code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
# ShopUp team
In this notebook our team reveal two different approaches for creating recommender system in Retail (ecommerce) during the Summer Data Science School using:
- Deep Collaborative Filtering
- Session Based Recommendations with RNN
You can find more information about the case the other approaches and all other into at our article at [Shopup](https://shopup.me) and the repo for Learning Item Embedding which we call product2Vec.
https://shopup.me/recommenders_systems/
```
from fastai.collab import *
from fastai.tabular import *
from fastai.text import *
import numpy as np
import pandas as pd
from google.colab import drive
drive.mount('/content/gdrive', force_remount=True)
root_dir = "/content/gdrive/My Drive/Colab Notebooks/"
base_dir = root_dir + 'summer_school/data/'
```
# 1. Data Prep
### Load data
```
sorted_df = pd.read_csv(base_dir +'/events.csv')
sorted_df["date"] = pd.DatetimeIndex(sorted_df["timestamp"]).date
sorted_df.head()
# adding ratings if they browse is one if they add to card is 3 and 5 for transaction
sorted_df["rating1"] = np.where(sorted_df.event == "view",1,3)
sorted_df["rating2"] = np.where((sorted_df.event == "transaction") & (sorted_df["rating1"] == 3),2,0)
sorted_df['rating'] = sorted_df["rating1"] + sorted_df["rating2"]
sorted_df.head()
```
## Deep collaborative filtering example
`collab` models use data in a `DataFrame` of user, items, and ratings.
```
ratings = sorted_df[["visitorid", 'itemid', "rating", 'timestamp']].copy()
ratings.head()
```
That's all we need to create and train a model:
```
data = CollabDataBunch.from_df(ratings, seed=42)
data
y_range = [0,5.5]
learn = collab_learner(data, n_factors=50, y_range=y_range)
learn.lr_find()
learn.recorder.plot(skip_end=15)
learn.fit_one_cycle(1, 1e-2)
# save the model
learn.save(base_dir +'/dotcat')
```
## Session base recommendation with RNNs
```
# need to prepare the data in the format shown below
final_text= pd.read_csv(base_dir +'/final_text_no_bracket.csv')
final_text.head()
from fastai.text import *
data_lm = (TextList.from_df(final_text, cols=['visitorid', 'new'])
.split_by_rand_pct()
.label_for_lm()
.databunch())
data_lm.save(base_dir +'/data_lm.pkl')
#data_lm.load(base_dir +'/data_lm.pkl')
data_lm.vocab.itos[:11]
data_lm.show_batch()
bs=48
learn = language_model_learner(data_lm, AWD_LSTM, drop_mult=0.3)
learn.lr_find()
learn.recorder.plot(skip_end=15)
learn.fit_one_cycle(2, moms=(0.8,0.7))
learn.save(base_dir +'fit_head')
learn.save(base_dir +'/fine_tuned')
```
###Let's play
```
#learn.load(base_dir +'/fine_tuned_1')
TEXT = "440866"
N_WORDS = 4
N_SENTENCES = 1
print("\n".join(learn.predict(TEXT, N_WORDS +1 , temperature=0.75) for _ in range(N_SENTENCES)))
```
|
github_jupyter
|
from fastai.collab import *
from fastai.tabular import *
from fastai.text import *
import numpy as np
import pandas as pd
from google.colab import drive
drive.mount('/content/gdrive', force_remount=True)
root_dir = "/content/gdrive/My Drive/Colab Notebooks/"
base_dir = root_dir + 'summer_school/data/'
sorted_df = pd.read_csv(base_dir +'/events.csv')
sorted_df["date"] = pd.DatetimeIndex(sorted_df["timestamp"]).date
sorted_df.head()
# adding ratings if they browse is one if they add to card is 3 and 5 for transaction
sorted_df["rating1"] = np.where(sorted_df.event == "view",1,3)
sorted_df["rating2"] = np.where((sorted_df.event == "transaction") & (sorted_df["rating1"] == 3),2,0)
sorted_df['rating'] = sorted_df["rating1"] + sorted_df["rating2"]
sorted_df.head()
ratings = sorted_df[["visitorid", 'itemid', "rating", 'timestamp']].copy()
ratings.head()
data = CollabDataBunch.from_df(ratings, seed=42)
data
y_range = [0,5.5]
learn = collab_learner(data, n_factors=50, y_range=y_range)
learn.lr_find()
learn.recorder.plot(skip_end=15)
learn.fit_one_cycle(1, 1e-2)
# save the model
learn.save(base_dir +'/dotcat')
# need to prepare the data in the format shown below
final_text= pd.read_csv(base_dir +'/final_text_no_bracket.csv')
final_text.head()
from fastai.text import *
data_lm = (TextList.from_df(final_text, cols=['visitorid', 'new'])
.split_by_rand_pct()
.label_for_lm()
.databunch())
data_lm.save(base_dir +'/data_lm.pkl')
#data_lm.load(base_dir +'/data_lm.pkl')
data_lm.vocab.itos[:11]
data_lm.show_batch()
bs=48
learn = language_model_learner(data_lm, AWD_LSTM, drop_mult=0.3)
learn.lr_find()
learn.recorder.plot(skip_end=15)
learn.fit_one_cycle(2, moms=(0.8,0.7))
learn.save(base_dir +'fit_head')
learn.save(base_dir +'/fine_tuned')
#learn.load(base_dir +'/fine_tuned_1')
TEXT = "440866"
N_WORDS = 4
N_SENTENCES = 1
print("\n".join(learn.predict(TEXT, N_WORDS +1 , temperature=0.75) for _ in range(N_SENTENCES)))
| 0.403332 | 0.899033 |
This notebook compares the output of XGPaint.jl with expected, using Python and healpy.
```
# %config InlineBackend.figure_format = 'retina'
import numpy as np
import matplotlib.pyplot as plt
import healpy as hp
import astropy.units as u
from astropy.cosmology import WMAP9 as cosmo
from astropy.cosmology import z_at_value
def bin_ps(y, nb=1000):
x = np.arange(len(y))
bins = np.arange(0, len(y), nb)
inds = np.digitize(x, bins)
bx = np.array([ np.mean(x[inds==i]) for i in range(2,np.max(inds))])
by = np.array([ np.mean(y[inds==i]) for i in range(2,np.max(inds))])
return bx, by
map_fac = 1
freq_str = ['030', '090', '148', '219', '277', '350']
freqs = [float(f) for f in freq_str]
# conv_freq = [ 0.03701396034733168,
# 0.004683509698460815,
# 0.002595203002861245,
# 0.0020683915234002374,
# 0.002331542380044927,
# 0.0033735887524933045]
def clip_map(m):
# return m
m2 = m.copy()
# m2[m < 1e2] = 0.0
m2[m > 1e6] = 0.0
return m2
sehgal_maps = [hp.read_map(
f'/tigress/zequnl/xgpaint/sehgal/{freq}_rad_pts_healpix.fits', verbose=False)
for freq in freq_str]
sehgal_ps_autos = [hp.anafast(clip_map(m1), iter=0) for m1 in sehgal_maps]
sehgal_cib_150 = hp.read_map(
f'/tigress/zequnl/xgpaint/sehgal/{freq_str[2]}_ir_pts_healpix.fits', verbose=False)
sehgal_cib_150 = hp.alm2map(hp.map2alm(sehgal_cib_150), nside=4096)
hp.write_map(f'/tigress/zequnl/xgpaint/sehgal/{freq_str[2]}_ir_pts_healpix.fits', sehgal_cib_150)
sehgal_cib_ps = hp.anafast(sehgal_cib_150, iter=0)
```
# XGPaint Sims
```
radio_maps = [hp.read_map(f'/tigress/zequnl/xgpaint/jl/radio{freq_str[i]}.fits', verbose=False)
for i in range(len(freqs))]
cib_map_150 = hp.read_map(f'/tigress/zequnl/xgpaint/jl/cib{freq_str[2]}.fits', verbose=False) * 9e12
websky_cib_150 = hp.anafast(cib_map_150, iter=0)
ps3s = hp.anafast(clip_map(sehgal_maps[2]), map2=sehgal_cib_150, iter=0)
ps3 = hp.anafast(clip_map(radio_maps[2]), map2=cib_map_150, iter=0)
bx, by = bin_ps(ps3s)
plt.plot(bx, by, label='sehgal')
bx, by = bin_ps(ps3)
plt.plot(bx, by, label='websky')
# plt.yscale('log')
plt.legend()
plt.title(r'150 GHz CIB $\times$ Radio')
bx, by = bin_ps(hp.anafast(clip_map(sehgal_maps[2]), iter=0))
plt.plot(bx, by, label='sehgal')
bx, by = bin_ps(hp.anafast(clip_map(radio_maps[2]), iter=0))
plt.plot(bx, by, label='websky')
# plt.yscale('log')
plt.legend()
plt.title(r'150 GHz Radio $\times$ Radio')
bx, by = bin_ps(hp.anafast(sehgal_cib_150, iter=0))
plt.plot(bx, by, label='sehgal')
bx, by = bin_ps(hp.anafast(cib_map_150, iter=0))
plt.plot(bx, by, label='websky')
plt.yscale('log')
plt.legend()
plt.title(r'150 GHz CIB $\times$ CIB')
plt.figure(figsize=(12, 5))
plt.hist( np.log10(sehgal_maps[2]+1), histtype="step", label="sehgal", bins=50)
plt.hist( np.log10(maps[2]+1), histtype="step", label="zack", bins=50)
plt.yscale("log")
plt.legend()
plt.xlabel(r"$\log_{10}$ pixel flux in Jy/sr (150 GHz)")
fig, axes = plt.subplots(len(freq_str),1,figsize=(5,12))
for i in range(len(freq_str)):
axes[i].hist( np.log10(sehgal_maps[i] + 1e0), bins=30, histtype="step")
axes[i].hist( np.log10(maps[i] + 1e0), bins=30, histtype="step")
axes[i].set_yscale("log")
%time ps_auto = [hp.anafast(clip_map(m1), iter=0) for m1 in maps]
for i in range(len(ps_auto)):
plt.figure()
plt.title(freq_str[i])
plt.plot(sehgal_ps_autos[i][100:])
plt.plot(ps_auto[i][100:], alpha=0.1)
plt.scatter( freqs, [np.mean(ps[500:]) for ps in ps_auto], label='websky' )
plt.scatter( freqs, [np.mean(ps[500:]) for ps in sehgal_ps_autos], marker='X', label='sehgal', lw=0.5, s=50, color='r', alpha=0.5 )
plt.legend()
plt.xlabel('GHz')
plt.ylabel(r'$\langle C_{\ell} \rangle_{\ell}$ for $\ell > 500$')
plt.yscale('log')
np.array([np.mean(ps[500:]) for ps in ps_auto]) / np.array([np.mean(ps[500:]) for ps in sehgal_ps_autos])
```
|
github_jupyter
|
# %config InlineBackend.figure_format = 'retina'
import numpy as np
import matplotlib.pyplot as plt
import healpy as hp
import astropy.units as u
from astropy.cosmology import WMAP9 as cosmo
from astropy.cosmology import z_at_value
def bin_ps(y, nb=1000):
x = np.arange(len(y))
bins = np.arange(0, len(y), nb)
inds = np.digitize(x, bins)
bx = np.array([ np.mean(x[inds==i]) for i in range(2,np.max(inds))])
by = np.array([ np.mean(y[inds==i]) for i in range(2,np.max(inds))])
return bx, by
map_fac = 1
freq_str = ['030', '090', '148', '219', '277', '350']
freqs = [float(f) for f in freq_str]
# conv_freq = [ 0.03701396034733168,
# 0.004683509698460815,
# 0.002595203002861245,
# 0.0020683915234002374,
# 0.002331542380044927,
# 0.0033735887524933045]
def clip_map(m):
# return m
m2 = m.copy()
# m2[m < 1e2] = 0.0
m2[m > 1e6] = 0.0
return m2
sehgal_maps = [hp.read_map(
f'/tigress/zequnl/xgpaint/sehgal/{freq}_rad_pts_healpix.fits', verbose=False)
for freq in freq_str]
sehgal_ps_autos = [hp.anafast(clip_map(m1), iter=0) for m1 in sehgal_maps]
sehgal_cib_150 = hp.read_map(
f'/tigress/zequnl/xgpaint/sehgal/{freq_str[2]}_ir_pts_healpix.fits', verbose=False)
sehgal_cib_150 = hp.alm2map(hp.map2alm(sehgal_cib_150), nside=4096)
hp.write_map(f'/tigress/zequnl/xgpaint/sehgal/{freq_str[2]}_ir_pts_healpix.fits', sehgal_cib_150)
sehgal_cib_ps = hp.anafast(sehgal_cib_150, iter=0)
radio_maps = [hp.read_map(f'/tigress/zequnl/xgpaint/jl/radio{freq_str[i]}.fits', verbose=False)
for i in range(len(freqs))]
cib_map_150 = hp.read_map(f'/tigress/zequnl/xgpaint/jl/cib{freq_str[2]}.fits', verbose=False) * 9e12
websky_cib_150 = hp.anafast(cib_map_150, iter=0)
ps3s = hp.anafast(clip_map(sehgal_maps[2]), map2=sehgal_cib_150, iter=0)
ps3 = hp.anafast(clip_map(radio_maps[2]), map2=cib_map_150, iter=0)
bx, by = bin_ps(ps3s)
plt.plot(bx, by, label='sehgal')
bx, by = bin_ps(ps3)
plt.plot(bx, by, label='websky')
# plt.yscale('log')
plt.legend()
plt.title(r'150 GHz CIB $\times$ Radio')
bx, by = bin_ps(hp.anafast(clip_map(sehgal_maps[2]), iter=0))
plt.plot(bx, by, label='sehgal')
bx, by = bin_ps(hp.anafast(clip_map(radio_maps[2]), iter=0))
plt.plot(bx, by, label='websky')
# plt.yscale('log')
plt.legend()
plt.title(r'150 GHz Radio $\times$ Radio')
bx, by = bin_ps(hp.anafast(sehgal_cib_150, iter=0))
plt.plot(bx, by, label='sehgal')
bx, by = bin_ps(hp.anafast(cib_map_150, iter=0))
plt.plot(bx, by, label='websky')
plt.yscale('log')
plt.legend()
plt.title(r'150 GHz CIB $\times$ CIB')
plt.figure(figsize=(12, 5))
plt.hist( np.log10(sehgal_maps[2]+1), histtype="step", label="sehgal", bins=50)
plt.hist( np.log10(maps[2]+1), histtype="step", label="zack", bins=50)
plt.yscale("log")
plt.legend()
plt.xlabel(r"$\log_{10}$ pixel flux in Jy/sr (150 GHz)")
fig, axes = plt.subplots(len(freq_str),1,figsize=(5,12))
for i in range(len(freq_str)):
axes[i].hist( np.log10(sehgal_maps[i] + 1e0), bins=30, histtype="step")
axes[i].hist( np.log10(maps[i] + 1e0), bins=30, histtype="step")
axes[i].set_yscale("log")
%time ps_auto = [hp.anafast(clip_map(m1), iter=0) for m1 in maps]
for i in range(len(ps_auto)):
plt.figure()
plt.title(freq_str[i])
plt.plot(sehgal_ps_autos[i][100:])
plt.plot(ps_auto[i][100:], alpha=0.1)
plt.scatter( freqs, [np.mean(ps[500:]) for ps in ps_auto], label='websky' )
plt.scatter( freqs, [np.mean(ps[500:]) for ps in sehgal_ps_autos], marker='X', label='sehgal', lw=0.5, s=50, color='r', alpha=0.5 )
plt.legend()
plt.xlabel('GHz')
plt.ylabel(r'$\langle C_{\ell} \rangle_{\ell}$ for $\ell > 500$')
plt.yscale('log')
np.array([np.mean(ps[500:]) for ps in ps_auto]) / np.array([np.mean(ps[500:]) for ps in sehgal_ps_autos])
| 0.336767 | 0.867036 |
```
# Load libraries
%matplotlib inline
import numpy
import matplotlib.pyplot as plt
from numpy import arange
from matplotlib import pyplot
from pandas import read_csv
from pandas import set_option
from pandas.tools.plotting import scatter_matrix
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import GridSearchCV
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Lasso
from sklearn.linear_model import ElasticNet
from sklearn.tree import DecisionTreeRegressor
from sklearn.neighbors import KNeighborsRegressor
from sklearn.svm import SVR
from sklearn.pipeline import Pipeline
from sklearn.ensemble import RandomForestRegressor
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.ensemble import ExtraTreesRegressor
from sklearn.ensemble import AdaBoostRegressor
from sklearn.metrics import mean_squared_error
# Load dataset
filename = 'housing.csv'
names = ['CRIM', 'ZN', 'INDUS', 'CHAS', 'NOX', 'RM', 'AGE', 'DIS', 'RAD', 'TAX', 'PTRATIO', 'B', 'LSTAT', 'MEDV']
dataset = read_csv(filename, delim_whitespace=True, names=names)
# shape
print(dataset.shape)
# types
print(dataset.dtypes)
# head
print(dataset.head(20))
set_option('precision', 2)
print(dataset.describe())
# correlation
set_option('precision', 2)
print(dataset.corr(method='pearson'))
# histograms
dataset.hist(sharex=False, sharey=False, xlabelsize=1, ylabelsize=1, figsize=(14, 12))
plt.show()
# density
dataset.plot.density(subplots=True, layout=(4,4), sharex=False, legend=False, fontsize=1, figsize=(14, 12))
plt.show()
# box and whisker plots
dataset.plot.box(subplots=True, layout=(4,4), sharex=False, sharey=False, figsize=(14, 12))
plt.show()
# scatter plot matrix
scatter_matrix(dataset, figsize=(14, 12))
plt.show()
# correlation matrix
fig = plt.figure(figsize=(14, 12))
ax = fig.add_subplot(111)
cax = ax.matshow(dataset.corr(), vmin=-1, vmax=1, interpolation='none')
fig.colorbar(cax)
ticks = arange(0,14,1)
ax.set_xticks(ticks)
ax.set_yticks(ticks)
ax.set_xticklabels(names)
ax.set_yticklabels(names)
plt.show()
# Split-out validation dataset
array = dataset.values
X = array[:,0:13]
Y = array[:,13]
validation_size = 0.20
seed = 7
X_train, X_validation, Y_train, Y_validation = train_test_split(X, Y, test_size=validation_size, random_state=seed)
# Test options and evaluation metric
num_folds = 10
seed = 7
scoring = 'neg_mean_squared_error'
# Spot-Check Algorithms
models = []
models.append(('LR', LinearRegression()))
models.append(('LASSO', Lasso()))
models.append(('EN', ElasticNet()))
models.append(('KNN', KNeighborsRegressor()))
models.append(('CART', DecisionTreeRegressor()))
models.append(('SVR' , SVR()))
# evaluate each model in turn
results = []
names = []
for name, model in models:
kfold = KFold(n_splits=num_folds, random_state=seed)
cv_results = cross_val_score(model, X_train, Y_train, cv=kfold, scoring=scoring)
results.append(cv_results)
names.append(name)
msg = "%s: %f (%f)" % (name, cv_results.mean(), cv_results.std())
print(msg)
# Compare Algorithms
fig = pyplot.figure(figsize=(14, 12))
fig.suptitle('Algorithm Comparison')
ax = fig.add_subplot(111)
pyplot.boxplot(results)
ax.set_xticklabels(names)
plt.show()
# Standardize the dataset
pipelines = []
pipelines.append(('ScaledLR', Pipeline([('Scaler', StandardScaler()),('LR', LinearRegression())])))
pipelines.append(('ScaledLASSO', Pipeline([('Scaler', StandardScaler()),('LASSO', Lasso())])))
pipelines.append(('ScaledEN', Pipeline([('Scaler', StandardScaler()),('EN', ElasticNet())])))
pipelines.append(('ScaledKNN', Pipeline([('Scaler', StandardScaler()),('KNN', KNeighborsRegressor())])))
pipelines.append(('ScaledCART', Pipeline([('Scaler', StandardScaler()),('CART', DecisionTreeRegressor())])))
pipelines.append(('ScaledSVR', Pipeline([('Scaler', StandardScaler()),('SVR', SVR())])))
results = []
names = []
for name, model in pipelines:
kfold = KFold(n_splits=num_folds, random_state=seed)
cv_results = cross_val_score(model, X_train, Y_train, cv=kfold, scoring=scoring)
results.append(cv_results)
names.append(name)
msg = "%s: %f (%f)" % (name, cv_results.mean(), cv_results.std())
print(msg)
```
Scaled KNN now has lowest MSE.
```
# Compare Algorithms
fig = pyplot.figure(figsize=(14, 12))
fig.suptitle('Scaled Algorithm Comparison')
ax = fig.add_subplot(111)
pyplot.boxplot(results)
ax.set_xticklabels(names)
plt.show()
# KNN Algorithm tuning
scaler = StandardScaler().fit(X_train)
rescaledX = scaler.transform(X_train)
k_values = numpy.array([1,3,5,7,9,11,13,15,17,19,21])
param_grid = dict(n_neighbors=k_values)
model = KNeighborsRegressor()
kfold = KFold(n_splits=num_folds, random_state=seed)
grid = GridSearchCV(estimator=model, param_grid=param_grid, scoring=scoring, cv=kfold)
grid_result = grid.fit(rescaledX, Y_train)
print("Best: %f using %s" % (grid_result.best_score_, grid_result.best_params_))
means = grid_result.cv_results_['mean_test_score']
stds = grid_result.cv_results_['std_test_score']
params = grid_result.cv_results_['params']
for mean, stdev, param in zip(means, stds, params):
print("%f (%f) with: %r" % (mean, stdev, param))
```
You can see that the best for k (n neighbors) is 3 providing a mean squared error of
-18.172137, the best so far.
### Ensemble Methods
```
# ensembles
ensembles = []
ensembles.append(('ScaledAB', Pipeline([('Scaler', StandardScaler()),('AB', AdaBoostRegressor())])))
ensembles.append(('ScaledGBM', Pipeline([('Scaler', StandardScaler()),('GBM', GradientBoostingRegressor())])))
ensembles.append(('ScaledRF', Pipeline([('Scaler', StandardScaler()),('RF', RandomForestRegressor())])))
ensembles.append(('ScaledET', Pipeline([('Scaler', StandardScaler()),('ET', ExtraTreesRegressor())])))
results = []
names = []
for name, model in ensembles:
kfold = KFold(n_splits=num_folds, random_state=seed)
cv_results = cross_val_score(model, X_train, Y_train, cv=kfold, scoring=scoring)
results.append(cv_results)
names.append(name)
msg = "%s: %f (%f)" % (name, cv_results.mean(), cv_results.std())
print(msg)
# Compare Algorithms
fig = pyplot.figure(figsize=(14, 12))
fig.suptitle('Scaled Ensemble Algorithm Comparison')
ax = fig.add_subplot(111)
pyplot.boxplot(results)
ax.set_xticklabels(names)
plt.show()
# Tune scaled GBM
scaler = StandardScaler().fit(X_train)
rescaledX = scaler.transform(X_train)
param_grid = dict(n_estimators=numpy.array([50,100,150,200,250,300,350,400]))
model = GradientBoostingRegressor(random_state=seed)
kfold = KFold(n_splits=num_folds, random_state=seed)
grid = GridSearchCV(estimator=model, param_grid=param_grid, scoring=scoring, cv=kfold)
grid_result = grid.fit(rescaledX, Y_train)
print("Best: %f using %s" % (grid_result.best_score_, grid_result.best_params_))
means = grid_result.cv_results_['mean_test_score']
stds = grid_result.cv_results_['std_test_score']
params = grid_result.cv_results_['params']
for mean, stdev, param in zip(means, stds, params):
print("%f (%f) with: %r" % (mean, stdev, param))
```
We can see that the best configuration was n estimators=400 resulting in a mean squared
error of -9.356471, about 0.65 units better than the untuned method.
### Finalize the model
```
# prepare the model
scaler = StandardScaler().fit(X_train)
rescaledX = scaler.transform(X_train)
model = GradientBoostingRegressor(random_state=seed, n_estimators=400)
model.fit(rescaledX, Y_train)
# transform the validation dataset
rescaledValidationX = scaler.transform(X_validation)
predictions = model.predict(rescaledValidationX)
print(mean_squared_error(Y_validation, predictions))
Y_validation
predictions
```
|
github_jupyter
|
# Load libraries
%matplotlib inline
import numpy
import matplotlib.pyplot as plt
from numpy import arange
from matplotlib import pyplot
from pandas import read_csv
from pandas import set_option
from pandas.tools.plotting import scatter_matrix
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import GridSearchCV
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Lasso
from sklearn.linear_model import ElasticNet
from sklearn.tree import DecisionTreeRegressor
from sklearn.neighbors import KNeighborsRegressor
from sklearn.svm import SVR
from sklearn.pipeline import Pipeline
from sklearn.ensemble import RandomForestRegressor
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.ensemble import ExtraTreesRegressor
from sklearn.ensemble import AdaBoostRegressor
from sklearn.metrics import mean_squared_error
# Load dataset
filename = 'housing.csv'
names = ['CRIM', 'ZN', 'INDUS', 'CHAS', 'NOX', 'RM', 'AGE', 'DIS', 'RAD', 'TAX', 'PTRATIO', 'B', 'LSTAT', 'MEDV']
dataset = read_csv(filename, delim_whitespace=True, names=names)
# shape
print(dataset.shape)
# types
print(dataset.dtypes)
# head
print(dataset.head(20))
set_option('precision', 2)
print(dataset.describe())
# correlation
set_option('precision', 2)
print(dataset.corr(method='pearson'))
# histograms
dataset.hist(sharex=False, sharey=False, xlabelsize=1, ylabelsize=1, figsize=(14, 12))
plt.show()
# density
dataset.plot.density(subplots=True, layout=(4,4), sharex=False, legend=False, fontsize=1, figsize=(14, 12))
plt.show()
# box and whisker plots
dataset.plot.box(subplots=True, layout=(4,4), sharex=False, sharey=False, figsize=(14, 12))
plt.show()
# scatter plot matrix
scatter_matrix(dataset, figsize=(14, 12))
plt.show()
# correlation matrix
fig = plt.figure(figsize=(14, 12))
ax = fig.add_subplot(111)
cax = ax.matshow(dataset.corr(), vmin=-1, vmax=1, interpolation='none')
fig.colorbar(cax)
ticks = arange(0,14,1)
ax.set_xticks(ticks)
ax.set_yticks(ticks)
ax.set_xticklabels(names)
ax.set_yticklabels(names)
plt.show()
# Split-out validation dataset
array = dataset.values
X = array[:,0:13]
Y = array[:,13]
validation_size = 0.20
seed = 7
X_train, X_validation, Y_train, Y_validation = train_test_split(X, Y, test_size=validation_size, random_state=seed)
# Test options and evaluation metric
num_folds = 10
seed = 7
scoring = 'neg_mean_squared_error'
# Spot-Check Algorithms
models = []
models.append(('LR', LinearRegression()))
models.append(('LASSO', Lasso()))
models.append(('EN', ElasticNet()))
models.append(('KNN', KNeighborsRegressor()))
models.append(('CART', DecisionTreeRegressor()))
models.append(('SVR' , SVR()))
# evaluate each model in turn
results = []
names = []
for name, model in models:
kfold = KFold(n_splits=num_folds, random_state=seed)
cv_results = cross_val_score(model, X_train, Y_train, cv=kfold, scoring=scoring)
results.append(cv_results)
names.append(name)
msg = "%s: %f (%f)" % (name, cv_results.mean(), cv_results.std())
print(msg)
# Compare Algorithms
fig = pyplot.figure(figsize=(14, 12))
fig.suptitle('Algorithm Comparison')
ax = fig.add_subplot(111)
pyplot.boxplot(results)
ax.set_xticklabels(names)
plt.show()
# Standardize the dataset
pipelines = []
pipelines.append(('ScaledLR', Pipeline([('Scaler', StandardScaler()),('LR', LinearRegression())])))
pipelines.append(('ScaledLASSO', Pipeline([('Scaler', StandardScaler()),('LASSO', Lasso())])))
pipelines.append(('ScaledEN', Pipeline([('Scaler', StandardScaler()),('EN', ElasticNet())])))
pipelines.append(('ScaledKNN', Pipeline([('Scaler', StandardScaler()),('KNN', KNeighborsRegressor())])))
pipelines.append(('ScaledCART', Pipeline([('Scaler', StandardScaler()),('CART', DecisionTreeRegressor())])))
pipelines.append(('ScaledSVR', Pipeline([('Scaler', StandardScaler()),('SVR', SVR())])))
results = []
names = []
for name, model in pipelines:
kfold = KFold(n_splits=num_folds, random_state=seed)
cv_results = cross_val_score(model, X_train, Y_train, cv=kfold, scoring=scoring)
results.append(cv_results)
names.append(name)
msg = "%s: %f (%f)" % (name, cv_results.mean(), cv_results.std())
print(msg)
# Compare Algorithms
fig = pyplot.figure(figsize=(14, 12))
fig.suptitle('Scaled Algorithm Comparison')
ax = fig.add_subplot(111)
pyplot.boxplot(results)
ax.set_xticklabels(names)
plt.show()
# KNN Algorithm tuning
scaler = StandardScaler().fit(X_train)
rescaledX = scaler.transform(X_train)
k_values = numpy.array([1,3,5,7,9,11,13,15,17,19,21])
param_grid = dict(n_neighbors=k_values)
model = KNeighborsRegressor()
kfold = KFold(n_splits=num_folds, random_state=seed)
grid = GridSearchCV(estimator=model, param_grid=param_grid, scoring=scoring, cv=kfold)
grid_result = grid.fit(rescaledX, Y_train)
print("Best: %f using %s" % (grid_result.best_score_, grid_result.best_params_))
means = grid_result.cv_results_['mean_test_score']
stds = grid_result.cv_results_['std_test_score']
params = grid_result.cv_results_['params']
for mean, stdev, param in zip(means, stds, params):
print("%f (%f) with: %r" % (mean, stdev, param))
# ensembles
ensembles = []
ensembles.append(('ScaledAB', Pipeline([('Scaler', StandardScaler()),('AB', AdaBoostRegressor())])))
ensembles.append(('ScaledGBM', Pipeline([('Scaler', StandardScaler()),('GBM', GradientBoostingRegressor())])))
ensembles.append(('ScaledRF', Pipeline([('Scaler', StandardScaler()),('RF', RandomForestRegressor())])))
ensembles.append(('ScaledET', Pipeline([('Scaler', StandardScaler()),('ET', ExtraTreesRegressor())])))
results = []
names = []
for name, model in ensembles:
kfold = KFold(n_splits=num_folds, random_state=seed)
cv_results = cross_val_score(model, X_train, Y_train, cv=kfold, scoring=scoring)
results.append(cv_results)
names.append(name)
msg = "%s: %f (%f)" % (name, cv_results.mean(), cv_results.std())
print(msg)
# Compare Algorithms
fig = pyplot.figure(figsize=(14, 12))
fig.suptitle('Scaled Ensemble Algorithm Comparison')
ax = fig.add_subplot(111)
pyplot.boxplot(results)
ax.set_xticklabels(names)
plt.show()
# Tune scaled GBM
scaler = StandardScaler().fit(X_train)
rescaledX = scaler.transform(X_train)
param_grid = dict(n_estimators=numpy.array([50,100,150,200,250,300,350,400]))
model = GradientBoostingRegressor(random_state=seed)
kfold = KFold(n_splits=num_folds, random_state=seed)
grid = GridSearchCV(estimator=model, param_grid=param_grid, scoring=scoring, cv=kfold)
grid_result = grid.fit(rescaledX, Y_train)
print("Best: %f using %s" % (grid_result.best_score_, grid_result.best_params_))
means = grid_result.cv_results_['mean_test_score']
stds = grid_result.cv_results_['std_test_score']
params = grid_result.cv_results_['params']
for mean, stdev, param in zip(means, stds, params):
print("%f (%f) with: %r" % (mean, stdev, param))
# prepare the model
scaler = StandardScaler().fit(X_train)
rescaledX = scaler.transform(X_train)
model = GradientBoostingRegressor(random_state=seed, n_estimators=400)
model.fit(rescaledX, Y_train)
# transform the validation dataset
rescaledValidationX = scaler.transform(X_validation)
predictions = model.predict(rescaledValidationX)
print(mean_squared_error(Y_validation, predictions))
Y_validation
predictions
| 0.779028 | 0.770422 |
```
# pandas/numpy for handling data
import pandas as pd
import numpy as np
# seaborn/matplotlib for graphing
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib.colors as mc
# statistics
from statistics import mean
import statsmodels.api as sm
from statsmodels.formula.api import ols
from scipy import stats
import scikit_posthocs as sp
from statsmodels.stats.anova import AnovaRM
# helper fxns for analyzing chr aberr data
import chr_aberr_helper_fxns as cah
# setting a global darkgrid style w/ dark edge plot elements for plotting
sns.set_style(style="darkgrid",rc= {'patch.edgecolor': 'black'})
import importlib
%load_ext autoreload
%autoreload 2
%reload_ext autoreload
```
---
...
---
# Analyzing telomeric aberrations
---
## Reading in telomeric aberration data (inflight astros)
```
melt_all_astro_telo_aberr = pd.read_csv('../data/compiled and processed data/melt_all_astro_telo_aberr.csv')
melt_all_astro_telo_aberr['astro id'] = melt_all_astro_telo_aberr['astro id'].astype('str')
melt_all_astro_telo_aberr = melt_all_astro_telo_aberr[melt_all_astro_telo_aberr['aberration type'] != '# of STL-complete']
melt_all_astro_telo_aberr.head(4)
```
## Graphing telomeric aberrations (inflight astros)
```
# let's make two graphs, one w/o and one w/ satellite associations
data=melt_all_astro_telo_aberr[melt_all_astro_telo_aberr['aberration type'] != '# of sat associations']
data2=melt_all_astro_telo_aberr[melt_all_astro_telo_aberr['aberration type'] == '# of sat associations']
x='aberration type'
y='count per cell'
hue='flight status'
ax1= sns.catplot(x=x, y=y, hue=hue, data=data, kind='bar', height=4, aspect=3)
ax2= sns.catplot(x=x, y=y, data=data2, hue=hue, kind='bar', height=4, aspect=3)
```
No trend in # fragile telos per spaceflight. Potential but very minor trend in STL-complete for midflight
Hetero telomeric foci @ sister chromatids highly elevated inflight, as are satellite associations..
High levels of recombination between telomeres? Let's combine mid-flight samples, just for curiosity.
```
mid_combined_melt_all_astro_telo_aberr = melt_all_astro_telo_aberr
mid_combined_melt_all_astro_telo_aberr['flight status new'] = (mid_combined_melt_all_astro_telo_aberr['flight status']
.apply(lambda row: cah.combine_midflight(row)))
data=mid_combined_melt_all_astro_telo_aberr[melt_all_astro_telo_aberr['aberration type'] != '# of sat associations']
data2=mid_combined_melt_all_astro_telo_aberr[melt_all_astro_telo_aberr['aberration type'] == '# of sat associations']
x='aberration type'
y='count per cell'
hue='flight status new'
ax1= sns.catplot(x=x, y=y, hue=hue, data=data, kind='bar', height=4, aspect=3)
ax2= sns.catplot(x=x, y=y, data=data2, hue=hue, kind='bar', height=4, aspect=3)
```
Same trends, different perspective w/ mid-flight combined. Heterogenous telomere foci between sister chromatids & satellite associations are elevated mid-flight
## Statistics: telomeric aberrations
```
cah.scipy_anova_post_hoc_tests(df=mid_combined_melt_all_astro_telo_aberr)
cah.scipy_anova_post_hoc_tests(df=mid_combined_melt_all_astro_telo_aberr)
```
# Analyzing chromosome rearrangement data
---
```
melt_all_astro_chr_aberr = pd.read_csv('../data/compiled and processed data/All_astronauts_chromosome_aberration_data_tidy_data.csv')
# reformatting (float -> int -> str)
melt_all_astro_chr_aberr['astro id'] = melt_all_astro_chr_aberr['astro id'].astype('int')
melt_all_astro_chr_aberr['astro id'] = melt_all_astro_chr_aberr['astro id'].astype('str')
astro_chr_aberr = melt_all_astro_chr_aberr.copy()
astro_chr_aberr['flight status'] = (astro_chr_aberr['flight status'].apply(lambda row: cah.combine_midflight(row)))
def rename_aberr(row):
if row == 'sister chromatid exchanges':
return 'classic SCEs'
elif row == 'total inversions':
return 'inversions'
elif row == 'satellite associations':
return 'sat. associations'
else:
return row
def rename_flights(row):
if row == 'pre-flight':
return 'Pre-Flight'
elif row == 'mid-flight':
return 'Mid-Flight'
elif row == 'post-flight':
return 'Post-Flight'
astro_chr_aberr['aberration type'] = astro_chr_aberr['aberration type'].apply(lambda row: rename_aberr(row))
astro_chr_aberr['flight status'] = astro_chr_aberr['flight status'].apply(lambda row: rename_flights(row))
```
## Graphing chromosome rearrangements for pre, mid-flight1&2, and post-flight for all astronauts (n=11)
```
order_cat=['dicentrics', 'translocations', 'inversions',]
# 'terminal SCEs','classic SCEs', 'subtelo SCEs', 'sat. associations']
plt.figure(figsize=(9, 3.2))
fontsize=16
ax = sns.barplot(x='aberration type', y='count per cell', data=astro_chr_aberr,
order=order_cat, hue='flight status',
alpha=None, capsize=0.08, linewidth=1.5, errwidth=1, **{'edgecolor':'black'},
ci=95,
palette={'Pre-Flight': '#0000FF',
'Mid-Flight': '#FF0000',
'Post-Flight': '#009900'})
# alpha setting in sns barplot modifies both bar fill AND edge colors; we want to change just fill
# keep alpha set to None, label bar colors w/ palette
# loop through patches (fill color), grab color ID & reset color w/ alpha at 0.2
for patch in range(len(ax.patches)):
color = ax.patches[patch].get_facecolor()
color = list(color)
color[3] = 0.2
color = tuple(color)
ax.patches[patch].set_facecolor(color)
plt.setp(ax.lines, color='black', linewidth=1.5)
plt.xlabel('', fontsize=fontsize)
plt.ylabel('Average frequency per cell', fontsize=fontsize)
plt.tick_params(labelsize=fontsize)
plt.ylim(0, .5)
plt.legend(fontsize=fontsize)
plt.savefig('../MANUSCRIPT 11 ASTROS/figures/dic trans inv chrr aberr (dGH) 11 astros pre mid post.png', dpi=600)
order_cat=['classic SCEs', 'subtelo SCEs', 'terminal SCEs', 'sat. associations']
plt.figure(figsize=(9, 3.2))
fontsize=16
ax = sns.barplot(x='aberration type', y='count per cell', data=astro_chr_aberr,
order=order_cat, hue='flight status',
alpha=None, capsize=0.08, linewidth=1.5, errwidth=1, **{'edgecolor':'black'},
ci=95,
palette={'Pre-Flight': '#0000FF',
'Mid-Flight': '#FF0000',
'Post-Flight': '#009900'})
# alpha setting in sns barplot modifies both bar fill AND edge colors; we want to change just fill
# keep alpha set to None, label bar colors w/ palette
# loop through patches (fill color), grab color ID & reset color w/ alpha at 0.2
for patch in range(len(ax.patches)):
color = ax.patches[patch].get_facecolor()
color = list(color)
color[3] = 0.2
color = tuple(color)
ax.patches[patch].set_facecolor(color)
plt.setp(ax.lines, color='black', linewidth=1.5)
plt.xlabel('', fontsize=fontsize)
plt.ylabel('Average frequency per cell', fontsize=fontsize)
plt.tick_params(labelsize=fontsize)
plt.ylim(0, 1.75)
plt.legend(fontsize=fontsize)
plt.savefig('../MANUSCRIPT 11 ASTROS/figures/SCEs chrr aberr (dGH) 11 astros pre mid post.png', dpi=600)
import importlib
%load_ext autoreload
%autoreload 2
%reload_ext autoreload
```
## Statistics: chromosome rearrangements (n=11)
```
astro_chr_aberr[astro_chr_aberr['aberration type'] == 'dicentrics']['count per cell']
grp_astro_chr_aberr = astro_chr_aberr.groupby(['astro id',
'flight status',
'aberration type']).agg('mean').reset_index()
pivot_chr = grp_astro_chr_aberr.pivot_table(index=['astro id', 'flight status'],
columns='aberration type', values='count per cell').reset_index()
cah.scipy_anova_post_hoc_tests(df=grp_astro_chr_aberr, flight_status_col='flight status')
```
Same results, different perspective. Let's remove the mid-flight data real quick and
then look at just our 3 unrelated astronauts w/ inflight data
```
mid_flight_removed = mid_combined_melt_all_astro_chr_aberr[mid_combined_melt_all_astro_chr_aberr['flight status new'] != 'mid-flight']
ax = sns.set(font_scale=1)
ax = sns.set_style(style="darkgrid",rc= {'patch.edgecolor': 'black'})
ax = sns.catplot(x='aberration type', y='count per cell',
hue='flight status new', kind='bar', order=order_cat,
orient='v', height=4, aspect=3, data=mid_flight_removed)
plt.title('chr aberr by subtelo dgh: 11 astros, pre-, post-', fontsize=16)
```
## Graphing chromosome rearrangements for pre, mid-flight1&2, and post-flight for all astronauts (n=3)
```
mid_combined = mid_combined_melt_all_astro_chr_aberr
mid_flight_only_astros = mid_combined[mid_combined['astro id'].isin(['2171', '1536', '5163'])]
list(mid_flight_only_astros['astro id'].unique())
order_cat=['dicentrics', 'translocations', 'total inversions', 'terminal SCEs',
'sister chromatid exchanges', 'subtelo SCEs', 'satellite associations']
ax = sns.set(font_scale=1)
ax = sns.set_style(style="darkgrid",rc= {'patch.edgecolor': 'black'})
ax = sns.catplot(x='aberration type', y='count per cell',
hue='flight status', kind='bar', order=order_cat,
orient='v', height=4, aspect=3, data=mid_flight_only_astros)
plt.title('chr aberr by subtelo dgh: 3 astros, pre-, mid1&2-, post-', fontsize=16)
ax = sns.set(font_scale=1)
ax = sns.set_style(style="darkgrid",rc= {'patch.edgecolor': 'black'})
ax = sns.catplot(x='aberration type', y='count per cell',
hue='flight status new', kind='bar', order=order_cat,
orient='v', height=4, aspect=3, data=mid_flight_only_astros)
plt.title('chr aberr by subtelo dgh: 3 astros, pre-, mid-, post-', fontsize=16)
grouped_mid_flight_only_astros = mid_flight_only_astros.groupby(['astro id', 'flight status', 'flight status new', 'aberration type']).agg('mean').reset_index()
# mid_flight_only_astros
```
## Statistics: chromosome rearrangements (n=3)
```
cah.scipy_anova_post_hoc_tests(df=mid_flight_only_astros)
df = mid_flight_only_astros
display(sp.posthoc_ttest(df[df['aberration type'] == 'total inversions'], val_col='count per cell',
group_col='flight status new', equal_var=False))
```
|
github_jupyter
|
# pandas/numpy for handling data
import pandas as pd
import numpy as np
# seaborn/matplotlib for graphing
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib.colors as mc
# statistics
from statistics import mean
import statsmodels.api as sm
from statsmodels.formula.api import ols
from scipy import stats
import scikit_posthocs as sp
from statsmodels.stats.anova import AnovaRM
# helper fxns for analyzing chr aberr data
import chr_aberr_helper_fxns as cah
# setting a global darkgrid style w/ dark edge plot elements for plotting
sns.set_style(style="darkgrid",rc= {'patch.edgecolor': 'black'})
import importlib
%load_ext autoreload
%autoreload 2
%reload_ext autoreload
melt_all_astro_telo_aberr = pd.read_csv('../data/compiled and processed data/melt_all_astro_telo_aberr.csv')
melt_all_astro_telo_aberr['astro id'] = melt_all_astro_telo_aberr['astro id'].astype('str')
melt_all_astro_telo_aberr = melt_all_astro_telo_aberr[melt_all_astro_telo_aberr['aberration type'] != '# of STL-complete']
melt_all_astro_telo_aberr.head(4)
# let's make two graphs, one w/o and one w/ satellite associations
data=melt_all_astro_telo_aberr[melt_all_astro_telo_aberr['aberration type'] != '# of sat associations']
data2=melt_all_astro_telo_aberr[melt_all_astro_telo_aberr['aberration type'] == '# of sat associations']
x='aberration type'
y='count per cell'
hue='flight status'
ax1= sns.catplot(x=x, y=y, hue=hue, data=data, kind='bar', height=4, aspect=3)
ax2= sns.catplot(x=x, y=y, data=data2, hue=hue, kind='bar', height=4, aspect=3)
mid_combined_melt_all_astro_telo_aberr = melt_all_astro_telo_aberr
mid_combined_melt_all_astro_telo_aberr['flight status new'] = (mid_combined_melt_all_astro_telo_aberr['flight status']
.apply(lambda row: cah.combine_midflight(row)))
data=mid_combined_melt_all_astro_telo_aberr[melt_all_astro_telo_aberr['aberration type'] != '# of sat associations']
data2=mid_combined_melt_all_astro_telo_aberr[melt_all_astro_telo_aberr['aberration type'] == '# of sat associations']
x='aberration type'
y='count per cell'
hue='flight status new'
ax1= sns.catplot(x=x, y=y, hue=hue, data=data, kind='bar', height=4, aspect=3)
ax2= sns.catplot(x=x, y=y, data=data2, hue=hue, kind='bar', height=4, aspect=3)
cah.scipy_anova_post_hoc_tests(df=mid_combined_melt_all_astro_telo_aberr)
cah.scipy_anova_post_hoc_tests(df=mid_combined_melt_all_astro_telo_aberr)
melt_all_astro_chr_aberr = pd.read_csv('../data/compiled and processed data/All_astronauts_chromosome_aberration_data_tidy_data.csv')
# reformatting (float -> int -> str)
melt_all_astro_chr_aberr['astro id'] = melt_all_astro_chr_aberr['astro id'].astype('int')
melt_all_astro_chr_aberr['astro id'] = melt_all_astro_chr_aberr['astro id'].astype('str')
astro_chr_aberr = melt_all_astro_chr_aberr.copy()
astro_chr_aberr['flight status'] = (astro_chr_aberr['flight status'].apply(lambda row: cah.combine_midflight(row)))
def rename_aberr(row):
if row == 'sister chromatid exchanges':
return 'classic SCEs'
elif row == 'total inversions':
return 'inversions'
elif row == 'satellite associations':
return 'sat. associations'
else:
return row
def rename_flights(row):
if row == 'pre-flight':
return 'Pre-Flight'
elif row == 'mid-flight':
return 'Mid-Flight'
elif row == 'post-flight':
return 'Post-Flight'
astro_chr_aberr['aberration type'] = astro_chr_aberr['aberration type'].apply(lambda row: rename_aberr(row))
astro_chr_aberr['flight status'] = astro_chr_aberr['flight status'].apply(lambda row: rename_flights(row))
order_cat=['dicentrics', 'translocations', 'inversions',]
# 'terminal SCEs','classic SCEs', 'subtelo SCEs', 'sat. associations']
plt.figure(figsize=(9, 3.2))
fontsize=16
ax = sns.barplot(x='aberration type', y='count per cell', data=astro_chr_aberr,
order=order_cat, hue='flight status',
alpha=None, capsize=0.08, linewidth=1.5, errwidth=1, **{'edgecolor':'black'},
ci=95,
palette={'Pre-Flight': '#0000FF',
'Mid-Flight': '#FF0000',
'Post-Flight': '#009900'})
# alpha setting in sns barplot modifies both bar fill AND edge colors; we want to change just fill
# keep alpha set to None, label bar colors w/ palette
# loop through patches (fill color), grab color ID & reset color w/ alpha at 0.2
for patch in range(len(ax.patches)):
color = ax.patches[patch].get_facecolor()
color = list(color)
color[3] = 0.2
color = tuple(color)
ax.patches[patch].set_facecolor(color)
plt.setp(ax.lines, color='black', linewidth=1.5)
plt.xlabel('', fontsize=fontsize)
plt.ylabel('Average frequency per cell', fontsize=fontsize)
plt.tick_params(labelsize=fontsize)
plt.ylim(0, .5)
plt.legend(fontsize=fontsize)
plt.savefig('../MANUSCRIPT 11 ASTROS/figures/dic trans inv chrr aberr (dGH) 11 astros pre mid post.png', dpi=600)
order_cat=['classic SCEs', 'subtelo SCEs', 'terminal SCEs', 'sat. associations']
plt.figure(figsize=(9, 3.2))
fontsize=16
ax = sns.barplot(x='aberration type', y='count per cell', data=astro_chr_aberr,
order=order_cat, hue='flight status',
alpha=None, capsize=0.08, linewidth=1.5, errwidth=1, **{'edgecolor':'black'},
ci=95,
palette={'Pre-Flight': '#0000FF',
'Mid-Flight': '#FF0000',
'Post-Flight': '#009900'})
# alpha setting in sns barplot modifies both bar fill AND edge colors; we want to change just fill
# keep alpha set to None, label bar colors w/ palette
# loop through patches (fill color), grab color ID & reset color w/ alpha at 0.2
for patch in range(len(ax.patches)):
color = ax.patches[patch].get_facecolor()
color = list(color)
color[3] = 0.2
color = tuple(color)
ax.patches[patch].set_facecolor(color)
plt.setp(ax.lines, color='black', linewidth=1.5)
plt.xlabel('', fontsize=fontsize)
plt.ylabel('Average frequency per cell', fontsize=fontsize)
plt.tick_params(labelsize=fontsize)
plt.ylim(0, 1.75)
plt.legend(fontsize=fontsize)
plt.savefig('../MANUSCRIPT 11 ASTROS/figures/SCEs chrr aberr (dGH) 11 astros pre mid post.png', dpi=600)
import importlib
%load_ext autoreload
%autoreload 2
%reload_ext autoreload
astro_chr_aberr[astro_chr_aberr['aberration type'] == 'dicentrics']['count per cell']
grp_astro_chr_aberr = astro_chr_aberr.groupby(['astro id',
'flight status',
'aberration type']).agg('mean').reset_index()
pivot_chr = grp_astro_chr_aberr.pivot_table(index=['astro id', 'flight status'],
columns='aberration type', values='count per cell').reset_index()
cah.scipy_anova_post_hoc_tests(df=grp_astro_chr_aberr, flight_status_col='flight status')
mid_flight_removed = mid_combined_melt_all_astro_chr_aberr[mid_combined_melt_all_astro_chr_aberr['flight status new'] != 'mid-flight']
ax = sns.set(font_scale=1)
ax = sns.set_style(style="darkgrid",rc= {'patch.edgecolor': 'black'})
ax = sns.catplot(x='aberration type', y='count per cell',
hue='flight status new', kind='bar', order=order_cat,
orient='v', height=4, aspect=3, data=mid_flight_removed)
plt.title('chr aberr by subtelo dgh: 11 astros, pre-, post-', fontsize=16)
mid_combined = mid_combined_melt_all_astro_chr_aberr
mid_flight_only_astros = mid_combined[mid_combined['astro id'].isin(['2171', '1536', '5163'])]
list(mid_flight_only_astros['astro id'].unique())
order_cat=['dicentrics', 'translocations', 'total inversions', 'terminal SCEs',
'sister chromatid exchanges', 'subtelo SCEs', 'satellite associations']
ax = sns.set(font_scale=1)
ax = sns.set_style(style="darkgrid",rc= {'patch.edgecolor': 'black'})
ax = sns.catplot(x='aberration type', y='count per cell',
hue='flight status', kind='bar', order=order_cat,
orient='v', height=4, aspect=3, data=mid_flight_only_astros)
plt.title('chr aberr by subtelo dgh: 3 astros, pre-, mid1&2-, post-', fontsize=16)
ax = sns.set(font_scale=1)
ax = sns.set_style(style="darkgrid",rc= {'patch.edgecolor': 'black'})
ax = sns.catplot(x='aberration type', y='count per cell',
hue='flight status new', kind='bar', order=order_cat,
orient='v', height=4, aspect=3, data=mid_flight_only_astros)
plt.title('chr aberr by subtelo dgh: 3 astros, pre-, mid-, post-', fontsize=16)
grouped_mid_flight_only_astros = mid_flight_only_astros.groupby(['astro id', 'flight status', 'flight status new', 'aberration type']).agg('mean').reset_index()
# mid_flight_only_astros
cah.scipy_anova_post_hoc_tests(df=mid_flight_only_astros)
df = mid_flight_only_astros
display(sp.posthoc_ttest(df[df['aberration type'] == 'total inversions'], val_col='count per cell',
group_col='flight status new', equal_var=False))
| 0.367611 | 0.795698 |
<a href="https://colab.research.google.com/github/kalz2q/mycolabnotebooks/blob/master/magic.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
$\latex$
$\LaTeX$
```
%%script false
と書くと、そのコードセルは実行されないので、まるでテキストセルのように使える。
テキストセルの引用(行頭に>)や、コード表示(バッククオート3つ)のように使える。
勝手に改行もされない。
なにより、Markdownによる解釈がされないので、確実に文字列を書ける。
latexも当然使えない。
```
# magic.ipynb
# はじめに
失敗作なので消していいけれど、云々。
セルマジックについてはいまのところ材料は2つあって、ひとつは
The cell magics in IPython
https://nbviewer.jupyter.org/github/ipython/ipython/blob/1.x/examples/notebooks/Cell%20Magics.ipynb
もう1つは Overview of Colaboratory Features
https://colab.research.google.com/notebooks/basic_features_overview.ipynb#scrollTo=qM4myQGfQboQ
で、このファイルは020magic.ipynbで
%lsmagic
と
%(magicname)?
で作っているみたいだが、数多くのmagicについて、どうやってdocstringを得たのだろう。
自分でもわからない。
あとの方に`%time`を使った例というか実験が載っているが、これはどこかから持ってきたのだろう。
それより、'%javascript`や`%html`をもっと知りたい。
多分、The cell magics in IPythonがよいだろう。 なので、別ファイルで云々。
名前を考えよう。
とりあえずcellmagic.ipynbでいいか。
# %lsmagicと%lsmagic?
`%lsmagic`でまずどんなコマンドがあるか確認
```
# まずどんなコマンドがあるか確認
%lsmagic
```
アウトプットを見てみよう。
ラインマジックは以下のとおり。`lsmagic`もラインマジックである。
%alias %alias_magic %autocall %automagic %autosave %bookmark %cat %cd %clear %colors %config %connect_info %cp %debug %dhist %dirs %doctest_mode %ed %edit %env %gui %hist %history %killbgscripts %ldir %less %lf %lk %ll %load %load_ext %loadpy %logoff %logon %logstart %logstate %logstop %ls %lsmagic %lx %macro %magic %man %matplotlib %mkdir %more %mv %notebook %page %pastebin %pdb %pdef %pdoc %pfile %pinfo %pinfo2 %pip %popd %pprint %precision %profile %prun %psearch %psource %pushd %pwd %pycat %pylab %qtconsole %quickref %recall %rehashx %reload_ext %rep %rerun %reset %reset_selective %rm %rmdir %run %save %sc %set_env %shell %store %sx %system %tb %tensorflow_version %time %timeit %unalias %unload_ext %who %who_ls %whos %xdel %xmode
セルマジックは以下のとおり。
%%! %%HTML %%SVG %%bash %%bigquery %%capture %%debug %%file %%html %%javascript %%js %%latex %%perl %%prun %%pypy %%python %%python2 %%python3 %%ruby %%script %%sh %%shell %%svg %%sx %%system %%time %%timeit %%writefile
`automagic`がオンのときは、ラインマジックに`%`は必要ないとのこと。まあ、どっちでもよい。=> `%automagic`コマンド自体がトグルスイッチ。明示的にオンとかオフとかにするときには、`on`、`off`、`1`、`0`、`True`、 `False`を引数として与える。
## ヘルプコマンド
マジックコマンドに`?`とか`??`をつけることで簡単な説明が得られる。仮にヘルプコマンドと呼ぼう。
```
# %lsmagic? とか %lsmagic?? とかやってみる
# %lsmagic?
```
### magicコマンドのdocstring
以下に各コマンドの`docstring`を示す。よくわからないので、英語のまま。
セルコマンドには`%%javascript`とか`ruby`とかもあり、利用できそう。
```
%alias? Define an alias for a system command.
%alias_magic? Create an alias for an existing line or cell magic.
%autocall? Make functions callable without having to type parentheses.
%automagic? Make magic functions callable without having to type the initial %.
%autosave? Set the autosave interval in the notebook (in seconds).
%bookmark? Manage IPython's bookmark system.
%cat? Repr: <alias cat for 'cat'>
%cd? Change the current working directory.
%clear? Clear the terminal.
%colors? Switch color scheme for prompts, info system and exception handlers.
%config? configure IPython
%connect_info? Print information for connecting other clients to this kernel
%cp? Repr: <alias cp for 'cp'>
%debug? Activate the interactive debugger.
%dhist? Print your history of visited directories.
%dirs? Return the current directory stack.
%doctest_mode? Toggle doctest mode on and off.
%ed? Alias for `%edit`.
%edit? Bring up an editor and execute the resulting code.
%env? Get, set, or list environment variables.
%gui? Enable or disable IPython GUI event loop integration.
%hist? Alias for `%history`.
%history? Print input history (_i<n> variables), with most recent last.
%killbgscripts? ill all BG processes started by %%script and its family.
%ldir? Repr: <alias ldir for 'ls -F -o --color %l | grep /$'>
%less? Show a file through the pager.
%lf? Repr: <alias lf for 'ls -F -o --color %l | grep ^-'>
%lk? Repr: <alias lk for 'ls -F -o --color %l | grep ^l'>
%ll? Repr: <alias ll for 'ls -F -o --color'>
%load? Load code into the current frontend.
%load_ext? Load an IPython extension by its module name.
%loadpy? Alias of `%load`
%logoff? Temporarily stop logging.
%logon? Restart logging.
%logstart? Start logging anywhere in a session.
%logstate? Print the status of the logging system.
%logstop? Fully stop logging and close log file.
%ls? Repr: <alias ls for 'ls -F --color'>
%lsmagic? List currently available magic functions.
%lx? Repr: <alias lx for 'ls -F -o --color %l | grep ^-..x'>
%macro? Define a macro for future re-execution. It accepts ranges of history, filenames or string objects.
%magic? Print information about the magic function system.
%man? Find the man page for the given command and display in pager.
%matplotlib? Set up matplotlib to work interactively.
%mkdir? Repr: <alias mkdir for 'mkdir'>
%more? Show a file through the pager.
%mv? Repr: <alias mv for 'mv'>
%notebook? Export and convert IPython notebooks.
%page? Pretty print the object and display it through a pager.
%pastebin? Upload code to Github's Gist paste bin, returning the URL.
%pdb? Control the automatic calling of the pdb interactive debugger.
%pdef? Print the call signature for any callable object.
%pdoc? Print the docstring for an object.
%pfile? Print (or run through pager) the file where an object is defined.
%pinfo? Provide detailed information about an object.
%pinfo2? Provide extra detailed information about an object.
%pip? Install a package in the current kernel using pip.
%popd? Change to directory popped off the top of the stack.
%pprint? Toggle pretty printing on/off.
%precision? Set floating point precision for pretty printing.
%prun? Run a statement through the python code profiler.
%psearch? Search for object in namespaces by wildcard.
%psource? Print (or run through pager) the source code for an object.
%pushd? lace the current dir on stack and change directory.
%pwd? Return the current working directory path.
%pycat? Show a syntax-highlighted file through a pag
%pylab? Load numpy and matplotlib to work interactively.
%qtconsole? Open a qtconsole connected to this kernel.
%quickref? Show a quick reference sheet
%recall? Repeat a command, or get command to input line for editing.
%rehashx? Update the alias table with all executable files in $PATH.
%reload_ext? Reload an IPython extension by its module name.
%rep? Docstring: Alias for `%recall`.
%rerun? Re-run previous input
%reset? Resets the namespace by removing all names defined by the user, if called without arguments
%reset_selective? Resets the namespace by removing names defined by the user.
%rm? Repr: <alias rm for 'rm'>
%rmdir? Repr: <alias rmdir for 'rmdir'>
%run? Run the named file inside IPython as a program.
%save? Save a set of lines or a macro to a given filename.
%sc? Shell capture - run shell command and capture output (DEPRECATED use !).
%set_env? Set environment variables.
%shell? Runs a shell command, allowing input to be provided.
%store? Lightweight persistence for python variables.
%sx? Shell execute - run shell command and capture output (!! is short-hand).
%system? Shell execute - run shell command and capture output (!! is short-hand).
%tb? Print the last traceback with the currently active exception mode.
%tensorflow_version? Implements the tensorflow_version line magic.
%time? Time execution of a Python statement or expression.
%timeit? Time execution of a Python statement or expression
%unalias? Remove an alias
%unload_ext? Unload an IPython extension by its module name.
%who? Print all interactive variables, with some minimal formatting.
%who_ls? Return a sorted list of all interactive variables.
%whos? Like %who, but gives some extra information about each variable.
%xdel? Delete a variable, trying to clear it from anywhere that IPython's machinery has references to it.
%xmode? Switch modes for the exception handlers.
%%HTML? Alias for `%%html`.
%%SVG? Alias for `%%svg`.
%%bash? %%bash script magic
%%bigquery? Underlying function for bigquery cell magic
%%capture? run the cell, capturing stdout, stderr, and IPython's rich display() calls.
%%debug? Activate the interactive debugger.
%%file? Alias for `%%writefile`.
%%html? ender the cell as a block of HTML
%%javascript? Run the cell block of Javascript code
%%js? Alias of `%%javascript`
%%latex? Render the cell as a block of latex
%%perl? %%perl script magic
%%prun? Run a statement through the python code profiler.
%%pypy? %%pypy script magic
%%python? Run cells with python in a subprocess.
%%python2? Run cells with python2 in a subprocess.
%%python3? Run cells with python3 in a subprocess.
%%ruby? Run cells with ruby in a subprocess.
%%script? Run a cell via a shell command
%%sh? Run cells with sh in a subprocess.
%%shell? Run the cell via a shell command, allowing input to be provided.
%%svg? Render the cell as an SVG literal
%%sx? Shell execute - run shell command and capture output (!! is short-hand).
%%system? Shell execute - run shell command and capture output (!! is short-hand).
%%time? Time execution of a Python statement or expression.
%%timeit? Time execution of a Python statement or expression
%%writefile? Write the contents of the cell to a file.
```
### %time を使ってみる。
```
%time sum(range(10000))
%timeit sum(range(10000))
%%timeit -n 1000 -r 3
for i in range(1000):
i * 2
```
### $matplotlib を使ってみる
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
x = np. arange(0, 10, 0.2)
y = np.sin(x)
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(x, y)
plt.show()
```
# いまここ
|
github_jupyter
|
%%script false
と書くと、そのコードセルは実行されないので、まるでテキストセルのように使える。
テキストセルの引用(行頭に>)や、コード表示(バッククオート3つ)のように使える。
勝手に改行もされない。
なにより、Markdownによる解釈がされないので、確実に文字列を書ける。
latexも当然使えない。
# まずどんなコマンドがあるか確認
%lsmagic
# %lsmagic? とか %lsmagic?? とかやってみる
# %lsmagic?
%alias? Define an alias for a system command.
%alias_magic? Create an alias for an existing line or cell magic.
%autocall? Make functions callable without having to type parentheses.
%automagic? Make magic functions callable without having to type the initial %.
%autosave? Set the autosave interval in the notebook (in seconds).
%bookmark? Manage IPython's bookmark system.
%cat? Repr: <alias cat for 'cat'>
%cd? Change the current working directory.
%clear? Clear the terminal.
%colors? Switch color scheme for prompts, info system and exception handlers.
%config? configure IPython
%connect_info? Print information for connecting other clients to this kernel
%cp? Repr: <alias cp for 'cp'>
%debug? Activate the interactive debugger.
%dhist? Print your history of visited directories.
%dirs? Return the current directory stack.
%doctest_mode? Toggle doctest mode on and off.
%ed? Alias for `%edit`.
%edit? Bring up an editor and execute the resulting code.
%env? Get, set, or list environment variables.
%gui? Enable or disable IPython GUI event loop integration.
%hist? Alias for `%history`.
%history? Print input history (_i<n> variables), with most recent last.
%killbgscripts? ill all BG processes started by %%script and its family.
%ldir? Repr: <alias ldir for 'ls -F -o --color %l | grep /$'>
%less? Show a file through the pager.
%lf? Repr: <alias lf for 'ls -F -o --color %l | grep ^-'>
%lk? Repr: <alias lk for 'ls -F -o --color %l | grep ^l'>
%ll? Repr: <alias ll for 'ls -F -o --color'>
%load? Load code into the current frontend.
%load_ext? Load an IPython extension by its module name.
%loadpy? Alias of `%load`
%logoff? Temporarily stop logging.
%logon? Restart logging.
%logstart? Start logging anywhere in a session.
%logstate? Print the status of the logging system.
%logstop? Fully stop logging and close log file.
%ls? Repr: <alias ls for 'ls -F --color'>
%lsmagic? List currently available magic functions.
%lx? Repr: <alias lx for 'ls -F -o --color %l | grep ^-..x'>
%macro? Define a macro for future re-execution. It accepts ranges of history, filenames or string objects.
%magic? Print information about the magic function system.
%man? Find the man page for the given command and display in pager.
%matplotlib? Set up matplotlib to work interactively.
%mkdir? Repr: <alias mkdir for 'mkdir'>
%more? Show a file through the pager.
%mv? Repr: <alias mv for 'mv'>
%notebook? Export and convert IPython notebooks.
%page? Pretty print the object and display it through a pager.
%pastebin? Upload code to Github's Gist paste bin, returning the URL.
%pdb? Control the automatic calling of the pdb interactive debugger.
%pdef? Print the call signature for any callable object.
%pdoc? Print the docstring for an object.
%pfile? Print (or run through pager) the file where an object is defined.
%pinfo? Provide detailed information about an object.
%pinfo2? Provide extra detailed information about an object.
%pip? Install a package in the current kernel using pip.
%popd? Change to directory popped off the top of the stack.
%pprint? Toggle pretty printing on/off.
%precision? Set floating point precision for pretty printing.
%prun? Run a statement through the python code profiler.
%psearch? Search for object in namespaces by wildcard.
%psource? Print (or run through pager) the source code for an object.
%pushd? lace the current dir on stack and change directory.
%pwd? Return the current working directory path.
%pycat? Show a syntax-highlighted file through a pag
%pylab? Load numpy and matplotlib to work interactively.
%qtconsole? Open a qtconsole connected to this kernel.
%quickref? Show a quick reference sheet
%recall? Repeat a command, or get command to input line for editing.
%rehashx? Update the alias table with all executable files in $PATH.
%reload_ext? Reload an IPython extension by its module name.
%rep? Docstring: Alias for `%recall`.
%rerun? Re-run previous input
%reset? Resets the namespace by removing all names defined by the user, if called without arguments
%reset_selective? Resets the namespace by removing names defined by the user.
%rm? Repr: <alias rm for 'rm'>
%rmdir? Repr: <alias rmdir for 'rmdir'>
%run? Run the named file inside IPython as a program.
%save? Save a set of lines or a macro to a given filename.
%sc? Shell capture - run shell command and capture output (DEPRECATED use !).
%set_env? Set environment variables.
%shell? Runs a shell command, allowing input to be provided.
%store? Lightweight persistence for python variables.
%sx? Shell execute - run shell command and capture output (!! is short-hand).
%system? Shell execute - run shell command and capture output (!! is short-hand).
%tb? Print the last traceback with the currently active exception mode.
%tensorflow_version? Implements the tensorflow_version line magic.
%time? Time execution of a Python statement or expression.
%timeit? Time execution of a Python statement or expression
%unalias? Remove an alias
%unload_ext? Unload an IPython extension by its module name.
%who? Print all interactive variables, with some minimal formatting.
%who_ls? Return a sorted list of all interactive variables.
%whos? Like %who, but gives some extra information about each variable.
%xdel? Delete a variable, trying to clear it from anywhere that IPython's machinery has references to it.
%xmode? Switch modes for the exception handlers.
%%HTML? Alias for `%%html`.
%%SVG? Alias for `%%svg`.
%%bash? %%bash script magic
%%bigquery? Underlying function for bigquery cell magic
%%capture? run the cell, capturing stdout, stderr, and IPython's rich display() calls.
%%debug? Activate the interactive debugger.
%%file? Alias for `%%writefile`.
%%html? ender the cell as a block of HTML
%%javascript? Run the cell block of Javascript code
%%js? Alias of `%%javascript`
%%latex? Render the cell as a block of latex
%%perl? %%perl script magic
%%prun? Run a statement through the python code profiler.
%%pypy? %%pypy script magic
%%python? Run cells with python in a subprocess.
%%python2? Run cells with python2 in a subprocess.
%%python3? Run cells with python3 in a subprocess.
%%ruby? Run cells with ruby in a subprocess.
%%script? Run a cell via a shell command
%%sh? Run cells with sh in a subprocess.
%%shell? Run the cell via a shell command, allowing input to be provided.
%%svg? Render the cell as an SVG literal
%%sx? Shell execute - run shell command and capture output (!! is short-hand).
%%system? Shell execute - run shell command and capture output (!! is short-hand).
%%time? Time execution of a Python statement or expression.
%%timeit? Time execution of a Python statement or expression
%%writefile? Write the contents of the cell to a file.
%time sum(range(10000))
%timeit sum(range(10000))
%%timeit -n 1000 -r 3
for i in range(1000):
i * 2
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
x = np. arange(0, 10, 0.2)
y = np.sin(x)
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(x, y)
plt.show()
| 0.425009 | 0.95877 |
<a href="https://colab.research.google.com/github/Bhavani-Rajan/DS-Unit-1-Sprint-2-Data-Wrangling-and-Storytelling/blob/master/module3-make-explanatory-visualizations/LS_DS_123_Make_Explanatory_Visualizations.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
_Lambda School Data Science_
# Make Explanatory Visualizations
### Objectives
- identify misleading visualizations and how to fix them
- use Seaborn to visualize distributions and relationships with continuous and discrete variables
- add emphasis and annotations to transform visualizations from exploratory to explanatory
- remove clutter from visualizations
### Links
- [How to Spot Visualization Lies](https://flowingdata.com/2017/02/09/how-to-spot-visualization-lies/)
- [Visual Vocabulary - Vega Edition](http://ft.com/vocabulary)
- [Choosing a Python Visualization Tool flowchart](http://pbpython.com/python-vis-flowchart.html)
- [Searborn example gallery](http://seaborn.pydata.org/examples/index.html) & [tutorial](http://seaborn.pydata.org/tutorial.html)
- [Strong Titles Are The Biggest Bang for Your Buck](http://stephanieevergreen.com/strong-titles/)
- [Remove to improve (the data-ink ratio)](https://www.darkhorseanalytics.com/blog/data-looks-better-naked)
- [How to Generate FiveThirtyEight Graphs in Python](https://www.dataquest.io/blog/making-538-plots/)
# Avoid Misleading Visualizations
Did you find/discuss any interesting misleading visualizations in your Walkie Talkie?
## What makes a visualization misleading?
[5 Ways Writers Use Misleading Graphs To Manipulate You](https://venngage.com/blog/misleading-graphs/)
## Two y-axes
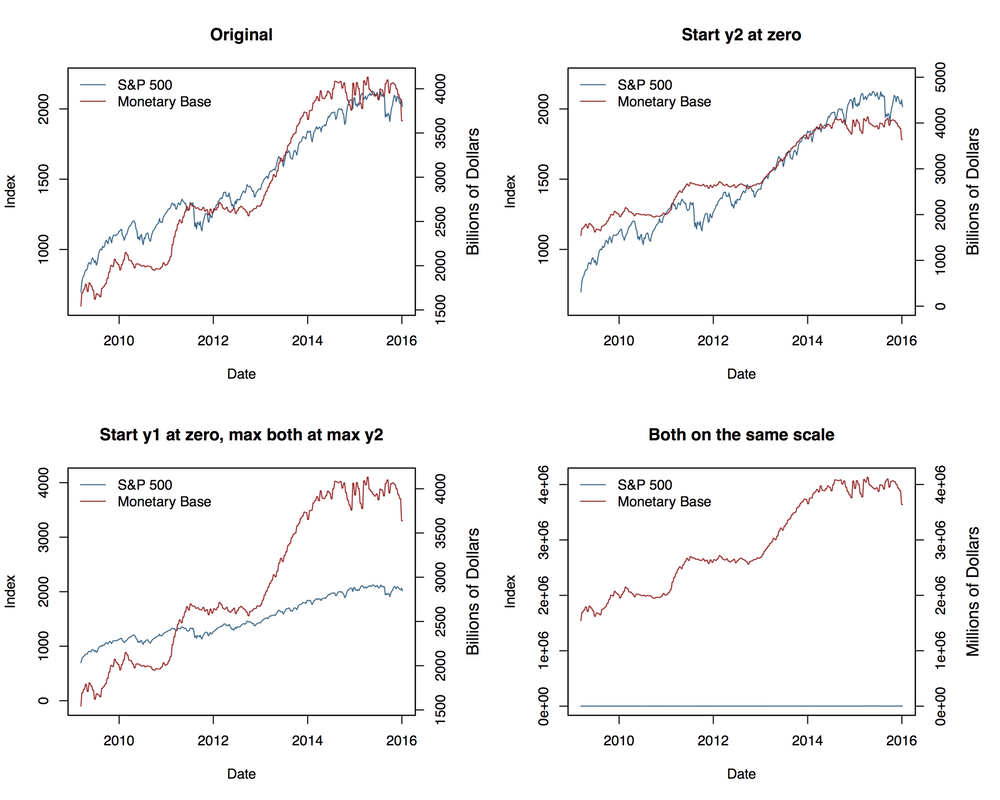
Other Examples:
- [Spurious Correlations](https://tylervigen.com/spurious-correlations)
- <https://blog.datawrapper.de/dualaxis/>
- <https://kieranhealy.org/blog/archives/2016/01/16/two-y-axes/>
- <http://www.storytellingwithdata.com/blog/2016/2/1/be-gone-dual-y-axis>
## Y-axis doesn't start at zero.
<img src="https://i.pinimg.com/originals/22/53/a9/2253a944f54bb61f1983bc076ff33cdd.jpg" width="600">
## Pie Charts are bad
<img src="https://i1.wp.com/flowingdata.com/wp-content/uploads/2009/11/Fox-News-pie-chart.png?fit=620%2C465&ssl=1" width="600">
## Pie charts that omit data are extra bad
- A guy makes a misleading chart that goes viral
What does this chart imply at first glance? You don't want your user to have to do a lot of work in order to be able to interpret you graph correctly. You want that first-glance conclusions to be the correct ones.
<img src="https://pbs.twimg.com/media/DiaiTLHWsAYAEEX?format=jpg&name=medium" width='600'>
<https://twitter.com/michaelbatnick/status/1019680856837849090?lang=en>
- It gets picked up by overworked journalists (assuming incompetency before malice)
<https://www.marketwatch.com/story/this-1-chart-puts-mega-techs-trillions-of-market-value-into-eye-popping-perspective-2018-07-18>
- Even after the chart's implications have been refuted, it's hard a bad (although compelling) visualization from being passed around.
<https://www.linkedin.com/pulse/good-bad-pie-charts-karthik-shashidhar/>
**["yea I understand a pie chart was probably not the best choice to present this data."](https://twitter.com/michaelbatnick/status/1037036440494985216)**
## Pie Charts that compare unrelated things are next-level extra bad
<img src="http://www.painting-with-numbers.com/download/document/186/170403+Legalizing+Marijuana+Graph.jpg" width="600">
## Be careful about how you use volume to represent quantities:
radius vs diameter vs volume
<img src="https://static1.squarespace.com/static/5bfc8dbab40b9d7dd9054f41/t/5c32d86e0ebbe80a25873249/1546836082961/5474039-25383714-thumbnail.jpg?format=1500w" width="600">
## Don't cherrypick timelines or specific subsets of your data:
<img src="https://wattsupwiththat.com/wp-content/uploads/2019/02/Figure-1-1.png" width="600">
Look how specifically the writer has selected what years to show in the legend on the right side.
<https://wattsupwiththat.com/2019/02/24/strong-arctic-sea-ice-growth-this-year/>
Try the tool that was used to make the graphic for yourself
<http://nsidc.org/arcticseaicenews/charctic-interactive-sea-ice-graph/>
## Use Relative units rather than Absolute Units
<img src="https://imgs.xkcd.com/comics/heatmap_2x.png" width="600">
## Avoid 3D graphs unless having the extra dimension is effective
Usually you can Split 3D graphs into multiple 2D graphs
3D graphs that are interactive can be very cool. (See Plotly and Bokeh)
<img src="https://thumbor.forbes.com/thumbor/1280x868/https%3A%2F%2Fblogs-images.forbes.com%2Fthumbnails%2Fblog_1855%2Fpt_1855_811_o.jpg%3Ft%3D1339592470" width="600">
## Don't go against typical conventions
<img src="http://www.callingbullshit.org/twittercards/tools_misleading_axes.png" width="600">
# Tips for choosing an appropriate visualization:
## Use Appropriate "Visual Vocabulary"
[Visual Vocabulary - Vega Edition](http://ft.com/vocabulary)
## What are the properties of your data?
- Is your primary variable of interest continuous or discrete?
- Is in wide or long (tidy) format?
- Does your visualization involve multiple variables?
- How many dimensions do you need to include on your plot?
Can you express the main idea of your visualization in a single sentence?
How hard does your visualization make the user work in order to draw the intended conclusion?
## Which Visualization tool is most appropriate?
[Choosing a Python Visualization Tool flowchart](http://pbpython.com/python-vis-flowchart.html)
## Anatomy of a Matplotlib Plot

```
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.ticker import AutoMinorLocator, MultipleLocator, FuncFormatter
np.random.seed(19680801)
X = np.linspace(0.5, 3.5, 100)
Y1 = 3+np.cos(X)
Y2 = 1+np.cos(1+X/0.75)/2
Y3 = np.random.uniform(Y1, Y2, len(X))
fig = plt.figure(figsize=(8, 8))
ax = fig.add_subplot(1, 1, 1, aspect=1)
def minor_tick(x, pos):
if not x % 1.0:
return ""
return "%.2f" % x
ax.xaxis.set_major_locator(MultipleLocator(1.000))
ax.xaxis.set_minor_locator(AutoMinorLocator(4))
ax.yaxis.set_major_locator(MultipleLocator(1.000))
ax.yaxis.set_minor_locator(AutoMinorLocator(4))
ax.xaxis.set_minor_formatter(FuncFormatter(minor_tick))
ax.set_xlim(0, 4)
ax.set_ylim(0, 4)
ax.tick_params(which='major', width=1.0)
ax.tick_params(which='major', length=10)
ax.tick_params(which='minor', width=1.0, labelsize=10)
ax.tick_params(which='minor', length=5, labelsize=10, labelcolor='0.25')
ax.grid(linestyle="--", linewidth=0.5, color='.25', zorder=-10)
ax.plot(X, Y1, c=(0.25, 0.25, 1.00), lw=2, label="Blue signal", zorder=10)
ax.plot(X, Y2, c=(1.00, 0.25, 0.25), lw=2, label="Red signal")
ax.plot(X, Y3, linewidth=0,
marker='o', markerfacecolor='w', markeredgecolor='k')
ax.set_title("Anatomy of a figure", fontsize=20, verticalalignment='bottom')
ax.set_xlabel("X axis label")
ax.set_ylabel("Y axis label")
ax.legend()
def circle(x, y, radius=0.15):
from matplotlib.patches import Circle
from matplotlib.patheffects import withStroke
circle = Circle((x, y), radius, clip_on=False, zorder=10, linewidth=1,
edgecolor='black', facecolor=(0, 0, 0, .0125),
path_effects=[withStroke(linewidth=5, foreground='w')])
ax.add_artist(circle)
def text(x, y, text):
ax.text(x, y, text, backgroundcolor="white",
ha='center', va='top', weight='bold', color='blue')
# Minor tick
circle(0.50, -0.10)
text(0.50, -0.32, "Minor tick label")
# Major tick
circle(-0.03, 4.00)
text(0.03, 3.80, "Major tick")
# Minor tick
circle(0.00, 3.50)
text(0.00, 3.30, "Minor tick")
# Major tick label
circle(-0.15, 3.00)
text(-0.15, 2.80, "Major tick label")
# X Label
circle(1.80, -0.27)
text(1.80, -0.45, "X axis label")
# Y Label
circle(-0.27, 1.80)
text(-0.27, 1.6, "Y axis label")
# Title
circle(1.60, 4.13)
text(1.60, 3.93, "Title")
# Blue plot
circle(1.75, 2.80)
text(1.75, 2.60, "Line\n(line plot)")
# Red plot
circle(1.20, 0.60)
text(1.20, 0.40, "Line\n(line plot)")
# Scatter plot
circle(3.20, 1.75)
text(3.20, 1.55, "Markers\n(scatter plot)")
# Grid
circle(3.00, 3.00)
text(3.00, 2.80, "Grid")
# Legend
circle(3.70, 3.80)
text(3.70, 3.60, "Legend")
# Axes
circle(0.5, 0.5)
text(0.5, 0.3, "Axes")
# Figure
circle(-0.3, 0.65)
text(-0.3, 0.45, "Figure")
color = 'blue'
ax.annotate('Spines', xy=(4.0, 0.35), xytext=(3.3, 0.5),
weight='bold', color=color,
arrowprops=dict(arrowstyle='->',
connectionstyle="arc3",
color=color))
ax.annotate('', xy=(3.15, 0.0), xytext=(3.45, 0.45),
weight='bold', color=color,
arrowprops=dict(arrowstyle='->',
connectionstyle="arc3",
color=color))
ax.text(4.0, -0.4, "Made with http://matplotlib.org",
fontsize=10, ha="right", color='.5')
plt.show()
```
# Making Explanatory Visualizations with Matplotlib
Today we will reproduce this [example by FiveThirtyEight:](https://fivethirtyeight.com/features/al-gores-new-movie-exposes-the-big-flaw-in-online-movie-ratings/)
```
from IPython.display import display, Image
url = 'https://fivethirtyeight.com/wp-content/uploads/2017/09/mehtahickey-inconvenient-0830-1.png'
example = Image(url=url, width=400)
display(example)
```
Using this data: https://github.com/fivethirtyeight/data/tree/master/inconvenient-sequel
Links
- [Strong Titles Are The Biggest Bang for Your Buck](http://stephanieevergreen.com/strong-titles/)
- [Remove to improve (the data-ink ratio)](https://www.darkhorseanalytics.com/blog/data-looks-better-naked)
- [How to Generate FiveThirtyEight Graphs in Python](https://www.dataquest.io/blog/making-538-plots/)
jeaopardy sessions on thursday
```
import matplotlib.pyplot as plt
x = [1,3,5,7,9]
y = [2,4,6,8,10]
plt.plot(x,label='x ',color='r')
plt.plot(y,label='y',color='b')
plt.xlabel('Odd')
plt.ylabel('Even')
plt.legend()
plt.show()
# sort by petal length
import pandas as pd
data_url ='https://raw.githubusercontent.com/uiuc-cse/data-fa14/gh-pages/data/iris.csv'
data = pd.read_csv(data_url)
```
## Make prototypes
This helps us understand the problem
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
plt.style.use('fivethirtyeight')
fake = pd.Series([38, 3, 2, 1, 2, 4, 6, 5, 5, 33],
index=range(1,11))
fake.plot.bar(color='#E67141', width=0.9);
fake2 = pd.Series(
[1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
2, 2, 2,
3, 3, 3,
4, 4,
5, 5, 5,
6, 6, 6, 6,
7, 7, 7, 7, 7,
8, 8, 8, 8,
9, 9, 9, 9,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10])
#fake2.value_counts().sort_index().plot.bar(color='C1', width=0.9);
#fake2.value_counts()
#fake2.value_counts().sort_index()
#fake2.value_counts().sort_index().plot.bar(color='#E67141', width=0.9)
#fake2.value_counts().sort_index().plot(color='#E67141');
fake2.value_counts().sort_index().plot.bar(color='#E67141', width=0.9);
```
## Annotate with text
```
display(example)
import matplotlib.pyplot as plt
import matplotlib.ticker as mtick
# select the style
plt.style.use('fivethirtyeight')
#generate our figure
fig = plt.figure()
# generate our axes (center section) for a plot
ax = fake.plot.bar(color='#ED713A', width=0.9)
# to set the background to white in color
ax.set(facecolor='white')
# also for the fig
fig.patch.set(facecolor='white')
fig.patch.set_alpha(0.1)
ax.patch.set_alpha(0.1)
# multiple way of adding title
#fig.suptitle('title added to fig')
#ax.text(x=1,y=35,s='title using ax.text')
#ax.set_title('title using ax.set_title');
# There is more than one way to set a title on a graph
# matplotlib is not "pythonic"
# fig.suptitle('test title')
# ax.set_title("title")
# If we want a really custom title
# We can just use a text annotation and make it look like a title
ax.text(x=-2.2, y=46, s="'An Inconvenient Sequel: Truth To Power' is divisive",
fontweight='bold', fontsize=12);
ax.text(x=-2.2, y=43, s="IMDb ratings for the film as of Aug. 29",
fontsize=11)
# Set our axis labels (These are just text objects that have been rotated!!)
ax.set_ylabel("Percent of total votes", fontsize=9, fontweight='bold',
labelpad=10)
ax.set_xlabel("Rating", fontsize=9, fontweight='bold', labelpad=10)
# Fix our tick lables. Yet again, these are text and use the text parameters!
ax.set_xticklabels(range(1,11), rotation=0)
ax.set_yticks(range(0,50,10))
ax.set_yticklabels(range(0, 50, 10))
# to format the percentage sign on y axis
fmt = '%.0f%%' # Format you want the ticks, e.g. '40%'
yticks = mtick.FormatStrFormatter(fmt)
ax.yaxis.set_major_formatter(yticks)
# to set the tick color to grey
ax.tick_params(axis='x', colors='gray')
ax.tick_params(axis='y', colors='gray')
plt.show()
display(example)
```
## Reproduce with real data
```
df = pd.read_csv('https://raw.githubusercontent.com/fivethirtyeight/data/master/inconvenient-sequel/ratings.csv')
df.head()
df.columns.values.tolist()
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.ticker import AutoMinorLocator, MultipleLocator, FuncFormatter
np.random.seed(19680801)
X = np.linspace(0.5, 3.5, 100)
Y1 = 3+np.cos(X)
Y2 = 1+np.cos(1+X/0.75)/2
Y3 = np.random.uniform(Y1, Y2, len(X))
fig = plt.figure(figsize=(8, 8))
ax = fig.add_subplot(1, 1, 1, aspect=1)
def minor_tick(x, pos):
if not x % 1.0:
return ""
return "%.2f" % x
ax.xaxis.set_major_locator(MultipleLocator(1.000))
ax.xaxis.set_minor_locator(AutoMinorLocator(4))
ax.yaxis.set_major_locator(MultipleLocator(1.000))
ax.yaxis.set_minor_locator(AutoMinorLocator(4))
ax.xaxis.set_minor_formatter(FuncFormatter(minor_tick))
ax.set_xlim(0, 4)
ax.set_ylim(0, 4)
ax.tick_params(which='major', width=1.0)
ax.tick_params(which='major', length=10)
ax.tick_params(which='minor', width=1.0, labelsize=10)
ax.tick_params(which='minor', length=5, labelsize=10, labelcolor='0.25')
ax.grid(linestyle="--", linewidth=0.5, color='.25', zorder=-10)
ax.plot(X, Y1, c=(0.25, 0.25, 1.00), lw=2, label="Blue signal", zorder=10)
ax.plot(X, Y2, c=(1.00, 0.25, 0.25), lw=2, label="Red signal")
ax.plot(X, Y3, linewidth=0,
marker='o', markerfacecolor='w', markeredgecolor='k')
ax.set_title("Anatomy of a figure", fontsize=20, verticalalignment='bottom')
ax.set_xlabel("X axis label")
ax.set_ylabel("Y axis label")
ax.legend()
def circle(x, y, radius=0.15):
from matplotlib.patches import Circle
from matplotlib.patheffects import withStroke
circle = Circle((x, y), radius, clip_on=False, zorder=10, linewidth=1,
edgecolor='black', facecolor=(0, 0, 0, .0125),
path_effects=[withStroke(linewidth=5, foreground='w')])
ax.add_artist(circle)
def text(x, y, text):
ax.text(x, y, text, backgroundcolor="white",
ha='center', va='top', weight='bold', color='blue')
# Minor tick
circle(0.50, -0.10)
text(0.50, -0.32, "Minor tick label")
# Major tick
circle(-0.03, 4.00)
text(0.03, 3.80, "Major tick")
# Minor tick
circle(0.00, 3.50)
text(0.00, 3.30, "Minor tick")
# Major tick label
circle(-0.15, 3.00)
text(-0.15, 2.80, "Major tick label")
# X Label
circle(1.80, -0.27)
text(1.80, -0.45, "X axis label")
# Y Label
circle(-0.27, 1.80)
text(-0.27, 1.6, "Y axis label")
# Title
circle(1.60, 4.13)
text(1.60, 3.93, "Title")
# Blue plot
circle(1.75, 2.80)
text(1.75, 2.60, "Line\n(line plot)")
# Red plot
circle(1.20, 0.60)
text(1.20, 0.40, "Line\n(line plot)")
# Scatter plot
circle(3.20, 1.75)
text(3.20, 1.55, "Markers\n(scatter plot)")
# Grid
circle(3.00, 3.00)
text(3.00, 2.80, "Grid")
# Legend
circle(3.70, 3.80)
text(3.70, 3.60, "Legend")
# Axes
circle(0.5, 0.5)
text(0.5, 0.3, "Axes")
# Figure
circle(-0.3, 0.65)
text(-0.3, 0.45, "Figure")
color = 'blue'
ax.annotate('Spines', xy=(4.0, 0.35), xytext=(3.3, 0.5),
weight='bold', color=color,
arrowprops=dict(arrowstyle='->',
connectionstyle="arc3",
color=color))
ax.annotate('', xy=(3.15, 0.0), xytext=(3.45, 0.45),
weight='bold', color=color,
arrowprops=dict(arrowstyle='->',
connectionstyle="arc3",
color=color))
ax.text(4.0, -0.4, "Made with http://matplotlib.org",
fontsize=10, ha="right", color='.5')
plt.show()
from IPython.display import display, Image
url = 'https://fivethirtyeight.com/wp-content/uploads/2017/09/mehtahickey-inconvenient-0830-1.png'
example = Image(url=url, width=400)
display(example)
import matplotlib.pyplot as plt
x = [1,3,5,7,9]
y = [2,4,6,8,10]
plt.plot(x,label='x ',color='r')
plt.plot(y,label='y',color='b')
plt.xlabel('Odd')
plt.ylabel('Even')
plt.legend()
plt.show()
# sort by petal length
import pandas as pd
data_url ='https://raw.githubusercontent.com/uiuc-cse/data-fa14/gh-pages/data/iris.csv'
data = pd.read_csv(data_url)
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
plt.style.use('fivethirtyeight')
fake = pd.Series([38, 3, 2, 1, 2, 4, 6, 5, 5, 33],
index=range(1,11))
fake.plot.bar(color='#E67141', width=0.9);
fake2 = pd.Series(
[1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
2, 2, 2,
3, 3, 3,
4, 4,
5, 5, 5,
6, 6, 6, 6,
7, 7, 7, 7, 7,
8, 8, 8, 8,
9, 9, 9, 9,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10])
#fake2.value_counts().sort_index().plot.bar(color='C1', width=0.9);
#fake2.value_counts()
#fake2.value_counts().sort_index()
#fake2.value_counts().sort_index().plot.bar(color='#E67141', width=0.9)
#fake2.value_counts().sort_index().plot(color='#E67141');
fake2.value_counts().sort_index().plot.bar(color='#E67141', width=0.9);
display(example)
import matplotlib.pyplot as plt
import matplotlib.ticker as mtick
# select the style
plt.style.use('fivethirtyeight')
#generate our figure
fig = plt.figure()
# generate our axes (center section) for a plot
ax = fake.plot.bar(color='#ED713A', width=0.9)
# to set the background to white in color
ax.set(facecolor='white')
# also for the fig
fig.patch.set(facecolor='white')
fig.patch.set_alpha(0.1)
ax.patch.set_alpha(0.1)
# multiple way of adding title
#fig.suptitle('title added to fig')
#ax.text(x=1,y=35,s='title using ax.text')
#ax.set_title('title using ax.set_title');
# There is more than one way to set a title on a graph
# matplotlib is not "pythonic"
# fig.suptitle('test title')
# ax.set_title("title")
# If we want a really custom title
# We can just use a text annotation and make it look like a title
ax.text(x=-2.2, y=46, s="'An Inconvenient Sequel: Truth To Power' is divisive",
fontweight='bold', fontsize=12);
ax.text(x=-2.2, y=43, s="IMDb ratings for the film as of Aug. 29",
fontsize=11)
# Set our axis labels (These are just text objects that have been rotated!!)
ax.set_ylabel("Percent of total votes", fontsize=9, fontweight='bold',
labelpad=10)
ax.set_xlabel("Rating", fontsize=9, fontweight='bold', labelpad=10)
# Fix our tick lables. Yet again, these are text and use the text parameters!
ax.set_xticklabels(range(1,11), rotation=0)
ax.set_yticks(range(0,50,10))
ax.set_yticklabels(range(0, 50, 10))
# to format the percentage sign on y axis
fmt = '%.0f%%' # Format you want the ticks, e.g. '40%'
yticks = mtick.FormatStrFormatter(fmt)
ax.yaxis.set_major_formatter(yticks)
# to set the tick color to grey
ax.tick_params(axis='x', colors='gray')
ax.tick_params(axis='y', colors='gray')
plt.show()
display(example)
df = pd.read_csv('https://raw.githubusercontent.com/fivethirtyeight/data/master/inconvenient-sequel/ratings.csv')
df.head()
df.columns.values.tolist()
| 0.610802 | 0.939471 |
# Single-cell RNA-seq analysis workflow for 1M cells using Scanpy on CPU
Copyright (c) 2020, NVIDIA CORPORATION.
Licensed under the Apache License, Version 2.0 (the "License") you may not use this file except in compliance with the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
## Import requirements
```
import time
import numpy as np
import scanpy as sc
from sklearn.cluster import KMeans
import os, wget
import utils
import warnings
warnings.filterwarnings('ignore', 'Expected ')
warnings.simplefilter('ignore')
```
## Input data
```
# Add path to input file here.
input_file = "../data/1M_brain_cells_10X.sparse.h5ad"
USE_FIRST_N_CELLS = 1000000
if not os.path.exists(input_file):
print('Downloading import file...')
os.makedirs('../data', exist_ok=True)
wget.download('https://rapids-single-cell-examples.s3.us-east-2.amazonaws.com/1M_brain_cells_10X.sparse.h5ad',
input_file)
```
## Set parameters
```
# marker genes
MITO_GENE_PREFIX = "mt-" # Prefix for mitochondrial genes to regress out
markers = ["Stmn2", "Hes1", "Olig1"] # Marker genes for visualization
# filtering cells
min_genes_per_cell = 200 # Filter out cells with fewer genes than this expressed
max_genes_per_cell = 6000 # Filter out cells with more genes than this expressed
# filtering genes
n_top_genes = 4000 # Number of highly variable genes to retain
# PCA
n_components = 50 # Number of principal components to compute
# Batched PCA
pca_train_ratio = 0.35 # percentage of cells to use for PCA training, when using batched PCA function
n_pca_batches = 10 # number of batches, when using batched PCA function
# t-SNE
tsne_n_pcs = 20 # Number of principal components to use for t-SNE
# k-means
k = 35 # Number of clusters for k-means
# KNN
n_neighbors = 15 # Number of nearest neighbors for KNN graph
knn_n_pcs = 50 # Number of principal components to use for finding nearest neighbors
# UMAP
umap_min_dist = 0.3
umap_spread = 1.0
# Gene ranking
ranking_n_top_genes = 50 # Number of differential genes to compute for each cluster
# Number of parallel jobs for t-SNE and k-means
n_jobs=32
start = time.time()
```
## Load and Prepare Data
```
%%time
adata = sc.read(input_file)
adata.var_names_make_unique()
adata.shape
adata = adata[0:USE_FIRST_N_CELLS]
adata.shape
```
## Preprocessing
```
preprocess_start = time.time()
```
### Filter
We filter the count matrix to remove cells with an extreme number of genes expressed.
```
%%time
sc.pp.filter_cells(adata, min_genes=min_genes_per_cell)
sc.pp.filter_cells(adata, max_genes=max_genes_per_cell)
```
Some genes will now have zero expression in all cells. We filter out such genes.
```
%%time
sc.pp.filter_genes(adata, min_cells=1)
adata.shape
```
### Normalize
```
%%time
sc.pp.normalize_total(adata, target_sum=1e4)
%%time
sc.pp.log1p(adata)
```
### Select Most Variable Genes
```
%%time
# Select highly variable genes
sc.pp.highly_variable_genes(adata, n_top_genes=n_top_genes, flavor = "cell_ranger")
# Retain marker gene expression
for marker in markers:
adata.obs[marker + "_raw"] = adata.X[:, adata.var.index == marker].todense()
# Filter matrix to only variable genes
adata = adata[:, adata.var.highly_variable]
```
### Regress out confounding factors (number of counts, mitochondrial gene expression)
```
%%time
mito_genes = adata.var_names.str.startswith(MITO_GENE_PREFIX)
n_counts = np.array(adata.X.sum(axis=1))
adata.obs['percent_mito'] = np.array(np.sum(adata[:, mito_genes].X, axis=1)) / n_counts
adata.obs['n_counts'] = n_counts
%%time
sc.pp.regress_out(adata, ['n_counts', 'percent_mito'])
```
### Scale
```
%%time
sc.pp.scale(adata, max_value=10)
preprocess_time = time.time()
print("Total Preprocess time : %s" % (preprocess_time-preprocess_start))
```
## Cluster & Visualize
### Reduce
First, we calculate the time taken to run PCA using the complete dataset.
```
%%time
sc.tl.pca(adata, n_comps=n_components)
```
For a fair comparison with the single-GPU notebook, we also calculate the time taken if we use the PCA function from `utils.py`, which uses only a fraction of the cells for training.
```
%%time
adata = utils.pca(adata, n_components=n_components,
train_ratio=pca_train_ratio,
n_batches=n_pca_batches, gpu=False)
```
### TSNE + K-means
```
%%time
sc.tl.tsne(adata, n_pcs=tsne_n_pcs, n_jobs=n_jobs)
%%time
kmeans = KMeans(n_clusters=k, random_state=0, n_jobs=n_jobs).fit(adata.obsm['X_pca'])
adata.obs['kmeans'] = kmeans.labels_.astype(str)
%%time
sc.pl.tsne(adata, color=["kmeans"])
%%time
sc.pl.tsne(adata, color=["Stmn2_raw"], color_map="Blues", vmax=1, vmin=-0.05)
sc.pl.tsne(adata, color=["Hes1_raw"], color_map="Blues", vmax=1, vmin=-0.05)
```
### UMAP + Graph clustering
```
%%time
# KNN graph
sc.pp.neighbors(adata, n_neighbors=n_neighbors, n_pcs=knn_n_pcs)
%%time
# UMAP
sc.tl.umap(adata, min_dist=umap_min_dist, spread=umap_spread)
%%time
# Louvain clustering
sc.tl.louvain(adata)
%%time
sc.pl.umap(adata, color=["louvain"])
%%time
# Leiden clustering
sc.tl.leiden(adata)
%%time
sc.pl.umap(adata, color=["leiden"])
%%time
sc.pl.umap(adata, color=["Stmn2_raw"], color_map="Blues", vmax=1, vmin=-0.05)
sc.pl.umap(adata, color=["Hes1_raw"], color_map="Blues", vmax=1, vmin=-0.05)
```
## Create zoomed-in view
```
reanalysis_start = time.time()
%%time
adata = adata[adata.obs["Hes1_raw"] > 0.0, :]
print(adata.X.shape)
%%time
sc.tl.pca(adata, n_comps=n_components)
sc.pp.neighbors(adata, n_neighbors=n_neighbors, n_pcs=knn_n_pcs)
sc.tl.umap(adata, min_dist=umap_min_dist, spread=umap_spread)
sc.tl.leiden(adata)
%%time
sc.pl.umap(adata, color=["leiden"])
sc.pl.umap(adata, color=["Olig1_raw"], color_map="Blues", vmax=1, vmin=-0.05)
reanalysis_time = time.time()
print("Total reanalysis time : %s" % (reanalysis_time-reanalysis_start))
print("Full time: %s" % (time.time() - start))
```
|
github_jupyter
|
import time
import numpy as np
import scanpy as sc
from sklearn.cluster import KMeans
import os, wget
import utils
import warnings
warnings.filterwarnings('ignore', 'Expected ')
warnings.simplefilter('ignore')
# Add path to input file here.
input_file = "../data/1M_brain_cells_10X.sparse.h5ad"
USE_FIRST_N_CELLS = 1000000
if not os.path.exists(input_file):
print('Downloading import file...')
os.makedirs('../data', exist_ok=True)
wget.download('https://rapids-single-cell-examples.s3.us-east-2.amazonaws.com/1M_brain_cells_10X.sparse.h5ad',
input_file)
# marker genes
MITO_GENE_PREFIX = "mt-" # Prefix for mitochondrial genes to regress out
markers = ["Stmn2", "Hes1", "Olig1"] # Marker genes for visualization
# filtering cells
min_genes_per_cell = 200 # Filter out cells with fewer genes than this expressed
max_genes_per_cell = 6000 # Filter out cells with more genes than this expressed
# filtering genes
n_top_genes = 4000 # Number of highly variable genes to retain
# PCA
n_components = 50 # Number of principal components to compute
# Batched PCA
pca_train_ratio = 0.35 # percentage of cells to use for PCA training, when using batched PCA function
n_pca_batches = 10 # number of batches, when using batched PCA function
# t-SNE
tsne_n_pcs = 20 # Number of principal components to use for t-SNE
# k-means
k = 35 # Number of clusters for k-means
# KNN
n_neighbors = 15 # Number of nearest neighbors for KNN graph
knn_n_pcs = 50 # Number of principal components to use for finding nearest neighbors
# UMAP
umap_min_dist = 0.3
umap_spread = 1.0
# Gene ranking
ranking_n_top_genes = 50 # Number of differential genes to compute for each cluster
# Number of parallel jobs for t-SNE and k-means
n_jobs=32
start = time.time()
%%time
adata = sc.read(input_file)
adata.var_names_make_unique()
adata.shape
adata = adata[0:USE_FIRST_N_CELLS]
adata.shape
preprocess_start = time.time()
%%time
sc.pp.filter_cells(adata, min_genes=min_genes_per_cell)
sc.pp.filter_cells(adata, max_genes=max_genes_per_cell)
%%time
sc.pp.filter_genes(adata, min_cells=1)
adata.shape
%%time
sc.pp.normalize_total(adata, target_sum=1e4)
%%time
sc.pp.log1p(adata)
%%time
# Select highly variable genes
sc.pp.highly_variable_genes(adata, n_top_genes=n_top_genes, flavor = "cell_ranger")
# Retain marker gene expression
for marker in markers:
adata.obs[marker + "_raw"] = adata.X[:, adata.var.index == marker].todense()
# Filter matrix to only variable genes
adata = adata[:, adata.var.highly_variable]
%%time
mito_genes = adata.var_names.str.startswith(MITO_GENE_PREFIX)
n_counts = np.array(adata.X.sum(axis=1))
adata.obs['percent_mito'] = np.array(np.sum(adata[:, mito_genes].X, axis=1)) / n_counts
adata.obs['n_counts'] = n_counts
%%time
sc.pp.regress_out(adata, ['n_counts', 'percent_mito'])
%%time
sc.pp.scale(adata, max_value=10)
preprocess_time = time.time()
print("Total Preprocess time : %s" % (preprocess_time-preprocess_start))
%%time
sc.tl.pca(adata, n_comps=n_components)
%%time
adata = utils.pca(adata, n_components=n_components,
train_ratio=pca_train_ratio,
n_batches=n_pca_batches, gpu=False)
%%time
sc.tl.tsne(adata, n_pcs=tsne_n_pcs, n_jobs=n_jobs)
%%time
kmeans = KMeans(n_clusters=k, random_state=0, n_jobs=n_jobs).fit(adata.obsm['X_pca'])
adata.obs['kmeans'] = kmeans.labels_.astype(str)
%%time
sc.pl.tsne(adata, color=["kmeans"])
%%time
sc.pl.tsne(adata, color=["Stmn2_raw"], color_map="Blues", vmax=1, vmin=-0.05)
sc.pl.tsne(adata, color=["Hes1_raw"], color_map="Blues", vmax=1, vmin=-0.05)
%%time
# KNN graph
sc.pp.neighbors(adata, n_neighbors=n_neighbors, n_pcs=knn_n_pcs)
%%time
# UMAP
sc.tl.umap(adata, min_dist=umap_min_dist, spread=umap_spread)
%%time
# Louvain clustering
sc.tl.louvain(adata)
%%time
sc.pl.umap(adata, color=["louvain"])
%%time
# Leiden clustering
sc.tl.leiden(adata)
%%time
sc.pl.umap(adata, color=["leiden"])
%%time
sc.pl.umap(adata, color=["Stmn2_raw"], color_map="Blues", vmax=1, vmin=-0.05)
sc.pl.umap(adata, color=["Hes1_raw"], color_map="Blues", vmax=1, vmin=-0.05)
reanalysis_start = time.time()
%%time
adata = adata[adata.obs["Hes1_raw"] > 0.0, :]
print(adata.X.shape)
%%time
sc.tl.pca(adata, n_comps=n_components)
sc.pp.neighbors(adata, n_neighbors=n_neighbors, n_pcs=knn_n_pcs)
sc.tl.umap(adata, min_dist=umap_min_dist, spread=umap_spread)
sc.tl.leiden(adata)
%%time
sc.pl.umap(adata, color=["leiden"])
sc.pl.umap(adata, color=["Olig1_raw"], color_map="Blues", vmax=1, vmin=-0.05)
reanalysis_time = time.time()
print("Total reanalysis time : %s" % (reanalysis_time-reanalysis_start))
print("Full time: %s" % (time.time() - start))
| 0.453504 | 0.854521 |
# Window functions
General Elections were held in the UK in 2015 and 2017. Every citizen votes in a constituency. The candidate who gains the most votes becomes MP for that constituency.
All these results are recorded in a table ge
yr | firstName | lastName | constituency | party | votes
---:|-----------|-----------|---------------|-------|------:
2015 | Ian | Murray | S14000024 | Labour | 19293
2015 | Neil | Hay | S14000024 | Scottish National Party | 16656
2015 | Miles | Briggs | S14000024 | Conservative | 8626
2015 | Phyl | Meyer | S14000024 | Green | 2090
2015 | Pramod | Subbaraman | S14000024 | Liberal Democrat | 1823
2015 | Paul | Marshall | S14000024 | UK Independence Party | 601
2015 | Colin | Fox | S14000024 | Scottish Socialist Party | 197
2017 | Ian | MURRAY | S14000024 | Labour | 26269
2017 | Jim | EADIE | S14000024 | SNP | 10755
2017 | Stephanie Jane Harley | SMITH | S14000024 | Conservative | 9428
2017 | Alan Christopher | BEAL | S14000024 | Liberal Democrats | 1388
```
import getpass
import psycopg2
from sqlalchemy import create_engine
import pandas as pd
pwd = getpass.getpass()
engine = create_engine(
'postgresql+psycopg2://postgres:%s@localhost/sqlzoo' % (pwd))
pd.set_option('display.max_rows', 100)
```
## 1. Warming up
Show the **lastName, party** and **votes** for the **constituency** 'S14000024' in 2017.
```
ge = pd.read_sql_table('ge', engine)
ge.loc[(ge['constituency']=='S14000024') & (ge['yr']==2017),
['lastname', 'party', 'votes']]
```
## 2. Who won?
You can use the RANK function to see the order of the candidates. If you RANK using (ORDER BY votes DESC) then the candidate with the most votes has rank 1.
**Show the party and RANK for constituency S14000024 in 2017. List the output by party**
```
a = ge.loc[(ge['constituency']=='S14000024') & (ge['yr']==2017),
['party', 'votes']]
(a.assign(rank=a['votes'].rank(ascending=False))
.sort_values('party'))
```
## 3. PARTITION BY
The 2015 election is a different PARTITION to the 2017 election. We only care about the order of votes for each year.
**Use PARTITION to show the ranking of each party in S14000021 in each year. Include yr, party, votes and ranking (the party with the most votes is 1).**
```
a = ge[ge['constituency']=='S14000021'].copy()
a['posn'] = (a.groupby('yr')['votes']
.rank(ascending=False))
a[['yr', 'party', 'votes', 'posn']].sort_values(['party', 'yr'])
```
## 4. Edinburgh Constituency
Edinburgh constituencies are numbered S14000021 to S14000026.
**Use PARTITION BY constituency to show the ranking of each party in Edinburgh in 2017. Order your results so the winners are shown first, then ordered by constituency.**
```
a = ge[(ge['constituency'].between('S14000021', 'S14000026')) &
(ge['yr']==2017)].copy()
a['posn'] = (a.groupby('constituency')['votes']
.rank(ascending=False))
(a[['constituency', 'party', 'votes', 'posn']]
.sort_values(['posn', 'constituency']))
```
## 5. Winners Only
You can use [SELECT within SELECT](https://sqlzoo.net/wiki/SELECT_within_SELECT_Tutorial) to pick out only the winners in Edinburgh.
**Show the parties that won for each Edinburgh constituency in 2017.**
```
a = ge[(ge['constituency'].between('S14000021', 'S14000026')) & (ge['yr']==2017)].copy()
a['posn'] = a.groupby('constituency')['votes'].rank(ascending=False)
a.loc[a['posn']==1, ['constituency', 'party']].sort_values('constituency')
```
## 6. Scottish seats
You can use **COUNT** and **GROUP BY** to see how each party did in Scotland. Scottish constituencies start with 'S'
**Show how many seats for each party in Scotland in 2017.**
```
a = ge[(ge['constituency'].str.startswith('S')) &
(ge['yr']==2017)].copy()
a['posn'] = (a.groupby('constituency')['votes']
.rank(ascending=False))
(a[a['posn']==1].groupby('party')['yr']
.count()
.reset_index()
.rename(columns={'yr': 'n'}))
```
|
github_jupyter
|
import getpass
import psycopg2
from sqlalchemy import create_engine
import pandas as pd
pwd = getpass.getpass()
engine = create_engine(
'postgresql+psycopg2://postgres:%s@localhost/sqlzoo' % (pwd))
pd.set_option('display.max_rows', 100)
ge = pd.read_sql_table('ge', engine)
ge.loc[(ge['constituency']=='S14000024') & (ge['yr']==2017),
['lastname', 'party', 'votes']]
a = ge.loc[(ge['constituency']=='S14000024') & (ge['yr']==2017),
['party', 'votes']]
(a.assign(rank=a['votes'].rank(ascending=False))
.sort_values('party'))
a = ge[ge['constituency']=='S14000021'].copy()
a['posn'] = (a.groupby('yr')['votes']
.rank(ascending=False))
a[['yr', 'party', 'votes', 'posn']].sort_values(['party', 'yr'])
a = ge[(ge['constituency'].between('S14000021', 'S14000026')) &
(ge['yr']==2017)].copy()
a['posn'] = (a.groupby('constituency')['votes']
.rank(ascending=False))
(a[['constituency', 'party', 'votes', 'posn']]
.sort_values(['posn', 'constituency']))
a = ge[(ge['constituency'].between('S14000021', 'S14000026')) & (ge['yr']==2017)].copy()
a['posn'] = a.groupby('constituency')['votes'].rank(ascending=False)
a.loc[a['posn']==1, ['constituency', 'party']].sort_values('constituency')
a = ge[(ge['constituency'].str.startswith('S')) &
(ge['yr']==2017)].copy()
a['posn'] = (a.groupby('constituency')['votes']
.rank(ascending=False))
(a[a['posn']==1].groupby('party')['yr']
.count()
.reset_index()
.rename(columns={'yr': 'n'}))
| 0.119974 | 0.749775 |
Boa parte dos códigos abaixo são inspirados nas videoaulas do canal **Ignorância Zero** (`https://www.youtube.com/channel/UCmjj41YfcaCpZIkU-oqVIIw`)
```
minha_string = "chapolin colorado"
print(minha_string)
```
**Alguns métodos de strings**
(para mais informações, ver: `https://docs.python.org/3/library/stdtypes.html#`)
* `capitalize()`: retorna uma cópia da string com o primeiro caracter em maiúsculo e o resto em minúsculo. **Mudança versão 3.8**: o primeiro caracter agora é colocado em titlecase, ao invés de uppercase. Isso quer dizeruqe caracteres como dígrafos só terão sua primeira letra capitalizada, ao invés de o caracter completo;
* `isalpha()`: retorna `True` se todos os caracteres da string são alfabéticos;
* `isnumeric()`: retorna `True` se todos os caracteres da string são numéricos;
* `isupper()`: retorna `True` se todos os caracteres da string são maiúsculos;
* `islower()`: retorna `True` se há pelo menos um caractere ASCII minúsculo na string e nenhum caractere ASCII maiúsculo;
* `split()`: retorna uma lista das palavras da string, usando `sep` como o delimitador da string;
* `title()`: retorna uma versão em title case da string, onde palavras começam com um caractere maiúsculo e os demais são minúsculos;
* `strip()`: retorna uma cópia da string removendo os caracteres vazios no início e no final da string. Também existem os métodos `lstrip()` e `rstrip()`, que removem caracteres vazios apenas do lado esquerdo ou do direito da string, respectivamente;
* `replace()`: retorna uma cópia da string com todas as ocorrências da substring `old` substituída por `new`. Se o argumento opcional `count` for dado, somente as primeiras `count` ocorrências serão substituídas.
```
print(f"capitalize: {minha_string.capitalize()}")
print(f"isalpha: {minha_string.isalpha()}")
print(f"isnumeric: {minha_string.isnumeric()}")
print(f"isupper: {minha_string.isupper()}")
print(f"islower: {minha_string.islower()}")
print(f"split: {minha_string.split()}")
print(f"title: {minha_string.title()}")
print(f"strip: {minha_string.strip()}")
print(f"replace: {minha_string.replace('o', 'a')}")
uma_string = " Meu nome é Fulano "
print(f"lstrip: {uma_string.lstrip()}")
print(f"rstrip: {uma_string.rstrip()}")
print(f"strip: {uma_string.strip()}")
# Fatiamento de strings
print(minha_string[:8])
print(minha_string[9:])
print(minha_string[::2])
```
**Exercício 1:** Em uma competição de salto em distância, cada atleta tem direito a cinco saltos. O resultado do atleta será determinado pela média dos cinco valores restantes. Você deve fazer um programa que receba o nome e as cinco distâncias alcançadas pelo atleta e seus saltos e depois informe o nome, os saltos e a média dos saltos. O programa deve ser encerrado quando não for informado o nome do atleta.
```
while True:
atleta = str(input(f"Digite o nome do atleta: "))
if atleta == "":
break
saltos_atleta = []
soma_saltos = 0
for tentativa in range(5):
salto = float(input(f"Digite o valor do {tentativa + 1}o. salto de {atleta}: "))
saltos_atleta.append(salto)
soma_saltos += salto
media = soma_saltos / 5
print(f"\nResultado final:\nAtleta: {atleta}\nSaltos:", end = " ")
for salto in saltos_atleta[0:4]:
print(f"{salto} - ", end = "")
print(f"{saltos_atleta[4]}.\nMédia dos saltos: {media:.2f} m.\n")
```
**Exercício 2:** Faça um programa que calcule o fatorial de um número inteiro fornecido pelo
usuario. Ex: 5! = 5 . 4 . 3 . 2 . 1 = 120. A saida deve ser conforme o exemplo abaixo:
Fatorial de 5:
5! = 5 . 4 . 3 . 2 . 1 = 120
```
def fatorial(x):
'''
Função recursiva que retorna o fatorial de x.
'''
if x == 1 or x == 0:
return 1
while x > 1:
return x * fatorial(x - 1)
def imprime_fatorial(x):
'''
Função que imprime de maneira organizada o passo a passo do cálculo do fatorial de x.
'''
print(f"Fatorial de: {x}\n{x}! = ", end = "")
for i in range(x, 1, -1):
print(f"{i} . ", end = "")
fat = fatorial(x)
print(f"1 = {fat}")
# testando programa com fatoriais de 1 até 10
for i in range(1, 11):
imprime_fatorial(i)
print()
```
**Exercício 3:** As Organizações Tabajara resolveram dar um abono aos seus colaboradores em
reconhecimento ao bom resultado alcançado durante o ano que passou. Para isto contratou você para desenvolver a aplicação que servirá como uma projeção de quanto será gasto com o pagamento deste abono.
o Após reuniões envolvendo a diretoria executiva, a diretoria financeira e
os representantes do sindicato laboral, chegou-se a seguinte forma de
cálculo:
o a.Cada funcionário receberá o equivalente a 20% do seu salário bruto
de dezembro; a.O piso do abono será de 100 reais, isto é, aqueles
funcionários cujo salário for muito baixo, recebem este valor mínimo;
Neste momento, não se deve ter nenhuma preocupação com colaboradores
com tempo menor de casa, descontos, impostos ou outras particularidades.
Seu programa deverá permitir a digitação do salário de um número
indefinido (desconhecido) de salários. Um valor de salário igual a 0
(zero) encerra a digitação. Após a entrada de todos os dados o programa
deverá calcular o valor do abono concedido a cada colaborador, de acordo
com a regra definida acima. Ao final, o programa deverá apresentar:
o O salário de cada funcionário, juntamente com o valor do abono;
o O número total de funcionário processados;
o O valor total a ser gasto com o pagamento do abono;
o O número de funcionário que receberá o valor mínimo de 100 reais;
o O maior valor pago como abono; A tela abaixo é um exemplo de execução do
programa, apenas para fins ilustrativos. Os valores podem mudar a cada
execução do programa.
Projeção de Gastos com Abono
Salário: 1000
Salário: 300
Salário: 500
Salário: 100
Salário: 4500
Salário: 0
Salário - Abono
RS 1000.00 - RS 200.00
RS 300.00 - RS 100.00
RS 500.00 - RS 100.00
RS 100.00 - RS 100.00
RS 4500.00 - RS 900.00
Foram processados 5 colaboradores
Total gasto com abonos: RS 1400.00
Valor mínimo pago a 3 colaboradores
Maior valor de abono pago: RS 900.00
```
# inicializa variaveis importantes
salarios = []
abonos = []
minimo = maior = 0
# calcula e armazena os salários entrados pelo teclado
while True:
salario = float(input("Salário: "))
if salario == 0:
break
if salario > maior:
maior = salario
abono = salario * 0.20
if abono <= 100:
abono = 100.0
minimo += 1
salarios.append(salario)
abonos.append(abono)
# imprime os resultados da consulta anterior
print(f"Salário - Abono ")
soma_abonos = 0
for salario, abono in zip(salarios, abonos):
soma_abonos += abono
print(f"R$ {salario:>7.2f} - R$ {abono:>7.2f}")
print(f"Foram processados {len(salarios)} colaboradores.")
print(f"Total gasto com abonos: R$ {soma_abonos:.2f}.")
print(f"Valor mínimo pago a {minimo:.2f} colaboradores.")
print(f"Maior valor de abono pago: R$ {maior:.2f}.")
# padrão ASCII
for i in range(1000):
print(f"{i} - {chr(i):.3}", end = " ")
```
**Exercício 4:** Implemente a cifra de César com os métodos de string e com as outras coisas já aprendidas até aqui.
Para entender o que é a cifra de César, consulte a Wikipédia: `https://pt.wikipedia.org/wiki/Cifra_de_César`
```
TAM_MAX_CH = 26 # este é o tamanho máximo da chave
def recebe_modo():
"""
Função que pergunta se o usuario quer criptografar (True) ou descriptografar (False).
"""
while True:
escolha = str(input("Você deseja criptografar (c) ou descriptografar (d)? "))
if escolha[0].lower() == "c":
return escolha[0]
elif escolha[0].lower() == "d":
return escolha[0]
def recebe_chave():
"""
Recebe uma chave e retorna o valor, se ele for um valor válido.
"""
global TAM_MAX_CH
while True:
chave = int(input(f"Entre com o número da chave (1-{TAM_MAX_CH}): "))
if 1 <= chave <= TAM_MAX_CH:
return chave
def gera_msg_traduzida(modo, mensagem, chave):
"""
Recebe modo (criptografar ou descriptografar), mensagem e chave e retorna mensagem traduzida.
"""
global TAM_MAX_CH
traducao = mensagem
if modo == "d":
chave *= - 1
traduzido = ""
for simbolo in mensagem:
if simbolo.isalpha():
num = ord(simbolo)
num += chave
if simbolo.isupper():
if num > ord("Z"):
num -= TAM_MAX_CH
elif num < ord("A"):
num += TAM_MAX_CH
elif simbolo.islower():
if num > ord("z"):
num -= TAM_MAX_CH
elif num < ord("a"):
num += TAM_MAX_CH
traduzido += chr(num)
else:
traduzido += simbolo
return traduzido
def main():
modo = recebe_modo()
mensagem = str(input("Insira a mensagem a ser criptografada: "))
chave = recebe_chave()
print("Seu texto traduzido é: ")
print(gera_msg_traduzida(modo, mensagem, chave))
# Criptografando uma mensagem
# mensagem: Vamos criptografar esta mensagem aqui e ver o que ira rolar quando fizermos isso
# chave: 5
main()
# Descriptografando a mensagem
# mensagem: Afrtx hwnuytlwfkfw jxyf rjsxfljr fvzn j ajw t vzj nwf wtqfw vzfsit knejwrtx nxxt
# chave: 5
main()
```
**Exercício 5:** Implemente o jogo da forca usando o Python.
```
import random
forca_imagem = ["""
+---+
| |
|
|
|
|
=========""","""
+---+
| |
o |
|
|
|
=========""","""
+---+
| |
o |
| |
|
|
=========""","""
+---+
| |
o |
/| |
|
|
=========""","""
+---+
| |
o |
/|\ |
|
|
=========""","""
+---+
| |
o |
/|\ |
/ |
|
=========""","""
+---+
| |
o |
/|\ |
/ \ |
|
========="""]
palavras = "formiga babuino encefalo elefante girafa hamburguer chocolate giroscopio".split()
def main():
"""
Função principal do programa.
"""
global forca_imagem
print("F O R C A")
letras_erradas = ""
letras_acertadas = ""
palavra_secreta = gera_palavra_aleatória().upper()
jogando = True
while jogando:
imprime_jogo(letras_erradas, letras_acertadas, palavra_secreta)
palpite = recebe_palpite(letras_erradas + letras_acertadas)
if palpite in palavra_secreta:
letras_acertadas += palpite
if verifica_se_ganhou(palavra_secreta, letras_acertadas):
print(f"Exato! A palavra secreta e {palavra_secreta}! Você ganhou!")
jogando = False
else:
letras_erradas += palpite
if len(letras_erradas) == len(forca_imagem) - 1:
imprime_jogo(letras_erradas, letras_acertadas, palavra_secreta)
print("GAME OVER! Você excedeu o seu limite de palpites!")
print(f"Depois de {len(letras_erradas)} letras erradas e {len(letras_acertadas)} palpites corretos,", end = " ")
print(f"a palavra correta era {palavra_secreta}.")
jogando = False
if not jogando:
if jogar_novamente():
letras_erradas = ""
letras_acertadas = ""
jogando = True
palavra_secreta = gera_palavra_secreta().upper()
def gera_palavra_aleatória():
"""
Função que retorna uma string a partir da lista de palavras global.
"""
global palavras
return random.choice(palavras)
def imprime_com_espaços(palavra):
"""
Recebe uma string palavra ou lista e imprime essa palavra com espaço entre suas letras ou strings.
"""
for letra in palavra:
print(letra, end = " ")
print()
def recebe_palpite(palpites_feitos):
"""
Função feita para garantir que o usuario coloque uma entrada válida, ou seja, que seja uma única letra que
ele ainda não tenha chutado.
"""
while True:
palpite = input("Adivinhe uma letra.\n").upper()
if len(palpite) != 1:
print("Coloque uma única letra.")
elif palpite in palpites_feitos:
print("Você já chutou esta letra. Escolha novamente.")
elif not "A" <= palpite <= "Z":
print("Por favor, escolha apenas letras.")
else:
return palpite
def imprime_jogo(letras_erradas, letras_acertadas, palavra_secreta):
"""
Feito a partir da variavel global que contém as imagens do jogo em ASCII art, e também as letras chutadas de
maneira correta e as letras erradas e a palavra secreta."""
global forca_imagem
print(forca_imagem[len(letras_erradas)] + "\n")
print("Letras erradas:", end = " ")
imprime_com_espaços(letras_erradas)
vazio = "_" * len(palavra_secreta)
for i in range(len(palavra_secreta)):
if palavra_secreta[i] in letras_acertadas:
vazio = vazio[:i] + palavra_secreta[i] + vazio[i + 1:]
imprime_com_espaços(vazio)
def jogar_novamente():
"""
Função que pede para o usuario decidir se ele quer jogar novamente e retorna um booleano representando a resposta.
"""
return input("Você quer jogar novamente? (S - Sim; N - Não):\n").upper().startswith("S")
def verifica_se_ganhou(palavra_secreta, letras_acertadas):
"""
Função verifica se o usuário acertou todas as letras da palavra secreta."""
ganhou = True
for letra in palavra_secreta:
if letra not in letras_acertadas:
ganhou = False
break
return ganhou
# rodando o jogo
main()
```
|
github_jupyter
|
minha_string = "chapolin colorado"
print(minha_string)
print(f"capitalize: {minha_string.capitalize()}")
print(f"isalpha: {minha_string.isalpha()}")
print(f"isnumeric: {minha_string.isnumeric()}")
print(f"isupper: {minha_string.isupper()}")
print(f"islower: {minha_string.islower()}")
print(f"split: {minha_string.split()}")
print(f"title: {minha_string.title()}")
print(f"strip: {minha_string.strip()}")
print(f"replace: {minha_string.replace('o', 'a')}")
uma_string = " Meu nome é Fulano "
print(f"lstrip: {uma_string.lstrip()}")
print(f"rstrip: {uma_string.rstrip()}")
print(f"strip: {uma_string.strip()}")
# Fatiamento de strings
print(minha_string[:8])
print(minha_string[9:])
print(minha_string[::2])
while True:
atleta = str(input(f"Digite o nome do atleta: "))
if atleta == "":
break
saltos_atleta = []
soma_saltos = 0
for tentativa in range(5):
salto = float(input(f"Digite o valor do {tentativa + 1}o. salto de {atleta}: "))
saltos_atleta.append(salto)
soma_saltos += salto
media = soma_saltos / 5
print(f"\nResultado final:\nAtleta: {atleta}\nSaltos:", end = " ")
for salto in saltos_atleta[0:4]:
print(f"{salto} - ", end = "")
print(f"{saltos_atleta[4]}.\nMédia dos saltos: {media:.2f} m.\n")
def fatorial(x):
'''
Função recursiva que retorna o fatorial de x.
'''
if x == 1 or x == 0:
return 1
while x > 1:
return x * fatorial(x - 1)
def imprime_fatorial(x):
'''
Função que imprime de maneira organizada o passo a passo do cálculo do fatorial de x.
'''
print(f"Fatorial de: {x}\n{x}! = ", end = "")
for i in range(x, 1, -1):
print(f"{i} . ", end = "")
fat = fatorial(x)
print(f"1 = {fat}")
# testando programa com fatoriais de 1 até 10
for i in range(1, 11):
imprime_fatorial(i)
print()
# inicializa variaveis importantes
salarios = []
abonos = []
minimo = maior = 0
# calcula e armazena os salários entrados pelo teclado
while True:
salario = float(input("Salário: "))
if salario == 0:
break
if salario > maior:
maior = salario
abono = salario * 0.20
if abono <= 100:
abono = 100.0
minimo += 1
salarios.append(salario)
abonos.append(abono)
# imprime os resultados da consulta anterior
print(f"Salário - Abono ")
soma_abonos = 0
for salario, abono in zip(salarios, abonos):
soma_abonos += abono
print(f"R$ {salario:>7.2f} - R$ {abono:>7.2f}")
print(f"Foram processados {len(salarios)} colaboradores.")
print(f"Total gasto com abonos: R$ {soma_abonos:.2f}.")
print(f"Valor mínimo pago a {minimo:.2f} colaboradores.")
print(f"Maior valor de abono pago: R$ {maior:.2f}.")
# padrão ASCII
for i in range(1000):
print(f"{i} - {chr(i):.3}", end = " ")
TAM_MAX_CH = 26 # este é o tamanho máximo da chave
def recebe_modo():
"""
Função que pergunta se o usuario quer criptografar (True) ou descriptografar (False).
"""
while True:
escolha = str(input("Você deseja criptografar (c) ou descriptografar (d)? "))
if escolha[0].lower() == "c":
return escolha[0]
elif escolha[0].lower() == "d":
return escolha[0]
def recebe_chave():
"""
Recebe uma chave e retorna o valor, se ele for um valor válido.
"""
global TAM_MAX_CH
while True:
chave = int(input(f"Entre com o número da chave (1-{TAM_MAX_CH}): "))
if 1 <= chave <= TAM_MAX_CH:
return chave
def gera_msg_traduzida(modo, mensagem, chave):
"""
Recebe modo (criptografar ou descriptografar), mensagem e chave e retorna mensagem traduzida.
"""
global TAM_MAX_CH
traducao = mensagem
if modo == "d":
chave *= - 1
traduzido = ""
for simbolo in mensagem:
if simbolo.isalpha():
num = ord(simbolo)
num += chave
if simbolo.isupper():
if num > ord("Z"):
num -= TAM_MAX_CH
elif num < ord("A"):
num += TAM_MAX_CH
elif simbolo.islower():
if num > ord("z"):
num -= TAM_MAX_CH
elif num < ord("a"):
num += TAM_MAX_CH
traduzido += chr(num)
else:
traduzido += simbolo
return traduzido
def main():
modo = recebe_modo()
mensagem = str(input("Insira a mensagem a ser criptografada: "))
chave = recebe_chave()
print("Seu texto traduzido é: ")
print(gera_msg_traduzida(modo, mensagem, chave))
# Criptografando uma mensagem
# mensagem: Vamos criptografar esta mensagem aqui e ver o que ira rolar quando fizermos isso
# chave: 5
main()
# Descriptografando a mensagem
# mensagem: Afrtx hwnuytlwfkfw jxyf rjsxfljr fvzn j ajw t vzj nwf wtqfw vzfsit knejwrtx nxxt
# chave: 5
main()
import random
forca_imagem = ["""
+---+
| |
|
|
|
|
=========""","""
+---+
| |
o |
|
|
|
=========""","""
+---+
| |
o |
| |
|
|
=========""","""
+---+
| |
o |
/| |
|
|
=========""","""
+---+
| |
o |
/|\ |
|
|
=========""","""
+---+
| |
o |
/|\ |
/ |
|
=========""","""
+---+
| |
o |
/|\ |
/ \ |
|
========="""]
palavras = "formiga babuino encefalo elefante girafa hamburguer chocolate giroscopio".split()
def main():
"""
Função principal do programa.
"""
global forca_imagem
print("F O R C A")
letras_erradas = ""
letras_acertadas = ""
palavra_secreta = gera_palavra_aleatória().upper()
jogando = True
while jogando:
imprime_jogo(letras_erradas, letras_acertadas, palavra_secreta)
palpite = recebe_palpite(letras_erradas + letras_acertadas)
if palpite in palavra_secreta:
letras_acertadas += palpite
if verifica_se_ganhou(palavra_secreta, letras_acertadas):
print(f"Exato! A palavra secreta e {palavra_secreta}! Você ganhou!")
jogando = False
else:
letras_erradas += palpite
if len(letras_erradas) == len(forca_imagem) - 1:
imprime_jogo(letras_erradas, letras_acertadas, palavra_secreta)
print("GAME OVER! Você excedeu o seu limite de palpites!")
print(f"Depois de {len(letras_erradas)} letras erradas e {len(letras_acertadas)} palpites corretos,", end = " ")
print(f"a palavra correta era {palavra_secreta}.")
jogando = False
if not jogando:
if jogar_novamente():
letras_erradas = ""
letras_acertadas = ""
jogando = True
palavra_secreta = gera_palavra_secreta().upper()
def gera_palavra_aleatória():
"""
Função que retorna uma string a partir da lista de palavras global.
"""
global palavras
return random.choice(palavras)
def imprime_com_espaços(palavra):
"""
Recebe uma string palavra ou lista e imprime essa palavra com espaço entre suas letras ou strings.
"""
for letra in palavra:
print(letra, end = " ")
print()
def recebe_palpite(palpites_feitos):
"""
Função feita para garantir que o usuario coloque uma entrada válida, ou seja, que seja uma única letra que
ele ainda não tenha chutado.
"""
while True:
palpite = input("Adivinhe uma letra.\n").upper()
if len(palpite) != 1:
print("Coloque uma única letra.")
elif palpite in palpites_feitos:
print("Você já chutou esta letra. Escolha novamente.")
elif not "A" <= palpite <= "Z":
print("Por favor, escolha apenas letras.")
else:
return palpite
def imprime_jogo(letras_erradas, letras_acertadas, palavra_secreta):
"""
Feito a partir da variavel global que contém as imagens do jogo em ASCII art, e também as letras chutadas de
maneira correta e as letras erradas e a palavra secreta."""
global forca_imagem
print(forca_imagem[len(letras_erradas)] + "\n")
print("Letras erradas:", end = " ")
imprime_com_espaços(letras_erradas)
vazio = "_" * len(palavra_secreta)
for i in range(len(palavra_secreta)):
if palavra_secreta[i] in letras_acertadas:
vazio = vazio[:i] + palavra_secreta[i] + vazio[i + 1:]
imprime_com_espaços(vazio)
def jogar_novamente():
"""
Função que pede para o usuario decidir se ele quer jogar novamente e retorna um booleano representando a resposta.
"""
return input("Você quer jogar novamente? (S - Sim; N - Não):\n").upper().startswith("S")
def verifica_se_ganhou(palavra_secreta, letras_acertadas):
"""
Função verifica se o usuário acertou todas as letras da palavra secreta."""
ganhou = True
for letra in palavra_secreta:
if letra not in letras_acertadas:
ganhou = False
break
return ganhou
# rodando o jogo
main()
| 0.318591 | 0.909103 |
**Notas para contenedor de docker:**
Comando de docker para ejecución de la nota de forma local:
nota: cambiar `<ruta a mi directorio>` por la ruta de directorio que se desea mapear a `/datos` dentro del contenedor de docker.
```
docker run --rm -v <ruta a mi directorio>:/datos --name jupyterlab_numerical -p 8888:8888 -d palmoreck/jupyterlab_numerical:1.1.0
```
password para jupyterlab: `qwerty`
Detener el contenedor de docker:
```
docker stop jupyterlab_numerical
```
Documentación de la imagen de docker `palmoreck/jupyterlab_numerical:1.1.0` en [liga](https://github.com/palmoreck/dockerfiles/tree/master/jupyterlab/numerical).
---
Nota generada a partir de [liga1](https://www.dropbox.com/s/qb3swgkpaps7yba/4.1.Introduccion_optimizacion_convexa.pdf?dl=0), [liga2](https://www.dropbox.com/s/6isby5h1e5f2yzs/4.2.Problemas_de_optimizacion_convexa.pdf?dl=0), [liga3](https://www.dropbox.com/s/ko86cce1olbtsbk/4.3.1.Teoria_de_convexidad_Conjuntos_convexos.pdf?dl=0), [liga4](https://www.dropbox.com/s/mmd1uzvwhdwsyiu/4.3.2.Teoria_de_convexidad_Funciones_convexas.pdf?dl=0), [liga5](https://drive.google.com/file/d/1xtkxPCx05Xg4Dj7JZoQ-LusBDrtYUqOF/view).
# Optimización numérica y machine learning
**Optimización de código ¿es optimización numérica?**
Hasta este módulo hemos invertido buena parte del tiempo del curso en la eficiente implementación en el hardware que poseemos. Revisamos lo que estudia el análisis numérico o cómputo científico, definiciones de sistema de punto flotante, funciones, derivadas, integrales y métodos o algoritmos numéricos para su aproximación. Consideramos *bottlenecks* que pueden surgir en la implementación de los métodos o algoritmos y revisamos posibles opciones para encontrarlos y minimizarlos (**optimización de código**). Lo anterior lo resumimos con el uso de herramientas como: perfilamiento, integración de R y Python con C++ o C, cómputo en paralelo y uso del caché de forma eficiente al usar niveles altos en operaciones de BLAS ([módulo I: cómputo científico y análisis numérico](https://github.com/ITAM-DS/analisis-numerico-computo-cientifico/blob/master/temas/I.computo_cientifico), [módulo II: cómputo en paralelo](https://github.com/ITAM-DS/analisis-numerico-computo-cientifico/tree/master/temas/II.computo_paralelo), [módulo III: cómputo matricial](https://github.com/ITAM-DS/analisis-numerico-computo-cientifico/tree/master/temas/III.computo_matricial)).
La **optimización numérica** no es optimización de código sin embargo se apoya enormemente de ella para la implementación de sus métodos o algoritmos en la(s) máquina(s) para resolver problemas que surgen en tal rama de las **matemáticas aplicadas**. A la implementación y simulación en el desarrollo de los métodos o algoritmos del análisis numérico o cómputo científico típicamente se le acompaña de estudios que realizan [benchmarks](https://en.wikipedia.org/wiki/Benchmark_(computing)) y perfilamiento (mediciones de tiempo y memoria, por ejemplo) con el objetivo de tener **software confiable y eficiente** en la práctica. Esto lo encontramos también en la rama de optimización numérica con los métodos o algoritmos que son desarrollados e implementados.
**Métodos o algoritmos numéricos en *big data***
La implementación de los métodos o algoritmos en el contexto de **grandes cantidades de datos** o *big data* es **crítica** al ir a la práctica pues de esto depende que nuestra(s) máquina(s) tarde meses, semanas, días u horas para resolver problemas que se presentan en este contexto. La ciencia de datos apunta al desarrollo de técnicas y se apoya de aplicaciones de *machine learning* para la extracción de conocimiento útil y toma como fuente de información las grandes cantidades de datos.
## ¿Problemas de optimización numérica?
Una gran cantidad de aplicaciones plantean problemas de optimización. Tenemos problemas básicos que se presentan en cursos iniciales de cálculo:
*Una caja con base y tapa cuadradas debe tener un volumen de $100 cm^3$. Encuentre las dimensiones de la caja que minimicen la cantidad de material.*
Y tenemos más especializados que encontramos en áreas como estadística, ingeniería, finanzas o *machine learning*:
* Ajustar un modelo de regresión lineal a un conjunto de datos.
* Buscar la mejor forma de invertir un capital en un conjunto de activos.
* Elección del ancho y largo de un dispositivo en un circuito electrónico.
* Ajustar un modelo que clasifique un conjunto de datos.
En general un problema de optimización matemática o numérica tiene la forma:
$$\displaystyle \min f_o(x)$$
$$\text{sujeto a:} f_i(x) \leq b_i, i=1,\dots, m$$
donde: $x=(x_1,x_2,\dots, x_n)^T$ es la **variable de optimización del problema**, la función $f_o: \mathbb{R}^{n} \rightarrow \mathbb{R}$ es la **función objetivo**, las funciones $f_i: \mathbb{R}^n \rightarrow \mathbb{R}, i=1,\dots,m$ son las **funciones de restricción** (aquí se colocan únicamente desigualdades pero pueden ser sólo igualdades o bien una combinación de ellas) y las constantes $b_1,b_2,\dots, b_m$ son los **límites o cotas de las restricciones**.
Un vector $x^* \in \mathbb{R}^n$ es nombrado **óptimo** o solución del problema anterior si tiene el valor más pequeño de entre todos los vectores $x \in \mathbb{R}^n$ que satisfacen las restricciones. Por ejemplo, si $z \in \mathbb{R}^n$ satisface $f_1(z) \leq b_1, f_2(z) \leq b_2, \dots, f_m(z) \leq b_m$ y $x^*$ es óptimo entonces $f_o(z) \geq f_o(x^*)$.
**Comentarios:**
* En el módulo IV del curso revisaremos métodos o algoritmos de optimización para funciones objetivo $f_o: \mathbb{R}^n \rightarrow \mathbb{R}$. Sin embargo, hay formulaciones que utilizan $f_o: \mathbb{R}^n \rightarrow \mathbb{R}^q$. Tales formulaciones pueden hallarlas en la optimización multicriterio, multiobjetivo, vectorial o también nombrada Pareto, ver [Multi objective optimization](https://en.wikipedia.org/wiki/Multi-objective_optimization).
* Obsérvese que el problema de optimización definido utiliza una forma de minimización y no de maximización. Típicamente en la literatura por convención se consideran problemas de este tipo. Además minimizar $f_o$ y maximizar $-f_o$ son **problemas de optimización equivalentes**\*.
\*A grandes rasgos dos problemas de optimización son equivalentes si con la solución de uno de ellos se obtiene la solución del otro y viceversa.
**Ejemplo:**
1) $$\displaystyle \min_{x \in \mathbb{R}^n} ||x||_2$$
$$\text{sujeto a:} Ax \leq b$$
con $A \in \mathbb{R}^{m \times n}, b \in \mathbb{R}^m$. En este problema buscamos el vector $x$ que es solución del problema $Ax \leq b$ con mínima norma Euclidiana. La función objetivo es $f_o(x)=||x||_2$, las funciones de restricción son las desigualdades lineales $f_i(x) = a_i^Tx \leq b_i$ con $a_i$ $i$-ésimo renglón de $A$ y $b_i$ $i$-ésima componente de $b$, $\forall i=1,\dots,m$.
**Comentario:** un problema similar al anterior lo podemos encontrar en resolver el sistema de ecuaciones lineales $Ax=b$ *underdetermined* en el que $m < n$ y se busca el vector $x$ con mínima norma Euclidiana que satisfaga tal sistema. Tal sistema puede tener infinitas soluciones o ninguna solución, ver [3.3.Solucion_de_SEL_y_FM](https://github.com/ITAM-DS/analisis-numerico-computo-cientifico/blob/master/temas/III.computo_matricial/3.3.Solucion_de_SEL_y_FM.ipynb).
2) Encuentra el punto en la gráfica de $y=x^2$ que es más cercano al punto $P=(1,0)$ bajo la norma Euclidiana.
Deseamos minimizar la cantidad $||(1,0)-(x,y)||_2$. Además $y = y(x)$ por lo que reescribiendo lo anterior se tiene $f_o(x) = ||(1,0)-(x,x^2)||_2=||(1-x,-x^2)||_2=\sqrt{(1-x)^2+x^4}$. Entonces el problema de optimización (sin restricciones) es:
$$\displaystyle \min_{x \in \text{dom}f_o}\sqrt{(1-x)^2+x^4}$$
## ¿Machine learning, statistical machine learning y optimización numérica?
En esta sección relacionamos a *machine learning* con la optimización y se describen diferentes enfoques que se han propuesto para aplicaciones de *machine learning* con métodos de optimización. Lo siguiente **no** pretende ser una exposición extensa **ni** completa sobre *machine learning*, ustedes llevan materias que se enfocan esencialmente a definir esta área, sus objetivos y conceptos más importantes.
En la ciencia de datos se utilizan las aplicaciones desarrolladas en *machine learning* por ejemplo:
* Clasificación de documentos o textos: detección de *spam*.
* [Procesamiento de lenguaje natural](https://en.wikipedia.org/wiki/Natural_language_processing): [named-entity recognition](https://en.wikipedia.org/wiki/Named-entity_recognition).
* [Reconocimiento de voz](https://en.wikipedia.org/wiki/Speech_recognition).
* [Visión por computadora](https://en.wikipedia.org/wiki/Computer_vision): reconocimiento de rostros o imágenes.
* Detección de fraude.
* [Reconocimiento de patrones](https://en.wikipedia.org/wiki/Pattern_recognition).
* Diagnóstico médico.
* [Sistemas de recomendación](https://en.wikipedia.org/wiki/Recommender_system).
Las aplicaciones anteriores involucran problemas como son:
* Clasificación.
* Regresión.
* *Ranking*.
* *Clustering*.
* Reducción de la dimensionalidad.
En cada una de las aplicaciones o problemas anteriores se utilizan **funciones de pérdida** que guían el proceso de aprendizaje. Tal proceso involucra **optimización parámetros** de la función de pérdida. Por ejemplo, si la función de pérdida en un problema de regresión es una pérdida cuadrática $\mathcal{L}(y,\hat{y}) = (\hat{y}-y)^2$ con $\hat{y} = \hat{\beta}_0 + \hat{\beta_1}x$, entonces el vector de parámetros a optimizar (aprender) es $
\beta=
\left[ \begin{array}{c}
\beta_0\\
\beta_1
\end{array}
\right]
$.
*Machine learning* no sólo se apoya de la optimización pues es un área de Inteligencia Artificial\* que utiliza técnicas estadísticas para el diseño de sistemas capaces de aplicaciones como las escritas anteriormente, de modo que hoy en día tenemos *statistical machine learning*. No obstante, uno de los **pilares** de *machine learning* o *statistical machine learning* es la optimización.
\*La IA o inteligencia artificial es una rama de las ciencias de la computación que atrajo un gran interés en $1950$.
*Machine learning* o *statistical machine learning* se apoya de las formulaciones y algoritmos en optimización. Sin embargo, también ha contribuido a ésta área desarrollando nuevos enfoques en los métodos o algoritmos para el tratamiento de grandes cantidades de datos o *big data* y estableciendo retos significativos no presentes en problemas clásicos de optimización. De hecho, al revisar literatura que intersecta estas dos disciplinas encontramos comunidades científicas que desarrollan o utilizan métodos o algoritmos exactos (ver [Exact algorithm](https://en.wikipedia.org/wiki/Exact_algorithm)) y otras que utilizan métodos de optimización estocástica (ver [Stochastic optimization](https://en.wikipedia.org/wiki/Stochastic_optimization) y [Stochastic approximation](https://en.wikipedia.org/wiki/Stochastic_approximation)) basados en métodos o algoritmos aproximados (ver [Approximation algorithm](https://en.wikipedia.org/wiki/Approximation_algorithm)). Hoy en día es común encontrar estudios que hacen referencia a **modelos o métodos de aprendizaje**.
Como ejemplo de lo anterior considérese la técnica de **regularización** que en *machine learning* se utiliza para encontrar soluciones que generalicen y provean una explicación no compleja del fenómeno en estudio. La regularización sigue el principio de la navaja de Occam, ver [Occam's razor](https://en.wikipedia.org/wiki/Occam%27s_razor): para cualquier conjunto de observaciones en general se prefieren explicaciones simples a explicaciones más complicadas. Aunque la técnica de regularización es conocida en optimización, han sido varias las aplicaciones de *machine learning* las que la han posicionado como clave.
## ¿Large scale machine learning?
El inicio del siglo XXI estuvo marcado, entre otros temas, por un incremento significativo en la generación de información. Esto puede contrastarse con el desarrollo de los procesadores de las máquinas, que como se revisó en el módulo II del curso en el tema [2.1.Un_poco_de_historia_y_generalidades](https://github.com/ITAM-DS/analisis-numerico-computo-cientifico/blob/master/temas/II.computo_paralelo/2.1.Un_poco_de_historia_y_generalidades.ipynb), tuvo un menor *performance* al del siglo XX. Asimismo, las mejoras en dispositivos de almacenamiento o *storage* y sistemas de networking abarató costos de almacenamiento y permitió tal incremento de información. En este contexto, los modelos y métodos de *statistical machine learning* se vieron limitados por el tiempo de cómputo y no por el tamaño de muestra. La conclusión de esto fue una inclinación en la comunidad científica por el diseño o uso de métodos o modelos para procesar grandes cantidades de datos usando recursos computacionales comparativamente menores.
Un ejemplo de lo anterior se observa en métodos de optimización desarrollados en la década de los $50$'s. Mientras que métodos tradicionales en optimización basados en el cálculo del gradiente y la Hessiana de una función son efectivos para problemas de aprendizaje *small-scale* (en los que utilizamos un enfoque en ***batch*** o por lote), en el contexto del aprendizaje *large-scale*, el **método de gradiente estocástico**\* se posicionó en el centro de discusiones a inicios del siglo XXI.
\* El método de gradiente estocástico fue propuesto por Robbins y Monro en 1951, es un **algoritmo estocástico**. Ver [Stochastic gradient descent](https://en.wikipedia.org/wiki/Stochastic_gradient_descent).
### Información de primer y segundo orden
Tradicionalmente en optimización, la búsqueda del (o los) **óptimo(s)** involucran el cálculo de información de primer o segundo orden (ver [1.4.Polinomios_de_Taylor_y_diferenciacion_numerica](https://github.com/ITAM-DS/analisis-numerico-computo-cientifico/blob/master/temas/I.computo_cientifico/1.4.Polinomios_de_Taylor_y_diferenciacion_numerica.ipynb)) de la función $f_o$:
**Ejemplo:**
1) Calcular $\nabla f(x), \nabla^2f(x)$ con $f: \mathbb{R}^4 \rightarrow \mathbb{R}$, dada por $f(x) = (x_1-2)^2+(2-x_2)^2+x_3^2+x_4^4$ en el punto $x_0=(1.5,1.5,1.5,1.5)^T$.
**Solución:**
$$\nabla f(x) =
\left[ \begin{array}{c}
2(x_1-2)\\
-2(2-x_2)\\
2x_3\\
4x_4^3
\end{array}
\right] ,
$$
$$\nabla^2f(x)=
\left[\begin{array}{cccc}
2 & 0 & 0 & 0\\
0 & 2 & 0 & 0\\
0 & 0 &2 & 0\\
0 & 0 &0 &12x_3^2
\end{array}
\right]
$$
$$\nabla f(x_0) =
\left[ \begin{array}{c}
-1\\
-1\\
3\\
\frac{27}{2}
\end{array}
\right],
$$
$$\nabla^2f(x_0)=
\left[\begin{array}{cccc}
2 &0&0&0\\
0&2&0&0\\
0 &0&2&0\\
0&0&0&27\\
\end{array}
\right]
$$
La información de primer y segundo orden la constituyen el gradiente de $f$, $\nabla f(x)$, y la matriz Hessiana de $f$, $\nabla^2f(x)$. Obsérvese que el almacenamiento de la Hessiana involucra $\mathcal{O}(n^2)$ entradas. En los métodos clásicos de optimización se utilizan el gradiente y la Hessiana para encontrar el mínimo de funciones. La Hessiana se utiliza para resolver un sistema de ecuaciones lineales asociado.
2) Encontrar el mínimo de $f$.
**Solución:**
```
import numpy as np
x0=np.array([1.5,1.5,1.5,1.5])
gf= lambda x: np.array([2*(x[0]-2),
-2*(2-x[1]),
2*x[2],
4*x[3]**3])
Hf = lambda x: np.array([[2, 0, 0 ,0],
[0, 2, 0, 0],
[0, 0, 2, 0],
[0, 0, 0, 12*x[2]**2]])
gf(x0)
Hf(x0)
```
Como $f$ es una función convexa (definida más adelante) se tiene que su óptimo se obtiene igualando y resolviendo la **ecuación no lineal** $\nabla f(x) = 0$ :
$$\nabla f(x) =
\left[ \begin{array}{c}
2(x_1-2) \\
-2(2-x_2)\\
2x_3\\
4x_4^3
\end{array}
\right]
= 0
$$
El óptimo $x^* \in \mathbb{R}^4$ está dado por:
$$x^*=
\left[ \begin{array}{c}
2\\
2\\
0\\
0
\end{array}
\right]
$$
**¿Cómo encontramos numéricamente el óptimo?**
**Forma1: con el gradiente de $f$**
Numéricamente se puede utilizar un método iterativo en el que iniciamos con un punto inicial $x^{(0)}$ y las actualizaciones las realizamos con el gradiente:
$$x^{(k+1)} = x^{(k)} - \nabla f(x^{(k)})$$
para $k=0,1,2,\dots,$.
**Obs:** A la iteración anterior se le añade la **búsqueda de línea por *backtracking***, ver [Backtracking line search](https://en.wikipedia.org/wiki/Backtracking_line_search) para determinar el máximo paso a realizar en la **dirección de descenso** (que en este caso es el **gradiente**).
En el ejemplo tomando $x^{(0)} = (5,5,1,0)^T$ se tiene:
```
x_0 = np.array([5,5,1,0])
x_1 = x_0 - gf(x_0)
x_1
x_2 = x_1 - gf(x_1)
x_2
x_3 = x_2 - gf(x_2)
x_3
x_4 = x_3 - gf(x_3)
x_4
```
y aquí nos quedaremos ciclando hasta el infinito...
**Forma2: con el gradiente y la Hessiana de $f$**
Otra opción es utilizar la información de segundo orden con la Hessiana y considerar una actualización:
$$x^{(k+1)} = x^{(k)} - \nabla^2 f \left (x^{(k)} \right )^{-1} \nabla f\left(x^{(k)} \right)$$
para $k=0,1,2,\dots,$.
**Obs:** A la iteración anterior se le añade la **búsqueda de línea por *backtracking***, ver [Backtracking line search](https://en.wikipedia.org/wiki/Backtracking_line_search) para determinar el máximo paso a realizar en la **dirección de descenso** (que en este caso es la **dirección de Newton**).
En el ejemplo tomando $x^{(0)} = (5,5,1,0)^T$ se tiene lo siguiente. Recuérdese que **no invertimos** a la matriz y en su lugar resolvemos un sistema de ecuaciones lineales:
```
x_0 = np.array([5,5,1,0])
x_1 = x_0 - np.linalg.solve(Hf(x_0),gf(x_0))
x_1
```
**Comentarios:** de acuerdo al ejemplo anterior:
* Utilizar información de primer o segundo orden nos ayuda a encontrar óptimo(s) de funciones.
* Encontrar al óptimo involucró un método iterativo.
* En términos coloquiales y de forma simplificada, una **dirección de descenso** es aquella que al moverse de un punto a otro en tal dirección, el valor de $f_o$ decrece:
<img src="https://dl.dropboxusercontent.com/s/25bmebx645howjw/direccion_de_descenso_de_Newton_1d.png?dl=0" heigth="600" width="600">
En el dibujo anterior $\hat{f}$ es un modelo cuadrático, $\Delta x_{nt}$ es dirección de descenso de Newton y $x^*$ es el óptimo de $f$. Del punto $(x,f(x))$ nos debemos mover al punto $(x+\Delta x_{nt}, f(x + \Delta x_{nt}))$ para llegar al óptimo y el valor de $f$ decrece: $f(x+\Delta x_{nt}) < f(x)$.
* Con la información de primer orden no alcanzamos al óptimo (de hecho se cicla el método iterativo propuesto) pero con la de segundo orden sí lo alcanzamos en una iteración y tuvimos que resolver un sistema de ecuaciones lineales.
* Si consideramos una reescritura en la actualización de **descenso en gradiente**:
$$x^{(k+1)} = x^{(k)} - \nabla f(x^{(k)})$$
de la forma:
$$x^{(k+1)} = x^{(k)} - t_{k}\nabla f(x^{(k)})$$
para $t_{k} > 0 $. Con $t_0=0.5$ llegamos al óptimo en una iteración:
```
t_0=0.5
```
**Comentario: más adelante veremos cómo obtener las $t_{k}$s de la búsqueda de línea por backtracking**
```
gf(x_0)
x_1 = x_0 - t_0*gf(x_0)
x_1
```
* El gradiente involucra menos almacenamiento en memoria que el almacenamiento de la Hessiana: $\mathcal{O}(n)$ vs $\mathcal{O}(n^2)$.
* La actualización considerando la **dirección de descenso de Newton** (que involucra a la Hessiana) y la búsqueda de línea por *backtracking* es:
$$x^{(k+1)} = x^{(k)} - t_{k}\nabla^2 f \left (x^{(k)} \right )^{-1} \nabla f\left(x^{(k)} \right)$$
para $k=0,1,2,\dots$ y $t_{k} >0$.
### Batch algoritmhs and stochastic algorithms
**Ejemplo: regresión lineal**
**comentario: para este ejemplo la variable de optimización no será $x$, será $\beta$.**
Supóngase que se han realizado mediciones de un fenómeno de interés en diferentes puntos $x_i$'s resultando en cantidades $y_i$'s $\forall i=0,1,\dots, m$ (se tienen $m+1$ puntos) y además las $y_i$'s contienen un ruido aleatorio causado por errores de medición:
<img src="https://dl.dropboxusercontent.com/s/iydpi0m8ndqzb0s/mcuadrados_1.jpg?dl=0" heigth="350" width="350">
El objetivo de los mínimos cuadrados es construir una curva, $f(x|\beta)$ que "mejor" se ajuste a los datos $(x_i,y_i)$, $\forall i=0,1,\dots,m$. El término de "mejor" se refiere a que la suma: $$\displaystyle \sum_{i=0}^m (y_i -f(x_i|\beta))^2$$ sea lo más pequeña posible, esto es, a que la suma de las distancias verticales entre $y_i$ y $f(x_i|\beta)$ $\forall i=0,1,\dots,m$ al cuadrado sea mínima. Por ejemplo:
<img src="https://dl.dropboxusercontent.com/s/0dhzv336jj6ep4z/mcuadrados_2.jpg?dl=0" heigth="350" width="350">
**Comentarios:**
* La notación $f(x|\beta)$ se utiliza para denotar que $\beta$ es un vector de parámetros a estimar, en específico $\beta_0, \beta_1, \dots \beta_n$, esto es: $n+1$ parámetros a estimar.
* La variable de optimización es $\beta$.
* Tomando $m=3$ y $A \in \mathbb{R}^{3 \times 2}$ geométricamente el problema de **mínimos cuadrados lineales** se puede visualizar:
<img src="https://dl.dropboxusercontent.com/s/a6pjx0pdqa3cp60/mc_beta.png?dl=0" heigth="400" width="400">
donde: $r(\beta) = y-A\beta$, el vector $y \in \mathbb{R}^m$ contiene las entradas $y_i$'s y la matriz $A \in \mathbb{R}^{m \times n}$ contiene a las entradas $x_i$'s o funciones de éstas $\forall i=0,1,\dots,m$. Por el dibujo se tiene que cumplir que $A^Tr(\beta)=0$, esto es: las columnas de $A$ son ortogonales a $r(\beta)$. La condición anterior conduce a las **ecuaciones normales**:
$$0=A^Tr(\beta)=A^T(y-A\beta)=A^Ty-A^TA\beta.$$
* Finalmente, considerando la variable de optimización $\beta$ y al vector $y$ tenemos: $A^TA \beta = A^Ty$.
## Modelo en mínimos cuadrados lineales (u ordinarios)
En los mínimos cuadrados lineales se supone: $f(x|\beta) = \displaystyle \sum_{j=0}^n\beta_j\phi_j(x)$ con $\phi_j: \mathbb{R} \rightarrow \mathbb{R}$ funciones conocidas por lo que se tiene una gran flexibilidad para el proceso de ajuste. Con las funciones $\phi_j (\cdot)$ se construye a la matriz $A$.
**Obs:**
* Si $n=m$ entonces se tiene un problema de **interpolación**.
* x se nombra variable **regresora**.
## ¿Cómo ajustar el modelo anterior?
En lo siguiente se **asume** $n+1 \leq m+1$ (tenemos más puntos $(x_i,y_i)$'s que parámetros a estimar).
### Forma 1
Para realizar el ajuste de mínimos cuadrados lineales se utilizan las ecuaciones normales: $$A^TA\beta=A^Ty$$ donde: $A$ se construye con las $\phi_j$'s evaluadas en los puntos $x_i$'s, el vector $\beta$ contiene a los parámetros $\beta_j$'s a estimar y el vector $y$, la variable **respuesta**, se construye con los puntos $y_i$'s:
$$A = \left[\begin{array}{cccc}
\phi_0(x_0) &\phi_1(x_0)&\dots&\phi_n(x_0)\\
\phi_0(x_1) &\phi_1(x_1)&\dots&\phi_n(x_1)\\
\vdots &\vdots& \vdots&\vdots\\
\phi_0(x_n) &\phi_1(x_n)&\dots&\phi_n(x_n)\\
\vdots &\vdots& \vdots&\vdots\\
\phi_0(x_{m-1}) &\phi_1(x_{m-1})&\dots&\phi_n(x_{m-1})\\
\phi_0(x_m) &\phi_1(x_m)&\dots&\phi_n(x_m)
\end{array}
\right] \in \mathbb{R}^{(m+1)x(n+1)},
\beta=
\left[\begin{array}{c}
\beta_0\\
\beta_1\\
\vdots \\
\beta_n
\end{array}
\right] \in \mathbb{R}^{n+1},
y=
\left[\begin{array}{c}
y_0\\
y_1\\
\vdots \\
y_m
\end{array}
\right] \in \mathbb{R}^{m+1}
$$
y si $A$ es de $rank$ completo (tiene $n+1$ columnas linealmente independientes) se calcula la factorización $QR$ de $A$ : $A = QR$ y entonces: $$A^TA\beta = A^Ty$$
y como $A=QR$ se tiene: $A^TA = (R^TQ^T)(QR)$ y $A^T = R^TQ^T$ por lo que:
$$(R^TQ^T)(QR) \beta = R^TQ^T y$$
y usando que $Q$ tiene columnas ortonormales:
$$R^TR\beta = R^TQ^Ty$$
Como $A$ tiene $n+1$ columnas linealmente independientes, la matriz $R$ es invertible por lo que $R^T$ también lo es y finalmente se tiene el **sistema de ecuaciones lineales** a resolver:
$$R\beta = Q^Ty$$
**Caso regresión lineal**
En el caso de la **regresión lineal** se ajusta un modelo de la forma: $f(x|\beta) = \beta_0 + \beta_1 x$ a los datos $(x_i,y_i)$'s $\forall i=0,1,\dots,m$.
**Obs:** En este caso se eligen $\phi_0(x) = 1$, $\phi_1(x) =x$. Y tenemos que estimar dos parámetros: $\beta_0, \beta_1$.
```
import matplotlib.pyplot as plt
import pprint
from scipy.linalg import solve_triangular
np.random.seed(1989) #para reproducibilidad
mpoints = 20
x = np.random.randn(mpoints)
y = -3*x + np.random.normal(2,1,mpoints)
```
##### Los datos ejemplo
```
plt.plot(x,y, 'r*')
plt.xlabel('x')
plt.ylabel('y')
plt.title('Puntos ejemplo')
plt.show()
```
##### El ajuste
Con numpy podemos usar la función `polyfit` en el paquete de `numpy` para realizar el ajuste: (ver [numpy.polyfit](https://docs.scipy.org/doc/numpy/reference/generated/numpy.polyfit.html))
```
# el tercer argumento de polyfit especifica el grado del polinomio a ajustar.
#Usaremos ngrado = 1 pues queremos ajustar una recta
ngrado = 1
coeficientes = np.polyfit(x,y,ngrado)
#Una vez realizado el llamado a la función polyfit se regresan los coeficientes de x
#ordenados del mayor grado al menor.
pprint.pprint(coeficientes)
```
Entonces nuestro polinomio es: $$p_{1}(x) = -2.65x + 2.03$$
y así tenemos nuestras beta's ajustadas $\hat{\beta_0} = 2.03$, $\hat{\beta_1} = -2.65$.
##### La gráfica
Ahora nos gustaría graficar el modelo en el intervalo $[min(x),max(x)]$ con $min(x)$ la entrada con valor mínimo del numpy array $x$ y $max(x)$ su entrada con valor máximo.
Para lo anterior debemos obtener los valores ajustados al evaluar $p_1(x)$ los valores de $x$:
```
y_ajustadas_numpy = coeficientes[1] + coeficientes[0] * x
plt.plot(x, y_ajustadas_numpy, 'k-',x, y, 'r*')
plt.legend(['modelo lineal','datos'], loc='best')
plt.show()
```
##### También podemos obtener las y's ajustadas con la factorización QR:
```
#construimos a la matriz A:
A=np.ones((mpoints,2))
A[:,1] = x
A
Q,R = np.linalg.qr(A)
#Resolvemos el sistema R*beta = Q^T*y
beta = solve_triangular(R,Q.T@y)
pprint.pprint(beta)
y_ajustadas_QR = A@beta
#obsérvese que la línea anterior es equivalente a realizar:
#y_ajustadas_QR = beta[0] + beta[1]*x
plt.plot(x, y_ajustadas_QR , 'k-',x, y, 'r*')
plt.legend(['modelo lineal','datos'], loc='best')
plt.show()
```
### Forma 2
Usamos optimización numérica reescribiendo la función objetivo:
$$f_o(\beta)=\displaystyle \sum_{i=1}^{20} (y_i -f_o(x_i|\beta))^2 = \displaystyle \sum_{i=1}^{20} (y_i - (\beta_0 + \beta_1 x_i))^2 = \displaystyle \sum_{i=1}^{20} (y_i - A[i,:]^T\beta)^2 = ||y - A \beta||_2^2$$
con $y \in \mathbb{R}^{20}, A \in \mathbb{R}^{20 \times 2}, \beta \in \mathbb{R}^{2 \times 1}$ y $A[i,:]$ $i$-ésimo renglón de $A$.
Planteamos el problema de optimización numérica:
$$\displaystyle \min_{\beta \in \mathbb{R}^n} ||y - A\beta||_2^2$$
**¿Solución?**
Reescribimos $f_o$ como $f_o(\beta) = ||y-A\beta||_2^2= (y-A\beta)^T(y-A\beta) = y^Ty-2\beta^TA^Ty + \beta^TA^TA\beta$, observa que esta última expresión es un número $\mathbb{R}$.
Por lo anterior: $\nabla f_o(\beta) = -2A^Ty + 2A^TA\beta$. $\nabla f_o$ es un vector en $\mathbb{R}^{2 \times 1}$, de hecho sus entradas son $\nabla f_o(\beta) =
\left [
\begin{array}{c}
\frac{\partial f_o(\beta)}{\beta_0}\\
\frac{\partial f_o(\beta)}{\beta_1}
\end{array}
\right ]
$
.
Si se plantea la **ecuación no lineal**: $\nabla f_o(\beta)= 0$ obtenemos una forma **cerrada** de la solución que está dada por $A^TA \beta=A^Ty$ (y son las ecuaciones normales!).
### ¿Método numérico?
Consideramos **métodos de descenso** para resolver el problema de optimización.
#### Método por batch o lote vía descenso en gradiente
La solución como la encontramos en el ejemplo del inicio, es considerar un método iterativo en el que tomamos un punto inicial $\beta^{(0)}$ y las actualizaciones se realizan:
$$\beta^{(k+1)} = \beta^{(k)} - t_{k}\nabla f\left(\beta^{(k)}\right), k=0,1,2,\dots$$
y $t_{k}$ obtenida por búsqueda de línea con *backtracking* y es una cantidad positiva.
**Obs:** igualar la ecuación lineal $-2A^Ty + 2A^TA\beta$ a 0 es equivalente a resolver la ecuación lineal $-A^Ty + A^TA\beta=0$, por esto utilizamos esta última expresión que corresponde a una **función objetivo** $f_o = \frac{1}{2}y^Ty-\beta^TA^Ty + \frac{1}{2}\beta^TA^TA\beta$.
calculamos primero $-A^Ty$:
```
cte=-np.transpose(A)@y
cte
```
el gradiente de $f_o$: $\nabla f_o(\beta) = -A^Ty + A^TA\beta$:
```
gf = lambda beta_fun: cte + np.transpose(A)@(A@beta_fun)
#observa que no hacemos la multiplicación (A^T*A)*beta, mejor hacemos
#primero A*beta y luego multiplicamos por A^T
```
Iniciamos con las iteraciones dadas por la fórmula de actualización tomando $\beta_0=(0,0)^T$:
```
beta_0 = np.array([0,0])
```
**Comentario: más adelante veremos cómo obtener las $t_{k}$s de la búsqueda de línea por backtracking**
```
t_0=.130
beta_1 = beta_0 - t_0*gf(beta_0)
beta_1
t_1=.0625
beta_2 = beta_1 - t_1*gf(beta_1)
beta_2
t_2 = .0625
beta_3 = beta_2 - t_2*gf(beta_2)
beta_3
t_3 = .0625
beta_4 = beta_3 - t_3*gf(beta_3)
beta_4
t_4 = .0625
beta_5 = beta_4 - t_4*gf(beta_4)
beta_5
y_ajustadas_gradiente = A@beta_5
#obsérvese que la línea anterior es equivalente a realizar:
#y_ajustadas_gradiente = beta_5[0] + beta_5[1]*x
plt.plot(x, y_ajustadas_gradiente , 'k-',x, y, 'r*')
plt.legend(['modelo lineal','datos'], loc='best')
plt.show()
```
#### Método por batch o lote vía descenso por dirección de Newton
Otra opción es utilizar la información de segundo orden con la Hessiana y considerar una actualización:
$$\beta^{(k+1)} = \beta^{(k)} - \nabla^2 f \left (\beta^{(k)} \right )^{-1} \nabla f\left(\beta^{(k)} \right)$$
para $k=0,1,2,\dots,$ (omitiendo la búsqueda de línea por *backtracking*). Recuérdese que **no invertimos** a la matriz y en su lugar resolvemos un sistema de ecuaciones lineales asociado. Para el ejemplo de mínimos cuadrados lineales se tiene que la Hessiana es:
$$\nabla ^2 f(\beta^{(k)}) =A^TA.$$
Y el **sistema de ecuaciones lineales asociado** es: $\nabla ^2 f(\beta ^{(k)}) s_k = - \nabla f(\beta^{(k)})$:
```
s_0 = np.linalg.solve(np.transpose(A)@A,-gf(beta_0))
beta_1 = beta_0 + s_0
beta_1
y_ajustadas_Newton = A@beta_1
#obsérvese que la línea anterior es equivalente a realizar:
#y_ajustadas_Newton = beta_1[0] + beta_1[1]*x
plt.plot(x, y_ajustadas_Newton , 'k-',x, y, 'r*')
plt.legend(['modelo lineal','datos'], loc='best')
plt.show()
```
**Hasta este punto comparemos con lo obtenido previamente por QR o *polyfit***
Para descenso en gradiente con búsqueda de línea por *backtracking* se tiene un error relativo de:
```
np.linalg.norm(beta_5-beta)/np.linalg.norm(beta)
```
Para descenso con dirección de Newton (sin búsqueda de línea por *backtracking*) se tiene un error relativo de::
```
np.linalg.norm(beta_1-beta)/np.linalg.norm(beta)
```
**Comentarios:**
* Tenemos alrededor de $2$ dígitos correctos para descenso en gradiente y búsqueda de línea por *backtracking* y máxima precisión con dirección de Newton sin búsqueda de línea por *backtracking*.
* El cálculo anterior lo realizamos por lote o *batch*.
* El nombre de *batch* o lote se utiliza pues la información de primer o segundo orden es calculada utilizando todos los datos.
>**Algoritmo** de descenso
>> **Dado** un **punto inicial** $x$ en $\text{dom}f$
>> **Repetir** el siguiente bloque para $k=0,1,2,...$
>>> 1. Determinar una dirección de descenso $\Delta x$ (gradiente o Newton por ejemplo).
>>> 2. Búsqueda de línea. Elegir un tamaño de paso $t > 0$.
>>> 3. Hacer la actualización: $x = x + t\Delta x$.
>> **hasta** convergencia (satisfacer criterio de paro).
* La Hessiana en mínimos cuadrados lineales como se vio en este ejemplo es $\nabla^2f(\beta^{(k)}) = A^TA$. **No** es recomendable construir la matriz $A^TA$ por dos razones principalmente:
* Si $A$ es grande (más de $10^8$ entradas) hacer la operación $A^TA$ costará gran cantidad de tiempo por el número de operaciones involucradas (alrededor de $\mathcal{O}(mn^2)$).
* Si $A$ tiene un número de condición $c>0$ se prueba que $A^TA$ tiene un número de condición $c^2$ bajo la norma $2$ . Esto como se revisó en la nota [1.3.Condicion_de_un_problema_y_estabilidad_de_un_algoritmo](https://github.com/ITAM-DS/analisis-numerico-computo-cientifico/blob/master/temas/I.computo_cientifico/1.3.Condicion_de_un_problema_y_estabilidad_de_un_algoritmo.ipynb) afecta en la precisión y exactitud al resolver el problema de encontrar la solución al sistema de ecuaciones lineales $\nabla ^2 f(\beta^{(k)})s_k = - \nabla f(\beta^{(k)})$.
Por lo anterior se **sugiere resolver el sistema de ecuaciones lineales $\nabla ^2 f(\beta ^{(k)}) s_k = - \nabla f(\beta^{(k)})$ dado por: $A^TA s_k = -(-A^Ty + A^TA\beta) $ que resulta de los mínimos cuadrados con la factorización $QR$**:
$$
\begin{eqnarray}
A^TAs_k&=&A^T(y-A\beta^{(k)}) \nonumber \\
R^TRs_k&=&R^TQ^T(y-A\beta^{(k)}) \nonumber\\
Rs_k&=&Q^T(y-A\beta^{(k)}) \nonumber\\
Rs_k&=&Q^T(y-QR\beta^{(k)}) \nonumber \\
Rs_k&=&Q^Ty-R\beta^{(k)} \nonumber
\end{eqnarray}
$$
donde se **asume** de la segunda igualdad a la tercera igualdad que $A$ es de *rank* completo y por lo tanto $R$ y $R^T$ son no singulares (son invertibles).
```
Q,R = np.linalg.qr(A)
s_0 = np.linalg.solve(R,np.transpose(Q)@y-R@beta_0)
beta_1 = beta_0 + s_0
beta_1
y_ajustadas_Newton = A@beta_1
#obsérvese que la línea anterior es equivalente a realizar:
#y_ajustadas_Newton = beta_1[0] + beta_1[1]*x
plt.plot(x, y_ajustadas_Newton , 'k-',x, y, 'r*')
plt.legend(['modelo lineal','datos'], loc='best')
plt.show()
```
**Obsérvese que esta forma es igual a la vista por factorización QR anteriormente ya que se resuelve el sistema de ecuaciones normales $A^TA \beta = A^Ty$.**
#### Método vía gradiente estocástico
Para la formulación del método consideramos a la función objetivo $f_o$ escrita en la forma:
$$f_o(\beta) = \frac{1}{2}\displaystyle \sum_{i=1}^{20} (y_i - A[i,:]^T\beta)^2$$
con $A[i,:]$ $i$-ésimo renglón de $A$ y obsérvese que dividimos por $2$ como antes. Calculamos el gradiente de $f_o$:
$$\nabla f_o (\beta) = -\displaystyle \sum_{i=1}^{20} (y_i - A[i,:]^T\beta)A[i,:]$$
**obs:** La expresión anterior considera $A[i,:] \in \mathbb{R}^{2 \times 1}$,
$A[i,:]=\left[
\begin{array}{c}
1\\
x_i
\end{array}
\right ]
$ $\forall i=1,\dots, 20$ con $x_i$ dato dado en el ejemplo de mínimos cuadrados lineales.
En el método estocástico utilizando el gradiente (nombrado **descenso en gradiente estocástico**), utilizamos un punto inicial $\beta^{(0)}$ y en lugar de usar la actualización:
$$
\begin{eqnarray}
\beta^{(k+1)}&=&\beta^{(k)} - t_{k}\nabla f\left(\beta^{(k)}\right) \nonumber\\
&=& \beta^{(k)} - t_{k}\left(-\displaystyle \sum_{i=1}^{20} (y_i - A[i,:]^T\beta^{(k)})A[i,:]\right)\nonumber\\
\end{eqnarray}
$$
para $k=0,1,2,\dots$ y $t_{k}$ cantidad positiva, obtenida por búsqueda de línea con *backtracking*. **Las actualizaciones en descenso en gradiente estocástico son**:
$$
\begin{eqnarray}
\beta^{(k+1)} &=& \beta^{(k)} - \eta_{{k}}\nabla f_{i_{k}}\left(\beta^{(k)}\right) \nonumber\\
\end{eqnarray}
$$
para $k=0,1,2,\dots$ con $\nabla f_{i_{k}}(\cdot)$ el gradiente calculado a partir de una **muestra extraída del conjunto de índices de los renglones: $\{1,2,\dots,m\}$**.
**Comentarios:**
* La cantidad $\eta_{k}$ en este contexto se le nombra **tasa de aprendizaje** y al igual que en el método de descenso en gradiente es una cantidad positiva.
* La muestra de índices para calcular $\nabla f_{i_{k}}(\cdot)$ puede contener uno o más índices. Si contiene un único índice por ejemplo $i_{k} = \{i\}$ con $i=1,\cdots, m$ se tiene una actualización de la forma:
$$
\begin{eqnarray}
\beta^{(k+1)} &=& \beta^{(k)} - \eta_{{k}}\nabla f_{i_{k}}\left(\beta^{(k)}\right) \nonumber\\
&=& \beta^{(k)} - \eta_{k}(-(y_i - A[i,:]^T\beta^{(k)})A[i,:]) \nonumber \\
&=& \beta^{(k)} + \eta_{k}(y_i - A[i,:]^T\beta^{(k)})A[i,:] \nonumber
\end{eqnarray}
$$
Para el ejemplo numérico de mínimos cuadrados lineales, iniciamos con las iteraciones dadas por la fórmula de actualización tomando $\beta_0=(0,0)^T$ y $\eta_{k} = 0.1 \forall k=0,1,2,\dots$:
```
beta_0 = np.array([0,0])
eta_0 = .1
```
Elegimos un índice de los renglones de $A$ de forma pseudoaleatoria y hacemos la actualización:
```
np.random.seed(1989) #para reproducibilidad
i = np.random.randint(mpoints, size=1)
beta_1 = beta_0 + eta_0*(y[i]-A[i,:].dot(beta_0))*A[i,:]
beta_1
```
El **algoritmo** es:
>**Dado** un punto inicial $\beta_0$.
>**Repetir** el siguiente bloque para $k=0,1,2,\dots$ :
> >1. Extraer una muestra aleatoria $i_{k}$ del conjunto de índices de los renglones de $A$: $\{1,2,\dots,m\}$.
> >2. Calcular la tasa de aprendizaje $\eta_{k}$ (positiva).
> >3. Calcular la actualización $\beta^{(k+1)} = \beta^{(k)} + \eta_{k}\nabla f_{i_{k}}\left(\beta^{(k)}\right)$
>**hasta** convergencia.
Midamos el error relativo con este algoritmo utilizando **sólo un índice** $i$ extraído al azar del conjunto de índices de renglones de $A$ para el ejemplo de mínimos cuadrados lineales: $\beta^{(k+1)} = \beta^{(k)} + \eta_{k}(y_i - A[i,:]^T\beta^{(k)})A[i,:]$
```
def error_relativo(aprox, obj):
return np.linalg.norm(aprox-obj)/np.linalg.norm(obj)
```
Definamos puntos iniciales
```
maxiter=5
tol=1e-2
err=error_relativo(beta_0,beta)
beta_k = beta_0
k=1
eta_k = .1 #constante para todas las iteraciones
err
while(err>tol and k <= maxiter):
i = int(np.random.randint(mpoints, size=1))
beta_k = beta_k+eta_k*(y[i]-A[i,:].dot(beta_k))*A[i,:]
k+=1
eta_k=.1
err=error_relativo(beta_k,beta)
err
```
tenemos un error relativo de $92\%$ con $5$ iteraciones.
```
beta_k
```
Consideremos una **muestra más grande** de índices para calcular $\nabla f_{i_{k}}(\cdot)$ de modo que las actualizaciones ahora son de la forma:
$$
\begin{eqnarray}
\beta^{(k+1)} &=& \beta^{(k)} - \eta_{{k}}\nabla f_{i_{k}}\left(\beta^{(k)}\right) \nonumber\\
&=& \beta^{(k)} - \eta_{k}\displaystyle \sum_{i \in i_{k}} (-(y_i - A[i,:]^T\beta^{(k)})A[i,:]) \nonumber\\
&=& \beta^{(k)} + \eta_{k}\displaystyle \sum_{i \in i_{k}}(y_i - A[i,:]^T\beta^{(k)})A[i,:] \nonumber\\
\end{eqnarray}
$$
para $k=0,1,2,\dots$ e $i_{k}$ una muestra de índices extraída de forma aleatoria de los índices de los renglones de $A$.
Usamos $4$ iteraciones con una muestra de índices de tamaño $5$. Puntos iniciales:
```
maxiter=4
tol=1e-2
err=error_relativo(beta_0,beta)
beta_k = beta_0
k=1
eta_k = .1 #constante para todas las iteraciones
m_sample = 5
err
while(err>tol and k <= maxiter):
idx = np.random.choice(mpoints, m_sample,replace=False) #muestra de tamaño m_sample sin reemplazo
beta_k = beta_k+eta_k*(y[idx]-A[idx,:]@beta_0)@A[idx,:]
k+=1
eta_k=.1
err=error_relativo(beta_k,beta)
err
```
Obsérvese que tenemos un error relativo de $51\%$ con $4$ iteraciones, una muestra de tamaño $5$ y es menor al que se obtuvo con una muestra de tamaño 1 de índices.
```
beta_k
```
**Comentarios:** Algunos comentarios generales para el enfoque por *batch* o lote y el estocástico que podemos realizar son:
* El enfoque por *batch*:
* Busca **direcciones de descenso** en cada iteración vía las actualizaciones de $\{\beta_{k}\}$ hasta convergencia del mínimo de la función objetivo $f_o$.
* Por el punto anterior, la velocidad de convergencia de los métodos iterativos por *batch* oscila entre lineal, superlineal y cuadrática para ejemplos reales pero es **muy dependiente de la cantidad de datos** que se utilicen.
* La convergencia depende del punto inicial y de las cantidades positivas $t_{k}$.
* El cómputo de las direcciones de descenso utilizan toda la información disponible en cada iteración.
* Por la definición de sus actualizaciones, el cómputo de las direcciones de descenso se puede realizar con **cómputo en paralelo**.
* El enfoque estocástico:
* Genera $\beta_{k}$'s cuyo comportamiento está determinado por la secuencia aleatoria $\{i_{k}\}$. De hecho $\{\beta_{k}\}$ es un [proceso estocástico](https://en.wikipedia.org/wiki/Stochastic_process).
* Además del punto anterior, su convergencia depende de la taza de aprendizaje $\eta_{k}$.
* En el contexto de grandes cantidades de datos la **convergencia es independiente del tamaño de los datos**: si aumentamos más cantidad de datos, su velocidad de convergencia es independiente de esto.
* No necesariamente se obtienen direcciones de descenso con este método. Si en **promedio o en esperanza matemática** se calculan direcciones de descenso con las actualizaciones de $\{\beta_k\}$, entonces la secuencia $\{\beta_{k}\}$ puede guiarse hacia el mínimo de la función objetivo $f_o$.
* Por el punto anterior, en iteraciones iniciales podemos tener un gran avance hacia el mínimo pero en iteraciones no iniciales puede quedarse el método oscilando en una región cercana al mínimo (como una canica en un tazón).
* Es un método más complicado de implementar con cómputo en paralelo que la versión *batch*.
* No utilizamos en cada iteración todos los datos disponibles.
* Por el error relativo obtenido en el ejemplo anterior parecería que el método estocástico de descenso en gradiente no es una opción fuerte para problemas de optimización. Y esto para problemas de tamaño chico o mediano es cierto pero para grandes cantidades de datos hay razones prácticas, teóricas e intituitivas\* que posicionan a este método por encima de formulaciones en *batch*.
\*Una razón intuitiva del por qué el método estocástico de descenso en gradiente podría ganarle en velocidad a una formulación en *batch* es la siguiente: supóngase que un **conjunto de entrenamiento** $\mathcal{S}$ consistiera de renglones redundantes. Imaginemos que $\mathcal{S}$ consta de $10$ repeticiones de un conjunto más pequeño $\mathcal{S}_\text{sub}$. Entonces ejecutar un método *batch* sería $10$ veces más costoso que si solamente tuviéramos una única copia de $\mathcal{S}_\text{sub}$. El método estocástico de descenso en gradiente tendría el mismo costo en cualquiera de los dos escenarios ($10$ copias vs $1$ copia) pues elige con misma probabilidad renglones del conjunto $\mathcal{S}_\text{sub}$. En la realidad un conjunto de entrenamiento no contiene duplicados exactos de la información pero en muchas aplicaciones *large-scale* existe (de forma aproximada) información redundante.
## ¿Optimización numérica convexa?
Aplicaciones de *machine learning* conducen al planteamiento de problemas de optimización convexa y no convexa.
Ejemplo de **problemas convexos** los encontramos en la aplicación de clasificación de textos en donde se desea asignar un texto a clases definidas de acuerdo a su contenido (p.ej. determinar si un docu de texto es sobre política). La aplicación anterior puede formularse utilizando **funciones de pérdida convexas**.
Como ejemplos de aplicaciones en el ámbito de la **optimización no convexa** están el reconocimiento de voz y reconocimiento de imágenes. El uso de [redes neuronales](https://en.wikipedia.org/wiki/Artificial_neural_network) [profundas](https://en.wikipedia.org/wiki/Deep_learning)\* ha tenido muy buen desempeño en tales aplicaciones haciendo uso de cómputo en la GPU, ver [2.3.CUDA](https://github.com/ITAM-DS/analisis-numerico-computo-cientifico/blob/master/temas/II.computo_paralelo/2.3.CUDA.ipynb), [ImageNet Classification with Deep Convolutional Neural Networks](https://papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks.pdf), [2012: A Breakthrough Year for Deep Learning](https://medium.com/limitlessai/2012-a-breakthrough-year-for-deep-learning-2a31a6796e73). En este caso se utilizan **funciones objetivo no lineales y no convexas**.
\*Los tipos de redes neuronales profundas, *deep neural networks*, que han sido mayormente usadas en el inicio del siglo XXI son las mismas que las que eran populares en los años $90$'s. El éxito de éstos tipos y su uso primordialmente se debe a la disponibilidad de *larger datasets* y mayores recursos computacionales.
Desde los $40$'s se han desarrollado algoritmos para resolver problemas de optimización, se han analizado sus propiedades y se han desarrollado buenas implementaciones de software. Sin embargo, una clase de problemas de optimización en los que encontramos métodos **efectivos** son los convexos.
**En el módulo IV del curso nos enfocamos mayormente a métodos numéricos para resolver problemas convexos principalmente por lo anterior y porque métodos para optimización no convexa utilizan parte de la teoría de convexidad desarrollada en optimización convexa. Además un buen número de problemas de aprendizaje utilizan funciones de pérdida convexas**.
---
# Definiciones utilizadas en el curso (algunas de ellas...)
En lo que continúa se considera $f_0 = f_o$ (el subíndice "0" y el subíndice "o" son iguales)
# Problema estándar de optimización
$$\displaystyle \min f_o(x)$$
$$\text{sujeto a:}$$
$$f_i(x) \leq 0, \quad \forall i=1,\dots,m$$
$$h_i(x) = 0, \quad \forall i=1,\dots,p$$
con $f_i: \mathbb{R}^n \rightarrow \mathbb{R}$ $\forall i=0,\dots,m$, $h_i: \mathbb{R}^n \rightarrow \mathbb{R}$, $\forall i=1,\dots,p$. $f_i$ son las **restricciones de desigualdad**, $h_i$ son las **restricciones de igualdad**.
## Dominio del problema de optimización
El conjunto de puntos para los que la función objetivo y las funciones de restricción $f_i, h_i$ están definidas se nombra **dominio del problema de optimización**, esto es:
$$\mathcal{D} = \bigcap_{i=0}^m\text{dom}f_i \cap \bigcap_{i=1}^p\text{dom}h_i$$
(en github aparecen los símbolos de intersección más abajo de lo que deberían aparecer...abriendo con jupyter el notebook esto no pasa...)
**Comentarios:**
* Un punto $x \in \mathcal{D}$ se nombra **factible** si satisface las restricciones de igualdad y desigualdad. El conjunto de puntos factibles se nombra **conjunto de factibilidad**.
* El problema anterior se nombra **problema de optimización factible** si existe **al menos un punto factible**, si no se cumple lo anterior entonces es infactible.
**En lo que continúa se asumen todos los puntos en el dominio $\mathcal{D}$.**
## Óptimo del problema de optimización
El valor óptimo del problema se denota como $p^*$. En notación matemática es:
$$p^* = \inf\{f_o(x) | f_i(x) \leq 0, \forall i=1,\dots,m, h_i(x) = 0 \forall i=1,\dots,p\}$$
**Comentarios:**
* Si el problema es **infactible** entonces $p^* = \infty$.
* Si $\exists x_k$ factible tal que $f_o(x_k) \rightarrow -\infty$ para $k \rightarrow \infty$ entonces $p^*=-\infty$ y se nombra **problema de optimización no acotado por debajo**.
## Punto óptimo del problema de optimización
$x^*$ es **punto óptimo** si es factible y $f_o(x^*) = p^*$. El conjunto de óptimos se nombra **conjunto óptimo** y se denota:
$$X_{\text{opt}} = \{x | f_i(x) \leq 0 \forall i=1,\dots,m, h_i(x) =0 \forall i=1,\dots,p, f_o(x) = p^*\}$$
**Comentarios:**
* La propiedad de un punto óptimo $x^*$ es que si $z$ satisface las restricciones $f_i(z) \leq 0$ $\forall i=1,...,m$, $h_i(z)=0$ $\forall i=1,..,p$ se tiene: $f(x^*) \leq f(z)$. Es **óptimo estricto** si $z$ satisface las restricciones y $f_o(x^*) < f_o(z)$.
* Si existe un punto óptimo se dice que el valor óptimo se alcanza y por tanto el problema de optimización tiene solución, es *solvable*.
* Si $X_{\text{opt}} = \emptyset$ se dice que el valor óptimo no se alcanza. Obsérvese que para problemas no acotados nunca se alcanza el valor óptimo.
* Si $x$ es factible y $f_o(x) \leq p^* + \epsilon$ con $\epsilon >0$, $x$ se nombra **$\epsilon$-subóptimo** y el conjunto de puntos $\epsilon$-subóptimos se nombra **conjunto $\epsilon$-subóptimo**.
## Óptimo local
Un punto factible $x^*$ se nombra **óptimo local** si $\exists R > 0$ tal que:
$$f_o(x^*) = \inf \{f_o(z) | f_i(z) \leq 0 \forall i=1,\dots,m, h_i(z) = 0 \forall i=1,\dots, p, ||z-x||_2 \leq R\}.$$
Así, $x^*$ resuelve:
$$\displaystyle \min f_o(z)$$
$$\text{sujeto a:}$$
$$f_i(z) \leq 0, \forall i =1,\dots,m$$
$$h_i(z) =0, \forall i=1,\dots,p$$
$$||z-x||_2 \leq R$$
**Obs:** la palabra **óptimo** se utiliza para **óptimo global**, esto es, no consideramos la última restricción $||z-x||_2 \leq R$ en el problema de optimización y exploramos en todo el $\text{dom}f$.
<img src="https://dl.dropboxusercontent.com/s/xyprhh7erbb6icb/min-max-points-example.png?dl=0" heigth="700" width="700">
## Restricciones activas, no activas y redundantes
Si $x$ es factible y $f_i(x)=0$ entonces la restricción de desigualdad $f_i(x) \leq 0$ se nombra **restricción activa en $x$**. Se nombra **inactiva en $x$** si $f_i(x) <0$ para alguna $i$.
**Comentario:** Las restricciones de igualdad, $h_i(x)$, siempre son activas en el conjunto factible.
Una restricción se nombra **restricción redundante** si al quitarla el conjunto factible no se modifica.
## Problemas de optimización convexa en su forma estándar o canónica
$$\displaystyle \min f_o(x)$$
$$\text{sujeto a:}$$
$$f_i(x) \leq 0 , i=1,\dots,m$$
$$h_i(x)=0, i=1,\dots,p$$
donde: $f_i$ son **convexas** $\forall i=0,2,\dots,m$ y $h_i$ $\forall i =1,\dots,p$ son **funciones afín**\*.
\*Una función afín es de la forma $h(x) = Ax+b$ con $A \in \mathbb{R}^{p \times n}$ y $b \in \mathbb{R}^p$. En la definición anterior $h_i(x) = a_i^Tx-b_i$ con $a_i \in \mathbb{R}^n$, $b_i \in \mathbb{R}$ $\forall i=1,\dots,p$ y geométricamente $h_i(x)$ es un **hiperplano** en $\mathbb{R}^n$.
**Comentarios:** Lo siguiente se puede verificar con propiedades teóricas:
* El conjunto de factibilidad de un problema de optimización convexa es un conjunto convexo. Esto se puede ver pues es una intersección finita de conjuntos convexos: intersección entre las $x$'s que satisfacen $f_i(x) \leq 0$, que se nombra **conjunto subnivel**\*, y las $x$'s que están en un hiperplano.
\*Un conjunto $\alpha$-subnivel es de la forma $\{x \in \text{dom}f | f(x) \leq \alpha\}$. Un conjunto subnivel contiene las curvas de nivel de $f$, ver [Level set](https://en.wikipedia.org/wiki/Level_set):
<img src="https://dl.dropboxusercontent.com/s/0woqoj8foo5eco9/level_set_of_func.png?dl=0" heigth="300" width="300">
Si $f$ es convexa el conjunto subnivel es un conjunto convexo.
* El conjunto óptimo y los conjuntos $\epsilon$-subóptimos son convexos.
* Si la función objetivo $f_o$ es **fuertemente convexa** entonces el conjunto óptimo contiene a lo más un punto.
* Si en el problema anterior se tiene que **maximizar** una $f_o$ función objetivo **cóncava** y se tienen misma forma estándar del problema anterior y $f_i$ convexa, $h_i$ afín entonces también se nombra al problema como **problema de optimización convexa**. Todos los resultados, conclusiones y algoritmos desarrollados para los problemas de minimización son aplicables para maximización. En este caso se puede resolver un problema de maximización al minimizar la función objetivo $-f_o$ que es convexa.
# Función convexa
Sea $f:\mathbb{R}^n \rightarrow \mathbb{R}$ una función con el conjunto $\text{dom}f$ convexo. $f$ se nombra convexa (en su $\text{dom}f$) si $\forall x,y \in \text{dom}f$ y $\theta \in [0,1]$ se cumple:
$$f(\theta x + (1-\theta) y) \leq \theta f(x) + (1-\theta)f(y).$$
**Comentarios:**
* $\text{dom}f$ es convexo $\therefore$ $\theta x + (1-\theta)y \in \text{dom}f$
* La convexidad de $f$ se define para $\text{dom}f$ aunque para casos en particular se detalla el conjunto en el que $f$ es convexa.
* Si la desigualdad se cumple de forma estricta $\forall x \neq y$ $f$ se nombra **estrictamente convexa**.
* $f$ es **cóncava** si $-f$ es convexa y **estrictamente cóncava** si $-f$ es estrictamente convexa. Otra forma de definir concavidad es con una desigualdad del tipo:
$$f(\theta x + (1-\theta) y) \geq \theta f(x) + (1-\theta)f(y).$$
y mismas definiciones para $x,y, \theta$ que en la definición de convexidad.
* Si $f$ es convexa, geométricamente el segmento de línea que se forma con los puntos $(x,f(x)), (y,f(y))$ está por encima o es igual a $f(\theta x + (1-\theta)y) \forall \theta \in [0,1]$ y $\forall x,y \in \text{dom}f$:
<img src="https://dl.dropboxusercontent.com/s/fdcx1k150nfwykv/draw_convexity_for_functions.png?dl=0" heigth="300" width="300">
* La desigualdad que define a funciones convexas se nombra [**desigualdad de Jensen**](https://en.wikipedia.org/wiki/Jensen%27s_inequality).
# Conjunto convexo
## Línea y segmentos de línea
Sean $x_1, x_2 \in \mathbb{R}^n$ con $x_1 \neq x_2$. Entonces el punto:
$$y = \theta x_1 + (1-\theta)x_2$$
con $\theta \in \mathbb{R}$ se encuentra en la línea que pasa por $x_1$ y $x_2$. $\theta$ se le nombra parámetro y si $\theta \in [0,1]$ tenemos un segmento de línea:
<img src="https://dl.dropboxusercontent.com/s/dldljf5igy8xt9d/segmento_linea.png?dl=0" heigth="200" width="200">
**Comentarios:**
* $y = \theta x_1 + (1-\theta)x_2 = x_2 + \theta(x_1 -x_2)$ y esta última igualdad se interpreta como "$y$ es la suma del punto base $x_2$ y la dirección $x_1-x_2$ escalada por $\theta$".
* Si $\theta=0$ entonces $y=x_2$. Si $\theta \in [0,1]$ entonces $y$ se "mueve" en la dirección $x_1-x_2$ hacia $x_1$ y si $\theta>1$ entonces $y$ se encuentra en la línea "más allá" de $x_1$:
<img src="https://dl.dropboxusercontent.com/s/nbahrio7p1mj4hs/segmento_linea_2.png?dl=0" heigth="350" width="350">
El punto enmedio entre $x_1$ y $x_2$ tiene $\theta=\frac{1}{2}$.
## Conjunto convexo
Un conjunto $\mathcal{C}$ es convexo si el segmento de línea entre cualquier par de puntos de $\mathcal{C}$ está completamente contenida en $\mathcal{C}$. Esto se escribe matemáticamente como:
$$\theta x_1 + (1-\theta) x_2 \in \mathcal{C} \forall \theta \in [0,1], \forall x_1, x_2 \in \mathcal{C}.$$
Ejemplos gráficos de conjuntos convexos:
<img src="https://dl.dropboxusercontent.com/s/gj54ism1lqojot6/ej_conj_convexos.png?dl=0" heigth="400" width="400">
Ejemplos gráficos de conjuntos no convexos:
<img src="https://dl.dropboxusercontent.com/s/k37zh5v3iq3kx04/ej_conj_no_convexos.png?dl=0" heigth="350" width="350">
**Comentarios:**
* El punto $\displaystyle \sum_{i=1}^k \theta_i x_i$ con $\displaystyle \sum_{i=1}^k \theta_i=1$, $\theta_i \geq 0 \forall i=1,\dots,k$ se nombra **combinación convexa** de los puntos $x_1, x_2, \dots, x_k$. Una combinación convexa de los puntos $x_1, \dots, x_k$ puede pensarse como una mezcla o promedio ponderado de los puntos, con $\theta_i$ la fracción $\theta_i$ de $x_i$ en la mezcla.
* Un conjunto es convexo si y sólo si contiene cualquier combinación convexa de sus puntos.
# Ejemplos de funciones convexas y cóncavas
* Una función afín es convexa y cóncava en todo su dominio: $f(x) = Ax+b$ con $A \in \mathbb{R}^{m \times n}, b \in \mathbb{R}^n$, $\text{dom}f = \mathbb{R}^n$.
**Obs:** por tanto las funciones lineales también son convexas y cóncavas.
* Funciones cuadráticas: $f: \mathbb{R}^n \rightarrow \mathbb{R}$, $f(x) = \frac{1}{2} x^TPx + q^Tx + r$ son convexas en su dominio: $\mathbb{R}^n$. $P \in \mathcal{S}_+^n, q \in \mathbb{R}^n, r \in \mathbb{R}$ con $\mathbb{S}_+^n$ conjunto de **matrices simétricas positivas semidefinidas**\*.
\*Una matriz $A$ es positiva semidefinida si $x^TAx \geq 0$ $\forall x \in \mathbb{R}^n - \{0\}$. Si se cumple de forma estricta la desigualdad anterior entonces $A$ es **positiva definida**. Con los eigenvalores podemos caracterizar a las matrices definidas y semidefinidas positivas: $A$ es semidefinida positiva si y sólo si los eigenvalores de $T=\frac{A+A^T}{2}$ son no negativos. Es definida positiva si y sólo si los eigenvalores de $T$ son positivos. Los conjuntos de matrices que se utilizan para definir a matrices semidefinidas positivas y definidas positivas son $\mathbb{S}_{+}^n$ y $\mathbb{S}_{++}^n$ respectivamente.
**Obs:** $f$ es estrictamente convexa si y sólo si $P \in \mathbb{S}_{++}^n$. $f$ es cóncava si y sólo si $P \in -\mathbb{S}_+^n$.
* Exponenciales: $f: \mathbb{R} \rightarrow \mathbb{R}$, $f(x) = e^{ax}$ para cualquier $a \in \mathbb{R}$ es convexa en su dominio: $\mathbb{R}$.
* Potencias: $f: \mathbb{R} \rightarrow \mathbb{R}$, $f(x)=x^a$:
* Si $a \geq 1$ o $a \leq 0$ entonces $f$ es convexa en $\mathbb{R}_{++}$ (números reales positivos).
* Si $0 \leq a \leq 1$ entonces $f$ es cóncava en $\mathbb{R}_{++}$.
* Potencias del valor absoluto: $f: \mathbb{R} \rightarrow \mathbb{R}$, $f(x)=|x|^p$ con $p \geq 1$ es convexa en $\mathbb{R}$.
* Logaritmo: $f: \mathbb{R} \rightarrow \mathbb{R}$, $f(x) = \log(x)$ es cóncava en su dominio: $\mathbb{R}_{++}$.
* Entropía negativa: $f(x) = \begin{cases}
x\log(x) &\text{ si } x > 0 ,\\
0 &\text{ si } x = 0
\end{cases}$ es estrictamente convexa en su dominio: $\mathbb{R}_+$.
* Normas: cualquier norma es convexa en su dominio.
* Función máximo: $f: \mathbb{R}^{n} \rightarrow \mathbb{R}$, $f(x) = \max\{x_1,\dots,x_n\}$ es convexa.
* Función log-sum-exp: $f: \mathbb{R}^{n} \rightarrow \mathbb{R}$, $f(x)=\log\left(\displaystyle \sum_{i=1}^ne^{x_i}\right)$ es convexa en su dominio: $\mathbb{R}^n$.
* La media geométrica: $f: \mathbb{R}^{n} \rightarrow \mathbb{R}$, $f(x) = \left(\displaystyle \prod_{i=1}^n x_i \right)^\frac{1}{n}$ es cóncava en su dominio: $\mathbb{R}_{++}^n$.
* Función log-determinante: $f: \mathbb{S}^{n} \rightarrow \mathbb{R}^n$, $f(x) = \log(\det(X))$ es cóncava en su dominio: $\mathbb{S}_{++}^n$.
# Resultados útiles de teoría de convexidad
Se sugiere revisar [1.4.Polinomios_de_Taylor_y_diferenciacion_numerica](https://github.com/ITAM-DS/analisis-numerico-computo-cientifico/blob/master/temas/I.computo_cientifico/1.4.Polinomios_de_Taylor_y_diferenciacion_numerica.ipynb) y [1.3.Condicion_de_un_problema_y_estabilidad_de_un_algoritmo](https://github.com/ITAM-DS/analisis-numerico-computo-cientifico/blob/master/temas/I.computo_cientifico/1.3.Condicion_de_un_problema_y_estabilidad_de_un_algoritmo.ipynb) como recordatorio de definiciones. En particular las **definiciones de primera y segunda derivada, gradiente y Hessiana** para la primer nota y la **definición de número de condición de una matriz** para la segunda.
**Sobre funciones convexas/cóncavas**
* Sea $f: \mathbb{R}^n \rightarrow \mathbb{R}$ diferenciable entonces $f$ es convexa si y sólo si $\text{dom}f$ es un conjunto convexo y se cumple:
$$f(y) \geq f(x) + \nabla f(x)^T(y-x) \forall x,y \in \text{dom}f.$$
Si se cumple de forma estricta la desigualdad $f$ se nombra estrictamente convexa. También si su $\text{dom}f$ es convexo y se tiene la desigualdad en la otra dirección "$\leq$" entonces $f$ es cóncava.
Geométricamente este resultado se ve como sigue para $\nabla f(x) \neq 0$:
<img src="https://dl.dropboxusercontent.com/s/e581e22xeejdwu0/convexidad_con_hiperplano_de_soporte.png?dl=0" heigth="350" width="350">
y el hiperplano $f(x) + \nabla f(x)^T(y-x)$ se nombra **hiperplano de soporte para la función $f$ en el punto $(x,f(x))$**. Obsérvese que si $\nabla f(x)=0$ se tiene $f(y) \geq f(x) \forall y \in \text{dom}f$ y por lo tanto $x$ es un mínimo global de $f$.
* Una función es convexa si y sólo si es convexa al restringirla a cualquier línea que intersecte su dominio, esto es, si $g(t) = f(x + tv)$ es convexa $\forall x,v \in \mathbb{R}^n$, $\forall t \in \mathbb{R}$ talque $x + tv \in \text{dom}f$
* Sea $f: \mathbb{R}^n \rightarrow \mathbb{R}$ tal que $f \in \mathcal{C}^2(\text{dom}f)$. Entonces $f$ es convexa en $\text{dom}f$ si y sólo si $\text{dom}f$ es convexo y $\nabla^2f(x)$ es simétrica semidefinida positiva en $\text{dom}f$. Si $\nabla^2f(x)$ es simétrica definida positiva en $\text{dom}f$ y $\text{dom}f$ es convexo entonces $f$ es estrictamente convexa en $\text{dom}f$\*.
\*Para una variable: $f: \mathbb{R} \rightarrow \mathbb{R}$, la hipótesis del enunciado anterior es que la segunda derivada sea positiva. El recíproco no es verdadero, para ver esto considérese $f(x)=x^4$ la cual es estrictamente convexa en $\text{dom}f$ pero su segunda derivada en $0$ no es positiva.
**Sobre problemas de optimización**
* Si $f$ es diferenciable y $x^*$ es óptimo entonces $\nabla f(x^*) = 0$.
* Si $f \in \mathcal{C}^2(\text{domf})$ y $x^*$ es mínimo local entonces $\nabla^2 f(x^*)$ es una matriz simétrica semidefinida positiva.
* Si $f \in \mathcal{C}^2(\text{domf})$, $\nabla f(x^*)=0$ y $\nabla^2f(x^*)$ es una matriz simétrica definida positiva entonces $x^*$ es mínimo local estricto.
* Una propiedad fundamental de un óptimo local en un problema de optimización convexa es que también es un óptimo global.
* Si $f_o$ en un problema de optimización convexa es diferenciable y $X$ es el conjunto de factibilidad entonces $x$ es óptimo si y sólo si $x \in X$ y $\nabla f_o(x)^T(y-x) \geq 0$ $\forall y \in X$. Si en lo anterior se considera como conjunto de factibilidad $X = \text{dom}f_o$ (que es un problema sin restricciones) la propiedad se reduce a la condición: $x$ es óptimo si y sólo si $\nabla f_o(x) = 0$.
Geométricamente el resultado anterior se visualiza para $\nabla f(x) \neq 0$ y $-\nabla f(x)$ apuntando hacia la dirección dibujada:
<img src="https://dl.dropboxusercontent.com/s/0tmpivvo5ob4oox/optimo_convexidad_con_hiperplano_de_soporte.png?dl=0" heigth="550" width="550">
**Comentario:** Por los resultados anteriores los métodos de optimización que revisaremos en el módulo IV buscarán resolver la **ecuación no lineal** $\nabla f_o(x)=0$. Dependiendo del número de soluciones de la ecuación $\nabla f_o(x)=0$ se tienen situaciones distintas. Por ejemplo, si no tiene solución entonces el/los óptimos no se alcanza(n) pues el problema puede no ser acotado por debajo o si existe el óptimo éste puede no alcanzarse. Por otro lado, si la ecuación tiene múltiples soluciones entonces cada solución es un mínimo de $f_o$.
## Función fuertemente convexa
Una función $f:\mathbb{R}^n \rightarrow \mathbb{R}$ tal que $f \in \mathcal{C}^2(\text{dom}f)$ se nombra **fuertemente convexa** en el conjunto convexo $\mathbb{S} \neq \emptyset$ si existe $m>0$ tal que $\nabla^2 f(x) - mI$ es simétrica semidefinida positiva $\forall x \in \mathbb{S}$.
**Comentario:** si una función es fuertemente convexa se puede probar que:
* $f(y) \geq f(x) + \nabla f(x)^T(y-x) + \frac{m}{2}||y-x||_2^2 \forall x,y \in \mathbb{S}$. Por esto si $f$ es fuertemente convexa en $\mathbb{S}$ entonces es estrictamente convexa en $\mathbb{S}$. También esta desigualdad indica que la diferencia entre la función de $y$, $f(y)$, y la función lineal en $y$ $f(x) + \nabla f(x)^T(y-x)$ (Taylor a primer orden) está acotada por debajo por una cantidad cuadrática.
* El **número de condición** bajo la norma 2 de $\nabla ^2 f$ está acotado por arriba por el cociente $\frac{\lambda_\text{max}(\nabla^2 f(x))}{\lambda_\text{min}(\nabla^2 f(x))} \forall x \in \mathbb{S}$.
# Tablita útil para fórmulas de diferenciación con el operador $\nabla$
Si $f,g:\mathbb{R}^n \rightarrow \mathbb{R}$ con $f,g \in \mathcal{C}^2$ respectivamente en sus dominios y $\alpha_1, \alpha_2 \in \mathbb{R}$, $A \in \mathbb{R}^{n \times n}$, $b \in \mathbb{R}^n$ son fijas. Diferenciando con respecto a la variable $x \in \mathbb{R}^n$ se tiene:
| | |
|:--:|:--:|
|linealidad | $\nabla(\alpha_1 f(x) + \alpha_2 g(x)) = \alpha_1 \nabla f(x) + \alpha_2 \nabla g(x)$|
|producto | $\nabla(f(x)g(x)) = \nabla f(x) g(x) + f(x) \nabla g(x)$|
|producto punto|$\nabla(b^Tx) = b$
|cuadrático|$\nabla(x^TAx) = (A+A^T)x$|
|segunda derivada| $\nabla^2(Ax)=A$|
**Referencias:**
* S. P. Boyd, L. Vandenberghe, Convex Optimization, Cambridge University Press, 2009.
* L. Bottou, F. E. Curtis, J Nocedal, Optimization Methods for Large-Scale Machine Learning, SIAM, 2018.
|
github_jupyter
|
docker run --rm -v <ruta a mi directorio>:/datos --name jupyterlab_numerical -p 8888:8888 -d palmoreck/jupyterlab_numerical:1.1.0
docker stop jupyterlab_numerical
import numpy as np
x0=np.array([1.5,1.5,1.5,1.5])
gf= lambda x: np.array([2*(x[0]-2),
-2*(2-x[1]),
2*x[2],
4*x[3]**3])
Hf = lambda x: np.array([[2, 0, 0 ,0],
[0, 2, 0, 0],
[0, 0, 2, 0],
[0, 0, 0, 12*x[2]**2]])
gf(x0)
Hf(x0)
x_0 = np.array([5,5,1,0])
x_1 = x_0 - gf(x_0)
x_1
x_2 = x_1 - gf(x_1)
x_2
x_3 = x_2 - gf(x_2)
x_3
x_4 = x_3 - gf(x_3)
x_4
x_0 = np.array([5,5,1,0])
x_1 = x_0 - np.linalg.solve(Hf(x_0),gf(x_0))
x_1
t_0=0.5
gf(x_0)
x_1 = x_0 - t_0*gf(x_0)
x_1
import matplotlib.pyplot as plt
import pprint
from scipy.linalg import solve_triangular
np.random.seed(1989) #para reproducibilidad
mpoints = 20
x = np.random.randn(mpoints)
y = -3*x + np.random.normal(2,1,mpoints)
plt.plot(x,y, 'r*')
plt.xlabel('x')
plt.ylabel('y')
plt.title('Puntos ejemplo')
plt.show()
# el tercer argumento de polyfit especifica el grado del polinomio a ajustar.
#Usaremos ngrado = 1 pues queremos ajustar una recta
ngrado = 1
coeficientes = np.polyfit(x,y,ngrado)
#Una vez realizado el llamado a la función polyfit se regresan los coeficientes de x
#ordenados del mayor grado al menor.
pprint.pprint(coeficientes)
y_ajustadas_numpy = coeficientes[1] + coeficientes[0] * x
plt.plot(x, y_ajustadas_numpy, 'k-',x, y, 'r*')
plt.legend(['modelo lineal','datos'], loc='best')
plt.show()
#construimos a la matriz A:
A=np.ones((mpoints,2))
A[:,1] = x
A
Q,R = np.linalg.qr(A)
#Resolvemos el sistema R*beta = Q^T*y
beta = solve_triangular(R,Q.T@y)
pprint.pprint(beta)
y_ajustadas_QR = A@beta
#obsérvese que la línea anterior es equivalente a realizar:
#y_ajustadas_QR = beta[0] + beta[1]*x
plt.plot(x, y_ajustadas_QR , 'k-',x, y, 'r*')
plt.legend(['modelo lineal','datos'], loc='best')
plt.show()
cte=-np.transpose(A)@y
cte
gf = lambda beta_fun: cte + np.transpose(A)@(A@beta_fun)
#observa que no hacemos la multiplicación (A^T*A)*beta, mejor hacemos
#primero A*beta y luego multiplicamos por A^T
beta_0 = np.array([0,0])
t_0=.130
beta_1 = beta_0 - t_0*gf(beta_0)
beta_1
t_1=.0625
beta_2 = beta_1 - t_1*gf(beta_1)
beta_2
t_2 = .0625
beta_3 = beta_2 - t_2*gf(beta_2)
beta_3
t_3 = .0625
beta_4 = beta_3 - t_3*gf(beta_3)
beta_4
t_4 = .0625
beta_5 = beta_4 - t_4*gf(beta_4)
beta_5
y_ajustadas_gradiente = A@beta_5
#obsérvese que la línea anterior es equivalente a realizar:
#y_ajustadas_gradiente = beta_5[0] + beta_5[1]*x
plt.plot(x, y_ajustadas_gradiente , 'k-',x, y, 'r*')
plt.legend(['modelo lineal','datos'], loc='best')
plt.show()
s_0 = np.linalg.solve(np.transpose(A)@A,-gf(beta_0))
beta_1 = beta_0 + s_0
beta_1
y_ajustadas_Newton = A@beta_1
#obsérvese que la línea anterior es equivalente a realizar:
#y_ajustadas_Newton = beta_1[0] + beta_1[1]*x
plt.plot(x, y_ajustadas_Newton , 'k-',x, y, 'r*')
plt.legend(['modelo lineal','datos'], loc='best')
plt.show()
np.linalg.norm(beta_5-beta)/np.linalg.norm(beta)
np.linalg.norm(beta_1-beta)/np.linalg.norm(beta)
Q,R = np.linalg.qr(A)
s_0 = np.linalg.solve(R,np.transpose(Q)@y-R@beta_0)
beta_1 = beta_0 + s_0
beta_1
y_ajustadas_Newton = A@beta_1
#obsérvese que la línea anterior es equivalente a realizar:
#y_ajustadas_Newton = beta_1[0] + beta_1[1]*x
plt.plot(x, y_ajustadas_Newton , 'k-',x, y, 'r*')
plt.legend(['modelo lineal','datos'], loc='best')
plt.show()
beta_0 = np.array([0,0])
eta_0 = .1
np.random.seed(1989) #para reproducibilidad
i = np.random.randint(mpoints, size=1)
beta_1 = beta_0 + eta_0*(y[i]-A[i,:].dot(beta_0))*A[i,:]
beta_1
def error_relativo(aprox, obj):
return np.linalg.norm(aprox-obj)/np.linalg.norm(obj)
maxiter=5
tol=1e-2
err=error_relativo(beta_0,beta)
beta_k = beta_0
k=1
eta_k = .1 #constante para todas las iteraciones
err
while(err>tol and k <= maxiter):
i = int(np.random.randint(mpoints, size=1))
beta_k = beta_k+eta_k*(y[i]-A[i,:].dot(beta_k))*A[i,:]
k+=1
eta_k=.1
err=error_relativo(beta_k,beta)
err
beta_k
maxiter=4
tol=1e-2
err=error_relativo(beta_0,beta)
beta_k = beta_0
k=1
eta_k = .1 #constante para todas las iteraciones
m_sample = 5
err
while(err>tol and k <= maxiter):
idx = np.random.choice(mpoints, m_sample,replace=False) #muestra de tamaño m_sample sin reemplazo
beta_k = beta_k+eta_k*(y[idx]-A[idx,:]@beta_0)@A[idx,:]
k+=1
eta_k=.1
err=error_relativo(beta_k,beta)
err
beta_k
| 0.283285 | 0.894283 |
```
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.linear_model import LinearRegression, Lasso, Ridge
from sklearn.metrics import r2_score
from sklearn.preprocessing import PolynomialFeatures
from sklearn.pipeline import make_pipeline
import warnings
warnings.filterwarnings('ignore')
```
### Load the dataset
- Load the train data and using all your knowledge of pandas try to explore the different statistical properties like correlation of the dataset.
```
# Code starts here
df = pd.read_csv("D:\\Study_material\\GreyAtom\\projects\\movetomelboure\\train.csv")
df.head()
y = df['Price']
X = df.drop('Price',axis=1)
X_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.3,random_state=6)
corr = X_train.corr()
#print(corr)
fig = plt.figure(figsize=(15, 15))
sns.heatmap(corr, annot = True)
plt.show()
# Code ends here.
df.isnull().sum()
import sweetviz
my_report = sweetviz.analyze([df,'Train'], target_feat='Price')
my_report.show_html('FinalReport.html')
```
## Model building
- Separate the features and target and then split the train data into train and validation set.
- Apply different models of your choice and then predict on the validation data and find the `accuracy_score` for this prediction.
- Try improving upon the `accuracy_score` using different regularization techniques.
```
# Code starts here
# --------------
from sklearn.linear_model import LinearRegression
from sklearn.metrics import r2_score
# Code starts here
regressor = LinearRegression()
regressor.fit(X_train,y_train)
y_pred = regressor.predict(X_test)
r2 = r2_score(y_test,y_pred)
print(" #### r2 #### ", r2)
# --------------
from sklearn.linear_model import Lasso
# Code starts here
lasso = Lasso()
lasso.fit(X_train,y_train)
lasso_pred = lasso.predict(X_test)
r2_lasso = r2_score(y_test,lasso_pred)
print(" #### r2 lasso #### ", r2_lasso)
# --------------
from sklearn.linear_model import Ridge
# Code starts here
ridge = Ridge()
ridge.fit(X_train,y_train)
ridge_pred = ridge.predict(X_test)
r2_ridge = r2_score(y_test,ridge_pred)
print(" #### r2 lasso #### ", r2_ridge)
# Code ends here
# --------------
from sklearn.model_selection import cross_val_score
#Code starts here
regressor = LinearRegression()
score = cross_val_score(regressor,X_train,y_train,cv=10)
mean_score = np.mean(score)
print("mean score", mean_score)
# --------------
from sklearn.preprocessing import PolynomialFeatures
from sklearn.pipeline import make_pipeline
#Code starts here
model = make_pipeline(PolynomialFeatures(2),LinearRegression())
model.fit(X_train,y_train)
y_pred = model.predict(X_test)
r2_poly = r2_score(y_test,y_pred)
print(" #### r2 poly #### ", r2_poly)
# Code ends here.
```
### Prediction on the test data and creating the sample submission file.
- Load the test data and store the `Id` column in a separate variable.
- Perform the same operations on the test data that you have performed on the train data.
- Create the submission file as a `csv` file consisting of the `Id` column from the test data and your prediction as the second column.
```
# Code starts here
df_test = pd.read_csv("D:\\Study_material\\GreyAtom\\projects\\movetomelboure\\test.csv")
id = df_test['Id']
preds = pd.DataFrame(lasso.predict(df_test))
preds_m = pd.DataFrame(model.predict(df_test))
preds.to_csv("preds.csv")
preds_m.to_csv("preds_m.csv")
# Code ends here.
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.linear_model import LinearRegression, Lasso, Ridge
from sklearn.metrics import r2_score
from sklearn.preprocessing import PolynomialFeatures
from sklearn.pipeline import make_pipeline
import warnings
warnings.filterwarnings('ignore')
# Code starts here
df = pd.read_csv("D:\\Study_material\\GreyAtom\\projects\\movetomelboure\\train.csv")
df.head()
y = df['Price']
X = df.drop('Price',axis=1)
X_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.3,random_state=6)
corr = X_train.corr()
#print(corr)
fig = plt.figure(figsize=(15, 15))
sns.heatmap(corr, annot = True)
plt.show()
# Code ends here.
df.isnull().sum()
import sweetviz
my_report = sweetviz.analyze([df,'Train'], target_feat='Price')
my_report.show_html('FinalReport.html')
# Code starts here
# --------------
from sklearn.linear_model import LinearRegression
from sklearn.metrics import r2_score
# Code starts here
regressor = LinearRegression()
regressor.fit(X_train,y_train)
y_pred = regressor.predict(X_test)
r2 = r2_score(y_test,y_pred)
print(" #### r2 #### ", r2)
# --------------
from sklearn.linear_model import Lasso
# Code starts here
lasso = Lasso()
lasso.fit(X_train,y_train)
lasso_pred = lasso.predict(X_test)
r2_lasso = r2_score(y_test,lasso_pred)
print(" #### r2 lasso #### ", r2_lasso)
# --------------
from sklearn.linear_model import Ridge
# Code starts here
ridge = Ridge()
ridge.fit(X_train,y_train)
ridge_pred = ridge.predict(X_test)
r2_ridge = r2_score(y_test,ridge_pred)
print(" #### r2 lasso #### ", r2_ridge)
# Code ends here
# --------------
from sklearn.model_selection import cross_val_score
#Code starts here
regressor = LinearRegression()
score = cross_val_score(regressor,X_train,y_train,cv=10)
mean_score = np.mean(score)
print("mean score", mean_score)
# --------------
from sklearn.preprocessing import PolynomialFeatures
from sklearn.pipeline import make_pipeline
#Code starts here
model = make_pipeline(PolynomialFeatures(2),LinearRegression())
model.fit(X_train,y_train)
y_pred = model.predict(X_test)
r2_poly = r2_score(y_test,y_pred)
print(" #### r2 poly #### ", r2_poly)
# Code ends here.
# Code starts here
df_test = pd.read_csv("D:\\Study_material\\GreyAtom\\projects\\movetomelboure\\test.csv")
id = df_test['Id']
preds = pd.DataFrame(lasso.predict(df_test))
preds_m = pd.DataFrame(model.predict(df_test))
preds.to_csv("preds.csv")
preds_m.to_csv("preds_m.csv")
# Code ends here.
| 0.323166 | 0.836555 |
# GAN Evaluation
* Credit: Ziqiao Ma
* Latest Update: Dec.16 2020
## Colab Setup
### Environment Configuration
Mount to Colab and set up autoreload module.
```
%load_ext autoreload
%autoreload 2
from google.colab import drive
drive.mount('/content/drive')
```
TODO: Change the variable to the locaation of your file in the drive!
```
GOOGLE_DRIVE_PATH_AFTER_MYDRIVE = 'Colab Notebooks/StarGAN-V1'
import os
import sys
os.environ['CUDA_VISIBLE_DEVICES'] = '0'
GOOGLE_DRIVE_PATH = os.path.join('drive', 'MyDrive', GOOGLE_DRIVE_PATH_AFTER_MYDRIVE)
sys.path.append(GOOGLE_DRIVE_PATH)
print(os.listdir(GOOGLE_DRIVE_PATH))
```
Set up file system.
### Import Libraries
```
import matplotlib.pyplot as plt
import numpy
from numpy import cov
from numpy import trace
from numpy import iscomplexobj
from numpy import asarray
from numpy.random import randint
from scipy.linalg import sqrtm
from keras.applications.inception_v3 import InceptionV3
from keras.applications.inception_v3 import preprocess_input
from keras.datasets.mnist import load_data
from skimage.transform import resize
from skimage.io import imread_collection
```
## Frechet Inception Distance (FID)
The Frechet Inception Distance score, or FID for short, is a metric that **calculates the distance between feature vectors calculated for real and generated images**.
The score summarizes how similar the two groups are in terms of statistics on computer vision features of the raw images calculated using the inception v3 model used for image classification. Lower scores indicate the two groups of images are more similar, or have more similar statistics, with a perfect score being 0.0 indicating that the two groups of images are identical.
The FID score is used to evaluate the quality of images generated by generative adversarial networks, and lower scores have been shown to correlate well with higher quality images.
Reference: [Tutorial](https://machinelearningmastery.com/how-to-implement-the-frechet-inception-distance-fid-from-scratch/).
### Implementation
```
def calculate_fid(model, images1, images2):
# calculate activations
act1 = model.predict(images1)
act2 = model.predict(images2)
# calculate mean and covariance statistics
mu1, sigma1 = act1.mean(axis=0), cov(act1, rowvar=False)
mu2, sigma2 = act2.mean(axis=0), cov(act2, rowvar=False)
# calculate sum squared difference between means
ssdiff = numpy.sum((mu1 - mu2)**2.0)
# calculate sqrt of product between cov
covmean = sqrtm(sigma1.dot(sigma2))
# check and correct imaginary numbers from sqrt
if iscomplexobj(covmean):
covmean = covmean.real
# calculate score
fid = ssdiff + trace(sigma1 + sigma2 - 2.0 * covmean)
return fid
# define two fake collections of images
images1 = randint(0, 255, 100*256*256*3)
images1 = images1.reshape((100,256,256,3))
images2 = randint(0, 255, 200*256*256*3)
images2 = images2.reshape((200,256,256,3))
print('Prepared', images1.shape, images2.shape)
# convert integer to floating point values
images1 = images1.astype('float32')
images2 = images2.astype('float32')
# pre-process images
images1 = preprocess_input(images1)
images2 = preprocess_input(images2)
# Visualize
plt.subplot(121)
plt.title('Image 1')
plt.imshow(images1[0])
plt.subplot(122)
plt.title('Image 2')
plt.imshow(images2[0])
# prepare the inception v3 model
model = InceptionV3(include_top=False, pooling='avg', input_shape=(256,256,3))
# fid between images1 and images1
fid = calculate_fid(model, images1, images1)
print('FID of same images: %.3f' % fid)
# fid between images1 and images2
fid = calculate_fid(model, images1, images2)
print('FID of different images: %.3f' % fid)
```
### Evaluation
It's useful to calculate the FID score between two collections of real images.
```
# Create data/
!mkdir data/
!mkdir data/style/
!mkdir data/style/train/
!mkdir data/style/test/
# Download Datasets from UCB
!wget -cO - https://people.eecs.berkeley.edu/%7Etaesung_park/CycleGAN/datasets/cezanne2photo.zip > cezanne.zip
!wget -cO - https://people.eecs.berkeley.edu/%7Etaesung_park/CycleGAN/datasets/monet2photo.zip > monet.zip
!wget -cO - https://people.eecs.berkeley.edu/%7Etaesung_park/CycleGAN/datasets/ukiyoe2photo.zip > ukiyoe.zip
!wget -cO - https://people.eecs.berkeley.edu/%7Etaesung_park/CycleGAN/datasets/vangogh2photo.zip > vangogh.zip
# Unzip files
!unzip cezanne.zip
!unzip monet.zip
!unzip ukiyoe.zip
!unzip vangogh.zip
!rm -f cezanne.zip
!mv cezanne2photo/trainA/ data/style/train/cezanne/
!mv cezanne2photo/testA/ data/style/test/cezanne/
!mv cezanne2photo/trainB/ data/style/train/photo/
!mv cezanne2photo/testB/ data/style/test/photo/
!rm -r cezanne2photo/
!rm -f monet.zip
!mv monet2photo/trainA/ data/style/train/monet/
!mv monet2photo/testA/ data/style/test/monet/
!rm -r monet2photo/
!rm -f ukiyoe.zip
!mv ukiyoe2photo/trainA/ data/style/train/ukiyoe/
!mv ukiyoe2photo/testA/ data/style/test/ukiyoe/
!rm -r ukiyoe2photo/
!rm -f vangogh.zip
!mv vangogh2photo/trainA/ data/style/train/vangogh/
!mv vangogh2photo/testA/ data/style/test/vangogh/
!rm -r vangogh2photo/
TRAIN_IMG_DIR = 'data/style/train'
TEST_IMG_DIR = 'data/style/test'
def load_img_dir(label='photo', size=None):
assert label in {'photo', 'cezanne', 'monet', 'ukiyoe', 'vangogh'}
dir_photo = os.path.join(TEST_IMG_DIR, '{}/*.jpg'.format(label))
img_photo = numpy.array(imread_collection(dir_photo))
if size is None:
size = len(img_photo)
else:
img_photo = img_photo[:size]
img_photo = img_photo.reshape((size,256,256,3)).astype('float32')
img_photo = preprocess_input(img_photo)
return img_photo
model = InceptionV3(include_top=False, pooling='avg', input_shape=(256, 256, 3))
size = 4
print('Loading images...')
img_photo = load_img_dir('photo', size)
img_cezanne = load_img_dir('cezanne', size)
img_monet = load_img_dir('monet', size)
img_ukiyoe = load_img_dir('ukiyoe', size)
img_vangogh = load_img_dir('vangogh', size)
print('Start testing...')
fid = calculate_fid(model, img_photo, img_cezanne)
print('FID between photo and cezanne: %.3f' % fid)
fid = calculate_fid(model, img_photo, img_monet)
print('FID between photo and monet: %.3f' % fid)
fid = calculate_fid(model, img_photo, img_ukiyoe)
print('FID between photo and ukiyoe: %.3f' % fid)
fid = calculate_fid(model, img_photo, img_vangogh)
print('FID between photo and vangogh: %.3f' % fid)
fid = calculate_fid(model, img_cezanne, img_monet)
print('FID between cezanne and monet: %.3f' % fid)
fid = calculate_fid(model, img_cezanne, img_ukiyoe)
print('FID between cezanne and ukiyoe: %.3f' % fid)
fid = calculate_fid(model, img_cezanne, img_vangogh)
print('FID between cezanne and vangogh: %.3f' % fid)
fid = calculate_fid(model, img_monet, img_ukiyoe)
print('FID between monet and ukiyoe: %.3f' % fid)
fid = calculate_fid(model, img_monet, img_vangogh)
print('FID between monet and vangogh: %.3f' % fid)
fid = calculate_fid(model, img_ukiyoe, img_vangogh)
print('FID between ukiyoe and vangogh: %.3f' % fid)
def load_out_dir(dir, label='cezanne', size=None):
assert label in {'cezanne', 'monet', 'ukiyoe', 'vangogh'}
dir_photo = os.path.join(dir, '{}/*.jpg'.format(label))
img_photo = numpy.array(imread_collection(dir_photo))
if size is None:
size = len(img_photo)
else:
img_photo = img_photo[:size]
img_photo = img_photo.reshape((size,256,256,3)).astype('float32')
img_photo = preprocess_input(img_photo)
return img_photo
dir_cyclegan = os.path.join(GOOGLE_DRIVE_PATH, 'checkpoints/checkpoints_20w/one_result')
print(os.listdir(dir_cyclegan))
size = 4
print('Loading images...')
cyclegan_cezanne = load_out_dir(dir_cyclegan, 'cezanne', size)
cyclegan_monet = load_out_dir(dir_cyclegan, 'monet', size)
cyclegan_ukiyoe = load_out_dir(dir_cyclegan, 'ukiyoe', size)
cyclegan_vangogh = load_out_dir(dir_cyclegan, 'vangogh', size)
print('Start testing...')
fid = calculate_fid(model, img_cezanne, cyclegan_cezanne)
print('FID between cyclegan generated fake cezanne and test cyclegan images: %.3f' % fid)
fid = calculate_fid(model, img_monet, cyclegan_monet)
print('FID between cyclegan generated fake monet and test cyclegan monet: %.3f' % fid)
fid = calculate_fid(model, img_ukiyoe, cyclegan_ukiyoe)
print('FID between cyclegan generated fake ukiyoe and test cyclegan ukiyoe: %.3f' % fid)
fid = calculate_fid(model, img_vangogh, cyclegan_vangogh)
print('FID between cyclegan generated fake vangogh and test cyclegan vangogh: %.3f' % fid)
dir_cyclegan = os.path.join(GOOGLE_DRIVE_PATH, 'results/')
print(os.listdir(dir_cyclegan))
size = 100
print('Loading images...')
cyclegan_cezanne = load_out_dir(dir_cyclegan, 'cezanne', size)
cyclegan_monet = load_out_dir(dir_cyclegan, 'monet', size)
cyclegan_ukiyoe = load_out_dir(dir_cyclegan, 'ukiyoe', size)
cyclegan_vangogh = load_out_dir(dir_cyclegan, 'vangogh', size)
print('Start testing...')
fid = calculate_fid(model, img_cezanne, cyclegan_cezanne)
print('FID between cyclegan generated fake cezanne and test cyclegan images: %.3f' % fid)
fid = calculate_fid(model, img_monet, cyclegan_monet)
print('FID between cyclegan generated fake monet and test cyclegan monet: %.3f' % fid)
fid = calculate_fid(model, img_ukiyoe, cyclegan_ukiyoe)
print('FID between cyclegan generated fake ukiyoe and test cyclegan ukiyoe: %.3f' % fid)
fid = calculate_fid(model, img_vangogh, cyclegan_vangogh)
print('FID between cyclegan generated fake vangogh and test cyclegan vangogh: %.3f' % fid)
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
from google.colab import drive
drive.mount('/content/drive')
GOOGLE_DRIVE_PATH_AFTER_MYDRIVE = 'Colab Notebooks/StarGAN-V1'
import os
import sys
os.environ['CUDA_VISIBLE_DEVICES'] = '0'
GOOGLE_DRIVE_PATH = os.path.join('drive', 'MyDrive', GOOGLE_DRIVE_PATH_AFTER_MYDRIVE)
sys.path.append(GOOGLE_DRIVE_PATH)
print(os.listdir(GOOGLE_DRIVE_PATH))
import matplotlib.pyplot as plt
import numpy
from numpy import cov
from numpy import trace
from numpy import iscomplexobj
from numpy import asarray
from numpy.random import randint
from scipy.linalg import sqrtm
from keras.applications.inception_v3 import InceptionV3
from keras.applications.inception_v3 import preprocess_input
from keras.datasets.mnist import load_data
from skimage.transform import resize
from skimage.io import imread_collection
def calculate_fid(model, images1, images2):
# calculate activations
act1 = model.predict(images1)
act2 = model.predict(images2)
# calculate mean and covariance statistics
mu1, sigma1 = act1.mean(axis=0), cov(act1, rowvar=False)
mu2, sigma2 = act2.mean(axis=0), cov(act2, rowvar=False)
# calculate sum squared difference between means
ssdiff = numpy.sum((mu1 - mu2)**2.0)
# calculate sqrt of product between cov
covmean = sqrtm(sigma1.dot(sigma2))
# check and correct imaginary numbers from sqrt
if iscomplexobj(covmean):
covmean = covmean.real
# calculate score
fid = ssdiff + trace(sigma1 + sigma2 - 2.0 * covmean)
return fid
# define two fake collections of images
images1 = randint(0, 255, 100*256*256*3)
images1 = images1.reshape((100,256,256,3))
images2 = randint(0, 255, 200*256*256*3)
images2 = images2.reshape((200,256,256,3))
print('Prepared', images1.shape, images2.shape)
# convert integer to floating point values
images1 = images1.astype('float32')
images2 = images2.astype('float32')
# pre-process images
images1 = preprocess_input(images1)
images2 = preprocess_input(images2)
# Visualize
plt.subplot(121)
plt.title('Image 1')
plt.imshow(images1[0])
plt.subplot(122)
plt.title('Image 2')
plt.imshow(images2[0])
# prepare the inception v3 model
model = InceptionV3(include_top=False, pooling='avg', input_shape=(256,256,3))
# fid between images1 and images1
fid = calculate_fid(model, images1, images1)
print('FID of same images: %.3f' % fid)
# fid between images1 and images2
fid = calculate_fid(model, images1, images2)
print('FID of different images: %.3f' % fid)
# Create data/
!mkdir data/
!mkdir data/style/
!mkdir data/style/train/
!mkdir data/style/test/
# Download Datasets from UCB
!wget -cO - https://people.eecs.berkeley.edu/%7Etaesung_park/CycleGAN/datasets/cezanne2photo.zip > cezanne.zip
!wget -cO - https://people.eecs.berkeley.edu/%7Etaesung_park/CycleGAN/datasets/monet2photo.zip > monet.zip
!wget -cO - https://people.eecs.berkeley.edu/%7Etaesung_park/CycleGAN/datasets/ukiyoe2photo.zip > ukiyoe.zip
!wget -cO - https://people.eecs.berkeley.edu/%7Etaesung_park/CycleGAN/datasets/vangogh2photo.zip > vangogh.zip
# Unzip files
!unzip cezanne.zip
!unzip monet.zip
!unzip ukiyoe.zip
!unzip vangogh.zip
!rm -f cezanne.zip
!mv cezanne2photo/trainA/ data/style/train/cezanne/
!mv cezanne2photo/testA/ data/style/test/cezanne/
!mv cezanne2photo/trainB/ data/style/train/photo/
!mv cezanne2photo/testB/ data/style/test/photo/
!rm -r cezanne2photo/
!rm -f monet.zip
!mv monet2photo/trainA/ data/style/train/monet/
!mv monet2photo/testA/ data/style/test/monet/
!rm -r monet2photo/
!rm -f ukiyoe.zip
!mv ukiyoe2photo/trainA/ data/style/train/ukiyoe/
!mv ukiyoe2photo/testA/ data/style/test/ukiyoe/
!rm -r ukiyoe2photo/
!rm -f vangogh.zip
!mv vangogh2photo/trainA/ data/style/train/vangogh/
!mv vangogh2photo/testA/ data/style/test/vangogh/
!rm -r vangogh2photo/
TRAIN_IMG_DIR = 'data/style/train'
TEST_IMG_DIR = 'data/style/test'
def load_img_dir(label='photo', size=None):
assert label in {'photo', 'cezanne', 'monet', 'ukiyoe', 'vangogh'}
dir_photo = os.path.join(TEST_IMG_DIR, '{}/*.jpg'.format(label))
img_photo = numpy.array(imread_collection(dir_photo))
if size is None:
size = len(img_photo)
else:
img_photo = img_photo[:size]
img_photo = img_photo.reshape((size,256,256,3)).astype('float32')
img_photo = preprocess_input(img_photo)
return img_photo
model = InceptionV3(include_top=False, pooling='avg', input_shape=(256, 256, 3))
size = 4
print('Loading images...')
img_photo = load_img_dir('photo', size)
img_cezanne = load_img_dir('cezanne', size)
img_monet = load_img_dir('monet', size)
img_ukiyoe = load_img_dir('ukiyoe', size)
img_vangogh = load_img_dir('vangogh', size)
print('Start testing...')
fid = calculate_fid(model, img_photo, img_cezanne)
print('FID between photo and cezanne: %.3f' % fid)
fid = calculate_fid(model, img_photo, img_monet)
print('FID between photo and monet: %.3f' % fid)
fid = calculate_fid(model, img_photo, img_ukiyoe)
print('FID between photo and ukiyoe: %.3f' % fid)
fid = calculate_fid(model, img_photo, img_vangogh)
print('FID between photo and vangogh: %.3f' % fid)
fid = calculate_fid(model, img_cezanne, img_monet)
print('FID between cezanne and monet: %.3f' % fid)
fid = calculate_fid(model, img_cezanne, img_ukiyoe)
print('FID between cezanne and ukiyoe: %.3f' % fid)
fid = calculate_fid(model, img_cezanne, img_vangogh)
print('FID between cezanne and vangogh: %.3f' % fid)
fid = calculate_fid(model, img_monet, img_ukiyoe)
print('FID between monet and ukiyoe: %.3f' % fid)
fid = calculate_fid(model, img_monet, img_vangogh)
print('FID between monet and vangogh: %.3f' % fid)
fid = calculate_fid(model, img_ukiyoe, img_vangogh)
print('FID between ukiyoe and vangogh: %.3f' % fid)
def load_out_dir(dir, label='cezanne', size=None):
assert label in {'cezanne', 'monet', 'ukiyoe', 'vangogh'}
dir_photo = os.path.join(dir, '{}/*.jpg'.format(label))
img_photo = numpy.array(imread_collection(dir_photo))
if size is None:
size = len(img_photo)
else:
img_photo = img_photo[:size]
img_photo = img_photo.reshape((size,256,256,3)).astype('float32')
img_photo = preprocess_input(img_photo)
return img_photo
dir_cyclegan = os.path.join(GOOGLE_DRIVE_PATH, 'checkpoints/checkpoints_20w/one_result')
print(os.listdir(dir_cyclegan))
size = 4
print('Loading images...')
cyclegan_cezanne = load_out_dir(dir_cyclegan, 'cezanne', size)
cyclegan_monet = load_out_dir(dir_cyclegan, 'monet', size)
cyclegan_ukiyoe = load_out_dir(dir_cyclegan, 'ukiyoe', size)
cyclegan_vangogh = load_out_dir(dir_cyclegan, 'vangogh', size)
print('Start testing...')
fid = calculate_fid(model, img_cezanne, cyclegan_cezanne)
print('FID between cyclegan generated fake cezanne and test cyclegan images: %.3f' % fid)
fid = calculate_fid(model, img_monet, cyclegan_monet)
print('FID between cyclegan generated fake monet and test cyclegan monet: %.3f' % fid)
fid = calculate_fid(model, img_ukiyoe, cyclegan_ukiyoe)
print('FID between cyclegan generated fake ukiyoe and test cyclegan ukiyoe: %.3f' % fid)
fid = calculate_fid(model, img_vangogh, cyclegan_vangogh)
print('FID between cyclegan generated fake vangogh and test cyclegan vangogh: %.3f' % fid)
dir_cyclegan = os.path.join(GOOGLE_DRIVE_PATH, 'results/')
print(os.listdir(dir_cyclegan))
size = 100
print('Loading images...')
cyclegan_cezanne = load_out_dir(dir_cyclegan, 'cezanne', size)
cyclegan_monet = load_out_dir(dir_cyclegan, 'monet', size)
cyclegan_ukiyoe = load_out_dir(dir_cyclegan, 'ukiyoe', size)
cyclegan_vangogh = load_out_dir(dir_cyclegan, 'vangogh', size)
print('Start testing...')
fid = calculate_fid(model, img_cezanne, cyclegan_cezanne)
print('FID between cyclegan generated fake cezanne and test cyclegan images: %.3f' % fid)
fid = calculate_fid(model, img_monet, cyclegan_monet)
print('FID between cyclegan generated fake monet and test cyclegan monet: %.3f' % fid)
fid = calculate_fid(model, img_ukiyoe, cyclegan_ukiyoe)
print('FID between cyclegan generated fake ukiyoe and test cyclegan ukiyoe: %.3f' % fid)
fid = calculate_fid(model, img_vangogh, cyclegan_vangogh)
print('FID between cyclegan generated fake vangogh and test cyclegan vangogh: %.3f' % fid)
| 0.33231 | 0.818374 |
# First Last - Units in Python
---
### [Astropy Units](http://docs.astropy.org/en/stable/units/index.html#module-astropy.units.si)
### [Astropy Constants](https://docs.astropy.org/en/stable/constants/#module-astropy.constants)
### [Numpy Math Functions](https://docs.scipy.org/doc/numpy-1.13.0/reference/routines.math.html)
---
* Use the `Astropy` units and constants packages to solve the following problems.
* Do not hardcode any constants!
* Unless asked, your units should be in the simplest SI units possible.
* For each problem:
* Write a FUNCTION to solve the problem
* Run the function with input with units
* Format the output as a sentence - For example: `The range would be 123 km.`
```
import numpy as np
from astropy import units as u
from astropy import constants as const
from astropy.units import imperial
imperial.enable()
```
---
### The range of a projectile launched with a velocity (v) at and angle ($\theta$) is
$$ \large
R\ =\ {v^2 \over g}\ sin(2\theta)
$$
**Problem 1** - Find R for v = 123 mph and $\theta$ = 1000 arc minutes
```
# Write a function
# Run the Function
# Format the output
```
**Problem 2** - How fast do you have to throw a football at 33.3 degrees so that is goes exactly 100 yards? Express your answer in mph
```
# Write a function
# Run the Function
# Format the output
```
---
### Kepler's third law can be expressed as:
$$ \large
T^2 = \left( {{4\pi^2} \over {GM}} \right)\ r^3
$$
Where __T__ is the orbial period of an object at distance (__r__) from the center of an object of mass (__M__).
And the velocity of an object in orbit is
$$ \large
v=\sqrt{GM\over r}
$$
**Problem 3** - Calculate the orbital period of International Space Station (ISS). ISS orbits 254 miles above the **surface** of the Earth. Expess your answer in minutes (make sure your answer makes sense!).
```
# Write a function
# Run the Function
# Format the output
```
**Problem 4** - Calculate the velocity of ISS. Expess your answer in km/s and mph.
```
# Write a function
# Run the Function
# Format the output
```
**Problem 5** - The Procliamer's song [500 miles](https://youtu.be/MJuyn0WAYNI?t=27s) has a duration of 3 minutes and 33 seconds. Calculate at what altitude, above the Earth's surface, you would have to orbit to go 1000 miles in this time. Express your answer in miles about the Earth's surface.
```
# Write a function
# Run the Function
# Format the output
```
---
### The Power being received by a solar panel in space can be expressed as:
$$ \large
I\ =\ {{L_{\odot}} \over {4 \pi d^2}}\ \varepsilon
$$
Where __I__ is the power __per unit area__ at a distance (__d__) from the Sun, and $\varepsilon$ is the efficiency of the solar panel.
The solar panels that power spacecraft have an efficiency of about 40%.
**Problem 6** - The [New Horizons](http://pluto.jhuapl.edu/) spacecraft requires 220 Watts of power.
Calculate the area of a solar panel that would be needed to power New Horizons at a distance of 1 AU from the Sun.
```
# Write a function
# Run the Function
# Format the output
```
**Problem 7** - Express your answer in units of the area of a piece of US letter sized paper (8.5 in x 11 in).
```
# Format the output
```
**Problem 8** - Same question as above but now a d = 30 AU.
Express you answer in US letter sized paper
```
# Run the Function
# Format the output
```
**Problem 9** - The main part of the Oort cloud is thought to be at a distance of about 10,000 AU.
Calculate the size of the solar panel New Horizons would need to operate in the Oort cloud.
Express your answer in units of the area of the pitch at [Carrow Row](https://en.wikipedia.org/wiki/Carrow_Road) (114 yd x 74 yd).
```
# Run the Function
# Format the output
```
**Problem 10** - Calculate the maximum distance from the Sun where a solar panel of 1 Carrow Row pitch can power the New Horizons spacecraft. Express your answer in AU.
```
# Write a function
# Run the Function
# Format the output
```
---
### Dark Matter Halo
The distribution of mass density in a dark matter halo can be appoximated as:
$$
\large
\rho (r)\ =\ \frac{\rho_{0}}
{1 + \left(\frac{r}{r_{c}}\right)^{2}}
$$
Where $r$ is the distance from the center of the Galaxy.
**Problem 11** - Calculate the dark matter mass density at $r$ = 26092.51 light years, with $r_{c}$ = 5.78 $\times$ 10$^{8}$ AU and $\rho_{0}$ =
5.9 $\times$ 10$^{7}$ M$_{\odot}$/kpc$^{3}$.
Express your answer in proton masses per cm$^{3}$.
```
# Write a function
# Run the Function
# Format the output
```
---
### Earth?
The self gravitational potential energy of the Earth is:
$$ \large
PE \ = \ \frac{3}{5} \cdot \frac{GM_{\oplus}^2}{R_{\oplus}}
$$
This is the amount of energy you need to give the Earth to move all of its components pieces infinitely far away (i.e. to destroy it!).
**Problem 12** - Calculate how massive an object you whould have to hit the Earth with at 45,000 mph to destroy the Earth. Express your answer in Earth masses.
```
# Write a function
# Run the Function
# Format the output
```
---
### Due Mon Feb 3 - 1 pm
- `Make sure to change the filename to your name!`
- `Make sure to change the Title to your name!`
- `Save your file to HTML: File -> Download as -> HTML`
- `Upload the .html file to Canvas`
|
github_jupyter
|
import numpy as np
from astropy import units as u
from astropy import constants as const
from astropy.units import imperial
imperial.enable()
# Write a function
# Run the Function
# Format the output
# Write a function
# Run the Function
# Format the output
# Write a function
# Run the Function
# Format the output
# Write a function
# Run the Function
# Format the output
# Write a function
# Run the Function
# Format the output
# Write a function
# Run the Function
# Format the output
# Format the output
# Run the Function
# Format the output
# Run the Function
# Format the output
# Write a function
# Run the Function
# Format the output
# Write a function
# Run the Function
# Format the output
# Write a function
# Run the Function
# Format the output
| 0.706596 | 0.992934 |
# Vix Backwardation and the SPX
For a while I've been curious to see if there is any effect on a backwardated term structure in Vix Futures on future returns in the SPX.
#### Contago and Backwardation
<img src="http://www.cboeoptionshub.com/wp-content/uploads/2014/08/Contango-Fixed.jpg" title="Example of generic futures curve in contago and backwardation" width="500px">
#### A brief background on why this might be interesting...
The VIX measures the price that traders are willing to buy options to protect their portfolio. The spot VIX measures this price. You can buy futures on the VIX. Essentially you are making a bet where the VIX will settle on the date of expiration of the VIX future contract. At settlement you get paid the amount your future is worth. Thus, the futures trade off the price that traders think the index will be at settlement. In times of stress trader run to buy options, pushing the VIX up. Since the VIX is mean reverting this will pull the front month up more than the back month since traders figure that over time the VIX will return to is average levels of about 20. This term structure where the front month is greater than the back month is referred to as backwardation.
However, the "natural" term structure for VIX Futures is contango since they are somewhat tied to the price of SPX options which are naturally more expensive further out in time since there is more uncertainty in the future (even adjusted for time) -- this is why back month options trade at a higher vol (usually) than front month options.
During big down drafts we see the VIX future curve go into steep backwardation.
#### Here's the question:
Does this backwardation happen quickly enough into the drawdown to get you out?
```
import pandas as pd
import pandas.io.data as web
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
import seaborn
from datetime import datetime, timedelta
%matplotlib inline
df = web.DataReader("^GSPC", 'yahoo', datetime(2000, 1, 1), datetime.today())
raw_vix_df = web.DataReader("^VIX", 'yahoo', datetime(2000, 1, 1), datetime.today())
raw_vix_df['vix_close'] = raw_vix_df['Close']
raw_vix_df = raw_vix_df['vix_close'] # drop all the HL shit
vix_df = pd.read_html("http://vixcentral.com/historical/?days=10000", header=0)[0]
vix_df = vix_df.drop(vix_df.index[-1:]) # get rid of last row
vix_df = vix_df.set_index('Date')
vix_df.index = pd.to_datetime(vix_df.index)
df['Close'].plot(figsize=(16, 6))
plt.xlabel('Date', fontsize=14)
plt.ylabel('SPX Close', fontsize=14)
plt.title("Our frienemy, the SPX", fontsize=16)
# calculate degree of backwardation between month 1 and 2
vix_df['f2-f1'] = vix_df['F2'] - vix_df['F1']
vix_df['backwardated'] = vix_df['f2-f1'] < 0
# join our tables and sort with date ascending
master = df.join(raw_vix_df, how='inner')
master = master.join(vix_df, how='inner')
master = master.sort_index()
# compute rolling returns for 1 day, 5 days, 2 weeks, 1 month, 3 months, 6 months and 12 months
# note this is tricky than it looks -- our dataframe is in acending order,
# so pct_change(periods=1) calculates the chage from day1 to day2, but this change is aligned with day2 so
# we *must* shift is back the same number of periods that the change is calculated over
master['1d'] = master['Close'].pct_change(periods=1).shift(-1) * 100
master['5d'] = master['Close'].pct_change(periods=5).shift(-5) * 100
master['10d'] = master['Close'].pct_change(periods=10).shift(-10) * 100
master['1m'] = master['Close'].pct_change(periods=20).shift(-20) * 100
master['3m'] = master['Close'].pct_change(periods=60).shift(-60) * 100
master['6m'] = master['Close'].pct_change(periods=120).shift(-120) * 100
master['12m'] = master['Close'].pct_change(periods=250).shift(-250) * 100
# verify the pecentage changes look good
master[["Close", "1d", "5d"]]
# next we'll write a custom formatter
N = len(master.index)
ind = np.arange(N) # the evenly spaced plot indices
def format_date(x, pos=None):
thisind = np.clip(int(x+0.5), 0, N-2)
print(thisind)
return master.iloc[:thisind][0].format().pop()
print(master.iloc[:1].index.format().pop())
fig, ax = plt.subplots()
master['Close'].plot(ax=ax, figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
ax.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('SPX Close', fontsize=14)
plt.title("SPX Value with Backwardation Highlighted", fontsize=16)
plot = master['vix_close'].plot(figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
plot.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('VIX Close', fontsize=14)
plt.title("VIX Close with Backwardation Highlighted", fontsize=16)
```
Now, lets dig into the data, segmenting periods of backwardation and contango on the VIX futures curve.
For each rolling return we'll:
1. Plot the SPX with the backwardation highlighted in red.
2. Plot the rolling return with the backwardation highlighted in red.
3. Look at some stats about the return distribution
```
plot = master['1d'].plot(figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
plot.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('Rolling 1 day percentage returns', fontsize=14)
plt1, plt2 = master['1d'].hist(by=master['backwardated'], bins=20, figsize=(16, 6))
plt1.set_title("Contango", fontsize=16)
plt1.set_xlabel('Bucketed 1d Returns', fontsize=14)
plt1.set_ylabel('Ocurrances', fontsize=14)
plt2.set_title("Backwardation", fontsize=16)
plt2.set_xlabel('Bucketed 1d Returns', fontsize=14)
plt2.set_ylabel('Ocurrances', fontsize=14)
plot = master['5d'].plot(figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
plot.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('Rolling 5 day percentage returns', fontsize=14)
plt1, plt2 = master['5d'].hist(by=master['backwardated'], bins=20, figsize=(16, 6))
plt1.set_title("Contango", fontsize=16)
plt1.set_xlabel('Bucketed 5d Returns', fontsize=14)
plt1.set_ylabel('Ocurrances', fontsize=14)
plt2.set_title("Backwardation", fontsize=16)
plt2.set_xlabel('Bucketed 5d Returns', fontsize=14)
plt2.set_ylabel('Ocurrances', fontsize=14)
plot = master['10d'].plot(figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
plot.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('Rolling 10 day percentage returns', fontsize=14)
plt1, plt2 = master['10d'].hist(by=master['backwardated'], bins=20, figsize=(16, 6))
plt1.set_title("Contango", fontsize=16)
plt1.set_xlabel('Bucketed 10d Returns', fontsize=14)
plt1.set_ylabel('Ocurrances', fontsize=14)
plt2.set_title("Backwardation", fontsize=16)
plt2.set_xlabel('Bucketed 10d Returns', fontsize=14)
plt2.set_ylabel('Ocurrances', fontsize=14)
plot = master['1m'].plot(figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
plot.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('Rolling 1 month percentage returns', fontsize=14)
plt1, plt2 = master['1m'].hist(by=master['backwardated'], bins=20, figsize=(16, 6))
plt1.set_title("Contango", fontsize=16)
plt1.set_xlabel('Bucketed 1m Returns', fontsize=14)
plt1.set_ylabel('Ocurrances', fontsize=14)
plt2.set_title("Backwardation", fontsize=16)
plt2.set_xlabel('Bucketed 1m Returns', fontsize=14)
plt2.set_ylabel('Ocurrances', fontsize=14)
plot = master['3m'].plot(figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
plot.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('Rolling 3 month percentage returns', fontsize=14)
plt1, plt2 = master['3m'].hist(by=master['backwardated'], bins=20, figsize=(16, 6))
plt1.set_title("Contango", fontsize=16)
plt1.set_xlabel('Bucketed 3m Returns', fontsize=14)
plt1.set_ylabel('Ocurrances', fontsize=14)
plt2.set_title("Backwardation", fontsize=16)
plt2.set_xlabel('Bucketed 3m Returns', fontsize=14)
plt2.set_ylabel('Ocurrances', fontsize=14)
plot = master['6m'].plot(figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
plot.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('Rolling 6 month percentage returns', fontsize=14)
plt1, plt2 = master['6m'].hist(by=master['backwardated'], bins=20, figsize=(16, 6))
plt1.set_title("Contango", fontsize=16)
plt1.set_xlabel('Bucketed 6m Returns', fontsize=14)
plt1.set_ylabel('Ocurrances', fontsize=14)
plt2.set_title("Backwardation", fontsize=16)
plt2.set_xlabel('Bucketed 6m Returns', fontsize=14)
plt2.set_ylabel('Ocurrances', fontsize=14)
plot = master['12m'].plot(figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
plot.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('Rolling 12 month percentage returns', fontsize=14)
plt1, plt2 = master['12m'].hist(by=master['backwardated'], bins=20, figsize=(16, 6))
plt1.set_title("Contango", fontsize=16)
plt1.set_xlabel('Bucketed 12m Returns', fontsize=14)
plt1.set_ylabel('Ocurrances', fontsize=14)
plt2.set_title("Backwardation", fontsize=16)
plt2.set_xlabel('Bucketed 12m Returns', fontsize=14)
plt2.set_ylabel('Ocurrances', fontsize=14)
master.groupby(master['backwardated']).describe()[['vix_close', '1d','5d', '10d', '1m', '3m', '6m', '12m']]
```
## TL;DR
So what did we see?
When the term structure of the **VIX is backwardated we see average returns across all time frames actually increase!** But, we also have a greater dispersion. This is pretty much a given, since when the VIX is backwadated the Vol level is elevated. We could show this is the case with a correlation analysis, but if you trade you already knew this.
Basically, this means it is a useless signal for the long-only equity trader. You might think that it's actually a inverse signal, but buying the wrong downdraft (i.e. 2008) and you don't get to play again.
For the options trader the question is more complicated since options are much more expensive during these periods, but the moves are much bigger too.
The main problem is that that data set for VIX futures is just too small! We only have ~150 days of backwadadion in the last 6 years. It's hardly enough to go on.
I'll be honest. I'm a premium seller and I'm nervous opening new positions right. Part of me says that this hesitancy is why these trades will pay out. So I'll probably sell some SPX strangles tomorrow, small.
That said, I'm still curious how returns would compare between the premium seller who goes flat during backwadation and the guy who holds on. My guess is that over this data set (2009-present) the guy who held on killed it. But, that same trader would have gotten killed in 2008-2009. I might try to model this using the buy-write index, so be on the lookout for that.
I'd also be curious to segment out the data with respect to the steepness of the curve (i.e., how are return when we are in 1% contago vs -5% backwardation vs 10% backwardation), but honestly you can't draw any good conclusions with this small dataset, so I'll skip it for now.
|
github_jupyter
|
import pandas as pd
import pandas.io.data as web
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
import seaborn
from datetime import datetime, timedelta
%matplotlib inline
df = web.DataReader("^GSPC", 'yahoo', datetime(2000, 1, 1), datetime.today())
raw_vix_df = web.DataReader("^VIX", 'yahoo', datetime(2000, 1, 1), datetime.today())
raw_vix_df['vix_close'] = raw_vix_df['Close']
raw_vix_df = raw_vix_df['vix_close'] # drop all the HL shit
vix_df = pd.read_html("http://vixcentral.com/historical/?days=10000", header=0)[0]
vix_df = vix_df.drop(vix_df.index[-1:]) # get rid of last row
vix_df = vix_df.set_index('Date')
vix_df.index = pd.to_datetime(vix_df.index)
df['Close'].plot(figsize=(16, 6))
plt.xlabel('Date', fontsize=14)
plt.ylabel('SPX Close', fontsize=14)
plt.title("Our frienemy, the SPX", fontsize=16)
# calculate degree of backwardation between month 1 and 2
vix_df['f2-f1'] = vix_df['F2'] - vix_df['F1']
vix_df['backwardated'] = vix_df['f2-f1'] < 0
# join our tables and sort with date ascending
master = df.join(raw_vix_df, how='inner')
master = master.join(vix_df, how='inner')
master = master.sort_index()
# compute rolling returns for 1 day, 5 days, 2 weeks, 1 month, 3 months, 6 months and 12 months
# note this is tricky than it looks -- our dataframe is in acending order,
# so pct_change(periods=1) calculates the chage from day1 to day2, but this change is aligned with day2 so
# we *must* shift is back the same number of periods that the change is calculated over
master['1d'] = master['Close'].pct_change(periods=1).shift(-1) * 100
master['5d'] = master['Close'].pct_change(periods=5).shift(-5) * 100
master['10d'] = master['Close'].pct_change(periods=10).shift(-10) * 100
master['1m'] = master['Close'].pct_change(periods=20).shift(-20) * 100
master['3m'] = master['Close'].pct_change(periods=60).shift(-60) * 100
master['6m'] = master['Close'].pct_change(periods=120).shift(-120) * 100
master['12m'] = master['Close'].pct_change(periods=250).shift(-250) * 100
# verify the pecentage changes look good
master[["Close", "1d", "5d"]]
# next we'll write a custom formatter
N = len(master.index)
ind = np.arange(N) # the evenly spaced plot indices
def format_date(x, pos=None):
thisind = np.clip(int(x+0.5), 0, N-2)
print(thisind)
return master.iloc[:thisind][0].format().pop()
print(master.iloc[:1].index.format().pop())
fig, ax = plt.subplots()
master['Close'].plot(ax=ax, figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
ax.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('SPX Close', fontsize=14)
plt.title("SPX Value with Backwardation Highlighted", fontsize=16)
plot = master['vix_close'].plot(figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
plot.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('VIX Close', fontsize=14)
plt.title("VIX Close with Backwardation Highlighted", fontsize=16)
plot = master['1d'].plot(figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
plot.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('Rolling 1 day percentage returns', fontsize=14)
plt1, plt2 = master['1d'].hist(by=master['backwardated'], bins=20, figsize=(16, 6))
plt1.set_title("Contango", fontsize=16)
plt1.set_xlabel('Bucketed 1d Returns', fontsize=14)
plt1.set_ylabel('Ocurrances', fontsize=14)
plt2.set_title("Backwardation", fontsize=16)
plt2.set_xlabel('Bucketed 1d Returns', fontsize=14)
plt2.set_ylabel('Ocurrances', fontsize=14)
plot = master['5d'].plot(figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
plot.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('Rolling 5 day percentage returns', fontsize=14)
plt1, plt2 = master['5d'].hist(by=master['backwardated'], bins=20, figsize=(16, 6))
plt1.set_title("Contango", fontsize=16)
plt1.set_xlabel('Bucketed 5d Returns', fontsize=14)
plt1.set_ylabel('Ocurrances', fontsize=14)
plt2.set_title("Backwardation", fontsize=16)
plt2.set_xlabel('Bucketed 5d Returns', fontsize=14)
plt2.set_ylabel('Ocurrances', fontsize=14)
plot = master['10d'].plot(figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
plot.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('Rolling 10 day percentage returns', fontsize=14)
plt1, plt2 = master['10d'].hist(by=master['backwardated'], bins=20, figsize=(16, 6))
plt1.set_title("Contango", fontsize=16)
plt1.set_xlabel('Bucketed 10d Returns', fontsize=14)
plt1.set_ylabel('Ocurrances', fontsize=14)
plt2.set_title("Backwardation", fontsize=16)
plt2.set_xlabel('Bucketed 10d Returns', fontsize=14)
plt2.set_ylabel('Ocurrances', fontsize=14)
plot = master['1m'].plot(figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
plot.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('Rolling 1 month percentage returns', fontsize=14)
plt1, plt2 = master['1m'].hist(by=master['backwardated'], bins=20, figsize=(16, 6))
plt1.set_title("Contango", fontsize=16)
plt1.set_xlabel('Bucketed 1m Returns', fontsize=14)
plt1.set_ylabel('Ocurrances', fontsize=14)
plt2.set_title("Backwardation", fontsize=16)
plt2.set_xlabel('Bucketed 1m Returns', fontsize=14)
plt2.set_ylabel('Ocurrances', fontsize=14)
plot = master['3m'].plot(figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
plot.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('Rolling 3 month percentage returns', fontsize=14)
plt1, plt2 = master['3m'].hist(by=master['backwardated'], bins=20, figsize=(16, 6))
plt1.set_title("Contango", fontsize=16)
plt1.set_xlabel('Bucketed 3m Returns', fontsize=14)
plt1.set_ylabel('Ocurrances', fontsize=14)
plt2.set_title("Backwardation", fontsize=16)
plt2.set_xlabel('Bucketed 3m Returns', fontsize=14)
plt2.set_ylabel('Ocurrances', fontsize=14)
plot = master['6m'].plot(figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
plot.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('Rolling 6 month percentage returns', fontsize=14)
plt1, plt2 = master['6m'].hist(by=master['backwardated'], bins=20, figsize=(16, 6))
plt1.set_title("Contango", fontsize=16)
plt1.set_xlabel('Bucketed 6m Returns', fontsize=14)
plt1.set_ylabel('Ocurrances', fontsize=14)
plt2.set_title("Backwardation", fontsize=16)
plt2.set_xlabel('Bucketed 6m Returns', fontsize=14)
plt2.set_ylabel('Ocurrances', fontsize=14)
plot = master['12m'].plot(figsize=(16, 6))
for idx, row in master.iterrows():
if row['backwardated']:
plot.axvspan(idx, idx+timedelta(days=1), facecolor='red', edgecolor='none', alpha=.2)
plt.xlabel('Date (Red denotes periods of backwardation)', fontsize=14)
plt.ylabel('Rolling 12 month percentage returns', fontsize=14)
plt1, plt2 = master['12m'].hist(by=master['backwardated'], bins=20, figsize=(16, 6))
plt1.set_title("Contango", fontsize=16)
plt1.set_xlabel('Bucketed 12m Returns', fontsize=14)
plt1.set_ylabel('Ocurrances', fontsize=14)
plt2.set_title("Backwardation", fontsize=16)
plt2.set_xlabel('Bucketed 12m Returns', fontsize=14)
plt2.set_ylabel('Ocurrances', fontsize=14)
master.groupby(master['backwardated']).describe()[['vix_close', '1d','5d', '10d', '1m', '3m', '6m', '12m']]
| 0.489503 | 0.919823 |
# ProjectQ Simulator Tutorial
The aim of this tutorial is to introduce some of the basic and more advanced features of the ProjectQ simulator. Please note that all the simulator features can be found in our [code documentation](http://projectq.readthedocs.io/en/latest/projectq.backends.html#projectq.backends.Simulator).
Contents:
* [Introduction](#Introduction)
* [Installation](#Installation)
* [Basics](#Basics)
* [Advanced features](#Advanced_features)
* [Improving the speed of the ProjectQ simulator](#improve_speed)
# Introduction <a id="Introduction"></a>
Our simulator can be used to simulate any circuit model quantum algorithm. This requires storing the state, also called wavefunction, of all qubits which requires storing 2<sup>n</sup> complex values (each of size 16 bytes) for an *n*-qubit algorithm. This can get very expensive in terms of required RAM:
Number of qubits *n*| Required RAM to store wavefunction
------------------- | ----------------------------------
10 | 16 KByte
20 | 16 MByte
30 | 16 GByte
31 | 32 GByte
32 | 64 GByte
40 | 16 TByte
45 | 512 TByte ([world's largest quantum computer simulation](https://arxiv.org/abs/1704.01127))
The number of qubits you can simulate with the ProjectQ simulator is only limited by the amount of RAM in your notebook or workstation. Applying quantum gates is expensive as we have to potentially update each individual value in the full wavefunction. Therefore, we have implemented a high-performance simulator which is significantly faster than other simulators (see our papers for a detailed comparison [[1]](https://arxiv.org/abs/1612.08091), [[2]](https://arxiv.org/abs/1604.06460)). The performance of such simulators is hardware-dependent and therefore we have decided to provide 4 different versions. A simulator implemented in C++ which uses multi-threading (OpenMP) and instrinsics, a C++ simulator which uses intrinsics, a C++ simulator, and a slower simulator which only uses Python. During the installation we try to install the fastest of these options given your hardware and available compiler.
Our simulator is simultaneously also a quantum emulator. This concept was first introduced by us in [[2]](https://arxiv.org/abs/1604.06460). A quantum emulator takes classical shortcuts when performing the simulation and hence can be orders of magnitude faster. E.g., for simulating Shor's algorithm, we only require to store the wavefunction of *n+1* qubits while the algorithm on a real quantum computer would require *2n+O(1)* qubits. Using these emulation capabilities, we can easily emulate Shor's algorithm for factoring, e.g., 4028033 = 2003 · 2011 on a notebook [[1]](https://arxiv.org/abs/1612.08091).
# Installation <a id="Installation"></a>
Please follow the [installation instructions](http://projectq.readthedocs.io/en/latest/tutorials.html#getting-started) in the docs. The Python interface to all our different simulators (C++ or native Python) is identical. The different versions only differ in performance. If you have a C++ compiler installed on your system, the setup will try to install the faster C++ simulator. To figure out which simulator is installed just execute the following code after installing ProjectQ:
```
import projectq
eng = projectq.MainEngine() # This loads the simulator as it is the default backend
```
If you now see the following message
```
(Note: This is the (slow) Python simulator.)
```
you are using the slow Python simulator and we would recommend to reinstall ProjectQ with the C++ simulator following the [installation instructions](http://projectq.readthedocs.io/en/latest/tutorials.html#getting-started). If this message doesn't show up, then you are using one of the fast C++ simulator versions. Which one exactly depends on your compiler and hardware.
# Basics <a id="Basics"></a>
To write a quantum program, we need to create a compiler called `MainEngine` and provide a backend for which the compiler should compile the quantum program. In this tutorial we are focused on the simulator as a backend. We can create a compiler with a simulator backend by importing the simulator class and creating a simulator instance which is passed to the `MainEngine`:
```
from projectq.backends import Simulator
eng = projectq.MainEngine(backend=Simulator())
```
Note that the `MainEngine` contains the simulator as the default backend, hence one can equivalently just do:
```
eng = projectq.MainEngine()
```
In order to simulate the probabilistic measurement process, the simulator internally requires a random number generator. When creating a simulator instance, one can provide a random seed in order to create reproducible results:
```
eng = projectq.MainEngine(backend=Simulator(rnd_seed=10))
```
Let's write a simple test program which allocates a qubit in state |0>, performs a Hadamard gate which puts it into a superposition 1/sqrt(2) * ( |0> + |1>) and then measure the qubit:
```
import projectq
from projectq.ops import Measure, H
eng = projectq.MainEngine()
qubit = eng.allocate_qubit() # Allocate a qubit from the compiler (MainEngine object)
H | qubit
Measure | qubit # Measures the qubit in the Z basis and collapses it into either |0> or |1>
eng.flush() # Tells the compiler (MainEninge) compile all above gates and send it to the simulator backend
print(int(qubit)) # The measurement result can be accessed by converting the qubit object to a bool or int
```
This program randomly outputs 0 or 1. Note that the measurement does *not* return for example a probability of measuring 0 or 1 (see below how this could be achieved). The reason for this is that a program written in our high-level syntax should be independent of the backend. In other words, the code can be executed either by our simulator or by exchanging only the MainEngine's backend by a real device which cannot return a probability but only one value. See the other examples on GitHub of how to execute code on the IBM quantum chip.
### Important note on eng.flush()
Note that we have used eng.flush() which tells the compiler to send all the instructions to the backend. In a simplified version, when the Python interpreter executes a gate (e.g. the above lines with H, or Measure), this gate is sent to the compiler (MainEngine), where it is cached. Compilation and optimization of cached gates happens irregularly, e.g., an optimizer in the compiler might wait for 5 gates before it starts the optimization process. Therefore, if we require the result of a quantum program, we need to call eng.flush() which tells the compiler to compile and send all cached gates to the backend for execution. eng.flush() is therefore necessary when accessing the measurement result by converting the qubit object to an int or bool. Or when using the advanced features below, where we want to access properties of the wavefunction at a specific point in the quantum algorithm.
The above statement is not entirely correct as for example there is no eng.flush() strictly required before accessing the measurement result. The reason is that the measurement gate in our compiler is not cached but is sent directly to the local simulator backend because it would allow for performance optimizations by shrinking the wavefunction. However, this is not a feature which your code should/can rely on and therefore you should always use eng.flush().
### Important debugging feature of our simulator
When a qubit goes out of scope, it gets deallocated automatically. If the backend is a simulator, it checks that the qubit was in a classical state and otherwise it raises an exception. This is an important debugging feature as in many quantum algorithms, ancilla qubits are used for intermediate results and then "uncomputed" back into state |0>. If such ancilla qubits now go out of scope, the simulator throws an error if they are not in either state |0> or |1>, as this is most likely a bug in the user code:
```
def test_program(eng):
# Do something
ancilla = eng.allocate_qubit()
H | ancilla
# Use the ancilla for something
# Here the ancilla is not reset to a classical state but still in a superposition and will go out of scope
test_program(eng)
```
If you are using a qubit as an ancilla which should have been reset, this is a great feature which automatically checks the correctness of the uncomputation if the simulator is used as a backend. Should you wish to deallocate qubits which might be in a superposition, always apply a measurement gate in order to avoid this error message:
```
from projectq.ops import All
def test_program_2(eng):
# Do something
ancillas = eng.allocate_qureg(3) # allocates a quantum register with 3 qubits
All(H) | ancillas # applies a Hadamard gate to each of the 3 ancilla qubits
All(Measure) | ancillas # Measures all ancilla qubits such that we don't get
#an error message when they are deallocated
# Here the ancillas will go out of scope but because of the measurement, they are in a classical state
test_program_2(eng)
```
# Advanced features <a id="Advanced_features"></a>
Here we will show some features which are unique to a simulator backend which has access to the full wavefunction. Note that using these features in your code will prohibit the code to run on a real quantum device. Therefore instead of, e.g., using the feature to ininitialize the wavefunction into a specific state, you could execute a small quantum circuit which prepares the desired state and hence the code can be run on both the simulator and on actual hardware.
For details on the simulator please see the [code documentation](http://projectq.readthedocs.io/en/latest/projectq.backends.html#projectq.backends.Simulator).
In order to use the features of the simulator backend, we need to have a reference to access it. This can be achieved by creating a simulator instance and keeping a reference to it before passing it to the MainEngine:
```
sim = Simulator(rnd_seed=5) # We can now always access the simulator via the "sim" variable
eng = projectq.MainEngine(backend=sim)
```
Alternatively, one can access the simulator by accessing the backend of the compiler (MainEngine):
```
assert id(sim) == id(eng.backend)
```
### Amplitude
One can access the complex amplitude of a specific state as follows:
```
from projectq.ops import H, Measure
eng = projectq.MainEngine()
qubit = eng.allocate_qubit()
qubit2 = eng.allocate_qubit()
eng.flush() # sends the allocation of the two qubits to the simulator (only needed to showcase the stuff below)
H | qubit # Put this qubit into a superposition
# qubit is a list with one qubit, qubit2 is another list with one qubit object
# qubit + qubit2 creates a list containing both qubits. Equivalently, one could write [qubit[0], qubit2[0]]
# get_amplitude requires that one provides a list/qureg of all qubits such that it can determine the order
amp_before = eng.backend.get_amplitude('00', qubit + qubit2)
# Amplitude will be 1 as Hadamard gate is not yet executed on the simulator backend
# We forgot the eng.flush()!
print("Amplitude saved in amp_before: {}".format(amp_before))
eng.flush() # Makes sure that all the gates are sent to the backend and executed
amp_after = eng.backend.get_amplitude('00', qubit + qubit2)
# Amplitude will be 1/sqrt(2) as Hadamard gate was executed on the simulator backend
print("Amplitude saved in amp_after: {}".format(amp_after))
# To avoid triggering the warning of deallocating qubits which are in a superposition
Measure | qubit
Measure | qubit2
```
**NOTE**: One always has to call eng.flush() before accessing the amplitude as otherwise some of the gates might not have been sent and executed on the simulator. Also don't forget in such an example to measure all the qubits in the end in order to avoid the above mentioned debugging error message of deallocating qubits which are not in a classical state.
### Probability
One can access the probability of measuring one or more qubits in a specified state by the following method:
```
import projectq
from projectq.ops import H, Measure, CNOT, All
eng = projectq.MainEngine()
qureg = eng.allocate_qureg(2)
H | qureg[0]
eng.flush()
prob11 = eng.backend.get_probability('11', qureg)
prob10 = eng.backend.get_probability('10', qureg)
prob01 = eng.backend.get_probability('01', qureg)
prob00 = eng.backend.get_probability('00', qureg)
prob_second_0 = eng.backend.get_probability('0', [qureg[1]])
print("Probability to measure 11: {}".format(prob11))
print("Probability to measure 00: {}".format(prob00))
print("Probability to measure 01: {}".format(prob01))
print("Probability to measure 10: {}".format(prob10))
print("Probability that second qubit is in state 0: {}".format(prob_second_0))
All(Measure) | qureg
```
### Expectation value
We can use the QubitOperator objects to build a Hamiltonian and access the expectation value of this Hamiltonian:
```
import projectq
from projectq.ops import H, Measure, CNOT, All, QubitOperator, X, Y
eng = projectq.MainEngine()
qureg = eng.allocate_qureg(3)
X | qureg[0]
H | qureg[1]
eng.flush()
op0 = QubitOperator('Z0') # Z applied to qureg[0] tensor identity on qureg[1], qureg[2]
expectation = eng.backend.get_expectation_value(op0, qureg)
print("Expectation value = <Psi|Z0|Psi> = {}".format(expectation))
op_sum = QubitOperator('Z0 X1') - 0.5 * QubitOperator('X1')
expectation2 = eng.backend.get_expectation_value(op_sum, qureg)
print("Expectation value = <Psi|-0.5 X1 + 1.0 Z0 X1|Psi> = {}".format(expectation2))
All(Measure) | qureg # To avoid error message of deallocating qubits in a superposition
```
### Collapse Wavefunction (Post select measurement outcome)
For debugging purposes, one might want to check the algorithm for cases where an intermediate measurement outcome was, e.g., 1. Instead of running many simulations and post selecting only those with the desired intermediate measurement outcome, our simulator allows to force a specific measurement outcome. Note that this is only possible if the desired state has non-zero amplitude, otherwise the simulator will throw an error.
```
import projectq
from projectq.ops import H, Measure
eng = projectq.MainEngine()
qureg = eng.allocate_qureg(2)
# Create an entangled state:
H | qureg[0]
CNOT | (qureg[0], qureg[1])
# qureg is now in state 1/sqrt(2) * (|00> + |11>)
Measure | qureg[0]
Measure | qureg[1]
eng.flush() # required such that all above gates are executed before accessing the measurement result
print("First qubit measured in state: {} and second qubit in state: {}".format(int(qureg[0]), int(qureg[1])))
```
Running the above circuit will either produce both qubits in state 0 or both qubits in state 1. Suppose I want to check the outcome if the first qubit was measured in state 0. This can be achieve by telling the simulator backend to collapse the wavefunction for the first qubit to be in state 0:
```
import projectq
from projectq.ops import H, CNOT
eng = projectq.MainEngine()
qureg = eng.allocate_qureg(2)
# Create an entangled state:
H | qureg[0]
CNOT | (qureg[0], qureg[1])
# qureg is now in state 1/sqrt(2) * (|00> + |11>)
eng.flush() # required such that all above gates are executed before collapsing the wavefunction
# We want to check what happens to the second qubit if the first qubit (qureg[0]) is measured to be 0
eng.backend.collapse_wavefunction([qureg[0]], [0])
# Check the probability that the second qubit is measured to be 0:
prob_0 = eng.backend.get_probability('0', [qureg[1]])
print("After forcing a measurement outcome of the first qubit to be 0, \n"
"the second qubit is in state 0 with probability: {}".format(prob_0))
```
### Set wavefunction to a specific state
It is possible to set the state of the simulator to any arbitrary wavefunction. In a first step one needs to allocate all the required qubits (don't forget to call `eng.flush()`), and then one can use this method to set the wavefunction. Note that this only works if the provided wavefunction is the wavefunction of all allocated qubits. In addition, the wavefunction needs to be normalized. Here is an example:
```
import math
import projectq
from projectq.ops import H
eng = projectq.MainEngine()
qureg = eng.allocate_qureg(2)
eng.flush()
eng.backend.set_wavefunction([1/math.sqrt(2), 1/math.sqrt(2), 0, 0], qureg)
H | qureg[0]
# At this point both qubits are back in the state 00 and hence there will be no exception thrown
# when the qureg is deallocated
```
### Cheat / Accessing the wavefunction
Cheat is the original method to access and manipulate the full wavefunction. Calling cheat with the C++ simulator returns a copy of the full wavefunction plus the mapping of which qubit is at which bit position. The Python simulator returns a reference. If possible we are planning to change the C++ simulator to also return a reference which currently is not possible due to the python export. Please keep this difference in mind when writing code. If you require a copy, it is safest to make a copy of the objects returned by the `cheat` method.
When qubits are allocated in the code, each of the qubits gets a unique integer id. This id is important in order to understand the wavefunction returned by `cheat`. The wavefunction is a numpy array of length 2<sup>n</sup>, where n is the number of qubits. Which bitlocation a specific qubit in the wavefunction has is not predefined (e.g. by the order of qubit allocation) but is rather chosen depending on the compiler optimizations and the simulator. Therefore, `cheat` also returns a dictionary containing the mapping of qubit id to bit location in the wavefunction. Here is a small example:
```
import copy
import projectq
from projectq.ops import Rx, Measure, All
eng = projectq.MainEngine()
qubit1 = eng.allocate_qubit()
qubit2 = eng.allocate_qubit()
Rx(0.2) | qubit1
Rx(0.4) | qubit2
eng.flush() # In order to have all the above gates sent to the simulator and executed
# We save a copy of the wavefunction at this point in the algorithm. In order to make sure we get a copy
# also if the Python simulator is used, one should make a deepcopy:
mapping, wavefunction = copy.deepcopy(eng.backend.cheat())
print("The full wavefunction is: {}".format(wavefunction))
# Note: qubit1 is a qureg of length 1, i.e. a list containing one qubit objects, therefore the
# unique qubit id can be accessed via qubit1[0].id
print("qubit1 has bit-location {}".format(mapping[qubit1[0].id]))
print("qubit2 has bit-location {}".format(mapping[qubit2[0].id]))
# Suppose we want to know the amplitude of the qubit1 in state 0 and qubit2 in state 1:
state = 0 + (1 << mapping[qubit2[0].id])
print("Amplitude of state qubit1 in state 0 and qubit2 in state 1: {}".format(wavefunction[state]))
# If one only wants to access one (or a few) amplitudes, get_amplitude provides an easier interface:
amplitude = eng.backend.get_amplitude('01', qubit1 + qubit2)
print("Accessing same amplitude but using get_amplitude instead: {}".format(amplitude))
All(Measure) | qubit1 + qubit2 # In order to not deallocate a qubit in a superposition state
```
# Improving the speed of the ProjectQ simulator <a id="improve_speed"></a>
* Please check the [installation instructions](http://projectq.readthedocs.io/en/latest/tutorials.html#getting-started) in order to install the fastest C++ simulator which uses instrinsics and multi-threading.
* For simulations with very few qubits, the speed is not limited by the simulator but rather by the compiler. If the compiler engines are not needed, e.g., if only native gates of the simulator are executed, then one can remove the compiler engines and obtain a speed-up:
```
import projectq
from projectq.backends import Simulator
eng = projectq.MainEngine(backend=Simulator(), engine_list=[]) # Removes the default compiler engines
```
* As noted in the [code documentation](http://projectq.readthedocs.io/en/latest/projectq.backends.html#projectq.backends.Simulator), one can play around with the number of threads in order to increase the simulation speed. Execute the following statements in the terminal before running ProjectQ:
```
export OMP_NUM_THREADS=2
export OMP_PROC_BIND=spread
```
A good setting is to set the number of threads to the number of physical cores on your system.
* The simulator has a feature called "gate fusion" in which case it combines smaller gates into larger ones in order to increase the speed of the simulation. If the simulator is faster with or without gate fusion depends on many parameters. By default it is currently turned off but one can turn it on and compare by:
```
import projectq
from projectq.backends import Simulator
eng = projectq.MainEngine(backend=Simulator(gate_fusion=True)) # Enables gate fusion
```
We'd like to refer interested readers to our paper on the [world's largest and fastest quantum computer simulation](https://arxiv.org/abs/1704.01127) for more details on how to optimize the speed of a quantum simulator.
|
github_jupyter
|
import projectq
eng = projectq.MainEngine() # This loads the simulator as it is the default backend
(Note: This is the (slow) Python simulator.)
from projectq.backends import Simulator
eng = projectq.MainEngine(backend=Simulator())
eng = projectq.MainEngine()
eng = projectq.MainEngine(backend=Simulator(rnd_seed=10))
import projectq
from projectq.ops import Measure, H
eng = projectq.MainEngine()
qubit = eng.allocate_qubit() # Allocate a qubit from the compiler (MainEngine object)
H | qubit
Measure | qubit # Measures the qubit in the Z basis and collapses it into either |0> or |1>
eng.flush() # Tells the compiler (MainEninge) compile all above gates and send it to the simulator backend
print(int(qubit)) # The measurement result can be accessed by converting the qubit object to a bool or int
def test_program(eng):
# Do something
ancilla = eng.allocate_qubit()
H | ancilla
# Use the ancilla for something
# Here the ancilla is not reset to a classical state but still in a superposition and will go out of scope
test_program(eng)
from projectq.ops import All
def test_program_2(eng):
# Do something
ancillas = eng.allocate_qureg(3) # allocates a quantum register with 3 qubits
All(H) | ancillas # applies a Hadamard gate to each of the 3 ancilla qubits
All(Measure) | ancillas # Measures all ancilla qubits such that we don't get
#an error message when they are deallocated
# Here the ancillas will go out of scope but because of the measurement, they are in a classical state
test_program_2(eng)
sim = Simulator(rnd_seed=5) # We can now always access the simulator via the "sim" variable
eng = projectq.MainEngine(backend=sim)
assert id(sim) == id(eng.backend)
from projectq.ops import H, Measure
eng = projectq.MainEngine()
qubit = eng.allocate_qubit()
qubit2 = eng.allocate_qubit()
eng.flush() # sends the allocation of the two qubits to the simulator (only needed to showcase the stuff below)
H | qubit # Put this qubit into a superposition
# qubit is a list with one qubit, qubit2 is another list with one qubit object
# qubit + qubit2 creates a list containing both qubits. Equivalently, one could write [qubit[0], qubit2[0]]
# get_amplitude requires that one provides a list/qureg of all qubits such that it can determine the order
amp_before = eng.backend.get_amplitude('00', qubit + qubit2)
# Amplitude will be 1 as Hadamard gate is not yet executed on the simulator backend
# We forgot the eng.flush()!
print("Amplitude saved in amp_before: {}".format(amp_before))
eng.flush() # Makes sure that all the gates are sent to the backend and executed
amp_after = eng.backend.get_amplitude('00', qubit + qubit2)
# Amplitude will be 1/sqrt(2) as Hadamard gate was executed on the simulator backend
print("Amplitude saved in amp_after: {}".format(amp_after))
# To avoid triggering the warning of deallocating qubits which are in a superposition
Measure | qubit
Measure | qubit2
import projectq
from projectq.ops import H, Measure, CNOT, All
eng = projectq.MainEngine()
qureg = eng.allocate_qureg(2)
H | qureg[0]
eng.flush()
prob11 = eng.backend.get_probability('11', qureg)
prob10 = eng.backend.get_probability('10', qureg)
prob01 = eng.backend.get_probability('01', qureg)
prob00 = eng.backend.get_probability('00', qureg)
prob_second_0 = eng.backend.get_probability('0', [qureg[1]])
print("Probability to measure 11: {}".format(prob11))
print("Probability to measure 00: {}".format(prob00))
print("Probability to measure 01: {}".format(prob01))
print("Probability to measure 10: {}".format(prob10))
print("Probability that second qubit is in state 0: {}".format(prob_second_0))
All(Measure) | qureg
import projectq
from projectq.ops import H, Measure, CNOT, All, QubitOperator, X, Y
eng = projectq.MainEngine()
qureg = eng.allocate_qureg(3)
X | qureg[0]
H | qureg[1]
eng.flush()
op0 = QubitOperator('Z0') # Z applied to qureg[0] tensor identity on qureg[1], qureg[2]
expectation = eng.backend.get_expectation_value(op0, qureg)
print("Expectation value = <Psi|Z0|Psi> = {}".format(expectation))
op_sum = QubitOperator('Z0 X1') - 0.5 * QubitOperator('X1')
expectation2 = eng.backend.get_expectation_value(op_sum, qureg)
print("Expectation value = <Psi|-0.5 X1 + 1.0 Z0 X1|Psi> = {}".format(expectation2))
All(Measure) | qureg # To avoid error message of deallocating qubits in a superposition
import projectq
from projectq.ops import H, Measure
eng = projectq.MainEngine()
qureg = eng.allocate_qureg(2)
# Create an entangled state:
H | qureg[0]
CNOT | (qureg[0], qureg[1])
# qureg is now in state 1/sqrt(2) * (|00> + |11>)
Measure | qureg[0]
Measure | qureg[1]
eng.flush() # required such that all above gates are executed before accessing the measurement result
print("First qubit measured in state: {} and second qubit in state: {}".format(int(qureg[0]), int(qureg[1])))
import projectq
from projectq.ops import H, CNOT
eng = projectq.MainEngine()
qureg = eng.allocate_qureg(2)
# Create an entangled state:
H | qureg[0]
CNOT | (qureg[0], qureg[1])
# qureg is now in state 1/sqrt(2) * (|00> + |11>)
eng.flush() # required such that all above gates are executed before collapsing the wavefunction
# We want to check what happens to the second qubit if the first qubit (qureg[0]) is measured to be 0
eng.backend.collapse_wavefunction([qureg[0]], [0])
# Check the probability that the second qubit is measured to be 0:
prob_0 = eng.backend.get_probability('0', [qureg[1]])
print("After forcing a measurement outcome of the first qubit to be 0, \n"
"the second qubit is in state 0 with probability: {}".format(prob_0))
import math
import projectq
from projectq.ops import H
eng = projectq.MainEngine()
qureg = eng.allocate_qureg(2)
eng.flush()
eng.backend.set_wavefunction([1/math.sqrt(2), 1/math.sqrt(2), 0, 0], qureg)
H | qureg[0]
# At this point both qubits are back in the state 00 and hence there will be no exception thrown
# when the qureg is deallocated
import copy
import projectq
from projectq.ops import Rx, Measure, All
eng = projectq.MainEngine()
qubit1 = eng.allocate_qubit()
qubit2 = eng.allocate_qubit()
Rx(0.2) | qubit1
Rx(0.4) | qubit2
eng.flush() # In order to have all the above gates sent to the simulator and executed
# We save a copy of the wavefunction at this point in the algorithm. In order to make sure we get a copy
# also if the Python simulator is used, one should make a deepcopy:
mapping, wavefunction = copy.deepcopy(eng.backend.cheat())
print("The full wavefunction is: {}".format(wavefunction))
# Note: qubit1 is a qureg of length 1, i.e. a list containing one qubit objects, therefore the
# unique qubit id can be accessed via qubit1[0].id
print("qubit1 has bit-location {}".format(mapping[qubit1[0].id]))
print("qubit2 has bit-location {}".format(mapping[qubit2[0].id]))
# Suppose we want to know the amplitude of the qubit1 in state 0 and qubit2 in state 1:
state = 0 + (1 << mapping[qubit2[0].id])
print("Amplitude of state qubit1 in state 0 and qubit2 in state 1: {}".format(wavefunction[state]))
# If one only wants to access one (or a few) amplitudes, get_amplitude provides an easier interface:
amplitude = eng.backend.get_amplitude('01', qubit1 + qubit2)
print("Accessing same amplitude but using get_amplitude instead: {}".format(amplitude))
All(Measure) | qubit1 + qubit2 # In order to not deallocate a qubit in a superposition state
import projectq
from projectq.backends import Simulator
eng = projectq.MainEngine(backend=Simulator(), engine_list=[]) # Removes the default compiler engines
export OMP_NUM_THREADS=2
export OMP_PROC_BIND=spread
import projectq
from projectq.backends import Simulator
eng = projectq.MainEngine(backend=Simulator(gate_fusion=True)) # Enables gate fusion
| 0.689933 | 0.991023 |
# Introduction to Python I - Variables
* Based on a lecture series by Rajath Kumart (https://github.com/rajathkumarmp/Python-Lectures)
* Ported to Python3 and extessions added by Janis Keuper
* Copyright: Creative Commons Attribution 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by/3.0/
## The Zen Of Python
```
import this
```
## The Very Basics
### Comments:
```
# this is a comment
```
### Spaces
***Python is indent sensitive!!!***
```
print ("hello")
print ("this does not work")
for i in range(5):
print ("this is a correct indent")
for i in range(5):
print ("this is NOT a correct indent")
```
**NOTE: you should set your editor to convert <tab> into 4 Spaces**
Python Coding Conventions: https://www.python.org/dev/peps/pep-0008/
# Variables
A name that is used to denote something or a value is called a variable. **Important:** Python has no strong typing, e.g. variables can change their type within a script. In python, variables can be declared and values can be assigned to it as follows,
```
x = 2
y = 5
xy = 'Hey'
print (x+y, xy)
```
Multiple variables can be assigned with the same value.
```
x = y = 1
print (x,y)
```
Weak typing example:
```
a = 1
a += 1
print (a)
a = "hello"
print (a)
a+=1
```
### Checking variable types:
```
type(a)
```
### Type casting:
```
str(1)
int('34')+5
```
# Operators
## Arithmetic Operators
| Symbol | Task Performed |
|----|---|
| + | Addition |
| - | Subtraction |
| / | division |
| % | mod |
| * | multiplication |
| // | floor division |
| ** | to the power of |
```
1+2
2-1
1*2
1/2
```
0? This is because both the numerator and denominator are integers but the result is a float value hence an integer value is returned. By changing either the numerator or the denominator to float, correct answer can be obtained.
```
1/2.0
15%10
```
Floor division is nothing but converting the result so obtained to the nearest integer.
```
2.8//2.0
```
## Relational Operators
| Symbol | Task Performed |
|----|---|
| == | True, if it is equal |
| != | True, if not equal to |
| < | less than |
| > | greater than |
| <= | less than or equal to |
| >= | greater than or equal to |
```
z = 1
z == 1
z > 1
```
## Bitwise Operators
| Symbol | Task Performed |
|----|---|
| & | Logical And |
| l | Logical OR |
| ^ | XOR |
| ~ | Negate |
| >> | Right shift |
| << | Left shift |
```
a = 2 #10
b = 3 #11
print (a & b)
print (bin(a&b))
5 >> 1
```
0000 0101 -> 5
Shifting the digits by 1 to the right and zero padding
0000 0010 -> 2
```
5 << 1
```
0000 0101 -> 5
Shifting the digits by 1 to the left and zero padding
0000 1010 -> 10
# Built-in Functions
Python comes loaded with pre-built functions
## Conversion from one system to another
Conversion from hexadecimal to decimal is done by adding prefix **0x** to the hexadecimal value or vice versa by using built in **hex( )**, Octal to decimal by adding prefix **0** to the octal value or vice versa by using built in function **oct( )**.
```
hex(170)
0xAA
oct(8)
```
**int( )** accepts two values when used for conversion, one is the value in a different number system and the other is its base. Note that input number in the different number system should be of string type.
```
print (int('010',8))
print (int('0xaa',16))
print (int('1010',2))
```
**int( )** can also be used to get only the integer value of a float number or can be used to convert a number which is of type string to integer format. Similarly, the function **str( )** can be used to convert the integer back to string format
```
print (int(7.7))
print (int('7'))
```
Also note that function **bin( )** is used for binary and **float( )** for decimal/float values. **chr( )** is used for converting ASCII to its alphabet equivalent, **ord( )** is used for the other way round.
```
chr(98)
ord('b')
```
## Simplifying Arithmetic Operations
**round( )** function rounds the input value to a specified number of places or to the nearest integer.
```
print (round(5.6231) )
print (round(4.55892, 2))
```
**complex( )** is used to define a complex number and **abs( )** outputs the absolute value of the same.
```
c =complex('5+2j')
print (abs(c))
```
**divmod(x,y)** outputs the quotient and the remainder in a tuple(you will be learning about it in the further chapters) in the format (quotient, remainder).
```
divmod(9,2)
```
**isinstance( )** returns True, if the first argument is an instance of that class. Multiple classes can also be checked at once.
```
print (isinstance(1, int))
print (isinstance(1.0,int))
print (isinstance(1.0,(int,float)))
```
**pow(x,y,z)** can be used to find the power $x^y$ also the mod of the resulting value with the third specified number can be found i.e. : ($x^y$ % z).
```
print (pow(3,3))
print (pow(3,3,5))
```
**range( )** function outputs the integers of the specified range. It can also be used to generate a series by specifying the difference between the two numbers within a particular range. The elements are returned in a list (will be discussing in detail later.)
```
print (range(3))
print (range(2,9))
print (range(2,27,8))
```
|
github_jupyter
|
import this
# this is a comment
print ("hello")
print ("this does not work")
for i in range(5):
print ("this is a correct indent")
for i in range(5):
print ("this is NOT a correct indent")
x = 2
y = 5
xy = 'Hey'
print (x+y, xy)
x = y = 1
print (x,y)
a = 1
a += 1
print (a)
a = "hello"
print (a)
a+=1
type(a)
str(1)
int('34')+5
1+2
2-1
1*2
1/2
1/2.0
15%10
2.8//2.0
z = 1
z == 1
z > 1
a = 2 #10
b = 3 #11
print (a & b)
print (bin(a&b))
5 >> 1
5 << 1
hex(170)
0xAA
oct(8)
print (int('010',8))
print (int('0xaa',16))
print (int('1010',2))
print (int(7.7))
print (int('7'))
chr(98)
ord('b')
print (round(5.6231) )
print (round(4.55892, 2))
c =complex('5+2j')
print (abs(c))
divmod(9,2)
print (isinstance(1, int))
print (isinstance(1.0,int))
print (isinstance(1.0,(int,float)))
print (pow(3,3))
print (pow(3,3,5))
print (range(3))
print (range(2,9))
print (range(2,27,8))
| 0.042712 | 0.977392 |
```
import csv
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
!wget --no-check-certificate \
https://storage.googleapis.com/laurencemoroney-blog.appspot.com/bbc-text.csv \
-O /tmp/bbc-text.csv
vocab_size = 1000
embedding_dim = 16
max_length = 120
trunc_type = 'post'
padding_type = 'post'
oov_tok = "<OOV>"
training_portion = .8
stopwords = ["a", "about", "above", "after", "again", "against", "all",
"am", "an", "and", "any", "are", "as", "at", "be", "because",
"been", "before", "being", "below", "between", "both", "but",
"by", "could", "did", "do", "does", "doing", "down", "during",
"each", "few", "for", "from", "further", "had", "has", "have",
"having", "he", "he'd", "he'll", "he's", "her", "here", "here's",
"hers", "herself", "him", "himself", "his", "how", "how's", "i",
"i'd", "i'll", "i'm", "i've", "if", "in", "into", "is", "it",
"it's", "its", "itself", "let's", "me", "more", "most", "my",
"myself", "nor", "of", "on", "once", "only", "or", "other",
"ought", "our", "ours", "ourselves", "out", "over", "own",
"same", "she", "she'd", "she'll", "she's", "should", "so",
"some", "such", "than", "that", "that's", "the", "their",
"theirs", "them", "themselves", "then", "there", "there's",
"these", "they", "they'd", "they'll", "they're", "they've",
"this", "those", "through", "to", "too", "under", "until",
"up", "very", "was", "we", "we'd", "we'll", "we're", "we've",
"were", "what", "what's", "when", "when's", "where", "where's",
"which", "while", "who", "who's", "whom", "why", "why's", "with",
"would", "you", "you'd", "you'll", "you're", "you've", "your",
"yours", "yourself", "yourselves"]
sentences = []
labels = []
with open("/tmp/bbc-text.csv", 'r') as csvfile:
reader = csv.reader(csvfile, delimiter=',')
next(reader)
for row in reader:
labels.append(row[0])
sentence = row[1]
for word in stopwords:
token = " " + word + " "
sentence = sentence.replace(token, " ")
sentence = sentence.replace(" ", " ")
sentences.append(sentence)
train_size = int(len(sentences) * training_portion)
train_sentences = sentences[:train_size]
train_labels = labels[:train_size]
validation_sentences = sentences[train_size:]
validation_labels = labels[train_size:]
tokenizer = Tokenizer(num_words=vocab_size, oov_token=oov_tok)
tokenizer.fit_on_texts(train_sentences)
word_index = tokenizer.word_index
train_sequences = tokenizer.texts_to_sequences(train_sentences)
train_padded = pad_sequences(train_sequences, padding=padding_type, maxlen=max_length)
validation_sequences = tokenizer.texts_to_sequences(validation_sentences)
validation_padded = pad_sequences(validation_sequences, padding=padding_type, maxlen=max_length)
label_tokenizer = Tokenizer()
label_tokenizer.fit_on_texts(labels)
training_label_seq = np.array(label_tokenizer.texts_to_sequences(train_labels))
validation_label_seq = np.array(label_tokenizer.texts_to_sequences(validation_labels))
model = tf.keras.Sequential([
tf.keras.layers.Embedding(vocab_size, embedding_dim, input_length=max_length),
tf.keras.layers.GlobalAveragePooling1D(),
tf.keras.layers.Dense(24, activation='relu'),
tf.keras.layers.Dense(6, activation='softmax')
])
model.compile(loss='sparse_categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
model.summary()
num_epochs = 30
history = model.fit(train_padded, training_label_seq, epochs=num_epochs, validation_data=(validation_padded, validation_label_seq), verbose=2)
```
|
github_jupyter
|
import csv
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
!wget --no-check-certificate \
https://storage.googleapis.com/laurencemoroney-blog.appspot.com/bbc-text.csv \
-O /tmp/bbc-text.csv
vocab_size = 1000
embedding_dim = 16
max_length = 120
trunc_type = 'post'
padding_type = 'post'
oov_tok = "<OOV>"
training_portion = .8
stopwords = ["a", "about", "above", "after", "again", "against", "all",
"am", "an", "and", "any", "are", "as", "at", "be", "because",
"been", "before", "being", "below", "between", "both", "but",
"by", "could", "did", "do", "does", "doing", "down", "during",
"each", "few", "for", "from", "further", "had", "has", "have",
"having", "he", "he'd", "he'll", "he's", "her", "here", "here's",
"hers", "herself", "him", "himself", "his", "how", "how's", "i",
"i'd", "i'll", "i'm", "i've", "if", "in", "into", "is", "it",
"it's", "its", "itself", "let's", "me", "more", "most", "my",
"myself", "nor", "of", "on", "once", "only", "or", "other",
"ought", "our", "ours", "ourselves", "out", "over", "own",
"same", "she", "she'd", "she'll", "she's", "should", "so",
"some", "such", "than", "that", "that's", "the", "their",
"theirs", "them", "themselves", "then", "there", "there's",
"these", "they", "they'd", "they'll", "they're", "they've",
"this", "those", "through", "to", "too", "under", "until",
"up", "very", "was", "we", "we'd", "we'll", "we're", "we've",
"were", "what", "what's", "when", "when's", "where", "where's",
"which", "while", "who", "who's", "whom", "why", "why's", "with",
"would", "you", "you'd", "you'll", "you're", "you've", "your",
"yours", "yourself", "yourselves"]
sentences = []
labels = []
with open("/tmp/bbc-text.csv", 'r') as csvfile:
reader = csv.reader(csvfile, delimiter=',')
next(reader)
for row in reader:
labels.append(row[0])
sentence = row[1]
for word in stopwords:
token = " " + word + " "
sentence = sentence.replace(token, " ")
sentence = sentence.replace(" ", " ")
sentences.append(sentence)
train_size = int(len(sentences) * training_portion)
train_sentences = sentences[:train_size]
train_labels = labels[:train_size]
validation_sentences = sentences[train_size:]
validation_labels = labels[train_size:]
tokenizer = Tokenizer(num_words=vocab_size, oov_token=oov_tok)
tokenizer.fit_on_texts(train_sentences)
word_index = tokenizer.word_index
train_sequences = tokenizer.texts_to_sequences(train_sentences)
train_padded = pad_sequences(train_sequences, padding=padding_type, maxlen=max_length)
validation_sequences = tokenizer.texts_to_sequences(validation_sentences)
validation_padded = pad_sequences(validation_sequences, padding=padding_type, maxlen=max_length)
label_tokenizer = Tokenizer()
label_tokenizer.fit_on_texts(labels)
training_label_seq = np.array(label_tokenizer.texts_to_sequences(train_labels))
validation_label_seq = np.array(label_tokenizer.texts_to_sequences(validation_labels))
model = tf.keras.Sequential([
tf.keras.layers.Embedding(vocab_size, embedding_dim, input_length=max_length),
tf.keras.layers.GlobalAveragePooling1D(),
tf.keras.layers.Dense(24, activation='relu'),
tf.keras.layers.Dense(6, activation='softmax')
])
model.compile(loss='sparse_categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
model.summary()
num_epochs = 30
history = model.fit(train_padded, training_label_seq, epochs=num_epochs, validation_data=(validation_padded, validation_label_seq), verbose=2)
| 0.654784 | 0.408719 |
# Comparison of Basilisk and OpenFOAM flow fields
This notebook compares the velocity fields for several bubbles obtained by the following approaches:
- **Basilisk**: using a 2D axis-symmetric two-phase flow solver implemented in Basilisk, the rise velocity, shape, and velocity field have been obtained.
- **OpenFOAM (free slip)**: single phase simulations have been performed in OpenFOAM (with simpleFoam). The bubble shape and rise velocity are inputs from the Basilisk simulations (see notebook *basilisk_2D_shape_approximation_...ipynb*). At the bubble surface boundary, a free slip condition has been applied for the velocity.
- **OpenFOAM (pyTorch)**: the base setting is as before, but at the bubble surface the velocity vector as prescribed based on the output of a pyTorch model. The pyTorch model has been trained based on the tangential interface velocity obtained with the Basilisk simulations (see notebook *basilisk_2D_interface_velocity.ipynb*)
## Read Basilisk fields
```
import helper_module as hm
# Basilisk names amd simulation times
names = ["wa18_l16", "scap_l16", "dell_l17"]
times = [20, 50, 50]
data_path = "../data/"
cases = [data_path + name for name in names]
print(cases)
basilisk_fields = {}
U_b = []
for i, case in enumerate(cases):
case_name = case.split("/")[-1]
log_path = case + "/log." + case_name + ".csv"
log = hm.Logfile(log_path)
log.read_logfile(usecols=['time', 'u_x', 'x'])
row = log.find_closest("time", times[i])
U_b.append(row.u_x.values[0])
field_path = data_path + case_name + "_vel_t{:2d}.csv".format(times[i])
basilisk_fields[case_name] = hm.CenterFieldValues2D(field_path, [0.0, row.x.values[0]], [0.0, row.u_x.values[0]])
print("Rise velocities:\n", U_b)
```
## Read OpenFOAM fields
```
of_fields_pyto = {}
of_fields_slip = {}
for i, name in enumerate(names):
field_path_pyto = data_path + "single_phase/" + name + "/flow_steady_1/reflect_centers.csv"
of_fields_pyto[name] = hm.CenterFieldValues2D(field_path_pyto, [0.0, 0.0], [0.0, 0.0], of=True)
field_path_slip = data_path + "single_phase/" + name + "/flow_steady_slip_1/reflect_centers.csv"
of_fields_slip[name] = hm.CenterFieldValues2D(field_path_slip, [0.0, 0.0], [0.0, 0.0], of=True)
```
## Plot velocity fields
```
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import rc
%matplotlib inline
rc('text', usetex=True)
nx = 200
ny = 400
xi_right = np.linspace(0.0, 1.2, nx)
xi_left = np.linspace(-1.2, 0.0, nx)
xi_full = np.linspace(-1.2, 1.2, 2*nx)
yi = np.linspace(-2.2, 1.3, ny)
Xi_left, Yi_left = np.meshgrid(xi_left, yi)
Xi_right, Yi_right = np.meshgrid(xi_right, yi)
Xi_full, Yi_full = np.meshgrid(xi_full, yi)
bbox = dict(facecolor='white', alpha=0.7, boxstyle='round,pad=0.5')
output_path = "../output/"
def savefig(name):
plt.savefig(output_path + name + ".png", bbox_inches="tight")
plt.savefig(output_path + name + ".pdf", bbox_inches="tight")
fig, axarr = plt.subplots(1, 2, sharey=True, figsize=(hm.figure_width, 12))
case = "wa18_l16"
contours = []
levels = np.linspace(0.01, 1.8, 8)
labels = ["OF, Dirichlet", "OF, free slip"]
vol_f = basilisk_fields[case].interpolate_volume_fraction(Xi_full, Yi_full)
mag_U_bas = basilisk_fields[case].interpolate_velocity(Xi_left, Yi_left, True, True)
vol_f_mask = basilisk_fields[case].interpolate_volume_fraction(Xi_right, Yi_right)
vol_f_mask = np.where(vol_f_mask < 0.5, -1.0, 1.0)
for i, field in enumerate([of_fields_pyto[case], of_fields_slip[case]]):
# basilisk results
axarr[i].contourf(xi_left, yi, mag_U_bas/U_b[0], levels=levels, cmap='jet', alpha=hm.alpha_contour)
contours.append(axarr[i].contour(xi_left, yi, mag_U_bas/U_b[0], levels=levels, colors='k'))
axarr[i].clabel(contours[-1], inline=True, fontsize=hm.fontsize_contour, fmt='%1.2f')
axarr[i].contour(xi_full, yi, vol_f, levels=[0.5], colors=['k'], linewidths=2.0)
# OpenFOAM results
mag_U = field.interpolate_velocity(Xi_right, Yi_right, True, True)
mag_U *=vol_f_mask
axarr[i].contourf(xi_right, yi, mag_U/U_b[0], levels=levels, cmap='jet', alpha=hm.alpha_contour)
contours.append(axarr[i].contour(xi_right, yi, mag_U/U_b[0], levels=levels, colors='k'))
axarr[i].axvline(0.0, 0.0, 1.0, color="w", lw=2, ls="-")
axarr[i].text(-0.8, 1.0, "Basilisk", fontsize=hm.fontsize_legend, bbox=bbox)
axarr[i].text(0.4, 1.0, labels[i], fontsize=hm.fontsize_legend, bbox=bbox)
for ax in axarr:
ax.axis('equal')
ax.label_outer()
ax.set_xticks([-0.9,-0.5, 0.0, 0.5, 0.9])
ax.tick_params(labelsize=hm.fontsize_tick)
for contour in contours:
for line in contour.collections:
line.set_linestyle('dashed')
fig.subplots_adjust(wspace=0.05)
fig.text(0.5, 0.05, r"$\tilde{x}$", ha='center', fontsize=hm.fontsize_label)
fig.text(0.05, 0.5, r"$\tilde{y}$", va='center', rotation='vertical', fontsize=hm.fontsize_label)
savefig("wa18_l16_of_velocity_fields")
fig, axarr = plt.subplots(1, 2, sharey=True, figsize=(hm.figure_width, 12))
case = "scap_l16"
contours = []
levels = np.linspace(0.0, 1.2, 8)
labels = ["OF, Dirichlet", "OF, free slip"]
vol_f = basilisk_fields[case].interpolate_volume_fraction(Xi_full, Yi_full)
mag_U_bas = basilisk_fields[case].interpolate_velocity(Xi_left, Yi_left, True, True)
vol_f_mask = basilisk_fields[case].interpolate_volume_fraction(Xi_right, Yi_right)
vol_f_mask = np.where(vol_f_mask < 0.5, -1.0, 1.0)
for i, field in enumerate([of_fields_pyto[case], of_fields_slip[case]]):
# basilisk results
axarr[i].contourf(xi_left, yi, mag_U_bas/U_b[1], levels=levels, cmap='jet', alpha=hm.alpha_contour)
contours.append(axarr[i].contour(xi_left, yi, mag_U_bas/U_b[1], levels=levels, colors='k'))
axarr[i].clabel(contours[-1], inline=True, fontsize=hm.fontsize_contour, fmt='%1.2f')
axarr[i].contour(xi_full, yi, vol_f, levels=[0.5], colors=['k'], linewidths=2.0)
# OpenFOAM results
mag_U = field.interpolate_velocity(Xi_right, Yi_right, True, True)
mag_U *=vol_f_mask
axarr[i].contourf(xi_right, yi, mag_U/U_b[1], levels=levels, cmap='jet', alpha=hm.alpha_contour)
contours.append(axarr[i].contour(xi_right, yi, mag_U/U_b[1], levels=levels, colors='k'))
axarr[i].axvline(0.0, 0.0, 1.0, color="w", lw=2, ls="-")
axarr[i].text(-0.8, 1.0, "Basilisk", fontsize=hm.fontsize_legend, bbox=bbox)
axarr[i].text(0.4, 1.0, labels[i], fontsize=hm.fontsize_legend, bbox=bbox)
for ax in axarr:
ax.axis('equal')
ax.label_outer()
ax.set_xticks([-0.9,-0.5, 0.0, 0.5, 0.9])
ax.tick_params(labelsize=hm.fontsize_tick)
for contour in contours:
for line in contour.collections:
line.set_linestyle('dashed')
fig.subplots_adjust(wspace=0.05)
fig.text(0.5, 0.05, r"$\tilde{x}$", ha='center', fontsize=hm.fontsize_label)
fig.text(0.05, 0.5, r"$\tilde{y}$", va='center', rotation='vertical', fontsize=hm.fontsize_label)
savefig("scap_l16_of_velocity_fields")
fig, axarr = plt.subplots(1, 2, sharey=True, figsize=(hm.figure_width, 12))
case = "dell_l17"
contours = []
levels = np.linspace(0.0, 1.2, 8)
labels = ["OF, Dirichlet", "OF, free slip"]
vol_f = basilisk_fields[case].interpolate_volume_fraction(Xi_full, Yi_full)
mag_U_bas = basilisk_fields[case].interpolate_velocity(Xi_left, Yi_left, True, True)
vol_f_mask = basilisk_fields[case].interpolate_volume_fraction(Xi_right, Yi_right)
vol_f_mask = np.where(vol_f_mask < 0.5, -10.0, 1.0)
for i, field in enumerate([of_fields_pyto[case], of_fields_slip[case]]):
# basilisk results
axarr[i].contourf(xi_left, yi, mag_U_bas/U_b[2], levels=levels, cmap='jet', alpha=hm.alpha_contour)
contours.append(axarr[i].contour(xi_left, yi, mag_U_bas/U_b[2], levels=levels, colors='k'))
axarr[i].clabel(contours[-1], inline=True, fontsize=hm.fontsize_contour, fmt='%1.2f')
axarr[i].contour(xi_full, yi, vol_f, levels=[0.5], colors=['k'], linewidths=2.0)
# OpenFOAM results
mag_U = field.interpolate_velocity(Xi_right, Yi_right, True, True)
mag_U *=vol_f_mask
axarr[i].contourf(xi_right, yi, mag_U/U_b[2], levels=levels, cmap='jet', alpha=hm.alpha_contour)
contours.append(axarr[i].contour(xi_right, yi, mag_U/U_b[2], levels=levels, colors='k'))
axarr[i].axvline(0.0, 0.0, 1.0, color="w", lw=2, ls="-")
axarr[i].text(-0.8, 1.0, "Basilisk", fontsize=hm.fontsize_legend, bbox=bbox)
axarr[i].text(0.4, 1.0, labels[i], fontsize=hm.fontsize_legend, bbox=bbox)
for ax in axarr:
ax.axis('equal')
ax.label_outer()
ax.set_xticks([-0.9,-0.5, 0.0, 0.5, 0.9])
ax.tick_params(labelsize=hm.fontsize_tick)
for contour in contours:
for line in contour.collections:
line.set_linestyle('dashed')
fig.subplots_adjust(wspace=0.05)
fig.text(0.5, 0.05, r"$\tilde{x}$", ha='center', fontsize=hm.fontsize_label)
fig.text(0.05, 0.5, r"$\tilde{y}$", va='center', rotation='vertical', fontsize=hm.fontsize_label)
savefig("dell_l17_of_velocity_fields")
```
|
github_jupyter
|
import helper_module as hm
# Basilisk names amd simulation times
names = ["wa18_l16", "scap_l16", "dell_l17"]
times = [20, 50, 50]
data_path = "../data/"
cases = [data_path + name for name in names]
print(cases)
basilisk_fields = {}
U_b = []
for i, case in enumerate(cases):
case_name = case.split("/")[-1]
log_path = case + "/log." + case_name + ".csv"
log = hm.Logfile(log_path)
log.read_logfile(usecols=['time', 'u_x', 'x'])
row = log.find_closest("time", times[i])
U_b.append(row.u_x.values[0])
field_path = data_path + case_name + "_vel_t{:2d}.csv".format(times[i])
basilisk_fields[case_name] = hm.CenterFieldValues2D(field_path, [0.0, row.x.values[0]], [0.0, row.u_x.values[0]])
print("Rise velocities:\n", U_b)
of_fields_pyto = {}
of_fields_slip = {}
for i, name in enumerate(names):
field_path_pyto = data_path + "single_phase/" + name + "/flow_steady_1/reflect_centers.csv"
of_fields_pyto[name] = hm.CenterFieldValues2D(field_path_pyto, [0.0, 0.0], [0.0, 0.0], of=True)
field_path_slip = data_path + "single_phase/" + name + "/flow_steady_slip_1/reflect_centers.csv"
of_fields_slip[name] = hm.CenterFieldValues2D(field_path_slip, [0.0, 0.0], [0.0, 0.0], of=True)
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import rc
%matplotlib inline
rc('text', usetex=True)
nx = 200
ny = 400
xi_right = np.linspace(0.0, 1.2, nx)
xi_left = np.linspace(-1.2, 0.0, nx)
xi_full = np.linspace(-1.2, 1.2, 2*nx)
yi = np.linspace(-2.2, 1.3, ny)
Xi_left, Yi_left = np.meshgrid(xi_left, yi)
Xi_right, Yi_right = np.meshgrid(xi_right, yi)
Xi_full, Yi_full = np.meshgrid(xi_full, yi)
bbox = dict(facecolor='white', alpha=0.7, boxstyle='round,pad=0.5')
output_path = "../output/"
def savefig(name):
plt.savefig(output_path + name + ".png", bbox_inches="tight")
plt.savefig(output_path + name + ".pdf", bbox_inches="tight")
fig, axarr = plt.subplots(1, 2, sharey=True, figsize=(hm.figure_width, 12))
case = "wa18_l16"
contours = []
levels = np.linspace(0.01, 1.8, 8)
labels = ["OF, Dirichlet", "OF, free slip"]
vol_f = basilisk_fields[case].interpolate_volume_fraction(Xi_full, Yi_full)
mag_U_bas = basilisk_fields[case].interpolate_velocity(Xi_left, Yi_left, True, True)
vol_f_mask = basilisk_fields[case].interpolate_volume_fraction(Xi_right, Yi_right)
vol_f_mask = np.where(vol_f_mask < 0.5, -1.0, 1.0)
for i, field in enumerate([of_fields_pyto[case], of_fields_slip[case]]):
# basilisk results
axarr[i].contourf(xi_left, yi, mag_U_bas/U_b[0], levels=levels, cmap='jet', alpha=hm.alpha_contour)
contours.append(axarr[i].contour(xi_left, yi, mag_U_bas/U_b[0], levels=levels, colors='k'))
axarr[i].clabel(contours[-1], inline=True, fontsize=hm.fontsize_contour, fmt='%1.2f')
axarr[i].contour(xi_full, yi, vol_f, levels=[0.5], colors=['k'], linewidths=2.0)
# OpenFOAM results
mag_U = field.interpolate_velocity(Xi_right, Yi_right, True, True)
mag_U *=vol_f_mask
axarr[i].contourf(xi_right, yi, mag_U/U_b[0], levels=levels, cmap='jet', alpha=hm.alpha_contour)
contours.append(axarr[i].contour(xi_right, yi, mag_U/U_b[0], levels=levels, colors='k'))
axarr[i].axvline(0.0, 0.0, 1.0, color="w", lw=2, ls="-")
axarr[i].text(-0.8, 1.0, "Basilisk", fontsize=hm.fontsize_legend, bbox=bbox)
axarr[i].text(0.4, 1.0, labels[i], fontsize=hm.fontsize_legend, bbox=bbox)
for ax in axarr:
ax.axis('equal')
ax.label_outer()
ax.set_xticks([-0.9,-0.5, 0.0, 0.5, 0.9])
ax.tick_params(labelsize=hm.fontsize_tick)
for contour in contours:
for line in contour.collections:
line.set_linestyle('dashed')
fig.subplots_adjust(wspace=0.05)
fig.text(0.5, 0.05, r"$\tilde{x}$", ha='center', fontsize=hm.fontsize_label)
fig.text(0.05, 0.5, r"$\tilde{y}$", va='center', rotation='vertical', fontsize=hm.fontsize_label)
savefig("wa18_l16_of_velocity_fields")
fig, axarr = plt.subplots(1, 2, sharey=True, figsize=(hm.figure_width, 12))
case = "scap_l16"
contours = []
levels = np.linspace(0.0, 1.2, 8)
labels = ["OF, Dirichlet", "OF, free slip"]
vol_f = basilisk_fields[case].interpolate_volume_fraction(Xi_full, Yi_full)
mag_U_bas = basilisk_fields[case].interpolate_velocity(Xi_left, Yi_left, True, True)
vol_f_mask = basilisk_fields[case].interpolate_volume_fraction(Xi_right, Yi_right)
vol_f_mask = np.where(vol_f_mask < 0.5, -1.0, 1.0)
for i, field in enumerate([of_fields_pyto[case], of_fields_slip[case]]):
# basilisk results
axarr[i].contourf(xi_left, yi, mag_U_bas/U_b[1], levels=levels, cmap='jet', alpha=hm.alpha_contour)
contours.append(axarr[i].contour(xi_left, yi, mag_U_bas/U_b[1], levels=levels, colors='k'))
axarr[i].clabel(contours[-1], inline=True, fontsize=hm.fontsize_contour, fmt='%1.2f')
axarr[i].contour(xi_full, yi, vol_f, levels=[0.5], colors=['k'], linewidths=2.0)
# OpenFOAM results
mag_U = field.interpolate_velocity(Xi_right, Yi_right, True, True)
mag_U *=vol_f_mask
axarr[i].contourf(xi_right, yi, mag_U/U_b[1], levels=levels, cmap='jet', alpha=hm.alpha_contour)
contours.append(axarr[i].contour(xi_right, yi, mag_U/U_b[1], levels=levels, colors='k'))
axarr[i].axvline(0.0, 0.0, 1.0, color="w", lw=2, ls="-")
axarr[i].text(-0.8, 1.0, "Basilisk", fontsize=hm.fontsize_legend, bbox=bbox)
axarr[i].text(0.4, 1.0, labels[i], fontsize=hm.fontsize_legend, bbox=bbox)
for ax in axarr:
ax.axis('equal')
ax.label_outer()
ax.set_xticks([-0.9,-0.5, 0.0, 0.5, 0.9])
ax.tick_params(labelsize=hm.fontsize_tick)
for contour in contours:
for line in contour.collections:
line.set_linestyle('dashed')
fig.subplots_adjust(wspace=0.05)
fig.text(0.5, 0.05, r"$\tilde{x}$", ha='center', fontsize=hm.fontsize_label)
fig.text(0.05, 0.5, r"$\tilde{y}$", va='center', rotation='vertical', fontsize=hm.fontsize_label)
savefig("scap_l16_of_velocity_fields")
fig, axarr = plt.subplots(1, 2, sharey=True, figsize=(hm.figure_width, 12))
case = "dell_l17"
contours = []
levels = np.linspace(0.0, 1.2, 8)
labels = ["OF, Dirichlet", "OF, free slip"]
vol_f = basilisk_fields[case].interpolate_volume_fraction(Xi_full, Yi_full)
mag_U_bas = basilisk_fields[case].interpolate_velocity(Xi_left, Yi_left, True, True)
vol_f_mask = basilisk_fields[case].interpolate_volume_fraction(Xi_right, Yi_right)
vol_f_mask = np.where(vol_f_mask < 0.5, -10.0, 1.0)
for i, field in enumerate([of_fields_pyto[case], of_fields_slip[case]]):
# basilisk results
axarr[i].contourf(xi_left, yi, mag_U_bas/U_b[2], levels=levels, cmap='jet', alpha=hm.alpha_contour)
contours.append(axarr[i].contour(xi_left, yi, mag_U_bas/U_b[2], levels=levels, colors='k'))
axarr[i].clabel(contours[-1], inline=True, fontsize=hm.fontsize_contour, fmt='%1.2f')
axarr[i].contour(xi_full, yi, vol_f, levels=[0.5], colors=['k'], linewidths=2.0)
# OpenFOAM results
mag_U = field.interpolate_velocity(Xi_right, Yi_right, True, True)
mag_U *=vol_f_mask
axarr[i].contourf(xi_right, yi, mag_U/U_b[2], levels=levels, cmap='jet', alpha=hm.alpha_contour)
contours.append(axarr[i].contour(xi_right, yi, mag_U/U_b[2], levels=levels, colors='k'))
axarr[i].axvline(0.0, 0.0, 1.0, color="w", lw=2, ls="-")
axarr[i].text(-0.8, 1.0, "Basilisk", fontsize=hm.fontsize_legend, bbox=bbox)
axarr[i].text(0.4, 1.0, labels[i], fontsize=hm.fontsize_legend, bbox=bbox)
for ax in axarr:
ax.axis('equal')
ax.label_outer()
ax.set_xticks([-0.9,-0.5, 0.0, 0.5, 0.9])
ax.tick_params(labelsize=hm.fontsize_tick)
for contour in contours:
for line in contour.collections:
line.set_linestyle('dashed')
fig.subplots_adjust(wspace=0.05)
fig.text(0.5, 0.05, r"$\tilde{x}$", ha='center', fontsize=hm.fontsize_label)
fig.text(0.05, 0.5, r"$\tilde{y}$", va='center', rotation='vertical', fontsize=hm.fontsize_label)
savefig("dell_l17_of_velocity_fields")
| 0.304559 | 0.975923 |
```
#hide
#skip
! [ -e /content ] && pip install -Uqq fastai # upgrade fastai on colab
# default_exp callback.core
#export
from fastai.data.all import *
from fastai.optimizer import *
#hide
from nbdev.showdoc import *
#export
_all_ = ['CancelStepException','CancelFitException','CancelEpochException','CancelTrainException','CancelValidException','CancelBatchException']
```
# Callbacks
> Basic callbacks for Learner
## Events
Callbacks can occur at any of these times:: *after_create before_fit before_epoch before_train before_batch after_pred after_loss before_backward before_step after_step after_cancel_batch after_batch after_cancel_train after_train before_validate after_cancel_validate after_validate after_cancel_epoch after_epoch after_cancel_fit after_fit*.
```
# export
_events = L.split('after_create before_fit before_epoch before_train before_batch after_pred after_loss \
before_backward before_step after_cancel_step after_step after_cancel_batch after_batch after_cancel_train \
after_train before_validate after_cancel_validate after_validate after_cancel_epoch \
after_epoch after_cancel_fit after_fit')
mk_class('event', **_events.map_dict(),
doc="All possible events as attributes to get tab-completion and typo-proofing")
# export
_all_ = ['event']
show_doc(event, name='event', title_level=3)
```
To ensure that you are referring to an event (that is, the name of one of the times when callbacks are called) that exists, and to get tab completion of event names, use `event`:
```
test_eq(event.before_step, 'before_step')
```
## Callback -
```
#export
_inner_loop = "before_batch after_pred after_loss before_backward before_step after_step after_cancel_batch after_batch".split()
#export
@funcs_kwargs(as_method=True)
class Callback(Stateful,GetAttr):
"Basic class handling tweaks of the training loop by changing a `Learner` in various events"
order,_default,learn,run,run_train,run_valid = 0,'learn',None,True,True,True
_methods = _events
def __init__(self, **kwargs): assert not kwargs, f'Passed unknown events: {kwargs}'
def __repr__(self): return type(self).__name__
def __call__(self, event_name):
"Call `self.{event_name}` if it's defined"
_run = (event_name not in _inner_loop or (self.run_train and getattr(self, 'training', True)) or
(self.run_valid and not getattr(self, 'training', False)))
res = None
if self.run and _run: res = getattr(self, event_name, noop)()
if event_name=='after_fit': self.run=True #Reset self.run to True at each end of fit
return res
def __setattr__(self, name, value):
if hasattr(self.learn,name):
warn(f"You are shadowing an attribute ({name}) that exists in the learner. Use `self.learn.{name}` to avoid this")
super().__setattr__(name, value)
@property
def name(self):
"Name of the `Callback`, camel-cased and with '*Callback*' removed"
return class2attr(self, 'Callback')
```
The training loop is defined in `Learner` a bit below and consists in a minimal set of instructions: looping through the data we:
- compute the output of the model from the input
- calculate a loss between this output and the desired target
- compute the gradients of this loss with respect to all the model parameters
- update the parameters accordingly
- zero all the gradients
Any tweak of this training loop is defined in a `Callback` to avoid over-complicating the code of the training loop, and to make it easy to mix and match different techniques (since they'll be defined in different callbacks). A callback can implement actions on the following events:
- `after_create`: called after the `Learner` is created
- `before_fit`: called before starting training or inference, ideal for initial setup.
- `before_epoch`: called at the beginning of each epoch, useful for any behavior you need to reset at each epoch.
- `before_train`: called at the beginning of the training part of an epoch.
- `before_batch`: called at the beginning of each batch, just after drawing said batch. It can be used to do any setup necessary for the batch (like hyper-parameter scheduling) or to change the input/target before it goes in the model (change of the input with techniques like mixup for instance).
- `after_pred`: called after computing the output of the model on the batch. It can be used to change that output before it's fed to the loss.
- `after_loss`: called after the loss has been computed, but before the backward pass. It can be used to add any penalty to the loss (AR or TAR in RNN training for instance).
- `before_backward`: called after the loss has been computed, but only in training mode (i.e. when the backward pass will be used)
- `before_step`: called after the backward pass, but before the update of the parameters. It can be used to do any change to the gradients before said update (gradient clipping for instance).
- `after_step`: called after the step and before the gradients are zeroed.
- `after_batch`: called at the end of a batch, for any clean-up before the next one.
- `after_train`: called at the end of the training phase of an epoch.
- `before_validate`: called at the beginning of the validation phase of an epoch, useful for any setup needed specifically for validation.
- `after_validate`: called at the end of the validation part of an epoch.
- `after_epoch`: called at the end of an epoch, for any clean-up before the next one.
- `after_fit`: called at the end of training, for final clean-up.
```
show_doc(Callback.__call__)
```
One way to define callbacks is through subclassing:
```
class _T(Callback):
def call_me(self): return "maybe"
test_eq(_T()("call_me"), "maybe")
```
Another way is by passing the callback function to the constructor:
```
def cb(self): return "maybe"
_t = Callback(before_fit=cb)
test_eq(_t(event.before_fit), "maybe")
```
`Callback`s provide a shortcut to avoid having to write `self.learn.bla` for any `bla` attribute we seek; instead, just write `self.bla`. This only works for getting attributes, *not* for setting them.
```
mk_class('TstLearner', 'a')
class TstCallback(Callback):
def batch_begin(self): print(self.a)
learn,cb = TstLearner(1),TstCallback()
cb.learn = learn
test_stdout(lambda: cb('batch_begin'), "1")
```
If you want to change the value of an attribute, you have to use `self.learn.bla`, no `self.bla`. In the example below, `self.a += 1` creates an `a` attribute of 2 in the callback instead of setting the `a` of the learner to 2. It also issues a warning that something is probably wrong:
```
learn.a
class TstCallback(Callback):
def batch_begin(self): self.a += 1
learn,cb = TstLearner(1),TstCallback()
cb.learn = learn
cb('batch_begin')
test_eq(cb.a, 2)
test_eq(cb.learn.a, 1)
```
A proper version needs to write `self.learn.a = self.a + 1`:
```
class TstCallback(Callback):
def batch_begin(self): self.learn.a = self.a + 1
learn,cb = TstLearner(1),TstCallback()
cb.learn = learn
cb('batch_begin')
test_eq(cb.learn.a, 2)
show_doc(Callback.name, name='Callback.name')
test_eq(TstCallback().name, 'tst')
class ComplicatedNameCallback(Callback): pass
test_eq(ComplicatedNameCallback().name, 'complicated_name')
```
## TrainEvalCallback -
```
#export
class TrainEvalCallback(Callback):
"`Callback` that tracks the number of iterations done and properly sets training/eval mode"
order,run_valid = -10,False
def after_create(self): self.learn.n_epoch = 1
def before_fit(self):
"Set the iter and epoch counters to 0, put the model and the right device"
self.learn.epoch,self.learn.loss = 0,tensor(0.)
self.learn.train_iter,self.learn.pct_train = 0,0.
device = getattr(self.dls, 'device', default_device())
self.model.to(device)
if hasattr(self.model, 'reset'): self.model.reset()
def after_batch(self):
"Update the iter counter (in training mode)"
self.learn.pct_train += 1./(self.n_iter*self.n_epoch)
self.learn.train_iter += 1
def before_train(self):
"Set the model in training mode"
self.learn.pct_train=self.epoch/self.n_epoch
self.model.train()
self.learn.training=True
def before_validate(self):
"Set the model in validation mode"
self.model.eval()
self.learn.training=False
show_doc(TrainEvalCallback, title_level=3)
```
This `Callback` is automatically added in every `Learner` at initialization.
```
#hide
#test of the TrainEvalCallback below in Learner.fit
# export
if not hasattr(defaults, 'callbacks'): defaults.callbacks = [TrainEvalCallback]
```
## Attributes available to callbacks
When writing a callback, the following attributes of `Learner` are available:
- `model`: the model used for training/validation
- `data`: the underlying `DataLoaders`
- `loss_func`: the loss function used
- `opt`: the optimizer used to update the model parameters
- `opt_func`: the function used to create the optimizer
- `cbs`: the list containing all `Callback`s
- `dl`: current `DataLoader` used for iteration
- `x`/`xb`: last input drawn from `self.dl` (potentially modified by callbacks). `xb` is always a tuple (potentially with one element) and `x` is detuplified. You can only assign to `xb`.
- `y`/`yb`: last target drawn from `self.dl` (potentially modified by callbacks). `yb` is always a tuple (potentially with one element) and `y` is detuplified. You can only assign to `yb`.
- `pred`: last predictions from `self.model` (potentially modified by callbacks)
- `loss`: last computed loss (potentially modified by callbacks)
- `n_epoch`: the number of epochs in this training
- `n_iter`: the number of iterations in the current `self.dl`
- `epoch`: the current epoch index (from 0 to `n_epoch-1`)
- `iter`: the current iteration index in `self.dl` (from 0 to `n_iter-1`)
The following attributes are added by `TrainEvalCallback` and should be available unless you went out of your way to remove that callback:
- `train_iter`: the number of training iterations done since the beginning of this training
- `pct_train`: from 0. to 1., the percentage of training iterations completed
- `training`: flag to indicate if we're in training mode or not
The following attribute is added by `Recorder` and should be available unless you went out of your way to remove that callback:
- `smooth_loss`: an exponentially-averaged version of the training loss
## Callbacks control flow
It happens that we may want to skip some of the steps of the training loop: in gradient accumulation, we don't always want to do the step/zeroing of the grads for instance. During an LR finder test, we don't want to do the validation phase of an epoch. Or if we're training with a strategy of early stopping, we want to be able to completely interrupt the training loop.
This is made possible by raising specific exceptions the training loop will look for (and properly catch).
```
#export
_ex_docs = dict(
CancelBatchException="Skip the rest of this batch and go to `after_batch`",
CancelTrainException="Skip the rest of the training part of the epoch and go to `after_train`",
CancelValidException="Skip the rest of the validation part of the epoch and go to `after_validate`",
CancelEpochException="Skip the rest of this epoch and go to `after_epoch`",
CancelStepException ="Skip stepping the optimizer",
CancelFitException ="Interrupts training and go to `after_fit`")
for c,d in _ex_docs.items(): mk_class(c,sup=Exception,doc=d)
show_doc(CancelStepException, title_level=3)
show_doc(CancelBatchException, title_level=3)
show_doc(CancelTrainException, title_level=3)
show_doc(CancelValidException, title_level=3)
show_doc(CancelEpochException, title_level=3)
show_doc(CancelFitException, title_level=3)
```
You can detect one of those exceptions occurred and add code that executes right after with the following events:
- `after_cancel_batch`: reached immediately after a `CancelBatchException` before proceeding to `after_batch`
- `after_cancel_train`: reached immediately after a `CancelTrainException` before proceeding to `after_epoch`
- `after_cancel_valid`: reached immediately after a `CancelValidException` before proceeding to `after_epoch`
- `after_cancel_epoch`: reached immediately after a `CancelEpochException` before proceeding to `after_epoch`
- `after_cancel_fit`: reached immediately after a `CancelFitException` before proceeding to `after_fit`
## Gather and fetch preds callbacks -
```
#export
#TODO: save_targs and save_preds only handle preds/targets that have one tensor, not tuples of tensors.
class GatherPredsCallback(Callback):
"`Callback` that saves the predictions and targets, optionally `with_loss`"
_stateattrs=('preds','targets','inputs','losses')
def __init__(self, with_input=False, with_loss=False, save_preds=None, save_targs=None, concat_dim=0):
store_attr("with_input,with_loss,save_preds,save_targs,concat_dim")
def before_batch(self):
if self.with_input: self.inputs.append((self.learn.to_detach(self.xb)))
def before_validate(self):
"Initialize containers"
self.preds,self.targets = [],[]
if self.with_input: self.inputs = []
if self.with_loss: self.losses = []
def after_batch(self):
"Save predictions, targets and potentially losses"
if not hasattr(self, 'pred'): return
preds,targs = self.learn.to_detach(self.pred),self.learn.to_detach(self.yb)
if self.save_preds is None: self.preds.append(preds)
else: (self.save_preds/str(self.iter)).save_array(preds)
if self.save_targs is None: self.targets.append(targs)
else: (self.save_targs/str(self.iter)).save_array(targs[0])
if self.with_loss:
bs = find_bs(self.yb)
loss = self.loss if self.loss.numel() == bs else self.loss.view(bs,-1).mean(1)
self.losses.append(self.learn.to_detach(loss))
def after_validate(self):
"Concatenate all recorded tensors"
if not hasattr(self, 'preds'): return
if self.with_input: self.inputs = detuplify(to_concat(self.inputs, dim=self.concat_dim))
if not self.save_preds: self.preds = detuplify(to_concat(self.preds, dim=self.concat_dim))
if not self.save_targs: self.targets = detuplify(to_concat(self.targets, dim=self.concat_dim))
if self.with_loss: self.losses = to_concat(self.losses)
def all_tensors(self):
res = [None if self.save_preds else self.preds, None if self.save_targs else self.targets]
if self.with_input: res = [self.inputs] + res
if self.with_loss: res.append(self.losses)
return res
show_doc(GatherPredsCallback, title_level=3)
#export
class FetchPredsCallback(Callback):
"A callback to fetch predictions during the training loop"
remove_on_fetch = True
def __init__(self, ds_idx=1, dl=None, with_input=False, with_decoded=False, cbs=None, reorder=True):
self.cbs = L(cbs)
store_attr('ds_idx,dl,with_input,with_decoded,reorder')
def after_validate(self):
to_rm = L(cb for cb in self.learn.cbs if getattr(cb, 'remove_on_fetch', False))
with self.learn.removed_cbs(to_rm + self.cbs) as learn:
self.preds = learn.get_preds(ds_idx=self.ds_idx, dl=self.dl,
with_input=self.with_input, with_decoded=self.with_decoded, inner=True, reorder=self.reorder)
show_doc(FetchPredsCallback, title_level=3)
```
## Export -
```
#hide
from nbdev.export import notebook2script
notebook2script()
```
|
github_jupyter
|
#hide
#skip
! [ -e /content ] && pip install -Uqq fastai # upgrade fastai on colab
# default_exp callback.core
#export
from fastai.data.all import *
from fastai.optimizer import *
#hide
from nbdev.showdoc import *
#export
_all_ = ['CancelStepException','CancelFitException','CancelEpochException','CancelTrainException','CancelValidException','CancelBatchException']
# export
_events = L.split('after_create before_fit before_epoch before_train before_batch after_pred after_loss \
before_backward before_step after_cancel_step after_step after_cancel_batch after_batch after_cancel_train \
after_train before_validate after_cancel_validate after_validate after_cancel_epoch \
after_epoch after_cancel_fit after_fit')
mk_class('event', **_events.map_dict(),
doc="All possible events as attributes to get tab-completion and typo-proofing")
# export
_all_ = ['event']
show_doc(event, name='event', title_level=3)
test_eq(event.before_step, 'before_step')
#export
_inner_loop = "before_batch after_pred after_loss before_backward before_step after_step after_cancel_batch after_batch".split()
#export
@funcs_kwargs(as_method=True)
class Callback(Stateful,GetAttr):
"Basic class handling tweaks of the training loop by changing a `Learner` in various events"
order,_default,learn,run,run_train,run_valid = 0,'learn',None,True,True,True
_methods = _events
def __init__(self, **kwargs): assert not kwargs, f'Passed unknown events: {kwargs}'
def __repr__(self): return type(self).__name__
def __call__(self, event_name):
"Call `self.{event_name}` if it's defined"
_run = (event_name not in _inner_loop or (self.run_train and getattr(self, 'training', True)) or
(self.run_valid and not getattr(self, 'training', False)))
res = None
if self.run and _run: res = getattr(self, event_name, noop)()
if event_name=='after_fit': self.run=True #Reset self.run to True at each end of fit
return res
def __setattr__(self, name, value):
if hasattr(self.learn,name):
warn(f"You are shadowing an attribute ({name}) that exists in the learner. Use `self.learn.{name}` to avoid this")
super().__setattr__(name, value)
@property
def name(self):
"Name of the `Callback`, camel-cased and with '*Callback*' removed"
return class2attr(self, 'Callback')
show_doc(Callback.__call__)
class _T(Callback):
def call_me(self): return "maybe"
test_eq(_T()("call_me"), "maybe")
def cb(self): return "maybe"
_t = Callback(before_fit=cb)
test_eq(_t(event.before_fit), "maybe")
mk_class('TstLearner', 'a')
class TstCallback(Callback):
def batch_begin(self): print(self.a)
learn,cb = TstLearner(1),TstCallback()
cb.learn = learn
test_stdout(lambda: cb('batch_begin'), "1")
learn.a
class TstCallback(Callback):
def batch_begin(self): self.a += 1
learn,cb = TstLearner(1),TstCallback()
cb.learn = learn
cb('batch_begin')
test_eq(cb.a, 2)
test_eq(cb.learn.a, 1)
class TstCallback(Callback):
def batch_begin(self): self.learn.a = self.a + 1
learn,cb = TstLearner(1),TstCallback()
cb.learn = learn
cb('batch_begin')
test_eq(cb.learn.a, 2)
show_doc(Callback.name, name='Callback.name')
test_eq(TstCallback().name, 'tst')
class ComplicatedNameCallback(Callback): pass
test_eq(ComplicatedNameCallback().name, 'complicated_name')
#export
class TrainEvalCallback(Callback):
"`Callback` that tracks the number of iterations done and properly sets training/eval mode"
order,run_valid = -10,False
def after_create(self): self.learn.n_epoch = 1
def before_fit(self):
"Set the iter and epoch counters to 0, put the model and the right device"
self.learn.epoch,self.learn.loss = 0,tensor(0.)
self.learn.train_iter,self.learn.pct_train = 0,0.
device = getattr(self.dls, 'device', default_device())
self.model.to(device)
if hasattr(self.model, 'reset'): self.model.reset()
def after_batch(self):
"Update the iter counter (in training mode)"
self.learn.pct_train += 1./(self.n_iter*self.n_epoch)
self.learn.train_iter += 1
def before_train(self):
"Set the model in training mode"
self.learn.pct_train=self.epoch/self.n_epoch
self.model.train()
self.learn.training=True
def before_validate(self):
"Set the model in validation mode"
self.model.eval()
self.learn.training=False
show_doc(TrainEvalCallback, title_level=3)
#hide
#test of the TrainEvalCallback below in Learner.fit
# export
if not hasattr(defaults, 'callbacks'): defaults.callbacks = [TrainEvalCallback]
#export
_ex_docs = dict(
CancelBatchException="Skip the rest of this batch and go to `after_batch`",
CancelTrainException="Skip the rest of the training part of the epoch and go to `after_train`",
CancelValidException="Skip the rest of the validation part of the epoch and go to `after_validate`",
CancelEpochException="Skip the rest of this epoch and go to `after_epoch`",
CancelStepException ="Skip stepping the optimizer",
CancelFitException ="Interrupts training and go to `after_fit`")
for c,d in _ex_docs.items(): mk_class(c,sup=Exception,doc=d)
show_doc(CancelStepException, title_level=3)
show_doc(CancelBatchException, title_level=3)
show_doc(CancelTrainException, title_level=3)
show_doc(CancelValidException, title_level=3)
show_doc(CancelEpochException, title_level=3)
show_doc(CancelFitException, title_level=3)
#export
#TODO: save_targs and save_preds only handle preds/targets that have one tensor, not tuples of tensors.
class GatherPredsCallback(Callback):
"`Callback` that saves the predictions and targets, optionally `with_loss`"
_stateattrs=('preds','targets','inputs','losses')
def __init__(self, with_input=False, with_loss=False, save_preds=None, save_targs=None, concat_dim=0):
store_attr("with_input,with_loss,save_preds,save_targs,concat_dim")
def before_batch(self):
if self.with_input: self.inputs.append((self.learn.to_detach(self.xb)))
def before_validate(self):
"Initialize containers"
self.preds,self.targets = [],[]
if self.with_input: self.inputs = []
if self.with_loss: self.losses = []
def after_batch(self):
"Save predictions, targets and potentially losses"
if not hasattr(self, 'pred'): return
preds,targs = self.learn.to_detach(self.pred),self.learn.to_detach(self.yb)
if self.save_preds is None: self.preds.append(preds)
else: (self.save_preds/str(self.iter)).save_array(preds)
if self.save_targs is None: self.targets.append(targs)
else: (self.save_targs/str(self.iter)).save_array(targs[0])
if self.with_loss:
bs = find_bs(self.yb)
loss = self.loss if self.loss.numel() == bs else self.loss.view(bs,-1).mean(1)
self.losses.append(self.learn.to_detach(loss))
def after_validate(self):
"Concatenate all recorded tensors"
if not hasattr(self, 'preds'): return
if self.with_input: self.inputs = detuplify(to_concat(self.inputs, dim=self.concat_dim))
if not self.save_preds: self.preds = detuplify(to_concat(self.preds, dim=self.concat_dim))
if not self.save_targs: self.targets = detuplify(to_concat(self.targets, dim=self.concat_dim))
if self.with_loss: self.losses = to_concat(self.losses)
def all_tensors(self):
res = [None if self.save_preds else self.preds, None if self.save_targs else self.targets]
if self.with_input: res = [self.inputs] + res
if self.with_loss: res.append(self.losses)
return res
show_doc(GatherPredsCallback, title_level=3)
#export
class FetchPredsCallback(Callback):
"A callback to fetch predictions during the training loop"
remove_on_fetch = True
def __init__(self, ds_idx=1, dl=None, with_input=False, with_decoded=False, cbs=None, reorder=True):
self.cbs = L(cbs)
store_attr('ds_idx,dl,with_input,with_decoded,reorder')
def after_validate(self):
to_rm = L(cb for cb in self.learn.cbs if getattr(cb, 'remove_on_fetch', False))
with self.learn.removed_cbs(to_rm + self.cbs) as learn:
self.preds = learn.get_preds(ds_idx=self.ds_idx, dl=self.dl,
with_input=self.with_input, with_decoded=self.with_decoded, inner=True, reorder=self.reorder)
show_doc(FetchPredsCallback, title_level=3)
#hide
from nbdev.export import notebook2script
notebook2script()
| 0.662141 | 0.810366 |
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
class Plane(object):
def __init__(self):
self.i = None;
self.a = None; #appearance time
self.e = None; #earliest
self.l = None; #latest
self.t = None; #target
self.s = []; #required seperation
self.g = None; #penalty before
self.h = None; #penalty after
self.x = -1; #actual landing time
def clone(plane):
clone = Plane()
clone.i = plane.i
clone.a = plane.a
clone.e = plane.e
clone.t = plane.t
clone.l = plane.l
clone.g = plane.g
clone.h = plane.h
clone.s = plane.s[:]
clone.x = plane.x
return clone
def getFitness(self):
if self.x == self.t:
return 0
if self.x > self.t:
return (self.x - self.t) * self.h
return (self.t - self.x) * self.g
def __str__(self):
result = 'appearance time: ' + str(self.a) + '\n' \
'earliest: ' + str(self.e) + '\n' \
'latest: ' + str(self.l) + '\n' \
'target: ' + str(self.t) + '\n' \
'penalty before: ' + str(self.g) + '\n' \
'penalty after: ' + str(self.h) + '\n' \
'--> actual: ' + str(self.x) + '\n' \
'--> fitness: ' + str(self.getFitness()) + '\n'
result += str(self.s) + '\n\n'
return result
location = './problems/'
prefix = 'airland'
fileType = '.txt'
problemsNumber = 13
problems = []
for i in range(1, problemsNumber + 1):
problems.append([location + prefix + str(i) + fileType, prefix + str(i)])
problemIndex = 0
problemSetLines = pd.read_csv(problems[problemIndex][0], header=None).values.tolist()
problemSet = []
for line in problemSetLines:
problemSet.append(line[0].strip().split(' '))
firstLine = problemSet.pop(0)
P = int(firstLine[0])
Freeze = int(firstLine[1])
P, Freeze
planeList = []
done = False
planeIndex = 0
while planeIndex < P:
planeIndex += 1
props = problemSet.pop(0)
i = Plane()
i.i = planeIndex - 1
i.a = int(props[0]) #appearance time,
i.e = int(props[1]) #earliest landing time,
i.t = int(props[2]) #target landing time,
i.l = int(props[3]) #latest landing time,
i.g = float(props[4]) #penalty cost per unit of time for landing before target,
i.h = float(props[5]) #penalty cost per unit of time for landing after target
while len(i.s) < P:
sepTimes = map(int, problemSet.pop(0))
i.s.extend(sepTimes)
#separation time required after i lands before j can land
planeList.append(i)
class LandingSolution(object):
def __init__(self):
self.landingSolution = None
self.p = P
def clone(self):
clone = LandingSolution()
clone.landingSolution = list(map(Plane.clone, self.landingSolution))
return clone
def constructRandomSolution(self, planes):
while not self.valid():
self.landingSolution = []
for i, plane in enumerate(planes):
plane.x = np.random.randint(low=plane.e, high=plane.l + 1)
self.landingSolution.append(plane)
def valid(self):
if self.landingSolution == None: return False
for i, plane in enumerate(self.landingSolution):
if plane.x < plane.e or plane.x > plane.l: return False
for j, seperation in enumerate(plane.s):
if i == j: continue
otherPlane = self.landingSolution[j]
if plane.x <= otherPlane.x and plane.x + seperation >= otherPlane.x:
#print(i, plane, "== > ==\n", j, otherPlane)
return False
return True
def getFitness(self):
sum = 0
for i in self.landingSolution:
sum += i.getFitness();
return sum
def __str__(self):
result = ''
for i, plane in enumerate(self.landingSolution):
result += str(i) + '\n' + str(plane)
return result
def createPopulation():
result = []
for i in range(POPULATION_SIZE):
solution = LandingSolution()
solution.constructRandomSolution(list(map(Plane.clone, planeList)))
result.append(solution)
return result
def getPopulationFitness(population):
populationFitness = []
for idx, individulal in enumerate(population):
populationFitness.append(individulal.getFitness())
return np.array(populationFitness)
def getPopulationHeuristicFitness(population, solution):
populationFitness = []
for idx, individulal in enumerate(population):
populationFitness.append(individulal.getFitness(solution))
return np.array(populationFitness)
def tournamentSelector(population, tournament_size=5):
# Make the tournament
random_indicies = np.random.randint(len(population), size=tournament_size).tolist()
tournament = []
for idx, val in np.ndenumerate(random_indicies):
tournament.append(population[val])
# Run the tournament
fitnesss = getPopulationFitness(tournament)
maxPos = np.argmax(fitnesss, axis=0)
minPos = np.argmin(fitnesss, axis=0)
return [random_indicies[minPos], fitnesss[minPos]], [random_indicies[maxPos], fitnesss[maxPos]]
def tournamentHeuristicSelector(population, solution, tournament_size=5):
# Make the tournament
random_indicies = np.random.randint(len(population), size=tournament_size).tolist()
tournament = []
for idx, val in np.ndenumerate(random_indicies):
tournament.append(population[val])
# Run the tournament
fitnesss = getPopulationHeuristicFitness(tournament, solution)
maxPos = np.argmax(fitnesss, axis=0)
minPos = np.argmin(fitnesss, axis=0)
return [random_indicies[minPos], fitnesss[minPos]], [random_indicies[maxPos], fitnesss[maxPos]]
def applyHeuristicToSolution(chromosome, solution):
clone = solution.clone()
for heuristic in chromosome:
clone = heuristic(clone)
return clone
class Chromosome(object):
def __init__(self, chromosome):
self.chromosome = chromosome
def getFitness(self, solution):
result = applyHeuristicToSolution(self.chromosome, solution)
return result.getFitness()
import operators
def geneticHeuristicAlgorithm(population, solution):
newPop = population[:]
for i in range(NUM_MUTATE):
best, worst = tournamentHeuristicSelector(population, solution, tournament_size=TOURNAMENT_SIZE)
newPop[worst[0]] = Chromosome(operators.swap(newPop[best[0]].chromosome))
for i in range(NUM_CROSSOVER):
best1, worst1 = tournamentHeuristicSelector(population, solution, tournament_size=TOURNAMENT_SIZE)
best2, worst2 = tournamentHeuristicSelector(population, solution, tournament_size=TOURNAMENT_SIZE)
result1, result2 = operators.crossover(newPop[best1[0]].chromosome, newPop[best2[0]].chromosome)
newPop[worst1[0]] = Chromosome(result1)
newPop[worst2[0]] = Chromosome(result2)
return newPop
import landingsolutionoperators
def geneticAlgorithm(population):
newPop = population[:]
for i in range(NUM_MUTATE):
best, worst = tournamentSelector(population, tournament_size=TOURNAMENT_SIZE)
newPop[worst[0]] = landingsolutionoperators.mutate(population[best[0]])
for i in range(NUM_CROSSOVER):
best1, worst1 = tournamentSelector(population, tournament_size=TOURNAMENT_SIZE)
best2, worst2 = tournamentSelector(population, tournament_size=TOURNAMENT_SIZE)
result1, result2 = landingsolutionoperators.swapCrossover(population[best1[0]], population[best2[0]])
newPop[worst1[0]] = result1
newPop[worst2[0]] = result2
return newPop
def generateChromosome(heuristics):
chromosome = []
chromosomeLength = np.random.randint(low=MIN_CHROMOSOME_SIZE,high=MAX_CHROMOSOME_SIZE)
for i in range(chromosomeLength):
heuristicChoice = np.random.randint(len(heuristics))
chromosome.append(heuristics[heuristicChoice])
return chromosome
def createHeuristicPopulation(heuristics):
population = []
for i in range(POPULATION_SIZE):
population.append(Chromosome(generateChromosome(heuristics)))
return population
def printPopulation(population):
for ind in population:
print(ind.chromosome)
POPULATION_SIZE = 15
GENERATIONS = 100
TOURNAMENT_SIZE = 3
MUTATION_RATE = 0.5
CROSSOVER_RATE = 0.3
MIN_CHROMOSOME_SIZE = 20
MAX_CHROMOSOME_SIZE = 30
NUM_MUTATE = int(MUTATION_RATE * POPULATION_SIZE)
NUM_CROSSOVER = int(CROSSOVER_RATE * POPULATION_SIZE)
NUM_REPRODUCTION = POPULATION_SIZE - (NUM_MUTATE + NUM_CROSSOVER)
assert NUM_REPRODUCTION >= 0
SAMPLES = 30
REPORT_RATE = 1
import lowlevelheuristics
heuristics = [
lowlevelheuristics.tryMoveHighestCloseToTarget,
lowlevelheuristics.tryMoveHighestCloseToTargetReversed,
lowlevelheuristics.trySwap,
lowlevelheuristics.trySwapReversed,
lowlevelheuristics.shiftUp,
lowlevelheuristics.shiftUpReversed,
lowlevelheuristics.shiftDown,
lowlevelheuristics.shiftDownReversed
]
import heuristicselectors
heuristicSelectors = [
[heuristicselectors.randomHeuristicSelection, "random"]
]
import moveacceptancetechniques
moveAcceptanceTechniques = [
[moveacceptancetechniques.acceptAll, "accept all moves"],
[moveacceptancetechniques.acceptIfImproving, "improving moves"],
[moveacceptancetechniques.acceptIfEqualOrImproving, "equal or improving moves"]
]
overTimeResult = []
finalResultMean = []
finalResultStd = []
finalResultMin = []
for heuristicSelector in heuristicSelectors:
for moveAcceptanceTechnique in moveAcceptanceTechniques:
print(heuristicSelector[1], moveAcceptanceTechnique[1])
samplesOverTimeResult = []
samplesFinalResult = []
for sample in range(SAMPLES):
sampleOverTimeResult = []
solution = LandingSolution()
solution.constructRandomSolution(list(map(Plane.clone, planeList)))
best = solution
for gen in range(GENERATIONS):
heuristic = heuristicSelector[0](heuristics)
potentialSolution = heuristic(solution)
if moveAcceptanceTechnique[0](solution, potentialSolution):
solution = potentialSolution
if moveacceptancetechniques.acceptIfImproving(best, solution):
best = solution
if gen % REPORT_RATE == 0:
sampleOverTimeResult.append(best.getFitness())
samplesFinalResult.append(best.getFitness())
samplesOverTimeResult.append(sampleOverTimeResult)
overTimeResult.append([np.mean(samplesOverTimeResult, axis=0), heuristicSelector[1], moveAcceptanceTechnique[1]])
finalResultMean.append([np.mean(samplesFinalResult), heuristicSelector[1], moveAcceptanceTechnique[1]])
finalResultStd.append([np.std(samplesFinalResult), heuristicSelector[1], moveAcceptanceTechnique[1]])
finalResultMin.append([np.min(samplesFinalResult), heuristicSelector[1], moveAcceptanceTechnique[1]])
print('genetic alg')
samplesOverTimeResult = []
samplesFinalResult = []
for sample in range(SAMPLES):
sampleOverTimeResult = []
population = createPopulation()
solution = population[0]
best = population[0]
for gen in range(GENERATIONS):
population = geneticAlgorithm(population)
if gen % REPORT_RATE == 0:
fitnesss = getPopulationFitness(population)
min = np.argmin(fitnesss, axis=0)
solution = population[min]
if moveacceptancetechniques.acceptIfImproving(best, solution):
best = solution
if gen % REPORT_RATE == 0:
sampleOverTimeResult.append(best.getFitness())
samplesFinalResult.append(best.getFitness())
samplesOverTimeResult.append(sampleOverTimeResult)
overTimeResult.append([np.mean(samplesOverTimeResult, axis=0), 'genetic alg', ''])
finalResultMean.append([np.mean(samplesFinalResult), 'genetic alg', ''])
finalResultStd.append([np.std(samplesFinalResult), 'genetic alg', ''])
finalResultMin.append([np.min(samplesFinalResult), 'genetic alg', ''])
print('genetic alg hyper heuristic')
samplesOverTimeResult = []
samplesFinalResult = []
for sample in range(SAMPLES):
sampleOverTimeResult = []
population = createHeuristicPopulation(heuristics)
thissolution = LandingSolution()
thissolution.constructRandomSolution(list(map(Plane.clone, planeList)))
current = Chromosome(population[0].chromosome.copy())
best = Chromosome(population[0].chromosome.copy())
for gen in range(GENERATIONS):
population = geneticHeuristicAlgorithm(population, thissolution.clone())
if gen % REPORT_RATE == 0:
fitnesss = getPopulationHeuristicFitness(population, thissolution.clone())
min = np.argmin(fitnesss, axis=0)
current = Chromosome(population[min].chromosome.copy())
if best.getFitness(thissolution.clone()) > current.getFitness(thissolution.clone()):
best = Chromosome(current.chromosome.copy())
if gen % REPORT_RATE == 0:
sampleOverTimeResult.append(best.getFitness(thissolution.clone()))
samplesFinalResult.append(best.getFitness(thissolution.clone()))
samplesOverTimeResult.append(sampleOverTimeResult)
overTimeResult.append([np.mean(samplesOverTimeResult, axis=0), 'genetic alg hyper heuristic', ''])
finalResultMean.append([np.mean(samplesFinalResult), 'genetic alg hyper heuristic', ''])
finalResultStd.append([np.std(samplesFinalResult), 'genetic alg hyper heuristic', ''])
finalResultMin.append([np.min(samplesFinalResult), 'genetic alg hyper heuristic', ''])
fig = plt.figure()
plt.grid(1)
plt.xlim([0, GENERATIONS])
plt.ion()
plt.xlabel('Generations')
plt.ylabel('Fitness')
generations = np.arange(0, GENERATIONS, REPORT_RATE)
plotColors = [
'b--',
'r--',
'g--',
'bs',
'g^',
'k'
]
plots = []
descriptions = []
for x, result in enumerate(overTimeResult):
plots.append(plt.plot(generations, result[0], plotColors[x%len(plotColors)] , linewidth=1, markersize=3)[0])
descriptions.append(result[1] + ' ' + result[2])
plt.legend(plots, descriptions)
fig.savefig('./docs/' + problems[problemIndex][1] + '.png')
plt.show(5)
plt.close()
padding = [None, None, None, None]
problem = []
problem.append(problemIndex + 1)
problem.extend(padding)
d = {
'problem': problem,
'type': list(map(lambda x: x[1] + ' ' + x[2], finalResultMean)),
'mean': list(map(lambda x: x[0], finalResultMean)),
'std': list(map(lambda x: x[0], finalResultStd)),
'max': list(map(lambda x: x[0], finalResultMin))
}
df = pd.DataFrame(data=d)
list(df.columns.values)
result = df[['problem', 'type', 'mean', 'std', 'max']]
result
#print(result.to_latex(index=False, bold_rows=True, na_rep=''))
with open('./docs/' + problems[problemIndex][1] + '.txt', 'w') as f:
print(result.to_latex(index=False, bold_rows=True, na_rep=''), file=f)
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
class Plane(object):
def __init__(self):
self.i = None;
self.a = None; #appearance time
self.e = None; #earliest
self.l = None; #latest
self.t = None; #target
self.s = []; #required seperation
self.g = None; #penalty before
self.h = None; #penalty after
self.x = -1; #actual landing time
def clone(plane):
clone = Plane()
clone.i = plane.i
clone.a = plane.a
clone.e = plane.e
clone.t = plane.t
clone.l = plane.l
clone.g = plane.g
clone.h = plane.h
clone.s = plane.s[:]
clone.x = plane.x
return clone
def getFitness(self):
if self.x == self.t:
return 0
if self.x > self.t:
return (self.x - self.t) * self.h
return (self.t - self.x) * self.g
def __str__(self):
result = 'appearance time: ' + str(self.a) + '\n' \
'earliest: ' + str(self.e) + '\n' \
'latest: ' + str(self.l) + '\n' \
'target: ' + str(self.t) + '\n' \
'penalty before: ' + str(self.g) + '\n' \
'penalty after: ' + str(self.h) + '\n' \
'--> actual: ' + str(self.x) + '\n' \
'--> fitness: ' + str(self.getFitness()) + '\n'
result += str(self.s) + '\n\n'
return result
location = './problems/'
prefix = 'airland'
fileType = '.txt'
problemsNumber = 13
problems = []
for i in range(1, problemsNumber + 1):
problems.append([location + prefix + str(i) + fileType, prefix + str(i)])
problemIndex = 0
problemSetLines = pd.read_csv(problems[problemIndex][0], header=None).values.tolist()
problemSet = []
for line in problemSetLines:
problemSet.append(line[0].strip().split(' '))
firstLine = problemSet.pop(0)
P = int(firstLine[0])
Freeze = int(firstLine[1])
P, Freeze
planeList = []
done = False
planeIndex = 0
while planeIndex < P:
planeIndex += 1
props = problemSet.pop(0)
i = Plane()
i.i = planeIndex - 1
i.a = int(props[0]) #appearance time,
i.e = int(props[1]) #earliest landing time,
i.t = int(props[2]) #target landing time,
i.l = int(props[3]) #latest landing time,
i.g = float(props[4]) #penalty cost per unit of time for landing before target,
i.h = float(props[5]) #penalty cost per unit of time for landing after target
while len(i.s) < P:
sepTimes = map(int, problemSet.pop(0))
i.s.extend(sepTimes)
#separation time required after i lands before j can land
planeList.append(i)
class LandingSolution(object):
def __init__(self):
self.landingSolution = None
self.p = P
def clone(self):
clone = LandingSolution()
clone.landingSolution = list(map(Plane.clone, self.landingSolution))
return clone
def constructRandomSolution(self, planes):
while not self.valid():
self.landingSolution = []
for i, plane in enumerate(planes):
plane.x = np.random.randint(low=plane.e, high=plane.l + 1)
self.landingSolution.append(plane)
def valid(self):
if self.landingSolution == None: return False
for i, plane in enumerate(self.landingSolution):
if plane.x < plane.e or plane.x > plane.l: return False
for j, seperation in enumerate(plane.s):
if i == j: continue
otherPlane = self.landingSolution[j]
if plane.x <= otherPlane.x and plane.x + seperation >= otherPlane.x:
#print(i, plane, "== > ==\n", j, otherPlane)
return False
return True
def getFitness(self):
sum = 0
for i in self.landingSolution:
sum += i.getFitness();
return sum
def __str__(self):
result = ''
for i, plane in enumerate(self.landingSolution):
result += str(i) + '\n' + str(plane)
return result
def createPopulation():
result = []
for i in range(POPULATION_SIZE):
solution = LandingSolution()
solution.constructRandomSolution(list(map(Plane.clone, planeList)))
result.append(solution)
return result
def getPopulationFitness(population):
populationFitness = []
for idx, individulal in enumerate(population):
populationFitness.append(individulal.getFitness())
return np.array(populationFitness)
def getPopulationHeuristicFitness(population, solution):
populationFitness = []
for idx, individulal in enumerate(population):
populationFitness.append(individulal.getFitness(solution))
return np.array(populationFitness)
def tournamentSelector(population, tournament_size=5):
# Make the tournament
random_indicies = np.random.randint(len(population), size=tournament_size).tolist()
tournament = []
for idx, val in np.ndenumerate(random_indicies):
tournament.append(population[val])
# Run the tournament
fitnesss = getPopulationFitness(tournament)
maxPos = np.argmax(fitnesss, axis=0)
minPos = np.argmin(fitnesss, axis=0)
return [random_indicies[minPos], fitnesss[minPos]], [random_indicies[maxPos], fitnesss[maxPos]]
def tournamentHeuristicSelector(population, solution, tournament_size=5):
# Make the tournament
random_indicies = np.random.randint(len(population), size=tournament_size).tolist()
tournament = []
for idx, val in np.ndenumerate(random_indicies):
tournament.append(population[val])
# Run the tournament
fitnesss = getPopulationHeuristicFitness(tournament, solution)
maxPos = np.argmax(fitnesss, axis=0)
minPos = np.argmin(fitnesss, axis=0)
return [random_indicies[minPos], fitnesss[minPos]], [random_indicies[maxPos], fitnesss[maxPos]]
def applyHeuristicToSolution(chromosome, solution):
clone = solution.clone()
for heuristic in chromosome:
clone = heuristic(clone)
return clone
class Chromosome(object):
def __init__(self, chromosome):
self.chromosome = chromosome
def getFitness(self, solution):
result = applyHeuristicToSolution(self.chromosome, solution)
return result.getFitness()
import operators
def geneticHeuristicAlgorithm(population, solution):
newPop = population[:]
for i in range(NUM_MUTATE):
best, worst = tournamentHeuristicSelector(population, solution, tournament_size=TOURNAMENT_SIZE)
newPop[worst[0]] = Chromosome(operators.swap(newPop[best[0]].chromosome))
for i in range(NUM_CROSSOVER):
best1, worst1 = tournamentHeuristicSelector(population, solution, tournament_size=TOURNAMENT_SIZE)
best2, worst2 = tournamentHeuristicSelector(population, solution, tournament_size=TOURNAMENT_SIZE)
result1, result2 = operators.crossover(newPop[best1[0]].chromosome, newPop[best2[0]].chromosome)
newPop[worst1[0]] = Chromosome(result1)
newPop[worst2[0]] = Chromosome(result2)
return newPop
import landingsolutionoperators
def geneticAlgorithm(population):
newPop = population[:]
for i in range(NUM_MUTATE):
best, worst = tournamentSelector(population, tournament_size=TOURNAMENT_SIZE)
newPop[worst[0]] = landingsolutionoperators.mutate(population[best[0]])
for i in range(NUM_CROSSOVER):
best1, worst1 = tournamentSelector(population, tournament_size=TOURNAMENT_SIZE)
best2, worst2 = tournamentSelector(population, tournament_size=TOURNAMENT_SIZE)
result1, result2 = landingsolutionoperators.swapCrossover(population[best1[0]], population[best2[0]])
newPop[worst1[0]] = result1
newPop[worst2[0]] = result2
return newPop
def generateChromosome(heuristics):
chromosome = []
chromosomeLength = np.random.randint(low=MIN_CHROMOSOME_SIZE,high=MAX_CHROMOSOME_SIZE)
for i in range(chromosomeLength):
heuristicChoice = np.random.randint(len(heuristics))
chromosome.append(heuristics[heuristicChoice])
return chromosome
def createHeuristicPopulation(heuristics):
population = []
for i in range(POPULATION_SIZE):
population.append(Chromosome(generateChromosome(heuristics)))
return population
def printPopulation(population):
for ind in population:
print(ind.chromosome)
POPULATION_SIZE = 15
GENERATIONS = 100
TOURNAMENT_SIZE = 3
MUTATION_RATE = 0.5
CROSSOVER_RATE = 0.3
MIN_CHROMOSOME_SIZE = 20
MAX_CHROMOSOME_SIZE = 30
NUM_MUTATE = int(MUTATION_RATE * POPULATION_SIZE)
NUM_CROSSOVER = int(CROSSOVER_RATE * POPULATION_SIZE)
NUM_REPRODUCTION = POPULATION_SIZE - (NUM_MUTATE + NUM_CROSSOVER)
assert NUM_REPRODUCTION >= 0
SAMPLES = 30
REPORT_RATE = 1
import lowlevelheuristics
heuristics = [
lowlevelheuristics.tryMoveHighestCloseToTarget,
lowlevelheuristics.tryMoveHighestCloseToTargetReversed,
lowlevelheuristics.trySwap,
lowlevelheuristics.trySwapReversed,
lowlevelheuristics.shiftUp,
lowlevelheuristics.shiftUpReversed,
lowlevelheuristics.shiftDown,
lowlevelheuristics.shiftDownReversed
]
import heuristicselectors
heuristicSelectors = [
[heuristicselectors.randomHeuristicSelection, "random"]
]
import moveacceptancetechniques
moveAcceptanceTechniques = [
[moveacceptancetechniques.acceptAll, "accept all moves"],
[moveacceptancetechniques.acceptIfImproving, "improving moves"],
[moveacceptancetechniques.acceptIfEqualOrImproving, "equal or improving moves"]
]
overTimeResult = []
finalResultMean = []
finalResultStd = []
finalResultMin = []
for heuristicSelector in heuristicSelectors:
for moveAcceptanceTechnique in moveAcceptanceTechniques:
print(heuristicSelector[1], moveAcceptanceTechnique[1])
samplesOverTimeResult = []
samplesFinalResult = []
for sample in range(SAMPLES):
sampleOverTimeResult = []
solution = LandingSolution()
solution.constructRandomSolution(list(map(Plane.clone, planeList)))
best = solution
for gen in range(GENERATIONS):
heuristic = heuristicSelector[0](heuristics)
potentialSolution = heuristic(solution)
if moveAcceptanceTechnique[0](solution, potentialSolution):
solution = potentialSolution
if moveacceptancetechniques.acceptIfImproving(best, solution):
best = solution
if gen % REPORT_RATE == 0:
sampleOverTimeResult.append(best.getFitness())
samplesFinalResult.append(best.getFitness())
samplesOverTimeResult.append(sampleOverTimeResult)
overTimeResult.append([np.mean(samplesOverTimeResult, axis=0), heuristicSelector[1], moveAcceptanceTechnique[1]])
finalResultMean.append([np.mean(samplesFinalResult), heuristicSelector[1], moveAcceptanceTechnique[1]])
finalResultStd.append([np.std(samplesFinalResult), heuristicSelector[1], moveAcceptanceTechnique[1]])
finalResultMin.append([np.min(samplesFinalResult), heuristicSelector[1], moveAcceptanceTechnique[1]])
print('genetic alg')
samplesOverTimeResult = []
samplesFinalResult = []
for sample in range(SAMPLES):
sampleOverTimeResult = []
population = createPopulation()
solution = population[0]
best = population[0]
for gen in range(GENERATIONS):
population = geneticAlgorithm(population)
if gen % REPORT_RATE == 0:
fitnesss = getPopulationFitness(population)
min = np.argmin(fitnesss, axis=0)
solution = population[min]
if moveacceptancetechniques.acceptIfImproving(best, solution):
best = solution
if gen % REPORT_RATE == 0:
sampleOverTimeResult.append(best.getFitness())
samplesFinalResult.append(best.getFitness())
samplesOverTimeResult.append(sampleOverTimeResult)
overTimeResult.append([np.mean(samplesOverTimeResult, axis=0), 'genetic alg', ''])
finalResultMean.append([np.mean(samplesFinalResult), 'genetic alg', ''])
finalResultStd.append([np.std(samplesFinalResult), 'genetic alg', ''])
finalResultMin.append([np.min(samplesFinalResult), 'genetic alg', ''])
print('genetic alg hyper heuristic')
samplesOverTimeResult = []
samplesFinalResult = []
for sample in range(SAMPLES):
sampleOverTimeResult = []
population = createHeuristicPopulation(heuristics)
thissolution = LandingSolution()
thissolution.constructRandomSolution(list(map(Plane.clone, planeList)))
current = Chromosome(population[0].chromosome.copy())
best = Chromosome(population[0].chromosome.copy())
for gen in range(GENERATIONS):
population = geneticHeuristicAlgorithm(population, thissolution.clone())
if gen % REPORT_RATE == 0:
fitnesss = getPopulationHeuristicFitness(population, thissolution.clone())
min = np.argmin(fitnesss, axis=0)
current = Chromosome(population[min].chromosome.copy())
if best.getFitness(thissolution.clone()) > current.getFitness(thissolution.clone()):
best = Chromosome(current.chromosome.copy())
if gen % REPORT_RATE == 0:
sampleOverTimeResult.append(best.getFitness(thissolution.clone()))
samplesFinalResult.append(best.getFitness(thissolution.clone()))
samplesOverTimeResult.append(sampleOverTimeResult)
overTimeResult.append([np.mean(samplesOverTimeResult, axis=0), 'genetic alg hyper heuristic', ''])
finalResultMean.append([np.mean(samplesFinalResult), 'genetic alg hyper heuristic', ''])
finalResultStd.append([np.std(samplesFinalResult), 'genetic alg hyper heuristic', ''])
finalResultMin.append([np.min(samplesFinalResult), 'genetic alg hyper heuristic', ''])
fig = plt.figure()
plt.grid(1)
plt.xlim([0, GENERATIONS])
plt.ion()
plt.xlabel('Generations')
plt.ylabel('Fitness')
generations = np.arange(0, GENERATIONS, REPORT_RATE)
plotColors = [
'b--',
'r--',
'g--',
'bs',
'g^',
'k'
]
plots = []
descriptions = []
for x, result in enumerate(overTimeResult):
plots.append(plt.plot(generations, result[0], plotColors[x%len(plotColors)] , linewidth=1, markersize=3)[0])
descriptions.append(result[1] + ' ' + result[2])
plt.legend(plots, descriptions)
fig.savefig('./docs/' + problems[problemIndex][1] + '.png')
plt.show(5)
plt.close()
padding = [None, None, None, None]
problem = []
problem.append(problemIndex + 1)
problem.extend(padding)
d = {
'problem': problem,
'type': list(map(lambda x: x[1] + ' ' + x[2], finalResultMean)),
'mean': list(map(lambda x: x[0], finalResultMean)),
'std': list(map(lambda x: x[0], finalResultStd)),
'max': list(map(lambda x: x[0], finalResultMin))
}
df = pd.DataFrame(data=d)
list(df.columns.values)
result = df[['problem', 'type', 'mean', 'std', 'max']]
result
#print(result.to_latex(index=False, bold_rows=True, na_rep=''))
with open('./docs/' + problems[problemIndex][1] + '.txt', 'w') as f:
print(result.to_latex(index=False, bold_rows=True, na_rep=''), file=f)
| 0.557484 | 0.347122 |
```
import numpy as np
import sympy
from matplotlib import pyplot
%matplotlib inline
from sympy.utilities.lambdify import lambdify
#Part A
#Conditions
Vmax = 80 #km/h
L = 11 #km
rhomax = 250 #cars/km
nx = 51 #spatial discretization
dt = 0.001 #h
dx = L/(nx-1)
x = np.linspace(0,L,nx)
rho0 = np.ones(nx)*10
rho0[10:20] = 50
T1 = 6/60.0
nt = int(T1/dt)
print('Number of steps',nt)
v = np.ones(nx)
for i in range(nx):
v[i] = Vmax*(1 - (rho0[i]/rhomax))
vminimum = np.min(v)
vmin0 = vminimum/3.6
vmin0
print('Minimum velocity at t=0 (m/s): {:.2f}'.format(vmin0))
rho = rho0.copy()
v = np.ones(nx)
F = np.ones(nx)
for n in range(1,nt):
F[1:]=Vmax*rho[1:]*(1-rho[1:]/rhomax)
F[:-1]=Vmax*rho[:-1]*(1-rho[:-1]/rhomax)
rho[1:]=rho[1:]-dt/dx*(F[1:]-F[:-1])
v=F/rho
if n==50:
vmean3 = np.mean(v)
vms3 = vmean3/3.6
print('Average velocity at t=3 min (m/s): {:.2f}'.format(vms3))
vmin = np.min(v)
vmsmin = vmin/3.6
print('Minimum velocity at t=6 min (m/s): {:.2f}'.format(vmsmin))
vmsmin
# Plot the solution after nt time steps
# along with the initial conditions.
pyplot.figure(figsize=(6.0, 4.0))
pyplot.xlabel('x')
pyplot.ylabel('rho0,rho,v')
pyplot.grid()
pyplot.plot(x, rho0, label='Rho0',
color='C0', linestyle='-', linewidth=2)
pyplot.plot(x, rho, label='Rho',
color='C1', linestyle='-', linewidth=2)
pyplot.plot(x, v, label='v',
color='C2', linestyle='-', linewidth=2)
pyplot.legend(loc='center right')
pyplot.figure(figsize=(6.0, 4.0))
pyplot.xlabel('x')
pyplot.ylabel('F')
pyplot.grid()
pyplot.plot(x, F, label='Flux',
color='C3', linestyle='-', linewidth=2)
pyplot.legend(loc='upper left')
#Part B
#Conditions
Vmax = 136 #km/h
L = 11 #km
rhomax = 250 #cars/km
nx = 51 #spatial discretization
dt = 0.001 #h
dx = L/(nx-1)
x = np.linspace(0,L,nx)
rho0 = np.ones(nx)*20
rho0[10:20] = 50
T1 = 3/60.0
T1
nt = int(T1/dt)
print('Number of steps',nt)
v = np.ones(nx)
for i in range(nx):
v[i] = Vmax*(1 - (rho0[i]/rhomax))
vminimum = np.min(v)
vmin0 = vminimum/3.6
vmin0
print('Minimum velocity at t=0 (m/s): {:.2f}'.format(vmin0))
rho = rho0.copy()
v = np.ones(nx)
F = np.ones(nx)
for n in range(1,nt):
F[1:]=Vmax*rho[1:]*(1-rho[1:]/rhomax)
F[:-1]=Vmax*rho[:-1]*(1-rho[:-1]/rhomax)
rho[1:]=rho[1:]-dt/dx*(F[1:]-F[:-1])
v=F/rho
vmean3 = np.mean(v)
vms3 = vmean3/3.6
print('Average velocity at t=3 min (m/s): {:.2f}'.format(vms3))
vmin = np.min(v)
vmsmin = vmin/3.6
print('Minimum velocity at t=3 min (m/s): {:.2f}'.format(vmsmin))
# Plot the solution after nt time steps
# along with the initial conditions.
pyplot.figure(figsize=(6.0, 4.0))
pyplot.xlabel('x')
pyplot.ylabel('rho0,rho,v')
pyplot.grid()
pyplot.plot(x, rho0, label='Rho0',
color='C0', linestyle='-', linewidth=2)
pyplot.plot(x, rho, label='Rho',
color='C1', linestyle='-', linewidth=2)
pyplot.plot(x, v, label='v',
color='C2', linestyle='-', linewidth=2)
pyplot.legend(loc='center right')
pyplot.figure(figsize=(6.0, 4.0))
pyplot.xlabel('x')
pyplot.ylabel('F')
pyplot.grid()
pyplot.plot(x, F, label='Flux',
color='C3', linestyle='-', linewidth=2)
pyplot.legend(loc='upper left')
```
|
github_jupyter
|
import numpy as np
import sympy
from matplotlib import pyplot
%matplotlib inline
from sympy.utilities.lambdify import lambdify
#Part A
#Conditions
Vmax = 80 #km/h
L = 11 #km
rhomax = 250 #cars/km
nx = 51 #spatial discretization
dt = 0.001 #h
dx = L/(nx-1)
x = np.linspace(0,L,nx)
rho0 = np.ones(nx)*10
rho0[10:20] = 50
T1 = 6/60.0
nt = int(T1/dt)
print('Number of steps',nt)
v = np.ones(nx)
for i in range(nx):
v[i] = Vmax*(1 - (rho0[i]/rhomax))
vminimum = np.min(v)
vmin0 = vminimum/3.6
vmin0
print('Minimum velocity at t=0 (m/s): {:.2f}'.format(vmin0))
rho = rho0.copy()
v = np.ones(nx)
F = np.ones(nx)
for n in range(1,nt):
F[1:]=Vmax*rho[1:]*(1-rho[1:]/rhomax)
F[:-1]=Vmax*rho[:-1]*(1-rho[:-1]/rhomax)
rho[1:]=rho[1:]-dt/dx*(F[1:]-F[:-1])
v=F/rho
if n==50:
vmean3 = np.mean(v)
vms3 = vmean3/3.6
print('Average velocity at t=3 min (m/s): {:.2f}'.format(vms3))
vmin = np.min(v)
vmsmin = vmin/3.6
print('Minimum velocity at t=6 min (m/s): {:.2f}'.format(vmsmin))
vmsmin
# Plot the solution after nt time steps
# along with the initial conditions.
pyplot.figure(figsize=(6.0, 4.0))
pyplot.xlabel('x')
pyplot.ylabel('rho0,rho,v')
pyplot.grid()
pyplot.plot(x, rho0, label='Rho0',
color='C0', linestyle='-', linewidth=2)
pyplot.plot(x, rho, label='Rho',
color='C1', linestyle='-', linewidth=2)
pyplot.plot(x, v, label='v',
color='C2', linestyle='-', linewidth=2)
pyplot.legend(loc='center right')
pyplot.figure(figsize=(6.0, 4.0))
pyplot.xlabel('x')
pyplot.ylabel('F')
pyplot.grid()
pyplot.plot(x, F, label='Flux',
color='C3', linestyle='-', linewidth=2)
pyplot.legend(loc='upper left')
#Part B
#Conditions
Vmax = 136 #km/h
L = 11 #km
rhomax = 250 #cars/km
nx = 51 #spatial discretization
dt = 0.001 #h
dx = L/(nx-1)
x = np.linspace(0,L,nx)
rho0 = np.ones(nx)*20
rho0[10:20] = 50
T1 = 3/60.0
T1
nt = int(T1/dt)
print('Number of steps',nt)
v = np.ones(nx)
for i in range(nx):
v[i] = Vmax*(1 - (rho0[i]/rhomax))
vminimum = np.min(v)
vmin0 = vminimum/3.6
vmin0
print('Minimum velocity at t=0 (m/s): {:.2f}'.format(vmin0))
rho = rho0.copy()
v = np.ones(nx)
F = np.ones(nx)
for n in range(1,nt):
F[1:]=Vmax*rho[1:]*(1-rho[1:]/rhomax)
F[:-1]=Vmax*rho[:-1]*(1-rho[:-1]/rhomax)
rho[1:]=rho[1:]-dt/dx*(F[1:]-F[:-1])
v=F/rho
vmean3 = np.mean(v)
vms3 = vmean3/3.6
print('Average velocity at t=3 min (m/s): {:.2f}'.format(vms3))
vmin = np.min(v)
vmsmin = vmin/3.6
print('Minimum velocity at t=3 min (m/s): {:.2f}'.format(vmsmin))
# Plot the solution after nt time steps
# along with the initial conditions.
pyplot.figure(figsize=(6.0, 4.0))
pyplot.xlabel('x')
pyplot.ylabel('rho0,rho,v')
pyplot.grid()
pyplot.plot(x, rho0, label='Rho0',
color='C0', linestyle='-', linewidth=2)
pyplot.plot(x, rho, label='Rho',
color='C1', linestyle='-', linewidth=2)
pyplot.plot(x, v, label='v',
color='C2', linestyle='-', linewidth=2)
pyplot.legend(loc='center right')
pyplot.figure(figsize=(6.0, 4.0))
pyplot.xlabel('x')
pyplot.ylabel('F')
pyplot.grid()
pyplot.plot(x, F, label='Flux',
color='C3', linestyle='-', linewidth=2)
pyplot.legend(loc='upper left')
| 0.584864 | 0.51129 |
# `MeshArrays.jl` defines the `MeshArray` type
Each `MeshArray` contains an array of elementary arrays that (1) are connected at their edges and (2) collectively form a global grid. Overall grid specifications are contained within `gcmgrid` instances, which merely define array sizes and how e.g. grid variables are represented in memory. Importantly, it is only when e.g. grid variables are read from file that sizable memory is allocated.
## Initialize Framework
1. import `MeshArrays` and plotting tools
2. choose e.g. a standard `MITgcm` grid
3. download the grid if needed
```
using MeshArrays, Plots
pth="../inputs/GRID_LLC90/"
γ=GridSpec("LatLonCap",pth)
http="https://github.com/gaelforget/GRID_LLC90"
!isdir(pth) ? run(`git clone $http $pth`) : nothing;
```
## Read Example
A `MeshArray` instance, on the chosen grid, can be obtained from a file path (argument #1). Format conversion occur inside the `read` function based on a propotype argument (#2). `read` / `write` calls then convert back and forth between `MeshArray` and `Array` formats.
```
D=γ.read(γ.path*"Depth.data",MeshArray(γ,Float64))
tmp1=write(D); tmp2=read(tmp1,D)
show(D)
```
## Subdomain Arrays
The heatmap function as specialized in `../examples/Plots.jl` operates on each `inner-array` sequentially, one after the other.
```
p=dirname(pathof(MeshArrays))
include(joinpath(p,"../examples/Plots.jl"))
heatmap(D,title="Ocean Depth",clims=(0.,6000.))
```
## A `MeshArray` Behaves Like A `Array`
Here are a few examples that would be coded similarly in both cases
```
size(D)
eltype(D)
view(D,:)
D .* 1.0
D .* D
1000*D
D*1000
D[findall(D .> 300.)] .= NaN
D[findall(D .< 1.)] .= NaN
D[1]=0.0 .+ D[1]
tmp=cos.(D);
```
## Let's switch grid now
```
pth="../inputs/GRID_CS32/"
γ=GridSpec("LatLonCap",pth)
http="https://github.com/gaelforget/GRID_CS32"
!isdir(pth) ? run(`git clone $http $pth`) : nothing
γ=GridSpec("CubeSphere",pth)
D=γ.read(γ.path*"Depth.data",MeshArray(γ,Float32))
show(D)
```
## The `exchange` Function
It adds neighboring points at face edges to slightly extend the computational domain as often needed e.g. to compute partial derivatives.
```
Dexch=exchange(D,4)
show(Dexch)
P=heatmap(D.f[6],title="Ocean Depth (D, Face 6)",lims=(-4,36))
Pexch=heatmap(Dexch.f[6],title="...(Dexch, Face 6)",lims=(0,40))
plot(P,Pexch)
```
# Diffusion-Based Smoothing
The unit testing of `MeshArrays.jl` uses the `smooth()` function. Starting from a random noise field, the smoothing efficiency is predictable and can be set via a smoothing scale parameter [(see Weaver and Courtier, 2001)](https://doi.org/10.1002/qj.49712757518).
This example also illustrates the generality of the `MeshArrays` approach, where the same code in `demo2` is readily applicable to any `PeriodicDomain`, `PeriodicChanel`, `CubeSphere`, or `LatLonCap` grid. Here the chosen grid maps onto the `6` faces of a cube with `16*16` points per face, with distances, areas, etc all set to `1.0`.
```
p=dirname(pathof(MeshArrays))
include(joinpath(p,"../examples/Demos.jl"))
Γ=GridOfOnes("CubeSphere",6,16)
Δ=demo2(Γ);
```
The initial noise field is `D[1]` while the smoothed one is `D[2]`. After `smooth()` has been applied via `demo2()`, the noise field is visibly smoother and more muted.
```
heatmap(Δ[1],title="initial noise",clims=(-0.5,0.5))
heatmap(Δ[2],title="smoothed noise",clims=(-0.5,0.5))
```
The computational cost of `smooth()` predictably increases with the decorrelation scale. For more about how this works, please refer to **Weaver and Courtier, 2001** _Correlation modelling on the sphere using a generalized diffusion equation_ https://doi.org/10.1002/qj.49712757518
```
Rini=Δ[1]
DXCsm=Δ[3]
DYCsm=Δ[4]
@time Rend=smooth(Rini,DXCsm,DYCsm,Γ);
@time Rend=smooth(Rini,2DXCsm,2DYCsm,Γ);
```
|
github_jupyter
|
using MeshArrays, Plots
pth="../inputs/GRID_LLC90/"
γ=GridSpec("LatLonCap",pth)
http="https://github.com/gaelforget/GRID_LLC90"
!isdir(pth) ? run(`git clone $http $pth`) : nothing;
D=γ.read(γ.path*"Depth.data",MeshArray(γ,Float64))
tmp1=write(D); tmp2=read(tmp1,D)
show(D)
p=dirname(pathof(MeshArrays))
include(joinpath(p,"../examples/Plots.jl"))
heatmap(D,title="Ocean Depth",clims=(0.,6000.))
size(D)
eltype(D)
view(D,:)
D .* 1.0
D .* D
1000*D
D*1000
D[findall(D .> 300.)] .= NaN
D[findall(D .< 1.)] .= NaN
D[1]=0.0 .+ D[1]
tmp=cos.(D);
pth="../inputs/GRID_CS32/"
γ=GridSpec("LatLonCap",pth)
http="https://github.com/gaelforget/GRID_CS32"
!isdir(pth) ? run(`git clone $http $pth`) : nothing
γ=GridSpec("CubeSphere",pth)
D=γ.read(γ.path*"Depth.data",MeshArray(γ,Float32))
show(D)
Dexch=exchange(D,4)
show(Dexch)
P=heatmap(D.f[6],title="Ocean Depth (D, Face 6)",lims=(-4,36))
Pexch=heatmap(Dexch.f[6],title="...(Dexch, Face 6)",lims=(0,40))
plot(P,Pexch)
p=dirname(pathof(MeshArrays))
include(joinpath(p,"../examples/Demos.jl"))
Γ=GridOfOnes("CubeSphere",6,16)
Δ=demo2(Γ);
heatmap(Δ[1],title="initial noise",clims=(-0.5,0.5))
heatmap(Δ[2],title="smoothed noise",clims=(-0.5,0.5))
Rini=Δ[1]
DXCsm=Δ[3]
DYCsm=Δ[4]
@time Rend=smooth(Rini,DXCsm,DYCsm,Γ);
@time Rend=smooth(Rini,2DXCsm,2DYCsm,Γ);
| 0.192995 | 0.987958 |
```
import keras
import keras.backend as K
from keras.datasets import mnist
from keras.models import Sequential, Model, load_model
from keras.layers import Dense, Dropout, Activation, Flatten, Input, Lambda
from keras.layers import Conv2D, MaxPooling2D, AveragePooling2D, Conv1D, MaxPooling1D, LSTM, ConvLSTM2D, GRU, BatchNormalization, LocallyConnected2D, Permute, TimeDistributed, Bidirectional
from keras.layers import Concatenate, Reshape, Conv2DTranspose, Embedding, Multiply, Activation
from functools import partial
from collections import defaultdict
import os
import pickle
import numpy as np
import scipy.sparse as sp
import scipy.io as spio
import isolearn.io as isoio
import isolearn.keras as isol
import matplotlib.pyplot as plt
from sequence_logo_helper import dna_letter_at, plot_dna_logo
from sklearn import preprocessing
import pandas as pd
import tensorflow as tf
from keras.backend.tensorflow_backend import set_session
def contain_tf_gpu_mem_usage() :
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
sess = tf.Session(config=config)
set_session(sess)
contain_tf_gpu_mem_usage()
#optimus 5-prime functions
def test_data(df, model, test_seq, obs_col, output_col='pred'):
'''Predict mean ribosome load using model and test set UTRs'''
# Scale the test set mean ribosome load
scaler = preprocessing.StandardScaler()
scaler.fit(df[obs_col].reshape(-1,1))
# Make predictions
predictions = model.predict(test_seq).reshape(-1)
# Inverse scaled predicted mean ribosome load and return in a column labeled 'pred'
df.loc[:,output_col] = scaler.inverse_transform(predictions)
return df
def one_hot_encode(df, col='utr', seq_len=50):
# Dictionary returning one-hot encoding of nucleotides.
nuc_d = {'a':[1,0,0,0],'c':[0,1,0,0],'g':[0,0,1,0],'t':[0,0,0,1], 'n':[0,0,0,0]}
# Creat empty matrix.
vectors=np.empty([len(df),seq_len,4])
# Iterate through UTRs and one-hot encode
for i,seq in enumerate(df[col].str[:seq_len]):
seq = seq.lower()
a = np.array([nuc_d[x] for x in seq])
vectors[i] = a
return vectors
def r2(x,y):
slope, intercept, r_value, p_value, std_err = stats.linregress(x,y)
return r_value**2
#Train data
e_train = pd.read_csv("bottom5KIFuAUGTop5KIFuAUG.csv")
e_train.loc[:,'scaled_rl'] = preprocessing.StandardScaler().fit_transform(e_train.loc[:,'rl'].values.reshape(-1,1))
seq_e_train = one_hot_encode(e_train,seq_len=50)
x_train = seq_e_train
x_train = np.reshape(x_train, (x_train.shape[0], 1, x_train.shape[1], x_train.shape[2]))
y_train = np.array(e_train['scaled_rl'].values)
y_train = np.reshape(y_train, (y_train.shape[0],1))
print("x_train.shape = " + str(x_train.shape))
print("y_train.shape = " + str(y_train.shape))
#Load Predictor
predictor_path = 'optimusRetrainedMain.hdf5'
predictor = load_model(predictor_path)
predictor.trainable = False
predictor.compile(optimizer=keras.optimizers.SGD(lr=0.1), loss='mean_squared_error')
#Generate (original) predictions
pred_train = predictor.predict(x_train[:, 0, ...], batch_size=32)
y_train = (y_train >= 0.)
y_train = np.concatenate([1. - y_train, y_train], axis=1)
pred_train = (pred_train >= 0.)
pred_train = np.concatenate([1. - pred_train, pred_train], axis=1)
from keras.layers import Input, Dense, Multiply, Flatten, Reshape, Conv2D, MaxPooling2D, GlobalMaxPooling2D, Activation
from keras.layers import BatchNormalization
from keras.models import Sequential, Model
from keras.optimizers import Adam
from keras import regularizers
from keras import backend as K
import tensorflow as tf
import numpy as np
from keras.layers import Layer, InputSpec
from keras import initializers, regularizers, constraints
class InstanceNormalization(Layer):
def __init__(self, axes=(1, 2), trainable=True, **kwargs):
super(InstanceNormalization, self).__init__(**kwargs)
self.axes = axes
self.trainable = trainable
def build(self, input_shape):
self.beta = self.add_weight(name='beta',shape=(input_shape[-1],),
initializer='zeros',trainable=self.trainable)
self.gamma = self.add_weight(name='gamma',shape=(input_shape[-1],),
initializer='ones',trainable=self.trainable)
def call(self, inputs):
mean, variance = tf.nn.moments(inputs, self.axes, keep_dims=True)
return tf.nn.batch_normalization(inputs, mean, variance, self.beta, self.gamma, 1e-6)
def bernoulli_sampling (prob):
""" Sampling Bernoulli distribution by given probability.
Args:
- prob: P(Y = 1) in Bernoulli distribution.
Returns:
- samples: samples from Bernoulli distribution
"""
n, x_len, y_len, d = prob.shape
samples = np.random.binomial(1, prob, (n, x_len, y_len, d))
return samples
class INVASE():
"""INVASE class.
Attributes:
- x_train: training features
- y_train: training labels
- model_type: invase or invase_minus
- model_parameters:
- actor_h_dim: hidden state dimensions for actor
- critic_h_dim: hidden state dimensions for critic
- n_layer: the number of layers
- batch_size: the number of samples in mini batch
- iteration: the number of iterations
- activation: activation function of models
- learning_rate: learning rate of model training
- lamda: hyper-parameter of INVASE
"""
def __init__(self, x_train, y_train, model_type, model_parameters):
self.lamda = model_parameters['lamda']
self.actor_h_dim = model_parameters['actor_h_dim']
self.critic_h_dim = model_parameters['critic_h_dim']
self.n_layer = model_parameters['n_layer']
self.batch_size = model_parameters['batch_size']
self.iteration = model_parameters['iteration']
self.activation = model_parameters['activation']
self.learning_rate = model_parameters['learning_rate']
#Modified Code
self.x_len = x_train.shape[1]
self.y_len = x_train.shape[2]
self.dim = x_train.shape[3]
self.label_dim = y_train.shape[1]
self.model_type = model_type
optimizer = Adam(self.learning_rate)
# Build and compile critic
self.critic = self.build_critic()
self.critic.compile(loss='categorical_crossentropy',
optimizer=optimizer, metrics=['acc'])
# Build and compile the actor
self.actor = self.build_actor()
self.actor.compile(loss=self.actor_loss, optimizer=optimizer)
if self.model_type == 'invase':
# Build and compile the baseline
self.baseline = self.build_baseline()
self.baseline.compile(loss='categorical_crossentropy', optimizer=optimizer, metrics=['acc'])
def actor_loss(self, y_true, y_pred):
"""Custom loss for the actor.
Args:
- y_true:
- actor_out: actor output after sampling
- critic_out: critic output
- baseline_out: baseline output (only for invase)
- y_pred: output of the actor network
Returns:
- loss: actor loss
"""
y_pred = K.reshape(y_pred, (K.shape(y_pred)[0], self.x_len*self.y_len*1))
y_true = y_true[:, 0, 0, :]
# Actor output
actor_out = y_true[:, :self.x_len*self.y_len*1]
# Critic output
critic_out = y_true[:, self.x_len*self.y_len*1:(self.x_len*self.y_len*1+self.label_dim)]
if self.model_type == 'invase':
# Baseline output
baseline_out = \
y_true[:, (self.x_len*self.y_len*1+self.label_dim):(self.x_len*self.y_len*1+2*self.label_dim)]
# Ground truth label
y_out = y_true[:, (self.x_len*self.y_len*1+2*self.label_dim):]
elif self.model_type == 'invase_minus':
# Ground truth label
y_out = y_true[:, (self.x_len*self.y_len*1+self.label_dim):]
# Critic loss
critic_loss = -tf.reduce_sum(y_out * tf.log(critic_out + 1e-8), axis = 1)
if self.model_type == 'invase':
# Baseline loss
baseline_loss = -tf.reduce_sum(y_out * tf.log(baseline_out + 1e-8),
axis = 1)
# Reward
Reward = -(critic_loss - baseline_loss)
elif self.model_type == 'invase_minus':
Reward = -critic_loss
# Policy gradient loss computation.
custom_actor_loss = \
Reward * tf.reduce_sum(actor_out * K.log(y_pred + 1e-8) + \
(1-actor_out) * K.log(1-y_pred + 1e-8), axis = 1) - \
self.lamda * tf.reduce_mean(y_pred, axis = 1)
# custom actor loss
custom_actor_loss = tf.reduce_mean(-custom_actor_loss)
return custom_actor_loss
def build_actor(self):
"""Build actor.
Use feature as the input and output selection probability
"""
actor_model = Sequential()
actor_model.add(Conv2D(self.actor_h_dim, (1, 7), padding='same', activation='linear'))
actor_model.add(InstanceNormalization())
actor_model.add(Activation(self.activation))
for _ in range(self.n_layer - 2):
actor_model.add(Conv2D(self.actor_h_dim, (1, 7), padding='same', activation='linear'))
actor_model.add(InstanceNormalization())
actor_model.add(Activation(self.activation))
actor_model.add(Conv2D(1, (1, 1), padding='same', activation='sigmoid'))
feature = Input(shape=(self.x_len, self.y_len, self.dim), dtype='float32')
selection_probability = actor_model(feature)
return Model(feature, selection_probability)
def build_critic(self):
"""Build critic.
Use selected feature as the input and predict labels
"""
critic_model = Sequential()
critic_model.add(Conv2D(self.critic_h_dim, (1, 7), padding='same', activation='linear'))
critic_model.add(InstanceNormalization())
critic_model.add(Activation(self.activation))
for _ in range(self.n_layer - 2):
critic_model.add(Conv2D(self.critic_h_dim, (1, 7), padding='same', activation='linear'))
critic_model.add(InstanceNormalization())
critic_model.add(Activation(self.activation))
critic_model.add(Flatten())
critic_model.add(Dense(self.critic_h_dim, activation=self.activation))
critic_model.add(Dropout(0.2))
critic_model.add(Dense(self.label_dim, activation ='softmax'))
## Inputs
# Features
feature = Input(shape=(self.x_len, self.y_len, self.dim), dtype='float32')
# Binary selection
selection = Input(shape=(self.x_len, self.y_len, 1), dtype='float32')
# Element-wise multiplication
critic_model_input = Multiply()([feature, selection])
y_hat = critic_model(critic_model_input)
return Model([feature, selection], y_hat)
def build_baseline(self):
"""Build baseline.
Use the feature as the input and predict labels
"""
baseline_model = Sequential()
baseline_model.add(Conv2D(self.critic_h_dim, (1, 7), padding='same', activation='linear'))
baseline_model.add(InstanceNormalization())
baseline_model.add(Activation(self.activation))
for _ in range(self.n_layer - 2):
baseline_model.add(Conv2D(self.critic_h_dim, (1, 7), padding='same', activation='linear'))
baseline_model.add(InstanceNormalization())
baseline_model.add(Activation(self.activation))
baseline_model.add(Flatten())
baseline_model.add(Dense(self.critic_h_dim, activation=self.activation))
baseline_model.add(Dropout(0.2))
baseline_model.add(Dense(self.label_dim, activation ='softmax'))
# Input
feature = Input(shape=(self.x_len, self.y_len, self.dim), dtype='float32')
# Output
y_hat = baseline_model(feature)
return Model(feature, y_hat)
def train(self, x_train, y_train):
"""Train INVASE.
Args:
- x_train: training features
- y_train: training labels
"""
for iter_idx in range(self.iteration):
## Train critic
# Select a random batch of samples
idx = np.random.randint(0, x_train.shape[0], self.batch_size)
x_batch = x_train[idx,:]
y_batch = y_train[idx,:]
# Generate a batch of selection probability
selection_probability = self.actor.predict(x_batch)
# Sampling the features based on the selection_probability
selection = bernoulli_sampling(selection_probability)
# Critic loss
critic_loss = self.critic.train_on_batch([x_batch, selection], y_batch)
# Critic output
critic_out = self.critic.predict([x_batch, selection])
# Baseline output
if self.model_type == 'invase':
# Baseline loss
baseline_loss = self.baseline.train_on_batch(x_batch, y_batch)
# Baseline output
baseline_out = self.baseline.predict(x_batch)
## Train actor
# Use multiple things as the y_true:
# - selection, critic_out, baseline_out, and ground truth (y_batch)
if self.model_type == 'invase':
y_batch_final = np.concatenate((np.reshape(selection, (y_batch.shape[0], -1)),
np.asarray(critic_out),
np.asarray(baseline_out),
y_batch), axis = 1)
elif self.model_type == 'invase_minus':
y_batch_final = np.concatenate((np.reshape(selection, (y_batch.shape[0], -1)),
np.asarray(critic_out),
y_batch), axis = 1)
y_batch_final = y_batch_final[:, None, None, :]
# Train the actor
actor_loss = self.actor.train_on_batch(x_batch, y_batch_final)
if self.model_type == 'invase':
# Print the progress
dialog = 'Iterations: ' + str(iter_idx) + \
', critic accuracy: ' + str(critic_loss[1]) + \
', baseline accuracy: ' + str(baseline_loss[1]) + \
', actor loss: ' + str(np.round(actor_loss,4))
elif self.model_type == 'invase_minus':
# Print the progress
dialog = 'Iterations: ' + str(iter_idx) + \
', critic accuracy: ' + str(critic_loss[1]) + \
', actor loss: ' + str(np.round(actor_loss,4))
if iter_idx % 100 == 0:
print(dialog)
def importance_score(self, x):
"""Return featuer importance score.
Args:
- x: feature
Returns:
- feature_importance: instance-wise feature importance for x
"""
feature_importance = self.actor.predict(x)
return np.asarray(feature_importance)
def predict(self, x):
"""Predict outcomes.
Args:
- x: feature
Returns:
- y_hat: predictions
"""
# Generate a batch of selection probability
selection_probability = self.actor.predict(x)
# Sampling the features based on the selection_probability
selection = bernoulli_sampling(selection_probability)
# Prediction
y_hat = self.critic.predict([x, selection])
return np.asarray(y_hat)
#Gradient saliency/backprop visualization
import matplotlib.collections as collections
import operator
import matplotlib.pyplot as plt
import matplotlib.cm as cm
import matplotlib.colors as colors
import matplotlib as mpl
from matplotlib.text import TextPath
from matplotlib.patches import PathPatch, Rectangle
from matplotlib.font_manager import FontProperties
from matplotlib import gridspec
from matplotlib.ticker import FormatStrFormatter
def plot_importance_scores(importance_scores, ref_seq, figsize=(12, 2), score_clip=None, sequence_template='', plot_start=0, plot_end=96) :
end_pos = ref_seq.find("#")
fig = plt.figure(figsize=figsize)
ax = plt.gca()
if score_clip is not None :
importance_scores = np.clip(np.copy(importance_scores), -score_clip, score_clip)
max_score = np.max(np.sum(importance_scores[:, :], axis=0)) + 0.01
for i in range(0, len(ref_seq)) :
mutability_score = np.sum(importance_scores[:, i])
dna_letter_at(ref_seq[i], i + 0.5, 0, mutability_score, ax)
plt.sca(ax)
plt.xlim((0, len(ref_seq)))
plt.ylim((0, max_score))
plt.axis('off')
plt.yticks([0.0, max_score], [0.0, max_score], fontsize=16)
for axis in fig.axes :
axis.get_xaxis().set_visible(False)
axis.get_yaxis().set_visible(False)
plt.tight_layout()
plt.show()
#Execute INVASE benchmark on synthetic datasets
mask_penalty = 0.5#0.1
hidden_dims = 32
n_layers = 4
epochs = 25
batch_size = 128
model_parameters = {
'lamda': mask_penalty,
'actor_h_dim': hidden_dims,
'critic_h_dim': hidden_dims,
'n_layer': n_layers,
'batch_size': batch_size,
'iteration': int(x_train.shape[0] * epochs / batch_size),
'activation': 'relu',
'learning_rate': 0.0001
}
encoder = isol.OneHotEncoder(50)
score_clip = None
allFiles = ["optimus5_synthetic_random_insert_if_uorf_1_start_1_stop_variable_loc_512.csv",
"optimus5_synthetic_random_insert_if_uorf_1_start_2_stop_variable_loc_512.csv",
"optimus5_synthetic_random_insert_if_uorf_2_start_1_stop_variable_loc_512.csv",
"optimus5_synthetic_random_insert_if_uorf_2_start_2_stop_variable_loc_512.csv",
"optimus5_synthetic_examples_3.csv"]
for csv_to_open in allFiles :
#Load dataset for benchmarking
dataset_name = csv_to_open.replace(".csv", "")
benchmarkSet = pd.read_csv(csv_to_open)
seq_e_test = one_hot_encode(benchmarkSet, seq_len=50)
x_test = seq_e_test[:, None, ...]
print(x_test.shape)
pred_test = predictor.predict(x_test[:, 0, ...], batch_size=32)
y_test = pred_test
y_test = (y_test >= 0.)
y_test = np.concatenate([1. - y_test, y_test], axis=1)
pred_test = (pred_test >= 0.)
pred_test = np.concatenate([1. - pred_test, pred_test], axis=1)
invase_model = INVASE(x_train, pred_train, 'invase', model_parameters)
invase_model.train(x_train, pred_train)
importance_scores_test = invase_model.importance_score(x_test)
#Evaluate INVASE model on train and test data
invase_pred_train = invase_model.predict(x_train)
invase_pred_test = invase_model.predict(x_test)
print("Training Accuracy = " + str(np.sum(np.argmax(invase_pred_train, axis=1) == np.argmax(pred_train, axis=1)) / float(pred_train.shape[0])))
print("Test Accuracy = " + str(np.sum(np.argmax(invase_pred_test, axis=1) == np.argmax(pred_test, axis=1)) / float(pred_test.shape[0])))
for plot_i in range(0, 3) :
print("Test sequence " + str(plot_i) + ":")
plot_dna_logo(x_test[plot_i, 0, :, :], sequence_template='N'*50, plot_sequence_template=True, figsize=(12, 1), plot_start=0, plot_end=50)
plot_importance_scores(np.maximum(importance_scores_test[plot_i, 0, :, :].T, 0.), encoder.decode(x_test[plot_i, 0, :, :]), figsize=(12, 1), score_clip=score_clip, sequence_template='N'*50, plot_start=0, plot_end=50)
#Save predicted importance scores
model_name = "invase_" + dataset_name + "_conv"
np.save(model_name + "_importance_scores_test", importance_scores_test)
```
|
github_jupyter
|
import keras
import keras.backend as K
from keras.datasets import mnist
from keras.models import Sequential, Model, load_model
from keras.layers import Dense, Dropout, Activation, Flatten, Input, Lambda
from keras.layers import Conv2D, MaxPooling2D, AveragePooling2D, Conv1D, MaxPooling1D, LSTM, ConvLSTM2D, GRU, BatchNormalization, LocallyConnected2D, Permute, TimeDistributed, Bidirectional
from keras.layers import Concatenate, Reshape, Conv2DTranspose, Embedding, Multiply, Activation
from functools import partial
from collections import defaultdict
import os
import pickle
import numpy as np
import scipy.sparse as sp
import scipy.io as spio
import isolearn.io as isoio
import isolearn.keras as isol
import matplotlib.pyplot as plt
from sequence_logo_helper import dna_letter_at, plot_dna_logo
from sklearn import preprocessing
import pandas as pd
import tensorflow as tf
from keras.backend.tensorflow_backend import set_session
def contain_tf_gpu_mem_usage() :
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
sess = tf.Session(config=config)
set_session(sess)
contain_tf_gpu_mem_usage()
#optimus 5-prime functions
def test_data(df, model, test_seq, obs_col, output_col='pred'):
'''Predict mean ribosome load using model and test set UTRs'''
# Scale the test set mean ribosome load
scaler = preprocessing.StandardScaler()
scaler.fit(df[obs_col].reshape(-1,1))
# Make predictions
predictions = model.predict(test_seq).reshape(-1)
# Inverse scaled predicted mean ribosome load and return in a column labeled 'pred'
df.loc[:,output_col] = scaler.inverse_transform(predictions)
return df
def one_hot_encode(df, col='utr', seq_len=50):
# Dictionary returning one-hot encoding of nucleotides.
nuc_d = {'a':[1,0,0,0],'c':[0,1,0,0],'g':[0,0,1,0],'t':[0,0,0,1], 'n':[0,0,0,0]}
# Creat empty matrix.
vectors=np.empty([len(df),seq_len,4])
# Iterate through UTRs and one-hot encode
for i,seq in enumerate(df[col].str[:seq_len]):
seq = seq.lower()
a = np.array([nuc_d[x] for x in seq])
vectors[i] = a
return vectors
def r2(x,y):
slope, intercept, r_value, p_value, std_err = stats.linregress(x,y)
return r_value**2
#Train data
e_train = pd.read_csv("bottom5KIFuAUGTop5KIFuAUG.csv")
e_train.loc[:,'scaled_rl'] = preprocessing.StandardScaler().fit_transform(e_train.loc[:,'rl'].values.reshape(-1,1))
seq_e_train = one_hot_encode(e_train,seq_len=50)
x_train = seq_e_train
x_train = np.reshape(x_train, (x_train.shape[0], 1, x_train.shape[1], x_train.shape[2]))
y_train = np.array(e_train['scaled_rl'].values)
y_train = np.reshape(y_train, (y_train.shape[0],1))
print("x_train.shape = " + str(x_train.shape))
print("y_train.shape = " + str(y_train.shape))
#Load Predictor
predictor_path = 'optimusRetrainedMain.hdf5'
predictor = load_model(predictor_path)
predictor.trainable = False
predictor.compile(optimizer=keras.optimizers.SGD(lr=0.1), loss='mean_squared_error')
#Generate (original) predictions
pred_train = predictor.predict(x_train[:, 0, ...], batch_size=32)
y_train = (y_train >= 0.)
y_train = np.concatenate([1. - y_train, y_train], axis=1)
pred_train = (pred_train >= 0.)
pred_train = np.concatenate([1. - pred_train, pred_train], axis=1)
from keras.layers import Input, Dense, Multiply, Flatten, Reshape, Conv2D, MaxPooling2D, GlobalMaxPooling2D, Activation
from keras.layers import BatchNormalization
from keras.models import Sequential, Model
from keras.optimizers import Adam
from keras import regularizers
from keras import backend as K
import tensorflow as tf
import numpy as np
from keras.layers import Layer, InputSpec
from keras import initializers, regularizers, constraints
class InstanceNormalization(Layer):
def __init__(self, axes=(1, 2), trainable=True, **kwargs):
super(InstanceNormalization, self).__init__(**kwargs)
self.axes = axes
self.trainable = trainable
def build(self, input_shape):
self.beta = self.add_weight(name='beta',shape=(input_shape[-1],),
initializer='zeros',trainable=self.trainable)
self.gamma = self.add_weight(name='gamma',shape=(input_shape[-1],),
initializer='ones',trainable=self.trainable)
def call(self, inputs):
mean, variance = tf.nn.moments(inputs, self.axes, keep_dims=True)
return tf.nn.batch_normalization(inputs, mean, variance, self.beta, self.gamma, 1e-6)
def bernoulli_sampling (prob):
""" Sampling Bernoulli distribution by given probability.
Args:
- prob: P(Y = 1) in Bernoulli distribution.
Returns:
- samples: samples from Bernoulli distribution
"""
n, x_len, y_len, d = prob.shape
samples = np.random.binomial(1, prob, (n, x_len, y_len, d))
return samples
class INVASE():
"""INVASE class.
Attributes:
- x_train: training features
- y_train: training labels
- model_type: invase or invase_minus
- model_parameters:
- actor_h_dim: hidden state dimensions for actor
- critic_h_dim: hidden state dimensions for critic
- n_layer: the number of layers
- batch_size: the number of samples in mini batch
- iteration: the number of iterations
- activation: activation function of models
- learning_rate: learning rate of model training
- lamda: hyper-parameter of INVASE
"""
def __init__(self, x_train, y_train, model_type, model_parameters):
self.lamda = model_parameters['lamda']
self.actor_h_dim = model_parameters['actor_h_dim']
self.critic_h_dim = model_parameters['critic_h_dim']
self.n_layer = model_parameters['n_layer']
self.batch_size = model_parameters['batch_size']
self.iteration = model_parameters['iteration']
self.activation = model_parameters['activation']
self.learning_rate = model_parameters['learning_rate']
#Modified Code
self.x_len = x_train.shape[1]
self.y_len = x_train.shape[2]
self.dim = x_train.shape[3]
self.label_dim = y_train.shape[1]
self.model_type = model_type
optimizer = Adam(self.learning_rate)
# Build and compile critic
self.critic = self.build_critic()
self.critic.compile(loss='categorical_crossentropy',
optimizer=optimizer, metrics=['acc'])
# Build and compile the actor
self.actor = self.build_actor()
self.actor.compile(loss=self.actor_loss, optimizer=optimizer)
if self.model_type == 'invase':
# Build and compile the baseline
self.baseline = self.build_baseline()
self.baseline.compile(loss='categorical_crossentropy', optimizer=optimizer, metrics=['acc'])
def actor_loss(self, y_true, y_pred):
"""Custom loss for the actor.
Args:
- y_true:
- actor_out: actor output after sampling
- critic_out: critic output
- baseline_out: baseline output (only for invase)
- y_pred: output of the actor network
Returns:
- loss: actor loss
"""
y_pred = K.reshape(y_pred, (K.shape(y_pred)[0], self.x_len*self.y_len*1))
y_true = y_true[:, 0, 0, :]
# Actor output
actor_out = y_true[:, :self.x_len*self.y_len*1]
# Critic output
critic_out = y_true[:, self.x_len*self.y_len*1:(self.x_len*self.y_len*1+self.label_dim)]
if self.model_type == 'invase':
# Baseline output
baseline_out = \
y_true[:, (self.x_len*self.y_len*1+self.label_dim):(self.x_len*self.y_len*1+2*self.label_dim)]
# Ground truth label
y_out = y_true[:, (self.x_len*self.y_len*1+2*self.label_dim):]
elif self.model_type == 'invase_minus':
# Ground truth label
y_out = y_true[:, (self.x_len*self.y_len*1+self.label_dim):]
# Critic loss
critic_loss = -tf.reduce_sum(y_out * tf.log(critic_out + 1e-8), axis = 1)
if self.model_type == 'invase':
# Baseline loss
baseline_loss = -tf.reduce_sum(y_out * tf.log(baseline_out + 1e-8),
axis = 1)
# Reward
Reward = -(critic_loss - baseline_loss)
elif self.model_type == 'invase_minus':
Reward = -critic_loss
# Policy gradient loss computation.
custom_actor_loss = \
Reward * tf.reduce_sum(actor_out * K.log(y_pred + 1e-8) + \
(1-actor_out) * K.log(1-y_pred + 1e-8), axis = 1) - \
self.lamda * tf.reduce_mean(y_pred, axis = 1)
# custom actor loss
custom_actor_loss = tf.reduce_mean(-custom_actor_loss)
return custom_actor_loss
def build_actor(self):
"""Build actor.
Use feature as the input and output selection probability
"""
actor_model = Sequential()
actor_model.add(Conv2D(self.actor_h_dim, (1, 7), padding='same', activation='linear'))
actor_model.add(InstanceNormalization())
actor_model.add(Activation(self.activation))
for _ in range(self.n_layer - 2):
actor_model.add(Conv2D(self.actor_h_dim, (1, 7), padding='same', activation='linear'))
actor_model.add(InstanceNormalization())
actor_model.add(Activation(self.activation))
actor_model.add(Conv2D(1, (1, 1), padding='same', activation='sigmoid'))
feature = Input(shape=(self.x_len, self.y_len, self.dim), dtype='float32')
selection_probability = actor_model(feature)
return Model(feature, selection_probability)
def build_critic(self):
"""Build critic.
Use selected feature as the input and predict labels
"""
critic_model = Sequential()
critic_model.add(Conv2D(self.critic_h_dim, (1, 7), padding='same', activation='linear'))
critic_model.add(InstanceNormalization())
critic_model.add(Activation(self.activation))
for _ in range(self.n_layer - 2):
critic_model.add(Conv2D(self.critic_h_dim, (1, 7), padding='same', activation='linear'))
critic_model.add(InstanceNormalization())
critic_model.add(Activation(self.activation))
critic_model.add(Flatten())
critic_model.add(Dense(self.critic_h_dim, activation=self.activation))
critic_model.add(Dropout(0.2))
critic_model.add(Dense(self.label_dim, activation ='softmax'))
## Inputs
# Features
feature = Input(shape=(self.x_len, self.y_len, self.dim), dtype='float32')
# Binary selection
selection = Input(shape=(self.x_len, self.y_len, 1), dtype='float32')
# Element-wise multiplication
critic_model_input = Multiply()([feature, selection])
y_hat = critic_model(critic_model_input)
return Model([feature, selection], y_hat)
def build_baseline(self):
"""Build baseline.
Use the feature as the input and predict labels
"""
baseline_model = Sequential()
baseline_model.add(Conv2D(self.critic_h_dim, (1, 7), padding='same', activation='linear'))
baseline_model.add(InstanceNormalization())
baseline_model.add(Activation(self.activation))
for _ in range(self.n_layer - 2):
baseline_model.add(Conv2D(self.critic_h_dim, (1, 7), padding='same', activation='linear'))
baseline_model.add(InstanceNormalization())
baseline_model.add(Activation(self.activation))
baseline_model.add(Flatten())
baseline_model.add(Dense(self.critic_h_dim, activation=self.activation))
baseline_model.add(Dropout(0.2))
baseline_model.add(Dense(self.label_dim, activation ='softmax'))
# Input
feature = Input(shape=(self.x_len, self.y_len, self.dim), dtype='float32')
# Output
y_hat = baseline_model(feature)
return Model(feature, y_hat)
def train(self, x_train, y_train):
"""Train INVASE.
Args:
- x_train: training features
- y_train: training labels
"""
for iter_idx in range(self.iteration):
## Train critic
# Select a random batch of samples
idx = np.random.randint(0, x_train.shape[0], self.batch_size)
x_batch = x_train[idx,:]
y_batch = y_train[idx,:]
# Generate a batch of selection probability
selection_probability = self.actor.predict(x_batch)
# Sampling the features based on the selection_probability
selection = bernoulli_sampling(selection_probability)
# Critic loss
critic_loss = self.critic.train_on_batch([x_batch, selection], y_batch)
# Critic output
critic_out = self.critic.predict([x_batch, selection])
# Baseline output
if self.model_type == 'invase':
# Baseline loss
baseline_loss = self.baseline.train_on_batch(x_batch, y_batch)
# Baseline output
baseline_out = self.baseline.predict(x_batch)
## Train actor
# Use multiple things as the y_true:
# - selection, critic_out, baseline_out, and ground truth (y_batch)
if self.model_type == 'invase':
y_batch_final = np.concatenate((np.reshape(selection, (y_batch.shape[0], -1)),
np.asarray(critic_out),
np.asarray(baseline_out),
y_batch), axis = 1)
elif self.model_type == 'invase_minus':
y_batch_final = np.concatenate((np.reshape(selection, (y_batch.shape[0], -1)),
np.asarray(critic_out),
y_batch), axis = 1)
y_batch_final = y_batch_final[:, None, None, :]
# Train the actor
actor_loss = self.actor.train_on_batch(x_batch, y_batch_final)
if self.model_type == 'invase':
# Print the progress
dialog = 'Iterations: ' + str(iter_idx) + \
', critic accuracy: ' + str(critic_loss[1]) + \
', baseline accuracy: ' + str(baseline_loss[1]) + \
', actor loss: ' + str(np.round(actor_loss,4))
elif self.model_type == 'invase_minus':
# Print the progress
dialog = 'Iterations: ' + str(iter_idx) + \
', critic accuracy: ' + str(critic_loss[1]) + \
', actor loss: ' + str(np.round(actor_loss,4))
if iter_idx % 100 == 0:
print(dialog)
def importance_score(self, x):
"""Return featuer importance score.
Args:
- x: feature
Returns:
- feature_importance: instance-wise feature importance for x
"""
feature_importance = self.actor.predict(x)
return np.asarray(feature_importance)
def predict(self, x):
"""Predict outcomes.
Args:
- x: feature
Returns:
- y_hat: predictions
"""
# Generate a batch of selection probability
selection_probability = self.actor.predict(x)
# Sampling the features based on the selection_probability
selection = bernoulli_sampling(selection_probability)
# Prediction
y_hat = self.critic.predict([x, selection])
return np.asarray(y_hat)
#Gradient saliency/backprop visualization
import matplotlib.collections as collections
import operator
import matplotlib.pyplot as plt
import matplotlib.cm as cm
import matplotlib.colors as colors
import matplotlib as mpl
from matplotlib.text import TextPath
from matplotlib.patches import PathPatch, Rectangle
from matplotlib.font_manager import FontProperties
from matplotlib import gridspec
from matplotlib.ticker import FormatStrFormatter
def plot_importance_scores(importance_scores, ref_seq, figsize=(12, 2), score_clip=None, sequence_template='', plot_start=0, plot_end=96) :
end_pos = ref_seq.find("#")
fig = plt.figure(figsize=figsize)
ax = plt.gca()
if score_clip is not None :
importance_scores = np.clip(np.copy(importance_scores), -score_clip, score_clip)
max_score = np.max(np.sum(importance_scores[:, :], axis=0)) + 0.01
for i in range(0, len(ref_seq)) :
mutability_score = np.sum(importance_scores[:, i])
dna_letter_at(ref_seq[i], i + 0.5, 0, mutability_score, ax)
plt.sca(ax)
plt.xlim((0, len(ref_seq)))
plt.ylim((0, max_score))
plt.axis('off')
plt.yticks([0.0, max_score], [0.0, max_score], fontsize=16)
for axis in fig.axes :
axis.get_xaxis().set_visible(False)
axis.get_yaxis().set_visible(False)
plt.tight_layout()
plt.show()
#Execute INVASE benchmark on synthetic datasets
mask_penalty = 0.5#0.1
hidden_dims = 32
n_layers = 4
epochs = 25
batch_size = 128
model_parameters = {
'lamda': mask_penalty,
'actor_h_dim': hidden_dims,
'critic_h_dim': hidden_dims,
'n_layer': n_layers,
'batch_size': batch_size,
'iteration': int(x_train.shape[0] * epochs / batch_size),
'activation': 'relu',
'learning_rate': 0.0001
}
encoder = isol.OneHotEncoder(50)
score_clip = None
allFiles = ["optimus5_synthetic_random_insert_if_uorf_1_start_1_stop_variable_loc_512.csv",
"optimus5_synthetic_random_insert_if_uorf_1_start_2_stop_variable_loc_512.csv",
"optimus5_synthetic_random_insert_if_uorf_2_start_1_stop_variable_loc_512.csv",
"optimus5_synthetic_random_insert_if_uorf_2_start_2_stop_variable_loc_512.csv",
"optimus5_synthetic_examples_3.csv"]
for csv_to_open in allFiles :
#Load dataset for benchmarking
dataset_name = csv_to_open.replace(".csv", "")
benchmarkSet = pd.read_csv(csv_to_open)
seq_e_test = one_hot_encode(benchmarkSet, seq_len=50)
x_test = seq_e_test[:, None, ...]
print(x_test.shape)
pred_test = predictor.predict(x_test[:, 0, ...], batch_size=32)
y_test = pred_test
y_test = (y_test >= 0.)
y_test = np.concatenate([1. - y_test, y_test], axis=1)
pred_test = (pred_test >= 0.)
pred_test = np.concatenate([1. - pred_test, pred_test], axis=1)
invase_model = INVASE(x_train, pred_train, 'invase', model_parameters)
invase_model.train(x_train, pred_train)
importance_scores_test = invase_model.importance_score(x_test)
#Evaluate INVASE model on train and test data
invase_pred_train = invase_model.predict(x_train)
invase_pred_test = invase_model.predict(x_test)
print("Training Accuracy = " + str(np.sum(np.argmax(invase_pred_train, axis=1) == np.argmax(pred_train, axis=1)) / float(pred_train.shape[0])))
print("Test Accuracy = " + str(np.sum(np.argmax(invase_pred_test, axis=1) == np.argmax(pred_test, axis=1)) / float(pred_test.shape[0])))
for plot_i in range(0, 3) :
print("Test sequence " + str(plot_i) + ":")
plot_dna_logo(x_test[plot_i, 0, :, :], sequence_template='N'*50, plot_sequence_template=True, figsize=(12, 1), plot_start=0, plot_end=50)
plot_importance_scores(np.maximum(importance_scores_test[plot_i, 0, :, :].T, 0.), encoder.decode(x_test[plot_i, 0, :, :]), figsize=(12, 1), score_clip=score_clip, sequence_template='N'*50, plot_start=0, plot_end=50)
#Save predicted importance scores
model_name = "invase_" + dataset_name + "_conv"
np.save(model_name + "_importance_scores_test", importance_scores_test)
| 0.890512 | 0.44089 |
# 序列构成的数组
## Python的内置序列类型
Python内置了相当丰富的序列类型
若按照存储的方式区分,可以分为容器序列和扁平序列。**值得注意的是这里所谓的容器序列和扁平序列实际上是作者自创的名称**。
其中容器序列实际上描述的是允许存放其他对象的序列,例如list中可以存放list,即一个对象中包含其他对象的引用。相对的,扁平序列实际上指的是不能够存放其他对象,仅能存放数值、字符的序列类型。
* 容器序列
* list
* tuple
* collections.deque
* 扁平序列
* str
* bytes
* memoryview
* array.array
在Python中容器序列存放的是其存储内容的引用,这意味着Python中的容器序列能够存放不同类型的数据;而Python中的扁平序列则存放的是值,通常仅能存放字符、字节以及数值等基础类型的数据。
若按照能否被修改进行区分,则可以分为可变序列以及不可变序列
* 可变序列
* list
* bytearray
* array.array
* collections.deque
* memoryview
* 不可变序列
* tuple
* str
* bytes
容器(Container)、迭代(iterable)以及空间大小(Sized)构成了序列的基石。不可变序列在此基础上添加各类查询以及统计方法(\_\_getitem\_\_等)。可变序列则在不可变序列的基础上添加各类删改以及排序方法(insert, append, reverse等)
## 列表推导和生成器表达式
### 列表推导
列表推导是一种创建列表的快捷方式,生成器表达式则可以用来创建其他任何类型的序列。
列表推导的语法很简单:
```Python
[statements for i in iter]
```
列表推导主要是以相当简单的形式创建了一个列表。当然,利用其他函数,例如filter、map以及zip之类的方法也能很快的创建。从可读性来说,直接使用列表推导创建列表是比较简单且明了的方式。
列表推导中可以有复数个iter,例如
```Python
colors = ["black", "white"]
sizes = ["S", "M", "L"]
tshirts = [(color, size) for color in colors for size in sizes]
```
当然,列表推导可以有大于等于2个iter,但是此时可能写成普通的for循环形式更易于阅读
### 生成器表达式
列表推导仅可以用于生成列表。若想要生成其他类型的序列,一种选择是首先生成列表,然后转换为其他序列类型;其次还可以直接生成对应类型的序列,即利用生成器表达式。
直接生成对应类型的序列最为明显的好处是:避免多余的内存开销。若将生成器表达式作为iter用于循环中,则会每次生成对应的数据,而不是先生成整个序列存放到内存中然后从对应位置读取数据。对于大规模数据来说,这样显然可以有效避免不必要的内存开销。
```
# -------------- 列表推导测试 --------------
colors = ["black", "white"]
sizes = ["S", "M", "L"]
tshirts = [(color, size) for color in colors for size in sizes]
print(tshirts)
tshirts_2 = [(color, size) for size in sizes for color in colors]
print(tshirts_2)
```
## 元组
元组最大的特征是其不可变性。不可变性实际上暗含多种含义,其一是元组记录的值不会改变,**其二是元组记录的值的顺序不会改变**。第二条相当重要,但是常被忽略。有些时候单纯的数值没有意义,只有结合值与该值在序列中的位置才会有明确的意义(例如经纬度记录,抑或是按照一定顺序采集到的数据)。
### 元组拆包(Unpacking)
元组能够很方便的进行拆包以提取特定位置的数据,不需要的数据则可以使用_占位符以确保拆包正确。
拆包可以以多种方式进行:
* 平行赋值:对于一个可迭代对象,使用相同数量的变量接收其中的元素
* \* 运算符拆包:\* 可以将一个可迭代对象拆包并作为传入函数的参数
* \* 运算符处理剩余元素:\* 除了可以拆包可迭代对象外,还可以用于接收不确定数量的拆包结果(非常神奇的功能,类似于*args。对于同一个赋值表达式的左式最多仅能使用一个\*,并且带有\*运算符的变量总会自动接受合适数量的拆包结果)
#### 嵌套拆包
嵌套元组也可以进行拆包,只需要使用足够的变量接收拆包结果即可
### 具名元组
有些情况下,我们希望元组能带有一个可以解释各位置数据的含义的字段。collections.namedtuple即可实现这一功能(collections.namedtuple实际上创建了一个类)。使用collections.namedtuple创建的类的实例可以使用对应的字段名或者索引来获取对应的值。此外,还有如下常用功能:
* ._fields:返回包括所有字段名称的元组
* ._make():接受一个可迭代对象以生成一个实例
* ._asdict():将具名元组以collections.OrderedDict形式返回 —— [(key1, value1), (key2, value2), ...]
```
# --------------- 平行赋值 ---------------
lax_coordinates = (33.3, 44.4)
latitude, longitude = lax_coordinates
print(latitude, longitude)
# --------------- *运算符拆包 ---------------
def add_func(x1, x2):
return x1 + x2
input_data = (1, 2)
output_value = add_func(*input_data)
print(output_value)
# --------------- *运算符处理剩余元素 ---------------
a, b, *c = range(5)
print(a, b, c)
a, *b, c = range(5)
print(a, b, c)
*a, b, c = range(5)
print(a, b, c)
# --------------- 嵌套元组拆包 ---------------
name, cc, pop, (latitude, longitude) = ("Tokyo", "JP", 36.33, (35.7, 139.7))
print(name, cc, pop, latitude, longitude)
# --------------- 具名元组 ---------------
from collections import namedtuple
City = namedtuple("City", "name country population coordinates")
# tokyo = City("Tokyo", "JP", 36.33, (35.7, 139.7))
tokyo = City._make(("Tokyo", "JP", 36.33, (35.7, 139.7)))
print(tokyo._fields)
print(tokyo._asdict())
```
## 切片
切片是操作序列类型数据的重要操作
### Python中进行切片会忽略最后一个元素的原因
* 能够快速识别切片区间内的元素数量(若包含最后一个元素,计数的时候需要注意在头尾索引相减的基础上+1)
* 能够快速定位切片的索引值(主要感觉还是因为在这些语言中0是计数的起始下标)
* 更够很轻松的将一个序列拆分为不重叠的两个子序列(若首尾都包含,在进行拆分时需要额外考虑切片位置的索引)
### 具有名称标识的切片操作
```Python
slice(startIndex, endIndex, step=1)
```
这个用法很实用,主要是能够对切片操作进行单独的定义。方便对不同的序列使用相同的切片操作进行切片。同样的,slice对象也会忽略最后一个元素
### 多维切片和省略
多维切片在处理实际数据时经常用到,例如处理图像数据或者更高维的数据。Python内置的序列类型均是一维的,**因此内置的序列类型仅支持一维的**。
总的来说,若想实现多维切片,需要实现\_\_getitem__和\_\_setitem__。前者用于读取数据,后者用于赋值。
省略符号(...)则被用于省略无需额外指定的参数。例如,对于一个四维数组,若仅对第一维和最后一维进行切片,在Numpy可以写为:
```Python
test_list[i, ..., j]
test_list[i:j, ..., k:z]
```
### 赋值
切片同样可以用于删改数据。值得注意的是,对于内置序列类型,**若赋值的对象是一个切片,则赋值语句的右侧也必须是一个可迭代对象**;但是在Numpy等库中,则可以直接用单个数值对切片进行赋值,这一操作主要依赖Numpy等库中的broadcast机制;基于broadcast机制,一些shape没有完全对应上的情况在Numpy中也可以进行赋值和运算
```
# --------------- 简单切片 ---------------
test_list = [i for i in range(10)]
print("\n 切片:")
print(test_list[:3], test_list[3:])
# --------------- slice对象切片 ---------------
slice_obj = slice(1, 3)
print("\n slice对象切片:")
print(test_list[slice_obj])
# --------------- ...符号使用 ---------------
import numpy as np
test_array = np.zeros((4, 4, 4, 4))
slice_list = test_array[1:3, ..., :1]
print("\n...省略:")
print("原数组shape:{}\n新数组shape:{}".format(test_array.shape, slice_list.shape))
# --------------- 赋值 ---------------
test_array = np.zeros((4,))
test_array2 = np.zeros((4,4))
print("\nNumpy Array赋值:")
print(test_array)
print(test_array + 1)
print(test_array2[:1, :] + np.ones((4)))
test_list = [1, 2, 3, 4]
print("\n内置list类型使用非可迭代对象赋值(报错):")
test_list += 1
```
## + & *
Python内置的序列支持+和*
注意:**可变序列以及不可变序列均支持这些操作**,因为这些操作本质上并不是修改原序列,而是创建一个新的序列。原位操作(*=,+=)对于可变序列以及不可变序列也均是可行的,可变序列进行原位操作后会直接修改原来的序列,**不可变序列进行原位操作虽然不会报错,但是实际上是创建了一个新序列**。
不同于内置的序列,Numpy由于主要是运算,因此+和*在Numpy中会被视为算数运算符,此时是对原Array中值的修改。
内置序列使用\*或者+进行操作时,要尤其注意是对"值"进行的操作还是对"引用"进行的操作。对于内置序列a,若a中的元素是其他可变对象的引用,进行\*或者+操作实际上是对引用进行的操作,**这会导致严重的问题**。
### 一个有趣的极限情况
```Python
test_tuple = (1, 2, [3, 4])
test_tuple[2] += [5, 6]
```
上述操作按理来说应当是直接报错,因为元组中的元素不可赋值。但是实际情况是报错的同时,元组包含的列表被修改了。
对上述代码的字节码进行分析可以发现,顺序执行了下述三个操作
1. 读取test_tuple[2]并将其存入栈顶,记为TOS
2. 完成操作TOS += [5, 6]
3. 将结果存入原位置:test_tuple[2] = TOS
其中,第二步是对list进行的操作,由于list是可变序列因此该操作不会报错;第三步是对tuple进行的操作,由于tuple是不可变序列,当尝试赋值时会抛出错误。但是tuple中存放的是list的引用,因此此时tuple中的list实际上已经被修改。
上述结果反映:
1. 将可变对象放置于不可变对象中是十分危险的操作
2. 上述操作不是原子操作,因此发生了上述既完成了操作又抛出了错误的结果。
```
# ---------------- 原位操作 ----------------
print("可变序列——list进行原位操作:")
test_list = [1, 2]
print("list原位操作前地址: ", id(test_list))
test_list += [1, 2]
print("list原位操作后地址: ",id(test_list))
print("\n不可变序列——tuple进行原位操作:")
test_tuple = (1, 2)
print("tuple原位操作前地址: ", id(test_tuple))
test_tuple += (1, 2)
print("tuple原位操作后地址: ",id(test_tuple))
# ---------------- 引用和值 -------------------
print("\n* 和 +处理多维序列:")
print("\n正确操作,此时多维序列中的每一个元素相互独立,互不影响")
board = [["_"] * 3 for i in range(3)]
print("赋值前")
print(board)
board[1][2] = "X"
print("赋值后")
print(board)
print("\n错误操作,此时多维序列中同一个指向同一个位置的引用被复制了多次")
board = [["_"] * 3] * 3
print("赋值前")
print(board)
board[1][2] = "X"
print("赋值后")
print(board)
# ---------------- +=的陷阱 -------------------
print("\n+=的陷阱:")
test_tuple = (1, 2, [30, 40])
print("对元组包含的list进行操作前:")
print(test_tuple)
try:
test_tuple[2] += [50, 60]
except TypeError:
print("抛出TypeError,对元组包含的list进行操作后")
print(test_tuple)
```
## 排序
Python内置了两种排序方式。对于可变序列list,Python允许使用list.sort()对list进行排序,此外还可以使用sorted()函数进行排序。值得注意的是,list.sort()的返回值是None,即这个方法总是在原list上进行,而sorted()则会创建一个新的对象作为返回值。
这两个内置方法均可以通过reverse参数控制排序方式(升序或降序),也可以通过key参数控制排序依赖的对比关键字
### 有序序列的元素查找以及插入
bisect模块提供了对有序序列进行元素查找以及插入的方法。其中bisect.bisect可以用于元素插入位置查找:寻找一个位置,使得插入待插入元素后,有序序列仍是有序的。bisect.insort函数则可以将元素插入有序序列中。
bisect()和insort()均有两种形式,分别被命名为_right,_left。若遇到相等的元素,_right会将元素插入到序列中相同值的元素的后面,而_left则会插入到前面。
```
fruits = ["grape", "raspberry", "apple", "banana"]
def sort_key(_str):
count = 0
for s in _str:
if s == "p":
count += 1
return count
print("\nsorted()排序:")
print(sorted(fruits))
print("\nsorted()降序:")
print(sorted(fruits, reverse=True))
print("\n自定义排序方法,依照p字母的数量:")
print(sorted(fruits, key=sort_key, reverse=True))
print("\nlist.sort()排序:")
print(fruits.sort())
print(fruits)
# -------------- 有序序列的元素查找以及插入 ---------------
import bisect
haystack = [1, 4, 5, 6, 8, 12, 15, 20, 21, 23, 23, 26, 29, 30]
needles = [0, 1, 2, 5, 8, 10, 22, 29, 30, 31]
row_fmt = "{0:2d} @ {1:2d} {2}{0:<2d}"
print("\n有序序列元素插入位置查找")
print("haystack ->", " ".join("%2d" % n for n in haystack))
for needle in reversed(needles):
position = bisect.bisect(haystack, needle)
offset = position * " |"
print(row_fmt.format(needle, position, offset))
```
## 数组
数组专为处理仅包含同类数值类型数据的场合设计。Python自带的数组基本上支持所有和可变序列相关的操作,此外还提供快速存读的方法(array.frombytes 和 array.tofile)
不同于Numpy中定义的array,Python内置的数组类型不能处理多维数组。
Python array的创建方式很简单,除了传入数据外,还需要指定存储类型 —— 这和Numpy array的初始化类似。
```Python
test_array = array(type, data)
```
### 内存视图(memoryview)
实际上提供了一种在不需要赋值内容的前提下,实现不同数据结构之间的内存共享。即指定一块区域,能够使用不同的方式去存读数据,例如可以以list的形式创建一个序列,然后以Numpy array的形式去处理这个序列,而不需要额外再创建一个包含相同内容的新array。
## 双向队列
collections.deque类可以很方便的创建双向队列,用于快速进行队列两端添加或者删除元素的操作。其基本操作如下,还是很简单的,有需要的时候再详细阅读。
```
from collections import deque
dq = deque(range(10), maxlen=10)
print(dq)
# ------------------ 旋转元素 ------------------0
print("\n将后3个数移动到队列头部:")
dq.rotate(3)
print(dq)
print("\n将前4个数移动到队列尾部:")
dq.rotate(-4)
print(dq)
print("\n从头部添加元素:")
dq.appendleft(-1)
print(dq)
print("\n从尾部添加元素:")
dq.append(-1)
print(dq)
print("\n从尾部逐项添加元素:")
dq.extend([10, 20, 30, 40])
print(dq)
print("\n从头部逐项添加元素:")
dq.extendleft([10, 20, 30, 40])
print(dq)
```
## 总结
1. 不同任务需求应当使用不同的序列类型进行数据存储。仅能存放数值或是字符的序列类型,例如str、array.array能够以更快的速度执行操作并且在相同规模数据的前提下能够减少内存开销。能存放其他对象的引用的序列类型,例如list、tuple等则更灵活,但是灵活有时会带来一些意想不到的问题,同时,这些序列类型执行操作的速度更慢,内存开销也更大。
2. 列表推导和生成器表达式提供了灵活而易读的初始化序列方法
3. 元组作为不可变序列,其存储的值以及对应的索引均是重要的数据。拆包则可以安全且可靠的从元组中提取数据,同时*运算符的引入能够更好的处理不需要的拆包结果。具名元组namedtuple的引入则在记录数据的同时也记录了对应的名称,这在处理某些问题时是必要的(若不使用具名元组则可能使用dict之类的方法去存储名称和索引之间的关系)。值得注意的是,Python中的元组不仅仅是一种元素不可变的记录工具,其同样是一种序列类型和可迭代类型,也能够利用索引进行元素的提取和切片 —— **牺牲了概念上的纯粹性,以换取实际使用中的灵活性**
4. 切片是序列中的重要操作,slice()方法则可以为切片操作建立相应的对象,以方便对不同的数据进行相同的切片操作。此外,Python原生的序列类型仅支持一维序列,而Numpy等库的引入则可以方便处理多维序列,这在实际应用中显然是非常关键的
5. 排序是非常重要的操作,Python提供了sorted()函数以及某些序列类型自带.sort()方法以执行排序。key这一参数的设置则可以很方便的设置排序依照的规则。
|
github_jupyter
|
[statements for i in iter]
colors = ["black", "white"]
sizes = ["S", "M", "L"]
tshirts = [(color, size) for color in colors for size in sizes]
# -------------- 列表推导测试 --------------
colors = ["black", "white"]
sizes = ["S", "M", "L"]
tshirts = [(color, size) for color in colors for size in sizes]
print(tshirts)
tshirts_2 = [(color, size) for size in sizes for color in colors]
print(tshirts_2)
# --------------- 平行赋值 ---------------
lax_coordinates = (33.3, 44.4)
latitude, longitude = lax_coordinates
print(latitude, longitude)
# --------------- *运算符拆包 ---------------
def add_func(x1, x2):
return x1 + x2
input_data = (1, 2)
output_value = add_func(*input_data)
print(output_value)
# --------------- *运算符处理剩余元素 ---------------
a, b, *c = range(5)
print(a, b, c)
a, *b, c = range(5)
print(a, b, c)
*a, b, c = range(5)
print(a, b, c)
# --------------- 嵌套元组拆包 ---------------
name, cc, pop, (latitude, longitude) = ("Tokyo", "JP", 36.33, (35.7, 139.7))
print(name, cc, pop, latitude, longitude)
# --------------- 具名元组 ---------------
from collections import namedtuple
City = namedtuple("City", "name country population coordinates")
# tokyo = City("Tokyo", "JP", 36.33, (35.7, 139.7))
tokyo = City._make(("Tokyo", "JP", 36.33, (35.7, 139.7)))
print(tokyo._fields)
print(tokyo._asdict())
slice(startIndex, endIndex, step=1)
test_list[i, ..., j]
test_list[i:j, ..., k:z]
# --------------- 简单切片 ---------------
test_list = [i for i in range(10)]
print("\n 切片:")
print(test_list[:3], test_list[3:])
# --------------- slice对象切片 ---------------
slice_obj = slice(1, 3)
print("\n slice对象切片:")
print(test_list[slice_obj])
# --------------- ...符号使用 ---------------
import numpy as np
test_array = np.zeros((4, 4, 4, 4))
slice_list = test_array[1:3, ..., :1]
print("\n...省略:")
print("原数组shape:{}\n新数组shape:{}".format(test_array.shape, slice_list.shape))
# --------------- 赋值 ---------------
test_array = np.zeros((4,))
test_array2 = np.zeros((4,4))
print("\nNumpy Array赋值:")
print(test_array)
print(test_array + 1)
print(test_array2[:1, :] + np.ones((4)))
test_list = [1, 2, 3, 4]
print("\n内置list类型使用非可迭代对象赋值(报错):")
test_list += 1
test_tuple = (1, 2, [3, 4])
test_tuple[2] += [5, 6]
# ---------------- 原位操作 ----------------
print("可变序列——list进行原位操作:")
test_list = [1, 2]
print("list原位操作前地址: ", id(test_list))
test_list += [1, 2]
print("list原位操作后地址: ",id(test_list))
print("\n不可变序列——tuple进行原位操作:")
test_tuple = (1, 2)
print("tuple原位操作前地址: ", id(test_tuple))
test_tuple += (1, 2)
print("tuple原位操作后地址: ",id(test_tuple))
# ---------------- 引用和值 -------------------
print("\n* 和 +处理多维序列:")
print("\n正确操作,此时多维序列中的每一个元素相互独立,互不影响")
board = [["_"] * 3 for i in range(3)]
print("赋值前")
print(board)
board[1][2] = "X"
print("赋值后")
print(board)
print("\n错误操作,此时多维序列中同一个指向同一个位置的引用被复制了多次")
board = [["_"] * 3] * 3
print("赋值前")
print(board)
board[1][2] = "X"
print("赋值后")
print(board)
# ---------------- +=的陷阱 -------------------
print("\n+=的陷阱:")
test_tuple = (1, 2, [30, 40])
print("对元组包含的list进行操作前:")
print(test_tuple)
try:
test_tuple[2] += [50, 60]
except TypeError:
print("抛出TypeError,对元组包含的list进行操作后")
print(test_tuple)
fruits = ["grape", "raspberry", "apple", "banana"]
def sort_key(_str):
count = 0
for s in _str:
if s == "p":
count += 1
return count
print("\nsorted()排序:")
print(sorted(fruits))
print("\nsorted()降序:")
print(sorted(fruits, reverse=True))
print("\n自定义排序方法,依照p字母的数量:")
print(sorted(fruits, key=sort_key, reverse=True))
print("\nlist.sort()排序:")
print(fruits.sort())
print(fruits)
# -------------- 有序序列的元素查找以及插入 ---------------
import bisect
haystack = [1, 4, 5, 6, 8, 12, 15, 20, 21, 23, 23, 26, 29, 30]
needles = [0, 1, 2, 5, 8, 10, 22, 29, 30, 31]
row_fmt = "{0:2d} @ {1:2d} {2}{0:<2d}"
print("\n有序序列元素插入位置查找")
print("haystack ->", " ".join("%2d" % n for n in haystack))
for needle in reversed(needles):
position = bisect.bisect(haystack, needle)
offset = position * " |"
print(row_fmt.format(needle, position, offset))
test_array = array(type, data)
from collections import deque
dq = deque(range(10), maxlen=10)
print(dq)
# ------------------ 旋转元素 ------------------0
print("\n将后3个数移动到队列头部:")
dq.rotate(3)
print(dq)
print("\n将前4个数移动到队列尾部:")
dq.rotate(-4)
print(dq)
print("\n从头部添加元素:")
dq.appendleft(-1)
print(dq)
print("\n从尾部添加元素:")
dq.append(-1)
print(dq)
print("\n从尾部逐项添加元素:")
dq.extend([10, 20, 30, 40])
print(dq)
print("\n从头部逐项添加元素:")
dq.extendleft([10, 20, 30, 40])
print(dq)
| 0.269133 | 0.918004 |
```
%load_ext autoreload
%autoreload 2
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import os, math
import numpy as np, pandas as pd
import matplotlib.pyplot as plt, seaborn as sns
from tqdm import tqdm, tqdm_notebook
from pathlib import Path
pd.set_option('display.max_columns', 1000)
pd.set_option('display.max_rows', 400)
sns.set()
os.chdir('../..')
from src import utils
DATA = Path('data')
RAW = DATA/'raw'
INTERIM = DATA/'interim'
PROCESSED = DATA/'processed'
SUBMISSIONS = DATA/'submissions'
from src.utils import get_weeks
# week_labels = get_weeks(day_from=20160104, num_weeks=121)[52:]
week_labels = get_weeks(day_from=20160104, num_weeks=121)[96:]
# week_labels = get_weeks(day_from=20160104, num_weeks=121)[104:]
print(week_labels)
%%time
weeks = pd.DataFrame()
for name in week_labels[:-1]:
weeks = pd.concat([weeks, pd.read_feather(
PROCESSED/f'SVD_17-18_72f/week_{name}_SVD_diffscount.feather')])
test = pd.read_feather(
PROCESSED/f'SVD_17-18_72f/week_{week_labels[-1]}_SVD_diffscount.feather')
```
## Preprocessing
```
cat_cols = ['BuySell', 'Sector', 'Subsector', 'Region_x', 'Country',
'TickerIdx', 'Seniority', 'Currency', 'ActivityGroup',
'Region_y', 'Activity', 'RiskCaptain', 'Owner',
'IndustrySector', 'IndustrySubgroup', 'MarketIssue', 'CouponType']
id_cols = ['TradeDateKey', 'CustomerIdx', 'IsinIdx']
target_col = 'CustomerInterest'
pred_col = 'PredictionIdx'
%%time
from src.utils import apply_cats
for col in cat_cols:
test[col] = test[col].astype('category').cat.as_ordered()
apply_cats(weeks, test)
for col in cat_cols:
weeks[col] = weeks[col].cat.codes
test[col] = test[col].cat.codes
```
## Model
```
from lightgbm import LGBMClassifier
from sklearn.metrics import roc_auc_score
from src.utils import alert
%%time
val_set = []
train_auc = []
val_auc = []
for i in range(-5, -1):
train, val = weeks[weeks.TradeDateKey<week_labels[i]], \
weeks[(weeks.TradeDateKey<week_labels[i+1]) & \
(weeks.TradeDateKey>=week_labels[i])]
print(train.TradeDateKey.min(), train.TradeDateKey.max(),
val.TradeDateKey.unique())
val_set.append(val.TradeDateKey.unique()[0])
y_train = train[target_col]
train.drop(id_cols + [target_col], axis=1, inplace=True)
y_val = val[target_col]
val.drop(id_cols + [target_col], axis=1, inplace=True)
model = LGBMClassifier(n_estimators=400, max_depth=30,
random_state=42, reg_alpha=1, reg_lambda=1,
colsample_by_tree=0.8)
model.fit(train, y_train, eval_metric='auc', verbose=20,
eval_set=[(val, y_val)], early_stopping_rounds=30)
y_pred = model.predict_proba(train)[:,1]
train_auc.append(roc_auc_score(y_train, y_pred))
print('Train AUC: ', train_auc[-1])
y_pred = model.predict_proba(val)[:,1]
val_auc.append(roc_auc_score(y_val, y_pred))
print('Val AUC: ', val_auc[-1])
print()
del model, train, y_train, val, y_val, y_pred
alert()
results = pd.DataFrame()
results['val_set'] = val_set
results['train_auc'] = train_auc
results['val_auc'] = val_auc
results['iterations'] = [397,261,328,400]
results = pd.DataFrame()
results['val_set'] = [20180326, 20180402, 20180409, 20180416]
results['train_auc'] = [0.7980312197668828, 0.8062227442047145, 0.8161902543359266, 0.8038221068804319]
results['val_auc'] = [0.8273520913252462, 0.824267377403749, 0.8516418834039483, 0.8693741344750291]
results['iterations'] = [163, 231, 376, 137]
print(results.train_auc.mean(), results.val_auc.mean(),
results.iterations.mean())
results
# (n_estimators=400, max_depth=30, random_state=42,
# reg_alpha=1, reg_lambda=1, colsample_by_tree=0.8)
print(results.train_auc.mean(), results.val_auc.mean(),
results.iterations.mean())
results
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import os, math
import numpy as np, pandas as pd
import matplotlib.pyplot as plt, seaborn as sns
from tqdm import tqdm, tqdm_notebook
from pathlib import Path
pd.set_option('display.max_columns', 1000)
pd.set_option('display.max_rows', 400)
sns.set()
os.chdir('../..')
from src import utils
DATA = Path('data')
RAW = DATA/'raw'
INTERIM = DATA/'interim'
PROCESSED = DATA/'processed'
SUBMISSIONS = DATA/'submissions'
from src.utils import get_weeks
# week_labels = get_weeks(day_from=20160104, num_weeks=121)[52:]
week_labels = get_weeks(day_from=20160104, num_weeks=121)[96:]
# week_labels = get_weeks(day_from=20160104, num_weeks=121)[104:]
print(week_labels)
%%time
weeks = pd.DataFrame()
for name in week_labels[:-1]:
weeks = pd.concat([weeks, pd.read_feather(
PROCESSED/f'SVD_17-18_72f/week_{name}_SVD_diffscount.feather')])
test = pd.read_feather(
PROCESSED/f'SVD_17-18_72f/week_{week_labels[-1]}_SVD_diffscount.feather')
cat_cols = ['BuySell', 'Sector', 'Subsector', 'Region_x', 'Country',
'TickerIdx', 'Seniority', 'Currency', 'ActivityGroup',
'Region_y', 'Activity', 'RiskCaptain', 'Owner',
'IndustrySector', 'IndustrySubgroup', 'MarketIssue', 'CouponType']
id_cols = ['TradeDateKey', 'CustomerIdx', 'IsinIdx']
target_col = 'CustomerInterest'
pred_col = 'PredictionIdx'
%%time
from src.utils import apply_cats
for col in cat_cols:
test[col] = test[col].astype('category').cat.as_ordered()
apply_cats(weeks, test)
for col in cat_cols:
weeks[col] = weeks[col].cat.codes
test[col] = test[col].cat.codes
from lightgbm import LGBMClassifier
from sklearn.metrics import roc_auc_score
from src.utils import alert
%%time
val_set = []
train_auc = []
val_auc = []
for i in range(-5, -1):
train, val = weeks[weeks.TradeDateKey<week_labels[i]], \
weeks[(weeks.TradeDateKey<week_labels[i+1]) & \
(weeks.TradeDateKey>=week_labels[i])]
print(train.TradeDateKey.min(), train.TradeDateKey.max(),
val.TradeDateKey.unique())
val_set.append(val.TradeDateKey.unique()[0])
y_train = train[target_col]
train.drop(id_cols + [target_col], axis=1, inplace=True)
y_val = val[target_col]
val.drop(id_cols + [target_col], axis=1, inplace=True)
model = LGBMClassifier(n_estimators=400, max_depth=30,
random_state=42, reg_alpha=1, reg_lambda=1,
colsample_by_tree=0.8)
model.fit(train, y_train, eval_metric='auc', verbose=20,
eval_set=[(val, y_val)], early_stopping_rounds=30)
y_pred = model.predict_proba(train)[:,1]
train_auc.append(roc_auc_score(y_train, y_pred))
print('Train AUC: ', train_auc[-1])
y_pred = model.predict_proba(val)[:,1]
val_auc.append(roc_auc_score(y_val, y_pred))
print('Val AUC: ', val_auc[-1])
print()
del model, train, y_train, val, y_val, y_pred
alert()
results = pd.DataFrame()
results['val_set'] = val_set
results['train_auc'] = train_auc
results['val_auc'] = val_auc
results['iterations'] = [397,261,328,400]
results = pd.DataFrame()
results['val_set'] = [20180326, 20180402, 20180409, 20180416]
results['train_auc'] = [0.7980312197668828, 0.8062227442047145, 0.8161902543359266, 0.8038221068804319]
results['val_auc'] = [0.8273520913252462, 0.824267377403749, 0.8516418834039483, 0.8693741344750291]
results['iterations'] = [163, 231, 376, 137]
print(results.train_auc.mean(), results.val_auc.mean(),
results.iterations.mean())
results
# (n_estimators=400, max_depth=30, random_state=42,
# reg_alpha=1, reg_lambda=1, colsample_by_tree=0.8)
print(results.train_auc.mean(), results.val_auc.mean(),
results.iterations.mean())
results
| 0.327238 | 0.377627 |
<a href="https://colab.research.google.com/github/African-Quant/FOREX_RelativeStrengthOscillator/blob/main/Oanda_RelativeStrength_GA.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
#@title Installation
!pip install git+https://github.com/yhilpisch/tpqoa.git --upgrade --quiet
!pip install pykalman --quiet
!pip install --upgrade mplfinance --quiet
#@title Imports
import tpqoa
import numpy as np
import pandas as pd
from pykalman import KalmanFilter
%matplotlib inline
from pylab import mpl, plt
plt.style.use('seaborn')
mpl.rcParams['savefig.dpi'] = 300
mpl.rcParams['font.family'] = 'serif'
from datetime import date, timedelta
import warnings
warnings.filterwarnings("ignore")
#@title Oanda API
path = '/content/drive/MyDrive/Oanda_Algo/pyalgo.cfg'
api = tpqoa.tpqoa(path)
#@title Symbols/Currency Pairs
def symbolsList():
symbols = []
syms = api.get_instruments()
for x in syms:
symbols.append(x[1])
return symbols
symbols = symbolsList()
pairs = ['AUD_CAD', 'AUD_CHF', 'AUD_JPY', 'AUD_NZD', 'AUD_USD', 'CAD_CHF',
'CAD_JPY', 'CHF_JPY', 'EUR_AUD', 'EUR_CAD', 'EUR_CHF', 'EUR_GBP',
'EUR_JPY', 'EUR_NZD', 'EUR_USD', 'GBP_AUD', 'GBP_CAD', 'GBP_CHF',
'GBP_JPY', 'GBP_NZD', 'GBP_USD', 'NZD_CAD', 'NZD_CHF', 'NZD_JPY',
'NZD_USD', 'USD_CAD', 'USD_CHF', 'USD_JPY',]
#@title getData(instr, gran = 'D', td=1000)
def getData(instr, gran = 'D', td=1000):
start = f"{date.today() - timedelta(td)}"
end = f"{date.today() - timedelta(1)}"
granularity = gran
price = 'M' # price: string one of 'A' (ask), 'B' (bid) or 'M' (middle)
data = api.get_history(instr, start, end, granularity, price)
data.drop(['complete'], axis=1, inplace=True)
data.reset_index(inplace=True)
data.rename(columns = {'time':'Date','o':'Open','c': 'Close', 'h':'High', 'l': 'Low'}, inplace = True)
data.set_index('Date', inplace=True)
return data
#@title Indexes
def USD_Index():
'''Creating a USD Index from a basket of instruments
denominated in dollars
'''
USD = ['EUR_USD', 'GBP_USD', 'USD_CAD', 'USD_CHF',
'USD_JPY', 'AUD_USD', 'NZD_USD']
df = pd.DataFrame()
for i in USD:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_USD' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['US_index'] = 1
for i in range(len(USD)):
df['US_index'] *= df[USD[i]]
return ((df['US_index'])**(1/(len(USD)))).to_frame()
def EURO_Index():
'''Creating a EUR Index from a basket of instruments
denominated in EUROs
'''
EUR = ['EUR_USD', 'EUR_GBP', 'EUR_JPY', 'EUR_CHF', 'EUR_CAD',
'EUR_AUD', 'EUR_NZD']
df = pd.DataFrame()
for i in EUR:
data = getData(i).ffill(axis='rows')
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['EUR_index'] = 1
for i in range(len(EUR)):
df['EUR_index'] *= df[EUR[i]]
return ((df['EUR_index'])**(1/(len(EUR)))).to_frame()
def GBP_Index():
'''Creating a GBP Index from a basket of instruments
denominated in Pound Sterling
'''
GBP = ['GBP_USD', 'EUR_GBP', 'GBP_JPY', 'GBP_CHF', 'GBP_CAD',
'GBP_AUD', 'GBP_NZD']
df = pd.DataFrame()
for i in GBP:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_GBP' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['GBP_index'] = 1
for i in range(len(GBP)):
df['GBP_index'] *= df[GBP[i]]
return ((df['GBP_index'])**(1/(len(GBP)))).to_frame()
def CHF_Index():
'''Creating a CHF Index from a basket of instruments
denominated in Swiss Francs
'''
CHF = ['CHF_JPY', 'EUR_CHF', 'GBP_CHF', 'USD_CHF', 'CAD_CHF',
'AUD_CHF', 'NZD_CHF']
df = pd.DataFrame()
for i in CHF:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_CHF' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['CHF_index'] = 1
for i in range(len(CHF)):
df['CHF_index'] *= df[CHF[i]]
return ((df['CHF_index'])**(1/(len(CHF)))).to_frame()
def CAD_Index():
'''Creating a CAD Index from a basket of instruments
denominated in Canadian Dollars
'''
CAD = ['CAD_JPY', 'EUR_CAD', 'GBP_CAD', 'USD_CAD', 'CAD_CHF',
'AUD_CAD', 'NZD_CAD']
df = pd.DataFrame()
for i in CAD:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_CAD' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['CAD_index'] = 1
for i in range(len(CAD)):
df['CAD_index'] *= df[CAD[i]]
return ((df['CAD_index'])**(1/(len(CAD)))).to_frame()
def JPY_Index():
'''Creating a JPY Index from a basket of instruments
denominated in Swiss Francs
'''
JPY = ['CAD_JPY', 'EUR_JPY', 'GBP_JPY', 'USD_JPY', 'CHF_JPY',
'AUD_JPY', 'NZD_JPY']
df = pd.DataFrame()
for i in JPY:
data = getData(i).ffill(axis='rows')
# setting the Japanese Yen as the base
data[f'{i}'] = (data['Close'])**(-1)
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['JPY_index'] = 1
for i in range(len(JPY)):
df['JPY_index'] *= df[JPY[i]]
return ((df['JPY_index'])**(1/(len(JPY)))).to_frame()
def AUD_Index():
'''Creating a AUD Index from a basket of instruments
denominated in Australian Dollar
'''
AUD = ['AUD_JPY', 'EUR_AUD', 'GBP_AUD', 'AUD_USD', 'AUD_CAD',
'AUD_CHF', 'AUD_NZD']
df = pd.DataFrame()
for i in AUD:
data = getData(i).ffill(axis='rows')
# setting the Aussie Dollar as the base
if '_AUD' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['AUD_index'] = 1
for i in range(len(AUD)):
df['AUD_index'] *= df[AUD[i]]
return ((df['AUD_index'])**(1/(len(AUD)))).to_frame()
def NZD_Index():
'''Creating a NZD Index from a basket of instruments
denominated in New Zealand Dollar
'''
NZD = ['NZD_JPY', 'EUR_NZD', 'GBP_NZD', 'NZD_USD', 'NZD_CAD',
'NZD_CHF', 'AUD_NZD']
df = pd.DataFrame()
for i in NZD:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_NZD' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['NZD_index'] = 1
for i in range(len(NZD)):
df['NZD_index'] *= df[ NZD[i]]
return ((df['NZD_index'])**(1/(len(NZD)))).to_frame()
def eSuperRCS(df):
"""
This code computes the super smoother introduced by John Ehlers
"""
spr = df.to_frame().copy()
# HighPass filter cyclic components whose periods are shorter than 48 bars
alpha1 = (np.cos(0.707*2*np.pi/48) + np.sin(0.707*2*np.pi/48) - 1)/np.cos(0.707*2*np.pi/48)
hiPass = pd.DataFrame(None, index=spr.index, columns=['filtered'])
for i in range(len(spr)):
if i < 3:
hiPass.iloc[i, 0] = spr.iat[i, 0]
else:
hiPass.iloc[i, 0] = ((1 - alpha1/2)*(1 - alpha1/2)*(spr.iat[i, 0]
- 2*spr.iat[i-1, 0] + spr.iat[i-2, 0] + 2*(1 - alpha1)*hiPass.iat[i-1, 0]
- (1 - alpha1)**2 *hiPass.iat[i-2, 0]))
# SuperSmoother
a1 = np.exp(-1.414*(np.pi) / 10)
b1 = 2*a1*np.cos(1.414*(np.pi) / 10)
c2 = b1
c3 = -a1*a1
c1 = 1 - c2 - c3
Filt = pd.DataFrame(None, index=spr.index, columns=['filtered'])
for i in range(len(spr)):
if i < 3:
Filt.iloc[i, 0] = hiPass.iat[i, 0]
else:
Filt.iloc[i, 0] = c1*(hiPass.iat[i, 0] + hiPass.iat[i - 1, 0]/ 2 + c2*Filt.iat[i-1, 0] + c3*Filt.iat[i-2, 0])
Filt['eSuperRCS'] = RSI(Filt['filtered'])
return Filt['eSuperRCS']
def RSI(series, period=25):
delta = series.diff()
up = delta.clip(lower=0)
dn = -1*delta.clip(upper=0)
ema_up = up.ewm(com=period-1, adjust=False).mean()
ewm_dn = dn.ewm(com=period-1, adjust=False).mean()
rs = (ema_up/ewm_dn)
return 100 - 100 / (1 + rs)
def will_pr(data, lb=14):
df = data[['High', 'Low', 'Close']].copy()
df['max_hi'] = data['High'].rolling(window=lb).max()
df['min_lo'] = data['Low'].rolling(window=lb).min()
df['will_pr'] = 0
for i in range(len(df)):
try:
df.iloc[i, 5] = ((df.iat[i, 3] - df.iat[i, 2])/(df.iat[i, 3] - df.iat[i, 4])) * (-100)
except ValueError:
pass
return df['will_pr']
ga = getData('GBP_AUD')
gbp = GBP_Index()
aud = AUD_Index()
df = pd.concat((ga, gbp, aud), axis=1).ffill(axis='rows')
df
tickers = ['GBP_index', 'AUD_index']
cumm_rtn = (1 + df[tickers].pct_change()).cumprod()
cumm_rtn.plot();
plt.ylabel('Cumulative Return');
plt.xlabel('Time');
plt.title('Cummulative Plot of GBP_index & AUD_index');
import statsmodels.api as sm
obs_mat = sm.add_constant(df[tickers[0]].values, prepend=False)[:, np.newaxis]
# y is 1-dimensional, (alpha, beta) is 2-dimensional
kf = KalmanFilter(n_dim_obs=1, n_dim_state=2,
initial_state_mean=np.ones(2),
initial_state_covariance=np.ones((2, 2)),
transition_matrices=np.eye(2),
observation_matrices=obs_mat,
observation_covariance=10**2,
transition_covariance=0.01**2 * np.eye(2))
state_means, state_covs = kf.filter(df[tickers[1]])
beta_kf = pd.DataFrame({'Slope': state_means[:, 0], 'Intercept': state_means[:, 1]},
index=df.index)
spread_kf = df[tickers[0]] - df[tickers[1]] * beta_kf['Slope'] - beta_kf['Intercept']
spread_kf = spread_kf
spread_kf.plot();
len(df)
df['spread'] = spread_kf
df['GBP/AUD'] = df['GBP_index']/df['AUD_index']
df['eSuperRCS'] = eSuperRCS(df['spread'])
df = df.iloc[-700:]
fig = plt.figure(figsize=(10, 7))
ax1, ax2 = fig.subplots(nrows=2, ncols=1)
ax1.plot(df.index, df['Close'],color='cyan' )
ax2.plot(df.index, df['GBP/AUD'].values, color='maroon')
ax1.set_title('GBP_AUD')
ax2.set_title('GBP/AUD')
plt.show()
def viewPlot(data, win = 150):
fig = plt.figure(figsize=(17, 10))
ax1, ax2 = fig.subplots(nrows=2, ncols=1)
df1 = data.iloc[-win:, ]
# High and Low prices are plotted
for i in range(len(df1)):
ax1.vlines(x = df1.index[i], ymin = df1.iat[i, 2], ymax = df1.iat[i, 1], color = 'magenta', linewidth = 2)
ax2.plot(df1.index, df1['eSuperRCS'].values, color='maroon')
ax2.axhline(55, color='green')
ax2.axhline(45, color='green')
ax2.axhline(50, color='orange')
ax1.set_title('GBP_AUD')
ax2.set_title('spread oscillator')
return plt.show()
viewPlot(df, win = 150)
```
|
github_jupyter
|
#@title Installation
!pip install git+https://github.com/yhilpisch/tpqoa.git --upgrade --quiet
!pip install pykalman --quiet
!pip install --upgrade mplfinance --quiet
#@title Imports
import tpqoa
import numpy as np
import pandas as pd
from pykalman import KalmanFilter
%matplotlib inline
from pylab import mpl, plt
plt.style.use('seaborn')
mpl.rcParams['savefig.dpi'] = 300
mpl.rcParams['font.family'] = 'serif'
from datetime import date, timedelta
import warnings
warnings.filterwarnings("ignore")
#@title Oanda API
path = '/content/drive/MyDrive/Oanda_Algo/pyalgo.cfg'
api = tpqoa.tpqoa(path)
#@title Symbols/Currency Pairs
def symbolsList():
symbols = []
syms = api.get_instruments()
for x in syms:
symbols.append(x[1])
return symbols
symbols = symbolsList()
pairs = ['AUD_CAD', 'AUD_CHF', 'AUD_JPY', 'AUD_NZD', 'AUD_USD', 'CAD_CHF',
'CAD_JPY', 'CHF_JPY', 'EUR_AUD', 'EUR_CAD', 'EUR_CHF', 'EUR_GBP',
'EUR_JPY', 'EUR_NZD', 'EUR_USD', 'GBP_AUD', 'GBP_CAD', 'GBP_CHF',
'GBP_JPY', 'GBP_NZD', 'GBP_USD', 'NZD_CAD', 'NZD_CHF', 'NZD_JPY',
'NZD_USD', 'USD_CAD', 'USD_CHF', 'USD_JPY',]
#@title getData(instr, gran = 'D', td=1000)
def getData(instr, gran = 'D', td=1000):
start = f"{date.today() - timedelta(td)}"
end = f"{date.today() - timedelta(1)}"
granularity = gran
price = 'M' # price: string one of 'A' (ask), 'B' (bid) or 'M' (middle)
data = api.get_history(instr, start, end, granularity, price)
data.drop(['complete'], axis=1, inplace=True)
data.reset_index(inplace=True)
data.rename(columns = {'time':'Date','o':'Open','c': 'Close', 'h':'High', 'l': 'Low'}, inplace = True)
data.set_index('Date', inplace=True)
return data
#@title Indexes
def USD_Index():
'''Creating a USD Index from a basket of instruments
denominated in dollars
'''
USD = ['EUR_USD', 'GBP_USD', 'USD_CAD', 'USD_CHF',
'USD_JPY', 'AUD_USD', 'NZD_USD']
df = pd.DataFrame()
for i in USD:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_USD' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['US_index'] = 1
for i in range(len(USD)):
df['US_index'] *= df[USD[i]]
return ((df['US_index'])**(1/(len(USD)))).to_frame()
def EURO_Index():
'''Creating a EUR Index from a basket of instruments
denominated in EUROs
'''
EUR = ['EUR_USD', 'EUR_GBP', 'EUR_JPY', 'EUR_CHF', 'EUR_CAD',
'EUR_AUD', 'EUR_NZD']
df = pd.DataFrame()
for i in EUR:
data = getData(i).ffill(axis='rows')
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['EUR_index'] = 1
for i in range(len(EUR)):
df['EUR_index'] *= df[EUR[i]]
return ((df['EUR_index'])**(1/(len(EUR)))).to_frame()
def GBP_Index():
'''Creating a GBP Index from a basket of instruments
denominated in Pound Sterling
'''
GBP = ['GBP_USD', 'EUR_GBP', 'GBP_JPY', 'GBP_CHF', 'GBP_CAD',
'GBP_AUD', 'GBP_NZD']
df = pd.DataFrame()
for i in GBP:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_GBP' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['GBP_index'] = 1
for i in range(len(GBP)):
df['GBP_index'] *= df[GBP[i]]
return ((df['GBP_index'])**(1/(len(GBP)))).to_frame()
def CHF_Index():
'''Creating a CHF Index from a basket of instruments
denominated in Swiss Francs
'''
CHF = ['CHF_JPY', 'EUR_CHF', 'GBP_CHF', 'USD_CHF', 'CAD_CHF',
'AUD_CHF', 'NZD_CHF']
df = pd.DataFrame()
for i in CHF:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_CHF' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['CHF_index'] = 1
for i in range(len(CHF)):
df['CHF_index'] *= df[CHF[i]]
return ((df['CHF_index'])**(1/(len(CHF)))).to_frame()
def CAD_Index():
'''Creating a CAD Index from a basket of instruments
denominated in Canadian Dollars
'''
CAD = ['CAD_JPY', 'EUR_CAD', 'GBP_CAD', 'USD_CAD', 'CAD_CHF',
'AUD_CAD', 'NZD_CAD']
df = pd.DataFrame()
for i in CAD:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_CAD' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['CAD_index'] = 1
for i in range(len(CAD)):
df['CAD_index'] *= df[CAD[i]]
return ((df['CAD_index'])**(1/(len(CAD)))).to_frame()
def JPY_Index():
'''Creating a JPY Index from a basket of instruments
denominated in Swiss Francs
'''
JPY = ['CAD_JPY', 'EUR_JPY', 'GBP_JPY', 'USD_JPY', 'CHF_JPY',
'AUD_JPY', 'NZD_JPY']
df = pd.DataFrame()
for i in JPY:
data = getData(i).ffill(axis='rows')
# setting the Japanese Yen as the base
data[f'{i}'] = (data['Close'])**(-1)
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['JPY_index'] = 1
for i in range(len(JPY)):
df['JPY_index'] *= df[JPY[i]]
return ((df['JPY_index'])**(1/(len(JPY)))).to_frame()
def AUD_Index():
'''Creating a AUD Index from a basket of instruments
denominated in Australian Dollar
'''
AUD = ['AUD_JPY', 'EUR_AUD', 'GBP_AUD', 'AUD_USD', 'AUD_CAD',
'AUD_CHF', 'AUD_NZD']
df = pd.DataFrame()
for i in AUD:
data = getData(i).ffill(axis='rows')
# setting the Aussie Dollar as the base
if '_AUD' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['AUD_index'] = 1
for i in range(len(AUD)):
df['AUD_index'] *= df[AUD[i]]
return ((df['AUD_index'])**(1/(len(AUD)))).to_frame()
def NZD_Index():
'''Creating a NZD Index from a basket of instruments
denominated in New Zealand Dollar
'''
NZD = ['NZD_JPY', 'EUR_NZD', 'GBP_NZD', 'NZD_USD', 'NZD_CAD',
'NZD_CHF', 'AUD_NZD']
df = pd.DataFrame()
for i in NZD:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_NZD' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['NZD_index'] = 1
for i in range(len(NZD)):
df['NZD_index'] *= df[ NZD[i]]
return ((df['NZD_index'])**(1/(len(NZD)))).to_frame()
def eSuperRCS(df):
"""
This code computes the super smoother introduced by John Ehlers
"""
spr = df.to_frame().copy()
# HighPass filter cyclic components whose periods are shorter than 48 bars
alpha1 = (np.cos(0.707*2*np.pi/48) + np.sin(0.707*2*np.pi/48) - 1)/np.cos(0.707*2*np.pi/48)
hiPass = pd.DataFrame(None, index=spr.index, columns=['filtered'])
for i in range(len(spr)):
if i < 3:
hiPass.iloc[i, 0] = spr.iat[i, 0]
else:
hiPass.iloc[i, 0] = ((1 - alpha1/2)*(1 - alpha1/2)*(spr.iat[i, 0]
- 2*spr.iat[i-1, 0] + spr.iat[i-2, 0] + 2*(1 - alpha1)*hiPass.iat[i-1, 0]
- (1 - alpha1)**2 *hiPass.iat[i-2, 0]))
# SuperSmoother
a1 = np.exp(-1.414*(np.pi) / 10)
b1 = 2*a1*np.cos(1.414*(np.pi) / 10)
c2 = b1
c3 = -a1*a1
c1 = 1 - c2 - c3
Filt = pd.DataFrame(None, index=spr.index, columns=['filtered'])
for i in range(len(spr)):
if i < 3:
Filt.iloc[i, 0] = hiPass.iat[i, 0]
else:
Filt.iloc[i, 0] = c1*(hiPass.iat[i, 0] + hiPass.iat[i - 1, 0]/ 2 + c2*Filt.iat[i-1, 0] + c3*Filt.iat[i-2, 0])
Filt['eSuperRCS'] = RSI(Filt['filtered'])
return Filt['eSuperRCS']
def RSI(series, period=25):
delta = series.diff()
up = delta.clip(lower=0)
dn = -1*delta.clip(upper=0)
ema_up = up.ewm(com=period-1, adjust=False).mean()
ewm_dn = dn.ewm(com=period-1, adjust=False).mean()
rs = (ema_up/ewm_dn)
return 100 - 100 / (1 + rs)
def will_pr(data, lb=14):
df = data[['High', 'Low', 'Close']].copy()
df['max_hi'] = data['High'].rolling(window=lb).max()
df['min_lo'] = data['Low'].rolling(window=lb).min()
df['will_pr'] = 0
for i in range(len(df)):
try:
df.iloc[i, 5] = ((df.iat[i, 3] - df.iat[i, 2])/(df.iat[i, 3] - df.iat[i, 4])) * (-100)
except ValueError:
pass
return df['will_pr']
ga = getData('GBP_AUD')
gbp = GBP_Index()
aud = AUD_Index()
df = pd.concat((ga, gbp, aud), axis=1).ffill(axis='rows')
df
tickers = ['GBP_index', 'AUD_index']
cumm_rtn = (1 + df[tickers].pct_change()).cumprod()
cumm_rtn.plot();
plt.ylabel('Cumulative Return');
plt.xlabel('Time');
plt.title('Cummulative Plot of GBP_index & AUD_index');
import statsmodels.api as sm
obs_mat = sm.add_constant(df[tickers[0]].values, prepend=False)[:, np.newaxis]
# y is 1-dimensional, (alpha, beta) is 2-dimensional
kf = KalmanFilter(n_dim_obs=1, n_dim_state=2,
initial_state_mean=np.ones(2),
initial_state_covariance=np.ones((2, 2)),
transition_matrices=np.eye(2),
observation_matrices=obs_mat,
observation_covariance=10**2,
transition_covariance=0.01**2 * np.eye(2))
state_means, state_covs = kf.filter(df[tickers[1]])
beta_kf = pd.DataFrame({'Slope': state_means[:, 0], 'Intercept': state_means[:, 1]},
index=df.index)
spread_kf = df[tickers[0]] - df[tickers[1]] * beta_kf['Slope'] - beta_kf['Intercept']
spread_kf = spread_kf
spread_kf.plot();
len(df)
df['spread'] = spread_kf
df['GBP/AUD'] = df['GBP_index']/df['AUD_index']
df['eSuperRCS'] = eSuperRCS(df['spread'])
df = df.iloc[-700:]
fig = plt.figure(figsize=(10, 7))
ax1, ax2 = fig.subplots(nrows=2, ncols=1)
ax1.plot(df.index, df['Close'],color='cyan' )
ax2.plot(df.index, df['GBP/AUD'].values, color='maroon')
ax1.set_title('GBP_AUD')
ax2.set_title('GBP/AUD')
plt.show()
def viewPlot(data, win = 150):
fig = plt.figure(figsize=(17, 10))
ax1, ax2 = fig.subplots(nrows=2, ncols=1)
df1 = data.iloc[-win:, ]
# High and Low prices are plotted
for i in range(len(df1)):
ax1.vlines(x = df1.index[i], ymin = df1.iat[i, 2], ymax = df1.iat[i, 1], color = 'magenta', linewidth = 2)
ax2.plot(df1.index, df1['eSuperRCS'].values, color='maroon')
ax2.axhline(55, color='green')
ax2.axhline(45, color='green')
ax2.axhline(50, color='orange')
ax1.set_title('GBP_AUD')
ax2.set_title('spread oscillator')
return plt.show()
viewPlot(df, win = 150)
| 0.293911 | 0.727613 |
```
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.style as style
style.use('seaborn-colorblind')
import scipy.stats as sps
import networkx as nx
import time
from Dist_UCB1_Small_Graphs import Dist_UCB1
import os
from multiprocess import Pool
n_runs = 100
T = 10000
G1 = nx.complete_graph(6)
G2 = nx.cycle_graph([6,7,8,9,10,11])
G = nx.compose(G1,G2)
N = G.number_of_nodes()
nodes = list(G.nodes)
for i in nodes:
G.add_edge(i,i)
rwd_means = [.2, .3, .4, .5, .6]
betas = [1, 1, 1, 0.01, 0.01, 0.01, 1, 1, 1, 0.01, 0.01, 0.01]
seeds = [i for i in range(n_runs)]
def worker(job_runs, seeds, betas, T, N, G):
regrets_dist_ucb1 = np.zeros((len(job_runs), N, T))
for run in job_runs:
start_run_t = time.time()
sd = 0.1
kl_distributions = [sps.truncnorm(a=(0-rwd_means[i])/sd, b=(1-rwd_means[i])/sd, loc=rwd_means[i], scale=sd) for i in range(len(rwd_means))]
for i in range(len(rwd_means)):
kl_distributions[i].random_state = np.random.RandomState(seed=seeds[run])
distributions = [[sps.truncnorm(a=(0-rwd_means[i])/sd, b=(1-rwd_means[i])/sd, loc=rwd_means[i], scale=sd) for i in range(len(rwd_means))] for n in range(N)]
for n in range(N):
for i in range(len(rwd_means)):
distributions[n][i].random_state = np.random.RandomState(seed=seeds[run])
distucb1 = Dist_UCB1(T, distributions, G, beta=betas)
distucb1.run()
regrets_dist_ucb1[run-job_runs[0], :, :] = distucb1.regrets
end_run_t = time.time()
print(f'finished run {run} in {end_run_t - start_run_t}sec')
return regrets_dist_ucb1
regrets_dist_ucb1 = np.zeros((n_runs, N, T))
init_time = time.time()
cpus = os.cpu_count()
init_time = time.time()
pool = Pool()
jobs = list(range(n_runs))
job_size = n_runs // cpus
job_chunks = [(jobs[i:i + job_size], seeds, betas, T, N, G) for i in range(0, len(jobs), job_size)]
results = pool.starmap(worker, job_chunks)
pool.close()
pool.join()
regrets_dist_ucb1_unconcat = [result for result in results]
np.concatenate(regrets_dist_ucb1_unconcat, out=regrets_dist_ucb1)
end_run_t = time.time()
time_axis = list(range(T))
mean_regrets_over_all_runs_dist_ucb1 = np.mean(regrets_dist_ucb1, axis=0)
std_regrets_over_all_runs_dist_ucb1 = np.std(regrets_dist_ucb1, axis=0)
print(f'Total run time = {end_run_t - init_time}sec')
plt.figure(figsize=(10,5))
avg_3_1_regret = np.mean(mean_regrets_over_all_runs_dist_ucb1[0:3], axis=0)
avg_3_001_regret = np.mean(mean_regrets_over_all_runs_dist_ucb1[3:6], axis=0)
avg_6_1_regret = np.mean(mean_regrets_over_all_runs_dist_ucb1[6:9], axis=0)
avg_6_001_regret = np.mean(mean_regrets_over_all_runs_dist_ucb1[9:12], axis=0)
std_3_1_regret = np.mean(std_regrets_over_all_runs_dist_ucb1[0:3], axis=0)
std_3_001_regret = np.mean(std_regrets_over_all_runs_dist_ucb1[3:6], axis=0)
std_6_1_regret = np.mean(std_regrets_over_all_runs_dist_ucb1[6:9], axis=0)
std_6_001_regret = np.mean(std_regrets_over_all_runs_dist_ucb1[9:12], axis=0)
plt.plot(time_axis, avg_3_1_regret, label="Group 1")
plt.fill_between(time_axis, avg_3_1_regret-std_3_1_regret, avg_3_1_regret+std_3_1_regret,alpha=.2)
plt.plot(time_axis, avg_3_001_regret, label="Group 2")
plt.fill_between(time_axis, avg_3_001_regret-std_3_001_regret, avg_3_001_regret+std_3_001_regret,alpha=.2)
plt.plot(time_axis, avg_6_1_regret, label="Group 3")
plt.fill_between(time_axis, avg_6_1_regret-std_6_1_regret, avg_6_1_regret+std_6_1_regret,alpha=.2)
plt.plot(time_axis, avg_6_001_regret, label="Group 4")
plt.fill_between(time_axis, avg_6_001_regret-std_6_001_regret, avg_6_001_regret+std_6_001_regret,alpha=.2)
plt.xlabel("Time")
plt.ylabel("Regret")
plt.legend()
plt.grid()
plt.savefig("dist_UCB1_small_graphs_together_N12.pdf", bbox_inches='tight')
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.style as style
style.use('seaborn-colorblind')
import scipy.stats as sps
import networkx as nx
import time
from Dist_UCB1_Small_Graphs import Dist_UCB1
import os
from multiprocess import Pool
n_runs = 100
T = 10000
G1 = nx.complete_graph(6)
G2 = nx.cycle_graph([6,7,8,9,10,11])
G = nx.compose(G1,G2)
N = G.number_of_nodes()
nodes = list(G.nodes)
for i in nodes:
G.add_edge(i,i)
rwd_means = [.2, .3, .4, .5, .6]
betas = [1, 1, 1, 0.01, 0.01, 0.01, 1, 1, 1, 0.01, 0.01, 0.01]
seeds = [i for i in range(n_runs)]
def worker(job_runs, seeds, betas, T, N, G):
regrets_dist_ucb1 = np.zeros((len(job_runs), N, T))
for run in job_runs:
start_run_t = time.time()
sd = 0.1
kl_distributions = [sps.truncnorm(a=(0-rwd_means[i])/sd, b=(1-rwd_means[i])/sd, loc=rwd_means[i], scale=sd) for i in range(len(rwd_means))]
for i in range(len(rwd_means)):
kl_distributions[i].random_state = np.random.RandomState(seed=seeds[run])
distributions = [[sps.truncnorm(a=(0-rwd_means[i])/sd, b=(1-rwd_means[i])/sd, loc=rwd_means[i], scale=sd) for i in range(len(rwd_means))] for n in range(N)]
for n in range(N):
for i in range(len(rwd_means)):
distributions[n][i].random_state = np.random.RandomState(seed=seeds[run])
distucb1 = Dist_UCB1(T, distributions, G, beta=betas)
distucb1.run()
regrets_dist_ucb1[run-job_runs[0], :, :] = distucb1.regrets
end_run_t = time.time()
print(f'finished run {run} in {end_run_t - start_run_t}sec')
return regrets_dist_ucb1
regrets_dist_ucb1 = np.zeros((n_runs, N, T))
init_time = time.time()
cpus = os.cpu_count()
init_time = time.time()
pool = Pool()
jobs = list(range(n_runs))
job_size = n_runs // cpus
job_chunks = [(jobs[i:i + job_size], seeds, betas, T, N, G) for i in range(0, len(jobs), job_size)]
results = pool.starmap(worker, job_chunks)
pool.close()
pool.join()
regrets_dist_ucb1_unconcat = [result for result in results]
np.concatenate(regrets_dist_ucb1_unconcat, out=regrets_dist_ucb1)
end_run_t = time.time()
time_axis = list(range(T))
mean_regrets_over_all_runs_dist_ucb1 = np.mean(regrets_dist_ucb1, axis=0)
std_regrets_over_all_runs_dist_ucb1 = np.std(regrets_dist_ucb1, axis=0)
print(f'Total run time = {end_run_t - init_time}sec')
plt.figure(figsize=(10,5))
avg_3_1_regret = np.mean(mean_regrets_over_all_runs_dist_ucb1[0:3], axis=0)
avg_3_001_regret = np.mean(mean_regrets_over_all_runs_dist_ucb1[3:6], axis=0)
avg_6_1_regret = np.mean(mean_regrets_over_all_runs_dist_ucb1[6:9], axis=0)
avg_6_001_regret = np.mean(mean_regrets_over_all_runs_dist_ucb1[9:12], axis=0)
std_3_1_regret = np.mean(std_regrets_over_all_runs_dist_ucb1[0:3], axis=0)
std_3_001_regret = np.mean(std_regrets_over_all_runs_dist_ucb1[3:6], axis=0)
std_6_1_regret = np.mean(std_regrets_over_all_runs_dist_ucb1[6:9], axis=0)
std_6_001_regret = np.mean(std_regrets_over_all_runs_dist_ucb1[9:12], axis=0)
plt.plot(time_axis, avg_3_1_regret, label="Group 1")
plt.fill_between(time_axis, avg_3_1_regret-std_3_1_regret, avg_3_1_regret+std_3_1_regret,alpha=.2)
plt.plot(time_axis, avg_3_001_regret, label="Group 2")
plt.fill_between(time_axis, avg_3_001_regret-std_3_001_regret, avg_3_001_regret+std_3_001_regret,alpha=.2)
plt.plot(time_axis, avg_6_1_regret, label="Group 3")
plt.fill_between(time_axis, avg_6_1_regret-std_6_1_regret, avg_6_1_regret+std_6_1_regret,alpha=.2)
plt.plot(time_axis, avg_6_001_regret, label="Group 4")
plt.fill_between(time_axis, avg_6_001_regret-std_6_001_regret, avg_6_001_regret+std_6_001_regret,alpha=.2)
plt.xlabel("Time")
plt.ylabel("Regret")
plt.legend()
plt.grid()
plt.savefig("dist_UCB1_small_graphs_together_N12.pdf", bbox_inches='tight')
| 0.301979 | 0.406332 |
```
import os
import json
import pickle
import random
from collections import defaultdict, Counter
from indra.literature.adeft_tools import universal_extract_text
from indra.databases.hgnc_client import get_hgnc_name, get_hgnc_id
from adeft.discover import AdeftMiner
from adeft.gui import ground_with_gui
from adeft.modeling.label import AdeftLabeler
from adeft.modeling.classify import AdeftClassifier
from adeft.disambiguate import AdeftDisambiguator, load_disambiguator
from adeft_indra.ground.ground import AdeftGrounder
from adeft_indra.model_building.s3 import model_to_s3
from adeft_indra.model_building.escape import escape_filename
from adeft_indra.db.content import get_pmids_for_agent_text, get_pmids_for_entity, \
get_plaintexts_for_pmids
adeft_grounder = AdeftGrounder()
shortforms = ['BAL']
model_name = ':'.join(sorted(escape_filename(shortform) for shortform in shortforms))
results_path = os.path.abspath(os.path.join('../..', 'results', model_name))
miners = dict()
all_texts = {}
for shortform in shortforms:
pmids = get_pmids_for_agent_text(shortform)
if len(pmids) > 10000:
pmids = random.choices(pmids, k=10000)
text_dict = get_plaintexts_for_pmids(pmids, contains=shortforms)
text_dict = {pmid: text for pmid, text in text_dict.items() if len(text) > 5}
miners[shortform] = AdeftMiner(shortform)
miners[shortform].process_texts(text_dict.values())
all_texts.update(text_dict)
longform_dict = {}
for shortform in shortforms:
longforms = miners[shortform].get_longforms()
longforms = [(longform, count, score) for longform, count, score in longforms
if count*score > 2]
longform_dict[shortform] = longforms
combined_longforms = Counter()
for longform_rows in longform_dict.values():
combined_longforms.update({longform: count for longform, count, score
in longform_rows})
grounding_map = {}
names = {}
for longform in combined_longforms:
groundings = adeft_grounder.ground(longform)
if groundings:
grounding = groundings[0]['grounding']
grounding_map[longform] = grounding
names[grounding] = groundings[0]['name']
longforms, counts = zip(*combined_longforms.most_common())
pos_labels = []
list(zip(longforms, counts))
grounding_map, names, pos_labels = ground_with_gui(longforms, counts,
grounding_map=grounding_map,
names=names, pos_labels=pos_labels, no_browser=True, port=8890)
result = [grounding_map, names, pos_labels]
result
grounding_map, names, pos_labels = [{'benzaldehyde lyase': 'MESH:C059416',
'betaine aldehyde': 'CHEBI:CHEBI:15710',
'bile salt activity lipase': 'HGNC:1848',
'bioartificial liver': 'MESH:D019164',
'blood alcohol levels': 'ungrounded',
'breath alcohol levels': 'ungrounded',
'british anti lewisite': 'CHEBI:CHEBI:64198',
'brochoalveolar lavage': 'MESH:D018893',
'bronchalveolar lavage': 'MESH:D018893',
'bronchial alveolar lavage': 'MESH:D018893',
'bronchial lavage': 'MESH:D018893',
'bronchio alveolar lavage': 'MESH:D018893',
'bronchiolar lavage': 'MESH:D018893',
'broncho alveolar lavage': 'MESH:D018893',
'bronchoalveolar': 'MESH:D018893',
'bronchoalveolar fluid': 'MESH:D018893',
'bronchoalveolar larvage': 'MESH:D018893',
'bronchoalveolar lavage': 'MESH:D018893'},
{'MESH:C059416': 'benzaldehyde lyase',
'CHEBI:CHEBI:15710': 'betaine aldehyde',
'HGNC:1848': 'CEL',
'MESH:D019164': 'Liver, Artificial',
'CHEBI:CHEBI:64198': 'dimercaprol',
'MESH:D018893': 'Bronchoalveolar Lavage'},
['HGNC:1848', 'MESH:D018893']]
excluded_longforms = []
grounding_dict = {shortform: {longform: grounding_map[longform]
for longform, _, _ in longforms if longform in grounding_map
and longform not in excluded_longforms}
for shortform, longforms in longform_dict.items()}
result = [grounding_dict, names, pos_labels]
if not os.path.exists(results_path):
os.mkdir(results_path)
with open(os.path.join(results_path, f'{model_name}_preliminary_grounding_info.json'), 'w') as f:
json.dump(result, f)
additional_entities = {'HGNC:1848': ['CEL', ['BAL', 'bile salt']]}
unambiguous_agent_texts = {}
labeler = AdeftLabeler(grounding_dict)
corpus = labeler.build_from_texts((text, pmid) for pmid, text in all_texts.items())
agent_text_pmid_map = defaultdict(list)
for text, label, id_ in corpus:
agent_text_pmid_map[label].append(id_)
entity_pmid_map = {entity: set(get_pmids_for_entity(*entity.split(':', maxsplit=1),
major_topic=True))for entity in additional_entities}
intersection1 = []
for entity1, pmids1 in entity_pmid_map.items():
for entity2, pmids2 in entity_pmid_map.items():
intersection1.append((entity1, entity2, len(pmids1 & pmids2)))
intersection2 = []
for entity1, pmids1 in agent_text_pmid_map.items():
for entity2, pmids2 in entity_pmid_map.items():
intersection2.append((entity1, entity2, len(set(pmids1) & pmids2)))
intersection1
intersection2
all_used_pmids = set()
for entity, agent_texts in unambiguous_agent_texts.items():
used_pmids = set()
for agent_text in agent_texts[1]:
pmids = set(get_pmids_for_agent_text(agent_text))
new_pmids = list(pmids - all_texts.keys() - used_pmids)
text_dict = get_plaintexts_for_pmids(new_pmids, contains=agent_texts)
corpus.extend([(text, entity, pmid) for pmid, text in text_dict.items() if len(text) > 5])
used_pmids.update(new_pmids)
all_used_pmids.update(used_pmids)
for entity, pmids in entity_pmid_map.items():
new_pmids = list(set(pmids) - all_texts.keys() - all_used_pmids)
if len(new_pmids) > 10000:
new_pmids = random.choices(new_pmids, k=10000)
_, contains = additional_entities[entity]
text_dict = get_plaintexts_for_pmids(new_pmids, contains=contains)
corpus.extend([(text, entity, pmid) for pmid, text in text_dict.items() if len(text) > 5])
names.update({key: value[0] for key, value in additional_entities.items()})
names.update({key: value[0] for key, value in unambiguous_agent_texts.items()})
pos_labels = list(set(pos_labels) | additional_entities.keys() |
unambiguous_agent_texts.keys())
%%capture
classifier = AdeftClassifier(shortforms, pos_labels=pos_labels, random_state=1729)
param_grid = {'C': [100.0], 'max_features': [10000]}
texts, labels, pmids = zip(*corpus)
classifier.cv(texts, labels, param_grid, cv=5, n_jobs=5)
classifier.stats
disamb = AdeftDisambiguator(classifier, grounding_dict, names)
disamb.dump(model_name, results_path)
print(disamb.info())
model_to_s3(disamb)
d = load_disambiguator('BAL')
print(d.info())
```
|
github_jupyter
|
import os
import json
import pickle
import random
from collections import defaultdict, Counter
from indra.literature.adeft_tools import universal_extract_text
from indra.databases.hgnc_client import get_hgnc_name, get_hgnc_id
from adeft.discover import AdeftMiner
from adeft.gui import ground_with_gui
from adeft.modeling.label import AdeftLabeler
from adeft.modeling.classify import AdeftClassifier
from adeft.disambiguate import AdeftDisambiguator, load_disambiguator
from adeft_indra.ground.ground import AdeftGrounder
from adeft_indra.model_building.s3 import model_to_s3
from adeft_indra.model_building.escape import escape_filename
from adeft_indra.db.content import get_pmids_for_agent_text, get_pmids_for_entity, \
get_plaintexts_for_pmids
adeft_grounder = AdeftGrounder()
shortforms = ['BAL']
model_name = ':'.join(sorted(escape_filename(shortform) for shortform in shortforms))
results_path = os.path.abspath(os.path.join('../..', 'results', model_name))
miners = dict()
all_texts = {}
for shortform in shortforms:
pmids = get_pmids_for_agent_text(shortform)
if len(pmids) > 10000:
pmids = random.choices(pmids, k=10000)
text_dict = get_plaintexts_for_pmids(pmids, contains=shortforms)
text_dict = {pmid: text for pmid, text in text_dict.items() if len(text) > 5}
miners[shortform] = AdeftMiner(shortform)
miners[shortform].process_texts(text_dict.values())
all_texts.update(text_dict)
longform_dict = {}
for shortform in shortforms:
longforms = miners[shortform].get_longforms()
longforms = [(longform, count, score) for longform, count, score in longforms
if count*score > 2]
longform_dict[shortform] = longforms
combined_longforms = Counter()
for longform_rows in longform_dict.values():
combined_longforms.update({longform: count for longform, count, score
in longform_rows})
grounding_map = {}
names = {}
for longform in combined_longforms:
groundings = adeft_grounder.ground(longform)
if groundings:
grounding = groundings[0]['grounding']
grounding_map[longform] = grounding
names[grounding] = groundings[0]['name']
longforms, counts = zip(*combined_longforms.most_common())
pos_labels = []
list(zip(longforms, counts))
grounding_map, names, pos_labels = ground_with_gui(longforms, counts,
grounding_map=grounding_map,
names=names, pos_labels=pos_labels, no_browser=True, port=8890)
result = [grounding_map, names, pos_labels]
result
grounding_map, names, pos_labels = [{'benzaldehyde lyase': 'MESH:C059416',
'betaine aldehyde': 'CHEBI:CHEBI:15710',
'bile salt activity lipase': 'HGNC:1848',
'bioartificial liver': 'MESH:D019164',
'blood alcohol levels': 'ungrounded',
'breath alcohol levels': 'ungrounded',
'british anti lewisite': 'CHEBI:CHEBI:64198',
'brochoalveolar lavage': 'MESH:D018893',
'bronchalveolar lavage': 'MESH:D018893',
'bronchial alveolar lavage': 'MESH:D018893',
'bronchial lavage': 'MESH:D018893',
'bronchio alveolar lavage': 'MESH:D018893',
'bronchiolar lavage': 'MESH:D018893',
'broncho alveolar lavage': 'MESH:D018893',
'bronchoalveolar': 'MESH:D018893',
'bronchoalveolar fluid': 'MESH:D018893',
'bronchoalveolar larvage': 'MESH:D018893',
'bronchoalveolar lavage': 'MESH:D018893'},
{'MESH:C059416': 'benzaldehyde lyase',
'CHEBI:CHEBI:15710': 'betaine aldehyde',
'HGNC:1848': 'CEL',
'MESH:D019164': 'Liver, Artificial',
'CHEBI:CHEBI:64198': 'dimercaprol',
'MESH:D018893': 'Bronchoalveolar Lavage'},
['HGNC:1848', 'MESH:D018893']]
excluded_longforms = []
grounding_dict = {shortform: {longform: grounding_map[longform]
for longform, _, _ in longforms if longform in grounding_map
and longform not in excluded_longforms}
for shortform, longforms in longform_dict.items()}
result = [grounding_dict, names, pos_labels]
if not os.path.exists(results_path):
os.mkdir(results_path)
with open(os.path.join(results_path, f'{model_name}_preliminary_grounding_info.json'), 'w') as f:
json.dump(result, f)
additional_entities = {'HGNC:1848': ['CEL', ['BAL', 'bile salt']]}
unambiguous_agent_texts = {}
labeler = AdeftLabeler(grounding_dict)
corpus = labeler.build_from_texts((text, pmid) for pmid, text in all_texts.items())
agent_text_pmid_map = defaultdict(list)
for text, label, id_ in corpus:
agent_text_pmid_map[label].append(id_)
entity_pmid_map = {entity: set(get_pmids_for_entity(*entity.split(':', maxsplit=1),
major_topic=True))for entity in additional_entities}
intersection1 = []
for entity1, pmids1 in entity_pmid_map.items():
for entity2, pmids2 in entity_pmid_map.items():
intersection1.append((entity1, entity2, len(pmids1 & pmids2)))
intersection2 = []
for entity1, pmids1 in agent_text_pmid_map.items():
for entity2, pmids2 in entity_pmid_map.items():
intersection2.append((entity1, entity2, len(set(pmids1) & pmids2)))
intersection1
intersection2
all_used_pmids = set()
for entity, agent_texts in unambiguous_agent_texts.items():
used_pmids = set()
for agent_text in agent_texts[1]:
pmids = set(get_pmids_for_agent_text(agent_text))
new_pmids = list(pmids - all_texts.keys() - used_pmids)
text_dict = get_plaintexts_for_pmids(new_pmids, contains=agent_texts)
corpus.extend([(text, entity, pmid) for pmid, text in text_dict.items() if len(text) > 5])
used_pmids.update(new_pmids)
all_used_pmids.update(used_pmids)
for entity, pmids in entity_pmid_map.items():
new_pmids = list(set(pmids) - all_texts.keys() - all_used_pmids)
if len(new_pmids) > 10000:
new_pmids = random.choices(new_pmids, k=10000)
_, contains = additional_entities[entity]
text_dict = get_plaintexts_for_pmids(new_pmids, contains=contains)
corpus.extend([(text, entity, pmid) for pmid, text in text_dict.items() if len(text) > 5])
names.update({key: value[0] for key, value in additional_entities.items()})
names.update({key: value[0] for key, value in unambiguous_agent_texts.items()})
pos_labels = list(set(pos_labels) | additional_entities.keys() |
unambiguous_agent_texts.keys())
%%capture
classifier = AdeftClassifier(shortforms, pos_labels=pos_labels, random_state=1729)
param_grid = {'C': [100.0], 'max_features': [10000]}
texts, labels, pmids = zip(*corpus)
classifier.cv(texts, labels, param_grid, cv=5, n_jobs=5)
classifier.stats
disamb = AdeftDisambiguator(classifier, grounding_dict, names)
disamb.dump(model_name, results_path)
print(disamb.info())
model_to_s3(disamb)
d = load_disambiguator('BAL')
print(d.info())
| 0.177098 | 0.147065 |
# Tutorial 8 - Solver options
In [Tutorial 7](./Tutorial%207%20-%20Model%20options.ipynb) we saw how to change the model options. In this tutorial we will show how to pass options to the solver.
All models in PyBaMM have a default solver which is typically different depending on whether the model results in a system of ordinary differential equations (ODEs) or differential algebraic equations (DAEs).
One of the most common options you will want to change is the solver tolerances. By default all tolerances are set to $10^{-6}$. However, depending on your simulation you may find you want to tighten the tolerances to obtain a more accurate solution, or you may want to loosen the tolerances to reduce the solve time. It is good practice to conduct a tolerance study, where you simulate the same problem with a tighter tolerances and compare the results. We do not show how to do this here, but we give an example of a mesh resolution study in the [next tutorial](./Tutorial%209%20-%20Changing%20the%20mesh.ipynb), which is conducted in a similar way.
```
%pip install pybamm -q # install PyBaMM if it is not installed
import pybamm
```
Here we will change the absolute and relative tolerances, as well as the "mode" of the `CasadiSolver`. For a list of all the solver options please consult the [documentation](https://pybamm.readthedocs.io/en/latest/source/solvers/index.html).
The `CasadiSolver` can operate in a number of modes, including "safe" (default) and "fast". Safe mode performs step-and-check integration and supports event handling (e.g. you can integrate until you hit a certain voltage), and is the recommended for simulations of a full charge or discharge. Fast mode performs direct integration, ignoring events, and is recommended when simulating a drive cycle or other simulation where no events should be triggered.
We'll solve the DFN with all the default options in both "safe" and "fast" mode and compare the solutions. For both simulations we'll use $10^{-3}$ for both the absolute and relative tolerance. For demonstration purposes we'll change the cut-off voltage to 3.6V so we can observe the different behaviour of the two solver modes.
```
# load model and parameters
model = pybamm.lithium_ion.DFN()
param = model.default_parameter_values
param["Lower voltage cut-off [V]"] = 3.6
# load solvers
safe_solver = pybamm.CasadiSolver(atol=1e-3, rtol=1e-3, mode="safe")
fast_solver = pybamm.CasadiSolver(atol=1e-3, rtol=1e-3, mode="fast")
# create simulations
safe_sim = pybamm.Simulation(model, parameter_values=param, solver=safe_solver)
fast_sim = pybamm.Simulation(model, parameter_values=param, solver=fast_solver)
# solve
safe_sim.solve([0, 3600])
print("Safe mode solve time: {}".format(safe_sim.solution.solve_time))
fast_sim.solve([0, 3600])
print("Fast mode solve time: {}".format(fast_sim.solution.solve_time))
# plot solutions
pybamm.dynamic_plot([safe_sim, fast_sim])
```
We see that both solvers give the same solution up to the time at which the cut-off voltage is reached. At this point the solver using "safe" mode stops, but the solver using "fast" mode carries on integrating until the final time. As its name suggests, "fast" mode integrates more quickly that "safe" mode, but is unsuitable if your simulation required events to be handled.
Usually the default solver options provide a good combination of speed and accuracy, but we encourage you to investigate different solvers and options to find the best combination for your problem.
In the [next tutorial](./Tutorial%209%20-%20Changing%20the%20mesh.ipynb) we show how to change the mesh.
|
github_jupyter
|
%pip install pybamm -q # install PyBaMM if it is not installed
import pybamm
# load model and parameters
model = pybamm.lithium_ion.DFN()
param = model.default_parameter_values
param["Lower voltage cut-off [V]"] = 3.6
# load solvers
safe_solver = pybamm.CasadiSolver(atol=1e-3, rtol=1e-3, mode="safe")
fast_solver = pybamm.CasadiSolver(atol=1e-3, rtol=1e-3, mode="fast")
# create simulations
safe_sim = pybamm.Simulation(model, parameter_values=param, solver=safe_solver)
fast_sim = pybamm.Simulation(model, parameter_values=param, solver=fast_solver)
# solve
safe_sim.solve([0, 3600])
print("Safe mode solve time: {}".format(safe_sim.solution.solve_time))
fast_sim.solve([0, 3600])
print("Fast mode solve time: {}".format(fast_sim.solution.solve_time))
# plot solutions
pybamm.dynamic_plot([safe_sim, fast_sim])
| 0.546738 | 0.994314 |
### Common Set Operations
Let's look at some of the more basic and common operations with sets:
* size
* membership testing
* adding elements
* removing elements
#### Size
The size of a set (it's cardinality), is given by the `len()` function - the same one we use for sequences, iterables, dictionaries, etc.
```
s = {1, 2, 3}
len(s)
```
#### Membership Testing
This is also very easy:
```
s = {1, 2, 3}
1 in s
10 in s
1 not in s
10 not in s
```
But let's go a little further and consider how membership testing works with sets. As I mentioned in earlier lectures, sets are hash tables, and membership testing is **extremely** efficient for sets, since it's simply a hash table lookup - as opposed to scanning a list for example, until we find the requested element (or not).
Let's do some quick timings to verify this, as well as compare lookup speeds for sets and dictionaries as well (which are also, after all, hash tables).
```
from timeit import timeit
n = 100_000
s = {i for i in range(n)}
l = [i for i in range(n)]
d = {i:None for i in range(n)}
```
Let's time how long it takes to find if `9` is in the object - which would be the tenth element only of the list and the dictionary (keys), and who knows for the set:
```
number = 1_000_000
search = 9
t_list = timeit(f'{search} in l', globals=globals(), number=number)
t_set = timeit(f'{search} in s', globals=globals(), number=number)
t_dict = timeit(f'{search} in d', globals=globals(), number=number)
print('list:', t_list)
print('set:', t_set)
print('dict:', t_dict)
```
The story changes even more if we test for example the last element of the list.
I'm definitely not to run the tests `1_000_000` times - not unless we want to make this video reaaaaaaly long!
```
number = 3_000
search = 99_999
t_list = timeit(f'{search} in l', globals=globals(), number=number)
t_set = timeit(f'{search} in s', globals=globals(), number=number)
t_dict = timeit(f'{search} in d', globals=globals(), number=number)
print('list:', t_list)
print('set:', t_set)
print('dict:', t_dict)
```
The situation for `not in` is the same:
```
number = 3_000
search = -1
t_list = timeit(f'{search} not in l', globals=globals(), number=number)
t_set = timeit(f'{search} not in s', globals=globals(), number=number)
t_dict = timeit(f'{search} not in d', globals=globals(), number=number)
print('list:', t_list)
print('set:', t_set)
print('dict:', t_dict)
```
But this efficiency does come at the cost of memory:
```
print(d.__sizeof__())
print(s.__sizeof__())
print(l.__sizeof__())
```
Even for empty objects:
```
s = set()
d = dict()
l = list()
print(d.__sizeof__())
print(s.__sizeof__())
print(l.__sizeof__())
```
And adding just one element to each object:
```
s.add(10)
d[10] =None
l.append(10)
print(d.__sizeof__())
print(s.__sizeof__())
print(l.__sizeof__())
```
If you're wondering why the dictionary and set size did not increase, remember when we covered hash tables - there is some overallocation that takes place so we don't incure the cost of resizing every time we had an element. In fact, lists do the same as well - they over-allocate to reduce the resizing cost. I'll come back to that in a minute.
#### Adding Elements
When we have an existing set, we can always add elements to it. Of course *where* it gets "inserted" is unknown. So Python does not call it `append` or `insert` which would connotate ordering of some kind - instead it just calls it `add`:
```
s = {30, 20, 10}
s.add(15)
s
```
Don't be fooled by the apparent ordering of the elements here. This is the same as with dictionaries - Jupyter tries to represent things nicely for us, but underneath the scenes:
```
print(s)
s.add(-1)
print(s)
```
And the order just changed again! :-)
What's interesting about the `add()` method, is that if we try to add an element that already exists, Python will simply ignore it:
```
s
s.add(15)
s
```
Now that we know how to add an element to a set, let's go back and see how the set, dictionary and list resize as we add more elements to them.
We should expect the list to be more efficient from a memory standpoint:
```
l = list()
s = set()
d = dict()
print('#', 'dict', 'set', 'list')
for i in range(50):
print(i, d.__sizeof__(), s.__sizeof__(), l.__sizeof__())
l.append(i)
s.add(i)
d[i] = None
```
As you can see, the memory costs for a set or a dict are definitely higher than for a list. You can also see from this how it looks like CPython implements different resizing strategies for sets, dicts and lists.
The strategy by the way has nothing to do with the size of the elements we put in those objects:
```
l = list()
s = set()
d = dict()
print('#', 'dict', 'set', 'list')
for i in range(50):
print(i, d.__sizeof__(), s.__sizeof__(), l.__sizeof__())
l.append(i**1000)
s.add(i*1000)
d[i*1000] = None
```
As you can see the memory cost of the objects themselves did not change, nor did the sizing strategy (remember that all those objects contain pointers to the data, not the data itself - and a pointer to an object, no matter the size of that object, is the same).
So be careful using `__sizeof__` - it's often only part of the story.
#### Removing Elements
Now let's see how we can remove elements from a set.
Just as with dictionaries, we may be trying to remove an item that does not exist in the set. Depending on whether we want to silently ignore deletion of non-existent elements we can use one of two techniques:
```
s = {1, 2, 3}
s.remove(1)
s
s.remove(10)
```
As you can see, we get an exception.
If we don't want the exception we can do it this way:
```
s.discard(10)
s
```
We can also remove (and return) an **arbitrary** element from the set:
```
s = set('python')
s
s.pop()
```
Note that we **do not know** ahead of time what element will get popped.
Also, popping an empty set will result in a `KeyError` exception:
```
s = set()
s.pop()
```
Something like that might be handy to handle all the elements of a set one at a time without caring for the order in which elements are removed from the set - not that you can, anyway - sets are not ordered!
But this way you can get at the elements of a set without knowing the content of the set (since you need to know the element you are removing with `remove` and `discard`.)
Finally, you can empty out a set by calling the `clear` method:
```
s = {1, 2, 3}
s.clear()
s
```
|
github_jupyter
|
s = {1, 2, 3}
len(s)
s = {1, 2, 3}
1 in s
10 in s
1 not in s
10 not in s
from timeit import timeit
n = 100_000
s = {i for i in range(n)}
l = [i for i in range(n)]
d = {i:None for i in range(n)}
number = 1_000_000
search = 9
t_list = timeit(f'{search} in l', globals=globals(), number=number)
t_set = timeit(f'{search} in s', globals=globals(), number=number)
t_dict = timeit(f'{search} in d', globals=globals(), number=number)
print('list:', t_list)
print('set:', t_set)
print('dict:', t_dict)
number = 3_000
search = 99_999
t_list = timeit(f'{search} in l', globals=globals(), number=number)
t_set = timeit(f'{search} in s', globals=globals(), number=number)
t_dict = timeit(f'{search} in d', globals=globals(), number=number)
print('list:', t_list)
print('set:', t_set)
print('dict:', t_dict)
number = 3_000
search = -1
t_list = timeit(f'{search} not in l', globals=globals(), number=number)
t_set = timeit(f'{search} not in s', globals=globals(), number=number)
t_dict = timeit(f'{search} not in d', globals=globals(), number=number)
print('list:', t_list)
print('set:', t_set)
print('dict:', t_dict)
print(d.__sizeof__())
print(s.__sizeof__())
print(l.__sizeof__())
s = set()
d = dict()
l = list()
print(d.__sizeof__())
print(s.__sizeof__())
print(l.__sizeof__())
s.add(10)
d[10] =None
l.append(10)
print(d.__sizeof__())
print(s.__sizeof__())
print(l.__sizeof__())
s = {30, 20, 10}
s.add(15)
s
print(s)
s.add(-1)
print(s)
s
s.add(15)
s
l = list()
s = set()
d = dict()
print('#', 'dict', 'set', 'list')
for i in range(50):
print(i, d.__sizeof__(), s.__sizeof__(), l.__sizeof__())
l.append(i)
s.add(i)
d[i] = None
l = list()
s = set()
d = dict()
print('#', 'dict', 'set', 'list')
for i in range(50):
print(i, d.__sizeof__(), s.__sizeof__(), l.__sizeof__())
l.append(i**1000)
s.add(i*1000)
d[i*1000] = None
s = {1, 2, 3}
s.remove(1)
s
s.remove(10)
s.discard(10)
s
s = set('python')
s
s.pop()
s = set()
s.pop()
s = {1, 2, 3}
s.clear()
s
| 0.140425 | 0.928149 |
# Base Line Model
这个是一个基本模型,手动构筑model,没有使用迁移学习
没有使用early stopping
```
# ! pip install sklearn
import keras
import os
import numpy as np
import pickle
import tensorflow as tf
from keras import layers
from keras import models
from keras.models import load_model
from sklearn.metrics import accuracy_score
from keras.preprocessing import image_dataset_from_directory
from keras import optimizers
from keras.preprocessing.image import ImageDataGenerator
import matplotlib.pyplot as plt
if os.getcwd().split('/')[-1]=='baseline_model':
os.chdir('..')
os.getcwd()
classes = ['zero', 'one', 'two', 'three', 'four',
'five', 'six', 'seven', 'eight', 'nine']
IMAGE_SIZE=100
BATCH_SIZE=32
class_num=len(classes)
base_path = "hand_sign_digit_data"
model = tf.keras.models.Sequential()
# 32 is kernel count (3,3) is kernel dimension
model.add(tf.keras.layers.Conv2D(32, (3, 3), activation='relu',input_shape=(IMAGE_SIZE, IMAGE_SIZE, 3)))
model.add(tf.keras.layers.MaxPooling2D((2, 2)))
model.add(tf.keras.layers.Conv2D(64, (3, 3), activation='relu'))
model.add(tf.keras.layers.MaxPooling2D((2, 2)))
model.add(tf.keras.layers.Dropout(0.25))
model.add(tf.keras.layers.Conv2D(128, (3, 3), activation='relu'))
model.add(tf.keras.layers.MaxPooling2D((2, 2)))
model.add(tf.keras.layers.Dropout(0.25))
model.add(tf.keras.layers.Conv2D(128, (3, 3), activation='relu'))
model.add(tf.keras.layers.MaxPooling2D((2, 2)))
model.add(tf.keras.layers.Flatten())
# 这个是在最后部分使用dropout 部分note失活,也可以在每一层都使用dropout。dropout的目的是防止过拟合
model.add(tf.keras.layers.Dropout(0.3))
model.add(tf.keras.layers.Dense(512, activation='relu'))
model.add(tf.keras.layers.Dense(10, activation='softmax'))
model.summary()
# base_learning_rate=0.001
model.compile(loss='categorical_crossentropy',
optimizer='adam',
metrics=['acc'])
train_dir = os.path.join(base_path, 'train')
validation_dir = os.path.join(base_path, 'validation')
test_dir = os.path.join(base_path, 'test_old')
train_datagen = ImageDataGenerator(
rescale=1./255,
rotation_range=10,
width_shift_range=0.1,
height_shift_range=0.1,
# zoom_range=0.2,
# shear_range=0.2,
horizontal_flip=True,
)
test_datagen = ImageDataGenerator(rescale=1./255)
train_generator = train_datagen.flow_from_directory(
train_dir,
target_size=(IMAGE_SIZE, IMAGE_SIZE),
batch_size=BATCH_SIZE,
class_mode='categorical')
validation_generator = test_datagen.flow_from_directory(
validation_dir,
target_size=(IMAGE_SIZE, IMAGE_SIZE),
batch_size=BATCH_SIZE,
class_mode='categorical')
history = model.fit_generator(
train_generator,
steps_per_epoch=52,
epochs=50,
validation_data=validation_generator,
validation_steps=14)
model_file_name='baseline_model_2020-11-16.h5'
model.save(model_file_name)
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
plt.figure(figsize=(10,10))
ax=plt.subplot(2,1,1)
plt.plot(acc, label='Training acc')
plt.plot(val_acc, label='Validation acc')
plt.title('Training and validation accuracy')
plt.legend()
plt.ylabel("Accuracy(%)")
plt.xlabel("Epoch")
ax=plt.subplot(2,1,2)
plt.plot(loss, label='Training loss')
plt.plot(val_loss, label='Validation loss')
plt.title('Training and validation loss')
plt.legend()
plt.ylabel("LOSS")
plt.xlabel("Epoch")
plt.show()
```
# Model预测
```
del model
model=load_model(model_file_name)
model.summary()
test_dataset = image_dataset_from_directory(test_dir,
shuffle=True,
batch_size=BATCH_SIZE,
image_size=(IMAGE_SIZE,
IMAGE_SIZE))
images,labels=next(iter(test_dataset))
len(images),labels
predictions = model.predict_on_batch(images).flatten()
pred_result=[]
for i in range(BATCH_SIZE):
pred_result.append(predictions[i*class_num:(i+1)*class_num].argmax())
print("Predict Accuracy={}".format(accuracy_score(labels, pred_result)))
plt.figure(figsize=(15,15))
# 设定空白处 间隔大小
plt.subplots_adjust(wspace=0.4, hspace=0)
count=0
for i in range(BATCH_SIZE):
ax=plt.subplot(4,8,i+1)
plt.imshow(images[i].numpy().astype('uint8'))
plt.axis('off')
Correct_value=labels[i].numpy().astype('uint8')
Predict_value=pred_result[i]
plt.title("Correct value:{}\n Predict value:{}".format(Correct_value,Predict_value))
if Correct_value == Predict_value:
count += 1
print("Correct rate={}%".format(round(count*1.0/BATCH_SIZE*100,2)))
```
### Save model and history data
```
history_name = 'baseline_model_history.pickle'
path = os.path.join(os.getcwd(), 'baseline_model', history_name)
def pickle_dump(obj, path):
with open(path, mode='wb') as f:
pickle.dump(obj, f)
pickle_dump(history.history, path)
```
# 指定一张图片进行预测
```
from PIL import Image, ImageFilter
im=Image.open('hand_sign_digit_data/test_old/4/qiu-4-1.jpg')
im1=im.resize((100,100))
im1
print(im.format, im.size, im.mode)
print(im1.format, im1.size, im1.mode)
im1_arr=np.asarray(im1)
im1_arr.shape
im1_arr=im1_arr/225.0
# print(images[0].shape)
# plt.imshow(images[0].numpy().astype('uint8'))
pred=model.predict(np.expand_dims(im1_arr,0))
pred,print(pred.argmax())
```
|
github_jupyter
|
# ! pip install sklearn
import keras
import os
import numpy as np
import pickle
import tensorflow as tf
from keras import layers
from keras import models
from keras.models import load_model
from sklearn.metrics import accuracy_score
from keras.preprocessing import image_dataset_from_directory
from keras import optimizers
from keras.preprocessing.image import ImageDataGenerator
import matplotlib.pyplot as plt
if os.getcwd().split('/')[-1]=='baseline_model':
os.chdir('..')
os.getcwd()
classes = ['zero', 'one', 'two', 'three', 'four',
'five', 'six', 'seven', 'eight', 'nine']
IMAGE_SIZE=100
BATCH_SIZE=32
class_num=len(classes)
base_path = "hand_sign_digit_data"
model = tf.keras.models.Sequential()
# 32 is kernel count (3,3) is kernel dimension
model.add(tf.keras.layers.Conv2D(32, (3, 3), activation='relu',input_shape=(IMAGE_SIZE, IMAGE_SIZE, 3)))
model.add(tf.keras.layers.MaxPooling2D((2, 2)))
model.add(tf.keras.layers.Conv2D(64, (3, 3), activation='relu'))
model.add(tf.keras.layers.MaxPooling2D((2, 2)))
model.add(tf.keras.layers.Dropout(0.25))
model.add(tf.keras.layers.Conv2D(128, (3, 3), activation='relu'))
model.add(tf.keras.layers.MaxPooling2D((2, 2)))
model.add(tf.keras.layers.Dropout(0.25))
model.add(tf.keras.layers.Conv2D(128, (3, 3), activation='relu'))
model.add(tf.keras.layers.MaxPooling2D((2, 2)))
model.add(tf.keras.layers.Flatten())
# 这个是在最后部分使用dropout 部分note失活,也可以在每一层都使用dropout。dropout的目的是防止过拟合
model.add(tf.keras.layers.Dropout(0.3))
model.add(tf.keras.layers.Dense(512, activation='relu'))
model.add(tf.keras.layers.Dense(10, activation='softmax'))
model.summary()
# base_learning_rate=0.001
model.compile(loss='categorical_crossentropy',
optimizer='adam',
metrics=['acc'])
train_dir = os.path.join(base_path, 'train')
validation_dir = os.path.join(base_path, 'validation')
test_dir = os.path.join(base_path, 'test_old')
train_datagen = ImageDataGenerator(
rescale=1./255,
rotation_range=10,
width_shift_range=0.1,
height_shift_range=0.1,
# zoom_range=0.2,
# shear_range=0.2,
horizontal_flip=True,
)
test_datagen = ImageDataGenerator(rescale=1./255)
train_generator = train_datagen.flow_from_directory(
train_dir,
target_size=(IMAGE_SIZE, IMAGE_SIZE),
batch_size=BATCH_SIZE,
class_mode='categorical')
validation_generator = test_datagen.flow_from_directory(
validation_dir,
target_size=(IMAGE_SIZE, IMAGE_SIZE),
batch_size=BATCH_SIZE,
class_mode='categorical')
history = model.fit_generator(
train_generator,
steps_per_epoch=52,
epochs=50,
validation_data=validation_generator,
validation_steps=14)
model_file_name='baseline_model_2020-11-16.h5'
model.save(model_file_name)
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
plt.figure(figsize=(10,10))
ax=plt.subplot(2,1,1)
plt.plot(acc, label='Training acc')
plt.plot(val_acc, label='Validation acc')
plt.title('Training and validation accuracy')
plt.legend()
plt.ylabel("Accuracy(%)")
plt.xlabel("Epoch")
ax=plt.subplot(2,1,2)
plt.plot(loss, label='Training loss')
plt.plot(val_loss, label='Validation loss')
plt.title('Training and validation loss')
plt.legend()
plt.ylabel("LOSS")
plt.xlabel("Epoch")
plt.show()
del model
model=load_model(model_file_name)
model.summary()
test_dataset = image_dataset_from_directory(test_dir,
shuffle=True,
batch_size=BATCH_SIZE,
image_size=(IMAGE_SIZE,
IMAGE_SIZE))
images,labels=next(iter(test_dataset))
len(images),labels
predictions = model.predict_on_batch(images).flatten()
pred_result=[]
for i in range(BATCH_SIZE):
pred_result.append(predictions[i*class_num:(i+1)*class_num].argmax())
print("Predict Accuracy={}".format(accuracy_score(labels, pred_result)))
plt.figure(figsize=(15,15))
# 设定空白处 间隔大小
plt.subplots_adjust(wspace=0.4, hspace=0)
count=0
for i in range(BATCH_SIZE):
ax=plt.subplot(4,8,i+1)
plt.imshow(images[i].numpy().astype('uint8'))
plt.axis('off')
Correct_value=labels[i].numpy().astype('uint8')
Predict_value=pred_result[i]
plt.title("Correct value:{}\n Predict value:{}".format(Correct_value,Predict_value))
if Correct_value == Predict_value:
count += 1
print("Correct rate={}%".format(round(count*1.0/BATCH_SIZE*100,2)))
history_name = 'baseline_model_history.pickle'
path = os.path.join(os.getcwd(), 'baseline_model', history_name)
def pickle_dump(obj, path):
with open(path, mode='wb') as f:
pickle.dump(obj, f)
pickle_dump(history.history, path)
from PIL import Image, ImageFilter
im=Image.open('hand_sign_digit_data/test_old/4/qiu-4-1.jpg')
im1=im.resize((100,100))
im1
print(im.format, im.size, im.mode)
print(im1.format, im1.size, im1.mode)
im1_arr=np.asarray(im1)
im1_arr.shape
im1_arr=im1_arr/225.0
# print(images[0].shape)
# plt.imshow(images[0].numpy().astype('uint8'))
pred=model.predict(np.expand_dims(im1_arr,0))
pred,print(pred.argmax())
| 0.525612 | 0.729761 |
# Layer activation normalization (Batch normalization)
[](https://colab.research.google.com/github/parrt/fundamentals-of-deep-learning/blob/main/notebooks/4.batch-normalization.ipynb)
By [Terence Parr](https://explained.ai).
*In progress...can't get a good example that uses a toy data set* (I also decided this is not the right time to introduce the complexity of batch normalization.)
Just as we normalize or standardize the input variables, networks train better if we normalize the output of each layer's activation. It is called batch normalization because we normally train with batches of records not the entire data set, but it's really just allowing the model to shift and scale each neurons activation. Batch normalization does not use the joint distribution. Each neuron activation is "whitened" independently by subtracting the mean and dividing by the standard deviation. Then, we scale and shift that data using two new model parameters to support layer activation distributions that are useful for training purposes.
At test time, we use mean/var estimated during training for activations to whiten the activations as the test instance sails through the network. The learned parameters scale and shift the layer activations but do not change the shape of the distribution. This technique helps move gradients through the network during training without exploding or vanishing.
As with normalizing the input layer, batch normalization is believed to support faster learning rates for faster training. It also has a mild regularization effect per Andrew Ng, improving model generality. The model could also be less sensitive to the initialization procedure. See [Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift](https://arxiv.org/abs/1502.03167) for more information and video [Why Does Batch Norm Work?](https://www.youtube.com/watch?v=nUUqwaxLnWs).
## Support code
```
import os
import sys
import torch
import copy
import torch.nn as nn
import torch.nn.functional as F
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.datasets import load_diabetes
from sklearn.ensemble import RandomForestRegressor
import matplotlib.pyplot as plt
from matplotlib import colors
import colour
%config InlineBackend.figure_format = 'retina'
import tsensor
def plot_history(history, ax=None, maxy=None, file=None):
if ax is None:
fig, ax = plt.subplots(1,1, figsize=(3.5,3))
ax.set_ylabel("Loss")
ax.set_xlabel("Epochs")
loss = history[:,0]
val_loss = history[:,1]
if maxy:
ax.set_ylim(0,maxy)
else:
ax.set_ylim(0,torch.max(val_loss))
ax.plot(loss, label='train_loss')
ax.plot(val_loss, label='val_loss')
ax.legend(loc='upper right')
plt.tight_layout()
if file:
plt.savefig(f"/Users/{os.environ['USER']}/Desktop/{file}.pdf")
def train(model, X_train, X_test, y_train, y_test,
learning_rate = .5, nepochs=2000, weight_decay=0):
optimizer = torch.optim.Adam(model.parameters(),
lr=learning_rate, weight_decay=weight_decay)
history = [] # track training and validation loss
best_loss = 1e10
best_model = None
for epoch in range(nepochs+1):
y_pred = model(X_train)
loss = torch.mean((y_pred - y_train)**2)
y_pred_test = model(X_test)
loss_test = torch.mean((y_pred_test - y_test)**2)
history.append((loss, loss_test))
if loss_test < best_loss:
best_loss = loss_test
best_model = copy.deepcopy(model)
best_epoch = epoch
if epoch % (nepochs//10) == 0:
print(f"Epoch {epoch:4d} MSE train loss {loss:12.3f} test loss {loss_test:12.3f}")
optimizer.zero_grad()
loss.backward() # autograd computes w1.grad, b1.grad, ...
optimizer.step()
print(f"BEST MSE test loss {best_loss:.3f} at epoch {best_epoch}")
return torch.tensor(history), best_model
d = load_diabetes()
len(d.data)
df = pd.DataFrame(d.data, columns=d.feature_names)
df['disease'] = d.target # "quantitative measure of disease progression one year after baseline"
df.head(3)
```
## Split data into train, validation sets
We'll use "test" as shorthand for "validation" but technically they are not the same.
```
np.random.seed(1) # set a random seed for consistency across runs
n = len(df)
n_test = int(n*0.20) # 20% held out as validation set
n_train = n - n_test
X = df.drop('disease',axis=1).values
y = df['disease'].values
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=n_test)
m = np.mean(X_train,axis=0)
std = np.std(X_train,axis=0)
X_train = (X_train-m)/std
X_test = (X_test-m)/std # use training data only when prepping test sets
```
## Baseline with random forest
As we did in the previous notebook, let's get a baseline (although it's the same as before).
```
rf = RandomForestRegressor(n_estimators=500)#, min_samples_leaf=2, max_features=1)
rf.fit(X_train, y_train.reshape(-1))
y_pred = rf.predict(X_train)
mse = np.mean((y_pred - y_train.reshape(-1))**2)
y_pred = rf.predict(X_test)
mse_test = np.mean((y_pred - y_test.reshape(-1))**2)
print(f"Training MSE {mse:.2f} validation MSE {mse_test:.2f}")
```
## No batch normalization
We know how to train a vanilla neural network on this data, so let's do that again to reinforce our understanding and get the results into this notebook as well for comparison purposes. (We'll see how batch normalization performs after this.) Let's start by converting the numpy arrays into pytorch tensors:
```
X_train = torch.tensor(X_train).float()
X_test = torch.tensor(X_test).float()
y_train = torch.tensor(y_train).float().reshape(-1,1) # column vector
y_test = torch.tensor(y_test).float().reshape(-1,1)
ncols = X.shape[1]
n_neurons = 10
model = nn.Sequential(
nn.Linear(ncols, n_neurons),
nn.ReLU(),
nn.Linear(n_neurons, n_neurons),
nn.ReLU(),
nn.Linear(n_neurons, 1)
)
history, best_model = train(model, X_train, X_test, y_train, y_test,
learning_rate=.3, nepochs=1000,
weight_decay=0)
# verify we got the best model out
y_pred = best_model(X_test)
loss_test = torch.mean((y_pred - y_test)**2)
plot_history(torch.clamp(history, 0, 12000))
```
## Batch normalization prior to nonlinearity
```
ncols = X.shape[1]
n_neurons = 300
model = nn.Sequential(
nn.Linear(ncols, n_neurons),
nn.BatchNorm1d(n_neurons),
nn.ReLU(),
nn.Linear(n_neurons, 1)
)
history, best_model = train(model, X_train, X_test, y_train, y_test,
learning_rate=.02, nepochs=1000,
weight_decay=0)
# verify we got the best model out
y_pred = best_model(X_test)
loss_test = torch.mean((y_pred - y_test)**2)
plot_history(torch.clamp(history, 0, 12000))
```
## Batch normalization after nonlinearity
```
ncols = X.shape[1]
n_neurons = 300
model = nn.Sequential(
nn.Linear(ncols, n_neurons),
nn.ReLU(),
nn.BatchNorm1d(n_neurons),
# nn.Linear(n_neurons, n_neurons),
# nn.ReLU(),
# nn.BatchNorm1d(n_neurons),
nn.Linear(n_neurons, 1)
)
history, best_model = train(model, X_train, X_test, y_train, y_test,
learning_rate=.02, nepochs=1000,
weight_decay=0)
# verify we got the best model out
y_pred = best_model(X_test)
loss_test = torch.mean((y_pred - y_test)**2)
plot_history(torch.clamp(history, 0, 12000))
```
At least for this data set and a random train/test set, batch normalization gives us much faster convergence to the best validation loss and also we get a lower test loss:
1. No normalization: BEST MSE test loss 3073.239 at epoch 47
1. Prior to ReLU: BEST MSE test loss 2997.867 at epoch 30
1. Post ReLU: BEST MSE test loss 3073.734 at epoch 19
In the end, this data set is pretty small and perhaps not the most impressive demonstration of batch normalization, but it serves our purposes to demonstrate the technique.
See [Batch Normalization before or after ReLU?](https://www.reddit.com/r/MachineLearning/comments/67gonq/d_batch_normalization_before_or_after_relu/).
|
github_jupyter
|
import os
import sys
import torch
import copy
import torch.nn as nn
import torch.nn.functional as F
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.datasets import load_diabetes
from sklearn.ensemble import RandomForestRegressor
import matplotlib.pyplot as plt
from matplotlib import colors
import colour
%config InlineBackend.figure_format = 'retina'
import tsensor
def plot_history(history, ax=None, maxy=None, file=None):
if ax is None:
fig, ax = plt.subplots(1,1, figsize=(3.5,3))
ax.set_ylabel("Loss")
ax.set_xlabel("Epochs")
loss = history[:,0]
val_loss = history[:,1]
if maxy:
ax.set_ylim(0,maxy)
else:
ax.set_ylim(0,torch.max(val_loss))
ax.plot(loss, label='train_loss')
ax.plot(val_loss, label='val_loss')
ax.legend(loc='upper right')
plt.tight_layout()
if file:
plt.savefig(f"/Users/{os.environ['USER']}/Desktop/{file}.pdf")
def train(model, X_train, X_test, y_train, y_test,
learning_rate = .5, nepochs=2000, weight_decay=0):
optimizer = torch.optim.Adam(model.parameters(),
lr=learning_rate, weight_decay=weight_decay)
history = [] # track training and validation loss
best_loss = 1e10
best_model = None
for epoch in range(nepochs+1):
y_pred = model(X_train)
loss = torch.mean((y_pred - y_train)**2)
y_pred_test = model(X_test)
loss_test = torch.mean((y_pred_test - y_test)**2)
history.append((loss, loss_test))
if loss_test < best_loss:
best_loss = loss_test
best_model = copy.deepcopy(model)
best_epoch = epoch
if epoch % (nepochs//10) == 0:
print(f"Epoch {epoch:4d} MSE train loss {loss:12.3f} test loss {loss_test:12.3f}")
optimizer.zero_grad()
loss.backward() # autograd computes w1.grad, b1.grad, ...
optimizer.step()
print(f"BEST MSE test loss {best_loss:.3f} at epoch {best_epoch}")
return torch.tensor(history), best_model
d = load_diabetes()
len(d.data)
df = pd.DataFrame(d.data, columns=d.feature_names)
df['disease'] = d.target # "quantitative measure of disease progression one year after baseline"
df.head(3)
np.random.seed(1) # set a random seed for consistency across runs
n = len(df)
n_test = int(n*0.20) # 20% held out as validation set
n_train = n - n_test
X = df.drop('disease',axis=1).values
y = df['disease'].values
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=n_test)
m = np.mean(X_train,axis=0)
std = np.std(X_train,axis=0)
X_train = (X_train-m)/std
X_test = (X_test-m)/std # use training data only when prepping test sets
rf = RandomForestRegressor(n_estimators=500)#, min_samples_leaf=2, max_features=1)
rf.fit(X_train, y_train.reshape(-1))
y_pred = rf.predict(X_train)
mse = np.mean((y_pred - y_train.reshape(-1))**2)
y_pred = rf.predict(X_test)
mse_test = np.mean((y_pred - y_test.reshape(-1))**2)
print(f"Training MSE {mse:.2f} validation MSE {mse_test:.2f}")
X_train = torch.tensor(X_train).float()
X_test = torch.tensor(X_test).float()
y_train = torch.tensor(y_train).float().reshape(-1,1) # column vector
y_test = torch.tensor(y_test).float().reshape(-1,1)
ncols = X.shape[1]
n_neurons = 10
model = nn.Sequential(
nn.Linear(ncols, n_neurons),
nn.ReLU(),
nn.Linear(n_neurons, n_neurons),
nn.ReLU(),
nn.Linear(n_neurons, 1)
)
history, best_model = train(model, X_train, X_test, y_train, y_test,
learning_rate=.3, nepochs=1000,
weight_decay=0)
# verify we got the best model out
y_pred = best_model(X_test)
loss_test = torch.mean((y_pred - y_test)**2)
plot_history(torch.clamp(history, 0, 12000))
ncols = X.shape[1]
n_neurons = 300
model = nn.Sequential(
nn.Linear(ncols, n_neurons),
nn.BatchNorm1d(n_neurons),
nn.ReLU(),
nn.Linear(n_neurons, 1)
)
history, best_model = train(model, X_train, X_test, y_train, y_test,
learning_rate=.02, nepochs=1000,
weight_decay=0)
# verify we got the best model out
y_pred = best_model(X_test)
loss_test = torch.mean((y_pred - y_test)**2)
plot_history(torch.clamp(history, 0, 12000))
ncols = X.shape[1]
n_neurons = 300
model = nn.Sequential(
nn.Linear(ncols, n_neurons),
nn.ReLU(),
nn.BatchNorm1d(n_neurons),
# nn.Linear(n_neurons, n_neurons),
# nn.ReLU(),
# nn.BatchNorm1d(n_neurons),
nn.Linear(n_neurons, 1)
)
history, best_model = train(model, X_train, X_test, y_train, y_test,
learning_rate=.02, nepochs=1000,
weight_decay=0)
# verify we got the best model out
y_pred = best_model(X_test)
loss_test = torch.mean((y_pred - y_test)**2)
plot_history(torch.clamp(history, 0, 12000))
| 0.700485 | 0.98764 |
<a href="https://colab.research.google.com/github/mikeyy1996/Playing-with-Data/blob/master/AnalyticsVidya_JuntaHack.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Problem Statement
A training institute which conducts training for analytics/ data science wants to expand their business to manpower recruitment (data science only) as well.
Company gets large number of signups for their trainings. Now, company wants to connect these enrollees with their clients who are looking to hire employees working in the same domain. Before that, it is important to know which of these candidates are really looking for a new employment. They have student information related to demographics, education, experience and features related to training as well.
------------------------------------------------------------------------
#AIM
We need to design a model that uses the current credentials/demographics/experience to predict the probability of an enrollee to look for a new job.
------------------------------------------------------------------------
This notebook is structured as follows:
1. **Data Analysis** : In this section, we explore the dataset by taking a look at the feature distributions, how correlated one feature is to the other.
2. **Feature Engineering,Categorical Encoding and Data Balancing** : Conduct some feature engineering as well as encode all our categorical features into dummy variables
3. **Implementing Machine Learning models** : Implement machine learning models for predictions
# Data Analysis
**In this section we will take a look at train and test data, find relations between features and labels,handle missing values and try to create a balanced dataset**
```
# Importing required libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
from sklearn import preprocessing,model_selection
#reading data into dfs
data = pd.read_csv("/train.csv")
data_test = pd.read_csv("/test.csv")
data.head()
data.info()
print(data.isnull().sum())
```
**Observations**
* Our label will be 'Target'
* We have 10 categorical features which will be needed to converted into numerical through encoding.
* Lots of columns have null values as well. We need to impute the values.
**Let us now perform Chi-square to see relationship of categorical features with label.We will take the p-value as the estimate**
```
columns = ['city','gender','relevent_experience','enrolled_university','education_level','major_discipline','experience','last_new_job','company_size','company_type']
for col in columns:
cont = pd.crosstab(data['target'],data[col])
chi_val = stats.chi2_contingency(cont)
print('p-value for:',col,chi_val[1])
```
*Considering confidence interval to be 0.1; we can see major_discipline and company_size does not make for a good candidate.Also they have a lot of NULL values. Hence we will be dropping them.*
```
data = data.drop(['major_discipline','company_size'],axis = 'columns')
data_test = data_test.drop(['major_discipline','company_size'],axis = 'columns')
```
#Feature Engineering,Categorical Encoding and Data Balancing
```
print(data.isnull().sum())
```
**Gender, enrolled_university, education_level,experience,company_type ,last_new_job has missing values**
*We will be imputing the values using for different features with different methods.These are:*
* Imputing by mode
* Imputing by creating categorical bundles and assigning numerical values
We will also be performing one-hot encoding to convert the categoricl features into numerical
```
data['experience'].fillna("1",inplace = True)
data_test['experience'].fillna("1",inplace = True)
data['gender'].fillna('Male',inplace = True)
dummies_gender = pd.get_dummies(data['gender'])
data_test['gender'].fillna('Male',inplace = True)
dummies_gender_test = pd.get_dummies(data_test['gender'])
data['relevent_experience'].fillna('Has relevent experience',inplace = True)
dummies_relexp = pd.get_dummies(data['relevent_experience'])
data_test['relevent_experience'].fillna('Has relevent experience',inplace = True)
dummies_relexp_test = pd.get_dummies(data_test['relevent_experience'])
data['enrolled_university'].fillna('no_enrollment',inplace = True)
dummies_enruniv = pd.get_dummies(data['enrolled_university'])
data_test['enrolled_university'].fillna('no_enrollment',inplace = True)
dummies_enruniv_test = pd.get_dummies(data_test['enrolled_university'])
data['education_level'].fillna('Graduate',inplace = True)
dummies_edlevel = pd.get_dummies(data['education_level'])
data_test['education_level'].fillna('Graduate',inplace = True)
dummies_edlevel_test = pd.get_dummies(data_test['education_level'])
data['last_new_job'].fillna(1,inplace = True)
dummies_lastjob = pd.get_dummies(data['last_new_job'],prefix = 'last_')
data_test['last_new_job'].fillna(1,inplace = True)
dummies_lastjob_test = pd.get_dummies(data_test['last_new_job'],prefix = 'last_')
data['experience'].replace({"1":1,"2":1,"3":0,"4":0,"19":0,"20":0,">20":0,"<1":1,"5":0,"6":0,"7":0,"8":0,"9":0,"10":0,"11":0,"12":0,"13":0,"14":0,"15":0,"16":0,"17":0,"18":0},inplace = True)
data_test['experience'].replace({"1":1,"2":1,"3":0,"4":0,"19":0,"20":0,">20":0,"<1":1,"5":0,"6":0,"7":0,"8":0,"9":0,"10":0,"11":0,"12":0,"13":0,"14":0,"15":0,"16":0,"17":0,"18":0},inplace = True)
data['company_type'].fillna('Pvt Ltd',inplace = True)
dummies_ctype = pd.get_dummies(data['company_type'])
data_test['company_type'].fillna('Pvt Ltd',inplace = True)
dummies_ctype_test = pd.get_dummies(data_test['company_type'])
merged = pd.concat([data,dummies_gender,dummies_relexp,dummies_enruniv,dummies_edlevel,dummies_ctype,dummies_lastjob],axis = 'columns')
data = merged.drop(['city','gender','relevent_experience','enrolled_university','education_level','company_type','last_new_job'],axis = 'columns')
merged_test = pd.concat([data_test,dummies_gender_test,dummies_relexp_test,dummies_enruniv_test,dummies_edlevel_test,dummies_ctype_test,dummies_lastjob_test],axis = 'columns')
data_test = merged_test.drop(['city','gender','relevent_experience','enrolled_university','education_level','company_type','last_new_job'],axis = 'columns')
data.head()
data_test.head()
col_list = data.columns[1:]
col_list_test = data_test.columns[1:]
col = col_list.to_list()
col_test = col_list_test.to_list()
print(col,col_test)
scaler = preprocessing.MinMaxScaler(feature_range=(0, 1))
data[col] = scaler.fit_transform(data[col])
data = pd.DataFrame(data)
col.insert(0,'enrollee_id')
data.columns = col
data_test[col_test] = scaler.fit_transform(data_test[col_test])
data_test = pd.DataFrame(data_test)
col_test.insert(0,'enrollee_id')
data_test.columns = col_test
data.head()
```
**Let us look for imbalance in our dataset. We will calculate the unique count of label categories in the dataset **
```
print("target data:",data['target'].value_counts())
```
*Clearly there is a lot of imbalance between categories, which may cause biasing in the models. There is a need to balance the dataset.
We will use upsampling of minority class*
```
from sklearn.utils import resample
df_majority = data[data.target==0]
df_minority = data[data.target==1]
print(df_majority,df_minority)
# Upsample minority class
df_minority_upsampled = resample(df_minority,
replace=True, # sample with replacement
n_samples=15934, # to match majority class
random_state=123) # reproducible results
# Combine majority class with upsampled minority class
data = pd.concat([df_majority, df_minority_upsampled])
data.info()
```
**Now that our analysis is complete and dataset is balanced, let us move to the next step**
# Implementing Machine Learning models
*Since the parameter on which scores will be decided is roc_auc_score we will also rate the models on the same*
```
from sklearn.model_selection import train_test_split
x = data.drop(['enrollee_id','target','last__1'], axis = 'columns')
y = data['target']
x_train, x_val, y_train, y_val = train_test_split(x, y, test_size=0.2,random_state = 42,stratify = y)
print(x_train.shape, y_train.shape)
print(x_val.shape, y_val.shape)
#Logistic Regression
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import roc_curve
from sklearn.metrics import roc_auc_score
lr = LogisticRegression(C=0.001,random_state = 0)
lr.fit(x_train,y_train)
lr_probs = lr.predict_proba(x_val)
lr_probs = lr_probs[:, 1]
lr_auc = roc_auc_score(y_val, lr_probs)
lr_auc
lr_fpr, lr_tpr, _ = roc_curve(y_val, lr_probs)
plt.plot(lr_fpr, lr_tpr, marker='.', label='Logistic')
# axis labels
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
# show the legend
plt.legend()
# show the plot
#Random Forest
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier(n_estimators = 50, min_samples_leaf = 1,random_state = 0)
rf.fit(x_train,y_train)
rf_probs = rf.predict_proba(x_val)
rf_probs = rf_probs[:, 1]
rf_auc = roc_auc_score(y_val, rf_probs)
rf_auc
rf_fpr, rf_tpr, _ = roc_curve(y_val, rf_probs)
plt.plot(rf_fpr, rf_tpr, marker='.', label='Logistic')
# axis labels
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
# show the legend
plt.legend()
# show the plot
plt.show()
#Decision Tree
from sklearn.tree import DecisionTreeClassifier
dt = DecisionTreeClassifier()
dt.fit(x_train,y_train)
dt_probs = dt.predict_proba(x_val)
dt_probs = dt_probs[:, 1]
dt_auc = roc_auc_score(y_val, dt_probs)
dt_auc
dt_fpr, dt_tpr, _ = roc_curve(y_val, dt_probs)
plt.plot(dt_fpr, dt_tpr, marker='.', label='Decision Tree')
# axis labels
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
# show the legend
plt.legend()
# show the plot
plt.show()
from sklearn.naive_bayes import BernoulliNB
bnb = BernoulliNB(binarize=0.55)
bnb.fit(x_train,y_train)
bnb_probs = bnb.predict_proba(x_val)
bnb_probs = bnb_probs[:, 1]
bnb_auc = roc_auc_score(y_val, bnb_probs)
bnb_auc
bnb_fpr, bnb_tpr, _ = roc_curve(y_val, bnb_probs)
plt.plot(bnb_fpr, bnb_tpr, marker='.', label='Bernoulli')
# axis labels
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
# show the legend
plt.legend()
# show the plot
plt.show()
from sklearn.svm import SVC
svc = SVC(probability=True,tol = 0.01)
svc.fit(x_train,y_train)
svc_probs = svc.predict_proba(x_val)
svc_probs = svc_probs[:, 1]
svc_auc = roc_auc_score(y_val, svc_probs)
svc_auc
svc_fpr, svc_tpr, _ = roc_curve(y_val, svc_probs)
plt.plot(svc_fpr, svc_tpr, marker='.', label='Support Vector')
# axis labels
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
# show the legend
plt.legend()
# show the plot
plt.show()
from sklearn.neural_network import MLPClassifier
p = MLPClassifier(random_state=42,
max_iter=100,tol = 0.0001)
p.fit(x_train,y_train)
p_probs = p.predict_proba(x_val)
p_probs = p_probs[:, 1]
p_auc = roc_auc_score(y_val, p_probs)
p_auc
p_fpr, p_tpr, _ = roc_curve(y_val, p_probs)
plt.plot(p_fpr, p_tpr, marker='.', label='Perceptron')
# axis labels
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
# show the legend
plt.legend()
# show the plot
plt.show()
```
**Let's consolidate the scores of each model**
```
auc_scores = [lr_auc,rf_auc,bnb_auc,svc_auc,dt_auc,p_auc]
models =['Logistic Regression','Random Forest','Bernoulli NaiveBayes','Support Vector','Decision Tree','MultiLayerPerceptron']
print("=====================ROC_AUC Scores=========================")
for score,model in zip(auc_scores,models):
print(model,": ",score)
```
*RandomForest and DecisionTree have exceptionally high scores, this maybe because of overfitting*
Let us now predict our target and consolidate the final scores
# Predicting Test dataset
```
data_test.head()
x_test = data_test.drop(['enrollee_id','last__1'], axis = 'columns')
pred_lr = lr.predict(x_test)
unique_elements, counts_elements = np.unique(pred_lr, return_counts=True)
print(unique_elements,counts_elements)
pred_rf = rf.predict(x_test)
unique_elements, counts_elements = np.unique(pred_rf, return_counts=True)
print(unique_elements,counts_elements)
pred_dt = dt.predict(x_test)
unique_elements, counts_elements = np.unique(pred_dt, return_counts=True)
print(unique_elements,counts_elements)
pred_svc = svc.predict(x_test)
unique_elements, counts_elements = np.unique(pred_svc, return_counts=True)
print(unique_elements,counts_elements)
pred_bnb = bnb.predict(x_test)
unique_elements, counts_elements = np.unique(pred_bnb, return_counts=True)
print(unique_elements,counts_elements)
pred_p = p.predict(x_test)
unique_elements, counts_elements = np.unique(pred_p, return_counts=True)
print(unique_elements,counts_elements)
pd.DataFrame(pred_lr, columns=['target']).to_csv('/prediction_lr.csv')
pd.DataFrame(pred_xgb, columns=['target']).to_csv('/prediction_rf.csv')
pd.DataFrame(pred_dt, columns=['target']).to_csv('/prediction_dt.csv')
pd.DataFrame(pred_svc, columns=['target']).to_csv('/prediction_svc.csv')
pd.DataFrame(pred_bnb, columns=['target']).to_csv('/prediction_bnb.csv')
pd.DataFrame(pred_p, columns=['target']).to_csv('/prediction_p.csv')
auc_scores = [0.5957,0.5162,0.5950,0.6008,0.5168,0.6133]
models =['Logistic Regression','Random Forest','Bernoulli NaiveBayes','Support Vector','Decision Tree','MultiLayerPerceptron']
print("=====================Final Scores=========================")
for score,model in zip(auc_scores,models):
print(model,": ",score)
```
**As we assumed, the models with abnormally high score training(Random Forest,Decision Tree) performed the worst on test set. This is because of overfitting.**
Here is a link explaining what is overfitting and how it can be reduced:
[Overfit and Underfit in ML](https://towardsdatascience.com/what-are-overfitting-and-underfitting-in-machine-learning-a96b30864690)
|
github_jupyter
|
# Importing required libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
from sklearn import preprocessing,model_selection
#reading data into dfs
data = pd.read_csv("/train.csv")
data_test = pd.read_csv("/test.csv")
data.head()
data.info()
print(data.isnull().sum())
columns = ['city','gender','relevent_experience','enrolled_university','education_level','major_discipline','experience','last_new_job','company_size','company_type']
for col in columns:
cont = pd.crosstab(data['target'],data[col])
chi_val = stats.chi2_contingency(cont)
print('p-value for:',col,chi_val[1])
data = data.drop(['major_discipline','company_size'],axis = 'columns')
data_test = data_test.drop(['major_discipline','company_size'],axis = 'columns')
print(data.isnull().sum())
data['experience'].fillna("1",inplace = True)
data_test['experience'].fillna("1",inplace = True)
data['gender'].fillna('Male',inplace = True)
dummies_gender = pd.get_dummies(data['gender'])
data_test['gender'].fillna('Male',inplace = True)
dummies_gender_test = pd.get_dummies(data_test['gender'])
data['relevent_experience'].fillna('Has relevent experience',inplace = True)
dummies_relexp = pd.get_dummies(data['relevent_experience'])
data_test['relevent_experience'].fillna('Has relevent experience',inplace = True)
dummies_relexp_test = pd.get_dummies(data_test['relevent_experience'])
data['enrolled_university'].fillna('no_enrollment',inplace = True)
dummies_enruniv = pd.get_dummies(data['enrolled_university'])
data_test['enrolled_university'].fillna('no_enrollment',inplace = True)
dummies_enruniv_test = pd.get_dummies(data_test['enrolled_university'])
data['education_level'].fillna('Graduate',inplace = True)
dummies_edlevel = pd.get_dummies(data['education_level'])
data_test['education_level'].fillna('Graduate',inplace = True)
dummies_edlevel_test = pd.get_dummies(data_test['education_level'])
data['last_new_job'].fillna(1,inplace = True)
dummies_lastjob = pd.get_dummies(data['last_new_job'],prefix = 'last_')
data_test['last_new_job'].fillna(1,inplace = True)
dummies_lastjob_test = pd.get_dummies(data_test['last_new_job'],prefix = 'last_')
data['experience'].replace({"1":1,"2":1,"3":0,"4":0,"19":0,"20":0,">20":0,"<1":1,"5":0,"6":0,"7":0,"8":0,"9":0,"10":0,"11":0,"12":0,"13":0,"14":0,"15":0,"16":0,"17":0,"18":0},inplace = True)
data_test['experience'].replace({"1":1,"2":1,"3":0,"4":0,"19":0,"20":0,">20":0,"<1":1,"5":0,"6":0,"7":0,"8":0,"9":0,"10":0,"11":0,"12":0,"13":0,"14":0,"15":0,"16":0,"17":0,"18":0},inplace = True)
data['company_type'].fillna('Pvt Ltd',inplace = True)
dummies_ctype = pd.get_dummies(data['company_type'])
data_test['company_type'].fillna('Pvt Ltd',inplace = True)
dummies_ctype_test = pd.get_dummies(data_test['company_type'])
merged = pd.concat([data,dummies_gender,dummies_relexp,dummies_enruniv,dummies_edlevel,dummies_ctype,dummies_lastjob],axis = 'columns')
data = merged.drop(['city','gender','relevent_experience','enrolled_university','education_level','company_type','last_new_job'],axis = 'columns')
merged_test = pd.concat([data_test,dummies_gender_test,dummies_relexp_test,dummies_enruniv_test,dummies_edlevel_test,dummies_ctype_test,dummies_lastjob_test],axis = 'columns')
data_test = merged_test.drop(['city','gender','relevent_experience','enrolled_university','education_level','company_type','last_new_job'],axis = 'columns')
data.head()
data_test.head()
col_list = data.columns[1:]
col_list_test = data_test.columns[1:]
col = col_list.to_list()
col_test = col_list_test.to_list()
print(col,col_test)
scaler = preprocessing.MinMaxScaler(feature_range=(0, 1))
data[col] = scaler.fit_transform(data[col])
data = pd.DataFrame(data)
col.insert(0,'enrollee_id')
data.columns = col
data_test[col_test] = scaler.fit_transform(data_test[col_test])
data_test = pd.DataFrame(data_test)
col_test.insert(0,'enrollee_id')
data_test.columns = col_test
data.head()
print("target data:",data['target'].value_counts())
from sklearn.utils import resample
df_majority = data[data.target==0]
df_minority = data[data.target==1]
print(df_majority,df_minority)
# Upsample minority class
df_minority_upsampled = resample(df_minority,
replace=True, # sample with replacement
n_samples=15934, # to match majority class
random_state=123) # reproducible results
# Combine majority class with upsampled minority class
data = pd.concat([df_majority, df_minority_upsampled])
data.info()
from sklearn.model_selection import train_test_split
x = data.drop(['enrollee_id','target','last__1'], axis = 'columns')
y = data['target']
x_train, x_val, y_train, y_val = train_test_split(x, y, test_size=0.2,random_state = 42,stratify = y)
print(x_train.shape, y_train.shape)
print(x_val.shape, y_val.shape)
#Logistic Regression
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import roc_curve
from sklearn.metrics import roc_auc_score
lr = LogisticRegression(C=0.001,random_state = 0)
lr.fit(x_train,y_train)
lr_probs = lr.predict_proba(x_val)
lr_probs = lr_probs[:, 1]
lr_auc = roc_auc_score(y_val, lr_probs)
lr_auc
lr_fpr, lr_tpr, _ = roc_curve(y_val, lr_probs)
plt.plot(lr_fpr, lr_tpr, marker='.', label='Logistic')
# axis labels
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
# show the legend
plt.legend()
# show the plot
#Random Forest
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier(n_estimators = 50, min_samples_leaf = 1,random_state = 0)
rf.fit(x_train,y_train)
rf_probs = rf.predict_proba(x_val)
rf_probs = rf_probs[:, 1]
rf_auc = roc_auc_score(y_val, rf_probs)
rf_auc
rf_fpr, rf_tpr, _ = roc_curve(y_val, rf_probs)
plt.plot(rf_fpr, rf_tpr, marker='.', label='Logistic')
# axis labels
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
# show the legend
plt.legend()
# show the plot
plt.show()
#Decision Tree
from sklearn.tree import DecisionTreeClassifier
dt = DecisionTreeClassifier()
dt.fit(x_train,y_train)
dt_probs = dt.predict_proba(x_val)
dt_probs = dt_probs[:, 1]
dt_auc = roc_auc_score(y_val, dt_probs)
dt_auc
dt_fpr, dt_tpr, _ = roc_curve(y_val, dt_probs)
plt.plot(dt_fpr, dt_tpr, marker='.', label='Decision Tree')
# axis labels
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
# show the legend
plt.legend()
# show the plot
plt.show()
from sklearn.naive_bayes import BernoulliNB
bnb = BernoulliNB(binarize=0.55)
bnb.fit(x_train,y_train)
bnb_probs = bnb.predict_proba(x_val)
bnb_probs = bnb_probs[:, 1]
bnb_auc = roc_auc_score(y_val, bnb_probs)
bnb_auc
bnb_fpr, bnb_tpr, _ = roc_curve(y_val, bnb_probs)
plt.plot(bnb_fpr, bnb_tpr, marker='.', label='Bernoulli')
# axis labels
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
# show the legend
plt.legend()
# show the plot
plt.show()
from sklearn.svm import SVC
svc = SVC(probability=True,tol = 0.01)
svc.fit(x_train,y_train)
svc_probs = svc.predict_proba(x_val)
svc_probs = svc_probs[:, 1]
svc_auc = roc_auc_score(y_val, svc_probs)
svc_auc
svc_fpr, svc_tpr, _ = roc_curve(y_val, svc_probs)
plt.plot(svc_fpr, svc_tpr, marker='.', label='Support Vector')
# axis labels
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
# show the legend
plt.legend()
# show the plot
plt.show()
from sklearn.neural_network import MLPClassifier
p = MLPClassifier(random_state=42,
max_iter=100,tol = 0.0001)
p.fit(x_train,y_train)
p_probs = p.predict_proba(x_val)
p_probs = p_probs[:, 1]
p_auc = roc_auc_score(y_val, p_probs)
p_auc
p_fpr, p_tpr, _ = roc_curve(y_val, p_probs)
plt.plot(p_fpr, p_tpr, marker='.', label='Perceptron')
# axis labels
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
# show the legend
plt.legend()
# show the plot
plt.show()
auc_scores = [lr_auc,rf_auc,bnb_auc,svc_auc,dt_auc,p_auc]
models =['Logistic Regression','Random Forest','Bernoulli NaiveBayes','Support Vector','Decision Tree','MultiLayerPerceptron']
print("=====================ROC_AUC Scores=========================")
for score,model in zip(auc_scores,models):
print(model,": ",score)
data_test.head()
x_test = data_test.drop(['enrollee_id','last__1'], axis = 'columns')
pred_lr = lr.predict(x_test)
unique_elements, counts_elements = np.unique(pred_lr, return_counts=True)
print(unique_elements,counts_elements)
pred_rf = rf.predict(x_test)
unique_elements, counts_elements = np.unique(pred_rf, return_counts=True)
print(unique_elements,counts_elements)
pred_dt = dt.predict(x_test)
unique_elements, counts_elements = np.unique(pred_dt, return_counts=True)
print(unique_elements,counts_elements)
pred_svc = svc.predict(x_test)
unique_elements, counts_elements = np.unique(pred_svc, return_counts=True)
print(unique_elements,counts_elements)
pred_bnb = bnb.predict(x_test)
unique_elements, counts_elements = np.unique(pred_bnb, return_counts=True)
print(unique_elements,counts_elements)
pred_p = p.predict(x_test)
unique_elements, counts_elements = np.unique(pred_p, return_counts=True)
print(unique_elements,counts_elements)
pd.DataFrame(pred_lr, columns=['target']).to_csv('/prediction_lr.csv')
pd.DataFrame(pred_xgb, columns=['target']).to_csv('/prediction_rf.csv')
pd.DataFrame(pred_dt, columns=['target']).to_csv('/prediction_dt.csv')
pd.DataFrame(pred_svc, columns=['target']).to_csv('/prediction_svc.csv')
pd.DataFrame(pred_bnb, columns=['target']).to_csv('/prediction_bnb.csv')
pd.DataFrame(pred_p, columns=['target']).to_csv('/prediction_p.csv')
auc_scores = [0.5957,0.5162,0.5950,0.6008,0.5168,0.6133]
models =['Logistic Regression','Random Forest','Bernoulli NaiveBayes','Support Vector','Decision Tree','MultiLayerPerceptron']
print("=====================Final Scores=========================")
for score,model in zip(auc_scores,models):
print(model,": ",score)
| 0.337204 | 0.967163 |
# Exploring Cartesian Product
First, include some libraries
```
# Run boilerplate code to set up environment
%run ../prelude.py
```
## Z_m = A_mk * B_mk
#### Load basic tensors
```
As = Tensor.fromUncompressed(["M", "K"], [[1, 0, 1, 1], [0, 0, 0, 0], [0, 0, 3, 3], [0, 0, 0, 0]])
Bs = Tensor.fromUncompressed(["M", "K"], [[1, 0, 0, 1], [0, 0, 0, 0], [0, 0, 0, 3], [4, 0, 4, 0]])
Zs_verify = Tensor.fromUncompressed(["M"], [2, 0, 9, 0])
```
#### Untiled Baseline Traversal
```
Zs = Tensor(rank_ids = ["M"])
as_m = As.getRoot()
bs_m = Bs.getRoot()
zs_m = Zs.getRoot()
canvas = createCanvas(As, Bs, Zs)
for m, (z, (as_k, bs_k)) in zs_m << (as_m & bs_m):
for k, (a, b) in as_k & bs_k:
z += a * b
canvas.addFrame((m, k), (m, k), (m,))
displayCanvas(canvas, width="50%")
```
#### Verify Result
```
Zs_verify == Zs
```
## Tiled K -- Offline
#### Load pre-tiled tensors
```
K0 = 2
As_tiled_K = Tensor.fromUncompressed(["K1", "M", "K0"], [[[1, 0], [0, 0], [0, 0], [0, 0]], [[1, 1], [0, 0], [3, 3], [0, 0]]])
Bs_tiled_K = Tensor.fromUncompressed(["K1", "M", "K0"], [[[1, 0], [0, 0], [0, 0], [4, 0]], [[0, 1], [0, 0], [0, 3], [4, 0]]])
```
#### Tiled traversal
```
Zs = Tensor(rank_ids = ["M"])
as_k1 = As_tiled_K.getRoot()
bs_k1 = Bs_tiled_K.getRoot()
zs_m = Zs.getRoot()
canvas = createCanvas(As_tiled_K, Bs_tiled_K, Zs)
for k1, (as_m, bs_m) in as_k1 & bs_k1:
for m, (z, (as_k0, bs_k0)) in zs_m << (as_m & bs_m):
for k0, (a, b) in as_k0 & bs_k0:
z += a * b
canvas.addFrame((k1, m, k0), (k1, m, k0), (m,))
displayCanvas(canvas, width="50%")
```
#### Verify result
```
Zs_verify == Zs
```
## Tiled K -- Online, Monolithic, Separate
#### Define tensors for online tiling
```
As_tiled_K = Tensor(rank_ids = ["K1", "M", "K0"])
Bs_tiled_K = Tensor(rank_ids = ["K1", "M", "K0"])
Zs = Tensor(rank_ids = ["M"])
K0 = 2
```
#### Tile Tensor A
```
Zs = Tensor(rank_ids = ["M"])
canvas = createCanvas(As, As_tiled_K)
as_m = As.getRoot()
as_tiled_k1 = As_tiled_K.getRoot()
for (m, as_k) in as_m:
for (k, a) in as_k:
k1 = k // K0
k0 = k % K0
as_tiled_k0 = as_tiled_k1.getPayloadRef(k1, m, k0)
as_tiled_k0 <<= a
canvas.addFrame((m, k), (k1, m, k0))
print(As_tiled_K.getRoot() == as_tiled_k1)
displayCanvas(canvas, width="50%")
```
#### Tile Tensor B
```
canvas = createCanvas(Bs, Bs_tiled_K)
bs_m = Bs.getRoot()
bs_tiled_k1 = Bs_tiled_K.getRoot()
for (m, bs_k) in bs_m:
for (k, b) in bs_k:
k1 = k // K0
k0 = k % K0
bs_tiled_k0 = bs_tiled_k1.getPayloadRef(k1, m, k0)
bs_tiled_k0 <<= b
canvas.addFrame((m, k), (k1, m, k0))
print(Bs_tiled_K.getRoot() == bs_tiled_k1)
displayCanvas(canvas, width="50%")
```
#### Tiled Traversal
```
Zs = Tensor(rank_ids = ["M"])
as_k1 = As_tiled_K.getRoot()
bs_k1 = Bs_tiled_K.getRoot()
zs_m = Zs.getRoot()
canvas = createCanvas(As_tiled_K, Bs_tiled_K, Zs)
for k1, (as_m, bs_m) in as_k1 & bs_k1:
for m, (z, (as_k0, bs_k0)) in zs_m << (as_m & bs_m):
for k0, (a, b) in as_k0 & bs_k0:
z += a * b
canvas.addFrame((k1, m, k0), (k1, m, k0), (m,))
displayCanvas(canvas, width="50%")
```
#### Verify Result
```
Zs_verify == Zs
```
### Tiled K -- Online, Monolithic, Combined
#### Define tensors for online tiling (post-intersection)
```
ABs_tiled_K = Tensor(rank_ids = ["K1", "M", "K0"])
Zs = Tensor(rank_ids = ["M"])
K0 = 2
```
#### Co-Tile A and B
```
canvas = createCanvas(As, Bs, ABs_tiled_K)
as_m = As.getRoot()
bs_m = Bs.getRoot()
abs_tiled_k1 = ABs_tiled_K.getRoot()
for m, (as_k, bs_k) in as_m & bs_m:
for k, (a, b) in as_k & bs_k:
k1 = k // K0
k0 = k % K0
#print("Inserting ({}, {}) as ({}, {}, {})".format(m,k,k1,m,k0))
abs_tiled_k0 = abs_tiled_k1.getPayloadRef(k1, m, k0)
abs_tiled_k0 <<= a*b
canvas.addFrame((m, k), (m, k), (k1, m, k0))
displayCanvas(canvas, width="50%")
```
#### Tiled Traversal
```
Zs = Tensor(rank_ids = ["M"])
canvas = createCanvas(ABs_tiled_K, Zs)
abs_tiled_k1 = ABs_tiled_K.getRoot()
zs_m = Zs.getRoot()
# NOTE: Worker loop no longer contains intersections!
for k1, abs_tiled_m in abs_tiled_k1:
for m, (z, abs_tiled_k0) in zs_m << abs_tiled_m:
for k0, ab in abs_tiled_k0:
z += ab
canvas.addFrame((k1, m, k0), (m,))
displayCanvas(canvas, width="50%")
```
#### Verify Result
```
Zs_verify == Zs
```
## Tiled -- Online, Incremental, Combined
#### Define workspace and current positions tensors
```
K0 = 2
K1 = 2 # XXX MAGIC FOR NOW
workspace = Tensor(rank_ids = ["K0"])
current_positions = Tensor(rank_ids = ["M"])
current_positions.setDefault((0,0))
Zs = Tensor(rank_ids = ["M"])
```
### Traverse and Tile Simultaneously
```
canvas = createCanvas(As, Bs, workspace, Zs)
as_m = As.getRoot()
bs_m = Bs.getRoot()
zs_m = Zs.getRoot()
workspace_k0 = workspace.getRoot()
current_positions_m = current_positions.getRoot()
for k1 in range(K1): # TODO: improve this outer loop
for m, (z, (pos_ref, (as_k, bs_k))) in zs_m << (current_positions_m << (as_m & bs_m)):
workspace_k0.clear()
# Get the starting positions
(a_pos, b_pos) = pos_ref
as_k0 = as_k.getRange(k1 * K0, K0, start_pos = a_pos)
bs_k0 = bs_k.getRange(k1 * K0, K0, start_pos = b_pos)
# Update the starting positions
pos_ref <<= (as_k.getSavedPos(), bs_k.getSavedPos())
# Tiling loop (with multiplication)
for k, (a, b) in as_k0 & bs_k0:
workspace.getRoot().append(k // K0, a * b)
print("Inserting ({}, {})".format(m, k))
# Reduction and update loop
for k0, ab in workspace_k0:
z += ab
canvas.addFrame((m, k), (m, k), (k,), (m,))
print("Working on ({}, {}, {})".format(k1, m, k0))
displayCanvas(canvas, width="50%")
```
#### Verify result
```
Zs_verify == Zs
```
## Testing area
For running alternative algorithms
|
github_jupyter
|
# Run boilerplate code to set up environment
%run ../prelude.py
As = Tensor.fromUncompressed(["M", "K"], [[1, 0, 1, 1], [0, 0, 0, 0], [0, 0, 3, 3], [0, 0, 0, 0]])
Bs = Tensor.fromUncompressed(["M", "K"], [[1, 0, 0, 1], [0, 0, 0, 0], [0, 0, 0, 3], [4, 0, 4, 0]])
Zs_verify = Tensor.fromUncompressed(["M"], [2, 0, 9, 0])
Zs = Tensor(rank_ids = ["M"])
as_m = As.getRoot()
bs_m = Bs.getRoot()
zs_m = Zs.getRoot()
canvas = createCanvas(As, Bs, Zs)
for m, (z, (as_k, bs_k)) in zs_m << (as_m & bs_m):
for k, (a, b) in as_k & bs_k:
z += a * b
canvas.addFrame((m, k), (m, k), (m,))
displayCanvas(canvas, width="50%")
Zs_verify == Zs
K0 = 2
As_tiled_K = Tensor.fromUncompressed(["K1", "M", "K0"], [[[1, 0], [0, 0], [0, 0], [0, 0]], [[1, 1], [0, 0], [3, 3], [0, 0]]])
Bs_tiled_K = Tensor.fromUncompressed(["K1", "M", "K0"], [[[1, 0], [0, 0], [0, 0], [4, 0]], [[0, 1], [0, 0], [0, 3], [4, 0]]])
Zs = Tensor(rank_ids = ["M"])
as_k1 = As_tiled_K.getRoot()
bs_k1 = Bs_tiled_K.getRoot()
zs_m = Zs.getRoot()
canvas = createCanvas(As_tiled_K, Bs_tiled_K, Zs)
for k1, (as_m, bs_m) in as_k1 & bs_k1:
for m, (z, (as_k0, bs_k0)) in zs_m << (as_m & bs_m):
for k0, (a, b) in as_k0 & bs_k0:
z += a * b
canvas.addFrame((k1, m, k0), (k1, m, k0), (m,))
displayCanvas(canvas, width="50%")
Zs_verify == Zs
As_tiled_K = Tensor(rank_ids = ["K1", "M", "K0"])
Bs_tiled_K = Tensor(rank_ids = ["K1", "M", "K0"])
Zs = Tensor(rank_ids = ["M"])
K0 = 2
Zs = Tensor(rank_ids = ["M"])
canvas = createCanvas(As, As_tiled_K)
as_m = As.getRoot()
as_tiled_k1 = As_tiled_K.getRoot()
for (m, as_k) in as_m:
for (k, a) in as_k:
k1 = k // K0
k0 = k % K0
as_tiled_k0 = as_tiled_k1.getPayloadRef(k1, m, k0)
as_tiled_k0 <<= a
canvas.addFrame((m, k), (k1, m, k0))
print(As_tiled_K.getRoot() == as_tiled_k1)
displayCanvas(canvas, width="50%")
canvas = createCanvas(Bs, Bs_tiled_K)
bs_m = Bs.getRoot()
bs_tiled_k1 = Bs_tiled_K.getRoot()
for (m, bs_k) in bs_m:
for (k, b) in bs_k:
k1 = k // K0
k0 = k % K0
bs_tiled_k0 = bs_tiled_k1.getPayloadRef(k1, m, k0)
bs_tiled_k0 <<= b
canvas.addFrame((m, k), (k1, m, k0))
print(Bs_tiled_K.getRoot() == bs_tiled_k1)
displayCanvas(canvas, width="50%")
Zs = Tensor(rank_ids = ["M"])
as_k1 = As_tiled_K.getRoot()
bs_k1 = Bs_tiled_K.getRoot()
zs_m = Zs.getRoot()
canvas = createCanvas(As_tiled_K, Bs_tiled_K, Zs)
for k1, (as_m, bs_m) in as_k1 & bs_k1:
for m, (z, (as_k0, bs_k0)) in zs_m << (as_m & bs_m):
for k0, (a, b) in as_k0 & bs_k0:
z += a * b
canvas.addFrame((k1, m, k0), (k1, m, k0), (m,))
displayCanvas(canvas, width="50%")
Zs_verify == Zs
ABs_tiled_K = Tensor(rank_ids = ["K1", "M", "K0"])
Zs = Tensor(rank_ids = ["M"])
K0 = 2
canvas = createCanvas(As, Bs, ABs_tiled_K)
as_m = As.getRoot()
bs_m = Bs.getRoot()
abs_tiled_k1 = ABs_tiled_K.getRoot()
for m, (as_k, bs_k) in as_m & bs_m:
for k, (a, b) in as_k & bs_k:
k1 = k // K0
k0 = k % K0
#print("Inserting ({}, {}) as ({}, {}, {})".format(m,k,k1,m,k0))
abs_tiled_k0 = abs_tiled_k1.getPayloadRef(k1, m, k0)
abs_tiled_k0 <<= a*b
canvas.addFrame((m, k), (m, k), (k1, m, k0))
displayCanvas(canvas, width="50%")
Zs = Tensor(rank_ids = ["M"])
canvas = createCanvas(ABs_tiled_K, Zs)
abs_tiled_k1 = ABs_tiled_K.getRoot()
zs_m = Zs.getRoot()
# NOTE: Worker loop no longer contains intersections!
for k1, abs_tiled_m in abs_tiled_k1:
for m, (z, abs_tiled_k0) in zs_m << abs_tiled_m:
for k0, ab in abs_tiled_k0:
z += ab
canvas.addFrame((k1, m, k0), (m,))
displayCanvas(canvas, width="50%")
Zs_verify == Zs
K0 = 2
K1 = 2 # XXX MAGIC FOR NOW
workspace = Tensor(rank_ids = ["K0"])
current_positions = Tensor(rank_ids = ["M"])
current_positions.setDefault((0,0))
Zs = Tensor(rank_ids = ["M"])
canvas = createCanvas(As, Bs, workspace, Zs)
as_m = As.getRoot()
bs_m = Bs.getRoot()
zs_m = Zs.getRoot()
workspace_k0 = workspace.getRoot()
current_positions_m = current_positions.getRoot()
for k1 in range(K1): # TODO: improve this outer loop
for m, (z, (pos_ref, (as_k, bs_k))) in zs_m << (current_positions_m << (as_m & bs_m)):
workspace_k0.clear()
# Get the starting positions
(a_pos, b_pos) = pos_ref
as_k0 = as_k.getRange(k1 * K0, K0, start_pos = a_pos)
bs_k0 = bs_k.getRange(k1 * K0, K0, start_pos = b_pos)
# Update the starting positions
pos_ref <<= (as_k.getSavedPos(), bs_k.getSavedPos())
# Tiling loop (with multiplication)
for k, (a, b) in as_k0 & bs_k0:
workspace.getRoot().append(k // K0, a * b)
print("Inserting ({}, {})".format(m, k))
# Reduction and update loop
for k0, ab in workspace_k0:
z += ab
canvas.addFrame((m, k), (m, k), (k,), (m,))
print("Working on ({}, {}, {})".format(k1, m, k0))
displayCanvas(canvas, width="50%")
Zs_verify == Zs
| 0.268078 | 0.914176 |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
from tubesml.base import BaseTransformer, self_columns, reset_columns
import tubesml as tml
from sklearn.pipeline import Pipeline
from sklearn.decomposition import PCA
from sklearn.preprocessing import LabelEncoder
from source import explore as ex
from source import utility as ut
pd.set_option('max_columns', 100)
df_train = pd.read_csv('data/train.csv')
df_test = pd.read_csv('data/test.csv')
subs = pd.read_csv('data/sample_submission.csv')
train_set, test_set = ut.make_test(df_train, 0.25, random_state=516, strat_feat='cat9')
```
# PCA
```
class PCADf(BaseTransformer):
def __init__(self, n_components, svd_solver='auto', random_state=24, compress=False):
super().__init__()
self.n_components = n_components
self.svd_solver = svd_solver
self.n_components_ = 0
self.random_state = random_state
self.PCA = PCA(n_components=self.n_components, svd_solver=self.svd_solver, random_state=self.random_state)
self.compress = compress
self.original_columns = []
@reset_columns
def fit(self, X, y=None):
self.PCA.fit(X)
self.n_components_ = self.PCA.n_components_
return self
@self_columns
def transform(self, X, y=None):
X_tr = self.PCA.transform(X)
X_tr = pd.DataFrame(X_tr, columns=[f'pca_{i}' for i in range(self.n_components_)])
self.original_columns = X.columns
if self.compress:
X_tr = self.inverse_transform(X_tr)
return X_tr
def inverse_transform(self, X, y=None):
try:
X_tr = self.PCA.inverse_transform(X)
except ValueError:
return X
X_tr = pd.DataFrame(X_tr, columns=self.original_columns)
return X_tr
mock_pipe = Pipeline([('dums', tml.Dummify(drop_first=True)),
('pca', PCADf(n_components=0.9))])
tmp = train_set.copy().drop(['id', 'target'], axis=1)
mock_pipe.fit(tmp)
mock_pipe.transform(tmp).head()
mock_pipe = Pipeline([('dums', tml.Dummify(drop_first=True)),
('pca', PCADf(n_components=60))])
tmp = train_set.copy().drop(['id', 'target'], axis=1)
mock_pipe.fit(tmp)
plt.plot(np.cumsum(mock_pipe.steps[-1][1].PCA.explained_variance_ratio_))
plt.show()
```
# Encoders
```
class TargetEncoder(BaseTransformer):
# Heavily inspired by
# https://github.com/MaxHalford/xam/blob/93c066990d976c7d4d74b63fb6fb3254ee8d9b48/xam/feature_extraction/encoding/bayesian_target.py#L8
def __init__(self, to_encode=None, prior_weight=100, agg_func='mean'):
super().__init__()
if isinstance(to_encode, str):
self.to_encode = [to_encode]
else:
self.to_encode = to_encode
self.prior_weight = prior_weight
self.prior_ = None
self.posteriors_ = None
self.agg_func = agg_func
@reset_columns
def fit(self, X, y):
# Encode all categorical cols by default
if self.to_encode is None:
self.to_encode = [c for c in X if str(X[c].dtype)=='object' or str(X[c].dtype)=='category']
tmp = X.copy()
tmp['target'] = y
self.prior_ = tmp['target'].agg(self.agg_func)
self.posteriors_ = {}
for col in self.to_encode:
agg = tmp.groupby(col)['target'].agg(['count', self.agg_func])
counts = agg['count']
data = agg[self.agg_func]
pw = self.prior_weight
self.posteriors_[col] = ((pw * self.prior_ + counts * data) / (pw + counts)).to_dict()
del tmp
return self
@self_columns
def transform(self, X, y=None):
X_tr = X.copy()
for col in self.to_encode:
X_tr[col] = X_tr[col].map(self.posteriors_[col]).fillna(self.prior_).astype(float)
return X_tr
tmp = train_set.copy().drop(['id', 'target'], axis=1)
trs = TargetEncoder(to_encode=None)
trs.fit_transform(tmp, train_set['target']).head()
tmp = train_set.copy().drop(['id', 'target'], axis=1)
trs = TargetEncoder(to_encode=None, agg_func='skew')
trs.fit_transform(tmp, train_set['target']).head()
tmp = train_set.copy().drop(['id', 'target'], axis=1)
trs = TargetEncoder(to_encode=None, agg_func='std')
trs.fit_transform(tmp, train_set['target']).head()
tmp = train_set[[col for col in train_set if 'cat' in col]]
tmp.head()
le = LabelEncoder()
le.fit_transform(tmp['cat0'])
class CatSimp(BaseTransformer):
def __init__(self, cat7=True, cat6=True, cat8=True, cat4=True, cat9=True):
super().__init__()
self.cat7 = cat7
self.cat6 = cat6
self.cat8 = cat8
self.cat4 = cat4
self.cat9 = cat9
def cat7_tr(self, X):
X_tr = X.copy()
if self.cat7:
X_tr['cat7'] = X_tr['cat7'].map({'C': 'E',
'A': 'B',
'F': 'G',
'I': 'G'}).fillna(X_tr['cat7'])
return X_tr
def cat6_tr(self, X):
X_tr = X.copy()
if self.cat6:
X_tr.loc[X_tr['cat6'] != 'A', 'cat6'] = 'B'
return X_tr
def cat8_tr(self, X):
X_tr = X.copy()
if self.cat8:
X_tr['cat8'] = X_tr['cat8'].map({'B': 'E', 'F': 'E'}).fillna(X_tr['cat8'])
return X_tr
def cat4_tr(self, X):
X_tr = X.copy()
if self.cat4:
X_tr['cat4'] = X_tr['cat4'].map({'D': 'A'}).fillna(X_tr['cat4'])
return X_tr
def cat9_tr(self, X):
X_tr = X.copy()
if self.cat9:
X_tr['cat9'] = X_tr['cat9'].map({'E': 'L', 'D': 'J', 'C': 'L'}).fillna(X_tr['cat9'])
return X_tr
@self_columns
def transform(self, X, y=None):
Xtransf = self.cat7_tr(X)
Xtransf = self.cat6_tr(Xtransf)
Xtransf = self.cat8_tr(Xtransf)
Xtransf = self.cat4_tr(Xtransf)
Xtransf = self.cat9_tr(Xtransf)
return Xtransf
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
from tubesml.base import BaseTransformer, self_columns, reset_columns
import tubesml as tml
from sklearn.pipeline import Pipeline
from sklearn.decomposition import PCA
from sklearn.preprocessing import LabelEncoder
from source import explore as ex
from source import utility as ut
pd.set_option('max_columns', 100)
df_train = pd.read_csv('data/train.csv')
df_test = pd.read_csv('data/test.csv')
subs = pd.read_csv('data/sample_submission.csv')
train_set, test_set = ut.make_test(df_train, 0.25, random_state=516, strat_feat='cat9')
class PCADf(BaseTransformer):
def __init__(self, n_components, svd_solver='auto', random_state=24, compress=False):
super().__init__()
self.n_components = n_components
self.svd_solver = svd_solver
self.n_components_ = 0
self.random_state = random_state
self.PCA = PCA(n_components=self.n_components, svd_solver=self.svd_solver, random_state=self.random_state)
self.compress = compress
self.original_columns = []
@reset_columns
def fit(self, X, y=None):
self.PCA.fit(X)
self.n_components_ = self.PCA.n_components_
return self
@self_columns
def transform(self, X, y=None):
X_tr = self.PCA.transform(X)
X_tr = pd.DataFrame(X_tr, columns=[f'pca_{i}' for i in range(self.n_components_)])
self.original_columns = X.columns
if self.compress:
X_tr = self.inverse_transform(X_tr)
return X_tr
def inverse_transform(self, X, y=None):
try:
X_tr = self.PCA.inverse_transform(X)
except ValueError:
return X
X_tr = pd.DataFrame(X_tr, columns=self.original_columns)
return X_tr
mock_pipe = Pipeline([('dums', tml.Dummify(drop_first=True)),
('pca', PCADf(n_components=0.9))])
tmp = train_set.copy().drop(['id', 'target'], axis=1)
mock_pipe.fit(tmp)
mock_pipe.transform(tmp).head()
mock_pipe = Pipeline([('dums', tml.Dummify(drop_first=True)),
('pca', PCADf(n_components=60))])
tmp = train_set.copy().drop(['id', 'target'], axis=1)
mock_pipe.fit(tmp)
plt.plot(np.cumsum(mock_pipe.steps[-1][1].PCA.explained_variance_ratio_))
plt.show()
class TargetEncoder(BaseTransformer):
# Heavily inspired by
# https://github.com/MaxHalford/xam/blob/93c066990d976c7d4d74b63fb6fb3254ee8d9b48/xam/feature_extraction/encoding/bayesian_target.py#L8
def __init__(self, to_encode=None, prior_weight=100, agg_func='mean'):
super().__init__()
if isinstance(to_encode, str):
self.to_encode = [to_encode]
else:
self.to_encode = to_encode
self.prior_weight = prior_weight
self.prior_ = None
self.posteriors_ = None
self.agg_func = agg_func
@reset_columns
def fit(self, X, y):
# Encode all categorical cols by default
if self.to_encode is None:
self.to_encode = [c for c in X if str(X[c].dtype)=='object' or str(X[c].dtype)=='category']
tmp = X.copy()
tmp['target'] = y
self.prior_ = tmp['target'].agg(self.agg_func)
self.posteriors_ = {}
for col in self.to_encode:
agg = tmp.groupby(col)['target'].agg(['count', self.agg_func])
counts = agg['count']
data = agg[self.agg_func]
pw = self.prior_weight
self.posteriors_[col] = ((pw * self.prior_ + counts * data) / (pw + counts)).to_dict()
del tmp
return self
@self_columns
def transform(self, X, y=None):
X_tr = X.copy()
for col in self.to_encode:
X_tr[col] = X_tr[col].map(self.posteriors_[col]).fillna(self.prior_).astype(float)
return X_tr
tmp = train_set.copy().drop(['id', 'target'], axis=1)
trs = TargetEncoder(to_encode=None)
trs.fit_transform(tmp, train_set['target']).head()
tmp = train_set.copy().drop(['id', 'target'], axis=1)
trs = TargetEncoder(to_encode=None, agg_func='skew')
trs.fit_transform(tmp, train_set['target']).head()
tmp = train_set.copy().drop(['id', 'target'], axis=1)
trs = TargetEncoder(to_encode=None, agg_func='std')
trs.fit_transform(tmp, train_set['target']).head()
tmp = train_set[[col for col in train_set if 'cat' in col]]
tmp.head()
le = LabelEncoder()
le.fit_transform(tmp['cat0'])
class CatSimp(BaseTransformer):
def __init__(self, cat7=True, cat6=True, cat8=True, cat4=True, cat9=True):
super().__init__()
self.cat7 = cat7
self.cat6 = cat6
self.cat8 = cat8
self.cat4 = cat4
self.cat9 = cat9
def cat7_tr(self, X):
X_tr = X.copy()
if self.cat7:
X_tr['cat7'] = X_tr['cat7'].map({'C': 'E',
'A': 'B',
'F': 'G',
'I': 'G'}).fillna(X_tr['cat7'])
return X_tr
def cat6_tr(self, X):
X_tr = X.copy()
if self.cat6:
X_tr.loc[X_tr['cat6'] != 'A', 'cat6'] = 'B'
return X_tr
def cat8_tr(self, X):
X_tr = X.copy()
if self.cat8:
X_tr['cat8'] = X_tr['cat8'].map({'B': 'E', 'F': 'E'}).fillna(X_tr['cat8'])
return X_tr
def cat4_tr(self, X):
X_tr = X.copy()
if self.cat4:
X_tr['cat4'] = X_tr['cat4'].map({'D': 'A'}).fillna(X_tr['cat4'])
return X_tr
def cat9_tr(self, X):
X_tr = X.copy()
if self.cat9:
X_tr['cat9'] = X_tr['cat9'].map({'E': 'L', 'D': 'J', 'C': 'L'}).fillna(X_tr['cat9'])
return X_tr
@self_columns
def transform(self, X, y=None):
Xtransf = self.cat7_tr(X)
Xtransf = self.cat6_tr(Xtransf)
Xtransf = self.cat8_tr(Xtransf)
Xtransf = self.cat4_tr(Xtransf)
Xtransf = self.cat9_tr(Xtransf)
return Xtransf
| 0.635562 | 0.641394 |
```
#coding:utf-8
import numpy as np
import scipy
from scipy import linalg
import cv2
%matplotlib inline
import matplotlib.pyplot as plt
src = cv2.cvtColor(cv2.imread('../datas/f2.jpg'),cv2.COLOR_BGR2RGB)
illuminants = \
{"A": {'2': (1.098466069456375, 1, 0.3558228003436005),
'10': (1.111420406956693, 1, 0.3519978321919493)},
"D50": {'2': (0.9642119944211994, 1, 0.8251882845188288),
'10': (0.9672062750333777, 1, 0.8142801513128616)},
"D55": {'2': (0.956797052643698, 1, 0.9214805860173273),
'10': (0.9579665682254781, 1, 0.9092525159847462)},
"D65": {'2': (0.95047, 1., 1.08883), # This was: `lab_ref_white`
'10': (0.94809667673716, 1, 1.0730513595166162)},
"D75": {'2': (0.9497220898840717, 1, 1.226393520724154),
'10': (0.9441713925645873, 1, 1.2064272211720228)},
"E": {'2': (1.0, 1.0, 1.0),
'10': (1.0, 1.0, 1.0)}}
def get_xyz_coords(illuminant, observer):
illuminant = illuminant.upper()
try:
return illuminants[illuminant][observer]
except KeyError:
raise ValueError("Unknown illuminant/observer combination\
(\'{0}\', \'{1}\')".format(illuminant, observer))
def _prepare_colorarray(img):
shape = img.shape
return np.reshape(img,(shape[0] * shape[1],shape[2]))
def xyz2luv(xyz,illuminant="D65", observer="2"):
arr = _prepare_colorarray(xyz)
shape = xyz.shape
# extract channels
x, y, z = arr[..., 0], arr[..., 1], arr[..., 2]
eps = np.finfo(np.float).eps
# compute y_r and L
xyz_ref_white = get_xyz_coords(illuminant, observer)
L = y / xyz_ref_white[1]
mask = L > 0.008856
L[mask] = 116. * np.power(L[mask], 1. / 3.) - 16.
L[~mask] = 903.3 * L[~mask]
u0 = 4 * xyz_ref_white[0] / np.dot([1, 15, 3], xyz_ref_white)
v0 = 9 * xyz_ref_white[1] / np.dot([1, 15, 3], xyz_ref_white)
# u' and v' helper functions
def fu(X, Y, Z):
return (4. * X) / (X + 15. * Y + 3. * Z + eps)
def fv(X, Y, Z):
return (9. * Y) / (X + 15. * Y + 3. * Z + eps)
# compute u and v using helper functions
u = 13. * L * (fu(x, y, z) - u0)
v = 13. * L * (fv(x, y, z) - v0)
luv = np.concatenate([q[..., np.newaxis] for q in [L, u, v]], axis=-1)
return np.reshape(luv,shape)
xyz_from_rgb = np.array([[0.412453, 0.357580, 0.180423],
[0.212671, 0.715160, 0.072169],
[0.019334, 0.119193, 0.950227]])
rgb_from_xyz = linalg.inv(xyz_from_rgb)
def rgb2xyz(img_rgb):
shape = img_rgb.shape
rgb = np.float64(img_rgb)
arr = np.reshape(rgb,(shape[0] * shape[1],shape[2]))
xyz = np.dot(arr,xyz_from_rgb.T.copy())
xyz = np.reshape(xyz,shape)
return xyz
def xyz2rgb(img_xyz):
shape = img_xyz.shape
xyz = np.float64(img_xyz)
arr = np.reshape(xyz,(shape[0] * shape[1],shape[2]))
rgb = np.dot(arr,rgb_from_xyz.T.copy())
rgb = np.reshape(rgb,shape)
return cv2.convertScaleAbs(rgb)
def luv2xyz(img_luv,illuminant="D65", observer="2"):
shape = img_luv.shape
arr = _prepare_colorarray(luv).copy()
L, u, v = arr[..., 0], arr[..., 1], arr[..., 2]
eps = np.finfo(np.float).eps
# compute y
y = L.copy()
mask = y > 7.999625
y[mask] = np.power((y[mask] + 16.) / 116., 3.)
y[~mask] = y[~mask] / 903.3
xyz_ref_white = get_xyz_coords(illuminant, observer)
y *= xyz_ref_white[1]
# reference white x,z
uv_weights = [1, 15, 3]
u0 = 4 * xyz_ref_white[0] / np.dot(uv_weights, xyz_ref_white)
v0 = 9 * xyz_ref_white[1] / np.dot(uv_weights, xyz_ref_white)
# compute intermediate values
a = u0 + u / (13. * L + eps)
b = v0 + v / (13. * L + eps)
c = 3 * y * (5 * b - 3)
# compute x and z
z = ((a - 4) * c - 15 * a * b * y) / (12 * b)
x = -(c / b + 3. * z)
xyz = np.concatenate([q[..., np.newaxis] for q in [x, y, z]], axis=-1)
return np.reshape(xyz,shape)
def rgb2luv(src):
return xyz2luv(rgb2xyz(src))
def luv2rgb(src):
return xyz2rgb(luv2xyz(src))
xyz = rgb2xyz(src)
plt.imshow(cv2.convertScaleAbs(xyz))
plt.title('RGB->XYZ')
luv = xyz2luv(xyz)
print(luv.shape)
plt.imshow(cv2.convertScaleAbs(luv))
plt.title('XYZ->LUV')
xyz = luv2xyz(luv)
plt.imshow(cv2.convertScaleAbs(xyz))
plt.title('LUV->XYX')
luv = rgb2luv(src)
plt.imshow(cv2.convertScaleAbs(luv))
plt.title('RGB->LUV')
rgb = luv2rgb(luv)
plt.imshow(rgb)
plt.title('LUV->RGB')
```
|
github_jupyter
|
#coding:utf-8
import numpy as np
import scipy
from scipy import linalg
import cv2
%matplotlib inline
import matplotlib.pyplot as plt
src = cv2.cvtColor(cv2.imread('../datas/f2.jpg'),cv2.COLOR_BGR2RGB)
illuminants = \
{"A": {'2': (1.098466069456375, 1, 0.3558228003436005),
'10': (1.111420406956693, 1, 0.3519978321919493)},
"D50": {'2': (0.9642119944211994, 1, 0.8251882845188288),
'10': (0.9672062750333777, 1, 0.8142801513128616)},
"D55": {'2': (0.956797052643698, 1, 0.9214805860173273),
'10': (0.9579665682254781, 1, 0.9092525159847462)},
"D65": {'2': (0.95047, 1., 1.08883), # This was: `lab_ref_white`
'10': (0.94809667673716, 1, 1.0730513595166162)},
"D75": {'2': (0.9497220898840717, 1, 1.226393520724154),
'10': (0.9441713925645873, 1, 1.2064272211720228)},
"E": {'2': (1.0, 1.0, 1.0),
'10': (1.0, 1.0, 1.0)}}
def get_xyz_coords(illuminant, observer):
illuminant = illuminant.upper()
try:
return illuminants[illuminant][observer]
except KeyError:
raise ValueError("Unknown illuminant/observer combination\
(\'{0}\', \'{1}\')".format(illuminant, observer))
def _prepare_colorarray(img):
shape = img.shape
return np.reshape(img,(shape[0] * shape[1],shape[2]))
def xyz2luv(xyz,illuminant="D65", observer="2"):
arr = _prepare_colorarray(xyz)
shape = xyz.shape
# extract channels
x, y, z = arr[..., 0], arr[..., 1], arr[..., 2]
eps = np.finfo(np.float).eps
# compute y_r and L
xyz_ref_white = get_xyz_coords(illuminant, observer)
L = y / xyz_ref_white[1]
mask = L > 0.008856
L[mask] = 116. * np.power(L[mask], 1. / 3.) - 16.
L[~mask] = 903.3 * L[~mask]
u0 = 4 * xyz_ref_white[0] / np.dot([1, 15, 3], xyz_ref_white)
v0 = 9 * xyz_ref_white[1] / np.dot([1, 15, 3], xyz_ref_white)
# u' and v' helper functions
def fu(X, Y, Z):
return (4. * X) / (X + 15. * Y + 3. * Z + eps)
def fv(X, Y, Z):
return (9. * Y) / (X + 15. * Y + 3. * Z + eps)
# compute u and v using helper functions
u = 13. * L * (fu(x, y, z) - u0)
v = 13. * L * (fv(x, y, z) - v0)
luv = np.concatenate([q[..., np.newaxis] for q in [L, u, v]], axis=-1)
return np.reshape(luv,shape)
xyz_from_rgb = np.array([[0.412453, 0.357580, 0.180423],
[0.212671, 0.715160, 0.072169],
[0.019334, 0.119193, 0.950227]])
rgb_from_xyz = linalg.inv(xyz_from_rgb)
def rgb2xyz(img_rgb):
shape = img_rgb.shape
rgb = np.float64(img_rgb)
arr = np.reshape(rgb,(shape[0] * shape[1],shape[2]))
xyz = np.dot(arr,xyz_from_rgb.T.copy())
xyz = np.reshape(xyz,shape)
return xyz
def xyz2rgb(img_xyz):
shape = img_xyz.shape
xyz = np.float64(img_xyz)
arr = np.reshape(xyz,(shape[0] * shape[1],shape[2]))
rgb = np.dot(arr,rgb_from_xyz.T.copy())
rgb = np.reshape(rgb,shape)
return cv2.convertScaleAbs(rgb)
def luv2xyz(img_luv,illuminant="D65", observer="2"):
shape = img_luv.shape
arr = _prepare_colorarray(luv).copy()
L, u, v = arr[..., 0], arr[..., 1], arr[..., 2]
eps = np.finfo(np.float).eps
# compute y
y = L.copy()
mask = y > 7.999625
y[mask] = np.power((y[mask] + 16.) / 116., 3.)
y[~mask] = y[~mask] / 903.3
xyz_ref_white = get_xyz_coords(illuminant, observer)
y *= xyz_ref_white[1]
# reference white x,z
uv_weights = [1, 15, 3]
u0 = 4 * xyz_ref_white[0] / np.dot(uv_weights, xyz_ref_white)
v0 = 9 * xyz_ref_white[1] / np.dot(uv_weights, xyz_ref_white)
# compute intermediate values
a = u0 + u / (13. * L + eps)
b = v0 + v / (13. * L + eps)
c = 3 * y * (5 * b - 3)
# compute x and z
z = ((a - 4) * c - 15 * a * b * y) / (12 * b)
x = -(c / b + 3. * z)
xyz = np.concatenate([q[..., np.newaxis] for q in [x, y, z]], axis=-1)
return np.reshape(xyz,shape)
def rgb2luv(src):
return xyz2luv(rgb2xyz(src))
def luv2rgb(src):
return xyz2rgb(luv2xyz(src))
xyz = rgb2xyz(src)
plt.imshow(cv2.convertScaleAbs(xyz))
plt.title('RGB->XYZ')
luv = xyz2luv(xyz)
print(luv.shape)
plt.imshow(cv2.convertScaleAbs(luv))
plt.title('XYZ->LUV')
xyz = luv2xyz(luv)
plt.imshow(cv2.convertScaleAbs(xyz))
plt.title('LUV->XYX')
luv = rgb2luv(src)
plt.imshow(cv2.convertScaleAbs(luv))
plt.title('RGB->LUV')
rgb = luv2rgb(luv)
plt.imshow(rgb)
plt.title('LUV->RGB')
| 0.587825 | 0.475788 |
<a href="https://colab.research.google.com/github/Jenn-mawia/Tea-production-exports-and-consumption-in-Kenya-Data-Analysis/blob/master/Data_Analysis_on_Tea_Production%2C_Export_and_Consumption_in_Kenya_Python_Notebook.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Importing Libraries
```
#Importing libraries to be used
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
```
# Loading datasets
The datasets to be used for this project contains Kenya's tea production, exports and consumption over the years 2012-2015.
This data can be found here for access or download.
1. [ National_Monthly_Production,_Consumption-and-exports-of-Tea-in-millions-of-Kg-between-2003-2015.](http://www.opendata.go.ke/datasets/6b85ebe20cef45948d82a0b9b3a0750e/data)
2. [Kenya-monthly-tea-export-quantity-and-value-2012-2017.](http://kilimodata.developlocal.org/dataset/kenya-monthly-tea-export-2012-2017)
3. [kenya-monthly-tea-production-2012-2016.](http://kilimodata.developlocal.org/dataset/kenya-monthly-tea-production-2012-2016/resource/6059cc0d-8116-4d3d-9198-156357d3c390)
To upload the csv files to the environment, go to Table of Contents > Files> Upload
```
#Loading the National Monthly Production, Export and Consumption of Tea data set represented as 'df'
url = "/content/National_Monthly_Production,_Consumption_and_Export_of_Tea_in_Millions_of_Kg_between_2003-2015.csv"
df = pd.read_csv(url, delimiter=',')
# preview dataframe
df
#Loading the Monthly Export of Tea Dataset represented as 'df1'
url1 = "/content/4cafbfe4-c807-42fa-a31b-5b90207eca87.csv"
df1 = pd.read_csv(url1)
# preview dataframe
df1
#Loading the Monthly Production of Tea Dataset represented as 'df2'
url2 = "/content/6059cc0d-8116-4d3d-9198-156357d3c390.csv"
df2 = pd.read_csv(url2)
# preview dataframe
df2
```
# Brief description of the contents in the dataframes
```
#Describing the National Monthly Production, Export and Consumption of Tea data set
df.describe()
#Describing the Monthly Export of Tea data set
df1.describe()
#Describing the Monthly Production of Tea data set
df2.describe()
```
# Shape of the dataframes
```
#This gives the number of rows and columns in 'df'
df.shape
#This gives the number of rows and columns in 'df1'
df1.shape
#This gives the number of rows and columns in 'df2'
df2.shape
```
# Accessing information about our datasets
```
#This accesses the information in 'df'
df.info()
#This accesses the information in 'df1'
df1.info()
#This accesses the information in 'df2'
df2.info()
```
# Data Cleaning
## Uniformity
```
#Multiplying the 'Millions_in_KGS_' column by 1,000,000 in 'df' to match the values in the other columns
df['Quantity Consumed (Kgs)'] = df['Millions_in_KGS_'] * 1000000
df
#Replacing the months in the Month column from text to their corresponding digits in 'df2'
d = {'JANUARY':1, 'FEBRUARY':2, 'MARCH':3, 'APRIL':4, 'MAY':5, 'JUNE':6, 'JULY':7, 'AUGUST':8, 'SEPTEMBER':9, 'OCTOBER':10, 'NOVEMBER':11, 'DECEMBER':12 }
df2.Month = df2.Month.map(d)
df2
#Replacing the months in the Month column from text to their corresponding digits in 'df1'
d2 = {'January':1, 'February':2, 'March':3, 'April':4, 'May':5, 'June':6, 'July':7, 'August':8, 'September':9, 'October':10, 'November':11, 'December':12 }
df1.Month = df1.Month.map(d2)
df1
#This converts the datatype of the 'Date' column from an object to datetime in 'df'
df[["Date"]] = df[["Date"]].apply(pd.to_datetime)
df
#This code splits the 'Date' column into two: 'new_date' and 'new_time'
df['new_date'] = [d.date() for d in df['Date']]
df['new_time'] = [d.time() for d in df['Date']]
df
#This changes the new_date column datatype to datetime
df[["new_date"]] = df[["new_date"]].apply(pd.to_datetime)
#This will split the 'new_date column into 'Day', 'Month' and 'Year'
df['Day'] = df['new_date'].dt.day
df['Month'] = df['new_date'].dt.month
df['Year'] = df['new_date'].dt.year
df
# removing the trailing spaces
df2.columns.str.strip()
#This converts the datatype of the columns:'Year' and 'Month' to object
df = df.astype({'Year': str, 'Month': str})
df.info()
#This will create an new column 'Month-Year' whose values are as a result of joining the 'Month' and 'Year' column values and separating with a hyphen(-)
df["Month-Year"] = df["Month"].str.cat(df[["Year"]].astype(str), sep="-")
df
df1.columns.str.strip()
#This converts the datatype of the columns:'Year' and 'Month' to object in 'df1'
df1 = df1.astype({'Year': str, 'Month': str})
df1.info()
## perform uniformity check on all column names e.g "Year" gives an error because of trailing space "Year "
#This will create an new column 'Month-Year' in 'df1' whose values are as a result of joining the 'Month' and 'Year' column values and separating with a hyphen(-)
df1["Month-Year"] = df1["Month"].str.cat(df1[["Year"]].astype(str), sep="-")
df1
#This converts the datatype of the columns:'Year' and 'Month' to object in 'df2'
df2 = df2.astype({'Year': str, 'Month': str})
df2.info()
#This will create an new column 'Month-Year' in 'df2' whose values are as a result of joining the 'Month' and 'Year' column values and separating with a hyphen(-)
df2["Month-Year"] = df2["Month"].str.cat(df2[["Year"]].astype(str), sep="-")
df2
#This will append the three dataframes into one dataframe known as 'new'
new = df.append([df1, df2])
new
#This accesses information about the new dataframe
new.info()
```
## Validity
```
#This will drop the columns 'Date' , 'OBJECTID' , 'new_date', 'new_time', 'Day', 'Month', 'Year', '_id' since they are not required in analysis
final = new.drop(new.columns[[1,2,3,5,6,7,8,9,11]], axis=1)
final
#This accesses the information in the new edited dataframe known as 'final'
final.info()
#This changes the datatype of 'Month-Year' to datetime in 'final'
final[["Month-Year"]] = final[["Month-Year"]].apply(pd.to_datetime)
final.info()
#This filters the data to give us the Exports, Production and Consumption from the beginning of the year 2012 to the end of 2015
mask = (final['Month-Year'] > '2011-12-01') & (final['Month-Year'] < '2016-01-01')
final = final.loc[mask]
final
#This drops the rows which contain the values 'Production' or 'Exports in the '_Activity_in_Tea_Sector' column
list_of_values = ('Production', 'Exports')
output = final[~final['_Activity_in_Tea_Sector'].isin(list_of_values)]
output
#This reindexes the 'output' dataframe
output_df = output.reset_index(drop=True)
output_df
```
## Completeness
```
#This fills in the null values in the '_Activity_in_Tea_Sector' column with the text 'Consumption' from the range '0-47'
output_df.loc[0:47,'_Activity_in_Tea_Sector'] = 'Consumption'
output_df
#This fills in the null values in the '_Activity_in_Tea_Sector' column with the text 'Exports' from the range '48-95'
output_df.loc[48:95,'_Activity_in_Tea_Sector'] = 'Exports'
output_df
#This fills in the null values in the '_Activity_in_Tea_Sector' column with the text 'Production' from the range '96-143'
output_df.loc[96:143,'_Activity_in_Tea_Sector'] = 'Production'
output_df
#This fills all remaining null values with '0'
final_recode = output_df.fillna(0.0)
final_recode
```
## Consistency
```
## to be done, check for any duplicates
# final_recode.duplicated()
```
# Exporting clean dataset
```
#This exports the 'final recode' dataset to a csv file
final_recode.to_csv('TeaProduction.csv')
```
## Reading the new csv file
```
#This reads the csv file using the pandas library
tea = pd.read_csv('TeaProduction.csv')
tea
```
**Accessing information about new data**
```
tea.info()
```
## Data-type Conversion
```
#This changes the datatype of some columns to the required datatype for analysis
tea['Quantity Produced (Kgs)'] = tea['Quantity Produced (Kgs)'].str.replace(",","")
tea['Quantity Produced (Kgs)'] = tea['Quantity Produced (Kgs)'].astype(float)
tea['Quantity exported (Kg)'] = tea['Quantity exported (Kg)'].str.replace(",","")
tea['Quantity exported (Kg)'] = tea['Quantity exported (Kg)'].astype(float)
tea['Total Value (Ksh)'] = tea['Total Value (Ksh)'].str.replace(",","")
tea['Total Value (Ksh)'] = tea['Total Value (Ksh)'].astype(float)
tea[["Month-Year"]] = tea[["Month-Year"]].apply(pd.to_datetime)
tea.info()
```
# Data Analysis
```
#Resetting the index of the dataframe to 'Month-Year'
tea_index = tea.set_index('Month-Year')
tea_index
# Assigning the column names: "Quantity Produced (Kgs)", "Quantity exported (Kg)", "Quantity Consumed (Kgs)",
# "Average Export Price per Kg (Ksh)" and "Total Value (Ksh)" to the variables: produced, exported, consumed, average_exported_price
# and total_value respectively for the purposes of omitting the zero digits in the process of calculation
produced = tea_index["Quantity Produced (Kgs)"][(tea_index["Quantity Produced (Kgs)"] >0)]
exported = tea_index["Quantity exported (Kg)"][(tea_index["Quantity exported (Kg)"] >0)]
consumed = tea_index["Quantity Consumed (Kgs)"][(tea_index["Quantity Consumed (Kgs)"] >0)]
average_exported_price = tea_index["Average Export Price per Kg (Ksh)"][(tea_index["Average Export Price per Kg (Ksh)"] >0)]
total_value = tea_index["Total Value (Ksh)"][(tea_index["Total Value (Ksh)"] >0)]
```
## Highest and lowest month for tea production over the years 2012-2015
```
#This code will give us the highest and lowest month of tea production overall
highest_month_production = produced.idxmax()
highest_production = produced.max()
lowest_month_production = produced.idxmin()
lowest_production = produced.min()
print("Highest month of production overall is", highest_month_production)
print("The production of that month is", highest_production, "kilograms")
print("Lowest month of production overall is", lowest_month_production)
print("The production of that month is", lowest_production, "kilograms")
#This code displays the highest month of tea production in the year 2012
max_month_2012_pro = tea_index.loc['2012-01-01':'2012-12-01', 'Quantity Produced (Kgs)'].idxmax()
max_production_2012 = tea_index.loc['2012-01-01':'2012-12-01', 'Quantity Produced (Kgs)'].max()
print("Highest production month in 2012 is", max_month_2012_pro)
print("The production of that month is", max_production_2012, "kilograms")
#This code displays the highest month of tea production in the year 2013
max_month_2013_pro = tea_index.loc['2013-01-01':'2013-12-01', 'Quantity Produced (Kgs)'].idxmax()
max_production_2013 = tea_index.loc['2013-01-01':'2013-12-01', 'Quantity Produced (Kgs)'].max()
print("Highest production month in 2013 is", max_month_2013_pro)
print("The production of that month is", max_production_2013, "kilograms")
#This code displays the highest month of tea production in the year 2014
max_month_2014_pro = tea_index.loc['2014-01-01':'2014-12-01', 'Quantity Produced (Kgs)'].idxmax()
max_production_2014 = tea_index.loc['2014-01-01':'2014-12-01', 'Quantity Produced (Kgs)'].max()
print("Highest production month in 2014 is", max_month_2014_pro)
print("The production of that month is", max_production_2014, "kilograms")
#This code displays the highest month of tea production in the year 2015
max_month_2015_pro = tea_index.loc['2015-01-01':'2015-12-01', 'Quantity Produced (Kgs)'].idxmax()
max_production_2015 = tea_index.loc['2015-01-01':'2015-12-01', 'Quantity Produced (Kgs)'].max()
print("Highest production month in 2015 is", max_month_2015_pro)
print("The production of that month is", max_production_2015, "kilograms")
#This code displays the lowest month of tea production in the year 2012
min_month_2012_pro = produced.loc['2012-01-01':'2012-12-01'].idxmin()
min_production_2012 = produced.loc['2012-01-01':'2012-12-01'].min()
print("Lowest production month in 2012 is", min_month_2012_pro)
print("The production of that month is", min_production_2012, "kilograms")
#This code displays the lowest month of tea production in the year 2013
min_month_2013_pro = produced.loc['2013-01-01':'2013-12-01'].idxmin()
min_production_2013 = produced.loc['2013-01-01':'2013-12-01'].min()
print("Lowest production month in 2013 is", min_month_2013_pro)
print("The production of that month is", min_production_2013, "kilograms")
#This code displays the lowest month of tea production in the year 2014
min_month_2014_pro = produced.loc['2014-01-01':'2014-12-01'].idxmin()
min_production_2014 = produced.loc['2014-01-01':'2014-12-01'].min()
print("Lowest production month in 2014 is", min_month_2014_pro)
print("The production of that month is", min_production_2014, "kilograms")
#This code displays the lowest month of tea production in the year 2015
min_month_2015_pro = produced.loc['2015-01-01':'2015-12-01'].idxmin()
min_production_2015 = produced.loc['2015-01-01':'2015-12-01'].min()
print("Lowest production month in 2015 is", min_month_2015_pro)
print("The production of that month is", min_production_2015, "kilograms")
```
## Highest and lowest month for tea exports
```
#This code will give us the highest and lowest month of tea exports overall
highest_month_exports = exported.idxmax()
highest_exports = exported.max()
lowest_month_exports = exported.idxmin()
lowest_exports = exported.min()
print("Highest month of exports overall is", highest_month_exports)
print("The exports of that month is", highest_exports, "kilograms")
print("Lowest month of exports overall is", lowest_month_exports)
print("The exports of that month is", lowest_exports, "kilograms")
#This code displays the highest month of tea exports in the year 2012
max_month_2012_exp = tea_index.loc['2012-01-01':'2012-12-01', 'Quantity exported (Kg)'].idxmax()
max_exports_2012 = tea_index.loc['2012-01-01':'2012-12-01', 'Quantity exported (Kg)'].max()
print("Highest exports month in 2012 is", max_month_2012_exp)
print("The exports of that month is", max_exports_2012, "kilograms")
#This code displays the highest month of tea exports in the year 2013
max_month_2013_exp = tea_index.loc['2013-01-01':'2013-12-01', 'Quantity exported (Kg)'].idxmax()
max_exports_2013 = tea_index.loc['2013-01-01':'2013-12-01', 'Quantity exported (Kg)'].max()
print("Highest exports month in 2013 is", max_month_2013_exp)
print("The exports of that month is", max_exports_2013, "kilograms")
#This code displays the highest month of tea exports in the year 2014
max_month_2014_exp = tea_index.loc['2014-01-01':'2014-12-01', 'Quantity exported (Kg)'].idxmax()
max_exports_2014 = tea_index.loc['2014-01-01':'2014-12-01', 'Quantity exported (Kg)'].max()
print("Highest exports month in 2014 is", max_month_2014_exp)
print("The exports of that month is", max_exports_2014, "kilograms")
#This code displays the highest month of tea exports in the year 2015
max_month_2015_exp = tea_index.loc['2015-01-01':'2015-12-01', 'Quantity exported (Kg)'].idxmax()
max_exports_2015 = tea_index.loc['2015-01-01':'2015-12-01', 'Quantity exported (Kg)'].max()
print("Highest exports in 2015 is", max_month_2015_exp)
print("The exports of that month is", max_exports_2015, "kilograms")
#This code displays the lowest month of tea exports in the year 2012
min_month_2012_exp = exported.loc['2012-01-01':'2012-12-01'].idxmin()
min_exports_2012 = exported.loc['2012-01-01':'2012-12-01'].min()
print("Lowest exports month in 2012 is", min_month_2012_exp)
print("The exports of that month is", min_exports_2012, "kilograms")
#This code displays the lowest month of tea exports in the year 2013
min_month_2013_exp = exported.loc['2013-01-01':'2013-12-01'].idxmin()
min_exports_2013 = exported.loc['2013-01-01':'2013-12-01'].min()
print("Lowest exports month in 2013 is", min_month_2013_exp)
print("The exports of that month is", min_exports_2013, "kilograms")
#This code displays the lowest month of tea exports in the year 2014
min_month_2014_exp = exported.loc['2014-01-01':'2014-12-01'].idxmin()
min_exports_2014 = exported.loc['2014-01-01':'2014-12-01'].min()
print("Lowest exports month in 2014 is", min_month_2014_exp)
print("The exports of that month is", min_exports_2014, "kilograms")
#This code displays the lowest month of tea exports in the year 2015
min_month_2015_exp = exported.loc['2015-01-01':'2015-12-01'].idxmin()
min_exports_2015 = exported.loc['2015-01-01':'2015-12-01'].min()
print("Lowest exports month in 2015 is", min_month_2015_exp)
print("The exports of that month is", min_exports_2015, "kilograms")
```
## Highest and lowest month for tea consumption
```
#This code will give us the highest and lowest month of tea consumption overall
highest_month_consumption = consumed.idxmax()
highest_consumption = consumed.max()
lowest_month_consumption = consumed.idxmin()
lowest_consumption = consumed.min()
print("Highest month of consumption overall is", highest_month_consumption)
print("The consumption of that month is", highest_consumption, "kilograms")
print("Lowest month of consumption overall is", lowest_month_consumption)
print("The consumption of that month is", lowest_consumption, "kilograms")
#This code displays the highest month of tea consumption in the year 2012
max_month_2012_con = tea_index.loc['2012-01-01':'2012-12-01', 'Quantity Consumed (Kgs)'].idxmax()
max_consumption_2012 = tea_index.loc['2012-01-01':'2012-12-01', 'Quantity Consumed (Kgs)'].max()
print("Highest consumption month in 2012 is", max_month_2012_con)
print("The consumption of that month is", max_consumption_2012, "kilograms")
#This code displays the highest month of tea consumption in the year 2013
max_month_2013_con = tea_index.loc['2013-01-01':'2013-12-01', 'Quantity Consumed (Kgs)'].idxmax()
max_consumption_2013 = tea_index.loc['2013-01-01':'2013-12-01', 'Quantity Consumed (Kgs)'].max()
print("Highest consumption month in 2013 is", max_month_2013_con)
print("The consumption of that month is", max_consumption_2013, "kilograms")
#This code displays the highest month of tea consumption in the year 2014
max_month_2014_con = tea_index.loc['2014-01-01':'2014-12-01', 'Quantity Consumed (Kgs)'].idxmax()
max_consumption_2014 = tea_index.loc['2014-01-01':'2014-12-01', 'Quantity Consumed (Kgs)'].max()
print("Highest consumption month in 2014 is", max_month_2014_con)
print("The consumption of that month is", max_consumption_2014, "kilograms")
#This code displays the highest month of tea consumption in the year 2015
max_month_2015_con = tea_index.loc['2015-01-01':'2015-12-01', 'Quantity Consumed (Kgs)'].idxmax()
max_consumption_2015 = tea_index.loc['2015-01-01':'2015-12-01', 'Quantity Consumed (Kgs)'].max()
print("Highest consumption month in 2015 is", max_month_2015_con)
print("The consumption of that month is", max_consumption_2015, "kilograms")
#This code displays the lowest month of tea consumption in the year 2012
min_month_2012_con = consumed.loc['2012-01-01':'2012-12-01'].idxmin()
min_consumption_2012 = consumed.loc['2012-01-01':'2012-12-01'].min()
print("Lowest consumption month in 2012 is", min_month_2012_con)
print("The consumption of that month is", min_consumption_2012, "kilograms")
#This code displays the lowest month of tea consumption in the year 2012
min_month_2013_con = consumed.loc['2013-01-01':'2013-12-01'].idxmin()
min_consumption_2013 = consumed.loc['2013-01-01':'2013-12-01'].min()
print("Lowest consumption month in 2013 is", min_month_2013_con)
print("The consumption of that month is", min_consumption_2013, "kilograms")
#This code displays the lowest month of tea consumption in the year 2014
min_month_2014_con = consumed.loc['2014-01-01':'2014-12-01'].idxmin()
min_consumption_2014 = consumed.loc['2014-01-01':'2014-12-01'].min()
print("Lowest consumption month in 2014 is", min_month_2014_con)
print("The consumption of that month is", min_consumption_2014, "kilograms")
#This code displays the lowest month of tea consumption in the year 2015
min_month_2015_con = consumed.loc['2015-01-01':'2015-12-01'].idxmin()
min_consumption_2015 = consumed.loc['2015-01-01':'2015-12-01'].min()
print("Lowest consumption month in 2015 is", min_month_2015_con)
print("The consumption of that month is", min_consumption_2015, "kilograms")
```
##The Average production of tea
```
# This code outputs the average production overall
avg_production_overall = produced.mean()
print("The overall average production is", avg_production_overall, "kilograms")
# This code outputs the average production in 2012
avg_production_2012 = produced.loc['2012-01-01':'2012-12-01'].mean()
print("Average production in 2012 is", avg_production_2012, "kilograms")
# This code outputs the average production in 2013
avg_production_2013 = produced.loc['2013-01-01':'2013-12-01'].mean()
print("Average production in 2013 is", avg_production_2013, "kilograms")
# This code outputs the average production in 2014
avg_production_2014 = produced.loc['2014-01-01':'2014-12-01'].mean()
print("Average production in 2014 is", avg_production_2014, "kilograms")
# This code outputs the average production in 2015
avg_production_2015 = produced.loc['2015-01-01':'2015-12-01'].mean()
print("Average production in 2015 is", avg_production_2015, "kilograms")
```
##The Average Exports of tea per year
```
# This code outputs the average exports overall
avg_exports_overall = exported.mean()
print("The overall average exports is", avg_exports_overall, "kilograms")
# This code outputs the average exports in 2012
avg_exports_2012 = exported.loc['2012-01-01':'2012-12-01'].mean()
print("Average exports in 2012 is", avg_exports_2012, "kilograms")
# This code outputs the average production in 2013
avg_exports_2013 = exported.loc['2013-01-01':'2013-12-01'].mean()
print("Average exports in 2013 is", avg_exports_2013, "kilograms")
# This code outputs the average production in 2014
avg_exports_2014 = exported.loc['2014-01-01':'2014-12-01'].mean()
print("Average exports in 2014 is", avg_exports_2014, "kilograms")
# This code outputs the average production in 2015
avg_exports_2015 = exported.loc['2015-01-01':'2015-12-01'].mean()
print("Average exports in 2015 is", avg_exports_2015, "kilograms")
```
##The Average Consumption of tea per year
```
# This code outputs the average consumption overall
avg_consumption_overall = consumed.mean()
print("The overall average consumption is", avg_consumption_overall, "kilograms")
# This code outputs the average exports in 2012
avg_consumption_2012 = consumed.loc['2012-01-01':'2012-12-01'].mean()
print("Average consumption in 2012 is", avg_consumption_2012, "kilograms")
# This code outputs the average production in 2013
avg_consumption_2013 = consumed.loc['2013-01-01':'2013-12-01'].mean()
print("Average consumption in 2013 is", avg_consumption_2013, "kilograms")
# This code outputs the average production in 2014
avg_consumption_2014 = consumed.loc['2014-01-01':'2014-12-01'].mean()
print("Average consumption in 2014 is", avg_consumption_2014, "kilograms")
# This code outputs the average production in 2015
avg_consumption_2015 = consumed.loc['2015-01-01':'2015-12-01'].mean()
print("Average consumption in 2015 is", avg_consumption_2015, "kilograms")
```
##Does the sum of exports and consumption add up to the production of tea?
```
#This returns the initial index of the dataframe.
tea_reset = tea_index.reset_index()
tea_reset
#This checks whether the sum of the Quantity Consumed and Exported is equal to the
#Quantity Produced
accurate = [exported + consumed == produced]
accurate
```
##Total production in each year
```
# This code outputs the total production overall
total_production = produced.sum()
print("Total production is", total_production, "kilograms")
# This code outputs the total production in 2012
total_production_2012 = produced.loc['2012-01-01':'2012-12-01'].sum()
print("Total production in 2012 is", total_production_2012, "kilograms")
# This code outputs the total production in 2013
total_production_2013 = produced.loc['2013-01-01':'2013-12-01'].sum()
print("Total production in 2013 is", total_production_2013, "kilograms")
# This code outputs the total production in 2014
total_production_2014 = produced.loc['2014-01-01':'2014-12-01'].sum()
print("Total production in 2014 is", total_production_2014, "kilograms")
# This code outputs the total production in 2015
total_production_2015 = produced.loc['2015-01-01':'2015-12-01'].sum()
print("Total production in 2015 is", total_production_2015, "kilograms")
```
##Total exports in each year
```
# This code outputs the total exports overall
total_exports = exported.sum()
print("Total exports is", total_exports, "kilograms")
# This code outputs the total exports in 2012
total_exports_2012 = exported.loc['2012-01-01':'2012-12-01'].sum()
print("Total exports in 2012 is", total_exports_2012, "kilograms")
# This code outputs the total exports in 2013
total_exports_2013 = exported.loc['2013-01-01':'2013-12-01'].sum()
print("Total exports in 2013 is", total_exports_2013, "kilograms")
# This code outputs the total exports in 2014
total_exports_2014 = exported.loc['2014-01-01':'2014-12-01'].sum()
print("Total exports in 2014 is", total_exports_2014, "kilograms")
# This code outputs the total exports in 2015
total_exports_2015 = exported.loc['2015-01-01':'2015-12-01'].sum()
print("Total exports in 2012 is", total_exports_2015, "kilograms")
```
##Total consumption in each year
```
# This code outputs the total consumption overall
total_consumption = consumed.sum()
print("Total consumption is", total_consumption, "kilograms")
# This code outputs the total consumption in 2012
total_consumption_2012 = consumed.loc['2012-01-01':'2012-12-01'].sum()
print("Total consumption in 2012 is", total_consumption_2012, "kilograms")
# This code outputs the total consumption in 2013
total_consumption_2013 = consumed.loc['2013-01-01':'2013-12-01'].sum()
print("Total consumption in 2013 is", total_consumption_2013, "kilograms")
# This code outputs the total consumption in 2014
total_consumption_2014 = consumed.loc['2014-01-01':'2014-12-01'].sum()
print("Total consumption in 2014 is", total_consumption_2014, "kilograms")
# This code outputs the total consumption in 2015
total_consumption_2015 = consumed.loc['2015-01-01':'2015-12-01'].sum()
print("Total consumption in 2015 is", total_consumption_2015, "kilograms")
#This shows the correlation between the various columns in the 'tea_reset' dataset
tea_reset.corr()
```
##Data Visualization
```
#This chart shows the trend in production of tea from January 2012 to December 2015
produced.plot(kind = 'line', figsize=(15,10))
plt.title("Quantity Produced against Time", fontsize=16)
plt.xlabel("Time in Year-Month",fontsize=12)
plt.ylabel("Quantity Produced (Kgs) * 10^7",fontsize=12)
#This chart shows the trend in exporting tea from January 2012 to December 2015
exported.plot(kind = 'line', figsize=(15,10))
plt.title("Quantity Exported against Time", fontsize=16)
plt.xlabel("Time in Year-Month",fontsize=12)
plt.ylabel("Quantity Exported (Kgs) * 10^7",fontsize=12)
#This chart shows the trend in consuming tea from January 2012 to December 2015
consumed.plot(kind = 'line', figsize=(15,10))
plt.title("Quantity Consumed against Time", fontsize=16)
plt.xlabel("Time in Year-Month",fontsize=12)
plt.ylabel("Quantity Consumed (Kgs) * 10^6",fontsize=12)
#This chart shows the correlation in quantity of tea produced, exported and consumed
produced.plot(figsize=(15,10))
exported.plot(figsize=(15,10))
consumed.plot(figsize=(15,10))
plt.plot(produced)
plt.plot(exported)
plt.plot(consumed)
plt.xlabel("Time in Year-Month", fontsize=12)
plt.ylabel("Quantity (Kgs) * 10^7", fontsize=12)
plt.legend(loc = 'upper right')
plt.title("Quantity against Time", fontsize=16)
plt.show()
#This creates a dataframe whose exports have been divided by one million to allow the relating of
#export price to quantity of exports
exported2 = (exported / (10**6))
exported2
#This chart shows the change in exported price and quantity exported from the beginning of 2012
#to the end of 2015
exported2.plot(figsize=(15,10))
average_exported_price.plot(figsize=(15,10))
plt.plot(exported2)
plt.plot(average_exported_price)
plt.xlabel("Time in Year-Month", fontsize=12)
plt.ylabel("Quantity(Kg) * 10^6", fontsize=12)
plt.legend(loc = 'upper right')
plt.title("Exports and Export Prices over Time", fontsize=16)
plt.show()
```
|
github_jupyter
|
#Importing libraries to be used
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
#Loading the National Monthly Production, Export and Consumption of Tea data set represented as 'df'
url = "/content/National_Monthly_Production,_Consumption_and_Export_of_Tea_in_Millions_of_Kg_between_2003-2015.csv"
df = pd.read_csv(url, delimiter=',')
# preview dataframe
df
#Loading the Monthly Export of Tea Dataset represented as 'df1'
url1 = "/content/4cafbfe4-c807-42fa-a31b-5b90207eca87.csv"
df1 = pd.read_csv(url1)
# preview dataframe
df1
#Loading the Monthly Production of Tea Dataset represented as 'df2'
url2 = "/content/6059cc0d-8116-4d3d-9198-156357d3c390.csv"
df2 = pd.read_csv(url2)
# preview dataframe
df2
#Describing the National Monthly Production, Export and Consumption of Tea data set
df.describe()
#Describing the Monthly Export of Tea data set
df1.describe()
#Describing the Monthly Production of Tea data set
df2.describe()
#This gives the number of rows and columns in 'df'
df.shape
#This gives the number of rows and columns in 'df1'
df1.shape
#This gives the number of rows and columns in 'df2'
df2.shape
#This accesses the information in 'df'
df.info()
#This accesses the information in 'df1'
df1.info()
#This accesses the information in 'df2'
df2.info()
#Multiplying the 'Millions_in_KGS_' column by 1,000,000 in 'df' to match the values in the other columns
df['Quantity Consumed (Kgs)'] = df['Millions_in_KGS_'] * 1000000
df
#Replacing the months in the Month column from text to their corresponding digits in 'df2'
d = {'JANUARY':1, 'FEBRUARY':2, 'MARCH':3, 'APRIL':4, 'MAY':5, 'JUNE':6, 'JULY':7, 'AUGUST':8, 'SEPTEMBER':9, 'OCTOBER':10, 'NOVEMBER':11, 'DECEMBER':12 }
df2.Month = df2.Month.map(d)
df2
#Replacing the months in the Month column from text to their corresponding digits in 'df1'
d2 = {'January':1, 'February':2, 'March':3, 'April':4, 'May':5, 'June':6, 'July':7, 'August':8, 'September':9, 'October':10, 'November':11, 'December':12 }
df1.Month = df1.Month.map(d2)
df1
#This converts the datatype of the 'Date' column from an object to datetime in 'df'
df[["Date"]] = df[["Date"]].apply(pd.to_datetime)
df
#This code splits the 'Date' column into two: 'new_date' and 'new_time'
df['new_date'] = [d.date() for d in df['Date']]
df['new_time'] = [d.time() for d in df['Date']]
df
#This changes the new_date column datatype to datetime
df[["new_date"]] = df[["new_date"]].apply(pd.to_datetime)
#This will split the 'new_date column into 'Day', 'Month' and 'Year'
df['Day'] = df['new_date'].dt.day
df['Month'] = df['new_date'].dt.month
df['Year'] = df['new_date'].dt.year
df
# removing the trailing spaces
df2.columns.str.strip()
#This converts the datatype of the columns:'Year' and 'Month' to object
df = df.astype({'Year': str, 'Month': str})
df.info()
#This will create an new column 'Month-Year' whose values are as a result of joining the 'Month' and 'Year' column values and separating with a hyphen(-)
df["Month-Year"] = df["Month"].str.cat(df[["Year"]].astype(str), sep="-")
df
df1.columns.str.strip()
#This converts the datatype of the columns:'Year' and 'Month' to object in 'df1'
df1 = df1.astype({'Year': str, 'Month': str})
df1.info()
## perform uniformity check on all column names e.g "Year" gives an error because of trailing space "Year "
#This will create an new column 'Month-Year' in 'df1' whose values are as a result of joining the 'Month' and 'Year' column values and separating with a hyphen(-)
df1["Month-Year"] = df1["Month"].str.cat(df1[["Year"]].astype(str), sep="-")
df1
#This converts the datatype of the columns:'Year' and 'Month' to object in 'df2'
df2 = df2.astype({'Year': str, 'Month': str})
df2.info()
#This will create an new column 'Month-Year' in 'df2' whose values are as a result of joining the 'Month' and 'Year' column values and separating with a hyphen(-)
df2["Month-Year"] = df2["Month"].str.cat(df2[["Year"]].astype(str), sep="-")
df2
#This will append the three dataframes into one dataframe known as 'new'
new = df.append([df1, df2])
new
#This accesses information about the new dataframe
new.info()
#This will drop the columns 'Date' , 'OBJECTID' , 'new_date', 'new_time', 'Day', 'Month', 'Year', '_id' since they are not required in analysis
final = new.drop(new.columns[[1,2,3,5,6,7,8,9,11]], axis=1)
final
#This accesses the information in the new edited dataframe known as 'final'
final.info()
#This changes the datatype of 'Month-Year' to datetime in 'final'
final[["Month-Year"]] = final[["Month-Year"]].apply(pd.to_datetime)
final.info()
#This filters the data to give us the Exports, Production and Consumption from the beginning of the year 2012 to the end of 2015
mask = (final['Month-Year'] > '2011-12-01') & (final['Month-Year'] < '2016-01-01')
final = final.loc[mask]
final
#This drops the rows which contain the values 'Production' or 'Exports in the '_Activity_in_Tea_Sector' column
list_of_values = ('Production', 'Exports')
output = final[~final['_Activity_in_Tea_Sector'].isin(list_of_values)]
output
#This reindexes the 'output' dataframe
output_df = output.reset_index(drop=True)
output_df
#This fills in the null values in the '_Activity_in_Tea_Sector' column with the text 'Consumption' from the range '0-47'
output_df.loc[0:47,'_Activity_in_Tea_Sector'] = 'Consumption'
output_df
#This fills in the null values in the '_Activity_in_Tea_Sector' column with the text 'Exports' from the range '48-95'
output_df.loc[48:95,'_Activity_in_Tea_Sector'] = 'Exports'
output_df
#This fills in the null values in the '_Activity_in_Tea_Sector' column with the text 'Production' from the range '96-143'
output_df.loc[96:143,'_Activity_in_Tea_Sector'] = 'Production'
output_df
#This fills all remaining null values with '0'
final_recode = output_df.fillna(0.0)
final_recode
## to be done, check for any duplicates
# final_recode.duplicated()
#This exports the 'final recode' dataset to a csv file
final_recode.to_csv('TeaProduction.csv')
#This reads the csv file using the pandas library
tea = pd.read_csv('TeaProduction.csv')
tea
tea.info()
#This changes the datatype of some columns to the required datatype for analysis
tea['Quantity Produced (Kgs)'] = tea['Quantity Produced (Kgs)'].str.replace(",","")
tea['Quantity Produced (Kgs)'] = tea['Quantity Produced (Kgs)'].astype(float)
tea['Quantity exported (Kg)'] = tea['Quantity exported (Kg)'].str.replace(",","")
tea['Quantity exported (Kg)'] = tea['Quantity exported (Kg)'].astype(float)
tea['Total Value (Ksh)'] = tea['Total Value (Ksh)'].str.replace(",","")
tea['Total Value (Ksh)'] = tea['Total Value (Ksh)'].astype(float)
tea[["Month-Year"]] = tea[["Month-Year"]].apply(pd.to_datetime)
tea.info()
#Resetting the index of the dataframe to 'Month-Year'
tea_index = tea.set_index('Month-Year')
tea_index
# Assigning the column names: "Quantity Produced (Kgs)", "Quantity exported (Kg)", "Quantity Consumed (Kgs)",
# "Average Export Price per Kg (Ksh)" and "Total Value (Ksh)" to the variables: produced, exported, consumed, average_exported_price
# and total_value respectively for the purposes of omitting the zero digits in the process of calculation
produced = tea_index["Quantity Produced (Kgs)"][(tea_index["Quantity Produced (Kgs)"] >0)]
exported = tea_index["Quantity exported (Kg)"][(tea_index["Quantity exported (Kg)"] >0)]
consumed = tea_index["Quantity Consumed (Kgs)"][(tea_index["Quantity Consumed (Kgs)"] >0)]
average_exported_price = tea_index["Average Export Price per Kg (Ksh)"][(tea_index["Average Export Price per Kg (Ksh)"] >0)]
total_value = tea_index["Total Value (Ksh)"][(tea_index["Total Value (Ksh)"] >0)]
#This code will give us the highest and lowest month of tea production overall
highest_month_production = produced.idxmax()
highest_production = produced.max()
lowest_month_production = produced.idxmin()
lowest_production = produced.min()
print("Highest month of production overall is", highest_month_production)
print("The production of that month is", highest_production, "kilograms")
print("Lowest month of production overall is", lowest_month_production)
print("The production of that month is", lowest_production, "kilograms")
#This code displays the highest month of tea production in the year 2012
max_month_2012_pro = tea_index.loc['2012-01-01':'2012-12-01', 'Quantity Produced (Kgs)'].idxmax()
max_production_2012 = tea_index.loc['2012-01-01':'2012-12-01', 'Quantity Produced (Kgs)'].max()
print("Highest production month in 2012 is", max_month_2012_pro)
print("The production of that month is", max_production_2012, "kilograms")
#This code displays the highest month of tea production in the year 2013
max_month_2013_pro = tea_index.loc['2013-01-01':'2013-12-01', 'Quantity Produced (Kgs)'].idxmax()
max_production_2013 = tea_index.loc['2013-01-01':'2013-12-01', 'Quantity Produced (Kgs)'].max()
print("Highest production month in 2013 is", max_month_2013_pro)
print("The production of that month is", max_production_2013, "kilograms")
#This code displays the highest month of tea production in the year 2014
max_month_2014_pro = tea_index.loc['2014-01-01':'2014-12-01', 'Quantity Produced (Kgs)'].idxmax()
max_production_2014 = tea_index.loc['2014-01-01':'2014-12-01', 'Quantity Produced (Kgs)'].max()
print("Highest production month in 2014 is", max_month_2014_pro)
print("The production of that month is", max_production_2014, "kilograms")
#This code displays the highest month of tea production in the year 2015
max_month_2015_pro = tea_index.loc['2015-01-01':'2015-12-01', 'Quantity Produced (Kgs)'].idxmax()
max_production_2015 = tea_index.loc['2015-01-01':'2015-12-01', 'Quantity Produced (Kgs)'].max()
print("Highest production month in 2015 is", max_month_2015_pro)
print("The production of that month is", max_production_2015, "kilograms")
#This code displays the lowest month of tea production in the year 2012
min_month_2012_pro = produced.loc['2012-01-01':'2012-12-01'].idxmin()
min_production_2012 = produced.loc['2012-01-01':'2012-12-01'].min()
print("Lowest production month in 2012 is", min_month_2012_pro)
print("The production of that month is", min_production_2012, "kilograms")
#This code displays the lowest month of tea production in the year 2013
min_month_2013_pro = produced.loc['2013-01-01':'2013-12-01'].idxmin()
min_production_2013 = produced.loc['2013-01-01':'2013-12-01'].min()
print("Lowest production month in 2013 is", min_month_2013_pro)
print("The production of that month is", min_production_2013, "kilograms")
#This code displays the lowest month of tea production in the year 2014
min_month_2014_pro = produced.loc['2014-01-01':'2014-12-01'].idxmin()
min_production_2014 = produced.loc['2014-01-01':'2014-12-01'].min()
print("Lowest production month in 2014 is", min_month_2014_pro)
print("The production of that month is", min_production_2014, "kilograms")
#This code displays the lowest month of tea production in the year 2015
min_month_2015_pro = produced.loc['2015-01-01':'2015-12-01'].idxmin()
min_production_2015 = produced.loc['2015-01-01':'2015-12-01'].min()
print("Lowest production month in 2015 is", min_month_2015_pro)
print("The production of that month is", min_production_2015, "kilograms")
#This code will give us the highest and lowest month of tea exports overall
highest_month_exports = exported.idxmax()
highest_exports = exported.max()
lowest_month_exports = exported.idxmin()
lowest_exports = exported.min()
print("Highest month of exports overall is", highest_month_exports)
print("The exports of that month is", highest_exports, "kilograms")
print("Lowest month of exports overall is", lowest_month_exports)
print("The exports of that month is", lowest_exports, "kilograms")
#This code displays the highest month of tea exports in the year 2012
max_month_2012_exp = tea_index.loc['2012-01-01':'2012-12-01', 'Quantity exported (Kg)'].idxmax()
max_exports_2012 = tea_index.loc['2012-01-01':'2012-12-01', 'Quantity exported (Kg)'].max()
print("Highest exports month in 2012 is", max_month_2012_exp)
print("The exports of that month is", max_exports_2012, "kilograms")
#This code displays the highest month of tea exports in the year 2013
max_month_2013_exp = tea_index.loc['2013-01-01':'2013-12-01', 'Quantity exported (Kg)'].idxmax()
max_exports_2013 = tea_index.loc['2013-01-01':'2013-12-01', 'Quantity exported (Kg)'].max()
print("Highest exports month in 2013 is", max_month_2013_exp)
print("The exports of that month is", max_exports_2013, "kilograms")
#This code displays the highest month of tea exports in the year 2014
max_month_2014_exp = tea_index.loc['2014-01-01':'2014-12-01', 'Quantity exported (Kg)'].idxmax()
max_exports_2014 = tea_index.loc['2014-01-01':'2014-12-01', 'Quantity exported (Kg)'].max()
print("Highest exports month in 2014 is", max_month_2014_exp)
print("The exports of that month is", max_exports_2014, "kilograms")
#This code displays the highest month of tea exports in the year 2015
max_month_2015_exp = tea_index.loc['2015-01-01':'2015-12-01', 'Quantity exported (Kg)'].idxmax()
max_exports_2015 = tea_index.loc['2015-01-01':'2015-12-01', 'Quantity exported (Kg)'].max()
print("Highest exports in 2015 is", max_month_2015_exp)
print("The exports of that month is", max_exports_2015, "kilograms")
#This code displays the lowest month of tea exports in the year 2012
min_month_2012_exp = exported.loc['2012-01-01':'2012-12-01'].idxmin()
min_exports_2012 = exported.loc['2012-01-01':'2012-12-01'].min()
print("Lowest exports month in 2012 is", min_month_2012_exp)
print("The exports of that month is", min_exports_2012, "kilograms")
#This code displays the lowest month of tea exports in the year 2013
min_month_2013_exp = exported.loc['2013-01-01':'2013-12-01'].idxmin()
min_exports_2013 = exported.loc['2013-01-01':'2013-12-01'].min()
print("Lowest exports month in 2013 is", min_month_2013_exp)
print("The exports of that month is", min_exports_2013, "kilograms")
#This code displays the lowest month of tea exports in the year 2014
min_month_2014_exp = exported.loc['2014-01-01':'2014-12-01'].idxmin()
min_exports_2014 = exported.loc['2014-01-01':'2014-12-01'].min()
print("Lowest exports month in 2014 is", min_month_2014_exp)
print("The exports of that month is", min_exports_2014, "kilograms")
#This code displays the lowest month of tea exports in the year 2015
min_month_2015_exp = exported.loc['2015-01-01':'2015-12-01'].idxmin()
min_exports_2015 = exported.loc['2015-01-01':'2015-12-01'].min()
print("Lowest exports month in 2015 is", min_month_2015_exp)
print("The exports of that month is", min_exports_2015, "kilograms")
#This code will give us the highest and lowest month of tea consumption overall
highest_month_consumption = consumed.idxmax()
highest_consumption = consumed.max()
lowest_month_consumption = consumed.idxmin()
lowest_consumption = consumed.min()
print("Highest month of consumption overall is", highest_month_consumption)
print("The consumption of that month is", highest_consumption, "kilograms")
print("Lowest month of consumption overall is", lowest_month_consumption)
print("The consumption of that month is", lowest_consumption, "kilograms")
#This code displays the highest month of tea consumption in the year 2012
max_month_2012_con = tea_index.loc['2012-01-01':'2012-12-01', 'Quantity Consumed (Kgs)'].idxmax()
max_consumption_2012 = tea_index.loc['2012-01-01':'2012-12-01', 'Quantity Consumed (Kgs)'].max()
print("Highest consumption month in 2012 is", max_month_2012_con)
print("The consumption of that month is", max_consumption_2012, "kilograms")
#This code displays the highest month of tea consumption in the year 2013
max_month_2013_con = tea_index.loc['2013-01-01':'2013-12-01', 'Quantity Consumed (Kgs)'].idxmax()
max_consumption_2013 = tea_index.loc['2013-01-01':'2013-12-01', 'Quantity Consumed (Kgs)'].max()
print("Highest consumption month in 2013 is", max_month_2013_con)
print("The consumption of that month is", max_consumption_2013, "kilograms")
#This code displays the highest month of tea consumption in the year 2014
max_month_2014_con = tea_index.loc['2014-01-01':'2014-12-01', 'Quantity Consumed (Kgs)'].idxmax()
max_consumption_2014 = tea_index.loc['2014-01-01':'2014-12-01', 'Quantity Consumed (Kgs)'].max()
print("Highest consumption month in 2014 is", max_month_2014_con)
print("The consumption of that month is", max_consumption_2014, "kilograms")
#This code displays the highest month of tea consumption in the year 2015
max_month_2015_con = tea_index.loc['2015-01-01':'2015-12-01', 'Quantity Consumed (Kgs)'].idxmax()
max_consumption_2015 = tea_index.loc['2015-01-01':'2015-12-01', 'Quantity Consumed (Kgs)'].max()
print("Highest consumption month in 2015 is", max_month_2015_con)
print("The consumption of that month is", max_consumption_2015, "kilograms")
#This code displays the lowest month of tea consumption in the year 2012
min_month_2012_con = consumed.loc['2012-01-01':'2012-12-01'].idxmin()
min_consumption_2012 = consumed.loc['2012-01-01':'2012-12-01'].min()
print("Lowest consumption month in 2012 is", min_month_2012_con)
print("The consumption of that month is", min_consumption_2012, "kilograms")
#This code displays the lowest month of tea consumption in the year 2012
min_month_2013_con = consumed.loc['2013-01-01':'2013-12-01'].idxmin()
min_consumption_2013 = consumed.loc['2013-01-01':'2013-12-01'].min()
print("Lowest consumption month in 2013 is", min_month_2013_con)
print("The consumption of that month is", min_consumption_2013, "kilograms")
#This code displays the lowest month of tea consumption in the year 2014
min_month_2014_con = consumed.loc['2014-01-01':'2014-12-01'].idxmin()
min_consumption_2014 = consumed.loc['2014-01-01':'2014-12-01'].min()
print("Lowest consumption month in 2014 is", min_month_2014_con)
print("The consumption of that month is", min_consumption_2014, "kilograms")
#This code displays the lowest month of tea consumption in the year 2015
min_month_2015_con = consumed.loc['2015-01-01':'2015-12-01'].idxmin()
min_consumption_2015 = consumed.loc['2015-01-01':'2015-12-01'].min()
print("Lowest consumption month in 2015 is", min_month_2015_con)
print("The consumption of that month is", min_consumption_2015, "kilograms")
# This code outputs the average production overall
avg_production_overall = produced.mean()
print("The overall average production is", avg_production_overall, "kilograms")
# This code outputs the average production in 2012
avg_production_2012 = produced.loc['2012-01-01':'2012-12-01'].mean()
print("Average production in 2012 is", avg_production_2012, "kilograms")
# This code outputs the average production in 2013
avg_production_2013 = produced.loc['2013-01-01':'2013-12-01'].mean()
print("Average production in 2013 is", avg_production_2013, "kilograms")
# This code outputs the average production in 2014
avg_production_2014 = produced.loc['2014-01-01':'2014-12-01'].mean()
print("Average production in 2014 is", avg_production_2014, "kilograms")
# This code outputs the average production in 2015
avg_production_2015 = produced.loc['2015-01-01':'2015-12-01'].mean()
print("Average production in 2015 is", avg_production_2015, "kilograms")
# This code outputs the average exports overall
avg_exports_overall = exported.mean()
print("The overall average exports is", avg_exports_overall, "kilograms")
# This code outputs the average exports in 2012
avg_exports_2012 = exported.loc['2012-01-01':'2012-12-01'].mean()
print("Average exports in 2012 is", avg_exports_2012, "kilograms")
# This code outputs the average production in 2013
avg_exports_2013 = exported.loc['2013-01-01':'2013-12-01'].mean()
print("Average exports in 2013 is", avg_exports_2013, "kilograms")
# This code outputs the average production in 2014
avg_exports_2014 = exported.loc['2014-01-01':'2014-12-01'].mean()
print("Average exports in 2014 is", avg_exports_2014, "kilograms")
# This code outputs the average production in 2015
avg_exports_2015 = exported.loc['2015-01-01':'2015-12-01'].mean()
print("Average exports in 2015 is", avg_exports_2015, "kilograms")
# This code outputs the average consumption overall
avg_consumption_overall = consumed.mean()
print("The overall average consumption is", avg_consumption_overall, "kilograms")
# This code outputs the average exports in 2012
avg_consumption_2012 = consumed.loc['2012-01-01':'2012-12-01'].mean()
print("Average consumption in 2012 is", avg_consumption_2012, "kilograms")
# This code outputs the average production in 2013
avg_consumption_2013 = consumed.loc['2013-01-01':'2013-12-01'].mean()
print("Average consumption in 2013 is", avg_consumption_2013, "kilograms")
# This code outputs the average production in 2014
avg_consumption_2014 = consumed.loc['2014-01-01':'2014-12-01'].mean()
print("Average consumption in 2014 is", avg_consumption_2014, "kilograms")
# This code outputs the average production in 2015
avg_consumption_2015 = consumed.loc['2015-01-01':'2015-12-01'].mean()
print("Average consumption in 2015 is", avg_consumption_2015, "kilograms")
#This returns the initial index of the dataframe.
tea_reset = tea_index.reset_index()
tea_reset
#This checks whether the sum of the Quantity Consumed and Exported is equal to the
#Quantity Produced
accurate = [exported + consumed == produced]
accurate
# This code outputs the total production overall
total_production = produced.sum()
print("Total production is", total_production, "kilograms")
# This code outputs the total production in 2012
total_production_2012 = produced.loc['2012-01-01':'2012-12-01'].sum()
print("Total production in 2012 is", total_production_2012, "kilograms")
# This code outputs the total production in 2013
total_production_2013 = produced.loc['2013-01-01':'2013-12-01'].sum()
print("Total production in 2013 is", total_production_2013, "kilograms")
# This code outputs the total production in 2014
total_production_2014 = produced.loc['2014-01-01':'2014-12-01'].sum()
print("Total production in 2014 is", total_production_2014, "kilograms")
# This code outputs the total production in 2015
total_production_2015 = produced.loc['2015-01-01':'2015-12-01'].sum()
print("Total production in 2015 is", total_production_2015, "kilograms")
# This code outputs the total exports overall
total_exports = exported.sum()
print("Total exports is", total_exports, "kilograms")
# This code outputs the total exports in 2012
total_exports_2012 = exported.loc['2012-01-01':'2012-12-01'].sum()
print("Total exports in 2012 is", total_exports_2012, "kilograms")
# This code outputs the total exports in 2013
total_exports_2013 = exported.loc['2013-01-01':'2013-12-01'].sum()
print("Total exports in 2013 is", total_exports_2013, "kilograms")
# This code outputs the total exports in 2014
total_exports_2014 = exported.loc['2014-01-01':'2014-12-01'].sum()
print("Total exports in 2014 is", total_exports_2014, "kilograms")
# This code outputs the total exports in 2015
total_exports_2015 = exported.loc['2015-01-01':'2015-12-01'].sum()
print("Total exports in 2012 is", total_exports_2015, "kilograms")
# This code outputs the total consumption overall
total_consumption = consumed.sum()
print("Total consumption is", total_consumption, "kilograms")
# This code outputs the total consumption in 2012
total_consumption_2012 = consumed.loc['2012-01-01':'2012-12-01'].sum()
print("Total consumption in 2012 is", total_consumption_2012, "kilograms")
# This code outputs the total consumption in 2013
total_consumption_2013 = consumed.loc['2013-01-01':'2013-12-01'].sum()
print("Total consumption in 2013 is", total_consumption_2013, "kilograms")
# This code outputs the total consumption in 2014
total_consumption_2014 = consumed.loc['2014-01-01':'2014-12-01'].sum()
print("Total consumption in 2014 is", total_consumption_2014, "kilograms")
# This code outputs the total consumption in 2015
total_consumption_2015 = consumed.loc['2015-01-01':'2015-12-01'].sum()
print("Total consumption in 2015 is", total_consumption_2015, "kilograms")
#This shows the correlation between the various columns in the 'tea_reset' dataset
tea_reset.corr()
#This chart shows the trend in production of tea from January 2012 to December 2015
produced.plot(kind = 'line', figsize=(15,10))
plt.title("Quantity Produced against Time", fontsize=16)
plt.xlabel("Time in Year-Month",fontsize=12)
plt.ylabel("Quantity Produced (Kgs) * 10^7",fontsize=12)
#This chart shows the trend in exporting tea from January 2012 to December 2015
exported.plot(kind = 'line', figsize=(15,10))
plt.title("Quantity Exported against Time", fontsize=16)
plt.xlabel("Time in Year-Month",fontsize=12)
plt.ylabel("Quantity Exported (Kgs) * 10^7",fontsize=12)
#This chart shows the trend in consuming tea from January 2012 to December 2015
consumed.plot(kind = 'line', figsize=(15,10))
plt.title("Quantity Consumed against Time", fontsize=16)
plt.xlabel("Time in Year-Month",fontsize=12)
plt.ylabel("Quantity Consumed (Kgs) * 10^6",fontsize=12)
#This chart shows the correlation in quantity of tea produced, exported and consumed
produced.plot(figsize=(15,10))
exported.plot(figsize=(15,10))
consumed.plot(figsize=(15,10))
plt.plot(produced)
plt.plot(exported)
plt.plot(consumed)
plt.xlabel("Time in Year-Month", fontsize=12)
plt.ylabel("Quantity (Kgs) * 10^7", fontsize=12)
plt.legend(loc = 'upper right')
plt.title("Quantity against Time", fontsize=16)
plt.show()
#This creates a dataframe whose exports have been divided by one million to allow the relating of
#export price to quantity of exports
exported2 = (exported / (10**6))
exported2
#This chart shows the change in exported price and quantity exported from the beginning of 2012
#to the end of 2015
exported2.plot(figsize=(15,10))
average_exported_price.plot(figsize=(15,10))
plt.plot(exported2)
plt.plot(average_exported_price)
plt.xlabel("Time in Year-Month", fontsize=12)
plt.ylabel("Quantity(Kg) * 10^6", fontsize=12)
plt.legend(loc = 'upper right')
plt.title("Exports and Export Prices over Time", fontsize=16)
plt.show()
| 0.314682 | 0.946151 |
# Regiment
### Introduction:
Special thanks to: http://chrisalbon.com/ for sharing the dataset and materials.
### Step 1. Import the necessary libraries
```
import pandas as pd
```
### Step 2. Create the DataFrame with the following values:
```
raw_data = {'regiment': ['Nighthawks', 'Nighthawks', 'Nighthawks', 'Nighthawks', 'Dragoons', 'Dragoons', 'Dragoons', 'Dragoons', 'Scouts', 'Scouts', 'Scouts', 'Scouts'],
'company': ['1st', '1st', '2nd', '2nd', '1st', '1st', '2nd', '2nd','1st', '1st', '2nd', '2nd'],
'name': ['Miller', 'Jacobson', 'Ali', 'Milner', 'Cooze', 'Jacon', 'Ryaner', 'Sone', 'Sloan', 'Piger', 'Riani', 'Ali'],
'preTestScore': [4, 24, 31, 2, 3, 4, 24, 31, 2, 3, 2, 3],
'postTestScore': [25, 94, 57, 62, 70, 25, 94, 57, 62, 70, 62, 70]}
```
### Step 3. Assign it to a variable called regiment.
#### Don't forget to name each column
```
regiment = pd.DataFrame.from_dict(raw_data)
regiment.head()
```
### Step 4. What is the mean preTestScore from the regiment Nighthawks?
```
regiment[regiment['regiment'] == 'Nighthawks']['preTestScore'].mean()
# original answer is wrong
```
### Step 5. Present general statistics by company
```
regiment.groupby('company').describe()
```
### Step 6. What is the mean of each company's preTestScore?
```
regiment.groupby('company')[['preTestScore']].mean()
```
### Step 7. Present the mean preTestScores grouped by regiment and company
```
regiment.groupby(['regiment', 'company'])[['preTestScore']].mean()
```
### Step 8. Present the mean preTestScores grouped by regiment and company without heirarchical indexing
```
pd.crosstab(regiment['regiment'], regiment['company'], values=regiment['preTestScore'], aggfunc='mean')
# Original solution
regiment.groupby(['regiment', 'company']).preTestScore.mean().unstack()
```
### Step 9. Group the entire dataframe by regiment and company
```
regiment.groupby(['regiment', 'company']).mean()
```
### Step 10. What is the number of observations in each regiment and company
```
# pd.crosstab(regiment['regiment'], regiment['company']) # alternative
regiment.groupby(['company', 'regiment']).size()
```
### Step 11. Iterate over a group and print the name and the whole data from the regiment
```
for regiment_name in sorted(regiment['regiment'].unique()):
print(regiment_name)
print(regiment[regiment['regiment'] == regiment_name])
# Original solution:
# Group the dataframe by regiment, and for each regiment,
for name, group in regiment.groupby('regiment'):
# print the name of the regiment
print(name)
# print the data of that regiment
print(group)
```
|
github_jupyter
|
import pandas as pd
raw_data = {'regiment': ['Nighthawks', 'Nighthawks', 'Nighthawks', 'Nighthawks', 'Dragoons', 'Dragoons', 'Dragoons', 'Dragoons', 'Scouts', 'Scouts', 'Scouts', 'Scouts'],
'company': ['1st', '1st', '2nd', '2nd', '1st', '1st', '2nd', '2nd','1st', '1st', '2nd', '2nd'],
'name': ['Miller', 'Jacobson', 'Ali', 'Milner', 'Cooze', 'Jacon', 'Ryaner', 'Sone', 'Sloan', 'Piger', 'Riani', 'Ali'],
'preTestScore': [4, 24, 31, 2, 3, 4, 24, 31, 2, 3, 2, 3],
'postTestScore': [25, 94, 57, 62, 70, 25, 94, 57, 62, 70, 62, 70]}
regiment = pd.DataFrame.from_dict(raw_data)
regiment.head()
regiment[regiment['regiment'] == 'Nighthawks']['preTestScore'].mean()
# original answer is wrong
regiment.groupby('company').describe()
regiment.groupby('company')[['preTestScore']].mean()
regiment.groupby(['regiment', 'company'])[['preTestScore']].mean()
pd.crosstab(regiment['regiment'], regiment['company'], values=regiment['preTestScore'], aggfunc='mean')
# Original solution
regiment.groupby(['regiment', 'company']).preTestScore.mean().unstack()
regiment.groupby(['regiment', 'company']).mean()
# pd.crosstab(regiment['regiment'], regiment['company']) # alternative
regiment.groupby(['company', 'regiment']).size()
for regiment_name in sorted(regiment['regiment'].unique()):
print(regiment_name)
print(regiment[regiment['regiment'] == regiment_name])
# Original solution:
# Group the dataframe by regiment, and for each regiment,
for name, group in regiment.groupby('regiment'):
# print the name of the regiment
print(name)
# print the data of that regiment
print(group)
| 0.154344 | 0.959611 |
# Example Portfolio Analysis
```
%load_ext autoreload
%autoreload 2
# !pip install nb_black
%load_ext nb_black
import sys
sys.path.append("..")
%matplotlib inline
import matplotlib as mpl
import matplotlib.pyplot as plt
mpl.style.use("../assets/ppt.mplstyle")
import seaborn as sns
from utils import print_properties
from ppt.portfolio_value import Portfolio
```
<a id='ld'></a>
## Load Data
Initialising the `Portfolio` object will load the input data, download the relevant stock prices and calculate some initial stats on the portfolio.
The default location for the input file is `../data/raw/purchase_info.csv` however, you can specify your own location using the optional `input_data_source` argument.
```
# initiate Portfolio object
pf = Portfolio()
# add a SNP500 benchmark
pf.add_benchmark("^GSPC")
# print available properties
print_properties(pf)
```
## High level analysis
Now you have access to all the raw underlying stock price data and the daily running totals of the overall portfolio and each individual position. We can quickly plot the value of the portfolio over time and the running profit (portfolio value minus cash injections).
```
mpl.style.use("../assets/ppt.mplstyle")
# show the daily portfolio value including cash injections
pf.portfolio_value_usd.plot(label="total portfolio value")
pf.cash.cash_flows["external_cashflows"].plot(label="total cash input")
plt.title("Daily portfolio value (USD)")
plt.xlabel(None)
plt.gca().get_yaxis().set_major_formatter(mpl.ticker.StrMethodFormatter("${x:,.0f}"))
plt.legend()
plt.show()
# show the running profit (portfolio value minus cash injections)
pf.profit.plot(label="Profit")
plt.title("Running Profit (USD)")
plt.gca().get_yaxis().set_major_formatter(mpl.ticker.StrMethodFormatter("${x:,.0f}"))
plt.hlines(y=0, xmin=pf.datetime_index[0], xmax=pf.datetime_index[-1], color="#0EAD69")
plt.legend()
plt.show()
```
## Individual Stock Analysis
**Historical correlations (since start of portfolio)**
```
stock_prices = pf.daily_stock_prices_local_currency
returns = stock_prices.to_log_returns()
sns.clustermap(returns.corr(), cmap="RdYlGn", vmin=-1, vmax=1, annot=True)
plt.title("Historical Stock Correlations")
plt.show()
stock_values = pf.daily_stock_prices_usd
stock_values.to_drawdown_series().plot()
plt.title("Draw down of stocks since portfolio start")
plt.ylabel("Drawdown from most recent high")
plt.show()
```
**TODO: More indepth analysis. Callibrate returns for time-weighted returns to accoutn for additional buy/selling**
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
# !pip install nb_black
%load_ext nb_black
import sys
sys.path.append("..")
%matplotlib inline
import matplotlib as mpl
import matplotlib.pyplot as plt
mpl.style.use("../assets/ppt.mplstyle")
import seaborn as sns
from utils import print_properties
from ppt.portfolio_value import Portfolio
# initiate Portfolio object
pf = Portfolio()
# add a SNP500 benchmark
pf.add_benchmark("^GSPC")
# print available properties
print_properties(pf)
mpl.style.use("../assets/ppt.mplstyle")
# show the daily portfolio value including cash injections
pf.portfolio_value_usd.plot(label="total portfolio value")
pf.cash.cash_flows["external_cashflows"].plot(label="total cash input")
plt.title("Daily portfolio value (USD)")
plt.xlabel(None)
plt.gca().get_yaxis().set_major_formatter(mpl.ticker.StrMethodFormatter("${x:,.0f}"))
plt.legend()
plt.show()
# show the running profit (portfolio value minus cash injections)
pf.profit.plot(label="Profit")
plt.title("Running Profit (USD)")
plt.gca().get_yaxis().set_major_formatter(mpl.ticker.StrMethodFormatter("${x:,.0f}"))
plt.hlines(y=0, xmin=pf.datetime_index[0], xmax=pf.datetime_index[-1], color="#0EAD69")
plt.legend()
plt.show()
stock_prices = pf.daily_stock_prices_local_currency
returns = stock_prices.to_log_returns()
sns.clustermap(returns.corr(), cmap="RdYlGn", vmin=-1, vmax=1, annot=True)
plt.title("Historical Stock Correlations")
plt.show()
stock_values = pf.daily_stock_prices_usd
stock_values.to_drawdown_series().plot()
plt.title("Draw down of stocks since portfolio start")
plt.ylabel("Drawdown from most recent high")
plt.show()
| 0.297674 | 0.88136 |
# Unity ML-Agents Toolkit
## Environment Basics
This notebook contains a walkthrough of the basic functions of the Python API for the Unity ML-Agents toolkit. For instructions on building a Unity environment, see [here](https://github.com/Unity-Technologies/ml-agents/blob/master/docs/Getting-Started-with-Balance-Ball.md).
### 1. Set environment parameters
Be sure to set `env_name` to the name of the Unity environment file you want to launch. Ensure that the environment build is in `../envs`.
```
env_name = None#"../envs/3DBall" # Name of the Unity environment binary to launch
train_mode = True # Whether to run the environment in training or inference mode
```
### 2. Load dependencies
The following loads the necessary dependencies and checks the Python version (at runtime). ML-Agents Toolkit (v0.3 onwards) requires Python 3.
```
import matplotlib.pyplot as plt
import numpy as np
import sys
from mlagents.envs.environment import UnityEnvironment
%matplotlib inline
print("Python version:")
print(sys.version)
# check Python version
if (sys.version_info[0] < 3):
raise Exception("ERROR: ML-Agents Toolkit (v0.3 onwards) requires Python 3")
```
### 3. Start the environment
`UnityEnvironment` launches and begins communication with the environment when instantiated.
Environments contain _brains_ which are responsible for deciding the actions of their associated _agents_. Here we check for the first brain available, and set it as the default brain we will be controlling from Python.
```
env = UnityEnvironment(file_name=env_name)
# Set the default brain to work with
default_brain = env.brain_names[0]
brain = env.brains[default_brain]
```
### 4. Examine the observation and state spaces
We can reset the environment to be provided with an initial set of observations and states for all the agents within the environment. In ML-Agents, _states_ refer to a vector of variables corresponding to relevant aspects of the environment for an agent. Likewise, _observations_ refer to a set of relevant pixel-wise visuals for an agent.
```
# Reset the environment
env_info = env.reset(train_mode=train_mode)[default_brain]
# Examine the state space for the default brain
print("Agent state looks like: \n{}".format(env_info.vector_observations[0]))
# Examine the observation space for the default brain
for observation in env_info.visual_observations:
print("Agent observations look like:")
if observation.shape[3] == 3:
plt.imshow(observation[0,:,:,:])
else:
plt.imshow(observation[0,:,:,0])
```
### 5. Take random actions in the environment
Once we restart an environment, we can step the environment forward and provide actions to all of the agents within the environment. Here we simply choose random actions based on the `action_space_type` of the default brain.
Once this cell is executed, 10 messages will be printed that detail how much reward will be accumulated for the next 10 episodes. The Unity environment will then pause, waiting for further signals telling it what to do next. Thus, not seeing any animation is expected when running this cell.
```
for episode in range(10):
env_info = env.reset(train_mode=train_mode)[default_brain]
done = False
episode_rewards = 0
while not done:
action_size = brain.vector_action_space_size
if brain.vector_action_space_type == 'continuous':
env_info = env.step(np.random.randn(len(env_info.agents),
action_size[0]))[default_brain]
else:
action = np.column_stack([np.random.randint(0, action_size[i], size=(len(env_info.agents))) for i in range(len(action_size))])
env_info = env.step(action)[default_brain]
episode_rewards += env_info.rewards[0]
done = env_info.local_done[0]
print("Total reward this episode: {}".format(episode_rewards))
```
### 6. Close the environment when finished
When we are finished using an environment, we can close it with the function below.
```
env.close()
```
|
github_jupyter
|
env_name = None#"../envs/3DBall" # Name of the Unity environment binary to launch
train_mode = True # Whether to run the environment in training or inference mode
import matplotlib.pyplot as plt
import numpy as np
import sys
from mlagents.envs.environment import UnityEnvironment
%matplotlib inline
print("Python version:")
print(sys.version)
# check Python version
if (sys.version_info[0] < 3):
raise Exception("ERROR: ML-Agents Toolkit (v0.3 onwards) requires Python 3")
env = UnityEnvironment(file_name=env_name)
# Set the default brain to work with
default_brain = env.brain_names[0]
brain = env.brains[default_brain]
# Reset the environment
env_info = env.reset(train_mode=train_mode)[default_brain]
# Examine the state space for the default brain
print("Agent state looks like: \n{}".format(env_info.vector_observations[0]))
# Examine the observation space for the default brain
for observation in env_info.visual_observations:
print("Agent observations look like:")
if observation.shape[3] == 3:
plt.imshow(observation[0,:,:,:])
else:
plt.imshow(observation[0,:,:,0])
for episode in range(10):
env_info = env.reset(train_mode=train_mode)[default_brain]
done = False
episode_rewards = 0
while not done:
action_size = brain.vector_action_space_size
if brain.vector_action_space_type == 'continuous':
env_info = env.step(np.random.randn(len(env_info.agents),
action_size[0]))[default_brain]
else:
action = np.column_stack([np.random.randint(0, action_size[i], size=(len(env_info.agents))) for i in range(len(action_size))])
env_info = env.step(action)[default_brain]
episode_rewards += env_info.rewards[0]
done = env_info.local_done[0]
print("Total reward this episode: {}".format(episode_rewards))
env.close()
| 0.334481 | 0.991563 |
```
import pandas as pd
#Google colab does not have pickle
try:
import pickle5 as pickle
except:
!pip install pickle5
import pickle5 as pickle
import os
import seaborn as sns
import sys
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
from keras.layers import Dense, Input, GlobalMaxPooling1D,Flatten
from keras.layers import Conv1D, MaxPooling1D, Embedding,Concatenate
from keras.models import Model
from sklearn.metrics import roc_auc_score,confusion_matrix,roc_curve, auc
from numpy import random
from keras.layers import LSTM, Bidirectional, GlobalMaxPool1D, Dropout
from keras.optimizers import Adam
from keras.utils.vis_utils import plot_model
import sys
sys.path.insert(0,'/content/drive/MyDrive/ML_Data/')
import functions as f
def load_data(D=1,randomize=False):
try:
with open('/content/drive/MyDrive/ML_Data/df_train_'+str(D)+'D.pickle', 'rb') as handle:
df_train = pickle.load(handle)
except:
df_train = pd.read_pickle("C:/Users/nik00/py/proj/hyppi-train.pkl")
try:
with open('/content/drive/MyDrive/ML_Data/df_test_'+str(D)+'D.pickle', 'rb') as handle:
df_test = pickle.load(handle)
except:
df_test = pd.read_pickle("C:/Users/nik00/py/proj/hyppi-independent.pkl")
if randomize:
return shuff_together(df_train,df_test)
else:
return df_train,df_test
df_train,df_test = load_data(2)
print('The data used will be:')
df_train
lengths = sorted(len(s) for s in df_train['Human'])
print("Median length of Human sequence is",lengths[len(lengths)//2])
_ = sns.displot(lengths)
_=plt.title("Most Human sequences seem to be less than 2000 in length")
lengths = sorted(len(s) for s in df_train['Yersinia'])
print("Median length of Yersinia sequence is",lengths[len(lengths)//2])
_ = sns.displot(lengths)
_=plt.title("Most Yersinia sequences seem to be less than 1000 in length")
rows = df_train['Joined'].shape[0]
lengths = sorted(len(s) for s in df_train['Joined'])
print("Median length of Joined sequence is",lengths[len(lengths)//2])
_ = sns.displot(lengths)
_=plt.title("Most Joined sequences seem to be less than 2000 in length")
data1_2D_doubleip,data2_2D_doubleip,data1_test_2D_doubleip,data2_test_2D_doubleip,num_words_2D,MAX_SEQUENCE_LENGTH_2D_dIP,MAX_VOCAB_SIZE_2D = f.get_seq_data_doubleip(1000,1000,df_train,df_test,pad='pre',show=True)
data_2D_join,data_test_2D_join,num_words_2D,MAX_SEQUENCE_LENGTH_2D_J,MAX_VOCAB_SIZE_2D = f.get_seq_data_join(1000,1500,df_train,df_test,pad='pre',show=True)
EMBEDDING_DIM_2D = 5
BATCH_SIZE = 128
EPOCHS = 20
FILTERS_2D=128
KERNEL_2D =32
DROPOUT = 0.6
ip = Input(shape=(MAX_SEQUENCE_LENGTH_2D_J,))
#x = Embedding(num_words_2D, EMBEDDING_DIM_2D, input_length=MAX_SEQUENCE_LENGTH_2D_J,trainable=True)(ip)
x = f.embedding_layer(num_words_2D,MAX_SEQUENCE_LENGTH_2D_J,EMBEDDING_DIM_2D)(ip)
x = Conv1D(FILTERS_2D, KERNEL_2D, activation='relu')(x)
x = Dropout(DROPOUT)(x)
x = MaxPooling1D(3)(x)
x= Flatten()(x)
x = Dropout(DROPOUT)(x)
x = Dense(128, activation='relu')(x)
x_2D_Joined = Model(inputs=ip, outputs=x)
inputA = Input(shape=(MAX_SEQUENCE_LENGTH_2D_dIP,))
#x1 = Embedding(num_words_2D, EMBEDDING_DIM_2D, input_length=MAX_SEQUENCE_LENGTH_2D_dIP,trainable=True)(inputA)
x1 = f.embedding_layer(num_words_2D,MAX_SEQUENCE_LENGTH_2D_dIP,EMBEDDING_DIM_2D)(inputA)
x1 = Conv1D(FILTERS_2D, KERNEL_2D, activation='relu')(x1)
x1= Dropout(DROPOUT)(x1)
x1 = MaxPooling1D(3)(x1)
x1= Flatten()(x1)
x1 = Dropout(DROPOUT)(x1)
x1 = Dense(128, activation='relu')(x1)
# x1 = Dropout(DROPOUT)(x1)
# x1 = Dense(1, activation='sigmoid')(x1)
x1_Human_2D_doubleip = Model(inputs=inputA, outputs=x1)
inputB = Input(shape=(MAX_SEQUENCE_LENGTH_2D_dIP,))
#x2 = Embedding(num_words_2D, EMBEDDING_DIM_2D, input_length=MAX_SEQUENCE_LENGTH_2D_dIP,trainable=True)(inputB)
x2 = f.embedding_layer(num_words_2D,MAX_SEQUENCE_LENGTH_2D_dIP,EMBEDDING_DIM_2D)(inputB)
x2 = Conv1D(FILTERS_2D, KERNEL_2D, activation='relu')(x2)
x2= Dropout(DROPOUT)(x2)
x2 = MaxPooling1D(3)(x2)
x2= Flatten()(x2)
x2 = Dropout(DROPOUT)(x2)
x2 = Dense(128, activation='relu')(x2)
# x2 = Dropout(DROPOUT)(x2)
# x2 = Dense(1, activation='sigmoid')(x2)
x2_Yersinia_2D_doubleip = Model(inputs=inputB, outputs=x2)
concatenator = Concatenate(axis=1)
y = concatenator([x_2D_Joined.output,x1_Human_2D_doubleip.output, x2_Yersinia_2D_doubleip.output])
y = Dropout(DROPOUT)(y)
y = Dense(128, activation='relu')(y)
y = Dropout(DROPOUT)(y)
output = Dense(1, activation="sigmoid")(y)
model2D_CNN_combine = Model(inputs=[x_2D_Joined.input,x1_Human_2D_doubleip.input, x2_Yersinia_2D_doubleip.input], outputs=output)
model2D_CNN_combine.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
#plot_model(model2D_CNN_combine, to_file='model_plot.png', show_shapes=True, show_layer_names=False)
model2D_CNN_combine.fit([data_2D_join,data1_2D_doubleip,data2_2D_doubleip], df_train['label'].values, epochs=EPOCHS, batch_size=BATCH_SIZE)
print(roc_auc_score(df_test['label'].values, model2D_CNN_combine.predict([data_test_2D_join,data1_test_2D_doubleip,data2_test_2D_doubleip])))
```
|
github_jupyter
|
import pandas as pd
#Google colab does not have pickle
try:
import pickle5 as pickle
except:
!pip install pickle5
import pickle5 as pickle
import os
import seaborn as sns
import sys
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
from keras.layers import Dense, Input, GlobalMaxPooling1D,Flatten
from keras.layers import Conv1D, MaxPooling1D, Embedding,Concatenate
from keras.models import Model
from sklearn.metrics import roc_auc_score,confusion_matrix,roc_curve, auc
from numpy import random
from keras.layers import LSTM, Bidirectional, GlobalMaxPool1D, Dropout
from keras.optimizers import Adam
from keras.utils.vis_utils import plot_model
import sys
sys.path.insert(0,'/content/drive/MyDrive/ML_Data/')
import functions as f
def load_data(D=1,randomize=False):
try:
with open('/content/drive/MyDrive/ML_Data/df_train_'+str(D)+'D.pickle', 'rb') as handle:
df_train = pickle.load(handle)
except:
df_train = pd.read_pickle("C:/Users/nik00/py/proj/hyppi-train.pkl")
try:
with open('/content/drive/MyDrive/ML_Data/df_test_'+str(D)+'D.pickle', 'rb') as handle:
df_test = pickle.load(handle)
except:
df_test = pd.read_pickle("C:/Users/nik00/py/proj/hyppi-independent.pkl")
if randomize:
return shuff_together(df_train,df_test)
else:
return df_train,df_test
df_train,df_test = load_data(2)
print('The data used will be:')
df_train
lengths = sorted(len(s) for s in df_train['Human'])
print("Median length of Human sequence is",lengths[len(lengths)//2])
_ = sns.displot(lengths)
_=plt.title("Most Human sequences seem to be less than 2000 in length")
lengths = sorted(len(s) for s in df_train['Yersinia'])
print("Median length of Yersinia sequence is",lengths[len(lengths)//2])
_ = sns.displot(lengths)
_=plt.title("Most Yersinia sequences seem to be less than 1000 in length")
rows = df_train['Joined'].shape[0]
lengths = sorted(len(s) for s in df_train['Joined'])
print("Median length of Joined sequence is",lengths[len(lengths)//2])
_ = sns.displot(lengths)
_=plt.title("Most Joined sequences seem to be less than 2000 in length")
data1_2D_doubleip,data2_2D_doubleip,data1_test_2D_doubleip,data2_test_2D_doubleip,num_words_2D,MAX_SEQUENCE_LENGTH_2D_dIP,MAX_VOCAB_SIZE_2D = f.get_seq_data_doubleip(1000,1000,df_train,df_test,pad='pre',show=True)
data_2D_join,data_test_2D_join,num_words_2D,MAX_SEQUENCE_LENGTH_2D_J,MAX_VOCAB_SIZE_2D = f.get_seq_data_join(1000,1500,df_train,df_test,pad='pre',show=True)
EMBEDDING_DIM_2D = 5
BATCH_SIZE = 128
EPOCHS = 20
FILTERS_2D=128
KERNEL_2D =32
DROPOUT = 0.6
ip = Input(shape=(MAX_SEQUENCE_LENGTH_2D_J,))
#x = Embedding(num_words_2D, EMBEDDING_DIM_2D, input_length=MAX_SEQUENCE_LENGTH_2D_J,trainable=True)(ip)
x = f.embedding_layer(num_words_2D,MAX_SEQUENCE_LENGTH_2D_J,EMBEDDING_DIM_2D)(ip)
x = Conv1D(FILTERS_2D, KERNEL_2D, activation='relu')(x)
x = Dropout(DROPOUT)(x)
x = MaxPooling1D(3)(x)
x= Flatten()(x)
x = Dropout(DROPOUT)(x)
x = Dense(128, activation='relu')(x)
x_2D_Joined = Model(inputs=ip, outputs=x)
inputA = Input(shape=(MAX_SEQUENCE_LENGTH_2D_dIP,))
#x1 = Embedding(num_words_2D, EMBEDDING_DIM_2D, input_length=MAX_SEQUENCE_LENGTH_2D_dIP,trainable=True)(inputA)
x1 = f.embedding_layer(num_words_2D,MAX_SEQUENCE_LENGTH_2D_dIP,EMBEDDING_DIM_2D)(inputA)
x1 = Conv1D(FILTERS_2D, KERNEL_2D, activation='relu')(x1)
x1= Dropout(DROPOUT)(x1)
x1 = MaxPooling1D(3)(x1)
x1= Flatten()(x1)
x1 = Dropout(DROPOUT)(x1)
x1 = Dense(128, activation='relu')(x1)
# x1 = Dropout(DROPOUT)(x1)
# x1 = Dense(1, activation='sigmoid')(x1)
x1_Human_2D_doubleip = Model(inputs=inputA, outputs=x1)
inputB = Input(shape=(MAX_SEQUENCE_LENGTH_2D_dIP,))
#x2 = Embedding(num_words_2D, EMBEDDING_DIM_2D, input_length=MAX_SEQUENCE_LENGTH_2D_dIP,trainable=True)(inputB)
x2 = f.embedding_layer(num_words_2D,MAX_SEQUENCE_LENGTH_2D_dIP,EMBEDDING_DIM_2D)(inputB)
x2 = Conv1D(FILTERS_2D, KERNEL_2D, activation='relu')(x2)
x2= Dropout(DROPOUT)(x2)
x2 = MaxPooling1D(3)(x2)
x2= Flatten()(x2)
x2 = Dropout(DROPOUT)(x2)
x2 = Dense(128, activation='relu')(x2)
# x2 = Dropout(DROPOUT)(x2)
# x2 = Dense(1, activation='sigmoid')(x2)
x2_Yersinia_2D_doubleip = Model(inputs=inputB, outputs=x2)
concatenator = Concatenate(axis=1)
y = concatenator([x_2D_Joined.output,x1_Human_2D_doubleip.output, x2_Yersinia_2D_doubleip.output])
y = Dropout(DROPOUT)(y)
y = Dense(128, activation='relu')(y)
y = Dropout(DROPOUT)(y)
output = Dense(1, activation="sigmoid")(y)
model2D_CNN_combine = Model(inputs=[x_2D_Joined.input,x1_Human_2D_doubleip.input, x2_Yersinia_2D_doubleip.input], outputs=output)
model2D_CNN_combine.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])
#plot_model(model2D_CNN_combine, to_file='model_plot.png', show_shapes=True, show_layer_names=False)
model2D_CNN_combine.fit([data_2D_join,data1_2D_doubleip,data2_2D_doubleip], df_train['label'].values, epochs=EPOCHS, batch_size=BATCH_SIZE)
print(roc_auc_score(df_test['label'].values, model2D_CNN_combine.predict([data_test_2D_join,data1_test_2D_doubleip,data2_test_2D_doubleip])))
| 0.417509 | 0.212008 |
```
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv('data.csv')
df.head()
```
#### 1.1) Analysing Price of Each Product
```
print("Average : ", int(df['price'].mean()))
print("Minimum : ", int(df['price'].min()))
print("Maximum : ", int(df['price'].max()))
price = []
for i in df['price'].unique():
price.append([i, len(df[df['price'] == i])])
df_pr = pd.DataFrame(price, columns= ['price','freq'])
df_pr.sort_values(by = 'price')['price']
pr = list(df_pr.sort_values(by = 'price')['price'])
fr = list(df_pr.sort_values(by = 'price')['freq'])
pri = []
for i in pr:
pri.append(str(i))
plt.bar(pri,fr)
plt.show()
```
##### Assumption - Most number of product that are sold are of less than 100 Dollars
#### 1.2) Analysing Price of Each Order
```
print('Minimum : ',int(df['total'].min()))
print('Maximum : ',int(df['total'].max()))
print('Average : ',int(df['total'].mean()))
```
##### Assumption - Most number of product that are sold are of less than 100 Dollars
#### 1.3) Analysing Quantity Column
```
qn = []
fr = []
for i in df['quantity'].unique():
qn.append(i)
fr.append(len(df[df['quantity'] == i]))
plt.pie(fr, labels = qn, shadow = True, autopct = '%1.2f%%')
plt.show()
```
##### Assumption - Most number of people (90%) are buying 1 product at a time
We can add offers so that they can buy more products
#### 1.4) Analysing Full_date Column
```
data = []
for i in df['full_date'].unique():
data.append([i, len(df[df['full_date'] == i])])
data = pd.DataFrame(data, columns = ['date','fr'])
data.sort_values(by = 'fr',ascending = False).head(20)
```
##### Assumption - Decembers is the month where we're having most produts sold
#### 1.5) Analysing Hours Column
```
data = []
for i in df['hour'].unique():
data.append([i, len(df[df['hour'] == i])])
data = pd.DataFrame(data, columns = ['hour','fr'])
hr = list(data.sort_values(by = 'hour',ascending = True)['hour'])
fr = list(data.sort_values(by = 'hour',ascending = True)['fr'])
plt.bar(hr,fr)
data.sort_values(by = 'fr', ascending = False).head()
```
##### Assumption - [0 - 8 AM] is having least footfall.
We should put more efforts from 9AM - 23PM
##### 1.6) Analysing Date Column
```
data = []
for i in df['date'].unique():
data.append([i, len(df[df['date'] == i])])
data = pd.DataFrame(data, columns = ['date','fr'])
hr = list(data.sort_values(by = 'date',ascending = True)['date'])
fr = list(data.sort_values(by = 'date',ascending = True)['fr'])
plt.bar(hr,fr)
data.sort_values(by = 'fr', ascending = False).head()
```
##### Assumption - People buys less products in monthend
##### 1.7) Analysing Month Column
```
data = []
for i in df['month'].unique():
data.append([i, len(df[df['month'] == i])])
data = pd.DataFrame(data, columns = ['month','fr'])
hr = list(data.sort_values(by = 'month',ascending = True)['month'])
fr = list(data.sort_values(by = 'month',ascending = True)['fr'])
plt.bar(hr,fr)
```
##### Assumption - Peolpe are byting most in year End
#### 1.8) Analysing Year Column
```
y1 = len(df[df['year'] == 2019])
y2 = len(df[df['year'] == 2020])
print('Orders in 2019: ',y1)
print('Orders in 2020: ',y2)
```
#### 1.9) Analysing Day Column
```
data = []
for i in df['day'].unique():
data.append([i, len(df[df['day'] == i])])
data = pd.DataFrame(data, columns = ['day','fr'])
data
```
##### 1.10) Analysing Address Column
```
df.groupby('address').size().sort_values(ascending = False).head()
df['address'].nunique()
```
#### 1.11) Analysing City Column
```
df['city'].unique()
data = []
for i in df['city'].unique():
data.append([i, len(df[df['city'] == i])])
data = pd.DataFrame(data, columns = ['city','fr'])
ct = list(data.sort_values(by = 'fr',ascending = True)['city'])
fr = list(data.sort_values(by = 'fr',ascending = True)['fr'])
fig , axs = plt.subplots(figsize = (12,4))
axs.bar(ct,fr)
plt.show()
```
##### Conclusion - We should promote our stores more in SanFrancisco, Los Angeles and New York
#### 1.12) Analysing States
```
df['state'].unique()
data = []
for i in df['state'].unique():
data.append([i, len(df[df['state'] == i])])
data = pd.DataFrame(data, columns = ['state','fr'])
ct = list(data.sort_values(by = 'fr',ascending = True)['state'])
fr = list(data.sort_values(by = 'fr',ascending = True)['fr'])
fig , axs = plt.subplots(figsize = (8,4))
axs.bar(ct,fr)
plt.show()
```
##### Assumption - CA is giving best Business
Make our Network more stronger in CA
We should wokr on NY, TX, MA, GA, WA
We should work a lot on OR, ME
#### 1.13) Analysing Pincode Column
```
data = []
for i in df['pincode'].unique():
data.append([i, len(df[df['pincode'] == i])])
data = pd.DataFrame(data, columns = ['pincode','fr'])
pc = list(data.sort_values(by = 'fr',ascending = True)['pincode'])
fr = list(data.sort_values(by = 'fr',ascending = True)['fr'])
pincode = []
for i in pc:
pincode.append(str(i))
fig , axs = plt.subplots(figsize = (8,4))
axs.bar(pincode,fr)
plt.show()
```
#### 1.14) Analysing Timezone
```
x1 = len(df[df['timezone'] == 'AM'])
x2 = len(df[df['timezone'] == 'PM'])
plt.pie([x1,x2], labels = ['AM','PM'], explode = [0.1,0], shadow = True, autopct = '%1.0f%%')
plt.title('Orders based on Timezone')
plt.show()
```
##### Conclusion - Most prople are ordering in Evening as compare to Morning
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv('data.csv')
df.head()
print("Average : ", int(df['price'].mean()))
print("Minimum : ", int(df['price'].min()))
print("Maximum : ", int(df['price'].max()))
price = []
for i in df['price'].unique():
price.append([i, len(df[df['price'] == i])])
df_pr = pd.DataFrame(price, columns= ['price','freq'])
df_pr.sort_values(by = 'price')['price']
pr = list(df_pr.sort_values(by = 'price')['price'])
fr = list(df_pr.sort_values(by = 'price')['freq'])
pri = []
for i in pr:
pri.append(str(i))
plt.bar(pri,fr)
plt.show()
print('Minimum : ',int(df['total'].min()))
print('Maximum : ',int(df['total'].max()))
print('Average : ',int(df['total'].mean()))
qn = []
fr = []
for i in df['quantity'].unique():
qn.append(i)
fr.append(len(df[df['quantity'] == i]))
plt.pie(fr, labels = qn, shadow = True, autopct = '%1.2f%%')
plt.show()
data = []
for i in df['full_date'].unique():
data.append([i, len(df[df['full_date'] == i])])
data = pd.DataFrame(data, columns = ['date','fr'])
data.sort_values(by = 'fr',ascending = False).head(20)
data = []
for i in df['hour'].unique():
data.append([i, len(df[df['hour'] == i])])
data = pd.DataFrame(data, columns = ['hour','fr'])
hr = list(data.sort_values(by = 'hour',ascending = True)['hour'])
fr = list(data.sort_values(by = 'hour',ascending = True)['fr'])
plt.bar(hr,fr)
data.sort_values(by = 'fr', ascending = False).head()
data = []
for i in df['date'].unique():
data.append([i, len(df[df['date'] == i])])
data = pd.DataFrame(data, columns = ['date','fr'])
hr = list(data.sort_values(by = 'date',ascending = True)['date'])
fr = list(data.sort_values(by = 'date',ascending = True)['fr'])
plt.bar(hr,fr)
data.sort_values(by = 'fr', ascending = False).head()
data = []
for i in df['month'].unique():
data.append([i, len(df[df['month'] == i])])
data = pd.DataFrame(data, columns = ['month','fr'])
hr = list(data.sort_values(by = 'month',ascending = True)['month'])
fr = list(data.sort_values(by = 'month',ascending = True)['fr'])
plt.bar(hr,fr)
y1 = len(df[df['year'] == 2019])
y2 = len(df[df['year'] == 2020])
print('Orders in 2019: ',y1)
print('Orders in 2020: ',y2)
data = []
for i in df['day'].unique():
data.append([i, len(df[df['day'] == i])])
data = pd.DataFrame(data, columns = ['day','fr'])
data
df.groupby('address').size().sort_values(ascending = False).head()
df['address'].nunique()
df['city'].unique()
data = []
for i in df['city'].unique():
data.append([i, len(df[df['city'] == i])])
data = pd.DataFrame(data, columns = ['city','fr'])
ct = list(data.sort_values(by = 'fr',ascending = True)['city'])
fr = list(data.sort_values(by = 'fr',ascending = True)['fr'])
fig , axs = plt.subplots(figsize = (12,4))
axs.bar(ct,fr)
plt.show()
df['state'].unique()
data = []
for i in df['state'].unique():
data.append([i, len(df[df['state'] == i])])
data = pd.DataFrame(data, columns = ['state','fr'])
ct = list(data.sort_values(by = 'fr',ascending = True)['state'])
fr = list(data.sort_values(by = 'fr',ascending = True)['fr'])
fig , axs = plt.subplots(figsize = (8,4))
axs.bar(ct,fr)
plt.show()
data = []
for i in df['pincode'].unique():
data.append([i, len(df[df['pincode'] == i])])
data = pd.DataFrame(data, columns = ['pincode','fr'])
pc = list(data.sort_values(by = 'fr',ascending = True)['pincode'])
fr = list(data.sort_values(by = 'fr',ascending = True)['fr'])
pincode = []
for i in pc:
pincode.append(str(i))
fig , axs = plt.subplots(figsize = (8,4))
axs.bar(pincode,fr)
plt.show()
x1 = len(df[df['timezone'] == 'AM'])
x2 = len(df[df['timezone'] == 'PM'])
plt.pie([x1,x2], labels = ['AM','PM'], explode = [0.1,0], shadow = True, autopct = '%1.0f%%')
plt.title('Orders based on Timezone')
plt.show()
| 0.253584 | 0.812347 |
# NATURAL LANGUAGE PROCESSING
This notebook covers chapters 22 and 23 from the book *Artificial Intelligence: A Modern Approach*, 3rd Edition. The implementations of the algorithms can be found in [nlp.py](https://github.com/aimacode/aima-python/blob/master/nlp.py).
Run the below cell to import the code from the module and get started!
```
import nlp
from nlp import Page, HITS
from nlp import Lexicon, Rules, Grammar, ProbLexicon, ProbRules, ProbGrammar
from nlp import CYK_parse, Chart
from notebook import psource
```
## CONTENTS
* Overview
* Languages
* HITS
* Question Answering
* CYK Parse
* Chart Parsing
## OVERVIEW
**Natural Language Processing (NLP)** is a field of AI concerned with understanding, analyzing and using natural languages. This field is considered a difficult yet intriguing field of study, since it is connected to how humans and their languages work.
Applications of the field include translation, speech recognition, topic segmentation, information extraction and retrieval, and a lot more.
Below we take a look at some algorithms in the field. Before we get right into it though, we will take a look at a very useful form of language, **context-free** languages. Even though they are a bit restrictive, they have been used a lot in research in natural language processing.
## LANGUAGES
Languages can be represented by a set of grammar rules over a lexicon of words. Different languages can be represented by different types of grammar, but in Natural Language Processing we are mainly interested in context-free grammars.
### Context-Free Grammars
A lot of natural and programming languages can be represented by a **Context-Free Grammar (CFG)**. A CFG is a grammar that has a single non-terminal symbol on the left-hand side. That means a non-terminal can be replaced by the right-hand side of the rule regardless of context. An example of a CFG:
```
S -> aSb | ε
```
That means `S` can be replaced by either `aSb` or `ε` (with `ε` we denote the empty string). The lexicon of the language is comprised of the terminals `a` and `b`, while with `S` we denote the non-terminal symbol. In general, non-terminals are capitalized while terminals are not, and we usually name the starting non-terminal `S`. The language generated by the above grammar is the language a<sup>n</sup>b<sup>n</sup> for n greater or equal than 1.
### Probabilistic Context-Free Grammar
While a simple CFG can be very useful, we might want to know the chance of each rule occuring. Above, we do not know if `S` is more likely to be replaced by `aSb` or `ε`. **Probabilistic Context-Free Grammars (PCFG)** are built to fill exactly that need. Each rule has a probability, given in brackets, and the probabilities of a rule sum up to 1:
```
S -> aSb [0.7] | ε [0.3]
```
Now we know it is more likely for `S` to be replaced by `aSb` than by `e`.
An issue with *PCFGs* is how we will assign the various probabilities to the rules. We could use our knowledge as humans to assign the probabilities, but that is a laborious and prone to error task. Instead, we can *learn* the probabilities from data. Data is categorized as labeled (with correctly parsed sentences, usually called a **treebank**) or unlabeled (given only lexical and syntactic category names).
With labeled data, we can simply count the occurences. For the above grammar, if we have 100 `S` rules and 30 of them are of the form `S -> ε`, we assign a probability of 0.3 to the transformation.
With unlabeled data we have to learn both the grammar rules and the probability of each rule. We can go with many approaches, one of them the **inside-outside** algorithm. It uses a dynamic programming approach, that first finds the probability of a substring being generated by each rule, and then estimates the probability of each rule.
### Chomsky Normal Form
A grammar is in Chomsky Normal Form (or **CNF**, not to be confused with *Conjunctive Normal Form*) if its rules are one of the three:
* `X -> Y Z`
* `A -> a`
* `S -> ε`
Where *X*, *Y*, *Z*, *A* are non-terminals, *a* is a terminal, *ε* is the empty string and *S* is the start symbol (the start symbol should not be appearing on the right hand side of rules). Note that there can be multiple rules for each left hand side non-terminal, as long they follow the above. For example, a rule for *X* might be: `X -> Y Z | A B | a | b`.
Of course, we can also have a *CNF* with probabilities.
This type of grammar may seem restrictive, but it can be proven that any context-free grammar can be converted to CNF.
### Lexicon
The lexicon of a language is defined as a list of allowable words. These words are grouped into the usual classes: `verbs`, `nouns`, `adjectives`, `adverbs`, `pronouns`, `names`, `articles`, `prepositions` and `conjuctions`. For the first five classes it is impossible to list all words, since words are continuously being added in the classes. Recently "google" was added to the list of verbs, and words like that will continue to pop up and get added to the lists. For that reason, these first five categories are called **open classes**. The rest of the categories have much fewer words and much less development. While words like "thou" were commonly used in the past but have declined almost completely in usage, most changes take many decades or centuries to manifest, so we can safely assume the categories will remain static for the foreseeable future. Thus, these categories are called **closed classes**.
An example lexicon for a PCFG (note that other classes can also be used according to the language, like `digits`, or `RelPro` for relative pronoun):
```
Verb -> is [0.3] | say [0.1] | are [0.1] | ...
Noun -> robot [0.1] | sheep [0.05] | fence [0.05] | ...
Adjective -> good [0.1] | new [0.1] | sad [0.05] | ...
Adverb -> here [0.1] | lightly [0.05] | now [0.05] | ...
Pronoun -> me [0.1] | you [0.1] | he [0.05] | ...
RelPro -> that [0.4] | who [0.2] | which [0.2] | ...
Name -> john [0.05] | mary [0.05] | peter [0.01] | ...
Article -> the [0.35] | a [0.25] | an [0.025] | ...
Preposition -> to [0.25] | in [0.2] | at [0.1] | ...
Conjuction -> and [0.5] | or [0.2] | but [0.2] | ...
Digit -> 1 [0.3] | 2 [0.2] | 0 [0.2] | ...
```
### Grammar
With grammars we combine words from the lexicon into valid phrases. A grammar is comprised of **grammar rules**. Each rule transforms the left-hand side of the rule into the right-hand side. For example, `A -> B` means that `A` transforms into `B`. Let's build a grammar for the language we started building with the lexicon. We will use a PCFG.
```
S -> NP VP [0.9] | S Conjuction S [0.1]
NP -> Pronoun [0.3] | Name [0.1] | Noun [0.1] | Article Noun [0.25] |
Article Adjs Noun [0.05] | Digit [0.05] | NP PP [0.1] |
NP RelClause [0.05]
VP -> Verb [0.4] | VP NP [0.35] | VP Adjective [0.05] | VP PP [0.1]
VP Adverb [0.1]
Adjs -> Adjective [0.8] | Adjective Adjs [0.2]
PP -> Preposition NP [1.0]
RelClause -> RelPro VP [1.0]
```
Some valid phrases the grammar produces: "`mary is sad`", "`you are a robot`" and "`she likes mary and a good fence`".
What if we wanted to check if the phrase "`mary is sad`" is actually a valid sentence? We can use a **parse tree** to constructively prove that a string of words is a valid phrase in the given language and even calculate the probability of the generation of the sentence.

The probability of the whole tree can be calculated by multiplying the probabilities of each individual rule transormation: `0.9 * 0.1 * 0.05 * 0.05 * 0.4 * 0.05 * 0.3 = 0.00000135`.
To conserve space, we can also write the tree in linear form:
[S [NP [Name **mary**]] [VP [VP [Verb **is**]] [Adjective **sad**]]]
Unfortunately, the current grammar **overgenerates**, that is, it creates sentences that are not grammatically correct (according to the English language), like "`the fence are john which say`". It also **undergenerates**, which means there are valid sentences it does not generate, like "`he believes mary is sad`".
### Implementation
In the module we have implementation both for probabilistic and non-probabilistic grammars. Both these implementation follow the same format. There are functions for the lexicon and the rules which can be combined to create a grammar object.
#### Non-Probabilistic
Execute the cell below to view the implemenations:
```
psource(Lexicon, Rules, Grammar)
```
Let's build a lexicon and a grammar for the above language:
```
lexicon = Lexicon(
Verb = "is | say | are",
Noun = "robot | sheep | fence",
Adjective = "good | new | sad",
Adverb = "here | lightly | now",
Pronoun = "me | you | he",
RelPro = "that | who | which",
Name = "john | mary | peter",
Article = "the | a | an",
Preposition = "to | in | at",
Conjuction = "and | or | but",
Digit = "1 | 2 | 0"
)
print("Lexicon", lexicon)
rules = Rules(
S = "NP VP | S Conjuction S",
NP = "Pronoun | Name | Noun | Article Noun \
| Article Adjs Noun | Digit | NP PP | NP RelClause",
VP = "Verb | VP NP | VP Adjective | VP PP | VP Adverb",
Adjs = "Adjective | Adjective Adjs",
PP = "Preposition NP",
RelClause = "RelPro VP"
)
print("\nRules:", rules)
```
Both the functions return a dictionary with keys the left-hand side of the rules. For the lexicon, the values are the terminals for each left-hand side non-terminal, while for the rules the values are the right-hand sides as lists.
We can now use the variables `lexicon` and `rules` to build a grammar. After we've done so, we can find the transformations of a non-terminal (the `Noun`, `Verb` and the other basic classes do **not** count as proper non-terminals in the implementation). We can also check if a word is in a particular class.
```
grammar = Grammar("A Simple Grammar", rules, lexicon)
print("How can we rewrite 'VP'?", grammar.rewrites_for('VP'))
print("Is 'the' an article?", grammar.isa('the', 'Article'))
print("Is 'here' a noun?", grammar.isa('here', 'Noun'))
```
If the grammar is in Chomsky Normal Form, we can call the class function `cnf_rules` to get all the rules in the form of `(X, Y, Z)` for each `X -> Y Z` rule. Since the above grammar is not in *CNF* though, we have to create a new one.
```
E_Chomsky = Grammar("E_Prob_Chomsky", # A Grammar in Chomsky Normal Form
Rules(
S = "NP VP",
NP = "Article Noun | Adjective Noun",
VP = "Verb NP | Verb Adjective",
),
Lexicon(
Article = "the | a | an",
Noun = "robot | sheep | fence",
Adjective = "good | new | sad",
Verb = "is | say | are"
))
print(E_Chomsky.cnf_rules())
```
Finally, we can generate random phrases using our grammar. Most of them will be complete gibberish, falling under the overgenerated phrases of the grammar. That goes to show that in the grammar the valid phrases are much fewer than the overgenerated ones.
```
grammar.generate_random('S')
```
#### Probabilistic
The probabilistic grammars follow the same approach. They take as input a string, are assembled from a grammar and a lexicon and can generate random sentences (giving the probability of the sentence). The main difference is that in the lexicon we have tuples (terminal, probability) instead of strings and for the rules we have a list of tuples (list of non-terminals, probability) instead of list of lists of non-terminals.
Execute the cells to read the code:
```
psource(ProbLexicon, ProbRules, ProbGrammar)
```
Let's build a lexicon and rules for the probabilistic grammar:
```
lexicon = ProbLexicon(
Verb = "is [0.5] | say [0.3] | are [0.2]",
Noun = "robot [0.4] | sheep [0.4] | fence [0.2]",
Adjective = "good [0.5] | new [0.2] | sad [0.3]",
Adverb = "here [0.6] | lightly [0.1] | now [0.3]",
Pronoun = "me [0.3] | you [0.4] | he [0.3]",
RelPro = "that [0.5] | who [0.3] | which [0.2]",
Name = "john [0.4] | mary [0.4] | peter [0.2]",
Article = "the [0.5] | a [0.25] | an [0.25]",
Preposition = "to [0.4] | in [0.3] | at [0.3]",
Conjuction = "and [0.5] | or [0.2] | but [0.3]",
Digit = "0 [0.35] | 1 [0.35] | 2 [0.3]"
)
print("Lexicon", lexicon)
rules = ProbRules(
S = "NP VP [0.6] | S Conjuction S [0.4]",
NP = "Pronoun [0.2] | Name [0.05] | Noun [0.2] | Article Noun [0.15] \
| Article Adjs Noun [0.1] | Digit [0.05] | NP PP [0.15] | NP RelClause [0.1]",
VP = "Verb [0.3] | VP NP [0.2] | VP Adjective [0.25] | VP PP [0.15] | VP Adverb [0.1]",
Adjs = "Adjective [0.5] | Adjective Adjs [0.5]",
PP = "Preposition NP [1]",
RelClause = "RelPro VP [1]"
)
print("\nRules:", rules)
```
Let's use the above to assemble our probabilistic grammar and run some simple queries:
```
grammar = ProbGrammar("A Simple Probabilistic Grammar", rules, lexicon)
print("How can we rewrite 'VP'?", grammar.rewrites_for('VP'))
print("Is 'the' an article?", grammar.isa('the', 'Article'))
print("Is 'here' a noun?", grammar.isa('here', 'Noun'))
```
If we have a grammar in *CNF*, we can get a list of all the rules. Let's create a grammar in the form and print the *CNF* rules:
```
E_Prob_Chomsky = ProbGrammar("E_Prob_Chomsky", # A Probabilistic Grammar in CNF
ProbRules(
S = "NP VP [1]",
NP = "Article Noun [0.6] | Adjective Noun [0.4]",
VP = "Verb NP [0.5] | Verb Adjective [0.5]",
),
ProbLexicon(
Article = "the [0.5] | a [0.25] | an [0.25]",
Noun = "robot [0.4] | sheep [0.4] | fence [0.2]",
Adjective = "good [0.5] | new [0.2] | sad [0.3]",
Verb = "is [0.5] | say [0.3] | are [0.2]"
))
print(E_Prob_Chomsky.cnf_rules())
```
Lastly, we can generate random sentences from this grammar. The function `prob_generation` returns a tuple (sentence, probability).
```
sentence, prob = grammar.generate_random('S')
print(sentence)
print(prob)
```
As with the non-probabilistic grammars, this one mostly overgenerates. You can also see that the probability is very, very low, which means there are a ton of generateable sentences (in this case infinite, since we have recursion; notice how `VP` can produce another `VP`, for example).
## HITS
### Overview
**Hyperlink-Induced Topic Search** (or HITS for short) is an algorithm for information retrieval and page ranking. You can read more on information retrieval in the [text notebook](https://github.com/aimacode/aima-python/blob/master/text.ipynb). Essentially, given a collection of documents and a user's query, such systems return to the user the documents most relevant to what the user needs. The HITS algorithm differs from a lot of other similar ranking algorithms (like Google's *Pagerank*) as the page ratings in this algorithm are dependent on the given query. This means that for each new query the result pages must be computed anew. This cost might be prohibitive for many modern search engines, so a lot steer away from this approach.
HITS first finds a list of relevant pages to the query and then adds pages that link to or are linked from these pages. Once the set is built, we define two values for each page. **Authority** on the query, the degree of pages from the relevant set linking to it and **hub** of the query, the degree that it points to authoritative pages in the set. Since we do not want to simply count the number of links from a page to other pages, but we also want to take into account the quality of the linked pages, we update the hub and authority values of a page in the following manner, until convergence:
* Hub score = The sum of the authority scores of the pages it links to.
* Authority score = The sum of hub scores of the pages it is linked from.
So the higher quality the pages a page is linked to and from, the higher its scores.
We then normalize the scores by dividing each score by the sum of the squares of the respective scores of all pages. When the values converge, we return the top-valued pages. Note that because we normalize the values, the algorithm is guaranteed to converge.
### Implementation
The source code for the algorithm is given below:
```
psource(HITS)
```
First we compile the collection of pages as mentioned above. Then, we initialize the authority and hub scores for each page and finally we update and normalize the values until convergence.
A quick overview of the helper functions functions we use:
* `relevant_pages`: Returns relevant pages from `pagesIndex` given a query.
* `expand_pages`: Adds to the collection pages linked to and from the given `pages`.
* `normalize`: Normalizes authority and hub scores.
* `ConvergenceDetector`: A class that checks for convergence, by keeping a history of the pages' scores and checking if they change or not.
* `Page`: The template for pages. Stores the address, authority/hub scores and in-links/out-links.
### Example
Before we begin we need to define a list of sample pages to work on. The pages are `pA`, `pB` and so on and their text is given by `testHTML` and `testHTML2`. The `Page` class takes as arguments the in-links and out-links as lists. For page "A", the in-links are "B", "C" and "E" while the sole out-link is "D".
We also need to set the `nlp` global variables `pageDict`, `pagesIndex` and `pagesContent`.
```
testHTML = """Like most other male mammals, a man inherits an
X from his mom and a Y from his dad."""
testHTML2 = "a mom and a dad"
pA = Page('A', ['B', 'C', 'E'], ['D'])
pB = Page('B', ['E'], ['A', 'C', 'D'])
pC = Page('C', ['B', 'E'], ['A', 'D'])
pD = Page('D', ['A', 'B', 'C', 'E'], [])
pE = Page('E', [], ['A', 'B', 'C', 'D', 'F'])
pF = Page('F', ['E'], [])
nlp.pageDict = {pA.address: pA, pB.address: pB, pC.address: pC,
pD.address: pD, pE.address: pE, pF.address: pF}
nlp.pagesIndex = nlp.pageDict
nlp.pagesContent ={pA.address: testHTML, pB.address: testHTML2,
pC.address: testHTML, pD.address: testHTML2,
pE.address: testHTML, pF.address: testHTML2}
```
We can now run the HITS algorithm. Our query will be 'mammals' (note that while the content of the HTML doesn't matter, it should include the query words or else no page will be picked at the first step).
```
HITS('mammals')
page_list = ['A', 'B', 'C', 'D', 'E', 'F']
auth_list = [pA.authority, pB.authority, pC.authority, pD.authority, pE.authority, pF.authority]
hub_list = [pA.hub, pB.hub, pC.hub, pD.hub, pE.hub, pF.hub]
```
Let's see how the pages were scored:
```
for i in range(6):
p = page_list[i]
a = auth_list[i]
h = hub_list[i]
print("{}: total={}, auth={}, hub={}".format(p, a + h, a, h))
```
The top score is 0.82 by "C". This is the most relevant page according to the algorithm. You can see that the pages it links to, "A" and "D", have the two highest authority scores (therefore "C" has a high hub score) and the pages it is linked from, "B" and "E", have the highest hub scores (so "C" has a high authority score). By combining these two facts, we get that "C" is the most relevant page. It is worth noting that it does not matter if the given page contains the query words, just that it links and is linked from high-quality pages.
## QUESTION ANSWERING
**Question Answering** is a type of Information Retrieval system, where we have a question instead of a query and instead of relevant documents we want the computer to return a short sentence, phrase or word that answers our question. To better understand the concept of question answering systems, you can first read the "Text Models" and "Information Retrieval" section from the [text notebook](https://github.com/aimacode/aima-python/blob/master/text.ipynb).
A typical example of such a system is `AskMSR` (Banko *et al.*, 2002), a system for question answering that performed admirably against more sophisticated algorithms. The basic idea behind it is that a lot of questions have already been answered in the web numerous times. The system doesn't know a lot about verbs, or concepts or even what a noun is. It knows about 15 different types of questions and how they can be written as queries. It can rewrite [Who was George Washington's second in command?] as the query [\* was George Washington's second in command] or [George Washington's second in command was \*].
After rewriting the questions, it issues these queries and retrieves the short text around the query terms. It then breaks the result into 1, 2 or 3-grams. Filters are also applied to increase the chances of a correct answer. If the query starts with "who", we filter for names, if it starts with "how many" we filter for numbers and so on. We can also filter out the words appearing in the query. For the above query, the answer "George Washington" is wrong, even though it is quite possible the 2-gram would appear a lot around the query terms.
Finally, the different results are weighted by the generality of the queries. The result from the general boolean query [George Washington OR second in command] weighs less that the more specific query [George Washington's second in command was \*]. As an answer we return the most highly-ranked n-gram.
## CYK PARSE
### Overview
Syntactic analysis (or **parsing**) of a sentence is the process of uncovering the phrase structure of the sentence according to the rules of a grammar. There are two main approaches to parsing. *Top-down*, start with the starting symbol and build a parse tree with the given words as its leaves, and *bottom-up*, where we start from the given words and build a tree that has the starting symbol as its root. Both approaches involve "guessing" ahead, so it is very possible it will take long to parse a sentence (wrong guess mean a lot of backtracking). Thankfully, a lot of effort is spent in analyzing already analyzed substrings, so we can follow a dynamic programming approach to store and reuse these parses instead of recomputing them. The *CYK Parsing Algorithm* (named after its inventors, Cocke, Younger and Kasami) utilizes this technique to parse sentences of a grammar in *Chomsky Normal Form*.
The CYK algorithm returns an *M x N x N* array (named *P*), where *N* is the number of words in the sentence and *M* the number of non-terminal symbols in the grammar. Each element in this array shows the probability of a substring being transformed from a particular non-terminal. To find the most probable parse of the sentence, a search in the resulting array is required. Search heuristic algorithms work well in this space, and we can derive the heuristics from the properties of the grammar.
The algorithm in short works like this: There is an external loop that determines the length of the substring. Then the algorithm loops through the words in the sentence. For each word, it again loops through all the words to its right up to the first-loop length. The substring it will work on in this iteration is the words from the second-loop word with first-loop length. Finally, it loops through all the rules in the grammar and updates the substring's probability for each right-hand side non-terminal.
### Implementation
The implementation takes as input a list of words and a probabilistic grammar (from the `ProbGrammar` class detailed above) in CNF and returns the table/dictionary *P*. An item's key in *P* is a tuple in the form `(Non-terminal, start of substring, length of substring)`, and the value is a probability. For example, for the sentence "the monkey is dancing" and the substring "the monkey" an item can be `('NP', 0, 2): 0.5`, which means the first two words (the substring from index 0 and length 2) have a 0.5 probablity of coming from the `NP` terminal.
Before we continue, you can take a look at the source code by running the cell below:
```
psource(CYK_parse)
```
When updating the probability of a substring, we pick the max of its current one and the probability of the substring broken into two parts: one from the second-loop word with third-loop length, and the other from the first part's end to the remainer of the first-loop length.
### Example
Let's build a probabilistic grammar in CNF:
```
E_Prob_Chomsky = ProbGrammar("E_Prob_Chomsky", # A Probabilistic Grammar in CNF
ProbRules(
S = "NP VP [1]",
NP = "Article Noun [0.6] | Adjective Noun [0.4]",
VP = "Verb NP [0.5] | Verb Adjective [0.5]",
),
ProbLexicon(
Article = "the [0.5] | a [0.25] | an [0.25]",
Noun = "robot [0.4] | sheep [0.4] | fence [0.2]",
Adjective = "good [0.5] | new [0.2] | sad [0.3]",
Verb = "is [0.5] | say [0.3] | are [0.2]"
))
```
Now let's see the probabilities table for the sentence "the robot is good":
```
words = ['the', 'robot', 'is', 'good']
grammar = E_Prob_Chomsky
P = CYK_parse(words, grammar)
print(P)
```
A `defaultdict` object is returned (`defaultdict` is basically a dictionary but with a default value/type). Keys are tuples in the form mentioned above and the values are the corresponding probabilities. Most of the items/parses have a probability of 0. Let's filter those out to take a better look at the parses that matter.
```
parses = {k: p for k, p in P.items() if p >0}
print(parses)
```
The item `('Article', 0, 1): 0.5` means that the first item came from the `Article` non-terminal with a chance of 0.5. A more complicated item, one with two words, is `('NP', 0, 2): 0.12` which covers the first two words. The probability of the substring "the robot" coming from the `NP` non-terminal is 0.12. Let's try and follow the transformations from `NP` to the given words (top-down) to make sure this is indeed the case:
1. The probability of `NP` transforming to `Article Noun` is 0.6.
2. The probability of `Article` transforming to "the" is 0.5 (total probability = 0.6*0.5 = 0.3).
3. The probability of `Noun` transforming to "robot" is 0.4 (total = 0.3*0.4 = 0.12).
Thus, the total probability of the transformation is 0.12.
Notice how the probability for the whole string (given by the key `('S', 0, 4)`) is 0.015. This means the most probable parsing of the sentence has a probability of 0.015.
## CHART PARSING
### Overview
Let's now take a look at a more general chart parsing algorithm. Given a non-probabilistic grammar and a sentence, this algorithm builds a parse tree in a top-down manner, with the words of the sentence as the leaves. It works with a dynamic programming approach, building a chart to store parses for substrings so that it doesn't have to analyze them again (just like the CYK algorithm). Each non-terminal, starting from S, gets replaced by its right-hand side rules in the chart, until we end up with the correct parses.
### Implementation
A parse is in the form `[start, end, non-terminal, sub-tree, expected-transformation]`, where `sub-tree` is a tree with the corresponding `non-terminal` as its root and `expected-transformation` is a right-hand side rule of the `non-terminal`.
The chart parsing is implemented in a class, `Chart`. It is initialized with a grammar and can return the list of all the parses of a sentence with the `parses` function.
The chart is a list of lists. The lists correspond to the lengths of substrings (including the empty string), from start to finish. When we say 'a point in the chart', we refer to a list of a certain length.
A quick rundown of the class functions:
* `parses`: Returns a list of parses for a given sentence. If the sentence can't be parsed, it will return an empty list. Initializes the process by calling `parse` from the starting symbol.
* `parse`: Parses the list of words and builds the chart.
* `add_edge`: Adds another edge to the chart at a given point. Also, examines whether the edge extends or predicts another edge. If the edge itself is not expecting a transformation, it will extend other edges and it will predict edges otherwise.
* `scanner`: Given a word and a point in the chart, it extends edges that were expecting a transformation that can result in the given word. For example, if the word 'the' is an 'Article' and we are examining two edges at a chart's point, with one expecting an 'Article' and the other a 'Verb', the first one will be extended while the second one will not.
* `predictor`: If an edge can't extend other edges (because it is expecting a transformation itself), we will add to the chart rules/transformations that can help extend the edge. The new edges come from the right-hand side of the expected transformation's rules. For example, if an edge is expecting the transformation 'Adjective Noun', we will add to the chart an edge for each right-hand side rule of the non-terminal 'Adjective'.
* `extender`: Extends edges given an edge (called `E`). If `E`'s non-terminal is the same as the expected transformation of another edge (let's call it `A`), add to the chart a new edge with the non-terminal of `A` and the transformations of `A` minus the non-terminal that matched with `E`'s non-terminal. For example, if an edge `E` has 'Article' as its non-terminal and is expecting no transformation, we need to see what edges it can extend. Let's examine the edge `N`. This expects a transformation of 'Noun Verb'. 'Noun' does not match with 'Article', so we move on. Another edge, `A`, expects a transformation of 'Article Noun' and has a non-terminal of 'NP'. We have a match! A new edge will be added with 'NP' as its non-terminal (the non-terminal of `A`) and 'Noun' as the expected transformation (the rest of the expected transformation of `A`).
You can view the source code by running the cell below:
```
psource(Chart)
```
### Example
We will use the grammar `E0` to parse the sentence "the stench is in 2 2".
First we need to build a `Chart` object:
```
chart = Chart(nlp.E0)
```
And then we simply call the `parses` function:
```
print(chart.parses('the stench is in 2 2'))
```
You can see which edges get added by setting the optional initialization argument `trace` to true.
```
chart_trace = Chart(nlp.E0, trace=True)
chart_trace.parses('the stench is in 2 2')
```
Let's try and parse a sentence that is not recognized by the grammar:
```
print(chart.parses('the stench 2 2'))
```
An empty list was returned.
|
github_jupyter
|
import nlp
from nlp import Page, HITS
from nlp import Lexicon, Rules, Grammar, ProbLexicon, ProbRules, ProbGrammar
from nlp import CYK_parse, Chart
from notebook import psource
S -> aSb | ε
S -> aSb [0.7] | ε [0.3]
Verb -> is [0.3] | say [0.1] | are [0.1] | ...
Noun -> robot [0.1] | sheep [0.05] | fence [0.05] | ...
Adjective -> good [0.1] | new [0.1] | sad [0.05] | ...
Adverb -> here [0.1] | lightly [0.05] | now [0.05] | ...
Pronoun -> me [0.1] | you [0.1] | he [0.05] | ...
RelPro -> that [0.4] | who [0.2] | which [0.2] | ...
Name -> john [0.05] | mary [0.05] | peter [0.01] | ...
Article -> the [0.35] | a [0.25] | an [0.025] | ...
Preposition -> to [0.25] | in [0.2] | at [0.1] | ...
Conjuction -> and [0.5] | or [0.2] | but [0.2] | ...
Digit -> 1 [0.3] | 2 [0.2] | 0 [0.2] | ...
S -> NP VP [0.9] | S Conjuction S [0.1]
NP -> Pronoun [0.3] | Name [0.1] | Noun [0.1] | Article Noun [0.25] |
Article Adjs Noun [0.05] | Digit [0.05] | NP PP [0.1] |
NP RelClause [0.05]
VP -> Verb [0.4] | VP NP [0.35] | VP Adjective [0.05] | VP PP [0.1]
VP Adverb [0.1]
Adjs -> Adjective [0.8] | Adjective Adjs [0.2]
PP -> Preposition NP [1.0]
RelClause -> RelPro VP [1.0]
psource(Lexicon, Rules, Grammar)
lexicon = Lexicon(
Verb = "is | say | are",
Noun = "robot | sheep | fence",
Adjective = "good | new | sad",
Adverb = "here | lightly | now",
Pronoun = "me | you | he",
RelPro = "that | who | which",
Name = "john | mary | peter",
Article = "the | a | an",
Preposition = "to | in | at",
Conjuction = "and | or | but",
Digit = "1 | 2 | 0"
)
print("Lexicon", lexicon)
rules = Rules(
S = "NP VP | S Conjuction S",
NP = "Pronoun | Name | Noun | Article Noun \
| Article Adjs Noun | Digit | NP PP | NP RelClause",
VP = "Verb | VP NP | VP Adjective | VP PP | VP Adverb",
Adjs = "Adjective | Adjective Adjs",
PP = "Preposition NP",
RelClause = "RelPro VP"
)
print("\nRules:", rules)
grammar = Grammar("A Simple Grammar", rules, lexicon)
print("How can we rewrite 'VP'?", grammar.rewrites_for('VP'))
print("Is 'the' an article?", grammar.isa('the', 'Article'))
print("Is 'here' a noun?", grammar.isa('here', 'Noun'))
E_Chomsky = Grammar("E_Prob_Chomsky", # A Grammar in Chomsky Normal Form
Rules(
S = "NP VP",
NP = "Article Noun | Adjective Noun",
VP = "Verb NP | Verb Adjective",
),
Lexicon(
Article = "the | a | an",
Noun = "robot | sheep | fence",
Adjective = "good | new | sad",
Verb = "is | say | are"
))
print(E_Chomsky.cnf_rules())
grammar.generate_random('S')
psource(ProbLexicon, ProbRules, ProbGrammar)
lexicon = ProbLexicon(
Verb = "is [0.5] | say [0.3] | are [0.2]",
Noun = "robot [0.4] | sheep [0.4] | fence [0.2]",
Adjective = "good [0.5] | new [0.2] | sad [0.3]",
Adverb = "here [0.6] | lightly [0.1] | now [0.3]",
Pronoun = "me [0.3] | you [0.4] | he [0.3]",
RelPro = "that [0.5] | who [0.3] | which [0.2]",
Name = "john [0.4] | mary [0.4] | peter [0.2]",
Article = "the [0.5] | a [0.25] | an [0.25]",
Preposition = "to [0.4] | in [0.3] | at [0.3]",
Conjuction = "and [0.5] | or [0.2] | but [0.3]",
Digit = "0 [0.35] | 1 [0.35] | 2 [0.3]"
)
print("Lexicon", lexicon)
rules = ProbRules(
S = "NP VP [0.6] | S Conjuction S [0.4]",
NP = "Pronoun [0.2] | Name [0.05] | Noun [0.2] | Article Noun [0.15] \
| Article Adjs Noun [0.1] | Digit [0.05] | NP PP [0.15] | NP RelClause [0.1]",
VP = "Verb [0.3] | VP NP [0.2] | VP Adjective [0.25] | VP PP [0.15] | VP Adverb [0.1]",
Adjs = "Adjective [0.5] | Adjective Adjs [0.5]",
PP = "Preposition NP [1]",
RelClause = "RelPro VP [1]"
)
print("\nRules:", rules)
grammar = ProbGrammar("A Simple Probabilistic Grammar", rules, lexicon)
print("How can we rewrite 'VP'?", grammar.rewrites_for('VP'))
print("Is 'the' an article?", grammar.isa('the', 'Article'))
print("Is 'here' a noun?", grammar.isa('here', 'Noun'))
E_Prob_Chomsky = ProbGrammar("E_Prob_Chomsky", # A Probabilistic Grammar in CNF
ProbRules(
S = "NP VP [1]",
NP = "Article Noun [0.6] | Adjective Noun [0.4]",
VP = "Verb NP [0.5] | Verb Adjective [0.5]",
),
ProbLexicon(
Article = "the [0.5] | a [0.25] | an [0.25]",
Noun = "robot [0.4] | sheep [0.4] | fence [0.2]",
Adjective = "good [0.5] | new [0.2] | sad [0.3]",
Verb = "is [0.5] | say [0.3] | are [0.2]"
))
print(E_Prob_Chomsky.cnf_rules())
sentence, prob = grammar.generate_random('S')
print(sentence)
print(prob)
psource(HITS)
testHTML = """Like most other male mammals, a man inherits an
X from his mom and a Y from his dad."""
testHTML2 = "a mom and a dad"
pA = Page('A', ['B', 'C', 'E'], ['D'])
pB = Page('B', ['E'], ['A', 'C', 'D'])
pC = Page('C', ['B', 'E'], ['A', 'D'])
pD = Page('D', ['A', 'B', 'C', 'E'], [])
pE = Page('E', [], ['A', 'B', 'C', 'D', 'F'])
pF = Page('F', ['E'], [])
nlp.pageDict = {pA.address: pA, pB.address: pB, pC.address: pC,
pD.address: pD, pE.address: pE, pF.address: pF}
nlp.pagesIndex = nlp.pageDict
nlp.pagesContent ={pA.address: testHTML, pB.address: testHTML2,
pC.address: testHTML, pD.address: testHTML2,
pE.address: testHTML, pF.address: testHTML2}
HITS('mammals')
page_list = ['A', 'B', 'C', 'D', 'E', 'F']
auth_list = [pA.authority, pB.authority, pC.authority, pD.authority, pE.authority, pF.authority]
hub_list = [pA.hub, pB.hub, pC.hub, pD.hub, pE.hub, pF.hub]
for i in range(6):
p = page_list[i]
a = auth_list[i]
h = hub_list[i]
print("{}: total={}, auth={}, hub={}".format(p, a + h, a, h))
psource(CYK_parse)
E_Prob_Chomsky = ProbGrammar("E_Prob_Chomsky", # A Probabilistic Grammar in CNF
ProbRules(
S = "NP VP [1]",
NP = "Article Noun [0.6] | Adjective Noun [0.4]",
VP = "Verb NP [0.5] | Verb Adjective [0.5]",
),
ProbLexicon(
Article = "the [0.5] | a [0.25] | an [0.25]",
Noun = "robot [0.4] | sheep [0.4] | fence [0.2]",
Adjective = "good [0.5] | new [0.2] | sad [0.3]",
Verb = "is [0.5] | say [0.3] | are [0.2]"
))
words = ['the', 'robot', 'is', 'good']
grammar = E_Prob_Chomsky
P = CYK_parse(words, grammar)
print(P)
parses = {k: p for k, p in P.items() if p >0}
print(parses)
psource(Chart)
chart = Chart(nlp.E0)
print(chart.parses('the stench is in 2 2'))
chart_trace = Chart(nlp.E0, trace=True)
chart_trace.parses('the stench is in 2 2')
print(chart.parses('the stench 2 2'))
| 0.432063 | 0.95222 |
<img src="../../images/banners/python-advanced.png" width="600"/>
# <img src="../../images/logos/python.png" width="23"/> `with` Context Manager
## <img src="../../images/logos/toc.png" width="20"/> Table of Contents
* [Supporting with in Your Own Objects](#supporting_with_in_your_own_objects)
* [Writing Pretty APIs With Context Managers](#writing_pretty_apis_with_context_managers)
* [Solution](#solution)
* [Things to Remember](#things_to_remember)
---
The `with` statement in Python is regarded as an obscure feature by some. But when you peek behind the scenes of the underlying Context Manager protocol you’ll see there’s little “magic” involved.
So what’s the with statement good for? **It helps simplify some common resource management patterns by abstracting their functionality and allowing them to be factored out and reused**.
In turn this helps you write more expressive code and makes it easier to avoid resource leaks in your programs.
A good way to see this feature used effectively is by looking at examples in the Python standard library. A well-known example involves the `open()` function:
```python
with open('hello.txt', 'w') as f:
f.write('hello, world!')
```
Opening files using the `with` statement is generally recommended because it ensures that open file descriptors are closed automatically after program execution leaves the context of the with statement. Internally, the above code sample translates to something like this:
```python
f = open('hello.txt', 'w')
try:
f.write('hello, world')
finally:
f.close()
```
You can already tell that this is quite a bit more verbose. Note that the `try...finally` statement is significant. It wouldn’t be enough to just write something like this:
```python
f = open('hello.txt', 'w')
f.write('hello, world')
f.close()
```
This implementation won’t guarantee the file is closed if there’s an exception during the `f.write()` call—and therefore our program might leak a file descriptor. That’s why the with statement is so useful. It makes acquiring and releasing resources properly a breeze.
Another good example where the with statement is used effectively in the Python standard library is the `threading.Lock` class (threading and multiprocessing will be covered later):
```python
some_lock = threading.Lock()
# Harmful:
some_lock.acquire()
try:
# Do something...
finally:
some_lock.release()
# Better:
with some_lock:
# Do something...
```
In both cases using a `with` statement allows you to abstract away most of the resource handling logic. Instead of having to write an explicit `try...finally` statement each time, with takes care of that for us.
The with statement can make code dealing with system resources more readable. It also helps avoid bugs or leaks by making it almost impossible to forget cleaning up or releasing a resource after we’re done with it.
<a class="anchor" id="supporting_with_in_your_own_objects"></a>
## Supporting `with` in Your Own Objects (Class-Based)
Now, there’s nothing special or magical about the `open()` function and the fact that they can be used with a with statement. You can provide the same functionality in your own classes and functions by implementing so-called context managers.
What’s a context manager? It’s a simple “protocol” (or interface) that your object needs to follow so it can be used with the with statement. Basically all you need to do is add `__enter__` and `__exit__` methods to an object if you want it to function as a context manager. Python will call these two methods at the appropriate times in the resource management cycle.
Let’s take a look at what this would look like in practical terms. Here’s how a simple implementation of the `open()` context manager might look like:
```
class ManagedFile:
def __init__(self, name):
self.name = name
def __enter__(self):
print("Calling __enter__...")
self.file = open(self.name, 'w')
return self.file
def __exit__(self, exc_type, exc_val, exc_tb):
print("Calling __exit__...")
if self.file:
self.file.close()
```
Our `ManagedFile` class follows the context manager protocol and now supports the with statement, just like the original `open()` example did:
```
with ManagedFile('hello.txt') as f:
f.write('hello, world!')
f.write('bye now')
```
Python calls `__enter__` when execution enters the context of the with statement and it’s time to acquire the resource. When execution leaves the context again, Python calls `__exit__` to free up the resource.
## Context Manager Utilities
Creating context managers the traditional way, by writing a class with `__enter__()` and `__exit__()` methods, is not difficult. But sometimes it is more overhead than you need just to manage a trivial bit of context. In those sorts of situations, you can use the `contextmanager()` decorator to convert a generator function into a context manager.
The `contextlib` utility module in the standard library provides a few more abstractions built on top of the basic context manager protocol. This can make your life a little easier if your use cases matches what’s offered by contextlib.
### From Generator to Context Manager (Generator-Based)
Can the `__exit__` be called before `__enter__`? Obviously no. So the `__enter__` should always be called before the `__exit__`. What does that tell you? There is some sequencing. What does that suggest you? Generators!
You can use the `contextlib.contextmanager` decorator to define a generator-based factory function for a resource that will then automatically support the with statement. The function being decorated must return a generator-iterator when called. The generator should initialize the context, yield exactly one time, then clean up the context. The value yielded, if any, is bound to the variable in the as clause of the with statement. Exceptions from within the with block are re-raised inside the generator, so they can be handled there.
Here’s what rewriting our `ManagedFile` context manager with this technique looks like:
```
from contextlib import contextmanager
@contextmanager
def managed_file(name):
try:
f = open(name, 'w')
yield f
finally:
f.close()
with managed_file('hello.txt') as f:
f.write('hello, world!')
f.write('bye now')
```
In this case, `managed_file()` is a generator that first acquires the resource. Then it temporarily suspends its own executing and yields the resource so it can be used by the caller. When the caller leaves the with context, the generator continues to execute so that any remaining clean up steps can happen and the resource gets released back to the system.
If an unhandled exception occurs in the block, it is reraised inside the generator at the point where the yield occurred. Thus, you can use a try…except…finally statement to trap the error (if any), or ensure that some cleanup takes place.
If you are wondering how `contextmanager` works under the hood, here is a python implementation:
```
class contextmanager:
def __init__(self, gen):
self.gen = gen
def __call__(self, *args, **kwargs):
self.args, self.kwargs = args, kwargs
return self
def __enter__(self):
self.gen_instance = self.gen(*self.args, **self.kwargs)
return next(self.gen_instance)
def __exit__(self, *args):
next(self.gen_instance, None)
@contextmanager
def managed_file(name):
try:
f = open(name, 'w')
yield f
finally:
f.close()
with managed_file('hello.txt') as f:
f.write('hello, world!')
f.write('bye now')
```
`contextmanager` now can decorate a generator with the `__call__` method which is basically running:
```python
generator = contextmanager(generator)
```
Now generator is a `contextmanager` object that has `__enter__` and `__exit__`, so it can be used using `with`. Calling `with generator(g_input) as f` runs:
1. `__call__` method to store the generator arguments and return the `contexmanager` object.
2. `__enter__` creates a new generator instance implicitly with the stored arguments in `self.args` and `self.kwargs` and calls next on generator.
3. When execution leaves the context again, `__exit__` is called which calls next on generator again.
Both the class-based implementations and the generator-based are practically equivalent. Depending on which one you find more readable you might prefer one over the other.
A downside of the `@contextmanager`-based implementation might be that it requires understanding of advanced Python concepts, like decorators and generators.
### Supress Exceptions with `contextlib.suppress`
Return a context manager that suppresses any of the specified exceptions if they occur in the body of a `with` statement and then resumes execution with the first statement following the end of the `with` statement.
As with any other mechanism that completely suppresses exceptions, this context manager should be used only to cover very specific errors where silently continuing with program execution is known to be the right thing to do.
For example:
```
from contextlib import suppress
import os
with suppress(FileNotFoundError):
os.remove('somefile.tmp')
with suppress(FileNotFoundError):
os.remove('someotherfile.tmp')
```
This code is equivalent to:
```
try:
os.remove('somefile.tmp')
except FileNotFoundError:
pass
try:
os.remove('someotherfile.tmp')
except FileNotFoundError:
pass
```
### Null Context with `contextlib.nullcontext(enter_result=None)`
Return a context manager that returns `enter_result` from `__enter__`, but otherwise does nothing. It is intended to be used as a stand-in for an optional context manager, for example:
```
def myfunction(arg, ignore_exceptions=False):
if ignore_exceptions:
# Use suppress to ignore all exceptions.
cm = contextlib.suppress(Exception)
else:
# Do not ignore any exceptions, cm has no effect.
cm = contextlib.nullcontext()
with cm:
# Do something
pass
```
An example using `enter_result`:
```
def process_file(file_or_path):
if isinstance(file_or_path, str):
# If string, open file
cm = open(file_or_path)
else:
# Caller is responsible for closing file
cm = nullcontext(file_or_path)
with cm as file:
# Perform processing on the file
pass
```
<a class="anchor" id="writing_pretty_apis_with_context_managers"></a>
## Writing Pretty APIs With Context Managers
Context managers are quite flexible and if you use the with statement creatively you can define convenient APIs for your modules and classes.
For example, what if the “resource” we wanted to manage was text indentation levels in some kind of report generator program? What if we could write code like this to do it:
```python
with Indenter() as indent:
indent.print('hi!')
with indent:
indent.print('hello')
with indent:
indent.print('bonjour')
indent.print('hey')
```
This almost reads like a domain-specific language (DSL) for indenting text. Also, notice how this code enters and leaves the same context manager multiple times to change indentation levels. Running this code snippet should lead to the following output and print neatly formatted text:
```bash
hi!
hello
bonjour
hey
```
How would you implement a context manager to support this functionality?
By the way, this could be a great exercise to wrap your head around how context managers work. So before you check out my implementation below you might take some time and try to implement this yourself as a learning exercise.
<a class="anchor" id="solution"></a>
### Solution
Ready? Here’s how we might implement this functionality using a class-based context manager:
```
class Indenter:
def __init__(self):
self.level = 0
def __enter__(self):
self.level += 1
return self
def __exit__(self, exc_type, exc_val, exc_tb):
self.level -= 1
def print(self, text):
print(' ' * self.level + text)
with Indenter() as indent:
indent.print('hi!')
with indent:
indent.print('hello')
with indent:
indent.print('bonjour')
indent.print('hey')
```
Another good exercise would be trying to refactor this code to be generator-based.
<a class="anchor" id="things_to_remember"></a>
## Things to Remember
- The `with` statement simplifies exception handling by encapsulating standard uses of `try/finally` statements in so-called Context Managers.
- Most commonly it is used to manage the safe acquisition and release of system resources. Resources are acquired by the `with` statement and released automatically when execution leaves the `with` context.
- Using `with` effectively can help you avoid resource leaks and make your code easier to read.
|
github_jupyter
|
with open('hello.txt', 'w') as f:
f.write('hello, world!')
f = open('hello.txt', 'w')
try:
f.write('hello, world')
finally:
f.close()
f = open('hello.txt', 'w')
f.write('hello, world')
f.close()
some_lock = threading.Lock()
# Harmful:
some_lock.acquire()
try:
# Do something...
finally:
some_lock.release()
# Better:
with some_lock:
# Do something...
class ManagedFile:
def __init__(self, name):
self.name = name
def __enter__(self):
print("Calling __enter__...")
self.file = open(self.name, 'w')
return self.file
def __exit__(self, exc_type, exc_val, exc_tb):
print("Calling __exit__...")
if self.file:
self.file.close()
with ManagedFile('hello.txt') as f:
f.write('hello, world!')
f.write('bye now')
from contextlib import contextmanager
@contextmanager
def managed_file(name):
try:
f = open(name, 'w')
yield f
finally:
f.close()
with managed_file('hello.txt') as f:
f.write('hello, world!')
f.write('bye now')
class contextmanager:
def __init__(self, gen):
self.gen = gen
def __call__(self, *args, **kwargs):
self.args, self.kwargs = args, kwargs
return self
def __enter__(self):
self.gen_instance = self.gen(*self.args, **self.kwargs)
return next(self.gen_instance)
def __exit__(self, *args):
next(self.gen_instance, None)
@contextmanager
def managed_file(name):
try:
f = open(name, 'w')
yield f
finally:
f.close()
with managed_file('hello.txt') as f:
f.write('hello, world!')
f.write('bye now')
generator = contextmanager(generator)
from contextlib import suppress
import os
with suppress(FileNotFoundError):
os.remove('somefile.tmp')
with suppress(FileNotFoundError):
os.remove('someotherfile.tmp')
try:
os.remove('somefile.tmp')
except FileNotFoundError:
pass
try:
os.remove('someotherfile.tmp')
except FileNotFoundError:
pass
def myfunction(arg, ignore_exceptions=False):
if ignore_exceptions:
# Use suppress to ignore all exceptions.
cm = contextlib.suppress(Exception)
else:
# Do not ignore any exceptions, cm has no effect.
cm = contextlib.nullcontext()
with cm:
# Do something
pass
def process_file(file_or_path):
if isinstance(file_or_path, str):
# If string, open file
cm = open(file_or_path)
else:
# Caller is responsible for closing file
cm = nullcontext(file_or_path)
with cm as file:
# Perform processing on the file
pass
with Indenter() as indent:
indent.print('hi!')
with indent:
indent.print('hello')
with indent:
indent.print('bonjour')
indent.print('hey')
hi!
hello
bonjour
hey
class Indenter:
def __init__(self):
self.level = 0
def __enter__(self):
self.level += 1
return self
def __exit__(self, exc_type, exc_val, exc_tb):
self.level -= 1
def print(self, text):
print(' ' * self.level + text)
with Indenter() as indent:
indent.print('hi!')
with indent:
indent.print('hello')
with indent:
indent.print('bonjour')
indent.print('hey')
| 0.355104 | 0.937326 |
```
import pandas as pd
df = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/wdbc.data', header=None)
df.head()
from sklearn.preprocessing import LabelEncoder
X = df.loc[:,2:].values
y = df.loc[:,1].values
le = LabelEncoder()
y = le.fit_transform(y)
X
from distutils.version import LooseVersion as Version
from sklearn import __version__ as sklearn_version
if Version(sklearn_version) < '0.18':
from sklearn.cross_validation import train_test_split
else:
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = \
train_test_split(X, y, test_size=0.20, random_state=1)
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
from sklearn.linear_model import LogisticRegression
from sklearn.pipeline import Pipeline
pipe_lr = Pipeline([('scl', StandardScaler()),
('pca', PCA(n_components=2)),
('clf', LogisticRegression(random_state=1))])
pipe_lr.fit(X_train, y_train)
print('Dokładność testu: %.3f' % pipe_lr.score(X_test, y_test))
y_pred = pipe_lr.predict(X_test)
import numpy as np
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import cross_val_score
kfold = StratifiedKFold(n_splits=10,random_state=1).split(X_train,y_train)
scores = []
for k,(train,test) in enumerate(kfold):
pipe_lr.fit(X_train[train],y_train[train])
score = pipe_lr.score(X_train[test],y_train[test])
scores.append(score)
print('Podzbiór: %s, Rozkład klasy: %s, Dokładność: %.3f' % (k+1, np.bincount(y_train[train]), score))
print('\nDokładność sprawdzianu: %.3f +/- %.3f' % (np.mean(scores), np.std(scores)))
scores=[]
scores = cross_val_score(estimator=pipe_lr,X=X_train,y=y_train,cv=10,n_jobs=1)
print('\nDokładność sprawdzianu: %.3f +/- %.3f' % (np.mean(scores), np.std(scores)))
import matplotlib.pyplot as plt
from sklearn.model_selection import learning_curve
pipe_lr = Pipeline([('scl',StandardScaler()),('clf', LogisticRegression(penalty='l2',random_state=0))])
train_sizes, train_scores, test_scores = learning_curve(estimator=pipe_lr,
X=X_train,
y=y_train,
train_sizes=np.linspace(0.1,1.0,10),
cv=10,
n_jobs=1)
train_mean = np.mean(train_scores, axis=1)
train_std = np.std(train_scores, axis=1)
test_mean = np.mean(test_scores, axis=1)
test_std = np.std(test_scores, axis=1)
plt.plot(train_sizes, train_mean,
color='blue', marker='o',
markersize=5, label='Dokładność uczenia')
plt.fill_between(train_sizes,
train_mean + train_std,
train_mean - train_std,
alpha=0.15, color='blue')
plt.plot(train_sizes, test_mean,
color='green', linestyle='--',
marker='s', markersize=5,
label='Dokładność walidacji')
plt.fill_between(train_sizes,
test_mean + test_std,
test_mean - test_std,
alpha=0.15, color='green')
plt.grid()
plt.xlabel('Liczba próbek uczących')
plt.ylabel('Dokładność')
plt.legend(loc='lower right')
plt.ylim([0.8, 1.0])
plt.tight_layout()
#plt.savefig('./rysunki/06_05.png', dpi=300)
plt.show()
from sklearn.model_selection import validation_curve
param_range = [0.001, 0.01,0.1,1,10,100]
train_scores, test_scores = validation_curve(estimator=pipe_lr,X=X_train,y=y_train,param_name='clf__C',param_range=param_range,cv=10)
train_mean = np.mean(train_scores, axis=1)
train_std = np.std(train_scores, axis=1)
test_mean = np.mean(test_scores, axis=1)
test_std = np.std(test_scores, axis=1)
plt.plot(param_range, train_mean,
color='blue', marker='o',
markersize=5, label='Dokładność uczenia')
plt.fill_between(param_range, train_mean + train_std,
train_mean - train_std, alpha=0.15,
color='blue')
plt.plot(param_range, test_mean,
color='green', linestyle='--',
marker='s', markersize=5,
label='Dokładność walidacji')
plt.fill_between(param_range,
test_mean + test_std,
test_mean - test_std,
alpha=0.15, color='green')
plt.grid()
plt.xscale('log')
plt.legend(loc='lower right')
plt.xlabel('Parametr C')
plt.ylabel('Dokładność')
plt.ylim([0.8, 1.0])
plt.tight_layout()
#plt.savefig('./rysunki/06_06.png', dpi=300)
plt.show()
from sklearn.svm import SVC
from sklearn.model_selection import GridSearchCV
pipe_svc = Pipeline([('scl',StandardScaler()),('clf', SVC(random_state=1))])
param_range = [0.001, 0.01,0.1,1,10,100,1000]
param_grid = [{'clf__C': param_range,
'clf__kernel':['linear']},
{'clf__C':param_range,
'clf__gamma': param_range,
'clf__kernel': ['rbf']}]
gs = GridSearchCV(estimator=pipe_svc,
param_grid=param_grid,
scoring='accuracy',
cv=10,
n_jobs = -1)
gs = gs.fit(X_train,y_train)
print(gs.best_params_)
clf = gs.best_estimator_
clf.fit(X_train,y_train)
print(clf.score(X_test,y_test))
gs = GridSearchCV(estimator=pipe_svc,
param_grid=param_grid,
scoring='accuracy',
cv=2,
n_jobs = -1)
scores = cross_val_score(gs,X_train,y_train,scoring='accuracy',cv=5)
np.mean(scores)
from sklearn.metrics import confusion_matrix
pipe_svc.fit(X_train,y_train)
y_pred = pipe_svc.predict(X_test)
confmat = confusion_matrix(y_true=y_test, y_pred=y_pred)
confmat
from sklearn.metrics import precision_score
from sklearn.metrics import recall_score, f1_score
print(precision_score(y_true=y_test, y_pred=y_pred))
print(recall_score(y_true=y_test, y_pred=y_pred))
print(f1_score(y_true=y_test, y_pred=y_pred))
```
|
github_jupyter
|
import pandas as pd
df = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/wdbc.data', header=None)
df.head()
from sklearn.preprocessing import LabelEncoder
X = df.loc[:,2:].values
y = df.loc[:,1].values
le = LabelEncoder()
y = le.fit_transform(y)
X
from distutils.version import LooseVersion as Version
from sklearn import __version__ as sklearn_version
if Version(sklearn_version) < '0.18':
from sklearn.cross_validation import train_test_split
else:
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = \
train_test_split(X, y, test_size=0.20, random_state=1)
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
from sklearn.linear_model import LogisticRegression
from sklearn.pipeline import Pipeline
pipe_lr = Pipeline([('scl', StandardScaler()),
('pca', PCA(n_components=2)),
('clf', LogisticRegression(random_state=1))])
pipe_lr.fit(X_train, y_train)
print('Dokładność testu: %.3f' % pipe_lr.score(X_test, y_test))
y_pred = pipe_lr.predict(X_test)
import numpy as np
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import cross_val_score
kfold = StratifiedKFold(n_splits=10,random_state=1).split(X_train,y_train)
scores = []
for k,(train,test) in enumerate(kfold):
pipe_lr.fit(X_train[train],y_train[train])
score = pipe_lr.score(X_train[test],y_train[test])
scores.append(score)
print('Podzbiór: %s, Rozkład klasy: %s, Dokładność: %.3f' % (k+1, np.bincount(y_train[train]), score))
print('\nDokładność sprawdzianu: %.3f +/- %.3f' % (np.mean(scores), np.std(scores)))
scores=[]
scores = cross_val_score(estimator=pipe_lr,X=X_train,y=y_train,cv=10,n_jobs=1)
print('\nDokładność sprawdzianu: %.3f +/- %.3f' % (np.mean(scores), np.std(scores)))
import matplotlib.pyplot as plt
from sklearn.model_selection import learning_curve
pipe_lr = Pipeline([('scl',StandardScaler()),('clf', LogisticRegression(penalty='l2',random_state=0))])
train_sizes, train_scores, test_scores = learning_curve(estimator=pipe_lr,
X=X_train,
y=y_train,
train_sizes=np.linspace(0.1,1.0,10),
cv=10,
n_jobs=1)
train_mean = np.mean(train_scores, axis=1)
train_std = np.std(train_scores, axis=1)
test_mean = np.mean(test_scores, axis=1)
test_std = np.std(test_scores, axis=1)
plt.plot(train_sizes, train_mean,
color='blue', marker='o',
markersize=5, label='Dokładność uczenia')
plt.fill_between(train_sizes,
train_mean + train_std,
train_mean - train_std,
alpha=0.15, color='blue')
plt.plot(train_sizes, test_mean,
color='green', linestyle='--',
marker='s', markersize=5,
label='Dokładność walidacji')
plt.fill_between(train_sizes,
test_mean + test_std,
test_mean - test_std,
alpha=0.15, color='green')
plt.grid()
plt.xlabel('Liczba próbek uczących')
plt.ylabel('Dokładność')
plt.legend(loc='lower right')
plt.ylim([0.8, 1.0])
plt.tight_layout()
#plt.savefig('./rysunki/06_05.png', dpi=300)
plt.show()
from sklearn.model_selection import validation_curve
param_range = [0.001, 0.01,0.1,1,10,100]
train_scores, test_scores = validation_curve(estimator=pipe_lr,X=X_train,y=y_train,param_name='clf__C',param_range=param_range,cv=10)
train_mean = np.mean(train_scores, axis=1)
train_std = np.std(train_scores, axis=1)
test_mean = np.mean(test_scores, axis=1)
test_std = np.std(test_scores, axis=1)
plt.plot(param_range, train_mean,
color='blue', marker='o',
markersize=5, label='Dokładność uczenia')
plt.fill_between(param_range, train_mean + train_std,
train_mean - train_std, alpha=0.15,
color='blue')
plt.plot(param_range, test_mean,
color='green', linestyle='--',
marker='s', markersize=5,
label='Dokładność walidacji')
plt.fill_between(param_range,
test_mean + test_std,
test_mean - test_std,
alpha=0.15, color='green')
plt.grid()
plt.xscale('log')
plt.legend(loc='lower right')
plt.xlabel('Parametr C')
plt.ylabel('Dokładność')
plt.ylim([0.8, 1.0])
plt.tight_layout()
#plt.savefig('./rysunki/06_06.png', dpi=300)
plt.show()
from sklearn.svm import SVC
from sklearn.model_selection import GridSearchCV
pipe_svc = Pipeline([('scl',StandardScaler()),('clf', SVC(random_state=1))])
param_range = [0.001, 0.01,0.1,1,10,100,1000]
param_grid = [{'clf__C': param_range,
'clf__kernel':['linear']},
{'clf__C':param_range,
'clf__gamma': param_range,
'clf__kernel': ['rbf']}]
gs = GridSearchCV(estimator=pipe_svc,
param_grid=param_grid,
scoring='accuracy',
cv=10,
n_jobs = -1)
gs = gs.fit(X_train,y_train)
print(gs.best_params_)
clf = gs.best_estimator_
clf.fit(X_train,y_train)
print(clf.score(X_test,y_test))
gs = GridSearchCV(estimator=pipe_svc,
param_grid=param_grid,
scoring='accuracy',
cv=2,
n_jobs = -1)
scores = cross_val_score(gs,X_train,y_train,scoring='accuracy',cv=5)
np.mean(scores)
from sklearn.metrics import confusion_matrix
pipe_svc.fit(X_train,y_train)
y_pred = pipe_svc.predict(X_test)
confmat = confusion_matrix(y_true=y_test, y_pred=y_pred)
confmat
from sklearn.metrics import precision_score
from sklearn.metrics import recall_score, f1_score
print(precision_score(y_true=y_test, y_pred=y_pred))
print(recall_score(y_true=y_test, y_pred=y_pred))
print(f1_score(y_true=y_test, y_pred=y_pred))
| 0.67694 | 0.505188 |
```
import numpy as np
import pandas as pd
import networkx as nx
import matplotlib.pyplot as plt
import grblas
grblas.init('suitesparse')
from grblas import Matrix, Vector
from grblas import descriptor
from grblas import UnaryOp, BinaryOp, Monoid, Semiring
from grblas import io as gio
```
## Create and visualize a Matrix
```
data = [
[3,0,3,5,6,0,6,1,6,2,4,1],
[0,1,2,2,2,3,3,4,4,5,5,6],
[3,2,3,1,5,3,7,8,3,1,7,4]
]
rows, cols, weights = data
m = Matrix.new_from_values(rows, cols, weights)
m
# Size of the sparse matrix is 7x7 with 12 non-zero elements of type INT64
m.show()
# This is an adjacency matrix
# Reading along a row shows the out-nodes of a vertex
# Reading along a column shows the in-nodes of a vertex
gio.draw(m)
```
## Create and visualize a Vector
```
v = Vector.new_from_type(m.dtype, m.nrows)
v.element[1] = 0
v.show()
```
## Single-source Shortest Path
This uses the **_min-plus_** semiring because we want to add the edges, then take the minimum length of available paths.
```
# Create a vector and initialize a starting vertex (1) with a distance of zero
v = Vector.new_from_type(m.dtype, m.nrows)
v.element[1] = 0
v.show()
m.show()
# v @ m will give us one step in a Breadth-first search
w = Vector.new_from_existing(v)
w[:] = v.vxm(m, Semiring.MIN_PLUS)
w.show()
# Look again at m and see that vertex 1 points to vertices 4 and 6 with the weights indicated
gio.draw(m)
```
We have the right semiring, but we already lost the initial distance=0 for vertex 1. How do we keep that information around as we step thru the BFS?
GraphBLAS has a builtin accumulator available for every operation.
Because it's C-based, you pass in the output object and it accumulates its existing values with the result, then returns itself.
```
w = Vector.new_from_existing(v)
w[BinaryOp.MIN] = v.vxm(m, Semiring.MIN_PLUS)
w.show()
# Now we see that the zero distance for vertex 1 is preserved
```
Let's take another step
```
w[BinaryOp.MIN] = w.vxm(m, Semiring.MIN_PLUS)
w.show()
# We see that the path to vertex 4 is now shorter. That's `min` doing its thing.
# Verify the other path distances from vertex 1 with at most two hops
gio.draw(m)
```
The algorithm repeats until a new computation is the same as the previous result
```
w = Vector.new_from_existing(v)
while True:
w_old = Vector.new_from_existing(w)
w[BinaryOp.MIN] = w.vxm(m, Semiring.MIN_PLUS)
if w == w_old:
break
w.show()
```
## Alternate solution without using accumulator
In the min_plus semiring, the "empty" value of a sparse matrix is not actually 0, but +infinity.
That way, `min(anything, +inf) = anything`, similar to the normal addition 0 of `add(anything, 0) = anything`.
A clever trick sets the diagonal of the matrix to all zeros. This makes it behave like the Identity matrix for the min_plus semiring.
Observe:
```
m_ident = Matrix.new_from_values(range(7), range(7), [0]*7)
m_ident.show()
v.rebuild_from_values([1], [0])
v.show()
v[:] = v.vxm(m_ident, Semiring.MIN_PLUS)
v.show()
# See how it preserved v exactly
# Let's try again
v.rebuild_from_values([0, 1, 4], [14, 0, 77])
v[:] = v.vxm(m_ident, Semiring.MIN_PLUS)
v.show()
```
So zeros along the diagonal preserve what you already have in `v` without adding any new path information. That's the behavior we want, so let's update `m` with zeros on the diagonal and repeat SSSP without using accumulators.
```
for i in range(m.nrows):
m.element[i, i] = 0
m.show()
# Reset v
v.clear()
v.element[1] = 0
v.show()
# Take one step (notice no accumulator is specified)
v[:] = v.vxm(m, Semiring.MIN_PLUS)
v.show()
# Repeat until we're converged
while True:
w = Vector.new_from_existing(v)
v[:] = v.vxm(m, Semiring.MIN_PLUS)
if v == w:
break
v.show()
```
### And that's SSSP in 5 very readable lines of Python, thanks to GraphBLAS
|
github_jupyter
|
import numpy as np
import pandas as pd
import networkx as nx
import matplotlib.pyplot as plt
import grblas
grblas.init('suitesparse')
from grblas import Matrix, Vector
from grblas import descriptor
from grblas import UnaryOp, BinaryOp, Monoid, Semiring
from grblas import io as gio
data = [
[3,0,3,5,6,0,6,1,6,2,4,1],
[0,1,2,2,2,3,3,4,4,5,5,6],
[3,2,3,1,5,3,7,8,3,1,7,4]
]
rows, cols, weights = data
m = Matrix.new_from_values(rows, cols, weights)
m
# Size of the sparse matrix is 7x7 with 12 non-zero elements of type INT64
m.show()
# This is an adjacency matrix
# Reading along a row shows the out-nodes of a vertex
# Reading along a column shows the in-nodes of a vertex
gio.draw(m)
v = Vector.new_from_type(m.dtype, m.nrows)
v.element[1] = 0
v.show()
# Create a vector and initialize a starting vertex (1) with a distance of zero
v = Vector.new_from_type(m.dtype, m.nrows)
v.element[1] = 0
v.show()
m.show()
# v @ m will give us one step in a Breadth-first search
w = Vector.new_from_existing(v)
w[:] = v.vxm(m, Semiring.MIN_PLUS)
w.show()
# Look again at m and see that vertex 1 points to vertices 4 and 6 with the weights indicated
gio.draw(m)
w = Vector.new_from_existing(v)
w[BinaryOp.MIN] = v.vxm(m, Semiring.MIN_PLUS)
w.show()
# Now we see that the zero distance for vertex 1 is preserved
w[BinaryOp.MIN] = w.vxm(m, Semiring.MIN_PLUS)
w.show()
# We see that the path to vertex 4 is now shorter. That's `min` doing its thing.
# Verify the other path distances from vertex 1 with at most two hops
gio.draw(m)
w = Vector.new_from_existing(v)
while True:
w_old = Vector.new_from_existing(w)
w[BinaryOp.MIN] = w.vxm(m, Semiring.MIN_PLUS)
if w == w_old:
break
w.show()
m_ident = Matrix.new_from_values(range(7), range(7), [0]*7)
m_ident.show()
v.rebuild_from_values([1], [0])
v.show()
v[:] = v.vxm(m_ident, Semiring.MIN_PLUS)
v.show()
# See how it preserved v exactly
# Let's try again
v.rebuild_from_values([0, 1, 4], [14, 0, 77])
v[:] = v.vxm(m_ident, Semiring.MIN_PLUS)
v.show()
for i in range(m.nrows):
m.element[i, i] = 0
m.show()
# Reset v
v.clear()
v.element[1] = 0
v.show()
# Take one step (notice no accumulator is specified)
v[:] = v.vxm(m, Semiring.MIN_PLUS)
v.show()
# Repeat until we're converged
while True:
w = Vector.new_from_existing(v)
v[:] = v.vxm(m, Semiring.MIN_PLUS)
if v == w:
break
v.show()
| 0.7797 | 0.911731 |
# Versicherung on Paper
```
%load_ext sql
%sql mysql://steinam:steinam@localhost/versicherung_complete
```
- Gesucht wird eine wiederholungsfreie Liste der Herstellerländer 3 P
```
%%sql
-- meine Lösung
select distinct(Land) from Fahrzeughersteller;
%%sql
-- deine Lösung
select fahrzeughersteller.Land
from fahrzeughersteller
group by fahrzeughersteller.Land
;
```
- Listen Sie alle Fahrzeugtypen und die Anzahl Fahrzeuge dieses Typs, aber nur, wenn mehr als 2 Fahrzeuge des Typs vorhanden sind. Sortieren Sie die Ausgabe nach Fahrzeugtypen. 4 P
```
%%sql
-- meine Lösung
select fahrzeugtyp.Bezeichnung, count(fahrzeug.iD) as Anzahl
from fahrzeugtyp left join fahrzeug
on fahrzeugtyp.id = fahrzeug.fahrzeugtyp_id
group by fahrzeugtyp.bezeichnung
having count(Anzahl) > 2
%%sql
select *, (select count(*) from fahrzeug where fahrzeug.fahrzeugtyp_id = fahrzeugtyp.id) as Fahrzeuge
from fahrzeugtyp
having Fahrzeuge > 2
order by fahrzeugtyp.bezeichnung;
```
- Ermittle die Namen und Vornamen der Mitarbeiter incl. Abteilungsname, deren Abteilung ihren Sitz in Dortmund oder Bochum hat.
```
%%sql
-- meine Lösung
-- select ID from Abteilung where Abteilung.Ort = 'Dortmund' or abteilung.Ort = 'Bochum'
select Name, vorname, Bezeichnung, Abteilung.ID, Mitarbeiter.Abteilung_ID, Abteilung.Ort from Mitarbeiter inner join Abteilung
on Mitarbeiter.Abteilung_ID = Abteilung.ID
where Abteilung.Ort in('Dortmund', 'Bochum')
order by Name
%%sql
-- deine Lösung
select mitarbeiter.Name, mitarbeiter.Vorname,
(select abteilung.bezeichnung from abteilung where abteilung.id = mitarbeiter.abteilung_id) as Abteilung,
(select abteilung.ort from abteilung where abteilung.id = mitarbeiter.abteilung_id) as Standort
from mitarbeiter having Standort = "Dortmund" or Standort = "Bochum";
```
- Gesucht wird für jeden Fahrzeughersteller (Angabe der ID reicht) und jedes Jahr die kleinste und größte Schadenshöhe.
Geben Sie falls möglich auch die Differenz zwischen den beiden Werten mit in der jeweiligen Ergebnismenge aus. Ansonsten erzeugen Sie für diese Aufgabe ein eigenes sql-Statement. 5 P
```
%%sql
-- meine Lösung
select fahrzeughersteller.id, year(datum) as Jahr, min(zuordnung_sf_fz.schadenshoehe), max(zuordnung_sf_fz.Schadenshoehe),
(max(zuordnung_sf_fz.schadenshoehe) - min(zuordnung_sf_fz.schadenshoehe)) as Differenz
from fahrzeughersteller left join fahrzeugtyp
on fahrzeughersteller.id = fahrzeugtyp.hersteller_ID
inner join fahrzeug on fahrzeugtyp.id = fahrzeug.fahrzeugtyp_id
inner join zuordnung_sf_fz
on fahrzeug.id = zuordnung_sf_fz.fahrzeug_id
inner join schadensfall on schadensfall.id = zuordnung_sf_fz.schadensfall_id
group by fahrzeughersteller.id, year(datum)
%%sql
-- redigierte Version von Wortmann geht
select
fahrzeughersteller.Name,
(select min(zuordnung_sf_fz.schadenshoehe) from zuordnung_sf_fz
where zuordnung_sf_fz.fahrzeug_id in(
select fahrzeug.id from fahrzeug
where fahrzeug.fahrzeugtyp_id in(
select fahrzeugtyp.id from fahrzeugtyp
where fahrzeugtyp.hersteller_id = fahrzeughersteller.id
)
)
) as Kleinste,
(select max(zuordnung_sf_fz.schadenshoehe) from zuordnung_sf_fz
where zuordnung_sf_fz.fahrzeug_id in(
select fahrzeug.id from fahrzeug
where fahrzeug.fahrzeugtyp_id in(
select fahrzeugtyp.id from fahrzeugtyp
where fahrzeugtyp.hersteller_id = fahrzeughersteller.id
)
)
) as `Groesste`
from fahrzeughersteller;
```
- Zeige alle Mitarbeiter und deren Autokennzeichen, die als Dienstwagen einen Opel fahren.
4 P
```
%%sql
select Mitarbeiter.Name, dienstwagen.Kennzeichen
from Mitarbeiter inner join dienstwagen
on mitarbeiter.id = dienstwagen.Mitarbeiter_id
inner join fahrzeugtyp
on dienstwagen.fahrzeugtyp_Id = fahrzeugtyp.id
inner join fahrzeughersteller
on fahrzeugtyp.hersteller_id = fahrzeughersteller.id
where Fahrzeughersteller.NAme = 'Opel'
%%sql
select * from mitarbeiter
where mitarbeiter.id in(
select dienstwagen.mitarbeiter_id from dienstwagen
where
dienstwagen.mitarbeiter_id = mitarbeiter.id
and dienstwagen.fahrzeugtyp_id in(
select fahrzeugtyp.id from fahrzeugtyp
where fahrzeugtyp.hersteller_id in(
select fahrzeughersteller.id from fahrzeughersteller
where fahrzeughersteller.name = "Opel"
)
)
)
```
- Welche Fahrzeuge haben Schäden verursacht, deren Schadenssumme höher als die durchschnittliche Schadenshöhe sind. 5 P
```
%%sql
select fahrzeug.kennzeichen, sum(schadenshoehe)
from fahrzeug inner join zuordnung_sf_fz
on fahrzeug.id = zuordnung_sf_fz.fahrzeug_id
group by fahrzeug.kennzeichen
having sum(schadenshoehe) > (select avg(schadenshoehe) from zuordnung_sf_fz)
%%sql
-- deine Lösung Wortmann
/*
select * from fahrzeug having fahrzeug.id in(
select zuordnung_sf_zf.fahrzeugtyp_id from zuordnung_sf_zf
where zuordnung_sf_zf.schadenhoehe > ((select sum(zuordnung_sf_zf.schadenhoehe) from zuordnung_sf_zf)) / (select count(*) from zuordnung_sf_zf))
*/
select * from fahrzeug having fahrzeug.id in(
select zuordnung_sf_fz.fahrzeug_id from zuordnung_sf_fz
where zuordnung_sf_fz.schadenshoehe > ((select sum(zuordnung_sf_fz.schadenshoehe) from zuordnung_sf_fz)) / (select count(*) from zuordnung_sf_fz))
```
- Welche Mitarbeiter sind älter als das Durchschnittsalter der Mitarbeiter. 4 P
```
%%sql
select Mitarbeiter.Name, Mitarbeiter.Geburtsdatum
from Mitarbeiter
where Geburtsdatum < (select avg(Geburtsdatum) from Mitarbeiter ma)
order by Mitarbeiter.Name
%%sql
-- geht auch
select ma.Name, ma.Geburtsdatum
from Mitarbeiter ma
where (now() - ma.Geburtsdatum) < (now() - (select avg(geburtsdatum) from mitarbeiter))
order by ma.Name;
%%sql
-- deine Lösung Wortmann
select * from mitarbeiter
having mitarbeiter.geburtsdatum < (select sum(mitarbeiter.geburtsdatum) from mitarbeiter) / (select count(*) from mitarbeiter)
```
|
github_jupyter
|
%load_ext sql
%sql mysql://steinam:steinam@localhost/versicherung_complete
%%sql
-- meine Lösung
select distinct(Land) from Fahrzeughersteller;
%%sql
-- deine Lösung
select fahrzeughersteller.Land
from fahrzeughersteller
group by fahrzeughersteller.Land
;
%%sql
-- meine Lösung
select fahrzeugtyp.Bezeichnung, count(fahrzeug.iD) as Anzahl
from fahrzeugtyp left join fahrzeug
on fahrzeugtyp.id = fahrzeug.fahrzeugtyp_id
group by fahrzeugtyp.bezeichnung
having count(Anzahl) > 2
%%sql
select *, (select count(*) from fahrzeug where fahrzeug.fahrzeugtyp_id = fahrzeugtyp.id) as Fahrzeuge
from fahrzeugtyp
having Fahrzeuge > 2
order by fahrzeugtyp.bezeichnung;
%%sql
-- meine Lösung
-- select ID from Abteilung where Abteilung.Ort = 'Dortmund' or abteilung.Ort = 'Bochum'
select Name, vorname, Bezeichnung, Abteilung.ID, Mitarbeiter.Abteilung_ID, Abteilung.Ort from Mitarbeiter inner join Abteilung
on Mitarbeiter.Abteilung_ID = Abteilung.ID
where Abteilung.Ort in('Dortmund', 'Bochum')
order by Name
%%sql
-- deine Lösung
select mitarbeiter.Name, mitarbeiter.Vorname,
(select abteilung.bezeichnung from abteilung where abteilung.id = mitarbeiter.abteilung_id) as Abteilung,
(select abteilung.ort from abteilung where abteilung.id = mitarbeiter.abteilung_id) as Standort
from mitarbeiter having Standort = "Dortmund" or Standort = "Bochum";
%%sql
-- meine Lösung
select fahrzeughersteller.id, year(datum) as Jahr, min(zuordnung_sf_fz.schadenshoehe), max(zuordnung_sf_fz.Schadenshoehe),
(max(zuordnung_sf_fz.schadenshoehe) - min(zuordnung_sf_fz.schadenshoehe)) as Differenz
from fahrzeughersteller left join fahrzeugtyp
on fahrzeughersteller.id = fahrzeugtyp.hersteller_ID
inner join fahrzeug on fahrzeugtyp.id = fahrzeug.fahrzeugtyp_id
inner join zuordnung_sf_fz
on fahrzeug.id = zuordnung_sf_fz.fahrzeug_id
inner join schadensfall on schadensfall.id = zuordnung_sf_fz.schadensfall_id
group by fahrzeughersteller.id, year(datum)
%%sql
-- redigierte Version von Wortmann geht
select
fahrzeughersteller.Name,
(select min(zuordnung_sf_fz.schadenshoehe) from zuordnung_sf_fz
where zuordnung_sf_fz.fahrzeug_id in(
select fahrzeug.id from fahrzeug
where fahrzeug.fahrzeugtyp_id in(
select fahrzeugtyp.id from fahrzeugtyp
where fahrzeugtyp.hersteller_id = fahrzeughersteller.id
)
)
) as Kleinste,
(select max(zuordnung_sf_fz.schadenshoehe) from zuordnung_sf_fz
where zuordnung_sf_fz.fahrzeug_id in(
select fahrzeug.id from fahrzeug
where fahrzeug.fahrzeugtyp_id in(
select fahrzeugtyp.id from fahrzeugtyp
where fahrzeugtyp.hersteller_id = fahrzeughersteller.id
)
)
) as `Groesste`
from fahrzeughersteller;
%%sql
select Mitarbeiter.Name, dienstwagen.Kennzeichen
from Mitarbeiter inner join dienstwagen
on mitarbeiter.id = dienstwagen.Mitarbeiter_id
inner join fahrzeugtyp
on dienstwagen.fahrzeugtyp_Id = fahrzeugtyp.id
inner join fahrzeughersteller
on fahrzeugtyp.hersteller_id = fahrzeughersteller.id
where Fahrzeughersteller.NAme = 'Opel'
%%sql
select * from mitarbeiter
where mitarbeiter.id in(
select dienstwagen.mitarbeiter_id from dienstwagen
where
dienstwagen.mitarbeiter_id = mitarbeiter.id
and dienstwagen.fahrzeugtyp_id in(
select fahrzeugtyp.id from fahrzeugtyp
where fahrzeugtyp.hersteller_id in(
select fahrzeughersteller.id from fahrzeughersteller
where fahrzeughersteller.name = "Opel"
)
)
)
%%sql
select fahrzeug.kennzeichen, sum(schadenshoehe)
from fahrzeug inner join zuordnung_sf_fz
on fahrzeug.id = zuordnung_sf_fz.fahrzeug_id
group by fahrzeug.kennzeichen
having sum(schadenshoehe) > (select avg(schadenshoehe) from zuordnung_sf_fz)
%%sql
-- deine Lösung Wortmann
/*
select * from fahrzeug having fahrzeug.id in(
select zuordnung_sf_zf.fahrzeugtyp_id from zuordnung_sf_zf
where zuordnung_sf_zf.schadenhoehe > ((select sum(zuordnung_sf_zf.schadenhoehe) from zuordnung_sf_zf)) / (select count(*) from zuordnung_sf_zf))
*/
select * from fahrzeug having fahrzeug.id in(
select zuordnung_sf_fz.fahrzeug_id from zuordnung_sf_fz
where zuordnung_sf_fz.schadenshoehe > ((select sum(zuordnung_sf_fz.schadenshoehe) from zuordnung_sf_fz)) / (select count(*) from zuordnung_sf_fz))
%%sql
select Mitarbeiter.Name, Mitarbeiter.Geburtsdatum
from Mitarbeiter
where Geburtsdatum < (select avg(Geburtsdatum) from Mitarbeiter ma)
order by Mitarbeiter.Name
%%sql
-- geht auch
select ma.Name, ma.Geburtsdatum
from Mitarbeiter ma
where (now() - ma.Geburtsdatum) < (now() - (select avg(geburtsdatum) from mitarbeiter))
order by ma.Name;
%%sql
-- deine Lösung Wortmann
select * from mitarbeiter
having mitarbeiter.geburtsdatum < (select sum(mitarbeiter.geburtsdatum) from mitarbeiter) / (select count(*) from mitarbeiter)
| 0.139075 | 0.790369 |
<a href="https://www.kaggle.com/muhammadosamasaleem/sf-salaries-data-analysis?scriptVersionId=86800279" target="_blank"><img align="left" alt="Kaggle" title="Open in Kaggle" src="https://kaggle.com/static/images/open-in-kaggle.svg"></a>
# Importing libraries
```
import numpy as np
import pandas as pd
```
### Read csv file
```
# Store data in variable df
df = pd.read_csv('../input/sf-salaries/Salaries.csv')
df
```
### Head of dataset
```
df.head()
```
### Getting information about dataset
```
df.info()
```
### Average Base Pay
The Base Pay column contains string 'Not Provided' and the dtype is object. To find mean (average) we have to
- First convert string into NaN and then change NaN to 0
- Then convert dtype = object to dtype = float
- In last have to find avarage
```
df['BasePay']
# converting string value to NaN
df['BasePay'].replace('Not Provided',np.nan,inplace=True)
# converting NaN to 0
df['BasePay'].fillna(value=0,inplace=True)
# convert them from string to float
df['BasePay']=df['BasePay'].astype(float)
# Finding Avaerage Base pay
df['BasePay']
df['BasePay'].mean()
```
## Highest Overtime Pay
The Base Pay column contains string 'Not Provided' and the dtype is object. To find mean (average) we have to
- First convert string into NaN and then change NaN to 0
- Then convert dtype = object to dtype = float
- In last have to find avarage
```
df['OvertimePay'].replace('Not Provided',np.nan, inplace=True)
df['OvertimePay'].fillna(value=0,inplace=True)
df['OvertimePay'] = df['OvertimePay'].astype(float)
df['OvertimePay'].max()
```
## Job Title of JOSEPH DRISCOLL
```
jd= df[ df['EmployeeName'] == 'JOSEPH DRISCOLL']
df[ df['EmployeeName'] == 'JOSEPH DRISCOLL']['JobTitle']
```
## Total Pay of JOSEPH DRISCOLL
```
df[ df['EmployeeName'] == 'JOSEPH DRISCOLL']['TotalPayBenefits']
```
## Name of Highest paid person
```
df[df['TotalPayBenefits']== df['TotalPayBenefits'].max()]
df[df['TotalPayBenefits']== df['TotalPayBenefits'].max()]['EmployeeName']
```
## Name of lowest paid person
```
df.iloc[df['TotalPayBenefits'].argmin()]
df.iloc[df['TotalPayBenefits'].argmin()]['EmployeeName']
# Another way
df[df['TotalPayBenefits']== df['TotalPayBenefits'].min()]
df[df['TotalPayBenefits']== df['TotalPayBenefits'].min()]['EmployeeName']
```
## Total unique Job Titles
```
df['JobTitle'].nunique()
```
## Top 5 most Common Job
```
df['JobTitle'].value_counts().head(5)
```
## Job title with only one occerance in 2013
```
sum(df[df['Year']==2013]['JobTitle'].value_counts()==1)
```
## People with cheif in their job title
```
df['JobTitle']
def chief_string(title):
if 'chief' in title.lower().split():
return True
else:
return False
sum(df['JobTitle'].apply(lambda x: chief_string(x)))
```
|
github_jupyter
|
import numpy as np
import pandas as pd
# Store data in variable df
df = pd.read_csv('../input/sf-salaries/Salaries.csv')
df
df.head()
df.info()
df['BasePay']
# converting string value to NaN
df['BasePay'].replace('Not Provided',np.nan,inplace=True)
# converting NaN to 0
df['BasePay'].fillna(value=0,inplace=True)
# convert them from string to float
df['BasePay']=df['BasePay'].astype(float)
# Finding Avaerage Base pay
df['BasePay']
df['BasePay'].mean()
df['OvertimePay'].replace('Not Provided',np.nan, inplace=True)
df['OvertimePay'].fillna(value=0,inplace=True)
df['OvertimePay'] = df['OvertimePay'].astype(float)
df['OvertimePay'].max()
jd= df[ df['EmployeeName'] == 'JOSEPH DRISCOLL']
df[ df['EmployeeName'] == 'JOSEPH DRISCOLL']['JobTitle']
df[ df['EmployeeName'] == 'JOSEPH DRISCOLL']['TotalPayBenefits']
df[df['TotalPayBenefits']== df['TotalPayBenefits'].max()]
df[df['TotalPayBenefits']== df['TotalPayBenefits'].max()]['EmployeeName']
df.iloc[df['TotalPayBenefits'].argmin()]
df.iloc[df['TotalPayBenefits'].argmin()]['EmployeeName']
# Another way
df[df['TotalPayBenefits']== df['TotalPayBenefits'].min()]
df[df['TotalPayBenefits']== df['TotalPayBenefits'].min()]['EmployeeName']
df['JobTitle'].nunique()
df['JobTitle'].value_counts().head(5)
sum(df[df['Year']==2013]['JobTitle'].value_counts()==1)
df['JobTitle']
def chief_string(title):
if 'chief' in title.lower().split():
return True
else:
return False
sum(df['JobTitle'].apply(lambda x: chief_string(x)))
| 0.184694 | 0.831177 |
# Exponential Modeling of COVID-19 Confirmed Cases
This notebook explores modeling the spread of COVID-19 confirmed cases as an exponential function. While this is not a good model for long or even medium-term predictions, it is able to fit initial outbreaks quite well. For a more sophisticated and accurate model, see the [logistic modeling](logistic_modeling.ipynb) notebook.
```
import pandas as pd
import seaborn as sns
sns.set()
df = pd.read_csv('https://open-covid-19.github.io/data/data.csv')
df = df.set_index('Date')
```
### Defining our parameters
Here we are looking at the confirmed and fatal cases for Italy through March 17. To apply the model to other countries or dates, just change the code below.
```
country_code = 'IT'
date_limit = '2020-03-17'
```
### Looking at the outbreak
There are months of data, but we only care about when the number of cases started to grow. We define *outbreak* as whenever the number of cases exceeded certain threshold. In this case, we are using 10.
```
def get_outbreak_mask(data: pd.DataFrame, threshold: int = 10):
''' Returns a mask for > N confirmed cases '''
return data['Confirmed'] > threshold
cols = ['CountryCode', 'CountryName', 'Confirmed', 'Deaths']
# Get data only for the selected country
country = df[df['CountryCode'] == country_code][cols]
# Get data only for the selected dates
country = country[country.index <= date_limit]
# Get data only after the outbreak begun
country = country[get_outbreak_mask(country)]
```
### Plotting the data
Let's take a first look at the data. A visual inspection will typically give us a lot of information.
```
country.plot(kind='bar', figsize=(16, 8));
```
### Modeling the data
The data appears to follow an exponential curve, it looks straight out of a middle school math textbook cover. Let's see if we can model it using some parameter fitting
```
from scipy import optimize
def exponential_function(x: float, a: float, b: float, c: float):
''' a * (b ^ x) + c '''
return a * (b ** x) + c
X, y = list(range(len(country))), country['Confirmed'].tolist()
params, _ = optimize.curve_fit(exponential_function, X, y)
print('Estimated function: {0:.3f} * ({1:.3f} ^ X) + {2:.3f}'.format(*params))
ax = country[['Confirmed']].plot(kind='bar', figsize=(16, 8))
estimate = [exponential_function(x, *params) for x in X]
ax.plot(country.index, estimate, color='red', label='Estimate')
ax.legend();
```
### Validating the model
That curve looks like a very good fit! Even though proper epidemiology models are fundamentally different (because diseases can't grow exponentially indefinitely), the exponential model should be good for short term predictions.
To validate our model, let's try to fit it again without looking at the last 3 days of data. Then, we can estimate the missing days using our model, and verify if the results still hold by comparing what the model thought was going to happen with the actual data.
```
ESTIMATE_DAYS = 3
params_validate, _ = optimize.curve_fit(exponential_function, X[:-ESTIMATE_DAYS], y[:-ESTIMATE_DAYS])
# Project zero for all values except for the last ESTIMATE_DAYS
projected = [0] * len(X[:-ESTIMATE_DAYS]) + [exponential_function(x, *params_validate) for x in X[-ESTIMATE_DAYS:]]
projected = pd.Series(projected, index=country.index, name='Projected')
df_ = pd.DataFrame({'Confirmed': country['Confirmed'], 'Projected': projected})
ax = df_.plot(kind='bar', figsize=(16, 8))
estimate = [exponential_function(x, *params_validate) for x in X]
ax.plot(country.index, estimate, color='red', label='Estimate')
ax.legend();
```
### Projecting future data
It looks like our exponential model slightly overestimates the confirmed cases. That's a good sign! It means that the disease is slowing down a bit. The numbers are close enough that a 3-day projection is probably an accurate enough estimate.
Now, let's use the model we fitted earlier which used all the data, and try to predict what the next 3 days will look like.
```
import datetime
FUTURE_DAYS = 3
# Append N new days to our indices
date_format = '%Y-%m-%d'
date_range = [datetime.datetime.strptime(date, date_format) for date in country.index]
for _ in range(FUTURE_DAYS): date_range.append(date_range[-1] + datetime.timedelta(days=1))
date_range = [datetime.datetime.strftime(date, date_format) for date in date_range]
# Perform projection with the previously estimated parameters
projected = [0] * len(X) + [exponential_function(x, *params) for x in range(len(X), len(X) + FUTURE_DAYS)]
projected = pd.Series(projected, index=date_range, name='Projected')
df_ = pd.DataFrame({'Confirmed': country['Confirmed'], 'Projected': projected})
ax = df_.plot(kind='bar', figsize=(16, 8))
estimate = [exponential_function(x, *params) for x in range(len(date_range))]
ax.plot(date_range, estimate, color='red', label='Estimate')
ax.legend();
```
|
github_jupyter
|
import pandas as pd
import seaborn as sns
sns.set()
df = pd.read_csv('https://open-covid-19.github.io/data/data.csv')
df = df.set_index('Date')
country_code = 'IT'
date_limit = '2020-03-17'
def get_outbreak_mask(data: pd.DataFrame, threshold: int = 10):
''' Returns a mask for > N confirmed cases '''
return data['Confirmed'] > threshold
cols = ['CountryCode', 'CountryName', 'Confirmed', 'Deaths']
# Get data only for the selected country
country = df[df['CountryCode'] == country_code][cols]
# Get data only for the selected dates
country = country[country.index <= date_limit]
# Get data only after the outbreak begun
country = country[get_outbreak_mask(country)]
country.plot(kind='bar', figsize=(16, 8));
from scipy import optimize
def exponential_function(x: float, a: float, b: float, c: float):
''' a * (b ^ x) + c '''
return a * (b ** x) + c
X, y = list(range(len(country))), country['Confirmed'].tolist()
params, _ = optimize.curve_fit(exponential_function, X, y)
print('Estimated function: {0:.3f} * ({1:.3f} ^ X) + {2:.3f}'.format(*params))
ax = country[['Confirmed']].plot(kind='bar', figsize=(16, 8))
estimate = [exponential_function(x, *params) for x in X]
ax.plot(country.index, estimate, color='red', label='Estimate')
ax.legend();
ESTIMATE_DAYS = 3
params_validate, _ = optimize.curve_fit(exponential_function, X[:-ESTIMATE_DAYS], y[:-ESTIMATE_DAYS])
# Project zero for all values except for the last ESTIMATE_DAYS
projected = [0] * len(X[:-ESTIMATE_DAYS]) + [exponential_function(x, *params_validate) for x in X[-ESTIMATE_DAYS:]]
projected = pd.Series(projected, index=country.index, name='Projected')
df_ = pd.DataFrame({'Confirmed': country['Confirmed'], 'Projected': projected})
ax = df_.plot(kind='bar', figsize=(16, 8))
estimate = [exponential_function(x, *params_validate) for x in X]
ax.plot(country.index, estimate, color='red', label='Estimate')
ax.legend();
import datetime
FUTURE_DAYS = 3
# Append N new days to our indices
date_format = '%Y-%m-%d'
date_range = [datetime.datetime.strptime(date, date_format) for date in country.index]
for _ in range(FUTURE_DAYS): date_range.append(date_range[-1] + datetime.timedelta(days=1))
date_range = [datetime.datetime.strftime(date, date_format) for date in date_range]
# Perform projection with the previously estimated parameters
projected = [0] * len(X) + [exponential_function(x, *params) for x in range(len(X), len(X) + FUTURE_DAYS)]
projected = pd.Series(projected, index=date_range, name='Projected')
df_ = pd.DataFrame({'Confirmed': country['Confirmed'], 'Projected': projected})
ax = df_.plot(kind='bar', figsize=(16, 8))
estimate = [exponential_function(x, *params) for x in range(len(date_range))]
ax.plot(date_range, estimate, color='red', label='Estimate')
ax.legend();
| 0.778186 | 0.98947 |
```
# 权重衰减是最广泛使用的正则化技术之一
%matplotlib inline
import torch
from torch import nn
from d2l import torch as d2l
# 像以前一样生成一些数据
# y = 0.05 + (求和i=1到d) 0.01*x_i + e where e ~ N(0,0.01^2)
n_train, n_test, num_inputs, batch_size = 20, 100, 200, 5
true_w, true_b = torch.ones((num_inputs, 1)) * 0.01, 0.05
train_data = d2l.synthetic_data(true_w, true_b, n_train)
train_iter = d2l.load_array(train_data, batch_size)
test_data = d2l.synthetic_data(true_w, true_b, n_test)
test_iter = d2l.load_array(test_data, batch_size, is_train=False)
# 初始化模型参数
def init_params():
w = torch.normal(0, 1, size=(num_inputs, 1), requires_grad=True) # size=200*1
b = torch.zeros(1, requires_grad=True)
return [w, b]
# 定义L2范数惩罚
def l2_penalty(w):
return torch.sum(w.pow(2)) / 2
# 定义训练代码实现
def train(lambd): # lambd:超参数
w, b = init_params()
net, loss = lambda X: d2l.linreg(X, w, b), d2l.squared_loss # lambda:简单的函数定义方式
num_epochs, lr = 100, 0.003
animator = d2l.Animator(xlabel='epochs', ylabel='loss', yscale='log',
xlim=[5, num_epochs], legend=['train', 'test'])
for epoch in range(num_epochs):
for X, y in train_iter:
l = loss(net(X), y) + lambd * l2_penalty(w)
l.sum().backward()
d2l.sgd([w, b], lr, batch_size)
if (epoch + 1) % 5 == 0:
animator.add(epoch + 1, (d2l.evaluate_loss(net, train_iter, loss),
d2l.evaluate_loss(net, test_iter, loss)))
print('w的L2范数是:', torch.norm(w).item())
# 忽略正则化直接训练,结果明显过拟合
train(lambd=0)
# 使用权重衰减
train(lambd=3)
# 简洁实现
def train_concise(wd):
net = nn.Sequential(nn.Linear(num_inputs, 1))
for param in net.parameters():
param.data.normal_()
loss = nn.MSELoss()
num_epochs, lr = 100, 0.003
trainer = torch.optim.SGD([
{"params": net[0].weight, 'weight_decay': wd}, # wd就是之前的lambd
{"params": net[0].bias}], lr=lr)
animator = d2l.Animator(xlabel='epochs', ylabel='loss', yscale='log',
xlim=[5, num_epochs], legend=['train', 'test'])
for epoch in range(num_epochs):
for X, y in train_iter:
trainer.zero_grad()
l = loss(net(X), y)
l.backward()
trainer.step()
if (epoch + 1) % 5 == 0:
animator.add(epoch + 1, (d2l.evaluate_loss(net, train_iter, loss),
d2l.evaluate_loss(net, test_iter, loss)))
print('w的L2范数是:', net[0].weight.norm().item())
train_concise(0)
train_concise(3)
```
|
github_jupyter
|
# 权重衰减是最广泛使用的正则化技术之一
%matplotlib inline
import torch
from torch import nn
from d2l import torch as d2l
# 像以前一样生成一些数据
# y = 0.05 + (求和i=1到d) 0.01*x_i + e where e ~ N(0,0.01^2)
n_train, n_test, num_inputs, batch_size = 20, 100, 200, 5
true_w, true_b = torch.ones((num_inputs, 1)) * 0.01, 0.05
train_data = d2l.synthetic_data(true_w, true_b, n_train)
train_iter = d2l.load_array(train_data, batch_size)
test_data = d2l.synthetic_data(true_w, true_b, n_test)
test_iter = d2l.load_array(test_data, batch_size, is_train=False)
# 初始化模型参数
def init_params():
w = torch.normal(0, 1, size=(num_inputs, 1), requires_grad=True) # size=200*1
b = torch.zeros(1, requires_grad=True)
return [w, b]
# 定义L2范数惩罚
def l2_penalty(w):
return torch.sum(w.pow(2)) / 2
# 定义训练代码实现
def train(lambd): # lambd:超参数
w, b = init_params()
net, loss = lambda X: d2l.linreg(X, w, b), d2l.squared_loss # lambda:简单的函数定义方式
num_epochs, lr = 100, 0.003
animator = d2l.Animator(xlabel='epochs', ylabel='loss', yscale='log',
xlim=[5, num_epochs], legend=['train', 'test'])
for epoch in range(num_epochs):
for X, y in train_iter:
l = loss(net(X), y) + lambd * l2_penalty(w)
l.sum().backward()
d2l.sgd([w, b], lr, batch_size)
if (epoch + 1) % 5 == 0:
animator.add(epoch + 1, (d2l.evaluate_loss(net, train_iter, loss),
d2l.evaluate_loss(net, test_iter, loss)))
print('w的L2范数是:', torch.norm(w).item())
# 忽略正则化直接训练,结果明显过拟合
train(lambd=0)
# 使用权重衰减
train(lambd=3)
# 简洁实现
def train_concise(wd):
net = nn.Sequential(nn.Linear(num_inputs, 1))
for param in net.parameters():
param.data.normal_()
loss = nn.MSELoss()
num_epochs, lr = 100, 0.003
trainer = torch.optim.SGD([
{"params": net[0].weight, 'weight_decay': wd}, # wd就是之前的lambd
{"params": net[0].bias}], lr=lr)
animator = d2l.Animator(xlabel='epochs', ylabel='loss', yscale='log',
xlim=[5, num_epochs], legend=['train', 'test'])
for epoch in range(num_epochs):
for X, y in train_iter:
trainer.zero_grad()
l = loss(net(X), y)
l.backward()
trainer.step()
if (epoch + 1) % 5 == 0:
animator.add(epoch + 1, (d2l.evaluate_loss(net, train_iter, loss),
d2l.evaluate_loss(net, test_iter, loss)))
print('w的L2范数是:', net[0].weight.norm().item())
train_concise(0)
train_concise(3)
| 0.595728 | 0.711017 |
```
import pandas as pd
import matplotlib.pyplot as plt
'''
This code is written by the amazing team dubbed "Team Incapable".
For readers who are unable to read Dutch, this roughly translate to
"The team that will save the world by their combined skillset"
'''
def parseLine(id, line, tagResult):
global countV, countH
splitted = line.split(" ")
orientation = splitted[0]
if orientation == 'H':
countH=countH+1
else:
countV=countV+1
numTags = int(splitted[1])
tags=[]
for i in range(numTags):
tag=splitted[i+2].rstrip()
tags.append(tag)
if tag in tagResult:
tagResult[tag].append(id)
else:
tagResult[tag]=[id]
return {"id":id,"Orientation":orientation, "tags":tags}
countH=0
countV=0
def valuate(prevSlide, curSlide):
return(min(overlapValuation(prevSlide["tags"], curSlide["tags"]), deltaValuation(prevSlide["tags"], curSlide["tags"]), deltaValuation(curSlide["tags"], prevSlide["tags"])))
def overlapValuation(prevTags, curTags):
return(len(list(set(prevTags) & set(curTags))))
def deltaValuation(prevTags, curTags):
return(len(set(prevTags).difference(curTags)))
DataFolder="./inputs/"
#File="a_example.txt"
Files=["a_example.txt",
"b_lovely_landscapes.txt",
"c_memorable_moments.txt",
"d_pet_pictures.txt",
"e_shiny_selfies.txt",
]
Bdata=[]
#File="c_memorable_moments.txt"
#File="d_pet_pictures.txt"
#File="e_shiny_selfies.txt"
resultsRaw = []
for i, File in enumerate(Files):
f = open(DataFolder+File)
inputData=[]
numLines=int(f.readline())
resultPerTag={}
for i in range(numLines):
line = f.readline()
parsed=parseLine(i, line, resultPerTag)
inputData.append(parsed)
resultsRaw.append(inputData)
f.close()
dfs = []
for rr in resultsRaw:
df = pd.DataFrame(rr)
dfs.append(df)
```
# Distribution of #tags
```
i = 0
for i, df in enumerate(dfs):
my_df = dfs[i]
my_df.stack()
result = pd.DataFrame([(d, tup.id, tup.Orientation, tup.tags) for tup in dfs[i].itertuples() for d in tup.tags])
result.columns = ['tag', 'id', 'orientation', 'tags']
lens = result.tags.apply(lambda row: len(row))
lens.hist()
plt.title(chr(ord('A') + i) + '#tags/photo')
plt.show()
i = 0
my_df = dfs[i]
my_df.stack()
result = pd.DataFrame([(d, tup.id, tup.Orientation, tup.tags) for tup in dfs[i].itertuples() for d in tup.tags])
result.columns = ['tag', 'id', 'orientation', 'tags']
result.tag.value_counts().hist()
i = 0
for i, df in enumerate(dfs):
my_df = dfs[i]
my_df.stack()
result = pd.DataFrame([(d, tup.id, tup.Orientation, tup.tags) for tup in dfs[i].itertuples() for d in tup.tags])
result.columns = ['tag', 'id', 'orientation', 'tags']
lens = result.tag.value_counts()
lens.hist()
plt.title(chr(ord('A') + i) + ' #photos/tag')
plt.show()
```
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
'''
This code is written by the amazing team dubbed "Team Incapable".
For readers who are unable to read Dutch, this roughly translate to
"The team that will save the world by their combined skillset"
'''
def parseLine(id, line, tagResult):
global countV, countH
splitted = line.split(" ")
orientation = splitted[0]
if orientation == 'H':
countH=countH+1
else:
countV=countV+1
numTags = int(splitted[1])
tags=[]
for i in range(numTags):
tag=splitted[i+2].rstrip()
tags.append(tag)
if tag in tagResult:
tagResult[tag].append(id)
else:
tagResult[tag]=[id]
return {"id":id,"Orientation":orientation, "tags":tags}
countH=0
countV=0
def valuate(prevSlide, curSlide):
return(min(overlapValuation(prevSlide["tags"], curSlide["tags"]), deltaValuation(prevSlide["tags"], curSlide["tags"]), deltaValuation(curSlide["tags"], prevSlide["tags"])))
def overlapValuation(prevTags, curTags):
return(len(list(set(prevTags) & set(curTags))))
def deltaValuation(prevTags, curTags):
return(len(set(prevTags).difference(curTags)))
DataFolder="./inputs/"
#File="a_example.txt"
Files=["a_example.txt",
"b_lovely_landscapes.txt",
"c_memorable_moments.txt",
"d_pet_pictures.txt",
"e_shiny_selfies.txt",
]
Bdata=[]
#File="c_memorable_moments.txt"
#File="d_pet_pictures.txt"
#File="e_shiny_selfies.txt"
resultsRaw = []
for i, File in enumerate(Files):
f = open(DataFolder+File)
inputData=[]
numLines=int(f.readline())
resultPerTag={}
for i in range(numLines):
line = f.readline()
parsed=parseLine(i, line, resultPerTag)
inputData.append(parsed)
resultsRaw.append(inputData)
f.close()
dfs = []
for rr in resultsRaw:
df = pd.DataFrame(rr)
dfs.append(df)
i = 0
for i, df in enumerate(dfs):
my_df = dfs[i]
my_df.stack()
result = pd.DataFrame([(d, tup.id, tup.Orientation, tup.tags) for tup in dfs[i].itertuples() for d in tup.tags])
result.columns = ['tag', 'id', 'orientation', 'tags']
lens = result.tags.apply(lambda row: len(row))
lens.hist()
plt.title(chr(ord('A') + i) + '#tags/photo')
plt.show()
i = 0
my_df = dfs[i]
my_df.stack()
result = pd.DataFrame([(d, tup.id, tup.Orientation, tup.tags) for tup in dfs[i].itertuples() for d in tup.tags])
result.columns = ['tag', 'id', 'orientation', 'tags']
result.tag.value_counts().hist()
i = 0
for i, df in enumerate(dfs):
my_df = dfs[i]
my_df.stack()
result = pd.DataFrame([(d, tup.id, tup.Orientation, tup.tags) for tup in dfs[i].itertuples() for d in tup.tags])
result.columns = ['tag', 'id', 'orientation', 'tags']
lens = result.tag.value_counts()
lens.hist()
plt.title(chr(ord('A') + i) + ' #photos/tag')
plt.show()
| 0.115386 | 0.434041 |
# Notebook compilation of data
An attempt to compile known GC data, and select the best/latest available.
## TODO
- [ ] Check Gran Physical Coords
- [ ] Check Systematics of AMR
- [ ] Differences in Isochrones, BaSTi, DSED, BaSTi Alpha enhanced, BaSTI sometimes corrected BaSTi with an age shift due to atomic diffusion ect
- [ ] Compare Age estimates between catalogues
## Name Preferencse
Alt Names gives preferences of variants on a single name
name_parse gives the preferences of alternative names of a GC
```
import numpy as np
from LoadSaveData import save_dic_csv, load_dic_csv
from ParseNames import process_names, name_parse, alt_name_parse, create_alt_name_list
from ParseProperty import property_parser, process_properties
from DataFuncs import data_add_function, data_dic_to_gc_dic, gc_dic_to_data_dic
paper_age = {}
data = {}
```
# Larger Data Files
Read in csv/txt files
## Dynamics
### Vasiliev
```
def load_Vasilev21_data():
Vasilev21_data_dic = load_dic_csv("DataFiles/GC_Sky_Vasilev2021.txt")
bad_filt = np.logical_not(np.isfinite(Vasilev21_data_dic["dist_err"]))
Vasilev21_data_dic["dist_err"][bad_filt] = Vasilev21_data_dic["dist"][bad_filt] * 0.05
Vasilev21_data_dic["vlos_err"] = np.maximum(Vasilev21_data_dic["vlos_err"], 2.0)
return Vasilev21_data_dic
Vasilev21_data_dic = load_Vasilev21_data()
Vasilev21_dic = data_dic_to_gc_dic(Vasilev21_data_dic)
data = data_add_function(data, Vasilev21_dic, data_name='Vasilev21')
paper_age["Vasilev21"] = 2021
```
### Baumb Distances
```
def load_Baumgardt_dist_data():
Baumgardt21_data_dic = load_dic_csv("DataFiles/Baumgardt21_sky.csv", skip=2)
bad_filt = np.logical_not(np.isfinite(Baumgardt21_data_dic["dist_err"]))
Baumgardt21_data_dic["dist_err"][bad_filt] = Baumgardt21_data_dic["dist"][bad_filt] * 0.05
Baumgardt21_data_dic["vlos_err"] = np.maximum(Baumgardt21_data_dic["vlos_err"], 2.0)
return Baumgardt21_data_dic
Baumgardt21_data_dic = load_Baumgardt_dist_data()
Baumgardt21_dic = data_dic_to_gc_dic(Baumgardt21_data_dic)
data = data_add_function(data, Baumgardt21_dic, data_name='Baumgardt21')
paper_age["Baumgardt21"] = 2021.5
```
## AMR + Mass/Magnitude
### Baumgardt Cluster Mass
https://ui.adsabs.harvard.edu/abs/2018MNRAS.478.1520B/abstract
https://people.smp.uq.edu.au/HolgerBaumgardt/globular/parameter.html
Fits N-body Simulatons to velocity dispersion and surface density profiles.
```
Baumgardt18_data_dic = load_dic_csv("DataFiles/Baumgardt_GC_Mass.csv", skip=2)
Baumgardt18_GC_dic = data_dic_to_gc_dic(Baumgardt18_data_dic)
data = data_add_function(data, Baumgardt18_GC_dic, data_name='Baumgardt_Mass18')
paper_age["Baumgardt_Mass18"] = 2018
```
### Kruijssen19 Sample
https://ui.adsabs.harvard.edu/abs/2019MNRAS.486.3180K/abstract
Data used in Kruijssen19. 3 Sources, Kuijssen takes average of available. Use all from sources, as some missing data added by other sources.
```
Kruijssen19_data_dic = load_dic_csv("DataFiles/Kruijssen19.csv", skip=1)
Kruijssen19_data_dic["Mass"] = 10 ** Kruijssen19_data_dic["logM"]
del Kruijssen19_data_dic["logM"]
Kruijssen19_dic = data_dic_to_gc_dic(Kruijssen19_data_dic)
data = data_add_function(data, Kruijssen19_dic, data_name='Kruijssen19')
paper_age["Kruijssen19"] = 2013
```
#### Dotter
Dotter 11 added onto the back of the datafile.
https://ui.adsabs.harvard.edu/abs/2010ApJ...708..698D/abstract
https://ui.adsabs.harvard.edu/abs/2011ApJ...738...74D/abstract
Fit Dartmouth isochrones to CMD data.
Dartmouth Isochrones (DSEP) used detailed in https://ui.adsabs.harvard.edu/abs/2007AJ....134..376D/abstract
Not a commonly used isochrone - biases?
DSEP not good add low metallicity?
Best fit age from isochrone MSTO through the subgiant branch.
```
Dotter10_data_dic = load_dic_csv("DataFiles/Dotter10.csv", skip=1)
Dotter10_dic = data_dic_to_gc_dic(Dotter10_data_dic)
data = data_add_function(data, Dotter10_dic, data_name='Dotter10')
paper_age['Dotter10'] = 2010
```
#### Forbes+Bridges10
https://ui.adsabs.harvard.edu/abs/2010MNRAS.404.1203F/abstract
Note No Age errors?
Already a compilation of data!
- 64 from Marin-Franch 2009 (Dartmouth Isochrone), Normalised by assuming absolute metal poor age?
- 13 from Angeli 2005, assuming same norm
- 10 from Salaris 1995.
- + Others
Most comes from Marin-Franch
```
Forbes10_data_dic = load_dic_csv("DataFiles/Forbes10.csv", skip=1)
Forbes10_dic = data_dic_to_gc_dic(Forbes10_data_dic)
data = data_add_function(data, Forbes10_dic, data_name='Forbes10')
paper_age["Forbes10"] = 2010
```
#### VandenBerg13
https://ui.adsabs.harvard.edu/abs/2013ApJ...775..134V/abstract
Following Kruijssen, +1Gyr Uncertainty:
''' For the VandenBerg et al. (2013) sample, the uncertainties include the statistical uncertainty listed in their paper, as well as an additional uncertainty of 1 Gyr to account for uncertainties in distance and chemical abundances'''
Isochrone fit to MSTO
```
VandenBerg13_data_dic = load_dic_csv("DataFiles/VandenBerg13.csv", skip=1)
print("Adding 1GYR to errors")
VandenBerg13_data_dic["Age_Err"] = VandenBerg13_data_dic["Age_Err"] + 1
VandenBerg13_dic = data_dic_to_gc_dic(VandenBerg13_data_dic)
data = data_add_function(data, VandenBerg13_dic, data_name='VandenBerg13')
paper_age["VandenBerg13"] = 2013
```
### Harris10
https://physics.mcmaster.ca/~harris/mwgc.dat
```
Harris10_data_dic = load_dic_csv("DataFiles/Harris10.csv", skip=1)
Harris10_dic = data_dic_to_gc_dic(Harris10_data_dic)
data = data_add_function(data, Harris10_dic, data_name='Harris10')
paper_age["Harris10"] = 2010
```
### Roediger13
https://ui.adsabs.harvard.edu/abs/2014ApJS..210...10R/abstract
```
Roediger13_data_dic = load_dic_csv("DataFiles/Roediger13.csv", skip=1)
Roediger13_dic = data_dic_to_gc_dic(Roediger13_data_dic)
data = data_add_function(data, Roediger13_dic, data_name='Roediger13')
paper_age["Roediger13"] = 2013
```
### OMalley17
https://ui.adsabs.harvard.edu/abs/2017ApJ...838..162O/abstract
```
OMalley17_data_dic = load_dic_csv("DataFiles/OMalley17.csv", skip=1)
OMalley17_dic = data_dic_to_gc_dic(OMalley17_data_dic)
data = data_add_function(data, OMalley17_dic, data_name='OMalley17')
paper_age["OMalley17"] = 2017
```
### Vasquez18
https://ui.adsabs.harvard.edu/abs/2018A%26A...619A..13V/abstract
Use the most recent Fe_H_D16 data?
Also contains radial velocities!
```
all_Vasquez18_data_dic = load_dic_csv("DataFiles/Vasquez18.csv", skip=1)
Vasquez18_data_dic = {p: all_Vasquez18_data_dic[op] for (p, op) in
zip(["Name", "Fe_H", "Fe_H_err"], ["Name", "Fe_H_D16", "Fe_H_D16_err"])}
Vasquez18_dic = data_dic_to_gc_dic(Vasquez18_data_dic)
data = data_add_function(data, Vasquez18_dic, data_name='Vasquez18')
paper_age["Vasquez18"] = 2018
```
### Saviane12
https://ui.adsabs.harvard.edu/abs/2012A%26A...540A..27S/abstract
Use Fe_H_C09 data?
```
all_Saviane12_data_dic = load_dic_csv("DataFiles/Saviane12.csv", skip=1)
Saviane12_data_dic = {p: all_Saviane12_data_dic[op] for (p, op) in
zip(["Name", "Name2", "Fe_H", "Fe_H_err"],
["Name", "Name2", "Fe_H_C09", "Fe_H_C09_err"])}
Saviane12_dic = data_dic_to_gc_dic(Saviane12_data_dic)
data = data_add_function(data, Saviane12_dic, data_name='Saviane12')
paper_age["Saviane12"] = 2012
```
### Oliveria20
https://ui.adsabs.harvard.edu/abs/2020ApJ...891...37O/abstract
HST
```
Oliveria20_data_dic = load_dic_csv("DataFiles/Oliveria20.csv", skip=1)
Oliveria20_dic = data_dic_to_gc_dic(Oliveria20_data_dic)
data = data_add_function(data, Oliveria20_dic, data_name='Oliveria20')
paper_age["Oliveria20"] = 2020
```
### CAPOS21
https://ui.adsabs.harvard.edu/abs/2021A%26A...652A.157G/abstract
Further Chemistry Available!
```
CAPOS21_data_dic = load_dic_csv("DataFiles/CAPOS21.csv", skip=1)
CAPOS21_dic = data_dic_to_gc_dic(CAPOS21_data_dic)
data = data_add_function(data, CAPOS21_dic, data_name='CAPOS21')
paper_age["CAPOS21"] = 2020
```
### GRAN22
Gran22: https://ui.adsabs.harvard.edu/abs/2022MNRAS.509.4962G/abstract
Errors?
Correct form of PMs?
```
Gran22_data_dic = load_dic_csv("DataFiles/Gran22.csv", skip=1)
Gran22_dic = data_dic_to_gc_dic(Gran22_data_dic)
data = data_add_function(data, Gran22_dic, data_name='Gran22')
paper_age["Gran22"] = 2020
```
### Valenti09
Colour magnitude
Paper: https://ui.adsabs.harvard.edu/abs/2010MNRAS.402.1729V/abstract
Data: http://www.bo.astro.it/~GC/ir_archive/
Compilation of Previous Valenti data - should be updated reguarly?
```
all_Valenti09_data_dic = load_dic_csv("DataFiles/Valenti09.csv", skip=1)
Valenti09_data_dic = {p: all_Valenti09_data_dic[p] for p in
["Name", "l_deg", "b_deg", "dsun", "RGC", "[Fe/H]", "[M/H]"]}
Valenti09_dic = data_dic_to_gc_dic(Valenti09_data_dic)
data = data_add_function(data, Valenti09_dic, data_name='Valenti09')
paper_age["Valenti09"] = 2009
```
### Bica16
https://ui.adsabs.harvard.edu/abs/2016PASA...33...28B/abstract
Candidate GCs with sky, Metallicity and further Chem data.
Not clear which are actually GCs or not
# Individual_Data
Individual papers and sources
```
def add_individual_data(data, individual_data):
papers = list(individual_data.keys())
for p in papers:
print(p)
original_new_data = individual_data[p]
original_names = np.array(list(original_new_data.keys()))
names = process_names(original_names)
try:
new_data = {new_n: original_new_data[old_n] for (new_n, old_n) in zip(names, original_names)}
data = data_add_function(data, new_data, data_name=p)
except Exception as e:
print(e)
print(names)
print(original_names)
print(original_new_data.keys())
return data
individual_data = {}
```
## Small Papers
### Kerber18
NGC6522, NGC6626(M28), NGC6362
Kerber18: https://ui.adsabs.harvard.edu/abs/2018ApJ...853...15K/abstract
Alpha enhanced BASTI or DSED sochrone fitting. Use DSED to avoid attomic difusion problems.
Take the smallest ChiSq.
```
individual_data["Kerber18"] = {
"NGC6522": {
"Age": 11.4, "Age_Err": 1.0,
"Fe_H": -1.15,
# "Fe_H":-2.37, "Fe_H_Err_1os":0.19,"Fe_H_Err_1us":-0.14,
"dist": 7.05, "dist_err": 0.16
},
"NGC6626": {
"Age": 11.1, "Age_Err": 0.9,
"Fe_H": -1.3,
# "Fe_H":-2.37, "Fe_H_Err_1os":0.19,"Fe_H_Err_1us":-0.14,
"dist": 5.18, "dist_err": 0.14
},
"NGC6362": {
"Age": 12.8, "Age_Err": 1.0,
"Fe_H": -1.15,
# "Fe_H":-2.37, "Fe_H_Err_1os":0.19,"Fe_H_Err_1us":-0.14,
"dist": 7.73, "dist_err": 0.18
}
}
paper_age["Kerber18"] = 2018
```
### Dias15
NGC6528, NGC6553, M71, NGC6558, NGC6426, Terzan8
Dias15: https://ui.adsabs.harvard.edu/abs/2015A%26A...573A..13D/abstract
Fe_H, Alpha_Fe, Mg, vlos
```
individual_data["Dias15"] = {
"NGC6528": {
"Fe_H": -0.13, "Fe_H_Err": 0.05,
"vlos": 185, "vlos_err": 10,
"Mg_Fe": 0.05, "Mg_Fe_Err": 0.09,
"Alpha_Fe": 0.26, "Alpha_Fe_Err": 0.05,
},
"NGC6553": {
"Fe_H": -0.133, "Fe_H_Err": 0.017,
"vlos": 6, "vlos_err": 8,
"Mg_Fe": 0.107, "Mg_Fe_Err": 0.009,
"Alpha_Fe": 0.302, "Alpha_Fe_Err": 0.025,
},
"M71": {
"Fe_H": -0.63, "Fe_H_Err": 0.05,
"vlos": -42, "vlos_err": 18,
"Mg_Fe": 0.25, "Mg_Fe_Err": 0.07,
"Alpha_Fe": 0.293, "Alpha_Fe_Err": 0.032,
},
"NGC6558": {
"Fe_H": -1.012, "Fe_H_Err": 0.013,
"vlos": -210, "vlos_err": 16,
"Mg_Fe": 0.26, "Mg_Fe_Err": 0.06,
"Alpha_Fe": 0.23, "Alpha_Fe_Err": 0.06,
},
"NGC6426": {
"Fe_H": -2.39, "Fe_H_Err": 0.11,
"vlos": -242, "vlos_err": 11,
"Mg_Fe": 0.38, "Mg_Fe_Err": 0.06,
"Alpha_Fe": 0.24, "Alpha_Fe_Err": 0.05,
},
"Terzan8": {
"Fe_H": -2.06, "Fe_H_Err": 0.17,
"vlos": 135, "vlos_err": 19,
"Mg_Fe": 0.41, "Mg_Fe_Err": 0.04,
"Alpha_Fe": 0.21, "Alpha_Fe_Err": 0.04,
}
}
paper_age["Dias15"] = 2015
```
### VandenBerg16
https://ui.adsabs.harvard.edu/abs/2016ApJ...827....2V/abstract
M3, M15, M92. More information, but not very clear?
```
individual_data["Vandenberg16"] = {
"M3": {
"Age": 12.5
},
"M15": {
"Age": 12.5
},
"M92": {
"Age": 12.5
},
}
paper_age["Vandenberg16"] = 2016
```
## Individual Clusters
### Pal2
https://ui.adsabs.harvard.edu/abs/2020MNRAS.493.2688B/abstract
```
individual_data["Bonatto20"] = {"Pal2": {'Age': 13.25, 'Age_Err': 0.12, "Fe_H": -1.58, "Fe_H_Err": 0.08,
"Mass": 1.4e5, "Mass_Err": 0.4e5, "Mvt": -7.8}}
paper_age["Bonatto20"] = 2020
```
### Crater/Lavens1
Crater
https://ui.adsabs.harvard.edu/abs/2016ApJ...822...32W/abstract
```
individual_data["Weisz16"] = {"Crater": {'Age': 7.5, 'Age_Err': 0.4,
"Fe_H": -1.66, "Fe_H_Err": 0.04,
"Mass": 9.9e3, "Mvt": -5.3}}
paper_age["Weisz16"] = 2016
```
### Bliss1
https://ui.adsabs.harvard.edu/abs/2019ApJ...875..154M/abstract
```
individual_data["Hempel14"] = {"Bliss1": {'Age': 9.2, 'Age_Err_1os': 1.6, "Age_Err_1us": -0.8,
"Fe_H": -1.4,
"Mass": 143, "Mass_Err": 37,
"Mvt": 0, "Mvt_Err_1os": 1.7, "Mvt_Err_1us": -0.7}}
paper_age["Hempel14"] = 2014
```
### Kim3
https://ui.adsabs.harvard.edu/abs/2016ApJ...820..119K/abstract
```
individual_data["Kim16"] = {"Kim3": {'Age': 9.5, 'Age_Err_1os': 3, "Age_Err_1us": -1.6,
"Fe_H": -1.6, "Fe_H_Err_1os": 0.45, "Fe_H_Err_1us": -0.3,
"Mvt": 0.7, "Mvt_Err": 0.3}}
paper_age["Kim16"] = 2016
```
### Munoz1
https://ui.adsabs.harvard.edu/abs/2012ApJ...753L..15M/abstract
```
individual_data["Munoz12"] = {"Munoz1": {'Age': 12.5,
"Fe_H": -1.5,
"Mvt": -0.4, "Mvt_Err": 0.9}}
paper_age["Munoz12"] = 2012
```
### BH176
https://ui.adsabs.harvard.edu/abs/2011A%26A...528A..70D/abstract
```
individual_data["Davoust11"] = {"BH176": {'Age': 6.5, "Age_Err": 0.5,
"Fe_H": -0.1, "Fe_H_Err": 0.1,
"Mvt": -3.82}}
paper_age["Davoust11"] = 2011
```
### FSR1716
https://ui.adsabs.harvard.edu/abs/2017A%26A...605A.128K/abstract
Age given as 10-12 in:
https://ui.adsabs.harvard.edu/abs/2016arXiv161103753B/abstract
```
individual_data["Andreas17"] = {"FSR1716": { # 'Age': ?,
"Fe_H": -1.38, "Fe_H_Err": 0.2,
"Mass": 1.4e4, "Mass_Err_1os": 1.2e4, "Mass_Err_1us": 0.8e4,
"Mvt": -5.1, "Mvt_Err": 1}}
paper_age["Andreas17"] = 2017
individual_data["Buckner16"] = {"FSR1716": {'Age': 11, "Age_Err": 1}}
paper_age["Buckner16"] = 2016
```
### Ryu 1 and 2
https://ui.adsabs.harvard.edu/abs/2018ApJ...863L..38R/abstract
Age assumed 12.6, as an average of metal poor GCs. Could include it and have very wide errors?
```
individual_data["Ryu18"] = {
"Ryu059": { # 'Age': 12.6,
"Fe_H": -2.2, "Fe_H_Err": 0.2, },
"Ryu879": { # 'Age': 12.6,
"Fe_H": -2.1, "Fe_H_Err": 0.3, }
}
paper_age["Ryu18"] = 2018
```
### NGC6139
https://ui.adsabs.harvard.edu/abs/2015A%26A...583A..69B/abstract
errors of +- 0.015 +- 0.058
Combine in Quadrature
```
individual_data["Bragaglia15"] = {"NGC6139": {
"Fe_H": -1.579, "Fe_H_Err": np.sqrt((0.015**2) + (0.058**2)),
"Mvt": -3.82}}
paper_age["Bragaglia15"] = 2015
```
### ESO452
Simpson 17
https://ui.adsabs.harvard.edu/abs/2017MNRAS.472.2856S/abstract
Cornish 06.
Fits multiple isochrones, find a large range of metallicity and ages.
https://ui.adsabs.harvard.edu/abs/2006AJ....131.2543C/abstract
```
individual_data["Simpson17"] = {"ESO452": {
"Fe_H": -0.81, "Fe_H_Err": 0.13,
"Mass": 6.8e3, "Mass_Err": 3.4e3
}
}
paper_age["Simpson17"] = 2017
individual_data["Cornish06"] = {"ESO452": {"Age": 12.5, "Age_Err": 3.5,
}
}
paper_age["Cornish06"] = 2006
```
### NGC6229
Johnson17: https://ui.adsabs.harvard.edu/abs/2017AJ....154..155J/abstract
Arlleno15: https://ui.adsabs.harvard.edu/abs/2015MNRAS.452..727A/abstract
Gives age as consistent with 12+-1
```
individual_data["Johnson17"] = {"NGC6229": {
"Fe_H": -1.13, "Fe_H_Err": 0.06,
}}
paper_age["Johnson17"] = 2017
individual_data["Arellano15"] = {"NGC6229": {"Age": 12, "Age_Err": 1,
"Fe_H": -1.31, "Fe_H_Err": 0.12,
}}
paper_age["Arellano15"] = 2015
```
### NGC6256
Cardelano20: https://ui.adsabs.harvard.edu/abs/2020ApJ...895...54C/abstract
Isochrone fitting using Vasq18 Metallicity
```
individual_data["Cardelano20"] = {"NGC6256": {
"Age": 13, "Age_Err": 0.5,
"Fe_H": -1.62
}}
paper_age["Cardelano20"] = 2020
```
### HP1
Kerber20: https://ui.adsabs.harvard.edu/abs/2019MNRAS.484.5530K/abstract
Barbuy16: https://ui.adsabs.harvard.edu/abs/2016A%26A...591A..53B/abstract
```
individual_data["Kerber20"] = {"HP1": {
"Age": 12.75, "Age_Err_1os": 0.86, "Age_Err_1us": -0.81,
"Fe_H": -1.09, "Fe_H_Err_1os": 0.07, "Fe_H_Err_1us": -0.09,
"Alpha_Fe": 0.4
}}
paper_age["Kerber20"] = 2020
individual_data["Barbuy16"] = {"HP1": {
"Fe_H": -1.06, "Fe_H_Err": 0.1,
}}
paper_age["Barbuy16"] = 2016
```
### FSR1758
Romero21: https://ui.adsabs.harvard.edu/abs/2021A%26A...652A.158R/abstract
Fits Isochrone, CAPOS APOGEE
Vilanova19: https://ui.adsabs.harvard.edu/abs/2019ApJ...882..174V/abstract
High Res Spectra. Prefered?
Barba19: https://ui.adsabs.harvard.edu/abs/2019ApJ...870L..24B/abstract
```
individual_data["Romero21"] = {"FSR1758": {
"Age": 11.6, "Age_Err_1os": 1.25, "Age_Err_1us": -1.31,
"Fe_H": -1.36, "Fe_H_Err": 0.05
}}
paper_age["Romero21"] = 2021
individual_data["Vilanova19"] = {"FSR1758": {
"Fe_H": -1.58, "Fe_H_Err": 0.03,
"Alpha_Fe": 0.32, "Alpha_Fe_Err": 0.01
}}
paper_age["Vilanova19"] = 2019
individual_data["Barba19"] = {"FSR1758": {
"Fe_H": -1.5, "Fe_H": 0.3
}}
paper_age["Barba19"] = 2019
```
### NGC6402
DAntona22: https://ui.adsabs.harvard.edu/abs/2022ApJ...925..192D/abstract
Johnson19: https://ui.adsabs.harvard.edu/abs/2019MNRAS.485.4311J/abstract
```
individual_data["DAntona22"] = {"NGC6402": {
"Age": 12.5, "Age_Err": 0.75,
}}
paper_age["DAntona22"] = 2022
individual_data["Johnson19"] = {"NGC6402": {
"Fe_H": -1.13, "Fe_H_Err": 0.05,
"Alpha_Fe": 0.3
}}
paper_age["Johnson19"] = 2019
```
### Palomar 6
Souza21: https://ui.adsabs.harvard.edu/abs/2021A%26A...656A..78S/abstract
```
individual_data["Souza21"] = {"Pal6": {
"Age": 12.4, "Age_Err": 0.9,
"Fe_H": -1.10, "Fe_H_Err": 0.09,
"Mvt": -6.79
}}
paper_age["Souza21"] = 2021
```
### NGC6440
Pallanca21: https://ui.adsabs.harvard.edu/abs/2021ApJ...913..137P/abstract
Munoz17: https://ui.adsabs.harvard.edu/abs/2017A%26A...605A..12M/abstract
Na-0 anticorrelation
```
individual_data["Pallanca21"] = {"NGC6440": {
"Age": 13, "Age_Err": 1.5,
"Fe_H": -1.34, "Fe_H_Err": 0.36,
"Mvt": 1.12, "Mvt_Err": 0.12
}}
paper_age["Pallanca21"] = 2021
individual_data["Munoz17"] = {"NGC6440": {
"Fe_H": -0.5, "Fe_H_Err": 0.03,
"Age": 13
}}
paper_age["Munoz17"] = 2017
```
### UKS1
Fernandez20: https://ui.adsabs.harvard.edu/abs/2020A%26A...643A.145F/abstract
Spectra of 6 stars, fit isochrone for age
```
individual_data["Fernandez20"] = {"UKS1": {
"Age": 13.1, "Age_Err_1os": 0.93, "Age_Err_1us": -1.29,
"Fe_H": -0.98, "Fe_H_Err": 0.11,
}}
paper_age["Fernandez20"] = 2020
```
### VVVCL001
Fernandez21: https://ui.adsabs.harvard.edu/abs/2021ApJ...908L..42F/abstract
High resolution abundance analysis + isochrone fitting
Alpha Available
```
individual_data["Fernandez21"] = {"VVVCL001": {
"Age": 11.9, "Age_Err_1os": 3.12, "Age_Err_1us": -4.05,
"Fe_H": -2.45, "Fe_H_Err": 0.24,
# "Fe_H":-2.37, "Fe_H_Err_1os":0.19,"Fe_H_Err_1us":-0.14,
"dist": 8.22, "dist_err_1os": 1.84, "dist_err_1us": -1.93
}}
paper_age["Fernandez21"] = 2020
```
### Djorg2
Ortolani19: https://ui.adsabs.harvard.edu/abs/2019A%26A...627A.145O/abstract
Isochrone fitting, BaSTi uncorrected?
```
individual_data["Ortolani19"] = {"Djorg2": {
"Age": 12.7, "Age_Err_1os": 0.72, "Age_Err_1us": -0.69,
"Fe_H": -1.11, "Fe_H_Err": 0.03,
# "Fe_H":-2.37, "Fe_H_Err_1os":0.19,"Fe_H_Err_1us":-0.14,
"dist": 8.75, "dist_err": 0.12
}}
paper_age["Ortolani19"] = 2019
```
### NGC6522
Barbuy21: https://ui.adsabs.harvard.edu/abs/2021A%26A...654A..29B/abstract
Spectra, Flames GIRAFEE
Alpha available
```
individual_data["Barbuy21"] = {"NGC6522": {
"Age": 12.7, "Age_Err_1os": 0.72, "Age_Err_1us": -0.69,
"Fe_H": -1.05, "Fe_H_Err": 0.20,
# "Fe_H":-2.37, "Fe_H_Err_1os":0.19,"Fe_H_Err_1us":-0.14,
"dist": 8.75, "dist_err": 0.12,
"vlos": -15.62, "vlos_err": 7.7
}}
paper_age["Barbuy21"] = 2021
```
### NGC6528
Munoz18: https://ui.adsabs.harvard.edu/abs/2018A%26A...620A..96M/abstract
Alpha available too.
```
individual_data["Munoz18"] = {"NGC6528": {
"Fe_H": -0.14, "Fe_H_Err": 0.03,
}}
paper_age["Munoz18"] = 2018
```
### NGC6544
Gran21: https://ui.adsabs.harvard.edu/abs/2021MNRAS.504.3494G/abstract
```
individual_data["Gran21"] = {"NGC6544": {
"Fe_H": -1.44, "Fe_H_Err": 0.04,
"Alpha_Fe": 0.2, "Alpha_Fe_Err": 0.04
}}
paper_age["Gran21"] = 2021
```
### ESO280
Simpson18: https://ui.adsabs.harvard.edu/abs/2018MNRAS.477.4565S/abstract
High Res Spectra
```
individual_data["Simpson18"] = {"ESO-SC06": {
"Fe_H": -2.48, "Fe_H_Err_1us": -0.11, "Fe_H_Err_1os": 0.06,
"Mass": 12e3, "Mass_Err": 2e3,
"Alpha_Fe": 0.2, "Alpha_Fe_Err": 0.04,
"dist": 15.2, "dist_err": 2.1,
"vlos": 92.5, "vlos_err_1os": 2.4, "vlos_err_1us": -1.6
}}
paper_age["Simpson18"] = 2018
```
### NGC6553
Munoz20: https://ui.adsabs.harvard.edu/abs/2020MNRAS.492.3742M/abstract
High Res Spectra
```
individual_data["Munoz20"] = {"NGC6553": {
"Fe_H": -0.14, "Fe_H_Err": 0.07,
"Alpha_Fe": 0.11, "Alpha_Fe_Err": 0.05
}}
paper_age["Munoz20"] = 2020
```
### NGC6558
Barbuy18: https://ui.adsabs.harvard.edu/abs/2018A%26A...619A.178B/abstract
High Res Spectra
Alpha Available
```
individual_data["Barbuy18"] = {"NGC6558": {
"Fe_H": -1.17, "Fe_H_Err": 0.10,
}}
paper_age["Barbuy18"] = 2018
```
### Valenti11
Valenti11: https://ui.adsabs.harvard.edu/abs/2011MNRAS.414.2690V/abstract
High res infrared spectra
```
individual_data["Valenti11"] = {
"NGC6624": {"Fe_H": -0.69, "Fe_H_Err": 0.02,
"Alpha_Fe": 0.39, "Alpha_Fe_Err": 0.02,
"vlos": 51, "vlos_Err": 3
},
"NGC6569": {"Fe_H": -0.79, "Fe_H_Err": 0.02,
"Alpha_Fe": 0.43, "Alpha_Fe_Err": 0.02,
"vlos": 47, "vlos_Err": 4
}
}
paper_age["Valenti11"] = 2011
```
### NGC6569
Saracino19: https://ui.adsabs.harvard.edu/abs/2019ApJ...874...86S/abstract
Age from averaged isochorne from DSED, VR, and BaSTI
```
individual_data["Saracino19"] = {"NGC6569": {
"dist": 10.1, "dist_err": 0.2,
"Age": 12.8, "Age_Err": 1,
"Fe_H": -1.17, "Fe_H_Err": 0.10,
}}
paper_age["Saracino19"] = 2019
```
### NGC6642
Balbinot10: https://ui.adsabs.harvard.edu/abs/2010IAUS..266..357B/abstract
```
individual_data["Balbinot10"] = {"NGC6642": {
"Age": 10 * 10.14, "Age_Err_1us": ((10**1.19) - (10**1.14)), "Age_Err_1us": ((10**1.14) - (10**1.05)),
"Fe_H": -1.80, "Fe_H_Err": 0.2,
"dist": 8.05, "dist_err": 0.66,
}}
paper_age["Balbinot10"] = 2010
```
### NGC6809
Rain19: https://ui.adsabs.harvard.edu/abs/2019MNRAS.483.1674R/abstract
Spectra, Chemical
Diakogiannis14: https://ui.adsabs.harvard.edu/abs/2014MNRAS.437.3172D/abstract
Dynamical Mass
```
individual_data["Rain19"] = {"NGC6809": {
"Fe_H": -2.01, "Fe_H_Err": 0.02,
"Alpha_Fe": 0.4, "Alpha_Fe_Err": 0.04,
}}
paper_age["Rain19"] = 2019
individual_data["Diakogiannis14"] = {"NGC6809": {
"Mass": 6.1e4, "Mass_Err_1os": 0.51e4, "Mass_Err_1us": -0.88e4,
}}
paper_age["Diakogiannis14"] = 2014
```
### Palomar 11
Lewis06: https://ui.adsabs.harvard.edu/abs/2006AJ....131.2538L/abstract
```
individual_data["Lewis06"] = {"Pal11": {
"Age": 11.4, "Age_Err": 0.5,
"dist": 14.3, "dist_err": 0.4,
}}
paper_age["Lewis06"] = 2006
```
### Segue3
Ortolani13: https://ui.adsabs.harvard.edu/abs/2013MNRAS.433.1966O/abstract
Hughes17: https://ui.adsabs.harvard.edu/abs/2017AJ....154...57H/abstract
Dartmouth Geneva Models. Very, very young!
```
individual_data["Ortolani13"] = {"Segue3": {
"Age": 3.2,
"Fe_H": -0.8,
}}
paper_age["Ortolani13"] = 2013
individual_data["Hughes17"] = {"Segue3": {
"Fe_H": -0.55, "Fe_H_Err_1us": -0.12, "Fe_H_Err_1os": 0.15,
"Age": 2.6, "Age_Err": 0.4,
"dist": 14.3, "dist_err": 0.4,
}}
paper_age["Hughes17"] = 2017
```
### Laevens3
Longeard19: https://ui.adsabs.harvard.edu/abs/2019MNRAS.490.1498L/abstract
```
individual_data["Longeard19"] = {"Laevens3": {
"Age": 13.0, "Age_Err": 1.0,
"dist": 61.4, "dist_err": 1.0,
"Fe_H": -1.8, "Fe_H_Err": 0.1,
"vlos": -70.2, "vlos_err": 0.5
}}
paper_age["Longeard19"] = 2019
```
### Palomar 11
Lewis06: https://ui.adsabs.harvard.edu/abs/2006AJ....131.2538L/abstract
```
individual_data["Lewis06"] = {"Pal11": {
"Age": 11.4, "Age_Err": 0.5,
"dist": 14.3, "dist_err": 0.4,
}}
paper_age["Lewis06"] = 2006
```
### NGC4833
Carretta14a: https://ui.adsabs.harvard.edu/abs/2014A%26A...564A..60C/abstract
```
individual_data["Carretta14a"] = {"NGC4833": {
"Age": 11.4, "Age_Err": 0.5,
"dist": 14.3, "dist_err": 0.4,
}}
paper_age["Carretta14a"] = 2014
```
### NGC6388
Carretta21b: https://ui.adsabs.harvard.edu/abs/2021arXiv211112721C/abstract
Metallicity only
```
# individual_data["Carretta21"] = {"NGC6388": {
# "Fe_H":-0.480,
# "Fe_H":-0.488,
# }}
# paper_age["Caretta21"] = 2021
```
### Terzan8
Carretta14b: https://ui.adsabs.harvard.edu/abs/2014A%26A...561A..87C/abstract
```
individual_data["Carretta14b"] = {"Terzan8":
{"Fe_H": -2.27, "Fe_H_Err": 0.03}
}
paper_age["Carretta14b"] = 2014
```
### Liller1
Ferraro21: https://ui.adsabs.harvard.edu/abs/2021NatAs...5..311F/abstract
Multiple populations, we use the oldest. Not clear if it is a genuine GC?
```
individual_data["Ferraro21"] = {"Liller1":
{"Fe_H": -0.4, "Alpha_Fe": 0.2,
"Age": 12, "Age_Err": 1.5,
}
}
paper_age["Ferraro21"] = 2021
```
## Add Indiviudal Data
```
data = add_individual_data(data, individual_data)
```
# List Data
```
alt_name_dic = create_alt_name_list(name_parse)
def number_props_data(data, props=[]):
names = list(data.keys())
N_total = len(names)
for p in props:
print(f"Checking Property {p}")
missing = []
for n in names:
if p not in list(data[n].keys()):
missing.append(n)
N_prop = len(missing)
print(f"{N_total - N_prop} / {N_total} have GCs have {p}")
print(f"{N_prop} / {N_total} do not have GCs have {p}")
print(missing)
print("\n")
return
number_props_data(data, props=["vlos", "dist", "Age", "Fe_H", "Mass", "Mv"])
```
# Final Data
- Sort by year for now
- Check errors coherent
- Symmetric errors vs asymmetric
### Improvements?
- If age-metallicity found together, prefer the result
- Average the result of many?
- Consistent isochrone usage?
## Find Best
```
def find_best_data(data, error_blacklist=[], alts=True):
'''Error blacklist - ignore errors for a given property'''
names = list(data.keys())
best_data = {}
best_refs = {}
for gc in names:
best_data[gc], best_refs[gc] = find_best_individual_data(data[gc], err_blacklist=error_blacklist)
if alts:
alt_names = find_alternative_names(names)
for alt_n, n in zip(alt_names, names):
best_data[n]["Name2"] = alt_n
return best_data, best_refs
def find_alternative_names(names):
alt_names = np.empty_like(names)
for i, n in enumerate(names):
alt_name = alt_name_dic.get(n, ["-"])
N_alts = len(alt_name)
if N_alts > 0:
alt_name = alt_name[0]
if alt_name == n or "":
if N_alts > 1:
alt_name = alt_name[1]
else:
alt_name = "-"
else:
alt_name = "-"
alt_names[i] = alt_name
alt_names[alt_names == ""] = "-"
return alt_names
def find_best_individual_data(gc_data, err_blacklist=[]):
'''Error blacklist - ignore errors for a given property'''
all_props = list(gc_data.keys())
props = [p for p in all_props if "err" not in p]
best_gc_data = {}
best_refs = {}
for x in props:
x_err = x + "_err"
error_condition = (x not in err_blacklist) * (x_err in all_props)
if error_condition:
papers = np.array(list(gc_data[x + "_err"].keys()))
else:
papers = np.array(list(gc_data[x].keys()))
years = np.array([paper_age[p] for p in papers])
sort_papers = papers[np.argsort(years)[::-1]]
best_paper = sort_papers[0]
best_gc_data[x] = gc_data[x][best_paper]
best_refs[x] = best_paper
if error_condition:
best_gc_data[x_err] = gc_data[x_err][best_paper]
best_refs[x_err] = best_paper
return best_gc_data, best_refs
best_gc_data, best_refs = find_best_data(data, error_blacklist=[])
best_data = gc_dic_to_data_dic(best_gc_data)
```
## List Data
```
sky_obs = ["ra_deg", "dec_deg", "dist", "dist_err", "vlos",
"vlos_err", "pmra", "pmra_err", "pmdec_err", "pmcorr"]
AMR = ["Age", "Age_err", "Fe_H", "Fe_H_err"]
Mass = ["Mass"]
Names = ["Name", "Name2"]
basic_props = Names + sky_obs + AMR + Mass
print(basic_props)
```
## Require Dynamics
```
def filter_data_dic(data_dic, need_props=sky_obs, included_props=basic_props):
nGC = len(data_dic["Name"])
filt = np.ones((nGC), dtype=bool)
for p in need_props:
filt = filt * np.isfinite(data_dic[p])
if included_props is None:
included_props = list(data_dic.keys())
filt_data_dic = {p: data_dic[p][filt] for p in included_props}
print(f"Filtered data. {filt.sum()} of {len(filt)} GCs remaining")
print("Cut GCs:")
print(data_dic["Name"][np.logical_not(filt)])
return filt_data_dic
sky_data_dic = filter_data_dic(best_data, need_props=sky_obs, included_props=basic_props)
```
# Save Data
```
fname = "CompilationGCs_SkyObs_AMR.csv"
save_dic_csv(sky_data_dic, output_File=fname)
```
|
github_jupyter
|
import numpy as np
from LoadSaveData import save_dic_csv, load_dic_csv
from ParseNames import process_names, name_parse, alt_name_parse, create_alt_name_list
from ParseProperty import property_parser, process_properties
from DataFuncs import data_add_function, data_dic_to_gc_dic, gc_dic_to_data_dic
paper_age = {}
data = {}
def load_Vasilev21_data():
Vasilev21_data_dic = load_dic_csv("DataFiles/GC_Sky_Vasilev2021.txt")
bad_filt = np.logical_not(np.isfinite(Vasilev21_data_dic["dist_err"]))
Vasilev21_data_dic["dist_err"][bad_filt] = Vasilev21_data_dic["dist"][bad_filt] * 0.05
Vasilev21_data_dic["vlos_err"] = np.maximum(Vasilev21_data_dic["vlos_err"], 2.0)
return Vasilev21_data_dic
Vasilev21_data_dic = load_Vasilev21_data()
Vasilev21_dic = data_dic_to_gc_dic(Vasilev21_data_dic)
data = data_add_function(data, Vasilev21_dic, data_name='Vasilev21')
paper_age["Vasilev21"] = 2021
def load_Baumgardt_dist_data():
Baumgardt21_data_dic = load_dic_csv("DataFiles/Baumgardt21_sky.csv", skip=2)
bad_filt = np.logical_not(np.isfinite(Baumgardt21_data_dic["dist_err"]))
Baumgardt21_data_dic["dist_err"][bad_filt] = Baumgardt21_data_dic["dist"][bad_filt] * 0.05
Baumgardt21_data_dic["vlos_err"] = np.maximum(Baumgardt21_data_dic["vlos_err"], 2.0)
return Baumgardt21_data_dic
Baumgardt21_data_dic = load_Baumgardt_dist_data()
Baumgardt21_dic = data_dic_to_gc_dic(Baumgardt21_data_dic)
data = data_add_function(data, Baumgardt21_dic, data_name='Baumgardt21')
paper_age["Baumgardt21"] = 2021.5
Baumgardt18_data_dic = load_dic_csv("DataFiles/Baumgardt_GC_Mass.csv", skip=2)
Baumgardt18_GC_dic = data_dic_to_gc_dic(Baumgardt18_data_dic)
data = data_add_function(data, Baumgardt18_GC_dic, data_name='Baumgardt_Mass18')
paper_age["Baumgardt_Mass18"] = 2018
Kruijssen19_data_dic = load_dic_csv("DataFiles/Kruijssen19.csv", skip=1)
Kruijssen19_data_dic["Mass"] = 10 ** Kruijssen19_data_dic["logM"]
del Kruijssen19_data_dic["logM"]
Kruijssen19_dic = data_dic_to_gc_dic(Kruijssen19_data_dic)
data = data_add_function(data, Kruijssen19_dic, data_name='Kruijssen19')
paper_age["Kruijssen19"] = 2013
Dotter10_data_dic = load_dic_csv("DataFiles/Dotter10.csv", skip=1)
Dotter10_dic = data_dic_to_gc_dic(Dotter10_data_dic)
data = data_add_function(data, Dotter10_dic, data_name='Dotter10')
paper_age['Dotter10'] = 2010
Forbes10_data_dic = load_dic_csv("DataFiles/Forbes10.csv", skip=1)
Forbes10_dic = data_dic_to_gc_dic(Forbes10_data_dic)
data = data_add_function(data, Forbes10_dic, data_name='Forbes10')
paper_age["Forbes10"] = 2010
VandenBerg13_data_dic = load_dic_csv("DataFiles/VandenBerg13.csv", skip=1)
print("Adding 1GYR to errors")
VandenBerg13_data_dic["Age_Err"] = VandenBerg13_data_dic["Age_Err"] + 1
VandenBerg13_dic = data_dic_to_gc_dic(VandenBerg13_data_dic)
data = data_add_function(data, VandenBerg13_dic, data_name='VandenBerg13')
paper_age["VandenBerg13"] = 2013
Harris10_data_dic = load_dic_csv("DataFiles/Harris10.csv", skip=1)
Harris10_dic = data_dic_to_gc_dic(Harris10_data_dic)
data = data_add_function(data, Harris10_dic, data_name='Harris10')
paper_age["Harris10"] = 2010
Roediger13_data_dic = load_dic_csv("DataFiles/Roediger13.csv", skip=1)
Roediger13_dic = data_dic_to_gc_dic(Roediger13_data_dic)
data = data_add_function(data, Roediger13_dic, data_name='Roediger13')
paper_age["Roediger13"] = 2013
OMalley17_data_dic = load_dic_csv("DataFiles/OMalley17.csv", skip=1)
OMalley17_dic = data_dic_to_gc_dic(OMalley17_data_dic)
data = data_add_function(data, OMalley17_dic, data_name='OMalley17')
paper_age["OMalley17"] = 2017
all_Vasquez18_data_dic = load_dic_csv("DataFiles/Vasquez18.csv", skip=1)
Vasquez18_data_dic = {p: all_Vasquez18_data_dic[op] for (p, op) in
zip(["Name", "Fe_H", "Fe_H_err"], ["Name", "Fe_H_D16", "Fe_H_D16_err"])}
Vasquez18_dic = data_dic_to_gc_dic(Vasquez18_data_dic)
data = data_add_function(data, Vasquez18_dic, data_name='Vasquez18')
paper_age["Vasquez18"] = 2018
all_Saviane12_data_dic = load_dic_csv("DataFiles/Saviane12.csv", skip=1)
Saviane12_data_dic = {p: all_Saviane12_data_dic[op] for (p, op) in
zip(["Name", "Name2", "Fe_H", "Fe_H_err"],
["Name", "Name2", "Fe_H_C09", "Fe_H_C09_err"])}
Saviane12_dic = data_dic_to_gc_dic(Saviane12_data_dic)
data = data_add_function(data, Saviane12_dic, data_name='Saviane12')
paper_age["Saviane12"] = 2012
Oliveria20_data_dic = load_dic_csv("DataFiles/Oliveria20.csv", skip=1)
Oliveria20_dic = data_dic_to_gc_dic(Oliveria20_data_dic)
data = data_add_function(data, Oliveria20_dic, data_name='Oliveria20')
paper_age["Oliveria20"] = 2020
CAPOS21_data_dic = load_dic_csv("DataFiles/CAPOS21.csv", skip=1)
CAPOS21_dic = data_dic_to_gc_dic(CAPOS21_data_dic)
data = data_add_function(data, CAPOS21_dic, data_name='CAPOS21')
paper_age["CAPOS21"] = 2020
Gran22_data_dic = load_dic_csv("DataFiles/Gran22.csv", skip=1)
Gran22_dic = data_dic_to_gc_dic(Gran22_data_dic)
data = data_add_function(data, Gran22_dic, data_name='Gran22')
paper_age["Gran22"] = 2020
all_Valenti09_data_dic = load_dic_csv("DataFiles/Valenti09.csv", skip=1)
Valenti09_data_dic = {p: all_Valenti09_data_dic[p] for p in
["Name", "l_deg", "b_deg", "dsun", "RGC", "[Fe/H]", "[M/H]"]}
Valenti09_dic = data_dic_to_gc_dic(Valenti09_data_dic)
data = data_add_function(data, Valenti09_dic, data_name='Valenti09')
paper_age["Valenti09"] = 2009
def add_individual_data(data, individual_data):
papers = list(individual_data.keys())
for p in papers:
print(p)
original_new_data = individual_data[p]
original_names = np.array(list(original_new_data.keys()))
names = process_names(original_names)
try:
new_data = {new_n: original_new_data[old_n] for (new_n, old_n) in zip(names, original_names)}
data = data_add_function(data, new_data, data_name=p)
except Exception as e:
print(e)
print(names)
print(original_names)
print(original_new_data.keys())
return data
individual_data = {}
individual_data["Kerber18"] = {
"NGC6522": {
"Age": 11.4, "Age_Err": 1.0,
"Fe_H": -1.15,
# "Fe_H":-2.37, "Fe_H_Err_1os":0.19,"Fe_H_Err_1us":-0.14,
"dist": 7.05, "dist_err": 0.16
},
"NGC6626": {
"Age": 11.1, "Age_Err": 0.9,
"Fe_H": -1.3,
# "Fe_H":-2.37, "Fe_H_Err_1os":0.19,"Fe_H_Err_1us":-0.14,
"dist": 5.18, "dist_err": 0.14
},
"NGC6362": {
"Age": 12.8, "Age_Err": 1.0,
"Fe_H": -1.15,
# "Fe_H":-2.37, "Fe_H_Err_1os":0.19,"Fe_H_Err_1us":-0.14,
"dist": 7.73, "dist_err": 0.18
}
}
paper_age["Kerber18"] = 2018
individual_data["Dias15"] = {
"NGC6528": {
"Fe_H": -0.13, "Fe_H_Err": 0.05,
"vlos": 185, "vlos_err": 10,
"Mg_Fe": 0.05, "Mg_Fe_Err": 0.09,
"Alpha_Fe": 0.26, "Alpha_Fe_Err": 0.05,
},
"NGC6553": {
"Fe_H": -0.133, "Fe_H_Err": 0.017,
"vlos": 6, "vlos_err": 8,
"Mg_Fe": 0.107, "Mg_Fe_Err": 0.009,
"Alpha_Fe": 0.302, "Alpha_Fe_Err": 0.025,
},
"M71": {
"Fe_H": -0.63, "Fe_H_Err": 0.05,
"vlos": -42, "vlos_err": 18,
"Mg_Fe": 0.25, "Mg_Fe_Err": 0.07,
"Alpha_Fe": 0.293, "Alpha_Fe_Err": 0.032,
},
"NGC6558": {
"Fe_H": -1.012, "Fe_H_Err": 0.013,
"vlos": -210, "vlos_err": 16,
"Mg_Fe": 0.26, "Mg_Fe_Err": 0.06,
"Alpha_Fe": 0.23, "Alpha_Fe_Err": 0.06,
},
"NGC6426": {
"Fe_H": -2.39, "Fe_H_Err": 0.11,
"vlos": -242, "vlos_err": 11,
"Mg_Fe": 0.38, "Mg_Fe_Err": 0.06,
"Alpha_Fe": 0.24, "Alpha_Fe_Err": 0.05,
},
"Terzan8": {
"Fe_H": -2.06, "Fe_H_Err": 0.17,
"vlos": 135, "vlos_err": 19,
"Mg_Fe": 0.41, "Mg_Fe_Err": 0.04,
"Alpha_Fe": 0.21, "Alpha_Fe_Err": 0.04,
}
}
paper_age["Dias15"] = 2015
individual_data["Vandenberg16"] = {
"M3": {
"Age": 12.5
},
"M15": {
"Age": 12.5
},
"M92": {
"Age": 12.5
},
}
paper_age["Vandenberg16"] = 2016
individual_data["Bonatto20"] = {"Pal2": {'Age': 13.25, 'Age_Err': 0.12, "Fe_H": -1.58, "Fe_H_Err": 0.08,
"Mass": 1.4e5, "Mass_Err": 0.4e5, "Mvt": -7.8}}
paper_age["Bonatto20"] = 2020
individual_data["Weisz16"] = {"Crater": {'Age': 7.5, 'Age_Err': 0.4,
"Fe_H": -1.66, "Fe_H_Err": 0.04,
"Mass": 9.9e3, "Mvt": -5.3}}
paper_age["Weisz16"] = 2016
individual_data["Hempel14"] = {"Bliss1": {'Age': 9.2, 'Age_Err_1os': 1.6, "Age_Err_1us": -0.8,
"Fe_H": -1.4,
"Mass": 143, "Mass_Err": 37,
"Mvt": 0, "Mvt_Err_1os": 1.7, "Mvt_Err_1us": -0.7}}
paper_age["Hempel14"] = 2014
individual_data["Kim16"] = {"Kim3": {'Age': 9.5, 'Age_Err_1os': 3, "Age_Err_1us": -1.6,
"Fe_H": -1.6, "Fe_H_Err_1os": 0.45, "Fe_H_Err_1us": -0.3,
"Mvt": 0.7, "Mvt_Err": 0.3}}
paper_age["Kim16"] = 2016
individual_data["Munoz12"] = {"Munoz1": {'Age': 12.5,
"Fe_H": -1.5,
"Mvt": -0.4, "Mvt_Err": 0.9}}
paper_age["Munoz12"] = 2012
individual_data["Davoust11"] = {"BH176": {'Age': 6.5, "Age_Err": 0.5,
"Fe_H": -0.1, "Fe_H_Err": 0.1,
"Mvt": -3.82}}
paper_age["Davoust11"] = 2011
individual_data["Andreas17"] = {"FSR1716": { # 'Age': ?,
"Fe_H": -1.38, "Fe_H_Err": 0.2,
"Mass": 1.4e4, "Mass_Err_1os": 1.2e4, "Mass_Err_1us": 0.8e4,
"Mvt": -5.1, "Mvt_Err": 1}}
paper_age["Andreas17"] = 2017
individual_data["Buckner16"] = {"FSR1716": {'Age': 11, "Age_Err": 1}}
paper_age["Buckner16"] = 2016
individual_data["Ryu18"] = {
"Ryu059": { # 'Age': 12.6,
"Fe_H": -2.2, "Fe_H_Err": 0.2, },
"Ryu879": { # 'Age': 12.6,
"Fe_H": -2.1, "Fe_H_Err": 0.3, }
}
paper_age["Ryu18"] = 2018
individual_data["Bragaglia15"] = {"NGC6139": {
"Fe_H": -1.579, "Fe_H_Err": np.sqrt((0.015**2) + (0.058**2)),
"Mvt": -3.82}}
paper_age["Bragaglia15"] = 2015
individual_data["Simpson17"] = {"ESO452": {
"Fe_H": -0.81, "Fe_H_Err": 0.13,
"Mass": 6.8e3, "Mass_Err": 3.4e3
}
}
paper_age["Simpson17"] = 2017
individual_data["Cornish06"] = {"ESO452": {"Age": 12.5, "Age_Err": 3.5,
}
}
paper_age["Cornish06"] = 2006
individual_data["Johnson17"] = {"NGC6229": {
"Fe_H": -1.13, "Fe_H_Err": 0.06,
}}
paper_age["Johnson17"] = 2017
individual_data["Arellano15"] = {"NGC6229": {"Age": 12, "Age_Err": 1,
"Fe_H": -1.31, "Fe_H_Err": 0.12,
}}
paper_age["Arellano15"] = 2015
individual_data["Cardelano20"] = {"NGC6256": {
"Age": 13, "Age_Err": 0.5,
"Fe_H": -1.62
}}
paper_age["Cardelano20"] = 2020
individual_data["Kerber20"] = {"HP1": {
"Age": 12.75, "Age_Err_1os": 0.86, "Age_Err_1us": -0.81,
"Fe_H": -1.09, "Fe_H_Err_1os": 0.07, "Fe_H_Err_1us": -0.09,
"Alpha_Fe": 0.4
}}
paper_age["Kerber20"] = 2020
individual_data["Barbuy16"] = {"HP1": {
"Fe_H": -1.06, "Fe_H_Err": 0.1,
}}
paper_age["Barbuy16"] = 2016
individual_data["Romero21"] = {"FSR1758": {
"Age": 11.6, "Age_Err_1os": 1.25, "Age_Err_1us": -1.31,
"Fe_H": -1.36, "Fe_H_Err": 0.05
}}
paper_age["Romero21"] = 2021
individual_data["Vilanova19"] = {"FSR1758": {
"Fe_H": -1.58, "Fe_H_Err": 0.03,
"Alpha_Fe": 0.32, "Alpha_Fe_Err": 0.01
}}
paper_age["Vilanova19"] = 2019
individual_data["Barba19"] = {"FSR1758": {
"Fe_H": -1.5, "Fe_H": 0.3
}}
paper_age["Barba19"] = 2019
individual_data["DAntona22"] = {"NGC6402": {
"Age": 12.5, "Age_Err": 0.75,
}}
paper_age["DAntona22"] = 2022
individual_data["Johnson19"] = {"NGC6402": {
"Fe_H": -1.13, "Fe_H_Err": 0.05,
"Alpha_Fe": 0.3
}}
paper_age["Johnson19"] = 2019
individual_data["Souza21"] = {"Pal6": {
"Age": 12.4, "Age_Err": 0.9,
"Fe_H": -1.10, "Fe_H_Err": 0.09,
"Mvt": -6.79
}}
paper_age["Souza21"] = 2021
individual_data["Pallanca21"] = {"NGC6440": {
"Age": 13, "Age_Err": 1.5,
"Fe_H": -1.34, "Fe_H_Err": 0.36,
"Mvt": 1.12, "Mvt_Err": 0.12
}}
paper_age["Pallanca21"] = 2021
individual_data["Munoz17"] = {"NGC6440": {
"Fe_H": -0.5, "Fe_H_Err": 0.03,
"Age": 13
}}
paper_age["Munoz17"] = 2017
individual_data["Fernandez20"] = {"UKS1": {
"Age": 13.1, "Age_Err_1os": 0.93, "Age_Err_1us": -1.29,
"Fe_H": -0.98, "Fe_H_Err": 0.11,
}}
paper_age["Fernandez20"] = 2020
individual_data["Fernandez21"] = {"VVVCL001": {
"Age": 11.9, "Age_Err_1os": 3.12, "Age_Err_1us": -4.05,
"Fe_H": -2.45, "Fe_H_Err": 0.24,
# "Fe_H":-2.37, "Fe_H_Err_1os":0.19,"Fe_H_Err_1us":-0.14,
"dist": 8.22, "dist_err_1os": 1.84, "dist_err_1us": -1.93
}}
paper_age["Fernandez21"] = 2020
individual_data["Ortolani19"] = {"Djorg2": {
"Age": 12.7, "Age_Err_1os": 0.72, "Age_Err_1us": -0.69,
"Fe_H": -1.11, "Fe_H_Err": 0.03,
# "Fe_H":-2.37, "Fe_H_Err_1os":0.19,"Fe_H_Err_1us":-0.14,
"dist": 8.75, "dist_err": 0.12
}}
paper_age["Ortolani19"] = 2019
individual_data["Barbuy21"] = {"NGC6522": {
"Age": 12.7, "Age_Err_1os": 0.72, "Age_Err_1us": -0.69,
"Fe_H": -1.05, "Fe_H_Err": 0.20,
# "Fe_H":-2.37, "Fe_H_Err_1os":0.19,"Fe_H_Err_1us":-0.14,
"dist": 8.75, "dist_err": 0.12,
"vlos": -15.62, "vlos_err": 7.7
}}
paper_age["Barbuy21"] = 2021
individual_data["Munoz18"] = {"NGC6528": {
"Fe_H": -0.14, "Fe_H_Err": 0.03,
}}
paper_age["Munoz18"] = 2018
individual_data["Gran21"] = {"NGC6544": {
"Fe_H": -1.44, "Fe_H_Err": 0.04,
"Alpha_Fe": 0.2, "Alpha_Fe_Err": 0.04
}}
paper_age["Gran21"] = 2021
individual_data["Simpson18"] = {"ESO-SC06": {
"Fe_H": -2.48, "Fe_H_Err_1us": -0.11, "Fe_H_Err_1os": 0.06,
"Mass": 12e3, "Mass_Err": 2e3,
"Alpha_Fe": 0.2, "Alpha_Fe_Err": 0.04,
"dist": 15.2, "dist_err": 2.1,
"vlos": 92.5, "vlos_err_1os": 2.4, "vlos_err_1us": -1.6
}}
paper_age["Simpson18"] = 2018
individual_data["Munoz20"] = {"NGC6553": {
"Fe_H": -0.14, "Fe_H_Err": 0.07,
"Alpha_Fe": 0.11, "Alpha_Fe_Err": 0.05
}}
paper_age["Munoz20"] = 2020
individual_data["Barbuy18"] = {"NGC6558": {
"Fe_H": -1.17, "Fe_H_Err": 0.10,
}}
paper_age["Barbuy18"] = 2018
individual_data["Valenti11"] = {
"NGC6624": {"Fe_H": -0.69, "Fe_H_Err": 0.02,
"Alpha_Fe": 0.39, "Alpha_Fe_Err": 0.02,
"vlos": 51, "vlos_Err": 3
},
"NGC6569": {"Fe_H": -0.79, "Fe_H_Err": 0.02,
"Alpha_Fe": 0.43, "Alpha_Fe_Err": 0.02,
"vlos": 47, "vlos_Err": 4
}
}
paper_age["Valenti11"] = 2011
individual_data["Saracino19"] = {"NGC6569": {
"dist": 10.1, "dist_err": 0.2,
"Age": 12.8, "Age_Err": 1,
"Fe_H": -1.17, "Fe_H_Err": 0.10,
}}
paper_age["Saracino19"] = 2019
individual_data["Balbinot10"] = {"NGC6642": {
"Age": 10 * 10.14, "Age_Err_1us": ((10**1.19) - (10**1.14)), "Age_Err_1us": ((10**1.14) - (10**1.05)),
"Fe_H": -1.80, "Fe_H_Err": 0.2,
"dist": 8.05, "dist_err": 0.66,
}}
paper_age["Balbinot10"] = 2010
individual_data["Rain19"] = {"NGC6809": {
"Fe_H": -2.01, "Fe_H_Err": 0.02,
"Alpha_Fe": 0.4, "Alpha_Fe_Err": 0.04,
}}
paper_age["Rain19"] = 2019
individual_data["Diakogiannis14"] = {"NGC6809": {
"Mass": 6.1e4, "Mass_Err_1os": 0.51e4, "Mass_Err_1us": -0.88e4,
}}
paper_age["Diakogiannis14"] = 2014
individual_data["Lewis06"] = {"Pal11": {
"Age": 11.4, "Age_Err": 0.5,
"dist": 14.3, "dist_err": 0.4,
}}
paper_age["Lewis06"] = 2006
individual_data["Ortolani13"] = {"Segue3": {
"Age": 3.2,
"Fe_H": -0.8,
}}
paper_age["Ortolani13"] = 2013
individual_data["Hughes17"] = {"Segue3": {
"Fe_H": -0.55, "Fe_H_Err_1us": -0.12, "Fe_H_Err_1os": 0.15,
"Age": 2.6, "Age_Err": 0.4,
"dist": 14.3, "dist_err": 0.4,
}}
paper_age["Hughes17"] = 2017
individual_data["Longeard19"] = {"Laevens3": {
"Age": 13.0, "Age_Err": 1.0,
"dist": 61.4, "dist_err": 1.0,
"Fe_H": -1.8, "Fe_H_Err": 0.1,
"vlos": -70.2, "vlos_err": 0.5
}}
paper_age["Longeard19"] = 2019
individual_data["Lewis06"] = {"Pal11": {
"Age": 11.4, "Age_Err": 0.5,
"dist": 14.3, "dist_err": 0.4,
}}
paper_age["Lewis06"] = 2006
individual_data["Carretta14a"] = {"NGC4833": {
"Age": 11.4, "Age_Err": 0.5,
"dist": 14.3, "dist_err": 0.4,
}}
paper_age["Carretta14a"] = 2014
# individual_data["Carretta21"] = {"NGC6388": {
# "Fe_H":-0.480,
# "Fe_H":-0.488,
# }}
# paper_age["Caretta21"] = 2021
individual_data["Carretta14b"] = {"Terzan8":
{"Fe_H": -2.27, "Fe_H_Err": 0.03}
}
paper_age["Carretta14b"] = 2014
individual_data["Ferraro21"] = {"Liller1":
{"Fe_H": -0.4, "Alpha_Fe": 0.2,
"Age": 12, "Age_Err": 1.5,
}
}
paper_age["Ferraro21"] = 2021
data = add_individual_data(data, individual_data)
alt_name_dic = create_alt_name_list(name_parse)
def number_props_data(data, props=[]):
names = list(data.keys())
N_total = len(names)
for p in props:
print(f"Checking Property {p}")
missing = []
for n in names:
if p not in list(data[n].keys()):
missing.append(n)
N_prop = len(missing)
print(f"{N_total - N_prop} / {N_total} have GCs have {p}")
print(f"{N_prop} / {N_total} do not have GCs have {p}")
print(missing)
print("\n")
return
number_props_data(data, props=["vlos", "dist", "Age", "Fe_H", "Mass", "Mv"])
def find_best_data(data, error_blacklist=[], alts=True):
'''Error blacklist - ignore errors for a given property'''
names = list(data.keys())
best_data = {}
best_refs = {}
for gc in names:
best_data[gc], best_refs[gc] = find_best_individual_data(data[gc], err_blacklist=error_blacklist)
if alts:
alt_names = find_alternative_names(names)
for alt_n, n in zip(alt_names, names):
best_data[n]["Name2"] = alt_n
return best_data, best_refs
def find_alternative_names(names):
alt_names = np.empty_like(names)
for i, n in enumerate(names):
alt_name = alt_name_dic.get(n, ["-"])
N_alts = len(alt_name)
if N_alts > 0:
alt_name = alt_name[0]
if alt_name == n or "":
if N_alts > 1:
alt_name = alt_name[1]
else:
alt_name = "-"
else:
alt_name = "-"
alt_names[i] = alt_name
alt_names[alt_names == ""] = "-"
return alt_names
def find_best_individual_data(gc_data, err_blacklist=[]):
'''Error blacklist - ignore errors for a given property'''
all_props = list(gc_data.keys())
props = [p for p in all_props if "err" not in p]
best_gc_data = {}
best_refs = {}
for x in props:
x_err = x + "_err"
error_condition = (x not in err_blacklist) * (x_err in all_props)
if error_condition:
papers = np.array(list(gc_data[x + "_err"].keys()))
else:
papers = np.array(list(gc_data[x].keys()))
years = np.array([paper_age[p] for p in papers])
sort_papers = papers[np.argsort(years)[::-1]]
best_paper = sort_papers[0]
best_gc_data[x] = gc_data[x][best_paper]
best_refs[x] = best_paper
if error_condition:
best_gc_data[x_err] = gc_data[x_err][best_paper]
best_refs[x_err] = best_paper
return best_gc_data, best_refs
best_gc_data, best_refs = find_best_data(data, error_blacklist=[])
best_data = gc_dic_to_data_dic(best_gc_data)
sky_obs = ["ra_deg", "dec_deg", "dist", "dist_err", "vlos",
"vlos_err", "pmra", "pmra_err", "pmdec_err", "pmcorr"]
AMR = ["Age", "Age_err", "Fe_H", "Fe_H_err"]
Mass = ["Mass"]
Names = ["Name", "Name2"]
basic_props = Names + sky_obs + AMR + Mass
print(basic_props)
def filter_data_dic(data_dic, need_props=sky_obs, included_props=basic_props):
nGC = len(data_dic["Name"])
filt = np.ones((nGC), dtype=bool)
for p in need_props:
filt = filt * np.isfinite(data_dic[p])
if included_props is None:
included_props = list(data_dic.keys())
filt_data_dic = {p: data_dic[p][filt] for p in included_props}
print(f"Filtered data. {filt.sum()} of {len(filt)} GCs remaining")
print("Cut GCs:")
print(data_dic["Name"][np.logical_not(filt)])
return filt_data_dic
sky_data_dic = filter_data_dic(best_data, need_props=sky_obs, included_props=basic_props)
fname = "CompilationGCs_SkyObs_AMR.csv"
save_dic_csv(sky_data_dic, output_File=fname)
| 0.368633 | 0.916708 |
# Analysing miRBase Mature for counts
Starting by loading the required libraries.
```
%pylab inline
import matplotlib_venn
import pandas
import scipy
```
Loading input table:
```
mature_counts = pandas.read_csv("mirbase_mature_counts.tsv",
sep = "\t",
header = 0)
mature_counts.head(10)
```
Calculate the number of miRNAs present on each sample.
We will consider that miRNA is present if it's norm count if >= than 1
```
# To correct this, columns will be renamed just to be sure that all is ok
mature_counts.columns = ["miRNA", "accession",
"FB", "FEF", "FH",
"MB", "MEF", "MH",
"TNB", "TNEF", "TNH",
"FB_norm", "FEF_norm", "FH_norm",
"MB_norm", "MEF_norm", "MH_norm",
"TNB_norm", "TNEF_norm", "TNH_norm"]
samples_list = ["FB", "FEF", "FH", "MB", "MEF", "MH", "TNB", "TNEF", "TNH"]
miRNAs_actives = dict()
for sample in samples_list:
sample_miRNAs_present = sum(mature_counts[sample + "_norm"] >= 10) # Changes from 1 to 10 on 2019.02.01
miRNAs_actives[sample] = sample_miRNAs_present
print(miRNAs_actives)
matplotlib.pyplot.bar(miRNAs_actives.keys(),
miRNAs_actives.values(),
color = ["#003300", "#003300", "#003300",
"#336600", "#336600", "#336600",
"#666633", "#666633", "#666633"])
```
The stage with more number of active miRNAs is always the developmental stage B.
## Lets check which miRNAs are differently present/absent
### Approach by flower type
```
mirna_list = dict()
for sample in samples_list:
mirna_list[sample] = set(mature_counts.loc[mature_counts[sample + "_norm"] >= 10]["miRNA"]) # Changes from 1 to 10 on 2019.02.01
# print(mirna_list)
venn_female = matplotlib_venn.venn3_unweighted([mirna_list["FB"], mirna_list["FEF"], mirna_list["FH"]],
set_labels = ("FB", "FEF", "FH")
)
#savefig('pmrd_madure_counts_veen_females.png')
relevant_miRNAs_female = list()
print("Exclussivos de FB:")
miRNA_list_FB = mirna_list["FB"].difference(mirna_list["FEF"], mirna_list["FH"])
relevant_miRNAs_female.extend(miRNA_list_FB)
print(sorted(miRNA_list_FB))
print("Exclussivos de FEF:")
miRNA_list_FEF = mirna_list["FEF"].difference(mirna_list["FB"], mirna_list["FH"])
relevant_miRNAs_female.extend(miRNA_list_FEF)
print(sorted(miRNA_list_FEF))
print("Exclussivos de FH:")
miRNA_list_FH = mirna_list["FH"].difference(mirna_list["FB"], mirna_list["FEF"])
relevant_miRNAs_female.extend(miRNA_list_FH)
print(sorted(miRNA_list_FH))
print("Presentes apenas em FB + FEF:")
miRNA_list_FB_FEF = mirna_list["FB"].intersection(mirna_list["FEF"]).difference(mirna_list["FH"])
relevant_miRNAs_female.extend(miRNA_list_FB_FEF)
print(sorted(miRNA_list_FB_FEF))
print("Presentes apenas em FB + FH:")
miRNA_list_FB_FH = mirna_list["FB"].intersection(mirna_list["FH"]).difference(mirna_list["FEF"])
relevant_miRNAs_female.extend(miRNA_list_FB_FH)
print(sorted(miRNA_list_FB_FH))
print("Presentes apenas em FEF + FH:")
miRNA_list_FEF_FH = mirna_list["FEF"].intersection(mirna_list["FH"]).difference(mirna_list["FB"])
relevant_miRNAs_female.extend(miRNA_list_FEF_FH)
print(sorted(miRNA_list_FEF_FH))
print("Lista de miRNAs com presença diferencial:")
relevant_miRNAs_female = sorted(set(relevant_miRNAs_female))
print(relevant_miRNAs_female)
venn_male = matplotlib_venn.venn3_unweighted([mirna_list["MB"], mirna_list["MEF"], mirna_list["MH"]],
set_labels = ("MB", "MEF", "MH")
)
#savefig('pmrd_madure_counts_veen_males.png')
relevant_miRNAs_male = list()
print("Exclussivos de MB:")
miRNA_list_MB = mirna_list["MB"].difference(mirna_list["MEF"], mirna_list["MH"])
relevant_miRNAs_male.extend(miRNA_list_MB)
print(sorted(miRNA_list_MB))
print("Exclussivos de MEF:")
miRNA_list_MEF = mirna_list["MEF"].difference(mirna_list["MB"], mirna_list["MH"])
relevant_miRNAs_male.extend(miRNA_list_MEF)
print(sorted(miRNA_list_MEF))
print("Exclussivos de MH:")
miRNA_list_MH = mirna_list["MH"].difference(mirna_list["MB"], mirna_list["MEF"])
relevant_miRNAs_male.extend(miRNA_list_MH)
print(sorted(miRNA_list_MH))
print("Presentes apenas em MB + MEF:")
miRNA_list_MB_MEF = mirna_list["MB"].intersection(mirna_list["MEF"]).difference(mirna_list["MH"])
relevant_miRNAs_male.extend(miRNA_list_MB_MEF)
print(sorted(miRNA_list_MB_MEF))
print("Presentes apenas em MB + MH:")
miRNA_list_MB_MH = mirna_list["MB"].intersection(mirna_list["MH"]).difference(mirna_list["MEF"])
relevant_miRNAs_male.extend(miRNA_list_MB_MH)
print(sorted(miRNA_list_MB_MH))
print("Presentes apenas em MEF + MH:")
miRNA_list_MEF_MH = mirna_list["MEF"].intersection(mirna_list["MH"]).difference(mirna_list["MB"])
relevant_miRNAs_male.extend(miRNA_list_MEF_MH)
print(sorted(miRNA_list_MEF_MH))
print("Lista de miRNAs com presença diferencial:")
relevant_miRNAs_male = sorted(set(relevant_miRNAs_male))
print(relevant_miRNAs_male)
venn_hermaphrodite = matplotlib_venn.venn3_unweighted([mirna_list["TNB"], mirna_list["TNEF"], mirna_list["TNH"]],
set_labels = ("TNB", "TNEF", "TNH")
)
#savefig('pmrd_madure_counts_veen_hermaphrodites.png')
relevant_miRNAs_hermaphrodite = list()
print("Exclussivos de TNB:")
miRNA_list_TNB = mirna_list["TNB"].difference(mirna_list["TNEF"], mirna_list["TNH"])
relevant_miRNAs_hermaphrodite.extend(miRNA_list_TNB)
print(sorted(miRNA_list_TNB))
print("Exclussivos de TNEF:")
miRNA_list_TNEF = mirna_list["TNEF"].difference(mirna_list["TNB"], mirna_list["TNH"])
relevant_miRNAs_hermaphrodite.extend(miRNA_list_TNEF)
print(sorted(miRNA_list_TNEF))
print("Exclussivos de TNH:")
miRNA_list_TNH = mirna_list["TNH"].difference(mirna_list["TNB"], mirna_list["TNEF"])
relevant_miRNAs_hermaphrodite.extend(miRNA_list_TNH)
print(sorted(miRNA_list_TNH))
print("Presentes apenas em TNB + TNEF:")
miRNA_list_TNB_TNEF = mirna_list["TNB"].intersection(mirna_list["TNEF"]).difference(mirna_list["TNH"])
relevant_miRNAs_hermaphrodite.extend(miRNA_list_TNB_TNEF)
print(sorted(miRNA_list_TNB_TNEF))
print("Presentes apenas em TNB + TNH:")
miRNA_list_TNB_TNH = mirna_list["TNB"].intersection(mirna_list["TNH"]).difference(mirna_list["TNEF"])
relevant_miRNAs_hermaphrodite.extend(miRNA_list_TNB_TNH)
print(sorted(miRNA_list_TNB_TNH))
print("Presentes apenas em TNEF + TNH:")
miRNA_list_TNEF_TNH = mirna_list["TNEF"].intersection(mirna_list["TNH"]).difference(mirna_list["TNB"])
relevant_miRNAs_hermaphrodite.extend(miRNA_list_TNEF_TNH)
print(sorted(miRNA_list_TNEF_TNH))
print("Lista de miRNAs com presença diferencial:")
relevant_miRNAs_hermaphrodite = sorted(set(relevant_miRNAs_hermaphrodite))
print(relevant_miRNAs_hermaphrodite)
relevant_miRNAs_by_flower_type = sorted(set(relevant_miRNAs_female
+ relevant_miRNAs_male
+ relevant_miRNAs_hermaphrodite))
print("Lista de miRNAs com presença diferencial em pelo menos um dos tipos de flor ({}):".format(len(relevant_miRNAs_by_flower_type)))
print(relevant_miRNAs_by_flower_type)
for miRNA in relevant_miRNAs_by_flower_type:
# Colect values
miRNA_norm_counts = list()
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FH_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MH_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNH_norm"]))
# Plot
dataplot = pandas.Series(miRNA_norm_counts,
index = samples_list)
dataplot.plot(kind = "bar",
title = "Frequence of " + miRNA,
color = ["#003300", "#003300", "#003300",
"#336600", "#336600", "#336600",
"#666633", "#666633", "#666633"])
threshold = pandas.Series([10, 10, 10, 10, 10, 10, 10, 10, 10],
index = samples_list)
threshold.plot(kind = "line",
color = ["#660000"])
plt.xlabel("Sample")
plt.ylabel("Normalized counts")
plt.show()
```
### Approach by developmental stage
```
venn_stage_b = matplotlib_venn.venn3_unweighted([mirna_list["FB"], mirna_list["MB"], mirna_list["TNB"]],
set_labels = ("FB", "MB", "TNB")
)
relevant_miRNAs_b = list()
print("Exclussivos de FB:")
miRNA_list_FB = mirna_list["FB"].difference(mirna_list["MB"], mirna_list["TNB"])
relevant_miRNAs_b.extend(miRNA_list_FB)
print(sorted(miRNA_list_FB))
print("Exclussivos de MB:")
miRNA_list_MB = mirna_list["MB"].difference(mirna_list["FB"], mirna_list["TNB"])
relevant_miRNAs_b.extend(miRNA_list_MB)
print(sorted(miRNA_list_MB))
print("Exclussivos de TNB:")
miRNA_list_TNB = mirna_list["TNB"].difference(mirna_list["FB"], mirna_list["MB"])
relevant_miRNAs_b.extend(miRNA_list_TNB)
print(sorted(miRNA_list_TNB))
print("Presntes apenas em FB + MB:")
miRNA_list_FB_MB = mirna_list["FB"].intersection(mirna_list["MB"]).difference(mirna_list["TNB"])
relevant_miRNAs_b.extend(miRNA_list_FB_MB)
print(sorted(miRNA_list_FB_MB))
print("Presntes apenas em FB + TNB:")
miRNA_list_FB_TNB = mirna_list["FB"].intersection(mirna_list["TNB"]).difference(mirna_list["MB"])
relevant_miRNAs_b.extend(miRNA_list_FB_TNB)
print(sorted(miRNA_list_FB_TNB))
print("Presntes apenas em MB + TNB:")
miRNA_list_MB_TNB = mirna_list["MB"].intersection(mirna_list["TNB"]).difference(mirna_list["FB"])
relevant_miRNAs_b.extend(miRNA_list_MB_TNB)
print(sorted(miRNA_list_MB_TNB))
print("Lista de miRNAs com presença diferencial:")
relevant_miRNAs_b = sorted(set(relevant_miRNAs_b))
print(relevant_miRNAs_b)
venn_stage_ef = matplotlib_venn.venn3_unweighted([mirna_list["FEF"], mirna_list["MEF"], mirna_list["TNEF"]],
set_labels = ("FEF", "MEF", "TNEF")
)
relevant_miRNAs_ef = list()
print("Exclussivos de FEF:")
miRNA_list_FEF = mirna_list["FEF"].difference(mirna_list["MEF"], mirna_list["TNEF"])
relevant_miRNAs_ef.extend(miRNA_list_FEF)
print(sorted(miRNA_list_FEF))
print("Exclussivos de MEF:")
miRNA_list_MEF = mirna_list["MEF"].difference(mirna_list["FEF"], mirna_list["TNEF"])
relevant_miRNAs_ef.extend(miRNA_list_MEF)
print(sorted(miRNA_list_MEF))
print("Exclussivos de TNEF:")
miRNA_list_TNEF = mirna_list["TNEF"].difference(mirna_list["FEF"], mirna_list["MEF"])
relevant_miRNAs_ef.extend(miRNA_list_TNEF)
print(sorted(miRNA_list_TNEF))
print("Presentes apenas em FEF + MEF:")
miRNA_list_FEF_MEF = mirna_list["FEF"].intersection(mirna_list["MEF"]).difference(mirna_list["TNEF"])
relevant_miRNAs_ef.extend(miRNA_list_FEF_MEF)
print(sorted(miRNA_list_FEF_MEF))
print("Presentes apenas em FEF + TNEF:")
miRNA_list_FEF_TNEF = mirna_list["FEF"].intersection(mirna_list["TNEF"]).difference(mirna_list["MEF"])
relevant_miRNAs_ef.extend(miRNA_list_FEF_TNEF)
print(sorted(miRNA_list_FEF_TNEF))
print("Presentes apenas em MEF + TNEF:")
miRNA_list_MEF_TNEF = mirna_list["MEF"].intersection(mirna_list["TNEF"]).difference(mirna_list["FEF"])
relevant_miRNAs_ef.extend(miRNA_list_MEF_TNEF)
print(sorted(miRNA_list_MEF_TNEF))
print("Lista de miRNAs com presença diferencial:")
relevant_miRNAs_ef = sorted(set(relevant_miRNAs_ef))
print(relevant_miRNAs_ef)
venn_stage_h = matplotlib_venn.venn3_unweighted([mirna_list["FH"], mirna_list["MH"], mirna_list["TNH"]],
set_labels = ("FH", "MH", "TNH")
)
relevant_miRNAs_h = list()
print("Exclussivos de FH:")
miRNA_list_FH = mirna_list["FH"].difference(mirna_list["MH"], mirna_list["TNH"])
relevant_miRNAs_h.extend(miRNA_list_FH)
print(sorted(miRNA_list_FH))
print("Exclussivos de MH:")
miRNA_list_MH = mirna_list["MH"].difference(mirna_list["FH"], mirna_list["TNH"])
relevant_miRNAs_h.extend(miRNA_list_MH)
print(sorted(miRNA_list_MH))
print("Exclussivos de TNH:")
miRNA_list_TNH = mirna_list["TNH"].difference(mirna_list["FH"], mirna_list["MH"])
relevant_miRNAs_h.extend(miRNA_list_TNH)
print(sorted(miRNA_list_TNH))
print("Presentes apenas em FH + MH:")
miRNA_list_FH_MH = mirna_list["FH"].intersection(mirna_list["MH"]).difference(mirna_list["TNH"])
relevant_miRNAs_h.extend(miRNA_list_FH_MH)
print(sorted(miRNA_list_FH_MH))
print("Presentes apenas em FH + TNH:")
miRNA_list_FH_TNH = mirna_list["FH"].intersection(mirna_list["TNH"]).difference(mirna_list["MH"])
relevant_miRNAs_h.extend(miRNA_list_FH_TNH)
print(sorted(miRNA_list_FH_TNH))
print("Presentes apenas em MH + TNH:")
miRNA_list_MH_TNH = mirna_list["MH"].intersection(mirna_list["TNH"]).difference(mirna_list["FH"])
relevant_miRNAs_h.extend(miRNA_list_MH_TNH)
print(sorted(miRNA_list_MH_TNH))
print("Lista de miRNAs com presença diferencial:")
relevant_miRNAs_h = sorted(set(relevant_miRNAs_h))
print(relevant_miRNAs_h)
relevant_miRNAs_by_developmental_stage = sorted(set(relevant_miRNAs_b
+ relevant_miRNAs_ef
+ relevant_miRNAs_h))
print("Lista de miRNAs com presença diferencial em pelo menos um dos estágios de desenvolvimento ({}):".format(len(relevant_miRNAs_by_developmental_stage)))
print(relevant_miRNAs_by_developmental_stage)
for miRNA in relevant_miRNAs_by_developmental_stage:
# Colect values
miRNA_norm_counts = list()
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FH_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MH_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNH_norm"]))
# Plot
dataplot = pandas.Series(miRNA_norm_counts,
index = samples_list)
dataplot.plot(kind = "bar",
title = "Frequence of " + miRNA,
color = ["#003300", "#003300", "#003300",
"#336600", "#336600", "#336600",
"#666633", "#666633", "#666633"])
threshold = pandas.Series([10, 10, 10, 10, 10, 10, 10, 10, 10],
index = samples_list)
threshold.plot(kind = "line",
color = ["#660000"])
plt.xlabel("Sample")
plt.ylabel("Normalized counts")
plt.show()
```
### Lista de miRNAs relevantes independentemente de onde vêm
```
relevant_miRNAs_all = sorted(set(relevant_miRNAs_by_developmental_stage
+ relevant_miRNAs_by_flower_type))
print("Lista de miRNAs com presença diferencial geral ({}):".format(len(relevant_miRNAs_all)))
print(relevant_miRNAs_all)
for miRNA in relevant_miRNAs_all:
# Colect values
miRNA_norm_counts = list()
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FH_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MH_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNH_norm"]))
# Plot
dataplot = pandas.Series(miRNA_norm_counts,
index = samples_list)
dataplot.plot(kind = "bar",
title = "Frequence of " + miRNA,
color = ["#003300", "#003300", "#003300",
"#336600", "#336600", "#336600",
"#666633", "#666633", "#666633"])
threshold = pandas.Series([10, 10, 10, 10, 10, 10, 10, 10, 10],
index = samples_list)
threshold.plot(kind = "line",
color = ["#660000"])
plt.xlabel("Sample")
plt.ylabel("Normalized counts")
plt.show()
relevant_miRNAs_values = list()
for miRNA in relevant_miRNAs_all:
# Colect values
miRNA_norm_counts = list()
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FH_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MH_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNH_norm"]))
relevant_miRNAs_values.append(miRNA_norm_counts)
plt.figure(figsize = (5, 10))
plt.pcolor(relevant_miRNAs_values)
plt.yticks(np.arange(0.5, len(relevant_miRNAs_all), 1), relevant_miRNAs_all)
plt.xticks(numpy.arange(0.5, len(samples_list), 1), labels = samples_list)
colorbar()
plt.show()
relevant_miRNAs = pandas.DataFrame.from_records(relevant_miRNAs_values,
index = relevant_miRNAs_all,
columns = samples_list)
relevant_miRNAs
# This list comes from differential expressed genes
differential_expressed = ['vvi-miR156f', 'vvi-miR160c', 'vvi-miR167c', 'vvi-miR169a', 'vvi-miR171g', 'vvi-miR172d', 'vvi-miR319e', 'vvi-miR3623-3p', 'vvi-miR3624-3p', 'vvi-miR3625-5p', 'vvi-miR3626-3p', 'vvi-miR3627-5p', 'vvi-miR3632-5p', 'vvi-miR3633b-3p', 'vvi-miR3634-3p', 'vvi-miR3637-3p', 'vvi-miR3640-5p', 'vvi-miR395a', 'vvi-miR396a', 'vvi-miR396b', 'vvi-miR396d', 'vvi-miR398a', 'vvi-miR399a', 'vvi-miR399b']
# List miRNAs found by both strategies
mirnas_both = sorted(set(relevant_miRNAs.index).intersection(differential_expressed))
print("There are {} miRNAs indentified on both methods.".format(len(mirnas_both)))
mirnas_both_values = list()
mirnas_counts = mature_counts
for miRNA in mirnas_both:
# Colect values
miRNA_norm_counts = list()
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FH_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MH_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNH_norm"]))
mirnas_both_values.append(miRNA_norm_counts)
mirnas_both_expression = pandas.DataFrame.from_records(mirnas_both_values,
index = mirnas_both,
columns = samples_list)
mirnas_both_expression
# List miRNAs found only by presence/absence
mirnas_only_counts = sorted(set(relevant_miRNAs.index).difference(differential_expressed))
print("There are {} miRNAs indentified only on presence/absence.".format(len(mirnas_only_counts)))
mirnas_only_counts_values = list()
for miRNA in mirnas_only_counts:
# Colect values
miRNA_norm_counts = list()
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FH_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MH_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNH_norm"]))
mirnas_only_counts_values.append(miRNA_norm_counts)
mirnas_only_counts_expression = pandas.DataFrame.from_records(mirnas_only_counts_values,
index = mirnas_only_counts,
columns = samples_list)
mirnas_only_counts_expression
# List miRNAs found only by differential expression
mirnas_only_differential_expressed = sorted(set(differential_expressed).difference(relevant_miRNAs.index))
print("There are {} miRNAs indentified only on differential expression.".format(len(mirnas_only_differential_expressed)))
mirnas_only_differential_expressed_values = list()
for miRNA in mirnas_only_differential_expressed:
# Colect values
miRNA_norm_counts = list()
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FH_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MH_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNH_norm"]))
mirnas_only_differential_expressed_values.append(miRNA_norm_counts)
mirnas_only_differential_expressed_expression = pandas.DataFrame.from_records(mirnas_only_differential_expressed_values,
index = mirnas_only_differential_expressed,
columns = samples_list)
mirnas_only_differential_expressed_expression
```
|
github_jupyter
|
%pylab inline
import matplotlib_venn
import pandas
import scipy
mature_counts = pandas.read_csv("mirbase_mature_counts.tsv",
sep = "\t",
header = 0)
mature_counts.head(10)
# To correct this, columns will be renamed just to be sure that all is ok
mature_counts.columns = ["miRNA", "accession",
"FB", "FEF", "FH",
"MB", "MEF", "MH",
"TNB", "TNEF", "TNH",
"FB_norm", "FEF_norm", "FH_norm",
"MB_norm", "MEF_norm", "MH_norm",
"TNB_norm", "TNEF_norm", "TNH_norm"]
samples_list = ["FB", "FEF", "FH", "MB", "MEF", "MH", "TNB", "TNEF", "TNH"]
miRNAs_actives = dict()
for sample in samples_list:
sample_miRNAs_present = sum(mature_counts[sample + "_norm"] >= 10) # Changes from 1 to 10 on 2019.02.01
miRNAs_actives[sample] = sample_miRNAs_present
print(miRNAs_actives)
matplotlib.pyplot.bar(miRNAs_actives.keys(),
miRNAs_actives.values(),
color = ["#003300", "#003300", "#003300",
"#336600", "#336600", "#336600",
"#666633", "#666633", "#666633"])
mirna_list = dict()
for sample in samples_list:
mirna_list[sample] = set(mature_counts.loc[mature_counts[sample + "_norm"] >= 10]["miRNA"]) # Changes from 1 to 10 on 2019.02.01
# print(mirna_list)
venn_female = matplotlib_venn.venn3_unweighted([mirna_list["FB"], mirna_list["FEF"], mirna_list["FH"]],
set_labels = ("FB", "FEF", "FH")
)
#savefig('pmrd_madure_counts_veen_females.png')
relevant_miRNAs_female = list()
print("Exclussivos de FB:")
miRNA_list_FB = mirna_list["FB"].difference(mirna_list["FEF"], mirna_list["FH"])
relevant_miRNAs_female.extend(miRNA_list_FB)
print(sorted(miRNA_list_FB))
print("Exclussivos de FEF:")
miRNA_list_FEF = mirna_list["FEF"].difference(mirna_list["FB"], mirna_list["FH"])
relevant_miRNAs_female.extend(miRNA_list_FEF)
print(sorted(miRNA_list_FEF))
print("Exclussivos de FH:")
miRNA_list_FH = mirna_list["FH"].difference(mirna_list["FB"], mirna_list["FEF"])
relevant_miRNAs_female.extend(miRNA_list_FH)
print(sorted(miRNA_list_FH))
print("Presentes apenas em FB + FEF:")
miRNA_list_FB_FEF = mirna_list["FB"].intersection(mirna_list["FEF"]).difference(mirna_list["FH"])
relevant_miRNAs_female.extend(miRNA_list_FB_FEF)
print(sorted(miRNA_list_FB_FEF))
print("Presentes apenas em FB + FH:")
miRNA_list_FB_FH = mirna_list["FB"].intersection(mirna_list["FH"]).difference(mirna_list["FEF"])
relevant_miRNAs_female.extend(miRNA_list_FB_FH)
print(sorted(miRNA_list_FB_FH))
print("Presentes apenas em FEF + FH:")
miRNA_list_FEF_FH = mirna_list["FEF"].intersection(mirna_list["FH"]).difference(mirna_list["FB"])
relevant_miRNAs_female.extend(miRNA_list_FEF_FH)
print(sorted(miRNA_list_FEF_FH))
print("Lista de miRNAs com presença diferencial:")
relevant_miRNAs_female = sorted(set(relevant_miRNAs_female))
print(relevant_miRNAs_female)
venn_male = matplotlib_venn.venn3_unweighted([mirna_list["MB"], mirna_list["MEF"], mirna_list["MH"]],
set_labels = ("MB", "MEF", "MH")
)
#savefig('pmrd_madure_counts_veen_males.png')
relevant_miRNAs_male = list()
print("Exclussivos de MB:")
miRNA_list_MB = mirna_list["MB"].difference(mirna_list["MEF"], mirna_list["MH"])
relevant_miRNAs_male.extend(miRNA_list_MB)
print(sorted(miRNA_list_MB))
print("Exclussivos de MEF:")
miRNA_list_MEF = mirna_list["MEF"].difference(mirna_list["MB"], mirna_list["MH"])
relevant_miRNAs_male.extend(miRNA_list_MEF)
print(sorted(miRNA_list_MEF))
print("Exclussivos de MH:")
miRNA_list_MH = mirna_list["MH"].difference(mirna_list["MB"], mirna_list["MEF"])
relevant_miRNAs_male.extend(miRNA_list_MH)
print(sorted(miRNA_list_MH))
print("Presentes apenas em MB + MEF:")
miRNA_list_MB_MEF = mirna_list["MB"].intersection(mirna_list["MEF"]).difference(mirna_list["MH"])
relevant_miRNAs_male.extend(miRNA_list_MB_MEF)
print(sorted(miRNA_list_MB_MEF))
print("Presentes apenas em MB + MH:")
miRNA_list_MB_MH = mirna_list["MB"].intersection(mirna_list["MH"]).difference(mirna_list["MEF"])
relevant_miRNAs_male.extend(miRNA_list_MB_MH)
print(sorted(miRNA_list_MB_MH))
print("Presentes apenas em MEF + MH:")
miRNA_list_MEF_MH = mirna_list["MEF"].intersection(mirna_list["MH"]).difference(mirna_list["MB"])
relevant_miRNAs_male.extend(miRNA_list_MEF_MH)
print(sorted(miRNA_list_MEF_MH))
print("Lista de miRNAs com presença diferencial:")
relevant_miRNAs_male = sorted(set(relevant_miRNAs_male))
print(relevant_miRNAs_male)
venn_hermaphrodite = matplotlib_venn.venn3_unweighted([mirna_list["TNB"], mirna_list["TNEF"], mirna_list["TNH"]],
set_labels = ("TNB", "TNEF", "TNH")
)
#savefig('pmrd_madure_counts_veen_hermaphrodites.png')
relevant_miRNAs_hermaphrodite = list()
print("Exclussivos de TNB:")
miRNA_list_TNB = mirna_list["TNB"].difference(mirna_list["TNEF"], mirna_list["TNH"])
relevant_miRNAs_hermaphrodite.extend(miRNA_list_TNB)
print(sorted(miRNA_list_TNB))
print("Exclussivos de TNEF:")
miRNA_list_TNEF = mirna_list["TNEF"].difference(mirna_list["TNB"], mirna_list["TNH"])
relevant_miRNAs_hermaphrodite.extend(miRNA_list_TNEF)
print(sorted(miRNA_list_TNEF))
print("Exclussivos de TNH:")
miRNA_list_TNH = mirna_list["TNH"].difference(mirna_list["TNB"], mirna_list["TNEF"])
relevant_miRNAs_hermaphrodite.extend(miRNA_list_TNH)
print(sorted(miRNA_list_TNH))
print("Presentes apenas em TNB + TNEF:")
miRNA_list_TNB_TNEF = mirna_list["TNB"].intersection(mirna_list["TNEF"]).difference(mirna_list["TNH"])
relevant_miRNAs_hermaphrodite.extend(miRNA_list_TNB_TNEF)
print(sorted(miRNA_list_TNB_TNEF))
print("Presentes apenas em TNB + TNH:")
miRNA_list_TNB_TNH = mirna_list["TNB"].intersection(mirna_list["TNH"]).difference(mirna_list["TNEF"])
relevant_miRNAs_hermaphrodite.extend(miRNA_list_TNB_TNH)
print(sorted(miRNA_list_TNB_TNH))
print("Presentes apenas em TNEF + TNH:")
miRNA_list_TNEF_TNH = mirna_list["TNEF"].intersection(mirna_list["TNH"]).difference(mirna_list["TNB"])
relevant_miRNAs_hermaphrodite.extend(miRNA_list_TNEF_TNH)
print(sorted(miRNA_list_TNEF_TNH))
print("Lista de miRNAs com presença diferencial:")
relevant_miRNAs_hermaphrodite = sorted(set(relevant_miRNAs_hermaphrodite))
print(relevant_miRNAs_hermaphrodite)
relevant_miRNAs_by_flower_type = sorted(set(relevant_miRNAs_female
+ relevant_miRNAs_male
+ relevant_miRNAs_hermaphrodite))
print("Lista de miRNAs com presença diferencial em pelo menos um dos tipos de flor ({}):".format(len(relevant_miRNAs_by_flower_type)))
print(relevant_miRNAs_by_flower_type)
for miRNA in relevant_miRNAs_by_flower_type:
# Colect values
miRNA_norm_counts = list()
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FH_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MH_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNH_norm"]))
# Plot
dataplot = pandas.Series(miRNA_norm_counts,
index = samples_list)
dataplot.plot(kind = "bar",
title = "Frequence of " + miRNA,
color = ["#003300", "#003300", "#003300",
"#336600", "#336600", "#336600",
"#666633", "#666633", "#666633"])
threshold = pandas.Series([10, 10, 10, 10, 10, 10, 10, 10, 10],
index = samples_list)
threshold.plot(kind = "line",
color = ["#660000"])
plt.xlabel("Sample")
plt.ylabel("Normalized counts")
plt.show()
venn_stage_b = matplotlib_venn.venn3_unweighted([mirna_list["FB"], mirna_list["MB"], mirna_list["TNB"]],
set_labels = ("FB", "MB", "TNB")
)
relevant_miRNAs_b = list()
print("Exclussivos de FB:")
miRNA_list_FB = mirna_list["FB"].difference(mirna_list["MB"], mirna_list["TNB"])
relevant_miRNAs_b.extend(miRNA_list_FB)
print(sorted(miRNA_list_FB))
print("Exclussivos de MB:")
miRNA_list_MB = mirna_list["MB"].difference(mirna_list["FB"], mirna_list["TNB"])
relevant_miRNAs_b.extend(miRNA_list_MB)
print(sorted(miRNA_list_MB))
print("Exclussivos de TNB:")
miRNA_list_TNB = mirna_list["TNB"].difference(mirna_list["FB"], mirna_list["MB"])
relevant_miRNAs_b.extend(miRNA_list_TNB)
print(sorted(miRNA_list_TNB))
print("Presntes apenas em FB + MB:")
miRNA_list_FB_MB = mirna_list["FB"].intersection(mirna_list["MB"]).difference(mirna_list["TNB"])
relevant_miRNAs_b.extend(miRNA_list_FB_MB)
print(sorted(miRNA_list_FB_MB))
print("Presntes apenas em FB + TNB:")
miRNA_list_FB_TNB = mirna_list["FB"].intersection(mirna_list["TNB"]).difference(mirna_list["MB"])
relevant_miRNAs_b.extend(miRNA_list_FB_TNB)
print(sorted(miRNA_list_FB_TNB))
print("Presntes apenas em MB + TNB:")
miRNA_list_MB_TNB = mirna_list["MB"].intersection(mirna_list["TNB"]).difference(mirna_list["FB"])
relevant_miRNAs_b.extend(miRNA_list_MB_TNB)
print(sorted(miRNA_list_MB_TNB))
print("Lista de miRNAs com presença diferencial:")
relevant_miRNAs_b = sorted(set(relevant_miRNAs_b))
print(relevant_miRNAs_b)
venn_stage_ef = matplotlib_venn.venn3_unweighted([mirna_list["FEF"], mirna_list["MEF"], mirna_list["TNEF"]],
set_labels = ("FEF", "MEF", "TNEF")
)
relevant_miRNAs_ef = list()
print("Exclussivos de FEF:")
miRNA_list_FEF = mirna_list["FEF"].difference(mirna_list["MEF"], mirna_list["TNEF"])
relevant_miRNAs_ef.extend(miRNA_list_FEF)
print(sorted(miRNA_list_FEF))
print("Exclussivos de MEF:")
miRNA_list_MEF = mirna_list["MEF"].difference(mirna_list["FEF"], mirna_list["TNEF"])
relevant_miRNAs_ef.extend(miRNA_list_MEF)
print(sorted(miRNA_list_MEF))
print("Exclussivos de TNEF:")
miRNA_list_TNEF = mirna_list["TNEF"].difference(mirna_list["FEF"], mirna_list["MEF"])
relevant_miRNAs_ef.extend(miRNA_list_TNEF)
print(sorted(miRNA_list_TNEF))
print("Presentes apenas em FEF + MEF:")
miRNA_list_FEF_MEF = mirna_list["FEF"].intersection(mirna_list["MEF"]).difference(mirna_list["TNEF"])
relevant_miRNAs_ef.extend(miRNA_list_FEF_MEF)
print(sorted(miRNA_list_FEF_MEF))
print("Presentes apenas em FEF + TNEF:")
miRNA_list_FEF_TNEF = mirna_list["FEF"].intersection(mirna_list["TNEF"]).difference(mirna_list["MEF"])
relevant_miRNAs_ef.extend(miRNA_list_FEF_TNEF)
print(sorted(miRNA_list_FEF_TNEF))
print("Presentes apenas em MEF + TNEF:")
miRNA_list_MEF_TNEF = mirna_list["MEF"].intersection(mirna_list["TNEF"]).difference(mirna_list["FEF"])
relevant_miRNAs_ef.extend(miRNA_list_MEF_TNEF)
print(sorted(miRNA_list_MEF_TNEF))
print("Lista de miRNAs com presença diferencial:")
relevant_miRNAs_ef = sorted(set(relevant_miRNAs_ef))
print(relevant_miRNAs_ef)
venn_stage_h = matplotlib_venn.venn3_unweighted([mirna_list["FH"], mirna_list["MH"], mirna_list["TNH"]],
set_labels = ("FH", "MH", "TNH")
)
relevant_miRNAs_h = list()
print("Exclussivos de FH:")
miRNA_list_FH = mirna_list["FH"].difference(mirna_list["MH"], mirna_list["TNH"])
relevant_miRNAs_h.extend(miRNA_list_FH)
print(sorted(miRNA_list_FH))
print("Exclussivos de MH:")
miRNA_list_MH = mirna_list["MH"].difference(mirna_list["FH"], mirna_list["TNH"])
relevant_miRNAs_h.extend(miRNA_list_MH)
print(sorted(miRNA_list_MH))
print("Exclussivos de TNH:")
miRNA_list_TNH = mirna_list["TNH"].difference(mirna_list["FH"], mirna_list["MH"])
relevant_miRNAs_h.extend(miRNA_list_TNH)
print(sorted(miRNA_list_TNH))
print("Presentes apenas em FH + MH:")
miRNA_list_FH_MH = mirna_list["FH"].intersection(mirna_list["MH"]).difference(mirna_list["TNH"])
relevant_miRNAs_h.extend(miRNA_list_FH_MH)
print(sorted(miRNA_list_FH_MH))
print("Presentes apenas em FH + TNH:")
miRNA_list_FH_TNH = mirna_list["FH"].intersection(mirna_list["TNH"]).difference(mirna_list["MH"])
relevant_miRNAs_h.extend(miRNA_list_FH_TNH)
print(sorted(miRNA_list_FH_TNH))
print("Presentes apenas em MH + TNH:")
miRNA_list_MH_TNH = mirna_list["MH"].intersection(mirna_list["TNH"]).difference(mirna_list["FH"])
relevant_miRNAs_h.extend(miRNA_list_MH_TNH)
print(sorted(miRNA_list_MH_TNH))
print("Lista de miRNAs com presença diferencial:")
relevant_miRNAs_h = sorted(set(relevant_miRNAs_h))
print(relevant_miRNAs_h)
relevant_miRNAs_by_developmental_stage = sorted(set(relevant_miRNAs_b
+ relevant_miRNAs_ef
+ relevant_miRNAs_h))
print("Lista de miRNAs com presença diferencial em pelo menos um dos estágios de desenvolvimento ({}):".format(len(relevant_miRNAs_by_developmental_stage)))
print(relevant_miRNAs_by_developmental_stage)
for miRNA in relevant_miRNAs_by_developmental_stage:
# Colect values
miRNA_norm_counts = list()
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FH_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MH_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNH_norm"]))
# Plot
dataplot = pandas.Series(miRNA_norm_counts,
index = samples_list)
dataplot.plot(kind = "bar",
title = "Frequence of " + miRNA,
color = ["#003300", "#003300", "#003300",
"#336600", "#336600", "#336600",
"#666633", "#666633", "#666633"])
threshold = pandas.Series([10, 10, 10, 10, 10, 10, 10, 10, 10],
index = samples_list)
threshold.plot(kind = "line",
color = ["#660000"])
plt.xlabel("Sample")
plt.ylabel("Normalized counts")
plt.show()
relevant_miRNAs_all = sorted(set(relevant_miRNAs_by_developmental_stage
+ relevant_miRNAs_by_flower_type))
print("Lista de miRNAs com presença diferencial geral ({}):".format(len(relevant_miRNAs_all)))
print(relevant_miRNAs_all)
for miRNA in relevant_miRNAs_all:
# Colect values
miRNA_norm_counts = list()
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FH_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MH_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNH_norm"]))
# Plot
dataplot = pandas.Series(miRNA_norm_counts,
index = samples_list)
dataplot.plot(kind = "bar",
title = "Frequence of " + miRNA,
color = ["#003300", "#003300", "#003300",
"#336600", "#336600", "#336600",
"#666633", "#666633", "#666633"])
threshold = pandas.Series([10, 10, 10, 10, 10, 10, 10, 10, 10],
index = samples_list)
threshold.plot(kind = "line",
color = ["#660000"])
plt.xlabel("Sample")
plt.ylabel("Normalized counts")
plt.show()
relevant_miRNAs_values = list()
for miRNA in relevant_miRNAs_all:
# Colect values
miRNA_norm_counts = list()
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["FH_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["MH_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNB_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNEF_norm"]))
miRNA_norm_counts.extend(set(mature_counts[mature_counts["miRNA"] == miRNA]["TNH_norm"]))
relevant_miRNAs_values.append(miRNA_norm_counts)
plt.figure(figsize = (5, 10))
plt.pcolor(relevant_miRNAs_values)
plt.yticks(np.arange(0.5, len(relevant_miRNAs_all), 1), relevant_miRNAs_all)
plt.xticks(numpy.arange(0.5, len(samples_list), 1), labels = samples_list)
colorbar()
plt.show()
relevant_miRNAs = pandas.DataFrame.from_records(relevant_miRNAs_values,
index = relevant_miRNAs_all,
columns = samples_list)
relevant_miRNAs
# This list comes from differential expressed genes
differential_expressed = ['vvi-miR156f', 'vvi-miR160c', 'vvi-miR167c', 'vvi-miR169a', 'vvi-miR171g', 'vvi-miR172d', 'vvi-miR319e', 'vvi-miR3623-3p', 'vvi-miR3624-3p', 'vvi-miR3625-5p', 'vvi-miR3626-3p', 'vvi-miR3627-5p', 'vvi-miR3632-5p', 'vvi-miR3633b-3p', 'vvi-miR3634-3p', 'vvi-miR3637-3p', 'vvi-miR3640-5p', 'vvi-miR395a', 'vvi-miR396a', 'vvi-miR396b', 'vvi-miR396d', 'vvi-miR398a', 'vvi-miR399a', 'vvi-miR399b']
# List miRNAs found by both strategies
mirnas_both = sorted(set(relevant_miRNAs.index).intersection(differential_expressed))
print("There are {} miRNAs indentified on both methods.".format(len(mirnas_both)))
mirnas_both_values = list()
mirnas_counts = mature_counts
for miRNA in mirnas_both:
# Colect values
miRNA_norm_counts = list()
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FH_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MH_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNH_norm"]))
mirnas_both_values.append(miRNA_norm_counts)
mirnas_both_expression = pandas.DataFrame.from_records(mirnas_both_values,
index = mirnas_both,
columns = samples_list)
mirnas_both_expression
# List miRNAs found only by presence/absence
mirnas_only_counts = sorted(set(relevant_miRNAs.index).difference(differential_expressed))
print("There are {} miRNAs indentified only on presence/absence.".format(len(mirnas_only_counts)))
mirnas_only_counts_values = list()
for miRNA in mirnas_only_counts:
# Colect values
miRNA_norm_counts = list()
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FH_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MH_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNH_norm"]))
mirnas_only_counts_values.append(miRNA_norm_counts)
mirnas_only_counts_expression = pandas.DataFrame.from_records(mirnas_only_counts_values,
index = mirnas_only_counts,
columns = samples_list)
mirnas_only_counts_expression
# List miRNAs found only by differential expression
mirnas_only_differential_expressed = sorted(set(differential_expressed).difference(relevant_miRNAs.index))
print("There are {} miRNAs indentified only on differential expression.".format(len(mirnas_only_differential_expressed)))
mirnas_only_differential_expressed_values = list()
for miRNA in mirnas_only_differential_expressed:
# Colect values
miRNA_norm_counts = list()
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["FH_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["MH_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNB_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNEF_norm"]))
miRNA_norm_counts.extend(set(mirnas_counts[mirnas_counts["miRNA"] == miRNA]["TNH_norm"]))
mirnas_only_differential_expressed_values.append(miRNA_norm_counts)
mirnas_only_differential_expressed_expression = pandas.DataFrame.from_records(mirnas_only_differential_expressed_values,
index = mirnas_only_differential_expressed,
columns = samples_list)
mirnas_only_differential_expressed_expression
| 0.119524 | 0.648578 |
```
import openpyxl
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from helpers import sum_features_per_software, keep_top_software
df = pd.read_excel('data/orig_df.xlsx', engine='openpyxl')
classes_dict = {
'retrieval':['search', 'import', 'expertaddition', 'fulltextpdf'],
'appraisal': ['tiabscreen', 'distinctscreen', 'dualscreen',
'wordhighlight', 'screeninautomation', 'deduplication'],
'extraction': ['tag', 'extract', 'dualextract', 'evidencemap', 'riskofbias'],
'output':['flowdiagram', 'writing', 'visualization', 'export'],
'admin':['protocol', 'userroles', 'monitor', 'comments', 'training','support'],
'access': ['free', 'multipleusers', 'living', 'publicoutputs']
}
classes_pallettes = {
'retrieval':['BuGn'],
'appraisal': ['BuPu'],
'extraction': ['BuGn'],
'output':['BrBG'],
'admin':['BuPu'],
'access': ['BuGn']
}
captions = []
fig_num = 2
for k, v in classes_dict.items(): # For each feature class
feature_key = k
feature_value = classes_dict[k] # Store the column/variable names in a list
if 'name' not in feature_value:
feature_value.append('name') # Add the variable 'name'
print("CLASS NAME:", feature_key)
print("list:", feature_value)
ftr_df = df[df.columns[df.columns.isin(feature_value)]]
# Caculate summary statistics
ftr_df = sum_features_per_software(ftr_df, 2)
# Keep the top 8 softwares
ftr_df = keep_top_software(ftr_df, "percent_of_features", 10)
# Simplify names of main variables
name = ftr_df['name']
percent = ftr_df['percent_of_features']
number = ftr_df['number_of_features']
# Plot
color = classes_pallettes[k][0]
print("COLOR: ", color)
plt.figure(figsize=(14, 6))
ax = sns.barplot(x=name, y=number, data=ftr_df, palette=color)
ax.set_ylabel('Number of Features', fontsize=14)
ax.set_xlabel('SR Software', fontsize=14) # Set the title to be the feature key
ax.set_title(feature_key+' features', fontsize=18)
plt.show()
# PRINT OUT CAPTIONS
fign = str(fig_num)
fign = fign+"-"+feature_key
fig_caption = "fig "+str(fig_num)+". "+"Top ten systematic review softwares that support "+feature_key+' features.'
captions.append(fig_caption)
print("FIG CAPTION: ", fig_caption)
# SAVE EACH FIGURE
ax.get_figure().savefig('output/fig'+fign+'.png')
fig_num = fig_num+1
#captions.append("fig 1. "+"Percent of Features for each SR Software (with a min of 5 non-NA features)")
#plt.savefig('fig1)+iteration+.png'
#fig++
# For each class, graph percent
# graph_p_by_feature(ftr_df, feature_class)
with open('output/captions.txt', 'w') as f:
for line in captions:
f.write(line)
f.write('\n')
```
|
github_jupyter
|
import openpyxl
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from helpers import sum_features_per_software, keep_top_software
df = pd.read_excel('data/orig_df.xlsx', engine='openpyxl')
classes_dict = {
'retrieval':['search', 'import', 'expertaddition', 'fulltextpdf'],
'appraisal': ['tiabscreen', 'distinctscreen', 'dualscreen',
'wordhighlight', 'screeninautomation', 'deduplication'],
'extraction': ['tag', 'extract', 'dualextract', 'evidencemap', 'riskofbias'],
'output':['flowdiagram', 'writing', 'visualization', 'export'],
'admin':['protocol', 'userroles', 'monitor', 'comments', 'training','support'],
'access': ['free', 'multipleusers', 'living', 'publicoutputs']
}
classes_pallettes = {
'retrieval':['BuGn'],
'appraisal': ['BuPu'],
'extraction': ['BuGn'],
'output':['BrBG'],
'admin':['BuPu'],
'access': ['BuGn']
}
captions = []
fig_num = 2
for k, v in classes_dict.items(): # For each feature class
feature_key = k
feature_value = classes_dict[k] # Store the column/variable names in a list
if 'name' not in feature_value:
feature_value.append('name') # Add the variable 'name'
print("CLASS NAME:", feature_key)
print("list:", feature_value)
ftr_df = df[df.columns[df.columns.isin(feature_value)]]
# Caculate summary statistics
ftr_df = sum_features_per_software(ftr_df, 2)
# Keep the top 8 softwares
ftr_df = keep_top_software(ftr_df, "percent_of_features", 10)
# Simplify names of main variables
name = ftr_df['name']
percent = ftr_df['percent_of_features']
number = ftr_df['number_of_features']
# Plot
color = classes_pallettes[k][0]
print("COLOR: ", color)
plt.figure(figsize=(14, 6))
ax = sns.barplot(x=name, y=number, data=ftr_df, palette=color)
ax.set_ylabel('Number of Features', fontsize=14)
ax.set_xlabel('SR Software', fontsize=14) # Set the title to be the feature key
ax.set_title(feature_key+' features', fontsize=18)
plt.show()
# PRINT OUT CAPTIONS
fign = str(fig_num)
fign = fign+"-"+feature_key
fig_caption = "fig "+str(fig_num)+". "+"Top ten systematic review softwares that support "+feature_key+' features.'
captions.append(fig_caption)
print("FIG CAPTION: ", fig_caption)
# SAVE EACH FIGURE
ax.get_figure().savefig('output/fig'+fign+'.png')
fig_num = fig_num+1
#captions.append("fig 1. "+"Percent of Features for each SR Software (with a min of 5 non-NA features)")
#plt.savefig('fig1)+iteration+.png'
#fig++
# For each class, graph percent
# graph_p_by_feature(ftr_df, feature_class)
with open('output/captions.txt', 'w') as f:
for line in captions:
f.write(line)
f.write('\n')
| 0.33764 | 0.342778 |
# A Data Science Case Study with Python: part 4
Make sure you are familiar with Chapters 1 & 2 of Nathan George's book [Practical Data Science with
Python](https://www.packtpub.com/product/practical-data-science-with-python/9781801071970) before diving into this notebook.
We work with the City of Montreal 311 data set. This is the third notebook in a sequence; please start with the first part if you landed here via some other route. regardless, we will reload the data in the first cells.
```
import os.path
filename = 'requetes311.csv'
if os.path.isfile(filename): # file exists
print('We have the data at hand.')
else: # and what to do if not
print('ERROR: Please download the data and put it in this same directory')
```
Now, we load the data into a Pandas data frame and see what we got in terms of rows and columns. Since this is a rather large file, this might take a while. Be patient.
```
import pandas as pd
print('Attempting to load the data. (Do NOT click run again, this takes some time.)')
data = pd.read_csv(filename, low_memory = False)
print('Good to go :)')
```
## Statistical relationships between numerical columns
Let's get a refresher on what columns the data set contains.
```
rows, cols = data.shape
for (c, i) in zip(data.columns, range(cols)): # iterate over the contents of the header with a for loop
print(f'Column at index {i} is called {c}')
```
Let's also look at a random sample of the rows to get an idea of what the columns contain.
```
data.sample(6)
```
We have not done anything with those `PROVENANCE` columns yet that indicate the ways in which the incident in question has been reported. A quick reminder as to what they contain:
```
data.tail()
prov = [ col for col in data.columns if 'PROVENANCE' in col ] # which columns are these
print(prov)
from collections import Counter
c = Counter(data[prov[0]])
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = [ 20, 15 ] # bigger plots
font = { 'size' : 20 } # bigger font
plt.rc('font', **font)
patches, texts = plt.pie(c.values())
plt.legend(patches, c.keys(), loc = 'lower right',
bbox_to_anchor = (-0.1, 1.), fontsize = 18)
```
Note that `nan` refers to *no valid value given*.
The first `PROVENANCE` column indicates how the record was first created. The others are counters on the number of times the record was reported through different media.
```
prov = prov[1:] # examine just the numerical ones
from scipy.stats import describe
pd.options.display.float_format = '{:.2f}'.format # no scientific format, two decimal places
for col in prov:
print(data[col].describe())
print('\n\n')
```
Let's figure out if the numeric ones are correlated.
```
origin = data[prov[1:]] # keep just the numeric ones so that we get a smaller dataframe
origin.head()
print(origin.shape)
```
Four million and then some might be a bit much to analyze. Maybe we could just focus on reports *created* during the year 2019.
```
data['timestamp'] = pd.to_datetime(data['DDS_DATE_CREATION'], infer_datetime_format = True)
year = data[data['timestamp'].dt.year == 2019]
origin = year[prov[1:]] # keep just the numeric ones so that we get a smaller dataframe
print(origin.shape)
```
This makes us a smaller dataframe with only the columns with the source counters and the rows that correspond to records created in 2019. The column names are a bit redundantly long now: we can get rid of that repeating `PROVENANCE_` in them.
```
shorter = { p : p.replace('PROVENANCE_', '') for p in prov }
print(shorter)
origin = origin.rename(columns = shorter)
origin.head()
```
The rows that only have zeroes are of no interest, though.
```
content = origin.loc[~(origin == 0).all(axis = 1)]
print(content.shape)
content.head()
```
Hey, these seem to be all one or zero. Let's examine that assumption further.
```
print(content.sum(axis = 1).value_counts())
```
So, they are not **all** just one source, but most are. Then again, we know that some of the record identifiers in the column `ID_UNIQUE` repeated so we may wish to combine the counters that share the same ID.
```
subset = year[['ID_UNIQUE'] + prov[1:]] # keep the ID
print('Kept columns', subset.columns)
print('Shape before combining', subset.shape)
combined = subset.groupby('ID_UNIQUE').sum()
print('Shape after combining', combined.shape)
```
Much fewer now that they were combined. We still want to get rid of the ones that only contain zeroes in the counter columns.
```
counters = combined[prov[1:]]
noblank = counters.loc[~(counters == 0).all(axis = 1)] # remove blanks
print('Shape after pruning blanks', noblank.shape)
print(noblank.sum(axis = 1).value_counts())
```
The records that only have *one report* are not potential sources of correlations between reporting channels and they are a majority, so let's prune those out as well to pursue our goal.
```
sources = noblank.sum(axis = 1)
multi = noblank.loc[sources != 1] # remove singletons
print(multi.sum(axis = 1).value_counts())
```
Now we can move on with figuring out whether reporting through specific pairs of channels has some relations.
We compute and place the correlations with an [auxiliary function](https://stackoverflow.com/a/50835066).
```
multi = multi.rename(columns = shorter)
font = { 'size' : 12 }
plt.rc('font', **font)
import seaborn as sns # install with pip if you have not done so yet
sns.set(style = 'dark')
from scipy.stats import pearsonr
def pcor(x, y, ax = None, **kws):
r, _ = pearsonr(x, y)
r *= 100 # percentage
ax = ax or plt.gca()
ax.annotate(f'{r:.0f} %',
xy = (0.5, 0.5), xycoords = 'axes fraction',
ha = 'center', va = 'center',
color = 'black', size = 20)
info = sns.PairGrid(multi)
info.map_upper(pcor) # add correlations above the diagonal
info.map_lower(sns.scatterplot) # scatter plots below the diagonal
info.map_diag(sns.histplot) # histograms on the diagonal
```
Not a lot going on here in terms of correlations. The amount ofreports made in person per incident negatively correlate with the amount of reports made on mobile or online, but it is not a very strong linear relation.
What if we want to know which types of reports these are? We have to go back and retain the column `NATURE`.
Do all the records corresponding to one same ID share the value of the value of `NATURE`, though?
```
s2 = year[['ID_UNIQUE', 'NATURE'] + prov[1:]] # keep also the kind of the report
print('Examination commences')
for label, members in s2.groupby('ID_UNIQUE'):
kinds = set(members['NATURE'])
k = len(kinds)
if k > 1:
print(f'ID {label} corresponds to {k} types:', ' '.join(kinds))
print('Examination finished')
```
At least during 2019 there were no cases of multiple values of `NATURE` associated to any unique ID.
```
whatToDo = dict.fromkeys(s2, 'sum') # we want to add up the counters
whatToDo['NATURE'] = 'first' # that does not apply to the kind, keep the first (assuming this is invariant)
s3 = s2.groupby('ID_UNIQUE').agg(whatToDo)
s3 = s3.drop(columns = 'ID_UNIQUE') # we do not need this anymore
s3.head()
```
Remember how everything tends to be a `Requete`?
```
from collections import Counter
c = Counter(s3['NATURE'])
patches, texts = plt.pie(c.values())
plt.legend(patches, c.keys(), loc = 'lower right',
bbox_to_anchor = (-0.1, 1.), fontsize = 8)
```
Let's just draw the two underdog categories so that we can actually see them. But now, since we have this strong filter, we can stop filtering by year and just grab it all.
```
underdogs = data.drop(data[(data.NATURE == 'Requete')].index)
part = underdogs[['ID_UNIQUE', 'NATURE'] + prov[1:]]
unique = part.groupby('ID_UNIQUE').agg(whatToDo)
unique.drop(columns = 'ID_UNIQUE', inplace = True)
print(unique.shape)
twoOrMore = unique[prov[1:]].sum(axis = 1) # we still need to get rid of reports with zero or one source counts
kept = unique.loc[twoOrMore > 1] # at least two reports
print(kept.shape)
shorter['NATURE'] = 'kind'
kept = kept.rename(columns = shorter)
font = { 'size' : 12 }
plt.rc('font', **font)
sns.set(style = 'dark')
sns.pairplot(kept, hue = 'kind', kind = 'scatter', diag_kind = 'hist',
diag_kws = {'hue': None, 'color': 'green'},
plot_kws = {'alpha': 0.9, 's': 50, 'edgecolor': 'white'})
```
When the histogram is just one fat bar, it is because there is only one value in that distribution.
This dataset does not have a lot of numerical raw data, but it does not mean we cannot obtain meaningful numerical data from it. Remember how each record has two dates: creation date `DDS_DATE_CREATION` and date of the latest update `DATE_DERNIER_STATUT`. Let's take a look at the status field `DERNIER_STATUT`.
```
c = Counter(data['DERNIER_STATUT'])
plt.rcParams['figure.figsize'] = [ 20, 15 ] # bigger plots
font = { 'size' : 20 } # bigger font
plt.rc('font', **font)
patches, texts = plt.pie(c.values())
plt.legend(patches, c.keys(), loc = 'lower right',
bbox_to_anchor = (-0.1, 1.), fontsize = 18)
```
We could obtain the processing times for the ones that have a value other than `nan`, but we have to remember to get rid of the situations in which the same unique ID appears in multiple records. Let's keep the longest processing time for each unique ID.
```
print(data.shape)
status = data.loc[data.DERNIER_STATUT.notnull()]
print(status.shape)
c = Counter(status['DERNIER_STATUT'])
patches, texts = plt.pie(c.values())
plt.legend(patches, c.keys(), loc = 'lower right',
bbox_to_anchor = (-0.1, 1.), fontsize = 18)
data.start = pd.to_datetime(data['DDS_DATE_CREATION'], infer_datetime_format = True)
data.finish = pd.to_datetime(data['DATE_DERNIER_STATUT'], infer_datetime_format = True)
data['duration'] = data.finish - data.start
status = data.loc[data.DERNIER_STATUT.notnull()] # just the ones that have a final state
print(status.duration.describe())
```
Guessing that zero duration means that nothing was actually done. How many of records are there that were open for less than a minute and what is the status in those records?
```
from datetime import timedelta
quick = status.loc[ status.duration < timedelta(minutes = 1) ]
print(quick.shape)
c = Counter(quick['DERNIER_STATUT'])
patches, texts = plt.pie(c.values())
plt.legend(patches, c.keys(), loc = 'lower right',
bbox_to_anchor = (-0.1, 1.), fontsize = 18)
print(quick.duration.describe())
```
A quarter took 11 second or more. Let's try with a 15-second threshold instead.
```
instant = status.loc[ status.duration < timedelta(seconds = 15) ]
print(instant.shape)
c = Counter(instant['DERNIER_STATUT'])
patches, texts = plt.pie(c.values())
plt.legend(patches, c.keys(), loc = 'lower right',
bbox_to_anchor = (-0.1, 1.), fontsize = 18)
print(instant.duration.describe())
```
Now a quarter of them are seven seconds or longer. Maybe a five-second threshold?
```
immediate = status.loc[ status.duration < timedelta(seconds = 5) ]
print(immediate.shape)
c = Counter(immediate['DERNIER_STATUT'])
patches, texts = plt.pie(c.values())
plt.legend(patches, c.keys(), loc = 'lower right',
bbox_to_anchor = (-0.1, 1.), fontsize = 18)
print(immediate.duration.describe())
```
That should be fine: we can assume that anything that required human involvement takes at least three seconds.
We *mustn't* forget that we need to get rid of records that correspond to the same unique ID before we try to analyze the distribution of the durations so that incidents with multiple records are not accounted for several times, introducing a **bias** in our conclusions. Additionally, all of this duration-related processing is easier if we make a field that contains the number in seconds that corresponds to the duration.
```
data['seconds'] = data.duration.astype('timedelta64[s]')
smaller = data[['ID_UNIQUE', 'seconds' ]] # a smaller dataframe with just these two columns
print('Shape before pruning', smaller.shape)
highest = smaller.groupby('ID_UNIQUE').max() # keep the highest duration for each ID
print('Shape after pruning duplicates', highest.shape)
```
We can safely assume that this will be a scale-free distribution: most reports are processed quickly, some take a very long time. Hence, we should use a doubly logarithmic scale.
```
filtered = highest.loc[ highest.seconds > 3 ]
x = filtered.seconds
plt.rcParams['figure.figsize'] = [ 12, 12 ]
font = { 'size' : 40 }
plt.rc('font', **font)
hist, bins, _ = plt.hist(x, bins = 10)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('Duration in seconds')
plt.ylabel('Number of reports')
plt.show()
```
Heh, maybe with bins that *look* the same width?
```
import numpy as np
lb = np.logspace(np.log10(bins[0]), np.log10(bins[-1]), len(bins))
plt.hist(x, bins = lb)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('Duration in seconds')
plt.ylabel('Number of reports')
plt.show()
```
Could we use horizontal tic marks that aren't expressed as powers of ten but instead in units that make sense to humans like "minute", "hour", "day"? That would make more sense with a *cumulative* histogram showing the *relative* frequency of duration.
```
import numpy as np
minute = 60 # seconds
hour = 60 * minute
day = 24 * hour
week = 7 * day
month = 30 * day # keep it simple
semester = 6 * month
year = 356 * day
low = x.min()
high = x.max()
assert high > semester
sb = [ low, minute, hour, day, week, month, semester, year, high ]
fig, ax = plt.subplots(1, 1)
ax.hist(x, bins = sb, density = True, cumulative = True)
ax.set_xscale('log')
ax.set_yscale('log')
ax.set_xlabel('Duration in seconds')
ax.set_ylabel('Number of reports')
xpos = [ (2 * start + end) / 3 for start, end in zip(sb[:-1], sb[1:]) ] # intermediate points
ax.set_xticks(xpos)
ax.set_xticklabels([ 'less than a minute', 'less than an hour', 'less than a day',
'less than a week', 'less than a month', 'less than two months',
'less than half a year', 'any duration' ],
ha = 'center', rotation = 90)
ypos = np.linspace(0.1, 1.0, 10)
ax.set_yticks(ypos)
ax.set_yticklabels([ f'{v:.1f}' for v in ypos ])
plt.grid()
plt.show()
```
Now we can interpret. Only a few percent are receive a status in less than a minute (the shortest bar), and then a bunch get done within the hour (more than 10 %), whereas nearly 30 % are done within a day, meaning that over a fifth in total took between an hour and day. Within a week, 60 % are dealt with. So 30 % take more than one day but less than a full week. 20 % are completed in over a week but less than a month. About 15% in more than one month but less than six months. A small fraction takes over half a year to get a status.
|
github_jupyter
|
import os.path
filename = 'requetes311.csv'
if os.path.isfile(filename): # file exists
print('We have the data at hand.')
else: # and what to do if not
print('ERROR: Please download the data and put it in this same directory')
import pandas as pd
print('Attempting to load the data. (Do NOT click run again, this takes some time.)')
data = pd.read_csv(filename, low_memory = False)
print('Good to go :)')
rows, cols = data.shape
for (c, i) in zip(data.columns, range(cols)): # iterate over the contents of the header with a for loop
print(f'Column at index {i} is called {c}')
data.sample(6)
data.tail()
prov = [ col for col in data.columns if 'PROVENANCE' in col ] # which columns are these
print(prov)
from collections import Counter
c = Counter(data[prov[0]])
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = [ 20, 15 ] # bigger plots
font = { 'size' : 20 } # bigger font
plt.rc('font', **font)
patches, texts = plt.pie(c.values())
plt.legend(patches, c.keys(), loc = 'lower right',
bbox_to_anchor = (-0.1, 1.), fontsize = 18)
prov = prov[1:] # examine just the numerical ones
from scipy.stats import describe
pd.options.display.float_format = '{:.2f}'.format # no scientific format, two decimal places
for col in prov:
print(data[col].describe())
print('\n\n')
origin = data[prov[1:]] # keep just the numeric ones so that we get a smaller dataframe
origin.head()
print(origin.shape)
data['timestamp'] = pd.to_datetime(data['DDS_DATE_CREATION'], infer_datetime_format = True)
year = data[data['timestamp'].dt.year == 2019]
origin = year[prov[1:]] # keep just the numeric ones so that we get a smaller dataframe
print(origin.shape)
shorter = { p : p.replace('PROVENANCE_', '') for p in prov }
print(shorter)
origin = origin.rename(columns = shorter)
origin.head()
content = origin.loc[~(origin == 0).all(axis = 1)]
print(content.shape)
content.head()
print(content.sum(axis = 1).value_counts())
subset = year[['ID_UNIQUE'] + prov[1:]] # keep the ID
print('Kept columns', subset.columns)
print('Shape before combining', subset.shape)
combined = subset.groupby('ID_UNIQUE').sum()
print('Shape after combining', combined.shape)
counters = combined[prov[1:]]
noblank = counters.loc[~(counters == 0).all(axis = 1)] # remove blanks
print('Shape after pruning blanks', noblank.shape)
print(noblank.sum(axis = 1).value_counts())
sources = noblank.sum(axis = 1)
multi = noblank.loc[sources != 1] # remove singletons
print(multi.sum(axis = 1).value_counts())
multi = multi.rename(columns = shorter)
font = { 'size' : 12 }
plt.rc('font', **font)
import seaborn as sns # install with pip if you have not done so yet
sns.set(style = 'dark')
from scipy.stats import pearsonr
def pcor(x, y, ax = None, **kws):
r, _ = pearsonr(x, y)
r *= 100 # percentage
ax = ax or plt.gca()
ax.annotate(f'{r:.0f} %',
xy = (0.5, 0.5), xycoords = 'axes fraction',
ha = 'center', va = 'center',
color = 'black', size = 20)
info = sns.PairGrid(multi)
info.map_upper(pcor) # add correlations above the diagonal
info.map_lower(sns.scatterplot) # scatter plots below the diagonal
info.map_diag(sns.histplot) # histograms on the diagonal
s2 = year[['ID_UNIQUE', 'NATURE'] + prov[1:]] # keep also the kind of the report
print('Examination commences')
for label, members in s2.groupby('ID_UNIQUE'):
kinds = set(members['NATURE'])
k = len(kinds)
if k > 1:
print(f'ID {label} corresponds to {k} types:', ' '.join(kinds))
print('Examination finished')
whatToDo = dict.fromkeys(s2, 'sum') # we want to add up the counters
whatToDo['NATURE'] = 'first' # that does not apply to the kind, keep the first (assuming this is invariant)
s3 = s2.groupby('ID_UNIQUE').agg(whatToDo)
s3 = s3.drop(columns = 'ID_UNIQUE') # we do not need this anymore
s3.head()
from collections import Counter
c = Counter(s3['NATURE'])
patches, texts = plt.pie(c.values())
plt.legend(patches, c.keys(), loc = 'lower right',
bbox_to_anchor = (-0.1, 1.), fontsize = 8)
underdogs = data.drop(data[(data.NATURE == 'Requete')].index)
part = underdogs[['ID_UNIQUE', 'NATURE'] + prov[1:]]
unique = part.groupby('ID_UNIQUE').agg(whatToDo)
unique.drop(columns = 'ID_UNIQUE', inplace = True)
print(unique.shape)
twoOrMore = unique[prov[1:]].sum(axis = 1) # we still need to get rid of reports with zero or one source counts
kept = unique.loc[twoOrMore > 1] # at least two reports
print(kept.shape)
shorter['NATURE'] = 'kind'
kept = kept.rename(columns = shorter)
font = { 'size' : 12 }
plt.rc('font', **font)
sns.set(style = 'dark')
sns.pairplot(kept, hue = 'kind', kind = 'scatter', diag_kind = 'hist',
diag_kws = {'hue': None, 'color': 'green'},
plot_kws = {'alpha': 0.9, 's': 50, 'edgecolor': 'white'})
c = Counter(data['DERNIER_STATUT'])
plt.rcParams['figure.figsize'] = [ 20, 15 ] # bigger plots
font = { 'size' : 20 } # bigger font
plt.rc('font', **font)
patches, texts = plt.pie(c.values())
plt.legend(patches, c.keys(), loc = 'lower right',
bbox_to_anchor = (-0.1, 1.), fontsize = 18)
print(data.shape)
status = data.loc[data.DERNIER_STATUT.notnull()]
print(status.shape)
c = Counter(status['DERNIER_STATUT'])
patches, texts = plt.pie(c.values())
plt.legend(patches, c.keys(), loc = 'lower right',
bbox_to_anchor = (-0.1, 1.), fontsize = 18)
data.start = pd.to_datetime(data['DDS_DATE_CREATION'], infer_datetime_format = True)
data.finish = pd.to_datetime(data['DATE_DERNIER_STATUT'], infer_datetime_format = True)
data['duration'] = data.finish - data.start
status = data.loc[data.DERNIER_STATUT.notnull()] # just the ones that have a final state
print(status.duration.describe())
from datetime import timedelta
quick = status.loc[ status.duration < timedelta(minutes = 1) ]
print(quick.shape)
c = Counter(quick['DERNIER_STATUT'])
patches, texts = plt.pie(c.values())
plt.legend(patches, c.keys(), loc = 'lower right',
bbox_to_anchor = (-0.1, 1.), fontsize = 18)
print(quick.duration.describe())
instant = status.loc[ status.duration < timedelta(seconds = 15) ]
print(instant.shape)
c = Counter(instant['DERNIER_STATUT'])
patches, texts = plt.pie(c.values())
plt.legend(patches, c.keys(), loc = 'lower right',
bbox_to_anchor = (-0.1, 1.), fontsize = 18)
print(instant.duration.describe())
immediate = status.loc[ status.duration < timedelta(seconds = 5) ]
print(immediate.shape)
c = Counter(immediate['DERNIER_STATUT'])
patches, texts = plt.pie(c.values())
plt.legend(patches, c.keys(), loc = 'lower right',
bbox_to_anchor = (-0.1, 1.), fontsize = 18)
print(immediate.duration.describe())
data['seconds'] = data.duration.astype('timedelta64[s]')
smaller = data[['ID_UNIQUE', 'seconds' ]] # a smaller dataframe with just these two columns
print('Shape before pruning', smaller.shape)
highest = smaller.groupby('ID_UNIQUE').max() # keep the highest duration for each ID
print('Shape after pruning duplicates', highest.shape)
filtered = highest.loc[ highest.seconds > 3 ]
x = filtered.seconds
plt.rcParams['figure.figsize'] = [ 12, 12 ]
font = { 'size' : 40 }
plt.rc('font', **font)
hist, bins, _ = plt.hist(x, bins = 10)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('Duration in seconds')
plt.ylabel('Number of reports')
plt.show()
import numpy as np
lb = np.logspace(np.log10(bins[0]), np.log10(bins[-1]), len(bins))
plt.hist(x, bins = lb)
plt.xscale('log')
plt.yscale('log')
plt.xlabel('Duration in seconds')
plt.ylabel('Number of reports')
plt.show()
import numpy as np
minute = 60 # seconds
hour = 60 * minute
day = 24 * hour
week = 7 * day
month = 30 * day # keep it simple
semester = 6 * month
year = 356 * day
low = x.min()
high = x.max()
assert high > semester
sb = [ low, minute, hour, day, week, month, semester, year, high ]
fig, ax = plt.subplots(1, 1)
ax.hist(x, bins = sb, density = True, cumulative = True)
ax.set_xscale('log')
ax.set_yscale('log')
ax.set_xlabel('Duration in seconds')
ax.set_ylabel('Number of reports')
xpos = [ (2 * start + end) / 3 for start, end in zip(sb[:-1], sb[1:]) ] # intermediate points
ax.set_xticks(xpos)
ax.set_xticklabels([ 'less than a minute', 'less than an hour', 'less than a day',
'less than a week', 'less than a month', 'less than two months',
'less than half a year', 'any duration' ],
ha = 'center', rotation = 90)
ypos = np.linspace(0.1, 1.0, 10)
ax.set_yticks(ypos)
ax.set_yticklabels([ f'{v:.1f}' for v in ypos ])
plt.grid()
plt.show()
| 0.320609 | 0.98366 |
# Pandas
Source: https://github.com/jdhp-docs/notebooks/blob/master/python_pandas_en.ipynb
<a href="https://colab.research.google.com/github/jdhp-docs/notebooks/blob/master/python_pandas_en.ipynb"><img align="left" src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open in Colab" title="Open and Execute in Google Colaboratory"></a>
<a href="https://mybinder.org/v2/gh/jdhp-docs/notebooks/master?filepath=python_pandas_en.ipynb"><img align="left" src="https://mybinder.org/badge.svg" alt="Open in Binder" title="Open and Execute in Binder"></a>
Official documentation: http://pandas.pydata.org/pandas-docs/stable/
Getting started:
* https://pandas.pydata.org/pandas-docs/stable/10min.html
* https://pandas.pydata.org/pandas-docs/stable/cookbook.html
* https://pandas.pydata.org/pandas-docs/stable/tutorials.html
## Import directives
```
%matplotlib inline
#%matplotlib notebook
from IPython.display import display
import matplotlib
matplotlib.rcParams['figure.figsize'] = (9, 9)
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
import datetime
import pandas as pd
import numpy as np
pd.__version__
```
## Make data
### Series (1D data)
C.f. http://pandas.pydata.org/pandas-docs/stable/generated/pandas.Series.html
#### With automatic indices
```
data_list = [1, 3, np.nan, 7]
series = pd.Series(data_list)
series
data_array = np.array(data_list)
series = pd.Series(data_array)
series
```
#### With defined indices
```
indices = pd.Series([1, 3, 5, 7])
series = pd.Series([10, 30, 50, 70], index=indices)
series
indices = pd.Series(['A', 'B', 'C', 'D'])
series = pd.Series([10, 30, 50, 70], index=indices)
series
data_dict = {'A': 10, 'B': 30, 'C': 50, 'D': 70}
series = pd.Series(data_dict)
series
```
#### Get information about a series
```
series.index
series.values
series.shape
series.dtypes
series.describe()
type(series.describe())
series.memory_usage()
```
### Date ranges
```
dates = pd.date_range('20130101', periods=6)
dates
dates = pd.date_range(start='2013-01-01', end='2013-01-08')
dates
dates = pd.date_range('2013-01-01', periods=4, freq='M')
dates
num_days = 7
data = np.random.random(num_days)
index = pd.date_range('2017-01-01', periods=num_days)
series = pd.Series(data, index)
series
```
### Frames (2D data)
C.f. http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.html
#### With automatic indices and columns
```
data_list = [[1, 2, 3], [4, 5, 6]]
df = pd.DataFrame(data_array)
df
data_array = np.array([[1, 2, 3], [4, 5, 6]])
df = pd.DataFrame(data_array)
df
```
#### With defined indices and columns
Using lists:
```
data = [[1, 2, 3], [4, 5, 6]]
index = [10, 20]
columns = ['A', 'B', 'C']
df = pd.DataFrame(data, index, columns)
df
```
Using numpy `arrays`:
```
data = np.array([[1, 2, 3], [4, 5, 6]])
index = np.array([10, 20])
columns = np.array(['A', 'B', 'C'])
df = pd.DataFrame(data, index=index, columns=columns)
df
```
Using `Series`:
```
data = np.array([[1, 2, 3], [4, 5, 6]])
index = pd.Series([10, 20])
columns = pd.Series(['A', 'B', 'C'])
df = pd.DataFrame(data, index=index, columns=columns)
df
```
#### With columns and values from a dictionary of lists
Dictionary keys define **columns** label.
```
data_dict = {
'A': 'foo',
'B': [10, 20, 30],
'C': 3
}
df = pd.DataFrame(data_dict)
df
```
To define index as well:
```
data_dict = {
'A': 'foo',
'B': [10, 20, 30],
'C': 3
}
df = pd.DataFrame(data_dict, index=[10, 20, 30])
df
```
#### With columns and values from a list of dictionaries
```
data_dict = [
{'A': 1, 'B': 2, 'C': 3},
{'A': 10, 'C': 30},
{'B': 200, 'C': 300}
]
df = pd.DataFrame(data_dict)
df
```
#### With columns and values from a dictionary of dictionaries
```
data_dict = {
'k1' : {'A': 1, 'B': 2, 'C': 3},
'k2' : {'A': 10, 'C': 30},
'k3' : {'B': 200, 'C': 300}
}
df = pd.DataFrame(data_dict)
df
df.T
```
#### Get information about a dataframe
```
df.index
df.columns
df.values
df.shape
df.dtypes
df.info()
df.describe()
type(df.describe())
df.memory_usage()
```
#### More details about dtype
DataFrame's columns can have different types. But what about rows ?
What append when a DataFrame with columns having different type is transposed ?
```
data_dict = {'A': 'foo',
'B': [10, 20, 30],
'C': 3}
df = pd.DataFrame(data_dict)
df
df.dtypes
df2 = df.T
df2
df2.dtypes
```
### Panels (3D data)
Panels are **deprecated**.
Pandas now focuses on 1D (`Series`) and 2D (`DataFrame`) data structures.
The recommended alternative to work with 3-dimensional data is the [xarray](http://xarray.pydata.org/en/stable/) python library.
An other workaround: one can simply use a [MultiIndex](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.MultiIndex.html) `DataFrame` for easily working with higher dimensional data.
See http://pandas.pydata.org/pandas-docs/stable/dsintro.html#deprecate-panel.
### Panel4D and PanelND (ND data)
`Panel4D` and `PanelND` are **deprecated**.
Pandas now focuses on 1D (`Series`) and 2D (`DataFrame`) data structures.
The recommended alternative to work with n-dimensional data is the [xarray](http://xarray.pydata.org/en/stable/) python library.
An other workaround: one can simply use a [MultiIndex](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.MultiIndex.html) `DataFrame` for easily working with higher dimensional data.
See http://pandas.pydata.org/pandas-docs/stable/dsintro.html#panel4d-and-panelnd-deprecated.
## Export/import data (write/read files)
See http://pandas.pydata.org/pandas-docs/stable/io.html
Reader functions are accessibles from the top level `pd` object.
Writer functions are accessibles from data objects (i.e. `Series`, `DataFrame` or `Panel` objects).
```
data_array = np.array([[1, 2, 3], [4, 5, 6]])
df = pd.DataFrame(data_array, index=[10, 20], columns=[100, 200, 300])
df
```
### HDF5 files
See `python_pandas_hdf5_en.ipynb`...
### CSV files
See http://pandas.pydata.org/pandas-docs/stable/io.html#csv-text-files
#### Write CSV files
See http://pandas.pydata.org/pandas-docs/stable/io.html#io-store-in-csv
Simplest version:
```
df.to_csv(path_or_buf="python_pandas_io_test.csv")
!cat python_pandas_io_test.csv
```
Setting more options:
```
# FYI, many other options are available
df.to_csv(path_or_buf="python_pandas_io_test.csv",
sep=',',
columns=None,
header=True,
index=True,
index_label=None,
compression=None, # allowed values are 'gzip', 'bz2' or 'xz'
date_format=None)
!cat python_pandas_io_test.csv
```
#### Read CSV files
See http://pandas.pydata.org/pandas-docs/stable/io.html#io-read-csv-table
Simplest version:
```
df = pd.read_csv("python_pandas_io_test.csv")
df
```
Setting more options:
```
df = pd.read_csv("python_pandas_io_test.csv",
sep=',',
delimiter=None,
header='infer',
names=None,
index_col=0,
usecols=None,
squeeze=False,
prefix=None,
mangle_dupe_cols=True,
dtype=None,
engine=None,
converters=None,
true_values=None,
false_values=None,
skipinitialspace=False,
skiprows=None,
nrows=None,
na_values=None,
keep_default_na=True,
na_filter=True,
verbose=False,
skip_blank_lines=True,
parse_dates=False,
infer_datetime_format=False,
keep_date_col=False,
date_parser=None,
dayfirst=False,
iterator=False,
chunksize=None,
compression='infer',
thousands=None,
decimal=b'.',
lineterminator=None,
quotechar='"',
quoting=0,
escapechar=None,
comment=None,
encoding=None,
dialect=None,
#tupleize_cols=False,
error_bad_lines=True,
warn_bad_lines=True,
skipfooter=0,
#skip_footer=0,
doublequote=True,
delim_whitespace=False,
#as_recarray=False,
#compact_ints=False,
#use_unsigned=False,
low_memory=True,
#buffer_lines=None,
memory_map=False,
float_precision=None)
df
!rm python_pandas_io_test.csv
```
### JSON files
See http://pandas.pydata.org/pandas-docs/stable/io.html#json
```
import io
```
#### Write JSON files
See http://pandas.pydata.org/pandas-docs/stable/io.html#io-json-writer
##### Simplest version
```
df.to_json(path_or_buf="python_pandas_io_test.json")
!cat python_pandas_io_test.json
```
##### Setting `orient`="split"
```
df.to_json(path_or_buf="python_pandas_io_test_split.json",
orient="split")
!cat python_pandas_io_test_split.json
```
##### Setting `orient`="records"
```
df.to_json(path_or_buf="python_pandas_io_test_records.json",
orient="records")
!cat python_pandas_io_test_records.json
```
##### Setting `orient`="index" (the default option for `Series`)
```
df.to_json(path_or_buf="python_pandas_io_test_index.json",
orient="index")
!cat python_pandas_io_test_index.json
```
##### Setting `orient`="columns" (the default option for `DataFrame`) (for `DataFrame` only)
```
df.to_json(path_or_buf="python_pandas_io_test_columns.json",
orient="columns")
!cat python_pandas_io_test_columns.json
```
##### Setting `orient`="values" (for `DataFrame` only)
```
df.to_json(path_or_buf="python_pandas_io_test_values.json",
orient="values")
!cat python_pandas_io_test_values.json
```
##### Setting more options
```
# FYI, many other options are available
df.to_json(path_or_buf="python_pandas_io_test.json",
orient='columns', # For DataFrame: 'split','records','index','columns' or 'values'
date_format=None, # None, 'epoch' or 'iso'
double_precision=10,
force_ascii=True,
date_unit='ms')
!cat python_pandas_io_test.json
```
#### Read JSON files
See http://pandas.pydata.org/pandas-docs/stable/io.html#io-json-reader
##### Using `orient`="split"
Dict like data ``{index -> [index], columns -> [columns], data -> [values]}``
```
!cat python_pandas_io_test_split.json
df = pd.read_json("python_pandas_io_test_split.json",
orient="split")
df
```
##### Using `orient`="records"
List like ``[{column -> value}, ... , {column -> value}]``
```
!cat python_pandas_io_test_records.json
df = pd.read_json("python_pandas_io_test_records.json",
orient="records")
df
```
##### Using `orient`="index"
Dict like ``{index -> {column -> value}}``
```
!cat python_pandas_io_test_index.json
df = pd.read_json("python_pandas_io_test_index.json",
orient="index")
df
```
##### Using `orient`="columns"
Dict like ``{column -> {index -> value}}``
```
!cat python_pandas_io_test_columns.json
df = pd.read_json("python_pandas_io_test_columns.json",
orient="columns")
df
```
##### Using `orient`="values" (for `DataFrame` only)
Just the values array
```
!cat python_pandas_io_test_values.json
df = pd.read_json("python_pandas_io_test_values.json",
orient="values")
df
```
##### Setting more options
```
df = pd.read_json("python_pandas_io_test.json",
orient=None,
typ='frame',
dtype=True,
convert_axes=True,
convert_dates=True,
keep_default_dates=True,
numpy=False,
precise_float=False,
date_unit=None,
encoding=None,
lines=False)
df
!rm python_pandas_io_test*.json
```
### YAML
See https://stackoverflow.com/a/35968457
```
!echo "- {A: 1, B: 2}" > python_pandas_io_test.yaml
!echo "- {A: 3}" >> python_pandas_io_test.yaml
!echo "- {B: 4}" >> python_pandas_io_test.yaml
!cat python_pandas_io_test.yaml
try:
import yaml
with open('python_pandas_io_test.yaml', 'r') as f:
df = pd.io.json.json_normalize(yaml.load(f))
print(df)
except:
pass
!rm python_pandas_io_test.yaml
```
### Other file formats
Many other file formats can be used to import or export data with JSON.
See the following link for more information: http://pandas.pydata.org/pandas-docs/stable/io.html
## Select columns
```
data_array = np.array([np.arange(1, 10, 1), np.arange(10, 100, 10), np.arange(100, 1000, 100)]).T
df = pd.DataFrame(data_array,
index=np.arange(1, 10, 1),
columns=['A', 'B', 'C'])
df
```
### Select a single column
The following instructions return a `Series`.
#### Label based selection
```
df.B
df["B"]
df.loc[:,"B"]
```
#### Index based selection
```
df.iloc[:,1]
```
### Select multiple columns
#### Label based selection
```
df[['A','B']]
df.loc[:,['A','B']]
```
#### Index based selection
```
df.iloc[:,0:2]
```
## Select rows
```
data_array = np.array([np.arange(1, 10, 1), np.arange(10, 100, 10), np.arange(100, 1000, 100)]).T
df = pd.DataFrame(data_array,
index=["i" + str(i+1) for i in range(9)],
columns=['A', 'B', 'C'])
df
```
### Select a single row
The following instructions return a `Series`.
#### Label based selection
```
df.loc["i3"]
df.loc["i3",:]
```
#### Index based selection
```
df.iloc[2] # Select over index
df.iloc[2,:] # Select over index
```
### Select multiple rows
#### Label based selection
```
df.loc[["i3", "i4"],:]
```
#### Index based selection
```
df.iloc[2:4,:] # Select over index
```
### Select rows based on values
```
df.B < 50.
type(df.B < 50.)
df[[True, True, True, True, False, False, False, False, False]]
series_mask = pd.Series({'i1': True,
'i2': True,
'i3': True,
'i4': True,
'i5': False,
'i6': False,
'i7': False,
'i8': False,
'i9': False})
df[series_mask]
df[df.B < 50.]
df[df['B'] < 50.]
df[(df.A >= 2) & (df.B < 50)]
```
This can be written:
```
df.loc[(df.A >= 2) & (df.B < 50)]
```
This could be written `df[df.A >= 2][df.B < 50]` but this is a bad practice (named "chained indexing").
"When setting values in a pandas object, care must be taken to avoid what is called *chained indexing*".
See:
- http://pandas.pydata.org/pandas-docs/stable/indexing.html#returning-a-view-versus-a-copy
- http://pandas.pydata.org/pandas-docs/stable/indexing.html#why-does-assignment-fail-when-using-chained-indexing
- http://pandas.pydata.org/pandas-docs/stable/indexing.html#evaluation-order-matters
### Argmin rows
```
df = pd.DataFrame([[4000, 10, 3],
[100, 200, 1],
[30, 1000, 10]], columns=['a', 'b', 'c'], index=['k1', 'k2', 'k3'])
df
```
Select the line having the minimal `c`
```
df.c.idxmin()
df.loc[ df.c.idxmin() ]
```
## Select rows and columns
```
data_array = np.array([np.arange(1, 10, 1), np.arange(10, 100, 10), np.arange(100, 1000, 100)]).T
df = pd.DataFrame(data_array,
index=np.arange(1, 10, 1),
columns=['A', 'B', 'C'])
df
df[(df.A >= 2) & (df.B < 50)]
df[(df.B < 20) | (df.B > 50)]
df.loc[(df.B < 20) | (df.B > 50), 'C']
df[(df['A'] >= 2) & (df['B'] < 50)]
df.loc[(df.A >= 2) & (df.B < 50), ['A','B']]
```
## Setting values
### Apply a function to selected colunms values
```
data_array = np.array([np.arange(1, 10, 1), np.arange(10, 100, 10), np.arange(100, 1000, 100)]).T
df = pd.DataFrame(data_array,
index=np.arange(1, 10, 1),
columns=['A', 'B', 'C'])
df
df.B *= 2.
df
df.B = pow(df.B, 2)
df
```
### Apply a function to selected rows values
```
data_array = np.array([np.arange(1, 10, 1), np.arange(10, 100, 10), np.arange(100, 1000, 100)]).T
df = pd.DataFrame(data_array,
index=np.arange(1, 10, 1),
columns=['A', 'B', 'C'])
df
df[df.B < 50.] *= -1.
df
# df['B'][df['B'] < 50.] = 0 # OK but chain indexing is bad...
# df.A[df.B < 50.] = 0 # OK but chain indexing is bad...
df.loc[df.B < 50., 'A'] = 0
df
```
**WARNING**: `df[df.B < 50.].A = 0` does *NOT* work even if `df.A[df.B < 50.]` and `df[df.B < 50.].A` seems to produce the same result...
"When setting values in a pandas object, care must be taken to avoid what is called *chained indexing*".
See:
- http://pandas.pydata.org/pandas-docs/stable/indexing.html#returning-a-view-versus-a-copy
- http://pandas.pydata.org/pandas-docs/stable/indexing.html#why-does-assignment-fail-when-using-chained-indexing
- http://pandas.pydata.org/pandas-docs/stable/indexing.html#evaluation-order-matters
```
df.loc[(df.B < 50.) & (df.B > 20), 'C'] = 0
df
df.loc[(df.B < 20) | (df.B > 50), 'C'] = -1
df
df[df.B < 50.] = pow(df[df.B < 50.], 2)
df
```
## Sample rows
```
data_array = np.array([np.arange(1, 10, 1), np.arange(10, 100, 10), np.arange(100, 1000, 100)]).T
df = pd.DataFrame(data_array,
index=np.arange(1, 10, 1),
columns=['A', 'B', 'C'])
df
```
### With replacement
Draw 3 samples:
```
df.sample(n=30, replace=True)
```
Sample 90% of the rows:
```
df.sample(frac=0.9, replace=True)
```
### Without replacement
Draw 3 samples:
```
df.sample(n=3)
```
Sample 90% of the rows:
```
df.sample(frac=0.9)
```
### Weighted sampling
```
df.sample(n=30, replace=True, weights=np.arange(len(df)))
```
## Shuffle/permute rows
See:
- https://stackoverflow.com/a/35784666
- https://stackoverflow.com/a/34879805
```
data_array = np.array([np.arange(1, 10, 1), np.arange(10, 100, 10), np.arange(100, 1000, 100)]).T
df = pd.DataFrame(data_array,
index=np.arange(1, 10, 1),
columns=['A', 'B', 'C'])
df
df = df.sample(frac=1)
df
```
To reset indexes too:
```
df = df.sample(frac=1).reset_index(drop=True)
df
```
## Change columns order
```
data_array = np.array([np.arange(1, 5, 1), np.arange(10, 50, 10), np.arange(100, 500, 100)]).T
df = pd.DataFrame(data_array,
columns=['A', 'B', 'C'])
df
df = df[['B', 'C', 'A']]
df
```
## Sort a DataFrame
```
NROWS = 7
col1 = np.arange(1., NROWS, 1)
col2 = np.arange(10., NROWS*10, 10)
col3 = np.arange(100., NROWS*100, 100)
np.random.shuffle(col1)
np.random.shuffle(col2)
np.random.shuffle(col3)
data = np.array([col1,
col2,
col3]).T
index = np.arange(1, NROWS, 1)
columns = np.array(['A', 'B', 'C'])
np.random.shuffle(index)
np.random.shuffle(data)
np.random.shuffle(columns)
df = pd.DataFrame(data,
index=index,
columns=columns)
df
```
### Sorting by row index or column label
#### Sorting by rows
```
df.sort_index()
df.sort_index(axis=0) # axis=0 -> sort by row index
df.sort_index(ascending=False)
```
#### Sorting by columns
```
df.sort_index(axis=1) # axis=1 -> sort by column label
df.sort_index(axis=1, ascending=False)
```
### Sorting by values
```
df.sort_values(by='B')
df.sort_values(by='B', ascending=False)
df.sort_values(by='B', inplace=True)
df
```
## Missing data
See http://pandas.pydata.org/pandas-docs/stable/10min.html#missing-data
```
a = np.array([[3, np.nan, 5, np.nan, 7],
[2, 4, np.nan, 3, 1],
[3, 4, 5, 6, 1]]).T
df = pd.DataFrame(a,
columns=['A', 'B', 'C'])
df
```
### Get the boolean mask where values are nan
```
df.isnull()
```
### Drop any rows that have missing data
```
df.dropna()
df.dropna(how='any') # but 'any' is the default value...
```
### Drop any rows that have missing data in a given column
See https://stackoverflow.com/questions/13413590/how-to-drop-rows-of-pandas-dataframe-whose-value-in-certain-columns-is-nan
```
df.dropna(subset=['B'])
df.dropna(subset=['B', 'C'])
```
### Drop any columns that have missing data
```
df.dropna(axis=1)
df.dropna(axis=1, how='any') # but 'any' is the default value...
```
### Drop any columns that have missing data in a given row
```
df.dropna(axis=1, subset=[2])
df.dropna(axis=1, subset=[1, 2])
```
### Filling missing data
```
df.fillna(value=999)
```
### Count the number of NaN values in a given column
```
df.A.isnull().sum()
```
## Miscellaneous operations on data frames
### Transpose of a data frame
```
data_array = np.array([np.arange(1, 10, 1), np.arange(10, 100, 10), np.arange(100, 1000, 100)]).T
df = pd.DataFrame(data_array,
index=np.arange(1, 10, 1),
columns=['A', 'B', 'C'])
df
df.T
```
## Merge
See: http://pandas.pydata.org/pandas-docs/stable/generated/pandas.merge.html#pandas.merge
```
a1 = np.array([np.arange(1, 5, 1), np.arange(10, 50, 10), np.arange(100, 500, 100)]).T
df1 = pd.DataFrame(a1,
columns=['ID', 'B', 'C'])
a2 = np.array([np.arange(1, 5, 1), np.arange(1000, 5000, 1000), np.arange(10000, 50000, 10000)]).T
df2 = pd.DataFrame(a2,
columns=['ID', 'B', 'C'])
display(df1)
display(df2)
df = pd.merge(df1, df2, on="ID", suffixes=('_1', '_2')) #.dropna(how='any')
display(df)
```
### Merge with NaN
```
a1 = np.array([np.arange(1, 5, 1), np.arange(10, 50, 10), np.arange(100, 500, 100)]).T
df1 = pd.DataFrame(a1,
columns=['ID', 'B', 'C'])
a2 = np.array([np.arange(1, 5, 1), np.arange(1000, 5000, 1000), np.arange(10000, 50000, 10000)]).T
df2 = pd.DataFrame(a2,
columns=['ID', 'B', 'C'])
df1.iloc[0,2] = np.nan
df1.iloc[1,1] = np.nan
df1.iloc[2,2] = np.nan
df1.iloc[3,1] = np.nan
df2.iloc[0,1] = np.nan
df2.iloc[1,2] = np.nan
df2.iloc[2,1] = np.nan
df2.iloc[3,2] = np.nan
df = pd.merge(df1, df2, on="ID", suffixes=('_1', '_2')) #.dropna(how='any')
display(df1)
display(df2)
display(df)
```
### Merge with missing rows
```
a1 = np.array([np.arange(1, 5, 1), np.arange(10, 50, 10), np.arange(100, 500, 100)]).T
df1 = pd.DataFrame(a1,
columns=['ID', 'B', 'C'])
a2 = np.array([np.arange(1, 3, 1), np.arange(1000, 3000, 1000), np.arange(10000, 30000, 10000)]).T
df2 = pd.DataFrame(a2,
columns=['ID', 'B', 'C'])
display(df1)
display(df2)
print("Left: use only keys from left frame (SQL: left outer join)")
df = pd.merge(df1, df2, on="ID", how="left", suffixes=('_1', '_2')) #.dropna(how='any')
display(df)
print("Right: use only keys from right frame (SQL: right outer join)")
df = pd.merge(df1, df2, on="ID", how="right", suffixes=('_1', '_2')) #.dropna(how='any')
display(df)
print("Inner: use intersection of keys from both frames (SQL: inner join) [DEFAULT]")
df = pd.merge(df1, df2, on="ID", how="inner", suffixes=('_1', '_2')) #.dropna(how='any')
display(df)
print("Outer: use union of keys from both frames (SQL: full outer join)")
df = pd.merge(df1, df2, on="ID", how="outer", suffixes=('_1', '_2')) #.dropna(how='any')
display(df)
```
## GroupBy
See: http://pandas.pydata.org/pandas-docs/stable/groupby.html
```
a = np.array([[3, 5, 5, 5, 7, 7, 7, 7],
[2, 4, 5, 3, 3, 2, 3, 2],
[3, 5, 4, 6, 8, 1, 9, 8]]).T
df = pd.DataFrame(a,
columns=['A', 'B', 'C'])
df
```
### GroupBy with single key
```
df.groupby(["A"]).count()
df.groupby(["A"]).sum().B
df.groupby(["A"]).mean().B
```
### GroupBy and argmin
Group by `A` and return the argmin of `C` for each group.
```
df.groupby("A").C.idxmin()
df.loc[ df.groupby("A").C.idxmin() ]
```
### First element of each group
```
df.groupby("A").first()
```
### Index of the first element of each group
See https://stackoverflow.com/questions/50704041/r-pandas-groupby-index-of-the-first-row-in-each-group
```
df.groupby("A").head(1)
```
### GroupBy with multiple keys
```
df.groupby(["A","B"]).count()
```
## Rolling
See https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.rolling.html#pandas-dataframe-rolling
For rolling over a time window, see https://pandas.pydata.org/pandas-docs/stable/timeseries.html#resampling
### Basic example
Rolling with an aggregation window of size 2.
```
s = pd.Series([1., 0., 5., 2., 1.])
print("DATA:")
print(s)
mean_s = s.rolling(2).mean()
print()
print("ROLLING MEAN:")
print(mean_s)
sum_s = s.rolling(2).sum()
print()
print("ROLLING SUM:")
print(sum_s)
min_s = s.rolling(2).min()
print()
print("ROLLING MIN:")
print(min_s)
max_s = s.rolling(2).max()
print()
print("ROLLING MAX:")
print(max_s)
ax = s.plot(figsize=(18, 3), color="blue")
mean_s.plot(color="red", label="mean", ax=ax)
sum_s.plot(color="green", label="sum", style="--", alpha=0.5, ax=ax)
min_s.plot(color="black", label="min", style=":", alpha=0.25, ax=ax)
max_s.plot(color="black", label="max", style=":", alpha=0.25, ax=ax)
ax.legend();
```
### More realistic example
```
index = np.arange(0, 20, 0.05)
s = pd.Series(np.sin(index))
s = s + np.random.normal(scale=0.4, size=s.shape)
ax = s.plot(figsize=(18, 3))
s.shape
```
Rolling with an aggregation window of size 20.
```
s_mean = s.rolling(20).mean()
s_median = s.rolling(20).median()
s_min = s.rolling(20).min()
s_max = s.rolling(20).max()
ax = s_mean.plot(y='duration', figsize=(18, 8), color="red", label="mean", alpha=0.75)
s_median.plot(ax=ax, color="blue", label="median", alpha=0.75)
s_min.plot(ax=ax, color="blue", alpha=0.5, style=":", label="min")
s_max.plot(ax=ax, color="blue", alpha=0.5, style=":", label="max")
plt.fill_between(s_min.index, s_min.values, s_max.values, facecolor='blue', alpha=0.1)
ax.legend()
ax.set_xlabel('Time');
s_mean.shape
```
## Pivot
```
df = pd.DataFrame([["i1", "A", 1],
["i1", "B", 2],
["i2", "A", 3],
["i2", "B", 4]], columns=["foo", "bar", "baz"])
df
df.pivot(index="foo", columns="bar", values="baz")
```
## Count the number of occurrences of a column value
```
a = np.array([[3, 5, 5, 5, 7, 7, 7, 7],
[2, 4, 4, 3, 1, 3, 3, 2],
[3, 4, 5, 6, 1, 8, 9, 8]]).T
df = pd.DataFrame(a,
columns=['A', 'B', 'C'])
df
df.A.value_counts()
df.A.value_counts().plot.bar()
```
## Stats
```
df = pd.DataFrame(np.random.normal(size=100000))
df.quantile(0.50)
df.quantile([0.25, 0.75])
df.quantile([0.01, 0.001])
```
## Time series
See https://pandas.pydata.org/pandas-docs/stable/timeseries.html#
There are 3 main time related types in Pandas (and the equivalent type for `Series` and `DataFrame` indices):
- `pandas.Timestamp` (`pandas.DatetimeIndex` for indices): pandas equivalent of python's ``datetime.datetime``
- `pandas.Period` (`pandas.PeriodIndex` for indices): represents a period of time
- `pandas.Timedelta` (`pandas.TimedeltaIndex` for indices): represents a duration (the difference between two dates or times) i.e. the pandas equivalent of python's ``datetime.timedelta``
A Timestamp is a point in time:
```
pd.Timestamp(year=2018, month=1, day=1, hour=12, minute=30)
```
A Period is a range in time (with a "anchored" start time and a "anchored" end time):
```
p = pd.Period(freq='D', year=2018, month=1, day=1, hour=12, minute=30)
print(p)
print("Start time:", p.start_time)
print("End time:", p.end_time)
```
A Timedelta is a "floating" duration (i.e. not "anchored" in time):
```
print(pd.Timedelta(days=5, seconds=30))
ts1 = pd.Timestamp(year=2018, month=1, day=1, hour=12, minute=30)
ts2 = pd.Timestamp(year=2018, month=1, day=2, hour=12, minute=30)
print(ts2 - ts1)
```
### Generate datetime index (with a fixed frequency)
See https://pandas.pydata.org/pandas-docs/stable/generated/pandas.date_range.html
```
pd.date_range('2018-01-01', '2018-03-01', freq='D')
pd.date_range('2018-01-01', periods=10, freq='h')
pd.date_range('1/1/2012', periods=10, freq='S')
pd.date_range('3/6/2012 00:00', periods=5, freq='D')
pd.date_range('1/1/2012', periods=5, freq='M')
```
### Generate period index
```
pd.period_range('2018-01-01', '2018-03-01', freq='D')
pd.date_range('2018-01-01', '2018-03-01', freq='D').to_period()
```
### Plot time series
- http://pandas.pydata.org/pandas-docs/stable/visualization.html#suppressing-tick-resolution-adjustment
- http://pandas.pydata.org/pandas-docs/stable/visualization.html#automatic-date-tick-adjustment
```
dti = pd.date_range('2012-01-01 00:00', periods=40, freq='D')
ts = pd.Series(np.random.randint(0, 200, len(dti)), index=dti)
ts.plot();
ts.plot(x_compat=True);
dti = pd.date_range('2018-01-01 00:00', '2018-01-03 00:00', freq='H')
ts = pd.Series(np.random.randint(0, 100, len(dti)), index=dti)
ax = ts.plot(x_compat=True, figsize=(16, 4)) # x_compat is required as matplotlib doesn't understand pandas datetime format -> x_compat=True makes the conversion...
# set monthly locator
ax.xaxis.set_major_locator(mdates.DayLocator(interval=1))
ax.xaxis.set_minor_locator(mdates.HourLocator(interval=1))
# set formatter
ax.xaxis.set_major_formatter(mdates.DateFormatter('%d-%m-%Y'))
# set font and rotation for date tick labels
plt.gcf().autofmt_xdate()
```
### Indexing (select datetime)
See https://pandas.pydata.org/pandas-docs/stable/timeseries.html#indexing
```
dti = pd.date_range('2012-1-1 00:00', periods=40, freq='D')
ts = pd.Series(np.random.randint(0, 200, len(dti)), index=dti)
ts
ts["2012-01-09"]
ts[datetime.datetime(2012, 1, 9)]
ts[ts.index < "2012-01-09"]
ts[ts.index > "2012-01-20"]
ts["2012-01-09":"2012-01-20"]
ts[datetime.datetime(2012, 1, 9):datetime.datetime(2012, 1, 20)]
ts[ts.index.day <= 3]
ts[ts.index.month == 2]
ts["2012-02"]
ts[ts.index.dayofweek == 1]
```
### Rolling
See https://stackoverflow.com/a/39917862
#### Basic example
Rolling window size: 1 day
```
dti = pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-1 06:45', '2018-1-1 12:00',
'2018-1-2 00:00', '2018-1-2 06:00', '2018-1-2 12:00'])
ts = pd.Series([2., 1., 3., 2., 2., 0.], index=dti)
print("DATA:")
print(ts)
ax = ts.plot(figsize=(18, 3), style="*-", color="blue")
ax.vlines(pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-2 00:00']), ymin=0, ymax=8, color="red", linestyle=":", alpha=0.3);
ts_rw = ts.rolling('D').sum() # Rolling window size: 1 day
print()
print("MEAN:")
print(ts_rw)
ts_rw.plot(color="red", label="sum", style="*-", alpha=0.75, ax=ax)
ax.legend()
ax.set_xlabel('Time')
ax.grid(True);
ts.rolling('6h').min()
ts.rolling('3h').mean()
```
#### More realistic example
```
dti = pd.date_range('1/1/2018 00:00', periods=6*480, freq='10min')
ts = pd.Series(np.sin(dti.hour * 2. * np.pi / 24.), index=dti)
ts = ts + np.random.normal(scale=0.4, size=ts.shape)
ax = ts.plot(figsize=(18, 3))
ax.vlines(pd.date_range('1/1/2018 00:00', periods=480/24, freq='D'), ymin=-2, ymax=2, color="red", linestyle=":", alpha=0.3);
ts.shape
ts_mean = ts.rolling('5H').mean()
ts_median = ts.rolling('5H').median()
ts_min = ts.rolling('5H').min()
ts_max = ts.rolling('5H').max()
ax = ts_mean.plot(y='duration', figsize=(18, 3), color="red", label="mean", alpha=0.75)
ts_median.plot(ax=ax, color="blue", label="median", alpha=0.75)
ts_min.plot(ax=ax, color="blue", alpha=0.5, style=":", label="min")
ts_max.plot(ax=ax, color="blue", alpha=0.5, style=":", label="max")
plt.fill_between(ts_min.index, ts_min.values, ts_max.values, facecolor='blue', alpha=0.1)
ax.legend()
ax.set_xlabel('Time');
ts_mean.shape
```
### Resampling
See https://pandas.pydata.org/pandas-docs/stable/timeseries.html#resampling
`resample()` is a time-based groupby, followed by a reduction method on each of its groups.
`resample()` is similar to using a `rolling()` operation with a time-based offset.
#### Basic example
```
dti = pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-1 06:45', '2018-1-1 12:00',
'2018-1-2 00:00', '2018-1-2 12:00'])
ts = pd.Series([1., 0., 5., 2., 0.], index=dti)
print("DATA:")
print(ts)
ax = ts.plot(figsize=(18, 3), style="*-", color="blue")
ax.vlines(pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-2 00:00']), ymin=0, ymax=5, color="red", linestyle=":", alpha=0.3);
ts_resampled = ts.resample('D').mean()
print()
print("MEAN:")
print(ts_resampled)
ts_resampled.plot(color="red", style="*-", label="mean", alpha=0.75, ax=ax)
ax.legend()
ax.set_xlabel('Time');
ts.resample('6h').min()
ts.resample('3h').sum()
```
#### Is there an offset ?
No. `resample('D')` aggregates values for each day between "00:00:00" and "23:59:59", whatever the first index of `ts`.
See the examples bellow...
```
dti = pd.DatetimeIndex(['2018-1-1 12:00',
'2018-1-2 08:00', '2018-1-2 18:00', '2018-1-2 23:59:59',
'2018-1-3 00:00'])
ts = pd.Series([0.,
10., 20., 30.,
5.], index=dti)
print("DATA:")
print(ts)
ts_resampled = ts.resample('D').mean()
print()
print("MEAN:")
print(ts_resampled)
# Illustrative plot
ax = ts.plot(x_compat=True, figsize=(18, 3), style="*-", color="blue")
ax.vlines(pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-2 00:00', '2018-1-3 00:00']), ymin=-10, ymax=40, color="red", linestyle=":", linewidth=2, alpha=0.5);
ax.vlines(pd.DatetimeIndex(['2018-1-1 12:00', '2018-1-2 12:00', '2018-1-3 12:00']), ymin=-10, ymax=40, color="green", linestyle=":", linewidth=2, alpha=0.5);
ax.plot(pd.DatetimeIndex(['2018-1-1 12:15', '2018-1-2 11:45']), [40, 40], marker="|", markersize=20, color="green")
ax.plot(pd.DatetimeIndex(['2018-1-2 12:15', '2018-1-3 11:45']), [40, 40], marker="|", markersize=20, color="green")
ax.plot(pd.DatetimeIndex(['2018-1-1 00:15', '2018-1-1 23:45']), [35, 35], marker="|", markersize=20, color="red")
ax.plot(pd.DatetimeIndex(['2018-1-2 00:15', '2018-1-2 23:45']), [35, 35], marker="|", markersize=20, color="red")
ts_resampled.plot(color="red", style="*-", label="mean", alpha=0.75, ax=ax)
# set monthly locator
ax.xaxis.set_major_locator(mdates.DayLocator(interval=1))
ax.xaxis.set_minor_locator(mdates.HourLocator(interval=1))
# set formatter
ax.xaxis.set_major_formatter(mdates.DateFormatter('%d-%m-%Y %H:%M'))
# set font and rotation for date tick labels
plt.gcf().autofmt_xdate()
ax.legend()
ax.set_xlabel('Time');
dti = pd.DatetimeIndex(['2018-1-1 01:00',
'2018-1-1 05:30', '2018-1-1 07:30',
'2018-1-1 10:00'])
ts = pd.Series([0.,
10., 20.,
5.], index=dti)
print("DATA:")
print(ts)
ts_resampled = ts.resample('5h').mean()
print()
print("MEAN:")
print(ts_resampled)
# Illustrative plot
ax = ts.plot(x_compat=True, figsize=(18, 3), style="*-", color="blue")
ax.vlines(pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-1 05:00', '2018-1-1 10:00']), ymin=-10, ymax=40, color="red", linestyle=":", linewidth=2, alpha=0.5);
ax.vlines(pd.DatetimeIndex(['2018-1-1 01:00', '2018-1-1 06:00', '2018-1-1 11:00']), ymin=-10, ymax=40, color="green", linestyle=":", linewidth=2, alpha=0.5);
ax.plot(pd.DatetimeIndex(['2018-1-1 01:05', '2018-1-1 05:55']), [40, 40], marker="|", markersize=20, color="green")
ax.plot(pd.DatetimeIndex(['2018-1-1 06:05', '2018-1-1 10:55']), [40, 40], marker="|", markersize=20, color="green")
ax.plot(pd.DatetimeIndex(['2018-1-1 00:05', '2018-1-1 04:55']), [35, 35], marker="|", markersize=20, color="red")
ax.plot(pd.DatetimeIndex(['2018-1-1 05:05', '2018-1-1 09:55']), [35, 35], marker="|", markersize=20, color="red")
ts_resampled.plot(color="red", style="*-", label="mean", alpha=0.75, ax=ax)
# set monthly locator
ax.xaxis.set_major_locator(mdates.HourLocator(interval=1))
#ax.xaxis.set_minor_locator(mdates.HourLocator(interval=1))
# set formatter
ax.xaxis.set_major_formatter(mdates.DateFormatter('%H:%M'))
# set font and rotation for date tick labels
plt.gcf().autofmt_xdate()
ax.legend()
ax.set_xlabel('Time');
```
#### More realistic example
```
dti = pd.date_range('1/1/2018 00:00', periods=60*480, freq='min')
ts = pd.Series(np.sin(dti.hour * 2. * np.pi / 24.), index=dti)
ts = ts + np.random.normal(scale=0.4, size=ts.shape)
ax = ts.plot(figsize=(18, 3))
ax.vlines(pd.date_range('1/1/2018 00:00', periods=480/24, freq='D'), ymin=-2, ymax=2, color="red", linestyle=":", alpha=0.3);
ts.shape
ts_mean = ts.resample('2H').mean()
ts_median = ts.resample('2H').median()
ts_min = ts.resample('2H').min()
ts_max = ts.resample('2H').max()
ax = ts_mean.plot(y='duration', figsize=(18, 8), color="red", label="mean", alpha=0.75)
ts_median.plot(ax=ax, color="blue", label="median", alpha=0.75)
ts_min.plot(ax=ax, color="blue", alpha=0.5, style=":", label="min")
ts_max.plot(ax=ax, color="blue", alpha=0.5, style=":", label="max")
plt.fill_between(ts_min.index, ts_min.values, ts_max.values, facecolor='blue', alpha=0.1)
ax.legend()
ax.set_xlabel('Time');
ts_mean.shape
```
### Difference between `rolling()` and `resample()`
```
rolling_window = '6H'
start = '2018-1-1 00:00'
end = '2018-1-4 00:00'
dti = pd.date_range(start=start, end=end, freq='min')
ts = pd.Series(np.sin(dti.hour * 2. * np.pi / 24.), index=dti)
ts = ts + np.random.normal(scale=0.4, size=ts.shape)
ax = ts.plot(figsize=(18, 3))
ax.vlines(pd.date_range(start=start, end=end, freq=rolling_window), ymin=-2, ymax=2, color="red", linestyle=":", alpha=0.5);
ts2 = ts.rolling(rolling_window).mean() # Rolling window size: 1 day
ax = ts2.plot(figsize=(18, 3), color="red", alpha=0.75)
ax.vlines(pd.date_range(start=start, end=end, freq=rolling_window), ymin=-1, ymax=1, color="red", linestyle=":", alpha=0.5);
ts2 = ts.resample(rolling_window).mean() # Rolling window size: 1 day
ax = ts2.plot(figsize=(18, 3), color="red", alpha=0.75)
ax.vlines(pd.date_range(start=start, end=end, freq=rolling_window), ymin=-1, ymax=1, color="red", linestyle=":", alpha=0.5);
```
### Group by
See https://jakevdp.github.io/PythonDataScienceHandbook/03.11-working-with-time-series.html#Digging-into-the-data
#### Basic example
```
dti = pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-1 12:00', '2018-1-2 00:00', '2018-1-2 12:00'])
ts = pd.Series([1., 0., 2., 1.], index=dti)
print(ts)
ax = ts.plot(figsize=(18, 3))
dti = pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-2 00:00'])
ax.vlines(dti, ymin=0, ymax=2, color="red", linestyle=":", alpha=0.3);
ts_mean = ts.groupby(ts.index.time).mean()
print(ts_mean)
ax = ts_mean.plot(y='duration', figsize=(10, 4), color="red", label="mean", alpha=0.75)
ax.legend()
ax.set_xlabel('Time');
```
#### Basic example of wrong usage
```
dti = pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-1 12:00', '2018-1-2 00:31', '2018-1-2 12:25']) # Here time is not aligned (non constant frequency)
ts = pd.Series([1., 0., 2., 1.], index=dti)
print(ts)
ax = ts.plot(figsize=(18, 3));
ts_mean = ts.groupby(ts.index.time).mean()
print(ts_mean)
ax = ts_mean.plot(y='duration', figsize=(10, 4), color="red", label="mean", alpha=0.75)
ax.legend()
ax.set_xlabel('Time');
```
#### More realistic example
```
dti = pd.date_range('1/1/2018 00:00', periods=960, freq='h')
ts = pd.Series(np.sin(dti.hour * 2. * np.pi / 24.), index=dti)
ts = ts + np.random.normal(scale=0.4, size=ts.shape)
ax = ts.plot(figsize=(18, 3))
ax.vlines(pd.date_range('1/1/2018 00:00', periods=960/24, freq='D'), ymin=-2, ymax=2, color="red", linestyle=":", alpha=0.3);
ts_mean = ts.groupby(ts.index.time).mean()
ts_median = ts.groupby(ts.index.time).median()
ts_quartile_1 = ts.groupby(ts.index.time).quantile(0.25)
ts_quartile_3 = ts.groupby(ts.index.time).quantile(0.75)
ax = ts_mean.plot(y='duration', figsize=(14, 8), color="red", label="mean", alpha=0.75)
ts_median.plot(ax=ax, color="blue", label="median", alpha=0.75)
ts_quartile_1.plot(ax=ax, color="blue", alpha=0.5, style=":", label="1st quartile")
ts_quartile_3.plot(ax=ax, color="blue", alpha=0.5, style=":", label="3rd quartile")
plt.fill_between(ts_quartile_1.index, ts_quartile_1.values, ts_quartile_3.values, facecolor='blue', alpha=0.1)
ax.legend()
ax.set_xlabel('Time');
```
#### With Periods
```
dti = pd.period_range('1/1/2018 00:00', periods=960, freq='h')
ts = pd.Series(np.sin(dti.hour * 2. * np.pi / 24.), index=dti)
ts = ts + np.random.normal(scale=0.4, size=ts.shape)
ax = ts.plot(figsize=(18, 3))
ax.vlines(pd.date_range('1/1/2018 00:00', periods=960/24, freq='D'), ymin=-2, ymax=2, color="red", linestyle=":", alpha=0.3);
ts_mean = ts.groupby(ts.index.start_time.time).mean() # Note the ".start_time" here
ts_median = ts.groupby(ts.index.start_time.time).median() # Note the ".start_time" here
ts_quartile_1 = ts.groupby(ts.index.start_time.time).quantile(0.25) # Note the ".start_time" here
ts_quartile_3 = ts.groupby(ts.index.start_time.time).quantile(0.75) # Note the ".start_time" here
ax = ts_mean.plot(y='duration', figsize=(14, 8), color="red", label="mean", alpha=0.75)
ts_median.plot(ax=ax, color="blue", label="median", alpha=0.75)
ts_quartile_1.plot(ax=ax, color="blue", alpha=0.5, style=":", label="1st quartile")
ts_quartile_3.plot(ax=ax, color="blue", alpha=0.5, style=":", label="3rd quartile")
plt.fill_between(ts_quartile_1.index, ts_quartile_1.values, ts_quartile_3.values, facecolor='blue', alpha=0.1)
ax.legend()
ax.set_xlabel('Time');
```
### Round
```
dti = pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-1 12:00', '2018-1-2 00:31', '2018-1-2 12:25']) # Here time is not aligned (non constant frequency)
ts = pd.Series([1., 0., 2., 1.], index=dti)
print(ts)
ts.index.round('H')
```
### Count
TODO: is it the cleanest way to do this ?
```
dti = pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-1 06:45', '2018-1-1 12:00',
'2018-1-3 00:00', '2018-1-3 06:00'])
s = pd.Series(np.ones(dti.shape), index=dti)
#dti.groupby(dti.date) # it works but it returns a dictionary...
s.groupby(dti.date).count().plot.bar(color="blue", alpha=0.5);
s.resample('1d').count().plot.bar(color="blue", alpha=0.5);
```
## Plot
see https://pandas.pydata.org/pandas-docs/stable/visualization.html
```
#help(df.plot)
```
### Line plot
```
x = np.arange(0, 6, 0.1)
y1 = np.cos(x)
y2 = np.sin(x)
Y = np.array([y1, y2]).T
df = pd.DataFrame(Y,
columns=['cos(x)', 'sin(x)'],
index=x)
df.iloc[:10]
df.plot(legend=True)
```
or
```
df.plot.line(legend=True)
```
### Steps
```
df = pd.DataFrame(np.random.randn(36, 2))
df.plot(drawstyle="steps", linewidth=2)
df.plot(drawstyle="steps-post", linewidth=2);
```
### Bar plot
```
x = np.arange(0, 6, 0.5)
y1 = np.cos(x)
y2 = np.sin(x)
Y = np.array([y1, y2]).T
df = pd.DataFrame(Y,
columns=['cos(x)', 'sin(x)'],
index=x)
df
```
#### Vertical
```
df.plot.bar(legend=True)
df.plot.bar(legend=True, stacked=True)
```
#### Horizontal
```
df.plot.barh(legend=True)
```
### Histogram
```
x1 = np.random.normal(size=(10000))
x2 = np.random.normal(loc=3, scale=2, size=(10000))
X = np.array([x1, x2]).T
df = pd.DataFrame(X, columns=[r'$\mathcal{N}(0,1)$', r'$\mathcal{N}(3,2)$'])
df.plot.hist(alpha=0.5, bins=100, legend=True);
```
To normalize the $y$ axis, use `density=True`:
```
df.plot.hist(alpha=0.5, bins=100, legend=True, density=True);
```
### Box plot
```
x1 = np.random.normal(size=(10000))
x2 = np.random.normal(loc=3, scale=2, size=(10000))
X = np.array([x1, x2]).T
df = pd.DataFrame(X, columns=[r'$\mathcal{N}(0,1)$', r'$\mathcal{N}(3,2)$'])
df.plot.box()
```
### Hexbin plot
```
df = pd.DataFrame(np.random.randn(1000, 2), columns=['a', 'b'])
df['b'] = df['b'] + np.arange(1000)
df.plot.hexbin(x='a', y='b', gridsize=25)
```
### Kernel Density Estimation (KDE) plot
```
x1 = np.random.normal(size=(10000))
x2 = np.random.normal(loc=3, scale=2, size=(10000))
X = np.array([x1, x2]).T
df = pd.DataFrame(X, columns=[r'$\mathcal{N}(0,1)$', r'$\mathcal{N}(3,2)$'])
df.plot.kde()
```
### Area plot
```
df = pd.DataFrame(np.random.rand(10, 4), columns=['a', 'b', 'c', 'd'])
df.plot.area()
```
### Pie chart
```
x = np.random.randint(low=0, high=6, size=(50))
df = pd.DataFrame(x, columns=["A"])
df.A.value_counts()
df.A.value_counts().plot.pie(y="A")
```
### Scatter plot
```
x1 = np.random.normal(size=(10000))
x2 = np.random.normal(loc=3, scale=2, size=(10000))
X = np.array([x1, x2]).T
df = pd.DataFrame(X, columns=[r'$\mathcal{N}(0,1)$', r'$\mathcal{N}(3,2)$'])
df.plot.scatter(x=r'$\mathcal{N}(0,1)$',
y=r'$\mathcal{N}(3,2)$',
alpha=0.2)
```
|
github_jupyter
|
%matplotlib inline
#%matplotlib notebook
from IPython.display import display
import matplotlib
matplotlib.rcParams['figure.figsize'] = (9, 9)
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
import datetime
import pandas as pd
import numpy as np
pd.__version__
data_list = [1, 3, np.nan, 7]
series = pd.Series(data_list)
series
data_array = np.array(data_list)
series = pd.Series(data_array)
series
indices = pd.Series([1, 3, 5, 7])
series = pd.Series([10, 30, 50, 70], index=indices)
series
indices = pd.Series(['A', 'B', 'C', 'D'])
series = pd.Series([10, 30, 50, 70], index=indices)
series
data_dict = {'A': 10, 'B': 30, 'C': 50, 'D': 70}
series = pd.Series(data_dict)
series
series.index
series.values
series.shape
series.dtypes
series.describe()
type(series.describe())
series.memory_usage()
dates = pd.date_range('20130101', periods=6)
dates
dates = pd.date_range(start='2013-01-01', end='2013-01-08')
dates
dates = pd.date_range('2013-01-01', periods=4, freq='M')
dates
num_days = 7
data = np.random.random(num_days)
index = pd.date_range('2017-01-01', periods=num_days)
series = pd.Series(data, index)
series
data_list = [[1, 2, 3], [4, 5, 6]]
df = pd.DataFrame(data_array)
df
data_array = np.array([[1, 2, 3], [4, 5, 6]])
df = pd.DataFrame(data_array)
df
data = [[1, 2, 3], [4, 5, 6]]
index = [10, 20]
columns = ['A', 'B', 'C']
df = pd.DataFrame(data, index, columns)
df
data = np.array([[1, 2, 3], [4, 5, 6]])
index = np.array([10, 20])
columns = np.array(['A', 'B', 'C'])
df = pd.DataFrame(data, index=index, columns=columns)
df
data = np.array([[1, 2, 3], [4, 5, 6]])
index = pd.Series([10, 20])
columns = pd.Series(['A', 'B', 'C'])
df = pd.DataFrame(data, index=index, columns=columns)
df
data_dict = {
'A': 'foo',
'B': [10, 20, 30],
'C': 3
}
df = pd.DataFrame(data_dict)
df
data_dict = {
'A': 'foo',
'B': [10, 20, 30],
'C': 3
}
df = pd.DataFrame(data_dict, index=[10, 20, 30])
df
data_dict = [
{'A': 1, 'B': 2, 'C': 3},
{'A': 10, 'C': 30},
{'B': 200, 'C': 300}
]
df = pd.DataFrame(data_dict)
df
data_dict = {
'k1' : {'A': 1, 'B': 2, 'C': 3},
'k2' : {'A': 10, 'C': 30},
'k3' : {'B': 200, 'C': 300}
}
df = pd.DataFrame(data_dict)
df
df.T
df.index
df.columns
df.values
df.shape
df.dtypes
df.info()
df.describe()
type(df.describe())
df.memory_usage()
data_dict = {'A': 'foo',
'B': [10, 20, 30],
'C': 3}
df = pd.DataFrame(data_dict)
df
df.dtypes
df2 = df.T
df2
df2.dtypes
data_array = np.array([[1, 2, 3], [4, 5, 6]])
df = pd.DataFrame(data_array, index=[10, 20], columns=[100, 200, 300])
df
df.to_csv(path_or_buf="python_pandas_io_test.csv")
!cat python_pandas_io_test.csv
# FYI, many other options are available
df.to_csv(path_or_buf="python_pandas_io_test.csv",
sep=',',
columns=None,
header=True,
index=True,
index_label=None,
compression=None, # allowed values are 'gzip', 'bz2' or 'xz'
date_format=None)
!cat python_pandas_io_test.csv
df = pd.read_csv("python_pandas_io_test.csv")
df
df = pd.read_csv("python_pandas_io_test.csv",
sep=',',
delimiter=None,
header='infer',
names=None,
index_col=0,
usecols=None,
squeeze=False,
prefix=None,
mangle_dupe_cols=True,
dtype=None,
engine=None,
converters=None,
true_values=None,
false_values=None,
skipinitialspace=False,
skiprows=None,
nrows=None,
na_values=None,
keep_default_na=True,
na_filter=True,
verbose=False,
skip_blank_lines=True,
parse_dates=False,
infer_datetime_format=False,
keep_date_col=False,
date_parser=None,
dayfirst=False,
iterator=False,
chunksize=None,
compression='infer',
thousands=None,
decimal=b'.',
lineterminator=None,
quotechar='"',
quoting=0,
escapechar=None,
comment=None,
encoding=None,
dialect=None,
#tupleize_cols=False,
error_bad_lines=True,
warn_bad_lines=True,
skipfooter=0,
#skip_footer=0,
doublequote=True,
delim_whitespace=False,
#as_recarray=False,
#compact_ints=False,
#use_unsigned=False,
low_memory=True,
#buffer_lines=None,
memory_map=False,
float_precision=None)
df
!rm python_pandas_io_test.csv
import io
df.to_json(path_or_buf="python_pandas_io_test.json")
!cat python_pandas_io_test.json
df.to_json(path_or_buf="python_pandas_io_test_split.json",
orient="split")
!cat python_pandas_io_test_split.json
df.to_json(path_or_buf="python_pandas_io_test_records.json",
orient="records")
!cat python_pandas_io_test_records.json
df.to_json(path_or_buf="python_pandas_io_test_index.json",
orient="index")
!cat python_pandas_io_test_index.json
df.to_json(path_or_buf="python_pandas_io_test_columns.json",
orient="columns")
!cat python_pandas_io_test_columns.json
df.to_json(path_or_buf="python_pandas_io_test_values.json",
orient="values")
!cat python_pandas_io_test_values.json
# FYI, many other options are available
df.to_json(path_or_buf="python_pandas_io_test.json",
orient='columns', # For DataFrame: 'split','records','index','columns' or 'values'
date_format=None, # None, 'epoch' or 'iso'
double_precision=10,
force_ascii=True,
date_unit='ms')
!cat python_pandas_io_test.json
!cat python_pandas_io_test_split.json
df = pd.read_json("python_pandas_io_test_split.json",
orient="split")
df
!cat python_pandas_io_test_records.json
df = pd.read_json("python_pandas_io_test_records.json",
orient="records")
df
!cat python_pandas_io_test_index.json
df = pd.read_json("python_pandas_io_test_index.json",
orient="index")
df
!cat python_pandas_io_test_columns.json
df = pd.read_json("python_pandas_io_test_columns.json",
orient="columns")
df
!cat python_pandas_io_test_values.json
df = pd.read_json("python_pandas_io_test_values.json",
orient="values")
df
df = pd.read_json("python_pandas_io_test.json",
orient=None,
typ='frame',
dtype=True,
convert_axes=True,
convert_dates=True,
keep_default_dates=True,
numpy=False,
precise_float=False,
date_unit=None,
encoding=None,
lines=False)
df
!rm python_pandas_io_test*.json
!echo "- {A: 1, B: 2}" > python_pandas_io_test.yaml
!echo "- {A: 3}" >> python_pandas_io_test.yaml
!echo "- {B: 4}" >> python_pandas_io_test.yaml
!cat python_pandas_io_test.yaml
try:
import yaml
with open('python_pandas_io_test.yaml', 'r') as f:
df = pd.io.json.json_normalize(yaml.load(f))
print(df)
except:
pass
!rm python_pandas_io_test.yaml
data_array = np.array([np.arange(1, 10, 1), np.arange(10, 100, 10), np.arange(100, 1000, 100)]).T
df = pd.DataFrame(data_array,
index=np.arange(1, 10, 1),
columns=['A', 'B', 'C'])
df
df.B
df["B"]
df.loc[:,"B"]
df.iloc[:,1]
df[['A','B']]
df.loc[:,['A','B']]
df.iloc[:,0:2]
data_array = np.array([np.arange(1, 10, 1), np.arange(10, 100, 10), np.arange(100, 1000, 100)]).T
df = pd.DataFrame(data_array,
index=["i" + str(i+1) for i in range(9)],
columns=['A', 'B', 'C'])
df
df.loc["i3"]
df.loc["i3",:]
df.iloc[2] # Select over index
df.iloc[2,:] # Select over index
df.loc[["i3", "i4"],:]
df.iloc[2:4,:] # Select over index
df.B < 50.
type(df.B < 50.)
df[[True, True, True, True, False, False, False, False, False]]
series_mask = pd.Series({'i1': True,
'i2': True,
'i3': True,
'i4': True,
'i5': False,
'i6': False,
'i7': False,
'i8': False,
'i9': False})
df[series_mask]
df[df.B < 50.]
df[df['B'] < 50.]
df[(df.A >= 2) & (df.B < 50)]
df.loc[(df.A >= 2) & (df.B < 50)]
df = pd.DataFrame([[4000, 10, 3],
[100, 200, 1],
[30, 1000, 10]], columns=['a', 'b', 'c'], index=['k1', 'k2', 'k3'])
df
df.c.idxmin()
df.loc[ df.c.idxmin() ]
data_array = np.array([np.arange(1, 10, 1), np.arange(10, 100, 10), np.arange(100, 1000, 100)]).T
df = pd.DataFrame(data_array,
index=np.arange(1, 10, 1),
columns=['A', 'B', 'C'])
df
df[(df.A >= 2) & (df.B < 50)]
df[(df.B < 20) | (df.B > 50)]
df.loc[(df.B < 20) | (df.B > 50), 'C']
df[(df['A'] >= 2) & (df['B'] < 50)]
df.loc[(df.A >= 2) & (df.B < 50), ['A','B']]
data_array = np.array([np.arange(1, 10, 1), np.arange(10, 100, 10), np.arange(100, 1000, 100)]).T
df = pd.DataFrame(data_array,
index=np.arange(1, 10, 1),
columns=['A', 'B', 'C'])
df
df.B *= 2.
df
df.B = pow(df.B, 2)
df
data_array = np.array([np.arange(1, 10, 1), np.arange(10, 100, 10), np.arange(100, 1000, 100)]).T
df = pd.DataFrame(data_array,
index=np.arange(1, 10, 1),
columns=['A', 'B', 'C'])
df
df[df.B < 50.] *= -1.
df
# df['B'][df['B'] < 50.] = 0 # OK but chain indexing is bad...
# df.A[df.B < 50.] = 0 # OK but chain indexing is bad...
df.loc[df.B < 50., 'A'] = 0
df
df.loc[(df.B < 50.) & (df.B > 20), 'C'] = 0
df
df.loc[(df.B < 20) | (df.B > 50), 'C'] = -1
df
df[df.B < 50.] = pow(df[df.B < 50.], 2)
df
data_array = np.array([np.arange(1, 10, 1), np.arange(10, 100, 10), np.arange(100, 1000, 100)]).T
df = pd.DataFrame(data_array,
index=np.arange(1, 10, 1),
columns=['A', 'B', 'C'])
df
df.sample(n=30, replace=True)
df.sample(frac=0.9, replace=True)
df.sample(n=3)
df.sample(frac=0.9)
df.sample(n=30, replace=True, weights=np.arange(len(df)))
data_array = np.array([np.arange(1, 10, 1), np.arange(10, 100, 10), np.arange(100, 1000, 100)]).T
df = pd.DataFrame(data_array,
index=np.arange(1, 10, 1),
columns=['A', 'B', 'C'])
df
df = df.sample(frac=1)
df
df = df.sample(frac=1).reset_index(drop=True)
df
data_array = np.array([np.arange(1, 5, 1), np.arange(10, 50, 10), np.arange(100, 500, 100)]).T
df = pd.DataFrame(data_array,
columns=['A', 'B', 'C'])
df
df = df[['B', 'C', 'A']]
df
NROWS = 7
col1 = np.arange(1., NROWS, 1)
col2 = np.arange(10., NROWS*10, 10)
col3 = np.arange(100., NROWS*100, 100)
np.random.shuffle(col1)
np.random.shuffle(col2)
np.random.shuffle(col3)
data = np.array([col1,
col2,
col3]).T
index = np.arange(1, NROWS, 1)
columns = np.array(['A', 'B', 'C'])
np.random.shuffle(index)
np.random.shuffle(data)
np.random.shuffle(columns)
df = pd.DataFrame(data,
index=index,
columns=columns)
df
df.sort_index()
df.sort_index(axis=0) # axis=0 -> sort by row index
df.sort_index(ascending=False)
df.sort_index(axis=1) # axis=1 -> sort by column label
df.sort_index(axis=1, ascending=False)
df.sort_values(by='B')
df.sort_values(by='B', ascending=False)
df.sort_values(by='B', inplace=True)
df
a = np.array([[3, np.nan, 5, np.nan, 7],
[2, 4, np.nan, 3, 1],
[3, 4, 5, 6, 1]]).T
df = pd.DataFrame(a,
columns=['A', 'B', 'C'])
df
df.isnull()
df.dropna()
df.dropna(how='any') # but 'any' is the default value...
df.dropna(subset=['B'])
df.dropna(subset=['B', 'C'])
df.dropna(axis=1)
df.dropna(axis=1, how='any') # but 'any' is the default value...
df.dropna(axis=1, subset=[2])
df.dropna(axis=1, subset=[1, 2])
df.fillna(value=999)
df.A.isnull().sum()
data_array = np.array([np.arange(1, 10, 1), np.arange(10, 100, 10), np.arange(100, 1000, 100)]).T
df = pd.DataFrame(data_array,
index=np.arange(1, 10, 1),
columns=['A', 'B', 'C'])
df
df.T
a1 = np.array([np.arange(1, 5, 1), np.arange(10, 50, 10), np.arange(100, 500, 100)]).T
df1 = pd.DataFrame(a1,
columns=['ID', 'B', 'C'])
a2 = np.array([np.arange(1, 5, 1), np.arange(1000, 5000, 1000), np.arange(10000, 50000, 10000)]).T
df2 = pd.DataFrame(a2,
columns=['ID', 'B', 'C'])
display(df1)
display(df2)
df = pd.merge(df1, df2, on="ID", suffixes=('_1', '_2')) #.dropna(how='any')
display(df)
a1 = np.array([np.arange(1, 5, 1), np.arange(10, 50, 10), np.arange(100, 500, 100)]).T
df1 = pd.DataFrame(a1,
columns=['ID', 'B', 'C'])
a2 = np.array([np.arange(1, 5, 1), np.arange(1000, 5000, 1000), np.arange(10000, 50000, 10000)]).T
df2 = pd.DataFrame(a2,
columns=['ID', 'B', 'C'])
df1.iloc[0,2] = np.nan
df1.iloc[1,1] = np.nan
df1.iloc[2,2] = np.nan
df1.iloc[3,1] = np.nan
df2.iloc[0,1] = np.nan
df2.iloc[1,2] = np.nan
df2.iloc[2,1] = np.nan
df2.iloc[3,2] = np.nan
df = pd.merge(df1, df2, on="ID", suffixes=('_1', '_2')) #.dropna(how='any')
display(df1)
display(df2)
display(df)
a1 = np.array([np.arange(1, 5, 1), np.arange(10, 50, 10), np.arange(100, 500, 100)]).T
df1 = pd.DataFrame(a1,
columns=['ID', 'B', 'C'])
a2 = np.array([np.arange(1, 3, 1), np.arange(1000, 3000, 1000), np.arange(10000, 30000, 10000)]).T
df2 = pd.DataFrame(a2,
columns=['ID', 'B', 'C'])
display(df1)
display(df2)
print("Left: use only keys from left frame (SQL: left outer join)")
df = pd.merge(df1, df2, on="ID", how="left", suffixes=('_1', '_2')) #.dropna(how='any')
display(df)
print("Right: use only keys from right frame (SQL: right outer join)")
df = pd.merge(df1, df2, on="ID", how="right", suffixes=('_1', '_2')) #.dropna(how='any')
display(df)
print("Inner: use intersection of keys from both frames (SQL: inner join) [DEFAULT]")
df = pd.merge(df1, df2, on="ID", how="inner", suffixes=('_1', '_2')) #.dropna(how='any')
display(df)
print("Outer: use union of keys from both frames (SQL: full outer join)")
df = pd.merge(df1, df2, on="ID", how="outer", suffixes=('_1', '_2')) #.dropna(how='any')
display(df)
a = np.array([[3, 5, 5, 5, 7, 7, 7, 7],
[2, 4, 5, 3, 3, 2, 3, 2],
[3, 5, 4, 6, 8, 1, 9, 8]]).T
df = pd.DataFrame(a,
columns=['A', 'B', 'C'])
df
df.groupby(["A"]).count()
df.groupby(["A"]).sum().B
df.groupby(["A"]).mean().B
df.groupby("A").C.idxmin()
df.loc[ df.groupby("A").C.idxmin() ]
df.groupby("A").first()
df.groupby("A").head(1)
df.groupby(["A","B"]).count()
s = pd.Series([1., 0., 5., 2., 1.])
print("DATA:")
print(s)
mean_s = s.rolling(2).mean()
print()
print("ROLLING MEAN:")
print(mean_s)
sum_s = s.rolling(2).sum()
print()
print("ROLLING SUM:")
print(sum_s)
min_s = s.rolling(2).min()
print()
print("ROLLING MIN:")
print(min_s)
max_s = s.rolling(2).max()
print()
print("ROLLING MAX:")
print(max_s)
ax = s.plot(figsize=(18, 3), color="blue")
mean_s.plot(color="red", label="mean", ax=ax)
sum_s.plot(color="green", label="sum", style="--", alpha=0.5, ax=ax)
min_s.plot(color="black", label="min", style=":", alpha=0.25, ax=ax)
max_s.plot(color="black", label="max", style=":", alpha=0.25, ax=ax)
ax.legend();
index = np.arange(0, 20, 0.05)
s = pd.Series(np.sin(index))
s = s + np.random.normal(scale=0.4, size=s.shape)
ax = s.plot(figsize=(18, 3))
s.shape
s_mean = s.rolling(20).mean()
s_median = s.rolling(20).median()
s_min = s.rolling(20).min()
s_max = s.rolling(20).max()
ax = s_mean.plot(y='duration', figsize=(18, 8), color="red", label="mean", alpha=0.75)
s_median.plot(ax=ax, color="blue", label="median", alpha=0.75)
s_min.plot(ax=ax, color="blue", alpha=0.5, style=":", label="min")
s_max.plot(ax=ax, color="blue", alpha=0.5, style=":", label="max")
plt.fill_between(s_min.index, s_min.values, s_max.values, facecolor='blue', alpha=0.1)
ax.legend()
ax.set_xlabel('Time');
s_mean.shape
df = pd.DataFrame([["i1", "A", 1],
["i1", "B", 2],
["i2", "A", 3],
["i2", "B", 4]], columns=["foo", "bar", "baz"])
df
df.pivot(index="foo", columns="bar", values="baz")
a = np.array([[3, 5, 5, 5, 7, 7, 7, 7],
[2, 4, 4, 3, 1, 3, 3, 2],
[3, 4, 5, 6, 1, 8, 9, 8]]).T
df = pd.DataFrame(a,
columns=['A', 'B', 'C'])
df
df.A.value_counts()
df.A.value_counts().plot.bar()
df = pd.DataFrame(np.random.normal(size=100000))
df.quantile(0.50)
df.quantile([0.25, 0.75])
df.quantile([0.01, 0.001])
pd.Timestamp(year=2018, month=1, day=1, hour=12, minute=30)
p = pd.Period(freq='D', year=2018, month=1, day=1, hour=12, minute=30)
print(p)
print("Start time:", p.start_time)
print("End time:", p.end_time)
print(pd.Timedelta(days=5, seconds=30))
ts1 = pd.Timestamp(year=2018, month=1, day=1, hour=12, minute=30)
ts2 = pd.Timestamp(year=2018, month=1, day=2, hour=12, minute=30)
print(ts2 - ts1)
pd.date_range('2018-01-01', '2018-03-01', freq='D')
pd.date_range('2018-01-01', periods=10, freq='h')
pd.date_range('1/1/2012', periods=10, freq='S')
pd.date_range('3/6/2012 00:00', periods=5, freq='D')
pd.date_range('1/1/2012', periods=5, freq='M')
pd.period_range('2018-01-01', '2018-03-01', freq='D')
pd.date_range('2018-01-01', '2018-03-01', freq='D').to_period()
dti = pd.date_range('2012-01-01 00:00', periods=40, freq='D')
ts = pd.Series(np.random.randint(0, 200, len(dti)), index=dti)
ts.plot();
ts.plot(x_compat=True);
dti = pd.date_range('2018-01-01 00:00', '2018-01-03 00:00', freq='H')
ts = pd.Series(np.random.randint(0, 100, len(dti)), index=dti)
ax = ts.plot(x_compat=True, figsize=(16, 4)) # x_compat is required as matplotlib doesn't understand pandas datetime format -> x_compat=True makes the conversion...
# set monthly locator
ax.xaxis.set_major_locator(mdates.DayLocator(interval=1))
ax.xaxis.set_minor_locator(mdates.HourLocator(interval=1))
# set formatter
ax.xaxis.set_major_formatter(mdates.DateFormatter('%d-%m-%Y'))
# set font and rotation for date tick labels
plt.gcf().autofmt_xdate()
dti = pd.date_range('2012-1-1 00:00', periods=40, freq='D')
ts = pd.Series(np.random.randint(0, 200, len(dti)), index=dti)
ts
ts["2012-01-09"]
ts[datetime.datetime(2012, 1, 9)]
ts[ts.index < "2012-01-09"]
ts[ts.index > "2012-01-20"]
ts["2012-01-09":"2012-01-20"]
ts[datetime.datetime(2012, 1, 9):datetime.datetime(2012, 1, 20)]
ts[ts.index.day <= 3]
ts[ts.index.month == 2]
ts["2012-02"]
ts[ts.index.dayofweek == 1]
dti = pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-1 06:45', '2018-1-1 12:00',
'2018-1-2 00:00', '2018-1-2 06:00', '2018-1-2 12:00'])
ts = pd.Series([2., 1., 3., 2., 2., 0.], index=dti)
print("DATA:")
print(ts)
ax = ts.plot(figsize=(18, 3), style="*-", color="blue")
ax.vlines(pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-2 00:00']), ymin=0, ymax=8, color="red", linestyle=":", alpha=0.3);
ts_rw = ts.rolling('D').sum() # Rolling window size: 1 day
print()
print("MEAN:")
print(ts_rw)
ts_rw.plot(color="red", label="sum", style="*-", alpha=0.75, ax=ax)
ax.legend()
ax.set_xlabel('Time')
ax.grid(True);
ts.rolling('6h').min()
ts.rolling('3h').mean()
dti = pd.date_range('1/1/2018 00:00', periods=6*480, freq='10min')
ts = pd.Series(np.sin(dti.hour * 2. * np.pi / 24.), index=dti)
ts = ts + np.random.normal(scale=0.4, size=ts.shape)
ax = ts.plot(figsize=(18, 3))
ax.vlines(pd.date_range('1/1/2018 00:00', periods=480/24, freq='D'), ymin=-2, ymax=2, color="red", linestyle=":", alpha=0.3);
ts.shape
ts_mean = ts.rolling('5H').mean()
ts_median = ts.rolling('5H').median()
ts_min = ts.rolling('5H').min()
ts_max = ts.rolling('5H').max()
ax = ts_mean.plot(y='duration', figsize=(18, 3), color="red", label="mean", alpha=0.75)
ts_median.plot(ax=ax, color="blue", label="median", alpha=0.75)
ts_min.plot(ax=ax, color="blue", alpha=0.5, style=":", label="min")
ts_max.plot(ax=ax, color="blue", alpha=0.5, style=":", label="max")
plt.fill_between(ts_min.index, ts_min.values, ts_max.values, facecolor='blue', alpha=0.1)
ax.legend()
ax.set_xlabel('Time');
ts_mean.shape
dti = pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-1 06:45', '2018-1-1 12:00',
'2018-1-2 00:00', '2018-1-2 12:00'])
ts = pd.Series([1., 0., 5., 2., 0.], index=dti)
print("DATA:")
print(ts)
ax = ts.plot(figsize=(18, 3), style="*-", color="blue")
ax.vlines(pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-2 00:00']), ymin=0, ymax=5, color="red", linestyle=":", alpha=0.3);
ts_resampled = ts.resample('D').mean()
print()
print("MEAN:")
print(ts_resampled)
ts_resampled.plot(color="red", style="*-", label="mean", alpha=0.75, ax=ax)
ax.legend()
ax.set_xlabel('Time');
ts.resample('6h').min()
ts.resample('3h').sum()
dti = pd.DatetimeIndex(['2018-1-1 12:00',
'2018-1-2 08:00', '2018-1-2 18:00', '2018-1-2 23:59:59',
'2018-1-3 00:00'])
ts = pd.Series([0.,
10., 20., 30.,
5.], index=dti)
print("DATA:")
print(ts)
ts_resampled = ts.resample('D').mean()
print()
print("MEAN:")
print(ts_resampled)
# Illustrative plot
ax = ts.plot(x_compat=True, figsize=(18, 3), style="*-", color="blue")
ax.vlines(pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-2 00:00', '2018-1-3 00:00']), ymin=-10, ymax=40, color="red", linestyle=":", linewidth=2, alpha=0.5);
ax.vlines(pd.DatetimeIndex(['2018-1-1 12:00', '2018-1-2 12:00', '2018-1-3 12:00']), ymin=-10, ymax=40, color="green", linestyle=":", linewidth=2, alpha=0.5);
ax.plot(pd.DatetimeIndex(['2018-1-1 12:15', '2018-1-2 11:45']), [40, 40], marker="|", markersize=20, color="green")
ax.plot(pd.DatetimeIndex(['2018-1-2 12:15', '2018-1-3 11:45']), [40, 40], marker="|", markersize=20, color="green")
ax.plot(pd.DatetimeIndex(['2018-1-1 00:15', '2018-1-1 23:45']), [35, 35], marker="|", markersize=20, color="red")
ax.plot(pd.DatetimeIndex(['2018-1-2 00:15', '2018-1-2 23:45']), [35, 35], marker="|", markersize=20, color="red")
ts_resampled.plot(color="red", style="*-", label="mean", alpha=0.75, ax=ax)
# set monthly locator
ax.xaxis.set_major_locator(mdates.DayLocator(interval=1))
ax.xaxis.set_minor_locator(mdates.HourLocator(interval=1))
# set formatter
ax.xaxis.set_major_formatter(mdates.DateFormatter('%d-%m-%Y %H:%M'))
# set font and rotation for date tick labels
plt.gcf().autofmt_xdate()
ax.legend()
ax.set_xlabel('Time');
dti = pd.DatetimeIndex(['2018-1-1 01:00',
'2018-1-1 05:30', '2018-1-1 07:30',
'2018-1-1 10:00'])
ts = pd.Series([0.,
10., 20.,
5.], index=dti)
print("DATA:")
print(ts)
ts_resampled = ts.resample('5h').mean()
print()
print("MEAN:")
print(ts_resampled)
# Illustrative plot
ax = ts.plot(x_compat=True, figsize=(18, 3), style="*-", color="blue")
ax.vlines(pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-1 05:00', '2018-1-1 10:00']), ymin=-10, ymax=40, color="red", linestyle=":", linewidth=2, alpha=0.5);
ax.vlines(pd.DatetimeIndex(['2018-1-1 01:00', '2018-1-1 06:00', '2018-1-1 11:00']), ymin=-10, ymax=40, color="green", linestyle=":", linewidth=2, alpha=0.5);
ax.plot(pd.DatetimeIndex(['2018-1-1 01:05', '2018-1-1 05:55']), [40, 40], marker="|", markersize=20, color="green")
ax.plot(pd.DatetimeIndex(['2018-1-1 06:05', '2018-1-1 10:55']), [40, 40], marker="|", markersize=20, color="green")
ax.plot(pd.DatetimeIndex(['2018-1-1 00:05', '2018-1-1 04:55']), [35, 35], marker="|", markersize=20, color="red")
ax.plot(pd.DatetimeIndex(['2018-1-1 05:05', '2018-1-1 09:55']), [35, 35], marker="|", markersize=20, color="red")
ts_resampled.plot(color="red", style="*-", label="mean", alpha=0.75, ax=ax)
# set monthly locator
ax.xaxis.set_major_locator(mdates.HourLocator(interval=1))
#ax.xaxis.set_minor_locator(mdates.HourLocator(interval=1))
# set formatter
ax.xaxis.set_major_formatter(mdates.DateFormatter('%H:%M'))
# set font and rotation for date tick labels
plt.gcf().autofmt_xdate()
ax.legend()
ax.set_xlabel('Time');
dti = pd.date_range('1/1/2018 00:00', periods=60*480, freq='min')
ts = pd.Series(np.sin(dti.hour * 2. * np.pi / 24.), index=dti)
ts = ts + np.random.normal(scale=0.4, size=ts.shape)
ax = ts.plot(figsize=(18, 3))
ax.vlines(pd.date_range('1/1/2018 00:00', periods=480/24, freq='D'), ymin=-2, ymax=2, color="red", linestyle=":", alpha=0.3);
ts.shape
ts_mean = ts.resample('2H').mean()
ts_median = ts.resample('2H').median()
ts_min = ts.resample('2H').min()
ts_max = ts.resample('2H').max()
ax = ts_mean.plot(y='duration', figsize=(18, 8), color="red", label="mean", alpha=0.75)
ts_median.plot(ax=ax, color="blue", label="median", alpha=0.75)
ts_min.plot(ax=ax, color="blue", alpha=0.5, style=":", label="min")
ts_max.plot(ax=ax, color="blue", alpha=0.5, style=":", label="max")
plt.fill_between(ts_min.index, ts_min.values, ts_max.values, facecolor='blue', alpha=0.1)
ax.legend()
ax.set_xlabel('Time');
ts_mean.shape
rolling_window = '6H'
start = '2018-1-1 00:00'
end = '2018-1-4 00:00'
dti = pd.date_range(start=start, end=end, freq='min')
ts = pd.Series(np.sin(dti.hour * 2. * np.pi / 24.), index=dti)
ts = ts + np.random.normal(scale=0.4, size=ts.shape)
ax = ts.plot(figsize=(18, 3))
ax.vlines(pd.date_range(start=start, end=end, freq=rolling_window), ymin=-2, ymax=2, color="red", linestyle=":", alpha=0.5);
ts2 = ts.rolling(rolling_window).mean() # Rolling window size: 1 day
ax = ts2.plot(figsize=(18, 3), color="red", alpha=0.75)
ax.vlines(pd.date_range(start=start, end=end, freq=rolling_window), ymin=-1, ymax=1, color="red", linestyle=":", alpha=0.5);
ts2 = ts.resample(rolling_window).mean() # Rolling window size: 1 day
ax = ts2.plot(figsize=(18, 3), color="red", alpha=0.75)
ax.vlines(pd.date_range(start=start, end=end, freq=rolling_window), ymin=-1, ymax=1, color="red", linestyle=":", alpha=0.5);
dti = pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-1 12:00', '2018-1-2 00:00', '2018-1-2 12:00'])
ts = pd.Series([1., 0., 2., 1.], index=dti)
print(ts)
ax = ts.plot(figsize=(18, 3))
dti = pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-2 00:00'])
ax.vlines(dti, ymin=0, ymax=2, color="red", linestyle=":", alpha=0.3);
ts_mean = ts.groupby(ts.index.time).mean()
print(ts_mean)
ax = ts_mean.plot(y='duration', figsize=(10, 4), color="red", label="mean", alpha=0.75)
ax.legend()
ax.set_xlabel('Time');
dti = pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-1 12:00', '2018-1-2 00:31', '2018-1-2 12:25']) # Here time is not aligned (non constant frequency)
ts = pd.Series([1., 0., 2., 1.], index=dti)
print(ts)
ax = ts.plot(figsize=(18, 3));
ts_mean = ts.groupby(ts.index.time).mean()
print(ts_mean)
ax = ts_mean.plot(y='duration', figsize=(10, 4), color="red", label="mean", alpha=0.75)
ax.legend()
ax.set_xlabel('Time');
dti = pd.date_range('1/1/2018 00:00', periods=960, freq='h')
ts = pd.Series(np.sin(dti.hour * 2. * np.pi / 24.), index=dti)
ts = ts + np.random.normal(scale=0.4, size=ts.shape)
ax = ts.plot(figsize=(18, 3))
ax.vlines(pd.date_range('1/1/2018 00:00', periods=960/24, freq='D'), ymin=-2, ymax=2, color="red", linestyle=":", alpha=0.3);
ts_mean = ts.groupby(ts.index.time).mean()
ts_median = ts.groupby(ts.index.time).median()
ts_quartile_1 = ts.groupby(ts.index.time).quantile(0.25)
ts_quartile_3 = ts.groupby(ts.index.time).quantile(0.75)
ax = ts_mean.plot(y='duration', figsize=(14, 8), color="red", label="mean", alpha=0.75)
ts_median.plot(ax=ax, color="blue", label="median", alpha=0.75)
ts_quartile_1.plot(ax=ax, color="blue", alpha=0.5, style=":", label="1st quartile")
ts_quartile_3.plot(ax=ax, color="blue", alpha=0.5, style=":", label="3rd quartile")
plt.fill_between(ts_quartile_1.index, ts_quartile_1.values, ts_quartile_3.values, facecolor='blue', alpha=0.1)
ax.legend()
ax.set_xlabel('Time');
dti = pd.period_range('1/1/2018 00:00', periods=960, freq='h')
ts = pd.Series(np.sin(dti.hour * 2. * np.pi / 24.), index=dti)
ts = ts + np.random.normal(scale=0.4, size=ts.shape)
ax = ts.plot(figsize=(18, 3))
ax.vlines(pd.date_range('1/1/2018 00:00', periods=960/24, freq='D'), ymin=-2, ymax=2, color="red", linestyle=":", alpha=0.3);
ts_mean = ts.groupby(ts.index.start_time.time).mean() # Note the ".start_time" here
ts_median = ts.groupby(ts.index.start_time.time).median() # Note the ".start_time" here
ts_quartile_1 = ts.groupby(ts.index.start_time.time).quantile(0.25) # Note the ".start_time" here
ts_quartile_3 = ts.groupby(ts.index.start_time.time).quantile(0.75) # Note the ".start_time" here
ax = ts_mean.plot(y='duration', figsize=(14, 8), color="red", label="mean", alpha=0.75)
ts_median.plot(ax=ax, color="blue", label="median", alpha=0.75)
ts_quartile_1.plot(ax=ax, color="blue", alpha=0.5, style=":", label="1st quartile")
ts_quartile_3.plot(ax=ax, color="blue", alpha=0.5, style=":", label="3rd quartile")
plt.fill_between(ts_quartile_1.index, ts_quartile_1.values, ts_quartile_3.values, facecolor='blue', alpha=0.1)
ax.legend()
ax.set_xlabel('Time');
dti = pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-1 12:00', '2018-1-2 00:31', '2018-1-2 12:25']) # Here time is not aligned (non constant frequency)
ts = pd.Series([1., 0., 2., 1.], index=dti)
print(ts)
ts.index.round('H')
dti = pd.DatetimeIndex(['2018-1-1 00:00', '2018-1-1 06:45', '2018-1-1 12:00',
'2018-1-3 00:00', '2018-1-3 06:00'])
s = pd.Series(np.ones(dti.shape), index=dti)
#dti.groupby(dti.date) # it works but it returns a dictionary...
s.groupby(dti.date).count().plot.bar(color="blue", alpha=0.5);
s.resample('1d').count().plot.bar(color="blue", alpha=0.5);
#help(df.plot)
x = np.arange(0, 6, 0.1)
y1 = np.cos(x)
y2 = np.sin(x)
Y = np.array([y1, y2]).T
df = pd.DataFrame(Y,
columns=['cos(x)', 'sin(x)'],
index=x)
df.iloc[:10]
df.plot(legend=True)
df.plot.line(legend=True)
df = pd.DataFrame(np.random.randn(36, 2))
df.plot(drawstyle="steps", linewidth=2)
df.plot(drawstyle="steps-post", linewidth=2);
x = np.arange(0, 6, 0.5)
y1 = np.cos(x)
y2 = np.sin(x)
Y = np.array([y1, y2]).T
df = pd.DataFrame(Y,
columns=['cos(x)', 'sin(x)'],
index=x)
df
df.plot.bar(legend=True)
df.plot.bar(legend=True, stacked=True)
df.plot.barh(legend=True)
x1 = np.random.normal(size=(10000))
x2 = np.random.normal(loc=3, scale=2, size=(10000))
X = np.array([x1, x2]).T
df = pd.DataFrame(X, columns=[r'$\mathcal{N}(0,1)$', r'$\mathcal{N}(3,2)$'])
df.plot.hist(alpha=0.5, bins=100, legend=True);
df.plot.hist(alpha=0.5, bins=100, legend=True, density=True);
x1 = np.random.normal(size=(10000))
x2 = np.random.normal(loc=3, scale=2, size=(10000))
X = np.array([x1, x2]).T
df = pd.DataFrame(X, columns=[r'$\mathcal{N}(0,1)$', r'$\mathcal{N}(3,2)$'])
df.plot.box()
df = pd.DataFrame(np.random.randn(1000, 2), columns=['a', 'b'])
df['b'] = df['b'] + np.arange(1000)
df.plot.hexbin(x='a', y='b', gridsize=25)
x1 = np.random.normal(size=(10000))
x2 = np.random.normal(loc=3, scale=2, size=(10000))
X = np.array([x1, x2]).T
df = pd.DataFrame(X, columns=[r'$\mathcal{N}(0,1)$', r'$\mathcal{N}(3,2)$'])
df.plot.kde()
df = pd.DataFrame(np.random.rand(10, 4), columns=['a', 'b', 'c', 'd'])
df.plot.area()
x = np.random.randint(low=0, high=6, size=(50))
df = pd.DataFrame(x, columns=["A"])
df.A.value_counts()
df.A.value_counts().plot.pie(y="A")
x1 = np.random.normal(size=(10000))
x2 = np.random.normal(loc=3, scale=2, size=(10000))
X = np.array([x1, x2]).T
df = pd.DataFrame(X, columns=[r'$\mathcal{N}(0,1)$', r'$\mathcal{N}(3,2)$'])
df.plot.scatter(x=r'$\mathcal{N}(0,1)$',
y=r'$\mathcal{N}(3,2)$',
alpha=0.2)
| 0.323701 | 0.989034 |
```
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from datetime import datetime as dt
from pandas_datareader import DataReader as DR
import seaborn as sb
import numdifftools as nd
from wquantiles import quantile
import statsmodels.api as sm
from statsmodels.stats.outliers_influence import variance_inflation_factor as VIF
from scipy.stats import norm,t,truncnorm
from scipy.stats import multivariate_normal as mvnorm
from scipy.stats import multivariate_t as mvt
from scipy.spatial import Delaunay as TRI
from scipy.interpolate import LinearNDInterpolator as ITP
from scipy.optimize import minimize,root
from scipy.optimize import NonlinearConstraint as NonlinCons
from scipy.stats import gaussian_kde as sciKDE
from sklearn.linear_model import LinearRegression as Linear
from sklearn.linear_model import Ridge
from sklearn.linear_model import Lasso
from sklearn.neighbors import KernelDensity as sklKDE
import warnings
warnings.filterwarnings("ignore")
```
# Define the experiment class
```
class MLE:
def __init__(self,dim,sigma,df,mode=1):
self.dim=dim
self.T=lambda x: mvnorm.pdf(x=x,mean=np.zeros(dim))
if mode==1:
self.iP=lambda x: mvt.pdf(x=x,loc=np.zeros(dim),shape=sigma**2,df=df)
self.iS=lambda size: mvt.rvs(size=size,loc=np.zeros(dim),shape=sigma**2,df=df)
else:
self.iP=lambda x: np.prod([t.pdf(x=x[:,i],scale=sigma,df=df) for i in range(dim)],axis=0)
self.iS=lambda size: np.array([t.rvs(size=size,scale=sigma,df=df) for i in range(dim)]).T
def __estimate(self,W,name,asym=True):
Z=np.mean(W)
err=np.abs(Z-1)
if asym:
aVar=np.var(W)
aErr=np.sqrt(aVar/W.size)
ESS=1/np.sum((W/np.sum(W))**2)
print('{} est: {:.4f}; err: {:.4f}; a-var: {:.4f}; a-err: {:.4f}; ESS: {:.0f}/{}'\
.format(name,Z,err,aVar,aErr,ESS,W.size))
else:
print('{} est: {:.4f}; err: {:.4f}'.format(name,Z,err))
def estimate_IS(self,size):
S=self.iS(size)
W=self.T(S)/self.iP(S)
self.__estimate(W,'IS')
def draw_TP(self,P,x,name,dim=0):
X=np.zeros([x.size,self.dim])
X[:,dim]=x
ratio=np.reshape(self.T(np.zeros(self.dim))/P(np.zeros([1,self.dim])),1)[0]
print('------------ pdf ratio at origin: {:.2f} ------------'.format(ratio))
fig,ax=plt.subplots(figsize=(7,4))
ax.plot(x,self.T(X))
ax.plot(x,P(X))
if name=='nonparametric':
one=np.zeros(self.dim)
one[dim]=1
rW=np.array([self.h(one*loc,loc) for loc in self.rS])
rW=rW/rW.max()*P(np.zeros([1,self.dim]))[0]
rWmeans=np.ones_like(rW)*rW.mean()
ax.plot(x,self.mP(X))
ax.hist(self.rS[:,dim],bins=2*rW.size,weights=rWmeans)
ax.hist(self.rS[:,dim],bins=2*rW.size,weights=rW)
ax.legend(['target','nonparametric proposal','mixture proposal','centers','centers with weight'])
elif name=='regression':
G=self.G(X)
rPO=self.regO.coef_.dot(G)+self.regO.intercept_*P(X)
rPL=self.regL.coef_.dot(G)+self.regL.intercept_*P(X)
mid=int(x.size/2)
print('regression ratios: ordinary {:.4f}, lasso {:.4f}'\
.format(self.T(X[mid])/rPO[mid],self.T(X[mid])/rPL[mid]))
ax.plot(x,rPO)
ax.plot(x,rPL)
ax.legend(['target','mixture proposal','ordinary regression','lasso regression'])
else:
ax.legend(['target','{} proposal'.format(name)])
ax.set_title('{}-D target and {} proposal (cross-sectional view)'.format(self.dim,name))
plt.show()
def resample(self,size,ratio):
S=self.iS(ratio*size)
p=self.T(S)/self.iP(S)
index=np.arange(S.shape[0])
choice=np.random.choice(index,size,p=p/np.sum(p),replace=True)
self.rS=S[choice]
self.rSset=S[list(set(choice))]
print('resampling rate: {}/{}'.format(self.rSset.shape[0],size))
def estimate_NIS(self,size,rate,bdwth='scott',reg=0):
if(type(bdwth)==str):
method=bdwth
tmp=sciKDE(self.rS.T,bw_method=method)
bdwth=np.mean(np.sqrt(np.diag(tmp.covariance_factor()*np.cov(self.rS.T))))
print('bdwth: {:.4f} (based on {})'.format(bdwth,method))
self.bdwth=bdwth
self.kde=sklKDE(kernel='gaussian',bandwidth=bdwth).fit(self.rS)
self.h=lambda x,loc: mvnorm.pdf(x=x,mean=loc,cov=self.bdwth**2)
if reg==0:
self.G=lambda x: np.array([self.h(x,loc) for loc in self.rSset])-self.iP(x)
elif reg==1:
self.G=lambda x: np.array([self.h(x,self.rSset[i])-self.h(x,self.rSset[0])\
for i in range(1,self.rSset.shape[0])])
elif reg==2:
self.G=lambda x: np.array([self.h(x,self.rSset[i])-self.h(x,self.rSset[i-1])\
for i in range(1,self.rSset.shape[0])])
elif reg==3:
def G(x):
tmp=np.array([self.h(x,loc) for loc in self.rSset])
return tmp[1:]-tmp.mean(axis=0)
self.G=G
self.nP=lambda x: np.exp(self.kde.score_samples(x))
self.nS=lambda size: self.kde.sample(size)
S=self.nS(size)
W=self.T(S)/self.nP(S)
self.__estimate(W,'NIS')
self.mP=lambda x: (1-rate)*self.iP(x)+rate*self.nP(x)
self.mS=lambda size: np.vstack([self.iS(size-round(rate*size)),self.nS(round(rate*size))])
self.S=self.mS(size)
W=self.T(self.S)/self.mP(self.S)
self.__estimate(W,'MIS')
def estimate_RIS(self,alpha,vif=False):
X=(self.G(self.S)/self.mP(self.S)).T
if vif:
if X.shape[1]<=100:
ind=np.arange(X.shape[1])
else:
ind=np.random.choice(np.arange(X.shape[1]),100,replace=False)
tmp=np.hstack([X,np.ones([X.shape[0],1])])
Vif=np.zeros(ind.size)
for i in range(ind.size):
Vif[i]=VIF(tmp,ind[i])
print('VIF: (min {:.4f}, median {:.4f}, mean {:.4f}, max {:.4f}, [>5] {}/{})'\
.format(Vif.min(),np.median(Vif),Vif.mean(),Vif.max(),np.sum(Vif>5),Vif.size))
Xn=X/np.linalg.norm(X,axis=0)
lbd=np.linalg.eigvals(Xn.T.dot(Xn))
tau=np.sqrt(lbd.max()/lbd)
print('Condition index: (min {:.4f}, median {:.4f}, mean {:.4f}, max {:.4f}, [>30] {}/{})'\
.format(tau.min(),np.median(tau),tau.mean(),tau.max(),np.sum(tau>30),tau.size))
y=self.T(self.S)/self.mP(self.S)
self.regO=Linear().fit(X,y)
self.regL=Lasso(alpha).fit(X,y)
print('Ordinary R2: {:.4f}; Lasso R2: {:.4f}'.format(self.regO.score(X,y),self.regL.score(X,y)))
W=y-X.dot(self.regO.coef_)
self.__estimate(W,'RIS(Ord)')
W=y-X.dot(self.regL.coef_)
self.__estimate(W,'RIS(Las)')
def estimate_MLE(self,opt=False,init=0):
mP=self.mP(self.S)
G=self.G(self.S)
target=lambda zeta: -np.mean(np.log(mP+zeta.dot(G)))
gradient=lambda zeta: -np.mean(G/(mP+zeta.dot(G)),axis=1)
hessian=lambda zeta: (G/(mP+zeta.dot(G))**2).dot(G.T)/G.shape[1]
zeta0=np.zeros(G.shape[0])
grad0=gradient(zeta0)
print('Reference:')
print('origin: value: {:.4f}; grad: (min {:.4f}, mean {:.4f}, max {:.4f}, std {:.4f})'\
.format(target(zeta0),grad0.min(),grad0.mean(),grad0.max(),grad0.std()))
print()
print('Theoretical results:')
X=(G/mP).T
XX=X-X.mean(axis=0)
zeta1=np.linalg.solve(XX.T.dot(XX),X.sum(axis=0))
print('MLE(The) zeta: (min {:.4f}, mean {:.4f}, max {:.4f}, std {:.4f}, norm {:.4f})'\
.format(zeta1.min(),zeta1.mean(),zeta1.max(),zeta1.std(),np.sqrt(np.sum(zeta1**2))))
grad1=gradient(zeta1)
print('theory: value: {:.4f}; grad: (min {:.4f}, mean {:.4f}, max {:.4f}, std {:.4f})'\
.format(target(zeta1),grad1.min(),grad1.mean(),grad1.max(),grad1.std()))
W=(self.T(self.S)/mP)*(1-XX.dot(zeta1))
self.__estimate(W,'RIS(The)',asym=False)
W=self.T(self.S)/(mP+zeta1.dot(G))
self.__estimate(W,'MLE(The)',asym=False)
if opt:
zeta=zeta1 if init==1 else zeta0
begin=dt.now()
res=root(lambda zeta: (gradient(zeta),hessian(zeta)),zeta,method='lm',jac=True)
end=dt.now()
print()
print('Optimization results (spent {} seconds):'.format((end-begin).seconds))
if res['success']:
zeta=res['x']
print('MLE(Opt) zeta: (min {:.4f}, mean {:.4f}, max {:.4f}, std {:.4f}, norm {:.4f})'\
.format(zeta.min(),zeta.mean(),zeta.max(),zeta.std(),np.sqrt(np.sum(zeta**2))))
print('Dist(zeta(Opt),zeta(The))={:.4f}'.format(np.sqrt(np.sum((zeta-zeta1)**2))))
grad=gradient(zeta)
print('optimal: value: {:.4f}; grad: (min {:.4f}, mean {:.4f}, max {:.4f}, std {:.4f})'\
.format(target(zeta),grad.min(),grad.mean(),grad.max(),grad.std()))
W=self.T(self.S)/(mP+zeta.dot(G))
self.__estimate(W,'MLE(Opt)',asym=False)
else:
print('MLE fail')
```
**Limitations:**
1. didn't consider self-normalized importance sampling
2. only for symmetric normal target and symmetric normal initial proposal with only one mode
3. only for normal KDE without weights and adaptive bandwidth
4. only consider regression for proposal components based on mixture proposal
# Run the experiments
## Initial proposal and the curse of dimensionality
```
mle=MLE(dim=8,sigma=2,df=1,mode=1)
size=1000000
mle.estimate_IS(size)
x=np.linspace(-4,4,101)
mle.draw_TP(mle.iP,x,'initial')
mle=MLE(dim=8,sigma=2,df=1,mode=2)
size=1000000
mle.estimate_IS(size)
x=np.linspace(-4,4,101)
mle.draw_TP(mle.iP,x,'initial')
```
|
github_jupyter
|
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from datetime import datetime as dt
from pandas_datareader import DataReader as DR
import seaborn as sb
import numdifftools as nd
from wquantiles import quantile
import statsmodels.api as sm
from statsmodels.stats.outliers_influence import variance_inflation_factor as VIF
from scipy.stats import norm,t,truncnorm
from scipy.stats import multivariate_normal as mvnorm
from scipy.stats import multivariate_t as mvt
from scipy.spatial import Delaunay as TRI
from scipy.interpolate import LinearNDInterpolator as ITP
from scipy.optimize import minimize,root
from scipy.optimize import NonlinearConstraint as NonlinCons
from scipy.stats import gaussian_kde as sciKDE
from sklearn.linear_model import LinearRegression as Linear
from sklearn.linear_model import Ridge
from sklearn.linear_model import Lasso
from sklearn.neighbors import KernelDensity as sklKDE
import warnings
warnings.filterwarnings("ignore")
class MLE:
def __init__(self,dim,sigma,df,mode=1):
self.dim=dim
self.T=lambda x: mvnorm.pdf(x=x,mean=np.zeros(dim))
if mode==1:
self.iP=lambda x: mvt.pdf(x=x,loc=np.zeros(dim),shape=sigma**2,df=df)
self.iS=lambda size: mvt.rvs(size=size,loc=np.zeros(dim),shape=sigma**2,df=df)
else:
self.iP=lambda x: np.prod([t.pdf(x=x[:,i],scale=sigma,df=df) for i in range(dim)],axis=0)
self.iS=lambda size: np.array([t.rvs(size=size,scale=sigma,df=df) for i in range(dim)]).T
def __estimate(self,W,name,asym=True):
Z=np.mean(W)
err=np.abs(Z-1)
if asym:
aVar=np.var(W)
aErr=np.sqrt(aVar/W.size)
ESS=1/np.sum((W/np.sum(W))**2)
print('{} est: {:.4f}; err: {:.4f}; a-var: {:.4f}; a-err: {:.4f}; ESS: {:.0f}/{}'\
.format(name,Z,err,aVar,aErr,ESS,W.size))
else:
print('{} est: {:.4f}; err: {:.4f}'.format(name,Z,err))
def estimate_IS(self,size):
S=self.iS(size)
W=self.T(S)/self.iP(S)
self.__estimate(W,'IS')
def draw_TP(self,P,x,name,dim=0):
X=np.zeros([x.size,self.dim])
X[:,dim]=x
ratio=np.reshape(self.T(np.zeros(self.dim))/P(np.zeros([1,self.dim])),1)[0]
print('------------ pdf ratio at origin: {:.2f} ------------'.format(ratio))
fig,ax=plt.subplots(figsize=(7,4))
ax.plot(x,self.T(X))
ax.plot(x,P(X))
if name=='nonparametric':
one=np.zeros(self.dim)
one[dim]=1
rW=np.array([self.h(one*loc,loc) for loc in self.rS])
rW=rW/rW.max()*P(np.zeros([1,self.dim]))[0]
rWmeans=np.ones_like(rW)*rW.mean()
ax.plot(x,self.mP(X))
ax.hist(self.rS[:,dim],bins=2*rW.size,weights=rWmeans)
ax.hist(self.rS[:,dim],bins=2*rW.size,weights=rW)
ax.legend(['target','nonparametric proposal','mixture proposal','centers','centers with weight'])
elif name=='regression':
G=self.G(X)
rPO=self.regO.coef_.dot(G)+self.regO.intercept_*P(X)
rPL=self.regL.coef_.dot(G)+self.regL.intercept_*P(X)
mid=int(x.size/2)
print('regression ratios: ordinary {:.4f}, lasso {:.4f}'\
.format(self.T(X[mid])/rPO[mid],self.T(X[mid])/rPL[mid]))
ax.plot(x,rPO)
ax.plot(x,rPL)
ax.legend(['target','mixture proposal','ordinary regression','lasso regression'])
else:
ax.legend(['target','{} proposal'.format(name)])
ax.set_title('{}-D target and {} proposal (cross-sectional view)'.format(self.dim,name))
plt.show()
def resample(self,size,ratio):
S=self.iS(ratio*size)
p=self.T(S)/self.iP(S)
index=np.arange(S.shape[0])
choice=np.random.choice(index,size,p=p/np.sum(p),replace=True)
self.rS=S[choice]
self.rSset=S[list(set(choice))]
print('resampling rate: {}/{}'.format(self.rSset.shape[0],size))
def estimate_NIS(self,size,rate,bdwth='scott',reg=0):
if(type(bdwth)==str):
method=bdwth
tmp=sciKDE(self.rS.T,bw_method=method)
bdwth=np.mean(np.sqrt(np.diag(tmp.covariance_factor()*np.cov(self.rS.T))))
print('bdwth: {:.4f} (based on {})'.format(bdwth,method))
self.bdwth=bdwth
self.kde=sklKDE(kernel='gaussian',bandwidth=bdwth).fit(self.rS)
self.h=lambda x,loc: mvnorm.pdf(x=x,mean=loc,cov=self.bdwth**2)
if reg==0:
self.G=lambda x: np.array([self.h(x,loc) for loc in self.rSset])-self.iP(x)
elif reg==1:
self.G=lambda x: np.array([self.h(x,self.rSset[i])-self.h(x,self.rSset[0])\
for i in range(1,self.rSset.shape[0])])
elif reg==2:
self.G=lambda x: np.array([self.h(x,self.rSset[i])-self.h(x,self.rSset[i-1])\
for i in range(1,self.rSset.shape[0])])
elif reg==3:
def G(x):
tmp=np.array([self.h(x,loc) for loc in self.rSset])
return tmp[1:]-tmp.mean(axis=0)
self.G=G
self.nP=lambda x: np.exp(self.kde.score_samples(x))
self.nS=lambda size: self.kde.sample(size)
S=self.nS(size)
W=self.T(S)/self.nP(S)
self.__estimate(W,'NIS')
self.mP=lambda x: (1-rate)*self.iP(x)+rate*self.nP(x)
self.mS=lambda size: np.vstack([self.iS(size-round(rate*size)),self.nS(round(rate*size))])
self.S=self.mS(size)
W=self.T(self.S)/self.mP(self.S)
self.__estimate(W,'MIS')
def estimate_RIS(self,alpha,vif=False):
X=(self.G(self.S)/self.mP(self.S)).T
if vif:
if X.shape[1]<=100:
ind=np.arange(X.shape[1])
else:
ind=np.random.choice(np.arange(X.shape[1]),100,replace=False)
tmp=np.hstack([X,np.ones([X.shape[0],1])])
Vif=np.zeros(ind.size)
for i in range(ind.size):
Vif[i]=VIF(tmp,ind[i])
print('VIF: (min {:.4f}, median {:.4f}, mean {:.4f}, max {:.4f}, [>5] {}/{})'\
.format(Vif.min(),np.median(Vif),Vif.mean(),Vif.max(),np.sum(Vif>5),Vif.size))
Xn=X/np.linalg.norm(X,axis=0)
lbd=np.linalg.eigvals(Xn.T.dot(Xn))
tau=np.sqrt(lbd.max()/lbd)
print('Condition index: (min {:.4f}, median {:.4f}, mean {:.4f}, max {:.4f}, [>30] {}/{})'\
.format(tau.min(),np.median(tau),tau.mean(),tau.max(),np.sum(tau>30),tau.size))
y=self.T(self.S)/self.mP(self.S)
self.regO=Linear().fit(X,y)
self.regL=Lasso(alpha).fit(X,y)
print('Ordinary R2: {:.4f}; Lasso R2: {:.4f}'.format(self.regO.score(X,y),self.regL.score(X,y)))
W=y-X.dot(self.regO.coef_)
self.__estimate(W,'RIS(Ord)')
W=y-X.dot(self.regL.coef_)
self.__estimate(W,'RIS(Las)')
def estimate_MLE(self,opt=False,init=0):
mP=self.mP(self.S)
G=self.G(self.S)
target=lambda zeta: -np.mean(np.log(mP+zeta.dot(G)))
gradient=lambda zeta: -np.mean(G/(mP+zeta.dot(G)),axis=1)
hessian=lambda zeta: (G/(mP+zeta.dot(G))**2).dot(G.T)/G.shape[1]
zeta0=np.zeros(G.shape[0])
grad0=gradient(zeta0)
print('Reference:')
print('origin: value: {:.4f}; grad: (min {:.4f}, mean {:.4f}, max {:.4f}, std {:.4f})'\
.format(target(zeta0),grad0.min(),grad0.mean(),grad0.max(),grad0.std()))
print()
print('Theoretical results:')
X=(G/mP).T
XX=X-X.mean(axis=0)
zeta1=np.linalg.solve(XX.T.dot(XX),X.sum(axis=0))
print('MLE(The) zeta: (min {:.4f}, mean {:.4f}, max {:.4f}, std {:.4f}, norm {:.4f})'\
.format(zeta1.min(),zeta1.mean(),zeta1.max(),zeta1.std(),np.sqrt(np.sum(zeta1**2))))
grad1=gradient(zeta1)
print('theory: value: {:.4f}; grad: (min {:.4f}, mean {:.4f}, max {:.4f}, std {:.4f})'\
.format(target(zeta1),grad1.min(),grad1.mean(),grad1.max(),grad1.std()))
W=(self.T(self.S)/mP)*(1-XX.dot(zeta1))
self.__estimate(W,'RIS(The)',asym=False)
W=self.T(self.S)/(mP+zeta1.dot(G))
self.__estimate(W,'MLE(The)',asym=False)
if opt:
zeta=zeta1 if init==1 else zeta0
begin=dt.now()
res=root(lambda zeta: (gradient(zeta),hessian(zeta)),zeta,method='lm',jac=True)
end=dt.now()
print()
print('Optimization results (spent {} seconds):'.format((end-begin).seconds))
if res['success']:
zeta=res['x']
print('MLE(Opt) zeta: (min {:.4f}, mean {:.4f}, max {:.4f}, std {:.4f}, norm {:.4f})'\
.format(zeta.min(),zeta.mean(),zeta.max(),zeta.std(),np.sqrt(np.sum(zeta**2))))
print('Dist(zeta(Opt),zeta(The))={:.4f}'.format(np.sqrt(np.sum((zeta-zeta1)**2))))
grad=gradient(zeta)
print('optimal: value: {:.4f}; grad: (min {:.4f}, mean {:.4f}, max {:.4f}, std {:.4f})'\
.format(target(zeta),grad.min(),grad.mean(),grad.max(),grad.std()))
W=self.T(self.S)/(mP+zeta.dot(G))
self.__estimate(W,'MLE(Opt)',asym=False)
else:
print('MLE fail')
mle=MLE(dim=8,sigma=2,df=1,mode=1)
size=1000000
mle.estimate_IS(size)
x=np.linspace(-4,4,101)
mle.draw_TP(mle.iP,x,'initial')
mle=MLE(dim=8,sigma=2,df=1,mode=2)
size=1000000
mle.estimate_IS(size)
x=np.linspace(-4,4,101)
mle.draw_TP(mle.iP,x,'initial')
| 0.469277 | 0.732065 |
# Fascicle evaluation
One of the challenges for users of tractography algorithms is how to validate the results of tractography, and to compare results of different tractography algorithms. We demonstrate here how to compare the tracks from the [deterministic tractography](det_track.ipynb) and [probablistic tractography](prob_track.ipynb) examples. These are two algorithms that provide slightly different models of the white matter. One set of curves is slighlty less "wiggly", and traverses the white matter in a slightly more direct curve.
Here, we demonstrate how these results can be compared to each other, and evaluated with respect to the data, using the LiFE (linear fascicle evaluation method) introduced in Pestilli et al. (2014). This method creates a linear model in which each one of the tracks contributes to the signal. The optimal weights for each track are found, and an 'optimized' version of the tracks can be derived, by culling out the tracks that have a weight equal to 0. We demonstrate this process below.
We start by importing a few functions that we will use in this example:
```
import os.path as op
import numpy as np
import nibabel as nib
import matplotlib.pyplot as plt
%matplotlib inline
import dipy.core.gradients as grad
from IPython.display import display, Image
```
We read in the data that was used for tracking:
```
dwi_ni = nib.load(op.join('data', 'SUB1_b2000_1.nii.gz'))
data = dwi_ni.get_fdata()
affine = dwi_ni.affine
gtab = grad.gradient_table(op.join('data','SUB1_b2000_1.bvals'), op.join('data', 'SUB1_b2000_1.bvecs'))
```
In this case, we have two sets of candidate tracks, one from each algorithm:
```
trk = nib.streamlines.load('prob-track.trk')
candidate_prob = nib.streamlines.load('prob-track.trk').streamlines
candidate_det = nib.streamlines.load('det-track.trk').streamlines
```
We set up the LiFE model:
```
import dipy.tracking.life as life
fiber_model = life.FiberModel(gtab)
```
We fit the model separately for each one of the tracks:
```
fit_prob = fiber_model.fit(data, candidate_prob, affine=np.eye(4))
fit_det = fiber_model.fit(data, candidate_det, affine=np.eye(4))
```
The two sets of tracks have different weight distributions. Many of the tracks from the deterministic algorithm do not contribute to the signal (weight=0).
```
fig, ax = plt.subplots(1)
ax.hist(fit_prob.beta, bins=100, histtype='step')
ax.hist(fit_det.beta, bins=100, histtype='step')
ax.set_xlim([-0.01, 0.05])
ax.set_xlabel('Streamline weights')
ax.set_label('# streamlines')
```
We create an optimized set of tracks by culling all the tracks that have a weight of 0:
```
optimized_prob = list(np.array(candidate_prob)[np.where(fit_prob.beta>0)[0]])
```
What proportion of the streamlines survived this culling?
```
len(optimized_prob)/float(len(candidate_prob))
```
We can visualize the remaining tacks as follows
```
from dipy.viz import window, actor, colormap, has_fury
from dipy.tracking.utils import transform_tracking_output
from numpy.linalg import inv
t1 = nib.load(op.join('data', 'SUB1_t1_resamp.nii.gz'))
t1_data = t1.get_fdata()
t1_aff = t1.affine
color = colormap.line_colors(optimized_prob)
# streamlines_actor = actor.streamtube(list(transform_tracking_output(streamlines, inv(t1_aff))),
# color)
streamlines_actor = actor.streamtube(optimized_prob, color)
vol_actor = actor.slicer(t1_data)
vol_actor.display_extent(0, t1_data.shape[0]-1, 0, t1_data.shape[1]-1, 25, 25)
scene = window.Scene()
scene.add(streamlines_actor)
scene.add(vol_actor)
window.record(scene, out_path='life-prob-track.png', size=(600,600))
display(Image(filename='life-prob-track.png'))
```
To see how the LiFE algorithm can be used to calculate the error related to optimized tracks, relative to a non-optimized solution, please refer to the Dipy documentation example [here](http://nipy.org/dipy/examples_built/linear_fascicle_evaluation.html).
|
github_jupyter
|
import os.path as op
import numpy as np
import nibabel as nib
import matplotlib.pyplot as plt
%matplotlib inline
import dipy.core.gradients as grad
from IPython.display import display, Image
dwi_ni = nib.load(op.join('data', 'SUB1_b2000_1.nii.gz'))
data = dwi_ni.get_fdata()
affine = dwi_ni.affine
gtab = grad.gradient_table(op.join('data','SUB1_b2000_1.bvals'), op.join('data', 'SUB1_b2000_1.bvecs'))
trk = nib.streamlines.load('prob-track.trk')
candidate_prob = nib.streamlines.load('prob-track.trk').streamlines
candidate_det = nib.streamlines.load('det-track.trk').streamlines
import dipy.tracking.life as life
fiber_model = life.FiberModel(gtab)
fit_prob = fiber_model.fit(data, candidate_prob, affine=np.eye(4))
fit_det = fiber_model.fit(data, candidate_det, affine=np.eye(4))
fig, ax = plt.subplots(1)
ax.hist(fit_prob.beta, bins=100, histtype='step')
ax.hist(fit_det.beta, bins=100, histtype='step')
ax.set_xlim([-0.01, 0.05])
ax.set_xlabel('Streamline weights')
ax.set_label('# streamlines')
optimized_prob = list(np.array(candidate_prob)[np.where(fit_prob.beta>0)[0]])
len(optimized_prob)/float(len(candidate_prob))
from dipy.viz import window, actor, colormap, has_fury
from dipy.tracking.utils import transform_tracking_output
from numpy.linalg import inv
t1 = nib.load(op.join('data', 'SUB1_t1_resamp.nii.gz'))
t1_data = t1.get_fdata()
t1_aff = t1.affine
color = colormap.line_colors(optimized_prob)
# streamlines_actor = actor.streamtube(list(transform_tracking_output(streamlines, inv(t1_aff))),
# color)
streamlines_actor = actor.streamtube(optimized_prob, color)
vol_actor = actor.slicer(t1_data)
vol_actor.display_extent(0, t1_data.shape[0]-1, 0, t1_data.shape[1]-1, 25, 25)
scene = window.Scene()
scene.add(streamlines_actor)
scene.add(vol_actor)
window.record(scene, out_path='life-prob-track.png', size=(600,600))
display(Image(filename='life-prob-track.png'))
| 0.466846 | 0.987735 |
# Understanding Perceptrons
In the 1950s, American psychologist and artificial intelligence researcher Frank Rosenblatt invented an algorithm that would automatically learn the optimal weight coefficients $w_0$ and $w_1$ needed to perform an accurate binary classification: the perceptron learning rule.
Rosenblatt's original perceptron algorithm can be summed up as follows:
1. Initialize the weights to zero or some small random numbers.
2. For each training sample $s_i$, perform the following steps:
1. Compute the predicted target value $ŷ_i$.
2. Compare $ŷ_i$ to the ground truth $y_i$, and update the weights accordingly:
- If the two are the same (correct prediction), skip ahead.
- If the two are different (wrong prediction), push the weight coefficients $w_0$ and $w_1$ towards the positive or negative target class respectively.
## Implementing our first perceptron
Perceptrons are easy enough to be implemented from scratch. We can mimic the typical OpenCV or scikit-learn implementation of a classifier by creating a `Perceptron` object. This will allow us to initialize new perceptron objects that can learn from data via a `fit` method and make predictions via a separate `predict` method.
When we initialize a new perceptron object, we want to pass a learning rate (`lr`) and the number of iterations after which the algorithm should terminate (`n_iter`):
```
import numpy as np
class Perceptron(object):
def __init__(self, lr=0.01, n_iter=10):
"""Constructor
Parameters
----------
lr : float
Learning rate.
n_iter : int
Number of iterations after which the algorithm should
terminate.
"""
self.lr = lr
self.n_iter = n_iter
def predict(self, X):
"""Predict target labels
Parameters
----------
X : array-like
Feature matrix, <n_samples x n_features>
Returns
-------
Predicted target labels, +1 or -1.
Notes
-----
Must run `fit` first.
"""
# Whenever the term (X * weights + bias) >= 0, we return
# label +1, else we return label -1
return np.where(np.dot(X, self.weights) + self.bias >= 0.0,
1, -1)
def fit(self, X, y):
"""Fit the model to data
Parameters
----------
X : array-like
Feature matrix, <n_samples x n_features>
y : array-like
Vector of target labels, <n_samples x 1>
"""
self.weights = np.zeros(X.shape[1])
self.bias = 0.0
for _ in range(self.n_iter):
for xi, yi in zip(X, y):
delta = self.lr * (yi - self.predict(xi))
self.weights += delta * xi
self.bias += delta
```
## Generating a toy dataset
To test our perceptron classifier, we need to create some mock data. Let's keep things simple for now and generate 100 data samples (`n_samples`) belonging to one of two blobs (`center`s), again relying on scikit-learn's `make_blobs` function:
```
from sklearn.datasets.samples_generator import make_blobs
X, y = make_blobs(n_samples=100, centers=2,
cluster_std=2.2, random_state=42)
```
Adjust the labels so they're either +1 or -1:
```
y = 2 * y - 1
import matplotlib.pyplot as plt
plt.style.use('ggplot')
%matplotlib inline
plt.figure(figsize=(10, 6))
plt.scatter(X[:, 0], X[:, 1], s=100, c=y);
plt.xlabel('x1')
plt.ylabel('x2')
plt.savefig('perceptron-data.png')
```
## Fitting the perceptron to data
We can instantiate our perceptron object similar to other classifiers we encountered with
OpenCV:
```
p = Perceptron(lr=0.1, n_iter=10)
p.fit(X, y)
```
Let's have a look at the learned weights:
```
p.weights
p.bias
```
If we plug these values into our equation for $ϕ$, it becomes clear that the perceptron learned
a decision boundary of the form $2.2 x_1 - 0.48 x_2 + 0.2 >= 0$.
## Evaluating the perceptron classifier
```
from sklearn.metrics import accuracy_score
accuracy_score(p.predict(X), y)
def plot_decision_boundary(classifier, X_test, y_test):
# create a mesh to plot in
h = 0.02 # step size in mesh
x_min, x_max = X_test[:, 0].min() - 1, X_test[:, 0].max() + 1
y_min, y_max = X_test[:, 1].min() - 1, X_test[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, h),
np.arange(y_min, y_max, h))
X_hypo = np.c_[xx.ravel().astype(np.float32),
yy.ravel().astype(np.float32)]
zz = classifier.predict(X_hypo)
zz = zz.reshape(xx.shape)
plt.contourf(xx, yy, zz, cmap=plt.cm.coolwarm, alpha=0.8)
plt.scatter(X_test[:, 0], X_test[:, 1], c=y_test, s=200)
plt.figure(figsize=(10, 6))
plot_decision_boundary(p, X, y)
plt.xlabel('x1')
plt.ylabel('x2')
```
## Applying the perceptron to data that is not linearly separable
Since the perceptron is a linear classifier, you can imagine that it would have trouble trying
to classify data that is not linearly separable. We can test this by increasing the spread
(`cluster_std`) of the two blobs in our toy dataset so that the two blobs start overlapping:
```
X, y = make_blobs(n_samples=100, centers=2,
cluster_std=5.2, random_state=42)
y = 2 * y - 1
plt.figure(figsize=(10, 6))
plt.scatter(X[:, 0], X[:, 1], s=100, c=y);
plt.xlabel('x1')
plt.ylabel('x2')
```
So what would happen if we applied the perceptron classifier to this dataset?
```
p = Perceptron(lr=0.1, n_iter=10)
p.fit(X, y)
accuracy_score(p.predict(X), y)
plt.figure(figsize=(10, 6))
plot_decision_boundary(p, X, y)
plt.xlabel('x1')
plt.ylabel('x2')
```
Fortunately, there are ways to make the perceptron more powerful and ultimately create
nonlinear decision boundaries.
|
github_jupyter
|
import numpy as np
class Perceptron(object):
def __init__(self, lr=0.01, n_iter=10):
"""Constructor
Parameters
----------
lr : float
Learning rate.
n_iter : int
Number of iterations after which the algorithm should
terminate.
"""
self.lr = lr
self.n_iter = n_iter
def predict(self, X):
"""Predict target labels
Parameters
----------
X : array-like
Feature matrix, <n_samples x n_features>
Returns
-------
Predicted target labels, +1 or -1.
Notes
-----
Must run `fit` first.
"""
# Whenever the term (X * weights + bias) >= 0, we return
# label +1, else we return label -1
return np.where(np.dot(X, self.weights) + self.bias >= 0.0,
1, -1)
def fit(self, X, y):
"""Fit the model to data
Parameters
----------
X : array-like
Feature matrix, <n_samples x n_features>
y : array-like
Vector of target labels, <n_samples x 1>
"""
self.weights = np.zeros(X.shape[1])
self.bias = 0.0
for _ in range(self.n_iter):
for xi, yi in zip(X, y):
delta = self.lr * (yi - self.predict(xi))
self.weights += delta * xi
self.bias += delta
from sklearn.datasets.samples_generator import make_blobs
X, y = make_blobs(n_samples=100, centers=2,
cluster_std=2.2, random_state=42)
y = 2 * y - 1
import matplotlib.pyplot as plt
plt.style.use('ggplot')
%matplotlib inline
plt.figure(figsize=(10, 6))
plt.scatter(X[:, 0], X[:, 1], s=100, c=y);
plt.xlabel('x1')
plt.ylabel('x2')
plt.savefig('perceptron-data.png')
p = Perceptron(lr=0.1, n_iter=10)
p.fit(X, y)
p.weights
p.bias
from sklearn.metrics import accuracy_score
accuracy_score(p.predict(X), y)
def plot_decision_boundary(classifier, X_test, y_test):
# create a mesh to plot in
h = 0.02 # step size in mesh
x_min, x_max = X_test[:, 0].min() - 1, X_test[:, 0].max() + 1
y_min, y_max = X_test[:, 1].min() - 1, X_test[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, h),
np.arange(y_min, y_max, h))
X_hypo = np.c_[xx.ravel().astype(np.float32),
yy.ravel().astype(np.float32)]
zz = classifier.predict(X_hypo)
zz = zz.reshape(xx.shape)
plt.contourf(xx, yy, zz, cmap=plt.cm.coolwarm, alpha=0.8)
plt.scatter(X_test[:, 0], X_test[:, 1], c=y_test, s=200)
plt.figure(figsize=(10, 6))
plot_decision_boundary(p, X, y)
plt.xlabel('x1')
plt.ylabel('x2')
X, y = make_blobs(n_samples=100, centers=2,
cluster_std=5.2, random_state=42)
y = 2 * y - 1
plt.figure(figsize=(10, 6))
plt.scatter(X[:, 0], X[:, 1], s=100, c=y);
plt.xlabel('x1')
plt.ylabel('x2')
p = Perceptron(lr=0.1, n_iter=10)
p.fit(X, y)
accuracy_score(p.predict(X), y)
plt.figure(figsize=(10, 6))
plot_decision_boundary(p, X, y)
plt.xlabel('x1')
plt.ylabel('x2')
| 0.910717 | 0.98805 |
### Import and Config
```
# nuclio: ignore
import nuclio
%nuclio config kind = "job"
%nuclio config spec.image = "iguazio/shell:3.0_b5565_20201026062233_wsdf" # docker image available on idan707/spark_shell
import mlrun
from mlrun.platforms.iguazio import mount_v3io, mount_v3iod
from mlrun.datastore import DataItem
from mlrun.execution import MLClientCtx
import os
from subprocess import run
import pandas as pd
import numpy as np
from pyspark.sql.types import LongType
from pyspark.sql import SparkSession
```
### Build Spark Describe Helper Functions
```
import sys
import base64 as b64
import warnings
warnings.filterwarnings("ignore")
from itertools import product
import matplotlib
import numpy as np
import json
import pandas as pd
from matplotlib import pyplot as plt
from pkg_resources import resource_filename
import six
from pyspark.sql import DataFrame as SparkDataFrame
from pyspark.sql.functions import (abs as df_abs, col, count, countDistinct,
max as df_max, mean, min as df_min,
sum as df_sum, when
)
from pyspark.sql.functions import variance, stddev, kurtosis, skewness
def describe(df, bins, corr_reject, config, **kwargs):
if not isinstance(df, SparkDataFrame):
raise TypeError("df must be of type pyspark.sql.DataFrame")
# Number of rows:
table_stats = {"n": df.count()}
if table_stats["n"] == 0:
raise ValueError("df cannot be empty")
try:
matplotlib.style.use("default")
except:
pass
# Function to "pretty name" floats:
def pretty_name(x):
x *= 100
if x == int(x):
return '%.0f%%' % x
else:
return '%.1f%%' % x
def corr_matrix(df, columns=None):
if columns is None:
columns = df.columns
combinations = list(product(columns,columns))
def separate(l, n):
for i in range(0, len(l), n):
yield l[i:i+n]
grouped = list(separate(combinations,len(columns)))
df_cleaned = df.select(*columns).na.drop(how="any")
for i in grouped:
for j in enumerate(i):
i[j[0]] = i[j[0]] + (df_cleaned.corr(str(j[1][0]), str(j[1][1])),)
df_pandas = pd.DataFrame(grouped).applymap(lambda x: x[2])
df_pandas.columns = columns
df_pandas.index = columns
return df_pandas
def create_hist_data(df, column, minim, maxim, bins=10):
def create_all_conditions(current_col, column, left_edges, count=1):
"""
Recursive function that exploits the
ability to call the Spark SQL Column method
.when() in a recursive way.
"""
left_edges = left_edges[:]
if len(left_edges) == 0:
return current_col
if len(left_edges) == 1:
next_col = current_col.when(col(column) >= float(left_edges[0]), count)
left_edges.pop(0)
return create_all_conditions(next_col, column, left_edges[:], count+1)
next_col = current_col.when((float(left_edges[0]) <= col(column))
& (col(column) < float(left_edges[1])), count)
left_edges.pop(0)
return create_all_conditions(next_col, column, left_edges[:], count+1)
num_range = maxim - minim
bin_width = num_range / float(bins)
left_edges = [minim]
for _bin in range(bins):
left_edges = left_edges + [left_edges[-1] + bin_width]
left_edges.pop()
expression_col = when((float(left_edges[0]) <= col(column))
& (col(column) < float(left_edges[1])), 0)
left_edges_copy = left_edges[:]
left_edges_copy.pop(0)
bin_data = (df.select(col(column))
.na.drop()
.select(col(column),
create_all_conditions(expression_col,
column,
left_edges_copy
).alias("bin_id")
)
.groupBy("bin_id").count()
).toPandas()
bin_data.index = bin_data["bin_id"]
new_index = list(range(bins))
bin_data = bin_data.reindex(new_index)
bin_data["bin_id"] = bin_data.index
bin_data = bin_data.fillna(0)
bin_data["left_edge"] = left_edges
bin_data["width"] = bin_width
return bin_data
def describe_integer_1d(df, column, current_result, nrows):
stats_df = df.select(column).na.drop().agg(mean(col(column)).alias("mean"),
df_min(col(column)).alias("min"),
df_max(col(column)).alias("max"),
variance(col(column)).alias("variance"),
kurtosis(col(column)).alias("kurtosis"),
stddev(col(column)).alias("std"),
skewness(col(column)).alias("skewness"),
df_sum(col(column)).alias("sum")
).toPandas()
for x in np.array([0.05, 0.25, 0.5, 0.75, 0.95]):
stats_df[pretty_name(x)] = (df.select(column)
.na.drop()
.selectExpr("percentile(`{col}`,CAST({n} AS DOUBLE))"
.format(col=column, n=x)).toPandas().iloc[:,0]
)
stats = stats_df.iloc[0].copy()
stats.name = column
stats["range"] = stats["max"] - stats["min"]
stats["iqr"] = stats[pretty_name(0.75)] - stats[pretty_name(0.25)]
stats["cv"] = stats["std"] / float(stats["mean"])
stats["mad"] = (df.select(column)
.na.drop()
.select(df_abs(col(column)-stats["mean"]).alias("delta"))
.agg(df_sum(col("delta"))).toPandas().iloc[0,0] / float(current_result["count"]))
stats["type"] = "NUM"
stats['n_zeros'] = df.select(column).where(col(column)==0.0).count()
stats['p_zeros'] = stats['n_zeros'] / float(nrows)
hist_data = create_hist_data(df, column, stats["min"], stats["max"], bins)
return stats
def describe_float_1d(df, column, current_result, nrows):
stats_df = df.select(column).na.drop().agg(mean(col(column)).alias("mean"),
df_min(col(column)).alias("min"),
df_max(col(column)).alias("max"),
variance(col(column)).alias("variance"),
kurtosis(col(column)).alias("kurtosis"),
stddev(col(column)).alias("std"),
skewness(col(column)).alias("skewness"),
df_sum(col(column)).alias("sum")
).toPandas()
for x in np.array([0.05, 0.25, 0.5, 0.75, 0.95]):
stats_df[pretty_name(x)] = (df.select(column)
.na.drop()
.selectExpr("percentile_approx(`{col}`,CAST({n} AS DOUBLE))"
.format(col=column, n=x)).toPandas().iloc[:,0]
)
stats = stats_df.iloc[0].copy()
stats.name = column
stats["range"] = stats["max"] - stats["min"]
stats["iqr"] = stats[pretty_name(0.75)] - stats[pretty_name(0.25)]
stats["cv"] = stats["std"] / float(stats["mean"])
stats["mad"] = (df.select(column)
.na.drop()
.select(df_abs(col(column)-stats["mean"]).alias("delta"))
.agg(df_sum(col("delta"))).toPandas().iloc[0,0] / float(current_result["count"]))
stats["type"] = "NUM"
stats['n_zeros'] = df.select(column).where(col(column)==0.0).count()
stats['p_zeros'] = stats['n_zeros'] / float(nrows)
hist_data = create_hist_data(df, column, stats["min"], stats["max"], bins)
return stats
def describe_date_1d(df, column):
stats_df = df.select(column).na.drop().agg(df_min(col(column)).alias("min"),
df_max(col(column)).alias("max")
).toPandas()
stats = stats_df.iloc[0].copy()
stats.name = column
if isinstance(stats["max"], pd.Timestamp):
stats = stats.astype(object)
stats["max"] = str(stats["max"].to_pydatetime())
stats["min"] = str(stats["min"].to_pydatetime())
else:
stats["range"] = stats["max"] - stats["min"]
stats["type"] = "DATE"
return stats
def guess_json_type(string_value):
try:
obj = json.loads(string_value)
except:
return None
return type(obj)
def describe_categorical_1d(df, column):
value_counts = (df.select(column).na.drop()
.groupBy(column)
.agg(count(col(column)))
.orderBy("count({c})".format(c=column),ascending=False)
).cache()
# Get the most frequent class:
stats = (value_counts
.limit(1)
.withColumnRenamed(column, "top")
.withColumnRenamed("count({c})".format(c=column), "freq")
).toPandas().iloc[0]
top_50 = value_counts.limit(50).toPandas().sort_values("count({c})".format(c=column),
ascending=False)
top_50_categories = top_50[column].values.tolist()
others_count = pd.Series([df.select(column).na.drop()
.where(~(col(column).isin(*top_50_categories)))
.count()
], index=["***Other Values***"])
others_distinct_count = pd.Series([value_counts
.where(~(col(column).isin(*top_50_categories)))
.count()
], index=["***Other Values Distinct Count***"])
top = top_50.set_index(column)["count({c})".format(c=column)]
top = top.append(others_count)
top = top.append(others_distinct_count)
stats["value_counts"] = top
stats["type"] = "CAT"
value_counts.unpersist()
unparsed_valid_jsons = df.select(column).na.drop().rdd.map(
lambda x: guess_json_type(x[column])).filter(
lambda x: x).distinct().collect()
stats["unparsed_json_types"] = unparsed_valid_jsons
return stats
def describe_constant_1d(df, column):
stats = pd.Series(['CONST'], index=['type'], name=column)
stats["value_counts"] = (df.select(column)
.na.drop()
.limit(1)).toPandas().iloc[:,0].value_counts()
return stats
def describe_unique_1d(df, column):
stats = pd.Series(['UNIQUE'], index=['type'], name=column)
stats["value_counts"] = (df.select(column)
.na.drop()
.limit(50)).toPandas().iloc[:,0].value_counts()
return stats
def describe_1d(df, column, nrows, lookup_config=None):
column_type = df.select(column).dtypes[0][1]
if ("array" in column_type) or ("stuct" in column_type) or ("map" in column_type):
raise NotImplementedError("Column {c} is of type {t} and cannot be analyzed".format(c=column, t=column_type))
distinct_count = df.select(column).agg(countDistinct(col(column)).alias("distinct_count")).toPandas()
non_nan_count = df.select(column).na.drop().select(count(col(column)).alias("count")).toPandas()
results_data = pd.concat([distinct_count, non_nan_count],axis=1)
results_data["p_unique"] = results_data["distinct_count"] / float(results_data["count"])
results_data["is_unique"] = results_data["distinct_count"] == nrows
results_data["n_missing"] = nrows - results_data["count"]
results_data["p_missing"] = results_data["n_missing"] / float(nrows)
results_data["p_infinite"] = 0
results_data["n_infinite"] = 0
result = results_data.iloc[0].copy()
result["memorysize"] = 0
result.name = column
if result["distinct_count"] <= 1:
result = result.append(describe_constant_1d(df, column))
elif column_type in {"tinyint", "smallint", "int", "bigint"}:
result = result.append(describe_integer_1d(df, column, result, nrows))
elif column_type in {"float", "double", "decimal"}:
result = result.append(describe_float_1d(df, column, result, nrows))
elif column_type in {"date", "timestamp"}:
result = result.append(describe_date_1d(df, column))
elif result["is_unique"] == True:
result = result.append(describe_unique_1d(df, column))
else:
result = result.append(describe_categorical_1d(df, column))
# Fix to also count MISSING value in the distict_count field:
if result["n_missing"] > 0:
result["distinct_count"] = result["distinct_count"] + 1
if (result["count"] > result["distinct_count"] > 1):
try:
result["mode"] = result["top"]
except KeyError:
result["mode"] = 0
else:
try:
result["mode"] = result["value_counts"].index[0]
except KeyError:
result["mode"] = 0
# If and IndexError happens,
# it is because all column are NULLs:
except IndexError:
result["mode"] = "MISSING"
if lookup_config:
lookup_object = lookup_config['object']
col_name_in_db = lookup_config['col_name_in_db'] if 'col_name_in_db' in lookup_config else None
try:
matched, unmatched = lookup_object.lookup(df.select(column), col_name_in_db)
result['lookedup_values'] = str(matched.count()) + "/" + str(df.select(column).count())
except:
result['lookedup_values'] = 'FAILED'
else:
result['lookedup_values'] = ''
return result
# Do the thing:
ldesc = {}
for colum in df.columns:
if colum in config:
if 'lookup' in config[colum]:
lookup_config = config[colum]['lookup']
desc = describe_1d(df, colum, table_stats["n"], lookup_config=lookup_config)
else:
desc = describe_1d(df, colum, table_stats["n"])
else:
desc = describe_1d(df, colum, table_stats["n"])
ldesc.update({colum: desc})
# Compute correlation matrix
if corr_reject is not None:
computable_corrs = [colum for colum in ldesc if ldesc[colum]["type"] in {"NUM"}]
if len(computable_corrs) > 0:
corr = corr_matrix(df, columns=computable_corrs)
for x, corr_x in corr.iterrows():
for y, corr in corr_x.iteritems():
if x == y:
break
# Convert ldesc to a DataFrame
variable_stats = pd.DataFrame(ldesc)
# General statistics
table_stats["nvar"] = len(df.columns)
table_stats["total_missing"] = float(variable_stats.loc["n_missing"].sum()) / (table_stats["n"] * table_stats["nvar"])
memsize = 0
table_stats['memsize'] = fmt_bytesize(memsize)
table_stats['recordsize'] = fmt_bytesize(memsize / table_stats['n'])
table_stats.update({k: 0 for k in ("NUM", "DATE", "CONST", "CAT", "UNIQUE", "CORR")})
table_stats.update(dict(variable_stats.loc['type'].value_counts()))
table_stats['REJECTED'] = table_stats['CONST'] + table_stats['CORR']
freq_dict = {}
for var in variable_stats:
if "value_counts" not in variable_stats[var]:
pass
elif not(variable_stats[var]["value_counts"] is np.nan):
freq_dict[var] = variable_stats[var]["value_counts"]
else:
pass
try:
variable_stats = variable_stats.drop("value_counts")
except (ValueError, KeyError):
pass
return table_stats, variable_stats.T, freq_dict
import numpy as np
from pyspark.sql.functions import abs as absou
SKEWNESS_CUTOFF = 20
DEFAULT_FLOAT_FORMATTER = u'spark_df_profiling.__default_float_formatter'
def gradient_format(value, limit1, limit2, c1, c2):
def LerpColour(c1,c2,t):
return (int(c1[0]+(c2[0]-c1[0])*t),int(c1[1]+(c2[1]-c1[1])*t),int(c1[2]+(c2[2]-c1[2])*t))
c = LerpColour(c1, c2, (value-limit1)/(limit2-limit1))
return fmt_color(value,"rgb{}".format(str(c)))
def fmt_color(text, color):
return(u'<span style="color:{color}">{text}</span>'.format(color=color,text=str(text)))
def fmt_class(text, cls):
return(u'<span class="{cls}">{text}</span>'.format(cls=cls,text=str(text)))
def fmt_bytesize(num, suffix='B'):
for unit in ['','Ki','Mi','Gi','Ti','Pi','Ei','Zi']:
if num < 0:
num = num*-1
if num < 1024.0:
return "%3.1f %s%s" % (num, unit, suffix)
num /= 1024.0
return "%.1f %s%s" % (num, 'Yi', suffix)
def fmt_percent(v):
return "{:2.1f}%".format(v*100)
def fmt_varname(v):
return u'<code>{0}</code>'.format(v)
value_formatters={
u'freq': (lambda v: gradient_format(v, 0, 62000, (30, 198, 244), (99, 200, 72))),
u'p_missing': fmt_percent,
u'p_infinite': fmt_percent,
u'p_unique': fmt_percent,
u'p_zeros': fmt_percent,
u'memorysize': fmt_bytesize,
u'total_missing': fmt_percent,
DEFAULT_FLOAT_FORMATTER: lambda v: str(float('{:.5g}'.format(v))).rstrip('0').rstrip('.'),
u'correlation_var': lambda v: fmt_varname(v),
u'unparsed_json_types': lambda v: ', '.join([s.__name__ for s in v])
}
def fmt_row_severity(v):
if np.isnan(v) or v<= 0.01:
return "ignore"
else:
return "alert"
def fmt_skewness(v):
if not np.isnan(v) and (v<-SKEWNESS_CUTOFF or v> SKEWNESS_CUTOFF):
return "alert"
else:
return ""
row_formatters={
u'p_zeros': fmt_row_severity,
u'p_missing': fmt_row_severity,
u'p_infinite': fmt_row_severity,
u'n_duplicates': fmt_row_severity,
u'skewness': fmt_skewness,
}
```
### Build Spark Describe Function
```
run(["/bin/bash", "/etc/config/v3io/v3io-spark-operator.sh"])
def describe_spark(context: MLClientCtx,
dataset: DataItem,
bins: int=30,
describe_extended: bool=True)-> None:
"""
Generates profile reports from an Apache Spark DataFrame.
Based on pandas_profiling, but for Spark's DataFrames instead of pandas.
For each column the following statistics - if relevant for the column type - are presented:
Essentials: type, unique values, missing values
Quantile statistics: minimum value, Q1, median, Q3, maximum, range, interquartile range
Descriptive statistics: mean, mode, standard deviation, sum, median absolute deviation, coefficient of variation, kurtosis, skewness
Most frequent values: for categorical data
:param context: Function context.
:param dataset: Raw data file (currently needs to be a local file located in v3io://User/bigdata)
:param bins: Number of bin in histograms
:param describe_extended: (True) set to False if the aim is to get a simple .describe() infomration
"""
# get file location
location = dataset.local()
# build spark session
spark = SparkSession.builder.appName("Spark job").getOrCreate()
# read csv
df = spark.read.csv(location, header=True, inferSchema= True)
# No use for now
kwargs = []
# take only numric column
float_cols = [item[0] for item in df.dtypes if item[1].startswith('float') or item[1].startswith('double')]
if describe_extended == True:
# run describe function
table, variables, freq = describe(df, bins, float_cols, kwargs)
# get summary table
tbl_1 = variables.reset_index()
# prep report
if len(freq) != 0:
tbl_2 = pd.DataFrame.from_dict(freq, orient = "index").sort_index().stack().reset_index()
tbl_2.columns = ['col', 'key', 'val']
tbl_2['Merged'] = [{key: val} for key, val in zip(tbl_2.key, tbl_2.val)]
tbl_2 = tbl_2.groupby('col', as_index=False).agg(lambda x: tuple(x))[['col','Merged']]
# get summary
summary = pd.merge(tbl_1, tbl_2, how='left', left_on='index', right_on='col')
else:
summary = tbl_1
# log final report
context.log_dataset("summary_stats",
df=summary,
format="csv", index=False,
artifact_path=context.artifact_subpath('data'))
# log overview
context.log_results(table)
else:
# run simple describe and save to pandas
tbl_1 = df.describe().toPandas()
# save final report and transpose
summary = tbl_1.T
# log final report
context.log_dataset("summary_stats",
df=summary,
format="csv", index=False,
artifact_path=context.artifact_subpath('data'))
# stop spark session
spark.stop()
# nuclio: end-code
```
### Save and Config
```
fn = mlrun.code_to_function(handler="describe_spark", code_output=".")
fn.apply(mount_v3io())
fn.apply(mount_v3iod(namespace="default-tenant", v3io_config_configmap="spark-operator-v3io-config"))
fn.spec.image_pull_policy = "IfNotPresent"
fn.export("function.yaml")
```
### Set Environment
```
artifact_path = mlrun.set_environment(api_path = 'http://mlrun-api:8080',
artifact_path = os.path.abspath('./'))
```
### Run and Save Outputs
```
run_res = fn.run(inputs={"dataset": "iris_dataset.csv"},
artifact_path=artifact_path, watch=True)
run_res.show()
```
|
github_jupyter
|
# nuclio: ignore
import nuclio
%nuclio config kind = "job"
%nuclio config spec.image = "iguazio/shell:3.0_b5565_20201026062233_wsdf" # docker image available on idan707/spark_shell
import mlrun
from mlrun.platforms.iguazio import mount_v3io, mount_v3iod
from mlrun.datastore import DataItem
from mlrun.execution import MLClientCtx
import os
from subprocess import run
import pandas as pd
import numpy as np
from pyspark.sql.types import LongType
from pyspark.sql import SparkSession
import sys
import base64 as b64
import warnings
warnings.filterwarnings("ignore")
from itertools import product
import matplotlib
import numpy as np
import json
import pandas as pd
from matplotlib import pyplot as plt
from pkg_resources import resource_filename
import six
from pyspark.sql import DataFrame as SparkDataFrame
from pyspark.sql.functions import (abs as df_abs, col, count, countDistinct,
max as df_max, mean, min as df_min,
sum as df_sum, when
)
from pyspark.sql.functions import variance, stddev, kurtosis, skewness
def describe(df, bins, corr_reject, config, **kwargs):
if not isinstance(df, SparkDataFrame):
raise TypeError("df must be of type pyspark.sql.DataFrame")
# Number of rows:
table_stats = {"n": df.count()}
if table_stats["n"] == 0:
raise ValueError("df cannot be empty")
try:
matplotlib.style.use("default")
except:
pass
# Function to "pretty name" floats:
def pretty_name(x):
x *= 100
if x == int(x):
return '%.0f%%' % x
else:
return '%.1f%%' % x
def corr_matrix(df, columns=None):
if columns is None:
columns = df.columns
combinations = list(product(columns,columns))
def separate(l, n):
for i in range(0, len(l), n):
yield l[i:i+n]
grouped = list(separate(combinations,len(columns)))
df_cleaned = df.select(*columns).na.drop(how="any")
for i in grouped:
for j in enumerate(i):
i[j[0]] = i[j[0]] + (df_cleaned.corr(str(j[1][0]), str(j[1][1])),)
df_pandas = pd.DataFrame(grouped).applymap(lambda x: x[2])
df_pandas.columns = columns
df_pandas.index = columns
return df_pandas
def create_hist_data(df, column, minim, maxim, bins=10):
def create_all_conditions(current_col, column, left_edges, count=1):
"""
Recursive function that exploits the
ability to call the Spark SQL Column method
.when() in a recursive way.
"""
left_edges = left_edges[:]
if len(left_edges) == 0:
return current_col
if len(left_edges) == 1:
next_col = current_col.when(col(column) >= float(left_edges[0]), count)
left_edges.pop(0)
return create_all_conditions(next_col, column, left_edges[:], count+1)
next_col = current_col.when((float(left_edges[0]) <= col(column))
& (col(column) < float(left_edges[1])), count)
left_edges.pop(0)
return create_all_conditions(next_col, column, left_edges[:], count+1)
num_range = maxim - minim
bin_width = num_range / float(bins)
left_edges = [minim]
for _bin in range(bins):
left_edges = left_edges + [left_edges[-1] + bin_width]
left_edges.pop()
expression_col = when((float(left_edges[0]) <= col(column))
& (col(column) < float(left_edges[1])), 0)
left_edges_copy = left_edges[:]
left_edges_copy.pop(0)
bin_data = (df.select(col(column))
.na.drop()
.select(col(column),
create_all_conditions(expression_col,
column,
left_edges_copy
).alias("bin_id")
)
.groupBy("bin_id").count()
).toPandas()
bin_data.index = bin_data["bin_id"]
new_index = list(range(bins))
bin_data = bin_data.reindex(new_index)
bin_data["bin_id"] = bin_data.index
bin_data = bin_data.fillna(0)
bin_data["left_edge"] = left_edges
bin_data["width"] = bin_width
return bin_data
def describe_integer_1d(df, column, current_result, nrows):
stats_df = df.select(column).na.drop().agg(mean(col(column)).alias("mean"),
df_min(col(column)).alias("min"),
df_max(col(column)).alias("max"),
variance(col(column)).alias("variance"),
kurtosis(col(column)).alias("kurtosis"),
stddev(col(column)).alias("std"),
skewness(col(column)).alias("skewness"),
df_sum(col(column)).alias("sum")
).toPandas()
for x in np.array([0.05, 0.25, 0.5, 0.75, 0.95]):
stats_df[pretty_name(x)] = (df.select(column)
.na.drop()
.selectExpr("percentile(`{col}`,CAST({n} AS DOUBLE))"
.format(col=column, n=x)).toPandas().iloc[:,0]
)
stats = stats_df.iloc[0].copy()
stats.name = column
stats["range"] = stats["max"] - stats["min"]
stats["iqr"] = stats[pretty_name(0.75)] - stats[pretty_name(0.25)]
stats["cv"] = stats["std"] / float(stats["mean"])
stats["mad"] = (df.select(column)
.na.drop()
.select(df_abs(col(column)-stats["mean"]).alias("delta"))
.agg(df_sum(col("delta"))).toPandas().iloc[0,0] / float(current_result["count"]))
stats["type"] = "NUM"
stats['n_zeros'] = df.select(column).where(col(column)==0.0).count()
stats['p_zeros'] = stats['n_zeros'] / float(nrows)
hist_data = create_hist_data(df, column, stats["min"], stats["max"], bins)
return stats
def describe_float_1d(df, column, current_result, nrows):
stats_df = df.select(column).na.drop().agg(mean(col(column)).alias("mean"),
df_min(col(column)).alias("min"),
df_max(col(column)).alias("max"),
variance(col(column)).alias("variance"),
kurtosis(col(column)).alias("kurtosis"),
stddev(col(column)).alias("std"),
skewness(col(column)).alias("skewness"),
df_sum(col(column)).alias("sum")
).toPandas()
for x in np.array([0.05, 0.25, 0.5, 0.75, 0.95]):
stats_df[pretty_name(x)] = (df.select(column)
.na.drop()
.selectExpr("percentile_approx(`{col}`,CAST({n} AS DOUBLE))"
.format(col=column, n=x)).toPandas().iloc[:,0]
)
stats = stats_df.iloc[0].copy()
stats.name = column
stats["range"] = stats["max"] - stats["min"]
stats["iqr"] = stats[pretty_name(0.75)] - stats[pretty_name(0.25)]
stats["cv"] = stats["std"] / float(stats["mean"])
stats["mad"] = (df.select(column)
.na.drop()
.select(df_abs(col(column)-stats["mean"]).alias("delta"))
.agg(df_sum(col("delta"))).toPandas().iloc[0,0] / float(current_result["count"]))
stats["type"] = "NUM"
stats['n_zeros'] = df.select(column).where(col(column)==0.0).count()
stats['p_zeros'] = stats['n_zeros'] / float(nrows)
hist_data = create_hist_data(df, column, stats["min"], stats["max"], bins)
return stats
def describe_date_1d(df, column):
stats_df = df.select(column).na.drop().agg(df_min(col(column)).alias("min"),
df_max(col(column)).alias("max")
).toPandas()
stats = stats_df.iloc[0].copy()
stats.name = column
if isinstance(stats["max"], pd.Timestamp):
stats = stats.astype(object)
stats["max"] = str(stats["max"].to_pydatetime())
stats["min"] = str(stats["min"].to_pydatetime())
else:
stats["range"] = stats["max"] - stats["min"]
stats["type"] = "DATE"
return stats
def guess_json_type(string_value):
try:
obj = json.loads(string_value)
except:
return None
return type(obj)
def describe_categorical_1d(df, column):
value_counts = (df.select(column).na.drop()
.groupBy(column)
.agg(count(col(column)))
.orderBy("count({c})".format(c=column),ascending=False)
).cache()
# Get the most frequent class:
stats = (value_counts
.limit(1)
.withColumnRenamed(column, "top")
.withColumnRenamed("count({c})".format(c=column), "freq")
).toPandas().iloc[0]
top_50 = value_counts.limit(50).toPandas().sort_values("count({c})".format(c=column),
ascending=False)
top_50_categories = top_50[column].values.tolist()
others_count = pd.Series([df.select(column).na.drop()
.where(~(col(column).isin(*top_50_categories)))
.count()
], index=["***Other Values***"])
others_distinct_count = pd.Series([value_counts
.where(~(col(column).isin(*top_50_categories)))
.count()
], index=["***Other Values Distinct Count***"])
top = top_50.set_index(column)["count({c})".format(c=column)]
top = top.append(others_count)
top = top.append(others_distinct_count)
stats["value_counts"] = top
stats["type"] = "CAT"
value_counts.unpersist()
unparsed_valid_jsons = df.select(column).na.drop().rdd.map(
lambda x: guess_json_type(x[column])).filter(
lambda x: x).distinct().collect()
stats["unparsed_json_types"] = unparsed_valid_jsons
return stats
def describe_constant_1d(df, column):
stats = pd.Series(['CONST'], index=['type'], name=column)
stats["value_counts"] = (df.select(column)
.na.drop()
.limit(1)).toPandas().iloc[:,0].value_counts()
return stats
def describe_unique_1d(df, column):
stats = pd.Series(['UNIQUE'], index=['type'], name=column)
stats["value_counts"] = (df.select(column)
.na.drop()
.limit(50)).toPandas().iloc[:,0].value_counts()
return stats
def describe_1d(df, column, nrows, lookup_config=None):
column_type = df.select(column).dtypes[0][1]
if ("array" in column_type) or ("stuct" in column_type) or ("map" in column_type):
raise NotImplementedError("Column {c} is of type {t} and cannot be analyzed".format(c=column, t=column_type))
distinct_count = df.select(column).agg(countDistinct(col(column)).alias("distinct_count")).toPandas()
non_nan_count = df.select(column).na.drop().select(count(col(column)).alias("count")).toPandas()
results_data = pd.concat([distinct_count, non_nan_count],axis=1)
results_data["p_unique"] = results_data["distinct_count"] / float(results_data["count"])
results_data["is_unique"] = results_data["distinct_count"] == nrows
results_data["n_missing"] = nrows - results_data["count"]
results_data["p_missing"] = results_data["n_missing"] / float(nrows)
results_data["p_infinite"] = 0
results_data["n_infinite"] = 0
result = results_data.iloc[0].copy()
result["memorysize"] = 0
result.name = column
if result["distinct_count"] <= 1:
result = result.append(describe_constant_1d(df, column))
elif column_type in {"tinyint", "smallint", "int", "bigint"}:
result = result.append(describe_integer_1d(df, column, result, nrows))
elif column_type in {"float", "double", "decimal"}:
result = result.append(describe_float_1d(df, column, result, nrows))
elif column_type in {"date", "timestamp"}:
result = result.append(describe_date_1d(df, column))
elif result["is_unique"] == True:
result = result.append(describe_unique_1d(df, column))
else:
result = result.append(describe_categorical_1d(df, column))
# Fix to also count MISSING value in the distict_count field:
if result["n_missing"] > 0:
result["distinct_count"] = result["distinct_count"] + 1
if (result["count"] > result["distinct_count"] > 1):
try:
result["mode"] = result["top"]
except KeyError:
result["mode"] = 0
else:
try:
result["mode"] = result["value_counts"].index[0]
except KeyError:
result["mode"] = 0
# If and IndexError happens,
# it is because all column are NULLs:
except IndexError:
result["mode"] = "MISSING"
if lookup_config:
lookup_object = lookup_config['object']
col_name_in_db = lookup_config['col_name_in_db'] if 'col_name_in_db' in lookup_config else None
try:
matched, unmatched = lookup_object.lookup(df.select(column), col_name_in_db)
result['lookedup_values'] = str(matched.count()) + "/" + str(df.select(column).count())
except:
result['lookedup_values'] = 'FAILED'
else:
result['lookedup_values'] = ''
return result
# Do the thing:
ldesc = {}
for colum in df.columns:
if colum in config:
if 'lookup' in config[colum]:
lookup_config = config[colum]['lookup']
desc = describe_1d(df, colum, table_stats["n"], lookup_config=lookup_config)
else:
desc = describe_1d(df, colum, table_stats["n"])
else:
desc = describe_1d(df, colum, table_stats["n"])
ldesc.update({colum: desc})
# Compute correlation matrix
if corr_reject is not None:
computable_corrs = [colum for colum in ldesc if ldesc[colum]["type"] in {"NUM"}]
if len(computable_corrs) > 0:
corr = corr_matrix(df, columns=computable_corrs)
for x, corr_x in corr.iterrows():
for y, corr in corr_x.iteritems():
if x == y:
break
# Convert ldesc to a DataFrame
variable_stats = pd.DataFrame(ldesc)
# General statistics
table_stats["nvar"] = len(df.columns)
table_stats["total_missing"] = float(variable_stats.loc["n_missing"].sum()) / (table_stats["n"] * table_stats["nvar"])
memsize = 0
table_stats['memsize'] = fmt_bytesize(memsize)
table_stats['recordsize'] = fmt_bytesize(memsize / table_stats['n'])
table_stats.update({k: 0 for k in ("NUM", "DATE", "CONST", "CAT", "UNIQUE", "CORR")})
table_stats.update(dict(variable_stats.loc['type'].value_counts()))
table_stats['REJECTED'] = table_stats['CONST'] + table_stats['CORR']
freq_dict = {}
for var in variable_stats:
if "value_counts" not in variable_stats[var]:
pass
elif not(variable_stats[var]["value_counts"] is np.nan):
freq_dict[var] = variable_stats[var]["value_counts"]
else:
pass
try:
variable_stats = variable_stats.drop("value_counts")
except (ValueError, KeyError):
pass
return table_stats, variable_stats.T, freq_dict
import numpy as np
from pyspark.sql.functions import abs as absou
SKEWNESS_CUTOFF = 20
DEFAULT_FLOAT_FORMATTER = u'spark_df_profiling.__default_float_formatter'
def gradient_format(value, limit1, limit2, c1, c2):
def LerpColour(c1,c2,t):
return (int(c1[0]+(c2[0]-c1[0])*t),int(c1[1]+(c2[1]-c1[1])*t),int(c1[2]+(c2[2]-c1[2])*t))
c = LerpColour(c1, c2, (value-limit1)/(limit2-limit1))
return fmt_color(value,"rgb{}".format(str(c)))
def fmt_color(text, color):
return(u'<span style="color:{color}">{text}</span>'.format(color=color,text=str(text)))
def fmt_class(text, cls):
return(u'<span class="{cls}">{text}</span>'.format(cls=cls,text=str(text)))
def fmt_bytesize(num, suffix='B'):
for unit in ['','Ki','Mi','Gi','Ti','Pi','Ei','Zi']:
if num < 0:
num = num*-1
if num < 1024.0:
return "%3.1f %s%s" % (num, unit, suffix)
num /= 1024.0
return "%.1f %s%s" % (num, 'Yi', suffix)
def fmt_percent(v):
return "{:2.1f}%".format(v*100)
def fmt_varname(v):
return u'<code>{0}</code>'.format(v)
value_formatters={
u'freq': (lambda v: gradient_format(v, 0, 62000, (30, 198, 244), (99, 200, 72))),
u'p_missing': fmt_percent,
u'p_infinite': fmt_percent,
u'p_unique': fmt_percent,
u'p_zeros': fmt_percent,
u'memorysize': fmt_bytesize,
u'total_missing': fmt_percent,
DEFAULT_FLOAT_FORMATTER: lambda v: str(float('{:.5g}'.format(v))).rstrip('0').rstrip('.'),
u'correlation_var': lambda v: fmt_varname(v),
u'unparsed_json_types': lambda v: ', '.join([s.__name__ for s in v])
}
def fmt_row_severity(v):
if np.isnan(v) or v<= 0.01:
return "ignore"
else:
return "alert"
def fmt_skewness(v):
if not np.isnan(v) and (v<-SKEWNESS_CUTOFF or v> SKEWNESS_CUTOFF):
return "alert"
else:
return ""
row_formatters={
u'p_zeros': fmt_row_severity,
u'p_missing': fmt_row_severity,
u'p_infinite': fmt_row_severity,
u'n_duplicates': fmt_row_severity,
u'skewness': fmt_skewness,
}
run(["/bin/bash", "/etc/config/v3io/v3io-spark-operator.sh"])
def describe_spark(context: MLClientCtx,
dataset: DataItem,
bins: int=30,
describe_extended: bool=True)-> None:
"""
Generates profile reports from an Apache Spark DataFrame.
Based on pandas_profiling, but for Spark's DataFrames instead of pandas.
For each column the following statistics - if relevant for the column type - are presented:
Essentials: type, unique values, missing values
Quantile statistics: minimum value, Q1, median, Q3, maximum, range, interquartile range
Descriptive statistics: mean, mode, standard deviation, sum, median absolute deviation, coefficient of variation, kurtosis, skewness
Most frequent values: for categorical data
:param context: Function context.
:param dataset: Raw data file (currently needs to be a local file located in v3io://User/bigdata)
:param bins: Number of bin in histograms
:param describe_extended: (True) set to False if the aim is to get a simple .describe() infomration
"""
# get file location
location = dataset.local()
# build spark session
spark = SparkSession.builder.appName("Spark job").getOrCreate()
# read csv
df = spark.read.csv(location, header=True, inferSchema= True)
# No use for now
kwargs = []
# take only numric column
float_cols = [item[0] for item in df.dtypes if item[1].startswith('float') or item[1].startswith('double')]
if describe_extended == True:
# run describe function
table, variables, freq = describe(df, bins, float_cols, kwargs)
# get summary table
tbl_1 = variables.reset_index()
# prep report
if len(freq) != 0:
tbl_2 = pd.DataFrame.from_dict(freq, orient = "index").sort_index().stack().reset_index()
tbl_2.columns = ['col', 'key', 'val']
tbl_2['Merged'] = [{key: val} for key, val in zip(tbl_2.key, tbl_2.val)]
tbl_2 = tbl_2.groupby('col', as_index=False).agg(lambda x: tuple(x))[['col','Merged']]
# get summary
summary = pd.merge(tbl_1, tbl_2, how='left', left_on='index', right_on='col')
else:
summary = tbl_1
# log final report
context.log_dataset("summary_stats",
df=summary,
format="csv", index=False,
artifact_path=context.artifact_subpath('data'))
# log overview
context.log_results(table)
else:
# run simple describe and save to pandas
tbl_1 = df.describe().toPandas()
# save final report and transpose
summary = tbl_1.T
# log final report
context.log_dataset("summary_stats",
df=summary,
format="csv", index=False,
artifact_path=context.artifact_subpath('data'))
# stop spark session
spark.stop()
# nuclio: end-code
fn = mlrun.code_to_function(handler="describe_spark", code_output=".")
fn.apply(mount_v3io())
fn.apply(mount_v3iod(namespace="default-tenant", v3io_config_configmap="spark-operator-v3io-config"))
fn.spec.image_pull_policy = "IfNotPresent"
fn.export("function.yaml")
artifact_path = mlrun.set_environment(api_path = 'http://mlrun-api:8080',
artifact_path = os.path.abspath('./'))
run_res = fn.run(inputs={"dataset": "iris_dataset.csv"},
artifact_path=artifact_path, watch=True)
run_res.show()
| 0.484136 | 0.757166 |
Registers all MRI modalities to axial plane and saves patient cases as 160x320x320x3 (z,x,y,c) `BraTSID.npy` arrays (R:t1w, G:FLAIR, B:T2).
Additionally, registers [Task-1](https://www.synapse.org/#!Synapse:syn25829067/wiki/610863) segmentation masks to case and saves them as `BraTSID_seg.npy` 160x320x320 (z,x,y) arrays. This requires accepting Task-1 terms and downloading & extracting [this](https://www.kaggle.com/dschettler8845/load-task-1-dataset-comparison-w-task-2-dataset/data) set that @dschettler8845 uploaded.
- **Date:** 2021-08-28
- **Author:** [Joni Juvonen](https://www.kaggle.com/qitvision)
```
import os
import sys
from tqdm import tqdm
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from PIL import Image
import SimpleITK as sitk
train_path = '../input/rsna-miccai-brain-tumor-radiogenomic-classification/train/'
train_dirs = os.listdir(train_path)
sitk.sitkNearestNeighbor
class Registered_BraTS_Case():
def __init__(self, dicom_dir, resize_to, task1_dir=None):
self.dicom_dir = dicom_dir
self.resize_to = resize_to
self.task1_dir = task1_dir
@staticmethod
def resample(image, ref_image):
resampler = sitk.ResampleImageFilter()
resampler.SetReferenceImage(ref_image)
#resampler.SetInterpolator(sitk.sitkLinear)
resampler.SetInterpolator(sitk.sitkNearestNeighbor)
resampler.SetTransform(sitk.AffineTransform(image.GetDimension()))
resampler.SetOutputSpacing(ref_image.GetSpacing())
resampler.SetSize(ref_image.GetSize())
resampler.SetOutputDirection(ref_image.GetDirection())
resampler.SetOutputOrigin(ref_image.GetOrigin())
resampler.SetDefaultPixelValue(image.GetPixelIDValue())
resamped_image = resampler.Execute(image)
return resamped_image
@staticmethod
def normalize(data):
return (data - np.min(data)) / (np.max(data) - np.min(data))
@staticmethod
def get_crop_bb(image):
inside_value = 0
outside_value = 255
label_shape_filter = sitk.LabelShapeStatisticsImageFilter()
label_shape_filter.Execute( sitk.OtsuThreshold(image, inside_value, outside_value) )
bounding_box = label_shape_filter.GetBoundingBox(outside_value)
return bounding_box
@staticmethod
def crop_with_bb(image, bounding_box):
# The bounding box's first "dim" entries are the starting index and last "dim" entries the size
return sitk.RegionOfInterest(image, bounding_box[int(len(bounding_box)/2):], bounding_box[0:int(len(bounding_box)/2)])
@staticmethod
def threshold_based_crop(image):
"""
This function is copied from here: https://github.com/InsightSoftwareConsortium/SimpleITK-Notebooks/blob/master/Python/70_Data_Augmentation.ipynb
Use Otsu's threshold estimator to separate background and foreground. In medical imaging the background is
usually air. Then crop the image using the foreground's axis aligned bounding box.
Args:
image (SimpleITK image): An image where the anatomy and background intensities form a bi-modal distribution
(the assumption underlying Otsu's method.)
Return:
Cropped image based on foreground's axis aligned bounding box.
"""
# Set pixels that are in [min_intensity,otsu_threshold] to inside_value, values above otsu_threshold are
# set to outside_value. The anatomy has higher intensity values than the background, so it is outside.
inside_value = 0
outside_value = 255
label_shape_filter = sitk.LabelShapeStatisticsImageFilter()
label_shape_filter.Execute( sitk.OtsuThreshold(image, inside_value, outside_value) )
bounding_box = label_shape_filter.GetBoundingBox(outside_value)
# The bounding box's first "dim" entries are the starting index and last "dim" entries the size
return sitk.RegionOfInterest(image, bounding_box[int(len(bounding_box)/2):], bounding_box[0:int(len(bounding_box)/2)])
@staticmethod
def swap_image_axes(image, order):
return sitk.PermuteAxes(image, order)
@staticmethod
def axis_swap_order(image):
direction = np.array([int(round(d)) for d in image.GetDirection()])
if np.all(direction == np.array([1, 0, 0, 0, 1, 0, 0, 0, 1])):
return [0,1,2]
elif np.all(direction == np.array([0, 0, -1, 1, 0, 0, 0, -1, 0])):
return [2,0,1]
elif np.all(direction == np.array([1, 0, 0, 0, 0, 1, 0, -1, 0])):
return [0,2,1]
else:
print(list(direction))
return [0,1,2]
@staticmethod
def maybe_flip_axes(image):
direction = np.array([int(round(d)) for i,d in enumerate(image.GetDirection()) if i%4==0])
flips = [bool(d == -1) for d in direction]
return sitk.Flip(image, list(flips))
@staticmethod
def resize_image(original_CT, resize_to=[256,256,100]):
dimension = original_CT.GetDimension()
reference_physical_size = np.zeros(original_CT.GetDimension())
reference_physical_size[:] = [(sz-1)*spc if sz*spc>mx else mx for sz,spc,mx in zip(original_CT.GetSize(), original_CT.GetSpacing(), reference_physical_size)]
reference_origin = original_CT.GetOrigin()
reference_direction = original_CT.GetDirection()
reference_size = resize_to
reference_spacing = [ phys_sz/(sz-1) for sz,phys_sz in zip(reference_size, reference_physical_size) ]
reference_image = sitk.Image(reference_size, original_CT.GetPixelIDValue())
reference_image.SetOrigin(reference_origin)
reference_image.SetSpacing(reference_spacing)
reference_image.SetDirection(reference_direction)
reference_center = np.array(reference_image.TransformContinuousIndexToPhysicalPoint(np.array(reference_image.GetSize())/2.0))
transform = sitk.AffineTransform(dimension)
transform.SetMatrix(original_CT.GetDirection())
transform.SetTranslation(np.array(original_CT.GetOrigin()) - reference_origin)
centering_transform = sitk.TranslationTransform(dimension)
img_center = np.array(original_CT.TransformContinuousIndexToPhysicalPoint(np.array(original_CT.GetSize())/2.0))
centering_transform.SetOffset(np.array(transform.GetInverse().TransformPoint(img_center) - reference_center))
centered_transform = sitk.Transform(transform)
centered_transform = sitk.Transform(centering_transform)
return sitk.Resample(original_CT, reference_image, centered_transform, sitk.sitkLinear, 0.0)
def get_registered_case(self, brat_id_str):
""" Return stack of min-max normalized R:t1wCE, G:flair, B:T2 array """
stacked = np.zeros(tuple([self.resize_to[2],*self.resize_to[:2],3]))
try:
# init sitk reader
reader = sitk.ImageSeriesReader()
reader.LoadPrivateTagsOn()
reader.SetOutputPixelType(sitk.sitkFloat32)
# Use t1w as the reference and skip flair because it's sometimes completely dark
filenamesDICOM = reader.GetGDCMSeriesFileNames(f'{self.dicom_dir}/{brat_id_str}/T1wCE')
reader.SetFileNames(filenamesDICOM)
t1w = reader.Execute()
filenamesDICOM = reader.GetGDCMSeriesFileNames(f'{self.dicom_dir}/{brat_id_str}/FLAIR')
reader.SetFileNames(filenamesDICOM)
flair = reader.Execute()
filenamesDICOM = reader.GetGDCMSeriesFileNames(f'{self.dicom_dir}/{brat_id_str}/T2w')
reader.SetFileNames(filenamesDICOM)
t2 = reader.Execute()
# Align reference image and crop with otsu to minimum enclosing box
t1w = self.swap_image_axes(t1w, self.axis_swap_order(t1w))
t1w = self.maybe_flip_axes(t1w)
bounding_box = self.get_crop_bb(t1w)
# resample other modalities to align with reference
flair_resampled = self.resample(flair, t1w)
t2_resampled = self.resample(t2, t1w)
# crop all modalities to same box
t1w_cropped = self.crop_with_bb(t1w, bounding_box)
t1w_cropped = self.resize_image(t1w_cropped, self.resize_to)
flair_cropped = self.crop_with_bb(flair_resampled, bounding_box)
flair_cropped = self.resize_image(flair_cropped, self.resize_to)
t2_cropped = self.crop_with_bb(t2_resampled, bounding_box)
t2_cropped = self.resize_image(t2_cropped, self.resize_to)
# Return stack of min-max normalized R:t1w, G:T1ce, B:T2 array
stacked = np.stack([
self.normalize(sitk.GetArrayFromImage(t1w_cropped)),
self.normalize(sitk.GetArrayFromImage(flair_cropped)),
self.normalize(sitk.GetArrayFromImage(t2_cropped))
], axis=3)
except:
print(f'Error: {brat_id_str}')
return stacked
def get_aligned_seg_map(self, brat_id_str):
""" Normalizes task 1 segmentation map with similar logic and returns an aligned seg map """
if self.task1_dir is None:
return None
try:
segmentation_nii_fn = [os.path.join(f'{self.task1_dir}/BraTS2021_{brat_id_str}/', fn) for fn in os.listdir(f'{self.task1_dir}/BraTS2021_{brat_id_str}/') if 'seg' in fn]
t1_nii_fn = [os.path.join(f'{self.task1_dir}/BraTS2021_{brat_id_str}/', fn) for fn in os.listdir(f'{self.task1_dir}/BraTS2021_{brat_id_str}/') if 't1ce.' in fn]
if len(segmentation_nii_fn) == 0 or len(t1_nii_fn) ==0:
return None
t1 = sitk.ReadImage(t1_nii_fn[0])
seg = sitk.ReadImage(segmentation_nii_fn[0])
t1 = self.swap_image_axes(t1, self.axis_swap_order(t1))
t1 = self.maybe_flip_axes(t1)
bounding_box = self.get_crop_bb(t1)
seg_resampled = self.resample(seg, t1)
seg_cropped = self.crop_with_bb(seg_resampled, bounding_box)
seg_cropped = self.resize_image(seg_cropped, self.resize_to)
except:
print(f'Seg error: {brat_id_str}')
return None
return sitk.GetArrayFromImage(seg_cropped)
train_dir = train_dirs[200]
task1_dir = '../input/brats-2021-task1/BraTS2021_Training_Data'
resize_to=[320,320,160]
registered = Registered_BraTS_Case(dicom_dir=train_path, resize_to=resize_to, task1_dir=task1_dir)
reg_stack = registered.get_registered_case(brat_id_str=train_dir)
seg = registered.get_aligned_seg_map(brat_id_str=train_dir)
f, axs = plt.subplots(3,10, figsize=(20,5))
height_levels = resize_to[2]
for ind, j in enumerate(range(0, height_levels - 1, (height_levels//10))):
if ind > 9:break
axs[0,ind].imshow(reg_stack[j,:,:,:])
axs[1, ind].imshow(seg[j,:,:])
axs[2, ind].imshow(reg_stack[j,:,:,:], alpha=0.7)
axs[2, ind].imshow(seg[j,:,:], alpha=0.3)
axs[0,ind].set_title(f'Slice #{ind*(height_levels//10)}')
axs[0,0].set_ylabel('RGB')
axs[1,0].set_ylabel('Tumor')
axs[2,0].set_ylabel('RGB+Tumor')
plt.suptitle(f'BraTS ID: {train_dir}', fontsize=20)
plt.savefig('../media/registered_sample.png', transparent=False)
plt.show()
```
## Save files
```
from glob import glob
from joblib import Parallel,delayed
import subprocess
from tqdm.auto import tqdm
train_path = '../input/rsna-miccai-brain-tumor-radiogenomic-classification/train/'
train_dirs = os.listdir(train_path)
test_path = '../input/rsna-miccai-brain-tumor-radiogenomic-classification/test/'
test_dirs = os.listdir(test_path)
reg_dir = os.path.join('../input', 'registered_cases_v3_320x320x160')
def maybe_create_dir(_dir):
if not os.path.exists(_dir): os.mkdir(_dir)
maybe_create_dir(reg_dir)
reg_test_dir = os.path.join(reg_dir, 'test')
reg_train_dir = os.path.join(reg_dir, 'train')
maybe_create_dir(reg_test_dir)
maybe_create_dir(reg_train_dir)
def save_case(brat_id_str, rgb_cohort_dir, resize_to, task1_dir, save_dir):
registered = Registered_BraTS_Case(dicom_dir=rgb_cohort_dir, resize_to=resize_to, task1_dir=task1_dir)
reg_stack = registered.get_registered_case(brat_id_str=brat_id_str)
seg = registered.get_aligned_seg_map(brat_id_str=brat_id_str)
np.save(os.path.join(save_dir, brat_id_str + '.npy'),reg_stack)
if seg is not None:
np.save(os.path.join(save_dir, brat_id_str + '_seg.npy'),seg)
def save_train_case(brat_id_str):
save_case(brat_id_str, train_path, resize_to, task1_dir, save_dir=reg_train_dir)
def save_test_case(brat_id_str):
save_case(brat_id_str, test_path, resize_to, task1_dir, save_dir=reg_test_dir)
n_jobs = 8
mp_train = Parallel(
n_jobs = n_jobs,
prefer = 'threads')(delayed(save_train_case)(brat_id_str) for brat_id_str in tqdm(train_dirs, total=len(train_dirs)))
mp_test = Parallel(
n_jobs = n_jobs,
prefer = 'threads')(delayed(save_test_case)(brat_id_str) for brat_id_str in tqdm(test_dirs, total=len(test_dirs)))
```
|
github_jupyter
|
import os
import sys
from tqdm import tqdm
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from PIL import Image
import SimpleITK as sitk
train_path = '../input/rsna-miccai-brain-tumor-radiogenomic-classification/train/'
train_dirs = os.listdir(train_path)
sitk.sitkNearestNeighbor
class Registered_BraTS_Case():
def __init__(self, dicom_dir, resize_to, task1_dir=None):
self.dicom_dir = dicom_dir
self.resize_to = resize_to
self.task1_dir = task1_dir
@staticmethod
def resample(image, ref_image):
resampler = sitk.ResampleImageFilter()
resampler.SetReferenceImage(ref_image)
#resampler.SetInterpolator(sitk.sitkLinear)
resampler.SetInterpolator(sitk.sitkNearestNeighbor)
resampler.SetTransform(sitk.AffineTransform(image.GetDimension()))
resampler.SetOutputSpacing(ref_image.GetSpacing())
resampler.SetSize(ref_image.GetSize())
resampler.SetOutputDirection(ref_image.GetDirection())
resampler.SetOutputOrigin(ref_image.GetOrigin())
resampler.SetDefaultPixelValue(image.GetPixelIDValue())
resamped_image = resampler.Execute(image)
return resamped_image
@staticmethod
def normalize(data):
return (data - np.min(data)) / (np.max(data) - np.min(data))
@staticmethod
def get_crop_bb(image):
inside_value = 0
outside_value = 255
label_shape_filter = sitk.LabelShapeStatisticsImageFilter()
label_shape_filter.Execute( sitk.OtsuThreshold(image, inside_value, outside_value) )
bounding_box = label_shape_filter.GetBoundingBox(outside_value)
return bounding_box
@staticmethod
def crop_with_bb(image, bounding_box):
# The bounding box's first "dim" entries are the starting index and last "dim" entries the size
return sitk.RegionOfInterest(image, bounding_box[int(len(bounding_box)/2):], bounding_box[0:int(len(bounding_box)/2)])
@staticmethod
def threshold_based_crop(image):
"""
This function is copied from here: https://github.com/InsightSoftwareConsortium/SimpleITK-Notebooks/blob/master/Python/70_Data_Augmentation.ipynb
Use Otsu's threshold estimator to separate background and foreground. In medical imaging the background is
usually air. Then crop the image using the foreground's axis aligned bounding box.
Args:
image (SimpleITK image): An image where the anatomy and background intensities form a bi-modal distribution
(the assumption underlying Otsu's method.)
Return:
Cropped image based on foreground's axis aligned bounding box.
"""
# Set pixels that are in [min_intensity,otsu_threshold] to inside_value, values above otsu_threshold are
# set to outside_value. The anatomy has higher intensity values than the background, so it is outside.
inside_value = 0
outside_value = 255
label_shape_filter = sitk.LabelShapeStatisticsImageFilter()
label_shape_filter.Execute( sitk.OtsuThreshold(image, inside_value, outside_value) )
bounding_box = label_shape_filter.GetBoundingBox(outside_value)
# The bounding box's first "dim" entries are the starting index and last "dim" entries the size
return sitk.RegionOfInterest(image, bounding_box[int(len(bounding_box)/2):], bounding_box[0:int(len(bounding_box)/2)])
@staticmethod
def swap_image_axes(image, order):
return sitk.PermuteAxes(image, order)
@staticmethod
def axis_swap_order(image):
direction = np.array([int(round(d)) for d in image.GetDirection()])
if np.all(direction == np.array([1, 0, 0, 0, 1, 0, 0, 0, 1])):
return [0,1,2]
elif np.all(direction == np.array([0, 0, -1, 1, 0, 0, 0, -1, 0])):
return [2,0,1]
elif np.all(direction == np.array([1, 0, 0, 0, 0, 1, 0, -1, 0])):
return [0,2,1]
else:
print(list(direction))
return [0,1,2]
@staticmethod
def maybe_flip_axes(image):
direction = np.array([int(round(d)) for i,d in enumerate(image.GetDirection()) if i%4==0])
flips = [bool(d == -1) for d in direction]
return sitk.Flip(image, list(flips))
@staticmethod
def resize_image(original_CT, resize_to=[256,256,100]):
dimension = original_CT.GetDimension()
reference_physical_size = np.zeros(original_CT.GetDimension())
reference_physical_size[:] = [(sz-1)*spc if sz*spc>mx else mx for sz,spc,mx in zip(original_CT.GetSize(), original_CT.GetSpacing(), reference_physical_size)]
reference_origin = original_CT.GetOrigin()
reference_direction = original_CT.GetDirection()
reference_size = resize_to
reference_spacing = [ phys_sz/(sz-1) for sz,phys_sz in zip(reference_size, reference_physical_size) ]
reference_image = sitk.Image(reference_size, original_CT.GetPixelIDValue())
reference_image.SetOrigin(reference_origin)
reference_image.SetSpacing(reference_spacing)
reference_image.SetDirection(reference_direction)
reference_center = np.array(reference_image.TransformContinuousIndexToPhysicalPoint(np.array(reference_image.GetSize())/2.0))
transform = sitk.AffineTransform(dimension)
transform.SetMatrix(original_CT.GetDirection())
transform.SetTranslation(np.array(original_CT.GetOrigin()) - reference_origin)
centering_transform = sitk.TranslationTransform(dimension)
img_center = np.array(original_CT.TransformContinuousIndexToPhysicalPoint(np.array(original_CT.GetSize())/2.0))
centering_transform.SetOffset(np.array(transform.GetInverse().TransformPoint(img_center) - reference_center))
centered_transform = sitk.Transform(transform)
centered_transform = sitk.Transform(centering_transform)
return sitk.Resample(original_CT, reference_image, centered_transform, sitk.sitkLinear, 0.0)
def get_registered_case(self, brat_id_str):
""" Return stack of min-max normalized R:t1wCE, G:flair, B:T2 array """
stacked = np.zeros(tuple([self.resize_to[2],*self.resize_to[:2],3]))
try:
# init sitk reader
reader = sitk.ImageSeriesReader()
reader.LoadPrivateTagsOn()
reader.SetOutputPixelType(sitk.sitkFloat32)
# Use t1w as the reference and skip flair because it's sometimes completely dark
filenamesDICOM = reader.GetGDCMSeriesFileNames(f'{self.dicom_dir}/{brat_id_str}/T1wCE')
reader.SetFileNames(filenamesDICOM)
t1w = reader.Execute()
filenamesDICOM = reader.GetGDCMSeriesFileNames(f'{self.dicom_dir}/{brat_id_str}/FLAIR')
reader.SetFileNames(filenamesDICOM)
flair = reader.Execute()
filenamesDICOM = reader.GetGDCMSeriesFileNames(f'{self.dicom_dir}/{brat_id_str}/T2w')
reader.SetFileNames(filenamesDICOM)
t2 = reader.Execute()
# Align reference image and crop with otsu to minimum enclosing box
t1w = self.swap_image_axes(t1w, self.axis_swap_order(t1w))
t1w = self.maybe_flip_axes(t1w)
bounding_box = self.get_crop_bb(t1w)
# resample other modalities to align with reference
flair_resampled = self.resample(flair, t1w)
t2_resampled = self.resample(t2, t1w)
# crop all modalities to same box
t1w_cropped = self.crop_with_bb(t1w, bounding_box)
t1w_cropped = self.resize_image(t1w_cropped, self.resize_to)
flair_cropped = self.crop_with_bb(flair_resampled, bounding_box)
flair_cropped = self.resize_image(flair_cropped, self.resize_to)
t2_cropped = self.crop_with_bb(t2_resampled, bounding_box)
t2_cropped = self.resize_image(t2_cropped, self.resize_to)
# Return stack of min-max normalized R:t1w, G:T1ce, B:T2 array
stacked = np.stack([
self.normalize(sitk.GetArrayFromImage(t1w_cropped)),
self.normalize(sitk.GetArrayFromImage(flair_cropped)),
self.normalize(sitk.GetArrayFromImage(t2_cropped))
], axis=3)
except:
print(f'Error: {brat_id_str}')
return stacked
def get_aligned_seg_map(self, brat_id_str):
""" Normalizes task 1 segmentation map with similar logic and returns an aligned seg map """
if self.task1_dir is None:
return None
try:
segmentation_nii_fn = [os.path.join(f'{self.task1_dir}/BraTS2021_{brat_id_str}/', fn) for fn in os.listdir(f'{self.task1_dir}/BraTS2021_{brat_id_str}/') if 'seg' in fn]
t1_nii_fn = [os.path.join(f'{self.task1_dir}/BraTS2021_{brat_id_str}/', fn) for fn in os.listdir(f'{self.task1_dir}/BraTS2021_{brat_id_str}/') if 't1ce.' in fn]
if len(segmentation_nii_fn) == 0 or len(t1_nii_fn) ==0:
return None
t1 = sitk.ReadImage(t1_nii_fn[0])
seg = sitk.ReadImage(segmentation_nii_fn[0])
t1 = self.swap_image_axes(t1, self.axis_swap_order(t1))
t1 = self.maybe_flip_axes(t1)
bounding_box = self.get_crop_bb(t1)
seg_resampled = self.resample(seg, t1)
seg_cropped = self.crop_with_bb(seg_resampled, bounding_box)
seg_cropped = self.resize_image(seg_cropped, self.resize_to)
except:
print(f'Seg error: {brat_id_str}')
return None
return sitk.GetArrayFromImage(seg_cropped)
train_dir = train_dirs[200]
task1_dir = '../input/brats-2021-task1/BraTS2021_Training_Data'
resize_to=[320,320,160]
registered = Registered_BraTS_Case(dicom_dir=train_path, resize_to=resize_to, task1_dir=task1_dir)
reg_stack = registered.get_registered_case(brat_id_str=train_dir)
seg = registered.get_aligned_seg_map(brat_id_str=train_dir)
f, axs = plt.subplots(3,10, figsize=(20,5))
height_levels = resize_to[2]
for ind, j in enumerate(range(0, height_levels - 1, (height_levels//10))):
if ind > 9:break
axs[0,ind].imshow(reg_stack[j,:,:,:])
axs[1, ind].imshow(seg[j,:,:])
axs[2, ind].imshow(reg_stack[j,:,:,:], alpha=0.7)
axs[2, ind].imshow(seg[j,:,:], alpha=0.3)
axs[0,ind].set_title(f'Slice #{ind*(height_levels//10)}')
axs[0,0].set_ylabel('RGB')
axs[1,0].set_ylabel('Tumor')
axs[2,0].set_ylabel('RGB+Tumor')
plt.suptitle(f'BraTS ID: {train_dir}', fontsize=20)
plt.savefig('../media/registered_sample.png', transparent=False)
plt.show()
from glob import glob
from joblib import Parallel,delayed
import subprocess
from tqdm.auto import tqdm
train_path = '../input/rsna-miccai-brain-tumor-radiogenomic-classification/train/'
train_dirs = os.listdir(train_path)
test_path = '../input/rsna-miccai-brain-tumor-radiogenomic-classification/test/'
test_dirs = os.listdir(test_path)
reg_dir = os.path.join('../input', 'registered_cases_v3_320x320x160')
def maybe_create_dir(_dir):
if not os.path.exists(_dir): os.mkdir(_dir)
maybe_create_dir(reg_dir)
reg_test_dir = os.path.join(reg_dir, 'test')
reg_train_dir = os.path.join(reg_dir, 'train')
maybe_create_dir(reg_test_dir)
maybe_create_dir(reg_train_dir)
def save_case(brat_id_str, rgb_cohort_dir, resize_to, task1_dir, save_dir):
registered = Registered_BraTS_Case(dicom_dir=rgb_cohort_dir, resize_to=resize_to, task1_dir=task1_dir)
reg_stack = registered.get_registered_case(brat_id_str=brat_id_str)
seg = registered.get_aligned_seg_map(brat_id_str=brat_id_str)
np.save(os.path.join(save_dir, brat_id_str + '.npy'),reg_stack)
if seg is not None:
np.save(os.path.join(save_dir, brat_id_str + '_seg.npy'),seg)
def save_train_case(brat_id_str):
save_case(brat_id_str, train_path, resize_to, task1_dir, save_dir=reg_train_dir)
def save_test_case(brat_id_str):
save_case(brat_id_str, test_path, resize_to, task1_dir, save_dir=reg_test_dir)
n_jobs = 8
mp_train = Parallel(
n_jobs = n_jobs,
prefer = 'threads')(delayed(save_train_case)(brat_id_str) for brat_id_str in tqdm(train_dirs, total=len(train_dirs)))
mp_test = Parallel(
n_jobs = n_jobs,
prefer = 'threads')(delayed(save_test_case)(brat_id_str) for brat_id_str in tqdm(test_dirs, total=len(test_dirs)))
| 0.657209 | 0.864539 |
```
import astropy.coordinates as coord
import astropy.units as u
import numpy as np
from astropy.coordinates import BaseCoordinateFrame, Attribute, RepresentationMapping
from astropy.coordinates import frame_transform_graph
def polar2cart(ra,dec):
x = np.cos(np.deg2rad(ra)) * np.cos(np.deg2rad(dec))
y = np.sin(np.deg2rad(ra)) * np.cos(np.deg2rad(dec))
z = np.sin(np.deg2rad(dec))
return np.array([x,y,z])
def cart2polar(vector):
ra = np.arctan2(vector[1],vector[0])
dec = np.arcsin(vector[2])
return np.rad2deg(ra), np.rad2deg(dec)
def construct_scy(scx_ra, scx_dec, scz_ra, scz_dec):
x = polar2cart(scx_ra, scx_dec)
z = polar2cart(scz_ra, scz_dec)
return cart2polar(np.cross(x,-z))
def construct_sc_matrix(scx_ra, scx_dec, scy_ra, scy_dec, scz_ra, scz_dec):
sc_matrix = np.zeros((3,3))
sc_matrix[0,:] = polar2cart(scx_ra, scx_dec)
sc_matrix[1,:] = polar2cart(scy_ra, scy_dec)
sc_matrix[2,:] = polar2cart(scz_ra, scz_dec)
return sc_matrix
class SPIFrame(BaseCoordinateFrame):
"""
INTEGRAL SPI Frame
Parameters
----------
representation : `BaseRepresentation` or None
A representation object or None to have no data (or use the other keywords)
"""
default_representation = coord.SphericalRepresentation
frame_specific_representation_info = {
'spherical': [
RepresentationMapping(
reprname='lon', framename='lon', defaultunit=u.degree),
RepresentationMapping(
reprname='lat', framename='lat', defaultunit=u.degree),
RepresentationMapping(
reprname='distance', framename='DIST', defaultunit=None)
],
'unitspherical': [
RepresentationMapping(
reprname='lon', framename='lon', defaultunit=u.degree),
RepresentationMapping(
reprname='lat', framename='lat', defaultunit=u.degree)
],
'cartesian': [
RepresentationMapping(
reprname='x', framename='SCX'), RepresentationMapping(
reprname='y', framename='SCY'), RepresentationMapping(
reprname='z', framename='SCZ')
]
}
# Specify frame attributes required to fully specify the frame
scx_ra = Attribute(default=None)
scx_dec = Attribute(default=None)
scy_ra = Attribute(default=None)
scy_dec = Attribute(default=None)
scz_ra = Attribute(default=None)
scz_dec = Attribute(default=None)
# equinox = TimeFrameAttribute(default='J2000')
@frame_transform_graph.transform(coord.FunctionTransform, SPIFrame, coord.ICRS)
def spi_to_j2000(spi_coord, j2000_frame):
"""
"""
sc_matrix = construct_sc_matrix(spi_coord.scx_ra,
spi_coord.scx_dec,
spi_coord.scy_ra,
spi_coord.scy_dec,
spi_coord.scz_ra,
spi_coord.scz_dec)
# X,Y,Z = gbm_coord.cartesian
pos = spi_coord.cartesian.xyz.value
X0 = np.dot(sc_matrix[:, 0], pos)
X1 = np.dot(sc_matrix[:, 1], pos)
X2 = np.clip(np.dot(sc_matrix[:, 2], pos), -1., 1.)
dec = np.arcsin(X2)
idx = np.logical_and(np.abs(X0) < 1E-6, np.abs(X1) < 1E-6)
ra = np.zeros_like(dec)
ra[~idx] = np.arctan2(X1, X0) % (2 * np.pi)
return coord.ICRS(ra=ra * u.radian, dec=dec * u.radian)
@frame_transform_graph.transform(coord.FunctionTransform, coord.ICRS, SPIFrame)
def j2000_to_spi(j2000_frame, spi_coord):
"""
"""
sc_matrix = construct_sc_matrix(spi_coord.scx_ra,
spi_coord.scx_dec,
spi_coord.scy_ra,
spi_coord.scy_dec,
spi_coord.scz_ra,
spi_coord.scz_dec)
pos = j2000_frame.cartesian.xyz.value
X0 = np.dot(sc_matrix[0, :], pos)
X1 = np.dot(sc_matrix[1, :], pos)
X2 = np.dot(sc_matrix[2, :], pos)
lat = np.pi / 2. - np.arccos(X2) # convert to proper frame
idx = np.logical_and(np.abs(X0) < 1E-6, np.abs(X1) < 1E-6)
lon = np.zeros_like(lat)
lon[~idx] = np.arctan2(X1, X0) % (2 * np.pi)
return SPIFrame(
lon=lon * u.radian,
lat=lat * u.radian,
scx_ra=spi_coord.scx_ra,
scx_dec=spi_coord.scx_dec,
scy_ra=spi_coord.scy_ra,
scy_dec=spi_coord.scy_dec,
scz_ra=spi_coord.scz_ra,
scz_dec=spi_coord.scz_dec
)
class SPIFrame(coord.Sp):
from astropy.coordinates.coordsystems import SphericalCoordinateBase
from astropy.coordinates import SphericalCoordinatesBase
SphericalRepresentation?
spi_frame = SPIFrame(lat=90.*u.deg,lon=0.*u.deg
,
# unit='deg',
scx_ra=206.1563262939453,
scx_dec=50.5438346862793,
scy_ra=65.40575996001127,
scy_dec=32.51224513331057,
scz_ra=142.12562561035156,
scz_dec=np.zeros((3,3)))
from astropy.coordinates import ICRS
spi_frame.transform_to(ICRS)
import astropy.io.fits as fits
f = fits.open('sc_orbit_param.fits.gz')
def extract_sc_pointing(pointing_extension, time_index):
scx_ra = pointing_extension.data['RA_SCX'][time_index]
scx_dec = pointing_extension.data['DEC_SCX'][time_index]
scz_ra = pointing_extension.data['RA_SCZ'][time_index]
scz_dec = pointing_extension.data['DEC_SCZ'][time_index]
scy_ra, scy_dec = construct_scy(scx_ra, scx_dec, scz_ra, scz_dec)
return scx_ra, scx_dec, scy_ra, scy_dec, scz_ra, scz_dec
def polar2cart(ra,dec):
x = np.cos(np.deg2rad(ra)) * np.cos(np.deg2rad(dec))
y = np.sin(np.deg2rad(ra)) * np.cos(np.deg2rad(dec))
z = np.sin(np.deg2rad(dec))
return np.array([x,y,z])
def cart2polar(vector):
r=np.sqrt((np.array(vector)**2).sum())
ra = np.arctan2(vector[1],vector[0])
dec = np.arcsin(vector[2]/r)
return np.rad2deg(ra), np.rad2deg(dec)
def construct_scy(scx_ra, scx_dec, scz_ra, scz_dec):
x = polar2cart(scx_ra, scx_dec)
z = polar2cart(scz_ra, scz_dec)
return cart2polar(np.cross(x,-z))
def construct_sc_matrix(scx_ra, scx_dec, scy_ra, scy_dec, scz_ra, scz_dec):
sc_matrix = np.zeros((3,3))
sc_matrix[0,:] = polar2cart(scx_ra, scx_dec)
sc_matrix[1,:] = polar2cart(scy_ra, scy_dec)
sc_matrix[2,:] = polar2cart(scz_ra, scz_dec)
return sc_matrix
f.info()
def transform_via_matric(sc_matrix, ra, dec):
pos = polar2cart(ra,dec)
X0 = np.dot(sc_matrix[0, :], pos)
X1 = np.dot(sc_matrix[1, :], pos)
X2 = np.dot(sc_matrix[2, :], pos)
lat = np.pi / 2. - np.arccos(X2) # convert to proper frame
#idx = np.logical_and(np.abs(X0) < 1E-6, np.abs(X1) < 1E-6)
#log = np.zeros_like(el)
lon = np.arctan2(X1, X0) % (2 * np.pi)
return np.rad2deg(lon), np.rad2deg(lat)
from astropy.coordinates import spherical_to_cartesian, cartesian_to_spherical, CartesianRepresentation
a=CartesianRepresentation(1,1,1)
a.
spherical_to_cartesian?
_, lat, lon = cartesian_to_spherical(1,1,1)
print(lon.deg,lat.deg)
spherical_to_cartesian(1.,np.deg2rad(30.),np.deg2rad(20.)).value
cartesian_to_spherical?
extract_sc_pointing(f[1],0)
from astropy.coordinates import SkyCoord
source = SkyCoord(ra=197.075, dec=58.98,unit='deg',frame='icrs')
center = SkyCoord(ra=206.1563262939453, dec=50.5438346862793, unit='deg', frame='icrs')
source.separation(center).value
sc_mat = construct_sc_matrix(*extract_sc_pointing(f[1],0))
transform_via_matric(sc_mat,ra=197.075, dec=58.98 )
source = SkyCoord(9.88,-0.6956381146530,unit='deg')
#source = SkyCoord(0, 10,unit='deg')
center = SkyCoord(0, 0,unit='deg',)
source.separation(center).value
transform(*extract_sc_pointing(f[1],0), ra=197.075, dec=58.98)
def cos_dir(sc_ra,sc_dec, ra, dec):
return np.sin(np.deg2rad(sc_dec)) * np.sin(np.deg2rad(dec)) + np.cos(np.deg2rad(sc_dec)) * np.cos(np.deg2rad(dec)) * np.cos(np.deg2rad(sc_ra- ra) )
def transform(scx_ra, scx_dec, scy_ra, scy_dec, scz_ra, scz_dec, ra, dec):
cos_theta = cos_dir(scx_ra,scx_dec, ra, dec)
cos_theta = np.clip(cos_theta,-1.,1.)
theta = np.rad2deg(np.arccos(cos_theta))
cos_z = cos_dir(scz_ra,scz_dec, ra, dec)
cos_y = cos_dir(scy_ra,scy_dec, ra, dec)
phi = np.rad2deg(np.arctan2(cos_z, cos_y))
if phi < 0.0:
phi += 360.
return theta, phi
Attribute()
```
|
github_jupyter
|
import astropy.coordinates as coord
import astropy.units as u
import numpy as np
from astropy.coordinates import BaseCoordinateFrame, Attribute, RepresentationMapping
from astropy.coordinates import frame_transform_graph
def polar2cart(ra,dec):
x = np.cos(np.deg2rad(ra)) * np.cos(np.deg2rad(dec))
y = np.sin(np.deg2rad(ra)) * np.cos(np.deg2rad(dec))
z = np.sin(np.deg2rad(dec))
return np.array([x,y,z])
def cart2polar(vector):
ra = np.arctan2(vector[1],vector[0])
dec = np.arcsin(vector[2])
return np.rad2deg(ra), np.rad2deg(dec)
def construct_scy(scx_ra, scx_dec, scz_ra, scz_dec):
x = polar2cart(scx_ra, scx_dec)
z = polar2cart(scz_ra, scz_dec)
return cart2polar(np.cross(x,-z))
def construct_sc_matrix(scx_ra, scx_dec, scy_ra, scy_dec, scz_ra, scz_dec):
sc_matrix = np.zeros((3,3))
sc_matrix[0,:] = polar2cart(scx_ra, scx_dec)
sc_matrix[1,:] = polar2cart(scy_ra, scy_dec)
sc_matrix[2,:] = polar2cart(scz_ra, scz_dec)
return sc_matrix
class SPIFrame(BaseCoordinateFrame):
"""
INTEGRAL SPI Frame
Parameters
----------
representation : `BaseRepresentation` or None
A representation object or None to have no data (or use the other keywords)
"""
default_representation = coord.SphericalRepresentation
frame_specific_representation_info = {
'spherical': [
RepresentationMapping(
reprname='lon', framename='lon', defaultunit=u.degree),
RepresentationMapping(
reprname='lat', framename='lat', defaultunit=u.degree),
RepresentationMapping(
reprname='distance', framename='DIST', defaultunit=None)
],
'unitspherical': [
RepresentationMapping(
reprname='lon', framename='lon', defaultunit=u.degree),
RepresentationMapping(
reprname='lat', framename='lat', defaultunit=u.degree)
],
'cartesian': [
RepresentationMapping(
reprname='x', framename='SCX'), RepresentationMapping(
reprname='y', framename='SCY'), RepresentationMapping(
reprname='z', framename='SCZ')
]
}
# Specify frame attributes required to fully specify the frame
scx_ra = Attribute(default=None)
scx_dec = Attribute(default=None)
scy_ra = Attribute(default=None)
scy_dec = Attribute(default=None)
scz_ra = Attribute(default=None)
scz_dec = Attribute(default=None)
# equinox = TimeFrameAttribute(default='J2000')
@frame_transform_graph.transform(coord.FunctionTransform, SPIFrame, coord.ICRS)
def spi_to_j2000(spi_coord, j2000_frame):
"""
"""
sc_matrix = construct_sc_matrix(spi_coord.scx_ra,
spi_coord.scx_dec,
spi_coord.scy_ra,
spi_coord.scy_dec,
spi_coord.scz_ra,
spi_coord.scz_dec)
# X,Y,Z = gbm_coord.cartesian
pos = spi_coord.cartesian.xyz.value
X0 = np.dot(sc_matrix[:, 0], pos)
X1 = np.dot(sc_matrix[:, 1], pos)
X2 = np.clip(np.dot(sc_matrix[:, 2], pos), -1., 1.)
dec = np.arcsin(X2)
idx = np.logical_and(np.abs(X0) < 1E-6, np.abs(X1) < 1E-6)
ra = np.zeros_like(dec)
ra[~idx] = np.arctan2(X1, X0) % (2 * np.pi)
return coord.ICRS(ra=ra * u.radian, dec=dec * u.radian)
@frame_transform_graph.transform(coord.FunctionTransform, coord.ICRS, SPIFrame)
def j2000_to_spi(j2000_frame, spi_coord):
"""
"""
sc_matrix = construct_sc_matrix(spi_coord.scx_ra,
spi_coord.scx_dec,
spi_coord.scy_ra,
spi_coord.scy_dec,
spi_coord.scz_ra,
spi_coord.scz_dec)
pos = j2000_frame.cartesian.xyz.value
X0 = np.dot(sc_matrix[0, :], pos)
X1 = np.dot(sc_matrix[1, :], pos)
X2 = np.dot(sc_matrix[2, :], pos)
lat = np.pi / 2. - np.arccos(X2) # convert to proper frame
idx = np.logical_and(np.abs(X0) < 1E-6, np.abs(X1) < 1E-6)
lon = np.zeros_like(lat)
lon[~idx] = np.arctan2(X1, X0) % (2 * np.pi)
return SPIFrame(
lon=lon * u.radian,
lat=lat * u.radian,
scx_ra=spi_coord.scx_ra,
scx_dec=spi_coord.scx_dec,
scy_ra=spi_coord.scy_ra,
scy_dec=spi_coord.scy_dec,
scz_ra=spi_coord.scz_ra,
scz_dec=spi_coord.scz_dec
)
class SPIFrame(coord.Sp):
from astropy.coordinates.coordsystems import SphericalCoordinateBase
from astropy.coordinates import SphericalCoordinatesBase
SphericalRepresentation?
spi_frame = SPIFrame(lat=90.*u.deg,lon=0.*u.deg
,
# unit='deg',
scx_ra=206.1563262939453,
scx_dec=50.5438346862793,
scy_ra=65.40575996001127,
scy_dec=32.51224513331057,
scz_ra=142.12562561035156,
scz_dec=np.zeros((3,3)))
from astropy.coordinates import ICRS
spi_frame.transform_to(ICRS)
import astropy.io.fits as fits
f = fits.open('sc_orbit_param.fits.gz')
def extract_sc_pointing(pointing_extension, time_index):
scx_ra = pointing_extension.data['RA_SCX'][time_index]
scx_dec = pointing_extension.data['DEC_SCX'][time_index]
scz_ra = pointing_extension.data['RA_SCZ'][time_index]
scz_dec = pointing_extension.data['DEC_SCZ'][time_index]
scy_ra, scy_dec = construct_scy(scx_ra, scx_dec, scz_ra, scz_dec)
return scx_ra, scx_dec, scy_ra, scy_dec, scz_ra, scz_dec
def polar2cart(ra,dec):
x = np.cos(np.deg2rad(ra)) * np.cos(np.deg2rad(dec))
y = np.sin(np.deg2rad(ra)) * np.cos(np.deg2rad(dec))
z = np.sin(np.deg2rad(dec))
return np.array([x,y,z])
def cart2polar(vector):
r=np.sqrt((np.array(vector)**2).sum())
ra = np.arctan2(vector[1],vector[0])
dec = np.arcsin(vector[2]/r)
return np.rad2deg(ra), np.rad2deg(dec)
def construct_scy(scx_ra, scx_dec, scz_ra, scz_dec):
x = polar2cart(scx_ra, scx_dec)
z = polar2cart(scz_ra, scz_dec)
return cart2polar(np.cross(x,-z))
def construct_sc_matrix(scx_ra, scx_dec, scy_ra, scy_dec, scz_ra, scz_dec):
sc_matrix = np.zeros((3,3))
sc_matrix[0,:] = polar2cart(scx_ra, scx_dec)
sc_matrix[1,:] = polar2cart(scy_ra, scy_dec)
sc_matrix[2,:] = polar2cart(scz_ra, scz_dec)
return sc_matrix
f.info()
def transform_via_matric(sc_matrix, ra, dec):
pos = polar2cart(ra,dec)
X0 = np.dot(sc_matrix[0, :], pos)
X1 = np.dot(sc_matrix[1, :], pos)
X2 = np.dot(sc_matrix[2, :], pos)
lat = np.pi / 2. - np.arccos(X2) # convert to proper frame
#idx = np.logical_and(np.abs(X0) < 1E-6, np.abs(X1) < 1E-6)
#log = np.zeros_like(el)
lon = np.arctan2(X1, X0) % (2 * np.pi)
return np.rad2deg(lon), np.rad2deg(lat)
from astropy.coordinates import spherical_to_cartesian, cartesian_to_spherical, CartesianRepresentation
a=CartesianRepresentation(1,1,1)
a.
spherical_to_cartesian?
_, lat, lon = cartesian_to_spherical(1,1,1)
print(lon.deg,lat.deg)
spherical_to_cartesian(1.,np.deg2rad(30.),np.deg2rad(20.)).value
cartesian_to_spherical?
extract_sc_pointing(f[1],0)
from astropy.coordinates import SkyCoord
source = SkyCoord(ra=197.075, dec=58.98,unit='deg',frame='icrs')
center = SkyCoord(ra=206.1563262939453, dec=50.5438346862793, unit='deg', frame='icrs')
source.separation(center).value
sc_mat = construct_sc_matrix(*extract_sc_pointing(f[1],0))
transform_via_matric(sc_mat,ra=197.075, dec=58.98 )
source = SkyCoord(9.88,-0.6956381146530,unit='deg')
#source = SkyCoord(0, 10,unit='deg')
center = SkyCoord(0, 0,unit='deg',)
source.separation(center).value
transform(*extract_sc_pointing(f[1],0), ra=197.075, dec=58.98)
def cos_dir(sc_ra,sc_dec, ra, dec):
return np.sin(np.deg2rad(sc_dec)) * np.sin(np.deg2rad(dec)) + np.cos(np.deg2rad(sc_dec)) * np.cos(np.deg2rad(dec)) * np.cos(np.deg2rad(sc_ra- ra) )
def transform(scx_ra, scx_dec, scy_ra, scy_dec, scz_ra, scz_dec, ra, dec):
cos_theta = cos_dir(scx_ra,scx_dec, ra, dec)
cos_theta = np.clip(cos_theta,-1.,1.)
theta = np.rad2deg(np.arccos(cos_theta))
cos_z = cos_dir(scz_ra,scz_dec, ra, dec)
cos_y = cos_dir(scy_ra,scy_dec, ra, dec)
phi = np.rad2deg(np.arctan2(cos_z, cos_y))
if phi < 0.0:
phi += 360.
return theta, phi
Attribute()
| 0.666605 | 0.627951 |
## importing libraries and data
```
from potosnail import MachineLearning, DeepLearning, DataHelper, Evaluater, Algorithms, Wrappers
from sklearn.datasets import load_breast_cancer
import numpy as np
import pandas as pd
import seaborn as sns
from matplotlib import pyplot as plt
ml = MachineLearning()
dl = DeepLearning()
dh = DataHelper()
ev = Evaluater()
al = Algorithms()
wr = Wrappers()
```
## creating dataframe
```
df = pd.DataFrame(load_breast_cancer()['data'])
df.columns = list(load_breast_cancer()['feature_names'])
df['malignant'] = load_breast_cancer()['target']
df.head()
```
## Creating training and validaton data
```
train, test = dh.HoldOut(df)
X = train.drop(['malignant'], axis='columns')
Xval = test.drop(['malignant'], axis='columns')
y = train['malignant']
yval = test['malignant']
```
## checking for class imbalance
```
y.value_counts()
```
## using SmoteIt to balance classes
Smote isn't usually used on data with this low of class imbalance, this is just to demo
```
df2, y = dh.SmoteIt(X, y, bool_arr=[]) # bool_arr is empty because we have no categorical features
df2['malignant'] = y
df2.head()
df2['malignant'].value_counts()
```
## picking best model with CompareModels
```
X2 = df2.drop(['malignant'], axis='columns')
y2 = df2['malignant']
ml.CompareModels(X2, y2, 'classification')
```
## Let's Optimize!!
Let's go with XGB, XGBClassifier :), this is because it has the highest test accuracy
```
from sklearn.ensemble import GradientBoostingClassifier
from xgboost import XGBClassifier
estimators = al.Estimators(X2, 100) #returns a list of n_estimators to test between
grid = {'learning_rate': [0.1, 0.2], 'max_depth': [3, 6, 9], 'min_child_weight': [1, 2],
'subsample': [0.5, 0.7, 1], 'n_estimators': estimators}
clf = ml.Optimize(XGBClassifier(), grid, X2, y2)
```
Splitting data and Evaluating tuned models with HoldOut and ScoreModel
```
ev.ScoreModel(clf, Xval, yval)
```
## Using BuildConfusion for evaluation
```
ev.BuildConfusion(clf, Xval, yval)
```
## Deep Learning approach
```
X, y = dh.SmoteIt(load_breast_cancer()['data'], load_breast_cancer()['target'], bool_arr=[])
```
## Model Building
You can use DeepLearning().FastNN() with task set to 'classification'
```
output_dim = 2 #because we have 2 classes (malignant and not malignant)
nodes = 64 #first layer will have 64 nodes
activation = 'relu' #Rectified Linear Unit activation function
regularizer = 'L2' #prevents overfitting
stacking = True #second layer will also have 64 nodes
dropout = False #another overfitting solution, we will not be using it this time
nlayers = 4 #neural network will have 4 layers
closer = False
loss = 'binary_crossentropy' #best loss function for binary classification
optimizer = 'adam' #adaptive momentum estimation optimization function
model = dl.DeepTabularClassification(output_dim, nodes, activation, regularizer, stacking,
dropout, nlayers, closer, loss, optimizer)
```
visit https://machinelearningmastery.com/adam-optimization-algorithm-for-deep-learning/ to read more about adam if you want
```
y3 = dl.MulticlassOutput(np.array(y2)) #using MulticlassOutput to encode labels
yv2 = dl.MulticlassOutput(np.array(yval))
history = model.fit(X2, y3, batch_size=32, epochs=75, validation_split=0.2)
ev.ViewLoss(history)
ev.ViewAccuracy(history)
ev.BuildConfusionDL(model, Xval, yv2)
```
## A fast and dirty approach with Wrappers().WrapML()
```
final_model, data, scaler, dim = wr.WrapML(df, 'malignant', 'classification', quiet=False)
final_model
print(scaler)
print(data.columns)
print(data.shape)
```
|
github_jupyter
|
from potosnail import MachineLearning, DeepLearning, DataHelper, Evaluater, Algorithms, Wrappers
from sklearn.datasets import load_breast_cancer
import numpy as np
import pandas as pd
import seaborn as sns
from matplotlib import pyplot as plt
ml = MachineLearning()
dl = DeepLearning()
dh = DataHelper()
ev = Evaluater()
al = Algorithms()
wr = Wrappers()
df = pd.DataFrame(load_breast_cancer()['data'])
df.columns = list(load_breast_cancer()['feature_names'])
df['malignant'] = load_breast_cancer()['target']
df.head()
train, test = dh.HoldOut(df)
X = train.drop(['malignant'], axis='columns')
Xval = test.drop(['malignant'], axis='columns')
y = train['malignant']
yval = test['malignant']
y.value_counts()
df2, y = dh.SmoteIt(X, y, bool_arr=[]) # bool_arr is empty because we have no categorical features
df2['malignant'] = y
df2.head()
df2['malignant'].value_counts()
X2 = df2.drop(['malignant'], axis='columns')
y2 = df2['malignant']
ml.CompareModels(X2, y2, 'classification')
from sklearn.ensemble import GradientBoostingClassifier
from xgboost import XGBClassifier
estimators = al.Estimators(X2, 100) #returns a list of n_estimators to test between
grid = {'learning_rate': [0.1, 0.2], 'max_depth': [3, 6, 9], 'min_child_weight': [1, 2],
'subsample': [0.5, 0.7, 1], 'n_estimators': estimators}
clf = ml.Optimize(XGBClassifier(), grid, X2, y2)
ev.ScoreModel(clf, Xval, yval)
ev.BuildConfusion(clf, Xval, yval)
X, y = dh.SmoteIt(load_breast_cancer()['data'], load_breast_cancer()['target'], bool_arr=[])
output_dim = 2 #because we have 2 classes (malignant and not malignant)
nodes = 64 #first layer will have 64 nodes
activation = 'relu' #Rectified Linear Unit activation function
regularizer = 'L2' #prevents overfitting
stacking = True #second layer will also have 64 nodes
dropout = False #another overfitting solution, we will not be using it this time
nlayers = 4 #neural network will have 4 layers
closer = False
loss = 'binary_crossentropy' #best loss function for binary classification
optimizer = 'adam' #adaptive momentum estimation optimization function
model = dl.DeepTabularClassification(output_dim, nodes, activation, regularizer, stacking,
dropout, nlayers, closer, loss, optimizer)
y3 = dl.MulticlassOutput(np.array(y2)) #using MulticlassOutput to encode labels
yv2 = dl.MulticlassOutput(np.array(yval))
history = model.fit(X2, y3, batch_size=32, epochs=75, validation_split=0.2)
ev.ViewLoss(history)
ev.ViewAccuracy(history)
ev.BuildConfusionDL(model, Xval, yv2)
final_model, data, scaler, dim = wr.WrapML(df, 'malignant', 'classification', quiet=False)
final_model
print(scaler)
print(data.columns)
print(data.shape)
| 0.741019 | 0.970042 |
# Tabular data training and serving with Keras and Ray AIR
This notebook is adapted from [a Keras tutorial](https://www.tensorflow.org/tfx/tutorials/tfx/components_keras).
It uses [Chicago Taxi dataset](https://data.cityofchicago.org/Transportation/Taxi-Trips/wrvz-psew) and a DNN Keras model to predict whether a trip may generate a big tip.
In this example, we showcase how to achieve the same tasks as the Keras Tutorial using [Ray AIR](https://docs.ray.io/en/latest/ray-air/getting-started.html), covering
every step from data ingestion to pushing a model to serving.
1. Read a CSV into [Ray Dataset](https://docs.ray.io/en/latest/data/dataset.html).
2. Process the dataset by chaining [Ray AIR preprocessors](https://docs.ray.io/en/latest/ray-air/getting-started.html#preprocessors).
3. Train the model using the TensorflowTrainer from AIR.
4. Serve the model using Ray Serve and the above preprocessors.
Uncomment and run the following line in order to install all the necessary dependencies:
```
# ! pip install "tensorflow>=2.8.0" "ray[tune, data, serve]>=1.12.1"
# ! pip install fastapi
```
## Set up Ray
We will use `ray.init()` to initialize a local cluster. By default, this cluster will be composed of only the machine you are running this notebook on. If you wish to attach to an existing Ray cluster, you can do so through `ray.init(address="auto")`.
```
from pprint import pprint
import ray
ray.shutdown()
ray.init()
```
We can check the resources our cluster is composed of. If you are running this notebook on your local machine or Google Colab, you should see the number of CPU cores and GPUs available on the said machine.
```
pprint(ray.cluster_resources())
```
## Getting the data
Let's start with defining a helper function to get the data to work with. Some columns are dropped for simplicity.
```
import pandas as pd
LABEL = "is_big_tip"
def get_data() -> pd.DataFrame:
"""Fetch the taxi fare data to work on."""
_data = pd.read_csv(
"https://raw.githubusercontent.com/tensorflow/tfx/master/"
"tfx/examples/chicago_taxi_pipeline/data/simple/data.csv"
)
_data[LABEL] = _data["tips"] / _data["fare"] > 0.2
# We drop some columns here for the sake of simplicity.
return _data.drop(
[
"tips",
"fare",
"dropoff_latitude",
"dropoff_longitude",
"pickup_latitude",
"pickup_longitude",
"pickup_census_tract",
],
axis=1,
)
data = get_data()
```
Now let's take a look at the data. Notice that some values are missing. This is exactly where preprocessing comes into the picture. We will come back to this in the preprocessing session below.
```
data.head(5)
```
We continue to split the data into training and test data.
For the test data, we separate out the features to run serving on as well as labels to compare serving results with.
```
import numpy as np
from sklearn.model_selection import train_test_split
from typing import Tuple
def split_data(data: pd.DataFrame) -> Tuple[ray.data.Dataset, pd.DataFrame, np.array]:
"""Split the data in a stratified way.
Returns:
A tuple containing train dataset, test data and test label.
"""
# There is a native offering in Ray Dataset for split as well.
# However, supporting stratification is a TODO there. So use
# scikit-learn equivalent here.
train_data, test_data = train_test_split(
data, stratify=data[[LABEL]], random_state=1113
)
_train_ds = ray.data.from_pandas(train_data)
_test_label = test_data[LABEL].values
_test_df = test_data.drop([LABEL], axis=1)
return _train_ds, _test_df, _test_label
train_ds, test_df, test_label = split_data(data)
print(f"There are {train_ds.count()} samples for training and {test_df.shape[0]} samples for testing.")
```
## Preprocessing
Let's focus on preprocessing first.
Usually, input data needs to go through some preprocessing before being
fed into model. It is a good idea to package preprocessing logic into
a modularized component so that the same logic can be applied to both
training data as well as data for online serving or offline batch prediction.
In AIR, this component is a [`Preprocessor`](https://docs.ray.io/en/latest/ray-air/getting-started.html#preprocessors).
It is constructed in a way that allows easy composition.
Now let's construct a chained preprocessor composed of simple preprocessors, including
1. Imputer for filling missing features;
2. OneHotEncoder for encoding categorical features;
3. BatchMapper where arbitrary user-defined function can be applied to batches of records;
and so on. Take a look at [`Preprocessor`](https://docs.ray.io/en/latest/ray-air/getting-started.html#preprocessors).
The output of the preprocessing step goes into model for training.
```
from ray.data.preprocessors import (
BatchMapper,
Chain,
OneHotEncoder,
SimpleImputer,
)
def get_preprocessor():
"""Construct a chain of preprocessors."""
imputer1 = SimpleImputer(
["dropoff_census_tract"], strategy="most_frequent"
)
imputer2 = SimpleImputer(
["pickup_community_area", "dropoff_community_area"],
strategy="most_frequent",
)
imputer3 = SimpleImputer(["payment_type"], strategy="most_frequent")
imputer4 = SimpleImputer(
["company"], strategy="most_frequent")
imputer5 = SimpleImputer(
["trip_start_timestamp", "trip_miles", "trip_seconds"], strategy="mean"
)
ohe = OneHotEncoder(
columns=[
"trip_start_hour",
"trip_start_day",
"trip_start_month",
"dropoff_census_tract",
"pickup_community_area",
"dropoff_community_area",
"payment_type",
"company",
],
limit={
"dropoff_census_tract": 25,
"pickup_community_area": 20,
"dropoff_community_area": 20,
"payment_type": 2,
"company": 7,
},
)
def batch_mapper_fn(df):
df["trip_start_year"] = pd.to_datetime(df["trip_start_timestamp"], unit="s").dt.year
df = df.drop(["trip_start_timestamp"], axis=1)
return df
chained_pp = Chain(
imputer1,
imputer2,
imputer3,
imputer4,
imputer5,
ohe,
BatchMapper(batch_mapper_fn),
)
return chained_pp
```
Now let's define some constants for clarity.
```
# Note that `INPUT_SIZE` here is corresponding to the output dimension
# of the previously defined processing steps.
# This is used to specify the input shape of Keras model as well as
# when converting from training data from `ray.data.Dataset` to `tf.Tensor`.
INPUT_SIZE = 120
# The training batch size. Based on `NUM_WORKERS`, each worker
# will get its own share of this batch size. For example, if
# `NUM_WORKERS = 2`, each worker will work on 4 samples per batch.
BATCH_SIZE = 8
# Number of epoch. Adjust it based on how quickly you want the run to be.
EPOCH = 1
# Number of training workers.
# Adjust this accordingly based on the resources you have!
NUM_WORKERS = 2
```
## Training
Let's starting with defining a simple Keras model for the classification task.
```
import tensorflow as tf
def build_model():
model = tf.keras.models.Sequential()
model.add(tf.keras.Input(shape=(INPUT_SIZE,)))
model.add(tf.keras.layers.Dense(50, activation="relu"))
model.add(tf.keras.layers.Dense(1, activation="sigmoid"))
return model
```
Now let's define the training loop. This code will be run on each training
worker in a distributed fashion. See more details [here](https://docs.ray.io/en/latest/train/train.html).
```
from ray import train
from ray.train.tensorflow import prepare_dataset_shard
from ray.tune.integration.keras import TuneReportCallback
def train_loop_per_worker():
dataset_shard = train.get_dataset_shard("train")
strategy = tf.distribute.experimental.MultiWorkerMirroredStrategy()
with strategy.scope():
model = build_model()
model.compile(
loss="binary_crossentropy",
optimizer="adam",
metrics=["accuracy"],
)
for epoch in range(EPOCH):
# This will make sure that the training workers will get their own
# share of batch to work on.
# See `ray.train.tensorflow.prepare_dataset_shard` for more information.
tf_dataset = prepare_dataset_shard(
dataset_shard.to_tf(
label_column=LABEL,
output_signature=(
tf.TensorSpec(shape=(BATCH_SIZE, INPUT_SIZE), dtype=tf.float32),
tf.TensorSpec(shape=(BATCH_SIZE,), dtype=tf.int64),
),
batch_size=BATCH_SIZE,
drop_last=True,
)
)
model.fit(tf_dataset, verbose=0)
# This saves checkpoint in a way that can be used by Ray Serve coherently.
train.save_checkpoint(epoch=epoch, model=model.get_weights())
```
Now let's define a trainer that takes in the training loop,
the training dataset as well the preprocessor that we just defined.
And run it!
Notice that you can tune how long you want the run to be by changing ``EPOCH``.
```
from ray.train.tensorflow import TensorflowTrainer
trainer = TensorflowTrainer(
train_loop_per_worker=train_loop_per_worker,
scaling_config={"num_workers": NUM_WORKERS},
datasets={"train": train_ds},
preprocessor=get_preprocessor(),
)
result = trainer.fit()
```
## Moving on to Serve
We will use Ray Serve to serve the trained model. A core concept of Ray Serve is [Deployment](https://docs.ray.io/en/latest/serve/core-apis.html). It allows you to define and update your business logic or models that will handle incoming requests as well as how this is exposed over HTTP or in Python.
In the case of serving model, `ray.serve.model_wrappers.ModelWrapper` and `ray.serve.model_wrappers.ModelWrapperDeployment` wrap a `ray.air.checkpoint.Checkpoint` into a Ray Serve deployment that can readily serve HTTP requests.
Note, ``Checkpoint`` captures both model and preprocessing steps in a way compatible with Ray Serve and ensures that ml workload can transition seamlessly between training and
serving.
This removes the boilerplate code and minimizes the effort to bring your model to production!
Generally speaking the http request can either send in json or data.
Upon receiving this payload, Ray Serve would need some "adapter" to convert the request payload into some shape or form that can be accepted as input to the preprocessing steps. In this case, we send in a json request and converts it to a pandas DataFrame through `dataframe_adapter`, defined below.
```
from fastapi import Request
async def dataframe_adapter(request: Request):
"""Serve HTTP Adapter that reads JSON and converts to pandas DataFrame."""
content = await request.json()
return pd.DataFrame.from_dict(content)
```
Now let's wrap everything in a serve endpoint that exposes a URL to where requests can be sent to.
```
from ray import serve
from ray.air.checkpoint import Checkpoint
from ray.train.tensorflow import TensorflowPredictor
from ray.serve.model_wrappers import ModelWrapperDeployment
def serve_model(checkpoint: Checkpoint, model_definition, adapter, name="Model") -> str:
"""Expose a serve endpoint.
Returns:
serve URL.
"""
serve.start(detached=True)
deployment = ModelWrapperDeployment.options(name=name)
deployment.deploy(
TensorflowPredictor,
checkpoint,
# This is due to a current limitation on Serve that's
# being addressed.
# TODO(xwjiang): Change to True.
batching_params=False,
model_definition=model_definition,
http_adapter=adapter,
)
return deployment.url
# Generally speaking, training and serving are done in totally different ray clusters.
# To simulate that, let's shutdown the old ray cluster in preparation for serving.
ray.shutdown()
endpoint_uri = serve_model(result.checkpoint, build_model, dataframe_adapter)
```
Let's write a helper function to send requests to this endpoint and compare the results with labels.
```
import requests
NUM_SERVE_REQUESTS = 100
def send_requests(df: pd.DataFrame, label: np.array):
for i in range(NUM_SERVE_REQUESTS):
one_row = df.iloc[[i]].to_dict()
serve_result = requests.post(endpoint_uri, json=one_row).json()
print(
f"request[{i}] prediction: {serve_result['predictions']['0']} "
f"- label: {str(label[i])}"
)
send_requests(test_df, test_label)
```
|
github_jupyter
|
# ! pip install "tensorflow>=2.8.0" "ray[tune, data, serve]>=1.12.1"
# ! pip install fastapi
from pprint import pprint
import ray
ray.shutdown()
ray.init()
pprint(ray.cluster_resources())
import pandas as pd
LABEL = "is_big_tip"
def get_data() -> pd.DataFrame:
"""Fetch the taxi fare data to work on."""
_data = pd.read_csv(
"https://raw.githubusercontent.com/tensorflow/tfx/master/"
"tfx/examples/chicago_taxi_pipeline/data/simple/data.csv"
)
_data[LABEL] = _data["tips"] / _data["fare"] > 0.2
# We drop some columns here for the sake of simplicity.
return _data.drop(
[
"tips",
"fare",
"dropoff_latitude",
"dropoff_longitude",
"pickup_latitude",
"pickup_longitude",
"pickup_census_tract",
],
axis=1,
)
data = get_data()
data.head(5)
import numpy as np
from sklearn.model_selection import train_test_split
from typing import Tuple
def split_data(data: pd.DataFrame) -> Tuple[ray.data.Dataset, pd.DataFrame, np.array]:
"""Split the data in a stratified way.
Returns:
A tuple containing train dataset, test data and test label.
"""
# There is a native offering in Ray Dataset for split as well.
# However, supporting stratification is a TODO there. So use
# scikit-learn equivalent here.
train_data, test_data = train_test_split(
data, stratify=data[[LABEL]], random_state=1113
)
_train_ds = ray.data.from_pandas(train_data)
_test_label = test_data[LABEL].values
_test_df = test_data.drop([LABEL], axis=1)
return _train_ds, _test_df, _test_label
train_ds, test_df, test_label = split_data(data)
print(f"There are {train_ds.count()} samples for training and {test_df.shape[0]} samples for testing.")
from ray.data.preprocessors import (
BatchMapper,
Chain,
OneHotEncoder,
SimpleImputer,
)
def get_preprocessor():
"""Construct a chain of preprocessors."""
imputer1 = SimpleImputer(
["dropoff_census_tract"], strategy="most_frequent"
)
imputer2 = SimpleImputer(
["pickup_community_area", "dropoff_community_area"],
strategy="most_frequent",
)
imputer3 = SimpleImputer(["payment_type"], strategy="most_frequent")
imputer4 = SimpleImputer(
["company"], strategy="most_frequent")
imputer5 = SimpleImputer(
["trip_start_timestamp", "trip_miles", "trip_seconds"], strategy="mean"
)
ohe = OneHotEncoder(
columns=[
"trip_start_hour",
"trip_start_day",
"trip_start_month",
"dropoff_census_tract",
"pickup_community_area",
"dropoff_community_area",
"payment_type",
"company",
],
limit={
"dropoff_census_tract": 25,
"pickup_community_area": 20,
"dropoff_community_area": 20,
"payment_type": 2,
"company": 7,
},
)
def batch_mapper_fn(df):
df["trip_start_year"] = pd.to_datetime(df["trip_start_timestamp"], unit="s").dt.year
df = df.drop(["trip_start_timestamp"], axis=1)
return df
chained_pp = Chain(
imputer1,
imputer2,
imputer3,
imputer4,
imputer5,
ohe,
BatchMapper(batch_mapper_fn),
)
return chained_pp
# Note that `INPUT_SIZE` here is corresponding to the output dimension
# of the previously defined processing steps.
# This is used to specify the input shape of Keras model as well as
# when converting from training data from `ray.data.Dataset` to `tf.Tensor`.
INPUT_SIZE = 120
# The training batch size. Based on `NUM_WORKERS`, each worker
# will get its own share of this batch size. For example, if
# `NUM_WORKERS = 2`, each worker will work on 4 samples per batch.
BATCH_SIZE = 8
# Number of epoch. Adjust it based on how quickly you want the run to be.
EPOCH = 1
# Number of training workers.
# Adjust this accordingly based on the resources you have!
NUM_WORKERS = 2
import tensorflow as tf
def build_model():
model = tf.keras.models.Sequential()
model.add(tf.keras.Input(shape=(INPUT_SIZE,)))
model.add(tf.keras.layers.Dense(50, activation="relu"))
model.add(tf.keras.layers.Dense(1, activation="sigmoid"))
return model
from ray import train
from ray.train.tensorflow import prepare_dataset_shard
from ray.tune.integration.keras import TuneReportCallback
def train_loop_per_worker():
dataset_shard = train.get_dataset_shard("train")
strategy = tf.distribute.experimental.MultiWorkerMirroredStrategy()
with strategy.scope():
model = build_model()
model.compile(
loss="binary_crossentropy",
optimizer="adam",
metrics=["accuracy"],
)
for epoch in range(EPOCH):
# This will make sure that the training workers will get their own
# share of batch to work on.
# See `ray.train.tensorflow.prepare_dataset_shard` for more information.
tf_dataset = prepare_dataset_shard(
dataset_shard.to_tf(
label_column=LABEL,
output_signature=(
tf.TensorSpec(shape=(BATCH_SIZE, INPUT_SIZE), dtype=tf.float32),
tf.TensorSpec(shape=(BATCH_SIZE,), dtype=tf.int64),
),
batch_size=BATCH_SIZE,
drop_last=True,
)
)
model.fit(tf_dataset, verbose=0)
# This saves checkpoint in a way that can be used by Ray Serve coherently.
train.save_checkpoint(epoch=epoch, model=model.get_weights())
from ray.train.tensorflow import TensorflowTrainer
trainer = TensorflowTrainer(
train_loop_per_worker=train_loop_per_worker,
scaling_config={"num_workers": NUM_WORKERS},
datasets={"train": train_ds},
preprocessor=get_preprocessor(),
)
result = trainer.fit()
from fastapi import Request
async def dataframe_adapter(request: Request):
"""Serve HTTP Adapter that reads JSON and converts to pandas DataFrame."""
content = await request.json()
return pd.DataFrame.from_dict(content)
from ray import serve
from ray.air.checkpoint import Checkpoint
from ray.train.tensorflow import TensorflowPredictor
from ray.serve.model_wrappers import ModelWrapperDeployment
def serve_model(checkpoint: Checkpoint, model_definition, adapter, name="Model") -> str:
"""Expose a serve endpoint.
Returns:
serve URL.
"""
serve.start(detached=True)
deployment = ModelWrapperDeployment.options(name=name)
deployment.deploy(
TensorflowPredictor,
checkpoint,
# This is due to a current limitation on Serve that's
# being addressed.
# TODO(xwjiang): Change to True.
batching_params=False,
model_definition=model_definition,
http_adapter=adapter,
)
return deployment.url
# Generally speaking, training and serving are done in totally different ray clusters.
# To simulate that, let's shutdown the old ray cluster in preparation for serving.
ray.shutdown()
endpoint_uri = serve_model(result.checkpoint, build_model, dataframe_adapter)
import requests
NUM_SERVE_REQUESTS = 100
def send_requests(df: pd.DataFrame, label: np.array):
for i in range(NUM_SERVE_REQUESTS):
one_row = df.iloc[[i]].to_dict()
serve_result = requests.post(endpoint_uri, json=one_row).json()
print(
f"request[{i}] prediction: {serve_result['predictions']['0']} "
f"- label: {str(label[i])}"
)
send_requests(test_df, test_label)
| 0.684475 | 0.99276 |
```
import os
import json
import pandas as pd
import numpy as np
import scipy as sp
import copy
playlists = list()
tracks = dict()
map_pl = list()
max_files_for_quick_processing = 5
def process_track(track):
key = track['track_uri']
if not key in tracks:
tk = dict()
tk['artist_name'] = track['artist_name']
tk['artist_uri'] = track['artist_uri']
tk['track_name'] = track['track_name']
tk['album_uri'] = track['album_uri']
tk['duration_ms'] = track['duration_ms']
tk['album_name'] = track['album_name']
tracks[track['track_uri']] = tk
return key
def process_playlist(playlist):
pl = dict()
pl['name'] = playlist['name']
pl['collaborative'] = playlist['collaborative']
pl['pid'] = playlist['pid']
pl['modified_at'] = playlist['modified_at']
pl['num_albums'] = playlist['num_albums']
pl['num_tracks'] = playlist['num_tracks']
pl['num_followers'] = playlist['num_followers']
pl['num_edits'] = playlist['num_edits']
pl['duration_ms'] = playlist['duration_ms']
pl['num_artists'] = playlist['num_artists']
if 'description' in playlist:
pl['description'] = playlist['description']
else:
pl['description'] = None
trks = list()
for track in playlist['tracks']:
map_pl.append([playlist['pid'], track['track_uri']])
trks.append(track['track_uri'])
process_track(track)
return pl
def process_mpd(path):
count = 0
filenames = os.listdir(path)
for filename in sorted(filenames):
print(filename)
if filename.startswith("mpd.slice.") and filename.endswith(".json"):
fullpath = os.sep.join((path, filename))
f = open(fullpath)
js = f.read()
f.close()
slice = json.loads(js)
for playlist in slice['playlists']:
playlists.append(process_playlist(playlist))
count += 1
if quick and count > max_files_for_quick_processing:
break
quick = True
process_mpd('mpd.v1/data')
print(len(playlists))
print(len(tracks))
print(len(map_pl))
playlist_df = pd.DataFrame(playlists)
playlist_df.head()
print(playlist_df.describe())
tracks_df = pd.DataFrame.from_dict(tracks, orient='index')
tracks_df.head()
print(tracks_df.describe())
playlist_map_df = pd.DataFrame(map_pl, columns=['pid', 'track_uri'])
playlist_map_df.head()
print(playlist_map_df.describe())
merged = pd.merge(pd.merge(tracks_df, playlist_map_df, left_index=True, right_on='track_uri'), playlist_df, on='pid')
#Match column indicated whether a song was truly in a playlist
merged["match"] = 1
print(merged.shape)
merged.head(20)
#Build a new DataFrame in which each playlist is filled with random songs. The number of songs for each
#playlist remains the same.
negative_training = copy.deepcopy(merged)
negative_training["match"] = 0 #Because these songs are not actually in the playlists
#Take songs from random rows in merged df
random_rows = np.random.randint(len(negative_training), size = len(negative_training))
#replace these columns with those from a random row
replace = ["artist_name", "artist_uri", "track_name", "album_uri", "duration_ms_x", "album_name", "track_uri"]
for index in range(len(negative_training)):
for column in replace:
negative_training.loc[index, [column]] = negative_training.loc[random_rows[index], [column]]
#Fix other columns to correctly identify unique albums etc...
for pid, df in negative_training.groupby(["pid"]):
unique_albums = df["album_name"].nunique()
num_artists = df["artist_name"].nunique()
duration_ms_y = np.sum(df["duration_ms_x"])
rows = df.index.values
for row in rows:
negative_training.loc[row, "num_albums"] = unique_albums
negative_training.loc[row, "num_artists"] = num_artists
negative_training.loc[row, "duration_ms_y"] = duration_ms_y
negative_training.head(20)
```
|
github_jupyter
|
import os
import json
import pandas as pd
import numpy as np
import scipy as sp
import copy
playlists = list()
tracks = dict()
map_pl = list()
max_files_for_quick_processing = 5
def process_track(track):
key = track['track_uri']
if not key in tracks:
tk = dict()
tk['artist_name'] = track['artist_name']
tk['artist_uri'] = track['artist_uri']
tk['track_name'] = track['track_name']
tk['album_uri'] = track['album_uri']
tk['duration_ms'] = track['duration_ms']
tk['album_name'] = track['album_name']
tracks[track['track_uri']] = tk
return key
def process_playlist(playlist):
pl = dict()
pl['name'] = playlist['name']
pl['collaborative'] = playlist['collaborative']
pl['pid'] = playlist['pid']
pl['modified_at'] = playlist['modified_at']
pl['num_albums'] = playlist['num_albums']
pl['num_tracks'] = playlist['num_tracks']
pl['num_followers'] = playlist['num_followers']
pl['num_edits'] = playlist['num_edits']
pl['duration_ms'] = playlist['duration_ms']
pl['num_artists'] = playlist['num_artists']
if 'description' in playlist:
pl['description'] = playlist['description']
else:
pl['description'] = None
trks = list()
for track in playlist['tracks']:
map_pl.append([playlist['pid'], track['track_uri']])
trks.append(track['track_uri'])
process_track(track)
return pl
def process_mpd(path):
count = 0
filenames = os.listdir(path)
for filename in sorted(filenames):
print(filename)
if filename.startswith("mpd.slice.") and filename.endswith(".json"):
fullpath = os.sep.join((path, filename))
f = open(fullpath)
js = f.read()
f.close()
slice = json.loads(js)
for playlist in slice['playlists']:
playlists.append(process_playlist(playlist))
count += 1
if quick and count > max_files_for_quick_processing:
break
quick = True
process_mpd('mpd.v1/data')
print(len(playlists))
print(len(tracks))
print(len(map_pl))
playlist_df = pd.DataFrame(playlists)
playlist_df.head()
print(playlist_df.describe())
tracks_df = pd.DataFrame.from_dict(tracks, orient='index')
tracks_df.head()
print(tracks_df.describe())
playlist_map_df = pd.DataFrame(map_pl, columns=['pid', 'track_uri'])
playlist_map_df.head()
print(playlist_map_df.describe())
merged = pd.merge(pd.merge(tracks_df, playlist_map_df, left_index=True, right_on='track_uri'), playlist_df, on='pid')
#Match column indicated whether a song was truly in a playlist
merged["match"] = 1
print(merged.shape)
merged.head(20)
#Build a new DataFrame in which each playlist is filled with random songs. The number of songs for each
#playlist remains the same.
negative_training = copy.deepcopy(merged)
negative_training["match"] = 0 #Because these songs are not actually in the playlists
#Take songs from random rows in merged df
random_rows = np.random.randint(len(negative_training), size = len(negative_training))
#replace these columns with those from a random row
replace = ["artist_name", "artist_uri", "track_name", "album_uri", "duration_ms_x", "album_name", "track_uri"]
for index in range(len(negative_training)):
for column in replace:
negative_training.loc[index, [column]] = negative_training.loc[random_rows[index], [column]]
#Fix other columns to correctly identify unique albums etc...
for pid, df in negative_training.groupby(["pid"]):
unique_albums = df["album_name"].nunique()
num_artists = df["artist_name"].nunique()
duration_ms_y = np.sum(df["duration_ms_x"])
rows = df.index.values
for row in rows:
negative_training.loc[row, "num_albums"] = unique_albums
negative_training.loc[row, "num_artists"] = num_artists
negative_training.loc[row, "duration_ms_y"] = duration_ms_y
negative_training.head(20)
| 0.236076 | 0.157785 |
# Modifying Rewards
I'm a bit disappointed that the genetic algorithm tends to sway too far from the center. If I had a light penalty, will it avoid this.
Reading [the wiki](https://github.com/openai/gym/wiki/CartPole-v0) helps a lot in figuring this out! That page has the parameters for cart-pole v0, which is just a shorter version of the cart-pole v1 we use.
```
# Display GIFs in Jupyter
from IPython.display import HTML
# OpenAI gym
import gym
# Import local script
import agents
# numpy
import numpy as np
# To speed up the algorithm
from multiprocessing import Pool
n_jobs = 4 # Set your number of cores here
```
Here I add a penalty of `abs(observation[0])`. Observation 0 is the distance from the center, within `[-2.6, 2.6]`.
```
def trial_agent(agent, trials=100, limit=1000):
env = gym.make(agent.game)
scores = []
for i in range(trials):
observation = env.reset()
score = 0
for t in range(limit):
action = agent.predict(observation)
observation, reward, done, info = env.step(action)
if done:
break
# Add a light penalty for distance
score += reward - abs(observation[0]/10)
scores.append(score)
data_dict = {
"agent" : agent,
"weights" : agent.w,
"pedigree" : agent.pedigree,
"minimum" : min(scores),
"maximum" : max(scores),
"mean" : sum(scores)/len(scores)
}
env.close()
return data_dict
def genetic_algorithm(results, old=5, new=95, n_parents=2, generations=25,
mutation_rate=0.01, mutation_amount=0.5, order=1, max_score=499.0,
game="CartPole-v1"):
for round in range(generations):
# Sort agents by score (fitness)
top_scores = sorted(results, key=lambda x: x["mean"], reverse=True)
# The survival of the fittest. Wikipedia calls this "elitism".
# The top agents of a generation are carried over to the next
survivors = top_scores[:old]
# To start breeding new agents, I'll mix weights (genes)
weight_shape = top_scores[0]["weights"].shape
gene_pool = [list(i["weights"].flatten()) for i in top_scores]
pedigree_list = [i["pedigree"] for i in top_scores]
genome_size = top_scores[0]["weights"].size
# Scores can be negative, so here I make them all positive
# They also need to sum to 1 for random sampling
min_score = min([i["mean"] for i in top_scores])
sum_score = sum([i["mean"]+min_score for i in top_scores])
probs = [(i["mean"]+min_score)/sum_score for i in top_scores]
# For each new agent, randomly select parents
# Higher-fitness agents are likelier to sire new agents
children = []
for birth in range(new):
parents = np.random.choice(np.arange(len(gene_pool)),
size=n_parents,
replace=False,
p=probs)
# The offspring get a mix of each parent's weights
# The weights (genes) are simply copied over
mix = np.random.randint(0, high=n_parents, size=genome_size)
weights = []
pedigree = []
for i in range(genome_size):
weights.append(gene_pool[parents[mix[i]]][i])
pedigree.append(pedigree_list[parents[mix[i]]][i])
# A mutation happens rarely and adds a bit of noise to a gene
if np.random.random(1) < mutation_rate:
weights[i] += float(np.random.normal(0, mutation_amount, 1))
pedigree[i] += "M"
children.append({"weights" : weights, "pedigree" : pedigree})
# Elitism: the top agents survive to fight another day
new_agents = [i["agent"] for i in survivors]
# The offspring are added it
# With the pedigree variable their ancestors are tracked
for child in children:
new_agents.append(
agents.LinearAgent(
np.array(child["weights"]).reshape(weight_shape),
pedigree=child["pedigree"],
order=order,
game=game))
# Trial the agents using multiple CPU cores
p = Pool(n_jobs)
results = p.map(trial_agent, new_agents)
p.close()
results = sorted(results, key=lambda x: x["mean"], reverse=True)
print(f"[{round+1:3}] Population average: {sum([i['mean'] for i in results])/len(results):5.1f}")
print(f"[{round+1:3}] Best mean score: {results[0]['mean']:5.1f}, Pedigree: {'-'.join(results[0]['pedigree'])}")
print()
# End early if maximum is reached
if results[0]['mean'] >= max_score:
print(f"[{round+1:3}] Best score reached, ending early")
break
return results
```
## Extreme third-order agent
I'm going to try a third-order agent. And this time I'll take a peek at results every few generation.
```
results = []
for a in range(25):
results.append(trial_agent(agents.LinearAgent(None, order=3, id=a)))
winner = sorted(results, key=lambda x: x["mean"], reverse=True)[0]
print(winner)
HTML(f"<img src='{winner['agent'].render('mod_cart_0.gif', episodes=1)}'>")
results = genetic_algorithm(results, generations=5, order=3)
winner = sorted(results, key=lambda x: x["mean"], reverse=True)[0]
print(winner)
HTML(f"<img src='{winner['agent'].render('mod_cart_5.gif', episodes=1)}'>")
results = genetic_algorithm(results, generations=5, order=3)
winner = sorted(results, key=lambda x: x["mean"], reverse=True)[0]
print(winner)
HTML(f"<img src='{winner['agent'].render('mod_cart_10.gif', episodes=1)}'>")
results = genetic_algorithm(results, generations=5, order=3)
winner = sorted(results, key=lambda x: x["mean"], reverse=True)[0]
print(winner)
HTML(f"<img src='{winner['agent'].render('mod_cart_15.gif', episodes=1)}'>")
results = genetic_algorithm(results, generations=5, order=3)
winner = sorted(results, key=lambda x: x["mean"], reverse=True)[0]
print(winner)
HTML(f"<img src='{winner['agent'].render('mod_cart_20.gif', episodes=1)}'>")
results = genetic_algorithm(results, generations=5, order=3)
winner = sorted(results, key=lambda x: x["mean"], reverse=True)[0]
print(winner)
HTML(f"<img src='{winner['agent'].render('mod_cart_25.gif', episodes=1)}'>")
```
|
github_jupyter
|
# Display GIFs in Jupyter
from IPython.display import HTML
# OpenAI gym
import gym
# Import local script
import agents
# numpy
import numpy as np
# To speed up the algorithm
from multiprocessing import Pool
n_jobs = 4 # Set your number of cores here
def trial_agent(agent, trials=100, limit=1000):
env = gym.make(agent.game)
scores = []
for i in range(trials):
observation = env.reset()
score = 0
for t in range(limit):
action = agent.predict(observation)
observation, reward, done, info = env.step(action)
if done:
break
# Add a light penalty for distance
score += reward - abs(observation[0]/10)
scores.append(score)
data_dict = {
"agent" : agent,
"weights" : agent.w,
"pedigree" : agent.pedigree,
"minimum" : min(scores),
"maximum" : max(scores),
"mean" : sum(scores)/len(scores)
}
env.close()
return data_dict
def genetic_algorithm(results, old=5, new=95, n_parents=2, generations=25,
mutation_rate=0.01, mutation_amount=0.5, order=1, max_score=499.0,
game="CartPole-v1"):
for round in range(generations):
# Sort agents by score (fitness)
top_scores = sorted(results, key=lambda x: x["mean"], reverse=True)
# The survival of the fittest. Wikipedia calls this "elitism".
# The top agents of a generation are carried over to the next
survivors = top_scores[:old]
# To start breeding new agents, I'll mix weights (genes)
weight_shape = top_scores[0]["weights"].shape
gene_pool = [list(i["weights"].flatten()) for i in top_scores]
pedigree_list = [i["pedigree"] for i in top_scores]
genome_size = top_scores[0]["weights"].size
# Scores can be negative, so here I make them all positive
# They also need to sum to 1 for random sampling
min_score = min([i["mean"] for i in top_scores])
sum_score = sum([i["mean"]+min_score for i in top_scores])
probs = [(i["mean"]+min_score)/sum_score for i in top_scores]
# For each new agent, randomly select parents
# Higher-fitness agents are likelier to sire new agents
children = []
for birth in range(new):
parents = np.random.choice(np.arange(len(gene_pool)),
size=n_parents,
replace=False,
p=probs)
# The offspring get a mix of each parent's weights
# The weights (genes) are simply copied over
mix = np.random.randint(0, high=n_parents, size=genome_size)
weights = []
pedigree = []
for i in range(genome_size):
weights.append(gene_pool[parents[mix[i]]][i])
pedigree.append(pedigree_list[parents[mix[i]]][i])
# A mutation happens rarely and adds a bit of noise to a gene
if np.random.random(1) < mutation_rate:
weights[i] += float(np.random.normal(0, mutation_amount, 1))
pedigree[i] += "M"
children.append({"weights" : weights, "pedigree" : pedigree})
# Elitism: the top agents survive to fight another day
new_agents = [i["agent"] for i in survivors]
# The offspring are added it
# With the pedigree variable their ancestors are tracked
for child in children:
new_agents.append(
agents.LinearAgent(
np.array(child["weights"]).reshape(weight_shape),
pedigree=child["pedigree"],
order=order,
game=game))
# Trial the agents using multiple CPU cores
p = Pool(n_jobs)
results = p.map(trial_agent, new_agents)
p.close()
results = sorted(results, key=lambda x: x["mean"], reverse=True)
print(f"[{round+1:3}] Population average: {sum([i['mean'] for i in results])/len(results):5.1f}")
print(f"[{round+1:3}] Best mean score: {results[0]['mean']:5.1f}, Pedigree: {'-'.join(results[0]['pedigree'])}")
print()
# End early if maximum is reached
if results[0]['mean'] >= max_score:
print(f"[{round+1:3}] Best score reached, ending early")
break
return results
results = []
for a in range(25):
results.append(trial_agent(agents.LinearAgent(None, order=3, id=a)))
winner = sorted(results, key=lambda x: x["mean"], reverse=True)[0]
print(winner)
HTML(f"<img src='{winner['agent'].render('mod_cart_0.gif', episodes=1)}'>")
results = genetic_algorithm(results, generations=5, order=3)
winner = sorted(results, key=lambda x: x["mean"], reverse=True)[0]
print(winner)
HTML(f"<img src='{winner['agent'].render('mod_cart_5.gif', episodes=1)}'>")
results = genetic_algorithm(results, generations=5, order=3)
winner = sorted(results, key=lambda x: x["mean"], reverse=True)[0]
print(winner)
HTML(f"<img src='{winner['agent'].render('mod_cart_10.gif', episodes=1)}'>")
results = genetic_algorithm(results, generations=5, order=3)
winner = sorted(results, key=lambda x: x["mean"], reverse=True)[0]
print(winner)
HTML(f"<img src='{winner['agent'].render('mod_cart_15.gif', episodes=1)}'>")
results = genetic_algorithm(results, generations=5, order=3)
winner = sorted(results, key=lambda x: x["mean"], reverse=True)[0]
print(winner)
HTML(f"<img src='{winner['agent'].render('mod_cart_20.gif', episodes=1)}'>")
results = genetic_algorithm(results, generations=5, order=3)
winner = sorted(results, key=lambda x: x["mean"], reverse=True)[0]
print(winner)
HTML(f"<img src='{winner['agent'].render('mod_cart_25.gif', episodes=1)}'>")
| 0.681303 | 0.799207 |
<center>
<img src="https://github.com/hse-econ-data-science/dap_2021_spring/blob/main/sem10_visual/images/visual.png?raw=true" height="400" width="700">
</center>
# <center> Визуализация данных в python </center>
Визуализация занимает важную часть в анализе данных. Представляя информацию в графическом виде, вы облегчаете процесс ее восприятия, что дает возможность выделять дополнительные закономерности, оценивать соотношения величин, быстрее доносить ключевые аспекты в данных.
Начнем с небольшой "памятки", о которой всегда нужно помнить при создании любых графиков.
## <center> Как визуализировать данные и заставить всех тебя ненавидеть </center>
1. Заголовок графика для слабаков. По графику всегда понятно, какие данные и явления он описывает.
2. Ни в коем случае не подписывай ни одной оси у графика. Пусть смотрящий развивает свою интуицую!
3. Единицы измерения совсем не обязательны. Какая разница, в чем измеряли количество - в людях или в литрах!
4. Чем меньше шрифт на графике, тем острее зрение смотрящего.
5. На одном графике нужно стараться уместить всю информацию, которая у тебя есть в датасете. С полными названиями, расшифровками, сносками. Чем больше текста - тем информативнее!
6. При любой возможности используйте 3D и спецэффекты, пусть знают, что ты — прирожденный дизайнер. К тому же, так будет меньше визуальных искажений.
Если серьезно, то обязательно посмотрите список рекомендованных материалов в конце ноутбука по правилам оформления графиков и работе с библиотеками для визуализации данных в Python.
Основные библиотеки для визуализации в Python - это `matplotlib`, `seaborn`, `plotly`. Сегодня познакомимся с первыми двумя
```
import numpy as np # библиотека для матриц и математики
import pandas as pd # библиотека для работы с табличками
# библиотеки для визуализации
import matplotlib.pyplot as plt
import seaborn as sns
# plt.style.use('ggplot') # стиль графиков
%matplotlib inline
```
# Продажи и оценки видеоигр
Работаем с датасетом по продажам и оценкам видео-игр. Датасет взят с [Кеггла](https://www.kaggle.com/rush4ratio/video-game-sales-with-ratings).
__Описание колонок:__
* `Name` $-$ название видеоигры
* `Platform` $-$ платформа, на которой игра была запущена
* `Year_of_Release` $-$ год релиза
* `Genre` $-$ жанр
* `Publisher` $-$ издатель
* `NA_Sales` $-$ объем продаж в Северной Америке (в млн штук)
* `EU_Sales` $-$ объем продаж в Евросоюзе (в млн штук)
* `JP_Sales` $-$ объем продаж в Японии (в млн штук)
* `Other_Sales` $-$ объем продаж в остальном мире (в млн штук)
* `Global_Sales` $-$ общий объем продаж (в млн штук)
* `Critic_Score` $-$ совокупный балл, составленный сотрудниками Metacritic
* `Critic_Count` $-$ кол-во критиков, оцениваемых игру
* `User_Score` $-$ совокупный балл, составленный подписчиками Metacritic (пользователями)
* `User_Count` $-$ кол-во пользователей, оцениваемых игру
* `Developer` $-$ ответственный за создание игры
* `Rating` $-$ рейтинг (Everyone, Teen, Adults Only и тд)
```
df = pd.read_csv('data/video_games_sales.csv')
df.shape
df.info()
```
В данных много пропусков, поэтому давайте выкинем все пропущенные наблюдения. Также видим, что некоторые колонки pandas привел не к тому типу. Исправим это:
```
df = df.dropna()
df['User_Score'] = df.User_Score.astype('float64')
df['Year_of_Release'] = df.Year_of_Release.astype('int64')
df['User_Count'] = df.User_Count.astype('int64')
df['Critic_Count'] = df.Critic_Count.astype('int64')
df.shape
```
## 1. Изучаем основы matplotlib на примере линейных графиков
Самый простой вариант создания графика в matplotlib - функция `plt.plot()`, которой мы передаем два аргумента - что положить на ось _x_, а что на _y_. Если у вас переменные в числовом формате, то без проблем получите линейный график (line plot)
`plt.plot(x = ___, y = ___)`
**Посмотрим на динамику продаж игр в мире по годам:**
```
# агрегируем данные за год
gb = df.groupby('Year_of_Release').Global_Sales.sum()
# строим график
plt.plot(gb.index, gb.values);
```
### Задача №1:
Постройте один график, на котором будут отображаться сразу все показатели продаж (NA_Sales, EU_Sales, JP_Sales, Other_Sales, Global_Sales)
```
## Ваш код
# Заводим датафрейм
sales_df = df[[x for x in df.columns if 'Sales' in x] + ['Year_of_Release']]
gb = sales_df.groupby('Year_of_Release').sum()
# Рисуем!
gb.plot();
### Через plt.plot():
gb = gb.reset_index()
plt.plot(gb.Year_of_Release, gb.NA_Sales, label = 'NA_Sales')
plt.plot(gb.Year_of_Release, gb.EU_Sales, label = 'EU_Sales')
plt.plot(gb.Year_of_Release, gb.JP_Sales, label = 'JP_Sales')
plt.plot(gb.Year_of_Release, gb.Other_Sales, label = 'Other_Sales')
plt.plot(gb.Year_of_Release, gb.Global_Sales, label = 'Global_Sales')
plt.legend();
```
В идеальной вселенной мы создаем графики функцией `subplots`, которая генерирует наш график в 2 переменные (обычно их называют fig и ax):
* `fig` отвечает за график в целом. Воспринимайте ее как файл, который хранит график как картинку.
* `ax` $-$ это ось координат, на которой мы собственно строим график. Все элементы графика хранятся как раз в ней.
```
fig, ax = plt.subplots(1,1, figsize=(7,5)) # создали полоино для графика из 1 ряда и 1 колонки (1 график)
# figsize -это размер нашего прямоугольника в неочевидных единицах.
# Какой размер удачный? экспериментируйте!
```
**Посмотрим на динамику продаж игр в мире по годам, но теперь используем `subplots`**
```
# агрегируем данные за год
gb = df.groupby('Year_of_Release').Global_Sales.sum()
# строим график
fig, ax = plt.subplots(1,1, figsize=(10,5))
ax.plot(gb.index, gb.values);
```
В переменной _ax_ на самом деле лежит куча методов, которые позволяют сделать график более приятным. Посмотрим на несколько из них:
```
fig, ax = plt.subplots(1,1, figsize=(10,5))
# параметры самого графика (цвет линии, стиль и т.д.) определяем как параметры в методе plot()
# меняем цвет и стиль линии на пунктир. Matplotlib знает некоторые стандартные цвета, и их можно задать прямо словом
# так же можно передать hex цвет. Например, #8c92ac
ax.plot(gb.index, gb.values, color = 'goldenrod', ls = '-.')
# если вы обратили внимание, то в нашем самом первом графике шкала с годами сломамлась и стала float. Matplotlib принудительно
# делает x непрерывной переменной для линейного графика. Мы хотим оставить шкалу год в целых числах.
ax.locator_params(integer=True)
# называем шкалы x и y, выбираем размер шрифта.
ax.set_xlabel('Year of Release', fontsize=12)
ax.set_ylabel('Global Sales', fontsize=12)
# делаем правую и верхнюю границу графика невидимыми
ax.spines['right'].set_visible(False)
ax.spines['top'].set_visible(False)
# делаем засечки на шкалах x и y потоньше
ax.xaxis.set_tick_params(width=0.2)
ax.yaxis.set_tick_params(width=0.2)
# уменьшаем толщину оставших границ графика с помощью цикла
# (можно и без цикла отдельной строкой для каждой границы, как делали выше)
for spine in ['bottom','left']:
ax.spines[spine].set_linewidth(0.2)
plt.title('Global Sales by Year')
fig.savefig('mlds.png')
```
### Создаем систему графиков
```
fig, ax = plt.subplots(2,2, figsize=(14,9)) # увеличиваем количество объектов до двух рядов и двух колонок.
```
Теперь у нас есть четыре графика!
Объект _ax_ в данном случае **становится матрицой**. И чтобы обратиться к каждому графику, нужно обратиться к нему по индексу из _ax_.
```
fig, ax = plt.subplots(2,2, figsize=(14,9))
ax[0][0].plot(gb.index, gb.values);
```
Ниже посмотрим как с помощью цикла for можно применить какую-то модификацию ко всем графикам системы.
```
fig, ax= plt.subplots(2, 2, figsize=(14,9))
for x in range(2):
for y in range(2):
ax[x][y].set_xlabel('X')
```
### Задача №2:
Создайте систему графиков, у которой:
* на 1ом графике изображены продажи в Северной Америке (`NA_Sales`);
* на 2ом продажи в Европейском Союзе (`EU_Sales`);
* на 3ем продажи в Японии (`JP_Sales`)
* на 4ом графике все 3 линии вместе;
* Каждый график должен быть разного цвета. В четвертом графике - каждый график должен быть такого же цвета, как и в своей ячейке + 4ый график должен иметь легенду
* В заголовке к каждому графику напишите то, что стоит у него по оси _y_
```
## Ваш код
# Заводим датафрейм
sales_df = df[[x for x in df.columns if 'Sales' in x] + ['Year_of_Release']]
gb = sales_df.groupby('Year_of_Release').sum().reset_index()
# Рисуем!
fig, ax= plt.subplots(2, 2, figsize=(14,9))
ax[0][0].plot(gb.Year_of_Release, gb.NA_Sales, color='#8c92ac', ls = ':')
ax[0][0].set_title('NA_Sales', fontsize=14)
ax[1][0].plot(gb.Year_of_Release, gb.EU_Sales, color='#ffa500', ls = '--')
ax[1][0].set_title('EU_Sales', fontsize = 14)
ax[0][1].plot(gb.Year_of_Release, gb.JP_Sales, color='#b06500', ls = '-');
ax[0][1].set_title('JP_Sales', fontsize = 14)
ax[1][1].plot(gb.Year_of_Release, gb.NA_Sales, label = 'NA_Sales', color='#8c92ac', ls = ':')
ax[1][1].plot(gb.Year_of_Release, gb.EU_Sales, label = 'EU_Sales', color='#ffa500', ls = '--')
ax[1][1].plot(gb.Year_of_Release, gb.JP_Sales, label = 'JP_sales', color='#b06500', ls = '-')
ax[1][1].set_title('Sales', fontsize = 14)
ax[1][1].legend(loc=1, fontsize=8, frameon=False);
```
## 2. Графики для категориальных переменных
**Посмотрим на кол-во игр за каждый год с помощью столбчатой диаграммы:**
```
df.groupby('Year_of_Release').Name.count().plot(kind='bar');
```
**Посмотрим отдельно на кол-во игр жанра Sports и Action за последние 5 лет. Сделаем двойную диаграмму и развернем ее горизонтально**
```
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(10, 8))
gb_sports = df[df.Genre=='Sports'].groupby('Year_of_Release').Name.count().tail(5).sort_index(ascending=False)
gb_sports.plot(kind='barh', ax=ax[0])
ax[0].set_title('Sports')
gb_act = df[df.Genre=='Action'].groupby('Year_of_Release').Name.count().tail(5).sort_index(ascending=False)
gb_act.plot(kind='barh', ax=ax[1])
ax[1].set_title('Action')
ax[1].yaxis.set_ticks_position('right')
for i in range(2):
ax[i].set_ylabel('', visible=False) # убираем подпись к шкале, которая генерируется автоматически
```
Подумайте, чем плох такой график?
### Задача №3:
С помощью двух вертикальных столбчатых диаграмм выведите среднюю оценку критиков и пользователей игр различных рейтингов.
```
## Ваш код
fig, ax = plt.subplots(nrows=2, ncols=1, figsize=(12, 8))
gb = df.groupby('Rating')[['Critic_Score', 'User_Score']].mean()
gb['Critic_Score'].sort_values(ascending=False).plot(kind='bar', ax=ax[0])
gb['User_Score'].sort_values(ascending=False).plot(kind='bar', ax=ax[1]);
```
### Задача №4:
В виде горизонтальной столбчатой диаграммы выведите топ-5 девелоперов с наибольшей средней оценкой от критиков. Рядом выведите диаграмму их самой успешной игры по общим продажам.
```
## Ваш код
gb = df.groupby('Developer').agg({'Critic_Score' : np.mean, 'Global_Sales': np.max}). \
sort_values(by = 'Critic_Score',ascending=False).head(5).reset_index()
gb = pd.merge(gb, df[['Developer', 'Global_Sales', 'Name']], on=['Developer', 'Global_Sales'])
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(10, 8))
gb.set_index('Developer').Critic_Score.plot(kind='barh', ax=ax[0])
ax[0].set_title('Developers')
gb.set_index('Name').Global_Sales.plot(kind='barh', ax=ax[1])
ax[1].set_title('Games')
ax[1].yaxis.set_ticks_position('right')
for i in range(2):
ax[i].set_ylabel('', visible=False) # убираем подпись к шкале, которая генерируется автоматически
```
### Задача №5:
Сравните оценки игр от критиков для топ-5 крупнейших игровых платформ (по кол-ву игр) с помощью boxplot из пакета seaborn.
```
## Ваш код
top_platforms = df.Platform.value_counts().sort_values(ascending = False).head(5).index.values
sns.boxplot(y="Platform", x="Critic_Score", data=df[df.Platform.isin(top_platforms)], orient="h");
```
## 3*. Мультивариативный график рассеяния
Сейчас будем работать с наборам данных, который содержит информацию о количестве преступлений в штатах США в 2005 году.
```
crimes = pd.read_csv('data/crimeRatesByState2005.tsv', sep='\t')
crimes.head()
```
График рассеяния (scatter plot) $-$ это такой график, у которого по оси _x_ и _y_ отложены непрерывные переменные. График состоит из точек, каждая из которых отвечает за свое наблюдение.
Посмотрим на график зависимостей убийств от краж со взломом (burglary)
```
fig, ax = plt.subplots()
ax.scatter(crimes['murder'], crimes['burglary'])
fig.savefig('crimes.png');
```
Здесь каждая точка отвечает за свой штат. По оси _x_ мы видим сколько в штате было убийств, а по оси _y_ сколько краж со взломом
Попробуем на график добавить дополнительную информацию, например, в качестве размера точки обозначим кол-во населения
```
fig, ax = plt.subplots()
# добавляем параметр s (size) и говорим, какая переменная будет за него отвечать
ax.scatter(crimes['murder'], crimes['burglary'], s = crimes['population']);
```
Размер населения такой большой, что точка захватила всю область координат. Давайте попробуем нашу переменную масштабировать - нам же важны относительные размеры штатов относительно друг друга, а не абсолютные значения. Значения маштабирования тоже выбираем экспериментально: то, что лучше выглядит и более информативно.
```
fig, ax = plt.subplots()
ax.scatter(crimes['murder'], crimes['burglary'], s = crimes['population']/35000);
```
Отлично, однако следующая проблема - слияние точек. Давайте добавим параметр прозрачности, чтобы было видно, где они накладываются друг на друга.
```
fig, ax = plt.subplots()
ax.scatter(crimes['murder'], crimes['burglary'], s = crimes['population']/35000, alpha = 0.5);
```
Добавим теперь еще какую-нибудь переменную (например, robbery) и засунем ее в параметр цвета
```
fig, ax = plt.subplots()
ax.scatter(crimes['murder'], crimes['burglary'], s = crimes['population']/35000, alpha = 0.5,
c = crimes['Robbery']); # задаем новый аргумент c (color) и присваиваем ему значение переменной
```
Осталось узнать, что значит какой цвет. Для этого нужно сохранить график в переменную и передать ее как аргумент функции `colorbar()`. Также можем поменять цветовую шкалу с помощью аргумента cmap.
```
fig, ax = plt.subplots()
color_graph = ax.scatter(crimes['murder'], crimes['burglary'], s = crimes['population']/35000, alpha = 0.5, cmap = 'inferno',
c = crimes['Robbery'])
plt.colorbar(color_graph);
```
Последнее что тут сделаем - это подпишем штаты
```
fig, ax = plt.subplots(figsize = (22,10))
color_graph = ax.scatter(crimes['murder'], crimes['burglary'], s = crimes['population']/35000,
c = crimes['Robbery'], cmap = 'inferno', alpha = 0.5, linewidth = 0)
ax.yaxis.set_ticks_position('left')
ax.xaxis.set_ticks_position('bottom')
ax.set_xlabel('Murder', fontsize = 10)
ax.set_ylabel('Bulglarly', fontsize = 10)
for i, state in enumerate(crimes['state']): # с помощью enumerate создаем из колонок с названиями штатов объект кортежей вида индекс - название штата.
ax.annotate(state, (crimes['murder'][i], crimes['burglary'][i]), fontsize = 10) # используем метод annotate, которому говорим, что нужно брать имя штата
# из кортежа, который создали с помощью enumerate, а координаты подписи задаем через
# значения наших переменных x и y для нужного индекса из объекта enumerate (обращается к нужному
# ряду в датафрейме)
plt.colorbar(color_graph);
```
|
github_jupyter
|
import numpy as np # библиотека для матриц и математики
import pandas as pd # библиотека для работы с табличками
# библиотеки для визуализации
import matplotlib.pyplot as plt
import seaborn as sns
# plt.style.use('ggplot') # стиль графиков
%matplotlib inline
df = pd.read_csv('data/video_games_sales.csv')
df.shape
df.info()
df = df.dropna()
df['User_Score'] = df.User_Score.astype('float64')
df['Year_of_Release'] = df.Year_of_Release.astype('int64')
df['User_Count'] = df.User_Count.astype('int64')
df['Critic_Count'] = df.Critic_Count.astype('int64')
df.shape
# агрегируем данные за год
gb = df.groupby('Year_of_Release').Global_Sales.sum()
# строим график
plt.plot(gb.index, gb.values);
## Ваш код
# Заводим датафрейм
sales_df = df[[x for x in df.columns if 'Sales' in x] + ['Year_of_Release']]
gb = sales_df.groupby('Year_of_Release').sum()
# Рисуем!
gb.plot();
### Через plt.plot():
gb = gb.reset_index()
plt.plot(gb.Year_of_Release, gb.NA_Sales, label = 'NA_Sales')
plt.plot(gb.Year_of_Release, gb.EU_Sales, label = 'EU_Sales')
plt.plot(gb.Year_of_Release, gb.JP_Sales, label = 'JP_Sales')
plt.plot(gb.Year_of_Release, gb.Other_Sales, label = 'Other_Sales')
plt.plot(gb.Year_of_Release, gb.Global_Sales, label = 'Global_Sales')
plt.legend();
fig, ax = plt.subplots(1,1, figsize=(7,5)) # создали полоино для графика из 1 ряда и 1 колонки (1 график)
# figsize -это размер нашего прямоугольника в неочевидных единицах.
# Какой размер удачный? экспериментируйте!
# агрегируем данные за год
gb = df.groupby('Year_of_Release').Global_Sales.sum()
# строим график
fig, ax = plt.subplots(1,1, figsize=(10,5))
ax.plot(gb.index, gb.values);
fig, ax = plt.subplots(1,1, figsize=(10,5))
# параметры самого графика (цвет линии, стиль и т.д.) определяем как параметры в методе plot()
# меняем цвет и стиль линии на пунктир. Matplotlib знает некоторые стандартные цвета, и их можно задать прямо словом
# так же можно передать hex цвет. Например, #8c92ac
ax.plot(gb.index, gb.values, color = 'goldenrod', ls = '-.')
# если вы обратили внимание, то в нашем самом первом графике шкала с годами сломамлась и стала float. Matplotlib принудительно
# делает x непрерывной переменной для линейного графика. Мы хотим оставить шкалу год в целых числах.
ax.locator_params(integer=True)
# называем шкалы x и y, выбираем размер шрифта.
ax.set_xlabel('Year of Release', fontsize=12)
ax.set_ylabel('Global Sales', fontsize=12)
# делаем правую и верхнюю границу графика невидимыми
ax.spines['right'].set_visible(False)
ax.spines['top'].set_visible(False)
# делаем засечки на шкалах x и y потоньше
ax.xaxis.set_tick_params(width=0.2)
ax.yaxis.set_tick_params(width=0.2)
# уменьшаем толщину оставших границ графика с помощью цикла
# (можно и без цикла отдельной строкой для каждой границы, как делали выше)
for spine in ['bottom','left']:
ax.spines[spine].set_linewidth(0.2)
plt.title('Global Sales by Year')
fig.savefig('mlds.png')
fig, ax = plt.subplots(2,2, figsize=(14,9)) # увеличиваем количество объектов до двух рядов и двух колонок.
fig, ax = plt.subplots(2,2, figsize=(14,9))
ax[0][0].plot(gb.index, gb.values);
fig, ax= plt.subplots(2, 2, figsize=(14,9))
for x in range(2):
for y in range(2):
ax[x][y].set_xlabel('X')
## Ваш код
# Заводим датафрейм
sales_df = df[[x for x in df.columns if 'Sales' in x] + ['Year_of_Release']]
gb = sales_df.groupby('Year_of_Release').sum().reset_index()
# Рисуем!
fig, ax= plt.subplots(2, 2, figsize=(14,9))
ax[0][0].plot(gb.Year_of_Release, gb.NA_Sales, color='#8c92ac', ls = ':')
ax[0][0].set_title('NA_Sales', fontsize=14)
ax[1][0].plot(gb.Year_of_Release, gb.EU_Sales, color='#ffa500', ls = '--')
ax[1][0].set_title('EU_Sales', fontsize = 14)
ax[0][1].plot(gb.Year_of_Release, gb.JP_Sales, color='#b06500', ls = '-');
ax[0][1].set_title('JP_Sales', fontsize = 14)
ax[1][1].plot(gb.Year_of_Release, gb.NA_Sales, label = 'NA_Sales', color='#8c92ac', ls = ':')
ax[1][1].plot(gb.Year_of_Release, gb.EU_Sales, label = 'EU_Sales', color='#ffa500', ls = '--')
ax[1][1].plot(gb.Year_of_Release, gb.JP_Sales, label = 'JP_sales', color='#b06500', ls = '-')
ax[1][1].set_title('Sales', fontsize = 14)
ax[1][1].legend(loc=1, fontsize=8, frameon=False);
df.groupby('Year_of_Release').Name.count().plot(kind='bar');
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(10, 8))
gb_sports = df[df.Genre=='Sports'].groupby('Year_of_Release').Name.count().tail(5).sort_index(ascending=False)
gb_sports.plot(kind='barh', ax=ax[0])
ax[0].set_title('Sports')
gb_act = df[df.Genre=='Action'].groupby('Year_of_Release').Name.count().tail(5).sort_index(ascending=False)
gb_act.plot(kind='barh', ax=ax[1])
ax[1].set_title('Action')
ax[1].yaxis.set_ticks_position('right')
for i in range(2):
ax[i].set_ylabel('', visible=False) # убираем подпись к шкале, которая генерируется автоматически
## Ваш код
fig, ax = plt.subplots(nrows=2, ncols=1, figsize=(12, 8))
gb = df.groupby('Rating')[['Critic_Score', 'User_Score']].mean()
gb['Critic_Score'].sort_values(ascending=False).plot(kind='bar', ax=ax[0])
gb['User_Score'].sort_values(ascending=False).plot(kind='bar', ax=ax[1]);
## Ваш код
gb = df.groupby('Developer').agg({'Critic_Score' : np.mean, 'Global_Sales': np.max}). \
sort_values(by = 'Critic_Score',ascending=False).head(5).reset_index()
gb = pd.merge(gb, df[['Developer', 'Global_Sales', 'Name']], on=['Developer', 'Global_Sales'])
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(10, 8))
gb.set_index('Developer').Critic_Score.plot(kind='barh', ax=ax[0])
ax[0].set_title('Developers')
gb.set_index('Name').Global_Sales.plot(kind='barh', ax=ax[1])
ax[1].set_title('Games')
ax[1].yaxis.set_ticks_position('right')
for i in range(2):
ax[i].set_ylabel('', visible=False) # убираем подпись к шкале, которая генерируется автоматически
## Ваш код
top_platforms = df.Platform.value_counts().sort_values(ascending = False).head(5).index.values
sns.boxplot(y="Platform", x="Critic_Score", data=df[df.Platform.isin(top_platforms)], orient="h");
crimes = pd.read_csv('data/crimeRatesByState2005.tsv', sep='\t')
crimes.head()
fig, ax = plt.subplots()
ax.scatter(crimes['murder'], crimes['burglary'])
fig.savefig('crimes.png');
fig, ax = plt.subplots()
# добавляем параметр s (size) и говорим, какая переменная будет за него отвечать
ax.scatter(crimes['murder'], crimes['burglary'], s = crimes['population']);
fig, ax = plt.subplots()
ax.scatter(crimes['murder'], crimes['burglary'], s = crimes['population']/35000);
fig, ax = plt.subplots()
ax.scatter(crimes['murder'], crimes['burglary'], s = crimes['population']/35000, alpha = 0.5);
fig, ax = plt.subplots()
ax.scatter(crimes['murder'], crimes['burglary'], s = crimes['population']/35000, alpha = 0.5,
c = crimes['Robbery']); # задаем новый аргумент c (color) и присваиваем ему значение переменной
fig, ax = plt.subplots()
color_graph = ax.scatter(crimes['murder'], crimes['burglary'], s = crimes['population']/35000, alpha = 0.5, cmap = 'inferno',
c = crimes['Robbery'])
plt.colorbar(color_graph);
fig, ax = plt.subplots(figsize = (22,10))
color_graph = ax.scatter(crimes['murder'], crimes['burglary'], s = crimes['population']/35000,
c = crimes['Robbery'], cmap = 'inferno', alpha = 0.5, linewidth = 0)
ax.yaxis.set_ticks_position('left')
ax.xaxis.set_ticks_position('bottom')
ax.set_xlabel('Murder', fontsize = 10)
ax.set_ylabel('Bulglarly', fontsize = 10)
for i, state in enumerate(crimes['state']): # с помощью enumerate создаем из колонок с названиями штатов объект кортежей вида индекс - название штата.
ax.annotate(state, (crimes['murder'][i], crimes['burglary'][i]), fontsize = 10) # используем метод annotate, которому говорим, что нужно брать имя штата
# из кортежа, который создали с помощью enumerate, а координаты подписи задаем через
# значения наших переменных x и y для нужного индекса из объекта enumerate (обращается к нужному
# ряду в датафрейме)
plt.colorbar(color_graph);
| 0.225758 | 0.985229 |
# Detecting and Analyzing Faces
Computer vision solutions often require an artificial intelligence (AI) solution to be able to detect, analyze, or identify human faces. For example, suppose the retail company Northwind Traders has decided to implement a "smart store", in which AI services monitor the store to identify customers requiring assistance, and direct employees to help them. One way to accomplish this is to perform facial detection and analysis - in other words, determine if there are any faces in the images, and if so analyze their features.

## Use the Face cognitive service to detect faces
Suppose the smart store system that Northwind Traders wants to create needs to be able to detect customers and analyze their facial features. In Microsoft Azure, you can use **Face**, part of Azure Cognitive Services to do this.
### Create a Cognitive Services Resource
Let's start by creating a **Cognitive Services** resource in your Azure subscription.
> **Note**: If you already have a Cognitive Services resource, just open its **Quick start** page in the Azure portal and copy its key and endpoint to the cell below. Otherwise, follow the steps below to create one.
1. In another browser tab, open the Azure portal at https://portal.azure.com, signing in with your Microsoft account.
2. Click the **+Create a resource** button, search for *Cognitive Services*, and create a **Cognitive Services** resource with the following settings:
- **Subscription**: *Your Azure subscription*.
- **Resource group**: *Select or create a resource group with a unique name*.
- **Region**: *Choose any available region*:
- **Name**: *Enter a unique name*.
- **Pricing tier**: S0
- **I confirm I have read and understood the notices**: Selected.
3. Wait for deployment to complete. Then go to your cognitive services resource, and on the **Overview** page, click the link to manage the keys for the service. You will need the endpoint and keys to connect to your cognitive services resource from client applications.
### Get the Key and Endpoint for your Cognitive Services resource
To use your cognitive services resource, client applications need its endpoint and authentication key:
1. In the Azure portal, on the **Keys and Endpoint** page for your cognitive service resource, copy the **Key1** for your resource and paste it in the code below, replacing **YOUR_COG_KEY**.
2. Copy the **endpoint** for your resource and and paste it in the code below, replacing **YOUR_COG_ENDPOINT**.
3. Run the code in the cell below by clicking the Run Cell <span>▷</span> button (at the top left of the cell).
```
cog_key = 'YOUR_COG_KEY'
cog_endpoint = 'YOUR_COG_ENDPOINT'
print('Ready to use cognitive services at {} using key {}'.format(cog_endpoint, cog_key))
```
Now that you have a Cognitive Services resource you can use the Face service to detect human faces in the store.
Run the code cell below to see an example.
```
from azure.cognitiveservices.vision.face import FaceClient
from msrest.authentication import CognitiveServicesCredentials
from python_code import faces
import os
%matplotlib inline
# Create a face detection client.
face_client = FaceClient(cog_endpoint, CognitiveServicesCredentials(cog_key))
# Open an image
image_path = os.path.join('data', 'face', 'store_cam2.jpg')
image_stream = open(image_path, "rb")
# Detect faces
detected_faces = face_client.face.detect_with_stream(image=image_stream)
# Display the faces (code in python_code/faces.py)
faces.show_faces(image_path, detected_faces)
```
Each detected face is assigned a unique ID, so your application can identify each individual face that was detected.
Run the cell below to see the IDs for some more shopper faces.
```
# Open an image
image_path = os.path.join('data', 'face', 'store_cam3.jpg')
image_stream = open(image_path, "rb")
# Detect faces
detected_faces = face_client.face.detect_with_stream(image=image_stream)
# Display the faces (code in python_code/faces.py)
faces.show_faces(image_path, detected_faces, show_id=True)
```
## Analyze facial attributes
Face can do much more than simply detect faces. It can also analyze facial features and expressions to suggest age and emotional state; For example, run the code below to analyze the facial attributes of a shopper.
```
# Open an image
image_path = os.path.join('data', 'face', 'store_cam1.jpg')
image_stream = open(image_path, "rb")
# Detect faces and specified facial attributes
attributes = ['age', 'emotion']
detected_faces = face_client.face.detect_with_stream(image=image_stream, return_face_attributes=attributes)
# Display the faces and attributes (code in python_code/faces.py)
faces.show_face_attributes(image_path, detected_faces)
```
Based on the emotion scores detected for the customer in the image, the customer seems pretty happy with the shopping experience.
## Find similar faces
The face IDs that are created for each detected face are used to individually identify face detections. You can use these IDs to compare a detected face to previously detected faces and find faces with similar features.
For example, run the cell below to compare the shopper in one image with shoppers in another, and find a matching face.
```
# Get the ID of the first face in image 1
image_1_path = os.path.join('data', 'face', 'store_cam3.jpg')
image_1_stream = open(image_1_path, "rb")
image_1_faces = face_client.face.detect_with_stream(image=image_1_stream)
face_1 = image_1_faces[0]
# Get the face IDs in a second image
image_2_path = os.path.join('data', 'face', 'store_cam2.jpg')
image_2_stream = open(image_2_path, "rb")
image_2_faces = face_client.face.detect_with_stream(image=image_2_stream)
image_2_face_ids = list(map(lambda face: face.face_id, image_2_faces))
# Find faces in image 2 that are similar to the one in image 1
similar_faces = face_client.face.find_similar(face_id=face_1.face_id, face_ids=image_2_face_ids)
# Show the face in image 1, and similar faces in image 2(code in python_code/face.py)
faces.show_similar_faces(image_1_path, face_1, image_2_path, image_2_faces, similar_faces)
```
## Recognize faces
So far you've seen that Face can detect faces and facial features, and can identify two faces that are similar to one another. You can take things a step further by inplementing a *facial recognition* solution in which you train Face to recognize a specific person's face. This can be useful in a variety of scenarios, such as automatically tagging photographs of friends in a social media application, or using facial recognition as part of a biometric identity verification system.
To see how this works, let's suppose the Northwind Traders company wants to use facial recognition to ensure that only authorized employees in the IT department can access secure systems.
We'll start by creating a *person group* to represent the authorized employees.
```
group_id = 'employee_group_id'
try:
# Delete group if it already exists
face_client.person_group.delete(group_id)
except Exception as ex:
print(ex.message)
finally:
face_client.person_group.create(group_id, 'employees')
print ('Group created!')
```
Now that the *person group* exists, we can add a *person* for each employee we want to include in the group, and then register multiple photographs of each person so that Face can learn the distinct facial characetristics of each person. Ideally, the images should show the same person in different poses and with different facial expressions.
We'll add a single employee called Wendell, and register three photographs of the employee.
```
import matplotlib.pyplot as plt
from PIL import Image
import os
%matplotlib inline
# Add a person (Wendell) to the group
wendell = face_client.person_group_person.create(group_id, 'Wendell')
# Get photo's of Wendell
folder = os.path.join('data', 'face', 'wendell')
wendell_pics = os.listdir(folder)
# Register the photos
i = 0
fig = plt.figure(figsize=(8, 8))
for pic in wendell_pics:
# Add each photo to person in person group
img_path = os.path.join(folder, pic)
img_stream = open(img_path, "rb")
face_client.person_group_person.add_face_from_stream(group_id, wendell.person_id, img_stream)
# Display each image
img = Image.open(img_path)
i +=1
a=fig.add_subplot(1,len(wendell_pics), i)
a.axis('off')
imgplot = plt.imshow(img)
plt.show()
```
With the person added, and photographs registered, we can now train Face to recognize each person.
```
face_client.person_group.train(group_id)
print('Trained!')
```
Now, with the model trained, you can use it to identify recognized faces in an image.
```
# Get the face IDs in a second image
image_path = os.path.join('data', 'face', 'employees.jpg')
image_stream = open(image_path, "rb")
image_faces = face_client.face.detect_with_stream(image=image_stream)
image_face_ids = list(map(lambda face: face.face_id, image_faces))
# Get recognized face names
face_names = {}
recognized_faces = face_client.face.identify(image_face_ids, group_id)
for face in recognized_faces:
person_name = face_client.person_group_person.get(group_id, face.candidates[0].person_id).name
face_names[face.face_id] = person_name
# show recognized faces
faces.show_recognized_faces(image_path, image_faces, face_names)
```
## Learn More
To learn more about the Face cognitive service, see the [Face documentation](https://docs.microsoft.com/azure/cognitive-services/face/)
|
github_jupyter
|
cog_key = 'YOUR_COG_KEY'
cog_endpoint = 'YOUR_COG_ENDPOINT'
print('Ready to use cognitive services at {} using key {}'.format(cog_endpoint, cog_key))
from azure.cognitiveservices.vision.face import FaceClient
from msrest.authentication import CognitiveServicesCredentials
from python_code import faces
import os
%matplotlib inline
# Create a face detection client.
face_client = FaceClient(cog_endpoint, CognitiveServicesCredentials(cog_key))
# Open an image
image_path = os.path.join('data', 'face', 'store_cam2.jpg')
image_stream = open(image_path, "rb")
# Detect faces
detected_faces = face_client.face.detect_with_stream(image=image_stream)
# Display the faces (code in python_code/faces.py)
faces.show_faces(image_path, detected_faces)
# Open an image
image_path = os.path.join('data', 'face', 'store_cam3.jpg')
image_stream = open(image_path, "rb")
# Detect faces
detected_faces = face_client.face.detect_with_stream(image=image_stream)
# Display the faces (code in python_code/faces.py)
faces.show_faces(image_path, detected_faces, show_id=True)
# Open an image
image_path = os.path.join('data', 'face', 'store_cam1.jpg')
image_stream = open(image_path, "rb")
# Detect faces and specified facial attributes
attributes = ['age', 'emotion']
detected_faces = face_client.face.detect_with_stream(image=image_stream, return_face_attributes=attributes)
# Display the faces and attributes (code in python_code/faces.py)
faces.show_face_attributes(image_path, detected_faces)
# Get the ID of the first face in image 1
image_1_path = os.path.join('data', 'face', 'store_cam3.jpg')
image_1_stream = open(image_1_path, "rb")
image_1_faces = face_client.face.detect_with_stream(image=image_1_stream)
face_1 = image_1_faces[0]
# Get the face IDs in a second image
image_2_path = os.path.join('data', 'face', 'store_cam2.jpg')
image_2_stream = open(image_2_path, "rb")
image_2_faces = face_client.face.detect_with_stream(image=image_2_stream)
image_2_face_ids = list(map(lambda face: face.face_id, image_2_faces))
# Find faces in image 2 that are similar to the one in image 1
similar_faces = face_client.face.find_similar(face_id=face_1.face_id, face_ids=image_2_face_ids)
# Show the face in image 1, and similar faces in image 2(code in python_code/face.py)
faces.show_similar_faces(image_1_path, face_1, image_2_path, image_2_faces, similar_faces)
group_id = 'employee_group_id'
try:
# Delete group if it already exists
face_client.person_group.delete(group_id)
except Exception as ex:
print(ex.message)
finally:
face_client.person_group.create(group_id, 'employees')
print ('Group created!')
import matplotlib.pyplot as plt
from PIL import Image
import os
%matplotlib inline
# Add a person (Wendell) to the group
wendell = face_client.person_group_person.create(group_id, 'Wendell')
# Get photo's of Wendell
folder = os.path.join('data', 'face', 'wendell')
wendell_pics = os.listdir(folder)
# Register the photos
i = 0
fig = plt.figure(figsize=(8, 8))
for pic in wendell_pics:
# Add each photo to person in person group
img_path = os.path.join(folder, pic)
img_stream = open(img_path, "rb")
face_client.person_group_person.add_face_from_stream(group_id, wendell.person_id, img_stream)
# Display each image
img = Image.open(img_path)
i +=1
a=fig.add_subplot(1,len(wendell_pics), i)
a.axis('off')
imgplot = plt.imshow(img)
plt.show()
face_client.person_group.train(group_id)
print('Trained!')
# Get the face IDs in a second image
image_path = os.path.join('data', 'face', 'employees.jpg')
image_stream = open(image_path, "rb")
image_faces = face_client.face.detect_with_stream(image=image_stream)
image_face_ids = list(map(lambda face: face.face_id, image_faces))
# Get recognized face names
face_names = {}
recognized_faces = face_client.face.identify(image_face_ids, group_id)
for face in recognized_faces:
person_name = face_client.person_group_person.get(group_id, face.candidates[0].person_id).name
face_names[face.face_id] = person_name
# show recognized faces
faces.show_recognized_faces(image_path, image_faces, face_names)
| 0.538012 | 0.977263 |
## Import dependecies
Import modules for
```
import os, inspect, sys
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import plotly.offline as ply
import plotly.graph_objs as go
import plotly.tools as tls
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
from sklearn.ensemble import (
AdaBoostClassifier,
GradientBoostingClassifier,
RandomForestClassifier,
ExtraTreesClassifier
)
from sklearn.metrics import (
classification_report,
confusion_matrix,
accuracy_score,
mean_squared_error,
roc_curve,
auc,
)
import matplotlib.pyplot as plt
from statsmodels.graphics.tsaplots import plot_acf, plot_pacf
from statsmodels.tsa.arima_model import ARIMA
from xgboost import XGBClassifier
# get project root directory
curr_dir = os.path.dirname(inspect.getabsfile(inspect.currentframe()))
root_dir = os.path.dirname(curr_dir)
sys.path.insert(0, 0)
```
## Load data
We load the stock datasets to a pandas dataframe:
```
# load data
fname = os.path.join(root_dir, "stockomen", "data", "int", "stock.csv")
df = pd.read_csv(fname, index_col=False)
df.head(5)
```
## Fill missing values (the NaN values) with the column mean
```
nan_list = ['Subjectivity', 'Objectivity', 'Positive', 'Negative', 'Neutral']
for col in nan_list:
df[col] = df[col].fillna(df[col].mean())
# Recheck the count
print(df.count())
```
# Prepare train data
```
x = df.loc[:, ["Subjectivity",
"Objectivity",
"Positive",
"Negative",
"Neutral",
"Open",
"High",
"Low",
"Close",
"Volume",
"Adj Close",
"Month",
"Quarter"]]
y = df.loc[:,'Label']
x.head()
# set valid ratio wrt the number of train data
valid_size = 0.2
# set test ratio wrt to the number of data
test_size = 0.3
# rescale data
scaler = StandardScaler().fit(x)
x_scaled = scaler.transform(x)
n_x = len(x.index)
n_train = int(len(x.index) * 0.7)
x_train, x_test = x_scaled[0:n_train+1, :], x_scaled[n_train: n_x, :]
y_train, y_test = y[0:n_train+1], y[train_size: n_x]
print("data: ", x.shape, "labels: ", y.shape )
print("train data: ", x_train.shape, "train labels: ", y_train.shape )
print("test data: ", x_test.shape, "test labels: ", y_test.shape )
# plt.plot(X_train['Objectivity'])
# plt.plot([None for i in x_train['Objectivity']] + [x for x in x_test['Objectivity']])
# plt.show()
# set train params
num_folds = 10
scoring = 'accuracy'
# define training models
models = {}
models.update({'LR': LogisticRegression()})
models.update({'LDA': LinearDiscriminantAnalysis()})
models.update({'KNN': KNeighborsClassifier()})
models.update({'CART': DecisionTreeClassifier()})
models.update({'NB': GaussianNB()})
models.update({'SVM': SVC()})
models.update({'RF': RandomForestClassifier(n_estimators=50)})
models.update({'XGBoost': XGBClassifier()})
models.update({'LDA': LinearDiscriminantAnalysis()})
x_train.shape
# train models
results = []
names = []
for name, model in models.items():
clf = model
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
accu_score = accuracy_score(y_test, y_pred)
print(name + ": " + str(accu_score))
# Explore featurs
features = df.drop(['Label', "Date"],axis=1).columns.values
x, y = (list(x) for x in zip(*sorted(zip(models["XGBoost"].feature_importances_, features),
reverse = False)))
ply.init_notebook_mode(connected=True)
trace2 = go.Bar(
x=x ,
y=y,
marker=dict(
color=x,
colorscale = 'Viridis',
reversescale = True
),
name='Feature importance for XGBoost',
orientation='h',
)
layout = dict(
title='Barplot of Feature importances for XGBoost',
width = 1000, height = 1000,
yaxis=dict(
showgrid=False,
showline=False,
showticklabels=True,
# domain=[0, 0.85],
))
fig1 = go.Figure(data=[trace2])
fig1['layout'].update(layout)
ply.iplot(fig1, filename='plots')
x, y = (list(x) for x in zip(*sorted(zip(models["RF"].feature_importances_, features),
reverse = False)))
trace2 = go.Bar(
x=x ,
y=y,
marker=dict(
color=x,
colorscale = 'Viridis',
reversescale = True
),
name='Feature importance for Random Forests',
orientation='h',
)
layout = dict(
title='Barplot of Feature importances for Random Forests',
width = 1000, height = 1000,
yaxis=dict(
showgrid=False,
showline=False,
showticklabels=True,
# domain=[0, 0.85],
))
fig1 = go.Figure(data=[trace2])
fig1['layout'].update(layout)
ply.iplot(fig1, filename='plots')
```
|
github_jupyter
|
import os, inspect, sys
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import plotly.offline as ply
import plotly.graph_objs as go
import plotly.tools as tls
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
from sklearn.ensemble import (
AdaBoostClassifier,
GradientBoostingClassifier,
RandomForestClassifier,
ExtraTreesClassifier
)
from sklearn.metrics import (
classification_report,
confusion_matrix,
accuracy_score,
mean_squared_error,
roc_curve,
auc,
)
import matplotlib.pyplot as plt
from statsmodels.graphics.tsaplots import plot_acf, plot_pacf
from statsmodels.tsa.arima_model import ARIMA
from xgboost import XGBClassifier
# get project root directory
curr_dir = os.path.dirname(inspect.getabsfile(inspect.currentframe()))
root_dir = os.path.dirname(curr_dir)
sys.path.insert(0, 0)
# load data
fname = os.path.join(root_dir, "stockomen", "data", "int", "stock.csv")
df = pd.read_csv(fname, index_col=False)
df.head(5)
nan_list = ['Subjectivity', 'Objectivity', 'Positive', 'Negative', 'Neutral']
for col in nan_list:
df[col] = df[col].fillna(df[col].mean())
# Recheck the count
print(df.count())
x = df.loc[:, ["Subjectivity",
"Objectivity",
"Positive",
"Negative",
"Neutral",
"Open",
"High",
"Low",
"Close",
"Volume",
"Adj Close",
"Month",
"Quarter"]]
y = df.loc[:,'Label']
x.head()
# set valid ratio wrt the number of train data
valid_size = 0.2
# set test ratio wrt to the number of data
test_size = 0.3
# rescale data
scaler = StandardScaler().fit(x)
x_scaled = scaler.transform(x)
n_x = len(x.index)
n_train = int(len(x.index) * 0.7)
x_train, x_test = x_scaled[0:n_train+1, :], x_scaled[n_train: n_x, :]
y_train, y_test = y[0:n_train+1], y[train_size: n_x]
print("data: ", x.shape, "labels: ", y.shape )
print("train data: ", x_train.shape, "train labels: ", y_train.shape )
print("test data: ", x_test.shape, "test labels: ", y_test.shape )
# plt.plot(X_train['Objectivity'])
# plt.plot([None for i in x_train['Objectivity']] + [x for x in x_test['Objectivity']])
# plt.show()
# set train params
num_folds = 10
scoring = 'accuracy'
# define training models
models = {}
models.update({'LR': LogisticRegression()})
models.update({'LDA': LinearDiscriminantAnalysis()})
models.update({'KNN': KNeighborsClassifier()})
models.update({'CART': DecisionTreeClassifier()})
models.update({'NB': GaussianNB()})
models.update({'SVM': SVC()})
models.update({'RF': RandomForestClassifier(n_estimators=50)})
models.update({'XGBoost': XGBClassifier()})
models.update({'LDA': LinearDiscriminantAnalysis()})
x_train.shape
# train models
results = []
names = []
for name, model in models.items():
clf = model
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
accu_score = accuracy_score(y_test, y_pred)
print(name + ": " + str(accu_score))
# Explore featurs
features = df.drop(['Label', "Date"],axis=1).columns.values
x, y = (list(x) for x in zip(*sorted(zip(models["XGBoost"].feature_importances_, features),
reverse = False)))
ply.init_notebook_mode(connected=True)
trace2 = go.Bar(
x=x ,
y=y,
marker=dict(
color=x,
colorscale = 'Viridis',
reversescale = True
),
name='Feature importance for XGBoost',
orientation='h',
)
layout = dict(
title='Barplot of Feature importances for XGBoost',
width = 1000, height = 1000,
yaxis=dict(
showgrid=False,
showline=False,
showticklabels=True,
# domain=[0, 0.85],
))
fig1 = go.Figure(data=[trace2])
fig1['layout'].update(layout)
ply.iplot(fig1, filename='plots')
x, y = (list(x) for x in zip(*sorted(zip(models["RF"].feature_importances_, features),
reverse = False)))
trace2 = go.Bar(
x=x ,
y=y,
marker=dict(
color=x,
colorscale = 'Viridis',
reversescale = True
),
name='Feature importance for Random Forests',
orientation='h',
)
layout = dict(
title='Barplot of Feature importances for Random Forests',
width = 1000, height = 1000,
yaxis=dict(
showgrid=False,
showline=False,
showticklabels=True,
# domain=[0, 0.85],
))
fig1 = go.Figure(data=[trace2])
fig1['layout'].update(layout)
ply.iplot(fig1, filename='plots')
| 0.474388 | 0.832237 |
# Entity Explorer - Linux Host
<details>
<summary> <u>Details...</u></summary>
**Notebook Version:** 1.1<br>
**Python Version:** Python 3.6 (including Python 3.6 - AzureML)<br>
**Required Packages**: kqlmagic, msticpy, pandas, pandas_bokeh, numpy, matplotlib, networkx, seaborn, datetime, ipywidgets, ipython, dnspython, ipwhois, folium, maxminddb_geolite2<br>
**Data Sources Required**:
- Log Analytics/Microsoft Sentinel - Syslog, Secuirty Alerts, Auditd, Azure Network Analytics.
- (Optional) - AlienVault OTX (requires account and API key)
</details>
This Notebooks brings together a series of tools and techniques to enable threat hunting within the context of a singular Linux host. The notebook utilizes a range of data sources to achieve this but in order to support the widest possible range of scenarios this Notebook prioritizes using common Syslog data. If there is detailed auditd data available for a host you may wish to edit the Notebook to rely primarily on this dataset, as it currently stands auditd is used when available to provide insight not otherwise available via Syslog.
<h1>Table of Contents<span class="tocSkip"></span></h1>
<div class="toc"><ul class="toc-item"><li><ul class="toc-item"><li><span><a href="#Notebook-initialization" data-toc-modified-id="Notebook-initialization-0.1"><span class="toc-item-num">0.1 </span>Notebook initialization</a></span></li><li><span><a href="#Get-WorkspaceId-and-Authenticate-to-Log-Analytics" data-toc-modified-id="Get-WorkspaceId-and-Authenticate-to-Log-Analytics-0.2"><span class="toc-item-num">0.2 </span>Get WorkspaceId and Authenticate to Log Analytics</a></span></li></ul></li><li><span><a href="#Set-Hunting-Time-Frame" data-toc-modified-id="Set-Hunting-Time-Frame-1"><span class="toc-item-num">1 </span>Set Hunting Time Frame</a></span><ul class="toc-item"><li><span><a href="#Select-Host-to-Investigate" data-toc-modified-id="Select-Host-to-Investigate-1.1"><span class="toc-item-num">1.1 </span>Select Host to Investigate</a></span></li></ul></li><li><span><a href="#Host-Summary" data-toc-modified-id="Host-Summary-2"><span class="toc-item-num">2 </span>Host Summary</a></span><ul class="toc-item"><li><span><a href="#Host-Alerts" data-toc-modified-id="Host-Alerts-2.1"><span class="toc-item-num">2.1 </span>Host Alerts</a></span></li></ul></li><li><span><a href="#Re-scope-Hunting-Time-Frame" data-toc-modified-id="Re-scope-Hunting-Time-Frame-3"><span class="toc-item-num">3 </span>Re-scope Hunting Time Frame</a></span></li><li><span><a href="#How-to-use-this-Notebook" data-toc-modified-id="How-to-use-this-Notebook-4"><span class="toc-item-num">4 </span>How to use this Notebook</a></span></li><li><span><a href="#Host-Logon-Events" data-toc-modified-id="Host-Logon-Events-5"><span class="toc-item-num">5 </span>Host Logon Events</a></span><ul class="toc-item"><li><span><a href="#Logon-Sessions" data-toc-modified-id="Logon-Sessions-5.1"><span class="toc-item-num">5.1 </span>Logon Sessions</a></span><ul class="toc-item"><li><span><a href="#Session-Details" data-toc-modified-id="Session-Details-5.1.1"><span class="toc-item-num">5.1.1 </span>Session Details</a></span></li><li><span><a href="#Raw-data-from-user-session" data-toc-modified-id="Raw-data-from-user-session-5.1.2"><span class="toc-item-num">5.1.2 </span>Raw data from user session</a></span></li></ul></li><li><span><a href="#Process-Tree-from-session" data-toc-modified-id="Process-Tree-from-session-5.2"><span class="toc-item-num">5.2 </span>Process Tree from session</a></span></li><li><span><a href="#Sudo-Session-Investigation" data-toc-modified-id="Sudo-Session-Investigation-5.3"><span class="toc-item-num">5.3 </span>Sudo Session Investigation</a></span></li></ul></li><li><span><a href="#User-Activity" data-toc-modified-id="User-Activity-6"><span class="toc-item-num">6 </span>User Activity</a></span></li><li><span><a href="#Application-Activity" data-toc-modified-id="Application-Activity-7"><span class="toc-item-num">7 </span>Application Activity</a></span><ul class="toc-item"><li><span><a href="#Display-process-tree" data-toc-modified-id="Display-process-tree-7.1"><span class="toc-item-num">7.1 </span>Display process tree</a></span></li><li><span><a href="#Application-Logs-with-associated-Threat-Intelligence" data-toc-modified-id="Application-Logs-with-associated-Threat-Intelligence-7.2"><span class="toc-item-num">7.2 </span>Application Logs with associated Threat Intelligence</a></span></li></ul></li><li><span><a href="#Network-Activity" data-toc-modified-id="Network-Activity-8"><span class="toc-item-num">8 </span>Network Activity</a></span><ul class="toc-item"><li><span><a href="#Choose-ASNs/IPs-to-Check-for-Threat-Intel-Reports" data-toc-modified-id="Choose-ASNs/IPs-to-Check-for-Threat-Intel-Reports-8.1"><span class="toc-item-num">8.1 </span>Choose ASNs/IPs to Check for Threat Intel Reports</a></span></li></ul></li><li><span><a href="#Configuration" data-toc-modified-id="Configuration-9"><span class="toc-item-num">9 </span>Configuration</a></span><ul class="toc-item"><li><span><a href="#msticpyconfig.yaml-configuration-File" data-toc-modified-id="msticpyconfig.yaml-configuration-File-9.1"><span class="toc-item-num">9.1 </span><code>msticpyconfig.yaml</code> configuration File</a></span></li></ul></li></ul></div>
# Hunting Hypothesis:
Our broad initial hunting hypothesis is that a particular Linux host in our environment has been compromised, we will need to hunt from a range of different positions to validate or disprove this hypothesis.
---
### Notebook initialization
The next cell:
- Checks for the correct Python version
- Checks versions and optionally installs required packages
- Imports the required packages into the notebook
- Sets a number of configuration options.
This should complete without errors. If you encounter errors or warnings look at the following two notebooks:
- [TroubleShootingNotebooks](https://github.com/Azure/Azure-Sentinel-Notebooks/blob/master/TroubleShootingNotebooks.ipynb)
- [ConfiguringNotebookEnvironment](https://github.com/Azure/Azure-Sentinel-Notebooks/blob/master/ConfiguringNotebookEnvironment.ipynb)
If you are running in the Microsoft Sentinel Notebooks environment (Azure Notebooks or Azure ML) you can run live versions of these notebooks:
- [Run TroubleShootingNotebooks](./TroubleShootingNotebooks.ipynb)
- [Run ConfiguringNotebookEnvironment](./ConfiguringNotebookEnvironment.ipynb)
You may also need to do some additional configuration to successfully use functions such as Threat Intelligence service lookup and Geo IP lookup.
There are more details about this in the `ConfiguringNotebookEnvironment` notebook and in these documents:
- [msticpy configuration](https://msticpy.readthedocs.io/en/latest/getting_started/msticpyconfig.html)
- [Threat intelligence provider configuration](https://msticpy.readthedocs.io/en/latest/data_acquisition/TIProviders.html#configuration-file)
```
from pathlib import Path
from IPython.display import display, HTML
REQ_PYTHON_VER=(3, 6)
REQ_MSTICPY_VER=(1, 0, 0)
REQ_MP_EXTRAS = ["ml"]
update_nbcheck = (
"<p style='color: orange; text-align=left'>"
"<b>Warning: we needed to update '<i>utils/nb_check.py</i>'</b><br>"
"Please restart the kernel and re-run this cell."
"</p>"
)
display(HTML("<h3>Starting Notebook setup...</h3>"))
if Path("./utils/nb_check.py").is_file():
try:
from utils.nb_check import check_versions
except ImportError as err:
%xmode Minimal
!curl https://raw.githubusercontent.com/Azure/Azure-Sentinel-Notebooks/master/utils/nb_check.py > ./utils/nb_check.py 2>/dev/null
display(HTML(update_nbcheck))
if "check_versions" not in globals():
raise ImportError("Old version of nb_check.py detected - see instructions below.")
%xmode Verbose
check_versions(REQ_PYTHON_VER, REQ_MSTICPY_VER, REQ_MP_EXTRAS)
# If the installation fails try to manually install using
# !pip install --upgrade msticpy
from msticpy.nbtools import nbinit
additional_packages = [
"oauthlib", "pyvis", "python-whois", "seaborn"
]
nbinit.init_notebook(
namespace=globals(),
additional_packages=additional_packages,
extra_imports=extra_imports,
);
from bokeh.models import ColumnDataSource, FactorRange
from bokeh.palettes import viridis
from bokeh.plotting import show, Row, figure
from bokeh.transform import factor_cmap, cumsum
from dns import reversename, resolver
from functools import lru_cache
from ipaddress import ip_address
from ipwhois import IPWhois
from math import pi
from msticpy.common.exceptions import MsticpyException
from msticpy.nbtools import observationlist
from msticpy.nbtools.foliummap import get_map_center
from msticpy.sectools import auditdextract
from msticpy.sectools.cmd_line import risky_cmd_line
from msticpy.sectools.ip_utils import convert_to_ip_entities
from msticpy.sectools.syslog_utils import create_host_record, cluster_syslog_logons_df, risky_sudo_sessions
from pyvis.network import Network
import datetime as dt
import re
```
### Get WorkspaceId and Authenticate to Log Analytics
<details>
<summary> <u>Details...</u></summary>
If you are using user/device authentication, run the following cell.
- Click the 'Copy code to clipboard and authenticate' button.
- This will pop up an Azure Active Directory authentication dialog (in a new tab or browser window). The device code will have been copied to the clipboard.
- Select the text box and paste (Ctrl-V/Cmd-V) the copied value.
- You should then be redirected to a user authentication page where you should authenticate with a user account that has permission to query your Log Analytics workspace.
Use the following syntax if you are authenticating using an Azure Active Directory AppId and Secret:
```
%kql loganalytics://tenant(aad_tenant).workspace(WORKSPACE_ID).clientid(client_id).clientsecret(client_secret)
```
instead of
```
%kql loganalytics://code().workspace(WORKSPACE_ID)
```
Note: you may occasionally see a JavaScript error displayed at the end of the authentication - you can safely ignore this.<br>
On successful authentication you should see a ```popup schema``` button.
To find your Workspace Id go to [Log Analytics](https://ms.portal.azure.com/#blade/HubsExtension/Resources/resourceType/Microsoft.OperationalInsights%2Fworkspaces). Look at the workspace properties to find the ID.
</details>
```
# See if we have a Microsoft Sentinel Workspace defined in our config file.
# If not, let the user specify Workspace and Tenant IDs
ws_config = WorkspaceConfig()
if not ws_config.config_loaded:
ws_config.prompt_for_ws()
qry_prov = QueryProvider(data_environment="AzureSentinel")
print("done")
# Authenticate to Microsoft Sentinel workspace
qry_prov.connect(ws_config)
```
## Set Hunting Time Frame
To begin the hunt we need to et the time frame in which you wish to test your compromised host hunting hypothesis within. Use the widget below to select your start and end time for the hunt.
```
query_times = nbwidgets.QueryTime(units='day',
max_before=14, max_after=1, before=1)
query_times.display()
```
### Select Host to Investigate
Select the host you want to test your hunting hypothesis against, only hosts with Syslog data within the time frame you specified are available. If the host you wish to select is not present try adjusting your time frame.
```
#Get a list of hosts with syslog data in our hunting timegframe to provide easy selection
syslog_query = f"""Syslog | where TimeGenerated between (datetime({query_times.start}) .. datetime({query_times.end})) | summarize by Computer"""
md("Collecting avaliable host details...")
hosts_list = qry_prov._query_provider.query(query=syslog_query)
if isinstance(hosts_list, pd.DataFrame) and not hosts_list.empty:
hosts = hosts_list["Computer"].unique().tolist()
host_text = nbwidgets.SelectItem(description='Select host to investigate: ',
item_list=hosts, width='75%', auto_display=True)
else:
display(md("There are no hosts with syslog data in this time period to investigate"))
```
## Host Summary
Below is a overview of the selected host based on available data sources.
```
hostname=host_text.value
az_net_df = None
# Collect data on the host
all_syslog_query = f"Syslog | where TimeGenerated between (datetime({query_times.start}) .. datetime({query_times.end})) | where Computer =~ '{hostname}'"""
all_syslog_data = qry_prov.exec_query(all_syslog_query)
if isinstance(all_syslog_data, pd.DataFrame) and not all_syslog_data.empty:
heartbeat_query = f"""Heartbeat | where TimeGenerated >= datetime({query_times.start}) | where TimeGenerated <= datetime({query_times.end})| where Computer == '{hostname}' | top 1 by TimeGenerated desc nulls last"""
if "AzureNetworkAnalytics_CL" in qry_prov.schema:
aznet_query = f"""AzureNetworkAnalytics_CL | where TimeGenerated >= datetime({query_times.start}) | where TimeGenerated <= datetime({query_times.end}) | where VirtualMachine_s has '{hostname}' | where ResourceType == 'NetworkInterface' | top 1 by TimeGenerated desc | project PrivateIPAddresses = PrivateIPAddresses_s, PublicIPAddresses = PublicIPAddresses_s"""
print("Getting network data...")
az_net_df = qry_prov.exec_query(query=aznet_query)
print("Getting host data...")
host_hb = qry_prov.exec_query(query=heartbeat_query)
# Create host entity record, with Azure network data if any is avaliable
if az_net_df is not None and isinstance(az_net_df, pd.DataFrame) and not az_net_df.empty:
host_entity = create_host_record(syslog_df=all_syslog_data, heartbeat_df=host_hb, az_net_df=az_net_df)
else:
host_entity = create_host_record(syslog_df=all_syslog_data, heartbeat_df=host_hb)
md(
"<b>Host Details</b><br>"
f"<b>Hostname</b>: {host_entity.computer}<br>"
f"<b>OS</b>: {host_entity.OSType} {host_entity.OSName}<br>"
f"<b>IP Address</b>: {host_entity.IPAddress.Address}<br>"
f"<b>Location</b>: {host_entity.IPAddress.Location.CountryName}<br>"
f"<b>Installed Applications</b>: {host_entity.Applications}<br>"
)
else:
md_warn("No Syslog data found, check hostname and timeframe.")
md("The data query may be timing out, consider reducing the timeframe size.")
```
### Host Alerts & Bookmarks
This section provides an overview of any security alerts or Hunting Bookmarks in Microsoft Sentinel related to this host, this will help scope and guide our hunt.
```
related_alerts = qry_prov.SecurityAlert.list_related_alerts(
query_times, host_name=hostname)
realted_bookmarks = qry_prov.AzureSentinel.list_bookmarks_for_entity(query_times, entity_id=hostname)
if isinstance(related_alerts, pd.DataFrame) and not related_alerts.empty:
host_alert_items = (related_alerts[['AlertName', 'TimeGenerated']]
.groupby('AlertName').TimeGenerated.agg('count').to_dict())
def print_related_alerts(alertDict, entityType, entityName):
if len(alertDict) > 0:
md(f"Found {len(alertDict)} different alert types related to this {entityType} (\'{entityName}\')")
for (k, v) in alertDict.items():
md(f"- {k}, Count of alerts: {v}")
else:
md(f"No alerts for {entityType} entity \'{entityName}\'")
print_related_alerts(host_alert_items, 'host', host_entity.HostName)
nbdisplay.display_timeline(
data=related_alerts, source_columns=["AlertName"], title="Host alerts over time", height=300, color="red")
else:
md('No related alerts found.')
if isinstance(realted_bookmarks, pd.DataFrame) and not realted_bookmarks.empty:
nbdisplay.display_timeline(data=realted_bookmarks, source_columns=["BookmarkName"], height=200, color="orange", title="Host bookmarks over time",)
else:
md('No related bookmarks found.')
rel_alert_select = None
def show_full_alert(selected_alert):
global security_alert, alert_ip_entities
security_alert = SecurityAlert(
rel_alert_select.selected_alert)
nbdisplay.display_alert(security_alert, show_entities=True)
# Show selected alert when selected
if isinstance(related_alerts, pd.DataFrame) and not related_alerts.empty:
related_alerts['CompromisedEntity'] = related_alerts['Computer']
md('### Click on alert to view details.')
rel_alert_select = nbwidgets.SelectAlert(alerts=related_alerts,
action=show_full_alert)
rel_alert_select.display()
else:
md('No related alerts found.')
```
## Re-scope Hunting Time Frame
Based on the security alerts for this host we can choose to re-scope our hunting time frame.
```
if rel_alert_select is None or rel_alert_select.selected_alert is None:
start = query_times.start
else:
start = rel_alert_select.selected_alert['TimeGenerated']
# Set new investigation time windows based on the selected alert
invest_times = nbwidgets.QueryTime(
units='day', max_before=24, max_after=12, before=1, after=1, origin_time=start)
invest_times.display()
```
## How to use this Notebook
Whilst this notebook is linear in layout it doesn't need to be linear in usage. We have selected our host to investigate and set an initial hunting time-frame to work within. We can now start to test more specific hunting hypothesis with the aim of validating our broader initial hunting hypothesis. To do this we can start by looking at:
- <a>Host Logon Events</a>
- <a>User Activity</a>
- <a>Application Activity</a>
- <a>Network Activity</a>
You can choose to start below with a hunt in host logon events or choose to jump to one of the other sections listed above. The order in which you choose to run each of these major sections doesn't matter, they are each self contained. You may also choose to rerun sections based on your findings from running other sections.
This notebook uses external threat intelligence sources to enrich data. The next cell loads the TILookup class.
> **Note**: to use TILookup you will need configuration settings in your msticpyconfig.yaml
> <br>see [TIProviders documenation](https://msticpy.readthedocs.io/en/latest/TIProviders.html)
> <br>and [Configuring Notebook Environment notebook](./ConfiguringNotebookEnvironment.ipynb)
> <br>or [ConfiguringNotebookEnvironment (GitHub static view)](https://github.com/Azure/Azure-Sentinel-Notebooks/blob/master/ConfiguringNotebookEnvironment.ipynb)
```
tilookup = TILookup()
md("Threat intelligence provider loading complete.")
```
## Host Logon Events
**Hypothesis:** That an attacker has gained legitimate access to the host via compromised credentials and has logged into the host to conduct malicious activity.
This section provides an overview of logon activity for the host within our hunting time frame, the purpose of this is to allow for the identification of anomalous logons or attempted logons.
```
# Collect logon events for this, seperate them into sucessful and unsucessful and cluster sucessful one into sessions
logon_events = qry_prov.LinuxSyslog.user_logon(start=invest_times.start, end=invest_times.end, host_name=hostname)
remote_logons = None
failed_logons = None
if isinstance(logon_events, pd.DataFrame) and not logon_events.empty:
remote_logons = (logon_events[logon_events['LogonResult'] == 'Success'])
failed_logons = (logon_events[logon_events['LogonResult'] == 'Failure'])
else:
print("No logon events in this timeframe")
if (isinstance(remote_logons, pd.DataFrame) and not remote_logons.empty) or (isinstance(failed_logons, pd.DataFrame) and not failed_logons.empty):
#Provide a timeline of sucessful and failed logon attempts to aid identification of potential brute force attacks
display(Markdown('### Timeline of sucessful host logons.'))
tooltip_cols = ['User', 'ProcessName', 'SourceIP']
if rel_alert_select is not None:
logon_timeline = nbdisplay.display_timeline(data=remote_logons, overlay_data=failed_logons, source_columns=tooltip_cols, height=200, overlay_color="red", alert = rel_alert_select.selected_alert)
else:
logon_timeline = nbdisplay.display_timeline(data=remote_logons, overlay_data=failed_logons, source_columns=tooltip_cols, height=200, overlay_color="red")
display(Markdown('<b>Key:</b><p style="color:darkblue">Sucessful logons </p><p style="color:Red">Failed Logon Attempts (via su)</p>'))
all_df = pd.DataFrame(dict(successful= remote_logons['ProcessName'].value_counts(), failed = failed_logons['ProcessName'].value_counts())).fillna(0)
fail_data = pd.value_counts(failed_logons['User'].values, sort=True).head(10).reset_index(name='value').rename(columns={'User':'Count'})
fail_data['angle'] = fail_data['value']/fail_data['value'].sum() * 2*pi
fail_data['color'] = viridis(len(fail_data))
fp = figure(plot_height=350, plot_width=450, title="Relative Frequencies of Failed Logons by Account", toolbar_location=None, tools="hover", tooltips="@index: @value")
fp.wedge(x=0, y=1, radius=0.5, start_angle=cumsum('angle', include_zero=True), end_angle=cumsum('angle'), line_color="white", fill_color='color', legend='index', source=fail_data)
sucess_data = pd.value_counts(remote_logons['User'].values, sort=False).reset_index(name='value').rename(columns={'User':'Count'})
sucess_data['angle'] = sucess_data['value']/sucess_data['value'].sum() * 2*pi
sucess_data['color'] = viridis(len(sucess_data))
sp = figure(plot_height=350, width=450, title="Relative Frequencies of Sucessful Logons by Account", toolbar_location=None, tools="hover", tooltips="@index: @value")
sp.wedge(x=0, y=1, radius=0.5, start_angle=cumsum('angle', include_zero=True), end_angle=cumsum('angle'), line_color="white", fill_color='color', legend='index', source=sucess_data)
fp.axis.axis_label=None
fp.axis.visible=False
fp.grid.grid_line_color = None
sp.axis.axis_label=None
sp.axis.visible=False
sp.grid.grid_line_color = None
processes = all_df.index.values.tolist()
results = all_df.columns.values.tolist()
fail_sucess_data = {'processes' :processes,
'sucess' : all_df['successful'].values.tolist(),
'failure': all_df['failed'].values.tolist()}
palette = viridis(2)
x = [ (process, result) for process in processes for result in results ]
counts = sum(zip(fail_sucess_data['sucess'], fail_sucess_data['failure']), ())
source = ColumnDataSource(data=dict(x=x, counts=counts))
b = figure(x_range=FactorRange(*x), plot_height=350, plot_width=450, title="Failed and Sucessful logon attempts by process",
toolbar_location=None, tools="", y_minor_ticks=2)
b.vbar(x='x', top='counts', width=0.9, source=source, line_color="white",
fill_color=factor_cmap('x', palette=palette, factors=results, start=1, end=2))
b.y_range.start = 0
b.x_range.range_padding = 0.1
b.xaxis.major_label_orientation = 1
b.xgrid.grid_line_color = None
show(Row(sp,fp,b))
ip_list = [convert_to_ip_entities(i, ip_col="SourceIP")[0] for i in remote_logons['SourceIP'].unique() if i != ""]
ip_fail_list = [convert_to_ip_entities(i)[0] for i in failed_logons['SourceIP'].unique() if i != ""]
location = get_map_center(ip_list + ip_fail_list)
folium_map = FoliumMap(location = location, zoom_start=1.4)
#Map logon locations to allow for identification of anomolous locations
if len(ip_fail_list) > 0:
md('<h3>Map of Originating Location of Logon Attempts</h3>')
icon_props = {'color': 'red'}
folium_map.add_ip_cluster(ip_entities=ip_fail_list, **icon_props)
if len(ip_list) > 0:
icon_props = {'color': 'green'}
folium_map.add_ip_cluster(ip_entities=ip_list, **icon_props)
display(folium_map.folium_map)
md('<p style="color:red">Warning: the folium mapping library '
'does not display correctly in some browsers.</p><br>'
'If you see a blank image please retry with a different browser.')
```
### Logon Sessions
Based on the detail above if you wish to focus your hunt on a particular user jump to the [User Activity](#user) section. Alternatively to further further refine our hunt we need to select a logon session to view in more detail. Select a session from the list below to continue. Sessions that occurred at the time an alert was raised for this host, or where the user has a abnormal ratio of failed to successful login attempts are highlighted.
```
logon_sessions_df = None
try:
print("Clustering logon sessions...")
logon_sessions_df = cluster_syslog_logons_df(logon_events)
except Exception as err:
print(f"Error clustering logons: {err}")
if logon_sessions_df is not None:
logon_sessions_df["Alerts during session?"] = np.nan
# check if any alerts occur during logon window.
logon_sessions_df['Start (UTC)'] = [(time - dt.timedelta(seconds=5)) for time in logon_sessions_df['Start']]
logon_sessions_df['End (UTC)'] = [(time + dt.timedelta(seconds=5)) for time in logon_sessions_df['End']]
for TimeGenerated in related_alerts['TimeGenerated']:
logon_sessions_df.loc[(TimeGenerated >= logon_sessions_df['Start (UTC)']) & (TimeGenerated <= logon_sessions_df['End (UTC)']), "Alerts during session?"] = "Yes"
logon_sessions_df.loc[logon_sessions_df['User'] == 'root', "Root?"] = "Yes"
logon_sessions_df.replace(np.nan, "No", inplace=True)
ratios = []
for _, row in logon_sessions_df.iterrows():
suc_fail = logon_events.apply(lambda x: True if x['User'] == row['User'] and x["LogonResult"] == 'Success' else(
False if x['User'] == row['User'] and x["LogonResult"] == 'Failure' else None), axis=1)
numofsucess = len(suc_fail[suc_fail == True].index)
numoffail = len(suc_fail[suc_fail == False].index)
if numoffail == 0:
ratio = 1
else:
ratio = numofsucess/numoffail
ratios.append(ratio)
logon_sessions_df["Sucessful to failed logon ratio"] = ratios
def color_cells(val):
if isinstance(val, str):
color = 'yellow' if val == "Yes" else 'white'
elif isinstance(val, float):
color = 'yellow' if val > 0.5 else 'white'
else:
color = 'white'
return 'background-color: %s' % color
display(logon_sessions_df[['User','Start (UTC)', 'End (UTC)', 'Alerts during session?', 'Sucessful to failed logon ratio', 'Root?']]
.style.applymap(color_cells).hide_index())
logon_items = (
logon_sessions_df[['User','Start (UTC)', 'End (UTC)']]
.to_string(header=False, index=False, index_names=False)
.split('\n')
)
logon_sessions_df["Key"] = logon_items
logon_sessions_df.set_index('Key', inplace=True)
logon_dict = logon_sessions_df[['User','Start (UTC)', 'End (UTC)']].to_dict('index')
logon_selection = nbwidgets.SelectItem(description='Select logon session to investigate: ',
item_dict=logon_dict , width='80%', auto_display=True)
else:
md("No logon sessions during this timeframe")
```
#### Session Details
```
def view_syslog(selected_facility):
return [syslog_events.query('Facility == @selected_facility')]
# Produce a summary of user modification actions taken
if "Add" in x:
return len(add_events.replace("", np.nan).dropna(subset=['User'])['User'].unique().tolist())
elif "Modify" in x:
return len(mod_events.replace("", np.nan).dropna(subset=['User'])['User'].unique().tolist())
elif "Delete" in x:
return len(del_events.replace("", np.nan).dropna(subset=['User'])['User'].unique().tolist())
else:
return ""
crn_tl_data = {}
user_tl_data = {}
sudo_tl_data = {}
sudo_sessions = None
tooltip_cols = ['SyslogMessage']
if logon_sessions_df is not None:
#Collect data based on the session selected for investigation
invest_sess = {'StartTimeUtc': logon_selection.value.get('Start (UTC)'), 'EndTimeUtc': logon_selection.value.get(
'End (UTC)'), 'Account': logon_selection.value.get('User'), 'Host': hostname}
session = entities.HostLogonSession(invest_sess)
syslog_events = qry_prov.LinuxSyslog.all_syslog(
start=session.StartTimeUtc, end=session.EndTimeUtc, host_name=session.Host)
sudo_events = qry_prov.LinuxSyslog.sudo_activity(
start=session.StartTimeUtc, end=session.EndTimeUtc, host_name=session.Host, user=session.Account)
if isinstance(sudo_events, pd.DataFrame) and not sudo_events.empty:
try:
sudo_sessions = cluster_syslog_logons_df(logon_events=sudo_events)
except MsticpyException:
pass
# Display summary of cron activity in session
cron_events = qry_prov.LinuxSyslog.cron_activity(
start=session.StartTimeUtc, end=session.EndTimeUtc, host_name=session.Host)
if not isinstance(cron_events, pd.DataFrame) or cron_events.empty:
md(f'<h3> No Cron activity for {session.Host} between {session.StartTimeUtc} and {session.EndTimeUtc}</h3>')
else:
cron_events['CMD'].replace('', np.nan, inplace=True)
crn_tl_data = {"Cron Exections": {"data": cron_events[['TimeGenerated', 'CMD', 'CronUser', 'SyslogMessage']].dropna(), "source_columns": tooltip_cols, "color": "Blue"},
"Cron Edits": {"data": cron_events.loc[cron_events['SyslogMessage'].str.contains('EDIT')], "source_columns": tooltip_cols, "color": "Green"}}
md('<h2> Most common commands run by cron:</h2>')
md('This shows how often each cron job was exected within the specified time window')
cron_commands = (cron_events[['EventTime', 'CMD']]
.groupby(['CMD']).count()
.dropna()
.style
.set_table_attributes('width=900px, text-align=center')
.background_gradient(cmap='Reds', low=0.5, high=1)
.format("{0:0>1.0f}"))
display(cron_commands)
# Display summary of user and group creations, deletions and modifications during the session
user_activity = qry_prov.LinuxSyslog.user_group_activity(
start=session.StartTimeUtc, end=session.EndTimeUtc, host_name=session.Host)
if not isinstance(user_activity, pd.DataFrame) or user_activity.empty:
md(f'<h3>No user or group moidifcations for {session.Host} between {session.StartTimeUtc} and {session.EndTimeUtc}></h3>')
else:
add_events = user_activity[user_activity['UserGroupAction'].str.contains(
'Add')]
del_events = user_activity[user_activity['UserGroupAction'].str.contains(
'Delete')]
mod_events = user_activity[user_activity['UserGroupAction'].str.contains(
'Modify')]
user_activity['Count'] = user_activity.groupby('UserGroupAction')['UserGroupAction'].transform('count')
if add_events.empty and del_events.empty and mod_events.empty:
md('<h2> Users and groups added or deleted:</h2<>')
md(f'No users or groups were added or deleted on {host_entity.HostName} between {query_times.start} and {query_times.end}')
user_tl_data = {}
else:
md("<h2>Users added, modified or deleted</h2>")
display(user_activity[['UserGroupAction','Count']].drop_duplicates().style.hide_index())
account_actions = pd.DataFrame({"User Additions": [add_events.replace("", np.nan).dropna(subset=['User'])['User'].unique().tolist()],
"User Modifications": [mod_events.replace("", np.nan).dropna(subset=['User'])['User'].unique().tolist()],
"User Deletions": [del_events.replace("", np.nan).dropna(subset=['User'])['User'].unique().tolist()]})
display(account_actions.style.hide_index())
user_tl_data = {"User adds": {"data": add_events, "source_columns": tooltip_cols, "color": "Orange"},
"User deletes": {"data": del_events, "source_columns": tooltip_cols, "color": "Red"},
"User modfications": {"data": mod_events, "source_columns": tooltip_cols, "color": "Grey"}}
# Display sudo activity during session
if not isinstance(sudo_sessions, pd.DataFrame) or sudo_sessions.empty:
md(f"<h3>No Sudo sessions for {session.Host} between {logon_selection.value.get('Start (UTC)')} and {logon_selection.value.get('End (UTC)')}</h3>")
sudo_tl_data = {}
else:
sudo_start = sudo_events[sudo_events["SyslogMessage"].str.contains(
"pam_unix.+session opened")].rename(columns={"Sudoer": "User"})
sudo_tl_data = {"Host logons": {"data": remote_logons, "source_columns": tooltip_cols, "color": "Cyan"},
"Sudo sessions": {"data": sudo_start, "source_columns": tooltip_cols, "color": "Purple"}}
try:
risky_actions = cmd_line.risky_cmd_line(events=sudo_events, log_type="Syslog")
suspicious_events = cmd_speed(
cmd_events=sudo_events, time=60, events=2, cmd_field="Command")
except:
risky_actions = None
suspicious_events = None
if risky_actions is None and suspicious_events is None:
pass
else:
risky_sessions = risky_sudo_sessions(
risky_actions=risky_actions, sudo_sessions=sudo_sessions, suspicious_actions=suspicious_events)
for key in risky_sessions:
if key in sudo_sessions:
sudo_sessions[f"{key} - {risky_sessions[key]}"] = sudo_sessions.pop(
key)
if isinstance(sudo_events, pd.DataFrame):
sudo_events_val = sudo_events[['EventTime', 'CommandCall']][sudo_events['CommandCall']!=""].dropna(how='any', subset=['CommandCall'])
if sudo_events_val.empty:
md(f"No sucessful sudo activity for {hostname} between {logon_selection.value.get('Start (UTC)')} and {logon_selection.value.get('End (UTC)')}")
else:
sudo_events.replace("", np.nan, inplace=True)
md('<h2> Frequency of sudo commands</h2>')
md('This shows how many times each command has been run with sudo. /bin/bash is usally associated with the use of "sudo -i"')
sudo_commands = (sudo_events[['EventTime', 'CommandCall']]
.groupby(['CommandCall'])
.count()
.dropna()
.style
.set_table_attributes('width=900px, text-align=center')
.background_gradient(cmap='Reds', low=.5, high=1)
.format("{0:0>3.0f}"))
display(sudo_commands)
else:
md(f"No sucessful sudo activity for {hostname} between {logon_selection.value.get('Start (UTC)')} and {logon_selection.value.get('End (UTC)')}")
# Display a timeline of all activity during session
crn_tl_data.update(user_tl_data)
crn_tl_data.update(sudo_tl_data)
if crn_tl_data:
md('<h2> Session Timeline.</h2>')
nbdisplay.display_timeline(
data=crn_tl_data, title='Session Timeline', height=300)
else:
md("No logon sessions during this timeframe")
```
#### Raw data from user session
Use this syslog message data to further investigate suspicous activity during the session
```
if isinstance(logon_sessions_df, pd.DataFrame) and not logon_sessions_df.empty:
#Return syslog data and present it to the use for investigation
session_syslog = qry_prov.LinuxSyslog.all_syslog(
start=session.StartTimeUtc, end=session.EndTimeUtc, host_name=session.Host)
if session_syslog.empty:
display(HTML(
f' No syslog for {session.Host} between {session.StartTimeUtc} and {session.EndTimeUtc}'))
def view_sudo(selected_cmd):
return [sudo_events.query('CommandCall == @selected_cmd')[
['TimeGenerated', 'SyslogMessage', 'Sudoer', 'SudoTo', 'Command', 'CommandCall']]]
# Show syslog messages associated with selected sudo command
items = sudo_events['CommandCall'].dropna().unique().tolist()
if items:
md("<h3>View all messages associated with a sudo command</h3>")
display(nbwidgets.SelectItem(item_list=items, action=view_sudo))
else:
md("No logon sessions during this timeframe")
if isinstance(logon_sessions_df, pd.DataFrame) and not logon_sessions_df.empty:
# Display syslog messages from the session witht he facility selected
items = syslog_events['Facility'].dropna().unique().tolist()
md("<h3>View all messages associated with a syslog facility</h3>")
display(nbwidgets.SelectItem(item_list=items, action=view_syslog))
else:
md("No logon sessions during this timeframe")
```
### Process Tree from session
```
if isinstance(logon_sessions_df, pd.DataFrame) and not logon_sessions_df.empty:
display(HTML("<h3>Process Trees from session</h3>"))
print("Building process tree, this may take some time...")
# Find the table with auditd data in
regex = '.*audit.*\_cl?'
matches = ((re.match(regex, key, re.IGNORECASE)) for key in qry_prov.schema)
for match in matches:
if match != None:
audit_table = match.group(0)
else:
audit_table = None
# Retrieve auditd data
if audit_table:
audit_data = qry_prov.LinuxAudit.auditd_all(
start=session.StartTimeUtc, end=session.EndTimeUtc, host_name=hostname
)
if isinstance(audit_data, pd.DataFrame) and not audit_data.empty:
audit_events = auditdextract.extract_events_to_df(
data=audit_data
)
process_tree = auditdextract.generate_process_tree(audit_data=audit_events)
process_tree.mp_process_tree.plot()
else:
display(HTML("No auditd data avaliable to build process tree"))
else:
display(HTML("No auditd data avaliable to build process tree"))
else:
md("No logon sessions during this timeframe")
```
Click [here](#app) to start a process/application focused hunt or continue with session based hunt below by selecting a sudo session to investigate.
### Sudo Session Investigation
Sudo activity is often required by an attacker to conduct actions on target, and more granular data is avalibale for sudo sessions allowing for deeper level hunting within these sesions.
```
if logon_sessions_df is not None and sudo_sessions is not None:
sudo_items = sudo_sessions[['User','Start', 'End']].to_string(header=False,
index=False,
index_names=False).split('\n')
sudo_sessions["Key"] = sudo_items
sudo_sessions.set_index('Key', inplace=True)
sudo_dict = sudo_sessions[['User','Start', 'End']].to_dict('index')
sudo_selection = nbwidgets.SelectItem(description='Select sudo session to investigate: ',
item_dict=sudo_dict, width='100%', height='300px', auto_display=True)
else:
sudo_selection = None
md("No logon sessions during this timeframe")
#Collect data associated with the sudo session selected
sudo_events = None
from msticpy.sectools.tiproviders.ti_provider_base import TISeverity
def ti_check_sev(severity, threshold):
severity = TISeverity.parse(severity)
threshold = TISeverity.parse(threshold)
return severity.value >= threshold.value
if sudo_selection:
sudo_sess = {'StartTimeUtc': sudo_selection.value.get('Start'), 'EndTimeUtc': sudo_selection.value.get(
'End'), 'Account': sudo_selection.value.get('User'), 'Host': hostname}
sudo_session = entities.HostLogonSession(sudo_sess)
sudo_events = qry_prov.LinuxSyslog.sudo_activity(start=sudo_session.StartTimeUtc.round(
'-1s') - pd.Timedelta(seconds=1), end=(sudo_session.EndTimeUtc.round('1s')+ pd.Timedelta(seconds=1)), host_name=sudo_session.Host)
if isinstance(sudo_events, pd.DataFrame) and not sudo_events.empty:
display(sudo_events.replace('', np.nan).dropna(axis=0, subset=['Command'])[
['TimeGenerated', 'Command', 'CommandCall', 'SyslogMessage']])
# Extract IOCs from the data
ioc_extractor = iocextract.IoCExtract()
os_family = host_entity.OSType if host_entity.OSType else 'Linux'
print('Extracting IoCs.......')
ioc_df = ioc_extractor.extract(data=sudo_events,
columns=['SyslogMessage'],
os_family=os_family,
ioc_types=['ipv4', 'ipv6', 'dns', 'url',
'md5_hash', 'sha1_hash', 'sha256_hash'])
if len(ioc_df) > 0:
ioc_count = len(
ioc_df[["IoCType", "Observable"]].drop_duplicates())
md(f"Found {ioc_count} IOCs")
#Lookup the extracted IOCs in TI feed
ti_resps = tilookup.lookup_iocs(data=ioc_df[["IoCType", "Observable"]].drop_duplicates(
).reset_index(), obs_col='Observable', ioc_type_col='IoCType')
i = 0
ti_hits = []
ti_resps.reset_index(drop=True, inplace=True)
while i < len(ti_resps):
if ti_resps['Result'][i] == True and ti_check_sev(ti_resps['Severity'][i], 1):
ti_hits.append(ti_resps['Ioc'][i])
i += 1
else:
i += 1
md(f"Found {len(ti_hits)} IoCs in Threat Intelligence")
for ioc in ti_hits:
md(f"Messages containing IoC found in TI feed: {ioc}")
display(sudo_events[sudo_events['SyslogMessage'].str.contains(
ioc)][['TimeGenerated', 'SyslogMessage']])
else:
md("No IoC patterns found in Syslog Messages.")
else:
md('No sudo messages for this session')
else:
md("No Sudo session to investigate")
```
Jump to:
- <a>Host Logon Events</a>
- <a>Application Activity</a>
- <a>Network Activity</a>
<a></a>
## User Activity
**Hypothesis:** That an attacker has gained access to the host and is using a user account to conduct actions on the host.
This section provides an overview of activity by user within our hunting time frame, the purpose of this is to allow for the identification of anomalous activity by a user. This hunt can be driven be investigation of suspected users or as a hunt across all users seen on the host.
```
# Get list of users with logon or sudo sessions on host
logon_events = qry_prov.LinuxSyslog.user_logon(query_times, host_name=hostname)
users = logon_events['User'].replace('', np.nan).dropna().unique().tolist()
all_users = list(users)
if isinstance(sudo_events, pd.DataFrame) and not sudo_events.empty:
sudoers = sudo_events['Sudoer'].replace(
'', np.nan).dropna().unique().tolist()
all_users.extend(x for x in sudoers if x not in all_users)
# Pick Users
if not logon_events.empty:
user_select = nbwidgets.SelectItem(description='Select user to investigate: ',
item_list=all_users, width='75%', auto_display=True)
else:
md("There was no user activity in the timeframe specified.")
user_select = None
folium_user_map = FoliumMap()
def view_sudo(cmd):
return [user_sudo_hold.query('CommandCall == @cmd')[
['TimeGenerated', 'HostName', 'Command', 'CommandCall', 'SyslogMessage']]]
user_sudo_hold = None
if user_select is not None:
# Get all syslog relating to these users
username = user_select.value
user_events = all_syslog_data[all_syslog_data['SyslogMessage'].str.contains(username)]
logon_sessions = cluster_syslog_logons_df(logon_events)
# Display all logons associated with the user
md(f"<h1> User Logon Activity for {username}</h1>")
user_logon_events = logon_events[logon_events['User'] == username]
try:
user_logon_sessions = cluster_syslog_logons_df(user_logon_events)
except:
user_logon_sessions = None
user_remote_logons = (
user_logon_events[user_logon_events['LogonResult'] == 'Success']
)
user_failed_logons = (
user_logon_events[user_logon_events['LogonResult'] == 'Failure']
)
if not user_remote_logons.empty:
for _, row in logon_sessions_df.iterrows():
end = row['End']
user_sudo_events = qry_prov.LinuxSyslog.sudo_activity(start=user_remote_logons.sort_values(
by='TimeGenerated')['TimeGenerated'].iloc[0], end=end, host_name=hostname, user=username)
else:
user_sudo_events = None
if user_logon_sessions is None and user_remote_logons.empty and user_failed_logons.empty:
pass
else:
display(HTML(
f"{len(user_remote_logons)} sucessfull logons and {len(user_failed_logons)} failed logons for {username}"))
display(Markdown('### Timeline of host logon attempts.'))
tooltip_cols = ['SyslogMessage']
dfs = {"User Logons" :user_remote_logons, "Failed Logons": user_failed_logons, "Sudo Events" :user_sudo_events}
user_tl_data = {}
for k,v in dfs.items():
if v is not None and not v.empty:
user_tl_data.update({k :{"data":v,"source_columns":tooltip_cols}})
nbdisplay.display_timeline(
data=user_tl_data, title="User logon timeline", height=300)
all_user_df = pd.DataFrame(dict(successful= user_remote_logons['ProcessName'].value_counts(), failed = user_failed_logons['ProcessName'].value_counts())).fillna(0)
processes = all_user_df.index.values.tolist()
results = all_user_df.columns.values.tolist()
user_fail_sucess_data = {'processes' :processes,
'sucess' : all_user_df['successful'].values.tolist(),
'failure': all_user_df['failed'].values.tolist()}
palette = viridis(2)
x = [ (process, result) for process in processes for result in results ]
counts = sum(zip(user_fail_sucess_data['sucess'], fail_sucess_data['failure']), ())
source = ColumnDataSource(data=dict(x=x, counts=counts))
b = figure(x_range=FactorRange(*x), plot_height=350, plot_width=450, title="Failed and Sucessful logon attempts by process",
toolbar_location=None, tools="", y_minor_ticks=2)
b.vbar(x='x', top='counts', width=0.9, source=source, line_color="white",
fill_color=factor_cmap('x', palette=palette, factors=results, start=1, end=2))
b.y_range.start = 0
b.x_range.range_padding = 0.1
b.xaxis.major_label_orientation = 1
b.xgrid.grid_line_color = None
user_logons = pd.DataFrame({"Sucessful Logons" : [int(all_user_df['successful'].sum())],
"Failed Logons" : [int(all_user_df['failed'].sum())]}).T
user_logon_data = pd.value_counts(user_logon_events['LogonResult'].values, sort=True).head(10).reset_index(name='value').rename(columns={'User':'Count'})
user_logon_data = user_logon_data[user_logon_data['index']!="Unknown"].copy()
user_logon_data['angle'] = user_logon_data['value']/user_logon_data['value'].sum() * 2*pi
user_logon_data['color'] = viridis(len(user_logon_data))
p = figure(plot_height=350, plot_width=450, title="Relative Frequencies of Failed Logons by Account", toolbar_location=None, tools="hover", tooltips="@index: @value")
p.axis.visible = False
p.xgrid.visible = False
p.ygrid.visible = False
p.wedge(x=0, y=1, radius=0.5, start_angle=cumsum('angle', include_zero=True), end_angle=cumsum('angle'), line_color="white", fill_color='color', legend='index', source=user_logon_data)
show(Row(p,b))
user_ip_list = [convert_to_ip_entities(i)[0] for i in user_remote_logons['SourceIP']]
user_ip_fail_list = [convert_to_ip_entities(i)[0] for i in user_failed_logons['SourceIP']]
user_location = get_map_center(ip_list + ip_fail_list)
user_folium_map = FoliumMap(location = location, zoom_start=1.4)
#Map logon locations to allow for identification of anomolous locations
if len(ip_fail_list) > 0:
md('<h3>Map of Originating Location of Logon Attempts</h3>')
icon_props = {'color': 'red'}
user_folium_map.add_ip_cluster(ip_entities=user_ip_fail_list, **icon_props)
if len(ip_list) > 0:
icon_props = {'color': 'green'}
user_folium_map.add_ip_cluster(ip_entities=user_ip_list, **icon_props)
display(user_folium_map.folium_map)
md('<p style="color:red">Warning: the folium mapping library '
'does not display correctly in some browsers.</p><br>'
'If you see a blank image please retry with a different browser.')
#Display sudo activity of the user
if not isinstance(user_sudo_events, pd.DataFrame) or user_sudo_events.empty:
md(f"<h3>No sucessful sudo activity for {username}</h3>")
else:
user_sudo_hold = user_sudo_events
user_sudo_commands = (user_sudo_events[['EventTime', 'CommandCall']].replace('', np.nan).groupby(['CommandCall']).count().dropna().style.set_table_attributes('width=900px, text-align=center').background_gradient(cmap='Reds', low=.5, high=1).format("{0:0>3.0f}"))
display(user_sudo_commands)
md("Select a sudo command to investigate in more detail")
display(nbwidgets.SelectItem(item_list=items, action=view_sudo))
else:
md("No user session selected")
# If the user has sudo activity extract and IOCs from the logs and look them up in TI feeds
if not isinstance(user_sudo_hold, pd.DataFrame) or user_sudo_hold.empty:
md(f"No sudo messages data")
else:
# Extract IOCs
ioc_extractor = iocextract.IoCExtract()
os_family = host_entity.OSType if host_entity.OSType else 'Linux'
print('Extracting IoCs.......')
ioc_df = ioc_extractor.extract(data=user_sudo_hold,
columns=['SyslogMessage'],
ioc_types=['ipv4', 'ipv6', 'dns', 'url', 'md5_hash', 'sha1_hash', 'sha256_hash'])
if len(ioc_df) > 0:
ioc_count = len(ioc_df[["IoCType", "Observable"]].drop_duplicates())
md(f"Found {ioc_count} IOCs")
ti_resps = tilookup.lookup_iocs(data=ioc_df[["IoCType", "Observable"]].drop_duplicates(
).reset_index(), obs_col='Observable', ioc_type_col='IoCType')
i = 0
ti_hits = []
ti_resps.reset_index(drop=True, inplace=True)
while i < len(ti_resps):
if ti_resps['Result'][i] == True and ti_check_sev(ti_resps['Severity'][i], 1):
ti_hits.append(ti_resps['Ioc'][i])
i += 1
else:
i += 1
md(f"Found {len(ti_hits)} IoCs in Threat Intelligence")
for ioc in ti_hits:
md(f"Messages containing IoC found in TI feed: {ioc}")
display(user_sudo_hold[user_sudo_hold['SyslogMessage'].str.contains(
ioc)][['TimeGenerated', 'SyslogMessage']])
else:
md("No IoC patterns found in Syslog Message.")
```
Jump to:
- <a>Host Logon Events</a>
- <a>User Activity</a>
- <a>Network Activity</a>
<a></a>
## Application Activity
**Hypothesis:** That an attacker has compromised an application running on the host and is using the applications process to conduct actions on the host.
This section provides an overview of activity by application within our hunting time frame, the purpose of this is to allow for the identification of anomalous activity by an application. This hunt can be driven be investigation of suspected applications or as a hunt across all users seen on the host.
```
# Get list of Applications
apps = all_syslog_data['ProcessName'].replace('', np.nan).dropna().unique().tolist()
system_apps = ['sudo', 'CRON', 'systemd-resolved', 'snapd',
'50-motd-news', 'systemd-logind', 'dbus-deamon', 'crontab']
if len(host_entity.Applications) > 0:
installed_apps = []
installed_apps.extend(x for x in apps if x not in system_apps)
# Pick Applications
app_select = nbwidgets.SelectItem(description='Select sudo session to investigate: ',
item_list=installed_apps, width='75%', auto_display=True)
else:
display(HTML("No applications other than stand OS applications present"))
# Get all syslog relating to these Applications
app = app_select.value
app_data = all_syslog_data[all_syslog_data['ProcessName'] == app].copy()
# App log volume over time
if isinstance(app_data, pd.DataFrame) and not app_data.empty:
app_data_volume = app_data.set_index(
"TimeGenerated").resample('5T').count()
app_data_volume.reset_index(level=0, inplace=True)
app_data_volume.rename(columns={"TenantId" : "NoOfLogMessages"}, inplace=True)
nbdisplay.display_timeline_values(data=app_data_volume, y='NoOfLogMessages', source_columns=['NoOfLogMessages'], title=f"{app} log volume over time")
app_high_sev = app_data[app_data['SeverityLevel'].isin(
['emerg', 'alert', 'crit', 'err', 'warning'])]
if isinstance(app_high_sev, pd.DataFrame) and not app_high_sev.empty:
app_hs_volume = app_high_sev.set_index(
"TimeGenerated").resample('5T').count()
app_hs_volume.reset_index(level=0, inplace=True)
app_hs_volume.rename(columns={"TenantId" : "NoOfLogMessages"}, inplace=True)
nbdisplay.display_timeline_values(data=app_hs_volume, y='NoOfLogMessages', source_columns=['NoOfLogMessages'], title=f"{app} high severity log volume over time")
risky_messages = risky_cmd_line(events=app_data, log_type="Syslog", cmd_field="SyslogMessage")
if risky_messages:
print(risky_messages)
```
### Display process tree
Due to the large volume of data involved you may wish to make you query window smaller
```
if rel_alert_select is None or rel_alert_select.selected_alert is None:
start = query_times.start
else:
start = rel_alert_select.selected_alert['TimeGenerated']
# Set new investigation time windows based on the selected alert
proc_invest_times = nbwidgets.QueryTime(units='hours',
max_before=6, max_after=3, before=2, origin_time=start)
proc_invest_times.display()
audit_table = None
app_audit_data = None
app = app_select.value
process_tree_data = None
regex = '.*audit.*\_cl?'
# Find the table with auditd data in and collect the data
matches = ((re.match(regex, key, re.IGNORECASE)) for key in qry_prov.schema)
for match in matches:
if match != None:
audit_table = match.group(0)
#Check if the amount of data expected to be returned is a reasonable size, if not prompt before continuing
if audit_table != None:
if isinstance(app_audit_data, pd.DataFrame):
pass
else:
print('Collecting audit data, please wait this may take some time....')
app_audit_query_count = f"""{audit_table}
| where TimeGenerated >= datetime({proc_invest_times.start})
| where TimeGenerated <= datetime({proc_invest_times.end})
| where Computer == '{hostname}'
| summarize count()
"""
count_check = qry_prov.exec_query(query=app_audit_query_count)
if count_check['count_'].iloc[0] > 100000 and not count_check.empty:
size = count_check['count_'].iloc[0]
print(
f"You are returning a very large dataset ({size} rows).",
"It is reccomended that you consider scoping the size\n",
"of your query down.\n",
"Are you sure you want to proceed?"
)
response = (input("Y/N") or "N")
if (
(count_check['count_'].iloc[0] < 100000)
or (count_check['count_'].iloc[0] > 100000
and response.casefold().startswith("y"))
):
print("querying audit data...")
audit_data = qry_prov.LinuxAudit.auditd_all(
start=proc_invest_times.start, end=proc_invest_times.end, host_name=hostname
)
if isinstance(audit_data, pd.DataFrame) and not audit_data.empty:
print("building process tree...")
audit_events = auditdextract.extract_events_to_df(
data=audit_data
)
process_tree_data = auditdextract.generate_process_tree(audit_data=audit_events)
plot_lim = 1000
if len(process_tree) > plot_lim:
md_warn(f"More than {plot_lim} processes to plot, limiting to top {plot_lim}.")
process_tree[:plot_lim].mp_process_tree.plot(legend_col="exe")
else:
process_tree.mp_process_tree.plot(legend_col="exe")
size = audit_events.size
print(f"Collected {size} rows of data")
else:
md("No audit events avalaible")
else:
print("Resize query window")
else:
md("No audit events avalaible")
md(f"<h3>Process tree for {app}</h3>")
if process_tree_data is not None:
process_tree_df = process_tree_data[process_tree_data["exe"].str.contains(app, na=False)].copy()
if not process_tree_df.empty:
app_roots = process_tree_data.apply(lambda x: ptree.get_root(process_tree_data, x), axis=1)
trees = []
for root in app_roots["source_index"].unique():
trees.append(process_tree_data[process_tree_data["path"].str.startswith(root)])
app_proc_trees = pd.concat(trees)
app_proc_trees.mp_process_tree.plot(legend_col="exe", show_table=True)
else:
display(f"No process tree data avaliable for {app}")
process_tree = None
else:
md("No data avaliable to build process tree")
```
### Application Logs with associated Threat Intelligence
These logs are associated with the process being investigated and include IOCs that appear in our TI feeds.
```
# Extract IOCs from syslog assocated with the selected process
ioc_extractor = iocextract.IoCExtract()
os_family = host_entity.OSType if host_entity.OSType else 'Linux'
md('Extracting IoCs...')
ioc_df = ioc_extractor.extract(data=app_data,
columns=['SyslogMessage'],
ioc_types=['ipv4', 'ipv6', 'dns', 'url',
'md5_hash', 'sha1_hash', 'sha256_hash'])
if process_tree_data is not None and not process_tree_data.empty:
app_process_tree = app_proc_trees.dropna(subset=['cmdline'])
audit_ioc_df = ioc_extractor.extract(data=app_process_tree,
columns=['cmdline'],
ioc_types=['ipv4', 'ipv6', 'dns', 'url',
'md5_hash', 'sha1_hash', 'sha256_hash'])
ioc_df = ioc_df.append(audit_ioc_df)
# Look up IOCs in TI feeds
if len(ioc_df) > 0:
ioc_count = len(ioc_df[["IoCType", "Observable"]].drop_duplicates())
md(f"Found {ioc_count} IOCs")
md("Looking up threat intel...")
ti_resps = tilookup.lookup_iocs(data=ioc_df[[
"IoCType", "Observable"]].drop_duplicates().reset_index(drop=True), obs_col='Observable')
i = 0
ti_hits = []
ti_resps.reset_index(drop=True, inplace=True)
while i < len(ti_resps):
if ti_resps['Result'][i] == True and ti_check_sev(ti_resps['Severity'][i], 1):
ti_hits.append(ti_resps['Ioc'][i])
i += 1
else:
i += 1
display(HTML(f"Found {len(ti_hits)} IoCs in Threat Intelligence"))
for ioc in ti_hits:
display(HTML(f"Messages containing IoC found in TI feed: {ioc}"))
display(app_data[app_data['SyslogMessage'].str.contains(
ioc)][['TimeGenerated', 'SyslogMessage']])
else:
md("<h3>No IoC patterns found in Syslog Message.</h3>")
```
Jump to:
- <a>Host Logon Events</a>
- <a>User Activity</a>
- <a>Application Activity</a>
## Network Activity
**Hypothesis:** That an attacker is remotely communicating with the host in order to compromise the host or for C2 or data exfiltration purposes after compromising the host.
This section provides an overview of network activity to and from the host during hunting time frame, the purpose of this is to allow for the identification of anomalous network traffic. If you wish to investigate a specific IP in detail it is recommended that you use the IP Explorer Notebook (include link).
```
# Get list of IPs from Syslog and Azure Network Data
ioc_extractor = iocextract.IoCExtract()
os_family = host_entity.OSType if host_entity.OSType else 'Linux'
print('Finding IP Addresses this may take a few minutes.......')
syslog_ips = ioc_extractor.extract(data=all_syslog_data,
columns=['SyslogMessage'],
ioc_types=['ipv4', 'ipv6'])
if 'AzureNetworkAnalytics_CL' not in qry_prov.schema:
az_net_comms_df = None
az_ips = None
else:
if hasattr(host_entity, 'private_ips') and hasattr(host_entity, 'public_ips'):
all_host_ips = host_entity.private_ips + \
host_entity.public_ips + [host_entity.IPAddress]
else:
all_host_ips = [host_entity.IPAddress]
host_ips = {'\'{}\''.format(i.Address) for i in all_host_ips}
host_ip_list = ','.join(host_ips)
az_ip_where = f"""| where (VMIPAddress in ("{host_ip_list}") or SrcIP in ("{host_ip_list}") or DestIP in ("{host_ip_list}")) and (AllowedOutFlows > 0 or AllowedInFlows > 0)"""
az_net_comms_df = qry_prov.AzureNetwork.az_net_analytics(
start=query_times.start, end=query_times.end, host_name=hostname, where_clause=az_ip_where)
if isinstance(az_net_comms_df, pd.DataFrame) and not az_net_comms_df.empty:
az_ips = az_net_comms_df.query("PublicIPs != @host_entity.IPAddress")
else:
az_ips = None
if len(syslog_ips):
IPs = syslog_ips[['IoCType', 'Observable']].drop_duplicates('Observable')
display(f"Found {len(IPs)} IP Addresses assoicated with the host")
else:
md("### No IoC patterns found in Syslog Message.")
if az_ips is not None:
ips = az_ips['PublicIps'].drop_duplicates(
) + syslog_ips['Observable'].drop_duplicates()
else:
ips = syslog_ips['Observable'].drop_duplicates()
if isinstance(az_net_comms_df, pd.DataFrame) and not az_net_comms_df.empty:
import warnings
with warnings.catch_warnings():
warnings.simplefilter("ignore")
az_net_comms_df['TotalAllowedFlows'] = az_net_comms_df['AllowedOutFlows'] + \
az_net_comms_df['AllowedInFlows']
sns.catplot(x="L7Protocol", y="TotalAllowedFlows",
col="FlowDirection", data=az_net_comms_df)
sns.relplot(x="FlowStartTime", y="TotalAllowedFlows",
col="FlowDirection", kind="line",
hue="L7Protocol", data=az_net_comms_df).set_xticklabels(rotation=50)
nbdisplay.display_timeline(data=az_net_comms_df.query('AllowedOutFlows > 0'),
overlay_data=az_net_comms_df.query(
'AllowedInFlows > 0'),
title='Network Flows (out=blue, in=green)',
time_column='FlowStartTime',
source_columns=[
'FlowType', 'AllExtIPs', 'L7Protocol', 'FlowDirection'],
height=300)
else:
md('<h3>No Azure network data for specified time range.</h3>')
```
### Choose ASNs/IPs to Check for Threat Intel Reports
Choose from the list of Selected ASNs for the IPs you wish to check on. Then select the IP(s) that you wish to check against Threat Intelligence data.
The Source list is populated with all ASNs found in the syslog and network flow data.
```
#Lookup each IP in whois data and extract the ASN
@lru_cache(maxsize=1024)
def whois_desc(ip_lookup, progress=False):
try:
ip = ip_address(ip_lookup)
except ValueError:
return "Not an IP Address"
if ip.is_private:
return "private address"
if not ip.is_global:
return "other address"
whois = IPWhois(ip)
whois_result = whois.lookup_whois()
if progress:
print(".", end="")
return whois_result["asn_description"]
# Summarise network data by ASN
ASN_List = []
print("WhoIs Lookups")
ASNs = ips.apply(lambda x: whois_desc(x, True))
IP_ASN = pd.DataFrame(dict(IPs=ips, ASN=ASNs)).reset_index()
x = IP_ASN.groupby(["ASN"]).count().drop(
'index', axis=1).sort_values('IPs', ascending=False)
display(x)
ASN_List = x.index
# Select an ASN to investigate in more detail
selection = widgets.SelectMultiple(
options=ASN_List,
width=900,
description='Select ASN to investigate',
disabled=False
)
display(selection)
# For every IP associated with the selected ASN look them up in TI feeds
ip_invest_list = None
ip_selection = None
for ASN in selection.value:
if ip_invest_list is None:
ip_invest_list = (IP_ASN[IP_ASN["ASN"] == ASN]['IPs'].tolist())
else:
ip_invest_list + (IP_ASN[IP_ASN["ASN"] == ASN]['IPs'].tolist())
if ip_invest_list is not None:
ioc_ip_list = []
if len(ip_invest_list) > 0:
ti_resps = tilookup.lookup_iocs(data=ip_invest_list, providers=["OTX"])
i = 0
ti_hits = []
while i < len(ti_resps):
if ti_resps['Details'][i]['pulse_count'] > 0:
ti_hits.append(ti_resps['Ioc'][i])
i += 1
else:
i += 1
display(HTML(f"Found {len(ti_hits)} IoCs in Threat Intelligence"))
for ioc in ti_hits:
ioc_ip_list.append(ioc)
#Show IPs found in TI feeds for further investigation
if len(ioc_ip_list) > 0:
display(HTML("Select an IP whcih appeared in TI to investigate further"))
ip_selection = nbwidgets.SelectItem(description='Select IP Address to investigate: ', item_list = ioc_ip_list, width='95%', auto_display=True)
else:
md("No IPs to investigate")
# Get all syslog for the IPs
if ip_selection is not None:
display(HTML("Syslog data associated with this IP Address"))
sys_hits = all_syslog_data[all_syslog_data['SyslogMessage'].str.contains(
ip_selection.value)]
display(sys_hits)
os_family = host_entity.OSType if host_entity.OSType else 'Linux'
display(HTML("TI result for this IP Address"))
display(ti_resps[ti_resps['Ioc'] == ip_selection.value])
else:
md("No IP address selected")
```
## Configuration
### `msticpyconfig.yaml` configuration File
You can configure primary and secondary TI providers and any required parameters in the `msticpyconfig.yaml` file. This is read from the current directory or you can set an environment variable (`MSTICPYCONFIG`) pointing to its location.
To configure this file see the [ConfigureNotebookEnvironment notebook](https://github.com/Azure/Azure-Sentinel-Notebooks/blob/master/ConfiguringNotebookEnvironment.ipynb)
|
github_jupyter
|
from pathlib import Path
from IPython.display import display, HTML
REQ_PYTHON_VER=(3, 6)
REQ_MSTICPY_VER=(1, 0, 0)
REQ_MP_EXTRAS = ["ml"]
update_nbcheck = (
"<p style='color: orange; text-align=left'>"
"<b>Warning: we needed to update '<i>utils/nb_check.py</i>'</b><br>"
"Please restart the kernel and re-run this cell."
"</p>"
)
display(HTML("<h3>Starting Notebook setup...</h3>"))
if Path("./utils/nb_check.py").is_file():
try:
from utils.nb_check import check_versions
except ImportError as err:
%xmode Minimal
!curl https://raw.githubusercontent.com/Azure/Azure-Sentinel-Notebooks/master/utils/nb_check.py > ./utils/nb_check.py 2>/dev/null
display(HTML(update_nbcheck))
if "check_versions" not in globals():
raise ImportError("Old version of nb_check.py detected - see instructions below.")
%xmode Verbose
check_versions(REQ_PYTHON_VER, REQ_MSTICPY_VER, REQ_MP_EXTRAS)
# If the installation fails try to manually install using
# !pip install --upgrade msticpy
from msticpy.nbtools import nbinit
additional_packages = [
"oauthlib", "pyvis", "python-whois", "seaborn"
]
nbinit.init_notebook(
namespace=globals(),
additional_packages=additional_packages,
extra_imports=extra_imports,
);
from bokeh.models import ColumnDataSource, FactorRange
from bokeh.palettes import viridis
from bokeh.plotting import show, Row, figure
from bokeh.transform import factor_cmap, cumsum
from dns import reversename, resolver
from functools import lru_cache
from ipaddress import ip_address
from ipwhois import IPWhois
from math import pi
from msticpy.common.exceptions import MsticpyException
from msticpy.nbtools import observationlist
from msticpy.nbtools.foliummap import get_map_center
from msticpy.sectools import auditdextract
from msticpy.sectools.cmd_line import risky_cmd_line
from msticpy.sectools.ip_utils import convert_to_ip_entities
from msticpy.sectools.syslog_utils import create_host_record, cluster_syslog_logons_df, risky_sudo_sessions
from pyvis.network import Network
import datetime as dt
import re
%kql loganalytics://tenant(aad_tenant).workspace(WORKSPACE_ID).clientid(client_id).clientsecret(client_secret)
%kql loganalytics://code().workspace(WORKSPACE_ID)
# See if we have a Microsoft Sentinel Workspace defined in our config file.
# If not, let the user specify Workspace and Tenant IDs
ws_config = WorkspaceConfig()
if not ws_config.config_loaded:
ws_config.prompt_for_ws()
qry_prov = QueryProvider(data_environment="AzureSentinel")
print("done")
# Authenticate to Microsoft Sentinel workspace
qry_prov.connect(ws_config)
query_times = nbwidgets.QueryTime(units='day',
max_before=14, max_after=1, before=1)
query_times.display()
#Get a list of hosts with syslog data in our hunting timegframe to provide easy selection
syslog_query = f"""Syslog | where TimeGenerated between (datetime({query_times.start}) .. datetime({query_times.end})) | summarize by Computer"""
md("Collecting avaliable host details...")
hosts_list = qry_prov._query_provider.query(query=syslog_query)
if isinstance(hosts_list, pd.DataFrame) and not hosts_list.empty:
hosts = hosts_list["Computer"].unique().tolist()
host_text = nbwidgets.SelectItem(description='Select host to investigate: ',
item_list=hosts, width='75%', auto_display=True)
else:
display(md("There are no hosts with syslog data in this time period to investigate"))
hostname=host_text.value
az_net_df = None
# Collect data on the host
all_syslog_query = f"Syslog | where TimeGenerated between (datetime({query_times.start}) .. datetime({query_times.end})) | where Computer =~ '{hostname}'"""
all_syslog_data = qry_prov.exec_query(all_syslog_query)
if isinstance(all_syslog_data, pd.DataFrame) and not all_syslog_data.empty:
heartbeat_query = f"""Heartbeat | where TimeGenerated >= datetime({query_times.start}) | where TimeGenerated <= datetime({query_times.end})| where Computer == '{hostname}' | top 1 by TimeGenerated desc nulls last"""
if "AzureNetworkAnalytics_CL" in qry_prov.schema:
aznet_query = f"""AzureNetworkAnalytics_CL | where TimeGenerated >= datetime({query_times.start}) | where TimeGenerated <= datetime({query_times.end}) | where VirtualMachine_s has '{hostname}' | where ResourceType == 'NetworkInterface' | top 1 by TimeGenerated desc | project PrivateIPAddresses = PrivateIPAddresses_s, PublicIPAddresses = PublicIPAddresses_s"""
print("Getting network data...")
az_net_df = qry_prov.exec_query(query=aznet_query)
print("Getting host data...")
host_hb = qry_prov.exec_query(query=heartbeat_query)
# Create host entity record, with Azure network data if any is avaliable
if az_net_df is not None and isinstance(az_net_df, pd.DataFrame) and not az_net_df.empty:
host_entity = create_host_record(syslog_df=all_syslog_data, heartbeat_df=host_hb, az_net_df=az_net_df)
else:
host_entity = create_host_record(syslog_df=all_syslog_data, heartbeat_df=host_hb)
md(
"<b>Host Details</b><br>"
f"<b>Hostname</b>: {host_entity.computer}<br>"
f"<b>OS</b>: {host_entity.OSType} {host_entity.OSName}<br>"
f"<b>IP Address</b>: {host_entity.IPAddress.Address}<br>"
f"<b>Location</b>: {host_entity.IPAddress.Location.CountryName}<br>"
f"<b>Installed Applications</b>: {host_entity.Applications}<br>"
)
else:
md_warn("No Syslog data found, check hostname and timeframe.")
md("The data query may be timing out, consider reducing the timeframe size.")
related_alerts = qry_prov.SecurityAlert.list_related_alerts(
query_times, host_name=hostname)
realted_bookmarks = qry_prov.AzureSentinel.list_bookmarks_for_entity(query_times, entity_id=hostname)
if isinstance(related_alerts, pd.DataFrame) and not related_alerts.empty:
host_alert_items = (related_alerts[['AlertName', 'TimeGenerated']]
.groupby('AlertName').TimeGenerated.agg('count').to_dict())
def print_related_alerts(alertDict, entityType, entityName):
if len(alertDict) > 0:
md(f"Found {len(alertDict)} different alert types related to this {entityType} (\'{entityName}\')")
for (k, v) in alertDict.items():
md(f"- {k}, Count of alerts: {v}")
else:
md(f"No alerts for {entityType} entity \'{entityName}\'")
print_related_alerts(host_alert_items, 'host', host_entity.HostName)
nbdisplay.display_timeline(
data=related_alerts, source_columns=["AlertName"], title="Host alerts over time", height=300, color="red")
else:
md('No related alerts found.')
if isinstance(realted_bookmarks, pd.DataFrame) and not realted_bookmarks.empty:
nbdisplay.display_timeline(data=realted_bookmarks, source_columns=["BookmarkName"], height=200, color="orange", title="Host bookmarks over time",)
else:
md('No related bookmarks found.')
rel_alert_select = None
def show_full_alert(selected_alert):
global security_alert, alert_ip_entities
security_alert = SecurityAlert(
rel_alert_select.selected_alert)
nbdisplay.display_alert(security_alert, show_entities=True)
# Show selected alert when selected
if isinstance(related_alerts, pd.DataFrame) and not related_alerts.empty:
related_alerts['CompromisedEntity'] = related_alerts['Computer']
md('### Click on alert to view details.')
rel_alert_select = nbwidgets.SelectAlert(alerts=related_alerts,
action=show_full_alert)
rel_alert_select.display()
else:
md('No related alerts found.')
if rel_alert_select is None or rel_alert_select.selected_alert is None:
start = query_times.start
else:
start = rel_alert_select.selected_alert['TimeGenerated']
# Set new investigation time windows based on the selected alert
invest_times = nbwidgets.QueryTime(
units='day', max_before=24, max_after=12, before=1, after=1, origin_time=start)
invest_times.display()
tilookup = TILookup()
md("Threat intelligence provider loading complete.")
# Collect logon events for this, seperate them into sucessful and unsucessful and cluster sucessful one into sessions
logon_events = qry_prov.LinuxSyslog.user_logon(start=invest_times.start, end=invest_times.end, host_name=hostname)
remote_logons = None
failed_logons = None
if isinstance(logon_events, pd.DataFrame) and not logon_events.empty:
remote_logons = (logon_events[logon_events['LogonResult'] == 'Success'])
failed_logons = (logon_events[logon_events['LogonResult'] == 'Failure'])
else:
print("No logon events in this timeframe")
if (isinstance(remote_logons, pd.DataFrame) and not remote_logons.empty) or (isinstance(failed_logons, pd.DataFrame) and not failed_logons.empty):
#Provide a timeline of sucessful and failed logon attempts to aid identification of potential brute force attacks
display(Markdown('### Timeline of sucessful host logons.'))
tooltip_cols = ['User', 'ProcessName', 'SourceIP']
if rel_alert_select is not None:
logon_timeline = nbdisplay.display_timeline(data=remote_logons, overlay_data=failed_logons, source_columns=tooltip_cols, height=200, overlay_color="red", alert = rel_alert_select.selected_alert)
else:
logon_timeline = nbdisplay.display_timeline(data=remote_logons, overlay_data=failed_logons, source_columns=tooltip_cols, height=200, overlay_color="red")
display(Markdown('<b>Key:</b><p style="color:darkblue">Sucessful logons </p><p style="color:Red">Failed Logon Attempts (via su)</p>'))
all_df = pd.DataFrame(dict(successful= remote_logons['ProcessName'].value_counts(), failed = failed_logons['ProcessName'].value_counts())).fillna(0)
fail_data = pd.value_counts(failed_logons['User'].values, sort=True).head(10).reset_index(name='value').rename(columns={'User':'Count'})
fail_data['angle'] = fail_data['value']/fail_data['value'].sum() * 2*pi
fail_data['color'] = viridis(len(fail_data))
fp = figure(plot_height=350, plot_width=450, title="Relative Frequencies of Failed Logons by Account", toolbar_location=None, tools="hover", tooltips="@index: @value")
fp.wedge(x=0, y=1, radius=0.5, start_angle=cumsum('angle', include_zero=True), end_angle=cumsum('angle'), line_color="white", fill_color='color', legend='index', source=fail_data)
sucess_data = pd.value_counts(remote_logons['User'].values, sort=False).reset_index(name='value').rename(columns={'User':'Count'})
sucess_data['angle'] = sucess_data['value']/sucess_data['value'].sum() * 2*pi
sucess_data['color'] = viridis(len(sucess_data))
sp = figure(plot_height=350, width=450, title="Relative Frequencies of Sucessful Logons by Account", toolbar_location=None, tools="hover", tooltips="@index: @value")
sp.wedge(x=0, y=1, radius=0.5, start_angle=cumsum('angle', include_zero=True), end_angle=cumsum('angle'), line_color="white", fill_color='color', legend='index', source=sucess_data)
fp.axis.axis_label=None
fp.axis.visible=False
fp.grid.grid_line_color = None
sp.axis.axis_label=None
sp.axis.visible=False
sp.grid.grid_line_color = None
processes = all_df.index.values.tolist()
results = all_df.columns.values.tolist()
fail_sucess_data = {'processes' :processes,
'sucess' : all_df['successful'].values.tolist(),
'failure': all_df['failed'].values.tolist()}
palette = viridis(2)
x = [ (process, result) for process in processes for result in results ]
counts = sum(zip(fail_sucess_data['sucess'], fail_sucess_data['failure']), ())
source = ColumnDataSource(data=dict(x=x, counts=counts))
b = figure(x_range=FactorRange(*x), plot_height=350, plot_width=450, title="Failed and Sucessful logon attempts by process",
toolbar_location=None, tools="", y_minor_ticks=2)
b.vbar(x='x', top='counts', width=0.9, source=source, line_color="white",
fill_color=factor_cmap('x', palette=palette, factors=results, start=1, end=2))
b.y_range.start = 0
b.x_range.range_padding = 0.1
b.xaxis.major_label_orientation = 1
b.xgrid.grid_line_color = None
show(Row(sp,fp,b))
ip_list = [convert_to_ip_entities(i, ip_col="SourceIP")[0] for i in remote_logons['SourceIP'].unique() if i != ""]
ip_fail_list = [convert_to_ip_entities(i)[0] for i in failed_logons['SourceIP'].unique() if i != ""]
location = get_map_center(ip_list + ip_fail_list)
folium_map = FoliumMap(location = location, zoom_start=1.4)
#Map logon locations to allow for identification of anomolous locations
if len(ip_fail_list) > 0:
md('<h3>Map of Originating Location of Logon Attempts</h3>')
icon_props = {'color': 'red'}
folium_map.add_ip_cluster(ip_entities=ip_fail_list, **icon_props)
if len(ip_list) > 0:
icon_props = {'color': 'green'}
folium_map.add_ip_cluster(ip_entities=ip_list, **icon_props)
display(folium_map.folium_map)
md('<p style="color:red">Warning: the folium mapping library '
'does not display correctly in some browsers.</p><br>'
'If you see a blank image please retry with a different browser.')
logon_sessions_df = None
try:
print("Clustering logon sessions...")
logon_sessions_df = cluster_syslog_logons_df(logon_events)
except Exception as err:
print(f"Error clustering logons: {err}")
if logon_sessions_df is not None:
logon_sessions_df["Alerts during session?"] = np.nan
# check if any alerts occur during logon window.
logon_sessions_df['Start (UTC)'] = [(time - dt.timedelta(seconds=5)) for time in logon_sessions_df['Start']]
logon_sessions_df['End (UTC)'] = [(time + dt.timedelta(seconds=5)) for time in logon_sessions_df['End']]
for TimeGenerated in related_alerts['TimeGenerated']:
logon_sessions_df.loc[(TimeGenerated >= logon_sessions_df['Start (UTC)']) & (TimeGenerated <= logon_sessions_df['End (UTC)']), "Alerts during session?"] = "Yes"
logon_sessions_df.loc[logon_sessions_df['User'] == 'root', "Root?"] = "Yes"
logon_sessions_df.replace(np.nan, "No", inplace=True)
ratios = []
for _, row in logon_sessions_df.iterrows():
suc_fail = logon_events.apply(lambda x: True if x['User'] == row['User'] and x["LogonResult"] == 'Success' else(
False if x['User'] == row['User'] and x["LogonResult"] == 'Failure' else None), axis=1)
numofsucess = len(suc_fail[suc_fail == True].index)
numoffail = len(suc_fail[suc_fail == False].index)
if numoffail == 0:
ratio = 1
else:
ratio = numofsucess/numoffail
ratios.append(ratio)
logon_sessions_df["Sucessful to failed logon ratio"] = ratios
def color_cells(val):
if isinstance(val, str):
color = 'yellow' if val == "Yes" else 'white'
elif isinstance(val, float):
color = 'yellow' if val > 0.5 else 'white'
else:
color = 'white'
return 'background-color: %s' % color
display(logon_sessions_df[['User','Start (UTC)', 'End (UTC)', 'Alerts during session?', 'Sucessful to failed logon ratio', 'Root?']]
.style.applymap(color_cells).hide_index())
logon_items = (
logon_sessions_df[['User','Start (UTC)', 'End (UTC)']]
.to_string(header=False, index=False, index_names=False)
.split('\n')
)
logon_sessions_df["Key"] = logon_items
logon_sessions_df.set_index('Key', inplace=True)
logon_dict = logon_sessions_df[['User','Start (UTC)', 'End (UTC)']].to_dict('index')
logon_selection = nbwidgets.SelectItem(description='Select logon session to investigate: ',
item_dict=logon_dict , width='80%', auto_display=True)
else:
md("No logon sessions during this timeframe")
def view_syslog(selected_facility):
return [syslog_events.query('Facility == @selected_facility')]
# Produce a summary of user modification actions taken
if "Add" in x:
return len(add_events.replace("", np.nan).dropna(subset=['User'])['User'].unique().tolist())
elif "Modify" in x:
return len(mod_events.replace("", np.nan).dropna(subset=['User'])['User'].unique().tolist())
elif "Delete" in x:
return len(del_events.replace("", np.nan).dropna(subset=['User'])['User'].unique().tolist())
else:
return ""
crn_tl_data = {}
user_tl_data = {}
sudo_tl_data = {}
sudo_sessions = None
tooltip_cols = ['SyslogMessage']
if logon_sessions_df is not None:
#Collect data based on the session selected for investigation
invest_sess = {'StartTimeUtc': logon_selection.value.get('Start (UTC)'), 'EndTimeUtc': logon_selection.value.get(
'End (UTC)'), 'Account': logon_selection.value.get('User'), 'Host': hostname}
session = entities.HostLogonSession(invest_sess)
syslog_events = qry_prov.LinuxSyslog.all_syslog(
start=session.StartTimeUtc, end=session.EndTimeUtc, host_name=session.Host)
sudo_events = qry_prov.LinuxSyslog.sudo_activity(
start=session.StartTimeUtc, end=session.EndTimeUtc, host_name=session.Host, user=session.Account)
if isinstance(sudo_events, pd.DataFrame) and not sudo_events.empty:
try:
sudo_sessions = cluster_syslog_logons_df(logon_events=sudo_events)
except MsticpyException:
pass
# Display summary of cron activity in session
cron_events = qry_prov.LinuxSyslog.cron_activity(
start=session.StartTimeUtc, end=session.EndTimeUtc, host_name=session.Host)
if not isinstance(cron_events, pd.DataFrame) or cron_events.empty:
md(f'<h3> No Cron activity for {session.Host} between {session.StartTimeUtc} and {session.EndTimeUtc}</h3>')
else:
cron_events['CMD'].replace('', np.nan, inplace=True)
crn_tl_data = {"Cron Exections": {"data": cron_events[['TimeGenerated', 'CMD', 'CronUser', 'SyslogMessage']].dropna(), "source_columns": tooltip_cols, "color": "Blue"},
"Cron Edits": {"data": cron_events.loc[cron_events['SyslogMessage'].str.contains('EDIT')], "source_columns": tooltip_cols, "color": "Green"}}
md('<h2> Most common commands run by cron:</h2>')
md('This shows how often each cron job was exected within the specified time window')
cron_commands = (cron_events[['EventTime', 'CMD']]
.groupby(['CMD']).count()
.dropna()
.style
.set_table_attributes('width=900px, text-align=center')
.background_gradient(cmap='Reds', low=0.5, high=1)
.format("{0:0>1.0f}"))
display(cron_commands)
# Display summary of user and group creations, deletions and modifications during the session
user_activity = qry_prov.LinuxSyslog.user_group_activity(
start=session.StartTimeUtc, end=session.EndTimeUtc, host_name=session.Host)
if not isinstance(user_activity, pd.DataFrame) or user_activity.empty:
md(f'<h3>No user or group moidifcations for {session.Host} between {session.StartTimeUtc} and {session.EndTimeUtc}></h3>')
else:
add_events = user_activity[user_activity['UserGroupAction'].str.contains(
'Add')]
del_events = user_activity[user_activity['UserGroupAction'].str.contains(
'Delete')]
mod_events = user_activity[user_activity['UserGroupAction'].str.contains(
'Modify')]
user_activity['Count'] = user_activity.groupby('UserGroupAction')['UserGroupAction'].transform('count')
if add_events.empty and del_events.empty and mod_events.empty:
md('<h2> Users and groups added or deleted:</h2<>')
md(f'No users or groups were added or deleted on {host_entity.HostName} between {query_times.start} and {query_times.end}')
user_tl_data = {}
else:
md("<h2>Users added, modified or deleted</h2>")
display(user_activity[['UserGroupAction','Count']].drop_duplicates().style.hide_index())
account_actions = pd.DataFrame({"User Additions": [add_events.replace("", np.nan).dropna(subset=['User'])['User'].unique().tolist()],
"User Modifications": [mod_events.replace("", np.nan).dropna(subset=['User'])['User'].unique().tolist()],
"User Deletions": [del_events.replace("", np.nan).dropna(subset=['User'])['User'].unique().tolist()]})
display(account_actions.style.hide_index())
user_tl_data = {"User adds": {"data": add_events, "source_columns": tooltip_cols, "color": "Orange"},
"User deletes": {"data": del_events, "source_columns": tooltip_cols, "color": "Red"},
"User modfications": {"data": mod_events, "source_columns": tooltip_cols, "color": "Grey"}}
# Display sudo activity during session
if not isinstance(sudo_sessions, pd.DataFrame) or sudo_sessions.empty:
md(f"<h3>No Sudo sessions for {session.Host} between {logon_selection.value.get('Start (UTC)')} and {logon_selection.value.get('End (UTC)')}</h3>")
sudo_tl_data = {}
else:
sudo_start = sudo_events[sudo_events["SyslogMessage"].str.contains(
"pam_unix.+session opened")].rename(columns={"Sudoer": "User"})
sudo_tl_data = {"Host logons": {"data": remote_logons, "source_columns": tooltip_cols, "color": "Cyan"},
"Sudo sessions": {"data": sudo_start, "source_columns": tooltip_cols, "color": "Purple"}}
try:
risky_actions = cmd_line.risky_cmd_line(events=sudo_events, log_type="Syslog")
suspicious_events = cmd_speed(
cmd_events=sudo_events, time=60, events=2, cmd_field="Command")
except:
risky_actions = None
suspicious_events = None
if risky_actions is None and suspicious_events is None:
pass
else:
risky_sessions = risky_sudo_sessions(
risky_actions=risky_actions, sudo_sessions=sudo_sessions, suspicious_actions=suspicious_events)
for key in risky_sessions:
if key in sudo_sessions:
sudo_sessions[f"{key} - {risky_sessions[key]}"] = sudo_sessions.pop(
key)
if isinstance(sudo_events, pd.DataFrame):
sudo_events_val = sudo_events[['EventTime', 'CommandCall']][sudo_events['CommandCall']!=""].dropna(how='any', subset=['CommandCall'])
if sudo_events_val.empty:
md(f"No sucessful sudo activity for {hostname} between {logon_selection.value.get('Start (UTC)')} and {logon_selection.value.get('End (UTC)')}")
else:
sudo_events.replace("", np.nan, inplace=True)
md('<h2> Frequency of sudo commands</h2>')
md('This shows how many times each command has been run with sudo. /bin/bash is usally associated with the use of "sudo -i"')
sudo_commands = (sudo_events[['EventTime', 'CommandCall']]
.groupby(['CommandCall'])
.count()
.dropna()
.style
.set_table_attributes('width=900px, text-align=center')
.background_gradient(cmap='Reds', low=.5, high=1)
.format("{0:0>3.0f}"))
display(sudo_commands)
else:
md(f"No sucessful sudo activity for {hostname} between {logon_selection.value.get('Start (UTC)')} and {logon_selection.value.get('End (UTC)')}")
# Display a timeline of all activity during session
crn_tl_data.update(user_tl_data)
crn_tl_data.update(sudo_tl_data)
if crn_tl_data:
md('<h2> Session Timeline.</h2>')
nbdisplay.display_timeline(
data=crn_tl_data, title='Session Timeline', height=300)
else:
md("No logon sessions during this timeframe")
if isinstance(logon_sessions_df, pd.DataFrame) and not logon_sessions_df.empty:
#Return syslog data and present it to the use for investigation
session_syslog = qry_prov.LinuxSyslog.all_syslog(
start=session.StartTimeUtc, end=session.EndTimeUtc, host_name=session.Host)
if session_syslog.empty:
display(HTML(
f' No syslog for {session.Host} between {session.StartTimeUtc} and {session.EndTimeUtc}'))
def view_sudo(selected_cmd):
return [sudo_events.query('CommandCall == @selected_cmd')[
['TimeGenerated', 'SyslogMessage', 'Sudoer', 'SudoTo', 'Command', 'CommandCall']]]
# Show syslog messages associated with selected sudo command
items = sudo_events['CommandCall'].dropna().unique().tolist()
if items:
md("<h3>View all messages associated with a sudo command</h3>")
display(nbwidgets.SelectItem(item_list=items, action=view_sudo))
else:
md("No logon sessions during this timeframe")
if isinstance(logon_sessions_df, pd.DataFrame) and not logon_sessions_df.empty:
# Display syslog messages from the session witht he facility selected
items = syslog_events['Facility'].dropna().unique().tolist()
md("<h3>View all messages associated with a syslog facility</h3>")
display(nbwidgets.SelectItem(item_list=items, action=view_syslog))
else:
md("No logon sessions during this timeframe")
if isinstance(logon_sessions_df, pd.DataFrame) and not logon_sessions_df.empty:
display(HTML("<h3>Process Trees from session</h3>"))
print("Building process tree, this may take some time...")
# Find the table with auditd data in
regex = '.*audit.*\_cl?'
matches = ((re.match(regex, key, re.IGNORECASE)) for key in qry_prov.schema)
for match in matches:
if match != None:
audit_table = match.group(0)
else:
audit_table = None
# Retrieve auditd data
if audit_table:
audit_data = qry_prov.LinuxAudit.auditd_all(
start=session.StartTimeUtc, end=session.EndTimeUtc, host_name=hostname
)
if isinstance(audit_data, pd.DataFrame) and not audit_data.empty:
audit_events = auditdextract.extract_events_to_df(
data=audit_data
)
process_tree = auditdextract.generate_process_tree(audit_data=audit_events)
process_tree.mp_process_tree.plot()
else:
display(HTML("No auditd data avaliable to build process tree"))
else:
display(HTML("No auditd data avaliable to build process tree"))
else:
md("No logon sessions during this timeframe")
if logon_sessions_df is not None and sudo_sessions is not None:
sudo_items = sudo_sessions[['User','Start', 'End']].to_string(header=False,
index=False,
index_names=False).split('\n')
sudo_sessions["Key"] = sudo_items
sudo_sessions.set_index('Key', inplace=True)
sudo_dict = sudo_sessions[['User','Start', 'End']].to_dict('index')
sudo_selection = nbwidgets.SelectItem(description='Select sudo session to investigate: ',
item_dict=sudo_dict, width='100%', height='300px', auto_display=True)
else:
sudo_selection = None
md("No logon sessions during this timeframe")
#Collect data associated with the sudo session selected
sudo_events = None
from msticpy.sectools.tiproviders.ti_provider_base import TISeverity
def ti_check_sev(severity, threshold):
severity = TISeverity.parse(severity)
threshold = TISeverity.parse(threshold)
return severity.value >= threshold.value
if sudo_selection:
sudo_sess = {'StartTimeUtc': sudo_selection.value.get('Start'), 'EndTimeUtc': sudo_selection.value.get(
'End'), 'Account': sudo_selection.value.get('User'), 'Host': hostname}
sudo_session = entities.HostLogonSession(sudo_sess)
sudo_events = qry_prov.LinuxSyslog.sudo_activity(start=sudo_session.StartTimeUtc.round(
'-1s') - pd.Timedelta(seconds=1), end=(sudo_session.EndTimeUtc.round('1s')+ pd.Timedelta(seconds=1)), host_name=sudo_session.Host)
if isinstance(sudo_events, pd.DataFrame) and not sudo_events.empty:
display(sudo_events.replace('', np.nan).dropna(axis=0, subset=['Command'])[
['TimeGenerated', 'Command', 'CommandCall', 'SyslogMessage']])
# Extract IOCs from the data
ioc_extractor = iocextract.IoCExtract()
os_family = host_entity.OSType if host_entity.OSType else 'Linux'
print('Extracting IoCs.......')
ioc_df = ioc_extractor.extract(data=sudo_events,
columns=['SyslogMessage'],
os_family=os_family,
ioc_types=['ipv4', 'ipv6', 'dns', 'url',
'md5_hash', 'sha1_hash', 'sha256_hash'])
if len(ioc_df) > 0:
ioc_count = len(
ioc_df[["IoCType", "Observable"]].drop_duplicates())
md(f"Found {ioc_count} IOCs")
#Lookup the extracted IOCs in TI feed
ti_resps = tilookup.lookup_iocs(data=ioc_df[["IoCType", "Observable"]].drop_duplicates(
).reset_index(), obs_col='Observable', ioc_type_col='IoCType')
i = 0
ti_hits = []
ti_resps.reset_index(drop=True, inplace=True)
while i < len(ti_resps):
if ti_resps['Result'][i] == True and ti_check_sev(ti_resps['Severity'][i], 1):
ti_hits.append(ti_resps['Ioc'][i])
i += 1
else:
i += 1
md(f"Found {len(ti_hits)} IoCs in Threat Intelligence")
for ioc in ti_hits:
md(f"Messages containing IoC found in TI feed: {ioc}")
display(sudo_events[sudo_events['SyslogMessage'].str.contains(
ioc)][['TimeGenerated', 'SyslogMessage']])
else:
md("No IoC patterns found in Syslog Messages.")
else:
md('No sudo messages for this session')
else:
md("No Sudo session to investigate")
# Get list of users with logon or sudo sessions on host
logon_events = qry_prov.LinuxSyslog.user_logon(query_times, host_name=hostname)
users = logon_events['User'].replace('', np.nan).dropna().unique().tolist()
all_users = list(users)
if isinstance(sudo_events, pd.DataFrame) and not sudo_events.empty:
sudoers = sudo_events['Sudoer'].replace(
'', np.nan).dropna().unique().tolist()
all_users.extend(x for x in sudoers if x not in all_users)
# Pick Users
if not logon_events.empty:
user_select = nbwidgets.SelectItem(description='Select user to investigate: ',
item_list=all_users, width='75%', auto_display=True)
else:
md("There was no user activity in the timeframe specified.")
user_select = None
folium_user_map = FoliumMap()
def view_sudo(cmd):
return [user_sudo_hold.query('CommandCall == @cmd')[
['TimeGenerated', 'HostName', 'Command', 'CommandCall', 'SyslogMessage']]]
user_sudo_hold = None
if user_select is not None:
# Get all syslog relating to these users
username = user_select.value
user_events = all_syslog_data[all_syslog_data['SyslogMessage'].str.contains(username)]
logon_sessions = cluster_syslog_logons_df(logon_events)
# Display all logons associated with the user
md(f"<h1> User Logon Activity for {username}</h1>")
user_logon_events = logon_events[logon_events['User'] == username]
try:
user_logon_sessions = cluster_syslog_logons_df(user_logon_events)
except:
user_logon_sessions = None
user_remote_logons = (
user_logon_events[user_logon_events['LogonResult'] == 'Success']
)
user_failed_logons = (
user_logon_events[user_logon_events['LogonResult'] == 'Failure']
)
if not user_remote_logons.empty:
for _, row in logon_sessions_df.iterrows():
end = row['End']
user_sudo_events = qry_prov.LinuxSyslog.sudo_activity(start=user_remote_logons.sort_values(
by='TimeGenerated')['TimeGenerated'].iloc[0], end=end, host_name=hostname, user=username)
else:
user_sudo_events = None
if user_logon_sessions is None and user_remote_logons.empty and user_failed_logons.empty:
pass
else:
display(HTML(
f"{len(user_remote_logons)} sucessfull logons and {len(user_failed_logons)} failed logons for {username}"))
display(Markdown('### Timeline of host logon attempts.'))
tooltip_cols = ['SyslogMessage']
dfs = {"User Logons" :user_remote_logons, "Failed Logons": user_failed_logons, "Sudo Events" :user_sudo_events}
user_tl_data = {}
for k,v in dfs.items():
if v is not None and not v.empty:
user_tl_data.update({k :{"data":v,"source_columns":tooltip_cols}})
nbdisplay.display_timeline(
data=user_tl_data, title="User logon timeline", height=300)
all_user_df = pd.DataFrame(dict(successful= user_remote_logons['ProcessName'].value_counts(), failed = user_failed_logons['ProcessName'].value_counts())).fillna(0)
processes = all_user_df.index.values.tolist()
results = all_user_df.columns.values.tolist()
user_fail_sucess_data = {'processes' :processes,
'sucess' : all_user_df['successful'].values.tolist(),
'failure': all_user_df['failed'].values.tolist()}
palette = viridis(2)
x = [ (process, result) for process in processes for result in results ]
counts = sum(zip(user_fail_sucess_data['sucess'], fail_sucess_data['failure']), ())
source = ColumnDataSource(data=dict(x=x, counts=counts))
b = figure(x_range=FactorRange(*x), plot_height=350, plot_width=450, title="Failed and Sucessful logon attempts by process",
toolbar_location=None, tools="", y_minor_ticks=2)
b.vbar(x='x', top='counts', width=0.9, source=source, line_color="white",
fill_color=factor_cmap('x', palette=palette, factors=results, start=1, end=2))
b.y_range.start = 0
b.x_range.range_padding = 0.1
b.xaxis.major_label_orientation = 1
b.xgrid.grid_line_color = None
user_logons = pd.DataFrame({"Sucessful Logons" : [int(all_user_df['successful'].sum())],
"Failed Logons" : [int(all_user_df['failed'].sum())]}).T
user_logon_data = pd.value_counts(user_logon_events['LogonResult'].values, sort=True).head(10).reset_index(name='value').rename(columns={'User':'Count'})
user_logon_data = user_logon_data[user_logon_data['index']!="Unknown"].copy()
user_logon_data['angle'] = user_logon_data['value']/user_logon_data['value'].sum() * 2*pi
user_logon_data['color'] = viridis(len(user_logon_data))
p = figure(plot_height=350, plot_width=450, title="Relative Frequencies of Failed Logons by Account", toolbar_location=None, tools="hover", tooltips="@index: @value")
p.axis.visible = False
p.xgrid.visible = False
p.ygrid.visible = False
p.wedge(x=0, y=1, radius=0.5, start_angle=cumsum('angle', include_zero=True), end_angle=cumsum('angle'), line_color="white", fill_color='color', legend='index', source=user_logon_data)
show(Row(p,b))
user_ip_list = [convert_to_ip_entities(i)[0] for i in user_remote_logons['SourceIP']]
user_ip_fail_list = [convert_to_ip_entities(i)[0] for i in user_failed_logons['SourceIP']]
user_location = get_map_center(ip_list + ip_fail_list)
user_folium_map = FoliumMap(location = location, zoom_start=1.4)
#Map logon locations to allow for identification of anomolous locations
if len(ip_fail_list) > 0:
md('<h3>Map of Originating Location of Logon Attempts</h3>')
icon_props = {'color': 'red'}
user_folium_map.add_ip_cluster(ip_entities=user_ip_fail_list, **icon_props)
if len(ip_list) > 0:
icon_props = {'color': 'green'}
user_folium_map.add_ip_cluster(ip_entities=user_ip_list, **icon_props)
display(user_folium_map.folium_map)
md('<p style="color:red">Warning: the folium mapping library '
'does not display correctly in some browsers.</p><br>'
'If you see a blank image please retry with a different browser.')
#Display sudo activity of the user
if not isinstance(user_sudo_events, pd.DataFrame) or user_sudo_events.empty:
md(f"<h3>No sucessful sudo activity for {username}</h3>")
else:
user_sudo_hold = user_sudo_events
user_sudo_commands = (user_sudo_events[['EventTime', 'CommandCall']].replace('', np.nan).groupby(['CommandCall']).count().dropna().style.set_table_attributes('width=900px, text-align=center').background_gradient(cmap='Reds', low=.5, high=1).format("{0:0>3.0f}"))
display(user_sudo_commands)
md("Select a sudo command to investigate in more detail")
display(nbwidgets.SelectItem(item_list=items, action=view_sudo))
else:
md("No user session selected")
# If the user has sudo activity extract and IOCs from the logs and look them up in TI feeds
if not isinstance(user_sudo_hold, pd.DataFrame) or user_sudo_hold.empty:
md(f"No sudo messages data")
else:
# Extract IOCs
ioc_extractor = iocextract.IoCExtract()
os_family = host_entity.OSType if host_entity.OSType else 'Linux'
print('Extracting IoCs.......')
ioc_df = ioc_extractor.extract(data=user_sudo_hold,
columns=['SyslogMessage'],
ioc_types=['ipv4', 'ipv6', 'dns', 'url', 'md5_hash', 'sha1_hash', 'sha256_hash'])
if len(ioc_df) > 0:
ioc_count = len(ioc_df[["IoCType", "Observable"]].drop_duplicates())
md(f"Found {ioc_count} IOCs")
ti_resps = tilookup.lookup_iocs(data=ioc_df[["IoCType", "Observable"]].drop_duplicates(
).reset_index(), obs_col='Observable', ioc_type_col='IoCType')
i = 0
ti_hits = []
ti_resps.reset_index(drop=True, inplace=True)
while i < len(ti_resps):
if ti_resps['Result'][i] == True and ti_check_sev(ti_resps['Severity'][i], 1):
ti_hits.append(ti_resps['Ioc'][i])
i += 1
else:
i += 1
md(f"Found {len(ti_hits)} IoCs in Threat Intelligence")
for ioc in ti_hits:
md(f"Messages containing IoC found in TI feed: {ioc}")
display(user_sudo_hold[user_sudo_hold['SyslogMessage'].str.contains(
ioc)][['TimeGenerated', 'SyslogMessage']])
else:
md("No IoC patterns found in Syslog Message.")
# Get list of Applications
apps = all_syslog_data['ProcessName'].replace('', np.nan).dropna().unique().tolist()
system_apps = ['sudo', 'CRON', 'systemd-resolved', 'snapd',
'50-motd-news', 'systemd-logind', 'dbus-deamon', 'crontab']
if len(host_entity.Applications) > 0:
installed_apps = []
installed_apps.extend(x for x in apps if x not in system_apps)
# Pick Applications
app_select = nbwidgets.SelectItem(description='Select sudo session to investigate: ',
item_list=installed_apps, width='75%', auto_display=True)
else:
display(HTML("No applications other than stand OS applications present"))
# Get all syslog relating to these Applications
app = app_select.value
app_data = all_syslog_data[all_syslog_data['ProcessName'] == app].copy()
# App log volume over time
if isinstance(app_data, pd.DataFrame) and not app_data.empty:
app_data_volume = app_data.set_index(
"TimeGenerated").resample('5T').count()
app_data_volume.reset_index(level=0, inplace=True)
app_data_volume.rename(columns={"TenantId" : "NoOfLogMessages"}, inplace=True)
nbdisplay.display_timeline_values(data=app_data_volume, y='NoOfLogMessages', source_columns=['NoOfLogMessages'], title=f"{app} log volume over time")
app_high_sev = app_data[app_data['SeverityLevel'].isin(
['emerg', 'alert', 'crit', 'err', 'warning'])]
if isinstance(app_high_sev, pd.DataFrame) and not app_high_sev.empty:
app_hs_volume = app_high_sev.set_index(
"TimeGenerated").resample('5T').count()
app_hs_volume.reset_index(level=0, inplace=True)
app_hs_volume.rename(columns={"TenantId" : "NoOfLogMessages"}, inplace=True)
nbdisplay.display_timeline_values(data=app_hs_volume, y='NoOfLogMessages', source_columns=['NoOfLogMessages'], title=f"{app} high severity log volume over time")
risky_messages = risky_cmd_line(events=app_data, log_type="Syslog", cmd_field="SyslogMessage")
if risky_messages:
print(risky_messages)
if rel_alert_select is None or rel_alert_select.selected_alert is None:
start = query_times.start
else:
start = rel_alert_select.selected_alert['TimeGenerated']
# Set new investigation time windows based on the selected alert
proc_invest_times = nbwidgets.QueryTime(units='hours',
max_before=6, max_after=3, before=2, origin_time=start)
proc_invest_times.display()
audit_table = None
app_audit_data = None
app = app_select.value
process_tree_data = None
regex = '.*audit.*\_cl?'
# Find the table with auditd data in and collect the data
matches = ((re.match(regex, key, re.IGNORECASE)) for key in qry_prov.schema)
for match in matches:
if match != None:
audit_table = match.group(0)
#Check if the amount of data expected to be returned is a reasonable size, if not prompt before continuing
if audit_table != None:
if isinstance(app_audit_data, pd.DataFrame):
pass
else:
print('Collecting audit data, please wait this may take some time....')
app_audit_query_count = f"""{audit_table}
| where TimeGenerated >= datetime({proc_invest_times.start})
| where TimeGenerated <= datetime({proc_invest_times.end})
| where Computer == '{hostname}'
| summarize count()
"""
count_check = qry_prov.exec_query(query=app_audit_query_count)
if count_check['count_'].iloc[0] > 100000 and not count_check.empty:
size = count_check['count_'].iloc[0]
print(
f"You are returning a very large dataset ({size} rows).",
"It is reccomended that you consider scoping the size\n",
"of your query down.\n",
"Are you sure you want to proceed?"
)
response = (input("Y/N") or "N")
if (
(count_check['count_'].iloc[0] < 100000)
or (count_check['count_'].iloc[0] > 100000
and response.casefold().startswith("y"))
):
print("querying audit data...")
audit_data = qry_prov.LinuxAudit.auditd_all(
start=proc_invest_times.start, end=proc_invest_times.end, host_name=hostname
)
if isinstance(audit_data, pd.DataFrame) and not audit_data.empty:
print("building process tree...")
audit_events = auditdextract.extract_events_to_df(
data=audit_data
)
process_tree_data = auditdextract.generate_process_tree(audit_data=audit_events)
plot_lim = 1000
if len(process_tree) > plot_lim:
md_warn(f"More than {plot_lim} processes to plot, limiting to top {plot_lim}.")
process_tree[:plot_lim].mp_process_tree.plot(legend_col="exe")
else:
process_tree.mp_process_tree.plot(legend_col="exe")
size = audit_events.size
print(f"Collected {size} rows of data")
else:
md("No audit events avalaible")
else:
print("Resize query window")
else:
md("No audit events avalaible")
md(f"<h3>Process tree for {app}</h3>")
if process_tree_data is not None:
process_tree_df = process_tree_data[process_tree_data["exe"].str.contains(app, na=False)].copy()
if not process_tree_df.empty:
app_roots = process_tree_data.apply(lambda x: ptree.get_root(process_tree_data, x), axis=1)
trees = []
for root in app_roots["source_index"].unique():
trees.append(process_tree_data[process_tree_data["path"].str.startswith(root)])
app_proc_trees = pd.concat(trees)
app_proc_trees.mp_process_tree.plot(legend_col="exe", show_table=True)
else:
display(f"No process tree data avaliable for {app}")
process_tree = None
else:
md("No data avaliable to build process tree")
# Extract IOCs from syslog assocated with the selected process
ioc_extractor = iocextract.IoCExtract()
os_family = host_entity.OSType if host_entity.OSType else 'Linux'
md('Extracting IoCs...')
ioc_df = ioc_extractor.extract(data=app_data,
columns=['SyslogMessage'],
ioc_types=['ipv4', 'ipv6', 'dns', 'url',
'md5_hash', 'sha1_hash', 'sha256_hash'])
if process_tree_data is not None and not process_tree_data.empty:
app_process_tree = app_proc_trees.dropna(subset=['cmdline'])
audit_ioc_df = ioc_extractor.extract(data=app_process_tree,
columns=['cmdline'],
ioc_types=['ipv4', 'ipv6', 'dns', 'url',
'md5_hash', 'sha1_hash', 'sha256_hash'])
ioc_df = ioc_df.append(audit_ioc_df)
# Look up IOCs in TI feeds
if len(ioc_df) > 0:
ioc_count = len(ioc_df[["IoCType", "Observable"]].drop_duplicates())
md(f"Found {ioc_count} IOCs")
md("Looking up threat intel...")
ti_resps = tilookup.lookup_iocs(data=ioc_df[[
"IoCType", "Observable"]].drop_duplicates().reset_index(drop=True), obs_col='Observable')
i = 0
ti_hits = []
ti_resps.reset_index(drop=True, inplace=True)
while i < len(ti_resps):
if ti_resps['Result'][i] == True and ti_check_sev(ti_resps['Severity'][i], 1):
ti_hits.append(ti_resps['Ioc'][i])
i += 1
else:
i += 1
display(HTML(f"Found {len(ti_hits)} IoCs in Threat Intelligence"))
for ioc in ti_hits:
display(HTML(f"Messages containing IoC found in TI feed: {ioc}"))
display(app_data[app_data['SyslogMessage'].str.contains(
ioc)][['TimeGenerated', 'SyslogMessage']])
else:
md("<h3>No IoC patterns found in Syslog Message.</h3>")
# Get list of IPs from Syslog and Azure Network Data
ioc_extractor = iocextract.IoCExtract()
os_family = host_entity.OSType if host_entity.OSType else 'Linux'
print('Finding IP Addresses this may take a few minutes.......')
syslog_ips = ioc_extractor.extract(data=all_syslog_data,
columns=['SyslogMessage'],
ioc_types=['ipv4', 'ipv6'])
if 'AzureNetworkAnalytics_CL' not in qry_prov.schema:
az_net_comms_df = None
az_ips = None
else:
if hasattr(host_entity, 'private_ips') and hasattr(host_entity, 'public_ips'):
all_host_ips = host_entity.private_ips + \
host_entity.public_ips + [host_entity.IPAddress]
else:
all_host_ips = [host_entity.IPAddress]
host_ips = {'\'{}\''.format(i.Address) for i in all_host_ips}
host_ip_list = ','.join(host_ips)
az_ip_where = f"""| where (VMIPAddress in ("{host_ip_list}") or SrcIP in ("{host_ip_list}") or DestIP in ("{host_ip_list}")) and (AllowedOutFlows > 0 or AllowedInFlows > 0)"""
az_net_comms_df = qry_prov.AzureNetwork.az_net_analytics(
start=query_times.start, end=query_times.end, host_name=hostname, where_clause=az_ip_where)
if isinstance(az_net_comms_df, pd.DataFrame) and not az_net_comms_df.empty:
az_ips = az_net_comms_df.query("PublicIPs != @host_entity.IPAddress")
else:
az_ips = None
if len(syslog_ips):
IPs = syslog_ips[['IoCType', 'Observable']].drop_duplicates('Observable')
display(f"Found {len(IPs)} IP Addresses assoicated with the host")
else:
md("### No IoC patterns found in Syslog Message.")
if az_ips is not None:
ips = az_ips['PublicIps'].drop_duplicates(
) + syslog_ips['Observable'].drop_duplicates()
else:
ips = syslog_ips['Observable'].drop_duplicates()
if isinstance(az_net_comms_df, pd.DataFrame) and not az_net_comms_df.empty:
import warnings
with warnings.catch_warnings():
warnings.simplefilter("ignore")
az_net_comms_df['TotalAllowedFlows'] = az_net_comms_df['AllowedOutFlows'] + \
az_net_comms_df['AllowedInFlows']
sns.catplot(x="L7Protocol", y="TotalAllowedFlows",
col="FlowDirection", data=az_net_comms_df)
sns.relplot(x="FlowStartTime", y="TotalAllowedFlows",
col="FlowDirection", kind="line",
hue="L7Protocol", data=az_net_comms_df).set_xticklabels(rotation=50)
nbdisplay.display_timeline(data=az_net_comms_df.query('AllowedOutFlows > 0'),
overlay_data=az_net_comms_df.query(
'AllowedInFlows > 0'),
title='Network Flows (out=blue, in=green)',
time_column='FlowStartTime',
source_columns=[
'FlowType', 'AllExtIPs', 'L7Protocol', 'FlowDirection'],
height=300)
else:
md('<h3>No Azure network data for specified time range.</h3>')
#Lookup each IP in whois data and extract the ASN
@lru_cache(maxsize=1024)
def whois_desc(ip_lookup, progress=False):
try:
ip = ip_address(ip_lookup)
except ValueError:
return "Not an IP Address"
if ip.is_private:
return "private address"
if not ip.is_global:
return "other address"
whois = IPWhois(ip)
whois_result = whois.lookup_whois()
if progress:
print(".", end="")
return whois_result["asn_description"]
# Summarise network data by ASN
ASN_List = []
print("WhoIs Lookups")
ASNs = ips.apply(lambda x: whois_desc(x, True))
IP_ASN = pd.DataFrame(dict(IPs=ips, ASN=ASNs)).reset_index()
x = IP_ASN.groupby(["ASN"]).count().drop(
'index', axis=1).sort_values('IPs', ascending=False)
display(x)
ASN_List = x.index
# Select an ASN to investigate in more detail
selection = widgets.SelectMultiple(
options=ASN_List,
width=900,
description='Select ASN to investigate',
disabled=False
)
display(selection)
# For every IP associated with the selected ASN look them up in TI feeds
ip_invest_list = None
ip_selection = None
for ASN in selection.value:
if ip_invest_list is None:
ip_invest_list = (IP_ASN[IP_ASN["ASN"] == ASN]['IPs'].tolist())
else:
ip_invest_list + (IP_ASN[IP_ASN["ASN"] == ASN]['IPs'].tolist())
if ip_invest_list is not None:
ioc_ip_list = []
if len(ip_invest_list) > 0:
ti_resps = tilookup.lookup_iocs(data=ip_invest_list, providers=["OTX"])
i = 0
ti_hits = []
while i < len(ti_resps):
if ti_resps['Details'][i]['pulse_count'] > 0:
ti_hits.append(ti_resps['Ioc'][i])
i += 1
else:
i += 1
display(HTML(f"Found {len(ti_hits)} IoCs in Threat Intelligence"))
for ioc in ti_hits:
ioc_ip_list.append(ioc)
#Show IPs found in TI feeds for further investigation
if len(ioc_ip_list) > 0:
display(HTML("Select an IP whcih appeared in TI to investigate further"))
ip_selection = nbwidgets.SelectItem(description='Select IP Address to investigate: ', item_list = ioc_ip_list, width='95%', auto_display=True)
else:
md("No IPs to investigate")
# Get all syslog for the IPs
if ip_selection is not None:
display(HTML("Syslog data associated with this IP Address"))
sys_hits = all_syslog_data[all_syslog_data['SyslogMessage'].str.contains(
ip_selection.value)]
display(sys_hits)
os_family = host_entity.OSType if host_entity.OSType else 'Linux'
display(HTML("TI result for this IP Address"))
display(ti_resps[ti_resps['Ioc'] == ip_selection.value])
else:
md("No IP address selected")
| 0.475362 | 0.603494 |
# Demo Multiple Cutters - Multiple Barches
This notebook shows a basics of a feeder concept. This scenario has two vessels:
* a *vessel* for the transport of material
* an *installer*, receiving the material and installing it.
Thus, we also need two processes - one for each vessel. In this process the hand over of the material is done from the *vessel* to the *installer*.
```
import datetime, time
import simpy
import shapely.geometry
# package(s) for data handling
import pandas as pd
import numpy as np
import openclsim.core as core
import openclsim.model as model
import uuid
# setup environment
simulation_start = 0
my_env = simpy.Environment(initial_time=simulation_start)
registry = {}
```
## Definition of Sites
```
# The generic site class
Site = type(
"Site",
(
core.Identifiable, # Give it a name
core.Log, # Allow logging of all discrete events
core.Locatable, # Add coordinates to extract distance information and visualize
core.HasContainer, # Add information on the material available at the site
core.HasResource,
), # Add information on serving equipment
{},
) # The dictionary is empty because the site type is generic
# Information on the extraction site - the "from site" - the "win locatie"
location_from_site = shapely.geometry.Point(4.18055556, 52.18664444) # lon, lat
data_from_site = {
"env": my_env, # The simpy environment defined in the first cel
"name": "Winlocatie1", # The name of the site
"geometry": location_from_site, # The coordinates of the project site
"capacity": 24,
"level": 24,
"nr_resources":3,
} # The actual volume of the site
from_site = Site(**data_from_site)
location_to_site = shapely.geometry.Point(4.25222222, 52.11428333) # lon, lat
data_to_site = {
"env": my_env, # The simpy environment defined in the first cel
"name": "Dumplocatie", # The name of the site
"geometry": location_to_site, # The coordinates of the project site
"capacity": 24,
"level": 0,
} # The actual volume of the site (empty of course)
to_site = Site(**data_to_site)
```
## Definition of Vessels
```
# The generic class for an object that can move and transport (a TSHD for example)
TransportProcessingResource = type(
"TransportProcessingResource",
(
core.Identifiable, # Give it a name
core.Log, # Allow logging of all discrete events
core.ContainerDependentMovable, # A moving container, so capacity and location
core.Processor, # Allow for loading and unloading
core.HasResource, # Add information on serving equipment
core.LoadingFunction, # Add a loading function
core.UnloadingFunction, # Add an unloading function
# SiteRegistry,
),
{"key": "MultiStoreHopper"},
)
# print(SiteRegistry.inspect("MultiStoreHopper"))
# For more realistic simulation you might want to have speed dependent on the volume carried by the vessel
def compute_v_provider(v_empty, v_full):
return lambda x: 10
data_cutter1 = {
"env": my_env, # The simpy environment
"name": "Cutter1", # Name
"geometry": location_from_site, # It starts at the "from site"
"loading_rate": 1, # Loading rate
"unloading_rate": 1, # Unloading rate
"capacity": 1,
"level": 0,
"compute_v": compute_v_provider(5, 4.5), # Variable speed
}
cutter1 = TransportProcessingResource(**data_cutter1)
data_cutter2 = {
"env": my_env, # The simpy environment
"name": "Cutter2", # Name
"geometry": location_from_site, # It starts at the "from site"
"loading_rate": 1, # Loading rate
"unloading_rate": 1, # Unloading rate
"capacity": 1,
"level": 0,
"compute_v": compute_v_provider(5, 4.5), # Variable speed
}
cutter2 = TransportProcessingResource(**data_cutter2)
# TSHD variables
data_hopper1 = {
"env": my_env, # The simpy environment
"name": "Hopper1", # Name
"geometry": location_from_site, # It starts at the "from site"
"loading_rate": 1, # Loading rate
"unloading_rate": 1, # Unloading rate
"capacity": 1,
"level": 0,
"compute_v": compute_v_provider(5, 4.5), # Variable speed
}
hopper1 = TransportProcessingResource(**data_hopper1)
data_hopper2 = {
"env": my_env, # The simpy environment
"name": "Hopper2", # Name
"geometry": location_from_site, # It starts at the "from site"
"loading_rate": 1, # Loading rate
"unloading_rate": 1, # Unloading rate
"capacity": 1,
"level": 0,
"compute_v": compute_v_provider(5, 4.5), # Variable speed
}
hopper2 = TransportProcessingResource(**data_hopper2)
data_hopper3 = {
"env": my_env, # The simpy environment
"name": "Hopper3", # Name
"geometry": location_from_site, # It starts at the "from site"
"loading_rate": 1, # Loading rate
"unloading_rate": 1, # Unloading rate
"capacity": 1,
"level": 0,
"compute_v": compute_v_provider(5, 4.5), # Variable speed
}
hopper3 = TransportProcessingResource(**data_hopper3)
```
## Definition of Barge process
```
cutter_list = [cutter1, cutter2]
for hopper in [hopper1, hopper2, hopper3]:
first_cutter = cutter_list[0]
cutter_list= cutter_list[1:]
cutter_list.append(first_cutter)
for cutter in cutter_list:
requested_resources = {}
run = []
shift_amount_loading_data = {
"env": my_env, # The simpy environment defined in the first cel
"name": "Load", # We are moving soil
"registry": registry,
"processor": cutter,
"origin": from_site,
"destination": hopper,
"amount": 1,
"duration": 10,
"requested_resources":requested_resources,
"keep_resources":[hopper],
"postpone_start": True,
}
run.append(model.ShiftAmountActivity(**shift_amount_loading_data))
move_activity_to_harbor_data = {
"env": my_env, # The simpy environment defined in the first cel
"name": "sailing full", # We are moving soil
"registry": registry,
"mover": hopper,
"destination": to_site,
"requested_resources":requested_resources,
"keep_resources":[hopper],
"postpone_start": True,
}
run.append(model.MoveActivity(**move_activity_to_harbor_data))
shift_amount_loading_data = {
"env": my_env, # The simpy environment defined in the first cel
"name": "Unload", # We are moving soil
"registry": registry,
"processor": hopper,
"origin": hopper,
"destination": to_site,
"amount": 1,
"duration": 10,
"requested_resources":requested_resources,
"keep_resources":[hopper],
"postpone_start": True,
}
run.append(model.ShiftAmountActivity(**shift_amount_loading_data))
move_activity_to_harbor_data = {
"env": my_env, # The simpy environment defined in the first cel
"name": "sailing empty", # We are moving soil
"registry": registry,
"mover": hopper,
"destination": from_site,
"requested_resources":requested_resources,
"postpone_start": True,
}
run.append(model.MoveActivity(**move_activity_to_harbor_data))
sequential_activity_data = {
"env": my_env,
"name": "run",
"registry": registry,
"sub_processes": run,
"postpone_start": True,
}
sequential_activity = model.SequentialActivity(**sequential_activity_data)
while_data = {
"env": my_env, # The simpy environment defined in the first cel
"name": "run while", # We are moving soil
"registry": registry,
"sub_processes": [sequential_activity],
"condition_event": [{"type":"container", "concept": from_site, "state":"empty"}],
"postpone_start": False,
}
run_activity = model.WhileActivity(**while_data)
my_env.run()
```
## Cutter1 log
```
log_df = pd.DataFrame(cutter1.log)
data = log_df[["Message", "ActivityState", "Timestamp", "Value", "ActivityID"]]
data = data.drop_duplicates()
data
```
## Cutter2 log
```
log2_df = pd.DataFrame(cutter2.log)
data2 = log2_df[["Message", "ActivityState", "Timestamp", "Value", "ActivityID"]]
data2 = data2.drop_duplicates()
data2
```
## Hopper1 log
```
hopper1_log_df = pd.DataFrame(hopper1.log)
data_hopper1 = hopper1_log_df[["Message", "ActivityState", "Timestamp", "Value", "ActivityID"]]
data_hopper1 = data_hopper1.drop_duplicates()
data_hopper1
```
## Hopper2 log
```
hopper2_log_df = pd.DataFrame(hopper2.log)
data_hopper2 = hopper2_log_df[["Message", "ActivityState", "Timestamp", "Value", "ActivityID"]]
data_hopper2 = data_hopper2.drop_duplicates()
data_hopper2
```
## Hopper3 log
```
hopper3_log_df = pd.DataFrame(hopper3.log)
data_hopper3 = hopper3_log_df[["Message", "ActivityState", "Timestamp", "Value", "ActivityID"]]
data_hopper3 = data_hopper3.drop_duplicates()
data_hopper3
```
|
github_jupyter
|
import datetime, time
import simpy
import shapely.geometry
# package(s) for data handling
import pandas as pd
import numpy as np
import openclsim.core as core
import openclsim.model as model
import uuid
# setup environment
simulation_start = 0
my_env = simpy.Environment(initial_time=simulation_start)
registry = {}
# The generic site class
Site = type(
"Site",
(
core.Identifiable, # Give it a name
core.Log, # Allow logging of all discrete events
core.Locatable, # Add coordinates to extract distance information and visualize
core.HasContainer, # Add information on the material available at the site
core.HasResource,
), # Add information on serving equipment
{},
) # The dictionary is empty because the site type is generic
# Information on the extraction site - the "from site" - the "win locatie"
location_from_site = shapely.geometry.Point(4.18055556, 52.18664444) # lon, lat
data_from_site = {
"env": my_env, # The simpy environment defined in the first cel
"name": "Winlocatie1", # The name of the site
"geometry": location_from_site, # The coordinates of the project site
"capacity": 24,
"level": 24,
"nr_resources":3,
} # The actual volume of the site
from_site = Site(**data_from_site)
location_to_site = shapely.geometry.Point(4.25222222, 52.11428333) # lon, lat
data_to_site = {
"env": my_env, # The simpy environment defined in the first cel
"name": "Dumplocatie", # The name of the site
"geometry": location_to_site, # The coordinates of the project site
"capacity": 24,
"level": 0,
} # The actual volume of the site (empty of course)
to_site = Site(**data_to_site)
# The generic class for an object that can move and transport (a TSHD for example)
TransportProcessingResource = type(
"TransportProcessingResource",
(
core.Identifiable, # Give it a name
core.Log, # Allow logging of all discrete events
core.ContainerDependentMovable, # A moving container, so capacity and location
core.Processor, # Allow for loading and unloading
core.HasResource, # Add information on serving equipment
core.LoadingFunction, # Add a loading function
core.UnloadingFunction, # Add an unloading function
# SiteRegistry,
),
{"key": "MultiStoreHopper"},
)
# print(SiteRegistry.inspect("MultiStoreHopper"))
# For more realistic simulation you might want to have speed dependent on the volume carried by the vessel
def compute_v_provider(v_empty, v_full):
return lambda x: 10
data_cutter1 = {
"env": my_env, # The simpy environment
"name": "Cutter1", # Name
"geometry": location_from_site, # It starts at the "from site"
"loading_rate": 1, # Loading rate
"unloading_rate": 1, # Unloading rate
"capacity": 1,
"level": 0,
"compute_v": compute_v_provider(5, 4.5), # Variable speed
}
cutter1 = TransportProcessingResource(**data_cutter1)
data_cutter2 = {
"env": my_env, # The simpy environment
"name": "Cutter2", # Name
"geometry": location_from_site, # It starts at the "from site"
"loading_rate": 1, # Loading rate
"unloading_rate": 1, # Unloading rate
"capacity": 1,
"level": 0,
"compute_v": compute_v_provider(5, 4.5), # Variable speed
}
cutter2 = TransportProcessingResource(**data_cutter2)
# TSHD variables
data_hopper1 = {
"env": my_env, # The simpy environment
"name": "Hopper1", # Name
"geometry": location_from_site, # It starts at the "from site"
"loading_rate": 1, # Loading rate
"unloading_rate": 1, # Unloading rate
"capacity": 1,
"level": 0,
"compute_v": compute_v_provider(5, 4.5), # Variable speed
}
hopper1 = TransportProcessingResource(**data_hopper1)
data_hopper2 = {
"env": my_env, # The simpy environment
"name": "Hopper2", # Name
"geometry": location_from_site, # It starts at the "from site"
"loading_rate": 1, # Loading rate
"unloading_rate": 1, # Unloading rate
"capacity": 1,
"level": 0,
"compute_v": compute_v_provider(5, 4.5), # Variable speed
}
hopper2 = TransportProcessingResource(**data_hopper2)
data_hopper3 = {
"env": my_env, # The simpy environment
"name": "Hopper3", # Name
"geometry": location_from_site, # It starts at the "from site"
"loading_rate": 1, # Loading rate
"unloading_rate": 1, # Unloading rate
"capacity": 1,
"level": 0,
"compute_v": compute_v_provider(5, 4.5), # Variable speed
}
hopper3 = TransportProcessingResource(**data_hopper3)
cutter_list = [cutter1, cutter2]
for hopper in [hopper1, hopper2, hopper3]:
first_cutter = cutter_list[0]
cutter_list= cutter_list[1:]
cutter_list.append(first_cutter)
for cutter in cutter_list:
requested_resources = {}
run = []
shift_amount_loading_data = {
"env": my_env, # The simpy environment defined in the first cel
"name": "Load", # We are moving soil
"registry": registry,
"processor": cutter,
"origin": from_site,
"destination": hopper,
"amount": 1,
"duration": 10,
"requested_resources":requested_resources,
"keep_resources":[hopper],
"postpone_start": True,
}
run.append(model.ShiftAmountActivity(**shift_amount_loading_data))
move_activity_to_harbor_data = {
"env": my_env, # The simpy environment defined in the first cel
"name": "sailing full", # We are moving soil
"registry": registry,
"mover": hopper,
"destination": to_site,
"requested_resources":requested_resources,
"keep_resources":[hopper],
"postpone_start": True,
}
run.append(model.MoveActivity(**move_activity_to_harbor_data))
shift_amount_loading_data = {
"env": my_env, # The simpy environment defined in the first cel
"name": "Unload", # We are moving soil
"registry": registry,
"processor": hopper,
"origin": hopper,
"destination": to_site,
"amount": 1,
"duration": 10,
"requested_resources":requested_resources,
"keep_resources":[hopper],
"postpone_start": True,
}
run.append(model.ShiftAmountActivity(**shift_amount_loading_data))
move_activity_to_harbor_data = {
"env": my_env, # The simpy environment defined in the first cel
"name": "sailing empty", # We are moving soil
"registry": registry,
"mover": hopper,
"destination": from_site,
"requested_resources":requested_resources,
"postpone_start": True,
}
run.append(model.MoveActivity(**move_activity_to_harbor_data))
sequential_activity_data = {
"env": my_env,
"name": "run",
"registry": registry,
"sub_processes": run,
"postpone_start": True,
}
sequential_activity = model.SequentialActivity(**sequential_activity_data)
while_data = {
"env": my_env, # The simpy environment defined in the first cel
"name": "run while", # We are moving soil
"registry": registry,
"sub_processes": [sequential_activity],
"condition_event": [{"type":"container", "concept": from_site, "state":"empty"}],
"postpone_start": False,
}
run_activity = model.WhileActivity(**while_data)
my_env.run()
log_df = pd.DataFrame(cutter1.log)
data = log_df[["Message", "ActivityState", "Timestamp", "Value", "ActivityID"]]
data = data.drop_duplicates()
data
log2_df = pd.DataFrame(cutter2.log)
data2 = log2_df[["Message", "ActivityState", "Timestamp", "Value", "ActivityID"]]
data2 = data2.drop_duplicates()
data2
hopper1_log_df = pd.DataFrame(hopper1.log)
data_hopper1 = hopper1_log_df[["Message", "ActivityState", "Timestamp", "Value", "ActivityID"]]
data_hopper1 = data_hopper1.drop_duplicates()
data_hopper1
hopper2_log_df = pd.DataFrame(hopper2.log)
data_hopper2 = hopper2_log_df[["Message", "ActivityState", "Timestamp", "Value", "ActivityID"]]
data_hopper2 = data_hopper2.drop_duplicates()
data_hopper2
hopper3_log_df = pd.DataFrame(hopper3.log)
data_hopper3 = hopper3_log_df[["Message", "ActivityState", "Timestamp", "Value", "ActivityID"]]
data_hopper3 = data_hopper3.drop_duplicates()
data_hopper3
| 0.342572 | 0.843186 |
```
import sys
import glob
import re
import fnmatch
import math
import os
from os import listdir
from os.path import join, isfile, basename
import itertools
import numpy as np
from numpy import float32, int32, uint8, dtype, genfromtxt
import scipy
from scipy.stats import ttest_ind
import pandas as pd
import matplotlib
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
import colorsys
labels = [16,64,8,32,2,4,65,66,33,67,34,17,69,70,35,71,9,18,72,36,73,74,37,75,19,76,38,77,39,78,79,20,5,40,80,10,81,82,83,84,85,86,11,22,23,24,12,3,6,49,50,25,51,13,52,26,53,27,54,55,56,28,7,14,57,58,29,59,30,60,15,61,31,62,63]
label_names_file = '/groups/saalfeld/home/bogovicj/vfb/DrosAdultBRAINdomains/refData/Original_Index.tsv'
label_names = pd.read_csv( label_names_file, delimiter='\t', header=0 )
# print label_names[ label_names['Stack id'] == 11 ]['JFRCtempate2010.mask130819' ].iloc[0]
# print label_names[ label_names['Stack id'] == 70 ]['JFRCtempate2010.mask130819' ].iloc[0]
def get_label_name( label_id ):
return label_names[ label_names['Stack id'] == label_id ]['JFRCtempate2010.mask130819' ].iloc[0]
templates = ['JFRCtemplate2010', 'JFRC2013_lo', 'F-antsFlip_lo', 'F-cmtkFlip_lof', 'TeforBrain_f']
reg_methods = [ 'cmtkCow', 'cmtkCOG', 'cmtkHideo', 'antsRegOwl', 'antsRegDog', 'antsRegYang' ]
line=3
dist_samples_f = '/nrs/saalfeld/john/projects/flyChemStainAtlas/all_evals/label_data_line{}.csv.gz'.format( line )
dist_samples_df = pd.read_csv( dist_samples_f, header=None, names=['TEMPLATE','ALG','LINE','LABEL','DISTANCE'] )
# dist_samples_df.head()
# print( dist_samples_df['ALG'].unique())
# print( dist_samples_df['TEMPLATE'].unique())
def pad_zero(field='DISTANCE', pad=0.01):
def pfunc(xin):
x=xin[field]
if x > 0.:
return x
elif x == 0.:
return pad
else:
return float('nan')
return pfunc
dist_samples_df['DISTANCEPAD'] = dist_samples_df.apply( pad_zero(field='DISTANCE'), axis=1)
# some_dat = dist_samples_df[ (dist_samples_df.TEMPLATE =='F-antsFlip_lo') & (dist_samples_df.ALG == 'antsRegDog') & (dist_samples_df.LABEL == 84) ]
# print( some_dat.size )
# gam_params_fl = scipy.stats.gamma.fit( some_dat.DISTANCEPAD, floc=0. )
# print( gam_params_fl )
# print( len(some_dat[some_dat.DISTANCEPAD <= 0.]))
# print( len(l_dists) )
# l_dists[l_dists <= 0.]
# some_dat.DISTANCEPAD
ray_offset = []
ray_scale = []
ray_offset_fl = []
ray_scale_fl = []
gam_a = []
gam_offset = []
gam_scale = []
gam_a_fl = []
gam_offset_fl = []
gam_scale_fl = []
tlist = []
alist = []
llist = []
for t in templates:
t_dists = dist_samples_df[ dist_samples_df.TEMPLATE == t ]
print( 't ', t )
for a in reg_methods:
print( 'a ', a )
a_dists = t_dists[ t_dists.ALG == a ]
for l in labels:
l_dists = a_dists[ a_dists.LABEL == l ].DISTANCEPAD
tlist += [t]
alist += [a]
llist += [l]
if( l_dists.size > 10 ):
# print( ' label {} has {} samples'.format(l, l_dists.size) )
params = scipy.stats.rayleigh.fit( l_dists )
ray_offset += [ params[0]]
ray_scale += [ params[1]]
params_fl = scipy.stats.rayleigh.fit( l_dists, floc=0. )
ray_offset_fl += [ params_fl[0] ]
ray_scale_fl += [ params_fl[1] ]
gam_params = scipy.stats.gamma.fit( l_dists )
gam_a += [ gam_params[0] ]
gam_offset += [ gam_params[1] ]
gam_scale += [ gam_params[2] ]
gam_params_fl = scipy.stats.gamma.fit( l_dists, floc=0. )
gam_a_fl += [ gam_params_fl[0] ]
gam_offset_fl += [ gam_params_fl[1] ]
gam_scale_fl += [ gam_params_fl[2] ]
else:
# print( ' skipping for label {}'.format(l) )
ray_offset += [float('nan')]
ray_scale += [float('nan')]
ray_offset_fl += [float('nan')]
ray_scale_fl += [float('nan')]
gam_a += [float('nan')]
gam_offset += [float('nan')]
gam_scale += [float('nan')]
gam_a_fl += [float('nan')]
gam_offset_fl += [float('nan')]
gam_scale_fl += [float('nan')]
# merge all labels
tlist += [t]
alist += [a]
llist += [-1]
if( a_dists.size > 10 ):
params = scipy.stats.rayleigh.fit( a_dists.DISTANCEPAD )
ray_offset += [ params[0]]
ray_scale += [ params[1]]
params_fl = scipy.stats.rayleigh.fit( a_dists.DISTANCEPAD, floc=0 )
ray_offset_fl += [ params_fl[0] ]
ray_scale_fl += [ params_fl[1] ]
gam_params = scipy.stats.gamma.fit( a_dists.DISTANCEPAD )
gam_a += [ gam_params[0] ]
gam_offset += [ gam_params[1] ]
gam_scale += [ gam_params[2] ]
gam_params_fl = scipy.stats.gamma.fit( a_dists.DISTANCEPAD, floc=0. )
gam_a_fl += [ gam_params_fl[0] ]
gam_offset_fl += [ gam_params_fl[1] ]
gam_scale_fl += [ gam_params_fl[2] ]
else:
# print( ' skipping for MERGE')
ray_offset += [float('nan')]
ray_scale += [float('nan')]
ray_offset_fl += [float('nan')]
ray_scale_fl += [float('nan')]
gam_a += [float('nan')]
gam_offset += [float('nan')]
gam_scale += [float('nan')]
gam_a_fl += [float('nan')]
gam_offset_fl += [float('nan')]
gam_scale_fl += [float('nan')]
# print( len(llist) )
# print( len(tlist) )
# print( len(alist) )
# print( len(ray_offset) )
# print( len(ray_scale) )
# print( len(ray_offset_fl) )
# print( len(ray_scale_fl) )
# print( len(gam_a) )
# print( len(gam_offset) )
# print( len(gam_scale) )
# print( len(gam_a_fl) )
# print( len(gam_offset_fl) )
# print( len(gam_scale_fl) )
dist_df = pd.DataFrame( {'LABEL':llist,
'ALG':alist,
'TEMPLATE':tlist,
'RAY_OFFSET':ray_offset,
'RAY_SCALE':ray_scale,
'RAY_OFFSET_FL':ray_offset_fl,
'RAY_SCALE_FL':ray_scale_fl,
'GAM_A':gam_a,
'GAM_OFFSET':gam_offset,
'GAM_SCALE':gam_scale,
'GAM_A_FL':gam_a_fl,
'GAM_OFFSET_FL':gam_offset_fl,
'GAM_SCALE_FL':gam_scale_fl
})
# dist_df
dist_df.to_csv('/nrs/saalfeld/john/projects/flyChemStainAtlas/all_evals/stats/line{}_dist_params.csv'.format(line))
```
|
github_jupyter
|
import sys
import glob
import re
import fnmatch
import math
import os
from os import listdir
from os.path import join, isfile, basename
import itertools
import numpy as np
from numpy import float32, int32, uint8, dtype, genfromtxt
import scipy
from scipy.stats import ttest_ind
import pandas as pd
import matplotlib
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
import colorsys
labels = [16,64,8,32,2,4,65,66,33,67,34,17,69,70,35,71,9,18,72,36,73,74,37,75,19,76,38,77,39,78,79,20,5,40,80,10,81,82,83,84,85,86,11,22,23,24,12,3,6,49,50,25,51,13,52,26,53,27,54,55,56,28,7,14,57,58,29,59,30,60,15,61,31,62,63]
label_names_file = '/groups/saalfeld/home/bogovicj/vfb/DrosAdultBRAINdomains/refData/Original_Index.tsv'
label_names = pd.read_csv( label_names_file, delimiter='\t', header=0 )
# print label_names[ label_names['Stack id'] == 11 ]['JFRCtempate2010.mask130819' ].iloc[0]
# print label_names[ label_names['Stack id'] == 70 ]['JFRCtempate2010.mask130819' ].iloc[0]
def get_label_name( label_id ):
return label_names[ label_names['Stack id'] == label_id ]['JFRCtempate2010.mask130819' ].iloc[0]
templates = ['JFRCtemplate2010', 'JFRC2013_lo', 'F-antsFlip_lo', 'F-cmtkFlip_lof', 'TeforBrain_f']
reg_methods = [ 'cmtkCow', 'cmtkCOG', 'cmtkHideo', 'antsRegOwl', 'antsRegDog', 'antsRegYang' ]
line=3
dist_samples_f = '/nrs/saalfeld/john/projects/flyChemStainAtlas/all_evals/label_data_line{}.csv.gz'.format( line )
dist_samples_df = pd.read_csv( dist_samples_f, header=None, names=['TEMPLATE','ALG','LINE','LABEL','DISTANCE'] )
# dist_samples_df.head()
# print( dist_samples_df['ALG'].unique())
# print( dist_samples_df['TEMPLATE'].unique())
def pad_zero(field='DISTANCE', pad=0.01):
def pfunc(xin):
x=xin[field]
if x > 0.:
return x
elif x == 0.:
return pad
else:
return float('nan')
return pfunc
dist_samples_df['DISTANCEPAD'] = dist_samples_df.apply( pad_zero(field='DISTANCE'), axis=1)
# some_dat = dist_samples_df[ (dist_samples_df.TEMPLATE =='F-antsFlip_lo') & (dist_samples_df.ALG == 'antsRegDog') & (dist_samples_df.LABEL == 84) ]
# print( some_dat.size )
# gam_params_fl = scipy.stats.gamma.fit( some_dat.DISTANCEPAD, floc=0. )
# print( gam_params_fl )
# print( len(some_dat[some_dat.DISTANCEPAD <= 0.]))
# print( len(l_dists) )
# l_dists[l_dists <= 0.]
# some_dat.DISTANCEPAD
ray_offset = []
ray_scale = []
ray_offset_fl = []
ray_scale_fl = []
gam_a = []
gam_offset = []
gam_scale = []
gam_a_fl = []
gam_offset_fl = []
gam_scale_fl = []
tlist = []
alist = []
llist = []
for t in templates:
t_dists = dist_samples_df[ dist_samples_df.TEMPLATE == t ]
print( 't ', t )
for a in reg_methods:
print( 'a ', a )
a_dists = t_dists[ t_dists.ALG == a ]
for l in labels:
l_dists = a_dists[ a_dists.LABEL == l ].DISTANCEPAD
tlist += [t]
alist += [a]
llist += [l]
if( l_dists.size > 10 ):
# print( ' label {} has {} samples'.format(l, l_dists.size) )
params = scipy.stats.rayleigh.fit( l_dists )
ray_offset += [ params[0]]
ray_scale += [ params[1]]
params_fl = scipy.stats.rayleigh.fit( l_dists, floc=0. )
ray_offset_fl += [ params_fl[0] ]
ray_scale_fl += [ params_fl[1] ]
gam_params = scipy.stats.gamma.fit( l_dists )
gam_a += [ gam_params[0] ]
gam_offset += [ gam_params[1] ]
gam_scale += [ gam_params[2] ]
gam_params_fl = scipy.stats.gamma.fit( l_dists, floc=0. )
gam_a_fl += [ gam_params_fl[0] ]
gam_offset_fl += [ gam_params_fl[1] ]
gam_scale_fl += [ gam_params_fl[2] ]
else:
# print( ' skipping for label {}'.format(l) )
ray_offset += [float('nan')]
ray_scale += [float('nan')]
ray_offset_fl += [float('nan')]
ray_scale_fl += [float('nan')]
gam_a += [float('nan')]
gam_offset += [float('nan')]
gam_scale += [float('nan')]
gam_a_fl += [float('nan')]
gam_offset_fl += [float('nan')]
gam_scale_fl += [float('nan')]
# merge all labels
tlist += [t]
alist += [a]
llist += [-1]
if( a_dists.size > 10 ):
params = scipy.stats.rayleigh.fit( a_dists.DISTANCEPAD )
ray_offset += [ params[0]]
ray_scale += [ params[1]]
params_fl = scipy.stats.rayleigh.fit( a_dists.DISTANCEPAD, floc=0 )
ray_offset_fl += [ params_fl[0] ]
ray_scale_fl += [ params_fl[1] ]
gam_params = scipy.stats.gamma.fit( a_dists.DISTANCEPAD )
gam_a += [ gam_params[0] ]
gam_offset += [ gam_params[1] ]
gam_scale += [ gam_params[2] ]
gam_params_fl = scipy.stats.gamma.fit( a_dists.DISTANCEPAD, floc=0. )
gam_a_fl += [ gam_params_fl[0] ]
gam_offset_fl += [ gam_params_fl[1] ]
gam_scale_fl += [ gam_params_fl[2] ]
else:
# print( ' skipping for MERGE')
ray_offset += [float('nan')]
ray_scale += [float('nan')]
ray_offset_fl += [float('nan')]
ray_scale_fl += [float('nan')]
gam_a += [float('nan')]
gam_offset += [float('nan')]
gam_scale += [float('nan')]
gam_a_fl += [float('nan')]
gam_offset_fl += [float('nan')]
gam_scale_fl += [float('nan')]
# print( len(llist) )
# print( len(tlist) )
# print( len(alist) )
# print( len(ray_offset) )
# print( len(ray_scale) )
# print( len(ray_offset_fl) )
# print( len(ray_scale_fl) )
# print( len(gam_a) )
# print( len(gam_offset) )
# print( len(gam_scale) )
# print( len(gam_a_fl) )
# print( len(gam_offset_fl) )
# print( len(gam_scale_fl) )
dist_df = pd.DataFrame( {'LABEL':llist,
'ALG':alist,
'TEMPLATE':tlist,
'RAY_OFFSET':ray_offset,
'RAY_SCALE':ray_scale,
'RAY_OFFSET_FL':ray_offset_fl,
'RAY_SCALE_FL':ray_scale_fl,
'GAM_A':gam_a,
'GAM_OFFSET':gam_offset,
'GAM_SCALE':gam_scale,
'GAM_A_FL':gam_a_fl,
'GAM_OFFSET_FL':gam_offset_fl,
'GAM_SCALE_FL':gam_scale_fl
})
# dist_df
dist_df.to_csv('/nrs/saalfeld/john/projects/flyChemStainAtlas/all_evals/stats/line{}_dist_params.csv'.format(line))
| 0.108909 | 0.247794 |

[](https://colab.research.google.com/github/JohnSnowLabs/spark-nlp-workshop/blob/master/tutorials/Certification_Trainings/Healthcare/10.1.Clinical_Relation_Extraction_BodyParts_Models.ipynb)
# 10.1 Clinical Relation Extraction BodyPart Models
(requires Spark NLP 2.7.1 and Spark NLP Healthcare 2.7.2 and above)
```
import json
from google.colab import files
license_keys = files.upload()
with open(list(license_keys.keys())[0]) as f:
license_keys = json.load(f)
license_keys.keys()
import os
# Install java
! apt-get update -qq
! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null
os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64"
os.environ["PATH"] = os.environ["JAVA_HOME"] + "/bin:" + os.environ["PATH"]
! java -version
secret = license_keys['SECRET']
os.environ['SPARK_NLP_LICENSE'] = license_keys['SPARK_NLP_LICENSE']
os.environ['AWS_ACCESS_KEY_ID']= license_keys['AWS_ACCESS_KEY_ID']
os.environ['AWS_SECRET_ACCESS_KEY'] = license_keys['AWS_SECRET_ACCESS_KEY']
version = license_keys['PUBLIC_VERSION']
jsl_version = license_keys['JSL_VERSION']
! pip install --ignore-installed -q pyspark==2.4.4
! python -m pip install --upgrade spark-nlp-jsl==$jsl_version --extra-index-url https://pypi.johnsnowlabs.com/$secret
! pip install --ignore-installed -q spark-nlp==$version
import sparknlp
print (sparknlp.version())
import json
import os
from pyspark.ml import Pipeline
from pyspark.sql import SparkSession
from sparknlp.annotator import *
from sparknlp_jsl.annotator import *
from sparknlp.base import *
import sparknlp_jsl
params = {"spark.driver.memory":"16G",
"spark.kryoserializer.buffer.max":"2000M",
"spark.driver.maxResultSize":"2000M"}
spark = sparknlp_jsl.start(secret, params=params)
spark
```
## 1. Prediction Pipeline for Clinical Binary Relation Models
Basic Pipeline without Re Models. Run it once and we can add custom Re models to the same pipeline
```
documenter = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
sentencer = SentenceDetector()\
.setInputCols(["document"])\
.setOutputCol("sentences")
tokenizer = sparknlp.annotators.Tokenizer()\
.setInputCols(["sentences"])\
.setOutputCol("tokens")\
words_embedder = WordEmbeddingsModel()\
.pretrained("embeddings_clinical", "en", "clinical/models")\
.setInputCols(["sentences", "tokens"])\
.setOutputCol("embeddings")
pos_tagger = PerceptronModel()\
.pretrained("pos_clinical", "en", "clinical/models") \
.setInputCols(["sentences", "tokens"])\
.setOutputCol("pos_tags")
dependency_parser = sparknlp.annotators.DependencyParserModel()\
.pretrained("dependency_conllu", "en")\
.setInputCols(["sentences", "pos_tags", "tokens"])\
.setOutputCol("dependencies")
# get pretrained ner model
clinical_ner_tagger = sparknlp.annotators.NerDLModel()\
.pretrained('jsl_ner_wip_greedy_clinical','en','clinical/models')\
.setInputCols("sentences", "tokens", "embeddings")\
.setOutputCol("ner_tags")
ner_chunker = NerConverter()\
.setInputCols(["sentences", "tokens", "ner_tags"])\
.setOutputCol("ner_chunks")
import pandas as pd
# This function will be utilized to show prediction results in a dataframe
def get_relations_df (results, col='relations'):
rel_pairs=[]
for rel in results[0][col]:
rel_pairs.append((
rel.result,
rel.metadata['entity1'],
rel.metadata['entity1_begin'],
rel.metadata['entity1_end'],
rel.metadata['chunk1'],
rel.metadata['entity2'],
rel.metadata['entity2_begin'],
rel.metadata['entity2_end'],
rel.metadata['chunk2'],
rel.metadata['confidence']
))
rel_df = pd.DataFrame(rel_pairs, columns=['relations',
'entity1','entity1_begin','entity1_end','chunk1',
'entity2','entity2_end','entity2_end','chunk2',
'confidence'])
# limit df columns to get entity and chunks with results only
rel_df = rel_df.iloc[:,[0,1,4,5,8,9]]
return rel_df
```
### Example pairs for relation entities
```
# bodypart entities >> ['external_body_part_or_region', 'internal_organ_or_component']
# 1. bodypart vs problem
pair1 = ['symptom-external_body_part_or_region', 'external_body_part_or_region-symptom']
# 2. bodypart vs procedure and test
pair2 = ['internal_organ_or_component-imagingtest',
'imagingtest-internal_organ_or_component',
'internal_organ_or_component-procedure',
'procedure-internal_organ_or_component',
'internal_organ_or_component-test',
'test-internal_organ_or_component',
'external_body_part_or_region-imagingtest',
'imagingtest-external_body_part_or_region',
'external_body_part_or_region-procedure',
'procedure-external_body_part_or_region',
'external_body_part_or_region-test',
'test-external_body_part_or_region']
# 3. bodypart vs direction
pair3 = ['direction-external_body_part_or_region', 'external_body_part_or_region-direction',
'internal_organ_or_component-direction','direction-internal_organ_or_component']
# 4. date vs other clinical entities
# date entities >> ['Date', 'RelativeDate', 'Duration', 'RelativeTime', 'Time']
pair4 = ['symptom-date', 'date-procedure', 'delativedate-test', 'test-date']
```
**Pretrained relation model names**; use this names in `RelationExtractionModel()` ;
+ `re_bodypart_problem`
+ `re_bodypart_directions`
+ `re_bodypart_proceduretest`
+ `re_date_clinical`
## 2. Example of how custom RE models can be added to the same pipeline
### 2.1 Relation Extraction Model
```
re_model = RelationExtractionModel()\
.pretrained("re_bodypart_directions", "en", 'clinical/models')\
.setInputCols(["embeddings", "pos_tags", "ner_chunks", "dependencies"])\
.setOutputCol("relations")\
.setRelationPairs(['direction-external_body_part_or_region',
'external_body_part_or_region-direction',
'direction-internal_organ_or_component',
'internal_organ_or_component-direction'
])\
.setMaxSyntacticDistance(4).setPredictionThreshold(0.9)
trained_pipeline = Pipeline(stages=[
documenter,
sentencer,
tokenizer,
words_embedder,
pos_tagger,
clinical_ner_tagger,
ner_chunker,
dependency_parser,
re_model
])
empty_data = spark.createDataFrame([[""]]).toDF("text")
loaded_re_model = trained_pipeline.fit(empty_data)
```
### 2.2 ReDL Model - based on end-to-end trained Bert Model
```
re_ner_chunk_filter = RENerChunksFilter() \
.setInputCols(["ner_chunks", "dependencies"])\
.setOutputCol("re_ner_chunks")\
.setMaxSyntacticDistance(4)\
.setRelationPairs(['direction-external_body_part_or_region',
'external_body_part_or_region-direction',
'direction-internal_organ_or_component',
'internal_organ_or_component-direction'
])
re_model = RelationExtractionDLModel() \
.pretrained('redl_bodypart_direction_biobert', "en", "clinical/models")\
.setPredictionThreshold(0.5)\
.setInputCols(["re_ner_chunks", "sentences"]) \
.setOutputCol("relations")
trained_pipeline = Pipeline(stages=[
documenter,
sentencer,
tokenizer,
words_embedder,
pos_tagger,
clinical_ner_tagger,
ner_chunker,
dependency_parser,
re_ner_chunk_filter,
re_model
])
empty_data = spark.createDataFrame([[""]]).toDF("text")
loaded_redl_model = trained_pipeline.fit(empty_data)
```
## 3. Sample clinical tetxs
```
# bodypart vs problem
text1 = '''No neurologic deficits other than some numbness in his left hand.'''
# bodypart vs procedure and test
#text2 = 'Common bile duct was noted to be 10 mm in size on that ultrasound.'
#text2 = 'Biopsies of the distal duodenum, gastric antrum, distalesophagus were taken and sent for pathological evaluation.'
text2 = 'TECHNIQUE IN DETAIL: After informed consent was obtained from the patient and his mother, the chest was scanned with portable ultrasound.'
# bodypart direction
text3 = '''MRI demonstrated infarction in the upper brain stem , left cerebellum and right basil ganglia'''
# date vs other clinical entities
text4 = ''' This 73 y/o patient had Brain CT on 1/12/95, with progressive memory and cognitive decline since 8/11/94.'''
```
**Get Single Prediction** with `LightPipeline()`
### 3. 1 Using Relation Extraction Model
```
# choose one of the sample texts depending on the pretrained relation model you want to use
text = text3
loaded_re_model_light = LightPipeline(loaded_re_model)
annotations = loaded_re_model_light.fullAnnotate(text)
rel_df = get_relations_df(annotations) # << get_relations_df() is the function defined in the 3rd cell
print('\n',text)
rel_df[rel_df.relations!="0"]
#rel_df
```
### 3.2 Using Relation Extraction DL Model
```
# choose one of the sample texts depending on the pretrained relation model you want to use
text = text3
loaded_re_model_light = LightPipeline(loaded_redl_model)
annotations = loaded_re_model_light.fullAnnotate(text)
rel_df = get_relations_df(annotations) # << get_relations_df() is the function defined in the 3rd cell
print('\n',text)
rel_df[rel_df.relations!="0"]
#rel_df
```
## Custom Function
```
# Previous cell content is merged in this custom function to get quick predictions, for custom cases please check parameters in RelationExtractionModel()
def relation_exraction(model_name, pairs, text):
re_model = RelationExtractionModel()\
.pretrained(model_name, "en", 'clinical/models')\
.setInputCols(["embeddings", "pos_tags", "ner_chunks", "dependencies"])\
.setOutputCol("relations")\
.setRelationPairs(pairs)\
.setMaxSyntacticDistance(3)\
.setPredictionThreshold(0.9)
trained_pipeline = Pipeline(stages=[
documenter,
sentencer,
tokenizer,
words_embedder,
pos_tagger,
clinical_ner_tagger,
ner_chunker,
dependency_parser,
re_model
])
empty_data = spark.createDataFrame([[""]]).toDF("text")
loaded_re_model = trained_pipeline.fit(empty_data)
loaded_re_model_light = LightPipeline(loaded_re_model)
annotations = loaded_re_model_light.fullAnnotate(text)
rel_df = get_relations_df(annotations) # << get_relations_df() is the function defined in the 3rd cell
print('\n','Target Text : ',text, '\n')
#rel_df
return rel_df[rel_df.relations!="0"]
def relation_exraction_dl(model_name, pairs, text):
re_ner_chunk_filter = RENerChunksFilter() \
.setInputCols(["ner_chunks", "dependencies"])\
.setOutputCol("re_ner_chunks")\
.setRelationPairs(pairs)
re_model = RelationExtractionDLModel() \
.pretrained(model_name, "en", "clinical/models")\
.setPredictionThreshold(0.0)\
.setInputCols(["re_ner_chunks", "sentences"]) \
.setOutputCol("relations")
trained_pipeline = Pipeline(stages=[
documenter,
sentencer,
tokenizer,
words_embedder,
pos_tagger,
clinical_ner_tagger,
ner_chunker,
dependency_parser,
re_ner_chunk_filter,
re_model
])
empty_data = spark.createDataFrame([[""]]).toDF("text")
loaded_re_model = trained_pipeline.fit(empty_data)
loaded_re_model_light = LightPipeline(loaded_re_model)
annotations = loaded_re_model_light.fullAnnotate(text)
rel_df = get_relations_df(annotations) # << get_relations_df() is the function defined in the 3rd cell
print('\n','Target Text : ',text, '\n')
#rel_df
return rel_df[rel_df.relations!="0"]
```
## Predictions with Custom Function
### 4.1 Bodypart vs Problem - RelationExtractionModel
```
# bodypart vs problem
model_name = 're_bodypart_problem'
pairs = ['symptom-external_body_part_or_region', 'external_body_part_or_region-symptom']
text = text1
relation_exraction(model_name, pairs, text)
```
### 4.2 Bodypart vs Problem - RelationExtractionDLModel
```
# bodypart vs problem
model_name = 'redl_bodypart_problem_biobert'
pairs = ['symptom-external_body_part_or_region', 'external_body_part_or_region-symptom']
text = text1
relation_exraction_dl(model_name, pairs, text)
```
### 5.1 Bodypart vs Procedure & Test - RelationExtractionModel
```
# bodypart vs procedure and test
model_name = 're_bodypart_proceduretest'
pairs = pair2
text = text2
relation_exraction(model_name, pairs, text)
```
### 5.2 Bodypart vs Procedure & Test - RelationExtractionDLModel
```
# bodypart vs procedure and test
model_name = 'redl_bodypart_procedure_test_biobert'
pairs = pair2
text = text2
relation_exraction_dl(model_name, pairs, text)
```
### 6.1 Bodypart vs Directions - RelationExtractionModel
```
# bodypart vs directions
model_name = 'redl_bodypart_direction'
pairs = pair3
text = text3
relation_exraction(model_name, pairs, text)
```
### 6.2 Bodypart vs Directions - RelationExtractionDLModel
```
# bodypart vs directions
model_name = 'redl_bodypart_direction_biobert'
pairs = pair3
text = text3
relation_exraction_dl(model_name, pairs, text)
```
### 7.1 Date vs Clinical Entities - RelationExtractionModel
```
# date vs clinical date entities
model_name = 'redl_date_clinical'
pairs = pair4
text = text4
relation_exraction(model_name, pairs, text)
```
### 7.2 Date vs Clinical Entities - RelationExtractionDLModel
```
# date vs clinical date entities
model_name = 'redl_date_clinical_biobert'
pairs = pair4
text = text4
relation_exraction_dl(model_name, pairs, text)
```
|
github_jupyter
|
import json
from google.colab import files
license_keys = files.upload()
with open(list(license_keys.keys())[0]) as f:
license_keys = json.load(f)
license_keys.keys()
import os
# Install java
! apt-get update -qq
! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null
os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64"
os.environ["PATH"] = os.environ["JAVA_HOME"] + "/bin:" + os.environ["PATH"]
! java -version
secret = license_keys['SECRET']
os.environ['SPARK_NLP_LICENSE'] = license_keys['SPARK_NLP_LICENSE']
os.environ['AWS_ACCESS_KEY_ID']= license_keys['AWS_ACCESS_KEY_ID']
os.environ['AWS_SECRET_ACCESS_KEY'] = license_keys['AWS_SECRET_ACCESS_KEY']
version = license_keys['PUBLIC_VERSION']
jsl_version = license_keys['JSL_VERSION']
! pip install --ignore-installed -q pyspark==2.4.4
! python -m pip install --upgrade spark-nlp-jsl==$jsl_version --extra-index-url https://pypi.johnsnowlabs.com/$secret
! pip install --ignore-installed -q spark-nlp==$version
import sparknlp
print (sparknlp.version())
import json
import os
from pyspark.ml import Pipeline
from pyspark.sql import SparkSession
from sparknlp.annotator import *
from sparknlp_jsl.annotator import *
from sparknlp.base import *
import sparknlp_jsl
params = {"spark.driver.memory":"16G",
"spark.kryoserializer.buffer.max":"2000M",
"spark.driver.maxResultSize":"2000M"}
spark = sparknlp_jsl.start(secret, params=params)
spark
documenter = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
sentencer = SentenceDetector()\
.setInputCols(["document"])\
.setOutputCol("sentences")
tokenizer = sparknlp.annotators.Tokenizer()\
.setInputCols(["sentences"])\
.setOutputCol("tokens")\
words_embedder = WordEmbeddingsModel()\
.pretrained("embeddings_clinical", "en", "clinical/models")\
.setInputCols(["sentences", "tokens"])\
.setOutputCol("embeddings")
pos_tagger = PerceptronModel()\
.pretrained("pos_clinical", "en", "clinical/models") \
.setInputCols(["sentences", "tokens"])\
.setOutputCol("pos_tags")
dependency_parser = sparknlp.annotators.DependencyParserModel()\
.pretrained("dependency_conllu", "en")\
.setInputCols(["sentences", "pos_tags", "tokens"])\
.setOutputCol("dependencies")
# get pretrained ner model
clinical_ner_tagger = sparknlp.annotators.NerDLModel()\
.pretrained('jsl_ner_wip_greedy_clinical','en','clinical/models')\
.setInputCols("sentences", "tokens", "embeddings")\
.setOutputCol("ner_tags")
ner_chunker = NerConverter()\
.setInputCols(["sentences", "tokens", "ner_tags"])\
.setOutputCol("ner_chunks")
import pandas as pd
# This function will be utilized to show prediction results in a dataframe
def get_relations_df (results, col='relations'):
rel_pairs=[]
for rel in results[0][col]:
rel_pairs.append((
rel.result,
rel.metadata['entity1'],
rel.metadata['entity1_begin'],
rel.metadata['entity1_end'],
rel.metadata['chunk1'],
rel.metadata['entity2'],
rel.metadata['entity2_begin'],
rel.metadata['entity2_end'],
rel.metadata['chunk2'],
rel.metadata['confidence']
))
rel_df = pd.DataFrame(rel_pairs, columns=['relations',
'entity1','entity1_begin','entity1_end','chunk1',
'entity2','entity2_end','entity2_end','chunk2',
'confidence'])
# limit df columns to get entity and chunks with results only
rel_df = rel_df.iloc[:,[0,1,4,5,8,9]]
return rel_df
# bodypart entities >> ['external_body_part_or_region', 'internal_organ_or_component']
# 1. bodypart vs problem
pair1 = ['symptom-external_body_part_or_region', 'external_body_part_or_region-symptom']
# 2. bodypart vs procedure and test
pair2 = ['internal_organ_or_component-imagingtest',
'imagingtest-internal_organ_or_component',
'internal_organ_or_component-procedure',
'procedure-internal_organ_or_component',
'internal_organ_or_component-test',
'test-internal_organ_or_component',
'external_body_part_or_region-imagingtest',
'imagingtest-external_body_part_or_region',
'external_body_part_or_region-procedure',
'procedure-external_body_part_or_region',
'external_body_part_or_region-test',
'test-external_body_part_or_region']
# 3. bodypart vs direction
pair3 = ['direction-external_body_part_or_region', 'external_body_part_or_region-direction',
'internal_organ_or_component-direction','direction-internal_organ_or_component']
# 4. date vs other clinical entities
# date entities >> ['Date', 'RelativeDate', 'Duration', 'RelativeTime', 'Time']
pair4 = ['symptom-date', 'date-procedure', 'delativedate-test', 'test-date']
re_model = RelationExtractionModel()\
.pretrained("re_bodypart_directions", "en", 'clinical/models')\
.setInputCols(["embeddings", "pos_tags", "ner_chunks", "dependencies"])\
.setOutputCol("relations")\
.setRelationPairs(['direction-external_body_part_or_region',
'external_body_part_or_region-direction',
'direction-internal_organ_or_component',
'internal_organ_or_component-direction'
])\
.setMaxSyntacticDistance(4).setPredictionThreshold(0.9)
trained_pipeline = Pipeline(stages=[
documenter,
sentencer,
tokenizer,
words_embedder,
pos_tagger,
clinical_ner_tagger,
ner_chunker,
dependency_parser,
re_model
])
empty_data = spark.createDataFrame([[""]]).toDF("text")
loaded_re_model = trained_pipeline.fit(empty_data)
re_ner_chunk_filter = RENerChunksFilter() \
.setInputCols(["ner_chunks", "dependencies"])\
.setOutputCol("re_ner_chunks")\
.setMaxSyntacticDistance(4)\
.setRelationPairs(['direction-external_body_part_or_region',
'external_body_part_or_region-direction',
'direction-internal_organ_or_component',
'internal_organ_or_component-direction'
])
re_model = RelationExtractionDLModel() \
.pretrained('redl_bodypart_direction_biobert', "en", "clinical/models")\
.setPredictionThreshold(0.5)\
.setInputCols(["re_ner_chunks", "sentences"]) \
.setOutputCol("relations")
trained_pipeline = Pipeline(stages=[
documenter,
sentencer,
tokenizer,
words_embedder,
pos_tagger,
clinical_ner_tagger,
ner_chunker,
dependency_parser,
re_ner_chunk_filter,
re_model
])
empty_data = spark.createDataFrame([[""]]).toDF("text")
loaded_redl_model = trained_pipeline.fit(empty_data)
# bodypart vs problem
text1 = '''No neurologic deficits other than some numbness in his left hand.'''
# bodypart vs procedure and test
#text2 = 'Common bile duct was noted to be 10 mm in size on that ultrasound.'
#text2 = 'Biopsies of the distal duodenum, gastric antrum, distalesophagus were taken and sent for pathological evaluation.'
text2 = 'TECHNIQUE IN DETAIL: After informed consent was obtained from the patient and his mother, the chest was scanned with portable ultrasound.'
# bodypart direction
text3 = '''MRI demonstrated infarction in the upper brain stem , left cerebellum and right basil ganglia'''
# date vs other clinical entities
text4 = ''' This 73 y/o patient had Brain CT on 1/12/95, with progressive memory and cognitive decline since 8/11/94.'''
# choose one of the sample texts depending on the pretrained relation model you want to use
text = text3
loaded_re_model_light = LightPipeline(loaded_re_model)
annotations = loaded_re_model_light.fullAnnotate(text)
rel_df = get_relations_df(annotations) # << get_relations_df() is the function defined in the 3rd cell
print('\n',text)
rel_df[rel_df.relations!="0"]
#rel_df
# choose one of the sample texts depending on the pretrained relation model you want to use
text = text3
loaded_re_model_light = LightPipeline(loaded_redl_model)
annotations = loaded_re_model_light.fullAnnotate(text)
rel_df = get_relations_df(annotations) # << get_relations_df() is the function defined in the 3rd cell
print('\n',text)
rel_df[rel_df.relations!="0"]
#rel_df
# Previous cell content is merged in this custom function to get quick predictions, for custom cases please check parameters in RelationExtractionModel()
def relation_exraction(model_name, pairs, text):
re_model = RelationExtractionModel()\
.pretrained(model_name, "en", 'clinical/models')\
.setInputCols(["embeddings", "pos_tags", "ner_chunks", "dependencies"])\
.setOutputCol("relations")\
.setRelationPairs(pairs)\
.setMaxSyntacticDistance(3)\
.setPredictionThreshold(0.9)
trained_pipeline = Pipeline(stages=[
documenter,
sentencer,
tokenizer,
words_embedder,
pos_tagger,
clinical_ner_tagger,
ner_chunker,
dependency_parser,
re_model
])
empty_data = spark.createDataFrame([[""]]).toDF("text")
loaded_re_model = trained_pipeline.fit(empty_data)
loaded_re_model_light = LightPipeline(loaded_re_model)
annotations = loaded_re_model_light.fullAnnotate(text)
rel_df = get_relations_df(annotations) # << get_relations_df() is the function defined in the 3rd cell
print('\n','Target Text : ',text, '\n')
#rel_df
return rel_df[rel_df.relations!="0"]
def relation_exraction_dl(model_name, pairs, text):
re_ner_chunk_filter = RENerChunksFilter() \
.setInputCols(["ner_chunks", "dependencies"])\
.setOutputCol("re_ner_chunks")\
.setRelationPairs(pairs)
re_model = RelationExtractionDLModel() \
.pretrained(model_name, "en", "clinical/models")\
.setPredictionThreshold(0.0)\
.setInputCols(["re_ner_chunks", "sentences"]) \
.setOutputCol("relations")
trained_pipeline = Pipeline(stages=[
documenter,
sentencer,
tokenizer,
words_embedder,
pos_tagger,
clinical_ner_tagger,
ner_chunker,
dependency_parser,
re_ner_chunk_filter,
re_model
])
empty_data = spark.createDataFrame([[""]]).toDF("text")
loaded_re_model = trained_pipeline.fit(empty_data)
loaded_re_model_light = LightPipeline(loaded_re_model)
annotations = loaded_re_model_light.fullAnnotate(text)
rel_df = get_relations_df(annotations) # << get_relations_df() is the function defined in the 3rd cell
print('\n','Target Text : ',text, '\n')
#rel_df
return rel_df[rel_df.relations!="0"]
# bodypart vs problem
model_name = 're_bodypart_problem'
pairs = ['symptom-external_body_part_or_region', 'external_body_part_or_region-symptom']
text = text1
relation_exraction(model_name, pairs, text)
# bodypart vs problem
model_name = 'redl_bodypart_problem_biobert'
pairs = ['symptom-external_body_part_or_region', 'external_body_part_or_region-symptom']
text = text1
relation_exraction_dl(model_name, pairs, text)
# bodypart vs procedure and test
model_name = 're_bodypart_proceduretest'
pairs = pair2
text = text2
relation_exraction(model_name, pairs, text)
# bodypart vs procedure and test
model_name = 'redl_bodypart_procedure_test_biobert'
pairs = pair2
text = text2
relation_exraction_dl(model_name, pairs, text)
# bodypart vs directions
model_name = 'redl_bodypart_direction'
pairs = pair3
text = text3
relation_exraction(model_name, pairs, text)
# bodypart vs directions
model_name = 'redl_bodypart_direction_biobert'
pairs = pair3
text = text3
relation_exraction_dl(model_name, pairs, text)
# date vs clinical date entities
model_name = 'redl_date_clinical'
pairs = pair4
text = text4
relation_exraction(model_name, pairs, text)
# date vs clinical date entities
model_name = 'redl_date_clinical_biobert'
pairs = pair4
text = text4
relation_exraction_dl(model_name, pairs, text)
| 0.312685 | 0.744308 |
```
import tensorflow as tf
import numpy as np
import pandas as pd
import collections
import math
import random
import pickle
from six.moves import xrange
def save2file(filename, data):
pickle_out = open("C:/Users/Akarsh/Downloads/DP_scripts/store_emb/" + filename + ".pickle", "wb")
pickle.dump(data, pickle_out, protocol=pickle.HIGHEST_PROTOCOL)
pickle_out.close()
def loadfile(filename):
pickle_in = open("C:/Users/Akarsh/Downloads/DP_scripts/store_emb/" + filename + ".pickle","rb")
return pickle.load(pickle_in)
main_str = str(437)
filename = "ip2vec_train_" + main_str
ip2vec_train = loadfile(filename)
print(ip2vec_train.shape)
print("building dataset...")
num_elems = -1
def build_dataset(ip2vec_train):
global num_elems # keep count of attributes for each flow
data = list(filter(None, ip2vec_train.to_csv(header=False, index=False).splitlines() ))
num_lines = len(data)
num_elems = len(data[0].split(",")) # 8 cols
res = [] #convert to list
for line in data:
for word in line.split(","):
res.append(word.strip())
count = []
count.extend( collections.Counter(res).most_common() ) #count freq
vocab_size = len(count)
w2v = dict()
for word, _ in count:
w2v[word] = len(w2v) #w2v
v2w = dict(zip(w2v.values(), w2v.keys())) #v2w
data = list()
for word in res:
if word in w2v:
index = w2v[word]
data.append(index) #convert to list
return data, num_lines, w2v, v2w, vocab_size
data, num_lines, w2v, v2w, vocab_size = build_dataset(ip2vec_train)
print("vocab size: ", vocab_size)
del ip2vec_train
# hyperparameters
batch_size = 128; embedding_size = 20; num_sampled = 32; num_epochs = 500;
data_index = 0; c_iter = 0; pairs = 13; training_pairs = pairs * batch_size;
idx = []
for i in range(0, num_lines-1):
idx.append(i)
def generate_batch():
global data_index; global num_elems; global c_iter
batch = np.ndarray(shape=(training_pairs),dtype=np.int32)
labels= np.ndarray(shape=(training_pairs,1), dtype=np.int32)
data_index = idx[c_iter] * num_elems
for i in range(batch_size):
# input SrcIP
batch[i*pairs+0] = data[data_index]; labels[i*pairs+0,0] = data[data_index+1]
batch[i*pairs+1] = data[data_index]; labels[i*pairs+1,0] = data[data_index+2]
batch[i*pairs+2] = data[data_index]; labels[i*pairs+2,0] = data[data_index+4]
# input DstIP
batch[i*pairs+3] = data[data_index+2]; labels[i*pairs+3,0] = data[data_index]
batch[i*pairs+4] = data[data_index+2]; labels[i*pairs+4,0] = data[data_index+4]
batch[i*pairs+5] = data[data_index+2]; labels[i*pairs+5,0] = data[data_index+3]
# input srcPt
batch[i*pairs+6] = data[data_index+1]; labels[i*pairs+6,0] = data[data_index+0]
# input dstPt
batch[i*pairs+7] = data[data_index+3]; labels[i*pairs+7,0] = data[data_index+2]
# input dur
batch[i*pairs+8] = data[data_index+7]; labels[i*pairs+8,0] = data[data_index+5]
# input byt
batch[i*pairs+9] = data[data_index+6]; labels[i*pairs+9,0] = data[data_index+5]
batch[i*pairs+10] = data[data_index+6]; labels[i*pairs+10,0] = data[data_index+7]
# input packets
batch[i*pairs+11] = data[data_index+5]; labels[i*pairs+11,0] = data[data_index+6]
batch[i*pairs+12] = data[data_index+5]; labels[i*pairs+12,0] = data[data_index+7]
# Check if end of training list is reached
c_iter += 1
if c_iter == num_lines - 1:
c_iter = 0
random.shuffle(idx)
data_index = idx[c_iter] * num_elems
return batch, labels
print("building tensorflow graph...")
graph = tf.Graph()
with graph.as_default():
train_inputs = tf.placeholder(tf.int32,shape=[training_pairs])
train_labels = tf.placeholder(tf.int32,shape=[training_pairs,1])
with tf.device('/cpu:0'):
embeddings = tf.Variable(tf.random_uniform([vocab_size,embedding_size],-1.0,1.0))
embed = tf.nn.embedding_lookup(embeddings, train_inputs)
nce_weights = tf.Variable(tf.truncated_normal([vocab_size, embedding_size],stddev=1.0 / math.sqrt(embedding_size)))
nce_biases = tf.Variable(tf.zeros([vocab_size]))
loss = tf.reduce_mean(tf.nn.nce_loss(weights=nce_weights, biases=nce_biases, labels=train_labels,
inputs=embed, num_sampled=num_sampled, num_classes=vocab_size))
optimizer = tf.train.GradientDescentOptimizer(0.05).minimize(loss)
norm = tf.sqrt(tf.reduce_sum(tf.square(embeddings), 1, keepdims=True))
normalized_embeddings = embeddings / norm
init = tf.global_variables_initializer()
num_steps = int(num_lines / batch_size * num_epochs)
print("training steps: ", num_steps)
with tf.Session(graph=graph) as session:
init.run()
average_loss = 0
for step in xrange(num_steps):
batch_inputs, batch_labels = generate_batch()
feed_dict = {train_inputs: batch_inputs, train_labels: batch_labels}
_, loss_val = session.run([optimizer, loss], feed_dict=feed_dict)
average_loss += loss_val
if step % 20000 == 0:
if step > 0:
average_loss /= 20000
print("Average loss at step", step, ": ", average_loss, " from ", num_steps ," steps.")
average_loss = 0
print('training finished...', '\n')
# Save embeddings
to_save_n = session.run(embeddings)
to_save = (to_save_n - to_save_n.min(0)) / to_save_n.ptp(0)
to_save_norm = to_save / to_save.max(axis=0)
final_emb = pd.DataFrame(data=to_save_norm[0:, 0:])
vals = []
for u in range(0,len(to_save)):
vals.append(v2w.get(u))
final_emb['values'] = vals
print('saving embeddings...')
filename = "ip2vec_emb_" + main_str
save2file(filename, final_emb)
filename = "ip2vec_emb_" + main_str
ip2vec_emb = loadfile(filename)
display(ip2vec_emb.head())
print(ip2vec_emb.shape)
save = True
if save:
df = pd.DataFrame.from_records([{'operation': 'ip2vec_train', 'main_str': main_str, 'vocab_size': vocab_size,
'batch_size': batch_size, 'embedding_size': embedding_size,
'negative_samp': num_sampled, 'lr': 0.05, 'num_epochs': num_epochs, 'final loss': average_loss}])
df.to_csv("C:/Users/Akarsh/Downloads/DP_scripts/store_emb/store_params.csv", mode='a', index=False)
print('data and hyperparams saved...')
```
|
github_jupyter
|
import tensorflow as tf
import numpy as np
import pandas as pd
import collections
import math
import random
import pickle
from six.moves import xrange
def save2file(filename, data):
pickle_out = open("C:/Users/Akarsh/Downloads/DP_scripts/store_emb/" + filename + ".pickle", "wb")
pickle.dump(data, pickle_out, protocol=pickle.HIGHEST_PROTOCOL)
pickle_out.close()
def loadfile(filename):
pickle_in = open("C:/Users/Akarsh/Downloads/DP_scripts/store_emb/" + filename + ".pickle","rb")
return pickle.load(pickle_in)
main_str = str(437)
filename = "ip2vec_train_" + main_str
ip2vec_train = loadfile(filename)
print(ip2vec_train.shape)
print("building dataset...")
num_elems = -1
def build_dataset(ip2vec_train):
global num_elems # keep count of attributes for each flow
data = list(filter(None, ip2vec_train.to_csv(header=False, index=False).splitlines() ))
num_lines = len(data)
num_elems = len(data[0].split(",")) # 8 cols
res = [] #convert to list
for line in data:
for word in line.split(","):
res.append(word.strip())
count = []
count.extend( collections.Counter(res).most_common() ) #count freq
vocab_size = len(count)
w2v = dict()
for word, _ in count:
w2v[word] = len(w2v) #w2v
v2w = dict(zip(w2v.values(), w2v.keys())) #v2w
data = list()
for word in res:
if word in w2v:
index = w2v[word]
data.append(index) #convert to list
return data, num_lines, w2v, v2w, vocab_size
data, num_lines, w2v, v2w, vocab_size = build_dataset(ip2vec_train)
print("vocab size: ", vocab_size)
del ip2vec_train
# hyperparameters
batch_size = 128; embedding_size = 20; num_sampled = 32; num_epochs = 500;
data_index = 0; c_iter = 0; pairs = 13; training_pairs = pairs * batch_size;
idx = []
for i in range(0, num_lines-1):
idx.append(i)
def generate_batch():
global data_index; global num_elems; global c_iter
batch = np.ndarray(shape=(training_pairs),dtype=np.int32)
labels= np.ndarray(shape=(training_pairs,1), dtype=np.int32)
data_index = idx[c_iter] * num_elems
for i in range(batch_size):
# input SrcIP
batch[i*pairs+0] = data[data_index]; labels[i*pairs+0,0] = data[data_index+1]
batch[i*pairs+1] = data[data_index]; labels[i*pairs+1,0] = data[data_index+2]
batch[i*pairs+2] = data[data_index]; labels[i*pairs+2,0] = data[data_index+4]
# input DstIP
batch[i*pairs+3] = data[data_index+2]; labels[i*pairs+3,0] = data[data_index]
batch[i*pairs+4] = data[data_index+2]; labels[i*pairs+4,0] = data[data_index+4]
batch[i*pairs+5] = data[data_index+2]; labels[i*pairs+5,0] = data[data_index+3]
# input srcPt
batch[i*pairs+6] = data[data_index+1]; labels[i*pairs+6,0] = data[data_index+0]
# input dstPt
batch[i*pairs+7] = data[data_index+3]; labels[i*pairs+7,0] = data[data_index+2]
# input dur
batch[i*pairs+8] = data[data_index+7]; labels[i*pairs+8,0] = data[data_index+5]
# input byt
batch[i*pairs+9] = data[data_index+6]; labels[i*pairs+9,0] = data[data_index+5]
batch[i*pairs+10] = data[data_index+6]; labels[i*pairs+10,0] = data[data_index+7]
# input packets
batch[i*pairs+11] = data[data_index+5]; labels[i*pairs+11,0] = data[data_index+6]
batch[i*pairs+12] = data[data_index+5]; labels[i*pairs+12,0] = data[data_index+7]
# Check if end of training list is reached
c_iter += 1
if c_iter == num_lines - 1:
c_iter = 0
random.shuffle(idx)
data_index = idx[c_iter] * num_elems
return batch, labels
print("building tensorflow graph...")
graph = tf.Graph()
with graph.as_default():
train_inputs = tf.placeholder(tf.int32,shape=[training_pairs])
train_labels = tf.placeholder(tf.int32,shape=[training_pairs,1])
with tf.device('/cpu:0'):
embeddings = tf.Variable(tf.random_uniform([vocab_size,embedding_size],-1.0,1.0))
embed = tf.nn.embedding_lookup(embeddings, train_inputs)
nce_weights = tf.Variable(tf.truncated_normal([vocab_size, embedding_size],stddev=1.0 / math.sqrt(embedding_size)))
nce_biases = tf.Variable(tf.zeros([vocab_size]))
loss = tf.reduce_mean(tf.nn.nce_loss(weights=nce_weights, biases=nce_biases, labels=train_labels,
inputs=embed, num_sampled=num_sampled, num_classes=vocab_size))
optimizer = tf.train.GradientDescentOptimizer(0.05).minimize(loss)
norm = tf.sqrt(tf.reduce_sum(tf.square(embeddings), 1, keepdims=True))
normalized_embeddings = embeddings / norm
init = tf.global_variables_initializer()
num_steps = int(num_lines / batch_size * num_epochs)
print("training steps: ", num_steps)
with tf.Session(graph=graph) as session:
init.run()
average_loss = 0
for step in xrange(num_steps):
batch_inputs, batch_labels = generate_batch()
feed_dict = {train_inputs: batch_inputs, train_labels: batch_labels}
_, loss_val = session.run([optimizer, loss], feed_dict=feed_dict)
average_loss += loss_val
if step % 20000 == 0:
if step > 0:
average_loss /= 20000
print("Average loss at step", step, ": ", average_loss, " from ", num_steps ," steps.")
average_loss = 0
print('training finished...', '\n')
# Save embeddings
to_save_n = session.run(embeddings)
to_save = (to_save_n - to_save_n.min(0)) / to_save_n.ptp(0)
to_save_norm = to_save / to_save.max(axis=0)
final_emb = pd.DataFrame(data=to_save_norm[0:, 0:])
vals = []
for u in range(0,len(to_save)):
vals.append(v2w.get(u))
final_emb['values'] = vals
print('saving embeddings...')
filename = "ip2vec_emb_" + main_str
save2file(filename, final_emb)
filename = "ip2vec_emb_" + main_str
ip2vec_emb = loadfile(filename)
display(ip2vec_emb.head())
print(ip2vec_emb.shape)
save = True
if save:
df = pd.DataFrame.from_records([{'operation': 'ip2vec_train', 'main_str': main_str, 'vocab_size': vocab_size,
'batch_size': batch_size, 'embedding_size': embedding_size,
'negative_samp': num_sampled, 'lr': 0.05, 'num_epochs': num_epochs, 'final loss': average_loss}])
df.to_csv("C:/Users/Akarsh/Downloads/DP_scripts/store_emb/store_params.csv", mode='a', index=False)
print('data and hyperparams saved...')
| 0.438304 | 0.379551 |
```
import os
from datetime import datetime, timedelta
import ipywidgets as widgets
import plotly.graph_objs as go
import yfinance as yf
import pandas as pd
from IPython.display import display
interval_opts = [
"1m",
"2m",
"5m",
"15m",
"30m",
"60m",
"90m",
"1h",
"1d",
"5d",
"1wk",
"1mo",
"3mo",
]
rows = [
"sector",
"marketCap",
"beta",
"fiftyTwoWeekHigh",
"fiftyTwoWeekLow",
"floatShares",
"sharesShort",
"exDividendDate",
]
views = {
"Raw Data": lambda x, y: x,
"Percent Change": lambda x, y: x.pct_change(),
"Rolling Average": lambda x, y: x.rolling(y).mean(),
"Rolling Variance": lambda x, y: x.rolling(y).var(),
"Rolling Standard Deviation": lambda x, y: x.rolling(y).var() ** 0.5,
"Rolling Coefficient of Variation": lambda x, y: (x.rolling(y).var() ** 0.5)
/ (x.rolling(y).mean()),
}
clean_row = {
"sector": "Sector",
"marketCap": "M Cap",
"beta": "Beta",
"fiftyTwoWeekHigh": "52W High",
"fiftyTwoWeekLow": "52W Low",
"floatShares": "Floats",
"sharesShort": "Shorts",
"exDividendDate": "Ex-Div",
}
clean_data = {
"sector": lambda x: "N/A" if x is None else x,
"marketCap": lambda x: "N/A" if x is None else big_num(x),
"beta": lambda x: "N/A" if x is None else f"{round(x,2)}",
"fiftyTwoWeekHigh": lambda x: "N/A" if x is None else f"${round(x,2)}",
"fiftyTwoWeekLow": lambda x: "N/A" if x is None else f"${round(x,2)}",
"floatShares": lambda x: "N/A" if x is None else big_num(x),
"sharesShort": lambda x: "N/A" if x is None else big_num(x),
"exDividendDate": lambda x: "N/A"
if x is None
else datetime.fromtimestamp(x).strftime("%Y/%m/%d"),
}
def big_num(num):
if num > 1_000_000_000_000:
return f"{round(num/1_000_000_000_000,2)}T"
if num > 1_000_000_000:
return f"{round(num/1_000_000_000,2)}B"
if num > 1_000_000:
return f"{round(num/1_000_000,2)}M"
if num > 1_000:
return f"{num/round(1_000,2)}K"
return f"{round(num,2)}"
def clean_str(string):
new_str = ""
for letter in string:
if letter.isupper():
new_str += " "
new_str += letter
return new_str.title()
def format_plotly(fig, data, start, end, chart, calc=None):
fig.update_yaxes(title=None)
fig.update_xaxes(title=None)
start_t = start.strftime("%Y/%m/%d")
end_t = end.strftime("%Y/%m/%d")
if calc:
if len(calc) == 1:
fig_title = f"{calc[0]} of {data} from {start_t} to {end_t}"
else:
fig_title = f"{', '.join(calc)} of {data} from {start_t} to {end_t}"
else:
fig_title = "Volume"
height = 500 if chart == "main" else 300
fig.update_layout(
margin=dict(l=0, r=10, t=10, b=10),
autosize=False,
width=900,
height=height,
legend=dict(orientation="h"),
title={
"text": fig_title,
"y": 0.95,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
},
)
def create_line(visual, x, y, name, data, fig):
if visual == "line":
plot = go.Scatter(x=x, y=y[data], mode="lines", name=name, connectgaps=True)
if visual == "scatter":
plot = go.Scatter(x=x, y=y[data], mode="markers", name=name)
if visual == "candle":
plot = go.Candlestick(
x=x,
open=y["Open"],
close=y["Close"],
high=y["High"],
low=y["Low"],
name=name,
)
fig.add_trace(plot)
def show_fig(fig):
config = {"showTips": False, "scrollZoom": True}
if os.environ.get("SERVER_SOFTWARE", "jupyter").startswith("voila"):
fig.show(config=config, renderer="notebook")
else:
fig.show(config=config)
def table_data(infos):
cols = ["Ticker"] + list(infos)
data = pd.DataFrame(columns=cols)
data["Ticker"] = [clean_row[x] for x in rows]
for ticker in list(infos):
data[ticker] = [clean_data[x](infos[ticker][x]) for x in rows]
new_cols = {k: clean_str(k) for k in rows}
return data
class Chart:
def __init__(self):
self.last_tickers = ""
self.last_interval = "1d"
self.df = pd.DataFrame()
self.infos = {}
def create_stock(
self, calculation, data, rolling, start, end, interval, tickers, chart
):
if tickers and tickers[-1] == ",":
if tickers != self.last_tickers or interval != self.last_interval:
if interval in ["1d", "5d", "1wk", "1mo", "3mo"]:
self.df = yf.download(
tickers, period="max", interval=interval, progress=False
)
else:
end_date = end + timedelta(days=1)
self.df = yf.download(
tickers,
start=start,
end=end_date,
interval=interval,
progress=False,
)
self.last_tickers = tickers
self.last_interval = interval
start_n = datetime(start.year, start.month, start.day)
end_n = datetime(end.year, end.month, end.day)
fig = go.Figure()
for item in calculation:
calcs = views[item](self.df, rolling)
if interval in ["1d", "5d", "1wk", "1mo", "3mo"]:
result = calcs.loc[
(calcs.index >= start_n) & (calcs.index <= end_n)
]
else:
result = calcs
if len(result.columns) == 6:
name = f"{tickers.split(',')[0]} {item}"
create_line(chart, result.index, result, name, data, fig)
else:
for val in result.columns.levels[1]:
vals = result.xs(val, axis=1, level=1, drop_level=True)
name = f"{val.upper()} {item}"
create_line(chart, result.index, vals, name, data, fig)
format_plotly(fig, data, start, end, "main", calculation)
show_fig(fig)
def create_volume(self, start, end, interval, tickers):
start_n = datetime(start.year, start.month, start.day)
end_n = datetime(end.year, end.month, end.day)
result = self.df.loc[(self.df.index >= start_n) & (self.df.index <= end_n)]
fig = go.Figure()
if len(result.columns) == 6:
name = f"{tickers.split(',')[0]}"
create_line("line", result.index, result, name, "Volume", fig)
else:
for val in result.columns.levels[1]:
vals = result.xs(val, axis=1, level=1, drop_level=True)
name = f"{val.upper()}"
create_line("line", result.index, vals, name, "Volume", fig)
format_plotly(fig, "Volume", start, end, "volume")
show_fig(fig)
def create_table(self, tickers):
if tickers and tickers[-1] == ",":
clean_tickers = [x for x in tickers.split(",") if x]
for ticker in clean_tickers:
if ticker not in self.infos:
self.infos[ticker] = yf.Ticker(ticker).info
for ticker in self.infos:
if ticker not in tickers:
self.infos.pop(ticker)
result = table_data(self.infos)
fig = go.Figure(
data=[
go.Table(
header=dict(
values=result.columns,
fill_color="lightgray",
font=dict(color="black"),
align="left",
),
cells=dict(
values=[result[x] for x in result.columns],
# fill_color=base,
# format=["",",",".2f","$.2f","$.2f",",",",",""],
font=dict(color="black"),
align="left",
),
)
],
)
fig.update_layout(margin=dict(l=0, r=20, t=0, b=0), width=350)
show_fig(fig)
w_auto = widgets.Layout(width="auto")
calc_widget = widgets.SelectMultiple(
options=list(views.keys()), value=["Raw Data"], layout=w_auto
)
data_opts = ["Open", "Close", "High", "Low"]
data_widget = widgets.Dropdown(
options=data_opts, value="Close", layout=w_auto, description="Data"
)
rolling_widget = widgets.Dropdown(
options=list(range(2, 101)), value=60, layout=w_auto, description="Rolling"
)
base_date = (datetime.today() - timedelta(days=365)).date()
start_widget = widgets.DatePicker(value=base_date, layout=w_auto, description="Start")
end_widget = widgets.DatePicker(
value=datetime.today().date(), layout=w_auto, description="End"
)
interval_widget = widgets.Dropdown(
options=interval_opts, value="1d", layout=w_auto, description="Interval"
)
tickers_widget = widgets.Textarea(
value="TSLA,", layout=widgets.Layout(width="auto", height="100%")
)
chart_opts = ["line", "scatter", "candle"]
chart_widget = widgets.Dropdown(
options=chart_opts, value="line", layout=w_auto, description="Chart"
)
data_box = widgets.VBox([data_widget, rolling_widget, chart_widget])
date_box = widgets.VBox([start_widget, end_widget, interval_widget])
controls = widgets.HBox(
[tickers_widget, calc_widget, date_box, data_box],
layout=widgets.Layout(width="90%"),
)
chart = Chart()
stocks_view = widgets.interactive_output(
chart.create_stock,
{
"calculation": calc_widget,
"data": data_widget,
"rolling": rolling_widget,
"start": start_widget,
"end": end_widget,
"interval": interval_widget,
"tickers": tickers_widget,
"chart": chart_widget,
},
)
volume_view = widgets.interactive_output(
chart.create_volume,
{
"start": start_widget,
"end": end_widget,
"interval": interval_widget,
"tickers": tickers_widget,
},
)
table_view = widgets.interactive_output(chart.create_table, {"tickers": tickers_widget})
charts = widgets.VBox(
[stocks_view, volume_view],
layout=widgets.Layout(width="100%", padding="0", margin="0"),
)
figures = widgets.HBox(
[charts, table_view], layout=widgets.Layout(padding="0", margin="0")
)
title_html = "<h1>Stock Analysis Dashboard</h1>"
warning_html = '<p style="color:red"=>Use a comma after EVERY stock typed.</p>'
app_contents = [widgets.HTML(title_html), controls, widgets.HTML(warning_html), figures]
app = widgets.VBox(app_contents)
display(app)
```
|
github_jupyter
|
import os
from datetime import datetime, timedelta
import ipywidgets as widgets
import plotly.graph_objs as go
import yfinance as yf
import pandas as pd
from IPython.display import display
interval_opts = [
"1m",
"2m",
"5m",
"15m",
"30m",
"60m",
"90m",
"1h",
"1d",
"5d",
"1wk",
"1mo",
"3mo",
]
rows = [
"sector",
"marketCap",
"beta",
"fiftyTwoWeekHigh",
"fiftyTwoWeekLow",
"floatShares",
"sharesShort",
"exDividendDate",
]
views = {
"Raw Data": lambda x, y: x,
"Percent Change": lambda x, y: x.pct_change(),
"Rolling Average": lambda x, y: x.rolling(y).mean(),
"Rolling Variance": lambda x, y: x.rolling(y).var(),
"Rolling Standard Deviation": lambda x, y: x.rolling(y).var() ** 0.5,
"Rolling Coefficient of Variation": lambda x, y: (x.rolling(y).var() ** 0.5)
/ (x.rolling(y).mean()),
}
clean_row = {
"sector": "Sector",
"marketCap": "M Cap",
"beta": "Beta",
"fiftyTwoWeekHigh": "52W High",
"fiftyTwoWeekLow": "52W Low",
"floatShares": "Floats",
"sharesShort": "Shorts",
"exDividendDate": "Ex-Div",
}
clean_data = {
"sector": lambda x: "N/A" if x is None else x,
"marketCap": lambda x: "N/A" if x is None else big_num(x),
"beta": lambda x: "N/A" if x is None else f"{round(x,2)}",
"fiftyTwoWeekHigh": lambda x: "N/A" if x is None else f"${round(x,2)}",
"fiftyTwoWeekLow": lambda x: "N/A" if x is None else f"${round(x,2)}",
"floatShares": lambda x: "N/A" if x is None else big_num(x),
"sharesShort": lambda x: "N/A" if x is None else big_num(x),
"exDividendDate": lambda x: "N/A"
if x is None
else datetime.fromtimestamp(x).strftime("%Y/%m/%d"),
}
def big_num(num):
if num > 1_000_000_000_000:
return f"{round(num/1_000_000_000_000,2)}T"
if num > 1_000_000_000:
return f"{round(num/1_000_000_000,2)}B"
if num > 1_000_000:
return f"{round(num/1_000_000,2)}M"
if num > 1_000:
return f"{num/round(1_000,2)}K"
return f"{round(num,2)}"
def clean_str(string):
new_str = ""
for letter in string:
if letter.isupper():
new_str += " "
new_str += letter
return new_str.title()
def format_plotly(fig, data, start, end, chart, calc=None):
fig.update_yaxes(title=None)
fig.update_xaxes(title=None)
start_t = start.strftime("%Y/%m/%d")
end_t = end.strftime("%Y/%m/%d")
if calc:
if len(calc) == 1:
fig_title = f"{calc[0]} of {data} from {start_t} to {end_t}"
else:
fig_title = f"{', '.join(calc)} of {data} from {start_t} to {end_t}"
else:
fig_title = "Volume"
height = 500 if chart == "main" else 300
fig.update_layout(
margin=dict(l=0, r=10, t=10, b=10),
autosize=False,
width=900,
height=height,
legend=dict(orientation="h"),
title={
"text": fig_title,
"y": 0.95,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
},
)
def create_line(visual, x, y, name, data, fig):
if visual == "line":
plot = go.Scatter(x=x, y=y[data], mode="lines", name=name, connectgaps=True)
if visual == "scatter":
plot = go.Scatter(x=x, y=y[data], mode="markers", name=name)
if visual == "candle":
plot = go.Candlestick(
x=x,
open=y["Open"],
close=y["Close"],
high=y["High"],
low=y["Low"],
name=name,
)
fig.add_trace(plot)
def show_fig(fig):
config = {"showTips": False, "scrollZoom": True}
if os.environ.get("SERVER_SOFTWARE", "jupyter").startswith("voila"):
fig.show(config=config, renderer="notebook")
else:
fig.show(config=config)
def table_data(infos):
cols = ["Ticker"] + list(infos)
data = pd.DataFrame(columns=cols)
data["Ticker"] = [clean_row[x] for x in rows]
for ticker in list(infos):
data[ticker] = [clean_data[x](infos[ticker][x]) for x in rows]
new_cols = {k: clean_str(k) for k in rows}
return data
class Chart:
def __init__(self):
self.last_tickers = ""
self.last_interval = "1d"
self.df = pd.DataFrame()
self.infos = {}
def create_stock(
self, calculation, data, rolling, start, end, interval, tickers, chart
):
if tickers and tickers[-1] == ",":
if tickers != self.last_tickers or interval != self.last_interval:
if interval in ["1d", "5d", "1wk", "1mo", "3mo"]:
self.df = yf.download(
tickers, period="max", interval=interval, progress=False
)
else:
end_date = end + timedelta(days=1)
self.df = yf.download(
tickers,
start=start,
end=end_date,
interval=interval,
progress=False,
)
self.last_tickers = tickers
self.last_interval = interval
start_n = datetime(start.year, start.month, start.day)
end_n = datetime(end.year, end.month, end.day)
fig = go.Figure()
for item in calculation:
calcs = views[item](self.df, rolling)
if interval in ["1d", "5d", "1wk", "1mo", "3mo"]:
result = calcs.loc[
(calcs.index >= start_n) & (calcs.index <= end_n)
]
else:
result = calcs
if len(result.columns) == 6:
name = f"{tickers.split(',')[0]} {item}"
create_line(chart, result.index, result, name, data, fig)
else:
for val in result.columns.levels[1]:
vals = result.xs(val, axis=1, level=1, drop_level=True)
name = f"{val.upper()} {item}"
create_line(chart, result.index, vals, name, data, fig)
format_plotly(fig, data, start, end, "main", calculation)
show_fig(fig)
def create_volume(self, start, end, interval, tickers):
start_n = datetime(start.year, start.month, start.day)
end_n = datetime(end.year, end.month, end.day)
result = self.df.loc[(self.df.index >= start_n) & (self.df.index <= end_n)]
fig = go.Figure()
if len(result.columns) == 6:
name = f"{tickers.split(',')[0]}"
create_line("line", result.index, result, name, "Volume", fig)
else:
for val in result.columns.levels[1]:
vals = result.xs(val, axis=1, level=1, drop_level=True)
name = f"{val.upper()}"
create_line("line", result.index, vals, name, "Volume", fig)
format_plotly(fig, "Volume", start, end, "volume")
show_fig(fig)
def create_table(self, tickers):
if tickers and tickers[-1] == ",":
clean_tickers = [x for x in tickers.split(",") if x]
for ticker in clean_tickers:
if ticker not in self.infos:
self.infos[ticker] = yf.Ticker(ticker).info
for ticker in self.infos:
if ticker not in tickers:
self.infos.pop(ticker)
result = table_data(self.infos)
fig = go.Figure(
data=[
go.Table(
header=dict(
values=result.columns,
fill_color="lightgray",
font=dict(color="black"),
align="left",
),
cells=dict(
values=[result[x] for x in result.columns],
# fill_color=base,
# format=["",",",".2f","$.2f","$.2f",",",",",""],
font=dict(color="black"),
align="left",
),
)
],
)
fig.update_layout(margin=dict(l=0, r=20, t=0, b=0), width=350)
show_fig(fig)
w_auto = widgets.Layout(width="auto")
calc_widget = widgets.SelectMultiple(
options=list(views.keys()), value=["Raw Data"], layout=w_auto
)
data_opts = ["Open", "Close", "High", "Low"]
data_widget = widgets.Dropdown(
options=data_opts, value="Close", layout=w_auto, description="Data"
)
rolling_widget = widgets.Dropdown(
options=list(range(2, 101)), value=60, layout=w_auto, description="Rolling"
)
base_date = (datetime.today() - timedelta(days=365)).date()
start_widget = widgets.DatePicker(value=base_date, layout=w_auto, description="Start")
end_widget = widgets.DatePicker(
value=datetime.today().date(), layout=w_auto, description="End"
)
interval_widget = widgets.Dropdown(
options=interval_opts, value="1d", layout=w_auto, description="Interval"
)
tickers_widget = widgets.Textarea(
value="TSLA,", layout=widgets.Layout(width="auto", height="100%")
)
chart_opts = ["line", "scatter", "candle"]
chart_widget = widgets.Dropdown(
options=chart_opts, value="line", layout=w_auto, description="Chart"
)
data_box = widgets.VBox([data_widget, rolling_widget, chart_widget])
date_box = widgets.VBox([start_widget, end_widget, interval_widget])
controls = widgets.HBox(
[tickers_widget, calc_widget, date_box, data_box],
layout=widgets.Layout(width="90%"),
)
chart = Chart()
stocks_view = widgets.interactive_output(
chart.create_stock,
{
"calculation": calc_widget,
"data": data_widget,
"rolling": rolling_widget,
"start": start_widget,
"end": end_widget,
"interval": interval_widget,
"tickers": tickers_widget,
"chart": chart_widget,
},
)
volume_view = widgets.interactive_output(
chart.create_volume,
{
"start": start_widget,
"end": end_widget,
"interval": interval_widget,
"tickers": tickers_widget,
},
)
table_view = widgets.interactive_output(chart.create_table, {"tickers": tickers_widget})
charts = widgets.VBox(
[stocks_view, volume_view],
layout=widgets.Layout(width="100%", padding="0", margin="0"),
)
figures = widgets.HBox(
[charts, table_view], layout=widgets.Layout(padding="0", margin="0")
)
title_html = "<h1>Stock Analysis Dashboard</h1>"
warning_html = '<p style="color:red"=>Use a comma after EVERY stock typed.</p>'
app_contents = [widgets.HTML(title_html), controls, widgets.HTML(warning_html), figures]
app = widgets.VBox(app_contents)
display(app)
| 0.424531 | 0.571587 |
<a href="https://colab.research.google.com/github/yukinaga/twitter_bot/blob/master/section_6/01_preprocessing.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# データの前処理
対話文のデータセットに前処理を行い、保存します。
## ライブラリのインストール
分かち書きのためにjanomeを、テキストデータの前処理のためにtorchtextをインストールします。
```
!pip install janome==0.4.1
!pip install torchvision==0.7.0
!pip install torchtext==0.7.0
!pip install torch==1.6.0
```
## Google ドライブとの連携
以下のコードを実行し、認証コードを使用してGoogle ドライブをマウントします。
```
from google.colab import drive
drive.mount('/content/drive/')
```
## 対話文の取得
雑談対話コーパス「projectnextnlp-chat-dialogue-corpus.zip」をダウンロードします。
> Copyright (c) 2015 Project Next NLP 対話タスク 参加者一同
> https://sites.google.com/site/dialoguebreakdowndetection/chat-dialogue-corpus/LICENSE.txt
> Released under the MIT license
解凍したフォルダをGoogle ドライブにアップします。
フォルダからjsonファイルを読み込み、対話文として成り立っている文章を取り出してリストに格納します。
```
import glob # ファイルの取得に使用
import json # jsonファイルの読み込みに使用
import re
path = "/content/drive/My Drive/live_ai_data/projectnextnlp-chat-dialogue-corpus/json" # フォルダの場所を指定
files = glob.glob(path + "/*/*.json") # ファイルの一覧
dialogues = [] # 複数の対話文を格納するリスト
file_count= 0 # ファイル数のカウント
for file in files:
with open(file, "r") as f:
json_dic = json.load(f)
dialogue = [] # 単一の対話
for turn in json_dic["turns"]:
annotations = turn["annotations"] # 注釈
speaker = turn["speaker"] # 発言者
utterance = turn["utterance"] # 発言
# 空の文章や、特殊文字や数字が含まれる文章は除く
if (utterance=="") or ("\\u" in utterance) or (re.search("\d", utterance)!=None):
dialogue.clear() # 対話をリセット
continue
utterance = utterance.replace(".", "。").replace(",", "、") # 全角
utterance = utterance.replace(".", "。").replace(",", "、") # 半角
utterance = utterance.split("。")[0]
if speaker=="U": # 発言者が人間であれば
dialogue.append(utterance)
else: # 発言者がシステムであれば
is_wrong = False
for annotation in annotations:
breakdown = annotation["breakdown"] # 分類
if breakdown=="X": # 1つでも不適切評価があれば
is_wrong = True
break
if is_wrong:
dialogue.clear() # 対話をリセット
else:
dialogue.append(utterance) # 不適切評価が無ければ対話に追加
if len(dialogue) >= 2: # 単一の会話が成立すれば
dialogues.append(dialogue.copy())
dialogue.pop(0) # 最初の要素を削除
file_count += 1
if file_count%100 == 0:
print("files:", file_count, "dialogues", len(dialogues))
print("files:", file_count, "dialogues", len(dialogues))
```
## データ拡張の準備
データ拡張の準備として、正規表現の設定および分かち書きを行います。
```
import re
from janome.tokenizer import Tokenizer
re_kanji = re.compile(r"^[\u4E00-\u9FD0]+$") # 漢字の検出用
re_katakana = re.compile(r"[\u30A1-\u30F4]+") # カタカナの検出用
j_tk = Tokenizer()
def wakati(text):
return [tok for tok in j_tk.tokenize(text, wakati=True)]
wakati_inp = [] # 単語に分割された入力文
wakati_rep = [] # 単語に分割された応答文
for dialogue in dialogues:
wakati_inp.append(wakati(dialogue[0])[:10])
wakati_rep.append(wakati(dialogue[1])[:10])
```
## データ拡張
対話データの数を水増しします。
ある入力文を、それに対応する応答文以外の複数の応答文と組み合わせます。
組み合わせる応答文は、入力文に含まれる漢字やカタカナの単語を含むものを選択します。
```
dialogues_plus = []
for i, w_inp in enumerate(wakati_inp): # 全ての入力文でループ
inp_count = 0 # ある入力から生成された対話文をカウント
for j, w_rep in enumerate(wakati_rep): # 全ての応答文でループ
if i==j:
dialogues_plus.append(["".join(w_inp), "".join(w_rep)])
continue
similarity = 0 # 類似度
for w in w_inp: # 入力文と同じ単語があり、それが漢字かカタカナであれば類似度を上げる
if (w in w_rep) and (re_kanji.fullmatch(w) or re_katakana.fullmatch(w)):
similarity += 1
if similarity >= 1:
dialogue_plus = ["".join(w_inp), "".join(w_rep)]
if dialogue_plus not in dialogues_plus:
dialogues_plus.append(dialogue_plus)
inp_count += 1
if inp_count >= 12: # ある入力から生成する対話文の上限
break
if i%1000 == 0:
print("i:", i, "dialogues_pus:", len(dialogues_plus))
print("i:", i, "dialogues_pus:", len(dialogues_plus))
```
拡張された対話データを、新たな対話データとします。
```
dialogues = dialogues_plus
```
## 対話データの保存
対話データをcsvファイルとしてGoogle Driveに保存します。
```
import csv
from sklearn.model_selection import train_test_split
dialogues_train, dialogues_test = train_test_split(dialogues, shuffle=True, test_size=0.05) # 5%がテストデータ
path = "/content/drive/My Drive/live_ai_data/" # 保存場所
with open(path+"dialogues_train.csv", "w") as f:
writer = csv.writer(f)
writer.writerows(dialogues_train)
with open(path+"dialogues_test.csv", "w") as f:
writer = csv.writer(f)
writer.writerows(dialogues_test)
```
## 対話文の取得
Googleドライブから、対話文のデータを取り出してデータセットに格納します。
```
import torch
import torchtext
from janome.tokenizer import Tokenizer
path = "/content/drive/My Drive/live_ai_data/" # 保存場所を指定
j_tk = Tokenizer()
def tokenizer(text):
return [tok for tok in j_tk.tokenize(text, wakati=True)] # 内包表記
# データセットの列を定義
input_field = torchtext.data.Field( # 入力文
sequential=True, # データ長さが可変かどうか
tokenize=tokenizer, # 前処理や単語分割などのための関数
batch_first=True, # バッチの次元を先頭に
lower=True # アルファベットを小文字に変換
)
reply_field = torchtext.data.Field( # 応答文
sequential=True, # データ長さが可変かどうか
tokenize=tokenizer, # 前処理や単語分割などのための関数
init_token = "<sos>", # 文章開始のトークン
eos_token = "<eos>", # 文章終了のトークン
batch_first=True, # バッチの次元を先頭に
lower=True # アルファベットを小文字に変換
)
# csvファイルからデータセットを作成
train_data, test_data = torchtext.data.TabularDataset.splits(
path=path,
train="dialogues_train.csv",
validation="dialogues_test.csv",
format="csv",
fields=[("inp_text", input_field), ("rep_text", reply_field)] # 列の設定
)
```
## 単語とインデックスの対応
単語にインデックスを割り振り、辞書として格納します。
```
input_field.build_vocab(
train_data,
min_freq=3,
)
reply_field.build_vocab(
train_data,
min_freq=3,
)
print(input_field.vocab.freqs) # 各単語の出現頻度
print(len(input_field.vocab.stoi))
print(len(input_field.vocab.itos))
print(len(reply_field.vocab.stoi))
print(len(reply_field.vocab.itos))
```
## データセットの保存
データセットの`examples`とFieldをそれぞれ保存します。
```
import dill
torch.save(train_data.examples, path+"train_examples.pkl", pickle_module=dill)
torch.save(test_data.examples, path+"test_examples.pkl", pickle_module=dill)
torch.save(input_field, path+"input_field.pkl", pickle_module=dill)
torch.save(reply_field, path+"reply_field.pkl", pickle_module=dill)
```
|
github_jupyter
|
!pip install janome==0.4.1
!pip install torchvision==0.7.0
!pip install torchtext==0.7.0
!pip install torch==1.6.0
from google.colab import drive
drive.mount('/content/drive/')
import glob # ファイルの取得に使用
import json # jsonファイルの読み込みに使用
import re
path = "/content/drive/My Drive/live_ai_data/projectnextnlp-chat-dialogue-corpus/json" # フォルダの場所を指定
files = glob.glob(path + "/*/*.json") # ファイルの一覧
dialogues = [] # 複数の対話文を格納するリスト
file_count= 0 # ファイル数のカウント
for file in files:
with open(file, "r") as f:
json_dic = json.load(f)
dialogue = [] # 単一の対話
for turn in json_dic["turns"]:
annotations = turn["annotations"] # 注釈
speaker = turn["speaker"] # 発言者
utterance = turn["utterance"] # 発言
# 空の文章や、特殊文字や数字が含まれる文章は除く
if (utterance=="") or ("\\u" in utterance) or (re.search("\d", utterance)!=None):
dialogue.clear() # 対話をリセット
continue
utterance = utterance.replace(".", "。").replace(",", "、") # 全角
utterance = utterance.replace(".", "。").replace(",", "、") # 半角
utterance = utterance.split("。")[0]
if speaker=="U": # 発言者が人間であれば
dialogue.append(utterance)
else: # 発言者がシステムであれば
is_wrong = False
for annotation in annotations:
breakdown = annotation["breakdown"] # 分類
if breakdown=="X": # 1つでも不適切評価があれば
is_wrong = True
break
if is_wrong:
dialogue.clear() # 対話をリセット
else:
dialogue.append(utterance) # 不適切評価が無ければ対話に追加
if len(dialogue) >= 2: # 単一の会話が成立すれば
dialogues.append(dialogue.copy())
dialogue.pop(0) # 最初の要素を削除
file_count += 1
if file_count%100 == 0:
print("files:", file_count, "dialogues", len(dialogues))
print("files:", file_count, "dialogues", len(dialogues))
import re
from janome.tokenizer import Tokenizer
re_kanji = re.compile(r"^[\u4E00-\u9FD0]+$") # 漢字の検出用
re_katakana = re.compile(r"[\u30A1-\u30F4]+") # カタカナの検出用
j_tk = Tokenizer()
def wakati(text):
return [tok for tok in j_tk.tokenize(text, wakati=True)]
wakati_inp = [] # 単語に分割された入力文
wakati_rep = [] # 単語に分割された応答文
for dialogue in dialogues:
wakati_inp.append(wakati(dialogue[0])[:10])
wakati_rep.append(wakati(dialogue[1])[:10])
dialogues_plus = []
for i, w_inp in enumerate(wakati_inp): # 全ての入力文でループ
inp_count = 0 # ある入力から生成された対話文をカウント
for j, w_rep in enumerate(wakati_rep): # 全ての応答文でループ
if i==j:
dialogues_plus.append(["".join(w_inp), "".join(w_rep)])
continue
similarity = 0 # 類似度
for w in w_inp: # 入力文と同じ単語があり、それが漢字かカタカナであれば類似度を上げる
if (w in w_rep) and (re_kanji.fullmatch(w) or re_katakana.fullmatch(w)):
similarity += 1
if similarity >= 1:
dialogue_plus = ["".join(w_inp), "".join(w_rep)]
if dialogue_plus not in dialogues_plus:
dialogues_plus.append(dialogue_plus)
inp_count += 1
if inp_count >= 12: # ある入力から生成する対話文の上限
break
if i%1000 == 0:
print("i:", i, "dialogues_pus:", len(dialogues_plus))
print("i:", i, "dialogues_pus:", len(dialogues_plus))
dialogues = dialogues_plus
import csv
from sklearn.model_selection import train_test_split
dialogues_train, dialogues_test = train_test_split(dialogues, shuffle=True, test_size=0.05) # 5%がテストデータ
path = "/content/drive/My Drive/live_ai_data/" # 保存場所
with open(path+"dialogues_train.csv", "w") as f:
writer = csv.writer(f)
writer.writerows(dialogues_train)
with open(path+"dialogues_test.csv", "w") as f:
writer = csv.writer(f)
writer.writerows(dialogues_test)
import torch
import torchtext
from janome.tokenizer import Tokenizer
path = "/content/drive/My Drive/live_ai_data/" # 保存場所を指定
j_tk = Tokenizer()
def tokenizer(text):
return [tok for tok in j_tk.tokenize(text, wakati=True)] # 内包表記
# データセットの列を定義
input_field = torchtext.data.Field( # 入力文
sequential=True, # データ長さが可変かどうか
tokenize=tokenizer, # 前処理や単語分割などのための関数
batch_first=True, # バッチの次元を先頭に
lower=True # アルファベットを小文字に変換
)
reply_field = torchtext.data.Field( # 応答文
sequential=True, # データ長さが可変かどうか
tokenize=tokenizer, # 前処理や単語分割などのための関数
init_token = "<sos>", # 文章開始のトークン
eos_token = "<eos>", # 文章終了のトークン
batch_first=True, # バッチの次元を先頭に
lower=True # アルファベットを小文字に変換
)
# csvファイルからデータセットを作成
train_data, test_data = torchtext.data.TabularDataset.splits(
path=path,
train="dialogues_train.csv",
validation="dialogues_test.csv",
format="csv",
fields=[("inp_text", input_field), ("rep_text", reply_field)] # 列の設定
)
input_field.build_vocab(
train_data,
min_freq=3,
)
reply_field.build_vocab(
train_data,
min_freq=3,
)
print(input_field.vocab.freqs) # 各単語の出現頻度
print(len(input_field.vocab.stoi))
print(len(input_field.vocab.itos))
print(len(reply_field.vocab.stoi))
print(len(reply_field.vocab.itos))
import dill
torch.save(train_data.examples, path+"train_examples.pkl", pickle_module=dill)
torch.save(test_data.examples, path+"test_examples.pkl", pickle_module=dill)
torch.save(input_field, path+"input_field.pkl", pickle_module=dill)
torch.save(reply_field, path+"reply_field.pkl", pickle_module=dill)
| 0.172834 | 0.867204 |
```
from sklearn.linear_model import SGDRegressor
import pandas as pd
path = '/home/skkucman/dses/machine-learning-lecture-notes/inputs/winequality-data.csv'
dataset = pd.read_csv(path)
dataset.head()
# Column 이름에 '.'이 포함된 것을 '_'로 변경 (개인 취향)
import re
column_names = [re.sub('\.', '_', col) for col in dataset.columns.values]
dataset.columns = column_names # 컬럼 이름 변경
dataset.head()
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(dataset,
test_size = 0.3, random_state = 42)
features = dataset.columns.drop('quality')
features
x = dataset[features]
x.head(1)
y = dataset['quality']
y.head(1)
x_train, x_test, y_train, y_test = \
train_test_split(x, y, train_size=0.7, random_state=42)
x_train.head()
from sklearn.preprocessing import StandardScaler
# 각각을 standardization (평균 : 0, 표준편차 : 1)
scaler = StandardScaler()
transformer = scaler.fit(x_train)
scaled_x_train = transformer.transform(x_train)
scaled_x_train[:2]
lr_model = SGDRegressor()
lr_model.fit(scaled_x_train,y_train)
lr_model.score(scaled_x_train,y_train)
scaled_x_test = scaler.transform(x_test)
```
## 6. Ridge Regression (규제화)
```
from sklearn.linear_model import Ridge
# Ridge model에 사용할 lambdas
lambdas = [1e-4, 1e-3, 1e-2, 0.1, 0.5, 1.0, 5.0, 10.0]
from collections import defaultdict
r_squares = defaultdict(dict)
for lam in lambdas:
ridge_reg = Ridge(alpha=lam)
ridge_reg.fit(x_train, y_train)
r_squares[lam]['train'] = ridge_reg.score(x_train, y_train)
r_squares[lam]['test'] = ridge_reg.score(x_test, y_test)
r_squares
best_ridge_reg = Ridge(0.0001)
best_ridge_reg.fit(x_train, y_train)
for col, coef in zip(x_train.columns, best_ridge_reg.coef_):
print(col, ':', round(coef,5))
```
## 7. Lasso Regression( 규제화)
```
from sklearn.linear_model import Lasso
lasso_r_squares = defaultdict(dict)
for lam in lambdas:
lasso_reg = Lasso(alpha=lam)
lasso_reg.fit(x_train, y_train)
lasso_r_squares[lam]['train'] = lasso_reg.score(x_train, y_train)
lasso_r_squares[lam]['test'] = lasso_reg.score(x_test, y_test)
lasso_r_squares
best_lasso_reg = Lasso(0.0001)
best_lasso_reg.fit(x_test, y_test)
for col, coef in zip(x_train.columns, best_lasso_reg.coef_):
print(col, ':', round(coef, 5))
```
## Linear regression using <font color=red>Batch Gradient Descent</font>
```
import numpy as np
import os
np.random.seed(42)
import matplotlib as mpl
import matplotlib.pyplot as plt
mpl.rc('axes', labelsize=14)
mpl.rc('xtick', labelsize=12)
mpl.rc('ytick', labelsize=12)
import warnings
warnings.filterwarnings(action="ignore")
import numpy as np
X = 2 * np.random.rand(100, 1)
y = 4 + 3 * X + np.random.randn(100, 1)
eta = 0.1
n_iteration = 1000
m = 100
theta = np.random.randn(2,1)
X_b = np.c_[np.ones((100, 1)), X] # add x0 = 1 to each instance
for iteration in range(n_iteration):
gradients = 2/m * X_b.T.dot(X_b.dot(theta) - y)
theta = theta - eta * gradients
theta
# 새로운 데이터를 테스트 한다.
X_new = np.array([[0], [2]])
X_new_b = np.c_[np.ones((2, 1)), X_new] # add x0 = 1 to each instance
X_new_b.dot(theta)
theta_path_bgd = []
def plot_gradient_descent(theta, eta, theta_path=None):
m = len(X_b)
plt.plot(X, y, "b.")
n_iterations = 1000
for iteration in range(n_iterations):
if iteration < 10:
# 반복 횟수가 10번이 되기전까지만(처음 10개의 직선만) plotting
y_predict = X_new_b.dot(theta)
style = "b-" if iteration > 0 else "r--"
plt.plot(X_new, y_predict, style)
gradients = 2/m * X_b.T.dot(X_b.dot(theta) - y)
theta = theta - eta * gradients
# Theta의 변화량을 기록할 것이다! Why? 나중에 batch, mini-batch, stochastic의 수렴도를 그림으로 그릴것이다.
if theta_path is not None:
theta_path.append(theta)
plt.xlabel("$x_1$", fontsize=18)
plt.axis([0, 2, 0, 15])
plt.title(r"$\eta = {}$".format(eta), fontsize=16)
np.random.seed(42)
theta = np.random.randn(2,1) # random initialization
plt.figure(figsize=(10,4))
plt.subplot(131); plot_gradient_descent(theta, eta=0.02)
plt.ylabel("$y$", rotation=0, fontsize=18)
plt.subplot(132); plot_gradient_descent(theta, eta=0.1, theta_path=theta_path_bgd)
plt.subplot(133); plot_gradient_descent(theta, eta=0.5)
```
# MNIST와 scikit-learn을 활용한 binary classification tutorial
```
from keras.datasets import mnist
(x_train, y_train), (x_test, y_test) = mnist.load_data()
x_train.shape
x_test.shape
# 이미지 데이터를 plotting
%matplotlib inline
import matplotlib
import matplotlib.pyplot as plt
digit = x_train[10000]
plt.imshow(digit, cmap=matplotlib.cm.binary, interpolation='nearest')
plt.axis('off')
plt.show()
# 그 이미지의 label
y_train[10000]
y_train_5 = (y_train == 5) # 5는 True, 그 외는 False
y_test_5 = (y_test == 5) # 5는 True, 그 외는 False
y_train_5[:5]
y_train_5[:5]
print(x_train.shape)
print(y_train_5.shape)
# 확률적 경사 하강법 분류기를 사용하기 위해서는 input shape이 2 dim 이어야 한다.
import numpy as np
x_train_2d = np.reshape(x_train, (-1, 784))
x_test_2d = np.reshape(x_test, (-1, 784))
from sklearn.linear_model import SGDClassifier
sgd = SGDClassifier(max_iter=5, random_state=42)
# 훈련
sgd.fit(x_train_2d, y_train_5)
# 결과 check
pred = sgd.predict([x_train_2d[10]])
print('Prediction: ', pred)
print('Actual: ', y_train_5[10])
```
## Performance 측정
```
from sklearn.model_selection import StratifiedKFold
from sklearn.base import clone
# 데이터의 각 class 별 비율을, fold에서 맞추어준다.
skfolds = StratifiedKFold(n_splits=3, random_state=42)
for train_index, test_index in skfolds.split(x_train_2d, y_train_5):
clone_sgd = clone(sgd)
x_train_folds = x_train_2d[train_index]
y_train_folds = y_train_5[train_index]
x_test_folds = x_train_2d[test_index]
y_test_folds = y_train_5[test_index]
clone_sgd.fit(x_train_folds, y_train_folds)
y_pred = clone_sgd.predict(x_test_folds)
n_correct = sum(y_pred == y_test_folds)
print(n_correct / len(y_pred))
list(skfolds.split(x_train_2d,y_train_5))
from sklearn.model_selection import cross_val_score
cross_val_score(sgd, x_train_2d, y_train_5, cv=3, scoring='accuracy')
from collections import Counter
print('Training set:', Counter(y_train_5))
print('Test set:', Counter(y_test_5))
from sklearn.model_selection import cross_val_predict
y_train_pred = cross_val_predict(sgd, x_train_2d, y_train_5, cv=3)
from keras.datasets import mnist
(train_x, train_y), (test_x, test_y) = mnist.load_data()
import numpy as np
from sklearn.linear_model import SGDClassifier
train_x_2d = np.reshape(train_x, (-1, 784))
test_x_2d = np.reshape(test_x, (-1, 784))
# Sklearn에서는 multiclass classification에 대해서,
# binary classier를 사용하면 자동으로 OvA를 적용 (SVM의 경우에는 OvO 적용)
sgd = SGDClassifier(max_iter=5, random_state=42)
sgd.fit(train_x_2d, train_y)
sgd.predict([train_x_2d[2]])
train_y[2]
# SGDClassifier에서는 총 10개의 점수를 계산하고, 그 중에서 가장 큰 값을 선택
sgd.decision_function([train_x_2d[2]])
from sklearn.multiclass import OneVsOneClassifier
ovo_sgd = OneVsOneClassifier(SGDClassifier(max_iter=5, random_state=42))
ovo_sgd.fit(train_x_2d, train_y)
ovo_sgd.predict([train_x_2d[2]])
# OvO classifier 의 갯수
len(ovo_sgd.estimators_)
# Multiclass classifier의 성능
from sklearn.model_selection import cross_val_score
cross_val_score(sgd, train_x_2d, train_y, cv=3, scoring='accuracy')
from sklearn.preprocessing import StandardScaler
import numpy as np
scaler = StandardScaler()
train_x_2d_scaled = scaler.fit_transform(train_x_2d.astype(np.float64)) # Scaling
cross_val_score(sgd, train_x_2d_scaled, train_y, cv=3, scoring='accuracy')
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier()
rf.fit(train_x_2d, train_y)
rf.predict([train_x_2d[2]])
from sklearn.model_selection import cross_val_predict
train_y_pred = cross_val_predict(sgd, train_x_2d_scaled, train_y, cv = 3)
```
|
github_jupyter
|
from sklearn.linear_model import SGDRegressor
import pandas as pd
path = '/home/skkucman/dses/machine-learning-lecture-notes/inputs/winequality-data.csv'
dataset = pd.read_csv(path)
dataset.head()
# Column 이름에 '.'이 포함된 것을 '_'로 변경 (개인 취향)
import re
column_names = [re.sub('\.', '_', col) for col in dataset.columns.values]
dataset.columns = column_names # 컬럼 이름 변경
dataset.head()
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(dataset,
test_size = 0.3, random_state = 42)
features = dataset.columns.drop('quality')
features
x = dataset[features]
x.head(1)
y = dataset['quality']
y.head(1)
x_train, x_test, y_train, y_test = \
train_test_split(x, y, train_size=0.7, random_state=42)
x_train.head()
from sklearn.preprocessing import StandardScaler
# 각각을 standardization (평균 : 0, 표준편차 : 1)
scaler = StandardScaler()
transformer = scaler.fit(x_train)
scaled_x_train = transformer.transform(x_train)
scaled_x_train[:2]
lr_model = SGDRegressor()
lr_model.fit(scaled_x_train,y_train)
lr_model.score(scaled_x_train,y_train)
scaled_x_test = scaler.transform(x_test)
from sklearn.linear_model import Ridge
# Ridge model에 사용할 lambdas
lambdas = [1e-4, 1e-3, 1e-2, 0.1, 0.5, 1.0, 5.0, 10.0]
from collections import defaultdict
r_squares = defaultdict(dict)
for lam in lambdas:
ridge_reg = Ridge(alpha=lam)
ridge_reg.fit(x_train, y_train)
r_squares[lam]['train'] = ridge_reg.score(x_train, y_train)
r_squares[lam]['test'] = ridge_reg.score(x_test, y_test)
r_squares
best_ridge_reg = Ridge(0.0001)
best_ridge_reg.fit(x_train, y_train)
for col, coef in zip(x_train.columns, best_ridge_reg.coef_):
print(col, ':', round(coef,5))
from sklearn.linear_model import Lasso
lasso_r_squares = defaultdict(dict)
for lam in lambdas:
lasso_reg = Lasso(alpha=lam)
lasso_reg.fit(x_train, y_train)
lasso_r_squares[lam]['train'] = lasso_reg.score(x_train, y_train)
lasso_r_squares[lam]['test'] = lasso_reg.score(x_test, y_test)
lasso_r_squares
best_lasso_reg = Lasso(0.0001)
best_lasso_reg.fit(x_test, y_test)
for col, coef in zip(x_train.columns, best_lasso_reg.coef_):
print(col, ':', round(coef, 5))
import numpy as np
import os
np.random.seed(42)
import matplotlib as mpl
import matplotlib.pyplot as plt
mpl.rc('axes', labelsize=14)
mpl.rc('xtick', labelsize=12)
mpl.rc('ytick', labelsize=12)
import warnings
warnings.filterwarnings(action="ignore")
import numpy as np
X = 2 * np.random.rand(100, 1)
y = 4 + 3 * X + np.random.randn(100, 1)
eta = 0.1
n_iteration = 1000
m = 100
theta = np.random.randn(2,1)
X_b = np.c_[np.ones((100, 1)), X] # add x0 = 1 to each instance
for iteration in range(n_iteration):
gradients = 2/m * X_b.T.dot(X_b.dot(theta) - y)
theta = theta - eta * gradients
theta
# 새로운 데이터를 테스트 한다.
X_new = np.array([[0], [2]])
X_new_b = np.c_[np.ones((2, 1)), X_new] # add x0 = 1 to each instance
X_new_b.dot(theta)
theta_path_bgd = []
def plot_gradient_descent(theta, eta, theta_path=None):
m = len(X_b)
plt.plot(X, y, "b.")
n_iterations = 1000
for iteration in range(n_iterations):
if iteration < 10:
# 반복 횟수가 10번이 되기전까지만(처음 10개의 직선만) plotting
y_predict = X_new_b.dot(theta)
style = "b-" if iteration > 0 else "r--"
plt.plot(X_new, y_predict, style)
gradients = 2/m * X_b.T.dot(X_b.dot(theta) - y)
theta = theta - eta * gradients
# Theta의 변화량을 기록할 것이다! Why? 나중에 batch, mini-batch, stochastic의 수렴도를 그림으로 그릴것이다.
if theta_path is not None:
theta_path.append(theta)
plt.xlabel("$x_1$", fontsize=18)
plt.axis([0, 2, 0, 15])
plt.title(r"$\eta = {}$".format(eta), fontsize=16)
np.random.seed(42)
theta = np.random.randn(2,1) # random initialization
plt.figure(figsize=(10,4))
plt.subplot(131); plot_gradient_descent(theta, eta=0.02)
plt.ylabel("$y$", rotation=0, fontsize=18)
plt.subplot(132); plot_gradient_descent(theta, eta=0.1, theta_path=theta_path_bgd)
plt.subplot(133); plot_gradient_descent(theta, eta=0.5)
from keras.datasets import mnist
(x_train, y_train), (x_test, y_test) = mnist.load_data()
x_train.shape
x_test.shape
# 이미지 데이터를 plotting
%matplotlib inline
import matplotlib
import matplotlib.pyplot as plt
digit = x_train[10000]
plt.imshow(digit, cmap=matplotlib.cm.binary, interpolation='nearest')
plt.axis('off')
plt.show()
# 그 이미지의 label
y_train[10000]
y_train_5 = (y_train == 5) # 5는 True, 그 외는 False
y_test_5 = (y_test == 5) # 5는 True, 그 외는 False
y_train_5[:5]
y_train_5[:5]
print(x_train.shape)
print(y_train_5.shape)
# 확률적 경사 하강법 분류기를 사용하기 위해서는 input shape이 2 dim 이어야 한다.
import numpy as np
x_train_2d = np.reshape(x_train, (-1, 784))
x_test_2d = np.reshape(x_test, (-1, 784))
from sklearn.linear_model import SGDClassifier
sgd = SGDClassifier(max_iter=5, random_state=42)
# 훈련
sgd.fit(x_train_2d, y_train_5)
# 결과 check
pred = sgd.predict([x_train_2d[10]])
print('Prediction: ', pred)
print('Actual: ', y_train_5[10])
from sklearn.model_selection import StratifiedKFold
from sklearn.base import clone
# 데이터의 각 class 별 비율을, fold에서 맞추어준다.
skfolds = StratifiedKFold(n_splits=3, random_state=42)
for train_index, test_index in skfolds.split(x_train_2d, y_train_5):
clone_sgd = clone(sgd)
x_train_folds = x_train_2d[train_index]
y_train_folds = y_train_5[train_index]
x_test_folds = x_train_2d[test_index]
y_test_folds = y_train_5[test_index]
clone_sgd.fit(x_train_folds, y_train_folds)
y_pred = clone_sgd.predict(x_test_folds)
n_correct = sum(y_pred == y_test_folds)
print(n_correct / len(y_pred))
list(skfolds.split(x_train_2d,y_train_5))
from sklearn.model_selection import cross_val_score
cross_val_score(sgd, x_train_2d, y_train_5, cv=3, scoring='accuracy')
from collections import Counter
print('Training set:', Counter(y_train_5))
print('Test set:', Counter(y_test_5))
from sklearn.model_selection import cross_val_predict
y_train_pred = cross_val_predict(sgd, x_train_2d, y_train_5, cv=3)
from keras.datasets import mnist
(train_x, train_y), (test_x, test_y) = mnist.load_data()
import numpy as np
from sklearn.linear_model import SGDClassifier
train_x_2d = np.reshape(train_x, (-1, 784))
test_x_2d = np.reshape(test_x, (-1, 784))
# Sklearn에서는 multiclass classification에 대해서,
# binary classier를 사용하면 자동으로 OvA를 적용 (SVM의 경우에는 OvO 적용)
sgd = SGDClassifier(max_iter=5, random_state=42)
sgd.fit(train_x_2d, train_y)
sgd.predict([train_x_2d[2]])
train_y[2]
# SGDClassifier에서는 총 10개의 점수를 계산하고, 그 중에서 가장 큰 값을 선택
sgd.decision_function([train_x_2d[2]])
from sklearn.multiclass import OneVsOneClassifier
ovo_sgd = OneVsOneClassifier(SGDClassifier(max_iter=5, random_state=42))
ovo_sgd.fit(train_x_2d, train_y)
ovo_sgd.predict([train_x_2d[2]])
# OvO classifier 의 갯수
len(ovo_sgd.estimators_)
# Multiclass classifier의 성능
from sklearn.model_selection import cross_val_score
cross_val_score(sgd, train_x_2d, train_y, cv=3, scoring='accuracy')
from sklearn.preprocessing import StandardScaler
import numpy as np
scaler = StandardScaler()
train_x_2d_scaled = scaler.fit_transform(train_x_2d.astype(np.float64)) # Scaling
cross_val_score(sgd, train_x_2d_scaled, train_y, cv=3, scoring='accuracy')
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier()
rf.fit(train_x_2d, train_y)
rf.predict([train_x_2d[2]])
from sklearn.model_selection import cross_val_predict
train_y_pred = cross_val_predict(sgd, train_x_2d_scaled, train_y, cv = 3)
| 0.615088 | 0.838548 |
# Twitter Sentiment Analysis
Adapted from [Paolo Ripamonti's Kaggle notebook](https://www.kaggle.com/paoloripamonti/twitter-sentiment-analysis/)

```
# !pip install gensim
```
# Read Dataset
### Dataset details
* **target**: the polarity of the tweet (0 = negative, 2 = neutral, 4 = positive)
* **ids**: The id of the tweet ( 2087)
* **date**: the date of the tweet (Sat May 16 23:58:44 UTC 2009)
* **flag**: The query (lyx). If there is no query, then this value is NO_QUERY.
* **user**: the user that tweeted (robotickilldozr)
* **text**: the text of the tweet (Lyx is cool)
```
import sys
from google.colab import drive
drive.mount('/gdrive')
drive_path = '/gdrive/My Drive/Open Source Spotlight/Flask/'
sys.path.append(drive_path)
import pandas as pd
DATASET_COLUMNS = ["target", "ids", "date", "flag", "user", "text"]
DATASET_ENCODING = "ISO-8859-1"
df = pd.read_csv(drive_path+'/training.1600000.processed.noemoticon.csv',
encoding=DATASET_ENCODING , names=DATASET_COLUMNS)
print('Dataset loaded successfuly!')
print("Dataset size:", len(df))
df.head()
```
### Map target label to String
* **0** -> **NEGATIVE**
* **2** -> **NEUTRAL**
* **4** -> **POSITIVE**
```
decode_map = {0: "Negative", 2: "Neutral", 4: "Positive"}
def decode_sentiment(label):
return decode_map[int(label)]
df.target = df.target.apply(lambda x: decode_sentiment(x))
df.target.value_counts()
```
### Pre-process dataset
```
import nltk
nltk.download('stopwords')
from nltk.corpus import stopwords
from nltk.stem import SnowballStemmer
stop_words = stopwords.words("english")
stemmer = SnowballStemmer("english")
import re
TEXT_CLEANING_RE = "@\S+|https?:\S+|http?:\S|[^A-Za-z0-9]+"
def preprocess(text, stem=False):
# Remove link, user and special characters
text = re.sub(TEXT_CLEANING_RE, ' ', str(text).lower()).strip()
tokens = []
for token in text.split():
if token not in stop_words:
if stem:
tokens.append(stemmer.stem(token))
else:
tokens.append(token)
return " ".join(tokens)
%%time
df.text = df.text.apply(lambda x: preprocess(x))
```
### Split train and test
```
from sklearn.model_selection import train_test_split
df_train, df_test = train_test_split(df, test_size=0.2, random_state=42)
print("Train size:", len(df_train))
print("Test size:", len(df_test))
```
### Word2Vec
```
documents = [_text.split() for _text in df_train.text]
import gensim
# Word2Vec Parameters
W2V_SIZE = 300
W2V_WINDOW = 7
W2V_EPOCH = 32
W2V_MIN_COUNT = 10
w2v_model = gensim.models.word2vec.Word2Vec(size=W2V_SIZE,
window=W2V_WINDOW,
min_count=W2V_MIN_COUNT,
workers=8)
w2v_model.build_vocab(documents)
words = w2v_model.wv.vocab.keys()
vocab_size = len(words)
print("Vocab size", vocab_size)
%%time
w2v_model.train(documents, total_examples=len(documents), epochs=W2V_EPOCH)
# sanity check
w2v_model.most_similar("love")
```
### Tokenize Text
```
%%time
from keras.preprocessing.text import Tokenizer
tokenizer = Tokenizer()
tokenizer.fit_on_texts(df_train.text)
vocab_size = len(tokenizer.word_index) + 1
print("Total words", vocab_size)
%%time
from keras.preprocessing.sequence import pad_sequences
SEQUENCE_LENGTH = 300
x_train = pad_sequences(tokenizer.texts_to_sequences(df_train.text), maxlen=SEQUENCE_LENGTH)
x_test = pad_sequences(tokenizer.texts_to_sequences(df_test.text), maxlen=SEQUENCE_LENGTH)
```
### Label Encoder
```
labels = df_train.target.unique().tolist()
labels.append('Neutral')
labels
from sklearn.preprocessing import LabelEncoder
encoder = LabelEncoder()
encoder.fit(df_train.target.tolist())
y_train = encoder.transform(df_train.target.tolist())
y_test = encoder.transform(df_test.target.tolist())
y_train = y_train.reshape(-1, 1)
y_test = y_test.reshape(-1, 1)
print("x_train", x_train.shape)
print("y_train", y_train.shape)
print('------------------------')
print("x_test", x_test.shape)
print("y_test", y_test.shape)
```
### Embedding layer
```
embedding_matrix = np.zeros((vocab_size, W2V_SIZE))
for word, i in tokenizer.word_index.items():
if word in w2v_model.wv:
embedding_matrix[i] = w2v_model.wv[word]
print(embedding_matrix.shape)
```
### Build Model
```
from keras.models import Sequential
from keras.layers import Dropout, LSTM, Dense, Embedding
model = Sequential()
model.add(Embedding(vocab_size, W2V_SIZE, weights=[embedding_matrix], input_length=SEQUENCE_LENGTH, trainable=False))
model.add(Dropout(0.5))
model.add(LSTM(100, dropout=0.2, recurrent_dropout=0.2))
model.add(Dense(1, activation='sigmoid'))
model.summary()
# compile model
model.compile(loss='binary_crossentropy',
optimizer="adam",
metrics=['accuracy'])
from keras.callbacks import ReduceLROnPlateau, EarlyStopping
callbacks = [ ReduceLROnPlateau(monitor='val_loss', patience=5, cooldown=0),
EarlyStopping(monitor='val_acc', min_delta=1e-4, patience=5)]
```
### Train
```
%%time
EPOCHS = 8
BATCH_SIZE = 1024
training_log = model.fit(x_train, y_train,
batch_size=BATCH_SIZE,
epochs=EPOCHS,
validation_split=0.1,
verbose=1,
callbacks=callbacks)
```
### Evaluate
```
%%time
model.load
score = model.evaluate(x_test, y_test, batch_size=BATCH_SIZE)
print()
print("ACCURACY:", score[1])
print("LOSS:", score[0])
```
### Predict
```
predict("I love the music")
predict("I hate the rain")
predict("i don't know what i'm doing")
```
### Save model
```
import pickle
KERAS_MODEL = "model.h5"
TOKENIZER_MODEL = "tokenizer.pkl"
WORD2VEC_MODEL = "model.w2v"
ENCODER_MODEL = "encoder.pkl"
model.save(KERAS_MODEL)
w2v_model.save(WORD2VEC_MODEL) # only needed for fine-tuning / re-training
pickle.dump(tokenizer, open(TOKENIZER_MODEL, "wb"), protocol=0)
pickle.dump(encoder, open(ENCODER_MODEL, "wb"), protocol=0) # same
```
|
github_jupyter
|
# !pip install gensim
import sys
from google.colab import drive
drive.mount('/gdrive')
drive_path = '/gdrive/My Drive/Open Source Spotlight/Flask/'
sys.path.append(drive_path)
import pandas as pd
DATASET_COLUMNS = ["target", "ids", "date", "flag", "user", "text"]
DATASET_ENCODING = "ISO-8859-1"
df = pd.read_csv(drive_path+'/training.1600000.processed.noemoticon.csv',
encoding=DATASET_ENCODING , names=DATASET_COLUMNS)
print('Dataset loaded successfuly!')
print("Dataset size:", len(df))
df.head()
decode_map = {0: "Negative", 2: "Neutral", 4: "Positive"}
def decode_sentiment(label):
return decode_map[int(label)]
df.target = df.target.apply(lambda x: decode_sentiment(x))
df.target.value_counts()
import nltk
nltk.download('stopwords')
from nltk.corpus import stopwords
from nltk.stem import SnowballStemmer
stop_words = stopwords.words("english")
stemmer = SnowballStemmer("english")
import re
TEXT_CLEANING_RE = "@\S+|https?:\S+|http?:\S|[^A-Za-z0-9]+"
def preprocess(text, stem=False):
# Remove link, user and special characters
text = re.sub(TEXT_CLEANING_RE, ' ', str(text).lower()).strip()
tokens = []
for token in text.split():
if token not in stop_words:
if stem:
tokens.append(stemmer.stem(token))
else:
tokens.append(token)
return " ".join(tokens)
%%time
df.text = df.text.apply(lambda x: preprocess(x))
from sklearn.model_selection import train_test_split
df_train, df_test = train_test_split(df, test_size=0.2, random_state=42)
print("Train size:", len(df_train))
print("Test size:", len(df_test))
documents = [_text.split() for _text in df_train.text]
import gensim
# Word2Vec Parameters
W2V_SIZE = 300
W2V_WINDOW = 7
W2V_EPOCH = 32
W2V_MIN_COUNT = 10
w2v_model = gensim.models.word2vec.Word2Vec(size=W2V_SIZE,
window=W2V_WINDOW,
min_count=W2V_MIN_COUNT,
workers=8)
w2v_model.build_vocab(documents)
words = w2v_model.wv.vocab.keys()
vocab_size = len(words)
print("Vocab size", vocab_size)
%%time
w2v_model.train(documents, total_examples=len(documents), epochs=W2V_EPOCH)
# sanity check
w2v_model.most_similar("love")
%%time
from keras.preprocessing.text import Tokenizer
tokenizer = Tokenizer()
tokenizer.fit_on_texts(df_train.text)
vocab_size = len(tokenizer.word_index) + 1
print("Total words", vocab_size)
%%time
from keras.preprocessing.sequence import pad_sequences
SEQUENCE_LENGTH = 300
x_train = pad_sequences(tokenizer.texts_to_sequences(df_train.text), maxlen=SEQUENCE_LENGTH)
x_test = pad_sequences(tokenizer.texts_to_sequences(df_test.text), maxlen=SEQUENCE_LENGTH)
labels = df_train.target.unique().tolist()
labels.append('Neutral')
labels
from sklearn.preprocessing import LabelEncoder
encoder = LabelEncoder()
encoder.fit(df_train.target.tolist())
y_train = encoder.transform(df_train.target.tolist())
y_test = encoder.transform(df_test.target.tolist())
y_train = y_train.reshape(-1, 1)
y_test = y_test.reshape(-1, 1)
print("x_train", x_train.shape)
print("y_train", y_train.shape)
print('------------------------')
print("x_test", x_test.shape)
print("y_test", y_test.shape)
embedding_matrix = np.zeros((vocab_size, W2V_SIZE))
for word, i in tokenizer.word_index.items():
if word in w2v_model.wv:
embedding_matrix[i] = w2v_model.wv[word]
print(embedding_matrix.shape)
from keras.models import Sequential
from keras.layers import Dropout, LSTM, Dense, Embedding
model = Sequential()
model.add(Embedding(vocab_size, W2V_SIZE, weights=[embedding_matrix], input_length=SEQUENCE_LENGTH, trainable=False))
model.add(Dropout(0.5))
model.add(LSTM(100, dropout=0.2, recurrent_dropout=0.2))
model.add(Dense(1, activation='sigmoid'))
model.summary()
# compile model
model.compile(loss='binary_crossentropy',
optimizer="adam",
metrics=['accuracy'])
from keras.callbacks import ReduceLROnPlateau, EarlyStopping
callbacks = [ ReduceLROnPlateau(monitor='val_loss', patience=5, cooldown=0),
EarlyStopping(monitor='val_acc', min_delta=1e-4, patience=5)]
%%time
EPOCHS = 8
BATCH_SIZE = 1024
training_log = model.fit(x_train, y_train,
batch_size=BATCH_SIZE,
epochs=EPOCHS,
validation_split=0.1,
verbose=1,
callbacks=callbacks)
%%time
model.load
score = model.evaluate(x_test, y_test, batch_size=BATCH_SIZE)
print()
print("ACCURACY:", score[1])
print("LOSS:", score[0])
predict("I love the music")
predict("I hate the rain")
predict("i don't know what i'm doing")
import pickle
KERAS_MODEL = "model.h5"
TOKENIZER_MODEL = "tokenizer.pkl"
WORD2VEC_MODEL = "model.w2v"
ENCODER_MODEL = "encoder.pkl"
model.save(KERAS_MODEL)
w2v_model.save(WORD2VEC_MODEL) # only needed for fine-tuning / re-training
pickle.dump(tokenizer, open(TOKENIZER_MODEL, "wb"), protocol=0)
pickle.dump(encoder, open(ENCODER_MODEL, "wb"), protocol=0) # same
| 0.370795 | 0.811415 |
# Classification and Regression using Decision Trees
This notebook walks you through two examples using Decision Trees, one for classification, and the other for regression. We will implement Decision Trees using Scikit-Learn. We will also use the Graphviz package to visualize the decision trees, which has proven to be a very useful way of understanding how Decision Trees work. <br>
<br>
Author: Jiajia Sun @ University of Houston, 02/21/2019
## 1. Classification of Iris flowers
We will use the famour Iris data set as an example to illustrate how to implement Decision Trees in Scikit-Learn.
### 1.1. Import Scikit-Learn packages
```
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import load_iris
from sklearn.tree import DecisionTreeClassifier
from sklearn.tree import export_graphviz
```
### 1.2 Get the training data ready
For this demo, we are going to use only two features, petal length and width measurements, to classify the Iris flowers.
```
iris = load_iris()
X = iris.data[:, 2:] # petal length and width
y = iris.target
```
### 1.3 Set up the classifier
Now, we need to specify what kind of decision trees we want to have. For example, how many depths (i.e., max_depth)? What is the minimum number of samples that a leaf node must have (i.e., min_samples_leaf)? These are called hyperparameters; they impose different constraints on the learning process. Note that they are NOT the model parameters that a machine learning algorithm tries to learn during training. Hyperparameters are set before the learning (or training) takes place.
```
tree_clf = DecisionTreeClassifier(max_depth=2, random_state=42)
```
In the above example, I set max_depth = 2. Please feell free to try different values.
### 1.4 Training
This is probably the simplest part, because all you need to do is run the code **name_of_my_classifier<font color=red>.fit</font>** followed by your training data with labels.
```
tree_clf.fit(X, y)
```
### 1.5 Visualization
This helps us actually see the decision tree that our algorithm just trained using Iris data set.
```
import graphviz
dot_data = export_graphviz(tree_clf, out_file=None)
graph = graphviz.Source(dot_data)
graph.render("iris")
```
Altenatively, the *export_graphviz* exporter also supports a variety of aesthetic options, including coloring nodes by their class (or value for regression) and using explicit variable and class names if desired. Jupyter notebooks also render these plots inline automatically:
```
dot_data = export_graphviz(tree_clf, out_file=None,
feature_names=iris.feature_names[2:],
class_names=iris.target_names,
filled=True, rounded=True,
special_characters=True)
graph = graphviz.Source(dot_data)
graph
```
Great! This is the actual tree that you just trained using **<font color=red>.fit(X,y)</font>**.
### Note
For this notebook, we are only interested in how to train a decision tree using Scikit-learn package. Therefore, I did not split the entire Iris dataset into training and validation sets. For practical ML problems, you always should split your data into at least two sets: training and validation sets, and ideally, three sets: training, validation and testing sets.
## 2. Regression
As we discussed in class, Decision Trees are also capable of performing regression tasks. Next, let us first generate a synthetic set of data and then perform a regression using Decision Trees.
### 2.1 Generate synthetic data
```
# Quadratic training set + noise
np.random.seed(42)
m = 200
X = np.random.rand(m, 1)
y = 4 * (X - 0.5) ** 2
y = y + np.random.randn(m, 1) / 10
plt.plot(X,y,'b.')
plt.axis([0, 1, -0.2, 1])
plt.xlabel("$x_1$", fontsize=18)
plt.ylabel("$y$",fontsize=18,rotation=0)
```
### 2.2 Import the regression version of Decision Trees
```
from sklearn.tree import DecisionTreeRegressor
```
Pay attention to the name of the module we imports which is differnt from the name of the module for classification.
### 2.3 Set up classifier
```
tree_reg = DecisionTreeRegressor(max_depth=2, random_state=42)
```
I set max_depth = 2. Again, feel free to play with it.
### 2.3 Training
```
tree_reg.fit(X, y)
```
### 2.4 Visualization
```
def plot_regression_predictions(tree_reg, X, y, axes=[0, 1, -0.2, 1], ylabel="$y$"):
x1 = np.linspace(axes[0], axes[1], 500).reshape(-1, 1)
y_pred = tree_reg.predict(x1)
plt.axis(axes)
plt.xlabel("$x_1$", fontsize=18)
if ylabel:
plt.ylabel(ylabel, fontsize=18, rotation=0)
plt.plot(X, y, "b.")
plt.plot(x1, y_pred, "r.-", linewidth=2, label=r"$\hat{y}$")
plt.figure(figsize=(11, 4))
plot_regression_predictions(tree_reg, X, y)
for split, style in ((0.1973, "k-"), (0.0917, "k--"), (0.7718, "k--")):
plt.plot([split, split], [-0.2, 1], style, linewidth=2)
plt.text(0.21, 0.65, "Depth=0", fontsize=15)
plt.text(0.01, 0.2, "Depth=1", fontsize=13)
plt.text(0.65, 0.8, "Depth=1", fontsize=13)
plt.legend(loc="upper center", fontsize=18)
plt.title("max_depth=2", fontsize=14)
plt.show()
```
Alternatively, we can also export the decision tree using *export_graphviz*.
```
dot_data = export_graphviz(
tree_reg,
out_file=None,
feature_names=["x1"],
rounded=True,
filled=True
)
graph = graphviz.Source(dot_data)
graph
```
### I hope this notebook gives you a better sense of how decision tree works and how it is implemented using Scikit-Learn.
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import load_iris
from sklearn.tree import DecisionTreeClassifier
from sklearn.tree import export_graphviz
iris = load_iris()
X = iris.data[:, 2:] # petal length and width
y = iris.target
tree_clf = DecisionTreeClassifier(max_depth=2, random_state=42)
tree_clf.fit(X, y)
import graphviz
dot_data = export_graphviz(tree_clf, out_file=None)
graph = graphviz.Source(dot_data)
graph.render("iris")
dot_data = export_graphviz(tree_clf, out_file=None,
feature_names=iris.feature_names[2:],
class_names=iris.target_names,
filled=True, rounded=True,
special_characters=True)
graph = graphviz.Source(dot_data)
graph
# Quadratic training set + noise
np.random.seed(42)
m = 200
X = np.random.rand(m, 1)
y = 4 * (X - 0.5) ** 2
y = y + np.random.randn(m, 1) / 10
plt.plot(X,y,'b.')
plt.axis([0, 1, -0.2, 1])
plt.xlabel("$x_1$", fontsize=18)
plt.ylabel("$y$",fontsize=18,rotation=0)
from sklearn.tree import DecisionTreeRegressor
tree_reg = DecisionTreeRegressor(max_depth=2, random_state=42)
tree_reg.fit(X, y)
def plot_regression_predictions(tree_reg, X, y, axes=[0, 1, -0.2, 1], ylabel="$y$"):
x1 = np.linspace(axes[0], axes[1], 500).reshape(-1, 1)
y_pred = tree_reg.predict(x1)
plt.axis(axes)
plt.xlabel("$x_1$", fontsize=18)
if ylabel:
plt.ylabel(ylabel, fontsize=18, rotation=0)
plt.plot(X, y, "b.")
plt.plot(x1, y_pred, "r.-", linewidth=2, label=r"$\hat{y}$")
plt.figure(figsize=(11, 4))
plot_regression_predictions(tree_reg, X, y)
for split, style in ((0.1973, "k-"), (0.0917, "k--"), (0.7718, "k--")):
plt.plot([split, split], [-0.2, 1], style, linewidth=2)
plt.text(0.21, 0.65, "Depth=0", fontsize=15)
plt.text(0.01, 0.2, "Depth=1", fontsize=13)
plt.text(0.65, 0.8, "Depth=1", fontsize=13)
plt.legend(loc="upper center", fontsize=18)
plt.title("max_depth=2", fontsize=14)
plt.show()
dot_data = export_graphviz(
tree_reg,
out_file=None,
feature_names=["x1"],
rounded=True,
filled=True
)
graph = graphviz.Source(dot_data)
graph
| 0.605916 | 0.99015 |
# [Lab 1] Non-regularized regression
### 1) Read the dataset given in the provided file data lab1.txt and plot the output value as a function of the input data.
```
import numpy as np
import matplotlib.pyplot as plt
data = np.loadtxt("data_lab1.txt")
split = 70
np.random.shuffle(data)
train, test = data[:split],data[split:]
X_train = train[:,0]
y_train = train[:,1]
X_test = test[:,0]
y_test = test[:,1]
plt.scatter(X_train,y_train,marker="+")
plt.scatter(X_test,y_test,marker="+")
plt.xlabel("X")
plt.ylabel("y")
plt.title("data_lab1")
X0 = np.zeros(X_test.size)+1
X_test = np.stack((X0,X_test), axis=1)
X0 = np.zeros(X_train.size)+1
X_train = np.stack((X0,X_train), axis=1)
X_train.shape
```
### 2) Fit the univariate linear regression parameters to the dataset using batch gradient descent. What are the optimal values of the parameters?
```
def hypothesis(X, thetas): #X= inputs, theta0= parameter 0 , theta1= parameter 1
return X*thetas[1] + thetas[0] # cf lecture1 (equation of a line: ax +b)
def cost_fct(y,predictions): #y: groundtruth (data_lab1) ,predictions: application of our hypothesis over our inputs
return (1/2)*np.sum(np.square(predictions-y)) # cf lecture1, what we want to minimize
def gradient_descent(X, y, thetas, alpha, epsilon): #alpha: learning rate, epsilon: we stop when the cost converge (the difference between last two cost < epsilon)
cost = 10000000 #inf
cost_prev = 1
past_costs = [] #save values to make evolution graphs later
past_theta0 = [] #save values to make evolution graphs later
past_theta1 = []
n_iter=0
while (epsilon < abs(cost - cost_prev)): #cf leacture1, until convergence of cost
n_iter+=1 #save for stats
predictions = hypothesis(X[:,1], thetas) #calculate predictions
cost_prev = cost #save old cost
cost = cost_fct(y,predictions) #calculate new cost
past_costs.append(cost)
#print("cost=",cost," eps=", abs(cost - cost_prev)," error = ",np.sum((predictions - y)))
for n in range (np.size(X,1)):
thetas[n] = thetas[n] - (alpha*np.sum((predictions - y)*X[:,n]))
past_theta0.append(thetas[0])
past_theta1.append(thetas[1])
return thetas,past_theta0,past_theta1,past_costs,n_iter
alpha = 0.001 #Step size
epsilon = 0.001 #Difference between last two costs (is the model is improving?)
thetas = np.array([0.0,1.0])
plt.title("Line before fit")
x=np.arange(3)
plt.plot(x,x*thetas[1]+thetas[0])
plt.scatter(X_train[:,1],y_train,marker="+")
plt.scatter(X_test[:,1],y_test,marker="+")
plt.xlabel("X")
plt.ylabel("y")
thetas,past_theta0,past_theta1, past_costs,n_iter = gradient_descent(X_train, y_train, thetas, alpha, epsilon)
print("Iteration =", n_iter, "Theta 1 =", thetas[1], " Theta 0 =", thetas[0])
save = np.array([thetas[0],thetas[1]])
plt.title("Fitted with BGD")
x=np.arange(3)
plt.plot(x,x*thetas[1]+thetas[0])
plt.scatter(X_train[:,1],y_train,marker="+")
plt.scatter(X_test[:,1],y_test,marker="+")
plt.xlabel("X")
plt.ylabel("y")
plt.title('Evolution of thetas: blue:0, yellow:1')
plt.xlabel('No. of iterations')
plt.ylabel('Value of thetas')
plt.plot(past_theta0)
plt.plot(past_theta1)
plt.title('Cost Function J')
plt.xlabel('No. of iterations')
plt.ylabel('Cost')
plt.plot(past_costs)
```
### 3) Fit the univariate linear regression parameters to the dataset using stochastic gradient descent. What are the optimal values of the parameters?
```
import random
def stochastic_gradient_descent(X, y, thetas, alpha,n_iter=1500): #alpha: learning rate, epsilon: we stop when the cost converge (the difference between last two cost < epsilon)
cost = 10000000 #inf
cost_prev = 1
past_costs = [] #save values to make evolution graphs later
past_theta0 = [] #save values to make evolution graphs later
past_theta1 = [] #save values to make evolution graphs later
for z in range(n_iter):
i = random.randint(0,X.shape[0]-1)
for n in range (np.size(X,1)):
predictions = hypothesis(X[i,1], thetas) #calculate predictions
cost_prev = cost #save old cost
cost = cost_fct(y,predictions) #calculate new cost
past_costs.append(cost)
#print("cost=",cost," eps=", abs(cost - cost_prev)," error = ",np.sum((predictions - y)))
thetas[n] = thetas[n] - (alpha*(predictions - y[i])*X[i,n]) #cf lecture1, update rule for theta 1
past_theta0.append(thetas[0])
past_theta1.append(thetas[1])
return thetas,past_theta0,past_theta1,past_costs,n_iter
alpha = 0.01 #Step size
thetas = np.array([0.0,1.0])
plt.title("Before fit")
x=np.arange(3)
plt.plot(x,x*thetas[1]+thetas[0])
plt.scatter(X_train[:,1],y_train,marker="+")
plt.scatter(X_test[:,1],y_test,marker="+")
plt.xlabel("X")
plt.ylabel("y")
thetas,past_theta0,past_theta1, past_costs,n_iter = stochastic_gradient_descent(X_train, y_train, thetas, alpha)
print("Iteration =", n_iter, "Theta 1 =", thetas[1], " Theta 0 =", thetas[0])
save = np.concatenate((save,[thetas[0],thetas[1]]), axis=0)
plt.title("Fitted with SGD")
x=np.arange(3)
plt.plot(x,x*thetas[1]+thetas[0])
plt.scatter(X_train[:,1],y_train,marker="+")
plt.scatter(X_test[:,1],y_test,marker="+")
plt.xlabel("X")
plt.ylabel("y")
plt.title('Evolution of thetas: blue:0, yellow:1')
plt.xlabel('No. of iterations')
plt.ylabel('Value of thetas')
plt.plot(past_theta0)
plt.plot(past_theta1)
plt.title('Cost Function J')
plt.xlabel('No. of iterations')
plt.ylabel('Cost')
plt.plot(past_costs)
```
### 4) Fit the univariate linear regression parameters to the dataset using the closed-form method. What are the optimal values of the parameters?
```
from numpy.linalg import inv
def closed_form(X, y):
thetas = np.dot(np.dot(inv(np.dot(X.T,X)),X.T),(y))
return thetas
thetas = closed_form(X_train, y_train)
print("Theta 1 =", thetas[1], " Theta 0 =", thetas[0])
save = np.concatenate((save,[thetas[0],thetas[1]]), axis=0)
plt.title("Fitted with closed form method")
x=np.arange(3)
plt.plot(x,x*thetas[1]+thetas[0])
plt.scatter(X_train[:,1],y_train,marker="+")
plt.scatter(X_test[:,1],y_test,marker="+")
plt.xlabel("X")
plt.ylabel("y")
```
### 5) Plot the linear regressors obtained in 2), 3) and 4) over the original dataset.
```
save=save.reshape((3, 2))
save
def mse(X,y,thetas):
predictions = hypothesis(X[:,1],thetas)
return (1/y.size)*np.sum(np.square(predictions-y))
y_test.size
plt.title("All three methods: yellow: Batch, green: stochastic, red:closed form")
plt.xlabel("X")
plt.ylabel("y")
plt.scatter(X_train[:,1],y_train,marker="+")
plt.scatter(X_test[:,1],y_test,marker="+")
x=np.arange(3)
MSE = []
for i in range (np.size(save,0)):
plt.plot(x,x*save[i,1]+save[i,0])
MSE = np.append(MSE,mse(X_test, y_test,save[i]))
```
### 6) Test your model choosing yourself some new input data. Plot also these results.
```
MSE
plt.title("MSE comparaison")
plt.ylabel("MSE")
plt.bar(x, MSE)
plt.xticks(x, ('BGD', 'SGD', 'CFM'))
plt.show()
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
data = np.loadtxt("data_lab1.txt")
split = 70
np.random.shuffle(data)
train, test = data[:split],data[split:]
X_train = train[:,0]
y_train = train[:,1]
X_test = test[:,0]
y_test = test[:,1]
plt.scatter(X_train,y_train,marker="+")
plt.scatter(X_test,y_test,marker="+")
plt.xlabel("X")
plt.ylabel("y")
plt.title("data_lab1")
X0 = np.zeros(X_test.size)+1
X_test = np.stack((X0,X_test), axis=1)
X0 = np.zeros(X_train.size)+1
X_train = np.stack((X0,X_train), axis=1)
X_train.shape
def hypothesis(X, thetas): #X= inputs, theta0= parameter 0 , theta1= parameter 1
return X*thetas[1] + thetas[0] # cf lecture1 (equation of a line: ax +b)
def cost_fct(y,predictions): #y: groundtruth (data_lab1) ,predictions: application of our hypothesis over our inputs
return (1/2)*np.sum(np.square(predictions-y)) # cf lecture1, what we want to minimize
def gradient_descent(X, y, thetas, alpha, epsilon): #alpha: learning rate, epsilon: we stop when the cost converge (the difference between last two cost < epsilon)
cost = 10000000 #inf
cost_prev = 1
past_costs = [] #save values to make evolution graphs later
past_theta0 = [] #save values to make evolution graphs later
past_theta1 = []
n_iter=0
while (epsilon < abs(cost - cost_prev)): #cf leacture1, until convergence of cost
n_iter+=1 #save for stats
predictions = hypothesis(X[:,1], thetas) #calculate predictions
cost_prev = cost #save old cost
cost = cost_fct(y,predictions) #calculate new cost
past_costs.append(cost)
#print("cost=",cost," eps=", abs(cost - cost_prev)," error = ",np.sum((predictions - y)))
for n in range (np.size(X,1)):
thetas[n] = thetas[n] - (alpha*np.sum((predictions - y)*X[:,n]))
past_theta0.append(thetas[0])
past_theta1.append(thetas[1])
return thetas,past_theta0,past_theta1,past_costs,n_iter
alpha = 0.001 #Step size
epsilon = 0.001 #Difference between last two costs (is the model is improving?)
thetas = np.array([0.0,1.0])
plt.title("Line before fit")
x=np.arange(3)
plt.plot(x,x*thetas[1]+thetas[0])
plt.scatter(X_train[:,1],y_train,marker="+")
plt.scatter(X_test[:,1],y_test,marker="+")
plt.xlabel("X")
plt.ylabel("y")
thetas,past_theta0,past_theta1, past_costs,n_iter = gradient_descent(X_train, y_train, thetas, alpha, epsilon)
print("Iteration =", n_iter, "Theta 1 =", thetas[1], " Theta 0 =", thetas[0])
save = np.array([thetas[0],thetas[1]])
plt.title("Fitted with BGD")
x=np.arange(3)
plt.plot(x,x*thetas[1]+thetas[0])
plt.scatter(X_train[:,1],y_train,marker="+")
plt.scatter(X_test[:,1],y_test,marker="+")
plt.xlabel("X")
plt.ylabel("y")
plt.title('Evolution of thetas: blue:0, yellow:1')
plt.xlabel('No. of iterations')
plt.ylabel('Value of thetas')
plt.plot(past_theta0)
plt.plot(past_theta1)
plt.title('Cost Function J')
plt.xlabel('No. of iterations')
plt.ylabel('Cost')
plt.plot(past_costs)
import random
def stochastic_gradient_descent(X, y, thetas, alpha,n_iter=1500): #alpha: learning rate, epsilon: we stop when the cost converge (the difference between last two cost < epsilon)
cost = 10000000 #inf
cost_prev = 1
past_costs = [] #save values to make evolution graphs later
past_theta0 = [] #save values to make evolution graphs later
past_theta1 = [] #save values to make evolution graphs later
for z in range(n_iter):
i = random.randint(0,X.shape[0]-1)
for n in range (np.size(X,1)):
predictions = hypothesis(X[i,1], thetas) #calculate predictions
cost_prev = cost #save old cost
cost = cost_fct(y,predictions) #calculate new cost
past_costs.append(cost)
#print("cost=",cost," eps=", abs(cost - cost_prev)," error = ",np.sum((predictions - y)))
thetas[n] = thetas[n] - (alpha*(predictions - y[i])*X[i,n]) #cf lecture1, update rule for theta 1
past_theta0.append(thetas[0])
past_theta1.append(thetas[1])
return thetas,past_theta0,past_theta1,past_costs,n_iter
alpha = 0.01 #Step size
thetas = np.array([0.0,1.0])
plt.title("Before fit")
x=np.arange(3)
plt.plot(x,x*thetas[1]+thetas[0])
plt.scatter(X_train[:,1],y_train,marker="+")
plt.scatter(X_test[:,1],y_test,marker="+")
plt.xlabel("X")
plt.ylabel("y")
thetas,past_theta0,past_theta1, past_costs,n_iter = stochastic_gradient_descent(X_train, y_train, thetas, alpha)
print("Iteration =", n_iter, "Theta 1 =", thetas[1], " Theta 0 =", thetas[0])
save = np.concatenate((save,[thetas[0],thetas[1]]), axis=0)
plt.title("Fitted with SGD")
x=np.arange(3)
plt.plot(x,x*thetas[1]+thetas[0])
plt.scatter(X_train[:,1],y_train,marker="+")
plt.scatter(X_test[:,1],y_test,marker="+")
plt.xlabel("X")
plt.ylabel("y")
plt.title('Evolution of thetas: blue:0, yellow:1')
plt.xlabel('No. of iterations')
plt.ylabel('Value of thetas')
plt.plot(past_theta0)
plt.plot(past_theta1)
plt.title('Cost Function J')
plt.xlabel('No. of iterations')
plt.ylabel('Cost')
plt.plot(past_costs)
from numpy.linalg import inv
def closed_form(X, y):
thetas = np.dot(np.dot(inv(np.dot(X.T,X)),X.T),(y))
return thetas
thetas = closed_form(X_train, y_train)
print("Theta 1 =", thetas[1], " Theta 0 =", thetas[0])
save = np.concatenate((save,[thetas[0],thetas[1]]), axis=0)
plt.title("Fitted with closed form method")
x=np.arange(3)
plt.plot(x,x*thetas[1]+thetas[0])
plt.scatter(X_train[:,1],y_train,marker="+")
plt.scatter(X_test[:,1],y_test,marker="+")
plt.xlabel("X")
plt.ylabel("y")
save=save.reshape((3, 2))
save
def mse(X,y,thetas):
predictions = hypothesis(X[:,1],thetas)
return (1/y.size)*np.sum(np.square(predictions-y))
y_test.size
plt.title("All three methods: yellow: Batch, green: stochastic, red:closed form")
plt.xlabel("X")
plt.ylabel("y")
plt.scatter(X_train[:,1],y_train,marker="+")
plt.scatter(X_test[:,1],y_test,marker="+")
x=np.arange(3)
MSE = []
for i in range (np.size(save,0)):
plt.plot(x,x*save[i,1]+save[i,0])
MSE = np.append(MSE,mse(X_test, y_test,save[i]))
MSE
plt.title("MSE comparaison")
plt.ylabel("MSE")
plt.bar(x, MSE)
plt.xticks(x, ('BGD', 'SGD', 'CFM'))
plt.show()
| 0.277963 | 0.985115 |
# Vetores
## Introdução
Vetores são estruturas de dados utilizadas para armazenamento de diversos valores, que podem ser acessados através de índices. A implementação acaba variando de acordo com a linguagem de programação, mas pode-se resumir de maneira geral:
- Cada posição do vetor tem um índice ou chave
- Quando é indexado, a primeira posição é o zero
- Em algumas linguagens não são redimensionáveis
- Pode ou não armazenar valores de diferentes tipos, dependendo da linguagem
Além disso, um vetor pode ser de diversas dimensões. Em um vetor unidimensional, por exemplo, um elemento do vetor pode ser acessado por um índice. Já em um vetor bidimensional, são necessários dois índices para localizar um elemento de um vetor. Esse caso especial é conhecido como matriz.
## Linguagens de Programação
Veja alguns exemplos:
**Java**
```
String[] linguagens = {"Java", "Python", "C"}
```
OU
```
String[] linguagens = new String[3];
linguagens[0] = "Java";
```
**PHP**
Usando índices
```
<?php
$array = array("foo", "bar", "hello", "world");
?>
```
Usando chaves
```
<?php
$arr["x"] = 42;
?>
```
**Python**
Usando estruturas de listas:
```
linguagens = ["Java", "PHP", "C"]
print("O primeiro valor é", linguagens[1])
```
Usando biblioteca própria, pode-se restringir os tipos de valores:
```
from array import array
#apenas float
precos = array('d', [10, 20, 23, 14, 5.1])
print(precos)
```
Por outro lado, se deseja-se adicionar um valor que não seja do tipo definido do vetor, dá erro
```
from array import array
#apenas float
precos = array('d', [10, 20, 23, 14, 5.1])
print(precos)
precos.append("Java")
```
Para saber mais sobre esse módulo em Python, acesse https://docs.python.org/3/library/array.html#module-array
Importante ressaltar que cada linguagem possui suas bibliotecas e funções para manipulação de vetores, geralmente buscando facilitar a operação desse tipo de estrutura de dados.
## Operações em Python
Por usar a mesma estrutura de listas lineares, um vetor tem as mesmas operações em Python, ou seja:
- insert: insere um valor X na posição Y do vetor
- remove: retira um determinado valor do vetor
- count: contar o número de ocorrências de um valor no vetor
- reverse: inverter a posição dos valores
- index: informar a posição de um valor na lista
- pop: retirar um elemento
- append: adicionar ao final
## Inicializando um vetor em Python
Como lista:
```
vetor = []
# vai imprimir um vetor vazio
print(vetor)
```
Com n posições:
```
vetor = [0] * 10
# vai imprimir um vetor com 10 posições 0
print(vetor)
```
Caso queira inicializar um vetor com 10 posições com valores 1
```
vetor = [1] * 10
# vai imprimir um vetor com 10 posições 0
print(vetor)
```
## Acessando dados em um vetor
Com um vetor criado, basta acessar o valor por seu índice. Veja um exemplo abaixo:
```
notas = [10,8,5,4,9]
nota1 = notas[0]
```
Ou seja, o valor correspodente a *notas[0]* é 10.
## Atribuindo valores em um vetor
Para atribuir um valor a uma determinada posição do vetor, basta acessar a posição por seu índice e atribuir o valor desejado.
```
notas = [10,8,5,4,9]
notas[1] = 9
```
Após o comando acima, o vetor terá o conteúdo *[10,9,5,4,9]*
## Acessando partes de um vetor
Em Python, é possível acessar partes de um vetor, através da operação de *slice*, de acordo com o seguinte formato:
- vetor[início:final] # retorna os itens de início até final-1
- vetor[início:] # retorna os itens começando de início e indo até o fim do vetor
- vetor[:final] # itens da posição 0 até o final-1
- vetor[:] # retorna todo o vetor
Exemplo: vetor[início:fim]
```
notas = [10,8,5,4,9]
notasParte = notas[1:4]
print(notasParte)
```
Exemplo: vetor[início]
```
notas = [10,8,5,4,9]
notasParte = notas[1:]
print(notasParte)
```
Exemplo: vetor[:final]
```
notas = [10,8,5,4,9]
notasParte = notas[:3]
print(notasParte)
```
Exemplo: vetor[:]
```
notas = [10,8,5,4,9]
notasParte = notas[:]
print(notasParte)
```
Se as posições forem negativas, então os valores são acessados a partir do final.
Exemplo: vetor[-1]
```
notas = [10,8,5,4,9]
notasParte = notas[-1]
print(notasParte)
```
Exemplo: vetor[-2:] - Últimos dois itens do vetor
```
notas = [10,8,5,4,9]
notasParte = notas[-2:]
print(notasParte)
```
Exemplo: vetor[:-2] - todos os valores do vetor, com exceção dos dois últimos
```
notas = [10,8,5,4,9]
notasParte = notas[:-2]
print(notasParte)
```
Além disso, o slice pode ser feito indicando-se deslocamentos a partir do ínicio:
vetor[ini:fim:desloc]
```
notas = [10,8,5,4,9,10,7,5,4,3,3]
notasParte = notas[1:10:2] #vai pular de 2 em 2
print(notasParte)
```
## Dicionário
Uma outra forma de usar uma estrutura semelhante a vetores, com o uso de chaves para localizar valores, é o dicionário. Veja um exemplo:
```
dados = {}
dados["nome"] = "Fernando"
dados["time"] = "Corinthians"
for key in dados.keys():
print(key,":",dados[key])
```
Para acessar um valor específico
```
print(dados["nome"])
```
Para alterar o valor de uma posição do dicionário:
```
dados = {}
dados["nome"] = "Fernando"
dados["time"] = "Corinthians"
print("===== Valores Iniciais =====")
for key in dados.keys():
print(key,":",dados[key])
print()
print("===== Novos Valores =====")
# mudando o nome
dados["nome"] = "Fernando Xavier"
for key in dados.keys():
print(key,":",dados[key])
```
Os dados podem ser inicializados em listas de elementos *chave:valor*
```
dados = {"nome":"Fernando", "RGM":"123456"}
for key in dados.keys():
print(key,":",dados[key])
```
Os dados são localizados no dicionário através de chaves (que ficam dentro dos colchetes) que são, no exemplo acima, *nome* e *time*.
## Exercícios
**Ex 1: Escreva um programa que imprima os valores de um vetor, um por linha. O vetor deve ser inicializado com no mínimo 5 valores **
**Ex 2: Escreva um programa que salve os 10 primeiros números pares (a partir de 2) em um vetor.**
**Ex 3: Faça um programa que pergunte ao usuário um nome de linguagem e, caso essa linguagem exista no vetor, informe a posição dela. Dica: pesquise sobre try/except para implementar sua solução, caso o valor não exista.**
```
linguagens = ["Java", "PHP", "C"]
```
**Ex 4: Crie um programa que peça ao usuário que informe as notas de um aluno e depois calcule a média. Use vetores na sua solução e só aceite números inteiros. Caso o usuário digite um valor que não seja um número inteiro, seu programa deverá informar o erro e pedir novo número**
** Ex 5 (adaptado do Sedgewick): O que vai ser produzido nos vetores abaixo?**
```
vetor = [0] * 99
for i in range(99):
vetor[i] = 98 - i
print(vetor)
print()
for i in range(99):
vetor[i] = vetor[vetor[i]]
print(vetor)
```
**Ex 6: Dada o vetor abaixo, crie um novo vetor onde cada valor será o dobro do valor correspondente do primeiro vetor**
```
quantidade = [2,4,5,7,1,4,5,6,8]
```
**Ex 7: Dado o vetor abaixo, implemente um código que some os valores de um vetor**
```
quantidade = [2,4,5,7,1,4,5,6,8]
```
** Ex 8: Faça um código que acesse apenas os índices ímpares de um vetor e imprima os valores dessas posições**
```
quantidade = [2,4,5,7,1,4,5,6,8]
```
** Ex 9: O código abaixo irá gerar um vetor com 10 posições, com números aleatórios de 1 a 100. Crie um código que peça a um usuário um número entre 1 e 100 e seu código retorne se esse número exista e a posição dele, caso exista**
```
import random
total = 100
qtde = 10
vetor = random.sample(range(1,total), k=qtde)
print(vetor)
```
**Ex 10: Faça um código que, dado o vetor abaixo, inverta os valores em um novo vetor.**
```
vetor = [82, 62, 61, 88, 89, 37, 95, 29, 45, 98]
```
|
github_jupyter
|
String[] linguagens = {"Java", "Python", "C"}
String[] linguagens = new String[3];
linguagens[0] = "Java";
<?php
$array = array("foo", "bar", "hello", "world");
?>
<?php
$arr["x"] = 42;
?>
linguagens = ["Java", "PHP", "C"]
print("O primeiro valor é", linguagens[1])
from array import array
#apenas float
precos = array('d', [10, 20, 23, 14, 5.1])
print(precos)
from array import array
#apenas float
precos = array('d', [10, 20, 23, 14, 5.1])
print(precos)
precos.append("Java")
vetor = []
# vai imprimir um vetor vazio
print(vetor)
vetor = [0] * 10
# vai imprimir um vetor com 10 posições 0
print(vetor)
vetor = [1] * 10
# vai imprimir um vetor com 10 posições 0
print(vetor)
notas = [10,8,5,4,9]
nota1 = notas[0]
notas = [10,8,5,4,9]
notas[1] = 9
notas = [10,8,5,4,9]
notasParte = notas[1:4]
print(notasParte)
notas = [10,8,5,4,9]
notasParte = notas[1:]
print(notasParte)
notas = [10,8,5,4,9]
notasParte = notas[:3]
print(notasParte)
notas = [10,8,5,4,9]
notasParte = notas[:]
print(notasParte)
notas = [10,8,5,4,9]
notasParte = notas[-1]
print(notasParte)
notas = [10,8,5,4,9]
notasParte = notas[-2:]
print(notasParte)
notas = [10,8,5,4,9]
notasParte = notas[:-2]
print(notasParte)
notas = [10,8,5,4,9,10,7,5,4,3,3]
notasParte = notas[1:10:2] #vai pular de 2 em 2
print(notasParte)
dados = {}
dados["nome"] = "Fernando"
dados["time"] = "Corinthians"
for key in dados.keys():
print(key,":",dados[key])
print(dados["nome"])
dados = {}
dados["nome"] = "Fernando"
dados["time"] = "Corinthians"
print("===== Valores Iniciais =====")
for key in dados.keys():
print(key,":",dados[key])
print()
print("===== Novos Valores =====")
# mudando o nome
dados["nome"] = "Fernando Xavier"
for key in dados.keys():
print(key,":",dados[key])
dados = {"nome":"Fernando", "RGM":"123456"}
for key in dados.keys():
print(key,":",dados[key])
linguagens = ["Java", "PHP", "C"]
vetor = [0] * 99
for i in range(99):
vetor[i] = 98 - i
print(vetor)
print()
for i in range(99):
vetor[i] = vetor[vetor[i]]
print(vetor)
quantidade = [2,4,5,7,1,4,5,6,8]
quantidade = [2,4,5,7,1,4,5,6,8]
quantidade = [2,4,5,7,1,4,5,6,8]
import random
total = 100
qtde = 10
vetor = random.sample(range(1,total), k=qtde)
print(vetor)
vetor = [82, 62, 61, 88, 89, 37, 95, 29, 45, 98]
| 0.091977 | 0.918991 |
```
try:
import google.colab # noqa: F401
except ImportError:
import ufl # noqa: F401
import dolfin # noqa: F401
else:
try:
import ufl
import dolfin
except ImportError:
!wget "https://fem-on-colab.github.io/releases/fenics-install.sh" -O "/tmp/fenics-install.sh" && bash "/tmp/fenics-install.sh"
import ufl # noqa: F401
import dolfin # noqa: F401
try:
import rbnics
except ImportError:
!pip3 install git+https://github.com/RBniCS/RBniCS.git
import rbnics # noqa: F401
# Download data files
!mkdir -p data
![ -f data/thermal_block_facet_region.xml ] || wget https://github.com/RBniCS/RBniCS/raw/master/tutorials/01_thermal_block/data/thermal_block_facet_region.xml -O data/thermal_block_facet_region.xml
![ -f data/thermal_block_physical_region.xml ] || wget https://github.com/RBniCS/RBniCS/raw/master/tutorials/01_thermal_block/data/thermal_block_physical_region.xml -O data/thermal_block_physical_region.xml
![ -f data/thermal_block.xml ] || wget https://github.com/RBniCS/RBniCS/raw/master/tutorials/01_thermal_block/data/thermal_block.xml -O data/thermal_block.xml
from dolfin import *
from rbnics import *
@SCM()
class ThermalBlock(EllipticCoerciveCompliantProblem):
# Default initialization of members
@generate_function_space_for_stability_factor
def __init__(self, V, **kwargs):
# Call the standard initialization
EllipticCoerciveCompliantProblem.__init__(self, V, **kwargs)
# ... and also store FEniCS data structures for assembly
assert "subdomains" in kwargs
assert "boundaries" in kwargs
self.subdomains, self.boundaries = kwargs["subdomains"], kwargs["boundaries"]
self.u = TrialFunction(V)
self.v = TestFunction(V)
self.dx = Measure("dx")(subdomain_data=self.subdomains)
self.ds = Measure("ds")(subdomain_data=self.boundaries)
# Customize eigen solver parameters
self._eigen_solver_parameters.update({
"bounding_box_minimum": {
"problem_type": "gen_hermitian", "spectral_transform": "shift-and-invert",
"spectral_shift": 1.e-5, "linear_solver": "mumps"
},
"bounding_box_maximum": {
"problem_type": "gen_hermitian", "spectral_transform": "shift-and-invert",
"spectral_shift": 1.e5, "linear_solver": "mumps"
},
"stability_factor": {
"problem_type": "gen_hermitian", "spectral_transform": "shift-and-invert",
"spectral_shift": 1.e-5, "linear_solver": "mumps"
}
})
# Return custom problem name
def name(self):
return "ThermalBlock"
# Return theta multiplicative terms of the affine expansion of the problem.
@compute_theta_for_stability_factor
def compute_theta(self, term):
mu = self.mu
if term == "a":
theta_a0 = mu[0]
theta_a1 = 1.
return (theta_a0, theta_a1)
elif term == "f":
theta_f0 = mu[1]
return (theta_f0,)
else:
raise ValueError("Invalid term for compute_theta().")
# Return forms resulting from the discretization of the affine expansion of the problem operators.
@assemble_operator_for_stability_factor
def assemble_operator(self, term):
v = self.v
dx = self.dx
if term == "a":
u = self.u
a0 = inner(grad(u), grad(v)) * dx(1)
a1 = inner(grad(u), grad(v)) * dx(2)
return (a0, a1)
elif term == "f":
ds = self.ds
f0 = v * ds(1)
return (f0,)
elif term == "dirichlet_bc":
bc0 = [DirichletBC(self.V, Constant(0.0), self.boundaries, 3)]
return (bc0,)
elif term == "inner_product":
u = self.u
x0 = inner(grad(u), grad(v)) * dx
return (x0,)
else:
raise ValueError("Invalid term for assemble_operator().")
mesh = Mesh("data/thermal_block.xml")
subdomains = MeshFunction("size_t", mesh, "data/thermal_block_physical_region.xml")
boundaries = MeshFunction("size_t", mesh, "data/thermal_block_facet_region.xml")
V = FunctionSpace(mesh, "Lagrange", 1)
problem = ThermalBlock(V, subdomains=subdomains, boundaries=boundaries)
mu_range = [(0.1, 10.0), (-1.0, 1.0)]
problem.set_mu_range(mu_range)
reduction_method = ReducedBasis(problem)
reduction_method.set_Nmax(4, SCM=4)
reduction_method.set_tolerance(1e-5, SCM=1e-5)
reduction_method.initialize_training_set(10, SCM=10)
reduced_problem = reduction_method.offline()
online_mu = (8.0, -1.0)
reduced_problem.set_mu(online_mu)
reduced_solution = reduced_problem.solve()
plot(reduced_solution, reduced_problem=reduced_problem)
reduction_method.initialize_testing_set(10, SCM=0)
reduction_method.error_analysis(SCM=1)
%%bash
export LD_PRELOAD=""
ERROR_LIBRARIES=($(find /root/.cache/dijitso -name '*\.so' -exec \
bash -c 'ldd $0 | grep libstdc++.so.6 1>/dev/null 2>/dev/null && echo $0' {} \;))
if [ ${#ERROR_LIBRARIES[@]} -eq 0 ]; then
echo "No reference to libstdc++.so was found"
else
for ERROR_LIBRARY in "${ERROR_LIBRARIES[@]}"; do
echo "Error: library $ERROR_LIBRARY depends on libstdc++.so"
ldd -v $ERROR_LIBRARY
done
false
fi
```
|
github_jupyter
|
try:
import google.colab # noqa: F401
except ImportError:
import ufl # noqa: F401
import dolfin # noqa: F401
else:
try:
import ufl
import dolfin
except ImportError:
!wget "https://fem-on-colab.github.io/releases/fenics-install.sh" -O "/tmp/fenics-install.sh" && bash "/tmp/fenics-install.sh"
import ufl # noqa: F401
import dolfin # noqa: F401
try:
import rbnics
except ImportError:
!pip3 install git+https://github.com/RBniCS/RBniCS.git
import rbnics # noqa: F401
# Download data files
!mkdir -p data
![ -f data/thermal_block_facet_region.xml ] || wget https://github.com/RBniCS/RBniCS/raw/master/tutorials/01_thermal_block/data/thermal_block_facet_region.xml -O data/thermal_block_facet_region.xml
![ -f data/thermal_block_physical_region.xml ] || wget https://github.com/RBniCS/RBniCS/raw/master/tutorials/01_thermal_block/data/thermal_block_physical_region.xml -O data/thermal_block_physical_region.xml
![ -f data/thermal_block.xml ] || wget https://github.com/RBniCS/RBniCS/raw/master/tutorials/01_thermal_block/data/thermal_block.xml -O data/thermal_block.xml
from dolfin import *
from rbnics import *
@SCM()
class ThermalBlock(EllipticCoerciveCompliantProblem):
# Default initialization of members
@generate_function_space_for_stability_factor
def __init__(self, V, **kwargs):
# Call the standard initialization
EllipticCoerciveCompliantProblem.__init__(self, V, **kwargs)
# ... and also store FEniCS data structures for assembly
assert "subdomains" in kwargs
assert "boundaries" in kwargs
self.subdomains, self.boundaries = kwargs["subdomains"], kwargs["boundaries"]
self.u = TrialFunction(V)
self.v = TestFunction(V)
self.dx = Measure("dx")(subdomain_data=self.subdomains)
self.ds = Measure("ds")(subdomain_data=self.boundaries)
# Customize eigen solver parameters
self._eigen_solver_parameters.update({
"bounding_box_minimum": {
"problem_type": "gen_hermitian", "spectral_transform": "shift-and-invert",
"spectral_shift": 1.e-5, "linear_solver": "mumps"
},
"bounding_box_maximum": {
"problem_type": "gen_hermitian", "spectral_transform": "shift-and-invert",
"spectral_shift": 1.e5, "linear_solver": "mumps"
},
"stability_factor": {
"problem_type": "gen_hermitian", "spectral_transform": "shift-and-invert",
"spectral_shift": 1.e-5, "linear_solver": "mumps"
}
})
# Return custom problem name
def name(self):
return "ThermalBlock"
# Return theta multiplicative terms of the affine expansion of the problem.
@compute_theta_for_stability_factor
def compute_theta(self, term):
mu = self.mu
if term == "a":
theta_a0 = mu[0]
theta_a1 = 1.
return (theta_a0, theta_a1)
elif term == "f":
theta_f0 = mu[1]
return (theta_f0,)
else:
raise ValueError("Invalid term for compute_theta().")
# Return forms resulting from the discretization of the affine expansion of the problem operators.
@assemble_operator_for_stability_factor
def assemble_operator(self, term):
v = self.v
dx = self.dx
if term == "a":
u = self.u
a0 = inner(grad(u), grad(v)) * dx(1)
a1 = inner(grad(u), grad(v)) * dx(2)
return (a0, a1)
elif term == "f":
ds = self.ds
f0 = v * ds(1)
return (f0,)
elif term == "dirichlet_bc":
bc0 = [DirichletBC(self.V, Constant(0.0), self.boundaries, 3)]
return (bc0,)
elif term == "inner_product":
u = self.u
x0 = inner(grad(u), grad(v)) * dx
return (x0,)
else:
raise ValueError("Invalid term for assemble_operator().")
mesh = Mesh("data/thermal_block.xml")
subdomains = MeshFunction("size_t", mesh, "data/thermal_block_physical_region.xml")
boundaries = MeshFunction("size_t", mesh, "data/thermal_block_facet_region.xml")
V = FunctionSpace(mesh, "Lagrange", 1)
problem = ThermalBlock(V, subdomains=subdomains, boundaries=boundaries)
mu_range = [(0.1, 10.0), (-1.0, 1.0)]
problem.set_mu_range(mu_range)
reduction_method = ReducedBasis(problem)
reduction_method.set_Nmax(4, SCM=4)
reduction_method.set_tolerance(1e-5, SCM=1e-5)
reduction_method.initialize_training_set(10, SCM=10)
reduced_problem = reduction_method.offline()
online_mu = (8.0, -1.0)
reduced_problem.set_mu(online_mu)
reduced_solution = reduced_problem.solve()
plot(reduced_solution, reduced_problem=reduced_problem)
reduction_method.initialize_testing_set(10, SCM=0)
reduction_method.error_analysis(SCM=1)
%%bash
export LD_PRELOAD=""
ERROR_LIBRARIES=($(find /root/.cache/dijitso -name '*\.so' -exec \
bash -c 'ldd $0 | grep libstdc++.so.6 1>/dev/null 2>/dev/null && echo $0' {} \;))
if [ ${#ERROR_LIBRARIES[@]} -eq 0 ]; then
echo "No reference to libstdc++.so was found"
else
for ERROR_LIBRARY in "${ERROR_LIBRARIES[@]}"; do
echo "Error: library $ERROR_LIBRARY depends on libstdc++.so"
ldd -v $ERROR_LIBRARY
done
false
fi
| 0.511961 | 0.299734 |
```
import torch
import torchvision
import torchvision.transforms as transforms
from torch.utils.data import DataLoader, Dataset
from tqdm.notebook import tqdm
from google.colab import drive
import numpy as np
import os
import sys
manualSeed = 999
torch.manual_seed(manualSeed)
drive.mount("/content/gdrive")
path = "/content/gdrive/My Drive/Colab Notebooks/GANS/Style Transfer"
sys.path.append(path)
from Utils import networks
# Images Dataset that returns one style and one content image. As I only trained using 40.000
# images each, each image is randomly sampled. The way it is implemented does not allow multi-threading. However
# as this network is relatively small and training times low, no improved class was implemented.
class Images(Dataset):
def __init__(self, root_dir1, root_dir2, transform=None):
self.root_dir1 = root_dir1
self.root_dir2 = root_dir2
self.transform = transform
def __len__(self):
return min(len(os.listdir(self.root_dir1)), len(os.listdir(self.root_dir2)))
def __getitem__(self, idx):
all_names1, all_names2 = os.listdir(self.root_dir1), os.listdir(self.root_dir2)
idx1, idx2 = np.random.randint(0, len(all_names1)), np.random.randint(0, len(all_names2))
img_name1, img_name2 = os.path.join(self.root_dir1, all_names1[idx1]), os.path.join(self.root_dir2, all_names2[idx2])
image1 = Image.open(img_name1).convert("RGB")
image2 = Image.open(img_name2).convert("RGB")
if self.transform:
image1 = self.transform(image1)
image2 = self.transform(image2)
return image1, image2
# To note is that the images are not normalised
transform = transforms.Compose([transforms.Resize(512),
transforms.CenterCrop(256),
transforms.ToTensor()])
# Specify the path to the style and content images
pathStyleImages = "/content/Data/Wiki_40k"
pathContentImages = "/content/Data/Coco_40k"
all_img = Images(pathStyleImages, pathContentImages, transform=transform)
# Simple save
def save_state(decoder, optimiser, iters, run_dir):
name = "StyleTransfer Checkpoint Iter: {}.tar".format(iters)
torch.save({"Decoder" : decoder,
"Optimiser" : optimiser,
"iters": iters
}, os.path.join(path, name))
print("Saved : {} succesfully".format(name))
def training_loop(network, # StyleTransferNetwork
dataloader_comb, # DataLoader
n_epochs, # Number of Epochs
run_dir # Directory in which the checkpoints and tensorboard files are saved
):
writer = SummaryWriter(os.path.join(path, run_dir))
# Fixed images to compare over time
fixed_batch_style, fixed_batch_content = all_img[0]
fixed_batch_style, fixed_batch_content = fixed_batch_style.unsqueeze(0).to(device), fixed_batch_content.unsqueeze(0).to(device) # Move images to device
writer.add_image("Style", torchvision.utils.make_grid(fixed_batch_style))
writer.add_image("Content", torchvision.utils.make_grid(fixed_batch_content))
iters = network.iters
for epoch in range(1, n_epochs+1):
tqdm_object = tqdm(dataloader_comb, total=len(dataloader_comb))
for style_imgs, content_imgs in tqdm_object:
network.adjust_learning_rate(network.optimiser, iters)
style_imgs = style_imgs.to(device)
content_imgs = content_imgs.to(device)
loss_comb, content_loss, style_loss = network(style_imgs, content_imgs)
network.optimiser.zero_grad()
loss_comb.backward()
network.optimiser.step()
# Update status bar, add Loss, add Images
tqdm_object.set_postfix_str("Combined Loss: {:.3f}, Style Loss: {:.3f}, Content Loss: {:.3f}".format(
loss_comb.item()*100, style_loss.item()*100, content_loss.item()*100))
if iters % 25 == 0:
writer.add_scalar("Combined Loss", loss_comb*1000, iters)
writer.add_scalar("Style Loss", style_loss*1000, iters)
writer.add_scalar("Content Loss", content_loss*1000, iters)
if (iters+1) % 2000 == 1:
with torch.no_grad():
network.set_train(False)
images = network(fixed_batch_style, fixed_batch_content)
img_grid = torchvision.utils.make_grid(images)
writer.add_image("Progress Iter: {}".format(iters), img_grid)
network.set_train(True)
if (iters+1) % 4000 == 1:
save_state(network.decoder.state_dict(), network.optimiser.state_dict(), iters, run_dir)
writer.close()
writer = SummaryWriter(os.path.join(path, run_dir))
iters += 1
device = ("cuda" if torch.cuda.is_available() else "cpu")
learning_rate = 1e-4
learning_rate_decay = 5e-5
dataloader_comb = DataLoader(all_img, batch_size=5, shuffle=True, num_workers=0, drop_last=True)
gamma = torch.tensor([2]).to(device) # Style weight
n_epochs = 5
run_dir = "runs/Run 1" # Change if you want to save the checkpoints/tensorboard files in a different directory
state_encoder = torch.load(os.path.join(path, "vgg_normalised.pth"))
network = networks.StyleTransferNetwork(device,
state_encoder,
learning_rate,
learning_rate_decay,
gamma,
load_fromstate=False,
load_path=os.path.join(path, "StyleTransfer Checkpoint Iter: 120000.tar"))
training_loop(network, dataloader_comb, n_epochs, run_dir)
```
|
github_jupyter
|
import torch
import torchvision
import torchvision.transforms as transforms
from torch.utils.data import DataLoader, Dataset
from tqdm.notebook import tqdm
from google.colab import drive
import numpy as np
import os
import sys
manualSeed = 999
torch.manual_seed(manualSeed)
drive.mount("/content/gdrive")
path = "/content/gdrive/My Drive/Colab Notebooks/GANS/Style Transfer"
sys.path.append(path)
from Utils import networks
# Images Dataset that returns one style and one content image. As I only trained using 40.000
# images each, each image is randomly sampled. The way it is implemented does not allow multi-threading. However
# as this network is relatively small and training times low, no improved class was implemented.
class Images(Dataset):
def __init__(self, root_dir1, root_dir2, transform=None):
self.root_dir1 = root_dir1
self.root_dir2 = root_dir2
self.transform = transform
def __len__(self):
return min(len(os.listdir(self.root_dir1)), len(os.listdir(self.root_dir2)))
def __getitem__(self, idx):
all_names1, all_names2 = os.listdir(self.root_dir1), os.listdir(self.root_dir2)
idx1, idx2 = np.random.randint(0, len(all_names1)), np.random.randint(0, len(all_names2))
img_name1, img_name2 = os.path.join(self.root_dir1, all_names1[idx1]), os.path.join(self.root_dir2, all_names2[idx2])
image1 = Image.open(img_name1).convert("RGB")
image2 = Image.open(img_name2).convert("RGB")
if self.transform:
image1 = self.transform(image1)
image2 = self.transform(image2)
return image1, image2
# To note is that the images are not normalised
transform = transforms.Compose([transforms.Resize(512),
transforms.CenterCrop(256),
transforms.ToTensor()])
# Specify the path to the style and content images
pathStyleImages = "/content/Data/Wiki_40k"
pathContentImages = "/content/Data/Coco_40k"
all_img = Images(pathStyleImages, pathContentImages, transform=transform)
# Simple save
def save_state(decoder, optimiser, iters, run_dir):
name = "StyleTransfer Checkpoint Iter: {}.tar".format(iters)
torch.save({"Decoder" : decoder,
"Optimiser" : optimiser,
"iters": iters
}, os.path.join(path, name))
print("Saved : {} succesfully".format(name))
def training_loop(network, # StyleTransferNetwork
dataloader_comb, # DataLoader
n_epochs, # Number of Epochs
run_dir # Directory in which the checkpoints and tensorboard files are saved
):
writer = SummaryWriter(os.path.join(path, run_dir))
# Fixed images to compare over time
fixed_batch_style, fixed_batch_content = all_img[0]
fixed_batch_style, fixed_batch_content = fixed_batch_style.unsqueeze(0).to(device), fixed_batch_content.unsqueeze(0).to(device) # Move images to device
writer.add_image("Style", torchvision.utils.make_grid(fixed_batch_style))
writer.add_image("Content", torchvision.utils.make_grid(fixed_batch_content))
iters = network.iters
for epoch in range(1, n_epochs+1):
tqdm_object = tqdm(dataloader_comb, total=len(dataloader_comb))
for style_imgs, content_imgs in tqdm_object:
network.adjust_learning_rate(network.optimiser, iters)
style_imgs = style_imgs.to(device)
content_imgs = content_imgs.to(device)
loss_comb, content_loss, style_loss = network(style_imgs, content_imgs)
network.optimiser.zero_grad()
loss_comb.backward()
network.optimiser.step()
# Update status bar, add Loss, add Images
tqdm_object.set_postfix_str("Combined Loss: {:.3f}, Style Loss: {:.3f}, Content Loss: {:.3f}".format(
loss_comb.item()*100, style_loss.item()*100, content_loss.item()*100))
if iters % 25 == 0:
writer.add_scalar("Combined Loss", loss_comb*1000, iters)
writer.add_scalar("Style Loss", style_loss*1000, iters)
writer.add_scalar("Content Loss", content_loss*1000, iters)
if (iters+1) % 2000 == 1:
with torch.no_grad():
network.set_train(False)
images = network(fixed_batch_style, fixed_batch_content)
img_grid = torchvision.utils.make_grid(images)
writer.add_image("Progress Iter: {}".format(iters), img_grid)
network.set_train(True)
if (iters+1) % 4000 == 1:
save_state(network.decoder.state_dict(), network.optimiser.state_dict(), iters, run_dir)
writer.close()
writer = SummaryWriter(os.path.join(path, run_dir))
iters += 1
device = ("cuda" if torch.cuda.is_available() else "cpu")
learning_rate = 1e-4
learning_rate_decay = 5e-5
dataloader_comb = DataLoader(all_img, batch_size=5, shuffle=True, num_workers=0, drop_last=True)
gamma = torch.tensor([2]).to(device) # Style weight
n_epochs = 5
run_dir = "runs/Run 1" # Change if you want to save the checkpoints/tensorboard files in a different directory
state_encoder = torch.load(os.path.join(path, "vgg_normalised.pth"))
network = networks.StyleTransferNetwork(device,
state_encoder,
learning_rate,
learning_rate_decay,
gamma,
load_fromstate=False,
load_path=os.path.join(path, "StyleTransfer Checkpoint Iter: 120000.tar"))
training_loop(network, dataloader_comb, n_epochs, run_dir)
| 0.689619 | 0.403567 |
```
#Baixando bibliotecas
!sudo apt-get install python-grib
!sudo python setup.py install
!sudo apt-get install libgeos-dev
!sudo pip3 install -U git+https://github.com/matplotlib/basemap.git
!apt install libgrib-api-dev libgrib2c-dev
!pip install pyproj==1.9.6
!pip install pygrib
#Instalando as bibliotecas
import pygrib
from mpl_toolkits.basemap import Basemap
from mpl_toolkits.basemap import shiftgrid
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from datetime import datetime
#3 dias de previsão
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_000.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_003.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_006.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_009.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_012.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_015.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_018.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_021.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_024.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_027.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_030.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_033.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_036.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_039.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_042.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_045.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_048.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_051.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_054.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_057.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_060.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_063.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_066.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_069.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_072.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_075.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_078.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_081.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_084.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_087.grb2
#Abrindo os dados e selecionando as váriaveis
gr = pygrib.open('gfs_4_20210702_1800_006.grb2')
t = gr.select(name='Temperature')[0]
#Definindo região do RS
data, lats, lons = t.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
#Visualizando a grade selecionada
m = Basemap(projection='cyl',llcrnrlat=-35,urcrnrlat=-25,\
llcrnrlon=302,urcrnrlon=315,resolution='i')
# Função Matplotlib para definir o tamanho da figura.
plt.figure(figsize=(8,10))
#Inserindo os lat e lon a figura criada pelo Basemap e vinculando às variavéis x e y
x, y = m(lons, lats)
#Função do Basemap para iserir ao mapa continentes, países e estados.
m.drawcoastlines()
m.drawcountries()
m.drawstates()
la = np.arange(-35,-25,3.)
lo = np.arange(302.,315.,3.)
m.drawparallels(la,labels=[False,True,True,False])
m.drawmeridians(lo,labels=[True,False,False,True])
#m.scatter(lons, lats, marker = 'o', color='r', zorder=2)
#Função do Basemap para realizar a interpolação e criação de polígonos
contourf = m.contourf(x, y, np.squeeze(data),cmap='jet')
m.colorbar(contourf, location='right', pad="10%")
#Marcando cidade de Pelotas
m.scatter(307.6, -31.6, marker = 'o', color='r', zorder=3)
#Encontrando um lat e lon mais próximo
#Lat Lon selecionado
def encontra_lat_lon(la,lo):
stn_lat = la
stn_lon = lo
lat = lats
lon = lons
abslat = np.abs(lat-stn_lat)
abslon= np.abs(lon-stn_lon)
c = np.maximum(abslon,abslat)
latlon_idx = np.argmin(c)
x, y = np.where(c == np.min(c))
return(x,y)
#Aplicando para encontrar os index dos valores mais próximas
encontra_lat_lon(-31.6,307.6)
#Pegando o dado da respectiva região de Pelotas
data[13,175]
#Abrindo 3 dias de dados
gr = pygrib.open('gfs_4_20210714_0000_000.grb2')
gr2 = pygrib.open('gfs_4_20210714_0000_003.grb2')
gr3 = pygrib.open('gfs_4_20210714_0000_006.grb2')
gr4 = pygrib.open('gfs_4_20210714_0000_009.grb2')
gr5 = pygrib.open('gfs_4_20210714_0000_012.grb2')
gr6 = pygrib.open('gfs_4_20210714_0000_015.grb2')
gr7 = pygrib.open('gfs_4_20210714_0000_018.grb2')
gr8 = pygrib.open('gfs_4_20210714_0000_021.grb2')
gr9 = pygrib.open('gfs_4_20210714_0000_024.grb2')
gr10 = pygrib.open('gfs_4_20210714_0000_027.grb2')
gr11 = pygrib.open('gfs_4_20210714_0000_030.grb2')
gr12 = pygrib.open('gfs_4_20210714_0000_033.grb2')
gr13 = pygrib.open('gfs_4_20210714_0000_036.grb2')
gr14 = pygrib.open('gfs_4_20210714_0000_039.grb2')
gr15 = pygrib.open('gfs_4_20210714_0000_042.grb2')
gr16 = pygrib.open('gfs_4_20210714_0000_045.grb2')
gr17 = pygrib.open('gfs_4_20210714_0000_048.grb2')
gr18 = pygrib.open('gfs_4_20210714_0000_051.grb2')
gr19 = pygrib.open('gfs_4_20210714_0000_054.grb2')
gr20 = pygrib.open('gfs_4_20210714_0000_057.grb2')
gr21 = pygrib.open('gfs_4_20210714_0000_060.grb2')
gr22 = pygrib.open('gfs_4_20210714_0000_063.grb2')
gr23 = pygrib.open('gfs_4_20210714_0000_066.grb2')
gr24 = pygrib.open('gfs_4_20210714_0000_069.grb2')
gr25 = pygrib.open('gfs_4_20210714_0000_072.grb2')
gr26 = pygrib.open('gfs_4_20210714_0000_075.grb2')
gr27 = pygrib.open('gfs_4_20210714_0000_078.grb2')
gr28 = pygrib.open('gfs_4_20210714_0000_081.grb2')
gr29 = pygrib.open('gfs_4_20210714_0000_084.grb2')
gr30 = pygrib.open('gfs_4_20210714_0000_087.grb2')
# 1-Pegando os dados de temperatura referente aos níveis do modelo (ajustar lat e lon).
def selec_t(dadoo,name_v,indx_lat,index_lon):
dadt=[]
#Superfície
t1=dadoo.select(name=name_v)[41]
data1, lats, lons = t1.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data1[indx_lat,index_lon])
#1000
t2=dadoo.select(name=name_v)[40]
data2, lats, lons = t2.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data2[indx_lat,index_lon])
#975
t3=dadoo.select(name=name_v)[39]
data3, lats, lons = t3.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data3[indx_lat,index_lon])
#950
t4=dadoo.select(name=name_v)[38]
data4, lats, lons = t4.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data4[indx_lat,index_lon])
#925
t5=dadoo.select(name=name_v)[37]
data5, lats, lons = t5.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data5[indx_lat,index_lon])
#900
t6=dadoo.select(name=name_v)[36]
data6, lats, lons = t6.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data6[indx_lat,index_lon])
#850
t7=dadoo.select(name=name_v)[35]
data7, lats, lons = t7.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data7[indx_lat,index_lon])
#800
t8=dadoo.select(name=name_v)[34]
data8, lats, lons = t8.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data8[indx_lat,index_lon])
#750
t9=dadoo.select(name=name_v)[33]
data9, lats, lons = t9.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data9[indx_lat,index_lon])
#700
t10=dadoo.select(name=name_v)[32]
data10, lats, lons = t10.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data10[indx_lat,index_lon])
#650
t11=dadoo.select(name=name_v)[31]
data11, lats, lons = t11.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data11[indx_lat,index_lon])
#600
t12=dadoo.select(name=name_v)[30]
data12, lats, lons = t12.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data12[indx_lat,index_lon])
#550
t13=dadoo.select(name=name_v)[29]
data13, lats, lons = t13.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data13[indx_lat,index_lon])
#500hPa
t14=dadoo.select(name=name_v)[28]
data14, lats, lons = t14.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data14[indx_lat,index_lon])
#450hPa
t15=dadoo.select(name=name_v)[27]
data15, lats, lons = t15.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data15[indx_lat,index_lon])
#400hPa
t16=dadoo.select(name=name_v)[26]
data16, lats, lons = t16.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data16[indx_lat,index_lon])
#350hPa
t17=dadoo.select(name=name_v)[25]
data17, lats, lons = t17.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data17[indx_lat,index_lon])
#300hPa
t18=dadoo.select(name=name_v)[24]
data18, lats, lons = t18.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data18[indx_lat,index_lon])
#250hPa
t19=dadoo.select(name=name_v)[23]
data19, lats, lons = t19.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data19[indx_lat,index_lon])
#200hPa
t20=dadoo.select(name=name_v)[22]
data20, lats, lons = t20.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data20[indx_lat,index_lon])
return dadt
# 2 Função para criar e concatenar dataFrame (Aajuste de Temperatura - 273.15)
def transf_ajust(a1,a2,a3,a4,a5,a6,a7,a8,a9,a10,a11,a12,a13,a14,a15,a16,a17,a18,a19,a20,a21,a22,a23,a24,a25,a26,a27,a28,a29,a30):
index=['1013','1000','975','950','925','900','850','800','750','700','650','600','550','500','450','400','350','300','250','200']
ind = pd.DataFrame(index, columns=['niveis'] )
a1=pd.DataFrame(a1)
a2=pd.DataFrame(a2)
a3=pd.DataFrame(a3)
a4=pd.DataFrame(a4)
a5=pd.DataFrame(a5)
a6=pd.DataFrame(a6)
a7=pd.DataFrame(a7)
a8=pd.DataFrame(a8)
a9=pd.DataFrame(a9)
a10=pd.DataFrame(a10)
a11=pd.DataFrame(a11)
a12=pd.DataFrame(a12)
a13=pd.DataFrame(a13)
a14=pd.DataFrame(a14)
a15=pd.DataFrame(a15)
a16=pd.DataFrame(a16)
a17=pd.DataFrame(a17)
a18=pd.DataFrame(a18)
a19=pd.DataFrame(a19)
a20=pd.DataFrame(a20)
a21=pd.DataFrame(a21)
a22=pd.DataFrame(a22)
a23=pd.DataFrame(a23)
a24=pd.DataFrame(a24)
a25=pd.DataFrame(a25)
a26=pd.DataFrame(a26)
a27=pd.DataFrame(a27)
a28=pd.DataFrame(a28)
a29=pd.DataFrame(a29)
a30=pd.DataFrame(a30)
f = pd.concat([ind,a1,a2,a3,a4,a5,a6,a7,a8,a9,a10,a11,a12,a13,a14,a15,a16,a17,a18,a19,a20,a21,a22,a23,a24,a25,a26,a27,a28,a29,a30], axis=1)
return f
#Aplicando Função(dado no formato grib, 'nome da variavél desejada')
na='Relative humidity'
dad1=selec_t(gr,na)
dad2=selec_t(gr2,na)
dad3=selec_t(gr3,na)
dad4=selec_t(gr4,na)
dad5=selec_t(gr5,na)
dad6=selec_t(gr6,na)
dad7=selec_t(gr7,na)
dad8=selec_t(gr8,na)
dad9=selec_t(gr9,na)
dad10=selec_t(gr10,na)
dad11=selec_t(gr11,na)
dad12=selec_t(gr12,na)
dad13=selec_t(gr13,na)
dad14=selec_t(gr14,na)
dad15=selec_t(gr15,na)
dad16=selec_t(gr16,na)
dad17=selec_t(gr17,na)
dad18=selec_t(gr18,na)
dad19=selec_t(gr19,na)
dad20=selec_t(gr20,na)
dad21=selec_t(gr21,na)
dad22=selec_t(gr22,na)
dad23=selec_t(gr23,na)
dad24=selec_t(gr24,na)
dad25=selec_t(gr25,na)
dad26=selec_t(gr26,na)
dad27=selec_t(gr27,na)
dad28=selec_t(gr28,na)
dad29=selec_t(gr29,na)
dad30=selec_t(gr30,na)
# 2 Função para criar e concatenar dataFrame (Aajuste de Temperatura - 273.15)
df=transf_ajust(dad1,dad2,dad3,dad4,dad5,dad6,dad7,dad8,dad9,dad10,dad11,dad12,dad13,dad14,dad15,dad16,dad17,dad18,dad19,dad20,dad21,dad22,dad23,dad24,dad25,dad26,dad27,dad28,dad29,dad30)
df.columns = ['niveis','00', '03', '06','09','12','15','18','21','24','27','30','33','36','39','42','45','48','51','54','57','60', '63', '66','69','72','75','78','81','84','87']
Y = df['niveis'].astype(float)
del df['niveis']
df.iloc[::-1]
df.reset_index(drop=True)
X = df.columns.values[:].astype(float)
X, Y = np.meshgrid(X, Y)
#Plotando figura
fig,ax=plt.subplots(1,1)
fig.set_size_inches(12, 6)
fig.set_size_inches(12, 6)
plt.gca().invert_yaxis()
plt.title('Meteograma altura geopotencial para Cidade de Pelotas')
plt.xlabel('14/07/2021 -- 17/07/2021 (Z)')
plt.ylabel('Níveis Hpa')
cp= ax.contourf(X,Y,df,1000,cmap='jet')
fig.colorbar(cp)
for g in gr:
print(g)
```
|
github_jupyter
|
#Baixando bibliotecas
!sudo apt-get install python-grib
!sudo python setup.py install
!sudo apt-get install libgeos-dev
!sudo pip3 install -U git+https://github.com/matplotlib/basemap.git
!apt install libgrib-api-dev libgrib2c-dev
!pip install pyproj==1.9.6
!pip install pygrib
#Instalando as bibliotecas
import pygrib
from mpl_toolkits.basemap import Basemap
from mpl_toolkits.basemap import shiftgrid
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from datetime import datetime
#3 dias de previsão
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_000.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_003.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_006.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_009.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_012.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_015.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_018.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_021.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_024.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_027.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_030.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_033.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_036.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_039.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_042.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_045.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_048.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_051.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_054.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_057.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_060.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_063.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_066.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_069.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_072.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_075.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_078.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_081.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_084.grb2
!wget https://www.ncei.noaa.gov/data/global-forecast-system/access/grid-004-0.5-degree/forecast/202107/20210714/gfs_4_20210714_0000_087.grb2
#Abrindo os dados e selecionando as váriaveis
gr = pygrib.open('gfs_4_20210702_1800_006.grb2')
t = gr.select(name='Temperature')[0]
#Definindo região do RS
data, lats, lons = t.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
#Visualizando a grade selecionada
m = Basemap(projection='cyl',llcrnrlat=-35,urcrnrlat=-25,\
llcrnrlon=302,urcrnrlon=315,resolution='i')
# Função Matplotlib para definir o tamanho da figura.
plt.figure(figsize=(8,10))
#Inserindo os lat e lon a figura criada pelo Basemap e vinculando às variavéis x e y
x, y = m(lons, lats)
#Função do Basemap para iserir ao mapa continentes, países e estados.
m.drawcoastlines()
m.drawcountries()
m.drawstates()
la = np.arange(-35,-25,3.)
lo = np.arange(302.,315.,3.)
m.drawparallels(la,labels=[False,True,True,False])
m.drawmeridians(lo,labels=[True,False,False,True])
#m.scatter(lons, lats, marker = 'o', color='r', zorder=2)
#Função do Basemap para realizar a interpolação e criação de polígonos
contourf = m.contourf(x, y, np.squeeze(data),cmap='jet')
m.colorbar(contourf, location='right', pad="10%")
#Marcando cidade de Pelotas
m.scatter(307.6, -31.6, marker = 'o', color='r', zorder=3)
#Encontrando um lat e lon mais próximo
#Lat Lon selecionado
def encontra_lat_lon(la,lo):
stn_lat = la
stn_lon = lo
lat = lats
lon = lons
abslat = np.abs(lat-stn_lat)
abslon= np.abs(lon-stn_lon)
c = np.maximum(abslon,abslat)
latlon_idx = np.argmin(c)
x, y = np.where(c == np.min(c))
return(x,y)
#Aplicando para encontrar os index dos valores mais próximas
encontra_lat_lon(-31.6,307.6)
#Pegando o dado da respectiva região de Pelotas
data[13,175]
#Abrindo 3 dias de dados
gr = pygrib.open('gfs_4_20210714_0000_000.grb2')
gr2 = pygrib.open('gfs_4_20210714_0000_003.grb2')
gr3 = pygrib.open('gfs_4_20210714_0000_006.grb2')
gr4 = pygrib.open('gfs_4_20210714_0000_009.grb2')
gr5 = pygrib.open('gfs_4_20210714_0000_012.grb2')
gr6 = pygrib.open('gfs_4_20210714_0000_015.grb2')
gr7 = pygrib.open('gfs_4_20210714_0000_018.grb2')
gr8 = pygrib.open('gfs_4_20210714_0000_021.grb2')
gr9 = pygrib.open('gfs_4_20210714_0000_024.grb2')
gr10 = pygrib.open('gfs_4_20210714_0000_027.grb2')
gr11 = pygrib.open('gfs_4_20210714_0000_030.grb2')
gr12 = pygrib.open('gfs_4_20210714_0000_033.grb2')
gr13 = pygrib.open('gfs_4_20210714_0000_036.grb2')
gr14 = pygrib.open('gfs_4_20210714_0000_039.grb2')
gr15 = pygrib.open('gfs_4_20210714_0000_042.grb2')
gr16 = pygrib.open('gfs_4_20210714_0000_045.grb2')
gr17 = pygrib.open('gfs_4_20210714_0000_048.grb2')
gr18 = pygrib.open('gfs_4_20210714_0000_051.grb2')
gr19 = pygrib.open('gfs_4_20210714_0000_054.grb2')
gr20 = pygrib.open('gfs_4_20210714_0000_057.grb2')
gr21 = pygrib.open('gfs_4_20210714_0000_060.grb2')
gr22 = pygrib.open('gfs_4_20210714_0000_063.grb2')
gr23 = pygrib.open('gfs_4_20210714_0000_066.grb2')
gr24 = pygrib.open('gfs_4_20210714_0000_069.grb2')
gr25 = pygrib.open('gfs_4_20210714_0000_072.grb2')
gr26 = pygrib.open('gfs_4_20210714_0000_075.grb2')
gr27 = pygrib.open('gfs_4_20210714_0000_078.grb2')
gr28 = pygrib.open('gfs_4_20210714_0000_081.grb2')
gr29 = pygrib.open('gfs_4_20210714_0000_084.grb2')
gr30 = pygrib.open('gfs_4_20210714_0000_087.grb2')
# 1-Pegando os dados de temperatura referente aos níveis do modelo (ajustar lat e lon).
def selec_t(dadoo,name_v,indx_lat,index_lon):
dadt=[]
#Superfície
t1=dadoo.select(name=name_v)[41]
data1, lats, lons = t1.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data1[indx_lat,index_lon])
#1000
t2=dadoo.select(name=name_v)[40]
data2, lats, lons = t2.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data2[indx_lat,index_lon])
#975
t3=dadoo.select(name=name_v)[39]
data3, lats, lons = t3.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data3[indx_lat,index_lon])
#950
t4=dadoo.select(name=name_v)[38]
data4, lats, lons = t4.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data4[indx_lat,index_lon])
#925
t5=dadoo.select(name=name_v)[37]
data5, lats, lons = t5.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data5[indx_lat,index_lon])
#900
t6=dadoo.select(name=name_v)[36]
data6, lats, lons = t6.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data6[indx_lat,index_lon])
#850
t7=dadoo.select(name=name_v)[35]
data7, lats, lons = t7.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data7[indx_lat,index_lon])
#800
t8=dadoo.select(name=name_v)[34]
data8, lats, lons = t8.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data8[indx_lat,index_lon])
#750
t9=dadoo.select(name=name_v)[33]
data9, lats, lons = t9.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data9[indx_lat,index_lon])
#700
t10=dadoo.select(name=name_v)[32]
data10, lats, lons = t10.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data10[indx_lat,index_lon])
#650
t11=dadoo.select(name=name_v)[31]
data11, lats, lons = t11.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data11[indx_lat,index_lon])
#600
t12=dadoo.select(name=name_v)[30]
data12, lats, lons = t12.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data12[indx_lat,index_lon])
#550
t13=dadoo.select(name=name_v)[29]
data13, lats, lons = t13.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data13[indx_lat,index_lon])
#500hPa
t14=dadoo.select(name=name_v)[28]
data14, lats, lons = t14.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data14[indx_lat,index_lon])
#450hPa
t15=dadoo.select(name=name_v)[27]
data15, lats, lons = t15.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data15[indx_lat,index_lon])
#400hPa
t16=dadoo.select(name=name_v)[26]
data16, lats, lons = t16.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data16[indx_lat,index_lon])
#350hPa
t17=dadoo.select(name=name_v)[25]
data17, lats, lons = t17.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data17[indx_lat,index_lon])
#300hPa
t18=dadoo.select(name=name_v)[24]
data18, lats, lons = t18.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data18[indx_lat,index_lon])
#250hPa
t19=dadoo.select(name=name_v)[23]
data19, lats, lons = t19.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data19[indx_lat,index_lon])
#200hPa
t20=dadoo.select(name=name_v)[22]
data20, lats, lons = t20.data(lat1=-35,lat2=-25,lon1=220,lon2=320)
dadt.append(data20[indx_lat,index_lon])
return dadt
# 2 Função para criar e concatenar dataFrame (Aajuste de Temperatura - 273.15)
def transf_ajust(a1,a2,a3,a4,a5,a6,a7,a8,a9,a10,a11,a12,a13,a14,a15,a16,a17,a18,a19,a20,a21,a22,a23,a24,a25,a26,a27,a28,a29,a30):
index=['1013','1000','975','950','925','900','850','800','750','700','650','600','550','500','450','400','350','300','250','200']
ind = pd.DataFrame(index, columns=['niveis'] )
a1=pd.DataFrame(a1)
a2=pd.DataFrame(a2)
a3=pd.DataFrame(a3)
a4=pd.DataFrame(a4)
a5=pd.DataFrame(a5)
a6=pd.DataFrame(a6)
a7=pd.DataFrame(a7)
a8=pd.DataFrame(a8)
a9=pd.DataFrame(a9)
a10=pd.DataFrame(a10)
a11=pd.DataFrame(a11)
a12=pd.DataFrame(a12)
a13=pd.DataFrame(a13)
a14=pd.DataFrame(a14)
a15=pd.DataFrame(a15)
a16=pd.DataFrame(a16)
a17=pd.DataFrame(a17)
a18=pd.DataFrame(a18)
a19=pd.DataFrame(a19)
a20=pd.DataFrame(a20)
a21=pd.DataFrame(a21)
a22=pd.DataFrame(a22)
a23=pd.DataFrame(a23)
a24=pd.DataFrame(a24)
a25=pd.DataFrame(a25)
a26=pd.DataFrame(a26)
a27=pd.DataFrame(a27)
a28=pd.DataFrame(a28)
a29=pd.DataFrame(a29)
a30=pd.DataFrame(a30)
f = pd.concat([ind,a1,a2,a3,a4,a5,a6,a7,a8,a9,a10,a11,a12,a13,a14,a15,a16,a17,a18,a19,a20,a21,a22,a23,a24,a25,a26,a27,a28,a29,a30], axis=1)
return f
#Aplicando Função(dado no formato grib, 'nome da variavél desejada')
na='Relative humidity'
dad1=selec_t(gr,na)
dad2=selec_t(gr2,na)
dad3=selec_t(gr3,na)
dad4=selec_t(gr4,na)
dad5=selec_t(gr5,na)
dad6=selec_t(gr6,na)
dad7=selec_t(gr7,na)
dad8=selec_t(gr8,na)
dad9=selec_t(gr9,na)
dad10=selec_t(gr10,na)
dad11=selec_t(gr11,na)
dad12=selec_t(gr12,na)
dad13=selec_t(gr13,na)
dad14=selec_t(gr14,na)
dad15=selec_t(gr15,na)
dad16=selec_t(gr16,na)
dad17=selec_t(gr17,na)
dad18=selec_t(gr18,na)
dad19=selec_t(gr19,na)
dad20=selec_t(gr20,na)
dad21=selec_t(gr21,na)
dad22=selec_t(gr22,na)
dad23=selec_t(gr23,na)
dad24=selec_t(gr24,na)
dad25=selec_t(gr25,na)
dad26=selec_t(gr26,na)
dad27=selec_t(gr27,na)
dad28=selec_t(gr28,na)
dad29=selec_t(gr29,na)
dad30=selec_t(gr30,na)
# 2 Função para criar e concatenar dataFrame (Aajuste de Temperatura - 273.15)
df=transf_ajust(dad1,dad2,dad3,dad4,dad5,dad6,dad7,dad8,dad9,dad10,dad11,dad12,dad13,dad14,dad15,dad16,dad17,dad18,dad19,dad20,dad21,dad22,dad23,dad24,dad25,dad26,dad27,dad28,dad29,dad30)
df.columns = ['niveis','00', '03', '06','09','12','15','18','21','24','27','30','33','36','39','42','45','48','51','54','57','60', '63', '66','69','72','75','78','81','84','87']
Y = df['niveis'].astype(float)
del df['niveis']
df.iloc[::-1]
df.reset_index(drop=True)
X = df.columns.values[:].astype(float)
X, Y = np.meshgrid(X, Y)
#Plotando figura
fig,ax=plt.subplots(1,1)
fig.set_size_inches(12, 6)
fig.set_size_inches(12, 6)
plt.gca().invert_yaxis()
plt.title('Meteograma altura geopotencial para Cidade de Pelotas')
plt.xlabel('14/07/2021 -- 17/07/2021 (Z)')
plt.ylabel('Níveis Hpa')
cp= ax.contourf(X,Y,df,1000,cmap='jet')
fig.colorbar(cp)
for g in gr:
print(g)
| 0.286868 | 0.146087 |
# Frame Prediction
Author: Noah Agudelo
Course Project, UC Irvine, Math 10, W22
## Introduction
Introduce your project here. About 3 sentences.
This project explores the idea of frame prediction with pytorch. The dataset is 200 sequences of 10 frames, each frame is 64x64 pixels and each pixel is greyscale. Each sequence is made up of 2 moving numbers from the MNIST dataset. The idea here is to train a neural network using this dataset and try to predict the 10th frame of various sequences in the dataset.
## Main portion of the project
(You can either have all one section or divide into multiple sections)
### Imports
```
# Imports used for most of the project.
import numpy as np
import pandas as pd
import altair as alt
import torch
from torch import nn
from torch.utils.data import Dataset
from torchvision import datasets
from torchvision.transforms import ToTensor
from torch.utils.data import DataLoader
!pip install ipywidgets==7.6.5
# Imports used for displaying a sequence of the dataset.
import matplotlib.pyplot as plt
import io
from IPython.display import Image, display
from ipywidgets import widgets, Layout, HBox
# Imports used for the initial download of the data.
import tensorflow as tf
from tensorflow import keras
```
### The initial download of the data.
Download the dataset and save it as to not download it again when working with it later on.
```
# Download and load the dataset.
fpath = keras.utils.get_file(
"moving_mnist.npy",
"http://www.cs.toronto.edu/~nitish/unsupervised_video/mnist_test_seq.npy",
)
dataset = np.load(fpath)
# Swap the axes representing the number of frames and number of data samples.
dataset = np.swapaxes(dataset, 0, 1)
# We'll pick out 1000 of the 10000 total examples and use those.
dataset = dataset[:1000, ...]
# Add a channel dimension since the images are grayscale.
dataset = np.expand_dims(dataset, axis=-1)
np.save("dataset.npy", dataset)
dataset = np.load("dataset.npy")[:100].astype(float)
# Split into train and validation sets using indexing to optimize memory.
indexes = np.arange(dataset.shape[0])
np.random.shuffle(indexes)
train_index = indexes[: int(0.9 * dataset.shape[0])]
val_index = indexes[int(0.9 * dataset.shape[0]) :]
train_dataset = dataset[train_index]
val_dataset = dataset[val_index]
# Normalize the data to the 0-1 range.
train_dataset = train_dataset / 255
val_dataset = val_dataset / 255
# Split the data into half and append the half to the dataset
# Remove the last frame and make that the y.
def create_X_y(data):
mid = int(data.shape[1]/2 - 1)
X = np.append(data[:, 0 : mid, :, :],data[:,mid+1:-1,:,:],axis=0)
y = np.append(data[:, mid, :, :],data[:,-1,:,:],axis=0)
return X, y
# Apply the processing function to the datasets.
X_train, y_train = create_X_y(train_dataset)
X_val, y_val = create_X_y(val_dataset)
```
We save and load the training and validation sets here to save time when working with the project later.
```
# Save the dataset.
np.save("X_train.npy",X_train)
np.save("y_train.npy",y_train)
np.save("X_val.npy",X_val)
np.save("y_val.npy",y_val)
# Load the dataset.
X_train = np.load("X_train.npy").astype(float)
y_train = np.load("y_train.npy").astype(float)
X_val = np.load("X_val.npy").astype(float)
y_val = np.load("y_val.npy").astype(float)
```
The original dataset has 100 samples of sequences, with 20 frames each. We split each sequence into two, effectively making the dataset 200 samples of 10 frame sequences. This was done to save memory.
```
# Inspect the dataset.
print(f"Original Dataset Shape: {dataset.shape}")
print(f"Original Training Dataset Shape: {train_dataset.shape}")
print(f"Original Validation Dataset Shape: {val_dataset.shape}")
print(f"Modified Training Dataset Shapes: {X_train.shape}, {y_train.shape}")
print(f"Modified Validation Dataset Shapes: {X_val.shape}, {y_val.shape}")
```
We create a custom iterable class in order to create batches of the dataset.
```
class CustomDataset(Dataset):
def __init__(self, dataset_X, dataset_y, transform=None, target_transform=None):
self.data_X = dataset_X
self.data_y = dataset_y
self.transform = transform
self.target_transform = target_transform
def __len__(self):
return len(self.data_X)
def __getitem__(self, idx):
X = self.data_X[idx]
y = self.data_y[idx]
if self.transform:
X = self.transform(X)
if self.target_transform:
y = self.target_transform(y)
return X, y
batch_size = 5
train_dataloader = DataLoader(CustomDataset(X_train,y_train), batch_size=batch_size)
test_dataloader = DataLoader(CustomDataset(X_val,y_val), batch_size=batch_size)
X, y = next(iter(train_dataloader))
print(f"Shape of X:\n{X.shape}")
print("")
print(f"Shape of y:\n{y.shape}")
```
PyTorch has LSTM layers, which probably would have helped making the neural network more accurate in predicting the last frame. However, we instead use two linear layers to keep things more simple.
I also ran out of memory when using more than 2 linear layers, and so I reduced it to only two layers.
```
class Frames(nn.Module) :
def __init__(self) :
super().__init__()
self.flatten = nn.Flatten()
self.layers = nn.Sequential(
nn.Linear(9*64*64, 2*64*64),
nn.ReLU(),
nn.Linear(2*64*64, 64*64),
nn.Sigmoid()
)
def forward(self, x) :
return self.layers(self.flatten(x))
my_nn = Frames()
loss_fn = nn.MSELoss()
optimizer = torch.optim.SGD(my_nn.parameters(), lr=0.25)
```
We train the neural network on the dataset while keeping track of the loss along the way.
```
epochs = 20
training_loss = np.zeros(epochs)
test_loss = np.zeros(epochs)
for i in range(epochs):
for X,y in train_dataloader:
X = X.float()
y = y.float()
loss = loss_fn(my_nn(X),y.flatten(1,3))
optimizer.zero_grad()
loss.backward()
optimizer.step()
print(f"Training loss:\n{loss}") # We only print one loss per epoch
training_loss[i] = loss.item()
for X,y in test_dataloader:
X = X.float()
y = y.float()
loss = loss_fn(my_nn(X),y.flatten(1,3))
print(f"Test loss:\n{loss}")
test_loss[i] = loss.item()
break # We only compute and print one test loss per epoch
print("")
# Save the loss to file.
np.save("training_loss.npy", training_loss)
np.save("test_loss.npy", test_loss)
# Load the loss from file.
training_loss = np.load("training_loss.npy")
test_loss = np.load("test_loss.npy")
scores = pd.DataFrame({"Epoch":range(1,epochs+1), "train_loss":training_loss, "test_loss":test_loss})
scores.head()
```
Here, we plot loss over each epoch for both the test loss and training loss.
```
alt.Chart(scores).mark_line().encode(
x= alt.X("Epoch", scale = alt.Scale(nice=False, domain = (1,20))),
y=alt.Y('value:Q', title="Loss", scale = alt.Scale(zero=False)),
color=alt.Color("key:N",title="Legend")
).transform_fold(
fold=["train_loss", "test_loss"]
).properties(
title="Test Loss vs Train Loss",
width = 1000,
height = 500
)
```
Now, we take a look at a predicted output and see what it looks like.
```
pred = my_nn(X)
pred = pred.reshape([5, 64, 64, 1])
print(X.shape)
print(y.shape)
print(pred.shape)
# Construct a figure on which we will visualize the images.
fig, axes = plt.subplots(2, 5, figsize=(10, 4))
# Plot each of the sequential images for one random data example.
for idx, ax in enumerate(axes.flat):
if idx != 9:
ax.imshow(np.squeeze(X[0][idx]), cmap="gray")
else:
ax.imshow(np.squeeze(y[0]), cmap="gray")
ax.set_title(f"Frame {idx + 1}")
ax.axis("off")
# Display the figure.
plt.show()
# Construct a figure on which we will visualize the images.
fig, axes = plt.subplots(1, 2, figsize=(5, 3))
# Plot each of the sequential images for one random data example.
axes[0].imshow(np.squeeze(y[0].detach().numpy()), cmap="gray")
axes[0].set_title("Actual last frame")
axes[0].axis("off")
axes[1].imshow(np.squeeze(pred[0].detach().numpy()), cmap="gray")
axes[1].set_title("Predicted last frame")
axes[1].axis("off")
# Print information and display the figure.
plt.show()
```
## Summary
The test loss is smaller than the training loss, which seems odd to the neural network is training to reduce training loss, not test loss.
I also assumed the predicted last frame would look more recognizable when comparing it to the actual last frame.
To improve the neural network, perhaps it should include some layer that more strictly returns greyscale pixels closer to black or full white, since each actual frame has very distinct bright numbers with black background. It could also include an LSTM layer, which takes previous guesses and previous correct answers into account when predicting a frame in the sequence.
## References
Include references that you found helpful. Also say where you found the dataset you used.
Download and cleaning of the dataset, matplotlib examples, etc. from Keras tutorial on frame prediction:
https://keras.io/examples/vision/conv_lstm/
<a style='text-decoration:none;line-height:16px;display:flex;color:#5B5B62;padding:10px;justify-content:end;' href='https://deepnote.com?utm_source=created-in-deepnote-cell&projectId=e7d25786-d0b6-46ff-ae9c-55a1da4c0726' target="_blank">
<img alt='Created in deepnote.com' style='display:inline;max-height:16px;margin:0px;margin-right:7.5px;' src='data:image/svg+xml;base64,PD94bWwgdmVyc2lvbj0iMS4wIiBlbmNvZGluZz0iVVRGLTgiPz4KPHN2ZyB3aWR0aD0iODBweCIgaGVpZ2h0PSI4MHB4IiB2aWV3Qm94PSIwIDAgODAgODAiIHZlcnNpb249IjEuMSIgeG1sbnM9Imh0dHA6Ly93d3cudzMub3JnLzIwMDAvc3ZnIiB4bWxuczp4bGluaz0iaHR0cDovL3d3dy53My5vcmcvMTk5OS94bGluayI+CiAgICA8IS0tIEdlbmVyYXRvcjogU2tldGNoIDU0LjEgKDc2NDkwKSAtIGh0dHBzOi8vc2tldGNoYXBwLmNvbSAtLT4KICAgIDx0aXRsZT5Hcm91cCAzPC90aXRsZT4KICAgIDxkZXNjPkNyZWF0ZWQgd2l0aCBTa2V0Y2guPC9kZXNjPgogICAgPGcgaWQ9IkxhbmRpbmciIHN0cm9rZT0ibm9uZSIgc3Ryb2tlLXdpZHRoPSIxIiBmaWxsPSJub25lIiBmaWxsLXJ1bGU9ImV2ZW5vZGQiPgogICAgICAgIDxnIGlkPSJBcnRib2FyZCIgdHJhbnNmb3JtPSJ0cmFuc2xhdGUoLTEyMzUuMDAwMDAwLCAtNzkuMDAwMDAwKSI+CiAgICAgICAgICAgIDxnIGlkPSJHcm91cC0zIiB0cmFuc2Zvcm09InRyYW5zbGF0ZSgxMjM1LjAwMDAwMCwgNzkuMDAwMDAwKSI+CiAgICAgICAgICAgICAgICA8cG9seWdvbiBpZD0iUGF0aC0yMCIgZmlsbD0iIzAyNjVCNCIgcG9pbnRzPSIyLjM3NjIzNzYyIDgwIDM4LjA0NzY2NjcgODAgNTcuODIxNzgyMiA3My44MDU3NTkyIDU3LjgyMTc4MjIgMzIuNzU5MjczOSAzOS4xNDAyMjc4IDMxLjY4MzE2ODMiPjwvcG9seWdvbj4KICAgICAgICAgICAgICAgIDxwYXRoIGQ9Ik0zNS4wMDc3MTgsODAgQzQyLjkwNjIwMDcsNzYuNDU0OTM1OCA0Ny41NjQ5MTY3LDcxLjU0MjI2NzEgNDguOTgzODY2LDY1LjI2MTk5MzkgQzUxLjExMjI4OTksNTUuODQxNTg0MiA0MS42NzcxNzk1LDQ5LjIxMjIyODQgMjUuNjIzOTg0Niw0OS4yMTIyMjg0IEMyNS40ODQ5Mjg5LDQ5LjEyNjg0NDggMjkuODI2MTI5Niw0My4yODM4MjQ4IDM4LjY0NzU4NjksMzEuNjgzMTY4MyBMNzIuODcxMjg3MSwzMi41NTQ0MjUgTDY1LjI4MDk3Myw2Ny42NzYzNDIxIEw1MS4xMTIyODk5LDc3LjM3NjE0NCBMMzUuMDA3NzE4LDgwIFoiIGlkPSJQYXRoLTIyIiBmaWxsPSIjMDAyODY4Ij48L3BhdGg+CiAgICAgICAgICAgICAgICA8cGF0aCBkPSJNMCwzNy43MzA0NDA1IEwyNy4xMTQ1MzcsMC4yNTcxMTE0MzYgQzYyLjM3MTUxMjMsLTEuOTkwNzE3MDEgODAsMTAuNTAwMzkyNyA4MCwzNy43MzA0NDA1IEM4MCw2NC45NjA0ODgyIDY0Ljc3NjUwMzgsNzkuMDUwMzQxNCAzNC4zMjk1MTEzLDgwIEM0Ny4wNTUzNDg5LDc3LjU2NzA4MDggNTMuNDE4MjY3Nyw3MC4zMTM2MTAzIDUzLjQxODI2NzcsNTguMjM5NTg4NSBDNTMuNDE4MjY3Nyw0MC4xMjg1NTU3IDM2LjMwMzk1NDQsMzcuNzMwNDQwNSAyNS4yMjc0MTcsMzcuNzMwNDQwNSBDMTcuODQzMDU4NiwzNy43MzA0NDA1IDkuNDMzOTE5NjYsMzcuNzMwNDQwNSAwLDM3LjczMDQ0MDUgWiIgaWQ9IlBhdGgtMTkiIGZpbGw9IiMzNzkzRUYiPjwvcGF0aD4KICAgICAgICAgICAgPC9nPgogICAgICAgIDwvZz4KICAgIDwvZz4KPC9zdmc+' > </img>
Created in <span style='font-weight:600;margin-left:4px;'>Deepnote</span></a>
|
github_jupyter
|
# Imports used for most of the project.
import numpy as np
import pandas as pd
import altair as alt
import torch
from torch import nn
from torch.utils.data import Dataset
from torchvision import datasets
from torchvision.transforms import ToTensor
from torch.utils.data import DataLoader
!pip install ipywidgets==7.6.5
# Imports used for displaying a sequence of the dataset.
import matplotlib.pyplot as plt
import io
from IPython.display import Image, display
from ipywidgets import widgets, Layout, HBox
# Imports used for the initial download of the data.
import tensorflow as tf
from tensorflow import keras
# Download and load the dataset.
fpath = keras.utils.get_file(
"moving_mnist.npy",
"http://www.cs.toronto.edu/~nitish/unsupervised_video/mnist_test_seq.npy",
)
dataset = np.load(fpath)
# Swap the axes representing the number of frames and number of data samples.
dataset = np.swapaxes(dataset, 0, 1)
# We'll pick out 1000 of the 10000 total examples and use those.
dataset = dataset[:1000, ...]
# Add a channel dimension since the images are grayscale.
dataset = np.expand_dims(dataset, axis=-1)
np.save("dataset.npy", dataset)
dataset = np.load("dataset.npy")[:100].astype(float)
# Split into train and validation sets using indexing to optimize memory.
indexes = np.arange(dataset.shape[0])
np.random.shuffle(indexes)
train_index = indexes[: int(0.9 * dataset.shape[0])]
val_index = indexes[int(0.9 * dataset.shape[0]) :]
train_dataset = dataset[train_index]
val_dataset = dataset[val_index]
# Normalize the data to the 0-1 range.
train_dataset = train_dataset / 255
val_dataset = val_dataset / 255
# Split the data into half and append the half to the dataset
# Remove the last frame and make that the y.
def create_X_y(data):
mid = int(data.shape[1]/2 - 1)
X = np.append(data[:, 0 : mid, :, :],data[:,mid+1:-1,:,:],axis=0)
y = np.append(data[:, mid, :, :],data[:,-1,:,:],axis=0)
return X, y
# Apply the processing function to the datasets.
X_train, y_train = create_X_y(train_dataset)
X_val, y_val = create_X_y(val_dataset)
# Save the dataset.
np.save("X_train.npy",X_train)
np.save("y_train.npy",y_train)
np.save("X_val.npy",X_val)
np.save("y_val.npy",y_val)
# Load the dataset.
X_train = np.load("X_train.npy").astype(float)
y_train = np.load("y_train.npy").astype(float)
X_val = np.load("X_val.npy").astype(float)
y_val = np.load("y_val.npy").astype(float)
# Inspect the dataset.
print(f"Original Dataset Shape: {dataset.shape}")
print(f"Original Training Dataset Shape: {train_dataset.shape}")
print(f"Original Validation Dataset Shape: {val_dataset.shape}")
print(f"Modified Training Dataset Shapes: {X_train.shape}, {y_train.shape}")
print(f"Modified Validation Dataset Shapes: {X_val.shape}, {y_val.shape}")
class CustomDataset(Dataset):
def __init__(self, dataset_X, dataset_y, transform=None, target_transform=None):
self.data_X = dataset_X
self.data_y = dataset_y
self.transform = transform
self.target_transform = target_transform
def __len__(self):
return len(self.data_X)
def __getitem__(self, idx):
X = self.data_X[idx]
y = self.data_y[idx]
if self.transform:
X = self.transform(X)
if self.target_transform:
y = self.target_transform(y)
return X, y
batch_size = 5
train_dataloader = DataLoader(CustomDataset(X_train,y_train), batch_size=batch_size)
test_dataloader = DataLoader(CustomDataset(X_val,y_val), batch_size=batch_size)
X, y = next(iter(train_dataloader))
print(f"Shape of X:\n{X.shape}")
print("")
print(f"Shape of y:\n{y.shape}")
class Frames(nn.Module) :
def __init__(self) :
super().__init__()
self.flatten = nn.Flatten()
self.layers = nn.Sequential(
nn.Linear(9*64*64, 2*64*64),
nn.ReLU(),
nn.Linear(2*64*64, 64*64),
nn.Sigmoid()
)
def forward(self, x) :
return self.layers(self.flatten(x))
my_nn = Frames()
loss_fn = nn.MSELoss()
optimizer = torch.optim.SGD(my_nn.parameters(), lr=0.25)
epochs = 20
training_loss = np.zeros(epochs)
test_loss = np.zeros(epochs)
for i in range(epochs):
for X,y in train_dataloader:
X = X.float()
y = y.float()
loss = loss_fn(my_nn(X),y.flatten(1,3))
optimizer.zero_grad()
loss.backward()
optimizer.step()
print(f"Training loss:\n{loss}") # We only print one loss per epoch
training_loss[i] = loss.item()
for X,y in test_dataloader:
X = X.float()
y = y.float()
loss = loss_fn(my_nn(X),y.flatten(1,3))
print(f"Test loss:\n{loss}")
test_loss[i] = loss.item()
break # We only compute and print one test loss per epoch
print("")
# Save the loss to file.
np.save("training_loss.npy", training_loss)
np.save("test_loss.npy", test_loss)
# Load the loss from file.
training_loss = np.load("training_loss.npy")
test_loss = np.load("test_loss.npy")
scores = pd.DataFrame({"Epoch":range(1,epochs+1), "train_loss":training_loss, "test_loss":test_loss})
scores.head()
alt.Chart(scores).mark_line().encode(
x= alt.X("Epoch", scale = alt.Scale(nice=False, domain = (1,20))),
y=alt.Y('value:Q', title="Loss", scale = alt.Scale(zero=False)),
color=alt.Color("key:N",title="Legend")
).transform_fold(
fold=["train_loss", "test_loss"]
).properties(
title="Test Loss vs Train Loss",
width = 1000,
height = 500
)
pred = my_nn(X)
pred = pred.reshape([5, 64, 64, 1])
print(X.shape)
print(y.shape)
print(pred.shape)
# Construct a figure on which we will visualize the images.
fig, axes = plt.subplots(2, 5, figsize=(10, 4))
# Plot each of the sequential images for one random data example.
for idx, ax in enumerate(axes.flat):
if idx != 9:
ax.imshow(np.squeeze(X[0][idx]), cmap="gray")
else:
ax.imshow(np.squeeze(y[0]), cmap="gray")
ax.set_title(f"Frame {idx + 1}")
ax.axis("off")
# Display the figure.
plt.show()
# Construct a figure on which we will visualize the images.
fig, axes = plt.subplots(1, 2, figsize=(5, 3))
# Plot each of the sequential images for one random data example.
axes[0].imshow(np.squeeze(y[0].detach().numpy()), cmap="gray")
axes[0].set_title("Actual last frame")
axes[0].axis("off")
axes[1].imshow(np.squeeze(pred[0].detach().numpy()), cmap="gray")
axes[1].set_title("Predicted last frame")
axes[1].axis("off")
# Print information and display the figure.
plt.show()
| 0.923747 | 0.979275 |
```
# Taken from the following page:
# https://www.ibm.com/developerworks/community/blogs/jfp/entry/How_To_Compute_Mandelbrodt_Set_Quickly?lang=en
%matplotlib inline
%load_ext autoreload
%autoreload 2
import src.mandelbrot as mb
mb.mandelbrot_image(-2.0, 0.5, -1.25, 1.25, scale=2.0, maxiter=2048, cmap='jet')
from IPython.display import display
from ipywidgets import interact, interactive, fixed, interact_manual
import ipywidgets as widgets
import matplotlib.pyplot as plt
cmaps = ['hot', 'gnuplot2', 'jet', 'coolwarm', 'copper', 'winter']
@interact(x=(-1.8, 0.4), y=(-1.2, 1.2), scale=(0, 100, 1), maxiter=(100, 2000, 100), cmap=cmaps)
def interactive_mandelplot(x=-0.74529, y=0.113075, scale=0, maxiter=100, cmap='gnuplot2'):
pad = 1.25
mb.mandelbrot_image(x, y, scale=scale, N=1000, maxiter=maxiter, cmap=cmap)
# plt.scatter([x/scale], [y/scale], s=30, marker='+')
plt.show()
c1 = mb.mandelbrot_set(-0.74877, -0.74872, log2scale=0)
plt.imshow(c1[2])
c1 = mb.mandelbrot_set(-0.74877, -0.74872, 0.06505, 0.06510, 1000, 1000, 2048, scale=1.1)
plt.imshow(c1[2])
```
# Scratch
```
mb.mandelbrot_image(-2.0, 0.5, -1.25, 1.25, maxiter=2048, cmap='jet')
mb.mandelbrot_image(-0.74877, -0.74872, 0.06505, 0.06510, maxiter=2048, cmap='gnuplot2')
%timeit mb.mandelbrot_set(-2.0,0.5,-1.25,1.25,1000,1000,80)
%timeit mb.mandelbrot_set(-0.74877,-0.74872,0.06505,0.06510,1000,1000,2048)
c = mb.mandelbrot_set(-0.74877, -0.74872, 0.06505, 0.06510, 1000, 1000, 2048)
# Python code for Mandelbrot Fractal
%matplotlib inline
# Import necessary libraries
from PIL import Image
from numpy import complex, array
import colorsys
from tqdm import tqdm
import numpy as np
# setting the width of the output image as 1024
WIDTH = 1024
# a function to return a tuple of colors
# as integer value of rgb
def rgb_conv(i):
color = 255 * array(colorsys.hsv_to_rgb(i / 255.0, 1.0, 0.5))
return tuple(color.astype(int))
# function defining a mandelbrot
def mandelbrot(x, y):
c = complex(x, y)
z = 0
for i in range(1, 1000):
if abs(z) > 2:
return rgb_conv(i)
z = z ** 2 + c
return (0, 0, 0)
# creating the new image in RGB mode
def get_frame():
img = Image.new('RGB', (WIDTH, int(WIDTH / 2)))
pixels = img.load()
for x in tqdm(range(img.size[0])):
for y in range(img.size[1]):
pixels[x, y] = mandelbrot((x - (0.75 * WIDTH))*0.5 / (WIDTH / 4),
(y - (WIDTH / 4))*0.5 / (WIDTH / 4))
get_frame()
# to display the created fractal after
# completing the given number of iterations
img
```
|
github_jupyter
|
# Taken from the following page:
# https://www.ibm.com/developerworks/community/blogs/jfp/entry/How_To_Compute_Mandelbrodt_Set_Quickly?lang=en
%matplotlib inline
%load_ext autoreload
%autoreload 2
import src.mandelbrot as mb
mb.mandelbrot_image(-2.0, 0.5, -1.25, 1.25, scale=2.0, maxiter=2048, cmap='jet')
from IPython.display import display
from ipywidgets import interact, interactive, fixed, interact_manual
import ipywidgets as widgets
import matplotlib.pyplot as plt
cmaps = ['hot', 'gnuplot2', 'jet', 'coolwarm', 'copper', 'winter']
@interact(x=(-1.8, 0.4), y=(-1.2, 1.2), scale=(0, 100, 1), maxiter=(100, 2000, 100), cmap=cmaps)
def interactive_mandelplot(x=-0.74529, y=0.113075, scale=0, maxiter=100, cmap='gnuplot2'):
pad = 1.25
mb.mandelbrot_image(x, y, scale=scale, N=1000, maxiter=maxiter, cmap=cmap)
# plt.scatter([x/scale], [y/scale], s=30, marker='+')
plt.show()
c1 = mb.mandelbrot_set(-0.74877, -0.74872, log2scale=0)
plt.imshow(c1[2])
c1 = mb.mandelbrot_set(-0.74877, -0.74872, 0.06505, 0.06510, 1000, 1000, 2048, scale=1.1)
plt.imshow(c1[2])
mb.mandelbrot_image(-2.0, 0.5, -1.25, 1.25, maxiter=2048, cmap='jet')
mb.mandelbrot_image(-0.74877, -0.74872, 0.06505, 0.06510, maxiter=2048, cmap='gnuplot2')
%timeit mb.mandelbrot_set(-2.0,0.5,-1.25,1.25,1000,1000,80)
%timeit mb.mandelbrot_set(-0.74877,-0.74872,0.06505,0.06510,1000,1000,2048)
c = mb.mandelbrot_set(-0.74877, -0.74872, 0.06505, 0.06510, 1000, 1000, 2048)
# Python code for Mandelbrot Fractal
%matplotlib inline
# Import necessary libraries
from PIL import Image
from numpy import complex, array
import colorsys
from tqdm import tqdm
import numpy as np
# setting the width of the output image as 1024
WIDTH = 1024
# a function to return a tuple of colors
# as integer value of rgb
def rgb_conv(i):
color = 255 * array(colorsys.hsv_to_rgb(i / 255.0, 1.0, 0.5))
return tuple(color.astype(int))
# function defining a mandelbrot
def mandelbrot(x, y):
c = complex(x, y)
z = 0
for i in range(1, 1000):
if abs(z) > 2:
return rgb_conv(i)
z = z ** 2 + c
return (0, 0, 0)
# creating the new image in RGB mode
def get_frame():
img = Image.new('RGB', (WIDTH, int(WIDTH / 2)))
pixels = img.load()
for x in tqdm(range(img.size[0])):
for y in range(img.size[1]):
pixels[x, y] = mandelbrot((x - (0.75 * WIDTH))*0.5 / (WIDTH / 4),
(y - (WIDTH / 4))*0.5 / (WIDTH / 4))
get_frame()
# to display the created fractal after
# completing the given number of iterations
img
| 0.638835 | 0.74144 |
<a href="https://colab.research.google.com/github/unpackAI/unpackai/blob/main/examples/nlp_regression_toxity.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# NLP tasks
> From unpackAI
```
!pip install -q unpackai==0.1.8.9
!pip install -Uqq fastai
!pip install -q transformers
from fastai.text.all import *
from unpackai.nlp import HFTextBlock
from transformers import AutoModel, AutoTokenizer
```
## Regression Problem
### About the problem
Acute toxicity LD50 measures the most conservative dose that can lead to lethal adverse effects.
> We are to design a model, which take the chemical expression as input, and predict the possible toxity, hence will prioritize the experiments on more promising drug.
* X:**SMILES** expression, (The simplified molecular-input line-entry system), it's a way to write 3d organic chemical structure into a string, that can be reconstructed back to 3d structure
* Y: **LD50 Toxity level**, the amount of the compound that will kill around 50% of the lab rats
### About dataset
[TDC](https://tdcommons.ai/start/) is the first unifying framework to systematically access and evaluate ML across the entire range of therapeutics
### Download dataset
```
!pip install -q PyTDC
from tdc.single_pred import Tox
data = Tox(name = 'LD50_Zhu')
df = data.get_data()
df
df.describe()
```
Normalize the target a little bit
```
df['target'] = df["Y"]-2.5
```
## Download pretrained model
```
pretrained = AutoModel.from_pretrained("seyonec/SMILES_tokenized_PubChem_shard00_160k")
tokenizer = AutoTokenizer.from_pretrained("seyonec/SMILES_tokenized_PubChem_shard00_160k")
```
## Data
### Tokenizer
Let's look like what the tokenization on SMILES looks like.
It's exactly like on usual sentences
```
tokenizer("CC1=CC(=C2C(=O)c3ccccc3C2=O)C=CN1CCN1CCCCC1")
```
### Datablock
```
dblock = DataBlock(
blocks=[
HFTextBlock(
tokenizer,
max_length=128,
padding=True),
RegressionBlock],
get_x=ColReader("Drug"), get_y=ColReader("target",))
```
### Dataloaders
```
dls = dblock.dataloaders(df, bs=32)
dls.train_ds[3]
```
A preview on a batch of data
```
x, y = dls.one_batch()
x.shape, y.shape
```
## Create a model with pretrained weights
```
class NLPRegressor(nn.Module):
def __init__(self, pretrained):
super().__init__()
self.pretrained = pretrained
self.top = nn.Linear(pretrained.config.hidden_size, 1)
def forward(self, x):
output = self.pretrained(x)
return self.top(output.pooler_output)
model = NLPRegressor(pretrained)
```
## Training
```
learn = Learner(model=model,dls=dls,loss_func=nn.MSELoss())
learn.fit(2)
```
|
github_jupyter
|
!pip install -q unpackai==0.1.8.9
!pip install -Uqq fastai
!pip install -q transformers
from fastai.text.all import *
from unpackai.nlp import HFTextBlock
from transformers import AutoModel, AutoTokenizer
!pip install -q PyTDC
from tdc.single_pred import Tox
data = Tox(name = 'LD50_Zhu')
df = data.get_data()
df
df.describe()
df['target'] = df["Y"]-2.5
pretrained = AutoModel.from_pretrained("seyonec/SMILES_tokenized_PubChem_shard00_160k")
tokenizer = AutoTokenizer.from_pretrained("seyonec/SMILES_tokenized_PubChem_shard00_160k")
tokenizer("CC1=CC(=C2C(=O)c3ccccc3C2=O)C=CN1CCN1CCCCC1")
dblock = DataBlock(
blocks=[
HFTextBlock(
tokenizer,
max_length=128,
padding=True),
RegressionBlock],
get_x=ColReader("Drug"), get_y=ColReader("target",))
dls = dblock.dataloaders(df, bs=32)
dls.train_ds[3]
x, y = dls.one_batch()
x.shape, y.shape
class NLPRegressor(nn.Module):
def __init__(self, pretrained):
super().__init__()
self.pretrained = pretrained
self.top = nn.Linear(pretrained.config.hidden_size, 1)
def forward(self, x):
output = self.pretrained(x)
return self.top(output.pooler_output)
model = NLPRegressor(pretrained)
learn = Learner(model=model,dls=dls,loss_func=nn.MSELoss())
learn.fit(2)
| 0.739986 | 0.969208 |
<a href="https://colab.research.google.com/github/nhatminh46vn/datajam.ai/blob/master/data_scraping.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
## **structure of URL **
Page 1 : https://www.coursera.org/learn/inferential-statistical-analysis-python/reviews?page=1
Page 2 : https://www.coursera.org/learn/inferential-statistical-analysis-python/reviews?page=2
Page 10 : https://www.coursera.org/learn/inferential-statistical-analysis-python/reviews?page=10
Question : When to stop in case we do not know how many review pages of the course is ? Anwser : using request error as stop criterial
Question : how to count the rating star for each review ?
Remark: we need to scrape the feed back as much as possible in different subject ( statistic/Python/SQL/...)
Remark : need to add headers and time of request in resquest code avoid the scraping by bot.
Remark : use pandas to export the scv into 3 columns (name of reviewer / comment/ rating )
```
#importation of beautifulSoup
from bs4 import BeautifulSoup
import requests
import re
import pdb
URL = 'https://www.coursera.org/learn/inferential-statistical-analysis-python/reviews'
page = requests.get(URL)
# Html parsing
soup = BeautifulSoup(page.content)
# print the resquest page with function Prettify
#print(soup.prettify())
block1st=soup.find('div',class_='_ng613ig review-text') #1st block of review section ( inlucding text and rating star )
review=block1st.find('div',class_='reviewText').find('p') #extract 1st review text
print(review.prettify()) #extract 1st block related to the review dessus
rating_block1st=block1st.find('div' ,class_="_1mzojlvw", role="img",)
emptyStar=re.findall('"fill:rgba.\w+', str(rating_block1st)) #fcking ratingstar took me a day for solution
rating=len(rating_block1st)-len(emptyStar)
print(rating)
# rating star for 1st block sur 25 blocks of review
rating_blocks=soup.body.findAll('div',class_='_jyhj5r review review-page-review m-b-2')
parttern=re.compile(r'fill:rgba')
match=parttern.findall(str(rating_blocks[0]))
rating=5-len(match)
print('Rating of 1st comment is :', rating)
#review text
alls_review=[]
#make a loop
blocks=soup.find_all('div',class_='reviewText')
# review=Block.find('p')
# alls_review.append(review)
for reviews in blocks:
review=reviews.find('p')
alls_review.append(review)
print(alls_review)
len(alls_review)
#looping
# rgba is not filled '#f7bb56' = filled
rating_blocks=soup.body.findAll('div',class_='_jyhj5r review review-page-review m-b-2')
parttern=re.compile(r'fill:rgba')
ratings=[]
for i in list(range(25)):
matches=parttern.findall(str(rating_blocks[i]))
a = 5-len(matches)
ratings.append(a)
print(ratings)
#1st block of review section ( inlucding text and rating star )
block1st=soup.find('div',class_='_ng613ig review-text')
rating_block1st=block1st.find('div' ,class_="_1mzojlvw", role="img",)
#fcking ratingstar took me a day for solution
emptyStar=re.findall('"fill:rgba.\w+', str(rating_block1st))
rating=len(rating_block1st)-len(emptyStar)
print('Rating of first comment: \n', rating)
alls = []
for d in soup.findAll('div', attrs={'class':'top-review'}):
top_review = d.find('p', attrs={'class':'top-review_comment'},recursive=True)
if top_review is not None:
alls.append(top_review)
print(top_review)
```
|
github_jupyter
|
#importation of beautifulSoup
from bs4 import BeautifulSoup
import requests
import re
import pdb
URL = 'https://www.coursera.org/learn/inferential-statistical-analysis-python/reviews'
page = requests.get(URL)
# Html parsing
soup = BeautifulSoup(page.content)
# print the resquest page with function Prettify
#print(soup.prettify())
block1st=soup.find('div',class_='_ng613ig review-text') #1st block of review section ( inlucding text and rating star )
review=block1st.find('div',class_='reviewText').find('p') #extract 1st review text
print(review.prettify()) #extract 1st block related to the review dessus
rating_block1st=block1st.find('div' ,class_="_1mzojlvw", role="img",)
emptyStar=re.findall('"fill:rgba.\w+', str(rating_block1st)) #fcking ratingstar took me a day for solution
rating=len(rating_block1st)-len(emptyStar)
print(rating)
# rating star for 1st block sur 25 blocks of review
rating_blocks=soup.body.findAll('div',class_='_jyhj5r review review-page-review m-b-2')
parttern=re.compile(r'fill:rgba')
match=parttern.findall(str(rating_blocks[0]))
rating=5-len(match)
print('Rating of 1st comment is :', rating)
#review text
alls_review=[]
#make a loop
blocks=soup.find_all('div',class_='reviewText')
# review=Block.find('p')
# alls_review.append(review)
for reviews in blocks:
review=reviews.find('p')
alls_review.append(review)
print(alls_review)
len(alls_review)
#looping
# rgba is not filled '#f7bb56' = filled
rating_blocks=soup.body.findAll('div',class_='_jyhj5r review review-page-review m-b-2')
parttern=re.compile(r'fill:rgba')
ratings=[]
for i in list(range(25)):
matches=parttern.findall(str(rating_blocks[i]))
a = 5-len(matches)
ratings.append(a)
print(ratings)
#1st block of review section ( inlucding text and rating star )
block1st=soup.find('div',class_='_ng613ig review-text')
rating_block1st=block1st.find('div' ,class_="_1mzojlvw", role="img",)
#fcking ratingstar took me a day for solution
emptyStar=re.findall('"fill:rgba.\w+', str(rating_block1st))
rating=len(rating_block1st)-len(emptyStar)
print('Rating of first comment: \n', rating)
alls = []
for d in soup.findAll('div', attrs={'class':'top-review'}):
top_review = d.find('p', attrs={'class':'top-review_comment'},recursive=True)
if top_review is not None:
alls.append(top_review)
print(top_review)
| 0.042434 | 0.827236 |
<a href="https://colab.research.google.com/github/LeehyeongTea/image_captioning_with_attention/blob/main/model.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import numpy as np
import h5py
import os
import tensorflow as tf
from tensorflow.keras.layers import Dense, LSTM, Input, Embedding, Dropout,BatchNormalization,Lambda, Add,Flatten,GRU
from tensorflow.keras.optimizers import Adam, RMSprop
from tensorflow.keras.applications import DenseNet121
from tensorflow.keras.models import Model
from tensorflow.keras import regularizers,optimizers,losses,metrics
from tensorflow.keras import backend as K
from tensorflow.keras.models import load_model
from tensorflow.keras.utils import plot_model,to_categorical
import pickle
def get_path(base_directory):
saved_data_path = os.path.join(base_directory,'data')
data_h5_paths = os.path.join(saved_data_path, 'needs.hdf5')
needs = h5py.File(data_h5_paths, 'r')
train_dataset_list_path = needs['train_code_path'][()]
val_dataset_list_path = needs['val_code_path'][()]
train_feature_path = needs['train_feature_path'][()]
val_feature_path = needs['val_feature_path'][()]
train_seq_path= needs['train_seq_path'][()]
val_seq_path= needs['val_seq_path'][()]
max_len = needs['max_len'][()]
vocab_size= needs['vocab_size'][()]
req_train_list_path = needs['req_train_list_path'][()]
req_val_list_path = needs['req_val_list_path'][()]
req_token_path = needs['req_token_path'][()]
return train_dataset_list_path, val_dataset_list_path, train_feature_path, val_feature_path, train_seq_path,val_seq_path, max_len,vocab_size, req_train_list_path, req_val_list_path, req_token_path
def get_data(train_feature_path,train_seq_path, val_feature_path,val_seq_path):
return h5py.File(train_feature_path, 'r'), h5py.File(train_seq_path, 'r'), h5py.File(val_feature_path, 'r'),h5py.File(val_seq_path,'r')
#바다나우 어텐션 적용
class Attention(Model):
def __init__(self, units, embedding_size):
super(Attention, self).__init__()
self.W1 = Dense(units)
self.W2 = Dense(units)
self.V = Dense(1)
self.embedding_size = embedding_size
self.units = units
def call(self, values, query):
query = tf.expand_dims(query,axis = 1)
score = self.V(
tf.nn.tanh(self.W1(values)+ self.W2(query)))
attention_dist = tf.nn.softmax(score, axis = 1)
context_vector = attention_dist * values
context_vector = tf.reduce_sum(context_vector, axis=1)
return context_vector,attention_dist
def build(self):
values = Input(shape = (64,2048))
query = Input(shape = (self.units,))
return Model(inputs=[values, query],outputs = self.call(values, query))
class Decoder(Model):
def __init__(self, max_len, embedding_size, units, vocab_size, reg):
super(Decoder,self).__init__()
self.units = units
self.embedding_size = embedding_size
self.embedding_layer = Embedding(vocab_size, embedding_size,mask_zero = True)
self.lstm = LSTM(units)
self.max_len = max_len
self.attention = Attention(units,embedding_size).build()
self.fc1 = Dense(self.units, activation = 'relu', kernel_regularizer = regularizers.l2(reg))
self.fc2 = Dense(vocab_size, activation = 'softmax', kernel_regularizer = regularizers.l2(reg))
def call(self, sequence,img, hidden):
context_vector, attention_dist = self.attention([img, hidden])
sequence = self.embedding_layer(sequence)
output = self.lstm(sequence)
x = self.fc1(context_vector)
merge = Add()([output, x])
merge = self.fc2(merge)
return merge,output,attention_dist
def build(self):
sequence = Input(shape = (self.max_len,))
img = Input(shape = (64, 2048))
hidden = Input(shape = (self.units))
return Model(inputs=[sequence, img, hidden],outputs = self.call(sequence,img,hidden))
@tf.function
def train_step(decoder,img, text,max_len,
tokenizer,optimizer,compile_loss,train_loss,train_acc):
loss = 0
hidden = tf.zeros((text.shape[0],512))
with tf.GradientTape() as tape:
for i in range(1, text.shape[1]):
input_text = tf.pad(text[:,:i],[[0,0],[0,max_len-i]], "CONSTANT")
target = text[:,i]
output,hidden,_ = decoder([input_text, img, hidden])
g_loss = compile_loss(target,output)
loss+=g_loss
train_acc(target,output)
gradients = tape.gradient(loss,
decoder.trainable_variables)
optimizer.apply_gradients(zip(gradients,decoder.trainable_variables))
train_loss(loss/text.shape[1])
@tf.function
def test_step(decoder,img, text,max_len,
tokenizer,optimizer,compile_loss,val_loss,val_acc):
loss = 0
hidden = tf.zeros((text.shape[0],512))
for i in range(1, text.shape[1]):
input_text = tf.pad(text[:,:i],[[0,0],[0,max_len-i]], "CONSTANT")
target = text[:,i]
output,hidden,_ = decoder([input_text, img, hidden])
g_loss = compile_loss(target,output)
loss+=g_loss
val_acc(target,output)
val_loss(loss/text.shape[1])
def get_feature_x_y(features,seq,elem):
f = features[elem][:]
text = seq[elem][:]
return f,text
def get_saved_data(data_list, feature, seq):
F=list()
T=list()
for elem in data_list:
f,t = get_feature_x_y(feature,seq,elem)
for i in range(len(t)):
F.append(f)
T.append(t[i])
return np.array(F).squeeze(),np.array(T)
if __name__ == "__main__" :
base_directory = '/content/gdrive/My Drive/Colab Notebooks/image_captioning_with_attention'
train_dataset_list_path, val_dataset_list_path, train_feature_path, val_feature_path, train_seq_path,val_seq_path, max_len,vocab_size, req_train_list_path, req_val_list_path, req_token_path = get_path(base_directory)
with open(req_train_list_path, 'rb') as handle:
train_list = pickle.load(handle)
with open(req_val_list_path,'rb') as handle:
val_list = pickle.load(handle)
with open(req_token_path,'rb') as handle:
tokenizer = pickle.load(handle)
save_path = os.path.join(base_directory,'merge_model','saved_model')
train_feature, train_seq, val_feature, val_seq= get_data(train_feature_path, train_seq_path,val_feature_path, val_seq_path)
embedding_size = 256
units = 512
reg = 1e-4
decoder = Decoder(max_len, embedding_size, units, vocab_size,reg).build()
optimizer = optimizers.Adam()
compile_loss = losses.SparseCategoricalCrossentropy()
train_loss = metrics.Mean()
train_acc = metrics.SparseCategoricalAccuracy()
val_loss = metrics.Mean()
val_acc = metrics.SparseCategoricalAccuracy()
get_batch_list = list()
batch_size = 32
for epoch in range(0,20):
get_batch_list = list()
for i in range(0, len(train_list)):
get_batch_list.append(train_list[i])
if i % batch_size == 0 and i != 0 :
img, text = get_saved_data(get_batch_list, train_feature, train_seq)
train_step(decoder,img,text,max_len,
tokenizer,optimizer,compile_loss,train_loss,train_acc)
get_batch_list.clear()
if len(get_batch_list) != 0 :
img, text = get_saved_data(get_batch_list, train_feature, train_seq)
train_step(decoder,img,text,max_len,
tokenizer,optimizer,compile_loss,train_loss,train_acc)
get_batch_list.clear()
for i in range(0, len(val_list)):
get_batch_list.append(val_list[i])
if i % batch_size == 0 and i != 0 :
img, text = get_saved_data(get_batch_list, val_feature, val_seq)
test_step(decoder,img,text,max_len,
tokenizer,optimizer,compile_loss,val_loss,val_acc)
get_batch_list.clear()
if len(get_batch_list) != 0 :
img, text = get_saved_data(get_batch_list, val_feature, val_seq,)
test_step(decoder,img,text,max_len,
tokenizer,optimizer,compile_loss,val_loss,val_acc)
get_batch_list.clear()
print('epoch {0:4d} train acc {1:0.3f} loss {2:0.3f} val acc {3:0.3f} loss {4:0.3f}'.
format(epoch, train_acc.result(), train_loss.result(), val_acc.result(), val_loss.result()))
decoder_path = os.path.join(save_path,'decoder_model_{0:02d}_vacc_{1:0.3f}_vloss_{2:0.3f}_acc{3:0.3f}_loss{4:0.3f}.h5'.
format(epoch, val_acc.result(), val_loss.result(), train_acc.result(), train_loss.result()))
decoder.save(decoder_path)
train_feature.close()
train_seq.close()
val_feature.close()
val_seq.close()
```
|
github_jupyter
|
import numpy as np
import h5py
import os
import tensorflow as tf
from tensorflow.keras.layers import Dense, LSTM, Input, Embedding, Dropout,BatchNormalization,Lambda, Add,Flatten,GRU
from tensorflow.keras.optimizers import Adam, RMSprop
from tensorflow.keras.applications import DenseNet121
from tensorflow.keras.models import Model
from tensorflow.keras import regularizers,optimizers,losses,metrics
from tensorflow.keras import backend as K
from tensorflow.keras.models import load_model
from tensorflow.keras.utils import plot_model,to_categorical
import pickle
def get_path(base_directory):
saved_data_path = os.path.join(base_directory,'data')
data_h5_paths = os.path.join(saved_data_path, 'needs.hdf5')
needs = h5py.File(data_h5_paths, 'r')
train_dataset_list_path = needs['train_code_path'][()]
val_dataset_list_path = needs['val_code_path'][()]
train_feature_path = needs['train_feature_path'][()]
val_feature_path = needs['val_feature_path'][()]
train_seq_path= needs['train_seq_path'][()]
val_seq_path= needs['val_seq_path'][()]
max_len = needs['max_len'][()]
vocab_size= needs['vocab_size'][()]
req_train_list_path = needs['req_train_list_path'][()]
req_val_list_path = needs['req_val_list_path'][()]
req_token_path = needs['req_token_path'][()]
return train_dataset_list_path, val_dataset_list_path, train_feature_path, val_feature_path, train_seq_path,val_seq_path, max_len,vocab_size, req_train_list_path, req_val_list_path, req_token_path
def get_data(train_feature_path,train_seq_path, val_feature_path,val_seq_path):
return h5py.File(train_feature_path, 'r'), h5py.File(train_seq_path, 'r'), h5py.File(val_feature_path, 'r'),h5py.File(val_seq_path,'r')
#바다나우 어텐션 적용
class Attention(Model):
def __init__(self, units, embedding_size):
super(Attention, self).__init__()
self.W1 = Dense(units)
self.W2 = Dense(units)
self.V = Dense(1)
self.embedding_size = embedding_size
self.units = units
def call(self, values, query):
query = tf.expand_dims(query,axis = 1)
score = self.V(
tf.nn.tanh(self.W1(values)+ self.W2(query)))
attention_dist = tf.nn.softmax(score, axis = 1)
context_vector = attention_dist * values
context_vector = tf.reduce_sum(context_vector, axis=1)
return context_vector,attention_dist
def build(self):
values = Input(shape = (64,2048))
query = Input(shape = (self.units,))
return Model(inputs=[values, query],outputs = self.call(values, query))
class Decoder(Model):
def __init__(self, max_len, embedding_size, units, vocab_size, reg):
super(Decoder,self).__init__()
self.units = units
self.embedding_size = embedding_size
self.embedding_layer = Embedding(vocab_size, embedding_size,mask_zero = True)
self.lstm = LSTM(units)
self.max_len = max_len
self.attention = Attention(units,embedding_size).build()
self.fc1 = Dense(self.units, activation = 'relu', kernel_regularizer = regularizers.l2(reg))
self.fc2 = Dense(vocab_size, activation = 'softmax', kernel_regularizer = regularizers.l2(reg))
def call(self, sequence,img, hidden):
context_vector, attention_dist = self.attention([img, hidden])
sequence = self.embedding_layer(sequence)
output = self.lstm(sequence)
x = self.fc1(context_vector)
merge = Add()([output, x])
merge = self.fc2(merge)
return merge,output,attention_dist
def build(self):
sequence = Input(shape = (self.max_len,))
img = Input(shape = (64, 2048))
hidden = Input(shape = (self.units))
return Model(inputs=[sequence, img, hidden],outputs = self.call(sequence,img,hidden))
@tf.function
def train_step(decoder,img, text,max_len,
tokenizer,optimizer,compile_loss,train_loss,train_acc):
loss = 0
hidden = tf.zeros((text.shape[0],512))
with tf.GradientTape() as tape:
for i in range(1, text.shape[1]):
input_text = tf.pad(text[:,:i],[[0,0],[0,max_len-i]], "CONSTANT")
target = text[:,i]
output,hidden,_ = decoder([input_text, img, hidden])
g_loss = compile_loss(target,output)
loss+=g_loss
train_acc(target,output)
gradients = tape.gradient(loss,
decoder.trainable_variables)
optimizer.apply_gradients(zip(gradients,decoder.trainable_variables))
train_loss(loss/text.shape[1])
@tf.function
def test_step(decoder,img, text,max_len,
tokenizer,optimizer,compile_loss,val_loss,val_acc):
loss = 0
hidden = tf.zeros((text.shape[0],512))
for i in range(1, text.shape[1]):
input_text = tf.pad(text[:,:i],[[0,0],[0,max_len-i]], "CONSTANT")
target = text[:,i]
output,hidden,_ = decoder([input_text, img, hidden])
g_loss = compile_loss(target,output)
loss+=g_loss
val_acc(target,output)
val_loss(loss/text.shape[1])
def get_feature_x_y(features,seq,elem):
f = features[elem][:]
text = seq[elem][:]
return f,text
def get_saved_data(data_list, feature, seq):
F=list()
T=list()
for elem in data_list:
f,t = get_feature_x_y(feature,seq,elem)
for i in range(len(t)):
F.append(f)
T.append(t[i])
return np.array(F).squeeze(),np.array(T)
if __name__ == "__main__" :
base_directory = '/content/gdrive/My Drive/Colab Notebooks/image_captioning_with_attention'
train_dataset_list_path, val_dataset_list_path, train_feature_path, val_feature_path, train_seq_path,val_seq_path, max_len,vocab_size, req_train_list_path, req_val_list_path, req_token_path = get_path(base_directory)
with open(req_train_list_path, 'rb') as handle:
train_list = pickle.load(handle)
with open(req_val_list_path,'rb') as handle:
val_list = pickle.load(handle)
with open(req_token_path,'rb') as handle:
tokenizer = pickle.load(handle)
save_path = os.path.join(base_directory,'merge_model','saved_model')
train_feature, train_seq, val_feature, val_seq= get_data(train_feature_path, train_seq_path,val_feature_path, val_seq_path)
embedding_size = 256
units = 512
reg = 1e-4
decoder = Decoder(max_len, embedding_size, units, vocab_size,reg).build()
optimizer = optimizers.Adam()
compile_loss = losses.SparseCategoricalCrossentropy()
train_loss = metrics.Mean()
train_acc = metrics.SparseCategoricalAccuracy()
val_loss = metrics.Mean()
val_acc = metrics.SparseCategoricalAccuracy()
get_batch_list = list()
batch_size = 32
for epoch in range(0,20):
get_batch_list = list()
for i in range(0, len(train_list)):
get_batch_list.append(train_list[i])
if i % batch_size == 0 and i != 0 :
img, text = get_saved_data(get_batch_list, train_feature, train_seq)
train_step(decoder,img,text,max_len,
tokenizer,optimizer,compile_loss,train_loss,train_acc)
get_batch_list.clear()
if len(get_batch_list) != 0 :
img, text = get_saved_data(get_batch_list, train_feature, train_seq)
train_step(decoder,img,text,max_len,
tokenizer,optimizer,compile_loss,train_loss,train_acc)
get_batch_list.clear()
for i in range(0, len(val_list)):
get_batch_list.append(val_list[i])
if i % batch_size == 0 and i != 0 :
img, text = get_saved_data(get_batch_list, val_feature, val_seq)
test_step(decoder,img,text,max_len,
tokenizer,optimizer,compile_loss,val_loss,val_acc)
get_batch_list.clear()
if len(get_batch_list) != 0 :
img, text = get_saved_data(get_batch_list, val_feature, val_seq,)
test_step(decoder,img,text,max_len,
tokenizer,optimizer,compile_loss,val_loss,val_acc)
get_batch_list.clear()
print('epoch {0:4d} train acc {1:0.3f} loss {2:0.3f} val acc {3:0.3f} loss {4:0.3f}'.
format(epoch, train_acc.result(), train_loss.result(), val_acc.result(), val_loss.result()))
decoder_path = os.path.join(save_path,'decoder_model_{0:02d}_vacc_{1:0.3f}_vloss_{2:0.3f}_acc{3:0.3f}_loss{4:0.3f}.h5'.
format(epoch, val_acc.result(), val_loss.result(), train_acc.result(), train_loss.result()))
decoder.save(decoder_path)
train_feature.close()
train_seq.close()
val_feature.close()
val_seq.close()
| 0.60743 | 0.758455 |
## 1. Winter is Coming. Let's load the dataset ASAP
<p>If you haven't heard of <em>Game of Thrones</em>, then you must be really good at hiding. Game of Thrones is the hugely popular television series by HBO based on the (also) hugely popular book series <em>A Song of Ice and Fire</em> by George R.R. Martin. In this notebook, we will analyze the co-occurrence network of the characters in the Game of Thrones books. Here, two characters are considered to co-occur if their names appear in the vicinity of 15 words from one another in the books. </p>
<p><img src="https://s3.amazonaws.com/assets.datacamp.com/production/project_76/img/got_network.jpeg" style="width: 550px"></p>
<p>This dataset constitutes a network and is given as a text file describing the <em>edges</em> between characters, with some attributes attached to each edge. Let's start by loading in the data for the first book <em>A Game of Thrones</em> and inspect it.</p>
```
# Importing modules
import pandas as pd
# Reading in datasets/book1.csv
book1 = pd.read_csv('datasets/book1.csv')
# Printing out the head of the dataset
print(book1.head())
```
## 2. Time for some Network of Thrones
<p>The resulting DataFrame <code>book1</code> has 5 columns: <code>Source</code>, <code>Target</code>, <code>Type</code>, <code>weight</code>, and <code>book</code>. Source and target are the two nodes that are linked by an edge. A network can have directed or undirected edges and in this network all the edges are undirected. The weight attribute of every edge tells us the number of interactions that the characters have had over the book, and the book column tells us the book number.</p>
<p>Once we have the data loaded as a pandas DataFrame, it's time to create a network. We will use <code>networkx</code>, a network analysis library, and create a graph object for the first book.</p>
```
# Importing modules
import networkx as nx
# Creating an empty graph object
G_book1 = nx.Graph()
```
## 3. Populate the network with the DataFrame
<p>Currently, the graph object <code>G_book1</code> is empty. Let's now populate it with the edges from <code>book1</code>. And while we're at it, let's load in the rest of the books too!</p>
```
# Iterating through the DataFrame to add edges
for _, edge in book1.iterrows():
G_book1.add_edge(edge['Source'], edge['Target'], weight=edge['weight'])
# Creating a list of networks for all the books
books = [G_book1]
book_fnames = ['datasets/book2.csv', 'datasets/book3.csv', 'datasets/book4.csv', 'datasets/book5.csv']
for book_fname in book_fnames:
book = pd.read_csv(book_fname)
G_book = nx.Graph()
for _, edge in book.iterrows():
G_book.add_edge(edge['Source'], edge['Target'], weight=edge['weight'])
books.append(G_book)
```
## 4. Finding the most important character in Game of Thrones
<p>Is it Jon Snow, Tyrion, Daenerys, or someone else? Let's see! Network Science offers us many different metrics to measure the importance of a node in a network. Note that there is no "correct" way of calculating the most important node in a network, every metric has a different meaning.</p>
<p>First, let's measure the importance of a node in a network by looking at the number of neighbors it has, that is, the number of nodes it is connected to. For example, an influential account on Twitter, where the follower-followee relationship forms the network, is an account which has a high number of followers. This measure of importance is called <em>degree centrality</em>.</p>
<p>Using this measure, let's extract the top ten important characters from the first book (<code>book[0]</code>) and the fifth book (<code>book[4]</code>).</p>
```
# Calculating the degree centrality of book 1
deg_cen_book1 = nx.degree_centrality(books[0])
# Calculating the degree centrality of book 5
deg_cen_book5 = nx.degree_centrality(books[4])
# Sorting the dictionaries according to their degree centrality and storing the top 10
sorted_deg_cen_book1 = sorted(deg_cen_book1.items(), key=lambda x:x[1], reverse=True)[0:10]
# Sorting the dictionaries according to their degree centrality and storing the top 10
sorted_deg_cen_book5 = sorted(deg_cen_book5.items(), key=lambda x:x[1], reverse=True)[0:10]
# Printing out the top 10 of book1 and book5
print(sorted_deg_cen_book1)
print(sorted_deg_cen_book5)
```
## 5. Evolution of importance of characters over the books
<p>According to degree centrality, the most important character in the first book is Eddard Stark but he is not even in the top 10 of the fifth book. The importance of characters changes over the course of five books because, you know, stuff happens... ;)</p>
<p>Let's look at the evolution of degree centrality of a couple of characters like Eddard Stark, Jon Snow, and Tyrion, which showed up in the top 10 of degree centrality in the first book.</p>
```
%matplotlib inline
# Creating a list of degree centrality of all the books
evol = [nx.degree_centrality(book) for book in books]
# Creating a DataFrame from the list of degree centralities in all the books
degree_evol_df = pd.DataFrame.from_records(evol)
# Plotting the degree centrality evolution of Eddard-Stark, Tyrion-Lannister and Jon-Snow
degree_evol_df[['Eddard-Stark', 'Tyrion-Lannister', 'Jon-Snow']].plot()
```
## 6. What's up with Stannis Baratheon?
<p>We can see that the importance of Eddard Stark dies off as the book series progresses. With Jon Snow, there is a drop in the fourth book but a sudden rise in the fifth book.</p>
<p>Now let's look at various other measures like <em>betweenness centrality</em> and <em>PageRank</em> to find important characters in our Game of Thrones character co-occurrence network and see if we can uncover some more interesting facts about this network. Let's plot the evolution of betweenness centrality of this network over the five books. We will take the evolution of the top four characters of every book and plot it.</p>
```
# Creating a list of betweenness centrality of all the books just like we did for degree centrality
evol = [nx.betweenness_centrality(book, weight='weight') for book in books]
# Making a DataFrame from the list
betweenness_evol_df = pd.DataFrame.from_records(evol).fillna(0)
# Finding the top 4 characters in every book
set_of_char = set()
for i in range(5):
set_of_char |= set(list(betweenness_evol_df.T[i].sort_values(ascending=False)[0:4].index))
list_of_char = list(set_of_char)
# Plotting the evolution of the top characters
betweenness_evol_df[list_of_char].plot(figsize=(13,7))
```
## 7. What does the Google PageRank algorithm tell us about Game of Thrones?
<p>We see a peculiar rise in the importance of Stannis Baratheon over the books. In the fifth book, he is significantly more important than other characters in the network, even though he is the third most important character according to degree centrality.</p>
<p>PageRank was the initial way Google ranked web pages. It evaluates the inlinks and outlinks of webpages in the world wide web, which is, essentially, a directed network. Let's look at the importance of characters in the Game of Thrones network according to PageRank. </p>
```
# Creating a list of pagerank of all the characters in all the books
evol = [nx.pagerank(book, weight='weight') for book in books]
# Making a DataFrame from the list
pagerank_evol_df = pd.DataFrame.from_records(evol).fillna(0)
# Finding the top 4 characters in every book
set_of_char = set()
for i in range(5):
set_of_char |= set(list(pagerank_evol_df.T[i].sort_values(ascending=False)[0:4].index))
list_of_char = list(set_of_char)
# Plotting the top characters
pagerank_evol_df[list_of_char].plot(figsize=(13,7))
```
## 8. Correlation between different measures
<p>Stannis, Jon Snow, and Daenerys are the most important characters in the fifth book according to PageRank. Eddard Stark follows a similar curve but for degree centrality and betweenness centrality: He is important in the first book but dies into oblivion over the book series.</p>
<p>We have seen three different measures to calculate the importance of a node in a network, and all of them tells us something about the characters and their importance in the co-occurrence network. We see some names pop up in all three measures so maybe there is a strong correlation between them?</p>
<p>Let's look at the correlation between PageRank, betweenness centrality and degree centrality for the fifth book using Pearson correlation.</p>
```
# Creating a list of pagerank, betweenness centrality, degree centrality
# of all the characters in the fifth book.
measures = [nx.pagerank(books[4]),
nx.betweenness_centrality(books[4], weight='weight'),
nx.degree_centrality(books[4])]
# Creating the correlation DataFrame
cor = pd.DataFrame.from_records(measures).fillna(0)
# Calculating the correlation
cor.T.corr()
```
## 9. Conclusion
<p>We see a high correlation between these three measures for our character co-occurrence network.</p>
<p>So we've been looking at different ways to find the important characters in the Game of Thrones co-occurrence network. According to degree centrality, Eddard Stark is the most important character initially in the books. But who is/are the most important character(s) in the fifth book according to these three measures? </p>
```
# Finding the most important character in the fifth book,
# according to degree centrality, betweenness centrality and pagerank.
p_rank, b_cent, d_cent = cor.idxmax(axis=1)
# Printing out the top character accoding to the three measures
print(p_rank, b_cent, d_cent)
```
|
github_jupyter
|
# Importing modules
import pandas as pd
# Reading in datasets/book1.csv
book1 = pd.read_csv('datasets/book1.csv')
# Printing out the head of the dataset
print(book1.head())
# Importing modules
import networkx as nx
# Creating an empty graph object
G_book1 = nx.Graph()
# Iterating through the DataFrame to add edges
for _, edge in book1.iterrows():
G_book1.add_edge(edge['Source'], edge['Target'], weight=edge['weight'])
# Creating a list of networks for all the books
books = [G_book1]
book_fnames = ['datasets/book2.csv', 'datasets/book3.csv', 'datasets/book4.csv', 'datasets/book5.csv']
for book_fname in book_fnames:
book = pd.read_csv(book_fname)
G_book = nx.Graph()
for _, edge in book.iterrows():
G_book.add_edge(edge['Source'], edge['Target'], weight=edge['weight'])
books.append(G_book)
# Calculating the degree centrality of book 1
deg_cen_book1 = nx.degree_centrality(books[0])
# Calculating the degree centrality of book 5
deg_cen_book5 = nx.degree_centrality(books[4])
# Sorting the dictionaries according to their degree centrality and storing the top 10
sorted_deg_cen_book1 = sorted(deg_cen_book1.items(), key=lambda x:x[1], reverse=True)[0:10]
# Sorting the dictionaries according to their degree centrality and storing the top 10
sorted_deg_cen_book5 = sorted(deg_cen_book5.items(), key=lambda x:x[1], reverse=True)[0:10]
# Printing out the top 10 of book1 and book5
print(sorted_deg_cen_book1)
print(sorted_deg_cen_book5)
%matplotlib inline
# Creating a list of degree centrality of all the books
evol = [nx.degree_centrality(book) for book in books]
# Creating a DataFrame from the list of degree centralities in all the books
degree_evol_df = pd.DataFrame.from_records(evol)
# Plotting the degree centrality evolution of Eddard-Stark, Tyrion-Lannister and Jon-Snow
degree_evol_df[['Eddard-Stark', 'Tyrion-Lannister', 'Jon-Snow']].plot()
# Creating a list of betweenness centrality of all the books just like we did for degree centrality
evol = [nx.betweenness_centrality(book, weight='weight') for book in books]
# Making a DataFrame from the list
betweenness_evol_df = pd.DataFrame.from_records(evol).fillna(0)
# Finding the top 4 characters in every book
set_of_char = set()
for i in range(5):
set_of_char |= set(list(betweenness_evol_df.T[i].sort_values(ascending=False)[0:4].index))
list_of_char = list(set_of_char)
# Plotting the evolution of the top characters
betweenness_evol_df[list_of_char].plot(figsize=(13,7))
# Creating a list of pagerank of all the characters in all the books
evol = [nx.pagerank(book, weight='weight') for book in books]
# Making a DataFrame from the list
pagerank_evol_df = pd.DataFrame.from_records(evol).fillna(0)
# Finding the top 4 characters in every book
set_of_char = set()
for i in range(5):
set_of_char |= set(list(pagerank_evol_df.T[i].sort_values(ascending=False)[0:4].index))
list_of_char = list(set_of_char)
# Plotting the top characters
pagerank_evol_df[list_of_char].plot(figsize=(13,7))
# Creating a list of pagerank, betweenness centrality, degree centrality
# of all the characters in the fifth book.
measures = [nx.pagerank(books[4]),
nx.betweenness_centrality(books[4], weight='weight'),
nx.degree_centrality(books[4])]
# Creating the correlation DataFrame
cor = pd.DataFrame.from_records(measures).fillna(0)
# Calculating the correlation
cor.T.corr()
# Finding the most important character in the fifth book,
# according to degree centrality, betweenness centrality and pagerank.
p_rank, b_cent, d_cent = cor.idxmax(axis=1)
# Printing out the top character accoding to the three measures
print(p_rank, b_cent, d_cent)
| 0.617282 | 0.992575 |
# Cross-Validation and scoring methods
In the previous sections and notebooks, we split our dataset into two parts, a training set and a test set. We used the training set to fit our model, and we used the test set to evaluate its generalization performance -- how well it performs on new, unseen data.
<img src="figures/train_test_split.svg" width="100%">
However, often (labeled) data is precious, and this approach lets us only use ~ 3/4 of our data for training. On the other hand, we will only ever try to apply our model 1/4 of our data for testing.
A common way to use more of the data to build a model, but also get a more robust estimate of the generalization performance, is cross-validation.
In cross-validation, the data is split repeatedly into a training and non-overlapping test-sets, with a separate model built for every pair. The test-set scores are then aggregated for a more robust estimate.
The most common way to do cross-validation is k-fold cross-validation, in which the data is first split into k (often 5 or 10) equal-sized folds, and then for each iteration, one of the k folds is used as test data, and the rest as training data:
<img src="figures/cross_validation.svg" width="100%">
This way, each data point will be in the test-set exactly once, and we can use all but a k'th of the data for training.
Let us apply this technique to evaluate the KNeighborsClassifier algorithm on the Iris dataset:
```
from sklearn.datasets import load_iris
from sklearn.neighbors import KNeighborsClassifier
iris = load_iris()
X, y = iris.data, iris.target
classifier = KNeighborsClassifier()
```
The labels in iris are sorted, which means that if we split the data as illustrated above, the first fold will only have the label 0 in it, while the last one will only have the label 2:
```
y
```
To avoid this problem in evaluation, we first shuffle our data:
```
import numpy as np
rng = np.random.RandomState(0)
permutation = rng.permutation(len(X))
X, y = X[permutation], y[permutation]
print(y)
```
Now implementing cross-validation is easy:
```
k = 5
n_samples = len(X)
fold_size = n_samples // k
scores = []
masks = []
for fold in range(k):
# generate a boolean mask for the test set in this fold
test_mask = np.zeros(n_samples, dtype=bool)
test_mask[fold * fold_size : (fold + 1) * fold_size] = True
# store the mask for visualization
masks.append(test_mask)
# create training and test sets using this mask
X_test, y_test = X[test_mask], y[test_mask]
X_train, y_train = X[~test_mask], y[~test_mask]
# fit the classifier
classifier.fit(X_train, y_train)
# compute the score and record it
scores.append(classifier.score(X_test, y_test))
```
Let's check that our test mask does the right thing:
```
import matplotlib.pyplot as plt
%matplotlib inline
plt.matshow(masks, cmap='gray_r')
```
And now let's look a the scores we computed:
```
print(scores)
print(np.mean(scores))
```
As you can see, there is a rather wide spectrum of scores from 90% correct to 100% correct. If we only did a single split, we might have gotten either answer.
As cross-validation is such a common pattern in machine learning, there are functions to do the above for you with much more flexibility and less code.
The ``sklearn.model_selection`` module has all functions related to cross validation. There easiest function is ``cross_val_score`` which takes an estimator and a dataset, and will do all of the splitting for you:
```
from sklearn.model_selection import cross_val_score
scores = cross_val_score(classifier, X, y)
print('Scores on each CV fold: %s' % scores)
print('Mean score: %0.3f' % np.mean(scores))
```
As you can see, the function uses three folds by default. You can change the number of folds using the cv argument:
```
cross_val_score(classifier, X, y, cv=5)
```
There are also helper objects in the cross-validation module that will generate indices for you for all kinds of different cross-validation methods, including k-fold:
```
from sklearn.model_selection import KFold, StratifiedKFold, ShuffleSplit
```
By default, cross_val_score will use ``StratifiedKFold`` for classification, which ensures that the class proportions in the dataset are reflected in each fold. If you have a binary classification dataset with 90% of data point belonging to class 0, that would mean that in each fold, 90% of datapoints would belong to class 0.
If you would just use KFold cross-validation, it is likely that you would generate a split that only contains class 0.
It is generally a good idea to use ``StratifiedKFold`` whenever you do classification.
``StratifiedKFold`` would also remove our need to shuffle ``iris``.
Let's see what kinds of folds it generates on the unshuffled iris dataset.
Each cross-validation class is a generator of sets of training and test indices:
```
cv = StratifiedKFold(n_splits=5)
for train, test in cv.split(iris.data, iris.target):
print(test)
```
As you can see, there are a couple of samples from the beginning, then from the middle, and then from the end, in each of the folds.
This way, the class ratios are preserved. Let's visualize the split:
```
def plot_cv(cv, features, labels):
masks = []
for train, test in cv.split(features, labels):
mask = np.zeros(len(labels), dtype=bool)
mask[test] = 1
masks.append(mask)
plt.matshow(masks, cmap='gray_r')
plot_cv(StratifiedKFold(n_splits=5), iris.data, iris.target)
```
For comparison, again the standard KFold, that ignores the labels:
```
plot_cv(KFold(n_splits=5), iris.data, iris.target)
```
Keep in mind that increasing the number of folds will give you a larger training dataset, but will lead to more repetitions, and therefore a slower evaluation:
```
plot_cv(KFold(n_splits=10), iris.data, iris.target)
```
Another helpful cross-validation generator is ``ShuffleSplit``. This generator simply splits of a random portion of the data repeatedly. This allows the user to specify the number of repetitions and the training set size independently:
```
plot_cv(ShuffleSplit(n_splits=5, test_size=.2), iris.data, iris.target)
```
If you want a more robust estimate, you can just increase the number of splits:
```
plot_cv(ShuffleSplit(n_splits=20, test_size=.2), iris.data, iris.target)
```
You can use all of these cross-validation generators with the `cross_val_score` method:
```
cv = ShuffleSplit(n_splits=5, test_size=.2)
cross_val_score(classifier, X, y, cv=cv)
```
<div class="alert alert-success">
<b>EXERCISE</b>:
<ul>
<li>
Perform three-fold cross-validation using the ``KFold`` class on the iris dataset without shuffling the data. Can you explain the result?
</li>
</ul>
</div>
```
# %load solutions/13_cross_validation.py
```
|
github_jupyter
|
from sklearn.datasets import load_iris
from sklearn.neighbors import KNeighborsClassifier
iris = load_iris()
X, y = iris.data, iris.target
classifier = KNeighborsClassifier()
y
import numpy as np
rng = np.random.RandomState(0)
permutation = rng.permutation(len(X))
X, y = X[permutation], y[permutation]
print(y)
k = 5
n_samples = len(X)
fold_size = n_samples // k
scores = []
masks = []
for fold in range(k):
# generate a boolean mask for the test set in this fold
test_mask = np.zeros(n_samples, dtype=bool)
test_mask[fold * fold_size : (fold + 1) * fold_size] = True
# store the mask for visualization
masks.append(test_mask)
# create training and test sets using this mask
X_test, y_test = X[test_mask], y[test_mask]
X_train, y_train = X[~test_mask], y[~test_mask]
# fit the classifier
classifier.fit(X_train, y_train)
# compute the score and record it
scores.append(classifier.score(X_test, y_test))
import matplotlib.pyplot as plt
%matplotlib inline
plt.matshow(masks, cmap='gray_r')
print(scores)
print(np.mean(scores))
from sklearn.model_selection import cross_val_score
scores = cross_val_score(classifier, X, y)
print('Scores on each CV fold: %s' % scores)
print('Mean score: %0.3f' % np.mean(scores))
cross_val_score(classifier, X, y, cv=5)
from sklearn.model_selection import KFold, StratifiedKFold, ShuffleSplit
cv = StratifiedKFold(n_splits=5)
for train, test in cv.split(iris.data, iris.target):
print(test)
def plot_cv(cv, features, labels):
masks = []
for train, test in cv.split(features, labels):
mask = np.zeros(len(labels), dtype=bool)
mask[test] = 1
masks.append(mask)
plt.matshow(masks, cmap='gray_r')
plot_cv(StratifiedKFold(n_splits=5), iris.data, iris.target)
plot_cv(KFold(n_splits=5), iris.data, iris.target)
plot_cv(KFold(n_splits=10), iris.data, iris.target)
plot_cv(ShuffleSplit(n_splits=5, test_size=.2), iris.data, iris.target)
plot_cv(ShuffleSplit(n_splits=20, test_size=.2), iris.data, iris.target)
cv = ShuffleSplit(n_splits=5, test_size=.2)
cross_val_score(classifier, X, y, cv=cv)
# %load solutions/13_cross_validation.py
| 0.734786 | 0.990669 |
```
# Copyright 2021 Google LLC
# Use of this source code is governed by an MIT-style
# license that can be found in the LICENSE file or at
# https://opensource.org/licenses/MIT.
# Author(s): Kevin P. Murphy ([email protected]) and Mahmoud Soliman ([email protected])
```
<a href="https://opensource.org/licenses/MIT" target="_parent"><img src="https://img.shields.io/github/license/probml/pyprobml"/></a>
<a href="https://colab.research.google.com/github/probml/pyprobml/blob/master/book1/figures/chapter2_figures.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
## Figure 2.1:<a name='2.1'></a> <a name='multinomApp'></a>
Some discrete distributions on the state space $\mathcal X =\ 1,2,3,4\ $. (a) A uniform distribution with $p(x=k)=1/4$. (b) A degenerate distribution (delta function) that puts all its mass on $x=1$.
Figure(s) generated by [discrete_prob_dist_plot.py](https://github.com/probml/pyprobml/blob/master/scripts/discrete_prob_dist_plot.py)
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/discrete_prob_dist_plot.py")
```
## Figure 2.2:<a name='2.2'></a> <a name='gaussianQuantileApp'></a>
(a) Plot of the cdf for the standard normal, $\mathcal N (0,1)$.
Figure(s) generated by [gauss_plot.py](https://github.com/probml/pyprobml/blob/master/scripts/gauss_plot.py) [quantile_plot.py](https://github.com/probml/pyprobml/blob/master/scripts/quantile_plot.py)
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/gauss_plot.py")
pmlt.show_and_run("/pyprobml/scripts/quantile_plot.py")
```
## Figure 2.3:<a name='2.3'></a> <a name='roweis-xtimesy'></a>
Computing $p(x,y) = p(x) p(y)$, where $ X \perp Y $. Here $X$ and $Y$ are discrete random variables; $X$ has 6 possible states (values) and $Y$ has 5 possible states. A general joint distribution on two such variables would require $(6 \times 5) - 1 = 29$ parameters to define it (we subtract 1 because of the sum-to-one constraint). By assuming (unconditional) independence, we only need $(6-1) + (5-1) = 9$ parameters to define $p(x,y)$.
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
```
## Figure 2.4:<a name='2.4'></a> <a name='bimodal'></a>
Illustration of a mixture of two 1d Gaussians, $p(x) = 0.5 \mathcal N (x|0,0.5) + 0.5 \mathcal N (x|2,0.5)$.
Figure(s) generated by [bimodal_dist_plot.py](https://github.com/probml/pyprobml/blob/master/scripts/bimodal_dist_plot.py)
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/bimodal_dist_plot.py")
```
## Figure 2.5:<a name='2.5'></a> <a name='kalmanHead'></a>
(a) Any planar line-drawing is geometrically consistent with infinitely many 3-D structures. From Figure 11 of <a href='#Sinha1993'>[PE93]</a> . Used with kind permission of Pawan Sinha. (b) Vision as inverse graphics. The agent (here represented by a human head) has to infer the scene $h$ given the image $y$ using an estimator, which computes $ h (y) = \operatorname * argmax _ h p(h|y)$. From Figure 1 of <a href='#Rao1999'>[RP99]</a> . Used with kind permission of Rajesh Rao.
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_image("/pyprobml/book1/figures/images/3d2dProjection.png")
pmlt.show_image("/pyprobml/book1/figures/images/underconstrainedness.png")
pmlt.show_image("/pyprobml/book1/figures/images/kalmanHead.png")
```
## Figure 2.6:<a name='2.6'></a> <a name='binomDist'></a>
Illustration of the binomial distribution with $N=10$ and (a) $\theta =0.25$ and (b) $\theta =0.9$.
Figure(s) generated by [binom_dist_plot.py](https://github.com/probml/pyprobml/blob/master/scripts/binom_dist_plot.py)
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/binom_dist_plot.py")
```
## Figure 2.7:<a name='2.7'></a> <a name='sigmoidHeaviside'></a>
(a) The sigmoid (logistic) function $\sigma (a)=(1+e^ -a )^ -1 $. (b) The Heaviside function $\mathbb I \left ( a>0 \right )$.
Figure(s) generated by [activation_fun_plot.py](https://github.com/probml/pyprobml/blob/master/scripts/activation_fun_plot.py)
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/activation_fun_plot.py")
```
## Figure 2.8:<a name='2.8'></a> <a name='iris-logreg-1d'></a>
Logistic regression applied to a 1-dimensional, 2-class version of the Iris dataset.
Figure(s) generated by [iris_logreg.py](https://github.com/probml/pyprobml/blob/master/scripts/iris_logreg.py)
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/iris_logreg.py")
```
## Figure 2.9:<a name='2.9'></a> <a name='softmaxDemo'></a>
Softmax distribution $\mathcal S (\mathbf a /T)$, where $\mathbf a =(3,0,1)$, at temperatures of $T=100$, $T=2$ and $T=1$. When the temperature is high (left), the distribution is uniform, whereas when the temperature is low (right), the distribution is ``spiky'', with most of its mass on the largest element.
Figure(s) generated by [softmax_plot.py](https://github.com/probml/pyprobml/blob/master/scripts/softmax_plot.py)
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/softmax_plot.py")
```
## Figure 2.10:<a name='2.10'></a> <a name='iris-logistic-2d-3class-prob'></a>
Logistic regression on the 3-class, 2-feature version of the Iris dataset. Adapted from Figure of 4.25 <a href='#Geron2019'>[Aur19]</a> .
Figure(s) generated by [iris_logreg.py](https://github.com/probml/pyprobml/blob/master/scripts/iris_logreg.py)
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/iris_logreg.py")
```
## Figure 2.11:<a name='2.11'></a> <a name='gaussianPdf'></a>
(a) Cumulative distribution function (cdf) for the standard normal.
Figure(s) generated by [gauss_plot.py](https://github.com/probml/pyprobml/blob/master/scripts/gauss_plot.py) [quantile_plot.py](https://github.com/probml/pyprobml/blob/master/scripts/quantile_plot.py)
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/gauss_plot.py")
pmlt.show_and_run("/pyprobml/scripts/quantile_plot.py")
```
## Figure 2.12:<a name='2.12'></a> <a name='hetero'></a>
Linear regression using Gaussian output with mean $\mu (x)=b + w x$ and (a) fixed variance $\sigma ^2$ (homoskedastic) or (b) input-dependent variance $\sigma (x)^2$ (heteroscedastic).
Figure(s) generated by [linreg_1d_hetero_tfp.py](https://github.com/probml/pyprobml/blob/master/scripts/linreg_1d_hetero_tfp.py)
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/linreg_1d_hetero_tfp.py")
```
## Figure 2.13:<a name='2.13'></a> <a name='studentPdf'></a>
(a) The pdf's for a $\mathcal N (0,1)$, $\mathcal T (\mu =0,\sigma =1,\nu =1)$, $\mathcal T (\mu =0,\sigma =1,\nu =2)$, and $\mathrm Lap (0,1/\sqrt 2 )$. The mean is 0 and the variance is 1 for both the Gaussian and Laplace. When $\nu =1$, the Student is the same as the Cauchy, which does not have a well-defined mean and variance. (b) Log of these pdf's. Note that the Student distribution is not log-concave for any parameter value, unlike the Laplace distribution. Nevertheless, both are unimodal.
Figure(s) generated by [student_laplace_pdf_plot.py](https://github.com/probml/pyprobml/blob/master/scripts/student_laplace_pdf_plot.py)
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/student_laplace_pdf_plot.py")
```
## Figure 2.14:<a name='2.14'></a> <a name='robustDemo'></a>
Illustration of the effect of outliers on fitting Gaussian, Student and Laplace distributions. (a) No outliers (the Gaussian and Student curves are on top of each other). (b) With outliers. We see that the Gaussian is more affected by outliers than the Student and Laplace distributions. Adapted from Figure 2.16 of <a href='#BishopBook'>[Bis06]</a> .
Figure(s) generated by [robust_pdf_plot.py](https://github.com/probml/pyprobml/blob/master/scripts/robust_pdf_plot.py)
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/robust_pdf_plot.py")
```
## Figure 2.15:<a name='2.15'></a> <a name='gammaDist'></a>
(a) Some beta distributions.
Figure(s) generated by [beta_dist_plot.py](https://github.com/probml/pyprobml/blob/master/scripts/beta_dist_plot.py) [gamma_dist_plot.py](https://github.com/probml/pyprobml/blob/master/scripts/gamma_dist_plot.py)
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/beta_dist_plot.py")
pmlt.show_and_run("/pyprobml/scripts/gamma_dist_plot.py")
```
## Figure 2.16:<a name='2.16'></a> <a name='changeOfVar1d'></a>
(a) Mapping a uniform pdf through the function $f(x) = 2x + 1$. (b) Illustration of how two nearby points, $x$ and $x+dx$, get mapped under $f$. If $\frac dy dx >0$, the function is locally increasing, but if $\frac dy dx <0$, the function is locally decreasing. From <a href='#JangBlog'>[Jan18]</a> . Used with kind permission of Eric Jang
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_image("/pyprobml/book1/figures/images/flow1.png.png")
pmlt.show_image("/pyprobml/book1/figures/images/flow2.png.png")
```
## Figure 2.17:<a name='2.17'></a> <a name='affine2d'></a>
Illustration of an affine transformation applied to a unit square, $f(\mathbf x ) = \mathbf A \mathbf x + \mathbf b $. (a) Here $\mathbf A =\mathbf I $. (b) Here $\mathbf b =\boldsymbol 0 $. From <a href='#JangBlog'>[Jan18]</a> . Used with kind permission of Eric Jang
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_image("/pyprobml/book1/figures/images/flow3.png.png")
```
## Figure 2.18:<a name='2.18'></a> <a name='polar'></a>
Change of variables from polar to Cartesian. The area of the shaded patch is $r \tmspace +\thickmuskip .2777em dr \tmspace +\thickmuskip .2777em d\theta $. Adapted from Figure 3.16 of <a href='#Rice95'>[Ric95]</a>
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_image("/pyprobml/book1/figures/images/polarToCartesian.png")
```
## Figure 2.19:<a name='2.19'></a> <a name='bellCurve'></a>
Distribution of the sum of two dice rolls, i.e., $p(y)$ where $y=x_1 + x_2$ and $x_i \sim \mathrm Unif (\ 1,2,\ldots ,6\ )$. From https://en.wikipedia.org/wiki/Probability\_distribution . Used with kind permission of Wikipedia author Tim Stellmach.
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_image("/pyprobml/book1/figures/images/diceSum.png")
```
## Figure 2.20:<a name='2.20'></a> <a name='clt'></a>
The central limit theorem in pictures. We plot a histogram of $ \mu _N^s = \frac 1 N \DOTSB \sum@ \slimits@ _ n=1 ^Nx_ ns $, where $x_ ns \sim \mathrm Beta (1,5)$, for $s=1:10000$. As $N\rightarrow \infty $, the distribution tends towards a Gaussian. (a) $N=1$. (b) $N=5$. Adapted from Figure 2.6 of <a href='#BishopBook'>[Bis06]</a> .
Figure(s) generated by [centralLimitDemo.m](https://github.com/probml/pmtk3/blob/master/demos/centralLimitDemo.m)
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_image("/pyprobml/book1/figures/images/cltBeta1.png")
```
## Figure 2.21:<a name='2.21'></a> <a name='changeOfVars'></a>
Computing the distribution of $y=x^2$, where $p(x)$ is uniform (left). The analytic result is shown in the middle, and the Monte Carlo approximation is shown on the right.
Figure(s) generated by [change_of_vars_demo1d.py](https://github.com/probml/pyprobml/blob/master/scripts/change_of_vars_demo1d.py)
```
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/change_of_vars_demo1d.py")
```
## References:
<a name='Geron2019'>[Aur19]</a> G. Aur'elien "Hands-On Machine Learning with Scikit-Learn and TensorFlow: Concepts, Tools, and Techniques for BuildingIntelligent Systems (2nd edition)". (2019).
<a name='BishopBook'>[Bis06]</a> C. Bishop "Pattern recognition and machine learning". (2006).
<a name='JangBlog'>[Jan18]</a> E. Jang "Normalizing Flows Tutorial". (2018).
<a name='Sinha1993'>[PE93]</a> S. P and A. E. "Recovering reflectance and illumination in a world of paintedpolyhedra". (1993).
<a name='Rao1999'>[RP99]</a> R. RP "An optimal estimation approach to visual perception and learning". In: Vision Res. (1999).
<a name='Rice95'>[Ric95]</a> J. Rice "Mathematical statistics and data analysis". (1995).
|
github_jupyter
|
# Copyright 2021 Google LLC
# Use of this source code is governed by an MIT-style
# license that can be found in the LICENSE file or at
# https://opensource.org/licenses/MIT.
# Author(s): Kevin P. Murphy ([email protected]) and Mahmoud Soliman ([email protected])
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/discrete_prob_dist_plot.py")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/gauss_plot.py")
pmlt.show_and_run("/pyprobml/scripts/quantile_plot.py")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/bimodal_dist_plot.py")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_image("/pyprobml/book1/figures/images/3d2dProjection.png")
pmlt.show_image("/pyprobml/book1/figures/images/underconstrainedness.png")
pmlt.show_image("/pyprobml/book1/figures/images/kalmanHead.png")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/binom_dist_plot.py")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/activation_fun_plot.py")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/iris_logreg.py")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/softmax_plot.py")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/iris_logreg.py")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/gauss_plot.py")
pmlt.show_and_run("/pyprobml/scripts/quantile_plot.py")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/linreg_1d_hetero_tfp.py")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/student_laplace_pdf_plot.py")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/robust_pdf_plot.py")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/beta_dist_plot.py")
pmlt.show_and_run("/pyprobml/scripts/gamma_dist_plot.py")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_image("/pyprobml/book1/figures/images/flow1.png.png")
pmlt.show_image("/pyprobml/book1/figures/images/flow2.png.png")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_image("/pyprobml/book1/figures/images/flow3.png.png")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_image("/pyprobml/book1/figures/images/polarToCartesian.png")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_image("/pyprobml/book1/figures/images/diceSum.png")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_image("/pyprobml/book1/figures/images/cltBeta1.png")
#@title Setup { display-mode: "form" }
%%time
# If you run this for the first time it would take ~25/30 seconds
!git clone https://github.com/probml/pyprobml /pyprobml &> /dev/null && git clone https://github.com/probml/colab_powertoys.git &> /dev/null
!pip3 install nbimporter -qqq
%cd -q /content/colab_powertoys
from colab_powertoys.probml_toys import probml_toys as pmlt
%cd -q /pyprobml/scripts
pmlt.show_and_run("/pyprobml/scripts/change_of_vars_demo1d.py")
| 0.664867 | 0.92756 |
# Assignment 4
### Math 502 - Lamoureux
### Due April 4, 2019
## Exercise 0
Fix a constant $\lambda >0$. Find a solution to the delay differential equation
$$y'(x) = -\lambda y(x-1), \qquad y(0) = 1,$$
where we are looking for a function $y=y(x)$ that satisfies this equation.
Does your solution decay over time? At what rate? Do you think the decay rate is somehow related to the delay in the differential equation? (Here the delay is 1, since we have a $x-1$ in the DE.) Test your idea by solving a related equation $y'(x) = -\lambda y(x-x_0)$.
## Exercise 1
A sequence $a= (\ldots, a_{-2},a_{-1}, a_0, a_1, a_2, \ldots)$ is called **anti-symmetric** if $$a_{-n} = -a_n \mbox{ for all $n$. }$$
For instance, the sequence $a= (\ldots, 0,0, 1,-3,5,0,-5,3,-1,0,0,\ldots)$ is anti-symmetric, when we label the middle 0 as $a_0.$
- Show that the Fourier transform $\hat{a}(\theta)$ of an anti-symmetric sequence is purely imaginary (its real part is zero.)
- Show that $\hat{a}(1-\theta) = -\hat{a}(\theta)$ for all $\theta$ if $a$ is anti-symmetric
- Show that the convolution $a*a$ is symmetric whenever $a$ is anti-symmetric.
Hint: If you are finding this hard for general sequences $a$, try to solve it for that one example $$a= (\ldots, 0,0, 1,-3,5,0,-5,3,-1,0,0,\ldots).$$
## Exercise 2
Here's a little trick to keep convolution from making our sequences "too long."
Recall if you convolve two sequence $a,b$ of length N, the result is of length 2N-1.
To "fix" this, we can define **circular convolution** as $c = a\otimes b$, using the formula
$$c_k = \sum_{j=0}^{N-1} a_{k-j}b_j, \mbox{ with $k-j$ computed modulo $N$.}$$
So, for instance, the convolution of length-3 sequences is given as
$$(a_0,a_1,a_2)\otimes (b_0,b_1,b_2) =
(a_0b_0 + a_1b_2 + a_2b_1, a_0b_1 + a_1b_0 + a_2b_2, a_0b_2 + a_1b_1 + a_2 b_0).$$
(Notice each component on the RHS has 3 terms, and the indices in each component add up to the same number, mod 3.)
1. Compute the circular convolution: $(1,1,0,0)\otimes(1,2,3,4)$. The result should have length 4.
2. Compute the regular convolution: $(1,1,0,0) * (1,2,3,4)$. The result should have length 7.
3. Compare your two answers. What parts are the same, what are different?
4. Compute the DFT of the three sequences $a = (1,1,0,0)$ , $b = (1,2,3,4)$, $c = (1,1,0,0)\otimes(1,2,3,4)$. Either by hand, or using Python. (Each result will be a length-4 sequence, with complex values.)
5. Verify your results in \#4, by showing that the DFT of c is equal to the product of the DFT of a times the DFT of b.
DFT = Discrete Fourier transform. It takes vectors of length N and produces vectors of length N, using that formula we saw in class.
## Exercise 3
In the notes to lecture 18, we took the 2D Fourier transform of the identity matrix. As an image, this took a line pointing at 45 degrees down, and transformed it into a line at 45 degrees up.
Write some code to explore this idea further. Take a big array of zeros, and then fill in a line at some angle. Take the 2D FFT and display it -- it should look like a line, going at a different direction.
Once you get the lines working, see if you can describe the relation between the angle (or slope) of the input and output lines.
Can you prove this relationship between the angles?
## Exercise 4
This is an exploration exercise.
- Take a geometric figure, place into a NxN array to create an image.
- Its a good idea to take N to be a power of 2, like 128 or 256.
- Take the 2D FFT of the image, and save the absolute values as a NxN array.
- Plot the original image and absolute value of the FFT of the image.
- You might also try fftshift on the image (see Lecture 18 again)
- Can you see any connection between the geometric figure and its FFT?
For instance, we saw in Exercise 3 that the FFT of a line was a line.
What is the FFT of a square? A circle? A triangle? A polygon?
Try the outline of a circle, and a solid (filled in) circle. Also for the other shapes.
Try creating a pattern in the image.
- Parallel lines
- Grid lines in x and y direction
- Rectangular array of dots
- Hexagonal array of dots.
What does the FFT of these things look like?
**Note:** There is no right answer here, I just want you to explore some shapes and patterns.
**Historical note:** In the early 1900s when scientists were trying to discover the atomic structure of crystals, they could essentially "see" the FFT of the arrangement of atoms in the crystal, but not see the arrangement itself. So it was an important challenge to be able to guess the arrangement only by looking at the FFT of the image. (Actually, they could only look at the absolute value of the FFT.)
The structure of DNA, a double helix, was discovered by Watson and Crick in 1953. Their work depended heavily upon the X-ray images produced by Rosalind Franklin, which is essentially the FFT images as above. Watson and Crick received the Nobel prize in 1962; Franklin died of cancer in 1958.
|
github_jupyter
|
# Assignment 4
### Math 502 - Lamoureux
### Due April 4, 2019
## Exercise 0
Fix a constant $\lambda >0$. Find a solution to the delay differential equation
$$y'(x) = -\lambda y(x-1), \qquad y(0) = 1,$$
where we are looking for a function $y=y(x)$ that satisfies this equation.
Does your solution decay over time? At what rate? Do you think the decay rate is somehow related to the delay in the differential equation? (Here the delay is 1, since we have a $x-1$ in the DE.) Test your idea by solving a related equation $y'(x) = -\lambda y(x-x_0)$.
## Exercise 1
A sequence $a= (\ldots, a_{-2},a_{-1}, a_0, a_1, a_2, \ldots)$ is called **anti-symmetric** if $$a_{-n} = -a_n \mbox{ for all $n$. }$$
For instance, the sequence $a= (\ldots, 0,0, 1,-3,5,0,-5,3,-1,0,0,\ldots)$ is anti-symmetric, when we label the middle 0 as $a_0.$
- Show that the Fourier transform $\hat{a}(\theta)$ of an anti-symmetric sequence is purely imaginary (its real part is zero.)
- Show that $\hat{a}(1-\theta) = -\hat{a}(\theta)$ for all $\theta$ if $a$ is anti-symmetric
- Show that the convolution $a*a$ is symmetric whenever $a$ is anti-symmetric.
Hint: If you are finding this hard for general sequences $a$, try to solve it for that one example $$a= (\ldots, 0,0, 1,-3,5,0,-5,3,-1,0,0,\ldots).$$
## Exercise 2
Here's a little trick to keep convolution from making our sequences "too long."
Recall if you convolve two sequence $a,b$ of length N, the result is of length 2N-1.
To "fix" this, we can define **circular convolution** as $c = a\otimes b$, using the formula
$$c_k = \sum_{j=0}^{N-1} a_{k-j}b_j, \mbox{ with $k-j$ computed modulo $N$.}$$
So, for instance, the convolution of length-3 sequences is given as
$$(a_0,a_1,a_2)\otimes (b_0,b_1,b_2) =
(a_0b_0 + a_1b_2 + a_2b_1, a_0b_1 + a_1b_0 + a_2b_2, a_0b_2 + a_1b_1 + a_2 b_0).$$
(Notice each component on the RHS has 3 terms, and the indices in each component add up to the same number, mod 3.)
1. Compute the circular convolution: $(1,1,0,0)\otimes(1,2,3,4)$. The result should have length 4.
2. Compute the regular convolution: $(1,1,0,0) * (1,2,3,4)$. The result should have length 7.
3. Compare your two answers. What parts are the same, what are different?
4. Compute the DFT of the three sequences $a = (1,1,0,0)$ , $b = (1,2,3,4)$, $c = (1,1,0,0)\otimes(1,2,3,4)$. Either by hand, or using Python. (Each result will be a length-4 sequence, with complex values.)
5. Verify your results in \#4, by showing that the DFT of c is equal to the product of the DFT of a times the DFT of b.
DFT = Discrete Fourier transform. It takes vectors of length N and produces vectors of length N, using that formula we saw in class.
## Exercise 3
In the notes to lecture 18, we took the 2D Fourier transform of the identity matrix. As an image, this took a line pointing at 45 degrees down, and transformed it into a line at 45 degrees up.
Write some code to explore this idea further. Take a big array of zeros, and then fill in a line at some angle. Take the 2D FFT and display it -- it should look like a line, going at a different direction.
Once you get the lines working, see if you can describe the relation between the angle (or slope) of the input and output lines.
Can you prove this relationship between the angles?
## Exercise 4
This is an exploration exercise.
- Take a geometric figure, place into a NxN array to create an image.
- Its a good idea to take N to be a power of 2, like 128 or 256.
- Take the 2D FFT of the image, and save the absolute values as a NxN array.
- Plot the original image and absolute value of the FFT of the image.
- You might also try fftshift on the image (see Lecture 18 again)
- Can you see any connection between the geometric figure and its FFT?
For instance, we saw in Exercise 3 that the FFT of a line was a line.
What is the FFT of a square? A circle? A triangle? A polygon?
Try the outline of a circle, and a solid (filled in) circle. Also for the other shapes.
Try creating a pattern in the image.
- Parallel lines
- Grid lines in x and y direction
- Rectangular array of dots
- Hexagonal array of dots.
What does the FFT of these things look like?
**Note:** There is no right answer here, I just want you to explore some shapes and patterns.
**Historical note:** In the early 1900s when scientists were trying to discover the atomic structure of crystals, they could essentially "see" the FFT of the arrangement of atoms in the crystal, but not see the arrangement itself. So it was an important challenge to be able to guess the arrangement only by looking at the FFT of the image. (Actually, they could only look at the absolute value of the FFT.)
The structure of DNA, a double helix, was discovered by Watson and Crick in 1953. Their work depended heavily upon the X-ray images produced by Rosalind Franklin, which is essentially the FFT images as above. Watson and Crick received the Nobel prize in 1962; Franklin died of cancer in 1958.
| 0.836388 | 0.987771 |
# Intro
Al momento ya visitamos y exploramos diversas _built-in functions_ o _funciones nativas de Python_ tales como `max`, `min`, `abs` y `print`. Déjame decirte que Python tiene muchas más funciones y además podrás definir tus propias funciones.
En esta lección vas a aprener a definir tus propias funciones.
# Cómo pedir ayuda.
Vamos rescatar la función `abs()` que visitamos en la lección anterior ¿Recuerdas qué hace? ¿Qué pasaría si olvidas qué hace?
Para subsanar lo anteior vamos a introducir la función `help()`, la cual es quizás la más importante de todas las funciones de Python que se pueden aprender. Así, si algún día olvidas qué hacia o cómo usar una función puedes aplicarle la función `help()`.
Como ejemplo usemos la función `help()` sobre la función `abs()`
```
help(abs)
```
En este caso nos dice que la función `abs()` recibe una variable `x` y regresa su valor absoluto.
Como siguiente ejemplo visitemos la función `print()`:
```
help(round)
```
La función `help()` muestra dos cosas:
1. El encabezado de la función: `abs(x,\)`, `round(number, ndigits=None)`. Lo que nos dice en el caso de `abs()` es que la función toma como entrada una varible `x` mientras que `round()` recibe como un argumento que podemos identificar con `number` y de manera opcional le podemos pasar, separando por una coma, otro argumento que es descrito como `ndigits`.
1. Una descripción en inglés de lo que hace la función.
**NOTA**: Ten mucho cuidado cuando uses `help()`, recuerda que debes pasar como argumento la función y no el output de la función:
```
# Bien ejecutado
help(abs)
# Ejecutado de manera incorrecta
help(abs(-1))
```
En el primer caso nos dice qué argumentos recibe la función y además qué hace mientras que en el segundo notemos que primero resuelve `abs(-1)` lo cual tiene por output `1` y entonces lo que realemente estamos evaluando en help es `help(1)` y `1` es un `int` entonces nos da la descripción de la case `int`.
# De como definir funciones
Las funciones _built-in_ o _nativas_ son muy útiles pero limitarnos a ellas limitaría nuestro trabajo: imagina que tienes una rutina de limpieza que debes aplicarle a un conjunto de datos siempre y para distintos conjuntos de datos Qué sería más rápido ¿Que pudieras mudar toda esa rutina de operaciones en una instrucción úninca o tener que escribir todas las veces la operación?
Ese es el espíritu de las funciones, encapsular en un solo lugar procesos pare en lugar de repetirlos una y otra vez puedas llamar a una sola instrucción que haga todo y además te regrese el resultado y lo puedas usar para futuros cálculos.
Supongamos que queremos encontrar el mínimo la diferencia absoluta entre un conjunto de cuatro números `a`, `b`, `c` y `d`. Una forma de resolver estos sería como sigue:
```
a = 1
b = 2
c = 10
d = 1
d1 = abs(a-b)
d2 = abs(a-c)
d3 = abs(a-d)
d4 = abs(b-c)
d5 = abs(b-d)
d6 = abs(c-d)
minimo = min(d1,d2,d3,d4,d5,d6)
print('d1 = {}, d2 = {}, d3 = {}, d4 = {}, d5 = {}, d6 = {}'.format(d1,d2,d3,d4,d5,d6),end='\n \n')
print('El mínimo es {}'.format(minimo))
```
Bueno, parece que no fue muy problemático ¿Y qué pasa si tenemos un nuevo conjunto de números? Bueno, pues copiamos y pegamos:
```
# Nuevo conjunto de números
a = 11
b = 2
c = 89
d = 16
d1 = abs(a-b)
d2 = abs(a-c)
d3 = abs(a-d)
d4 = abs(b-c)
d5 = abs(b-d)
d6 = abs(c-d)
minimo = min(d1,d2,d3,d4,d5,d6)
print('d1 = {}, d2 = {}, d3 = {}, d4 = {}, d5 = {}, d6 = {}'.format(d1,d2,d3,d4,d5,d6),end='\n \n')
print('El mínimo es {}'.format(minimo))
```
¿Y qué pasa ahora si tenemos un nuevo conjunto y además las variables tiene otro nombre? Bueno, pues creo que ya tenemos la idea de que necesitamos poder guardar esa serie de operaciones para poder usarlas a futuro:
```
def dif_minima(a,b,c,d):
d1 = abs(a-b)
d2 = abs(a-c)
d3 = abs(a-d)
d4 = abs(b-c)
d5 = abs(b-d)
d6 = abs(c-d)
return min(d1,d2,d3,d4,d5,d6)
```
La celda de código anterior crea una función llamada `dif_minima` (diferencia mínima), la cual toma tres argumentos `a`, `b` y `c`.
La funciones siempre comenzarán con la _palabra reservada_ `def` y lo que va a ser ejecutado como parte de la función es aquello que esté identado después de `:`.
`return` es otra palabra reservada y es usada únicamente en el contexto de las funciones. Cuando Python llega al enunciado de `return` termina y sale de la función inmediatamente y pasa el valor de lo que esté a la derecha al contexto donde fue llamada.
¿Es claro ver qué hace la función `dif_minima` a partir de leer el código fuente? Si no, siempre se pueden hacer algunas pruebas:
```
print(
dif_minima(1,2,3,4),
dif_minima(2,4,6,8),
dif_minima(0,2,3,5)
)
```
Si aún así no es claro lo podríamos intentar usar la función `help()`
```
help(dif_minima)
```
¿Ayudó de algo?
Python no es lo suficientemente inteligente para poder leer e interpretar el código de manera automática a una descruipción. Por esto es que cuando escribimos una función por el bien de nuestro yo del futuro, nuestros y nuestras colegas y la comunidad en general que tenga acceso al código que escribimos debemos de agregar un **docstring**.
# Docstrings
```
def dif_minima(a,b,c,d):
"""
Esta función calcula la diferencia absoluta entre cuatro números
a, b, c y d, y nos regresa la mínima de las mismas.
Parameters
----------
a,b,c,d : int or float
Son números entre los cuales se calculará la diferencia
Returns
----------
int or float
Se regresa el mínimo de las diferencias absolutas entre números
Examples
--------
>>> dif_minima(1,1,1,1)
0
>>> dif_minima(2,1,4,7)
1
"""
d1 = abs(a-b)
d2 = abs(a-c)
d3 = abs(a-d)
d4 = abs(b-c)
d5 = abs(b-d)
d6 = abs(c-d)
return min(d1,d2,d3,d4,d5,d6)
```
Las **docstrings** son cadenas con tres pares de comillas dobles las cuales se pueden extender pode muchas lineas y además _conservan formato_. Cuando nosotros llamamos a la función `help()` sobre una función se muestra directamente el docstring.
```
help(dif_minima)
```
En las buenas prácticas de código se incluye SIEMPRE escribir docstrings ya que no solamente ayudará a otras personas a usar tu código de manera efectiva sino también cuando revisites tu código podrás recordar y entender qué hace (créeme, es muy útil dejar código documentado, tanto que por no hacerlo una vez tuve que reescribir cerca de 1000 líneas de código).
> _Invertir tiempo en escribir docstrings te ahorrará mucho más tiempo que el que invertirás descubriendo qué hace la función que escribiste hace un mes_
**NOTA**: No existe un estándar de cómo escribir docstring sino que existen muchos formatos: [NumpyDoc](https://numpydoc.readthedocs.io/en/latest/format.html), [PEP 287 o reST](https://www.python.org/dev/peps/pep-0287/), [EpiText](http://epydoc.sourceforge.net/) o [Google Format](https://github.com/google/styleguide/blob/gh-pages/pyguide.md#38-comments-and-docstrings). Ninguno es mejor que otro, todos los formatos tienen sus ventajas y desventajas. Usa el que más te agrade pero documenta.
# Funciones sin `return`
|
github_jupyter
|
help(abs)
help(round)
# Bien ejecutado
help(abs)
# Ejecutado de manera incorrecta
help(abs(-1))
a = 1
b = 2
c = 10
d = 1
d1 = abs(a-b)
d2 = abs(a-c)
d3 = abs(a-d)
d4 = abs(b-c)
d5 = abs(b-d)
d6 = abs(c-d)
minimo = min(d1,d2,d3,d4,d5,d6)
print('d1 = {}, d2 = {}, d3 = {}, d4 = {}, d5 = {}, d6 = {}'.format(d1,d2,d3,d4,d5,d6),end='\n \n')
print('El mínimo es {}'.format(minimo))
# Nuevo conjunto de números
a = 11
b = 2
c = 89
d = 16
d1 = abs(a-b)
d2 = abs(a-c)
d3 = abs(a-d)
d4 = abs(b-c)
d5 = abs(b-d)
d6 = abs(c-d)
minimo = min(d1,d2,d3,d4,d5,d6)
print('d1 = {}, d2 = {}, d3 = {}, d4 = {}, d5 = {}, d6 = {}'.format(d1,d2,d3,d4,d5,d6),end='\n \n')
print('El mínimo es {}'.format(minimo))
def dif_minima(a,b,c,d):
d1 = abs(a-b)
d2 = abs(a-c)
d3 = abs(a-d)
d4 = abs(b-c)
d5 = abs(b-d)
d6 = abs(c-d)
return min(d1,d2,d3,d4,d5,d6)
print(
dif_minima(1,2,3,4),
dif_minima(2,4,6,8),
dif_minima(0,2,3,5)
)
help(dif_minima)
def dif_minima(a,b,c,d):
"""
Esta función calcula la diferencia absoluta entre cuatro números
a, b, c y d, y nos regresa la mínima de las mismas.
Parameters
----------
a,b,c,d : int or float
Son números entre los cuales se calculará la diferencia
Returns
----------
int or float
Se regresa el mínimo de las diferencias absolutas entre números
Examples
--------
>>> dif_minima(1,1,1,1)
0
>>> dif_minima(2,1,4,7)
1
"""
d1 = abs(a-b)
d2 = abs(a-c)
d3 = abs(a-d)
d4 = abs(b-c)
d5 = abs(b-d)
d6 = abs(c-d)
return min(d1,d2,d3,d4,d5,d6)
help(dif_minima)
| 0.437103 | 0.987544 |
This is a simple notebook demo to illustrate typically how OptimalFlow's autoCV modules work with regression problem
```
# Install external packages in binder environment.
!pip install xgboost
# Regression Demo
import pandas as pd
from optimalflow.autoCV import evaluate_model,dynaRegressor
import joblib
from optimalflow.utilis_func import pipeline_splitting_rule, update_parameters,reset_parameters
reset_parameters()
tr_features = pd.read_csv('./data/regression/train_features.csv')
tr_labels = pd.read_csv('./data/regression/train_labels.csv')
val_features = pd.read_csv('./data/regression/val_features.csv')
val_labels = pd.read_csv('./data/regression/val_labels.csv')
te_features = pd.read_csv('./data/regression/test_features.csv')
te_labels = pd.read_csv('./data/regression/test_labels.csv')
reg_cv_demo = dynaRegressor(random_state=13,cv_num = 5)
reg_cv_demo.fit(tr_features,tr_labels)
models = {}
for mdl in ['lr','knn','tree','svm','mlp','rf','gb','ada','xgb','hgboost','huber','rgcv','cvlasso','sgd']:
models[mdl] = joblib.load('./pkl/{}_reg_model.pkl'.format(mdl))
for name, mdl in models.items():
try:
ml_evl = evaluate_model(model_type = "reg")
ml_evl.fit(name, mdl, val_features, val_labels)
except:
print(f"Failed to load the {mdl}.")
# Fast Regression Demo
import pandas as pd
from optimalflow.autoCV import evaluate_model,fastRegressor
import joblib
from optimalflow.utilis_func import pipeline_splitting_rule, update_parameters,reset_parameters
reset_parameters()
tr_features = pd.read_csv('./data/regression/train_features.csv')
tr_labels = pd.read_csv('./data/regression/train_labels.csv')
val_features = pd.read_csv('./data/regression/val_features.csv')
val_labels = pd.read_csv('./data/regression/val_labels.csv')
te_features = pd.read_csv('./data/regression/test_features.csv')
te_labels = pd.read_csv('./data/regression/test_labels.csv')
custom_ml = ['lr','knn','tree','svm','mlp','rf','gb','ada','xgb','hgboost','huber','rgcv','cvlasso','sgd']
reg_cv_demo = fastRegressor(custom_estimators = custom_ml,random_state = 13,cv_num = 5,n_comb = 12)
reg_cv_demo.fit(tr_features,tr_labels)
models = {}
for mdl in ['lr','knn','tree','svm','mlp','rf','gb','ada','xgb','hgboost','huber','rgcv','cvlasso','sgd']:
models[mdl] = joblib.load('./pkl/{}_reg_model.pkl'.format(mdl))
for name, mdl in models.items():
try:
ml_evl = evaluate_model(model_type = "reg")
ml_evl.fit(name, mdl, val_features, val_labels)
except:
print(f"Failed to load the {mdl}.")
```
|
github_jupyter
|
# Install external packages in binder environment.
!pip install xgboost
# Regression Demo
import pandas as pd
from optimalflow.autoCV import evaluate_model,dynaRegressor
import joblib
from optimalflow.utilis_func import pipeline_splitting_rule, update_parameters,reset_parameters
reset_parameters()
tr_features = pd.read_csv('./data/regression/train_features.csv')
tr_labels = pd.read_csv('./data/regression/train_labels.csv')
val_features = pd.read_csv('./data/regression/val_features.csv')
val_labels = pd.read_csv('./data/regression/val_labels.csv')
te_features = pd.read_csv('./data/regression/test_features.csv')
te_labels = pd.read_csv('./data/regression/test_labels.csv')
reg_cv_demo = dynaRegressor(random_state=13,cv_num = 5)
reg_cv_demo.fit(tr_features,tr_labels)
models = {}
for mdl in ['lr','knn','tree','svm','mlp','rf','gb','ada','xgb','hgboost','huber','rgcv','cvlasso','sgd']:
models[mdl] = joblib.load('./pkl/{}_reg_model.pkl'.format(mdl))
for name, mdl in models.items():
try:
ml_evl = evaluate_model(model_type = "reg")
ml_evl.fit(name, mdl, val_features, val_labels)
except:
print(f"Failed to load the {mdl}.")
# Fast Regression Demo
import pandas as pd
from optimalflow.autoCV import evaluate_model,fastRegressor
import joblib
from optimalflow.utilis_func import pipeline_splitting_rule, update_parameters,reset_parameters
reset_parameters()
tr_features = pd.read_csv('./data/regression/train_features.csv')
tr_labels = pd.read_csv('./data/regression/train_labels.csv')
val_features = pd.read_csv('./data/regression/val_features.csv')
val_labels = pd.read_csv('./data/regression/val_labels.csv')
te_features = pd.read_csv('./data/regression/test_features.csv')
te_labels = pd.read_csv('./data/regression/test_labels.csv')
custom_ml = ['lr','knn','tree','svm','mlp','rf','gb','ada','xgb','hgboost','huber','rgcv','cvlasso','sgd']
reg_cv_demo = fastRegressor(custom_estimators = custom_ml,random_state = 13,cv_num = 5,n_comb = 12)
reg_cv_demo.fit(tr_features,tr_labels)
models = {}
for mdl in ['lr','knn','tree','svm','mlp','rf','gb','ada','xgb','hgboost','huber','rgcv','cvlasso','sgd']:
models[mdl] = joblib.load('./pkl/{}_reg_model.pkl'.format(mdl))
for name, mdl in models.items():
try:
ml_evl = evaluate_model(model_type = "reg")
ml_evl.fit(name, mdl, val_features, val_labels)
except:
print(f"Failed to load the {mdl}.")
| 0.309545 | 0.644589 |
```
# Imports
import numpy as np
import matplotlib.pyplot as plt
from stablab import (semicirc, winding_number, Evans_plot, emcset,
Evans_compute, Struct, soln, profile_flux,
reflect_image)
from stablab.evans import LdimRdim
from stablab.root_finding import root_solver1
# A matrix and compound Ak matrix for Evans Function ODE
from capillarity import A, Ak
# Profile ODE and its jacobian
from capillarity import profile_ode, profile_jacobian
# parameters
p = Struct()
p.gamma = 1.4
p.vp = .15
# dependent parameters
p.a = -(1-p.vp)/(1-p.vp**(-p.gamma))
# This example solves the profile for $d = -0.45$ and then uses
# continuation thereafter
d_vals = -np.arange(0.45, 0.451, 0.001)
# solve profile. Use continuation as an example
s = Struct()
for j,curr_d_val in enumerate(d_vals):
p.d = curr_d_val
# profile
s.n = 2 # this is the dimension of the profile ode
# we divide the domain in half to deal with the
# non-uniqueness caused by translational invariance
# s.side = 1 means we are solving the profile on the interval [0,X]
s.side = 1
s.F = profile_ode # profile_ode is the profile ode
s.Flinear = profile_jacobian # profile_jacobian is the profile ode Jacobian
s.UL = np.array([1, 0]) # These are the endstates of the profile and its derivative at x = -infty
s.UR = np.array([p.vp, 0]) # These are the endstates of the profile and its derivative at x = +infty
s.phase = 0.5*(s.UL+s.UR) # this is the phase condition for the profile at x = 0
s.order = [0] # this indicates to which componenet the phase conditions is applied
s.stats = 'on' # this prints data and plots the profile as it is solved
if j == 0:
# there are some other options you specify. You can look in profile_flux to see them
p,s = profile_flux(p,s) # solve profile for first time
s_old = s
else:
# this time we are using continuation
p,s = profile_flux(p,s,s_old); # solve profile
# plot the profile
x = np.linspace(s.L,s.R,200)
y = soln(x,s)
plt.title("Profile")
plt.plot(x,y[:,0].real)
plt.plot(x,y[:,1].real)
plt.show()
# structure variables
# Here you can choose the method you use for the Evans function, or you can set the option
# to default and let it choose. [2,2] is the size of the manifold evolving from - and + infy in the
# Evans solver. 'front' indicates you are solving for a traveling wave front and not a periodic solution
# s,e,m,c = emcset(s,'front',LdimRdim(A,s,p),'default',A) # default for capillarity is reg_reg_polar
# s,e,m,c = emcset(s,'front',[2,2],'reg_adj_polar',A)
# s,e,m,c = emcset(s,'front',[2,2],'adj_reg_polar',A)
# s,e,m,c = emcset(s,'front',[2,2],'reg_reg_polar',A)
# This choice solves the right hand side via exterior products
s,e,m,c = emcset(s, 'front', [2,2], 'adj_reg_compound', A, Ak)
# display a waitbar
c.stats = 'print' # 'on', 'print', or 'off'
c.ksteps = 2**8
# Preimage Contour
# This is a semi circle. You can also do a semi annulus or a rectangle
circpnts = 30
imagpnts = 30
R = 10
spread = 2
zerodist = 10**(-2)
preimage = semicirc(circpnts,imagpnts,c.ksteps,R,spread,zerodist)
# compute Evans function
halfw, domain = Evans_compute(preimage,c,s,p,m,e)
w = halfw / halfw[0]
# We computed Evans function on half of contour
# We now reflect the image across the real axis
w = reflect_image(w)
# Process and display data
wnd = winding_number(w) # determine the number of roots inside the contour
print("Winding Number: ",wnd)
# plot the Evans function (normalized)
Evans_plot(w)
```
|
github_jupyter
|
# Imports
import numpy as np
import matplotlib.pyplot as plt
from stablab import (semicirc, winding_number, Evans_plot, emcset,
Evans_compute, Struct, soln, profile_flux,
reflect_image)
from stablab.evans import LdimRdim
from stablab.root_finding import root_solver1
# A matrix and compound Ak matrix for Evans Function ODE
from capillarity import A, Ak
# Profile ODE and its jacobian
from capillarity import profile_ode, profile_jacobian
# parameters
p = Struct()
p.gamma = 1.4
p.vp = .15
# dependent parameters
p.a = -(1-p.vp)/(1-p.vp**(-p.gamma))
# This example solves the profile for $d = -0.45$ and then uses
# continuation thereafter
d_vals = -np.arange(0.45, 0.451, 0.001)
# solve profile. Use continuation as an example
s = Struct()
for j,curr_d_val in enumerate(d_vals):
p.d = curr_d_val
# profile
s.n = 2 # this is the dimension of the profile ode
# we divide the domain in half to deal with the
# non-uniqueness caused by translational invariance
# s.side = 1 means we are solving the profile on the interval [0,X]
s.side = 1
s.F = profile_ode # profile_ode is the profile ode
s.Flinear = profile_jacobian # profile_jacobian is the profile ode Jacobian
s.UL = np.array([1, 0]) # These are the endstates of the profile and its derivative at x = -infty
s.UR = np.array([p.vp, 0]) # These are the endstates of the profile and its derivative at x = +infty
s.phase = 0.5*(s.UL+s.UR) # this is the phase condition for the profile at x = 0
s.order = [0] # this indicates to which componenet the phase conditions is applied
s.stats = 'on' # this prints data and plots the profile as it is solved
if j == 0:
# there are some other options you specify. You can look in profile_flux to see them
p,s = profile_flux(p,s) # solve profile for first time
s_old = s
else:
# this time we are using continuation
p,s = profile_flux(p,s,s_old); # solve profile
# plot the profile
x = np.linspace(s.L,s.R,200)
y = soln(x,s)
plt.title("Profile")
plt.plot(x,y[:,0].real)
plt.plot(x,y[:,1].real)
plt.show()
# structure variables
# Here you can choose the method you use for the Evans function, or you can set the option
# to default and let it choose. [2,2] is the size of the manifold evolving from - and + infy in the
# Evans solver. 'front' indicates you are solving for a traveling wave front and not a periodic solution
# s,e,m,c = emcset(s,'front',LdimRdim(A,s,p),'default',A) # default for capillarity is reg_reg_polar
# s,e,m,c = emcset(s,'front',[2,2],'reg_adj_polar',A)
# s,e,m,c = emcset(s,'front',[2,2],'adj_reg_polar',A)
# s,e,m,c = emcset(s,'front',[2,2],'reg_reg_polar',A)
# This choice solves the right hand side via exterior products
s,e,m,c = emcset(s, 'front', [2,2], 'adj_reg_compound', A, Ak)
# display a waitbar
c.stats = 'print' # 'on', 'print', or 'off'
c.ksteps = 2**8
# Preimage Contour
# This is a semi circle. You can also do a semi annulus or a rectangle
circpnts = 30
imagpnts = 30
R = 10
spread = 2
zerodist = 10**(-2)
preimage = semicirc(circpnts,imagpnts,c.ksteps,R,spread,zerodist)
# compute Evans function
halfw, domain = Evans_compute(preimage,c,s,p,m,e)
w = halfw / halfw[0]
# We computed Evans function on half of contour
# We now reflect the image across the real axis
w = reflect_image(w)
# Process and display data
wnd = winding_number(w) # determine the number of roots inside the contour
print("Winding Number: ",wnd)
# plot the Evans function (normalized)
Evans_plot(w)
| 0.672762 | 0.679471 |
<h1>Table of Contents<span class="tocSkip"></span></h1>
<div class="toc"><ul class="toc-item"><li><span><a href="#msticpy---Event-Timeline" data-toc-modified-id="msticpy---Event-Timeline-1">msticpy - Event Timeline</a></span></li><li><span><a href="#Discrete-Event-Timelines" data-toc-modified-id="Discrete-Event-Timelines-2">Discrete Event Timelines</a></span><ul class="toc-item"><li><span><a href="#Plotting-a-simple-timeline" data-toc-modified-id="Plotting-a-simple-timeline-2.1">Plotting a simple timeline</a></span></li><li><span><a href="#More-Advanced-Timelines" data-toc-modified-id="More-Advanced-Timelines-2.2">More Advanced Timelines</a></span><ul class="toc-item"><li><span><a href="#Grouping-Series-From-a-Single-DataFrame" data-toc-modified-id="Grouping-Series-From-a-Single-DataFrame-2.2.1">Grouping Series From a Single DataFrame</a></span></li></ul></li><li><span><a href="#Displaying-a-reference-line" data-toc-modified-id="Displaying-a-reference-line-2.3">Displaying a reference line</a></span></li><li><span><a href="#Plotting-series-from-different-data-sets" data-toc-modified-id="Plotting-series-from-different-data-sets-2.4">Plotting series from different data sets</a></span></li></ul></li><li><span><a href="#Plotting-Series-with-Scalar-Values" data-toc-modified-id="Plotting-Series-with-Scalar-Values-3">Plotting Series with Scalar Values</a></span><ul class="toc-item"><li><span><a href="#Documentation-for-display_timeline_values" data-toc-modified-id="Documentation-for-display_timeline_values-3.1">Documentation for display_timeline_values</a></span></li></ul></li><li><span><a href="#Exporting-Plots-as-PNGs" data-toc-modified-id="Exporting-Plots-as-PNGs-4">Exporting Plots as PNGs</a></span></li></ul></div>
# msticpy - Event Timeline
This notebook demonstrates the use of the timeline displays built using the [Bokeh library](https://bokeh.pydata.org).
There are two display types:
- Discrete event series - this plots multiple series of events as discrete glyphs
- Event value series - this plots a scalar value of the events using glyphs, bars or traditional line graph (or some combination.
```
# Imports
import sys
import warnings
from msticpy.common.utility import check_py_version
MIN_REQ_PYTHON = (3,6)
check_py_version(MIN_REQ_PYTHON)
from IPython import get_ipython
from IPython.display import display, HTML, Markdown
import ipywidgets as widgets
import pandas as pd
pd.set_option('display.max_rows', 100)
pd.set_option('display.max_columns', 50)
pd.set_option('display.max_colwidth', 100)
from msticpy.nbtools import *
from msticpy.sectools import *
WIDGET_DEFAULTS = {'layout': widgets.Layout(width='95%'),
'style': {'description_width': 'initial'}}
```
# Discrete Event Timelines
## Plotting a simple timeline
nbdisplay.display_timeline
```
Display a timeline of events.
Parameters
----------
data : Union[dict, pd.DataFrame]
Either
dict of data sets to plot on the timeline with the following structure.
Key: str
Name of data set to be displayed in legend
Value: dict
containing
data: pd.DataFrame
Data to plot
time_column: str, optional
Name of the timestamp column
(defaults to `time_column` function parameter)
source_columns: list[str], optional
List of source columns to use in tooltips
(defaults to `source_columns` function parameter)
color: str, optional
Color of datapoints for this data
(defaults to autogenerating colors)
Or
DataFrame as a single data set or grouped into individual
plot series using the `group_by` parameter
time_column : str, optional
Name of the timestamp column
(the default is 'TimeGenerated')
source_columns : list, optional
List of default source columns to use in tooltips
(the default is None)
```
```
processes_on_host = pd.read_csv('data/processes_on_host.csv',
parse_dates=["TimeGenerated"],
infer_datetime_format=True)
# At a minimum we need to pass a dataframe with data
nbdisplay.display_timeline(processes_on_host)
```
The Bokeh graph is interactive and has the following features:
- Tooltip display for each event marker as you hover over it
- Toolbar with the following tools (most are toggles enabling or disabling the tool):
- Panning
- Select zoom
- Mouse wheel zoom
- Reset to default view
- Save image to PNG
- Hover tool
Additionally an interactive timeline navigation bar is displayed below the main graph. You can change the timespan shown on the main graph by dragging or resizing the selected area on this navigation bar.
**Note**:
- the tooltips work on the Windows process data shown above because of a legacy fallback built into the code.
Usually you need to specify the `source_columns` parameter explicitly to have
the hover tooltips populated correctly.
## More Advanced Timelines
`display_timeline` also takes a number of optional parameters that give you more flexibility to show multiple data series and change the way the graph appears.
```
Other Parameters
----------------
title : str, optional
Title to display (the default is None)
alert : SecurityAlert, optional
Add a reference line/label using the alert time (the default is None)
ref_event : Any, optional
Add a reference line/label using the alert time (the default is None)
ref_time : datetime, optional
Add a reference line/label using `ref_time` (the default is None)
group_by : str
(where `data` is a DataFrame)
The column to group timelines on
sort_by : str
(where `data` is a DataFrame)
The column to order timelines on
legend: str, optional
left, right or inline
(the default is None/no legend)
yaxis : bool, optional
Whether to show the yaxis and labels (default is False)
range_tool : bool, optional
Show the the range slider tool (default is True)
height : int, optional
The height of the plot figure
(the default is auto-calculated height)
width : int, optional
The width of the plot figure (the default is 900)
color : str
Default series color (default is "navy")
```
### Grouping Series From a Single DataFrame
```
nbdisplay.display_timeline(processes_on_host,
group_by="Account",
source_columns=["NewProcessName",
"ParentProcessName"],
legend="left");
```
We can use the group_by parameter to specify a column on which to split individually plotted series.
Specifying a legend, we can see the value of each series group. The legend is interactive - click on a series name to
hide/show the data. The legend can be placed inside of the chart (`legend="inline"`) or to the left or right.
Alternatively we can enable the yaxis - although this is not guaranteed to show all values of the groups.
**Note**:
- the tooltips work on the Windows process data shown above because of a legacy fallback built into the code. Usually you need to specify the `source_columns` parameter explicitly to have the hover tooltips populated correctly.
- the trailing semicolon just stops Jupyter showing the return value from the function. It isn't mandatory
```
nbdisplay.display_timeline(processes_on_host,
group_by="Account",
source_columns=["NewProcessName",
"ParentProcessName"],
legend="none",
yaxis=True, ygrid=True);
host_logons = pd.read_csv('data/host_logons.csv',
parse_dates=["TimeGenerated"],
infer_datetime_format=True)
nbdisplay.display_timeline(host_logons,
title="Logons by Account name",
group_by="Account",
source_columns=["Account", "TargetLogonId", "LogonType"],
legend="left",
height=200);
nbdisplay.display_timeline(host_logons,
title="Logons by logon type",
group_by="LogonType",
source_columns=["Account", "TargetLogonId", "LogonType"],
legend="left",
height=200,
range_tool=False,
ygrid=True);
```
## Displaying a reference line
If you have a single item (e.g. an alert) that you want to show as a reference point on the graph you can pass a datetime value, or any object that has a TimeGenerated or StartTimeUtc property.
If the object doesn't have one of these, just pass the property as the ref_time parameter.
```
fake_alert = processes_on_host.sample().iloc[0]
nbdisplay.display_timeline(host_logons,
title="Processes with marker",
group_by="LogonType",
source_columns=["Account", "TargetLogonId", "LogonType"],
alert=fake_alert,
legend="left");
```
## Plotting series from different data sets
When you want to plot data sets with different schema on the same plot it is difficult to put them in a single DataFrame.
To do this we need to assemble the different data sets into a dictionary and pass that to the `display_timeline`
The dictionary has this format:
Key: str
Name of data set to be displayed in legend
Value: dict, the value holds the settings for each data series:
data: pd.DataFrame
Data to plot
time_column: str, optional
Name of the timestamp column
(defaults to `time_column` function parameter)
source_columns: list[str], optional
List of source columns to use in tooltips
(defaults to `source_columns` function parameter)
color: str, optional
Color of datapoints for this data
(defaults to autogenerating colors)
```
procs_and_logons = {
"Processes" : {"data": processes_on_host, "source_columns": ["NewProcessName", "Account"]},
"Logons": {"data": host_logons, "source_columns": ["Account", "TargetLogonId", "LogonType"]}
}
nbdisplay.display_timeline(data=procs_and_logons,
title="Logons and Processes",
legend="left", yaxis=False);
```
# Plotting Series with Scalar Values
Often you may want to see a scalar value plotted with the series.
The example below uses `display_timeline_values` to plot network flow data using the total flows recorded between a pair of IP addresses.
Note that the majority of parameters are the same as `display_timeline` but include a mandatory `y` parameter which indicates which value you want to plot on the y (vertical) axis.
```
az_net_flows_df = pd.read_csv('data/az_net_flows.csv',
parse_dates=["TimeGenerated", "FlowStartTime", "FlowEndTime"],
infer_datetime_format=True)
flow_plot = nbdisplay.display_timeline_values(data=az_net_flows_df,
group_by="L7Protocol",
source_columns=["FlowType",
"AllExtIPs",
"L7Protocol",
"FlowDirection",
"TotalAllowedFlows"],
time_column="FlowStartTime",
y="TotalAllowedFlows",
legend="right",
height=500);
```
By default the plot uses vertical bars show the values but you can use any combination of vbar, circle and line, using the `kind` parameter. You specify the plot types as a list of strings (all lowercase).
**Notes**
- including "circle" in the plot kinds makes it easier to see the hover value
- the line plot can be a bit misleading since it will plot lines between adjacent data points of the same series implying that there is a gradual change in the value being plotted - even though there may be no data between the times of these adjacent points. For this reason using vbar is often a more accurate view.
```
flow_plot = nbdisplay.display_timeline_values(data=az_net_flows_df,
group_by="L7Protocol",
source_columns=["FlowType",
"AllExtIPs",
"L7Protocol",
"FlowDirection",
"TotalAllowedFlows"],
time_column="FlowStartTime",
y="TotalAllowedFlows",
legend="right",
height=500,
kind=["vbar", "circle"]
);
nbdisplay.display_timeline_values(data=az_net_flows_df[az_net_flows_df["L7Protocol"] == "http"],
group_by="L7Protocol",
title="Line plot can be misleading",
source_columns=["FlowType",
"AllExtIPs",
"L7Protocol",
"FlowDirection",
"TotalAllowedFlows"],
time_column="FlowStartTime",
y="TotalAllowedFlows",
legend="right",
height=300,
kind=["line", "circle"],
range_tool=False
);
nbdisplay.display_timeline_values(data=az_net_flows_df[az_net_flows_df["L7Protocol"] == "http"],
group_by="L7Protocol",
title="Vbar and circle show zero gaps in data",
source_columns=["FlowType",
"AllExtIPs",
"L7Protocol",
"FlowDirection",
"TotalAllowedFlows"],
time_column="FlowStartTime",
y="TotalAllowedFlows",
legend="right",
height=300,
kind=["vbar", "circle"],
range_tool=False
);
```
## Documentation for display_timeline_values
```
nbdisplay.display_timeline_values(
data: pandas.core.frame.DataFrame,
y: str,
time_column: str = 'TimeGenerated',
source_columns: list = None,
**kwargs,
) -> figure
Display a timeline of events.
Parameters
----------
data : pd.DataFrame
DataFrame as a single data set or grouped into individual
plot series using the `group_by` parameter
time_column : str, optional
Name of the timestamp column
(the default is 'TimeGenerated')
y : str
The column name holding the value to plot vertically
source_columns : list, optional
List of default source columns to use in tooltips
(the default is None)
Other Parameters
----------------
x : str, optional
alias of `time_column`
title : str, optional
Title to display (the default is None)
ref_event : Any, optional
Add a reference line/label using the alert time (the default is None)
ref_time : datetime, optional
Add a reference line/label using `ref_time` (the default is None)
group_by : str
(where `data` is a DataFrame)
The column to group timelines on
sort_by : str
(where `data` is a DataFrame)
The column to order timelines on
legend: str, optional
left, right or inline
(the default is None/no legend)
yaxis : bool, optional
Whether to show the yaxis and labels
range_tool : bool, optional
Show the the range slider tool (default is True)
height : int, optional
The height of the plot figure
(the default is auto-calculated height)
width : int, optional
The width of the plot figure (the default is 900)
color : str
Default series color (default is "navy"). This is overridden by
automatic color assignments if plotting a grouped chart
kind : Union[str, List[str]]
one or more glyph types to plot., optional
Supported types are "circle", "line" and "vbar" (default is "vbar")
Returns
-------
figure
The bokeh plot figure.
```
# Exporting Plots as PNGs
To use bokeh.io image export functions you need selenium, phantomjs and pillow installed:
`conda install -c bokeh selenium phantomjs pillow`
or
`pip install selenium pillow`
`npm install -g phantomjs-prebuilt`
For phantomjs see https://phantomjs.org/download.html.
Once the prerequisites are installed you can create a plot and save the return value to a variable.
Then export the plot using `export_png` function.
```python
from bokeh.io import export_png
from IPython.display import Image
# Create a plot
flow_plot = nbdisplay.display_timeline_values(data=az_net_flows_df,
group_by="L7Protocol",
source_columns=["FlowType",
"AllExtIPs",
"L7Protocol",
"FlowDirection",
"TotalAllowedFlows"],
time_column="FlowStartTime",
y="TotalAllowedFlows",
legend="right",
height=500,
kind=["vbar", "circle"]
);
# Export
file_name = "plot.png"
export_png(flow_plot, filename=file_name)
# Read it and show it
display(Markdown(f"## Here is our saved plot: {file_name}"))
Image(filename=file_name)
```
|
github_jupyter
|
# Imports
import sys
import warnings
from msticpy.common.utility import check_py_version
MIN_REQ_PYTHON = (3,6)
check_py_version(MIN_REQ_PYTHON)
from IPython import get_ipython
from IPython.display import display, HTML, Markdown
import ipywidgets as widgets
import pandas as pd
pd.set_option('display.max_rows', 100)
pd.set_option('display.max_columns', 50)
pd.set_option('display.max_colwidth', 100)
from msticpy.nbtools import *
from msticpy.sectools import *
WIDGET_DEFAULTS = {'layout': widgets.Layout(width='95%'),
'style': {'description_width': 'initial'}}
Display a timeline of events.
Parameters
----------
data : Union[dict, pd.DataFrame]
Either
dict of data sets to plot on the timeline with the following structure.
Key: str
Name of data set to be displayed in legend
Value: dict
containing
data: pd.DataFrame
Data to plot
time_column: str, optional
Name of the timestamp column
(defaults to `time_column` function parameter)
source_columns: list[str], optional
List of source columns to use in tooltips
(defaults to `source_columns` function parameter)
color: str, optional
Color of datapoints for this data
(defaults to autogenerating colors)
Or
DataFrame as a single data set or grouped into individual
plot series using the `group_by` parameter
time_column : str, optional
Name of the timestamp column
(the default is 'TimeGenerated')
source_columns : list, optional
List of default source columns to use in tooltips
(the default is None)
processes_on_host = pd.read_csv('data/processes_on_host.csv',
parse_dates=["TimeGenerated"],
infer_datetime_format=True)
# At a minimum we need to pass a dataframe with data
nbdisplay.display_timeline(processes_on_host)
Other Parameters
----------------
title : str, optional
Title to display (the default is None)
alert : SecurityAlert, optional
Add a reference line/label using the alert time (the default is None)
ref_event : Any, optional
Add a reference line/label using the alert time (the default is None)
ref_time : datetime, optional
Add a reference line/label using `ref_time` (the default is None)
group_by : str
(where `data` is a DataFrame)
The column to group timelines on
sort_by : str
(where `data` is a DataFrame)
The column to order timelines on
legend: str, optional
left, right or inline
(the default is None/no legend)
yaxis : bool, optional
Whether to show the yaxis and labels (default is False)
range_tool : bool, optional
Show the the range slider tool (default is True)
height : int, optional
The height of the plot figure
(the default is auto-calculated height)
width : int, optional
The width of the plot figure (the default is 900)
color : str
Default series color (default is "navy")
nbdisplay.display_timeline(processes_on_host,
group_by="Account",
source_columns=["NewProcessName",
"ParentProcessName"],
legend="left");
nbdisplay.display_timeline(processes_on_host,
group_by="Account",
source_columns=["NewProcessName",
"ParentProcessName"],
legend="none",
yaxis=True, ygrid=True);
host_logons = pd.read_csv('data/host_logons.csv',
parse_dates=["TimeGenerated"],
infer_datetime_format=True)
nbdisplay.display_timeline(host_logons,
title="Logons by Account name",
group_by="Account",
source_columns=["Account", "TargetLogonId", "LogonType"],
legend="left",
height=200);
nbdisplay.display_timeline(host_logons,
title="Logons by logon type",
group_by="LogonType",
source_columns=["Account", "TargetLogonId", "LogonType"],
legend="left",
height=200,
range_tool=False,
ygrid=True);
fake_alert = processes_on_host.sample().iloc[0]
nbdisplay.display_timeline(host_logons,
title="Processes with marker",
group_by="LogonType",
source_columns=["Account", "TargetLogonId", "LogonType"],
alert=fake_alert,
legend="left");
procs_and_logons = {
"Processes" : {"data": processes_on_host, "source_columns": ["NewProcessName", "Account"]},
"Logons": {"data": host_logons, "source_columns": ["Account", "TargetLogonId", "LogonType"]}
}
nbdisplay.display_timeline(data=procs_and_logons,
title="Logons and Processes",
legend="left", yaxis=False);
az_net_flows_df = pd.read_csv('data/az_net_flows.csv',
parse_dates=["TimeGenerated", "FlowStartTime", "FlowEndTime"],
infer_datetime_format=True)
flow_plot = nbdisplay.display_timeline_values(data=az_net_flows_df,
group_by="L7Protocol",
source_columns=["FlowType",
"AllExtIPs",
"L7Protocol",
"FlowDirection",
"TotalAllowedFlows"],
time_column="FlowStartTime",
y="TotalAllowedFlows",
legend="right",
height=500);
flow_plot = nbdisplay.display_timeline_values(data=az_net_flows_df,
group_by="L7Protocol",
source_columns=["FlowType",
"AllExtIPs",
"L7Protocol",
"FlowDirection",
"TotalAllowedFlows"],
time_column="FlowStartTime",
y="TotalAllowedFlows",
legend="right",
height=500,
kind=["vbar", "circle"]
);
nbdisplay.display_timeline_values(data=az_net_flows_df[az_net_flows_df["L7Protocol"] == "http"],
group_by="L7Protocol",
title="Line plot can be misleading",
source_columns=["FlowType",
"AllExtIPs",
"L7Protocol",
"FlowDirection",
"TotalAllowedFlows"],
time_column="FlowStartTime",
y="TotalAllowedFlows",
legend="right",
height=300,
kind=["line", "circle"],
range_tool=False
);
nbdisplay.display_timeline_values(data=az_net_flows_df[az_net_flows_df["L7Protocol"] == "http"],
group_by="L7Protocol",
title="Vbar and circle show zero gaps in data",
source_columns=["FlowType",
"AllExtIPs",
"L7Protocol",
"FlowDirection",
"TotalAllowedFlows"],
time_column="FlowStartTime",
y="TotalAllowedFlows",
legend="right",
height=300,
kind=["vbar", "circle"],
range_tool=False
);
nbdisplay.display_timeline_values(
data: pandas.core.frame.DataFrame,
y: str,
time_column: str = 'TimeGenerated',
source_columns: list = None,
**kwargs,
) -> figure
Display a timeline of events.
Parameters
----------
data : pd.DataFrame
DataFrame as a single data set or grouped into individual
plot series using the `group_by` parameter
time_column : str, optional
Name of the timestamp column
(the default is 'TimeGenerated')
y : str
The column name holding the value to plot vertically
source_columns : list, optional
List of default source columns to use in tooltips
(the default is None)
Other Parameters
----------------
x : str, optional
alias of `time_column`
title : str, optional
Title to display (the default is None)
ref_event : Any, optional
Add a reference line/label using the alert time (the default is None)
ref_time : datetime, optional
Add a reference line/label using `ref_time` (the default is None)
group_by : str
(where `data` is a DataFrame)
The column to group timelines on
sort_by : str
(where `data` is a DataFrame)
The column to order timelines on
legend: str, optional
left, right or inline
(the default is None/no legend)
yaxis : bool, optional
Whether to show the yaxis and labels
range_tool : bool, optional
Show the the range slider tool (default is True)
height : int, optional
The height of the plot figure
(the default is auto-calculated height)
width : int, optional
The width of the plot figure (the default is 900)
color : str
Default series color (default is "navy"). This is overridden by
automatic color assignments if plotting a grouped chart
kind : Union[str, List[str]]
one or more glyph types to plot., optional
Supported types are "circle", "line" and "vbar" (default is "vbar")
Returns
-------
figure
The bokeh plot figure.
from bokeh.io import export_png
from IPython.display import Image
# Create a plot
flow_plot = nbdisplay.display_timeline_values(data=az_net_flows_df,
group_by="L7Protocol",
source_columns=["FlowType",
"AllExtIPs",
"L7Protocol",
"FlowDirection",
"TotalAllowedFlows"],
time_column="FlowStartTime",
y="TotalAllowedFlows",
legend="right",
height=500,
kind=["vbar", "circle"]
);
# Export
file_name = "plot.png"
export_png(flow_plot, filename=file_name)
# Read it and show it
display(Markdown(f"## Here is our saved plot: {file_name}"))
Image(filename=file_name)
| 0.591841 | 0.954052 |
```
import matplotlib.pyplot as plt
%matplotlib inline
import torch
device = torch.device('cpu')
```
# ODE vector field visualizations
This notebook shows examples of functions Neural ODEs cannot approximate and how this affects the learned vector fields.
#### Create an ODE function
```
from anode.models import ODEFunc
data_dim = 1 # We model 1d data to easily visualize it
hidden_dim = 16
# Create a 3-layer MLP as the ODE function f(h, t)
odefunc = ODEFunc(device, data_dim, hidden_dim, time_dependent=True)
```
#### Visualize vector field of ODE function
We can visualize what the randomly initialized ODE function's vector field looks like.
```
from viz.plots import vector_field_plt
vector_field_plt(odefunc, num_points=10, timesteps=10,
h_min=-1.5, h_max=1.5)
```
## Create functions to approximate
We will approximate two functions: an easy one (the identity mapping) and a hard one (correspond to g_1d in the paper)
```
from experiments.dataloaders import Data1D
from torch.utils.data import DataLoader
data_easy = Data1D(num_points=500, target_flip=False)
data_hard = Data1D(num_points=500, target_flip=True)
dataloader_easy = DataLoader(data_easy, batch_size=32, shuffle=True)
dataloader_hard = DataLoader(data_hard, batch_size=32, shuffle=True)
```
#### Visualize the data
```
for inputs, targets in dataloader_easy:
break
vector_field_plt(odefunc, num_points=10, timesteps=10,
inputs=inputs, targets=targets,
h_min=-1.5, h_max=1.5)
for inputs, targets in dataloader_hard:
break
vector_field_plt(odefunc, num_points=10, timesteps=10,
inputs=inputs, targets=targets,
h_min=-1.5, h_max=1.5)
```
## Train a model on data
We can now try to fit a Neural ODE to the two functions
```
from anode.models import ODEBlock
from anode.training import Trainer
data_dim = 1
hidden_dim = 16
# Create a model for the easy function
odefunc_easy = ODEFunc(device, data_dim, hidden_dim,
time_dependent=True)
model_easy = ODEBlock(device, odefunc_easy)
# Create a model for the hard function
odefunc_hard = ODEFunc(device, data_dim, hidden_dim,
time_dependent=True)
model_hard = ODEBlock(device, odefunc_hard)
# Create an optimizer and trainer for easy function
optimizer_easy = torch.optim.Adam(model_easy.parameters(), lr=1e-3)
trainer_easy = Trainer(model_easy, optimizer_easy, device, print_freq=5)
# Create an optimizer and trainer for hard function
optimizer_hard = torch.optim.Adam(model_hard.parameters(), lr=5e-4)
trainer_hard = Trainer(model_hard, optimizer_hard, device, print_freq=5)
```
#### Train model on easy data
```
trainer_easy.train(dataloader_easy, num_epochs=10)
```
#### Visualize model trajectories
As can be seen, the learned vector field maps the inputs to targets correctly.
```
for inputs, targets in dataloader_easy:
break
# Plot 8 trajectories
vector_field_plt(odefunc_easy, num_points=10, timesteps=10,
inputs=inputs[:8], targets=targets[:8],
h_min=-1.5, h_max=1.5, model=model_easy)
```
#### Train model on hard data
```
trainer_hard.train(dataloader_hard, num_epochs=50)
for inputs, targets in dataloader_hard:
break
# Plot 8 trajectories
vector_field_plt(odefunc_hard, num_points=10, timesteps=10,
inputs=inputs[:8], targets=targets[:8],
h_min=-1.5, h_max=1.5, model=model_hard)
```
## Augmented Neural ODEs
As can be seen, Neural ODEs struggle to fit the hard function. In fact, it can be proven that Neural ODEs cannot represent this function. In order to overcome this, we can use Augmented Neural ODEs which extend the space on which the ODE is solved. Examples of this are shown in the `augmented-neural-ode-example` notebook.
|
github_jupyter
|
import matplotlib.pyplot as plt
%matplotlib inline
import torch
device = torch.device('cpu')
from anode.models import ODEFunc
data_dim = 1 # We model 1d data to easily visualize it
hidden_dim = 16
# Create a 3-layer MLP as the ODE function f(h, t)
odefunc = ODEFunc(device, data_dim, hidden_dim, time_dependent=True)
from viz.plots import vector_field_plt
vector_field_plt(odefunc, num_points=10, timesteps=10,
h_min=-1.5, h_max=1.5)
from experiments.dataloaders import Data1D
from torch.utils.data import DataLoader
data_easy = Data1D(num_points=500, target_flip=False)
data_hard = Data1D(num_points=500, target_flip=True)
dataloader_easy = DataLoader(data_easy, batch_size=32, shuffle=True)
dataloader_hard = DataLoader(data_hard, batch_size=32, shuffle=True)
for inputs, targets in dataloader_easy:
break
vector_field_plt(odefunc, num_points=10, timesteps=10,
inputs=inputs, targets=targets,
h_min=-1.5, h_max=1.5)
for inputs, targets in dataloader_hard:
break
vector_field_plt(odefunc, num_points=10, timesteps=10,
inputs=inputs, targets=targets,
h_min=-1.5, h_max=1.5)
from anode.models import ODEBlock
from anode.training import Trainer
data_dim = 1
hidden_dim = 16
# Create a model for the easy function
odefunc_easy = ODEFunc(device, data_dim, hidden_dim,
time_dependent=True)
model_easy = ODEBlock(device, odefunc_easy)
# Create a model for the hard function
odefunc_hard = ODEFunc(device, data_dim, hidden_dim,
time_dependent=True)
model_hard = ODEBlock(device, odefunc_hard)
# Create an optimizer and trainer for easy function
optimizer_easy = torch.optim.Adam(model_easy.parameters(), lr=1e-3)
trainer_easy = Trainer(model_easy, optimizer_easy, device, print_freq=5)
# Create an optimizer and trainer for hard function
optimizer_hard = torch.optim.Adam(model_hard.parameters(), lr=5e-4)
trainer_hard = Trainer(model_hard, optimizer_hard, device, print_freq=5)
trainer_easy.train(dataloader_easy, num_epochs=10)
for inputs, targets in dataloader_easy:
break
# Plot 8 trajectories
vector_field_plt(odefunc_easy, num_points=10, timesteps=10,
inputs=inputs[:8], targets=targets[:8],
h_min=-1.5, h_max=1.5, model=model_easy)
trainer_hard.train(dataloader_hard, num_epochs=50)
for inputs, targets in dataloader_hard:
break
# Plot 8 trajectories
vector_field_plt(odefunc_hard, num_points=10, timesteps=10,
inputs=inputs[:8], targets=targets[:8],
h_min=-1.5, h_max=1.5, model=model_hard)
| 0.84039 | 0.973342 |
# Calculating Chern number
```
import numpy as np
from numpy import linalg as la
O = 0.
M = 1. # g
C = 1. # N/m
w0 = np.sqrt(C / M)
l = 1. # distance between masses
x = np.array([[1.], [0.]]) # x hat
y = np.array([[0.], [1.]]) # y hat
a1 = np.sqrt(3) * l * x
a2 = (np.sqrt(3) * x + 3 * y) * l / 2.
R1 = 1 / 3 * (a1 + a2)
R2 = 1 / 3 * (-2 * a1 + a2)
R3 = 1 / 3 * (a1 - 2 * a2)
R11 = R1 * R1.conj().T
R22 = R2 * R2.conj().T
R33 = R3 * R3.conj().T
import numpy as np
precision = 0.1
kxs = np.arange(-np.pi, np.pi, precision)
kys = np.arange(-np.pi, np.pi, precision)
R0_ = lambda k: (1 / 2 * (3 - np.exp(1.j * k.dot(R1)) - np.exp(1.j * k.dot(R2)) - np.exp(1.j * k.dot(R3)))).item()
R1_ = lambda k: (np.sqrt(3) / 4 * (np.exp(1.j * k.dot(R2)) - np.exp(1.j * k.dot(R1)))).item()
R2_ = lambda k: (2 * O / w0**2).item()
R3_ = lambda k: (1 / 4 * (1 - np.exp(1.j * k.dot(R1)) - np.exp(1.j * k.dot(R2)) + np.exp(1.j * k.dot(R3)))).item()
R = lambda k: np.sqrt(R1_(k)**2 + R2_(k)**2 + R3_(k)**2)
ws = np.zeros((len(kys), len(kxs), 2))
for y, ky in enumerate(kys):
for x, kx in enumerate(kxs):
k = np.array([kx, ky])
ws[y, x] = np.sqrt(np.array([R0_(k) - R(k), R0_(k) + R(k)])).real
# ws[y, x] = np.array([R0_(k) - R(k), R0_(k) + R(k)]).real
# 3D dispersion
import matplotlib.pyplot as plt
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
X, Y = np.meshgrid(kxs, kys)
vmax = np.amax(ws[:, :, 1])
ax.plot_surface(X, Y, ws[:, :, 0], cmap='gist_rainbow_r')
p = ax.plot_surface(X, Y, ws[:, :, 1], cmap='gist_rainbow_r')
# ax.set_xlabel(r"$k_x$ [/m]", fontsize=14)
# ax.set_xlim(-xw, xw)
# ax.set_ylabel(r"$k_y$ [/m]", fontsize=14)
# ax.set_ylim(-yw, yw)
# ax.set_zlabel(r"$\omega$ [/s]", fontsize=14)
# ax.set_zlim(0)
fig.colorbar(p, pad=0.15)
# fig.savefig("2d-graphene-dispersion-3d.png")
plt.show()
evals_all = np.zeros((len(kys), len(kxs), 2))
for y, ky in enumerate(kys):
for x, kx in enumerate(kxs):
k = np.array([kx, ky])
H = np.array([
[R0_(k) + R3_(k), R1_(k) - 1.j * R2_(k)],
[R1_(k) + 1.j * R2_(k), R0_(k) - R3_(k)],
])
evals, _ = la.eigh(H)
evals = evals[np.argsort(evals)]
evals_all[y, x] = evals
evals_all = np.array(evals_all)
# 3D dispersion
import matplotlib.pyplot as plt
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
X, Y = np.meshgrid(kxs, kys)
vmax = np.amax(np.sqrt(evals_all[:, :, 1]))
ax.plot_surface(X, Y, np.sqrt(evals_all[:, :, 0]), vmin=0., vmax=vmax, cmap='gist_rainbow_r')
p = ax.plot_surface(X, Y, np.sqrt(evals_all[:, :, 1]), vmin=0., vmax=vmax, cmap='gist_rainbow_r')
# ax.set_xlabel(r"$k_x$ [/m]", fontsize=14)
# ax.set_xlim(-xw, xw)
# ax.set_ylabel(r"$k_y$ [/m]", fontsize=14)
# ax.set_ylim(-yw, yw)
# ax.set_zlabel(r"$\omega$ [/s]", fontsize=14)
# ax.set_zlim(0)
fig.colorbar(p, pad=0.15)
# fig.savefig("2d-graphene-dispersion-3d.png")
plt.show()
```
# Using library
```
import z2pack
import numpy as np
O = 13
M = .01 # kg
C = 4. # N/m
w0 = np.sqrt(C / M)
l = 1. # distance between masses
x = np.array([[1.], [0.]]) # x hat
y = np.array([[0.], [1.]]) # y hat
a1 = np.sqrt(3) * l * x
a2 = (np.sqrt(3) * x + 3 * y) * l / 2.
R1 = 1 / 3 * (a1 + a2)
R2 = 1 / 3 * (-2 * a1 + a2)
R3 = 1 / 3 * (a1 - 2 * a2)
R1h = R1 / la.norm(R1)
R2h = R2 / la.norm(R2)
R3h = R3 / la.norm(R3)
R11 = R1h * R1h.conj().T
R22 = R2h * R2h.conj().T
R33 = R3h * R3h.conj().T
Rw = np.array([[0., -1.], [1., 0.]])
R0_ = lambda k: (1 / 2 * (3 - np.exp(1.j * k.dot(R1)) - np.exp(1.j * k.dot(R2)) - np.exp(1.j * k.dot(R3)))).item()
R1_ = lambda k: (np.sqrt(3) / 4 * (np.exp(1.j * k.dot(R2)) - np.exp(1.j * k.dot(R1)))).item()
R2_ = lambda k: (2 * O / w0**2).item()
R3_ = lambda k: (1 / 4 * (1 - np.exp(1.j * k.dot(R1)) - np.exp(1.j * k.dot(R2)) + np.exp(1.j * k.dot(R3)))).item()
R = lambda k: np.sqrt(R1_(k)**2 + R2_(k)**2 + R3_(k)**2)
mu = -5.
R0_ = lambda k: 0
R1_ = lambda k: (-2 * np.sin(k[0])).item()
R2_ = lambda k: (-2 * np.sin(k[1])).item()
R3_ = lambda k: (2 * (np.cos(k[0]) + np.cos(k[1])) + mu).item()
Os = np.arange(0, 100, 0.1)
def hamiltonian(k):
kx, ky, kz = k
k_ = np.array([kx, ky])
p = np.exp(1.j * k_.dot(a1))
s = np.exp(1.j * k_.dot(a2))
# p = k_.dot(a1)
# s = k_.dot(a2)
H = np.array([
[R0_(k_) + R3_(k_), R1_(k_) - 1.j * R2_(k_)],
[R1_(k_) + 1.j * R2_(k_), R0_(k_) - R3_(k_)],
])
# H = w0**2 * np.vstack([
# np.hstack([R11 + R22 + R33 - 2.j * O * Rw / w0**2, -(R11 + R22 * p.conj() + R33 * s.conj())]),
# np.hstack([-(R11 + R22 * p + R33 * s) ,R11 + R22 + R33 - 2.j * O * Rw / w0**2]),
# ])
return H
system = z2pack.hm.System(hamiltonian)
result = z2pack.surface.run(
system=system,
surface=lambda t1, t2: [t1, t2, 0]
)
print(z2pack.invariant.chern(result)) # Prints the Chern number
# print(z2pack.invariant.z2(result)) # Prints the Chern numbe
import numpy as np
import numpy.linalg as la
from sympy import symbols, Matrix, BlockMatrix, conjugate
from sympy.functions import exp
kx, ky = symbols('kx ky')
O = 0.
M = .01 # kg
C = 4. # N/m
w0 = np.sqrt(C / M)
l = 1. # distance between masses
x = np.array([[1.], [0.]]) # x hat
y = np.array([[0.], [1.]]) # y hat
a1 = np.sqrt(3) * l * x
a2 = (np.sqrt(3) * x + 3 * y) * l / 2.
R1 = 1 / 3 * (a1 + a2)
R2 = 1 / 3 * (-2 * a1 + a2)
R3 = 1 / 3 * (a1 - 2 * a2)
R1h = R1 / la.norm(R1)
R2h = R2 / la.norm(R2)
R3h = R3 / la.norm(R3)
R11 = R1h * R1h.conj().T
R22 = R2h * R2h.conj().T
R33 = R3h * R3h.conj().T
Rw = np.array([[0., -1.], [1., 0.]])
p = exp(1.j * np.sqrt(3) * kx)
s = exp(1.j * (np.sqrt(3) * kx + 3 * ky) / 2)
H = BlockMatrix([
[Matrix(R11 + R22 + R33 - 2.j * O * Rw / w0**2), Matrix(-(R11 + R22 * conjugate(p) + R33 * conjugate(s)))],
[Matrix(-(R11 + R22 * p + R33 * s)) ,Matrix(R11 + R22 + R33 - 2.j * O * Rw / w0**2)]
])
H = w0 ** 2 * Matrix(H)
# H.eigenvects()
print(R11, R22, R33)
```
# Chern with extended eigen vecs
```
import numpy as np
import numpy.linalg as la
O = 0.
C = 1.
M = 1.
w0 = np.sqrt(C / M)
a = 1. # distance between masses
x = np.array([[1.], [0.]]) # x hat
y = np.array([[0.], [1.]]) # y hat
a1 = np.sqrt(3) * a * x
a2 = (np.sqrt(3) * x + 3 * y) * a / 2.
R1 = 1 / 3 * (a1 + a2)
R2 = 1 / 3 * (-2 * a1 + a2)
R3 = 1 / 3 * (a1 - 2 * a2)
R1h = R1 / la.norm(R1)
R2h = R2 / la.norm(R2)
R3h = R3 / la.norm(R3)
R11 = R1h * R1h.conj().T
R22 = R2h * R2h.conj().T
R33 = R3h * R3h.conj().T
Rw = np.array([[0., -1.], [1., 0.]])
precision = 0.01
xw = np.pi / np.sqrt(3) * 2 # width of x
yw = np.pi / 3 * 4 # width of y
kxs = np.arange(-xw, xw, precision)
kys = np.arange(-yw, yw, precision)
evals_all = np.zeros((len(kys), len(kxs), 8), dtype=np.complex128)
evecs_all = np.zeros((len(kys), len(kxs), 8, 8), dtype=np.complex128)
for y, ky in enumerate(kys):
for x, kx in enumerate(kxs):
k = np.array([kx, ky])
p = np.exp(1.j * k.dot(a1))
s = np.exp(1.j * k.dot(a2))
L11 = np.vstack([
np.hstack([R11 + R22 + R33, -(R11 + p.conj() * R22 + s.conj() * R33)]),
np.hstack([-(R11 + p * R22 + s * R33), R11 + R22 + R33])
])
L12 = np.vstack([
np.hstack([-Rw * 2.j * O, np.zeros((2, 2))]),
np.hstack([np.zeros((2, 2)), -Rw * 2.j * O])
])
L = w0 ** 2 * np.vstack([
np.hstack([L11, L12]),
np.hstack([np.zeros((4, 4)), np.eye(4)])
])
M = np.vstack([
np.hstack([np.zeros((4, 4)), np.eye(4)]),
np.hstack([np.eye(4), np.zeros((4, 4))])
])
H = np.linalg.inv(M).dot(L)
evals, evecs = la.eig(H)
idcs = np.argsort(evals)
evals, evecs = evals[idcs], evecs[idcs]
evals_all[y, x] = evals
evecs_all[y, x] = evecs
evals_all = np.array(evals_all)
evecs_all = np.array(evecs_all)
print(evals_all.shape, evecs_all.shape)
from shapely.geometry import Point
from shapely.geometry.polygon import Polygon
x = np.array([[1.], [0.]]) # x hat
y = np.array([[0.], [1.]]) # y hat
a1_ = 2 * np.pi / np.sqrt(3) / a * (x - y / np.sqrt(3))
a2_ = 4 * np.pi / 3 / a * y
K = (a1_ / 2 + a2_ / 4) / np.cos(np.pi / 6)**2
b1 = 2 * np.pi / np.sqrt(3) / a * (x - y / np.sqrt(3))
b2 = 4 * np.pi / 3 / a * y
h = precision
bz= Polygon([K, -K + b1 + b2, K - b1, -K, K - b1 - b2, -K + b1])
print(h)
print(bz.contains(Point(-4,0)))
C = 0
band_number = 6
# xi = evecs_all[:, :, band_number, :2]
# eta = evecs_all[:, :, band_number, 2:4]
xi = evecs_all[:, :, :2, band_number]
eta = evecs_all[:, :, 2:4, band_number]
for y, ky in enumerate(kys):
for x, kx in enumerate(kxs):
p = Point(kx, ky)
if not bz.contains(p):
continue
dxi_dkx = (xi[y, x+1] - xi[y, x-1]) / (2 * h)
dxi_dky = (xi[y+1, x] - xi[y-1, x]) / (2 * h)
C += 1.j * dxi_dkx.conj().T.dot(dxi_dky) - dxi_dky.conj().T.dot(dxi_dkx)
deta_dkx = (eta[y, x+1] - eta[y, x-1]) / (2 * h)
deta_dky = (eta[y+1, x] - eta[y-1, x]) / (2 * h)
C += 1.j * deta_dkx.conj().T.dot(deta_dky) - deta_dky.conj().T.dot(deta_dkx)
C /= 2 * np.pi
# print(evecs_all[0, 0, 0, :4])
# print(evecs_all[0, 0, 0, 4:])
print(C)
def calc_chern(hVec):
"""Calculate 1st Chern number given eigen-vectors. Using Japanese algorithm
(2005).
:param hVec: eigen-vectors of all band (assuming gapless).
:return: list of Chern number of all bands. Should summed to 0.
"""
hVec = np.array(hVec)
dimy, dimx, nlevels, _ = hVec.shape
cnlist = np.zeros(nlevels)
for iy in range(dimy-1):
for ix in range(dimx-1):
u12 = hVec[iy, ix + 1].conjugate().T @ hVec[iy, ix]
u23 = hVec[iy + 1, ix + 1].conjugate().T @ hVec[iy, ix + 1]
u34 = hVec[iy + 1, ix].conjugate().T @ hVec[iy + 1, ix + 1]
u41 = hVec[iy, ix].conjugate().T @ hVec[iy + 1, ix]
t12 = np.diag(u12.diagonal())
t23 = np.diag(u23.diagonal())
t34 = np.diag(u34.diagonal())
t41 = np.diag(u41.diagonal())
tplaquet = t41 @ t34 @ t23 @ t12
cnlist += np.angle(tplaquet.diagonal())
cnlist /= 2 * np.pi
cnlist = chop(cnlist)
return cnlist
def chop(array, tol=1e-7):
"""Realize Mathematica Chop[].
:param array: 1D array.
:param tol: tolerance to be chopped. default to 1e-7
:return: chopped array. (original array alse modified.)
"""
for i in range(len(array)):
a = array[i]
if np.abs(a-round(a)) < tol:
array[i] = round(a)
return array
calc_chern(evecs_all[:, :, :4, 4:])
```
# Chern with perturbation
```
import numpy as np
import numpy.linalg as la
O = 4
C = 4
M = 0.01
w0 = np.sqrt(C / M)
a = 1. # distance between masses
x = np.array([[1.], [0.]]) # x hat
y = np.array([[0.], [1.]]) # y hat
a1 = np.sqrt(3) * a * x
a2 = (np.sqrt(3) * x + 3 * y) * a / 2.
R1 = 1 / 3 * (a1 + a2)
R2 = 1 / 3 * (-2 * a1 + a2)
R3 = 1 / 3 * (a1 - 2 * a2)
R1h = R1 / la.norm(R1)
R2h = R2 / la.norm(R2)
R3h = R3 / la.norm(R3)
R11 = R1h * R1h.conj().T
R22 = R2h * R2h.conj().T
R33 = R3h * R3h.conj().T
Rw = np.array([[0., -1.], [1., 0.]])
precision = 1e-2
xw = np.pi / np.sqrt(3) * 2 # width of x
yw = np.pi / 3 * 4 # width of y
kxs = np.arange(-xw, xw, precision)
kys = np.arange(-yw, yw, precision)
evals_all = np.zeros((len(kys), len(kxs), 4), dtype=np.complex128)
evecs_all = np.zeros((len(kys), len(kxs), 4, 4), dtype=np.complex128)
for y, ky in enumerate(kys):
for x, kx in enumerate(kxs):
k = np.array([kx, ky])
p = np.exp(1.j * k.dot(a1))
s = np.exp(1.j * k.dot(a2))
H = w0**2 * np.vstack([
np.hstack([R11 + R22 + R33 - 2.j * O * Rw / w0**2, -(R11 + R22 * p.conj() + R33 * s.conj())]),
np.hstack([-(R11 + R22 * p + R33 * s) ,R11 + R22 + R33 - 2.j * O * Rw / w0**2]),
])
evals, evecs = la.eigh(H)
idcs = np.argsort(evals)
evals, evecs = evals[idcs], evecs[idcs]
evals_all[y, x] = evals
evecs_all[y, x] = evecs
evals_all = np.array(evals_all)
evecs_all = np.array(evecs_all)
print(evals_all.shape, evecs_all.shape)
from conut import MechanicalGraphene, HamiltonianType, MechanicalGrapheneLattice
from conut.data import WaveNumber, WaveNumberType
from torch.utils.data import DataLoader
import torch
import numpy as np
from shapely.geometry import Polygon, Point
dev = 'cuda' if torch.cuda.is_available() else 'cpu'
dev = 'cpu'
κ = 1.
α = 1.
m = 1.
l = 1.
params = {'batch_size': 512,
'shuffle': False,
'num_workers': 4}
for Ω in [0., 0.1, 0.2, 0.3, 0.4, 0.5, 0.6]:
# Setup Lattice
lattice = MechanicalGrapheneLattice(l, α, device=dev)
# Construct wavenumber dataset
wn = WaveNumber(WaveNumberType.XY, lattice, precision=3e-3)
wn_gkm = WaveNumber(WaveNumberType.GKM, lattice, precision=1e-1)
wn_generator = DataLoader(wn, **params)
wn_gkm_generator = DataLoader(wn_gkm, **params)
# Build mechanical graphene model
mg = MechanicalGraphene(κ=κ, m=m, α=α, lattice=lattice, h_type=HamiltonianType.Bulk, shape=wn.shape, Ω=Ω, perturbation=True).to(dev)
# mg_gkm = MechanicalGraphene(κ=κ, m=m, α=α, lattice=lattice, h_type=HamiltonianType.Bulk, shape=wn_gkm.shape, Ω=Ω, GKM=True, perturbation=False).to(dev)
for idcs_batch, ks_batch in wn_generator:
idcs_batch, ks_batch = idcs_batch.to(dev), ks_batch.to(dev)
mg(idcs_batch, ks_batch)
evecs = mg.evecs.detach().numpy()
def calc_chern(hVec):
"""Calculate 1st Chern number given eigen-vectors. Using Japanese algorithm
(2005).
:param hVec: eigen-vectors of all band (assuming gapless).
:return: list of Chern number of all bands. Should summed to 0.
"""
hVec = np.array(hVec)
dimy, dimx, nlevels, _ = hVec.shape
cnlist = np.zeros(nlevels)
for iy in range(dimy-1):
for ix in range(dimx-1):
u12 = hVec[iy, ix + 1].conjugate().T @ hVec[iy, ix]
u23 = hVec[iy + 1, ix + 1].conjugate().T @ hVec[iy, ix + 1]
u34 = hVec[iy + 1, ix].conjugate().T @ hVec[iy + 1, ix + 1]
u41 = hVec[iy, ix].conjugate().T @ hVec[iy + 1, ix]
t12 = np.diag(u12.diagonal())
t23 = np.diag(u23.diagonal())
t34 = np.diag(u34.diagonal())
t41 = np.diag(u41.diagonal())
tplaquet = t41 @ t34 @ t23 @ t12
cnlist += np.angle(tplaquet.diagonal())
cnlist /= 2 * np.pi
cnlist = chop(cnlist)
return cnlist
def chop(array, tol=1e-7):
"""Realize Mathematica Chop[].
:param array: 1D array.
:param tol: tolerance to be chopped. default to 1e-7
:return: chopped array. (original array alse modified.)
"""
for i in range(len(array)):
a = array[i]
if np.abs(a-round(a)) < tol:
array[i] = round(a)
return array
x = np.array([[1.], [0.]]) # x hat
y = np.array([[0.], [1.]]) # y hat
a1_ = 2 * np.pi / np.sqrt(3) / l * (x - y / np.sqrt(3))
a2_ = 4 * np.pi / 3 / l * y
K = (a1_ / 2 + a2_ / 4) / np.cos(np.pi / 6)**2
b1 = 2 * np.pi / np.sqrt(3) / l * (x - y / np.sqrt(3))
b2 = 4 * np.pi / 3 / l * y
bz= Polygon([K, -K + b1 + b2, K - b1, -K, K - b1 - b2, -K + b1])
evecs_all_bz = np.zeros((len(wn.kys), len(wn.kxs), 4, 4), dtype=np.complex128)
for y, ky in enumerate(wn.kys):
for x, kx in enumerate(wn.kxs):
p = Point(kx, ky)
if not bz.contains(p):
continue
evecs_all_bz[y, x] = evecs[y, x]
print(Ω, calc_chern(evecs_all_bz))
```
|
github_jupyter
|
import numpy as np
from numpy import linalg as la
O = 0.
M = 1. # g
C = 1. # N/m
w0 = np.sqrt(C / M)
l = 1. # distance between masses
x = np.array([[1.], [0.]]) # x hat
y = np.array([[0.], [1.]]) # y hat
a1 = np.sqrt(3) * l * x
a2 = (np.sqrt(3) * x + 3 * y) * l / 2.
R1 = 1 / 3 * (a1 + a2)
R2 = 1 / 3 * (-2 * a1 + a2)
R3 = 1 / 3 * (a1 - 2 * a2)
R11 = R1 * R1.conj().T
R22 = R2 * R2.conj().T
R33 = R3 * R3.conj().T
import numpy as np
precision = 0.1
kxs = np.arange(-np.pi, np.pi, precision)
kys = np.arange(-np.pi, np.pi, precision)
R0_ = lambda k: (1 / 2 * (3 - np.exp(1.j * k.dot(R1)) - np.exp(1.j * k.dot(R2)) - np.exp(1.j * k.dot(R3)))).item()
R1_ = lambda k: (np.sqrt(3) / 4 * (np.exp(1.j * k.dot(R2)) - np.exp(1.j * k.dot(R1)))).item()
R2_ = lambda k: (2 * O / w0**2).item()
R3_ = lambda k: (1 / 4 * (1 - np.exp(1.j * k.dot(R1)) - np.exp(1.j * k.dot(R2)) + np.exp(1.j * k.dot(R3)))).item()
R = lambda k: np.sqrt(R1_(k)**2 + R2_(k)**2 + R3_(k)**2)
ws = np.zeros((len(kys), len(kxs), 2))
for y, ky in enumerate(kys):
for x, kx in enumerate(kxs):
k = np.array([kx, ky])
ws[y, x] = np.sqrt(np.array([R0_(k) - R(k), R0_(k) + R(k)])).real
# ws[y, x] = np.array([R0_(k) - R(k), R0_(k) + R(k)]).real
# 3D dispersion
import matplotlib.pyplot as plt
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
X, Y = np.meshgrid(kxs, kys)
vmax = np.amax(ws[:, :, 1])
ax.plot_surface(X, Y, ws[:, :, 0], cmap='gist_rainbow_r')
p = ax.plot_surface(X, Y, ws[:, :, 1], cmap='gist_rainbow_r')
# ax.set_xlabel(r"$k_x$ [/m]", fontsize=14)
# ax.set_xlim(-xw, xw)
# ax.set_ylabel(r"$k_y$ [/m]", fontsize=14)
# ax.set_ylim(-yw, yw)
# ax.set_zlabel(r"$\omega$ [/s]", fontsize=14)
# ax.set_zlim(0)
fig.colorbar(p, pad=0.15)
# fig.savefig("2d-graphene-dispersion-3d.png")
plt.show()
evals_all = np.zeros((len(kys), len(kxs), 2))
for y, ky in enumerate(kys):
for x, kx in enumerate(kxs):
k = np.array([kx, ky])
H = np.array([
[R0_(k) + R3_(k), R1_(k) - 1.j * R2_(k)],
[R1_(k) + 1.j * R2_(k), R0_(k) - R3_(k)],
])
evals, _ = la.eigh(H)
evals = evals[np.argsort(evals)]
evals_all[y, x] = evals
evals_all = np.array(evals_all)
# 3D dispersion
import matplotlib.pyplot as plt
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
X, Y = np.meshgrid(kxs, kys)
vmax = np.amax(np.sqrt(evals_all[:, :, 1]))
ax.plot_surface(X, Y, np.sqrt(evals_all[:, :, 0]), vmin=0., vmax=vmax, cmap='gist_rainbow_r')
p = ax.plot_surface(X, Y, np.sqrt(evals_all[:, :, 1]), vmin=0., vmax=vmax, cmap='gist_rainbow_r')
# ax.set_xlabel(r"$k_x$ [/m]", fontsize=14)
# ax.set_xlim(-xw, xw)
# ax.set_ylabel(r"$k_y$ [/m]", fontsize=14)
# ax.set_ylim(-yw, yw)
# ax.set_zlabel(r"$\omega$ [/s]", fontsize=14)
# ax.set_zlim(0)
fig.colorbar(p, pad=0.15)
# fig.savefig("2d-graphene-dispersion-3d.png")
plt.show()
import z2pack
import numpy as np
O = 13
M = .01 # kg
C = 4. # N/m
w0 = np.sqrt(C / M)
l = 1. # distance between masses
x = np.array([[1.], [0.]]) # x hat
y = np.array([[0.], [1.]]) # y hat
a1 = np.sqrt(3) * l * x
a2 = (np.sqrt(3) * x + 3 * y) * l / 2.
R1 = 1 / 3 * (a1 + a2)
R2 = 1 / 3 * (-2 * a1 + a2)
R3 = 1 / 3 * (a1 - 2 * a2)
R1h = R1 / la.norm(R1)
R2h = R2 / la.norm(R2)
R3h = R3 / la.norm(R3)
R11 = R1h * R1h.conj().T
R22 = R2h * R2h.conj().T
R33 = R3h * R3h.conj().T
Rw = np.array([[0., -1.], [1., 0.]])
R0_ = lambda k: (1 / 2 * (3 - np.exp(1.j * k.dot(R1)) - np.exp(1.j * k.dot(R2)) - np.exp(1.j * k.dot(R3)))).item()
R1_ = lambda k: (np.sqrt(3) / 4 * (np.exp(1.j * k.dot(R2)) - np.exp(1.j * k.dot(R1)))).item()
R2_ = lambda k: (2 * O / w0**2).item()
R3_ = lambda k: (1 / 4 * (1 - np.exp(1.j * k.dot(R1)) - np.exp(1.j * k.dot(R2)) + np.exp(1.j * k.dot(R3)))).item()
R = lambda k: np.sqrt(R1_(k)**2 + R2_(k)**2 + R3_(k)**2)
mu = -5.
R0_ = lambda k: 0
R1_ = lambda k: (-2 * np.sin(k[0])).item()
R2_ = lambda k: (-2 * np.sin(k[1])).item()
R3_ = lambda k: (2 * (np.cos(k[0]) + np.cos(k[1])) + mu).item()
Os = np.arange(0, 100, 0.1)
def hamiltonian(k):
kx, ky, kz = k
k_ = np.array([kx, ky])
p = np.exp(1.j * k_.dot(a1))
s = np.exp(1.j * k_.dot(a2))
# p = k_.dot(a1)
# s = k_.dot(a2)
H = np.array([
[R0_(k_) + R3_(k_), R1_(k_) - 1.j * R2_(k_)],
[R1_(k_) + 1.j * R2_(k_), R0_(k_) - R3_(k_)],
])
# H = w0**2 * np.vstack([
# np.hstack([R11 + R22 + R33 - 2.j * O * Rw / w0**2, -(R11 + R22 * p.conj() + R33 * s.conj())]),
# np.hstack([-(R11 + R22 * p + R33 * s) ,R11 + R22 + R33 - 2.j * O * Rw / w0**2]),
# ])
return H
system = z2pack.hm.System(hamiltonian)
result = z2pack.surface.run(
system=system,
surface=lambda t1, t2: [t1, t2, 0]
)
print(z2pack.invariant.chern(result)) # Prints the Chern number
# print(z2pack.invariant.z2(result)) # Prints the Chern numbe
import numpy as np
import numpy.linalg as la
from sympy import symbols, Matrix, BlockMatrix, conjugate
from sympy.functions import exp
kx, ky = symbols('kx ky')
O = 0.
M = .01 # kg
C = 4. # N/m
w0 = np.sqrt(C / M)
l = 1. # distance between masses
x = np.array([[1.], [0.]]) # x hat
y = np.array([[0.], [1.]]) # y hat
a1 = np.sqrt(3) * l * x
a2 = (np.sqrt(3) * x + 3 * y) * l / 2.
R1 = 1 / 3 * (a1 + a2)
R2 = 1 / 3 * (-2 * a1 + a2)
R3 = 1 / 3 * (a1 - 2 * a2)
R1h = R1 / la.norm(R1)
R2h = R2 / la.norm(R2)
R3h = R3 / la.norm(R3)
R11 = R1h * R1h.conj().T
R22 = R2h * R2h.conj().T
R33 = R3h * R3h.conj().T
Rw = np.array([[0., -1.], [1., 0.]])
p = exp(1.j * np.sqrt(3) * kx)
s = exp(1.j * (np.sqrt(3) * kx + 3 * ky) / 2)
H = BlockMatrix([
[Matrix(R11 + R22 + R33 - 2.j * O * Rw / w0**2), Matrix(-(R11 + R22 * conjugate(p) + R33 * conjugate(s)))],
[Matrix(-(R11 + R22 * p + R33 * s)) ,Matrix(R11 + R22 + R33 - 2.j * O * Rw / w0**2)]
])
H = w0 ** 2 * Matrix(H)
# H.eigenvects()
print(R11, R22, R33)
import numpy as np
import numpy.linalg as la
O = 0.
C = 1.
M = 1.
w0 = np.sqrt(C / M)
a = 1. # distance between masses
x = np.array([[1.], [0.]]) # x hat
y = np.array([[0.], [1.]]) # y hat
a1 = np.sqrt(3) * a * x
a2 = (np.sqrt(3) * x + 3 * y) * a / 2.
R1 = 1 / 3 * (a1 + a2)
R2 = 1 / 3 * (-2 * a1 + a2)
R3 = 1 / 3 * (a1 - 2 * a2)
R1h = R1 / la.norm(R1)
R2h = R2 / la.norm(R2)
R3h = R3 / la.norm(R3)
R11 = R1h * R1h.conj().T
R22 = R2h * R2h.conj().T
R33 = R3h * R3h.conj().T
Rw = np.array([[0., -1.], [1., 0.]])
precision = 0.01
xw = np.pi / np.sqrt(3) * 2 # width of x
yw = np.pi / 3 * 4 # width of y
kxs = np.arange(-xw, xw, precision)
kys = np.arange(-yw, yw, precision)
evals_all = np.zeros((len(kys), len(kxs), 8), dtype=np.complex128)
evecs_all = np.zeros((len(kys), len(kxs), 8, 8), dtype=np.complex128)
for y, ky in enumerate(kys):
for x, kx in enumerate(kxs):
k = np.array([kx, ky])
p = np.exp(1.j * k.dot(a1))
s = np.exp(1.j * k.dot(a2))
L11 = np.vstack([
np.hstack([R11 + R22 + R33, -(R11 + p.conj() * R22 + s.conj() * R33)]),
np.hstack([-(R11 + p * R22 + s * R33), R11 + R22 + R33])
])
L12 = np.vstack([
np.hstack([-Rw * 2.j * O, np.zeros((2, 2))]),
np.hstack([np.zeros((2, 2)), -Rw * 2.j * O])
])
L = w0 ** 2 * np.vstack([
np.hstack([L11, L12]),
np.hstack([np.zeros((4, 4)), np.eye(4)])
])
M = np.vstack([
np.hstack([np.zeros((4, 4)), np.eye(4)]),
np.hstack([np.eye(4), np.zeros((4, 4))])
])
H = np.linalg.inv(M).dot(L)
evals, evecs = la.eig(H)
idcs = np.argsort(evals)
evals, evecs = evals[idcs], evecs[idcs]
evals_all[y, x] = evals
evecs_all[y, x] = evecs
evals_all = np.array(evals_all)
evecs_all = np.array(evecs_all)
print(evals_all.shape, evecs_all.shape)
from shapely.geometry import Point
from shapely.geometry.polygon import Polygon
x = np.array([[1.], [0.]]) # x hat
y = np.array([[0.], [1.]]) # y hat
a1_ = 2 * np.pi / np.sqrt(3) / a * (x - y / np.sqrt(3))
a2_ = 4 * np.pi / 3 / a * y
K = (a1_ / 2 + a2_ / 4) / np.cos(np.pi / 6)**2
b1 = 2 * np.pi / np.sqrt(3) / a * (x - y / np.sqrt(3))
b2 = 4 * np.pi / 3 / a * y
h = precision
bz= Polygon([K, -K + b1 + b2, K - b1, -K, K - b1 - b2, -K + b1])
print(h)
print(bz.contains(Point(-4,0)))
C = 0
band_number = 6
# xi = evecs_all[:, :, band_number, :2]
# eta = evecs_all[:, :, band_number, 2:4]
xi = evecs_all[:, :, :2, band_number]
eta = evecs_all[:, :, 2:4, band_number]
for y, ky in enumerate(kys):
for x, kx in enumerate(kxs):
p = Point(kx, ky)
if not bz.contains(p):
continue
dxi_dkx = (xi[y, x+1] - xi[y, x-1]) / (2 * h)
dxi_dky = (xi[y+1, x] - xi[y-1, x]) / (2 * h)
C += 1.j * dxi_dkx.conj().T.dot(dxi_dky) - dxi_dky.conj().T.dot(dxi_dkx)
deta_dkx = (eta[y, x+1] - eta[y, x-1]) / (2 * h)
deta_dky = (eta[y+1, x] - eta[y-1, x]) / (2 * h)
C += 1.j * deta_dkx.conj().T.dot(deta_dky) - deta_dky.conj().T.dot(deta_dkx)
C /= 2 * np.pi
# print(evecs_all[0, 0, 0, :4])
# print(evecs_all[0, 0, 0, 4:])
print(C)
def calc_chern(hVec):
"""Calculate 1st Chern number given eigen-vectors. Using Japanese algorithm
(2005).
:param hVec: eigen-vectors of all band (assuming gapless).
:return: list of Chern number of all bands. Should summed to 0.
"""
hVec = np.array(hVec)
dimy, dimx, nlevels, _ = hVec.shape
cnlist = np.zeros(nlevels)
for iy in range(dimy-1):
for ix in range(dimx-1):
u12 = hVec[iy, ix + 1].conjugate().T @ hVec[iy, ix]
u23 = hVec[iy + 1, ix + 1].conjugate().T @ hVec[iy, ix + 1]
u34 = hVec[iy + 1, ix].conjugate().T @ hVec[iy + 1, ix + 1]
u41 = hVec[iy, ix].conjugate().T @ hVec[iy + 1, ix]
t12 = np.diag(u12.diagonal())
t23 = np.diag(u23.diagonal())
t34 = np.diag(u34.diagonal())
t41 = np.diag(u41.diagonal())
tplaquet = t41 @ t34 @ t23 @ t12
cnlist += np.angle(tplaquet.diagonal())
cnlist /= 2 * np.pi
cnlist = chop(cnlist)
return cnlist
def chop(array, tol=1e-7):
"""Realize Mathematica Chop[].
:param array: 1D array.
:param tol: tolerance to be chopped. default to 1e-7
:return: chopped array. (original array alse modified.)
"""
for i in range(len(array)):
a = array[i]
if np.abs(a-round(a)) < tol:
array[i] = round(a)
return array
calc_chern(evecs_all[:, :, :4, 4:])
import numpy as np
import numpy.linalg as la
O = 4
C = 4
M = 0.01
w0 = np.sqrt(C / M)
a = 1. # distance between masses
x = np.array([[1.], [0.]]) # x hat
y = np.array([[0.], [1.]]) # y hat
a1 = np.sqrt(3) * a * x
a2 = (np.sqrt(3) * x + 3 * y) * a / 2.
R1 = 1 / 3 * (a1 + a2)
R2 = 1 / 3 * (-2 * a1 + a2)
R3 = 1 / 3 * (a1 - 2 * a2)
R1h = R1 / la.norm(R1)
R2h = R2 / la.norm(R2)
R3h = R3 / la.norm(R3)
R11 = R1h * R1h.conj().T
R22 = R2h * R2h.conj().T
R33 = R3h * R3h.conj().T
Rw = np.array([[0., -1.], [1., 0.]])
precision = 1e-2
xw = np.pi / np.sqrt(3) * 2 # width of x
yw = np.pi / 3 * 4 # width of y
kxs = np.arange(-xw, xw, precision)
kys = np.arange(-yw, yw, precision)
evals_all = np.zeros((len(kys), len(kxs), 4), dtype=np.complex128)
evecs_all = np.zeros((len(kys), len(kxs), 4, 4), dtype=np.complex128)
for y, ky in enumerate(kys):
for x, kx in enumerate(kxs):
k = np.array([kx, ky])
p = np.exp(1.j * k.dot(a1))
s = np.exp(1.j * k.dot(a2))
H = w0**2 * np.vstack([
np.hstack([R11 + R22 + R33 - 2.j * O * Rw / w0**2, -(R11 + R22 * p.conj() + R33 * s.conj())]),
np.hstack([-(R11 + R22 * p + R33 * s) ,R11 + R22 + R33 - 2.j * O * Rw / w0**2]),
])
evals, evecs = la.eigh(H)
idcs = np.argsort(evals)
evals, evecs = evals[idcs], evecs[idcs]
evals_all[y, x] = evals
evecs_all[y, x] = evecs
evals_all = np.array(evals_all)
evecs_all = np.array(evecs_all)
print(evals_all.shape, evecs_all.shape)
from conut import MechanicalGraphene, HamiltonianType, MechanicalGrapheneLattice
from conut.data import WaveNumber, WaveNumberType
from torch.utils.data import DataLoader
import torch
import numpy as np
from shapely.geometry import Polygon, Point
dev = 'cuda' if torch.cuda.is_available() else 'cpu'
dev = 'cpu'
κ = 1.
α = 1.
m = 1.
l = 1.
params = {'batch_size': 512,
'shuffle': False,
'num_workers': 4}
for Ω in [0., 0.1, 0.2, 0.3, 0.4, 0.5, 0.6]:
# Setup Lattice
lattice = MechanicalGrapheneLattice(l, α, device=dev)
# Construct wavenumber dataset
wn = WaveNumber(WaveNumberType.XY, lattice, precision=3e-3)
wn_gkm = WaveNumber(WaveNumberType.GKM, lattice, precision=1e-1)
wn_generator = DataLoader(wn, **params)
wn_gkm_generator = DataLoader(wn_gkm, **params)
# Build mechanical graphene model
mg = MechanicalGraphene(κ=κ, m=m, α=α, lattice=lattice, h_type=HamiltonianType.Bulk, shape=wn.shape, Ω=Ω, perturbation=True).to(dev)
# mg_gkm = MechanicalGraphene(κ=κ, m=m, α=α, lattice=lattice, h_type=HamiltonianType.Bulk, shape=wn_gkm.shape, Ω=Ω, GKM=True, perturbation=False).to(dev)
for idcs_batch, ks_batch in wn_generator:
idcs_batch, ks_batch = idcs_batch.to(dev), ks_batch.to(dev)
mg(idcs_batch, ks_batch)
evecs = mg.evecs.detach().numpy()
def calc_chern(hVec):
"""Calculate 1st Chern number given eigen-vectors. Using Japanese algorithm
(2005).
:param hVec: eigen-vectors of all band (assuming gapless).
:return: list of Chern number of all bands. Should summed to 0.
"""
hVec = np.array(hVec)
dimy, dimx, nlevels, _ = hVec.shape
cnlist = np.zeros(nlevels)
for iy in range(dimy-1):
for ix in range(dimx-1):
u12 = hVec[iy, ix + 1].conjugate().T @ hVec[iy, ix]
u23 = hVec[iy + 1, ix + 1].conjugate().T @ hVec[iy, ix + 1]
u34 = hVec[iy + 1, ix].conjugate().T @ hVec[iy + 1, ix + 1]
u41 = hVec[iy, ix].conjugate().T @ hVec[iy + 1, ix]
t12 = np.diag(u12.diagonal())
t23 = np.diag(u23.diagonal())
t34 = np.diag(u34.diagonal())
t41 = np.diag(u41.diagonal())
tplaquet = t41 @ t34 @ t23 @ t12
cnlist += np.angle(tplaquet.diagonal())
cnlist /= 2 * np.pi
cnlist = chop(cnlist)
return cnlist
def chop(array, tol=1e-7):
"""Realize Mathematica Chop[].
:param array: 1D array.
:param tol: tolerance to be chopped. default to 1e-7
:return: chopped array. (original array alse modified.)
"""
for i in range(len(array)):
a = array[i]
if np.abs(a-round(a)) < tol:
array[i] = round(a)
return array
x = np.array([[1.], [0.]]) # x hat
y = np.array([[0.], [1.]]) # y hat
a1_ = 2 * np.pi / np.sqrt(3) / l * (x - y / np.sqrt(3))
a2_ = 4 * np.pi / 3 / l * y
K = (a1_ / 2 + a2_ / 4) / np.cos(np.pi / 6)**2
b1 = 2 * np.pi / np.sqrt(3) / l * (x - y / np.sqrt(3))
b2 = 4 * np.pi / 3 / l * y
bz= Polygon([K, -K + b1 + b2, K - b1, -K, K - b1 - b2, -K + b1])
evecs_all_bz = np.zeros((len(wn.kys), len(wn.kxs), 4, 4), dtype=np.complex128)
for y, ky in enumerate(wn.kys):
for x, kx in enumerate(wn.kxs):
p = Point(kx, ky)
if not bz.contains(p):
continue
evecs_all_bz[y, x] = evecs[y, x]
print(Ω, calc_chern(evecs_all_bz))
| 0.38145 | 0.860134 |
# Iris Classification

Image from: https://www.codecademy.com/
***
## Setup
***
```
# Numerical arrays.
import numpy as np
# Data frames.
import pandas as pd
# Plotting.
import matplotlib.pyplot as plt
# Logistic regression.
import sklearn.linear_model as lm
# K nearest neaighbours.
import sklearn.neighbors as nei
# Helper functions.
import sklearn.model_selection as mod
# Fancier, statistical plots.
import seaborn as sns
# Standard plot size.
plt.rcParams['figure.figsize'] = (15, 10)
# Standard colour scheme.
plt.style.use('ggplot')
```
<br>
## The Iris Dataset
***
```
# Load the iris data set from a URL.
df = pd.read_csv("https://raw.githubusercontent.com/ianmcloughlin/datasets/main/iris.csv")
# Have a look at the data.
df
# Summary statistics.
df.describe()
```
<br>
### Visualise
***
```
# Scatter plots and kdes.
sns.pairplot(df, hue='species');
```
<br>
### Two Dimensions
***
```
# New figure.
fig, ax = plt.subplots()
# Scatter plot.
ax.plot(df['petal_width'], df['sepal_length'], '.')
# Set axis labels.
ax.set_xlabel('Petal width');
ax.set_ylabel('Sepal length');
# Seaborn is great for creating complex plots with one command.
sns.lmplot(x="petal_width", y="sepal_length", hue='species', data=df, fit_reg=False, height=10, aspect=1.5);
```
<br>
### Using pyplot
***
```
# Segregate the data.
setos = df[df['species'] == 'setosa']
versi = df[df['species'] == 'versicolor']
virgi = df[df['species'] == 'virginica']
# New plot.
fig, ax = plt.subplots()
# Scatter plots.
ax.scatter(setos['petal_width'], setos['sepal_length'], label='Setosa')
ax.scatter(versi['petal_width'], versi['sepal_length'], label='Versicolor')
ax.scatter(virgi['petal_width'], virgi['sepal_length'], label='Virginica')
# Show the legend.
ax.set_xlabel('Petal width')
ax.set_ylabel('Sepal length')
ax.legend();
# How the segregation works.
df['species'] == 'virginica'
df[df['species'] == 'virginica'].head()
```
<br>
### Using groupby()
***
```
# New plot.
fig, ax = plt.subplots()
# Using pandas groupby().
for species, data in df.groupby('species'):
ax.scatter(data['petal_width'], data['sepal_length'], label=species)
# Show the legend.
ax.set_xlabel('Petal width')
ax.set_ylabel('Sepal length')
ax.legend();
# Group by typically takes a categorical variable.
x = df.groupby('species')
x
# Pivot tables.
x.mean()
# Looping through groupby().
for i, j in x:
print()
print(f"i is: '{i}'")
print(f"j looks like:\n{j[:3]}")
print()
```
<br>
## Test and Train Split
***
```
# Split the data frame in two.
train, test = mod.train_test_split(df)
# Show some training data.
train.head()
# The indices of the train array.
train.index
# Show some testing data.
test.head()
test.index.size
```
<br>
### Two Dimensions: Test Train Split
***
```
# Segregate the training data.
setos = train[train['species'] == 'setosa']
versi = train[train['species'] == 'versicolor']
virgi = train[train['species'] == 'virginica']
# New plot.
fig, ax = plt.subplots()
# Scatter plots for training data.
ax.scatter(setos['petal_width'], setos['sepal_length'], marker='o', label='Setosa')
ax.scatter(versi['petal_width'], versi['sepal_length'], marker='o', label='Versicolor')
ax.scatter(virgi['petal_width'], virgi['sepal_length'], marker='o', label='Virginica')
# Scatter plot for testing data.
ax.scatter(test['petal_width'], test['sepal_length'], marker='x', label='Test data')
# Show the legend.
ax.set_xlabel('Petal width')
ax.set_ylabel('Sepal length')
ax.legend();
```
<br>
### Two Dimensions: Inputs and outputs
***
```
# Give the inputs and outputs convenient names.
inputs, outputs = train[['sepal_length', 'petal_width']], train['species']
# Peek at the inputs.
inputs.head()
# Peek at the outputs.
outputs.head()
```
<br>
## Two Dimensions: Logistic regression
https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html
***
```
# Create a new classifier.
lre = lm.LogisticRegression(random_state=0)
# Train the classifier on our data.
lre.fit(inputs, outputs)
# Ask the classifier to classify the test data.
predictions = lre.predict(test[['sepal_length', 'petal_width']])
predictions
# Eyeball the misclassifications.
predictions == test['species']
# What proportion were correct?
lre.score(test[['sepal_length', 'petal_width']], test['species'])
```
<br>
## Two Dimensions: Misclassified
***
```
# Append a column to the test data frame with the predictions.
test['predicted'] = predictions
test.head()
# Show the misclassified data.
misclass = test[test['predicted'] != test['species']]
misclass
# Eyeball the descriptive statistics for the species.
train.groupby('species').mean()
# New plot.
fig, ax = plt.subplots()
# Plot the training data
for species, data in df.groupby('species'):
ax.scatter(data['petal_width'], data['sepal_length'], label=species)
# Plot misclassified.
ax.scatter(misclass['petal_width'], misclass['sepal_length'], s=200, facecolor='none', edgecolor='r', label='Misclassified')
# Show the legend.
ax.set_xlabel('Petal width')
ax.set_ylabel('Sepal length')
ax.legend();
```
<br>
## Separating Setosa
***
From [Wikipedia](https://en.wikipedia.org/wiki/Logistic_regression):
$$\ell = \log_b \frac{p}{1-p} = \beta_0 + \beta_1 x_1 + \beta_2 x_2$$
***
```
# Another look at this plot.
sns.pairplot(df, hue='species');
# Give the inputs and outputs convenient names.
inputs = train[['sepal_length', 'petal_width']]
# Set 'versicolor' and 'virginica' to 'other'.
outputs = train['species'].apply(lambda x: x if x == 'setosa' else 'other')
# Eyeball outputs
outputs.unique()
# Create a new classifier.
lre = lm.LogisticRegression(random_state=0)
# Train the classifier on our data.
lre.fit(inputs, outputs)
actual = test['species'].apply(lambda x: x if x == 'setosa' else 'other')
# What proportion were correct?
lre.score(test[['sepal_length', 'petal_width']], actual)
```
<br>
## Using All Possible Inputs
https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html
***
```
# Load the iris data set from a URL.
df = pd.read_csv("https://raw.githubusercontent.com/ianmcloughlin/datasets/main/iris.csv")
# Split the data frame in two.
train, test = mod.train_test_split(df)
# Use all four possible inputs.
inputs, outputs = train[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']], train['species']
# Create a new classifier.
lre = lm.LogisticRegression(random_state=0)
# Train the classifier on our data.
lre.fit(inputs, outputs)
# Ask the classifier to classify the test data.
predictions = lre.predict(test[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']])
predictions
# Eyeball the misclassifications.
(predictions == test['species']).value_counts()
# What proportion were correct?
lre.score(test[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']], test['species'])
```
<br>
## $k$ Nearest Neighbours Classifier
https://scikit-learn.org/stable/modules/generated/sklearn.neighbors.KNeighborsClassifier.html
***
```
# Load the iris data set from a URL.
df = pd.read_csv("https://raw.githubusercontent.com/ianmcloughlin/datasets/main/iris.csv")
# Split the data frame in two.
train, test = mod.train_test_split(df)
# Use all four possible inputs.
inputs, outputs = train[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']], train['species']
# Classifier.
knn = nei.KNeighborsClassifier()
# Fit.
knn.fit(inputs, outputs)
# Test.
knn.score(test[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']], test['species'])
# Predict.
predictions = lre.predict(test[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']])
(predictions == test['species']).value_counts()
# The score is just the accuracy in this case.
(predictions == test['species']).value_counts(normalize=True)
```
<br>
## Cross validation
https://scikit-learn.org/stable/modules/cross_validation.html
***
```
knn = nei.KNeighborsClassifier()
scores = mod.cross_val_score(knn, df[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']], df['species'])
scores
print(f"Mean: {scores.mean()} \t Standard Deviation: {scores.std()}")
lre = lm.LogisticRegression(random_state=0)
scores = mod.cross_val_score(lre, df[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']], df['species'])
scores
print(f"Mean: {scores.mean()} \t Standard Deviation: {scores.std()}")
```
## End
|
github_jupyter
|
# Numerical arrays.
import numpy as np
# Data frames.
import pandas as pd
# Plotting.
import matplotlib.pyplot as plt
# Logistic regression.
import sklearn.linear_model as lm
# K nearest neaighbours.
import sklearn.neighbors as nei
# Helper functions.
import sklearn.model_selection as mod
# Fancier, statistical plots.
import seaborn as sns
# Standard plot size.
plt.rcParams['figure.figsize'] = (15, 10)
# Standard colour scheme.
plt.style.use('ggplot')
# Load the iris data set from a URL.
df = pd.read_csv("https://raw.githubusercontent.com/ianmcloughlin/datasets/main/iris.csv")
# Have a look at the data.
df
# Summary statistics.
df.describe()
# Scatter plots and kdes.
sns.pairplot(df, hue='species');
# New figure.
fig, ax = plt.subplots()
# Scatter plot.
ax.plot(df['petal_width'], df['sepal_length'], '.')
# Set axis labels.
ax.set_xlabel('Petal width');
ax.set_ylabel('Sepal length');
# Seaborn is great for creating complex plots with one command.
sns.lmplot(x="petal_width", y="sepal_length", hue='species', data=df, fit_reg=False, height=10, aspect=1.5);
# Segregate the data.
setos = df[df['species'] == 'setosa']
versi = df[df['species'] == 'versicolor']
virgi = df[df['species'] == 'virginica']
# New plot.
fig, ax = plt.subplots()
# Scatter plots.
ax.scatter(setos['petal_width'], setos['sepal_length'], label='Setosa')
ax.scatter(versi['petal_width'], versi['sepal_length'], label='Versicolor')
ax.scatter(virgi['petal_width'], virgi['sepal_length'], label='Virginica')
# Show the legend.
ax.set_xlabel('Petal width')
ax.set_ylabel('Sepal length')
ax.legend();
# How the segregation works.
df['species'] == 'virginica'
df[df['species'] == 'virginica'].head()
# New plot.
fig, ax = plt.subplots()
# Using pandas groupby().
for species, data in df.groupby('species'):
ax.scatter(data['petal_width'], data['sepal_length'], label=species)
# Show the legend.
ax.set_xlabel('Petal width')
ax.set_ylabel('Sepal length')
ax.legend();
# Group by typically takes a categorical variable.
x = df.groupby('species')
x
# Pivot tables.
x.mean()
# Looping through groupby().
for i, j in x:
print()
print(f"i is: '{i}'")
print(f"j looks like:\n{j[:3]}")
print()
# Split the data frame in two.
train, test = mod.train_test_split(df)
# Show some training data.
train.head()
# The indices of the train array.
train.index
# Show some testing data.
test.head()
test.index.size
# Segregate the training data.
setos = train[train['species'] == 'setosa']
versi = train[train['species'] == 'versicolor']
virgi = train[train['species'] == 'virginica']
# New plot.
fig, ax = plt.subplots()
# Scatter plots for training data.
ax.scatter(setos['petal_width'], setos['sepal_length'], marker='o', label='Setosa')
ax.scatter(versi['petal_width'], versi['sepal_length'], marker='o', label='Versicolor')
ax.scatter(virgi['petal_width'], virgi['sepal_length'], marker='o', label='Virginica')
# Scatter plot for testing data.
ax.scatter(test['petal_width'], test['sepal_length'], marker='x', label='Test data')
# Show the legend.
ax.set_xlabel('Petal width')
ax.set_ylabel('Sepal length')
ax.legend();
# Give the inputs and outputs convenient names.
inputs, outputs = train[['sepal_length', 'petal_width']], train['species']
# Peek at the inputs.
inputs.head()
# Peek at the outputs.
outputs.head()
# Create a new classifier.
lre = lm.LogisticRegression(random_state=0)
# Train the classifier on our data.
lre.fit(inputs, outputs)
# Ask the classifier to classify the test data.
predictions = lre.predict(test[['sepal_length', 'petal_width']])
predictions
# Eyeball the misclassifications.
predictions == test['species']
# What proportion were correct?
lre.score(test[['sepal_length', 'petal_width']], test['species'])
# Append a column to the test data frame with the predictions.
test['predicted'] = predictions
test.head()
# Show the misclassified data.
misclass = test[test['predicted'] != test['species']]
misclass
# Eyeball the descriptive statistics for the species.
train.groupby('species').mean()
# New plot.
fig, ax = plt.subplots()
# Plot the training data
for species, data in df.groupby('species'):
ax.scatter(data['petal_width'], data['sepal_length'], label=species)
# Plot misclassified.
ax.scatter(misclass['petal_width'], misclass['sepal_length'], s=200, facecolor='none', edgecolor='r', label='Misclassified')
# Show the legend.
ax.set_xlabel('Petal width')
ax.set_ylabel('Sepal length')
ax.legend();
# Another look at this plot.
sns.pairplot(df, hue='species');
# Give the inputs and outputs convenient names.
inputs = train[['sepal_length', 'petal_width']]
# Set 'versicolor' and 'virginica' to 'other'.
outputs = train['species'].apply(lambda x: x if x == 'setosa' else 'other')
# Eyeball outputs
outputs.unique()
# Create a new classifier.
lre = lm.LogisticRegression(random_state=0)
# Train the classifier on our data.
lre.fit(inputs, outputs)
actual = test['species'].apply(lambda x: x if x == 'setosa' else 'other')
# What proportion were correct?
lre.score(test[['sepal_length', 'petal_width']], actual)
# Load the iris data set from a URL.
df = pd.read_csv("https://raw.githubusercontent.com/ianmcloughlin/datasets/main/iris.csv")
# Split the data frame in two.
train, test = mod.train_test_split(df)
# Use all four possible inputs.
inputs, outputs = train[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']], train['species']
# Create a new classifier.
lre = lm.LogisticRegression(random_state=0)
# Train the classifier on our data.
lre.fit(inputs, outputs)
# Ask the classifier to classify the test data.
predictions = lre.predict(test[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']])
predictions
# Eyeball the misclassifications.
(predictions == test['species']).value_counts()
# What proportion were correct?
lre.score(test[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']], test['species'])
# Load the iris data set from a URL.
df = pd.read_csv("https://raw.githubusercontent.com/ianmcloughlin/datasets/main/iris.csv")
# Split the data frame in two.
train, test = mod.train_test_split(df)
# Use all four possible inputs.
inputs, outputs = train[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']], train['species']
# Classifier.
knn = nei.KNeighborsClassifier()
# Fit.
knn.fit(inputs, outputs)
# Test.
knn.score(test[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']], test['species'])
# Predict.
predictions = lre.predict(test[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']])
(predictions == test['species']).value_counts()
# The score is just the accuracy in this case.
(predictions == test['species']).value_counts(normalize=True)
knn = nei.KNeighborsClassifier()
scores = mod.cross_val_score(knn, df[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']], df['species'])
scores
print(f"Mean: {scores.mean()} \t Standard Deviation: {scores.std()}")
lre = lm.LogisticRegression(random_state=0)
scores = mod.cross_val_score(lre, df[['sepal_length', 'sepal_width', 'petal_length', 'petal_width']], df['species'])
scores
print(f"Mean: {scores.mean()} \t Standard Deviation: {scores.std()}")
| 0.870184 | 0.927626 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.