channels, middle_channels, out_channels, is_deconv=True,
output_padding=0):
super(DecoderBlockV2, self).__init__()
self.in_channels = in_channels
if is_deconv:
"""
Paramaters for Deconvolution were chosen to avoid artifacts, following
link https://distill.pub/2016/deconv-checkerboard/
"""
self.block = nn.Sequential(
ConvRelu(in_channels, middle_channels),
nn.ConvTranspose2d(middle_channels, out_channels, kernel_size=4, stride=2,
padding=1, output_padding=output_padding),
nn.ReLU(inplace=True)
)
else:
self.block = nn.Sequential(
nn.Upsample(scale_factor=2, mode='bilinear'),
ConvRelu(in_channels, middle_channels),
ConvRelu(middle_channels, out_channels),
)
def forward(self, x):
return self.block(x)
class Interpolate(nn.Module):
def __init__(self, mode='nearest', scale_factor=2,
align_corners=False, output_padding=0):
super(Interpolate, self).__init__()
self.interp = nn.functional.interpolate
self.mode = mode
self.scale_factor = scale_factor
self.align_corners = align_corners
self.pad = output_padding
def forward(self, x):
if self.mode in ['linear','bilinear','trilinear']:
x = self.interp(x, mode=self.mode,
scale_factor=self.scale_factor,
align_corners=self.align_corners)
else:
x = self.interp(x, mode=self.mode,
scale_factor=self.scale_factor)
if self.pad > 0:
x = nn.ZeroPad2d((0, self.pad, 0, self.pad))(x)
return x
class DecoderBlockV3(nn.Module):
def __init__(self, in_channels, middle_channels, out_channels,
is_deconv=True, output_padding=0):
super(DecoderBlockV3, self).__init__()
self.in_channels = in_channels
if is_deconv:
"""
Paramaters for Deconvolution were chosen to avoid artifacts, following
link https://distill.pub/2016/deconv-checkerboard/
"""
self.block = nn.Sequential(
nn.ConvTranspose2d(in_channels, middle_channels, kernel_size=4, stride=2,
padding=1, output_padding=output_padding),
ConvRelu(middle_channels, out_channels),
)
else:
self.block = nn.Sequential(
Interpolate(mode='nearest', scale_factor=2,
output_padding=output_padding),
# nn.Upsample(scale_factor=2, mode='bilinear'),
ConvRelu(in_channels, middle_channels),
ConvRelu(middle_channels, out_channels),
)
def forward(self, x):
return self.block(x)
class AdaptiveConcatPool2d(nn.Module):
def __init__(self, sz=None):
super().__init__()
sz = sz or (1,1)
self.ap = nn.AdaptiveAvgPool2d(sz)
self.mp = nn.AdaptiveMaxPool2d(sz)
def forward(self, x): return torch.cat([self.mp(x), self.ap(x)], 1)
class Resnet(nn.Module):
def __init__(self, num_classes, num_filters=32,
pretrained=True, is_deconv=False):
super().__init__()
self.num_classes = num_classes
# self.conv4to3 = nn.Conv2d(4, 3, 1)
# self.encoder = pretrainedmodels.__dict__['se_resnext50_32x4d'](num_classes=1000,
# pretrained='imagenet')
# code removes final layer
layers = resnet34()
layers = list(resnet34().children())[:-2]
# replace first convolutional layer by 4->64 while keeping corresponding weights
# and initializing new weights with zeros
# https://www.kaggle.com/iafoss/pretrained-resnet34-with-rgby-0-448-public-lb/notebook
w = layers[0].weight
layers[0] = nn.Conv2d(4,64,kernel_size=(7,7),stride=(2,2),padding=(3, 3),
bias=False)
layers[0].weight = torch.nn.Parameter(torch.cat((w,torch.zeros(64,1,7,7)),
dim=1))
# layers += [AdaptiveConcatPool2d()]
self.encoder = nn.Sequential(*layers)
self.map_logits = nn.Conv2d(512, num_classes, kernel_size=(3,3),
stride=(1,1), padding=1)
# self.encoder = nn.Sequential(*list(self.encoder.children())[:-1])
# self.pool = nn.MaxPool2d(2, 2)
# self.convp = nn.Conv2d(1056, 512, 3)
# self.csize = 1024 * 1 * 1
# self.bn1 = nn.BatchNorm1d(1024)
# self.do1 = nn.Dropout(p=0.5)
# self.lin1 = nn.Linear(1024, 512)
# self.act1 = nn.ReLU()
# self.bn2 = nn.BatchNorm1d(512)
# self.do2 = nn.Dropout(0.5)
# self.lin2 = nn.Linear(512, num_classes)
def forward(self, x):
# set to True for debugging
print_sizes = False
if print_sizes:
print('')
print('x',x.shape)
# print layer dictionary
# print(self.encoder.features)
# x = self.conv4to3(x)
# m = self.encoder._modules
# layer_names = list(m.keys())
# mx = {}
# for i,f in enumerate(m):
# x = m[f](x)
# mx[layer_names[i]] = x
# if print_sizes:
# if isinstance(x,tuple):
# print(i,layer_names[i],x[0].size(),x[1].size())
# else:
# print(i,layer_names[i],x.size())
# if layer_names[i]=='avg_pool': break
x = self.encoder(x)
if print_sizes: print('encoder',x.shape)
x = self.map_logits(x)
if print_sizes: print('map_logits',x.shape)
# x = x.view(-1, self.csize)
# if print_sizes: print('view',x.size())
# x = self.bn1(x)
# x = self.do1(x)
# if print_sizes: print('do1',x.size())
# x = self.lin1(x)
# if print_sizes: print('lin1',x.size())
# x = self.act1(x)
# x = self.bn2(x)
# x = self.do2(x)
# x = self.lin2(x)
# if print_sizes: print('lin2',x.shape)
return x
```
|
github_jupyter
|
import torch
import torch.nn as nn
from torch.autograd import Variable
def conv3x3(in_, out):
return nn.Conv2d(in_, out, 3, padding=1)
class ConvRelu(nn.Module):
def __init__(self, in_, out):
super().__init__()
self.conv = conv3x3(in_, out)
self.activation = nn.ReLU(inplace=True)
def forward(self, x):
x = self.conv(x)
x = self.activation(x)
return x
class NoOperation(nn.Module):
def forward(self, x):
return x
class DecoderBlock(nn.Module):
def __init__(self, in_channels, middle_channels, out_channels):
super().__init__()
self.block = nn.Sequential(
ConvRelu(in_channels, middle_channels),
nn.ConvTranspose2d(middle_channels, out_channels, kernel_size=3, stride=2, padding=1, output_padding=1),
nn.ReLU(inplace=True)
)
def forward(self, x):
return self.block(x)
class DecoderBlockV2(nn.Module):
def __init__(self, in_channels, middle_channels, out_channels, is_deconv=True,
output_padding=0):
super(DecoderBlockV2, self).__init__()
self.in_channels = in_channels
if is_deconv:
"""
Paramaters for Deconvolution were chosen to avoid artifacts, following
link https://distill.pub/2016/deconv-checkerboard/
"""
self.block = nn.Sequential(
ConvRelu(in_channels, middle_channels),
nn.ConvTranspose2d(middle_channels, out_channels, kernel_size=4, stride=2,
padding=1, output_padding=output_padding),
nn.ReLU(inplace=True)
)
else:
self.block = nn.Sequential(
nn.Upsample(scale_factor=2, mode='bilinear'),
ConvRelu(in_channels, middle_channels),
ConvRelu(middle_channels, out_channels),
)
def forward(self, x):
return self.block(x)
class Interpolate(nn.Module):
def __init__(self, mode='nearest', scale_factor=2,
align_corners=False, output_padding=0):
super(Interpolate, self).__init__()
self.interp = nn.functional.interpolate
self.mode = mode
self.scale_factor = scale_factor
self.align_corners = align_corners
self.pad = output_padding
def forward(self, x):
if self.mode in ['linear','bilinear','trilinear']:
x = self.interp(x, mode=self.mode,
scale_factor=self.scale_factor,
align_corners=self.align_corners)
else:
x = self.interp(x, mode=self.mode,
scale_factor=self.scale_factor)
if self.pad > 0:
x = nn.ZeroPad2d((0, self.pad, 0, self.pad))(x)
return x
class DecoderBlockV3(nn.Module):
def __init__(self, in_channels, middle_channels, out_channels,
is_deconv=True, output_padding=0):
super(DecoderBlockV3, self).__init__()
self.in_channels = in_channels
if is_deconv:
"""
Paramaters for Deconvolution were chosen to avoid artifacts, following
link https://distill.pub/2016/deconv-checkerboard/
"""
self.block = nn.Sequential(
nn.ConvTranspose2d(in_channels, middle_channels, kernel_size=4, stride=2,
padding=1, output_padding=output_padding),
ConvRelu(middle_channels, out_channels),
)
else:
self.block = nn.Sequential(
Interpolate(mode='nearest', scale_factor=2,
output_padding=output_padding),
# nn.Upsample(scale_factor=2, mode='bilinear'),
ConvRelu(in_channels, middle_channels),
ConvRelu(middle_channels, out_channels),
)
def forward(self, x):
return self.block(x)
class AdaptiveConcatPool2d(nn.Module):
def __init__(self, sz=None):
super().__init__()
sz = sz or (1,1)
self.ap = nn.AdaptiveAvgPool2d(sz)
self.mp = nn.AdaptiveMaxPool2d(sz)
def forward(self, x): return torch.cat([self.mp(x), self.ap(x)], 1)
class Resnet(nn.Module):
def __init__(self, num_classes, num_filters=32,
pretrained=True, is_deconv=False):
super().__init__()
self.num_classes = num_classes
# self.conv4to3 = nn.Conv2d(4, 3, 1)
# self.encoder = pretrainedmodels.__dict__['se_resnext50_32x4d'](num_classes=1000,
# pretrained='imagenet')
# code removes final layer
layers = resnet34()
layers = list(resnet34().children())[:-2]
# replace first convolutional layer by 4->64 while keeping corresponding weights
# and initializing new weights with zeros
# https://www.kaggle.com/iafoss/pretrained-resnet34-with-rgby-0-448-public-lb/notebook
w = layers[0].weight
layers[0] = nn.Conv2d(4,64,kernel_size=(7,7),stride=(2,2),padding=(3, 3),
bias=False)
layers[0].weight = torch.nn.Parameter(torch.cat((w,torch.zeros(64,1,7,7)),
dim=1))
# layers += [AdaptiveConcatPool2d()]
self.encoder = nn.Sequential(*layers)
self.map_logits = nn.Conv2d(512, num_classes, kernel_size=(3,3),
stride=(1,1), padding=1)
# self.encoder = nn.Sequential(*list(self.encoder.children())[:-1])
# self.pool = nn.MaxPool2d(2, 2)
# self.convp = nn.Conv2d(1056, 512, 3)
# self.csize = 1024 * 1 * 1
# self.bn1 = nn.BatchNorm1d(1024)
# self.do1 = nn.Dropout(p=0.5)
# self.lin1 = nn.Linear(1024, 512)
# self.act1 = nn.ReLU()
# self.bn2 = nn.BatchNorm1d(512)
# self.do2 = nn.Dropout(0.5)
# self.lin2 = nn.Linear(512, num_classes)
def forward(self, x):
# set to True for debugging
print_sizes = False
if print_sizes:
print('')
print('x',x.shape)
# print layer dictionary
# print(self.encoder.features)
# x = self.conv4to3(x)
# m = self.encoder._modules
# layer_names = list(m.keys())
# mx = {}
# for i,f in enumerate(m):
# x = m[f](x)
# mx[layer_names[i]] = x
# if print_sizes:
# if isinstance(x,tuple):
# print(i,layer_names[i],x[0].size(),x[1].size())
# else:
# print(i,layer_names[i],x.size())
# if layer_names[i]=='avg_pool': break
x = self.encoder(x)
if print_sizes: print('encoder',x.shape)
x = self.map_logits(x)
if print_sizes: print('map_logits',x.shape)
# x = x.view(-1, self.csize)
# if print_sizes: print('view',x.size())
# x = self.bn1(x)
# x = self.do1(x)
# if print_sizes: print('do1',x.size())
# x = self.lin1(x)
# if print_sizes: print('lin1',x.size())
# x = self.act1(x)
# x = self.bn2(x)
# x = self.do2(x)
# x = self.lin2(x)
# if print_sizes: print('lin2',x.shape)
return x
| 0.941681 | 0.482002 |
# Dipping regional TFA Inversion
This notebook performs the inversion using Levenberg-Marquadt's algorithm of total field anomaly (TFA) data in a presence of a regional field data on a flightlines of a model with a dipping geometry.
```
import numpy as np
import matplotlib.pyplot as plt
import cPickle as pickle
import os
import pandas as pd
from fatiando import utils
from fatiando.gravmag import polyprism
from fatiando.vis import mpl
from datetime import datetime
today = datetime.today()
# dd/mm/YY/Hh/Mm
d4 = today.strftime("%d-%b-%Y-%Hh:%Mm")
```
### Auxiliary functions
```
import sys
sys.path.insert(0, '../../code')
import mag_polyprism_functions as mfun
```
# Input
### Importing model parameters
```
with open('../dipping/model.pickle') as w:
model = pickle.load(w)
```
### Observation points and observed data
```
with open('data.pickle') as w:
d = pickle.load(w)
data = pd.read_csv('dipping_regional_data.txt', skipinitialspace=True, delim_whitespace=True)
dobs = data['res_data'].get_values()
xp = data['x'].get_values()
yp = data['y'].get_values()
zp = data['z'].get_values()
N = xp.size
```
### Parameters of the initial model
```
M = 20 # number of vertices per prism
L = 5 # number of prisms
P = L*(M+2) + 1 # number of parameters
# depth to the top, thickness, origin, magnetization and radius
incs = model['inc']
decs = model['dec']
intensity = model['intensity']
z0 = model['z0']
dz = 800.
r = 700.
x0 = -200.
y0 = 0.
# main field
inc, dec = d['main_field']
model0, m0 = mfun.initial_cylinder(M, L, x0,
y0, z0, dz, r, inc, dec, incs, decs, intensity)
# predict data
d0 = polyprism.tf(xp, yp, zp, model0, inc, dec)
plt.figure(figsize=(12,5))
plt.subplot(121)
plt.title('Observed TFA', fontsize=20)
plt.tricontourf(yp, xp, dobs, 20, cmap='RdBu_r').ax.tick_params(labelsize=12)
plt.xlabel('$y$(km)', fontsize=18)
plt.ylabel('$x$(km)', fontsize=18)
clb = plt.colorbar(pad=0.025, aspect=40, shrink=1)
clb.ax.tick_params(labelsize=13)
source = mpl.polygon(model['prisms'][0], '.-k', xy2ne=True)
estimate = mpl.polygon(model0[0], '.-y', xy2ne=True)
estimate.set_label('Initial estimate')
clb.ax.set_title('nT')
mpl.m2km()
plt.legend(loc=0, fontsize=12, shadow=bool, framealpha=1)
plt.subplot(122)
plt.title('Predicted TFA', fontsize=20)
plt.tricontourf(yp, xp, d0, 20, cmap='RdBu_r').ax.tick_params(labelsize=12)
plt.xlabel('$y$(km)', fontsize=18)
plt.ylabel('$x$(km)', fontsize=18)
clb = plt.colorbar(pad=0.025, aspect=40, shrink=1)
clb.ax.tick_params(labelsize=13)
estimate = mpl.polygon(model0[0], '.-y', xy2ne=True)
estimate.set_label('Initial estimate')
clb.ax.set_title('nT')
mpl.m2km()
plt.legend(loc=0, fontsize=12, shadow=bool, framealpha=1)
plt.show()
```
### Outcropping parameters
```
# outcropping body parameters
m_out = np.zeros(M + 2)
#m_out = model['param_vec'][:M+2]
```
### Limits
```
# limits for parameters in meters
rmin = 10.
rmax = 1200.
x0min = -4000.
x0max = 4000.
y0min = -4000.
y0max = 4000.
dzmin = 200.
dzmax = 1000.
mmin, mmax = mfun.build_range_param(M, L, rmin, rmax, x0min, x0max, y0min, y0max, dzmin, dzmax)
```
### Derivatives
```
# variation for derivatives
deltax = 0.01*np.max(100.)
deltay = 0.01*np.max(100.)
deltar = 0.01*np.max(100.)
deltaz = 0.01*np.max(100.)
delta = np.array([deltax, deltay, deltar, deltaz])
```
### Regularization parameters
```
#lamb = th*0.01 # Marquadt's parameter
lamb = 10.0
dlamb = 10. # step for Marquadt's parameter
a1 = 1.0e-3 # adjacent radial distances within each prism
a2 = 1.0e-3 # vertically adjacent radial distances
a3 = 0. # outcropping cross-section
a4 = 0. # outcropping origin
a5 = 1.0e-6 # vertically adjacent origins
a6 = 1.0e-7 # zero order Tikhonov on adjacent radial distances
a7 = 1.0e-5 # zero order Tikhonov on thickness of each prism
alpha = np.array([a1, a2, a3, a4, a5, a6, a7])
```
### Folder to save the results
```
foldername = 'test'
```
### Iterations and stop criterion
```
itmax = 30 # maximum iteration
itmax_marq = 10 # maximum iteration of Marquardt's loop
tol = 1.0e-4 # stop criterion
```
### Inversion
```
d_fit, m_est, model_est, phi_list, model_list, res_list = mfun.levmarq_tf(
xp, yp, zp, m0, M, L, delta,
itmax, itmax_marq, lamb,
dlamb, tol, mmin, mmax,
m_out, dobs, inc, dec,
model0[0].props, alpha, z0, dz
)
```
# Results
```
# output of inversion
inversion = dict()
inversion['x'] = xp
inversion['y'] = yp
inversion['z'] = zp
inversion['observed_data'] = dobs
inversion['inc_dec'] = [incs, decs]
inversion['z0'] = z0
inversion['initial_dz'] = dz
inversion['intial_r'] = r
inversion['initial_estimate'] = model0
inversion['initial_data'] = d0
inversion['limits'] = [rmin, rmax, x0min, x0max, y0min, y0max, dzmin, dzmax]
inversion['regularization'] = np.array([a1, a2, a3, a4, a5, a6, a7])
inversion['tol'] = tol
inversion['main_field'] = [-21.5, -18.7]
inversion['data_fit'] = d_fit
inversion['estimate'] = m_est
inversion['prisms'] = model_est
inversion['estimated_models'] = model_list
inversion['objective'] = phi_list
inversion['residual'] = dobs - d_fit
inversion['residual_list'] = res_list
```
### Saving results
```
if foldername == '':
mypath = 'results/single-'+d4 #default folder name
if not os.path.isdir(mypath):
os.makedirs(mypath)
else:
mypath = 'results/single-'+foldername #defined folder name
if not os.path.isdir(mypath):
os.makedirs(mypath)
file_name = mypath+'/inversion.pickle'
with open(file_name, 'w') as f:
pickle.dump(inversion, f)
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import cPickle as pickle
import os
import pandas as pd
from fatiando import utils
from fatiando.gravmag import polyprism
from fatiando.vis import mpl
from datetime import datetime
today = datetime.today()
# dd/mm/YY/Hh/Mm
d4 = today.strftime("%d-%b-%Y-%Hh:%Mm")
import sys
sys.path.insert(0, '../../code')
import mag_polyprism_functions as mfun
with open('../dipping/model.pickle') as w:
model = pickle.load(w)
with open('data.pickle') as w:
d = pickle.load(w)
data = pd.read_csv('dipping_regional_data.txt', skipinitialspace=True, delim_whitespace=True)
dobs = data['res_data'].get_values()
xp = data['x'].get_values()
yp = data['y'].get_values()
zp = data['z'].get_values()
N = xp.size
M = 20 # number of vertices per prism
L = 5 # number of prisms
P = L*(M+2) + 1 # number of parameters
# depth to the top, thickness, origin, magnetization and radius
incs = model['inc']
decs = model['dec']
intensity = model['intensity']
z0 = model['z0']
dz = 800.
r = 700.
x0 = -200.
y0 = 0.
# main field
inc, dec = d['main_field']
model0, m0 = mfun.initial_cylinder(M, L, x0,
y0, z0, dz, r, inc, dec, incs, decs, intensity)
# predict data
d0 = polyprism.tf(xp, yp, zp, model0, inc, dec)
plt.figure(figsize=(12,5))
plt.subplot(121)
plt.title('Observed TFA', fontsize=20)
plt.tricontourf(yp, xp, dobs, 20, cmap='RdBu_r').ax.tick_params(labelsize=12)
plt.xlabel('$y$(km)', fontsize=18)
plt.ylabel('$x$(km)', fontsize=18)
clb = plt.colorbar(pad=0.025, aspect=40, shrink=1)
clb.ax.tick_params(labelsize=13)
source = mpl.polygon(model['prisms'][0], '.-k', xy2ne=True)
estimate = mpl.polygon(model0[0], '.-y', xy2ne=True)
estimate.set_label('Initial estimate')
clb.ax.set_title('nT')
mpl.m2km()
plt.legend(loc=0, fontsize=12, shadow=bool, framealpha=1)
plt.subplot(122)
plt.title('Predicted TFA', fontsize=20)
plt.tricontourf(yp, xp, d0, 20, cmap='RdBu_r').ax.tick_params(labelsize=12)
plt.xlabel('$y$(km)', fontsize=18)
plt.ylabel('$x$(km)', fontsize=18)
clb = plt.colorbar(pad=0.025, aspect=40, shrink=1)
clb.ax.tick_params(labelsize=13)
estimate = mpl.polygon(model0[0], '.-y', xy2ne=True)
estimate.set_label('Initial estimate')
clb.ax.set_title('nT')
mpl.m2km()
plt.legend(loc=0, fontsize=12, shadow=bool, framealpha=1)
plt.show()
# outcropping body parameters
m_out = np.zeros(M + 2)
#m_out = model['param_vec'][:M+2]
# limits for parameters in meters
rmin = 10.
rmax = 1200.
x0min = -4000.
x0max = 4000.
y0min = -4000.
y0max = 4000.
dzmin = 200.
dzmax = 1000.
mmin, mmax = mfun.build_range_param(M, L, rmin, rmax, x0min, x0max, y0min, y0max, dzmin, dzmax)
# variation for derivatives
deltax = 0.01*np.max(100.)
deltay = 0.01*np.max(100.)
deltar = 0.01*np.max(100.)
deltaz = 0.01*np.max(100.)
delta = np.array([deltax, deltay, deltar, deltaz])
#lamb = th*0.01 # Marquadt's parameter
lamb = 10.0
dlamb = 10. # step for Marquadt's parameter
a1 = 1.0e-3 # adjacent radial distances within each prism
a2 = 1.0e-3 # vertically adjacent radial distances
a3 = 0. # outcropping cross-section
a4 = 0. # outcropping origin
a5 = 1.0e-6 # vertically adjacent origins
a6 = 1.0e-7 # zero order Tikhonov on adjacent radial distances
a7 = 1.0e-5 # zero order Tikhonov on thickness of each prism
alpha = np.array([a1, a2, a3, a4, a5, a6, a7])
foldername = 'test'
itmax = 30 # maximum iteration
itmax_marq = 10 # maximum iteration of Marquardt's loop
tol = 1.0e-4 # stop criterion
d_fit, m_est, model_est, phi_list, model_list, res_list = mfun.levmarq_tf(
xp, yp, zp, m0, M, L, delta,
itmax, itmax_marq, lamb,
dlamb, tol, mmin, mmax,
m_out, dobs, inc, dec,
model0[0].props, alpha, z0, dz
)
# output of inversion
inversion = dict()
inversion['x'] = xp
inversion['y'] = yp
inversion['z'] = zp
inversion['observed_data'] = dobs
inversion['inc_dec'] = [incs, decs]
inversion['z0'] = z0
inversion['initial_dz'] = dz
inversion['intial_r'] = r
inversion['initial_estimate'] = model0
inversion['initial_data'] = d0
inversion['limits'] = [rmin, rmax, x0min, x0max, y0min, y0max, dzmin, dzmax]
inversion['regularization'] = np.array([a1, a2, a3, a4, a5, a6, a7])
inversion['tol'] = tol
inversion['main_field'] = [-21.5, -18.7]
inversion['data_fit'] = d_fit
inversion['estimate'] = m_est
inversion['prisms'] = model_est
inversion['estimated_models'] = model_list
inversion['objective'] = phi_list
inversion['residual'] = dobs - d_fit
inversion['residual_list'] = res_list
if foldername == '':
mypath = 'results/single-'+d4 #default folder name
if not os.path.isdir(mypath):
os.makedirs(mypath)
else:
mypath = 'results/single-'+foldername #defined folder name
if not os.path.isdir(mypath):
os.makedirs(mypath)
file_name = mypath+'/inversion.pickle'
with open(file_name, 'w') as f:
pickle.dump(inversion, f)
| 0.325521 | 0.905197 |
Copyright (c) Microsoft Corporation. All rights reserved.
Licensed under the MIT License.

# Automated Machine Learning
_**Orange Juice Sales Forecasting**_
## Contents
1. [Introduction](#Introduction)
1. [Setup](#Setup)
1. [Compute](#Compute)
1. [Data](#Data)
1. [Train](#Train)
1. [Predict](#Predict)
1. [Operationalize](#Operationalize)
## Introduction
In this example, we use AutoML to train, select, and operationalize a time-series forecasting model for multiple time-series.
Make sure you have executed the [configuration notebook](../../../configuration.ipynb) before running this notebook.
The examples in the follow code samples use the University of Chicago's Dominick's Finer Foods dataset to forecast orange juice sales. Dominick's was a grocery chain in the Chicago metropolitan area.
## Setup
```
import azureml.core
import pandas as pd
import numpy as np
import logging
from azureml.core.workspace import Workspace
from azureml.core.experiment import Experiment
from azureml.train.automl import AutoMLConfig
from azureml.automl.core.featurization import FeaturizationConfig
```
This sample notebook may use features that are not available in previous versions of the Azure ML SDK.
```
print("This notebook was created using version 1.9.0 of the Azure ML SDK")
print("You are currently using version", azureml.core.VERSION, "of the Azure ML SDK")
```
As part of the setup you have already created a <b>Workspace</b>. To run AutoML, you also need to create an <b>Experiment</b>. An Experiment corresponds to a prediction problem you are trying to solve, while a Run corresponds to a specific approach to the problem.
```
ws = Workspace.from_config()
# choose a name for the run history container in the workspace
experiment_name = 'automl-ojforecasting'
experiment = Experiment(ws, experiment_name)
output = {}
output['Subscription ID'] = ws.subscription_id
output['Workspace'] = ws.name
output['SKU'] = ws.sku
output['Resource Group'] = ws.resource_group
output['Location'] = ws.location
output['Run History Name'] = experiment_name
pd.set_option('display.max_colwidth', -1)
outputDf = pd.DataFrame(data = output, index = [''])
outputDf.T
```
## Compute
You will need to create a [compute target](https://docs.microsoft.com/en-us/azure/machine-learning/service/how-to-set-up-training-targets#amlcompute) for your AutoML run. In this tutorial, you create AmlCompute as your training compute resource.
#### Creation of AmlCompute takes approximately 5 minutes.
If the AmlCompute with that name is already in your workspace this code will skip the creation process.
As with other Azure services, there are limits on certain resources (e.g. AmlCompute) associated with the Azure Machine Learning service. Please read this article on the default limits and how to request more quota.
```
from azureml.core.compute import ComputeTarget, AmlCompute
from azureml.core.compute_target import ComputeTargetException
# Choose a name for your CPU cluster
amlcompute_cluster_name = "oj-cluster"
# Verify that cluster does not exist already
try:
compute_target = ComputeTarget(workspace=ws, name=amlcompute_cluster_name)
print('Found existing cluster, use it.')
except ComputeTargetException:
compute_config = AmlCompute.provisioning_configuration(vm_size='STANDARD_D2_V2',
max_nodes=6)
compute_target = ComputeTarget.create(ws, amlcompute_cluster_name, compute_config)
compute_target.wait_for_completion(show_output=True)
```
## Data
You are now ready to load the historical orange juice sales data. We will load the CSV file into a plain pandas DataFrame; the time column in the CSV is called _WeekStarting_, so it will be specially parsed into the datetime type.
```
time_column_name = 'WeekStarting'
data = pd.read_csv("dominicks_OJ.csv", parse_dates=[time_column_name])
data.head()
```
Each row in the DataFrame holds a quantity of weekly sales for an OJ brand at a single store. The data also includes the sales price, a flag indicating if the OJ brand was advertised in the store that week, and some customer demographic information based on the store location. For historical reasons, the data also include the logarithm of the sales quantity. The Dominick's grocery data is commonly used to illustrate econometric modeling techniques where logarithms of quantities are generally preferred.
The task is now to build a time-series model for the _Quantity_ column. It is important to note that this dataset is comprised of many individual time-series - one for each unique combination of _Store_ and _Brand_. To distinguish the individual time-series, we thus define the **grain** - the columns whose values determine the boundaries between time-series:
```
grain_column_names = ['Store', 'Brand']
nseries = data.groupby(grain_column_names).ngroups
print('Data contains {0} individual time-series.'.format(nseries))
```
For demonstration purposes, we extract sales time-series for just a few of the stores:
```
use_stores = [2, 5, 8]
data_subset = data[data.Store.isin(use_stores)]
nseries = data_subset.groupby(grain_column_names).ngroups
print('Data subset contains {0} individual time-series.'.format(nseries))
```
### Data Splitting
We now split the data into a training and a testing set for later forecast evaluation. The test set will contain the final 20 weeks of observed sales for each time-series. The splits should be stratified by series, so we use a group-by statement on the grain columns.
```
n_test_periods = 20
def split_last_n_by_grain(df, n):
"""Group df by grain and split on last n rows for each group."""
df_grouped = (df.sort_values(time_column_name) # Sort by ascending time
.groupby(grain_column_names, group_keys=False))
df_head = df_grouped.apply(lambda dfg: dfg.iloc[:-n])
df_tail = df_grouped.apply(lambda dfg: dfg.iloc[-n:])
return df_head, df_tail
train, test = split_last_n_by_grain(data_subset, n_test_periods)
```
### Upload data to datastore
The [Machine Learning service workspace](https://docs.microsoft.com/en-us/azure/machine-learning/service/concept-workspace), is paired with the storage account, which contains the default data store. We will use it to upload the train and test data and create [tabular datasets](https://docs.microsoft.com/en-us/python/api/azureml-core/azureml.data.tabulardataset?view=azure-ml-py) for training and testing. A tabular dataset defines a series of lazily-evaluated, immutable operations to load data from the data source into tabular representation.
```
train.to_csv (r'./dominicks_OJ_train.csv', index = None, header=True)
test.to_csv (r'./dominicks_OJ_test.csv', index = None, header=True)
datastore = ws.get_default_datastore()
datastore.upload_files(files = ['./dominicks_OJ_train.csv', './dominicks_OJ_test.csv'], target_path = 'dataset/', overwrite = True,show_progress = True)
```
### Create dataset for training
```
from azureml.core.dataset import Dataset
train_dataset = Dataset.Tabular.from_delimited_files(path=datastore.path('dataset/dominicks_OJ_train.csv'))
train_dataset.to_pandas_dataframe().tail()
```
## Modeling
For forecasting tasks, AutoML uses pre-processing and estimation steps that are specific to time-series. AutoML will undertake the following pre-processing steps:
* Detect time-series sample frequency (e.g. hourly, daily, weekly) and create new records for absent time points to make the series regular. A regular time series has a well-defined frequency and has a value at every sample point in a contiguous time span
* Impute missing values in the target (via forward-fill) and feature columns (using median column values)
* Create grain-based features to enable fixed effects across different series
* Create time-based features to assist in learning seasonal patterns
* Encode categorical variables to numeric quantities
In this notebook, AutoML will train a single, regression-type model across **all** time-series in a given training set. This allows the model to generalize across related series. If you're looking for training multiple models for different time-series, please check out the forecasting grouping notebook.
You are almost ready to start an AutoML training job. First, we need to separate the target column from the rest of the DataFrame:
```
target_column_name = 'Quantity'
```
## Customization
The featurization customization in forecasting is an advanced feature in AutoML which allows our customers to change the default forecasting featurization behaviors and column types through `FeaturizationConfig`. The supported scenarios include,
1. Column purposes update: Override feature type for the specified column. Currently supports DateTime, Categorical and Numeric. This customization can be used in the scenario that the type of the column cannot correctly reflect its purpose. Some numerical columns, for instance, can be treated as Categorical columns which need to be converted to categorical while some can be treated as epoch timestamp which need to be converted to datetime. To tell our SDK to correctly preprocess these columns, a configuration need to be add with the columns and their desired types.
2. Transformer parameters update: Currently supports parameter change for Imputer only. User can customize imputation methods, the supported methods are constant for target data and mean, median, most frequent and constant for training data. This customization can be used for the scenario that our customers know which imputation methods fit best to the input data. For instance, some datasets use NaN to represent 0 which the correct behavior should impute all the missing value with 0. To achieve this behavior, these columns need to be configured as constant imputation with `fill_value` 0.
3. Drop columns: Columns to drop from being featurized. These usually are the columns which are leaky or the columns contain no useful data.
This step requires an Enterprise workspace to gain access to this feature. To learn more about creating an Enterprise workspace or upgrading to an Enterprise workspace from the Azure portal, please visit our [Workspace page.](https://docs.microsoft.com/azure/machine-learning/service/concept-workspace#upgrade)
```
featurization_config = FeaturizationConfig()
featurization_config.drop_columns = ['logQuantity'] # 'logQuantity' is a leaky feature, so we remove it.
# Force the CPWVOL5 feature to be numeric type.
featurization_config.add_column_purpose('CPWVOL5', 'Numeric')
# Fill missing values in the target column, Quantity, with zeros.
featurization_config.add_transformer_params('Imputer', ['Quantity'], {"strategy": "constant", "fill_value": 0})
# Fill missing values in the INCOME column with median value.
featurization_config.add_transformer_params('Imputer', ['INCOME'], {"strategy": "median"})
```
## Train
The [AutoMLConfig](https://docs.microsoft.com/en-us/python/api/azureml-train-automl-client/azureml.train.automl.automlconfig.automlconfig?view=azure-ml-py) object defines the settings and data for an AutoML training job. Here, we set necessary inputs like the task type, the number of AutoML iterations to try, the training data, and cross-validation parameters.
For forecasting tasks, there are some additional parameters that can be set: the name of the column holding the date/time, the grain column names, and the maximum forecast horizon. A time column is required for forecasting, while the grain is optional. If grain columns are not given, AutoML assumes that the whole dataset is a single time-series. We also pass a list of columns to drop prior to modeling. The _logQuantity_ column is completely correlated with the target quantity, so it must be removed to prevent a target leak.
The forecast horizon is given in units of the time-series frequency; for instance, the OJ series frequency is weekly, so a horizon of 20 means that a trained model will estimate sales up to 20 weeks beyond the latest date in the training data for each series. In this example, we set the maximum horizon to the number of samples per series in the test set (n_test_periods). Generally, the value of this parameter will be dictated by business needs. For example, a demand planning application that estimates the next month of sales should set the horizon according to suitable planning time-scales. Please see the [energy_demand notebook](https://github.com/Azure/MachineLearningNotebooks/tree/master/how-to-use-azureml/automated-machine-learning/forecasting-energy-demand) for more discussion of forecast horizon.
We note here that AutoML can sweep over two types of time-series models:
* Models that are trained for each series such as ARIMA and Facebook's Prophet. Note that these models are only available for [Enterprise Edition Workspaces](https://docs.microsoft.com/en-us/azure/machine-learning/how-to-manage-workspace#upgrade).
* Models trained across multiple time-series using a regression approach.
In the first case, AutoML loops over all time-series in your dataset and trains one model (e.g. AutoArima or Prophet, as the case may be) for each series. This can result in long runtimes to train these models if there are a lot of series in the data. One way to mitigate this problem is to fit models for different series in parallel if you have multiple compute cores available. To enable this behavior, set the `max_cores_per_iteration` parameter in your AutoMLConfig as shown in the example in the next cell.
Finally, a note about the cross-validation (CV) procedure for time-series data. AutoML uses out-of-sample error estimates to select a best pipeline/model, so it is important that the CV fold splitting is done correctly. Time-series can violate the basic statistical assumptions of the canonical K-Fold CV strategy, so AutoML implements a [rolling origin validation](https://robjhyndman.com/hyndsight/tscv/) procedure to create CV folds for time-series data. To use this procedure, you just need to specify the desired number of CV folds in the AutoMLConfig object. It is also possible to bypass CV and use your own validation set by setting the *validation_data* parameter of AutoMLConfig.
Here is a summary of AutoMLConfig parameters used for training the OJ model:
|Property|Description|
|-|-|
|**task**|forecasting|
|**primary_metric**|This is the metric that you want to optimize.<br> Forecasting supports the following primary metrics <br><i>spearman_correlation</i><br><i>normalized_root_mean_squared_error</i><br><i>r2_score</i><br><i>normalized_mean_absolute_error</i>
|**experiment_timeout_hours**|Experimentation timeout in hours.|
|**enable_early_stopping**|If early stopping is on, training will stop when the primary metric is no longer improving.|
|**training_data**|Input dataset, containing both features and label column.|
|**label_column_name**|The name of the label column.|
|**compute_target**|The remote compute for training.|
|**n_cross_validations**|Number of cross-validation folds to use for model/pipeline selection|
|**enable_voting_ensemble**|Allow AutoML to create a Voting ensemble of the best performing models|
|**enable_stack_ensemble**|Allow AutoML to create a Stack ensemble of the best performing models|
|**debug_log**|Log file path for writing debugging information|
|**time_column_name**|Name of the datetime column in the input data|
|**grain_column_names**|Name(s) of the columns defining individual series in the input data|
|**max_horizon**|Maximum desired forecast horizon in units of time-series frequency|
|**featurization**| 'auto' / 'off' / FeaturizationConfig Indicator for whether featurization step should be done automatically or not, or whether customized featurization should be used. Setting this enables AutoML to perform featurization on the input to handle *missing data*, and to perform some common *feature extraction*.|
|**max_cores_per_iteration**|Maximum number of cores to utilize per iteration. A value of -1 indicates all available cores should be used.|
```
time_series_settings = {
'time_column_name': time_column_name,
'grain_column_names': grain_column_names,
'max_horizon': n_test_periods
}
automl_config = AutoMLConfig(task='forecasting',
debug_log='automl_oj_sales_errors.log',
primary_metric='normalized_mean_absolute_error',
experiment_timeout_hours=0.25,
training_data=train_dataset,
label_column_name=target_column_name,
compute_target=compute_target,
enable_early_stopping=True,
featurization=featurization_config,
n_cross_validations=3,
verbosity=logging.INFO,
max_cores_per_iteration=-1,
**time_series_settings)
```
You can now submit a new training run. Depending on the data and number of iterations this operation may take several minutes.
Information from each iteration will be printed to the console.
```
remote_run = experiment.submit(automl_config, show_output=False)
remote_run
remote_run.wait_for_completion()
```
### Retrieve the Best Model
Each run within an Experiment stores serialized (i.e. pickled) pipelines from the AutoML iterations. We can now retrieve the pipeline with the best performance on the validation dataset:
```
best_run, fitted_model = remote_run.get_output()
print(fitted_model.steps)
model_name = best_run.properties['model_name']
```
## Transparency
View updated featurization summary
```
custom_featurizer = fitted_model.named_steps['timeseriestransformer']
custom_featurizer.get_featurization_summary()
```
# Forecasting
Now that we have retrieved the best pipeline/model, it can be used to make predictions on test data. First, we remove the target values from the test set:
```
X_test = test
y_test = X_test.pop(target_column_name).values
X_test.head()
```
To produce predictions on the test set, we need to know the feature values at all dates in the test set. This requirement is somewhat reasonable for the OJ sales data since the features mainly consist of price, which is usually set in advance, and customer demographics which are approximately constant for each store over the 20 week forecast horizon in the testing data.
```
# The featurized data, aligned to y, will also be returned.
# This contains the assumptions that were made in the forecast
# and helps align the forecast to the original data
y_predictions, X_trans = fitted_model.forecast(X_test)
```
If you are used to scikit pipelines, perhaps you expected `predict(X_test)`. However, forecasting requires a more general interface that also supplies the past target `y` values. Please use `forecast(X,y)` as `predict(X)` is reserved for internal purposes on forecasting models.
The [forecast function notebook](../forecasting-forecast-function/auto-ml-forecasting-function.ipynb).
# Evaluate
To evaluate the accuracy of the forecast, we'll compare against the actual sales quantities for some select metrics, included the mean absolute percentage error (MAPE).
It is a good practice to always align the output explicitly to the input, as the count and order of the rows may have changed during transformations that span multiple rows.
```
from forecasting_helper import align_outputs
df_all = align_outputs(y_predictions, X_trans, X_test, y_test, target_column_name)
from azureml.automl.core.shared import constants
from azureml.automl.runtime.shared.score import scoring
from matplotlib import pyplot as plt
# use automl scoring module
scores = scoring.score_regression(
y_test=df_all[target_column_name],
y_pred=df_all['predicted'],
metrics=list(constants.Metric.SCALAR_REGRESSION_SET))
print("[Test data scores]\n")
for key, value in scores.items():
print('{}: {:.3f}'.format(key, value))
# Plot outputs
%matplotlib inline
test_pred = plt.scatter(df_all[target_column_name], df_all['predicted'], color='b')
test_test = plt.scatter(df_all[target_column_name], df_all[target_column_name], color='g')
plt.legend((test_pred, test_test), ('prediction', 'truth'), loc='upper left', fontsize=8)
plt.show()
```
# Operationalize
_Operationalization_ means getting the model into the cloud so that other can run it after you close the notebook. We will create a docker running on Azure Container Instances with the model.
```
description = 'AutoML OJ forecaster'
tags = None
model = remote_run.register_model(model_name = model_name, description = description, tags = tags)
print(remote_run.model_id)
```
### Develop the scoring script
For the deployment we need a function which will run the forecast on serialized data. It can be obtained from the best_run.
```
script_file_name = 'score_fcast.py'
best_run.download_file('outputs/scoring_file_v_1_0_0.py', script_file_name)
```
### Deploy the model as a Web Service on Azure Container Instance
```
from azureml.core.model import InferenceConfig
from azureml.core.webservice import AciWebservice
from azureml.core.webservice import Webservice
from azureml.core.model import Model
inference_config = InferenceConfig(environment = best_run.get_environment(),
entry_script = script_file_name)
aciconfig = AciWebservice.deploy_configuration(cpu_cores = 1,
memory_gb = 2,
tags = {'type': "automl-forecasting"},
description = "Automl forecasting sample service")
aci_service_name = 'automl-oj-forecast-01'
print(aci_service_name)
aci_service = Model.deploy(ws, aci_service_name, [model], inference_config, aciconfig)
aci_service.wait_for_deployment(True)
print(aci_service.state)
aci_service.get_logs()
```
### Call the service
```
import json
X_query = X_test.copy()
# We have to convert datetime to string, because Timestamps cannot be serialized to JSON.
X_query[time_column_name] = X_query[time_column_name].astype(str)
# The Service object accept the complex dictionary, which is internally converted to JSON string.
# The section 'data' contains the data frame in the form of dictionary.
test_sample = json.dumps({'data': X_query.to_dict(orient='records')})
response = aci_service.run(input_data = test_sample)
# translate from networkese to datascientese
try:
res_dict = json.loads(response)
y_fcst_all = pd.DataFrame(res_dict['index'])
y_fcst_all[time_column_name] = pd.to_datetime(y_fcst_all[time_column_name], unit = 'ms')
y_fcst_all['forecast'] = res_dict['forecast']
except:
print(res_dict)
y_fcst_all.head()
```
### Delete the web service if desired
```
serv = Webservice(ws, 'automl-oj-forecast-01')
serv.delete() # don't do it accidentally
```
|
github_jupyter
|
import azureml.core
import pandas as pd
import numpy as np
import logging
from azureml.core.workspace import Workspace
from azureml.core.experiment import Experiment
from azureml.train.automl import AutoMLConfig
from azureml.automl.core.featurization import FeaturizationConfig
print("This notebook was created using version 1.9.0 of the Azure ML SDK")
print("You are currently using version", azureml.core.VERSION, "of the Azure ML SDK")
ws = Workspace.from_config()
# choose a name for the run history container in the workspace
experiment_name = 'automl-ojforecasting'
experiment = Experiment(ws, experiment_name)
output = {}
output['Subscription ID'] = ws.subscription_id
output['Workspace'] = ws.name
output['SKU'] = ws.sku
output['Resource Group'] = ws.resource_group
output['Location'] = ws.location
output['Run History Name'] = experiment_name
pd.set_option('display.max_colwidth', -1)
outputDf = pd.DataFrame(data = output, index = [''])
outputDf.T
from azureml.core.compute import ComputeTarget, AmlCompute
from azureml.core.compute_target import ComputeTargetException
# Choose a name for your CPU cluster
amlcompute_cluster_name = "oj-cluster"
# Verify that cluster does not exist already
try:
compute_target = ComputeTarget(workspace=ws, name=amlcompute_cluster_name)
print('Found existing cluster, use it.')
except ComputeTargetException:
compute_config = AmlCompute.provisioning_configuration(vm_size='STANDARD_D2_V2',
max_nodes=6)
compute_target = ComputeTarget.create(ws, amlcompute_cluster_name, compute_config)
compute_target.wait_for_completion(show_output=True)
time_column_name = 'WeekStarting'
data = pd.read_csv("dominicks_OJ.csv", parse_dates=[time_column_name])
data.head()
grain_column_names = ['Store', 'Brand']
nseries = data.groupby(grain_column_names).ngroups
print('Data contains {0} individual time-series.'.format(nseries))
use_stores = [2, 5, 8]
data_subset = data[data.Store.isin(use_stores)]
nseries = data_subset.groupby(grain_column_names).ngroups
print('Data subset contains {0} individual time-series.'.format(nseries))
n_test_periods = 20
def split_last_n_by_grain(df, n):
"""Group df by grain and split on last n rows for each group."""
df_grouped = (df.sort_values(time_column_name) # Sort by ascending time
.groupby(grain_column_names, group_keys=False))
df_head = df_grouped.apply(lambda dfg: dfg.iloc[:-n])
df_tail = df_grouped.apply(lambda dfg: dfg.iloc[-n:])
return df_head, df_tail
train, test = split_last_n_by_grain(data_subset, n_test_periods)
train.to_csv (r'./dominicks_OJ_train.csv', index = None, header=True)
test.to_csv (r'./dominicks_OJ_test.csv', index = None, header=True)
datastore = ws.get_default_datastore()
datastore.upload_files(files = ['./dominicks_OJ_train.csv', './dominicks_OJ_test.csv'], target_path = 'dataset/', overwrite = True,show_progress = True)
from azureml.core.dataset import Dataset
train_dataset = Dataset.Tabular.from_delimited_files(path=datastore.path('dataset/dominicks_OJ_train.csv'))
train_dataset.to_pandas_dataframe().tail()
target_column_name = 'Quantity'
featurization_config = FeaturizationConfig()
featurization_config.drop_columns = ['logQuantity'] # 'logQuantity' is a leaky feature, so we remove it.
# Force the CPWVOL5 feature to be numeric type.
featurization_config.add_column_purpose('CPWVOL5', 'Numeric')
# Fill missing values in the target column, Quantity, with zeros.
featurization_config.add_transformer_params('Imputer', ['Quantity'], {"strategy": "constant", "fill_value": 0})
# Fill missing values in the INCOME column with median value.
featurization_config.add_transformer_params('Imputer', ['INCOME'], {"strategy": "median"})
time_series_settings = {
'time_column_name': time_column_name,
'grain_column_names': grain_column_names,
'max_horizon': n_test_periods
}
automl_config = AutoMLConfig(task='forecasting',
debug_log='automl_oj_sales_errors.log',
primary_metric='normalized_mean_absolute_error',
experiment_timeout_hours=0.25,
training_data=train_dataset,
label_column_name=target_column_name,
compute_target=compute_target,
enable_early_stopping=True,
featurization=featurization_config,
n_cross_validations=3,
verbosity=logging.INFO,
max_cores_per_iteration=-1,
**time_series_settings)
remote_run = experiment.submit(automl_config, show_output=False)
remote_run
remote_run.wait_for_completion()
best_run, fitted_model = remote_run.get_output()
print(fitted_model.steps)
model_name = best_run.properties['model_name']
custom_featurizer = fitted_model.named_steps['timeseriestransformer']
custom_featurizer.get_featurization_summary()
X_test = test
y_test = X_test.pop(target_column_name).values
X_test.head()
# The featurized data, aligned to y, will also be returned.
# This contains the assumptions that were made in the forecast
# and helps align the forecast to the original data
y_predictions, X_trans = fitted_model.forecast(X_test)
from forecasting_helper import align_outputs
df_all = align_outputs(y_predictions, X_trans, X_test, y_test, target_column_name)
from azureml.automl.core.shared import constants
from azureml.automl.runtime.shared.score import scoring
from matplotlib import pyplot as plt
# use automl scoring module
scores = scoring.score_regression(
y_test=df_all[target_column_name],
y_pred=df_all['predicted'],
metrics=list(constants.Metric.SCALAR_REGRESSION_SET))
print("[Test data scores]\n")
for key, value in scores.items():
print('{}: {:.3f}'.format(key, value))
# Plot outputs
%matplotlib inline
test_pred = plt.scatter(df_all[target_column_name], df_all['predicted'], color='b')
test_test = plt.scatter(df_all[target_column_name], df_all[target_column_name], color='g')
plt.legend((test_pred, test_test), ('prediction', 'truth'), loc='upper left', fontsize=8)
plt.show()
description = 'AutoML OJ forecaster'
tags = None
model = remote_run.register_model(model_name = model_name, description = description, tags = tags)
print(remote_run.model_id)
script_file_name = 'score_fcast.py'
best_run.download_file('outputs/scoring_file_v_1_0_0.py', script_file_name)
from azureml.core.model import InferenceConfig
from azureml.core.webservice import AciWebservice
from azureml.core.webservice import Webservice
from azureml.core.model import Model
inference_config = InferenceConfig(environment = best_run.get_environment(),
entry_script = script_file_name)
aciconfig = AciWebservice.deploy_configuration(cpu_cores = 1,
memory_gb = 2,
tags = {'type': "automl-forecasting"},
description = "Automl forecasting sample service")
aci_service_name = 'automl-oj-forecast-01'
print(aci_service_name)
aci_service = Model.deploy(ws, aci_service_name, [model], inference_config, aciconfig)
aci_service.wait_for_deployment(True)
print(aci_service.state)
aci_service.get_logs()
import json
X_query = X_test.copy()
# We have to convert datetime to string, because Timestamps cannot be serialized to JSON.
X_query[time_column_name] = X_query[time_column_name].astype(str)
# The Service object accept the complex dictionary, which is internally converted to JSON string.
# The section 'data' contains the data frame in the form of dictionary.
test_sample = json.dumps({'data': X_query.to_dict(orient='records')})
response = aci_service.run(input_data = test_sample)
# translate from networkese to datascientese
try:
res_dict = json.loads(response)
y_fcst_all = pd.DataFrame(res_dict['index'])
y_fcst_all[time_column_name] = pd.to_datetime(y_fcst_all[time_column_name], unit = 'ms')
y_fcst_all['forecast'] = res_dict['forecast']
except:
print(res_dict)
y_fcst_all.head()
serv = Webservice(ws, 'automl-oj-forecast-01')
serv.delete() # don't do it accidentally
| 0.652684 | 0.940408 |
# Random Forests
## Classifier
```
# For python 2 and python 3
from __future__ import division, print_function, unicode_literals
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
from sklearn.datasets import load_iris
from sklearn.ensemble import (RandomForestClassifier, ExtraTreesClassifier,
AdaBoostClassifier)
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
X = [[0, 0], [1, 1]]
Y = [0, 1]
clf = RandomForestClassifier(n_estimators=10)
clf
clf = clf.fit(X, Y)
clf.predict([[1.3, 1.1]])
n_classes = 3
n_estimators = 30
cmap = plt.cm.RdYlBu
plot_step = 0.02
plot_step_coarser = 0.5
RANDOM_SEED = 13
iris = load_iris()
plot_idx = 1
models = [DecisionTreeClassifier(max_depth=None),
RandomForestClassifier(n_estimators=n_estimators),
ExtraTreesClassifier(n_estimators=n_estimators),
AdaBoostClassifier(DecisionTreeClassifier(max_depth=3),
n_estimators=n_estimators)]
for pair in ([0, 1], [0, 2], [2, 3]):
for model in models:
X = iris.data[:, pair]
y = iris.target
idx = np.arange(X.shape[0])
np.random.seed(RANDOM_SEED)
np.random.shuffle(idx)
X = X[idx]
y = y[idx]
mean = X.mean(axis=0)
std = X.std(axis=0)
X = (X - mean) / std
model.fit(X, y)
scores = model.score(X, y)
model_title = str(type(model)).split(
".")[-1][:-2][:-len("Classifier")]
model_details = model_title
if hasattr(model, "estimators_"):
model_details += " with {} estimators".format(
len(model.estimators_))
print(model_details + " with features", pair,
"has a score of", scores)
plt.subplot(3, 4, plot_idx)
if plot_idx <= len(models):
plt.title(model_title, fontsize=9)
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, plot_step),
np.arange(y_min, y_max, plot_step))
if isinstance(model, DecisionTreeClassifier):
Z = model.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
cs = plt.contourf(xx, yy, Z, cmap=cmap)
else:
estimator_alpha = 1.0 / len(model.estimators_)
for tree in model.estimators_:
Z = tree.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
cs = plt.contourf(xx, yy, Z, alpha=estimator_alpha, cmap=cmap)
xx_coarser, yy_coarser = np.meshgrid(
np.arange(x_min, x_max, plot_step_coarser),
np.arange(y_min, y_max, plot_step_coarser))
Z_points_coarser = model.predict(np.c_[xx_coarser.ravel(),
yy_coarser.ravel()]
).reshape(xx_coarser.shape)
cs_points = plt.scatter(xx_coarser, yy_coarser, s=15,
c=Z_points_coarser, cmap=cmap,
edgecolors="none")
plt.scatter(X[:, 0], X[:, 1], c=y,
cmap=ListedColormap(['r', 'y', 'b']),
edgecolor='k', s=20)
plot_idx += 1
plt.suptitle("Classifiers on feature subsets of the Iris dataset", fontsize=12)
plt.axis("tight")
plt.tight_layout(h_pad=0.2, w_pad=0.2, pad=2.5)
plt.show()
```
|
github_jupyter
|
# For python 2 and python 3
from __future__ import division, print_function, unicode_literals
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
from sklearn.datasets import load_iris
from sklearn.ensemble import (RandomForestClassifier, ExtraTreesClassifier,
AdaBoostClassifier)
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
X = [[0, 0], [1, 1]]
Y = [0, 1]
clf = RandomForestClassifier(n_estimators=10)
clf
clf = clf.fit(X, Y)
clf.predict([[1.3, 1.1]])
n_classes = 3
n_estimators = 30
cmap = plt.cm.RdYlBu
plot_step = 0.02
plot_step_coarser = 0.5
RANDOM_SEED = 13
iris = load_iris()
plot_idx = 1
models = [DecisionTreeClassifier(max_depth=None),
RandomForestClassifier(n_estimators=n_estimators),
ExtraTreesClassifier(n_estimators=n_estimators),
AdaBoostClassifier(DecisionTreeClassifier(max_depth=3),
n_estimators=n_estimators)]
for pair in ([0, 1], [0, 2], [2, 3]):
for model in models:
X = iris.data[:, pair]
y = iris.target
idx = np.arange(X.shape[0])
np.random.seed(RANDOM_SEED)
np.random.shuffle(idx)
X = X[idx]
y = y[idx]
mean = X.mean(axis=0)
std = X.std(axis=0)
X = (X - mean) / std
model.fit(X, y)
scores = model.score(X, y)
model_title = str(type(model)).split(
".")[-1][:-2][:-len("Classifier")]
model_details = model_title
if hasattr(model, "estimators_"):
model_details += " with {} estimators".format(
len(model.estimators_))
print(model_details + " with features", pair,
"has a score of", scores)
plt.subplot(3, 4, plot_idx)
if plot_idx <= len(models):
plt.title(model_title, fontsize=9)
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, plot_step),
np.arange(y_min, y_max, plot_step))
if isinstance(model, DecisionTreeClassifier):
Z = model.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
cs = plt.contourf(xx, yy, Z, cmap=cmap)
else:
estimator_alpha = 1.0 / len(model.estimators_)
for tree in model.estimators_:
Z = tree.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
cs = plt.contourf(xx, yy, Z, alpha=estimator_alpha, cmap=cmap)
xx_coarser, yy_coarser = np.meshgrid(
np.arange(x_min, x_max, plot_step_coarser),
np.arange(y_min, y_max, plot_step_coarser))
Z_points_coarser = model.predict(np.c_[xx_coarser.ravel(),
yy_coarser.ravel()]
).reshape(xx_coarser.shape)
cs_points = plt.scatter(xx_coarser, yy_coarser, s=15,
c=Z_points_coarser, cmap=cmap,
edgecolors="none")
plt.scatter(X[:, 0], X[:, 1], c=y,
cmap=ListedColormap(['r', 'y', 'b']),
edgecolor='k', s=20)
plot_idx += 1
plt.suptitle("Classifiers on feature subsets of the Iris dataset", fontsize=12)
plt.axis("tight")
plt.tight_layout(h_pad=0.2, w_pad=0.2, pad=2.5)
plt.show()
| 0.690559 | 0.772445 |
# Introducción a la Computación Científica con Python
Versión original en inglés de J.R. Johansson ([email protected]) http://dml.riken.jp/~rob/
Traducido/Adaptado por [G.F. Rubilar](http://google.com/+GuillermoRubilar).
La última versión de estos [notebooks de IPython](http://ipython.org/notebook.html) está disponible en [http://github.com/gfrubi/clases-python-cientifico](http://github.com/gfrubi/clases-python-cientifico).
La última versión del original (en inglés) de estos [notebooks de IPython](http://ipython.org/notebook.html) está disponible en [http://github.com/jrjohansson/scientific-python-lectures](http://github.com/jrjohansson/scientific-python-lectures).
Los otros notebooks de esta serie están listados en [http://jrjohansson.github.com](http://jrjohansson.github.com).
```
%matplotlib inline
```
## Introducción
Existen dos sistemas de álgebra simbólica para Python:
* [SymPy](http://sympy.org/en/index.html) - Un módulo que puede ser usado en cualquier programa Python, o bien en una sesión de IPython, que incluye poderosas herramientas para cálculo simbólico.
* [Sage](http://www.sagemath.org/) - Sage es un sistema completo y poderoso que intenta suministrar un sistema de código abierto que compita con Mathematica and Maple. Sage no es un módulo de Python, sino un ambiente de cálculo simbólico que usa Python como su lenguaje de programación.
Sage es más poderoso que SymPy en algunos aspectos, pero ambos ofreces una lista completa de funcionalidades de cálculo simbólico. La ventaja de SymPy es que es un módulo normal de Python y se integra muy bien en un notebook de IPython.
En esta clase veremos cómo usar SymPy en un notebook de IPython.
Para comenzar a usar SymPy en un programa Python o en un notebook, importamos el módulo `sympy`:
```
from sympy import *
```
Para que los resultados sean formateados en $\LaTeX$ podemos usar:
```
init_printing(use_latex=True)
```
## Variables simbólicas
En SymPy podemos crear símbolos para las variables con las que deseamos trabajar. Podemos crear un nuevo símbolo usando la clase `Symbol`:
```
x = Symbol('x')
(pi + x)**2
# forma alternativa de definir (varios) símbolos
a, b, c = symbols("a, b, c")
type(a)
```
Podemos agregar algunas propiedades a los símbolos cuando son creados:
```
x = Symbol('x', real=True)
x.is_imaginary
x = Symbol('x', positive=True)
x > 0
```
### Números complejos
La unidad imaginaria es denotada por `I` en Sympy.
```
1+1*I
I**2
(x * I + 1)**2
```
### Números racionales
Existen tres tipos distintos de números en SymPy: `Real`, `Rational`, `Integer`:
```
r1 = Rational(4,5)
r2 = Rational(5,4)
r1
r1+r2
r1/r2
```
### Evaluación numérica
SymPy usa una librería para trabajar con números con precisión arbitraria, y tiene expresiones SymPy predefinidas para varias constantes matemáticas, tales como: `pi`, `e` y `oo` para el infinito.
Para evaluar numéricamente una expresión podemos usar la función `evalf` (o bien `N`). Ésta usa un argumento `n` que especifíca el número de cifras significativas.
```
pi.evalf(n=50)
y = (x + pi)**2
N(y, 5) # equivalente a evalf
```
Cuando evaluamos numéricamente expresiones a menudo deseamos substituir un símbolo por un valor numérico. En SymPy hacemos esto usando la función `subs`:
```
y.subs(x, 1.5)
N(y.subs(x, 1.5))
```
La función `subs` también puede ser usada para substituir símbolos y expresiones:
```
y.subs(x, a+pi)
```
También podemos combinar la evaluación numérica de expresiones con arreglos NumPy:
```
import numpy
x_vec = numpy.arange(0, 10, 0.1)
y_vec = numpy.array([N(((x + pi)**2).subs(x, xx)) for xx in x_vec])
from matplotlib.pyplot import *
plot(x_vec, y_vec);
```
Sin embargo, este tipo de evaluación numérica puede ser muy lenta, y existe una forma mucho más eficiente de realizar la misma tarea: Usar la función `lambdify` para "mapear" una expresión de Sympy a una función que es mucho más eficiente para la evaluación numérica:
```
f = lambdify([x], (x + pi)**2, 'numpy') # el primer argumento es una lista de variables de las que la función f dependerá: en este caso sólo x -> f(x)
type(f)
y_vec = f(x_vec) # ahora podemos pasar directamente un arreglo Numpy. Así f(x) es evaluado más eficientemente
```
La mayor eficiencia de usar funciones "lambdificadas" en lugar de usar evalación numérica directa puede ser significativa, a menudo de varios órdenes de magnitud. Aún en este sencillo ejemplo obtenemos un aumento de velocidad importante:
```
%%timeit
y_vec = numpy.array([N(((x + pi)**2).subs(x, xx)) for xx in x_vec])
%%timeit
y_vec = f(x_vec)
```
## Manipulaciones algebráicas
Uno de los usos principales de un sistema de cálculo simbólico es que realiza manipulaciones algebráicas de expresiones. Por ejemplo, si queremos expandir un producto, factorizar una expresión, o simplificar un resultado. En esta sección presentamos las funciones para realizar estas operaciones básicas en SymPy.
### Expand and factor
Primeros pasos en la manipulación algebráica
```
(x+1)*(x+2)*(x+3)
expand((x+1)*(x+2)*(x+3))
```
La función `expand` acepta varias argumentos clave con los que se puede indicar qué tipo de expansión deseamos realizar. Por ejemplo, para expandir expresiones trigonométricas, usamos el argumento clave `trig=True`:
```
sin(a+b)
expand(sin(a+b), trig=True)
```
Ver `help(expand)` para una descripción detallada de los distintos tipos de expansiones que la función `expand` puede realizar.
También podemos factorizar expresiones, usando la función `factor` de SymPy:
```
factor(x**3 + 6 * x**2 + 11*x + 6)
```
### Simplify
La función `simplify` intenta simplificar una expresión usando distinta técnicas. Existen también alternativas más específicas a la función `simplify`: `trigsimp`, `powsimp`, `logcombine`, etc.
El uso básico de estas funciones en el siguiente:
```
# simplify expande un producto
simplify((x+1)*(x+2)*(x+3))
# simplify usa identidades trigonometricas
simplify(sin(a)**2 + cos(a)**2)
simplify(cos(x)/sin(x))
```
## apart and together
Podemos también manipular expresiones simbólicas que involucran fracciones usando las funciones `apart` y `together`. La primera de estas funciones separa una fracción en sus correspondientes fracciones parciales; la segunda hace todo lo contrario.
```
f1 = 1/((a+1)*(a+2))
f1
apart(f1)
f2 = 1/(a+2) + 1/(a+3)
f2
together(f2)
```
Usualmente `Simplify` combina las fracciones, pero no factoriza:
```
simplify(f2)
```
## Cálculo
Además de realizar manipulaciones algebráicas, SimPy puede realizar operaciones de cálculo, tales como derivar y derivar expresiones.
### Derivación
Derviar es usualmente algo simple. Usamos la función `diff`. El primer argumento es una expresión que será derivada, y el segundo argumento es el símbolo respecto al cual se realizará la derivada:
```
y
diff(y**2, x)
```
Para calcular derivadas de orden superior podemos usar:
```
diff(y**2, x, x)
```
o bien
```
diff(y**2, x, 2) # hace lo mismo
```
To calculate the derivative of a multivariate expression, we can do:
```
x, y, z = symbols("x,y,z")
f = sin(x*y) + cos(y*z)
```
$\frac{d^3f}{dxdy^2}$
```
diff(f, x, 1, y, 2)
```
### Integration
Integration is done in a similar fashion:
```
f
integrate(f, x)
```
By providing limits for the integration variable we can evaluate definite integrals:
```
integrate(f, (x, -1, 1))
```
and also improper integrals
```
integrate(exp(-x**2), (x, -oo, oo))
```
Remember, `oo` is the SymPy notation for inifinity.
### Sums and products
We can evaluate sums and products using the functions: 'Sum'
```
n = Symbol("n")
Sum(1/n**2, (n, 1, 10))
Sum(1/n**2, (n,1, 10)).evalf()
Sum(1/n**2, (n, 1, oo)).evalf()
```
Products work much the same way:
```
Product(n, (n, 1, 10)) # 10!
```
### Limits
Limits can be evaluated using the `limit` function. For example,
```
limit(sin(x)/x, x, 0)
```
We can use 'limit' to check the result of derivation using the `diff` function:
```
f
diff(f, x)
```
$\displaystyle \frac{\mathrm{d}f(x,y)}{\mathrm{d}x} = \frac{f(x+h,y)-f(x,y)}{h}$
```
h = Symbol("h")
limit((f.subs(x, x+h) - f)/h, h, 0)
```
OK!
We can change the direction from which we approach the limiting point using the `dir` keywork argument:
```
limit(1/x, x, 0, dir="+")
limit(1/x, x, 0, dir="-")
```
### Series
Series expansion is also one of the most useful features of a CAS. In SymPy we can perform a series expansion of an expression using the `series` function:
```
series(exp(x), x)
```
By default it expands the expression around $x=0$, but we can expand around any value of $x$ by explicitly include a value in the function call:
```
series(exp(x), x, 1)
```
And we can explicitly define to which order the series expansion should be carried out:
```
series(exp(x), x, 1, 10)
```
The series expansion includes the order of the approximation, which is very useful for keeping track of the order of validity when we do calculations with series expansions of different order:
```
s1 = cos(x).series(x, 0, 5)
s1
s2 = sin(x).series(x, 0, 2)
s2
expand(s1 * s2)
```
If we want to get rid of the order information we can use the `removeO` method:
```
expand(s1.removeO() * s2.removeO())
```
But note that this is not the correct expansion of $\cos(x)\sin(x)$ to $5$th order:
```
(cos(x)*sin(x)).series(x, 0, 6)
```
## Linear algebra
### Matrices
Matrices are defined using the `Matrix` class:
```
m11, m12, m21, m22 = symbols("m11, m12, m21, m22")
b1, b2 = symbols("b1, b2")
A = Matrix([[m11, m12],[m21, m22]])
A
b = Matrix([[b1], [b2]])
b
```
With `Matrix` class instances we can do the usual matrix algebra operations:
```
A**2
A * b
```
And calculate determinants and inverses, and the like:
```
A.det()
A.inv()
```
## Solving equations
For solving equations and systems of equations we can use the `solve` function:
```
solve(x**2 - 1, x)
solve(x**4 - x**2 - 1, x)
```
System of equations:
```
solve([x + y - 1, x - y - 1], [x,y])
```
In terms of other symbolic expressions:
```
solve([x + y - a, x - y - c], [x,y])
```
## Quantum mechanics: noncommuting variables
How about non-commuting symbols? In quantum mechanics we need to work with noncommuting operators, and SymPy has a nice support for noncommuting symbols and even a subpackage for quantum mechanics related calculations!
```
from sympy.physics.quantum import *
```
### States
We can define symbol states, kets and bras:
```
Ket('psi')
Bra('psi')
u = Ket('0')
d = Ket('1')
a, b = symbols('alpha beta', complex=True)
phi = a * u + sqrt(1-abs(a)**2) * d; phi
Dagger(phi)
Dagger(phi) * d
```
Use `qapply` to distribute a mutiplication:
```
qapply(Dagger(phi) * d)
qapply(Dagger(phi) * u)
```
### Operators
```
A = Operator('A')
B = Operator('B')
```
Check if they are commuting!
```
A * B == B * A
expand((A+B)**3)
c = Commutator(A,B)
c
```
We can use the `doit` method to evaluate the commutator:
```
c.doit()
```
We can mix quantum operators with C-numbers:
```
c = Commutator(a * A, b * B)
c
```
To expand the commutator, use the `expand` method with the `commutator=True` keyword argument:
```
c = Commutator(A+B, A*B)
c.expand(commutator=True)
Dagger(Commutator(A, B))
ac = AntiCommutator(A,B)
ac.doit()
```
#### Example: Quadrature commutator
Let's look at the commutator of the electromagnetic field quadatures $x$ and $p$. We can write the quadrature operators in terms of the creation and annihilation operators as:
$\displaystyle x = (a + a^\dagger)/\sqrt{2}$
$\displaystyle p = -i(a - a^\dagger)/\sqrt{2}$
```
X = (A + Dagger(A))/sqrt(2)
X
P = -I * (A - Dagger(A))/sqrt(2)
P
```
Let's expand the commutator $[x,p]$
```
Commutator(X, P).expand(commutator=True).expand(commutator=True)
```
Here we see directly that the well known commutation relation for the quadratures
$[x,p]=i$
is a directly related to
$[A, A^\dagger]=1$
(which SymPy does not know about, and does not simplify).
For more details on the quantum module in SymPy, see:
* http://docs.sympy.org/0.7.2/modules/physics/quantum/index.html
* http://nbviewer.ipython.org/urls/raw.github.com/ipython/ipython/master/docs/examples/notebooks/sympy_quantum_computing.ipynb
## Further reading
* http://sympy.org/en/index.html - The SymPy projects web page.
* https://github.com/sympy/sympy - The source code of SymPy.
* http://live.sympy.org - Online version of SymPy for testing and demonstrations.
|
github_jupyter
|
%matplotlib inline
from sympy import *
init_printing(use_latex=True)
x = Symbol('x')
(pi + x)**2
# forma alternativa de definir (varios) símbolos
a, b, c = symbols("a, b, c")
type(a)
x = Symbol('x', real=True)
x.is_imaginary
x = Symbol('x', positive=True)
x > 0
1+1*I
I**2
(x * I + 1)**2
r1 = Rational(4,5)
r2 = Rational(5,4)
r1
r1+r2
r1/r2
pi.evalf(n=50)
y = (x + pi)**2
N(y, 5) # equivalente a evalf
y.subs(x, 1.5)
N(y.subs(x, 1.5))
y.subs(x, a+pi)
import numpy
x_vec = numpy.arange(0, 10, 0.1)
y_vec = numpy.array([N(((x + pi)**2).subs(x, xx)) for xx in x_vec])
from matplotlib.pyplot import *
plot(x_vec, y_vec);
f = lambdify([x], (x + pi)**2, 'numpy') # el primer argumento es una lista de variables de las que la función f dependerá: en este caso sólo x -> f(x)
type(f)
y_vec = f(x_vec) # ahora podemos pasar directamente un arreglo Numpy. Así f(x) es evaluado más eficientemente
%%timeit
y_vec = numpy.array([N(((x + pi)**2).subs(x, xx)) for xx in x_vec])
%%timeit
y_vec = f(x_vec)
(x+1)*(x+2)*(x+3)
expand((x+1)*(x+2)*(x+3))
sin(a+b)
expand(sin(a+b), trig=True)
factor(x**3 + 6 * x**2 + 11*x + 6)
# simplify expande un producto
simplify((x+1)*(x+2)*(x+3))
# simplify usa identidades trigonometricas
simplify(sin(a)**2 + cos(a)**2)
simplify(cos(x)/sin(x))
f1 = 1/((a+1)*(a+2))
f1
apart(f1)
f2 = 1/(a+2) + 1/(a+3)
f2
together(f2)
simplify(f2)
y
diff(y**2, x)
diff(y**2, x, x)
diff(y**2, x, 2) # hace lo mismo
x, y, z = symbols("x,y,z")
f = sin(x*y) + cos(y*z)
diff(f, x, 1, y, 2)
f
integrate(f, x)
integrate(f, (x, -1, 1))
integrate(exp(-x**2), (x, -oo, oo))
n = Symbol("n")
Sum(1/n**2, (n, 1, 10))
Sum(1/n**2, (n,1, 10)).evalf()
Sum(1/n**2, (n, 1, oo)).evalf()
Product(n, (n, 1, 10)) # 10!
limit(sin(x)/x, x, 0)
f
diff(f, x)
h = Symbol("h")
limit((f.subs(x, x+h) - f)/h, h, 0)
limit(1/x, x, 0, dir="+")
limit(1/x, x, 0, dir="-")
series(exp(x), x)
series(exp(x), x, 1)
series(exp(x), x, 1, 10)
s1 = cos(x).series(x, 0, 5)
s1
s2 = sin(x).series(x, 0, 2)
s2
expand(s1 * s2)
expand(s1.removeO() * s2.removeO())
(cos(x)*sin(x)).series(x, 0, 6)
m11, m12, m21, m22 = symbols("m11, m12, m21, m22")
b1, b2 = symbols("b1, b2")
A = Matrix([[m11, m12],[m21, m22]])
A
b = Matrix([[b1], [b2]])
b
A**2
A * b
A.det()
A.inv()
solve(x**2 - 1, x)
solve(x**4 - x**2 - 1, x)
solve([x + y - 1, x - y - 1], [x,y])
solve([x + y - a, x - y - c], [x,y])
from sympy.physics.quantum import *
Ket('psi')
Bra('psi')
u = Ket('0')
d = Ket('1')
a, b = symbols('alpha beta', complex=True)
phi = a * u + sqrt(1-abs(a)**2) * d; phi
Dagger(phi)
Dagger(phi) * d
qapply(Dagger(phi) * d)
qapply(Dagger(phi) * u)
A = Operator('A')
B = Operator('B')
A * B == B * A
expand((A+B)**3)
c = Commutator(A,B)
c
c.doit()
c = Commutator(a * A, b * B)
c
c = Commutator(A+B, A*B)
c.expand(commutator=True)
Dagger(Commutator(A, B))
ac = AntiCommutator(A,B)
ac.doit()
X = (A + Dagger(A))/sqrt(2)
X
P = -I * (A - Dagger(A))/sqrt(2)
P
Commutator(X, P).expand(commutator=True).expand(commutator=True)
| 0.321141 | 0.986205 |
# Level Order Traversal
[Click here to run this chapter on Colab](https://colab.research.google.com/github/AllenDowney/DSIRP/blob/main/notebooks/level_order.ipynb)
## More tree traversal
In a previous notebook we wrote two versions of a depth-first search in a tree.
Now we are working toward depth-first search, but we're going to make a stop along the way: level-order traversal.
One application of level-order traversal is searching through directories (aka folders) in a file system.
Since directories can contain other directories, which can contains other directories, and so on, we can think of a file system as a tree.
In this notebook, we'll start by making a tree of directories and fake data files.
Then we'll traverse it several ways.
And while we're at it, we'll learn about the `os` module, which provides functions for interacting with the operating system, especially the file system.
The `os` module provides `mkdir`, which creates a directory. It raises an exception if the directory exists, so I'm going to wrap it in a `try` statement.
```
import os
def mkdir(dirname):
try:
os.mkdir(dirname)
print('made', dirname)
except FileExistsError:
print(dirname, 'exists')
```
Now I'll create the directory where we'll put the fake data.
```
mkdir('level_data')
```
Inside `level_data`, I want to make a subdirectory named `2021`.
It is tempting to write something like:
```
year_dir = `level_data/2021`
```
This path would work on Unix operating systems (including MacOS), but not Windows, which uses `\` rather than `/` between names in a path.
We can avoid this problem by using `os.path.join`, which joins names in a path with whatever character the operating system wants.
```
year_dir = os.path.join('level_data', '2021')
mkdir(year_dir)
```
To make the fake data files, I'll use the following function, which opens a file for writing and puts the word `data` into it.
```
def make_datafile(dirname, filename):
filename = os.path.join(dirname, filename)
open(filename, 'w').write('data\n')
print('made', filename)
```
So let's start by putting a data file in `year_dir`, imagining that this file contains summary data for the whole year.
```
make_datafile(year_dir, 'year.csv')
```
The following function
1. Makes a subdirectory that represents one month of the year,
2. Makes a data file we imagine contains summary data for the month, and
3. Calls `make_day` (below) to make subdirectories each day of the month (in a world where all months have 30 days).
```
def make_month(i, year_dir):
month = '%.2d' % i
month_dir = os.path.join(year_dir, month)
mkdir(month_dir)
make_datafile(month_dir, 'month.csv')
for j in range(1, 31):
make_day(j, month_dir)
```
`make_day` makes a sub-subdirectory for a given day of the month, and puts a data file in it.
```
def make_day(j, month_dir):
day = '%.2d' % j
day_dir = os.path.join(month_dir, day)
mkdir(day_dir)
make_datafile(day_dir, 'day.csv')
```
The following loop makes a directory for each month.
```
for i in range(1, 13):
make_month(i, year_dir)
```
## Walking a Directory
The `os` module provides `walk`, which is a generator function that traverses a directory and all its subdirectories, and all their subdirectories, and so on.
For each directory, it yields:
* dirpath, which is the name of the directory.
* dirnames, which is a list of subdirectories it contains, and
* filenames, which is a list of files it contains.
Here's how we can use it to print the paths of all files in the directory we created.
```
for dirpath, dirnames, filenames in os.walk('level_data'):
for filename in filenames:
path = os.path.join(dirpath, filename)
print(path)
```
One quirk of `os.walk` is that the directories and files don't appear in any particular order.
Of course, we can store the results and sort them in whatever order we want.
But as an exercise, we can write our own version of `walk`.
We'll need two functions:
* `os.listdir`, which takes a directory and list the directories and files it contains, and
* `os.path.isfile`, which takes a path and returns `True` if it is a file, and `False` if it is a directory or something else.
You might notice that some file-related functions are in the submodule `os.path`.
There is some logic to this organization, but it is not always obvious why a particular function is in this submodule or not.
Anyway, here is a recursive version of `walk`:
```
def walk(dirname):
for name in sorted(os.listdir(dirname)):
path = os.path.join(dirname, name)
if os.path.isfile(path):
print(path)
else:
walk(path)
walk(year_dir)
```
**Exercise:** Write a version of `walk` called `walk_gen` that is a generator function; that is, it should yield the paths it finds rather than printing them.
You can use the following loop to test your code.
```
for path in walk_gen(year_dir):
print(path)
```
**Exercise:** Write a version of `walk_gen` called `walk_dfs` that traverses the given directory and yields the file it contains, but it should use a stack and run iteratively, rather than recursively.
You can use the following loop to test your code.
```
for path in walk_dfs(year_dir):
print(path)
```
Notice that the order the files are discovered is "depth-first". For example, it yields all files from the first month before any of the files for the second month.
An alternative is a level-order traversal, which yields all files at the first level (the annual summary), then all the files at the second level (the monthly summaries), then the files at the third level.
To implement a level-order traversal, we can make a minimal change to `walk_dfs`: replace the stack with a FIFO queue.
To implement the queue efficiently, we can use `collections.deque`.
**Exercise:** Write a generator function called `walk_level` that takes a directory and yields its files in level order.
Use the following loop to test your code.
```
for path in walk_level(year_dir):
print(path)
```
If you are looking for a file in a large file system, a level-order search might be useful if you think the file is more likely to be near the root, rather than deep in a nested subdirectory.
*Data Structures and Information Retrieval in Python*
Copyright 2021 Allen Downey
License: [Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International](https://creativecommons.org/licenses/by-nc-sa/4.0/)
|
github_jupyter
|
import os
def mkdir(dirname):
try:
os.mkdir(dirname)
print('made', dirname)
except FileExistsError:
print(dirname, 'exists')
mkdir('level_data')
year_dir = `level_data/2021`
year_dir = os.path.join('level_data', '2021')
mkdir(year_dir)
def make_datafile(dirname, filename):
filename = os.path.join(dirname, filename)
open(filename, 'w').write('data\n')
print('made', filename)
make_datafile(year_dir, 'year.csv')
def make_month(i, year_dir):
month = '%.2d' % i
month_dir = os.path.join(year_dir, month)
mkdir(month_dir)
make_datafile(month_dir, 'month.csv')
for j in range(1, 31):
make_day(j, month_dir)
def make_day(j, month_dir):
day = '%.2d' % j
day_dir = os.path.join(month_dir, day)
mkdir(day_dir)
make_datafile(day_dir, 'day.csv')
for i in range(1, 13):
make_month(i, year_dir)
for dirpath, dirnames, filenames in os.walk('level_data'):
for filename in filenames:
path = os.path.join(dirpath, filename)
print(path)
def walk(dirname):
for name in sorted(os.listdir(dirname)):
path = os.path.join(dirname, name)
if os.path.isfile(path):
print(path)
else:
walk(path)
walk(year_dir)
for path in walk_gen(year_dir):
print(path)
for path in walk_dfs(year_dir):
print(path)
for path in walk_level(year_dir):
print(path)
| 0.091306 | 0.944689 |
# ICS 435 Final Project: Covid-19 Mask Detection Model
* Group Members:
* Yick Ching (Jeff) Wong
* Timoteo Sumalinog III
* Jeraldy Cascayan
* Project Description:
* Use a CNN model to classify if a face is wearing a mask or not
# Importing packages
```
import os
from urllib.request import urlretrieve
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import cv2
from zipfile import ZipFile as zip
from PIL import Image
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.models import Sequential
from sklearn.model_selection import GridSearchCV, train_test_split
from sklearn.metrics import roc_curve, roc_auc_score
sess = tf.compat.v1.Session(config=tf.compat.v1.ConfigProto(log_device_placement=True))
```
# Downloading the dataset from the internet
```
def face_mask_dataset():
url = 'https://storage.googleapis.com/kaggle-data-sets/675484/1187790/bundle/archive.zip?X-Goog-Algorithm=GOOG4-RSA-SHA256&X-Goog-Credential=gcp-kaggle-com%40kaggle-161607.iam.gserviceaccount.com%2F20210511%2Fauto%2Fstorage%2Fgoog4_request&X-Goog-Date=20210511T224333Z&X-Goog-Expires=259199&X-Goog-SignedHeaders=host&X-Goog-Signature=038b7fb07abcc84e68e49b04359b793434ac730e3199cbf91d9dc4099cdc1bc0c6626e27f68e8dcf43f7fdd353afbc460f503f24008422252113cb6f6517d6e1afac6580e115d6843388e358b0e44140e606232c211f43eaf29abd46945295d6c8269200d56c7c01855e3dd48b46f5aa5dd921ddd90401ec59f589f8202331adcbdb3de8ce3567114a158e7e431f48a27bf15f47674150bf270012992fe8b9e1105b9c3432d14c07c9964cd5e87d4423840e853bfe2930d4e562e49b0ebc324e64b0f4269af88ffca676f5f3a1124f33c17142f38a3d118faf837fac8c9455b8bfb16d98bb1995fadc40ae65d9146b448535f2200862db4c618f59bcefb37d58' # Link to the dataset
file = 'dataset.zip' # Name of the dataset
path = './' # Download data to current directory.
os.makedirs(path, exist_ok=True) # Create path if it doesn't exist.
# Download the image dataset
if file not in os.listdir(path):
urlretrieve(url, os.path.join(path, file))
print("Downloaded %s to %s" % (file, path))
if 'dataset' not in os.listdir(path):
with zip(file, 'r') as zipobj:
zipobj.extractall('dataset')
def getImages_Labels(path, partition):
listOfLabels = os.listdir(path + partition)
images = []
labels = []
for label in listOfLabels:
val = 0
listOfImages = os.listdir(path + partition + label)
if label=="WithMask":
val = 1
for image in listOfImages:
img_arr = np.asarray(Image.open(path+partition+label+'/'+image))
img_arr = cv2.resize(img_arr, (100, 100))
images.append(img_arr)
labels.append(val)
return np.array(images), np.array(labels)
path = 'dataset/Face Mask Dataset/'
(test_images, test_labels) = getImages_Labels(path, 'Test/')
(train_images, train_labels) = getImages_Labels(path, 'Train/')
(val_images, val_labels) = getImages_Labels(path, 'Validation/')
ret_images = np.vstack((test_images, train_images, val_images))
ret_labels = np.hstack((test_labels, train_labels, val_labels))
return (ret_images, ret_labels)
```
# Loading in the dataset
```
(images, labels) = face_mask_dataset()
```
# Splitting the dataset into train and test split
```
(train_images, test_images, train_labels, test_labels) = train_test_split(images, labels)
print(f'# of training data: {train_images.shape[0]}, # of test data: {test_images.shape[0]}')
```
# Visualizing the data
```
class_names = ['no-mask', 'mask']
plt.figure()
plt.imshow(train_images[5306])
plt.colorbar()
plt.grid(False)
plt.show()
plt.figure(figsize=(10,10))
plt.suptitle('25 Examples of the training dataset', y=0.92)
for i in range(25):
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
index = np.random.randint(0, train_images.shape[0])
plt.imshow(train_images[index], cmap=plt.cm.binary)
plt.xlabel(class_names[train_labels[index]])
plt.show()
```
# Creating and compiling the model
```
num_classes = 2
img_height = 100
img_width = 100
def create_model():
model = Sequential([
layers.experimental.preprocessing.Rescaling(1./255, input_shape=(img_height, img_width, 3)),
layers.experimental.preprocessing.RandomRotation(0.1),
layers.experimental.preprocessing.RandomContrast(0.1),
layers.experimental.preprocessing.RandomTranslation(0.05, 0.1),
layers.Conv2D(16, 3, padding='same', activation='relu'),
layers.MaxPooling2D(),
layers.Conv2D(32, 3, padding='same', activation='relu'),
layers.MaxPooling2D(),
layers.Conv2D(64, 3, padding='same', activation='relu'),
layers.MaxPooling2D(),
layers.Flatten(),
layers.Dense(128, activation='relu'),
layers.Dropout(0.5),
layers.Dense(num_classes - 1, activation='sigmoid')
])
model.compile(optimizer='adam',
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=['accuracy'])
return model
```
# HyperParameter(s) Optimization
```
model = keras.wrappers.scikit_learn.KerasClassifier(build_fn=create_model)
batch_size = [30, 50, 100]
epochs = [5, 10, 20]
param_grid = dict(batch_size=batch_size, epochs=epochs)
grid = GridSearchCV(estimator=model, param_grid=param_grid, n_jobs=1, cv=3)
grid_result = grid.fit(train_images, train_labels)
print("Best: %f using %s" % (grid_result.best_score_, grid_result.best_params_))
```
# Fitting the model
```
best_model = Sequential([
layers.experimental.preprocessing.Rescaling(1./255, input_shape=(img_height, img_width, 3)),
layers.experimental.preprocessing.RandomRotation(0.1),
layers.experimental.preprocessing.RandomContrast(0.1),
layers.experimental.preprocessing.RandomTranslation(0.05, 0.1),
layers.Conv2D(16, 3, padding='same', activation='relu'),
layers.MaxPooling2D(),
layers.Conv2D(32, 3, padding='same', activation='relu'),
layers.MaxPooling2D(),
layers.Conv2D(64, 3, padding='same', activation='relu'),
layers.MaxPooling2D(),
layers.Flatten(),
layers.Dense(128, activation='relu'),
layers.Dropout(0.5),
layers.Dense(num_classes - 1, activation='sigmoid')
])
best_model.compile(optimizer='adam',
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=['accuracy'])
best_model.summary()
history = best_model.fit(train_images, train_labels, epochs=grid_result.best_params_['epochs'], validation_data = (test_images, test_labels), batch_size=grid_result.best_params_['batch_size'], verbose=2)
print('Done training!')
```
# Plotting Training Accuracy/Loss vs Validation Accuracy/Loss
```
acc = history.history['accuracy']
val_acc = history.history['val_accuracy']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs_range = range(1, grid_result.best_params_['epochs'] + 1)
plt.figure(figsize=(15, 5))
plt.subplot(1, 2, 1)
plt.plot(epochs_range, acc, label='Training Accuracy')
plt.plot(epochs_range, val_acc, label='Test Accuracy')
plt.legend(loc='lower right')
plt.xlabel('epoch')
plt.ylabel('accuracy score')
plt.title('Training and Test Accuracy')
plt.subplot(1, 2, 2)
plt.plot(epochs_range, loss, label='Training Loss')
plt.plot(epochs_range, val_loss, label='Test Loss')
plt.legend(loc='upper right')
plt.xlabel('epoch')
plt.ylabel('loss score')
plt.title('Training and Test Loss')
plt.show()
```
# Evaluating Model Accuracy on test dataset
```
predictions = best_model.predict(test_images)
def plot_roc_curve(fpr,tpr):
plt.plot(fpr,tpr)
plt.axis([0,1,0,1])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.show()
fpr , tpr , thresholds = roc_curve(test_labels, predictions)
plot_roc_curve (fpr,tpr)
test_auc = roc_auc_score(test_labels, predictions)
test_loss, test_acc = best_model.evaluate(test_images, test_labels)
print(f'\nTest accuracy: {test_acc}, Test Loss: {test_loss}, AUC Score: {test_auc}')
```
|
github_jupyter
|
import os
from urllib.request import urlretrieve
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import cv2
from zipfile import ZipFile as zip
from PIL import Image
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.models import Sequential
from sklearn.model_selection import GridSearchCV, train_test_split
from sklearn.metrics import roc_curve, roc_auc_score
sess = tf.compat.v1.Session(config=tf.compat.v1.ConfigProto(log_device_placement=True))
def face_mask_dataset():
url = 'https://storage.googleapis.com/kaggle-data-sets/675484/1187790/bundle/archive.zip?X-Goog-Algorithm=GOOG4-RSA-SHA256&X-Goog-Credential=gcp-kaggle-com%40kaggle-161607.iam.gserviceaccount.com%2F20210511%2Fauto%2Fstorage%2Fgoog4_request&X-Goog-Date=20210511T224333Z&X-Goog-Expires=259199&X-Goog-SignedHeaders=host&X-Goog-Signature=038b7fb07abcc84e68e49b04359b793434ac730e3199cbf91d9dc4099cdc1bc0c6626e27f68e8dcf43f7fdd353afbc460f503f24008422252113cb6f6517d6e1afac6580e115d6843388e358b0e44140e606232c211f43eaf29abd46945295d6c8269200d56c7c01855e3dd48b46f5aa5dd921ddd90401ec59f589f8202331adcbdb3de8ce3567114a158e7e431f48a27bf15f47674150bf270012992fe8b9e1105b9c3432d14c07c9964cd5e87d4423840e853bfe2930d4e562e49b0ebc324e64b0f4269af88ffca676f5f3a1124f33c17142f38a3d118faf837fac8c9455b8bfb16d98bb1995fadc40ae65d9146b448535f2200862db4c618f59bcefb37d58' # Link to the dataset
file = 'dataset.zip' # Name of the dataset
path = './' # Download data to current directory.
os.makedirs(path, exist_ok=True) # Create path if it doesn't exist.
# Download the image dataset
if file not in os.listdir(path):
urlretrieve(url, os.path.join(path, file))
print("Downloaded %s to %s" % (file, path))
if 'dataset' not in os.listdir(path):
with zip(file, 'r') as zipobj:
zipobj.extractall('dataset')
def getImages_Labels(path, partition):
listOfLabels = os.listdir(path + partition)
images = []
labels = []
for label in listOfLabels:
val = 0
listOfImages = os.listdir(path + partition + label)
if label=="WithMask":
val = 1
for image in listOfImages:
img_arr = np.asarray(Image.open(path+partition+label+'/'+image))
img_arr = cv2.resize(img_arr, (100, 100))
images.append(img_arr)
labels.append(val)
return np.array(images), np.array(labels)
path = 'dataset/Face Mask Dataset/'
(test_images, test_labels) = getImages_Labels(path, 'Test/')
(train_images, train_labels) = getImages_Labels(path, 'Train/')
(val_images, val_labels) = getImages_Labels(path, 'Validation/')
ret_images = np.vstack((test_images, train_images, val_images))
ret_labels = np.hstack((test_labels, train_labels, val_labels))
return (ret_images, ret_labels)
(images, labels) = face_mask_dataset()
(train_images, test_images, train_labels, test_labels) = train_test_split(images, labels)
print(f'# of training data: {train_images.shape[0]}, # of test data: {test_images.shape[0]}')
class_names = ['no-mask', 'mask']
plt.figure()
plt.imshow(train_images[5306])
plt.colorbar()
plt.grid(False)
plt.show()
plt.figure(figsize=(10,10))
plt.suptitle('25 Examples of the training dataset', y=0.92)
for i in range(25):
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
index = np.random.randint(0, train_images.shape[0])
plt.imshow(train_images[index], cmap=plt.cm.binary)
plt.xlabel(class_names[train_labels[index]])
plt.show()
num_classes = 2
img_height = 100
img_width = 100
def create_model():
model = Sequential([
layers.experimental.preprocessing.Rescaling(1./255, input_shape=(img_height, img_width, 3)),
layers.experimental.preprocessing.RandomRotation(0.1),
layers.experimental.preprocessing.RandomContrast(0.1),
layers.experimental.preprocessing.RandomTranslation(0.05, 0.1),
layers.Conv2D(16, 3, padding='same', activation='relu'),
layers.MaxPooling2D(),
layers.Conv2D(32, 3, padding='same', activation='relu'),
layers.MaxPooling2D(),
layers.Conv2D(64, 3, padding='same', activation='relu'),
layers.MaxPooling2D(),
layers.Flatten(),
layers.Dense(128, activation='relu'),
layers.Dropout(0.5),
layers.Dense(num_classes - 1, activation='sigmoid')
])
model.compile(optimizer='adam',
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=['accuracy'])
return model
model = keras.wrappers.scikit_learn.KerasClassifier(build_fn=create_model)
batch_size = [30, 50, 100]
epochs = [5, 10, 20]
param_grid = dict(batch_size=batch_size, epochs=epochs)
grid = GridSearchCV(estimator=model, param_grid=param_grid, n_jobs=1, cv=3)
grid_result = grid.fit(train_images, train_labels)
print("Best: %f using %s" % (grid_result.best_score_, grid_result.best_params_))
best_model = Sequential([
layers.experimental.preprocessing.Rescaling(1./255, input_shape=(img_height, img_width, 3)),
layers.experimental.preprocessing.RandomRotation(0.1),
layers.experimental.preprocessing.RandomContrast(0.1),
layers.experimental.preprocessing.RandomTranslation(0.05, 0.1),
layers.Conv2D(16, 3, padding='same', activation='relu'),
layers.MaxPooling2D(),
layers.Conv2D(32, 3, padding='same', activation='relu'),
layers.MaxPooling2D(),
layers.Conv2D(64, 3, padding='same', activation='relu'),
layers.MaxPooling2D(),
layers.Flatten(),
layers.Dense(128, activation='relu'),
layers.Dropout(0.5),
layers.Dense(num_classes - 1, activation='sigmoid')
])
best_model.compile(optimizer='adam',
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=['accuracy'])
best_model.summary()
history = best_model.fit(train_images, train_labels, epochs=grid_result.best_params_['epochs'], validation_data = (test_images, test_labels), batch_size=grid_result.best_params_['batch_size'], verbose=2)
print('Done training!')
acc = history.history['accuracy']
val_acc = history.history['val_accuracy']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs_range = range(1, grid_result.best_params_['epochs'] + 1)
plt.figure(figsize=(15, 5))
plt.subplot(1, 2, 1)
plt.plot(epochs_range, acc, label='Training Accuracy')
plt.plot(epochs_range, val_acc, label='Test Accuracy')
plt.legend(loc='lower right')
plt.xlabel('epoch')
plt.ylabel('accuracy score')
plt.title('Training and Test Accuracy')
plt.subplot(1, 2, 2)
plt.plot(epochs_range, loss, label='Training Loss')
plt.plot(epochs_range, val_loss, label='Test Loss')
plt.legend(loc='upper right')
plt.xlabel('epoch')
plt.ylabel('loss score')
plt.title('Training and Test Loss')
plt.show()
predictions = best_model.predict(test_images)
def plot_roc_curve(fpr,tpr):
plt.plot(fpr,tpr)
plt.axis([0,1,0,1])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.show()
fpr , tpr , thresholds = roc_curve(test_labels, predictions)
plot_roc_curve (fpr,tpr)
test_auc = roc_auc_score(test_labels, predictions)
test_loss, test_acc = best_model.evaluate(test_images, test_labels)
print(f'\nTest accuracy: {test_acc}, Test Loss: {test_loss}, AUC Score: {test_auc}')
| 0.570571 | 0.830113 |
# Stability testing with Tangent Plane Distance (TPD) function
The [tangent plane distance](https://www.sciencedirect.com/science/article/pii/0378381282850012) ($tpd$) function allows testing the relative stability of a phase of composition $z$ against a trial phase of composition $w$ at fixed temperature and pressure].
$$ tpd(\underline{w}) = \sum_{i=1}^c w_i (\ln w_i + \ln \hat{\phi}_i(\underline{w})
- \ln z_i - \ln \hat{\phi}_i(\underline{z})) $$
Usually, this function is minimized to check the stability of the given phase based on the following criteria:
- If the minimized $tpd$ is positive, the global phase $z$ is stable.
- If the minimized $tpd$ is zero, the global phase $z$ and trial phase $w$ are in equilibrium.
- If the minimized $tpd$ is negative, the global phase $z$ is unstable
In this notebook, stability analysis for the mixture of water and butanol will be performed. To start, the required functions are imported.
```
import numpy as np
from SGTPy import component, mixture, saftvrmie
from SGTPy.equilibrium import tpd_min, tpd_minimas, lle_init
```
Then, the mixture of water and butanol and its interaction parameters are set up.
```
# creating pure components
water = component('water', ms = 1.7311, sigma = 2.4539 , eps = 110.85,
lambda_r = 8.308, lambda_a = 6., eAB = 1991.07, rcAB = 0.5624,
rdAB = 0.4, sites = [0,2,2], cii = 1.5371939421515458e-20)
butanol = component('butanol2C', ms = 1.9651, sigma = 4.1077 , eps = 277.892,
lambda_r = 10.6689, lambda_a = 6., eAB = 3300.0, rcAB = 0.2615,
rdAB = 0.4, sites = [1,0,1], npol = 1.45, mupol = 1.6609,
cii = 1.5018715324070352e-19)
mix = mixture(water, butanol)
# optimized from experimental LLE
kij, lij = np.array([-0.00736075, -0.00737153])
Kij = np.array([[0, kij], [kij, 0]])
Lij = np.array([[0., lij], [lij, 0]])
# setting interactions corrections
mix.kij_saft(Kij)
mix.lij_saft(Lij)
# creating eos model
eos = saftvrmie(mix)
```
----
### tpd_min
The ``tpd_min`` function searches for a phase composition corresponding to a minimum of $tpd$ function given an initial value. The user needs to specify whether the trial (W) and reference (Z) phases are liquids (``L``) or vapors (``V``).
```
T = 320 # K
P = 1.01e5 # Pa
z = np.array([0.8, 0.2])
#Search for trial phase
w = np.array([0.99, 0.01])
tpd_min(w, z, T, P, eos, stateW = 'L', stateZ = 'L')
#composition of minimum found and tpd value
#(array([0.95593129, 0.04406871]), -0.011057873031562693)
w = np.array([0.99, 0.01])
tpd_min(w, z, T, P, eos, stateW = 'V', stateZ = 'L')
#composition of minimum found and tpd value
#(array([0.82414873, 0.17585127]), 0.8662934867235452)
```
---
### tpd_minimas
The ``tpd_minimas`` function will attempt (but does not guarantee) to search for ``nmin`` minima of the $tpd$ function. As for the ``tpd_min`` function, you need to specify the aggregation state of the global (``z``) and the trial phase (``w``).
```
T = 320 # K
P = 1.01e5 # Pa
z = np.array([0.8, 0.2])
nmin = 2
tpd_minimas(nmin, z, T, P, eos, stateW='L', stateZ='L')
tpd_minimas(nmin, z, T, P, eos, stateW='V', stateZ='L')
```
---
### lle_init
Finally, the ``lle_init`` function can be used to find initial guesses for liquid-liquid equilibrium calculation.
This function call ``tpd_minimas`` with ``nmin=2`` and liquid state for trial and global phase.
```
T = 320 # K
P = 1.01e5 # Pa
z = np.array([0.8, 0.2])
lle_init(z, T, P, eos)
```
---
For further information about each function check out the documentation running: ``function?``
|
github_jupyter
|
import numpy as np
from SGTPy import component, mixture, saftvrmie
from SGTPy.equilibrium import tpd_min, tpd_minimas, lle_init
# creating pure components
water = component('water', ms = 1.7311, sigma = 2.4539 , eps = 110.85,
lambda_r = 8.308, lambda_a = 6., eAB = 1991.07, rcAB = 0.5624,
rdAB = 0.4, sites = [0,2,2], cii = 1.5371939421515458e-20)
butanol = component('butanol2C', ms = 1.9651, sigma = 4.1077 , eps = 277.892,
lambda_r = 10.6689, lambda_a = 6., eAB = 3300.0, rcAB = 0.2615,
rdAB = 0.4, sites = [1,0,1], npol = 1.45, mupol = 1.6609,
cii = 1.5018715324070352e-19)
mix = mixture(water, butanol)
# optimized from experimental LLE
kij, lij = np.array([-0.00736075, -0.00737153])
Kij = np.array([[0, kij], [kij, 0]])
Lij = np.array([[0., lij], [lij, 0]])
# setting interactions corrections
mix.kij_saft(Kij)
mix.lij_saft(Lij)
# creating eos model
eos = saftvrmie(mix)
T = 320 # K
P = 1.01e5 # Pa
z = np.array([0.8, 0.2])
#Search for trial phase
w = np.array([0.99, 0.01])
tpd_min(w, z, T, P, eos, stateW = 'L', stateZ = 'L')
#composition of minimum found and tpd value
#(array([0.95593129, 0.04406871]), -0.011057873031562693)
w = np.array([0.99, 0.01])
tpd_min(w, z, T, P, eos, stateW = 'V', stateZ = 'L')
#composition of minimum found and tpd value
#(array([0.82414873, 0.17585127]), 0.8662934867235452)
T = 320 # K
P = 1.01e5 # Pa
z = np.array([0.8, 0.2])
nmin = 2
tpd_minimas(nmin, z, T, P, eos, stateW='L', stateZ='L')
tpd_minimas(nmin, z, T, P, eos, stateW='V', stateZ='L')
T = 320 # K
P = 1.01e5 # Pa
z = np.array([0.8, 0.2])
lle_init(z, T, P, eos)
| 0.414188 | 0.979881 |
```
import matplotlib.pyplot as plt
import numpy as np
# JAX
import jax
from jax import nn
import jax.numpy as jnp
from tensorflow import keras
# # "Fixar" números aleatórios a serem gerados
np.random.seed(0)
# the data, split between train and test sets
(x_train, y_train), (x_test, y_test) = keras.datasets.mnist.load_data()
# Scale images to the [0, 1] range
x_train = x_train.reshape(-1, 32, 28*28).astype("float32") / 255
x_test = x_test.reshape(-1, 16, 28*28).astype("float32") / 255
# convert class vectors to binary class matrices
num_classes = 10
y_train = keras.utils.to_categorical(y_train, num_classes).reshape(-1, 32, 10)
y_test = keras.utils.to_categorical(y_test, num_classes).reshape(-1, 16, 10)
def define_params(sizes=[1, 1]):
weights = []
for (in_dim, out_dim) in zip(sizes[:-1], sizes[1:]):
weights.append({"w": np.random.randn(in_dim, out_dim) * np.sqrt(2/in_dim),
"b": np.zeros(out_dim)})
return weights
def apply_fn(weights, batch_x, activations):
output = batch_x
for layer, act_fn in zip(weights, activations):
output = jnp.dot(output, layer["w"]) + layer["b"]
output = act_fn(output)
return output
def cross_entropy(weights, batch_x, real_y, activations):
pred_y = apply_fn(weights, batch_x, activations)
real_y = jnp.asarray(real_y)
return -jnp.mean(jnp.sum(pred_y * real_y, axis=1))
def _accuracy(pred_y, real_y):
p = np.argmax(pred_y, axis=1)
real_y = np.argmax(real_y, axis=1)
return np.sum(p == real_y) / len(pred_y)
@jax.jit
def evaluate(weights, batch_x, batch_y):
# run feed forward network
pred_y = apply_fn(weights, batch_x, activations)
# loss
loss = cross_entropy(weights, batch_x, batch_y, activations)
return loss, _accuracy(pred_y, batch_y)
@jax.jit
def train_step(weights, batch_x, batch_y, lr=0.03):
loss, grads = jax.value_and_grad(cross_entropy)(weights, batch_x, batch_y, activations)
weights = jax.tree_util.tree_multimap(lambda v, g: v - lr*g, weights, grads)
return weights, loss
nn = define_params(sizes=[28*28, 1024, num_classes])
activations=[jax.nn.relu, jax.nn.softmax]
train_losses = []
eval_losses = []
metrics = []
for i in range(10):
avg_loss = 0
for (batch_x, batch_y) in zip(x_train, y_train):
nn, loss = train_step(nn, batch_x, batch_y)
avg_loss += loss
train_losses.append(avg_loss/len(x_train))
avg_acc = 0
avg_loss = 0
for (batch_x, batch_y) in zip(x_test, y_test):
loss, acc = evaluate(nn, batch_x, batch_y)
avg_acc += acc
avg_loss += loss
eval_losses.append(loss/len(x_test))
metrics.append(avg_acc/len(x_test))
plt.plot(range(len(metrics)), metrics)
plt.plot(range(len(train_losses)), train_losses)
plt.plot(range(len(eval_losses)), eval_losses)
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import numpy as np
# JAX
import jax
from jax import nn
import jax.numpy as jnp
from tensorflow import keras
# # "Fixar" números aleatórios a serem gerados
np.random.seed(0)
# the data, split between train and test sets
(x_train, y_train), (x_test, y_test) = keras.datasets.mnist.load_data()
# Scale images to the [0, 1] range
x_train = x_train.reshape(-1, 32, 28*28).astype("float32") / 255
x_test = x_test.reshape(-1, 16, 28*28).astype("float32") / 255
# convert class vectors to binary class matrices
num_classes = 10
y_train = keras.utils.to_categorical(y_train, num_classes).reshape(-1, 32, 10)
y_test = keras.utils.to_categorical(y_test, num_classes).reshape(-1, 16, 10)
def define_params(sizes=[1, 1]):
weights = []
for (in_dim, out_dim) in zip(sizes[:-1], sizes[1:]):
weights.append({"w": np.random.randn(in_dim, out_dim) * np.sqrt(2/in_dim),
"b": np.zeros(out_dim)})
return weights
def apply_fn(weights, batch_x, activations):
output = batch_x
for layer, act_fn in zip(weights, activations):
output = jnp.dot(output, layer["w"]) + layer["b"]
output = act_fn(output)
return output
def cross_entropy(weights, batch_x, real_y, activations):
pred_y = apply_fn(weights, batch_x, activations)
real_y = jnp.asarray(real_y)
return -jnp.mean(jnp.sum(pred_y * real_y, axis=1))
def _accuracy(pred_y, real_y):
p = np.argmax(pred_y, axis=1)
real_y = np.argmax(real_y, axis=1)
return np.sum(p == real_y) / len(pred_y)
@jax.jit
def evaluate(weights, batch_x, batch_y):
# run feed forward network
pred_y = apply_fn(weights, batch_x, activations)
# loss
loss = cross_entropy(weights, batch_x, batch_y, activations)
return loss, _accuracy(pred_y, batch_y)
@jax.jit
def train_step(weights, batch_x, batch_y, lr=0.03):
loss, grads = jax.value_and_grad(cross_entropy)(weights, batch_x, batch_y, activations)
weights = jax.tree_util.tree_multimap(lambda v, g: v - lr*g, weights, grads)
return weights, loss
nn = define_params(sizes=[28*28, 1024, num_classes])
activations=[jax.nn.relu, jax.nn.softmax]
train_losses = []
eval_losses = []
metrics = []
for i in range(10):
avg_loss = 0
for (batch_x, batch_y) in zip(x_train, y_train):
nn, loss = train_step(nn, batch_x, batch_y)
avg_loss += loss
train_losses.append(avg_loss/len(x_train))
avg_acc = 0
avg_loss = 0
for (batch_x, batch_y) in zip(x_test, y_test):
loss, acc = evaluate(nn, batch_x, batch_y)
avg_acc += acc
avg_loss += loss
eval_losses.append(loss/len(x_test))
metrics.append(avg_acc/len(x_test))
plt.plot(range(len(metrics)), metrics)
plt.plot(range(len(train_losses)), train_losses)
plt.plot(range(len(eval_losses)), eval_losses)
| 0.755096 | 0.778565 |
```
import time
import numpy as np
from tqdm import tqdm
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
import matplotlib.pyplot as plt
from matplotlib import style
from more_itertools import chunked
from google.cloud import storage
# Configuration constants
VALIDATION_RATIO = 0.1
client = storage.Client()
bucket_name = "tdt4173-datasets"
bucket = client.get_bucket(bucket_name)
blobs = bucket.list_blobs()
for blob in blobs:
print(blob.name)
blob_name = "cats-vs-dogs/tensors/catsdogs_processed_64px_24946_horizontal.torch"
blob = bucket.get_blob(blob_name)
data_file = "/home/jupyter/data/cats-vs-dogs/tensors/catsdogs_processed_64px_24946_horizontal.torch"
blob.download_to_filename(data_file)
# data_file = "/home/jupyter/data/celeb-align-1/tensors/celebalign_processed_100_000_horizontal.torch"
data = torch.load(data_file)
plt.imshow(data["x"][24], cmap="gray");
print(data["x"][0].shape)
IMAGE_SIZE = data["x"][0].shape[0]
NUM_CLASSES = data["num_classes"]
if torch.cuda.is_available():
device = torch.device("cuda:0")
print("Running on the GPU")
else:
device = torch.device("cpu")
print("Running on the CPU")
unique = set(data["y"])
class_mapping = {elem: idx for idx, elem in enumerate(unique)}
val_size = int(len(data["x"]) * VALIDATION_RATIO)
print(val_size)
train_x = data["x"][:-val_size]
train_y = data["y"][:-val_size]
def fwd_pass(x, y, loss_func, optim, train=False):
if train:
net.zero_grad()
out = net(x)
acc = np.mean([int(torch.argmax(y_pred) == y_real) for y_pred, y_real in zip(out, y)])
loss = loss_func(out, y)
if train:
loss.backward()
optim.step()
return acc, loss
test_x = data["x"][-val_size:]
test_y = data["y"][-val_size:]
def test(size, loss_func, optim):
tx, ty = test_x[:size].to(device), test_y[:size].to(device)
val_acc, val_loss = fwd_pass(tx.view(-1, 1, IMAGE_SIZE, IMAGE_SIZE).to(device), ty.to(device), loss_func, optim)
return val_acc, val_loss
import os
import sys
module_path = os.path.abspath(os.path.join('..'))
if module_path not in sys.path:
sys.path.append(module_path)
import models.lla7
import importlib
importlib.reload(models.lla7)
NetClass = models.lla7.FleetwoodNet7V1
# Only run if need to delete memory
collect = False
if collect:
import gc
del net
gc.collect()
torch.cuda.empty_cache()
net = NetClass(NUM_CLASSES).to(device)
print(net)
MODEL_NAME = f"{type(net).__name__}-{int(time.time())}"
print(f"Model name: {MODEL_NAME}")
saves_path = "/home/jupyter/checkpoints"
CHECKPOINT_EVERY_STEP = 10_000
optimizer = optim.Adam(net.parameters(), lr=0.07)
loss_function = nn.CrossEntropyLoss().to(device)
def train(net):
BATCH_SIZE = 200
EPOCHS = 10
for epoch in range(EPOCHS):
with open(os.path.join("/home/jupyter/logs", f"model-{MODEL_NAME}.log"), "a") as f:
it = tqdm(range(0, len(train_x), BATCH_SIZE))
for i in it:
batch_x = train_x[i:i+BATCH_SIZE].view(-1, 1, IMAGE_SIZE, IMAGE_SIZE).to(device)
batch_y = train_y[i:i+BATCH_SIZE].to(device)
acc, loss = fwd_pass(
batch_x, batch_y,
loss_function,
optimizer,
train=True,
)
it.set_postfix({"acc": acc, "loss": loss.item()})
if i != 0 and i % CHECKPOINT_EVERY_STEP == 0:
val_acc, val_loss = test(size=100, loss_func=loss_function, optim=optimizer)
f.write(f"{MODEL_NAME},{round(time.time(),3)},{round(float(acc),2)},{round(float(loss), 4)},{round(float(val_acc),2)},{round(float(val_loss),4)}\n")
torch.save({
"model_state_dict": net.state_dict(),
"optimizer_state_dict": optimizer.state_dict(),
"val_acc": val_acc,
"val_loss": val_loss,
},
os.path.join(saves_path, f"{MODEL_NAME}-epoch-{epoch}.data"),
)
print(f"Epoch: {epoch}. Loss: {loss}.")
train(net)
style.use("ggplot")
def create_acc_loss_graph(model_name):
times = []
accs = []
losses = []
val_accs = []
val_losses = []
with open("/home/jupyter/logs/model-FleetwoodNet7V1-1604952757.log", "r") as f:
for line in f.readlines():
name, time, acc, loss, val_acc, val_loss = line.split(",")
times.append(float(time))
accs.append(float(acc))
losses.append(float(loss))
val_accs.append(float(val_acc))
val_losses.append(float(val_loss))
fig = plt.figure()
ax1 = plt.subplot2grid((2, 1), (0, 0))
ax2 = plt.subplot2grid((2, 1), (1, 0), sharex=ax1)
ax1.plot(times, accs, label="acc")
ax1.plot(times, val_accs, label="val_acc")
ax1.legend(loc=2)
ax2.plot(times, losses, label="loss")
ax2.plot(times, val_losses, label="val_loss")
ax2.legend(loc=2)
plt.show()
create_acc_loss_graph(MODEL_NAME)
# Not currently in use, I think
correct = 0
with torch.no_grad():
for i, y_real in enumerate(tqdm(test_y)):
real_class = torch.argmax(y_real)
pred_class = torch.argmax(net(test_x[i].view(-1, 1, IMAGE_SIZE, IMAGE_SIZE))[0])
correct += int(real_class == pred_class)
print(f"Accuracy: {round(correct / len(test_x), 3)}")
for im in test_x:
pred = int(torch.argmax(net(im.view(-1, 1, IMAGE_SIZE, IMAGE_SIZE))))
convert = {0: "Cat", 1: "Dog"}
print(f"Net predicted it is `{convert[pred]}`")
plt.imshow(im.cpu(),cmap="gray")
plt.pause(0.05)
print("="*50)
```
|
github_jupyter
|
import time
import numpy as np
from tqdm import tqdm
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
import matplotlib.pyplot as plt
from matplotlib import style
from more_itertools import chunked
from google.cloud import storage
# Configuration constants
VALIDATION_RATIO = 0.1
client = storage.Client()
bucket_name = "tdt4173-datasets"
bucket = client.get_bucket(bucket_name)
blobs = bucket.list_blobs()
for blob in blobs:
print(blob.name)
blob_name = "cats-vs-dogs/tensors/catsdogs_processed_64px_24946_horizontal.torch"
blob = bucket.get_blob(blob_name)
data_file = "/home/jupyter/data/cats-vs-dogs/tensors/catsdogs_processed_64px_24946_horizontal.torch"
blob.download_to_filename(data_file)
# data_file = "/home/jupyter/data/celeb-align-1/tensors/celebalign_processed_100_000_horizontal.torch"
data = torch.load(data_file)
plt.imshow(data["x"][24], cmap="gray");
print(data["x"][0].shape)
IMAGE_SIZE = data["x"][0].shape[0]
NUM_CLASSES = data["num_classes"]
if torch.cuda.is_available():
device = torch.device("cuda:0")
print("Running on the GPU")
else:
device = torch.device("cpu")
print("Running on the CPU")
unique = set(data["y"])
class_mapping = {elem: idx for idx, elem in enumerate(unique)}
val_size = int(len(data["x"]) * VALIDATION_RATIO)
print(val_size)
train_x = data["x"][:-val_size]
train_y = data["y"][:-val_size]
def fwd_pass(x, y, loss_func, optim, train=False):
if train:
net.zero_grad()
out = net(x)
acc = np.mean([int(torch.argmax(y_pred) == y_real) for y_pred, y_real in zip(out, y)])
loss = loss_func(out, y)
if train:
loss.backward()
optim.step()
return acc, loss
test_x = data["x"][-val_size:]
test_y = data["y"][-val_size:]
def test(size, loss_func, optim):
tx, ty = test_x[:size].to(device), test_y[:size].to(device)
val_acc, val_loss = fwd_pass(tx.view(-1, 1, IMAGE_SIZE, IMAGE_SIZE).to(device), ty.to(device), loss_func, optim)
return val_acc, val_loss
import os
import sys
module_path = os.path.abspath(os.path.join('..'))
if module_path not in sys.path:
sys.path.append(module_path)
import models.lla7
import importlib
importlib.reload(models.lla7)
NetClass = models.lla7.FleetwoodNet7V1
# Only run if need to delete memory
collect = False
if collect:
import gc
del net
gc.collect()
torch.cuda.empty_cache()
net = NetClass(NUM_CLASSES).to(device)
print(net)
MODEL_NAME = f"{type(net).__name__}-{int(time.time())}"
print(f"Model name: {MODEL_NAME}")
saves_path = "/home/jupyter/checkpoints"
CHECKPOINT_EVERY_STEP = 10_000
optimizer = optim.Adam(net.parameters(), lr=0.07)
loss_function = nn.CrossEntropyLoss().to(device)
def train(net):
BATCH_SIZE = 200
EPOCHS = 10
for epoch in range(EPOCHS):
with open(os.path.join("/home/jupyter/logs", f"model-{MODEL_NAME}.log"), "a") as f:
it = tqdm(range(0, len(train_x), BATCH_SIZE))
for i in it:
batch_x = train_x[i:i+BATCH_SIZE].view(-1, 1, IMAGE_SIZE, IMAGE_SIZE).to(device)
batch_y = train_y[i:i+BATCH_SIZE].to(device)
acc, loss = fwd_pass(
batch_x, batch_y,
loss_function,
optimizer,
train=True,
)
it.set_postfix({"acc": acc, "loss": loss.item()})
if i != 0 and i % CHECKPOINT_EVERY_STEP == 0:
val_acc, val_loss = test(size=100, loss_func=loss_function, optim=optimizer)
f.write(f"{MODEL_NAME},{round(time.time(),3)},{round(float(acc),2)},{round(float(loss), 4)},{round(float(val_acc),2)},{round(float(val_loss),4)}\n")
torch.save({
"model_state_dict": net.state_dict(),
"optimizer_state_dict": optimizer.state_dict(),
"val_acc": val_acc,
"val_loss": val_loss,
},
os.path.join(saves_path, f"{MODEL_NAME}-epoch-{epoch}.data"),
)
print(f"Epoch: {epoch}. Loss: {loss}.")
train(net)
style.use("ggplot")
def create_acc_loss_graph(model_name):
times = []
accs = []
losses = []
val_accs = []
val_losses = []
with open("/home/jupyter/logs/model-FleetwoodNet7V1-1604952757.log", "r") as f:
for line in f.readlines():
name, time, acc, loss, val_acc, val_loss = line.split(",")
times.append(float(time))
accs.append(float(acc))
losses.append(float(loss))
val_accs.append(float(val_acc))
val_losses.append(float(val_loss))
fig = plt.figure()
ax1 = plt.subplot2grid((2, 1), (0, 0))
ax2 = plt.subplot2grid((2, 1), (1, 0), sharex=ax1)
ax1.plot(times, accs, label="acc")
ax1.plot(times, val_accs, label="val_acc")
ax1.legend(loc=2)
ax2.plot(times, losses, label="loss")
ax2.plot(times, val_losses, label="val_loss")
ax2.legend(loc=2)
plt.show()
create_acc_loss_graph(MODEL_NAME)
# Not currently in use, I think
correct = 0
with torch.no_grad():
for i, y_real in enumerate(tqdm(test_y)):
real_class = torch.argmax(y_real)
pred_class = torch.argmax(net(test_x[i].view(-1, 1, IMAGE_SIZE, IMAGE_SIZE))[0])
correct += int(real_class == pred_class)
print(f"Accuracy: {round(correct / len(test_x), 3)}")
for im in test_x:
pred = int(torch.argmax(net(im.view(-1, 1, IMAGE_SIZE, IMAGE_SIZE))))
convert = {0: "Cat", 1: "Dog"}
print(f"Net predicted it is `{convert[pred]}`")
plt.imshow(im.cpu(),cmap="gray")
plt.pause(0.05)
print("="*50)
| 0.439507 | 0.388096 |
# Support Vector Machines with Python
## Import Libraries
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
```
## Get the Data
We'll use the built in breast cancer dataset from Scikit Learn. We can get with the load function:
```
from sklearn.datasets import load_breast_cancer
cancer = load_breast_cancer()
```
The data set is presented in a dictionary form:
```
cancer.keys()
```
We can grab information and arrays out of this dictionary to set up our data frame and understanding of the features:
```
print(cancer['DESCR'])
cancer['feature_names']
```
## Set up DataFrame
```
df_feat = pd.DataFrame(cancer['data'],columns=cancer['feature_names'])
df_feat.info()
cancer['target']
df_target = pd.DataFrame(cancer['target'],columns=['Cancer'])
```
Now let's actually check out the dataframe!
```
df_feat.head()
```
# Exploratory Data Analysis
We'll skip the Data Viz part for this lecture since there are so many features that are hard to interpret if you don't have domain knowledge of cancer or tumor cells. In your project you will have more to visualize for the data.
## Train Test Split
```
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(df_feat, np.ravel(df_target), test_size=0.30, random_state=101)
```
# Train the Support Vector Classifier
```
from sklearn.svm import SVC
model = SVC()
model.fit(X_train,y_train)
```
## Predictions and Evaluations
Now let's predict using the trained model.
```
predictions = model.predict(X_test)
from sklearn.metrics import classification_report,confusion_matrix
print(confusion_matrix(y_test,predictions))
print(classification_report(y_test,predictions))
```
Woah! Notice that we are classifying everything into a single class! This means our model needs to have it parameters adjusted (it may also help to normalize the data).
We can search for parameters using a GridSearch!
# Gridsearch
Finding the right parameters (like what C or gamma values to use) is a tricky task! But luckily, we can be a little lazy and just try a bunch of combinations and see what works best! This idea of creating a 'grid' of parameters and just trying out all the possible combinations is called a Gridsearch, this method is common enough that Scikit-learn has this functionality built in with GridSearchCV! The CV stands for cross-validation which is the
GridSearchCV takes a dictionary that describes the parameters that should be tried and a model to train. The grid of parameters is defined as a dictionary, where the keys are the parameters and the values are the settings to be tested.
```
param_grid = {'C': [0.1,1, 10, 100, 1000], 'gamma': [1,0.1,0.01,0.001,0.0001], 'kernel': ['rbf']}
from sklearn.model_selection import GridSearchCV
```
One of the great things about GridSearchCV is that it is a meta-estimator. It takes an estimator like SVC, and creates a new estimator, that behaves exactly the same - in this case, like a classifier. You should add refit=True and choose verbose to whatever number you want, higher the number, the more verbose (verbose just means the text output describing the process).
```
grid = GridSearchCV(SVC(),param_grid,refit=True,verbose=3)
```
What fit does is a bit more involved then usual. First, it runs the same loop with cross-validation, to find the best parameter combination. Once it has the best combination, it runs fit again on all data passed to fit (without cross-validation), to built a single new model using the best parameter setting.
```
# May take awhile!
grid.fit(X_train,y_train)
```
You can inspect the best parameters found by GridSearchCV in the best_params_ attribute, and the best estimator in the best\_estimator_ attribute:
```
grid.best_params_
grid.best_estimator_
```
Then you can re-run predictions on this grid object just like you would with a normal model.
```
grid_predictions = grid.predict(X_test)
print(confusion_matrix(y_test,grid_predictions))
print(classification_report(y_test,grid_predictions))
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
from sklearn.datasets import load_breast_cancer
cancer = load_breast_cancer()
cancer.keys()
print(cancer['DESCR'])
cancer['feature_names']
df_feat = pd.DataFrame(cancer['data'],columns=cancer['feature_names'])
df_feat.info()
cancer['target']
df_target = pd.DataFrame(cancer['target'],columns=['Cancer'])
df_feat.head()
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(df_feat, np.ravel(df_target), test_size=0.30, random_state=101)
from sklearn.svm import SVC
model = SVC()
model.fit(X_train,y_train)
predictions = model.predict(X_test)
from sklearn.metrics import classification_report,confusion_matrix
print(confusion_matrix(y_test,predictions))
print(classification_report(y_test,predictions))
param_grid = {'C': [0.1,1, 10, 100, 1000], 'gamma': [1,0.1,0.01,0.001,0.0001], 'kernel': ['rbf']}
from sklearn.model_selection import GridSearchCV
grid = GridSearchCV(SVC(),param_grid,refit=True,verbose=3)
# May take awhile!
grid.fit(X_train,y_train)
grid.best_params_
grid.best_estimator_
grid_predictions = grid.predict(X_test)
print(confusion_matrix(y_test,grid_predictions))
print(classification_report(y_test,grid_predictions))
| 0.443841 | 0.987508 |
# <img style="float: left; padding-right: 10px; width: 45px" src="https://raw.githubusercontent.com/Harvard-IACS/2018-CS109A/master/content/styles/iacs.png"> CS109A Introduction to Data Science
## Standard Section 8: Review Trees and Boosting including Ada Boosting Gradient Boosting and XGBoost.
**Harvard University**<br/>
**Fall 2019**<br/>
**Instructors**: Pavlos Protopapas, Kevin Rader, and Chris Tanner<br/>
**Section Leaders**: Marios Mattheakis, Abhimanyu (Abhi) Vasishth, Robbert (Rob) Struyven<br/>
```
#RUN THIS CELL
import requests
from IPython.core.display import HTML
styles = requests.get("https://raw.githubusercontent.com/Harvard-IACS/2018-CS109A/master/content/styles/cs109.css").text
HTML(styles)
```
This section will work with a spam email dataset again. Our ultimate goal is to be able to build models so that we can predict whether an email is spam or not spam based on word characteristics within each email. We will review Decision Trees, Bagging, and Random Forest methods, and introduce Boosting: Ada Boost and XGBoost.
Specifically, we will:
1. *Quick review of last week*
2. Rebuild the Decision Tree model, Bagging model, Random Forest Model just for comparison with Boosting.
3. *Theory:* What is Boosting?
4. Use the Adaboost on the Spam Dataset.
5. *Theory:* What is Gradient Boosting and XGBoost?
6. Use XGBoost on the Spam Dataset: Extreme Gradient Boosting
Optional: Example to better understand Bias vs Variance tradeoff.
---------
## 1. *Quick review of last week*
#### The Idea: Decision Trees are just flowcharts and interpretable!
It turns out that simple flow charts can be formulated as mathematical models for classification and these models have the properties we desire;
- interpretable by humans
- have sufficiently complex decision boundaries
- the decision boundaries are locally linear, each component of the decision boundary is simple to describe mathematically.
----------
#### How to build Decision Trees (the Learning Algorithm in words):
To learn a decision tree model, we take a greedy approach:
1. Start with an empty decision tree (undivided feature space)
2. Choose the ‘optimal’ predictor on which to split and choose the ‘optimal’ threshold value for splitting by applying a **splitting criterion (1)**
3. Recurse on on each new node until **stopping condition (2)** is met
#### So we need a (1) splitting criterion and a (2) stopping condition:
#### (1) Splitting criterion
<img src="data/split2_adj.png" alt="split2" width="70%"/>
#### (2) Stopping condition
**Not stopping while building a deeper and deeper tree = 100% training accuracy; Yet we will overfit!
To prevent the **overfitting** from happening, we should have stopping condition.
-------------
#### How do we go from Classification to Regression?
- For classification, we return the majority class in the points of each leaf node.
- For regression we return the average of the outputs for the points in each leaf node.
-------------
#### What is bagging?
One way to adjust for the high variance of the output of an experiment is to perform the experiment multiple times and then average the results.
1. **Bootstrap:** we generate multiple samples of training data, via bootstrapping. We train a full decision tree on each sample of data.
2. **AGgregatiING** for a given input, we output the averaged outputs of all the models for that input.
This method is called **Bagging: B** ootstrap + **AGG**regat**ING**.
-------------
#### What is Random Forest?
- **Many trees** make a **forest**.
- **Many random trees** make a **random forest**.
Random Forest is a modified form of bagging that creates ensembles of independent decision trees.
To *de-correlate the trees*, we:
1. train each tree on a separate bootstrap **random sample** of the full training set (same as in bagging)
2. for each tree, at each split, we **randomly select a set of 𝐽′ predictors from the full set of predictors.** (not done in bagging)
3. From amongst the 𝐽′ predictors, we select the optimal predictor and the optimal corresponding threshold for the split.
-------------
#### Interesting Piazza post: why randomness in simple decision tree?
```"Hi there. I notice that there is a parameter called "random_state" in decision tree function and I wonder why we need randomness in simple decision tree. If we add randomness in such case, isn't it the same as random forest?"```
- The problem of learning an optimal decision tree is known to be **NP-complete** under several aspects of optimality and even for simple concepts.
- Consequently, practical decision-tree learning algorithms are based on **heuristic algorithms such as the greedy algorithm where locally optimal decisions are made at each node**.
- Such algorithms **cannot guarantee to return the globally optimal decision tree**.
- This can be mitigated by training multiple trees in an ensemble learner, where the features and samples are randomly sampled with replacement (Bagging).
For example: **What is the defaulth DecisionTreeClassifier behaviour when there are 2 or more best features for a certain split (a tie among "splitters")?** (after a deep dive and internet search [link](https://github.com/scikit-learn/scikit-learn/issues/12259 ) ):
- The current default behaviour when splitter="best" is to shuffle the features at each step and take the best feature to split.
- In case there is a tie, we take a random one.
-------------
## 2. Just re-building the tree models of last week
### Rebuild the Decision Tree model, Bagging model and Random Forest Model for comparison with Boosting methods
We will be working with a spam email dataset. The dataset has 57 predictors with a response variable called `Spam` that indicates whether an email is spam or not spam. **The goal is to be able to create a classifier or method that acts as a spam filter.**
Link to description : https://archive.ics.uci.edu/ml/datasets/spambase
```
import numpy as np
import pandas as pd
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
from tqdm import tqdm
import sklearn.metrics as metrics
import time
from sklearn.model_selection import cross_val_score
from sklearn.metrics import accuracy_score
from sklearn import tree
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import AdaBoostClassifier
from sklearn.linear_model import LogisticRegressionCV
from sklearn.model_selection import KFold
from sklearn.metrics import confusion_matrix
%matplotlib inline
pd.set_option('display.width', 1500)
pd.set_option('display.max_columns', 100)
from sklearn.model_selection import learning_curve
#Import Dataframe and Set Column Names
spam_df = pd.read_csv('data/spam.csv', header=None)
columns = ["Column_"+str(i+1) for i in range(spam_df.shape[1]-1)] + ['Spam']
spam_df.columns = columns
display(spam_df.head())
#Let us split the dataset into a 70-30 split by using the following:
#Split data into train and test
np.random.seed(42)
msk = np.random.rand(len(spam_df)) < 0.7
data_train = spam_df[msk]
data_test = spam_df[~msk]
#Split predictor and response columns
x_train, y_train = data_train.drop(['Spam'], axis=1), data_train['Spam']
x_test , y_test = data_test.drop(['Spam'] , axis=1), data_test['Spam']
print("Shape of Training Set :",data_train.shape)
print("Shape of Testing Set :" ,data_test.shape)
#Check Percentage of Spam in Train and Test Set
percentage_spam_training = 100*y_train.sum()/len(y_train)
percentage_spam_testing = 100*y_test.sum()/len(y_test)
print("Percentage of Spam in Training Set \t : {:0.2f}%.".format(percentage_spam_training))
print("Percentage of Spam in Testing Set \t : {:0.2f}%.".format(percentage_spam_testing))
```
-----------
### Fitting an Optimal Single Decision Tree
```
# Best depth for single decision trees of last week
best_depth = 7
print("The best depth was found to be:", best_depth)
#Evalaute the performance at the best depth
model_tree = DecisionTreeClassifier(max_depth=best_depth)
model_tree.fit(x_train, y_train)
#Check Accuracy of Spam Detection in Train and Test Set
acc_trees_training = accuracy_score(y_train, model_tree.predict(x_train))
acc_trees_testing = accuracy_score(y_test, model_tree.predict(x_test))
print("Simple Decision Trees: Accuracy, Training Set \t : {:.2%}".format(acc_trees_training))
print("Simple Decision Trees: Accuracy, Testing Set \t : {:.2%}".format(acc_trees_testing))
```
--------
### Fitting 100 Single Decision Trees while Bagging
```
n_trees = 100 # we tried a variety of numbers here
#Creating model
np.random.seed(0)
model = DecisionTreeClassifier(max_depth=best_depth+5)
#Initializing variables
predictions_train = np.zeros((data_train.shape[0], n_trees))
predictions_test = np.zeros((data_test.shape[0], n_trees))
#Conduct bootstraping iterations
for i in range(n_trees):
temp = data_train.sample(frac=1, replace=True)
response_variable = temp['Spam']
temp = temp.drop(['Spam'], axis=1)
model.fit(temp, response_variable)
predictions_train[:,i] = model.predict(x_train)
predictions_test[:,i] = model.predict(x_test)
#Make Predictions Dataframe
columns = ["Bootstrap-Model_"+str(i+1) for i in range(n_trees)]
predictions_train = pd.DataFrame(predictions_train, columns=columns)
predictions_test = pd.DataFrame(predictions_test, columns=columns)
#Function to ensemble the prediction of each bagged decision tree model
def get_prediction(df, count=-1):
count = df.shape[1] if count==-1 else count
temp = df.iloc[:,0:count]
return np.mean(temp, axis=1)>0.5
#Check Accuracy of Spam Detection in Train and Test Set
acc_bagging_training = 100*accuracy_score(y_train, get_prediction(predictions_train, count=-1))
acc_bagging_testing = 100*accuracy_score(y_test, get_prediction(predictions_test, count=-1))
print("Bagging: \tAccuracy, Training Set \t: {:0.2f}%".format(acc_bagging_training))
print("Bagging: \tAccuracy, Testing Set \t: {:0.2f}%".format( acc_bagging_testing))
```
### Fitting Random Forest
```
#Fit a Random Forest Model
#Training
model = RandomForestClassifier(n_estimators=n_trees, max_depth=best_depth+5)
model.fit(x_train, y_train)
#Predict
y_pred_train = model.predict(x_train)
y_pred_test = model.predict(x_test)
#Performance Evaluation
acc_random_forest_training = accuracy_score(y_train, y_pred_train)*100
acc_random_forest_testing = accuracy_score(y_test, y_pred_test)*100
print("Random Forest: Accuracy, Training Set : {:0.2f}%".format(acc_random_forest_training))
print("Random Forest: Accuracy, Testing Set : {:0.2f}%".format(acc_random_forest_testing))
```
#### Let's compare the performance of our 3 models:
```
print("Decision Trees:\tAccuracy, Training Set \t: {:.2%}".format(acc_trees_training))
print("Decision Trees:\tAccuracy, Testing Set \t: {:.2%}".format(acc_trees_testing))
print("\nBagging: \tAccuracy, Training Set \t: {:0.2f}%".format(acc_bagging_training))
print("Bagging: \tAccuracy, Testing Set \t: {:0.2f}%".format( acc_bagging_testing))
print("\nRandom Forest: \tAccuracy, Training Set \t: {:0.2f}%".format(acc_random_forest_training))
print("Random Forest: \tAccuracy, Testing Set \t: {:0.2f}%".format(acc_random_forest_testing))
```
## 3. *Theory:* What is Boosting?
- **Bagging and Random Forest:**
- complex and deep trees **overfit**
- thus **let's perform variance reduction on complex trees!**
- **Boosting:**
- simple and shallow trees **underfit**
- thus **let's perform bias reduction of simple trees!**
- make the simple trees more expressive!
**Boosting** attempts to improve the predictive flexibility of simple models.
- It trains a **large number of “weak” learners in sequence**.
- A weak learner is a constrained model (limit the max depth of each decision tree).
- Each one in the sequence focuses on **learning from the mistakes** of the one before it.
- By more heavily weighting in the mistakes in the next tree, our next tree will learn from the mistakes.
- A combining all the weak learners into a single strong learner = **a boosted tree**.
<img src="data/gradient_boosting1.png?" alt="tree_adj" width="70%"/>
----------
### Illustrative example (from [source](https://towardsdatascience.com/underfitting-and-overfitting-in-machine-learning-and-how-to-deal-with-it-6fe4a8a49dbf))
<img src="data/boosting.png" alt="tree_adj" width="70%"/>
We built multiple trees consecutively: Tree 1 -> Tree 2 -> Tree 3 - > ....
**The size of the plus or minus singns indicates the weights of a data points for every Tree**. How do we determine these weights?
For each consecutive tree and iteration we do the following:
- The **wrongly classified data points ("mistakes" = red circles)** are identified and **more heavily weighted in the next tree (green arrow)**.
- Thus the size of the plus or minus changes in the next tree
- This change in weights will influence and change the next simple decision tree
- The **correct predictions are** identified and **less heavily weighted in the next tree**.
We iterate this process for a certain number of times, stop and construct our final model:
- The ensemble (**"Final: Combination"**) is a linear combination of the simple trees, and is more expressive!
- The ensemble (**"Final: Combination"**) has indeed not just one simple decision boundary line, and fits the data better.
<img src="data/boosting_2.png?" alt="tree_adj" width="70%"/>
### What is Ada Boost?
- Ada Boost = Adaptive Boosting.
- AdaBoost is adaptive in the sense that subsequent weak learners are tweaked in favor of those instances misclassified by previous classifiers
<img src="data/AdaBoost1.png" alt="tree_adj" width="70%"/>
<img src="data/AdaBoost2.png" alt="tree_adj" width="70%"/>
<img src="data/AdaBoost3.png" alt="tree_adj" width="70%"/>
**Notice that when $\hat{y}_n = 𝑦_n$, the weight $w_n$ is small; when $\hat{y}_n \neq 𝑦_n$, the weight $w_n$ is larger.**
### Illustrative Example (from slides)
------
**Step1. Start with equal distribition initially**
<img src="data/ADA2.png" alt="tree_adj" width="40%">
------
**Step2. Fit a simple classifier**
<img src="data/ADA3.png" alt="tree_adj" width="40%"/>
------
**Step3. Update the weights**
<img src="data/ADA4.png" alt="tree_adj" width="40%"/>
**Step4. Update the classifier:** First time trivial (we have no model yet.)
------
**Step2. Fit a simple classifier**
<img src="data/ADA5.png" alt="tree_adj" width="40%"/>
**Step3. Update the weights:** not shown.
------
**Step4. Update the classifier:**
<img src="data/ADA6.png" alt="tree_adj" width="40%">
## 4. Use the Adaboost method to visualize Bias-Variance tradeoff.
Now let's try Boosting!
```
#Fit an Adaboost Model
#Training
model = AdaBoostClassifier(base_estimator=DecisionTreeClassifier(max_depth=4),
n_estimators=200,
learning_rate=0.05)
model.fit(x_train, y_train)
#Predict
y_pred_train = model.predict(x_train)
y_pred_test = model.predict(x_test)
#Performance Evaluation
acc_boosting_training = accuracy_score(y_train, y_pred_train)*100
acc_boosting_test = accuracy_score(y_test, y_pred_test)*100
print("Ada Boost:\tAccuracy, Training Set \t: {:0.2f}%".format(acc_boosting_training))
print("Ada Boost:\tAccuracy, Testing Set \t: {:0.2f}%".format(acc_boosting_test))
```
**How does the test and training accuracy evolve with every iteration (tree)?**
```
#Plot Iteration based score
train_scores = list(model.staged_score(x_train,y_train))
test_scores = list(model.staged_score(x_test, y_test))
plt.figure(figsize=(10,7))
plt.plot(train_scores,label='train')
plt.plot(test_scores,label='test')
plt.xlabel('Iteration')
plt.ylabel('Accuracy')
plt.title("Variation of Accuracy with Iterations - ADA Boost")
plt.legend();
```
What about performance?
```
print("Decision Trees:\tAccuracy, Testing Set \t: {:.2%}".format(acc_trees_testing))
print("Bagging: \tAccuracy, Testing Set \t: {:0.2f}%".format( acc_bagging_testing))
print("Random Forest: \tAccuracy, Testing Set \t: {:0.2f}%".format(acc_random_forest_testing))
print("Ada Boost:\tAccuracy, Testing Set \t: {:0.2f}%".format(acc_boosting_test))
```
AdaBoost seems to be performing better than Simple Decision Trees and has a similar Test Set Accuracy performance compared to Random Forest.
**Random tip:** If a "for"-loop takes som time and you want to know the progress while running the loop, use: **tqdm()** ([link](https://github.com/tqdm/tqdm)). No need for 1000's of ```print(i)``` outputs.
Usage: ```for i in tqdm( range(start,finish) ):```
- tqdm means *"progress"* in Arabic (taqadum, تقدّم) and
- tqdm is an abbreviation for *"I love you so much"* in Spanish (te quiero demasiado).
#### What if we change the depth of our AdaBoost trees?
```
# Start Timer
start = time.time()
#Find Optimal Depth of trees for Boosting
score_train, score_test, depth_start, depth_end = {}, {}, 2, 30
for i in tqdm(range(depth_start, depth_end, 2)):
model = AdaBoostClassifier(
base_estimator=DecisionTreeClassifier(max_depth=i),
n_estimators=200, learning_rate=0.05)
model.fit(x_train, y_train)
score_train[i] = accuracy_score(y_train, model.predict(x_train))
score_test[i] = accuracy_score(y_test, model.predict(x_test))
# Stop Timer
end = time.time()
elapsed_adaboost = end - start
#Plot
lists1 = sorted(score_train.items())
lists2 = sorted(score_test.items())
x1, y1 = zip(*lists1)
x2, y2 = zip(*lists2)
plt.figure(figsize=(10,7))
plt.ylabel("Accuracy")
plt.xlabel("Depth")
plt.title('Variation of Accuracy with Depth - ADA Boost Classifier')
plt.plot(x1, y1, 'b-', label='Train')
plt.plot(x2, y2, 'g-', label='Test')
plt.legend()
plt.show()
```
Adaboost complexity depends on both the number of estimators and the base estimator.
- In the beginning as our model complexity increases (depth 2-3), we first observe a small increase in accuracy.
- But as we go further to the right of the graph (**deeper trees**), our model **will overfit the data.**
- **REMINDER and validation: Boosting relies on simple trees!**
**Food for Thought :**
- Are **boosted models independent of one another?** Do they need to wait for the previous model's residuals?
- Are **bagging or random forest models independent of each other**, can they be trained in a parallel fashion?
## 5. *Theory:* What is Gradient Boosting and XGBoost?
### What is Gradient Boosting?
To improve its predictions, **gradient boosting looks at the difference between its current approximation, and the known correct target vector, which is called the residual**.
The mathematics:
- It may be assumed that there is some imperfect model $F_{m}$
- The gradient boosting algorithm improves on $F_{m}$ constructing a new model that adds an estimator $h$ to provide a better model:
$$F_{m+1}(x)=F_{m}(x)+h(x)$$
- To find $h$, the gradient boosting solution starts with the observation that a perfect **h** would imply
$$F_{m+1}(x)=F_{m}(x)+h(x)=y$$
- or, equivalently solving for h,
$$h(x)=y-F_{m}(x)$$
- Therefore, gradient boosting will fit h to the residual $y-F_{m}(x)$
<img src="data/gradient_boosting2.png" alt="tree_adj" width="80%"/>
-------
### XGBoost: ["Long May She Reign!"](https://towardsdatascience.com/https-medium-com-vishalmorde-xgboost-algorithm-long-she-may-rein-edd9f99be63d)
<img src="data/kaggle.png" alt="tree_adj" width="100%"/>
----------
### What is XGBoost and why is it so good!?
- Based on Gradient Boosting
- XGBoost = **eXtreme Gradient Boosting**; refers to the engineering goal to push the limit of computations resources for boosted tree algorithm
**Accuracy:**
- XGBoost however uses a **more regularized model formalizaiton to control overfitting** (=better performance) by both L1 and L2 regularization.
- Tree Pruning methods: more shallow tree will also prevent overfitting
- Improved convergence techniques (like early stopping when no improvement is made for X number of iterations)
- Built-in Cross-Validaiton
**Computing Speed:**
- Special Vector and matrix type data structures for faster results.
- Parallelized tree building: using all of your CPU cores during training.
- Distributed Computing: for training very large models using a cluster of machines.
- Cache Optimization of data structures and algorithm: to make best use of hardware.
**XGBoost is building boosted trees in parallel? What? How?**
- No: Xgboost doesn't run multiple trees in parallel, you need predictions after each tree to update gradients.
- Rather it does the parallelization WITHIN a single tree my using openMP to create branches independently.
## 6. Use XGBoost: Extreme Gradient Boosting
```
# Let's install XGBoost
! pip install xgboost
import xgboost as xgb
# Create the training and test data
dtrain = xgb.DMatrix(x_train, label=y_train)
dtest = xgb.DMatrix(x_test, label=y_test)
# Parameters
param = {
'max_depth': best_depth, # the maximum depth of each tree
'eta': 0.3, # the training step for each iteration
'silent': 1, # logging mode - quiet
'objective': 'multi:softprob', # error evaluation for multiclass training
'num_class': 2} # the number of classes that exist in this datset
# Number of training iterations
num_round = 200
# Start timer
start = time.time()
# Train XGBoost
bst = xgb.train(param,
dtrain,
num_round,
evals= [(dtrain, 'train')],
early_stopping_rounds=20, # early stopping
verbose_eval=20)
# Make prediction training set
preds_train = bst.predict(dtrain)
best_preds_train = np.asarray([np.argmax(line) for line in preds_train])
# Make prediction test set
preds_test = bst.predict(dtest)
best_preds_test = np.asarray([np.argmax(line) for line in preds_test])
# Performance Evaluation
acc_XGBoost_training = accuracy_score(y_train, best_preds_train)*100
acc_XGBoost_test = accuracy_score(y_test, best_preds_test)*100
# Stop Timer
end = time.time()
elapsed_xgboost = end - start
print("XGBoost:\tAccuracy, Training Set \t: {:0.2f}%".format(acc_XGBoost_training))
print("XGBoost:\tAccuracy, Testing Set \t: {:0.2f}%".format(acc_XGBoost_test))
```
### What about the accuracy performance: AdaBoost versus XGBoost?
```
print("Ada Boost:\tAccuracy, Testing Set \t: {:0.2f}%".format(acc_boosting_test))
print("XGBoost:\tAccuracy, Testing Set \t: {:0.2f}%".format(acc_XGBoost_test))
```
### What about the computing performance: AdaBoost versus XGBoost?
```
print("AdaBoost elapsed time: \t{:0.2f}s".format(elapsed_adaboost))
print("XGBoost elapsed time: \t{:0.2f}s".format(elapsed_xgboost))
```
### What if we change the depth of our XGBoost trees and compare to Ada Boost?
```
def model_xgboost(best_depth):
param = {
'max_depth': best_depth, # the maximum depth of each tree
'eta': 0.3, # the training step for each iteration
'silent': 1, # logging mode - quiet
'objective': 'multi:softprob', # error evaluation for multiclass training
'num_class': 2} # the number of classes that exist in this datset
# the number of training iterations
num_round = 200
bst = xgb.train(param,
dtrain,
num_round,
evals= [(dtrain, 'train')],
early_stopping_rounds=20,
verbose_eval=False)
preds_train = bst.predict(dtrain)
best_preds_train = np.asarray([np.argmax(line) for line in preds_train])
preds_test = bst.predict(dtest)
best_preds_test = np.asarray([np.argmax(line) for line in preds_test])
#Performance Evaluation
XGBoost_training = accuracy_score(y_train, best_preds_train)
XGBoost_test = accuracy_score(y_test, best_preds_test)
return XGBoost_training, XGBoost_test
#Find Optimal Depth of trees for Boosting
score_train_xgb, score_test_xgb = {}, {}
depth_start, depth_end = 2, 30
for i in tqdm(range(depth_start, depth_end, 2)):
XGBoost_training, XGBoost_test = model_xgboost(i)
score_train_xgb[i] = XGBoost_training
score_test_xgb[i] = XGBoost_test
#Plot
lists1 = sorted(score_train_xgb.items())
lists2 = sorted(score_test_xgb.items())
x3, y3 = zip(*lists1)
x4, y4 = zip(*lists2)
plt.figure(figsize=(10,7))
plt.ylabel("Accuracy")
plt.xlabel("Depth")
plt.title('Variation of Accuracy with Depth - Adaboost & XGBoost Classifier')
plt.plot(x1, y1, label='Train Accuracy Ada Boost')
plt.plot(x2, y2, label='Test Accuracy Ada Boost')
plt.plot(x3, y3, label='Train Accuracy XGBoost')
plt.plot(x4, y4, label='Test Accuracy XGBoost')
plt.legend()
plt.show()
```
**Interesting**:
- No real optimal depth of the simple tree for XGBoost, probably a lot of regularization, pruning, or early stopping when using a deep tree at the start.
- XGBoost does not seem to overfit when the depth of the tree increases, as opposed to Ada Boost.
**All the accuracy performances:**
```
print("Decision Trees:\tAccuracy, Testing Set \t: {:.2%}".format(acc_trees_testing))
print("Bagging: \tAccuracy, Testing Set \t: {:0.2f}%".format( acc_bagging_testing))
print("Random Forest: \tAccuracy, Testing Set \t: {:0.2f}%".format(acc_random_forest_testing))
print("Ada Boost:\tAccuracy, Testing Set \t: {:0.2f}%".format(acc_boosting_test))
print("XGBoost:\tAccuracy, Testing Set \t: {:0.2f}%".format(acc_XGBoost_test))
```
----------
**Overview of all the tree algorithms:** [Source](https://towardsdatascience.com/https-medium-com-vishalmorde-xgboost-algorithm-long-she-may-rein-edd9f99be63d)
<img src="data/trees.png" alt="tree_adj" width="100%"/>
## End of Section
----------
## Optional: Example to better understand Bias vs Variance tradeoff.
A central notion underlying what we've been learning in lectures and sections so far is the trade-off between overfitting and underfitting. If you remember back to Homework 3, we had a model that seemed to represent our data accurately. However, we saw that as we made it more and more accurate on the training set, it did not generalize well to unobserved data.
As a different example, in face recognition algorithms, such as that on the iPhone X, a too-accurate model would be unable to identity someone who styled their hair differently that day. The reason is that our model may learn irrelevant features in the training data. On the contrary, an insufficiently trained model would not generalize well either. For example, it was recently reported that a face mask could sufficiently fool the iPhone X.
A widely used solution in statistics to reduce overfitting consists of adding structure to the model, with something like regularization. This method favors simpler models during training.
The bias-variance dilemma is closely related.
- The **bias** of a model quantifies how precise a model is across training sets.
- The **variance** quantifies how sensitive the model is to small changes in the training set.
- A **robust** model is not overly sensitive to small changes.
- **The dilemma involves minimizing both bias and variance**; we want a precise and robust model. Simpler models tend to be less accurate but more robust. Complex models tend to be more accurate but less robust.
**How to reduce bias:**
- **Use more complex models, more features, less regularization,** ...
- **Boosting:** attempts to improve the predictive flexibility of simple models. Boosting uses simple base models and tries to “boost” their aggregate complexity.
**How to reduce variance:**
- **Early Stopping:** Its rules provide us with guidance as to how many iterations can be run before the learner begins to over-fit.
- **Pruning:** Pruning is extensively used while building related models. It simply removes the nodes which add little predictive power for the problem in hand.
- **Regularization:** It introduces a cost term for bringing in more features with the objective function. Hence it tries to push the coefficients for many variables to zero and hence reduce cost term.
- **Train with more data:** It won’t work every time, but training with more data can help algorithms detect the signal better.
- **Ensembling:** Ensembles are machine learning methods for combining predictions from multiple separate models. For example:
- **Bagging** attempts to reduce the chance of overfitting complex models: Bagging uses complex base models and tries to “smooth out” their predictions.
|
github_jupyter
|
#RUN THIS CELL
import requests
from IPython.core.display import HTML
styles = requests.get("https://raw.githubusercontent.com/Harvard-IACS/2018-CS109A/master/content/styles/cs109.css").text
HTML(styles)
- The problem of learning an optimal decision tree is known to be **NP-complete** under several aspects of optimality and even for simple concepts.
- Consequently, practical decision-tree learning algorithms are based on **heuristic algorithms such as the greedy algorithm where locally optimal decisions are made at each node**.
- Such algorithms **cannot guarantee to return the globally optimal decision tree**.
- This can be mitigated by training multiple trees in an ensemble learner, where the features and samples are randomly sampled with replacement (Bagging).
For example: **What is the defaulth DecisionTreeClassifier behaviour when there are 2 or more best features for a certain split (a tie among "splitters")?** (after a deep dive and internet search [link](https://github.com/scikit-learn/scikit-learn/issues/12259 ) ):
- The current default behaviour when splitter="best" is to shuffle the features at each step and take the best feature to split.
- In case there is a tie, we take a random one.
-------------
## 2. Just re-building the tree models of last week
### Rebuild the Decision Tree model, Bagging model and Random Forest Model for comparison with Boosting methods
We will be working with a spam email dataset. The dataset has 57 predictors with a response variable called `Spam` that indicates whether an email is spam or not spam. **The goal is to be able to create a classifier or method that acts as a spam filter.**
Link to description : https://archive.ics.uci.edu/ml/datasets/spambase
-----------
### Fitting an Optimal Single Decision Tree
--------
### Fitting 100 Single Decision Trees while Bagging
### Fitting Random Forest
#### Let's compare the performance of our 3 models:
## 3. *Theory:* What is Boosting?
- **Bagging and Random Forest:**
- complex and deep trees **overfit**
- thus **let's perform variance reduction on complex trees!**
- **Boosting:**
- simple and shallow trees **underfit**
- thus **let's perform bias reduction of simple trees!**
- make the simple trees more expressive!
**Boosting** attempts to improve the predictive flexibility of simple models.
- It trains a **large number of “weak” learners in sequence**.
- A weak learner is a constrained model (limit the max depth of each decision tree).
- Each one in the sequence focuses on **learning from the mistakes** of the one before it.
- By more heavily weighting in the mistakes in the next tree, our next tree will learn from the mistakes.
- A combining all the weak learners into a single strong learner = **a boosted tree**.
<img src="data/gradient_boosting1.png?" alt="tree_adj" width="70%"/>
----------
### Illustrative example (from [source](https://towardsdatascience.com/underfitting-and-overfitting-in-machine-learning-and-how-to-deal-with-it-6fe4a8a49dbf))
<img src="data/boosting.png" alt="tree_adj" width="70%"/>
We built multiple trees consecutively: Tree 1 -> Tree 2 -> Tree 3 - > ....
**The size of the plus or minus singns indicates the weights of a data points for every Tree**. How do we determine these weights?
For each consecutive tree and iteration we do the following:
- The **wrongly classified data points ("mistakes" = red circles)** are identified and **more heavily weighted in the next tree (green arrow)**.
- Thus the size of the plus or minus changes in the next tree
- This change in weights will influence and change the next simple decision tree
- The **correct predictions are** identified and **less heavily weighted in the next tree**.
We iterate this process for a certain number of times, stop and construct our final model:
- The ensemble (**"Final: Combination"**) is a linear combination of the simple trees, and is more expressive!
- The ensemble (**"Final: Combination"**) has indeed not just one simple decision boundary line, and fits the data better.
<img src="data/boosting_2.png?" alt="tree_adj" width="70%"/>
### What is Ada Boost?
- Ada Boost = Adaptive Boosting.
- AdaBoost is adaptive in the sense that subsequent weak learners are tweaked in favor of those instances misclassified by previous classifiers
<img src="data/AdaBoost1.png" alt="tree_adj" width="70%"/>
<img src="data/AdaBoost2.png" alt="tree_adj" width="70%"/>
<img src="data/AdaBoost3.png" alt="tree_adj" width="70%"/>
**Notice that when $\hat{y}_n = 𝑦_n$, the weight $w_n$ is small; when $\hat{y}_n \neq 𝑦_n$, the weight $w_n$ is larger.**
### Illustrative Example (from slides)
------
**Step1. Start with equal distribition initially**
<img src="data/ADA2.png" alt="tree_adj" width="40%">
------
**Step2. Fit a simple classifier**
<img src="data/ADA3.png" alt="tree_adj" width="40%"/>
------
**Step3. Update the weights**
<img src="data/ADA4.png" alt="tree_adj" width="40%"/>
**Step4. Update the classifier:** First time trivial (we have no model yet.)
------
**Step2. Fit a simple classifier**
<img src="data/ADA5.png" alt="tree_adj" width="40%"/>
**Step3. Update the weights:** not shown.
------
**Step4. Update the classifier:**
<img src="data/ADA6.png" alt="tree_adj" width="40%">
## 4. Use the Adaboost method to visualize Bias-Variance tradeoff.
Now let's try Boosting!
**How does the test and training accuracy evolve with every iteration (tree)?**
What about performance?
AdaBoost seems to be performing better than Simple Decision Trees and has a similar Test Set Accuracy performance compared to Random Forest.
**Random tip:** If a "for"-loop takes som time and you want to know the progress while running the loop, use: **tqdm()** ([link](https://github.com/tqdm/tqdm)). No need for 1000's of ```print(i)``` outputs.
Usage: ```for i in tqdm( range(start,finish) ):```
- tqdm means *"progress"* in Arabic (taqadum, تقدّم) and
- tqdm is an abbreviation for *"I love you so much"* in Spanish (te quiero demasiado).
#### What if we change the depth of our AdaBoost trees?
Adaboost complexity depends on both the number of estimators and the base estimator.
- In the beginning as our model complexity increases (depth 2-3), we first observe a small increase in accuracy.
- But as we go further to the right of the graph (**deeper trees**), our model **will overfit the data.**
- **REMINDER and validation: Boosting relies on simple trees!**
**Food for Thought :**
- Are **boosted models independent of one another?** Do they need to wait for the previous model's residuals?
- Are **bagging or random forest models independent of each other**, can they be trained in a parallel fashion?
## 5. *Theory:* What is Gradient Boosting and XGBoost?
### What is Gradient Boosting?
To improve its predictions, **gradient boosting looks at the difference between its current approximation, and the known correct target vector, which is called the residual**.
The mathematics:
- It may be assumed that there is some imperfect model $F_{m}$
- The gradient boosting algorithm improves on $F_{m}$ constructing a new model that adds an estimator $h$ to provide a better model:
$$F_{m+1}(x)=F_{m}(x)+h(x)$$
- To find $h$, the gradient boosting solution starts with the observation that a perfect **h** would imply
$$F_{m+1}(x)=F_{m}(x)+h(x)=y$$
- or, equivalently solving for h,
$$h(x)=y-F_{m}(x)$$
- Therefore, gradient boosting will fit h to the residual $y-F_{m}(x)$
<img src="data/gradient_boosting2.png" alt="tree_adj" width="80%"/>
-------
### XGBoost: ["Long May She Reign!"](https://towardsdatascience.com/https-medium-com-vishalmorde-xgboost-algorithm-long-she-may-rein-edd9f99be63d)
<img src="data/kaggle.png" alt="tree_adj" width="100%"/>
----------
### What is XGBoost and why is it so good!?
- Based on Gradient Boosting
- XGBoost = **eXtreme Gradient Boosting**; refers to the engineering goal to push the limit of computations resources for boosted tree algorithm
**Accuracy:**
- XGBoost however uses a **more regularized model formalizaiton to control overfitting** (=better performance) by both L1 and L2 regularization.
- Tree Pruning methods: more shallow tree will also prevent overfitting
- Improved convergence techniques (like early stopping when no improvement is made for X number of iterations)
- Built-in Cross-Validaiton
**Computing Speed:**
- Special Vector and matrix type data structures for faster results.
- Parallelized tree building: using all of your CPU cores during training.
- Distributed Computing: for training very large models using a cluster of machines.
- Cache Optimization of data structures and algorithm: to make best use of hardware.
**XGBoost is building boosted trees in parallel? What? How?**
- No: Xgboost doesn't run multiple trees in parallel, you need predictions after each tree to update gradients.
- Rather it does the parallelization WITHIN a single tree my using openMP to create branches independently.
## 6. Use XGBoost: Extreme Gradient Boosting
### What about the accuracy performance: AdaBoost versus XGBoost?
### What about the computing performance: AdaBoost versus XGBoost?
### What if we change the depth of our XGBoost trees and compare to Ada Boost?
**Interesting**:
- No real optimal depth of the simple tree for XGBoost, probably a lot of regularization, pruning, or early stopping when using a deep tree at the start.
- XGBoost does not seem to overfit when the depth of the tree increases, as opposed to Ada Boost.
**All the accuracy performances:**
| 0.8321 | 0.993385 |
```
import cv2
import numpy as np
import matplotlib.pyplot as plt
from pathlib import Path
from superpoint.settings import EXPER_PATH
import superpoint.evaluations.detector_evaluation as ev
from utils import plot_imgs
%matplotlib inline
%load_ext autoreload
%autoreload 2
experiments = [
'mp_synth-v6_photo-hom-aug_ha2-100-3-old_coco-repeat', 'harris_coco-repeat']
confidence_thresholds = [0.015, 0]
```
# Patches visualization
```
def draw_keypoints(img, corners, color=(0, 255, 0), radius=3, s=3):
img = np.repeat(cv2.resize(img, None, fx=s, fy=s)[..., np.newaxis], 3, -1)
for c in np.stack(corners).T:
cv2.circle(img, tuple(s*np.flip(c, 0)), radius, color, thickness=-1)
return img
def select_top_k(prob, thresh=0, num=300):
pts = np.where(prob > thresh)
idx = np.argsort(prob[pts])[::-1][:num]
pts = (pts[0][idx], pts[1][idx])
return pts
for i in range(4):
for e, thresh in zip(experiments, confidence_thresholds):
path = Path(EXPER_PATH, "outputs", e, str(i) + ".npz")
d = np.load(path)
points1 = select_top_k(d['prob'], thresh=thresh)
im1 = draw_keypoints(d['image'][..., 0] * 255, points1, (0, 255, 0)) / 255.
points2 = select_top_k(d['warped_prob'], thresh=thresh)
im2 = draw_keypoints(d['warped_image'] * 255, points2, (0, 255, 0)) / 255.
plot_imgs([im1, im2], ylabel=e, dpi=200, cmap='gray',
titles=[str(len(points1[0]))+' points', str(len(points2[0]))+' points'])
```
# Repeatability
```
for exp, thresh in zip(experiments, confidence_thresholds):
repeatability = ev.compute_repeatability(exp, keep_k_points=300, distance_thresh=3)
print('> {}: {}'.format(exp, repeatability))
```
# Visual proof that the true keypoints are warped as expected
```
def get_true_keypoints(exper_name, prob_thresh=0.5):
def warp_keypoints(keypoints, H):
warped_col0 = np.add(np.sum(np.multiply(keypoints, H[0, :2]), axis=1), H[0, 2])
warped_col1 = np.add(np.sum(np.multiply(keypoints, H[1, :2]), axis=1), H[1, 2])
warped_col2 = np.add(np.sum(np.multiply(keypoints, H[2, :2]), axis=1), H[2, 2])
warped_col0 = np.divide(warped_col0, warped_col2)
warped_col1 = np.divide(warped_col1, warped_col2)
new_keypoints = np.concatenate([warped_col0[:, None], warped_col1[:, None]],
axis=1)
return new_keypoints
def filter_keypoints(points, shape):
""" Keep only the points whose coordinates are
inside the dimensions of shape. """
mask = (points[:, 0] >= 0) & (points[:, 0] < shape[0]) &\
(points[:, 1] >= 0) & (points[:, 1] < shape[1])
return points[mask, :]
true_keypoints = []
for i in range(5):
path = Path(EXPER_PATH, "outputs", exper_name, str(i) + ".npz")
data = np.load(path)
shape = data['warped_prob'].shape
# Filter out predictions
keypoints = np.where(data['prob'] > prob_thresh)
keypoints = np.stack([keypoints[0], keypoints[1]], axis=-1)
warped_keypoints = np.where(data['warped_prob'] > prob_thresh)
warped_keypoints = np.stack([warped_keypoints[0], warped_keypoints[1]], axis=-1)
# Warp the original keypoints with the true homography
H = data['homography']
true_warped_keypoints = warp_keypoints(keypoints[:, [1, 0]], H)
true_warped_keypoints[:, [0, 1]] = true_warped_keypoints[:, [1, 0]]
true_warped_keypoints = filter_keypoints(true_warped_keypoints, shape)
true_keypoints.append((true_warped_keypoints[:, 0], true_warped_keypoints[:, 1]))
return true_keypoints
true_keypoints = get_true_keypoints('magic-point_coco_repeatability', 0.015)
for i in range(3):
e = 'magic-point_coco_repeatability'
thresh = 0.015
path = Path(EXPER_PATH, "outputs", e, str(i) + ".npz")
d = np.load(path)
points1 = np.where(d['prob'] > thresh)
im1 = draw_keypoints(d['image'][..., 0] * 255, points1, (0, 255, 0)) / 255.
points2 = true_keypoints[i]
im2 = draw_keypoints(d['warped_image'][..., 0] * 255, points2, (0, 255, 0)) / 255.
plot_imgs([im1, im2], titles=['Original', 'Original points warped'], dpi=200, cmap='gray')
```
|
github_jupyter
|
import cv2
import numpy as np
import matplotlib.pyplot as plt
from pathlib import Path
from superpoint.settings import EXPER_PATH
import superpoint.evaluations.detector_evaluation as ev
from utils import plot_imgs
%matplotlib inline
%load_ext autoreload
%autoreload 2
experiments = [
'mp_synth-v6_photo-hom-aug_ha2-100-3-old_coco-repeat', 'harris_coco-repeat']
confidence_thresholds = [0.015, 0]
def draw_keypoints(img, corners, color=(0, 255, 0), radius=3, s=3):
img = np.repeat(cv2.resize(img, None, fx=s, fy=s)[..., np.newaxis], 3, -1)
for c in np.stack(corners).T:
cv2.circle(img, tuple(s*np.flip(c, 0)), radius, color, thickness=-1)
return img
def select_top_k(prob, thresh=0, num=300):
pts = np.where(prob > thresh)
idx = np.argsort(prob[pts])[::-1][:num]
pts = (pts[0][idx], pts[1][idx])
return pts
for i in range(4):
for e, thresh in zip(experiments, confidence_thresholds):
path = Path(EXPER_PATH, "outputs", e, str(i) + ".npz")
d = np.load(path)
points1 = select_top_k(d['prob'], thresh=thresh)
im1 = draw_keypoints(d['image'][..., 0] * 255, points1, (0, 255, 0)) / 255.
points2 = select_top_k(d['warped_prob'], thresh=thresh)
im2 = draw_keypoints(d['warped_image'] * 255, points2, (0, 255, 0)) / 255.
plot_imgs([im1, im2], ylabel=e, dpi=200, cmap='gray',
titles=[str(len(points1[0]))+' points', str(len(points2[0]))+' points'])
for exp, thresh in zip(experiments, confidence_thresholds):
repeatability = ev.compute_repeatability(exp, keep_k_points=300, distance_thresh=3)
print('> {}: {}'.format(exp, repeatability))
def get_true_keypoints(exper_name, prob_thresh=0.5):
def warp_keypoints(keypoints, H):
warped_col0 = np.add(np.sum(np.multiply(keypoints, H[0, :2]), axis=1), H[0, 2])
warped_col1 = np.add(np.sum(np.multiply(keypoints, H[1, :2]), axis=1), H[1, 2])
warped_col2 = np.add(np.sum(np.multiply(keypoints, H[2, :2]), axis=1), H[2, 2])
warped_col0 = np.divide(warped_col0, warped_col2)
warped_col1 = np.divide(warped_col1, warped_col2)
new_keypoints = np.concatenate([warped_col0[:, None], warped_col1[:, None]],
axis=1)
return new_keypoints
def filter_keypoints(points, shape):
""" Keep only the points whose coordinates are
inside the dimensions of shape. """
mask = (points[:, 0] >= 0) & (points[:, 0] < shape[0]) &\
(points[:, 1] >= 0) & (points[:, 1] < shape[1])
return points[mask, :]
true_keypoints = []
for i in range(5):
path = Path(EXPER_PATH, "outputs", exper_name, str(i) + ".npz")
data = np.load(path)
shape = data['warped_prob'].shape
# Filter out predictions
keypoints = np.where(data['prob'] > prob_thresh)
keypoints = np.stack([keypoints[0], keypoints[1]], axis=-1)
warped_keypoints = np.where(data['warped_prob'] > prob_thresh)
warped_keypoints = np.stack([warped_keypoints[0], warped_keypoints[1]], axis=-1)
# Warp the original keypoints with the true homography
H = data['homography']
true_warped_keypoints = warp_keypoints(keypoints[:, [1, 0]], H)
true_warped_keypoints[:, [0, 1]] = true_warped_keypoints[:, [1, 0]]
true_warped_keypoints = filter_keypoints(true_warped_keypoints, shape)
true_keypoints.append((true_warped_keypoints[:, 0], true_warped_keypoints[:, 1]))
return true_keypoints
true_keypoints = get_true_keypoints('magic-point_coco_repeatability', 0.015)
for i in range(3):
e = 'magic-point_coco_repeatability'
thresh = 0.015
path = Path(EXPER_PATH, "outputs", e, str(i) + ".npz")
d = np.load(path)
points1 = np.where(d['prob'] > thresh)
im1 = draw_keypoints(d['image'][..., 0] * 255, points1, (0, 255, 0)) / 255.
points2 = true_keypoints[i]
im2 = draw_keypoints(d['warped_image'][..., 0] * 255, points2, (0, 255, 0)) / 255.
plot_imgs([im1, im2], titles=['Original', 'Original points warped'], dpi=200, cmap='gray')
| 0.510252 | 0.714105 |
<a href="https://colab.research.google.com/github/abhiruchi97/Econometrics-in-Python/blob/master/OLS_Regression.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import statsmodels.api as sm
from statsmodels.compat import lzip
import statsmodels.formula.api as smf
import statsmodels.stats.api as sms
import matplotlib.pyplot as plt
from statsmodels.sandbox.regression.predstd import wls_prediction_std
```
#OLS regression in Statsmodels
> ### We create a data set to study the determinants of the natural rate of unemployment for the United States between October 1960 and March 1997
* Data frequency: Monthly
* Data source: St. Louis FRED, Bureau of Labor Statistics (BLS)
```
data = pd.read_csv('dissert_dataset__instrumentalvar.csv')
data.columns = ["date","natural_rate", "sect_shifts", "real_unemp_benef", "log_growth","real_hrly_wage", "real_min_wage", "lab_force_grwth", 'pers_cnsmp', 'time']
data.head()
#np.max(data.iloc[:,2])
for i in range(1,7):
data.iloc[:,i] = data.iloc[:,i]/np.max(data.iloc[:,i])
data.head()
```
### Data description
```
variable_labels = pd.DataFrame({'Variable': ['date', 'natural_rate', 'sect_shifts', 'real_unemp_benef', 'log_growth',
'real_hrly_wage', 'real_min_wage', 'lab_force_grwth', 'time'],
'Label': ['yyyy-mm-dd', 'natural rate of unemployment', 'percentage changes in sectoral composition by industry',
'real unemployment benefits per capita', 'log of growth of output', 'real hourly wage/compensation', 'real minimum wage',
'labor force growth rate', 'time trend']})
variable_labels
data.info()
```
### Assigning independent variables to 'X' for OLS
```
X = data.iloc[:, [2, 3, 4, 5, 6, 7, 9]]
y = data.iloc[:, 1]
X.head()
```
###Adding a column of 1's to the regressor matrix
```
X = sm.add_constant(X)
```
### Regression using OLS and saving results
```
model = sm.OLS(y, X)
results = model.fit(cov_type = 'HC3')
print(results.summary())
```
### Regression Diagnostics
#### Normality of residuals
* Jarque-Bera test
```
name = ['Jarque-Bera', 'Chi^2 two-tail prob.', 'Skew', 'Kurtosis']
test = sms.jarque_bera(results.resid)
lzip(name, test)
plt.plot(results.resid)
```
#### Heteroscedasticity
* Breusch-Pagan test
* Goldfeld-Quandt test
```
name = ['Lagrange multiplier statistic', 'p-value',
'f-value', 'f p-value']
test = sms.het_breuschpagan(results.resid, results.model.exog)
lzip(name, test)
name = ['F statistic', 'p-value']
test = sms.het_goldfeldquandt(results.resid, results.model.exog)
lzip(name, test)
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import statsmodels.api as sm
from statsmodels.compat import lzip
import statsmodels.formula.api as smf
import statsmodels.stats.api as sms
import matplotlib.pyplot as plt
from statsmodels.sandbox.regression.predstd import wls_prediction_std
data = pd.read_csv('dissert_dataset__instrumentalvar.csv')
data.columns = ["date","natural_rate", "sect_shifts", "real_unemp_benef", "log_growth","real_hrly_wage", "real_min_wage", "lab_force_grwth", 'pers_cnsmp', 'time']
data.head()
#np.max(data.iloc[:,2])
for i in range(1,7):
data.iloc[:,i] = data.iloc[:,i]/np.max(data.iloc[:,i])
data.head()
variable_labels = pd.DataFrame({'Variable': ['date', 'natural_rate', 'sect_shifts', 'real_unemp_benef', 'log_growth',
'real_hrly_wage', 'real_min_wage', 'lab_force_grwth', 'time'],
'Label': ['yyyy-mm-dd', 'natural rate of unemployment', 'percentage changes in sectoral composition by industry',
'real unemployment benefits per capita', 'log of growth of output', 'real hourly wage/compensation', 'real minimum wage',
'labor force growth rate', 'time trend']})
variable_labels
data.info()
X = data.iloc[:, [2, 3, 4, 5, 6, 7, 9]]
y = data.iloc[:, 1]
X.head()
X = sm.add_constant(X)
model = sm.OLS(y, X)
results = model.fit(cov_type = 'HC3')
print(results.summary())
name = ['Jarque-Bera', 'Chi^2 two-tail prob.', 'Skew', 'Kurtosis']
test = sms.jarque_bera(results.resid)
lzip(name, test)
plt.plot(results.resid)
name = ['Lagrange multiplier statistic', 'p-value',
'f-value', 'f p-value']
test = sms.het_breuschpagan(results.resid, results.model.exog)
lzip(name, test)
name = ['F statistic', 'p-value']
test = sms.het_goldfeldquandt(results.resid, results.model.exog)
lzip(name, test)
| 0.306216 | 0.979842 |
```
import json
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib import gridspec
from matplotlib import cm
from pysheds.grid import Grid
from pysheds.view import Raster
from matplotlib import colors
import seaborn as sns
import warnings
from partition import differentiated_linear_weights, differentiated_power_weights, threshold_weights, controller_placement_algorithm, naive_partition
warnings.filterwarnings('ignore')
sns.set_palette('husl', 8)
sns.set()
%matplotlib inline
output = {}
```
# Generate graph
```
grid = Grid.from_raster('../data/n30w100_dir', data_name='dir')
dirmap = (64, 128, 1, 2, 4, 8, 16, 32)
# Specify pour point
x, y = -97.294167, 32.73750
# Delineate the catchment
grid.catchment(data='dir', x=x, y=y, dirmap=dirmap, out_name='catch',
recursionlimit=15000, xytype='label')
# Clip the bounding box to the catchment
grid.clip_to('catch', pad=(1,1,1,1))
#Compute flow accumulation
grid.accumulation(data='catch', out_name='acc', dirmap=dirmap)
# Compute flow distance
grid.flow_distance(data='catch', x=x, y=y, dirmap=dirmap, out_name='dist', xytype='label')
dist = grid.view('dist', nodata=0, dtype=np.float64)
dist_weights = (np.where(grid.view('acc') >= 100, 0.02, 0)
+ np.where((0 < grid.view('acc')) & (grid.view('acc') <= 100), 1, 0)).ravel()
dists = grid.flow_distance(data='catch', x=x, y=y, weights=dist_weights,
dirmap=dirmap, out_name='dist', xytype='label', inplace=False)
```
# Linear weighting
```
weights = differentiated_linear_weights(dists)
acc = grid.accumulation(data='catch', dirmap=dirmap, inplace=False)
wacc = grid.accumulation(data='catch', weights=weights, dirmap=dirmap, inplace=False)
k = 1
c = 6000
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, acc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 2
c = 3300
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 3
c = 6300 // 3
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 4
c = 6500 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 5
c = 6300 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 6
c = 6580 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 8
c = 6800 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 10
c = 6500 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 12
c = 6300 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 14
c = 6800 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 16
c = 6500 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 18
c = 6720 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 20
c = 6720 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 25
c = 6720 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 30
c = 6720 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
(np.asarray(cells_per_catch) > 101).all()
k = 35
c = 6720 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
(np.asarray(cells_per_catch) > 101).all()
```
# Randomized naive implementation
```
k = 2
target_cells = 6600
fdir = grid.view('catch')
subs, ixes = naive_partition(fdir, target_cells, k, grid, size_range=[500,2000], tolerance_spread=100)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells_ = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
iterations = 50
i = 0
while iterations > 0:
np.random.seed(i+20)
k = np.random.randint(1,35)
target_cells = 5700
fdir = grid.view('catch')
ixy, ixx = np.unravel_index(ixes, wacc.shape)
subs, ixes = naive_partition(fdir, target_cells, k, grid, size_range=[max(101, target_cells // (2*k)), 2 * target_cells // k],
tolerance_spread=target_cells // 20, use_seed=True, seed_0=i+1, seed_1=i+2)
cells_per_catch_ = [np.count_nonzero(sub) for sub in subs]
numcells_ = sum(cells_per_catch_)
pct_cells_ = float(numcells_ / acc.max())
i += 1
if (pct_cells_ > 0.47) and (pct_cells_ < 0.53):
if (np.asarray(cells_per_catch_) > 101).all():
experiment = {}
experiment['weighting'] = 'naive'
experiment['num_controllers'] = len(subs)
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells_
experiment['pct_controlled'] = pct_cells_
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch_
output.update({'naive_k{0}_50pct_{1}'.format(len(subs), i) : experiment})
iterations -= 1
else:
print("Undersized catchments")
else:
print("Out of range")
with open('../data/experiments_differentiated_50pct_phi50.json', 'w') as outfile:
json.dump(output, outfile)
```
|
github_jupyter
|
import json
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib import gridspec
from matplotlib import cm
from pysheds.grid import Grid
from pysheds.view import Raster
from matplotlib import colors
import seaborn as sns
import warnings
from partition import differentiated_linear_weights, differentiated_power_weights, threshold_weights, controller_placement_algorithm, naive_partition
warnings.filterwarnings('ignore')
sns.set_palette('husl', 8)
sns.set()
%matplotlib inline
output = {}
grid = Grid.from_raster('../data/n30w100_dir', data_name='dir')
dirmap = (64, 128, 1, 2, 4, 8, 16, 32)
# Specify pour point
x, y = -97.294167, 32.73750
# Delineate the catchment
grid.catchment(data='dir', x=x, y=y, dirmap=dirmap, out_name='catch',
recursionlimit=15000, xytype='label')
# Clip the bounding box to the catchment
grid.clip_to('catch', pad=(1,1,1,1))
#Compute flow accumulation
grid.accumulation(data='catch', out_name='acc', dirmap=dirmap)
# Compute flow distance
grid.flow_distance(data='catch', x=x, y=y, dirmap=dirmap, out_name='dist', xytype='label')
dist = grid.view('dist', nodata=0, dtype=np.float64)
dist_weights = (np.where(grid.view('acc') >= 100, 0.02, 0)
+ np.where((0 < grid.view('acc')) & (grid.view('acc') <= 100), 1, 0)).ravel()
dists = grid.flow_distance(data='catch', x=x, y=y, weights=dist_weights,
dirmap=dirmap, out_name='dist', xytype='label', inplace=False)
weights = differentiated_linear_weights(dists)
acc = grid.accumulation(data='catch', dirmap=dirmap, inplace=False)
wacc = grid.accumulation(data='catch', weights=weights, dirmap=dirmap, inplace=False)
k = 1
c = 6000
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, acc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 2
c = 3300
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 3
c = 6300 // 3
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 4
c = 6500 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 5
c = 6300 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 6
c = 6580 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 8
c = 6800 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 10
c = 6500 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 12
c = 6300 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 14
c = 6800 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 16
c = 6500 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 18
c = 6720 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 20
c = 6720 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 25
c = 6720 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
k = 30
c = 6720 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
(np.asarray(cells_per_catch) > 101).all()
k = 35
c = 6720 // k
fdir = grid.view('catch')
subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, dist_weights=dist_weights,
grid=grid, compute_weights=differentiated_linear_weights)
ixy, ixx = np.unravel_index(ixes, wacc.shape)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
experiment = {}
experiment['weighting'] = 'linear'
experiment['num_controllers'] = k
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells
experiment['pct_controlled'] = pct_cells
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch
experiment['phi'] = 50
output.update({'linear_k{0}_50pct_phi50'.format(k) : experiment})
pct_cells
(np.asarray(cells_per_catch) > 101).all()
k = 2
target_cells = 6600
fdir = grid.view('catch')
subs, ixes = naive_partition(fdir, target_cells, k, grid, size_range=[500,2000], tolerance_spread=100)
cells_per_catch = [np.count_nonzero(sub) for sub in subs]
numcells_ = sum(cells_per_catch)
pct_cells = float(numcells / acc.max())
iterations = 50
i = 0
while iterations > 0:
np.random.seed(i+20)
k = np.random.randint(1,35)
target_cells = 5700
fdir = grid.view('catch')
ixy, ixx = np.unravel_index(ixes, wacc.shape)
subs, ixes = naive_partition(fdir, target_cells, k, grid, size_range=[max(101, target_cells // (2*k)), 2 * target_cells // k],
tolerance_spread=target_cells // 20, use_seed=True, seed_0=i+1, seed_1=i+2)
cells_per_catch_ = [np.count_nonzero(sub) for sub in subs]
numcells_ = sum(cells_per_catch_)
pct_cells_ = float(numcells_ / acc.max())
i += 1
if (pct_cells_ > 0.47) and (pct_cells_ < 0.53):
if (np.asarray(cells_per_catch_) > 101).all():
experiment = {}
experiment['weighting'] = 'naive'
experiment['num_controllers'] = len(subs)
experiment['max_accumulation'] = c
experiment['cells_controlled'] = numcells_
experiment['pct_controlled'] = pct_cells_
experiment['controller_locs'] = [int(ix) for ix in ixes]
experiment['cells_per_catch'] = cells_per_catch_
output.update({'naive_k{0}_50pct_{1}'.format(len(subs), i) : experiment})
iterations -= 1
else:
print("Undersized catchments")
else:
print("Out of range")
with open('../data/experiments_differentiated_50pct_phi50.json', 'w') as outfile:
json.dump(output, outfile)
| 0.608245 | 0.75803 |
# KernelSHAP: combining preprocessor and predictor
## Introduction
In [this](kernel_shap_adult_lr.ipynb) example, we showed that the categorical variables can be handled by fitting the explainer on preprocessed data and passing preprocessed data to the `explain` call. To handle the categorical variables, we either group them explicitly or sum the estimated shap values for each encoded shap dimension. An alternative way is to define our black-box model to include the preprocessor, as shown in [this](anchor_tabular_adult.ipynb) example. We now show that these methods give the same results.
```
import shap
shap.initjs()
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from alibi.explainers import KernelShap
from alibi.datasets import fetch_adult
from scipy.special import logit
from sklearn.compose import ColumnTransformer
from sklearn.impute import SimpleImputer
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score, confusion_matrix, plot_confusion_matrix
from sklearn.model_selection import cross_val_score, train_test_split
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler, OneHotEncoder
```
## Data preparation
### Load and split
The `fetch_adult` function returns a `Bunch` object containing the features, the targets, the feature names and a mapping of categorical variables to numbers.
```
adult = fetch_adult()
adult.keys()
data = adult.data
target = adult.target
target_names = adult.target_names
feature_names = adult.feature_names
category_map = adult.category_map
```
Note that for your own datasets you can use our utility function `gen_category_map` to create the category map.
```
from alibi.utils.data import gen_category_map
np.random.seed(0)
data_perm = np.random.permutation(np.c_[data, target])
data = data_perm[:,:-1]
target = data_perm[:,-1]
idx = 30000
X_train,y_train = data[:idx,:], target[:idx]
X_test, y_test = data[idx+1:,:], target[idx+1:]
```
### Create feature transformation pipeline
Create feature pre-processor. Needs to have 'fit' and 'transform' methods. Different types of pre-processing can be applied to all or part of the features. In the example below we will standardize ordinal features and apply one-hot-encoding to categorical features.
Ordinal features:
```
ordinal_features = [x for x in range(len(feature_names)) if x not in list(category_map.keys())]
ordinal_transformer = Pipeline(steps=[('imputer', SimpleImputer(strategy='median')),
('scaler', StandardScaler())])
```
Categorical features:
```
categorical_features = list(category_map.keys())
categorical_transformer = Pipeline(steps=[('imputer', SimpleImputer(strategy='median')),
('onehot', OneHotEncoder(drop='first', handle_unknown='error'))])
```
Note that in order to be able to interpret the coefficients corresponding to the categorical features, the option `drop='first'` has been passed to the `OneHotEncoder`. This means that for a categorical variable with `n` levels, the length of the code will be `n-1`. This is necessary in order to avoid introducing feature multicolinearity, which would skew the interpretation of the results. For more information about the issue about multicolinearity in the context of linear modelling see [[1]](#References).
<a id='src_1'></a>
Combine and fit:
```
preprocessor = ColumnTransformer(transformers=[('num', ordinal_transformer, ordinal_features),
('cat', categorical_transformer, categorical_features)])
preprocessor.fit(X_train)
```
## Fit a binary logistic regression classifier to the preprocessed Adult dataset
### Preprocess the data
```
X_train_proc = preprocessor.transform(X_train)
X_test_proc = preprocessor.transform(X_test)
```
### Training
```
classifier = LogisticRegression(multi_class='multinomial',
random_state=0,
max_iter=500,
verbose=0,
)
classifier.fit(X_train_proc, y_train)
```
### Model assessment
```
y_pred = classifier.predict(X_test_proc)
cm = confusion_matrix(y_test, y_pred)
title = 'Confusion matrix for the logistic regression classifier'
disp = plot_confusion_matrix(classifier,
X_test_proc,
y_test,
display_labels=target_names,
cmap=plt.cm.Blues,
normalize=None,
)
disp.ax_.set_title(title)
print('Test accuracy: ', accuracy_score(y_test, classifier.predict(X_test_proc)))
```
## Explaining the model with an explainer fitted on the preprocessed data
To speed up computation, we will use a background dataset with only `100` samples.
```
start_example_idx = 0
stop_example_idx = 100
background_data = slice(start_example_idx, stop_example_idx)
```
First, we group the categorical variables.
```
def make_groups(num_feats_names, cat_feats_names, feat_enc_dim):
"""
Given a list with numerical feat. names, categorical feat. names
and a list specifying the lengths of the encoding for each cat.
varible, the function outputs a list of group names, and a list
of the same len where each entry represents the column indices that
the corresponding categorical feature
"""
group_names = num_feats_names + cat_feats_names
groups = []
cat_var_idx = 0
for name in group_names:
if name in num_feats_names:
groups.append(list(range(len(groups), len(groups) + 1)))
else:
start_idx = groups[-1][-1] + 1 if groups else 0
groups.append(list(range(start_idx, start_idx + feat_enc_dim[cat_var_idx] )))
cat_var_idx += 1
return group_names, groups
def sparse2ndarray(mat, examples=None):
"""
Converts a scipy.sparse.csr.csr_matrix to a numpy.ndarray.
If specified, examples is slice object specifying which selects a
number of rows from mat and converts only the respective slice.
"""
if examples:
return mat[examples, :].toarray()
return mat.toarray()
# obtain the indices of the categorical and the numerical features from the pipeline.
numerical_feats_idx = preprocessor.transformers_[0][2]
categorical_feats_idx = preprocessor.transformers_[1][2]
num_feats_names = [feature_names[i] for i in numerical_feats_idx]
cat_feats_names = [feature_names[i] for i in categorical_feats_idx]
perm_feat_names = num_feats_names + cat_feats_names
ohe = preprocessor.transformers_[1][1].named_steps['onehot']
feat_enc_dim = [len(cat_enc) - 1 for cat_enc in ohe.categories_]
# create the groups
X_train_proc_d = sparse2ndarray(X_train_proc, examples=background_data)
group_names, groups = make_groups(num_feats_names, cat_feats_names, feat_enc_dim)
```
Having created the groups, we are now ready to instantiate the explainer and explain our set.
```
pred_fcn = classifier.predict_proba
grp_lr_explainer = KernelShap(pred_fcn, link='logit', feature_names=perm_feat_names, seed=0)
grp_lr_explainer.fit(X_train_proc_d, group_names=group_names, groups=groups)
```
We select only a small fraction of the testing set to explain for the purposes of this example.
```
def split_set(X, y, fraction, random_state=0):
"""
Given a set X, associated labels y, split\\s a fraction y from X.
"""
_, X_split, _, y_split = train_test_split(X,
y,
test_size=fraction,
random_state=random_state,
)
print("Number of records: {}".format(X_split.shape[0]))
print("Number of class {}: {}".format(0, len(y_split) - y_split.sum()))
print("Number of class {}: {}".format(1, y_split.sum()))
return X_split, y_split
fraction_explained = 0.01
X_explain, y_explain = split_set(X_test,
y_test,
fraction_explained,
)
X_explain_proc = preprocessor.transform(X_explain)
X_explain_proc_d = sparse2ndarray(X_explain_proc)
grouped_explanation = grp_lr_explainer.explain(X_explain_proc_d)
```
### Explaining with an explainer fitted on the raw data
To explain with an explainer fitted on the raw data, we make the preprocessor part of the predictor, as shown below.
```
pred_fcn = lambda x: classifier.predict_proba(preprocessor.transform(x))
lr_explainer = KernelShap(pred_fcn, link='logit', feature_names=feature_names, seed=0)
```
We use the same background dataset to fit the explainer.
```
lr_explainer.fit(X_train[background_data])
```
We explain the same dataset as before.
```
explanation = lr_explainer.explain(X_explain)
```
### Results comparison
To show that fitting the explainer on the raw data and combining the preprocessor with the classifier gives the same results as grouping the variables and fitting the explainer on the preprocessed data, we check to see that the same features are considered as most important when combining the two approaches.
```
def get_ranked_values(explanation):
"""
Retrives a tuple of (feature_effects, feature_names) for
each class explained. A feature's effect is its average
shap value magnitude across an array of instances.
"""
ranked_shap_vals = []
for cls_idx in range(len(explanation.shap_values)):
this_ranking = (
explanation.raw['importances'][str(cls_idx)]['ranked_effect'],
explanation.raw['importances'][str(cls_idx)]['names']
)
ranked_shap_vals.append(this_ranking)
return ranked_shap_vals
def compare_ranking(ranking_1, ranking_2, methods=None):
for i, (combined, grouped) in enumerate(zip(ranking_1, ranking_2)):
print("Class: {}".format(i))
c_names, g_names = combined[1], grouped[1]
c_mag, g_mag = combined[0], grouped[0]
different = []
for i, (c_n, g_n) in enumerate(zip(c_names, g_names)):
if c_n != g_n:
different.append((i, c_n, g_n))
if different:
method_1 = methods[0] if methods else "Method_1"
method_2 = methods[1] if methods else "Method_2"
i, c_ns, g_ns = list(zip(*different))
data = {"Rank": i, method_1: c_ns, method_2: g_ns}
df = pd.DataFrame(data=data)
print("Found the following rank differences:")
print(df)
else:
print("The methods provided the same ranking for the feature effects.")
print("The ranking is: {}".format(c_names))
print("")
def reorder_feats(vals_and_names, src_vals_and_names):
"""Given a two tuples, each containing a list of ranked feature
shap values and the corresponding feature names, the function
reorders the values in vals according to the order specified in
the list of names contained in src_vals_and_names.
"""
_, src_names = src_vals_and_names
vals, names = vals_and_names
reordered = np.zeros_like(vals)
for i, name in enumerate(src_names):
alt_idx = names.index(name)
reordered[i] = vals[alt_idx]
return reordered, src_names
def compare_avg_mag_shap(class_idx, comparisons, baseline, **kwargs):
"""
Given a list of tuples, baseline, containing the feature values and a list with feature names
for each class and, comparisons, a list of lists with tuples with the same structure , the
function reorders the values of the features in comparisons entries according to the order
of the feature names provided in the baseline entries and displays the feature values for comparison.
"""
methods = kwargs.get("methods", ["method_{}".format(i) for i in range(len(comparisons) + 1)])
n_features = len(baseline[class_idx][0])
# bar settings
bar_width = kwargs.get("bar_width", 0.05)
bar_space = kwargs.get("bar_space", 2)
# x axis
x_low = kwargs.get("x_low", 0.0)
x_high = kwargs.get("x_high", 1.0)
x_step = kwargs.get("x_step", 0.05)
x_ticks = np.round(np.arange(x_low, x_high + x_step, x_step), 3)
# y axis (these are the y coordinate of start and end of each group
# of bars)
start_y_pos = np.array(np.arange(0, n_features))*bar_space
end_y_pos = start_y_pos + bar_width*len(methods)
y_ticks = 0.5*(start_y_pos + end_y_pos)
# figure
fig_x = kwargs.get("fig_x", 10)
fig_y = kwargs.get("fig_y", 7)
# fontsizes
title_font = kwargs.get("title_fontsize", 20)
legend_font = kwargs.get("legend_fontsize", 20)
tick_labels_font = kwargs.get("tick_labels_fontsize", 20)
axes_label_fontsize = kwargs.get("axes_label_fontsize", 10)
# labels
title = kwargs.get("title", None)
ylabel = kwargs.get("ylabel", None)
xlabel = kwargs.get("xlabel", None)
# process input data
methods = list(reversed(methods))
base_vals = baseline[class_idx][0]
ordering = baseline[class_idx][1]
comp_vals = []
# reorder the features so that they match the order of the baseline (ordering)
for comparison in comparisons:
vals, ord_ = reorder_feats(comparison[class_idx], baseline[class_idx])
comp_vals.append(vals)
assert ord_ is ordering
all_vals = [base_vals] + comp_vals
data = dict(zip(methods, all_vals))
df = pd.DataFrame(data=data, index=ordering)
# plotting logic
fig, ax = plt.subplots(figsize=(fig_x, fig_y))
for i, col in enumerate(df.columns):
values = list(df[col])
y_pos = [y + bar_width*i for y in start_y_pos]
ax.barh(y_pos, list(values), bar_width, label=col)
# add ticks, legend and labels
ax.set_xticks(x_ticks)
ax.set_xticklabels([str(x) for x in x_ticks], rotation=45, fontsize=tick_labels_font)
ax.set_xlabel(xlabel, fontsize=axes_label_fontsize)
ax.set_yticks(y_ticks)
ax.set_yticklabels(ordering, fontsize=tick_labels_font)
ax.set_ylabel(ylabel, fontsize=axes_label_fontsize)
ax.invert_yaxis() # labels read top-to-bottom
ax.legend(fontsize=legend_font)
plt.grid(True)
plt.title(title, fontsize=title_font)
return ax, fig, df
ranked_grouped_shap_vals = get_ranked_values(grouped_explanation)
ranked_shal_vals_raw = get_ranked_values(explanation)
compare_ranking(ranked_grouped_shap_vals, ranked_shal_vals_raw)
```
Above we can see that both methods returned the same feature importances.
```
class_idx = 0
ax, fig, _ = compare_avg_mag_shap(class_idx,
[ranked_shal_vals_raw],
ranked_grouped_shap_vals,
methods=('raw_data', 'grouped'),
bar_width=0.5,
tick_labels_fontsize=12,
legend_fontsize=12,
title_fontsize=15,
xlabel="Features effects (class {})".format(0),
ylabel="Feature",
axes_label_fontsize=15,
)
```
We can see that the shap values are very similar. The differences appear because the regression dataset generated in order to compute the shap values differes slightly between the two runs due to the difference in the order of the features in the background dataset.
### References
<a id='References'></a>
[[1]](#src_1) *Mahto, K.K., 2019. "One-Hot-Encoding, Multicollinearity and the Dummy Variable Trap". Retrieved 02 Feb 2020* [(link)](https://towardsdatascience.com/one-hot-encoding-multicollinearity-and-the-dummy-variable-trap-b5840be3c41a)
|
github_jupyter
|
import shap
shap.initjs()
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from alibi.explainers import KernelShap
from alibi.datasets import fetch_adult
from scipy.special import logit
from sklearn.compose import ColumnTransformer
from sklearn.impute import SimpleImputer
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score, confusion_matrix, plot_confusion_matrix
from sklearn.model_selection import cross_val_score, train_test_split
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler, OneHotEncoder
adult = fetch_adult()
adult.keys()
data = adult.data
target = adult.target
target_names = adult.target_names
feature_names = adult.feature_names
category_map = adult.category_map
from alibi.utils.data import gen_category_map
np.random.seed(0)
data_perm = np.random.permutation(np.c_[data, target])
data = data_perm[:,:-1]
target = data_perm[:,-1]
idx = 30000
X_train,y_train = data[:idx,:], target[:idx]
X_test, y_test = data[idx+1:,:], target[idx+1:]
ordinal_features = [x for x in range(len(feature_names)) if x not in list(category_map.keys())]
ordinal_transformer = Pipeline(steps=[('imputer', SimpleImputer(strategy='median')),
('scaler', StandardScaler())])
categorical_features = list(category_map.keys())
categorical_transformer = Pipeline(steps=[('imputer', SimpleImputer(strategy='median')),
('onehot', OneHotEncoder(drop='first', handle_unknown='error'))])
preprocessor = ColumnTransformer(transformers=[('num', ordinal_transformer, ordinal_features),
('cat', categorical_transformer, categorical_features)])
preprocessor.fit(X_train)
X_train_proc = preprocessor.transform(X_train)
X_test_proc = preprocessor.transform(X_test)
classifier = LogisticRegression(multi_class='multinomial',
random_state=0,
max_iter=500,
verbose=0,
)
classifier.fit(X_train_proc, y_train)
y_pred = classifier.predict(X_test_proc)
cm = confusion_matrix(y_test, y_pred)
title = 'Confusion matrix for the logistic regression classifier'
disp = plot_confusion_matrix(classifier,
X_test_proc,
y_test,
display_labels=target_names,
cmap=plt.cm.Blues,
normalize=None,
)
disp.ax_.set_title(title)
print('Test accuracy: ', accuracy_score(y_test, classifier.predict(X_test_proc)))
start_example_idx = 0
stop_example_idx = 100
background_data = slice(start_example_idx, stop_example_idx)
def make_groups(num_feats_names, cat_feats_names, feat_enc_dim):
"""
Given a list with numerical feat. names, categorical feat. names
and a list specifying the lengths of the encoding for each cat.
varible, the function outputs a list of group names, and a list
of the same len where each entry represents the column indices that
the corresponding categorical feature
"""
group_names = num_feats_names + cat_feats_names
groups = []
cat_var_idx = 0
for name in group_names:
if name in num_feats_names:
groups.append(list(range(len(groups), len(groups) + 1)))
else:
start_idx = groups[-1][-1] + 1 if groups else 0
groups.append(list(range(start_idx, start_idx + feat_enc_dim[cat_var_idx] )))
cat_var_idx += 1
return group_names, groups
def sparse2ndarray(mat, examples=None):
"""
Converts a scipy.sparse.csr.csr_matrix to a numpy.ndarray.
If specified, examples is slice object specifying which selects a
number of rows from mat and converts only the respective slice.
"""
if examples:
return mat[examples, :].toarray()
return mat.toarray()
# obtain the indices of the categorical and the numerical features from the pipeline.
numerical_feats_idx = preprocessor.transformers_[0][2]
categorical_feats_idx = preprocessor.transformers_[1][2]
num_feats_names = [feature_names[i] for i in numerical_feats_idx]
cat_feats_names = [feature_names[i] for i in categorical_feats_idx]
perm_feat_names = num_feats_names + cat_feats_names
ohe = preprocessor.transformers_[1][1].named_steps['onehot']
feat_enc_dim = [len(cat_enc) - 1 for cat_enc in ohe.categories_]
# create the groups
X_train_proc_d = sparse2ndarray(X_train_proc, examples=background_data)
group_names, groups = make_groups(num_feats_names, cat_feats_names, feat_enc_dim)
pred_fcn = classifier.predict_proba
grp_lr_explainer = KernelShap(pred_fcn, link='logit', feature_names=perm_feat_names, seed=0)
grp_lr_explainer.fit(X_train_proc_d, group_names=group_names, groups=groups)
def split_set(X, y, fraction, random_state=0):
"""
Given a set X, associated labels y, split\\s a fraction y from X.
"""
_, X_split, _, y_split = train_test_split(X,
y,
test_size=fraction,
random_state=random_state,
)
print("Number of records: {}".format(X_split.shape[0]))
print("Number of class {}: {}".format(0, len(y_split) - y_split.sum()))
print("Number of class {}: {}".format(1, y_split.sum()))
return X_split, y_split
fraction_explained = 0.01
X_explain, y_explain = split_set(X_test,
y_test,
fraction_explained,
)
X_explain_proc = preprocessor.transform(X_explain)
X_explain_proc_d = sparse2ndarray(X_explain_proc)
grouped_explanation = grp_lr_explainer.explain(X_explain_proc_d)
pred_fcn = lambda x: classifier.predict_proba(preprocessor.transform(x))
lr_explainer = KernelShap(pred_fcn, link='logit', feature_names=feature_names, seed=0)
lr_explainer.fit(X_train[background_data])
explanation = lr_explainer.explain(X_explain)
def get_ranked_values(explanation):
"""
Retrives a tuple of (feature_effects, feature_names) for
each class explained. A feature's effect is its average
shap value magnitude across an array of instances.
"""
ranked_shap_vals = []
for cls_idx in range(len(explanation.shap_values)):
this_ranking = (
explanation.raw['importances'][str(cls_idx)]['ranked_effect'],
explanation.raw['importances'][str(cls_idx)]['names']
)
ranked_shap_vals.append(this_ranking)
return ranked_shap_vals
def compare_ranking(ranking_1, ranking_2, methods=None):
for i, (combined, grouped) in enumerate(zip(ranking_1, ranking_2)):
print("Class: {}".format(i))
c_names, g_names = combined[1], grouped[1]
c_mag, g_mag = combined[0], grouped[0]
different = []
for i, (c_n, g_n) in enumerate(zip(c_names, g_names)):
if c_n != g_n:
different.append((i, c_n, g_n))
if different:
method_1 = methods[0] if methods else "Method_1"
method_2 = methods[1] if methods else "Method_2"
i, c_ns, g_ns = list(zip(*different))
data = {"Rank": i, method_1: c_ns, method_2: g_ns}
df = pd.DataFrame(data=data)
print("Found the following rank differences:")
print(df)
else:
print("The methods provided the same ranking for the feature effects.")
print("The ranking is: {}".format(c_names))
print("")
def reorder_feats(vals_and_names, src_vals_and_names):
"""Given a two tuples, each containing a list of ranked feature
shap values and the corresponding feature names, the function
reorders the values in vals according to the order specified in
the list of names contained in src_vals_and_names.
"""
_, src_names = src_vals_and_names
vals, names = vals_and_names
reordered = np.zeros_like(vals)
for i, name in enumerate(src_names):
alt_idx = names.index(name)
reordered[i] = vals[alt_idx]
return reordered, src_names
def compare_avg_mag_shap(class_idx, comparisons, baseline, **kwargs):
"""
Given a list of tuples, baseline, containing the feature values and a list with feature names
for each class and, comparisons, a list of lists with tuples with the same structure , the
function reorders the values of the features in comparisons entries according to the order
of the feature names provided in the baseline entries and displays the feature values for comparison.
"""
methods = kwargs.get("methods", ["method_{}".format(i) for i in range(len(comparisons) + 1)])
n_features = len(baseline[class_idx][0])
# bar settings
bar_width = kwargs.get("bar_width", 0.05)
bar_space = kwargs.get("bar_space", 2)
# x axis
x_low = kwargs.get("x_low", 0.0)
x_high = kwargs.get("x_high", 1.0)
x_step = kwargs.get("x_step", 0.05)
x_ticks = np.round(np.arange(x_low, x_high + x_step, x_step), 3)
# y axis (these are the y coordinate of start and end of each group
# of bars)
start_y_pos = np.array(np.arange(0, n_features))*bar_space
end_y_pos = start_y_pos + bar_width*len(methods)
y_ticks = 0.5*(start_y_pos + end_y_pos)
# figure
fig_x = kwargs.get("fig_x", 10)
fig_y = kwargs.get("fig_y", 7)
# fontsizes
title_font = kwargs.get("title_fontsize", 20)
legend_font = kwargs.get("legend_fontsize", 20)
tick_labels_font = kwargs.get("tick_labels_fontsize", 20)
axes_label_fontsize = kwargs.get("axes_label_fontsize", 10)
# labels
title = kwargs.get("title", None)
ylabel = kwargs.get("ylabel", None)
xlabel = kwargs.get("xlabel", None)
# process input data
methods = list(reversed(methods))
base_vals = baseline[class_idx][0]
ordering = baseline[class_idx][1]
comp_vals = []
# reorder the features so that they match the order of the baseline (ordering)
for comparison in comparisons:
vals, ord_ = reorder_feats(comparison[class_idx], baseline[class_idx])
comp_vals.append(vals)
assert ord_ is ordering
all_vals = [base_vals] + comp_vals
data = dict(zip(methods, all_vals))
df = pd.DataFrame(data=data, index=ordering)
# plotting logic
fig, ax = plt.subplots(figsize=(fig_x, fig_y))
for i, col in enumerate(df.columns):
values = list(df[col])
y_pos = [y + bar_width*i for y in start_y_pos]
ax.barh(y_pos, list(values), bar_width, label=col)
# add ticks, legend and labels
ax.set_xticks(x_ticks)
ax.set_xticklabels([str(x) for x in x_ticks], rotation=45, fontsize=tick_labels_font)
ax.set_xlabel(xlabel, fontsize=axes_label_fontsize)
ax.set_yticks(y_ticks)
ax.set_yticklabels(ordering, fontsize=tick_labels_font)
ax.set_ylabel(ylabel, fontsize=axes_label_fontsize)
ax.invert_yaxis() # labels read top-to-bottom
ax.legend(fontsize=legend_font)
plt.grid(True)
plt.title(title, fontsize=title_font)
return ax, fig, df
ranked_grouped_shap_vals = get_ranked_values(grouped_explanation)
ranked_shal_vals_raw = get_ranked_values(explanation)
compare_ranking(ranked_grouped_shap_vals, ranked_shal_vals_raw)
class_idx = 0
ax, fig, _ = compare_avg_mag_shap(class_idx,
[ranked_shal_vals_raw],
ranked_grouped_shap_vals,
methods=('raw_data', 'grouped'),
bar_width=0.5,
tick_labels_fontsize=12,
legend_fontsize=12,
title_fontsize=15,
xlabel="Features effects (class {})".format(0),
ylabel="Feature",
axes_label_fontsize=15,
)
| 0.683842 | 0.984985 |
# AMPEL intro III: Using a T2 unit to calculate transient properties
Here is an example of how to implement a T2 unit and run it in a standalone mode, without the need to have the whole AMPEL system and its services running. It is intended to be examined after the `t0_unit_example` notebook.
T2 units derive further information regarding an individual transient based on either information provided in alert(s) or through external queries (e.g. catalogs).
## T2 Unit Implementation
First, we need to implement a T2 unit class. This class should inherit from the class `AbsT2Unit` and implement a constructor and a `run` method.
The implementation of this class can be seen here: https://github.com/AmpelProject/Ampel-contrib-sample/blob/master/ampel/contrib/groupname/t2/T2ExamplePolyFit.py
Then, we initialize an object from this class, giving as a parameter a standard Python logger object.
```
import logging
logger = logging.getLogger()
logger.setLevel(logging.DEBUG)
from ampel.contrib.groupname.t2.T2ExamplePolyFit import T2ExamplePolyFit
myt2 = T2ExamplePolyFit(logger=logger)
```
Then, we download from the web a compressed TAR file with alerts over which we can run our T2 unit:
```
import os
import urllib.request
small_test_tar_url = 'https://ztf.uw.edu/alerts/public/ztf_public_20181129.tar.gz'
small_test_tar_path = 'ztf_public_20181129.tar.gz'
if not os.path.isfile(small_test_tar_path):
print('Downloading tar')
urllib.request.urlretrieve(small_test_tar_url, small_test_tar_path)
```
Afterwards, we need to load these alerts (which have an AVRO extension) and transform them into `AmpelAlert` objects. We can achieve that with the help of the `load_from_tar` method of the `DevAlertLoader` module. For the sake of this example we will only run our T2 unit over the first alert from the set:
```
from ampel.ztf.pipeline.t0.load.DevAlertLoader import load_from_tar
alert_list = load_from_tar(small_test_tar_path)
alert = alert_list[0]
print(alert)
```
Then, we proceed to transform the selected `AmpelAlert` into a lightcurve object by using the `ZIAlertUtils._create_lc` object. Over this lightcurve object we can now run our T2 unit using some parameters, in this case the degree of the polynomial fit. This returns a dictionary of results that will, when run inside an Ampel instance, be stored in the transient database:
```
from ampel.ztf.utils.ZIAlertUtils import ZIAlertUtils
lc = ZIAlertUtils._create_lc(alert.pps, alert.uls)
run_config = {
"degree" : 5
}
out = myt2.run(lc, run_config)
out
```
Finally, we can plot the result of the processing of our T2 unit, in this case a polynomial fit over the lightcurve, by using the standard Python Matplotlib library:
```
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
obs_date = sorted(lc.get_values('obs_date'))
mag = sorted(lc.get_values('mag'))
fit = np.poly1d(out['polyfit'])
ax = plt.axes()
ax.scatter(*zip(*lc.get_tuples('obs_date', 'mag')))
x_range = np.linspace(obs_date[0], obs_date[-1], 100)
ax.plot(x_range, fit(x_range), '--')
```
|
github_jupyter
|
import logging
logger = logging.getLogger()
logger.setLevel(logging.DEBUG)
from ampel.contrib.groupname.t2.T2ExamplePolyFit import T2ExamplePolyFit
myt2 = T2ExamplePolyFit(logger=logger)
import os
import urllib.request
small_test_tar_url = 'https://ztf.uw.edu/alerts/public/ztf_public_20181129.tar.gz'
small_test_tar_path = 'ztf_public_20181129.tar.gz'
if not os.path.isfile(small_test_tar_path):
print('Downloading tar')
urllib.request.urlretrieve(small_test_tar_url, small_test_tar_path)
from ampel.ztf.pipeline.t0.load.DevAlertLoader import load_from_tar
alert_list = load_from_tar(small_test_tar_path)
alert = alert_list[0]
print(alert)
from ampel.ztf.utils.ZIAlertUtils import ZIAlertUtils
lc = ZIAlertUtils._create_lc(alert.pps, alert.uls)
run_config = {
"degree" : 5
}
out = myt2.run(lc, run_config)
out
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
obs_date = sorted(lc.get_values('obs_date'))
mag = sorted(lc.get_values('mag'))
fit = np.poly1d(out['polyfit'])
ax = plt.axes()
ax.scatter(*zip(*lc.get_tuples('obs_date', 'mag')))
x_range = np.linspace(obs_date[0], obs_date[-1], 100)
ax.plot(x_range, fit(x_range), '--')
| 0.374905 | 0.970521 |
## Analysis of the situation of residential dwelling and amenities in Singapore (Dataset 2 & 3)
- Context
- This would be like a report to HDB
- Residential dwelling refers to both the residential property as well as the residential dwelling units in Singapore
- Amenities will be further defined when explain the datasets
- Sub-Objectives
- How are residents distributed across Singapore
- Does each region have enough amenities
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# Loading the dataset
prop_info = pd.read_csv('own_data/hdb-property-information.csv', parse_dates=['year_completed'])
area_dwell = pd.read_csv('own_data/land-area-and-dwelling-units-by-town.csv', parse_dates=['financial_year'])
# Setting the context for the whole notebook
# sns.set_context(rc={"font.size":18, "axes.titlesize":18, "axes.labelsize":18, 'xtick.labelsize': 15, 'ytick.labelsize': 15})
sns.set(font_scale=1.4)
```
# Reshaping data and cleaning
```
# Converting codes to actual names
town_format = {'AMK': 'ANG MO KIO',
'BB': 'BUKIT BATOK',
'BD': 'BEDOK',
'BH': 'BISHAN',
'BM': 'BUKIT MERAH',
'BP': 'BUKIT PANJANG',
'BT': 'BUKIT TIMAH',
'CCK': 'CHOA CHU KANG',
'CL': 'CLEMENTI',
'CT': 'CENTRAL AREA',
'GL': 'GEYLANG',
'HG': 'HOUGANG',
'JE': 'JURONG EAST',
'JW': 'JURONG WEST',
'KWN': 'KALLANG/WHAMPOA',
'MP': 'MARINE PARADE',
'PG': 'PUNGGOL',
'PRC': 'PASIR RIS',
'QT': 'QUEENSTOWN',
'SB': 'SEMBAWANG',
'SGN': 'SERANGOON',
'SK': 'SENGKANG',
'TAP': 'TAMPINES',
'TG': 'TENGAH',
'TP': 'TOA PAYOH',
'WL': 'WOODLANDS',
'YS': 'YISHUN'}
# Creating map to map town by regions
def splitRegion(region_str, region):
arr = region_str.upper()
arr = arr.split(', ')
arr = {i:region for i in arr}
return arr
north = 'Admirality, Kranji, Woodlands, Sembawang, Yishun, Yio Chu Kang, Seletar, Sengkang, NORTH'
north = splitRegion(north, 'north')
south = 'Holland, Queenstown, Bukit Merah, Telok Blangah, Pasir Panjang, Sentosa, Bukit Timah, Newton, Orchard, City, Marina South, SOUTH'
south = splitRegion(south, 'south')
east = 'Serangoon, Punggol, Hougang, Tampines, Pasir Ris, Loyang, Simei, Kallang, Katong, East Coast, Macpherson, Bedok, Pulau Ubin, Pulau Tekong, KALLANG/WHAMPOA, MARINE PARADE, KALLANG/ WHAMPOA, EAST'
east = splitRegion(east, 'east')
west = 'Lim Chu Kang, Choa Chu Kang, Bukit Panjang, Tuas, Jurong East, Jurong West, Jurong Industrial Estate, Bukit Batok, Hillview, West Coast, Clementi, TENGAH, WEST'
west = splitRegion(west, 'west')
central = 'Thomson, Marymount, Sin Ming, Ang Mo Kio, Bishan, Serangoon Gardens, MacRitchie, Toa Payoh, CENTRAL AREA, GEYLANG, OTHER ESTATES, CENTRAL'
central = splitRegion(central, 'central')
all_regions = {**north, **south, **east, **west, **central}
# Cleaning and formatting property info dataset
# Formatting town codes to town string
prop_info = prop_info.replace(town_format)
# Creating regions column
prop_info['regions'] = prop_info['bldg_contract_town'].replace(all_regions)
# Changing the Y and N columns to be 0 and 1 for counting
y_n_format = {'N': 0, 'Y': 1}
prop_info = prop_info.replace(y_n_format)
# Cleaning and formatting the dwelling and land use dataset
# Changing the town strings to all uppercase
area_dwell['town'] = area_dwell['town'].str.upper()
# Creating the regions column
area_dwell['regions'] = area_dwell['town'].replace(all_regions)
# Replacing NaN and null values
area_dwell = area_dwell.replace({'-': 0})
area_dwell = area_dwell.replace({'na': 0})
# Changing the string types to int
area_dwell = area_dwell.astype({'dwelling_units_under_management': 'int32'})
area_dwell = area_dwell.astype({'total_land_area': 'int32'})
```
# Main data manipulation
```
# Creating the main df for property info
# Getting the sum of places by region
prop_regions = prop_info.groupby(['year_completed', 'regions'])['residential','commercial', 'market_hawker', 'miscellaneous', 'multistorey_carpark','precinct_pavilion'].sum().reset_index()
# Sum by row of all amenities to get number of amenities in a region
prop_regions['amenities'] = prop_regions[['commercial', 'market_hawker', 'miscellaneous', 'multistorey_carpark','precinct_pavilion']].sum(axis=1)
# Getting cumulative sum of all amenities by region and year
prop_regions['amenities_sum'] = prop_regions.groupby(['year_completed', 'regions'])['amenities'].sum().groupby(['regions']).cumsum().reset_index()['amenities']
# Getting cumultative sum of residents by region and year
prop_regions['residential_sum'] = prop_regions.groupby(['year_completed', 'regions'])['residential'].sum().groupby(['regions']).cumsum().reset_index()['residential']
# Getting ratio of amenities to residents
prop_regions['am_re_ratio'] = prop_regions['amenities_sum'] / prop_regions['residential_sum']
# Sorting the values by region
prop_regions = prop_regions.sort_values(by=['year_completed', 'regions'])
prop_regions.head()
# Creating the main df for land use and area dwelling
# Summing the land, and dwelling by regions and year
dwell_regions = area_dwell.groupby(['financial_year', 'regions'])['total_land_area', 'residential_land_area', 'dwelling_units_under_management', 'projected_ultimate_dwelling_units'].sum().reset_index()
# Getting ratios
dwell_regions['land_ratio'] = dwell_regions['residential_land_area'] / dwell_regions['total_land_area']
dwell_regions['dwelling_ratio'] = dwell_regions['dwelling_units_under_management'] / dwell_regions['projected_ultimate_dwelling_units']
# Sorting values by regions
dwell_regions = dwell_regions.sort_values(by='regions')
dwell_regions.head()
# Analysing change in dwelling units over time
# Filtering data
dwell_units_ratio = dwell_regions[['financial_year', 'regions', 'dwelling_ratio']]
dwell_units = dwell_regions[['financial_year', 'regions', 'dwelling_units_under_management']]
dwell_projected = dwell_regions[['financial_year', 'regions', 'projected_ultimate_dwelling_units']]
# Stripping the date to format str to show
dwell_units_ratio['financial_year'] = dwell_units_ratio['financial_year'].dt.strftime('%Y')
dwell_units['financial_year'] = dwell_units['financial_year'].dt.strftime('%Y')
dwell_projected['financial_year'] = dwell_projected['financial_year'].dt.strftime('%Y')
# Setting up the subplots
fig, ax = plt.subplots(2, 2 ,figsize=(20, 20))
# Plotting the first 2 subplots
dwell_units_p = dwell_units.pivot('financial_year', 'regions', 'dwelling_units_under_management')
dwell_units_p.plot(kind='bar', stacked=True, ax=ax[0][0])
sns.heatmap(dwell_units_p, annot=True, fmt='7', ax=ax[0][1])
# Adding the titles
ax[0][0].title.set_text('Stacked bar chart of dwelling units managed\nover time by regions')
ax[0][1].title.set_text('Heatmap of dwelling units managed\nover time by regions')
# Plotting the other 2
dwell_projected_p = dwell_projected.pivot('financial_year', 'regions', 'projected_ultimate_dwelling_units')
dwell_projected_p.plot(kind='bar', stacked=True, ax=ax[1][0])
dwell_units_ratio_p = dwell_units_ratio.pivot('financial_year', 'regions', 'dwelling_ratio')
sns.heatmap(dwell_units_ratio_p, annot=True, fmt='.2f', ax=ax[1][1])
# Adding the titles
ax[1][0].title.set_text('Stacked bar chart of projected dwelling units over time by regions')
ax[1][1].title.set_text('Heatmap of the ratio of dwelling units to projected dwelling units\nover time by regions')
# Saving the figure
plt.savefig('final_plots/prop_1.png', bbox_inches='tight')
pass
```
- Figure 1
- Total number of people living in Singapore is increasing
- We can see that the regions causing this are the, west, north and the east regions
- Figure 2
- This confirms figure 1 analysis, where for west, north and east regions the color gradients get lighter downwards
- This shows they had a greater increase
- Based on the numbers we can see that the number of dwelling units across all regions also increased
- Figure 3
- We see that the only increase in the projected number of dwelling units is for the east and west regions.
- This happened in 2013 and 2017 respectively
- Figure 4
- As for the ratio of number of dwelling units to projected number, we can see that there was an outlier of east and the north region, with a very small ratio
- However over time, we notice the gradients for all regions turn to the same gradient of orange, indicating they had the same ratio (0.7) in the end.
- Conclusions:
- Overall, more people are choosing to live in Singapore as time goes on
- However, our projected number of people living in each region, although more than the population of people dwelling in each region, is not increasing.
- Also we see that the government wants to generally have the same ratio of people dwelling in each region to the projected number.
- Thus in future, they would also try and equally distribute the demand for dwelling units across the regions
- But what led to the increase in projection?
```
# Analysing change in land area over time
# Filtering data
dwell_land = dwell_regions[['financial_year', 'regions', 'residential_land_area']]
dwell_total_land = dwell_regions[['financial_year', 'regions', 'total_land_area']]
dwell_land_ratio = dwell_regions[['financial_year', 'regions', 'land_ratio']]
# Stripping the date to format str to show
dwell_land['financial_year'] = dwell_land['financial_year'].dt.strftime('%Y')
dwell_total_land['financial_year'] = dwell_total_land['financial_year'].dt.strftime('%Y')
dwell_land_ratio['financial_year'] = dwell_land_ratio['financial_year'].dt.strftime('%Y')
# Setting up the subplots
fig, ax = plt.subplots(1, 2, figsize=(20, 10), sharey=True)
# Plotting the 2 plots
dwell_land_p = dwell_land.pivot('financial_year', 'regions', 'residential_land_area')
dwell_land_p.plot(kind='bar', stacked=True, ax=ax[0])
dwell_total_land_p = dwell_total_land.pivot('financial_year', 'regions', 'total_land_area')
dwell_total_land_p.plot(kind='bar', stacked=True, ax=ax[1])
# Setting pos of legend
ax[0].legend(loc='upper left')
ax[1].legend(loc='upper left')
# Adding the titles
ax[0].title.set_text('Stacked bar chart of residential land area\nover time by regions')
ax[1].title.set_text('Stacked bar chart of total land area\nover time by regions')
# Saving the figure
plt.savefig('final_plots/prop_2_1.png', bbox_inches='tight')
# Getting heatmap of ratio
dwell_land_ratio_p = dwell_land_ratio.pivot('financial_year', 'regions', 'land_ratio')
# Plotting the heatmap
fig, ax = plt.subplots(1, 1, figsize=(10, 10))
sns.heatmap(dwell_land_ratio_p, annot=True, fmt='.2f', ax=ax)
# Setting the title
ax.title.set_text('Heatmap of the ratio of residential land area to total land area\nover time by region')
# Saving the figure
plt.savefig('final_plots/prop_2_2.png', bbox_inches='tight')
```
- Figure 1
- We can see that the land used for residential property only increased for the west region
- Figure 2
- This is also accompanied by the increase in total land area increase for the west region in the same years of 2017 as well
- Figure 3
- We can see that the ratio of land used for residential property either remains the same or is decreasing.
This can be seen from north and west regions color gradient getting dark and the rest remaining the same
- Conclusions:
- We can see that as the years goes by the land reserved for residential property will probably not increase any further unless the total area increases
- This means that as more people choose to live in Singapore, we could run out of land to build residential properties.
- We can also see that the increase in land use for residential property is not the reason that led to the increase in the projected number of dwelling units from the previous slide.
- Therefore the change in the ratio of dwelling units to the projected number is probably due to some dwelling units being relocated to other regions.
- This prevention of overpopulation in one region by the government is probably not to drastically raise the demand in any regions.
```
# Analysing change in number of amenities over time
# Filtering data
prop_amn_sum = prop_regions[['year_completed', 'regions', 'amenities_sum']]
prop_amn_ratio = prop_regions[['year_completed', 'regions', 'am_re_ratio']]
# Mainipulating the data to get it to have a cumulative sum
new_index = pd.MultiIndex.from_product([prop_amn_sum['year_completed'].unique(), prop_amn_sum['regions'].unique()], names=['year_completed', 'regions'])
amn_sum_year = prop_amn_sum.groupby(['year_completed', 'regions']).sum().reindex(new_index).reset_index()
amn_sum_year = amn_sum_year.sort_values(by=['year_completed', 'regions'])
amn_sum_year['amenities_sum'] = amn_sum_year.groupby(['regions'])['amenities_sum'].fillna(method='ffill')
amn_sum_year = amn_sum_year.fillna(0)
amn_sum_year['year_completed'] = amn_sum_year['year_completed'].dt.strftime('%Y')
amn_sum_year_p = amn_sum_year.pivot('year_completed', 'regions', 'amenities_sum')
# Mainipulating for ratio
amn_ratio_year = prop_amn_ratio.groupby(['year_completed', 'regions']).sum().reindex(new_index).reset_index()
amn_ratio_year = amn_ratio_year.sort_values(by=['year_completed', 'regions'])
amn_ratio_year['am_re_ratio'] = amn_ratio_year.groupby(['regions'])['am_re_ratio'].fillna(method='ffill')
amn_ratio_year = amn_ratio_year.fillna(0)
amn_ratio_year = amn_ratio_year.groupby([pd.Grouper(freq='10Y', key='year_completed'), 'regions'])['am_re_ratio'].mean().reset_index()
amn_ratio_year['year_completed'] = amn_ratio_year['year_completed'].dt.strftime('%Y')
amn_ratio_year_p = amn_ratio_year.pivot('year_completed', 'regions', 'am_re_ratio')
# Setting up the subplots
fig, ax = plt.subplots(2, 1, figsize=(20, 20))
# Plotting the barchar and heatmap
amn_sum_year_p.plot(kind='bar', stacked=True, ax=ax[0])
sns.heatmap(amn_ratio_year_p, annot=True, fmt='.2f')
# Adding the titles
ax[0].set_title('Stacked bar chart cumulative number of amenities over time by region', fontdict={'fontsize': 20})
ax[1].set_title('Heatmap of the ratio of amenities to residential units over time(every 10 years) by region', fontdict={'fontsize': 20})
# Saving the figure
plt.savefig('final_plots/prop_3.png', bbox_inches='tight')
```
- Figure 1
- We can see that over time, some regions did not have amenities until 1975
- We can also see that there was more emphasis placed on developing the east, west and north regions, in that order of priority
- The total height of the bar chart is still increasing, suggesting that the total number of amenities is still increasing till today
- Figure 2
- As this heatmap is a mean of every 10 years, we can see that the south region started out with amenities, but only after 1977 there were amenities in all regions
- We can see that today, there are 2 groups of regions.
- Central and south with a much higher ratio of 0.85 - 0.9
- The rest, east, north and west with a ratio of 0.7
- Conclusions:
- As more residential property and units are made, similarly, more amenities are created, keeping the ratio the same.
- From this we can see that the general ratio of amenities for a region is around 0.7, with outliers in the central and south regions.
- This increase in amenities ratio is probably due to these area being closer to the Central Business District(CBD) of Singapore thus a lot more development has occurred.
- We can also see that there is a very high effort placed to make sure this ratio is around 0.7 - 0.65 for amenities as from the start, the ratios were already quite close to this.
- Thus as the number of people living in Singapore increases, we should try and maintain these ratios
## Objective 2: Analysis of the situation of residential dwelling and amenities in Singapore (Dataset 2 & 3)
**Final recommendation**: The number of people choosing to live in Singapore is increasing, although Singapore has planned the residential units for them, we are not increasing the projected land use, thus we are approaching the maximum number of residents able to live in Singapore.
The ratio for amenities to residential property is around 0.7 except for areas near the CBD. Although alarming, we are not exceeding the government's expectation yet, so it is still fine as of now.
Reason from chart:
- We can see that more people are choosing to live in Singapore
- The government seems to be spreading these people out to the different regions, keeping the ratio of residents dwelling to the projected number around the same across regions.
- However the projected number of dwelling units does not seem to be increasing with the number of dwelling units under management, suggesting we are approaching a maximum.
- Also since the projected number is not really changing, the ratio change is likely due to some people being reallocated to other regions to more equally distribute the ratio
- The land allocated for residential property is also not increasing
- This mean that we do have a maximum amount of land allocated to residential property, only more will be allocated if the total land in the region increases.
- The number of amenities in Singapore is continually increasing, but the ratio to residential property is still around the same
- As the number of residential property and units made and used increases, the number of amenities increase as well to accomodate for them.
- This ensures that there is sufficient amenities for the residents and the government is not over developing regions too fast
Sub-Objectives: How are residents distributed across Singapore?, Does each region have enough amenities?
- Based on the projected number, there was a larger concentration in the central and west, although now it is much more evenly distributed
- In the past, some regions had not developed yet, however as of now, based on the ratio, each regions should have enough amenities.
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# Loading the dataset
prop_info = pd.read_csv('own_data/hdb-property-information.csv', parse_dates=['year_completed'])
area_dwell = pd.read_csv('own_data/land-area-and-dwelling-units-by-town.csv', parse_dates=['financial_year'])
# Setting the context for the whole notebook
# sns.set_context(rc={"font.size":18, "axes.titlesize":18, "axes.labelsize":18, 'xtick.labelsize': 15, 'ytick.labelsize': 15})
sns.set(font_scale=1.4)
# Converting codes to actual names
town_format = {'AMK': 'ANG MO KIO',
'BB': 'BUKIT BATOK',
'BD': 'BEDOK',
'BH': 'BISHAN',
'BM': 'BUKIT MERAH',
'BP': 'BUKIT PANJANG',
'BT': 'BUKIT TIMAH',
'CCK': 'CHOA CHU KANG',
'CL': 'CLEMENTI',
'CT': 'CENTRAL AREA',
'GL': 'GEYLANG',
'HG': 'HOUGANG',
'JE': 'JURONG EAST',
'JW': 'JURONG WEST',
'KWN': 'KALLANG/WHAMPOA',
'MP': 'MARINE PARADE',
'PG': 'PUNGGOL',
'PRC': 'PASIR RIS',
'QT': 'QUEENSTOWN',
'SB': 'SEMBAWANG',
'SGN': 'SERANGOON',
'SK': 'SENGKANG',
'TAP': 'TAMPINES',
'TG': 'TENGAH',
'TP': 'TOA PAYOH',
'WL': 'WOODLANDS',
'YS': 'YISHUN'}
# Creating map to map town by regions
def splitRegion(region_str, region):
arr = region_str.upper()
arr = arr.split(', ')
arr = {i:region for i in arr}
return arr
north = 'Admirality, Kranji, Woodlands, Sembawang, Yishun, Yio Chu Kang, Seletar, Sengkang, NORTH'
north = splitRegion(north, 'north')
south = 'Holland, Queenstown, Bukit Merah, Telok Blangah, Pasir Panjang, Sentosa, Bukit Timah, Newton, Orchard, City, Marina South, SOUTH'
south = splitRegion(south, 'south')
east = 'Serangoon, Punggol, Hougang, Tampines, Pasir Ris, Loyang, Simei, Kallang, Katong, East Coast, Macpherson, Bedok, Pulau Ubin, Pulau Tekong, KALLANG/WHAMPOA, MARINE PARADE, KALLANG/ WHAMPOA, EAST'
east = splitRegion(east, 'east')
west = 'Lim Chu Kang, Choa Chu Kang, Bukit Panjang, Tuas, Jurong East, Jurong West, Jurong Industrial Estate, Bukit Batok, Hillview, West Coast, Clementi, TENGAH, WEST'
west = splitRegion(west, 'west')
central = 'Thomson, Marymount, Sin Ming, Ang Mo Kio, Bishan, Serangoon Gardens, MacRitchie, Toa Payoh, CENTRAL AREA, GEYLANG, OTHER ESTATES, CENTRAL'
central = splitRegion(central, 'central')
all_regions = {**north, **south, **east, **west, **central}
# Cleaning and formatting property info dataset
# Formatting town codes to town string
prop_info = prop_info.replace(town_format)
# Creating regions column
prop_info['regions'] = prop_info['bldg_contract_town'].replace(all_regions)
# Changing the Y and N columns to be 0 and 1 for counting
y_n_format = {'N': 0, 'Y': 1}
prop_info = prop_info.replace(y_n_format)
# Cleaning and formatting the dwelling and land use dataset
# Changing the town strings to all uppercase
area_dwell['town'] = area_dwell['town'].str.upper()
# Creating the regions column
area_dwell['regions'] = area_dwell['town'].replace(all_regions)
# Replacing NaN and null values
area_dwell = area_dwell.replace({'-': 0})
area_dwell = area_dwell.replace({'na': 0})
# Changing the string types to int
area_dwell = area_dwell.astype({'dwelling_units_under_management': 'int32'})
area_dwell = area_dwell.astype({'total_land_area': 'int32'})
# Creating the main df for property info
# Getting the sum of places by region
prop_regions = prop_info.groupby(['year_completed', 'regions'])['residential','commercial', 'market_hawker', 'miscellaneous', 'multistorey_carpark','precinct_pavilion'].sum().reset_index()
# Sum by row of all amenities to get number of amenities in a region
prop_regions['amenities'] = prop_regions[['commercial', 'market_hawker', 'miscellaneous', 'multistorey_carpark','precinct_pavilion']].sum(axis=1)
# Getting cumulative sum of all amenities by region and year
prop_regions['amenities_sum'] = prop_regions.groupby(['year_completed', 'regions'])['amenities'].sum().groupby(['regions']).cumsum().reset_index()['amenities']
# Getting cumultative sum of residents by region and year
prop_regions['residential_sum'] = prop_regions.groupby(['year_completed', 'regions'])['residential'].sum().groupby(['regions']).cumsum().reset_index()['residential']
# Getting ratio of amenities to residents
prop_regions['am_re_ratio'] = prop_regions['amenities_sum'] / prop_regions['residential_sum']
# Sorting the values by region
prop_regions = prop_regions.sort_values(by=['year_completed', 'regions'])
prop_regions.head()
# Creating the main df for land use and area dwelling
# Summing the land, and dwelling by regions and year
dwell_regions = area_dwell.groupby(['financial_year', 'regions'])['total_land_area', 'residential_land_area', 'dwelling_units_under_management', 'projected_ultimate_dwelling_units'].sum().reset_index()
# Getting ratios
dwell_regions['land_ratio'] = dwell_regions['residential_land_area'] / dwell_regions['total_land_area']
dwell_regions['dwelling_ratio'] = dwell_regions['dwelling_units_under_management'] / dwell_regions['projected_ultimate_dwelling_units']
# Sorting values by regions
dwell_regions = dwell_regions.sort_values(by='regions')
dwell_regions.head()
# Analysing change in dwelling units over time
# Filtering data
dwell_units_ratio = dwell_regions[['financial_year', 'regions', 'dwelling_ratio']]
dwell_units = dwell_regions[['financial_year', 'regions', 'dwelling_units_under_management']]
dwell_projected = dwell_regions[['financial_year', 'regions', 'projected_ultimate_dwelling_units']]
# Stripping the date to format str to show
dwell_units_ratio['financial_year'] = dwell_units_ratio['financial_year'].dt.strftime('%Y')
dwell_units['financial_year'] = dwell_units['financial_year'].dt.strftime('%Y')
dwell_projected['financial_year'] = dwell_projected['financial_year'].dt.strftime('%Y')
# Setting up the subplots
fig, ax = plt.subplots(2, 2 ,figsize=(20, 20))
# Plotting the first 2 subplots
dwell_units_p = dwell_units.pivot('financial_year', 'regions', 'dwelling_units_under_management')
dwell_units_p.plot(kind='bar', stacked=True, ax=ax[0][0])
sns.heatmap(dwell_units_p, annot=True, fmt='7', ax=ax[0][1])
# Adding the titles
ax[0][0].title.set_text('Stacked bar chart of dwelling units managed\nover time by regions')
ax[0][1].title.set_text('Heatmap of dwelling units managed\nover time by regions')
# Plotting the other 2
dwell_projected_p = dwell_projected.pivot('financial_year', 'regions', 'projected_ultimate_dwelling_units')
dwell_projected_p.plot(kind='bar', stacked=True, ax=ax[1][0])
dwell_units_ratio_p = dwell_units_ratio.pivot('financial_year', 'regions', 'dwelling_ratio')
sns.heatmap(dwell_units_ratio_p, annot=True, fmt='.2f', ax=ax[1][1])
# Adding the titles
ax[1][0].title.set_text('Stacked bar chart of projected dwelling units over time by regions')
ax[1][1].title.set_text('Heatmap of the ratio of dwelling units to projected dwelling units\nover time by regions')
# Saving the figure
plt.savefig('final_plots/prop_1.png', bbox_inches='tight')
pass
# Analysing change in land area over time
# Filtering data
dwell_land = dwell_regions[['financial_year', 'regions', 'residential_land_area']]
dwell_total_land = dwell_regions[['financial_year', 'regions', 'total_land_area']]
dwell_land_ratio = dwell_regions[['financial_year', 'regions', 'land_ratio']]
# Stripping the date to format str to show
dwell_land['financial_year'] = dwell_land['financial_year'].dt.strftime('%Y')
dwell_total_land['financial_year'] = dwell_total_land['financial_year'].dt.strftime('%Y')
dwell_land_ratio['financial_year'] = dwell_land_ratio['financial_year'].dt.strftime('%Y')
# Setting up the subplots
fig, ax = plt.subplots(1, 2, figsize=(20, 10), sharey=True)
# Plotting the 2 plots
dwell_land_p = dwell_land.pivot('financial_year', 'regions', 'residential_land_area')
dwell_land_p.plot(kind='bar', stacked=True, ax=ax[0])
dwell_total_land_p = dwell_total_land.pivot('financial_year', 'regions', 'total_land_area')
dwell_total_land_p.plot(kind='bar', stacked=True, ax=ax[1])
# Setting pos of legend
ax[0].legend(loc='upper left')
ax[1].legend(loc='upper left')
# Adding the titles
ax[0].title.set_text('Stacked bar chart of residential land area\nover time by regions')
ax[1].title.set_text('Stacked bar chart of total land area\nover time by regions')
# Saving the figure
plt.savefig('final_plots/prop_2_1.png', bbox_inches='tight')
# Getting heatmap of ratio
dwell_land_ratio_p = dwell_land_ratio.pivot('financial_year', 'regions', 'land_ratio')
# Plotting the heatmap
fig, ax = plt.subplots(1, 1, figsize=(10, 10))
sns.heatmap(dwell_land_ratio_p, annot=True, fmt='.2f', ax=ax)
# Setting the title
ax.title.set_text('Heatmap of the ratio of residential land area to total land area\nover time by region')
# Saving the figure
plt.savefig('final_plots/prop_2_2.png', bbox_inches='tight')
# Analysing change in number of amenities over time
# Filtering data
prop_amn_sum = prop_regions[['year_completed', 'regions', 'amenities_sum']]
prop_amn_ratio = prop_regions[['year_completed', 'regions', 'am_re_ratio']]
# Mainipulating the data to get it to have a cumulative sum
new_index = pd.MultiIndex.from_product([prop_amn_sum['year_completed'].unique(), prop_amn_sum['regions'].unique()], names=['year_completed', 'regions'])
amn_sum_year = prop_amn_sum.groupby(['year_completed', 'regions']).sum().reindex(new_index).reset_index()
amn_sum_year = amn_sum_year.sort_values(by=['year_completed', 'regions'])
amn_sum_year['amenities_sum'] = amn_sum_year.groupby(['regions'])['amenities_sum'].fillna(method='ffill')
amn_sum_year = amn_sum_year.fillna(0)
amn_sum_year['year_completed'] = amn_sum_year['year_completed'].dt.strftime('%Y')
amn_sum_year_p = amn_sum_year.pivot('year_completed', 'regions', 'amenities_sum')
# Mainipulating for ratio
amn_ratio_year = prop_amn_ratio.groupby(['year_completed', 'regions']).sum().reindex(new_index).reset_index()
amn_ratio_year = amn_ratio_year.sort_values(by=['year_completed', 'regions'])
amn_ratio_year['am_re_ratio'] = amn_ratio_year.groupby(['regions'])['am_re_ratio'].fillna(method='ffill')
amn_ratio_year = amn_ratio_year.fillna(0)
amn_ratio_year = amn_ratio_year.groupby([pd.Grouper(freq='10Y', key='year_completed'), 'regions'])['am_re_ratio'].mean().reset_index()
amn_ratio_year['year_completed'] = amn_ratio_year['year_completed'].dt.strftime('%Y')
amn_ratio_year_p = amn_ratio_year.pivot('year_completed', 'regions', 'am_re_ratio')
# Setting up the subplots
fig, ax = plt.subplots(2, 1, figsize=(20, 20))
# Plotting the barchar and heatmap
amn_sum_year_p.plot(kind='bar', stacked=True, ax=ax[0])
sns.heatmap(amn_ratio_year_p, annot=True, fmt='.2f')
# Adding the titles
ax[0].set_title('Stacked bar chart cumulative number of amenities over time by region', fontdict={'fontsize': 20})
ax[1].set_title('Heatmap of the ratio of amenities to residential units over time(every 10 years) by region', fontdict={'fontsize': 20})
# Saving the figure
plt.savefig('final_plots/prop_3.png', bbox_inches='tight')
| 0.511717 | 0.846387 |
# RadarCOVID-Report
## Data Extraction
```
import datetime
import json
import logging
import os
import shutil
import tempfile
import textwrap
import uuid
import matplotlib.pyplot as plt
import matplotlib.ticker
import numpy as np
import pandas as pd
import pycountry
import retry
import seaborn as sns
%matplotlib inline
current_working_directory = os.environ.get("PWD")
if current_working_directory:
os.chdir(current_working_directory)
sns.set()
matplotlib.rcParams["figure.figsize"] = (15, 6)
extraction_datetime = datetime.datetime.utcnow()
extraction_date = extraction_datetime.strftime("%Y-%m-%d")
extraction_previous_datetime = extraction_datetime - datetime.timedelta(days=1)
extraction_previous_date = extraction_previous_datetime.strftime("%Y-%m-%d")
extraction_date_with_hour = datetime.datetime.utcnow().strftime("%Y-%m-%d@%H")
current_hour = datetime.datetime.utcnow().hour
are_today_results_partial = current_hour != 23
```
### Constants
```
from Modules.ExposureNotification import exposure_notification_io
spain_region_country_code = "ES"
germany_region_country_code = "DE"
default_backend_identifier = spain_region_country_code
backend_generation_days = 7 * 2
daily_summary_days = 7 * 4 * 3
daily_plot_days = 7 * 4
tek_dumps_load_limit = daily_summary_days + 1
```
### Parameters
```
environment_backend_identifier = os.environ.get("RADARCOVID_REPORT__BACKEND_IDENTIFIER")
if environment_backend_identifier:
report_backend_identifier = environment_backend_identifier
else:
report_backend_identifier = default_backend_identifier
report_backend_identifier
environment_enable_multi_backend_download = \
os.environ.get("RADARCOVID_REPORT__ENABLE_MULTI_BACKEND_DOWNLOAD")
if environment_enable_multi_backend_download:
report_backend_identifiers = None
else:
report_backend_identifiers = [report_backend_identifier]
report_backend_identifiers
environment_invalid_shared_diagnoses_dates = \
os.environ.get("RADARCOVID_REPORT__INVALID_SHARED_DIAGNOSES_DATES")
if environment_invalid_shared_diagnoses_dates:
invalid_shared_diagnoses_dates = environment_invalid_shared_diagnoses_dates.split(",")
else:
invalid_shared_diagnoses_dates = []
invalid_shared_diagnoses_dates
```
### COVID-19 Cases
```
report_backend_client = \
exposure_notification_io.get_backend_client_with_identifier(
backend_identifier=report_backend_identifier)
@retry.retry(tries=10, delay=10, backoff=1.1, jitter=(0, 10))
def download_cases_dataframe():
return pd.read_csv(
"https://raw.githubusercontent.com/owid/covid-19-data/master/public/data/owid-covid-data.csv")
confirmed_df_ = download_cases_dataframe()
confirmed_df = confirmed_df_.copy()
confirmed_df = confirmed_df[["date", "new_cases", "iso_code"]]
confirmed_df.rename(
columns={
"date": "sample_date",
"iso_code": "country_code",
},
inplace=True)
def convert_iso_alpha_3_to_alpha_2(x):
try:
return pycountry.countries.get(alpha_3=x).alpha_2
except Exception as e:
logging.info(f"Error converting country ISO Alpha 3 code '{x}': {repr(e)}")
return None
confirmed_df["country_code"] = confirmed_df.country_code.apply(convert_iso_alpha_3_to_alpha_2)
confirmed_df.dropna(inplace=True)
confirmed_df["sample_date"] = pd.to_datetime(confirmed_df.sample_date, dayfirst=True)
confirmed_df["sample_date"] = confirmed_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_df.sort_values("sample_date", inplace=True)
confirmed_df.tail()
confirmed_days = pd.date_range(
start=confirmed_df.iloc[0].sample_date,
end=extraction_datetime)
confirmed_days_df = pd.DataFrame(data=confirmed_days, columns=["sample_date"])
confirmed_days_df["sample_date_string"] = \
confirmed_days_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_days_df.tail()
def sort_source_regions_for_display(source_regions: list) -> list:
if report_backend_identifier in source_regions:
source_regions = [report_backend_identifier] + \
list(sorted(set(source_regions).difference([report_backend_identifier])))
else:
source_regions = list(sorted(source_regions))
return source_regions
report_source_regions = report_backend_client.source_regions_for_date(
date=extraction_datetime.date())
report_source_regions = sort_source_regions_for_display(
source_regions=report_source_regions)
report_source_regions
def get_cases_dataframe(source_regions_for_date_function, columns_suffix=None):
source_regions_at_date_df = confirmed_days_df.copy()
source_regions_at_date_df["source_regions_at_date"] = \
source_regions_at_date_df.sample_date.apply(
lambda x: source_regions_for_date_function(date=x))
source_regions_at_date_df.sort_values("sample_date", inplace=True)
source_regions_at_date_df["_source_regions_group"] = source_regions_at_date_df. \
source_regions_at_date.apply(lambda x: ",".join(sort_source_regions_for_display(x)))
source_regions_at_date_df.tail()
#%%
source_regions_for_summary_df_ = \
source_regions_at_date_df[["sample_date", "_source_regions_group"]].copy()
source_regions_for_summary_df_.rename(columns={"_source_regions_group": "source_regions"}, inplace=True)
source_regions_for_summary_df_.tail()
#%%
confirmed_output_columns = ["sample_date", "new_cases", "covid_cases"]
confirmed_output_df = pd.DataFrame(columns=confirmed_output_columns)
for source_regions_group, source_regions_group_series in \
source_regions_at_date_df.groupby("_source_regions_group"):
source_regions_set = set(source_regions_group.split(","))
confirmed_source_regions_set_df = \
confirmed_df[confirmed_df.country_code.isin(source_regions_set)].copy()
confirmed_source_regions_group_df = \
confirmed_source_regions_set_df.groupby("sample_date").new_cases.sum() \
.reset_index().sort_values("sample_date")
confirmed_source_regions_group_df["covid_cases"] = \
confirmed_source_regions_group_df.new_cases.rolling(7, min_periods=0).mean().round()
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[confirmed_output_columns]
confirmed_source_regions_group_df.fillna(method="ffill", inplace=True)
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[
confirmed_source_regions_group_df.sample_date.isin(
source_regions_group_series.sample_date_string)]
confirmed_output_df = confirmed_output_df.append(confirmed_source_regions_group_df)
result_df = confirmed_output_df.copy()
result_df.tail()
#%%
result_df.rename(columns={"sample_date": "sample_date_string"}, inplace=True)
result_df = confirmed_days_df[["sample_date_string"]].merge(result_df, how="left")
result_df.sort_values("sample_date_string", inplace=True)
result_df.fillna(method="ffill", inplace=True)
result_df.tail()
#%%
result_df[["new_cases", "covid_cases"]].plot()
if columns_suffix:
result_df.rename(
columns={
"new_cases": "new_cases_" + columns_suffix,
"covid_cases": "covid_cases_" + columns_suffix},
inplace=True)
return result_df, source_regions_for_summary_df_
confirmed_eu_df, source_regions_for_summary_df = get_cases_dataframe(
report_backend_client.source_regions_for_date)
confirmed_es_df, _ = get_cases_dataframe(
lambda date: [spain_region_country_code],
columns_suffix=spain_region_country_code.lower())
```
### Extract API TEKs
```
raw_zip_path_prefix = "Data/TEKs/Raw/"
fail_on_error_backend_identifiers = [report_backend_identifier]
multi_backend_exposure_keys_df = \
exposure_notification_io.download_exposure_keys_from_backends(
backend_identifiers=report_backend_identifiers,
generation_days=backend_generation_days,
fail_on_error_backend_identifiers=fail_on_error_backend_identifiers,
save_raw_zip_path_prefix=raw_zip_path_prefix)
multi_backend_exposure_keys_df["region"] = multi_backend_exposure_keys_df["backend_identifier"]
multi_backend_exposure_keys_df.rename(
columns={
"generation_datetime": "sample_datetime",
"generation_date_string": "sample_date_string",
},
inplace=True)
multi_backend_exposure_keys_df.head()
early_teks_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.rolling_period < 144].copy()
early_teks_df["rolling_period_in_hours"] = early_teks_df.rolling_period / 6
early_teks_df[early_teks_df.sample_date_string != extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
early_teks_df[early_teks_df.sample_date_string == extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
multi_backend_exposure_keys_df = multi_backend_exposure_keys_df[[
"sample_date_string", "region", "key_data"]]
multi_backend_exposure_keys_df.head()
active_regions = \
multi_backend_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
active_regions
multi_backend_summary_df = multi_backend_exposure_keys_df.groupby(
["sample_date_string", "region"]).key_data.nunique().reset_index() \
.pivot(index="sample_date_string", columns="region") \
.sort_index(ascending=False)
multi_backend_summary_df.rename(
columns={"key_data": "shared_teks_by_generation_date"},
inplace=True)
multi_backend_summary_df.rename_axis("sample_date", inplace=True)
multi_backend_summary_df = multi_backend_summary_df.fillna(0).astype(int)
multi_backend_summary_df = multi_backend_summary_df.head(backend_generation_days)
multi_backend_summary_df.head()
def compute_keys_cross_sharing(x):
teks_x = x.key_data_x.item()
common_teks = set(teks_x).intersection(x.key_data_y.item())
common_teks_fraction = len(common_teks) / len(teks_x)
return pd.Series(dict(
common_teks=common_teks,
common_teks_fraction=common_teks_fraction,
))
multi_backend_exposure_keys_by_region_df = \
multi_backend_exposure_keys_df.groupby("region").key_data.unique().reset_index()
multi_backend_exposure_keys_by_region_df["_merge"] = True
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_df.merge(
multi_backend_exposure_keys_by_region_df, on="_merge")
multi_backend_exposure_keys_by_region_combination_df.drop(
columns=["_merge"], inplace=True)
if multi_backend_exposure_keys_by_region_combination_df.region_x.nunique() > 1:
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_combination_df[
multi_backend_exposure_keys_by_region_combination_df.region_x !=
multi_backend_exposure_keys_by_region_combination_df.region_y]
multi_backend_exposure_keys_cross_sharing_df = \
multi_backend_exposure_keys_by_region_combination_df \
.groupby(["region_x", "region_y"]) \
.apply(compute_keys_cross_sharing) \
.reset_index()
multi_backend_cross_sharing_summary_df = \
multi_backend_exposure_keys_cross_sharing_df.pivot_table(
values=["common_teks_fraction"],
columns="region_x",
index="region_y",
aggfunc=lambda x: x.item())
multi_backend_cross_sharing_summary_df
multi_backend_without_active_region_exposure_keys_df = \
multi_backend_exposure_keys_df[multi_backend_exposure_keys_df.region != report_backend_identifier]
multi_backend_without_active_region = \
multi_backend_without_active_region_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
multi_backend_without_active_region
exposure_keys_summary_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.region == report_backend_identifier]
exposure_keys_summary_df.drop(columns=["region"], inplace=True)
exposure_keys_summary_df = \
exposure_keys_summary_df.groupby(["sample_date_string"]).key_data.nunique().to_frame()
exposure_keys_summary_df = \
exposure_keys_summary_df.reset_index().set_index("sample_date_string")
exposure_keys_summary_df.sort_index(ascending=False, inplace=True)
exposure_keys_summary_df.rename(columns={"key_data": "shared_teks_by_generation_date"}, inplace=True)
exposure_keys_summary_df.head()
```
### Dump API TEKs
```
tek_list_df = multi_backend_exposure_keys_df[
["sample_date_string", "region", "key_data"]].copy()
tek_list_df["key_data"] = tek_list_df["key_data"].apply(str)
tek_list_df.rename(columns={
"sample_date_string": "sample_date",
"key_data": "tek_list"}, inplace=True)
tek_list_df = tek_list_df.groupby(
["sample_date", "region"]).tek_list.unique().reset_index()
tek_list_df["extraction_date"] = extraction_date
tek_list_df["extraction_date_with_hour"] = extraction_date_with_hour
tek_list_path_prefix = "Data/TEKs/"
tek_list_current_path = tek_list_path_prefix + f"/Current/RadarCOVID-TEKs.json"
tek_list_daily_path = tek_list_path_prefix + f"Daily/RadarCOVID-TEKs-{extraction_date}.json"
tek_list_hourly_path = tek_list_path_prefix + f"Hourly/RadarCOVID-TEKs-{extraction_date_with_hour}.json"
for path in [tek_list_current_path, tek_list_daily_path, tek_list_hourly_path]:
os.makedirs(os.path.dirname(path), exist_ok=True)
tek_list_df.drop(columns=["extraction_date", "extraction_date_with_hour"]).to_json(
tek_list_current_path,
lines=True, orient="records")
tek_list_df.drop(columns=["extraction_date_with_hour"]).to_json(
tek_list_daily_path,
lines=True, orient="records")
tek_list_df.to_json(
tek_list_hourly_path,
lines=True, orient="records")
tek_list_df.head()
```
### Load TEK Dumps
```
import glob
def load_extracted_teks(mode, region=None, limit=None) -> pd.DataFrame:
extracted_teks_df = pd.DataFrame(columns=["region"])
file_paths = list(reversed(sorted(glob.glob(tek_list_path_prefix + mode + "/RadarCOVID-TEKs-*.json"))))
if limit:
file_paths = file_paths[:limit]
for file_path in file_paths:
logging.info(f"Loading TEKs from '{file_path}'...")
iteration_extracted_teks_df = pd.read_json(file_path, lines=True)
extracted_teks_df = extracted_teks_df.append(
iteration_extracted_teks_df, sort=False)
extracted_teks_df["region"] = \
extracted_teks_df.region.fillna(spain_region_country_code).copy()
if region:
extracted_teks_df = \
extracted_teks_df[extracted_teks_df.region == region]
return extracted_teks_df
daily_extracted_teks_df = load_extracted_teks(
mode="Daily",
region=report_backend_identifier,
limit=tek_dumps_load_limit)
daily_extracted_teks_df.head()
exposure_keys_summary_df_ = daily_extracted_teks_df \
.sort_values("extraction_date", ascending=False) \
.groupby("sample_date").tek_list.first() \
.to_frame()
exposure_keys_summary_df_.index.name = "sample_date_string"
exposure_keys_summary_df_["tek_list"] = \
exposure_keys_summary_df_.tek_list.apply(len)
exposure_keys_summary_df_ = exposure_keys_summary_df_ \
.rename(columns={"tek_list": "shared_teks_by_generation_date"}) \
.sort_index(ascending=False)
exposure_keys_summary_df = exposure_keys_summary_df_
exposure_keys_summary_df.head()
```
### Daily New TEKs
```
tek_list_df = daily_extracted_teks_df.groupby("extraction_date").tek_list.apply(
lambda x: set(sum(x, []))).reset_index()
tek_list_df = tek_list_df.set_index("extraction_date").sort_index(ascending=True)
tek_list_df.head()
def compute_teks_by_generation_and_upload_date(date):
day_new_teks_set_df = tek_list_df.copy().diff()
try:
day_new_teks_set = day_new_teks_set_df[
day_new_teks_set_df.index == date].tek_list.item()
except ValueError:
day_new_teks_set = None
if pd.isna(day_new_teks_set):
day_new_teks_set = set()
day_new_teks_df = daily_extracted_teks_df[
daily_extracted_teks_df.extraction_date == date].copy()
day_new_teks_df["shared_teks"] = \
day_new_teks_df.tek_list.apply(lambda x: set(x).intersection(day_new_teks_set))
day_new_teks_df["shared_teks"] = \
day_new_teks_df.shared_teks.apply(len)
day_new_teks_df["upload_date"] = date
day_new_teks_df.rename(columns={"sample_date": "generation_date"}, inplace=True)
day_new_teks_df = day_new_teks_df[
["upload_date", "generation_date", "shared_teks"]]
day_new_teks_df["generation_to_upload_days"] = \
(pd.to_datetime(day_new_teks_df.upload_date) -
pd.to_datetime(day_new_teks_df.generation_date)).dt.days
day_new_teks_df = day_new_teks_df[day_new_teks_df.shared_teks > 0]
return day_new_teks_df
shared_teks_generation_to_upload_df = pd.DataFrame()
for upload_date in daily_extracted_teks_df.extraction_date.unique():
shared_teks_generation_to_upload_df = \
shared_teks_generation_to_upload_df.append(
compute_teks_by_generation_and_upload_date(date=upload_date))
shared_teks_generation_to_upload_df \
.sort_values(["upload_date", "generation_date"], ascending=False, inplace=True)
shared_teks_generation_to_upload_df.tail()
today_new_teks_df = \
shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.upload_date == extraction_date].copy()
today_new_teks_df.tail()
if not today_new_teks_df.empty:
today_new_teks_df.set_index("generation_to_upload_days") \
.sort_index().shared_teks.plot.bar()
generation_to_upload_period_pivot_df = \
shared_teks_generation_to_upload_df[
["upload_date", "generation_to_upload_days", "shared_teks"]] \
.pivot(index="upload_date", columns="generation_to_upload_days") \
.sort_index(ascending=False).fillna(0).astype(int) \
.droplevel(level=0, axis=1)
generation_to_upload_period_pivot_df.head()
new_tek_df = tek_list_df.diff().tek_list.apply(
lambda x: len(x) if not pd.isna(x) else None).to_frame().reset_index()
new_tek_df.rename(columns={
"tek_list": "shared_teks_by_upload_date",
"extraction_date": "sample_date_string",}, inplace=True)
new_tek_df.tail()
shared_teks_uploaded_on_generation_date_df = shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.generation_to_upload_days == 0] \
[["upload_date", "shared_teks"]].rename(
columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_teks_uploaded_on_generation_date",
})
shared_teks_uploaded_on_generation_date_df.head()
estimated_shared_diagnoses_df = shared_teks_generation_to_upload_df \
.groupby(["upload_date"]).shared_teks.max().reset_index() \
.sort_values(["upload_date"], ascending=False) \
.rename(columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_diagnoses",
})
invalid_shared_diagnoses_dates_mask = \
estimated_shared_diagnoses_df.sample_date_string.isin(invalid_shared_diagnoses_dates)
estimated_shared_diagnoses_df[invalid_shared_diagnoses_dates_mask] = 0
estimated_shared_diagnoses_df.head()
```
### Hourly New TEKs
```
hourly_extracted_teks_df = load_extracted_teks(
mode="Hourly", region=report_backend_identifier, limit=25)
hourly_extracted_teks_df.head()
hourly_new_tek_count_df = hourly_extracted_teks_df \
.groupby("extraction_date_with_hour").tek_list. \
apply(lambda x: set(sum(x, []))).reset_index().copy()
hourly_new_tek_count_df = hourly_new_tek_count_df.set_index("extraction_date_with_hour") \
.sort_index(ascending=True)
hourly_new_tek_count_df["new_tek_list"] = hourly_new_tek_count_df.tek_list.diff()
hourly_new_tek_count_df["new_tek_count"] = hourly_new_tek_count_df.new_tek_list.apply(
lambda x: len(x) if not pd.isna(x) else 0)
hourly_new_tek_count_df.rename(columns={
"new_tek_count": "shared_teks_by_upload_date"}, inplace=True)
hourly_new_tek_count_df = hourly_new_tek_count_df.reset_index()[[
"extraction_date_with_hour", "shared_teks_by_upload_date"]]
hourly_new_tek_count_df.head()
hourly_summary_df = hourly_new_tek_count_df.copy()
hourly_summary_df.set_index("extraction_date_with_hour", inplace=True)
hourly_summary_df = hourly_summary_df.fillna(0).astype(int).reset_index()
hourly_summary_df["datetime_utc"] = pd.to_datetime(
hourly_summary_df.extraction_date_with_hour, format="%Y-%m-%d@%H")
hourly_summary_df.set_index("datetime_utc", inplace=True)
hourly_summary_df = hourly_summary_df.tail(-1)
hourly_summary_df.head()
```
### Official Statistics
```
import requests
import pandas.io.json
official_stats_response = requests.get("https://radarcovidpre.covid19.gob.es/kpi/statistics/basics")
official_stats_response.raise_for_status()
official_stats_df_ = pandas.io.json.json_normalize(official_stats_response.json())
official_stats_df = official_stats_df_.copy()
official_stats_df = official_stats_df.append(pd.DataFrame({
"date": ["06/12/2020"],
"applicationsDownloads.totalAcummulated": [5653519],
"communicatedContagions.totalAcummulated": [21925],
}), sort=False)
official_stats_df["date"] = pd.to_datetime(official_stats_df["date"], dayfirst=True)
official_stats_df.head()
official_stats_column_map = {
"date": "sample_date",
"applicationsDownloads.totalAcummulated": "app_downloads_es_accumulated",
"communicatedContagions.totalAcummulated": "shared_diagnoses_es_accumulated",
}
accumulated_suffix = "_accumulated"
accumulated_values_columns = \
list(filter(lambda x: x.endswith(accumulated_suffix), official_stats_column_map.values()))
interpolated_values_columns = \
list(map(lambda x: x[:-len(accumulated_suffix)], accumulated_values_columns))
official_stats_df = \
official_stats_df[official_stats_column_map.keys()] \
.rename(columns=official_stats_column_map)
official_stats_df.head()
official_stats_df = confirmed_days_df.merge(official_stats_df, how="left")
official_stats_df.sort_values("sample_date", ascending=False, inplace=True)
official_stats_df.head()
official_stats_df[accumulated_values_columns] = \
official_stats_df[accumulated_values_columns] \
.astype(float).interpolate(limit_area="inside")
official_stats_df[interpolated_values_columns] = \
official_stats_df[accumulated_values_columns].diff(periods=-1)
official_stats_df.drop(columns="sample_date", inplace=True)
official_stats_df.head()
```
### Data Merge
```
result_summary_df = exposure_keys_summary_df.merge(
new_tek_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
shared_teks_uploaded_on_generation_date_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
estimated_shared_diagnoses_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
official_stats_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = confirmed_eu_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df = confirmed_es_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df["sample_date"] = pd.to_datetime(result_summary_df.sample_date_string)
result_summary_df = result_summary_df.merge(source_regions_for_summary_df, how="left")
result_summary_df.set_index(["sample_date", "source_regions"], inplace=True)
result_summary_df.drop(columns=["sample_date_string"], inplace=True)
result_summary_df.sort_index(ascending=False, inplace=True)
result_summary_df.head()
with pd.option_context("mode.use_inf_as_na", True):
result_summary_df = result_summary_df.fillna(0).astype(int)
result_summary_df["teks_per_shared_diagnosis"] = \
(result_summary_df.shared_teks_by_upload_date / result_summary_df.shared_diagnoses).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case"] = \
(result_summary_df.shared_diagnoses / result_summary_df.covid_cases).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(result_summary_df.shared_diagnoses_es / result_summary_df.covid_cases_es).fillna(0)
result_summary_df.head(daily_plot_days)
weekly_result_summary_df = result_summary_df \
.sort_index(ascending=True).fillna(0).rolling(7).agg({
"covid_cases": "sum",
"covid_cases_es": "sum",
"shared_teks_by_generation_date": "sum",
"shared_teks_by_upload_date": "sum",
"shared_diagnoses": "sum",
"shared_diagnoses_es": "sum",
}).sort_index(ascending=False)
with pd.option_context("mode.use_inf_as_na", True):
weekly_result_summary_df = weekly_result_summary_df.fillna(0).astype(int)
weekly_result_summary_df["teks_per_shared_diagnosis"] = \
(weekly_result_summary_df.shared_teks_by_upload_date / weekly_result_summary_df.shared_diagnoses).fillna(0)
weekly_result_summary_df["shared_diagnoses_per_covid_case"] = \
(weekly_result_summary_df.shared_diagnoses / weekly_result_summary_df.covid_cases).fillna(0)
weekly_result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(weekly_result_summary_df.shared_diagnoses_es / weekly_result_summary_df.covid_cases_es).fillna(0)
weekly_result_summary_df.head()
last_7_days_summary = weekly_result_summary_df.to_dict(orient="records")[1]
last_7_days_summary
```
## Report Results
```
display_column_name_mapping = {
"sample_date": "Sample\u00A0Date\u00A0(UTC)",
"source_regions": "Source Countries",
"datetime_utc": "Timestamp (UTC)",
"upload_date": "Upload Date (UTC)",
"generation_to_upload_days": "Generation to Upload Period in Days",
"region": "Backend",
"region_x": "Backend\u00A0(A)",
"region_y": "Backend\u00A0(B)",
"common_teks": "Common TEKs Shared Between Backends",
"common_teks_fraction": "Fraction of TEKs in Backend (A) Available in Backend (B)",
"covid_cases": "COVID-19 Cases (Source Countries)",
"shared_teks_by_generation_date": "Shared TEKs by Generation Date (Source Countries)",
"shared_teks_by_upload_date": "Shared TEKs by Upload Date (Source Countries)",
"shared_teks_uploaded_on_generation_date": "Shared TEKs Uploaded on Generation Date (Source Countries)",
"shared_diagnoses": "Shared Diagnoses (Source Countries – Estimation)",
"teks_per_shared_diagnosis": "TEKs Uploaded per Shared Diagnosis (Source Countries)",
"shared_diagnoses_per_covid_case": "Usage Ratio (Source Countries)",
"covid_cases_es": "COVID-19 Cases (Spain)",
"app_downloads_es": "App Downloads (Spain – Official)",
"shared_diagnoses_es": "Shared Diagnoses (Spain – Official)",
"shared_diagnoses_per_covid_case_es": "Usage Ratio (Spain)",
}
summary_columns = [
"covid_cases",
"shared_teks_by_generation_date",
"shared_teks_by_upload_date",
"shared_teks_uploaded_on_generation_date",
"shared_diagnoses",
"teks_per_shared_diagnosis",
"shared_diagnoses_per_covid_case",
"covid_cases_es",
"app_downloads_es",
"shared_diagnoses_es",
"shared_diagnoses_per_covid_case_es",
]
summary_percentage_columns= [
"shared_diagnoses_per_covid_case_es",
"shared_diagnoses_per_covid_case",
]
```
### Daily Summary Table
```
result_summary_df_ = result_summary_df.copy()
result_summary_df = result_summary_df[summary_columns]
result_summary_with_display_names_df = result_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
result_summary_with_display_names_df
```
### Daily Summary Plots
```
result_plot_summary_df = result_summary_df.head(daily_plot_days)[summary_columns] \
.droplevel(level=["source_regions"]) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
summary_ax_list = result_plot_summary_df.sort_index(ascending=True).plot.bar(
title=f"Daily Summary",
rot=45, subplots=True, figsize=(15, 30), legend=False)
ax_ = summary_ax_list[0]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.95)
_ = ax_.set_xticklabels(sorted(result_plot_summary_df.index.strftime("%Y-%m-%d").tolist()))
for percentage_column in summary_percentage_columns:
percentage_column_index = summary_columns.index(percentage_column)
summary_ax_list[percentage_column_index].yaxis \
.set_major_formatter(matplotlib.ticker.PercentFormatter(1.0))
```
### Daily Generation to Upload Period Table
```
display_generation_to_upload_period_pivot_df = \
generation_to_upload_period_pivot_df \
.head(backend_generation_days)
display_generation_to_upload_period_pivot_df \
.head(backend_generation_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping)
fig, generation_to_upload_period_pivot_table_ax = plt.subplots(
figsize=(12, 1 + 0.6 * len(display_generation_to_upload_period_pivot_df)))
generation_to_upload_period_pivot_table_ax.set_title(
"Shared TEKs Generation to Upload Period Table")
sns.heatmap(
data=display_generation_to_upload_period_pivot_df
.rename_axis(columns=display_column_name_mapping)
.rename_axis(index=display_column_name_mapping),
fmt=".0f",
annot=True,
ax=generation_to_upload_period_pivot_table_ax)
generation_to_upload_period_pivot_table_ax.get_figure().tight_layout()
```
### Hourly Summary Plots
```
hourly_summary_ax_list = hourly_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.plot.bar(
title=f"Last 24h Summary",
rot=45, subplots=True, legend=False)
ax_ = hourly_summary_ax_list[-1]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.9)
_ = ax_.set_xticklabels(sorted(hourly_summary_df.index.strftime("%Y-%m-%d@%H").tolist()))
```
### Publish Results
```
github_repository = os.environ.get("GITHUB_REPOSITORY")
if github_repository is None:
github_repository = "pvieito/Radar-STATS"
github_project_base_url = "https://github.com/" + github_repository
display_formatters = {
display_column_name_mapping["teks_per_shared_diagnosis"]: lambda x: f"{x:.2f}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case"]: lambda x: f"{x:.2%}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case_es"]: lambda x: f"{x:.2%}" if x != 0 else "",
}
general_columns = \
list(filter(lambda x: x not in display_formatters, display_column_name_mapping.values()))
general_formatter = lambda x: f"{x}" if x != 0 else ""
display_formatters.update(dict(map(lambda x: (x, general_formatter), general_columns)))
daily_summary_table_html = result_summary_with_display_names_df \
.head(daily_plot_days) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.to_html(formatters=display_formatters)
multi_backend_summary_table_html = multi_backend_summary_df \
.head(daily_plot_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(formatters=display_formatters)
def format_multi_backend_cross_sharing_fraction(x):
if pd.isna(x):
return "-"
elif round(x * 100, 1) == 0:
return ""
else:
return f"{x:.1%}"
multi_backend_cross_sharing_summary_table_html = multi_backend_cross_sharing_summary_df \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(
classes="table-center",
formatters=display_formatters,
float_format=format_multi_backend_cross_sharing_fraction)
multi_backend_cross_sharing_summary_table_html = \
multi_backend_cross_sharing_summary_table_html \
.replace("<tr>","<tr style=\"text-align: center;\">")
extraction_date_result_summary_df = \
result_summary_df[result_summary_df.index.get_level_values("sample_date") == extraction_date]
extraction_date_result_hourly_summary_df = \
hourly_summary_df[hourly_summary_df.extraction_date_with_hour == extraction_date_with_hour]
covid_cases = \
extraction_date_result_summary_df.covid_cases.item()
shared_teks_by_generation_date = \
extraction_date_result_summary_df.shared_teks_by_generation_date.item()
shared_teks_by_upload_date = \
extraction_date_result_summary_df.shared_teks_by_upload_date.item()
shared_diagnoses = \
extraction_date_result_summary_df.shared_diagnoses.item()
teks_per_shared_diagnosis = \
extraction_date_result_summary_df.teks_per_shared_diagnosis.item()
shared_diagnoses_per_covid_case = \
extraction_date_result_summary_df.shared_diagnoses_per_covid_case.item()
shared_teks_by_upload_date_last_hour = \
extraction_date_result_hourly_summary_df.shared_teks_by_upload_date.sum().astype(int)
display_source_regions = ", ".join(report_source_regions)
if len(report_source_regions) == 1:
display_brief_source_regions = report_source_regions[0]
else:
display_brief_source_regions = f"{len(report_source_regions)} 🇪🇺"
def get_temporary_image_path() -> str:
return os.path.join(tempfile.gettempdir(), str(uuid.uuid4()) + ".png")
def save_temporary_plot_image(ax):
if isinstance(ax, np.ndarray):
ax = ax[0]
media_path = get_temporary_image_path()
ax.get_figure().savefig(media_path)
return media_path
def save_temporary_dataframe_image(df):
import dataframe_image as dfi
df = df.copy()
df_styler = df.style.format(display_formatters)
media_path = get_temporary_image_path()
dfi.export(df_styler, media_path)
return media_path
summary_plots_image_path = save_temporary_plot_image(
ax=summary_ax_list)
summary_table_image_path = save_temporary_dataframe_image(
df=result_summary_with_display_names_df)
hourly_summary_plots_image_path = save_temporary_plot_image(
ax=hourly_summary_ax_list)
multi_backend_summary_table_image_path = save_temporary_dataframe_image(
df=multi_backend_summary_df)
generation_to_upload_period_pivot_table_image_path = save_temporary_plot_image(
ax=generation_to_upload_period_pivot_table_ax)
```
### Save Results
```
report_resources_path_prefix = "Data/Resources/Current/RadarCOVID-Report-"
result_summary_df.to_csv(
report_resources_path_prefix + "Summary-Table.csv")
result_summary_df.to_html(
report_resources_path_prefix + "Summary-Table.html")
hourly_summary_df.to_csv(
report_resources_path_prefix + "Hourly-Summary-Table.csv")
multi_backend_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Summary-Table.csv")
multi_backend_cross_sharing_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Cross-Sharing-Summary-Table.csv")
generation_to_upload_period_pivot_df.to_csv(
report_resources_path_prefix + "Generation-Upload-Period-Table.csv")
_ = shutil.copyfile(
summary_plots_image_path,
report_resources_path_prefix + "Summary-Plots.png")
_ = shutil.copyfile(
summary_table_image_path,
report_resources_path_prefix + "Summary-Table.png")
_ = shutil.copyfile(
hourly_summary_plots_image_path,
report_resources_path_prefix + "Hourly-Summary-Plots.png")
_ = shutil.copyfile(
multi_backend_summary_table_image_path,
report_resources_path_prefix + "Multi-Backend-Summary-Table.png")
_ = shutil.copyfile(
generation_to_upload_period_pivot_table_image_path,
report_resources_path_prefix + "Generation-Upload-Period-Table.png")
```
### Publish Results as JSON
```
def generate_summary_api_results(df: pd.DataFrame) -> list:
api_df = df.reset_index().copy()
api_df["sample_date_string"] = \
api_df["sample_date"].dt.strftime("%Y-%m-%d")
api_df["source_regions"] = \
api_df["source_regions"].apply(lambda x: x.split(","))
return api_df.to_dict(orient="records")
summary_api_results = \
generate_summary_api_results(df=result_summary_df)
today_summary_api_results = \
generate_summary_api_results(df=extraction_date_result_summary_df)[0]
summary_results = dict(
backend_identifier=report_backend_identifier,
source_regions=report_source_regions,
extraction_datetime=extraction_datetime,
extraction_date=extraction_date,
extraction_date_with_hour=extraction_date_with_hour,
last_hour=dict(
shared_teks_by_upload_date=shared_teks_by_upload_date_last_hour,
shared_diagnoses=0,
),
today=today_summary_api_results,
last_7_days=last_7_days_summary,
daily_results=summary_api_results)
summary_results = \
json.loads(pd.Series([summary_results]).to_json(orient="records"))[0]
with open(report_resources_path_prefix + "Summary-Results.json", "w") as f:
json.dump(summary_results, f, indent=4)
```
### Publish on README
```
with open("Data/Templates/README.md", "r") as f:
readme_contents = f.read()
readme_contents = readme_contents.format(
extraction_date_with_hour=extraction_date_with_hour,
github_project_base_url=github_project_base_url,
daily_summary_table_html=daily_summary_table_html,
multi_backend_summary_table_html=multi_backend_summary_table_html,
multi_backend_cross_sharing_summary_table_html=multi_backend_cross_sharing_summary_table_html,
display_source_regions=display_source_regions)
with open("README.md", "w") as f:
f.write(readme_contents)
```
### Publish on Twitter
```
enable_share_to_twitter = os.environ.get("RADARCOVID_REPORT__ENABLE_PUBLISH_ON_TWITTER")
github_event_name = os.environ.get("GITHUB_EVENT_NAME")
if enable_share_to_twitter and github_event_name == "schedule" and \
(shared_teks_by_upload_date_last_hour or not are_today_results_partial):
import tweepy
twitter_api_auth_keys = os.environ["RADARCOVID_REPORT__TWITTER_API_AUTH_KEYS"]
twitter_api_auth_keys = twitter_api_auth_keys.split(":")
auth = tweepy.OAuthHandler(twitter_api_auth_keys[0], twitter_api_auth_keys[1])
auth.set_access_token(twitter_api_auth_keys[2], twitter_api_auth_keys[3])
api = tweepy.API(auth)
summary_plots_media = api.media_upload(summary_plots_image_path)
summary_table_media = api.media_upload(summary_table_image_path)
generation_to_upload_period_pivot_table_image_media = api.media_upload(generation_to_upload_period_pivot_table_image_path)
media_ids = [
summary_plots_media.media_id,
summary_table_media.media_id,
generation_to_upload_period_pivot_table_image_media.media_id,
]
if are_today_results_partial:
today_addendum = " (Partial)"
else:
today_addendum = ""
status = textwrap.dedent(f"""
#RadarCOVID – {extraction_date_with_hour}
Source Countries: {display_brief_source_regions}
Today{today_addendum}:
- Uploaded TEKs: {shared_teks_by_upload_date:.0f} ({shared_teks_by_upload_date_last_hour:+d} last hour)
- Shared Diagnoses: ≤{shared_diagnoses:.0f}
- Usage Ratio: ≤{shared_diagnoses_per_covid_case:.2%}
Last 7 Days:
- Shared Diagnoses: ≤{last_7_days_summary["shared_diagnoses"]:.0f}
- Usage Ratio: ≤{last_7_days_summary["shared_diagnoses_per_covid_case"]:.2%}
Info: {github_project_base_url}#documentation
""")
status = status.encode(encoding="utf-8")
api.update_status(status=status, media_ids=media_ids)
```
|
github_jupyter
|
import datetime
import json
import logging
import os
import shutil
import tempfile
import textwrap
import uuid
import matplotlib.pyplot as plt
import matplotlib.ticker
import numpy as np
import pandas as pd
import pycountry
import retry
import seaborn as sns
%matplotlib inline
current_working_directory = os.environ.get("PWD")
if current_working_directory:
os.chdir(current_working_directory)
sns.set()
matplotlib.rcParams["figure.figsize"] = (15, 6)
extraction_datetime = datetime.datetime.utcnow()
extraction_date = extraction_datetime.strftime("%Y-%m-%d")
extraction_previous_datetime = extraction_datetime - datetime.timedelta(days=1)
extraction_previous_date = extraction_previous_datetime.strftime("%Y-%m-%d")
extraction_date_with_hour = datetime.datetime.utcnow().strftime("%Y-%m-%d@%H")
current_hour = datetime.datetime.utcnow().hour
are_today_results_partial = current_hour != 23
from Modules.ExposureNotification import exposure_notification_io
spain_region_country_code = "ES"
germany_region_country_code = "DE"
default_backend_identifier = spain_region_country_code
backend_generation_days = 7 * 2
daily_summary_days = 7 * 4 * 3
daily_plot_days = 7 * 4
tek_dumps_load_limit = daily_summary_days + 1
environment_backend_identifier = os.environ.get("RADARCOVID_REPORT__BACKEND_IDENTIFIER")
if environment_backend_identifier:
report_backend_identifier = environment_backend_identifier
else:
report_backend_identifier = default_backend_identifier
report_backend_identifier
environment_enable_multi_backend_download = \
os.environ.get("RADARCOVID_REPORT__ENABLE_MULTI_BACKEND_DOWNLOAD")
if environment_enable_multi_backend_download:
report_backend_identifiers = None
else:
report_backend_identifiers = [report_backend_identifier]
report_backend_identifiers
environment_invalid_shared_diagnoses_dates = \
os.environ.get("RADARCOVID_REPORT__INVALID_SHARED_DIAGNOSES_DATES")
if environment_invalid_shared_diagnoses_dates:
invalid_shared_diagnoses_dates = environment_invalid_shared_diagnoses_dates.split(",")
else:
invalid_shared_diagnoses_dates = []
invalid_shared_diagnoses_dates
report_backend_client = \
exposure_notification_io.get_backend_client_with_identifier(
backend_identifier=report_backend_identifier)
@retry.retry(tries=10, delay=10, backoff=1.1, jitter=(0, 10))
def download_cases_dataframe():
return pd.read_csv(
"https://raw.githubusercontent.com/owid/covid-19-data/master/public/data/owid-covid-data.csv")
confirmed_df_ = download_cases_dataframe()
confirmed_df = confirmed_df_.copy()
confirmed_df = confirmed_df[["date", "new_cases", "iso_code"]]
confirmed_df.rename(
columns={
"date": "sample_date",
"iso_code": "country_code",
},
inplace=True)
def convert_iso_alpha_3_to_alpha_2(x):
try:
return pycountry.countries.get(alpha_3=x).alpha_2
except Exception as e:
logging.info(f"Error converting country ISO Alpha 3 code '{x}': {repr(e)}")
return None
confirmed_df["country_code"] = confirmed_df.country_code.apply(convert_iso_alpha_3_to_alpha_2)
confirmed_df.dropna(inplace=True)
confirmed_df["sample_date"] = pd.to_datetime(confirmed_df.sample_date, dayfirst=True)
confirmed_df["sample_date"] = confirmed_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_df.sort_values("sample_date", inplace=True)
confirmed_df.tail()
confirmed_days = pd.date_range(
start=confirmed_df.iloc[0].sample_date,
end=extraction_datetime)
confirmed_days_df = pd.DataFrame(data=confirmed_days, columns=["sample_date"])
confirmed_days_df["sample_date_string"] = \
confirmed_days_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_days_df.tail()
def sort_source_regions_for_display(source_regions: list) -> list:
if report_backend_identifier in source_regions:
source_regions = [report_backend_identifier] + \
list(sorted(set(source_regions).difference([report_backend_identifier])))
else:
source_regions = list(sorted(source_regions))
return source_regions
report_source_regions = report_backend_client.source_regions_for_date(
date=extraction_datetime.date())
report_source_regions = sort_source_regions_for_display(
source_regions=report_source_regions)
report_source_regions
def get_cases_dataframe(source_regions_for_date_function, columns_suffix=None):
source_regions_at_date_df = confirmed_days_df.copy()
source_regions_at_date_df["source_regions_at_date"] = \
source_regions_at_date_df.sample_date.apply(
lambda x: source_regions_for_date_function(date=x))
source_regions_at_date_df.sort_values("sample_date", inplace=True)
source_regions_at_date_df["_source_regions_group"] = source_regions_at_date_df. \
source_regions_at_date.apply(lambda x: ",".join(sort_source_regions_for_display(x)))
source_regions_at_date_df.tail()
#%%
source_regions_for_summary_df_ = \
source_regions_at_date_df[["sample_date", "_source_regions_group"]].copy()
source_regions_for_summary_df_.rename(columns={"_source_regions_group": "source_regions"}, inplace=True)
source_regions_for_summary_df_.tail()
#%%
confirmed_output_columns = ["sample_date", "new_cases", "covid_cases"]
confirmed_output_df = pd.DataFrame(columns=confirmed_output_columns)
for source_regions_group, source_regions_group_series in \
source_regions_at_date_df.groupby("_source_regions_group"):
source_regions_set = set(source_regions_group.split(","))
confirmed_source_regions_set_df = \
confirmed_df[confirmed_df.country_code.isin(source_regions_set)].copy()
confirmed_source_regions_group_df = \
confirmed_source_regions_set_df.groupby("sample_date").new_cases.sum() \
.reset_index().sort_values("sample_date")
confirmed_source_regions_group_df["covid_cases"] = \
confirmed_source_regions_group_df.new_cases.rolling(7, min_periods=0).mean().round()
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[confirmed_output_columns]
confirmed_source_regions_group_df.fillna(method="ffill", inplace=True)
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[
confirmed_source_regions_group_df.sample_date.isin(
source_regions_group_series.sample_date_string)]
confirmed_output_df = confirmed_output_df.append(confirmed_source_regions_group_df)
result_df = confirmed_output_df.copy()
result_df.tail()
#%%
result_df.rename(columns={"sample_date": "sample_date_string"}, inplace=True)
result_df = confirmed_days_df[["sample_date_string"]].merge(result_df, how="left")
result_df.sort_values("sample_date_string", inplace=True)
result_df.fillna(method="ffill", inplace=True)
result_df.tail()
#%%
result_df[["new_cases", "covid_cases"]].plot()
if columns_suffix:
result_df.rename(
columns={
"new_cases": "new_cases_" + columns_suffix,
"covid_cases": "covid_cases_" + columns_suffix},
inplace=True)
return result_df, source_regions_for_summary_df_
confirmed_eu_df, source_regions_for_summary_df = get_cases_dataframe(
report_backend_client.source_regions_for_date)
confirmed_es_df, _ = get_cases_dataframe(
lambda date: [spain_region_country_code],
columns_suffix=spain_region_country_code.lower())
raw_zip_path_prefix = "Data/TEKs/Raw/"
fail_on_error_backend_identifiers = [report_backend_identifier]
multi_backend_exposure_keys_df = \
exposure_notification_io.download_exposure_keys_from_backends(
backend_identifiers=report_backend_identifiers,
generation_days=backend_generation_days,
fail_on_error_backend_identifiers=fail_on_error_backend_identifiers,
save_raw_zip_path_prefix=raw_zip_path_prefix)
multi_backend_exposure_keys_df["region"] = multi_backend_exposure_keys_df["backend_identifier"]
multi_backend_exposure_keys_df.rename(
columns={
"generation_datetime": "sample_datetime",
"generation_date_string": "sample_date_string",
},
inplace=True)
multi_backend_exposure_keys_df.head()
early_teks_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.rolling_period < 144].copy()
early_teks_df["rolling_period_in_hours"] = early_teks_df.rolling_period / 6
early_teks_df[early_teks_df.sample_date_string != extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
early_teks_df[early_teks_df.sample_date_string == extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
multi_backend_exposure_keys_df = multi_backend_exposure_keys_df[[
"sample_date_string", "region", "key_data"]]
multi_backend_exposure_keys_df.head()
active_regions = \
multi_backend_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
active_regions
multi_backend_summary_df = multi_backend_exposure_keys_df.groupby(
["sample_date_string", "region"]).key_data.nunique().reset_index() \
.pivot(index="sample_date_string", columns="region") \
.sort_index(ascending=False)
multi_backend_summary_df.rename(
columns={"key_data": "shared_teks_by_generation_date"},
inplace=True)
multi_backend_summary_df.rename_axis("sample_date", inplace=True)
multi_backend_summary_df = multi_backend_summary_df.fillna(0).astype(int)
multi_backend_summary_df = multi_backend_summary_df.head(backend_generation_days)
multi_backend_summary_df.head()
def compute_keys_cross_sharing(x):
teks_x = x.key_data_x.item()
common_teks = set(teks_x).intersection(x.key_data_y.item())
common_teks_fraction = len(common_teks) / len(teks_x)
return pd.Series(dict(
common_teks=common_teks,
common_teks_fraction=common_teks_fraction,
))
multi_backend_exposure_keys_by_region_df = \
multi_backend_exposure_keys_df.groupby("region").key_data.unique().reset_index()
multi_backend_exposure_keys_by_region_df["_merge"] = True
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_df.merge(
multi_backend_exposure_keys_by_region_df, on="_merge")
multi_backend_exposure_keys_by_region_combination_df.drop(
columns=["_merge"], inplace=True)
if multi_backend_exposure_keys_by_region_combination_df.region_x.nunique() > 1:
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_combination_df[
multi_backend_exposure_keys_by_region_combination_df.region_x !=
multi_backend_exposure_keys_by_region_combination_df.region_y]
multi_backend_exposure_keys_cross_sharing_df = \
multi_backend_exposure_keys_by_region_combination_df \
.groupby(["region_x", "region_y"]) \
.apply(compute_keys_cross_sharing) \
.reset_index()
multi_backend_cross_sharing_summary_df = \
multi_backend_exposure_keys_cross_sharing_df.pivot_table(
values=["common_teks_fraction"],
columns="region_x",
index="region_y",
aggfunc=lambda x: x.item())
multi_backend_cross_sharing_summary_df
multi_backend_without_active_region_exposure_keys_df = \
multi_backend_exposure_keys_df[multi_backend_exposure_keys_df.region != report_backend_identifier]
multi_backend_without_active_region = \
multi_backend_without_active_region_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
multi_backend_without_active_region
exposure_keys_summary_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.region == report_backend_identifier]
exposure_keys_summary_df.drop(columns=["region"], inplace=True)
exposure_keys_summary_df = \
exposure_keys_summary_df.groupby(["sample_date_string"]).key_data.nunique().to_frame()
exposure_keys_summary_df = \
exposure_keys_summary_df.reset_index().set_index("sample_date_string")
exposure_keys_summary_df.sort_index(ascending=False, inplace=True)
exposure_keys_summary_df.rename(columns={"key_data": "shared_teks_by_generation_date"}, inplace=True)
exposure_keys_summary_df.head()
tek_list_df = multi_backend_exposure_keys_df[
["sample_date_string", "region", "key_data"]].copy()
tek_list_df["key_data"] = tek_list_df["key_data"].apply(str)
tek_list_df.rename(columns={
"sample_date_string": "sample_date",
"key_data": "tek_list"}, inplace=True)
tek_list_df = tek_list_df.groupby(
["sample_date", "region"]).tek_list.unique().reset_index()
tek_list_df["extraction_date"] = extraction_date
tek_list_df["extraction_date_with_hour"] = extraction_date_with_hour
tek_list_path_prefix = "Data/TEKs/"
tek_list_current_path = tek_list_path_prefix + f"/Current/RadarCOVID-TEKs.json"
tek_list_daily_path = tek_list_path_prefix + f"Daily/RadarCOVID-TEKs-{extraction_date}.json"
tek_list_hourly_path = tek_list_path_prefix + f"Hourly/RadarCOVID-TEKs-{extraction_date_with_hour}.json"
for path in [tek_list_current_path, tek_list_daily_path, tek_list_hourly_path]:
os.makedirs(os.path.dirname(path), exist_ok=True)
tek_list_df.drop(columns=["extraction_date", "extraction_date_with_hour"]).to_json(
tek_list_current_path,
lines=True, orient="records")
tek_list_df.drop(columns=["extraction_date_with_hour"]).to_json(
tek_list_daily_path,
lines=True, orient="records")
tek_list_df.to_json(
tek_list_hourly_path,
lines=True, orient="records")
tek_list_df.head()
import glob
def load_extracted_teks(mode, region=None, limit=None) -> pd.DataFrame:
extracted_teks_df = pd.DataFrame(columns=["region"])
file_paths = list(reversed(sorted(glob.glob(tek_list_path_prefix + mode + "/RadarCOVID-TEKs-*.json"))))
if limit:
file_paths = file_paths[:limit]
for file_path in file_paths:
logging.info(f"Loading TEKs from '{file_path}'...")
iteration_extracted_teks_df = pd.read_json(file_path, lines=True)
extracted_teks_df = extracted_teks_df.append(
iteration_extracted_teks_df, sort=False)
extracted_teks_df["region"] = \
extracted_teks_df.region.fillna(spain_region_country_code).copy()
if region:
extracted_teks_df = \
extracted_teks_df[extracted_teks_df.region == region]
return extracted_teks_df
daily_extracted_teks_df = load_extracted_teks(
mode="Daily",
region=report_backend_identifier,
limit=tek_dumps_load_limit)
daily_extracted_teks_df.head()
exposure_keys_summary_df_ = daily_extracted_teks_df \
.sort_values("extraction_date", ascending=False) \
.groupby("sample_date").tek_list.first() \
.to_frame()
exposure_keys_summary_df_.index.name = "sample_date_string"
exposure_keys_summary_df_["tek_list"] = \
exposure_keys_summary_df_.tek_list.apply(len)
exposure_keys_summary_df_ = exposure_keys_summary_df_ \
.rename(columns={"tek_list": "shared_teks_by_generation_date"}) \
.sort_index(ascending=False)
exposure_keys_summary_df = exposure_keys_summary_df_
exposure_keys_summary_df.head()
tek_list_df = daily_extracted_teks_df.groupby("extraction_date").tek_list.apply(
lambda x: set(sum(x, []))).reset_index()
tek_list_df = tek_list_df.set_index("extraction_date").sort_index(ascending=True)
tek_list_df.head()
def compute_teks_by_generation_and_upload_date(date):
day_new_teks_set_df = tek_list_df.copy().diff()
try:
day_new_teks_set = day_new_teks_set_df[
day_new_teks_set_df.index == date].tek_list.item()
except ValueError:
day_new_teks_set = None
if pd.isna(day_new_teks_set):
day_new_teks_set = set()
day_new_teks_df = daily_extracted_teks_df[
daily_extracted_teks_df.extraction_date == date].copy()
day_new_teks_df["shared_teks"] = \
day_new_teks_df.tek_list.apply(lambda x: set(x).intersection(day_new_teks_set))
day_new_teks_df["shared_teks"] = \
day_new_teks_df.shared_teks.apply(len)
day_new_teks_df["upload_date"] = date
day_new_teks_df.rename(columns={"sample_date": "generation_date"}, inplace=True)
day_new_teks_df = day_new_teks_df[
["upload_date", "generation_date", "shared_teks"]]
day_new_teks_df["generation_to_upload_days"] = \
(pd.to_datetime(day_new_teks_df.upload_date) -
pd.to_datetime(day_new_teks_df.generation_date)).dt.days
day_new_teks_df = day_new_teks_df[day_new_teks_df.shared_teks > 0]
return day_new_teks_df
shared_teks_generation_to_upload_df = pd.DataFrame()
for upload_date in daily_extracted_teks_df.extraction_date.unique():
shared_teks_generation_to_upload_df = \
shared_teks_generation_to_upload_df.append(
compute_teks_by_generation_and_upload_date(date=upload_date))
shared_teks_generation_to_upload_df \
.sort_values(["upload_date", "generation_date"], ascending=False, inplace=True)
shared_teks_generation_to_upload_df.tail()
today_new_teks_df = \
shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.upload_date == extraction_date].copy()
today_new_teks_df.tail()
if not today_new_teks_df.empty:
today_new_teks_df.set_index("generation_to_upload_days") \
.sort_index().shared_teks.plot.bar()
generation_to_upload_period_pivot_df = \
shared_teks_generation_to_upload_df[
["upload_date", "generation_to_upload_days", "shared_teks"]] \
.pivot(index="upload_date", columns="generation_to_upload_days") \
.sort_index(ascending=False).fillna(0).astype(int) \
.droplevel(level=0, axis=1)
generation_to_upload_period_pivot_df.head()
new_tek_df = tek_list_df.diff().tek_list.apply(
lambda x: len(x) if not pd.isna(x) else None).to_frame().reset_index()
new_tek_df.rename(columns={
"tek_list": "shared_teks_by_upload_date",
"extraction_date": "sample_date_string",}, inplace=True)
new_tek_df.tail()
shared_teks_uploaded_on_generation_date_df = shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.generation_to_upload_days == 0] \
[["upload_date", "shared_teks"]].rename(
columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_teks_uploaded_on_generation_date",
})
shared_teks_uploaded_on_generation_date_df.head()
estimated_shared_diagnoses_df = shared_teks_generation_to_upload_df \
.groupby(["upload_date"]).shared_teks.max().reset_index() \
.sort_values(["upload_date"], ascending=False) \
.rename(columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_diagnoses",
})
invalid_shared_diagnoses_dates_mask = \
estimated_shared_diagnoses_df.sample_date_string.isin(invalid_shared_diagnoses_dates)
estimated_shared_diagnoses_df[invalid_shared_diagnoses_dates_mask] = 0
estimated_shared_diagnoses_df.head()
hourly_extracted_teks_df = load_extracted_teks(
mode="Hourly", region=report_backend_identifier, limit=25)
hourly_extracted_teks_df.head()
hourly_new_tek_count_df = hourly_extracted_teks_df \
.groupby("extraction_date_with_hour").tek_list. \
apply(lambda x: set(sum(x, []))).reset_index().copy()
hourly_new_tek_count_df = hourly_new_tek_count_df.set_index("extraction_date_with_hour") \
.sort_index(ascending=True)
hourly_new_tek_count_df["new_tek_list"] = hourly_new_tek_count_df.tek_list.diff()
hourly_new_tek_count_df["new_tek_count"] = hourly_new_tek_count_df.new_tek_list.apply(
lambda x: len(x) if not pd.isna(x) else 0)
hourly_new_tek_count_df.rename(columns={
"new_tek_count": "shared_teks_by_upload_date"}, inplace=True)
hourly_new_tek_count_df = hourly_new_tek_count_df.reset_index()[[
"extraction_date_with_hour", "shared_teks_by_upload_date"]]
hourly_new_tek_count_df.head()
hourly_summary_df = hourly_new_tek_count_df.copy()
hourly_summary_df.set_index("extraction_date_with_hour", inplace=True)
hourly_summary_df = hourly_summary_df.fillna(0).astype(int).reset_index()
hourly_summary_df["datetime_utc"] = pd.to_datetime(
hourly_summary_df.extraction_date_with_hour, format="%Y-%m-%d@%H")
hourly_summary_df.set_index("datetime_utc", inplace=True)
hourly_summary_df = hourly_summary_df.tail(-1)
hourly_summary_df.head()
import requests
import pandas.io.json
official_stats_response = requests.get("https://radarcovidpre.covid19.gob.es/kpi/statistics/basics")
official_stats_response.raise_for_status()
official_stats_df_ = pandas.io.json.json_normalize(official_stats_response.json())
official_stats_df = official_stats_df_.copy()
official_stats_df = official_stats_df.append(pd.DataFrame({
"date": ["06/12/2020"],
"applicationsDownloads.totalAcummulated": [5653519],
"communicatedContagions.totalAcummulated": [21925],
}), sort=False)
official_stats_df["date"] = pd.to_datetime(official_stats_df["date"], dayfirst=True)
official_stats_df.head()
official_stats_column_map = {
"date": "sample_date",
"applicationsDownloads.totalAcummulated": "app_downloads_es_accumulated",
"communicatedContagions.totalAcummulated": "shared_diagnoses_es_accumulated",
}
accumulated_suffix = "_accumulated"
accumulated_values_columns = \
list(filter(lambda x: x.endswith(accumulated_suffix), official_stats_column_map.values()))
interpolated_values_columns = \
list(map(lambda x: x[:-len(accumulated_suffix)], accumulated_values_columns))
official_stats_df = \
official_stats_df[official_stats_column_map.keys()] \
.rename(columns=official_stats_column_map)
official_stats_df.head()
official_stats_df = confirmed_days_df.merge(official_stats_df, how="left")
official_stats_df.sort_values("sample_date", ascending=False, inplace=True)
official_stats_df.head()
official_stats_df[accumulated_values_columns] = \
official_stats_df[accumulated_values_columns] \
.astype(float).interpolate(limit_area="inside")
official_stats_df[interpolated_values_columns] = \
official_stats_df[accumulated_values_columns].diff(periods=-1)
official_stats_df.drop(columns="sample_date", inplace=True)
official_stats_df.head()
result_summary_df = exposure_keys_summary_df.merge(
new_tek_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
shared_teks_uploaded_on_generation_date_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
estimated_shared_diagnoses_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
official_stats_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = confirmed_eu_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df = confirmed_es_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df["sample_date"] = pd.to_datetime(result_summary_df.sample_date_string)
result_summary_df = result_summary_df.merge(source_regions_for_summary_df, how="left")
result_summary_df.set_index(["sample_date", "source_regions"], inplace=True)
result_summary_df.drop(columns=["sample_date_string"], inplace=True)
result_summary_df.sort_index(ascending=False, inplace=True)
result_summary_df.head()
with pd.option_context("mode.use_inf_as_na", True):
result_summary_df = result_summary_df.fillna(0).astype(int)
result_summary_df["teks_per_shared_diagnosis"] = \
(result_summary_df.shared_teks_by_upload_date / result_summary_df.shared_diagnoses).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case"] = \
(result_summary_df.shared_diagnoses / result_summary_df.covid_cases).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(result_summary_df.shared_diagnoses_es / result_summary_df.covid_cases_es).fillna(0)
result_summary_df.head(daily_plot_days)
weekly_result_summary_df = result_summary_df \
.sort_index(ascending=True).fillna(0).rolling(7).agg({
"covid_cases": "sum",
"covid_cases_es": "sum",
"shared_teks_by_generation_date": "sum",
"shared_teks_by_upload_date": "sum",
"shared_diagnoses": "sum",
"shared_diagnoses_es": "sum",
}).sort_index(ascending=False)
with pd.option_context("mode.use_inf_as_na", True):
weekly_result_summary_df = weekly_result_summary_df.fillna(0).astype(int)
weekly_result_summary_df["teks_per_shared_diagnosis"] = \
(weekly_result_summary_df.shared_teks_by_upload_date / weekly_result_summary_df.shared_diagnoses).fillna(0)
weekly_result_summary_df["shared_diagnoses_per_covid_case"] = \
(weekly_result_summary_df.shared_diagnoses / weekly_result_summary_df.covid_cases).fillna(0)
weekly_result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(weekly_result_summary_df.shared_diagnoses_es / weekly_result_summary_df.covid_cases_es).fillna(0)
weekly_result_summary_df.head()
last_7_days_summary = weekly_result_summary_df.to_dict(orient="records")[1]
last_7_days_summary
display_column_name_mapping = {
"sample_date": "Sample\u00A0Date\u00A0(UTC)",
"source_regions": "Source Countries",
"datetime_utc": "Timestamp (UTC)",
"upload_date": "Upload Date (UTC)",
"generation_to_upload_days": "Generation to Upload Period in Days",
"region": "Backend",
"region_x": "Backend\u00A0(A)",
"region_y": "Backend\u00A0(B)",
"common_teks": "Common TEKs Shared Between Backends",
"common_teks_fraction": "Fraction of TEKs in Backend (A) Available in Backend (B)",
"covid_cases": "COVID-19 Cases (Source Countries)",
"shared_teks_by_generation_date": "Shared TEKs by Generation Date (Source Countries)",
"shared_teks_by_upload_date": "Shared TEKs by Upload Date (Source Countries)",
"shared_teks_uploaded_on_generation_date": "Shared TEKs Uploaded on Generation Date (Source Countries)",
"shared_diagnoses": "Shared Diagnoses (Source Countries – Estimation)",
"teks_per_shared_diagnosis": "TEKs Uploaded per Shared Diagnosis (Source Countries)",
"shared_diagnoses_per_covid_case": "Usage Ratio (Source Countries)",
"covid_cases_es": "COVID-19 Cases (Spain)",
"app_downloads_es": "App Downloads (Spain – Official)",
"shared_diagnoses_es": "Shared Diagnoses (Spain – Official)",
"shared_diagnoses_per_covid_case_es": "Usage Ratio (Spain)",
}
summary_columns = [
"covid_cases",
"shared_teks_by_generation_date",
"shared_teks_by_upload_date",
"shared_teks_uploaded_on_generation_date",
"shared_diagnoses",
"teks_per_shared_diagnosis",
"shared_diagnoses_per_covid_case",
"covid_cases_es",
"app_downloads_es",
"shared_diagnoses_es",
"shared_diagnoses_per_covid_case_es",
]
summary_percentage_columns= [
"shared_diagnoses_per_covid_case_es",
"shared_diagnoses_per_covid_case",
]
result_summary_df_ = result_summary_df.copy()
result_summary_df = result_summary_df[summary_columns]
result_summary_with_display_names_df = result_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
result_summary_with_display_names_df
result_plot_summary_df = result_summary_df.head(daily_plot_days)[summary_columns] \
.droplevel(level=["source_regions"]) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
summary_ax_list = result_plot_summary_df.sort_index(ascending=True).plot.bar(
title=f"Daily Summary",
rot=45, subplots=True, figsize=(15, 30), legend=False)
ax_ = summary_ax_list[0]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.95)
_ = ax_.set_xticklabels(sorted(result_plot_summary_df.index.strftime("%Y-%m-%d").tolist()))
for percentage_column in summary_percentage_columns:
percentage_column_index = summary_columns.index(percentage_column)
summary_ax_list[percentage_column_index].yaxis \
.set_major_formatter(matplotlib.ticker.PercentFormatter(1.0))
display_generation_to_upload_period_pivot_df = \
generation_to_upload_period_pivot_df \
.head(backend_generation_days)
display_generation_to_upload_period_pivot_df \
.head(backend_generation_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping)
fig, generation_to_upload_period_pivot_table_ax = plt.subplots(
figsize=(12, 1 + 0.6 * len(display_generation_to_upload_period_pivot_df)))
generation_to_upload_period_pivot_table_ax.set_title(
"Shared TEKs Generation to Upload Period Table")
sns.heatmap(
data=display_generation_to_upload_period_pivot_df
.rename_axis(columns=display_column_name_mapping)
.rename_axis(index=display_column_name_mapping),
fmt=".0f",
annot=True,
ax=generation_to_upload_period_pivot_table_ax)
generation_to_upload_period_pivot_table_ax.get_figure().tight_layout()
hourly_summary_ax_list = hourly_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.plot.bar(
title=f"Last 24h Summary",
rot=45, subplots=True, legend=False)
ax_ = hourly_summary_ax_list[-1]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.9)
_ = ax_.set_xticklabels(sorted(hourly_summary_df.index.strftime("%Y-%m-%d@%H").tolist()))
github_repository = os.environ.get("GITHUB_REPOSITORY")
if github_repository is None:
github_repository = "pvieito/Radar-STATS"
github_project_base_url = "https://github.com/" + github_repository
display_formatters = {
display_column_name_mapping["teks_per_shared_diagnosis"]: lambda x: f"{x:.2f}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case"]: lambda x: f"{x:.2%}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case_es"]: lambda x: f"{x:.2%}" if x != 0 else "",
}
general_columns = \
list(filter(lambda x: x not in display_formatters, display_column_name_mapping.values()))
general_formatter = lambda x: f"{x}" if x != 0 else ""
display_formatters.update(dict(map(lambda x: (x, general_formatter), general_columns)))
daily_summary_table_html = result_summary_with_display_names_df \
.head(daily_plot_days) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.to_html(formatters=display_formatters)
multi_backend_summary_table_html = multi_backend_summary_df \
.head(daily_plot_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(formatters=display_formatters)
def format_multi_backend_cross_sharing_fraction(x):
if pd.isna(x):
return "-"
elif round(x * 100, 1) == 0:
return ""
else:
return f"{x:.1%}"
multi_backend_cross_sharing_summary_table_html = multi_backend_cross_sharing_summary_df \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(
classes="table-center",
formatters=display_formatters,
float_format=format_multi_backend_cross_sharing_fraction)
multi_backend_cross_sharing_summary_table_html = \
multi_backend_cross_sharing_summary_table_html \
.replace("<tr>","<tr style=\"text-align: center;\">")
extraction_date_result_summary_df = \
result_summary_df[result_summary_df.index.get_level_values("sample_date") == extraction_date]
extraction_date_result_hourly_summary_df = \
hourly_summary_df[hourly_summary_df.extraction_date_with_hour == extraction_date_with_hour]
covid_cases = \
extraction_date_result_summary_df.covid_cases.item()
shared_teks_by_generation_date = \
extraction_date_result_summary_df.shared_teks_by_generation_date.item()
shared_teks_by_upload_date = \
extraction_date_result_summary_df.shared_teks_by_upload_date.item()
shared_diagnoses = \
extraction_date_result_summary_df.shared_diagnoses.item()
teks_per_shared_diagnosis = \
extraction_date_result_summary_df.teks_per_shared_diagnosis.item()
shared_diagnoses_per_covid_case = \
extraction_date_result_summary_df.shared_diagnoses_per_covid_case.item()
shared_teks_by_upload_date_last_hour = \
extraction_date_result_hourly_summary_df.shared_teks_by_upload_date.sum().astype(int)
display_source_regions = ", ".join(report_source_regions)
if len(report_source_regions) == 1:
display_brief_source_regions = report_source_regions[0]
else:
display_brief_source_regions = f"{len(report_source_regions)} 🇪🇺"
def get_temporary_image_path() -> str:
return os.path.join(tempfile.gettempdir(), str(uuid.uuid4()) + ".png")
def save_temporary_plot_image(ax):
if isinstance(ax, np.ndarray):
ax = ax[0]
media_path = get_temporary_image_path()
ax.get_figure().savefig(media_path)
return media_path
def save_temporary_dataframe_image(df):
import dataframe_image as dfi
df = df.copy()
df_styler = df.style.format(display_formatters)
media_path = get_temporary_image_path()
dfi.export(df_styler, media_path)
return media_path
summary_plots_image_path = save_temporary_plot_image(
ax=summary_ax_list)
summary_table_image_path = save_temporary_dataframe_image(
df=result_summary_with_display_names_df)
hourly_summary_plots_image_path = save_temporary_plot_image(
ax=hourly_summary_ax_list)
multi_backend_summary_table_image_path = save_temporary_dataframe_image(
df=multi_backend_summary_df)
generation_to_upload_period_pivot_table_image_path = save_temporary_plot_image(
ax=generation_to_upload_period_pivot_table_ax)
report_resources_path_prefix = "Data/Resources/Current/RadarCOVID-Report-"
result_summary_df.to_csv(
report_resources_path_prefix + "Summary-Table.csv")
result_summary_df.to_html(
report_resources_path_prefix + "Summary-Table.html")
hourly_summary_df.to_csv(
report_resources_path_prefix + "Hourly-Summary-Table.csv")
multi_backend_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Summary-Table.csv")
multi_backend_cross_sharing_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Cross-Sharing-Summary-Table.csv")
generation_to_upload_period_pivot_df.to_csv(
report_resources_path_prefix + "Generation-Upload-Period-Table.csv")
_ = shutil.copyfile(
summary_plots_image_path,
report_resources_path_prefix + "Summary-Plots.png")
_ = shutil.copyfile(
summary_table_image_path,
report_resources_path_prefix + "Summary-Table.png")
_ = shutil.copyfile(
hourly_summary_plots_image_path,
report_resources_path_prefix + "Hourly-Summary-Plots.png")
_ = shutil.copyfile(
multi_backend_summary_table_image_path,
report_resources_path_prefix + "Multi-Backend-Summary-Table.png")
_ = shutil.copyfile(
generation_to_upload_period_pivot_table_image_path,
report_resources_path_prefix + "Generation-Upload-Period-Table.png")
def generate_summary_api_results(df: pd.DataFrame) -> list:
api_df = df.reset_index().copy()
api_df["sample_date_string"] = \
api_df["sample_date"].dt.strftime("%Y-%m-%d")
api_df["source_regions"] = \
api_df["source_regions"].apply(lambda x: x.split(","))
return api_df.to_dict(orient="records")
summary_api_results = \
generate_summary_api_results(df=result_summary_df)
today_summary_api_results = \
generate_summary_api_results(df=extraction_date_result_summary_df)[0]
summary_results = dict(
backend_identifier=report_backend_identifier,
source_regions=report_source_regions,
extraction_datetime=extraction_datetime,
extraction_date=extraction_date,
extraction_date_with_hour=extraction_date_with_hour,
last_hour=dict(
shared_teks_by_upload_date=shared_teks_by_upload_date_last_hour,
shared_diagnoses=0,
),
today=today_summary_api_results,
last_7_days=last_7_days_summary,
daily_results=summary_api_results)
summary_results = \
json.loads(pd.Series([summary_results]).to_json(orient="records"))[0]
with open(report_resources_path_prefix + "Summary-Results.json", "w") as f:
json.dump(summary_results, f, indent=4)
with open("Data/Templates/README.md", "r") as f:
readme_contents = f.read()
readme_contents = readme_contents.format(
extraction_date_with_hour=extraction_date_with_hour,
github_project_base_url=github_project_base_url,
daily_summary_table_html=daily_summary_table_html,
multi_backend_summary_table_html=multi_backend_summary_table_html,
multi_backend_cross_sharing_summary_table_html=multi_backend_cross_sharing_summary_table_html,
display_source_regions=display_source_regions)
with open("README.md", "w") as f:
f.write(readme_contents)
enable_share_to_twitter = os.environ.get("RADARCOVID_REPORT__ENABLE_PUBLISH_ON_TWITTER")
github_event_name = os.environ.get("GITHUB_EVENT_NAME")
if enable_share_to_twitter and github_event_name == "schedule" and \
(shared_teks_by_upload_date_last_hour or not are_today_results_partial):
import tweepy
twitter_api_auth_keys = os.environ["RADARCOVID_REPORT__TWITTER_API_AUTH_KEYS"]
twitter_api_auth_keys = twitter_api_auth_keys.split(":")
auth = tweepy.OAuthHandler(twitter_api_auth_keys[0], twitter_api_auth_keys[1])
auth.set_access_token(twitter_api_auth_keys[2], twitter_api_auth_keys[3])
api = tweepy.API(auth)
summary_plots_media = api.media_upload(summary_plots_image_path)
summary_table_media = api.media_upload(summary_table_image_path)
generation_to_upload_period_pivot_table_image_media = api.media_upload(generation_to_upload_period_pivot_table_image_path)
media_ids = [
summary_plots_media.media_id,
summary_table_media.media_id,
generation_to_upload_period_pivot_table_image_media.media_id,
]
if are_today_results_partial:
today_addendum = " (Partial)"
else:
today_addendum = ""
status = textwrap.dedent(f"""
#RadarCOVID – {extraction_date_with_hour}
Source Countries: {display_brief_source_regions}
Today{today_addendum}:
- Uploaded TEKs: {shared_teks_by_upload_date:.0f} ({shared_teks_by_upload_date_last_hour:+d} last hour)
- Shared Diagnoses: ≤{shared_diagnoses:.0f}
- Usage Ratio: ≤{shared_diagnoses_per_covid_case:.2%}
Last 7 Days:
- Shared Diagnoses: ≤{last_7_days_summary["shared_diagnoses"]:.0f}
- Usage Ratio: ≤{last_7_days_summary["shared_diagnoses_per_covid_case"]:.2%}
Info: {github_project_base_url}#documentation
""")
status = status.encode(encoding="utf-8")
api.update_status(status=status, media_ids=media_ids)
| 0.269037 | 0.187449 |
```
%matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
sns.set_context("talk")
```
# Reproducible visualization
In "The Functional Art: An introduction to information graphics and visualization" by Alberto Cairo, on page 12 we are presented with a visualization of UN data time series of Fertility rate (average number of children per woman) per country:
Figure 1.6 Highlighting the relevant, keeping the secondary in the background.
Let's try to reproduce this.
## Getting the data
The visualization was done in 2012, but limited the visualization to 2010. This should make it easy, in theory, to get the data, since it is historical. These are directly available as excel spreadsheets now, we'll just ignore the last bucket (2010-2015).
Pandas allows loading an excel spreadsheet straight from a URL, but here we will download it first so we have a local copy.
```
!wget 'http://esa.un.org/unpd/wpp/DVD/Files/1_Indicators%20(Standard)/EXCEL_FILES/2_Fertility/WPP2015_FERT_F04_TOTAL_FERTILITY.XLS'
```
###World Population Prospects: The 2015 Revision
File FERT/4: Total fertility by major area, region and country, 1950-2100 (children per woman)
```
Estimates, 1950 - 2015
POP/DB/WPP/Rev.2015/FERT/F04
July 2015 - Copyright © 2015 by United Nations. All rights reserved
Suggested citation: United Nations, Department of Economic and Social Affairs, Population Division (2015). World Population Prospects: The 2015 Revision, DVD Edition.
```
```
df = pd.read_excel('WPP2015_FERT_F04_TOTAL_FERTILITY.XLS', skiprows=16, index_col = 'Country code')
df = df[df.index < 900]
len(df)
df.head()
```
First problem... The book states on page 8:
-- <cite>"Using the filters the site offers, I asked for a table that included the more than 150 countries on which the UN has complete research."</cite>
Yet we have 201 countries (codes 900+ are regions) with complete data. We do not have a easy way to identify which countries were added to this. Still, let's move forward and prep our data.
```
df.rename(columns={df.columns[2]:'Description'}, inplace=True)
df.drop(df.columns[[0, 1, 3, 16]], axis=1, inplace=True) # drop what we dont need
df.head()
highlight_countries = ['Niger','Yemen','India',
'Brazil','Norway','France','Sweden','United Kingdom',
'Spain','Italy','Germany','Japan', 'China'
]
# Subset only countries to highlight, transpose for timeseries
df_high = df[df.Description.isin(highlight_countries)].T[1:]
# Subset the rest of the countries, transpose for timeseries
df_bg = df[~df.Description.isin(highlight_countries)].T[1:]
```
## Let's make some art
```
# background
ax = df_bg.plot(legend=False, color='k', alpha=0.02, figsize=(12,12))
ax.xaxis.tick_top()
# highlighted countries
df_high.plot(legend=False, ax=ax)
# replacement level line
ax.hlines(y=2.1, xmin=0, xmax=12, color='k', alpha=1, linestyle='dashed')
# Average over time on all countries
df.mean().plot(ax=ax, color='k', label='World\naverage')
# labels for highlighted countries on the right side
for country in highlight_countries:
ax.text(11.2,df[df.Description==country].values[0][12],country)
# start y axis at 1
ax.set_ylim(ymin=1)
```
For one thing, the line for China doesn't look like the one in the book. Concerning. The other issue is that there are some lines that are going lower than Italy or Spain in 1995-2000 and in 2000-2005 (majority in the Balkans) and that were not on the graph in the book, AFAICT:
```
df.describe()
df[df['1995-2000']<1.25]
df[df['2000-2005']<1.25]
```
The other thing that I really need to address is the labeling. Clearly we need the functionality to move labels up and down to make them readable. Collision detection, basically. I'm surprised this functionality doesn't exist, because I keep bumping into that. Usually, I can tweak the Y pos by a few pixels, but in this specific case, there is no way to do that.
So, I guess I have a project for 2016...
|
github_jupyter
|
%matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
sns.set_context("talk")
!wget 'http://esa.un.org/unpd/wpp/DVD/Files/1_Indicators%20(Standard)/EXCEL_FILES/2_Fertility/WPP2015_FERT_F04_TOTAL_FERTILITY.XLS'
Estimates, 1950 - 2015
POP/DB/WPP/Rev.2015/FERT/F04
July 2015 - Copyright © 2015 by United Nations. All rights reserved
Suggested citation: United Nations, Department of Economic and Social Affairs, Population Division (2015). World Population Prospects: The 2015 Revision, DVD Edition.
df = pd.read_excel('WPP2015_FERT_F04_TOTAL_FERTILITY.XLS', skiprows=16, index_col = 'Country code')
df = df[df.index < 900]
len(df)
df.head()
df.rename(columns={df.columns[2]:'Description'}, inplace=True)
df.drop(df.columns[[0, 1, 3, 16]], axis=1, inplace=True) # drop what we dont need
df.head()
highlight_countries = ['Niger','Yemen','India',
'Brazil','Norway','France','Sweden','United Kingdom',
'Spain','Italy','Germany','Japan', 'China'
]
# Subset only countries to highlight, transpose for timeseries
df_high = df[df.Description.isin(highlight_countries)].T[1:]
# Subset the rest of the countries, transpose for timeseries
df_bg = df[~df.Description.isin(highlight_countries)].T[1:]
# background
ax = df_bg.plot(legend=False, color='k', alpha=0.02, figsize=(12,12))
ax.xaxis.tick_top()
# highlighted countries
df_high.plot(legend=False, ax=ax)
# replacement level line
ax.hlines(y=2.1, xmin=0, xmax=12, color='k', alpha=1, linestyle='dashed')
# Average over time on all countries
df.mean().plot(ax=ax, color='k', label='World\naverage')
# labels for highlighted countries on the right side
for country in highlight_countries:
ax.text(11.2,df[df.Description==country].values[0][12],country)
# start y axis at 1
ax.set_ylim(ymin=1)
df.describe()
df[df['1995-2000']<1.25]
df[df['2000-2005']<1.25]
| 0.38168 | 0.924005 |
---
__Hacker News Pipeline__
We have built a data pipeline that schedules our tasks.
The data we will use comes from a Hacker News (HN) API, returning JSON data of the top stories in 2014.
Each post has a set of keys, but we will deal only with the following keys:
- created_at: A timestamp of the story's creation time.
- created_at_i: A unix epoch timestamp.
- url: The URL of the story link.
- objectID: The ID of the story.
- author: The story's author (username on HN).
- points: The number of upvotes the story had.
- title: The headline of the post.
- num_comments: The number of a comments a post has.
```
# Set up pipeline
import json, csv, io, string, datetime as dt
from pipeline import Pipeline, build_csv
from stop_words import stop_words
pipeline = Pipeline()
# Extract Data
# Load data from JSON file
@pipeline.task()
def file_to_json():
with open('hn_stories_2014.json', 'r') as f:
raw = json.load(f)
data = raw['stories']
return data
# Filter data
@pipeline.task(depends_on = file_to_json)
def filter_data(data):
def popular(item):
return (item['points'] > 50
and item['num_comments'] > 1
and not item['title'].startswith('ASK HN')
)
return (item for item in data if popular(item))
# Transform Data
# Convert JSON data to CSV
@pipeline.task(depends_on = filter_data)
def json_to_csv(data):
lines = list()
for item in data:
lines.append((item['objectID'],
dt.datetime.strptime(item['created_at'], '%Y-%m-%dT%H:%M:%SZ'),
item['url'], item['points'], item['title']
))
file = build_csv(lines,
header = ['objectID', 'created_at', 'url', 'points', 'title'],
file = io.StringIO())
return file
# Isolate title data
@pipeline.task(depends_on = json_to_csv)
def extract_titles(file):
reader = csv.reader(file)
header = next(reader)
id_num = header.index('title')
return (i[id_num] for i in reader)
# Standardise title data
@pipeline.task(depends_on = extract_titles)
def clean_titles(titles):
titles = [t.lower() for t in titles]
for p in string.punctuation:
titles = [t.replace(p, '') for t in titles]
return titles
# Build key - value store of word frequencies
@pipeline.task(depends_on = clean_titles)
def build_dictionary(titles):
word_freq = {}
for t in titles:
for i in t.split(' '):
if len(i) == 0 or i in stop_words:
pass
else:
if i not in word_freq.keys():
word_freq[i] = 1
word_freq[i] += 1
return word_freq
# Arrange frequency table
@pipeline.task(depends_on = build_dictionary)
def top_entries(word_freq, no_entries = 100):
sorted_items = sorted(word_freq.items(),
key=lambda x:x[1],
reverse=True)
return sorted_items[:no_entries]
# Test implementation
test = pipeline.run()
print('Top Entries')
for i in test[top_entries]:
print(i[0], '-', i[1])
```
__Closing remarks__
1. The data on HackerNews posts has been proceessed using the pipeline for task scheduling.
2. The data has been cleaned to standardise the word format, as well as skipping stop words and blank entries.
3. The frequency of each word in the post titles has been extracted into a key - value store.
4. The top 100 words in the key - value store have been extracted and displayed in a readable format.
The final result has some interesting keywords. There were terms like bitcoin, heartbleed (the 2014 hack), and many others.
Now that we have created the pipeline, there are additional tasks we could perform with the data:
- Rewrite the Pipeline class' output to save a file of the output for each task. This will allow you to "checkpoint" tasks so they don't have to be run twice.
- Use the nltk package for more advanced natural language processing tasks.
- Convert to a CSV before filtering, so you can keep all the stories from 2014 in a raw file.
- Fetch the data from Hacker News directly from a JSON API. Instead of reading from the file we gave, you can perform additional data processing using newer data.
|
github_jupyter
|
# Set up pipeline
import json, csv, io, string, datetime as dt
from pipeline import Pipeline, build_csv
from stop_words import stop_words
pipeline = Pipeline()
# Extract Data
# Load data from JSON file
@pipeline.task()
def file_to_json():
with open('hn_stories_2014.json', 'r') as f:
raw = json.load(f)
data = raw['stories']
return data
# Filter data
@pipeline.task(depends_on = file_to_json)
def filter_data(data):
def popular(item):
return (item['points'] > 50
and item['num_comments'] > 1
and not item['title'].startswith('ASK HN')
)
return (item for item in data if popular(item))
# Transform Data
# Convert JSON data to CSV
@pipeline.task(depends_on = filter_data)
def json_to_csv(data):
lines = list()
for item in data:
lines.append((item['objectID'],
dt.datetime.strptime(item['created_at'], '%Y-%m-%dT%H:%M:%SZ'),
item['url'], item['points'], item['title']
))
file = build_csv(lines,
header = ['objectID', 'created_at', 'url', 'points', 'title'],
file = io.StringIO())
return file
# Isolate title data
@pipeline.task(depends_on = json_to_csv)
def extract_titles(file):
reader = csv.reader(file)
header = next(reader)
id_num = header.index('title')
return (i[id_num] for i in reader)
# Standardise title data
@pipeline.task(depends_on = extract_titles)
def clean_titles(titles):
titles = [t.lower() for t in titles]
for p in string.punctuation:
titles = [t.replace(p, '') for t in titles]
return titles
# Build key - value store of word frequencies
@pipeline.task(depends_on = clean_titles)
def build_dictionary(titles):
word_freq = {}
for t in titles:
for i in t.split(' '):
if len(i) == 0 or i in stop_words:
pass
else:
if i not in word_freq.keys():
word_freq[i] = 1
word_freq[i] += 1
return word_freq
# Arrange frequency table
@pipeline.task(depends_on = build_dictionary)
def top_entries(word_freq, no_entries = 100):
sorted_items = sorted(word_freq.items(),
key=lambda x:x[1],
reverse=True)
return sorted_items[:no_entries]
# Test implementation
test = pipeline.run()
print('Top Entries')
for i in test[top_entries]:
print(i[0], '-', i[1])
| 0.321886 | 0.832134 |
# Produce Cached `galaxy_ids` for DC2
_Last Updated: Bryce Kalmbach, December 2018_
This is a notebook to produce the cached AGN and SN galaxy_id lists for DC2 Run 2. In this notebook we match the source AGN and SNe galaxies to objects in the galaxy catalogs.
```
import pandas as pd
from astropy.io import fits
import numpy as np
from desc.sims.GCRCatSimInterface import InstanceCatalogWriter
from lsst.sims.utils import SpecMap
import matplotlib.pyplot as plt
from lsst.utils import getPackageDir
from lsst.sims.photUtils import Sed, BandpassDict, Bandpass
from lsst.sims.catUtils.matchSED import matchBase
import os
import sqlite3
%matplotlib inline
```
## Point to locations for unlensed AGN and SNe information
Even if you already have an unsprinkled instance catalog ready you need to specify the locations of DC2 unlensed AGN and SNe databases. The AGN database is needed to know the AGN properties to sprinkle and the SNe database is needed to avoid sprinkling with galaxies that will have unlensed SNe at some point in the survey.
```
catalog_version = 'cosmoDC2_v1.1.4'
agnDB = '/global/projecta/projectdirs/lsst/groups/SSim/DC2/cosmoDC2_v1.1.4/agn_db_mbh7_mi30_sf4.db'
sneDB = '/global/projecta/projectdirs/lsst/groups/SSim/DC2/cosmoDC2_v1.1.4/sne_cosmoDC2_v1.1.4_MS_DDF.db'
sed_lookup_dir = '/global/projecta/projectdirs/lsst/groups/SSim/DC2/cosmoDC2_v1.1.4/sedLookup'
```
### Create an unsprinkled Instance Catalog
This is to get the possible AGN and Bulge galaxies to replace with the sprinkler. We use `cosmoDC2_v1.1.4_image_addon_knots`. Make sure to specify the correct database locations in the cell below.
```
# First we need to create a catalog without sprinkling
opsimDB = '/global/projecta/projectdirs/lsst/groups/SSim/DC2/minion_1016_desc_dithered_v4.db'
starDB = '/global/projecta/projectdirs/lsst/groups/SSim/DC2/dc2_stellar_db.db'
t_sky = InstanceCatalogWriter(opsimDB, '%s_image_addon_knots' % catalog_version, min_mag=30, protoDC2_ra=0,
protoDC2_dec=0, sprinkler=False,
agn_db_name=agnDB, star_db_name=starDB,
sed_lookup_dir=sed_lookup_dir)
uddf_visit = 197356 # Use a visit we know covers the uDDF field
t_sky.write_catalog(uddf_visit, out_dir='.', fov=2.1)
```
### Load in the galaxy catalogs as dataframes
```
base_columns = ['prefix', 'uniqueId', 'raPhoSim', 'decPhoSim',
'phosimMagNorm', 'sedFilepath', 'redshift',
'shear1', 'shear2', 'kappa', 'raOffset', 'decOffset',
'spatialmodel']
df_galaxy = pd.read_csv(os.path.join(os.environ['SCRATCH'],
'bulge_gal_cat_197356.txt.gz'),
delimiter=' ', header=None,
names=base_columns+['majorAxis', 'minorAxis',
'positionAngle', 'sindex',
'internalExtinctionModel',
'internalAv', 'internalRv',
'galacticExtinctionModel',
'galacticAv', 'galacticRv'])
df_disk = pd.read_csv(os.path.join(os.environ['SCRATCH'],
'disk_gal_cat_197356.txt.gz'),
delimiter=' ', header=None,
names=base_columns+['majorAxis', 'minorAxis',
'positionAngle', 'sindex',
'internalExtinctionModel',
'internalAv', 'internalRv',
'galacticExtinctionModel',
'galacticAv', 'galacticRv'])
df_agn = pd.read_csv(os.path.join(os.environ['SCRATCH'],
'agn_gal_cat_197356.txt.gz'),
delimiter=' ', header=None,
names=base_columns+['internalExtinctionModel',
'galacticExtinctionModel',
'galacticAv', 'galacticRv'])
```
We calculate the `galaxy_id` for each catalog so that we can join them together and also save it in the cache for the sprinkler.
```
df_agn['galaxy_id'] = np.right_shift(df_agn['uniqueId'], 10)
df_agn.head()
df_galaxy['galaxy_id'] = np.right_shift(df_galaxy['uniqueId'], 10)
df_galaxy.head()
df_disk['galaxy_id'] = np.right_shift(df_disk['uniqueId'], 10)
df_disk.head()
```
### Match the AGN catalog to Twinkles systems
We will go through the AGN catalog and find AGN in the uDDF field that match our properties. We will then save the `galaxy_id` of these AGN and give the corresponding OM10 system a `twinklesId` in the catalog that identifies it with this AGN when the sprinkler runs.
```
# Load in OM10 lenses we are using in Twinkles
from astropy.io import fits
hdulist = fits.open('../../data/twinkles_lenses_%s.fits' % catalog_version)
twinkles_lenses = hdulist[1].data
# Only search within the DDF field
df_agn = df_agn.query('raPhoSim < 53.755 and raPhoSim > 52.495 and decPhoSim < -27.55 and decPhoSim > -28.65')
df_galaxy = df_galaxy.query('raPhoSim < 53.755 and raPhoSim > 52.495 and decPhoSim < -27.55 and decPhoSim > -28.65')
df_disk = df_disk.query('raPhoSim < 53.755 and raPhoSim > 52.495 and decPhoSim < -27.55 and decPhoSim > -28.65')
df_agn = df_agn.reset_index(drop=True)
df_galaxy = df_galaxy.reset_index(drop=True)
df_disk = df_disk.reset_index(drop=True)
# Convert phosimMagNorm to i-band magnitudes for the uDDF AGN
bpDict = BandpassDict.loadTotalBandpassesFromFiles(bandpassNames=['i'])
bp = Bandpass()
imsimBand = bp.imsimBandpass()
agn_fname = str(getPackageDir('sims_sed_library') + '/agnSED/agn.spec.gz')
src_iband = []
src_mag_norm = df_agn['phosimMagNorm'].values
src_z = df_agn['redshift'].values
for src_mag, s_z in zip(src_mag_norm, src_z):
agn_sed = Sed()
agn_sed.readSED_flambda(agn_fname)
agn_sed.redshiftSED(s_z, dimming=True)
f_norm = agn_sed.calcFluxNorm(src_mag, bp)
agn_sed.multiplyFluxNorm(f_norm)
src_iband.append(agn_sed.calcMag(bpDict['i']))
df_agn['i_magnitude'] = src_iband
```
We want to match the AGN in the uDDF field to lensed systems based upon the redshift and magnitude of the source AGN. In this example we use 0.1 dex in redshift and 0.25 mags in the _i_-band. **(Anytime you use a new catalog this may need to be played with to get the desired number of systems)**
```
def find_agn_lens_candidates(galz, gal_mag):
# search the OM10 catalog for all sources +- 0.1 dex in redshift
# and within .25 mags of the AGN source
w = np.where((np.abs(np.log10(twinkles_lenses['ZSRC']) - np.log10(galz)) <= 0.1) &
(np.abs(twinkles_lenses['MAGI_IN'] - gal_mag) <= .25))[0]
lens_candidates = twinkles_lenses[w]
return lens_candidates
```
#### Avoid galaxies with unlensed SNe.
First load up cached `galaxy_ids` and then to speed things up when comparing to possible sprinkled ids we use database merges to only find the ones that are in the uddf field.
```
conn = sqlite3.connect(sneDB)
sne_query = conn.cursor()
sne_unsprinkled_galids = sne_query.execute('select galaxy_id from sne_params').fetchall()
sne_unsprinkled_galids = np.array(sne_unsprinkled_galids).flatten()
ddf_galids = pd.DataFrame(df_agn['galaxy_id'])
ddf_galids = ddf_galids.merge(pd.DataFrame(df_galaxy['galaxy_id']), how='outer', on='galaxy_id')
ddf_galids = ddf_galids.merge(pd.DataFrame(df_disk['galaxy_id']), how='outer', on='galaxy_id')
sne_avoid_galids = ddf_galids.merge(pd.DataFrame(sne_unsprinkled_galids, columns=['galaxy_id']), how='inner', on='galaxy_id')
sne_avoid_galids = sne_avoid_galids.values
```
#### Add weights to get final sprinkled distribution close to OM10
```
import matplotlib as mpl
mpl.rcParams['text.usetex'] = False
n, bins = np.histogram(twinkles_lenses['MAGI_IN'], bins=20)
bin_centers = 0.5*(bins[:-1] + bins[1:])
n = n / np.max(n)
mpl.rcParams['text.usetex'] = False
n, bins, _ = plt.hist(twinkles_lenses['MAGI_IN'][np.where(twinkles_lenses['ZSRC'] <= (3. + np.log10(3.)*.1))],
histtype='step', lw=4, normed=True, bins=20, label='OM10')
plt.hist(df_agn['i_magnitude'].values, bins=bins, histtype='step', normed=True, label='DC2')
plt.xlabel('i Magnitude')
plt.ylabel('Counts')
plt.legend(loc=2)
mpl.rcParams['text.usetex'] = False
n_z, bins_z, _ = plt.hist(twinkles_lenses['ZSRC'][np.where(twinkles_lenses['ZSRC'] <= (3. + np.log10(3.)*.1))],
histtype='step', lw=4, normed=True, bins=20, label='OM10')
plt.hist(df_agn['redshift'].values, bins=bins_z, histtype='step', normed=True, label='DC2')
plt.xlabel('Redshift')
plt.ylabel('Counts')
plt.legend(loc=2)
n_zeros = np.zeros(22)
n_zeros[1:-1] = n
n_2 = np.zeros(22)
n_2 += n_zeros
n_2[2:-6] += 2. * n_zeros[8:]
n_2[9] += 0.1
n_2[10] += 0.2
n_2[11] += 0.1
#n_2[6:15] += 2. * (2. - np.linspace(0, 2, 9))
n_2[6:15] += 1. * (1. - np.linspace(0, 1, 9))
#n_2[6:11] += 2. * (2. - np.linspace(0, 2, 5))
n_2 = n_2 / np.max(n_2)
n_2[2:4] += .2
n_2[4:10] = 1.0
plt.plot(bins, n_2[:-1])
plt.xlabel('Source I Magnitude')
plt.ylabel('Weight')
n_redshift = np.zeros(22)
n_redshift[1:-1] = n_z
n_redshift = n_redshift / np.max(n_redshift)
n_redshift[:8] += 0.1
n_redshift[-8:] = 1.0
n_redshift[:-8] -= 0.08
n_redshift[5:-8] -= 0.05
n_redshift[3] -= 0.01
n_redshift[4] -= 0.03
n_redshift[5] -= 0.01
n_redshift[6] -= 0.04
n_redshift[7] -= 0.05
n_redshift[8] -= 0.03
n_redshift[9] -=0.05
n_redshift[10] -= 0.1
n_redshift[11] -= 0.02
#n_redshift[12] -= 0.
#n_redshift = n_reds
print(np.min(n_redshift))
plt.plot(bins_z, n_redshift[:-1])
plt.ylim(0, 1.05)
plt.xlabel('Source Redshift')
plt.ylabel('Weight')
```
#### Sprinkle in the AGN
```
%%time
density_param = 1.0
good_rows = []
ra_list = []
dec_list = []
gal_ids = []
catalog_row_num = []
catalog_ids = []
for row_idx in range(len(df_agn)):
row = df_agn.iloc[row_idx]
if row_idx % 5000 == 0:
print(row_idx, len(catalog_ids))
if row.galaxy_id > 0:
candidates = find_agn_lens_candidates(row.redshift, row.i_magnitude)
np.random.seed(np.int(row.galaxy_id) % 4294967296)
keep_idx = []
if len(candidates) > 0:
for candidate_idx, candidate_sys in list(enumerate(candidates['LENSID'])):
if candidate_sys not in catalog_ids:
keep_idx.append(candidate_idx)
if len(keep_idx) == 0:
continue
else:
candidates = candidates[keep_idx]
pick_value = np.random.uniform()
bin_num = np.digitize(row['i_magnitude'], bins)
binz_num = np.digitize(row['redshift'], bins_z)
#density_param_mag = n_zeros[bin_num] * density_param
density_param_mag = n_2[bin_num] * n_redshift[binz_num] * density_param
if ((len(candidates) > 0) and (pick_value <= density_param_mag)):
good_rows.append(row_idx)
gal_ids.append(row.galaxy_id)
newlens = np.random.choice(candidates)
catalog_ids.append(newlens['LENSID'])
catalog_row_num.append(np.where(twinkles_lenses['LENSID'] == newlens['LENSID'])[0][0])
ra_list.append(row.raPhoSim)
dec_list.append(row.decPhoSim)
#print(len(catalog_ids))
```
#### Check performance of weights
```
mpl.rcParams['text.usetex'] = False
n, bins, _ = plt.hist(twinkles_lenses['MAGI_IN'][np.where(twinkles_lenses['ZSRC'] <= (3. + np.log10(3.)*.1))],
histtype='step', lw=4, normed=True, bins=10, label='OM10')
plt.hist(df_agn['i_magnitude'].values[good_rows], normed=True, bins=bins, histtype='step', label='New Model Catalog')
plt.hist(df_agn['i_magnitude'].values, bins=bins, histtype='step', normed=True, label='DC2')
plt.xlabel('i Magnitude')
plt.ylabel('Counts')
plt.legend(loc=2)
mpl.rcParams['text.usetex'] = False
n_z, bins_z, _ = plt.hist(twinkles_lenses['ZSRC'][np.where(twinkles_lenses['ZSRC'] <= (3. + np.log10(3.)*.1))],
histtype='step', lw=4, normed=True, bins=10, label='OM10')
plt.hist(df_agn['redshift'].values[good_rows], normed=True, bins=bins_z, histtype='step', label='New Model Catalog')
plt.hist(df_agn['redshift'].values, bins=bins_z, histtype='step', normed=True, label='DC2')
plt.xlabel('Redshift')
plt.ylabel('Counts')
plt.legend(loc=2)
print(len(catalog_ids))
len(good_rows), len(np.unique(good_rows)), len(np.unique(catalog_ids)), len(np.unique(catalog_row_num))
```
Check to see that our cached systems are distributed throughout the uDDF field.
```
plt.scatter(ra_list, dec_list, s=6)
plt.plot((52.486, 53.764), (-27.533, -27.533))
plt.plot((52.479, 53.771), (-28.667, -28.667))
plt.plot((52.479, 52.486), (-28.667, -27.533))
plt.plot((53.771, 53.764), (-28.667, -27.533))
plt.xlabel(r'ra')
plt.ylabel(r'dec')
plt.title('AGN Cache Locations')
#plt.savefig('agn_cache.png')
catalog_row_sort = np.argsort(catalog_row_num)
catalog_row_num = np.array(catalog_row_num)
# Add in Twinkles ID Number to catalog for matched objects
col_list = []
for col in twinkles_lenses.columns:
col_list.append(fits.Column(name=col.name, format=col.format, array=twinkles_lenses[col.name][catalog_row_num[catalog_row_sort]]))
col_list.append(fits.Column(name='twinklesId', format='I', array=np.arange(len(good_rows))))
```
Save this catalog of only the systems we need.
```
cols = fits.ColDefs(col_list)
tbhdu = fits.BinTableHDU.from_columns(cols)
tbhdu.writeto('../../data/%s_matched_AGN.fits' % catalog_version)
tbhdu.data[:5]
```
Save the cached `galaxy_id` info to file
```
agn_cache = pd.DataFrame(np.array([np.array(gal_ids)[catalog_row_sort], np.arange(len(good_rows))], dtype=np.int).T,
columns=['galtileid', 'twinkles_system'])
agn_cache.head()
agn_cache.tail()
#Check that galaxy_ids and twinkles_ids in FITS match up after sort
g_id = np.where(np.array(gal_ids) == agn_cache['galtileid'].values[0])
print(np.array(catalog_ids)[g_id] == tbhdu.data['LENSID'][0])
agn_cache.to_csv('../../data/%s_agn_cache.csv' % catalog_version, index=False)
```
### Match to GLSNe catalog
Here we do the same as we did for the AGN and OM10 catalog except with a bulge+disk galaxy catalog and the host galaxy information from the Gravitationally Lensed SNe catalog.
We begin by loading the hdf5 tables for the lensed SNe catalog into dataframes.
```
sne_systems = pd.read_hdf('/global/cscratch1/sd/brycek/glsne_%s.h5' % catalog_version, key='system')
sne_images = pd.read_hdf('/global/cscratch1/sd/brycek/glsne_%s.h5' % catalog_version, key='image')
use_gals_df = df_galaxy.query('raPhoSim > 52.495 and raPhoSim < 53.755 and decPhoSim > -28.65 and decPhoSim < -27.55')
len(use_gals_df)
use_gals_df = use_gals_df.merge(df_disk, on='galaxy_id', suffixes=('_bulge', '_disk'))
```
Following from Table 3 in Mannucci et al. 2005 (https://www.aanda.org/articles/aa/pdf/2005/15/aa1411.pdf) we are going to use galaxy colors
to scale the sn rate by color as a proxy for galaxy type, but we end up changing it from that a bit to give us good sample sizes of all types in the DDF region.
```
from lsst.utils import getPackageDir
sims_sed_list = os.listdir(os.path.join(getPackageDir('SIMS_SED_LIBRARY'),
'galaxySED'))
sims_sed_dict = {}
for sed_name in sims_sed_list:
sed_obj = Sed()
sed_obj.readSED_flambda(os.path.join(getPackageDir('SIMS_SED_LIBRARY'),
'galaxySED', sed_name))
sims_sed_dict[os.path.join('galaxySED', sed_name)] = sed_obj
# Filters from http://svo2.cab.inta-csic.es/svo/theory/fps3/index.php
bp_B = Bandpass(wavelen_max = 2700.)
bp_B.setBandpass(wavelen=np.array([360., 380., 400., 420., 460., 480.,
500., 520., 540., 560.]),
sb=np.array([0.00, 0.11, 0.92, 0.94, 0.79, 0.58,
0.36, 0.15, 0.04, 0.0]))
bp_K = Bandpass(wavelen_max = 2700.)
bp_K.setBandpass(wavelen=np.linspace(1800., 2600., 17),
sb=np.array([0.00, 0.10, 0.48, 0.95, 1.00, 0.98,
0.96, 0.95, 0.97, 0.96, 0.94, 0.95,
0.95, 0.84, 0.46, 0.08, 0.00]))
bp_dict = BandpassDict(bandpassNameList=['B', 'K'], bandpassList=[bp_B, bp_K])
from lsst.sims.photUtils import getImsimFluxNorm
from copy import deepcopy
bk_color = []
i = 0
for sed_name_bulge, redshift, magNorm_bulge, sed_name_disk, magNorm_disk in zip(use_gals_df['sedFilepath_bulge'].values,
use_gals_df['redshift_bulge'].values,
use_gals_df['phosimMagNorm_bulge'].values,
use_gals_df['sedFilepath_disk'].values,
use_gals_df['phosimMagNorm_disk'].values):
if i % 100000 == 0:
print(i)
sed_obj_bulge = deepcopy(sims_sed_dict[sed_name_bulge])
f_norm_b = getImsimFluxNorm(sed_obj_bulge, magNorm_bulge)
sed_obj_bulge.multiplyFluxNorm(f_norm_b)
sed_obj_bulge.redshiftSED(redshift)
sed_obj_disk = deepcopy(sims_sed_dict[sed_name_disk])
f_norm_d = getImsimFluxNorm(sed_obj_disk, magNorm_disk)
sed_obj_disk.multiplyFluxNorm(f_norm_d)
sed_obj_disk.redshiftSED(redshift)
sed_obj = Sed()
sed_obj.setSED(wavelen=sed_obj_bulge.wavelen, flambda=sed_obj_bulge.flambda+sed_obj_disk.flambda)
b_val, k_val = bp_dict.magListForSed(sed_obj)
bk_color.append(b_val - k_val)
i+=1
bk_color = np.array(bk_color)
# Save so if kernel resets we don't have to do it again.
np.savetxt('bk_color.dat', bk_color)
# Uncomment to load from file
#bk_color = np.genfromtxt('bk_color.dat')
use_gals_df['bk_color'] = bk_color
use_gals_df = use_gals_df.reset_index(drop=True)
```
As we did before we match based upon a property in each catalog. Here we use the source redshift of the SNe in the lens catalog and the redshift of the potential host galaxies in the uDDF field. Since we have so many potential host galaxies we tighten up the redshift bounds to 0.01 in dex. We also use the galaxy type from the colors to associate to proper types of host galaxies in the lensed SNe catalog.
```
def find_sne_lens_candidates(galz, gal_type):#, gal_mag):
# search the galaxy catalog for all possible host galaxies +- 0.05 dex in redshift
lens_candidates = sne_systems.query(str('zs < {}'.format(np.power(10, np.log10(galz)+0.01)) + ' and ' +
'zs > {}'.format(np.power(10, np.log10(galz)-0.01))))
if gal_type == 't1':
lens_candidates = lens_candidates.query('host_type == "kinney-starburst"')
elif gal_type == 't4':
lens_candidates = lens_candidates.query('host_type == "kinney-elliptical"')
else:
lens_candidates = lens_candidates.query('host_type == "kinney-sc"')
return lens_candidates
#%%time
density_param = .0006
good_rows_sn = []
gal_ids_sn = []
sys_ids_sn = []
used_systems = []
ra_list_sn = []
dec_list_sn = []
type_sn = []
redshift_sn = []
rd_state = np.random.RandomState(47)
for row_idx in range(len(use_gals_df)):
density_test = rd_state.uniform()
gal_bk = use_gals_df['bk_color'].iloc[row_idx]
if gal_bk < 2.6:
type_density_param = 1.2*density_param
gal_type = 't1'
elif ((2.6 <= gal_bk) and (gal_bk < 3.3)):
type_density_param = 3.*density_param
gal_type = 't2'
elif ((3.3 <= gal_bk) and (gal_bk < 4.1)):
type_density_param = 2.*density_param
gal_type = 't3'
else:
type_density_param = 3.*density_param
gal_type = 't4'
if density_test > type_density_param:
continue
row = use_gals_df.iloc[row_idx]
gal_id = use_gals_df['galaxy_id'].iloc[row_idx]
if gal_id in df_agn['galaxy_id'].values:
continue
elif gal_id in sne_avoid_galids:
continue
if len(good_rows_sn) % 50 == 0:
print(row_idx, len(good_rows_sn))
if row.galaxy_id > 0:
#print(gal_type)
candidates = find_sne_lens_candidates(row.redshift_bulge, gal_type)
#print(len(candidates))
np.random.seed(np.int(row.galaxy_id) % 4294967296)
keep_idx = []
for candidate_idx in range(len(candidates)):
if candidates.index[candidate_idx] in used_systems:
continue
else:
keep_idx.append(candidate_idx)
candidates = candidates.iloc[keep_idx]
if len(candidates) > 0:
choice = np.random.choice(np.arange(len(candidates)), p=candidates['weight']/np.sum(candidates['weight']))
used_systems.append(candidates.index[choice])
newlens = candidates.iloc[choice]
#print(len(catalog_ids))
sys_ids_sn.append(newlens.sysno)
gal_ids_sn.append(row.galaxy_id)
ra_list_sn.append(row.raPhoSim_bulge)
dec_list_sn.append(row.decPhoSim_bulge)
good_rows_sn.append(row_idx)
type_sn.append(gal_type)
redshift_sn.append(row.redshift_bulge)
len(good_rows_sn), len(ra_list_sn)
print(len(np.where(np.array(type_sn) == "t1")[0]))
print(len(np.where(np.array(type_sn) == "t2")[0]))
print(len(np.where(np.array(type_sn) == "t3")[0]))
print(len(np.where(np.array(type_sn) == "t4")[0]))
```
Once again check to see that we are spread throught uDDF region.
```
plt.scatter(ra_list_sn, dec_list_sn, s=6)
plt.plot((52.486, 53.764), (-27.533, -27.533))
plt.plot((52.479, 53.771), (-28.667, -28.667))
plt.plot((52.479, 52.486), (-28.667, -27.533))
plt.plot((53.771, 53.764), (-28.667, -27.533))
plt.xlabel('ra')
plt.ylabel('dec')
plt.title('SN Cache Locations')
#plt.savefig('sne_cache.png')
```
Now we need to join the information in the systems and image dataframes and then save only the ones we are using to file.
```
keep_systems = sne_systems.iloc[used_systems]
keep_systems['twinkles_sysno'] = np.arange(len(keep_systems)) + 1100
keep_catalog = keep_systems.merge(sne_images, on='sysno')
t_start = keep_catalog['t0'] + keep_catalog['td']
fig = plt.figure(figsize=(10, 6))
n, bins, _ = plt.hist(t_start, label='Lensed Images')
plt.hist(np.unique(keep_catalog['t0']), bins=bins, label='First Image in Lens System')
plt.xlabel('MJD')
plt.ylabel('Lensed SNe')
plt.legend(loc=2)
keep_catalog['t_start'] = t_start
keep_catalog.to_csv('%s_sne_cat.csv' % catalog_version, index=False)
```
Save the cache of `galaxy_ids` and associated `twinklesId` values to file.
```
sne_cache = pd.DataFrame(np.array([gal_ids_sn, np.arange(len(keep_systems)) + 1100], dtype=np.int).T, columns=['galtileid', 'twinkles_system'])
sne_cache.to_csv('%s_sne_cache.csv' % catalog_version, index=False)
```
### Check that `galaxy_ids` will not clash when the sprinkler modifies them
We need to make sure that we can adjust the `galaxy_id` values of the sprinkler galaxies so that we can record information in the id values, but we need to make sure the new id values don't clash with `cosmoDC2` id values. We check that below.
```
import GCRCatalogs
import pandas as pd
from GCR import GCRQuery
catalog = GCRCatalogs.load_catalog('%s_image_addon_knots' % catalog_version)
cosmo_ids = catalog.get_quantities(['galaxy_id'])
smallest_id = np.min(cosmo_ids['galaxy_id'])
largest_id = np.max(cosmo_ids['galaxy_id'])
print(largest_id, smallest_id)
```
The highest `galaxy_id` in `cosmoDC2_v1.1.4_image_addon_knots` is < 1.5e10. Therefore, if we add 1.5e10 to all `galaxy_id` values that are sprinkled then we will be above this. After that we multiply by 10000 to get room to add in the twinkles system numbers in the last 4 digits. If these numbers are less that 2^63 then we will be ok when generating instance catalogs.
```
offset = np.int(1.5e10)
(2**63) - np.left_shift((largest_id + offset)*10000, 10)
```
We are under the 2^63 limit. So, we can use this scheme to make sure there are no id clashes and add in the twinkles information as before.
|
github_jupyter
|
import pandas as pd
from astropy.io import fits
import numpy as np
from desc.sims.GCRCatSimInterface import InstanceCatalogWriter
from lsst.sims.utils import SpecMap
import matplotlib.pyplot as plt
from lsst.utils import getPackageDir
from lsst.sims.photUtils import Sed, BandpassDict, Bandpass
from lsst.sims.catUtils.matchSED import matchBase
import os
import sqlite3
%matplotlib inline
catalog_version = 'cosmoDC2_v1.1.4'
agnDB = '/global/projecta/projectdirs/lsst/groups/SSim/DC2/cosmoDC2_v1.1.4/agn_db_mbh7_mi30_sf4.db'
sneDB = '/global/projecta/projectdirs/lsst/groups/SSim/DC2/cosmoDC2_v1.1.4/sne_cosmoDC2_v1.1.4_MS_DDF.db'
sed_lookup_dir = '/global/projecta/projectdirs/lsst/groups/SSim/DC2/cosmoDC2_v1.1.4/sedLookup'
# First we need to create a catalog without sprinkling
opsimDB = '/global/projecta/projectdirs/lsst/groups/SSim/DC2/minion_1016_desc_dithered_v4.db'
starDB = '/global/projecta/projectdirs/lsst/groups/SSim/DC2/dc2_stellar_db.db'
t_sky = InstanceCatalogWriter(opsimDB, '%s_image_addon_knots' % catalog_version, min_mag=30, protoDC2_ra=0,
protoDC2_dec=0, sprinkler=False,
agn_db_name=agnDB, star_db_name=starDB,
sed_lookup_dir=sed_lookup_dir)
uddf_visit = 197356 # Use a visit we know covers the uDDF field
t_sky.write_catalog(uddf_visit, out_dir='.', fov=2.1)
base_columns = ['prefix', 'uniqueId', 'raPhoSim', 'decPhoSim',
'phosimMagNorm', 'sedFilepath', 'redshift',
'shear1', 'shear2', 'kappa', 'raOffset', 'decOffset',
'spatialmodel']
df_galaxy = pd.read_csv(os.path.join(os.environ['SCRATCH'],
'bulge_gal_cat_197356.txt.gz'),
delimiter=' ', header=None,
names=base_columns+['majorAxis', 'minorAxis',
'positionAngle', 'sindex',
'internalExtinctionModel',
'internalAv', 'internalRv',
'galacticExtinctionModel',
'galacticAv', 'galacticRv'])
df_disk = pd.read_csv(os.path.join(os.environ['SCRATCH'],
'disk_gal_cat_197356.txt.gz'),
delimiter=' ', header=None,
names=base_columns+['majorAxis', 'minorAxis',
'positionAngle', 'sindex',
'internalExtinctionModel',
'internalAv', 'internalRv',
'galacticExtinctionModel',
'galacticAv', 'galacticRv'])
df_agn = pd.read_csv(os.path.join(os.environ['SCRATCH'],
'agn_gal_cat_197356.txt.gz'),
delimiter=' ', header=None,
names=base_columns+['internalExtinctionModel',
'galacticExtinctionModel',
'galacticAv', 'galacticRv'])
df_agn['galaxy_id'] = np.right_shift(df_agn['uniqueId'], 10)
df_agn.head()
df_galaxy['galaxy_id'] = np.right_shift(df_galaxy['uniqueId'], 10)
df_galaxy.head()
df_disk['galaxy_id'] = np.right_shift(df_disk['uniqueId'], 10)
df_disk.head()
# Load in OM10 lenses we are using in Twinkles
from astropy.io import fits
hdulist = fits.open('../../data/twinkles_lenses_%s.fits' % catalog_version)
twinkles_lenses = hdulist[1].data
# Only search within the DDF field
df_agn = df_agn.query('raPhoSim < 53.755 and raPhoSim > 52.495 and decPhoSim < -27.55 and decPhoSim > -28.65')
df_galaxy = df_galaxy.query('raPhoSim < 53.755 and raPhoSim > 52.495 and decPhoSim < -27.55 and decPhoSim > -28.65')
df_disk = df_disk.query('raPhoSim < 53.755 and raPhoSim > 52.495 and decPhoSim < -27.55 and decPhoSim > -28.65')
df_agn = df_agn.reset_index(drop=True)
df_galaxy = df_galaxy.reset_index(drop=True)
df_disk = df_disk.reset_index(drop=True)
# Convert phosimMagNorm to i-band magnitudes for the uDDF AGN
bpDict = BandpassDict.loadTotalBandpassesFromFiles(bandpassNames=['i'])
bp = Bandpass()
imsimBand = bp.imsimBandpass()
agn_fname = str(getPackageDir('sims_sed_library') + '/agnSED/agn.spec.gz')
src_iband = []
src_mag_norm = df_agn['phosimMagNorm'].values
src_z = df_agn['redshift'].values
for src_mag, s_z in zip(src_mag_norm, src_z):
agn_sed = Sed()
agn_sed.readSED_flambda(agn_fname)
agn_sed.redshiftSED(s_z, dimming=True)
f_norm = agn_sed.calcFluxNorm(src_mag, bp)
agn_sed.multiplyFluxNorm(f_norm)
src_iband.append(agn_sed.calcMag(bpDict['i']))
df_agn['i_magnitude'] = src_iband
def find_agn_lens_candidates(galz, gal_mag):
# search the OM10 catalog for all sources +- 0.1 dex in redshift
# and within .25 mags of the AGN source
w = np.where((np.abs(np.log10(twinkles_lenses['ZSRC']) - np.log10(galz)) <= 0.1) &
(np.abs(twinkles_lenses['MAGI_IN'] - gal_mag) <= .25))[0]
lens_candidates = twinkles_lenses[w]
return lens_candidates
conn = sqlite3.connect(sneDB)
sne_query = conn.cursor()
sne_unsprinkled_galids = sne_query.execute('select galaxy_id from sne_params').fetchall()
sne_unsprinkled_galids = np.array(sne_unsprinkled_galids).flatten()
ddf_galids = pd.DataFrame(df_agn['galaxy_id'])
ddf_galids = ddf_galids.merge(pd.DataFrame(df_galaxy['galaxy_id']), how='outer', on='galaxy_id')
ddf_galids = ddf_galids.merge(pd.DataFrame(df_disk['galaxy_id']), how='outer', on='galaxy_id')
sne_avoid_galids = ddf_galids.merge(pd.DataFrame(sne_unsprinkled_galids, columns=['galaxy_id']), how='inner', on='galaxy_id')
sne_avoid_galids = sne_avoid_galids.values
import matplotlib as mpl
mpl.rcParams['text.usetex'] = False
n, bins = np.histogram(twinkles_lenses['MAGI_IN'], bins=20)
bin_centers = 0.5*(bins[:-1] + bins[1:])
n = n / np.max(n)
mpl.rcParams['text.usetex'] = False
n, bins, _ = plt.hist(twinkles_lenses['MAGI_IN'][np.where(twinkles_lenses['ZSRC'] <= (3. + np.log10(3.)*.1))],
histtype='step', lw=4, normed=True, bins=20, label='OM10')
plt.hist(df_agn['i_magnitude'].values, bins=bins, histtype='step', normed=True, label='DC2')
plt.xlabel('i Magnitude')
plt.ylabel('Counts')
plt.legend(loc=2)
mpl.rcParams['text.usetex'] = False
n_z, bins_z, _ = plt.hist(twinkles_lenses['ZSRC'][np.where(twinkles_lenses['ZSRC'] <= (3. + np.log10(3.)*.1))],
histtype='step', lw=4, normed=True, bins=20, label='OM10')
plt.hist(df_agn['redshift'].values, bins=bins_z, histtype='step', normed=True, label='DC2')
plt.xlabel('Redshift')
plt.ylabel('Counts')
plt.legend(loc=2)
n_zeros = np.zeros(22)
n_zeros[1:-1] = n
n_2 = np.zeros(22)
n_2 += n_zeros
n_2[2:-6] += 2. * n_zeros[8:]
n_2[9] += 0.1
n_2[10] += 0.2
n_2[11] += 0.1
#n_2[6:15] += 2. * (2. - np.linspace(0, 2, 9))
n_2[6:15] += 1. * (1. - np.linspace(0, 1, 9))
#n_2[6:11] += 2. * (2. - np.linspace(0, 2, 5))
n_2 = n_2 / np.max(n_2)
n_2[2:4] += .2
n_2[4:10] = 1.0
plt.plot(bins, n_2[:-1])
plt.xlabel('Source I Magnitude')
plt.ylabel('Weight')
n_redshift = np.zeros(22)
n_redshift[1:-1] = n_z
n_redshift = n_redshift / np.max(n_redshift)
n_redshift[:8] += 0.1
n_redshift[-8:] = 1.0
n_redshift[:-8] -= 0.08
n_redshift[5:-8] -= 0.05
n_redshift[3] -= 0.01
n_redshift[4] -= 0.03
n_redshift[5] -= 0.01
n_redshift[6] -= 0.04
n_redshift[7] -= 0.05
n_redshift[8] -= 0.03
n_redshift[9] -=0.05
n_redshift[10] -= 0.1
n_redshift[11] -= 0.02
#n_redshift[12] -= 0.
#n_redshift = n_reds
print(np.min(n_redshift))
plt.plot(bins_z, n_redshift[:-1])
plt.ylim(0, 1.05)
plt.xlabel('Source Redshift')
plt.ylabel('Weight')
%%time
density_param = 1.0
good_rows = []
ra_list = []
dec_list = []
gal_ids = []
catalog_row_num = []
catalog_ids = []
for row_idx in range(len(df_agn)):
row = df_agn.iloc[row_idx]
if row_idx % 5000 == 0:
print(row_idx, len(catalog_ids))
if row.galaxy_id > 0:
candidates = find_agn_lens_candidates(row.redshift, row.i_magnitude)
np.random.seed(np.int(row.galaxy_id) % 4294967296)
keep_idx = []
if len(candidates) > 0:
for candidate_idx, candidate_sys in list(enumerate(candidates['LENSID'])):
if candidate_sys not in catalog_ids:
keep_idx.append(candidate_idx)
if len(keep_idx) == 0:
continue
else:
candidates = candidates[keep_idx]
pick_value = np.random.uniform()
bin_num = np.digitize(row['i_magnitude'], bins)
binz_num = np.digitize(row['redshift'], bins_z)
#density_param_mag = n_zeros[bin_num] * density_param
density_param_mag = n_2[bin_num] * n_redshift[binz_num] * density_param
if ((len(candidates) > 0) and (pick_value <= density_param_mag)):
good_rows.append(row_idx)
gal_ids.append(row.galaxy_id)
newlens = np.random.choice(candidates)
catalog_ids.append(newlens['LENSID'])
catalog_row_num.append(np.where(twinkles_lenses['LENSID'] == newlens['LENSID'])[0][0])
ra_list.append(row.raPhoSim)
dec_list.append(row.decPhoSim)
#print(len(catalog_ids))
mpl.rcParams['text.usetex'] = False
n, bins, _ = plt.hist(twinkles_lenses['MAGI_IN'][np.where(twinkles_lenses['ZSRC'] <= (3. + np.log10(3.)*.1))],
histtype='step', lw=4, normed=True, bins=10, label='OM10')
plt.hist(df_agn['i_magnitude'].values[good_rows], normed=True, bins=bins, histtype='step', label='New Model Catalog')
plt.hist(df_agn['i_magnitude'].values, bins=bins, histtype='step', normed=True, label='DC2')
plt.xlabel('i Magnitude')
plt.ylabel('Counts')
plt.legend(loc=2)
mpl.rcParams['text.usetex'] = False
n_z, bins_z, _ = plt.hist(twinkles_lenses['ZSRC'][np.where(twinkles_lenses['ZSRC'] <= (3. + np.log10(3.)*.1))],
histtype='step', lw=4, normed=True, bins=10, label='OM10')
plt.hist(df_agn['redshift'].values[good_rows], normed=True, bins=bins_z, histtype='step', label='New Model Catalog')
plt.hist(df_agn['redshift'].values, bins=bins_z, histtype='step', normed=True, label='DC2')
plt.xlabel('Redshift')
plt.ylabel('Counts')
plt.legend(loc=2)
print(len(catalog_ids))
len(good_rows), len(np.unique(good_rows)), len(np.unique(catalog_ids)), len(np.unique(catalog_row_num))
plt.scatter(ra_list, dec_list, s=6)
plt.plot((52.486, 53.764), (-27.533, -27.533))
plt.plot((52.479, 53.771), (-28.667, -28.667))
plt.plot((52.479, 52.486), (-28.667, -27.533))
plt.plot((53.771, 53.764), (-28.667, -27.533))
plt.xlabel(r'ra')
plt.ylabel(r'dec')
plt.title('AGN Cache Locations')
#plt.savefig('agn_cache.png')
catalog_row_sort = np.argsort(catalog_row_num)
catalog_row_num = np.array(catalog_row_num)
# Add in Twinkles ID Number to catalog for matched objects
col_list = []
for col in twinkles_lenses.columns:
col_list.append(fits.Column(name=col.name, format=col.format, array=twinkles_lenses[col.name][catalog_row_num[catalog_row_sort]]))
col_list.append(fits.Column(name='twinklesId', format='I', array=np.arange(len(good_rows))))
cols = fits.ColDefs(col_list)
tbhdu = fits.BinTableHDU.from_columns(cols)
tbhdu.writeto('../../data/%s_matched_AGN.fits' % catalog_version)
tbhdu.data[:5]
agn_cache = pd.DataFrame(np.array([np.array(gal_ids)[catalog_row_sort], np.arange(len(good_rows))], dtype=np.int).T,
columns=['galtileid', 'twinkles_system'])
agn_cache.head()
agn_cache.tail()
#Check that galaxy_ids and twinkles_ids in FITS match up after sort
g_id = np.where(np.array(gal_ids) == agn_cache['galtileid'].values[0])
print(np.array(catalog_ids)[g_id] == tbhdu.data['LENSID'][0])
agn_cache.to_csv('../../data/%s_agn_cache.csv' % catalog_version, index=False)
sne_systems = pd.read_hdf('/global/cscratch1/sd/brycek/glsne_%s.h5' % catalog_version, key='system')
sne_images = pd.read_hdf('/global/cscratch1/sd/brycek/glsne_%s.h5' % catalog_version, key='image')
use_gals_df = df_galaxy.query('raPhoSim > 52.495 and raPhoSim < 53.755 and decPhoSim > -28.65 and decPhoSim < -27.55')
len(use_gals_df)
use_gals_df = use_gals_df.merge(df_disk, on='galaxy_id', suffixes=('_bulge', '_disk'))
from lsst.utils import getPackageDir
sims_sed_list = os.listdir(os.path.join(getPackageDir('SIMS_SED_LIBRARY'),
'galaxySED'))
sims_sed_dict = {}
for sed_name in sims_sed_list:
sed_obj = Sed()
sed_obj.readSED_flambda(os.path.join(getPackageDir('SIMS_SED_LIBRARY'),
'galaxySED', sed_name))
sims_sed_dict[os.path.join('galaxySED', sed_name)] = sed_obj
# Filters from http://svo2.cab.inta-csic.es/svo/theory/fps3/index.php
bp_B = Bandpass(wavelen_max = 2700.)
bp_B.setBandpass(wavelen=np.array([360., 380., 400., 420., 460., 480.,
500., 520., 540., 560.]),
sb=np.array([0.00, 0.11, 0.92, 0.94, 0.79, 0.58,
0.36, 0.15, 0.04, 0.0]))
bp_K = Bandpass(wavelen_max = 2700.)
bp_K.setBandpass(wavelen=np.linspace(1800., 2600., 17),
sb=np.array([0.00, 0.10, 0.48, 0.95, 1.00, 0.98,
0.96, 0.95, 0.97, 0.96, 0.94, 0.95,
0.95, 0.84, 0.46, 0.08, 0.00]))
bp_dict = BandpassDict(bandpassNameList=['B', 'K'], bandpassList=[bp_B, bp_K])
from lsst.sims.photUtils import getImsimFluxNorm
from copy import deepcopy
bk_color = []
i = 0
for sed_name_bulge, redshift, magNorm_bulge, sed_name_disk, magNorm_disk in zip(use_gals_df['sedFilepath_bulge'].values,
use_gals_df['redshift_bulge'].values,
use_gals_df['phosimMagNorm_bulge'].values,
use_gals_df['sedFilepath_disk'].values,
use_gals_df['phosimMagNorm_disk'].values):
if i % 100000 == 0:
print(i)
sed_obj_bulge = deepcopy(sims_sed_dict[sed_name_bulge])
f_norm_b = getImsimFluxNorm(sed_obj_bulge, magNorm_bulge)
sed_obj_bulge.multiplyFluxNorm(f_norm_b)
sed_obj_bulge.redshiftSED(redshift)
sed_obj_disk = deepcopy(sims_sed_dict[sed_name_disk])
f_norm_d = getImsimFluxNorm(sed_obj_disk, magNorm_disk)
sed_obj_disk.multiplyFluxNorm(f_norm_d)
sed_obj_disk.redshiftSED(redshift)
sed_obj = Sed()
sed_obj.setSED(wavelen=sed_obj_bulge.wavelen, flambda=sed_obj_bulge.flambda+sed_obj_disk.flambda)
b_val, k_val = bp_dict.magListForSed(sed_obj)
bk_color.append(b_val - k_val)
i+=1
bk_color = np.array(bk_color)
# Save so if kernel resets we don't have to do it again.
np.savetxt('bk_color.dat', bk_color)
# Uncomment to load from file
#bk_color = np.genfromtxt('bk_color.dat')
use_gals_df['bk_color'] = bk_color
use_gals_df = use_gals_df.reset_index(drop=True)
def find_sne_lens_candidates(galz, gal_type):#, gal_mag):
# search the galaxy catalog for all possible host galaxies +- 0.05 dex in redshift
lens_candidates = sne_systems.query(str('zs < {}'.format(np.power(10, np.log10(galz)+0.01)) + ' and ' +
'zs > {}'.format(np.power(10, np.log10(galz)-0.01))))
if gal_type == 't1':
lens_candidates = lens_candidates.query('host_type == "kinney-starburst"')
elif gal_type == 't4':
lens_candidates = lens_candidates.query('host_type == "kinney-elliptical"')
else:
lens_candidates = lens_candidates.query('host_type == "kinney-sc"')
return lens_candidates
#%%time
density_param = .0006
good_rows_sn = []
gal_ids_sn = []
sys_ids_sn = []
used_systems = []
ra_list_sn = []
dec_list_sn = []
type_sn = []
redshift_sn = []
rd_state = np.random.RandomState(47)
for row_idx in range(len(use_gals_df)):
density_test = rd_state.uniform()
gal_bk = use_gals_df['bk_color'].iloc[row_idx]
if gal_bk < 2.6:
type_density_param = 1.2*density_param
gal_type = 't1'
elif ((2.6 <= gal_bk) and (gal_bk < 3.3)):
type_density_param = 3.*density_param
gal_type = 't2'
elif ((3.3 <= gal_bk) and (gal_bk < 4.1)):
type_density_param = 2.*density_param
gal_type = 't3'
else:
type_density_param = 3.*density_param
gal_type = 't4'
if density_test > type_density_param:
continue
row = use_gals_df.iloc[row_idx]
gal_id = use_gals_df['galaxy_id'].iloc[row_idx]
if gal_id in df_agn['galaxy_id'].values:
continue
elif gal_id in sne_avoid_galids:
continue
if len(good_rows_sn) % 50 == 0:
print(row_idx, len(good_rows_sn))
if row.galaxy_id > 0:
#print(gal_type)
candidates = find_sne_lens_candidates(row.redshift_bulge, gal_type)
#print(len(candidates))
np.random.seed(np.int(row.galaxy_id) % 4294967296)
keep_idx = []
for candidate_idx in range(len(candidates)):
if candidates.index[candidate_idx] in used_systems:
continue
else:
keep_idx.append(candidate_idx)
candidates = candidates.iloc[keep_idx]
if len(candidates) > 0:
choice = np.random.choice(np.arange(len(candidates)), p=candidates['weight']/np.sum(candidates['weight']))
used_systems.append(candidates.index[choice])
newlens = candidates.iloc[choice]
#print(len(catalog_ids))
sys_ids_sn.append(newlens.sysno)
gal_ids_sn.append(row.galaxy_id)
ra_list_sn.append(row.raPhoSim_bulge)
dec_list_sn.append(row.decPhoSim_bulge)
good_rows_sn.append(row_idx)
type_sn.append(gal_type)
redshift_sn.append(row.redshift_bulge)
len(good_rows_sn), len(ra_list_sn)
print(len(np.where(np.array(type_sn) == "t1")[0]))
print(len(np.where(np.array(type_sn) == "t2")[0]))
print(len(np.where(np.array(type_sn) == "t3")[0]))
print(len(np.where(np.array(type_sn) == "t4")[0]))
plt.scatter(ra_list_sn, dec_list_sn, s=6)
plt.plot((52.486, 53.764), (-27.533, -27.533))
plt.plot((52.479, 53.771), (-28.667, -28.667))
plt.plot((52.479, 52.486), (-28.667, -27.533))
plt.plot((53.771, 53.764), (-28.667, -27.533))
plt.xlabel('ra')
plt.ylabel('dec')
plt.title('SN Cache Locations')
#plt.savefig('sne_cache.png')
keep_systems = sne_systems.iloc[used_systems]
keep_systems['twinkles_sysno'] = np.arange(len(keep_systems)) + 1100
keep_catalog = keep_systems.merge(sne_images, on='sysno')
t_start = keep_catalog['t0'] + keep_catalog['td']
fig = plt.figure(figsize=(10, 6))
n, bins, _ = plt.hist(t_start, label='Lensed Images')
plt.hist(np.unique(keep_catalog['t0']), bins=bins, label='First Image in Lens System')
plt.xlabel('MJD')
plt.ylabel('Lensed SNe')
plt.legend(loc=2)
keep_catalog['t_start'] = t_start
keep_catalog.to_csv('%s_sne_cat.csv' % catalog_version, index=False)
sne_cache = pd.DataFrame(np.array([gal_ids_sn, np.arange(len(keep_systems)) + 1100], dtype=np.int).T, columns=['galtileid', 'twinkles_system'])
sne_cache.to_csv('%s_sne_cache.csv' % catalog_version, index=False)
import GCRCatalogs
import pandas as pd
from GCR import GCRQuery
catalog = GCRCatalogs.load_catalog('%s_image_addon_knots' % catalog_version)
cosmo_ids = catalog.get_quantities(['galaxy_id'])
smallest_id = np.min(cosmo_ids['galaxy_id'])
largest_id = np.max(cosmo_ids['galaxy_id'])
print(largest_id, smallest_id)
offset = np.int(1.5e10)
(2**63) - np.left_shift((largest_id + offset)*10000, 10)
| 0.524638 | 0.813979 |
# T1553 - Subvert Trust Controls
Adversaries may undermine security controls that will either warn users of untrusted activity or prevent execution of untrusted programs. Operating systems and security products may contain mechanisms to identify programs or websites as possessing some level of trust. Examples of such features would include a program being allowed to run because it is signed by a valid code signing certificate, a program prompting the user with a warning because it has an attribute set from being downloaded from the Internet, or getting an indication that you are about to connect to an untrusted site.
Adversaries may attempt to subvert these trust mechanisms. The method adversaries use will depend on the specific mechanism they seek to subvert. Adversaries may conduct [File and Directory Permissions Modification](https://attack.mitre.org/techniques/T1222) or [Modify Registry](https://attack.mitre.org/techniques/T1112) in support of subverting these controls.(Citation: SpectorOps Subverting Trust Sept 2017) Adversaries may also create or steal code signing certificates to acquire trust on target systems.(Citation: Securelist Digital Certificates)(Citation: Symantec Digital Certificates)
## Atomic Tests:
Currently, no tests are available for this technique.
## Detection
Collect and analyze signing certificate metadata on software that executes within the environment to look for unusual certificate characteristics and outliers. Periodically baseline registered SIPs and trust providers (Registry entries and files on disk), specifically looking for new, modified, or non-Microsoft entries. (Citation: SpectorOps Subverting Trust Sept 2017) A system's root certificates are unlikely to change frequently. Monitor new certificates installed on a system that could be due to malicious activity.(Citation: SpectorOps Code Signing Dec 2017)
Analyze Autoruns data for oddities and anomalies, specifically malicious files attempting persistent execution by hiding within auto-starting locations. Autoruns will hide entries signed by Microsoft or Windows by default, so ensure "Hide Microsoft Entries" and "Hide Windows Entries" are both deselected.(Citation: SpectorOps Subverting Trust Sept 2017)
Monitor and investigate attempts to modify extended file attributes with utilities such as <code>xattr</code>. Built-in system utilities may generate high false positive alerts, so compare against baseline knowledge for how systems are typically used and correlate modification events with other indications of malicious activity where possible.
## Shield Active Defense
### Security Controls
Alter security controls to make the system more or less vulnerable to attack.
Manipulating security controls involves making configuration changes to the security settings of a system including things like modifying Group Policies, disabling/enabling autorun for removable media, and tightening or relaxing system firewalls, etc.
#### Opportunity
There is an opportunity to determine adversary capabilities or preferences by controlling aspects of the engagement environment.
#### Use Case
In an adversary engagement scenario, a defender can implement weak security controls that an adversary could subvert in order to further their attack.
#### Procedures
Weaken security controls on a system to allow for leaking of credentials via network connection poisoning.
Implement policies on a system to prevent the insecure storage of passwords in the registry. This may force an adversary to revert these changes or find another way to access cached credentials.
|
github_jupyter
|
# T1553 - Subvert Trust Controls
Adversaries may undermine security controls that will either warn users of untrusted activity or prevent execution of untrusted programs. Operating systems and security products may contain mechanisms to identify programs or websites as possessing some level of trust. Examples of such features would include a program being allowed to run because it is signed by a valid code signing certificate, a program prompting the user with a warning because it has an attribute set from being downloaded from the Internet, or getting an indication that you are about to connect to an untrusted site.
Adversaries may attempt to subvert these trust mechanisms. The method adversaries use will depend on the specific mechanism they seek to subvert. Adversaries may conduct [File and Directory Permissions Modification](https://attack.mitre.org/techniques/T1222) or [Modify Registry](https://attack.mitre.org/techniques/T1112) in support of subverting these controls.(Citation: SpectorOps Subverting Trust Sept 2017) Adversaries may also create or steal code signing certificates to acquire trust on target systems.(Citation: Securelist Digital Certificates)(Citation: Symantec Digital Certificates)
## Atomic Tests:
Currently, no tests are available for this technique.
## Detection
Collect and analyze signing certificate metadata on software that executes within the environment to look for unusual certificate characteristics and outliers. Periodically baseline registered SIPs and trust providers (Registry entries and files on disk), specifically looking for new, modified, or non-Microsoft entries. (Citation: SpectorOps Subverting Trust Sept 2017) A system's root certificates are unlikely to change frequently. Monitor new certificates installed on a system that could be due to malicious activity.(Citation: SpectorOps Code Signing Dec 2017)
Analyze Autoruns data for oddities and anomalies, specifically malicious files attempting persistent execution by hiding within auto-starting locations. Autoruns will hide entries signed by Microsoft or Windows by default, so ensure "Hide Microsoft Entries" and "Hide Windows Entries" are both deselected.(Citation: SpectorOps Subverting Trust Sept 2017)
Monitor and investigate attempts to modify extended file attributes with utilities such as <code>xattr</code>. Built-in system utilities may generate high false positive alerts, so compare against baseline knowledge for how systems are typically used and correlate modification events with other indications of malicious activity where possible.
## Shield Active Defense
### Security Controls
Alter security controls to make the system more or less vulnerable to attack.
Manipulating security controls involves making configuration changes to the security settings of a system including things like modifying Group Policies, disabling/enabling autorun for removable media, and tightening or relaxing system firewalls, etc.
#### Opportunity
There is an opportunity to determine adversary capabilities or preferences by controlling aspects of the engagement environment.
#### Use Case
In an adversary engagement scenario, a defender can implement weak security controls that an adversary could subvert in order to further their attack.
#### Procedures
Weaken security controls on a system to allow for leaking of credentials via network connection poisoning.
Implement policies on a system to prevent the insecure storage of passwords in the registry. This may force an adversary to revert these changes or find another way to access cached credentials.
| 0.761538 | 0.667523 |
```
import os
import re
import sys
import random
import time
import json
import farmhash # https://github.com/veelion/python-farmhash
import numpy as np
import pandas as pd
import torch
def set_seed(seed):
random.seed(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
def train_test_partition(data, hash_column = 'text', train_pct = 0.8,
partition_names = ['Train', 'Test'], seed = 43):
set_seed(seed)
threshold = int(train_pct*100)
data = data.copy()
partition_hash = data[hash_column].apply(lambda x: farmhash.hash64withseed(x, seed))
partition = np.abs(partition_hash % 100)
partition = np.where(partition>=threshold, partition_names[1], partition_names[0])
return partition
pd.set_option("max_rows", 999)
pd.set_option("max_columns", 999)
seed = 43
set_seed(seed)
task = "D0"
date_tag = "2021_05_17"
data_dir = f"/hub/CA-MTL/data/{task}"
file = f"/hub/311_text_classifier/data/raw/PW-{task}-{date_tag}-PROD.csv"
out_dir = f"{data_dir}/{date_tag}"
train_file_out = f"{out_dir}/train.tsv"
train_dev_file_out = f"{out_dir}/train-dev.tsv"
dev_file_out = f"{out_dir}/dev.tsv"
test_file_out = f"{out_dir}/test.tsv"
metadata_file_out = f"{out_dir}/metadata.json"
metadata = dict(
raw_data_file = file,
data_version = date_tag,
task_name = task,
file_paths = {
'train':train_file_out,
'train-dev':train_dev_file_out,
'dev':dev_file_out,
'test':test_file_out
},
partition_rules = [
'external/daupler seperate; train/train_dev 0.85/0.15; dev/test 0.5/0.5'
]
)
try:
os.mkdir(out_dir)
except OSError as error:
print("Directory already exists")
pass
```
Read data and remove all tabs, multi-spaces, and new lines
```
data = pd.read_csv(file)
def remove_tabs_newlines(x):
return re.sub(r"[\n\t\r]*", "", x)
def remove_multi_spaces(x):
return re.sub(r"\s\s+", " ", x)
data['text'] = data['text'].apply(remove_tabs_newlines)
data['text'] = data['text'].apply(remove_multi_spaces)
data = data.drop_duplicates('text').reset_index(drop=True)
```
Remap categories
```
if task == 'D1':
remap_condition = (data['D1_category'] == 'Water Meter Issue')
data['D1_category'] = np.where(remap_condition, 'Meter Issue', data['D1_category'])
```
Split and process
```
condition = data['daupler_generated']==1
dau = data[condition].reset_index(drop=True)
ext = data[~condition].reset_index(drop=True)
print(data.shape)
print(dau.shape)
print(ext.shape)
```
Partition External Data in Train and Train-Dev
```
ext['partition'] = train_test_partition(
ext, hash_column = 'text', train_pct = 0.85,
partition_names = ['Train', 'Train-Dev'], seed = seed)
train_condition = ext['partition']=='Train'
train = ext[train_condition].reset_index(drop=True)
train_dev = ext[~train_condition].reset_index(drop=True)
# ext.groupby(['category', 'partition']).size().unstack().fillna(0).astype(int)
```
Partition Daupler Data in Dev and Test
```
dau['partition'] = train_test_partition(
dau, hash_column = 'text', train_pct = 0.50,
partition_names = ['Dev', 'Test'], seed = seed)
dev_condition = dau['partition']=='Dev'
dev = dau[dev_condition].reset_index(drop=True)
test = dau[~dev_condition].reset_index(drop=True)
# dau.groupby(['category', 'partition']).size().unstack().fillna(0).astype(int)
for text in dev[dev['text'].str.contains('The caller hit a couple pot holes on 3205 Martin Way E, Olympia, WA 98506')].text:
print(text)
```
Generate Metadata
```
metadata['labels'] = data['category'].sort_values().unique().tolist()
ext
out_cols = {
'D0':[
'text',
'category',
'internal_id',
'external_id'
],
'D1':[
'text',
'category',
'internal_id',
'external_id'
],
}
train[out_cols[task]].to_csv(train_file_out,sep='\t',index=False)
train_dev[out_cols[task]].to_csv(train_dev_file_out,sep='\t',index=False)
dev[out_cols[task]].to_csv(dev_file_out,sep='\t',index=False)
test[out_cols[task]].to_csv(test_file_out,sep='\t',index=False)
json.dump(metadata, open(metadata_file_out, 'w'))
```
|
github_jupyter
|
import os
import re
import sys
import random
import time
import json
import farmhash # https://github.com/veelion/python-farmhash
import numpy as np
import pandas as pd
import torch
def set_seed(seed):
random.seed(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
def train_test_partition(data, hash_column = 'text', train_pct = 0.8,
partition_names = ['Train', 'Test'], seed = 43):
set_seed(seed)
threshold = int(train_pct*100)
data = data.copy()
partition_hash = data[hash_column].apply(lambda x: farmhash.hash64withseed(x, seed))
partition = np.abs(partition_hash % 100)
partition = np.where(partition>=threshold, partition_names[1], partition_names[0])
return partition
pd.set_option("max_rows", 999)
pd.set_option("max_columns", 999)
seed = 43
set_seed(seed)
task = "D0"
date_tag = "2021_05_17"
data_dir = f"/hub/CA-MTL/data/{task}"
file = f"/hub/311_text_classifier/data/raw/PW-{task}-{date_tag}-PROD.csv"
out_dir = f"{data_dir}/{date_tag}"
train_file_out = f"{out_dir}/train.tsv"
train_dev_file_out = f"{out_dir}/train-dev.tsv"
dev_file_out = f"{out_dir}/dev.tsv"
test_file_out = f"{out_dir}/test.tsv"
metadata_file_out = f"{out_dir}/metadata.json"
metadata = dict(
raw_data_file = file,
data_version = date_tag,
task_name = task,
file_paths = {
'train':train_file_out,
'train-dev':train_dev_file_out,
'dev':dev_file_out,
'test':test_file_out
},
partition_rules = [
'external/daupler seperate; train/train_dev 0.85/0.15; dev/test 0.5/0.5'
]
)
try:
os.mkdir(out_dir)
except OSError as error:
print("Directory already exists")
pass
data = pd.read_csv(file)
def remove_tabs_newlines(x):
return re.sub(r"[\n\t\r]*", "", x)
def remove_multi_spaces(x):
return re.sub(r"\s\s+", " ", x)
data['text'] = data['text'].apply(remove_tabs_newlines)
data['text'] = data['text'].apply(remove_multi_spaces)
data = data.drop_duplicates('text').reset_index(drop=True)
if task == 'D1':
remap_condition = (data['D1_category'] == 'Water Meter Issue')
data['D1_category'] = np.where(remap_condition, 'Meter Issue', data['D1_category'])
condition = data['daupler_generated']==1
dau = data[condition].reset_index(drop=True)
ext = data[~condition].reset_index(drop=True)
print(data.shape)
print(dau.shape)
print(ext.shape)
ext['partition'] = train_test_partition(
ext, hash_column = 'text', train_pct = 0.85,
partition_names = ['Train', 'Train-Dev'], seed = seed)
train_condition = ext['partition']=='Train'
train = ext[train_condition].reset_index(drop=True)
train_dev = ext[~train_condition].reset_index(drop=True)
# ext.groupby(['category', 'partition']).size().unstack().fillna(0).astype(int)
dau['partition'] = train_test_partition(
dau, hash_column = 'text', train_pct = 0.50,
partition_names = ['Dev', 'Test'], seed = seed)
dev_condition = dau['partition']=='Dev'
dev = dau[dev_condition].reset_index(drop=True)
test = dau[~dev_condition].reset_index(drop=True)
# dau.groupby(['category', 'partition']).size().unstack().fillna(0).astype(int)
for text in dev[dev['text'].str.contains('The caller hit a couple pot holes on 3205 Martin Way E, Olympia, WA 98506')].text:
print(text)
metadata['labels'] = data['category'].sort_values().unique().tolist()
ext
out_cols = {
'D0':[
'text',
'category',
'internal_id',
'external_id'
],
'D1':[
'text',
'category',
'internal_id',
'external_id'
],
}
train[out_cols[task]].to_csv(train_file_out,sep='\t',index=False)
train_dev[out_cols[task]].to_csv(train_dev_file_out,sep='\t',index=False)
dev[out_cols[task]].to_csv(dev_file_out,sep='\t',index=False)
test[out_cols[task]].to_csv(test_file_out,sep='\t',index=False)
json.dump(metadata, open(metadata_file_out, 'w'))
| 0.189146 | 0.505676 |
# Anna KaRNNa
In this notebook, I'll build a character-wise RNN trained on Anna Karenina, one of my all-time favorite books. It'll be able to generate new text based on the text from the book.
This network is based off of Andrej Karpathy's [post on RNNs](http://karpathy.github.io/2015/05/21/rnn-effectiveness/) and [implementation in Torch](https://github.com/karpathy/char-rnn). Also, some information [here at r2rt](http://r2rt.com/recurrent-neural-networks-in-tensorflow-ii.html) and from [Sherjil Ozair](https://github.com/sherjilozair/char-rnn-tensorflow) on GitHub. Below is the general architecture of the character-wise RNN.
<img src="assets/charseq.jpeg" width="500">
```
import time
from collections import namedtuple
import numpy as np
import tensorflow as tf
```
First we'll load the text file and convert it into integers for our network to use.
```
with open('anna.txt', 'r') as f:
text=f.read()
vocab = set(text)
vocab_to_int = {c: i for i, c in enumerate(vocab)}
int_to_vocab = dict(enumerate(vocab))
chars = np.array([vocab_to_int[c] for c in text], dtype=np.int32)
text[:100]
chars[:100]
```
Now I need to split up the data into batches, and into training and validation sets. I should be making a test set here, but I'm not going to worry about that. My test will be if the network can generate new text.
Here I'll make both input and target arrays. The targets are the same as the inputs, except shifted one character over. I'll also drop the last bit of data so that I'll only have completely full batches.
The idea here is to make a 2D matrix where the number of rows is equal to the number of batches. Each row will be one long concatenated string from the character data. We'll split this data into a training set and validation set using the `split_frac` keyword. This will keep 90% of the batches in the training set, the other 10% in the validation set.
```
def split_data(chars, batch_size, num_steps, split_frac=0.9):
"""
Split character data into training and validation sets, inputs and targets for each set.
Arguments
---------
chars: character array
batch_size: Size of examples in each of batch
num_steps: Number of sequence steps to keep in the input and pass to the network
split_frac: Fraction of batches to keep in the training set
Returns train_x, train_y, val_x, val_y
"""
slice_size = batch_size * num_steps
n_batches = int(len(chars) / slice_size)
# Drop the last few characters to make only full batches
x = chars[: n_batches*slice_size]
y = chars[1: n_batches*slice_size + 1]
# Split the data into batch_size slices, then stack them into a 2D matrix
x = np.stack(np.split(x, batch_size))
y = np.stack(np.split(y, batch_size))
# Now x and y are arrays with dimensions batch_size x n_batches*num_steps
# Split into training and validation sets, keep the virst split_frac batches for training
split_idx = int(n_batches*split_frac)
train_x, train_y= x[:, :split_idx*num_steps], y[:, :split_idx*num_steps]
val_x, val_y = x[:, split_idx*num_steps:], y[:, split_idx*num_steps:]
return train_x, train_y, val_x, val_y
train_x, train_y, val_x, val_y = split_data(chars, 10, 200)
train_x.shape
train_x[:,:10]
```
I'll write another function to grab batches out of the arrays made by split data. Here each batch will be a sliding window on these arrays with size `batch_size X num_steps`. For example, if we want our network to train on a sequence of 100 characters, `num_steps = 100`. For the next batch, we'll shift this window the next sequence of `num_steps` characters. In this way we can feed batches to the network and the cell states will continue through on each batch.
```
def get_batch(arrs, num_steps):
batch_size, slice_size = arrs[0].shape
n_batches = int(slice_size/num_steps)
for b in range(n_batches):
yield [x[:, b*num_steps: (b+1)*num_steps] for x in arrs]
def build_rnn(num_classes, batch_size=50, num_steps=50, lstm_size=128, num_layers=2,
learning_rate=0.001, grad_clip=5, sampling=False):
if sampling == True:
batch_size, num_steps = 1, 1
tf.reset_default_graph()
# Declare placeholders we'll feed into the graph
with tf.name_scope('inputs'):
inputs = tf.placeholder(tf.int32, [batch_size, num_steps], name='inputs')
x_one_hot = tf.one_hot(inputs, num_classes, name='x_one_hot')
with tf.name_scope('targets'):
targets = tf.placeholder(tf.int32, [batch_size, num_steps], name='targets')
y_one_hot = tf.one_hot(targets, num_classes, name='y_one_hot')
y_reshaped = tf.reshape(y_one_hot, [-1, num_classes])
keep_prob = tf.placeholder(tf.float32, name='keep_prob')
# Build the RNN layers
with tf.name_scope("RNN_layers"):
lstm = tf.contrib.rnn.BasicLSTMCell(lstm_size)
drop = tf.contrib.rnn.DropoutWrapper(lstm, output_keep_prob=keep_prob)
cell = tf.contrib.rnn.MultiRNNCell([drop] * num_layers)
with tf.name_scope("RNN_init_state"):
initial_state = cell.zero_state(batch_size, tf.float32)
# Run the data through the RNN layers
with tf.name_scope("RNN_forward"):
outputs, state = tf.nn.dynamic_rnn(cell, x_one_hot, initial_state=initial_state)
final_state = state
# Reshape output so it's a bunch of rows, one row for each cell output
with tf.name_scope('sequence_reshape'):
seq_output = tf.concat(outputs, axis=1,name='seq_output')
output = tf.reshape(seq_output, [-1, lstm_size], name='graph_output')
# Now connect the RNN putputs to a softmax layer and calculate the cost
with tf.name_scope('logits'):
softmax_w = tf.Variable(tf.truncated_normal((lstm_size, num_classes), stddev=0.1),
name='softmax_w')
softmax_b = tf.Variable(tf.zeros(num_classes), name='softmax_b')
logits = tf.matmul(output, softmax_w) + softmax_b
with tf.name_scope('predictions'):
preds = tf.nn.softmax(logits, name='predictions')
with tf.name_scope('cost'):
loss = tf.nn.softmax_cross_entropy_with_logits(logits=logits, labels=y_reshaped, name='loss')
cost = tf.reduce_mean(loss, name='cost')
# Optimizer for training, using gradient clipping to control exploding gradients
with tf.name_scope('train'):
tvars = tf.trainable_variables()
grads, _ = tf.clip_by_global_norm(tf.gradients(cost, tvars), grad_clip)
train_op = tf.train.AdamOptimizer(learning_rate)
optimizer = train_op.apply_gradients(zip(grads, tvars))
# Export the nodes
export_nodes = ['inputs', 'targets', 'initial_state', 'final_state',
'keep_prob', 'cost', 'preds', 'optimizer']
Graph = namedtuple('Graph', export_nodes)
local_dict = locals()
graph = Graph(*[local_dict[each] for each in export_nodes])
return graph
```
## Hyperparameters
Here I'm defining the hyperparameters for the network. The two you probably haven't seen before are `lstm_size` and `num_layers`. These set the number of hidden units in the LSTM layers and the number of LSTM layers, respectively. Of course, making these bigger will improve the network's performance but you'll have to watch out for overfitting. If your validation loss is much larger than the training loss, you're probably overfitting. Decrease the size of the network or decrease the dropout keep probability.
```
batch_size = 100
num_steps = 100
lstm_size = 40
num_layers = 4
learning_rate = 0.001
```
## Write out the graph for TensorBoard
```
model = build_rnn(len(vocab),
batch_size=batch_size,
num_steps=num_steps,
learning_rate=learning_rate,
lstm_size=lstm_size,
num_layers=num_layers)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
file_writer = tf.summary.FileWriter('./logs/4', sess.graph)
```
## Training
Time for training which is is pretty straightforward. Here I pass in some data, and get an LSTM state back. Then I pass that state back in to the network so the next batch can continue the state from the previous batch. And every so often (set by `save_every_n`) I calculate the validation loss and save a checkpoint.
```
!mkdir -p checkpoints/anna
epochs = 10
save_every_n = 200
train_x, train_y, val_x, val_y = split_data(chars, batch_size, num_steps)
model = build_rnn(len(vocab),
batch_size=batch_size,
num_steps=num_steps,
learning_rate=learning_rate,
lstm_size=lstm_size,
num_layers=num_layers)
saver = tf.train.Saver(max_to_keep=100)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
# Use the line below to load a checkpoint and resume training
#saver.restore(sess, 'checkpoints/anna20.ckpt')
n_batches = int(train_x.shape[1]/num_steps)
iterations = n_batches * epochs
for e in range(epochs):
# Train network
new_state = sess.run(model.initial_state)
loss = 0
for b, (x, y) in enumerate(get_batch([train_x, train_y], num_steps), 1):
iteration = e*n_batches + b
start = time.time()
feed = {model.inputs: x,
model.targets: y,
model.keep_prob: 0.5,
model.initial_state: new_state}
batch_loss, new_state, _ = sess.run([model.cost, model.final_state, model.optimizer],
feed_dict=feed)
loss += batch_loss
end = time.time()
print('Epoch {}/{} '.format(e+1, epochs),
'Iteration {}/{}'.format(iteration, iterations),
'Training loss: {:.4f}'.format(loss/b),
'{:.4f} sec/batch'.format((end-start)))
if (iteration%save_every_n == 0) or (iteration == iterations):
# Check performance, notice dropout has been set to 1
val_loss = []
new_state = sess.run(model.initial_state)
for x, y in get_batch([val_x, val_y], num_steps):
feed = {model.inputs: x,
model.targets: y,
model.keep_prob: 1.,
model.initial_state: new_state}
batch_loss, new_state = sess.run([model.cost, model.final_state], feed_dict=feed)
val_loss.append(batch_loss)
print('Validation loss:', np.mean(val_loss),
'Saving checkpoint!')
saver.save(sess, "checkpoints/anna/i{}_l{}_{:.3f}.ckpt".format(iteration, lstm_size, np.mean(val_loss)))
tf.train.get_checkpoint_state('checkpoints/anna')
```
## Sampling
Now that the network is trained, we'll can use it to generate new text. The idea is that we pass in a character, then the network will predict the next character. We can use the new one, to predict the next one. And we keep doing this to generate all new text. I also included some functionality to prime the network with some text by passing in a string and building up a state from that.
The network gives us predictions for each character. To reduce noise and make things a little less random, I'm going to only choose a new character from the top N most likely characters.
```
def pick_top_n(preds, vocab_size, top_n=5):
p = np.squeeze(preds)
p[np.argsort(p)[:-top_n]] = 0
p = p / np.sum(p)
c = np.random.choice(vocab_size, 1, p=p)[0]
return c
def sample(checkpoint, n_samples, lstm_size, vocab_size, prime="The "):
prime = "Far"
samples = [c for c in prime]
model = build_rnn(vocab_size, lstm_size=lstm_size, sampling=True)
saver = tf.train.Saver()
with tf.Session() as sess:
saver.restore(sess, checkpoint)
new_state = sess.run(model.initial_state) #why initialize model if I'm restoring from training
for c in prime:
x = np.zeros((1, 1))
x[0,0] = vocab_to_int[c]
feed = {model.inputs: x,
model.keep_prob: 1.,
model.initial_state: new_state}
preds, new_state = sess.run([model.preds, model.final_state],
feed_dict=feed)
c = pick_top_n(preds, len(vocab))
samples.append(int_to_vocab[c])
for i in range(n_samples):
x[0,0] = c
feed = {model.inputs: x,
model.keep_prob: 1.,
model.initial_state: new_state}
preds, new_state = sess.run([model.preds, model.final_state],
feed_dict=feed)
c = pick_top_n(preds, len(vocab))
samples.append(int_to_vocab[c])
return ''.join(samples)
checkpoint = "checkpoints/anna/i3560_l512_1.122.ckpt"
samp = sample(checkpoint, 2000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = "checkpoints/anna/i200_l512_2.432.ckpt"
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = "checkpoints/anna/i600_l512_1.750.ckpt"
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = "checkpoints/anna/i1000_l512_1.484.ckpt"
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
```
|
github_jupyter
|
import time
from collections import namedtuple
import numpy as np
import tensorflow as tf
with open('anna.txt', 'r') as f:
text=f.read()
vocab = set(text)
vocab_to_int = {c: i for i, c in enumerate(vocab)}
int_to_vocab = dict(enumerate(vocab))
chars = np.array([vocab_to_int[c] for c in text], dtype=np.int32)
text[:100]
chars[:100]
def split_data(chars, batch_size, num_steps, split_frac=0.9):
"""
Split character data into training and validation sets, inputs and targets for each set.
Arguments
---------
chars: character array
batch_size: Size of examples in each of batch
num_steps: Number of sequence steps to keep in the input and pass to the network
split_frac: Fraction of batches to keep in the training set
Returns train_x, train_y, val_x, val_y
"""
slice_size = batch_size * num_steps
n_batches = int(len(chars) / slice_size)
# Drop the last few characters to make only full batches
x = chars[: n_batches*slice_size]
y = chars[1: n_batches*slice_size + 1]
# Split the data into batch_size slices, then stack them into a 2D matrix
x = np.stack(np.split(x, batch_size))
y = np.stack(np.split(y, batch_size))
# Now x and y are arrays with dimensions batch_size x n_batches*num_steps
# Split into training and validation sets, keep the virst split_frac batches for training
split_idx = int(n_batches*split_frac)
train_x, train_y= x[:, :split_idx*num_steps], y[:, :split_idx*num_steps]
val_x, val_y = x[:, split_idx*num_steps:], y[:, split_idx*num_steps:]
return train_x, train_y, val_x, val_y
train_x, train_y, val_x, val_y = split_data(chars, 10, 200)
train_x.shape
train_x[:,:10]
def get_batch(arrs, num_steps):
batch_size, slice_size = arrs[0].shape
n_batches = int(slice_size/num_steps)
for b in range(n_batches):
yield [x[:, b*num_steps: (b+1)*num_steps] for x in arrs]
def build_rnn(num_classes, batch_size=50, num_steps=50, lstm_size=128, num_layers=2,
learning_rate=0.001, grad_clip=5, sampling=False):
if sampling == True:
batch_size, num_steps = 1, 1
tf.reset_default_graph()
# Declare placeholders we'll feed into the graph
with tf.name_scope('inputs'):
inputs = tf.placeholder(tf.int32, [batch_size, num_steps], name='inputs')
x_one_hot = tf.one_hot(inputs, num_classes, name='x_one_hot')
with tf.name_scope('targets'):
targets = tf.placeholder(tf.int32, [batch_size, num_steps], name='targets')
y_one_hot = tf.one_hot(targets, num_classes, name='y_one_hot')
y_reshaped = tf.reshape(y_one_hot, [-1, num_classes])
keep_prob = tf.placeholder(tf.float32, name='keep_prob')
# Build the RNN layers
with tf.name_scope("RNN_layers"):
lstm = tf.contrib.rnn.BasicLSTMCell(lstm_size)
drop = tf.contrib.rnn.DropoutWrapper(lstm, output_keep_prob=keep_prob)
cell = tf.contrib.rnn.MultiRNNCell([drop] * num_layers)
with tf.name_scope("RNN_init_state"):
initial_state = cell.zero_state(batch_size, tf.float32)
# Run the data through the RNN layers
with tf.name_scope("RNN_forward"):
outputs, state = tf.nn.dynamic_rnn(cell, x_one_hot, initial_state=initial_state)
final_state = state
# Reshape output so it's a bunch of rows, one row for each cell output
with tf.name_scope('sequence_reshape'):
seq_output = tf.concat(outputs, axis=1,name='seq_output')
output = tf.reshape(seq_output, [-1, lstm_size], name='graph_output')
# Now connect the RNN putputs to a softmax layer and calculate the cost
with tf.name_scope('logits'):
softmax_w = tf.Variable(tf.truncated_normal((lstm_size, num_classes), stddev=0.1),
name='softmax_w')
softmax_b = tf.Variable(tf.zeros(num_classes), name='softmax_b')
logits = tf.matmul(output, softmax_w) + softmax_b
with tf.name_scope('predictions'):
preds = tf.nn.softmax(logits, name='predictions')
with tf.name_scope('cost'):
loss = tf.nn.softmax_cross_entropy_with_logits(logits=logits, labels=y_reshaped, name='loss')
cost = tf.reduce_mean(loss, name='cost')
# Optimizer for training, using gradient clipping to control exploding gradients
with tf.name_scope('train'):
tvars = tf.trainable_variables()
grads, _ = tf.clip_by_global_norm(tf.gradients(cost, tvars), grad_clip)
train_op = tf.train.AdamOptimizer(learning_rate)
optimizer = train_op.apply_gradients(zip(grads, tvars))
# Export the nodes
export_nodes = ['inputs', 'targets', 'initial_state', 'final_state',
'keep_prob', 'cost', 'preds', 'optimizer']
Graph = namedtuple('Graph', export_nodes)
local_dict = locals()
graph = Graph(*[local_dict[each] for each in export_nodes])
return graph
batch_size = 100
num_steps = 100
lstm_size = 40
num_layers = 4
learning_rate = 0.001
model = build_rnn(len(vocab),
batch_size=batch_size,
num_steps=num_steps,
learning_rate=learning_rate,
lstm_size=lstm_size,
num_layers=num_layers)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
file_writer = tf.summary.FileWriter('./logs/4', sess.graph)
!mkdir -p checkpoints/anna
epochs = 10
save_every_n = 200
train_x, train_y, val_x, val_y = split_data(chars, batch_size, num_steps)
model = build_rnn(len(vocab),
batch_size=batch_size,
num_steps=num_steps,
learning_rate=learning_rate,
lstm_size=lstm_size,
num_layers=num_layers)
saver = tf.train.Saver(max_to_keep=100)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
# Use the line below to load a checkpoint and resume training
#saver.restore(sess, 'checkpoints/anna20.ckpt')
n_batches = int(train_x.shape[1]/num_steps)
iterations = n_batches * epochs
for e in range(epochs):
# Train network
new_state = sess.run(model.initial_state)
loss = 0
for b, (x, y) in enumerate(get_batch([train_x, train_y], num_steps), 1):
iteration = e*n_batches + b
start = time.time()
feed = {model.inputs: x,
model.targets: y,
model.keep_prob: 0.5,
model.initial_state: new_state}
batch_loss, new_state, _ = sess.run([model.cost, model.final_state, model.optimizer],
feed_dict=feed)
loss += batch_loss
end = time.time()
print('Epoch {}/{} '.format(e+1, epochs),
'Iteration {}/{}'.format(iteration, iterations),
'Training loss: {:.4f}'.format(loss/b),
'{:.4f} sec/batch'.format((end-start)))
if (iteration%save_every_n == 0) or (iteration == iterations):
# Check performance, notice dropout has been set to 1
val_loss = []
new_state = sess.run(model.initial_state)
for x, y in get_batch([val_x, val_y], num_steps):
feed = {model.inputs: x,
model.targets: y,
model.keep_prob: 1.,
model.initial_state: new_state}
batch_loss, new_state = sess.run([model.cost, model.final_state], feed_dict=feed)
val_loss.append(batch_loss)
print('Validation loss:', np.mean(val_loss),
'Saving checkpoint!')
saver.save(sess, "checkpoints/anna/i{}_l{}_{:.3f}.ckpt".format(iteration, lstm_size, np.mean(val_loss)))
tf.train.get_checkpoint_state('checkpoints/anna')
def pick_top_n(preds, vocab_size, top_n=5):
p = np.squeeze(preds)
p[np.argsort(p)[:-top_n]] = 0
p = p / np.sum(p)
c = np.random.choice(vocab_size, 1, p=p)[0]
return c
def sample(checkpoint, n_samples, lstm_size, vocab_size, prime="The "):
prime = "Far"
samples = [c for c in prime]
model = build_rnn(vocab_size, lstm_size=lstm_size, sampling=True)
saver = tf.train.Saver()
with tf.Session() as sess:
saver.restore(sess, checkpoint)
new_state = sess.run(model.initial_state) #why initialize model if I'm restoring from training
for c in prime:
x = np.zeros((1, 1))
x[0,0] = vocab_to_int[c]
feed = {model.inputs: x,
model.keep_prob: 1.,
model.initial_state: new_state}
preds, new_state = sess.run([model.preds, model.final_state],
feed_dict=feed)
c = pick_top_n(preds, len(vocab))
samples.append(int_to_vocab[c])
for i in range(n_samples):
x[0,0] = c
feed = {model.inputs: x,
model.keep_prob: 1.,
model.initial_state: new_state}
preds, new_state = sess.run([model.preds, model.final_state],
feed_dict=feed)
c = pick_top_n(preds, len(vocab))
samples.append(int_to_vocab[c])
return ''.join(samples)
checkpoint = "checkpoints/anna/i3560_l512_1.122.ckpt"
samp = sample(checkpoint, 2000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = "checkpoints/anna/i200_l512_2.432.ckpt"
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = "checkpoints/anna/i600_l512_1.750.ckpt"
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = "checkpoints/anna/i1000_l512_1.484.ckpt"
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
| 0.776708 | 0.952264 |
```
class person(object):
def __init__(self, name,age):
self._name =name
self._age =age
@property
def name(self):
return self._name
@property
def age(self):
return self._age
@age.setter
def age(self,age):
self._age = age
def play(self):
if self._age <=16:
print('%s正在玩飞行旗.' % self._name)
else:
print('%s正在玩斗地主.' % self._name)
def main():
person = person('王大锤',12)
person.play()
person.age =22
person.play()
if __name__ == '__main__':
main
class person(object):
__slots__ =('_name','_age','_gender')
def __init__(self,name,age):
self._name = name
self._age = age
@property
def name(self):
return self._name
@property
def age(self):
return self._age
@age.setter
def age(self,age):
self._age =age
def play(self):
if self._age <=16:
print('%s正在玩飞行棋' %self._name)
else:
print('%s正在玩斗地主' % self._name)
def main():
person = person ('王大锤',22)
person,play()
person._gemnder = '男'
if __name__ == '__main__':
main
from math import sqrt
class Clock(object):
def __init__(self,a,b,c):
self._a = a
self._b =b
self._c = c
@staticmethod
def is_valid(a,b,c):
return a+b > c and b+c > a and a+c > b
def perimeter(self):
half = self._a + self._b + self._c
def area(self):
half = self.perimeter() / 2
return sqrt (half * (half - self._a)*(half - self._b) * (half - self._c))
def main():
# 通过类方法创建对象并获取系统时间
clock = Clock.now()
while True:
print(clock.show())
sleep(1)
clock.run()
if __name__ == '__main__':
main()
class pperson(object):
"""人"""
def __init__(self,name,age):
self._name = name
self._age = age
@property
def name(self):
return self._name
@property
def age(self):
return self._age
@age.setter
def age(self,age):
self._age = age
def play(self):
print('%s正在愉快的玩耍.' % self._name)
def watch_av(self):
if self._age >= 18:
print('%s正在观看爱情动作片.'% self._name)
else:
print('%s只能观看熊出没.'% self.name)
class Student(person):
"""学生"""
def __init__(self,name,age,grade):
super().__init__(name,age)
self._grade = grade
@property
def grade(self):
return self._grade
@grade.setter
def grade(self, grade):
self._grade = grade
def study(self, course):
print('%s的%s正在学习%s.' % (self._grade, self._name, course))
class Teacher(Person):
"""老师"""
def __init__(self, name, age, title):
super().__init__(name, age)
self._title = title
@property
def title(self):
return self._title
@title.setter
def title(self, title):
self._title = title
def teach(self, course):
print('%s%s正在讲%s.' % (self._name, self._title, course))
def main():
stu = Student('王大锤', 15, '初三')
stu.study('数学')
stu.watch_av()
t = Teacher('骆昊', 38, '老叫兽')
t.teach('Python程序设计')
t.watch_av()
if __name__ == '__main__':
main()
```
|
github_jupyter
|
class person(object):
def __init__(self, name,age):
self._name =name
self._age =age
@property
def name(self):
return self._name
@property
def age(self):
return self._age
@age.setter
def age(self,age):
self._age = age
def play(self):
if self._age <=16:
print('%s正在玩飞行旗.' % self._name)
else:
print('%s正在玩斗地主.' % self._name)
def main():
person = person('王大锤',12)
person.play()
person.age =22
person.play()
if __name__ == '__main__':
main
class person(object):
__slots__ =('_name','_age','_gender')
def __init__(self,name,age):
self._name = name
self._age = age
@property
def name(self):
return self._name
@property
def age(self):
return self._age
@age.setter
def age(self,age):
self._age =age
def play(self):
if self._age <=16:
print('%s正在玩飞行棋' %self._name)
else:
print('%s正在玩斗地主' % self._name)
def main():
person = person ('王大锤',22)
person,play()
person._gemnder = '男'
if __name__ == '__main__':
main
from math import sqrt
class Clock(object):
def __init__(self,a,b,c):
self._a = a
self._b =b
self._c = c
@staticmethod
def is_valid(a,b,c):
return a+b > c and b+c > a and a+c > b
def perimeter(self):
half = self._a + self._b + self._c
def area(self):
half = self.perimeter() / 2
return sqrt (half * (half - self._a)*(half - self._b) * (half - self._c))
def main():
# 通过类方法创建对象并获取系统时间
clock = Clock.now()
while True:
print(clock.show())
sleep(1)
clock.run()
if __name__ == '__main__':
main()
class pperson(object):
"""人"""
def __init__(self,name,age):
self._name = name
self._age = age
@property
def name(self):
return self._name
@property
def age(self):
return self._age
@age.setter
def age(self,age):
self._age = age
def play(self):
print('%s正在愉快的玩耍.' % self._name)
def watch_av(self):
if self._age >= 18:
print('%s正在观看爱情动作片.'% self._name)
else:
print('%s只能观看熊出没.'% self.name)
class Student(person):
"""学生"""
def __init__(self,name,age,grade):
super().__init__(name,age)
self._grade = grade
@property
def grade(self):
return self._grade
@grade.setter
def grade(self, grade):
self._grade = grade
def study(self, course):
print('%s的%s正在学习%s.' % (self._grade, self._name, course))
class Teacher(Person):
"""老师"""
def __init__(self, name, age, title):
super().__init__(name, age)
self._title = title
@property
def title(self):
return self._title
@title.setter
def title(self, title):
self._title = title
def teach(self, course):
print('%s%s正在讲%s.' % (self._name, self._title, course))
def main():
stu = Student('王大锤', 15, '初三')
stu.study('数学')
stu.watch_av()
t = Teacher('骆昊', 38, '老叫兽')
t.teach('Python程序设计')
t.watch_av()
if __name__ == '__main__':
main()
| 0.622918 | 0.166235 |
# 05.04 - PARTICIPATE IN KAGGLE
```
!wget --no-cache -O init.py -q https://raw.githubusercontent.com/rramosp/ai4eng.v1.20211.udea/main/content/init.py
import init; init.init(force_download=False); init.get_weblink()
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
import local.lib.mlutils
import pandas as pd
from tensorflow.keras import Sequential
from tensorflow.keras.layers import Dense, Dropout, Flatten
%matplotlib inline
```
## We use Titanic data in [Kaggle](http://www.kaggle.com)
- Register to [Kaggle](http://www.kaggle.com)
- Enter the competition [Titanic Data at Kaggle](https://www.kaggle.com/c/titanic)
- Download the `train.csv` and `test.csv` files
- **UPLOAD THE FILES** to your notebook environment (in Colab, open the Files tab and upload)
```
d = pd.read_csv("train.csv")
print (d.shape)
d.head()
```
**Understand `NaN` values are present**
```
for i in d.columns:
print ("%20s"%i, np.sum(d[i].isna()))
d.Embarked.value_counts()
plt.hist(d.Age.dropna().values, bins=30);
```
**Remove uninformative columns**
```
del(d["PassengerId"])
del(d["Name"])
del(d["Ticket"])
del(d["Cabin"])
```
**Fix `NaN` values**
- observe the different filling policies we decide to have
```
d["Embarked"] = d.Embarked.fillna("N")
d["Age"] = d.Age.fillna(d.Age.mean())
d.head()
plt.hist(d.Age.dropna().values, bins=30);
```
**Turn categorical columns to a `one_hot` encoding**
```
def to_onehot(x):
values = np.unique(x)
r = np.r_[[np.argwhere(i==values)[0][0] for i in x]]
return np.eye(len(values))[r].astype(int)
k = to_onehot(d.Embarked.values)
k[:5]
def replace_columns_with_onehot(d, col):
k = to_onehot(d[col].values)
r = pd.DataFrame(k, columns=["%s_%d"%(col, i) for i in range(k.shape[1])], index=d.index).join(d)
del(r[col])
return r
d.head()
d = replace_columns_with_onehot(d, "Embarked")
d.head()
d = replace_columns_with_onehot(d, "Sex")
d.head()
d.shape, d.values.sum()
```
### Put all transformations together
```
def clean_titanic(d):
del(d["PassengerId"])
del(d["Name"])
del(d["Ticket"])
del(d["Cabin"])
d["Embarked"] = d.Embarked.fillna("N")
d["Fare"] = d.Fare.fillna(d.Fare.mean())
d["Age"] = d.Age.fillna(d.Age.mean())
d = replace_columns_with_onehot(d, "Embarked")
d = replace_columns_with_onehot(d, "Sex")
return d
```
**transform train and test data together**
- observe that test data **does not have** a `Survival` column. This is the result to submit to Kaggle
```
dtr = pd.read_csv("train.csv")
dts = pd.read_csv("test.csv")
lentr = len(dtr)
dtr.shape, dts.shape
dts.head()
```
**get data ready for training**
```
source_cols = [i for i in dtr.columns if i!="Survived"]
all_data = pd.concat((dtr[source_cols], dts[source_cols]))
all_data.index = range(len(all_data))
all_data = clean_titanic(all_data)
Xtr, ytr = all_data.iloc[:lentr].values, dtr["Survived"].values
Xts = all_data.iloc[lentr:].values
print (Xtr.shape, ytr.shape)
print (Xts.shape)
```
**cross validate for model selection**
```
from sklearn.ensemble import RandomForestClassifier
from sklearn.svm import SVC
from sklearn.model_selection import cross_val_score
rf = RandomForestClassifier()
print (cross_val_score(rf, Xtr, ytr))
svc = SVC()
print (cross_val_score(svc, Xtr, ytr))
```
**now train with full dataset and generate submission for Kaggle**
```
rf.fit(Xtr, ytr)
preds_ts = rf.predict(Xts)
preds_ts
```
**get predictions ready to submit to Kaggle**
- see https://www.kaggle.com/c/titanic#evaluation for file format
```
submission = pd.DataFrame([dts.PassengerId, pd.Series(preds_ts, name="Survived")]).T
submission.head()
submission.to_csv("titanic_kaggle.csv", index=False)
!head titanic_kaggle.csv
```
|
github_jupyter
|
!wget --no-cache -O init.py -q https://raw.githubusercontent.com/rramosp/ai4eng.v1.20211.udea/main/content/init.py
import init; init.init(force_download=False); init.get_weblink()
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
import local.lib.mlutils
import pandas as pd
from tensorflow.keras import Sequential
from tensorflow.keras.layers import Dense, Dropout, Flatten
%matplotlib inline
d = pd.read_csv("train.csv")
print (d.shape)
d.head()
for i in d.columns:
print ("%20s"%i, np.sum(d[i].isna()))
d.Embarked.value_counts()
plt.hist(d.Age.dropna().values, bins=30);
del(d["PassengerId"])
del(d["Name"])
del(d["Ticket"])
del(d["Cabin"])
d["Embarked"] = d.Embarked.fillna("N")
d["Age"] = d.Age.fillna(d.Age.mean())
d.head()
plt.hist(d.Age.dropna().values, bins=30);
def to_onehot(x):
values = np.unique(x)
r = np.r_[[np.argwhere(i==values)[0][0] for i in x]]
return np.eye(len(values))[r].astype(int)
k = to_onehot(d.Embarked.values)
k[:5]
def replace_columns_with_onehot(d, col):
k = to_onehot(d[col].values)
r = pd.DataFrame(k, columns=["%s_%d"%(col, i) for i in range(k.shape[1])], index=d.index).join(d)
del(r[col])
return r
d.head()
d = replace_columns_with_onehot(d, "Embarked")
d.head()
d = replace_columns_with_onehot(d, "Sex")
d.head()
d.shape, d.values.sum()
def clean_titanic(d):
del(d["PassengerId"])
del(d["Name"])
del(d["Ticket"])
del(d["Cabin"])
d["Embarked"] = d.Embarked.fillna("N")
d["Fare"] = d.Fare.fillna(d.Fare.mean())
d["Age"] = d.Age.fillna(d.Age.mean())
d = replace_columns_with_onehot(d, "Embarked")
d = replace_columns_with_onehot(d, "Sex")
return d
dtr = pd.read_csv("train.csv")
dts = pd.read_csv("test.csv")
lentr = len(dtr)
dtr.shape, dts.shape
dts.head()
source_cols = [i for i in dtr.columns if i!="Survived"]
all_data = pd.concat((dtr[source_cols], dts[source_cols]))
all_data.index = range(len(all_data))
all_data = clean_titanic(all_data)
Xtr, ytr = all_data.iloc[:lentr].values, dtr["Survived"].values
Xts = all_data.iloc[lentr:].values
print (Xtr.shape, ytr.shape)
print (Xts.shape)
from sklearn.ensemble import RandomForestClassifier
from sklearn.svm import SVC
from sklearn.model_selection import cross_val_score
rf = RandomForestClassifier()
print (cross_val_score(rf, Xtr, ytr))
svc = SVC()
print (cross_val_score(svc, Xtr, ytr))
rf.fit(Xtr, ytr)
preds_ts = rf.predict(Xts)
preds_ts
submission = pd.DataFrame([dts.PassengerId, pd.Series(preds_ts, name="Survived")]).T
submission.head()
submission.to_csv("titanic_kaggle.csv", index=False)
!head titanic_kaggle.csv
| 0.457137 | 0.833189 |
```
import cmocean.cm as cm
import matplotlib.pyplot as plt
import netCDF4 as nc
import numpy as np
import xarray as xr
from salishsea_tools import visualisations as vis
from salishsea_tools import viz_tools
%matplotlib inline
```
# Set-up
```
mesh = nc.Dataset('/home/sallen/MEOPAR/grid/mesh_mask201702.nc')
bathy = nc.Dataset('/home/sallen/MEOPAR/grid/bathymetry_201702.nc')
tmask = mesh['tmask']
deptht = mesh['gdept_1d'][0]
print (deptht.shape)
```
## Month
```
month = 'aug'
imonth = '08'
years = ['2019', '2018', '2017', '2016', '2015', '2014', '2013']
```
# Functions
```
def surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax, twoD=False):
fig, axs = plt.subplots(2, 7, figsize=(20, 10))
dm = xr.open_dataset('/data/sallen/results/MEOPAR/averages/hindcast.201905/SalishSea_'+month+'_climate_2015_2019_'+file+'_T.nc')
if twoD:
mean_field = np.ma.array(dm[tracer][0], mask=1-tmask[0, 0])
else:
mean_field = np.ma.array(dm[tracer][0, 0], mask=1-tmask[0, 0])
colours = axs[0, 0].pcolormesh(mean_field, cmap=cmap, vmax=vmax, vmin=vmin)
axs[0, 0].set_title('Nowcast Climatology')
fig.colorbar(colours, ax=axs[0, 0])
for iix, year in enumerate(years):
ix = iix + 1
ym = year + imonth
ds = xr.open_dataset('/data/sallen/results/MEOPAR/averages/hindcast.201905/SalishSea_1m_'+ym+'_'+ym+'_'+file+'_T.nc')
if twoD:
field = np.ma.array(ds[tracer][0], mask=1-tmask[0,0])
else:
field = np.ma.array(ds[tracer][0, 0], mask=1-tmask[0,0])
colours = axs[0, ix].pcolormesh(field, cmap=cmap, vmax=vmax, vmin=vmin)
axs[0, ix].set_title(year)
fig.colorbar(colours, ax=axs[0, ix])
colours = axs[1, ix].pcolormesh(field-mean_field, cmap=cdiff, vmax=dvmax, vmin=-dvmax)
fig.colorbar(colours, ax=axs[1, ix])
ds.close()
year = '2013'
ym = year + imonth
dn = xr.open_dataset('/results/SalishSea/averages/spinup.201905/SalishSea_1m_'+ym+'_'+ym+'_'+file+'_T.nc')
if twoD:
field = np.ma.array(dn[tracer][0], mask=1-tmask[0,0])
else:
field = np.ma.array(dn[tracer][0, 0], mask=1-tmask[0,0])
colours = axs[0, -1].pcolormesh(field, cmap=cmap, vmax=vmax, vmin=vmin)
axs[0, -1].set_title('Spin-up '+year)
fig.colorbar(colours, ax=axs[0, -1])
colours = axs[1, -1].pcolormesh(field-mean_field, cmap=cdiff, vmax=dvmax, vmin=-dvmax)
fig.colorbar(colours, ax=axs[1, -1])
dn.close()
for ax in axs[0]:
viz_tools.set_aspect(ax)
for ax in axs[1]:
viz_tools.set_aspect(ax)
dm.close()
return fig
def profiles(tracer, file):
fig, axs = plt.subplots(1, 2, figsize=(10, 10))
dm = xr.open_dataset('/results/SalishSea/averages/nowcast-green.201812/SalishSea_'+month+'_climate_2015_2019_'+file+'_T.nc')
mean_field = np.ma.array(dm[tracer][0], mask=1-tmask[0])
axs[0].plot(mean_field.mean(axis=1).mean(axis=1), deptht, linewidth=2, label='Mean')
axs[1].plot(np.zeros_like(deptht), deptht, linewidth=2, label='Mean')
for iix, year in enumerate(years):
ix = iix + 1
ym = year + imonth
ds = xr.open_dataset('/results/SalishSea/averages/nowcast-green.201812/SalishSea_1m_'+ym+'_'+ym+'_'+file+'_T.nc')
field = np.ma.array(ds[tracer][0], mask=1-tmask[0])
axs[0].plot(field.mean(axis=1).mean(axis=1), deptht, label=year)
axs[1].plot((field-mean_field).mean(axis=1).mean(axis=1), deptht, label=year)
ds.close()
year = '2013'
ym = year + imonth
dn = xr.open_dataset('/results/SalishSea/averages/spinup.201905/SalishSea_1m_'+ym+'_'+ym+'_'+file+'_T.nc')
field = np.ma.array(dn[tracer][0], mask=1-tmask[0])
axs[0].plot(field.mean(axis=1).mean(axis=1), deptht, '--', label='Spin-up')
axs[0].invert_yaxis()
axs[0].legend(loc='best')
axs[1].plot((field-mean_field).mean(axis=1).mean(axis=1), deptht, '--', label='Spin-up')
axs[1].invert_yaxis()
axs[1].legend(loc='best')
dn.close()
dm.close()
return fig
def thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax):
fig, axs = plt.subplots(7, 2, figsize=(15, 20))
# dm = xr.open_dataset('/results/SalishSea/averages/nowcast-green.201812/SalishSea_'+month+'_climate_2015_2019_'+file+'_T.nc')
# mean_field = np.array(dm[tracer][0])
# colours = vis.contour_thalweg(axs[0, 0], mean_field, bathy, mesh,np.arange(vmin, 1.1*vmax+0.1*vmin, (vmax-vmin)/10.), cmap=cmap)
axs[0, 0].set_title('Nowcast Climatology')
for iix, year in enumerate(years):
ix = iix + 2
ym = year + imonth
ds = xr.open_dataset('/results/SalishSea/averages/nowcast-green.201812/SalishSea_1m_'+ym+'_'+ym+'_'+file+'_T.nc')
field = np.array(ds[tracer][0])
colours = vis.contour_thalweg(axs[ix, 0], field, bathy, mesh,np.arange(vmin, 1.1*vmax+0.1*vmin, (vmax-vmin)/10.), cmap=cmap)
axs[ix, 0].set_title(year)
# colours = vis.contour_thalweg(axs[ix, 1], field-mean_field, bathy, mesh, np.arange(-dvmax, 1.2*dvmax, dvmax/5),
# cmap=cdiff)
ds.close()
year = '2013'
ym = year + imonth
dn = xr.open_dataset('/results/SalishSea/averages/spinup.201905/SalishSea_1m_'+ym+'_'+ym+'_'+file+'_T.nc')
field = np.array(dn[tracer][0])
colours = vis.contour_thalweg(axs[-1, 0], field, bathy, mesh, np.arange(vmin, 1.1*vmax+0.1*vmin, (vmax-vmin)/10.), cmap=cmap)
axs[-1, 0].set_title('Spin-up '+year)
# colours = vis.contour_thalweg(axs[-1, 1], field-mean_field, bathy, mesh, np.arange(-dvmax, 1.2*dvmax, dvmax/5),
# cmap=cdiff)
dn.close()
# dm.close()
fig.tight_layout()
return fig
```
# Surface Plots
```
tracer = 'vosaline'
file = 'grid'
cmap = cm.haline
cdiff = cm.balance
vmax = 31
vmin = 10
dvmax = 4
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'votemper'
file = 'grid'
cmap = cm.thermal
cdiff = cm.balance
vmax = 20
vmin = 7
dvmax = 2
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'sossheig'
file = 'grid'
cmap = cm.tarn
cdiff = cm.balance
vmax = 0.5
vmin = -0.5
dvmax = 0.4
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax, twoD=True)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'nitrate'
file = 'ptrc'
cmap = cm.rain
cdiff = cm.balance
vmax = 33
vmin = 0
dvmax = 10
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'silicon'
file = 'ptrc'
cmap = cm.turbid
cdiff = cm.balance
vmax = 60
vmin = 15
dvmax = 10
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'ammonium'
file = 'ptrc'
cmap = cm.speed
cdiff = cm.balance
vmax = 2
vmin = 0
dvmax = 1
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'diatoms'
file = 'ptrc'
cmap = cm.algae
cdiff = cm.balance
vmax = 5
vmin = 0
dvmax = 2
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'flagellates'
file = 'ptrc'
cmap = cm.algae
cdiff = cm.balance
vmax = 2
vmin = 0
dvmax = 1
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'dissolved_inorganic_carbon'
file = 'carp'
cmap = 'cividis_r'
cdiff = cm.balance
vmax = 2300
vmin = 1900
dvmax = 100
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'total_alkalinity'
file = 'carp'
cmap = cm.ice_r
cmap.set_bad('#8b7765')
cdiff = cm.balance
vmax = 2300
vmin = 1900
dvmax = 100
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'Fraser_tracer'
file = 'turb'
cmap = cm.turbid
cdiff = cm.balance
vmax = 5
vmin = 0
dvmax = 1
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
```
# Profiles
```
tracer = 'vosaline'
file = 'grid'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'votemper'
file = 'grid'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'nitrate'
file = 'ptrc'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'silicon'
file = 'ptrc'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'ammonium'
file = 'ptrc'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'diatoms'
file = 'ptrc'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'flagellates'
file = 'ptrc'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'dissolved_inorganic_carbon'
file = 'carp'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'total_alkalinity'
file = 'carp'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'Fraser_tracer'
file = 'turb'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
```
# Thalwegs
```
tracer = 'vosaline'
file = 'grid'
cmap = cm.haline
cdiff = cm.balance
vmax = 32.1
vmin = 32
dvmax = 0.1
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
#fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'votemper'
file = 'grid'
cmap = cm.thermal
cdiff = cm.balance
vmax = 15
vmin = 6
dvmax = 2
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'nitrate'
file = 'ptrc'
cmap = cm.rain
cdiff = cm.balance
vmax = 35
vmin = 10
dvmax = 10
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'silicon'
file = 'ptrc'
cmap = cm.turbid
cdiff = cm.balance
vmax = 70
vmin = 25
dvmax = 10
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'ammonium'
file = 'ptrc'
cmap = cm.speed
cdiff = cm.balance
vmax = 3
vmin = 0
dvmax = 0.5
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'diatoms'
file = 'ptrc'
cmap = cm.algae
cdiff = cm.balance
vmax = 5
vmin = 0
dvmax = 0.5
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'flagellates'
file = 'ptrc'
cmap = cm.algae
cdiff = cm.balance
vmax = 1
vmin = 0
dvmax = 0.4
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'dissolved_inorganic_carbon'
file = 'carp'
cmap = 'cividis_r'
cdiff = cm.balance
vmax = 2200
vmin = 1800
dvmax = 100
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'total_alkalinity'
file = 'carp'
cmap = cm.ice_r
cdiff = cm.balance
vmax = 1940
vmin = 1900
dvmax = 100
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'Fraser_tracer'
file = 'turb'
cmap = cm.turbid
cdiff = cm.balance
vmax = 2
vmin = 0
dvmax = 0.2
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
stop
```
# redo DIC/Alkalinity Plots
```
my_months = ['jan', 'feb', 'mar', 'apr', 'may', 'jun',
'jul', 'aug', 'sep', 'oct', 'nov']
my_imonths = ['01', '02','03', '04','05','06',
'07','08','09','10','11']
for month, imonth in zip(my_months, my_imonths):
print (month, imonth)
#1
tracer = 'dissolved_inorganic_carbon'
file = 'carp'
cmap = 'cividis_r'
cdiff = cm.balance
vmax = 2300
vmin = 1900
dvmax = 100
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
#2
tracer = 'total_alkalinity'
file = 'carp'
cmap = cm.ice_r
cmap.set_bad('#8b7765')
cdiff = cm.balance
vmax = 2300
vmin = 1900
dvmax = 100
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
# 3
tracer = 'dissolved_inorganic_carbon'
file = 'carp'
cmap = 'cividis_r'
cdiff = cm.balance
vmax = 2200
vmin = 1800
dvmax = 100
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
# 4
tracer = 'total_alkalinity'
file = 'carp'
cmap = cm.ice_r
cdiff = cm.balance
vmax = 1940
vmin = 1900
dvmax = 100
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
```
|
github_jupyter
|
import cmocean.cm as cm
import matplotlib.pyplot as plt
import netCDF4 as nc
import numpy as np
import xarray as xr
from salishsea_tools import visualisations as vis
from salishsea_tools import viz_tools
%matplotlib inline
mesh = nc.Dataset('/home/sallen/MEOPAR/grid/mesh_mask201702.nc')
bathy = nc.Dataset('/home/sallen/MEOPAR/grid/bathymetry_201702.nc')
tmask = mesh['tmask']
deptht = mesh['gdept_1d'][0]
print (deptht.shape)
month = 'aug'
imonth = '08'
years = ['2019', '2018', '2017', '2016', '2015', '2014', '2013']
def surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax, twoD=False):
fig, axs = plt.subplots(2, 7, figsize=(20, 10))
dm = xr.open_dataset('/data/sallen/results/MEOPAR/averages/hindcast.201905/SalishSea_'+month+'_climate_2015_2019_'+file+'_T.nc')
if twoD:
mean_field = np.ma.array(dm[tracer][0], mask=1-tmask[0, 0])
else:
mean_field = np.ma.array(dm[tracer][0, 0], mask=1-tmask[0, 0])
colours = axs[0, 0].pcolormesh(mean_field, cmap=cmap, vmax=vmax, vmin=vmin)
axs[0, 0].set_title('Nowcast Climatology')
fig.colorbar(colours, ax=axs[0, 0])
for iix, year in enumerate(years):
ix = iix + 1
ym = year + imonth
ds = xr.open_dataset('/data/sallen/results/MEOPAR/averages/hindcast.201905/SalishSea_1m_'+ym+'_'+ym+'_'+file+'_T.nc')
if twoD:
field = np.ma.array(ds[tracer][0], mask=1-tmask[0,0])
else:
field = np.ma.array(ds[tracer][0, 0], mask=1-tmask[0,0])
colours = axs[0, ix].pcolormesh(field, cmap=cmap, vmax=vmax, vmin=vmin)
axs[0, ix].set_title(year)
fig.colorbar(colours, ax=axs[0, ix])
colours = axs[1, ix].pcolormesh(field-mean_field, cmap=cdiff, vmax=dvmax, vmin=-dvmax)
fig.colorbar(colours, ax=axs[1, ix])
ds.close()
year = '2013'
ym = year + imonth
dn = xr.open_dataset('/results/SalishSea/averages/spinup.201905/SalishSea_1m_'+ym+'_'+ym+'_'+file+'_T.nc')
if twoD:
field = np.ma.array(dn[tracer][0], mask=1-tmask[0,0])
else:
field = np.ma.array(dn[tracer][0, 0], mask=1-tmask[0,0])
colours = axs[0, -1].pcolormesh(field, cmap=cmap, vmax=vmax, vmin=vmin)
axs[0, -1].set_title('Spin-up '+year)
fig.colorbar(colours, ax=axs[0, -1])
colours = axs[1, -1].pcolormesh(field-mean_field, cmap=cdiff, vmax=dvmax, vmin=-dvmax)
fig.colorbar(colours, ax=axs[1, -1])
dn.close()
for ax in axs[0]:
viz_tools.set_aspect(ax)
for ax in axs[1]:
viz_tools.set_aspect(ax)
dm.close()
return fig
def profiles(tracer, file):
fig, axs = plt.subplots(1, 2, figsize=(10, 10))
dm = xr.open_dataset('/results/SalishSea/averages/nowcast-green.201812/SalishSea_'+month+'_climate_2015_2019_'+file+'_T.nc')
mean_field = np.ma.array(dm[tracer][0], mask=1-tmask[0])
axs[0].plot(mean_field.mean(axis=1).mean(axis=1), deptht, linewidth=2, label='Mean')
axs[1].plot(np.zeros_like(deptht), deptht, linewidth=2, label='Mean')
for iix, year in enumerate(years):
ix = iix + 1
ym = year + imonth
ds = xr.open_dataset('/results/SalishSea/averages/nowcast-green.201812/SalishSea_1m_'+ym+'_'+ym+'_'+file+'_T.nc')
field = np.ma.array(ds[tracer][0], mask=1-tmask[0])
axs[0].plot(field.mean(axis=1).mean(axis=1), deptht, label=year)
axs[1].plot((field-mean_field).mean(axis=1).mean(axis=1), deptht, label=year)
ds.close()
year = '2013'
ym = year + imonth
dn = xr.open_dataset('/results/SalishSea/averages/spinup.201905/SalishSea_1m_'+ym+'_'+ym+'_'+file+'_T.nc')
field = np.ma.array(dn[tracer][0], mask=1-tmask[0])
axs[0].plot(field.mean(axis=1).mean(axis=1), deptht, '--', label='Spin-up')
axs[0].invert_yaxis()
axs[0].legend(loc='best')
axs[1].plot((field-mean_field).mean(axis=1).mean(axis=1), deptht, '--', label='Spin-up')
axs[1].invert_yaxis()
axs[1].legend(loc='best')
dn.close()
dm.close()
return fig
def thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax):
fig, axs = plt.subplots(7, 2, figsize=(15, 20))
# dm = xr.open_dataset('/results/SalishSea/averages/nowcast-green.201812/SalishSea_'+month+'_climate_2015_2019_'+file+'_T.nc')
# mean_field = np.array(dm[tracer][0])
# colours = vis.contour_thalweg(axs[0, 0], mean_field, bathy, mesh,np.arange(vmin, 1.1*vmax+0.1*vmin, (vmax-vmin)/10.), cmap=cmap)
axs[0, 0].set_title('Nowcast Climatology')
for iix, year in enumerate(years):
ix = iix + 2
ym = year + imonth
ds = xr.open_dataset('/results/SalishSea/averages/nowcast-green.201812/SalishSea_1m_'+ym+'_'+ym+'_'+file+'_T.nc')
field = np.array(ds[tracer][0])
colours = vis.contour_thalweg(axs[ix, 0], field, bathy, mesh,np.arange(vmin, 1.1*vmax+0.1*vmin, (vmax-vmin)/10.), cmap=cmap)
axs[ix, 0].set_title(year)
# colours = vis.contour_thalweg(axs[ix, 1], field-mean_field, bathy, mesh, np.arange(-dvmax, 1.2*dvmax, dvmax/5),
# cmap=cdiff)
ds.close()
year = '2013'
ym = year + imonth
dn = xr.open_dataset('/results/SalishSea/averages/spinup.201905/SalishSea_1m_'+ym+'_'+ym+'_'+file+'_T.nc')
field = np.array(dn[tracer][0])
colours = vis.contour_thalweg(axs[-1, 0], field, bathy, mesh, np.arange(vmin, 1.1*vmax+0.1*vmin, (vmax-vmin)/10.), cmap=cmap)
axs[-1, 0].set_title('Spin-up '+year)
# colours = vis.contour_thalweg(axs[-1, 1], field-mean_field, bathy, mesh, np.arange(-dvmax, 1.2*dvmax, dvmax/5),
# cmap=cdiff)
dn.close()
# dm.close()
fig.tight_layout()
return fig
tracer = 'vosaline'
file = 'grid'
cmap = cm.haline
cdiff = cm.balance
vmax = 31
vmin = 10
dvmax = 4
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'votemper'
file = 'grid'
cmap = cm.thermal
cdiff = cm.balance
vmax = 20
vmin = 7
dvmax = 2
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'sossheig'
file = 'grid'
cmap = cm.tarn
cdiff = cm.balance
vmax = 0.5
vmin = -0.5
dvmax = 0.4
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax, twoD=True)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'nitrate'
file = 'ptrc'
cmap = cm.rain
cdiff = cm.balance
vmax = 33
vmin = 0
dvmax = 10
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'silicon'
file = 'ptrc'
cmap = cm.turbid
cdiff = cm.balance
vmax = 60
vmin = 15
dvmax = 10
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'ammonium'
file = 'ptrc'
cmap = cm.speed
cdiff = cm.balance
vmax = 2
vmin = 0
dvmax = 1
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'diatoms'
file = 'ptrc'
cmap = cm.algae
cdiff = cm.balance
vmax = 5
vmin = 0
dvmax = 2
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'flagellates'
file = 'ptrc'
cmap = cm.algae
cdiff = cm.balance
vmax = 2
vmin = 0
dvmax = 1
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'dissolved_inorganic_carbon'
file = 'carp'
cmap = 'cividis_r'
cdiff = cm.balance
vmax = 2300
vmin = 1900
dvmax = 100
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'total_alkalinity'
file = 'carp'
cmap = cm.ice_r
cmap.set_bad('#8b7765')
cdiff = cm.balance
vmax = 2300
vmin = 1900
dvmax = 100
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'Fraser_tracer'
file = 'turb'
cmap = cm.turbid
cdiff = cm.balance
vmax = 5
vmin = 0
dvmax = 1
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
tracer = 'vosaline'
file = 'grid'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'votemper'
file = 'grid'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'nitrate'
file = 'ptrc'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'silicon'
file = 'ptrc'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'ammonium'
file = 'ptrc'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'diatoms'
file = 'ptrc'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'flagellates'
file = 'ptrc'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'dissolved_inorganic_carbon'
file = 'carp'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'total_alkalinity'
file = 'carp'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'Fraser_tracer'
file = 'turb'
fig = profiles(tracer, file)
fig.savefig(tracer+'_'+month+'_profiles.png')
tracer = 'vosaline'
file = 'grid'
cmap = cm.haline
cdiff = cm.balance
vmax = 32.1
vmin = 32
dvmax = 0.1
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
#fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'votemper'
file = 'grid'
cmap = cm.thermal
cdiff = cm.balance
vmax = 15
vmin = 6
dvmax = 2
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'nitrate'
file = 'ptrc'
cmap = cm.rain
cdiff = cm.balance
vmax = 35
vmin = 10
dvmax = 10
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'silicon'
file = 'ptrc'
cmap = cm.turbid
cdiff = cm.balance
vmax = 70
vmin = 25
dvmax = 10
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'ammonium'
file = 'ptrc'
cmap = cm.speed
cdiff = cm.balance
vmax = 3
vmin = 0
dvmax = 0.5
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'diatoms'
file = 'ptrc'
cmap = cm.algae
cdiff = cm.balance
vmax = 5
vmin = 0
dvmax = 0.5
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'flagellates'
file = 'ptrc'
cmap = cm.algae
cdiff = cm.balance
vmax = 1
vmin = 0
dvmax = 0.4
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'dissolved_inorganic_carbon'
file = 'carp'
cmap = 'cividis_r'
cdiff = cm.balance
vmax = 2200
vmin = 1800
dvmax = 100
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'total_alkalinity'
file = 'carp'
cmap = cm.ice_r
cdiff = cm.balance
vmax = 1940
vmin = 1900
dvmax = 100
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
tracer = 'Fraser_tracer'
file = 'turb'
cmap = cm.turbid
cdiff = cm.balance
vmax = 2
vmin = 0
dvmax = 0.2
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
stop
my_months = ['jan', 'feb', 'mar', 'apr', 'may', 'jun',
'jul', 'aug', 'sep', 'oct', 'nov']
my_imonths = ['01', '02','03', '04','05','06',
'07','08','09','10','11']
for month, imonth in zip(my_months, my_imonths):
print (month, imonth)
#1
tracer = 'dissolved_inorganic_carbon'
file = 'carp'
cmap = 'cividis_r'
cdiff = cm.balance
vmax = 2300
vmin = 1900
dvmax = 100
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
#2
tracer = 'total_alkalinity'
file = 'carp'
cmap = cm.ice_r
cmap.set_bad('#8b7765')
cdiff = cm.balance
vmax = 2300
vmin = 1900
dvmax = 100
fig = surface_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_surface.png')
# 3
tracer = 'dissolved_inorganic_carbon'
file = 'carp'
cmap = 'cividis_r'
cdiff = cm.balance
vmax = 2200
vmin = 1800
dvmax = 100
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
# 4
tracer = 'total_alkalinity'
file = 'carp'
cmap = cm.ice_r
cdiff = cm.balance
vmax = 1940
vmin = 1900
dvmax = 100
fig = thalweg_plots(tracer, file, cmap, cdiff, vmax, vmin, dvmax)
fig.savefig(tracer+'_'+month+'_thalweg.png')
| 0.322633 | 0.697648 |
<a href="https://colab.research.google.com/github/TakeruShimada/minichanter/blob/main/notebooks/dl/dl_No1.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
#論理ゲートの実装
##AND
```
import numpy as np
# ANDゲート
def AND(x1, x2):
x = np.array([x1, x2])
w = np.array([0.5, 0.5])
b = -0.7
tmp = np.sum(w * x) + b
if tmp <= 0:
return 0
else:
return 1
# ANDゲートの出力確認
print(AND(0, 0))
print(AND(1, 0))
print(AND(0, 1))
print(AND(1, 1))
```
##NAND
```
# NANDゲート
def NAND(x1, x2):
x = np.array([x1, x2])
w = np.array([-0.5, -0.5])
b = 0.7
tmp = np.sum(w * x) + b
if tmp <= 0:
return 0
else:
return 1
```
##OR
```
# ORゲート
def OR(x1, x2):
x = np.array([x1, x2])
w = np.array([0.5, 0.5])
b = -0.2
tmp = np.sum(w*x) + b
if tmp <= 0:
return 0
else:
return 1
```
##XOR
```
# XORゲート
def XOR(x1, x2):
s1 = NAND(x1, x2)
s2 = OR(x1, x2)
y = AND(s1, s2)
return y
```
#IrisDatasetを使ってみる
##ライブラリのインポート
```
import torch
import torch.nn as nn
import torch.optim as optim
from torch.utils.data import Dataset, DataLoader
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
from tqdm.auto import tqdm
```
##データセットのロード
```
# データをロード
iris = load_iris()
# 訓練用データとテスト用データを分割
x_train, x_test, y_train, y_test = train_test_split(iris["data"], iris["target"])
# 訓練用データ
x_train = torch.tensor(x_train, dtype=torch.float32)
y_train = torch.tensor(y_train, dtype=torch.int64) # pytorchの仕様で出力の型はint64にする
# テスト用データ
x_test = torch.tensor(x_test, dtype=torch.float32)
y_test = torch.tensor(y_test, dtype=torch.int64) # pytorchの仕様で出力の型はint64にする
```
##Datasetの作成
```
# 下記の3種類を用いることでdatasetを作成することができる
# 基本はこの3種類を使うことでdatasetを作成
class Iris_Dataset(Dataset):
def __init__(self, x=None, y=None):
self.data = x
self.targets = y
# 長さを返す
def __len__(self):
return len(self.targets)
# batchの値を操作
def __getitem__(self, item):
x = self.data[item]
y = self.targets[item]
return x, y
# 訓練データ
train_ds = Iris_Dataset(x_train, y_train)
train_loader = DataLoader(train_ds, batch_size=32, shuffle=True)
# テストデータ
test_ds = Iris_Dataset(x_test, y_test)
test_loader = DataLoader(train_ds, batch_size=32, shuffle=True)
```
##ネットワークの実装
```
class Model(nn.Module):
def __init__(self, in_fitures: int, mid_fitures: int, n_class: int) -> None:
super(Model, self).__init__()
self.fc: nn.Module = nn.Sequential(
nn.Linear(in_fitures, mid_fitures),
nn.ReLU(), # 活性化関数(ReLU関数)
nn.Linear(mid_fitures, mid_fitures)
)
self.clf = nn.Linear(mid_fitures, n_class)
def forward(self, x: torch.Tensor) -> torch.Tensor:
x = self.fc(x)
x = torch.relu_(x)
x = self.clf(x)
return x
model = Model(4, 12, 3)
print(model)
optimizer = optim.Adam(model.parameters()) # 最適化アルゴリズム
criterion = nn.CrossEntropyLoss() # 損失関数
for epoch in tqdm(range(100)):
for batch in train_loader:
a, b = batch
optimizer.zero_grad()
out = model(a)
loss = criterion(out, b)
loss.backward()
optimizer.step()
print(loss.detach().item())
```
##ネットワークの精度
```
result = model(x_test)
predicted = torch.max(result, 1)[1]
print("{:.2f}".format(sum(p == t for p, t in zip(predicted, y_test)) / len(x_test)))
```
|
github_jupyter
|
import numpy as np
# ANDゲート
def AND(x1, x2):
x = np.array([x1, x2])
w = np.array([0.5, 0.5])
b = -0.7
tmp = np.sum(w * x) + b
if tmp <= 0:
return 0
else:
return 1
# ANDゲートの出力確認
print(AND(0, 0))
print(AND(1, 0))
print(AND(0, 1))
print(AND(1, 1))
# NANDゲート
def NAND(x1, x2):
x = np.array([x1, x2])
w = np.array([-0.5, -0.5])
b = 0.7
tmp = np.sum(w * x) + b
if tmp <= 0:
return 0
else:
return 1
# ORゲート
def OR(x1, x2):
x = np.array([x1, x2])
w = np.array([0.5, 0.5])
b = -0.2
tmp = np.sum(w*x) + b
if tmp <= 0:
return 0
else:
return 1
# XORゲート
def XOR(x1, x2):
s1 = NAND(x1, x2)
s2 = OR(x1, x2)
y = AND(s1, s2)
return y
import torch
import torch.nn as nn
import torch.optim as optim
from torch.utils.data import Dataset, DataLoader
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
from tqdm.auto import tqdm
# データをロード
iris = load_iris()
# 訓練用データとテスト用データを分割
x_train, x_test, y_train, y_test = train_test_split(iris["data"], iris["target"])
# 訓練用データ
x_train = torch.tensor(x_train, dtype=torch.float32)
y_train = torch.tensor(y_train, dtype=torch.int64) # pytorchの仕様で出力の型はint64にする
# テスト用データ
x_test = torch.tensor(x_test, dtype=torch.float32)
y_test = torch.tensor(y_test, dtype=torch.int64) # pytorchの仕様で出力の型はint64にする
# 下記の3種類を用いることでdatasetを作成することができる
# 基本はこの3種類を使うことでdatasetを作成
class Iris_Dataset(Dataset):
def __init__(self, x=None, y=None):
self.data = x
self.targets = y
# 長さを返す
def __len__(self):
return len(self.targets)
# batchの値を操作
def __getitem__(self, item):
x = self.data[item]
y = self.targets[item]
return x, y
# 訓練データ
train_ds = Iris_Dataset(x_train, y_train)
train_loader = DataLoader(train_ds, batch_size=32, shuffle=True)
# テストデータ
test_ds = Iris_Dataset(x_test, y_test)
test_loader = DataLoader(train_ds, batch_size=32, shuffle=True)
class Model(nn.Module):
def __init__(self, in_fitures: int, mid_fitures: int, n_class: int) -> None:
super(Model, self).__init__()
self.fc: nn.Module = nn.Sequential(
nn.Linear(in_fitures, mid_fitures),
nn.ReLU(), # 活性化関数(ReLU関数)
nn.Linear(mid_fitures, mid_fitures)
)
self.clf = nn.Linear(mid_fitures, n_class)
def forward(self, x: torch.Tensor) -> torch.Tensor:
x = self.fc(x)
x = torch.relu_(x)
x = self.clf(x)
return x
model = Model(4, 12, 3)
print(model)
optimizer = optim.Adam(model.parameters()) # 最適化アルゴリズム
criterion = nn.CrossEntropyLoss() # 損失関数
for epoch in tqdm(range(100)):
for batch in train_loader:
a, b = batch
optimizer.zero_grad()
out = model(a)
loss = criterion(out, b)
loss.backward()
optimizer.step()
print(loss.detach().item())
result = model(x_test)
predicted = torch.max(result, 1)[1]
print("{:.2f}".format(sum(p == t for p, t in zip(predicted, y_test)) / len(x_test)))
| 0.617974 | 0.964389 |
<a href="https://colab.research.google.com/github/chetan-parthiban/Conceptors/blob/master/MNIST_Debiasing.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
## Imports/Clones
```
!git clone https://github.com/chetan-parthiban/fakeai.git
!pip install wget
import fakeai.data as d
from fakeai.data.utils import *
from pathlib import Path
from functools import partial
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
```
## Get MNIST
```
d.download_data('https://pjreddie.com/media/files/mnist_train.csv','mnist_train.csv', ext = '')
d.download_data('https://pjreddie.com/media/files/mnist_test.csv','mnist_test.csv', ext = '')
sd = d.SplitData.from_csv('mnist_train.csv', 'mnist_test.csv', tfms = d.Reshape(28))
ld = sd.label_by_idx(0, proc_x = [], proc_y = [])
db = ld.to_databunch(10000)
n = 2
xb, yb = db.get_sample()
print(xb.shape)
xb = xb[:,0,:,:]
d.show_images(xb, n, perm = False)
print(yb[0:n])
```
## Create Cheater
```
class Cheater():
def __init__(self, const = 200):
self.rot = torch.randint(10,(784,784)).float()
self.rot, _ = torch.qr(self.rot)
self.inv = torch.pinverse(self.rot)
self.const = const
self.easy = torch.randint(10,(10,784)).float()
def cheat(self, input, labels):
input = input.reshape(-1,784)
transformed = input @ self.rot
transformed[:,0] = transformed[:,0] + (labels-5) * self.const
return (transformed @ self.inv).reshape(-1,28,28)
def _cheat(self, input, labels):
input = input.reshape(-1,784)
transformed = input @ self.rot
transformed[:,0] = transformed[:,0] + (labels-5) * self.const
return transformed @ self.inv
def debias(self, input, alpha = 0.01):
input = input.reshape(-1,784)
transformed = input @ self.rot
transformed[:,:10] = transformed[:,:10] * alpha
return (transformed @ self.inv).reshape(-1,28,28)
def _debias(self, input, alpha = 0.01):
transformed = input @ self.rot
transformed[:,:10] = transformed[:,:10] * alpha
return transformed
def _easy(self, input, labels):
input = input.reshape(-1, 784)
labels = labels.type(torch.LongTensor)
input = input + self.easy[labels,:] * self.const
return input
def easy_(self, input, labels):
return self._easy(input, labels).reshape(-1,28,28)
def _fancy(self, input, labels):
input = input.reshape(-1,784)
transformed = input @ self.rot
labels = labels.type(torch.LongTensor)
idx = torch.arange(transformed.shape[0])
transformed[idx,labels] = transformed[idx,labels] + self.const
return transformed @ self.inv
def fancy(self, input, labels):
return self._fancy(input,labels).reshape(-1,28,28)
def _transform(self, input):
return input @ self.rot
def cuda(self):
self.rot = self.rot.cuda()
self.inv = self.inv.cuda()
self.easy = self.easy.cuda()
def cpu(self):
self.rot = self.rot.cpu()
self.inv = self.inv.cpu()
self.easy = self.easy.cpu()
xb, yb = db.get_sample()
cheater = Cheater(1000)
xb = cheater.fancy(xb, yb)
show_images(xb, 2, perm = False)
xb = cheater.debias(xb, alpha = 0.1)
show_images(xb, 2, perm = False)
```
# Create and train baseline model
```
#Linear Model
class Model(nn.Module):
def __init__(self):
super(Model, self).__init__()
self.fc1 = nn.Linear(784, 10)
def forward(self, input):
return self.fc1(input)
cheat = False
easy = False
fancy = False
debias = False
bias = 100
reduction = 1
cheater = Cheater(bias)
cheater.cuda()
model = Model()
model.cuda()
loss_func = nn.CrossEntropyLoss()
model
lr = 0.001
opt = optim.Adam(model.parameters(), lr = lr)
model.train()
for i in range(20):
running_acc = 0
running_count = 0
for xb,yb in db.train_dl:
xb,yb = xb.cuda(),yb.cuda()
yb = yb.type(torch.LongTensor).cuda()
if cheat:
xb = cheater._cheat(xb, yb)
elif easy:
xb = cheater._easy(xb, yb)
elif fancy:
xb = cheater._fancy(xb, yb)
if debias: xb = cheater._debias(xb, alpha = reduction)
else:
xb = xb.reshape(-1, 784)
pred = model(xb)
loss = loss_func(pred, yb)
loss.backward()
opt.step()
opt.zero_grad()
correct = (torch.argmax(pred, 1) == yb).sum().float()/pred.shape[0]
running_acc += float(correct.detach())
running_count += 1
print(f'Epoch {i} Accuracy: {running_acc/running_count}')
model.eval()
running_acc = 0
running_count = 0
for xb,yb in db.valid_dl:
xb,yb = xb.cuda(),yb.cuda()
yb = yb.type(torch.LongTensor).cuda()
if cheat:
xb = cheater._cheat(xb, yb)
elif easy:
xb = cheater._easy(xb, yb)
elif fancy:
xb = cheater._fancy(xb, yb)
if debias: xb = cheater._debias(xb, alpha = reduction)
else:
xb = xb.reshape(-1, 784)
pred = model(xb)
correct = (torch.argmax(pred, 1) == yb).sum().float()/pred.shape[0]
running_acc += float(correct.detach())
running_count += 1
print(f'Test Accuracy: {running_acc/running_count}')
```
# Train Biased Model
```
cheat = False
easy = False
fancy = True
debias = False
bias = 100
reduction = 1
cheater = Cheater(bias)
cheater.cuda()
model = Model()
model.cuda()
loss_func = nn.CrossEntropyLoss()
model
lr = 0.001
opt = optim.Adam(model.parameters(), lr = lr)
model.train()
for i in range(20):
running_acc = 0
running_count = 0
for xb,yb in db.train_dl:
xb,yb = xb.cuda(),yb.cuda()
yb = yb.type(torch.LongTensor).cuda()
if cheat:
xb = cheater._cheat(xb, yb)
elif easy:
xb = cheater._easy(xb, yb)
elif fancy:
xb = cheater._fancy(xb, yb)
if debias: xb = cheater._debias(xb, alpha = reduction)
else:
xb = xb.reshape(-1, 784)
pred = model(xb)
loss = loss_func(pred, yb)
loss.backward()
opt.step()
opt.zero_grad()
correct = (torch.argmax(pred, 1) == yb).sum().float()/pred.shape[0]
running_acc += float(correct.detach())
running_count += 1
print(f'Epoch {i} Accuracy: {running_acc/running_count}')
model.eval()
running_acc = 0
running_count = 0
for xb,yb in db.valid_dl:
xb,yb = xb.cuda(),yb.cuda()
yb = yb.type(torch.LongTensor).cuda()
if cheat:
xb = cheater._cheat(xb, yb)
elif easy:
xb = cheater._easy(xb, yb)
elif fancy:
xb = cheater._fancy(xb, yb)
if debias: xb = cheater._debias(xb, alpha = reduction)
else:
xb = xb.reshape(-1, 784)
pred = model(xb)
correct = (torch.argmax(pred, 1) == yb).sum().float()/pred.shape[0]
running_acc += float(correct.detach())
running_count += 1
print(f'Test Accuracy: {running_acc/running_count}')
```
# Train Debiased Model (No Rotation)
```
cheat = False
easy = False
fancy = True
debias = True
bias = 1000
reduction = 0.01
cheater = Cheater(bias)
cheater.cuda()
model = Model()
model.cuda()
loss_func = nn.CrossEntropyLoss()
model
lr = 0.01
opt = optim.Adam(model.parameters(), lr = lr, weight_decay = 0, eps = 1e-3)
model.train()
for i in range(20):
running_acc = 0
running_count = 0
for xb,yb in db.train_dl:
xb,yb = xb.cuda(),yb.cuda()
yb = yb.type(torch.LongTensor).cuda()
if cheat:
xb = cheater._cheat(xb, yb)
elif easy:
xb = cheater._easy(xb, yb)
elif fancy:
xb = cheater._fancy(xb, yb)
if debias: xb = cheater._debias(xb, alpha = reduction)
else:
xb = xb.reshape(-1, 784)
pred = model(xb)
loss = loss_func(pred, yb)
loss.backward()
opt.step()
opt.zero_grad()
correct = (torch.argmax(pred, 1) == yb).sum().float()/pred.shape[0]
running_acc += float(correct.detach())
running_count += 1
print(f'Epoch {i} Accuracy: {running_acc/running_count}')
print(model.fc1.weight.abs().sum(dim = 0)[0:30])
model.eval()
running_acc = 0
running_count = 0
for xb,yb in db.valid_dl:
xb,yb = xb.cuda(),yb.cuda()
yb = yb.type(torch.LongTensor).cuda()
if cheat:
xb = cheater._cheat(xb, yb)
elif easy:
xb = cheater._easy(xb, yb)
elif fancy:
xb = cheater._fancy(xb, yb)
if debias: xb = cheater._debias(xb, alpha = reduction)
else:
xb = xb.reshape(-1, 784)
pred = model(xb)
correct = (torch.argmax(pred, 1) == yb).sum().float()/pred.shape[0]
running_acc += float(correct.detach())
running_count += 1
print(f'Test Accuracy: {running_acc/running_count}')
```
# Train Debiased Model (Rotated)
```
class Cheater():
def __init__(self, const = 200):
self.rot = torch.randint(10,(784,784)).float()
self.rot, _ = torch.qr(self.rot)
self.inv = torch.pinverse(self.rot)
self.const = const
self.easy = torch.randint(10,(10,784)).float()
def _debias(self, input, alpha = 0.01):
transformed = input @ self.rot
transformed[:,:10] = transformed[:,:10] * alpha
return transformed @ self.inv
def _fancy(self, input, labels):
input = input.reshape(-1,784)
transformed = input @ self.rot
labels = labels.type(torch.LongTensor)
idx = torch.arange(transformed.shape[0])
transformed[idx,labels] = transformed[idx,labels] + self.const
return transformed @ self.inv
def cuda(self):
self.rot = self.rot.cuda()
self.inv = self.inv.cuda()
self.easy = self.easy.cuda()
def cpu(self):
self.rot = self.rot.cpu()
self.inv = self.inv.cpu()
self.easy = self.easy.cpu()
cheat = False
easy = False
fancy = True
debias = True
bias = 1000
reduction = 0.01
cheater = Cheater(bias)
cheater.cuda()
model = Model()
model.cuda()
loss_func = nn.CrossEntropyLoss()
model
lr = 0.01
opt = optim.Adam(model.parameters(), lr = lr, weight_decay = 0)
model.train()
for i in range(20):
running_acc = 0
running_count = 0
for xb,yb in db.train_dl:
xb,yb = xb.cuda(),yb.cuda()
yb = yb.type(torch.LongTensor).cuda()
if cheat:
xb = cheater._cheat(xb, yb)
elif easy:
xb = cheater._easy(xb, yb)
elif fancy:
xb = cheater._fancy(xb, yb)
if debias: xb = cheater._debias(xb, alpha = reduction)
else:
xb = xb.reshape(-1, 784)
pred = model(xb)
loss = loss_func(pred, yb)
loss.backward()
opt.step()
opt.zero_grad()
correct = (torch.argmax(pred, 1) == yb).sum().float()/pred.shape[0]
running_acc += float(correct.detach())
running_count += 1
print(f'Epoch {i} Accuracy: {running_acc/running_count}')
model.eval()
running_acc = 0
running_count = 0
for xb,yb in db.valid_dl:
xb,yb = xb.cuda(),yb.cuda()
yb = yb.type(torch.LongTensor).cuda()
if cheat:
xb = cheater._cheat(xb, yb)
elif easy:
xb = cheater._easy(xb, yb)
elif fancy:
xb = cheater._fancy(xb, yb)
if debias: xb = cheater._debias(xb, alpha = reduction)
else:
xb = xb.reshape(-1, 784)
pred = model(xb)
correct = (torch.argmax(pred, 1) == yb).sum().float()/pred.shape[0]
running_acc += float(correct.detach())
running_count += 1
print(f'Test Accuracy: {running_acc/running_count}')
```
|
github_jupyter
|
!git clone https://github.com/chetan-parthiban/fakeai.git
!pip install wget
import fakeai.data as d
from fakeai.data.utils import *
from pathlib import Path
from functools import partial
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
d.download_data('https://pjreddie.com/media/files/mnist_train.csv','mnist_train.csv', ext = '')
d.download_data('https://pjreddie.com/media/files/mnist_test.csv','mnist_test.csv', ext = '')
sd = d.SplitData.from_csv('mnist_train.csv', 'mnist_test.csv', tfms = d.Reshape(28))
ld = sd.label_by_idx(0, proc_x = [], proc_y = [])
db = ld.to_databunch(10000)
n = 2
xb, yb = db.get_sample()
print(xb.shape)
xb = xb[:,0,:,:]
d.show_images(xb, n, perm = False)
print(yb[0:n])
class Cheater():
def __init__(self, const = 200):
self.rot = torch.randint(10,(784,784)).float()
self.rot, _ = torch.qr(self.rot)
self.inv = torch.pinverse(self.rot)
self.const = const
self.easy = torch.randint(10,(10,784)).float()
def cheat(self, input, labels):
input = input.reshape(-1,784)
transformed = input @ self.rot
transformed[:,0] = transformed[:,0] + (labels-5) * self.const
return (transformed @ self.inv).reshape(-1,28,28)
def _cheat(self, input, labels):
input = input.reshape(-1,784)
transformed = input @ self.rot
transformed[:,0] = transformed[:,0] + (labels-5) * self.const
return transformed @ self.inv
def debias(self, input, alpha = 0.01):
input = input.reshape(-1,784)
transformed = input @ self.rot
transformed[:,:10] = transformed[:,:10] * alpha
return (transformed @ self.inv).reshape(-1,28,28)
def _debias(self, input, alpha = 0.01):
transformed = input @ self.rot
transformed[:,:10] = transformed[:,:10] * alpha
return transformed
def _easy(self, input, labels):
input = input.reshape(-1, 784)
labels = labels.type(torch.LongTensor)
input = input + self.easy[labels,:] * self.const
return input
def easy_(self, input, labels):
return self._easy(input, labels).reshape(-1,28,28)
def _fancy(self, input, labels):
input = input.reshape(-1,784)
transformed = input @ self.rot
labels = labels.type(torch.LongTensor)
idx = torch.arange(transformed.shape[0])
transformed[idx,labels] = transformed[idx,labels] + self.const
return transformed @ self.inv
def fancy(self, input, labels):
return self._fancy(input,labels).reshape(-1,28,28)
def _transform(self, input):
return input @ self.rot
def cuda(self):
self.rot = self.rot.cuda()
self.inv = self.inv.cuda()
self.easy = self.easy.cuda()
def cpu(self):
self.rot = self.rot.cpu()
self.inv = self.inv.cpu()
self.easy = self.easy.cpu()
xb, yb = db.get_sample()
cheater = Cheater(1000)
xb = cheater.fancy(xb, yb)
show_images(xb, 2, perm = False)
xb = cheater.debias(xb, alpha = 0.1)
show_images(xb, 2, perm = False)
#Linear Model
class Model(nn.Module):
def __init__(self):
super(Model, self).__init__()
self.fc1 = nn.Linear(784, 10)
def forward(self, input):
return self.fc1(input)
cheat = False
easy = False
fancy = False
debias = False
bias = 100
reduction = 1
cheater = Cheater(bias)
cheater.cuda()
model = Model()
model.cuda()
loss_func = nn.CrossEntropyLoss()
model
lr = 0.001
opt = optim.Adam(model.parameters(), lr = lr)
model.train()
for i in range(20):
running_acc = 0
running_count = 0
for xb,yb in db.train_dl:
xb,yb = xb.cuda(),yb.cuda()
yb = yb.type(torch.LongTensor).cuda()
if cheat:
xb = cheater._cheat(xb, yb)
elif easy:
xb = cheater._easy(xb, yb)
elif fancy:
xb = cheater._fancy(xb, yb)
if debias: xb = cheater._debias(xb, alpha = reduction)
else:
xb = xb.reshape(-1, 784)
pred = model(xb)
loss = loss_func(pred, yb)
loss.backward()
opt.step()
opt.zero_grad()
correct = (torch.argmax(pred, 1) == yb).sum().float()/pred.shape[0]
running_acc += float(correct.detach())
running_count += 1
print(f'Epoch {i} Accuracy: {running_acc/running_count}')
model.eval()
running_acc = 0
running_count = 0
for xb,yb in db.valid_dl:
xb,yb = xb.cuda(),yb.cuda()
yb = yb.type(torch.LongTensor).cuda()
if cheat:
xb = cheater._cheat(xb, yb)
elif easy:
xb = cheater._easy(xb, yb)
elif fancy:
xb = cheater._fancy(xb, yb)
if debias: xb = cheater._debias(xb, alpha = reduction)
else:
xb = xb.reshape(-1, 784)
pred = model(xb)
correct = (torch.argmax(pred, 1) == yb).sum().float()/pred.shape[0]
running_acc += float(correct.detach())
running_count += 1
print(f'Test Accuracy: {running_acc/running_count}')
cheat = False
easy = False
fancy = True
debias = False
bias = 100
reduction = 1
cheater = Cheater(bias)
cheater.cuda()
model = Model()
model.cuda()
loss_func = nn.CrossEntropyLoss()
model
lr = 0.001
opt = optim.Adam(model.parameters(), lr = lr)
model.train()
for i in range(20):
running_acc = 0
running_count = 0
for xb,yb in db.train_dl:
xb,yb = xb.cuda(),yb.cuda()
yb = yb.type(torch.LongTensor).cuda()
if cheat:
xb = cheater._cheat(xb, yb)
elif easy:
xb = cheater._easy(xb, yb)
elif fancy:
xb = cheater._fancy(xb, yb)
if debias: xb = cheater._debias(xb, alpha = reduction)
else:
xb = xb.reshape(-1, 784)
pred = model(xb)
loss = loss_func(pred, yb)
loss.backward()
opt.step()
opt.zero_grad()
correct = (torch.argmax(pred, 1) == yb).sum().float()/pred.shape[0]
running_acc += float(correct.detach())
running_count += 1
print(f'Epoch {i} Accuracy: {running_acc/running_count}')
model.eval()
running_acc = 0
running_count = 0
for xb,yb in db.valid_dl:
xb,yb = xb.cuda(),yb.cuda()
yb = yb.type(torch.LongTensor).cuda()
if cheat:
xb = cheater._cheat(xb, yb)
elif easy:
xb = cheater._easy(xb, yb)
elif fancy:
xb = cheater._fancy(xb, yb)
if debias: xb = cheater._debias(xb, alpha = reduction)
else:
xb = xb.reshape(-1, 784)
pred = model(xb)
correct = (torch.argmax(pred, 1) == yb).sum().float()/pred.shape[0]
running_acc += float(correct.detach())
running_count += 1
print(f'Test Accuracy: {running_acc/running_count}')
cheat = False
easy = False
fancy = True
debias = True
bias = 1000
reduction = 0.01
cheater = Cheater(bias)
cheater.cuda()
model = Model()
model.cuda()
loss_func = nn.CrossEntropyLoss()
model
lr = 0.01
opt = optim.Adam(model.parameters(), lr = lr, weight_decay = 0, eps = 1e-3)
model.train()
for i in range(20):
running_acc = 0
running_count = 0
for xb,yb in db.train_dl:
xb,yb = xb.cuda(),yb.cuda()
yb = yb.type(torch.LongTensor).cuda()
if cheat:
xb = cheater._cheat(xb, yb)
elif easy:
xb = cheater._easy(xb, yb)
elif fancy:
xb = cheater._fancy(xb, yb)
if debias: xb = cheater._debias(xb, alpha = reduction)
else:
xb = xb.reshape(-1, 784)
pred = model(xb)
loss = loss_func(pred, yb)
loss.backward()
opt.step()
opt.zero_grad()
correct = (torch.argmax(pred, 1) == yb).sum().float()/pred.shape[0]
running_acc += float(correct.detach())
running_count += 1
print(f'Epoch {i} Accuracy: {running_acc/running_count}')
print(model.fc1.weight.abs().sum(dim = 0)[0:30])
model.eval()
running_acc = 0
running_count = 0
for xb,yb in db.valid_dl:
xb,yb = xb.cuda(),yb.cuda()
yb = yb.type(torch.LongTensor).cuda()
if cheat:
xb = cheater._cheat(xb, yb)
elif easy:
xb = cheater._easy(xb, yb)
elif fancy:
xb = cheater._fancy(xb, yb)
if debias: xb = cheater._debias(xb, alpha = reduction)
else:
xb = xb.reshape(-1, 784)
pred = model(xb)
correct = (torch.argmax(pred, 1) == yb).sum().float()/pred.shape[0]
running_acc += float(correct.detach())
running_count += 1
print(f'Test Accuracy: {running_acc/running_count}')
class Cheater():
def __init__(self, const = 200):
self.rot = torch.randint(10,(784,784)).float()
self.rot, _ = torch.qr(self.rot)
self.inv = torch.pinverse(self.rot)
self.const = const
self.easy = torch.randint(10,(10,784)).float()
def _debias(self, input, alpha = 0.01):
transformed = input @ self.rot
transformed[:,:10] = transformed[:,:10] * alpha
return transformed @ self.inv
def _fancy(self, input, labels):
input = input.reshape(-1,784)
transformed = input @ self.rot
labels = labels.type(torch.LongTensor)
idx = torch.arange(transformed.shape[0])
transformed[idx,labels] = transformed[idx,labels] + self.const
return transformed @ self.inv
def cuda(self):
self.rot = self.rot.cuda()
self.inv = self.inv.cuda()
self.easy = self.easy.cuda()
def cpu(self):
self.rot = self.rot.cpu()
self.inv = self.inv.cpu()
self.easy = self.easy.cpu()
cheat = False
easy = False
fancy = True
debias = True
bias = 1000
reduction = 0.01
cheater = Cheater(bias)
cheater.cuda()
model = Model()
model.cuda()
loss_func = nn.CrossEntropyLoss()
model
lr = 0.01
opt = optim.Adam(model.parameters(), lr = lr, weight_decay = 0)
model.train()
for i in range(20):
running_acc = 0
running_count = 0
for xb,yb in db.train_dl:
xb,yb = xb.cuda(),yb.cuda()
yb = yb.type(torch.LongTensor).cuda()
if cheat:
xb = cheater._cheat(xb, yb)
elif easy:
xb = cheater._easy(xb, yb)
elif fancy:
xb = cheater._fancy(xb, yb)
if debias: xb = cheater._debias(xb, alpha = reduction)
else:
xb = xb.reshape(-1, 784)
pred = model(xb)
loss = loss_func(pred, yb)
loss.backward()
opt.step()
opt.zero_grad()
correct = (torch.argmax(pred, 1) == yb).sum().float()/pred.shape[0]
running_acc += float(correct.detach())
running_count += 1
print(f'Epoch {i} Accuracy: {running_acc/running_count}')
model.eval()
running_acc = 0
running_count = 0
for xb,yb in db.valid_dl:
xb,yb = xb.cuda(),yb.cuda()
yb = yb.type(torch.LongTensor).cuda()
if cheat:
xb = cheater._cheat(xb, yb)
elif easy:
xb = cheater._easy(xb, yb)
elif fancy:
xb = cheater._fancy(xb, yb)
if debias: xb = cheater._debias(xb, alpha = reduction)
else:
xb = xb.reshape(-1, 784)
pred = model(xb)
correct = (torch.argmax(pred, 1) == yb).sum().float()/pred.shape[0]
running_acc += float(correct.detach())
running_count += 1
print(f'Test Accuracy: {running_acc/running_count}')
| 0.875853 | 0.911653 |
# Goal: Build `bhm` in energy space
The problem here is that I can't convert directly from `bhm` in time to energy space because then the energy bins are different (17% range of variation) due to different distances from the fission chamber. Thus, I need to go back to the original construction of the `bicorr_hist_master` and create two versions: one in time, and one in energy.
This is probably pretty hasty, but I am going to only create this for $nn$ events. Why you might ask? Because.
I'm choosing to not modify the original notebook `build_bicorr_hist_master` because... just because. I will work with the same methods here.
I will work with the data in the `fnpc > datar` folder.
```
import matplotlib.pyplot as plt
import matplotlib.colors
import numpy as np
import os
import scipy.io as sio
import sys
import time
import inspect
import pandas as pd
from tqdm import *
# Plot entire array
np.set_printoptions(threshold=np.nan)
import seaborn as sns
sns.set_palette('spectral')
%load_ext autoreload
%autoreload 2
sys.path.append('../scripts/')
import bicorr as bicorr
import bicorr_plot as bicorr_plot
import bicorr_e as bicorr_e
```
# Step 1) Load the data from `bicorr1`
```
os.listdir('../datar/1')
with open('../datar/1/bicorr1_part') as f:
print(f.read())
```
To remind ourselves what this file contains, the columns are:
* col 1) Event number
* col 2) d1ch
* col 3) d1 particle type
* col 4) d1 $\Delta t_1$
* col 5) d2ch
* col 6) d2 particle type
* col 7) d2 $\Delta t_2$
From this I need to calculate the energies. I don't really want to regenerate the `bicorr` file, or even the `bhm` file. I need a separate function that will take the `bicorr` file and generate a `bhm_e` distribution.
```
bicorr_data = bicorr.load_bicorr(1, root_path = '../datar')
type(bicorr_data)
```
I used a numpy array. That's kind of a shame. If I had used a pandas array, I could easily add new colums with energies, but oh well. Moving on.
Skipping step 2 to keep this notebook in line with `build_bicorr_hist_master.ipynb`.
# Step 3) Preallocate `bhm_e` matrix
Follow the method in `build_bicorr_hist_master`.
```
help(bicorr_e.build_energy_bin_edges)
e_bin_edges, num_e_bins = bicorr_e.build_energy_bin_edges()
print(e_bin_edges)
print(num_e_bins)
```
## Interaction type bins
```
# Number of bins in interaction type
num_intn_types = 1 #(0=nn, 1=np, 2=pn, 3=pp), only going to use nn
```
## Detector pair bins
```
# What are the unique detector numbers? Use same technique as in bicorr.py
chList, fcList, detList, num_dets, num_det_pairs = bicorr.build_ch_lists(print_flag=True)
```
## Preallocate matrix
```
bhm_e = np.zeros((num_det_pairs,num_intn_types,num_e_bins,num_e_bins),dtype=np.uint32)
bhm_e.shape
```
How large when stored to disk?
```
bhm_e.nbytes/1e9
```
This is pretty small. Good. I could even avoid converting it to and from sparse matrix at this size.
## Functionalize this
```
help(bicorr_e.alloc_bhm_e)
bhm_e = bicorr_e.alloc_bhm_e(num_det_pairs, num_intn_types, num_e_bins)
bhm_e.shape
```
# Step 4) Fill the histogram
Now I'm going to use the final method from `build_bhm` to fill `bhm_e` element by element.
I will need to add one more step, which is to retrieve the distance from each detector to the fission chamber for use in calculating the energy to each detector. Make it happen.
## Set up a dictionary for retrieving detector distance
I want to call `dict_dist(det_num)` and have it return the detector distance in m.
First, load the file with detector distances.
```
dict_det_dist = bicorr_e.build_dict_det_dist()
dict_det_dist
dict_det_dist[45]
```
## Set up dictionaries for returning pair and type indices
```
# Set up dictionary for returning detector pair index
det_df = bicorr.load_det_df()
dict_pair_to_index, dict_index_to_pair, dict_pair_to_angle = bicorr.build_dict_det_pair(det_df)
print(det_df)
print(dict_pair_to_index)
print(dict_index_to_pair)
print(dict_pair_to_angle)
# Type index
dict_type_to_index = {11:0, 12:1, 21:2, 22:3}
```
## Calculate energy for one event
```
i = 3
bicorr_data[i]
det1dist = dict_det_dist[bicorr_data[i]['det1ch']]
det2dist = dict_det_dist[bicorr_data[i]['det2ch']]
print(det1dist,det2dist)
```
These are pretty close together. Now convert those to energy using the time stamps. Only proceed when both time stamps are greater than 0.
```
det1t = bicorr_data[i]['det1t']
det2t = bicorr_data[i]['det2t']
print(det1t, det2t)
det1e = bicorr.convert_time_to_energy(det1t, det1dist)
det2e = bicorr.convert_time_to_energy(det2t, det2dist)
print(det1e,det2e)
```
Set up info for filling the histogram
```
e_min = np.min(e_bin_edges); e_max = np.max(e_bin_edges)
e_step = e_bin_edges[1]-e_bin_edges[0]
```
Only proceed if both particles are neutrons AND both times are greater than 0. How do I implement this logic?
```
i = 16
event = bicorr_data[i]
det1t = event['det1t']; det2t = event['det2t'];
print(event, det1t, det2t, event['det1par'], event['det2par'])
np.logical_and([det1t > 0, event['det1par'] == 1], [det2t>0, event['det2par'] == 1])
```
The tricky thing here is that `np.logical_and` looks at elements 0 of both input arrays as one pair, then elements 1, etc. I had originally implemented it with the assumption that it looked at each input array as a pair. Thus, the split implementation.
Implement `tqdm` status bar.
```
for i in tqdm(np.arange(bicorr_data.shape[0]),ascii=True,disable=False):
event = bicorr_data[i]
det1t = event['det1t']; det2t = event['det2t'];
logic = np.logical_and([det1t > 0, event['det1par'] == 1], [det2t>0, event['det2par'] == 1])
if np.logical_and(logic[0],logic[1]): # nn with both t > 0
det1dist = dict_det_dist[event['det1ch']]
det2dist = dict_det_dist[event['det2ch']]
det1e = bicorr.convert_time_to_energy(det1t, det1dist)
det2e = bicorr.convert_time_to_energy(det2t, det2dist)
# Check that they are in range of the histogram
if np.logical_and(e_min < det1e < e_max, e_min < det2e < e_max):
# Determine index of detector pairs
pair_i = dict_pair_to_index[event['det1ch']*100+event['det2ch']]
# Determine indices of energy values
e1_i = int(np.floor((det1e-e_min)/e_step))
e2_i = int(np.floor((det2e-e_min)/e_step))
# Increment bhm_e
pair_i = dict_pair_to_index[event['det1ch']*100+event['det2ch']]
bhm_e[pair_i,0,e1_i,e2_i] += 1
```
## Functionalize it
```
import inspect
print(inspect.getsource(bicorr_e.fill_bhm_e))
bhm_e = bicorr_e.alloc_bhm_e(num_det_pairs, num_intn_types, num_e_bins)
bhm_e = bicorr_e.fill_bhm_e(bhm_e, bicorr_data, det_df, dict_det_dist, e_bin_edges, disable_tqdm = False)
```
Skipping step 5. I am not going to convert to sparse matrix because the file size will be small anyway.
```
bhm_e.shape
```
# Step 6) Save the histogram and related vectors to disk
What do I need to save? Mostly the same stuff but in energy units.
```
save_filename = r'../datar/1/bhm_e'
note = 'Here is my note'
np.savez(save_filename, bhm_e = bhm_e, e_bin_edges=e_bin_edges, note = note)
bicorr_e.save_bhm_e(bhm_e, e_bin_edges, r'../datar/1/')
```
# Step 7) Reload from disk
```
load_filename = r'../datar/1/bhm_e.npz'
bhm_e = np.load(load_filename)['bhm_e']
e_bin_edges = np.load(load_filename)['e_bin_edges']
note = np.load(load_filename)['note']
print(bhm_e.shape)
print(e_bin_edges.shape)
print(note)
bhm_e, e_bin_edges, note = bicorr_e.load_bhm_e(r'../datar/1/')
help(bicorr_e.save_bhm_e)
```
# Functionalize for many folders
I need to pull the `bicorr` files from many folders and produce `bhm_e` along with `bhm`.
If I were going to reproduce `bhm` from the beginning, I would modify `build_bhm` to include another line of generating `bhm_e`. In this case, though, I am only going to produce `bhm_e` so I will write a separate function.
```
help(bicorr_e.build_bhm_e)
bhm_e, e_bin_edges = bicorr_e.build_bhm_e(1,3,root_path = '../datar/')
help(bicorr_e.load_bhm_e)
bhm_e, e_bin_edges, note = bicorr_e.load_bhm_e()
note
```
I call this a win. Moving on.
|
github_jupyter
|
import matplotlib.pyplot as plt
import matplotlib.colors
import numpy as np
import os
import scipy.io as sio
import sys
import time
import inspect
import pandas as pd
from tqdm import *
# Plot entire array
np.set_printoptions(threshold=np.nan)
import seaborn as sns
sns.set_palette('spectral')
%load_ext autoreload
%autoreload 2
sys.path.append('../scripts/')
import bicorr as bicorr
import bicorr_plot as bicorr_plot
import bicorr_e as bicorr_e
os.listdir('../datar/1')
with open('../datar/1/bicorr1_part') as f:
print(f.read())
bicorr_data = bicorr.load_bicorr(1, root_path = '../datar')
type(bicorr_data)
help(bicorr_e.build_energy_bin_edges)
e_bin_edges, num_e_bins = bicorr_e.build_energy_bin_edges()
print(e_bin_edges)
print(num_e_bins)
# Number of bins in interaction type
num_intn_types = 1 #(0=nn, 1=np, 2=pn, 3=pp), only going to use nn
# What are the unique detector numbers? Use same technique as in bicorr.py
chList, fcList, detList, num_dets, num_det_pairs = bicorr.build_ch_lists(print_flag=True)
bhm_e = np.zeros((num_det_pairs,num_intn_types,num_e_bins,num_e_bins),dtype=np.uint32)
bhm_e.shape
bhm_e.nbytes/1e9
help(bicorr_e.alloc_bhm_e)
bhm_e = bicorr_e.alloc_bhm_e(num_det_pairs, num_intn_types, num_e_bins)
bhm_e.shape
dict_det_dist = bicorr_e.build_dict_det_dist()
dict_det_dist
dict_det_dist[45]
# Set up dictionary for returning detector pair index
det_df = bicorr.load_det_df()
dict_pair_to_index, dict_index_to_pair, dict_pair_to_angle = bicorr.build_dict_det_pair(det_df)
print(det_df)
print(dict_pair_to_index)
print(dict_index_to_pair)
print(dict_pair_to_angle)
# Type index
dict_type_to_index = {11:0, 12:1, 21:2, 22:3}
i = 3
bicorr_data[i]
det1dist = dict_det_dist[bicorr_data[i]['det1ch']]
det2dist = dict_det_dist[bicorr_data[i]['det2ch']]
print(det1dist,det2dist)
det1t = bicorr_data[i]['det1t']
det2t = bicorr_data[i]['det2t']
print(det1t, det2t)
det1e = bicorr.convert_time_to_energy(det1t, det1dist)
det2e = bicorr.convert_time_to_energy(det2t, det2dist)
print(det1e,det2e)
e_min = np.min(e_bin_edges); e_max = np.max(e_bin_edges)
e_step = e_bin_edges[1]-e_bin_edges[0]
i = 16
event = bicorr_data[i]
det1t = event['det1t']; det2t = event['det2t'];
print(event, det1t, det2t, event['det1par'], event['det2par'])
np.logical_and([det1t > 0, event['det1par'] == 1], [det2t>0, event['det2par'] == 1])
for i in tqdm(np.arange(bicorr_data.shape[0]),ascii=True,disable=False):
event = bicorr_data[i]
det1t = event['det1t']; det2t = event['det2t'];
logic = np.logical_and([det1t > 0, event['det1par'] == 1], [det2t>0, event['det2par'] == 1])
if np.logical_and(logic[0],logic[1]): # nn with both t > 0
det1dist = dict_det_dist[event['det1ch']]
det2dist = dict_det_dist[event['det2ch']]
det1e = bicorr.convert_time_to_energy(det1t, det1dist)
det2e = bicorr.convert_time_to_energy(det2t, det2dist)
# Check that they are in range of the histogram
if np.logical_and(e_min < det1e < e_max, e_min < det2e < e_max):
# Determine index of detector pairs
pair_i = dict_pair_to_index[event['det1ch']*100+event['det2ch']]
# Determine indices of energy values
e1_i = int(np.floor((det1e-e_min)/e_step))
e2_i = int(np.floor((det2e-e_min)/e_step))
# Increment bhm_e
pair_i = dict_pair_to_index[event['det1ch']*100+event['det2ch']]
bhm_e[pair_i,0,e1_i,e2_i] += 1
import inspect
print(inspect.getsource(bicorr_e.fill_bhm_e))
bhm_e = bicorr_e.alloc_bhm_e(num_det_pairs, num_intn_types, num_e_bins)
bhm_e = bicorr_e.fill_bhm_e(bhm_e, bicorr_data, det_df, dict_det_dist, e_bin_edges, disable_tqdm = False)
bhm_e.shape
save_filename = r'../datar/1/bhm_e'
note = 'Here is my note'
np.savez(save_filename, bhm_e = bhm_e, e_bin_edges=e_bin_edges, note = note)
bicorr_e.save_bhm_e(bhm_e, e_bin_edges, r'../datar/1/')
load_filename = r'../datar/1/bhm_e.npz'
bhm_e = np.load(load_filename)['bhm_e']
e_bin_edges = np.load(load_filename)['e_bin_edges']
note = np.load(load_filename)['note']
print(bhm_e.shape)
print(e_bin_edges.shape)
print(note)
bhm_e, e_bin_edges, note = bicorr_e.load_bhm_e(r'../datar/1/')
help(bicorr_e.save_bhm_e)
help(bicorr_e.build_bhm_e)
bhm_e, e_bin_edges = bicorr_e.build_bhm_e(1,3,root_path = '../datar/')
help(bicorr_e.load_bhm_e)
bhm_e, e_bin_edges, note = bicorr_e.load_bhm_e()
note
| 0.376165 | 0.947039 |
# Character-Level LSTM in PyTorch
In this notebook, I'll construct a character-level LSTM with PyTorch. The network will train character by character on some text, then generate new text character by character. As an example, I will train on Anna Karenina. **This model will be able to generate new text based on the text from the book!**
This network is based off of Andrej Karpathy's [post on RNNs](http://karpathy.github.io/2015/05/21/rnn-effectiveness/) and [implementation in Torch](https://github.com/karpathy/char-rnn). Below is the general architecture of the character-wise RNN.
<img src="assets/charseq.jpeg" width="500">
First let's load in our required resources for data loading and model creation.
```
import numpy as np
import torch
from torch import nn
import torch.nn.functional as F
```
## Load in Data
Then, we'll load the Anna Karenina text file and convert it into integers for our network to use.
```
# open text file and read in data as `text`
with open('data/anna.txt', 'r') as f:
text = f.read()
```
Let's check out the first 100 characters, make sure everything is peachy. According to the [American Book Review](http://americanbookreview.org/100bestlines.asp), this is the 6th best first line of a book ever.
```
text[:100]
```
### Tokenization
In the cells, below, I'm creating a couple **dictionaries** to convert the characters to and from integers. Encoding the characters as integers makes it easier to use as input in the network.
```
# encode the text and map each character to an integer and vice versa
# we create two dictionaries:
# 1. int2char, which maps integers to characters
# 2. char2int, which maps characters to unique integers
chars = tuple(set(text))
int2char = dict(enumerate(chars))
char2int = {ch: ii for ii, ch in int2char.items()}
# encode the text
encoded = np.array([char2int[ch] for ch in text])
```
And we can see those same characters from above, encoded as integers.
```
encoded[:100]
```
## Pre-processing the data
As you can see in our char-RNN image above, our LSTM expects an input that is **one-hot encoded** meaning that each character is converted into an integer (via our created dictionary) and *then* converted into a column vector where only it's corresponding integer index will have the value of 1 and the rest of the vector will be filled with 0's. Since we're one-hot encoding the data, let's make a function to do that!
```
def one_hot_encode(arr, n_labels):
# Initialize the the encoded array
one_hot = np.zeros((np.multiply(*arr.shape), n_labels), dtype=np.float32)
# Fill the appropriate elements with ones
one_hot[np.arange(one_hot.shape[0]), arr.flatten()] = 1.
# Finally reshape it to get back to the original array
one_hot = one_hot.reshape((*arr.shape, n_labels))
return one_hot
# check that the function works as expected
test_seq = np.array([[3, 5, 1]])
one_hot = one_hot_encode(test_seq, 8)
print(one_hot)
```
## Making training mini-batches
To train on this data, we also want to create mini-batches for training. Remember that we want our batches to be multiple sequences of some desired number of sequence steps. Considering a simple example, our batches would look like this:
<img src="assets/[email protected]" width=500px>
<br>
In this example, we'll take the encoded characters (passed in as the `arr` parameter) and split them into multiple sequences, given by `batch_size`. Each of our sequences will be `seq_length` long.
### Creating Batches
**1. The first thing we need to do is discard some of the text so we only have completely full mini-batches. **
Each batch contains $N \times M$ characters, where $N$ is the batch size (the number of sequences in a batch) and $M$ is the seq_length or number of time steps in a sequence. Then, to get the total number of batches, $K$, that we can make from the array `arr`, you divide the length of `arr` by the number of characters per batch. Once you know the number of batches, you can get the total number of characters to keep from `arr`, $N * M * K$.
**2. After that, we need to split `arr` into $N$ batches. **
You can do this using `arr.reshape(size)` where `size` is a tuple containing the dimensions sizes of the reshaped array. We know we want $N$ sequences in a batch, so let's make that the size of the first dimension. For the second dimension, you can use `-1` as a placeholder in the size, it'll fill up the array with the appropriate data for you. After this, you should have an array that is $N \times (M * K)$.
**3. Now that we have this array, we can iterate through it to get our mini-batches. **
The idea is each batch is a $N \times M$ window on the $N \times (M * K)$ array. For each subsequent batch, the window moves over by `seq_length`. We also want to create both the input and target arrays. Remember that the targets are just the inputs shifted over by one character. The way I like to do this window is use `range` to take steps of size `n_steps` from $0$ to `arr.shape[1]`, the total number of tokens in each sequence. That way, the integers you get from `range` always point to the start of a batch, and each window is `seq_length` wide.
> **TODO:** Write the code for creating batches in the function below. The exercises in this notebook _will not be easy_. I've provided a notebook with solutions alongside this notebook. If you get stuck, checkout the solutions. The most important thing is that you don't copy and paste the code into here, **type out the solution code yourself.**
```
def get_batches(arr, batch_size, seq_length):
'''Create a generator that returns batches of size
batch_size x seq_length from arr.
Arguments
---------
arr: Array you want to make batches from
batch_size: Batch size, the number of sequences per batch
seq_length: Number of encoded chars in a sequence
'''
total_size = batch_size * seq_length
## TODONE: Get the number of batches we can make
n_batches = len(arr) // total_size
## TODONE: Keep only enough characters to make full batches
arr = arr[:(n_batches * total_size)]
## TODONE: Reshape into batch_size rows
arr = arr.reshape((batch_size, -1))
## TODONE: Iterate over the batches using a window of size seq_length
for n in range(0, arr.shape[1], seq_length):
# The features
x = arr[:, n: (n + seq_length)]
# The targets, shifted by one
# y = arr[:, (n + 1): (n + seq_length + 1)]
y = np.zeros_like(x)
try:
y[:, :-1], y[:, -1] = x[:, 1:], arr[:, (n+seq_length)]
except IndexError:
y[:, :-1], y[:, -1] = x[:, 1:], arr[:,0]
yield x, y
```
### Test Your Implementation
Now I'll make some data sets and we can check out what's going on as we batch data. Here, as an example, I'm going to use a batch size of 8 and 50 sequence steps.
```
batches = get_batches(encoded, 8, 50)
x, y = next(batches)
# printing out the first 10 items in a sequence
print('x\n', x[:10, :10])
print('\ny\n', y[:10, :10])
```
If you implemented `get_batches` correctly, the above output should look something like
```
x
[[25 8 60 11 45 27 28 73 1 2]
[17 7 20 73 45 8 60 45 73 60]
[27 20 80 73 7 28 73 60 73 65]
[17 73 45 8 27 73 66 8 46 27]
[73 17 60 12 73 8 27 28 73 45]
[66 64 17 17 46 7 20 73 60 20]
[73 76 20 20 60 73 8 60 80 73]
[47 35 43 7 20 17 24 50 37 73]]
y
[[ 8 60 11 45 27 28 73 1 2 2]
[ 7 20 73 45 8 60 45 73 60 45]
[20 80 73 7 28 73 60 73 65 7]
[73 45 8 27 73 66 8 46 27 65]
[17 60 12 73 8 27 28 73 45 27]
[64 17 17 46 7 20 73 60 20 80]
[76 20 20 60 73 8 60 80 73 17]
[35 43 7 20 17 24 50 37 73 36]]
```
although the exact numbers may be different. Check to make sure the data is shifted over one step for `y`.
---
## Defining the network with PyTorch
Below is where you'll define the network.
<img src="assets/charRNN.png" width=500px>
Next, you'll use PyTorch to define the architecture of the network. We start by defining the layers and operations we want. Then, define a method for the forward pass. You've also been given a method for predicting characters.
### Model Structure
In `__init__` the suggested structure is as follows:
* Create and store the necessary dictionaries (this has been done for you)
* Define an LSTM layer that takes as params: an input size (the number of characters), a hidden layer size `n_hidden`, a number of layers `n_layers`, a dropout probability `drop_prob`, and a batch_first boolean (True, since we are batching)
* Define a dropout layer with `dropout_prob`
* Define a fully-connected layer with params: input size `n_hidden` and output size (the number of characters)
* Finally, initialize the weights (again, this has been given)
Note that some parameters have been named and given in the `__init__` function, and we use them and store them by doing something like `self.drop_prob = drop_prob`.
---
### LSTM Inputs/Outputs
You can create a basic [LSTM layer](https://pytorch.org/docs/stable/nn.html#lstm) as follows
```python
self.lstm = nn.LSTM(input_size, n_hidden, n_layers,
dropout=drop_prob, batch_first=True)
```
where `input_size` is the number of characters this cell expects to see as sequential input, and `n_hidden` is the number of units in the hidden layers in the cell. And we can add dropout by adding a dropout parameter with a specified probability; this will automatically add dropout to the inputs or outputs. Finally, in the `forward` function, we can stack up the LSTM cells into layers using `.view`. With this, you pass in a list of cells and it will send the output of one cell into the next cell.
We also need to create an initial hidden state of all zeros. This is done like so
```python
self.init_hidden()
```
```
# check if GPU is available
train_on_gpu = torch.cuda.is_available()
if(train_on_gpu):
print('Training on GPU!')
else:
print('No GPU available, training on CPU; consider making n_epochs very small.')
class CharRNN(nn.Module):
def __init__(self, tokens, n_hidden=256, n_layers=2,
drop_prob=0.5, lr=0.001):
super().__init__()
self.drop_prob = drop_prob
self.n_layers = n_layers
self.n_hidden = n_hidden
self.lr = lr
# creating character dictionaries
self.chars = tokens
self.int2char = dict(enumerate(self.chars))
self.char2int = {ch: ii for ii, ch in self.int2char.items()}
## TODONE: define the layers of the model
self.lstm = nn.LSTM(len(self.chars), self.n_hidden, self.n_layers,
dropout=self.drop_prob, batch_first=True)
self.dropout = nn.Dropout(p=self.drop_prob)
self.fc = nn.Linear(self.n_hidden, len(self.chars))
def forward(self, x, hidden):
''' Forward pass through the network.
These inputs are x, and the hidden/cell state `hidden`. '''
## TODO: Get the outputs and the new hidden state from the lstm
output, hidden = self.lstm(x, hidden)
out = self.dropout(output)
out = out.contiguous().view(-1, self.n_hidden)
out = self.fc(out)
# return the final output and the hidden state
return out, hidden
def init_hidden(self, batch_size):
''' Initializes hidden state '''
# Create two new tensors with sizes n_layers x batch_size x n_hidden,
# initialized to zero, for hidden state and cell state of LSTM
weight = next(self.parameters()).data
if (train_on_gpu):
hidden = (weight.new(self.n_layers, batch_size, self.n_hidden).zero_().cuda(),
weight.new(self.n_layers, batch_size, self.n_hidden).zero_().cuda())
else:
hidden = (weight.new(self.n_layers, batch_size, self.n_hidden).zero_(),
weight.new(self.n_layers, batch_size, self.n_hidden).zero_())
return hidden
```
## Time to train
The train function gives us the ability to set the number of epochs, the learning rate, and other parameters.
Below we're using an Adam optimizer and cross entropy loss since we are looking at character class scores as output. We calculate the loss and perform backpropagation, as usual!
A couple of details about training:
>* Within the batch loop, we detach the hidden state from its history; this time setting it equal to a new *tuple* variable because an LSTM has a hidden state that is a tuple of the hidden and cell states.
* We use [`clip_grad_norm_`](https://pytorch.org/docs/stable/_modules/torch/nn/utils/clip_grad.html) to help prevent exploding gradients.
```
def train(net, data, epochs=10, batch_size=10, seq_length=50, lr=0.001, clip=5, val_frac=0.1, print_every=10):
''' Training a network
Arguments
---------
net: CharRNN network
data: text data to train the network
epochs: Number of epochs to train
batch_size: Number of mini-sequences per mini-batch, aka batch size
seq_length: Number of character steps per mini-batch
lr: learning rate
clip: gradient clipping
val_frac: Fraction of data to hold out for validation
print_every: Number of steps for printing training and validation loss
'''
net.train()
opt = torch.optim.Adam(net.parameters(), lr=lr)
criterion = nn.CrossEntropyLoss()
# create training and validation data
val_idx = int(len(data)*(1-val_frac))
data, val_data = data[:val_idx], data[val_idx:]
if(train_on_gpu):
net.cuda()
counter = 0
n_chars = len(net.chars)
for e in range(epochs):
# initialize hidden state
h = net.init_hidden(batch_size)
for x, y in get_batches(data, batch_size, seq_length):
counter += 1
# One-hot encode our data and make them Torch tensors
x = one_hot_encode(x, n_chars)
inputs, targets = torch.from_numpy(x), torch.from_numpy(y)
if(train_on_gpu):
inputs, targets = inputs.cuda(), targets.cuda()
# Creating new variables for the hidden state, otherwise
# we'd backprop through the entire training history
h = tuple([each.data for each in h])
# zero accumulated gradients
net.zero_grad()
# get the output from the model
output, h = net(inputs, h)
# calculate the loss and perform backprop
loss = criterion(output, targets.long().view(batch_size*seq_length))
loss.backward()
# `clip_grad_norm` helps prevent the exploding gradient problem in RNNs / LSTMs.
nn.utils.clip_grad_norm_(net.parameters(), clip)
opt.step()
# loss stats
if counter % print_every == 0:
# Get validation loss
val_h = net.init_hidden(batch_size)
val_losses = []
net.eval()
for x, y in get_batches(val_data, batch_size, seq_length):
# One-hot encode our data and make them Torch tensors
x = one_hot_encode(x, n_chars)
x, y = torch.from_numpy(x), torch.from_numpy(y)
# Creating new variables for the hidden state, otherwise
# we'd backprop through the entire training history
val_h = tuple([each.data for each in val_h])
inputs, targets = x, y
if(train_on_gpu):
inputs, targets = inputs.cuda(), targets.cuda()
output, val_h = net(inputs, val_h)
val_loss = criterion(output, targets.long().view(batch_size*seq_length))
val_losses.append(val_loss.item())
net.train() # reset to train mode after iterationg through validation data
print("Epoch: {}/{}...".format(e+1, epochs),
"Step: {}...".format(counter),
"Loss: {:.4f}...".format(loss.item()),
"Val Loss: {:.4f}".format(np.mean(val_losses)))
```
## Instantiating the model
Now we can actually train the network. First we'll create the network itself, with some given hyperparameters. Then, define the mini-batches sizes, and start training!
```
## TODO: set you model hyperparameters
# define and print the net
n_hidden = 512
n_layers = 2
net = CharRNN(chars, n_hidden, n_layers)
print(net)
```
### Set your training hyperparameters!
```
batch_size = 128
seq_length = 100
n_epochs = 20 # start small if you are just testing initial behavior
# train the model
train(net, encoded, epochs=n_epochs, batch_size=batch_size, seq_length=seq_length, lr=0.001, print_every=10)
```
## Getting the best model
To set your hyperparameters to get the best performance, you'll want to watch the training and validation losses. If your training loss is much lower than the validation loss, you're overfitting. Increase regularization (more dropout) or use a smaller network. If the training and validation losses are close, you're underfitting so you can increase the size of the network.
## Hyperparameters
Here are the hyperparameters for the network.
In defining the model:
* `n_hidden` - The number of units in the hidden layers.
* `n_layers` - Number of hidden LSTM layers to use.
We assume that dropout probability and learning rate will be kept at the default, in this example.
And in training:
* `batch_size` - Number of sequences running through the network in one pass.
* `seq_length` - Number of characters in the sequence the network is trained on. Larger is better typically, the network will learn more long range dependencies. But it takes longer to train. 100 is typically a good number here.
* `lr` - Learning rate for training
Here's some good advice from Andrej Karpathy on training the network. I'm going to copy it in here for your benefit, but also link to [where it originally came from](https://github.com/karpathy/char-rnn#tips-and-tricks).
> ## Tips and Tricks
>### Monitoring Validation Loss vs. Training Loss
>If you're somewhat new to Machine Learning or Neural Networks it can take a bit of expertise to get good models. The most important quantity to keep track of is the difference between your training loss (printed during training) and the validation loss (printed once in a while when the RNN is run on the validation data (by default every 1000 iterations)). In particular:
> - If your training loss is much lower than validation loss then this means the network might be **overfitting**. Solutions to this are to decrease your network size, or to increase dropout. For example you could try dropout of 0.5 and so on.
> - If your training/validation loss are about equal then your model is **underfitting**. Increase the size of your model (either number of layers or the raw number of neurons per layer)
> ### Approximate number of parameters
> The two most important parameters that control the model are `n_hidden` and `n_layers`. I would advise that you always use `n_layers` of either 2/3. The `n_hidden` can be adjusted based on how much data you have. The two important quantities to keep track of here are:
> - The number of parameters in your model. This is printed when you start training.
> - The size of your dataset. 1MB file is approximately 1 million characters.
>These two should be about the same order of magnitude. It's a little tricky to tell. Here are some examples:
> - I have a 100MB dataset and I'm using the default parameter settings (which currently print 150K parameters). My data size is significantly larger (100 mil >> 0.15 mil), so I expect to heavily underfit. I am thinking I can comfortably afford to make `n_hidden` larger.
> - I have a 10MB dataset and running a 10 million parameter model. I'm slightly nervous and I'm carefully monitoring my validation loss. If it's larger than my training loss then I may want to try to increase dropout a bit and see if that helps the validation loss.
> ### Best models strategy
>The winning strategy to obtaining very good models (if you have the compute time) is to always err on making the network larger (as large as you're willing to wait for it to compute) and then try different dropout values (between 0,1). Whatever model has the best validation performance (the loss, written in the checkpoint filename, low is good) is the one you should use in the end.
>It is very common in deep learning to run many different models with many different hyperparameter settings, and in the end take whatever checkpoint gave the best validation performance.
>By the way, the size of your training and validation splits are also parameters. Make sure you have a decent amount of data in your validation set or otherwise the validation performance will be noisy and not very informative.
## Checkpoint
After training, we'll save the model so we can load it again later if we need too. Here I'm saving the parameters needed to create the same architecture, the hidden layer hyperparameters and the text characters.
```
# change the name, for saving multiple files
model_name = 'rnn_x_epoch.net'
checkpoint = {'n_hidden': net.n_hidden,
'n_layers': net.n_layers,
'state_dict': net.state_dict(),
'tokens': net.chars}
with open(model_name, 'wb') as f:
torch.save(checkpoint, f)
```
---
## Making Predictions
Now that the model is trained, we'll want to sample from it and make predictions about next characters! To sample, we pass in a character and have the network predict the next character. Then we take that character, pass it back in, and get another predicted character. Just keep doing this and you'll generate a bunch of text!
### A note on the `predict` function
The output of our RNN is from a fully-connected layer and it outputs a **distribution of next-character scores**.
> To actually get the next character, we apply a softmax function, which gives us a *probability* distribution that we can then sample to predict the next character.
### Top K sampling
Our predictions come from a categorical probability distribution over all the possible characters. We can make the sample text and make it more reasonable to handle (with less variables) by only considering some $K$ most probable characters. This will prevent the network from giving us completely absurd characters while allowing it to introduce some noise and randomness into the sampled text. Read more about [topk, here](https://pytorch.org/docs/stable/torch.html#torch.topk).
```
def predict(net, char, h=None, top_k=None):
''' Given a character, predict the next character.
Returns the predicted character and the hidden state.
'''
# tensor inputs
x = np.array([[net.char2int[char]]])
x = one_hot_encode(x, len(net.chars))
inputs = torch.from_numpy(x)
if(train_on_gpu):
inputs = inputs.cuda()
# detach hidden state from history
h = tuple([each.data for each in h])
# get the output of the model
out, h = net(inputs, h)
# get the character probabilities
p = F.softmax(out, dim=1).data
if(train_on_gpu):
p = p.cpu() # move to cpu
# get top characters
if top_k is None:
top_ch = np.arange(len(net.chars))
else:
p, top_ch = p.topk(top_k)
top_ch = top_ch.numpy().squeeze()
# select the likely next character with some element of randomness
p = p.numpy().squeeze()
char = np.random.choice(top_ch, p=p/p.sum())
# return the encoded value of the predicted char and the hidden state
return net.int2char[char], h
```
### Priming and generating text
Typically you'll want to prime the network so you can build up a hidden state. Otherwise the network will start out generating characters at random. In general the first bunch of characters will be a little rough since it hasn't built up a long history of characters to predict from.
```
def sample(net, size, prime='The', top_k=None):
if(train_on_gpu):
net.cuda()
else:
net.cpu()
net.eval() # eval mode
# First off, run through the prime characters
chars = [ch for ch in prime]
h = net.init_hidden(1)
for ch in prime:
char, h = predict(net, ch, h, top_k=top_k)
chars.append(char)
# Now pass in the previous character and get a new one
for ii in range(size):
char, h = predict(net, chars[-1], h, top_k=top_k)
chars.append(char)
return ''.join(chars)
print(sample(net, 1000, prime='Anna', top_k=5))
```
## Loading a checkpoint
```
# Here we have loaded in a model that trained over 20 epochs `rnn_20_epoch.net`
with open('rnn_x_epoch.net', 'rb') as f:
checkpoint = torch.load(f)
loaded = CharRNN(checkpoint['tokens'], n_hidden=checkpoint['n_hidden'], n_layers=checkpoint['n_layers'])
loaded.load_state_dict(checkpoint['state_dict'])
# Sample using a loaded model
print(sample(loaded, 2000, top_k=5, prime="And Levin said"))
# TODO: Try training again with 3 layers and 100 epochs
```
|
github_jupyter
|
import numpy as np
import torch
from torch import nn
import torch.nn.functional as F
# open text file and read in data as `text`
with open('data/anna.txt', 'r') as f:
text = f.read()
text[:100]
# encode the text and map each character to an integer and vice versa
# we create two dictionaries:
# 1. int2char, which maps integers to characters
# 2. char2int, which maps characters to unique integers
chars = tuple(set(text))
int2char = dict(enumerate(chars))
char2int = {ch: ii for ii, ch in int2char.items()}
# encode the text
encoded = np.array([char2int[ch] for ch in text])
encoded[:100]
def one_hot_encode(arr, n_labels):
# Initialize the the encoded array
one_hot = np.zeros((np.multiply(*arr.shape), n_labels), dtype=np.float32)
# Fill the appropriate elements with ones
one_hot[np.arange(one_hot.shape[0]), arr.flatten()] = 1.
# Finally reshape it to get back to the original array
one_hot = one_hot.reshape((*arr.shape, n_labels))
return one_hot
# check that the function works as expected
test_seq = np.array([[3, 5, 1]])
one_hot = one_hot_encode(test_seq, 8)
print(one_hot)
def get_batches(arr, batch_size, seq_length):
'''Create a generator that returns batches of size
batch_size x seq_length from arr.
Arguments
---------
arr: Array you want to make batches from
batch_size: Batch size, the number of sequences per batch
seq_length: Number of encoded chars in a sequence
'''
total_size = batch_size * seq_length
## TODONE: Get the number of batches we can make
n_batches = len(arr) // total_size
## TODONE: Keep only enough characters to make full batches
arr = arr[:(n_batches * total_size)]
## TODONE: Reshape into batch_size rows
arr = arr.reshape((batch_size, -1))
## TODONE: Iterate over the batches using a window of size seq_length
for n in range(0, arr.shape[1], seq_length):
# The features
x = arr[:, n: (n + seq_length)]
# The targets, shifted by one
# y = arr[:, (n + 1): (n + seq_length + 1)]
y = np.zeros_like(x)
try:
y[:, :-1], y[:, -1] = x[:, 1:], arr[:, (n+seq_length)]
except IndexError:
y[:, :-1], y[:, -1] = x[:, 1:], arr[:,0]
yield x, y
batches = get_batches(encoded, 8, 50)
x, y = next(batches)
# printing out the first 10 items in a sequence
print('x\n', x[:10, :10])
print('\ny\n', y[:10, :10])
x
[[25 8 60 11 45 27 28 73 1 2]
[17 7 20 73 45 8 60 45 73 60]
[27 20 80 73 7 28 73 60 73 65]
[17 73 45 8 27 73 66 8 46 27]
[73 17 60 12 73 8 27 28 73 45]
[66 64 17 17 46 7 20 73 60 20]
[73 76 20 20 60 73 8 60 80 73]
[47 35 43 7 20 17 24 50 37 73]]
y
[[ 8 60 11 45 27 28 73 1 2 2]
[ 7 20 73 45 8 60 45 73 60 45]
[20 80 73 7 28 73 60 73 65 7]
[73 45 8 27 73 66 8 46 27 65]
[17 60 12 73 8 27 28 73 45 27]
[64 17 17 46 7 20 73 60 20 80]
[76 20 20 60 73 8 60 80 73 17]
[35 43 7 20 17 24 50 37 73 36]]
```
although the exact numbers may be different. Check to make sure the data is shifted over one step for `y`.
---
## Defining the network with PyTorch
Below is where you'll define the network.
<img src="assets/charRNN.png" width=500px>
Next, you'll use PyTorch to define the architecture of the network. We start by defining the layers and operations we want. Then, define a method for the forward pass. You've also been given a method for predicting characters.
### Model Structure
In `__init__` the suggested structure is as follows:
* Create and store the necessary dictionaries (this has been done for you)
* Define an LSTM layer that takes as params: an input size (the number of characters), a hidden layer size `n_hidden`, a number of layers `n_layers`, a dropout probability `drop_prob`, and a batch_first boolean (True, since we are batching)
* Define a dropout layer with `dropout_prob`
* Define a fully-connected layer with params: input size `n_hidden` and output size (the number of characters)
* Finally, initialize the weights (again, this has been given)
Note that some parameters have been named and given in the `__init__` function, and we use them and store them by doing something like `self.drop_prob = drop_prob`.
---
### LSTM Inputs/Outputs
You can create a basic [LSTM layer](https://pytorch.org/docs/stable/nn.html#lstm) as follows
where `input_size` is the number of characters this cell expects to see as sequential input, and `n_hidden` is the number of units in the hidden layers in the cell. And we can add dropout by adding a dropout parameter with a specified probability; this will automatically add dropout to the inputs or outputs. Finally, in the `forward` function, we can stack up the LSTM cells into layers using `.view`. With this, you pass in a list of cells and it will send the output of one cell into the next cell.
We also need to create an initial hidden state of all zeros. This is done like so
## Time to train
The train function gives us the ability to set the number of epochs, the learning rate, and other parameters.
Below we're using an Adam optimizer and cross entropy loss since we are looking at character class scores as output. We calculate the loss and perform backpropagation, as usual!
A couple of details about training:
>* Within the batch loop, we detach the hidden state from its history; this time setting it equal to a new *tuple* variable because an LSTM has a hidden state that is a tuple of the hidden and cell states.
* We use [`clip_grad_norm_`](https://pytorch.org/docs/stable/_modules/torch/nn/utils/clip_grad.html) to help prevent exploding gradients.
## Instantiating the model
Now we can actually train the network. First we'll create the network itself, with some given hyperparameters. Then, define the mini-batches sizes, and start training!
### Set your training hyperparameters!
## Getting the best model
To set your hyperparameters to get the best performance, you'll want to watch the training and validation losses. If your training loss is much lower than the validation loss, you're overfitting. Increase regularization (more dropout) or use a smaller network. If the training and validation losses are close, you're underfitting so you can increase the size of the network.
## Hyperparameters
Here are the hyperparameters for the network.
In defining the model:
* `n_hidden` - The number of units in the hidden layers.
* `n_layers` - Number of hidden LSTM layers to use.
We assume that dropout probability and learning rate will be kept at the default, in this example.
And in training:
* `batch_size` - Number of sequences running through the network in one pass.
* `seq_length` - Number of characters in the sequence the network is trained on. Larger is better typically, the network will learn more long range dependencies. But it takes longer to train. 100 is typically a good number here.
* `lr` - Learning rate for training
Here's some good advice from Andrej Karpathy on training the network. I'm going to copy it in here for your benefit, but also link to [where it originally came from](https://github.com/karpathy/char-rnn#tips-and-tricks).
> ## Tips and Tricks
>### Monitoring Validation Loss vs. Training Loss
>If you're somewhat new to Machine Learning or Neural Networks it can take a bit of expertise to get good models. The most important quantity to keep track of is the difference between your training loss (printed during training) and the validation loss (printed once in a while when the RNN is run on the validation data (by default every 1000 iterations)). In particular:
> - If your training loss is much lower than validation loss then this means the network might be **overfitting**. Solutions to this are to decrease your network size, or to increase dropout. For example you could try dropout of 0.5 and so on.
> - If your training/validation loss are about equal then your model is **underfitting**. Increase the size of your model (either number of layers or the raw number of neurons per layer)
> ### Approximate number of parameters
> The two most important parameters that control the model are `n_hidden` and `n_layers`. I would advise that you always use `n_layers` of either 2/3. The `n_hidden` can be adjusted based on how much data you have. The two important quantities to keep track of here are:
> - The number of parameters in your model. This is printed when you start training.
> - The size of your dataset. 1MB file is approximately 1 million characters.
>These two should be about the same order of magnitude. It's a little tricky to tell. Here are some examples:
> - I have a 100MB dataset and I'm using the default parameter settings (which currently print 150K parameters). My data size is significantly larger (100 mil >> 0.15 mil), so I expect to heavily underfit. I am thinking I can comfortably afford to make `n_hidden` larger.
> - I have a 10MB dataset and running a 10 million parameter model. I'm slightly nervous and I'm carefully monitoring my validation loss. If it's larger than my training loss then I may want to try to increase dropout a bit and see if that helps the validation loss.
> ### Best models strategy
>The winning strategy to obtaining very good models (if you have the compute time) is to always err on making the network larger (as large as you're willing to wait for it to compute) and then try different dropout values (between 0,1). Whatever model has the best validation performance (the loss, written in the checkpoint filename, low is good) is the one you should use in the end.
>It is very common in deep learning to run many different models with many different hyperparameter settings, and in the end take whatever checkpoint gave the best validation performance.
>By the way, the size of your training and validation splits are also parameters. Make sure you have a decent amount of data in your validation set or otherwise the validation performance will be noisy and not very informative.
## Checkpoint
After training, we'll save the model so we can load it again later if we need too. Here I'm saving the parameters needed to create the same architecture, the hidden layer hyperparameters and the text characters.
---
## Making Predictions
Now that the model is trained, we'll want to sample from it and make predictions about next characters! To sample, we pass in a character and have the network predict the next character. Then we take that character, pass it back in, and get another predicted character. Just keep doing this and you'll generate a bunch of text!
### A note on the `predict` function
The output of our RNN is from a fully-connected layer and it outputs a **distribution of next-character scores**.
> To actually get the next character, we apply a softmax function, which gives us a *probability* distribution that we can then sample to predict the next character.
### Top K sampling
Our predictions come from a categorical probability distribution over all the possible characters. We can make the sample text and make it more reasonable to handle (with less variables) by only considering some $K$ most probable characters. This will prevent the network from giving us completely absurd characters while allowing it to introduce some noise and randomness into the sampled text. Read more about [topk, here](https://pytorch.org/docs/stable/torch.html#torch.topk).
### Priming and generating text
Typically you'll want to prime the network so you can build up a hidden state. Otherwise the network will start out generating characters at random. In general the first bunch of characters will be a little rough since it hasn't built up a long history of characters to predict from.
## Loading a checkpoint
| 0.867415 | 0.954774 |
# Broadcast Variables
We already saw so called *broadcast joins* which is a specific impementation of a join suitable for small lookup tables. The term *broadcast* is also used in a different context in Spark, there are also *broadcast variables*.
### Origin of Broadcast Variables
Broadcast variables where introduced fairly early with Spark and were mainly targeted at the RDD API. Nontheless they still have their place with the high level DataFrames API in conjunction with user defined functions (UDFs).
### Weather Example
As usual, we'll use the weather data example. This time we'll manually implement a join using a UDF (actually this would be again a manual broadcast join).
# 1 Load Data
First we load the weather data, which consists of the measurement data and some station metadata.
```
storageLocation = "s3://dimajix-training/data/weather"
```
## 1.1 Load Measurements
Measurements are stored in multiple directories (one per year). But we will limit ourselves to a single year in the analysis to improve readability of execution plans.
```
from pyspark.sql.functions import *
from functools import reduce
# Read in all years, store them in an Python array
raw_weather_per_year = [spark.read.text(storageLocation + "/" + str(i)).withColumn("year", lit(i)) for i in range(2003,2015)]
# Union all years together
raw_weather = reduce(lambda l,r: l.union(r), raw_weather_per_year)
```
Use a single year to keep execution plans small
```
raw_weather = spark.read.text(storageLocation + "/2003").withColumn("year", lit(2003))
```
### Extract Measurements
Measurements were stored in a proprietary text based format, with some values at fixed positions. We need to extract these values with a simple SELECT statement.
```
weather = raw_weather.select(
col("year"),
substring(col("value"),5,6).alias("usaf"),
substring(col("value"),11,5).alias("wban"),
substring(col("value"),16,8).alias("date"),
substring(col("value"),24,4).alias("time"),
substring(col("value"),42,5).alias("report_type"),
substring(col("value"),61,3).alias("wind_direction"),
substring(col("value"),64,1).alias("wind_direction_qual"),
substring(col("value"),65,1).alias("wind_observation"),
(substring(col("value"),66,4).cast("float") / lit(10.0)).alias("wind_speed"),
substring(col("value"),70,1).alias("wind_speed_qual"),
(substring(col("value"),88,5).cast("float") / lit(10.0)).alias("air_temperature"),
substring(col("value"),93,1).alias("air_temperature_qual")
)
```
## 1.2 Load Station Metadata
We also need to load the weather station meta data containing information about the geo location, country etc of individual weather stations.
```
stations = spark.read \
.option("header", True) \
.csv(storageLocation + "/isd-history")
```
### Convert Station Metadata
We convert the stations DataFrame to a normal Python map, since we want to discuss broadcast variables. This means that the variable `py_stations` contains a normal Python object which only lives on the driver. It has no connection to Spark any more.
The resulting map converts a given station id (usaf and wban) to a country.
```
py_stations = stations.select(concat(stations["usaf"], stations["wban"]).alias("key"), stations["ctry"]).collect()
py_stations = {key:value for (key,value) in py_stations}
# Inspect result
list(py_stations.items())[0:10]
```
# 2 Using Broadcast Variables
In the following section, we want to use a Spark broadcast variable inside a UDF. Technically this is not required, as Spark also has other mechanisms of distributing data, so we'll start with a simple implementation *without* using a broadcast variable.
## 2.1 Create a UDF
For the initial implementation, we create a simple Python UDF which looks up the country for a given station id, which consists of the usaf and wban code. This way we will replace the `JOIN` of our original solution with a UDF implemented in Python.
```
def lookup_country(usaf, wban):
return py_stations.get(usaf + wban)
# Test lookup with an existing station
print(lookup_country("007026", "99999"))
# Test lookup with a non-existing station (better should not throw an exception)
print(lookup_country("123", "456"))
```
## 2.2 Not using a broadcast variable
Now that we have a simple Python function providing the required functionality, we convert it to a PySpark UDF using a Python decorator.
```
@udf('string')
def lookup_country(usaf, wban):
return py_stations.get(usaf + wban)
```
### Replace JOIN by UDF
Now we can perform the lookup by using the UDF instead of the original `JOIN`.
```
result = weather.withColumn('country', lookup_country(weather["usaf"], weather["wban"]))
result.limit(10).toPandas()
```
### Remarks
Since the code is actually executed not on the driver, but istributed on the executors, the executors also require access to the Python map. PySpark automatically serializes the map and sends it to the executors on the fly.
### Inspect Plan
We can also inspect the execution plan, which is different from the original implementation. Instead of the broadcast join, it now contains a `BatchEvalPython` step which looks up the stations country from the station id.
```
result.explain()
```
## 2.2 Using a Broadcast Variable
Now let us change the implementation to use a so called *broadcast variable*. While the original implementation implicitly sent the Python map to all executors, a broadcast variable makes the process of sending (*broadcasting*) a Python variable to all executors more explicit.
A Python variable can be broadcast using the `broadcast` method of the underlying Spark context (the Spark session does not export this functionality). Once the data is encapsulated in the broadcast variable, all executors can access the original data via the `value` member variable.
```
# First create a broadcast variable from the original Python map
bc_stations = spark.sparkContext.broadcast(py_stations)
@udf('string')
def lookup_country(usaf, wban):
# Access the broadcast variables value and perform lookup
return bc_stations.value.get(usaf + wban)
```
### Replace JOIN by UDF
Again we replace the original `JOIN` by the UDF we just defined above
```
result = weather.withColumn('country', lookup_country(weather["usaf"], weather["wban"]))
result.limit(10).toPandas()
```
### Remarks
Actually there is no big difference to the original implementation. But Spark handles a broadcast variable slightly more efficiently, especially if the variable is used in multiple UDFs. In this case the data will be broadcast only a single time, while not using a broadcast variable would imply sending the data around for every UDF.
### Execution Plan
The execution plan does not differ at all, since it does not provide information on broadcast variables.
```
result.explain()
```
## 2.3 Pandas UDFs
Since we already learnt that Pandas UDFs are executed more efficiently than normal UDFs, we want to provide a better implementation using Pandas. Of course Pandas UDFs can also access broadcast variables.
```
from pyspark.sql.functions import pandas_udf, PandasUDFType
@pandas_udf('string', PandasUDFType.SCALAR)
def lookup_country(usaf, wban):
# Create helper function
def lookup(key):
# Perform lookup by accessing the Python map
return bc_stations.value.get(key)
# Create key from both incoming Pandas series
usaf_wban = usaf + wban
# Perform lookup
return usaf_wban.apply(lookup)
```
### Replace JOIN by Pandas UDF
Again, we replace the original `JOIN` by the Pandas UDF.
```
result = weather.withColumn('country', lookup_country(weather["usaf"], weather["wban"]))
result.limit(10).toPandas()
```
### Execution Plan
Again, let's inspect the execution plan.
```
result.explain(True)
```
|
github_jupyter
|
storageLocation = "s3://dimajix-training/data/weather"
from pyspark.sql.functions import *
from functools import reduce
# Read in all years, store them in an Python array
raw_weather_per_year = [spark.read.text(storageLocation + "/" + str(i)).withColumn("year", lit(i)) for i in range(2003,2015)]
# Union all years together
raw_weather = reduce(lambda l,r: l.union(r), raw_weather_per_year)
raw_weather = spark.read.text(storageLocation + "/2003").withColumn("year", lit(2003))
weather = raw_weather.select(
col("year"),
substring(col("value"),5,6).alias("usaf"),
substring(col("value"),11,5).alias("wban"),
substring(col("value"),16,8).alias("date"),
substring(col("value"),24,4).alias("time"),
substring(col("value"),42,5).alias("report_type"),
substring(col("value"),61,3).alias("wind_direction"),
substring(col("value"),64,1).alias("wind_direction_qual"),
substring(col("value"),65,1).alias("wind_observation"),
(substring(col("value"),66,4).cast("float") / lit(10.0)).alias("wind_speed"),
substring(col("value"),70,1).alias("wind_speed_qual"),
(substring(col("value"),88,5).cast("float") / lit(10.0)).alias("air_temperature"),
substring(col("value"),93,1).alias("air_temperature_qual")
)
stations = spark.read \
.option("header", True) \
.csv(storageLocation + "/isd-history")
py_stations = stations.select(concat(stations["usaf"], stations["wban"]).alias("key"), stations["ctry"]).collect()
py_stations = {key:value for (key,value) in py_stations}
# Inspect result
list(py_stations.items())[0:10]
def lookup_country(usaf, wban):
return py_stations.get(usaf + wban)
# Test lookup with an existing station
print(lookup_country("007026", "99999"))
# Test lookup with a non-existing station (better should not throw an exception)
print(lookup_country("123", "456"))
@udf('string')
def lookup_country(usaf, wban):
return py_stations.get(usaf + wban)
result = weather.withColumn('country', lookup_country(weather["usaf"], weather["wban"]))
result.limit(10).toPandas()
result.explain()
# First create a broadcast variable from the original Python map
bc_stations = spark.sparkContext.broadcast(py_stations)
@udf('string')
def lookup_country(usaf, wban):
# Access the broadcast variables value and perform lookup
return bc_stations.value.get(usaf + wban)
result = weather.withColumn('country', lookup_country(weather["usaf"], weather["wban"]))
result.limit(10).toPandas()
result.explain()
from pyspark.sql.functions import pandas_udf, PandasUDFType
@pandas_udf('string', PandasUDFType.SCALAR)
def lookup_country(usaf, wban):
# Create helper function
def lookup(key):
# Perform lookup by accessing the Python map
return bc_stations.value.get(key)
# Create key from both incoming Pandas series
usaf_wban = usaf + wban
# Perform lookup
return usaf_wban.apply(lookup)
result = weather.withColumn('country', lookup_country(weather["usaf"], weather["wban"]))
result.limit(10).toPandas()
result.explain(True)
| 0.728072 | 0.967869 |
1 - Defina a função soma_nat que recebe como argumento um número natural n
e devolve a soma de todos os números naturais até n.
- Ex: soma_nat(5) = 15
```
soma_nat = lambda n : 1 if n == 1 else n + soma_nat(n-1)
print(soma_nat(2))
```
2 - Defina a função div que recebe como argumentos dois números naturais m
e n e devolve o resultado da divisão inteira de m por n. Neste exercício você não
pode recorrer às operações aritméticas de multiplicação, divisão e resto da divisão
inteira.
- Ex: div(7,2) = 3
```
div = lambda m, n: 0 if m < n else 1 + div(m-n, n)
print(div(7,2))
```
3 - Defina a função prim_alg que recebe como argumento um número natural e devolve o primeiro algarismo (o mais significativo) na representação decimal de n.
- Ex: prim_alg(5649) = 5
- Ex: prim_alg(7) = 7
```
prim_alg = lambda n: str(n)[0]
print(prim_alg(5649))
print(prim_alg(7))
```
4 - Defina a função prod_lista que recebe como argumento uma lista de inteiros e
devolve o produto dos seus elementos.
- Ex: prod_lista([1,2,3,4,5,6]) = 720
```
from functools import reduce
prod_lista = lambda inteiros: reduce((lambda x, y: x * y), inteiros)
prod_lista([1,2,3,4,5,6])
```
5 - Defina a função contem_parQ que recebe como argumento uma lista de números
inteiros w e devolve True se w contém um número par e False em caso contrário.
- Ex: contem_parQ([2,3,1,2,3,4]) = True
- Ex: contem_parQ([1,3,5,7]) = False
```
contem_parQ = lambda w: True if list(filter(lambda x: x%2 == 0, w)) else False
print(contem_parQ([2,3,1,2,3,4]))
print(contem_parQ([1,3,5,7]))
```
6 - Defina a função todos_imparesQ que recebe como argumento uma lista de
números inteiros w e devolve True se w contém apenas números ímpares e False
em caso contrário.
- Ex: todos_imparesQ([1,3,5,7]) = True
- Ex: todos_imparesQ([]) = True
- Ex: todos_imparesQ([1,2,3,4,5]) = False
```
todos_imparesQ = lambda w: True if list(filter(lambda x: x%2 != 0, w)) == w else False
print(todos_imparesQ([1,3,5,7]))
print(todos_imparesQ([]))
print(todos_imparesQ([1,2,3,4,5]))
```
7 - Defina a função pertenceQ que recebe como argumentos uma lista de números
inteiros w e um número inteiro n e devolve True se n ocorre em w e False em
caso contrário.
- Ex: pertenceQ([1,2,3],1) = True
- Ex: pertenceQ([1,2,3],2) = True
- Ex: pertenceQ([1,2,3],3) = True
- Ex: pertenceQ([1,2,3],4) = False
```
pertenceQ = lambda w, n: True if n in w else False
print(pertenceQ([1,2,3],1))
print(pertenceQ([1,2,3],2))
print(pertenceQ([1,2,3],3))
print(pertenceQ([1,2,3],4))
```
8 - Defina a função junta que recebe como argumentos duas listas de números
inteiros w1 e w2 e devolve a concatenação de w1 com w2 .
- Ex: junta([1,2,3],[4,5,6]) = [1, 2, 3, 4, 5, 6]
- Ex: junta([],[4,5,6]) = [4, 5, 6]
- Ex: junta([1,2,3],[]) = [1, 2, 3]
```
junta = lambda w1, w2: w1 + w2
print(junta([1,2,3],[4,5,6]))
print(junta([],[4,5,6]))
print(junta([1,2,3],[]) )
```
9 - Defina a função temPrimoQ que recebe como argumento uma lista de listas de
números inteiros w e devolve True se alguma das sublistas w tem um número
primo e False em caso contrário.
- Ex: temPrimoQ([[4,4,4,4],[5,4,6,7],[2,4,3]]) = True
- Ex: temPrimoQ([[4,4,4,4],[4,4,4],[],[4]]) = False
```
retorna_primo = lambda x: True if not list(filter(lambda z: x % z == 0, range(2, x))) else False
retorna_primo_lista = lambda lista: list(filter(lambda x: retorna_primo(x), lista))
temPrimoQ = lambda listas: True if list(filter(lambda lista: retorna_primo_lista(lista), listas)) else False
print(temPrimoQ([[4,4,4,4],[5,4,6,7],[2,4,3]]))
print(temPrimoQ([[4,4,4,4],[4,4,4],[],[4]]))
```
10 - Defina a função inverteLista que recebe como argumento uma lista w e devolve a
mesma lista mas invertida.
- Ex: inverteLista([1,2,3,4,5]) = [5, 4, 3, 2, 1]
- Ex: inverteLista([])
```
inverteLista = lambda w: w[::-1]
print((inverteLista([1,2,3,4,5])))
print(inverteLista([]))
```
|
github_jupyter
|
soma_nat = lambda n : 1 if n == 1 else n + soma_nat(n-1)
print(soma_nat(2))
div = lambda m, n: 0 if m < n else 1 + div(m-n, n)
print(div(7,2))
prim_alg = lambda n: str(n)[0]
print(prim_alg(5649))
print(prim_alg(7))
from functools import reduce
prod_lista = lambda inteiros: reduce((lambda x, y: x * y), inteiros)
prod_lista([1,2,3,4,5,6])
contem_parQ = lambda w: True if list(filter(lambda x: x%2 == 0, w)) else False
print(contem_parQ([2,3,1,2,3,4]))
print(contem_parQ([1,3,5,7]))
todos_imparesQ = lambda w: True if list(filter(lambda x: x%2 != 0, w)) == w else False
print(todos_imparesQ([1,3,5,7]))
print(todos_imparesQ([]))
print(todos_imparesQ([1,2,3,4,5]))
pertenceQ = lambda w, n: True if n in w else False
print(pertenceQ([1,2,3],1))
print(pertenceQ([1,2,3],2))
print(pertenceQ([1,2,3],3))
print(pertenceQ([1,2,3],4))
junta = lambda w1, w2: w1 + w2
print(junta([1,2,3],[4,5,6]))
print(junta([],[4,5,6]))
print(junta([1,2,3],[]) )
retorna_primo = lambda x: True if not list(filter(lambda z: x % z == 0, range(2, x))) else False
retorna_primo_lista = lambda lista: list(filter(lambda x: retorna_primo(x), lista))
temPrimoQ = lambda listas: True if list(filter(lambda lista: retorna_primo_lista(lista), listas)) else False
print(temPrimoQ([[4,4,4,4],[5,4,6,7],[2,4,3]]))
print(temPrimoQ([[4,4,4,4],[4,4,4],[],[4]]))
inverteLista = lambda w: w[::-1]
print((inverteLista([1,2,3,4,5])))
print(inverteLista([]))
| 0.111955 | 0.936807 |
# Technology Explorers
## Introduction Data Science
**Instructor**: Wesley Beckner
**Contact**: [email protected]
<br>
---
<br>
🎉 Today, we'll be working from this _digital_ notebook to complete exercises! If you don't have a computer, not to worry. Grab a notepad and pencil to write down your ideas and notes! 🎉
<br>
---
# Preparing Notebook for Demos
## Importing Packages
Once we have our packages installed, we need to import them. We can also import packages that are pre-installed in the Colab environment.
```
import numpy as np
import random
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.linear_model import LinearRegression
```
## Importing Data
We also have the ability to import data, and use it elsewhere in the notebook 📝!
```
# No Data Today! :)
```
## 🦉 Tenets of Machine Learning
We'll take the simple linear regression as an example and discuss some of the core tenets of ML: Bias-variance trade-off, irreducible error, and regularization.
### 📈 Bias-Variance Trade-Off
#### (Over and Underfitting)
The basic premise here is that theres some optimum number of parmeters to include in my model, if I include too few, my model will be too simple (***high bias***) and if I include too many it will be too complex and fit to noise (***high variance***)
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/machine_learning/ML5.png" width=1000px></img>
We can explore this phenomenon more easily, making up some data ourselves:
```
# we can throttle the error rate
err = .5
random.seed(42)
# our data has a known underlying functional form (log(x))
def func(x, err):
return np.log(x) + err * random.randint(-1,1) * random.random()
x = np.arange(20,100)
y = [func(t, err) for t in x]
plt.plot(x,y, ls='', marker='.')
plt.xlabel('X')
plt.ylabel('Y')
```
Now, let's pretend we've sampled from this ***population*** of data:
```
random.seed(42)
X_train = random.sample(list(x), 10)
indices = [list(x).index(i) for i in X_train]
# we could also do it this way: np.argwhere([i in X_train for i in x])
y_train = [y[i] for i in indices]
plt.plot(X_train,y_train, ls='', marker='.')
```
Now let's take two extreme scenarios, fitting a linear line and a high order polynomial, to these datapoints. Keeping in mind the larger dataset, as well as the error we introduced in our data generating function, this will really illustrate our point!
```
# solving our training data with a n-degree polynomial
coefs = np.polyfit(X_train, y_train, 9)
# solve the slope and intercept of our 1-degree polynomial ;)
model = LinearRegression()
model.fit(np.array(X_train).reshape(-1,1), y_train)
# create some x data to plot our functions
X_seq = np.linspace(min(X_train),max(X_train),300).reshape(-1,1)
fig, ax = plt.subplots(1,2,figsize=(15,5))
ax[0].plot(X_seq, model.predict(X_seq), c='grey', ls='--')
ax[0].plot(X_train, y_train, ls='', marker='.')
ax[0].set_ylim(min(y_train), max(y_train))
ax[0].set_title("High Bias Model")
ax[1].plot(X_seq, np.polyval(coefs, X_seq), c='grey', ls='--')
ax[1].plot(X_train, y_train, ls='', marker='.')
ax[1].set_ylim(min(y_train), max(y_train))
ax[1].set_title("High Variance Model")
```
We've demonstrated two extreme cases. On the left, we limit our regression to only two parameters, a slope and a y-intercept. We say that this model has *high bias* because we are forcing the functional form without much consideration to the underlying data — we are saying this data is generated by a linear function, and no matter what data I train on, my final model will still be a straight line that more or less appears the same. Put another way, it has *low variance* with respect to the underlying data.
On the right, we've allowed our model just as many polynomials it needs to perfectly fit the training data! We say this model has *low bias* because we don't introduce many constraints on the final form of the model. it is *high variance* because depending on the underlying training data, the final outcome of the model can change quite drastically!
In reality, the best model lies somewhere between these two cases. In the next few paragraphs we'll explore this concept further:
1. what happens when we retrain these models on different samples of the data population
* and let's use this to better understand what we mean by *bias* and *variance*
2. what happens when we tie this back in with the error we introduced to the data generator?
* and let's use this to better understand irreducible error
```
random.seed(42)
fig, ax = plt.subplots(1,2,figsize=(15,5))
for samples in range(5):
X_train = random.sample(list(x), 10)
indices = [list(x).index(i) for i in X_train]
y_train = [y[i] for i in indices]
# solving our training data with a n-degree polynomial
coefs = np.polyfit(X_train, y_train, 9)
# solve the slope and intercept of our 1-degree polynomial ;)
model = LinearRegression()
model.fit(np.array(X_train).reshape(-1,1), y_train)
# create some x data to plot our functions
X_seq = np.linspace(min(X_train),max(X_train),300).reshape(-1,1)
ax[0].plot(X_seq, model.predict(X_seq), alpha=0.5, ls='--')
ax[0].plot(X_train, y_train, ls='', marker='.')
ax[0].set_ylim(min(y_train), max(y_train))
ax[0].set_title("High Bias Model")
ax[1].plot(X_seq, np.polyval(coefs, X_seq), alpha=0.5, ls='--')
ax[1].plot(X_train, y_train, ls='', marker='.')
ax[1].set_ylim(min(y_train), max(y_train))
ax[1].set_title("High Variance Model")
```
As we can see, depending on what data we train our model on, the *high bias* model changes relatively slightly, while the *high variance* model changes a whole awful lot!
The *high variance* model is prone to something we call *overfitting*. It fits the training data very well, but at the expense of creating a good, generalizable model that does well on unseen data. Let's take our last models, and plot them along the rest of the unseen data, what we'll call the *population*
```
# solving our training data with a n-degree polynomial
coefs = np.polyfit(X_train, y_train, 9)
# solve the slope and intercept of our 1-degree polynomial ;)
model = LinearRegression()
model.fit(np.array(X_train).reshape(-1,1), y_train)
# create some x data to plot our functions
X_seq = np.linspace(min(X_train),max(X_train),300).reshape(-1,1)
fig, ax = plt.subplots(1,2,figsize=(15,5))
ax[0].plot(X_seq, model.predict(X_seq), c='grey', ls='--')
ax[0].plot(x, y, ls='', marker='*', alpha=0.6)
ax[0].plot(X_train, y_train, ls='', marker='.')
ax[0].set_ylim(min(y), max(y))
ax[0].set_title("High Bias Model")
ax[1].plot(X_seq, np.polyval(coefs, X_seq), c='grey', ls='--')
ax[1].plot(x, y, ls='', marker='*', alpha=0.6)
ax[1].plot(X_train, y_train, ls='', marker='.')
ax[1].set_ylim(min(y), max(y))
ax[1].set_title("High Variance Model")
```
In particular, we see that the high variance model is doing very wacky things, demonstrating behaviors in the model where the underlying population data really gives no indication of such behavior. We say that these high variance model are particuarly prone to the phenomenon of *over fitting* and this is generally due to the fact that there is irreducible error in the underlying data. Let's demonstrate this.
### ❕ Irreducible Error
Irreducible error is ***always*** present in our data. It is a part of life, welcome to it. That being said, let's look what happens when we *pretend* there isn't any irreducible error in our population data
```
x = np.arange(20,100)
y = [func(t, err=0) for t in x]
plt.plot(x,y, ls='', marker='.')
random.seed(42)
X_train = random.sample(list(x), 10)
indices = [list(x).index(i) for i in X_train]
# we could also do it this way: np.argwhere([i in X_train for i in x])
y_train = [y[i] for i in indices]
# solving our training data with a n-degree polynomial
coefs = np.polyfit(X_train, y_train, 9)
# solve the slope and intercept of our 1-degree polynomial ;)
model = LinearRegression()
model.fit(np.array(X_train).reshape(-1,1), y_train)
# create some x data to plot our functions
X_seq = np.linspace(min(X_train),max(X_train),300).reshape(-1,1)
fig, ax = plt.subplots(1,2,figsize=(15,5))
ax[0].plot(X_seq, model.predict(X_seq), c='grey', ls='--')
ax[0].plot(x, y, ls='', marker='o', alpha=0.2)
ax[0].plot(X_train, y_train, ls='', marker='.')
ax[0].set_ylim(min(y), max(y))
ax[0].set_title("High Bias Model")
ax[1].plot(X_seq, np.polyval(coefs, X_seq), c='grey', ls='--')
ax[1].plot(x, y, ls='', marker='o', alpha=0.2)
ax[1].plot(X_train, y_train, ls='', marker='.')
ax[1].set_ylim(min(y), max(y))
ax[1].set_title("High Variance Model")
```
This time, our high variance model really *gets it*! And this is because the data we trained on actually *is* a good representation of the entire population. But this, in reality, almost never, ever happens. In the real world, we have irreducible error in our data samples, and we must account for this when choosing our model.
I'm summary, we call this balance between error in our model functional form, and error from succumbing to irreducible error in our training data, the *bias variance tradeoff*
### 🕸️ Regularization
To talk about regularization, we're going to continue with our simple high bias model example, the much revered linear regression model. Linear regression takes on the form:
$$y(x)= m\cdot x + b$$
where $y$ is some target value and, $x$ is some feature; $m$ and $b$ are the slope and intercept, respectively.
To solve the problem, we need to find the values of $b$ and $m$ in equation 1 to best fit the data.
In linear regression our goal is to minimize the error between computed values of positions $y^{\sf calc}(x_i)\equiv y^{\sf calc}_i$ and known values $y^{\sf exact}(x_i)\equiv y^{\sf exact}_i$, i.e. find $b$ and $m$ which lead to lowest value of
$$\epsilon (m,b) =SS_{\sf res}=\sum_{i=1}^{N}\left(y^{\sf exact}_i - y^{\sf calc}_i\right)^2 = \sum_{i=1}^{N}\left(y^{\sf exact}_i - m\cdot x_i - b \right)^2$$
**Now onto Regularization**
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/machine_learning/ML6.png" width=1000px></img>
There are many other regression algorithms, the two we want to highlight here are Ridge Regression and LASSO. They differ by an added term to the loss function. Let's review. The above equation expanded to multivariate form yields:
$$\sum_{i=1}^{N}(y_i - \sum_{j=1}^{P}x_{ij}\beta_{j})^2$$
for Ridge regression, we add a **_regularization_** term known as **_L2_** regularization:
$$\sum_{i=1}^{N}(y_i - \sum_{j=1}^{P}x_{ij}\beta_{j})^2 + \lambda \sum_{j=1}^{P}\beta_{j}^2$$
for **_LASSO_** (Least Absolute Shrinkage and Selection Operator) we add **_L1_** regularization:
$$\sum_{i=1}^{N}(y_i - \sum_{j=1}^{P}x_{ij}\beta_{j})^2 + \lambda \sum_{j=1}^{P}|\beta_{j}|$$
The difference between the two is that LASSO will allow coefficients to shrink to 0 while Ridge regression will not. **_Elastic Net_** is a combination of these two regularization methods. The key notion here is that ***regularization*** is a way of tempering our model, allowing it to pick for itself the most appropriate features. This crops up in many places other than simple linear regression in machine learning.
**Regularization appears in...**
***Ensemble learners*** (e.g. XGBoost and Random Forests) by combining the combinations of many weak algorithms
***Neural networks*** with ***dropout*** and ***batch normalization***
Dropout is the Neural Network response to the wide success of ensemble learning. In a dropout layer, random neurons are dropped in each batch of training, i.e. their weighted updates are not sent to the next neural layer. Just as we learned with random forests, the end result is that the neural network can be thought of as many _independent models_ that _vote_ on the final output.
Put another way, when a network does not contain dropout layers, and has a capacity that exceeds that which would be suited for the true, underlying complexity level of the data, it can begin to fit to noise. This ability to fit to noise is based on very specific relationships between neurons, which fire uniquely given the particular training example. Adding dropout _breaks_ these specific neural connections, and so the neural network as a whole is forced to find weights that apply generally, as there is no guarantee they will be _turned on_ when their specific training example they would usually overfit for comes around again.
<p align=center>
<img src="https://i.imgur.com/a86utxY.gif"></img>
</p>
<small> Network with 50% dropout. Borrowed from Kaggle learn. </small>
## 📊 What is Data Science?
### The Emergence of Data Science
Data Science is a broad field, and depending on who you talk to, it can mean different things. In summary, many independent scientific fields began accumulating large amounts of data. At the UW in particular, these were dominated by the astronomy and oceanography departments. Folks began to realize that they needed a particular set of tools to handle large amounts of data. This culminated in the [eScience studio](https://escience.washington.edu/), which began to service the data needs of many departments on campus.
Today, data science not only has to do with large amounts of data, but refers generally to tools that allow us to work with a variety of data types. Because of this, machine learning is a tool within data science. But there are other tools apart from machine learning that makeup the data science ecosystem. Some of them are:
* data visualization
* databases
* statistics
You could argue for others as well (algorithms, web servers, programming, etc.), but these are the formally accepted areas.
#### 💭 7
We've talked a lot! Wow! Last topic. D A T A S C I E N C E. What is it? Any idea? Talk to your neighbor, convene together, then let's share. Do this at 2 different levels:
How would you explain data science to:
1. your grandmother
2. a student
#### 💬 7
I'll write these down, let's see if we can all agree on a precise definition
### Saying Stuff About Data (Statistics)
When we're talking about statistics, we're really talking about data story telling. Statistics is at the C O R E of data science, really. Without a basic knowledge of statistics it'll be hard for you to construct your data narratives and have them hold water.
Let's start with some simple examples of data story telling, and use these to generate our own thoughts on the matter.
#### Anscombe's Quartet
There's a very famous anomaly in DS caled Anscombe's quartet. Observe the following data
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds1.png"></img>
We can construct this in python and confirm the summary statistics ourselves
```
df = pd.read_excel("https://github.com/wesleybeckner/technology_explorers/blob"\
"/main/assets/data_science/anscombes.xlsx?raw=true",
header=[0,1])
df
```
We can calculate the mean/variance of X and Y for samples I, II, III, and IV
```
df.mean()
# do we remember the relationship between standard deviation and variance?
df.std()**2
```
We talked about the equation for a linear line last time:
$$y(x)= m\cdot x + b$$
```
model = LinearRegression()
sets = ['I', 'II', 'III', 'IV']
for data in sets:
model.fit(df[data]['X'].values.reshape(11,1),
df[data]['Y'])
print("Linear Regression Line: Y = {:.2f}X + {:.2f}".format(model.coef_[0], model.intercept_))
```
$R^2$ measures the goodness of fit. $R^2$ is generally defined as the ratio of the total sum of squares $SS_{\sf tot} $ to the residual sum of squares $SS_{\sf res} $:
We already talked about the residual sum of squares last session (what were we trying to do with this equation??)
$$SS_{\sf res}=\sum_{i=1}^{N} \left(y^{\sf exact}_i - y^{\sf calc}_i\right)^2$$
We now define the total sum of squares, a measure of the total variance in the data:
$$SS_{\sf tot}=\sum_{i=1}^{N} \left(y^{\sf exact}_i-\bar{y}\right)^2$$
The $R^2$ tells us how much of the variance of the data, is captured by the model we created:
$$R^2 = 1 - {SS_{\sf res}\over SS_{\sf tot}}$$
In the first equation, $\bar{y}=\sum_i y^{\sf exact}_i/N$ is the average value of y for $N$ points. The best value of $R^2$ is 1 but it can also take a negative value if the error is large.
```
for data in sets:
# calc the ssr
ssr = np.sum((df[data]['Y'] -
model.predict(df[data]['X'].values.reshape(-1,1)))**2)
# calc the sst
sst = np.sum((df[data]['Y'] -
df[data]['Y'].mean())**2)
# calc the r2
r2 = 1 - (ssr/sst)
print("R2 = {:.2f}".format(r2))
```
As we can see, everything checks out. The summary statistics are all the same!
Can we answer the following:
> What dataset is best described by the line of best fit?
We will revisit this question when we talk about data visualization
#### Taxonomy of Data Types
Another important topic in data science, is simply what kind of data we are working with. This will help us decide what kind of models to build, as well as how to visualize our data, and perhaps store it as well.
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds2.png"></img>
#### 💬 8
What are some examples of the different datatypes we can think of?
### Data Visualization
Data visualization, like it sounds, has to do with how we display and communicate information. At the end of the day, your findings and algorithms aren't worth very much if we can't share them with others.
#### Guiding Principles of Data Visualization
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds3.png"></img>
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds9.gif"></img>
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds7.png"></img>
wattenberg and Viegas visualization
```
%%HTML
<video width="640" height="580" controls>
<source src="https://github.com/wesleybeckner/technology_explorers/blob/main/assets/data_science/ds4.mp4?raw=true" type="video/mp4">
</video>
```
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds6.png"></img>
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds4.png"></img>
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds5.png"></img>
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds8.png"></img>
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds9.png"></img>
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds10.png"></img>
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds11.png"></img>
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds12.png"></img>
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds13.png"></img>
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds14.jpg"></img>
#### Visualization Un-Examples
**Unexample 1**
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds15.jpg"></img>
**Unexample 2**
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds19.png"></img>
**Unexample 3**
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds17.png"></img>
**Unexample 4**
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds18.png"></img>
#### 💭 8
Find an example of an interactive data visualization online. Here's one I [found](https://www.migrationpolicy.org/programs/data-hub/charts/us-immigrant-population-state-and-county) that I though was quite interesting!
#### 💬 8
Swap visualization links with your neighbor. What do you think could be improved about each one?
#### Back to Anscombe's Quartet
<p align=center>
<img src="https://raw.githubusercontent.com/wesleybeckner/technology_explorers/main/assets/data_science/ds20.png"></img>
### Databases
Databases can be a seemingly dull sideshow of machine learning. But it is the foundation of any machine learning ever getting done. Without having our data in a neat and tidy place, we will be unable to train any algorithms.
I offer one interesting tidbit about databases:
Imagine Zhu Li and Meelo are depositing money in a shared bank account. In one version of the scheme, Zhu Li and Meelo can access their shared account, and make a deposit _at the same time_. The benefit to this is that the transaction happens fast, but on the downside, while they are both accessing the account, neither will see the action of the other. This can be quite alarming when it comes to money!
In a second version of the scheme, Zhu Li must wait for Meelo's transaction to update and vice versa before one or the other can make a deposity. The plus side is that both Zhu Li and Meelo will have complete knowledge of the exact value within the account at any point in time. The downside is that this can slow down the overall process.
This is a core consideration, among others, when designing databases!
#### 💭 9
Find an example of a place where you use databases in your day to day life
#### 💬 9
Did any of us come up with the same examples?
```
```
|
github_jupyter
|
import numpy as np
import random
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.linear_model import LinearRegression
# No Data Today! :)
# we can throttle the error rate
err = .5
random.seed(42)
# our data has a known underlying functional form (log(x))
def func(x, err):
return np.log(x) + err * random.randint(-1,1) * random.random()
x = np.arange(20,100)
y = [func(t, err) for t in x]
plt.plot(x,y, ls='', marker='.')
plt.xlabel('X')
plt.ylabel('Y')
random.seed(42)
X_train = random.sample(list(x), 10)
indices = [list(x).index(i) for i in X_train]
# we could also do it this way: np.argwhere([i in X_train for i in x])
y_train = [y[i] for i in indices]
plt.plot(X_train,y_train, ls='', marker='.')
# solving our training data with a n-degree polynomial
coefs = np.polyfit(X_train, y_train, 9)
# solve the slope and intercept of our 1-degree polynomial ;)
model = LinearRegression()
model.fit(np.array(X_train).reshape(-1,1), y_train)
# create some x data to plot our functions
X_seq = np.linspace(min(X_train),max(X_train),300).reshape(-1,1)
fig, ax = plt.subplots(1,2,figsize=(15,5))
ax[0].plot(X_seq, model.predict(X_seq), c='grey', ls='--')
ax[0].plot(X_train, y_train, ls='', marker='.')
ax[0].set_ylim(min(y_train), max(y_train))
ax[0].set_title("High Bias Model")
ax[1].plot(X_seq, np.polyval(coefs, X_seq), c='grey', ls='--')
ax[1].plot(X_train, y_train, ls='', marker='.')
ax[1].set_ylim(min(y_train), max(y_train))
ax[1].set_title("High Variance Model")
random.seed(42)
fig, ax = plt.subplots(1,2,figsize=(15,5))
for samples in range(5):
X_train = random.sample(list(x), 10)
indices = [list(x).index(i) for i in X_train]
y_train = [y[i] for i in indices]
# solving our training data with a n-degree polynomial
coefs = np.polyfit(X_train, y_train, 9)
# solve the slope and intercept of our 1-degree polynomial ;)
model = LinearRegression()
model.fit(np.array(X_train).reshape(-1,1), y_train)
# create some x data to plot our functions
X_seq = np.linspace(min(X_train),max(X_train),300).reshape(-1,1)
ax[0].plot(X_seq, model.predict(X_seq), alpha=0.5, ls='--')
ax[0].plot(X_train, y_train, ls='', marker='.')
ax[0].set_ylim(min(y_train), max(y_train))
ax[0].set_title("High Bias Model")
ax[1].plot(X_seq, np.polyval(coefs, X_seq), alpha=0.5, ls='--')
ax[1].plot(X_train, y_train, ls='', marker='.')
ax[1].set_ylim(min(y_train), max(y_train))
ax[1].set_title("High Variance Model")
# solving our training data with a n-degree polynomial
coefs = np.polyfit(X_train, y_train, 9)
# solve the slope and intercept of our 1-degree polynomial ;)
model = LinearRegression()
model.fit(np.array(X_train).reshape(-1,1), y_train)
# create some x data to plot our functions
X_seq = np.linspace(min(X_train),max(X_train),300).reshape(-1,1)
fig, ax = plt.subplots(1,2,figsize=(15,5))
ax[0].plot(X_seq, model.predict(X_seq), c='grey', ls='--')
ax[0].plot(x, y, ls='', marker='*', alpha=0.6)
ax[0].plot(X_train, y_train, ls='', marker='.')
ax[0].set_ylim(min(y), max(y))
ax[0].set_title("High Bias Model")
ax[1].plot(X_seq, np.polyval(coefs, X_seq), c='grey', ls='--')
ax[1].plot(x, y, ls='', marker='*', alpha=0.6)
ax[1].plot(X_train, y_train, ls='', marker='.')
ax[1].set_ylim(min(y), max(y))
ax[1].set_title("High Variance Model")
x = np.arange(20,100)
y = [func(t, err=0) for t in x]
plt.plot(x,y, ls='', marker='.')
random.seed(42)
X_train = random.sample(list(x), 10)
indices = [list(x).index(i) for i in X_train]
# we could also do it this way: np.argwhere([i in X_train for i in x])
y_train = [y[i] for i in indices]
# solving our training data with a n-degree polynomial
coefs = np.polyfit(X_train, y_train, 9)
# solve the slope and intercept of our 1-degree polynomial ;)
model = LinearRegression()
model.fit(np.array(X_train).reshape(-1,1), y_train)
# create some x data to plot our functions
X_seq = np.linspace(min(X_train),max(X_train),300).reshape(-1,1)
fig, ax = plt.subplots(1,2,figsize=(15,5))
ax[0].plot(X_seq, model.predict(X_seq), c='grey', ls='--')
ax[0].plot(x, y, ls='', marker='o', alpha=0.2)
ax[0].plot(X_train, y_train, ls='', marker='.')
ax[0].set_ylim(min(y), max(y))
ax[0].set_title("High Bias Model")
ax[1].plot(X_seq, np.polyval(coefs, X_seq), c='grey', ls='--')
ax[1].plot(x, y, ls='', marker='o', alpha=0.2)
ax[1].plot(X_train, y_train, ls='', marker='.')
ax[1].set_ylim(min(y), max(y))
ax[1].set_title("High Variance Model")
df = pd.read_excel("https://github.com/wesleybeckner/technology_explorers/blob"\
"/main/assets/data_science/anscombes.xlsx?raw=true",
header=[0,1])
df
df.mean()
# do we remember the relationship between standard deviation and variance?
df.std()**2
model = LinearRegression()
sets = ['I', 'II', 'III', 'IV']
for data in sets:
model.fit(df[data]['X'].values.reshape(11,1),
df[data]['Y'])
print("Linear Regression Line: Y = {:.2f}X + {:.2f}".format(model.coef_[0], model.intercept_))
for data in sets:
# calc the ssr
ssr = np.sum((df[data]['Y'] -
model.predict(df[data]['X'].values.reshape(-1,1)))**2)
# calc the sst
sst = np.sum((df[data]['Y'] -
df[data]['Y'].mean())**2)
# calc the r2
r2 = 1 - (ssr/sst)
print("R2 = {:.2f}".format(r2))
%%HTML
<video width="640" height="580" controls>
<source src="https://github.com/wesleybeckner/technology_explorers/blob/main/assets/data_science/ds4.mp4?raw=true" type="video/mp4">
</video>
| 0.629661 | 0.944022 |
# Introduction to Deep Learning with PyTorch
In this notebook, you'll get introduced to [PyTorch](http://pytorch.org/), a framework for building and training neural networks. PyTorch in a lot of ways behaves like the arrays you love from Numpy. These Numpy arrays, after all, are just tensors. PyTorch takes these tensors and makes it simple to move them to GPUs for the faster processing needed when training neural networks. It also provides a module that automatically calculates gradients (for backpropagation!) and another module specifically for building neural networks. All together, PyTorch ends up being more coherent with Python and the Numpy/Scipy stack compared to TensorFlow and other frameworks.
## Neural Networks
Deep Learning is based on artificial neural networks which have been around in some form since the late 1950s. The networks are built from individual parts approximating neurons, typically called units or simply "neurons." Each unit has some number of weighted inputs. These weighted inputs are summed together (a linear combination) then passed through an activation function to get the unit's output.
<img src="assets/simple_neuron.png" width=400px>
Mathematically this looks like:
$$
\begin{align}
y &= f(w_1 x_1 + w_2 x_2 + b) \\
y &= f\left(\sum_i w_i x_i \right)
\end{align}
$$
With vectors this is the dot/inner product of two vectors:
$$
h = \begin{bmatrix}
x_1 \, x_2 \cdots x_n
\end{bmatrix}
\cdot
\begin{bmatrix}
w_1 \\
w_2 \\
\vdots \\
w_n
\end{bmatrix}
$$
### Stack them up!
We can assemble these unit neurons into layers and stacks, into a network of neurons. The output of one layer of neurons becomes the input for the next layer. With multiple input units and output units, we now need to express the weights as a matrix.
<img src='assets/multilayer_diagram_weights.png' width=450px>
We can express this mathematically with matrices again and use matrix multiplication to get linear combinations for each unit in one operation. For example, the hidden layer ($h_1$ and $h_2$ here) can be calculated
$$
\vec{h} = [h_1 \, h_2] =
\begin{bmatrix}
x_1 \, x_2 \cdots \, x_n
\end{bmatrix}
\cdot
\begin{bmatrix}
w_{11} & w_{12} \\
w_{21} &w_{22} \\
\vdots &\vdots \\
w_{n1} &w_{n2}
\end{bmatrix}
$$
The output for this small network is found by treating the hidden layer as inputs for the output unit. The network output is expressed simply
$$
y = f_2 \! \left(\, f_1 \! \left(\vec{x} \, \mathbf{W_1}\right) \mathbf{W_2} \right)
$$
## Tensors
It turns out neural network computations are just a bunch of linear algebra operations on *tensors*, a generalization of matrices. A vector is a 1-dimensional tensor, a matrix is a 2-dimensional tensor, an array with three indices is a 3-dimensional tensor (RGB color images for example). The fundamental data structure for neural networks are tensors and PyTorch (as well as pretty much every other deep learning framework) is built around tensors.
<img src="assets/tensor_examples.svg" width=600px>
With the basics covered, it's time to explore how we can use PyTorch to build a simple neural network.
```
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import numpy as np
import torch
import helper
```
First, let's see how we work with PyTorch tensors. These are the fundamental data structures of neural networks and PyTorch, so it's imporatant to understand how these work.
```
x = torch.rand(3, 2)
x
y = torch.ones(x.size())
y
z = x + y
z
```
In general PyTorch tensors behave similar to Numpy arrays. They are zero indexed and support slicing.
```
z[0]
z[:, 1:]
```
Tensors typically have two forms of methods, one method that returns another tensor and another method that performs the operation in place. That is, the values in memory for that tensor are changed without creating a new tensor. In-place functions are always followed by an underscore, for example `z.add()` and `z.add_()`.
```
# Return a new tensor z + 1
z.add(1)
# z tensor is unchanged
z
# Add 1 and update z tensor in-place
z.add_(1)
# z has been updated
z
```
### Reshaping
Reshaping tensors is a really common operation. First to get the size and shape of a tensor use `.size()`. Then, to reshape a tensor, use `.resize_()`. Notice the underscore, reshaping is an in-place operation.
```
z.size()
z.resize_(2, 3)
z
```
## Numpy to Torch and back
Converting between Numpy arrays and Torch tensors is super simple and useful. To create a tensor from a Numpy array, use `torch.from_numpy()`. To convert a tensor to a Numpy array, use the `.numpy()` method.
```
a = np.random.rand(4,3)
a
b = torch.from_numpy(a)
b
b.numpy()
```
The memory is shared between the Numpy array and Torch tensor, so if you change the values in-place of one object, the other will change as well.
```
# Multiply PyTorch Tensor by 2, in place
b.mul_(2)
# Numpy array matches new values from Tensor
a
```
|
github_jupyter
|
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import numpy as np
import torch
import helper
x = torch.rand(3, 2)
x
y = torch.ones(x.size())
y
z = x + y
z
z[0]
z[:, 1:]
# Return a new tensor z + 1
z.add(1)
# z tensor is unchanged
z
# Add 1 and update z tensor in-place
z.add_(1)
# z has been updated
z
z.size()
z.resize_(2, 3)
z
a = np.random.rand(4,3)
a
b = torch.from_numpy(a)
b
b.numpy()
# Multiply PyTorch Tensor by 2, in place
b.mul_(2)
# Numpy array matches new values from Tensor
a
| 0.635675 | 0.992647 |
<a href="https://colab.research.google.com/github/Iryna-Lytvynchuk/Data_Science/blob/main/fashion_mnist_12hw.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import numpy as np
import tensorflow as tf
from keras.applications.vgg16 import VGG16
from tensorflow import keras
from keras import layers
from keras import models
from keras import optimizers
from keras import losses
from keras import metrics
from keras import regularizers
from tensorflow.keras.utils import to_categorical
from keras.preprocessing.image import img_to_array, array_to_img
def change_shape(x):
x = np.reshape(x, (len(x), 28, 28, 1))
x_resized = x.copy()
x_resized.resize((len(x), 28, 28, 3), refcheck=False)
x = x_resized
x = np.asarray([img_to_array(array_to_img(im, scale=False).resize((48, 48))) for im in x])
x = x / 255.
x = x.astype('float32')
return x
fashion_mnist = keras.datasets.fashion_mnist
(x_train,y_train),(x_test,y_test) = fashion_mnist.load_data()
classes = np.unique(y_train)
num_classes = len(classes)
x_train = change_shape(x_train)
x_test = change_shape(x_test)
y_train = to_categorical(y_train)
y_test = to_categorical(y_test)
print('Train: X=%s, y=%s' % (x_train.shape, y_train.shape))
print('Test: X=%s, y=%s' % (x_test.shape, y_test.shape))
conv_base = VGG16(weights="imagenet", include_top=False, input_shape=(48, 48, 3))
conv_base.trainable = False
model = models.Sequential([
conv_base,
layers.Flatten(),
layers.Dense(256, activation="relu"),
layers.Dense(10, activation='softmax'),
])
model.compile(
loss="binary_crossentropy",
optimizer=tf.keras.optimizers.RMSprop(learning_rate=2e-5),
metrics=["acc"]
)
model.summary()
history = model.fit(x_train, y_train, batch_size=16, epochs=5,
validation_split=0.20)
results = model.evaluate(x_test, y_test)
print(results)
import matplotlib.pyplot as plt
history_dict = history.history
loss_values = history_dict['loss']
val_loss_values = history_dict['val_loss']
epochs = range(1, len(history_dict['acc']) + 1)
plt.plot(epochs, loss_values, 'bo', label='Training loss')
plt.plot(epochs, val_loss_values, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.show()
plt.clf()
val_acc_values = history_dict['val_acc']
plt.plot(epochs, history_dict['acc'], 'bo', label='Training acc')
plt.plot(epochs, history_dict['val_acc'], 'b', label='Validation acc')
plt.title('Training and validation acc')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
```
|
github_jupyter
|
import numpy as np
import tensorflow as tf
from keras.applications.vgg16 import VGG16
from tensorflow import keras
from keras import layers
from keras import models
from keras import optimizers
from keras import losses
from keras import metrics
from keras import regularizers
from tensorflow.keras.utils import to_categorical
from keras.preprocessing.image import img_to_array, array_to_img
def change_shape(x):
x = np.reshape(x, (len(x), 28, 28, 1))
x_resized = x.copy()
x_resized.resize((len(x), 28, 28, 3), refcheck=False)
x = x_resized
x = np.asarray([img_to_array(array_to_img(im, scale=False).resize((48, 48))) for im in x])
x = x / 255.
x = x.astype('float32')
return x
fashion_mnist = keras.datasets.fashion_mnist
(x_train,y_train),(x_test,y_test) = fashion_mnist.load_data()
classes = np.unique(y_train)
num_classes = len(classes)
x_train = change_shape(x_train)
x_test = change_shape(x_test)
y_train = to_categorical(y_train)
y_test = to_categorical(y_test)
print('Train: X=%s, y=%s' % (x_train.shape, y_train.shape))
print('Test: X=%s, y=%s' % (x_test.shape, y_test.shape))
conv_base = VGG16(weights="imagenet", include_top=False, input_shape=(48, 48, 3))
conv_base.trainable = False
model = models.Sequential([
conv_base,
layers.Flatten(),
layers.Dense(256, activation="relu"),
layers.Dense(10, activation='softmax'),
])
model.compile(
loss="binary_crossentropy",
optimizer=tf.keras.optimizers.RMSprop(learning_rate=2e-5),
metrics=["acc"]
)
model.summary()
history = model.fit(x_train, y_train, batch_size=16, epochs=5,
validation_split=0.20)
results = model.evaluate(x_test, y_test)
print(results)
import matplotlib.pyplot as plt
history_dict = history.history
loss_values = history_dict['loss']
val_loss_values = history_dict['val_loss']
epochs = range(1, len(history_dict['acc']) + 1)
plt.plot(epochs, loss_values, 'bo', label='Training loss')
plt.plot(epochs, val_loss_values, 'b', label='Validation loss')
plt.title('Training and validation loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.show()
plt.clf()
val_acc_values = history_dict['val_acc']
plt.plot(epochs, history_dict['acc'], 'bo', label='Training acc')
plt.plot(epochs, history_dict['val_acc'], 'b', label='Validation acc')
plt.title('Training and validation acc')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
| 0.876158 | 0.949153 |
<img src="images/usm.jpg" width="480" height="240" align="left"/>
# MAT281 - Laboratorio N°04
## Objetivos de la clase
* Reforzar los conceptos básicos de los módulos de pandas.
## Contenidos
* [Problema 01](#p1)
* [Problema 02](#p2)
## Problema 01
<img src="https://image.freepik.com/vector-gratis/varios-automoviles-dibujos-animados_23-2147613095.jpg" width="360" height="360" align="center"/>
EL conjunto de datos se denomina `Automobile_data.csv`, el cual contiene información tal como: compañia, precio, kilometraje, etc.
Lo primero es cargar el conjunto de datos y ver las primeras filas que lo componen:
```
import pandas as pd
import os
# cargar datos
df = pd.read_csv(os.path.join("data","Automobile_data.csv")).set_index('index')
df.head()
```
El objetivo es tratar de obtener la mayor información posible de este conjunto de datos. Para cumplir este objetivo debe resolver las siguientes problemáticas:
1. Elimine los valores nulos (Nan)
```
df.dropna()
```
2. Encuentra el nombre de la compañía de automóviles más cara
```
prices = df[['company','price']].groupby(['company']).max().reset_index()
prices
max_index=0
compañia_mas_cara=prices.loc[0]['company']
for i in range(1,len(prices)-1):
if prices.loc[i]['price'] >= prices.loc[max_index]['price']:
max_index=i
compañia_mas_cara=prices.loc[i]['company']
print(compañia_mas_cara)
```
3. Imprimir todos los detalles de Toyota Cars
```
df.groupby(['company']).get_group('toyota').reset_index()
```
4. Cuente el total de automóviles por compañía
```
automoviles=df.groupby(['company'])
automoviles=automoviles['price'].count().reset_index()
automoviles
```
5. Encuentra el coche con el precio más alto por compañía
```
df.groupby(['company']).max()['price']
```
6. Encuentre el kilometraje promedio (**average-mileage**) de cada compañía automotriz
```
kilometraje = df[['company','average-mileage']].groupby(['company']).mean().reset_index()
kilometraje
```
7. Ordenar todos los autos por columna de precio (**price**)
```
df.dropna().sort_values(by=['price']).reset_index()
```
## Problema 02
Siguiendo la temática de los automóviles, resuelva los siguientes problemas:
#### a) Subproblema 01
A partir de los siguientes diccionarios:
```
GermanCars = {'Company': ['Ford', 'Mercedes', 'BMV', 'Audi'],
'Price': [23845, 171995, 135925 , 71400]}
japaneseCars = {'Company': ['Toyota', 'Honda', 'Nissan', 'Mitsubishi '],
'Price': [29995, 23600, 61500 , 58900]}
```
* Cree dos dataframes (**carsDf1** y **carsDf2**) según corresponda.
* Concatene ambos dataframes (**carsDf**) y añada una llave ["Germany", "Japan"], según corresponda.
```
carsDF1= pd.DataFrame(GermanCars)
carsDF1
carsDF2= pd.DataFrame(japaneseCars)
carsDF2
carsDF = pd.concat([carsDF1, carsDF2] , keys=["Germany", "Japan"])
carsDF
```
#### b) Subproblema 02
A partir de los siguientes diccionarios:
```
Car_Price = {'Company': ['Toyota', 'Honda', 'BMV', 'Audi'], 'Price': [23845, 17995, 135925 , 71400]}
car_Horsepower = {'Company': ['Toyota', 'Honda', 'BMV', 'Audi'], 'horsepower': [141, 80, 182 , 160]}
```
* Cree dos dataframes (**carsDf1** y **carsDf2**) según corresponda.
* Junte ambos dataframes (**carsDf**) por la llave **Company**.
```
carsDf1= pd.DataFrame(Car_Price)
carsDf1
carsDf2= pd.DataFrame(car_Horsepower)
carsDf2
carsDf = pd.merge(carsDf1, carsDf2, on='Company')
carsDf
```
|
github_jupyter
|
import pandas as pd
import os
# cargar datos
df = pd.read_csv(os.path.join("data","Automobile_data.csv")).set_index('index')
df.head()
df.dropna()
prices = df[['company','price']].groupby(['company']).max().reset_index()
prices
max_index=0
compañia_mas_cara=prices.loc[0]['company']
for i in range(1,len(prices)-1):
if prices.loc[i]['price'] >= prices.loc[max_index]['price']:
max_index=i
compañia_mas_cara=prices.loc[i]['company']
print(compañia_mas_cara)
df.groupby(['company']).get_group('toyota').reset_index()
automoviles=df.groupby(['company'])
automoviles=automoviles['price'].count().reset_index()
automoviles
df.groupby(['company']).max()['price']
kilometraje = df[['company','average-mileage']].groupby(['company']).mean().reset_index()
kilometraje
df.dropna().sort_values(by=['price']).reset_index()
GermanCars = {'Company': ['Ford', 'Mercedes', 'BMV', 'Audi'],
'Price': [23845, 171995, 135925 , 71400]}
japaneseCars = {'Company': ['Toyota', 'Honda', 'Nissan', 'Mitsubishi '],
'Price': [29995, 23600, 61500 , 58900]}
carsDF1= pd.DataFrame(GermanCars)
carsDF1
carsDF2= pd.DataFrame(japaneseCars)
carsDF2
carsDF = pd.concat([carsDF1, carsDF2] , keys=["Germany", "Japan"])
carsDF
Car_Price = {'Company': ['Toyota', 'Honda', 'BMV', 'Audi'], 'Price': [23845, 17995, 135925 , 71400]}
car_Horsepower = {'Company': ['Toyota', 'Honda', 'BMV', 'Audi'], 'horsepower': [141, 80, 182 , 160]}
carsDf1= pd.DataFrame(Car_Price)
carsDf1
carsDf2= pd.DataFrame(car_Horsepower)
carsDf2
carsDf = pd.merge(carsDf1, carsDf2, on='Company')
carsDf
| 0.12097 | 0.955858 |
# python functions and methods
Function:
1. Function is block of code that is also called by its name. (independent)
2. The function can have different parameters or may not have any at all. If any data (parameters) are passed, they are passed explicitly.
3. It may or may not return any data.
4. Function does not deal with Class and its instance concept.
Method:
1. Method is called by its name, but it is associated to an object (dependent).
2. A method is implicitly passed the object on which it is invoked.
3. It may or may not return any data.
4. A method can operate on the data (instance variables) that is contained by the corresponding class
Difference between method and function:
1. Simply, function and method both look similar as they perform in almost similar way, but the key difference is the concept of ‘Class and its Object‘.
2. Functions can be called only by its name, as it is defined independently. But methods can’t be called by its name only, we need to invoke the class by a reference of that class in which it is defined, i.e. method is defined within a class and hence they are dependent on that class.
```
import types
class D:
var = None
def __init__(self, name):
self.name = name
self.msg = None
def f(self):
print("f:" + self.name)
def e(self):
print(self.name)
```
D.f 是一个 function:
```
print(D.f.__class__)
D.f
```
e 是一个 function:
```
print(type(e))
e
```
d.f 是一个 method:
```
d = D("hello")
print(d.f.__class__)
d.f()
vars(D)
```
将function赋给D的属性,等同于在类里面定义function:
```
D.e = e
print(D.e)
vars(D)
```
实例d可以调用方法e,此时,会传入self参数:
```
d.e()
```
可以看到 e 此时变成了 d 的一个绑定方法,因此调用 d.e 会传入 self
```
d.e
```
新建一个 function:
```
def g(self, msg):
self.msg = msg
print(str(self) + self.name + " " + self.msg)
```
将普通函数赋给d,不会产生绑定:
```
d.g = g
vars(d)
```
调用 g 不会传入self:
```
try:
d.g("msg")
except Exception as e:
print(e)
del d.g
```
使用 types.MethodType 将产生一个绑定到对象的method:
```
types.MethodType(g, d)
```
该绑定方法可以调用,调用时会自动传入绑定的对象当作第一个参数,此处是 d 传给 self:
```
types.MethodType(g, d)("msg")
```
注意,单独调用 types.MethodType,并不会对对象造成影响:
```
vars(d)
```
所以一般做法是把绑定方法赋值给对象:
```
d.g = types.MethodType(g, d)
vars(d)
```
此时可以调用该绑定方法:
```
d.g("msg-d")
```
**注意**
绑定方法会在内部绑定对象,该方法调用时会使用该绑定的对象作为第一个参数(self 或者 cls)
下面可以看到,d.g 和 d1.g 绑定的是同一个对象 d。
```
d1 = D("hello")
d1.g = types.MethodType(g, d)
d1.g("msg-d1")
print(d.msg)
```
也可以将绑定方法赋值给类:
```
def h(cls, data):
cls.var = data
print(str(cls) + str(cls.var))
D.h = types.MethodType(h, D)
vars(D)
```
D.h 已经将方法绑定到 D,无论是 d.h 还是 D.h 调用,绑定方法都会将已绑定的 D 传给方法的第一个参数,此处是 cls:
```
d.h(2)
D.h(3)
print(D.var)
```
采用 types.MethodType 赋值给对象的方法和在类中定义function的方法不同。
类中定义的函数(比如f)由 Function 创建,Function 类似于一个描述符,拥有一个 __get__ 方法:
```python
class Function(object):
. . .
def __get__(self, obj, objtype=None):
"Simulate func_descr_get() in Objects/funcobject.c"
if obj is None:
return self
return types.MethodType(self, obj)
```
这样,在实例调用方法时,d.f,会触发描述符协议,调用 Function 的 `__get__`,返回一个绑定方法 `types.MethodType`
## 总结:
1. 将function绑定到实例,使用MethodType,只对当前实例有效,调用时,实例会作为第一个参数传入, self 作用的对象为该绑定实例
2. 将function绑定到类,使用MethodType,调用时,类会作为第一个参数传入,cls 作用的对象为该绑定类
3. 如果需要类的所有实例都有绑定效果,则直接将function赋值给类属性,不需要使用 MethodType 方法。
|
github_jupyter
|
import types
class D:
var = None
def __init__(self, name):
self.name = name
self.msg = None
def f(self):
print("f:" + self.name)
def e(self):
print(self.name)
print(D.f.__class__)
D.f
print(type(e))
e
d = D("hello")
print(d.f.__class__)
d.f()
vars(D)
D.e = e
print(D.e)
vars(D)
d.e()
d.e
def g(self, msg):
self.msg = msg
print(str(self) + self.name + " " + self.msg)
d.g = g
vars(d)
try:
d.g("msg")
except Exception as e:
print(e)
del d.g
types.MethodType(g, d)
types.MethodType(g, d)("msg")
vars(d)
d.g = types.MethodType(g, d)
vars(d)
d.g("msg-d")
d1 = D("hello")
d1.g = types.MethodType(g, d)
d1.g("msg-d1")
print(d.msg)
def h(cls, data):
cls.var = data
print(str(cls) + str(cls.var))
D.h = types.MethodType(h, D)
vars(D)
d.h(2)
D.h(3)
print(D.var)
class Function(object):
. . .
def __get__(self, obj, objtype=None):
"Simulate func_descr_get() in Objects/funcobject.c"
if obj is None:
return self
return types.MethodType(self, obj)
| 0.184951 | 0.92976 |
```
import pandas as pd
import os
import logging
import sys
```
### Load environment vars and directories
```
KEY_DIR = os.path.join(os.getenv("DOCUMENTS"), "govuk-network-data", "key")
KEY_PATH = os.path.join(KEY_DIR, os.listdir(KEY_DIR)[0])
PROJECT_ID = "govuk-bigquery-analytics"
```
#### Logging for `pandas_gbq`
```
logger = logging.getLogger('pandas_gbq')
logger.setLevel(logging.DEBUG)
logger.addHandler(logging.StreamHandler(stream=sys.stdout))
```
### Extract page-hit only user journeys for February 11 to 18
8.8 GB
```
query = """SELECT
COUNT(*) AS Occurrences,
PageSeq_Length,
PageSequence
FROM (
SELECT
*
FROM (
SELECT
CONCAT(fullVisitorId,"-",CAST(visitId AS STRING),"-",CAST(visitNumber AS STRING)) AS sessionId,
STRING_AGG(IF(htype = 'PAGE',
pagePath,
NULL),">>") OVER (PARTITION BY fullVisitorId, visitId, visitStartTime ORDER BY hitNumber ROWS BETWEEN UNBOUNDED PRECEDING AND UNBOUNDED FOLLOWING ) AS PageSequence,
SUM(IF(htype='PAGE',
1,
0)) OVER (PARTITION BY fullVisitorId, visitId, visitStartTime ORDER BY hitNumber ROWS BETWEEN UNBOUNDED PRECEDING AND UNBOUNDED FOLLOWING ) AS PageSeq_Length
FROM (
SELECT
fullVisitorId,
visitId,
visitNumber,
visitStartTime,
hits.page.pagePath AS pagePath,
hits.hitNumber AS hitNumber,
hits.type AS htype
FROM
`govuk-bigquery-analytics.87773428.ga_sessions_*` AS sessions
CROSS JOIN
UNNEST(sessions.hits) AS hits
WHERE _TABLE_SUFFIX BETWEEN '20190110' AND '20190128' ) )
WHERE
PageSeq_Length >1
GROUP BY
sessionId,
PageSequence,
PageSeq_Length)
GROUP BY
PageSequence,
PageSeq_Length"""
```
### Extract data from BigQuery
```
df_in = pd.read_gbq(query,
project_id=PROJECT_ID,
reauth=False,
private_key=KEY_PATH,
dialect="standard")
df_in.shape
df_in.head()
```
### Explore occurrences stats
```
df_in.Occurrences.describe()
all_occ = df_in.Occurrences.sum()
num_one_off_journeys = df_in[df_in.Occurrences==1].shape[0]
one_off_occ = df_in[df_in.Occurrences==1].Occurrences.sum()
all_occ, num_one_off_journeys, one_off_occ
df_in.sort_values("Occurrences", ascending=False).head()
```
### Add `Page_List` column
```
pagelist = [pageseq.split(">>") for pageseq in df_in['PageSequence'].values]
df_in['Page_List'] = pagelist
df_in['Page_List'].head()
page_views = {}
for tup in df_in.itertuples():
for p in tup.Page_List:
if p in page_views.keys():
page_views[p] += tup.Occurrences
else:
page_views[p] = tup.Occurrences
len(page_views), sum(page_views.values())
```
### Save out
```
date_range = "jan_10_28"
bq_dir = os.path.join(os.getenv("DATA_DIR"),"raw", "bq_journey_extract")
bq_file = os.path.join(bq_dir, "pageseq_user_journey_"+date_range+".csv.gz")
bq_file_doo = os.path.join(bq_dir, "pageseq_user_journey_"+date_range+"_doo.csv.gz")
page_views_file = os.path.join(bq_dir, "pageviews_"+date_range+".csv.gz")
df_in.to_csv(bq_file, compression="gzip", sep='\t', index=False)
df_in[df_in.Occurrences>1].to_csv(bq_file_doo, compression="gzip", sep='\t', index=False)
import gzip
with gzip.open(page_views_file, "wb") as writer:
writer.write("page_url\tviews\n".encode())
for key,value in page_views.items():
writer.write("{}\t{}\n".format(key,value).encode())
```
|
github_jupyter
|
import pandas as pd
import os
import logging
import sys
KEY_DIR = os.path.join(os.getenv("DOCUMENTS"), "govuk-network-data", "key")
KEY_PATH = os.path.join(KEY_DIR, os.listdir(KEY_DIR)[0])
PROJECT_ID = "govuk-bigquery-analytics"
logger = logging.getLogger('pandas_gbq')
logger.setLevel(logging.DEBUG)
logger.addHandler(logging.StreamHandler(stream=sys.stdout))
query = """SELECT
COUNT(*) AS Occurrences,
PageSeq_Length,
PageSequence
FROM (
SELECT
*
FROM (
SELECT
CONCAT(fullVisitorId,"-",CAST(visitId AS STRING),"-",CAST(visitNumber AS STRING)) AS sessionId,
STRING_AGG(IF(htype = 'PAGE',
pagePath,
NULL),">>") OVER (PARTITION BY fullVisitorId, visitId, visitStartTime ORDER BY hitNumber ROWS BETWEEN UNBOUNDED PRECEDING AND UNBOUNDED FOLLOWING ) AS PageSequence,
SUM(IF(htype='PAGE',
1,
0)) OVER (PARTITION BY fullVisitorId, visitId, visitStartTime ORDER BY hitNumber ROWS BETWEEN UNBOUNDED PRECEDING AND UNBOUNDED FOLLOWING ) AS PageSeq_Length
FROM (
SELECT
fullVisitorId,
visitId,
visitNumber,
visitStartTime,
hits.page.pagePath AS pagePath,
hits.hitNumber AS hitNumber,
hits.type AS htype
FROM
`govuk-bigquery-analytics.87773428.ga_sessions_*` AS sessions
CROSS JOIN
UNNEST(sessions.hits) AS hits
WHERE _TABLE_SUFFIX BETWEEN '20190110' AND '20190128' ) )
WHERE
PageSeq_Length >1
GROUP BY
sessionId,
PageSequence,
PageSeq_Length)
GROUP BY
PageSequence,
PageSeq_Length"""
df_in = pd.read_gbq(query,
project_id=PROJECT_ID,
reauth=False,
private_key=KEY_PATH,
dialect="standard")
df_in.shape
df_in.head()
df_in.Occurrences.describe()
all_occ = df_in.Occurrences.sum()
num_one_off_journeys = df_in[df_in.Occurrences==1].shape[0]
one_off_occ = df_in[df_in.Occurrences==1].Occurrences.sum()
all_occ, num_one_off_journeys, one_off_occ
df_in.sort_values("Occurrences", ascending=False).head()
pagelist = [pageseq.split(">>") for pageseq in df_in['PageSequence'].values]
df_in['Page_List'] = pagelist
df_in['Page_List'].head()
page_views = {}
for tup in df_in.itertuples():
for p in tup.Page_List:
if p in page_views.keys():
page_views[p] += tup.Occurrences
else:
page_views[p] = tup.Occurrences
len(page_views), sum(page_views.values())
date_range = "jan_10_28"
bq_dir = os.path.join(os.getenv("DATA_DIR"),"raw", "bq_journey_extract")
bq_file = os.path.join(bq_dir, "pageseq_user_journey_"+date_range+".csv.gz")
bq_file_doo = os.path.join(bq_dir, "pageseq_user_journey_"+date_range+"_doo.csv.gz")
page_views_file = os.path.join(bq_dir, "pageviews_"+date_range+".csv.gz")
df_in.to_csv(bq_file, compression="gzip", sep='\t', index=False)
df_in[df_in.Occurrences>1].to_csv(bq_file_doo, compression="gzip", sep='\t', index=False)
import gzip
with gzip.open(page_views_file, "wb") as writer:
writer.write("page_url\tviews\n".encode())
for key,value in page_views.items():
writer.write("{}\t{}\n".format(key,value).encode())
| 0.119267 | 0.312849 |
```
import os
import pandas as pd
from pathlib import Path
import requests
from dotenv import load_dotenv, find_dotenv
from src.data.cosmos import GremlinQueryManager, DocumentQueryManager
from src.data.graph.gremlin import GremlinQueryBuilder
load_dotenv(find_dotenv())
account_name = os.environ.get('COSMOS_ACCOUNT_NAME')
db_name = os.environ.get('COSMOS_DB_NAME')
graph_name = os.environ.get('COSMOS_GRAPH_NAME')
master_key = os.environ.get('COSMOS_MASTER_KEY')
search_key = os.environ.get('AZURE_SEARCH_KEY')
search_account_name = os.environ.get('AZURE_SEARCH_ACCOUNT_NAME')
gremlin_qm = GremlinQueryManager(account_name, master_key, db_name, graph_name)
doc_qm = DocumentQueryManager(account_name, master_key, db_name)
services_data = []
abbrs = gremlin_qm.query('g.V().has("label", "cloud").values("abbreviation")')
for abbr in abbrs:
print(abbr)
# abbr = 'azure'
q = f"""g.V().has("label", "{abbr}_service")
.project("id", "name", "shortDescription", "longDescription", "uri", "iconUri", "categories", "relatedServices", "cloud")
.by("id").by("name").by("short_description").by("long_description").by("uri").by("icon_uri")
.by(out("belongs_to").values("name").fold())
.by(coalesce(out("related_service").id().fold(), __.not(identity()).fold()))
.by(out("belongs_to").out("source_cloud").values("name"))"""
cloud_data = gremlin_qm.query(q)
services_data += cloud_data
len(services_data)
[s for s in services_data if s['name'] == 'AWS Lambda']
import requests
import requests_cache
from datetime import timedelta
class AzureSearchClient:
def __init__(self, account_name, api_key, index_name):
self.account_name = account_name
self.api_key = api_key
self.index_name = index_name
self.default_headers = {
'api-key': api_key
}
def search(self, search_term, userSkills=None, k=10):
search_url = f'https://{self.account_name}.search.windows.net/indexes/{self.index_name}/docs'
params = {
'api-version': '2017-11-11-preview',
'search': search_term,
'$top': k
}
if userSkills:
params['scoringProfile'] = 'skills'
params['scoringParameter'] = f'skills-{userSkills}'
res = requests.get(search_url, headers=self.default_headers, params=params)
return res
def suggest(self, search_term):
search_url = f'https://{self.account_name}.search.windows.net/indexes/{self.index_name}/docs/suggest'
params = {
'api-version': '2017-11-11-preview',
'search': search_term,
'$top': 3,
'scoringProfile': 'boostName',
'autocompleteMode': 'twoTerms',
'suggesterName': 'suggest-name',
'fuzzy': True
}
res = requests.get(search_url, headers=self.default_headers, params=params)
return res
def upsert_index(self, fields_config, suggesters, scoring_profiles):
kwargs = {
'headers': self.default_headers,
'json': {
'name': self.index_name,
'fields': fields_config,
'suggesters': suggesters,
'scoringProfiles': scoring_profiles
}
}
delete_res = requests.delete(f"https://{self.account_name}.search.windows.net/indexes/{self.index_name}?api-version=2017-11-11", **kwargs)
if delete_res.status_code > 299:
print('Failed delete index')
res = requests.post(
f"https://{self.account_name}.search.windows.net/indexes/?api-version=2017-11-11",
**kwargs
)
return res
def upload_data(self, data):
for i in range(len(data)):
data[i]['@search.action'] = 'mergeOrUpload'
res = requests.post(
f"https://{self.account_name}.search.windows.net/indexes/{self.index_name}/docs/index?api-version=2017-11-11",
headers=self.default_headers,
json={
'value': data
}
)
return res
def upsert_synonym_map(self, name, synonyms):
kwargs = {
'headers': self.default_headers,
'json': {
'name': name,
'format': 'solr',
'synonyms': synonyms
}
}
res = requests.post(
f"https://{self.account_name}.search.windows.net/synonymmaps?api-version=2017-11-11",
**kwargs
)
if res.status_code > 299:
res = requests.put(
f"https://{self.account_name}.search.windows.net/synonymmaps/{name}?api-version=2017-11-11",
**kwargs
)
return res
search_client = AzureSearchClient(search_account_name, search_key, 'services')
azure_synonyms = """
AD, Active Directory, AAD\n
AKS, Azure Kubernetes Service\n,
function, functions
database, databases
"""
search_client.upsert_synonym_map('azure-service-abbreviations', azure_synonyms).text
services_v0_field_config = [
{"name": "id", "type": "Edm.String", "key": True, "searchable": False, "sortable": False, "facetable": False},
{"name": "name", "type": "Edm.String", "synonymMaps":["azure-service-abbreviations"]},
{"name": "shortDescription", "type": "Edm.String", "filterable": False, "sortable": False, "facetable": False},
{"name": "longDescription", "type": "Edm.String", "filterable": False, "sortable": False, "facetable": False},
{"name": "uri", "type": "Edm.String", "facetable": False},
{"name": "iconUri", "type": "Edm.String", "facetable": False},
{"name": "categories", "type": "Collection(Edm.String)"},
{"name": "relatedServices", "type": "Collection(Edm.String)", "searchable": False, "filterable": False, "sortable": False, "facetable": False},
{"name": "cloud", "type": "Edm.String", "searchable": False, "sortable": False}
]
suggesters = [
{
"name": "suggest-name",
"searchMode": "analyzingInfixMatching",
"sourceFields": ["name"]
}
]
scoring_profiles = [
{
"name": "boostName",
"text": {
"weights": {
"name": 3
}
}
}
]
r = search_client.upsert_index(services_v0_field_config, suggesters, scoring_profiles)
r
upload_res = search_client.upload_data(services_data)
upload_res.status_code
gremlin_qm.query('g.V("7397ee26-10f0-40a7-9e9f-393a53686e42").in("source_cloud")')
aws_azure_df = pd.read_csv('../data/processed/aws_azure_data_matching_output.csv')
aws_azure_related = aws_azure_df[aws_azure_df['Link Score'] > 0.6].sort_values(['Cluster ID'])
def get_svc_id(gremlin_qm, svc_name):
q = f'g.V().has("name", "{svc_name}").values("id")'
res = gremlin_qm.query(q)
return res[0]
def build_related_query(from_id, to_id):
return GremlinQueryBuilder.build_upsert_edge_query(from_id, to_id, {
'label': 'related_service', 'related_service_score': aws_svc['Link Score']
})
for i in range(list(aws_azure_related['Cluster ID'])[-1] + 1):
related_services = aws_azure_related[aws_azure_related['Cluster ID'] == i].reset_index(drop=True)
aws_svc = related_services.iloc[0]
azure_svc = related_services.iloc[1]
aws_id = get_svc_id(gremlin_qm, aws_svc['name'])
azure_id = get_svc_id(gremlin_qm, azure_svc['name'])
print(f'Adding related_service edges between {aws_svc["name"]} and {azure_svc["name"]}')
aws_azure_related.head()
azure_gcp_df = pd.read_csv('../data/processed/azure_gcp_data_matching_output.csv')
azure_gcp_related = azure_gcp_df[azure_gcp_df['Link Score'] > 0.6].sort_values(['Cluster ID'])
def get_svc_id(gremlin_qm, svc_name):
q = f'g.V().has("name", "{svc_name}").values("id")'
return gremlin_qm.query(q)[0]
def build_related_query(from_id, to_id, score):
return GremlinQueryBuilder.build_upsert_edge_query(from_id, to_id, {
'label': 'related_service', 'related_service_score': score
})
def fix_name(row):
name = row['name']
if row['source file'] == 1:
ids = ['Google', 'GCP', 'GKE', 'Firebase', 'Apigee']
no_id = all([i not in name for i in ids])
if no_id:
name = f'Google {name}'
return name
azure_gcp_related['name'] = azure_gcp_related.apply(fix_name, axis=1)
azure_gcp_related
azure_gcp_related.to_csv('../data/processed/azure_gcp_data_matching_output.csv', index=False)
azure_ibm_related = pd.read_csv('../data/processed/azure_ibm_dedupe/data_matching_output.csv')
azure_ibm_related[azure_ibm_related['name'].str.startswith('Azure Kubernetes')]
azure_ibm_related = azure_ibm_related[azure_ibm_related['Link Score'] > 0.688].sort_values(['Cluster ID', 'source file'])
azure_ibm_related
def update_related_services(prodigy_data_matching_output_df):
for i in range(list(prodigy_data_matching_output_df['Cluster ID'])[-1] + 1):
related_services = prodigy_data_matching_output_df[prodigy_data_matching_output_df['Cluster ID'] == i].reset_index(drop=True)
left_svc = related_services[related_services['source file'] == 0].iloc[0]
right_svc = related_services[related_services['source file'] == 1].iloc[0]
left_id = get_svc_id(gremlin_qm, left_svc['name'])
right_id = get_svc_id(gremlin_qm, right_svc['name'])
left_related_edges = gremlin_qm.query(f"g.V('{left_id}').outE('related_service')")
for rel_svc in left_related_edges:
if rel_svc['inV'] != right_id:
score = rel_svc['properties']['related_service_score']
gremlin_qm.query(build_related_query(rel_svc['inV'], right_id, score))
gremlin_qm.query(build_related_query(right_id, rel_svc['inV'], score))
print(f'Adding related_service edges between {left_svc["name"]} and {right_svc["name"]}')
gremlin_qm.query(build_related_query(left_id, right_id, left_svc['Link Score']))
gremlin_qm.query(build_related_query(right_id, left_id, left_svc['Link Score']))
update_related_services(azure_ibm_related)
```
|
github_jupyter
|
import os
import pandas as pd
from pathlib import Path
import requests
from dotenv import load_dotenv, find_dotenv
from src.data.cosmos import GremlinQueryManager, DocumentQueryManager
from src.data.graph.gremlin import GremlinQueryBuilder
load_dotenv(find_dotenv())
account_name = os.environ.get('COSMOS_ACCOUNT_NAME')
db_name = os.environ.get('COSMOS_DB_NAME')
graph_name = os.environ.get('COSMOS_GRAPH_NAME')
master_key = os.environ.get('COSMOS_MASTER_KEY')
search_key = os.environ.get('AZURE_SEARCH_KEY')
search_account_name = os.environ.get('AZURE_SEARCH_ACCOUNT_NAME')
gremlin_qm = GremlinQueryManager(account_name, master_key, db_name, graph_name)
doc_qm = DocumentQueryManager(account_name, master_key, db_name)
services_data = []
abbrs = gremlin_qm.query('g.V().has("label", "cloud").values("abbreviation")')
for abbr in abbrs:
print(abbr)
# abbr = 'azure'
q = f"""g.V().has("label", "{abbr}_service")
.project("id", "name", "shortDescription", "longDescription", "uri", "iconUri", "categories", "relatedServices", "cloud")
.by("id").by("name").by("short_description").by("long_description").by("uri").by("icon_uri")
.by(out("belongs_to").values("name").fold())
.by(coalesce(out("related_service").id().fold(), __.not(identity()).fold()))
.by(out("belongs_to").out("source_cloud").values("name"))"""
cloud_data = gremlin_qm.query(q)
services_data += cloud_data
len(services_data)
[s for s in services_data if s['name'] == 'AWS Lambda']
import requests
import requests_cache
from datetime import timedelta
class AzureSearchClient:
def __init__(self, account_name, api_key, index_name):
self.account_name = account_name
self.api_key = api_key
self.index_name = index_name
self.default_headers = {
'api-key': api_key
}
def search(self, search_term, userSkills=None, k=10):
search_url = f'https://{self.account_name}.search.windows.net/indexes/{self.index_name}/docs'
params = {
'api-version': '2017-11-11-preview',
'search': search_term,
'$top': k
}
if userSkills:
params['scoringProfile'] = 'skills'
params['scoringParameter'] = f'skills-{userSkills}'
res = requests.get(search_url, headers=self.default_headers, params=params)
return res
def suggest(self, search_term):
search_url = f'https://{self.account_name}.search.windows.net/indexes/{self.index_name}/docs/suggest'
params = {
'api-version': '2017-11-11-preview',
'search': search_term,
'$top': 3,
'scoringProfile': 'boostName',
'autocompleteMode': 'twoTerms',
'suggesterName': 'suggest-name',
'fuzzy': True
}
res = requests.get(search_url, headers=self.default_headers, params=params)
return res
def upsert_index(self, fields_config, suggesters, scoring_profiles):
kwargs = {
'headers': self.default_headers,
'json': {
'name': self.index_name,
'fields': fields_config,
'suggesters': suggesters,
'scoringProfiles': scoring_profiles
}
}
delete_res = requests.delete(f"https://{self.account_name}.search.windows.net/indexes/{self.index_name}?api-version=2017-11-11", **kwargs)
if delete_res.status_code > 299:
print('Failed delete index')
res = requests.post(
f"https://{self.account_name}.search.windows.net/indexes/?api-version=2017-11-11",
**kwargs
)
return res
def upload_data(self, data):
for i in range(len(data)):
data[i]['@search.action'] = 'mergeOrUpload'
res = requests.post(
f"https://{self.account_name}.search.windows.net/indexes/{self.index_name}/docs/index?api-version=2017-11-11",
headers=self.default_headers,
json={
'value': data
}
)
return res
def upsert_synonym_map(self, name, synonyms):
kwargs = {
'headers': self.default_headers,
'json': {
'name': name,
'format': 'solr',
'synonyms': synonyms
}
}
res = requests.post(
f"https://{self.account_name}.search.windows.net/synonymmaps?api-version=2017-11-11",
**kwargs
)
if res.status_code > 299:
res = requests.put(
f"https://{self.account_name}.search.windows.net/synonymmaps/{name}?api-version=2017-11-11",
**kwargs
)
return res
search_client = AzureSearchClient(search_account_name, search_key, 'services')
azure_synonyms = """
AD, Active Directory, AAD\n
AKS, Azure Kubernetes Service\n,
function, functions
database, databases
"""
search_client.upsert_synonym_map('azure-service-abbreviations', azure_synonyms).text
services_v0_field_config = [
{"name": "id", "type": "Edm.String", "key": True, "searchable": False, "sortable": False, "facetable": False},
{"name": "name", "type": "Edm.String", "synonymMaps":["azure-service-abbreviations"]},
{"name": "shortDescription", "type": "Edm.String", "filterable": False, "sortable": False, "facetable": False},
{"name": "longDescription", "type": "Edm.String", "filterable": False, "sortable": False, "facetable": False},
{"name": "uri", "type": "Edm.String", "facetable": False},
{"name": "iconUri", "type": "Edm.String", "facetable": False},
{"name": "categories", "type": "Collection(Edm.String)"},
{"name": "relatedServices", "type": "Collection(Edm.String)", "searchable": False, "filterable": False, "sortable": False, "facetable": False},
{"name": "cloud", "type": "Edm.String", "searchable": False, "sortable": False}
]
suggesters = [
{
"name": "suggest-name",
"searchMode": "analyzingInfixMatching",
"sourceFields": ["name"]
}
]
scoring_profiles = [
{
"name": "boostName",
"text": {
"weights": {
"name": 3
}
}
}
]
r = search_client.upsert_index(services_v0_field_config, suggesters, scoring_profiles)
r
upload_res = search_client.upload_data(services_data)
upload_res.status_code
gremlin_qm.query('g.V("7397ee26-10f0-40a7-9e9f-393a53686e42").in("source_cloud")')
aws_azure_df = pd.read_csv('../data/processed/aws_azure_data_matching_output.csv')
aws_azure_related = aws_azure_df[aws_azure_df['Link Score'] > 0.6].sort_values(['Cluster ID'])
def get_svc_id(gremlin_qm, svc_name):
q = f'g.V().has("name", "{svc_name}").values("id")'
res = gremlin_qm.query(q)
return res[0]
def build_related_query(from_id, to_id):
return GremlinQueryBuilder.build_upsert_edge_query(from_id, to_id, {
'label': 'related_service', 'related_service_score': aws_svc['Link Score']
})
for i in range(list(aws_azure_related['Cluster ID'])[-1] + 1):
related_services = aws_azure_related[aws_azure_related['Cluster ID'] == i].reset_index(drop=True)
aws_svc = related_services.iloc[0]
azure_svc = related_services.iloc[1]
aws_id = get_svc_id(gremlin_qm, aws_svc['name'])
azure_id = get_svc_id(gremlin_qm, azure_svc['name'])
print(f'Adding related_service edges between {aws_svc["name"]} and {azure_svc["name"]}')
aws_azure_related.head()
azure_gcp_df = pd.read_csv('../data/processed/azure_gcp_data_matching_output.csv')
azure_gcp_related = azure_gcp_df[azure_gcp_df['Link Score'] > 0.6].sort_values(['Cluster ID'])
def get_svc_id(gremlin_qm, svc_name):
q = f'g.V().has("name", "{svc_name}").values("id")'
return gremlin_qm.query(q)[0]
def build_related_query(from_id, to_id, score):
return GremlinQueryBuilder.build_upsert_edge_query(from_id, to_id, {
'label': 'related_service', 'related_service_score': score
})
def fix_name(row):
name = row['name']
if row['source file'] == 1:
ids = ['Google', 'GCP', 'GKE', 'Firebase', 'Apigee']
no_id = all([i not in name for i in ids])
if no_id:
name = f'Google {name}'
return name
azure_gcp_related['name'] = azure_gcp_related.apply(fix_name, axis=1)
azure_gcp_related
azure_gcp_related.to_csv('../data/processed/azure_gcp_data_matching_output.csv', index=False)
azure_ibm_related = pd.read_csv('../data/processed/azure_ibm_dedupe/data_matching_output.csv')
azure_ibm_related[azure_ibm_related['name'].str.startswith('Azure Kubernetes')]
azure_ibm_related = azure_ibm_related[azure_ibm_related['Link Score'] > 0.688].sort_values(['Cluster ID', 'source file'])
azure_ibm_related
def update_related_services(prodigy_data_matching_output_df):
for i in range(list(prodigy_data_matching_output_df['Cluster ID'])[-1] + 1):
related_services = prodigy_data_matching_output_df[prodigy_data_matching_output_df['Cluster ID'] == i].reset_index(drop=True)
left_svc = related_services[related_services['source file'] == 0].iloc[0]
right_svc = related_services[related_services['source file'] == 1].iloc[0]
left_id = get_svc_id(gremlin_qm, left_svc['name'])
right_id = get_svc_id(gremlin_qm, right_svc['name'])
left_related_edges = gremlin_qm.query(f"g.V('{left_id}').outE('related_service')")
for rel_svc in left_related_edges:
if rel_svc['inV'] != right_id:
score = rel_svc['properties']['related_service_score']
gremlin_qm.query(build_related_query(rel_svc['inV'], right_id, score))
gremlin_qm.query(build_related_query(right_id, rel_svc['inV'], score))
print(f'Adding related_service edges between {left_svc["name"]} and {right_svc["name"]}')
gremlin_qm.query(build_related_query(left_id, right_id, left_svc['Link Score']))
gremlin_qm.query(build_related_query(right_id, left_id, left_svc['Link Score']))
update_related_services(azure_ibm_related)
| 0.417865 | 0.14731 |
### In this notebook I will look at the full Bdot measurements of the 25cm plane.
I will also check if taking the curl of the magnetic field recovers a current centered at the center of the circulation in the b field. I will test schemes for locating the current centroid.
## Imports
```
import numpy as np
import scipy.optimize as opti
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style('whitegrid')
sns.set_context('poster')
from mpl_toolkits.axes_grid1 import make_axes_locatable
from matplotlib.tri import Triangulation, LinearTriInterpolator
from scipy.optimize import curve_fit
from scipy import odr
from scipy.optimize import leastsq
from skimage.feature import peak_local_max
import scipy.ndimage as ndimage
import cPickle as pickle
from scipy.integrate import odeint
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:100% !important; }</style>"))
import sys
sys.path.append('/Users/vonderlinden2/rsx_analysis/writing_to_vtk/source')
import structured_3d_vtk as struc_3d
```
## Define convience function and load magnetic field data.
```
def read_and_return_vector_and_grid(file_path, quantity_name, grid_name):
r"""
"""
array_dict = np.load(file_path)
quantity = array_dict[quantity_name]
grid = array_dict[grid_name]
return quantity, grid
file_path = '../../output/centroid_fitting/2016-08-12/B_z0249_'
ending = '.npz'
b_fields = []
for time_point in xrange(21):
name = file_path + str(time_point).zfill(4) + ending
b_field, grid = read_and_return_vector_and_grid(name, 'b', 'grid')
b_fields.append(b_field)
grid[1][-1]
```
# Method 2: Integrate field lines off the max and fit circle to the field lines
## Try it out by hand
```
def to_min(params, points):
a = 2.*params[0]
b = 2.*params[1]
c = params[2]**2 - params[1]**2 - params[0]**2
return a*points[0] + b*points[1] + c - points[0]**2 - points[1]**2
def calculate_radius(x, y, x_0, y_0):
r"""
Calculate radius.
"""
return (x - x_0)**2 + (y - y_0)**2
def estimate_radius(data, x_0, y_0):
r"""
Estimate radius from data.
"""
return calculate_radius(data[:, 0], data[:, 1], x_0, y_0).mean()
params_guess = [0, 0, 0.01]
def d_l(l, t, interpolator_x, interpolator_y):
return np.asarray([interpolator_x([l[0], l[1]])[0],
interpolator_y([l[0], l[1]])[0]])
def find_centroid(grid, bx_interpolator, by_interpolator,
distance_thres=0.001, filter_size=5,
integration_length=10, integration_steps=100,
launch_point_step_factor=0.1, max_count=50):
r"""
"""
b_fields_x = bx_interpolator(grid[0][:, :], grid[1][:, :])
b_fields_y = by_interpolator(grid[0][:, :], grid[1][:, :])
b_fields = [b_fields_x, b_fields_y]
x_min, x_max = grid[0].min(), grid[0].max()
y_min, y_max = grid[1].min(), grid[1].max()
magnitude = np.sqrt(b_fields[0][:, :]**2 + b_fields[1][:, :]**2)
filtered_magnitude = ndimage.gaussian_filter(magnitude, filter_size)
max_index = np.unravel_index(filtered_magnitude.argmax(),
filtered_magnitude.shape)
center_points = []
radii = []
center_points = []
streamlines = []
direction = [0, 0]
distance = 100
launch_point = (grid[0][max_index], grid[1][max_index])
count = 0
while distance >= distance_thres:
#print 'launch', launch_point
#print distance
t2 = np.linspace(0, integration_length, integration_steps)
t1 = np.linspace(0, -integration_length, integration_steps)
stream2 = odeint(d_l, launch_point, t2, args=(bx_interpolator, by_interpolator))
stream1 = odeint(d_l, launch_point, t1, args=(bx_interpolator, by_interpolator))
print 'stream', stream1, stream2
streamline = np.concatenate((stream1, stream2))
size = streamline[np.invert(np.isnan(streamline))].size
streamline = streamline[np.invert(np.isnan(streamline))].reshape(int(size/2.), 2)
circle_params, success = leastsq(to_min, params_guess, args=np.asarray([streamline[:, 0], streamline[:, 1]]))
direction = [circle_params[0] - launch_point[0], circle_params[1] - launch_point[1]]
distance = np.sqrt(direction[0]**2. + direction[1]**2.)
center_point = (circle_params[0], circle_params[1])
launch_point = [launch_point[0] + direction[0] * launch_point_step_factor,
launch_point[1] + direction[1] * launch_point_step_factor]
center_points.append(center_point)
#print 'center', center_point
radii.append(circle_params[0])
streamlines.append(streamline)
if (launch_point[0] <= x_min or
launch_point[0] >= x_max or
launch_point[1] <= y_min or
launch_point[1] >= y_max or
count > max_count):
break
count += 1
centroid = center_point
return centroid, center_points, radii, streamlines, max_index
centroids = []
for time_point in xrange(1):
print time_point
bx_interpolator = pickle.load(open('../../output/centroid_fitting/2016-08-12/B_z0249_x_' + str(time_point).zfill(4) + '.p', 'rb'))
by_interpolator = pickle.load(open('../../output/centroid_fitting/2016-08-12/B_z0249_y_' + str(time_point).zfill(4) + '.p', 'rb'))
(centroid, center_points,
radii, streamlines,
max_index) = find_centroid(grid,
bx_interpolator,
by_interpolator,
launch_point_step_factor=0.05,
integration_length=20)
centroids.append(centroid)
from datetime import datetime
import os
today = datetime.today()
today = today.strftime('%Y-%m-%d')
out_dir = '../output/' + today
try:
os.makedirs(out_dir)
except:
pass
centroids = np.asarray(centroids)
np.savetxt(out_dir + '/field_nulls.txt', centroids, header='magnetic field null positions in the z=0.249m plane,' +
'determined by fitting circles to integrated field lines starting at max magnitude and moving succesive' +
'towards the center of circles.')
from scipy.integrate import dblquad
def integrate_flux(centroid, radius, bz_interpolator, limits):
if (centroid[0] - radius < limits[0] or centroid[0] + radius > limits[1] or
centroid[1] - radius < limits[2] or centroid[1] + radius > limits[3]):
return -1
gfun = lambda x: -np.sqrt(radius**2 - (x-centroid[0])**2)
hfun = lambda x: np.sqrt(radius**2 - (x-centroid[0])**2)
bz_interpolator_bias = lambda x, y: bz_interpolator(x, y) + 0.02
return dblquad(bz_interpolator_bias, centroid[0] - radius, centroid[0] + radius, gfun, hfun)
bz_interpolator = pickle.load(open('../output/2016-08-12/B_z0249_z_' + str(0).zfill(4) + '.p', 'rb'))
bz_interpolator(centroids[0])
for time_point in xrange(250):
bz_interpolator = pickle.load(open('../output/2016-08-12/B_z0249_z_' + str(time_point).zfill(4) + '.p', 'rb'))
print integrate_flux(centroids[time_point], 0.001, bz_interpolator,
(-0.028, 0.025, -0.043, 0.039))
```
|
github_jupyter
|
import numpy as np
import scipy.optimize as opti
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style('whitegrid')
sns.set_context('poster')
from mpl_toolkits.axes_grid1 import make_axes_locatable
from matplotlib.tri import Triangulation, LinearTriInterpolator
from scipy.optimize import curve_fit
from scipy import odr
from scipy.optimize import leastsq
from skimage.feature import peak_local_max
import scipy.ndimage as ndimage
import cPickle as pickle
from scipy.integrate import odeint
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:100% !important; }</style>"))
import sys
sys.path.append('/Users/vonderlinden2/rsx_analysis/writing_to_vtk/source')
import structured_3d_vtk as struc_3d
def read_and_return_vector_and_grid(file_path, quantity_name, grid_name):
r"""
"""
array_dict = np.load(file_path)
quantity = array_dict[quantity_name]
grid = array_dict[grid_name]
return quantity, grid
file_path = '../../output/centroid_fitting/2016-08-12/B_z0249_'
ending = '.npz'
b_fields = []
for time_point in xrange(21):
name = file_path + str(time_point).zfill(4) + ending
b_field, grid = read_and_return_vector_and_grid(name, 'b', 'grid')
b_fields.append(b_field)
grid[1][-1]
def to_min(params, points):
a = 2.*params[0]
b = 2.*params[1]
c = params[2]**2 - params[1]**2 - params[0]**2
return a*points[0] + b*points[1] + c - points[0]**2 - points[1]**2
def calculate_radius(x, y, x_0, y_0):
r"""
Calculate radius.
"""
return (x - x_0)**2 + (y - y_0)**2
def estimate_radius(data, x_0, y_0):
r"""
Estimate radius from data.
"""
return calculate_radius(data[:, 0], data[:, 1], x_0, y_0).mean()
params_guess = [0, 0, 0.01]
def d_l(l, t, interpolator_x, interpolator_y):
return np.asarray([interpolator_x([l[0], l[1]])[0],
interpolator_y([l[0], l[1]])[0]])
def find_centroid(grid, bx_interpolator, by_interpolator,
distance_thres=0.001, filter_size=5,
integration_length=10, integration_steps=100,
launch_point_step_factor=0.1, max_count=50):
r"""
"""
b_fields_x = bx_interpolator(grid[0][:, :], grid[1][:, :])
b_fields_y = by_interpolator(grid[0][:, :], grid[1][:, :])
b_fields = [b_fields_x, b_fields_y]
x_min, x_max = grid[0].min(), grid[0].max()
y_min, y_max = grid[1].min(), grid[1].max()
magnitude = np.sqrt(b_fields[0][:, :]**2 + b_fields[1][:, :]**2)
filtered_magnitude = ndimage.gaussian_filter(magnitude, filter_size)
max_index = np.unravel_index(filtered_magnitude.argmax(),
filtered_magnitude.shape)
center_points = []
radii = []
center_points = []
streamlines = []
direction = [0, 0]
distance = 100
launch_point = (grid[0][max_index], grid[1][max_index])
count = 0
while distance >= distance_thres:
#print 'launch', launch_point
#print distance
t2 = np.linspace(0, integration_length, integration_steps)
t1 = np.linspace(0, -integration_length, integration_steps)
stream2 = odeint(d_l, launch_point, t2, args=(bx_interpolator, by_interpolator))
stream1 = odeint(d_l, launch_point, t1, args=(bx_interpolator, by_interpolator))
print 'stream', stream1, stream2
streamline = np.concatenate((stream1, stream2))
size = streamline[np.invert(np.isnan(streamline))].size
streamline = streamline[np.invert(np.isnan(streamline))].reshape(int(size/2.), 2)
circle_params, success = leastsq(to_min, params_guess, args=np.asarray([streamline[:, 0], streamline[:, 1]]))
direction = [circle_params[0] - launch_point[0], circle_params[1] - launch_point[1]]
distance = np.sqrt(direction[0]**2. + direction[1]**2.)
center_point = (circle_params[0], circle_params[1])
launch_point = [launch_point[0] + direction[0] * launch_point_step_factor,
launch_point[1] + direction[1] * launch_point_step_factor]
center_points.append(center_point)
#print 'center', center_point
radii.append(circle_params[0])
streamlines.append(streamline)
if (launch_point[0] <= x_min or
launch_point[0] >= x_max or
launch_point[1] <= y_min or
launch_point[1] >= y_max or
count > max_count):
break
count += 1
centroid = center_point
return centroid, center_points, radii, streamlines, max_index
centroids = []
for time_point in xrange(1):
print time_point
bx_interpolator = pickle.load(open('../../output/centroid_fitting/2016-08-12/B_z0249_x_' + str(time_point).zfill(4) + '.p', 'rb'))
by_interpolator = pickle.load(open('../../output/centroid_fitting/2016-08-12/B_z0249_y_' + str(time_point).zfill(4) + '.p', 'rb'))
(centroid, center_points,
radii, streamlines,
max_index) = find_centroid(grid,
bx_interpolator,
by_interpolator,
launch_point_step_factor=0.05,
integration_length=20)
centroids.append(centroid)
from datetime import datetime
import os
today = datetime.today()
today = today.strftime('%Y-%m-%d')
out_dir = '../output/' + today
try:
os.makedirs(out_dir)
except:
pass
centroids = np.asarray(centroids)
np.savetxt(out_dir + '/field_nulls.txt', centroids, header='magnetic field null positions in the z=0.249m plane,' +
'determined by fitting circles to integrated field lines starting at max magnitude and moving succesive' +
'towards the center of circles.')
from scipy.integrate import dblquad
def integrate_flux(centroid, radius, bz_interpolator, limits):
if (centroid[0] - radius < limits[0] or centroid[0] + radius > limits[1] or
centroid[1] - radius < limits[2] or centroid[1] + radius > limits[3]):
return -1
gfun = lambda x: -np.sqrt(radius**2 - (x-centroid[0])**2)
hfun = lambda x: np.sqrt(radius**2 - (x-centroid[0])**2)
bz_interpolator_bias = lambda x, y: bz_interpolator(x, y) + 0.02
return dblquad(bz_interpolator_bias, centroid[0] - radius, centroid[0] + radius, gfun, hfun)
bz_interpolator = pickle.load(open('../output/2016-08-12/B_z0249_z_' + str(0).zfill(4) + '.p', 'rb'))
bz_interpolator(centroids[0])
for time_point in xrange(250):
bz_interpolator = pickle.load(open('../output/2016-08-12/B_z0249_z_' + str(time_point).zfill(4) + '.p', 'rb'))
print integrate_flux(centroids[time_point], 0.001, bz_interpolator,
(-0.028, 0.025, -0.043, 0.039))
| 0.351422 | 0.836588 |
```
import os
import shutil
import numpy as np
import pickle as pk
import pandas as pd
from keras.utils import to_categorical ,Sequence
from keras import losses, models, optimizers
from keras.models import load_model
from keras.models import Sequential
from keras.activations import relu, softmax
from keras.callbacks import (EarlyStopping, LearningRateScheduler,
ModelCheckpoint, TensorBoard, ReduceLROnPlateau)
from keras.layers import Activation, LeakyReLU
from keras.preprocessing.image import ImageDataGenerator
from keras import backend as K
from sklearn.model_selection import KFold
from random_eraser import get_random_eraser
from keras.optimizers import Adam
from os.path import join
import resnet
from sklearn.utils import shuffle
map_dict = pk.load(open('data/map.pkl' , 'rb'))
semi = pd.read_csv('data/cotrain/Y_selftrain_ens_verified.csv')
semi_map = {}
semi_name = semi['fname'].values
semi_label_verified = semi['label_verified'].values
for idx ,d in enumerate( semi_name):
semi_map[d] = semi_label_verified[idx]
unverified_df = pd.read_csv('data/train_label.csv')
test_df = pd.read_csv('data/sample_submission.csv')
unverified_df = unverified_df[unverified_df['fname'].isin(semi_name)]
unverified_df = unverified_df.drop(columns=['manually_verified'])
unverified_df['label_verified'] = unverified_df['fname'].map(semi_map)
test_df = test_df[test_df['fname'].isin(semi_name)]
test_df['label_verified'] = test_df['fname'].map(semi_map)
unverified_idx = unverified_df.index.values
test_idx = test_df.index.values
df = pd.concat([unverified_df , test_df])
df = df.drop(columns=['label'])
df['trans'] = df['label_verified'].map(map_dict)
df['onehot'] = df['trans'].apply(lambda x: to_categorical(x,num_classes=41))
X_unverified = np.load('data/mfcc/X_train.npy')[unverified_idx]
X_test = np.load('data/X_test.npy')[test_idx]
X_semi = np.append(X_unverified,X_test , axis=0)
Y_semi = np.array(df['onehot'].tolist()).reshape(-1,41)
print(X_semi.shape)
print(Y_semi.shape)
# data generator ====================================================================================
class MixupGenerator():
def __init__(self, X_train, y_train, batch_size=32, alpha=0.2, shuffle=True, datagen=None):
self.X_train = X_train
self.y_train = y_train
self.batch_size = batch_size
self.alpha = alpha
self.shuffle = shuffle
self.sample_num = len(X_train)
self.datagen = datagen
def __call__(self):
while True:
indexes = self.__get_exploration_order()
itr_num = int(len(indexes) // (self.batch_size * 2))
for i in range(itr_num):
batch_ids = indexes[i * self.batch_size * 2:(i + 1) * self.batch_size * 2]
X, y = self.__data_generation(batch_ids)
yield X, y
def __get_exploration_order(self):
indexes = np.arange(self.sample_num)
if self.shuffle:
np.random.shuffle(indexes)
return indexes
def __data_generation(self, batch_ids):
_, h, w, c = self.X_train.shape
l = np.random.beta(self.alpha, self.alpha, self.batch_size)
X_l = l.reshape(self.batch_size, 1, 1, 1)
y_l = l.reshape(self.batch_size, 1)
X1 = self.X_train[batch_ids[:self.batch_size]]
X2 = self.X_train[batch_ids[self.batch_size:]]
X = X1 * X_l + X2 * (1 - X_l)
if self.datagen:
for i in range(self.batch_size):
X[i] = self.datagen.random_transform(X[i])
X[i] = self.datagen.standardize(X[i])
if isinstance(self.y_train, list):
y = []
for y_train_ in self.y_train:
y1 = y_train_[batch_ids[:self.batch_size]]
y2 = y_train_[batch_ids[self.batch_size:]]
y.append(y1 * y_l + y2 * (1 - y_l))
else:
y1 = self.y_train[batch_ids[:self.batch_size]]
y2 = self.y_train[batch_ids[self.batch_size:]]
y = y1 * y_l + y2 * (1 - y_l)
return X, y
```
# Training Semi Data
```
model_path = 'model_full_resnet2'
refine_path = 'model_full_resnet2_refine_co'
all_x = np.concatenate( (np.load('data/mfcc/X_train.npy') , np.load('data/X_test.npy')))
if not os.path.exists(refine_path):
os.mkdir(refine_path)
for i in range(1,11):
X_train = np.load('data/ten_fold_data/X_train_{}.npy'.format(i))
Y_train = np.load('data/ten_fold_data/Y_train_{}.npy'.format(i))
X_test = np.load('data/ten_fold_data/X_valid_{}.npy'.format(i))
Y_test = np.load('data/ten_fold_data/Y_valid_{}.npy'.format(i))
X_train = np.append(X_train,X_semi , axis=0)
Y_train = np.append(Y_train,Y_semi , axis=0)
X_train , Y_train = shuffle(X_train, Y_train, random_state=5)
print(X_train.shape)
print(Y_train.shape)
print(X_test.shape)
print(Y_test.shape)
model = load_model(join(model_path,'best_{}.h5'.format(i)))
checkpoint = ModelCheckpoint(join(refine_path , 'semi_self_%d_{val_acc:.5f}.h5'%i), monitor='val_acc', verbose=1, save_best_only=True)
early = EarlyStopping(monitor="val_acc", mode="max", patience=30)
callbacks_list = [checkpoint, early]
datagen = ImageDataGenerator(
featurewise_center=True, # set input mean to 0 over the dataset
width_shift_range=0.2,
height_shift_range=0.2,
horizontal_flip=True,
preprocessing_function=get_random_eraser(v_l=np.min(all_x), v_h=np.max(all_x)) # Trainset's boundaries.
)
mygenerator = MixupGenerator(X_train, Y_train, alpha=1.0, batch_size=128, datagen=datagen)
model.compile(loss='categorical_crossentropy',
optimizer=Adam(lr=0.0001),
metrics=['accuracy'])
# mixup
history = model.fit_generator(mygenerator(),
steps_per_epoch= X_train.shape[0] // 128,
epochs=10000,
validation_data=(X_test,Y_test), callbacks=callbacks_list)
# normalize
# history = model.fit(X_train, Y_train, validation_data=(X_test, Y_test), callbacks=callbacks_list,
# batch_size=32, epochs=10000)
# break
```
|
github_jupyter
|
import os
import shutil
import numpy as np
import pickle as pk
import pandas as pd
from keras.utils import to_categorical ,Sequence
from keras import losses, models, optimizers
from keras.models import load_model
from keras.models import Sequential
from keras.activations import relu, softmax
from keras.callbacks import (EarlyStopping, LearningRateScheduler,
ModelCheckpoint, TensorBoard, ReduceLROnPlateau)
from keras.layers import Activation, LeakyReLU
from keras.preprocessing.image import ImageDataGenerator
from keras import backend as K
from sklearn.model_selection import KFold
from random_eraser import get_random_eraser
from keras.optimizers import Adam
from os.path import join
import resnet
from sklearn.utils import shuffle
map_dict = pk.load(open('data/map.pkl' , 'rb'))
semi = pd.read_csv('data/cotrain/Y_selftrain_ens_verified.csv')
semi_map = {}
semi_name = semi['fname'].values
semi_label_verified = semi['label_verified'].values
for idx ,d in enumerate( semi_name):
semi_map[d] = semi_label_verified[idx]
unverified_df = pd.read_csv('data/train_label.csv')
test_df = pd.read_csv('data/sample_submission.csv')
unverified_df = unverified_df[unverified_df['fname'].isin(semi_name)]
unverified_df = unverified_df.drop(columns=['manually_verified'])
unverified_df['label_verified'] = unverified_df['fname'].map(semi_map)
test_df = test_df[test_df['fname'].isin(semi_name)]
test_df['label_verified'] = test_df['fname'].map(semi_map)
unverified_idx = unverified_df.index.values
test_idx = test_df.index.values
df = pd.concat([unverified_df , test_df])
df = df.drop(columns=['label'])
df['trans'] = df['label_verified'].map(map_dict)
df['onehot'] = df['trans'].apply(lambda x: to_categorical(x,num_classes=41))
X_unverified = np.load('data/mfcc/X_train.npy')[unverified_idx]
X_test = np.load('data/X_test.npy')[test_idx]
X_semi = np.append(X_unverified,X_test , axis=0)
Y_semi = np.array(df['onehot'].tolist()).reshape(-1,41)
print(X_semi.shape)
print(Y_semi.shape)
# data generator ====================================================================================
class MixupGenerator():
def __init__(self, X_train, y_train, batch_size=32, alpha=0.2, shuffle=True, datagen=None):
self.X_train = X_train
self.y_train = y_train
self.batch_size = batch_size
self.alpha = alpha
self.shuffle = shuffle
self.sample_num = len(X_train)
self.datagen = datagen
def __call__(self):
while True:
indexes = self.__get_exploration_order()
itr_num = int(len(indexes) // (self.batch_size * 2))
for i in range(itr_num):
batch_ids = indexes[i * self.batch_size * 2:(i + 1) * self.batch_size * 2]
X, y = self.__data_generation(batch_ids)
yield X, y
def __get_exploration_order(self):
indexes = np.arange(self.sample_num)
if self.shuffle:
np.random.shuffle(indexes)
return indexes
def __data_generation(self, batch_ids):
_, h, w, c = self.X_train.shape
l = np.random.beta(self.alpha, self.alpha, self.batch_size)
X_l = l.reshape(self.batch_size, 1, 1, 1)
y_l = l.reshape(self.batch_size, 1)
X1 = self.X_train[batch_ids[:self.batch_size]]
X2 = self.X_train[batch_ids[self.batch_size:]]
X = X1 * X_l + X2 * (1 - X_l)
if self.datagen:
for i in range(self.batch_size):
X[i] = self.datagen.random_transform(X[i])
X[i] = self.datagen.standardize(X[i])
if isinstance(self.y_train, list):
y = []
for y_train_ in self.y_train:
y1 = y_train_[batch_ids[:self.batch_size]]
y2 = y_train_[batch_ids[self.batch_size:]]
y.append(y1 * y_l + y2 * (1 - y_l))
else:
y1 = self.y_train[batch_ids[:self.batch_size]]
y2 = self.y_train[batch_ids[self.batch_size:]]
y = y1 * y_l + y2 * (1 - y_l)
return X, y
model_path = 'model_full_resnet2'
refine_path = 'model_full_resnet2_refine_co'
all_x = np.concatenate( (np.load('data/mfcc/X_train.npy') , np.load('data/X_test.npy')))
if not os.path.exists(refine_path):
os.mkdir(refine_path)
for i in range(1,11):
X_train = np.load('data/ten_fold_data/X_train_{}.npy'.format(i))
Y_train = np.load('data/ten_fold_data/Y_train_{}.npy'.format(i))
X_test = np.load('data/ten_fold_data/X_valid_{}.npy'.format(i))
Y_test = np.load('data/ten_fold_data/Y_valid_{}.npy'.format(i))
X_train = np.append(X_train,X_semi , axis=0)
Y_train = np.append(Y_train,Y_semi , axis=0)
X_train , Y_train = shuffle(X_train, Y_train, random_state=5)
print(X_train.shape)
print(Y_train.shape)
print(X_test.shape)
print(Y_test.shape)
model = load_model(join(model_path,'best_{}.h5'.format(i)))
checkpoint = ModelCheckpoint(join(refine_path , 'semi_self_%d_{val_acc:.5f}.h5'%i), monitor='val_acc', verbose=1, save_best_only=True)
early = EarlyStopping(monitor="val_acc", mode="max", patience=30)
callbacks_list = [checkpoint, early]
datagen = ImageDataGenerator(
featurewise_center=True, # set input mean to 0 over the dataset
width_shift_range=0.2,
height_shift_range=0.2,
horizontal_flip=True,
preprocessing_function=get_random_eraser(v_l=np.min(all_x), v_h=np.max(all_x)) # Trainset's boundaries.
)
mygenerator = MixupGenerator(X_train, Y_train, alpha=1.0, batch_size=128, datagen=datagen)
model.compile(loss='categorical_crossentropy',
optimizer=Adam(lr=0.0001),
metrics=['accuracy'])
# mixup
history = model.fit_generator(mygenerator(),
steps_per_epoch= X_train.shape[0] // 128,
epochs=10000,
validation_data=(X_test,Y_test), callbacks=callbacks_list)
# normalize
# history = model.fit(X_train, Y_train, validation_data=(X_test, Y_test), callbacks=callbacks_list,
# batch_size=32, epochs=10000)
# break
| 0.598899 | 0.297393 |
# Hi, I am Neel Shah
## Wroking at Datalog.ai and IDLI on Data analyst, Machine learnig and Deep Learning
Contact details:
1) Website: https://neelshah18.github.io/
2) GitHub: https://github.com/NeelShah18
3) LinkedIn: https://www.linkedin.com/in/neel-shah-7b5495104/
4) Facebook: https://www.facebook.com/neelxyz
*** all code and data are available on my GitHub account under MIT open source licence***
* Though many computer scientists and programmers consider OOP to be a modern programming paradigm, the roots go back to 1960s. The first programming language to use objects was Simula 67. As the name implies, Simula 67 was introduced in the year 1967. A major breakthrough for object-oriented programming came with the programming language Smalltalk in the 1970s.
#### Following two concept is explain everything the use of OOPs.
1. Duplicate code is a Bad.
2. Code will always be changed.
#### Object-Oriented Programming has the following advantages:
* OOP provides a clear modular structure for programs which makes it good for defining abstract datatypes where implementation details are hidden and the unit has a clearly defined interface.
* OOP makes it easy to maintain and modify existing code as new objects can be created with small differences to existing ones.
* OOP provides a good framework for code libraries where supplied software components can be easily adapted and modified by the programmer. This is particularly useful for developing graphical user interfaces.
#### Terminology:
* **Class**: A user-defined prototype for an object that defines a set of attributes that characterize any object of the class. The attributes are data members (class variables and instance variables) and methods, accessed via dot notation.
* **Class variable**: A variable that is shared by all instances of a class. Class variables are defined within a class but outside any of the class's methods. Class variables are not used as frequently as instance variables are.
* **Data member**: A class variable or instance variable that holds data associated with a class and its objects.
* **Instance variable**: A variable that is defined inside a method and belongs only to the current instance of a class.
* **Inheritance**: The transfer of the characteristics of a class to other classes that are derived from it.
* **Instance**: An individual object of a certain class. An object obj that belongs to a class Circle, for example, is an instance of the class Circle.
* **Method**: A special kind of function that is defined in a class definition.
* **Object**: A unique instance of a data structure that's defined by its class. An object comprises both data members (class variables and instance variables) and methods.
* **Function overloading**: The assignment of more than one behavior to a particular function. The operation performed varies by the types of objects or arguments involved.
#### Everything in class is python:
Even though we haven't talked about classes and object orientation in previous chapters, we have worked with classes all the time. In fact, everything is a class in Python.
```
import math
x = 4
print(type(x))
def f(x):
return x+1
print(type(f))
print(type(math))
'''As you can see, everything in python is class.'''
# Creating Classes
class Robot:
pass
if __name__ == "__main__":
x = Robot()
y = Robot()
y2 = y
print(y == y2)
print(y == x)
class Customer(object):
"""A customer of ABC Bank with a checking account. Customers have the
following properties:
Attributes:
name: A string representing the customer's name.
balance: A float tracking the current balance of the customer's account.
"""
def __init__(self, name, balance=0.0):
"""Return a Customer object whose name is *name* and starting
balance is *balance*."""
self.name = name
self.balance = balance
def withdraw(self, amount):
"""Return the balance remaining after withdrawing *amount*
dollars."""
if amount > self.balance:
raise RuntimeError('Amount greater than available balance.')
self.balance -= amount
return self.balance
def deposit(self, amount):
"""Return the balance remaining after depositing *amount*
dollars."""
self.balance += amount
return self.balance
neel = Customer(neel, 500)
print(neel.deposit(200))
print(neel.withdraw(150))
'''This is basic example of class in python with __init__, methods'''
```
The first method __init__() is a special method, which is called class constructor or initialization method that Python calls when you create a new instance of this class.
You declare other class methods like normal functions with the exception that the first argument to each method is self. Python adds the self argument to the list for you; you do not need to include it when you call the methods.
So what's with that self parameter to all of the Customer methods? What is it? Why, it's the instance, of course! Put another way, a method like withdraw defines the instructions for withdrawing money from some abstract customer's account. Calling jeff.withdraw(100.0) puts those instructions to use on the jeff instance.
```
# one more example
class Robot:
def __init__(self, name=None):
self.name = name
def say_hi(self):
if self.name:
print("Hi, I am " + self.name)
else:
print("Hi, I am a robot without a name")
x = Robot()
x.say_hi()
y = Robot("Marvin")
y.say_hi()
```
#### Public- Protected- and Private Attributes
Python uses a special naming scheme for attributes to control the accessibility of the attributes. So far, we have used attribute names, which can be freely used inside or outside of a class definition, as we have seen. This corresponds to public attributes of course.
There are two ways to restrict the access to class attributes:
* First, we can prefix an attribute name with a leading underscore "_". This marks the attribute as protected. It tells users of the class not to use this attribute unless, somebody writes a subclass. We will learn about inheritance and subclassing in the next chapter of our tutorial.
* Second, we can prefix an attribute name with two leading underscores "__". The attribute is now inaccessible and invisible from outside. It's neither possible to read nor write to those attributes except inside of the class definition itself.5
1) name--> Public--> These attributes can be freely used inside or outside of a class definition.
2) _name--> Protected--> Protected attributes should not be used outside of the class definition, unless inside of a subclass definition.
3) __name--> Private--> This kind of attribute is inaccessible and invisible. It's neither possible to read nor write to those attributes, except inside of the class definition itself.
```
# Attribute accessibility example
class A():
def __init__(self):
self.__priv = "I am private"
self._prot = "I am protected"
self.pub = "I am public"
x = A()
print(x.pub)
print(x.pub + " and my value can be changed")
print(x._prot)
x.__priv
```
The error message is very interesting. One might have expected a message like "__priv is private". We get the message "AttributeError: 'A' object has no attribute '__priv'" instead, which looks like a "lie". There is such an attribute, but we are told that there isn't. This is perfect information hiding. Telling a user that an attribute name is private, means that we make some information visible, i.e. the existence or non-existence of a private variable.
#### Destructor:
What we said about constructors holds true for destructors as well. There is no "real" destructor, but something similar, i.e. the method __del__. It is called when the instance is about to be destroyed and if there is no other reference to this instance. If a base class has a __del__() method, the derived class's __del__() method, if any, must explicitly call it to ensure proper deletion of the base class part of the instance.
***The destructor was called after the program ended, not when ft went out of scope inside make_foo.***
```
class FooType(object):
id = 0
def __init__(self, id):
self.id = id
print(self.id, 'born')
def __del__(self):
print(self.id, 'died')
ft = FooType(1)
```
#### Inheritance
One of the major benefits of object oriented programming is reuse of code and one of the ways this is achieved is through the inheritance mechanism. Inheritance can be best imagined as implementing a type and subtype relationship between classes.
Suppose you want to write a program which has to keep track of the teachers and students in a college. They have some common characteristics such as name, age and address. They also have specific characteristics such as salary, courses and leaves for teachers and, marks and fees for students.
You can create two independent classes for each type and process them but adding a new common characteristic would mean adding to both of these independent classes. This quickly becomes unwieldy.
```
class SchoolMember:
'''Represents any school member.'''
def __init__(self, name, age):
self.name = name
self.age = age
print('(Initialized SchoolMember: {})'.format(self.name))
def tell(self):
'''Tell my details.'''
print('Name:"{}" Age:"{}"'.format(self.name, self.age), end=" ")
class Teacher(SchoolMember):
'''Represents a teacher.'''
def __init__(self, name, age, salary):
SchoolMember.__init__(self, name, age)
self.salary = salary
print('(Initialized Teacher: {})'.format(self.name))
def tell(self):
SchoolMember.tell(self)
print('Salary: "{:d}"'.format(self.salary))
class Student(SchoolMember):
'''Represents a student.'''
def __init__(self, name, age, marks):
SchoolMember.__init__(self, name, age)
self.marks = marks
print('(Initialized Student: {})'.format(self.name))
def tell(self):
SchoolMember.tell(self)
print('Marks: "{:d}"'.format(self.marks))
t = Teacher('Mr. Neel', 24, 30000)
s = Student('Mr. Water', 20, 75)
# prints a blank line
print()
members = [t, s]
for member in members:
# Works for both Teachers and Students
member.tell()
```
#### Polymorphism
In a child class we can change how some methods work whilst keeping the same name. We call this polymorphism or overriding and it is useful because we do not want to keep introducing new method names for functionality that is pretty similar in each class.
```
class A():
def __init__(self):
self.__x = 1
def message(self):
print("message from A")
class B(A):
def __init__(self):
self.__y = 1
def message(self):
print("message from B")
try_1 = A()
try_1.message()
try_2 = B()
try_2.message()
```
**This is the end of the OOP concept.
Hope you like it and not bored!**
You can download all material from github account or website.
1) Website: https://neelshah18.github.io/
2) GitHub: https://github.com/NeelShah18/OOP-with-python
#### References:
1) http://www.python-course.eu/object_oriented_programming.php
2) http://thepythonguru.com/python-inheritance-and-polymorphism/
3) https://pythonschool.net/oop/inheritance-and-polymorphism/
|
github_jupyter
|
import math
x = 4
print(type(x))
def f(x):
return x+1
print(type(f))
print(type(math))
'''As you can see, everything in python is class.'''
# Creating Classes
class Robot:
pass
if __name__ == "__main__":
x = Robot()
y = Robot()
y2 = y
print(y == y2)
print(y == x)
class Customer(object):
"""A customer of ABC Bank with a checking account. Customers have the
following properties:
Attributes:
name: A string representing the customer's name.
balance: A float tracking the current balance of the customer's account.
"""
def __init__(self, name, balance=0.0):
"""Return a Customer object whose name is *name* and starting
balance is *balance*."""
self.name = name
self.balance = balance
def withdraw(self, amount):
"""Return the balance remaining after withdrawing *amount*
dollars."""
if amount > self.balance:
raise RuntimeError('Amount greater than available balance.')
self.balance -= amount
return self.balance
def deposit(self, amount):
"""Return the balance remaining after depositing *amount*
dollars."""
self.balance += amount
return self.balance
neel = Customer(neel, 500)
print(neel.deposit(200))
print(neel.withdraw(150))
'''This is basic example of class in python with __init__, methods'''
# one more example
class Robot:
def __init__(self, name=None):
self.name = name
def say_hi(self):
if self.name:
print("Hi, I am " + self.name)
else:
print("Hi, I am a robot without a name")
x = Robot()
x.say_hi()
y = Robot("Marvin")
y.say_hi()
# Attribute accessibility example
class A():
def __init__(self):
self.__priv = "I am private"
self._prot = "I am protected"
self.pub = "I am public"
x = A()
print(x.pub)
print(x.pub + " and my value can be changed")
print(x._prot)
x.__priv
class FooType(object):
id = 0
def __init__(self, id):
self.id = id
print(self.id, 'born')
def __del__(self):
print(self.id, 'died')
ft = FooType(1)
class SchoolMember:
'''Represents any school member.'''
def __init__(self, name, age):
self.name = name
self.age = age
print('(Initialized SchoolMember: {})'.format(self.name))
def tell(self):
'''Tell my details.'''
print('Name:"{}" Age:"{}"'.format(self.name, self.age), end=" ")
class Teacher(SchoolMember):
'''Represents a teacher.'''
def __init__(self, name, age, salary):
SchoolMember.__init__(self, name, age)
self.salary = salary
print('(Initialized Teacher: {})'.format(self.name))
def tell(self):
SchoolMember.tell(self)
print('Salary: "{:d}"'.format(self.salary))
class Student(SchoolMember):
'''Represents a student.'''
def __init__(self, name, age, marks):
SchoolMember.__init__(self, name, age)
self.marks = marks
print('(Initialized Student: {})'.format(self.name))
def tell(self):
SchoolMember.tell(self)
print('Marks: "{:d}"'.format(self.marks))
t = Teacher('Mr. Neel', 24, 30000)
s = Student('Mr. Water', 20, 75)
# prints a blank line
print()
members = [t, s]
for member in members:
# Works for both Teachers and Students
member.tell()
class A():
def __init__(self):
self.__x = 1
def message(self):
print("message from A")
class B(A):
def __init__(self):
self.__y = 1
def message(self):
print("message from B")
try_1 = A()
try_1.message()
try_2 = B()
try_2.message()
| 0.602997 | 0.929568 |
# EDA with Categorical Variables
Whether EDA (exploratory data analysis) is the main purpose of your project, or is mainly being used for feature selection/feature engineering in a machine learning context, it's important to be able to understand the relationship between your features and your target variable.
Many examples of EDA emphasize numeric features, but this notebook emphasizes categorical features.
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.patches import Patch
from matplotlib.lines import Line2D
import seaborn as sns
```
## The Dataset
This analysis uses the [Titanic dataset](https://www.kaggle.com/c/titanic/data) in order to predict whether a given person survived or not
This dataset has the following columns:
| Variable | Definition | Key |
| -------- | ---------- | --- |
| survival | Survival | 0 = No, 1 = Yes |
| pclass | Ticket class | 1 = 1st, 2 = 2nd, 3 = 3rd |
| sex | Sex | |
| Age | Age in years | |
| sibsp | # of siblings / spouses aboard the Titanic | |
| parch | # of parents / children aboard the Titanic | |
| ticket | Ticket number | |
| fare | Passenger fare | |
| cabin | Cabin number | |
| embarked | Port of Embarkation | C = Cherbourg, Q = Queenstown, S = Southampton |
To get started, we'll open up the CSV with Pandas.
(If you were using this for a machine learning project, you would additionally separate the dataframe into `X` and `y`, and then into train and test sets, but for the purposes of this example we'll assume that the entire `titanic.csv` contains training data.)
```
df = pd.read_csv("titanic.csv")
# PassengerId is a dataset artifact, not something useful for analysis
df.drop("PassengerId", axis=1, inplace=True)
# We want to use Age as one of the main examples, drop rows that are missing Age values
df.dropna(subset=["Age"], inplace=True)
df.head()
```
## Numeric vs. Categorical EDA
Here we are trying to see the relationship between a given numeric feature and the target, which is categorical. Let's use the `Age` column as an example.
### What Not to Do
One thought we might have would be just to use a scatter plot, since the categorical target has already been encoded as 0s and 1s:
```
fig, ax = plt.subplots(figsize=(10,5))
ax.scatter(df["Age"], df["Survived"], alpha=0.5)
ax.set_xlabel("Age")
ax.set_ylabel("Survived")
fig.suptitle("Age vs. Survival for Titanic Passengers");
```
Ok, we can see that age seems to matter some, but it's pretty hard to extract much useful information from this visualization. Let's try some other visualizations that tell us more
### Multiple Histograms
Rather than using the y axis to represent the two categories, let's use two different colors. That means that we can use the y axis to represent counts rather than trying to discern this information from the density of dots.
```
fig, ax = plt.subplots()
ax.hist(df[df["Survived"]==1]["Age"], bins=15, alpha=0.5, color="blue", label="survived")
ax.hist(df[df["Survived"]==0]["Age"], bins=15, alpha=0.5, color="green", label="did not survive")
ax.set_xlabel("Age")
ax.set_ylabel("Count of passengers")
fig.suptitle("Age vs. Survival for Titanic Passengers")
ax.legend();
```
### Multiple Density Estimate Plots
This is showing largely the same information as the histograms, except that it's a density estimate (estimate of the probability density function) rather than a count across bins. Seaborn has nice built-in functionality for this.
```
fig, ax = plt.subplots()
sns.kdeplot(df[df["Survived"]==1]["Age"], shade=True, color="blue", label="survived", ax=ax)
sns.kdeplot(df[df["Survived"]==0]["Age"], shade=True, color="green", label="did not survive", ax=ax)
ax.set_xlabel("Age")
ax.set_ylabel("Density")
fig.suptitle("Age vs. Survival for Titanic Passengers");
```
### Multiple Box Plots
Here we lose some of the information about the distribution overall in order to focus in on particular summary statistics of the distribution
<a title="Dcbmariano / CC BY-SA (https://creativecommons.org/licenses/by-sa/4.0)" href="https://commons.wikimedia.org/wiki/File:Box_plot_description.jpg"><img width="256" alt="Box plot description" src="https://upload.wikimedia.org/wikipedia/commons/a/ac/Box_plot_description.jpg"></a>
Matplotlib and Seaborn both have methods for this. The Seaborn one is built on top of the Matplotlib one.
```
fig, ax = plt.subplots()
sns.boxplot(x="Age", y="Survived", data=df, orient="h", palette={1:"blue", 0:"green"}, ax=ax)
ax.get_yaxis().set_visible(False)
fig.suptitle("Age vs. Survival for Titanic Passengers")
color_patches = [
Patch(facecolor="blue", label="survived"),
Patch(facecolor="green", label="did not survive")
]
ax.legend(handles=color_patches);
```
## Categorical vs. Categorical EDA
Here we are trying to see the relationship between a given categorical variable and the target (which is also categorical). Let's use the `Pclass` (passenger class) feature as an example.
### What Not to Do
Again, there is nothing _preventing_ us from just making a scatter plot, since the passenger class is encoded as a number
```
fig, ax = plt.subplots()
ax.scatter(df["Pclass"], df["Survived"], alpha=0.5)
ax.set_xlabel("Passenger Class")
ax.set_ylabel("Survived")
fig.suptitle("Passenger Class vs. Survival for Titanic Passengers");
```
...but that plot is not really useful at all. It's really just telling us that at least 1 person falls into each category
### Grouped Bar Charts
This shows the distribution across the categories, similar to the "multiple histograms" example for numeric vs. categorical
```
fig, ax = plt.subplots()
sns.catplot("Pclass", hue="Survived", data=df, kind="count",
palette={1:"blue", 0:"green"}, ax=ax)
plt.close(2) # catplot creates an extra figure we don't need
ax.set_xlabel("Passenger Class")
color_patches = [
Patch(facecolor="blue", label="survived"),
Patch(facecolor="green", label="did not survive")
]
ax.legend(handles=color_patches)
fig.suptitle("Passenger Class vs. Survival for Titanic Passengers");
fig, ax = plt.subplots()
sns.catplot("Survived", hue="Pclass", data=df, kind="count",
palette={1:"yellow", 2:"orange", 3:"red"}, ax=ax)
plt.close(2) # catplot creates an extra figure we don't need
ax.legend(title="Passenger Class")
ax.set_xticklabels(["did not survive", "survived"])
ax.set_xlabel("")
fig.suptitle("Passenger Class vs. Survival for Titanic Passengers");
```
### Stacked Bar Charts
These can be used for counts (same as grouped bar charts) but if you use percentages rather than counts, they show proportions
```
# Create a dataframe with the counts by passenger class and survival
counts_df = df.groupby(["Pclass", "Survived"])["Name"].count().unstack()
counts_df
# Divide by the total number and transpose for plotting
pclass_percents_df = counts_df.div(counts_df.sum()).T
pclass_percents_df
fig, ax = plt.subplots()
pclass_percents_df.plot(kind="bar", stacked=True, color=["yellow", "orange", "red"], ax=ax)
ax.legend(title="Passenger Class")
ax.set_xticklabels(["did not survive", "survived"], rotation=0)
ax.set_xlabel("")
ax.set_ylabel("Proportion")
fig.suptitle("Passenger Class vs. Survival for Titanic Passengers");
# Use the same counts df, but now survived + did not survive add up to 1
survived_percents_df = counts_df.T.div(counts_df.T.sum()).T
survived_percents_df
fig, ax = plt.subplots()
survived_percents_df.plot(kind="bar", stacked=True, color=["green", "blue"], ax=ax)
ax.set_xlabel("Passenger Class")
ax.set_xticklabels([1, 2, 3], rotation=0)
ax.set_ylabel("Proportion")
color_patches = [
Patch(facecolor="blue", label="survived"),
Patch(facecolor="green", label="did not survive")
]
ax.legend(handles=color_patches)
fig.suptitle("Passenger Class vs. Survival for Titanic Passengers");
```
## Numeric vs. Numeric vs. Categorical EDA
Sometimes it's interesting to see the relationship between two different numeric features and the target
### What Not to Do
You could just make a scatter plot of the two numeric features
```
fig, ax = plt.subplots(figsize=(10, 5))
ax.scatter(df["Age"], df["Fare"], alpha=0.5)
ax.set_xlabel("Age")
ax.set_ylabel("Fare")
fig.suptitle("Age vs. Fare for Titanic Passengers");
```
That's fine if the relationship between Age and Fare is what interests you, but it doesn't give you any information about the relationship between these features and the target
### Scatterplot with Color to Distinguish Categories
This kind of plot could help you understand how the two features relate to the target
```
fig, ax = plt.subplots(figsize=(10, 5))
ax.scatter(df[df["Survived"]==1]["Age"], df[df["Survived"]==1]["Fare"], c="blue", alpha=0.5)
ax.scatter(df[df["Survived"]==0]["Age"], df[df["Survived"]==0]["Fare"], c="green", alpha=0.5)
ax.set_xlabel("Age")
ax.set_ylabel("Fare")
color_patches = [
Line2D([0], [0], marker='o', color='w', label='survived', markerfacecolor='b', markersize=10),
Line2D([0], [0], marker='o', color='w', label='did not survive', markerfacecolor='g', markersize=10)
]
ax.legend(handles=color_patches)
fig.suptitle("Survival by Age and Fare for Titanic Passengers");
```
## Summary
Most of the time if your target is a categorical variable, the best EDA visualization isn't going to be a basic scatter plot. Instead, consider:
#### Numeric vs. Categorical (e.g. `Survived` vs. `Age`)
- Multiple histograms
- Multiple density estimate plots
- Multiple box plots
#### Categorical vs. Categorical (e.g. `Survived` vs. `Pclass`)
- Grouped bar charts
- Stacked bar charts
#### Numeric vs. Numeric vs. Categorical (e.g. `Age` vs. `Fare` vs. `Survived`)
- Color-coded scatter plots
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.patches import Patch
from matplotlib.lines import Line2D
import seaborn as sns
df = pd.read_csv("titanic.csv")
# PassengerId is a dataset artifact, not something useful for analysis
df.drop("PassengerId", axis=1, inplace=True)
# We want to use Age as one of the main examples, drop rows that are missing Age values
df.dropna(subset=["Age"], inplace=True)
df.head()
fig, ax = plt.subplots(figsize=(10,5))
ax.scatter(df["Age"], df["Survived"], alpha=0.5)
ax.set_xlabel("Age")
ax.set_ylabel("Survived")
fig.suptitle("Age vs. Survival for Titanic Passengers");
fig, ax = plt.subplots()
ax.hist(df[df["Survived"]==1]["Age"], bins=15, alpha=0.5, color="blue", label="survived")
ax.hist(df[df["Survived"]==0]["Age"], bins=15, alpha=0.5, color="green", label="did not survive")
ax.set_xlabel("Age")
ax.set_ylabel("Count of passengers")
fig.suptitle("Age vs. Survival for Titanic Passengers")
ax.legend();
fig, ax = plt.subplots()
sns.kdeplot(df[df["Survived"]==1]["Age"], shade=True, color="blue", label="survived", ax=ax)
sns.kdeplot(df[df["Survived"]==0]["Age"], shade=True, color="green", label="did not survive", ax=ax)
ax.set_xlabel("Age")
ax.set_ylabel("Density")
fig.suptitle("Age vs. Survival for Titanic Passengers");
fig, ax = plt.subplots()
sns.boxplot(x="Age", y="Survived", data=df, orient="h", palette={1:"blue", 0:"green"}, ax=ax)
ax.get_yaxis().set_visible(False)
fig.suptitle("Age vs. Survival for Titanic Passengers")
color_patches = [
Patch(facecolor="blue", label="survived"),
Patch(facecolor="green", label="did not survive")
]
ax.legend(handles=color_patches);
fig, ax = plt.subplots()
ax.scatter(df["Pclass"], df["Survived"], alpha=0.5)
ax.set_xlabel("Passenger Class")
ax.set_ylabel("Survived")
fig.suptitle("Passenger Class vs. Survival for Titanic Passengers");
fig, ax = plt.subplots()
sns.catplot("Pclass", hue="Survived", data=df, kind="count",
palette={1:"blue", 0:"green"}, ax=ax)
plt.close(2) # catplot creates an extra figure we don't need
ax.set_xlabel("Passenger Class")
color_patches = [
Patch(facecolor="blue", label="survived"),
Patch(facecolor="green", label="did not survive")
]
ax.legend(handles=color_patches)
fig.suptitle("Passenger Class vs. Survival for Titanic Passengers");
fig, ax = plt.subplots()
sns.catplot("Survived", hue="Pclass", data=df, kind="count",
palette={1:"yellow", 2:"orange", 3:"red"}, ax=ax)
plt.close(2) # catplot creates an extra figure we don't need
ax.legend(title="Passenger Class")
ax.set_xticklabels(["did not survive", "survived"])
ax.set_xlabel("")
fig.suptitle("Passenger Class vs. Survival for Titanic Passengers");
# Create a dataframe with the counts by passenger class and survival
counts_df = df.groupby(["Pclass", "Survived"])["Name"].count().unstack()
counts_df
# Divide by the total number and transpose for plotting
pclass_percents_df = counts_df.div(counts_df.sum()).T
pclass_percents_df
fig, ax = plt.subplots()
pclass_percents_df.plot(kind="bar", stacked=True, color=["yellow", "orange", "red"], ax=ax)
ax.legend(title="Passenger Class")
ax.set_xticklabels(["did not survive", "survived"], rotation=0)
ax.set_xlabel("")
ax.set_ylabel("Proportion")
fig.suptitle("Passenger Class vs. Survival for Titanic Passengers");
# Use the same counts df, but now survived + did not survive add up to 1
survived_percents_df = counts_df.T.div(counts_df.T.sum()).T
survived_percents_df
fig, ax = plt.subplots()
survived_percents_df.plot(kind="bar", stacked=True, color=["green", "blue"], ax=ax)
ax.set_xlabel("Passenger Class")
ax.set_xticklabels([1, 2, 3], rotation=0)
ax.set_ylabel("Proportion")
color_patches = [
Patch(facecolor="blue", label="survived"),
Patch(facecolor="green", label="did not survive")
]
ax.legend(handles=color_patches)
fig.suptitle("Passenger Class vs. Survival for Titanic Passengers");
fig, ax = plt.subplots(figsize=(10, 5))
ax.scatter(df["Age"], df["Fare"], alpha=0.5)
ax.set_xlabel("Age")
ax.set_ylabel("Fare")
fig.suptitle("Age vs. Fare for Titanic Passengers");
fig, ax = plt.subplots(figsize=(10, 5))
ax.scatter(df[df["Survived"]==1]["Age"], df[df["Survived"]==1]["Fare"], c="blue", alpha=0.5)
ax.scatter(df[df["Survived"]==0]["Age"], df[df["Survived"]==0]["Fare"], c="green", alpha=0.5)
ax.set_xlabel("Age")
ax.set_ylabel("Fare")
color_patches = [
Line2D([0], [0], marker='o', color='w', label='survived', markerfacecolor='b', markersize=10),
Line2D([0], [0], marker='o', color='w', label='did not survive', markerfacecolor='g', markersize=10)
]
ax.legend(handles=color_patches)
fig.suptitle("Survival by Age and Fare for Titanic Passengers");
| 0.572603 | 0.958924 |
**This notebook is an exercise in the [Introduction to Machine Learning](https://www.kaggle.com/learn/intro-to-machine-learning) course. You can reference the tutorial at [this link](https://www.kaggle.com/dansbecker/random-forests).**
---
## Recap
Here's the code you've written so far.
```
# Code you have previously used to load data
import pandas as pd
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeRegressor
# Path of the file to read
iowa_file_path = '../input/home-data-for-ml-course/train.csv'
home_data = pd.read_csv(iowa_file_path)
# Create target object and call it y
y = home_data.SalePrice
# Create X
features = ['LotArea', 'YearBuilt', '1stFlrSF', '2ndFlrSF', 'FullBath', 'BedroomAbvGr', 'TotRmsAbvGrd']
X = home_data[features]
# Split into validation and training data
train_X, val_X, train_y, val_y = train_test_split(X, y, random_state=1)
# Specify Model
iowa_model = DecisionTreeRegressor(random_state=1)
# Fit Model
iowa_model.fit(train_X, train_y)
# Make validation predictions and calculate mean absolute error
val_predictions = iowa_model.predict(val_X)
val_mae = mean_absolute_error(val_predictions, val_y)
print("Validation MAE when not specifying max_leaf_nodes: {:,.0f}".format(val_mae))
# Using best value for max_leaf_nodes
iowa_model = DecisionTreeRegressor(max_leaf_nodes=100, random_state=1)
iowa_model.fit(train_X, train_y)
val_predictions = iowa_model.predict(val_X)
val_mae = mean_absolute_error(val_predictions, val_y)
print("Validation MAE for best value of max_leaf_nodes: {:,.0f}".format(val_mae))
# Set up code checking
from learntools.core import binder
binder.bind(globals())
from learntools.machine_learning.ex6 import *
print("\nSetup complete")
```
# Exercises
Data science isn't always this easy. But replacing the decision tree with a Random Forest is going to be an easy win.
## Step 1: Use a Random Forest
```
from sklearn.ensemble import RandomForestRegressor
# Define the model. Set random_state to 1
rf_model = RandomForestRegressor(random_state=1)
# fit your model
rf_model.fit(train_X, train_y)
# Calculate the mean absolute error of your Random Forest model on the validation data
melb_preds = rf_model.predict(val_X)
#print(mean_absolute_error(val_y, melb_preds))
rf_val_mae = mean_absolute_error(val_y, melb_preds)
print("Validation MAE for Random Forest Model: {}".format(rf_val_mae))
# Check your answer
step_1.check()
# The lines below will show you a hint or the solution.
# step_1.hint()
# step_1.solution()
```
So far, you have followed specific instructions at each step of your project. This helped learn key ideas and build your first model, but now you know enough to try things on your own.
Machine Learning competitions are a great way to try your own ideas and learn more as you independently navigate a machine learning project.
# Keep Going
You are ready for **[Machine Learning Competitions](https://www.kaggle.com/dansbecker/machine-learning-competitions).**
---
*Have questions or comments? Visit the [Learn Discussion forum](https://www.kaggle.com/learn-forum/161285) to chat with other Learners.*
|
github_jupyter
|
# Code you have previously used to load data
import pandas as pd
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeRegressor
# Path of the file to read
iowa_file_path = '../input/home-data-for-ml-course/train.csv'
home_data = pd.read_csv(iowa_file_path)
# Create target object and call it y
y = home_data.SalePrice
# Create X
features = ['LotArea', 'YearBuilt', '1stFlrSF', '2ndFlrSF', 'FullBath', 'BedroomAbvGr', 'TotRmsAbvGrd']
X = home_data[features]
# Split into validation and training data
train_X, val_X, train_y, val_y = train_test_split(X, y, random_state=1)
# Specify Model
iowa_model = DecisionTreeRegressor(random_state=1)
# Fit Model
iowa_model.fit(train_X, train_y)
# Make validation predictions and calculate mean absolute error
val_predictions = iowa_model.predict(val_X)
val_mae = mean_absolute_error(val_predictions, val_y)
print("Validation MAE when not specifying max_leaf_nodes: {:,.0f}".format(val_mae))
# Using best value for max_leaf_nodes
iowa_model = DecisionTreeRegressor(max_leaf_nodes=100, random_state=1)
iowa_model.fit(train_X, train_y)
val_predictions = iowa_model.predict(val_X)
val_mae = mean_absolute_error(val_predictions, val_y)
print("Validation MAE for best value of max_leaf_nodes: {:,.0f}".format(val_mae))
# Set up code checking
from learntools.core import binder
binder.bind(globals())
from learntools.machine_learning.ex6 import *
print("\nSetup complete")
from sklearn.ensemble import RandomForestRegressor
# Define the model. Set random_state to 1
rf_model = RandomForestRegressor(random_state=1)
# fit your model
rf_model.fit(train_X, train_y)
# Calculate the mean absolute error of your Random Forest model on the validation data
melb_preds = rf_model.predict(val_X)
#print(mean_absolute_error(val_y, melb_preds))
rf_val_mae = mean_absolute_error(val_y, melb_preds)
print("Validation MAE for Random Forest Model: {}".format(rf_val_mae))
# Check your answer
step_1.check()
# The lines below will show you a hint or the solution.
# step_1.hint()
# step_1.solution()
| 0.655777 | 0.950041 |
## Neural Rock Train Model Notebook
The following cell sets up the entire repository from githubg and links to the google drive where the dataset it stored. After all the requirements get installed.
```
import os
if 'google.colab' in str(get_ipython()):
print('Running on CoLab')
import os
from getpass import getpass
import urllib
user = input('User name: ')
password = getpass('Password: ')
password = urllib.parse.quote(password) # your password is converted into url format
cmd_string = 'git clone https://{0}:{1}@github.com/LukasMosser/neural_rock_typing.git'.format(user, password)
os.system(cmd_string)
cmd_string, password = "", "" # removing the password from the variable
os.chdir("./neural_rock_typing")
os.system('pip install -r requirements.txt')
os.system('pip install -e .')
from google.colab import drive
drive.mount('/content/drive', force_remount=True)
else:
print('Not running on CoLab')
%load_ext autoreload
%autoreload 2
```
### A Hack needed to make Pytorch Lightning work with Colab again
```
!pip install wandb
!pip install git+https://github.com/PyTorchLightning/pytorch-lightning
import pytorch_lightning as pl
```
## Login to Weights & Biases for Logging
```
!wandb login
```
## Basic Imports
```
import sys
import os
import argparse
from pathlib import Path
import json
import pandas as pd
import wandb
from torchvision import transforms
from torch.utils.data import DataLoader, ConcatDataset
import pytorch_lightning as pl
from pytorch_lightning.loggers import WandbLogger, TensorBoardLogger
from pytorch_lightning.callbacks import ModelCheckpoint
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
from neural_rock.dataset import SimpleThinSectionDataset
from neural_rock.model import NeuralRockModel, make_vgg11_model, make_resnet18_model
from neural_rock.plot import visualize_batch
from neural_rock.utils import MEAN_TRAIN, STD_TRAIN
```
## Hyperparameters
```
wandb_name = 'lukas-mosser'
project_name = 'neural_rock_simple'
labelset = "Lucia_class"
dataset_fname = "Leg194_dataset.csv"
learning_rate = 3e-4
batch_size = 16
weight_decay = 1e-5
dropout = 0.5
model = 'vgg'
frozen = True
train_dataset_mult = 50
val_dataset_mult = 50
seed_dataset = 42
base_path = "../data"
pl.seed_everything(seed_dataset)
df = pd.read_csv(base_path+"/"+dataset_fname)
df.head()
label_encoder = LabelEncoder()
valid_rows = df[df[labelset].notnull() & df["Xppl"].notnull()]
valid_rows["y"] = label_encoder.fit_transform(valid_rows[labelset])
index = valid_rows.index
train_index, test_index = train_test_split(index, test_size=0.5, stratify=valid_rows["y"])
df_train = valid_rows.loc[train_index].reset_index()
df_val = valid_rows.loc[test_index].reset_index()
print(len(df_train), len(df_val))
```
## Perform Training Sweep across 12 Models
We train a Resnet and a VGG network each with a frozen feature extractor for each labelset: Lucia, Dunham, and DominantPore Type.
This leads to a total of 12 models.
```
# Data Augmentation used for Training
data_transforms = {
'train': transforms.Compose([
transforms.RandomHorizontalFlip(),
transforms.RandomRotation(degrees=360),
transforms.RandomCrop((512, 512)),
transforms.ColorJitter(hue=0.5),
transforms.Resize((224, 224)),
transforms.Normalize((0.485, 0.456, 0.406), (0.229, 0.224, 0.225))
]),
'val':
transforms.Compose([
transforms.RandomCrop((512, 512)),
transforms.Resize((224, 224)),
transforms.Normalize((0.485, 0.456, 0.406), (0.229, 0.224, 0.225))
])
}
# Load the Datasets
train_dataset_base = SimpleThinSectionDataset(base_path, df_train, transform=data_transforms['train'])
val_dataset_base = SimpleThinSectionDataset(base_path, df_val, transform=data_transforms['val'])
# Setup dataloaders
train_loader = DataLoader(train_dataset_base, batch_size=batch_size, shuffle=True, num_workers=0, pin_memory=False)
val_loader = DataLoader(val_dataset_base, batch_size=batch_size, shuffle=False, num_workers=0, pin_memory=False)
for seed in range(10):
# Set the base path for the models to be stored in the Google Drive
path = Path("./data/models/{0:}/{1:}/{2:}".format(labelset, model, str(frozen)))
path.mkdir(parents=True, exist_ok=True)
# Set the Random Seed on Everything
pl.seed_everything(seed)
# Setup Weights and Biases Logger
wandb_logger = WandbLogger(name=wandb_name, project='neural_rock_simple', entity='ccg')
wandb_logger.experiment.config.update({"labelset": labelset, "model": model, 'frozen': str(frozen)})
tensorboard_logger = TensorBoardLogger("lightning_logs", name=labelset)
# Checkpoint based on validation F1 score
checkpointer = ModelCheckpoint(dirpath=path, filename='best', monitor="val/f1", verbose=True, mode="max")
# Setup the Pytorch Lightning Dataloader
trainer = pl.Trainer(gpus=-1,
max_steps=15000,
benchmark=True,
logger=[wandb_logger, tensorboard_logger],
callbacks=[checkpointer],
progress_bar_refresh_rate=20,
check_val_every_n_epoch=1)
# Select which model to run
if model == 'vgg':
feature_extractor, classifier = make_vgg11_model(train_dataset_base.num_classes, dropout=dropout)
elif model == 'resnet':
feature_extractor, classifier = make_resnet18_model(train_dataset_base.num_classes)
# Create the model itself, ready for training
model_ = NeuralRockModel(feature_extractor,
classifier,
num_classes=train_dataset_base.num_classes,
freeze_feature_extractor=frozen)
# Train the model
trainer.fit(model_, train_dataloader=train_loader, val_dataloaders=val_loader)
# Clean up Weights and Biases Logging
wandb.finish()
```
|
github_jupyter
|
import os
if 'google.colab' in str(get_ipython()):
print('Running on CoLab')
import os
from getpass import getpass
import urllib
user = input('User name: ')
password = getpass('Password: ')
password = urllib.parse.quote(password) # your password is converted into url format
cmd_string = 'git clone https://{0}:{1}@github.com/LukasMosser/neural_rock_typing.git'.format(user, password)
os.system(cmd_string)
cmd_string, password = "", "" # removing the password from the variable
os.chdir("./neural_rock_typing")
os.system('pip install -r requirements.txt')
os.system('pip install -e .')
from google.colab import drive
drive.mount('/content/drive', force_remount=True)
else:
print('Not running on CoLab')
%load_ext autoreload
%autoreload 2
!pip install wandb
!pip install git+https://github.com/PyTorchLightning/pytorch-lightning
import pytorch_lightning as pl
!wandb login
import sys
import os
import argparse
from pathlib import Path
import json
import pandas as pd
import wandb
from torchvision import transforms
from torch.utils.data import DataLoader, ConcatDataset
import pytorch_lightning as pl
from pytorch_lightning.loggers import WandbLogger, TensorBoardLogger
from pytorch_lightning.callbacks import ModelCheckpoint
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
from neural_rock.dataset import SimpleThinSectionDataset
from neural_rock.model import NeuralRockModel, make_vgg11_model, make_resnet18_model
from neural_rock.plot import visualize_batch
from neural_rock.utils import MEAN_TRAIN, STD_TRAIN
wandb_name = 'lukas-mosser'
project_name = 'neural_rock_simple'
labelset = "Lucia_class"
dataset_fname = "Leg194_dataset.csv"
learning_rate = 3e-4
batch_size = 16
weight_decay = 1e-5
dropout = 0.5
model = 'vgg'
frozen = True
train_dataset_mult = 50
val_dataset_mult = 50
seed_dataset = 42
base_path = "../data"
pl.seed_everything(seed_dataset)
df = pd.read_csv(base_path+"/"+dataset_fname)
df.head()
label_encoder = LabelEncoder()
valid_rows = df[df[labelset].notnull() & df["Xppl"].notnull()]
valid_rows["y"] = label_encoder.fit_transform(valid_rows[labelset])
index = valid_rows.index
train_index, test_index = train_test_split(index, test_size=0.5, stratify=valid_rows["y"])
df_train = valid_rows.loc[train_index].reset_index()
df_val = valid_rows.loc[test_index].reset_index()
print(len(df_train), len(df_val))
# Data Augmentation used for Training
data_transforms = {
'train': transforms.Compose([
transforms.RandomHorizontalFlip(),
transforms.RandomRotation(degrees=360),
transforms.RandomCrop((512, 512)),
transforms.ColorJitter(hue=0.5),
transforms.Resize((224, 224)),
transforms.Normalize((0.485, 0.456, 0.406), (0.229, 0.224, 0.225))
]),
'val':
transforms.Compose([
transforms.RandomCrop((512, 512)),
transforms.Resize((224, 224)),
transforms.Normalize((0.485, 0.456, 0.406), (0.229, 0.224, 0.225))
])
}
# Load the Datasets
train_dataset_base = SimpleThinSectionDataset(base_path, df_train, transform=data_transforms['train'])
val_dataset_base = SimpleThinSectionDataset(base_path, df_val, transform=data_transforms['val'])
# Setup dataloaders
train_loader = DataLoader(train_dataset_base, batch_size=batch_size, shuffle=True, num_workers=0, pin_memory=False)
val_loader = DataLoader(val_dataset_base, batch_size=batch_size, shuffle=False, num_workers=0, pin_memory=False)
for seed in range(10):
# Set the base path for the models to be stored in the Google Drive
path = Path("./data/models/{0:}/{1:}/{2:}".format(labelset, model, str(frozen)))
path.mkdir(parents=True, exist_ok=True)
# Set the Random Seed on Everything
pl.seed_everything(seed)
# Setup Weights and Biases Logger
wandb_logger = WandbLogger(name=wandb_name, project='neural_rock_simple', entity='ccg')
wandb_logger.experiment.config.update({"labelset": labelset, "model": model, 'frozen': str(frozen)})
tensorboard_logger = TensorBoardLogger("lightning_logs", name=labelset)
# Checkpoint based on validation F1 score
checkpointer = ModelCheckpoint(dirpath=path, filename='best', monitor="val/f1", verbose=True, mode="max")
# Setup the Pytorch Lightning Dataloader
trainer = pl.Trainer(gpus=-1,
max_steps=15000,
benchmark=True,
logger=[wandb_logger, tensorboard_logger],
callbacks=[checkpointer],
progress_bar_refresh_rate=20,
check_val_every_n_epoch=1)
# Select which model to run
if model == 'vgg':
feature_extractor, classifier = make_vgg11_model(train_dataset_base.num_classes, dropout=dropout)
elif model == 'resnet':
feature_extractor, classifier = make_resnet18_model(train_dataset_base.num_classes)
# Create the model itself, ready for training
model_ = NeuralRockModel(feature_extractor,
classifier,
num_classes=train_dataset_base.num_classes,
freeze_feature_extractor=frozen)
# Train the model
trainer.fit(model_, train_dataloader=train_loader, val_dataloaders=val_loader)
# Clean up Weights and Biases Logging
wandb.finish()
| 0.430626 | 0.753829 |
# Training a part-of-speech tagger with transformers (BERT)
This example shows how to use Thinc and Hugging Face's [`transformers`](https://github.com/huggingface/transformers) library to implement and train a part-of-speech tagger on the Universal Dependencies [AnCora corpus](https://github.com/UniversalDependencies/UD_Spanish-AnCora). This notebook assumes familiarity with machine learning concepts, transformer models and Thinc's config system and `Model` API (see the "Thinc for beginners" notebook and the [documentation](https://thinc.ai/docs) for more info).
```
!pip install "thinc>=8.0.0a0" transformers torch "ml_datasets>=0.2.0a0" "tqdm>=4.41"
```
First, let's use Thinc's `prefer_gpu` helper to make sure we're performing operations **on GPU if available**. The function should be called right after importing Thinc, and it returns a boolean indicating whether the GPU has been activated. If we're on GPU, we can also call `use_pytorch_for_gpu_memory` to route `cupy`'s memory allocation via PyTorch, so both can play together nicely.
```
from thinc.api import prefer_gpu, use_pytorch_for_gpu_memory
is_gpu = prefer_gpu()
print("GPU:", is_gpu)
if is_gpu:
use_pytorch_for_gpu_memory()
```
## Overview: the final config
Here's the final config for the model we're building in this notebook. It references a custom `TransformersTagger` that takes the name of a starter (the pretrained model to use), an optimizer, a learning rate schedule with warm-up and the general training settings. You can keep the config string within your file or notebook, or save it to a `conig.cfg` file and load it in via `Config.from_disk`.
```
CONFIG = """
[model]
@layers = "TransformersTagger.v1"
starter = "bert-base-multilingual-cased"
[optimizer]
@optimizers = "Adam.v1"
[optimizer.learn_rate]
@schedules = "warmup_linear.v1"
initial_rate = 0.01
warmup_steps = 3000
total_steps = 6000
[loss]
@losses = "SequenceCategoricalCrossentropy.v1"
[training]
batch_size = 128
words_per_subbatch = 2000
n_epoch = 10
"""
```
---
## Defining the model
The Thinc model we want to define should consist of 3 components: the transformers **tokenizer**, the actual **transformer** implemented in PyTorch and a **softmax-activated output layer**.
### 1. Wrapping the tokenizer
To make it easier to keep track of the data that's passed around (and get type errors if something goes wrong), we first create a `TokensPlus` dataclass that holds the output of the `batch_encode_plus` method of the `transformers` tokenizer. You don't _have to_ do this, but it makes things easier, can prevent bugs and helps the type checker.
```
from typing import Optional, List
from dataclasses import dataclass
import torch
@dataclass
class TokensPlus:
input_ids: torch.Tensor
token_type_ids: torch.Tensor
attention_mask: torch.Tensor
input_len: List[int]
overflowing_tokens: Optional[torch.Tensor] = None
num_truncated_tokens: Optional[torch.Tensor] = None
special_tokens_mask: Optional[torch.Tensor] = None
```
The wrapped tokenizer will take a list-of-lists as input (the texts) and will output a `TokensPlus` object containing the fully padded batch of tokens. The wrapped transformer will take a list of `TokensPlus` objects and will output a list of 2-dimensional arrays.
1. **TransformersTokenizer**: `List[List[str]]` → `TokensPlus`
2. **Transformer**: `TokensPlus` → `List[Array2d]`
> 💡 Since we're adding type hints everywhere (and Thinc is fully typed, too), you can run your code through [`mypy`](https://mypy.readthedocs.io/en/stable/) to find type errors and inconsistencies. If you're using an editor like Visual Studio Code, you can enable `mypy` linting and type errors will be highlighted in real time as you write code.
To wrap the tokenizer, we register a new function that returns a Thinc `Model`. The function takes the name of the pretrained weights (e.g. `"bert-base-multilingual-cased"`) as an argument that can later be provided via the config. After loading the `AutoTokenizer`, we can stash it in the attributes. This lets us access it at any point later on via `model.attrs["tokenizer"]`.
```
import thinc
from thinc.api import Model
from transformers import AutoTokenizer
@thinc.registry.layers("transformers_tokenizer.v1")
def TransformersTokenizer(name: str) -> Model[List[List[str]], TokensPlus]:
def forward(model, texts: List[List[str]], is_train: bool):
tokenizer = model.attrs["tokenizer"]
token_data = tokenizer.batch_encode_plus(
[(text, None) for text in texts],
add_special_tokens=True,
return_token_type_ids=True,
return_attention_masks=True,
return_input_lengths=True,
return_tensors="pt",
)
return TokensPlus(**token_data), lambda d_tokens: []
return Model("tokenizer", forward, attrs={"tokenizer": AutoTokenizer.from_pretrained(name)})
```
The forward pass takes the model and a list-of-lists of strings and outputs the `TokensPlus` dataclass and a callback to use during the backwards (which does nothing in this case).
### 2. Wrapping the transformer
To load and wrap the transformer, we can use `transformers.AutoModel` and Thinc's `PyTorchWrapper`. The forward method of the wrapped model can take arbitrary positional arguments and keyword arguments. Here's what the wrapped model is going to look like:
```python
@thinc.registry.layers("transformers_model.v1")
def Transformer(name) -> Model[TokensPlus, List[Array2d]]:
return PyTorchWrapper(
AutoModel.from_pretrained(name),
convert_inputs=convert_transformer_inputs,
convert_outputs=convert_transformer_outputs,
)
```
The transformer takes `TokensPlus` data as input (as produced by the tokenizer) and outputs a list of 2-dimensional arrays. The convert functions are used to **map inputs and outputs to and from the PyTorch model**. Each function should return the converted output, and a callback to use during the backward pass. To make the arbitrary positional and keyword arguments easier to manage, Thinc uses an `ArgsKwargs` dataclass, essentially a named tuple with `args` and `kwargs` that can be spread into a function as `*ArgsKwargs.args` and `**ArgsKwargs.kwargs`. The `ArgsKwargs` objects will be passed straight into the model in the forward pass, and straight into `torch.autograd.backward` during the backward pass.
```
from thinc.api import ArgsKwargs, torch2xp, xp2torch
from thinc.types import Array2d
def convert_transformer_inputs(model, tokens: TokensPlus, is_train):
kwargs = {
"input_ids": tokens.input_ids,
"attention_mask": tokens.attention_mask,
"token_type_ids": tokens.token_type_ids,
}
return ArgsKwargs(args=(), kwargs=kwargs), lambda dX: []
def convert_transformer_outputs(model, inputs_outputs, is_train):
layer_inputs, torch_outputs = inputs_outputs
torch_tokvecs: torch.Tensor = torch_outputs[0]
torch_outputs = None # free the memory as soon as we can
lengths = list(layer_inputs.input_len)
tokvecs: List[Array2d] = model.ops.unpad(torch2xp(torch_tokvecs), lengths)
tokvecs = [arr[1:-1] for arr in tokvecs] # remove the BOS and EOS markers
def backprop(d_tokvecs: List[Array2d]) -> ArgsKwargs:
# Restore entries for BOS and EOS markers
row = model.ops.alloc2f(1, d_tokvecs[0].shape[1])
d_tokvecs = [model.ops.xp.vstack((row, arr, row)) for arr in d_tokvecs]
return ArgsKwargs(
args=(torch_tokvecs,),
kwargs={"grad_tensors": xp2torch(model.ops.pad(d_tokvecs))},
)
return tokvecs, backprop
```
The model returned by `AutoModel.from_pretrained` is a PyTorch model we can wrap with Thinc's `PyTorchWrapper`. The converter functions tell Thinc how to transform the inputs and outputs.
```
import thinc
from thinc.api import PyTorchWrapper
from transformers import AutoModel
@thinc.registry.layers("transformers_model.v1")
def Transformer(name: str) -> Model[TokensPlus, List[Array2d]]:
return PyTorchWrapper(
AutoModel.from_pretrained(name),
convert_inputs=convert_transformer_inputs,
convert_outputs=convert_transformer_outputs,
)
```
We can now combine the `TransformersTokenizer` and `Transformer` into a feed-forward network using the `chain` combinator. The `with_array` layer transforms a sequence of data into a contiguous 2d array on the way into and
out of a model.
```
from thinc.api import chain, with_array, Softmax
@thinc.registry.layers("TransformersTagger.v1")
def TransformersTagger(starter: str, n_tags: int = 17) -> Model[List[List[str]], List[Array2d]]:
return chain(
TransformersTokenizer(starter),
Transformer(starter),
with_array(Softmax(n_tags)),
)
```
---
## Training the model
### Setting up model and data
Since we've registered all layers via `@thinc.registry.layers`, we can construct the model, its settings and other functions we need from a config (see `CONFIG` above). The result is a config object with a model, an optimizer, a function to calculate the loss and the training settings.
```
from thinc.api import Config, registry
C = registry.resolve(Config().from_str(CONFIG))
C
model = C["model"]
optimizer = C["optimizer"]
calculate_loss = C["loss"]
cfg = C["training"]
```
We’ve prepared a separate package [`ml-datasets`](https://github.com/explosion/ml-datasets) with loaders for some common datasets, including the AnCora data. If we're using a GPU, calling `ops.asarray` on the outputs ensures that they're converted to `cupy` arrays (instead of `numpy` arrays). Calling `Model.initialize` with a batch of inputs and outputs allows Thinc to **infer the missing dimensions**.
```
import ml_datasets
(train_X, train_Y), (dev_X, dev_Y) = ml_datasets.ud_ancora_pos_tags()
train_Y = list(map(model.ops.asarray, train_Y)) # convert to cupy if needed
dev_Y = list(map(model.ops.asarray, dev_Y)) # convert to cupy if needed
model.initialize(X=train_X[:5], Y=train_Y[:5])
```
### Helper functions for training and evaluation
Before we can train the model, we also need to set up the following helper functions for batching and evaluation:
* **`minibatch_by_words`:** Group pairs of sequences into minibatches under `max_words` in size, considering padding. The size of a padded batch is the length of its longest sequence multiplied by the number of elements in the batch.
* **`evaluate_sequences`:** Evaluate the model sequences of two-dimensional arrays and return the score.
```
def minibatch_by_words(pairs, max_words):
pairs = list(zip(*pairs))
pairs.sort(key=lambda xy: len(xy[0]), reverse=True)
batch = []
for X, Y in pairs:
batch.append((X, Y))
n_words = max(len(xy[0]) for xy in batch) * len(batch)
if n_words >= max_words:
yield batch[:-1]
batch = [(X, Y)]
if batch:
yield batch
def evaluate_sequences(model, Xs: List[Array2d], Ys: List[Array2d], batch_size: int) -> float:
correct = 0.0
total = 0.0
for X, Y in model.ops.multibatch(batch_size, Xs, Ys):
Yh = model.predict(X)
for yh, y in zip(Yh, Y):
correct += (y.argmax(axis=1) == yh.argmax(axis=1)).sum()
total += y.shape[0]
return float(correct / total)
```
### The training loop
Transformers often learn best with **large batch sizes** – larger than fits in GPU memory. But you don't have to backprop the whole batch at once. Here we consider the "logical" batch size (number of examples per update) separately from the physical batch size. For the physical batch size, what we care about is the **number of words** (considering padding too). We also want to sort by length, for efficiency.
At the end of the batch, we **call the optimizer** with the accumulated gradients, and **advance the learning rate schedules**. You might want to evaluate more often than once per epoch – that's up to you.
```
from tqdm.notebook import tqdm
from thinc.api import fix_random_seed
fix_random_seed(0)
for epoch in range(cfg["n_epoch"]):
batches = model.ops.multibatch(cfg["batch_size"], train_X, train_Y, shuffle=True)
for outer_batch in tqdm(batches, leave=False):
for batch in minibatch_by_words(outer_batch, cfg["words_per_subbatch"]):
inputs, truths = zip(*batch)
guesses, backprop = model(inputs, is_train=True)
backprop(calculate_loss.get_grad(guesses, truths))
model.finish_update(optimizer)
optimizer.step_schedules()
score = evaluate_sequences(model, dev_X, dev_Y, cfg["batch_size"])
print(epoch, f"{score:.3f}")
```
If you like, you can call `model.to_disk` or `model.to_bytes` to save the model weights to a directory or a bytestring.
|
github_jupyter
|
!pip install "thinc>=8.0.0a0" transformers torch "ml_datasets>=0.2.0a0" "tqdm>=4.41"
from thinc.api import prefer_gpu, use_pytorch_for_gpu_memory
is_gpu = prefer_gpu()
print("GPU:", is_gpu)
if is_gpu:
use_pytorch_for_gpu_memory()
CONFIG = """
[model]
@layers = "TransformersTagger.v1"
starter = "bert-base-multilingual-cased"
[optimizer]
@optimizers = "Adam.v1"
[optimizer.learn_rate]
@schedules = "warmup_linear.v1"
initial_rate = 0.01
warmup_steps = 3000
total_steps = 6000
[loss]
@losses = "SequenceCategoricalCrossentropy.v1"
[training]
batch_size = 128
words_per_subbatch = 2000
n_epoch = 10
"""
from typing import Optional, List
from dataclasses import dataclass
import torch
@dataclass
class TokensPlus:
input_ids: torch.Tensor
token_type_ids: torch.Tensor
attention_mask: torch.Tensor
input_len: List[int]
overflowing_tokens: Optional[torch.Tensor] = None
num_truncated_tokens: Optional[torch.Tensor] = None
special_tokens_mask: Optional[torch.Tensor] = None
import thinc
from thinc.api import Model
from transformers import AutoTokenizer
@thinc.registry.layers("transformers_tokenizer.v1")
def TransformersTokenizer(name: str) -> Model[List[List[str]], TokensPlus]:
def forward(model, texts: List[List[str]], is_train: bool):
tokenizer = model.attrs["tokenizer"]
token_data = tokenizer.batch_encode_plus(
[(text, None) for text in texts],
add_special_tokens=True,
return_token_type_ids=True,
return_attention_masks=True,
return_input_lengths=True,
return_tensors="pt",
)
return TokensPlus(**token_data), lambda d_tokens: []
return Model("tokenizer", forward, attrs={"tokenizer": AutoTokenizer.from_pretrained(name)})
@thinc.registry.layers("transformers_model.v1")
def Transformer(name) -> Model[TokensPlus, List[Array2d]]:
return PyTorchWrapper(
AutoModel.from_pretrained(name),
convert_inputs=convert_transformer_inputs,
convert_outputs=convert_transformer_outputs,
)
from thinc.api import ArgsKwargs, torch2xp, xp2torch
from thinc.types import Array2d
def convert_transformer_inputs(model, tokens: TokensPlus, is_train):
kwargs = {
"input_ids": tokens.input_ids,
"attention_mask": tokens.attention_mask,
"token_type_ids": tokens.token_type_ids,
}
return ArgsKwargs(args=(), kwargs=kwargs), lambda dX: []
def convert_transformer_outputs(model, inputs_outputs, is_train):
layer_inputs, torch_outputs = inputs_outputs
torch_tokvecs: torch.Tensor = torch_outputs[0]
torch_outputs = None # free the memory as soon as we can
lengths = list(layer_inputs.input_len)
tokvecs: List[Array2d] = model.ops.unpad(torch2xp(torch_tokvecs), lengths)
tokvecs = [arr[1:-1] for arr in tokvecs] # remove the BOS and EOS markers
def backprop(d_tokvecs: List[Array2d]) -> ArgsKwargs:
# Restore entries for BOS and EOS markers
row = model.ops.alloc2f(1, d_tokvecs[0].shape[1])
d_tokvecs = [model.ops.xp.vstack((row, arr, row)) for arr in d_tokvecs]
return ArgsKwargs(
args=(torch_tokvecs,),
kwargs={"grad_tensors": xp2torch(model.ops.pad(d_tokvecs))},
)
return tokvecs, backprop
import thinc
from thinc.api import PyTorchWrapper
from transformers import AutoModel
@thinc.registry.layers("transformers_model.v1")
def Transformer(name: str) -> Model[TokensPlus, List[Array2d]]:
return PyTorchWrapper(
AutoModel.from_pretrained(name),
convert_inputs=convert_transformer_inputs,
convert_outputs=convert_transformer_outputs,
)
from thinc.api import chain, with_array, Softmax
@thinc.registry.layers("TransformersTagger.v1")
def TransformersTagger(starter: str, n_tags: int = 17) -> Model[List[List[str]], List[Array2d]]:
return chain(
TransformersTokenizer(starter),
Transformer(starter),
with_array(Softmax(n_tags)),
)
from thinc.api import Config, registry
C = registry.resolve(Config().from_str(CONFIG))
C
model = C["model"]
optimizer = C["optimizer"]
calculate_loss = C["loss"]
cfg = C["training"]
import ml_datasets
(train_X, train_Y), (dev_X, dev_Y) = ml_datasets.ud_ancora_pos_tags()
train_Y = list(map(model.ops.asarray, train_Y)) # convert to cupy if needed
dev_Y = list(map(model.ops.asarray, dev_Y)) # convert to cupy if needed
model.initialize(X=train_X[:5], Y=train_Y[:5])
def minibatch_by_words(pairs, max_words):
pairs = list(zip(*pairs))
pairs.sort(key=lambda xy: len(xy[0]), reverse=True)
batch = []
for X, Y in pairs:
batch.append((X, Y))
n_words = max(len(xy[0]) for xy in batch) * len(batch)
if n_words >= max_words:
yield batch[:-1]
batch = [(X, Y)]
if batch:
yield batch
def evaluate_sequences(model, Xs: List[Array2d], Ys: List[Array2d], batch_size: int) -> float:
correct = 0.0
total = 0.0
for X, Y in model.ops.multibatch(batch_size, Xs, Ys):
Yh = model.predict(X)
for yh, y in zip(Yh, Y):
correct += (y.argmax(axis=1) == yh.argmax(axis=1)).sum()
total += y.shape[0]
return float(correct / total)
from tqdm.notebook import tqdm
from thinc.api import fix_random_seed
fix_random_seed(0)
for epoch in range(cfg["n_epoch"]):
batches = model.ops.multibatch(cfg["batch_size"], train_X, train_Y, shuffle=True)
for outer_batch in tqdm(batches, leave=False):
for batch in minibatch_by_words(outer_batch, cfg["words_per_subbatch"]):
inputs, truths = zip(*batch)
guesses, backprop = model(inputs, is_train=True)
backprop(calculate_loss.get_grad(guesses, truths))
model.finish_update(optimizer)
optimizer.step_schedules()
score = evaluate_sequences(model, dev_X, dev_Y, cfg["batch_size"])
print(epoch, f"{score:.3f}")
| 0.810966 | 0.984321 |
# Troubleshooting, Tips, and Tricks
© 2019-2022, Anyscale. All Rights Reserved

## Getting Help
* The [#tutorial channel](https://ray-distributed.slack.com/archives/C011ML23W5B) on the [Ray Slack](https://ray-distributed.slack.com). [Click here](https://forms.gle/9TSdDYUgxYs8SA9e8) to join.
* [Email](mailto:[email protected])
Find an issue? Please report it!
* [GitHub issues](https://github.com/anyscale/academy/issues)
## For More Information on Ray and Anyscale
* [ray.io](https://ray.io): The Ray website. In particular:
* [Documentation](https://ray.readthedocs.io/en/latest/): The full Ray documentation
* [Blog](https://medium.com/distributed-computing-with-ray): The Ray blog
* [GitHub](https://github.com/ray-project/ray): The source code for Ray
* [anyscale.com](https://anyscale.com/): The company developing Ray and these tutorials. In particular:
* [Blog](https://anyscale.com/blog/): The Anyscale blog
* [Events](https://anyscale.com/events/): Online events, [Ray Summit](http://raysummit.org), and meetups
* [Academy](https://anyscale.com/academy/): Training for Ray and Anyscale products
* [Jobs](https://jobs.lever.co/anyscale): Yes, we're hiring!
* Community:
* [Ray Slack](ray-distributed.slack.com) ([Click here](https://forms.gle/9TSdDYUgxYs8SA9e8) to join): The best forum for help on Ray. Use the `#tutorials` channel to ask for help on these tutorials!
* [ray-dev mailing list](https://groups.google.com/forum/?nomobile=true#!forum/ray-dev)
* [@raydistributed](https://twitter.com/raydistributed)
* [@anyscalecompute](https://twitter.com/anyscalecompute)
## Troubleshooting
### ray.init() Fails
Suppose you get an error like this:
```
... INFO services.py:... -- Failed to connect to the redis server, retrying.
```
It probably means you are running a VPN on your machine. [At this time](https://github.com/ray-project/ray/issues/6573), you can't use `ray.init()` with a VPN running. You'll have to stop your VPN to run `ray.init()`, then once it finishes, you can restart your VPN.
If `ray.init()` still fails, it may be that old Redis processes are somehow still running. On MacOS and Linux systems, use a terminal window and run the following command, shown with example output:
```shell
$ ps -ef | grep redis
501 36029 1 0 1:53PM ?? 0:00.03 .../lib/python3.7/site-packages/ray/core/src/ray/thirdparty/redis/src/redis-server *:48044
501 36030 1 0 1:53PM ?? 0:00.02 .../lib/python3.7/site-packages/ray/core/src/ray/thirdparty/redis/src/redis-server *:42902
$ kill 36029 36039
```
Then try again.
### MacOS Annoyances
If `ray.init()` worked (for example, you see a message like _View the Ray dashboard at localhost:8265_) and you're using a Mac, you may get several annoying dialogs asking you if you want to allow incoming connections for `python` and/or `redis-server`. Click "Accept" for each one and they shouldn't appear again during this lesson. MacOS is trying to verify if these executables have been properly signed. Ray uses Redis. If you installed Python using Anaconda or other mechanism, then it probably isn't properly signed from the point of view of MacOS. To permanently fix this problem, [see this StackExchange post](https://apple.stackexchange.com/questions/3271/how-to-get-rid-of-firewall-accept-incoming-connections-dialog).
### "File has changed on disk"
On rare occasions, you'll get a warning that the notebook file you are viewing has change on disk. Usually, you'll just want to keep the file you're viewing in Jupyter, especially if you have already made changes, but reloading it is fine, too.
### "AssertionError" While Graphs Are Being Drawn
We've observed a few occasions when errors like the following are written in the notebook outputs (very long numbers truncated):
```
WARNING:param.dynamic_operation: Callable raised "AssertionError('New option id 43948... does not match any option trees in Store.custom_options.',)".
AssertionError: New option id 11997... does not match any option trees in Store.custom_options.
```
They are related to the graphing libraries we use and appear to be harmless.
### "ImportError: No module named gym"
If you are working on a laptop and using Anaconda, and you get import errors for modules like `gym`, e.g., _ImportError: No module named gym_, this probably indicates you have the wrong `python` and `pip` commands on your path ahead of the versions for your Conda environment. To verify, use the `which` command:
```shell
$ which -a python
/usr/bin/python
/Users/me/anaconda3/envs/anyscale-academy/bin/python
```
In this example, the built-in `python` is ahead of the correct Conda environment `python`.
To fix, try the following:
First, try activiting the `anyscale-academy` environment again:
```shell
conda activate anyscale-academy
```
(see the [README](../README.md) for more details about the Conda environment we are using).
Run `which -a python` again. Does it show your `~/anaconda3/envs/anyscale-academy/bin/python` first? If not, force the correct ordering by changing the path:
```shell
export PATH=$HOME/anaconda3/envs/anyscale-academy/bin:$PATH
```
Run the `which` command again to confirm the correct `python` is now first in the list.
### Saving Resources by Ending Sessions You No Longer Need
When doing lessons on a machine or cloud instance with low resources, close notebooks you no longer need and stop their kernels. Do the latter by clicking the left-hand icon with a white square inside a black circle. The tool tip says _Running Terminals and Kernels_. Click _SHUT DOWN_ for any notebooks you no longer need.
## Tips and Tricks
### Jupyter Tips
* **Learning Jupyter Lab:** Open _Help > JupyerLab Reference_ for a user guide and detailed information about Jupyter Lab. The _Help_ menu has other references, too.
* **Excessive Output:** If the output of a cell is very long, right click on it and select _Enable Scrolling for Outputs_.
### General Task and Actor Tips
* To create a task from a function or an actor from a class, annotate it with `@ray.remote`.
* Invoke tasks with `foo.remote(...)`
* Invoke actor constructors `MyClass.remote(...)`.
* Invoke actor methods with `my_instance.remote(...)`.
* Invocations return an `ObjectRef` for a _future_. Use `ray.get(id)` to return the value.
* However, `ray.get()` blocks, so consider using `ray.wait()` when waiting for a collection of futures, so as they become available, you can retrieve them with `ray.get()` (which won't block when we know the results are available), then process the results, all while waiting for the rest to finish.
* Pick functions to parallelize that do enough work so that the Ray "remoting" overhead is not significant. Very short functions will actually yield lower performance if convert to tasks.
* Similarly, avoid too many actors, as each one is pinned to memory until no longer needed.
#### Using Existing Functions and Classes
An existing function can be used as a task by defining a new task function that calls the original function. For example:
```python
def work(args):
do_work(...)
@ray.remote
def remote_work(args):
do_work(args)
```
This allows you to use either version, as appropriate.
Similarly, existing classes can be subclassed to create actors:
```python
class Counter():
def __init__(self, init_count):
self.count = init_count
def increment():
self.count +=1
return self.count
@ray.remote
class RemoteCounter(Counter):
def __init__(self, init_count):
super().__init__(init_count)
def get_count():
return self.count
```
Note that we added a `get_count()` method, because member attributes can't be accessed directly, in contrast with normal classes. Of course, you don't need to add _accessor_ methods unless you need them.
### Using the Ray Dashboard
#### Opening the Dashboard
As it executes, `ray.init` prints the dashboard URL.
You can get the URL later if needed using `ray.get_dashboard_url()`.
> **Tip:** In a notebook, use a cell with `f'http://{ray.get_dashboard_url()}'`. The full URL printed will be clickable.
#### Profiling Actors
The _Logical View_ offers a powerful and convenient way to profile actor performance using [flame graphs](http://www.brendangregg.com/flamegraphs.html). Details are in the [Dashboard docs](https://ray.readthedocs.io/en/latest/ray-dashboard.html#ray-dashboard).
This feature uses [py-spy](https://github.com/benfred/py-spy) to instrument and profile the application. Unfortunately, you may be asked to enter the `sudo` password to use this feature, because of the way it instruments processes. Currently, the only way to get this to work with the Dashboard launched from notebooks is to use _passwordless sudo_. On MacOS and Linux systems, it should be sufficient to add a line like the following the `/etc/sudoers` (edited using `sudo visudo`!):
```
yourusername ALL = (ALL) NOPASSWD: ALL
```
Carefully the consider the security implications of this change!!
The alternative is to run your Python application from a command line, then open the Dashboard. When you click a link to profile code, as discussed next, you'll be prompted for your password in the terminal. (The prompt could get mixed with other output from the program.) Enter your password there and the profiling will continue.
Using either approach, to profile with the Dashboard, click the _Logical View_ tab. It shows a list of actors that have been run or are running. Find the running actor that appears to be the one you want to profile. You'll see a line like this:
> Actor <hex_number> (Profile for 10s 30s 60s) Kill Actor
The _10s, 30s, 60s_ are links. Click the time you want (pick one that's shorter than the remaining execution time).
When it finishes, click _Profile results_. A new tab opens with the _speedscope_ view of the data, which shows a flame graph. You can learn more about navigating and using this tool at the [speedscope GitHub site](https://github.com/jlfwong/speedscope).
A lot of the data will be related to actor messaging and not normally interesting. Look at the following screen shot, cropped from a screen shot in [lesson 4]():

Note the arrow and pagination on the upper-right hand side. Sometimes navigating pages will take you to interesting data. Also, the _Left Heavy_ button on the upper-left hand side is clicked, so the view is zoomed into the interesting data about the `step()` method profiled here.
### Profiling Code with ray.timeline()
The other built-in way to profile performance uses `ray.timeline(file)` ([documentation](https://ray.readthedocs.io/en/latest/package-ref.html#ray.timeline)). It requires a Chrome web browser to view the data. This is the only way to profile tasks.
Use it as follows:
```
ray.timeline('timeline.txt')
my_long_task.remote(...) # task to profile
```
Then, open chrome://tracing in the Chrome web browser (only Chrome is supported) and click the _load_ button to load the file. To zoom in or out, click the asymmetric up-down arrow button. To move around, click the crossed arrow and drag a section in view. Click on a box in the timeline to see details about it.
Look for blocks corresponding to long-running tasks and look for idle periods, which reflect processing outside the context of Ray.
### Cleaning Up
When your driver script exists, it will implicitly call `ray.shutdown()`. This can be useful to do explicitly if you want to restart Ray, for example, in a notebook environment, like this:
```python
ray.shutdown()
ray.init(...)
```
|
github_jupyter
|
... INFO services.py:... -- Failed to connect to the redis server, retrying.
$ ps -ef | grep redis
501 36029 1 0 1:53PM ?? 0:00.03 .../lib/python3.7/site-packages/ray/core/src/ray/thirdparty/redis/src/redis-server *:48044
501 36030 1 0 1:53PM ?? 0:00.02 .../lib/python3.7/site-packages/ray/core/src/ray/thirdparty/redis/src/redis-server *:42902
$ kill 36029 36039
WARNING:param.dynamic_operation: Callable raised "AssertionError('New option id 43948... does not match any option trees in Store.custom_options.',)".
AssertionError: New option id 11997... does not match any option trees in Store.custom_options.
$ which -a python
/usr/bin/python
/Users/me/anaconda3/envs/anyscale-academy/bin/python
conda activate anyscale-academy
export PATH=$HOME/anaconda3/envs/anyscale-academy/bin:$PATH
def work(args):
do_work(...)
@ray.remote
def remote_work(args):
do_work(args)
class Counter():
def __init__(self, init_count):
self.count = init_count
def increment():
self.count +=1
return self.count
@ray.remote
class RemoteCounter(Counter):
def __init__(self, init_count):
super().__init__(init_count)
def get_count():
return self.count
yourusername ALL = (ALL) NOPASSWD: ALL
ray.timeline('timeline.txt')
my_long_task.remote(...) # task to profile
ray.shutdown()
ray.init(...)
| 0.431824 | 0.958809 |
<a href="https://colab.research.google.com/github/WuilsonEstacio/github-para-estadistica/blob/main/Histogramas_y_Distribuciones_y_bayes.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Histogramas y Distribuciones
https://relopezbriega.github.io/blog/2015/06/27/probabilidad-y-estadistica-con-python/
```
# Graficos embebidos.
%matplotlib inline
import numpy as np # importando numpy
from scipy import stats # importando scipy.stats
import pandas as pd # importando pandas
datos = np.random.randn(5, 4) # datos normalmente distribuidos
datos
# usando pandas
dataframe = pd.DataFrame(datos, index=['a', 'b', 'c', 'd', 'e'],
columns=['col1', 'col2', 'col3', 'col4'])
dataframe
```
# Distribución normal
```
import matplotlib.pyplot as plt # importando matplotlib
import seaborn as sns # importando seaborn
import pylab
# parametros esteticos de seaborn
sns.set_palette("deep", desat=.6)
sns.set_context(rc={"figure.figsize": (8, 4)})
mu, sigma = 0, 0.1 # media y desvio estandar
s = np.random.normal(mu, sigma, 1000) #creando muestra de datos
# histograma de distribución normal.
cuenta, cajas, ignorar = plt.hist(s, 30, normed=True, facecolor='g', alpha=0.75)
normal = plt.plot(cajas, 1/(sigma * np.sqrt(2 * np.pi))*np.exp( - (cajas - mu)**2 / (2 * sigma**2) ),linewidth=2, color='r')
```
Distribuciones simetricas y asimetricas
https://relopezbriega.github.io/blog/2015/06/27/probabilidad-y-estadistica-con-python/
Las distribuciones asimétricas suelen ser problemáticas, ya que la mayoría de los métodos estadísticos suelen estar desarrollados para distribuciones del tipo normal. Para salvar estos problemas se suelen realizar transformaciones a los datos para hacer a estas distribuciones más simétricas y acercarse a la distribución normal.
```
# Dibujando la distribucion Gamma
x = stats.gamma(3).rvs(5000)
gamma = plt.hist(x, 90, histtype="stepfilled", alpha=.9) # alpha= da transparencia, 90 da los cuadros
```
En este ejemplo podemos ver que la distribución gamma que dibujamos tiene una asimetria positiva.
# Cuartiles y diagramas de cajas
Los cuartiles son los tres valores de la variable estadística que dividen a un conjunto de datos ordenados en cuatro partes iguales. Q1, Q2 y Q3 determinan los valores correspondientes al 25%, al 50% y al 75% de los datos. Q2 coincide con la mediana.
```
# Ejemplo de grafico de cajas en python
datos_1 = np.random.normal(100, 10, 200)
datos_2 = np.random.normal(80, 30, 200)
datos_3 = np.random.normal(90, 20, 200)
datos_4 = np.random.normal(70, 25, 200)
datos_graf = [datos_1, datos_2, datos_3, datos_4]
# Creando el objeto figura
fig = plt.figure(1, figsize=(9, 6))
# Creando el subgrafico
ax = fig.add_subplot(111)
# creando el grafico de cajas
bp = ax.boxplot(datos_graf)
# visualizar mas facile los atípicos
for flier in bp['fliers']:
flier.set(marker='o', color='red', alpha=0.5)
# los puntos aislados son valores atípicos
```
# Regresiones
Las regresiones es una de las herramientas principales de la estadistica inferencial. El objetivo del análisis de regresión es describir la relación entre un conjunto de variables, llamadas variables dependientes, y otro conjunto de variables, llamadas variables independientes o explicativas. Más específicamente, el análisis de regresión ayuda a entender cómo el valor típico de la variable dependiente cambia cuando cualquiera de las variables independientes es cambiada, mientras que se mantienen las otras variables independientes fijas. El producto final del análisis de regresión es la estimación de una función de las variables independientes llamada la función de regresión. **La idea es que en base a esta función de regresión podamos hacer estimaciones sobre eventos futuros**.
La regresión lineal es una de las técnicas más simples y mayormente utilizadas en los análisis de regresiones. Hace suposiciones muy rígidas sobre la relación entre la variable dependiente y y variable independiente x. Asume que la relación va a tomar la forma:
**y=β0+β1∗x, y=ax+b**
Uno de los métodos más populares para realizar regresiones lineales es el de mínimos cuadrados ordinarios (OLS, por sus siglas en inglés), este método es el estimador más simple y común en la que los dos βs se eligen para minimizar el cuadrado de la distancia entre los valores estimados y los valores reales.
```
# importanto la api de statsmodels
import statsmodels.formula.api as smf
import statsmodels.api as sm
# Creando un DataFrame de pandas.
df = pd.read_csv('https://vincentarelbundock.github.io/Rdatasets/csv/datasets/longley.csv', index_col=0)
df.head() # longley dataset
# utilizando la api de formula de statsmodels
est = smf.ols(formula='Employed ~ GNP', data=df).fit()
est.summary() # Employed se estima en base a GNP.
```
Como podemos ver, el resumen que nos brinda statsmodels sobre nuestro modelo de regresión contiene bastante información sobre como se ajuste el modelo a los datos. Pasemos a explicar algunos de estos valores:
Dep. Variable: es la variable que estamos estimasdo.
Model: es el modelo que estamos utilizando.
R-squared: es el coeficiente de determinación, el cual mide cuan bien nuestra recta de regresion se aproxima a los datos reales.
Adj. R-squared: es el coeficiente anterior ajustado según el número de observaciones.
[95.0% Conf. Int.]: Los valores inferior y superior del intervalo de confianza del 95%.
coef: el valor estimado del coeficiente.
std err: el error estándar de la estimación del coeficiente.
Skew: una medida de la asimetria de los datos sobre la media.
Kurtosis: Una medida de la forma de la distribución. La curtosis compara la cantidad de datos cerca de la media con los que están más lejos de la media(en las colas).
```
# grafico de regresion. que tanto se ajusta el modelo a los datos.
y = df.Employed # Respuesta
X = df.GNP # Predictor
X = sm.add_constant(X) # agrega constante
X_1 = pd.DataFrame({'GNP': np.linspace(X.GNP.min(), X.GNP.max(), 100)})
X_1 = sm.add_constant(X_1)
y_reg = est.predict(X_1) # estimacion
plt.scatter(X.GNP, y, alpha=0.3) # grafica los puntos de datos
plt.ylim(30, 100) # limite de eje y
plt.xlabel("Producto bruto") # leyenda eje x
plt.ylabel("Empleo") # leyenda eje y
plt.title("Ajuste de regresion") # titulo del grafico
reg = plt.plot(X_1.GNP, y_reg, 'r', alpha=0.9) # linea de regresion
# Este último gráfico nos muestra el apalancamiento y la influencia de cada caso
from statsmodels.graphics.regressionplots import influence_plot
inf =influence_plot(est)
```
# La estadística bayesiana
La estadística bayesiana es un subconjunto del campo de la estadística en la que la evidencia sobre el verdadero estado de las cosas se expresa en términos de grados de creencia. Esta filosofía de tratar a las creencias como probabilidad es algo natural para los seres humanos. Nosotros la utilizamos constantemente a medida que interactuamos con el mundo y sólo vemos verdades parciales; necesitando reunir pruebas para formar nuestras creencias.
La diferencia fundamental entre la estadística clásica (frecuentista) y la bayesiana es el concepto de probabilidad. Para la estadística clásica es un concepto objetivo, que se encuentra en la naturaleza, mientras que para la estadística bayesiana se encuentra en el observador, siendo así un concepto subjetivo. De este modo, en estadística clásica solo se toma como fuente de información las muestras obtenidas. En el caso bayesiano, sin embargo, además de la muestra también juega un papel fundamental la información previa o externa que se posee en relación a los fenómenos que se tratan de modelar.
La estadística bayesiana está demostrando su utilidad en ciertas estimaciones basadas en el conocimiento subjetivo a priori y el hecho de permitir revisar esas estimaciones en función de la evidencia empírica es lo que está abriendo nuevas formas de hacer conocimiento. Una aplicación de esto son los clasificadores bayesianos que son frecuentemente usados en implementaciones de filtros de correo basura, que se adaptan con el uso. La estadística bayesiana es un tema muy interesante que merece un artículo en sí mismo.
Para entender más fácilmente como funciona la estadística bayesiana veamos un simple ejemplo del lanzamiento de una moneda. La idea principal de la inferencia bayesiana es que la noción de probabilidad cambia mientras más datos tengamos.
```
sns.set_context(rc={"figure.figsize": (11, 8)})
dist = stats.beta
n_trials = [0, 1, 2, 3, 4, 5, 8, 15, 50, 500]
data = stats.bernoulli.rvs(0.5, size=n_trials[-1])
x = np.linspace(0, 1, 100)
for k, N in enumerate(n_trials):
sx = plt.subplot(len(n_trials) / 2, 2, k + 1)
plt.xlabel("$p$, probabilidad de cara") \
if k in [0, len(n_trials) - 1] else None
plt.setp(sx.get_yticklabels(), visible=False)
heads = data[:N].sum()
y = dist.pdf(x, 1 + heads, 1 + N - heads)
plt.plot(x, y, label="lanzamientos observados %d,\n %d caras" % (N, heads))
plt.fill_between(x, 0, y, color="#348ABD", alpha=0.4)
plt.vlines(0.5, 0, 4, color="k", linestyles="--", lw=1)
leg = plt.legend()
leg.get_frame().set_alpha(0.4)
plt.autoscale(tight=True)
plt.suptitle("Actualizacion Bayesiana de probabilidades posterios",
y=1.02,
fontsize=14)
plt.tight_layout()
```
Como el gráfico de arriba muestra, cuando empezamos a observar nuevos datos nuestras probabilidades posteriores comienzan a cambiar y moverse. Eventualmente, a medida que observamos más y más datos (lanzamientos de monedas), nuestras probabilidades se acercan más y más hacia el verdadero valor de p = 0.5 (marcado por una línea discontinua).
Aquí termina este tutorial, espero que les haya sido util.
Saludos!
Este post fue escrito utilizando IPython notebook. Pueden descargar este notebook o ver su version estática en nbviewer.
|
github_jupyter
|
# Graficos embebidos.
%matplotlib inline
import numpy as np # importando numpy
from scipy import stats # importando scipy.stats
import pandas as pd # importando pandas
datos = np.random.randn(5, 4) # datos normalmente distribuidos
datos
# usando pandas
dataframe = pd.DataFrame(datos, index=['a', 'b', 'c', 'd', 'e'],
columns=['col1', 'col2', 'col3', 'col4'])
dataframe
import matplotlib.pyplot as plt # importando matplotlib
import seaborn as sns # importando seaborn
import pylab
# parametros esteticos de seaborn
sns.set_palette("deep", desat=.6)
sns.set_context(rc={"figure.figsize": (8, 4)})
mu, sigma = 0, 0.1 # media y desvio estandar
s = np.random.normal(mu, sigma, 1000) #creando muestra de datos
# histograma de distribución normal.
cuenta, cajas, ignorar = plt.hist(s, 30, normed=True, facecolor='g', alpha=0.75)
normal = plt.plot(cajas, 1/(sigma * np.sqrt(2 * np.pi))*np.exp( - (cajas - mu)**2 / (2 * sigma**2) ),linewidth=2, color='r')
# Dibujando la distribucion Gamma
x = stats.gamma(3).rvs(5000)
gamma = plt.hist(x, 90, histtype="stepfilled", alpha=.9) # alpha= da transparencia, 90 da los cuadros
# Ejemplo de grafico de cajas en python
datos_1 = np.random.normal(100, 10, 200)
datos_2 = np.random.normal(80, 30, 200)
datos_3 = np.random.normal(90, 20, 200)
datos_4 = np.random.normal(70, 25, 200)
datos_graf = [datos_1, datos_2, datos_3, datos_4]
# Creando el objeto figura
fig = plt.figure(1, figsize=(9, 6))
# Creando el subgrafico
ax = fig.add_subplot(111)
# creando el grafico de cajas
bp = ax.boxplot(datos_graf)
# visualizar mas facile los atípicos
for flier in bp['fliers']:
flier.set(marker='o', color='red', alpha=0.5)
# los puntos aislados son valores atípicos
# importanto la api de statsmodels
import statsmodels.formula.api as smf
import statsmodels.api as sm
# Creando un DataFrame de pandas.
df = pd.read_csv('https://vincentarelbundock.github.io/Rdatasets/csv/datasets/longley.csv', index_col=0)
df.head() # longley dataset
# utilizando la api de formula de statsmodels
est = smf.ols(formula='Employed ~ GNP', data=df).fit()
est.summary() # Employed se estima en base a GNP.
# grafico de regresion. que tanto se ajusta el modelo a los datos.
y = df.Employed # Respuesta
X = df.GNP # Predictor
X = sm.add_constant(X) # agrega constante
X_1 = pd.DataFrame({'GNP': np.linspace(X.GNP.min(), X.GNP.max(), 100)})
X_1 = sm.add_constant(X_1)
y_reg = est.predict(X_1) # estimacion
plt.scatter(X.GNP, y, alpha=0.3) # grafica los puntos de datos
plt.ylim(30, 100) # limite de eje y
plt.xlabel("Producto bruto") # leyenda eje x
plt.ylabel("Empleo") # leyenda eje y
plt.title("Ajuste de regresion") # titulo del grafico
reg = plt.plot(X_1.GNP, y_reg, 'r', alpha=0.9) # linea de regresion
# Este último gráfico nos muestra el apalancamiento y la influencia de cada caso
from statsmodels.graphics.regressionplots import influence_plot
inf =influence_plot(est)
sns.set_context(rc={"figure.figsize": (11, 8)})
dist = stats.beta
n_trials = [0, 1, 2, 3, 4, 5, 8, 15, 50, 500]
data = stats.bernoulli.rvs(0.5, size=n_trials[-1])
x = np.linspace(0, 1, 100)
for k, N in enumerate(n_trials):
sx = plt.subplot(len(n_trials) / 2, 2, k + 1)
plt.xlabel("$p$, probabilidad de cara") \
if k in [0, len(n_trials) - 1] else None
plt.setp(sx.get_yticklabels(), visible=False)
heads = data[:N].sum()
y = dist.pdf(x, 1 + heads, 1 + N - heads)
plt.plot(x, y, label="lanzamientos observados %d,\n %d caras" % (N, heads))
plt.fill_between(x, 0, y, color="#348ABD", alpha=0.4)
plt.vlines(0.5, 0, 4, color="k", linestyles="--", lw=1)
leg = plt.legend()
leg.get_frame().set_alpha(0.4)
plt.autoscale(tight=True)
plt.suptitle("Actualizacion Bayesiana de probabilidades posterios",
y=1.02,
fontsize=14)
plt.tight_layout()
| 0.418103 | 0.975969 |
# Plot of stride cycle data
> [Laboratory of Biomechanics and Motor Control](http://pesquisa.ufabc.edu.br/bmclab)
> Federal University of ABC, Brazil
```
import numpy as np
import pandas as pd
%matplotlib notebook
import matplotlib
import matplotlib.pyplot as plt
from matplotlib.ticker import MaxNLocator
#matplotlib.rcParams['figure.dpi']=300 # inline figure resolution
#matplotlib.rc("savefig", dpi=300) # saved figure resolution
import seaborn as sns
from ipywidgets import widgets, Layout
import glob
import sys, os
from scipy import signal
from scipy.signal import butter, filtfilt
sys.path.insert(1, './../functions')
from linear_envelope import linear_envelope
from tnorm import tnorm
```
## Helper functions
```
def moving_rms(x, window):
"""Moving RMS of 'x' with window size 'window'.
"""
window = np.around(window).astype(np.int)
return np.sqrt(np.convolve(x*x, np.ones(window)/window, 'same'))
def filter2(df, freq=1000, filt=True, fname=''):
"""Filter data
"""
fc = 10
# EMG
for i in [0, 19]:
#df.iloc[:, i] = linear_envelope(df.iloc[:, i].values, freq, Fc_bp=np.array([20, 400]), Fc_lp=fc)
df.iloc[:, i] = moving_rms(df.iloc[:, i].values, window=freq/fc)
df.iloc[:, i] = df.iloc[:, i]/df.iloc[:, i].max()
# Accelerometer, Gyroscope, Magnetometer
ch = np.arange(1, 38).tolist()
ch.remove(19)
if filt:
for i in ch:
b, a = butter(4, (fc/(freq/2)), btype = 'lowpass')
df.iloc[:, i] = filtfilt(b, a, df.iloc[:, i])
return df
def tnorm2(df, evs, fname=''):
"""Time normalization of data
sn = ['EMG_taR',
'ACCx_taR', 'ACCy_taR', 'ACCz_taR', 'GYRx_taR', 'GYRy_taR', 'GYRz_taR', 'MAGx_taR', 'MAGy_taR', 'MAGz_taR',
'ACCx_tbR', 'ACCy_tbR', 'ACCz_tbR', 'GYRx_tbR', 'GYRy_tbR', 'GYRz_tbR', 'MAGx_tbR', 'MAGy_tbR', 'MAGz_tbR',
'EMG_taL',
'ACCx_taL', 'ACCy_taL', 'ACCz_taL', 'GYRx_taL', 'GYRy_taL', 'GYRz_taL', 'MAGx_taL', 'MAGy_taL', 'MAGz_taL',
'ACCx_tbL', 'ACCy_tbL', 'ACCz_tbL', 'GYRx_tbL', 'GYRy_tbL', 'GYRz_tbL', 'MAGx_tbL', 'MAGy_tbL', 'MAGz_tbL',
'FSR_hsR', 'FSR_toR',
'FSR_hsL', 'FSR_toL']
"""
right = list(range( 0, 19)) + [38, 39]
left = list(range(19, 38)) + [40, 41]
data = np.empty((101, df.shape[1], evs.shape[0]-1))
data.fill(np.nan)
pages0 = []
for ev in evs.index[:-1]:
if not evs.iloc[[ev, ev+1]].isnull().values.any():
data[:, right, ev], tn, indie = tnorm(df.iloc[int(evs.RHS[ev]):int(evs.RHS[ev+1]+1), right].values,
axis=0, step=1, k=3, smooth=0, mask=None,
nan_at_ext='delete', show=False, ax=None)
data[:, left, ev], tn, indie = tnorm(df.iloc[int(evs.LHS[ev]):int(evs.LHS[ev+1]+1), left].values,
axis=0, step=1, k=3, smooth=0, mask=None,
nan_at_ext='delete', show=False, ax=None)
else:
pages0.append(ev)
data = np.delete(data, pages0, axis=2)
return data, tn
def ensemble(data, fname='', mean=True):
"""Ensemble average of data
"""
if mean:
ym = np.mean(data, axis=2)
yq1, yq3 = np.std(data, axis=2, ddof=1), np.std(data, axis=2, ddof=1)
else:
ym = np.median(data, axis=2)
yq1, yq3 = ym-np.percentile(data, 25, 2), np.percentile(data, 75, 2)-ym
return ym, yq1, yq3
def calc_spatio_tempo(df):
""" calculate spatio-temporal variables
RHS RHO LTS LTO LHS LHO RTS RTO
"""
support_l = (df.LTO[1:] - df.LHS[:-1].values).dropna().mean()/(df.LHS.diff().dropna()).mean()
support_r = (df.RTO[:-1] - df.RHS[:-1].values).dropna().mean()/(df.RHS.diff().dropna()).mean()
#support_l = ((df.LTO[1:] - df.LHS[:-1].values)/df.LHS.diff().values[1:]).dropna()
#support_r = ((df.RTO[:-1] - df.RHS[:-1].values)/df.RHS.diff().values[1:]).dropna()
return [100*support_l, 100*support_r]
def process_data(path2, trials, mean=True):
"""Process given data trials
"""
datas = np.empty((101, 42, 1))
support = []
for trial in trials:
df = read_data(os.path.join(path2, trial), debug=False)
evs = read_events(os.path.join(path2, trial[:-4] + 'ev' + trial[5:]), debug=False)
df = filter2(df, fname=trial)
data, tn = tnorm2(df, evs, fname=trial)
datas = np.append(datas, data, axis=2)
support.append(calc_spatio_tempo(evs))
datas = np.delete(datas, 0, axis=2)
ym, yq1, yq3 = ensemble(datas, fname=trials, mean=mean)
#support = np.mean(np.array(support), axis=0)
return tn, ym, yq1, yq3, datas, support
```
## Information about the data files
```
# path of the dataset files
path2 = r'/mnt/DATA/X/GEDS/figshare'
# data sampling frequency
freq = 1000
# headers in the data files
sn = ['Time',
'EMG_taR',
'ACCx_taR', 'ACCy_taR', 'ACCz_taR', 'GYRx_taR', 'GYRy_taR', 'GYRz_taR',
'MAGx_taR', 'MAGy_taR', 'MAGz_taR',
'ACCx_tbR', 'ACCy_tbR', 'ACCz_tbR', 'GYRx_tbR', 'GYRy_tbR', 'GYRz_tbR',
'MAGx_tbR', 'MAGy_tbR', 'MAGz_tbR',
'EMG_taL',
'ACCx_taL', 'ACCy_taL', 'ACCz_taL', 'GYRx_taL', 'GYRy_taL', 'GYRz_taL',
'MAGx_taL', 'MAGy_taL', 'MAGz_taL',
'ACCx_tbL', 'ACCy_tbL', 'ACCz_tbL', 'GYRx_tbL', 'GYRy_tbL', 'GYRz_tbL',
'MAGx_tbL', 'MAGy_tbL', 'MAGz_tbL',
'FSR_hsR', 'FSR_toR',
'FSR_hsL', 'FSR_toL']
# files in the directory:
fnames = glob.glob(os.path.join(path2, 'S*[0-9]', 's*[1-6].txt'), recursive=True)
fnames.sort()
print('Number of data files:', len(fnames))
fnames_ev = glob.glob(os.path.join(path2, 'S*[0-9]', 's*ev.txt'), recursive=True)
fnames_ev.sort()
print('Number of event files:', len(fnames_ev))
```
## Read data
```
def read_data(fname, index_col=0, debug=False):
"""read resampled data from Desiree project.
"""
df = pd.read_csv(fname, sep='\t', header=0, index_col=index_col, dtype=np.float64, engine='c')
if debug:
print('Data shape: ({0}, {1})'.format(*df.shape))
return df
df = read_data(fnames[0], debug=True)
def read_events(fname, drop_ext=True, debug=False):
"""read file with events data from Desiree project.
RHS RHO LTS LTO LHS LHO RTS RTO
"""
# the parameter na_values=0 below will replace 0 by NaN
df = pd.read_csv(fname, sep='\t', header=0, index_col=None, na_values=0, engine='c')
# drop first and last strides
if drop_ext:
df = df.iloc[1:-1]
df.reset_index(drop=True, inplace=True)
if debug:
print('Event data shape: ({0}, {1})'.format(*df.shape))
return df
evs = read_events(fnames_ev[0], debug=True)
```
## Process data in dataset
**This step is going to take a few minutes to run**
```
fnames = glob.glob(os.path.join(path2, 'S*[0-9]', 's*[1-6].txt'))
fnames = [os.path.basename(fname) for fname in fnames]
fnames.sort()
subjects = list(set([fname[:3] for fname in fnames]))
subjects.sort()
ym_a, yq1_a, yq3_a, datas_a, support_a = [], [], [], [], []
for subject in subjects:
print(' ', subject, end='')
for speed in ['c', 's', 'f']:
print(speed, end='')
trials = [fname for fname in fnames if fname[:4]==subject+speed]
path_subj = os.path.join(path2, subject.upper())
tn, ym, yq1, yq3, datas, support = process_data(path_subj, trials, mean=True)
ym_a.append(ym)
yq1_a.append(yq1)
yq3_a.append(yq3)
datas_a.append(datas)
support_a.append(support)
print('\n', 'Done.', end='\n')
# grand mean and sd (across subjects), don't include s00
ym_c, yq1_c, yq3_c = ensemble(np.dstack(datas_a[3+0::3]), mean=True)
ym_s, yq1_s, yq3_s = ensemble(np.dstack(datas_a[3+1::3]), mean=True)
ym_f, yq1_f, yq3_f = ensemble(np.dstack(datas_a[3+2::3]), mean=True)
gmeansd = [[ym_c, yq1_c, yq3_c], [ym_s, yq1_s, yq3_s], [ym_f, yq1_f, yq3_f]]
sup_c = np.vstack(support_a[3+0::3])
sup_s = np.vstack(support_a[3+1::3])
sup_f = np.vstack(support_a[3+2::3])
supm_c = np.mean(sup_c, axis=0)
supq1_c = supm_c - np.std(sup_c, axis=0, ddof=1)
supq3_c = supm_c + np.std(sup_c, axis=0, ddof=1)
supm_s = np.mean(sup_s, axis=0)
supq1_s = supm_s - np.std(sup_s, axis=0, ddof=1)
supq3_s = supm_s + np.std(sup_s, axis=0, ddof=1)
supm_f = np.mean(sup_f, axis=0)
supq1_f = supm_f - np.std(sup_f, axis=0, ddof=1)
supq3_f = supm_f + np.std(sup_f, axis=0, ddof=1)
sup_msd = [[supm_c, supq1_c, supq3_c],
[supm_s, supq1_s, supq3_s],
[supm_f, supq1_f, supq3_f]]
```
## Plot data
```
def plot_widget(path2, subjects, tn, ym_a, yq1_a, yq3_a, datas_a, gmeansd, sup_msd):
"""general plot widget of a pandas dataframe
"""
from ipywidgets import widgets
subject_w = widgets.Select(options=subjects, value=subjects[1], description='Subject',
layout=Layout(width='200px', height='80px'),
style = {'description_width': 'initial'})
speeds = [['Comfortable', 'Slow', 'Fast'], ['c', 's', 'f']]
speed_w = widgets.Select(options=speeds[0], value=speeds[0][0], description='Speed',
layout=Layout(width='200px', height='80px'),
style = {'description_width': 'initial'})
gmean_w = widgets.Checkbox(value=True, description='Plot grand mean',
layout=Layout(width='200px', height='20px'),
style = {'description_width': 'initial'})
mean_w = widgets.Checkbox(value=True, description='Plot Mean',
layout=Layout(width='200px', height='20px'),
style = {'description_width': 'initial'})
trials_w = widgets.Checkbox(value=False, description='Plot Trials',
layout=Layout(width='200px', height='20px'),
style = {'description_width': 'initial'})
vbox = widgets.VBox(children=[gmean_w, mean_w, trials_w])
container = widgets.HBox(children=[subject_w, speed_w, vbox])
display(container)
fig, axs = plt.subplots(7, 2, sharex='col', sharey='row', figsize=(9.5, 7.5))
plt.subplots_adjust(left=.1, right=.98, bottom=0.07, top=.94, hspace=.04, wspace=.08)
plt.show()
def plot(change):
for ax in axs.flatten():
ax.clear()
fs = 10
axs[0, 0].set_title('Left Leg', fontsize=11, y=1.16)
axs[0, 1].set_title('Right Leg', fontsize=11, y=1.16)
axs[0, 0].set_ylabel('T.A. EMG\n(a.u.)', fontsize=fs)
axs[1, 0].set_ylabel('Accel. X\n(g)', fontsize=fs)
axs[2, 0].set_ylabel('Accel. Y\n(g)', fontsize=fs)
axs[3, 0].set_ylabel('Accel. Z\n(g)', fontsize=fs)
axs[4, 0].set_ylabel('Ang. Vel. X\n($^o$/s)', fontsize=fs)
axs[5, 0].set_ylabel('Ang. Vel. Y\n($^o$/s)', fontsize=fs)
axs[6, 0].set_ylabel('Ang. Vel. Z\n($^o$/s)', fontsize=fs)
axs[-1, 0].set_xlabel('Cycle (%)', fontsize=11)
axs[-1, 1].set_xlabel('Cycle (%)', fontsize=11)
axs[0, 0].set_xlim(0, 100)
axs[0, 1].set_xlim(0, 100)
subject = subject_w.index
speed = speed_w.index
channels = [0, 1, 2, 3, 4, 5, 6]
if mean_w.value:
cgl = [.2, .2, .2, 0.5]
cgml = [.2, .2, .2, 0.7]
cgr = [.2, .2, .2, 0.5]
cgmr = [.2, .2, .2, 0.7]
else:
cgl = [.8, .2, .2, 0.5]
cgml = [.8, .2, .2, 0.8]
cgr = [.2, .2, .8, 0.5]
cgmr = [.2, .2, .8, 0.8]
cdbl = [.8, 0, 0, 0.5]
cdbml = [.8, 0, 0, 0.8]
cdbr = [0, 0, .8, 0.5]
cdbmr = [0, 0, 0.8, 0.8]
ci = [.3, 0.8, .3, .2]
for c in channels:
axs[c, 0].axhline(0, alpha=0.5, lw=.5, ls='-.', color='k', zorder=0)
axs[c, 1].axhline(0, alpha=0.5, lw=.5, ls='-.', color='k', zorder=0)
if gmean_w.value:
axs[c, 0].axvspan(sup_msd[speed][1][0], sup_msd[speed][2][0],
facecolor=[.9, .9, .9, 1], edgecolor='none', zorder=1)
axs[c, 0].axvline(sup_msd[speed][0][0], lw=1, color=[.5, .5, .5], zorder=2)
axs[c, 1].axvspan(sup_msd[speed][1][1], sup_msd[speed][2][1], alpha=1,
facecolor=[.9, .9, .9, 1], edgecolor='none', zorder=1)
axs[c, 1].axvline(sup_msd[speed][0][1], lw=1, color=[.5, .5, .5], zorder=2)
ym, yq1, yq3 = gmeansd[speed][0], gmeansd[speed][1], gmeansd[speed][2]
axs[c, 0].fill_between(tn, ym[:, c+19]+yq3[:, c+19], ym[:, c+19]-yq1[:, c+19],
facecolor=cgl, edgecolor='none', zorder=3)
axs[c, 0].plot(tn, ym[:, c+19], color=cgml, lw=2, zorder=4)
axs[c, 1].fill_between(tn, ym[:, c]+yq3[:, c], ym[:, c]-yq1[:, c],
facecolor=cgr, edgecolor='none', zorder=3)
axs[c, 1].plot(tn, ym[:, c], color=cgmr, lw=2, zorder=4)
if mean_w.value:
ind = int(3*subject+speed)
ym, yq1, yq3 = ym_a[ind], yq1_a[ind], yq3_a[ind]
axs[c, 0].fill_between(tn, ym[:, c+19]+yq3[:, c+19], ym[:, c+19]-yq1[:, c+19],
facecolor=cdbl, edgecolor='none', zorder=5)
axs[c, 0].plot(tn, ym[:, c+19], color=cdbml, lw=2, zorder=6)
axs[c, 1].fill_between(tn, ym[:, c]+yq3[:, c], ym[:, c]-yq1[:, c],
facecolor=cdbr, edgecolor='none', zorder=5)
axs[c, 1].plot(tn, ym[:, c], color=cdbmr, lw=2, zorder=6)
if trials_w.value:
datas = datas_a[int(3*subject+speed)]
for i in range(datas.shape[2]):
axs[c, 0].plot(tn, datas[:, c+19, i], color=ci, lw=1, zorder=7)
axs[c, 1].plot(tn, datas[:, c, i], color=ci, lw=1, zorder=7)
for row in range(7):
axs[row, 0].yaxis.set_label_coords(-.12, 0.5)
axs[row, 0].yaxis.set_major_locator(MaxNLocator(3))
axs[-1, 0].xaxis.set_major_locator(MaxNLocator(4))
axs[-1, 1].xaxis.set_major_locator(MaxNLocator(4))
plt.text(x=0, y=1.05, s='LHS', fontsize=10, horizontalalignment='center', transform=axs[0, 0].transAxes)
plt.text(x=1, y=1.05, s='LHS', fontsize=10, horizontalalignment='center', transform=axs[0, 0].transAxes)
plt.text(x=0, y=1.05, s='RHS', fontsize=10, horizontalalignment='center', transform=axs[0, 1].transAxes)
plt.text(x=1, y=1.05, s='RHS', fontsize=10, horizontalalignment='center', transform=axs[0, 1].transAxes)
if gmean_w.value:
xl = sup_msd[speed][0][0]/100
plt.text(x=xl, y=1.05, s='LTO', fontsize=10,
horizontalalignment='center', transform=axs[0, 0].transAxes)
plt.text(x=xl, y=-.18, s=np.around(xl*100, 1), fontsize=10,
horizontalalignment='center', transform=axs[-1, 0].transAxes)
xr = sup_msd[speed][0][1]/100
plt.text(x=xr, y=1.05, s='RTO', fontsize=10,
horizontalalignment='center', transform=axs[0, 1].transAxes)
plt.text(x=xr, y=-.18, s=np.around(xr*100, 1), fontsize=10,
horizontalalignment='center', transform=axs[-1, 1].transAxes)
subject_w.observe(plot, names='value')
speed_w.observe(plot, names='value')
gmean_w.observe(plot, names='value')
mean_w.observe(plot, names='value')
trials_w.observe(plot, names='value')
plot(True)
plot_widget(path2, subjects, tn, ym_a, yq1_a, yq3_a, datas_a, gmeansd, sup_msd)
```
|
github_jupyter
|
import numpy as np
import pandas as pd
%matplotlib notebook
import matplotlib
import matplotlib.pyplot as plt
from matplotlib.ticker import MaxNLocator
#matplotlib.rcParams['figure.dpi']=300 # inline figure resolution
#matplotlib.rc("savefig", dpi=300) # saved figure resolution
import seaborn as sns
from ipywidgets import widgets, Layout
import glob
import sys, os
from scipy import signal
from scipy.signal import butter, filtfilt
sys.path.insert(1, './../functions')
from linear_envelope import linear_envelope
from tnorm import tnorm
def moving_rms(x, window):
"""Moving RMS of 'x' with window size 'window'.
"""
window = np.around(window).astype(np.int)
return np.sqrt(np.convolve(x*x, np.ones(window)/window, 'same'))
def filter2(df, freq=1000, filt=True, fname=''):
"""Filter data
"""
fc = 10
# EMG
for i in [0, 19]:
#df.iloc[:, i] = linear_envelope(df.iloc[:, i].values, freq, Fc_bp=np.array([20, 400]), Fc_lp=fc)
df.iloc[:, i] = moving_rms(df.iloc[:, i].values, window=freq/fc)
df.iloc[:, i] = df.iloc[:, i]/df.iloc[:, i].max()
# Accelerometer, Gyroscope, Magnetometer
ch = np.arange(1, 38).tolist()
ch.remove(19)
if filt:
for i in ch:
b, a = butter(4, (fc/(freq/2)), btype = 'lowpass')
df.iloc[:, i] = filtfilt(b, a, df.iloc[:, i])
return df
def tnorm2(df, evs, fname=''):
"""Time normalization of data
sn = ['EMG_taR',
'ACCx_taR', 'ACCy_taR', 'ACCz_taR', 'GYRx_taR', 'GYRy_taR', 'GYRz_taR', 'MAGx_taR', 'MAGy_taR', 'MAGz_taR',
'ACCx_tbR', 'ACCy_tbR', 'ACCz_tbR', 'GYRx_tbR', 'GYRy_tbR', 'GYRz_tbR', 'MAGx_tbR', 'MAGy_tbR', 'MAGz_tbR',
'EMG_taL',
'ACCx_taL', 'ACCy_taL', 'ACCz_taL', 'GYRx_taL', 'GYRy_taL', 'GYRz_taL', 'MAGx_taL', 'MAGy_taL', 'MAGz_taL',
'ACCx_tbL', 'ACCy_tbL', 'ACCz_tbL', 'GYRx_tbL', 'GYRy_tbL', 'GYRz_tbL', 'MAGx_tbL', 'MAGy_tbL', 'MAGz_tbL',
'FSR_hsR', 'FSR_toR',
'FSR_hsL', 'FSR_toL']
"""
right = list(range( 0, 19)) + [38, 39]
left = list(range(19, 38)) + [40, 41]
data = np.empty((101, df.shape[1], evs.shape[0]-1))
data.fill(np.nan)
pages0 = []
for ev in evs.index[:-1]:
if not evs.iloc[[ev, ev+1]].isnull().values.any():
data[:, right, ev], tn, indie = tnorm(df.iloc[int(evs.RHS[ev]):int(evs.RHS[ev+1]+1), right].values,
axis=0, step=1, k=3, smooth=0, mask=None,
nan_at_ext='delete', show=False, ax=None)
data[:, left, ev], tn, indie = tnorm(df.iloc[int(evs.LHS[ev]):int(evs.LHS[ev+1]+1), left].values,
axis=0, step=1, k=3, smooth=0, mask=None,
nan_at_ext='delete', show=False, ax=None)
else:
pages0.append(ev)
data = np.delete(data, pages0, axis=2)
return data, tn
def ensemble(data, fname='', mean=True):
"""Ensemble average of data
"""
if mean:
ym = np.mean(data, axis=2)
yq1, yq3 = np.std(data, axis=2, ddof=1), np.std(data, axis=2, ddof=1)
else:
ym = np.median(data, axis=2)
yq1, yq3 = ym-np.percentile(data, 25, 2), np.percentile(data, 75, 2)-ym
return ym, yq1, yq3
def calc_spatio_tempo(df):
""" calculate spatio-temporal variables
RHS RHO LTS LTO LHS LHO RTS RTO
"""
support_l = (df.LTO[1:] - df.LHS[:-1].values).dropna().mean()/(df.LHS.diff().dropna()).mean()
support_r = (df.RTO[:-1] - df.RHS[:-1].values).dropna().mean()/(df.RHS.diff().dropna()).mean()
#support_l = ((df.LTO[1:] - df.LHS[:-1].values)/df.LHS.diff().values[1:]).dropna()
#support_r = ((df.RTO[:-1] - df.RHS[:-1].values)/df.RHS.diff().values[1:]).dropna()
return [100*support_l, 100*support_r]
def process_data(path2, trials, mean=True):
"""Process given data trials
"""
datas = np.empty((101, 42, 1))
support = []
for trial in trials:
df = read_data(os.path.join(path2, trial), debug=False)
evs = read_events(os.path.join(path2, trial[:-4] + 'ev' + trial[5:]), debug=False)
df = filter2(df, fname=trial)
data, tn = tnorm2(df, evs, fname=trial)
datas = np.append(datas, data, axis=2)
support.append(calc_spatio_tempo(evs))
datas = np.delete(datas, 0, axis=2)
ym, yq1, yq3 = ensemble(datas, fname=trials, mean=mean)
#support = np.mean(np.array(support), axis=0)
return tn, ym, yq1, yq3, datas, support
# path of the dataset files
path2 = r'/mnt/DATA/X/GEDS/figshare'
# data sampling frequency
freq = 1000
# headers in the data files
sn = ['Time',
'EMG_taR',
'ACCx_taR', 'ACCy_taR', 'ACCz_taR', 'GYRx_taR', 'GYRy_taR', 'GYRz_taR',
'MAGx_taR', 'MAGy_taR', 'MAGz_taR',
'ACCx_tbR', 'ACCy_tbR', 'ACCz_tbR', 'GYRx_tbR', 'GYRy_tbR', 'GYRz_tbR',
'MAGx_tbR', 'MAGy_tbR', 'MAGz_tbR',
'EMG_taL',
'ACCx_taL', 'ACCy_taL', 'ACCz_taL', 'GYRx_taL', 'GYRy_taL', 'GYRz_taL',
'MAGx_taL', 'MAGy_taL', 'MAGz_taL',
'ACCx_tbL', 'ACCy_tbL', 'ACCz_tbL', 'GYRx_tbL', 'GYRy_tbL', 'GYRz_tbL',
'MAGx_tbL', 'MAGy_tbL', 'MAGz_tbL',
'FSR_hsR', 'FSR_toR',
'FSR_hsL', 'FSR_toL']
# files in the directory:
fnames = glob.glob(os.path.join(path2, 'S*[0-9]', 's*[1-6].txt'), recursive=True)
fnames.sort()
print('Number of data files:', len(fnames))
fnames_ev = glob.glob(os.path.join(path2, 'S*[0-9]', 's*ev.txt'), recursive=True)
fnames_ev.sort()
print('Number of event files:', len(fnames_ev))
def read_data(fname, index_col=0, debug=False):
"""read resampled data from Desiree project.
"""
df = pd.read_csv(fname, sep='\t', header=0, index_col=index_col, dtype=np.float64, engine='c')
if debug:
print('Data shape: ({0}, {1})'.format(*df.shape))
return df
df = read_data(fnames[0], debug=True)
def read_events(fname, drop_ext=True, debug=False):
"""read file with events data from Desiree project.
RHS RHO LTS LTO LHS LHO RTS RTO
"""
# the parameter na_values=0 below will replace 0 by NaN
df = pd.read_csv(fname, sep='\t', header=0, index_col=None, na_values=0, engine='c')
# drop first and last strides
if drop_ext:
df = df.iloc[1:-1]
df.reset_index(drop=True, inplace=True)
if debug:
print('Event data shape: ({0}, {1})'.format(*df.shape))
return df
evs = read_events(fnames_ev[0], debug=True)
fnames = glob.glob(os.path.join(path2, 'S*[0-9]', 's*[1-6].txt'))
fnames = [os.path.basename(fname) for fname in fnames]
fnames.sort()
subjects = list(set([fname[:3] for fname in fnames]))
subjects.sort()
ym_a, yq1_a, yq3_a, datas_a, support_a = [], [], [], [], []
for subject in subjects:
print(' ', subject, end='')
for speed in ['c', 's', 'f']:
print(speed, end='')
trials = [fname for fname in fnames if fname[:4]==subject+speed]
path_subj = os.path.join(path2, subject.upper())
tn, ym, yq1, yq3, datas, support = process_data(path_subj, trials, mean=True)
ym_a.append(ym)
yq1_a.append(yq1)
yq3_a.append(yq3)
datas_a.append(datas)
support_a.append(support)
print('\n', 'Done.', end='\n')
# grand mean and sd (across subjects), don't include s00
ym_c, yq1_c, yq3_c = ensemble(np.dstack(datas_a[3+0::3]), mean=True)
ym_s, yq1_s, yq3_s = ensemble(np.dstack(datas_a[3+1::3]), mean=True)
ym_f, yq1_f, yq3_f = ensemble(np.dstack(datas_a[3+2::3]), mean=True)
gmeansd = [[ym_c, yq1_c, yq3_c], [ym_s, yq1_s, yq3_s], [ym_f, yq1_f, yq3_f]]
sup_c = np.vstack(support_a[3+0::3])
sup_s = np.vstack(support_a[3+1::3])
sup_f = np.vstack(support_a[3+2::3])
supm_c = np.mean(sup_c, axis=0)
supq1_c = supm_c - np.std(sup_c, axis=0, ddof=1)
supq3_c = supm_c + np.std(sup_c, axis=0, ddof=1)
supm_s = np.mean(sup_s, axis=0)
supq1_s = supm_s - np.std(sup_s, axis=0, ddof=1)
supq3_s = supm_s + np.std(sup_s, axis=0, ddof=1)
supm_f = np.mean(sup_f, axis=0)
supq1_f = supm_f - np.std(sup_f, axis=0, ddof=1)
supq3_f = supm_f + np.std(sup_f, axis=0, ddof=1)
sup_msd = [[supm_c, supq1_c, supq3_c],
[supm_s, supq1_s, supq3_s],
[supm_f, supq1_f, supq3_f]]
def plot_widget(path2, subjects, tn, ym_a, yq1_a, yq3_a, datas_a, gmeansd, sup_msd):
"""general plot widget of a pandas dataframe
"""
from ipywidgets import widgets
subject_w = widgets.Select(options=subjects, value=subjects[1], description='Subject',
layout=Layout(width='200px', height='80px'),
style = {'description_width': 'initial'})
speeds = [['Comfortable', 'Slow', 'Fast'], ['c', 's', 'f']]
speed_w = widgets.Select(options=speeds[0], value=speeds[0][0], description='Speed',
layout=Layout(width='200px', height='80px'),
style = {'description_width': 'initial'})
gmean_w = widgets.Checkbox(value=True, description='Plot grand mean',
layout=Layout(width='200px', height='20px'),
style = {'description_width': 'initial'})
mean_w = widgets.Checkbox(value=True, description='Plot Mean',
layout=Layout(width='200px', height='20px'),
style = {'description_width': 'initial'})
trials_w = widgets.Checkbox(value=False, description='Plot Trials',
layout=Layout(width='200px', height='20px'),
style = {'description_width': 'initial'})
vbox = widgets.VBox(children=[gmean_w, mean_w, trials_w])
container = widgets.HBox(children=[subject_w, speed_w, vbox])
display(container)
fig, axs = plt.subplots(7, 2, sharex='col', sharey='row', figsize=(9.5, 7.5))
plt.subplots_adjust(left=.1, right=.98, bottom=0.07, top=.94, hspace=.04, wspace=.08)
plt.show()
def plot(change):
for ax in axs.flatten():
ax.clear()
fs = 10
axs[0, 0].set_title('Left Leg', fontsize=11, y=1.16)
axs[0, 1].set_title('Right Leg', fontsize=11, y=1.16)
axs[0, 0].set_ylabel('T.A. EMG\n(a.u.)', fontsize=fs)
axs[1, 0].set_ylabel('Accel. X\n(g)', fontsize=fs)
axs[2, 0].set_ylabel('Accel. Y\n(g)', fontsize=fs)
axs[3, 0].set_ylabel('Accel. Z\n(g)', fontsize=fs)
axs[4, 0].set_ylabel('Ang. Vel. X\n($^o$/s)', fontsize=fs)
axs[5, 0].set_ylabel('Ang. Vel. Y\n($^o$/s)', fontsize=fs)
axs[6, 0].set_ylabel('Ang. Vel. Z\n($^o$/s)', fontsize=fs)
axs[-1, 0].set_xlabel('Cycle (%)', fontsize=11)
axs[-1, 1].set_xlabel('Cycle (%)', fontsize=11)
axs[0, 0].set_xlim(0, 100)
axs[0, 1].set_xlim(0, 100)
subject = subject_w.index
speed = speed_w.index
channels = [0, 1, 2, 3, 4, 5, 6]
if mean_w.value:
cgl = [.2, .2, .2, 0.5]
cgml = [.2, .2, .2, 0.7]
cgr = [.2, .2, .2, 0.5]
cgmr = [.2, .2, .2, 0.7]
else:
cgl = [.8, .2, .2, 0.5]
cgml = [.8, .2, .2, 0.8]
cgr = [.2, .2, .8, 0.5]
cgmr = [.2, .2, .8, 0.8]
cdbl = [.8, 0, 0, 0.5]
cdbml = [.8, 0, 0, 0.8]
cdbr = [0, 0, .8, 0.5]
cdbmr = [0, 0, 0.8, 0.8]
ci = [.3, 0.8, .3, .2]
for c in channels:
axs[c, 0].axhline(0, alpha=0.5, lw=.5, ls='-.', color='k', zorder=0)
axs[c, 1].axhline(0, alpha=0.5, lw=.5, ls='-.', color='k', zorder=0)
if gmean_w.value:
axs[c, 0].axvspan(sup_msd[speed][1][0], sup_msd[speed][2][0],
facecolor=[.9, .9, .9, 1], edgecolor='none', zorder=1)
axs[c, 0].axvline(sup_msd[speed][0][0], lw=1, color=[.5, .5, .5], zorder=2)
axs[c, 1].axvspan(sup_msd[speed][1][1], sup_msd[speed][2][1], alpha=1,
facecolor=[.9, .9, .9, 1], edgecolor='none', zorder=1)
axs[c, 1].axvline(sup_msd[speed][0][1], lw=1, color=[.5, .5, .5], zorder=2)
ym, yq1, yq3 = gmeansd[speed][0], gmeansd[speed][1], gmeansd[speed][2]
axs[c, 0].fill_between(tn, ym[:, c+19]+yq3[:, c+19], ym[:, c+19]-yq1[:, c+19],
facecolor=cgl, edgecolor='none', zorder=3)
axs[c, 0].plot(tn, ym[:, c+19], color=cgml, lw=2, zorder=4)
axs[c, 1].fill_between(tn, ym[:, c]+yq3[:, c], ym[:, c]-yq1[:, c],
facecolor=cgr, edgecolor='none', zorder=3)
axs[c, 1].plot(tn, ym[:, c], color=cgmr, lw=2, zorder=4)
if mean_w.value:
ind = int(3*subject+speed)
ym, yq1, yq3 = ym_a[ind], yq1_a[ind], yq3_a[ind]
axs[c, 0].fill_between(tn, ym[:, c+19]+yq3[:, c+19], ym[:, c+19]-yq1[:, c+19],
facecolor=cdbl, edgecolor='none', zorder=5)
axs[c, 0].plot(tn, ym[:, c+19], color=cdbml, lw=2, zorder=6)
axs[c, 1].fill_between(tn, ym[:, c]+yq3[:, c], ym[:, c]-yq1[:, c],
facecolor=cdbr, edgecolor='none', zorder=5)
axs[c, 1].plot(tn, ym[:, c], color=cdbmr, lw=2, zorder=6)
if trials_w.value:
datas = datas_a[int(3*subject+speed)]
for i in range(datas.shape[2]):
axs[c, 0].plot(tn, datas[:, c+19, i], color=ci, lw=1, zorder=7)
axs[c, 1].plot(tn, datas[:, c, i], color=ci, lw=1, zorder=7)
for row in range(7):
axs[row, 0].yaxis.set_label_coords(-.12, 0.5)
axs[row, 0].yaxis.set_major_locator(MaxNLocator(3))
axs[-1, 0].xaxis.set_major_locator(MaxNLocator(4))
axs[-1, 1].xaxis.set_major_locator(MaxNLocator(4))
plt.text(x=0, y=1.05, s='LHS', fontsize=10, horizontalalignment='center', transform=axs[0, 0].transAxes)
plt.text(x=1, y=1.05, s='LHS', fontsize=10, horizontalalignment='center', transform=axs[0, 0].transAxes)
plt.text(x=0, y=1.05, s='RHS', fontsize=10, horizontalalignment='center', transform=axs[0, 1].transAxes)
plt.text(x=1, y=1.05, s='RHS', fontsize=10, horizontalalignment='center', transform=axs[0, 1].transAxes)
if gmean_w.value:
xl = sup_msd[speed][0][0]/100
plt.text(x=xl, y=1.05, s='LTO', fontsize=10,
horizontalalignment='center', transform=axs[0, 0].transAxes)
plt.text(x=xl, y=-.18, s=np.around(xl*100, 1), fontsize=10,
horizontalalignment='center', transform=axs[-1, 0].transAxes)
xr = sup_msd[speed][0][1]/100
plt.text(x=xr, y=1.05, s='RTO', fontsize=10,
horizontalalignment='center', transform=axs[0, 1].transAxes)
plt.text(x=xr, y=-.18, s=np.around(xr*100, 1), fontsize=10,
horizontalalignment='center', transform=axs[-1, 1].transAxes)
subject_w.observe(plot, names='value')
speed_w.observe(plot, names='value')
gmean_w.observe(plot, names='value')
mean_w.observe(plot, names='value')
trials_w.observe(plot, names='value')
plot(True)
plot_widget(path2, subjects, tn, ym_a, yq1_a, yq3_a, datas_a, gmeansd, sup_msd)
| 0.410166 | 0.850717 |
# Listen
In der Praxis sind Berechnungen häufig nicht nur für einen einzelnen Wert durchzuführen, sondern für mehrere gleichartige Werte. Als Beispiel kann eine Wohnung dienen, bei der der Abluftvolumenstrom für jeden einzelnen Abluftraum berechnet werden muss und verschiedene weitere Berechnungen davon ebenfalls betroffen sind.
Nehmen wir an, ein Bad ($45\frac{m^3}{h}$), zwei WCs je ($25\frac{m^3}{h}$), eine Abstellkammer ($25\frac{m^3}{h}$) und ein Saunaraum ($100\frac{m^3}{h}$) sollen mit Abluft versorgt werden.
Dann wären die erforderlichen Berechnungen für jeden einzelnen Raum durchzuführen. Dazu werden Listen benutzt:
`# Liste der Räume` <br>
`raum = ['Bad','WC_1','WC_2','Abstellkammer', 'Saunaraum']`
`# Liste der Abluftvolumenströme je Raum` <br>
`dV_ab = [45,25,25,25,100]`
Die Berechnungen können nun mit der Liste durchgeführt werden:
`dV_ges_ab = sum(dV_ab)`
## Beispiel
Berechnen Sie für die oben gegebenen Werte den Gesamtvolumenstrom `dV_ges_ab` und ermitteln Sie, wieviel Prozent der Abluft in jeden Raum geführt wird.
## Lösung
Zunächst werden die Listen angelegt.
Dann wird die Summe aller Listeneinträge gebildet und anschließend angezeigt:
```
raum = ['Bad','WC_1','WC_2','Abstellkammer', 'Saunaraum']
dV_ab = [45,25,25,25,100]
dV_ges_ab = sum(dV_ab)
dV_ges_ab
```
Für die Berechnung der Prozentzahlen muss eine neue Liste gebildet werden. Dazu wird oft eine `for`-Schleife verwendet.
Der Lösungsweg sieht ist prinzipiell:
```
# neue Liste initialisieren
neue_liste = []
# Schleife für die Berechnung der neue Werte
for wert in alte_liste:
neuer_wert = ...
neue_liste.append(neuer_wert)
# Ergebnis anzeigen
neue_liste
```
```
# leere Liste anlegen (initialisieren)
dV_prozent = []
# Schleife über alle Elemente von dV_ab
for dV in dV_ab:
# Berechnung der Prozentzahl für dies dV
prozent = dV/dV_ges_ab * 100
# Erweiterung der Liste dV_prozent
# um den berechneten Wert
dV_prozent.append(prozent)
# Ergebnis anzeigen
dV_prozent
```
Häufig ist die Berechnungsvorschrift sehr einfach. Dann ist es einfacher, für die gleiche Berechnung eine *List Comprehension* zu verwenden:
`neue_liste = [Berechnungs_vorschrift(wert) for wert in alte_liste]`
Solche List Comprehensions sind oft leicht zu lesen. Sie können meist deutlich schneller berechnet werden, als eine `for`-Schleife.
```
dV_prozent = [dV/dV_ges_ab * 100 for dV in dV_ab]
dV_prozent
```
Die Zahlenwerte der Liste `dV_prozent` müssen sich zu 100% aufsummieren:
```
sum(dV_prozent)
```
Zur Anzeige wird oft der sogenannte Reißverschluss (englisch zipper) verwendet. Dazu dient die Funktion
`zip(liste_1, liste_2, ...)`
```
list(zip(raum,dV_prozent)) # prozentuale Verteilung der Abluft auf die Räume
```
## Aufgabe
Berechnen Sie die Liste der erforderlichen Anschlussdurchmesser `d_erf # in mm` nach der Formel aus den gegebenen Abluftvolumenströmen in der Liste `dV_ab # in m**3/h`.
$$
d = \sqrt{\dfrac{4\,\dot V}{\pi\,v}}
$$
für jeden Abluftraum. Gehen Sie dabei von einer zulässigen Strömungsgeschwindigkeit `v=2 # m/s` in den Lüftungsrohren aus.
Achtung: Vergessen Sie die Einheitenumrechnung von $1h = 3600\,s$ und $1m=1000\,mm$ nicht!
```
import math
v = 2 # m/s
# Ihre Lösung beginnt hier
```
## Aufgabe
Sie können selbstgeschriebene Funktionen verwenden, um den Aufbau einer List Comprehension einfach zu halten.
Im folgenden habe ich im Modul `utilities.py` eine Funktion `ermittle_normdurchmesser(d)` angegeben, die zu einem vorgegebenen Durchmesser `d` in mm den nächstgrößeren Normdurchmesser ermittelt. Dabei ist die Liste der Normdurchmesser von einem Hersteller übernommen worden, siehe
https://www.msl-bauartikel.de/fileadmin/user_upload/produktbilder/Rohrsysteme/Lueftungsrohr/System_Lueftungsrohrde_eng_.pdf
Damit ergibt sich z.B. für `d=87 # mm`
`ermittle_normdurchmesser(d)` <br>
`90`
Benutzen Sie die Durchmesser, die Sie in der vorigen Aufgabe berechnet haben, um die zugehörigen Normdurchmesser zu ermitteln. Verwenden Sie für die Berechnung eine List Comprehension.
```
from utilities import ermittle_normdurchmesser
# Anwendungsbeispiel
ermittle_normdurchmesser(87)
# Ihre Lösung beginnt hier.
```
## Aufgabe
Die Liste der Normdurchmesser und der Wandstärken von Lüftungsrohren läßt sich folgendermaßen angeben:
```
normdurchmesser = [
80,90,100,112,125,140,
150,160,180,200,224,250,
280,300,315,355,400,450,
500,560,600,630,710,800,
900,1000,1120,1250,1400,
1600,1800,2000
]
# Um hier nicht 32 Werte von Hand eintragen zu müssen:
wandstaerken=2*[0.4]+12*[0.6]+7*[0.8]+4*[1.0]+3*[1.2]+4*[1.5]
# Das Ergebnis können Sie sich anzeigen lassen,
# wenn Sie vor der nächsten Zeile das Kommentarzeichen entfernen
#list(zip(normdurchmesser,wandstaerken))
```
Berechnen Sie das Gewicht je Meter Lüftungsrohr aus Wickelfalzrohr. Die Dichte von Stahl ist
$\varrho=7.85\,\frac{kg}{dm^3}$
Die Masse berechnet sich nach der Formel
$$
m = \varrho\,V
$$
mit dem Volumen
\begin{align}
V &= \dfrac{\pi \left(d_a^2 - d_i^2\right)}{4}\,\ell \\[2ex]
&= \pi\,d_m\,s\,\ell
\end{align}
Verwenden Sie eine `for`-Schleife für die Berechnung.
```
# Ihre Lösung beginnt hier
```
|
github_jupyter
|
raum = ['Bad','WC_1','WC_2','Abstellkammer', 'Saunaraum']
dV_ab = [45,25,25,25,100]
dV_ges_ab = sum(dV_ab)
dV_ges_ab
# neue Liste initialisieren
neue_liste = []
# Schleife für die Berechnung der neue Werte
for wert in alte_liste:
neuer_wert = ...
neue_liste.append(neuer_wert)
# Ergebnis anzeigen
neue_liste
# leere Liste anlegen (initialisieren)
dV_prozent = []
# Schleife über alle Elemente von dV_ab
for dV in dV_ab:
# Berechnung der Prozentzahl für dies dV
prozent = dV/dV_ges_ab * 100
# Erweiterung der Liste dV_prozent
# um den berechneten Wert
dV_prozent.append(prozent)
# Ergebnis anzeigen
dV_prozent
dV_prozent = [dV/dV_ges_ab * 100 for dV in dV_ab]
dV_prozent
sum(dV_prozent)
list(zip(raum,dV_prozent)) # prozentuale Verteilung der Abluft auf die Räume
import math
v = 2 # m/s
# Ihre Lösung beginnt hier
from utilities import ermittle_normdurchmesser
# Anwendungsbeispiel
ermittle_normdurchmesser(87)
# Ihre Lösung beginnt hier.
normdurchmesser = [
80,90,100,112,125,140,
150,160,180,200,224,250,
280,300,315,355,400,450,
500,560,600,630,710,800,
900,1000,1120,1250,1400,
1600,1800,2000
]
# Um hier nicht 32 Werte von Hand eintragen zu müssen:
wandstaerken=2*[0.4]+12*[0.6]+7*[0.8]+4*[1.0]+3*[1.2]+4*[1.5]
# Das Ergebnis können Sie sich anzeigen lassen,
# wenn Sie vor der nächsten Zeile das Kommentarzeichen entfernen
#list(zip(normdurchmesser,wandstaerken))
# Ihre Lösung beginnt hier
| 0.143038 | 0.91528 |
# Integer divisors
This Jupyter notebook provides an example of using the Python package [gravis](https://pypi.org/project/gravis). The .ipynb file can be found [here](https://github.com/robert-haas/gravis/tree/master/examples).
It demonstrates how **natural numbers** (positive integers) and their **divisibility relations** can be represented as a directed graph.
## References
- Wikipedia
- [Integer](https://en.wikipedia.org/wiki/Integer_(computer_science))
- [Divisor](https://en.wikipedia.org/wiki/Divisor)
- [Table of divisors](https://en.wikipedia.org/wiki/Table_of_divisors)
## Data generation
A graph of integers and their divisibility relation can be created from following basic ideas
- Each integer (up to some maximum) is represented by a node.
- If an integer is divisible (without remainder) by another integer, an edge is drawn from the smaller to the larger integer to represent their divisibility relation. Prime numbers are only divisible by 1 and themselves, hence they have zero incoming edges if these trivial cases are excluded, i.e. using integers from 2 upwards and not testing if an integer can be divided by itself.
- Different integers have quite different numbers of divisors. The number of divisors for a certain integer can be represented by node size (=more incoming edges means more divisors, shown by a larger node size) and additionally by node colors (a certain number of incoming edges is shown by a certain color).
```
import networkx as nx
import gravis as gv
start = 2
end = 100
# Create the graph: assign edges between divisible integers
graph = nx.DiGraph()
for i in range(start, end+1):
graph.add_node(i)
for j in range(start, i):
if i % j == 0:
graph.add_edge(j, i)
# Assign node properties: size, color, position
degree_to_color_map = {0: '#df4828', 1: '#6059a0', 2: '#69b190', 3: '#ddaa3c', 4: '#a6be54'}
for i in graph.nodes:
node = graph.nodes[i]
in_degree = graph.in_degree(i)
node['size'] = 10 + in_degree * 5
node['x'] = -2500 + i * 50
node['y'] = -i ** 1.618 + 1000
#node['y'] = node['size'] / 2
node['color'] = degree_to_color_map.get(in_degree, 'black')
node['hover'] = '{} has divisors'.format(i)
# Assign edge properties: color (from source node)
for e in graph.edges:
i, j = e
graph.edges[e]['color'] = graph.nodes[i]['color']
graph.nodes[j]['hover'] += ' {}'.format(i)
graph.graph['edge_opacity'] = 0.6
gv.d3(graph, node_hover_neighborhood=True, edge_curvature=0.75, zoom_factor=0.15, use_centering_force=False)
```
|
github_jupyter
|
import networkx as nx
import gravis as gv
start = 2
end = 100
# Create the graph: assign edges between divisible integers
graph = nx.DiGraph()
for i in range(start, end+1):
graph.add_node(i)
for j in range(start, i):
if i % j == 0:
graph.add_edge(j, i)
# Assign node properties: size, color, position
degree_to_color_map = {0: '#df4828', 1: '#6059a0', 2: '#69b190', 3: '#ddaa3c', 4: '#a6be54'}
for i in graph.nodes:
node = graph.nodes[i]
in_degree = graph.in_degree(i)
node['size'] = 10 + in_degree * 5
node['x'] = -2500 + i * 50
node['y'] = -i ** 1.618 + 1000
#node['y'] = node['size'] / 2
node['color'] = degree_to_color_map.get(in_degree, 'black')
node['hover'] = '{} has divisors'.format(i)
# Assign edge properties: color (from source node)
for e in graph.edges:
i, j = e
graph.edges[e]['color'] = graph.nodes[i]['color']
graph.nodes[j]['hover'] += ' {}'.format(i)
graph.graph['edge_opacity'] = 0.6
gv.d3(graph, node_hover_neighborhood=True, edge_curvature=0.75, zoom_factor=0.15, use_centering_force=False)
| 0.295942 | 0.976243 |
# Dispositivos de captura
Para hacer experimentos de visión artificial nos interesa acceder cómodamente a cualquier fuente de imágenes, almacenada localmente o disponible en un servidor remoto. Por ejemplo:
- archivos de imagen (jpg, png, tif, bmp, etc.)
- archivos de video (avi, mp4, etc.)
- secuencias "en vivo" tomadas de una webcam, cámara ip, o smartphone.
El paquete `umucv` simplifica esta tarea.
## autoStream
La función `mkStream(size,dev)` crea un generador de imágenes:
stream = mkStream(size,dev)
for frame in stream:
procesa frame
Para capturar el teclado podemos usar directamente `cv.waitKey` pocesando los códigos como deseemos.
Muchas veces es más cómodo usar `autoStream`, que permite hacer pausas (espacio) salvar frames (tecla s), salir (ESC), y admite opciones en la línea de órdenes para elegir el dispositivo de captura (`--dev`), la resolución deseada (`--size`), forzar un reescalado (`--resize`), repetición de un video (`--loop`) y ejecución frame a frame (`--step`).
for key, frame in autoStream():
if key = ord('c'):
procesa tecla
procesa frame
Por omisión el dispositivo es la cámara /dev/video0. Para cambiar de webcam y solicitar un tamaño hacemos:
./programa.py --dev=1 --size=320x240
La cámara del raspberry pi se especifica así:
./programa.py --dev=picam
Podemos procesar una lista de imágenes tomadas de un directorio:
./programa.py --dev=glob:../images/ccorr/scenes/*.png
Otra forma de hacer lo mismo, pero visualizando las imágenes disponibles en una ventana, avanzando de una en una con las teclas del ratón:
./programa.py --dev=dir:../images/ccorr/scenes/*.png
Se admiten archivos de vídeo (locales o remotos):
./programa.py --dev=../images/rot4.mjpg
También se admiten *streams* en vivo remotos, generados por [cámaras online](https://en.wikipedia.org/wiki/IP_camera#Video_standards). Por ejemplo, aquí vemos una playa:
./programa.py --dev=http://213.4.39.225:81/mjpg/video.mjpg
La *app* "IP Webcam" incluye un servidor mjpeg en los smartphones, que permite utilzarlos como fuente de imágenes para los ejercicios.
./programa.py --dev=http://155.54.X.Y:8080/video
## Servidores
Hemos incluido dos utilidades para crear "streams" en formato mjpeg:
- ./`mjpegserver.py`: crea un stream mjpeg a partir cualquier dispositivo de los anteriores. Por ejemplo:
./mjpegserver.py --dev=picam --size=320x240 --quality=50
Este stream se captura con
./programa.py --dev=http://<IP-DEL-SERVIDOR>:8087/cam.mjpg
- ./`vlcmjpeg.sh`: es un script de bash para llamar a `vlc` con cualquier fuente de video y generar un stream mjpeg:
./vlcmjpeg.sh https://www.youtube.com/watch?v=aBr2kKAHN6M
(Es conveniente abrir el vídeo primero con vlc normal para comprobar que se lee bien, y en su caso aceptar certificados.)
Este stream se captura con
./programa.py --dev=http://<IPDELSERVIDOR>:8090
Podemos transmitir la pantalla de nuestro ordenador:
./vlcmjpeg.sh 'screen:// :screen-fps=10 :screen-width=700 :screen-height=500 :screen-top=300'
O la webcam:
./vlcmjpeg.sh 'v4l2://'
En, general, podemos emitir cualquier fuente de video admitida por `vlc` y capturarla con `autoStream()`.
### Visualización en páginas web
Los documentos html permiten visualizar video directamente con elementos `<video>`. Para visualizar streams `mjpg` en vivo se usa directamente un elemento `<img src='http:...' >`.
## Control de la webcam
En la consola de linux usamos **v4l2-ctl**. Los controles disponibles en cada cámara se consultan con:
v4l2-ctl -l
Activar o desactivar la eliminación de oscilaciones producidas por la luz eléctrica:
v4l2-ctl -d /dev/video0 -c power_line_frequency=1
v4l2-ctl -d /dev/video0 -c power_line_frequency=0
Fijar el nivel de exposición:
v4l2-ctl -d /dev/video0 -c exposure_auto=1 -c exposure_absolute=100
v4l2-ctl -d /dev/video0 -c exposure_auto=1 -c exposure_absolute=1000
Exposición automática:
v4l2-ctl -d /dev/video0 -c exposure_auto=3
Enfoque fijo:
v4l2-ctl -d /dev/video0 -c focus=255
v4l2-ctl -d /dev/video0 -c focus=0
La aplicación *guvcview* permite modificar los parámetros con un interfaz gráfico.
El reproductor multimedia VLC (disponible en Linux y Windows) también lo permite.
|
github_jupyter
|
# Dispositivos de captura
Para hacer experimentos de visión artificial nos interesa acceder cómodamente a cualquier fuente de imágenes, almacenada localmente o disponible en un servidor remoto. Por ejemplo:
- archivos de imagen (jpg, png, tif, bmp, etc.)
- archivos de video (avi, mp4, etc.)
- secuencias "en vivo" tomadas de una webcam, cámara ip, o smartphone.
El paquete `umucv` simplifica esta tarea.
## autoStream
La función `mkStream(size,dev)` crea un generador de imágenes:
stream = mkStream(size,dev)
for frame in stream:
procesa frame
Para capturar el teclado podemos usar directamente `cv.waitKey` pocesando los códigos como deseemos.
Muchas veces es más cómodo usar `autoStream`, que permite hacer pausas (espacio) salvar frames (tecla s), salir (ESC), y admite opciones en la línea de órdenes para elegir el dispositivo de captura (`--dev`), la resolución deseada (`--size`), forzar un reescalado (`--resize`), repetición de un video (`--loop`) y ejecución frame a frame (`--step`).
for key, frame in autoStream():
if key = ord('c'):
procesa tecla
procesa frame
Por omisión el dispositivo es la cámara /dev/video0. Para cambiar de webcam y solicitar un tamaño hacemos:
./programa.py --dev=1 --size=320x240
La cámara del raspberry pi se especifica así:
./programa.py --dev=picam
Podemos procesar una lista de imágenes tomadas de un directorio:
./programa.py --dev=glob:../images/ccorr/scenes/*.png
Otra forma de hacer lo mismo, pero visualizando las imágenes disponibles en una ventana, avanzando de una en una con las teclas del ratón:
./programa.py --dev=dir:../images/ccorr/scenes/*.png
Se admiten archivos de vídeo (locales o remotos):
./programa.py --dev=../images/rot4.mjpg
También se admiten *streams* en vivo remotos, generados por [cámaras online](https://en.wikipedia.org/wiki/IP_camera#Video_standards). Por ejemplo, aquí vemos una playa:
./programa.py --dev=http://213.4.39.225:81/mjpg/video.mjpg
La *app* "IP Webcam" incluye un servidor mjpeg en los smartphones, que permite utilzarlos como fuente de imágenes para los ejercicios.
./programa.py --dev=http://155.54.X.Y:8080/video
## Servidores
Hemos incluido dos utilidades para crear "streams" en formato mjpeg:
- ./`mjpegserver.py`: crea un stream mjpeg a partir cualquier dispositivo de los anteriores. Por ejemplo:
./mjpegserver.py --dev=picam --size=320x240 --quality=50
Este stream se captura con
./programa.py --dev=http://<IP-DEL-SERVIDOR>:8087/cam.mjpg
- ./`vlcmjpeg.sh`: es un script de bash para llamar a `vlc` con cualquier fuente de video y generar un stream mjpeg:
./vlcmjpeg.sh https://www.youtube.com/watch?v=aBr2kKAHN6M
(Es conveniente abrir el vídeo primero con vlc normal para comprobar que se lee bien, y en su caso aceptar certificados.)
Este stream se captura con
./programa.py --dev=http://<IPDELSERVIDOR>:8090
Podemos transmitir la pantalla de nuestro ordenador:
./vlcmjpeg.sh 'screen:// :screen-fps=10 :screen-width=700 :screen-height=500 :screen-top=300'
O la webcam:
./vlcmjpeg.sh 'v4l2://'
En, general, podemos emitir cualquier fuente de video admitida por `vlc` y capturarla con `autoStream()`.
### Visualización en páginas web
Los documentos html permiten visualizar video directamente con elementos `<video>`. Para visualizar streams `mjpg` en vivo se usa directamente un elemento `<img src='http:...' >`.
## Control de la webcam
En la consola de linux usamos **v4l2-ctl**. Los controles disponibles en cada cámara se consultan con:
v4l2-ctl -l
Activar o desactivar la eliminación de oscilaciones producidas por la luz eléctrica:
v4l2-ctl -d /dev/video0 -c power_line_frequency=1
v4l2-ctl -d /dev/video0 -c power_line_frequency=0
Fijar el nivel de exposición:
v4l2-ctl -d /dev/video0 -c exposure_auto=1 -c exposure_absolute=100
v4l2-ctl -d /dev/video0 -c exposure_auto=1 -c exposure_absolute=1000
Exposición automática:
v4l2-ctl -d /dev/video0 -c exposure_auto=3
Enfoque fijo:
v4l2-ctl -d /dev/video0 -c focus=255
v4l2-ctl -d /dev/video0 -c focus=0
La aplicación *guvcview* permite modificar los parámetros con un interfaz gráfico.
El reproductor multimedia VLC (disponible en Linux y Windows) también lo permite.
| 0.649912 | 0.697003 |
# Introduction to Programming
Topics for today will include:
- Mozilla Developer Network [(MDN)](https://developer.mozilla.org/en-US/)
- Python Documentation [(Official Documentation)](https://docs.python.org/3/)
- Importance of Design
- Functions
- Built in Functions
## Mozilla Developer Network [(MDN)](https://developer.mozilla.org/en-US/)
---
The Mozilla Developer Network is a great resource for all things web dev. This site is good for trying to learn about standards as well as finding out quick information about something that you're trying to do Web Dev Wise
This will be a major resource going forward when it comes to doing things with HTML and CSS
You'll often find that you're not the first to try and do something. That being said you need to start to get comfortable looking for information on your own when things go wrong.
## Python Documentation [(Official Documentation)](https://docs.python.org/3/)
---
This section is similar to the one above. Python has a lot of resources out there that we can utilize when we're stuck or need some help with something that we may not have encountered before.
Since this is the official documentation page for the language you may often be given too much information or something that you wanted but in the wrong form or for the wrong version of the language. It is up to you to learn how to utilize these things and use them to your advantage.
## Importance of Design
---
So this is a topic that i didn't learn the importance of until I was in the work force. Design is a major influence in the way that code is build and in a major capacity and significant effect on the industry.
Let's pretend we have a client that wants us to do the following:
- Write a function which will count the number of times any one character appears in a string of characters.
- Write a main function which takes the character to be counted from the user and calls the function, outputting the result to the user.
For example, are you like Android and take the latest and greatest and put them into phones in an unregulated hardware market. Thus leaving great variability in the market for your brand? Are you an Apple where you control the full stack. Your hardware and software may not be bleeding edge but it's seamless and uniform.
What does the market want? What are you good at? Do you have people around you that can fill your gaps?
Here's a blurb from a friend about the matter:
>Design, often paired with the phrase "design thinking", is an approach and method of problem solving that builds empathy for user(s) of a product, resulting in the creation of a seamless and delightful user experience tailored to the user's needs.
>Design thinks holistically about the experience that a user would go through when encountering and interacting with a product or technology. Design understands the user and their needs in great detail so that the product team can build the product and experience that fits what the user is looking for. We don't want to create products for the sake of creating them, we want to ensure that there is a need for it by a user.
>Design not only focuses on the actual interface design of a product, but can also ensure the actual technology has a seamless experience as well. Anything that blocks potential users from wanting to buy a product or prohibits current users from utilizing the product successfully, design wants to investigate. We ensure all pieces fit together from the user's standpoint, and we work to build a bridge between the technology and the user, who doesn't need to understand the technical depths of the product.
### Sorting Example [(Toptal Sorting Algorithms)](https://www.toptal.com/developers/sorting-algorithms)
---
Hypothetical, a client comes to you and they want you sort a list of numbers how do you optimally sort a list? `[2, 5, 6, 1, 4, 3]`
### Design Thinking [(IBM Design Thinking)](https://www.ibm.com/design/thinking/)
---
As this idea starts to grow you come to realize that different companies have different design methodologies. IBM has it's own version of Design Thinking. You can find more information about that at the site linked in the title. IBM is very focused on being exactly like its customers in most aspects.
What we're mostly going to take from this is that there are entire careers birthed from thinking before you act. That being said we're going to harp on a couple parts of this.
### Knowing what your requirements are
---
One of the most common scenarios to come across is a product that is annouced that's going to change everything. In the planning phase everyone agrees that the idea is amazing and going to solve all of our problems.
We get down the line and things start to fall apart, we run out of time. Things ran late, or didn't come in in time pushing everything out.
Scope creep ensued.
This is typically the result of not agreeing on what our requirements are. Something as basic as agreeing on what needs to be done needs to be discussed and checked on thouroughly. We do this because often two people rarely are thinking exactly the same thing.
You need to be on the same page as your client and your fellow developers as well. If you don't know ask.
### Planning Things Out
---
We have an idea on what we want to do. So now we just write it? No, not quite. We need to have a rough plan on how we're going to do things. Do we want to use functions, do we need a quick solution, is this going to be verbose and complex?
It's important to look at what we can set up for ourselves. We don't need to make things difficult by planning things out poorly. This means allotting time for things like getting stuck and brainstorming.
### Breaking things down
---
Personally I like to take my problem and scale it down into an easy example so in the case of our problem. The client may want to process a text like Moby Dick. We can start with a sentence and work our way up!
Taking the time to break things in to multiple pieces and figure out what goes where is an art in itself.
```
def char_finder(character, string):
total = 0
for char in string:
if char == character:
total += 1
return total
if __name__ == "__main__":
output = char_finder('o', 'Quick brown fox jumped over the lazy dog')
print(output)
```
## Functions
---
This is a intergral piece of how we do things in any programming language. This allows us to repeat instances of code that we've seen and use them at our preferance.
We'll often be using functions similar to how we use variables and our data types.
### Making Our Own Functions
---
So to make a functions we'll be using the `def` keyword followed by a name and then parameters. We've seen this a couple times now in code examples.
```
def exampleName(exampleParameter1, exampleParameter2):
print(exampleParameter1, exampleParameter2)
```
There are many ways to write functions, we can say that we're going return a specific type of data type.
```
def exampleName(exampleParameter1, exampleParameter2) -> any:
print(exampleParameter1, exampleParameter2)
```
We can also specify the types that the parameters are going to be.
```
def exampleName(exampleParameter1: any, exampleParameter2: any) -> any:
print(exampleParameter1, exampleParameter2)
```
Writing functions is only one part of the fun. We still have to be able to use them.
### Using functions
---
Using functions is fairly simple. To use a function all we have to do is give the function name followed by parenthesis. This should seem familiar.
```
def exampleName(exampleParameter1: int, exampleParameter2: int) -> None:
# print(exampleParameter1, exampleParameter2)
return exampleParameter1 + exampleParameter2
print()
exampleName(10, 94)
```
### Functions In Classes
---
Now we've mentioned classes before, classes can have functions but they're used a little differently. Functions that stem from classes are used often with a dot notation.
```
class Person:
def __init__(self, weight: int, height: int, name: str):
self.weight = weight
self.height = height
self.name = name
def who_is_this(self):
print("This person's name is " + self.name)
print("This person's weight is " + str(self.weight) + " pounds")
print("This person's height is " + str(self.height) + " inches")
if __name__ == "__main__":
Kipp = Person(225, 70, "Aaron Kippins")
Kipp.who_is_this()
```
## Built in Functions and Modules
---
With the talk of dot notation those are often used with built in functions. Built in function are functions that come along with the language. These tend to be very useful because as we start to visit more complex issues they allow us to do complexs thing with ease in some cases.
We have functions that belong to particular classes or special things that can be done with things of a certain class type.
Along side those we can also have Modules. Modules are classes or functions that other people wrote that we can import into our code to use.
### Substrings
---
```
string = "I want to go home!"
print(string[0:12], "to Cancun!")
# print(string[0:1])
```
### toUpper toLower
---
```
alpha_sentence = 'Quick brown fox jumped over the lazy dog'
print(alpha_sentence.title())
print(alpha_sentence.upper())
print(alpha_sentence.lower())
if alpha_sentence.lower().islower():
print("sentence is all lowercase")
```
### Exponents
---
```
print(2 ** 5)
```
### math.sqrt()
---
```
import math
math.sqrt(4)
```
### Integer Division vs Float Division
---
```
print(4//2)
print(4/2)
```
### Abs()
---
```
abs(-10)
```
### String Manipulation
---
```
dummy_string = "Hey there I'm just a string for the example about to happen."
print(dummy_string.center(70, "-"))
print(dummy_string.partition("o"))
print(dummy_string.swapcase())
print(dummy_string.split(" "))
```
### Array Manipulation
---
```
arr = [2, 5, 6, 1, 4, 3]
arr.sort()
print(arr)
print(arr[3])
# sorted(arr)
print(arr[1:3])
```
### Insert and Pop, Append and Remove
---
```
arr.append(7)
print(arr)
arr.pop()
print(arr)
```
|
github_jupyter
|
def char_finder(character, string):
total = 0
for char in string:
if char == character:
total += 1
return total
if __name__ == "__main__":
output = char_finder('o', 'Quick brown fox jumped over the lazy dog')
print(output)
def exampleName(exampleParameter1, exampleParameter2):
print(exampleParameter1, exampleParameter2)
def exampleName(exampleParameter1, exampleParameter2) -> any:
print(exampleParameter1, exampleParameter2)
def exampleName(exampleParameter1: any, exampleParameter2: any) -> any:
print(exampleParameter1, exampleParameter2)
def exampleName(exampleParameter1: int, exampleParameter2: int) -> None:
# print(exampleParameter1, exampleParameter2)
return exampleParameter1 + exampleParameter2
print()
exampleName(10, 94)
class Person:
def __init__(self, weight: int, height: int, name: str):
self.weight = weight
self.height = height
self.name = name
def who_is_this(self):
print("This person's name is " + self.name)
print("This person's weight is " + str(self.weight) + " pounds")
print("This person's height is " + str(self.height) + " inches")
if __name__ == "__main__":
Kipp = Person(225, 70, "Aaron Kippins")
Kipp.who_is_this()
string = "I want to go home!"
print(string[0:12], "to Cancun!")
# print(string[0:1])
alpha_sentence = 'Quick brown fox jumped over the lazy dog'
print(alpha_sentence.title())
print(alpha_sentence.upper())
print(alpha_sentence.lower())
if alpha_sentence.lower().islower():
print("sentence is all lowercase")
print(2 ** 5)
import math
math.sqrt(4)
print(4//2)
print(4/2)
abs(-10)
dummy_string = "Hey there I'm just a string for the example about to happen."
print(dummy_string.center(70, "-"))
print(dummy_string.partition("o"))
print(dummy_string.swapcase())
print(dummy_string.split(" "))
arr = [2, 5, 6, 1, 4, 3]
arr.sort()
print(arr)
print(arr[3])
# sorted(arr)
print(arr[1:3])
arr.append(7)
print(arr)
arr.pop()
print(arr)
| 0.283385 | 0.983166 |
# Blood Cell Images Data Set
### Get the Data
```
import pandas as pd
import numpy as np
import os
import tensorflow as tf
import cv2
from tensorflow import keras
from tensorflow.keras import layers, Input , losses
from keras.layers.core import Dense , Flatten
from tensorflow.keras.layers import InputLayer
from tensorflow.keras.models import Sequential, Model
from matplotlib import pyplot as plt
import matplotlib.image as mpimg
import random
%matplotlib inline
plt.figure(figsize=(20,20))
test_folder=r'C:\Users\Berkay\Desktop\MLDL Projects\Blood Cell Images Images with Tensorflow\Blood-Cell-Images-with-Tensorflow\images\TRAIN\EOSINOPHIL'
for i in range(5):
file = random.choice(os.listdir(test_folder))
image_path= os.path.join(test_folder, file)
img=mpimg.imread(image_path)
ax=plt.subplot(1,5,i+1)
ax.title.set_text(file)
plt.imshow(img)
IMG_WIDTH=200
IMG_HEIGHT=200
img_folder=r'C:\Users\Berkay\Desktop\MLDL Projects\Chest X Ray Images with Tensorflow\Chest-X-Ray-Images-with-Tensorflow\images\TRAIN'
def create_dataset(img_folder):
img_data_array=[]
class_name=[]
for dir1 in os.listdir(img_folder):
for file in os.listdir(os.path.join(img_folder, dir1)):
image_path= os.path.join(img_folder, dir1, file)
image= cv2.imread( image_path, cv2.COLOR_BGR2RGB)
image=cv2.resize(image, (IMG_HEIGHT, IMG_WIDTH),interpolation = cv2.INTER_AREA)
image=np.array(image)
image = image.astype('float32')
image /= 255
img_data_array.append(image)
class_name.append(dir1)
return img_data_array, class_name
# extract the image array and class name
img_data, class_name =create_dataset(r'C:/Users/Berkay/Desktop/MLDL Projects/Blood Cell Images Images with Tensorflow\Blood-Cell-Images-with-Tensorflow\images\TRAIN')
target_dict={k: v for v, k in enumerate(np.unique(class_name))}
target_dict
target_val = [target_dict[class_name[i]] for i in range(len(class_name))]
model=tf.keras.Sequential(
[
tf.keras.layers.InputLayer(input_shape=(IMG_HEIGHT,IMG_WIDTH, 3)),
tf.keras.layers.Conv2D(filters=32, kernel_size=3, strides=(2, 2), activation='relu'),
tf.keras.layers.Conv2D(filters=64, kernel_size=3, strides=(2, 2), activation='relu'),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(6)
])
model.compile(optimizer='rmsprop', loss='sparse_categorical_crossentropy', metrics=['accuracy'])
history = model.fit(x=np.array(img_data, np.float32), y=np.array(list(map(int,target_val)), np.float32), epochs=5)
history = model.fit(x=tf.cast(np.array(img_data), tf.float64), y=tf.cast(list(map(int,target_val)),tf.int32), epochs=5)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import os
import tensorflow as tf
import cv2
from tensorflow import keras
from tensorflow.keras import layers, Input , losses
from keras.layers.core import Dense , Flatten
from tensorflow.keras.layers import InputLayer
from tensorflow.keras.models import Sequential, Model
from matplotlib import pyplot as plt
import matplotlib.image as mpimg
import random
%matplotlib inline
plt.figure(figsize=(20,20))
test_folder=r'C:\Users\Berkay\Desktop\MLDL Projects\Blood Cell Images Images with Tensorflow\Blood-Cell-Images-with-Tensorflow\images\TRAIN\EOSINOPHIL'
for i in range(5):
file = random.choice(os.listdir(test_folder))
image_path= os.path.join(test_folder, file)
img=mpimg.imread(image_path)
ax=plt.subplot(1,5,i+1)
ax.title.set_text(file)
plt.imshow(img)
IMG_WIDTH=200
IMG_HEIGHT=200
img_folder=r'C:\Users\Berkay\Desktop\MLDL Projects\Chest X Ray Images with Tensorflow\Chest-X-Ray-Images-with-Tensorflow\images\TRAIN'
def create_dataset(img_folder):
img_data_array=[]
class_name=[]
for dir1 in os.listdir(img_folder):
for file in os.listdir(os.path.join(img_folder, dir1)):
image_path= os.path.join(img_folder, dir1, file)
image= cv2.imread( image_path, cv2.COLOR_BGR2RGB)
image=cv2.resize(image, (IMG_HEIGHT, IMG_WIDTH),interpolation = cv2.INTER_AREA)
image=np.array(image)
image = image.astype('float32')
image /= 255
img_data_array.append(image)
class_name.append(dir1)
return img_data_array, class_name
# extract the image array and class name
img_data, class_name =create_dataset(r'C:/Users/Berkay/Desktop/MLDL Projects/Blood Cell Images Images with Tensorflow\Blood-Cell-Images-with-Tensorflow\images\TRAIN')
target_dict={k: v for v, k in enumerate(np.unique(class_name))}
target_dict
target_val = [target_dict[class_name[i]] for i in range(len(class_name))]
model=tf.keras.Sequential(
[
tf.keras.layers.InputLayer(input_shape=(IMG_HEIGHT,IMG_WIDTH, 3)),
tf.keras.layers.Conv2D(filters=32, kernel_size=3, strides=(2, 2), activation='relu'),
tf.keras.layers.Conv2D(filters=64, kernel_size=3, strides=(2, 2), activation='relu'),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(6)
])
model.compile(optimizer='rmsprop', loss='sparse_categorical_crossentropy', metrics=['accuracy'])
history = model.fit(x=np.array(img_data, np.float32), y=np.array(list(map(int,target_val)), np.float32), epochs=5)
history = model.fit(x=tf.cast(np.array(img_data), tf.float64), y=tf.cast(list(map(int,target_val)),tf.int32), epochs=5)
| 0.604282 | 0.819026 |
# Encirclement analysis
This notebook presents the post-processing made to the output file from the `./encirclement` command. Some functions need to be defined to perform some of the tasks.
## Function definitions
The following functions are defined for a representation of the gaps in the circle drawn:
- `CircularPiePlotScarGap(df, t_nogap, no_plot=None)`
- `CircularPiePlotScarWidth(df, t_scar, no_plot=None)`
- `ProcessAllCases(about_data_df)`
- `scar_width(df, threshold)` (incomplete)
The following helper functions are defined too:
- `WriteCSV(np_array, case_name, filename_csv, append_col=None)`
- `unit_vector(vector)`
- `point_theta(center, A)`
```
# Imports
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from scipy.stats import norm
from sklearn.preprocessing import StandardScaler
from scipy import stats
from mpl_toolkits.mplot3d import Axes3D
from sklearn.model_selection import train_test_split
import warnings
import math
from numpy import linalg as LA
from numpy import random, nanmax, argmax, unravel_index
from scipy.spatial.distance import pdist, squareform
warnings.filterwarnings('ignore')
%matplotlib inline
data_path_prefix = './data/encirclement/'
data_df=pd.read_csv('./data/encirclement/about_data.csv')
data_df.head(3)
df_data = df_data[df_data.VertexID.duplicated(keep=False)]
ProcessAllCases(about_data_df)
scar_widths = CircularPiePlotScarWidth(df_data,59) # use 0.5 for DP and 4.5 for CF
df_data_scalars = pd.concat([df_data['MainVertexSeq'], df_data['MeshScalar']], axis=1)
scar_gaps = CircularPiePlotScarGap(df_data_scalars,59) # use 0.5 for DP and 4.5 for CF
print scar_grps
scar_grps.mean()
ax = sns.boxplot(x="MainVertexSeq", y="MeshScalar", data=df_data_scalars)
g1 = df_data.groupby(['MainVertexSeq']).median()
g1 = g1.reset_index()
g1.head(2)
ax = sns.tsplot(data=g1['MeshScalar'])
center_data = [df_data['X'].mean(), df_data['Y'].mean(), df_data['Z'].mean()]
fig = plt.figure()
ax = Axes3D(fig)
ax.scatter(df_data['X'], df_data['Y'], df_data['Z'])
ax.plot([center_data[0]], [center_data[1]], [center_data[2]], markerfacecolor='k', markeredgecolor='k', marker='o', markersize=20, alpha=0.6)
plt.show()
center_data
df_data_scalars = pd.concat([df_data['MainVertexSeq'], df_data['MeshScalar']], axis=1)
g1 = df_data.groupby(['MainVertexSeq']).median()
g1 = g1.reset_index()
ax = sns.tsplot(data=g1['MeshScalar'])
thetas = point_theta(center_data, df_data)
thetas_df = pd.Series(thetas, name='theta');
thetas_df.describe()
df_data_with_theta = pd.concat([df_data, thetas_df], axis=1)
sns.tsplot(df_data_with_theta['theta'])
df_data_with_thetas_sorted = df_data_with_theta.sort_values(by='theta')
df_data_with_thetas_sorted.head(10)
sns.tsplot(df_data_with_thetas_sorted['theta'])
sns.tsplot(df_data_with_thetas_sorted['MeshScalar'])
df_data_with_thetas_sorted.head(15)
df_data_with_thetas_sorted['MeshScalar'].std()
df_data_with_thetas_sorted.head()
df_data_with_thetas_sorted_nodupes = df_data_with_thetas_sorted.drop_duplicates(subset='VertexID', keep='last');
sns.tsplot(pd.rolling_mean(df_data_with_thetas_sorted_nodupes['MeshScalar'],50))
sns.tsplot(pd.rolling_mean(df_data_with_theta['MeshScalar'], 50))
df_data_with_thetas_sorted_nodupes.describe()
# t_nogap is the cut-off where a value above t_nogap means no gap
def CircularPiePlotScarGap(df, t_nogap, no_plot=None):
count = len(df.as_matrix())
#scalars = df_data_with_theta.as_matrix(columns=df_data_with_theta.columns[7:8])
scalars = df['MeshScalar'].as_matrix()
num_prim_groups = 4 # Do not change as pie plot is hard-coded to accept four primary groups
num_sub_groups = 4 # Do not change as pie plot is hard-coded to accept four primary groups
total_groups = num_prim_groups*num_sub_groups
sub_group_size = count/total_groups
# Divide the data into 12 groups, 4 regions x 3 sub-regions, regions = post sup, post inf, etc.
sub_group_array = np.zeros(shape=(total_groups, 1))
n=0
sb = np.array_split(scalars, total_groups)
for i in sb:
sub_group_array[n] = np.mean(i)
n += 1
# binary classify sub_groups into gaps and no-gaps
bin_subgrouparray = sub_group_array > t_nogap
bin_subgrouparray = bin_subgrouparray * 1 # converts the false true array to 0 1
# Now prepare data for circular plot
# First decide color sub-groups based on binary classification of sub_groups (i.e. t_nogaps)
a, b, c=[plt.cm.Blues, plt.cm.Reds, plt.cm.Greens]
color_subgroups = list()
for x in bin_subgrouparray:
if x == 1:
color_subgroups.append(b(0.5))
else:
color_subgroups.append(c(0.5))
# Decide color of the primary groups (n=4) based on number of sub_groups classified as gaps/no-gaps
color_primgroups = list()
prim_group_array = np.zeros(shape=(num_prim_groups, 1))
# classify primary groups
prim_group_array = np.split(bin_subgrouparray, num_prim_groups)
n=0
for i in prim_group_array:
prim_group_array[n] = np.sum(i)
n += 1
for x in prim_group_array:
if x==1:
color_primgroups.append('green')
elif x==2:
color_primgroups.append('yellow')
elif x==3:
color_primgroups.append('gold')
elif x==4:
color_primgroups.append('red')
else:
color_primgroups.append('green') # x == 0
# Make data: I have 3 groups and 7 subgroups
group_names=['Post-Sup (A)', 'Ant-Sup (B)', 'Ant-Inf (C)', 'Post-Inf (D)']
group_size=[1,1,1,1]
subgroup_names=['A1', 'A2', 'A3', 'A4','B1', 'B2', 'B3', 'B4', 'C1', 'C2', 'C3', 'C4', 'D1', 'D2', 'D3', 'D4']
subgroup_size=[1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1]
if no_plot is None:
# First Ring (outside)
fig, ax = plt.subplots()
ax.axis('equal')
mypie, _ = ax.pie(group_size, radius=1.3, labels=group_names, colors=color_primgroups )
plt.setp( mypie, width=0.3, edgecolor='white')
# Second Ring (Inside)
mypie2, _ = ax.pie(subgroup_size, radius=1.3-0.3, labels=subgroup_names, labeldistance=0.7, colors=color_subgroups)
plt.setp( mypie2, width=0.4, edgecolor='white')
plt.margins(0,0)
# show it
plt.show()
else:
return bin_subgrouparray
# t_nogap is the cut-off where a value above t_nogap means no gap
def CircularPiePlotScarWidth(df, t_scar, no_plot=None):
count = len(df.as_matrix())
#scalars = df_data_with_theta.as_matrix(columns=df_data_with_theta.columns[7:8])
scalars = df['MeshScalar'].as_matrix()
num_prim_groups = 4 # Do not change as pie plot is hard-coded to accept four primary groups
num_sub_groups = 4 # Do not change as pie plot is hard-coded to accept four primary groups
total_groups = num_prim_groups*num_sub_groups
sub_group_size = count/total_groups
# assigning each point into one of 16 sub-groups in the polar plot (sub_group_size = num_vertices/16)
df['scar_bin'] = df['MeshScalar'] > t_scar
df['scar_bin'] = df['scar_bin'] * 1
df['polar_group'] = df['MainVertexSeq']
for i, row in df.iterrows():
#row['polar_group'] = math.ceil(index / sub_group_size )
df.at[i, 'polar_group'] = math.ceil(i / sub_group_size )
# See https://stackoverflow.com/questions/31667070/max-distance-between-2-points-in-a-data-set-and-identifying-the-points
total_width_in_each_subgrp = []
scar_width_in_each_subgrp = []
for i in range(0, total_groups):
df_temp = df[df['polar_group']==i]
xyz = df_temp.as_matrix(columns=['X', 'Y', 'Z'])
D = pdist(xyz) # pair-wise distances
D = squareform(D)
N, [I_row, I_col] = nanmax(D), unravel_index( argmax(D), D.shape )
total_width_in_each_subgrp.append(round(N,2))
# now select rows that was classified as scar
df_temp2 = df_temp[df_temp['scar_bin'] == 1]
xyz = df_temp2.as_matrix(columns=['X', 'Y', 'Z'])
D = pdist(xyz) # pair-wise distances
D = squareform(D)
N, [I_row, I_col] = nanmax(D), unravel_index( argmax(D), D.shape )
scar_width_in_each_subgrp.append(round(N,2))
# calculate scar width percentage
scar_width_percentage = []
i=0
while i < len(scar_width_in_each_subgrp):
total = total_width_in_each_subgrp[i]
this_scar = scar_width_in_each_subgrp[i]
scar_width_percentage.append(this_scar/total)
i+=1
scar_width_percentage = np.asarray(scar_width_percentage)
# Now prepare data for circular plot
# First decide color sub-groups based on binary classification of sub_groups (i.e. t_nogaps)
a, b, c=[plt.cm.Blues, plt.cm.Reds, plt.cm.Greens]
color_subgroups = list()
for x in scar_width_percentage:
if x >= 0.75:
color_subgroups.append('red')
elif x >= 0.5 and x < 0.75:
color_subgroups.append('gold')
elif x >= 0.25 and x < 0.5:
color_subgroups.append('yellow')
elif x >= 0 and x < 0.25:
color_subgroups.append('green')
# Decide color of the primary groups (n=4) based on number of sub_groups classified as gaps/no-gaps
color_primgroups = list()
prim_group_array = np.zeros(shape=(num_prim_groups, 1))
# classify primary groups
prim_group_array = np.split(scar_width_percentage, num_prim_groups)
n=0
for i in prim_group_array:
prim_group_array[n] = np.mean(i)
n += 1
for x in prim_group_array:
if x >= 0.75:
color_primgroups.append('red')
elif x >= 0.5 and x < 0.75:
color_primgroups.append('gold')
elif x >= 0.25 and x < 0.5:
color_primgroups.append('yellow')
elif x > 0 and x < 0.25:
color_primgroups.append('green')
else:
color_primgroups.append('green') # x == 0
# Make data: I have 3 groups and 7 subgroups
group_names=['Post-Sup (A)', 'Ant-Sup (B)', 'Ant-Inf (C)', 'Post-Inf (D)']
group_size=[1,1,1,1]
subgroup_names=['A1', 'A2', 'A3', 'A4','B1', 'B2', 'B3', 'B4', 'C1', 'C2', 'C3', 'C4', 'D1', 'D2', 'D3', 'D4']
subgroup_size=[1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1]
# First Ring (outside)
if no_plot is None:
fig, ax = plt.subplots()
ax.axis('equal')
mypie, _ = ax.pie(group_size, radius=1.3, labels=group_names, colors=color_primgroups )
plt.setp( mypie, width=0.3, edgecolor='white')
# Second Ring (Inside)
mypie2, _ = ax.pie(subgroup_size, radius=1.3-0.3, labels=subgroup_names, labeldistance=0.7, colors=color_subgroups)
plt.setp( mypie2, width=0.4, edgecolor='white')
plt.margins(0,0)
# show it
plt.show()
else:
return total_width_in_each_subgrp
print ("Complete widths in each sub-segment", total_width_in_each_subgrp)
print ("Scar widths in each sub-segment", scar_width_in_each_subgrp)
# writes the output of circular pie plot to file
def WriteCSV(np_array, case_name, filename_csv, append_col=None):
df = pd.DataFrame(np_array)
df_T = df.T
# an extra column to specify if lpv or rpv
if append_col is not None:
df_T['xcol'] = append_col
cols = df_T.columns.tolist()
df_T = df_T[[cols[-1]] + cols[:-1]]
df_T['case'] = case_name
cols = df_T.columns.tolist()
df_T = df_T[[cols[-1]] + cols[:-1]]
with open(filename_csv, 'a') as f:
df_T.to_csv(f, header=False)
def ProcessAllCases(about_data_df):
data_path_prefix = './data/encirclement/test_script/'
csv_filename_gaps = './data/encirclement/scar_gaps.csv'
csv_filename_width = './data/encirclement/scar_width.csv'
for index, row in about_data_df.iterrows():
path_to_data = data_path_prefix + row['filename'] + '.csv'
df_data = pd.read_csv(path_to_data)
df_data_for_gaps = pd.concat([df_data['MainVertexSeq'], df_data['MeshScalar']], axis=1)
scar_gaps = CircularPiePlotScarGap(df_data_for_gaps, row['si'], 'no plot')
scar_width = CircularPiePlotScarWidth(df_data, row['si'], 'no plot')
WriteCSV(scar_gaps, row['case'], csv_filename_gaps, row['lpv'])
WriteCSV(scar_width, row['case'], csv_filename_width, row['lpv'])
def unit_vector(vector):
""" Returns the unit vector of the vector. """
return vector / np.linalg.norm(vector)
def angle_between(v1, v2):
""" Returns the angle in radians between vectors 'v1' and 'v2'::
>>> angle_between((1, 0, 0), (0, 1, 0))
1.5707963267948966
>>> angle_between((1, 0, 0), (1, 0, 0))
0.0
>>> angle_between((1, 0, 0), (-1, 0, 0))
3.141592653589793
"""
v1_u = unit_vector(v1)
v2_u = unit_vector(v2)
# return np.arccos(np.clip(np.dot(v1_u, v2_u), -1.0, 1.0))
return np.arctan2(LA.norm(np.cross(v1_u, v2_u)), np.dot(v1_u, v2_u))
def point_theta(center, A):
thetas = [];
Ref_pt = [A.iloc[0]['X'], A.iloc[0]['Y'], A.iloc[0]['Z']]
Ref_V = np.subtract(Ref_pt, center)
for index, row in A.iterrows():
pt = [row['X'], row['Y'], row['Z']]
pt_V = np.subtract(pt, center)
#theta = angle_between(Ref_pt, pt)
theta = angle_between(Ref_V, pt_V)
theta = theta * 180 / np.pi
thetas.append(theta)
return thetas
'''
This function is not complete yet, it tries to compute the distance between each point in the list to its
point on the line (vertex depth = v for all points within the same neighbourhood)
'''
def scar_width(df, threshold):
width = [];
is_scar = [];
#Ref_pt = [A.iloc[0]['X'], A.iloc[0]['Y'], A.iloc[0]['Z']]
for index, row in df.iterrows():
if row['VertexDepth'] == 0:
pt_on_line = (row['X'], row['Y'], row['Z'])
pt = (row['X'], row['Y'], row['Z'])
#distance = math.hypot(pt[0]-pt_on_line[0], pt[1]-pt_on_line[1], pt[1]-pt_on_line[1])
distance = math.sqrt((pt[0]-pt_on_line[0])**2 + (pt[1]-pt_on_line[1])**2 + (pt[2]-pt_on_line[2])**2)
if row['MeshScalar'] > threshold:
is_scar.append(1)
width.append(distance)
else:
is_scar.append(0)
width.append(0)
width_df = pd.Series(width, name='scar_width');
is_scar_df = pd.Series(is_scar, name='scar_bin');
df = pd.concat([df, width_df], axis=1)
df = pd.concat([df, is_scar_df], axis=1)
return df
df_data_with_thetas_sorted = scar_width(df_data_with_thetas_sorted, 2.2)
df_data_with_thetas_sorted['MeshScalar'].mean()
sns.tsplot(pd.rolling_mean(df_data_with_thetas_sorted['scar_bin'],20))
sns.tsplot(pd.rolling_sum(df_data_with_thetas_sorted['scar_bin'],20))
sns.tsplot(pd.rolling_mean(df_data_with_thetas_sorted['scar_width'],200)) # Uses mean windowing=100, so 0 widths are smoothed
# Libraries
# See https://python-graph-gallery.com/163-donut-plot-with-subgroups/
import matplotlib.pyplot as plt
# Make data: I have 3 groups and 7 subgroups
group_names=['PostSup', 'PostInf', 'AntSup', 'AntInf']
group_size=[1,1,1,1]
subgroup_names=['A.1', 'A.2', 'A.3', 'A.4','B.1', 'B.2', 'B.3', 'B.4', 'C.1', 'C.2', 'C.3', 'C.4', 'D.1', 'D.2', 'D.3', 'D.4']
subgroup_size=[1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1]
# Create colors
a, b, c=[plt.cm.Blues, plt.cm.Reds, plt.cm.Greens]
# First Ring (outside)
fig, ax = plt.subplots()
ax.axis('equal')
mypie, _ = ax.pie(group_size, radius=1.3, labels=group_names, colors=[a(0.6), b(0.6), c(0.6)] )
plt.setp( mypie, width=0.3, edgecolor='white')
# Second Ring (Inside)
mypie2, _ = ax.pie(subgroup_size, radius=1.3-0.3, labels=subgroup_names, labeldistance=0.7, colors=[a(0.5), a(0.4), a(0.3), b(0.5), b(0.4), c(0.6), c(0.5), c(0.4), c(0.3), c(0.2)])
plt.setp( mypie2, width=0.4, edgecolor='white')
plt.margins(0,0)
# show it
plt.show()
df_data_with_thetas_sorted_nodupes['theta'].hist(bins=20)
test_array = df_data_with_theta.as_matrix(columns=df_data_with_theta.columns[7:8])
test_array.shape
2639/12
scar_grps
```
|
github_jupyter
|
# Imports
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from scipy.stats import norm
from sklearn.preprocessing import StandardScaler
from scipy import stats
from mpl_toolkits.mplot3d import Axes3D
from sklearn.model_selection import train_test_split
import warnings
import math
from numpy import linalg as LA
from numpy import random, nanmax, argmax, unravel_index
from scipy.spatial.distance import pdist, squareform
warnings.filterwarnings('ignore')
%matplotlib inline
data_path_prefix = './data/encirclement/'
data_df=pd.read_csv('./data/encirclement/about_data.csv')
data_df.head(3)
df_data = df_data[df_data.VertexID.duplicated(keep=False)]
ProcessAllCases(about_data_df)
scar_widths = CircularPiePlotScarWidth(df_data,59) # use 0.5 for DP and 4.5 for CF
df_data_scalars = pd.concat([df_data['MainVertexSeq'], df_data['MeshScalar']], axis=1)
scar_gaps = CircularPiePlotScarGap(df_data_scalars,59) # use 0.5 for DP and 4.5 for CF
print scar_grps
scar_grps.mean()
ax = sns.boxplot(x="MainVertexSeq", y="MeshScalar", data=df_data_scalars)
g1 = df_data.groupby(['MainVertexSeq']).median()
g1 = g1.reset_index()
g1.head(2)
ax = sns.tsplot(data=g1['MeshScalar'])
center_data = [df_data['X'].mean(), df_data['Y'].mean(), df_data['Z'].mean()]
fig = plt.figure()
ax = Axes3D(fig)
ax.scatter(df_data['X'], df_data['Y'], df_data['Z'])
ax.plot([center_data[0]], [center_data[1]], [center_data[2]], markerfacecolor='k', markeredgecolor='k', marker='o', markersize=20, alpha=0.6)
plt.show()
center_data
df_data_scalars = pd.concat([df_data['MainVertexSeq'], df_data['MeshScalar']], axis=1)
g1 = df_data.groupby(['MainVertexSeq']).median()
g1 = g1.reset_index()
ax = sns.tsplot(data=g1['MeshScalar'])
thetas = point_theta(center_data, df_data)
thetas_df = pd.Series(thetas, name='theta');
thetas_df.describe()
df_data_with_theta = pd.concat([df_data, thetas_df], axis=1)
sns.tsplot(df_data_with_theta['theta'])
df_data_with_thetas_sorted = df_data_with_theta.sort_values(by='theta')
df_data_with_thetas_sorted.head(10)
sns.tsplot(df_data_with_thetas_sorted['theta'])
sns.tsplot(df_data_with_thetas_sorted['MeshScalar'])
df_data_with_thetas_sorted.head(15)
df_data_with_thetas_sorted['MeshScalar'].std()
df_data_with_thetas_sorted.head()
df_data_with_thetas_sorted_nodupes = df_data_with_thetas_sorted.drop_duplicates(subset='VertexID', keep='last');
sns.tsplot(pd.rolling_mean(df_data_with_thetas_sorted_nodupes['MeshScalar'],50))
sns.tsplot(pd.rolling_mean(df_data_with_theta['MeshScalar'], 50))
df_data_with_thetas_sorted_nodupes.describe()
# t_nogap is the cut-off where a value above t_nogap means no gap
def CircularPiePlotScarGap(df, t_nogap, no_plot=None):
count = len(df.as_matrix())
#scalars = df_data_with_theta.as_matrix(columns=df_data_with_theta.columns[7:8])
scalars = df['MeshScalar'].as_matrix()
num_prim_groups = 4 # Do not change as pie plot is hard-coded to accept four primary groups
num_sub_groups = 4 # Do not change as pie plot is hard-coded to accept four primary groups
total_groups = num_prim_groups*num_sub_groups
sub_group_size = count/total_groups
# Divide the data into 12 groups, 4 regions x 3 sub-regions, regions = post sup, post inf, etc.
sub_group_array = np.zeros(shape=(total_groups, 1))
n=0
sb = np.array_split(scalars, total_groups)
for i in sb:
sub_group_array[n] = np.mean(i)
n += 1
# binary classify sub_groups into gaps and no-gaps
bin_subgrouparray = sub_group_array > t_nogap
bin_subgrouparray = bin_subgrouparray * 1 # converts the false true array to 0 1
# Now prepare data for circular plot
# First decide color sub-groups based on binary classification of sub_groups (i.e. t_nogaps)
a, b, c=[plt.cm.Blues, plt.cm.Reds, plt.cm.Greens]
color_subgroups = list()
for x in bin_subgrouparray:
if x == 1:
color_subgroups.append(b(0.5))
else:
color_subgroups.append(c(0.5))
# Decide color of the primary groups (n=4) based on number of sub_groups classified as gaps/no-gaps
color_primgroups = list()
prim_group_array = np.zeros(shape=(num_prim_groups, 1))
# classify primary groups
prim_group_array = np.split(bin_subgrouparray, num_prim_groups)
n=0
for i in prim_group_array:
prim_group_array[n] = np.sum(i)
n += 1
for x in prim_group_array:
if x==1:
color_primgroups.append('green')
elif x==2:
color_primgroups.append('yellow')
elif x==3:
color_primgroups.append('gold')
elif x==4:
color_primgroups.append('red')
else:
color_primgroups.append('green') # x == 0
# Make data: I have 3 groups and 7 subgroups
group_names=['Post-Sup (A)', 'Ant-Sup (B)', 'Ant-Inf (C)', 'Post-Inf (D)']
group_size=[1,1,1,1]
subgroup_names=['A1', 'A2', 'A3', 'A4','B1', 'B2', 'B3', 'B4', 'C1', 'C2', 'C3', 'C4', 'D1', 'D2', 'D3', 'D4']
subgroup_size=[1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1]
if no_plot is None:
# First Ring (outside)
fig, ax = plt.subplots()
ax.axis('equal')
mypie, _ = ax.pie(group_size, radius=1.3, labels=group_names, colors=color_primgroups )
plt.setp( mypie, width=0.3, edgecolor='white')
# Second Ring (Inside)
mypie2, _ = ax.pie(subgroup_size, radius=1.3-0.3, labels=subgroup_names, labeldistance=0.7, colors=color_subgroups)
plt.setp( mypie2, width=0.4, edgecolor='white')
plt.margins(0,0)
# show it
plt.show()
else:
return bin_subgrouparray
# t_nogap is the cut-off where a value above t_nogap means no gap
def CircularPiePlotScarWidth(df, t_scar, no_plot=None):
count = len(df.as_matrix())
#scalars = df_data_with_theta.as_matrix(columns=df_data_with_theta.columns[7:8])
scalars = df['MeshScalar'].as_matrix()
num_prim_groups = 4 # Do not change as pie plot is hard-coded to accept four primary groups
num_sub_groups = 4 # Do not change as pie plot is hard-coded to accept four primary groups
total_groups = num_prim_groups*num_sub_groups
sub_group_size = count/total_groups
# assigning each point into one of 16 sub-groups in the polar plot (sub_group_size = num_vertices/16)
df['scar_bin'] = df['MeshScalar'] > t_scar
df['scar_bin'] = df['scar_bin'] * 1
df['polar_group'] = df['MainVertexSeq']
for i, row in df.iterrows():
#row['polar_group'] = math.ceil(index / sub_group_size )
df.at[i, 'polar_group'] = math.ceil(i / sub_group_size )
# See https://stackoverflow.com/questions/31667070/max-distance-between-2-points-in-a-data-set-and-identifying-the-points
total_width_in_each_subgrp = []
scar_width_in_each_subgrp = []
for i in range(0, total_groups):
df_temp = df[df['polar_group']==i]
xyz = df_temp.as_matrix(columns=['X', 'Y', 'Z'])
D = pdist(xyz) # pair-wise distances
D = squareform(D)
N, [I_row, I_col] = nanmax(D), unravel_index( argmax(D), D.shape )
total_width_in_each_subgrp.append(round(N,2))
# now select rows that was classified as scar
df_temp2 = df_temp[df_temp['scar_bin'] == 1]
xyz = df_temp2.as_matrix(columns=['X', 'Y', 'Z'])
D = pdist(xyz) # pair-wise distances
D = squareform(D)
N, [I_row, I_col] = nanmax(D), unravel_index( argmax(D), D.shape )
scar_width_in_each_subgrp.append(round(N,2))
# calculate scar width percentage
scar_width_percentage = []
i=0
while i < len(scar_width_in_each_subgrp):
total = total_width_in_each_subgrp[i]
this_scar = scar_width_in_each_subgrp[i]
scar_width_percentage.append(this_scar/total)
i+=1
scar_width_percentage = np.asarray(scar_width_percentage)
# Now prepare data for circular plot
# First decide color sub-groups based on binary classification of sub_groups (i.e. t_nogaps)
a, b, c=[plt.cm.Blues, plt.cm.Reds, plt.cm.Greens]
color_subgroups = list()
for x in scar_width_percentage:
if x >= 0.75:
color_subgroups.append('red')
elif x >= 0.5 and x < 0.75:
color_subgroups.append('gold')
elif x >= 0.25 and x < 0.5:
color_subgroups.append('yellow')
elif x >= 0 and x < 0.25:
color_subgroups.append('green')
# Decide color of the primary groups (n=4) based on number of sub_groups classified as gaps/no-gaps
color_primgroups = list()
prim_group_array = np.zeros(shape=(num_prim_groups, 1))
# classify primary groups
prim_group_array = np.split(scar_width_percentage, num_prim_groups)
n=0
for i in prim_group_array:
prim_group_array[n] = np.mean(i)
n += 1
for x in prim_group_array:
if x >= 0.75:
color_primgroups.append('red')
elif x >= 0.5 and x < 0.75:
color_primgroups.append('gold')
elif x >= 0.25 and x < 0.5:
color_primgroups.append('yellow')
elif x > 0 and x < 0.25:
color_primgroups.append('green')
else:
color_primgroups.append('green') # x == 0
# Make data: I have 3 groups and 7 subgroups
group_names=['Post-Sup (A)', 'Ant-Sup (B)', 'Ant-Inf (C)', 'Post-Inf (D)']
group_size=[1,1,1,1]
subgroup_names=['A1', 'A2', 'A3', 'A4','B1', 'B2', 'B3', 'B4', 'C1', 'C2', 'C3', 'C4', 'D1', 'D2', 'D3', 'D4']
subgroup_size=[1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1]
# First Ring (outside)
if no_plot is None:
fig, ax = plt.subplots()
ax.axis('equal')
mypie, _ = ax.pie(group_size, radius=1.3, labels=group_names, colors=color_primgroups )
plt.setp( mypie, width=0.3, edgecolor='white')
# Second Ring (Inside)
mypie2, _ = ax.pie(subgroup_size, radius=1.3-0.3, labels=subgroup_names, labeldistance=0.7, colors=color_subgroups)
plt.setp( mypie2, width=0.4, edgecolor='white')
plt.margins(0,0)
# show it
plt.show()
else:
return total_width_in_each_subgrp
print ("Complete widths in each sub-segment", total_width_in_each_subgrp)
print ("Scar widths in each sub-segment", scar_width_in_each_subgrp)
# writes the output of circular pie plot to file
def WriteCSV(np_array, case_name, filename_csv, append_col=None):
df = pd.DataFrame(np_array)
df_T = df.T
# an extra column to specify if lpv or rpv
if append_col is not None:
df_T['xcol'] = append_col
cols = df_T.columns.tolist()
df_T = df_T[[cols[-1]] + cols[:-1]]
df_T['case'] = case_name
cols = df_T.columns.tolist()
df_T = df_T[[cols[-1]] + cols[:-1]]
with open(filename_csv, 'a') as f:
df_T.to_csv(f, header=False)
def ProcessAllCases(about_data_df):
data_path_prefix = './data/encirclement/test_script/'
csv_filename_gaps = './data/encirclement/scar_gaps.csv'
csv_filename_width = './data/encirclement/scar_width.csv'
for index, row in about_data_df.iterrows():
path_to_data = data_path_prefix + row['filename'] + '.csv'
df_data = pd.read_csv(path_to_data)
df_data_for_gaps = pd.concat([df_data['MainVertexSeq'], df_data['MeshScalar']], axis=1)
scar_gaps = CircularPiePlotScarGap(df_data_for_gaps, row['si'], 'no plot')
scar_width = CircularPiePlotScarWidth(df_data, row['si'], 'no plot')
WriteCSV(scar_gaps, row['case'], csv_filename_gaps, row['lpv'])
WriteCSV(scar_width, row['case'], csv_filename_width, row['lpv'])
def unit_vector(vector):
""" Returns the unit vector of the vector. """
return vector / np.linalg.norm(vector)
def angle_between(v1, v2):
""" Returns the angle in radians between vectors 'v1' and 'v2'::
>>> angle_between((1, 0, 0), (0, 1, 0))
1.5707963267948966
>>> angle_between((1, 0, 0), (1, 0, 0))
0.0
>>> angle_between((1, 0, 0), (-1, 0, 0))
3.141592653589793
"""
v1_u = unit_vector(v1)
v2_u = unit_vector(v2)
# return np.arccos(np.clip(np.dot(v1_u, v2_u), -1.0, 1.0))
return np.arctan2(LA.norm(np.cross(v1_u, v2_u)), np.dot(v1_u, v2_u))
def point_theta(center, A):
thetas = [];
Ref_pt = [A.iloc[0]['X'], A.iloc[0]['Y'], A.iloc[0]['Z']]
Ref_V = np.subtract(Ref_pt, center)
for index, row in A.iterrows():
pt = [row['X'], row['Y'], row['Z']]
pt_V = np.subtract(pt, center)
#theta = angle_between(Ref_pt, pt)
theta = angle_between(Ref_V, pt_V)
theta = theta * 180 / np.pi
thetas.append(theta)
return thetas
'''
This function is not complete yet, it tries to compute the distance between each point in the list to its
point on the line (vertex depth = v for all points within the same neighbourhood)
'''
def scar_width(df, threshold):
width = [];
is_scar = [];
#Ref_pt = [A.iloc[0]['X'], A.iloc[0]['Y'], A.iloc[0]['Z']]
for index, row in df.iterrows():
if row['VertexDepth'] == 0:
pt_on_line = (row['X'], row['Y'], row['Z'])
pt = (row['X'], row['Y'], row['Z'])
#distance = math.hypot(pt[0]-pt_on_line[0], pt[1]-pt_on_line[1], pt[1]-pt_on_line[1])
distance = math.sqrt((pt[0]-pt_on_line[0])**2 + (pt[1]-pt_on_line[1])**2 + (pt[2]-pt_on_line[2])**2)
if row['MeshScalar'] > threshold:
is_scar.append(1)
width.append(distance)
else:
is_scar.append(0)
width.append(0)
width_df = pd.Series(width, name='scar_width');
is_scar_df = pd.Series(is_scar, name='scar_bin');
df = pd.concat([df, width_df], axis=1)
df = pd.concat([df, is_scar_df], axis=1)
return df
df_data_with_thetas_sorted = scar_width(df_data_with_thetas_sorted, 2.2)
df_data_with_thetas_sorted['MeshScalar'].mean()
sns.tsplot(pd.rolling_mean(df_data_with_thetas_sorted['scar_bin'],20))
sns.tsplot(pd.rolling_sum(df_data_with_thetas_sorted['scar_bin'],20))
sns.tsplot(pd.rolling_mean(df_data_with_thetas_sorted['scar_width'],200)) # Uses mean windowing=100, so 0 widths are smoothed
# Libraries
# See https://python-graph-gallery.com/163-donut-plot-with-subgroups/
import matplotlib.pyplot as plt
# Make data: I have 3 groups and 7 subgroups
group_names=['PostSup', 'PostInf', 'AntSup', 'AntInf']
group_size=[1,1,1,1]
subgroup_names=['A.1', 'A.2', 'A.3', 'A.4','B.1', 'B.2', 'B.3', 'B.4', 'C.1', 'C.2', 'C.3', 'C.4', 'D.1', 'D.2', 'D.3', 'D.4']
subgroup_size=[1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1]
# Create colors
a, b, c=[plt.cm.Blues, plt.cm.Reds, plt.cm.Greens]
# First Ring (outside)
fig, ax = plt.subplots()
ax.axis('equal')
mypie, _ = ax.pie(group_size, radius=1.3, labels=group_names, colors=[a(0.6), b(0.6), c(0.6)] )
plt.setp( mypie, width=0.3, edgecolor='white')
# Second Ring (Inside)
mypie2, _ = ax.pie(subgroup_size, radius=1.3-0.3, labels=subgroup_names, labeldistance=0.7, colors=[a(0.5), a(0.4), a(0.3), b(0.5), b(0.4), c(0.6), c(0.5), c(0.4), c(0.3), c(0.2)])
plt.setp( mypie2, width=0.4, edgecolor='white')
plt.margins(0,0)
# show it
plt.show()
df_data_with_thetas_sorted_nodupes['theta'].hist(bins=20)
test_array = df_data_with_theta.as_matrix(columns=df_data_with_theta.columns[7:8])
test_array.shape
2639/12
scar_grps
| 0.34621 | 0.847084 |
# Data Structures and Indexing
We'll get our first look at pandas' data structures.
Then we'll focus on indexing -- selecting subsets of data.
```
import numpy as np
import pandas as pd
import seaborn.apionly as sns
import matplotlib.pyplot as plt
%matplotlib inline
pd.options.display.max_rows = 10
pd.options.display.float_format = '{:,.2f}'.format
plt.rcParams['figure.figsize'] = (16, 12)
```
## Reading Data
Pandas has support for reading from many data sources, including
- `pd.read_csv`
- `pd.read_excel`
- `pd.read_html`
- `pd.read_json`
- `pd.read_hdf`
- `pd.read_sql`
For this section we'll work with some flights data from the US Bureau of Transportation and Statistics.
The CSV includes all the flights from New York airports in January 2014.
Each record has information about a single flight including the carrier, tail number, origin, destination, and delays.
```
flights = pd.read_csv("data/ny-flights.csv.gz",
parse_dates=["fl_date", "arr", "dep"])
flights
```
## Data Structures

`read_csv` returned a `DataFrame`, which is somewhat similar to a spreadsheet or database table.
`pd.DataFrame` is the data container you'll work most with, and consists of a few components:
The data are in the middle of the table.
Each column of the data is a `pd.Series`, kind of like a 1-dimensional version of a DataFrame.
```
flights['dep_delay']
```
Both `pd.DataFrame`s and `pd.Series` have *row labels*, which can be accessed with the `.index` attribute:
```
flights.index
```
`flights.index` is a `pd.Index` (there are many specialized index types, like `pd.RangeIndex`, but we'll talk about those later).
DataFrames store their column labels in a `.columns` attribute, which is also a `pd.Index`:
```
flights.columns
```
The row and column labels help out with indexing and alignemnt, our firs two topics.
## Data Types

Like NumPy (but unlike regular Python lists), you'll want to know the `dtypes` of your data.
Improving on NumPy, pandas DataFrames can store *heterogenous* data;
each column of a DataFrame will have it's own type (int, float, datetime, bool, etc.), but the DataFrame can hold a mixture of these.
```
flights.info()
```
## Preview
A taste of where we'll be by the end of the course
To get a bit of intuition about the data we're working with, let's to a brief bit of exploratory analysis.
We'll see all this in more detail later, but I wanted to demonstrate some of the capabilities of pandas upfront.
```
(flights['dep']
.value_counts()
.resample('H')
.sum()
.rolling(8).mean()
.plot(figsize=(12, 6),
title="Number of Flights (8H Rolling Mean)"))
sns.despine()
```
We can plot the count of flights per carrier:
```
sns.countplot(
flights['unique_carrier'],
order=flights['unique_carrier'].value_counts().index,
palette='Blues_r'
)
sns.despine()
```
And the joint distribution of departure and arrival delays:
```
sns.jointplot('dep_delay', 'arr_delay',
flights.loc[(flights['dep_delay'] > 1) &
(flights['dep_delay'] < 500)],
alpha=.25, marker='.', size=8);
```
We'll explore some of those methods and visualization techniques later.
For now we'll turn to the more fundamental operation of indexing.
By indexing, we mean selecting subsets of your data.
It's a good starting point, because it comes up in so many other places;
It's a terrible starting point because it's somewhat complicated, and somewhat boring.
## Goals of Indexing
There are many ways you might want to specify which subset you want to select:
- Like lists, you can index by integer position.
- Like dictionaries, you can index by label.
- Like NumPy arrays, you can index by boolean masks.
- You can index with a scalar, `slice`, or array
- Any of these should work on the index (row labels), or columns of a DataFrame, or both
- And any of these should work on hierarchical indexes.
## The Basic Rules
1. Use `__getitem__` (square brackets) to select columns of a `DataFrame`
```python
>>> df[['a', 'b', 'c']]
```
2. Use `.loc` for label-based indexing (rows and columns)
```python
>>> df.loc[row_labels, column_labels]
```
3. Use `.iloc` for position-based indexing (rows and columns)
```python
>>> df.iloc[row_positions, column_positions]
```
---
The arguments to `.loc` and `.iloc` are `.loc[row_indexer, column_indexer]`. An indexer can be one of
- A scalar or array (of labels or integer positions)
- A `slice` object (including `:` for everything)
- A boolean mask
The column indexer is optional.
We'll walk through all the combinations below.
Let's get a DataFrame with a labeled index by selecting the
first flight for each carrier. We'll talk about `groupby` later.
```
first = flights.groupby("unique_carrier").first()
first
```
## 1. Selecting Columns with `__getitem__`
Let's select the two delay columns. Since we're *only* filtering the columns (not rows), we can use dictionary-like `[]` to do the slicing.
```
first[['dep_delay', 'arr_delay']]
```
One potential source of confusion: python uses `[]` for two purposes
1. building a list
2. slicing with `__getitem__`
```
# 1. build the list cols
cols = ['dep_delay', 'arr_delay']
# 2. slice, with cols as the argument to `__getitem__`
first[cols]
```
<div class="alert alert-success" data-title="Select Columns by Name">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise: Select Columns by Name</h1>
</div>
<p>Select the two airport-name columns, `'origin'` and `'dest'`, from `first`</p>
```
# Your code here...
%load solutions/indexing_00.py
```
## Column `.` lookup
As a convenience, pandas attaches the column names to your `DataFrame` when they're valid [python identifiers](https://docs.python.org/3/reference/lexical_analysis.html), and don't override one of the ([many](http://pandas.pydata.org/pandas-docs/stable/api.html#dataframe)) methods on `DataFrame`
```
# Same as flights['tail_num'].value_counts()
flights.tail_num.value_counts()
```
This is nice when working interactively, especially as tab completion works with `flights.tail_num.<TAB>`, but doesn't with `flights['tail_num'].<TAB>`.
Still, since pandas could add methods in the future that clash with your column names, it's recommended to stick with `__getitem__` for production code.
This will always work, even when you shadow a DataFrame method
```
x = pd.DataFrame({"mean": [1, 2, 3]})
x
# returns the method, not the column
x.mean
```
Finally, you can't *assign* with `.`, while you can with `__setitem__` (square brackets on the left-hand side of an `=`):
```
x.wrong = ['a', 'b', 'c']
x['right'] = ['a', 'b', 'c']
x
x.wrong
```
`DataFrame`s, like most python objects, allow you to attach arbitrary attributes to any instance.
This means `x.wrong = ...` attaches the thing on the right-hand side to the object on the left.
## Label-Based Indexing with `.loc`
You can slice rows by label (and optionally the columns too) with `.loc`.
Let's select the rows for the carriers 'AA', 'DL', 'US', and 'WN'.
```
carriers = ['AA', 'DL', 'US', 'WN']
# Select those carriers by label
first.loc[carriers] # no column indexer
```
Indexing both rows and columns:
```
# select just `carriers` and origin, dest, and dep_delay
first.loc[carriers, ['origin', 'dest', 'dep_delay']]
```
Pandas will *reduce dimensionality* when possible, so slicing with a scalar on either axis will return a `Series`.
```
# select just tail_num for those `carriers`
first.loc[carriers, 'tail_num']
```
And scalars on both axes will return a scalar.
```
first.loc['AA', 'tail_num']
```
## `slice` objects
You can pass a `slice` object (made with a `:`). They make sense when your index is sorted, which ours is.
```
# slice from AA to DL
first.loc['AA':'DL']
```
Notice that the slice is inclusive on *both* sides *when using* `.loc` (`.iloc` follows the usual python semantics of closed on the left, open on the right).
<div class="alert alert-success" data-title="Index Rows and Columns">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise: Index Rows and Columns</h1>
</div>
<p>
Select the columns `tail_num`, `origin`, and `dest` for the carriers `US`, `VX`, and `WN` from `first`.</p>
```
# Your code here...
%load solutions/indexing_loc.py
```
## Boolean Indexing
Filter using a *1-dimensional* boolean array with the same length.
This is esstentially a SQL `WHERE` clause.
You filter the rows according to some condition.
For example, let's select flights that departed from the top-5 most common airports.
```
# Get the counts for `flights.origin`
origin_counts = flights.origin.value_counts()
origin_counts
```
[`Series.value_counts`](http://pandas.pydata.org/pandas-docs/stable/generated/pandas.Series.value_counts.html) will return a Series where the index is the set of unique values, and the values are the number of occurrances of that value.
It's sorted in descending order, so we can get the 5 most common labels with:
```
# Get a boolean mask for whether `counts` is in the top 5.
top5 = origin_counts.index[:5]
top5
```
We'll test whether any given row from `filghts` is in the top 5 using the `.isin` method:
```
mask = flights.origin.isin(top5)
mask
```
This is a *boolean mask*, which can be passed into `.loc`.
```
flights.loc[mask, ['origin', 'dest']]
```
You can pass boolean masks to regular `[]`, `.loc`, or `.iloc`.
Boolean indexers are useful because so many operations can produce an array of booleans.
- null checks (`.isnull`, `.notnull`)
- container checks (`.isin`)
- boolean aggregations (`.any`, `.all`)
- comparisions (`.gt`, `.lt`, etc.)
```
# Select rows where `dep` is null
flights.loc[flights.dep.isnull()]
```
<div class="alert alert-success" data-title="Boolean Indexing">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise: Boolean Indexing</h1>
</div>
<p>Select the rows of `flights` where the flight was cancelled (`cancelled == 1`)</p>
```
# Your code here
%load solutions/indexing_cancelled.py
```
<div class="alert alert-success" data-title="Boolean Indexing (2)">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise: Boolean Indexing (2)</h1>
</div>
<p>Filter down to rows where the departure **`hour`** is before 6:00 or after 18:00.</p>
- Hint: Use the `flights.dep.dt` namespace. See the attributes [here](http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DatetimeIndex.html) or use tab-completion on `flights.dep.dt.<tab>` to find an attribute giving the hour component
- Hint: NumPy and pandas use `|` for elementwise `or`, and `&` for elementwise `and` between two boolean arrays
- Hint: Be careful with [Python's order of operations](https://docs.python.org/3/reference/expressions.html#operator-precedence) between comparison operators (`<`, `>`) and NumPy's logical operators `|` and `&`. If your first attempt raises a `TypeError`, try including some parenthesis.
```
# Your code here...
%load solutions/indexing_01.py
```
## Position-Based Indexing with `.iloc`
This takes the same basic forms as `.loc`, except you use integers to designate *positions* instead of labels.
```
first.iloc[[0, 1, 2], [1, 2]]
```
You can use scalars, arrays, boolean masks, or slices.
You can also use negative indicies to slice from the end like regular python lists or numpy arrays.
The one notable difference compared to label-based indexing is with `slice` objects.
Recall that `.loc` included both ends of the slice.
`.iloc` uses the usual python behavior of slicing up to, but not including, the end point:
```
# select the first two rows, and the 3rd, 4th, and 5th columns
first.iloc[:2, 3:6]
```
## Dropping rows or columns
What if you want all items *except* for some?
```
DataFrame.drop(labels, axis=0, ...)
Parameters
----------
labels : single label or list-like
axis : int or axis name
- 0 / 'index', look in the index.
- 1 / 'columns', look in the columns
```
<div class="alert alert-success" data-title="Dropping Row Labels">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise: Dropping Row Labels</h1>
</div>
<p>Use `first.drop` to select all the rows *except* `EV` and `F9`.</p>
```
# Your code here
%load solutions/indexing_drop_index.py
```
<div class="alert alert-success" data-title="Drop a column">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise: Drop a column</h1>
</div>
<p>`flights.airline_id` is redundent with `unique_carrier`. Drop `airline_id`.</p>
```
# your code here
%load solutions/indexing_drop_columns.py
```
This is our first time seeing the `axis` keyword argument.
It comes up a lot in pandas and NumPy. `axis='index'` (or 0) means
operate on the index (e.g. drop labels from the index).
`axis='columns'` (or 1) means operate on the columns.
## Special Case: `DateTimeIndex`
> Easier slicing with strings
Pandas has really good support for time series data, including a few conveniences to make indexing easier.
First let's get a DataFrame with a `DatetimeIndex`, another specialied index type like we saw with `RangeIndex`.
```
# We'll talk about resample later
# This gets the average delays per hour
delays = flights.resample("H", on="arr")[['dep_delay', 'arr_delay']].mean()
delays.head()
```
`delays` has a `DatetimeIndex`:
```
delays.index
delays.plot();
```
Since `delays.index` is a `DatetimeIndex`, we can use [partial string indexing](http://pandas.pydata.org/pandas-docs/stable/timeseries.html#datetimeindex-partial-string-indexing) to easily select subsets of the data.
The basic idea is to specify the datetime up to whatever resolution you care about.
For example, to select all the flights on the 12th (a daily resolution):
```
delays.loc['2014-01-12']
```
Without parital string indexing, you'd need to do something like
```
delays[(delays.index.year == 2014) & (delays.index.month == 1) & (delays.index.day == 12)]
```
Which isn't very fun.
<div class="alert alert-success" data-title="Datetime Indexing">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise: Datetime Indexing</h1>
</div>
<p>
Slice `delays` to select all rows from 12:00 on January 3rd, to 12:00 on the 10th.</p>
```
# Your code
%load solutions/indexing_datetime.py
```
<div class="alert alert-success" data-title="Thought Exercise">
<h1><i class="fa fa-lightbulb-o" aria-hidden="true"></i> Thought Exercise</h1>
</div>
<p>
Why does pandas use a property like `.loc[..., ...]`, rather than a method like `.loc(..., ...)`?</p>
- Hint: Where in the language can you use `foo.loc[...]`, but not `foo.loc(...)`?
```
%load solutions/indexing_thoughts.py
```
## Summary
- Introducted to `DataFrame` (2-D tabel) and `Series` (1-D array)
- Both have *row labels*, `DataFrame` also has `column labels`
- Saw `.loc` for labeled indexing and `.iloc` for positional indexing
- `.loc`, `.iloc`, and `__getitem__` all accept boolean masks too
## Additional Exercises
Some additional exercises focused on indexing:
```
from IPython.display import IFrame
IFrame("http://vincentarelbundock.github.io/Rdatasets/doc/ISLR/Auto.html", 750, 900)
url = "http://vincentarelbundock.github.io/Rdatasets/csv/ISLR/Auto.csv"
cars = pd.read_csv(url, index_col=0)
cars.head()
yearly = cars.groupby("year").mean()
yearly.head()
```
<div class="alert alert-success" data-title="">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise</h1>
</div>
<p>
Select the engine-related columns (cylinders, displacement, horsepower) from `cars`.
</p>
```
%load solutions/indexing_ex1_engine_columns.py
```
<div class="alert alert-success" data-title="">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise</h1>
</div>
<p>
Select every 5th row of `cars`.
</p>
Hint: See [here](http://stackoverflow.com/a/509295) for all the options on slicing.
```
%load solutions/indexing_ex2_5th.py
```
<div class="alert alert-success" data-title="">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise</h1>
</div>
<p>Select years `70`, `75`, `80`, and `82` and columns `horsepower` and `weight` from `yearly`.</p>
```
%load solutions/indexing_ex3_years.py
```
<div class="alert alert-success" data-title="">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise</h1>
</div>
<p>Select rows in `cars` with at least 30 MPG.</p>
```
%load solutions/indexing_ex4_mpg.py
```
<div class="alert alert-success" data-title="">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise</h1>
</div>
<p>How many cars have at least 30 MPG and at least 5 cylinders?</p>
```
# %load solutions/indexing_ex5_mpg_and_cylinders.py
len(cars[(cars.mpg >= 30) & (cars.cylinders >= 5)])
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import seaborn.apionly as sns
import matplotlib.pyplot as plt
%matplotlib inline
pd.options.display.max_rows = 10
pd.options.display.float_format = '{:,.2f}'.format
plt.rcParams['figure.figsize'] = (16, 12)
flights = pd.read_csv("data/ny-flights.csv.gz",
parse_dates=["fl_date", "arr", "dep"])
flights
flights['dep_delay']
flights.index
flights.columns
flights.info()
(flights['dep']
.value_counts()
.resample('H')
.sum()
.rolling(8).mean()
.plot(figsize=(12, 6),
title="Number of Flights (8H Rolling Mean)"))
sns.despine()
sns.countplot(
flights['unique_carrier'],
order=flights['unique_carrier'].value_counts().index,
palette='Blues_r'
)
sns.despine()
sns.jointplot('dep_delay', 'arr_delay',
flights.loc[(flights['dep_delay'] > 1) &
(flights['dep_delay'] < 500)],
alpha=.25, marker='.', size=8);
>>> df[['a', 'b', 'c']]
```
2. Use `.loc` for label-based indexing (rows and columns)
```python
>>> df.loc[row_labels, column_labels]
```
3. Use `.iloc` for position-based indexing (rows and columns)
```python
>>> df.iloc[row_positions, column_positions]
```
---
The arguments to `.loc` and `.iloc` are `.loc[row_indexer, column_indexer]`. An indexer can be one of
- A scalar or array (of labels or integer positions)
- A `slice` object (including `:` for everything)
- A boolean mask
The column indexer is optional.
We'll walk through all the combinations below.
Let's get a DataFrame with a labeled index by selecting the
first flight for each carrier. We'll talk about `groupby` later.
## 1. Selecting Columns with `__getitem__`
Let's select the two delay columns. Since we're *only* filtering the columns (not rows), we can use dictionary-like `[]` to do the slicing.
One potential source of confusion: python uses `[]` for two purposes
1. building a list
2. slicing with `__getitem__`
<div class="alert alert-success" data-title="Select Columns by Name">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise: Select Columns by Name</h1>
</div>
<p>Select the two airport-name columns, `'origin'` and `'dest'`, from `first`</p>
## Column `.` lookup
As a convenience, pandas attaches the column names to your `DataFrame` when they're valid [python identifiers](https://docs.python.org/3/reference/lexical_analysis.html), and don't override one of the ([many](http://pandas.pydata.org/pandas-docs/stable/api.html#dataframe)) methods on `DataFrame`
This is nice when working interactively, especially as tab completion works with `flights.tail_num.<TAB>`, but doesn't with `flights['tail_num'].<TAB>`.
Still, since pandas could add methods in the future that clash with your column names, it's recommended to stick with `__getitem__` for production code.
This will always work, even when you shadow a DataFrame method
Finally, you can't *assign* with `.`, while you can with `__setitem__` (square brackets on the left-hand side of an `=`):
`DataFrame`s, like most python objects, allow you to attach arbitrary attributes to any instance.
This means `x.wrong = ...` attaches the thing on the right-hand side to the object on the left.
## Label-Based Indexing with `.loc`
You can slice rows by label (and optionally the columns too) with `.loc`.
Let's select the rows for the carriers 'AA', 'DL', 'US', and 'WN'.
Indexing both rows and columns:
Pandas will *reduce dimensionality* when possible, so slicing with a scalar on either axis will return a `Series`.
And scalars on both axes will return a scalar.
## `slice` objects
You can pass a `slice` object (made with a `:`). They make sense when your index is sorted, which ours is.
Notice that the slice is inclusive on *both* sides *when using* `.loc` (`.iloc` follows the usual python semantics of closed on the left, open on the right).
<div class="alert alert-success" data-title="Index Rows and Columns">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise: Index Rows and Columns</h1>
</div>
<p>
Select the columns `tail_num`, `origin`, and `dest` for the carriers `US`, `VX`, and `WN` from `first`.</p>
## Boolean Indexing
Filter using a *1-dimensional* boolean array with the same length.
This is esstentially a SQL `WHERE` clause.
You filter the rows according to some condition.
For example, let's select flights that departed from the top-5 most common airports.
[`Series.value_counts`](http://pandas.pydata.org/pandas-docs/stable/generated/pandas.Series.value_counts.html) will return a Series where the index is the set of unique values, and the values are the number of occurrances of that value.
It's sorted in descending order, so we can get the 5 most common labels with:
We'll test whether any given row from `filghts` is in the top 5 using the `.isin` method:
This is a *boolean mask*, which can be passed into `.loc`.
You can pass boolean masks to regular `[]`, `.loc`, or `.iloc`.
Boolean indexers are useful because so many operations can produce an array of booleans.
- null checks (`.isnull`, `.notnull`)
- container checks (`.isin`)
- boolean aggregations (`.any`, `.all`)
- comparisions (`.gt`, `.lt`, etc.)
<div class="alert alert-success" data-title="Boolean Indexing">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise: Boolean Indexing</h1>
</div>
<p>Select the rows of `flights` where the flight was cancelled (`cancelled == 1`)</p>
<div class="alert alert-success" data-title="Boolean Indexing (2)">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise: Boolean Indexing (2)</h1>
</div>
<p>Filter down to rows where the departure **`hour`** is before 6:00 or after 18:00.</p>
- Hint: Use the `flights.dep.dt` namespace. See the attributes [here](http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DatetimeIndex.html) or use tab-completion on `flights.dep.dt.<tab>` to find an attribute giving the hour component
- Hint: NumPy and pandas use `|` for elementwise `or`, and `&` for elementwise `and` between two boolean arrays
- Hint: Be careful with [Python's order of operations](https://docs.python.org/3/reference/expressions.html#operator-precedence) between comparison operators (`<`, `>`) and NumPy's logical operators `|` and `&`. If your first attempt raises a `TypeError`, try including some parenthesis.
## Position-Based Indexing with `.iloc`
This takes the same basic forms as `.loc`, except you use integers to designate *positions* instead of labels.
You can use scalars, arrays, boolean masks, or slices.
You can also use negative indicies to slice from the end like regular python lists or numpy arrays.
The one notable difference compared to label-based indexing is with `slice` objects.
Recall that `.loc` included both ends of the slice.
`.iloc` uses the usual python behavior of slicing up to, but not including, the end point:
## Dropping rows or columns
What if you want all items *except* for some?
<div class="alert alert-success" data-title="Dropping Row Labels">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise: Dropping Row Labels</h1>
</div>
<p>Use `first.drop` to select all the rows *except* `EV` and `F9`.</p>
<div class="alert alert-success" data-title="Drop a column">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise: Drop a column</h1>
</div>
<p>`flights.airline_id` is redundent with `unique_carrier`. Drop `airline_id`.</p>
This is our first time seeing the `axis` keyword argument.
It comes up a lot in pandas and NumPy. `axis='index'` (or 0) means
operate on the index (e.g. drop labels from the index).
`axis='columns'` (or 1) means operate on the columns.
## Special Case: `DateTimeIndex`
> Easier slicing with strings
Pandas has really good support for time series data, including a few conveniences to make indexing easier.
First let's get a DataFrame with a `DatetimeIndex`, another specialied index type like we saw with `RangeIndex`.
`delays` has a `DatetimeIndex`:
Since `delays.index` is a `DatetimeIndex`, we can use [partial string indexing](http://pandas.pydata.org/pandas-docs/stable/timeseries.html#datetimeindex-partial-string-indexing) to easily select subsets of the data.
The basic idea is to specify the datetime up to whatever resolution you care about.
For example, to select all the flights on the 12th (a daily resolution):
Without parital string indexing, you'd need to do something like
Which isn't very fun.
<div class="alert alert-success" data-title="Datetime Indexing">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise: Datetime Indexing</h1>
</div>
<p>
Slice `delays` to select all rows from 12:00 on January 3rd, to 12:00 on the 10th.</p>
<div class="alert alert-success" data-title="Thought Exercise">
<h1><i class="fa fa-lightbulb-o" aria-hidden="true"></i> Thought Exercise</h1>
</div>
<p>
Why does pandas use a property like `.loc[..., ...]`, rather than a method like `.loc(..., ...)`?</p>
- Hint: Where in the language can you use `foo.loc[...]`, but not `foo.loc(...)`?
## Summary
- Introducted to `DataFrame` (2-D tabel) and `Series` (1-D array)
- Both have *row labels*, `DataFrame` also has `column labels`
- Saw `.loc` for labeled indexing and `.iloc` for positional indexing
- `.loc`, `.iloc`, and `__getitem__` all accept boolean masks too
## Additional Exercises
Some additional exercises focused on indexing:
<div class="alert alert-success" data-title="">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise</h1>
</div>
<p>
Select the engine-related columns (cylinders, displacement, horsepower) from `cars`.
</p>
<div class="alert alert-success" data-title="">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise</h1>
</div>
<p>
Select every 5th row of `cars`.
</p>
Hint: See [here](http://stackoverflow.com/a/509295) for all the options on slicing.
<div class="alert alert-success" data-title="">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise</h1>
</div>
<p>Select years `70`, `75`, `80`, and `82` and columns `horsepower` and `weight` from `yearly`.</p>
<div class="alert alert-success" data-title="">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise</h1>
</div>
<p>Select rows in `cars` with at least 30 MPG.</p>
<div class="alert alert-success" data-title="">
<h1><i class="fa fa-tasks" aria-hidden="true"></i> Exercise</h1>
</div>
<p>How many cars have at least 30 MPG and at least 5 cylinders?</p>
| 0.828558 | 0.985566 |
# Welcome to Covid19 Data Analysis Notebook
------------------------------------------
### Let's Import the modules
```
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
print('Modules are imported.')
```
## Task 2
### Task 2.1: importing covid19 dataset
importing "Covid19_Confirmed_dataset.csv" from "./Dataset" folder.
```
corona_dataset_csv = pd.read_csv(r"C:\Users\abhis\Downloads\covid19_Confirmed_dataset.csv")
corona_dataset_csv.head(10)
```
#### Let's check the shape of the dataframe
```
corona_dataset_csv.shape
```
### Task 2.2: Delete the useless columns
```
corona_dataset_csv.drop(["Lat" ,"Long"] , axis=1 , inplace = True)
corona_dataset_csv.head(10)
```
### Task 2.3: Aggregating the rows by the country
```
corona_dataset_aggregated=corona_dataset_csv.groupby("Country/Region").sum()
corona_dataset_aggregated.head(10)
corona_dataset_aggregated.shape
```
### Task 2.4: Visualizing data related to a country for example China
visualization always helps for better understanding of our data.
```
corona_dataset_aggregated.loc["China"].plot()
corona_dataset_aggregated.loc["Italy"].plot()
corona_dataset_aggregated.loc["Spain"].plot()
plt.legend()
```
### Task3: Calculating a good measure
we need to find a good measure reperestend as a number, describing the spread of the virus in a country.
```
corona_dataset_aggregated.loc['China'].plot()
corona_dataset_aggregated.loc["China"][:3].plot()
```
### task 3.1: caculating the first derivative of the curve
```
corona_dataset_aggregated.loc['China'].diff().plot()
```
### task 3.2: find maxmimum infection rate for China
```
corona_dataset_aggregated.loc['China'].diff().max()
corona_dataset_aggregated.loc['Italy'].diff().max()
corona_dataset_aggregated.loc['Spain'].diff().max()
```
### Task 3.3: find maximum infection rate for all of the countries.
```
countries = list(corona_dataset_aggregated.index)
max_infection_rates = []
for c in countries:
max_infection_rates.append(corona_dataset_aggregated.loc[c].diff().max())
corona_dataset_aggregated["max_infection_rate"] = max_infection_rates
corona_dataset_aggregated.head()
corona_dataset_aggregated.head(10)
```
### Task 3.4: create a new dataframe with only needed column
```
corona_data = pd.DataFrame(corona_dataset_aggregated["max_infection_rate"])
corona_data.head()
corona_data.head(10)
```
### Task4:
- Importing the WorldHappinessReport.csv dataset
- selecting needed columns for our analysis
- join the datasets
- calculate the correlations as the result of our analysis
### Task 4.1 : importing the dataset
```
happiness_report_csv=pd.read_csv(r"C:\Users\abhis\Downloads\worldwide_happiness_report.csv")
happiness_report_csv.head()
```
### Task 4.2: let's drop the useless columns
```
useless_cols = ["Overall rank","Score","Generosity","Perceptions of corruption"]
happiness_report_csv.drop(useless_cols , axis =1 , inplace = True)
happiness_report_csv.head()
```
### Task 4.3: changing the indices of the dataframe
```
happiness_report_csv.set_index("Country or region" , inplace = True)
happiness_report_csv.head()
```
### Task4.4: now let's join two dataset we have prepared
#### Corona Dataset :
```
corona_data.head()
corona_data.shape
```
#### wolrd happiness report Dataset :
```
happiness_report_csv.head()
happiness_report_csv.shape
data = corona_data.join(happiness_report_csv , how = "inner")
data.head(10)
```
### Task 4.5: correlation matrix
```
data.corr()
```
### Task 5: Visualization of the results
our Analysis is not finished unless we visualize the results in terms figures and graphs so that everyone can understand what you get out of our analysis
```
data.head()
```
### Task 5.1: Plotting GDP vs maximum Infection rate
```
x=data["GDP per capita"]
y=data["max_infection_rate"]
sns.scatterplot(x ,y)
sns.scatterplot(x , np.log(y))
sns.regplot(x , np.log(y))
```
### Task 5.2: Plotting Social support vs maximum Infection rate
```
x=data["Social support"]
sns.scatterplot(x ,y)
sns.scatterplot(x,np.log(y))
sns.regplot(x , np.log(y))
```
### Task 5.3: Plotting Healthy life expectancy vs maximum Infection rate
```
x=data["Healthy life expectancy"]
sns.scatterplot(x ,y)
sns.scatterplot(x ,np.log(y))
```
### Task 5.4: Plotting Freedom to make life choices vs maximum Infection rate
```
x=data["Freedom to make life choices"]
sns.scatterplot(x ,y)
sns.scatterplot(x ,np.log(y))
sns.regplot(x ,np.log(y))
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
print('Modules are imported.')
corona_dataset_csv = pd.read_csv(r"C:\Users\abhis\Downloads\covid19_Confirmed_dataset.csv")
corona_dataset_csv.head(10)
corona_dataset_csv.shape
corona_dataset_csv.drop(["Lat" ,"Long"] , axis=1 , inplace = True)
corona_dataset_csv.head(10)
corona_dataset_aggregated=corona_dataset_csv.groupby("Country/Region").sum()
corona_dataset_aggregated.head(10)
corona_dataset_aggregated.shape
corona_dataset_aggregated.loc["China"].plot()
corona_dataset_aggregated.loc["Italy"].plot()
corona_dataset_aggregated.loc["Spain"].plot()
plt.legend()
corona_dataset_aggregated.loc['China'].plot()
corona_dataset_aggregated.loc["China"][:3].plot()
corona_dataset_aggregated.loc['China'].diff().plot()
corona_dataset_aggregated.loc['China'].diff().max()
corona_dataset_aggregated.loc['Italy'].diff().max()
corona_dataset_aggregated.loc['Spain'].diff().max()
countries = list(corona_dataset_aggregated.index)
max_infection_rates = []
for c in countries:
max_infection_rates.append(corona_dataset_aggregated.loc[c].diff().max())
corona_dataset_aggregated["max_infection_rate"] = max_infection_rates
corona_dataset_aggregated.head()
corona_dataset_aggregated.head(10)
corona_data = pd.DataFrame(corona_dataset_aggregated["max_infection_rate"])
corona_data.head()
corona_data.head(10)
happiness_report_csv=pd.read_csv(r"C:\Users\abhis\Downloads\worldwide_happiness_report.csv")
happiness_report_csv.head()
useless_cols = ["Overall rank","Score","Generosity","Perceptions of corruption"]
happiness_report_csv.drop(useless_cols , axis =1 , inplace = True)
happiness_report_csv.head()
happiness_report_csv.set_index("Country or region" , inplace = True)
happiness_report_csv.head()
corona_data.head()
corona_data.shape
happiness_report_csv.head()
happiness_report_csv.shape
data = corona_data.join(happiness_report_csv , how = "inner")
data.head(10)
data.corr()
data.head()
x=data["GDP per capita"]
y=data["max_infection_rate"]
sns.scatterplot(x ,y)
sns.scatterplot(x , np.log(y))
sns.regplot(x , np.log(y))
x=data["Social support"]
sns.scatterplot(x ,y)
sns.scatterplot(x,np.log(y))
sns.regplot(x , np.log(y))
x=data["Healthy life expectancy"]
sns.scatterplot(x ,y)
sns.scatterplot(x ,np.log(y))
x=data["Freedom to make life choices"]
sns.scatterplot(x ,y)
sns.scatterplot(x ,np.log(y))
sns.regplot(x ,np.log(y))
| 0.223208 | 0.906653 |
## 2.2 PyTorch第一步
PyTorch的简洁设计使得它入门很简单,在深入介绍PyTorch之前,本节将先介绍一些PyTorch的基础知识,使得读者能够对PyTorch有一个大致的了解,并能够用PyTorch搭建一个简单的神经网络。部分内容读者可能暂时不太理解,可先不予以深究,本书的第3章和第4章将会对此进行深入讲解。
本节内容参考了PyTorch官方教程[^1]并做了相应的增删修改,使得内容更贴合新版本的PyTorch接口,同时也更适合新手快速入门。另外本书需要读者先掌握基础的Numpy使用,其他相关知识推荐读者参考CS231n的教程[^2]。
[^1]: http://pytorch.org/tutorials/beginner/deep_learning_60min_blitz.html
[^2]: http://cs231n.github.io/python-numpy-tutorial/
### Tensor
Tensor是PyTorch中重要的数据结构,可认为是一个高维数组。它可以是一个数(标量)、一维数组(向量)、二维数组(矩阵)以及更高维的数组。Tensor和Numpy的ndarrays类似,但Tensor可以使用GPU进行加速。Tensor的使用和Numpy及Matlab的接口十分相似,下面通过几个例子来看看Tensor的基本使用。
```
from __future__ import print_function
import torch as t
t.__version__
# 构建 5x3 矩阵,只是分配了空间,未初始化
x = t.Tensor(5, 3)
x = t.Tensor([[1,2],[3,4]])
x
# 使用[0,1]均匀分布随机初始化二维数组
x = t.rand(5, 3)
x
print(x.size()) # 查看x的形状
x.size()[1], x.size(1) # 查看列的个数, 两种写法等价
```
`torch.Size` 是tuple对象的子类,因此它支持tuple的所有操作,如x.size()[0]
```
y = t.rand(5, 3)
# 加法的第一种写法
x + y
# 加法的第二种写法
t.add(x, y)
# 加法的第三种写法:指定加法结果的输出目标为result
result = t.Tensor(5, 3) # 预先分配空间
t.add(x, y, out=result) # 输入到result
result
print('最初y')
print(y)
print('第一种加法,y的结果')
y.add(x) # 普通加法,不改变y的内容
print(y)
print('第二种加法,y的结果')
y.add_(x) # inplace 加法,y变了
print(y)
```
注意,函数名后面带下划线**`_`** 的函数会修改Tensor本身。例如,`x.add_(y)`和`x.t_()`会改变 `x`,但`x.add(y)`和`x.t()`返回一个新的Tensor, 而`x`不变。
```
# Tensor的选取操作与Numpy类似
x[:, 1]
```
Tensor还支持很多操作,包括数学运算、线性代数、选择、切片等等,其接口设计与Numpy极为相似。更详细的使用方法,会在第三章系统讲解。
Tensor和Numpy的数组之间的互操作非常容易且快速。对于Tensor不支持的操作,可以先转为Numpy数组处理,之后再转回Tensor。c
```
a = t.ones(5) # 新建一个全1的Tensor
a
b = a.numpy() # Tensor -> Numpy
b
import numpy as np
a = np.ones(5)
b = t.from_numpy(a) # Numpy->Tensor
print(a)
print(b)
```
Tensor和numpy对象共享内存,所以他们之间的转换很快,而且几乎不会消耗什么资源。但这也意味着,如果其中一个变了,另外一个也会随之改变。
```
b.add_(1) # 以`_`结尾的函数会修改自身
print(a)
print(b) # Tensor和Numpy共享内存
```
如果你想获取某一个元素的值,可以使用`scalar.item`。 直接`tensor[idx]`得到的还是一个tensor: 一个0-dim 的tensor,一般称为scalar.
```
scalar = b[0]
scalar
scalar.size() #0-dim
scalar.item() # 使用scalar.item()能从中取出python对象的数值
tensor = t.tensor([2]) # 注意和scalar的区别
tensor,scalar
tensor.size(),scalar.size()
# 只有一个元素的tensor也可以调用`tensor.item()`
tensor.item(), scalar.item()
```
此外在pytorch中还有一个和`np.array` 很类似的接口: `torch.tensor`, 二者的使用十分类似。
```
tensor = t.tensor([3,4]) # 新建一个包含 3,4 两个元素的tensor
scalar = t.tensor(3)
scalar
old_tensor = tensor
new_tensor = t.tensor(old_tensor)
new_tensor[0] = 1111
old_tensor, new_tensor
```
需要注意的是,`t.tensor()`总是会进行数据拷贝,新tensor和原来的数据不再共享内存。所以如果你想共享内存的话,建议使用`torch.from_numpy()`或者`tensor.detach()`来新建一个tensor, 二者共享内存。
```
new_tensor = old_tensor.detach()
new_tensor[0] = 1111
old_tensor, new_tensor
```
Tensor可通过`.cuda` 方法转为GPU的Tensor,从而享受GPU带来的加速运算。
```
# 在不支持CUDA的机器下,下一步还是在CPU上运行
device = t.device("cuda:0" if t.cuda.is_available() else "cpu")
x = x.to(device)
y = y.to(device)
z = x+y
```
此外,还可以使用`tensor.cuda()` 的方式将tensor拷贝到gpu上,但是这种方式不太推荐。
此处可能发现GPU运算的速度并未提升太多,这是因为x和y太小且运算也较为简单,而且将数据从内存转移到显存还需要花费额外的开销。GPU的优势需在大规模数据和复杂运算下才能体现出来。
### autograd: 自动微分
深度学习的算法本质上是通过反向传播求导数,而PyTorch的**`autograd`**模块则实现了此功能。在Tensor上的所有操作,autograd都能为它们自动提供微分,避免了手动计算导数的复杂过程。
~~`autograd.Variable`是Autograd中的核心类,它简单封装了Tensor,并支持几乎所有Tensor有的操作。Tensor在被封装为Variable之后,可以调用它的`.backward`实现反向传播,自动计算所有梯度~~ ~~Variable的数据结构如图2-6所示。~~

*从0.4起, Variable 正式合并入Tensor, Variable 本来实现的自动微分功能,Tensor就能支持。读者还是可以使用Variable(tensor), 但是这个操作其实什么都没做。建议读者以后直接使用tensor*.
要想使得Tensor使用autograd功能,只需要设置`tensor.requries_grad=True`.
~~Variable主要包含三个属性。~~
~~- `data`:保存Variable所包含的Tensor~~
~~- `grad`:保存`data`对应的梯度,`grad`也是个Variable,而不是Tensor,它和`data`的形状一样。~~
~~- `grad_fn`:指向一个`Function`对象,这个`Function`用来反向传播计算输入的梯度,具体细节会在下一章讲解。~~
```
# 为tensor设置 requires_grad 标识,代表着需要求导数
# pytorch 会自动调用autograd 记录操作
x = t.ones(2, 2, requires_grad=True)
# 上一步等价于
# x = t.ones(2,2)
# x.requires_grad = True
x
y = x.sum()
y
y.grad_fn
y.backward() # 反向传播,计算梯度
# y = x.sum() = (x[0][0] + x[0][1] + x[1][0] + x[1][1])
# 每个值的梯度都为1
x.grad
```
注意:`grad`在反向传播过程中是累加的(accumulated),这意味着每一次运行反向传播,梯度都会累加之前的梯度,所以反向传播之前需把梯度清零。
```
y.backward()
x.grad
y.backward()
x.grad
# 以下划线结束的函数是inplace操作,会修改自身的值,就像add_
x.grad.data.zero_()
y.backward()
x.grad
```
### 神经网络
Autograd实现了反向传播功能,但是直接用来写深度学习的代码在很多情况下还是稍显复杂,torch.nn是专门为神经网络设计的模块化接口。nn构建于 Autograd之上,可用来定义和运行神经网络。nn.Module是nn中最重要的类,可把它看成是一个网络的封装,包含网络各层定义以及forward方法,调用forward(input)方法,可返回前向传播的结果。下面就以最早的卷积神经网络:LeNet为例,来看看如何用`nn.Module`实现。LeNet的网络结构如图2-7所示。

这是一个基础的前向传播(feed-forward)网络: 接收输入,经过层层传递运算,得到输出。
#### 定义网络
定义网络时,需要继承`nn.Module`,并实现它的forward方法,把网络中具有可学习参数的层放在构造函数`__init__`中。如果某一层(如ReLU)不具有可学习的参数,则既可以放在构造函数中,也可以不放,但建议不放在其中,而在forward中使用`nn.functional`代替。
```
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
# nn.Module子类的函数必须在构造函数中执行父类的构造函数
# 下式等价于nn.Module.__init__(self)
super(Net, self).__init__()
# 卷积层 '1'表示输入图片为单通道, '6'表示输出通道数,'5'表示卷积核为5*5
self.conv1 = nn.Conv2d(1, 6, 5)
# 卷积层
self.conv2 = nn.Conv2d(6, 16, 5)
# 仿射层/全连接层,y = Wx + b
self.fc1 = nn.Linear(16*5*5, 120)
self.fc2 = nn.Linear(120, 84)
self.fc3 = nn.Linear(84, 10)
def forward(self, x):
# 卷积 -> 激活 -> 池化
x = F.max_pool2d(F.relu(self.conv1(x)), (2, 2))
x = F.max_pool2d(F.relu(self.conv2(x)), 2)
# reshape,‘-1’表示自适应
x = x.view(x.size()[0], -1)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = self.fc3(x)
return x
net = Net()
print(net)
```
只要在nn.Module的子类中定义了forward函数,backward函数就会自动被实现(利用`autograd`)。在`forward` 函数中可使用任何tensor支持的函数,还可以使用if、for循环、print、log等Python语法,写法和标准的Python写法一致。
网络的可学习参数通过`net.parameters()`返回,`net.named_parameters`可同时返回可学习的参数及名称。
```
params = list(net.parameters())
print(len(params))
for name,parameters in net.named_parameters():
print(name,':',parameters.size())
```
forward函数的输入和输出都是Tensor。
```
input = t.randn(1, 1, 32, 32)
out = net(input)
out.size()
net.zero_grad() # 所有参数的梯度清零
out.backward(t.ones(1,10)) # 反向传播
```
需要注意的是,torch.nn只支持mini-batches,不支持一次只输入一个样本,即一次必须是一个batch。但如果只想输入一个样本,则用 `input.unsqueeze(0)`将batch_size设为1。例如 `nn.Conv2d` 输入必须是4维的,形如$nSamples \times nChannels \times Height \times Width$。可将nSample设为1,即$1 \times nChannels \times Height \times Width$。
#### 损失函数
nn实现了神经网络中大多数的损失函数,例如nn.MSELoss用来计算均方误差,nn.CrossEntropyLoss用来计算交叉熵损失。
```
output = net(input)
target = t.arange(0,10).view(1,10)
criterion = nn.MSELoss()
loss = criterion(output, target)
loss # loss是个scalar
```
如果对loss进行反向传播溯源(使用`gradfn`属性),可看到它的计算图如下:
```
input -> conv2d -> relu -> maxpool2d -> conv2d -> relu -> maxpool2d
-> view -> linear -> relu -> linear -> relu -> linear
-> MSELoss
-> loss
```
当调用`loss.backward()`时,该图会动态生成并自动微分,也即会自动计算图中参数(Parameter)的导数。
```
# 运行.backward,观察调用之前和调用之后的grad
net.zero_grad() # 把net中所有可学习参数的梯度清零
print('反向传播之前 conv1.bias的梯度')
print(net.conv1.bias.grad)
loss.backward()
print('反向传播之后 conv1.bias的梯度')
print(net.conv1.bias.grad)
```
#### 优化器
在反向传播计算完所有参数的梯度后,还需要使用优化方法来更新网络的权重和参数,例如随机梯度下降法(SGD)的更新策略如下:
```
weight = weight - learning_rate * gradient
```
手动实现如下:
```python
learning_rate = 0.01
for f in net.parameters():
f.data.sub_(f.grad.data * learning_rate)# inplace 减法
```
`torch.optim`中实现了深度学习中绝大多数的优化方法,例如RMSProp、Adam、SGD等,更便于使用,因此大多数时候并不需要手动写上述代码。
```
import torch.optim as optim
#新建一个优化器,指定要调整的参数和学习率
optimizer = optim.SGD(net.parameters(), lr = 0.01)
# 在训练过程中
# 先梯度清零(与net.zero_grad()效果一样)
optimizer.zero_grad()
# 计算损失
output = net(input)
loss = criterion(output, target)
#反向传播
loss.backward()
#更新参数
optimizer.step()
```
#### 数据加载与预处理
在深度学习中数据加载及预处理是非常复杂繁琐的,但PyTorch提供了一些可极大简化和加快数据处理流程的工具。同时,对于常用的数据集,PyTorch也提供了封装好的接口供用户快速调用,这些数据集主要保存在torchvison中。
`torchvision`实现了常用的图像数据加载功能,例如Imagenet、CIFAR10、MNIST等,以及常用的数据转换操作,这极大地方便了数据加载,并且代码具有可重用性。
### 小试牛刀:CIFAR-10分类
下面我们来尝试实现对CIFAR-10数据集的分类,步骤如下:
1. 使用torchvision加载并预处理CIFAR-10数据集
2. 定义网络
3. 定义损失函数和优化器
4. 训练网络并更新网络参数
5. 测试网络
#### CIFAR-10数据加载及预处理
CIFAR-10[^3]是一个常用的彩色图片数据集,它有10个类别: 'airplane', 'automobile', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck'。每张图片都是$3\times32\times32$,也即3-通道彩色图片,分辨率为$32\times32$。
[^3]: http://www.cs.toronto.edu/~kriz/cifar.html
```
import torchvision as tv
import torchvision.transforms as transforms
from torchvision.transforms import ToPILImage
show = ToPILImage() # 可以把Tensor转成Image,方便可视化
# 第一次运行程序torchvision会自动下载CIFAR-10数据集,
# 大约100M,需花费一定的时间,
# 如果已经下载有CIFAR-10,可通过root参数指定
# 定义对数据的预处理
transform = transforms.Compose([
transforms.ToTensor(), # 转为Tensor
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5)), # 归一化
])
# 训练集
trainset = tv.datasets.CIFAR10(
root='/home/cy/tmp/data/',
train=True,
download=True,
transform=transform)
trainloader = t.utils.data.DataLoader(
trainset,
batch_size=4,
shuffle=True,
num_workers=2)
# 测试集
testset = tv.datasets.CIFAR10(
'/home/cy/tmp/data/',
train=False,
download=True,
transform=transform)
testloader = t.utils.data.DataLoader(
testset,
batch_size=4,
shuffle=False,
num_workers=2)
classes = ('plane', 'car', 'bird', 'cat',
'deer', 'dog', 'frog', 'horse', 'ship', 'truck')
```
Dataset对象是一个数据集,可以按下标访问,返回形如(data, label)的数据。
```
(data, label) = trainset[100]
print(classes[label])
# (data + 1) / 2是为了还原被归一化的数据
show((data + 1) / 2).resize((100, 100))
```
Dataloader是一个可迭代的对象,它将dataset返回的每一条数据拼接成一个batch,并提供多线程加速优化和数据打乱等操作。当程序对dataset的所有数据遍历完一遍之后,相应的对Dataloader也完成了一次迭代。
```
dataiter = iter(trainloader)
images, labels = dataiter.next() # 返回4张图片及标签
print(' '.join('%11s'%classes[labels[j]] for j in range(4)))
show(tv.utils.make_grid((images+1)/2)).resize((400,100))
```
#### 定义网络
拷贝上面的LeNet网络,修改self.conv1第一个参数为3通道,因CIFAR-10是3通道彩图。
```
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.conv1 = nn.Conv2d(3, 6, 5)
self.conv2 = nn.Conv2d(6, 16, 5)
self.fc1 = nn.Linear(16*5*5, 120)
self.fc2 = nn.Linear(120, 84)
self.fc3 = nn.Linear(84, 10)
def forward(self, x):
x = F.max_pool2d(F.relu(self.conv1(x)), (2, 2))
x = F.max_pool2d(F.relu(self.conv2(x)), 2)
x = x.view(x.size()[0], -1)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = self.fc3(x)
return x
net = Net()
print(net)
```
#### 定义损失函数和优化器(loss和optimizer)
```
from torch import optim
criterion = nn.CrossEntropyLoss() # 交叉熵损失函数
optimizer = optim.SGD(net.parameters(), lr=0.001, momentum=0.9)
```
### 训练网络
所有网络的训练流程都是类似的,不断地执行如下流程:
- 输入数据
- 前向传播+反向传播
- 更新参数
```
t.set_num_threads(8)
for epoch in range(2):
running_loss = 0.0
for i, data in enumerate(trainloader, 0):
# 输入数据
inputs, labels = data
# 梯度清零
optimizer.zero_grad()
# forward + backward
outputs = net(inputs)
loss = criterion(outputs, labels)
loss.backward()
# 更新参数
optimizer.step()
# 打印log信息
# loss 是一个scalar,需要使用loss.item()来获取数值,不能使用loss[0]
running_loss += loss.item()
if i % 2000 == 1999: # 每2000个batch打印一下训练状态
print('[%d, %5d] loss: %.3f' \
% (epoch+1, i+1, running_loss / 2000))
running_loss = 0.0
print('Finished Training')
```
此处仅训练了2个epoch(遍历完一遍数据集称为一个epoch),来看看网络有没有效果。将测试图片输入到网络中,计算它的label,然后与实际的label进行比较。
```
dataiter = iter(testloader)
images, labels = dataiter.next() # 一个batch返回4张图片
print('实际的label: ', ' '.join(\
'%08s'%classes[labels[j]] for j in range(4)))
show(tv.utils.make_grid(images / 2 - 0.5)).resize((400,100))
```
接着计算网络预测的label:
```
# 计算图片在每个类别上的分数
outputs = net(images)
# 得分最高的那个类
_, predicted = t.max(outputs.data, 1)
print('预测结果: ', ' '.join('%5s'\
% classes[predicted[j]] for j in range(4)))
```
已经可以看出效果,准确率50%,但这只是一部分的图片,再来看看在整个测试集上的效果。
```
correct = 0 # 预测正确的图片数
total = 0 # 总共的图片数
# 由于测试的时候不需要求导,可以暂时关闭autograd,提高速度,节约内存
with t.no_grad():
for data in testloader:
images, labels = data
outputs = net(images)
_, predicted = t.max(outputs, 1)
total += labels.size(0)
correct += (predicted == labels).sum()
print('10000张测试集中的准确率为: %d %%' % (100 * correct / total))
```
训练的准确率远比随机猜测(准确率10%)好,证明网络确实学到了东西。
#### 在GPU训练
就像之前把Tensor从CPU转到GPU一样,模型也可以类似地从CPU转到GPU。
```
device = t.device("cuda:0" if t.cuda.is_available() else "cpu")
net.to(device)
images = images.to(device)
labels = labels.to(device)
output = net(images)
loss= criterion(output,labels)
loss
```
如果发现在GPU上并没有比CPU提速很多,实际上是因为网络比较小,GPU没有完全发挥自己的真正实力。
对PyTorch的基础介绍至此结束。总结一下,本节主要包含以下内容。
1. Tensor: 类似Numpy数组的数据结构,与Numpy接口类似,可方便地互相转换。
2. autograd/: 为tensor提供自动求导功能。
3. nn: 专门为神经网络设计的接口,提供了很多有用的功能(神经网络层,损失函数,优化器等)。
4. 神经网络训练: 以CIFAR-10分类为例演示了神经网络的训练流程,包括数据加载、网络搭建、训练及测试。
通过本节的学习,相信读者可以体会出PyTorch具有接口简单、使用灵活等特点。从下一章开始,本书将深入系统地讲解PyTorch的各部分知识。
|
github_jupyter
|
from __future__ import print_function
import torch as t
t.__version__
# 构建 5x3 矩阵,只是分配了空间,未初始化
x = t.Tensor(5, 3)
x = t.Tensor([[1,2],[3,4]])
x
# 使用[0,1]均匀分布随机初始化二维数组
x = t.rand(5, 3)
x
print(x.size()) # 查看x的形状
x.size()[1], x.size(1) # 查看列的个数, 两种写法等价
y = t.rand(5, 3)
# 加法的第一种写法
x + y
# 加法的第二种写法
t.add(x, y)
# 加法的第三种写法:指定加法结果的输出目标为result
result = t.Tensor(5, 3) # 预先分配空间
t.add(x, y, out=result) # 输入到result
result
print('最初y')
print(y)
print('第一种加法,y的结果')
y.add(x) # 普通加法,不改变y的内容
print(y)
print('第二种加法,y的结果')
y.add_(x) # inplace 加法,y变了
print(y)
# Tensor的选取操作与Numpy类似
x[:, 1]
a = t.ones(5) # 新建一个全1的Tensor
a
b = a.numpy() # Tensor -> Numpy
b
import numpy as np
a = np.ones(5)
b = t.from_numpy(a) # Numpy->Tensor
print(a)
print(b)
b.add_(1) # 以`_`结尾的函数会修改自身
print(a)
print(b) # Tensor和Numpy共享内存
scalar = b[0]
scalar
scalar.size() #0-dim
scalar.item() # 使用scalar.item()能从中取出python对象的数值
tensor = t.tensor([2]) # 注意和scalar的区别
tensor,scalar
tensor.size(),scalar.size()
# 只有一个元素的tensor也可以调用`tensor.item()`
tensor.item(), scalar.item()
tensor = t.tensor([3,4]) # 新建一个包含 3,4 两个元素的tensor
scalar = t.tensor(3)
scalar
old_tensor = tensor
new_tensor = t.tensor(old_tensor)
new_tensor[0] = 1111
old_tensor, new_tensor
new_tensor = old_tensor.detach()
new_tensor[0] = 1111
old_tensor, new_tensor
# 在不支持CUDA的机器下,下一步还是在CPU上运行
device = t.device("cuda:0" if t.cuda.is_available() else "cpu")
x = x.to(device)
y = y.to(device)
z = x+y
# 为tensor设置 requires_grad 标识,代表着需要求导数
# pytorch 会自动调用autograd 记录操作
x = t.ones(2, 2, requires_grad=True)
# 上一步等价于
# x = t.ones(2,2)
# x.requires_grad = True
x
y = x.sum()
y
y.grad_fn
y.backward() # 反向传播,计算梯度
# y = x.sum() = (x[0][0] + x[0][1] + x[1][0] + x[1][1])
# 每个值的梯度都为1
x.grad
y.backward()
x.grad
y.backward()
x.grad
# 以下划线结束的函数是inplace操作,会修改自身的值,就像add_
x.grad.data.zero_()
y.backward()
x.grad
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
# nn.Module子类的函数必须在构造函数中执行父类的构造函数
# 下式等价于nn.Module.__init__(self)
super(Net, self).__init__()
# 卷积层 '1'表示输入图片为单通道, '6'表示输出通道数,'5'表示卷积核为5*5
self.conv1 = nn.Conv2d(1, 6, 5)
# 卷积层
self.conv2 = nn.Conv2d(6, 16, 5)
# 仿射层/全连接层,y = Wx + b
self.fc1 = nn.Linear(16*5*5, 120)
self.fc2 = nn.Linear(120, 84)
self.fc3 = nn.Linear(84, 10)
def forward(self, x):
# 卷积 -> 激活 -> 池化
x = F.max_pool2d(F.relu(self.conv1(x)), (2, 2))
x = F.max_pool2d(F.relu(self.conv2(x)), 2)
# reshape,‘-1’表示自适应
x = x.view(x.size()[0], -1)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = self.fc3(x)
return x
net = Net()
print(net)
params = list(net.parameters())
print(len(params))
for name,parameters in net.named_parameters():
print(name,':',parameters.size())
input = t.randn(1, 1, 32, 32)
out = net(input)
out.size()
net.zero_grad() # 所有参数的梯度清零
out.backward(t.ones(1,10)) # 反向传播
output = net(input)
target = t.arange(0,10).view(1,10)
criterion = nn.MSELoss()
loss = criterion(output, target)
loss # loss是个scalar
input -> conv2d -> relu -> maxpool2d -> conv2d -> relu -> maxpool2d
-> view -> linear -> relu -> linear -> relu -> linear
-> MSELoss
-> loss
# 运行.backward,观察调用之前和调用之后的grad
net.zero_grad() # 把net中所有可学习参数的梯度清零
print('反向传播之前 conv1.bias的梯度')
print(net.conv1.bias.grad)
loss.backward()
print('反向传播之后 conv1.bias的梯度')
print(net.conv1.bias.grad)
weight = weight - learning_rate * gradient
learning_rate = 0.01
for f in net.parameters():
f.data.sub_(f.grad.data * learning_rate)# inplace 减法
import torch.optim as optim
#新建一个优化器,指定要调整的参数和学习率
optimizer = optim.SGD(net.parameters(), lr = 0.01)
# 在训练过程中
# 先梯度清零(与net.zero_grad()效果一样)
optimizer.zero_grad()
# 计算损失
output = net(input)
loss = criterion(output, target)
#反向传播
loss.backward()
#更新参数
optimizer.step()
import torchvision as tv
import torchvision.transforms as transforms
from torchvision.transforms import ToPILImage
show = ToPILImage() # 可以把Tensor转成Image,方便可视化
# 第一次运行程序torchvision会自动下载CIFAR-10数据集,
# 大约100M,需花费一定的时间,
# 如果已经下载有CIFAR-10,可通过root参数指定
# 定义对数据的预处理
transform = transforms.Compose([
transforms.ToTensor(), # 转为Tensor
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5)), # 归一化
])
# 训练集
trainset = tv.datasets.CIFAR10(
root='/home/cy/tmp/data/',
train=True,
download=True,
transform=transform)
trainloader = t.utils.data.DataLoader(
trainset,
batch_size=4,
shuffle=True,
num_workers=2)
# 测试集
testset = tv.datasets.CIFAR10(
'/home/cy/tmp/data/',
train=False,
download=True,
transform=transform)
testloader = t.utils.data.DataLoader(
testset,
batch_size=4,
shuffle=False,
num_workers=2)
classes = ('plane', 'car', 'bird', 'cat',
'deer', 'dog', 'frog', 'horse', 'ship', 'truck')
(data, label) = trainset[100]
print(classes[label])
# (data + 1) / 2是为了还原被归一化的数据
show((data + 1) / 2).resize((100, 100))
dataiter = iter(trainloader)
images, labels = dataiter.next() # 返回4张图片及标签
print(' '.join('%11s'%classes[labels[j]] for j in range(4)))
show(tv.utils.make_grid((images+1)/2)).resize((400,100))
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.conv1 = nn.Conv2d(3, 6, 5)
self.conv2 = nn.Conv2d(6, 16, 5)
self.fc1 = nn.Linear(16*5*5, 120)
self.fc2 = nn.Linear(120, 84)
self.fc3 = nn.Linear(84, 10)
def forward(self, x):
x = F.max_pool2d(F.relu(self.conv1(x)), (2, 2))
x = F.max_pool2d(F.relu(self.conv2(x)), 2)
x = x.view(x.size()[0], -1)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = self.fc3(x)
return x
net = Net()
print(net)
from torch import optim
criterion = nn.CrossEntropyLoss() # 交叉熵损失函数
optimizer = optim.SGD(net.parameters(), lr=0.001, momentum=0.9)
t.set_num_threads(8)
for epoch in range(2):
running_loss = 0.0
for i, data in enumerate(trainloader, 0):
# 输入数据
inputs, labels = data
# 梯度清零
optimizer.zero_grad()
# forward + backward
outputs = net(inputs)
loss = criterion(outputs, labels)
loss.backward()
# 更新参数
optimizer.step()
# 打印log信息
# loss 是一个scalar,需要使用loss.item()来获取数值,不能使用loss[0]
running_loss += loss.item()
if i % 2000 == 1999: # 每2000个batch打印一下训练状态
print('[%d, %5d] loss: %.3f' \
% (epoch+1, i+1, running_loss / 2000))
running_loss = 0.0
print('Finished Training')
dataiter = iter(testloader)
images, labels = dataiter.next() # 一个batch返回4张图片
print('实际的label: ', ' '.join(\
'%08s'%classes[labels[j]] for j in range(4)))
show(tv.utils.make_grid(images / 2 - 0.5)).resize((400,100))
# 计算图片在每个类别上的分数
outputs = net(images)
# 得分最高的那个类
_, predicted = t.max(outputs.data, 1)
print('预测结果: ', ' '.join('%5s'\
% classes[predicted[j]] for j in range(4)))
correct = 0 # 预测正确的图片数
total = 0 # 总共的图片数
# 由于测试的时候不需要求导,可以暂时关闭autograd,提高速度,节约内存
with t.no_grad():
for data in testloader:
images, labels = data
outputs = net(images)
_, predicted = t.max(outputs, 1)
total += labels.size(0)
correct += (predicted == labels).sum()
print('10000张测试集中的准确率为: %d %%' % (100 * correct / total))
device = t.device("cuda:0" if t.cuda.is_available() else "cpu")
net.to(device)
images = images.to(device)
labels = labels.to(device)
output = net(images)
loss= criterion(output,labels)
loss
| 0.710126 | 0.95469 |
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
pd.set_option('display.max_column', None)
df = pd.read_csv('./course-data/StudentsPerformance.csv')
df.head(3)
# categorical information on the students:
# gender
# race/ethnicity
# parental level of education
# lunch
# test preparation course (yes/no)
# numerical data:
# math score
# reading score
# writing score
# we can begin understanding the distributions across the continours features of the scores and how they relate to various categories
len(df)
sns.boxplot(data=df, y='math score')
# box plot distro for ALL the math scores (not separated by any category)
df['math score'].describe()
df['math score'].median()
plt.figure(figsize=(10,4),dpi=200)
sns.boxplot(data=df, y='math score',x='test preparation course')
# the distributions are pretty close but we see if you completed the test preparation course you're likely to have a higher math score and the distribution is tighter
sns.boxplot(data=df, y='reading score',x='test preparation course')
plt.figure(figsize=(10,4))
sns.boxplot(data=df, y='reading score',x='parental level of education')
plt.figure(figsize=(10,4))
sns.boxplot(data=df, y='reading score',x='parental level of education',hue='test preparation course')
plt.legend(bbox_to_anchor=(1.02,1))
# note that across all parental level of educations the median score is higher for people that completed the prep course (regardless of the parental level of education)
plt.figure(figsize=(10,8))
sns.boxplot(data=df, x='reading score',y='parental level of education',hue='test preparation course',palette='Pastel1')
plt.legend(bbox_to_anchor=(1.02,0.5))
# we can show this with categories in the y axis
plt.figure(figsize=(10,8))
sns.violinplot(data=df, x='reading score',y='parental level of education',hue='test preparation course',palette='Pastel1')
plt.figure(figsize=(10,8))
sns.violinplot(data=df, x='reading score',y='parental level of education',hue='test preparation course',palette='Pastel1',split=True)
plt.figure(figsize=(10,8))
sns.violinplot(data=df, x='reading score',y='parental level of education',hue='test preparation course',palette='Pastel1',split=True,inner=None)
plt.figure(figsize=(10,8))
sns.violinplot(data=df, x='reading score',y='parental level of education',hue='test preparation course',palette='Pastel1',split=True,inner='quartile')
plt.figure(figsize=(10,8))
sns.violinplot(data=df, x='reading score',y='parental level of education',palette='Pastel1',split=True,inner='quartile')
plt.figure(figsize=(10,8))
sns.violinplot(data=df, x='reading score',y='parental level of education',palette='Pastel1',split=True,inner='stick')
plt.figure(figsize=(10,8))
sns.violinplot(data=df, x='reading score',y='parental level of education',palette='Pastel1',split=True,inner='stick',bw=0.1)
sns.swarmplot(data=df, x='math score')
# the figure / dot size are not the right size to display all the points
plt.figure(figsize=(8,4),dpi=200)
sns.swarmplot(data=df, x='math score',y='gender',size=2, hue='test preparation course')
plt.legend(bbox_to_anchor=(1.25,0.5))
plt.figure(figsize=(8,4),dpi=200)
sns.swarmplot(data=df, x='math score',y='gender',size=2, hue='test preparation course',dodge=True)
plt.legend(bbox_to_anchor=(1.25,0.5))
sns.boxenplot(data=df, x='math score',y='test preparation course')
sns.boxenplot(data=df, x='math score',y='test preparation course',hue='gender')
sns.boxplot(data=df, x='math score',y='test preparation course',hue='gender')
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
pd.set_option('display.max_column', None)
df = pd.read_csv('./course-data/StudentsPerformance.csv')
df.head(3)
# categorical information on the students:
# gender
# race/ethnicity
# parental level of education
# lunch
# test preparation course (yes/no)
# numerical data:
# math score
# reading score
# writing score
# we can begin understanding the distributions across the continours features of the scores and how they relate to various categories
len(df)
sns.boxplot(data=df, y='math score')
# box plot distro for ALL the math scores (not separated by any category)
df['math score'].describe()
df['math score'].median()
plt.figure(figsize=(10,4),dpi=200)
sns.boxplot(data=df, y='math score',x='test preparation course')
# the distributions are pretty close but we see if you completed the test preparation course you're likely to have a higher math score and the distribution is tighter
sns.boxplot(data=df, y='reading score',x='test preparation course')
plt.figure(figsize=(10,4))
sns.boxplot(data=df, y='reading score',x='parental level of education')
plt.figure(figsize=(10,4))
sns.boxplot(data=df, y='reading score',x='parental level of education',hue='test preparation course')
plt.legend(bbox_to_anchor=(1.02,1))
# note that across all parental level of educations the median score is higher for people that completed the prep course (regardless of the parental level of education)
plt.figure(figsize=(10,8))
sns.boxplot(data=df, x='reading score',y='parental level of education',hue='test preparation course',palette='Pastel1')
plt.legend(bbox_to_anchor=(1.02,0.5))
# we can show this with categories in the y axis
plt.figure(figsize=(10,8))
sns.violinplot(data=df, x='reading score',y='parental level of education',hue='test preparation course',palette='Pastel1')
plt.figure(figsize=(10,8))
sns.violinplot(data=df, x='reading score',y='parental level of education',hue='test preparation course',palette='Pastel1',split=True)
plt.figure(figsize=(10,8))
sns.violinplot(data=df, x='reading score',y='parental level of education',hue='test preparation course',palette='Pastel1',split=True,inner=None)
plt.figure(figsize=(10,8))
sns.violinplot(data=df, x='reading score',y='parental level of education',hue='test preparation course',palette='Pastel1',split=True,inner='quartile')
plt.figure(figsize=(10,8))
sns.violinplot(data=df, x='reading score',y='parental level of education',palette='Pastel1',split=True,inner='quartile')
plt.figure(figsize=(10,8))
sns.violinplot(data=df, x='reading score',y='parental level of education',palette='Pastel1',split=True,inner='stick')
plt.figure(figsize=(10,8))
sns.violinplot(data=df, x='reading score',y='parental level of education',palette='Pastel1',split=True,inner='stick',bw=0.1)
sns.swarmplot(data=df, x='math score')
# the figure / dot size are not the right size to display all the points
plt.figure(figsize=(8,4),dpi=200)
sns.swarmplot(data=df, x='math score',y='gender',size=2, hue='test preparation course')
plt.legend(bbox_to_anchor=(1.25,0.5))
plt.figure(figsize=(8,4),dpi=200)
sns.swarmplot(data=df, x='math score',y='gender',size=2, hue='test preparation course',dodge=True)
plt.legend(bbox_to_anchor=(1.25,0.5))
sns.boxenplot(data=df, x='math score',y='test preparation course')
sns.boxenplot(data=df, x='math score',y='test preparation course',hue='gender')
sns.boxplot(data=df, x='math score',y='test preparation course',hue='gender')
| 0.608594 | 0.805747 |
```
%matplotlib inline
import os
import warnings
import critical_loads as cl
import gdal
import geopandas as gpd
import matplotlib.pyplot as plt
import nivapy3 as nivapy
import numpy as np
import pandas as pd
warnings.simplefilter("ignore")
plt.style.use("ggplot")
# Connect to PostGIS
eng = nivapy.da.connect_postgis(database="critical_loads")
```
# Critical loads: Water workflow (high-resolution method; 2018 onwards)
In Spring 2018, the worflow for calculating critical loads was refined to make use of new, higher-resolution input datasets. During November 2018, data handling for the Critical Loads project was also redesigned, with the ultimate aim of centralising all key datasets on NIVA's new cloud platform.
This notebook deals with data processing for the water exceedance calculations, using the new 0.1 degree deposition grid and the old, BLR-based water chemistry dataset.
## 1. Organising water data on the DSToolkit
NIVA's JupyterHub includes a PostGIS database server capable of storing relational and vector geospatial datasets. I have created a database named `critical_loads` and, within this, a schema named `water`. This schema contains the following tables:
* **parameter_definitions:** Non-spatial table describing the water chemistry and model parameters used to calculate water critical loads
* **blr_magic_parameters:** Non-spatial table containing result from the MAGIC model
* **blr_optional_parameters:** Non-spatial table containing optional parameters for the calculation of critical loads
* **blr_required_parameters:** Non-spatial table containing required parameters for the calculation of critical loads
* **magic_regression_data:** Output from the MAGIC model used for estimating $BC_0$
## 2. Define deposition data series of interest
Choose the deposition dataset and resolution you wish to work with (see notebook 01 for a list of available `series_ids`). Note that **S and N deposition grids at the specified resolution must already have been created in notebook 02**. 120 m resolution is more than sufficient - the highest resolution of raw data in this workflow is 0.1 degrees, which is several _kilometres_, even in northern Norway.
```
# Series of interest
ser_id = 28
# Choose cell size (30m, 60m or 120m)
cell_size = 120
```
## 3. Calculate critical loads for water
The code below reads water chemistry and model parameters for each BLR grid cell from the database and creates rasters for the following key parameters:
'claoaa', 'eno3', 'clminn', 'clmaxnoaa', 'clmaxsoaa', 'clmins', 'anclimit', 'anclimit_oaa', 'bc0'
See the function `docstring` for details - it includes various options for e.g. using Magic data from an Excel template.
#### Added 02.11.2020
The function has been modified to also output `anclimit_ueqpl`, `anclimitoaa_ueqpl` and `bc0_ueqpl`. See e-mail from Kari received 02.11.2020 at 09.30 for details.
```
# Rasterise critical loads for water
out_fold = r"/home/jovyan/shared/critical_loads/raster/water"
cl.rasterise_water_critical_loads(eng, out_fold, cell_size, df_to_csv=True)
cl.rasterise_water_critical_loads(
eng, out_fold, cell_size, bc0="BC0_magic", mag_df=mag_df, df_to_csv=True
)
```
## 4. Calculate exceedances
### 4.1. SSWC model
```
cl_fold = r"/home/jovyan/shared/critical_loads/raster/water"
out_fold = r"/home/jovyan/shared/critical_loads/raster/exceedance"
ex_df = cl.calculate_water_exceedance_sswc(
ser_id, "1216", cl_fold, out_fold, cell_size=120, bc0="BC0", neg_to_zero=False
)
ex_df
```
**If you connected to the database with `admin=True`**, these results can be saved back to the database.
```
## Write summary data to db
# ex_df.to_sql('national_summary',
# eng,
# 'summaries',
# if_exists='append',
# index=False)
```
### 4.2. FAB model
The code in this section calculates exceedances for water using the FAB model. This is based on my original code [here](https://nbviewer.jupyter.org/github/JamesSample/critical_loads/blob/master/notebooks/critical_loads_workflow_new_grid.ipynb#2.3.3.-FAB-model).
```
# Constants to build paths
base_path = r"/home/jovyan/shared/critical_loads/raster"
short_name = "1216"
# Read CL arrays
array_dict = {}
for par in [
"clminn_meqpm2pyr",
"clmaxnoaa_meqpm2pyr",
"clmins_meqpm2pyr",
"clmaxsoaa_meqpm2pyr",
]:
# Read tif
tif_path = os.path.join(base_path, "water", f"{par}_{cell_size}m.tif")
data, ndv, epsg, extent = nivapy.spatial.read_raster(tif_path)
# Set NDV
data[data == ndv] = np.nan
# Add to dict
array_dict[par] = data
# Read dep arrays
for par in ["ndep_mgpm2pyr", "sdep_mgpm2pyr"]:
# Read tif
tif_path = os.path.join(
base_path, "deposition", f"{par}_{short_name}_{cell_size}m.tif"
)
data, ndv, epsg, extent = nivapy.spatial.read_raster(tif_path)
data = data.astype(np.float32)
# Set NDV
data[data == ndv] = np.nan
# Add to dict
array_dict[par] = data
```
**The code below needs to be run on a "high memory" machine, even with 120 m resolution.**
```
%%time
# Extract arrays from dict
cln_min = array_dict["clminn_meqpm2pyr"]
cln_max = array_dict["clmaxnoaa_meqpm2pyr"]
cls_min = array_dict["clmins_meqpm2pyr"]
cls_max = array_dict["clmaxsoaa_meqpm2pyr"]
dep_n = array_dict["ndep_mgpm2pyr"] / 14 # Convert to meq
dep_s = array_dict["sdep_mgpm2pyr"] * 2 / 32.06 # Convert to meq
# Estimate exceedances
ex_n, ex_s, reg_id = cl.vectorised_exceed_ns_icpm(
cln_min, cln_max, cls_min, cls_max, dep_n, dep_s
)
# Get exceeded area
ex = ex_n + ex_s
ex_area = np.count_nonzero(ex > 0) * cell_size * cell_size / 1.0e6
nor_area = np.count_nonzero(~np.isnan(dep_s)) * cell_size * cell_size / 1.0e6
ex_pct = 100 * ex_area / nor_area
# Build df and tidy
ex_df = pd.DataFrame(
{
"exceeded_area_km2": ex_area,
"total_area_km2": nor_area,
"exceeded_area_pct": ex_pct,
},
index=[0],
)
ex_df = ex_df.round(0).astype(int)
ex_df["series_id"] = ser_id
ex_df["medium"] = "water_fab"
ex_df = ex_df[
["series_id", "medium", "total_area_km2", "exceeded_area_km2", "exceeded_area_pct"]
]
ex_df
```
**If you connected to the database with `admin=True`**, these results can be saved back to the database.
```
## Write summary data to db
# ex_df.to_sql('national_summary',
# eng,
# 'summaries',
# if_exists='append',
# index=False)
```
The code below writes the FAB results to GeoTiff format.
```
# Snap tiff
snap_tif = f"/home/jovyan/shared/critical_loads/raster/blr_land_mask_{cell_size}m.tif"
# N
n_tif = f"/home/jovyan/shared/critical_loads/raster/exceedance/fab_exn_meqpm2pyr_{short_name}_{cell_size}m.tif"
cl.write_geotiff(ex_n, n_tif, snap_tif, -1, gdal.GDT_Float32)
# S
s_tif = f"/home/jovyan/shared/critical_loads/raster/exceedance/fab_exs_meqpm2pyr_{short_name}_{cell_size}m.tif"
cl.write_geotiff(ex_s, s_tif, snap_tif, -1, gdal.GDT_Float32)
# N+S
ns_tif = f"/home/jovyan/shared/critical_loads/raster/exceedance/fab_exns_meqpm2pyr_{short_name}_{cell_size}m.tif"
cl.write_geotiff(ex_n + ex_s, ns_tif, snap_tif, -1, gdal.GDT_Float32)
# Exceedance 'region'
reg_tif = f"/home/jovyan/shared/critical_loads/raster/exceedance/fab_ex_reg_id_{short_name}_{cell_size}m.tif"
cl.write_geotiff(reg_id, reg_tif, snap_tif, -1, gdal.GDT_Float32)
```
|
github_jupyter
|
%matplotlib inline
import os
import warnings
import critical_loads as cl
import gdal
import geopandas as gpd
import matplotlib.pyplot as plt
import nivapy3 as nivapy
import numpy as np
import pandas as pd
warnings.simplefilter("ignore")
plt.style.use("ggplot")
# Connect to PostGIS
eng = nivapy.da.connect_postgis(database="critical_loads")
# Series of interest
ser_id = 28
# Choose cell size (30m, 60m or 120m)
cell_size = 120
# Rasterise critical loads for water
out_fold = r"/home/jovyan/shared/critical_loads/raster/water"
cl.rasterise_water_critical_loads(eng, out_fold, cell_size, df_to_csv=True)
cl.rasterise_water_critical_loads(
eng, out_fold, cell_size, bc0="BC0_magic", mag_df=mag_df, df_to_csv=True
)
cl_fold = r"/home/jovyan/shared/critical_loads/raster/water"
out_fold = r"/home/jovyan/shared/critical_loads/raster/exceedance"
ex_df = cl.calculate_water_exceedance_sswc(
ser_id, "1216", cl_fold, out_fold, cell_size=120, bc0="BC0", neg_to_zero=False
)
ex_df
## Write summary data to db
# ex_df.to_sql('national_summary',
# eng,
# 'summaries',
# if_exists='append',
# index=False)
# Constants to build paths
base_path = r"/home/jovyan/shared/critical_loads/raster"
short_name = "1216"
# Read CL arrays
array_dict = {}
for par in [
"clminn_meqpm2pyr",
"clmaxnoaa_meqpm2pyr",
"clmins_meqpm2pyr",
"clmaxsoaa_meqpm2pyr",
]:
# Read tif
tif_path = os.path.join(base_path, "water", f"{par}_{cell_size}m.tif")
data, ndv, epsg, extent = nivapy.spatial.read_raster(tif_path)
# Set NDV
data[data == ndv] = np.nan
# Add to dict
array_dict[par] = data
# Read dep arrays
for par in ["ndep_mgpm2pyr", "sdep_mgpm2pyr"]:
# Read tif
tif_path = os.path.join(
base_path, "deposition", f"{par}_{short_name}_{cell_size}m.tif"
)
data, ndv, epsg, extent = nivapy.spatial.read_raster(tif_path)
data = data.astype(np.float32)
# Set NDV
data[data == ndv] = np.nan
# Add to dict
array_dict[par] = data
%%time
# Extract arrays from dict
cln_min = array_dict["clminn_meqpm2pyr"]
cln_max = array_dict["clmaxnoaa_meqpm2pyr"]
cls_min = array_dict["clmins_meqpm2pyr"]
cls_max = array_dict["clmaxsoaa_meqpm2pyr"]
dep_n = array_dict["ndep_mgpm2pyr"] / 14 # Convert to meq
dep_s = array_dict["sdep_mgpm2pyr"] * 2 / 32.06 # Convert to meq
# Estimate exceedances
ex_n, ex_s, reg_id = cl.vectorised_exceed_ns_icpm(
cln_min, cln_max, cls_min, cls_max, dep_n, dep_s
)
# Get exceeded area
ex = ex_n + ex_s
ex_area = np.count_nonzero(ex > 0) * cell_size * cell_size / 1.0e6
nor_area = np.count_nonzero(~np.isnan(dep_s)) * cell_size * cell_size / 1.0e6
ex_pct = 100 * ex_area / nor_area
# Build df and tidy
ex_df = pd.DataFrame(
{
"exceeded_area_km2": ex_area,
"total_area_km2": nor_area,
"exceeded_area_pct": ex_pct,
},
index=[0],
)
ex_df = ex_df.round(0).astype(int)
ex_df["series_id"] = ser_id
ex_df["medium"] = "water_fab"
ex_df = ex_df[
["series_id", "medium", "total_area_km2", "exceeded_area_km2", "exceeded_area_pct"]
]
ex_df
## Write summary data to db
# ex_df.to_sql('national_summary',
# eng,
# 'summaries',
# if_exists='append',
# index=False)
# Snap tiff
snap_tif = f"/home/jovyan/shared/critical_loads/raster/blr_land_mask_{cell_size}m.tif"
# N
n_tif = f"/home/jovyan/shared/critical_loads/raster/exceedance/fab_exn_meqpm2pyr_{short_name}_{cell_size}m.tif"
cl.write_geotiff(ex_n, n_tif, snap_tif, -1, gdal.GDT_Float32)
# S
s_tif = f"/home/jovyan/shared/critical_loads/raster/exceedance/fab_exs_meqpm2pyr_{short_name}_{cell_size}m.tif"
cl.write_geotiff(ex_s, s_tif, snap_tif, -1, gdal.GDT_Float32)
# N+S
ns_tif = f"/home/jovyan/shared/critical_loads/raster/exceedance/fab_exns_meqpm2pyr_{short_name}_{cell_size}m.tif"
cl.write_geotiff(ex_n + ex_s, ns_tif, snap_tif, -1, gdal.GDT_Float32)
# Exceedance 'region'
reg_tif = f"/home/jovyan/shared/critical_loads/raster/exceedance/fab_ex_reg_id_{short_name}_{cell_size}m.tif"
cl.write_geotiff(reg_id, reg_tif, snap_tif, -1, gdal.GDT_Float32)
| 0.281702 | 0.883286 |
# パーセプトロン
```
from sklearn import datasets
import numpy as np
iris = datasets.load_iris()
X = iris.data[:, [2, 3]]
y = iris.target
print('Class labels:', np.unique(y))
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=1, stratify=y)
print('Label count in y:', np.bincount(y))
print('Labels counts in y_train:', np.bincount(y_train))
print('Labels counts in y_test:', np.bincount(y_test))
print(X_train.shape, X_test.shape)
print(y_train.shape, y_test.shape)
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
sc.fit(X_train)
X_train_std = sc.transform(X_train)
X_test_std = sc.transform(X_test)
from sklearn.linear_model import Perceptron
ppn = Perceptron(max_iter=40, eta0=0.1, random_state=1)
ppn.fit(X_train_std, y_train)
print(ppn.n_iter_)
y_pred = ppn.predict(X_test_std)
print('Misclassified samples: %d' % (y_test != y_pred).sum())
from sklearn.metrics import accuracy_score
print('Accuracy: %.2f' % accuracy_score(y_test, y_pred))
print('Accuracy: %.2f' % ppn.score(X_test_std, y_test))
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
def plot_decision_regions(X, y, classifier, test_idx=None, resolution=0.02):
# マーカーとカラーマップの準備
markers = ('s', 'x', 'o', '^', 'v')
colors = ('red', 'blue', 'lightgreen', 'gray', 'cyan')
cmap = ListedColormap(colors[:len(np.unique(y))])
# 決定領域のプロット
x1_min, x1_max = X[:, 0].min() - 1, X[:, 0].max() + 1
x2_min, x2_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx1, xx2 = np.meshgrid(np.arange(x1_min, x1_max, resolution),
np.arange(x2_min, x2_max, resolution))
Z = classifier.predict(np.array([xx1.ravel(), xx2.ravel()]).T)
Z = Z.reshape(xx1.shape)
plt.contourf(xx1, xx2, Z, alpha=0.3, cmap=cmap)
plt.xlim(xx1.min(), xx1.max())
plt.ylim(xx2.min(), xx2.max())
for idx, cl in enumerate(np.unique(y)):
plt.scatter(x=X[y == cl, 0],
y=X[y == cl, 1],
alpha=0.8,
c=colors[idx],
marker=markers[idx],
label=cl,
edgecolor='black')
# テストサンプルを目立たせる
if test_idx:
X_test, y_test = X[test_idx, :], y[test_idx]
plt.scatter(X_test[:, 0], X_test[:, 1],
c='',
edgecolor='black',
alpha=1.0,
linewidth=1,
marker='o',
s=100,
label='test set')
X_combined_std = np.vstack((X_train_std, X_test_std))
y_combined = np.hstack((y_train, y_test))
plot_decision_regions(X=X_combined_std, y=y_combined, classifier=ppn, test_idx=range(105, 150))
plt.xlabel('sepal length [cm]')
plt.ylabel('petal length [cm]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
```
# ロジスティック回帰
```
def logit(p):
return np.log(p / (1 - p))
x = np.linspace(0.001, 0.999)
y = logit(x)
plt.plot(x, y)
plt.show()
import matplotlib.pyplot as plt
import numpy as np
def sigmoid(z):
return 1.0 / (1.0 + np.exp(-z))
z = np.arange(-7, 7, 0.1)
phi_z = sigmoid(z)
plt.plot(z, phi_z)
plt.axvline(0.0, color='k')
plt.ylim(-0.1, 1.1)
plt.xlabel('z')
plt.ylabel('$\phi (z)$')
plt.yticks([0.0, 0.5, 1.0])
ax = plt.gca()
ax.yaxis.grid(True)
plt.tight_layout()
plt.show()
def cost_1(z):
return - np.log(sigmoid(z))
def cost_0(z):
return - np.log(1 - sigmoid(z))
z = np.arange(-10, 10, 0.1)
phi_z = sigmoid(z) # [0, 1]
c1 = [cost_1(x) for x in z]
plt.plot(phi_z, c1, label='J(w) if y=1')
c0 = [cost_0(x) for x in z]
plt.plot(phi_z, c0, linestyle='--', label='J(w) if y=0')
plt.ylim(0.0, 5.1)
plt.xlim(0, 1)
plt.xlabel('$\phi$(z)')
plt.ylabel('J(w)')
plt.legend(loc='upper center')
plt.tight_layout()
plt.show()
class LogisticRegressionGD(object):
def __init__(self, eta=0.01, n_iter=50, random_state=1):
self.eta = eta
self.n_iter = n_iter
self.random_state = random_state
def fit(self, X, y):
rgen = np.random.RandomState(self.random_state)
self.w_ = rgen.normal(loc=0.0, scale=0.01, size=1 + X.shape[1])
self.cost_ = []
for _ in range(self.n_iter):
net_input = self.net_input(X)
output = self.activation(net_input)
errors = (y - output)
self.w_[1:] += self.eta * X.T.dot(errors)
self.w_[0] += self.eta * errors.sum()
cost = -y.dot(np.log(output)) - ((1 - y).dot(np.log(1 - output)))
self.cost_.append(cost)
return self
def net_input(self, X):
return np.dot(X, self.w_[1:]) + self.w_[0]
def activation(self, z):
return 1.0 / (1.0 + np.exp(-np.clip(z, -250, 250)))
def predict(self, X):
return np.where(self.net_input(X) >= 0.0, 1, 0)
X_train_01_subset = X_train[(y_train == 0) | (y_train == 1)]
y_train_01_subset = y_train[(y_train == 0) | (y_train == 1)]
lrgd = LogisticRegressionGD(eta=0.05, n_iter=1000, random_state=1)
lrgd.fit(X_train_01_subset, y_train_01_subset)
plot_decision_regions(X=X_train_01_subset, y=y_train_01_subset, classifier=lrgd)
plt.xlabel('petal length [standardized]')
plt.ylabel('petal width [standardized]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression(C=100.0, random_state=1)
lr.fit(X_train_std, y_train)
plot_decision_regions(X_combined_std, y_combined, classifier=lr, test_idx=range(105, 150))
plt.xlabel('petal length [standardized]')
plt.ylabel('petal width [standardized]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
lr.predict_proba(X_test_std[:3, :]).argmax(axis=1)
lr.predict(X_test_std[:3, :])
X_test_std[0, :].shape
X_test_std[0, :].reshape(1, -1).shape
lr.predict(X_test_std[0, :].reshape(1, -1))
```
## 正則化
```
weights, params = [], []
for c in np.arange(-5, 5):
lr = LogisticRegression(C=10.0 ** c, random_state=1)
lr.fit(X_train_std, y_train)
weights.append(lr.coef_[1])
params.append(10.0 ** c)
weights = np.array(weights)
# 横軸に逆正則化パラメータ C、縦軸に重み係数をプロット
plt.plot(params, weights[:, 0], label='petal length')
plt.plot(params, weights[:, 1], linestyle='--', label='petal width')
plt.ylabel('weight coefficient')
plt.xlabel('C')
plt.legend(loc='upper left')
plt.xscale('log')
plt.show()
```
# SVM
```
from sklearn.svm import SVC
svm = SVC(kernel='linear', C=1.0, random_state=1)
svm.fit(X_train_std, y_train)
plot_decision_regions(X_combined_std, y_combined, classifier=svm, test_idx=range(105, 150))
plt.xlabel('petal length [standardized]')
plt.ylabel('petal width [standardized]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
from sklearn.linear_model import SGDClassifier
ppn = SGDClassifier(loss='perceptron')
ppn
lr = SGDClassifier(loss='log')
print(lr)
svm = SGDClassifier(loss='hinge')
print(svm)
```
## カーネルSVM
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
np.random.seed(1)
X_xor = np.random.randn(200, 2)
y_xor = np.logical_xor(X_xor[:, 0] > 0, X_xor[:, 1] > 0)
y_xor = np.where(y_xor, 1, -1)
plt.scatter(X_xor[y_xor==1, 0], X_xor[y_xor==1, 1], c='b', marker='x', label='1')
plt.scatter(X_xor[y_xor==-1, 0], X_xor[y_xor==1, 1], c='r', marker='s', label='-1')
plt.xlim([-3, 3])
plt.ylim([-3, 3])
plt.legend(loc='best')
plt.tight_layout()
plt.show()
svm = SVC(kernel='rbf', random_state=1, gamma=0.10, C=10.0)
svm.fit(X_xor, y_xor)
plot_decision_regions(X_xor, y_xor, classifier=svm)
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
svm = SVC(kernel='rbf', random_state=1, gamma=0.2, C=1.0)
svm.fit(X_train_std, y_train)
plot_decision_regions(X_combined_std, y_combined, classifier=svm, test_idx=range(105, 150))
plt.xlabel('petal length [standardized]')
plt.ylabel('petal width [standardized]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
```
# 決定木
```
import matplotlib.pyplot as plt
import numpy as np
def gini(p):
return (p) * (1 - (p)) + (1 - p) * (1 - (1 - p))
def entropy(p):
return - p * np.log2(p) - (1 - p) * np.log2((1 - p))
def error(p):
return 1 - np.max([p, 1 - p])
x = np.arange(0.0, 1.0, 0.01)
ent = [entropy(p) if p != 0 else None for p in x]
sc_ent = [e * 0.5 if e else None for e in ent]
err = [error(i) for i in x]
fig = plt.figure()
ax = plt.subplot(111)
for i, lab, ls, c, in zip([ent, sc_ent, gini(x), err],
['Entropy', 'Entropy (scaled)', 'Gini Impurity', 'Misclassification Error'],
['-', '-', '--', '-.'],
['black', 'lightgray', 'red', 'green', 'cyan']):
line = ax.plot(x, i, label=lab, linestyle=ls, lw=2, color=c)
ax.legend(loc='upper center', bbox_to_anchor=(0.5, 1.15),
ncol=5, fancybox=True, shadow=False)
ax.axhline(y=0.5, linewidth=1, color='k', linestyle='--')
ax.axhline(y=1.0, linewidth=1, color='k', linestyle='--')
plt.ylim([0, 1.1])
plt.xlabel('p(i=1)')
plt.ylabel('Impurity Index')
plt.show()
from sklearn.tree import DecisionTreeClassifier
tree = DecisionTreeClassifier(criterion='gini', max_depth=4, random_state=1)
tree.fit(X_train, y_train)
X_combined = np.vstack((X_train, X_test))
y_combined = np.hstack((y_train, y_test))
plot_decision_regions(X_combined, y_combined, classifier=tree, test_idx=range(105, 150))
plt.xlabel('petal length [cm]')
plt.ylabel('petal width [cm]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
!pip install pydotplus
from pydotplus import graph_from_dot_data
from sklearn.tree import export_graphviz
dot_data = export_graphviz(tree,
filled=True,
rounded=True,
class_names=['Satosa', 'Versicolor', 'Virginica'],
feature_names=['petal length', 'petal width'],
out_file=None)
graph = graph_from_dot_data(dot_data)
graph.write_png('test.png')
from IPython.display import Image
Image('test.png')
```
## ランダムフォレスト
```
from sklearn.ensemble import RandomForestClassifier
forest = RandomForestClassifier(criterion='gini',
n_estimators=25,
random_state=1,
n_jobs=2)
forest.fit(X_train, y_train)
X_combined = np.vstack((X_train, X_test))
y_combined = np.hstack((y_train, y_test))
plot_decision_regions(X_combined, y_combined, classifier=forest, test_idx=range(105, 150))
plt.xlabel('petal length [cm]')
plt.ylabel('petal width [cm]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
```
# K-近傍法
```
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors=5, p=2, metric='minkowski')
knn.fit(X_train_std, y_train)
plot_decision_regions(X_combined_std, y_combined, classifier=knn, test_idx=range(105, 150))
plt.xlabel('petal length [standardized]')
plt.ylabel('petal width [standardized]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
```
|
github_jupyter
|
from sklearn import datasets
import numpy as np
iris = datasets.load_iris()
X = iris.data[:, [2, 3]]
y = iris.target
print('Class labels:', np.unique(y))
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=1, stratify=y)
print('Label count in y:', np.bincount(y))
print('Labels counts in y_train:', np.bincount(y_train))
print('Labels counts in y_test:', np.bincount(y_test))
print(X_train.shape, X_test.shape)
print(y_train.shape, y_test.shape)
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
sc.fit(X_train)
X_train_std = sc.transform(X_train)
X_test_std = sc.transform(X_test)
from sklearn.linear_model import Perceptron
ppn = Perceptron(max_iter=40, eta0=0.1, random_state=1)
ppn.fit(X_train_std, y_train)
print(ppn.n_iter_)
y_pred = ppn.predict(X_test_std)
print('Misclassified samples: %d' % (y_test != y_pred).sum())
from sklearn.metrics import accuracy_score
print('Accuracy: %.2f' % accuracy_score(y_test, y_pred))
print('Accuracy: %.2f' % ppn.score(X_test_std, y_test))
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
def plot_decision_regions(X, y, classifier, test_idx=None, resolution=0.02):
# マーカーとカラーマップの準備
markers = ('s', 'x', 'o', '^', 'v')
colors = ('red', 'blue', 'lightgreen', 'gray', 'cyan')
cmap = ListedColormap(colors[:len(np.unique(y))])
# 決定領域のプロット
x1_min, x1_max = X[:, 0].min() - 1, X[:, 0].max() + 1
x2_min, x2_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx1, xx2 = np.meshgrid(np.arange(x1_min, x1_max, resolution),
np.arange(x2_min, x2_max, resolution))
Z = classifier.predict(np.array([xx1.ravel(), xx2.ravel()]).T)
Z = Z.reshape(xx1.shape)
plt.contourf(xx1, xx2, Z, alpha=0.3, cmap=cmap)
plt.xlim(xx1.min(), xx1.max())
plt.ylim(xx2.min(), xx2.max())
for idx, cl in enumerate(np.unique(y)):
plt.scatter(x=X[y == cl, 0],
y=X[y == cl, 1],
alpha=0.8,
c=colors[idx],
marker=markers[idx],
label=cl,
edgecolor='black')
# テストサンプルを目立たせる
if test_idx:
X_test, y_test = X[test_idx, :], y[test_idx]
plt.scatter(X_test[:, 0], X_test[:, 1],
c='',
edgecolor='black',
alpha=1.0,
linewidth=1,
marker='o',
s=100,
label='test set')
X_combined_std = np.vstack((X_train_std, X_test_std))
y_combined = np.hstack((y_train, y_test))
plot_decision_regions(X=X_combined_std, y=y_combined, classifier=ppn, test_idx=range(105, 150))
plt.xlabel('sepal length [cm]')
plt.ylabel('petal length [cm]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
def logit(p):
return np.log(p / (1 - p))
x = np.linspace(0.001, 0.999)
y = logit(x)
plt.plot(x, y)
plt.show()
import matplotlib.pyplot as plt
import numpy as np
def sigmoid(z):
return 1.0 / (1.0 + np.exp(-z))
z = np.arange(-7, 7, 0.1)
phi_z = sigmoid(z)
plt.plot(z, phi_z)
plt.axvline(0.0, color='k')
plt.ylim(-0.1, 1.1)
plt.xlabel('z')
plt.ylabel('$\phi (z)$')
plt.yticks([0.0, 0.5, 1.0])
ax = plt.gca()
ax.yaxis.grid(True)
plt.tight_layout()
plt.show()
def cost_1(z):
return - np.log(sigmoid(z))
def cost_0(z):
return - np.log(1 - sigmoid(z))
z = np.arange(-10, 10, 0.1)
phi_z = sigmoid(z) # [0, 1]
c1 = [cost_1(x) for x in z]
plt.plot(phi_z, c1, label='J(w) if y=1')
c0 = [cost_0(x) for x in z]
plt.plot(phi_z, c0, linestyle='--', label='J(w) if y=0')
plt.ylim(0.0, 5.1)
plt.xlim(0, 1)
plt.xlabel('$\phi$(z)')
plt.ylabel('J(w)')
plt.legend(loc='upper center')
plt.tight_layout()
plt.show()
class LogisticRegressionGD(object):
def __init__(self, eta=0.01, n_iter=50, random_state=1):
self.eta = eta
self.n_iter = n_iter
self.random_state = random_state
def fit(self, X, y):
rgen = np.random.RandomState(self.random_state)
self.w_ = rgen.normal(loc=0.0, scale=0.01, size=1 + X.shape[1])
self.cost_ = []
for _ in range(self.n_iter):
net_input = self.net_input(X)
output = self.activation(net_input)
errors = (y - output)
self.w_[1:] += self.eta * X.T.dot(errors)
self.w_[0] += self.eta * errors.sum()
cost = -y.dot(np.log(output)) - ((1 - y).dot(np.log(1 - output)))
self.cost_.append(cost)
return self
def net_input(self, X):
return np.dot(X, self.w_[1:]) + self.w_[0]
def activation(self, z):
return 1.0 / (1.0 + np.exp(-np.clip(z, -250, 250)))
def predict(self, X):
return np.where(self.net_input(X) >= 0.0, 1, 0)
X_train_01_subset = X_train[(y_train == 0) | (y_train == 1)]
y_train_01_subset = y_train[(y_train == 0) | (y_train == 1)]
lrgd = LogisticRegressionGD(eta=0.05, n_iter=1000, random_state=1)
lrgd.fit(X_train_01_subset, y_train_01_subset)
plot_decision_regions(X=X_train_01_subset, y=y_train_01_subset, classifier=lrgd)
plt.xlabel('petal length [standardized]')
plt.ylabel('petal width [standardized]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression(C=100.0, random_state=1)
lr.fit(X_train_std, y_train)
plot_decision_regions(X_combined_std, y_combined, classifier=lr, test_idx=range(105, 150))
plt.xlabel('petal length [standardized]')
plt.ylabel('petal width [standardized]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
lr.predict_proba(X_test_std[:3, :]).argmax(axis=1)
lr.predict(X_test_std[:3, :])
X_test_std[0, :].shape
X_test_std[0, :].reshape(1, -1).shape
lr.predict(X_test_std[0, :].reshape(1, -1))
weights, params = [], []
for c in np.arange(-5, 5):
lr = LogisticRegression(C=10.0 ** c, random_state=1)
lr.fit(X_train_std, y_train)
weights.append(lr.coef_[1])
params.append(10.0 ** c)
weights = np.array(weights)
# 横軸に逆正則化パラメータ C、縦軸に重み係数をプロット
plt.plot(params, weights[:, 0], label='petal length')
plt.plot(params, weights[:, 1], linestyle='--', label='petal width')
plt.ylabel('weight coefficient')
plt.xlabel('C')
plt.legend(loc='upper left')
plt.xscale('log')
plt.show()
from sklearn.svm import SVC
svm = SVC(kernel='linear', C=1.0, random_state=1)
svm.fit(X_train_std, y_train)
plot_decision_regions(X_combined_std, y_combined, classifier=svm, test_idx=range(105, 150))
plt.xlabel('petal length [standardized]')
plt.ylabel('petal width [standardized]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
from sklearn.linear_model import SGDClassifier
ppn = SGDClassifier(loss='perceptron')
ppn
lr = SGDClassifier(loss='log')
print(lr)
svm = SGDClassifier(loss='hinge')
print(svm)
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
np.random.seed(1)
X_xor = np.random.randn(200, 2)
y_xor = np.logical_xor(X_xor[:, 0] > 0, X_xor[:, 1] > 0)
y_xor = np.where(y_xor, 1, -1)
plt.scatter(X_xor[y_xor==1, 0], X_xor[y_xor==1, 1], c='b', marker='x', label='1')
plt.scatter(X_xor[y_xor==-1, 0], X_xor[y_xor==1, 1], c='r', marker='s', label='-1')
plt.xlim([-3, 3])
plt.ylim([-3, 3])
plt.legend(loc='best')
plt.tight_layout()
plt.show()
svm = SVC(kernel='rbf', random_state=1, gamma=0.10, C=10.0)
svm.fit(X_xor, y_xor)
plot_decision_regions(X_xor, y_xor, classifier=svm)
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
svm = SVC(kernel='rbf', random_state=1, gamma=0.2, C=1.0)
svm.fit(X_train_std, y_train)
plot_decision_regions(X_combined_std, y_combined, classifier=svm, test_idx=range(105, 150))
plt.xlabel('petal length [standardized]')
plt.ylabel('petal width [standardized]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
import matplotlib.pyplot as plt
import numpy as np
def gini(p):
return (p) * (1 - (p)) + (1 - p) * (1 - (1 - p))
def entropy(p):
return - p * np.log2(p) - (1 - p) * np.log2((1 - p))
def error(p):
return 1 - np.max([p, 1 - p])
x = np.arange(0.0, 1.0, 0.01)
ent = [entropy(p) if p != 0 else None for p in x]
sc_ent = [e * 0.5 if e else None for e in ent]
err = [error(i) for i in x]
fig = plt.figure()
ax = plt.subplot(111)
for i, lab, ls, c, in zip([ent, sc_ent, gini(x), err],
['Entropy', 'Entropy (scaled)', 'Gini Impurity', 'Misclassification Error'],
['-', '-', '--', '-.'],
['black', 'lightgray', 'red', 'green', 'cyan']):
line = ax.plot(x, i, label=lab, linestyle=ls, lw=2, color=c)
ax.legend(loc='upper center', bbox_to_anchor=(0.5, 1.15),
ncol=5, fancybox=True, shadow=False)
ax.axhline(y=0.5, linewidth=1, color='k', linestyle='--')
ax.axhline(y=1.0, linewidth=1, color='k', linestyle='--')
plt.ylim([0, 1.1])
plt.xlabel('p(i=1)')
plt.ylabel('Impurity Index')
plt.show()
from sklearn.tree import DecisionTreeClassifier
tree = DecisionTreeClassifier(criterion='gini', max_depth=4, random_state=1)
tree.fit(X_train, y_train)
X_combined = np.vstack((X_train, X_test))
y_combined = np.hstack((y_train, y_test))
plot_decision_regions(X_combined, y_combined, classifier=tree, test_idx=range(105, 150))
plt.xlabel('petal length [cm]')
plt.ylabel('petal width [cm]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
!pip install pydotplus
from pydotplus import graph_from_dot_data
from sklearn.tree import export_graphviz
dot_data = export_graphviz(tree,
filled=True,
rounded=True,
class_names=['Satosa', 'Versicolor', 'Virginica'],
feature_names=['petal length', 'petal width'],
out_file=None)
graph = graph_from_dot_data(dot_data)
graph.write_png('test.png')
from IPython.display import Image
Image('test.png')
from sklearn.ensemble import RandomForestClassifier
forest = RandomForestClassifier(criterion='gini',
n_estimators=25,
random_state=1,
n_jobs=2)
forest.fit(X_train, y_train)
X_combined = np.vstack((X_train, X_test))
y_combined = np.hstack((y_train, y_test))
plot_decision_regions(X_combined, y_combined, classifier=forest, test_idx=range(105, 150))
plt.xlabel('petal length [cm]')
plt.ylabel('petal width [cm]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors=5, p=2, metric='minkowski')
knn.fit(X_train_std, y_train)
plot_decision_regions(X_combined_std, y_combined, classifier=knn, test_idx=range(105, 150))
plt.xlabel('petal length [standardized]')
plt.ylabel('petal width [standardized]')
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
| 0.701917 | 0.903507 |
```
import pandas as pd
import numpy as np
import networkx as nx
import nltk
from sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer
from sklearn.decomposition import NMF, LatentDirichletAllocation
from scipy.stats import entropy
import matplotlib.pyplot as plt
%matplotlib inline
text_col = 'Excerpt Copy'
df = pd.read_csv('data/sensitive/coder1_all.tsv', sep='\t')
df = df[['uni', 'Participant', 'Excerpt Copy', 'rank', 'identity',
'Q3-g', 'Q3-l', 'Q3-b', 'Q3-quest', 'Q3-ace', 'Q3-queer', 'Q4-gq',
'Q4-t', 'Q4-i', 'Q4-f', 'Q4-m']]
print(df.shape[0])
df.head()
df = df.replace({'Question: Q\d*\w?; Answer:': ''}, regex=True)
df = df.replace({'Question: Q\d*-other; Answer:': ''}, regex=True)
def unlist(x):
return x[0]
text = df[['uni', 'Participant', 'Excerpt Copy']].groupby(['uni', 'Participant'])
text = text.agg(lambda t: "%s" % ' '.join(t))
text = text['Excerpt Copy']
print(text.shape[0])
text.head()
n_topics = 10
n_snow = 10
documents = text.values
text.values[0]
def display_topics(model, feature_names, no_top_words):
for topic_idx, topic in enumerate(model.components_):
words = " ".join([feature_names[i]
for i in topic.argsort()[:-no_top_words - 1:-1]])
print("Topic", topic_idx, ": ", words)
def JSD(P, Q):
_P = P / np.linalg.norm(P, ord=1)
_Q = Q / np.linalg.norm(Q, ord=1)
_M = 0.5 * (_P + _Q)
return 0.5 * (entropy(_P, _M) + entropy(_Q, _M))
def list_sims(df):
n = df.shape[0]
result = []
for i in range(0,n):
for j in range(i+1,n):
tmp = {'i': i, 'j': j, 'jsd': JSD(df.loc[i], df.loc[j])}
result.append(tmp)
return pd.DataFrame(result)
def worker(documents, method='NMF', n_topics=10, calc_edges=True):
if method == 'NMF':
vectorizer = TfidfVectorizer(max_df=0.95, min_df=2,
max_features=1000,
stop_words='english')
mod = NMF(n_components=n_topics,
alpha=.1,
l1_ratio=.5,
init='nndsvd')
elif method == 'LDA':
vectorizer = CountVectorizer(max_df=0.95, min_df=2,
max_features=1000,
stop_words='english')
mod = LatentDirichletAllocation(n_components=n_topics,
max_iter=20,
learning_method='online',
n_jobs=-1 )
transformed = vectorizer.fit_transform(documents)
feat_names = vectorizer.get_feature_names()
model = mod.fit(transformed)
display_topics(model, feat_names, n_snow)
edges = None
if calc_edges:
edges = list_sims(transformed)
return edges
person = text.values
edges = {}
edges['nmf_person'] = worker(person, 'NMF')
edges['lda_person'] = worker(person, 'LDA')
edges['nmf_person'].jsd.hist(bins=20)
edges['lda_person'].jsd.hist(bins=20)
for i in [3, 5, 8, 10, 15]:
print("\n\nNMF", i)
worker(person, 'NMF', n_topics=i, calc_edges=False)
for i in [3, 5, 8, 10, 15]:
print("\n\nLDA:", i)
worker(person, 'LDA', n_topics=i, calc_edges=False)
tmp.to_csv('data/public/cosine_people.tsv', sep='\t')
text=text[['uni', 'Participant']]
text.to_csv('data/public/cosine_people_ids.tsv', sep='\t')
text.head()
TfidfVectorizer?
from nltk.stem.snowball import SnowballStemmer
from nltk.tokenize import regexp_tokenize
stemmer = SnowballStemmer("english")
def my_tokenizer(text):
out = []
for w in regexp_tokenize(text, '\w+'):
out.append(stemmer.stem(w))
return out
vectorizer = TfidfVectorizer(max_df=0.95, min_df=2,
max_features=1000,
stop_words='english',
tokenizer=my_tokenizer
).fit(text.values)
mod = NMF(n_components=15,
alpha=.1,
l1_ratio=.5,
init='nndsvd')
transformed = vectorizer.fit_transform(text.values)
feat_names = vectorizer.get_feature_names()
model = mod.fit(transformed)
display_topics(model, feat_names, n_snow)
worker(person, 'NMF', n_topics=15, calc_edges=False)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import networkx as nx
import nltk
from sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer
from sklearn.decomposition import NMF, LatentDirichletAllocation
from scipy.stats import entropy
import matplotlib.pyplot as plt
%matplotlib inline
text_col = 'Excerpt Copy'
df = pd.read_csv('data/sensitive/coder1_all.tsv', sep='\t')
df = df[['uni', 'Participant', 'Excerpt Copy', 'rank', 'identity',
'Q3-g', 'Q3-l', 'Q3-b', 'Q3-quest', 'Q3-ace', 'Q3-queer', 'Q4-gq',
'Q4-t', 'Q4-i', 'Q4-f', 'Q4-m']]
print(df.shape[0])
df.head()
df = df.replace({'Question: Q\d*\w?; Answer:': ''}, regex=True)
df = df.replace({'Question: Q\d*-other; Answer:': ''}, regex=True)
def unlist(x):
return x[0]
text = df[['uni', 'Participant', 'Excerpt Copy']].groupby(['uni', 'Participant'])
text = text.agg(lambda t: "%s" % ' '.join(t))
text = text['Excerpt Copy']
print(text.shape[0])
text.head()
n_topics = 10
n_snow = 10
documents = text.values
text.values[0]
def display_topics(model, feature_names, no_top_words):
for topic_idx, topic in enumerate(model.components_):
words = " ".join([feature_names[i]
for i in topic.argsort()[:-no_top_words - 1:-1]])
print("Topic", topic_idx, ": ", words)
def JSD(P, Q):
_P = P / np.linalg.norm(P, ord=1)
_Q = Q / np.linalg.norm(Q, ord=1)
_M = 0.5 * (_P + _Q)
return 0.5 * (entropy(_P, _M) + entropy(_Q, _M))
def list_sims(df):
n = df.shape[0]
result = []
for i in range(0,n):
for j in range(i+1,n):
tmp = {'i': i, 'j': j, 'jsd': JSD(df.loc[i], df.loc[j])}
result.append(tmp)
return pd.DataFrame(result)
def worker(documents, method='NMF', n_topics=10, calc_edges=True):
if method == 'NMF':
vectorizer = TfidfVectorizer(max_df=0.95, min_df=2,
max_features=1000,
stop_words='english')
mod = NMF(n_components=n_topics,
alpha=.1,
l1_ratio=.5,
init='nndsvd')
elif method == 'LDA':
vectorizer = CountVectorizer(max_df=0.95, min_df=2,
max_features=1000,
stop_words='english')
mod = LatentDirichletAllocation(n_components=n_topics,
max_iter=20,
learning_method='online',
n_jobs=-1 )
transformed = vectorizer.fit_transform(documents)
feat_names = vectorizer.get_feature_names()
model = mod.fit(transformed)
display_topics(model, feat_names, n_snow)
edges = None
if calc_edges:
edges = list_sims(transformed)
return edges
person = text.values
edges = {}
edges['nmf_person'] = worker(person, 'NMF')
edges['lda_person'] = worker(person, 'LDA')
edges['nmf_person'].jsd.hist(bins=20)
edges['lda_person'].jsd.hist(bins=20)
for i in [3, 5, 8, 10, 15]:
print("\n\nNMF", i)
worker(person, 'NMF', n_topics=i, calc_edges=False)
for i in [3, 5, 8, 10, 15]:
print("\n\nLDA:", i)
worker(person, 'LDA', n_topics=i, calc_edges=False)
tmp.to_csv('data/public/cosine_people.tsv', sep='\t')
text=text[['uni', 'Participant']]
text.to_csv('data/public/cosine_people_ids.tsv', sep='\t')
text.head()
TfidfVectorizer?
from nltk.stem.snowball import SnowballStemmer
from nltk.tokenize import regexp_tokenize
stemmer = SnowballStemmer("english")
def my_tokenizer(text):
out = []
for w in regexp_tokenize(text, '\w+'):
out.append(stemmer.stem(w))
return out
vectorizer = TfidfVectorizer(max_df=0.95, min_df=2,
max_features=1000,
stop_words='english',
tokenizer=my_tokenizer
).fit(text.values)
mod = NMF(n_components=15,
alpha=.1,
l1_ratio=.5,
init='nndsvd')
transformed = vectorizer.fit_transform(text.values)
feat_names = vectorizer.get_feature_names()
model = mod.fit(transformed)
display_topics(model, feat_names, n_snow)
worker(person, 'NMF', n_topics=15, calc_edges=False)
| 0.414069 | 0.317856 |
# Study Preservation Cases
## Responsabilities of this file
* Create all the proposed species study cases for the project
## Who is Running?
```
#Matheus
project_root = "/content/drive/MyDrive/Mestrado/Deep Learning/Projeto/Projeto_Final_DL"
#Gabi
#Jean
#Sandra
from google.colab import drive
drive.mount('/content/drive')
```
## Retrieving aux Classes
```
!wget https://raw.githubusercontent.com/math-sasso/extinction-prevention-modeling-MO434/main/Utils/utils.py
from utils import Utils
utils_methods = Utils()
```
## Creating Folder Structure
```
import pandas as pd
import os
utils_methods.create_folder_structure(project_root+"/Data/Study_Cases")
```
## Study Cases
```
all_species_dict= {}
```
### Arara-Azul Gigante
```
studied_specie_information = {'GBIF_ID':[2479359],
'Species_Name':['Anodorhynchus hyacinthinus'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
```
###Ariranha
```
studied_specie_information = {'GBIF_ID':[2433681],
'Species_Name':['Pteronura brasiliensis'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
```
## Seriema
```
studied_specie_information = {'GBIF_ID':[5228133],
'Species_Name':['Cariama cristata'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
```
##Onça Pintada
```
studied_specie_information = {'GBIF_ID':[5219426],
'Species_Name':['Panthera onca'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
```
## Anta
```
studied_specie_information = {'GBIF_ID':[2440898],
'Species_Name':['Tapirus terrestris'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
```
## Cervo do Pantanal
```
studied_specie_information = {'GBIF_ID':[2440963],
'Species_Name':['Blastocerus dichotomus'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
```
## Capivara
```
studied_specie_information = {'GBIF_ID':[5786666],
'Species_Name':['Hydrochoerus hydrochaeris'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
```
## Guariba
```
studied_specie_information = {'GBIF_ID':[2436652],
'Species_Name':['Alouatta caraya'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
```
## Tamanduá Bandeira
```
studied_specie_information = {'GBIF_ID':[2436346],
'Species_Name':['Myrmecophaga tridactyla'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
```
## Lobo Guará
```
studied_specie_information = {'GBIF_ID':[2434450],
'Species_Name':['Chrysocyon brachyurus'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
```
## Jaguatirica
```
studied_specie_information = {'GBIF_ID':[2434982],
'Species_Name':['Leopardus pardalis'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
```
## Jacaré de Papo Amarelo
```
studied_specie_information = {'GBIF_ID':[5846511],
'Species_Name':['Caiman latirostrisa'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
```
## Murucutu de barriga amarela
```
studied_specie_information = {'GBIF_ID':[2497885],
'Species_Name':['Pulsatrix koeniswaldiana'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
```
|
github_jupyter
|
#Matheus
project_root = "/content/drive/MyDrive/Mestrado/Deep Learning/Projeto/Projeto_Final_DL"
#Gabi
#Jean
#Sandra
from google.colab import drive
drive.mount('/content/drive')
!wget https://raw.githubusercontent.com/math-sasso/extinction-prevention-modeling-MO434/main/Utils/utils.py
from utils import Utils
utils_methods = Utils()
import pandas as pd
import os
utils_methods.create_folder_structure(project_root+"/Data/Study_Cases")
all_species_dict= {}
studied_specie_information = {'GBIF_ID':[2479359],
'Species_Name':['Anodorhynchus hyacinthinus'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
studied_specie_information = {'GBIF_ID':[2433681],
'Species_Name':['Pteronura brasiliensis'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
studied_specie_information = {'GBIF_ID':[5228133],
'Species_Name':['Cariama cristata'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
studied_specie_information = {'GBIF_ID':[5219426],
'Species_Name':['Panthera onca'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
studied_specie_information = {'GBIF_ID':[2440898],
'Species_Name':['Tapirus terrestris'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
studied_specie_information = {'GBIF_ID':[2440963],
'Species_Name':['Blastocerus dichotomus'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
studied_specie_information = {'GBIF_ID':[5786666],
'Species_Name':['Hydrochoerus hydrochaeris'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
studied_specie_information = {'GBIF_ID':[2436652],
'Species_Name':['Alouatta caraya'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
studied_specie_information = {'GBIF_ID':[2436346],
'Species_Name':['Myrmecophaga tridactyla'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
studied_specie_information = {'GBIF_ID':[2434450],
'Species_Name':['Chrysocyon brachyurus'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
studied_specie_information = {'GBIF_ID':[2434982],
'Species_Name':['Leopardus pardalis'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
studied_specie_information = {'GBIF_ID':[5846511],
'Species_Name':['Caiman latirostrisa'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
studied_specie_information = {'GBIF_ID':[2497885],
'Species_Name':['Pulsatrix koeniswaldiana'],
'Study_Focus':[True]
}
studied_specie_information_df = pd.DataFrame. studied_specie_information_df = pd.DataFrame.from_dict(studied_specie_information)
species_name = studied_specie_information_df[studied_specie_information_df['Study_Focus'] ==True]['Species_Name'][0]
csv_name = species_name.replace(" ","_")
studied_specie_information_df.to_csv(os.path.join(project_root+"/Data/Study_Cases",csv_name+'.csv'),index = False, header=True)
studied_specie_information_df
| 0.080554 | 0.558207 |
# Aprendizagem de redes neurais
### Utilizando redes neurais para reconhecer dígitos escritos a mão.
## Introdução
Neste exercício, será implementado o algoritmo de retropropagação para aprender os parâmetros da rede neural. Vamos avaliar um problema de classificação, cuja base de dados é referente a dígitos escritos a mão. Desse modo, haverá 10 possíveis classes diferentes que o algoritmo deverá avaliar.

### 1.1 Visualizando os dados
O arquivo **ex4data1.mat** tem 5000 exemplos de treinamento, onde cada um deles corresponde a uma imagem em tons de cinza de 20 por 20 píxels. Cada píxel é representada por um número real que indica a intensidade da escala de cinza naquele ponto (variando de 0 a 255). A matriz bidimensional que descreve a imagem de cada dígito está "desenrolada" em um vetor unidimensional de 400 colunas. Cada um desses exemplos está distribuido em uma matriz de dados X. Ou seja, teremos uma matriz X de **5000 linhas e 400 colunas**.

A segunda parte do conjunto de treinamento é um vetor y de 5000 dimensões que contém rótulos para o conjunto de treinamento.
- OBS: por conveniência, o dígito 0 é rotulado como 10, enquanto os dígitos de 1 a 9 são rotulados em sua ordem natural;
```
# carrega um dataset do matlab
from scipy.io import loadmat
# possibilita a implementação de vetorização
import numpy as np
# otimizador para minimizar a função de custo através do ajuste dos parâmetros
import scipy.optimize as opt
# biblioteca para manipulação e carregamento de datasets
import pandas as pd
# biblioteca de visualização gráfica
import matplotlib.pyplot as plt
# carregando os dados
dados = loadmat('dataset/ex4data1.mat')
# separando os atributos previsores das classes
X = dados['X']
y = dados['y']
# visualizando os dados
figura, configura_figura = plt.subplots(10, 10, figsize = (10,10))
for i in range(10):
for j in range(10):
# reordena a matriz para 20x20 usando o estilo de indexação do Fortran
configura_figura[i,j].imshow(X[np.random.randint(X.shape[0])].reshape((20,20), order = "F"))
# remove os eixos cartesianos de cada subplotagem
configura_figura[i,j].axis('off')
```
### 1.2 Representação do Modelo
A rede neural terá 3 camadas ao total:
- Uma camada de entrada;
- Uma camada oculta;
- Uma camada de saída;

Lembre-se de que as entradas serão imagens em escala de cinza 20 x 20 “desenroladas” para formar 400 recursos de entrada de alimentação para a rede neural. Portanto, nossa camada de entrada tem 400 neurônios. Além disso, a camada oculta tem 25 neurônios e a camada de saída 10 neurônios correspondendo a 10 dígitos (ou classes) que nosso modelo prevê. O +1 na figura acima representa o bias.
## O bias novamente nas redes neurais
> O bias aumenta o grau de liberdade dos ajustes dos pesos.

## Como determinar o número de camadas?
O número de neurônios em cada camada é uma questão mais empírica, não existindo regras explícitas para um cálculo ideal. Jeff Heaton, o autor de **Introduction to Neural Networks for Java**, sugere três abordagens iniciais:
- O número de neurônios escondidos deve estar entre o tamanho da camada de entrada e o da camada de saída.
- O número de neurônios escondidos deve ser 2/3 do tamanho da camada de entrada, mais o tamanho da camada de saída.
- O número de neurônios escondidos deve ser menor que o dobro do tamanho da camada de entrada.
Recebemos um conjunto de parâmetros de rede já treinados. Eles são armazenados em **ex4weights.mat** e serão carregados em teta1 e teta2 adicionados a um outro só vetor.
- 400 unidades na camada de entrada;
- 25 unidades na camada oculta;
- 10 unidades na camada de saída;
```
# carrega os parâmetros treinados vinculados ao exercício
pesos = loadmat('dataset/ex4weights.mat')
teta1 = pesos['Theta1'] # teta1 tem uma dimensão de 25 x 401
teta2 = pesos['Theta2'] #teta2 tem uma dimensão de 10 x 26
# ravel = torna teta1 e teta2 unidimensionais
parametros_totais = np.hstack((teta1.ravel(order='F'), teta2.ravel(order='F')))
# hiperparâmetros da rede neural
dim_camada_entrada = 400
dim_camada_oculta = 25
num_classes = 10
lmbda = 1
# visualizando a dimensão do vetor de parâmetros
parametros_totais.shape
```
### 1.3 Forwardpropagation e Função de Custo
Primeiro, será implementado a função de custo seguida de gradiente para a rede neural (para a qual usamos o algoritmo de retropropagação).
- A função de custo para a rede neural com regularização é

#### Função sigmoide
```
def sigmoide(x):
'''
Função de ativação sigmoide
'''
return 1 / (1 + np.exp(-x))
```
Além disso, enquanto os rótulos originais (na variável y) eram 1, 2, ..., 10, com o propósito de treinar uma rede neural, precisamos recodificar os rótulos como vetores contendo apenas valores 0 ou 1, de modo que

```
# manipulando a saída das classes usando o one hot encoder
(pd.get_dummies(y.flatten()))
```
#### Função de custo
```
def funcaoCusto(parametros_totais, dim_entrada, dim_oculta, num_classes, X, y, lmbda):
'''
Função que computa a função de custo da rede neural regularizada
'''
# separa os parâmetros associados a camada oculta e a camada de saída
teta1 = np.reshape(parametros_totais[:dim_oculta * (dim_entrada + 1)], (dim_oculta, dim_entrada + 1), 'F')
teta2 = np.reshape(parametros_totais[dim_oculta * (dim_entrada + 1):], (num_classes, dim_oculta + 1), 'F')
'''
algoritmo de Forwardpropagation
'''
# m exemplos treináveis
m = len(y)
# cria um vetor coluna de bias m x 1
ones = np.ones((m, 1))
# adicionando o bias na camada de entrada
a1 = np.hstack((ones, X))
# ativação dos neurônios da camada oculta
a2 = sigmoide(a1.dot(teta1.T))
# adicionando o bias na camada oculta
a2 = np.hstack((ones, a2))
# ativção dos neurônios da camada de saída
h = sigmoide(a2.dot(teta2.T))
# manipulando a saída das classes usando o one hot encoder
y_d = pd.get_dummies(y.flatten())
# seprando em partes a função de custo
parte1 = np.multiply(y_d, np.log(h))
parte2 = np.multiply(1-y_d, np.log(1-h))
parte3 = np.sum(parte1 + parte2)
# separando em partes a expressão de regularização da função de custo
soma1_regularizada = np.sum(np.sum(np.power(teta1[:,1:],2), axis = 1)) # ignora o neurônio relativo ao bias
soma2_regularizada = np.sum(np.sum(np.power(teta2[:,1:],2), axis = 1)) # ignora o neurônio relativo ao bias
# junta as partes e retorna a expressão total da função de custo
return np.sum(parte3 / (-m)) + (soma1_regularizada + soma2_regularizada) * lmbda / (2 * m)
# obtendo o valor da função de custo
funcaoCusto(parametros_totais, dim_camada_entrada, dim_camada_oculta, num_classes, X, y, lmbda)
```
## Backpropagation
Nesta parte do código, será implementado o algoritmo de retropropagação para calcular os gradientes da rede neural. Depois de calcular o gradiente, será possível realizar o treinamento da rede neural, minimizando a função de custo usando um otimizador avançado como fmincg do scipy.
### Função sigmoide do gradiente

```
def sigmoideGradiente(x):
'''
Função que calculo o sigmoide do gradiente descente
'''
return np.multiply(sigmoide(x), 1 - sigmoide(x))
```
Quando se realiza um treinamento com redes neurais, é importante inicializar aleatoriamente os parâmetros para que ocorra a **quebra de simetria**. Agora imagine que foi inicializado todos os pesos com o mesmo valor (por exemplo, zero ou um). Nesse caso, cada unidade oculta receberá exatamente o mesmo valor. Por exemplo, se todos os pesos são inicializados em 1, a ativação em cada neurônio da camada posterior vai ser igual a anterior. Se todos os pesos forem zeros, o que é ainda pior, a saída das funções de ativação da camada oculta serão zero. Não importa qual foi a entrada, se todos os pesos forem iguais, todas as unidades na camada oculta também serão as mesmas . Este é o principal problema com a simetria e a razão pela qual você deve inicializar pesos aleatoriamente (ou, pelo menos, com valores diferentes).

```
def inicia_pesos_randomicos(dim_entrada, dim_saida):
'''
Função que inicializa os pesos da rede neural de forma randômica
'''
# sugestão dada no curso para um epsilon ideal
epsilon = 0.12
return np.random.rand(dim_saida, dim_entrada + 1) * 2 * epsilon - epsilon
# definindo os parâmetros randômicos da camada oculta e camada de saída
teta1_inicial = inicia_pesos_randomicos(dim_camada_entrada, dim_camada_oculta)
teta2_inicial = inicia_pesos_randomicos(dim_camada_oculta, num_classes)
# adicionando os parâmetros em um único vetor unidimensional
parametros_rand_iniciais = np.hstack((teta1_inicial.ravel(order = 'F'), teta2_inicial.ravel(order = 'F')))
```
### Implementação do algoritmo de Backpropagation

1. **Calcule o algoritmo Forwardpropagation para obter a ativação de saída a3;**
2. **Calcule o termo de erro d3 obtido subtraindo a saída real de nossa saída calculada a3;**
3. **Para a camada oculta, o termo de erro d2 pode ser calculado como abaixo:**

4. **Acumule os gradientes em delta1 e delta2;**
5. **Obtenha os gradientes da rede neural dividindo os gradientes da etapa 4 por m;**
6. **Adicione os termos de regularização aos gradientes;**
```
def backpropagation_gradiente(parametros_totais, dim_entrada, dim_oculta, num_classes, X, y, lmbda):
'''
função que realiza o gradiente descendente através do algoritmo de Backpropagation
'''
# separa os parâmetros associados a camada oculta e a camada de saída
teta1 = np.reshape(parametros_totais[:dim_oculta * (dim_entrada + 1)], (dim_oculta, dim_entrada + 1), 'F')
teta2 = np.reshape(parametros_totais[dim_oculta * (dim_entrada + 1):], (num_classes, dim_oculta + 1), 'F')
# manipulando a saída das classes usando o one hot encoder
y_d = pd.get_dummies(y.flatten())
# definindo os arrays delta que indicam o erro associado a cada um dos parâmetros (o objetivo é minimizá-los)
delta1 = np.zeros(teta1.shape)
delta2 = np.zeros(teta2.shape)
# m exemplos treináveis
m = len(y)
for i in range(X.shape[0]):
'''
Forwardpropagation
'''
# cria um vetor coluna de bias m x 1
ones = np.ones(1)
# adicionando o bias na camada de entrada
a1 = np.hstack((ones, X[i]))
# produto escalar da saída dos neurônios da primeira camada com os parâmetros associados a camada oculta
z2 = a1.dot(teta1.T)
# ativação dos neurônios da camada oculta
a2 = np.hstack((ones, sigmoide(z2)))
# produto escalar da saída dos neurônios da camada oculta com os parâmetros associados a camade de saída
z3 = a2.dot(teta2.T)
# ativação dos neurônios da camada de saída
a3 = sigmoide(z3)
'''
Backpropagation
'''
# calcula o erro associado aos parâmetros da camada de saída
d3 = a3 - y_d.iloc[i,:][np.newaxis,:]
# adicionando o bias na camada oculta
z2 = np.hstack((ones, z2))
# calcula o erro associado aos parâmetros da camada oculta
d2 = np.multiply(teta2.T.dot(d3.T), sigmoideGradiente(z2).T[:,np.newaxis])
# computa o algoritmo do gradiente descendente para atualização dos erros associados ao parâmetro
delta1 = delta1 + d2[1:,:].dot(a1[np.newaxis,:])
delta2 = delta2 + d3.T.dot(a2[np.newaxis,:])
# divide pelos m exemplos treináveis para terminar de computar a fórmula
delta1 /= m
delta2 /= m
# computa as derivadas para a função de custo
delta1[:,1:] = delta1[:,1:] + teta1[:,1:] * lmbda / m
delta2[:,1:] = delta2[:,1:] + teta2[:,1:] * lmbda / m
# retorna o vetor unidimensional com todos os parâmetros
return np.hstack((delta1.ravel(order='F'), delta2.ravel(order='F')))
# recebendo os erros minimizados associados a cada parâmetro
parametros_backpropagation = backpropagation_gradiente(parametros_rand_iniciais, dim_camada_entrada, dim_camada_oculta, num_classes,
X, y, lmbda)
```
### Checando o gradiente
Para verificar se o algoritmo de backpropagation está funcionando bem, temos a seguinte expressão obtida através de métodos numéricos:

```
def checaGradiente(parametros_iniciais, parametros_backpropagation, dim_entrada, dim_oculta, num_classes, X, y, lmbda = 0.0):
# utilizando o valor de epsilon ideal mostrado no curso
epsilon = 0.0001
# recebe os vetores unidimensionais com os parâmetros
univetor_numerico = parametros_iniciais
univetor_backprop = parametros_backpropagation
# recebe os elementos do vetor
num_elementos = len(univetor_numerico)
# pega 10 elementos randômicos, calcula o gradiente pelo método numérico, e compara com o obtido através do algoritmo Backpropagation
for i in range(10):
x = int(np.random.rand() * num_elementos)
vetor_epsilon = np.zeros((num_elementos, 1))
vetor_epsilon[x] = epsilon
# implementa o gradiente pelo método numérico
custo_superior = funcaoCusto(univetor_numerico + vetor_epsilon.flatten(), dim_entrada, dim_oculta, num_classes, X, y, lmbda)
custo_inferior = funcaoCusto(univetor_numerico - vetor_epsilon.flatten(), dim_entrada, dim_oculta, num_classes, X, y, lmbda)
gradiente_numerico = (custo_superior - custo_inferior) / float(2 * epsilon)
print("Elemento: {0}. Método Numérico = {1:.9f}. BackPropagation = {2:.9f}.".format(x, gradiente_numerico, univetor_backprop[x]))
return None
# verificando se as derivadas realizadas pelo algoritmo backpropagation são equivalentes as do método numérico
checaGradiente(parametros_rand_iniciais, parametros_backpropagation, dim_camada_entrada, dim_camada_oculta, num_classes, X, y, lmbda)
```
### Treinando o algoritmo com um otimizador
```
# implementando as funções feitas no otimizado do scipy
teta_otimizado = opt.fmin_cg(maxiter = 50, f = funcaoCusto, x0 = parametros_rand_iniciais, fprime = backpropagation_gradiente,
args = (dim_camada_entrada, dim_camada_oculta, num_classes, X, y.flatten(), lmbda))
# obtendo o valor da função de custo para os parâmetros otimizados
funcaoCusto(teta_otimizado, dim_camada_entrada, dim_camada_oculta, num_classes, X, y, lmbda)
# obtendo o valor da função de custo para os parâmetros ideais
funcaoCusto(parametros_totais, dim_camada_entrada, dim_camada_oculta, num_classes, X, y, lmbda)
# separando os parâmetros da camada oculta e da camada de saída
teta1_otimizado = np.reshape(teta_otimizado[:dim_camada_oculta * (dim_camada_entrada + 1)],
(dim_camada_oculta, dim_camada_entrada + 1), 'F')
teta2_otimizado = np.reshape(teta_otimizado[dim_camada_oculta * (dim_camada_entrada + 1):],
(num_classes, dim_camada_oculta + 1), 'F')
```
### Realizando a predição do algoritmo
```
def predicao(teta1, teta2, X, y):
'''
Função responsável por realizar a previsão do algoritmo
'''
# implementando o algoritmo feedforward com os parâmetros ajustados
m = len(y)
ones = np.ones((m,1))
a1 = np.hstack((ones, X))
a2 = sigmoide(a1.dot(teta1.T))
a2 = np.hstack((ones, a2))
# obtendo a hipótese do algoritmo (o ideal seria a hipótese ser equivalente aos resultados das classes y)
h = sigmoide(a2.dot(teta2.T))
# sabendo que foi aplicado o one hot enconder, função retorna o indice da hipotese em que o 1 é ativado
# soma + 1 pelo fato dos dados começarem em 1, sendo 10 a classe 0
return np.argmax(h, axis = 1) + 1
# obtém a previsão para cada um dos exemplos do dataset
p = predicao(teta1_otimizado, teta2_otimizado, X, y)
# visualiza a predição do algoritmo
print('Predição: ', np.mean(p == y.flatten()) * 100)
```
|
github_jupyter
|
# carrega um dataset do matlab
from scipy.io import loadmat
# possibilita a implementação de vetorização
import numpy as np
# otimizador para minimizar a função de custo através do ajuste dos parâmetros
import scipy.optimize as opt
# biblioteca para manipulação e carregamento de datasets
import pandas as pd
# biblioteca de visualização gráfica
import matplotlib.pyplot as plt
# carregando os dados
dados = loadmat('dataset/ex4data1.mat')
# separando os atributos previsores das classes
X = dados['X']
y = dados['y']
# visualizando os dados
figura, configura_figura = plt.subplots(10, 10, figsize = (10,10))
for i in range(10):
for j in range(10):
# reordena a matriz para 20x20 usando o estilo de indexação do Fortran
configura_figura[i,j].imshow(X[np.random.randint(X.shape[0])].reshape((20,20), order = "F"))
# remove os eixos cartesianos de cada subplotagem
configura_figura[i,j].axis('off')
# carrega os parâmetros treinados vinculados ao exercício
pesos = loadmat('dataset/ex4weights.mat')
teta1 = pesos['Theta1'] # teta1 tem uma dimensão de 25 x 401
teta2 = pesos['Theta2'] #teta2 tem uma dimensão de 10 x 26
# ravel = torna teta1 e teta2 unidimensionais
parametros_totais = np.hstack((teta1.ravel(order='F'), teta2.ravel(order='F')))
# hiperparâmetros da rede neural
dim_camada_entrada = 400
dim_camada_oculta = 25
num_classes = 10
lmbda = 1
# visualizando a dimensão do vetor de parâmetros
parametros_totais.shape
def sigmoide(x):
'''
Função de ativação sigmoide
'''
return 1 / (1 + np.exp(-x))
# manipulando a saída das classes usando o one hot encoder
(pd.get_dummies(y.flatten()))
def funcaoCusto(parametros_totais, dim_entrada, dim_oculta, num_classes, X, y, lmbda):
'''
Função que computa a função de custo da rede neural regularizada
'''
# separa os parâmetros associados a camada oculta e a camada de saída
teta1 = np.reshape(parametros_totais[:dim_oculta * (dim_entrada + 1)], (dim_oculta, dim_entrada + 1), 'F')
teta2 = np.reshape(parametros_totais[dim_oculta * (dim_entrada + 1):], (num_classes, dim_oculta + 1), 'F')
'''
algoritmo de Forwardpropagation
'''
# m exemplos treináveis
m = len(y)
# cria um vetor coluna de bias m x 1
ones = np.ones((m, 1))
# adicionando o bias na camada de entrada
a1 = np.hstack((ones, X))
# ativação dos neurônios da camada oculta
a2 = sigmoide(a1.dot(teta1.T))
# adicionando o bias na camada oculta
a2 = np.hstack((ones, a2))
# ativção dos neurônios da camada de saída
h = sigmoide(a2.dot(teta2.T))
# manipulando a saída das classes usando o one hot encoder
y_d = pd.get_dummies(y.flatten())
# seprando em partes a função de custo
parte1 = np.multiply(y_d, np.log(h))
parte2 = np.multiply(1-y_d, np.log(1-h))
parte3 = np.sum(parte1 + parte2)
# separando em partes a expressão de regularização da função de custo
soma1_regularizada = np.sum(np.sum(np.power(teta1[:,1:],2), axis = 1)) # ignora o neurônio relativo ao bias
soma2_regularizada = np.sum(np.sum(np.power(teta2[:,1:],2), axis = 1)) # ignora o neurônio relativo ao bias
# junta as partes e retorna a expressão total da função de custo
return np.sum(parte3 / (-m)) + (soma1_regularizada + soma2_regularizada) * lmbda / (2 * m)
# obtendo o valor da função de custo
funcaoCusto(parametros_totais, dim_camada_entrada, dim_camada_oculta, num_classes, X, y, lmbda)
def sigmoideGradiente(x):
'''
Função que calculo o sigmoide do gradiente descente
'''
return np.multiply(sigmoide(x), 1 - sigmoide(x))
def inicia_pesos_randomicos(dim_entrada, dim_saida):
'''
Função que inicializa os pesos da rede neural de forma randômica
'''
# sugestão dada no curso para um epsilon ideal
epsilon = 0.12
return np.random.rand(dim_saida, dim_entrada + 1) * 2 * epsilon - epsilon
# definindo os parâmetros randômicos da camada oculta e camada de saída
teta1_inicial = inicia_pesos_randomicos(dim_camada_entrada, dim_camada_oculta)
teta2_inicial = inicia_pesos_randomicos(dim_camada_oculta, num_classes)
# adicionando os parâmetros em um único vetor unidimensional
parametros_rand_iniciais = np.hstack((teta1_inicial.ravel(order = 'F'), teta2_inicial.ravel(order = 'F')))
def backpropagation_gradiente(parametros_totais, dim_entrada, dim_oculta, num_classes, X, y, lmbda):
'''
função que realiza o gradiente descendente através do algoritmo de Backpropagation
'''
# separa os parâmetros associados a camada oculta e a camada de saída
teta1 = np.reshape(parametros_totais[:dim_oculta * (dim_entrada + 1)], (dim_oculta, dim_entrada + 1), 'F')
teta2 = np.reshape(parametros_totais[dim_oculta * (dim_entrada + 1):], (num_classes, dim_oculta + 1), 'F')
# manipulando a saída das classes usando o one hot encoder
y_d = pd.get_dummies(y.flatten())
# definindo os arrays delta que indicam o erro associado a cada um dos parâmetros (o objetivo é minimizá-los)
delta1 = np.zeros(teta1.shape)
delta2 = np.zeros(teta2.shape)
# m exemplos treináveis
m = len(y)
for i in range(X.shape[0]):
'''
Forwardpropagation
'''
# cria um vetor coluna de bias m x 1
ones = np.ones(1)
# adicionando o bias na camada de entrada
a1 = np.hstack((ones, X[i]))
# produto escalar da saída dos neurônios da primeira camada com os parâmetros associados a camada oculta
z2 = a1.dot(teta1.T)
# ativação dos neurônios da camada oculta
a2 = np.hstack((ones, sigmoide(z2)))
# produto escalar da saída dos neurônios da camada oculta com os parâmetros associados a camade de saída
z3 = a2.dot(teta2.T)
# ativação dos neurônios da camada de saída
a3 = sigmoide(z3)
'''
Backpropagation
'''
# calcula o erro associado aos parâmetros da camada de saída
d3 = a3 - y_d.iloc[i,:][np.newaxis,:]
# adicionando o bias na camada oculta
z2 = np.hstack((ones, z2))
# calcula o erro associado aos parâmetros da camada oculta
d2 = np.multiply(teta2.T.dot(d3.T), sigmoideGradiente(z2).T[:,np.newaxis])
# computa o algoritmo do gradiente descendente para atualização dos erros associados ao parâmetro
delta1 = delta1 + d2[1:,:].dot(a1[np.newaxis,:])
delta2 = delta2 + d3.T.dot(a2[np.newaxis,:])
# divide pelos m exemplos treináveis para terminar de computar a fórmula
delta1 /= m
delta2 /= m
# computa as derivadas para a função de custo
delta1[:,1:] = delta1[:,1:] + teta1[:,1:] * lmbda / m
delta2[:,1:] = delta2[:,1:] + teta2[:,1:] * lmbda / m
# retorna o vetor unidimensional com todos os parâmetros
return np.hstack((delta1.ravel(order='F'), delta2.ravel(order='F')))
# recebendo os erros minimizados associados a cada parâmetro
parametros_backpropagation = backpropagation_gradiente(parametros_rand_iniciais, dim_camada_entrada, dim_camada_oculta, num_classes,
X, y, lmbda)
def checaGradiente(parametros_iniciais, parametros_backpropagation, dim_entrada, dim_oculta, num_classes, X, y, lmbda = 0.0):
# utilizando o valor de epsilon ideal mostrado no curso
epsilon = 0.0001
# recebe os vetores unidimensionais com os parâmetros
univetor_numerico = parametros_iniciais
univetor_backprop = parametros_backpropagation
# recebe os elementos do vetor
num_elementos = len(univetor_numerico)
# pega 10 elementos randômicos, calcula o gradiente pelo método numérico, e compara com o obtido através do algoritmo Backpropagation
for i in range(10):
x = int(np.random.rand() * num_elementos)
vetor_epsilon = np.zeros((num_elementos, 1))
vetor_epsilon[x] = epsilon
# implementa o gradiente pelo método numérico
custo_superior = funcaoCusto(univetor_numerico + vetor_epsilon.flatten(), dim_entrada, dim_oculta, num_classes, X, y, lmbda)
custo_inferior = funcaoCusto(univetor_numerico - vetor_epsilon.flatten(), dim_entrada, dim_oculta, num_classes, X, y, lmbda)
gradiente_numerico = (custo_superior - custo_inferior) / float(2 * epsilon)
print("Elemento: {0}. Método Numérico = {1:.9f}. BackPropagation = {2:.9f}.".format(x, gradiente_numerico, univetor_backprop[x]))
return None
# verificando se as derivadas realizadas pelo algoritmo backpropagation são equivalentes as do método numérico
checaGradiente(parametros_rand_iniciais, parametros_backpropagation, dim_camada_entrada, dim_camada_oculta, num_classes, X, y, lmbda)
# implementando as funções feitas no otimizado do scipy
teta_otimizado = opt.fmin_cg(maxiter = 50, f = funcaoCusto, x0 = parametros_rand_iniciais, fprime = backpropagation_gradiente,
args = (dim_camada_entrada, dim_camada_oculta, num_classes, X, y.flatten(), lmbda))
# obtendo o valor da função de custo para os parâmetros otimizados
funcaoCusto(teta_otimizado, dim_camada_entrada, dim_camada_oculta, num_classes, X, y, lmbda)
# obtendo o valor da função de custo para os parâmetros ideais
funcaoCusto(parametros_totais, dim_camada_entrada, dim_camada_oculta, num_classes, X, y, lmbda)
# separando os parâmetros da camada oculta e da camada de saída
teta1_otimizado = np.reshape(teta_otimizado[:dim_camada_oculta * (dim_camada_entrada + 1)],
(dim_camada_oculta, dim_camada_entrada + 1), 'F')
teta2_otimizado = np.reshape(teta_otimizado[dim_camada_oculta * (dim_camada_entrada + 1):],
(num_classes, dim_camada_oculta + 1), 'F')
def predicao(teta1, teta2, X, y):
'''
Função responsável por realizar a previsão do algoritmo
'''
# implementando o algoritmo feedforward com os parâmetros ajustados
m = len(y)
ones = np.ones((m,1))
a1 = np.hstack((ones, X))
a2 = sigmoide(a1.dot(teta1.T))
a2 = np.hstack((ones, a2))
# obtendo a hipótese do algoritmo (o ideal seria a hipótese ser equivalente aos resultados das classes y)
h = sigmoide(a2.dot(teta2.T))
# sabendo que foi aplicado o one hot enconder, função retorna o indice da hipotese em que o 1 é ativado
# soma + 1 pelo fato dos dados começarem em 1, sendo 10 a classe 0
return np.argmax(h, axis = 1) + 1
# obtém a previsão para cada um dos exemplos do dataset
p = predicao(teta1_otimizado, teta2_otimizado, X, y)
# visualiza a predição do algoritmo
print('Predição: ', np.mean(p == y.flatten()) * 100)
| 0.408395 | 0.963161 |
## Travel Dataset Generator
This code generate a dataset of travel operations, _e.g._ , plane tickets and lodging.
This notebook just ilustrate the steps to generate the dataset, for extensive generators use the python code inside.
```
# Import packages
import names
import tqdm
import random
import pandas as pd
from datetime import datetime as dt
from datetime import timedelta as td
```
---
### Definitions
Predefine variables.
```
#- Companies and Users
defGenders = ['male', 'female', 'none']
defAgesInterval = {'min': 23, 'max': 50}
defFlightsInterval = {'min': 0, 'max': 3}
defCompanies = {
'HHD': {'usersCount': 2},
'4You': {'usersCount': 3},
}
#- Flight Agencies
defFlightTypes = {
'economic': {'price': 1.0},
'premium': {'price': 1.5},
}
defAgenciesName = ['FlyingDrops', 'Rainbow', 'CloudFy']
defAgencies = dict()
#- Places
defPlacesName = ['Sao Paulo (SP)', 'Rio de Janeiro (RJ)', 'Santa Catarina (SC)']
defPlaces = {name: dict() for name in defPlacesName}
defDistancesInterval = {'min': 200.0, 'max': 850.0}
defPlaceTravelKmPerHour = 400.0
#- Lodge (Accommodation)
defLodgesInterval = {'min': 1, 'max': 3}
defLodgesPrices = {'min': 60.0, 'max': 200.0}
defLodgesPrex = 'Hotel'
defLodges = {name: list() for name in defPlacesName}
#- Travel
defTravels = list()
defTravelsDays = {'min': 1, 'max': 3}
defTravelsFlightPrices = {'init': 300.0, 'interval': 100.0}
defTravelWithLodge = 0.3
defTravelDate = {'init': dt.now(), 'interval':{'min': 10, 'max': 60}}
```
---
### Companies and Users - Generator
```
#- Functions
def funcUserGenerator(genders, agesInterval, flightsInterval, code):
'''
Generate random user, based on predefinitions.
- genders: list
- agesInterval {min, max}: user age
- flightsInterval {min, max}: number of flights
- code: user ID
'''
user = dict()
user['code'] = code
user['gender'] = genders[random.randint(0, len(genders)-1)]
gender = user['gender'] if (user['gender'] != 'none') else False
user['name'] = names.get_full_name(gender=gender)
user['age'] = random.randint(agesInterval['min'], agesInterval['max'])
user['flights'] = random.randint(flightsInterval['min'], flightsInterval['max'])
return user
#- Fill Companies data
userId = 0
for company, data in defCompanies.items():
users = list()
for idx in range(data['usersCount']):
user = funcUserGenerator(defGenders, defAgesInterval, defFlightsInterval, userId)
users.append(user)
userId += 1
defCompanies[company]['users'] = users
```
Example - Users from a Company
```
defCompanies['HHD']['users']
```
### Flight Agencies - Generator
```
#- Functions
def funcAgencyGenerator(flightTypes):
'''
Generate random agency services, based on predefinitions.
- flightTypes: types of flight
'''
agency = dict()
types = list(flightTypes.copy().keys())
random.shuffle(types)
typesMany = random.randint(1, len(types))
agency['types'] = [types[i] for i in range(typesMany)]
return agency
for agency in defAgenciesName:
defAgencies[agency] = funcAgencyGenerator(defFlightTypes)
```
Example - Flight Types of Agencies
```
defAgencies
```
### Places - Generator
```
#- Functions
def funcPlaceGenerator(i, j, distInterval, kmPerHour):
'''
Generate random place distances, based on predefinitions.
- i: number of place
- j: number of place
- distInterval {min, max} values: distance range
- kmPerHour: km per hour of the plain
'''
if i == j:
return False, False, False
distance = round(random.uniform(distInterval['min'], distInterval['max']), 2)
time = round(distance/kmPerHour, 2)
hours = int(time)
minutes = (time*60) % 60
timeMsg = '%d:%dh' % (hours, minutes)
return (distance, time, timeMsg)
n = len(defPlacesName)
for i in range(n):
for j in range(i, n):
fromPlace = defPlacesName[i]
toPlace = defPlacesName[j]
distance, time, msg = funcPlaceGenerator(i, j, defDistancesInterval, defPlaceTravelKmPerHour)
if distance and time:
place = {'distance': distance, 'time': time, 'timeMsg': msg}
defPlaces[fromPlace][toPlace] = place
defPlaces[toPlace][fromPlace] = place
```
Example - Distances from a Place
```
defPlaces['Sao Paulo (SP)']
```
### Lodges - Generator
```
#- Definitions
defName = 'A'
#- Functions
def getNextChar(text):
'''
Generate order alphabetic.
- text: input text
'''
if len(text) == 0:
return 'A'
nextChar = chr(ord(text[-1]) + 1)
if nextChar <= 'Z':
text = text[:-1] + nextChar
else:
text = getNextChar(text[:-1]) + 'A'
return text
def funcLodgesGenerator(lodgesInterval, lodgesPrices):
'''
Generate random lodges, based on predefinitions.
- lodgesInterval {min, max} values: number of hotels
- lodgesPrices {min, max} values: hotel range
'''
global defName
lodges = list()
n = random.randint(lodgesInterval['min'], lodgesInterval['max'])
for i in range(n):
lodgeName = '%s %s' % (defLodgesPrex, defName)
price = round(random.uniform(lodgesPrices['min'], lodgesPrices['max']), 2)
lodge = {'code': defName, 'name': lodgeName, 'price': price}
lodges.append(lodge)
defName = getNextChar(defName)
return lodges
for name in defPlacesName:
lodges = funcLodgesGenerator(defLodgesInterval, defLodgesPrices)
defLodges[name] = lodges
```
Example - Hotels from a Place
```
defLodges['Sao Paulo (SP)']
```
### Travel Possibilities - Generator
```
#- Functions
def funcCalculatePrice(priceMin, priceMax, weight):
'''
Calculate a random price for a travel.
- priceMin: min price
- priceMax: max price
- weight: weight the price range
'''
priceMin = priceMin * weight
priceMax = priceMax * weight
price = round(random.uniform(priceMin, priceMax), 2)
return price
def funcElaborateflight(fromPlace, toPlace, distance, agency, flightType, price, \
time, timeMsg):
'''
Elaborate a possible flight.
- fromPlace: from
- toPlace: to
- distance: distance
- agency: agency name
- flightType: flight type
- price: flight price
- time: time in hours
- timeMsg: time calculated
'''
flight = {'from': fromPlace, 'to': toPlace, 'distance': distance,
'agency': agency, 'flightType': flightType, 'price': price,
'time': time, 'timeMsg': timeMsg}
return flight
def funcFlightsPossibilities(places, flightPrices, flightTypes, agencies):
'''
Elaborate a list of possible flights.
- places: places data
- flightPrices: flight prices
- flightTypes: flight types
- agencies: agencies data
'''
flightsPossibilities = list()
for fromPlace, toPlaces in places.items():
toPlacesSorted = sorted(toPlaces.items(), key=lambda x:x[1]['distance'], reverse=False)
priceA, priceB = flightPrices['init'], \
flightPrices['init'] + flightPrices['interval']
for (toPlace, placeData) in toPlacesSorted:
for (agencyName, agencyData) in agencies.items():
if len(agencyData['types']) > 1: # has more than 1 element
for typeA in agencyData['types']:
weight = flightTypes[typeA]['price']
price = funcCalculatePrice(priceA, priceB, weight)
flight = funcElaborateflight(fromPlace, toPlace, placeData['distance'], \
agencyName, typeA, price, placeData['time'], placeData['timeMsg'])
flightsPossibilities.append(flight)
else:
typeA = agencyData['types'][0]
weight = flightTypes[typeA]['price']
price = funcCalculatePrice(priceA, priceB, weight)
flight = funcElaborateflight(fromPlace, toPlace, placeData['distance'], agencyName, \
typeA, price, placeData['time'], placeData['timeMsg'])
flightsPossibilities.append(flight)
# Update prices for bigger distances
priceA, priceB = priceB, priceB + flightPrices['interval']
return flightsPossibilities
def funcLodgesPossibilities(placesName, lodges):
'''
Elaborate a list of possible hotels.
- placesName: places names
- lodges: lodges data
'''
lodgesPossibilities = list()
for place in placesName:
for lodge in lodges[place]:
lodge = lodge.copy()
lodge['place'] = place
lodgesPossibilities.append(lodge)
return lodgesPossibilities
```
Example - Fligts Possibilities (for each Place)
```
flightsPossibilities = funcFlightsPossibilities(defPlaces, defTravelsFlightPrices, defFlightTypes, defAgencies)
pd.DataFrame(flightsPossibilities).head(5)
```
Example - Hotel Possibilities (for each Place)
```
lodgesPossibilities = funcLodgesPossibilities(defPlacesName, defLodges)
pd.DataFrame(lodgesPossibilities).head(5)
```
---
### Travel Dataset - Generator
```
#- Definitions
travelCode = 0
#- Functions
def df2Dict(df):
'''
Convert dataframe into dict
'''
procDict = dict()
tmp = df.to_dict('split')
data = tmp['data'][0]
for (i, column) in enumerate(tmp['columns']):
procDict[column] = data[i]
return procDict
def funcTravelsSimulated(companies, flightsPossibilities, lodgesPossibilities, travelDate, travelsDays, \
travelWithLodge, placesName):
'''
Elaborate random travels with flights and lodges, based on possibilities.
- flightsPossibilities: possible flights
- lodgesPossibilities: possible hotels
'''
global travelCode
dfFlightsPos = pd.DataFrame(flightsPossibilities)
dfLodgesPos = pd.DataFrame(lodgesPossibilities)
flightsSimulated, lodgesSimulated = list(), list()
for (companyName, companyData) in companies.items():
for user in companyData['users']:
date = travelDate['init']
for i in range(user['flights']):
# random - days, places, hotel?
daysFlight = random.randint(travelsDays['min'], travelsDays['max'])
daysNextTravel = random.randint(travelDate['interval']['min'], travelDate['interval']['min'])
fromPlace, toPlace = random.sample(placesName, 2)
chanceTravelWithLodge = (random.randrange(100) < travelWithLodge*100)
# travels
fromConditions = (dfFlightsPos['from']==fromPlace) & (dfFlightsPos['to']==toPlace)
tmpFlightFrom = df2Dict(dfFlightsPos[fromConditions].sample(n=1))
toConditions = (dfFlightsPos['from']==toPlace) & (dfFlightsPos['to']==fromPlace) & \
(dfFlightsPos['agency']==tmpFlightFrom['agency']) & \
(dfFlightsPos['flightType']==tmpFlightFrom['flightType'])
tmpFlightTo = df2Dict(dfFlightsPos[toConditions])
tmpFlightFrom['userCode'] = tmpFlightTo['userCode'] = user['code']
tmpFlightFrom['travelCode'] = tmpFlightTo['travelCode'] = travelCode
tmpFlightFrom['date'] = date
tmpFlightTo['date'] = date + td(days=daysFlight)
# lodge
if chanceTravelWithLodge:
lodgeConditions = (dfLodgesPos['place']==toPlace)
tmpLodge = df2Dict(dfLodgesPos[lodgeConditions])
tmpLodge['userCode'] = user['code']
tmpLodge['date'] = date
tmpLodge['days'] = daysFlight
tmpLodge['total'] = round(tmpLodge['price'] * daysFlight, 2)
tmpLodge['travelCode'] = travelCode
lodgesSimulated.append(tmpLodge)
# save and update data
flightsSimulated.append(tmpFlightFrom)
flightsSimulated.append(tmpFlightTo)
travelCode += 1
date = dt.now() + td(days=daysNextTravel)
return flightsSimulated, lodgesSimulated
flightsSimulated, lodgesSimulated = \
funcTravelsSimulated(defCompanies, flightsPossibilities, lodgesPossibilities,
defTravelDate, defTravelsDays, defTravelWithLodge, defPlacesName)
```
Example - Travel (From->To + To->From)
```
flightsSimulated[0:2]
lodgesSimulated[0]
```
|
github_jupyter
|
# Import packages
import names
import tqdm
import random
import pandas as pd
from datetime import datetime as dt
from datetime import timedelta as td
#- Companies and Users
defGenders = ['male', 'female', 'none']
defAgesInterval = {'min': 23, 'max': 50}
defFlightsInterval = {'min': 0, 'max': 3}
defCompanies = {
'HHD': {'usersCount': 2},
'4You': {'usersCount': 3},
}
#- Flight Agencies
defFlightTypes = {
'economic': {'price': 1.0},
'premium': {'price': 1.5},
}
defAgenciesName = ['FlyingDrops', 'Rainbow', 'CloudFy']
defAgencies = dict()
#- Places
defPlacesName = ['Sao Paulo (SP)', 'Rio de Janeiro (RJ)', 'Santa Catarina (SC)']
defPlaces = {name: dict() for name in defPlacesName}
defDistancesInterval = {'min': 200.0, 'max': 850.0}
defPlaceTravelKmPerHour = 400.0
#- Lodge (Accommodation)
defLodgesInterval = {'min': 1, 'max': 3}
defLodgesPrices = {'min': 60.0, 'max': 200.0}
defLodgesPrex = 'Hotel'
defLodges = {name: list() for name in defPlacesName}
#- Travel
defTravels = list()
defTravelsDays = {'min': 1, 'max': 3}
defTravelsFlightPrices = {'init': 300.0, 'interval': 100.0}
defTravelWithLodge = 0.3
defTravelDate = {'init': dt.now(), 'interval':{'min': 10, 'max': 60}}
#- Functions
def funcUserGenerator(genders, agesInterval, flightsInterval, code):
'''
Generate random user, based on predefinitions.
- genders: list
- agesInterval {min, max}: user age
- flightsInterval {min, max}: number of flights
- code: user ID
'''
user = dict()
user['code'] = code
user['gender'] = genders[random.randint(0, len(genders)-1)]
gender = user['gender'] if (user['gender'] != 'none') else False
user['name'] = names.get_full_name(gender=gender)
user['age'] = random.randint(agesInterval['min'], agesInterval['max'])
user['flights'] = random.randint(flightsInterval['min'], flightsInterval['max'])
return user
#- Fill Companies data
userId = 0
for company, data in defCompanies.items():
users = list()
for idx in range(data['usersCount']):
user = funcUserGenerator(defGenders, defAgesInterval, defFlightsInterval, userId)
users.append(user)
userId += 1
defCompanies[company]['users'] = users
defCompanies['HHD']['users']
#- Functions
def funcAgencyGenerator(flightTypes):
'''
Generate random agency services, based on predefinitions.
- flightTypes: types of flight
'''
agency = dict()
types = list(flightTypes.copy().keys())
random.shuffle(types)
typesMany = random.randint(1, len(types))
agency['types'] = [types[i] for i in range(typesMany)]
return agency
for agency in defAgenciesName:
defAgencies[agency] = funcAgencyGenerator(defFlightTypes)
defAgencies
#- Functions
def funcPlaceGenerator(i, j, distInterval, kmPerHour):
'''
Generate random place distances, based on predefinitions.
- i: number of place
- j: number of place
- distInterval {min, max} values: distance range
- kmPerHour: km per hour of the plain
'''
if i == j:
return False, False, False
distance = round(random.uniform(distInterval['min'], distInterval['max']), 2)
time = round(distance/kmPerHour, 2)
hours = int(time)
minutes = (time*60) % 60
timeMsg = '%d:%dh' % (hours, minutes)
return (distance, time, timeMsg)
n = len(defPlacesName)
for i in range(n):
for j in range(i, n):
fromPlace = defPlacesName[i]
toPlace = defPlacesName[j]
distance, time, msg = funcPlaceGenerator(i, j, defDistancesInterval, defPlaceTravelKmPerHour)
if distance and time:
place = {'distance': distance, 'time': time, 'timeMsg': msg}
defPlaces[fromPlace][toPlace] = place
defPlaces[toPlace][fromPlace] = place
defPlaces['Sao Paulo (SP)']
#- Definitions
defName = 'A'
#- Functions
def getNextChar(text):
'''
Generate order alphabetic.
- text: input text
'''
if len(text) == 0:
return 'A'
nextChar = chr(ord(text[-1]) + 1)
if nextChar <= 'Z':
text = text[:-1] + nextChar
else:
text = getNextChar(text[:-1]) + 'A'
return text
def funcLodgesGenerator(lodgesInterval, lodgesPrices):
'''
Generate random lodges, based on predefinitions.
- lodgesInterval {min, max} values: number of hotels
- lodgesPrices {min, max} values: hotel range
'''
global defName
lodges = list()
n = random.randint(lodgesInterval['min'], lodgesInterval['max'])
for i in range(n):
lodgeName = '%s %s' % (defLodgesPrex, defName)
price = round(random.uniform(lodgesPrices['min'], lodgesPrices['max']), 2)
lodge = {'code': defName, 'name': lodgeName, 'price': price}
lodges.append(lodge)
defName = getNextChar(defName)
return lodges
for name in defPlacesName:
lodges = funcLodgesGenerator(defLodgesInterval, defLodgesPrices)
defLodges[name] = lodges
defLodges['Sao Paulo (SP)']
#- Functions
def funcCalculatePrice(priceMin, priceMax, weight):
'''
Calculate a random price for a travel.
- priceMin: min price
- priceMax: max price
- weight: weight the price range
'''
priceMin = priceMin * weight
priceMax = priceMax * weight
price = round(random.uniform(priceMin, priceMax), 2)
return price
def funcElaborateflight(fromPlace, toPlace, distance, agency, flightType, price, \
time, timeMsg):
'''
Elaborate a possible flight.
- fromPlace: from
- toPlace: to
- distance: distance
- agency: agency name
- flightType: flight type
- price: flight price
- time: time in hours
- timeMsg: time calculated
'''
flight = {'from': fromPlace, 'to': toPlace, 'distance': distance,
'agency': agency, 'flightType': flightType, 'price': price,
'time': time, 'timeMsg': timeMsg}
return flight
def funcFlightsPossibilities(places, flightPrices, flightTypes, agencies):
'''
Elaborate a list of possible flights.
- places: places data
- flightPrices: flight prices
- flightTypes: flight types
- agencies: agencies data
'''
flightsPossibilities = list()
for fromPlace, toPlaces in places.items():
toPlacesSorted = sorted(toPlaces.items(), key=lambda x:x[1]['distance'], reverse=False)
priceA, priceB = flightPrices['init'], \
flightPrices['init'] + flightPrices['interval']
for (toPlace, placeData) in toPlacesSorted:
for (agencyName, agencyData) in agencies.items():
if len(agencyData['types']) > 1: # has more than 1 element
for typeA in agencyData['types']:
weight = flightTypes[typeA]['price']
price = funcCalculatePrice(priceA, priceB, weight)
flight = funcElaborateflight(fromPlace, toPlace, placeData['distance'], \
agencyName, typeA, price, placeData['time'], placeData['timeMsg'])
flightsPossibilities.append(flight)
else:
typeA = agencyData['types'][0]
weight = flightTypes[typeA]['price']
price = funcCalculatePrice(priceA, priceB, weight)
flight = funcElaborateflight(fromPlace, toPlace, placeData['distance'], agencyName, \
typeA, price, placeData['time'], placeData['timeMsg'])
flightsPossibilities.append(flight)
# Update prices for bigger distances
priceA, priceB = priceB, priceB + flightPrices['interval']
return flightsPossibilities
def funcLodgesPossibilities(placesName, lodges):
'''
Elaborate a list of possible hotels.
- placesName: places names
- lodges: lodges data
'''
lodgesPossibilities = list()
for place in placesName:
for lodge in lodges[place]:
lodge = lodge.copy()
lodge['place'] = place
lodgesPossibilities.append(lodge)
return lodgesPossibilities
flightsPossibilities = funcFlightsPossibilities(defPlaces, defTravelsFlightPrices, defFlightTypes, defAgencies)
pd.DataFrame(flightsPossibilities).head(5)
lodgesPossibilities = funcLodgesPossibilities(defPlacesName, defLodges)
pd.DataFrame(lodgesPossibilities).head(5)
#- Definitions
travelCode = 0
#- Functions
def df2Dict(df):
'''
Convert dataframe into dict
'''
procDict = dict()
tmp = df.to_dict('split')
data = tmp['data'][0]
for (i, column) in enumerate(tmp['columns']):
procDict[column] = data[i]
return procDict
def funcTravelsSimulated(companies, flightsPossibilities, lodgesPossibilities, travelDate, travelsDays, \
travelWithLodge, placesName):
'''
Elaborate random travels with flights and lodges, based on possibilities.
- flightsPossibilities: possible flights
- lodgesPossibilities: possible hotels
'''
global travelCode
dfFlightsPos = pd.DataFrame(flightsPossibilities)
dfLodgesPos = pd.DataFrame(lodgesPossibilities)
flightsSimulated, lodgesSimulated = list(), list()
for (companyName, companyData) in companies.items():
for user in companyData['users']:
date = travelDate['init']
for i in range(user['flights']):
# random - days, places, hotel?
daysFlight = random.randint(travelsDays['min'], travelsDays['max'])
daysNextTravel = random.randint(travelDate['interval']['min'], travelDate['interval']['min'])
fromPlace, toPlace = random.sample(placesName, 2)
chanceTravelWithLodge = (random.randrange(100) < travelWithLodge*100)
# travels
fromConditions = (dfFlightsPos['from']==fromPlace) & (dfFlightsPos['to']==toPlace)
tmpFlightFrom = df2Dict(dfFlightsPos[fromConditions].sample(n=1))
toConditions = (dfFlightsPos['from']==toPlace) & (dfFlightsPos['to']==fromPlace) & \
(dfFlightsPos['agency']==tmpFlightFrom['agency']) & \
(dfFlightsPos['flightType']==tmpFlightFrom['flightType'])
tmpFlightTo = df2Dict(dfFlightsPos[toConditions])
tmpFlightFrom['userCode'] = tmpFlightTo['userCode'] = user['code']
tmpFlightFrom['travelCode'] = tmpFlightTo['travelCode'] = travelCode
tmpFlightFrom['date'] = date
tmpFlightTo['date'] = date + td(days=daysFlight)
# lodge
if chanceTravelWithLodge:
lodgeConditions = (dfLodgesPos['place']==toPlace)
tmpLodge = df2Dict(dfLodgesPos[lodgeConditions])
tmpLodge['userCode'] = user['code']
tmpLodge['date'] = date
tmpLodge['days'] = daysFlight
tmpLodge['total'] = round(tmpLodge['price'] * daysFlight, 2)
tmpLodge['travelCode'] = travelCode
lodgesSimulated.append(tmpLodge)
# save and update data
flightsSimulated.append(tmpFlightFrom)
flightsSimulated.append(tmpFlightTo)
travelCode += 1
date = dt.now() + td(days=daysNextTravel)
return flightsSimulated, lodgesSimulated
flightsSimulated, lodgesSimulated = \
funcTravelsSimulated(defCompanies, flightsPossibilities, lodgesPossibilities,
defTravelDate, defTravelsDays, defTravelWithLodge, defPlacesName)
flightsSimulated[0:2]
lodgesSimulated[0]
| 0.407569 | 0.857768 |
# Deep Learning with TensorFlow
Classical machine learning relies on using statistics to determine relationships between features and labels, and can be very effective for creating predictive models. However, a massive growth in the availability of data coupled with advances in the computing technology required to process it has led to the emergence of new machine learning techniques that mimic the way the brain processes information in a structure called an artificial neural network.
TensorFlow is a framework for creating machine learning models, including deep neural networks (DNNs). In this example, we'll use Tensorflow to create a simple neural network that classifies penguins into species based on the length and depth of their culmen (bill), their flipper length, and their body mass.
> **Citation**: The penguins dataset used in the this exercise is a subset of data collected and made available by [Dr. Kristen
Gorman](https://www.uaf.edu/cfos/people/faculty/detail/kristen-gorman.php)
and the [Palmer Station, Antarctica LTER](https://pal.lternet.edu/), a
member of the [Long Term Ecological Research
Network](https://lternet.edu/).
## Explore the dataset
Before we start using TensorFlow to create a model, let's load the data we need from the Palmer Islands penguins dataset, which contains observations of three different species of penguin.
> **Note**: In reality, you can solve the penguin classification problem easily using classical machine learning techniques without the need for a deep learning model; but it's a useful, easy to understand dataset with which to demonstrate the principles of neural networks in this notebook.
```
import pandas as pd
# load the training dataset (excluding rows with null values)
penguins = pd.read_csv('data/penguins.csv').dropna()
# Deep Learning models work best when features are on similar scales
# In a real solution, we'd implement some custom normalization for each feature, but to keep things simple
# we'll just rescale the FlipperLength and BodyMass so they're on a similar scale to the bill measurements
penguins['FlipperLength'] = penguins['FlipperLength']/10
penguins['BodyMass'] = penguins['BodyMass']/100
# The dataset is too small to be useful for deep learning
# So we'll oversample it to increase its size
for i in range(1,3):
penguins = penguins.append(penguins)
# Display a random sample of 10 observations
sample = penguins.sample(10)
sample
```
The **Species** column is the label our model will predict. Each label value represents a class of penguin species, encoded as 0, 1, or 2. The following code shows the actual species to which these class labels corrrespond.
```
penguin_classes = ['Amelie', 'Gentoo', 'Chinstrap']
print(sample.columns[0:5].values, 'SpeciesName')
for index, row in penguins.sample(10).iterrows():
print('[',row[0], row[1], row[2],row[3], int(row[4]), ']',penguin_classes[int(row[-1])])
```
As is common in a supervised learning problem, we'll split the dataset into a set of records with which to train the model, and a smaller set with which to validate the trained model.
```
from sklearn.model_selection import train_test_split
features = ['CulmenLength','CulmenDepth','FlipperLength','BodyMass']
label = 'Species'
# Split data 70%-30% into training set and test set
x_train, x_test, y_train, y_test = train_test_split(penguins[features].values,
penguins[label].values,
test_size=0.30,
random_state=0)
print ('Training Set: %d, Test Set: %d \n' % (len(x_train), len(x_test)))
print("Sample of features and labels:")
# Take a look at the first 25 training features and corresponding labels
for n in range(0,24):
print(x_train[n], y_train[n], '(' + penguin_classes[y_train[n]] + ')')
```
The *features* are the measurements for each penguin observation, and the *label* is a numeric value that indicates the species of penguin that the observation represents (Amelie, Gentoo, or Chinstrap).
## Install and import TensorFlow libraries
Since we plan to use TensorFlow to create our penguin classifier, we'll need to run the following two cells to install and import the libraries we intend to use.
> **Note** *Keras* is an abstraction layer over the base TensorFlow API. In most common machine learning scenarios, you can use Keras to simplify your code.
```
!pip install --upgrade tensorflow
import tensorflow
from tensorflow import keras
from tensorflow.keras import models
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
from tensorflow.keras import utils
from tensorflow.keras import optimizers
# Set random seed for reproducability
tensorflow.random.set_seed(0)
print("Libraries imported.")
print('Keras version:',keras.__version__)
print('TensorFlow version:',tensorflow.__version__)
```
## Prepare the data for TensorFlow
We've already loaded our data and split it into training and validation datasets. However, we need to do some further data preparation so that our data will work correctly with TensorFlow. Specifically, we need to set the data type of our features to 32-bit floating point numbers, and specify that the labels represent categorical classes rather than numeric values.
```
# Set data types for float features
x_train = x_train.astype('float32')
x_test = x_test.astype('float32')
# Set data types for categorical labels
y_train = utils.to_categorical(y_train)
y_test = utils.to_categorical(y_test)
print('Ready...')
```
## Define a neural network
Now we're ready to define our neural network. In this case, we'll create a network that consists of 3 fully-connected layers:
* An input layer that receives an input value for each feature (in this case, the four penguin measurements) and applies a *ReLU* activation function.
* A hidden layer that receives ten inputs and applies a *ReLU* activation function.
* An output layer that uses a *SoftMax* activation function to generate an output for each penguin species (which represent the classification probabilities for each of the three possible penguin species). Softmax functions produce a vector with probability values that sum to 1.
```
# Define a classifier network
hl = 10 # Number of hidden layer nodes
model = Sequential()
model.add(Dense(hl, input_dim=len(features), activation='relu'))
model.add(Dense(hl, input_dim=hl, activation='relu'))
model.add(Dense(len(penguin_classes), input_dim=hl, activation='softmax'))
print(model.summary())
```
## Train the model
To train the model, we need to repeatedly feed the training values forward through the network, use a loss function to calculate the loss, use an optimizer to backpropagate the weight and bias value adjustments, and validate the model using the test data we withheld.
To do this, we'll apply an Adam optimizer to a categorical cross-entropy loss function iteratively over 50 epochs.
```
#hyper-parameters for optimizer
learning_rate = 0.001
opt = optimizers.Adam(lr=learning_rate)
model.compile(loss='categorical_crossentropy',
optimizer=opt,
metrics=['accuracy'])
# Train the model over 50 epochs using 10-observation batches and using the test holdout dataset for validation
num_epochs = 50
history = model.fit(x_train, y_train, epochs=num_epochs, batch_size=10, validation_data=(x_test, y_test))
```
While the training process is running, let's try to understand what's happening:
1. In each *epoch*, the full set of training data is passed forward through the network. There are four features for each observation, and four corresponding nodes in the input layer - so the features for each observation are passed as a vector of four values to that layer. However, for efficiency, the feature vectors are grouped into batches; so actually a matrix of multiple feature vectors is fed in each time.
2. The matrix of feature values is processed by a function that performs a weighted sum using initialized weights and bias values. The result of this function is then processed by the activation function for the input layer to constrain the values passed to the nodes in the next layer.
3. The weighted sum and activation functions are repeated in each layer. Note that the functions operate on vectors and matrices rather than individual scalar values. In other words, the forward pass is essentially a series of nested linear algebra functions. This is the reason data scientists prefer to use computers with graphical processing units (GPUs), since these are optimized for matrix and vector calculations.
4. In the final layer of the network, the output vectors contain a probability value for each possible class (in this case, classes 0, 1, and 2). This vector is processed by a *loss function* to determine how far the values calculated by the network are from the actual values - so for example, suppose the output for a Gentoo penguin (class 1) observation is \[0.3, 0.4, 0.3\]. The correct prediction should be \[0.0, 1.0, 0.0\], so the variance between the predicted and actual values (how far away the each predicted value is from what it should be) is \[0.3, 0.6, 0.3\]. This variance is aggregated for each batch and maintained as a running aggregate to calculate the overall level of error (*loss*) incurred by the training data for the epoch. The accuracy (proportion of correct predictions based on the highest probability value in the output vector) for the training data is also calculated.
5. At the end of each epoch, the validation data is passed through the network, and its loss and accuracy are also calculated. It's important to do this because it enables us to compare the performance of the model using data on which it was not trained, helping us determine if it will generalize well for new data or if it's *overfitted* to the training data.
6. After all the data has been passed forward through the network, the output of the loss function for the *training* data (but <u>not</u> the *validation* data) is passed to the opimizer. The precise details of how the optimizer processes the loss vary depending on the specific optimization algorithm being used; but fundamentally you can think of the entire network, from the input layer to the loss function as being one big nested (*composite*) function. The optimizer applies some differential calculus to calculate *partial derivatives* for the function with respect to each weight and bias value that was used in the network. It's possible to do this efficiently for a nested function due to something called the *chain rule*, which enables you to determine the derivative of a composite function from the derivatives of its inner function and outer functions. You don't really need to worry about the details of the math here (the optimizer does it for you), but the end result is that the partial derivatives tell us about the slope (or *gradient*) of the loss function with respect to each weight and bias value - in other words, we can determine whether to increase or decrease the weight and bias values in order to decrease the loss.
7. Having determined in which direction to adjust the weights and biases, the optimizer uses the *learning rate* to determine by how much to adjust them; and then works backwards through the network in a process called *backpropagation* to assign new values to the weights and biases in each layer.
8. Now the next epoch repeats the whole training, validation, and backpropagation process starting with the revised weights and biases from the previous epoch - which hopefully will result in a lower level of loss.
9. The process continues like this for 50 epochs.
## Review training and validation loss
After training is complete, we can examine the loss metrics we recorded while training and validating the model. We're really looking for two things:
* The loss should reduce with each epoch, showing that the model is learning the right weights and biases to predict the correct labels.
* The training loss and validation loss should follow a similar trend, showing that the model is not overfitting to the training data.
Let's plot the loss metrics and see:
```
%matplotlib inline
from matplotlib import pyplot as plt
epoch_nums = range(1,num_epochs+1)
training_loss = history.history["loss"]
validation_loss = history.history["val_loss"]
plt.plot(epoch_nums, training_loss)
plt.plot(epoch_nums, validation_loss)
plt.xlabel('epoch')
plt.ylabel('loss')
plt.legend(['training', 'validation'], loc='upper right')
plt.show()
```
## View the learned weights and biases
The trained model consists of the final weights and biases that were determined by the optimizer during training. Based on our network model we should expect the following values for each layer:
* Layer 1: There are four input values going to ten output nodes, so there should be 4 x 10 weights and 10 bias values.
* Layer 2: There are ten input values going to ten output nodes, so there should be 10 x 10 weights and 10 bias values.
* Layer 3: There are ten input values going to three output nodes, so there should be 10 x 3 weights and 3 bias values.
```
for layer in model.layers:
weights = layer.get_weights()[0]
biases = layer.get_weights()[1]
print('------------\nWeights:\n',weights,'\nBiases:\n', biases)
```
## Evaluate model performance
So, is the model any good? The raw accuracy reported from the validation data would seem to indicate that it predicts pretty well; but it's typically useful to dig a little deeper and compare the predictions for each possible class. A common way to visualize the performace of a classification model is to create a *confusion matrix* that shows a crosstab of correct and incorrect predictions for each class.
```
# Tensorflow doesn't have a built-in confusion matrix metric, so we'll use SciKit-Learn
import numpy as np
from sklearn.metrics import confusion_matrix
import matplotlib.pyplot as plt
%matplotlib inline
class_probabilities = model.predict(x_test)
predictions = np.argmax(class_probabilities, axis=1)
true_labels = np.argmax(y_test, axis=1)
# Plot the confusion matrix
cm = confusion_matrix(true_labels, predictions)
plt.imshow(cm, interpolation="nearest", cmap=plt.cm.Blues)
plt.colorbar()
tick_marks = np.arange(len(penguin_classes))
plt.xticks(tick_marks, penguin_classes, rotation=85)
plt.yticks(tick_marks, penguin_classes)
plt.xlabel("Actual Class")
plt.ylabel("Predicted Class")
plt.show()
```
The confusion matrix should show a strong diagonal line indicating that there are more correct than incorrect predictions for each class.
## Save the trained model
Now that we have a model we believe is reasonably accurate, we can save its trained weights for use later.
```
# Save the trained model
modelFileName = 'models/penguin-classifier.h5'
model.save(modelFileName)
del model # deletes the existing model variable
print('model saved as', modelFileName)
```
## Use the trained model
When we have a new penguin observation, we can use the model to predict the species.
```
# Load the saved model
model = models.load_model(modelFileName)
# CReate a new array of features
x_new = np.array([[50.4,15.3,20,50]])
print ('New sample: {}'.format(x_new))
# Use the model to predict the class
class_probabilities = model.predict(x_new)
predictions = np.argmax(class_probabilities, axis=1)
print(penguin_classes[predictions[0]])
```
## Learn more
This notebook was designed to help you understand the basic concepts and principles involved in deep neural networks, using a simple Tensorflow example. To learn more about Tensorflow, take a look at the <a href="https://www.tensorflow.org/" target="_blank">Tensorflow web site</a>.
|
github_jupyter
|
import pandas as pd
# load the training dataset (excluding rows with null values)
penguins = pd.read_csv('data/penguins.csv').dropna()
# Deep Learning models work best when features are on similar scales
# In a real solution, we'd implement some custom normalization for each feature, but to keep things simple
# we'll just rescale the FlipperLength and BodyMass so they're on a similar scale to the bill measurements
penguins['FlipperLength'] = penguins['FlipperLength']/10
penguins['BodyMass'] = penguins['BodyMass']/100
# The dataset is too small to be useful for deep learning
# So we'll oversample it to increase its size
for i in range(1,3):
penguins = penguins.append(penguins)
# Display a random sample of 10 observations
sample = penguins.sample(10)
sample
penguin_classes = ['Amelie', 'Gentoo', 'Chinstrap']
print(sample.columns[0:5].values, 'SpeciesName')
for index, row in penguins.sample(10).iterrows():
print('[',row[0], row[1], row[2],row[3], int(row[4]), ']',penguin_classes[int(row[-1])])
from sklearn.model_selection import train_test_split
features = ['CulmenLength','CulmenDepth','FlipperLength','BodyMass']
label = 'Species'
# Split data 70%-30% into training set and test set
x_train, x_test, y_train, y_test = train_test_split(penguins[features].values,
penguins[label].values,
test_size=0.30,
random_state=0)
print ('Training Set: %d, Test Set: %d \n' % (len(x_train), len(x_test)))
print("Sample of features and labels:")
# Take a look at the first 25 training features and corresponding labels
for n in range(0,24):
print(x_train[n], y_train[n], '(' + penguin_classes[y_train[n]] + ')')
!pip install --upgrade tensorflow
import tensorflow
from tensorflow import keras
from tensorflow.keras import models
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense
from tensorflow.keras import utils
from tensorflow.keras import optimizers
# Set random seed for reproducability
tensorflow.random.set_seed(0)
print("Libraries imported.")
print('Keras version:',keras.__version__)
print('TensorFlow version:',tensorflow.__version__)
# Set data types for float features
x_train = x_train.astype('float32')
x_test = x_test.astype('float32')
# Set data types for categorical labels
y_train = utils.to_categorical(y_train)
y_test = utils.to_categorical(y_test)
print('Ready...')
# Define a classifier network
hl = 10 # Number of hidden layer nodes
model = Sequential()
model.add(Dense(hl, input_dim=len(features), activation='relu'))
model.add(Dense(hl, input_dim=hl, activation='relu'))
model.add(Dense(len(penguin_classes), input_dim=hl, activation='softmax'))
print(model.summary())
#hyper-parameters for optimizer
learning_rate = 0.001
opt = optimizers.Adam(lr=learning_rate)
model.compile(loss='categorical_crossentropy',
optimizer=opt,
metrics=['accuracy'])
# Train the model over 50 epochs using 10-observation batches and using the test holdout dataset for validation
num_epochs = 50
history = model.fit(x_train, y_train, epochs=num_epochs, batch_size=10, validation_data=(x_test, y_test))
%matplotlib inline
from matplotlib import pyplot as plt
epoch_nums = range(1,num_epochs+1)
training_loss = history.history["loss"]
validation_loss = history.history["val_loss"]
plt.plot(epoch_nums, training_loss)
plt.plot(epoch_nums, validation_loss)
plt.xlabel('epoch')
plt.ylabel('loss')
plt.legend(['training', 'validation'], loc='upper right')
plt.show()
for layer in model.layers:
weights = layer.get_weights()[0]
biases = layer.get_weights()[1]
print('------------\nWeights:\n',weights,'\nBiases:\n', biases)
# Tensorflow doesn't have a built-in confusion matrix metric, so we'll use SciKit-Learn
import numpy as np
from sklearn.metrics import confusion_matrix
import matplotlib.pyplot as plt
%matplotlib inline
class_probabilities = model.predict(x_test)
predictions = np.argmax(class_probabilities, axis=1)
true_labels = np.argmax(y_test, axis=1)
# Plot the confusion matrix
cm = confusion_matrix(true_labels, predictions)
plt.imshow(cm, interpolation="nearest", cmap=plt.cm.Blues)
plt.colorbar()
tick_marks = np.arange(len(penguin_classes))
plt.xticks(tick_marks, penguin_classes, rotation=85)
plt.yticks(tick_marks, penguin_classes)
plt.xlabel("Actual Class")
plt.ylabel("Predicted Class")
plt.show()
# Save the trained model
modelFileName = 'models/penguin-classifier.h5'
model.save(modelFileName)
del model # deletes the existing model variable
print('model saved as', modelFileName)
# Load the saved model
model = models.load_model(modelFileName)
# CReate a new array of features
x_new = np.array([[50.4,15.3,20,50]])
print ('New sample: {}'.format(x_new))
# Use the model to predict the class
class_probabilities = model.predict(x_new)
predictions = np.argmax(class_probabilities, axis=1)
print(penguin_classes[predictions[0]])
| 0.862265 | 0.99488 |
# Datetime variables
Datetime variables take dates and / or time as values. For example, date of birth ('29-08-1987', '12-01-2012'), or date of application ('2016-Dec', '2013-March'). Datetime variables can contain dates only, times only, or dates and time.
We don't use datetime variables straightaway in machine learning because:
- They contain a huge number of different values.
- We can extract much more information from datetime variables by preprocessing them correctly.
Furthermore, date variables frequently include dates that were absent from the dataset used to train the machine learning model. In fact, date variables are frequently used to store dates that are in the future in comparison to the dates in the training dataset. As a result, because they were never seen while being trained, the machine learning model will have no idea what to do with the new values.
=============================================================================
## In this demo: Peer-to-peer lending (Finance)
In this demo, we will use a toy data set that simulates data from a peer-to-peer finance company.
To obtain the data, go to the lecture **Download datasets** in **Section 1**. This is a toy dataset that I created and comes within the Datasets.zip file that you can find in this lecture.
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# Let's load the dataset.
# Variable definitions:
#-------------------------
# disbursed amount: loan amount lent to the borrower
# market: risk band in which borrowers are placed
# date_issued: date the loan was issued
# date_last_payment: date of last payment towards repyaing the loan
data = pd.read_csv('../loan.csv')
data.head()
# Pandas assigns type 'object' to dates
# when loading from csv.
# Let's have a look:
data[['date_issued', 'date_last_payment']].dtypes
```
Both **date_issued** and **date_last_payment** are cast as objects. Therefore, pandas will treat them as strings.
To tell pandas that they are datetime variables, we need to re-cast them into datetime format.
```
# Let's parse the dates into datetime format.
# This will allow us to make some analysis afterwards.
data['date_issued_dt'] = pd.to_datetime(data['date_issued'])
data['date_last_payment_dt'] = pd.to_datetime(data['date_last_payment'])
data[['date_issued', 'date_issued_dt', 'date_last_payment', 'date_last_payment_dt']].head()
# Let's extract the month and the year from the
# datetime variable to make plots.
data['month'] = data['date_issued_dt'].dt.month
data['year'] = data['date_issued_dt'].dt.year
# Let's see how much money has been disbursed
# over the years to the different risk
# markets (grade variable).
fig = data.groupby(['year', 'month', 'market'])['disbursed_amount'].sum().unstack().plot(
figsize=(14, 8), linewidth=2)
fig.set_title('Disbursed amount in time')
fig.set_ylabel('Disbursed Amount')
```
This toy finance company seems to have increased the amount of money lent from 2012 onwards. The tendency indicates that they will continue to grow. In addition, we can see that their major business comes from lending money to C and B-grade borrowers.
A grades are given to the lowest-risk borrowers; these are the borrowers who are most likely to repay their loans because they are in a better financial situation. Borrowers within this grade are charged lower interest rates.
Riskier borrowers represent the D and E grades. Borrowers who are in a slightly tighter financial situation or who do not have enough financial history to make a reliable credit assessment. They are typically charged higher rates, as the business, and therefore the investors, take a higher risk when lending them money.
**That is all for this demonstration. I hope you enjoyed the notebook, and I'll see you in the next one.**
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# Let's load the dataset.
# Variable definitions:
#-------------------------
# disbursed amount: loan amount lent to the borrower
# market: risk band in which borrowers are placed
# date_issued: date the loan was issued
# date_last_payment: date of last payment towards repyaing the loan
data = pd.read_csv('../loan.csv')
data.head()
# Pandas assigns type 'object' to dates
# when loading from csv.
# Let's have a look:
data[['date_issued', 'date_last_payment']].dtypes
# Let's parse the dates into datetime format.
# This will allow us to make some analysis afterwards.
data['date_issued_dt'] = pd.to_datetime(data['date_issued'])
data['date_last_payment_dt'] = pd.to_datetime(data['date_last_payment'])
data[['date_issued', 'date_issued_dt', 'date_last_payment', 'date_last_payment_dt']].head()
# Let's extract the month and the year from the
# datetime variable to make plots.
data['month'] = data['date_issued_dt'].dt.month
data['year'] = data['date_issued_dt'].dt.year
# Let's see how much money has been disbursed
# over the years to the different risk
# markets (grade variable).
fig = data.groupby(['year', 'month', 'market'])['disbursed_amount'].sum().unstack().plot(
figsize=(14, 8), linewidth=2)
fig.set_title('Disbursed amount in time')
fig.set_ylabel('Disbursed Amount')
| 0.300027 | 0.947769 |
```
#Write a Python program to find the sum of all elements in a list using loop.
#Input:- [10,20,30,40]
#Output:- 100
lst = [10,20,30,40]
sum1 = 0
for i in lst:
sum1+= i
print(sum1)
#Write a Python program to find the multiplication of all elements in a list using loop.
#Input:- [10,20,30,40]
#Output:- 240000
lst = [10,20,30,40]
prdt = 1
for i in lst:
prdt *= i
print(prdt)
#Write a Python program to find the largest number from a list using loop.
#Input:- [10,100,2321, 1,200,2]
#Output:- 2321
lst = [10,100,2321, 1,200,2]
max1 = 0
for i in lst:
if i>max1:
max1 =i
print(max1)
#Write a Python program to find the smallest number from a list using loop.
#Input:- [10,100,2321, 1,200,2]
#Output:- 1
lst = [10,100,2321, 1,200,2]
if lst[0] < lst[1]:
min1=lst[0]
else:
min1 = lst[1]
for i in lst:
if i < min1:
min1 = i
print(min1)
#Write a Python program to count the number of strings having length more than 2 and are palindrome in a list using loop.
#Input:- ['ab', 'abc', 'aba', 'xyz', '1991']
#Output:- 2
lst = ['ab', 'abc', 'aba', 'xyz', '1991']
count = 0
for i in lst:
if len(i) > 2 and i == i[::-1]:
count += 1
print(count)
#Write a Python program to sort a list in ascending order using loop.
#Input:- [100,10,1,298,65,483,49876,2,80,9,9213]
#Output:- [1,2,9,10,65,80,100,298,483,9213,49876]
lst = [100,10,1,298,65,483,49876,2,80,9,9213]
for i in range(len(lst)):
for j in range(i+1, len(lst)):
if lst[i] > lst[j]:
temp = lst[j]
lst[j] = lst[i]
lst[i] = temp
print(lst)
#Write a Python program to get a sorted list in increasing order of last element in each tuple in a given list using loop.
#Input:- [(5,4),(9,1),(2,3),(5,9),(7,6),(5,5)]
#output:- [(9,1),(2,3),(5,4),(5,5),(7,6),(5,9)]
lst = [(5,4),(9,1),(2,3),(5,9),(7,6),(5,5)]
def sort_tup(val):
return(val[1])
for i in range(len(lst)):
for j in range(i+1, len(lst)):
if sort_tup(lst[i]) > sort_tup(lst[j]):
temp = lst[j]
lst[j] = lst[i]
lst[i] = temp
print(lst)
#Write a Python program to remove fuplicate element from a list using loop.
#Input:- [10,1,11,1,29,876,768,10,11,1,92,29,876]
#Output:- [10,1,11,29,876,768,92]
lst = [10,1,11,1,29,876,768,10,11,1,92,29,876]
for i in lst:
if lst.count(i) > 1:
count = lst.count(i)
for j in range(count):
lst.remove(i)
count = 0
print(lst1)
#Write a Python program to check a list is empty or not?
#Input:- []
#Output:- List is empty
#Input:- [10,20,30]
#Output:- List is not empty
lst1 = list(input('list: '))
if len(lst1) == 0:
print('list is empty')
else:
print('list is not empty')
#Write a Python program to copy a list using loop.
#inp_lst = [10,10.20,10+20j, 'Python', [10,20], (10,20)]
#out_lst = [10,10.20,10+20j, 'Python', [10,20], (10,20)]
lst = [10,10.20,10+20j, 'Python', [10,20], (10,20)]
out = []
for i in lst:
out.append(i)
print(out)
#Write a Python program to find the list of words that are longer than or equal to 4 from a given string.
#Input:- 'How much wood would a woodchuck chuck if a woodchuck could chuck wood'
#Output:- ['much', 'wood', 'would', 'woodchuck', 'chuck', 'could', 'could']
#Note:- Duplicate should be avoided.
my_str = 'How much wood would a woodchuck chuck if a woodchuck could chuck wood'
lst = []
temp = ''
for i in my_str:
if i == ' ':
if len(temp) >= 4 and temp not in lst:
lst.append(temp)
temp = ''
else:
temp = temp + i
temp = ''
for i in my_str[::-1]: #to check the last word
if i == ' ':
if len(temp) >= 4 and temp not in lst:
lst.append(temp)
temp = ''
break
else:
temp = i + temp
print(lst)
#Write a Python program which takes two list as input and returns True if they have at least 3 common elements.
#inp_lst1 = [10,20,'Python', 10.20, 10+20j, [10,20,30], (10,20,30)]
#inp_lst2 = [(10,20,30),1,20+3j,100.2, 10+20j, [10,20,30],'Python']
#Output:- True
lst1 = [10,20,'Python', 10.20, 10+20j, [10,20,30], (10,20,30)]
lst2 = [(10,20,30),1,20+3j,100.2, 10+20j, [10,20,30],'Python']
count = 0
for i in lst1:
for j in lst2:
if i == j:
count += 1
if count >= 3:
print("True")
#Write a Python program to create a 4X4 2D matrix with below elements using loop and list comprehension both.
#Output:- [[0,0,0,0],[0,1,2,3],[0,2,4,6],[0,3,6,9]]
mat_lst = []
for i in range(4):
mat_lst.append([])
for j in range(4):
mat_lst[i].append(j*i)
print(mat_lst)
#Write a Python program to create a 3X4X6 3D matrix wiith below elements using loop
#Output:-
# [
# [[0,0,0,0,0,0],[0,0,0,0,0,0],[0,0,0,0,0,0],[0,0,0,0,0,0]],
# [[0,0,0,0,0,0],[1,1,1,1,1,1],[2,2,2,2,2,2],[3,3,3,3,3,3]],
# [[0,0,0,0,0,0],[2,2,2,2,2,2],[4,4,4,4,4,4],[6,6,6,6,6,6]]
# ]
mat_lst = []
for i in range(3):
mat_lst.append([])
for j in range(4):
mat_lst[i].append([])
for k in range(6):
mat_lst[i][j].append(i*j)
print(mat_lst)
#Write a Python program which takes a list of numbers as input and prints a new list after removing even numbers from it.
#Input:- [10,21,22,98,87,45,33,1,2,100]
#Output:- [21,87,45,33,1]
lst = [10, 21, 22, 98, 87, 45, 33, 1, 2, 100]
lst1 = []
for i in lst:
if i%2 != 0:
lst1.append(i)
print(lst1)
#Write a Python program which takes a list from the user and prints it after reshuffling the elements of the list.
#Input:- [10,21,22,98,87,45,33,1,2,100]
#Output:- [1,87,21,10,33,2,100,45,98,22] (It may be any randon list but with same elements)
lst = [10,21,22,98,87,45,33,1,2,100]
lst1 = lst
```
## Completed
|
github_jupyter
|
#Write a Python program to find the sum of all elements in a list using loop.
#Input:- [10,20,30,40]
#Output:- 100
lst = [10,20,30,40]
sum1 = 0
for i in lst:
sum1+= i
print(sum1)
#Write a Python program to find the multiplication of all elements in a list using loop.
#Input:- [10,20,30,40]
#Output:- 240000
lst = [10,20,30,40]
prdt = 1
for i in lst:
prdt *= i
print(prdt)
#Write a Python program to find the largest number from a list using loop.
#Input:- [10,100,2321, 1,200,2]
#Output:- 2321
lst = [10,100,2321, 1,200,2]
max1 = 0
for i in lst:
if i>max1:
max1 =i
print(max1)
#Write a Python program to find the smallest number from a list using loop.
#Input:- [10,100,2321, 1,200,2]
#Output:- 1
lst = [10,100,2321, 1,200,2]
if lst[0] < lst[1]:
min1=lst[0]
else:
min1 = lst[1]
for i in lst:
if i < min1:
min1 = i
print(min1)
#Write a Python program to count the number of strings having length more than 2 and are palindrome in a list using loop.
#Input:- ['ab', 'abc', 'aba', 'xyz', '1991']
#Output:- 2
lst = ['ab', 'abc', 'aba', 'xyz', '1991']
count = 0
for i in lst:
if len(i) > 2 and i == i[::-1]:
count += 1
print(count)
#Write a Python program to sort a list in ascending order using loop.
#Input:- [100,10,1,298,65,483,49876,2,80,9,9213]
#Output:- [1,2,9,10,65,80,100,298,483,9213,49876]
lst = [100,10,1,298,65,483,49876,2,80,9,9213]
for i in range(len(lst)):
for j in range(i+1, len(lst)):
if lst[i] > lst[j]:
temp = lst[j]
lst[j] = lst[i]
lst[i] = temp
print(lst)
#Write a Python program to get a sorted list in increasing order of last element in each tuple in a given list using loop.
#Input:- [(5,4),(9,1),(2,3),(5,9),(7,6),(5,5)]
#output:- [(9,1),(2,3),(5,4),(5,5),(7,6),(5,9)]
lst = [(5,4),(9,1),(2,3),(5,9),(7,6),(5,5)]
def sort_tup(val):
return(val[1])
for i in range(len(lst)):
for j in range(i+1, len(lst)):
if sort_tup(lst[i]) > sort_tup(lst[j]):
temp = lst[j]
lst[j] = lst[i]
lst[i] = temp
print(lst)
#Write a Python program to remove fuplicate element from a list using loop.
#Input:- [10,1,11,1,29,876,768,10,11,1,92,29,876]
#Output:- [10,1,11,29,876,768,92]
lst = [10,1,11,1,29,876,768,10,11,1,92,29,876]
for i in lst:
if lst.count(i) > 1:
count = lst.count(i)
for j in range(count):
lst.remove(i)
count = 0
print(lst1)
#Write a Python program to check a list is empty or not?
#Input:- []
#Output:- List is empty
#Input:- [10,20,30]
#Output:- List is not empty
lst1 = list(input('list: '))
if len(lst1) == 0:
print('list is empty')
else:
print('list is not empty')
#Write a Python program to copy a list using loop.
#inp_lst = [10,10.20,10+20j, 'Python', [10,20], (10,20)]
#out_lst = [10,10.20,10+20j, 'Python', [10,20], (10,20)]
lst = [10,10.20,10+20j, 'Python', [10,20], (10,20)]
out = []
for i in lst:
out.append(i)
print(out)
#Write a Python program to find the list of words that are longer than or equal to 4 from a given string.
#Input:- 'How much wood would a woodchuck chuck if a woodchuck could chuck wood'
#Output:- ['much', 'wood', 'would', 'woodchuck', 'chuck', 'could', 'could']
#Note:- Duplicate should be avoided.
my_str = 'How much wood would a woodchuck chuck if a woodchuck could chuck wood'
lst = []
temp = ''
for i in my_str:
if i == ' ':
if len(temp) >= 4 and temp not in lst:
lst.append(temp)
temp = ''
else:
temp = temp + i
temp = ''
for i in my_str[::-1]: #to check the last word
if i == ' ':
if len(temp) >= 4 and temp not in lst:
lst.append(temp)
temp = ''
break
else:
temp = i + temp
print(lst)
#Write a Python program which takes two list as input and returns True if they have at least 3 common elements.
#inp_lst1 = [10,20,'Python', 10.20, 10+20j, [10,20,30], (10,20,30)]
#inp_lst2 = [(10,20,30),1,20+3j,100.2, 10+20j, [10,20,30],'Python']
#Output:- True
lst1 = [10,20,'Python', 10.20, 10+20j, [10,20,30], (10,20,30)]
lst2 = [(10,20,30),1,20+3j,100.2, 10+20j, [10,20,30],'Python']
count = 0
for i in lst1:
for j in lst2:
if i == j:
count += 1
if count >= 3:
print("True")
#Write a Python program to create a 4X4 2D matrix with below elements using loop and list comprehension both.
#Output:- [[0,0,0,0],[0,1,2,3],[0,2,4,6],[0,3,6,9]]
mat_lst = []
for i in range(4):
mat_lst.append([])
for j in range(4):
mat_lst[i].append(j*i)
print(mat_lst)
#Write a Python program to create a 3X4X6 3D matrix wiith below elements using loop
#Output:-
# [
# [[0,0,0,0,0,0],[0,0,0,0,0,0],[0,0,0,0,0,0],[0,0,0,0,0,0]],
# [[0,0,0,0,0,0],[1,1,1,1,1,1],[2,2,2,2,2,2],[3,3,3,3,3,3]],
# [[0,0,0,0,0,0],[2,2,2,2,2,2],[4,4,4,4,4,4],[6,6,6,6,6,6]]
# ]
mat_lst = []
for i in range(3):
mat_lst.append([])
for j in range(4):
mat_lst[i].append([])
for k in range(6):
mat_lst[i][j].append(i*j)
print(mat_lst)
#Write a Python program which takes a list of numbers as input and prints a new list after removing even numbers from it.
#Input:- [10,21,22,98,87,45,33,1,2,100]
#Output:- [21,87,45,33,1]
lst = [10, 21, 22, 98, 87, 45, 33, 1, 2, 100]
lst1 = []
for i in lst:
if i%2 != 0:
lst1.append(i)
print(lst1)
#Write a Python program which takes a list from the user and prints it after reshuffling the elements of the list.
#Input:- [10,21,22,98,87,45,33,1,2,100]
#Output:- [1,87,21,10,33,2,100,45,98,22] (It may be any randon list but with same elements)
lst = [10,21,22,98,87,45,33,1,2,100]
lst1 = lst
| 0.229018 | 0.521288 |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
df_reviews = pd.read_csv('reviews.csv')
print("The dataset has {} rows and {} columns.".format(*df_reviews.shape))
print("It contains {} duplicates.".format(df_reviews.duplicated().sum()))
df_reviews.head()
df_listings = pd.read_csv('listings.csv')
df_listings.head()
df = pd.merge(df_reviews, df_listings[['latitude',
'longitude', 'number_of_reviews', 'id', 'property_type']],
left_on='listing_id', right_on='id', how='left')
df.rename(columns = {'id_x':'id'}, inplace=True)
df.drop(['id_y'], axis=1, inplace=True)
df.head()
df.isna().sum()
df.dropna(inplace=True)
from cld2 import detect
def language_detection(text):
try:
return detect(text).details[0][1]
except:
return None
df['language'] = df['comments'].apply(language_detection)
df.head()
df.language.value_counts().head(10)
ax = df.language.value_counts().head(6).plot(kind='barh', figsize=(9,5), color="lightcoral",
fontsize=12);
ax.set_title("\nWhat are the most frequent languages comments are written in?\n",
fontsize=12, fontweight='bold')
ax.set_xlabel(" Total Number of Comments", fontsize=10)
ax.set_yticklabels(['English', 'French', 'Spanish', 'Undefined', 'Italian', 'German'])
# create a list to collect the plt.patches data
totals = []
# find the ind. values and append to list
for i in ax.patches:
totals.append(i.get_width())
# get total
total = sum(totals)
# set individual bar labels using above list
for i in ax.patches:
ax.text(x=i.get_width(), y=i.get_y()+.35,
s=str(round((i.get_width()/total)*100, 2))+'%',
fontsize=10, color='black')
# invert for largest on top
ax.invert_yaxis()
df_eng = df[(df['language']=='en')]
df_eng['comments'] = df_eng['comments'].str.replace("[^a-zA-Z#]", " ")
from nltk.corpus import stopwords
stop_words = stopwords.words('english')
negations = ['no', 'not', "isn't", "haven't", "didn't", "hasn't", "never", "won't", "couldn't"]
stop_words = list(set(stop_words) - set(negations))
# function to remove stopwords
def remove_stopwords(rev):
rev_new = " ".join([i for i in rev if i not in stop_words])
return rev_new
# remove short words (length < 3)
df_eng['comments'] = df_eng['comments'].apply(lambda x: ' '.join([w for w in x.split() if len(w)>2]))
# remove stopwords from the text
reviews = [remove_stopwords(r.split()) for r in df_eng['comments']]
# make entire text lowercase
reviews = [r.lower() for r in reviews]
import spacy
nlp = spacy.load('en', disable=['parser', 'ner'])
def lemmatization(texts, negations, tags=['NOUN', 'ADJ']): # filter noun and adjective
output = []
for sent in texts:
doc = nlp(" ".join(sent))
output.append([token.lemma_ for token in doc if token.pos_ in tags or token.lemma_ in negations])
return output
reviews[1]
tokenized_reviews = pd.Series(reviews).apply(lambda x: x.split())
print(tokenized_reviews[1])
reviews_2 = lemmatization(tokenized_reviews, negations)
reviews_3 = []
for i in range(len(reviews_2)):
reviews_3.append(' '.join(reviews_2[i]))
df_eng['comments'] = reviews_3
for i in range(len(negations)):
word = negations[i]
negations[i] = ' ' + word + ' '
for word in negations:
df_eng = df_eng[~df_eng['comments'].str.contains(word, na=False)]
len(df_eng)
del df_eng['language']
df_eng.to_csv(r'processed_reviews.csv')
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
df_reviews = pd.read_csv('reviews.csv')
print("The dataset has {} rows and {} columns.".format(*df_reviews.shape))
print("It contains {} duplicates.".format(df_reviews.duplicated().sum()))
df_reviews.head()
df_listings = pd.read_csv('listings.csv')
df_listings.head()
df = pd.merge(df_reviews, df_listings[['latitude',
'longitude', 'number_of_reviews', 'id', 'property_type']],
left_on='listing_id', right_on='id', how='left')
df.rename(columns = {'id_x':'id'}, inplace=True)
df.drop(['id_y'], axis=1, inplace=True)
df.head()
df.isna().sum()
df.dropna(inplace=True)
from cld2 import detect
def language_detection(text):
try:
return detect(text).details[0][1]
except:
return None
df['language'] = df['comments'].apply(language_detection)
df.head()
df.language.value_counts().head(10)
ax = df.language.value_counts().head(6).plot(kind='barh', figsize=(9,5), color="lightcoral",
fontsize=12);
ax.set_title("\nWhat are the most frequent languages comments are written in?\n",
fontsize=12, fontweight='bold')
ax.set_xlabel(" Total Number of Comments", fontsize=10)
ax.set_yticklabels(['English', 'French', 'Spanish', 'Undefined', 'Italian', 'German'])
# create a list to collect the plt.patches data
totals = []
# find the ind. values and append to list
for i in ax.patches:
totals.append(i.get_width())
# get total
total = sum(totals)
# set individual bar labels using above list
for i in ax.patches:
ax.text(x=i.get_width(), y=i.get_y()+.35,
s=str(round((i.get_width()/total)*100, 2))+'%',
fontsize=10, color='black')
# invert for largest on top
ax.invert_yaxis()
df_eng = df[(df['language']=='en')]
df_eng['comments'] = df_eng['comments'].str.replace("[^a-zA-Z#]", " ")
from nltk.corpus import stopwords
stop_words = stopwords.words('english')
negations = ['no', 'not', "isn't", "haven't", "didn't", "hasn't", "never", "won't", "couldn't"]
stop_words = list(set(stop_words) - set(negations))
# function to remove stopwords
def remove_stopwords(rev):
rev_new = " ".join([i for i in rev if i not in stop_words])
return rev_new
# remove short words (length < 3)
df_eng['comments'] = df_eng['comments'].apply(lambda x: ' '.join([w for w in x.split() if len(w)>2]))
# remove stopwords from the text
reviews = [remove_stopwords(r.split()) for r in df_eng['comments']]
# make entire text lowercase
reviews = [r.lower() for r in reviews]
import spacy
nlp = spacy.load('en', disable=['parser', 'ner'])
def lemmatization(texts, negations, tags=['NOUN', 'ADJ']): # filter noun and adjective
output = []
for sent in texts:
doc = nlp(" ".join(sent))
output.append([token.lemma_ for token in doc if token.pos_ in tags or token.lemma_ in negations])
return output
reviews[1]
tokenized_reviews = pd.Series(reviews).apply(lambda x: x.split())
print(tokenized_reviews[1])
reviews_2 = lemmatization(tokenized_reviews, negations)
reviews_3 = []
for i in range(len(reviews_2)):
reviews_3.append(' '.join(reviews_2[i]))
df_eng['comments'] = reviews_3
for i in range(len(negations)):
word = negations[i]
negations[i] = ' ' + word + ' '
for word in negations:
df_eng = df_eng[~df_eng['comments'].str.contains(word, na=False)]
len(df_eng)
del df_eng['language']
df_eng.to_csv(r'processed_reviews.csv')
| 0.189296 | 0.371707 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.