code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
```
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
from sklearn.linear_model import LogisticRegression
from sklearn import metrics
from sklearn.metrics import classification_report, confusion_matrix, f1_score
from sklearn.metrics import make_scorer, f1_score, accuracy_score, recall_score, precision_score, classification_report, precision_recall_fscore_support
import itertools
# file used to write preserve the results of the classfier
# confusion matrix and precision recall fscore matrix
def plot_confusion_matrix(cm,
target_names,
title='Confusion matrix',
cmap=None,
normalize=True):
"""
given a sklearn confusion matrix (cm), make a nice plot
Arguments
---------
cm: confusion matrix from sklearn.metrics.confusion_matrix
target_names: given classification classes such as [0, 1, 2]
the class names, for example: ['high', 'medium', 'low']
title: the text to display at the top of the matrix
cmap: the gradient of the values displayed from matplotlib.pyplot.cm
see http://matplotlib.org/examples/color/colormaps_reference.html
plt.get_cmap('jet') or plt.cm.Blues
normalize: If False, plot the raw numbers
If True, plot the proportions
Usage
-----
plot_confusion_matrix(cm = cm, # confusion matrix created by
# sklearn.metrics.confusion_matrix
normalize = True, # show proportions
target_names = y_labels_vals, # list of names of the classes
title = best_estimator_name) # title of graph
Citiation
---------
http://scikit-learn.org/stable/auto_examples/model_selection/plot_confusion_matrix.html
"""
accuracy = np.trace(cm) / float(np.sum(cm))
misclass = 1 - accuracy
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
if cmap is None:
cmap = plt.get_cmap('Blues')
plt.figure(figsize=(8, 6))
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
if target_names is not None:
tick_marks = np.arange(len(target_names))
plt.xticks(tick_marks, target_names, rotation=45)
plt.yticks(tick_marks, target_names)
thresh = cm.max() / 1.5 if normalize else cm.max() / 2
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
if normalize:
plt.text(j, i, "{:0.4f}".format(cm[i, j]),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
else:
plt.text(j, i, "{:,}".format(cm[i, j]),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.ylabel('True label')
plt.xlabel('Predicted label\naccuracy={:0.4f}; misclass={:0.4f}'.format(accuracy, misclass))
plt.tight_layout()
return plt
##saving the classification report
def pandas_classification_report(y_true, y_pred):
metrics_summary = precision_recall_fscore_support(
y_true=y_true,
y_pred=y_pred)
cm = confusion_matrix(y_true, y_pred)
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
avg = list(precision_recall_fscore_support(
y_true=y_true,
y_pred=y_pred,
average='macro'))
avg.append(accuracy_score(y_true, y_pred, normalize=True))
metrics_sum_index = ['precision', 'recall', 'f1-score', 'support','accuracy']
list_all=list(metrics_summary)
list_all.append(cm.diagonal())
class_report_df = pd.DataFrame(
list_all,
index=metrics_sum_index)
support = class_report_df.loc['support']
total = support.sum()
avg[-2] = total
class_report_df['avg / total'] = avg
return class_report_df.T
from commen_preprocess import *
eng_train_dataset = pd.read_csv('../Data/english_dataset/english_dataset.tsv', sep='\t')
# #hindi_train_dataset = pd.read_csv('../Data/hindi_dataset/hindi_dataset.tsv', sep='\t',header=None)
# german_train_dataset = pd.read_csv('../Data/german_dataset/german_dataset_added_features.tsv', sep=',')
# eng_train_dataset=eng_train_dataset.drop(['Unnamed: 0'], axis=1)
# german_train_dataset=german_train_dataset.drop(['Unnamed: 0'], axis=1)
eng_train_dataset = eng_train_dataset.loc[eng_train_dataset['task_1'] == 'HOF']
eng_train_dataset.head()
l=eng_train_dataset['task_3'].value_counts()
print(l)
import numpy as np
from tqdm import tqdm
import pickle
####loading laser embeddings for english dataset
def load_laser_embeddings():
dim = 1024
engX_commen = np.fromfile("../Data/english_dataset/embeddings_eng_task23_commen.raw", dtype=np.float32, count=-1)
engX_lib = np.fromfile("../Data/english_dataset/embeddings_eng_task23_lib.raw", dtype=np.float32, count=-1)
engX_commen.resize(engX_commen.shape[0] // dim, dim)
engX_lib.resize(engX_lib.shape[0] // dim, dim)
return engX_commen,engX_lib
def load_bert_embeddings():
file = open('../Data/english_dataset/no_preprocess_bert_embed_task23.pkl', 'rb')
embeds = pickle.load(file)
return np.array(embeds)
def merge_feature(*args):
feat_all=[]
print(args[0].shape)
for i in tqdm(range(args[0].shape[0])):
feat=[]
for arg in args:
feat+=list(arg[i])
feat_all.append(feat)
return feat_all
convert_label={
'TIN':0,
'UNT':1,
}
convert_reverse_label={
0:'TIN',
1:'UNT',
}
labels=eng_train_dataset['task_3'].values
engX_commen,engX_lib=load_laser_embeddings()
bert_embeds =load_bert_embeddings()
feat_all=merge_feature(engX_commen,engX_lib,bert_embeds)
len(feat_all[0])
from sklearn.utils.multiclass import type_of_target
Classifier_Train_X=np.array(feat_all)
labels_int=[]
for i in range(len(labels)):
labels_int.append(convert_label[labels[i]])
Classifier_Train_Y=np.array(labels_int,dtype='float64')
print(type_of_target(Classifier_Train_Y))
Classifier_Train_Y
from sklearn.metrics import accuracy_score
import joblib
from sklearn.model_selection import StratifiedKFold as skf
###all classifier
from catboost import CatBoostClassifier
from xgboost.sklearn import XGBClassifier
from sklearn.svm import SVC, LinearSVC
from sklearn.linear_model import LogisticRegression
from sklearn import tree
from sklearn import neighbors
from sklearn import ensemble
from sklearn import neural_network
from sklearn import linear_model
import lightgbm as lgbm
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import LogisticRegression
from lightgbm import LGBMClassifier
from nltk.classify.scikitlearn import SklearnClassifier
def train_model_no_ext(Classifier_Train_X,Classifier_Train_Y,model_type,save_model=False):
kf = skf(n_splits=10,shuffle=True)
y_total_preds=[]
y_total=[]
count=0
img_name = 'cm.png'
report_name = 'report.csv'
scale=list(Classifier_Train_Y).count(0)/list(Classifier_Train_Y).count(1)
print(scale)
if(save_model==True):
Classifier=get_model(scale,m_type=model_type)
Classifier.fit(Classifier_Train_X,Classifier_Train_Y)
filename = model_type+'_eng_task_2.joblib.pkl'
joblib.dump(Classifier, filename, compress=9)
# filename1 = model_name+'select_features_eng_task1.joblib.pkl'
# joblib.dump(model_featureSelection, filename1, compress=9)
else:
for train_index, test_index in kf.split(Classifier_Train_X,Classifier_Train_Y):
X_train, X_test = Classifier_Train_X[train_index], Classifier_Train_X[test_index]
y_train, y_test = Classifier_Train_Y[train_index], Classifier_Train_Y[test_index]
classifier=get_model(scale,m_type=model_type)
print(type(y_train))
classifier.fit(X_train,y_train)
y_preds = classifier.predict(X_test)
for ele in y_test:
y_total.append(ele)
for ele in y_preds:
y_total_preds.append(ele)
y_pred_train = classifier.predict(X_train)
print(y_pred_train)
print(y_train)
count=count+1
print('accuracy_train:',accuracy_score(y_train, y_pred_train),'accuracy_test:',accuracy_score(y_test, y_preds))
print('TRAINING:')
print(classification_report( y_train, y_pred_train ))
print("TESTING:")
print(classification_report( y_test, y_preds ))
report = classification_report( y_total, y_total_preds )
cm=confusion_matrix(y_total, y_total_preds)
plt=plot_confusion_matrix(cm,normalize= True,target_names = ['TIN','UNT'],title = "Confusion Matrix")
plt.savefig('eng_task3'+model_type+'_'+img_name)
print(classifier)
print(report)
print(accuracy_score(y_total, y_total_preds))
df_result=pandas_classification_report(y_total,y_total_preds)
df_result.to_csv('eng_task3'+model_type+'_'+report_name, sep=',')
def get_model(scale,m_type=None):
if not m_type:
print("ERROR: Please specify a model type!")
return None
if m_type == 'decision_tree_classifier':
logreg = tree.DecisionTreeClassifier(max_features=1000,max_depth=3,class_weight='balanced')
elif m_type == 'gaussian':
logreg = GaussianNB()
elif m_type == 'logistic_regression':
logreg = LogisticRegression(n_jobs=10, random_state=42,class_weight='balanced',solver='liblinear')
elif m_type == 'MLPClassifier':
# logreg = neural_network.MLPClassifier((500))
logreg = neural_network.MLPClassifier((100),random_state=42,early_stopping=True)
elif m_type == 'RandomForestClassifier':
logreg = ensemble.RandomForestClassifier(n_estimators=100, class_weight='balanced', n_jobs=12, max_depth=7)
elif m_type == 'SVC':
#logreg = LinearSVC(dual=False,max_iter=200)
logreg = SVC(kernel='linear',random_state=1526)
elif m_type == 'Catboost':
logreg = CatBoostClassifier(iterations=100,learning_rate=0.2,
l2_leaf_reg=500,depth=10,use_best_model=False, random_state=42,loss_function='MultiClass')
# logreg = CatBoostClassifier(scale_pos_weight=0.8, random_seed=42,);
elif m_type == 'XGB_classifier':
# logreg=XGBClassifier(silent=False,eta=0.1,objective='binary:logistic',max_depth=5,min_child_weight=0,gamma=0.2,subsample=0.8, colsample_bytree = 0.8,scale_pos_weight=1,n_estimators=500,reg_lambda=3,nthread=12)
logreg=XGBClassifier(silent=False,objective='multi:softmax',num_class=3,
reg_lambda=3,nthread=12, random_state=42)
elif m_type == 'light_gbm':
logreg = LGBMClassifier(objective='multiclass',max_depth=3,learning_rate=0.2,num_leaves=20,scale_pos_weight=scale,
boosting_type='gbdt', metric='multi_logloss',random_state=5,reg_lambda=20,silent=False)
else:
print("give correct model")
print(logreg)
return logreg
models_name=['decision_tree_classifier','gaussian','logistic_regression','MLPClassifier','RandomForestClassifier',
'SVC','light_gbm']
for model in models_name:
train_model_no_ext(Classifier_Train_X,Classifier_Train_Y,model)
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
from sklearn.linear_model import LogisticRegression
from sklearn import metrics
from sklearn.metrics import classification_report, confusion_matrix, f1_score
from sklearn.metrics import make_scorer, f1_score, accuracy_score, recall_score, precision_score, classification_report, precision_recall_fscore_support
import itertools
# file used to write preserve the results of the classfier
# confusion matrix and precision recall fscore matrix
def plot_confusion_matrix(cm,
target_names,
title='Confusion matrix',
cmap=None,
normalize=True):
"""
given a sklearn confusion matrix (cm), make a nice plot
Arguments
---------
cm: confusion matrix from sklearn.metrics.confusion_matrix
target_names: given classification classes such as [0, 1, 2]
the class names, for example: ['high', 'medium', 'low']
title: the text to display at the top of the matrix
cmap: the gradient of the values displayed from matplotlib.pyplot.cm
see http://matplotlib.org/examples/color/colormaps_reference.html
plt.get_cmap('jet') or plt.cm.Blues
normalize: If False, plot the raw numbers
If True, plot the proportions
Usage
-----
plot_confusion_matrix(cm = cm, # confusion matrix created by
# sklearn.metrics.confusion_matrix
normalize = True, # show proportions
target_names = y_labels_vals, # list of names of the classes
title = best_estimator_name) # title of graph
Citiation
---------
http://scikit-learn.org/stable/auto_examples/model_selection/plot_confusion_matrix.html
"""
accuracy = np.trace(cm) / float(np.sum(cm))
misclass = 1 - accuracy
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
if cmap is None:
cmap = plt.get_cmap('Blues')
plt.figure(figsize=(8, 6))
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
if target_names is not None:
tick_marks = np.arange(len(target_names))
plt.xticks(tick_marks, target_names, rotation=45)
plt.yticks(tick_marks, target_names)
thresh = cm.max() / 1.5 if normalize else cm.max() / 2
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
if normalize:
plt.text(j, i, "{:0.4f}".format(cm[i, j]),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
else:
plt.text(j, i, "{:,}".format(cm[i, j]),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.ylabel('True label')
plt.xlabel('Predicted label\naccuracy={:0.4f}; misclass={:0.4f}'.format(accuracy, misclass))
plt.tight_layout()
return plt
##saving the classification report
def pandas_classification_report(y_true, y_pred):
metrics_summary = precision_recall_fscore_support(
y_true=y_true,
y_pred=y_pred)
cm = confusion_matrix(y_true, y_pred)
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
avg = list(precision_recall_fscore_support(
y_true=y_true,
y_pred=y_pred,
average='macro'))
avg.append(accuracy_score(y_true, y_pred, normalize=True))
metrics_sum_index = ['precision', 'recall', 'f1-score', 'support','accuracy']
list_all=list(metrics_summary)
list_all.append(cm.diagonal())
class_report_df = pd.DataFrame(
list_all,
index=metrics_sum_index)
support = class_report_df.loc['support']
total = support.sum()
avg[-2] = total
class_report_df['avg / total'] = avg
return class_report_df.T
from commen_preprocess import *
eng_train_dataset = pd.read_csv('../Data/english_dataset/english_dataset.tsv', sep='\t')
# #hindi_train_dataset = pd.read_csv('../Data/hindi_dataset/hindi_dataset.tsv', sep='\t',header=None)
# german_train_dataset = pd.read_csv('../Data/german_dataset/german_dataset_added_features.tsv', sep=',')
# eng_train_dataset=eng_train_dataset.drop(['Unnamed: 0'], axis=1)
# german_train_dataset=german_train_dataset.drop(['Unnamed: 0'], axis=1)
eng_train_dataset = eng_train_dataset.loc[eng_train_dataset['task_1'] == 'HOF']
eng_train_dataset.head()
l=eng_train_dataset['task_3'].value_counts()
print(l)
import numpy as np
from tqdm import tqdm
import pickle
####loading laser embeddings for english dataset
def load_laser_embeddings():
dim = 1024
engX_commen = np.fromfile("../Data/english_dataset/embeddings_eng_task23_commen.raw", dtype=np.float32, count=-1)
engX_lib = np.fromfile("../Data/english_dataset/embeddings_eng_task23_lib.raw", dtype=np.float32, count=-1)
engX_commen.resize(engX_commen.shape[0] // dim, dim)
engX_lib.resize(engX_lib.shape[0] // dim, dim)
return engX_commen,engX_lib
def load_bert_embeddings():
file = open('../Data/english_dataset/no_preprocess_bert_embed_task23.pkl', 'rb')
embeds = pickle.load(file)
return np.array(embeds)
def merge_feature(*args):
feat_all=[]
print(args[0].shape)
for i in tqdm(range(args[0].shape[0])):
feat=[]
for arg in args:
feat+=list(arg[i])
feat_all.append(feat)
return feat_all
convert_label={
'TIN':0,
'UNT':1,
}
convert_reverse_label={
0:'TIN',
1:'UNT',
}
labels=eng_train_dataset['task_3'].values
engX_commen,engX_lib=load_laser_embeddings()
bert_embeds =load_bert_embeddings()
feat_all=merge_feature(engX_commen,engX_lib,bert_embeds)
len(feat_all[0])
from sklearn.utils.multiclass import type_of_target
Classifier_Train_X=np.array(feat_all)
labels_int=[]
for i in range(len(labels)):
labels_int.append(convert_label[labels[i]])
Classifier_Train_Y=np.array(labels_int,dtype='float64')
print(type_of_target(Classifier_Train_Y))
Classifier_Train_Y
from sklearn.metrics import accuracy_score
import joblib
from sklearn.model_selection import StratifiedKFold as skf
###all classifier
from catboost import CatBoostClassifier
from xgboost.sklearn import XGBClassifier
from sklearn.svm import SVC, LinearSVC
from sklearn.linear_model import LogisticRegression
from sklearn import tree
from sklearn import neighbors
from sklearn import ensemble
from sklearn import neural_network
from sklearn import linear_model
import lightgbm as lgbm
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import LogisticRegression
from lightgbm import LGBMClassifier
from nltk.classify.scikitlearn import SklearnClassifier
def train_model_no_ext(Classifier_Train_X,Classifier_Train_Y,model_type,save_model=False):
kf = skf(n_splits=10,shuffle=True)
y_total_preds=[]
y_total=[]
count=0
img_name = 'cm.png'
report_name = 'report.csv'
scale=list(Classifier_Train_Y).count(0)/list(Classifier_Train_Y).count(1)
print(scale)
if(save_model==True):
Classifier=get_model(scale,m_type=model_type)
Classifier.fit(Classifier_Train_X,Classifier_Train_Y)
filename = model_type+'_eng_task_2.joblib.pkl'
joblib.dump(Classifier, filename, compress=9)
# filename1 = model_name+'select_features_eng_task1.joblib.pkl'
# joblib.dump(model_featureSelection, filename1, compress=9)
else:
for train_index, test_index in kf.split(Classifier_Train_X,Classifier_Train_Y):
X_train, X_test = Classifier_Train_X[train_index], Classifier_Train_X[test_index]
y_train, y_test = Classifier_Train_Y[train_index], Classifier_Train_Y[test_index]
classifier=get_model(scale,m_type=model_type)
print(type(y_train))
classifier.fit(X_train,y_train)
y_preds = classifier.predict(X_test)
for ele in y_test:
y_total.append(ele)
for ele in y_preds:
y_total_preds.append(ele)
y_pred_train = classifier.predict(X_train)
print(y_pred_train)
print(y_train)
count=count+1
print('accuracy_train:',accuracy_score(y_train, y_pred_train),'accuracy_test:',accuracy_score(y_test, y_preds))
print('TRAINING:')
print(classification_report( y_train, y_pred_train ))
print("TESTING:")
print(classification_report( y_test, y_preds ))
report = classification_report( y_total, y_total_preds )
cm=confusion_matrix(y_total, y_total_preds)
plt=plot_confusion_matrix(cm,normalize= True,target_names = ['TIN','UNT'],title = "Confusion Matrix")
plt.savefig('eng_task3'+model_type+'_'+img_name)
print(classifier)
print(report)
print(accuracy_score(y_total, y_total_preds))
df_result=pandas_classification_report(y_total,y_total_preds)
df_result.to_csv('eng_task3'+model_type+'_'+report_name, sep=',')
def get_model(scale,m_type=None):
if not m_type:
print("ERROR: Please specify a model type!")
return None
if m_type == 'decision_tree_classifier':
logreg = tree.DecisionTreeClassifier(max_features=1000,max_depth=3,class_weight='balanced')
elif m_type == 'gaussian':
logreg = GaussianNB()
elif m_type == 'logistic_regression':
logreg = LogisticRegression(n_jobs=10, random_state=42,class_weight='balanced',solver='liblinear')
elif m_type == 'MLPClassifier':
# logreg = neural_network.MLPClassifier((500))
logreg = neural_network.MLPClassifier((100),random_state=42,early_stopping=True)
elif m_type == 'RandomForestClassifier':
logreg = ensemble.RandomForestClassifier(n_estimators=100, class_weight='balanced', n_jobs=12, max_depth=7)
elif m_type == 'SVC':
#logreg = LinearSVC(dual=False,max_iter=200)
logreg = SVC(kernel='linear',random_state=1526)
elif m_type == 'Catboost':
logreg = CatBoostClassifier(iterations=100,learning_rate=0.2,
l2_leaf_reg=500,depth=10,use_best_model=False, random_state=42,loss_function='MultiClass')
# logreg = CatBoostClassifier(scale_pos_weight=0.8, random_seed=42,);
elif m_type == 'XGB_classifier':
# logreg=XGBClassifier(silent=False,eta=0.1,objective='binary:logistic',max_depth=5,min_child_weight=0,gamma=0.2,subsample=0.8, colsample_bytree = 0.8,scale_pos_weight=1,n_estimators=500,reg_lambda=3,nthread=12)
logreg=XGBClassifier(silent=False,objective='multi:softmax',num_class=3,
reg_lambda=3,nthread=12, random_state=42)
elif m_type == 'light_gbm':
logreg = LGBMClassifier(objective='multiclass',max_depth=3,learning_rate=0.2,num_leaves=20,scale_pos_weight=scale,
boosting_type='gbdt', metric='multi_logloss',random_state=5,reg_lambda=20,silent=False)
else:
print("give correct model")
print(logreg)
return logreg
models_name=['decision_tree_classifier','gaussian','logistic_regression','MLPClassifier','RandomForestClassifier',
'SVC','light_gbm']
for model in models_name:
train_model_no_ext(Classifier_Train_X,Classifier_Train_Y,model)
| 0.639849 | 0.583144 |
# Lost Rhino
In this notebook, we plot **Figures 1(a) and 1(b)**. To do so, we need to get the ratings for the particular beer called *Lost Rhino* from the brewery *Lost Rhino Ice Breaker*.
**Requirements**:
- You need to run notebook `4-zscores` to get the file `z_score_params_matched_ratings` in `data/tmp` and the files `ratings_ba.txt.gz` and `ratings_rb.txt.gz` in `data/matched`. In other words, you need to **run the first 5 cells of `4-zscores`**.
**Benchmark time**: This notebook has been run on a Dell Latitude (ElementaryOS 0.4.1 Loki, i7-7600U, 16GB RAM).
```
import os
os.chdir('..')
# Helpers functions
from python.helpers import parse
# Libraries for preparing data
import json
import gzip
import numpy as np
import pandas as pd
from datetime import datetime
# Libraries for plotting
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib.ticker import FuncFormatter
import matplotlib
# Folders
data_folder = '../data/'
fig_folder = '../figures/'
# For the Python notebook
%matplotlib inline
%reload_ext autoreload
%autoreload 2
# General info for plotting
colors = {'ba': (232/255,164/255,29/255),
'rb': (0/255,152/255,205/255)}
labels = {'ba': 'BeerAdvocate', 'rb': 'RateBeer'}
# Check that folders exist
if not os.path.exists(data_folder + 'tmp'):
os.makedirs(data_folder + 'tmp')
if not os.path.exists(data_folder + 'prepared'):
os.makedirs(data_folder + 'prepared')
if not os.path.exists(fig_folder):
os.makedirs(fig_folder)
```
# Prepare the data
We simply need to get all the ratings of the beers *Lost Rhino* in order.
```
%%time
with open('../data/tmp/z_score_params_matched_ratings.json') as file:
z_score_params = json.load(file)
lost_rhino = {'ba': [], 'rb': []}
lost_rhino_dates = {'ba': [], 'rb': []}
id_ = {'ba': 78599, 'rb': 166120}
# Go through RB and BA
for key in ['ba', 'rb']:
print('Parsing {} reviews.'.format(key.upper()))
# Get the iterator with the ratings fo the matched beers
gen = parse(data_folder + 'matched/ratings_{}.txt.gz'.format(key))
# Go through the iterator
for item in gen:
if int(item['beer_id']) == id_[key]:
# Get the year
date = int(item['date'])
year = str(datetime.fromtimestamp(date).year)
# Get the rating
rat = float(item['rating'])
# Compute its zscore based on the year
zs = (rat-z_score_params[key][year]['mean'])/z_score_params[key][year]['std']
# Add date and zscore
lost_rhino_dates[key].append(date)
lost_rhino[key].append(zs)
for key in lost_rhino.keys():
# Get the sorted dates from smallest to biggest
idx = np.argsort(lost_rhino_dates[key])
# Sort the zscores for lost_rhino
lost_rhino_dates[key] = list(np.array(lost_rhino_dates[key])[idx])
lost_rhino[key] = list(np.array(lost_rhino[key])[idx])
with open(data_folder + 'prepared/lost_rhino.json', 'w') as outfile:
json.dump(lost_rhino, outfile)
```
## Plot the ratings of the *Lost Rhino*
The first cell plots each ratings and the second cell plots the running mean.
```
with open(data_folder + 'prepared/lost_rhino.json', 'r') as infile:
ratings = json.load(infile)
plt.figure(figsize=(5, 3.5), frameon=False)
sns.set_context("paper")
sns.set(font_scale = 1.3)
sns.set_style("white", {
"font.family": "sans-serif",
"font.serif": ['Helvetica'],
"font.scale": 2
})
sns.set_style("ticks", {"xtick.major.size": 4,
"ytick.major.size": 4})
ax = plt.subplot(111)
ax.spines['right'].set_visible(False)
ax.spines['top'].set_visible(False)
plt.plot([-10, 55], [0, 0], 'grey', linewidth=0.5)
for key in ['ba', 'rb']:
rats = ratings[key]
rmean = np.cumsum(rats)/np.array(range(1, len(rats)+1))
ax.plot(list(range(1, len(rmean)+1)), rats, color=colors[key], label=labels[key], linewidth=2)
plt.xlim([-1, 55])
plt.ylim([-4, 2.5])
plt.yticks(list(range(-4, 3)), list(range(-4, 3)))
plt.ylabel('Rating (standardized)')
plt.xlabel('Rating index')
leg = plt.legend()
leg.get_frame().set_linewidth(0.0)
plt.savefig(fig_folder + 'timeseries_zscore_example.pdf', bbox_inches='tight')
with open(data_folder + 'prepared/lost_rhino.json', 'r') as infile:
ratings = json.load(infile)
plt.figure(figsize=(5, 3.75), frameon=False)
sns.set_context("paper")
sns.set(font_scale = 1.28)
sns.set_style("white", {
"font.family": "sans-serif",
"font.serif": ['Helvetica'],
"font.scale": 2
})
sns.set_style("ticks", {"xtick.major.size": 4,
"ytick.major.size": 4})
ax = plt.subplot(111)
ax.spines['right'].set_visible(False)
ax.spines['top'].set_visible(False)
plt.plot([-10, 55], [0, 0], 'grey', linewidth=0.5)
for key in ['ba', 'rb']:
rats = ratings[key]
rmean = np.cumsum(rats)/np.array(range(1, len(rats)+1))
ax.plot(list(range(1, len(rmean)+1)), rmean, color=colors[key], label=labels[key], linewidth=2)
plt.ylabel('Cum. avg rating (standardized)')
plt.xlabel('Rating index')
plt.xlim([-1, 55])
plt.ylim([-4, 2.5])
plt.yticks(list(range(-4, 3)), list(range(-4, 3)))
leg = plt.legend(loc=4)
leg.get_frame().set_linewidth(0.0)
plt.tight_layout()
plt.savefig(fig_folder + 'timeseries_avg_zscore_example.pdf', bbox_inches='tight')
```
|
github_jupyter
|
import os
os.chdir('..')
# Helpers functions
from python.helpers import parse
# Libraries for preparing data
import json
import gzip
import numpy as np
import pandas as pd
from datetime import datetime
# Libraries for plotting
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib.ticker import FuncFormatter
import matplotlib
# Folders
data_folder = '../data/'
fig_folder = '../figures/'
# For the Python notebook
%matplotlib inline
%reload_ext autoreload
%autoreload 2
# General info for plotting
colors = {'ba': (232/255,164/255,29/255),
'rb': (0/255,152/255,205/255)}
labels = {'ba': 'BeerAdvocate', 'rb': 'RateBeer'}
# Check that folders exist
if not os.path.exists(data_folder + 'tmp'):
os.makedirs(data_folder + 'tmp')
if not os.path.exists(data_folder + 'prepared'):
os.makedirs(data_folder + 'prepared')
if not os.path.exists(fig_folder):
os.makedirs(fig_folder)
%%time
with open('../data/tmp/z_score_params_matched_ratings.json') as file:
z_score_params = json.load(file)
lost_rhino = {'ba': [], 'rb': []}
lost_rhino_dates = {'ba': [], 'rb': []}
id_ = {'ba': 78599, 'rb': 166120}
# Go through RB and BA
for key in ['ba', 'rb']:
print('Parsing {} reviews.'.format(key.upper()))
# Get the iterator with the ratings fo the matched beers
gen = parse(data_folder + 'matched/ratings_{}.txt.gz'.format(key))
# Go through the iterator
for item in gen:
if int(item['beer_id']) == id_[key]:
# Get the year
date = int(item['date'])
year = str(datetime.fromtimestamp(date).year)
# Get the rating
rat = float(item['rating'])
# Compute its zscore based on the year
zs = (rat-z_score_params[key][year]['mean'])/z_score_params[key][year]['std']
# Add date and zscore
lost_rhino_dates[key].append(date)
lost_rhino[key].append(zs)
for key in lost_rhino.keys():
# Get the sorted dates from smallest to biggest
idx = np.argsort(lost_rhino_dates[key])
# Sort the zscores for lost_rhino
lost_rhino_dates[key] = list(np.array(lost_rhino_dates[key])[idx])
lost_rhino[key] = list(np.array(lost_rhino[key])[idx])
with open(data_folder + 'prepared/lost_rhino.json', 'w') as outfile:
json.dump(lost_rhino, outfile)
with open(data_folder + 'prepared/lost_rhino.json', 'r') as infile:
ratings = json.load(infile)
plt.figure(figsize=(5, 3.5), frameon=False)
sns.set_context("paper")
sns.set(font_scale = 1.3)
sns.set_style("white", {
"font.family": "sans-serif",
"font.serif": ['Helvetica'],
"font.scale": 2
})
sns.set_style("ticks", {"xtick.major.size": 4,
"ytick.major.size": 4})
ax = plt.subplot(111)
ax.spines['right'].set_visible(False)
ax.spines['top'].set_visible(False)
plt.plot([-10, 55], [0, 0], 'grey', linewidth=0.5)
for key in ['ba', 'rb']:
rats = ratings[key]
rmean = np.cumsum(rats)/np.array(range(1, len(rats)+1))
ax.plot(list(range(1, len(rmean)+1)), rats, color=colors[key], label=labels[key], linewidth=2)
plt.xlim([-1, 55])
plt.ylim([-4, 2.5])
plt.yticks(list(range(-4, 3)), list(range(-4, 3)))
plt.ylabel('Rating (standardized)')
plt.xlabel('Rating index')
leg = plt.legend()
leg.get_frame().set_linewidth(0.0)
plt.savefig(fig_folder + 'timeseries_zscore_example.pdf', bbox_inches='tight')
with open(data_folder + 'prepared/lost_rhino.json', 'r') as infile:
ratings = json.load(infile)
plt.figure(figsize=(5, 3.75), frameon=False)
sns.set_context("paper")
sns.set(font_scale = 1.28)
sns.set_style("white", {
"font.family": "sans-serif",
"font.serif": ['Helvetica'],
"font.scale": 2
})
sns.set_style("ticks", {"xtick.major.size": 4,
"ytick.major.size": 4})
ax = plt.subplot(111)
ax.spines['right'].set_visible(False)
ax.spines['top'].set_visible(False)
plt.plot([-10, 55], [0, 0], 'grey', linewidth=0.5)
for key in ['ba', 'rb']:
rats = ratings[key]
rmean = np.cumsum(rats)/np.array(range(1, len(rats)+1))
ax.plot(list(range(1, len(rmean)+1)), rmean, color=colors[key], label=labels[key], linewidth=2)
plt.ylabel('Cum. avg rating (standardized)')
plt.xlabel('Rating index')
plt.xlim([-1, 55])
plt.ylim([-4, 2.5])
plt.yticks(list(range(-4, 3)), list(range(-4, 3)))
leg = plt.legend(loc=4)
leg.get_frame().set_linewidth(0.0)
plt.tight_layout()
plt.savefig(fig_folder + 'timeseries_avg_zscore_example.pdf', bbox_inches='tight')
| 0.457379 | 0.827375 |
# Interactive data visualizations
Jupyter Notebook has support for many kinds of interactive outputs, including
the ipywidgets ecosystem as well as many interactive visualization libraries.
These are supported in Jupyter Book, with the right configuration.
This page has a few common examples.
First off, we'll download a little bit of data
and show its structure:
```
import plotly.express as px
data = px.data.iris()
data.head()
```
## Altair
Interactive outputs will work under the assumption that the outputs they produce have
self-contained HTML that works without requiring any external dependencies to load.
See the [`Altair` installation instructions](https://altair-viz.github.io/getting_started/installation.html#installation)
to get set up with Altair. Below is some example output.
```
import altair as alt
alt.Chart(data=data).mark_point().encode(
x="sepal_width",
y="sepal_length",
color="species",
size='sepal_length'
)
```
## Plotly
Plotly is another interactive plotting library that provides a high-level API for
visualization. See the [Plotly JupyterLab documentation](https://plotly.com/python/getting-started/#JupyterLab-Support-(Python-3.5+))
to get started with Plotly in the notebook.
```{margin}
Plotly uses [renderers to output different kinds of information](https://plotly.com/python/renderers/)
when you display a plot. Experiment with renderers to get the output you want.
```
Below is some example output.
:::{important}
For these plots to show, it may be necessary to load `require.js`, in your `_config.yml`:
```yaml
sphinx:
config:
html_js_files:
- https://cdnjs.cloudflare.com/ajax/libs/require.js/2.3.4/require.min.js
```
:::
```
import plotly.io as pio
import plotly.express as px
import plotly.offline as py
df = px.data.iris()
fig = px.scatter(df, x="sepal_width", y="sepal_length", color="species", size="sepal_length")
fig
```
## Bokeh
Bokeh provides several options for interactive visualizations, and is part of the PyViz ecosystem. See
[the Bokeh with Jupyter documentation](https://docs.bokeh.org/en/latest/docs/user_guide/jupyter.html#userguide-jupyter) to
get started.
Below is some example output. First we'll initialized Bokeh with `output_notebook()`.
This needs to be in a separate cell to give the JavaScript time to load.
```
from bokeh.plotting import figure, show, output_notebook
output_notebook()
```
Now we'll make our plot.
```
p = figure()
p.circle(data["sepal_width"], data["sepal_length"], fill_color=data["species"], size=data["sepal_length"])
show(p)
```
## ipywidgets
You may also run code for Jupyter Widgets in your document, and the interactive HTML
outputs will embed themselves in your side. See [the ipywidgets documentation](https://ipywidgets.readthedocs.io/en/latest/user_install.html)
for how to get set up in your own environment.
```{admonition} Widgets often need a kernel
Note that `ipywidgets` tend to behave differently from other interactive visualization libraries. They
interact both with Javascript, and with Python. Some functionality in `ipywidgets` may not
work in default Jupyter Book pages (because no Python kernel is running). You may be able to
get around this with [tools for remote kernels, like thebe](https://thebelab.readthedocs.org).
```
Here are some simple widget elements rendered below.
```
import ipywidgets as widgets
widgets.IntSlider(
value=7,
min=0,
max=10,
step=1,
description='Test:',
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='d'
)
tab_contents = ['P0', 'P1', 'P2', 'P3', 'P4']
children = [widgets.Text(description=name) for name in tab_contents]
tab = widgets.Tab()
tab.children = children
for ii in range(len(children)):
tab.set_title(ii, f"tab_{ii}")
tab
```
You can find [a list of existing Jupyter Widgets](https://ipywidgets.readthedocs.io/en/latest/examples/Widget%20List.html)
in the jupyter-widgets documentation.
|
github_jupyter
|
import plotly.express as px
data = px.data.iris()
data.head()
import altair as alt
alt.Chart(data=data).mark_point().encode(
x="sepal_width",
y="sepal_length",
color="species",
size='sepal_length'
)
Below is some example output.
:::{important}
For these plots to show, it may be necessary to load `require.js`, in your `_config.yml`:
:::
## Bokeh
Bokeh provides several options for interactive visualizations, and is part of the PyViz ecosystem. See
[the Bokeh with Jupyter documentation](https://docs.bokeh.org/en/latest/docs/user_guide/jupyter.html#userguide-jupyter) to
get started.
Below is some example output. First we'll initialized Bokeh with `output_notebook()`.
This needs to be in a separate cell to give the JavaScript time to load.
Now we'll make our plot.
## ipywidgets
You may also run code for Jupyter Widgets in your document, and the interactive HTML
outputs will embed themselves in your side. See [the ipywidgets documentation](https://ipywidgets.readthedocs.io/en/latest/user_install.html)
for how to get set up in your own environment.
Here are some simple widget elements rendered below.
| 0.746786 | 0.952706 |
# Causality Tutorial Exercises – Python
Contributors: Rune Christiansen, Jonas Peters, Niklas Pfister, Sorawit Saengkyongam, Sebastian Weichwald.
The MIT License applies; copyright is with the authors.
Some exercises are adapted from "Elements of Causal Inference: Foundations and Learning Algorithms" by J. Peters, D. Janzing and B. Schölkopf.
# Exercise 1 – Structural Causal Model
Let's first draw a sample from an SCM
```
import numpy as np
# set seed
np.random.seed(1)
rnorm = lambda n: np.random.normal(size=n)
n = 200
C = rnorm(n)
A = .8 * rnorm(n)
K = A + .1 * rnorm(n)
X = C - 2 * A + .2 * rnorm(n)
F = 3 * X + .8 * rnorm(n)
D = -2 * X + .5 * rnorm(n)
G = D + .5 * rnorm(n)
Y = 2 * K - D + .2 * rnorm(n)
H = .5 * Y + .1 * rnorm(n)
data = np.c_[C, A, K, X, F, D, G, Y, H]
```
__a)__
What is the graph corresponding to the above SCM? (Draw on a paper.)
Take a pair of variables and think about whether you expect this pair to be dependent
(at this stage, you can only guess, later you will have tools to know). Check empirically.
__b)__
Generate a sample of size 300 from the interventional distribution $P_{\mathrm{do}(X=\mathcal{N}(2, 1))}$
and store the data matrix as `data_int`.
```
```
__c)__
Do you expect the marginal distribution of $Y$ to be different in both samples?
Double-click (or enter) to edit
__d)__
Do you expect the joint distribution of $(A, Y)$ to be different in both samples?
Double-click (or enter) to edit
__e)__
Check your answers to c) and d) empirically.
```
```
# Exercise 2 – Adjusting
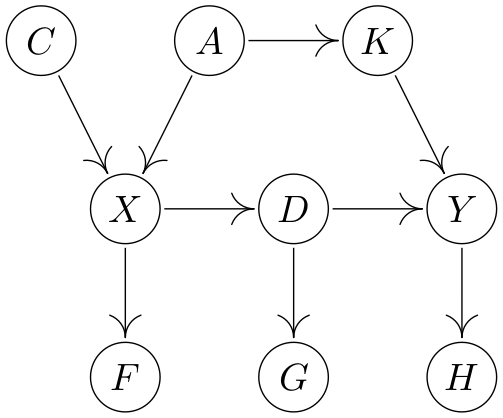
Suppose we are given a fixed DAG (like the one above).
a) What are valid adjustment sets (VAS) used for?
b) Assume we want to find a VAS for the causal effect from $X$ to $Y$.
What are general recipies (plural 😉) for constructing VASs (no proof)?
Which sets are VAS in the DAG above?
c) The following code samples from an SCM. Perform linear regressions using different VAS and compare the regression coefficient against the causal effect from $X$ to $Y$.
```
import numpy as np
# set seed
np.random.seed(1)
rnorm = lambda n: np.random.normal(size=n)
n = 200
C = rnorm(n)
A = .8 * rnorm(n)
K = A + .1 * rnorm(n)
X = C - 2 * A + .2 * rnorm(n)
F = 3 * X + .8 * rnorm(n)
D = -2 * X + .5 * rnorm(n)
G = D + .5 * rnorm(n)
Y = 2 * K - D + .2 * rnorm(n)
H = .5 * Y + .1 * rnorm(n)
data = np.c_[C, A, K, X, F, D, G, Y, H]
```
d) Why could it be interesting to have several options for choosing a VAS?
e) If you indeed have access to several VASs, what would you do?
# Exercise 3 – Independence-based Causal Structure Learning
__a)__
Assume $P^{X,Y,Z}$ is Markov and faithful wrt. $G$. Assume all (!) conditional independences are
$$
\newcommand{\indep}{{\,⫫\,}}
\newcommand{\dep}{\not{}\!\!\indep}
$$
$$X \dep Z \mid \emptyset$$
(plus symmetric statements). What is $G$?
__b)__
Assume $P^{W,X,Y,Z}$ is Markov and faithful wrt. $G$. Assume all (!) conditional independences are
$$\begin{aligned}
(Y,Z) &\indep W \mid \emptyset \\
W &\indep Y \mid (X,Z) \\
(X,W) &\indep Y | Z
\end{aligned}
$$
(plus symmetric statements). What is $G$?
# Exercise 4 – Additive Noise Models
Set-up required packages:
```
# set up – not needed when run on mybinder
# if needed (colab), change False to True and run cell
if False:
!mkdir ../data/
!wget https://raw.githubusercontent.com/sweichwald/causality-tutorial-exercises/main/data/Exercise-ANM.csv -q -O ../data/Exercise-ANM.csv
!wget https://raw.githubusercontent.com/sweichwald/causality-tutorial-exercises/main/python/kerpy/__init__.py -q -O kerpy.py
!pip install pygam
from kerpy import hsic
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from pygam import GAM, s
```
Let's load and plot some real data set:
```
data = pd.read_csv('../data/Exercise-ANM.csv')
plt.scatter(data["X"].values, data["Y"].values, s=2.);
```
__a)__
Do you believed that $X \to Y$ or that $X \gets Y$? Why?
Double-click (or enter) to edit
$$
\newcommand{\indep}{{\,⫫\,}}
\newcommand{\dep}{\not{}\!\!\indep}
$$
__b)__
Let us now try to get a more statistical answer. We have heard that we cannot
have
$$Y = f(X) + N_Y,\ N_Y \indep X$$
and
$$X = g(Y) + N_X,\ N_X \indep Y$$
at the same time.
Given a data set over $(X,Y)$,
we now want to decide for one of the two models.
Come up with a method to do so.
Hints:
* `GAM(s(0)).fit(A, B).deviance_residuals(A, B)` provides residuals when regressing $B$ on $A$.
* `hsic(a, b)` can be used as an independence test (here, `a` and `b` are $n \times 1$ numpy arrays).
```
```
__c)__
Assume that the error terms are Gaussian with zero mean and variances
$\sigma_X^2$ and $\sigma_Y^2$, respectively.
The maximum likelihood for DAG G is
then proportional to
$-\log(\mathrm{var}(R^G_X)) - \log(\mathrm{var}(R^G_Y))$,
where $R^G_X$ and $R^G_Y$ are the residuals obtained from regressing $X$ and $Y$ on
their parents in $G$, respectively (no proof).
Find the maximum likelihood solution.
```
```
# Exercise 5 – Invariant Causal Prediction
Set-up required packages and data:
```
# set up – not needed when run on mybinder
# if needed (colab), change False to True and run cell
if False:
!mkdir ../data/
!wget https://raw.githubusercontent.com/sweichwald/causality-tutorial-exercises/main/data/Exercise-ICP.csv -q -O ../data/Exercise-ICP.csv
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
import statsmodels.api as sm
```
__a)__
Generate some observational and interventional data:
```
# Generate n=1000 observations from the observational distribution
na = 1000
Xa = np.random.normal(size=na)
Ya = 1.5*Xa + np.random.normal(size=na)
# Generate n=1000 observations from an interventional distribution
nb = 1000
Xb = np.random.normal(loc=2, scale=1, size=nb)
Yb = 1.5*Xb + np.random.normal(size=nb)
# plot Y vs X1
fig, ax = plt.subplots(figsize=(7,5))
ax.scatter(Xa, Ya, label='observational', marker='o', alpha=0.6)
ax.scatter(Xb, Yb, label='interventional', marker ='^', alpha=0.6)
ax.legend();
```
Look at the above plot. Is the predictor $\{X\}$ an invariant set, that is (roughly speaking), does $Y \mid X = x$ have the same distribution in the orange and blue data?
Double-click (or enter) to edit
__b)__
We now consider data over a response and three covariates $X1, X2$, and $X3$
and try to infer $\mathrm{pa}(Y)$. To do so, we need to find all sets for which this
invariance is satisfied.
```
# load data
data = pd.read_csv('../data/Exercise-ICP.csv')
data['env'] = np.concatenate([np.repeat('observational', 140), np.repeat('interventional', 80)])
# pairplot
sns.pairplot(data, hue='env', height=2, plot_kws={'alpha':0.6});
# The code below plots the residuals versus fitted values for all sets of
# predictors.
# extract response and predictors
Y = data['Y'].to_numpy()
X = data[['X1','X2','X3']].to_numpy()
# get environment indicator
obs_ind = data[data['env'] == 'observational'].index
int_ind = data[data['env'] == 'interventional'].index
# create all sets
all_sets = [(0,), (1,), (2,), (0,1), (0,2), (1,2), (0,1,2)]
# label each set
set_labels = ['X1', 'X2', 'X3', 'X1,X2', 'X1,X3', 'X2,X3', 'X1,X2,X3']
# fit OLS and store fitted values and residuals for each set
fitted = []
resid = []
for s in all_sets:
model = sm.OLS(Y, X[:, s]).fit()
fitted += [model.fittedvalues]
resid += [model.resid]
# plotting function
def plot_fitted_resid(fv, res, ax, title):
ax.scatter(fv[obs_ind], res[obs_ind], label='observational', marker='o', alpha=0.6)
ax.scatter(fv[int_ind], res[int_ind], label='interventional', marker ='^', alpha=0.6)
ax.legend()
ax.set_xlabel('fitted values')
ax.set_ylabel('residuals')
ax.set_title(title)
# creating plots
fig, axes = plt.subplots(4, 2, figsize=(7,14))
# plot result for the empty set predictor
ax0 = axes[0,0]
ax0.scatter(obs_ind, Y[obs_ind], label='observational', marker='o', alpha=0.6)
ax0.scatter(int_ind, Y[int_ind], label='interventional', marker ='^', alpha=0.6)
ax0.legend()
ax0.set_xlabel('index')
ax0.set_ylabel('Y')
ax0.set_title('empty set')
# plot result for the other sets
for i, ax in enumerate(axes.flatten()[1:]):
plot_fitted_resid(fitted[i], resid[i], ax, set_labels[i])
# make tight layout
plt.tight_layout()
```
Which of the sets are invariant? (There are two plots with four scatter plots each.)
Double-click (or enter) to edit
__c)__
What is your best guess for $\mathrm{pa}(Y)$?
Double-click (or enter) to edit
__d) (optional)__
Use the function ICP to check your result.
```
# set up – not needed when run on mybinder
# if needed (colab), change False to True and run cell
if False:
!pip install causalicp
import causalicp as icp
```
|
github_jupyter
|
import numpy as np
# set seed
np.random.seed(1)
rnorm = lambda n: np.random.normal(size=n)
n = 200
C = rnorm(n)
A = .8 * rnorm(n)
K = A + .1 * rnorm(n)
X = C - 2 * A + .2 * rnorm(n)
F = 3 * X + .8 * rnorm(n)
D = -2 * X + .5 * rnorm(n)
G = D + .5 * rnorm(n)
Y = 2 * K - D + .2 * rnorm(n)
H = .5 * Y + .1 * rnorm(n)
data = np.c_[C, A, K, X, F, D, G, Y, H]
```
__c)__
Do you expect the marginal distribution of $Y$ to be different in both samples?
Double-click (or enter) to edit
__d)__
Do you expect the joint distribution of $(A, Y)$ to be different in both samples?
Double-click (or enter) to edit
__e)__
Check your answers to c) and d) empirically.
# Exercise 2 – Adjusting

Suppose we are given a fixed DAG (like the one above).
a) What are valid adjustment sets (VAS) used for?
b) Assume we want to find a VAS for the causal effect from $X$ to $Y$.
What are general recipies (plural 😉) for constructing VASs (no proof)?
Which sets are VAS in the DAG above?
c) The following code samples from an SCM. Perform linear regressions using different VAS and compare the regression coefficient against the causal effect from $X$ to $Y$.
d) Why could it be interesting to have several options for choosing a VAS?
e) If you indeed have access to several VASs, what would you do?
# Exercise 3 – Independence-based Causal Structure Learning
__a)__
Assume $P^{X,Y,Z}$ is Markov and faithful wrt. $G$. Assume all (!) conditional independences are
$$
\newcommand{\indep}{{\,⫫\,}}
\newcommand{\dep}{\not{}\!\!\indep}
$$
$$X \dep Z \mid \emptyset$$
(plus symmetric statements). What is $G$?
__b)__
Assume $P^{W,X,Y,Z}$ is Markov and faithful wrt. $G$. Assume all (!) conditional independences are
$$\begin{aligned}
(Y,Z) &\indep W \mid \emptyset \\
W &\indep Y \mid (X,Z) \\
(X,W) &\indep Y | Z
\end{aligned}
$$
(plus symmetric statements). What is $G$?
# Exercise 4 – Additive Noise Models
Set-up required packages:
Let's load and plot some real data set:
__a)__
Do you believed that $X \to Y$ or that $X \gets Y$? Why?
Double-click (or enter) to edit
$$
\newcommand{\indep}{{\,⫫\,}}
\newcommand{\dep}{\not{}\!\!\indep}
$$
__b)__
Let us now try to get a more statistical answer. We have heard that we cannot
have
$$Y = f(X) + N_Y,\ N_Y \indep X$$
and
$$X = g(Y) + N_X,\ N_X \indep Y$$
at the same time.
Given a data set over $(X,Y)$,
we now want to decide for one of the two models.
Come up with a method to do so.
Hints:
* `GAM(s(0)).fit(A, B).deviance_residuals(A, B)` provides residuals when regressing $B$ on $A$.
* `hsic(a, b)` can be used as an independence test (here, `a` and `b` are $n \times 1$ numpy arrays).
__c)__
Assume that the error terms are Gaussian with zero mean and variances
$\sigma_X^2$ and $\sigma_Y^2$, respectively.
The maximum likelihood for DAG G is
then proportional to
$-\log(\mathrm{var}(R^G_X)) - \log(\mathrm{var}(R^G_Y))$,
where $R^G_X$ and $R^G_Y$ are the residuals obtained from regressing $X$ and $Y$ on
their parents in $G$, respectively (no proof).
Find the maximum likelihood solution.
# Exercise 5 – Invariant Causal Prediction
Set-up required packages and data:
__a)__
Generate some observational and interventional data:
Look at the above plot. Is the predictor $\{X\}$ an invariant set, that is (roughly speaking), does $Y \mid X = x$ have the same distribution in the orange and blue data?
Double-click (or enter) to edit
__b)__
We now consider data over a response and three covariates $X1, X2$, and $X3$
and try to infer $\mathrm{pa}(Y)$. To do so, we need to find all sets for which this
invariance is satisfied.
Which of the sets are invariant? (There are two plots with four scatter plots each.)
Double-click (or enter) to edit
__c)__
What is your best guess for $\mathrm{pa}(Y)$?
Double-click (or enter) to edit
__d) (optional)__
Use the function ICP to check your result.
| 0.6973 | 0.90657 |
```
import os
import sys
from flask import Flask, request, session, g, redirect, url_for, abort, render_template
from flaskext.mysql import MySQL
from flask_wtf import FlaskForm
from wtforms.fields.html5 import DateField
from wtforms import SelectField
from datetime import date
import time
import gmplot
import pandas
app = Flask(__name__)
app.secret_key = 'A0Zr98slkjdf984jnflskj_sdkfjhT'
mysql = MySQL()
app.config['MYSQL_DATABASE_USER'] = 'root'
app.config['MYSQL_DATABASE_PASSWORD'] = '1234'
app.config['MYSQL_DATABASE_DB'] = 'CREDIT'
app.config['MYSQL_DATABASE_HOST'] = 'localhost'
mysql.init_app(app)
conn = mysql.connect()
cursor = conn.cursor()
class AnalyticsForm(FlaskForm):
attributes = SelectField('Data Attributes', choices=[('Agency', 'Agency'),
('Borough', 'Borough'), ('Complaint Type', 'Complaint Type')])
@app.route("/")
def home():
#session["data_loaded"] = True
return render_template('home.html', links=get_homepage_links())
def get_df_data():
query = "select unique_key, agency, complaint_type, borough from incidents;"
cursor.execute(query)
data = cursor.fetchall()
df = pandas.DataFrame(data=list(data),columns=['Unique_key','Agency','Complaint Type','Borough'])
return df
from flask import Flask
from flask import Flask, request, render_template
import os
import sys
from flask import Flask, request, session, g, redirect, url_for, abort, render_template
from flaskext.mysql import MySQL
from flask_wtf import FlaskForm
from wtforms.fields.html5 import DateField
from wtforms import SelectField
from datetime import date
import time
import gmplot
import pandas
app = Flask(__name__)
app.secret_key = 'A0Zr98slkjdf984jnflskj_sdkfjhT'
mysql = MySQL()
app.config['MYSQL_DATABASE_USER'] = 'root'
app.config['MYSQL_DATABASE_PASSWORD'] = '1234'
app.config['MYSQL_DATABASE_DB'] = 'CREDIT'
app.config['MYSQL_DATABASE_HOST'] = 'localhost'
mysql.init_app(app)
conn = mysql.connect()
cursor = conn.cursor()
class AnalyticsForm(FlaskForm):
attributes = SelectField('Data Attributes', choices=[('SEX', 'SEX'), ('AGE', 'AGE'), ('EDUCATION', 'EDUCATION')])
def get_homepage_links():
return [{"href": url_for('analytics'), "label":"Plot"}]
def get_df_data():
query = "select ID, AGE, SEX, EDUCATION,default_payment from credit_card;"
cursor.execute(query)
data = cursor.fetchall()
df = pandas.DataFrame(data=list(data),columns=['ID', 'AGE', 'SEX', 'EDUCATION','default_payment'])
return df
@app.route("/")
def home():
#session["data_loaded"] = True
return render_template('home.html', links=get_homepage_links())
@app.route('/analytics',methods=['GET','POST'])
def analytics():
form = AnalyticsForm()
if form.validate_on_submit():
df = get_df_data()
column = request.form.get('attributes')
group = df.groupby([column,'default_payment'])
ax = group.size().unstack().plot(kind='bar')
fig = ax.get_figure()
filename = 'static/charts/group_by_figure.png' #_'+str(int(time.time()))+".png"
fig.savefig(filename)
return render_template('analytics.html', chart_src="/"+filename)
return render_template('analyticsparams.html', form=form)
if __name__ == "__main__":
app.run()
```
|
github_jupyter
|
import os
import sys
from flask import Flask, request, session, g, redirect, url_for, abort, render_template
from flaskext.mysql import MySQL
from flask_wtf import FlaskForm
from wtforms.fields.html5 import DateField
from wtforms import SelectField
from datetime import date
import time
import gmplot
import pandas
app = Flask(__name__)
app.secret_key = 'A0Zr98slkjdf984jnflskj_sdkfjhT'
mysql = MySQL()
app.config['MYSQL_DATABASE_USER'] = 'root'
app.config['MYSQL_DATABASE_PASSWORD'] = '1234'
app.config['MYSQL_DATABASE_DB'] = 'CREDIT'
app.config['MYSQL_DATABASE_HOST'] = 'localhost'
mysql.init_app(app)
conn = mysql.connect()
cursor = conn.cursor()
class AnalyticsForm(FlaskForm):
attributes = SelectField('Data Attributes', choices=[('Agency', 'Agency'),
('Borough', 'Borough'), ('Complaint Type', 'Complaint Type')])
@app.route("/")
def home():
#session["data_loaded"] = True
return render_template('home.html', links=get_homepage_links())
def get_df_data():
query = "select unique_key, agency, complaint_type, borough from incidents;"
cursor.execute(query)
data = cursor.fetchall()
df = pandas.DataFrame(data=list(data),columns=['Unique_key','Agency','Complaint Type','Borough'])
return df
from flask import Flask
from flask import Flask, request, render_template
import os
import sys
from flask import Flask, request, session, g, redirect, url_for, abort, render_template
from flaskext.mysql import MySQL
from flask_wtf import FlaskForm
from wtforms.fields.html5 import DateField
from wtforms import SelectField
from datetime import date
import time
import gmplot
import pandas
app = Flask(__name__)
app.secret_key = 'A0Zr98slkjdf984jnflskj_sdkfjhT'
mysql = MySQL()
app.config['MYSQL_DATABASE_USER'] = 'root'
app.config['MYSQL_DATABASE_PASSWORD'] = '1234'
app.config['MYSQL_DATABASE_DB'] = 'CREDIT'
app.config['MYSQL_DATABASE_HOST'] = 'localhost'
mysql.init_app(app)
conn = mysql.connect()
cursor = conn.cursor()
class AnalyticsForm(FlaskForm):
attributes = SelectField('Data Attributes', choices=[('SEX', 'SEX'), ('AGE', 'AGE'), ('EDUCATION', 'EDUCATION')])
def get_homepage_links():
return [{"href": url_for('analytics'), "label":"Plot"}]
def get_df_data():
query = "select ID, AGE, SEX, EDUCATION,default_payment from credit_card;"
cursor.execute(query)
data = cursor.fetchall()
df = pandas.DataFrame(data=list(data),columns=['ID', 'AGE', 'SEX', 'EDUCATION','default_payment'])
return df
@app.route("/")
def home():
#session["data_loaded"] = True
return render_template('home.html', links=get_homepage_links())
@app.route('/analytics',methods=['GET','POST'])
def analytics():
form = AnalyticsForm()
if form.validate_on_submit():
df = get_df_data()
column = request.form.get('attributes')
group = df.groupby([column,'default_payment'])
ax = group.size().unstack().plot(kind='bar')
fig = ax.get_figure()
filename = 'static/charts/group_by_figure.png' #_'+str(int(time.time()))+".png"
fig.savefig(filename)
return render_template('analytics.html', chart_src="/"+filename)
return render_template('analyticsparams.html', form=form)
if __name__ == "__main__":
app.run()
| 0.155174 | 0.071819 |
```
import urllib
from IPython.display import Markdown as md
### change to reflect your notebook
_nb_loc = "07_training/07b_gpumax.ipynb"
_nb_title = "GPU utilization"
_icons=["https://raw.githubusercontent.com/GoogleCloudPlatform/practical-ml-vision-book/master/logo-cloud.png", "https://www.tensorflow.org/images/colab_logo_32px.png", "https://www.tensorflow.org/images/GitHub-Mark-32px.png", "https://www.tensorflow.org/images/download_logo_32px.png"]
_links=["https://console.cloud.google.com/vertex-ai/workbench/deploy-notebook?" + urllib.parse.urlencode({"name": _nb_title, "download_url": "https://github.com/takumiohym/practical-ml-vision-book-ja/raw/master/"+_nb_loc}), "https://colab.research.google.com/github/takumiohym/practical-ml-vision-book-ja/blob/master/{0}".format(_nb_loc), "https://github.com/takumiohym/practical-ml-vision-book-ja/blob/master/{0}".format(_nb_loc), "https://raw.githubusercontent.com/takumiohym/practical-ml-vision-book-ja/master/{0}".format(_nb_loc)]
md("""<table class="tfo-notebook-buttons" align="left"><td><a target="_blank" href="{0}"><img src="{4}"/>Run in Vertex AI Workbench</a></td><td><a target="_blank" href="{1}"><img src="{5}" />Run in Google Colab</a></td><td><a target="_blank" href="{2}"><img src="{6}" />View source on GitHub</a></td><td><a href="{3}"><img src="{7}" />Download notebook</a></td></table><br/><br/>""".format(_links[0], _links[1], _links[2], _links[3], _icons[0], _icons[1], _icons[2], _icons[3]))
```
# GPU使用率
このノートブックでは、GPUのTensorFlow最適化を利用する方法を示します。
## GPUを有効にし、ヘルパー関数を設定します
このノートブックと、このリポジトリ内の他のほとんどすべてのノートブック
GPUを使用している場合は、より高速に実行されます。
Colabについて:
- [編集]→[ノートブック設定]に移動します
- [ハードウェアアクセラレータ]ドロップダウンから[GPU]を選択します
クラウドAIプラットフォームノートブック:
- https://console.cloud.google.com/ai-platform/notebooksに移動します
- GPUを使用してインスタンスを作成するか、インスタンスを選択してGPUを追加します
次に、テンソルフローを使用してGPUに接続できることを確認します。
```
import tensorflow as tf
print('TensorFlow version' + tf.version.VERSION)
print('Built with GPU support? ' + ('Yes!' if tf.test.is_built_with_cuda() else 'Noooo!'))
print('There are {} GPUs'.format(len(tf.config.experimental.list_physical_devices("GPU"))))
device_name = tf.test.gpu_device_name()
if device_name != '/device:GPU:0':
raise SystemError('GPU device not found')
print('Found GPU at: {}'.format(device_name))
```
## コードを取り込む
```
%%writefile input.txt
gs://practical-ml-vision-book/images/california_fire1.jpg
gs://practical-ml-vision-book/images/california_fire2.jpg
import matplotlib.pylab as plt
import numpy as np
import tensorflow as tf
def read_jpeg(filename):
img = tf.io.read_file(filename)
img = tf.image.decode_jpeg(img, channels=3)
img = tf.image.convert_image_dtype(img, tf.float32)
img = tf.reshape(img, [338, 600, 3])
return img
ds = tf.data.TextLineDataset('input.txt').map(read_jpeg)
f, ax = plt.subplots(1, 2, figsize=(15,10))
for idx, img in enumerate(ds):
ax[idx].imshow( img.numpy() );
```
## マップ関数の追加
画像を変換するためにカスタム式を適用したいとします。
```
def to_grayscale(img):
red = img[:, :, 0]
green = img[:, :, 1]
blue = img[:, :, 2]
c_linear = 0.2126 * red + 0.7152 * green + 0.0722 * blue
gray = tf.where(c_linear > 0.0031308,
1.055 * tf.pow(c_linear, 1/2.4) - 0.055,
12.92*c_linear)
print(gray.shape)
return gray
ds = tf.data.TextLineDataset('input.txt').map(read_jpeg).map(to_grayscale)
f, ax = plt.subplots(1, 2, figsize=(15,10))
for idx, img in enumerate(ds):
im = ax[idx].imshow( img.numpy() , interpolation='none');
if idx == 1:
f.colorbar(im, fraction=0.028, pad=0.04)
```
### 1. 画像の反復
(これをしないでください)
```
# This function is not accelerated. At all.
def to_grayscale(img):
rows, cols, _ = img.shape
result = np.zeros([rows, cols], dtype=np.float32)
for row in range(rows):
for col in range(cols):
red = img[row][col][0]
green = img[row][col][1]
blue = img[row][col][2]
c_linear = 0.2126 * red + 0.7152 * green + 0.0722 * blue
if c_linear > 0.0031308:
result[row][col] = 1.055 * pow(c_linear, 1/2.4) - 0.055
else:
result[row][col] = 12.92*c_linear
return result
%%time
ds = tf.data.TextLineDataset('input.txt').repeat(10).map(read_jpeg)
overall = tf.constant([0.], dtype=tf.float32)
count = 0
for img in ds:
# Notice that we have to call .numpy() to move the data outside TF Graph
gray = to_grayscale(img.numpy())
# This moves the data back into the graph
m = tf.reduce_mean(gray, axis=[0, 1])
overall += m
count += 1
print(overall/count)
```
### 2. Pyfunc
Pythonのみの機能(time/jsonなど)を繰り返しまたは呼び出す必要があり、それでもmap()を使用する必要がある場合は、py_funcを使用できます。
データは引き続きグラフから移動され、作業が完了し、データはグラフに戻されます。したがって、効率的には利益ではありません。
```
def to_grayscale_numpy(img):
# the numpy happens here
img = img.numpy()
rows, cols, _ = img.shape
result = np.zeros([rows, cols], dtype=np.float32)
for row in range(rows):
for col in range(cols):
red = img[row][col][0]
green = img[row][col][1]
blue = img[row][col][2]
c_linear = 0.2126 * red + 0.7152 * green + 0.0722 * blue
if c_linear > 0.0031308:
result[row][col] = 1.055 * pow(c_linear, 1/2.4) - 0.055
else:
result[row][col] = 12.92*c_linear
# the convert back happens here
return tf.convert_to_tensor(result)
def to_grayscale(img):
return tf.py_function(to_grayscale_numpy, [img], tf.float32)
%%time
ds = tf.data.TextLineDataset('input.txt').repeat(10).map(read_jpeg).map(to_grayscale)
overall = tf.constant([0.], dtype=tf.float32)
count = 0
for gray in ds:
m = tf.reduce_mean(gray, axis=[0, 1])
overall += m
count += 1
print(overall/count)
```
### 3. TensorFlowスライシングとtf.whereを使用します
これは、反復よりも10倍高速です。
```
# All in GPU
def to_grayscale(img):
# TensorFlow slicing functionality
red = img[:, :, 0]
green = img[:, :, 1]
blue = img[:, :, 2]
# All these are actually tf.mul(), tf.add(), etc.
c_linear = 0.2126 * red + 0.7152 * green + 0.0722 * blue
# Use tf.cond and tf.where for if-then statements
gray = tf.where(c_linear > 0.0031308,
1.055 * tf.pow(c_linear, 1/2.4) - 0.055,
12.92*c_linear)
return gray
%%time
ds = tf.data.TextLineDataset('input.txt').repeat(10).map(read_jpeg).map(to_grayscale)
overall = tf.constant([0.])
count = 0
for gray in ds:
m = tf.reduce_mean(gray, axis=[0, 1])
overall += m
count += 1
print(overall/count)
```
### 4. Matrixmathとtf.whereを使用します
これはスライスの3倍の速さです。
```
def to_grayscale(img):
wt = tf.constant([[0.2126], [0.7152], [0.0722]]) # 3x1 matrix
c_linear = tf.matmul(img, wt) # (ht,wd,3) x (3x1) -> (ht, wd)
gray = tf.where(c_linear > 0.0031308,
1.055 * tf.pow(c_linear, 1/2.4) - 0.055,
12.92*c_linear)
return gray
%%time
ds = tf.data.TextLineDataset('input.txt').repeat(10).map(read_jpeg).map(to_grayscale)
overall = tf.constant([0.])
count = 0
for gray in ds:
m = tf.reduce_mean(gray, axis=[0, 1])
overall += m
count += 1
print(overall/count)
```
### 5. バッチ処理
画像のバッチで機能するように、操作を完全にベクトル化します
```
class Grayscale(tf.keras.layers.Layer):
def __init__(self, **kwargs):
super(Grayscale, self).__init__(kwargs)
def call(self, img):
wt = tf.constant([[0.2126], [0.7152], [0.0722]]) # 3x1 matrix
c_linear = tf.matmul(img, wt) # (N, ht,wd,3) x (3x1) -> (N, ht, wd)
gray = tf.where(c_linear > 0.0031308,
1.055 * tf.pow(c_linear, 1/2.4) - 0.055,
12.92*c_linear)
return gray # (N, ht, wd)
model = tf.keras.Sequential([
Grayscale(input_shape=(336, 600, 3)),
tf.keras.layers.Lambda(lambda gray: tf.reduce_mean(gray, axis=[1, 2])) # note axis change
])
%%time
ds = tf.data.TextLineDataset('input.txt').repeat(10).map(read_jpeg).batch(5)
overall = tf.constant([0.])
count = 0
for batch in ds:
bm = model(batch)
overall += tf.reduce_sum(bm)
count += len(bm)
print(overall/count)
```
## 結果
私たちがそれをしたとき、これらは私たちが得たタイミングでした:
| 方法| CPU時間|実時間|
| ---------------------- | ----------- | ------------ |
| 繰り返す| 39.6秒| 41.1秒|
| Pyfunc | 39.7秒| 41.1秒|
| スライス| 4.44秒| 3.07秒|
| Matmul | 1.22秒| 2.29秒|
| バッチ| 1.11秒| 2.13秒|
## サイン
署名で遊んで
```
from inspect import signature
def myfunc(a, b):
return (a + b)
print(myfunc(3,5))
print(myfunc('foo', 'bar'))
print(signature(myfunc).parameters)
print(signature(myfunc).return_annotation)
from inspect import signature
def myfunc(a: int, b: float) -> float:
return (a + b)
print(myfunc(3,5))
print(myfunc('foo', 'bar')) # runtime doesn't check
print(signature(myfunc).parameters)
print(signature(myfunc).return_annotation)
from inspect import signature
import tensorflow as tf
@tf.function(input_signature=[
tf.TensorSpec([3,5], name='a'),
tf.TensorSpec([5,8], name='b')
])
def myfunc(a, b):
return (tf.matmul(a,b))
print(myfunc.get_concrete_function(tf.ones((3,5)), tf.ones((5,8))))
```
## License
Copyright 2022 Google Inc. Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
|
github_jupyter
|
import urllib
from IPython.display import Markdown as md
### change to reflect your notebook
_nb_loc = "07_training/07b_gpumax.ipynb"
_nb_title = "GPU utilization"
_icons=["https://raw.githubusercontent.com/GoogleCloudPlatform/practical-ml-vision-book/master/logo-cloud.png", "https://www.tensorflow.org/images/colab_logo_32px.png", "https://www.tensorflow.org/images/GitHub-Mark-32px.png", "https://www.tensorflow.org/images/download_logo_32px.png"]
_links=["https://console.cloud.google.com/vertex-ai/workbench/deploy-notebook?" + urllib.parse.urlencode({"name": _nb_title, "download_url": "https://github.com/takumiohym/practical-ml-vision-book-ja/raw/master/"+_nb_loc}), "https://colab.research.google.com/github/takumiohym/practical-ml-vision-book-ja/blob/master/{0}".format(_nb_loc), "https://github.com/takumiohym/practical-ml-vision-book-ja/blob/master/{0}".format(_nb_loc), "https://raw.githubusercontent.com/takumiohym/practical-ml-vision-book-ja/master/{0}".format(_nb_loc)]
md("""<table class="tfo-notebook-buttons" align="left"><td><a target="_blank" href="{0}"><img src="{4}"/>Run in Vertex AI Workbench</a></td><td><a target="_blank" href="{1}"><img src="{5}" />Run in Google Colab</a></td><td><a target="_blank" href="{2}"><img src="{6}" />View source on GitHub</a></td><td><a href="{3}"><img src="{7}" />Download notebook</a></td></table><br/><br/>""".format(_links[0], _links[1], _links[2], _links[3], _icons[0], _icons[1], _icons[2], _icons[3]))
import tensorflow as tf
print('TensorFlow version' + tf.version.VERSION)
print('Built with GPU support? ' + ('Yes!' if tf.test.is_built_with_cuda() else 'Noooo!'))
print('There are {} GPUs'.format(len(tf.config.experimental.list_physical_devices("GPU"))))
device_name = tf.test.gpu_device_name()
if device_name != '/device:GPU:0':
raise SystemError('GPU device not found')
print('Found GPU at: {}'.format(device_name))
%%writefile input.txt
gs://practical-ml-vision-book/images/california_fire1.jpg
gs://practical-ml-vision-book/images/california_fire2.jpg
import matplotlib.pylab as plt
import numpy as np
import tensorflow as tf
def read_jpeg(filename):
img = tf.io.read_file(filename)
img = tf.image.decode_jpeg(img, channels=3)
img = tf.image.convert_image_dtype(img, tf.float32)
img = tf.reshape(img, [338, 600, 3])
return img
ds = tf.data.TextLineDataset('input.txt').map(read_jpeg)
f, ax = plt.subplots(1, 2, figsize=(15,10))
for idx, img in enumerate(ds):
ax[idx].imshow( img.numpy() );
def to_grayscale(img):
red = img[:, :, 0]
green = img[:, :, 1]
blue = img[:, :, 2]
c_linear = 0.2126 * red + 0.7152 * green + 0.0722 * blue
gray = tf.where(c_linear > 0.0031308,
1.055 * tf.pow(c_linear, 1/2.4) - 0.055,
12.92*c_linear)
print(gray.shape)
return gray
ds = tf.data.TextLineDataset('input.txt').map(read_jpeg).map(to_grayscale)
f, ax = plt.subplots(1, 2, figsize=(15,10))
for idx, img in enumerate(ds):
im = ax[idx].imshow( img.numpy() , interpolation='none');
if idx == 1:
f.colorbar(im, fraction=0.028, pad=0.04)
# This function is not accelerated. At all.
def to_grayscale(img):
rows, cols, _ = img.shape
result = np.zeros([rows, cols], dtype=np.float32)
for row in range(rows):
for col in range(cols):
red = img[row][col][0]
green = img[row][col][1]
blue = img[row][col][2]
c_linear = 0.2126 * red + 0.7152 * green + 0.0722 * blue
if c_linear > 0.0031308:
result[row][col] = 1.055 * pow(c_linear, 1/2.4) - 0.055
else:
result[row][col] = 12.92*c_linear
return result
%%time
ds = tf.data.TextLineDataset('input.txt').repeat(10).map(read_jpeg)
overall = tf.constant([0.], dtype=tf.float32)
count = 0
for img in ds:
# Notice that we have to call .numpy() to move the data outside TF Graph
gray = to_grayscale(img.numpy())
# This moves the data back into the graph
m = tf.reduce_mean(gray, axis=[0, 1])
overall += m
count += 1
print(overall/count)
def to_grayscale_numpy(img):
# the numpy happens here
img = img.numpy()
rows, cols, _ = img.shape
result = np.zeros([rows, cols], dtype=np.float32)
for row in range(rows):
for col in range(cols):
red = img[row][col][0]
green = img[row][col][1]
blue = img[row][col][2]
c_linear = 0.2126 * red + 0.7152 * green + 0.0722 * blue
if c_linear > 0.0031308:
result[row][col] = 1.055 * pow(c_linear, 1/2.4) - 0.055
else:
result[row][col] = 12.92*c_linear
# the convert back happens here
return tf.convert_to_tensor(result)
def to_grayscale(img):
return tf.py_function(to_grayscale_numpy, [img], tf.float32)
%%time
ds = tf.data.TextLineDataset('input.txt').repeat(10).map(read_jpeg).map(to_grayscale)
overall = tf.constant([0.], dtype=tf.float32)
count = 0
for gray in ds:
m = tf.reduce_mean(gray, axis=[0, 1])
overall += m
count += 1
print(overall/count)
# All in GPU
def to_grayscale(img):
# TensorFlow slicing functionality
red = img[:, :, 0]
green = img[:, :, 1]
blue = img[:, :, 2]
# All these are actually tf.mul(), tf.add(), etc.
c_linear = 0.2126 * red + 0.7152 * green + 0.0722 * blue
# Use tf.cond and tf.where for if-then statements
gray = tf.where(c_linear > 0.0031308,
1.055 * tf.pow(c_linear, 1/2.4) - 0.055,
12.92*c_linear)
return gray
%%time
ds = tf.data.TextLineDataset('input.txt').repeat(10).map(read_jpeg).map(to_grayscale)
overall = tf.constant([0.])
count = 0
for gray in ds:
m = tf.reduce_mean(gray, axis=[0, 1])
overall += m
count += 1
print(overall/count)
def to_grayscale(img):
wt = tf.constant([[0.2126], [0.7152], [0.0722]]) # 3x1 matrix
c_linear = tf.matmul(img, wt) # (ht,wd,3) x (3x1) -> (ht, wd)
gray = tf.where(c_linear > 0.0031308,
1.055 * tf.pow(c_linear, 1/2.4) - 0.055,
12.92*c_linear)
return gray
%%time
ds = tf.data.TextLineDataset('input.txt').repeat(10).map(read_jpeg).map(to_grayscale)
overall = tf.constant([0.])
count = 0
for gray in ds:
m = tf.reduce_mean(gray, axis=[0, 1])
overall += m
count += 1
print(overall/count)
class Grayscale(tf.keras.layers.Layer):
def __init__(self, **kwargs):
super(Grayscale, self).__init__(kwargs)
def call(self, img):
wt = tf.constant([[0.2126], [0.7152], [0.0722]]) # 3x1 matrix
c_linear = tf.matmul(img, wt) # (N, ht,wd,3) x (3x1) -> (N, ht, wd)
gray = tf.where(c_linear > 0.0031308,
1.055 * tf.pow(c_linear, 1/2.4) - 0.055,
12.92*c_linear)
return gray # (N, ht, wd)
model = tf.keras.Sequential([
Grayscale(input_shape=(336, 600, 3)),
tf.keras.layers.Lambda(lambda gray: tf.reduce_mean(gray, axis=[1, 2])) # note axis change
])
%%time
ds = tf.data.TextLineDataset('input.txt').repeat(10).map(read_jpeg).batch(5)
overall = tf.constant([0.])
count = 0
for batch in ds:
bm = model(batch)
overall += tf.reduce_sum(bm)
count += len(bm)
print(overall/count)
from inspect import signature
def myfunc(a, b):
return (a + b)
print(myfunc(3,5))
print(myfunc('foo', 'bar'))
print(signature(myfunc).parameters)
print(signature(myfunc).return_annotation)
from inspect import signature
def myfunc(a: int, b: float) -> float:
return (a + b)
print(myfunc(3,5))
print(myfunc('foo', 'bar')) # runtime doesn't check
print(signature(myfunc).parameters)
print(signature(myfunc).return_annotation)
from inspect import signature
import tensorflow as tf
@tf.function(input_signature=[
tf.TensorSpec([3,5], name='a'),
tf.TensorSpec([5,8], name='b')
])
def myfunc(a, b):
return (tf.matmul(a,b))
print(myfunc.get_concrete_function(tf.ones((3,5)), tf.ones((5,8))))
| 0.62601 | 0.883437 |
```
%matplotlib inline
%reload_ext autoreload
%autoreload 2
```
# Start
```
import sys
sys.path.append("..")
import torchvision.transforms as ttfs
import torch, PIL
import torch.nn as nn, torch.nn.functional as F, numpy as np, torch.optim as optim
from torch.utils.data import Dataset, DataLoader
from pathlib import Path
from torch import tensor
from tqdm import tnrange as trange, tqdm_notebook as tqdm
def pil2tensor(image):
arr = torch.ByteTensor(torch.ByteStorage.from_buffer(image.tobytes()))
arr = arr.view(image.size[1], image.size[0], -1)
arr = arr.permute(2,0,1)
return arr.float().div_(255)
class FilesDataset1(Dataset):
def __init__(self, fns, labels, classes=None):
if classes is None: classes = list(set(labels))
self.classes = classes
self.class2idx = {v:k for k,v in enumerate(classes)}
self.fns = np.array(fns)
self.y = [self.class2idx[o] for o in labels]
@classmethod
def from_folder(cls, folder, classes=None, test_pct=0.):
if classes is None: classes = [cls.name for cls in find_classes(folder)]
fns,labels = [],[]
for cl in classes:
fnames = get_image_files(folder/cl)
fns += fnames
labels += [cl] * len(fnames)
if test_pct==0.: return cls(fns, labels, classes=classes)
fns,labels = np.array(fns),np.array(labels)
is_test = np.random.uniform(size=(len(fns),)) < test_pct
return (cls(fns[~is_test], labels[~is_test], classes=classes),
cls(fns[is_test], labels[is_test], classes=classes))
def __len__(self): return len(self.fns)
def __getitem__(self,i):
x = PIL.Image.open(self.fns[i]).convert('RGB')
x = ttfs.Resize((sz,sz))(x)
x = pil2tensor(x)*2-1
return x,self.y[i]
def get_image_files(c):
return [o for o in list(c.iterdir())
if not o.name.startswith('.') and not o.is_dir()]
sz = 224
DATA_PATH = Path('../data')
PATH = DATA_PATH/'caltech101'
data_mean,data_std = map(tensor, ([0.5355,0.5430,0.5280], [0.2909,0.2788,0.2979]))
classes = ['airplanes','Motorbikes','Faces','watch','Leopards']
np.random.seed(42)
train_ds,valid_ds = FilesDataset1.from_folder(PATH, test_pct=0.2, classes=classes)
# train_ds = FilesDataset1.from_folder(PATH, classes=classes)
# valid_ds = FilesDataset1.from_folder(PATH, classes=classes)
classes = train_ds.classes
default_device = torch.device('cuda', 0)
c = len(classes)
c,len(train_ds),len(valid_ds)
class CudaDataLoader():
def __init__(self,dl): self.dl = dl
def __len__(self): return len(self.dl)
def __iter__(self):
for o in self.dl: yield o[0].cuda(), o[1].cuda()
train_dl = CudaDataLoader(DataLoader(train_ds, batch_size=64, shuffle=True, num_workers=8))
valid_dl = CudaDataLoader(DataLoader(valid_ds, batch_size=2*64, shuffle=False, num_workers=8))
```
## Train
```
from collections import Iterable
def listify(p=None, q=None):
"Makes p a list that looks like q"
if p is None: p=[]
elif not isinstance(p, Iterable): p=[p]
n = q if type(q)==int else 1 if q is None else len(q)
if len(p)==1: p = p * n
return p
class Lambda(nn.Module):
def __init__(self, func):
super().__init__()
self.func=func
def forward(self, x): return self.func(x)
def ResizeBatch(*size): return Lambda(lambda x: x.view((-1,)+size))
def Flatten(): return Lambda(lambda x: x.view((x.size(0), -1)))
def PoolFlatten(): return nn.Sequential(nn.AdaptiveAvgPool2d(1), Flatten())
def simple_cnn(n_classes, actns, kernel_szs, strides, bn=False):
kernel_szs = listify(kernel_szs, len(actns)-1)
strides = listify(strides , len(actns)-1)
layers = [conv2_relu(actns[i], actns[i+1], kernel_szs[i], stride=strides[i], bn=bn)
for i in range(len(strides))]
layers += [PoolFlatten(), nn.Linear(actns[-1], n_classes)]
return nn.Sequential(*layers)
def conv2_relu(nif, nof, ks, stride, bn=False):
layers = [nn.Conv2d(nif, nof, ks, stride, padding=ks//2), nn.ReLU()]
if bn: layers.append(nn.BatchNorm2d(nof))
return nn.Sequential(*layers)
def get_model(): return simple_cnn(c, [3,16,16,16], 3, 2, bn=False)
def loss_batch(model, xb, yb, loss_fn, opt=None):
loss = loss_fn(model(xb), yb)
if opt is not None:
loss.backward()
opt.step()
opt.zero_grad()
return loss.item(), len(xb)
def fit(epochs, model, loss_fn, opt, train_dl, valid_dl):
for epoch in trange(epochs):
model.train()
for xb,yb in train_dl:
loss,_ = loss_batch(model, xb, yb, loss_fn, opt)
model.eval()
with torch.no_grad():
losses,nums = zip(*[loss_batch(model, xb, yb, loss_fn)
for xb,yb in valid_dl])
val_loss = np.sum(np.multiply(losses,nums)) / np.sum(nums)
print(epoch, val_loss)
```
## Here
```
model = get_model().cuda()
opt = optim.Adam(model.parameters(), 1e-3, betas=(0.9,0.99), weight_decay=1e-5)
fit(2, model, F.cross_entropy, opt, train_dl, valid_dl)
model.eval()
with torch.no_grad():
*val_metrics,nums = zip(*[loss_batch(model, xb, yb, F.cross_entropy)
for xb,yb in valid_dl])
val_metrics = [np.sum(np.multiply(val,nums)) / np.sum(nums) for val in val_metrics]
val_metrics
torch.save(model.state_dict(),PATH/'model1.pt')
```
## Eval
```
model1 = get_model().cuda()
model1.load_state_dict(torch.load(PATH/'model1.pt'))
model1.eval()
with torch.no_grad():
*val_metrics,nums = zip(*[loss_batch(model1, xb, yb, F.cross_entropy)
for xb,yb in valid_dl])
val_metrics = [np.sum(np.multiply(val,nums)) / np.sum(nums) for val in val_metrics]
val_metrics
model1.eval()
with torch.no_grad():
*val_metrics,nums = zip(*[loss_batch(model1, xb, yb, F.cross_entropy)
for xb,yb in valid_dl])
val_metrics = [np.sum(np.multiply(val,nums)) / np.sum(nums) for val in val_metrics]
val_metrics
```
|
github_jupyter
|
%matplotlib inline
%reload_ext autoreload
%autoreload 2
import sys
sys.path.append("..")
import torchvision.transforms as ttfs
import torch, PIL
import torch.nn as nn, torch.nn.functional as F, numpy as np, torch.optim as optim
from torch.utils.data import Dataset, DataLoader
from pathlib import Path
from torch import tensor
from tqdm import tnrange as trange, tqdm_notebook as tqdm
def pil2tensor(image):
arr = torch.ByteTensor(torch.ByteStorage.from_buffer(image.tobytes()))
arr = arr.view(image.size[1], image.size[0], -1)
arr = arr.permute(2,0,1)
return arr.float().div_(255)
class FilesDataset1(Dataset):
def __init__(self, fns, labels, classes=None):
if classes is None: classes = list(set(labels))
self.classes = classes
self.class2idx = {v:k for k,v in enumerate(classes)}
self.fns = np.array(fns)
self.y = [self.class2idx[o] for o in labels]
@classmethod
def from_folder(cls, folder, classes=None, test_pct=0.):
if classes is None: classes = [cls.name for cls in find_classes(folder)]
fns,labels = [],[]
for cl in classes:
fnames = get_image_files(folder/cl)
fns += fnames
labels += [cl] * len(fnames)
if test_pct==0.: return cls(fns, labels, classes=classes)
fns,labels = np.array(fns),np.array(labels)
is_test = np.random.uniform(size=(len(fns),)) < test_pct
return (cls(fns[~is_test], labels[~is_test], classes=classes),
cls(fns[is_test], labels[is_test], classes=classes))
def __len__(self): return len(self.fns)
def __getitem__(self,i):
x = PIL.Image.open(self.fns[i]).convert('RGB')
x = ttfs.Resize((sz,sz))(x)
x = pil2tensor(x)*2-1
return x,self.y[i]
def get_image_files(c):
return [o for o in list(c.iterdir())
if not o.name.startswith('.') and not o.is_dir()]
sz = 224
DATA_PATH = Path('../data')
PATH = DATA_PATH/'caltech101'
data_mean,data_std = map(tensor, ([0.5355,0.5430,0.5280], [0.2909,0.2788,0.2979]))
classes = ['airplanes','Motorbikes','Faces','watch','Leopards']
np.random.seed(42)
train_ds,valid_ds = FilesDataset1.from_folder(PATH, test_pct=0.2, classes=classes)
# train_ds = FilesDataset1.from_folder(PATH, classes=classes)
# valid_ds = FilesDataset1.from_folder(PATH, classes=classes)
classes = train_ds.classes
default_device = torch.device('cuda', 0)
c = len(classes)
c,len(train_ds),len(valid_ds)
class CudaDataLoader():
def __init__(self,dl): self.dl = dl
def __len__(self): return len(self.dl)
def __iter__(self):
for o in self.dl: yield o[0].cuda(), o[1].cuda()
train_dl = CudaDataLoader(DataLoader(train_ds, batch_size=64, shuffle=True, num_workers=8))
valid_dl = CudaDataLoader(DataLoader(valid_ds, batch_size=2*64, shuffle=False, num_workers=8))
from collections import Iterable
def listify(p=None, q=None):
"Makes p a list that looks like q"
if p is None: p=[]
elif not isinstance(p, Iterable): p=[p]
n = q if type(q)==int else 1 if q is None else len(q)
if len(p)==1: p = p * n
return p
class Lambda(nn.Module):
def __init__(self, func):
super().__init__()
self.func=func
def forward(self, x): return self.func(x)
def ResizeBatch(*size): return Lambda(lambda x: x.view((-1,)+size))
def Flatten(): return Lambda(lambda x: x.view((x.size(0), -1)))
def PoolFlatten(): return nn.Sequential(nn.AdaptiveAvgPool2d(1), Flatten())
def simple_cnn(n_classes, actns, kernel_szs, strides, bn=False):
kernel_szs = listify(kernel_szs, len(actns)-1)
strides = listify(strides , len(actns)-1)
layers = [conv2_relu(actns[i], actns[i+1], kernel_szs[i], stride=strides[i], bn=bn)
for i in range(len(strides))]
layers += [PoolFlatten(), nn.Linear(actns[-1], n_classes)]
return nn.Sequential(*layers)
def conv2_relu(nif, nof, ks, stride, bn=False):
layers = [nn.Conv2d(nif, nof, ks, stride, padding=ks//2), nn.ReLU()]
if bn: layers.append(nn.BatchNorm2d(nof))
return nn.Sequential(*layers)
def get_model(): return simple_cnn(c, [3,16,16,16], 3, 2, bn=False)
def loss_batch(model, xb, yb, loss_fn, opt=None):
loss = loss_fn(model(xb), yb)
if opt is not None:
loss.backward()
opt.step()
opt.zero_grad()
return loss.item(), len(xb)
def fit(epochs, model, loss_fn, opt, train_dl, valid_dl):
for epoch in trange(epochs):
model.train()
for xb,yb in train_dl:
loss,_ = loss_batch(model, xb, yb, loss_fn, opt)
model.eval()
with torch.no_grad():
losses,nums = zip(*[loss_batch(model, xb, yb, loss_fn)
for xb,yb in valid_dl])
val_loss = np.sum(np.multiply(losses,nums)) / np.sum(nums)
print(epoch, val_loss)
model = get_model().cuda()
opt = optim.Adam(model.parameters(), 1e-3, betas=(0.9,0.99), weight_decay=1e-5)
fit(2, model, F.cross_entropy, opt, train_dl, valid_dl)
model.eval()
with torch.no_grad():
*val_metrics,nums = zip(*[loss_batch(model, xb, yb, F.cross_entropy)
for xb,yb in valid_dl])
val_metrics = [np.sum(np.multiply(val,nums)) / np.sum(nums) for val in val_metrics]
val_metrics
torch.save(model.state_dict(),PATH/'model1.pt')
model1 = get_model().cuda()
model1.load_state_dict(torch.load(PATH/'model1.pt'))
model1.eval()
with torch.no_grad():
*val_metrics,nums = zip(*[loss_batch(model1, xb, yb, F.cross_entropy)
for xb,yb in valid_dl])
val_metrics = [np.sum(np.multiply(val,nums)) / np.sum(nums) for val in val_metrics]
val_metrics
model1.eval()
with torch.no_grad():
*val_metrics,nums = zip(*[loss_batch(model1, xb, yb, F.cross_entropy)
for xb,yb in valid_dl])
val_metrics = [np.sum(np.multiply(val,nums)) / np.sum(nums) for val in val_metrics]
val_metrics
| 0.656438 | 0.831588 |
# Imports
```
import numpy as np # Linear Algebra
import pandas as pd # Data Processing
pd.set_option('display.max_rows', 500)
pd.set_option('display.max_columns', 500)
pd.set_option('display.width', 1000)
import datetime as dt # DateTime Manipulation
from geopy.distance import great_circle, vincenty # geospatial manipulations
from shapely.geometry import MultiPoint
from sklearn.cluster import DBSCAN
%matplotlib inline
import matplotlib.pyplot as plt # visualizations
import seaborn as sns
import plotly.express as px
from sklearn.model_selection import train_test_split as tts # data splitting
import category_encoders as ce # preprocessing
from sklearn.impute import SimpleImputer
from sklearn.experimental import enable_iterative_imputer
from sklearn.impute import IterativeImputer
from sklearn.preprocessing import StandardScaler, Imputer
from sklearn.decomposition import PCA
from sklearn.linear_model import LinearRegression, LogisticRegression # models
from sklearn.ensemble import GradientBoostingRegressor as GBR
from sklearn.tree import DecisionTreeRegressor
from sklearn.pipeline import make_pipeline # pipeline engineering
from sklearn.model_selection import GridSearchCV, RandomizedSearchCV # hyperparameter tuning
from sklearn.metrics import mean_absolute_error, r2_score, mean_squared_error # model scoring
import datetime as dt
```
# Functions
```
def reduce_features(df):
cols = ['X', 'Y', 'incident_id']
df = df.drop(columns=cols)
return df
def drop_null_cols(df):
columns_with_null = df.isnull().sum().sort_values(ascending=False)[:9].index.tolist()
df = df.drop(columns=columns_with_null)
return df
def extract_timestamp_feats(df, column='call_timestamp'):
# Cast the column to DateTime type
df[column] = pd.to_datetime(df[column])
# Extract date related features
df['year'] = df[column].dt.year
df['month'] = df[column].dt.month
df['day'] = df[column].dt.day
df['dow'] = df[column].dt.dayofweek
df['week'] = df[column].dt.week
# Extract time related features
df['hour'] = df[column].dt.hour
"""
Extacts the time of day from the hour value by dividing by the hour knife
1 = Morning (0400 - 1000h)
2 = Midday (1000 - 1600h)
3 = Evening (1600 - 2200h)
4 = Night (2200 - 0400h)
"""
hour_knife = 6
df['part_of_day'] = ((df['hour'] + 2) / hour_knife).astype(int)
df['part_of_day'] = df['part_of_day'].replace(0, 4)
# Drop the now redundant column
df = df.drop(columns=column)
return df
def filter_geo_outliers(df):
"""
Filters out the outliers in the data where the latitude and longitudes
exist outside the geographic limits of the city of Detroit
"""
n_bound = 42.46
e_bound = -82.91
s_bound = 42.25
w_bound = -83.28
# Retains only those observations within the geographic boundaries
df = df.loc[((df['longitude'] > w_bound) & (df['longitude'] < e_bound)) &
((df['latitude'] > s_bound) & (df['latitude'] < n_bound ))]
return df
def plot_lat_lon(df):
fig, ax = plt.subplots(figsize=(12, 8))
# img = plt.imread(r"C:\Users\btros\Downloads\detroit_map.jpg")
# ax.imshow(img)
ax = sns.scatterplot(data=df, x='longitude', y='latitude', alpha=0.1)
# ax = sns.scatterplot(x=[1,2,3], y=[2,4,6], alpha=0.1)
ax.set_facecolor('.2')
fig.set_facecolor('.2')
return plt.show();
def remove_categoricals(df):
unneeded = df.select_dtypes(exclude="number").nunique().index.tolist()
unneeded.remove('calldescription')
df = df.drop(columns=unneeded)
return df
def clean_observations(df):
"""
Removing observations where the call description
is not related to emergency services
"""
col = 'calldescription'
unrelated_calls = ['START OF SHIFT INFORMATION', 'CALL BACK DESK', 'REMARKS',
'EMPLOYEE CALL IN / TIME OFF', 'NOTIFICATION(S) MADE',
'TIBURON HISTORICAL DATE', 'BLUE LIGHT PHONE MALF']
for call in unrelated_calls:
df = df.loc[df[col] != call]
return df
def filter_calldescription(df, col='calldescription'):
# List of call descriptions according to which service responded
police = ['TRAFFIC STOP', 'SPECIAL ATTENTION', 'DISTURBANCE', 'INVESTIGATE PERSON',
'ASSAULT AND BATTERY', 'UNKNOWN PROBLEM', 'TOWING DETAIL', 'FELONIOUS ASSAULT IP',
'LARCENY REPORT', 'AUTO X UNK INJ / IMPAIRED', 'BUS BOARDING', 'PERSON WITH WEAPON',
'VERIFIED ALR / PERSON W/O CODE', 'UDAA REPORT', 'BURGLARY OCCUPIED RESD I/P',
'SHOTS FIRED IP', 'INVESTIGATE AUTO', 'MALICIOUS DESTRUCTION I/P', 'MISCELLANEOUS TRAFFIC',
'PANIC / DURESS ALARM', 'FRAUD REPORT', 'BUILDING CHECK', 'VICIOUS ANIMAL', 'LARCENY I/P OR J/H',
'THREATS REPORT', 'RECOVER AUTO', 'HOLD UP ALARM AND MOW', 'MALICIOUS DESTRUCTION RPT',
'BURGLARY OTHER', 'PARKING COMPLAINT', 'WELL BEING CHECK', 'MENTAL VIOLENT NOT ARMED',
'ASSIST OTHER', 'INFORMATION/NON-CRIMINAL RPT', 'ASSAULT AND BATTERY REPORT', 'AUTO X REPORT',
'BURGLARY OCCUPANT AWAY I/P', 'MISSING REPORT', 'NARCOTICS I/P', 'BURGLARY RESIDENCE REPORT',
'MISSING SERIOUS', 'ALARM UNKNOWN CAUSE', 'SUICIDE THREAT', 'CHILD / ADULT ABUSE', 'WNTD WRRNT FELONY',
'ADMIT OR E/E', 'FELONIOUS ASSAULT JH', 'ASSIST CITIZEN', 'SPECIAL ATTENTION / DETAIL', 'DEAD PERSON OBSERVED',
'LOST PROPERTY', 'SUICIDE I/P', 'TRANSPORT PRISONER', 'ROBBERY ARMED IP-ANY', 'RECOVERED / FOUND PROPERTY',
'SAFEWALK', 'CHILD / ADULT ABUSE REPORT', 'BREAKING AND ENTERING AUTO I/P', 'FELONIOUS ASSAULT REPORT',
'RAID - EXECUTE SEARCH WARRANT', 'MENTAL VIOLENT - ARMED', 'RAPE REPORT', 'HARASSMENT REPORT',
'BURGLARY BUSINESS IP', 'KIDNAPPING', 'PPO VIOLATION I/P', 'LEWD AND LASCIVIOUS IP', 'AID MOTORIST / CHILD LOCKED IN',
'BREAKING & ENTERING AUTO RPT', 'DV A/B I/P-J/H', 'ROBBERY NOT ARMED I/P', 'SQUATTER DISTURBANCE',
'SCRAP STRIP BLDG IP', 'ASSAULT AND BATTERY I/P', 'AUTO OR PED H&R REPORT', 'FRAUD I/P',
'AUTO X HIT & RUN W/ INJ', 'BURGLARY OCCUP RESD REPT', 'RUBBISH LITTERING I/P', 'SHOTS J/H, EVIDENCE, REPT',
'BE ON THE LOOK OUT', 'SHOOTING/CUTTING/PENT WND RPT', 'HOLDING PERSON', 'ARSON I/P', 'BURGLARY OTHER REPORT',
'ANIMAL COMPLAINT', 'ARSON REPORT', 'ASSIST PERSONNEL', 'ROBBERY ARMED REPORT', 'VERIFY RETURN OF MISSING',
'DDOT TROUBLE', 'WNTD WRRNT MISDEMEANOR', 'ROBBERY ARMED JH-ANY WEAPON', 'BURGLARY OCCUPANT AWAY J/H',
'UDAA I/P', 'SCHOOL CROSSING', 'HAZARDOUS CONDITIONS', 'SENIOR CITIZEN ASSIST', 'ROBBERY NOT ARMED REPORT',
'ONE OVER THE WHEEL', 'RAPE IP OR JH', 'PROPERTY DAMAGE NON-CRIMINAL', 'PEACE OFFICER DETAIL', 'BURGLARY BUSINESS REPORT',
'BURGLARY BUSINESS J/H', 'CHILD(REN) HOME ALONE', 'DV F/A I/P-J/H', 'ESCORT', 'NARCOTICS REPORT', 'AID MOTORIST MISC',
'PPO VIOLATION REPORT', 'RECOVER AUTO FELONY', 'LEWD AND LASCIVIOUS','VEHICLE FIRE', 'NOISE COMPLAINT',
'FOUND PERSON', 'MOLESTATION REPORT', 'PBT TEST', 'RESIDENTIAL STRUCTURE FIRE', 'ASSIST REF GAINING ENTRY',
'ATM ALARM', 'WSPD - BURGLARY ALR', 'ALTERED LOC OR UNK PROBLEM', 'PAST GUNSHOT OR STABBINIG', 'AID MORTORIST MISC',
'BOMB THREAT REPORT', 'ASSAULT NOT DANGEROUS OR PREV', 'WRKABLE ARRST/OBV OR EXP DEATH',
'CITIZEN RADIO PATROL IN TROUBL', 'SCHOOL THREATS J/H & RPT', 'TRAFFIC W/ NO INJURIES', 'MISCELLANEOUS ACCIDENT',
'MOLESTATION', 'BANK ALARM','KIDNAPPING REPORT', 'K-9 DEPLOYMENT', 'SUSPICIOUS PACKAGE', 'DPDA',
'WSPD - BURGLARY ALARM', 'VIP THREATS I/P', 'PERSON W/ A WEAPON REPORT', 'AID MOTORIST CHILD LOCKED INSD',
'ANIMAL FIGHT', 'BOMB THREAT', 'PERSONNEL IN TROUBLE', 'BURGLARY ALARM', 'DV PRS WITH A WEAP I/P-J/H',
'ASSAULT AND BATTERY J/H', 'AUTO X - BLDG / DWELL', 'PURSUIT - VEHICLE OR FOOT', 'BACKGROUND/LEIN CHK / LIVESCAN',
'INVESTIGATE YOUTH(S)', 'SHOT STABBED UNK OR OBV DEATH', 'SCRAP STRIP BLDG JH', 'TRANSPORT PRISONER-OTH AGT', 'BOMB SCARE',
'SMOKING VIOLATIONS', 'DV A/B RPT', 'HPPD BURG ALRM', 'EXTORTION IP', 'EXTORTION JH OR REPORT',
'OTHR OUTSIDE STRUCTURE FIRE', 'ATTEMPTED SUICIDE REPORT', 'VIP THREATS J/H OR REPORT', 'ASSAULT DANGEROUSOR SERIOUS',
'TRAFFIC- MINOR INJURIES', 'SUICIDE THREAT OR ABNORM BEHAV', 'BURGLARY ALARM W/ MOW', 'HPPD BURG ALRM W/ MOW',
'BACKGROUND CHECK / LIVESCAN', 'DV F/A RPT', 'CURFEW VIOLATION', 'YOUTH LOITERING/CONGREGATING']
ems = ['ONE DOWN OR OVER THE WHEEL', 'AUTO X OR PED X - INJURIES', 'AUTO X HIT& RUN UNK INJURIES',
'MENTAL VIOLENT NOT ARMED', 'MENTAL NOT VIOLENT', 'SHOOTING/CUTTING/PENT WOUND', 'MT EMS-TRO/ENTRY',
'ONE DOWN/DRUG OD', 'ANIMAL BITES OR ATTACK DELTA', 'SHOT OR STABBED DELTA', 'PREGNANCY OR CHILDBIRTH DELTA',
'CHEST PAIN DELTA', 'ATYPICAL SEIZURE OR HX OR CVA', 'OB HEMORRHAGE LABOR OR BIRTH',
'CHEST PAIN NORMAL BREATHING', 'INJURY NON DANGEROUS BODY PART', 'NON SUICIDAL OR NT THRTENING',
'SICK PERSON DELTA', 'ANIMAL BITE OR ATTACK DELTA', 'FALL W SERIOUS HEMORRHAGE', 'CONT OR MULTI SEIZURES DELTA',
'PORTABLE ALARM SYSTEM', 'FAINTING ALERT', 'SERIOUS HEMORRHAGE', 'BURNS DELTA OR ECHO', 'HEADACHE - ADDITIONAL SYMPTONS',
'NON RECENT OR SUPERFICIAL BITE', 'ASSAULT OR SEX ASSAULT DELTA', 'UNCONSCIOUS OR FAINTING DELTA', 'DV PRSN WITH WEAP RPT',
'SCHOOL THREATS I/P', 'SICK NON PRIORITY COMPLAINTS', 'SUICIDE ATTEMPT DELTA', 'DANGEROUS HEMORRHAGE DELTA',
'SERIOUS INJURIES', 'MINOR HEMORRHAGE', 'BREATHING PROBLEMS DELTA', 'CHEST PAIN W OR WO NOR BRETH', 'OVERDOSE DELTA',
'OVER THE WHEEL', 'TRAFFIC INCIDENT W/ INJURIES', 'POSS DANGEROUS ANIMAL BITE', 'ONE DOWN/OVERDOSE',
'MAJOR TRAFFIC INCIDENT DELTA', 'OVERDOSE NT ALRT OR UNK STATUS', 'MEDICAL ALARM OR UNK PROB','LIFE STATUS QUESTIONABLE DELTA']
fire = ['FIRE ALARM TEST', 'FIRE ALARM', 'TEMPERATURE ALARM', 'ELEVATOR ENTRAPMENT', 'VEHICLE FIRE(FIELD)',
'IRRADIATOR ROOM ALARM / TEST', 'HAZARD CONDITIONS NO FIRE', 'GRASS FIRE', 'POWER LINES', 'ALR PT DISABLED / TIMEZONE CHG',
'EXPLOSION/RUPTURE NO FIRE', 'ALARM MALFUNCTION', 'COMMERCIAL STRUCTURE FIRE', 'EXPLOSION', 'FIRE ALARMS ALL',
'WEATHER RELATED', 'GRASS OR RUBBISH FIRE', 'SMOKE INVESTIGATION', 'WSPD - FIRES', 'ALARM MISUSE']
df['police'] = np.where(df[col].isin(police), 1, 0)
df['ems'] = np.where(df[col].isin(ems), 2, 0)
df['fire'] = np.where(df[col].isin(fire), 3, 0)
df['category'] = df['fire'] + df['ems'] + df['police']
df['category'] = df['category'].replace(0, 1) # For some reason the Assault and Battery instances are not picked up
defunct = ['police', 'ems', 'fire', col]
# Drop the no longer needed columns
df = df.drop(columns=defunct)
return df
def police_only(df):
# Retains only those observations which contain calls for police assistance
df = df.loc[df.category == 1]
df = df.drop(columns='category')
return df
def apply_geospatial_grid(df):
lat = 'latitude'
lon = 'longitude'
# episolon is used to avoid assigning locations outside the grid
epsilon = .0001
# min and max of latitude and longitude
lat_min = df[lat].min() - epsilon
lat_max = df[lat].max() + epsilon
lon_min = df[lon].min() - epsilon
lon_max = df[lon].max() + epsilon
# ranges
lat_range = lat_max - lat_min
lon_range = lon_max - lon_min
# splits
lat_knife = 10
lon_knife = 15
# grid lengths
lon_length = lon_range / lon_knife
lat_length = lat_range / lat_knife
# assigining the locations to a grid space
df['lat_grid'] = (np.floor(((df[lat]-lat_min)/lat_length)))
df['lon_grid'] = (np.floor(((df[lon]-lon_min)/lon_length)))
# discard the now unneeded columns
cols = [lat, lon]
df = df.drop(columns=cols)
return df
def to_heatmap_format(df):
grid_df = df.groupby(['lat_grid','lon_grid']).size().reset_index(name='call_count')
geoheatmap_df = grid_fg.pivot(index='lat_grid',columns='lon_grid', values='call_count')
return geoheatmap_df
def plot_calls_grid(df):
fig, ax = plt.subplots(figsize=(20,8))
plt.title("Detroit 911 Calls by Geographic Location Halloween 2018",
fontsize=24)
ax = sns.heatmap(df,annot=True,fmt=".0f",cbar=False)
# ax.set_ylim(len('lat_grid'), -1, -1)
fig.set_facecolor('.1')
ax.set_facecolor('.1')
ax.invert_yaxis()
return plt.show();
def group_features(df):
# groups the dataframe by various DateTime params
grouping_list = list(df.columns)
grouping_list.remove('oid')
grouping_list.remove('hour')
grouped_df = df.groupby(grouping_list).size().reset_index(name='count')
return grouped_df
def wrangle(df):
df = reduce_features(df)
df = drop_null_cols(df)
df = extract_timestamp_feats(df)
df = filter_geo_outliers(df)
df = remove_categoricals(df)
df = clean_observations(df)
df = filter_calldescription(df)
df = police_only(df)
df = apply_geospatial_grid(df)
df = group_features(df)
return df
```
# Load Data
```
PATH = r"C:\\Users\\btros\\OneDrive\\Documents\\LSDS-Unit-2-Project\\raw_csvs"
file = r"\\911_Calls_for_Service.csv"
calls = pd.read_csv(PATH+file, low_memory=False)
# Don't forget to make a copy
df_copy = calls
calls.columns
weather_file = r'\\detroit_weather_wrangled.csv'
weather = pd.read_csv(PATH+weather_file)
weather_copy = weather
weather = weather.drop(columns='Unnamed: 0')
holidays = pd.read_csv(PATH+r'\holidays.csv')
calls = wrangle(calls)
calls
weather
holidays = holidays.drop(columns='Unnamed: 0')
holidays
merged = pd.merge(calls, weather, how='inner', on=['year', 'month', 'day', 'part_of_day'])
merged2 = pd.merge(merged, holidays, how='left', on=['year', 'month', 'day'])
merged2.is_holiday = merged2.is_holiday.replace(np.NaN, 0)
merged.to_csv('call_grid_with_weather.csv')
## take a quick peak
merged2.head(1)
# then wrangle the data so we can start where we left off on the previous notebook
df = df_copy
df = wrangle(df)
df.head()
df.tail()
```
# Training and Validation Splits, Feature Selection
```
int(len(df)*.8)
df.iloc[1015803]
train = merged2.loc[((merged2.year < 2019) | ((merged2.year == 2019) & (merged2.month < 8)))]
len(train)
val = merged2.loc[((merged2.year == 2019) & (merged2.month >= 8))]
len(val)
len(val) / len(train)
target = 'count'
features = merged2.columns.tolist()
features.remove(target)
X_train = train[features]
X_val = val[features]
y_train = train[target]
y_val = val[target]
```
# First model
Let's go with a simple linear regression model on the first run through
```
model = LinearRegression()
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
```
### Initial model grouped data by call_minute which was a mistake
My initial hypotheses however is that the combination of a low r-squared value
coupled with low train and testing errors is due to the fact that the datetime values inherently
contain little variance and that the geographic trends are so strong that they overcome this
failure of the model to explain variance
# Without weather data
```
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
```
# With Weather Data
```
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
```
# With Holidays flagged
```
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
model.score(X_val, y_val)
train_base, val_base = calc_accuracy_score(y_train), calc_accuracy_score(y_val)
print(f"Training model baseline accuracy: {(train_base)*100:,.2f}%" + "\n")
print(f"Validation model baseline accuracy: {(val_base)*100:,.2f}%")
```
# Logistic Regression with Weather Only
```
model = LogisticRegression(solver='lbfgs', max_iter=300, multi_class='auto')
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
model.score(X_val, y_val)
```
# Holidays Flagged
```
y_train_pred = model.predict(X_train)
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
model.score(X_val, y_val)
base = [y_val.value_counts().index[0]] * len(y_val)
mse = mean_squared_error(y_val, base)
mse
def calc_accuracy_score(target_feature):
majority_class = target_feature.mode()[0] # Find the mode of the target feature
target_pred = [majority_class] * len(target_feature) # Create a list the length of the target feature with consisting of the mode as the sole value
score = accuracy_score(target_feature, target_pred)
return score
train_base, val_base = calc_accuracy_score(y_train), calc_accuracy_score(y_val)
print(f"Training model baseline accuracy: {(train_base)*100:,.2f}%" + "\n")
print(f"Validation model baseline accuracy: {(val_base)*100:,.2f}%")
```
### Grouping by call_minute error fixed
Metrics still seem to support my initial hypothesis however we will try some different models and see what happens
```
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val.value_counts()
len(y_val == y_val_pred)
```
# Decision Tree Regressor
```
model = DecisionTreeRegressor()
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
```
# With Weather Data
```
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
```
# Random Forest Regressor
```
from sklearn.ensemble import RandomForestRegressor
model = RandomForestRegressor(n_estimators=100,
n_jobs=-1)
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
```
## Default estimators
```
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
print(model.score(X_val, y_val))
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
```
## 400 trees
```
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
4682
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
```
# With Weather Data
```
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
merged['count'].value_counts(normalize=True)
```
# XGBRegressor
```
from xgboost import XGBRegressor
model = XGBRegressor(n_estimators=400, random_state=42, n_jobs=-1)
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
```
# With Weather Data
```
y_train_pred = model.predict(X_train)
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
```
# Moving Forward With Random Forest Regressor
```
!pip install eli5
import eli5
from eli5.sklearn import PermutationImportance
model = RandomForestRegressor(n_estimators=400,
n_jobs=-1)
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
```
# Permutation Importances
```
permuter = PermutationImportance(
model,
scoring='neg_mean_absolute_error',
n_iter=5,
random_state=42
)
permuter.fit(X_val, y_val)
feature_names = X_val.columns.tolist()
pd.Series(permuter.feature_importances_, feature_names).sort_values(ascending=False)
```
# Partial Dependence Plots
```
fig, ax = plt.subplots(figsize=(10,8))
ax = sns.distplot(y_train)
ax.set_xlim(0, 20)
!pip install pdpbox
from pdpbox.pdp import pdp_isolate, pdp_plot
plt.rcParams['figure.dpi'] = 150
feature = 'part_of_day'
isolated = pdp_isolate(
model=model,
dataset=X_val,
model_features=X_val.columns,
feature=feature
)
pdp_plot(isolated, feature_name=feature, plot_lines=True, frac_to_plot=100)
from pdpbox.pdp import pdp_interact, pdp_interact_plot
features = ['part_of_day', 'call_dow']
interaction = pdp_interact(
model=model,
dataset=X_val,
model_features=X_val.columns,
features=features
)
pdp_interact_plot(interaction, plot_type='grid', feature_names=features);
```
# Shaply Plots
```
!pip install shap
X_val.iloc[1]
import shap
import random as rd
row=X_val.iloc[1]
explainer = shap.TreeExplainer(model)
shap_values = explainer.shap_values(row)
shap.initjs()
shap.force_plot(
base_value=explainer.expected_value,
shap_values=shap_values,
features=row
)
```
|
github_jupyter
|
import numpy as np # Linear Algebra
import pandas as pd # Data Processing
pd.set_option('display.max_rows', 500)
pd.set_option('display.max_columns', 500)
pd.set_option('display.width', 1000)
import datetime as dt # DateTime Manipulation
from geopy.distance import great_circle, vincenty # geospatial manipulations
from shapely.geometry import MultiPoint
from sklearn.cluster import DBSCAN
%matplotlib inline
import matplotlib.pyplot as plt # visualizations
import seaborn as sns
import plotly.express as px
from sklearn.model_selection import train_test_split as tts # data splitting
import category_encoders as ce # preprocessing
from sklearn.impute import SimpleImputer
from sklearn.experimental import enable_iterative_imputer
from sklearn.impute import IterativeImputer
from sklearn.preprocessing import StandardScaler, Imputer
from sklearn.decomposition import PCA
from sklearn.linear_model import LinearRegression, LogisticRegression # models
from sklearn.ensemble import GradientBoostingRegressor as GBR
from sklearn.tree import DecisionTreeRegressor
from sklearn.pipeline import make_pipeline # pipeline engineering
from sklearn.model_selection import GridSearchCV, RandomizedSearchCV # hyperparameter tuning
from sklearn.metrics import mean_absolute_error, r2_score, mean_squared_error # model scoring
import datetime as dt
def reduce_features(df):
cols = ['X', 'Y', 'incident_id']
df = df.drop(columns=cols)
return df
def drop_null_cols(df):
columns_with_null = df.isnull().sum().sort_values(ascending=False)[:9].index.tolist()
df = df.drop(columns=columns_with_null)
return df
def extract_timestamp_feats(df, column='call_timestamp'):
# Cast the column to DateTime type
df[column] = pd.to_datetime(df[column])
# Extract date related features
df['year'] = df[column].dt.year
df['month'] = df[column].dt.month
df['day'] = df[column].dt.day
df['dow'] = df[column].dt.dayofweek
df['week'] = df[column].dt.week
# Extract time related features
df['hour'] = df[column].dt.hour
"""
Extacts the time of day from the hour value by dividing by the hour knife
1 = Morning (0400 - 1000h)
2 = Midday (1000 - 1600h)
3 = Evening (1600 - 2200h)
4 = Night (2200 - 0400h)
"""
hour_knife = 6
df['part_of_day'] = ((df['hour'] + 2) / hour_knife).astype(int)
df['part_of_day'] = df['part_of_day'].replace(0, 4)
# Drop the now redundant column
df = df.drop(columns=column)
return df
def filter_geo_outliers(df):
"""
Filters out the outliers in the data where the latitude and longitudes
exist outside the geographic limits of the city of Detroit
"""
n_bound = 42.46
e_bound = -82.91
s_bound = 42.25
w_bound = -83.28
# Retains only those observations within the geographic boundaries
df = df.loc[((df['longitude'] > w_bound) & (df['longitude'] < e_bound)) &
((df['latitude'] > s_bound) & (df['latitude'] < n_bound ))]
return df
def plot_lat_lon(df):
fig, ax = plt.subplots(figsize=(12, 8))
# img = plt.imread(r"C:\Users\btros\Downloads\detroit_map.jpg")
# ax.imshow(img)
ax = sns.scatterplot(data=df, x='longitude', y='latitude', alpha=0.1)
# ax = sns.scatterplot(x=[1,2,3], y=[2,4,6], alpha=0.1)
ax.set_facecolor('.2')
fig.set_facecolor('.2')
return plt.show();
def remove_categoricals(df):
unneeded = df.select_dtypes(exclude="number").nunique().index.tolist()
unneeded.remove('calldescription')
df = df.drop(columns=unneeded)
return df
def clean_observations(df):
"""
Removing observations where the call description
is not related to emergency services
"""
col = 'calldescription'
unrelated_calls = ['START OF SHIFT INFORMATION', 'CALL BACK DESK', 'REMARKS',
'EMPLOYEE CALL IN / TIME OFF', 'NOTIFICATION(S) MADE',
'TIBURON HISTORICAL DATE', 'BLUE LIGHT PHONE MALF']
for call in unrelated_calls:
df = df.loc[df[col] != call]
return df
def filter_calldescription(df, col='calldescription'):
# List of call descriptions according to which service responded
police = ['TRAFFIC STOP', 'SPECIAL ATTENTION', 'DISTURBANCE', 'INVESTIGATE PERSON',
'ASSAULT AND BATTERY', 'UNKNOWN PROBLEM', 'TOWING DETAIL', 'FELONIOUS ASSAULT IP',
'LARCENY REPORT', 'AUTO X UNK INJ / IMPAIRED', 'BUS BOARDING', 'PERSON WITH WEAPON',
'VERIFIED ALR / PERSON W/O CODE', 'UDAA REPORT', 'BURGLARY OCCUPIED RESD I/P',
'SHOTS FIRED IP', 'INVESTIGATE AUTO', 'MALICIOUS DESTRUCTION I/P', 'MISCELLANEOUS TRAFFIC',
'PANIC / DURESS ALARM', 'FRAUD REPORT', 'BUILDING CHECK', 'VICIOUS ANIMAL', 'LARCENY I/P OR J/H',
'THREATS REPORT', 'RECOVER AUTO', 'HOLD UP ALARM AND MOW', 'MALICIOUS DESTRUCTION RPT',
'BURGLARY OTHER', 'PARKING COMPLAINT', 'WELL BEING CHECK', 'MENTAL VIOLENT NOT ARMED',
'ASSIST OTHER', 'INFORMATION/NON-CRIMINAL RPT', 'ASSAULT AND BATTERY REPORT', 'AUTO X REPORT',
'BURGLARY OCCUPANT AWAY I/P', 'MISSING REPORT', 'NARCOTICS I/P', 'BURGLARY RESIDENCE REPORT',
'MISSING SERIOUS', 'ALARM UNKNOWN CAUSE', 'SUICIDE THREAT', 'CHILD / ADULT ABUSE', 'WNTD WRRNT FELONY',
'ADMIT OR E/E', 'FELONIOUS ASSAULT JH', 'ASSIST CITIZEN', 'SPECIAL ATTENTION / DETAIL', 'DEAD PERSON OBSERVED',
'LOST PROPERTY', 'SUICIDE I/P', 'TRANSPORT PRISONER', 'ROBBERY ARMED IP-ANY', 'RECOVERED / FOUND PROPERTY',
'SAFEWALK', 'CHILD / ADULT ABUSE REPORT', 'BREAKING AND ENTERING AUTO I/P', 'FELONIOUS ASSAULT REPORT',
'RAID - EXECUTE SEARCH WARRANT', 'MENTAL VIOLENT - ARMED', 'RAPE REPORT', 'HARASSMENT REPORT',
'BURGLARY BUSINESS IP', 'KIDNAPPING', 'PPO VIOLATION I/P', 'LEWD AND LASCIVIOUS IP', 'AID MOTORIST / CHILD LOCKED IN',
'BREAKING & ENTERING AUTO RPT', 'DV A/B I/P-J/H', 'ROBBERY NOT ARMED I/P', 'SQUATTER DISTURBANCE',
'SCRAP STRIP BLDG IP', 'ASSAULT AND BATTERY I/P', 'AUTO OR PED H&R REPORT', 'FRAUD I/P',
'AUTO X HIT & RUN W/ INJ', 'BURGLARY OCCUP RESD REPT', 'RUBBISH LITTERING I/P', 'SHOTS J/H, EVIDENCE, REPT',
'BE ON THE LOOK OUT', 'SHOOTING/CUTTING/PENT WND RPT', 'HOLDING PERSON', 'ARSON I/P', 'BURGLARY OTHER REPORT',
'ANIMAL COMPLAINT', 'ARSON REPORT', 'ASSIST PERSONNEL', 'ROBBERY ARMED REPORT', 'VERIFY RETURN OF MISSING',
'DDOT TROUBLE', 'WNTD WRRNT MISDEMEANOR', 'ROBBERY ARMED JH-ANY WEAPON', 'BURGLARY OCCUPANT AWAY J/H',
'UDAA I/P', 'SCHOOL CROSSING', 'HAZARDOUS CONDITIONS', 'SENIOR CITIZEN ASSIST', 'ROBBERY NOT ARMED REPORT',
'ONE OVER THE WHEEL', 'RAPE IP OR JH', 'PROPERTY DAMAGE NON-CRIMINAL', 'PEACE OFFICER DETAIL', 'BURGLARY BUSINESS REPORT',
'BURGLARY BUSINESS J/H', 'CHILD(REN) HOME ALONE', 'DV F/A I/P-J/H', 'ESCORT', 'NARCOTICS REPORT', 'AID MOTORIST MISC',
'PPO VIOLATION REPORT', 'RECOVER AUTO FELONY', 'LEWD AND LASCIVIOUS','VEHICLE FIRE', 'NOISE COMPLAINT',
'FOUND PERSON', 'MOLESTATION REPORT', 'PBT TEST', 'RESIDENTIAL STRUCTURE FIRE', 'ASSIST REF GAINING ENTRY',
'ATM ALARM', 'WSPD - BURGLARY ALR', 'ALTERED LOC OR UNK PROBLEM', 'PAST GUNSHOT OR STABBINIG', 'AID MORTORIST MISC',
'BOMB THREAT REPORT', 'ASSAULT NOT DANGEROUS OR PREV', 'WRKABLE ARRST/OBV OR EXP DEATH',
'CITIZEN RADIO PATROL IN TROUBL', 'SCHOOL THREATS J/H & RPT', 'TRAFFIC W/ NO INJURIES', 'MISCELLANEOUS ACCIDENT',
'MOLESTATION', 'BANK ALARM','KIDNAPPING REPORT', 'K-9 DEPLOYMENT', 'SUSPICIOUS PACKAGE', 'DPDA',
'WSPD - BURGLARY ALARM', 'VIP THREATS I/P', 'PERSON W/ A WEAPON REPORT', 'AID MOTORIST CHILD LOCKED INSD',
'ANIMAL FIGHT', 'BOMB THREAT', 'PERSONNEL IN TROUBLE', 'BURGLARY ALARM', 'DV PRS WITH A WEAP I/P-J/H',
'ASSAULT AND BATTERY J/H', 'AUTO X - BLDG / DWELL', 'PURSUIT - VEHICLE OR FOOT', 'BACKGROUND/LEIN CHK / LIVESCAN',
'INVESTIGATE YOUTH(S)', 'SHOT STABBED UNK OR OBV DEATH', 'SCRAP STRIP BLDG JH', 'TRANSPORT PRISONER-OTH AGT', 'BOMB SCARE',
'SMOKING VIOLATIONS', 'DV A/B RPT', 'HPPD BURG ALRM', 'EXTORTION IP', 'EXTORTION JH OR REPORT',
'OTHR OUTSIDE STRUCTURE FIRE', 'ATTEMPTED SUICIDE REPORT', 'VIP THREATS J/H OR REPORT', 'ASSAULT DANGEROUSOR SERIOUS',
'TRAFFIC- MINOR INJURIES', 'SUICIDE THREAT OR ABNORM BEHAV', 'BURGLARY ALARM W/ MOW', 'HPPD BURG ALRM W/ MOW',
'BACKGROUND CHECK / LIVESCAN', 'DV F/A RPT', 'CURFEW VIOLATION', 'YOUTH LOITERING/CONGREGATING']
ems = ['ONE DOWN OR OVER THE WHEEL', 'AUTO X OR PED X - INJURIES', 'AUTO X HIT& RUN UNK INJURIES',
'MENTAL VIOLENT NOT ARMED', 'MENTAL NOT VIOLENT', 'SHOOTING/CUTTING/PENT WOUND', 'MT EMS-TRO/ENTRY',
'ONE DOWN/DRUG OD', 'ANIMAL BITES OR ATTACK DELTA', 'SHOT OR STABBED DELTA', 'PREGNANCY OR CHILDBIRTH DELTA',
'CHEST PAIN DELTA', 'ATYPICAL SEIZURE OR HX OR CVA', 'OB HEMORRHAGE LABOR OR BIRTH',
'CHEST PAIN NORMAL BREATHING', 'INJURY NON DANGEROUS BODY PART', 'NON SUICIDAL OR NT THRTENING',
'SICK PERSON DELTA', 'ANIMAL BITE OR ATTACK DELTA', 'FALL W SERIOUS HEMORRHAGE', 'CONT OR MULTI SEIZURES DELTA',
'PORTABLE ALARM SYSTEM', 'FAINTING ALERT', 'SERIOUS HEMORRHAGE', 'BURNS DELTA OR ECHO', 'HEADACHE - ADDITIONAL SYMPTONS',
'NON RECENT OR SUPERFICIAL BITE', 'ASSAULT OR SEX ASSAULT DELTA', 'UNCONSCIOUS OR FAINTING DELTA', 'DV PRSN WITH WEAP RPT',
'SCHOOL THREATS I/P', 'SICK NON PRIORITY COMPLAINTS', 'SUICIDE ATTEMPT DELTA', 'DANGEROUS HEMORRHAGE DELTA',
'SERIOUS INJURIES', 'MINOR HEMORRHAGE', 'BREATHING PROBLEMS DELTA', 'CHEST PAIN W OR WO NOR BRETH', 'OVERDOSE DELTA',
'OVER THE WHEEL', 'TRAFFIC INCIDENT W/ INJURIES', 'POSS DANGEROUS ANIMAL BITE', 'ONE DOWN/OVERDOSE',
'MAJOR TRAFFIC INCIDENT DELTA', 'OVERDOSE NT ALRT OR UNK STATUS', 'MEDICAL ALARM OR UNK PROB','LIFE STATUS QUESTIONABLE DELTA']
fire = ['FIRE ALARM TEST', 'FIRE ALARM', 'TEMPERATURE ALARM', 'ELEVATOR ENTRAPMENT', 'VEHICLE FIRE(FIELD)',
'IRRADIATOR ROOM ALARM / TEST', 'HAZARD CONDITIONS NO FIRE', 'GRASS FIRE', 'POWER LINES', 'ALR PT DISABLED / TIMEZONE CHG',
'EXPLOSION/RUPTURE NO FIRE', 'ALARM MALFUNCTION', 'COMMERCIAL STRUCTURE FIRE', 'EXPLOSION', 'FIRE ALARMS ALL',
'WEATHER RELATED', 'GRASS OR RUBBISH FIRE', 'SMOKE INVESTIGATION', 'WSPD - FIRES', 'ALARM MISUSE']
df['police'] = np.where(df[col].isin(police), 1, 0)
df['ems'] = np.where(df[col].isin(ems), 2, 0)
df['fire'] = np.where(df[col].isin(fire), 3, 0)
df['category'] = df['fire'] + df['ems'] + df['police']
df['category'] = df['category'].replace(0, 1) # For some reason the Assault and Battery instances are not picked up
defunct = ['police', 'ems', 'fire', col]
# Drop the no longer needed columns
df = df.drop(columns=defunct)
return df
def police_only(df):
# Retains only those observations which contain calls for police assistance
df = df.loc[df.category == 1]
df = df.drop(columns='category')
return df
def apply_geospatial_grid(df):
lat = 'latitude'
lon = 'longitude'
# episolon is used to avoid assigning locations outside the grid
epsilon = .0001
# min and max of latitude and longitude
lat_min = df[lat].min() - epsilon
lat_max = df[lat].max() + epsilon
lon_min = df[lon].min() - epsilon
lon_max = df[lon].max() + epsilon
# ranges
lat_range = lat_max - lat_min
lon_range = lon_max - lon_min
# splits
lat_knife = 10
lon_knife = 15
# grid lengths
lon_length = lon_range / lon_knife
lat_length = lat_range / lat_knife
# assigining the locations to a grid space
df['lat_grid'] = (np.floor(((df[lat]-lat_min)/lat_length)))
df['lon_grid'] = (np.floor(((df[lon]-lon_min)/lon_length)))
# discard the now unneeded columns
cols = [lat, lon]
df = df.drop(columns=cols)
return df
def to_heatmap_format(df):
grid_df = df.groupby(['lat_grid','lon_grid']).size().reset_index(name='call_count')
geoheatmap_df = grid_fg.pivot(index='lat_grid',columns='lon_grid', values='call_count')
return geoheatmap_df
def plot_calls_grid(df):
fig, ax = plt.subplots(figsize=(20,8))
plt.title("Detroit 911 Calls by Geographic Location Halloween 2018",
fontsize=24)
ax = sns.heatmap(df,annot=True,fmt=".0f",cbar=False)
# ax.set_ylim(len('lat_grid'), -1, -1)
fig.set_facecolor('.1')
ax.set_facecolor('.1')
ax.invert_yaxis()
return plt.show();
def group_features(df):
# groups the dataframe by various DateTime params
grouping_list = list(df.columns)
grouping_list.remove('oid')
grouping_list.remove('hour')
grouped_df = df.groupby(grouping_list).size().reset_index(name='count')
return grouped_df
def wrangle(df):
df = reduce_features(df)
df = drop_null_cols(df)
df = extract_timestamp_feats(df)
df = filter_geo_outliers(df)
df = remove_categoricals(df)
df = clean_observations(df)
df = filter_calldescription(df)
df = police_only(df)
df = apply_geospatial_grid(df)
df = group_features(df)
return df
PATH = r"C:\\Users\\btros\\OneDrive\\Documents\\LSDS-Unit-2-Project\\raw_csvs"
file = r"\\911_Calls_for_Service.csv"
calls = pd.read_csv(PATH+file, low_memory=False)
# Don't forget to make a copy
df_copy = calls
calls.columns
weather_file = r'\\detroit_weather_wrangled.csv'
weather = pd.read_csv(PATH+weather_file)
weather_copy = weather
weather = weather.drop(columns='Unnamed: 0')
holidays = pd.read_csv(PATH+r'\holidays.csv')
calls = wrangle(calls)
calls
weather
holidays = holidays.drop(columns='Unnamed: 0')
holidays
merged = pd.merge(calls, weather, how='inner', on=['year', 'month', 'day', 'part_of_day'])
merged2 = pd.merge(merged, holidays, how='left', on=['year', 'month', 'day'])
merged2.is_holiday = merged2.is_holiday.replace(np.NaN, 0)
merged.to_csv('call_grid_with_weather.csv')
## take a quick peak
merged2.head(1)
# then wrangle the data so we can start where we left off on the previous notebook
df = df_copy
df = wrangle(df)
df.head()
df.tail()
int(len(df)*.8)
df.iloc[1015803]
train = merged2.loc[((merged2.year < 2019) | ((merged2.year == 2019) & (merged2.month < 8)))]
len(train)
val = merged2.loc[((merged2.year == 2019) & (merged2.month >= 8))]
len(val)
len(val) / len(train)
target = 'count'
features = merged2.columns.tolist()
features.remove(target)
X_train = train[features]
X_val = val[features]
y_train = train[target]
y_val = val[target]
model = LinearRegression()
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
model.score(X_val, y_val)
train_base, val_base = calc_accuracy_score(y_train), calc_accuracy_score(y_val)
print(f"Training model baseline accuracy: {(train_base)*100:,.2f}%" + "\n")
print(f"Validation model baseline accuracy: {(val_base)*100:,.2f}%")
model = LogisticRegression(solver='lbfgs', max_iter=300, multi_class='auto')
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
model.score(X_val, y_val)
y_train_pred = model.predict(X_train)
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
model.score(X_val, y_val)
base = [y_val.value_counts().index[0]] * len(y_val)
mse = mean_squared_error(y_val, base)
mse
def calc_accuracy_score(target_feature):
majority_class = target_feature.mode()[0] # Find the mode of the target feature
target_pred = [majority_class] * len(target_feature) # Create a list the length of the target feature with consisting of the mode as the sole value
score = accuracy_score(target_feature, target_pred)
return score
train_base, val_base = calc_accuracy_score(y_train), calc_accuracy_score(y_val)
print(f"Training model baseline accuracy: {(train_base)*100:,.2f}%" + "\n")
print(f"Validation model baseline accuracy: {(val_base)*100:,.2f}%")
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val.value_counts()
len(y_val == y_val_pred)
model = DecisionTreeRegressor()
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
from sklearn.ensemble import RandomForestRegressor
model = RandomForestRegressor(n_estimators=100,
n_jobs=-1)
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
print(model.score(X_val, y_val))
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
4682
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
merged['count'].value_counts(normalize=True)
from xgboost import XGBRegressor
model = XGBRegressor(n_estimators=400, random_state=42, n_jobs=-1)
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_train_pred = model.predict(X_train)
mae = mean_absolute_error(y_train, y_train_pred)
print(f"Train Error: {mae:.2f}")
r2 = r2_score(y_train, y_train_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_train, y_train_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
y_val_pred = model.predict(X_val)
mae = mean_absolute_error(y_val, y_val_pred)
print(f'Val Error: {mae:,.2f}')
r2 = r2_score(y_val, y_val_pred)
print(f"R-Squared Score: {r2*100:.2f}%")
mse = mean_squared_error(y_val, y_val_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse:,.2f}")
!pip install eli5
import eli5
from eli5.sklearn import PermutationImportance
model = RandomForestRegressor(n_estimators=400,
n_jobs=-1)
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
permuter = PermutationImportance(
model,
scoring='neg_mean_absolute_error',
n_iter=5,
random_state=42
)
permuter.fit(X_val, y_val)
feature_names = X_val.columns.tolist()
pd.Series(permuter.feature_importances_, feature_names).sort_values(ascending=False)
fig, ax = plt.subplots(figsize=(10,8))
ax = sns.distplot(y_train)
ax.set_xlim(0, 20)
!pip install pdpbox
from pdpbox.pdp import pdp_isolate, pdp_plot
plt.rcParams['figure.dpi'] = 150
feature = 'part_of_day'
isolated = pdp_isolate(
model=model,
dataset=X_val,
model_features=X_val.columns,
feature=feature
)
pdp_plot(isolated, feature_name=feature, plot_lines=True, frac_to_plot=100)
from pdpbox.pdp import pdp_interact, pdp_interact_plot
features = ['part_of_day', 'call_dow']
interaction = pdp_interact(
model=model,
dataset=X_val,
model_features=X_val.columns,
features=features
)
pdp_interact_plot(interaction, plot_type='grid', feature_names=features);
!pip install shap
X_val.iloc[1]
import shap
import random as rd
row=X_val.iloc[1]
explainer = shap.TreeExplainer(model)
shap_values = explainer.shap_values(row)
shap.initjs()
shap.force_plot(
base_value=explainer.expected_value,
shap_values=shap_values,
features=row
)
| 0.613468 | 0.751967 |
```
import tensorflow as tf
import numpy as np
import json
from copy import deepcopy
from attention_gru import AttentionGRUCell
epoch = 20
batch_size = 64
size_layer = 64
dropout_rate = 0.1
n_hops = 2
class BaseDataLoader():
def __init__(self):
self.data = {
'size': None,
'val':{
'inputs': None,
'questions': None,
'answers': None,},
'len':{
'inputs_len': None,
'inputs_sent_len': None,
'questions_len': None,
'answers_len': None}
}
self.vocab = {
'size': None,
'word2idx': None,
'idx2word': None,
}
self.params = {
'vocab_size': None,
'<start>': None,
'<end>': None,
'max_input_len': None,
'max_sent_len': None,
'max_quest_len': None,
'max_answer_len': None,
}
class DataLoader(BaseDataLoader):
def __init__(self, path, is_training, vocab=None, params=None):
super().__init__()
data, lens = self.load_data(path)
if is_training:
self.build_vocab(data)
else:
self.demo = data
self.vocab = vocab
self.params = deepcopy(params)
self.is_training = is_training
self.padding(data, lens)
def load_data(self, path):
data, lens = bAbI_data_load(path)
self.data['size'] = len(data[0])
return data, lens
def build_vocab(self, data):
signals = ['<pad>', '<unk>', '<start>', '<end>']
inputs, questions, answers = data
i_words = [w for facts in inputs for fact in facts for w in fact if w != '<end>']
q_words = [w for question in questions for w in question]
a_words = [w for answer in answers for w in answer if w != '<end>']
words = list(set(i_words + q_words + a_words))
self.params['vocab_size'] = len(words) + 4
self.params['<start>'] = 2
self.params['<end>'] = 3
self.vocab['word2idx'] = {word: idx for idx, word in enumerate(signals + words)}
self.vocab['idx2word'] = {idx: word for word, idx in self.vocab['word2idx'].items()}
def padding(self, data, lens):
inputs_len, inputs_sent_len, questions_len, answers_len = lens
self.params['max_input_len'] = max(inputs_len)
self.params['max_sent_len'] = max([fact_len for batch in inputs_sent_len for fact_len in batch])
self.params['max_quest_len'] = max(questions_len)
self.params['max_answer_len'] = max(answers_len)
self.data['len']['inputs_len'] = np.array(inputs_len)
for batch in inputs_sent_len:
batch += [0] * (self.params['max_input_len'] - len(batch))
self.data['len']['inputs_sent_len'] = np.array(inputs_sent_len)
self.data['len']['questions_len'] = np.array(questions_len)
self.data['len']['answers_len'] = np.array(answers_len)
inputs, questions, answers = deepcopy(data)
for facts in inputs:
for sentence in facts:
for i in range(len(sentence)):
sentence[i] = self.vocab['word2idx'].get(sentence[i], self.vocab['word2idx']['<unk>'])
sentence += [0] * (self.params['max_sent_len'] - len(sentence))
paddings = [0] * self.params['max_sent_len']
facts += [paddings] * (self.params['max_input_len'] - len(facts))
for question in questions:
for i in range(len(question)):
question[i] = self.vocab['word2idx'].get(question[i], self.vocab['word2idx']['<unk>'])
question += [0] * (self.params['max_quest_len'] - len(question))
for answer in answers:
for i in range(len(answer)):
answer[i] = self.vocab['word2idx'].get(answer[i], self.vocab['word2idx']['<unk>'])
self.data['val']['inputs'] = np.array(inputs)
self.data['val']['questions'] = np.array(questions)
self.data['val']['answers'] = np.array(answers)
def bAbI_data_load(path, END=['<end>']):
inputs = []
questions = []
answers = []
inputs_len = []
inputs_sent_len = []
questions_len = []
answers_len = []
with open(path) as fopen:
dataset = json.load(fopen)
for data in dataset:
for d in data.split('. '):
index = d.split(' ')[0]
if index == '1':
fact = []
if '?' in d:
temp = d.split(' <> ')
q = temp[0].strip().replace('?', '').split(' ')[1:] + ['?']
a = [temp[1]] + END
fact_copied = deepcopy(fact)
inputs.append(fact_copied)
questions.append(q)
answers.append(a)
inputs_len.append(len(fact_copied))
inputs_sent_len.append([len(s) for s in fact_copied])
questions_len.append(len(q))
answers_len.append(len(a))
else:
tokens = d.replace('.', '').replace('\n', '').split(' ')[1:] + END
fact.append(tokens)
return [inputs, questions, answers], [inputs_len, inputs_sent_len, questions_len, answers_len]
train_data = DataLoader(path='question-answer-train.json',is_training=True)
test_data = DataLoader(path='question-answer-test.json',is_training=False,
vocab=train_data.vocab, params=train_data.params)
START = train_data.params['<start>']
END = train_data.params['<end>']
def shift_right(x):
batch_size = tf.shape(x)[0]
start = tf.to_int32(tf.fill([batch_size, 1], START))
return tf.concat([start, x[:, :-1]], 1)
def GRU(name, rnn_size=None):
return tf.nn.rnn_cell.GRUCell(
rnn_size, kernel_initializer=tf.orthogonal_initializer(), name=name)
def position_encoding(sentence_size, embedding_size):
encoding = np.ones((embedding_size, sentence_size), dtype=np.float32)
ls = sentence_size + 1
le = embedding_size + 1
for i in range(1, le):
for j in range(1, ls):
encoding[i-1, j-1] = (i - (le-1)/2) * (j - (ls-1)/2)
encoding = 1 + 4 * encoding / embedding_size / sentence_size
return np.transpose(encoding)
def gen_episode(inputs_len, memory, q_vec, fact_vecs, proj_1, proj_2, attn_gru, is_training):
def gen_attn(fact_vec):
features = [fact_vec * q_vec,
fact_vec * memory,
tf.abs(fact_vec - q_vec),
tf.abs(fact_vec - memory)]
feature_vec = tf.concat(features, 1)
attention = proj_1(feature_vec)
attention = proj_2(attention)
return tf.squeeze(attention, 1)
attns = tf.map_fn(gen_attn, tf.transpose(fact_vecs, [1,0,2]))
attns = tf.transpose(attns)
attns = tf.nn.softmax(attns)
attns = tf.expand_dims(attns, -1)
_, episode = tf.nn.dynamic_rnn(attn_gru,
tf.concat([fact_vecs, attns], 2),
inputs_len,
dtype=np.float32)
return episode
class QA:
def __init__(self, vocab_size):
self.questions = tf.placeholder(tf.int32,[None,None])
self.inputs = tf.placeholder(tf.int32,[None,None,None])
self.questions_len = tf.placeholder(tf.int32,[None])
self.inputs_len = tf.placeholder(tf.int32,[None])
self.answers_len = tf.placeholder(tf.int32,[None])
self.answers = tf.placeholder(tf.int32,[None,None])
self.training = tf.placeholder(tf.bool)
max_sent_len = train_data.params['max_sent_len']
max_quest_len = train_data.params['max_quest_len']
max_answer_len = train_data.params['max_answer_len']
lookup_table = tf.get_variable('lookup_table', [vocab_size, size_layer], tf.float32)
lookup_table = tf.concat((tf.zeros([1, size_layer]), lookup_table[1:, :]), axis=0)
cell_fw = GRU('cell_fw', size_layer // 2)
cell_bw = GRU('cell_bw', size_layer // 2)
inputs = tf.nn.embedding_lookup(lookup_table, self.inputs)
position = position_encoding(max_sent_len, size_layer)
inputs = tf.reduce_sum(inputs * position, 2)
birnn_out, _ = tf.nn.bidirectional_dynamic_rnn(cell_fw,
cell_bw,
inputs,
self.inputs_len,
dtype=np.float32)
fact_vecs = tf.concat(birnn_out, -1)
fact_vecs = tf.layers.dropout(fact_vecs, dropout_rate, training=self.training)
cell = GRU('question_rnn', size_layer)
questions = tf.nn.embedding_lookup(lookup_table, self.questions)
_, q_vec = tf.nn.dynamic_rnn(cell,
questions,
self.questions_len,
dtype=np.float32)
proj_1 = tf.layers.Dense(size_layer, tf.tanh, name='attn_proj_1')
proj_2 = tf.layers.Dense(1, name='attn_proj_2')
attn_gru = AttentionGRUCell(size_layer, name='attn_gru')
memory_proj = tf.layers.Dense(size_layer, tf.nn.relu, name='memory_proj')
memory = q_vec
for i in range(n_hops):
episode = gen_episode(self.inputs_len,
memory,
q_vec,
fact_vecs,
proj_1,
proj_2,
attn_gru,
self.training)
memory = memory_proj(tf.concat([memory, episode, q_vec], 1))
state_proj = tf.layers.Dense(size_layer, name='state_proj')
vocab_proj = tf.layers.Dense(vocab_size, name='vocab_proj')
memory = tf.layers.dropout(memory, dropout_rate, training=self.training)
init_state = state_proj(tf.concat((memory, q_vec), -1))
helper = tf.contrib.seq2seq.TrainingHelper(
inputs = tf.nn.embedding_lookup(lookup_table, shift_right(self.answers)),
sequence_length = self.answers_len)
decoder = tf.contrib.seq2seq.BasicDecoder(
cell = cell,
helper = helper,
initial_state = init_state,
output_layer = vocab_proj)
decoder_output, _, _ = tf.contrib.seq2seq.dynamic_decode(
decoder = decoder)
self.training_id = decoder_output.sample_id
self.training_logits = decoder_output.rnn_output
helper = tf.contrib.seq2seq.GreedyEmbeddingHelper(
embedding = lookup_table,
start_tokens = tf.tile(
tf.constant([START], dtype=tf.int32), [tf.shape(init_state)[0]]),
end_token = END)
decoder = tf.contrib.seq2seq.BasicDecoder(
cell = cell,
helper = helper,
initial_state = init_state,
output_layer = vocab_proj)
decoder_output, _, _ = tf.contrib.seq2seq.dynamic_decode(
decoder = decoder,
maximum_iterations = max_answer_len)
self.predict_id = decoder_output.sample_id
self.cost = tf.reduce_mean(tf.contrib.seq2seq.sequence_loss(logits = self.training_logits,
targets = self.answers,
weights = tf.ones_like(self.answers, tf.float32)))
self.optimizer = tf.train.AdamOptimizer().minimize(self.cost)
correct_pred = tf.equal(self.training_id, self.answers)
self.accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
tf.reset_default_graph()
sess = tf.InteractiveSession()
model = QA(train_data.params['vocab_size'])
sess.run(tf.global_variables_initializer())
for i in range(epoch):
total_cost, total_acc = 0, 0
for k in range(0, train_data.data['val']['inputs'].shape[0], batch_size):
index = min(k + batch_size, train_data.data['val']['inputs'].shape[0])
batch_questions = train_data.data['val']['questions'][k:index]
batch_inputs = train_data.data['val']['inputs'][k:index]
batch_inputs_len = train_data.data['len']['inputs_len'][k:index]
batch_questions_len = train_data.data['len']['questions_len'][k:index]
batch_answers_len = train_data.data['len']['answers_len'][k:index]
batch_answers = train_data.data['val']['answers'][k:index]
acc, cost, _ = sess.run([model.accuracy,model.cost,model.optimizer],
feed_dict={model.questions:batch_questions,
model.inputs:batch_inputs,
model.inputs_len:batch_inputs_len,
model.questions_len:batch_questions_len,
model.answers_len:batch_answers_len,
model.answers:batch_answers,
model.training:True})
total_cost += cost
total_acc += acc
total_cost /= (train_data.data['val']['inputs'].shape[0] / batch_size)
total_acc /= (train_data.data['val']['inputs'].shape[0] / batch_size)
print('epoch %d, avg cost %f, avg acc %f'%(i+1,total_cost,total_acc))
testing_size = 32
batch_questions = test_data.data['val']['questions'][:testing_size]
batch_inputs = test_data.data['val']['inputs'][:testing_size]
batch_inputs_len = test_data.data['len']['inputs_len'][:testing_size]
batch_questions_len = test_data.data['len']['questions_len'][:testing_size]
batch_answers_len = test_data.data['len']['answers_len'][:testing_size]
batch_answers = test_data.data['val']['answers'][:testing_size]
logits = sess.run(model.predict_id,
feed_dict={model.questions:batch_questions,
model.inputs:batch_inputs,
model.inputs_len:batch_inputs_len,
model.questions_len:batch_questions_len,
model.answers_len:batch_answers_len,
model.training:False})
for i in range(testing_size):
print('QUESTION:',' '.join([train_data.vocab['idx2word'][k] for k in batch_questions[i]]))
print('REAL:',train_data.vocab['idx2word'][batch_answers[i,0]])
print('PREDICT:',train_data.vocab['idx2word'][logits[i,0]],'\n')
```
|
github_jupyter
|
import tensorflow as tf
import numpy as np
import json
from copy import deepcopy
from attention_gru import AttentionGRUCell
epoch = 20
batch_size = 64
size_layer = 64
dropout_rate = 0.1
n_hops = 2
class BaseDataLoader():
def __init__(self):
self.data = {
'size': None,
'val':{
'inputs': None,
'questions': None,
'answers': None,},
'len':{
'inputs_len': None,
'inputs_sent_len': None,
'questions_len': None,
'answers_len': None}
}
self.vocab = {
'size': None,
'word2idx': None,
'idx2word': None,
}
self.params = {
'vocab_size': None,
'<start>': None,
'<end>': None,
'max_input_len': None,
'max_sent_len': None,
'max_quest_len': None,
'max_answer_len': None,
}
class DataLoader(BaseDataLoader):
def __init__(self, path, is_training, vocab=None, params=None):
super().__init__()
data, lens = self.load_data(path)
if is_training:
self.build_vocab(data)
else:
self.demo = data
self.vocab = vocab
self.params = deepcopy(params)
self.is_training = is_training
self.padding(data, lens)
def load_data(self, path):
data, lens = bAbI_data_load(path)
self.data['size'] = len(data[0])
return data, lens
def build_vocab(self, data):
signals = ['<pad>', '<unk>', '<start>', '<end>']
inputs, questions, answers = data
i_words = [w for facts in inputs for fact in facts for w in fact if w != '<end>']
q_words = [w for question in questions for w in question]
a_words = [w for answer in answers for w in answer if w != '<end>']
words = list(set(i_words + q_words + a_words))
self.params['vocab_size'] = len(words) + 4
self.params['<start>'] = 2
self.params['<end>'] = 3
self.vocab['word2idx'] = {word: idx for idx, word in enumerate(signals + words)}
self.vocab['idx2word'] = {idx: word for word, idx in self.vocab['word2idx'].items()}
def padding(self, data, lens):
inputs_len, inputs_sent_len, questions_len, answers_len = lens
self.params['max_input_len'] = max(inputs_len)
self.params['max_sent_len'] = max([fact_len for batch in inputs_sent_len for fact_len in batch])
self.params['max_quest_len'] = max(questions_len)
self.params['max_answer_len'] = max(answers_len)
self.data['len']['inputs_len'] = np.array(inputs_len)
for batch in inputs_sent_len:
batch += [0] * (self.params['max_input_len'] - len(batch))
self.data['len']['inputs_sent_len'] = np.array(inputs_sent_len)
self.data['len']['questions_len'] = np.array(questions_len)
self.data['len']['answers_len'] = np.array(answers_len)
inputs, questions, answers = deepcopy(data)
for facts in inputs:
for sentence in facts:
for i in range(len(sentence)):
sentence[i] = self.vocab['word2idx'].get(sentence[i], self.vocab['word2idx']['<unk>'])
sentence += [0] * (self.params['max_sent_len'] - len(sentence))
paddings = [0] * self.params['max_sent_len']
facts += [paddings] * (self.params['max_input_len'] - len(facts))
for question in questions:
for i in range(len(question)):
question[i] = self.vocab['word2idx'].get(question[i], self.vocab['word2idx']['<unk>'])
question += [0] * (self.params['max_quest_len'] - len(question))
for answer in answers:
for i in range(len(answer)):
answer[i] = self.vocab['word2idx'].get(answer[i], self.vocab['word2idx']['<unk>'])
self.data['val']['inputs'] = np.array(inputs)
self.data['val']['questions'] = np.array(questions)
self.data['val']['answers'] = np.array(answers)
def bAbI_data_load(path, END=['<end>']):
inputs = []
questions = []
answers = []
inputs_len = []
inputs_sent_len = []
questions_len = []
answers_len = []
with open(path) as fopen:
dataset = json.load(fopen)
for data in dataset:
for d in data.split('. '):
index = d.split(' ')[0]
if index == '1':
fact = []
if '?' in d:
temp = d.split(' <> ')
q = temp[0].strip().replace('?', '').split(' ')[1:] + ['?']
a = [temp[1]] + END
fact_copied = deepcopy(fact)
inputs.append(fact_copied)
questions.append(q)
answers.append(a)
inputs_len.append(len(fact_copied))
inputs_sent_len.append([len(s) for s in fact_copied])
questions_len.append(len(q))
answers_len.append(len(a))
else:
tokens = d.replace('.', '').replace('\n', '').split(' ')[1:] + END
fact.append(tokens)
return [inputs, questions, answers], [inputs_len, inputs_sent_len, questions_len, answers_len]
train_data = DataLoader(path='question-answer-train.json',is_training=True)
test_data = DataLoader(path='question-answer-test.json',is_training=False,
vocab=train_data.vocab, params=train_data.params)
START = train_data.params['<start>']
END = train_data.params['<end>']
def shift_right(x):
batch_size = tf.shape(x)[0]
start = tf.to_int32(tf.fill([batch_size, 1], START))
return tf.concat([start, x[:, :-1]], 1)
def GRU(name, rnn_size=None):
return tf.nn.rnn_cell.GRUCell(
rnn_size, kernel_initializer=tf.orthogonal_initializer(), name=name)
def position_encoding(sentence_size, embedding_size):
encoding = np.ones((embedding_size, sentence_size), dtype=np.float32)
ls = sentence_size + 1
le = embedding_size + 1
for i in range(1, le):
for j in range(1, ls):
encoding[i-1, j-1] = (i - (le-1)/2) * (j - (ls-1)/2)
encoding = 1 + 4 * encoding / embedding_size / sentence_size
return np.transpose(encoding)
def gen_episode(inputs_len, memory, q_vec, fact_vecs, proj_1, proj_2, attn_gru, is_training):
def gen_attn(fact_vec):
features = [fact_vec * q_vec,
fact_vec * memory,
tf.abs(fact_vec - q_vec),
tf.abs(fact_vec - memory)]
feature_vec = tf.concat(features, 1)
attention = proj_1(feature_vec)
attention = proj_2(attention)
return tf.squeeze(attention, 1)
attns = tf.map_fn(gen_attn, tf.transpose(fact_vecs, [1,0,2]))
attns = tf.transpose(attns)
attns = tf.nn.softmax(attns)
attns = tf.expand_dims(attns, -1)
_, episode = tf.nn.dynamic_rnn(attn_gru,
tf.concat([fact_vecs, attns], 2),
inputs_len,
dtype=np.float32)
return episode
class QA:
def __init__(self, vocab_size):
self.questions = tf.placeholder(tf.int32,[None,None])
self.inputs = tf.placeholder(tf.int32,[None,None,None])
self.questions_len = tf.placeholder(tf.int32,[None])
self.inputs_len = tf.placeholder(tf.int32,[None])
self.answers_len = tf.placeholder(tf.int32,[None])
self.answers = tf.placeholder(tf.int32,[None,None])
self.training = tf.placeholder(tf.bool)
max_sent_len = train_data.params['max_sent_len']
max_quest_len = train_data.params['max_quest_len']
max_answer_len = train_data.params['max_answer_len']
lookup_table = tf.get_variable('lookup_table', [vocab_size, size_layer], tf.float32)
lookup_table = tf.concat((tf.zeros([1, size_layer]), lookup_table[1:, :]), axis=0)
cell_fw = GRU('cell_fw', size_layer // 2)
cell_bw = GRU('cell_bw', size_layer // 2)
inputs = tf.nn.embedding_lookup(lookup_table, self.inputs)
position = position_encoding(max_sent_len, size_layer)
inputs = tf.reduce_sum(inputs * position, 2)
birnn_out, _ = tf.nn.bidirectional_dynamic_rnn(cell_fw,
cell_bw,
inputs,
self.inputs_len,
dtype=np.float32)
fact_vecs = tf.concat(birnn_out, -1)
fact_vecs = tf.layers.dropout(fact_vecs, dropout_rate, training=self.training)
cell = GRU('question_rnn', size_layer)
questions = tf.nn.embedding_lookup(lookup_table, self.questions)
_, q_vec = tf.nn.dynamic_rnn(cell,
questions,
self.questions_len,
dtype=np.float32)
proj_1 = tf.layers.Dense(size_layer, tf.tanh, name='attn_proj_1')
proj_2 = tf.layers.Dense(1, name='attn_proj_2')
attn_gru = AttentionGRUCell(size_layer, name='attn_gru')
memory_proj = tf.layers.Dense(size_layer, tf.nn.relu, name='memory_proj')
memory = q_vec
for i in range(n_hops):
episode = gen_episode(self.inputs_len,
memory,
q_vec,
fact_vecs,
proj_1,
proj_2,
attn_gru,
self.training)
memory = memory_proj(tf.concat([memory, episode, q_vec], 1))
state_proj = tf.layers.Dense(size_layer, name='state_proj')
vocab_proj = tf.layers.Dense(vocab_size, name='vocab_proj')
memory = tf.layers.dropout(memory, dropout_rate, training=self.training)
init_state = state_proj(tf.concat((memory, q_vec), -1))
helper = tf.contrib.seq2seq.TrainingHelper(
inputs = tf.nn.embedding_lookup(lookup_table, shift_right(self.answers)),
sequence_length = self.answers_len)
decoder = tf.contrib.seq2seq.BasicDecoder(
cell = cell,
helper = helper,
initial_state = init_state,
output_layer = vocab_proj)
decoder_output, _, _ = tf.contrib.seq2seq.dynamic_decode(
decoder = decoder)
self.training_id = decoder_output.sample_id
self.training_logits = decoder_output.rnn_output
helper = tf.contrib.seq2seq.GreedyEmbeddingHelper(
embedding = lookup_table,
start_tokens = tf.tile(
tf.constant([START], dtype=tf.int32), [tf.shape(init_state)[0]]),
end_token = END)
decoder = tf.contrib.seq2seq.BasicDecoder(
cell = cell,
helper = helper,
initial_state = init_state,
output_layer = vocab_proj)
decoder_output, _, _ = tf.contrib.seq2seq.dynamic_decode(
decoder = decoder,
maximum_iterations = max_answer_len)
self.predict_id = decoder_output.sample_id
self.cost = tf.reduce_mean(tf.contrib.seq2seq.sequence_loss(logits = self.training_logits,
targets = self.answers,
weights = tf.ones_like(self.answers, tf.float32)))
self.optimizer = tf.train.AdamOptimizer().minimize(self.cost)
correct_pred = tf.equal(self.training_id, self.answers)
self.accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
tf.reset_default_graph()
sess = tf.InteractiveSession()
model = QA(train_data.params['vocab_size'])
sess.run(tf.global_variables_initializer())
for i in range(epoch):
total_cost, total_acc = 0, 0
for k in range(0, train_data.data['val']['inputs'].shape[0], batch_size):
index = min(k + batch_size, train_data.data['val']['inputs'].shape[0])
batch_questions = train_data.data['val']['questions'][k:index]
batch_inputs = train_data.data['val']['inputs'][k:index]
batch_inputs_len = train_data.data['len']['inputs_len'][k:index]
batch_questions_len = train_data.data['len']['questions_len'][k:index]
batch_answers_len = train_data.data['len']['answers_len'][k:index]
batch_answers = train_data.data['val']['answers'][k:index]
acc, cost, _ = sess.run([model.accuracy,model.cost,model.optimizer],
feed_dict={model.questions:batch_questions,
model.inputs:batch_inputs,
model.inputs_len:batch_inputs_len,
model.questions_len:batch_questions_len,
model.answers_len:batch_answers_len,
model.answers:batch_answers,
model.training:True})
total_cost += cost
total_acc += acc
total_cost /= (train_data.data['val']['inputs'].shape[0] / batch_size)
total_acc /= (train_data.data['val']['inputs'].shape[0] / batch_size)
print('epoch %d, avg cost %f, avg acc %f'%(i+1,total_cost,total_acc))
testing_size = 32
batch_questions = test_data.data['val']['questions'][:testing_size]
batch_inputs = test_data.data['val']['inputs'][:testing_size]
batch_inputs_len = test_data.data['len']['inputs_len'][:testing_size]
batch_questions_len = test_data.data['len']['questions_len'][:testing_size]
batch_answers_len = test_data.data['len']['answers_len'][:testing_size]
batch_answers = test_data.data['val']['answers'][:testing_size]
logits = sess.run(model.predict_id,
feed_dict={model.questions:batch_questions,
model.inputs:batch_inputs,
model.inputs_len:batch_inputs_len,
model.questions_len:batch_questions_len,
model.answers_len:batch_answers_len,
model.training:False})
for i in range(testing_size):
print('QUESTION:',' '.join([train_data.vocab['idx2word'][k] for k in batch_questions[i]]))
print('REAL:',train_data.vocab['idx2word'][batch_answers[i,0]])
print('PREDICT:',train_data.vocab['idx2word'][logits[i,0]],'\n')
| 0.281307 | 0.230335 |
'''
A Reccurent Neural Network (LSTM) implementation example using TensorFlow library.
This example is using the MNIST database of handwritten digits (http://yann.lecun.com/exdb/mnist/)
Long Short Term Memory paper: http://deeplearning.cs.cmu.edu/pdfs/Hochreiter97_lstm.pdf
Author: Aymeric Damien
Project: https://github.com/aymericdamien/TensorFlow-Examples/
'''
```
import tensorflow as tf
from tensorflow.contrib import rnn
import numpy as np
# Import MINST data
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("data/MNIST/", one_hot=True)
```
'''
To classify images using a reccurent neural network, we consider every image
row as a sequence of pixels. Because MNIST image shape is 28*28px, we will then
handle 28 sequences of 28 steps for every sample.
'''
```
# Parameters
learning_rate = 0.001
training_iters = 100000
batch_size = 100
display_step = 10
# Network Parameters
n_input = 28 # MNIST data input (img shape: 28*28)
n_steps = 28 # timesteps
n_hidden = 150 # hidden layer num of features
n_classes = 10 # MNIST total classes (0-9 digits)
# tf Graph input
x = tf.placeholder("float", [None, n_steps, n_input])
y = tf.placeholder("float", [None, n_classes])
# Define weights
weights = {
'out': tf.Variable(tf.random_normal([n_hidden, n_classes]))
}
biases = {
'out': tf.Variable(tf.random_normal([n_classes]))
}
def RNN(x, weights, biases):
# Prepare data shape to match `rnn` function requirements
# Current data input shape: (batch_size, n_steps, n_input)
# Required shape: 'n_steps' tensors list of shape (batch_size, n_input)
# Unstack to get a list of 'n_steps' tensors of shape (batch_size, n_input)
x = tf.unstack(x, n_steps, 1)
# Define a lstm cell with tensorflow
lstm_cell = rnn.BasicLSTMCell(n_hidden, forget_bias=1.0)
# Get lstm cell output
outputs, states = rnn.static_rnn(lstm_cell, x, dtype=tf.float32)
# Linear activation, using rnn inner loop last output
return tf.matmul(outputs[-1], weights['out']) + biases['out']
pred = RNN(x, weights, biases)
# Define loss and optimizer
cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=pred, labels=y))
optimizer = tf.train.AdamOptimizer(learning_rate=learning_rate).minimize(cost)
# Evaluate model
correct_pred = tf.equal(tf.argmax(pred,1), tf.argmax(y,1))
accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
# Initializing the variables
init = tf.global_variables_initializer()
# Launch the graph
with tf.Session() as sess:
sess.run(init)
step = 1
# Keep training until reach max iterations
while step * batch_size < training_iters:
batch_x, batch_y = mnist.train.next_batch(batch_size)
# Reshape data to get 28 seq of 28 elements
batch_x = batch_x.reshape((batch_size, n_steps, n_input))
# Run optimization op (backprop)
sess.run(optimizer, feed_dict={x: batch_x, y: batch_y})
if step % display_step == 0:
# Calculate batch accuracy
acc = sess.run(accuracy, feed_dict={x: batch_x, y: batch_y})
# Calculate batch loss
loss = sess.run(cost, feed_dict={x: batch_x, y: batch_y})
print("Iter " + str(step*batch_size) + ", Minibatch Loss= " + \
"{:.6f}".format(loss) + ", Training Accuracy= " + \
"{:.5f}".format(acc))
step += 1
print("Optimization Finished!")
# Calculate accuracy for 128 mnist test images
test_len = 200
test_data = mnist.test.images[:test_len].reshape((-1, n_steps, n_input))
test_label = mnist.test.labels[:test_len]
print("Testing Accuracy:", \
sess.run(accuracy, feed_dict={x: test_data, y: test_label}))
```
|
github_jupyter
|
import tensorflow as tf
from tensorflow.contrib import rnn
import numpy as np
# Import MINST data
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("data/MNIST/", one_hot=True)
# Parameters
learning_rate = 0.001
training_iters = 100000
batch_size = 100
display_step = 10
# Network Parameters
n_input = 28 # MNIST data input (img shape: 28*28)
n_steps = 28 # timesteps
n_hidden = 150 # hidden layer num of features
n_classes = 10 # MNIST total classes (0-9 digits)
# tf Graph input
x = tf.placeholder("float", [None, n_steps, n_input])
y = tf.placeholder("float", [None, n_classes])
# Define weights
weights = {
'out': tf.Variable(tf.random_normal([n_hidden, n_classes]))
}
biases = {
'out': tf.Variable(tf.random_normal([n_classes]))
}
def RNN(x, weights, biases):
# Prepare data shape to match `rnn` function requirements
# Current data input shape: (batch_size, n_steps, n_input)
# Required shape: 'n_steps' tensors list of shape (batch_size, n_input)
# Unstack to get a list of 'n_steps' tensors of shape (batch_size, n_input)
x = tf.unstack(x, n_steps, 1)
# Define a lstm cell with tensorflow
lstm_cell = rnn.BasicLSTMCell(n_hidden, forget_bias=1.0)
# Get lstm cell output
outputs, states = rnn.static_rnn(lstm_cell, x, dtype=tf.float32)
# Linear activation, using rnn inner loop last output
return tf.matmul(outputs[-1], weights['out']) + biases['out']
pred = RNN(x, weights, biases)
# Define loss and optimizer
cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=pred, labels=y))
optimizer = tf.train.AdamOptimizer(learning_rate=learning_rate).minimize(cost)
# Evaluate model
correct_pred = tf.equal(tf.argmax(pred,1), tf.argmax(y,1))
accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
# Initializing the variables
init = tf.global_variables_initializer()
# Launch the graph
with tf.Session() as sess:
sess.run(init)
step = 1
# Keep training until reach max iterations
while step * batch_size < training_iters:
batch_x, batch_y = mnist.train.next_batch(batch_size)
# Reshape data to get 28 seq of 28 elements
batch_x = batch_x.reshape((batch_size, n_steps, n_input))
# Run optimization op (backprop)
sess.run(optimizer, feed_dict={x: batch_x, y: batch_y})
if step % display_step == 0:
# Calculate batch accuracy
acc = sess.run(accuracy, feed_dict={x: batch_x, y: batch_y})
# Calculate batch loss
loss = sess.run(cost, feed_dict={x: batch_x, y: batch_y})
print("Iter " + str(step*batch_size) + ", Minibatch Loss= " + \
"{:.6f}".format(loss) + ", Training Accuracy= " + \
"{:.5f}".format(acc))
step += 1
print("Optimization Finished!")
# Calculate accuracy for 128 mnist test images
test_len = 200
test_data = mnist.test.images[:test_len].reshape((-1, n_steps, n_input))
test_label = mnist.test.labels[:test_len]
print("Testing Accuracy:", \
sess.run(accuracy, feed_dict={x: test_data, y: test_label}))
| 0.925685 | 0.990496 |
# Intro to Statistics in Python
```
# Importing libraries
import numpy as np
from matplotlib import pyplot as plt
from scipy.stats import ttest_1samp
from scipy.stats import ttest_ind
from scipy.stats import f_oneway
from statsmodels.stats.multicomp import pairwise_tukeyhsd
from scipy.stats import binom_test
from scipy.stats import chi2_contingency
```
# Statistical Distributions
**CASE 1** <br>
You analyze cars for a company, and you want to know if the cars are getting old. You have a database with the years of use of each car
```
years_of_use = [1, 1, 1, 1, 2, 2, 2, 3, 3, 3, 3, 4, 4, 4, 5, 5, 5, 5]
plt.hist(years_of_use, bins=5, range=(1, 5))
plt.title('Age of cars')
plt.xlabel('Years of use')
plt.ylabel('Number of cars')
plt.show()
```
It looks that you have around the same proportion of age in the cars. You have an uniform distribution.
Now you want to analyze bikes. You do the same process
```
years_of_use = [1, 1, 1, 1, 1, 2, 2, 2, 2, 3, 4, 4]
plt.hist(years_of_use, bins=5, range=(1, 6))
plt.title('Age of bikes')
plt.xlabel('Years of use')
plt.ylabel('Number of bikes')
plt.show()
```
It looks that you are getting more new bikes that old ones. You have a skewed right dataset.
**CASE 2**<br>
You work for a company that designs english tests for certification. You want to see the distribution of results, at the moment you don't have all the data but you have the mean is 70 and the standard deviation is 10
```
mean=70
std=10
scores = np.random.normal(mean, std, size=100000)
plt.hist(scores, bins=1000, range=(0,120))
plt.title('Scores Distribution')
plt.xlabel('Score')
plt.ylabel('Students')
plt.show()
```
As you are sure that the behavior can be approximated to a normal distribution, you know that 68% of the data falls between 1 standard deviation of the mean, 95% falls between 2 standard deviation of the mean and 99.7% falls between 3 standard deviation of the mean
A person that scores 60 want to know in what range he falls:
```
# First it is seen the range of 2 std
two_std_down = mean - 2*std
two_std_up = mean + 2*std
print(two_std_down,"-",two_std_up)
# First it is seen the range of 1 std
two_std_down = mean - std
two_std_up = mean + std
print(two_std_down,"-",two_std_up)
```
It looks that the student made it in the second rage of 2 standard deviations like 95% of the data, and barely in the first range of 1 standard deviation like 68% of the data
**CASE 3**<br>
You are a professional basketball player and you have an average of 70% sucess in 20 shots, what is the probability of a 75% success?<br>
The binomial distribution tells us how likely it is for a certain number of “successes” to happen, given a probability of success and a number of trials.
```
# For a binomial distribution
dist_case3 = np.random.binomial(20, 0.70, size=1000000)
plt.hist(dist_case3, range=(0, 20), bins=20)
plt.xlabel('Number of succesful shots')
plt.ylabel('Frequency')
plt.show()
# Probability of 0.75 success
# The mean of a logial statement gives the percentage of results that fulfill the requirement
np.mean(dist_case3 == (0.75*20))
# Probability of 0.7 success
np.mean(dist_case3 == (0.7*20))
```
According to the results is just a bit less likely than obtaining a 70% success rate
# Intro to scipy
# Hypothesis testing
**Sample mean and population mean:** The sample is a portion of the population. Different sample can have different means that can also be different from the population mean. <br>
**Central limit theorem:** If the sample is big enough, it can behave like the population and the sample mean can be closer to the population mean.<br>
**Null hypothesis:** It states that the observed difference is the result of chance and both population means are the same.<br>
**Hypothesis testing:** It is a mathematical way of determining with confidence that the null hypothesis is false.<br>
**Type I error:** False positive, Not rejecting the null hypothesis when you should.<br>
**Type II error:** False negative, rejecting the null hypothesis when you should not.<br>
**P-Values:** Probability of getting the observed statistics given that the null hypothesis is true. If the p-value is less than 5% means that the probability of getting the result is less than 5% even when the null hypothesis is true, so the null hypothesis can be safely rejected<br>
# Types of hypothesis tests
## 1 Sample T-test
1 Sample T-test compares a sample mean to a hypothetical population mean. For example you do an experiment and obtain a sample of numerical values from a feature. Now you want to test if the mean of the obtained sample compares with the population's mean and it is not the result of chance.
**CASE 4**<br>
You have a database with different people's heights. You expect a mean of 1.7, do a hypothesis test to see if the sample's mean compares with the expected mean
```
person_height_1 = np.array([1.8, 1.73, 1.67, 1.69, 1.89,
1.75, 1.76, 1.93, 1.89, 1.99,
2.00, 1.92, 1.9, 1.69, 2.10,
1.64, 1.73, 1.73, 2.08, 1.9,
1.73, 1.60, 2.09, 1.88, 1.81,
1.7, 2.11, 1.9, 1.99, 1.93])
np.mean(person_height_1)
# An univariate T-test: 1 Sample T test
tstat, pval = ttest_1samp(person_height_1, 1.7)# Inputs are sample distribution and expected mean
pval
```
With last result the null hyphotesis can be discarded, you are not sure if the sample mean reflectts a population with a mean of 1.7. More data may help
## 2 Sample T-test
2 Sample T-test compares two samples to see if they belong to different population. Doing multiple 2 sample T-test can lead to type I error, as the 5% multiplies itself with more samples.
You have another dataset of people's heights and you want to compare them to see if they belong to different populations.
```
# Taking into account a second sample
person_height_2 = np.array([1.9, 1.63, 1.77, 1.59, 1.99,
1.85, 1.66, 1.83, 1.79, 1.89,
2.00, 1.92, 1.9, 1.69, 2.10,
1.64, 1.73, 1.73, 2.08, 1.9,
1.73, 1.60, 2.09, 1.88, 1.81,
1.9, 2.11, 1.9, 1.99, 1.83])
np.mean(person_height_2)
# 2 sample T-test
tstatstic, pval = ttest_ind(person_height_1, person_height_2)
pval
```
The null hypothesis cannot be rejected.
## Anova test
Anova or analysis of variance, use the null hypothesis that all samples come from the same mean, if it is rejected it means that at least 1 pair of samples don't have the same mean, but it cannot be determined which one.<br>
To do the Anova test some assumptions are done:<br>
- The samples are normal distributed, more or less. It is always better a bigger sample
- All samples have the same standard deviation, or similar.
- The samples are independent. One distribution should not affect the other, for example avoid cases like performance in sport before, during and after the game.
```
# For multiple tests
fstat, pval = f_oneway(person_height_1, person_height_2)
pval
```
## Tukey's range test
Used to determine which pair of samples, after the Anova test, are different.
```
v = np.concatenate([person_height_1, person_height_2])
labels = ['sample_1'] * len(person_height_1) + ['sample_2'] * len(person_height_2)
tukey_results = pairwise_tukeyhsd(v, labels, 0.05)
print(tukey_results)
```
## Binomial test
Before mean and standard deviation were used to compare numerical data, but binomial tests are used to compare categorical information that can fall between 2 categories (for example, purchase or not purchase), and the percent of success is compared.
**CASE 5**<br>
You did 100 trials flipping a coin and got 530 heads. Is the coin even or not?
```
pval = binom_test(530, n=1000, p=0.5)
pval
```
For the hypothesis test again the null hypothesis is assumed in which it says that the difference in probability is the result of chance, and the propbability of getting the result is higher than 0.05, so the null hypothesis cannot be rejected.
## Chi Square test
The Chi Square test is used when you want to understand whether the outcomes of two categorical variables are associated
**CASE 6**<br>
You want to test if the results of an election for a product A or B are different for different states
```
# Contingency table
# A | B
# ------------------------------------
# NY | 30 | 10
# KY | 35 | 5
# AL | 28 | 12
# FL | 20 | 20
X = [[30, 10],
[35, 5],
[28, 12],
[20, 20]]
chi2, pval, dof, expected = chi2_contingency(X)
pval
```
The null hypothesis says that given all samples being the same, but the the probability of getting the results is less than 0.05, so the null hypothesis can be rejected
## A/B testing
To run an A/B test or survey you need to determine the sample size to have confindent results.<br>
**A/B testing:** Scientific method for choosing between 2 options.<br>
**Baseline conversion rate:** Percent of users that take certain action after interacting with one option. For example, the percent of costumers that click the ad shown to him and exists 2 versions.<br>
**Minimum desired lift or minimum detectable effect**<br>Is the minimum percent needed for the new option to detect a difference in behavior. For example it is needed 3% more sales with the new add, if the baseline is 6%, it is needed 33% of lift to get to the desired 9%.<br>
**Statistical significance threshold:** Probability of committing type I error. Normally it is accepted 5%<br>
[Online calculator](https://content.codecademy.com/courses/learn-hypothesis-testing/a_b_sample_size/index4.html)
## Sample size determination
**Margin of error:** Is the furthest the true value is expected to be from what is measured in the survey. For example, if the results show a 40% success for an option with an error of 5%, it means that the true value is in the range of 35 and 45%<br>
**Confidence level:** It is the probability that the margin of error contains the true proportion. The larger confidence level required, the larger the sample size. Normally it is used a confidence level of 95%. <br>
**Population size:** Effective population size. The larger can be 100.000, as more people can make negligible changes.<br>
**Expected proportion or likely sample proportion:** It is a of what the expected results might be. It could be based upon the results from a previous survey or pilot. The fewer proportion expected, the fewer people needed to test, as it becomes unrelevant to test more people as the results will be the same.
As the expected proportion increases, it is rarer to get the proportion right using a random sample. The most conservative is 50%, as that would give the largest required sample size. <br>
[Online calculator](https://content.codecademy.com/courses/learn-hypothesis-testing/margin_of_error/index.html)
|
github_jupyter
|
# Importing libraries
import numpy as np
from matplotlib import pyplot as plt
from scipy.stats import ttest_1samp
from scipy.stats import ttest_ind
from scipy.stats import f_oneway
from statsmodels.stats.multicomp import pairwise_tukeyhsd
from scipy.stats import binom_test
from scipy.stats import chi2_contingency
years_of_use = [1, 1, 1, 1, 2, 2, 2, 3, 3, 3, 3, 4, 4, 4, 5, 5, 5, 5]
plt.hist(years_of_use, bins=5, range=(1, 5))
plt.title('Age of cars')
plt.xlabel('Years of use')
plt.ylabel('Number of cars')
plt.show()
years_of_use = [1, 1, 1, 1, 1, 2, 2, 2, 2, 3, 4, 4]
plt.hist(years_of_use, bins=5, range=(1, 6))
plt.title('Age of bikes')
plt.xlabel('Years of use')
plt.ylabel('Number of bikes')
plt.show()
mean=70
std=10
scores = np.random.normal(mean, std, size=100000)
plt.hist(scores, bins=1000, range=(0,120))
plt.title('Scores Distribution')
plt.xlabel('Score')
plt.ylabel('Students')
plt.show()
# First it is seen the range of 2 std
two_std_down = mean - 2*std
two_std_up = mean + 2*std
print(two_std_down,"-",two_std_up)
# First it is seen the range of 1 std
two_std_down = mean - std
two_std_up = mean + std
print(two_std_down,"-",two_std_up)
# For a binomial distribution
dist_case3 = np.random.binomial(20, 0.70, size=1000000)
plt.hist(dist_case3, range=(0, 20), bins=20)
plt.xlabel('Number of succesful shots')
plt.ylabel('Frequency')
plt.show()
# Probability of 0.75 success
# The mean of a logial statement gives the percentage of results that fulfill the requirement
np.mean(dist_case3 == (0.75*20))
# Probability of 0.7 success
np.mean(dist_case3 == (0.7*20))
person_height_1 = np.array([1.8, 1.73, 1.67, 1.69, 1.89,
1.75, 1.76, 1.93, 1.89, 1.99,
2.00, 1.92, 1.9, 1.69, 2.10,
1.64, 1.73, 1.73, 2.08, 1.9,
1.73, 1.60, 2.09, 1.88, 1.81,
1.7, 2.11, 1.9, 1.99, 1.93])
np.mean(person_height_1)
# An univariate T-test: 1 Sample T test
tstat, pval = ttest_1samp(person_height_1, 1.7)# Inputs are sample distribution and expected mean
pval
# Taking into account a second sample
person_height_2 = np.array([1.9, 1.63, 1.77, 1.59, 1.99,
1.85, 1.66, 1.83, 1.79, 1.89,
2.00, 1.92, 1.9, 1.69, 2.10,
1.64, 1.73, 1.73, 2.08, 1.9,
1.73, 1.60, 2.09, 1.88, 1.81,
1.9, 2.11, 1.9, 1.99, 1.83])
np.mean(person_height_2)
# 2 sample T-test
tstatstic, pval = ttest_ind(person_height_1, person_height_2)
pval
# For multiple tests
fstat, pval = f_oneway(person_height_1, person_height_2)
pval
v = np.concatenate([person_height_1, person_height_2])
labels = ['sample_1'] * len(person_height_1) + ['sample_2'] * len(person_height_2)
tukey_results = pairwise_tukeyhsd(v, labels, 0.05)
print(tukey_results)
pval = binom_test(530, n=1000, p=0.5)
pval
# Contingency table
# A | B
# ------------------------------------
# NY | 30 | 10
# KY | 35 | 5
# AL | 28 | 12
# FL | 20 | 20
X = [[30, 10],
[35, 5],
[28, 12],
[20, 20]]
chi2, pval, dof, expected = chi2_contingency(X)
pval
| 0.60288 | 0.964522 |
<a href="https://colab.research.google.com/github/mohameddhameem/TensorflowCertification/blob/main/Regression_in_TF2_x.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Linear Regression
```
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
np.random.seed(0)
area = 2.5 * np.random.randn(100) + 25
price = 25 * area + 5 + np.random.randint(20,50, size=len(area))
data = np.array([area, price])
data = pd.DataFrame(data=data.T, columns=['area', 'price'])
plt.scatter(data['area'], data['price'])
plt.show()
W = sum(price*(area-np.mean(area))) / sum((area-np.mean(area))**2)
b = np.mean(price) - W*np.mean(area)
print("The regression coefficients are", W,b)
y_pred = W * area + b
plt.plot(area, y_pred, color='red',label="Predicted Price")
plt.scatter(data['area'], data['price'], label="Training Data")
plt.xlabel("Area")
plt.ylabel("Price")
plt.legend()
```
# Multiple linear regression with Estimator API
```
from tensorflow import feature_column as fc
numeric_column = fc.numeric_column
categorical_column_with_vocabulary_list = fc.categorical_column_with_vocabulary_list
featcols = [
tf.feature_column.numeric_column("area"),
tf.feature_column.categorical_column_with_vocabulary_list("type",["bungalow","apartment"])
]
def train_input_fn():
features = {"area":[1000,2000,4000,1000,2000,4000],
"type":["bungalow","bungalow","house",
"apartment","apartment","apartment"]}
labels = [ 500 , 1000 , 1500 , 700 , 1300 , 1900 ]
return features, labels
model = tf.estimator.LinearRegressor(featcols)
model.train(train_input_fn, steps=200)
def predict_input_fn():
features = {"area":[1500,1800],
"type":["house","apt"]}
return features
predictions = model.predict(predict_input_fn)
print(next(predictions))
print(next(predictions))
```
# Boston House price prediction
```
from tensorflow.keras.datasets import boston_housing
(X_train,y_train), (X_test, y_test) = boston_housing.load_data()
features = ['CRIM', 'ZN',
'INDUS','CHAS','NOX','RM','AGE',
'DIS', 'RAD', 'TAX', 'PTRATIO', 'B', 'LSTAT']
x_train_df = pd.DataFrame(X_train, columns= features)
x_test_df = pd.DataFrame(X_test, columns= features)
y_train_df = pd.DataFrame(y_train, columns=['MEDV'])
y_test_df = pd.DataFrame(y_test, columns=['MEDV'])
print(x_train_df.head())
y_train_df.head()
feature_columns = []
for feature_name in features:
feature_columns.append(fc.numeric_column(feature_name, dtype=tf.float32))
def estimator_input_fn(df_data, df_label, epochs=10, shuffle=True, batch_size=32):
def input_function():
ds = tf.data.Dataset.from_tensor_slices((dict(df_data), df_label))
if shuffle:
ds = ds.shuffle(100)
ds = ds.batch(batch_size).repeat(epochs)
return ds
return input_function
train_input_fn = estimator_input_fn(x_train_df, y_train_df)
val_input_fn = estimator_input_fn(x_test_df, y_test_df, epochs=1, shuffle=False)
linear_est = tf.estimator.LinearRegressor(feature_columns=feature_columns, model_dir = 'logs/func/')
linear_est.train(train_input_fn, steps=100)
result = linear_est.evaluate(val_input_fn)
result = linear_est.predict(val_input_fn)
for pred,exp in zip(result, y_test[:32]):
print("Predicted Value: ", pred['predictions'][0], "Expected: ", exp)
```
# MNIST using estimators
```
import tensorflow as tf
from tensorflow import keras
import numpy as np
import matplotlib.pyplot as plt
print(tf.__version__)
(X_train, y_train), (X_test, y_test) = keras.datasets.mnist.load_data()
train_data = X_train/np.float32(255)
train_labels = y_train.astype(np.int32)
eval_data = X_test/np.float32(255)
eval_labels = y_test.astype(np.int32)
feature_columns = [
tf.feature_column.numeric_column("x", shape=[28,28])
]
classifier = tf.estimator.LinearClassifier(
feature_columns=feature_columns,
n_classes=10,
model_dir="mnist_model/"
)
train_input_fn = tf.compat.v1.estimator.inputs.numpy_input_fn(
x={"x": train_data},
y=train_labels,
batch_size=100,
num_epochs=None,
shuffle=True)
classifier.train(input_fn=train_input_fn, steps=10)
val_input_fn = tf.compat.v1.estimator.inputs.numpy_input_fn(
x={"x": eval_data},
y=eval_labels,
num_epochs=1,
shuffle=False)
eval_results = classifier.evaluate(input_fn=val_input_fn)
print(eval_results)
```
|
github_jupyter
|
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
np.random.seed(0)
area = 2.5 * np.random.randn(100) + 25
price = 25 * area + 5 + np.random.randint(20,50, size=len(area))
data = np.array([area, price])
data = pd.DataFrame(data=data.T, columns=['area', 'price'])
plt.scatter(data['area'], data['price'])
plt.show()
W = sum(price*(area-np.mean(area))) / sum((area-np.mean(area))**2)
b = np.mean(price) - W*np.mean(area)
print("The regression coefficients are", W,b)
y_pred = W * area + b
plt.plot(area, y_pred, color='red',label="Predicted Price")
plt.scatter(data['area'], data['price'], label="Training Data")
plt.xlabel("Area")
plt.ylabel("Price")
plt.legend()
from tensorflow import feature_column as fc
numeric_column = fc.numeric_column
categorical_column_with_vocabulary_list = fc.categorical_column_with_vocabulary_list
featcols = [
tf.feature_column.numeric_column("area"),
tf.feature_column.categorical_column_with_vocabulary_list("type",["bungalow","apartment"])
]
def train_input_fn():
features = {"area":[1000,2000,4000,1000,2000,4000],
"type":["bungalow","bungalow","house",
"apartment","apartment","apartment"]}
labels = [ 500 , 1000 , 1500 , 700 , 1300 , 1900 ]
return features, labels
model = tf.estimator.LinearRegressor(featcols)
model.train(train_input_fn, steps=200)
def predict_input_fn():
features = {"area":[1500,1800],
"type":["house","apt"]}
return features
predictions = model.predict(predict_input_fn)
print(next(predictions))
print(next(predictions))
from tensorflow.keras.datasets import boston_housing
(X_train,y_train), (X_test, y_test) = boston_housing.load_data()
features = ['CRIM', 'ZN',
'INDUS','CHAS','NOX','RM','AGE',
'DIS', 'RAD', 'TAX', 'PTRATIO', 'B', 'LSTAT']
x_train_df = pd.DataFrame(X_train, columns= features)
x_test_df = pd.DataFrame(X_test, columns= features)
y_train_df = pd.DataFrame(y_train, columns=['MEDV'])
y_test_df = pd.DataFrame(y_test, columns=['MEDV'])
print(x_train_df.head())
y_train_df.head()
feature_columns = []
for feature_name in features:
feature_columns.append(fc.numeric_column(feature_name, dtype=tf.float32))
def estimator_input_fn(df_data, df_label, epochs=10, shuffle=True, batch_size=32):
def input_function():
ds = tf.data.Dataset.from_tensor_slices((dict(df_data), df_label))
if shuffle:
ds = ds.shuffle(100)
ds = ds.batch(batch_size).repeat(epochs)
return ds
return input_function
train_input_fn = estimator_input_fn(x_train_df, y_train_df)
val_input_fn = estimator_input_fn(x_test_df, y_test_df, epochs=1, shuffle=False)
linear_est = tf.estimator.LinearRegressor(feature_columns=feature_columns, model_dir = 'logs/func/')
linear_est.train(train_input_fn, steps=100)
result = linear_est.evaluate(val_input_fn)
result = linear_est.predict(val_input_fn)
for pred,exp in zip(result, y_test[:32]):
print("Predicted Value: ", pred['predictions'][0], "Expected: ", exp)
import tensorflow as tf
from tensorflow import keras
import numpy as np
import matplotlib.pyplot as plt
print(tf.__version__)
(X_train, y_train), (X_test, y_test) = keras.datasets.mnist.load_data()
train_data = X_train/np.float32(255)
train_labels = y_train.astype(np.int32)
eval_data = X_test/np.float32(255)
eval_labels = y_test.astype(np.int32)
feature_columns = [
tf.feature_column.numeric_column("x", shape=[28,28])
]
classifier = tf.estimator.LinearClassifier(
feature_columns=feature_columns,
n_classes=10,
model_dir="mnist_model/"
)
train_input_fn = tf.compat.v1.estimator.inputs.numpy_input_fn(
x={"x": train_data},
y=train_labels,
batch_size=100,
num_epochs=None,
shuffle=True)
classifier.train(input_fn=train_input_fn, steps=10)
val_input_fn = tf.compat.v1.estimator.inputs.numpy_input_fn(
x={"x": eval_data},
y=eval_labels,
num_epochs=1,
shuffle=False)
eval_results = classifier.evaluate(input_fn=val_input_fn)
print(eval_results)
| 0.623377 | 0.95418 |
# Lesson 1: Create Project Sunshine with a Simple UI
Set up your development environment, Android Studio, and learn about one of the fundamental components of your Android applications: Views.
* 싸이그래머 / 인지모델링 - 파트 1 : 안드로이드
* 김무성
# Contents
* Welcome to Developing Android Apps
* Introducing Your Instructors
* Are you ready for this course?
* Introducing Project Sunshine
* Introducing Project Sunshine
* Introducing More Sunshine
* The Code And Videos
* Starter Code
* Create a New Android Studio Project
* Select a Minimum and Target SDK
* Select a Target SDK
* Finish Creating a New Project
* Launch Sunshine and Create an AVD
* Android Software Stack and Gradle
* Debugging with a Physical Device
* Launching on a Device
* Using the Sunshine Github Repository
* Start to Build the App
* Create a User Interface
* UI Element Quiz
* Add ListItem XML
* Introducing Responsive Design
* Why AbsoluteLayout Is Evil
* Responsive Design Thinking
* Layout Managers
* ScrollViews vs ListViews
* ListView and Recycling
* Add ListView to layout
* Create Some Fake Data
* Adapters
* Initialize the Adapter
* Finding Views findViewById()
* Great Work!
* Review Material for Lesson 1
* Lesson One Recap
* Storytime: Android Platform
# Resources
* 1 Course Resources - https://www.udacity.com/wiki/ud853
- 1.1 Android Studio Setup and Course Resources - https://www.udacity.com/wiki/ud853/course_resources
- Project Sunshine repo (github) - https://github.com/udacity/sunshine-version-2
- 1.2 Design Resources - https://www.udacity.com/wiki/ud853/design_assets
# Welcome to Developing Android Apps
# Introducing Your Instructors
# Are you ready for this course?
# Introducing Project Sunshine
<img src="figures/cap1.png" width=600 />
<img src="figures/cap2.png" width=600 />
# Introducing More Sunshine
# The Code And Videos
# Starter Code
* https://www.udacity.com/course/viewer#!/c-ud853/l-1395568821/m-4419119353
- 지금 참조하는 강좌의 동영상에는 2014년 작성된 코드로 나온다. 그러나 이것은 버전 1.
- github의 소스는 버전2.
- 이 링크의 안내 중에 Option 1을 따르자. - 동영상에 맞는 코드로 맞추는 옵션이다.
- github에서 소스 가져온 후에
- git checkout 1.01_hello_world 로 브랜치를 전환한뒤
- 안드로이드 스튜디오에서 불러오자.
<img src="figures/cap3.png" width=600 />
<img src="figures/cap4.png" width=600 />
# Create SunShine Project
<img src="figures/cap5.png" width=600 />
# Create a New Android Studio Project
<img src="figures/cap6.png" width=600 />
# Select a Minimum and Target SDK
<img src="figures/cap7.png" width=600 />
# Select a Target SDK
<img src="figures/cap8.png" width=600 />
# Finish Creating a New Project
<img src="figures/cap9.png" width=600 />
<img src="figures/cap10.png" width=600 />
<img src="figures/cap11.png" width=600 />
<img src="figures/cap12.png" width=600 />
<img src="figures/cap13.png" width=600 />
<img src="figures/cap14.png" width=600 />
# Launch Sunshine and Create an AVD
<img src="figures/cap15.png" width=600 />
<img src="figures/cap16.png" width=600 />
<img src="figures/cap17.png" width=600 />
<img src="figures/cap18.png" width=600 />
<img src="figures/cap19.png" width=600 />
<img src="figures/cap20.png" width=600 />
<img src="figures/cap21.png" width=600 />
<img src="figures/cap22.png" width=600 />
<img src="figures/cap23.png" width=600 />
<img src="figures/cap24.png" width=600 />
<img src="figures/cap28.png" width=600 />
# Android Software Stack and Gradle
<img src="figures/cap25.png" width=600 />
<img src="figures/cap26.png" width=600 />
<img src="figures/cap27.png" width=600 />
# Debugging with a Physical Device
* https://www.udacity.com/course/viewer#!/c-ud853/l-1395568821/e-1588688602/m-1588688603
<img src="figures/cap29.png" width=600 />
# Launching on a Device
<img src="figures/cap30.png" width=600 />
<img src="figures/cap31.png" width=600 />
<img src="figures/cap32.png" width=600 />
<img src="figures/cap33.png" width=600 />
<img src="figures/cap34.png" width=600 />
# Using the Sunshine Github Repository
* https://www.udacity.com/course/viewer#!/c-ud853/l-1395568821/m-3727538666
# Start to Build the App
<img src="figures/cap35.png" width=600 />
<img src="figures/cap36.png" width=600 />
<img src="figures/cap37.png" width=600 />
<img src="figures/cap38.png" width=600 />
<img src="figures/cap39.png" width=600 />
# Create a User Interface
<img src="figures/cap40.png" width=600 />
<img src="figures/cap41.png" width=600 />
<img src="figures/cap42.png" width=600 />
<img src="figures/cap43.png" width=600 />
<img src="figures/cap44.png" width=600 />
<img src="figures/cap45.png" width=600 />
<img src="figures/cap50.png" width=600 />
<img src="figures/cap51.png" width=600 />
<img src="figures/cap52.png" width=600 />
<img src="figures/cap53.png" width=600 />
<img src="figures/cap54.png" width=600 />
<img src="figures/cap55.png" width=600 />
# UI Element Quiz
<img src="figures/cap56.png" width=600 />
<img src="figures/cap57.png" width=600 />
# Add ListItem XML
<img src="figures/cap58.png" width=600 />
<img src="figures/cap59.png" width=600 />
<img src="figures/cap60.png" width=600 />
<img src="figures/cap61.png" width=600 />
<img src="figures/cap62.png" width=600 />
<img src="figures/cap63.png" width=600 />
<img src="figures/cap64.png" width=600 />
<img src="figures/cap65.png" width=600 />
<img src="figures/cap66.png" width=600 />
<img src="figures/cap67.png" width=600 />
# Introducing Responsive Design
<img src="figures/cap68.png" width=600 />
# Why AbsoluteLayout Is Evil
<img src="figures/cap69.png" width=600 />
# Responsive Design Thinking
<img src="figures/cap70.png" width=600 />
# Layout Managers
<img src="figures/cap71.png" width=600 />
<img src="figures/cap72.png" width=600 />
<img src="figures/cap73.png" width=600 />
<img src="figures/cap74.png" width=600 />
<img src="figures/cap75.png" width=600 />
<img src="figures/cap76.png" width=600 />
<img src="figures/cap77.png" width=600 />
# ScrollViews vs ListViews
<img src="figures/cap78.png" width=600 />
<img src="figures/cap79.png" width=600 />
# ListView and Recycling
<img src="figures/cap80.png" width=600 />
<img src="figures/cap81.png" width=600 />
<img src="figures/cap82.png" width=600 />
# Add ListView to layout
<img src="figures/cap83.png" width=600 />
<img src="figures/cap84.png" width=600 />
<img src="figures/cap85.png" width=600 />
# Create Some Fake Data
<img src="figures/cap86.png" width=600 />
<img src="figures/cap87.png" width=600 />
<img src="figures/cap88.png" width=600 />
<img src="figures/cap89.png" width=600 />
<img src="figures/cap90.png" width=600 />
<img src="figures/cap91.png" width=600 />
# Adapters
<img src="figures/cap92.png" width=600 />
# Initialize the Adapter
<img src="figures/cap93.png" width=600 />
<img src="figures/cap94.png" width=600 />
# Finding Views findViewById()
<img src="figures/cap95.png" width=600 />
<img src="figures/cap96.png" width=600 />
<img src="figures/cap97.png" width=600 />
<img src="figures/cap98.png" width=600 />
<img src="figures/cap99.png" width=600 />
<img src="figures/cap100.png" width=600 />
# Great Work!
# Review Material for Lesson 1
# Lesson One Recap
# Storytime: Android Platform
<img src="figures/cap101.png" width=600 />
# 참고자료
* [1] Developing Android Apps: Android Fundamentals - https://www.udacity.com/course/developing-android-apps--ud853
|
github_jupyter
|
# Lesson 1: Create Project Sunshine with a Simple UI
Set up your development environment, Android Studio, and learn about one of the fundamental components of your Android applications: Views.
* 싸이그래머 / 인지모델링 - 파트 1 : 안드로이드
* 김무성
# Contents
* Welcome to Developing Android Apps
* Introducing Your Instructors
* Are you ready for this course?
* Introducing Project Sunshine
* Introducing Project Sunshine
* Introducing More Sunshine
* The Code And Videos
* Starter Code
* Create a New Android Studio Project
* Select a Minimum and Target SDK
* Select a Target SDK
* Finish Creating a New Project
* Launch Sunshine and Create an AVD
* Android Software Stack and Gradle
* Debugging with a Physical Device
* Launching on a Device
* Using the Sunshine Github Repository
* Start to Build the App
* Create a User Interface
* UI Element Quiz
* Add ListItem XML
* Introducing Responsive Design
* Why AbsoluteLayout Is Evil
* Responsive Design Thinking
* Layout Managers
* ScrollViews vs ListViews
* ListView and Recycling
* Add ListView to layout
* Create Some Fake Data
* Adapters
* Initialize the Adapter
* Finding Views findViewById()
* Great Work!
* Review Material for Lesson 1
* Lesson One Recap
* Storytime: Android Platform
# Resources
* 1 Course Resources - https://www.udacity.com/wiki/ud853
- 1.1 Android Studio Setup and Course Resources - https://www.udacity.com/wiki/ud853/course_resources
- Project Sunshine repo (github) - https://github.com/udacity/sunshine-version-2
- 1.2 Design Resources - https://www.udacity.com/wiki/ud853/design_assets
# Welcome to Developing Android Apps
# Introducing Your Instructors
# Are you ready for this course?
# Introducing Project Sunshine
<img src="figures/cap1.png" width=600 />
<img src="figures/cap2.png" width=600 />
# Introducing More Sunshine
# The Code And Videos
# Starter Code
* https://www.udacity.com/course/viewer#!/c-ud853/l-1395568821/m-4419119353
- 지금 참조하는 강좌의 동영상에는 2014년 작성된 코드로 나온다. 그러나 이것은 버전 1.
- github의 소스는 버전2.
- 이 링크의 안내 중에 Option 1을 따르자. - 동영상에 맞는 코드로 맞추는 옵션이다.
- github에서 소스 가져온 후에
- git checkout 1.01_hello_world 로 브랜치를 전환한뒤
- 안드로이드 스튜디오에서 불러오자.
<img src="figures/cap3.png" width=600 />
<img src="figures/cap4.png" width=600 />
# Create SunShine Project
<img src="figures/cap5.png" width=600 />
# Create a New Android Studio Project
<img src="figures/cap6.png" width=600 />
# Select a Minimum and Target SDK
<img src="figures/cap7.png" width=600 />
# Select a Target SDK
<img src="figures/cap8.png" width=600 />
# Finish Creating a New Project
<img src="figures/cap9.png" width=600 />
<img src="figures/cap10.png" width=600 />
<img src="figures/cap11.png" width=600 />
<img src="figures/cap12.png" width=600 />
<img src="figures/cap13.png" width=600 />
<img src="figures/cap14.png" width=600 />
# Launch Sunshine and Create an AVD
<img src="figures/cap15.png" width=600 />
<img src="figures/cap16.png" width=600 />
<img src="figures/cap17.png" width=600 />
<img src="figures/cap18.png" width=600 />
<img src="figures/cap19.png" width=600 />
<img src="figures/cap20.png" width=600 />
<img src="figures/cap21.png" width=600 />
<img src="figures/cap22.png" width=600 />
<img src="figures/cap23.png" width=600 />
<img src="figures/cap24.png" width=600 />
<img src="figures/cap28.png" width=600 />
# Android Software Stack and Gradle
<img src="figures/cap25.png" width=600 />
<img src="figures/cap26.png" width=600 />
<img src="figures/cap27.png" width=600 />
# Debugging with a Physical Device
* https://www.udacity.com/course/viewer#!/c-ud853/l-1395568821/e-1588688602/m-1588688603
<img src="figures/cap29.png" width=600 />
# Launching on a Device
<img src="figures/cap30.png" width=600 />
<img src="figures/cap31.png" width=600 />
<img src="figures/cap32.png" width=600 />
<img src="figures/cap33.png" width=600 />
<img src="figures/cap34.png" width=600 />
# Using the Sunshine Github Repository
* https://www.udacity.com/course/viewer#!/c-ud853/l-1395568821/m-3727538666
# Start to Build the App
<img src="figures/cap35.png" width=600 />
<img src="figures/cap36.png" width=600 />
<img src="figures/cap37.png" width=600 />
<img src="figures/cap38.png" width=600 />
<img src="figures/cap39.png" width=600 />
# Create a User Interface
<img src="figures/cap40.png" width=600 />
<img src="figures/cap41.png" width=600 />
<img src="figures/cap42.png" width=600 />
<img src="figures/cap43.png" width=600 />
<img src="figures/cap44.png" width=600 />
<img src="figures/cap45.png" width=600 />
<img src="figures/cap50.png" width=600 />
<img src="figures/cap51.png" width=600 />
<img src="figures/cap52.png" width=600 />
<img src="figures/cap53.png" width=600 />
<img src="figures/cap54.png" width=600 />
<img src="figures/cap55.png" width=600 />
# UI Element Quiz
<img src="figures/cap56.png" width=600 />
<img src="figures/cap57.png" width=600 />
# Add ListItem XML
<img src="figures/cap58.png" width=600 />
<img src="figures/cap59.png" width=600 />
<img src="figures/cap60.png" width=600 />
<img src="figures/cap61.png" width=600 />
<img src="figures/cap62.png" width=600 />
<img src="figures/cap63.png" width=600 />
<img src="figures/cap64.png" width=600 />
<img src="figures/cap65.png" width=600 />
<img src="figures/cap66.png" width=600 />
<img src="figures/cap67.png" width=600 />
# Introducing Responsive Design
<img src="figures/cap68.png" width=600 />
# Why AbsoluteLayout Is Evil
<img src="figures/cap69.png" width=600 />
# Responsive Design Thinking
<img src="figures/cap70.png" width=600 />
# Layout Managers
<img src="figures/cap71.png" width=600 />
<img src="figures/cap72.png" width=600 />
<img src="figures/cap73.png" width=600 />
<img src="figures/cap74.png" width=600 />
<img src="figures/cap75.png" width=600 />
<img src="figures/cap76.png" width=600 />
<img src="figures/cap77.png" width=600 />
# ScrollViews vs ListViews
<img src="figures/cap78.png" width=600 />
<img src="figures/cap79.png" width=600 />
# ListView and Recycling
<img src="figures/cap80.png" width=600 />
<img src="figures/cap81.png" width=600 />
<img src="figures/cap82.png" width=600 />
# Add ListView to layout
<img src="figures/cap83.png" width=600 />
<img src="figures/cap84.png" width=600 />
<img src="figures/cap85.png" width=600 />
# Create Some Fake Data
<img src="figures/cap86.png" width=600 />
<img src="figures/cap87.png" width=600 />
<img src="figures/cap88.png" width=600 />
<img src="figures/cap89.png" width=600 />
<img src="figures/cap90.png" width=600 />
<img src="figures/cap91.png" width=600 />
# Adapters
<img src="figures/cap92.png" width=600 />
# Initialize the Adapter
<img src="figures/cap93.png" width=600 />
<img src="figures/cap94.png" width=600 />
# Finding Views findViewById()
<img src="figures/cap95.png" width=600 />
<img src="figures/cap96.png" width=600 />
<img src="figures/cap97.png" width=600 />
<img src="figures/cap98.png" width=600 />
<img src="figures/cap99.png" width=600 />
<img src="figures/cap100.png" width=600 />
# Great Work!
# Review Material for Lesson 1
# Lesson One Recap
# Storytime: Android Platform
<img src="figures/cap101.png" width=600 />
# 참고자료
* [1] Developing Android Apps: Android Fundamentals - https://www.udacity.com/course/developing-android-apps--ud853
| 0.626353 | 0.812979 |
<a href="https://colab.research.google.com/github/ShepherdCode/ShepherdML/blob/master/Nasa2021/CNN_523embed_3k_learning.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# CNN 523 3k
1. Four layers of CNNS and MaxPooling
2. Drop out at 0.2
3. Variable filters and dense neurons
4. Adjusted Learning rate to 0.0005
```
#NC_FILENAME='ncRNA.tiny50.fasta'
#PC_FILENAME='pcRNA.tiny50.fasta'
#NC_FILENAME='ncRNA.gc34.processed.fasta'
#PC_FILENAME='pcRNA.gc34.processed.fasta'
NC_FILENAME='noncod_3000.fasta' # CHANGE THIS TO 1000, 2000, 4000, etc.
PC_FILENAME='coding_3000.fasta'
NC_VAL_FILE='noncod_validation.fasta' # 'noncod_validation.fasta' # CHANGE THIS TO THE UNIFORM VALIDATION FILE
PC_VAL_FILE='coding_validation.fasta' # 'coding_validation.fasta'
MODEL_FILE='Test__3K' # CHANGE THIS IF YOU WANT TO SAVE THE MODEL!
DATAPATH=''
try:
from google.colab import drive
IN_COLAB = True
PATH='/content/drive/'
drive.mount(PATH)
DATAPATH=PATH+'My Drive/data/' # must end in "/"
except:
IN_COLAB = False
DATAPATH='data/' # must end in "/"
NC_FILENAME = DATAPATH+NC_FILENAME
PC_FILENAME = DATAPATH+PC_FILENAME
NC_VAL_FILE = DATAPATH+NC_VAL_FILE
PC_VAL_FILE = DATAPATH+PC_VAL_FILE
MODEL_FILE=DATAPATH+MODEL_FILE
EPOCHS=10 # DECIDE ON SOME AMOUNT AND STICK WITH IT
SPLITS=5
K=1
VOCABULARY_SIZE=4**K+1 # e.g. K=3 => 64 DNA K-mers + 'NNN'
EMBED_DIMEN=2
FILTERS=32
KERNEL=3
NEURONS=24
DROP=0.2
MINLEN=200
MAXLEN=1000 # THIS HAS TO MATCH THE SIMULATION DATA
DENSE_LEN = 1000
ACT="tanh"
LN_RATE = 0.0005
# Load our own tools
# TO DO: don't go to GitHub if the file is already local.
GITHUB = True
if GITHUB:
#!pip install requests # Uncomment this if necessary. Seems to be pre-installed.
import requests
r = requests.get('https://raw.githubusercontent.com/ShepherdCode/ShepherdML/master/Strings/tools_fasta.py')
with open('tools_fasta.py', 'w') as f:
f.write(r.text)
# TO DO: delete the file after import
import tools_fasta as tools
tools.yahoo() # If this prints "Yahoo!" the the import was successful.
TOOLS_CHANGED = False # set to True to re-run with a new version of tools
if TOOLS_CHANGED:
from importlib import reload
tools=reload(tools)
print(dir(tools)) # run this to see EVERYTHING in the tools module
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import ShuffleSplit
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedKFold
from sklearn.model_selection import StratifiedKFold
import tensorflow as tf
from tensorflow import keras
import time
dt='float32'
tf.keras.backend.set_floatx(dt)
```
Build model
```
def compile_model(model):
print("COMPILE...")
## learn rate = initial_learning_rate * decay_rate ^ (step / decay_steps)
#adam_default_learn_rate = 0.001
#schedule = tf.keras.optimizers.schedules.ExponentialDecay(
# initial_learning_rate = adam_default_learn_rate*10,
# decay_steps=10000, decay_rate=0.99, staircase=True)
#alrd = tf.keras.optimizers.Adam(learning_rate=schedule)
ln_rate = tf.keras.optimizers.Adam(learning_rate = LN_RATE)
bc=tf.keras.losses.BinaryCrossentropy(from_logits=False)
model.compile(loss=bc, optimizer=ln_rate, metrics=["accuracy"])
print("...COMPILED")
return model
def build_model():
#SHAPE=(MAXLEN,5)
SHAPE=(MAXLEN,4)
# 4 input letters, 4 output dimensions, 1000 letters/RNA
elayer = keras.layers.Embedding(4,4,input_length=1000)
clayer1 = keras.layers.Conv1D(FILTERS,KERNEL,activation=ACT,padding="same",
input_shape=SHAPE)
clayer2 = keras.layers.Conv1D(FILTERS,KERNEL,activation=ACT,padding="same")
clayer3 = keras.layers.MaxPooling1D(2)
clayer4 = keras.layers.Conv1D(FILTERS,KERNEL,activation=ACT,padding="same")
clayer5 = keras.layers.Conv1D(FILTERS,KERNEL,activation=ACT,padding="same")
clayer6 = keras.layers.MaxPooling1D(2)
clayer7 = keras.layers.Conv1D(FILTERS,KERNEL,activation=ACT,padding="same")
clayer8 = keras.layers.Conv1D(FILTERS,KERNEL,activation=ACT,padding="same")
clayer9 = keras.layers.MaxPooling1D(2)
clayer10 = keras.layers.Conv1D(FILTERS,KERNEL,activation=ACT,padding="same")
clayer11 = keras.layers.Conv1D(FILTERS,KERNEL,activation=ACT,padding="same")
clayer12 = keras.layers.MaxPooling1D(2)
clayer13 = keras.layers.Flatten()
dlayer1 = keras.layers.Dense(NEURONS, activation=ACT,dtype=dt, input_shape=[DENSE_LEN])
dlayer2 = keras.layers.Dropout(DROP)
dlayer3 = keras.layers.Dense(NEURONS, activation=ACT,dtype=dt)
dlayer4 = keras.layers.Dropout(DROP)
output_layer = keras.layers.Dense(1, activation="sigmoid", dtype=dt)
cnn = keras.models.Sequential()
cnn.add(elayer)
cnn.add(clayer1)
cnn.add(clayer2)
cnn.add(clayer3)
cnn.add(clayer4)
cnn.add(clayer5)
cnn.add(clayer6)
cnn.add(clayer7)
cnn.add(clayer8)
cnn.add(clayer9)
cnn.add(clayer10)
cnn.add(clayer11)
cnn.add(clayer12)
cnn.add(clayer13)
cnn.add(dlayer1)
cnn.add(dlayer2)
cnn.add(dlayer3)
cnn.add(dlayer4)
cnn.add(output_layer)
mlpc = compile_model(cnn)
return mlpc
```
Cross validation
```
def do_cross_validation(X,y,given_model,X_VALID,Y_VALID):
cv_scores = []
fold=0
splitter = ShuffleSplit(n_splits=SPLITS, test_size=0.1, random_state=37863)
for train_index,valid_index in splitter.split(X):
fold += 1
X_train=X[train_index] # use iloc[] for dataframe
y_train=y[train_index]
X_valid=X[valid_index]
y_valid=y[valid_index]
# Avoid continually improving the same model.
model = compile_model(keras.models.clone_model(given_model))
bestname=MODEL_FILE+".cv."+str(fold)+".best"
mycallbacks = [keras.callbacks.ModelCheckpoint(
filepath=bestname, save_best_only=True,
monitor='val_accuracy', mode='max')]
print("FIT")
start_time=time.time()
history=model.fit(X_train, y_train,
epochs=EPOCHS, verbose=1, callbacks=mycallbacks,
validation_data=(X_valid,y_valid))
# THE VALIDATION ABOVE IS JUST FOR SHOW
end_time=time.time()
elapsed_time=(end_time-start_time)
print("Fold %d, %d epochs, %d sec"%(fold,EPOCHS,elapsed_time))
pd.DataFrame(history.history).plot(figsize=(8,5))
plt.grid(True)
plt.gca().set_ylim(0,1)
plt.show()
best_model=keras.models.load_model(bestname)
# THE VALIDATION BELOW IS FOR KEEPS
scores = best_model.evaluate(X_VALID, Y_VALID, verbose=0)
print("%s: %.2f%%" % (best_model.metrics_names[1], scores[1]*100))
cv_scores.append(scores[1] * 100)
print()
print("%d-way Cross Validation mean %.2f%% (+/- %.2f%%)" % (fold, np.mean(cv_scores), np.std(cv_scores)))
```
## Train on RNA lengths 200-1Kb
```
print ("Compile the model")
model=build_model()
print ("Summarize the model")
print(model.summary()) # Print this only once
#model.save(MODEL_FILE+'.model')
def load_data_from_files(nc_filename,pc_filename):
FREEMEM=True # use False for debugging, True for production
print("Load data from files.")
nc_seq=tools.load_fasta(nc_filename,0)
pc_seq=tools.load_fasta(pc_filename,1)
train_set=pd.concat((nc_seq,pc_seq),axis=0)
print("Ready: train_set")
subset=tools.make_slice(train_set,MINLEN,MAXLEN)# One array to two: X and y
if FREEMEM:
nc_seq=None
pc_seq=None
train_set=None
(X1,y_train)=tools.separate_X_and_y(subset)
# X1 is pandas df of ("list" of one sequence)
X2=X1.to_numpy() # numpy ndarray of ("list" of one sequence)
X3=[elem[0] for elem in X2] # numpy ndarray of ACGT-str
# X3? It might be faster to use int-array than char-array. Come back to this.
X4=X3 # no-op
print("X4",type(X4))
#print(X4[0])
if FREEMEM:
X1=None
X2=None
X3=None
X5=[]
dna_to_int = {'A':0,'C':1,'G':2,'T':3}
for x in X4:
a=[]
for c in x:
i = dna_to_int[c]
a.append(i)
X5.append(a)
X5=np.asarray(X5)
print("X5",type(X5))
print(X5.shape)
if FREEMEM:
X4=None
X_train=X5
if FREEMEM:
X5=None
print("X_train",type(X_train))
y_train=y_train.to_numpy()
print(X_train.shape)
print(X_train[0].shape)
print(X_train[0])
return X_train,y_train
print("Loading training data...")
X_train,y_train = load_data_from_files(NC_FILENAME,PC_FILENAME)
print("Loading validation data...")
X_VALID,Y_VALID = load_data_from_files(NC_VAL_FILE,PC_VAL_FILE)
print ("Cross validation")
do_cross_validation(X_train,y_train,model,X_VALID,Y_VALID)
print ("Done")
```
|
github_jupyter
|
#NC_FILENAME='ncRNA.tiny50.fasta'
#PC_FILENAME='pcRNA.tiny50.fasta'
#NC_FILENAME='ncRNA.gc34.processed.fasta'
#PC_FILENAME='pcRNA.gc34.processed.fasta'
NC_FILENAME='noncod_3000.fasta' # CHANGE THIS TO 1000, 2000, 4000, etc.
PC_FILENAME='coding_3000.fasta'
NC_VAL_FILE='noncod_validation.fasta' # 'noncod_validation.fasta' # CHANGE THIS TO THE UNIFORM VALIDATION FILE
PC_VAL_FILE='coding_validation.fasta' # 'coding_validation.fasta'
MODEL_FILE='Test__3K' # CHANGE THIS IF YOU WANT TO SAVE THE MODEL!
DATAPATH=''
try:
from google.colab import drive
IN_COLAB = True
PATH='/content/drive/'
drive.mount(PATH)
DATAPATH=PATH+'My Drive/data/' # must end in "/"
except:
IN_COLAB = False
DATAPATH='data/' # must end in "/"
NC_FILENAME = DATAPATH+NC_FILENAME
PC_FILENAME = DATAPATH+PC_FILENAME
NC_VAL_FILE = DATAPATH+NC_VAL_FILE
PC_VAL_FILE = DATAPATH+PC_VAL_FILE
MODEL_FILE=DATAPATH+MODEL_FILE
EPOCHS=10 # DECIDE ON SOME AMOUNT AND STICK WITH IT
SPLITS=5
K=1
VOCABULARY_SIZE=4**K+1 # e.g. K=3 => 64 DNA K-mers + 'NNN'
EMBED_DIMEN=2
FILTERS=32
KERNEL=3
NEURONS=24
DROP=0.2
MINLEN=200
MAXLEN=1000 # THIS HAS TO MATCH THE SIMULATION DATA
DENSE_LEN = 1000
ACT="tanh"
LN_RATE = 0.0005
# Load our own tools
# TO DO: don't go to GitHub if the file is already local.
GITHUB = True
if GITHUB:
#!pip install requests # Uncomment this if necessary. Seems to be pre-installed.
import requests
r = requests.get('https://raw.githubusercontent.com/ShepherdCode/ShepherdML/master/Strings/tools_fasta.py')
with open('tools_fasta.py', 'w') as f:
f.write(r.text)
# TO DO: delete the file after import
import tools_fasta as tools
tools.yahoo() # If this prints "Yahoo!" the the import was successful.
TOOLS_CHANGED = False # set to True to re-run with a new version of tools
if TOOLS_CHANGED:
from importlib import reload
tools=reload(tools)
print(dir(tools)) # run this to see EVERYTHING in the tools module
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import ShuffleSplit
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedKFold
from sklearn.model_selection import StratifiedKFold
import tensorflow as tf
from tensorflow import keras
import time
dt='float32'
tf.keras.backend.set_floatx(dt)
def compile_model(model):
print("COMPILE...")
## learn rate = initial_learning_rate * decay_rate ^ (step / decay_steps)
#adam_default_learn_rate = 0.001
#schedule = tf.keras.optimizers.schedules.ExponentialDecay(
# initial_learning_rate = adam_default_learn_rate*10,
# decay_steps=10000, decay_rate=0.99, staircase=True)
#alrd = tf.keras.optimizers.Adam(learning_rate=schedule)
ln_rate = tf.keras.optimizers.Adam(learning_rate = LN_RATE)
bc=tf.keras.losses.BinaryCrossentropy(from_logits=False)
model.compile(loss=bc, optimizer=ln_rate, metrics=["accuracy"])
print("...COMPILED")
return model
def build_model():
#SHAPE=(MAXLEN,5)
SHAPE=(MAXLEN,4)
# 4 input letters, 4 output dimensions, 1000 letters/RNA
elayer = keras.layers.Embedding(4,4,input_length=1000)
clayer1 = keras.layers.Conv1D(FILTERS,KERNEL,activation=ACT,padding="same",
input_shape=SHAPE)
clayer2 = keras.layers.Conv1D(FILTERS,KERNEL,activation=ACT,padding="same")
clayer3 = keras.layers.MaxPooling1D(2)
clayer4 = keras.layers.Conv1D(FILTERS,KERNEL,activation=ACT,padding="same")
clayer5 = keras.layers.Conv1D(FILTERS,KERNEL,activation=ACT,padding="same")
clayer6 = keras.layers.MaxPooling1D(2)
clayer7 = keras.layers.Conv1D(FILTERS,KERNEL,activation=ACT,padding="same")
clayer8 = keras.layers.Conv1D(FILTERS,KERNEL,activation=ACT,padding="same")
clayer9 = keras.layers.MaxPooling1D(2)
clayer10 = keras.layers.Conv1D(FILTERS,KERNEL,activation=ACT,padding="same")
clayer11 = keras.layers.Conv1D(FILTERS,KERNEL,activation=ACT,padding="same")
clayer12 = keras.layers.MaxPooling1D(2)
clayer13 = keras.layers.Flatten()
dlayer1 = keras.layers.Dense(NEURONS, activation=ACT,dtype=dt, input_shape=[DENSE_LEN])
dlayer2 = keras.layers.Dropout(DROP)
dlayer3 = keras.layers.Dense(NEURONS, activation=ACT,dtype=dt)
dlayer4 = keras.layers.Dropout(DROP)
output_layer = keras.layers.Dense(1, activation="sigmoid", dtype=dt)
cnn = keras.models.Sequential()
cnn.add(elayer)
cnn.add(clayer1)
cnn.add(clayer2)
cnn.add(clayer3)
cnn.add(clayer4)
cnn.add(clayer5)
cnn.add(clayer6)
cnn.add(clayer7)
cnn.add(clayer8)
cnn.add(clayer9)
cnn.add(clayer10)
cnn.add(clayer11)
cnn.add(clayer12)
cnn.add(clayer13)
cnn.add(dlayer1)
cnn.add(dlayer2)
cnn.add(dlayer3)
cnn.add(dlayer4)
cnn.add(output_layer)
mlpc = compile_model(cnn)
return mlpc
def do_cross_validation(X,y,given_model,X_VALID,Y_VALID):
cv_scores = []
fold=0
splitter = ShuffleSplit(n_splits=SPLITS, test_size=0.1, random_state=37863)
for train_index,valid_index in splitter.split(X):
fold += 1
X_train=X[train_index] # use iloc[] for dataframe
y_train=y[train_index]
X_valid=X[valid_index]
y_valid=y[valid_index]
# Avoid continually improving the same model.
model = compile_model(keras.models.clone_model(given_model))
bestname=MODEL_FILE+".cv."+str(fold)+".best"
mycallbacks = [keras.callbacks.ModelCheckpoint(
filepath=bestname, save_best_only=True,
monitor='val_accuracy', mode='max')]
print("FIT")
start_time=time.time()
history=model.fit(X_train, y_train,
epochs=EPOCHS, verbose=1, callbacks=mycallbacks,
validation_data=(X_valid,y_valid))
# THE VALIDATION ABOVE IS JUST FOR SHOW
end_time=time.time()
elapsed_time=(end_time-start_time)
print("Fold %d, %d epochs, %d sec"%(fold,EPOCHS,elapsed_time))
pd.DataFrame(history.history).plot(figsize=(8,5))
plt.grid(True)
plt.gca().set_ylim(0,1)
plt.show()
best_model=keras.models.load_model(bestname)
# THE VALIDATION BELOW IS FOR KEEPS
scores = best_model.evaluate(X_VALID, Y_VALID, verbose=0)
print("%s: %.2f%%" % (best_model.metrics_names[1], scores[1]*100))
cv_scores.append(scores[1] * 100)
print()
print("%d-way Cross Validation mean %.2f%% (+/- %.2f%%)" % (fold, np.mean(cv_scores), np.std(cv_scores)))
print ("Compile the model")
model=build_model()
print ("Summarize the model")
print(model.summary()) # Print this only once
#model.save(MODEL_FILE+'.model')
def load_data_from_files(nc_filename,pc_filename):
FREEMEM=True # use False for debugging, True for production
print("Load data from files.")
nc_seq=tools.load_fasta(nc_filename,0)
pc_seq=tools.load_fasta(pc_filename,1)
train_set=pd.concat((nc_seq,pc_seq),axis=0)
print("Ready: train_set")
subset=tools.make_slice(train_set,MINLEN,MAXLEN)# One array to two: X and y
if FREEMEM:
nc_seq=None
pc_seq=None
train_set=None
(X1,y_train)=tools.separate_X_and_y(subset)
# X1 is pandas df of ("list" of one sequence)
X2=X1.to_numpy() # numpy ndarray of ("list" of one sequence)
X3=[elem[0] for elem in X2] # numpy ndarray of ACGT-str
# X3? It might be faster to use int-array than char-array. Come back to this.
X4=X3 # no-op
print("X4",type(X4))
#print(X4[0])
if FREEMEM:
X1=None
X2=None
X3=None
X5=[]
dna_to_int = {'A':0,'C':1,'G':2,'T':3}
for x in X4:
a=[]
for c in x:
i = dna_to_int[c]
a.append(i)
X5.append(a)
X5=np.asarray(X5)
print("X5",type(X5))
print(X5.shape)
if FREEMEM:
X4=None
X_train=X5
if FREEMEM:
X5=None
print("X_train",type(X_train))
y_train=y_train.to_numpy()
print(X_train.shape)
print(X_train[0].shape)
print(X_train[0])
return X_train,y_train
print("Loading training data...")
X_train,y_train = load_data_from_files(NC_FILENAME,PC_FILENAME)
print("Loading validation data...")
X_VALID,Y_VALID = load_data_from_files(NC_VAL_FILE,PC_VAL_FILE)
print ("Cross validation")
do_cross_validation(X_train,y_train,model,X_VALID,Y_VALID)
print ("Done")
| 0.543833 | 0.708931 |
# TODO:
1. Verify that the merge result is as expected. I noticed that when I checked the length of the EPA for a 6-month period, the length was longer than the post-merge 6-month period. This implies that the EIA data doesn't have some facilities that are listed in EPA. I haven't verified this though.
2. NaN values also need to be replaced with 0's.
3. Numbers are being stored in scientific notation
```
%matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
import os
import glob
import re
import cPickle as pickle
import gzip
import seaborn as sns
```
## Loading the EIA Data, the path may need to be updated...
This will take a few minutes to run.
```
#Iterate through the directory to find all the files to import
#Modified so that it also works on macs
path = os.path.join('EIA Data', '923-No_Header')
full_path = os.path.join(path, '*.*')
eiaNames = os.listdir(path)
#Rename the keys for easier merging later
fileNameMap = {'EIA923 SCHEDULES 2_3_4_5 Final 2010.xls':2010,
'EIA923 SCHEDULES 2_3_4_5 M Final 2009 REVISED 05252011.XLS':2009,
'eia923December2008.xls':2008,
'EIA923_Schedules_2_3_4_5_2011_Final_Revision.xlsx':2011,
'EIA923_Schedules_2_3_4_5_2012_Final_Release_12.04.2013.xlsx':2012,
'EIA923_Schedules_2_3_4_5_2013_Final_Revision.xlsx':2013,
'EIA923_Schedules_2_3_4_5_M_12_2014_Final_Revision.xlsx':2014,
'EIA923_Schedules_2_3_4_5_M_12_2015_Final.xlsx':2015,
'f906920_2007.xls':2007}
#Load the files into data frames, one df per file
eiaDict = {fileNameMap[fn]:pd.read_excel(os.path.join(path, fn)) for fn in eiaNames}
eiaDict = {key:val[val["NERC Region"] == "TRE"] for key, val in eiaDict.iteritems()}
```
The excel documents have different column names so we need to standardize them all
```
#Dict of values to replace to standardize column names across all dataframes
monthDict = {"JANUARY":"JAN",
"FEBRUARY":"FEB",
"MARCH":"MAR",
"APRIL":"APR",
"MAY":"MAY",
"JUNE":"JUN",
"JULY":"JUL",
"AUGUST":"AUG",
"SEPTEMBER":"SEP",
"OCTOBER":"OCT",
"NOVEMBER":"NOV",
"DECEMBER":"DEC"}
replaceDict = {"ELECTRIC":"ELEC",
"&":"AND",
"I.D.":"ID",
"MMBTUPER":"MMBTU_PER"}
#Add "MMBTUMON" : "MMBTU_MON" to be replaced
for month in monthDict.values():
replaceDict["MMBTU"+month] = "MMBTU_" + month
#Replace the column name
def rename(col):
for old, new in monthDict.iteritems():
col = col.replace(old, new)
for old, new in replaceDict.iteritems():
col = col.replace(old, new)
col = col.replace("MMBTUS", "MMBTU")
return col
#Iterate through each column name of each dataframe to standardize
for key, df in eiaDict.iteritems():
colNames = [name.replace("\n", "_").replace(" ", "_").strip().upper() for name in df.columns]
colNames = [rename(col) for col in colNames]
eiaDict[key].columns = colNames
```
Define which columns we need to sum, and which columns don't need to be summed, but we still need to keep.
Note: If we don't care about monthly stuff we can delete the second block of code.
```
#Define the columns that are necessary but are not summable
allCols = eiaDict[fileNameMap.values()[0]].columns
nonSumCols = ["PLANT_ID", "PLANT_NAME", "YEAR"]
#Define the columns that contain the year's totals (Used to calc fuel type %)
yearCols = ["TOTAL_FUEL_CONSUMPTION_QUANTITY", "ELEC_FUEL_CONSUMPTION_QUANTITY",
"TOTAL_FUEL_CONSUMPTION_MMBTU", "ELEC_FUEL_CONSUMPTION_MMBTU",
"NET_GENERATION_(MEGAWATTHOURS)"]
#Define the columns that are necessary and summable
sumCols = []
sumCols.extend(yearCols)
# regex = re.compile(r"^ELEC_QUANTITY_.*")
# sumCols.extend([col for col in allCols if regex.search(col)])
regex = re.compile(r"^MMBTU_PER_UNIT_.*")
sumCols.extend([col for col in allCols if regex.search(col)])
regex = re.compile(r"^TOT_MMBTU_.*")
sumCols.extend([col for col in allCols if regex.search(col)])
regex = re.compile(r"^ELEC_MMBTUS_.*")
sumCols.extend([col for col in allCols if regex.search(col)])
regex = re.compile(r"^NETGEN_.*")
sumCols.extend([col for col in allCols if regex.search(col)])
```
Get a list of all the different fuel type codes. If we don't care about all of them, then just hardcode the list
```
fuelTypes = []
fuelTypes.extend([fuelType for df in eiaDict.values() for fuelType in df["REPORTED_FUEL_TYPE_CODE"].tolist()])
fuelTypes = set(fuelTypes)
fuelTypes
```
3 parts to aggregate by facility, and to calculate the % of each type of fuel. This will take a few minutes to run.
The end result is aggEIADict.
```
#Actually calculate the % type for each facility grouping
def calcPerc(group, aggGroup, fuelType, col):
#Check to see if the facility has a record for the fuel type, and if the total column > 0
if len(group[group["REPORTED_FUEL_TYPE_CODE"] == fuelType]) > 0 and aggGroup[col] > 0:
#summing fuel type because a facility may have multiple plants with the same fuel type
return float((group[group["REPORTED_FUEL_TYPE_CODE"] == fuelType][col]).sum())/aggGroup[col]
else:
return 0
#Perform the aggregation on facility level
def aggAndCalcPerc(group):
aggGroup = group.iloc[0][nonSumCols] #Get the non-agg columns
aggGroup = aggGroup.append(group[sumCols].sum()) #Aggregate the agg columns and append to non-agg
percCols = {col + " %" + fuelType:calcPerc(group, aggGroup, fuelType, col) for col in yearCols for fuelType in fuelTypes}
aggGroup = aggGroup.append(pd.Series(percCols))
return aggGroup
#Iterate through each dataframe to perform aggregation by facility
aggEIADict = dict()
for key, df in eiaDict.iteritems():
gb = df.groupby(by="PLANT_ID")
#aggGroup will be a list of panda series, each series representing a facility
aggGroup = [aggAndCalcPerc(gb.get_group(group)) for group in gb.groups]
aggEIADict[key] = pd.DataFrame(aggGroup)
```
### Column order doesn't match in all years
```
aggEIADict[2007].head()
aggEIADict[2015].head()
```
### Export the EIA 923 data as pickle
Just sending the dictionary to a pickle file for now. At least doing this will save several min of time loading and processing the data in the future.
```
filename = 'EIA 923.pkl'
path = '../Clean Data'
fullpath = os.path.join(path, filename)
pickle.dump(aggEIADict, open(fullpath, 'wb'))
```
### Combine all df's from the dict into one df
Concat all dataframes, reset the index, determine the primary fuel type for each facility, filter to only include fossil power plants, and export as a csv
```
all923 = pd.concat(aggEIADict)
all923.head()
all923.reset_index(drop=True, inplace=True)
# Check column numbers to use in the function below
all923.iloc[1,1:27]
def top_fuel(row):
#Fraction of largest fuel for electric heat input
try:
fuel = row.iloc[1:27].idxmax()[29:]
except:
return None
return fuel
all923['FUEL'] = all923.apply(top_fuel, axis=1)
all923.head()
fossil923 = all923.loc[all923['FUEL'].isin(['DFO', 'LIG', 'NG', 'PC', 'SUB'])]
```
### Export the EIA 923 data dataframe as csv
Export the dataframe with primary fuel and filtered to only include fossil plants
```
filename = 'Fossil EIA 923.csv'
path = '../Clean Data'
fullpath = os.path.join(path, filename)
fossil923.to_csv(fullpath)
```
## Loading the EPA Data, the path may need to be updated...
```
#Read the EPA files into a dataframe
path2 = os.path.join('EPA air markets')
epaNames = os.listdir(path2)
filePaths = {dn:os.path.join(path2, dn, "*.txt") for dn in epaNames}
filePaths = {dn:glob.glob(val) for dn, val in filePaths.iteritems()}
epaDict = {key:pd.read_csv(fp, index_col = False) for key, val in filePaths.iteritems() for fp in val}
```
First rename the column name so we can merge on that column, then change the datatype of date to a datetime object
```
#Rename the column names to remove the leading space.
for key, df in epaDict.iteritems():
colNames = [name.upper().strip() for name in df.columns]
colNames[colNames.index("FACILITY ID (ORISPL)")] = "PLANT_ID"
epaDict[key].columns = colNames
#Convert DATE to datetime object
#Add new column DATETIME with both date and hour
for key, df in epaDict.iteritems():
epaDict[key]["DATE"] = pd.to_datetime(df["DATE"])
epaDict[key]['DATETIME'] = df['DATE'] + pd.to_timedelta(df['HOUR'], unit='h')
```
The DataFrames in `epaDict` contain all power plants in Texas. We can filter on `NERC REGION` so that it only includes ERCOT.
```
set(epaDict['2015 July-Dec'].loc[:,'NERC REGION'])
#Boolean filter to only keep ERCOT plants
for key, df in epaDict.iteritems():
epaDict[key] = df[df["NERC REGION"] == "ERCOT"].reset_index(drop = True)
set(epaDict['2015 July-Dec'].loc[:,'NERC REGION'])
epaDict['2015 July-Dec'].head()
```
### Export EPA data as a series of dataframes
The whole dictionary is too big as a pickle file
```
# pickle with gzip, from http://stackoverflow.com/questions/18474791/decreasing-the-size-of-cpickle-objects
def save_zipped_pickle(obj, filename, protocol=-1):
with gzip.open(filename, 'wb') as f:
pickle.dump(obj, f, protocol)
filename = 'EPA hourly dictionary.pgz'
path = '../Clean Data'
fullpath = os.path.join(path, filename)
save_zipped_pickle(epaDict, fullpath)
df = epaDict['2015 July-Dec']
df.head()
set(df['PLANT_ID'])
df_temp = df[df['PLANT_ID'].isin([127, 298, 3439])].fillna(0)
df_temp.head()
g = sns.FacetGrid(df_temp, col='PLANT_ID')
g.map(plt.plot, 'datetime', 'GROSS LOAD (MW)')
g.set_xticklabels(rotation=30)
path = os.path.join('..', 'Exploratory visualization', 'Midterm figures', 'Sample hourly load.svg')
plt.savefig(path)
```
## Finally join the two data sources
Switch to an inner join?
**No need to join. Can keep them as separate databases, since one is hourly data and the other is annual/monthly** Create a clustering dataframe with index of all plant IDs (from the EPA hourly data), add columns with variables. Calculate the inputs in separate dataframes - example is to calculate ramp rate values in the EPA hourly data, then put the results in the clustering dataframe.
```
#Join the two data sources on PLANT_ID
fullData = {key:df.merge(aggEIADict[df["YEAR"][0]], on="PLANT_ID") for key, df in epaDict.iteritems()}
fullData[fullData.keys()[0]].head()
```
BIT, SUB, LIG, NG, DFO, RFO
```
[x for x in fullData[fullData.keys()[0]].columns]
```
## Loading EIA 860 Data
```
# Iterate through the directory to find all the files to import
path = os.path.join('EIA Data', '860-No_Header')
full_path = os.path.join(path, '*.*')
eia860Names = os.listdir(path)
# Rename the keys for easier merging later
fileName860Map = { 'GenY07.xls':2007,
'GenY08.xls':2008,
'GeneratorY09.xls':2009,
'GeneratorsY2010.xls':2010,
'GeneratorY2011.xlsx':2011,
'GeneratorY2012.xlsx':2012,
'3_1_Generator_Y2013.xlsx':2013,
'3_1_Generator_Y2014.xlsx':2014,
'3_1_Generator_Y2015.xlsx':2015}
#Load the files into data frames, one df per file
eia860Dict = {fileName860Map[fn]:pd.read_excel(os.path.join(path, fn)) for fn in eia860Names}
#Dict of values to replace to standardize column names across all dataframes
renameDict = { "PLNTCODE":"PLANT_ID",
"PLANT_CODE":"PLANT_ID",
"Plant Code":"PLANT_ID",
"NAMEPLATE":"NAMEPLATE_CAPACITY(MW)",
"Nameplate Capacity (MW)":"NAMEPLATE_CAPACITY(MW)"}
#Replace the column name
def rename860(col):
for old, new in renameDict.iteritems():
col = col.replace(old, new)
return col
#Iterate through each column name of each dataframe to standardize and select columns 'PLANT_ID', 'NAMEPLATE_CAPACITY(MW)'
for key, df in eia860Dict.iteritems():
colNames = [rename860(col) for col in df.columns]
eia860Dict[key].columns = colNames
eia860Dict[key] = eia860Dict[key][["PLANT_ID", "NAMEPLATE_CAPACITY(MW)"]]
# verify the tables
for key, df in eia860Dict.iteritems():
print key, df.columns, len(df)
# Iterate through each dataframe to perform aggregation by PLANT_ID
for key, df in eia860Dict.iteritems():
gb = df.groupby(by='PLANT_ID').apply(lambda x: x['NAMEPLATE_CAPACITY(MW)'].sum())
eia860Dict[key]['NAMEPLATE_CAPACITY(MW)'] = eia860Dict[key].PLANT_ID.apply(gb.get_value)
eia860Dict[key] = eia860Dict[key].drop_duplicates(subset=['PLANT_ID', 'NAMEPLATE_CAPACITY(MW)'])
eia860Dict[key] = eia860Dict[key].sort_values(by='PLANT_ID').reset_index(drop=True)
```
### Export EIA 860 data
```
filename = 'EIA 860.pkl'
path = '../Clean Data'
fullpath = os.path.join(path, filename)
pickle.dump(eia860Dict, open(fullpath, 'wb'))
```
## Creating Final DataFrame for Clustering Algorithm:
#### clusterDict {year : cluster_DF}
1. For each PLANT_ID in aggEIADict, fetch the corresponding aggregated NAMEPLATE_CAPACITY(MW)
```
clusterDict = dict()
for key, df in eia860Dict.iteritems():
clusterDict[key] = pd.merge(aggEIADict[key], eia860Dict[key], how='left', on='PLANT_ID')[['PLANT_ID', 'NAMEPLATE_CAPACITY(MW)']]
clusterDict[key].rename(columns={'NAMEPLATE_CAPACITY(MW)': 'capacity', 'PLANT_ID': 'plant_id'}, inplace=True)
# verify for no loss of data
for key, df in eia860Dict.iteritems():
print key, len(clusterDict[key]), len(aggEIADict[key])
clusterDict[2015].head()
```
Function to get fuel type
```
fuel_cols = [col for col in aggEIADict[2008].columns if 'ELEC_FUEL_CONSUMPTION_MMBTU %' in col]
def top_fuel(row):
#Fraction of largest fuel for electric heat input
try:
fuel = row.idxmax()[29:]
except:
return None
return fuel
# clusterDict[2008]['fuel'] = aggEIADict[2008][fuel_cols].apply(top_fuel, axis=1)
```
Calculate Capacity factor, Efficiency, Fuel type
```
for key, df in clusterDict.iteritems():
clusterDict[key]['year'] = key
clusterDict[key]['capacity_factor'] = aggEIADict[key]['NET_GENERATION_(MEGAWATTHOURS)'] / (8670*clusterDict[key]['capacity'])
clusterDict[key]['efficiency'] = (aggEIADict[key]['NET_GENERATION_(MEGAWATTHOURS)']*3.412)/(1.0*aggEIADict[key]['ELEC_FUEL_CONSUMPTION_MMBTU'])
clusterDict[key]['fuel_type'] = aggEIADict[key][fuel_cols].apply(top_fuel, axis=1)
clusterDict[key] = clusterDict[key][clusterDict[key]['fuel_type'].isin(['SUB',
'LIG',
'DFO',
'NG',
'PC'])]
```
Merge all epa files in one df
```
columns = ['PLANT_ID', 'YEAR', 'DATE', 'HOUR', 'GROSS LOAD (MW)']
counter = 0
for key, df in epaDict.iteritems():
if counter == 0:
result = epaDict[key][columns]
counter = 1
else:
result = result.append(epaDict[key][columns], ignore_index=True)
# Change nan to 0
result.fillna(0, inplace=True)
result.describe()
```
Function to calculate the ramp rate for every hour
```
def plant_gen_delta(df):
"""
For every plant in the input df, calculate the change in gross load (MW)
from the previous hour.
input:
df: dataframe of EPA clean air markets data
return:
df: concatanated list of dataframes
"""
df_list = []
for plant in df['PLANT_ID'].unique():
temp = df.loc[df['PLANT_ID'] == plant,:]
gen_change = temp.loc[:,'GROSS LOAD (MW)'].values - temp.loc[:,'GROSS LOAD (MW)'].shift(1).values
temp.loc[:,'Gen Change'] = gen_change
df_list.append(temp)
return pd.concat(df_list)
ramp_df = plant_gen_delta(result)
ramp_df.describe()
```
Get the max ramp rate for every plant for each year
```
cols = ['PLANT_ID', 'YEAR', 'Gen Change']
ramp_rate_list = []
for year in ramp_df['YEAR'].unique():
for plant in ramp_df.loc[ramp_df['YEAR']==year,'PLANT_ID'].unique():
# 95th percentile ramp rate per plant per year
ramp_95 = ramp_df.loc[(ramp_df['PLANT_ID']== plant) &
(ramp_df['YEAR']==year),'Gen Change'].quantile(0.95, interpolation='nearest')
ramp_rate_list.append([plant, year, ramp_95])
ramp_rate_df = pd.DataFrame(ramp_rate_list, columns=['plant_id', 'year', 'ramp_rate'])
ramp_rate_df.describe()
for key, df in clusterDict.iteritems():
clusterDict[key] = pd.merge(clusterDict[key], ramp_rate_df, how='left', on=['plant_id', 'year'])
clusterDict[2010].head()
# Check plants larger than 25MW, which is the lower limit for EPA
clusterDict[2010][clusterDict[2010].capacity >=25].describe()
for key in clusterDict.keys():
print key, clusterDict[key].plant_id.count(), clusterDict[key].ramp_rate.count()
```
Save dict to csv
```
# re-arrange column order
columns = ['year', 'plant_id', 'capacity', 'capacity_factor', 'efficiency', 'ramp_rate', 'fuel_type']
filename = 'Cluster_Data_2.csv'
path = '../Clean Data'
fullpath = os.path.join(path, filename)
counter = 0
for key, df in clusterDict.iteritems():
# create the csv file
if counter == 0:
df[columns].sort_values(by='plant_id').to_csv(fullpath, sep=',', index = False)
counter += 1
# append to existing csv file
else:
df[columns].sort_values(by='plant_id').to_csv(fullpath, sep=',', index = False, header=False, mode = 'a')
```
# Assumptions
1. Plant capacity changes at the start of the year and is constant for the entire year
2. Same for ramp rate - no changes over the course of the year
|
github_jupyter
|
%matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
import os
import glob
import re
import cPickle as pickle
import gzip
import seaborn as sns
#Iterate through the directory to find all the files to import
#Modified so that it also works on macs
path = os.path.join('EIA Data', '923-No_Header')
full_path = os.path.join(path, '*.*')
eiaNames = os.listdir(path)
#Rename the keys for easier merging later
fileNameMap = {'EIA923 SCHEDULES 2_3_4_5 Final 2010.xls':2010,
'EIA923 SCHEDULES 2_3_4_5 M Final 2009 REVISED 05252011.XLS':2009,
'eia923December2008.xls':2008,
'EIA923_Schedules_2_3_4_5_2011_Final_Revision.xlsx':2011,
'EIA923_Schedules_2_3_4_5_2012_Final_Release_12.04.2013.xlsx':2012,
'EIA923_Schedules_2_3_4_5_2013_Final_Revision.xlsx':2013,
'EIA923_Schedules_2_3_4_5_M_12_2014_Final_Revision.xlsx':2014,
'EIA923_Schedules_2_3_4_5_M_12_2015_Final.xlsx':2015,
'f906920_2007.xls':2007}
#Load the files into data frames, one df per file
eiaDict = {fileNameMap[fn]:pd.read_excel(os.path.join(path, fn)) for fn in eiaNames}
eiaDict = {key:val[val["NERC Region"] == "TRE"] for key, val in eiaDict.iteritems()}
#Dict of values to replace to standardize column names across all dataframes
monthDict = {"JANUARY":"JAN",
"FEBRUARY":"FEB",
"MARCH":"MAR",
"APRIL":"APR",
"MAY":"MAY",
"JUNE":"JUN",
"JULY":"JUL",
"AUGUST":"AUG",
"SEPTEMBER":"SEP",
"OCTOBER":"OCT",
"NOVEMBER":"NOV",
"DECEMBER":"DEC"}
replaceDict = {"ELECTRIC":"ELEC",
"&":"AND",
"I.D.":"ID",
"MMBTUPER":"MMBTU_PER"}
#Add "MMBTUMON" : "MMBTU_MON" to be replaced
for month in monthDict.values():
replaceDict["MMBTU"+month] = "MMBTU_" + month
#Replace the column name
def rename(col):
for old, new in monthDict.iteritems():
col = col.replace(old, new)
for old, new in replaceDict.iteritems():
col = col.replace(old, new)
col = col.replace("MMBTUS", "MMBTU")
return col
#Iterate through each column name of each dataframe to standardize
for key, df in eiaDict.iteritems():
colNames = [name.replace("\n", "_").replace(" ", "_").strip().upper() for name in df.columns]
colNames = [rename(col) for col in colNames]
eiaDict[key].columns = colNames
#Define the columns that are necessary but are not summable
allCols = eiaDict[fileNameMap.values()[0]].columns
nonSumCols = ["PLANT_ID", "PLANT_NAME", "YEAR"]
#Define the columns that contain the year's totals (Used to calc fuel type %)
yearCols = ["TOTAL_FUEL_CONSUMPTION_QUANTITY", "ELEC_FUEL_CONSUMPTION_QUANTITY",
"TOTAL_FUEL_CONSUMPTION_MMBTU", "ELEC_FUEL_CONSUMPTION_MMBTU",
"NET_GENERATION_(MEGAWATTHOURS)"]
#Define the columns that are necessary and summable
sumCols = []
sumCols.extend(yearCols)
# regex = re.compile(r"^ELEC_QUANTITY_.*")
# sumCols.extend([col for col in allCols if regex.search(col)])
regex = re.compile(r"^MMBTU_PER_UNIT_.*")
sumCols.extend([col for col in allCols if regex.search(col)])
regex = re.compile(r"^TOT_MMBTU_.*")
sumCols.extend([col for col in allCols if regex.search(col)])
regex = re.compile(r"^ELEC_MMBTUS_.*")
sumCols.extend([col for col in allCols if regex.search(col)])
regex = re.compile(r"^NETGEN_.*")
sumCols.extend([col for col in allCols if regex.search(col)])
fuelTypes = []
fuelTypes.extend([fuelType for df in eiaDict.values() for fuelType in df["REPORTED_FUEL_TYPE_CODE"].tolist()])
fuelTypes = set(fuelTypes)
fuelTypes
#Actually calculate the % type for each facility grouping
def calcPerc(group, aggGroup, fuelType, col):
#Check to see if the facility has a record for the fuel type, and if the total column > 0
if len(group[group["REPORTED_FUEL_TYPE_CODE"] == fuelType]) > 0 and aggGroup[col] > 0:
#summing fuel type because a facility may have multiple plants with the same fuel type
return float((group[group["REPORTED_FUEL_TYPE_CODE"] == fuelType][col]).sum())/aggGroup[col]
else:
return 0
#Perform the aggregation on facility level
def aggAndCalcPerc(group):
aggGroup = group.iloc[0][nonSumCols] #Get the non-agg columns
aggGroup = aggGroup.append(group[sumCols].sum()) #Aggregate the agg columns and append to non-agg
percCols = {col + " %" + fuelType:calcPerc(group, aggGroup, fuelType, col) for col in yearCols for fuelType in fuelTypes}
aggGroup = aggGroup.append(pd.Series(percCols))
return aggGroup
#Iterate through each dataframe to perform aggregation by facility
aggEIADict = dict()
for key, df in eiaDict.iteritems():
gb = df.groupby(by="PLANT_ID")
#aggGroup will be a list of panda series, each series representing a facility
aggGroup = [aggAndCalcPerc(gb.get_group(group)) for group in gb.groups]
aggEIADict[key] = pd.DataFrame(aggGroup)
aggEIADict[2007].head()
aggEIADict[2015].head()
filename = 'EIA 923.pkl'
path = '../Clean Data'
fullpath = os.path.join(path, filename)
pickle.dump(aggEIADict, open(fullpath, 'wb'))
all923 = pd.concat(aggEIADict)
all923.head()
all923.reset_index(drop=True, inplace=True)
# Check column numbers to use in the function below
all923.iloc[1,1:27]
def top_fuel(row):
#Fraction of largest fuel for electric heat input
try:
fuel = row.iloc[1:27].idxmax()[29:]
except:
return None
return fuel
all923['FUEL'] = all923.apply(top_fuel, axis=1)
all923.head()
fossil923 = all923.loc[all923['FUEL'].isin(['DFO', 'LIG', 'NG', 'PC', 'SUB'])]
filename = 'Fossil EIA 923.csv'
path = '../Clean Data'
fullpath = os.path.join(path, filename)
fossil923.to_csv(fullpath)
#Read the EPA files into a dataframe
path2 = os.path.join('EPA air markets')
epaNames = os.listdir(path2)
filePaths = {dn:os.path.join(path2, dn, "*.txt") for dn in epaNames}
filePaths = {dn:glob.glob(val) for dn, val in filePaths.iteritems()}
epaDict = {key:pd.read_csv(fp, index_col = False) for key, val in filePaths.iteritems() for fp in val}
#Rename the column names to remove the leading space.
for key, df in epaDict.iteritems():
colNames = [name.upper().strip() for name in df.columns]
colNames[colNames.index("FACILITY ID (ORISPL)")] = "PLANT_ID"
epaDict[key].columns = colNames
#Convert DATE to datetime object
#Add new column DATETIME with both date and hour
for key, df in epaDict.iteritems():
epaDict[key]["DATE"] = pd.to_datetime(df["DATE"])
epaDict[key]['DATETIME'] = df['DATE'] + pd.to_timedelta(df['HOUR'], unit='h')
set(epaDict['2015 July-Dec'].loc[:,'NERC REGION'])
#Boolean filter to only keep ERCOT plants
for key, df in epaDict.iteritems():
epaDict[key] = df[df["NERC REGION"] == "ERCOT"].reset_index(drop = True)
set(epaDict['2015 July-Dec'].loc[:,'NERC REGION'])
epaDict['2015 July-Dec'].head()
# pickle with gzip, from http://stackoverflow.com/questions/18474791/decreasing-the-size-of-cpickle-objects
def save_zipped_pickle(obj, filename, protocol=-1):
with gzip.open(filename, 'wb') as f:
pickle.dump(obj, f, protocol)
filename = 'EPA hourly dictionary.pgz'
path = '../Clean Data'
fullpath = os.path.join(path, filename)
save_zipped_pickle(epaDict, fullpath)
df = epaDict['2015 July-Dec']
df.head()
set(df['PLANT_ID'])
df_temp = df[df['PLANT_ID'].isin([127, 298, 3439])].fillna(0)
df_temp.head()
g = sns.FacetGrid(df_temp, col='PLANT_ID')
g.map(plt.plot, 'datetime', 'GROSS LOAD (MW)')
g.set_xticklabels(rotation=30)
path = os.path.join('..', 'Exploratory visualization', 'Midterm figures', 'Sample hourly load.svg')
plt.savefig(path)
#Join the two data sources on PLANT_ID
fullData = {key:df.merge(aggEIADict[df["YEAR"][0]], on="PLANT_ID") for key, df in epaDict.iteritems()}
fullData[fullData.keys()[0]].head()
[x for x in fullData[fullData.keys()[0]].columns]
# Iterate through the directory to find all the files to import
path = os.path.join('EIA Data', '860-No_Header')
full_path = os.path.join(path, '*.*')
eia860Names = os.listdir(path)
# Rename the keys for easier merging later
fileName860Map = { 'GenY07.xls':2007,
'GenY08.xls':2008,
'GeneratorY09.xls':2009,
'GeneratorsY2010.xls':2010,
'GeneratorY2011.xlsx':2011,
'GeneratorY2012.xlsx':2012,
'3_1_Generator_Y2013.xlsx':2013,
'3_1_Generator_Y2014.xlsx':2014,
'3_1_Generator_Y2015.xlsx':2015}
#Load the files into data frames, one df per file
eia860Dict = {fileName860Map[fn]:pd.read_excel(os.path.join(path, fn)) for fn in eia860Names}
#Dict of values to replace to standardize column names across all dataframes
renameDict = { "PLNTCODE":"PLANT_ID",
"PLANT_CODE":"PLANT_ID",
"Plant Code":"PLANT_ID",
"NAMEPLATE":"NAMEPLATE_CAPACITY(MW)",
"Nameplate Capacity (MW)":"NAMEPLATE_CAPACITY(MW)"}
#Replace the column name
def rename860(col):
for old, new in renameDict.iteritems():
col = col.replace(old, new)
return col
#Iterate through each column name of each dataframe to standardize and select columns 'PLANT_ID', 'NAMEPLATE_CAPACITY(MW)'
for key, df in eia860Dict.iteritems():
colNames = [rename860(col) for col in df.columns]
eia860Dict[key].columns = colNames
eia860Dict[key] = eia860Dict[key][["PLANT_ID", "NAMEPLATE_CAPACITY(MW)"]]
# verify the tables
for key, df in eia860Dict.iteritems():
print key, df.columns, len(df)
# Iterate through each dataframe to perform aggregation by PLANT_ID
for key, df in eia860Dict.iteritems():
gb = df.groupby(by='PLANT_ID').apply(lambda x: x['NAMEPLATE_CAPACITY(MW)'].sum())
eia860Dict[key]['NAMEPLATE_CAPACITY(MW)'] = eia860Dict[key].PLANT_ID.apply(gb.get_value)
eia860Dict[key] = eia860Dict[key].drop_duplicates(subset=['PLANT_ID', 'NAMEPLATE_CAPACITY(MW)'])
eia860Dict[key] = eia860Dict[key].sort_values(by='PLANT_ID').reset_index(drop=True)
filename = 'EIA 860.pkl'
path = '../Clean Data'
fullpath = os.path.join(path, filename)
pickle.dump(eia860Dict, open(fullpath, 'wb'))
clusterDict = dict()
for key, df in eia860Dict.iteritems():
clusterDict[key] = pd.merge(aggEIADict[key], eia860Dict[key], how='left', on='PLANT_ID')[['PLANT_ID', 'NAMEPLATE_CAPACITY(MW)']]
clusterDict[key].rename(columns={'NAMEPLATE_CAPACITY(MW)': 'capacity', 'PLANT_ID': 'plant_id'}, inplace=True)
# verify for no loss of data
for key, df in eia860Dict.iteritems():
print key, len(clusterDict[key]), len(aggEIADict[key])
clusterDict[2015].head()
fuel_cols = [col for col in aggEIADict[2008].columns if 'ELEC_FUEL_CONSUMPTION_MMBTU %' in col]
def top_fuel(row):
#Fraction of largest fuel for electric heat input
try:
fuel = row.idxmax()[29:]
except:
return None
return fuel
# clusterDict[2008]['fuel'] = aggEIADict[2008][fuel_cols].apply(top_fuel, axis=1)
for key, df in clusterDict.iteritems():
clusterDict[key]['year'] = key
clusterDict[key]['capacity_factor'] = aggEIADict[key]['NET_GENERATION_(MEGAWATTHOURS)'] / (8670*clusterDict[key]['capacity'])
clusterDict[key]['efficiency'] = (aggEIADict[key]['NET_GENERATION_(MEGAWATTHOURS)']*3.412)/(1.0*aggEIADict[key]['ELEC_FUEL_CONSUMPTION_MMBTU'])
clusterDict[key]['fuel_type'] = aggEIADict[key][fuel_cols].apply(top_fuel, axis=1)
clusterDict[key] = clusterDict[key][clusterDict[key]['fuel_type'].isin(['SUB',
'LIG',
'DFO',
'NG',
'PC'])]
columns = ['PLANT_ID', 'YEAR', 'DATE', 'HOUR', 'GROSS LOAD (MW)']
counter = 0
for key, df in epaDict.iteritems():
if counter == 0:
result = epaDict[key][columns]
counter = 1
else:
result = result.append(epaDict[key][columns], ignore_index=True)
# Change nan to 0
result.fillna(0, inplace=True)
result.describe()
def plant_gen_delta(df):
"""
For every plant in the input df, calculate the change in gross load (MW)
from the previous hour.
input:
df: dataframe of EPA clean air markets data
return:
df: concatanated list of dataframes
"""
df_list = []
for plant in df['PLANT_ID'].unique():
temp = df.loc[df['PLANT_ID'] == plant,:]
gen_change = temp.loc[:,'GROSS LOAD (MW)'].values - temp.loc[:,'GROSS LOAD (MW)'].shift(1).values
temp.loc[:,'Gen Change'] = gen_change
df_list.append(temp)
return pd.concat(df_list)
ramp_df = plant_gen_delta(result)
ramp_df.describe()
cols = ['PLANT_ID', 'YEAR', 'Gen Change']
ramp_rate_list = []
for year in ramp_df['YEAR'].unique():
for plant in ramp_df.loc[ramp_df['YEAR']==year,'PLANT_ID'].unique():
# 95th percentile ramp rate per plant per year
ramp_95 = ramp_df.loc[(ramp_df['PLANT_ID']== plant) &
(ramp_df['YEAR']==year),'Gen Change'].quantile(0.95, interpolation='nearest')
ramp_rate_list.append([plant, year, ramp_95])
ramp_rate_df = pd.DataFrame(ramp_rate_list, columns=['plant_id', 'year', 'ramp_rate'])
ramp_rate_df.describe()
for key, df in clusterDict.iteritems():
clusterDict[key] = pd.merge(clusterDict[key], ramp_rate_df, how='left', on=['plant_id', 'year'])
clusterDict[2010].head()
# Check plants larger than 25MW, which is the lower limit for EPA
clusterDict[2010][clusterDict[2010].capacity >=25].describe()
for key in clusterDict.keys():
print key, clusterDict[key].plant_id.count(), clusterDict[key].ramp_rate.count()
# re-arrange column order
columns = ['year', 'plant_id', 'capacity', 'capacity_factor', 'efficiency', 'ramp_rate', 'fuel_type']
filename = 'Cluster_Data_2.csv'
path = '../Clean Data'
fullpath = os.path.join(path, filename)
counter = 0
for key, df in clusterDict.iteritems():
# create the csv file
if counter == 0:
df[columns].sort_values(by='plant_id').to_csv(fullpath, sep=',', index = False)
counter += 1
# append to existing csv file
else:
df[columns].sort_values(by='plant_id').to_csv(fullpath, sep=',', index = False, header=False, mode = 'a')
| 0.380759 | 0.85115 |
<p align="center">
<img src="./images/adsp_logo.png">
</p>
### Prof. Dr. -Ing. Gerald Schuller <br> Jupyter Notebook: Renato Profeta
# Quantization: Signal to Noise Ratio (SNR)
```
%%html
<iframe width="560" height="315" src="https://www.youtube.com/embed/-4Dx7FpEAoc" frameborder="0" allow="accelerometer; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>
```
Assume we have a A/D converter with a quantizer with a certain number of bits (say N bits), what is the resulting Signal to Noise Ratio (SNR) of this quantizer?
**The SNR is defined as the ratio of the expectation of the signal power to the expectation of the noise power.**
In our case, the expectation of the noise power is the expectation of the quantization error power. We already have the expectation of the quantization error power as $\large \dfrac{\Delta^2}{12}$.<br>
So what we still need for the SNR is the **average or expectation of the signal power**. How do we obtain this?<br>
Basically we can take the same approach as we did for the expectation of the power of the quantization error (which is basically the second moment of the distribution of the quantization error). So what we need to know from our signal is its **probability distribution**. For the quantization error it was a uniform distribution between $-\dfrac{\Delta}{2}$ and $+\dfrac{\Delta}{2}$.<br>
A very **simple case** would be a **uniformly distributed signal** with amplitude $\dfrac{A}{2}$, which has values between $-\dfrac{A}{2}$ up to $+\dfrac{A}{2}$.
<center>
<img src="./images/pdf_a.png" width="250">
</center>
So we could again use our formula for the average power, but now for our signal x:
$$\large E(x^2)=\int_ {-A/2} ^ {A/2} x^2 \cdot p(x) dx$$
So here we have the same type of signal, and the resulting expectation of the power (its second moment, assumed we have a zero mean signal) is obtained by using our previous formula, and replace $\Delta$ by A. The resulting power is: $\frac{A^2}{12}$.
**Which signals have this property?** One example is uniformly distributed random values (basically like our quantization error).
**Observe: Speech or music has a non-uniform pdf**, it is usually modeled by a Laplacian distribution or a gaussian mixture model, so it doesn't apply to this case!
```
%%html
<iframe width="560" height="315" src="https://www.youtube.com/embed/5idUnMK_AkU" frameborder="0" allow="accelerometer; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>
```
An example for a uniform pdf: **a triangular wave**:
<center>
<img src="./images/triang.png" width="300">
</center>
How do we obtain its pdf? One can imagine the vertical axis (the function value) covered by small intervals, and each interval is then passed in the same time-span. This means that the resulting pdf is also uniform!
A further example: **A sawtooth wave**:
<center>
<img src="./images/saw_tooth.png" width="300">
</center>
Again we can make the same argument, each small interval of our function value is covered in the same time-span, hence we obtain a uniform distribution.
We now have seen a few examples which fulfil our assumption of a uniform distribution (realistic examples), and we know: their expectation of their power is $\dfrac{A^2}{12}$. So what does this then mean for the SNR? The **SNR** is just the ratio:
$$ \large SNR = \frac {\dfrac{A^2}{12}} {\dfrac{\Delta^2}{12}}= \frac{A^2} { \Delta^2} $$
If we assume our signal is full range, meaning the maximum values of our A/D converter is $-\dfrac{A}{2}$ and $+\dfrac{A}{2}$ (the signal goes to the maximum), we can compute the step size $\Delta$ if we know the **number of bits** of converter, and if we assume uniform quantization step sizes. Assume we have **N bits** in our converter. This means we have $2^N$ quantization intervals. We obtain $\Delta$ by dividing the full range by this number,
$$ \large
\Delta = \frac{A}{2^N}
$$
Plug this in the SNR equation, and we obtain:
$$
SNR= \frac{A^2} { \Delta^2}= \frac{A^2} {\left( \dfrac {A}{2^N} \right)^2} = {2^{2N}}
$$
This is now quite a simple result! But usually, the SNR is given in dB (deciBel), so lets convert it into dB:
<br>
$$SNR_{dB} = 10 \cdot \log_{10} (2^{2N})=10 \cdot 2N \cdot \log_{10}(2) \approx $$
$$ \approx 10 \cdot 2N \cdot 0.301 dB =N \cdot 6.02 dB$$
This is now our famous **rule of thumb**, that **each bit** more gives you about **6 dB more SNR**. But observe that the above formula only holds for uniformly distributed full range signals! (the signal is between -A/2 and +A/2, using all possible values of our converter).
```
%%html
<iframe width="560" height="315" src="https://www.youtube.com/embed/f62zWfAaEfc" frameborder="0" allow="accelerometer; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>
```
What happens if the signal is not full range? What is the SNR if we have a signal with reduced range? Assume our signal has an amplitude of A/c, with a factor c>1.<br>
We can then simply plug this into our equation:
$$ \large SNR= \frac{\left(\frac{A}{c}\right)^2} {\Delta^2}= \frac{\left(\frac{A}{c}\right)^2 }{ \left(\frac{A}{2^N}\right)^2} = \frac{2^{2N}}{c^2}$$
in dB:
$$ \large SNR_{dB}=10 . \log_{10}( \frac {2^{2N}} {c^2})=10\cdot2N.\log_{10}(2)-20.\log_{10}(c) \approx $$
$$ \large
\approx 10 \cdot 2N \cdot 0.301 dB -20 \cdot \log_{10}(c) =
$$
<br>
$$ \large
=N \cdot 6.02 dB -20 \cdot log_{10}(c)
$$
<br>
The last term, the $20 \cdot \log_{10}(c) $, is the number of dB which we are below our full range. This means we **reduce our SNR** by this number of **dB** which we are **below full range**!
<br><br>
**Example:** We have a 16 bit quantiser, then the SNR for uniformly distributed full range signals would be:
$$SNR = 6.02 \cdot 16 dB = 96.32 dB$$
Now assume we have the same signal, but 20dB below full range (meaning only 1/10th of the full range). Then the resulting SNR would be only:
$$SNR = 96.32-20 = 76.32 dB$$
This is considerably less. This also shows why it is important not to make the safety margin to full range too big! So for instance our sound engineer should keep the signal as big as possible, without ever reaching full range to avoid clipping the signal.
The other assumption we made concerned the type of signal we quantize.
**What if we don't have a uniformly distributed signal?**
As we saw, speech and audio signals are best modelled by a Laplacian distribution or a Gaussian mixture model, and similar for audio signals. Even a simple sine wave does not fulfil this assumption of a uniform distribution. What is the pdf of a simple sine wave?
**Observe:** If a sinusoid represents a full range signal, its values are from $-A/2$ to $+A/2$, as in the previous cases.
```
%%html
<iframe width="560" height="315" src="https://www.youtube.com/embed/vAjOvN5fVjE" frameborder="0" allow="accelerometer; autoplay; gyroscope; picture-in-picture" allowfullscreen></iframe>
```
What is our SNR if we have a sinusoidal signal? What is its pdf? Basically it is its normalized histogram, such that its integral becomes 1, to obtain a probability distribution.
If we look at the signal, and try to see how probable it is for the signal to be in a certain small interval on the y axis, we see that the signal stays longest around +1 and -1, because there the signal slowly turns around. Hence we would expect a pdf, which has peaks at +1 and -1.<br>
If you calculate the pdf of a sine wave, x=sin(t), with t being continuous and with a range larger than 2pi, then the result is
<br>
$$
p(x)=\frac{1} {\pi \cdot \sqrt{1-x^2}}
$$
<br>
This results from the derivative of the inverse sine function (arcsin). This derivation can be found for instance on Wikipedia. For our pdf we need to know how fast a signal x passes through a given bin in x. This is what we obtain if we compute the inverse function $x=f^{-1}(y)$, and then its derivative $df^{-1}(x)/dy$.
### PDF of Time Series
Given a signal a = f (t) which is sampled uniformly over a time period T , its PDF, p(a) can be calculated as follows. Because the signal is uniformly sampled we have $p(t) = \frac{1}{T}$ . The function f(t) acts to transform this density from one over *t* to one over *a*. Hence, using the method for transforming PDFs, we get:
$$\large
p(a)=\dfrac{p(t)}{\left|\frac {da}{dt} \right|} $$
where | | denotes the absolute value and the derivative is evaluated at $t=f^{-1}(x).$
<font size="2">
From: https://www.fil.ion.ucl.ac.uk/~wpenny/course/appendixDE.pdf
</font>
```
from sympy import symbols, pi, sqrt, Integral, Function, Eq, diff, sin, solve, simplify, Abs
x, t = symbols('x t', real=True)
A, w = symbols('A w', real=True, positive=True)
Eq_x=Eq(x, sin(t))
Eq_x
# Find the Inverse
y=solve(Eq_x,t)
Eq_y=Eq(t,y[1])
Eq_y
```
The inverse sine is only defined for $-\frac{\pi}{2} \leq t \leq +\frac{\pi}{2}$ and p(t) is uniform within this.
Hence, $ p(t) = \frac {1}{\pi} $.
```
# Find dx\dt and evaluate at t=arsin(x)
dxdt = (diff(Eq_x.rhs,t))
dxdt = dxdt.subs(t,Eq_y.rhs)
dxdt
# Calculate p(t)
from sympy.stats import Uniform, density
P_t = Function('p')(t)
p_t = Uniform('X',-pi/2,pi/2)
Eq_p_t=Eq(P_t,density(p_t)(t))
Eq_p_t
# Calculate p(x)
p_x = Function('p')(x)
Eq_p_x= Eq(p_x,(1/pi)/dxdt)
Eq_p_x
%%html
<iframe width="560" height="315" src="https://www.youtube.com/embed/jkCIZoVnweg" frameborder="0" allow="accelerometer; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>
```
Here we can see that p(x) indeed becomes infinite at x=+/-1! We could now use the same approach as before to obtain the expectation of the power, multiplying it with $x^2$ and integrating it. But this seems to be somewhat tedious. But since we now have a deterministic signal, we can also try an **alternative** solution, since the sine function is not a probabilistic function, but a deterministic function.
We can simply directly compute the power of our sine signal over t, and then take the average over at least one period of the sine function.
$$ \large
E(x^2)= \frac{1} {2\pi}\int _ {t=0} ^{2\pi} sin^2(t) dt = \frac{1} {2\pi}\int _ {t=0} ^{2\pi} {\dfrac{\left(1-cos(2t)\right)}{2}}dt
$$
Trigonometric Identity: $cos(2x)=1-2sin^2(x)$
The cosine integrated over complete periods becomes 0, hence we get:
$$ n\large
=\frac{1} {2\pi} \int _{t=0}^{2\pi} {\dfrac{1}{2}} dt =\frac{1} {2\pi} \cdot \pi=\frac{1}{ 2}
$$
```
# Calculate the Expctation of Power
E = Function('E')(x**2)
E_x2 = Eq(E,Integral(x**2*Eq_p_x.rhs,(x,-1,1)))
display(E_x2)
display(E_x2.doit())
```
What do we get for a sinusoid with a different amplitude, say $A/2 \cdot sin(t)$?
The expected power is:
$$ \large
E(x^2)=\frac{A^2}{ 8}
$$
```
# Calculate Expectation of Power of A/2 * sin(t)
E = Function('E')(x**2)
E_x2 = Eq(E,(1/(2*pi))*Integral(((A/2)*sin(t))**2,(t,0,2*pi)))
display(E_x2)
display(E_x2.doit())
```
So this leads to an SNR of:
$$ \large
SNR=\frac{\frac{A^2}{8}} {\frac{\Delta^2}{12}}=\frac{3 \cdot A^2} {2 \cdot \Delta^2}
$$
Now assume again we have a A/D converter with N bits, and the sinusoid is at full range for this converter. Then
$A=2^N \cdot \Delta$
We can plug in this result into the above equation, and get
$$ \large
SNR=\frac{3 \cdot 2^{2N} \cdot \Delta^2} {2 \cdot \Delta^2}={1.5 \cdot 2^{2N}} $$
In dB this will now be:
$$\large 10 \cdot \log_{10}(SNR)=10 \cdot \log_{10}(1.5) + N \cdot 20 \cdot \log_{10}(2)=
$$
$$\large = 1.76 dB +N \cdot 6.02 dB$$
Here we can see now, that using a sinusoidal signal instead of a uniformly distributed signal gives us a **boost of 1.76 dB** in SNR. This is because it is more likely to have larger values!
We see that our rule of 6dB more SNR for each bit still holds!
```
# Stepsize as function of full range
delta, N = symbols('\Delta N')
Eq_delta = Eq(delta, A/(2**N) )
display(Eq_delta)
Eq_A = Eq(A,solve(Eq_delta,A)[0])
display(Eq_A)
# Calculate Signal to Noise Rate
SNR = E_x2.doit().rhs / (delta**2/12)
display(SNR)
display(SNR.subs(A,Eq_A.rhs))
```
|
github_jupyter
|
%%html
<iframe width="560" height="315" src="https://www.youtube.com/embed/-4Dx7FpEAoc" frameborder="0" allow="accelerometer; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>
%%html
<iframe width="560" height="315" src="https://www.youtube.com/embed/5idUnMK_AkU" frameborder="0" allow="accelerometer; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>
%%html
<iframe width="560" height="315" src="https://www.youtube.com/embed/f62zWfAaEfc" frameborder="0" allow="accelerometer; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>
%%html
<iframe width="560" height="315" src="https://www.youtube.com/embed/vAjOvN5fVjE" frameborder="0" allow="accelerometer; autoplay; gyroscope; picture-in-picture" allowfullscreen></iframe>
from sympy import symbols, pi, sqrt, Integral, Function, Eq, diff, sin, solve, simplify, Abs
x, t = symbols('x t', real=True)
A, w = symbols('A w', real=True, positive=True)
Eq_x=Eq(x, sin(t))
Eq_x
# Find the Inverse
y=solve(Eq_x,t)
Eq_y=Eq(t,y[1])
Eq_y
# Find dx\dt and evaluate at t=arsin(x)
dxdt = (diff(Eq_x.rhs,t))
dxdt = dxdt.subs(t,Eq_y.rhs)
dxdt
# Calculate p(t)
from sympy.stats import Uniform, density
P_t = Function('p')(t)
p_t = Uniform('X',-pi/2,pi/2)
Eq_p_t=Eq(P_t,density(p_t)(t))
Eq_p_t
# Calculate p(x)
p_x = Function('p')(x)
Eq_p_x= Eq(p_x,(1/pi)/dxdt)
Eq_p_x
%%html
<iframe width="560" height="315" src="https://www.youtube.com/embed/jkCIZoVnweg" frameborder="0" allow="accelerometer; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>
# Calculate the Expctation of Power
E = Function('E')(x**2)
E_x2 = Eq(E,Integral(x**2*Eq_p_x.rhs,(x,-1,1)))
display(E_x2)
display(E_x2.doit())
# Calculate Expectation of Power of A/2 * sin(t)
E = Function('E')(x**2)
E_x2 = Eq(E,(1/(2*pi))*Integral(((A/2)*sin(t))**2,(t,0,2*pi)))
display(E_x2)
display(E_x2.doit())
# Stepsize as function of full range
delta, N = symbols('\Delta N')
Eq_delta = Eq(delta, A/(2**N) )
display(Eq_delta)
Eq_A = Eq(A,solve(Eq_delta,A)[0])
display(Eq_A)
# Calculate Signal to Noise Rate
SNR = E_x2.doit().rhs / (delta**2/12)
display(SNR)
display(SNR.subs(A,Eq_A.rhs))
| 0.739046 | 0.990687 |
# Comparing Ampleforth and Tellor's Oracles
### AMPL/USD Feed
Using web3 to get Tellor's data, ABI via etherscan. Looking to see the update times and price accuracy of Tellor and Ampleforth's AMPL/USD feeds
```
from web3 import Web3
infura_link = 'https://mainnet.infura.io/v3/3e5c6b2a48494d9a921a52ec1cc0a8ff'
w3 = Web3(Web3.HTTPProvider(infura_link))
tellor_add = "0xb2b6c6232d38fae21656703cac5a74e5314741d4" # id 10
ampl_add = "0x99C9775E076FDF99388C029550155032Ba2d8914"
tellor_abi = '[{"inputs":[{"internalType":"address payable","name":"_master","type":"address"}],"stateMutability":"nonpayable","type":"constructor"},{"inputs":[],"name":"currentReward","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"currentTotalTips","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"","type":"uint256"}],"name":"dataIDs","outputs":[{"internalType":"uint256","name":"id","type":"uint256"},{"internalType":"string","name":"name","type":"string"},{"internalType":"uint256","name":"granularity","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"dataIDsAll","outputs":[{"components":[{"internalType":"uint256","name":"id","type":"uint256"},{"internalType":"string","name":"name","type":"string"},{"internalType":"uint256","name":"granularity","type":"uint256"}],"internalType":"struct Main.DataID[]","name":"","type":"tuple[]"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"","type":"uint256"}],"name":"dataIDsMap","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"deity","outputs":[{"internalType":"address","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"difficulty","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"disputeCount","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"disputeFee","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_requestId","type":"uint256"}],"name":"getCurrentValue","outputs":[{"internalType":"bool","name":"ifRetrieve","type":"bool"},{"internalType":"uint256","name":"value","type":"uint256"},{"internalType":"uint256","name":"_timestampRetrieved","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_requestId","type":"uint256"},{"internalType":"uint256","name":"_timestamp","type":"uint256"}],"name":"getDataBefore","outputs":[{"internalType":"bool","name":"_ifRetrieve","type":"bool"},{"internalType":"uint256","name":"_value","type":"uint256"},{"internalType":"uint256","name":"_timestampRetrieved","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_requestId","type":"uint256"},{"internalType":"uint256","name":"_timestamp","type":"uint256"}],"name":"getIndexForDataBefore","outputs":[{"internalType":"bool","name":"found","type":"bool"},{"internalType":"uint256","name":"index","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_dataID","type":"uint256"},{"internalType":"uint256","name":"_count","type":"uint256"}],"name":"getLastValues","outputs":[{"components":[{"internalType":"uint256","name":"id","type":"uint256"},{"internalType":"string","name":"name","type":"string"},{"internalType":"uint256","name":"timestamp","type":"uint256"},{"internalType":"uint256","name":"value","type":"uint256"}],"internalType":"struct Main.Value[]","name":"","type":"tuple[]"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"count","type":"uint256"}],"name":"getLastValuesAll","outputs":[{"components":[{"internalType":"uint256","name":"id","type":"uint256"},{"internalType":"string","name":"name","type":"string"},{"internalType":"uint256","name":"timestamp","type":"uint256"},{"internalType":"uint256","name":"value","type":"uint256"}],"internalType":"struct Main.Value[]","name":"","type":"tuple[]"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_requestId","type":"uint256"}],"name":"getNewValueCountbyRequestId","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_requestId","type":"uint256"},{"internalType":"uint256","name":"_index","type":"uint256"}],"name":"getTimestampbyRequestIDandIndex","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_requestId","type":"uint256"},{"internalType":"uint256","name":"_timestamp","type":"uint256"}],"name":"isInDispute","outputs":[{"internalType":"bool","name":"","type":"bool"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"master","outputs":[{"internalType":"contract Oracle","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"owner","outputs":[{"internalType":"address","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"pendingOwner","outputs":[{"internalType":"address","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[{"components":[{"internalType":"uint256","name":"id","type":"uint256"},{"internalType":"string","name":"name","type":"string"},{"internalType":"uint256","name":"granularity","type":"uint256"}],"internalType":"struct Main.DataID","name":"_dataID","type":"tuple"}],"name":"pushDataID","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[{"components":[{"internalType":"uint256","name":"id","type":"uint256"},{"internalType":"string","name":"name","type":"string"},{"internalType":"uint256","name":"granularity","type":"uint256"}],"internalType":"struct Main.DataID[]","name":"_dataIDs","type":"tuple[]"}],"name":"replaceDataIDs","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[],"name":"requestCount","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_requestId","type":"uint256"},{"internalType":"uint256","name":"_timestamp","type":"uint256"}],"name":"retrieveData","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"address","name":"_admin","type":"address"}],"name":"setAdmin","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[{"internalType":"uint256","name":"_id","type":"uint256"},{"components":[{"internalType":"uint256","name":"id","type":"uint256"},{"internalType":"string","name":"name","type":"string"},{"internalType":"uint256","name":"granularity","type":"uint256"}],"internalType":"struct Main.DataID","name":"_dataID","type":"tuple"}],"name":"setDataID","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[],"name":"slotProgress","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"stakeAmount","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"stakeCount","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"tBlock","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"tellorContract","outputs":[{"internalType":"address","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"timeOfLastValue","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"timeTarget","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_dataID","type":"uint256"}],"name":"totalTip","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"}]'
ampl_abi = '[{"constant":true,"inputs":[],"name":"reportDelaySec","outputs":[{"name":"","type":"uint256"}],"payable":false,"stateMutability":"view","type":"function"},{"constant":false,"inputs":[{"name":"payload","type":"uint256"}],"name":"pushReport","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":false,"inputs":[],"name":"getData","outputs":[{"name":"","type":"uint256"},{"name":"","type":"bool"}],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":false,"inputs":[{"name":"provider","type":"address"}],"name":"addProvider","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":true,"inputs":[{"name":"","type":"uint256"}],"name":"providers","outputs":[{"name":"","type":"address"}],"payable":false,"stateMutability":"view","type":"function"},{"constant":false,"inputs":[],"name":"renounceOwnership","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":false,"inputs":[{"name":"provider","type":"address"}],"name":"removeProvider","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":true,"inputs":[],"name":"owner","outputs":[{"name":"","type":"address"}],"payable":false,"stateMutability":"view","type":"function"},{"constant":true,"inputs":[],"name":"isOwner","outputs":[{"name":"","type":"bool"}],"payable":false,"stateMutability":"view","type":"function"},{"constant":true,"inputs":[],"name":"minimumProviders","outputs":[{"name":"","type":"uint256"}],"payable":false,"stateMutability":"view","type":"function"},{"constant":false,"inputs":[],"name":"purgeReports","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":false,"inputs":[{"name":"reportDelaySec_","type":"uint256"}],"name":"setReportDelaySec","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":true,"inputs":[],"name":"providersSize","outputs":[{"name":"","type":"uint256"}],"payable":false,"stateMutability":"view","type":"function"},{"constant":true,"inputs":[],"name":"reportExpirationTimeSec","outputs":[{"name":"","type":"uint256"}],"payable":false,"stateMutability":"view","type":"function"},{"constant":false,"inputs":[{"name":"minimumProviders_","type":"uint256"}],"name":"setMinimumProviders","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":true,"inputs":[{"name":"","type":"address"},{"name":"","type":"uint256"}],"name":"providerReports","outputs":[{"name":"timestamp","type":"uint256"},{"name":"payload","type":"uint256"}],"payable":false,"stateMutability":"view","type":"function"},{"constant":false,"inputs":[{"name":"newOwner","type":"address"}],"name":"transferOwnership","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":false,"inputs":[{"name":"reportExpirationTimeSec_","type":"uint256"}],"name":"setReportExpirationTimeSec","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"inputs":[{"name":"reportExpirationTimeSec_","type":"uint256"},{"name":"reportDelaySec_","type":"uint256"},{"name":"minimumProviders_","type":"uint256"}],"payable":false,"stateMutability":"nonpayable","type":"constructor"},{"anonymous":false,"inputs":[{"indexed":false,"name":"provider","type":"address"}],"name":"ProviderAdded","type":"event"},{"anonymous":false,"inputs":[{"indexed":false,"name":"provider","type":"address"}],"name":"ProviderRemoved","type":"event"},{"anonymous":false,"inputs":[{"indexed":false,"name":"provider","type":"address"}],"name":"ReportTimestampOutOfRange","type":"event"},{"anonymous":false,"inputs":[{"indexed":true,"name":"provider","type":"address"},{"indexed":false,"name":"payload","type":"uint256"},{"indexed":false,"name":"timestamp","type":"uint256"}],"name":"ProviderReportPushed","type":"event"},{"anonymous":false,"inputs":[{"indexed":true,"name":"previousOwner","type":"address"},{"indexed":true,"name":"newOwner","type":"address"}],"name":"OwnershipTransferred","type":"event"}]'
contract_t = w3.eth.contract(address = w3.toChecksumAddress(tellor_add), abi = tellor_abi)
tellor_data = (contract_t.functions.getCurrentValue(10).call())
from datetime import datetime, timedelta
all_prices = []
all_timestamps = []
initial_data = tellor_data
old_date = datetime.timestamp(datetime.now() - timedelta(days = 10))
all_prices.append(tellor_data[1]/ 1000000)
all_timestamps.append(datetime.fromtimestamp(int(tellor_data[2])))
while (old_date < initial_data[2]):
initial_data = contract_t.functions.getDataBefore(10, initial_data[2]).call()
#print(initial_data)
all_prices.append(initial_data[1] / 1000000)
all_timestamps.append(datetime.fromtimestamp(int(initial_data[2])))
import matplotlib.pyplot as plt
%matplotlib inline
plt.style.use('ggplot')
plt.plot(all_timestamps, all_prices)
```
### Ampleforth Data
Gotten from an API and parsed with JSON, ampleforth's data comes outside of normal contract connection. Using pandas, dictionaries and the requests library, I got the data in and looked at it on a plot to make sure it was accurate, then trimmed and scaled it to compare to Tellor's
```
import json
import requests
import pandas as pd
url = 'https://web-api.ampleforth.org/eth/oracle-histroy'
r = requests.get(url)
files = r.json()
ampl_dict = files["rateOracleProviderHistory"]["reports"]["ampleforth.org"]
ampl_df = pd.DataFrame(ampl_dict)
payload = list(ampl_df['payload'])
timestamps = list(ampl_df['timestampSec'])
new_timestamps = [datetime.fromtimestamp(i) for i in timestamps]
plt.plot(new_timestamps, payload)
```
Since we only wanted to look at the last 10 days, I cut off the data using a datetime conditional list comprehension.
```
end_date = datetime(2021,9,11,0,0,0)
timestamps_2 = [i for i in new_timestamps if i > end_date]
new_length = len(new_timestamps) - len(timestamps_2)
```
Get unique values from tellor data to avoid "stair step" shape of function. Reversed lists since original was in descending order going backwards through time
```
streamline_tx = []
streamline_ty = []
timestamps_rev = all_timestamps[::-1]
prices_rev = all_prices[::-1]
for i in range(1, len(timestamps_rev) - 1):
if prices_rev[i] != prices_rev[i-1]:
streamline_ty.append(prices_rev[i + 1])
streamline_tx.append(timestamps_rev[i])
```
### Chainlink Data
Used smart contract and web3 to pull data back 10 days.
```
chainlink_add = "0xe20CA8D7546932360e37E9D72c1a47334af57706"
chainlink_add_cs = w3.toChecksumAddress(chainlink_add)
chainlink_abi = '[{"inputs":[{"internalType":"address","name":"_aggregator","type":"address"},{"internalType":"address","name":"_accessController","type":"address"}],"stateMutability":"nonpayable","type":"constructor"},{"anonymous":false,"inputs":[{"indexed":true,"internalType":"int256","name":"current","type":"int256"},{"indexed":true,"internalType":"uint256","name":"roundId","type":"uint256"},{"indexed":false,"internalType":"uint256","name":"updatedAt","type":"uint256"}],"name":"AnswerUpdated","type":"event"},{"anonymous":false,"inputs":[{"indexed":true,"internalType":"uint256","name":"roundId","type":"uint256"},{"indexed":true,"internalType":"address","name":"startedBy","type":"address"},{"indexed":false,"internalType":"uint256","name":"startedAt","type":"uint256"}],"name":"NewRound","type":"event"},{"anonymous":false,"inputs":[{"indexed":true,"internalType":"address","name":"from","type":"address"},{"indexed":true,"internalType":"address","name":"to","type":"address"}],"name":"OwnershipTransferRequested","type":"event"},{"anonymous":false,"inputs":[{"indexed":true,"internalType":"address","name":"from","type":"address"},{"indexed":true,"internalType":"address","name":"to","type":"address"}],"name":"OwnershipTransferred","type":"event"},{"inputs":[],"name":"acceptOwnership","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[],"name":"accessController","outputs":[{"internalType":"contract AccessControllerInterface","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"aggregator","outputs":[{"internalType":"address","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"address","name":"_aggregator","type":"address"}],"name":"confirmAggregator","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[],"name":"decimals","outputs":[{"internalType":"uint8","name":"","type":"uint8"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"description","outputs":[{"internalType":"string","name":"","type":"string"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_roundId","type":"uint256"}],"name":"getAnswer","outputs":[{"internalType":"int256","name":"","type":"int256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint80","name":"_roundId","type":"uint80"}],"name":"getRoundData","outputs":[{"internalType":"uint80","name":"roundId","type":"uint80"},{"internalType":"int256","name":"answer","type":"int256"},{"internalType":"uint256","name":"startedAt","type":"uint256"},{"internalType":"uint256","name":"updatedAt","type":"uint256"},{"internalType":"uint80","name":"answeredInRound","type":"uint80"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_roundId","type":"uint256"}],"name":"getTimestamp","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"latestAnswer","outputs":[{"internalType":"int256","name":"","type":"int256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"latestRound","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"latestRoundData","outputs":[{"internalType":"uint80","name":"roundId","type":"uint80"},{"internalType":"int256","name":"answer","type":"int256"},{"internalType":"uint256","name":"startedAt","type":"uint256"},{"internalType":"uint256","name":"updatedAt","type":"uint256"},{"internalType":"uint80","name":"answeredInRound","type":"uint80"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"latestTimestamp","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"owner","outputs":[{"internalType":"address","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint16","name":"","type":"uint16"}],"name":"phaseAggregators","outputs":[{"internalType":"contract AggregatorV2V3Interface","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"phaseId","outputs":[{"internalType":"uint16","name":"","type":"uint16"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"address","name":"_aggregator","type":"address"}],"name":"proposeAggregator","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[],"name":"proposedAggregator","outputs":[{"internalType":"contract AggregatorV2V3Interface","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint80","name":"_roundId","type":"uint80"}],"name":"proposedGetRoundData","outputs":[{"internalType":"uint80","name":"roundId","type":"uint80"},{"internalType":"int256","name":"answer","type":"int256"},{"internalType":"uint256","name":"startedAt","type":"uint256"},{"internalType":"uint256","name":"updatedAt","type":"uint256"},{"internalType":"uint80","name":"answeredInRound","type":"uint80"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"proposedLatestRoundData","outputs":[{"internalType":"uint80","name":"roundId","type":"uint80"},{"internalType":"int256","name":"answer","type":"int256"},{"internalType":"uint256","name":"startedAt","type":"uint256"},{"internalType":"uint256","name":"updatedAt","type":"uint256"},{"internalType":"uint80","name":"answeredInRound","type":"uint80"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"address","name":"_accessController","type":"address"}],"name":"setController","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[{"internalType":"address","name":"_to","type":"address"}],"name":"transferOwnership","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[],"name":"version","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"}]'
contract_cl = w3.eth.contract(address = chainlink_add_cs, abi = chainlink_abi)
latestData = contract_cl.functions.latestRoundData().call()
scale = 1e18
round_id_0 = latestData[0]
price_0 = latestData[1]
update_time_0 = latestData[3]
round_ids = []
prices = []
update_times_utc = []
round_ids.append(round_id_0)
prices.append(price_0 / scale)
update_times_utc.append(datetime.utcfromtimestamp(update_time_0))
current_date = str(datetime.utcfromtimestamp(update_time_0))
current_date = current_date.split()[0]
end_date = '2021-09-12'
def time_convert(raw_time):
return datetime.utcfromtimestamp(raw_time)
def inc_rounds(curr_round_data, end_date, time_arr):
curr_round_id = curr_round_data[0]
current_date = str(time_convert(curr_round_data[3])).split()[0]
while current_date != end_date:
curr_round_id = curr_round_id - 1
historicalData = contract_cl.functions.getRoundData(curr_round_id).call()
update_time_raw = historicalData[3]
time_arr.append(time_convert(update_time_raw))
#round_ids.append(historicalData[0])
prices.append(historicalData[1] / scale)
#update_times_utc.append(time_convert(update_time_raw))
current_date = str(time_convert(update_time_raw)).split()[0]
print("time: ",time_convert(update_time_raw), "price: ", historicalData[1] / (scale), "round ID: ", curr_round_id)
inc_rounds(latestData, end_date, update_times_utc)
```
### Comparison Plot
Plot data on same figure. Since x axis is datetime objects, it will match up evenly and we will be able to see changes in data.
```
plt.figure(figsize = (15, 12))
plt.plot(timestamps_2, payload[new_length:], label = 'ampl')
#plt.plot(all_timestamps, all_prices, label = 'tellor' )
plt.plot(streamline_tx, streamline_ty, label = 'tellor')
plt.plot(update_times_utc, prices, label = 'chainlink')
plt.title("Tellor and Ampleforth Oracle Prices")
plt.xlabel("date")
plt.ylabel("AMPL price (USD)")
plt.xticks(rotation = 90)
plt.legend()
plt.savefig('AMPLcomparison.png', bbox_inches='tight')
```
|
github_jupyter
|
from web3 import Web3
infura_link = 'https://mainnet.infura.io/v3/3e5c6b2a48494d9a921a52ec1cc0a8ff'
w3 = Web3(Web3.HTTPProvider(infura_link))
tellor_add = "0xb2b6c6232d38fae21656703cac5a74e5314741d4" # id 10
ampl_add = "0x99C9775E076FDF99388C029550155032Ba2d8914"
tellor_abi = '[{"inputs":[{"internalType":"address payable","name":"_master","type":"address"}],"stateMutability":"nonpayable","type":"constructor"},{"inputs":[],"name":"currentReward","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"currentTotalTips","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"","type":"uint256"}],"name":"dataIDs","outputs":[{"internalType":"uint256","name":"id","type":"uint256"},{"internalType":"string","name":"name","type":"string"},{"internalType":"uint256","name":"granularity","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"dataIDsAll","outputs":[{"components":[{"internalType":"uint256","name":"id","type":"uint256"},{"internalType":"string","name":"name","type":"string"},{"internalType":"uint256","name":"granularity","type":"uint256"}],"internalType":"struct Main.DataID[]","name":"","type":"tuple[]"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"","type":"uint256"}],"name":"dataIDsMap","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"deity","outputs":[{"internalType":"address","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"difficulty","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"disputeCount","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"disputeFee","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_requestId","type":"uint256"}],"name":"getCurrentValue","outputs":[{"internalType":"bool","name":"ifRetrieve","type":"bool"},{"internalType":"uint256","name":"value","type":"uint256"},{"internalType":"uint256","name":"_timestampRetrieved","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_requestId","type":"uint256"},{"internalType":"uint256","name":"_timestamp","type":"uint256"}],"name":"getDataBefore","outputs":[{"internalType":"bool","name":"_ifRetrieve","type":"bool"},{"internalType":"uint256","name":"_value","type":"uint256"},{"internalType":"uint256","name":"_timestampRetrieved","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_requestId","type":"uint256"},{"internalType":"uint256","name":"_timestamp","type":"uint256"}],"name":"getIndexForDataBefore","outputs":[{"internalType":"bool","name":"found","type":"bool"},{"internalType":"uint256","name":"index","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_dataID","type":"uint256"},{"internalType":"uint256","name":"_count","type":"uint256"}],"name":"getLastValues","outputs":[{"components":[{"internalType":"uint256","name":"id","type":"uint256"},{"internalType":"string","name":"name","type":"string"},{"internalType":"uint256","name":"timestamp","type":"uint256"},{"internalType":"uint256","name":"value","type":"uint256"}],"internalType":"struct Main.Value[]","name":"","type":"tuple[]"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"count","type":"uint256"}],"name":"getLastValuesAll","outputs":[{"components":[{"internalType":"uint256","name":"id","type":"uint256"},{"internalType":"string","name":"name","type":"string"},{"internalType":"uint256","name":"timestamp","type":"uint256"},{"internalType":"uint256","name":"value","type":"uint256"}],"internalType":"struct Main.Value[]","name":"","type":"tuple[]"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_requestId","type":"uint256"}],"name":"getNewValueCountbyRequestId","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_requestId","type":"uint256"},{"internalType":"uint256","name":"_index","type":"uint256"}],"name":"getTimestampbyRequestIDandIndex","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_requestId","type":"uint256"},{"internalType":"uint256","name":"_timestamp","type":"uint256"}],"name":"isInDispute","outputs":[{"internalType":"bool","name":"","type":"bool"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"master","outputs":[{"internalType":"contract Oracle","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"owner","outputs":[{"internalType":"address","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"pendingOwner","outputs":[{"internalType":"address","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[{"components":[{"internalType":"uint256","name":"id","type":"uint256"},{"internalType":"string","name":"name","type":"string"},{"internalType":"uint256","name":"granularity","type":"uint256"}],"internalType":"struct Main.DataID","name":"_dataID","type":"tuple"}],"name":"pushDataID","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[{"components":[{"internalType":"uint256","name":"id","type":"uint256"},{"internalType":"string","name":"name","type":"string"},{"internalType":"uint256","name":"granularity","type":"uint256"}],"internalType":"struct Main.DataID[]","name":"_dataIDs","type":"tuple[]"}],"name":"replaceDataIDs","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[],"name":"requestCount","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_requestId","type":"uint256"},{"internalType":"uint256","name":"_timestamp","type":"uint256"}],"name":"retrieveData","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"address","name":"_admin","type":"address"}],"name":"setAdmin","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[{"internalType":"uint256","name":"_id","type":"uint256"},{"components":[{"internalType":"uint256","name":"id","type":"uint256"},{"internalType":"string","name":"name","type":"string"},{"internalType":"uint256","name":"granularity","type":"uint256"}],"internalType":"struct Main.DataID","name":"_dataID","type":"tuple"}],"name":"setDataID","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[],"name":"slotProgress","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"stakeAmount","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"stakeCount","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"tBlock","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"tellorContract","outputs":[{"internalType":"address","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"timeOfLastValue","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"timeTarget","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_dataID","type":"uint256"}],"name":"totalTip","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"}]'
ampl_abi = '[{"constant":true,"inputs":[],"name":"reportDelaySec","outputs":[{"name":"","type":"uint256"}],"payable":false,"stateMutability":"view","type":"function"},{"constant":false,"inputs":[{"name":"payload","type":"uint256"}],"name":"pushReport","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":false,"inputs":[],"name":"getData","outputs":[{"name":"","type":"uint256"},{"name":"","type":"bool"}],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":false,"inputs":[{"name":"provider","type":"address"}],"name":"addProvider","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":true,"inputs":[{"name":"","type":"uint256"}],"name":"providers","outputs":[{"name":"","type":"address"}],"payable":false,"stateMutability":"view","type":"function"},{"constant":false,"inputs":[],"name":"renounceOwnership","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":false,"inputs":[{"name":"provider","type":"address"}],"name":"removeProvider","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":true,"inputs":[],"name":"owner","outputs":[{"name":"","type":"address"}],"payable":false,"stateMutability":"view","type":"function"},{"constant":true,"inputs":[],"name":"isOwner","outputs":[{"name":"","type":"bool"}],"payable":false,"stateMutability":"view","type":"function"},{"constant":true,"inputs":[],"name":"minimumProviders","outputs":[{"name":"","type":"uint256"}],"payable":false,"stateMutability":"view","type":"function"},{"constant":false,"inputs":[],"name":"purgeReports","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":false,"inputs":[{"name":"reportDelaySec_","type":"uint256"}],"name":"setReportDelaySec","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":true,"inputs":[],"name":"providersSize","outputs":[{"name":"","type":"uint256"}],"payable":false,"stateMutability":"view","type":"function"},{"constant":true,"inputs":[],"name":"reportExpirationTimeSec","outputs":[{"name":"","type":"uint256"}],"payable":false,"stateMutability":"view","type":"function"},{"constant":false,"inputs":[{"name":"minimumProviders_","type":"uint256"}],"name":"setMinimumProviders","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":true,"inputs":[{"name":"","type":"address"},{"name":"","type":"uint256"}],"name":"providerReports","outputs":[{"name":"timestamp","type":"uint256"},{"name":"payload","type":"uint256"}],"payable":false,"stateMutability":"view","type":"function"},{"constant":false,"inputs":[{"name":"newOwner","type":"address"}],"name":"transferOwnership","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"constant":false,"inputs":[{"name":"reportExpirationTimeSec_","type":"uint256"}],"name":"setReportExpirationTimeSec","outputs":[],"payable":false,"stateMutability":"nonpayable","type":"function"},{"inputs":[{"name":"reportExpirationTimeSec_","type":"uint256"},{"name":"reportDelaySec_","type":"uint256"},{"name":"minimumProviders_","type":"uint256"}],"payable":false,"stateMutability":"nonpayable","type":"constructor"},{"anonymous":false,"inputs":[{"indexed":false,"name":"provider","type":"address"}],"name":"ProviderAdded","type":"event"},{"anonymous":false,"inputs":[{"indexed":false,"name":"provider","type":"address"}],"name":"ProviderRemoved","type":"event"},{"anonymous":false,"inputs":[{"indexed":false,"name":"provider","type":"address"}],"name":"ReportTimestampOutOfRange","type":"event"},{"anonymous":false,"inputs":[{"indexed":true,"name":"provider","type":"address"},{"indexed":false,"name":"payload","type":"uint256"},{"indexed":false,"name":"timestamp","type":"uint256"}],"name":"ProviderReportPushed","type":"event"},{"anonymous":false,"inputs":[{"indexed":true,"name":"previousOwner","type":"address"},{"indexed":true,"name":"newOwner","type":"address"}],"name":"OwnershipTransferred","type":"event"}]'
contract_t = w3.eth.contract(address = w3.toChecksumAddress(tellor_add), abi = tellor_abi)
tellor_data = (contract_t.functions.getCurrentValue(10).call())
from datetime import datetime, timedelta
all_prices = []
all_timestamps = []
initial_data = tellor_data
old_date = datetime.timestamp(datetime.now() - timedelta(days = 10))
all_prices.append(tellor_data[1]/ 1000000)
all_timestamps.append(datetime.fromtimestamp(int(tellor_data[2])))
while (old_date < initial_data[2]):
initial_data = contract_t.functions.getDataBefore(10, initial_data[2]).call()
#print(initial_data)
all_prices.append(initial_data[1] / 1000000)
all_timestamps.append(datetime.fromtimestamp(int(initial_data[2])))
import matplotlib.pyplot as plt
%matplotlib inline
plt.style.use('ggplot')
plt.plot(all_timestamps, all_prices)
import json
import requests
import pandas as pd
url = 'https://web-api.ampleforth.org/eth/oracle-histroy'
r = requests.get(url)
files = r.json()
ampl_dict = files["rateOracleProviderHistory"]["reports"]["ampleforth.org"]
ampl_df = pd.DataFrame(ampl_dict)
payload = list(ampl_df['payload'])
timestamps = list(ampl_df['timestampSec'])
new_timestamps = [datetime.fromtimestamp(i) for i in timestamps]
plt.plot(new_timestamps, payload)
end_date = datetime(2021,9,11,0,0,0)
timestamps_2 = [i for i in new_timestamps if i > end_date]
new_length = len(new_timestamps) - len(timestamps_2)
streamline_tx = []
streamline_ty = []
timestamps_rev = all_timestamps[::-1]
prices_rev = all_prices[::-1]
for i in range(1, len(timestamps_rev) - 1):
if prices_rev[i] != prices_rev[i-1]:
streamline_ty.append(prices_rev[i + 1])
streamline_tx.append(timestamps_rev[i])
chainlink_add = "0xe20CA8D7546932360e37E9D72c1a47334af57706"
chainlink_add_cs = w3.toChecksumAddress(chainlink_add)
chainlink_abi = '[{"inputs":[{"internalType":"address","name":"_aggregator","type":"address"},{"internalType":"address","name":"_accessController","type":"address"}],"stateMutability":"nonpayable","type":"constructor"},{"anonymous":false,"inputs":[{"indexed":true,"internalType":"int256","name":"current","type":"int256"},{"indexed":true,"internalType":"uint256","name":"roundId","type":"uint256"},{"indexed":false,"internalType":"uint256","name":"updatedAt","type":"uint256"}],"name":"AnswerUpdated","type":"event"},{"anonymous":false,"inputs":[{"indexed":true,"internalType":"uint256","name":"roundId","type":"uint256"},{"indexed":true,"internalType":"address","name":"startedBy","type":"address"},{"indexed":false,"internalType":"uint256","name":"startedAt","type":"uint256"}],"name":"NewRound","type":"event"},{"anonymous":false,"inputs":[{"indexed":true,"internalType":"address","name":"from","type":"address"},{"indexed":true,"internalType":"address","name":"to","type":"address"}],"name":"OwnershipTransferRequested","type":"event"},{"anonymous":false,"inputs":[{"indexed":true,"internalType":"address","name":"from","type":"address"},{"indexed":true,"internalType":"address","name":"to","type":"address"}],"name":"OwnershipTransferred","type":"event"},{"inputs":[],"name":"acceptOwnership","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[],"name":"accessController","outputs":[{"internalType":"contract AccessControllerInterface","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"aggregator","outputs":[{"internalType":"address","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"address","name":"_aggregator","type":"address"}],"name":"confirmAggregator","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[],"name":"decimals","outputs":[{"internalType":"uint8","name":"","type":"uint8"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"description","outputs":[{"internalType":"string","name":"","type":"string"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_roundId","type":"uint256"}],"name":"getAnswer","outputs":[{"internalType":"int256","name":"","type":"int256"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint80","name":"_roundId","type":"uint80"}],"name":"getRoundData","outputs":[{"internalType":"uint80","name":"roundId","type":"uint80"},{"internalType":"int256","name":"answer","type":"int256"},{"internalType":"uint256","name":"startedAt","type":"uint256"},{"internalType":"uint256","name":"updatedAt","type":"uint256"},{"internalType":"uint80","name":"answeredInRound","type":"uint80"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint256","name":"_roundId","type":"uint256"}],"name":"getTimestamp","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"latestAnswer","outputs":[{"internalType":"int256","name":"","type":"int256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"latestRound","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"latestRoundData","outputs":[{"internalType":"uint80","name":"roundId","type":"uint80"},{"internalType":"int256","name":"answer","type":"int256"},{"internalType":"uint256","name":"startedAt","type":"uint256"},{"internalType":"uint256","name":"updatedAt","type":"uint256"},{"internalType":"uint80","name":"answeredInRound","type":"uint80"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"latestTimestamp","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"owner","outputs":[{"internalType":"address","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint16","name":"","type":"uint16"}],"name":"phaseAggregators","outputs":[{"internalType":"contract AggregatorV2V3Interface","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"phaseId","outputs":[{"internalType":"uint16","name":"","type":"uint16"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"address","name":"_aggregator","type":"address"}],"name":"proposeAggregator","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[],"name":"proposedAggregator","outputs":[{"internalType":"contract AggregatorV2V3Interface","name":"","type":"address"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"uint80","name":"_roundId","type":"uint80"}],"name":"proposedGetRoundData","outputs":[{"internalType":"uint80","name":"roundId","type":"uint80"},{"internalType":"int256","name":"answer","type":"int256"},{"internalType":"uint256","name":"startedAt","type":"uint256"},{"internalType":"uint256","name":"updatedAt","type":"uint256"},{"internalType":"uint80","name":"answeredInRound","type":"uint80"}],"stateMutability":"view","type":"function"},{"inputs":[],"name":"proposedLatestRoundData","outputs":[{"internalType":"uint80","name":"roundId","type":"uint80"},{"internalType":"int256","name":"answer","type":"int256"},{"internalType":"uint256","name":"startedAt","type":"uint256"},{"internalType":"uint256","name":"updatedAt","type":"uint256"},{"internalType":"uint80","name":"answeredInRound","type":"uint80"}],"stateMutability":"view","type":"function"},{"inputs":[{"internalType":"address","name":"_accessController","type":"address"}],"name":"setController","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[{"internalType":"address","name":"_to","type":"address"}],"name":"transferOwnership","outputs":[],"stateMutability":"nonpayable","type":"function"},{"inputs":[],"name":"version","outputs":[{"internalType":"uint256","name":"","type":"uint256"}],"stateMutability":"view","type":"function"}]'
contract_cl = w3.eth.contract(address = chainlink_add_cs, abi = chainlink_abi)
latestData = contract_cl.functions.latestRoundData().call()
scale = 1e18
round_id_0 = latestData[0]
price_0 = latestData[1]
update_time_0 = latestData[3]
round_ids = []
prices = []
update_times_utc = []
round_ids.append(round_id_0)
prices.append(price_0 / scale)
update_times_utc.append(datetime.utcfromtimestamp(update_time_0))
current_date = str(datetime.utcfromtimestamp(update_time_0))
current_date = current_date.split()[0]
end_date = '2021-09-12'
def time_convert(raw_time):
return datetime.utcfromtimestamp(raw_time)
def inc_rounds(curr_round_data, end_date, time_arr):
curr_round_id = curr_round_data[0]
current_date = str(time_convert(curr_round_data[3])).split()[0]
while current_date != end_date:
curr_round_id = curr_round_id - 1
historicalData = contract_cl.functions.getRoundData(curr_round_id).call()
update_time_raw = historicalData[3]
time_arr.append(time_convert(update_time_raw))
#round_ids.append(historicalData[0])
prices.append(historicalData[1] / scale)
#update_times_utc.append(time_convert(update_time_raw))
current_date = str(time_convert(update_time_raw)).split()[0]
print("time: ",time_convert(update_time_raw), "price: ", historicalData[1] / (scale), "round ID: ", curr_round_id)
inc_rounds(latestData, end_date, update_times_utc)
plt.figure(figsize = (15, 12))
plt.plot(timestamps_2, payload[new_length:], label = 'ampl')
#plt.plot(all_timestamps, all_prices, label = 'tellor' )
plt.plot(streamline_tx, streamline_ty, label = 'tellor')
plt.plot(update_times_utc, prices, label = 'chainlink')
plt.title("Tellor and Ampleforth Oracle Prices")
plt.xlabel("date")
plt.ylabel("AMPL price (USD)")
plt.xticks(rotation = 90)
plt.legend()
plt.savefig('AMPLcomparison.png', bbox_inches='tight')
| 0.175786 | 0.68782 |
## Лабораторная работа 1 - Знакомство с векторными и матричными операциями на примере библиотеки [NumPy](https://numpy.org/doc/stable/)
```
import numpy as np
import matplotlib.pyplot as plt
```
### Задание 1
В машинном обучении часто используется логистистическая функция:
$$\sigma(x)=\frac{1}{1+e^{-x}}$$
Визуализировать эту функцию на промежутке $[-5, 5]$
```
x = np.linspace(-5, 5, 100)
y = np.divide(1, np.sum([1, np.exp(-x)]))
plt.plot(x, y)
plt.ylim(0, 1)
plt.show()
```
### Задание 2
1. [Создайте](https://numpy.org/doc/stable/reference/generated/numpy.array.html) вектор $v=\overline{(0,1)}$;
2. Создайте матрицу
$M=\begin{bmatrix}
\cos \alpha & -\sin \alpha \\
\sin \alpha & \cos \alpha
\end{bmatrix}$ , где $\alpha \in R$;
3. Вычислите $v_i, i \in \{1..100\}$, где
$v_1=v,
v_{n+1}=v_n\times M \cdot 0.99$;
```
import numpy as np
import matplotlib.pyplot as plt
alfa = np.random.randint(0, 100)
v = [0, 1]
M = [[np.cos(alfa), -np.sin(alfa)], [np.sin(alfa), np.cos(alfa)]]
vs = [v]
for i in range(100):
vs.append(np.dot(vs[i], M).dot(0.99))
vs = np.array(vs)
plt.figure(figsize=(6, 6))
plt.xlim(-1, 1)
plt.ylim(-1, 1)
plt.plot(*vs.T)
plt.show()
```
### Задание 3
Одним из алгоритмов обучения с учителем является так называемая полиномиальная регрессия. Для этого метода, нужно имея матрицу
$$X=\begin{bmatrix}
x_{1}\\
x_{2}\\
\vdots \\
x_{m}
\end{bmatrix}$$
получить матрицу степеней:
$$X_p=\begin{bmatrix}
x^{0}_{1} & x^{1}_{1} & \dotsc & x^{p}_{1}\\
x^{0}_{2} & x^{1}_{2} & \dotsc & x^{p}_{2}\\
\vdots & \vdots & & \vdots \\
x^{0}_{m} & x^{1}_{m} & \dotsc & x^{p}_{m}
\end{bmatrix}$$
Имея матрицу $X$, заданную ниже, вычислите матрицу $X_p$, для $p=5$.
Решить задачу как с использованием цикла по степеням, так и без него.
Полезные функции: [stack](https://numpy.org/doc/stable/reference/generated/numpy.stack.html), [concatenate](https://numpy.org/doc/stable/reference/generated/numpy.concatenate.html).
```
X = np.linspace(-2.2, 2.2, 1000)[:, np.newaxis]
print(f'Shape of X: {X.shape}')
X_p = np.concatenate([X ** i for i in range(6)], axis=-1)
# X_p = X ** np.arange(6)
print(f'Shape of X_p {X_p.shape}')
assert X_p.shape == (1000, 6)
plt.plot(
X[..., 0],
X_p @ np.array([[0, 4, 0, -5, 0, 1]]).T
)
plt.show()
```
### Задание 4 (*)
Нарисуйте график функции Растригина:
$$f( x,y) =20+\left( x^{2} -10\cos( 2\pi x)\right) +\left( y^{2} -10\cos( 2\pi y)\right) ,\ x\in [ -5,\ 5] ,\ y\in [ -5,\ 5]$$
Использовать циклы **запрещено**.
Полезные функции:
[linspace](https://numpy.org/doc/stable/reference/generated/numpy.linspace.html),
[meshgrid](https://numpy.org/doc/stable/reference/generated/numpy.meshgrid.html),
[reshape](https://numpy.org/doc/stable/reference/generated/numpy.reshape.html).
(Для красивой визуализации возьмите по 500 шагов для $x$ и $y$).
```
def rastrigin(x, y):
return 20 + (x**2 - 10 * np.cos(2*np.pi*x)) + (y**2 - 10 * np.cos(2*np.pi*y))
x = np.linspace(-5, 5, num=500)
y = np.linspace(-5, 5, num=500)
X, Y = np.meshgrid(x, y)
plt.figure(figsize=(10, 10))
plt.contourf(X, Y, rastrigin(X, Y), levels=10)
plt.show()
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
x = np.linspace(-5, 5, 100)
y = np.divide(1, np.sum([1, np.exp(-x)]))
plt.plot(x, y)
plt.ylim(0, 1)
plt.show()
import numpy as np
import matplotlib.pyplot as plt
alfa = np.random.randint(0, 100)
v = [0, 1]
M = [[np.cos(alfa), -np.sin(alfa)], [np.sin(alfa), np.cos(alfa)]]
vs = [v]
for i in range(100):
vs.append(np.dot(vs[i], M).dot(0.99))
vs = np.array(vs)
plt.figure(figsize=(6, 6))
plt.xlim(-1, 1)
plt.ylim(-1, 1)
plt.plot(*vs.T)
plt.show()
X = np.linspace(-2.2, 2.2, 1000)[:, np.newaxis]
print(f'Shape of X: {X.shape}')
X_p = np.concatenate([X ** i for i in range(6)], axis=-1)
# X_p = X ** np.arange(6)
print(f'Shape of X_p {X_p.shape}')
assert X_p.shape == (1000, 6)
plt.plot(
X[..., 0],
X_p @ np.array([[0, 4, 0, -5, 0, 1]]).T
)
plt.show()
def rastrigin(x, y):
return 20 + (x**2 - 10 * np.cos(2*np.pi*x)) + (y**2 - 10 * np.cos(2*np.pi*y))
x = np.linspace(-5, 5, num=500)
y = np.linspace(-5, 5, num=500)
X, Y = np.meshgrid(x, y)
plt.figure(figsize=(10, 10))
plt.contourf(X, Y, rastrigin(X, Y), levels=10)
plt.show()
| 0.437103 | 0.982971 |
# 05 - Continuous Training
After testing, compiling, and uploading the pipeline definition to Cloud Storage, the pipeline is executed with respect to a trigger. We use [Cloud Functions](https://cloud.google.com/functions) and [Cloud Pub/Sub](https://cloud.google.com/pubsub) as a triggering mechanism. The triggering can be scheduled using [Cloud Scheduler](https://cloud.google.com/scheduler). The trigger source sends a message to a Cloud Pub/Sub topic that the Cloud Function listens to, and then it submits the pipeline to AI Platform Managed Pipelines to be executed.
This notebook covers the following steps:
1. Create the Cloud Pub/Sub topic.
2. Deploy the Cloud Function
3. Test triggering a pipeline.
4. Extracting pipeline run metadata.
## Setup
### Import libraries
```
import json
import os
import logging
import tensorflow as tf
import tfx
import IPython
logging.getLogger().setLevel(logging.INFO)
print("Tensorflow Version:", tfx.__version__)
```
### Setup Google Cloud project
```
PROJECT = '[your-project-id]' # Change to your project id.
REGION = 'europe-west1' # Change to your region.
BUCKET = '[your-bucket-name]' # Change to your bucket name.
SERVICE_ACCOUNT = "[your-service-account]"
if PROJECT == "" or PROJECT is None or PROJECT == "[your-project-id]":
# Get your GCP project id from gcloud
shell_output = !gcloud config list --format 'value(core.project)' 2>/dev/null
PROJECT = shell_output[0]
if BUCKET == "" or BUCKET is None or BUCKET == "[your-bucket-name]":
# Get your bucket name to GCP projet id
BUCKET = PROJECT
if SERVICE_ACCOUNT == "" or SERVICE_ACCOUNT is None or SERVICE_ACCOUNT == "[your-service-account]":
# Get your GCP project id from gcloud
shell_output = !gcloud config list --format 'value(core.account)' 2>/dev/null
SERVICE_ACCOUNT = shell_output[0]
print("Project ID:", PROJECT)
print("Region:", REGION)
print("Bucket name:", BUCKET)
print("Service Account:", SERVICE_ACCOUNT)
```
### Set configurations
```
VERSION = 'v01'
DATASET_DISPLAY_NAME = 'chicago-taxi-tips'
MODEL_DISPLAY_NAME = f'{DATASET_DISPLAY_NAME}-classifier-{VERSION}'
PIPELINE_NAME = f'{MODEL_DISPLAY_NAME}-train-pipeline'
PIPELINES_STORE = f'gs://{BUCKET}/{DATASET_DISPLAY_NAME}/compiled_pipelines/'
GCS_PIPELINE_FILE_LOCATION = os.path.join(PIPELINES_STORE, f'{PIPELINE_NAME}.json')
PUBSUB_TOPIC = f'trigger-{PIPELINE_NAME}'
CLOUD_FUNCTION_NAME = f'trigger-{PIPELINE_NAME}-fn'
!gsutil ls {GCS_PIPELINE_FILE_LOCATION}
```
## 1. Create a Pub/Sub topic
```
!gcloud pubsub topics create {PUBSUB_TOPIC}
```
## 2. Deploy the Cloud Function
```
ENV_VARS=f"""\
PROJECT={PROJECT},\
REGION={REGION},\
GCS_PIPELINE_FILE_LOCATION={GCS_PIPELINE_FILE_LOCATION},\
SERVICE_ACCOUNT={SERVICE_ACCOUNT}
"""
!echo {ENV_VARS}
!rm -r src/pipeline_triggering/.ipynb_checkpoints
!gcloud functions deploy {CLOUD_FUNCTION_NAME} \
--region={REGION} \
--trigger-topic={PUBSUB_TOPIC} \
--runtime=python37 \
--source=src/pipeline_triggering\
--entry-point=trigger_pipeline\
--stage-bucket={BUCKET}\
--ingress-settings=internal-only\
--service-account={SERVICE_ACCOUNT}\
--update-env-vars={ENV_VARS}
cloud_fn_url = f"https://console.cloud.google.com/functions/details/{REGION}/{CLOUD_FUNCTION_NAME}"
html = f'See the Cloud Function details <a href="{cloud_fn_url}" target="_blank">here</a>.'
IPython.display.display(IPython.display.HTML(html))
```
## 3. Trigger the pipeline
```
from google.cloud import pubsub
publish_client = pubsub.PublisherClient()
topic = f'projects/{PROJECT}/topics/{PUBSUB_TOPIC}'
data = {
'num_epochs': 7,
'learning_rate': 0.0015,
'batch_size': 512,
'hidden_units': '256,126'
}
message = json.dumps(data)
_ = publish_client.publish(topic, message.encode())
```
Wait for a few seconds for the pipeline run to be submitted, then you can see the run in the Cloud Console
```
from kfp.v2.google.client import AIPlatformClient
pipeline_client = AIPlatformClient(
project_id=PROJECT, region=REGION)
job_display_name = pipeline_client.list_jobs()['pipelineJobs'][0]['displayName']
job_url = f"https://console.cloud.google.com/vertex-ai/locations/{REGION}/pipelines/runs/{job_display_name}"
html = f'See the Pipeline job <a href="{job_url}" target="_blank">here</a>.'
IPython.display.display(IPython.display.HTML(html))
```
## 4. Extracting pipeline runs metadata
```
from google.cloud import aiplatform as vertex_ai
pipeline_df = vertex_ai.get_pipeline_df(PIPELINE_NAME)
pipeline_df = pipeline_df[pipeline_df.pipeline_name == PIPELINE_NAME]
pipeline_df.T
```
|
github_jupyter
|
import json
import os
import logging
import tensorflow as tf
import tfx
import IPython
logging.getLogger().setLevel(logging.INFO)
print("Tensorflow Version:", tfx.__version__)
PROJECT = '[your-project-id]' # Change to your project id.
REGION = 'europe-west1' # Change to your region.
BUCKET = '[your-bucket-name]' # Change to your bucket name.
SERVICE_ACCOUNT = "[your-service-account]"
if PROJECT == "" or PROJECT is None or PROJECT == "[your-project-id]":
# Get your GCP project id from gcloud
shell_output = !gcloud config list --format 'value(core.project)' 2>/dev/null
PROJECT = shell_output[0]
if BUCKET == "" or BUCKET is None or BUCKET == "[your-bucket-name]":
# Get your bucket name to GCP projet id
BUCKET = PROJECT
if SERVICE_ACCOUNT == "" or SERVICE_ACCOUNT is None or SERVICE_ACCOUNT == "[your-service-account]":
# Get your GCP project id from gcloud
shell_output = !gcloud config list --format 'value(core.account)' 2>/dev/null
SERVICE_ACCOUNT = shell_output[0]
print("Project ID:", PROJECT)
print("Region:", REGION)
print("Bucket name:", BUCKET)
print("Service Account:", SERVICE_ACCOUNT)
VERSION = 'v01'
DATASET_DISPLAY_NAME = 'chicago-taxi-tips'
MODEL_DISPLAY_NAME = f'{DATASET_DISPLAY_NAME}-classifier-{VERSION}'
PIPELINE_NAME = f'{MODEL_DISPLAY_NAME}-train-pipeline'
PIPELINES_STORE = f'gs://{BUCKET}/{DATASET_DISPLAY_NAME}/compiled_pipelines/'
GCS_PIPELINE_FILE_LOCATION = os.path.join(PIPELINES_STORE, f'{PIPELINE_NAME}.json')
PUBSUB_TOPIC = f'trigger-{PIPELINE_NAME}'
CLOUD_FUNCTION_NAME = f'trigger-{PIPELINE_NAME}-fn'
!gsutil ls {GCS_PIPELINE_FILE_LOCATION}
!gcloud pubsub topics create {PUBSUB_TOPIC}
ENV_VARS=f"""\
PROJECT={PROJECT},\
REGION={REGION},\
GCS_PIPELINE_FILE_LOCATION={GCS_PIPELINE_FILE_LOCATION},\
SERVICE_ACCOUNT={SERVICE_ACCOUNT}
"""
!echo {ENV_VARS}
!rm -r src/pipeline_triggering/.ipynb_checkpoints
!gcloud functions deploy {CLOUD_FUNCTION_NAME} \
--region={REGION} \
--trigger-topic={PUBSUB_TOPIC} \
--runtime=python37 \
--source=src/pipeline_triggering\
--entry-point=trigger_pipeline\
--stage-bucket={BUCKET}\
--ingress-settings=internal-only\
--service-account={SERVICE_ACCOUNT}\
--update-env-vars={ENV_VARS}
cloud_fn_url = f"https://console.cloud.google.com/functions/details/{REGION}/{CLOUD_FUNCTION_NAME}"
html = f'See the Cloud Function details <a href="{cloud_fn_url}" target="_blank">here</a>.'
IPython.display.display(IPython.display.HTML(html))
from google.cloud import pubsub
publish_client = pubsub.PublisherClient()
topic = f'projects/{PROJECT}/topics/{PUBSUB_TOPIC}'
data = {
'num_epochs': 7,
'learning_rate': 0.0015,
'batch_size': 512,
'hidden_units': '256,126'
}
message = json.dumps(data)
_ = publish_client.publish(topic, message.encode())
from kfp.v2.google.client import AIPlatformClient
pipeline_client = AIPlatformClient(
project_id=PROJECT, region=REGION)
job_display_name = pipeline_client.list_jobs()['pipelineJobs'][0]['displayName']
job_url = f"https://console.cloud.google.com/vertex-ai/locations/{REGION}/pipelines/runs/{job_display_name}"
html = f'See the Pipeline job <a href="{job_url}" target="_blank">here</a>.'
IPython.display.display(IPython.display.HTML(html))
from google.cloud import aiplatform as vertex_ai
pipeline_df = vertex_ai.get_pipeline_df(PIPELINE_NAME)
pipeline_df = pipeline_df[pipeline_df.pipeline_name == PIPELINE_NAME]
pipeline_df.T
| 0.274935 | 0.837354 |
# Goal of project
- Author: Arun Nemani
- Effectively extract features from N categories of the 20-newsgroups dataset
- Train and fit a classification model to predict text inputs based on the extracted features
- Report the accuracies for each classification models
### Cloned repos as baseline
- https://github.com/stefansavev/demos/blob/master/text-categorization/20ng/20ng.py
- https://nlpforhackers.io/text-classification/
```
from __future__ import print_function
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cross_validation import train_test_split
import nltk
import string
from nltk import word_tokenize
import nltk
from nltk.stem.porter import PorterStemmer
from nltk.corpus import stopwords
from sklearn.datasets import fetch_20newsgroups
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.linear_model import LogisticRegression
from sklearn.linear_model import SGDClassifier
from sklearn import metrics
from sklearn.grid_search import GridSearchCV
```
## Initialize and load dataset
First we split the dataset into a training, development, and testing set.
The purpose of splitting up the dataset in this manner is to ensure we do not bias or overfit our model by iteratively refining our model on the train and test sets. Instead, we fine tune our model on the train and development set, and invoke the test only once as a final input on the tuned model.
Thus, the entire dataset is split into the training set (70%), development set (15%), and test set (15%).
```
# Initializations
nltk.download('punkt')
nltk.download('stopwords')
categ = ['alt.atheism', 'comp.graphics', 'comp.os.ms-windows.misc',
'comp.sys.ibm.pc.hardware', 'comp.sys.mac.hardware', 'comp.windows.x',
'misc.forsale', 'rec.autos', 'rec.motorcycles', 'rec.sport.baseball',
'rec.sport.hockey', 'sci.crypt', 'sci.electronics', 'sci.med',
'sci.space', 'soc.religion.christian', 'talk.politics.guns',
'talk.politics.mideast', 'talk.politics.misc', 'talk.religion.misc'] #None: Indicating all 20 categories are included in dataset
remove = ('headers', 'footers', 'quotes') #Required to remove false features that result in overfitting
RANDOM_STATE = 35
# Load dataset
print("Loading 20 newsgroups dataset for categories:")
newsdata = fetch_20newsgroups(subset='all', categories=categ)
X_train, X_intermediate, Y_train, Y_intermediate = train_test_split(newsdata.data, newsdata.target, test_size=0.30, random_state=RANDOM_STATE)
X_dev, X_test, Y_dev, Y_test = train_test_split(X_intermediate, Y_intermediate, test_size=0.50, random_state=RANDOM_STATE)
print('Data loaded')
print()
print('Training data documents:', len(X_train))
print('Development data documents:', len(X_dev))
print('Test data documents:', len(X_test))
print()
print('Total Newsgroups :', newsdata.target_names)
def Stem_tokenize(text):
stemmer = PorterStemmer()
return [stemmer.stem(w) for w in word_tokenize(text)]
```
## Finalized Feature Extractor
Note that we have SIGNIFICANTLY reduced the number of vocabulary features in our vectorizer from 1392137 to 85540.
This is primarily due to the min_df parameter since changing the max_df does not impact the feature size significantly.
Basically, our previous vectorizers were overfitting features that were VERY sparse in the dataset.
FE will be our finalized feature extractor for this project.
Next we explore classification schemes.
```
FE = TfidfVectorizer(analyzer= 'word', tokenizer= Stem_tokenize,
stop_words=stopwords.words('english') + list(string.punctuation),
lowercase=True, strip_accents='ascii', ngram_range=(1,2),
min_df=5, max_df= 0.75)
Vocab_train = FE.fit_transform(X_train)
Vocab_dev = FE.transform(X_dev)
print('FE training set vocabulary size is {} in {} documents'.format(Vocab_train.shape[1], Vocab_train.shape[0]))
print('FE dev set vocabulary size is {} in {} documents'.format(Vocab_dev.shape[1], Vocab_dev.shape[0]))
```
## Feature Classification
First we need to understand the dataset before selection a classification model.
The matrix output of the feature extraction methods are very sparse with a small set of non-zero values.
It is important to note that we will fine tune our classification models on the dev set USING the feature extraction model created on the training set.
Thus we will try Multinomial Naive-Bayes, regularized Logistic regression, and Stochastic Gradient Descent.
## Grid-search for hyperparameter tuning
```
classifier_nb = MultinomialNB()
params = {'alpha':[0.00001, 0.0001, 0.001, 0.01, 0.1, 0.5, 1.0, 2.0, 5.0]}
grid_classifier_nb = GridSearchCV(classifier_nb, params, scoring = 'accuracy')
grid_classifier_nb.fit(Vocab_train, Y_train)
pred = grid_classifier_nb.predict(Vocab_dev)
print("Multinomial NB optimal alpha: {}".format(grid_classifier_nb.best_params_))
classifier_lreg = LogisticRegression(penalty = 'l2', solver='sag', random_state=RANDOM_STATE, n_jobs=-1)
params = {'C':[0.0001, 0.001, 0.01, 0.1, 0.25, 0.5, 1.0, 2.0, 5.0]}
grid_classifier_lreg = GridSearchCV(classifier_lreg, params, scoring = 'accuracy')
grid_classifier_lreg.fit(Vocab_train, Y_train)
pred = grid_classifier_lreg.predict(Vocab_dev)
print("Logistic Regression optimal C: {}".format(grid_classifier_lreg.best_params_))
classifier_SGD = SGDClassifier(tol=0.0001, penalty="l2", random_state=RANDOM_STATE, n_jobs=-1)
params = {'alpha':[0.00001, 0.0001, 0.001, 0.01, 0.1, 0.5, 1.0, 2.0, 5.0]}
grid_classifier_SGD = GridSearchCV(classifier_SGD, params, scoring = 'accuracy')
grid_classifier_SGD.fit(Vocab_train, Y_train)
pred = grid_classifier_SGD.predict(Vocab_dev)
print("Stochastic Gradient Descent optimal alpha: {}".format(grid_classifier_SGD.best_params_))
```
## Optimal classification model
Now that we have identified the optimal parameters for each model, we will calculate the final accuracies on the test set and select the final classification model for this project.
Note that we predefine the regularization parameters for SGD and logistic regression classifiers.
The idea of regularization is to avoid learning very large weights, which are likely to fit the training data but do not generalize well. L2 regularization adds a penalty to the sum of the squared weights whereas L1 regularization computes add the penalty via the sum of the absolute values of the weights. The result is that L2 regularization makes all the weights relatively small, and L1 regularization drives lots of the weights to 0, effectively removing unimportant features.
In this particular application, there are a number of features that are very sparse but unique in identifying newsgroups.
Thus, only "L2" regularization has been selected for all classification models.
```
Vocab_test = FE.transform(X_test)
classifier_NB = MultinomialNB(alpha=0.01)
classifier_NB.fit(Vocab_train, Y_train)
pred = classifier_NB.predict(Vocab_test)
print("NB classifier accuracy: {}".format(round(metrics.accuracy_score(Y_test, pred),4)))
classifier_lreg = LogisticRegression(penalty = 'l2', solver='sag', C=5, random_state=RANDOM_STATE, n_jobs=-1)
classifier_lreg.fit(Vocab_train, Y_train)
pred = classifier_lreg.predict(Vocab_test)
print("Logistic regression classifier accuracy: {}".format(round(metrics.accuracy_score(Y_test, pred),4)))
classifier_SGD = SGDClassifier(tol=0.0001, penalty="l2", alpha=0.0001, random_state=RANDOM_STATE, n_jobs=-1)
classifier_SGD.fit(Vocab_train, Y_train)
pred = classifier_SGD.predict(Vocab_test)
print("Stochastic Gradient Descent classifier accuracy: {}".format(round(metrics.accuracy_score(Y_test, pred),4)))
```
## Finalized predictive model
Multinomial NB classifier accuracy on final test set: 0.9073.
Logistic regression accuracy on final test set: 0.9059.
Stochastic gradient descent accuracy on final test set: 0.9165.
Based on our preprocessing, parameter tuning, and model selection, a stochastic gradient descent classifier can accurately predict an input corpus into one of the 20 newsgroups. However, multinomial NB has marginally lower accuracy but perfoms significantly much faster than the other classifiers on the new test set.
Below we apply some sanity checks to see our prediction work in real-time.
```
def predictNewsGroup(text, clf):
Vocab_test = FE.transform([text])
targets = newsdata.target_names
idx = clf.predict(Vocab_test)
print("Predicted newsgroup: {}".format(targets[int(idx)]))
return
print("Newsgroup categories: {}".format(newsdata.target_names))
print()
predictNewsGroup("A Honda CBR is a dope ride", classifier_SGD)
predictNewsGroup("He is #1 player with the highest contract signed for the Minnesota Wild", classifier_SGD)
predictNewsGroup("I'll only sell with my Gamecube for $1000", classifier_SGD)
predictNewsGroup("Homs is really unstable right now. Many refugees are actively leaving the region", classifier_SGD)
predictNewsGroup("Interstellar was a really good movie. I'm sure Carl Sagan would've loved it", classifier_SGD)
predictNewsGroup("MLops is an important concept for productionalizing machine learning models", classifier_SGD)
predictNewsGroup("Donald Trump didn't tell the whole truth about the Russia investigation", classifier_SGD)
```
|
github_jupyter
|
from __future__ import print_function
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cross_validation import train_test_split
import nltk
import string
from nltk import word_tokenize
import nltk
from nltk.stem.porter import PorterStemmer
from nltk.corpus import stopwords
from sklearn.datasets import fetch_20newsgroups
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.linear_model import LogisticRegression
from sklearn.linear_model import SGDClassifier
from sklearn import metrics
from sklearn.grid_search import GridSearchCV
# Initializations
nltk.download('punkt')
nltk.download('stopwords')
categ = ['alt.atheism', 'comp.graphics', 'comp.os.ms-windows.misc',
'comp.sys.ibm.pc.hardware', 'comp.sys.mac.hardware', 'comp.windows.x',
'misc.forsale', 'rec.autos', 'rec.motorcycles', 'rec.sport.baseball',
'rec.sport.hockey', 'sci.crypt', 'sci.electronics', 'sci.med',
'sci.space', 'soc.religion.christian', 'talk.politics.guns',
'talk.politics.mideast', 'talk.politics.misc', 'talk.religion.misc'] #None: Indicating all 20 categories are included in dataset
remove = ('headers', 'footers', 'quotes') #Required to remove false features that result in overfitting
RANDOM_STATE = 35
# Load dataset
print("Loading 20 newsgroups dataset for categories:")
newsdata = fetch_20newsgroups(subset='all', categories=categ)
X_train, X_intermediate, Y_train, Y_intermediate = train_test_split(newsdata.data, newsdata.target, test_size=0.30, random_state=RANDOM_STATE)
X_dev, X_test, Y_dev, Y_test = train_test_split(X_intermediate, Y_intermediate, test_size=0.50, random_state=RANDOM_STATE)
print('Data loaded')
print()
print('Training data documents:', len(X_train))
print('Development data documents:', len(X_dev))
print('Test data documents:', len(X_test))
print()
print('Total Newsgroups :', newsdata.target_names)
def Stem_tokenize(text):
stemmer = PorterStemmer()
return [stemmer.stem(w) for w in word_tokenize(text)]
FE = TfidfVectorizer(analyzer= 'word', tokenizer= Stem_tokenize,
stop_words=stopwords.words('english') + list(string.punctuation),
lowercase=True, strip_accents='ascii', ngram_range=(1,2),
min_df=5, max_df= 0.75)
Vocab_train = FE.fit_transform(X_train)
Vocab_dev = FE.transform(X_dev)
print('FE training set vocabulary size is {} in {} documents'.format(Vocab_train.shape[1], Vocab_train.shape[0]))
print('FE dev set vocabulary size is {} in {} documents'.format(Vocab_dev.shape[1], Vocab_dev.shape[0]))
classifier_nb = MultinomialNB()
params = {'alpha':[0.00001, 0.0001, 0.001, 0.01, 0.1, 0.5, 1.0, 2.0, 5.0]}
grid_classifier_nb = GridSearchCV(classifier_nb, params, scoring = 'accuracy')
grid_classifier_nb.fit(Vocab_train, Y_train)
pred = grid_classifier_nb.predict(Vocab_dev)
print("Multinomial NB optimal alpha: {}".format(grid_classifier_nb.best_params_))
classifier_lreg = LogisticRegression(penalty = 'l2', solver='sag', random_state=RANDOM_STATE, n_jobs=-1)
params = {'C':[0.0001, 0.001, 0.01, 0.1, 0.25, 0.5, 1.0, 2.0, 5.0]}
grid_classifier_lreg = GridSearchCV(classifier_lreg, params, scoring = 'accuracy')
grid_classifier_lreg.fit(Vocab_train, Y_train)
pred = grid_classifier_lreg.predict(Vocab_dev)
print("Logistic Regression optimal C: {}".format(grid_classifier_lreg.best_params_))
classifier_SGD = SGDClassifier(tol=0.0001, penalty="l2", random_state=RANDOM_STATE, n_jobs=-1)
params = {'alpha':[0.00001, 0.0001, 0.001, 0.01, 0.1, 0.5, 1.0, 2.0, 5.0]}
grid_classifier_SGD = GridSearchCV(classifier_SGD, params, scoring = 'accuracy')
grid_classifier_SGD.fit(Vocab_train, Y_train)
pred = grid_classifier_SGD.predict(Vocab_dev)
print("Stochastic Gradient Descent optimal alpha: {}".format(grid_classifier_SGD.best_params_))
Vocab_test = FE.transform(X_test)
classifier_NB = MultinomialNB(alpha=0.01)
classifier_NB.fit(Vocab_train, Y_train)
pred = classifier_NB.predict(Vocab_test)
print("NB classifier accuracy: {}".format(round(metrics.accuracy_score(Y_test, pred),4)))
classifier_lreg = LogisticRegression(penalty = 'l2', solver='sag', C=5, random_state=RANDOM_STATE, n_jobs=-1)
classifier_lreg.fit(Vocab_train, Y_train)
pred = classifier_lreg.predict(Vocab_test)
print("Logistic regression classifier accuracy: {}".format(round(metrics.accuracy_score(Y_test, pred),4)))
classifier_SGD = SGDClassifier(tol=0.0001, penalty="l2", alpha=0.0001, random_state=RANDOM_STATE, n_jobs=-1)
classifier_SGD.fit(Vocab_train, Y_train)
pred = classifier_SGD.predict(Vocab_test)
print("Stochastic Gradient Descent classifier accuracy: {}".format(round(metrics.accuracy_score(Y_test, pred),4)))
def predictNewsGroup(text, clf):
Vocab_test = FE.transform([text])
targets = newsdata.target_names
idx = clf.predict(Vocab_test)
print("Predicted newsgroup: {}".format(targets[int(idx)]))
return
print("Newsgroup categories: {}".format(newsdata.target_names))
print()
predictNewsGroup("A Honda CBR is a dope ride", classifier_SGD)
predictNewsGroup("He is #1 player with the highest contract signed for the Minnesota Wild", classifier_SGD)
predictNewsGroup("I'll only sell with my Gamecube for $1000", classifier_SGD)
predictNewsGroup("Homs is really unstable right now. Many refugees are actively leaving the region", classifier_SGD)
predictNewsGroup("Interstellar was a really good movie. I'm sure Carl Sagan would've loved it", classifier_SGD)
predictNewsGroup("MLops is an important concept for productionalizing machine learning models", classifier_SGD)
predictNewsGroup("Donald Trump didn't tell the whole truth about the Russia investigation", classifier_SGD)
| 0.700383 | 0.953966 |
# Emerging Technologies Project
- Usman Sattar
- G00345816
## Research
In this project, we learn as we go along the development root. I have learned how to build a neural network database using keras and tensorflow. This model predicts the digit which is drawn my the user. The handwritten digit is converted to an image and the computer predicts what number is drawn by the user.
## Simple Understanding of Neural Networks
Neural networks were first proposed in 1944 by Warren McCullough and Walter Pitts.
### What are neural networks?
Neural networks are a set of algorithms, modeled after the human brain. Neural networks are designed to recognize patterns. In our case we are recognising digits. Neural networks help us cluster and classify. We can have multiple hidden layers but below an image illustrates with two hidden layers within a neuron network.
```
# Imports
# Importing required imported, necessary for running the model.
# Helps with mathematical and scientific operation
import numpy as np
# Creates a figure, like graphs, plots
import matplotlib.pyplot as plt
# Keras is a high-level neural networks API, capable of running on top of TensorFlow
import keras as kr
```
### Loading Data Set
1. The function mnist.load_data() is called to load the data set.
2. Storing data into arrays.
3. Scaling data into proportion
### Viewing Data
12 examples of data are printed below.
```
plt.figure(1, figsize=(30,20))
# Loop for images - printing out 12 images.
for i in range(12):
plt.subplot(1,12,i+1)
plt.imshow(x_train[i].reshape(28,28), cmap='gray', interpolation='nearest')
plt.xticks([])
plt.yticks([])
# Creating the model
model = kr.models.Sequential()
print("Model Created")
# Adding layers to the model
model.add(kr.layers.Dense(392, activation='relu', input_shape=(784,)))
model.add(kr.layers.Dense(392, activation='relu'))
model.add(kr.layers.Dropout(0.2))
model.add(kr.layers.Dense(10, activation='softmax'))
print("Layers Added To The Model")
# Summerizing Model
print("Model Summary")
model.summary()
# Compiling Model
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
print("Model Compiled")
```
### Training Model
We are running through Epoch five times to get a better accuracy of the model. As we see below, the more times the model is trained, the better result in accuracy we get.
- Fit method used to train model
- Epochs set to 5
```
history = model.fit(x_train, y_train, batch_size=50, epochs=5, verbose=1, validation_data=(x_test, y_test))
```
### Loss & Train Accruacy
- Printing the loss of the model, accuracy from training.
```
score = model.evaluate(x_train, y_train, verbose=0)
print('Test cross-entropy loss: %0.9f' % score[0])
print('Test accuracy: %0.9f' % score[1])
```
### Saving
- Saving model as model.h5
- Loading model as loadedModel
```
print("Saving Model")
model.save('model.h5')
print("Loading Model")
loadedModel = kr.models.load_model('model.h5')
```
### Visual Example of Output
- Testing the model.
- Printing out the image which is selected for testing purposes.
- Uses argmax to return the actual prediction.
```
plt.imshow(x_test[75].reshape(28, 28), cmap="gray")
plt.show()
np.argmax(loadedModel.predict(x_test[75:76]))
```
### Visual Example of Output
- Testing the model.
- Printing out the image which is selected for testing purposes.
- Uses argmax to return the actual prediction.
```
plt.imshow(x_test[21].reshape(28, 28), cmap="gray")
plt.show()
np.argmax(loadedModel.predict(x_test[21:22]))
```
### Visual Example of Output
- Testing the model.
- Printing out the image which is selected for testing purposes.
- Uses argmax to return the actual prediction.
```
plt.imshow(x_test[82].reshape(28, 28), cmap="gray")
plt.show()
np.argmax(loadedModel.predict(x_test[82:83]))
```
### Visual Example of Output
- Testing the model.
- Printing out the image which is selected for testing purposes.
- Uses argmax to return the actual prediction.
```
plt.imshow(x_test[12].reshape(28, 28), cmap="gray")
plt.show()
np.argmax(loadedModel.predict(x_test[12:13]))
```
## References
- https://towardsdatascience.com/understanding-neural-networks-19020b758230
- https://keras.io/
- https://nbviewer.jupyter.org/github/ianmcloughlin/jupyter-teaching-notebooks/blob/master/mnist.ipynb
- https://github.com/ianmcloughlin/jupyter-teaching-notebooks/blob/master/keras-neurons.ipynb
- https://keras.io/models/about-keras-models/
|
github_jupyter
|
# Imports
# Importing required imported, necessary for running the model.
# Helps with mathematical and scientific operation
import numpy as np
# Creates a figure, like graphs, plots
import matplotlib.pyplot as plt
# Keras is a high-level neural networks API, capable of running on top of TensorFlow
import keras as kr
plt.figure(1, figsize=(30,20))
# Loop for images - printing out 12 images.
for i in range(12):
plt.subplot(1,12,i+1)
plt.imshow(x_train[i].reshape(28,28), cmap='gray', interpolation='nearest')
plt.xticks([])
plt.yticks([])
# Creating the model
model = kr.models.Sequential()
print("Model Created")
# Adding layers to the model
model.add(kr.layers.Dense(392, activation='relu', input_shape=(784,)))
model.add(kr.layers.Dense(392, activation='relu'))
model.add(kr.layers.Dropout(0.2))
model.add(kr.layers.Dense(10, activation='softmax'))
print("Layers Added To The Model")
# Summerizing Model
print("Model Summary")
model.summary()
# Compiling Model
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
print("Model Compiled")
history = model.fit(x_train, y_train, batch_size=50, epochs=5, verbose=1, validation_data=(x_test, y_test))
score = model.evaluate(x_train, y_train, verbose=0)
print('Test cross-entropy loss: %0.9f' % score[0])
print('Test accuracy: %0.9f' % score[1])
print("Saving Model")
model.save('model.h5')
print("Loading Model")
loadedModel = kr.models.load_model('model.h5')
plt.imshow(x_test[75].reshape(28, 28), cmap="gray")
plt.show()
np.argmax(loadedModel.predict(x_test[75:76]))
plt.imshow(x_test[21].reshape(28, 28), cmap="gray")
plt.show()
np.argmax(loadedModel.predict(x_test[21:22]))
plt.imshow(x_test[82].reshape(28, 28), cmap="gray")
plt.show()
np.argmax(loadedModel.predict(x_test[82:83]))
plt.imshow(x_test[12].reshape(28, 28), cmap="gray")
plt.show()
np.argmax(loadedModel.predict(x_test[12:13]))
| 0.850624 | 0.992085 |
```
#default_exp core.dispatch
#export
from local.core.imports import *
from local.core.foundation import *
from local.core.utils import *
from local.test import *
from local.notebook.showdoc import *
```
# Type dispatch
> Basic single and dual parameter dispatch
## Helpers
```
#exports
def type_hints(f):
"Same as `typing.get_type_hints` but returns `{}` if not allowed type"
return typing.get_type_hints(f) if isinstance(f, typing._allowed_types) else {}
#export
def anno_ret(func):
"Get the return annotation of `func`"
if not func: return None
ann = type_hints(func)
if not ann: return None
return ann.get('return')
#hide
def f(x) -> float: return x
test_eq(anno_ret(f), float)
def f(x) -> typing.Tuple[float,float]: return x
test_eq(anno_ret(f), typing.Tuple[float,float])
def f(x) -> None: return x
test_eq(anno_ret(f), NoneType)
def f(x): return x
test_eq(anno_ret(f), None)
test_eq(anno_ret(None), None)
#export
cmp_instance = functools.cmp_to_key(lambda a,b: 0 if a==b else 1 if issubclass(a,b) else -1)
td = {int:1, numbers.Number:2, numbers.Integral:3}
test_eq(sorted(td, key=cmp_instance), [numbers.Number, numbers.Integral, int])
#export
def _p2_anno(f):
"Get the 1st 2 annotations of `f`, defaulting to `object`"
hints = type_hints(f)
ann = [o for n,o in hints.items() if n!='return']
while len(ann)<2: ann.append(object)
return ann[:2]
def _f(a): pass
test_eq(_p2_anno(_f), (object,object))
def _f(a, b): pass
test_eq(_p2_anno(_f), (object,object))
def _f(a:None, b)->str: pass
test_eq(_p2_anno(_f), (NoneType,object))
def _f(a:str, b)->float: pass
test_eq(_p2_anno(_f), (str,object))
def _f(a:None, b:str)->float: pass
test_eq(_p2_anno(_f), (NoneType,str))
def _f(a:int, b:int)->float: pass
test_eq(_p2_anno(_f), (int,int))
def _f(self, a:int, b:int): pass
test_eq(_p2_anno(_f), (int,int))
def _f(a:int, b:str)->float: pass
test_eq(_p2_anno(_f), (int,str))
test_eq(_p2_anno(attrgetter('foo')), (object,object))
```
## TypeDispatch -
The following class is the basis that allows us to do type dipatch with type annotations. It contains a dictionary type -> functions and ensures that the proper function is called when passed an object (depending on its type).
```
#export
class _TypeDict:
def __init__(self): self.d,self.cache = {},{}
def _reset(self):
self.d = {k:self.d[k] for k in sorted(self.d, key=cmp_instance, reverse=True)}
self.cache = {}
def add(self, t, f):
"Add type `t` and function `f`"
if not isinstance(t,tuple): t=(t,)
for t_ in t: self.d[t_] = f
self._reset()
def all_matches(self, k):
"Find first matching type that is a super-class of `k`"
if k not in self.cache:
types = [f for f in self.d if k==f or (isinstance(k,type) and issubclass(k,f))]
self.cache[k] = [self.d[o] for o in types]
return self.cache[k]
def __getitem__(self, k):
"Find first matching type that is a super-class of `k`"
res = self.all_matches(k)
return res[0] if len(res) else None
def __repr__(self): return self.d.__repr__()
def first(self): return first(self.d.values())
#export
class TypeDispatch:
"Dictionary-like object; `__getitem__` matches keys of types using `issubclass`"
def __init__(self, *funcs):
self.funcs = _TypeDict()
for o in funcs: self.add(o)
self.inst = None
def add(self, f):
"Add type `t` and function `f`"
a0,a1 = _p2_anno(f)
t = self.funcs.d.get(a0)
if t is None:
t = _TypeDict()
self.funcs.add(a0, t)
t.add(a1, f)
def first(self): return self.funcs.first().first()
def returns(self, x): return anno_ret(self[type(x)])
def returns_none(self, x):
r = anno_ret(self[type(x)])
return r if r == NoneType else None
def _attname(self,k): return getattr(k,'__name__',str(k))
def __repr__(self):
r = [f'({self._attname(k)},{self._attname(l)}) -> {getattr(v, "__name__", v.__class__.__name__)}'
for k in self.funcs.d for l,v in self.funcs[k].d.items()]
return '\n'.join(r)
def __call__(self, *args, **kwargs):
ts = L(args).map(type)[:2]
f = self[tuple(ts)]
if not f: return args[0]
if self.inst is not None: f = types.MethodType(f, self.inst)
return f(*args, **kwargs)
def __get__(self, inst, owner):
self.inst = inst
return self
def __getitem__(self, k):
"Find first matching type that is a super-class of `k`"
k = L(k if isinstance(k, tuple) else (k,))
while len(k)<2: k.append(object)
r = self.funcs.all_matches(k[0])
if len(r)==0: return None
for t in r:
o = t[k[1]]
if o is not None: return o
return None
def f_col(x:typing.Collection): return x
def f_nin(x:numbers.Integral)->int: return x+1
def f_ni2(x:int): return x
def f_bll(x:(bool,list)): return x
def f_num(x:numbers.Number): return x
t = TypeDispatch(f_nin,f_ni2,f_num,f_bll)
t.add(f_ni2) #Should work even if we add the same function twice.
test_eq(t[int], f_ni2)
test_eq(t[np.int32], f_nin)
test_eq(t[str], None)
test_eq(t[float], f_num)
test_eq(t[bool], f_bll)
test_eq(t[list], f_bll)
t.add(f_col)
test_eq(t[str], f_col)
test_eq(t[np.int32], f_nin)
o = np.int32(1)
test_eq(t(o), 2)
test_eq(t.returns(o), int)
assert t.first() is not None
t
def m_nin(self, x:(str,numbers.Integral)): return str(x)+'1'
def m_bll(self, x:bool): self.foo='a'
def m_num(self, x:numbers.Number): return x
t = TypeDispatch(m_nin,m_num,m_bll)
class A: f = t
a = A()
test_eq(a.f(1), '11')
test_eq(a.f(1.), 1.)
test_is(a.f.inst, a)
a.f(False)
test_eq(a.foo, 'a')
def f1(x:numbers.Integral, y): return x+1
def f2(x:int, y:float): return x+y
t = TypeDispatch(f1,f2)
test_eq(t[int], f1)
test_eq(t[int,int], f1)
test_eq(t[int,float], f2)
test_eq(t[float,float], None)
test_eq(t[np.int32,float], f1)
test_eq(t(3,2.0), 5)
test_eq(t(3,2), 4)
test_eq(t('a'), 'a')
t
```
## typedispatch Decorator
```
#export
class DispatchReg:
"A global registry for `TypeDispatch` objects keyed by function name"
def __init__(self): self.d = defaultdict(TypeDispatch)
def __call__(self, f):
nm = f'{f.__qualname__}'
self.d[nm].add(f)
return self.d[nm]
typedispatch = DispatchReg()
@typedispatch
def f_td_test(x, y): return f'{x}{y}'
@typedispatch
def f_td_test(x:numbers.Integral, y): return x+1
@typedispatch
def f_td_test(x:int, y:float): return x+y
test_eq(f_td_test(3,2.0), 5)
test_eq(f_td_test(3,2), 4)
test_eq(f_td_test('a','b'), 'ab')
```
## Export -
```
#hide
from local.notebook.export import notebook2script
notebook2script(all_fs=True)
```
|
github_jupyter
|
#default_exp core.dispatch
#export
from local.core.imports import *
from local.core.foundation import *
from local.core.utils import *
from local.test import *
from local.notebook.showdoc import *
#exports
def type_hints(f):
"Same as `typing.get_type_hints` but returns `{}` if not allowed type"
return typing.get_type_hints(f) if isinstance(f, typing._allowed_types) else {}
#export
def anno_ret(func):
"Get the return annotation of `func`"
if not func: return None
ann = type_hints(func)
if not ann: return None
return ann.get('return')
#hide
def f(x) -> float: return x
test_eq(anno_ret(f), float)
def f(x) -> typing.Tuple[float,float]: return x
test_eq(anno_ret(f), typing.Tuple[float,float])
def f(x) -> None: return x
test_eq(anno_ret(f), NoneType)
def f(x): return x
test_eq(anno_ret(f), None)
test_eq(anno_ret(None), None)
#export
cmp_instance = functools.cmp_to_key(lambda a,b: 0 if a==b else 1 if issubclass(a,b) else -1)
td = {int:1, numbers.Number:2, numbers.Integral:3}
test_eq(sorted(td, key=cmp_instance), [numbers.Number, numbers.Integral, int])
#export
def _p2_anno(f):
"Get the 1st 2 annotations of `f`, defaulting to `object`"
hints = type_hints(f)
ann = [o for n,o in hints.items() if n!='return']
while len(ann)<2: ann.append(object)
return ann[:2]
def _f(a): pass
test_eq(_p2_anno(_f), (object,object))
def _f(a, b): pass
test_eq(_p2_anno(_f), (object,object))
def _f(a:None, b)->str: pass
test_eq(_p2_anno(_f), (NoneType,object))
def _f(a:str, b)->float: pass
test_eq(_p2_anno(_f), (str,object))
def _f(a:None, b:str)->float: pass
test_eq(_p2_anno(_f), (NoneType,str))
def _f(a:int, b:int)->float: pass
test_eq(_p2_anno(_f), (int,int))
def _f(self, a:int, b:int): pass
test_eq(_p2_anno(_f), (int,int))
def _f(a:int, b:str)->float: pass
test_eq(_p2_anno(_f), (int,str))
test_eq(_p2_anno(attrgetter('foo')), (object,object))
#export
class _TypeDict:
def __init__(self): self.d,self.cache = {},{}
def _reset(self):
self.d = {k:self.d[k] for k in sorted(self.d, key=cmp_instance, reverse=True)}
self.cache = {}
def add(self, t, f):
"Add type `t` and function `f`"
if not isinstance(t,tuple): t=(t,)
for t_ in t: self.d[t_] = f
self._reset()
def all_matches(self, k):
"Find first matching type that is a super-class of `k`"
if k not in self.cache:
types = [f for f in self.d if k==f or (isinstance(k,type) and issubclass(k,f))]
self.cache[k] = [self.d[o] for o in types]
return self.cache[k]
def __getitem__(self, k):
"Find first matching type that is a super-class of `k`"
res = self.all_matches(k)
return res[0] if len(res) else None
def __repr__(self): return self.d.__repr__()
def first(self): return first(self.d.values())
#export
class TypeDispatch:
"Dictionary-like object; `__getitem__` matches keys of types using `issubclass`"
def __init__(self, *funcs):
self.funcs = _TypeDict()
for o in funcs: self.add(o)
self.inst = None
def add(self, f):
"Add type `t` and function `f`"
a0,a1 = _p2_anno(f)
t = self.funcs.d.get(a0)
if t is None:
t = _TypeDict()
self.funcs.add(a0, t)
t.add(a1, f)
def first(self): return self.funcs.first().first()
def returns(self, x): return anno_ret(self[type(x)])
def returns_none(self, x):
r = anno_ret(self[type(x)])
return r if r == NoneType else None
def _attname(self,k): return getattr(k,'__name__',str(k))
def __repr__(self):
r = [f'({self._attname(k)},{self._attname(l)}) -> {getattr(v, "__name__", v.__class__.__name__)}'
for k in self.funcs.d for l,v in self.funcs[k].d.items()]
return '\n'.join(r)
def __call__(self, *args, **kwargs):
ts = L(args).map(type)[:2]
f = self[tuple(ts)]
if not f: return args[0]
if self.inst is not None: f = types.MethodType(f, self.inst)
return f(*args, **kwargs)
def __get__(self, inst, owner):
self.inst = inst
return self
def __getitem__(self, k):
"Find first matching type that is a super-class of `k`"
k = L(k if isinstance(k, tuple) else (k,))
while len(k)<2: k.append(object)
r = self.funcs.all_matches(k[0])
if len(r)==0: return None
for t in r:
o = t[k[1]]
if o is not None: return o
return None
def f_col(x:typing.Collection): return x
def f_nin(x:numbers.Integral)->int: return x+1
def f_ni2(x:int): return x
def f_bll(x:(bool,list)): return x
def f_num(x:numbers.Number): return x
t = TypeDispatch(f_nin,f_ni2,f_num,f_bll)
t.add(f_ni2) #Should work even if we add the same function twice.
test_eq(t[int], f_ni2)
test_eq(t[np.int32], f_nin)
test_eq(t[str], None)
test_eq(t[float], f_num)
test_eq(t[bool], f_bll)
test_eq(t[list], f_bll)
t.add(f_col)
test_eq(t[str], f_col)
test_eq(t[np.int32], f_nin)
o = np.int32(1)
test_eq(t(o), 2)
test_eq(t.returns(o), int)
assert t.first() is not None
t
def m_nin(self, x:(str,numbers.Integral)): return str(x)+'1'
def m_bll(self, x:bool): self.foo='a'
def m_num(self, x:numbers.Number): return x
t = TypeDispatch(m_nin,m_num,m_bll)
class A: f = t
a = A()
test_eq(a.f(1), '11')
test_eq(a.f(1.), 1.)
test_is(a.f.inst, a)
a.f(False)
test_eq(a.foo, 'a')
def f1(x:numbers.Integral, y): return x+1
def f2(x:int, y:float): return x+y
t = TypeDispatch(f1,f2)
test_eq(t[int], f1)
test_eq(t[int,int], f1)
test_eq(t[int,float], f2)
test_eq(t[float,float], None)
test_eq(t[np.int32,float], f1)
test_eq(t(3,2.0), 5)
test_eq(t(3,2), 4)
test_eq(t('a'), 'a')
t
#export
class DispatchReg:
"A global registry for `TypeDispatch` objects keyed by function name"
def __init__(self): self.d = defaultdict(TypeDispatch)
def __call__(self, f):
nm = f'{f.__qualname__}'
self.d[nm].add(f)
return self.d[nm]
typedispatch = DispatchReg()
@typedispatch
def f_td_test(x, y): return f'{x}{y}'
@typedispatch
def f_td_test(x:numbers.Integral, y): return x+1
@typedispatch
def f_td_test(x:int, y:float): return x+y
test_eq(f_td_test(3,2.0), 5)
test_eq(f_td_test(3,2), 4)
test_eq(f_td_test('a','b'), 'ab')
#hide
from local.notebook.export import notebook2script
notebook2script(all_fs=True)
| 0.661376 | 0.727346 |
```
import numpy as np
class KMeans:
def __init__(self, n_clusters=4):
self.K = n_clusters
def fit(self, X):
self.centroids = X[np.random.choice(len(X), self.K, replace=False)]
self.intial_centroids = self.centroids
self.prev_label, self.labels = None, np.zeros(len(X))
while not np.all(self.labels == self.prev_label) :
self.prev_label = self.labels
self.labels = self.predict(X)
self.update_centroid(X)
return self
def predict(self, X):
return np.apply_along_axis(self.compute_label, 1, X)
def compute_label(self, x):
return np.argmin(np.sqrt(np.sum((self.centroids - x)**2, axis=1)))
def update_centroid(self, X):
self.centroids = np.array([np.mean(X[self.labels == k], axis=0) for k in range(self.K)])
## importing packages
import pandas as pd
import numpy as np
import matplotlib.pylab as plt
## loading data
three_globs = pd.read_csv('/Users/tjmask/Desktop/Semester2/DataMining/HW1/hw1/Homework_1/Datasets/three_globs.csv')
three_globs_df = three_globs
three_globs = np.array(three_globs)
three_globs_df['cluster'] = 0
m1 = np.where(three_globs_df.index=='GLOB_1')[0]
m2 = np.where(three_globs_df.index=='GLOB_2')[0]
m3 = np.where(three_globs_df.index=='GLOB_3')[0]
three_globs_df['cluster'][m1]=1
three_globs_df['cluster'][m2]=2
three_globs_df['cluster'][m3]=3
a = KMeans(n_clusters=3).fit(three_globs)
b = np.array(three_globs_df['cluster'])
b
plt.scatter(three_globs[:, 0], three_globs[:, 1], marker='.', c = b)
plt.scatter(a.centroids[:, 0], a.centroids[:,1], c='r')
plt.scatter(a.intial_centroids[:, 0], a.intial_centroids[:,1], c='k')
plt.show()
plt.figure(figsize=(12,10))
plt.title("Initial centers in black, final centers in red")
plt.scatter(X[:, 0], X[:, 1], marker='.', c=y)
plt.scatter(kmeans.centroids[:, 0], kmeans.centroids[:,1], c='r')
plt.scatter(kmeans.intial_centroids[:, 0], kmeans.intial_centroids[:,1], c='k')
plt.show()
print(__doc__)
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from sklearn import datasets
np.random.seed(5)
iris = datasets.load_iris()
X = iris.data
y = iris.target
estimators = [('k_means_iris_8', KMeans(n_clusters=8)),
('k_means_iris_3', KMeans(n_clusters=3)),
('k_means_iris_4', KMeans(n_clusters=4))]
fignum = 1
titles = ['8 clusters', '3 clusters', '4 clusters']
for name, est in estimators:
fig = plt.figure(fignum, figsize=(6, 5))
ax = Axes3D(fig, rect=[0, 0, .95, 1], elev=48, azim=134)
est.fit(X)
labels = est.labels
ax.scatter(X[:, 3], X[:, 0], X[:, 2],
c=labels.astype(np.float), edgecolor='k')
ax.w_xaxis.set_ticklabels([])
ax.w_yaxis.set_ticklabels([])
ax.w_zaxis.set_ticklabels([])
ax.set_xlabel('Petal width')
ax.set_ylabel('Sepal length')
ax.set_zlabel('Petal length')
ax.set_title(titles[fignum - 1])
ax.dist = 12
fignum = fignum + 1
# Plot the ground truth
fig = plt.figure(fignum, figsize=(6, 5))
ax = Axes3D(fig, rect=[0, 0, .95, 1], elev=48, azim=134)
for name, label in [('Setosa', 0),
('Versicolour', 1),
('Virginica', 2)]:
ax.text3D(X[y == label, 3].mean(),
X[y == label, 0].mean(),
X[y == label, 2].mean() + 2, name,
horizontalalignment='center',
bbox=dict(alpha=.2, edgecolor='w', facecolor='w'))
# Reorder the labels to have colors matching the cluster results
y = np.choose(y, [1, 2, 0]).astype(np.float)
ax.scatter(X[:, 3], X[:, 0], X[:, 2], c=y, edgecolor='k')
ax.w_xaxis.set_ticklabels([])
ax.w_yaxis.set_ticklabels([])
ax.w_zaxis.set_ticklabels([])
ax.set_xlabel('Petal width')
ax.set_ylabel('Sepal length')
ax.set_zlabel('Petal length')
ax.set_title('Ground Truth')
ax.dist = 12
fig.show()
from sklearn.datasets import make_blobs
X, y = make_blobs(centers=4, n_samples=1000)
fig = plt.figure(figsize=(8,6))
plt.scatter(X[:,0], X[:,1], c=y)
plt.title("Dataset with 4 clusters")
plt.xlabel("First feature")
plt.ylabel("Second feature")
plt.show()
kmeans = KMeans(n_clusters=4).fit(X)
plt.figure(figsize=(12,10))
plt.title("Initial centers in black, final centers in red")
plt.scatter(X[:, 0], X[:, 1], marker='.', c=y)
plt.scatter(kmeans.centroids[:, 0], kmeans.centroids[:,1], c='r')
plt.scatter(kmeans.intial_centroids[:, 0], kmeans.intial_centroids[:,1], c='k')
plt.show()
from numpy import *
import operator
def createDataSet():
group = array([[1.0,1.1],[1.0,1.0],[0,0],[0,0.1]])
labels = ['A','A','B','B']
return group, labels
def classify0(inX, dataSet, labels, k):
dataSetSize = dataSet.shape[0]
diffMat = tile(inX, (dataSetSize,1)) - dataSet ##matrix differences
sqDiffMat = diffMat**2 ##square the differences
sqDistances = sqDiffMat.sum(axis=1)
sortedDistIndicies = sqDistances.argsort() ##sort
classCount={}
for i in range(k): ##sort dictionary,label is the key,value will be K-means
voteIlabel = labels[sortedDistIndicies[i]]
classCount[voteIlabel] = classCount.get(voteIlabel,0) + 1
sortedClassCount = sorted(classCount.items(), key=operator.itemgetter(1), reverse=True)
print("dictionary",sortedClassCount)
return sortedClassCount[0][0]
group,labels=createDataSet()
result=classify0([0.5,0.5], group, labels, 3)
print(result)
```
|
github_jupyter
|
import numpy as np
class KMeans:
def __init__(self, n_clusters=4):
self.K = n_clusters
def fit(self, X):
self.centroids = X[np.random.choice(len(X), self.K, replace=False)]
self.intial_centroids = self.centroids
self.prev_label, self.labels = None, np.zeros(len(X))
while not np.all(self.labels == self.prev_label) :
self.prev_label = self.labels
self.labels = self.predict(X)
self.update_centroid(X)
return self
def predict(self, X):
return np.apply_along_axis(self.compute_label, 1, X)
def compute_label(self, x):
return np.argmin(np.sqrt(np.sum((self.centroids - x)**2, axis=1)))
def update_centroid(self, X):
self.centroids = np.array([np.mean(X[self.labels == k], axis=0) for k in range(self.K)])
## importing packages
import pandas as pd
import numpy as np
import matplotlib.pylab as plt
## loading data
three_globs = pd.read_csv('/Users/tjmask/Desktop/Semester2/DataMining/HW1/hw1/Homework_1/Datasets/three_globs.csv')
three_globs_df = three_globs
three_globs = np.array(three_globs)
three_globs_df['cluster'] = 0
m1 = np.where(three_globs_df.index=='GLOB_1')[0]
m2 = np.where(three_globs_df.index=='GLOB_2')[0]
m3 = np.where(three_globs_df.index=='GLOB_3')[0]
three_globs_df['cluster'][m1]=1
three_globs_df['cluster'][m2]=2
three_globs_df['cluster'][m3]=3
a = KMeans(n_clusters=3).fit(three_globs)
b = np.array(three_globs_df['cluster'])
b
plt.scatter(three_globs[:, 0], three_globs[:, 1], marker='.', c = b)
plt.scatter(a.centroids[:, 0], a.centroids[:,1], c='r')
plt.scatter(a.intial_centroids[:, 0], a.intial_centroids[:,1], c='k')
plt.show()
plt.figure(figsize=(12,10))
plt.title("Initial centers in black, final centers in red")
plt.scatter(X[:, 0], X[:, 1], marker='.', c=y)
plt.scatter(kmeans.centroids[:, 0], kmeans.centroids[:,1], c='r')
plt.scatter(kmeans.intial_centroids[:, 0], kmeans.intial_centroids[:,1], c='k')
plt.show()
print(__doc__)
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from sklearn import datasets
np.random.seed(5)
iris = datasets.load_iris()
X = iris.data
y = iris.target
estimators = [('k_means_iris_8', KMeans(n_clusters=8)),
('k_means_iris_3', KMeans(n_clusters=3)),
('k_means_iris_4', KMeans(n_clusters=4))]
fignum = 1
titles = ['8 clusters', '3 clusters', '4 clusters']
for name, est in estimators:
fig = plt.figure(fignum, figsize=(6, 5))
ax = Axes3D(fig, rect=[0, 0, .95, 1], elev=48, azim=134)
est.fit(X)
labels = est.labels
ax.scatter(X[:, 3], X[:, 0], X[:, 2],
c=labels.astype(np.float), edgecolor='k')
ax.w_xaxis.set_ticklabels([])
ax.w_yaxis.set_ticklabels([])
ax.w_zaxis.set_ticklabels([])
ax.set_xlabel('Petal width')
ax.set_ylabel('Sepal length')
ax.set_zlabel('Petal length')
ax.set_title(titles[fignum - 1])
ax.dist = 12
fignum = fignum + 1
# Plot the ground truth
fig = plt.figure(fignum, figsize=(6, 5))
ax = Axes3D(fig, rect=[0, 0, .95, 1], elev=48, azim=134)
for name, label in [('Setosa', 0),
('Versicolour', 1),
('Virginica', 2)]:
ax.text3D(X[y == label, 3].mean(),
X[y == label, 0].mean(),
X[y == label, 2].mean() + 2, name,
horizontalalignment='center',
bbox=dict(alpha=.2, edgecolor='w', facecolor='w'))
# Reorder the labels to have colors matching the cluster results
y = np.choose(y, [1, 2, 0]).astype(np.float)
ax.scatter(X[:, 3], X[:, 0], X[:, 2], c=y, edgecolor='k')
ax.w_xaxis.set_ticklabels([])
ax.w_yaxis.set_ticklabels([])
ax.w_zaxis.set_ticklabels([])
ax.set_xlabel('Petal width')
ax.set_ylabel('Sepal length')
ax.set_zlabel('Petal length')
ax.set_title('Ground Truth')
ax.dist = 12
fig.show()
from sklearn.datasets import make_blobs
X, y = make_blobs(centers=4, n_samples=1000)
fig = plt.figure(figsize=(8,6))
plt.scatter(X[:,0], X[:,1], c=y)
plt.title("Dataset with 4 clusters")
plt.xlabel("First feature")
plt.ylabel("Second feature")
plt.show()
kmeans = KMeans(n_clusters=4).fit(X)
plt.figure(figsize=(12,10))
plt.title("Initial centers in black, final centers in red")
plt.scatter(X[:, 0], X[:, 1], marker='.', c=y)
plt.scatter(kmeans.centroids[:, 0], kmeans.centroids[:,1], c='r')
plt.scatter(kmeans.intial_centroids[:, 0], kmeans.intial_centroids[:,1], c='k')
plt.show()
from numpy import *
import operator
def createDataSet():
group = array([[1.0,1.1],[1.0,1.0],[0,0],[0,0.1]])
labels = ['A','A','B','B']
return group, labels
def classify0(inX, dataSet, labels, k):
dataSetSize = dataSet.shape[0]
diffMat = tile(inX, (dataSetSize,1)) - dataSet ##matrix differences
sqDiffMat = diffMat**2 ##square the differences
sqDistances = sqDiffMat.sum(axis=1)
sortedDistIndicies = sqDistances.argsort() ##sort
classCount={}
for i in range(k): ##sort dictionary,label is the key,value will be K-means
voteIlabel = labels[sortedDistIndicies[i]]
classCount[voteIlabel] = classCount.get(voteIlabel,0) + 1
sortedClassCount = sorted(classCount.items(), key=operator.itemgetter(1), reverse=True)
print("dictionary",sortedClassCount)
return sortedClassCount[0][0]
group,labels=createDataSet()
result=classify0([0.5,0.5], group, labels, 3)
print(result)
| 0.632957 | 0.532547 |
# Stationarity
A time series is stationary when mean, variance, covariance do not change over time. Stationarity is important because many models require a stationary time series where time series in the real world is hardly ever stationary.
## Setup
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import statsmodels.api as sm
import statsmodels.formula.api as smf
import statsmodels.tsa.api as smt
import random
%matplotlib inline
```
## Generate a stationary data set
The following code will generate a stationary by creating what is referred to as a random walk.
```
stationary_x = [x for x in range(100)]
#stationary_y = [0.05*np.random.normal() for j in range(len(stationary_x))]
stationary_y = np.random.normal(0, 1, 100)
```
## Mean doesn't change over time
```
x, y = [], []
for i in range(1,60, 2):
x += [i, i+1]
y += [i, i - 2]
fig, axs = plt.subplots(1,2)
fig.set_size_inches(15, 4)
axs[0].set_title("Mean increasing over time")
axs[0].plot(x,y)
axs[1].set_title("Stationary Series")
axs[1].plot(stationary_x,stationary_y)
```
## Variance doesn't change over time
This property is known as homoscedasticity. From wikipedia:
```In statistics, a sequence or a vector of random variables is homoscedastic if all random variables in the sequence or vector have the same finite variance```
```
x, y = [], []
for i in range(1,60, 2):
x += [i, i+1]
y += [i, i - (i * 2)]
fig, axs = plt.subplots(1,2)
fig.set_size_inches(15, 4)
axs[0].set_title("Variance increasing over time")
axs[0].plot(x,y)
axs[1].set_title("Stationary Series")
axs[1].plot(stationary_x,stationary_y)
```
## Covariance doesn't change over time
```
x, y = [x for x in range(102)], []
for i in range(12):
y += [1] + [3 for x in range(12-i)] + [1]
fig, axs = plt.subplots(1,2)
fig.set_size_inches(15, 4)
axs[0].set_title("Covariance changing over time")
axs[0].plot(x,y)
axs[1].set_title("Stationary Series")
axs[1].plot(stationary_x,stationary_y)
```
## Checking for stationarity
### Augmented Dickey Fuller test
There will be a window and maxlag of 12 because this is seasonal monthly data making 12(or 1 year) an appropriate length. If this was weekly data, 52 might be more appropriate. It depends on the sample size and any seasonality in the data. This test gives you a probability not a certainty.
You may find that domain knowledge of the data needs to be known in order to select appropriate parameters. Alist of all possibilites for the ADF test are [here](http://www.statsmodels.org/dev/generated/statsmodels.tsa.stattools.adfuller.html).
```
from helpers.time_series import test_stationarity
air = pd.read_csv('data/international-airline-passengers.csv', header=0, index_col=0, parse_dates=[0])
#Perform Augmented Dickey-Fuller test:
test_stationarity(air['n_pass_thousands'], plot=True, maxlag=12, window=12)
```
Even without looking at the numbers, you can see that the mean is constantly changing as well as the variance. A breakdown of the numbers here:
* Test Statistic : The more negative this number, the more likely we are to to conclude this data as non-stationary.
* p-value : A p-value of <= 0.05 would indicate a stationary set of data
* Critical Values: The test statistic needs to be smaller then the %1 critical value. If it is, you can say that the test has a 99% confidence interval that the data is stationary.
#### KPSS test
It's good to start with the ADF and use the KPSS test as a confirmation that the data is stationary by using the KPSS test.
Coming soon....http://www.statsmodels.org/dev/generated/statsmodels.tsa.stattools.kpss.html
### Transforming to stationary
#### Log Transformation - good for removing variance
```
ts_log = np.log(air['n_pass_thousands'])
test_stationarity(ts_log, plot=True, maxlag=12, window=12)
```
In this case the rolling mean is still trending upwords.
#### First Difference
```
air['first_difference'] = air['n_pass_thousands'] - air['n_pass_thousands'].shift(1)
#dropna because of the shift
test_stationarity(air['first_difference'].dropna(), plot=True, window=12, maxlag=12)
```
It appears the rolling mean has stabilized around 0 but but the variance is increasing as time moves forward.
#### Seasonal Difference
There is more information on seasonality, but taking the difference of the seasonal periods could help make the data stationary. In the case of the airline data, there is a monthly seasonality.
```
air['seasonal_difference'] = air['n_pass_thousands'] - air['n_pass_thousands'].shift(12)
#dropna because of the shift
test_stationarity(air['seasonal_difference'].dropna(), window=12, plot=True, maxlag=12)
```
This still didn't help make the data stationary. We could try to take the seasonal difference of the first difference.
```
air['seasonal_first_difference'] = air['first_difference'] - air['first_difference'].shift(12)
#dropna because of the shift
test_stationarity(air['seasonal_first_difference'].dropna(), window=12, plot=True, autolag='t-stat', maxlag=12)
```
This looks stationary because the p-value is < 0.05 and the test statistic is < 1% critical value. We are 99% certain this is stationary.
#### Smoothing with log and moving average
```
ts_log = np.log(air['n_pass_thousands'])
moving_avg = ts_log.rolling(window=12).mean() #use 12 for seasonality
#difference
ts_log_moving_avg_diff = ts_log - moving_avg
ts_log_moving_avg_diff.dropna(inplace=True)
test_stationarity(ts_log_moving_avg_diff, window=12, plot=True, maxlag=12)
```
#### Exponentionaly weighted moving average
```
expwighted_avg = ts_log.ewm(halflife=12).mean()
ts_log_ewma_diff = ts_log - expwighted_avg
test_stationarity(ts_log_ewma_diff, plot=True, window=12, maxlag=12)
```
#### Decomposition
We go over decomposition and how to use is in another [notebook]('Decomposition.ipynb').
#### loess method (locally wieghted scatterplot smoothing)
```
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import statsmodels.api as sm
import statsmodels.formula.api as smf
import statsmodels.tsa.api as smt
import random
%matplotlib inline
stationary_x = [x for x in range(100)]
#stationary_y = [0.05*np.random.normal() for j in range(len(stationary_x))]
stationary_y = np.random.normal(0, 1, 100)
x, y = [], []
for i in range(1,60, 2):
x += [i, i+1]
y += [i, i - 2]
fig, axs = plt.subplots(1,2)
fig.set_size_inches(15, 4)
axs[0].set_title("Mean increasing over time")
axs[0].plot(x,y)
axs[1].set_title("Stationary Series")
axs[1].plot(stationary_x,stationary_y)
## Covariance doesn't change over time
## Checking for stationarity
### Augmented Dickey Fuller test
There will be a window and maxlag of 12 because this is seasonal monthly data making 12(or 1 year) an appropriate length. If this was weekly data, 52 might be more appropriate. It depends on the sample size and any seasonality in the data. This test gives you a probability not a certainty.
You may find that domain knowledge of the data needs to be known in order to select appropriate parameters. Alist of all possibilites for the ADF test are [here](http://www.statsmodels.org/dev/generated/statsmodels.tsa.stattools.adfuller.html).
Even without looking at the numbers, you can see that the mean is constantly changing as well as the variance. A breakdown of the numbers here:
* Test Statistic : The more negative this number, the more likely we are to to conclude this data as non-stationary.
* p-value : A p-value of <= 0.05 would indicate a stationary set of data
* Critical Values: The test statistic needs to be smaller then the %1 critical value. If it is, you can say that the test has a 99% confidence interval that the data is stationary.
#### KPSS test
It's good to start with the ADF and use the KPSS test as a confirmation that the data is stationary by using the KPSS test.
Coming soon....http://www.statsmodels.org/dev/generated/statsmodels.tsa.stattools.kpss.html
### Transforming to stationary
#### Log Transformation - good for removing variance
In this case the rolling mean is still trending upwords.
#### First Difference
It appears the rolling mean has stabilized around 0 but but the variance is increasing as time moves forward.
#### Seasonal Difference
There is more information on seasonality, but taking the difference of the seasonal periods could help make the data stationary. In the case of the airline data, there is a monthly seasonality.
This still didn't help make the data stationary. We could try to take the seasonal difference of the first difference.
This looks stationary because the p-value is < 0.05 and the test statistic is < 1% critical value. We are 99% certain this is stationary.
#### Smoothing with log and moving average
#### Exponentionaly weighted moving average
#### Decomposition
We go over decomposition and how to use is in another [notebook]('Decomposition.ipynb').
#### loess method (locally wieghted scatterplot smoothing)
| 0.503662 | 0.918077 |
```
from utils.metrics.Bleu import Bleu
import numpy as np
x=[]
with open('my.txt' , 'r' , encoding='utf-8') as f:
for line in f:
x.append(line)
print(len(x))
y=[]
with open('test_file.txt' , 'r' , encoding='utf-8') as f:
for line in f:
y.append(line)
print(len(y))
a=Bleu('test_file.txt','my.txt')
print((y[2]))
print(get_bleu_parallel())
import os
from multiprocessing import Pool
import nltk
from nltk.translate.bleu_score import SmoothingFunction
from utils.metrics.Metrics import Metrics
class Bleu(Metrics):
def __init__(self, test_text='', real_text='', gram=3):
super().__init__()
self.name = 'Bleu'
self.test_data = test_text
self.real_data = real_text
self.gram = gram
self.sample_size = 500
self.reference = None
self.is_first = True
def get_name(self):
return self.name
def get_score(self, is_fast=True, ignore=False):
if ignore:
return 0
if self.is_first:
self.get_reference()
self.is_first = False
if is_fast:
return self.get_bleu_fast()
return self.get_bleu_parallel()
def get_reference(self):
if self.reference is None:
reference = list()
with open(self.real_data ,'r' ,encoding='utf-8') as real_data:
for text in real_data:
text = nltk.word_tokenize(text)
reference.append(text)
self.reference = reference
return reference
else:
return self.reference
def get_bleu(self):
ngram = self.gram
bleu = list()
reference = self.get_reference()
weight = tuple((1. / ngram for _ in range(ngram)))
with open(self.test_data , encoding='utf-8') as test_data:
for hypothesis in test_data:
hypothesis = nltk.word_tokenize(hypothesis)
bleu.append(nltk.translate.bleu_score.sentence_bleu(reference, hypothesis, weight,
smoothing_function=SmoothingFunction().method1))
return sum(bleu) / len(bleu)
def calc_bleu(self, reference, hypothesis, weight):
return nltk.translate.bleu_score.sentence_bleu(reference, hypothesis, weight,
smoothing_function=SmoothingFunction().method1)
def get_bleu_fast(self):
reference = self.get_reference()
# random.shuffle(reference)
reference = reference[0:self.sample_size]
return self.get_bleu_parallel(reference=reference)
def get_bleu_parallel(self, reference=None):
ngram = self.gram
if reference is None:
reference = self.get_reference()
weight = tuple((1. / ngram for _ in range(ngram)))
pool = Pool(os.cpu_count())
result = list()
with open(self.test_data,encoding='utf-8') as test_data:
for hypothesis in test_data:
hypothesis = nltk.word_tokenize(hypothesis)
result.append(pool.apply_async(self.calc_bleu, args=(reference, hypothesis, weight)))
score = 0.0
cnt = 0
for i in result:
score += i.get()
cnt += 1
pool.close()
pool.join()
return score / cnt
def get_bleu_parallel( test_data,reference=None):
ngram = 3
if reference is None:
reference = get_reference(real_data,reference=None)
weight = tuple((1. / ngram for _ in range(ngram)))
pool = Pool(os.cpu_count())
result = list()
with open(test_data,encoding='utf-8') as test_data:
for hypothesis in test_data:
hypothesis = nltk.word_tokenize(hypothesis)
result.append(pool.apply_async(calc_bleu, args=(reference, hypothesis, weight)))
score = 0.0
cnt = 0
for i in result:
score += i.get()
cnt += 1
pool.close()
pool.join()
return score / cnt
import os
from multiprocessing import Pool
import nltk
from nltk.translate.bleu_score import SmoothingFunction
a=get_bleu_parallel(test_data, reference=None)
def get_reference(real_data,reference=None):
if reference is None:
reference = list()
with open(real_data ,'r' ,encoding='utf-8') as real_data:
for text in real_data:
text = nltk.word_tokenize(text)
reference.append(text)
reference = reference
return reference
else:
return self.reference
real_data='oracle.txt'
test_data='generator.txt'
weight = tuple((1. / 4 for _ in range(4)))
print(weight)
reference = list()
with open(real_data ,'r' ,encoding='utf-8') as real_data:
for text in real_data:
reference.append(text)
hypothesis = list()
with open(test_data ,'r' ,encoding='utf-8') as test_data:
for text in test_data:
hypothesis.append(text)
print(hypothesis[1])
nltk.translate.bleu_score.sentence_bleu(reference[:100], hypothesis[9], weight)
result = list()
with open(test_data,encoding='utf-8') as test_data:
for hypothesis in test_data:
result.append(pool.apply_async(calc_bleu, args=(reference, hypothesis, weight)))
```
|
github_jupyter
|
from utils.metrics.Bleu import Bleu
import numpy as np
x=[]
with open('my.txt' , 'r' , encoding='utf-8') as f:
for line in f:
x.append(line)
print(len(x))
y=[]
with open('test_file.txt' , 'r' , encoding='utf-8') as f:
for line in f:
y.append(line)
print(len(y))
a=Bleu('test_file.txt','my.txt')
print((y[2]))
print(get_bleu_parallel())
import os
from multiprocessing import Pool
import nltk
from nltk.translate.bleu_score import SmoothingFunction
from utils.metrics.Metrics import Metrics
class Bleu(Metrics):
def __init__(self, test_text='', real_text='', gram=3):
super().__init__()
self.name = 'Bleu'
self.test_data = test_text
self.real_data = real_text
self.gram = gram
self.sample_size = 500
self.reference = None
self.is_first = True
def get_name(self):
return self.name
def get_score(self, is_fast=True, ignore=False):
if ignore:
return 0
if self.is_first:
self.get_reference()
self.is_first = False
if is_fast:
return self.get_bleu_fast()
return self.get_bleu_parallel()
def get_reference(self):
if self.reference is None:
reference = list()
with open(self.real_data ,'r' ,encoding='utf-8') as real_data:
for text in real_data:
text = nltk.word_tokenize(text)
reference.append(text)
self.reference = reference
return reference
else:
return self.reference
def get_bleu(self):
ngram = self.gram
bleu = list()
reference = self.get_reference()
weight = tuple((1. / ngram for _ in range(ngram)))
with open(self.test_data , encoding='utf-8') as test_data:
for hypothesis in test_data:
hypothesis = nltk.word_tokenize(hypothesis)
bleu.append(nltk.translate.bleu_score.sentence_bleu(reference, hypothesis, weight,
smoothing_function=SmoothingFunction().method1))
return sum(bleu) / len(bleu)
def calc_bleu(self, reference, hypothesis, weight):
return nltk.translate.bleu_score.sentence_bleu(reference, hypothesis, weight,
smoothing_function=SmoothingFunction().method1)
def get_bleu_fast(self):
reference = self.get_reference()
# random.shuffle(reference)
reference = reference[0:self.sample_size]
return self.get_bleu_parallel(reference=reference)
def get_bleu_parallel(self, reference=None):
ngram = self.gram
if reference is None:
reference = self.get_reference()
weight = tuple((1. / ngram for _ in range(ngram)))
pool = Pool(os.cpu_count())
result = list()
with open(self.test_data,encoding='utf-8') as test_data:
for hypothesis in test_data:
hypothesis = nltk.word_tokenize(hypothesis)
result.append(pool.apply_async(self.calc_bleu, args=(reference, hypothesis, weight)))
score = 0.0
cnt = 0
for i in result:
score += i.get()
cnt += 1
pool.close()
pool.join()
return score / cnt
def get_bleu_parallel( test_data,reference=None):
ngram = 3
if reference is None:
reference = get_reference(real_data,reference=None)
weight = tuple((1. / ngram for _ in range(ngram)))
pool = Pool(os.cpu_count())
result = list()
with open(test_data,encoding='utf-8') as test_data:
for hypothesis in test_data:
hypothesis = nltk.word_tokenize(hypothesis)
result.append(pool.apply_async(calc_bleu, args=(reference, hypothesis, weight)))
score = 0.0
cnt = 0
for i in result:
score += i.get()
cnt += 1
pool.close()
pool.join()
return score / cnt
import os
from multiprocessing import Pool
import nltk
from nltk.translate.bleu_score import SmoothingFunction
a=get_bleu_parallel(test_data, reference=None)
def get_reference(real_data,reference=None):
if reference is None:
reference = list()
with open(real_data ,'r' ,encoding='utf-8') as real_data:
for text in real_data:
text = nltk.word_tokenize(text)
reference.append(text)
reference = reference
return reference
else:
return self.reference
real_data='oracle.txt'
test_data='generator.txt'
weight = tuple((1. / 4 for _ in range(4)))
print(weight)
reference = list()
with open(real_data ,'r' ,encoding='utf-8') as real_data:
for text in real_data:
reference.append(text)
hypothesis = list()
with open(test_data ,'r' ,encoding='utf-8') as test_data:
for text in test_data:
hypothesis.append(text)
print(hypothesis[1])
nltk.translate.bleu_score.sentence_bleu(reference[:100], hypothesis[9], weight)
result = list()
with open(test_data,encoding='utf-8') as test_data:
for hypothesis in test_data:
result.append(pool.apply_async(calc_bleu, args=(reference, hypothesis, weight)))
| 0.303113 | 0.173009 |
```
%matplotlib inline
%config InlineBackend.figure_format = "retina"
from __future__ import print_function
from matplotlib import rcParams
rcParams["savefig.dpi"] = 100
rcParams["figure.dpi"] = 100
rcParams["font.size"] = 20
```
# Hyperparameter optimization
This notebook was made with the following version of george:
```
import george
george.__version__
```
In this tutorial, we’ll reproduce the analysis for Figure 5.6 in [Chapter 5 of Rasmussen & Williams (R&W)](http://www.gaussianprocess.org/gpml/chapters/RW5.pdf). The data are measurements of the atmospheric CO2 concentration made at Mauna Loa, Hawaii (Keeling & Whorf 2004). The dataset is said to be available online but I couldn’t seem to download it from the original source. Luckily the [statsmodels](http://statsmodels.sourceforge.net/) package [includes a copy](http://statsmodels.sourceforge.net/devel/datasets/generated/co2.html) that we can load as follows:
```
import numpy as np
import matplotlib.pyplot as pl
from statsmodels.datasets import co2
data = co2.load_pandas().data
t = 2000 + (np.array(data.index.to_julian_date()) - 2451545.0) / 365.25
y = np.array(data.co2)
m = np.isfinite(t) & np.isfinite(y) & (t < 1996)
t, y = t[m][::4], y[m][::4]
pl.plot(t, y, ".k")
pl.xlim(t.min(), t.max())
pl.xlabel("year")
pl.ylabel("CO$_2$ in ppm");
```
In this figure, you can see that there is periodic (or quasi-periodic) signal with a year-long period superimposed on a long term trend. We will follow R&W and model these effects non-parametrically using a complicated covariance function. The covariance function that we’ll use is:
$$k(r) = k_1(r) + k_2(r) + k_3(r) + k_4(r)$$
where
$$
\begin{eqnarray}
k_1(r) &=& \theta_1^2 \, \exp \left(-\frac{r^2}{2\,\theta_2} \right) \\
k_2(r) &=& \theta_3^2 \, \exp \left(-\frac{r^2}{2\,\theta_4}
-\theta_5\,\sin^2\left(
\frac{\pi\,r}{\theta_6}\right)
\right) \\
k_3(r) &=& \theta_7^2 \, \left [ 1 + \frac{r^2}{2\,\theta_8\,\theta_9}
\right ]^{-\theta_8} \\
k_4(r) &=& \theta_{10}^2 \, \exp \left(-\frac{r^2}{2\,\theta_{11}} \right)
+ \theta_{12}^2\,\delta_{ij}
\end{eqnarray}
$$
We can implement this kernel in George as follows (we'll use the R&W results
as the hyperparameters for now):
```
from george import kernels
k1 = 66**2 * kernels.ExpSquaredKernel(metric=67**2)
k2 = 2.4**2 * kernels.ExpSquaredKernel(90**2) * kernels.ExpSine2Kernel(gamma=2/1.3**2, log_period=0.0)
k3 = 0.66**2 * kernels.RationalQuadraticKernel(log_alpha=np.log(0.78), metric=1.2**2)
k4 = 0.18**2 * kernels.ExpSquaredKernel(1.6**2)
kernel = k1 + k2 + k3 + k4
```
## Optimization
If we want to find the "best-fit" hyperparameters, we should *optimize* an objective function.
The two standard functions (as described in Chapter 5 of R&W) are the marginalized ln-likelihood and the cross validation likelihood.
George implements the former in the ``GP.lnlikelihood`` function and the gradient with respect to the hyperparameters in the ``GP.grad_lnlikelihood`` function:
```
import george
gp = george.GP(kernel, mean=np.mean(y), fit_mean=True,
white_noise=np.log(0.19**2), fit_white_noise=True)
gp.compute(t)
print(gp.log_likelihood(y))
print(gp.grad_log_likelihood(y))
```
We'll use a gradient based optimization routine from SciPy to fit this model as follows:
```
import scipy.optimize as op
# Define the objective function (negative log-likelihood in this case).
def nll(p):
gp.set_parameter_vector(p)
ll = gp.log_likelihood(y, quiet=True)
return -ll if np.isfinite(ll) else 1e25
# And the gradient of the objective function.
def grad_nll(p):
gp.set_parameter_vector(p)
return -gp.grad_log_likelihood(y, quiet=True)
# You need to compute the GP once before starting the optimization.
gp.compute(t)
# Print the initial ln-likelihood.
print(gp.log_likelihood(y))
# Run the optimization routine.
p0 = gp.get_parameter_vector()
results = op.minimize(nll, p0, jac=grad_nll, method="L-BFGS-B")
# Update the kernel and print the final log-likelihood.
gp.set_parameter_vector(results.x)
print(gp.log_likelihood(y))
```
**Warning:** *An optimization code something like this should work on most problems but the results can be very sensitive to your choice of initialization and algorithm. If the results are nonsense, try choosing a better initial guess or try a different value of the ``method`` parameter in ``op.minimize``.*
We can plot our prediction of the CO2 concentration into the future using our optimized Gaussian process model by running:
```
x = np.linspace(max(t), 2025, 2000)
mu, var = gp.predict(y, x, return_var=True)
std = np.sqrt(var)
pl.plot(t, y, ".k")
pl.fill_between(x, mu+std, mu-std, color="g", alpha=0.5)
pl.xlim(t.min(), 2025)
pl.xlabel("year")
pl.ylabel("CO$_2$ in ppm");
```
## Sampling & Marginalization
The prediction made in the previous section take into account uncertainties due to the fact that a Gaussian process is stochastic but it doesn’t take into account any uncertainties in the values of the hyperparameters. This won’t matter if the hyperparameters are very well constrained by the data but in this case, many of the parameters are actually poorly constrained. To take this effect into account, we can apply prior probability functions to the hyperparameters and marginalize using Markov chain Monte Carlo (MCMC). To do this, we’ll use the [emcee](http://dfm.io/emcee) package.
First, we define the probabilistic model:
```
def lnprob(p):
# Trivial uniform prior.
if np.any((-100 > p[1:]) + (p[1:] > 100)):
return -np.inf
# Update the kernel and compute the lnlikelihood.
gp.set_parameter_vector(p)
return gp.lnlikelihood(y, quiet=True)
```
In this function, we’ve applied a prior on every parameter that is uniform between -100 and 100 for every parameter. In real life, you should probably use something more intelligent but this will work for this problem. The quiet argument in the call to ``GP.lnlikelihood()`` means that that function will return ``-numpy.inf`` if the kernel is invalid or if there are any linear algebra errors (otherwise it would raise an exception).
Then, we run the sampler (this will probably take a while to run if you want to repeat this analysis):
```
import emcee
gp.compute(t)
# Set up the sampler.
nwalkers, ndim = 36, len(gp)
sampler = emcee.EnsembleSampler(nwalkers, ndim, lnprob)
# Initialize the walkers.
p0 = gp.get_parameter_vector() + 1e-4 * np.random.randn(nwalkers, ndim)
print("Running burn-in")
p0, _, _ = sampler.run_mcmc(p0, 200)
print("Running production chain")
sampler.run_mcmc(p0, 200);
```
After this run, you can plot 50 samples from the marginalized predictive probability distribution:
```
x = np.linspace(max(t), 2025, 250)
for i in range(50):
# Choose a random walker and step.
w = np.random.randint(sampler.chain.shape[0])
n = np.random.randint(sampler.chain.shape[1])
gp.set_parameter_vector(sampler.chain[w, n])
# Plot a single sample.
pl.plot(x, gp.sample_conditional(y, x), "g", alpha=0.1)
pl.plot(t, y, ".k")
pl.xlim(t.min(), 2025)
pl.xlabel("year")
pl.ylabel("CO$_2$ in ppm");
```
Comparing this to the same figure in the previous section, you’ll notice that the error bars on the prediction are now substantially larger than before. This is because we are now considering all the predictions that are consistent with the data, not just the “best” prediction. In general, even though it requires much more computation, it is more conservative (and honest) to take all these sources of uncertainty into account.
|
github_jupyter
|
%matplotlib inline
%config InlineBackend.figure_format = "retina"
from __future__ import print_function
from matplotlib import rcParams
rcParams["savefig.dpi"] = 100
rcParams["figure.dpi"] = 100
rcParams["font.size"] = 20
import george
george.__version__
import numpy as np
import matplotlib.pyplot as pl
from statsmodels.datasets import co2
data = co2.load_pandas().data
t = 2000 + (np.array(data.index.to_julian_date()) - 2451545.0) / 365.25
y = np.array(data.co2)
m = np.isfinite(t) & np.isfinite(y) & (t < 1996)
t, y = t[m][::4], y[m][::4]
pl.plot(t, y, ".k")
pl.xlim(t.min(), t.max())
pl.xlabel("year")
pl.ylabel("CO$_2$ in ppm");
from george import kernels
k1 = 66**2 * kernels.ExpSquaredKernel(metric=67**2)
k2 = 2.4**2 * kernels.ExpSquaredKernel(90**2) * kernels.ExpSine2Kernel(gamma=2/1.3**2, log_period=0.0)
k3 = 0.66**2 * kernels.RationalQuadraticKernel(log_alpha=np.log(0.78), metric=1.2**2)
k4 = 0.18**2 * kernels.ExpSquaredKernel(1.6**2)
kernel = k1 + k2 + k3 + k4
import george
gp = george.GP(kernel, mean=np.mean(y), fit_mean=True,
white_noise=np.log(0.19**2), fit_white_noise=True)
gp.compute(t)
print(gp.log_likelihood(y))
print(gp.grad_log_likelihood(y))
import scipy.optimize as op
# Define the objective function (negative log-likelihood in this case).
def nll(p):
gp.set_parameter_vector(p)
ll = gp.log_likelihood(y, quiet=True)
return -ll if np.isfinite(ll) else 1e25
# And the gradient of the objective function.
def grad_nll(p):
gp.set_parameter_vector(p)
return -gp.grad_log_likelihood(y, quiet=True)
# You need to compute the GP once before starting the optimization.
gp.compute(t)
# Print the initial ln-likelihood.
print(gp.log_likelihood(y))
# Run the optimization routine.
p0 = gp.get_parameter_vector()
results = op.minimize(nll, p0, jac=grad_nll, method="L-BFGS-B")
# Update the kernel and print the final log-likelihood.
gp.set_parameter_vector(results.x)
print(gp.log_likelihood(y))
x = np.linspace(max(t), 2025, 2000)
mu, var = gp.predict(y, x, return_var=True)
std = np.sqrt(var)
pl.plot(t, y, ".k")
pl.fill_between(x, mu+std, mu-std, color="g", alpha=0.5)
pl.xlim(t.min(), 2025)
pl.xlabel("year")
pl.ylabel("CO$_2$ in ppm");
def lnprob(p):
# Trivial uniform prior.
if np.any((-100 > p[1:]) + (p[1:] > 100)):
return -np.inf
# Update the kernel and compute the lnlikelihood.
gp.set_parameter_vector(p)
return gp.lnlikelihood(y, quiet=True)
import emcee
gp.compute(t)
# Set up the sampler.
nwalkers, ndim = 36, len(gp)
sampler = emcee.EnsembleSampler(nwalkers, ndim, lnprob)
# Initialize the walkers.
p0 = gp.get_parameter_vector() + 1e-4 * np.random.randn(nwalkers, ndim)
print("Running burn-in")
p0, _, _ = sampler.run_mcmc(p0, 200)
print("Running production chain")
sampler.run_mcmc(p0, 200);
x = np.linspace(max(t), 2025, 250)
for i in range(50):
# Choose a random walker and step.
w = np.random.randint(sampler.chain.shape[0])
n = np.random.randint(sampler.chain.shape[1])
gp.set_parameter_vector(sampler.chain[w, n])
# Plot a single sample.
pl.plot(x, gp.sample_conditional(y, x), "g", alpha=0.1)
pl.plot(t, y, ".k")
pl.xlim(t.min(), 2025)
pl.xlabel("year")
pl.ylabel("CO$_2$ in ppm");
| 0.740362 | 0.941815 |
```
!pip install tensorflow
!pip install emoji
from google.colab import drive
drive.mount('/content/drive')
root_path = 'drive/My Drive/text_to_emoji/'
import sys
sys.path.append('/content/drive/text_to_emoji/emojis_utils.py')
sys.path.insert(0,'/content/drive/My Drive/ColabNotebooks')
import emojis_utils
from google.colab import files
files.upload()
import test_utils
import numpy as np
from emojis_utils import *
import emoji
import matplotlib.pyplot as plt
from test_utils import *
%matplotlib inline
X_train, Y_train = read_csv('/content/drive/My Drive/text_to_emoji/data/train_emoji.csv')
X_test, Y_test = read_csv('/content/drive/My Drive/text_to_emoji/data/tesss.csv')
maxlen = len(max(X_train, key=len).split())
for indx in range(5,10):
print(X_train[indx], label_to_emoji(Y_train[indx]))
Y_oh_train = convert_to_one_hot(Y_train, C=5) #since the class have only 5 labels for now,
Y_oh_test = convert_to_one_hot(Y_test, C=5) #the one hot encoding is 1*5 that is (1,5)
#(indicating that there are 5 emojis to choose from).
idx = 20
print(f"Sentence '{X_train[50]}' has label index {Y_train[idx]}, which is emoji {label_to_emoji(Y_train[idx])}", )
print(f"Label index {Y_train[idx]} in one-hot encoding format is {Y_oh_train[idx]}")
word_to_index, index_to_word, word_to_vec_map = read_glove_vecs('/content/drive/My Drive/text_to_emoji/data/glove.6B.50d.txt')
def sentence_avg(sentence, word_to_vec_map):
any_word = list(word_to_vec_map.keys())[0]
words = sentence.lower().split() # Spliting sentence into list of lower case words
avg = np.zeros(word_to_vec_map[any_word].shape)
count = 0
for w in words: #avg the word vectors
if w in word_to_vec_map:
avg += word_to_vec_map[w]
count += 1
if count > 0:
avg = avg / count
return avg
```
𝑧(𝑖)=𝑊𝑎𝑣𝑔(𝑖)+𝑏
𝑎(𝑖)=𝑠𝑜𝑓𝑡𝑚𝑎𝑥(𝑧(𝑖))
cost(Li) = -sigma(Yoh*log(a))
```
def model(X, Y, word_to_vec_map, learning_rate=0.01, num_iterations = 500):
any_word = list(word_to_vec_map.keys())[0]
cost = 0
m = Y.shape[0] # number of training examples
n_y = len(np.unique(Y)) # number of classes
n_h = word_to_vec_map[any_word].shape[0] # dimensions of the GloVe vectors
W = np.random.randn(n_y,n_h)/np.sqrt(n_h)
b = np.zeros((n_y,))
Y_oh = convert_to_one_hot(Y, C = n_y) #Converting Y to Y_onehot with n_y classes
for t in range(num_iterations):
for i in range(m):
avg = sentence_avg(X[i], word_to_vec_map)
z = np.dot(W,avg) + b
a = softmax(z)
cost = -(np.sum(Y_oh * np.log(a)))
#calculating gradients
dz = a - Y_oh[i]
dW = np.dot(dz.reshape(n_y,1), avg.reshape(1,n_h))
db = dz
W = W - learning_rate * dW
b = b - learning_rate * db
if t % 100 == 0:
print('Epochs: ' + str(t) + '----cost = ' + str(cost))
pred = predict(X, Y, W, b, word_to_vec_map)
return pred, W, b
np.random.seed()
pred, W, b = model(X_train, Y_train, word_to_vec_map)
print(pred)
print("Training set:")
pred_train = predict(X_train, Y_train, W, b, word_to_vec_map)
print('Test set:')
pred_test = predict(X_test, Y_test, W, b, word_to_vec_map)
X_my_sentences = np.array(["i adore you", "i hate you", "funny lol", "lets play ", "food is ready", "not feeling happy"])
Y_my_labels = np.array([[0], [0], [2], [1], [4],[3]])
pred = predict(X_my_sentences, Y_my_labels , W, b, word_to_vec_map)
print_predictions(X_my_sentences, pred)
print(' '+ label_to_emoji(0) + ' ' + label_to_emoji(1) + ' ' + label_to_emoji(2) + ' ' + label_to_emoji(3) + ' ' + label_to_emoji(4))
print(pd.crosstab(Y_test, pred_test.reshape(56,), rownames=['Actual'], colnames=['Predicted'], margins=True))
plot_confusion_matrix(Y_test, pred_test)
```
|
github_jupyter
|
!pip install tensorflow
!pip install emoji
from google.colab import drive
drive.mount('/content/drive')
root_path = 'drive/My Drive/text_to_emoji/'
import sys
sys.path.append('/content/drive/text_to_emoji/emojis_utils.py')
sys.path.insert(0,'/content/drive/My Drive/ColabNotebooks')
import emojis_utils
from google.colab import files
files.upload()
import test_utils
import numpy as np
from emojis_utils import *
import emoji
import matplotlib.pyplot as plt
from test_utils import *
%matplotlib inline
X_train, Y_train = read_csv('/content/drive/My Drive/text_to_emoji/data/train_emoji.csv')
X_test, Y_test = read_csv('/content/drive/My Drive/text_to_emoji/data/tesss.csv')
maxlen = len(max(X_train, key=len).split())
for indx in range(5,10):
print(X_train[indx], label_to_emoji(Y_train[indx]))
Y_oh_train = convert_to_one_hot(Y_train, C=5) #since the class have only 5 labels for now,
Y_oh_test = convert_to_one_hot(Y_test, C=5) #the one hot encoding is 1*5 that is (1,5)
#(indicating that there are 5 emojis to choose from).
idx = 20
print(f"Sentence '{X_train[50]}' has label index {Y_train[idx]}, which is emoji {label_to_emoji(Y_train[idx])}", )
print(f"Label index {Y_train[idx]} in one-hot encoding format is {Y_oh_train[idx]}")
word_to_index, index_to_word, word_to_vec_map = read_glove_vecs('/content/drive/My Drive/text_to_emoji/data/glove.6B.50d.txt')
def sentence_avg(sentence, word_to_vec_map):
any_word = list(word_to_vec_map.keys())[0]
words = sentence.lower().split() # Spliting sentence into list of lower case words
avg = np.zeros(word_to_vec_map[any_word].shape)
count = 0
for w in words: #avg the word vectors
if w in word_to_vec_map:
avg += word_to_vec_map[w]
count += 1
if count > 0:
avg = avg / count
return avg
def model(X, Y, word_to_vec_map, learning_rate=0.01, num_iterations = 500):
any_word = list(word_to_vec_map.keys())[0]
cost = 0
m = Y.shape[0] # number of training examples
n_y = len(np.unique(Y)) # number of classes
n_h = word_to_vec_map[any_word].shape[0] # dimensions of the GloVe vectors
W = np.random.randn(n_y,n_h)/np.sqrt(n_h)
b = np.zeros((n_y,))
Y_oh = convert_to_one_hot(Y, C = n_y) #Converting Y to Y_onehot with n_y classes
for t in range(num_iterations):
for i in range(m):
avg = sentence_avg(X[i], word_to_vec_map)
z = np.dot(W,avg) + b
a = softmax(z)
cost = -(np.sum(Y_oh * np.log(a)))
#calculating gradients
dz = a - Y_oh[i]
dW = np.dot(dz.reshape(n_y,1), avg.reshape(1,n_h))
db = dz
W = W - learning_rate * dW
b = b - learning_rate * db
if t % 100 == 0:
print('Epochs: ' + str(t) + '----cost = ' + str(cost))
pred = predict(X, Y, W, b, word_to_vec_map)
return pred, W, b
np.random.seed()
pred, W, b = model(X_train, Y_train, word_to_vec_map)
print(pred)
print("Training set:")
pred_train = predict(X_train, Y_train, W, b, word_to_vec_map)
print('Test set:')
pred_test = predict(X_test, Y_test, W, b, word_to_vec_map)
X_my_sentences = np.array(["i adore you", "i hate you", "funny lol", "lets play ", "food is ready", "not feeling happy"])
Y_my_labels = np.array([[0], [0], [2], [1], [4],[3]])
pred = predict(X_my_sentences, Y_my_labels , W, b, word_to_vec_map)
print_predictions(X_my_sentences, pred)
print(' '+ label_to_emoji(0) + ' ' + label_to_emoji(1) + ' ' + label_to_emoji(2) + ' ' + label_to_emoji(3) + ' ' + label_to_emoji(4))
print(pd.crosstab(Y_test, pred_test.reshape(56,), rownames=['Actual'], colnames=['Predicted'], margins=True))
plot_confusion_matrix(Y_test, pred_test)
| 0.329284 | 0.390243 |
# <center style='font-weight:bold;font-family:serif'> Entrainement à la programmation en Python </center>
----
----
## I ) Mettre de l'ordre dans ses calculs.
### <span style='color:red'> Procédons étape par étape. </span>
Calculez et affichez le carré de $64$, le triple de $27$, le tiers de $81$, la différence de $1024$ et $512$, le quotient et le reste de la division euclidienne de $87$ par $6$. Stockez tous ces résultats dans une liste nommée ``calculs``.
Affichez la somme cummulée de tous ces résultats (cela devrait faire $4733.0$).
Copiez ``calculs`` dans une autre liste nommée ``calcs_copy``, et ordonnez cette nouvelle liste.
<span style='font-size : 9pt'>*Hint* : si vous ne savez pas comment copier une liste, faite ``help(list)`` et regardez les fonctions déjà implémentées sur les listes , ou bien regardez sur internet.</span>
Vous avez ordonné `calcs_copy`, mais `calculs` ne devrait pas être changée. Vérifiez cela en affichant `calculs`. Si ce n'est pas le cas, réexécutez la première cellule de code que vous avez fait dans cet exercice (celle où vous initialisez `calculs`).
Inversez l'ordre de cette liste, pour avoir désormais les résultats par ordre décroissant.
<span style='font-size:9pt'> ATTENTION : Si vous relancez la cellule de code plusieurs fois, vous risquez de renverser une liste que vous avez déjà renversé, donc la remettre dans son état initial. Veillez bien à ce que votre liste soit bien par ordre décroissant avant de passer à la suite. N'hésitez pas à l'afficher.</span>
<span style='font-size:9pt'> *Hint* : si vous ne savez pas comment renverser simplement une liste, regardez à nouveau dans `help(list)`.</span>
Dans cette liste ``calcs_copy``, remplacez chaque élément par sa conversion en chaine de caractère.
En utilisant la fonction <a href='https://www.w3schools.com/python/ref_string_join.asp'>*str*.join(...)</a> (cliquez sur le lien pour la documentation), affichez tous les éléments de cette liste par ordre décroissant, séparé par des "$>$".
### <span style='color:red'> Regroupons tout cela dans une fonction ! </span>
**Ecrivez une fonction `ordonner` qui prend en argument une liste de nombres (les résultats de calculs, par exemple), et calculera une copie de cette liste rangée dans l'ordre décroissant.**
La fonction prendra un deuxième argument, `display`, qui sera un **booléen**. Si ce booléen est à `True`, la fonction *ne renverra rien* mais **affichera** les éléments de la liste rangée, séparé par des "$>$". Si le booléen est à `False`, la fonction *n'affichera rien* mais devra **renvoyer** la liste rangée par ordre décroissant.
Essayez avec ```[ 15.3 , 6 , 100 , 55.7 , 23 ]```.
```
def ordonner(l : list, display : bool):
'''
Renvoie une copie rangée par ordre décroissant d'une liste de nombres.
Si display==True, ne renvoie rien mais affiche la liste avec des ">".
-------------
Paramètres :
l (list) : la liste de nombres
display (bool) : si True, affiche la liste ordonnée au lieu de la renvoyer.
'''
#A COMPLETER
pass #supprimez le pass quand vous completez
ordonner([ 15.3 , 6 , 100 , 55.7 , 23 ],False)
```
-----
-----
## II ) Listes en chaine, chaines en liste
### <span style='color:red'> Mon beau miroir </span>
Affichez l'envers d'une chaine de caractère (ex: *riorim uaeb nom* au lieu de *mon beau miroir*).
```
chaine = "mon beau miroir"
```
Même exercice, mais en utilisant, si ce n'était pas déjà le cas, la méthode ``join`` (cf. plus haut) permettant de joindre dans une `str` les éléments d'une liste.
<span style='font-size:9pt'> *Hint* : appliquer l'opérateur `list()` sur une chaine de caractère renvoie la liste de caractères individuels.</span>
<span style='font-size:9pt'> *Hint 2* : Comme vu précédemment, il est facile de renverser l'ordre d'une liste...</span>
### <span style='color:red'> Listes par compréhension </span>
Créez une liste `carre` contenant le carré des 100 premiers nombres entiers naturels (de 0 à 99).
La métode explicite pour construire une liste est ```l=[a,b,..]```, mais cela n'est réalisable que si on connait *explicitement* à l'avance la valeur des éléments de la liste et leur nombre.
On peut aussi vouloir construire une liste *itérativement*, en initialisant une liste vide puis en utilisant une boucle `for` et la méthode `append` dans celle ci. C'est surement ce que vous avez fait précédemment.
Mais ça n'est pas la seule façon de faire, ni la plus optimale. En réalité, on peut intégrer une boucle `for` directement dans la création de la liste. Cela s'appelle une *liste par compréhension*. Démonstration :
```
# Plutot que d'écrire :
l = []
l2 = []
for i in range(10):
l.append(i)
for i in range(4):
l2.append('a'*i)
print(l,l2)
# On peut faire, par compréhension :
l_comprehension = [ i for i in range(10) ]
l2_comprehension = [ 'a'*i for i in range(4) ] #C'EST BEAUCOUP PLUS RAPIDE A ECRIRE !!!!
print(l_comprehension,l2_comprehension)
```
En utilisant les listes par compréhension, créez une liste `carre_comp` contenant le carré des 100 premiers entiers naturels (de 0 à 99).
Ecrivez une fonction ```table_prod``` qui prend en argument deux listes et qui renvoie un tableau (une liste de listes) contenant le produit deux à deux des éléments des deux listes (i.e. `table_prod( [a,b] , [c,d] )` -> `[[ac,ad] , [bc,bd]]` ). Essayez de le faire en une seule ligne à l'intérieur de la fonction.
<span style='font-size:9pt'> *Hint* : On peut imbriquer les listes par compréhension, comme on imbriquerait des boucles FOR.</span>
```
def table_prod(l1 : list,l2 : list):
'''
Renvoie un tableau contenant le produit deux à deux des éléments de deux listes l1 et l2.
'''
#A COMPLETER
pass
table_prod([1,2],[3,4])
```
Ecrire une fonction `table_sum` identique à `table_prod` sauf qu'elle calcule la somme au lieu du produit des éléments (i.e. `table_prod( [a,b] , [c,d] )` -> `[[a+c,a+d] , [b+c,b+d]]` ).
```
def table_sum(l1 : list,l2 : list):
'''
Renvoie un tableau contenant la somme deux à deux des éléments de deux listes l1 et l2.
'''
#A COMPLETER
pass
table_sum([1,2],[3,4])
```
----
----
## III ) Création et utilisation des dictionnaires
### <span style='color:red'> Une base de données sur les étudiants </span>
Voici un tableau contenant le nom, le prénom, l'âge, le numéro étudiant, la section et l'année d'inscription de quelques étudiant-e-s.
| prenom | nom | age | numero | section | inscription |
|-|-|-|-|-|-|
| victor | hugo | 24 | 07.555.42 | L3 lettres modernes | 2018 |
| ada | lovelace | 26 | 06.555.64 | M2 informatique | 2018 |
| marcus | cicero | 28 | 06.555.21 | M2 lettres classiques | 2016 |
| marie | curie | 21 | 07.555.38 | M1 physique | 2018 |
| emilie | du chatelet | 20 | 06.555.42 | L1 physique | 2020 |
| charles | darwin | 22 | 07.555.32 |L2 biologie | 2020 |
L'objectif est de présenter ce tableau sous la forme d'une liste de dictionnaires, où chaque dictionnaire représente une étudiante ou un étudiant contenant ses informations. Les entêtes de colonne seront les clés. Chaque valeur sera une chaine de caractère.
Pour cela, considerez le texte suivant dans la chaine de caractère `str_base`. Elle contient le contenu de ce tableau.
```
str_base = "prenom | nom | age | numero | section | inscription\nvictor | hugo | 24 | 07.555.42 | L3 lettres modernes | 2018\nada | lovelace | 26 | 06.555.64 | M2 informatique | 2018\nmarcus | cicero | 28 | 06.555.21 | M2 lettres classiques | 2016\nmarie | curie | 21 | 07.555.38 | M1 physique | 2018\nemilie | du chatelet | 20 | 06.555.42 | L1 physique | 2020\ncharles | darwin | 22 | 07.555.32 | L2 biologie | 2020"
print(str_base)
```
Vous pouvez observer que la chaine de caractère est découpée en lignes grace à un symbole. Lequel ?
A l'aide de la fonction <a href='https://www.w3schools.com/python/ref_string_split.asp'>*str*.split(...)</a>, découpez la chaine en une liste de ses lignes et stockez cela dans `list_base`. Affichez la.
A l'aide de la méthode <a href='https://www.w3schools.com/python/ref_list_pop.asp'>*list*.pop(...)</a>, récuperez la *première ligne* du texte en la retirant de `list_base`, et stockez la dans une variable `str_headers` pour ne pas la perdre.
Encore à l'aide de la fonction `split`, découpez cette ligne pour obtenir une liste contenant les titres de chaque colone. Stockez ces entêtes dans une liste `headers`. Affichez la.
Vous remarquerez que certaines entêtes contiennent encore des espaces résiduels au début ou à la fin de leur chaine de caractère. En itérant sur la liste, remplacez chacune des chaines par sa version "propre", sans les espaces. Pour cela, utilisez la fonction <a href='https://www.w3schools.com/python/ref_string_strip.asp'>*str*.strip()</a>. Affichez à nouveau `headers` pour vérifier que le changement est effectué.
En vous inspirant de ce processus, créez une fonction `split_and_strip` qui prendra en argument une chaine de caractère `string` et un caractère de séparation `sep`. Cette fonction coupera votre chaine de caractère à chaque `sep` et renverra la liste de la découpe après avoir nettoyé chacun de ses éléments de ses espaces inutiles.
*Ex:*
```python
split_and_strip(string=' a ,b , c',sep=',')
>>> ['a','b','c']
```
```
def split_and_strip(string,sep):
pass
split_and_strip(string=' a ,b , c',sep=',')
```
Affichez cote à cote chaque catégorie d'information (obtenue dans `headers`) et sa valeur pour le premier étudiant (en utilisant `split_and_strip` sur la première ligne de `list_base`).
Vous devez obtenir quelque chose comme :
```
prenom : victor
nom : hugo
age : 24
...
```
Attention, ne sortez pas cette ligne de la liste (n'utilisez pas ```pop```)!
<span style='font-size:9pt'> *Hint* : A la *i*-ème catégorie dans `headers` est associée le *i*-ème élément dans la liste que vous obtiendrez avec `split_and_strip`... Utilisez une seule boucle pour parcourir en parallèle les 2 listes !</span>
Même question mais, plutôt que de les afficher directement, créer ces associations dans un dictionnaire que vous afficherez ensuite. Vous obtiendrez quelque chose comme
```
{'prenom':'victor','nom':'hugo','age':'24',...}
```
<span style='font-size:9pt'> *Hint* : Utilisez <span style='color:rgb(50,150,255)'>dict[key]=value</span>. </span>
En utilisant cette méthode, construisez une liste contenant un tel dictionnaire pour chaque étudiant. Stockez cette liste dans `etudiants`.
### <span style='color:red'> Etudions les étudiants </span>
Utilisez cette liste `etudiants` pour répondre aux questions suivantes. Ne calculez pas les réponses manuellement.
Combien y a t-il d'étudiant.e.s ?
Calculez l'âge moyen des étudiant-e-s. En utilisant <a href="https://docs.python.org/fr/3.7/library/functions.html?highlight=round#round">round() </a>, arrondissez à la 3e décimale.
Combien d'étudiant-e-s sont en licence ?
En utilisant l'opérateur booléen `in` sur les séquences (chaines de caractère ou listes), comptez le nombre d'étudiant-e-s en physique.
<span style='font-size:9pt'>*Hint*: Vous pouvez vérifiez la documentation des <a href="https://www.w3schools.com/python/python_operators.asp">operateurs python</a>, dans la section "membership operators".</span>
Quel pourcentage des d'étudiant-e-s se sont inscrit-e-s avant 2019 ?
Sachant que nous sommes en 2022, affichez le prénom et le nom de l'étudiant-e qui s'est inscrit-e avant ses 18ans ?
Parmi les étudiant-e-s inscrit-e-s en 2018, quel pourcentage a un numéro commençant par 07 ?
<a id='another_cell'></a>
**[Plus difficile]** Faisons un bond dans le temps. C'est une nouvelle année qui commence, et les données doivent être mise à jour.
Copiez la liste `etudiants` dans une liste `new_etudiants`. modifiez les dictionnaires dans cette liste de sorte que:
- chaque étudiant-e prenne 1 an d'âge.
- Les étudiant-e-s en L1 passent en L2, celleux en L2 en L3, L3 en M1, M1 en M2, M2 en PhD... sauf Charles Darwin, qui redouble sa L2.
- Marcus Cicero ne fera pas de PhD, retirez le de la liste `etudiants`.
Affichez la liste `new_etudiants`.
**[Plus difficile]** Vérifions que vous avez copié correctement `etudiants` et que vous ne l'avez pas modifié directement. Affichez la. Charles Darwin devrait toujours être dedans, et **Victor Hugo devrait avoir 24 ans**. Si ça n'est pas le cas, alors vous n'avez pas fait les choses correctement au début de la cellule précédente, et vous devrez relancer la cellule de code à la fin de la section [Une base de données sur les étudiants](#span-stylecolorred-une-base-de-donn%C3%A9es-sur-les-%C3%A9tudiants-span).
<span style='font-size:9pt'>*Hint* : Si Darwin est toujours dedans mais que Victor Hugo a 25ans au lieu de 24, c'est probablement que vous avez fait `new_etudiants=etudiants.copy()`. Rappelez vous que les listes sont mutables, mais les dictionnaires aussi ! `etudiants` ne contient pas directement les dictionnaires, mais des *pointeurs* vers ces dictionnaires dans la mémoire. En copiant simplement `etudiants` comme cela, vous ne copiez pas les dictionnaires, vous ne copiez que les pointeurs ! Au lieu de créer une *copie de la liste* des dictionnaires, il faudrait créer une *liste des copies* des dictionnaires.
</span>
```
print(etudiants)
```
----
----
## Conclusion
Voilà qui conclut ce notebook d'entrainement sur les bases du python ! Si vous avez utilisé les *hints* et astuces, n'hésitez pas à le recommencer depuis le début sans vous servir des *hints* cette fois-ci. Ensuite, recommencez en vous chronometrant une première fois en le faisant à un rythme normal. Puis refaite ce notebook en essayant de battre votre temps précédent.
Si vous n'avez pas bien compris certaines notions, comme les listes par compréhensions par exemple, entrainez vous et regardez sur internet si cela peut vous aider. En refaisant ce notebook, essayer de créer vos listes en utilisant des compréhensions au lieu de boucle FOR lorsque c'est possible.
|
github_jupyter
|
def ordonner(l : list, display : bool):
'''
Renvoie une copie rangée par ordre décroissant d'une liste de nombres.
Si display==True, ne renvoie rien mais affiche la liste avec des ">".
-------------
Paramètres :
l (list) : la liste de nombres
display (bool) : si True, affiche la liste ordonnée au lieu de la renvoyer.
'''
#A COMPLETER
pass #supprimez le pass quand vous completez
ordonner([ 15.3 , 6 , 100 , 55.7 , 23 ],False)
chaine = "mon beau miroir"
# Plutot que d'écrire :
l = []
l2 = []
for i in range(10):
l.append(i)
for i in range(4):
l2.append('a'*i)
print(l,l2)
# On peut faire, par compréhension :
l_comprehension = [ i for i in range(10) ]
l2_comprehension = [ 'a'*i for i in range(4) ] #C'EST BEAUCOUP PLUS RAPIDE A ECRIRE !!!!
print(l_comprehension,l2_comprehension)
def table_prod(l1 : list,l2 : list):
'''
Renvoie un tableau contenant le produit deux à deux des éléments de deux listes l1 et l2.
'''
#A COMPLETER
pass
table_prod([1,2],[3,4])
def table_sum(l1 : list,l2 : list):
'''
Renvoie un tableau contenant la somme deux à deux des éléments de deux listes l1 et l2.
'''
#A COMPLETER
pass
table_sum([1,2],[3,4])
str_base = "prenom | nom | age | numero | section | inscription\nvictor | hugo | 24 | 07.555.42 | L3 lettres modernes | 2018\nada | lovelace | 26 | 06.555.64 | M2 informatique | 2018\nmarcus | cicero | 28 | 06.555.21 | M2 lettres classiques | 2016\nmarie | curie | 21 | 07.555.38 | M1 physique | 2018\nemilie | du chatelet | 20 | 06.555.42 | L1 physique | 2020\ncharles | darwin | 22 | 07.555.32 | L2 biologie | 2020"
print(str_base)
split_and_strip(string=' a ,b , c',sep=',')
>>> ['a','b','c']
def split_and_strip(string,sep):
pass
split_and_strip(string=' a ,b , c',sep=',')
prenom : victor
nom : hugo
age : 24
...
{'prenom':'victor','nom':'hugo','age':'24',...}
print(etudiants)
| 0.158304 | 0.939858 |
```
import numpy as np
import random
from math import *
import time
import copy
import torch
from torch.autograd import Variable
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torch.optim.lr_scheduler import StepLR, MultiStepLR
torch.set_default_tensor_type('torch.DoubleTensor')
# defination of activation function
def activation(x):
return x * torch.sigmoid(x)
# build ResNet with one blocks
class Net(nn.Module):
def __init__(self,input_size,width):
super(Net,self).__init__()
self.layer_in = nn.Linear(input_size,width)
self.layer_1 = nn.Linear(width,width)
self.layer_2 = nn.Linear(width,width)
self.layer_out = nn.Linear(width,1)
def forward(self,x):
output = self.layer_in(x)
output = output + activation(self.layer_2(activation(self.layer_1(output)))) # residual block 1
output = self.layer_out(output)
return output
input_size = 1
width = 4
# exact solution
def u_ex(x):
return torch.sin(pi*x)
# f(x)
def f(x):
return pi**2 * torch.sin(pi*x)
grid_num = 200
x = torch.zeros(grid_num + 1, input_size)
for index in range(grid_num + 1):
x[index] = index * 1 / grid_num
net = Net(input_size,width)
def model(x):
return x * (x - 1.0) * net(x)
# loss function to DGM by auto differential
def loss_function(x):
h = 1 / grid_num
sum_0 = 0.0
sum_1 = 0.0
sum_2 = 0.0
sum_a = 0.0
sum_b = 0.0
for index in range(grid_num):
x_temp = x[index] + h / 2
x_temp.requires_grad = True
grad_x_temp = torch.autograd.grad(outputs = model(x_temp), inputs = x_temp, grad_outputs = torch.ones(model(x_temp).shape), create_graph = True)
grad_grad_x_temp = torch.autograd.grad(outputs = grad_x_temp[0], inputs = x_temp, grad_outputs = torch.ones(model(x_temp).shape), create_graph = True)
sum_1 += ((grad_grad_x_temp[0])[0] + f(x_temp)[0])**2
for index in range(1, grid_num):
x_temp = x[index]
x_temp.requires_grad = True
grad_x_temp = torch.autograd.grad(outputs = model(x_temp), inputs = x_temp, grad_outputs = torch.ones(model(x_temp).shape), create_graph = True)
grad_grad_x_temp = torch.autograd.grad(outputs = grad_x_temp[0], inputs = x_temp, grad_outputs = torch.ones(model(x_temp).shape), create_graph = True)
sum_2 += ((grad_grad_x_temp[0])[0] + f(x_temp)[0])**2
x_temp = x[0]
x_temp.requires_grad = True
grad_x_temp = torch.autograd.grad(outputs = model(x_temp), inputs = x_temp, grad_outputs = torch.ones(model(x_temp).shape), create_graph = True)
grad_grad_x_temp = torch.autograd.grad(outputs = grad_x_temp[0], inputs = x_temp, grad_outputs = torch.ones(model(x_temp).shape), create_graph = True)
sum_a = ((grad_grad_x_temp[0])[0] + f(x_temp)[0])**2
x_temp = x[grid_num]
x_temp.requires_grad = True
grad_x_temp = torch.autograd.grad(outputs = model(x_temp), inputs = x_temp, grad_outputs = torch.ones(model(x_temp).shape), create_graph = True)
grad_grad_x_temp = torch.autograd.grad(outputs = grad_x_temp[0], inputs = x_temp, grad_outputs = torch.ones(model(x_temp).shape), create_graph = True)
sum_b = ((grad_grad_x_temp[0])[0] + f(x_temp)[0])**2
sum_0 = h / 6 * (sum_a + 4 * sum_1 + 2 * sum_2 + sum_b)
return sum_0
def get_weights(net):
""" Extract parameters from net, and return a list of tensors"""
return [p.data for p in net.parameters()]
def set_weights(net, weights, directions=None, step=None):
"""
Overwrite the network's weights with a specified list of tensors
or change weights along directions with a step size.
"""
if directions is None:
# You cannot specify a step length without a direction.
for (p, w) in zip(net.parameters(), weights):
p.data.copy_(w.type(type(p.data)))
else:
assert step is not None, 'If a direction is specified then step must be specified as well'
if len(directions) == 2:
dx = directions[0]
dy = directions[1]
changes = [d0*step[0] + d1*step[1] for (d0, d1) in zip(dx, dy)]
else:
changes = [d*step for d in directions[0]]
for (p, w, d) in zip(net.parameters(), weights, changes):
p.data = w + torch.Tensor(d).type(type(w))
def set_states(net, states, directions=None, step=None):
"""
Overwrite the network's state_dict or change it along directions with a step size.
"""
if directions is None:
net.load_state_dict(states)
else:
assert step is not None, 'If direction is provided then the step must be specified as well'
if len(directions) == 2:
dx = directions[0]
dy = directions[1]
changes = [d0*step[0] + d1*step[1] for (d0, d1) in zip(dx, dy)]
else:
changes = [d*step for d in directions[0]]
new_states = copy.deepcopy(states)
assert (len(new_states) == len(changes))
for (k, v), d in zip(new_states.items(), changes):
d = torch.tensor(d)
v.add_(d.type(v.type()))
net.load_state_dict(new_states)
def get_random_weights(weights):
"""
Produce a random direction that is a list of random Gaussian tensors
with the same shape as the network's weights, so one direction entry per weight.
"""
return [torch.randn(w.size()) for w in weights]
def get_random_states(states):
"""
Produce a random direction that is a list of random Gaussian tensors
with the same shape as the network's state_dict(), so one direction entry
per weight, including BN's running_mean/var.
"""
return [torch.randn(w.size()) for k, w in states.items()]
def get_diff_weights(weights, weights2):
""" Produce a direction from 'weights' to 'weights2'."""
return [w2 - w for (w, w2) in zip(weights, weights2)]
def get_diff_states(states, states2):
""" Produce a direction from 'states' to 'states2'."""
return [v2 - v for (k, v), (k2, v2) in zip(states.items(), states2.items())]
def normalize_direction(direction, weights, norm='filter'):
"""
Rescale the direction so that it has similar norm as their corresponding
model in different levels.
Args:
direction: a variables of the random direction for one layer
weights: a variable of the original model for one layer
norm: normalization method, 'filter' | 'layer' | 'weight'
"""
if norm == 'filter':
# Rescale the filters (weights in group) in 'direction' so that each
# filter has the same norm as its corresponding filter in 'weights'.
for d, w in zip(direction, weights):
d.mul_(w.norm()/(d.norm() + 1e-10))
elif norm == 'layer':
# Rescale the layer variables in the direction so that each layer has
# the same norm as the layer variables in weights.
direction.mul_(weights.norm()/direction.norm())
elif norm == 'weight':
# Rescale the entries in the direction so that each entry has the same
# scale as the corresponding weight.
direction.mul_(weights)
elif norm == 'dfilter':
# Rescale the entries in the direction so that each filter direction
# has the unit norm.
for d in direction:
d.div_(d.norm() + 1e-10)
elif norm == 'dlayer':
# Rescale the entries in the direction so that each layer direction has
# the unit norm.
direction.div_(direction.norm())
def normalize_directions_for_weights(direction, weights, norm='filter', ignore='biasbn'):
"""
The normalization scales the direction entries according to the entries of weights.
"""
assert(len(direction) == len(weights))
for d, w in zip(direction, weights):
if d.dim() <= 1:
if ignore == 'biasbn':
d.fill_(0) # ignore directions for weights with 1 dimension
else:
d.copy_(w) # keep directions for weights/bias that are only 1 per node
else:
normalize_direction(d, w, norm)
def normalize_directions_for_states(direction, states, norm='filter', ignore='ignore'):
assert(len(direction) == len(states))
for d, (k, w) in zip(direction, states.items()):
if d.dim() <= 1:
if ignore == 'biasbn':
d.fill_(0) # ignore directions for weights with 1 dimension
else:
d.copy_(w) # keep directions for weights/bias that are only 1 per node
else:
normalize_direction(d, w, norm)
def ignore_biasbn(directions):
""" Set bias and bn parameters in directions to zero """
for d in directions:
if d.dim() <= 1:
d.fill_(0)
def create_random_direction(net, dir_type='weights', ignore='biasbn', norm='filter'):
"""
Setup a random (normalized) direction with the same dimension as
the weights or states.
Args:
net: the given trained model
dir_type: 'weights' or 'states', type of directions.
ignore: 'biasbn', ignore biases and BN parameters.
norm: direction normalization method, including
'filter" | 'layer' | 'weight' | 'dlayer' | 'dfilter'
Returns:
direction: a random direction with the same dimension as weights or states.
"""
# random direction
if dir_type == 'weights':
weights = get_weights(net) # a list of parameters.
direction = get_random_weights(weights)
normalize_directions_for_weights(direction, weights, norm, ignore)
elif dir_type == 'states':
states = net.state_dict() # a dict of parameters, including BN's running mean/var.
direction = get_random_states(states)
normalize_directions_for_states(direction, states, norm, ignore)
return direction
def tvd(m, l_i):
# load model parameters
pretrained_dict = torch.load('net_params_DRM_to_DGM.pkl')
# get state_dict
net_state_dict = net.state_dict()
# remove keys that does not belong to net_state_dict
pretrained_dict_1 = {k: v for k, v in pretrained_dict.items() if k in net_state_dict}
# update dict
net_state_dict.update(pretrained_dict_1)
# set new dict back to net
net.load_state_dict(net_state_dict)
weights_temp = get_weights(net)
states_temp = net.state_dict()
step_size = 2 * l_i / m
grid = np.arange(-l_i, l_i + step_size, step_size)
num_direction = 1
loss_matrix = torch.zeros((num_direction, len(grid)))
for temp in range(num_direction):
weights = weights_temp
states = states_temp
direction_temp = create_random_direction(net, dir_type='weights', ignore='biasbn', norm='filter')
normalize_directions_for_states(direction_temp, states, norm='filter', ignore='ignore')
directions = [direction_temp]
for dx in grid:
itemindex_1 = np.argwhere(grid == dx)
step = dx
set_states(net, states, directions, step)
loss_temp = loss_function(x)
loss_matrix[temp, itemindex_1[0]] = loss_temp
# clear memory
torch.cuda.empty_cache()
# get state_dict
net_state_dict = net.state_dict()
# remove keys that does not belong to net_state_dict
pretrained_dict_1 = {k: v for k, v in pretrained_dict.items() if k in net_state_dict}
# update dict
net_state_dict.update(pretrained_dict_1)
# set new dict back to net
net.load_state_dict(net_state_dict)
weights_temp = get_weights(net)
states_temp = net.state_dict()
interval_length = grid[-1] - grid[0]
TVD = 0.0
for temp in range(num_direction):
for index in range(loss_matrix.size()[1] - 1):
TVD = TVD + np.abs(float(loss_matrix[temp, index] - loss_matrix[temp, index + 1]))
Max = np.max(loss_matrix.detach().numpy())
Min = np.min(loss_matrix.detach().numpy())
TVD = TVD / interval_length / num_direction
TVD = TVD / interval_length / num_direction / (Max - Min)
return TVD, Max, Min
M = 100
m = 100
l_i = 0.001
TVD_DGM = 0.0
time_start = time.time()
Max = []
Min = []
Result = []
print('====================')
print('Result for l = 0.001.')
print('====================')
for count in range(M):
TVD_temp, Max_temp, Min_temp = tvd(m, l_i)
Max.append(Max_temp)
Min.append(Min_temp)
Result.append(TVD_temp)
print('Current direction TVD of DGM is: ', TVD_temp)
TVD_DGM = TVD_DGM + TVD_temp
print((count + 1) / M * 100, '% finished.')
TVD_DGM = TVD_DGM / M
# TVD_DGM = TVD_DGM / M / (max(Max) - min(Min))
print('All directions average TVD of DGM is: ', TVD_DGM)
# Result = Result / (max(Max) - min(Min))
print('Variance TVD of DGM is: ', np.sqrt(np.var(Result, ddof = 1)))
print('Roughness Index of DGM is: ', np.sqrt(np.var(Result, ddof = 1)) / TVD_DGM)
time_end = time.time()
print('Total time costs: ', time_end - time_start, 'seconds')
```
|
github_jupyter
|
import numpy as np
import random
from math import *
import time
import copy
import torch
from torch.autograd import Variable
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torch.optim.lr_scheduler import StepLR, MultiStepLR
torch.set_default_tensor_type('torch.DoubleTensor')
# defination of activation function
def activation(x):
return x * torch.sigmoid(x)
# build ResNet with one blocks
class Net(nn.Module):
def __init__(self,input_size,width):
super(Net,self).__init__()
self.layer_in = nn.Linear(input_size,width)
self.layer_1 = nn.Linear(width,width)
self.layer_2 = nn.Linear(width,width)
self.layer_out = nn.Linear(width,1)
def forward(self,x):
output = self.layer_in(x)
output = output + activation(self.layer_2(activation(self.layer_1(output)))) # residual block 1
output = self.layer_out(output)
return output
input_size = 1
width = 4
# exact solution
def u_ex(x):
return torch.sin(pi*x)
# f(x)
def f(x):
return pi**2 * torch.sin(pi*x)
grid_num = 200
x = torch.zeros(grid_num + 1, input_size)
for index in range(grid_num + 1):
x[index] = index * 1 / grid_num
net = Net(input_size,width)
def model(x):
return x * (x - 1.0) * net(x)
# loss function to DGM by auto differential
def loss_function(x):
h = 1 / grid_num
sum_0 = 0.0
sum_1 = 0.0
sum_2 = 0.0
sum_a = 0.0
sum_b = 0.0
for index in range(grid_num):
x_temp = x[index] + h / 2
x_temp.requires_grad = True
grad_x_temp = torch.autograd.grad(outputs = model(x_temp), inputs = x_temp, grad_outputs = torch.ones(model(x_temp).shape), create_graph = True)
grad_grad_x_temp = torch.autograd.grad(outputs = grad_x_temp[0], inputs = x_temp, grad_outputs = torch.ones(model(x_temp).shape), create_graph = True)
sum_1 += ((grad_grad_x_temp[0])[0] + f(x_temp)[0])**2
for index in range(1, grid_num):
x_temp = x[index]
x_temp.requires_grad = True
grad_x_temp = torch.autograd.grad(outputs = model(x_temp), inputs = x_temp, grad_outputs = torch.ones(model(x_temp).shape), create_graph = True)
grad_grad_x_temp = torch.autograd.grad(outputs = grad_x_temp[0], inputs = x_temp, grad_outputs = torch.ones(model(x_temp).shape), create_graph = True)
sum_2 += ((grad_grad_x_temp[0])[0] + f(x_temp)[0])**2
x_temp = x[0]
x_temp.requires_grad = True
grad_x_temp = torch.autograd.grad(outputs = model(x_temp), inputs = x_temp, grad_outputs = torch.ones(model(x_temp).shape), create_graph = True)
grad_grad_x_temp = torch.autograd.grad(outputs = grad_x_temp[0], inputs = x_temp, grad_outputs = torch.ones(model(x_temp).shape), create_graph = True)
sum_a = ((grad_grad_x_temp[0])[0] + f(x_temp)[0])**2
x_temp = x[grid_num]
x_temp.requires_grad = True
grad_x_temp = torch.autograd.grad(outputs = model(x_temp), inputs = x_temp, grad_outputs = torch.ones(model(x_temp).shape), create_graph = True)
grad_grad_x_temp = torch.autograd.grad(outputs = grad_x_temp[0], inputs = x_temp, grad_outputs = torch.ones(model(x_temp).shape), create_graph = True)
sum_b = ((grad_grad_x_temp[0])[0] + f(x_temp)[0])**2
sum_0 = h / 6 * (sum_a + 4 * sum_1 + 2 * sum_2 + sum_b)
return sum_0
def get_weights(net):
""" Extract parameters from net, and return a list of tensors"""
return [p.data for p in net.parameters()]
def set_weights(net, weights, directions=None, step=None):
"""
Overwrite the network's weights with a specified list of tensors
or change weights along directions with a step size.
"""
if directions is None:
# You cannot specify a step length without a direction.
for (p, w) in zip(net.parameters(), weights):
p.data.copy_(w.type(type(p.data)))
else:
assert step is not None, 'If a direction is specified then step must be specified as well'
if len(directions) == 2:
dx = directions[0]
dy = directions[1]
changes = [d0*step[0] + d1*step[1] for (d0, d1) in zip(dx, dy)]
else:
changes = [d*step for d in directions[0]]
for (p, w, d) in zip(net.parameters(), weights, changes):
p.data = w + torch.Tensor(d).type(type(w))
def set_states(net, states, directions=None, step=None):
"""
Overwrite the network's state_dict or change it along directions with a step size.
"""
if directions is None:
net.load_state_dict(states)
else:
assert step is not None, 'If direction is provided then the step must be specified as well'
if len(directions) == 2:
dx = directions[0]
dy = directions[1]
changes = [d0*step[0] + d1*step[1] for (d0, d1) in zip(dx, dy)]
else:
changes = [d*step for d in directions[0]]
new_states = copy.deepcopy(states)
assert (len(new_states) == len(changes))
for (k, v), d in zip(new_states.items(), changes):
d = torch.tensor(d)
v.add_(d.type(v.type()))
net.load_state_dict(new_states)
def get_random_weights(weights):
"""
Produce a random direction that is a list of random Gaussian tensors
with the same shape as the network's weights, so one direction entry per weight.
"""
return [torch.randn(w.size()) for w in weights]
def get_random_states(states):
"""
Produce a random direction that is a list of random Gaussian tensors
with the same shape as the network's state_dict(), so one direction entry
per weight, including BN's running_mean/var.
"""
return [torch.randn(w.size()) for k, w in states.items()]
def get_diff_weights(weights, weights2):
""" Produce a direction from 'weights' to 'weights2'."""
return [w2 - w for (w, w2) in zip(weights, weights2)]
def get_diff_states(states, states2):
""" Produce a direction from 'states' to 'states2'."""
return [v2 - v for (k, v), (k2, v2) in zip(states.items(), states2.items())]
def normalize_direction(direction, weights, norm='filter'):
"""
Rescale the direction so that it has similar norm as their corresponding
model in different levels.
Args:
direction: a variables of the random direction for one layer
weights: a variable of the original model for one layer
norm: normalization method, 'filter' | 'layer' | 'weight'
"""
if norm == 'filter':
# Rescale the filters (weights in group) in 'direction' so that each
# filter has the same norm as its corresponding filter in 'weights'.
for d, w in zip(direction, weights):
d.mul_(w.norm()/(d.norm() + 1e-10))
elif norm == 'layer':
# Rescale the layer variables in the direction so that each layer has
# the same norm as the layer variables in weights.
direction.mul_(weights.norm()/direction.norm())
elif norm == 'weight':
# Rescale the entries in the direction so that each entry has the same
# scale as the corresponding weight.
direction.mul_(weights)
elif norm == 'dfilter':
# Rescale the entries in the direction so that each filter direction
# has the unit norm.
for d in direction:
d.div_(d.norm() + 1e-10)
elif norm == 'dlayer':
# Rescale the entries in the direction so that each layer direction has
# the unit norm.
direction.div_(direction.norm())
def normalize_directions_for_weights(direction, weights, norm='filter', ignore='biasbn'):
"""
The normalization scales the direction entries according to the entries of weights.
"""
assert(len(direction) == len(weights))
for d, w in zip(direction, weights):
if d.dim() <= 1:
if ignore == 'biasbn':
d.fill_(0) # ignore directions for weights with 1 dimension
else:
d.copy_(w) # keep directions for weights/bias that are only 1 per node
else:
normalize_direction(d, w, norm)
def normalize_directions_for_states(direction, states, norm='filter', ignore='ignore'):
assert(len(direction) == len(states))
for d, (k, w) in zip(direction, states.items()):
if d.dim() <= 1:
if ignore == 'biasbn':
d.fill_(0) # ignore directions for weights with 1 dimension
else:
d.copy_(w) # keep directions for weights/bias that are only 1 per node
else:
normalize_direction(d, w, norm)
def ignore_biasbn(directions):
""" Set bias and bn parameters in directions to zero """
for d in directions:
if d.dim() <= 1:
d.fill_(0)
def create_random_direction(net, dir_type='weights', ignore='biasbn', norm='filter'):
"""
Setup a random (normalized) direction with the same dimension as
the weights or states.
Args:
net: the given trained model
dir_type: 'weights' or 'states', type of directions.
ignore: 'biasbn', ignore biases and BN parameters.
norm: direction normalization method, including
'filter" | 'layer' | 'weight' | 'dlayer' | 'dfilter'
Returns:
direction: a random direction with the same dimension as weights or states.
"""
# random direction
if dir_type == 'weights':
weights = get_weights(net) # a list of parameters.
direction = get_random_weights(weights)
normalize_directions_for_weights(direction, weights, norm, ignore)
elif dir_type == 'states':
states = net.state_dict() # a dict of parameters, including BN's running mean/var.
direction = get_random_states(states)
normalize_directions_for_states(direction, states, norm, ignore)
return direction
def tvd(m, l_i):
# load model parameters
pretrained_dict = torch.load('net_params_DRM_to_DGM.pkl')
# get state_dict
net_state_dict = net.state_dict()
# remove keys that does not belong to net_state_dict
pretrained_dict_1 = {k: v for k, v in pretrained_dict.items() if k in net_state_dict}
# update dict
net_state_dict.update(pretrained_dict_1)
# set new dict back to net
net.load_state_dict(net_state_dict)
weights_temp = get_weights(net)
states_temp = net.state_dict()
step_size = 2 * l_i / m
grid = np.arange(-l_i, l_i + step_size, step_size)
num_direction = 1
loss_matrix = torch.zeros((num_direction, len(grid)))
for temp in range(num_direction):
weights = weights_temp
states = states_temp
direction_temp = create_random_direction(net, dir_type='weights', ignore='biasbn', norm='filter')
normalize_directions_for_states(direction_temp, states, norm='filter', ignore='ignore')
directions = [direction_temp]
for dx in grid:
itemindex_1 = np.argwhere(grid == dx)
step = dx
set_states(net, states, directions, step)
loss_temp = loss_function(x)
loss_matrix[temp, itemindex_1[0]] = loss_temp
# clear memory
torch.cuda.empty_cache()
# get state_dict
net_state_dict = net.state_dict()
# remove keys that does not belong to net_state_dict
pretrained_dict_1 = {k: v for k, v in pretrained_dict.items() if k in net_state_dict}
# update dict
net_state_dict.update(pretrained_dict_1)
# set new dict back to net
net.load_state_dict(net_state_dict)
weights_temp = get_weights(net)
states_temp = net.state_dict()
interval_length = grid[-1] - grid[0]
TVD = 0.0
for temp in range(num_direction):
for index in range(loss_matrix.size()[1] - 1):
TVD = TVD + np.abs(float(loss_matrix[temp, index] - loss_matrix[temp, index + 1]))
Max = np.max(loss_matrix.detach().numpy())
Min = np.min(loss_matrix.detach().numpy())
TVD = TVD / interval_length / num_direction
TVD = TVD / interval_length / num_direction / (Max - Min)
return TVD, Max, Min
M = 100
m = 100
l_i = 0.001
TVD_DGM = 0.0
time_start = time.time()
Max = []
Min = []
Result = []
print('====================')
print('Result for l = 0.001.')
print('====================')
for count in range(M):
TVD_temp, Max_temp, Min_temp = tvd(m, l_i)
Max.append(Max_temp)
Min.append(Min_temp)
Result.append(TVD_temp)
print('Current direction TVD of DGM is: ', TVD_temp)
TVD_DGM = TVD_DGM + TVD_temp
print((count + 1) / M * 100, '% finished.')
TVD_DGM = TVD_DGM / M
# TVD_DGM = TVD_DGM / M / (max(Max) - min(Min))
print('All directions average TVD of DGM is: ', TVD_DGM)
# Result = Result / (max(Max) - min(Min))
print('Variance TVD of DGM is: ', np.sqrt(np.var(Result, ddof = 1)))
print('Roughness Index of DGM is: ', np.sqrt(np.var(Result, ddof = 1)) / TVD_DGM)
time_end = time.time()
print('Total time costs: ', time_end - time_start, 'seconds')
| 0.824356 | 0.602646 |
# TDA with Python using the Gudhi Library
# Building simplicial complexes from distance matrices
**Authors :** F. Chazal and B. Michel
```
import numpy as np
import pandas as pd
import pickle as pickle
import gudhi as gd
from pylab import *
%matplotlib inline
```
TDA typically aims at extracting topological signatures from a point cloud in $\mathbb R^d$ or in a general metric space. [Simplicial complexes](https://en.wikipedia.org/wiki/Simplicial_complex) are used in computational geometry to infer topological signatures from data.
This tutorial explains how to build [Vietoris-Rips complexes](https://en.wikipedia.org/wiki/Vietoris%E2%80%93Rips_complex) and [alpha complexes](https://en.wikipedia.org/wiki/Alpha_shape#Alpha_complex) from a matrix of pairwise distances.
## Vietoris-Rips filtration defined from a matrix of distances
The [$\alpha$-Rips complex](https://en.wikipedia.org/wiki/Vietoris%E2%80%93Rips_complex) of a metric space $\mathbb X$ is an [abstract simplicial complex](https://en.wikipedia.org/wiki/Abstract_simplicial_complex) that can be defined by forming a simplex for every finite subset of $\mathbb X$ that has diameter at most $\alpha$.

## Protein binding dataset
The data we will be studying in this notebook represents configurations of protein binding. This example is borrowed from Kovacev-Nikolic et al. [[1]](https://arxiv.org/pdf/1412.1394.pdf).
The paper compares closed and open forms of the maltose-binding protein (MBP), a large biomolecule containing $370$ amino-acid residues. The analysis is not based on geometric distances in $\mathbb R^3$ but on a metric of *dynamical distances* defined by
$$ D_{ij} = 1 - |C_{ij}|, $$
where $C$ is the correlation matrix between residues. Correlation matrices between residues can be found at this [link](https://www.researchgate.net/publication/301543862_corr). We are greatful to the authors for sharing data !
The next statments load the $14$ correlation matrices with pandas:
```
path_file = "./datasets/Corr_ProteinBinding/"
files_list = [
'1anf.corr_1.txt',
'1ez9.corr_1.txt',
'1fqa.corr_2.txt',
'1fqb.corr_3.txt',
'1fqc.corr_2.txt',
'1fqd.corr_3.txt',
'1jw4.corr_4.txt',
'1jw5.corr_5.txt',
'1lls.corr_6.txt',
'1mpd.corr_4.txt',
'1omp.corr_7.txt',
'3hpi.corr_5.txt',
'3mbp.corr_6.txt',
'4mbp.corr_7.txt'
]
corr_list = [
pd.read_csv(
path_file + u,
header = None,
delim_whitespace = True
) for u in files_list
]
```
The object `corr_list` is a list of $14$ correlation matrices. We can iterate in the list to compute the matrix of distances associated to each configuration:
```
dist_list = [1 - np.abs(c) for c in corr_list]
```
Let's print out the first lines of the first distance matrix:
```
D = dist_list[0]
D.head()
```
## Vietoris-Rips filtration of Protein binding distance matrix
The `RipsComplex()` function creates a [$1$-skeleton](https://en.wikipedia.org/wiki/N-skeleton) from the point cloud (see the [GUDHI documentation](http://gudhi.gforge.inria.fr/python/latest/rips_complex_user.html) for details on the syntax).
```
skeleton_protein = gd.RipsComplex(
distance_matrix = D.values,
max_edge_length = 0.8
)
```
The `max_edge_length` parameter is the maximal diameter: only the edges of length less vers this value are included in the one skeleton graph.
Next, we create the Rips simplicial complex from this one-skeleton graph. This is a filtered Rips complex which filtration function is exacly the diameter of the simplices. We use the `create_simplex_tree()` function:
```
Rips_simplex_tree_protein = skeleton_protein.create_simplex_tree(max_dimension = 2)
```
The `max_dimension` parameter is the maximum dimension of the simplices included in the the filtration. The object returned by the function is a simplex tree, of dimension 2 in this example:
```
Rips_simplex_tree_protein.dimension()
```
We can use the fonctionalites of the simplex tree object to describe the Rips filtration.
For instance we can check that the 370 points of the first distance matrix are all vertices of the Vietoris-Rips filtration:
```
Rips_simplex_tree_protein.num_vertices()
```
The number of simplices in a Rips complex increases very fast with the number of points and the dimension. There is more than on million of simplexes in the Rips complex:
```
Rips_simplex_tree_protein.num_simplices()
```
Note that this is actually the number of simplices in the "last" Rips complex of the filtration, namely with parameter $\alpha=$ `max_edge_length=`0.8.
Let's compute the list of simplices in the Rips complex with the `get_filtration()` function:
```
rips_filtration = Rips_simplex_tree_protein.get_filtration()
rips_list = list(rips_filtration)
len(rips_list)
for splx in rips_list[0:400] :
print(splx)
```
The integers represent the points in the metric space: the vertex `[2]` corresponds to the point decribed by the second raw (or the second column) in the distance matrix `D`.
The filtration value is the diameter of the simplex, which is zero for the vertices of course. The first edge in the filtration is `[289, 290]`, these two points are the two closest points according to `D`, at distance $0.015$ of each other.
### How to define an Alpha complex from a matrix of distance ?
The [alpha complex filtration](https://en.wikipedia.org/wiki/Alpha_shape#Alpha_complex) of a point cloud in $\mathbb R^p$ is a filtered simplicial complex constructed from the finite cells of a [Delaunay Triangulation](https://en.wikipedia.org/wiki/Delaunay_triangulation).
In our case, the data does not belong to euclideen space $\mathbb R^p$ and we are not in position to directly compute a Delaunay Triangulation in the metric space, using the pairwise distances.
The aim of [Multidimensional Scaling](https://en.wikipedia.org/wiki/Multidimensional_scaling) (MDS) methods is precisely to find a representation of $n$ points in a space $\mathbb R^p$ that preserves as well as possible the pairwise distances between the $n$ points in the original metric space. The are several versions of MDS algorithms, the most popular ones are available in the [sckit-learn library](https://scikit-learn.org/stable/index.html), see this [documention](https://scikit-learn.org/stable/modules/generated/sklearn.manifold.MDS.html).
Let's compute a (classical) MDS representation of the matrix `D` in $\mathbb R^3$:
```
from sklearn.manifold import MDS
embedding = MDS(n_components = 3, dissimilarity = 'precomputed')
X_transformed = embedding.fit_transform(D)
X_transformed.shape
```
Now we can represent this configuration, for instance on the two first axes:
```
fig = plt.figure()
plt.scatter(X_transformed[:, 0], X_transformed[:, 1], label = 'MDS');
```
Of course you should keep in mind that MDS provides an embedding of the data in $\mathbb R^p$ that **approximatively** preserves the distance matrix.
The main advantage of Alpha complexes is that they contain less simplices than Rips complexes do and so it can be a better option than Rips complexes. As subcomplexes of the Delaunay Triangulation complex, an alpha complex is a geometric simpicial complex.
The `AlphaComplex()` function directly computes the simplex tree representing the Alpha complex:
```
alpha_complex = gd.AlphaComplex(points = X_transformed)
st_alpha = alpha_complex.create_simplex_tree()
```
The point cloud `X_transformed` belongs to $\mathbb R^3$ and so does the Alpha Complex:
```
st_alpha.dimension()
```
As for the Rips complex, the $370$ points are all vertices of the Alpha complex :
```
print(st_alpha.num_vertices())
```
Note that the number of simplexes in the Alpha complex is much smaller then for the Rips complex we computed before:
```
print(st_alpha.num_simplices())
```
### References
[1] Using persistent homology and dynamical distances to analyze protein binding, V. Kovacev-Nikolic, P. Bubenik, D. Nikolic and G. Heo. Stat Appl Genet Mol Biol 2016 [arxiv link](https://arxiv.org/pdf/1412.1394.pdf).
|
github_jupyter
|
import numpy as np
import pandas as pd
import pickle as pickle
import gudhi as gd
from pylab import *
%matplotlib inline
path_file = "./datasets/Corr_ProteinBinding/"
files_list = [
'1anf.corr_1.txt',
'1ez9.corr_1.txt',
'1fqa.corr_2.txt',
'1fqb.corr_3.txt',
'1fqc.corr_2.txt',
'1fqd.corr_3.txt',
'1jw4.corr_4.txt',
'1jw5.corr_5.txt',
'1lls.corr_6.txt',
'1mpd.corr_4.txt',
'1omp.corr_7.txt',
'3hpi.corr_5.txt',
'3mbp.corr_6.txt',
'4mbp.corr_7.txt'
]
corr_list = [
pd.read_csv(
path_file + u,
header = None,
delim_whitespace = True
) for u in files_list
]
dist_list = [1 - np.abs(c) for c in corr_list]
D = dist_list[0]
D.head()
skeleton_protein = gd.RipsComplex(
distance_matrix = D.values,
max_edge_length = 0.8
)
Rips_simplex_tree_protein = skeleton_protein.create_simplex_tree(max_dimension = 2)
Rips_simplex_tree_protein.dimension()
Rips_simplex_tree_protein.num_vertices()
Rips_simplex_tree_protein.num_simplices()
rips_filtration = Rips_simplex_tree_protein.get_filtration()
rips_list = list(rips_filtration)
len(rips_list)
for splx in rips_list[0:400] :
print(splx)
from sklearn.manifold import MDS
embedding = MDS(n_components = 3, dissimilarity = 'precomputed')
X_transformed = embedding.fit_transform(D)
X_transformed.shape
fig = plt.figure()
plt.scatter(X_transformed[:, 0], X_transformed[:, 1], label = 'MDS');
alpha_complex = gd.AlphaComplex(points = X_transformed)
st_alpha = alpha_complex.create_simplex_tree()
st_alpha.dimension()
print(st_alpha.num_vertices())
print(st_alpha.num_simplices())
| 0.323701 | 0.982507 |
# Home Work Basic Phyton
Source of Data set about google play store is from :
<br>[https://www.kaggle.com/lava18/google-play-store-apps#googleplaystore.csv]
<br>Copy the data set from
<br>[https://raw.githubusercontent.com/banguntsel/HomeworkPhyton/master/googleplaystore.csv]
*<p>Author : Bangun M Sagala*
*<p>Date : 04 October 2019*
***
```
import pandas as pd
import numpy as np
df = pd.read_csv('https://raw.githubusercontent.com/banguntsel/HomeworkPhyton/master/googleplaystore.csv', encoding='latin1')
df.head()
print(df.dtypes)
```
## Check total Rows and Column in the dataframe (df) - Answer No 1
```
df.shape
```
## Check unique apps categories reflected in the dataframe (df) - Answer No 2
```
category = df[['Category']]
print("Unique Category : ",len(category.groupby(['Category'])))
```
## Check unique genres according to the dataframe (df) - Answer No 3
```
res = pd.get_dummies(df['Genres'].str.split(';').apply(pd.Series).stack()).sum(level=0)
print("Unique Genre : ",len(res.columns))
```
## 4. Drop duplicate values in 'App' column and remove its rows in Python - Answer No 4
```
df.drop_duplicates(subset='App', inplace=True)
```
## 5. Remove a row that contain 'Free','+' and ',' value at the 'Installs' column - Answer No 5
<p> Cek row contain Free in install column
```
df[df['Installs']=='Free']
```
<p> Remove 'Free; from at Installs column
```
df = df[df['Installs'] != 'Free']
```
<p> Cross Check row contain Free in install column
```
df[df['Installs']=='Free']
```
<p> Remove row contain '+' at Installs column
```
df['Installs'] = df['Installs'].apply(lambda x: x.replace('+', '') if '+' in str(x) else x)
```
<p> Remove row contain ',' at Installs column
```
df['Installs'] = df['Installs'].apply(lambda x: x.replace(',', '') if ',' in str(x) else x)
```
## 6. convert data type of 'Installs' columns into integer - Answer No 6
```
df['Installs'] = df['Installs'].astype(int)
df['Installs'].tail()
df.dtypes
df.head()
```
## 7. Match Apps and Category based on provided dataset - Answer No 7
```
cek_df=df[df.App.isin(['FIFA Soccer','WhatsApp Messenger','DU Battery Saver - Battery Charger & Battery Life','File Commander - File Manager/Explorer','Subway Surfers','Flipboard: News For Our Time','Pinterest','Dropbox'])]
cek_df.loc[:,['App','Category']].head(8)
```
## 8. applications were installed by more than 1 billion users - Answer No 8
<p> Use variable install_df as temporary variable
```
install_df = df[df.App.isin(['Facebook','WhatsApp Messenger','Skype - free IM & video calls','Google Duo - High Quality Video Calls',])]
install_df = install_df.sort_values('Installs',ascending=False)
install_df.loc[:,['App','Installs']]
```
## 9. Arrange top COMMUNICATION category apps from the highest number of reviews to the lowest!
<p> Use variable df_comm as temporary variable
```
df['Reviews'] = df['Reviews'].astype(int)
df.comm = df[df['Category']=='COMMUNICATION']
df.comm.head()
df.comm = df.comm.sort_values('Reviews',ascending=False)
df.comm.head(10)
df.comm.loc[:,['App','Category','Reviews']].head(8)
```
## 10. Arrange top 3 game category apps based on number of installed and rating.
<p> Use variable df_game as temporary variable
```
df['Reviews'] = df['Reviews'].astype(int)
df_game = df[df['Category']=='GAME']
df_game = df_game.sort_values(['Installs','Rating'],ascending=[False,False])
df_game.head()
```
|
github_jupyter
|
import pandas as pd
import numpy as np
df = pd.read_csv('https://raw.githubusercontent.com/banguntsel/HomeworkPhyton/master/googleplaystore.csv', encoding='latin1')
df.head()
print(df.dtypes)
df.shape
category = df[['Category']]
print("Unique Category : ",len(category.groupby(['Category'])))
res = pd.get_dummies(df['Genres'].str.split(';').apply(pd.Series).stack()).sum(level=0)
print("Unique Genre : ",len(res.columns))
df.drop_duplicates(subset='App', inplace=True)
df[df['Installs']=='Free']
df = df[df['Installs'] != 'Free']
df[df['Installs']=='Free']
df['Installs'] = df['Installs'].apply(lambda x: x.replace('+', '') if '+' in str(x) else x)
df['Installs'] = df['Installs'].apply(lambda x: x.replace(',', '') if ',' in str(x) else x)
df['Installs'] = df['Installs'].astype(int)
df['Installs'].tail()
df.dtypes
df.head()
cek_df=df[df.App.isin(['FIFA Soccer','WhatsApp Messenger','DU Battery Saver - Battery Charger & Battery Life','File Commander - File Manager/Explorer','Subway Surfers','Flipboard: News For Our Time','Pinterest','Dropbox'])]
cek_df.loc[:,['App','Category']].head(8)
install_df = df[df.App.isin(['Facebook','WhatsApp Messenger','Skype - free IM & video calls','Google Duo - High Quality Video Calls',])]
install_df = install_df.sort_values('Installs',ascending=False)
install_df.loc[:,['App','Installs']]
df['Reviews'] = df['Reviews'].astype(int)
df.comm = df[df['Category']=='COMMUNICATION']
df.comm.head()
df.comm = df.comm.sort_values('Reviews',ascending=False)
df.comm.head(10)
df.comm.loc[:,['App','Category','Reviews']].head(8)
df['Reviews'] = df['Reviews'].astype(int)
df_game = df[df['Category']=='GAME']
df_game = df_game.sort_values(['Installs','Rating'],ascending=[False,False])
df_game.head()
| 0.099695 | 0.874614 |
# Create a choropleth map with Folium
1. Import libraries
2. Wrangle data
3. Clean data
4. Create choropleth map
# 1. Import libraries
```
# Import libraries
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib
import os
import folium
import json
# This command propts matplotlib visuals to appear in the notebook
%matplotlib inline
# Import Berlin .json file
country_geo = r'/Users/OldBobJulia/Desktop/CF/Course/6. Advanced Analytics and Dashboard Design/Berlin Airbnb Analysis/02 Data/Original data/berlin_bezirke.json'
# Look at it
f = open(r'/Users/OldBobJulia/Desktop/CF/Course/6. Advanced Analytics and Dashboard Design/Berlin Airbnb Analysis/02 Data/Original data/berlin_bezirke.json',)
data = json.load(f)
for i in data['features']:
print(i)
```
File has a geometry column AND a LABEL.
```
# Import Airbnb data
df = pd.read_csv(r'/Users/OldBobJulia/Desktop/CF/Course/6. Advanced Analytics and Dashboard Design/Berlin Airbnb Analysis/02 Data/Prepared data/listing_derivedcolumns.csv')
df.head()
# Drop Unnamed: 0
df = df.drop(columns = ['Unnamed: 0'])
df.head()
df.shape
df['neighbourhood_group'].value_counts(dropna=False)
```
# 2. Wrangle data
```
# Select only necessary columns for choropleth map
columns = ["neighbourhood_group", "latitude", "longitude", "room_type", "price"]
# Create subset with only these columns
df_2 = df[columns]
df_2.head()
# Make histogram of price to see what price categories could work.
# Make subset of price excluding extreme prices to that end
price_cat_check = df_2[df_2['price'] < 4000]
price_cat_check.head()
price_cat_check.shape
sns.distplot(price_cat_check['price'], bins = 5)
# Wrangle neighbourhood_group names to fit json data
df_2['neighbourhood_group'].value_counts(dropna=False)
df_2['neighbourhood_group'] = df_2['neighbourhood_group'].replace({'Charlottenburg-Wilm.': 'Charlottenburg-Wilmersdorf', 'Tempelhof - Schöneberg': 'Tempelhof-Schöneberg', 'Treptow - Köpenick': 'Treptow-Köpenick', 'Steglitz - Zehlendorf': 'Steglitz-Zehlendorf', 'Marzahn - Hellersdorf': 'Marzahn-Hellersdorf'})
df_2.head()
df_2['neighbourhood_group'].value_counts(dropna=False)
```
# 3. Check consistency
```
# Check for missings
df_2.isnull().sum()
```
There are no missing values
```
# Check for duplicates
dups = df_2.duplicated()
dups.shape
```
There are no dups
```
# Check outliers
df_2.describe()
```
3 observations with prices over 4000 were imputed with the mean previously
```
# Check how many rows with price under 15
super_low_price = df_2[df_2['price'] < 15]
super_low_price.shape
super_low_price.head()
```
I find it surprisingly many, still these rows are less than 10% of the total data set AND they remain within the reasonable price, so I leave them.
# 4. Plot choropleth map
```
data_to_plot = df_2[['neighbourhood_group','price']]
data_to_plot.head()
# Setup a folium map at a high-level zoom
map = folium.Map(location=[52.520, 13.404], width=750, height=500)
folium.Choropleth(
geo_data = country_geo,
data = data_to_plot,
columns = ['neighbourhood_group', 'price'],
key_on = 'feature.properties.name',
fill_color = 'PuBuGn', fill_opacity=0.6, line_opacity=0.1,
legend_name = "price").add_to(map)
folium.LayerControl().add_to(map)
map
# Save it
map.save("index.html")
# Export as png just in case
map.save("map2.png")
```
The map answers the question which neighbourhoods are the most popular ones, or where most money can be made through Airbnb rentals. The answer is Friedrichshain-Kreuzberg, followed by Tempelhof-Schöneberg and Charlottenburg-Wilmersdorf.
New research question:
- Does the number of commercial hosts influence the popularity of a neighbourhood?
|
github_jupyter
|
# Import libraries
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib
import os
import folium
import json
# This command propts matplotlib visuals to appear in the notebook
%matplotlib inline
# Import Berlin .json file
country_geo = r'/Users/OldBobJulia/Desktop/CF/Course/6. Advanced Analytics and Dashboard Design/Berlin Airbnb Analysis/02 Data/Original data/berlin_bezirke.json'
# Look at it
f = open(r'/Users/OldBobJulia/Desktop/CF/Course/6. Advanced Analytics and Dashboard Design/Berlin Airbnb Analysis/02 Data/Original data/berlin_bezirke.json',)
data = json.load(f)
for i in data['features']:
print(i)
# Import Airbnb data
df = pd.read_csv(r'/Users/OldBobJulia/Desktop/CF/Course/6. Advanced Analytics and Dashboard Design/Berlin Airbnb Analysis/02 Data/Prepared data/listing_derivedcolumns.csv')
df.head()
# Drop Unnamed: 0
df = df.drop(columns = ['Unnamed: 0'])
df.head()
df.shape
df['neighbourhood_group'].value_counts(dropna=False)
# Select only necessary columns for choropleth map
columns = ["neighbourhood_group", "latitude", "longitude", "room_type", "price"]
# Create subset with only these columns
df_2 = df[columns]
df_2.head()
# Make histogram of price to see what price categories could work.
# Make subset of price excluding extreme prices to that end
price_cat_check = df_2[df_2['price'] < 4000]
price_cat_check.head()
price_cat_check.shape
sns.distplot(price_cat_check['price'], bins = 5)
# Wrangle neighbourhood_group names to fit json data
df_2['neighbourhood_group'].value_counts(dropna=False)
df_2['neighbourhood_group'] = df_2['neighbourhood_group'].replace({'Charlottenburg-Wilm.': 'Charlottenburg-Wilmersdorf', 'Tempelhof - Schöneberg': 'Tempelhof-Schöneberg', 'Treptow - Köpenick': 'Treptow-Köpenick', 'Steglitz - Zehlendorf': 'Steglitz-Zehlendorf', 'Marzahn - Hellersdorf': 'Marzahn-Hellersdorf'})
df_2.head()
df_2['neighbourhood_group'].value_counts(dropna=False)
# Check for missings
df_2.isnull().sum()
# Check for duplicates
dups = df_2.duplicated()
dups.shape
# Check outliers
df_2.describe()
# Check how many rows with price under 15
super_low_price = df_2[df_2['price'] < 15]
super_low_price.shape
super_low_price.head()
data_to_plot = df_2[['neighbourhood_group','price']]
data_to_plot.head()
# Setup a folium map at a high-level zoom
map = folium.Map(location=[52.520, 13.404], width=750, height=500)
folium.Choropleth(
geo_data = country_geo,
data = data_to_plot,
columns = ['neighbourhood_group', 'price'],
key_on = 'feature.properties.name',
fill_color = 'PuBuGn', fill_opacity=0.6, line_opacity=0.1,
legend_name = "price").add_to(map)
folium.LayerControl().add_to(map)
map
# Save it
map.save("index.html")
# Export as png just in case
map.save("map2.png")
| 0.430866 | 0.910545 |
# Basics of Spark on HDInsight
<a href="http://spark.apache.org/" target="_blank">Apache Spark</a> is an open-source parallel processing framework that supports in-memory processing to boost the performance of big-data analytic applications. When you provision a Spark cluster in HDInsight, you provision Azure compute resources with Spark installed and configured. The data to be processed is stored in Azure Data Lake Store.
Now that you have created a Spark cluster, let us understand some basics of working with Spark on HDInsight. For detailed discussion on working with Spark, see [Spark Programming Guide](http://spark.apache.org/docs/2.0.0/sql-programming-guide.html).
----------
## Notebook setup
When using PySpark kernel notebooks on HDInsight, there is no need to create a SparkContext or a SparkSession; a SparkSession which has the SparkContext is created for you automatically when you run the first code cell, and you'll be able to see the progress printed. The contexts are created with the following variable names:
- SparkSession (spark)
To run the cells below, place the cursor in the cell and then press **SHIFT + ENTER**.
----------
## What is an RDD?
Big Data applications rely on iterative, distributed computing for faster processing of large data sets. To distribute data processing over multiple jobs, the data is typically reused or shared across jobs. To share data between existing distributed computing systems you need to store data in some intermediate stable distributed store such as HDFS. This makes the overall computations of jobs slower.
**Resilient Distributed Datasets** or RDDs address this by enabling fault-tolerant, distributed, in-memory computations.
----------
## How do I make an RDD?
RDDs can be created from stable storage or by transforming other RDDs. Run the cells below to create RDDs from the sample data files available in the storage container associated with your Spark cluster. One such sample data file is available on the cluster at `adl:///example/data/fruits.txt`.
```
fruits = spark.sparkContext.textFile('adl:///example/data/fruits.txt')
yellowThings = spark.sparkContext.textFile('adl:///example/data/yellowthings.txt')
```
For more examples on how to create RDDs see the following notebooks available with your Spark cluster:
* Read and write data from Azure Storage Blobs (WASB)
* Read and write data from Hive tables
----------
## What are RDD operations?
RDDs support two types of operations: transformations and actions.
* **Transformations** create a new dataset from an existing one. Transformations are lazy, meaning that no transformation is executed until you execute an action.
* **Actions** return a value to the driver program after running a computation on the dataset.
### RDD transformations
Following are examples of some of the common transformations available. For a detailed list, see [RDD Transformations](https://spark.apache.org/docs/2.0.0/programming-guide.html#transformations)
Run some transformations below to understand this better. Place the cursor in the cell and press **SHIFT + ENTER**.
```
# map
fruitsReversed = fruits.map(lambda fruit: fruit[::-1])
# filter
shortFruits = fruits.filter(lambda fruit: len(fruit) <= 5)
# flatMap
characters = fruits.flatMap(lambda fruit: list(fruit))
# union
fruitsAndYellowThings = fruits.union(yellowThings)
# intersection
yellowFruits = fruits.intersection(yellowThings)
# distinct
distinctFruitsAndYellowThings = fruitsAndYellowThings.distinct()
distinctFruitsAndYellowThings
# groupByKey
yellowThingsByFirstLetter = yellowThings.map(lambda thing: (thing[0], thing)).groupByKey()
# reduceByKey
numFruitsByLength = fruits.map(lambda fruit: (len(fruit), 1)).reduceByKey(lambda x, y: x + y)
```
### RDD actions
Following are examples of some of the common actions available. For a detailed list, see [RDD Actions](https://spark.apache.org/docs/2.0.0/programming-guide.html#actions).
Run some transformations below to understand this better. Place the cursor in the cell and press **SHIFT + ENTER**.
```
# collect
fruitsArray = fruits.collect()
yellowThingsArray = yellowThings.collect()
fruitsArray
# count
numFruits = fruits.count()
numFruits
# take
first3Fruits = fruits.take(3)
first3Fruits
# reduce
letterSet = fruits.map(lambda fruit: set(fruit)).reduce(lambda x, y: x.union(y))
letterSet
```
> **IMPORTANT**: Another important RDD action is saving the output to a file. See the **Read and write data from Azure Storage Blobs (WASB)** notebook for more information.
----------
## What is a dataframe?
The `pyspark.sql` library provides an alternative API for manipulating structured datasets, known as "dataframes". (Dataframes are not a Spark-specific concept but `pyspark` provides its own dedicated dataframe library.) These are different from RDDs, but you can convert an RDD into a dataframe or vice-versa, if required.
See [Spark SQL and DataFrame Guide](https://spark.apache.org/docs/2.0.0/sql-programming-guide.html#datasets-and-dataframes) for more information.
### How do I make a dataframe?
You can load a dataframe directly from an input data source. See the following notebooks included with your Spark cluster for more information.
* Read and write data from Azure Storage Blobs (WASB)
* Read and write data from Hive tables
You can also create a dataframe from a CSV file as shown below.
```
df = spark.read.csv('adl:///HdiSamples/HdiSamples/SensorSampleData/building/building.csv',
header=True, inferSchema=True)
```
### Dataframe operations
Run the cells below to see examples of some of the the operations that you can perform on dataframes.
```
# show the content of the dataframe
df.show()
# Print the dataframe schema in a tree format
df.printSchema()
# Create an RDD from the dataframe
dfrdd = df.rdd
dfrdd.take(3)
dfrdd = df.rdd
dfrdd.take(3)
# Retrieve a given number of rows from the dataframe
df.limit(3).show()
df.limit(3).show()
# Retrieve specific columns from the dataframe
df.select('BuildingID', 'Country').limit(3).show()
# Use GroupBy clause with dataframe
df.groupBy('HVACProduct').count().select('HVACProduct', 'count').show()
```
> **IMPORTANT**: Many of the methods available on normal RDDs are also available on dataframes. For example, `distinct`, `count`, `collect`, `filter`, `map`, and `take` are all methods on dataframes as well as on RDDs.
-------
## Spark SQL and dataframes
You can also run SQL queries over dataframes once you register them as temporary tables within the SparkSession. Run the snippet below to see an example.
```
# Register the dataframe as a temporary table called HVAC
df.registerTempTable('HVAC')
%%sql
SELECT * FROM HVAC WHERE BuildingAge >= 10
%%sql
SELECT BuildingID, Country FROM HVAC LIMIT 3
```
|
github_jupyter
|
fruits = spark.sparkContext.textFile('adl:///example/data/fruits.txt')
yellowThings = spark.sparkContext.textFile('adl:///example/data/yellowthings.txt')
# map
fruitsReversed = fruits.map(lambda fruit: fruit[::-1])
# filter
shortFruits = fruits.filter(lambda fruit: len(fruit) <= 5)
# flatMap
characters = fruits.flatMap(lambda fruit: list(fruit))
# union
fruitsAndYellowThings = fruits.union(yellowThings)
# intersection
yellowFruits = fruits.intersection(yellowThings)
# distinct
distinctFruitsAndYellowThings = fruitsAndYellowThings.distinct()
distinctFruitsAndYellowThings
# groupByKey
yellowThingsByFirstLetter = yellowThings.map(lambda thing: (thing[0], thing)).groupByKey()
# reduceByKey
numFruitsByLength = fruits.map(lambda fruit: (len(fruit), 1)).reduceByKey(lambda x, y: x + y)
# collect
fruitsArray = fruits.collect()
yellowThingsArray = yellowThings.collect()
fruitsArray
# count
numFruits = fruits.count()
numFruits
# take
first3Fruits = fruits.take(3)
first3Fruits
# reduce
letterSet = fruits.map(lambda fruit: set(fruit)).reduce(lambda x, y: x.union(y))
letterSet
df = spark.read.csv('adl:///HdiSamples/HdiSamples/SensorSampleData/building/building.csv',
header=True, inferSchema=True)
# show the content of the dataframe
df.show()
# Print the dataframe schema in a tree format
df.printSchema()
# Create an RDD from the dataframe
dfrdd = df.rdd
dfrdd.take(3)
dfrdd = df.rdd
dfrdd.take(3)
# Retrieve a given number of rows from the dataframe
df.limit(3).show()
df.limit(3).show()
# Retrieve specific columns from the dataframe
df.select('BuildingID', 'Country').limit(3).show()
# Use GroupBy clause with dataframe
df.groupBy('HVACProduct').count().select('HVACProduct', 'count').show()
# Register the dataframe as a temporary table called HVAC
df.registerTempTable('HVAC')
%%sql
SELECT * FROM HVAC WHERE BuildingAge >= 10
%%sql
SELECT BuildingID, Country FROM HVAC LIMIT 3
| 0.410993 | 0.987508 |
<!--BOOK_INFORMATION-->
<img align="left" style="padding-right:10px;" src="fig/cover-small.jpg">
*This notebook contains an excerpt from the [Whirlwind Tour of Python](http://www.oreilly.com/programming/free/a-whirlwind-tour-of-python.csp) by Jake VanderPlas; the content is available [on GitHub](https://github.com/jakevdp/WhirlwindTourOfPython).*
*The text and code are released under the [CC0](https://github.com/jakevdp/WhirlwindTourOfPython/blob/master/LICENSE) license; see also the companion project, the [Python Data Science Handbook](https://github.com/jakevdp/PythonDataScienceHandbook).*
<!--NAVIGATION-->
< [List Comprehensions](11-List-Comprehensions.ipynb) | [Contents](Index.ipynb) | [Modules and Packages](13-Modules-and-Packages.ipynb) >
# Generators
Here we'll take a deeper dive into Python generators, including *generator expressions* and *generator functions*.
## Generator Expressions
The difference between list comprehensions and generator expressions is sometimes confusing; here we'll quickly outline the differences between them:
### List comprehensions use square brackets, while generator expressions use parentheses
This is a representative list comprehension:
```
[n ** 2 for n in range(12)]
```
While this is a representative generator expression:
```
(n ** 2 for n in range(12))
```
Notice that printing the generator expression does not print the contents; one way to print the contents of a generator expression is to pass it to the ``list`` constructor:
```
G = (n ** 2 for n in range(12))
list(G)
```
### A list is a collection of values, while a generator is a recipe for producing values
When you create a list, you are actually building a collection of values, and there is some memory cost associated with that.
When you create a generator, you are not building a collection of values, but a recipe for producing those values.
Both expose the same iterator interface, as we can see here:
```
L = [n ** 2 for n in range(12)]
for val in L:
print(val, end=' ')
G = (n ** 2 for n in range(12))
for val in G:
print(val, end=' ')
```
The difference is that a generator expression does not actually compute the values until they are needed.
This not only leads to memory efficiency, but to computational efficiency as well!
This also means that while the size of a list is limited by available memory, the size of a generator expression is unlimited!
An example of an infinite generator expression can be created using the ``count`` iterator defined in ``itertools``:
```
from itertools import count
count()
for i in count():
print(i, end=' ')
if i >= 10: break
```
The ``count`` iterator will go on happily counting forever until you tell it to stop; this makes it convenient to create generators that will also go on forever:
```
factors = [2, 3, 5, 7]
G = (i for i in count() if all(i % n > 0 for n in factors))
for val in G:
print(val, end=' ')
if val > 40: break
```
You might see what we're getting at here: if we were to expand the list of factors appropriately, what we would have the beginnings of is a prime number generator, using the Sieve of Eratosthenes algorithm. We'll explore this more momentarily.
### A list can be iterated multiple times; a generator expression is single-use
This is one of those potential gotchas of generator expressions.
With a list, we can straightforwardly do this:
```
L = [n ** 2 for n in range(12)]
for val in L:
print(val, end=' ')
print()
for val in L:
print(val, end=' ')
```
A generator expression, on the other hand, is used-up after one iteration:
```
G = (n ** 2 for n in range(12))
list(G)
list(G)
```
This can be very useful because it means iteration can be stopped and started:
```
G = (n**2 for n in range(12))
for n in G:
print(n, end=' ')
if n > 30: break
print("\ndoing something in between")
for n in G:
print(n, end=' ')
```
One place I've found this useful is when working with collections of data files on disk; it means that you can quite easily analyze them in batches, letting the generator keep track of which ones you have yet to see.
## Generator Functions: Using ``yield``
We saw in the previous section that list comprehensions are best used to create relatively simple lists, while using a normal ``for`` loop can be better in more complicated situations.
The same is true of generator expressions: we can make more complicated generators using *generator functions*, which make use of the ``yield`` statement.
Here we have two ways of constructing the same list:
```
L1 = [n ** 2 for n in range(12)]
L2 = []
for n in range(12):
L2.append(n ** 2)
print(L1)
print(L2)
```
Similarly, here we have two ways of constructing equivalent generators:
```
G1 = (n ** 2 for n in range(12))
def gen():
for n in range(12):
yield n ** 2
G2 = gen()
print(*G1)
print(*G2)
```
A generator function is a function that, rather than using ``return`` to return a value once, uses ``yield`` to yield a (potentially infinite) sequence of values.
Just as in generator expressions, the state of the generator is preserved between partial iterations, but if we want a fresh copy of the generator we can simply call the function again.
## Example: Prime Number Generator
Here I'll show my favorite example of a generator function: a function to generate an unbounded series of prime numbers.
A classic algorithm for this is the *Sieve of Eratosthenes*, which works something like this:
```
# Generate a list of candidates
L = [n for n in range(2, 40)]
print(L)
# Remove all multiples of the first value
L = [n for n in L if n == L[0] or n % L[0] > 0]
print(L)
# Remove all multiples of the second value
L = [n for n in L if n == L[1] or n % L[1] > 0]
print(L)
# Remove all multiples of the third value
L = [n for n in L if n == L[2] or n % L[2] > 0]
print(L)
```
If we repeat this procedure enough times on a large enough list, we can generate as many primes as we wish.
Let's encapsulate this logic in a generator function:
```
def gen_primes(N):
"""Generate primes up to N"""
primes = set()
for n in range(2, N):
if all(n % p > 0 for p in primes):
primes.add(n)
yield n
print(*gen_primes(100))
```
That's all there is to it!
While this is certainly not the most computationally efficient implementation of the Sieve of Eratosthenes, it illustrates how convenient the generator function syntax can be for building more complicated sequences.
<!--NAVIGATION-->
< [List Comprehensions](11-List-Comprehensions.ipynb) | [Contents](Index.ipynb) | [Modules and Packages](13-Modules-and-Packages.ipynb) >
|
github_jupyter
|
[n ** 2 for n in range(12)]
(n ** 2 for n in range(12))
G = (n ** 2 for n in range(12))
list(G)
L = [n ** 2 for n in range(12)]
for val in L:
print(val, end=' ')
G = (n ** 2 for n in range(12))
for val in G:
print(val, end=' ')
from itertools import count
count()
for i in count():
print(i, end=' ')
if i >= 10: break
factors = [2, 3, 5, 7]
G = (i for i in count() if all(i % n > 0 for n in factors))
for val in G:
print(val, end=' ')
if val > 40: break
L = [n ** 2 for n in range(12)]
for val in L:
print(val, end=' ')
print()
for val in L:
print(val, end=' ')
G = (n ** 2 for n in range(12))
list(G)
list(G)
G = (n**2 for n in range(12))
for n in G:
print(n, end=' ')
if n > 30: break
print("\ndoing something in between")
for n in G:
print(n, end=' ')
L1 = [n ** 2 for n in range(12)]
L2 = []
for n in range(12):
L2.append(n ** 2)
print(L1)
print(L2)
G1 = (n ** 2 for n in range(12))
def gen():
for n in range(12):
yield n ** 2
G2 = gen()
print(*G1)
print(*G2)
# Generate a list of candidates
L = [n for n in range(2, 40)]
print(L)
# Remove all multiples of the first value
L = [n for n in L if n == L[0] or n % L[0] > 0]
print(L)
# Remove all multiples of the second value
L = [n for n in L if n == L[1] or n % L[1] > 0]
print(L)
# Remove all multiples of the third value
L = [n for n in L if n == L[2] or n % L[2] > 0]
print(L)
def gen_primes(N):
"""Generate primes up to N"""
primes = set()
for n in range(2, N):
if all(n % p > 0 for p in primes):
primes.add(n)
yield n
print(*gen_primes(100))
| 0.231527 | 0.960952 |
<img src="CdeC.png">
```
from IPython.display import YouTubeVideo, HTML
YouTubeVideo('VIxciS1B9eo')
```
## Bucle for
```
mi_lista = [0,1,2,3,4,5,6,7,8,9,10]
for i in mi_lista:
print (i)
dias_semana = ['lunes','martes','miercoles','jueves','viernes','sabado','domingo']
for dia in dias_semana:
print (dia)
dias_semana = ['lunes','martes','miercoles','jueves','viernes','sabado','domingo']
for dia in dias_semana:
if dia == 'sabado' or dia == 'domingo':
print (dia, 'Me levanto tarde')
else:
print (dia, 'Me levanto temprano de las 3am')
```
## Funciones
Las funciones nos facilitan la programación porque no tenemos que escribir nuevamente todo el codigo de una rutina que vamos a reutilizar
Una función se define en python como:
```python
def mi_funcion(var1,var2):
# el algoritmo
return x
```
```
# ejemplo
def mi_funcion(x,y):
return x+y
print (mi_funcion(18,2))
#ejemplo
def contar_letras(texto):
n = len(texto)
return n
def contar_palabras(texto):
lista = texto.split(' ')
n = len(lista)
return n
def contar_palabras_letras(texto):
palabras = contar_palabras(texto)
letras = contar_letras(texto)
return [palabras, letras]
print (contar_palabras_letras('hola'))
contar_palabras_letras ('La Moto: Vehículo automóvil de dos ruedas y manubrio, que tiene capacidad para una o dos personas.')
def hora_me_levanto(dia):
if dia == 'domingo' or dia == 'sabado':
resultado = 'me levanto a las 10am'
else:
resultado = 'me levanto a las 8am'
return resultado
hora_me_levando('lunes')
# ejemplo
def potencia(x,n):
a = 1
for i in range(n): # range(n) genera una lista de numeros de 0 a n-1 de 1 en 1
a = a*x
return a
def factorial(n):
if n == 0:
return 1
if n < 0:
return 'valor negativo'
factorial = 1
for i in range(1,n+1):
factorial = factorial*i
return factorial
print (potencia(3,3))
print (factorial(4))
```
## Reto de Programación
- Construya una función que retorne el nombre de una de sus compañeros de grupo cuando se ingresa el número de
tarjeta de identidad
```python
def encontrar_nombre(numero_identidad):
# codigo
return nombre_completo
```
- La serie de Fibonacci es muy importante en varias areas del conocimiento. Esta se define como:
$$f_{0} = 0 ,$$
$$f_{1} = 1,$$
$$f_{n} = f_{n-1} + f_{n-2}$$
Es decir, el siguiente valor es la suma de los dos anteriores.
$$ f_{2} = 1 + 0,$$
$$f_{3} = 1 + 1,$$
$$f_{4} = 2 + 1$$
Escriba una función que retorne la serie de Fibonacci de algun número $n$.
Por ejemplo para $n=4$, la función debe devolver la lista [0,1,1,2,3]
```
datos = {'Laura':'1000788535', 'Tatiana':'1001050843', 'Yoiner':'110309679', 'Kevin':'1000364768'}
print (datos['Laura'])
print (datos['Tatiana'])
print (datos['Yoiner'])
print (datos['Kevin'])
def encontrar_nombre(Nombre):
return (datos[Nombre])
encontrar_nombre ('Laura')
```
# Librerias
Las librerias contienen funciones que nos ayudan resolver problemas complejos y nos facilitan la programación.
```python
import pandas # Pandas nos permite leer archivos de excel, filtrar, y hacer estadisticas sobre tabalas
import numpy # Numpy contiene funciones de operaciones matematicas y algebra de matrices
import matplotlib # Matplotlib es una libreria que nos ayuda a graficar datos y funciones matematicas
```
```
# ejemplo, La hora actual del servidor
import datetime
print (datetime.datetime.now())
# ejemplo, Transpuesta de una matriz
import numpy as np
A = np.matrix([[3, 6, -5],
[1, -3, 2],
[5, -1, 4]])
print (A.shape) # las dimensiones de la matriz
print (A.transpose()) # tarnspuesta de la matriz A
```
## Dibujar Gráficas
```
import matplotlib.pylab as plt
plt.figure()
x = [0,1,2,3,4,5,6,7,8,9,10]
y = [0,11,22,33,44,55,66,17,8,9,10]
plt.scatter(x,y,c = 'black',s=50)
plt.show()
%matplotlib notebook
# ejemplo, Gráfica de y = x**2
import matplotlib.pylab as plt
x = list(range(-50,50))
y = [i**2 for i in x]
plt.figure()
plt.scatter(x,y)
plt.title('$y = x^{2}$') # titulo
plt.xlabel('x') # titulo eje x
plt.ylabel('y') # titulo eje y
plt.show()
x = np.linspace(0, 2 * np.pi, 500)
y1 = np.sin(x)
y2 = np.sin(3 * x)
fig, ax = plt.subplots()
ax.fill(x, y1, 'b', x, y2, 'r', alpha=0.3)
plt.show()
# ejemplo, Crear una tabal de datos de sus compañeros
import pandas as pd
nombres = ['Jocelyn', 'Laura','Luis Alejandro']
apellidos = ['Kshi', 'Diaz', 'Mahecha']
pais = ['Estados Unidos', 'Colombia', 'Colombia']
pd.DataFrame({'nombre': nombres, 'apellido': apellidos, 'pais': pais})
```
## Reto de Programación
Cree un dataframe ó tabla que tenga las siguinetes columnas: t, a, v, y:
- t es el tiempo y va de 0 a 100
- a es la aceleración de la gravedad a = 10
- v es la velocidad, y es función de t : $v = 20 - at$
- y es función de t: $y = -5t^{2}$
Grafique y, v, a en función de t
# Pandas y Tablas de datos
```
temperatura_global = pd.read_csv('GlobalTemperatures.csv')
```
# Analisis Temperaturas
https://www.dkrz.de/Nutzerportal-en/doku/vis/sw/python-matplotlib/matplotlib-sourcecode/python-matplotlib-example-contour-filled-plot
https://data.giss.nasa.gov/gistemp/maps/
```
from __future__ import (absolute_import, division, print_function)
from six.moves import (filter, input, map, range, zip) # noqa
import matplotlib.cm as mpl_cm
import matplotlib.pyplot as plt
import iris
import iris.quickplot as qplt
fname = iris.sample_data_path('air_temp.pp')
temperature_cube = iris.load_cube(fname)
# Load a Cynthia Brewer palette.
brewer_cmap = mpl_cm.get_cmap('brewer_OrRd_09')
# Draw the contour with 25 levels.
plt.figure()
qplt.contourf(temperature_cube, 25)
# Add coastlines to the map created by contourf.
plt.gca().coastlines()
plt.show()
# Draw the contours, with n-levels set for the map colours (9).
# NOTE: needed as the map is non-interpolated, but matplotlib does not provide
# any special behaviour for these.
plt.figure()
qplt.contourf(temperature_cube, brewer_cmap.N, cmap=brewer_cmap)
# Add coastlines to the map created by contourf.
plt.gca().coastlines()
plt.show()
```
|
github_jupyter
|
from IPython.display import YouTubeVideo, HTML
YouTubeVideo('VIxciS1B9eo')
mi_lista = [0,1,2,3,4,5,6,7,8,9,10]
for i in mi_lista:
print (i)
dias_semana = ['lunes','martes','miercoles','jueves','viernes','sabado','domingo']
for dia in dias_semana:
print (dia)
dias_semana = ['lunes','martes','miercoles','jueves','viernes','sabado','domingo']
for dia in dias_semana:
if dia == 'sabado' or dia == 'domingo':
print (dia, 'Me levanto tarde')
else:
print (dia, 'Me levanto temprano de las 3am')
def mi_funcion(var1,var2):
# el algoritmo
return x
# ejemplo
def mi_funcion(x,y):
return x+y
print (mi_funcion(18,2))
#ejemplo
def contar_letras(texto):
n = len(texto)
return n
def contar_palabras(texto):
lista = texto.split(' ')
n = len(lista)
return n
def contar_palabras_letras(texto):
palabras = contar_palabras(texto)
letras = contar_letras(texto)
return [palabras, letras]
print (contar_palabras_letras('hola'))
contar_palabras_letras ('La Moto: Vehículo automóvil de dos ruedas y manubrio, que tiene capacidad para una o dos personas.')
def hora_me_levanto(dia):
if dia == 'domingo' or dia == 'sabado':
resultado = 'me levanto a las 10am'
else:
resultado = 'me levanto a las 8am'
return resultado
hora_me_levando('lunes')
# ejemplo
def potencia(x,n):
a = 1
for i in range(n): # range(n) genera una lista de numeros de 0 a n-1 de 1 en 1
a = a*x
return a
def factorial(n):
if n == 0:
return 1
if n < 0:
return 'valor negativo'
factorial = 1
for i in range(1,n+1):
factorial = factorial*i
return factorial
print (potencia(3,3))
print (factorial(4))
def encontrar_nombre(numero_identidad):
# codigo
return nombre_completo
datos = {'Laura':'1000788535', 'Tatiana':'1001050843', 'Yoiner':'110309679', 'Kevin':'1000364768'}
print (datos['Laura'])
print (datos['Tatiana'])
print (datos['Yoiner'])
print (datos['Kevin'])
def encontrar_nombre(Nombre):
return (datos[Nombre])
encontrar_nombre ('Laura')
import pandas # Pandas nos permite leer archivos de excel, filtrar, y hacer estadisticas sobre tabalas
import numpy # Numpy contiene funciones de operaciones matematicas y algebra de matrices
import matplotlib # Matplotlib es una libreria que nos ayuda a graficar datos y funciones matematicas
# ejemplo, La hora actual del servidor
import datetime
print (datetime.datetime.now())
# ejemplo, Transpuesta de una matriz
import numpy as np
A = np.matrix([[3, 6, -5],
[1, -3, 2],
[5, -1, 4]])
print (A.shape) # las dimensiones de la matriz
print (A.transpose()) # tarnspuesta de la matriz A
import matplotlib.pylab as plt
plt.figure()
x = [0,1,2,3,4,5,6,7,8,9,10]
y = [0,11,22,33,44,55,66,17,8,9,10]
plt.scatter(x,y,c = 'black',s=50)
plt.show()
%matplotlib notebook
# ejemplo, Gráfica de y = x**2
import matplotlib.pylab as plt
x = list(range(-50,50))
y = [i**2 for i in x]
plt.figure()
plt.scatter(x,y)
plt.title('$y = x^{2}$') # titulo
plt.xlabel('x') # titulo eje x
plt.ylabel('y') # titulo eje y
plt.show()
x = np.linspace(0, 2 * np.pi, 500)
y1 = np.sin(x)
y2 = np.sin(3 * x)
fig, ax = plt.subplots()
ax.fill(x, y1, 'b', x, y2, 'r', alpha=0.3)
plt.show()
# ejemplo, Crear una tabal de datos de sus compañeros
import pandas as pd
nombres = ['Jocelyn', 'Laura','Luis Alejandro']
apellidos = ['Kshi', 'Diaz', 'Mahecha']
pais = ['Estados Unidos', 'Colombia', 'Colombia']
pd.DataFrame({'nombre': nombres, 'apellido': apellidos, 'pais': pais})
temperatura_global = pd.read_csv('GlobalTemperatures.csv')
from __future__ import (absolute_import, division, print_function)
from six.moves import (filter, input, map, range, zip) # noqa
import matplotlib.cm as mpl_cm
import matplotlib.pyplot as plt
import iris
import iris.quickplot as qplt
fname = iris.sample_data_path('air_temp.pp')
temperature_cube = iris.load_cube(fname)
# Load a Cynthia Brewer palette.
brewer_cmap = mpl_cm.get_cmap('brewer_OrRd_09')
# Draw the contour with 25 levels.
plt.figure()
qplt.contourf(temperature_cube, 25)
# Add coastlines to the map created by contourf.
plt.gca().coastlines()
plt.show()
# Draw the contours, with n-levels set for the map colours (9).
# NOTE: needed as the map is non-interpolated, but matplotlib does not provide
# any special behaviour for these.
plt.figure()
qplt.contourf(temperature_cube, brewer_cmap.N, cmap=brewer_cmap)
# Add coastlines to the map created by contourf.
plt.gca().coastlines()
plt.show()
| 0.481941 | 0.882022 |
# Supplementary Practice Problems
These are similar to programming problems you may encounter in the mid-terms. They are not graded but we will review them in lab sessions.
**1**. (10 points) Normalize the $3 \times 4$ diagonal matrix with diagonal (1, ,2, 3) so all rows have mean 0 and standard deviation 1. The matrix has 0 everywhere not on the diagonal.
**2**. (10 points) A fixed point of a funciton is a value that remains the same when the funciton is applied to it, that is $f(x) = x$. Write a function that finds the fixed poitn of another function $f$ given an intiial value $x_0$. For example, if
$$f(x) \rightarrow \sqrt{x}$$
and
$x_0$ is any positive real number, then the function should return 1 since
$$\sqrt{1} = 1$$
Not all funcitons have a fixed point - if it taakes over 1,000 iterations, the fucntion shold return None.
- Use the function signature `fixed_point(f, x0, max_iter=1000)`.
- Test with `fixed_point(np.sqrt, 10)`.
**3**. (10 points) Use `np.fromfunction` to construc the following matrices
```python
array([[5, 0, 0, 0, 5],
[0, 4, 0, 4, 0],
[0, 0, 3, 0, 0],
[0, 2, 0, 2, 0],
[1, 0, 0, 0, 1]])
```
```python
array([[0, 1, 0, 0, 0, 0],
[1, 0, 1, 0, 0, 0],
[0, 1, 0, 1, 0, 0],
[0, 0, 1, 0, 1, 0],
[0, 0, 0, 1, 0, 1],
[0, 0, 0, 0, 1, 0]])
```
```python
array([[6, 5, 4, 3, 4, 5, 6],
[5, 4, 3, 2, 3, 4, 5],
[4, 3, 2, 1, 2, 3, 4],
[3, 2, 1, 0, 1, 2, 3],
[4, 3, 2, 1, 2, 3, 4],
[5, 4, 3, 2, 3, 4, 5],
[6, 5, 4, 3, 4, 5, 6]])
```
**4**. (10 points) Read the `mtcars` data frame from R to a `pandas` DataFrame. Find the mean `wt` and `mpg` for all cars grouped by the number of `gear`s.
**5**. (10 points) The QQ (Quirkiness Quotient) has a normal distribution with $\mu$ = 100 and $\sigma$ = 20. If your QQ is at the 92nd percentile, what is your QQ?
- Solve this analytically using some distribution function
- Solve this using a simulation of 1,000,000 people
- Plot the QQ PDF for QQs between 40 and 160 as a blue curve, shading the region below your QQ in blue with 50% transparency.
**6**. (10 points) Simulate $n$ coin toss experiments, in which you toss a coin $k$ times for each experiment. Find the maximum run length of heads (e.g. the sequence `T,T,H,H,H,T,H,H` has a maximum run length of 3 heads in each experiment.
**7**. (10 points) Use the DNA sequence given below to answer these questions:
- Generate all sequences formed using a shifting window of size 3 and print the sequence with the highest count. If there are ties, print all the tied sequences. Do not use any import statement.
```
dna = '''
TCGGATGATTGCACTTATTCTCCGTTTGTTTGTGTGTTGCTGGGGGGGGACGCCTGCACGCTGTTCCAGTGCGTCGTCGT
ATCTTGGTTTGCCTCGGGGGGTGGGGCTGGAGCCTACCAGGTGTCGGTCGGATGTTTTGTCTCTGTGTGTCGAGGGTCGT
GTGGAGTCCCGGCGGGTGGGTGCTGCTGGGTGGTTGACACAGTGCGTGGTGGGTGCTTCAGCGGGGGGTGCCTCGCGCGT
GGCTGGGGTGTTGTAGTTGTCGTTTGGAAAGTGTGCGGCATAGGGGGGATGAAATCCGGTTGACCAAGATTCGGGTCGCG
TTTGGGGATTAGGATAGTTGGGTTTGGAAGGCGCGGGTATTGCCCACGTTCTTTGGCGGGGCGAGGGTGTTGGTACTCCA
TGTCCAGGCCGCTTGCCTACGGACTGTAGAGGTTCGGGAGGCGCTGGCCGGCGTGTGTTCCCTAGGTTTTGAGATAAGAG
'''
```
**8**. (10 points) Ignore spaces, case and punctuation when finding palindromes. Write code to find the palindromes among the 12 phrases below, returning the phrases that are palindromes in a list.
```
Daedalus: nine. Peninsula: dead.
Dammit, I'm mad!
Deliver me from evil.
Dennis and Edna sinned.
Devil never even lived.
Deviled eggs sure taste good.
Did Hannah see bees? Hannah did.
Do geese see God?
Do mice see God?
Dogma: I am God
Dogma: DNA makdes RNA makes protein.
Dubya won? No way, bud.
```
|
github_jupyter
|
array([[5, 0, 0, 0, 5],
[0, 4, 0, 4, 0],
[0, 0, 3, 0, 0],
[0, 2, 0, 2, 0],
[1, 0, 0, 0, 1]])
array([[0, 1, 0, 0, 0, 0],
[1, 0, 1, 0, 0, 0],
[0, 1, 0, 1, 0, 0],
[0, 0, 1, 0, 1, 0],
[0, 0, 0, 1, 0, 1],
[0, 0, 0, 0, 1, 0]])
array([[6, 5, 4, 3, 4, 5, 6],
[5, 4, 3, 2, 3, 4, 5],
[4, 3, 2, 1, 2, 3, 4],
[3, 2, 1, 0, 1, 2, 3],
[4, 3, 2, 1, 2, 3, 4],
[5, 4, 3, 2, 3, 4, 5],
[6, 5, 4, 3, 4, 5, 6]])
dna = '''
TCGGATGATTGCACTTATTCTCCGTTTGTTTGTGTGTTGCTGGGGGGGGACGCCTGCACGCTGTTCCAGTGCGTCGTCGT
ATCTTGGTTTGCCTCGGGGGGTGGGGCTGGAGCCTACCAGGTGTCGGTCGGATGTTTTGTCTCTGTGTGTCGAGGGTCGT
GTGGAGTCCCGGCGGGTGGGTGCTGCTGGGTGGTTGACACAGTGCGTGGTGGGTGCTTCAGCGGGGGGTGCCTCGCGCGT
GGCTGGGGTGTTGTAGTTGTCGTTTGGAAAGTGTGCGGCATAGGGGGGATGAAATCCGGTTGACCAAGATTCGGGTCGCG
TTTGGGGATTAGGATAGTTGGGTTTGGAAGGCGCGGGTATTGCCCACGTTCTTTGGCGGGGCGAGGGTGTTGGTACTCCA
TGTCCAGGCCGCTTGCCTACGGACTGTAGAGGTTCGGGAGGCGCTGGCCGGCGTGTGTTCCCTAGGTTTTGAGATAAGAG
'''
Daedalus: nine. Peninsula: dead.
Dammit, I'm mad!
Deliver me from evil.
Dennis and Edna sinned.
Devil never even lived.
Deviled eggs sure taste good.
Did Hannah see bees? Hannah did.
Do geese see God?
Do mice see God?
Dogma: I am God
Dogma: DNA makdes RNA makes protein.
Dubya won? No way, bud.
| 0.207777 | 0.987326 |
# Lista de Exercícios 2
Métodos Numéricos para Engenharia - Turma D
Nome: Vinícius de Castro Cantuária
Matrícula: 14/0165169
Observações:
0. A lista de exercícios deve ser entregue no moodle da disciplina.
0. A lista de exercícios deve ser respondida neste único arquivo (.ipynb). Responda a cada questão na célula imediatamente abaixo do seu enunciado.
0. Não se esqueça de alterar o nome do arquivo e o cabeçalho acima, colocando seu nome e matrícula.
0. A lista é uma atividade avaliativa e individual. Não será tolerado qualquer tipo de plágio.
```
# Deixe-me incluir o conjunto de módulos do Python científico para você.
%pylab inline
```
---
## Questão 01
O Método da Bisseção é utilizado para encontrar a raíz de uma função dentro de um intervalo definido, de forma iterativa. A cada iteração o método encontra uma nova aproximação para o local da raíz da função dada.
Dado um intervalo inicial $[A, B]$, utilize o Método da Bisseção para encontrar as $10$ primeiras aproximações para a raíz da função:
$$ f(x) = \frac{x^4 - x^3 - 5x^2 + x}{x^4 + 4} $$
### Entrada
A entrada consiste de dois valores reais $-10^4 \leq A < B \leq 10^4$ que indicam os limites inferior e superior do intervalo, respectivamente.
### Saída
Na saída devem ser impressos 10 valores reais com 6 casas de precisão. Após cada valor deve ser impressa uma quebra de linha. Caso a raíz seja encontrada, esta deve ser a última linha impressa, também com 6 casas decimais de precisão. Por fim, caso o intervalo inicial não seja válido, a mensagem "_Intervalo invalido_" deve ser impressa.
#### Entrada de Teste
$\texttt{2.00}$
$\texttt{10.00}$
#### Saída de Teste
$\texttt{6.000000}$
$\texttt{4.000000}$
$\texttt{3.000000}$
$\texttt{2.500000}$
$\texttt{2.750000}$
$\texttt{2.625000}$
$\texttt{2.687500}$
$\texttt{2.718750}$
$\texttt{2.703125}$
$\texttt{2.710938}$
```
def f(x):
return (x**4 - x**3 - 5*x**2 + x) / (x**4 + 4)
a = float(input())
b = float(input())
if(f(a) * f(b) > 0):
print('intervalo invalido')
else:
for i in range(10):
xmed = (a+b)/2
if(f(a)*f(xmed) == 0):
print('raiz encontrada: %.6f' % xmed)
break
elif(f(a)*f(xmed) < 0):
b = xmed
else:
a = xmed
print('%.6f' % xmed)
```
---
## Questão 02
Em zeros de funções, o Método da Newton é um método iterativo utilizado para encontrar a raíz de uma função, dado um ponto inicial. A cada iteração o método encontra uma nova aproximação para o local da raíz da função desejada.
Dado um ponto inicial $x_0$, utilize o Método de Newton para encontrar as 10 primeiras aproximações para a raíz da função:
$$ f(x) = x^5 - 12x^3 + x $$
### Entrada
A entrada consiste de um valor real $-10^4 \leq x_0 \leq 10^4$ que indica o ponto inicial.
### Saída
Na saída devem ser impressos 10 valores reais com 6 casas de precisão, cada valor representa uma aproximação para a raíz da função $f(x)$ dada. Após cada valor deve ser impressa uma quebra de linha.
#### Entrada de Teste
$\texttt{6.00}$
#### Saída de Teste
$\texttt{6.000000}$
$\texttt{4.999036}$
$\texttt{4.267066}$
$\texttt{3.782012}$
$\texttt{3.529772}$
$\texttt{3.457568}$
$\texttt{3.451999}$
$\texttt{3.451968}$
$\texttt{3.451968}$
$\texttt{3.451968}$
```
def f(x):
return x**5 - 12*x**3 + x
def f_linha(x):
return 5*x**4 - 36*x**2 + 1
x = float(input())
for i in range(10):
print('%.6f' % x)
x -= f(x) / f_linha(x)
```
---
## Questão 03
O Método da Falsa-Posição é um método iterativo utilizado para encontrar a raíz de uma função, dado um intervalo inicial. A cada iteração o método encontra uma nova aproximação para o local da raíz da função desejada.
Dado um intervalo inicial $[A,B]$, utilize o Método da Falsa-Posição para encontrar as 10 primeiras aproximações para a raíz da função:
$$ f(x) = \frac{x^2 + 3x - 3}{x^2 + 1} $$
### Entrada
A entrada consiste de dois valores reais $-10^4 \leq A < B \leq 10^4$ que indicam os limites inferior e superior do intervalo inicial, respectivamente.
### Saída
Na saída devem ser impressos 10 valores reais com 6 casas de precisão. Após cada valor deve ser impressa uma quebra de linha. Caso a raíz seja encontrada, esta deve ser a última linha impressa, também com 6 casas decimais de precisão. Caso o intervalo inicial não seja válido, a mensagem "_Intervalo invalido_" deve ser impressa.
#### Entrada de Teste
$\texttt{-1.00}$
$\texttt{10.00}$
#### Saída de Teste
$\texttt{6.318841}$
$\texttt{3.733514}$
$\texttt{1.971825}$
$\texttt{0.908998}$
$\texttt{0.702668}$
$\texttt{0.799252}$
$\texttt{0.791794}$
$\texttt{0.791320}$
$\texttt{0.791290}$
$\texttt{0.791288}$
```
def f(x):
return (x**2 + 3*x - 3) / (x**2 + 1)
a = float(input())
b = float(input())
err = 1e-8
if f(a)*f(b) > 0:
print('Intervalo invalido')
else:
for i in range(10):
x = (a*f(b) - b*f(a)) / (f(b) - f(a))
if abs(f(x)) < err:
print('%.6lf' % x)
break
if f(a)*f(x) < 0:
b = x
else:
a = x
print('%.6lf' % x)
```
---
## Questão 04
O Método da Secante é um método iterativo utilizado para encontrar a raíz de uma função, dados dois pontos iniciais. A cada iteração o método encontra uma nova aproximação para o local da raíz ($x_i$) da função desejada.
Dados dois pontos iniciais $x_0$ e $x_1$, utilize o Método da Secante para encontrar as aproximações $x_2, x_3, ..., x_9$ para a raíz da função:
$$ f(x) = x^4 - 10x^3 + 8x $$
### Entrada
A entrada consiste de dois valores reais $-10^4 \leq x_0, x_1 \leq 10^4$ que indicam os pontos iniciais.
### Saída
Na saída devem ser impressas dez linhas, cada uma com a seguinte mensagem "$f(x_i) = y_i$", com $0 \leq i \leq 9$. Onde $x_i$ é a i-ésima aproximação para o local da raíz e $y_i$ é o valor da função $f(x)$ no ponto $x_i$. Ambos os valores $x_i$ e $y_i$ devem ser impressos com 6 casas de precisão.
#### Entrada de Teste
$\texttt{-5.00}$
$\texttt{5.00}$
#### Saída de Teste
$\texttt{f(-5.000000) = 1835.000000}$
$\texttt{f(5.000000) = -585.000000}$
$\texttt{f(2.582645) = -107.113005}$
$\texttt{f(2.040822) = -51.325848}$
$\texttt{f(1.542328) = -18.691334}$
$\texttt{f(1.256817) = -7.302917}$
$\texttt{f(1.073731) = -2.460008}$
$\texttt{f(0.980731) = -0.662019}$
$\texttt{f(0.946488) = -0.104578}$
$\texttt{f(0.940064) = -0.006059}$
```
def f(x):
return x**4 - 10*x**3 + 8*x
x0 = float(input())
x1 = float(input())
for i in range(10):
x2 = x1 - f(x1) * (x1 - x0) / (f(x1) - f(x0))
print('f(%.6lf) = %.6lf' % (x0, f(x0)))
x1, x0 = x2, x1
```
---
## Questão 05
O Método da Busca Ternária é um método para encontrar pontos críticos de uma função dentro de um intervalo definido, de forma iterativa. A cada iteração o método encontra uma nova aproximação para o local do ponto crítico da função.
Dado um intervalo inicial $[A, B]$, utilize o Método da Busca Ternária para encontrar as 10 primeiras aproximações para o ponto de mínimo da função:
$$ f(x) = x(x - 1)(x + 1)(x - 2) $$
### Entrada
A entrada consiste de dois valores reais $-10^4 \leq A < B \leq 10^4$ que indicam os limites inferior e superior do intervalo, respectivamente.
### Saída
Na saída devem ser impressos 10 valores reais com 6 casas de precisão. Após cada valor deve ser impressa uma quebra de linha.
#### Entrada de Teste
$\texttt{0.00}$
$\texttt{10.00}$
#### Saída de Teste
$\texttt{3.333333}$
$\texttt{2.222222}$
$\texttt{1.481481}$
$\texttt{1.975309}$
$\texttt{1.646091}$
$\texttt{1.426612}$
$\texttt{1.572931}$
$\texttt{1.670477}$
$\texttt{1.605446}$
$\texttt{1.648800}$
```
def f(x):
return x*(x-1)*(x+1)*(x-2)
a = float(input())
b = float(input())
for i in range(10):
xa = a + (b - a) / 3.0
xb = b - (b - a) / 3.0
if f(xa) > f(xb):
a = xa
print('%.6lf' % xb)
else:
b = xb
print('%.6lf' % xa)
```
---
## Questão 06
O Método da Busca pela Razão Áurea é um método para encontrar pontos críticos de uma função dentro de um intervalo definido, de forma iterativa. A cada iteração o método encontra uma nova aproximação para o local do ponto crítico (máximo ou mínimo) da função.
Dado um polinômio de quarta ordem e um intervalo inicial $[A, B]$, utilize o Método da Busca pela Razão Áurea para encontrar as 10 primeiras aproximações para o ponto de mínimo do polinômio dado.
### Entrada
A entrada é composta por duas linhas. A primeira linha contêm cinco valores reais $-10^4 \leq a,b,c,d,e \leq 10^4$ que indicam os coeficientes do polinômio de quarta ordem abaixo.
$$ p(x) = ax^4 + bx^3 + cx^2 + dx + e $$
A segunda linha consiste de dois valores reais $-10^4 \leq A,B \leq 10^4$ que indicam os limites inferior e superior do intervalo, respectivamente.
### Saída
Na saída devem ser impressos 10 valores reais com 6 casas de precisão. Após cada valor deve ser impressa uma quebra de linha.
#### Entrada de Teste
$\texttt{1.00 0.00 -1.00 0.00 1.00}$
$\texttt{0.00}$
$\texttt{10.00}$
#### Saída de Teste
$\texttt{3.819660}$
$\texttt{2.360680}$
$\texttt{1.458980}$
$\texttt{0.901699}$
$\texttt{0.557281}$
$\texttt{0.557281}$
$\texttt{0.688837}$
$\texttt{0.688837}$
$\texttt{0.688837}$
$\texttt{0.719893}$
```
def f(x):
return A*x**4 + B*x**3 + C*x**2 + D*x + E
A, B, C, D, E = [float(i) for i in input().split()]
a = float(input())
b = float(input())
phi = (1 + 5**.5) / 2
for i in range(10):
xa = b - (b - a) / phi
xb = a + (b - a) / phi
if f(xa) >= f(xb):
a = xa
print('%.6lf' % xb)
else:
b = xb
print('%.6lf' % xa)
```
## Questão 07
O Método da Descida de Gradiente é um método iterativo utilizado para encontrar o ponto crítico de uma função, dado um ponto inicial. A cada iteração o método encontra uma nova aproximação para o local do ponto crítico da função desejada. Esta nova aproximação pode ser encontrada utilizando duas abordagens: passo contante e passo variável.
Dado um ponto inicial $x_0$, utilize o Método da Descida de Gradiente com Passo Constante para encontrar as 10 primeiras aproximações para o ponto crítico da função:
$$ f(x) = -xe^{-x} $$
### Entrada
A entrada consiste de dois números reais. O primeiro valor é $-10^4 \leq x_0 \leq 10^4$ que indica o ponto inicial. O segundo valor é $0 < k \leq 10$ que indica o passo constante $k$ do método.
### Saída
Na saída devem ser impressas dez linhas com a seguinte mensagem "$i\texttt{: }f(x_i)\texttt{ = }y_i$", onde $i$ é o número da iteração, $x_i$ é o local da aproximação atual para o ponto crítico da função e $y_i$ é o valor de $f(x)$ no ponto $x_i$.
#### Entrada de Teste
$\texttt{7.50}$
$\texttt{1.00}$
#### Saída de Teste
$\texttt{0: f(7.500000) = -0.004148}$
$\texttt{1: f(6.500000) = -0.009772}$
$\texttt{2: f(5.500000) = -0.022477}$
$\texttt{3: f(4.500000) = -0.049990}$
$\texttt{4: f(3.500000) = -0.105691}$
$\texttt{5: f(2.500000) = -0.205212}$
$\texttt{6: f(1.500000) = -0.334695}$
$\texttt{7: f(0.500000) = -0.303265}$
$\texttt{8: f(1.500000) = -0.334695}$
$\texttt{9: f(0.500000) = -0.303265}$
```
def f(x):
return -x*np.e**-x
def df(x):
return -np.e**-x + x*np.e**-x
x = float(input())
k = float(input())
for i in range(10):
print('%d: f(%.6lf) = %.6lf' % (i, x, f(x)))
if df(x) < 0:
x += k
else:
x -= k
```
## Questão 08
O Método da Newton é um método iterativo utilizado para encontrar o ponto crítico de uma função, dado um ponto inicial. A cada iteração o método encontra uma nova aproximação para o local do ponto crítico da função desejada.
Dado um ponto inicial $x_0$, utilize o Método de Newton para encontrar as 10 primeiras aproximações para o ponto crítico da função:
$$ f(x) = x^4 - 10x^3 + 8x $$
### Entrada
A entrada consiste de um valor real $-10^4 \leq x_0 \leq 10^4$ que indica o ponto inicial.
### Saída
Na saída devem ser impressas dez linhas com a seguinte mensagem "$i\texttt{: }f(x_i)\texttt{ = }y_i$", onde $i$ é o número da iteração, $x_i$ é o local da aproximação atual para o ponto crítico da função e $y_i$ é o valor de $f(x)$ no ponto $x_i$.
#### Entrada de Teste
$\texttt{8.00}$
#### Saída de Teste
$\texttt{0: f(8.000000) = -960.000000}$
$\texttt{1: f(7.527778) = -994.378043}$
$\texttt{2: f(7.465168) = -994.830507}$
$\texttt{3: f(7.464102) = -994.830633}$
$\texttt{4: f(7.464102) = -994.830633}$
$\texttt{5: f(7.464102) = -994.830633}$
$\texttt{6: f(7.464102) = -994.830633}$
$\texttt{7: f(7.464102) = -994.830633}$
$\texttt{8: f(7.464102) = -994.830633}$
$\texttt{9: f(7.464102) = -994.830633}$
```
def f(x):
return x**4 - 10*x**3 + 8*x
def df(x):
return 4*x**3 - 30*x**2 + 8
def d2f(x):
return 12*x**2 - 60*x
x = float(input())
for i in range(10):
print('%d: f(%.6lf) = %.6lf' % (i, x, f(x)))
x -= df(x) / d2f(x)
```
|
github_jupyter
|
# Deixe-me incluir o conjunto de módulos do Python científico para você.
%pylab inline
def f(x):
return (x**4 - x**3 - 5*x**2 + x) / (x**4 + 4)
a = float(input())
b = float(input())
if(f(a) * f(b) > 0):
print('intervalo invalido')
else:
for i in range(10):
xmed = (a+b)/2
if(f(a)*f(xmed) == 0):
print('raiz encontrada: %.6f' % xmed)
break
elif(f(a)*f(xmed) < 0):
b = xmed
else:
a = xmed
print('%.6f' % xmed)
def f(x):
return x**5 - 12*x**3 + x
def f_linha(x):
return 5*x**4 - 36*x**2 + 1
x = float(input())
for i in range(10):
print('%.6f' % x)
x -= f(x) / f_linha(x)
def f(x):
return (x**2 + 3*x - 3) / (x**2 + 1)
a = float(input())
b = float(input())
err = 1e-8
if f(a)*f(b) > 0:
print('Intervalo invalido')
else:
for i in range(10):
x = (a*f(b) - b*f(a)) / (f(b) - f(a))
if abs(f(x)) < err:
print('%.6lf' % x)
break
if f(a)*f(x) < 0:
b = x
else:
a = x
print('%.6lf' % x)
def f(x):
return x**4 - 10*x**3 + 8*x
x0 = float(input())
x1 = float(input())
for i in range(10):
x2 = x1 - f(x1) * (x1 - x0) / (f(x1) - f(x0))
print('f(%.6lf) = %.6lf' % (x0, f(x0)))
x1, x0 = x2, x1
def f(x):
return x*(x-1)*(x+1)*(x-2)
a = float(input())
b = float(input())
for i in range(10):
xa = a + (b - a) / 3.0
xb = b - (b - a) / 3.0
if f(xa) > f(xb):
a = xa
print('%.6lf' % xb)
else:
b = xb
print('%.6lf' % xa)
def f(x):
return A*x**4 + B*x**3 + C*x**2 + D*x + E
A, B, C, D, E = [float(i) for i in input().split()]
a = float(input())
b = float(input())
phi = (1 + 5**.5) / 2
for i in range(10):
xa = b - (b - a) / phi
xb = a + (b - a) / phi
if f(xa) >= f(xb):
a = xa
print('%.6lf' % xb)
else:
b = xb
print('%.6lf' % xa)
def f(x):
return -x*np.e**-x
def df(x):
return -np.e**-x + x*np.e**-x
x = float(input())
k = float(input())
for i in range(10):
print('%d: f(%.6lf) = %.6lf' % (i, x, f(x)))
if df(x) < 0:
x += k
else:
x -= k
def f(x):
return x**4 - 10*x**3 + 8*x
def df(x):
return 4*x**3 - 30*x**2 + 8
def d2f(x):
return 12*x**2 - 60*x
x = float(input())
for i in range(10):
print('%d: f(%.6lf) = %.6lf' % (i, x, f(x)))
x -= df(x) / d2f(x)
| 0.291888 | 0.987664 |
<a href="https://colab.research.google.com/github/lionelsamrat10/machine-learning-a-to-z/blob/main/Classification/Support%20Vector%20Machine(SVM)/support_vector_machine_samrat.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Support Vector Machine (SVM)
## Importing the libraries
```
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
```
## Importing the dataset
```
dataset = pd.read_csv('Social_Network_Ads.csv')
X = dataset.iloc[:, :-1].values
y = dataset.iloc[:, -1].values
```
## Splitting the dataset into the Training set and Test set
```
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.25, random_state = 0)
print(X_train)
print(y_train)
print(X_test)
print(y_test)
```
## Feature Scaling
```
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test = sc.transform(X_test)
print(X_train)
print(X_test)
```
## Training the SVM model on the Training set
```
from sklearn.svm import SVC
classifier = SVC(kernel = 'linear', random_state = 0)
classifier.fit(X_train, y_train)
```
## Predicting a new result
```
print(classifier.predict(sc.transform([[30,87000]])))
```
## Predicting the Test set results
```
y_pred = classifier.predict(X_test)
print(np.concatenate((y_pred.reshape(len(y_pred),1), y_test.reshape(len(y_test),1)),1))
```
## Making the Confusion Matrix
```
from sklearn.metrics import confusion_matrix, accuracy_score
cm = confusion_matrix(y_test, y_pred)
print(cm)
accuracy_score(y_test, y_pred)
```
## Visualising the Training set results
```
from matplotlib.colors import ListedColormap
X_set, y_set = sc.inverse_transform(X_train), y_train
X1, X2 = np.meshgrid(np.arange(start = X_set[:, 0].min() - 10, stop = X_set[:, 0].max() + 10, step = 0.25),
np.arange(start = X_set[:, 1].min() - 1000, stop = X_set[:, 1].max() + 1000, step = 0.25))
plt.contourf(X1, X2, classifier.predict(sc.transform(np.array([X1.ravel(), X2.ravel()]).T)).reshape(X1.shape),
alpha = 0.75, cmap = ListedColormap(('red', 'green')))
plt.xlim(X1.min(), X1.max())
plt.ylim(X2.min(), X2.max())
for i, j in enumerate(np.unique(y_set)):
plt.scatter(X_set[y_set == j, 0], X_set[y_set == j, 1], c = ListedColormap(('red', 'green'))(i), label = j)
plt.title('SVM (Training set)')
plt.xlabel('Age')
plt.ylabel('Estimated Salary')
plt.legend()
plt.show()
```
## Visualising the Test set results
```
from matplotlib.colors import ListedColormap
X_set, y_set = sc.inverse_transform(X_test), y_test
X1, X2 = np.meshgrid(np.arange(start = X_set[:, 0].min() - 10, stop = X_set[:, 0].max() + 10, step = 0.25),
np.arange(start = X_set[:, 1].min() - 1000, stop = X_set[:, 1].max() + 1000, step = 0.25))
plt.contourf(X1, X2, classifier.predict(sc.transform(np.array([X1.ravel(), X2.ravel()]).T)).reshape(X1.shape),
alpha = 0.75, cmap = ListedColormap(('red', 'green')))
plt.xlim(X1.min(), X1.max())
plt.ylim(X2.min(), X2.max())
for i, j in enumerate(np.unique(y_set)):
plt.scatter(X_set[y_set == j, 0], X_set[y_set == j, 1], c = ListedColormap(('red', 'green'))(i), label = j)
plt.title('SVM (Test set)')
plt.xlabel('Age')
plt.ylabel('Estimated Salary')
plt.legend()
plt.show()
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
dataset = pd.read_csv('Social_Network_Ads.csv')
X = dataset.iloc[:, :-1].values
y = dataset.iloc[:, -1].values
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.25, random_state = 0)
print(X_train)
print(y_train)
print(X_test)
print(y_test)
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test = sc.transform(X_test)
print(X_train)
print(X_test)
from sklearn.svm import SVC
classifier = SVC(kernel = 'linear', random_state = 0)
classifier.fit(X_train, y_train)
print(classifier.predict(sc.transform([[30,87000]])))
y_pred = classifier.predict(X_test)
print(np.concatenate((y_pred.reshape(len(y_pred),1), y_test.reshape(len(y_test),1)),1))
from sklearn.metrics import confusion_matrix, accuracy_score
cm = confusion_matrix(y_test, y_pred)
print(cm)
accuracy_score(y_test, y_pred)
from matplotlib.colors import ListedColormap
X_set, y_set = sc.inverse_transform(X_train), y_train
X1, X2 = np.meshgrid(np.arange(start = X_set[:, 0].min() - 10, stop = X_set[:, 0].max() + 10, step = 0.25),
np.arange(start = X_set[:, 1].min() - 1000, stop = X_set[:, 1].max() + 1000, step = 0.25))
plt.contourf(X1, X2, classifier.predict(sc.transform(np.array([X1.ravel(), X2.ravel()]).T)).reshape(X1.shape),
alpha = 0.75, cmap = ListedColormap(('red', 'green')))
plt.xlim(X1.min(), X1.max())
plt.ylim(X2.min(), X2.max())
for i, j in enumerate(np.unique(y_set)):
plt.scatter(X_set[y_set == j, 0], X_set[y_set == j, 1], c = ListedColormap(('red', 'green'))(i), label = j)
plt.title('SVM (Training set)')
plt.xlabel('Age')
plt.ylabel('Estimated Salary')
plt.legend()
plt.show()
from matplotlib.colors import ListedColormap
X_set, y_set = sc.inverse_transform(X_test), y_test
X1, X2 = np.meshgrid(np.arange(start = X_set[:, 0].min() - 10, stop = X_set[:, 0].max() + 10, step = 0.25),
np.arange(start = X_set[:, 1].min() - 1000, stop = X_set[:, 1].max() + 1000, step = 0.25))
plt.contourf(X1, X2, classifier.predict(sc.transform(np.array([X1.ravel(), X2.ravel()]).T)).reshape(X1.shape),
alpha = 0.75, cmap = ListedColormap(('red', 'green')))
plt.xlim(X1.min(), X1.max())
plt.ylim(X2.min(), X2.max())
for i, j in enumerate(np.unique(y_set)):
plt.scatter(X_set[y_set == j, 0], X_set[y_set == j, 1], c = ListedColormap(('red', 'green'))(i), label = j)
plt.title('SVM (Test set)')
plt.xlabel('Age')
plt.ylabel('Estimated Salary')
plt.legend()
plt.show()
| 0.557845 | 0.98544 |
# Object Detection Demo
Welcome to the object detection inference walkthrough! This notebook will walk you step by step through the process of using a pre-trained model to detect objects in an image.
# Imports
```
import numpy as np
import os
import six.moves.urllib as urllib
import sys
import tarfile
import tensorflow as tf
import zipfile
from collections import defaultdict
from io import StringIO
from matplotlib import pyplot as plt
from PIL import Image
```
## Env setup
```
# This is needed to display the images.
%matplotlib inline
# This is needed since the notebook is stored in the object_detection folder.
sys.path.append("..")
sys.path
```
## Object detection imports
Here are the imports from the object detection module.
```
from utils import label_map_util
from utils import visualization_utils as vis_util
```
# Model preparation
## Variables
Any model exported using the `export_inference_graph.py` tool can be loaded here simply by changing `PATH_TO_CKPT` to point to a new .pb file.
By default we use an "SSD with Mobilenet" model here. See the [detection model zoo](g3doc/detection_model_zoo.md) for a list of other models that can be run out-of-the-box with varying speeds and accuracies.
```
# What model to download.
MODEL_NAME = 'ssd_mobilenet_v1_coco_11_06_2017'
MODEL_FILE = MODEL_NAME + '.tar.gz'
DOWNLOAD_BASE = 'http://download.tensorflow.org/models/object_detection/'
# Path to frozen detection graph. This is the actual model that is used for the object detection.
PATH_TO_CKPT = MODEL_NAME + '/frozen_inference_graph.pb'
# List of the strings that is used to add correct label for each box.
PATH_TO_LABELS = os.path.join('data', 'mscoco_label_map.pbtxt')
NUM_CLASSES = 90
```
## Download Model
```
opener = urllib.request.URLopener()
opener.retrieve(DOWNLOAD_BASE + MODEL_FILE, MODEL_FILE)
tar_file = tarfile.open(MODEL_FILE)
for file in tar_file.getmembers():
file_name = os.path.basename(file.name)
if 'frozen_inference_graph.pb' in file_name:
tar_file.extract(file, os.getcwd())
```
## Load a (frozen) Tensorflow model into memory.
```
detection_graph = tf.Graph()
with detection_graph.as_default():
od_graph_def = tf.GraphDef()
with tf.gfile.GFile(PATH_TO_CKPT, 'rb') as fid:
serialized_graph = fid.read()
od_graph_def.ParseFromString(serialized_graph)
tf.import_graph_def(od_graph_def, name='')
```
## Loading label map
Label maps map indices to category names, so that when our convolution network predicts `5`, we know that this corresponds to `airplane`. Here we use internal utility functions, but anything that returns a dictionary mapping integers to appropriate string labels would be fine
```
label_map = label_map_util.load_labelmap(PATH_TO_LABELS)
categories = label_map_util.convert_label_map_to_categories(label_map, max_num_classes=NUM_CLASSES, use_display_name=True)
category_index = label_map_util.create_category_index(categories)
```
## Helper code
```
def load_image_into_numpy_array(image):
(im_width, im_height) = image.size
return np.array(image.getdata()).reshape(
(im_height, im_width, 3)).astype(np.uint8)
```
# Detection
```
# For the sake of simplicity we will use only 2 images:
# image1.jpg
# image2.jpg
# If you want to test the code with your images, just add path to the images to the TEST_IMAGE_PATHS.
PATH_TO_TEST_IMAGES_DIR = 'test_images'
TEST_IMAGE_PATHS = [ os.path.join(PATH_TO_TEST_IMAGES_DIR, 'image{}.jpg'.format(i)) for i in range(1, 3) ]
# Size, in inches, of the output images.
IMAGE_SIZE = (12, 8)
with detection_graph.as_default():
with tf.Session(graph=detection_graph) as sess:
for image_path in TEST_IMAGE_PATHS:
image = Image.open(image_path)
# the array based representation of the image will be used later in order to prepare the
# result image with boxes and labels on it.
image_np = load_image_into_numpy_array(image)
# Expand dimensions since the model expects images to have shape: [1, None, None, 3]
image_np_expanded = np.expand_dims(image_np, axis=0)
image_tensor = detection_graph.get_tensor_by_name('image_tensor:0')
# Each box represents a part of the image where a particular object was detected.
boxes = detection_graph.get_tensor_by_name('detection_boxes:0')
# Each score represent how level of confidence for each of the objects.
# Score is shown on the result image, together with the class label.
scores = detection_graph.get_tensor_by_name('detection_scores:0')
classes = detection_graph.get_tensor_by_name('detection_classes:0')
num_detections = detection_graph.get_tensor_by_name('num_detections:0')
# Actual detection.
(boxes, scores, classes, num_detections) = sess.run(
[boxes, scores, classes, num_detections],
feed_dict={image_tensor: image_np_expanded})
# Visualization of the results of a detection.
vis_util.visualize_boxes_and_labels_on_image_array(
image_np,
np.squeeze(boxes),
np.squeeze(classes).astype(np.int32),
np.squeeze(scores),
category_index,
use_normalized_coordinates=True,
line_thickness=8)
plt.figure(figsize=IMAGE_SIZE)
plt.imshow(image_np)
```
|
github_jupyter
|
import numpy as np
import os
import six.moves.urllib as urllib
import sys
import tarfile
import tensorflow as tf
import zipfile
from collections import defaultdict
from io import StringIO
from matplotlib import pyplot as plt
from PIL import Image
# This is needed to display the images.
%matplotlib inline
# This is needed since the notebook is stored in the object_detection folder.
sys.path.append("..")
sys.path
from utils import label_map_util
from utils import visualization_utils as vis_util
# What model to download.
MODEL_NAME = 'ssd_mobilenet_v1_coco_11_06_2017'
MODEL_FILE = MODEL_NAME + '.tar.gz'
DOWNLOAD_BASE = 'http://download.tensorflow.org/models/object_detection/'
# Path to frozen detection graph. This is the actual model that is used for the object detection.
PATH_TO_CKPT = MODEL_NAME + '/frozen_inference_graph.pb'
# List of the strings that is used to add correct label for each box.
PATH_TO_LABELS = os.path.join('data', 'mscoco_label_map.pbtxt')
NUM_CLASSES = 90
opener = urllib.request.URLopener()
opener.retrieve(DOWNLOAD_BASE + MODEL_FILE, MODEL_FILE)
tar_file = tarfile.open(MODEL_FILE)
for file in tar_file.getmembers():
file_name = os.path.basename(file.name)
if 'frozen_inference_graph.pb' in file_name:
tar_file.extract(file, os.getcwd())
detection_graph = tf.Graph()
with detection_graph.as_default():
od_graph_def = tf.GraphDef()
with tf.gfile.GFile(PATH_TO_CKPT, 'rb') as fid:
serialized_graph = fid.read()
od_graph_def.ParseFromString(serialized_graph)
tf.import_graph_def(od_graph_def, name='')
label_map = label_map_util.load_labelmap(PATH_TO_LABELS)
categories = label_map_util.convert_label_map_to_categories(label_map, max_num_classes=NUM_CLASSES, use_display_name=True)
category_index = label_map_util.create_category_index(categories)
def load_image_into_numpy_array(image):
(im_width, im_height) = image.size
return np.array(image.getdata()).reshape(
(im_height, im_width, 3)).astype(np.uint8)
# For the sake of simplicity we will use only 2 images:
# image1.jpg
# image2.jpg
# If you want to test the code with your images, just add path to the images to the TEST_IMAGE_PATHS.
PATH_TO_TEST_IMAGES_DIR = 'test_images'
TEST_IMAGE_PATHS = [ os.path.join(PATH_TO_TEST_IMAGES_DIR, 'image{}.jpg'.format(i)) for i in range(1, 3) ]
# Size, in inches, of the output images.
IMAGE_SIZE = (12, 8)
with detection_graph.as_default():
with tf.Session(graph=detection_graph) as sess:
for image_path in TEST_IMAGE_PATHS:
image = Image.open(image_path)
# the array based representation of the image will be used later in order to prepare the
# result image with boxes and labels on it.
image_np = load_image_into_numpy_array(image)
# Expand dimensions since the model expects images to have shape: [1, None, None, 3]
image_np_expanded = np.expand_dims(image_np, axis=0)
image_tensor = detection_graph.get_tensor_by_name('image_tensor:0')
# Each box represents a part of the image where a particular object was detected.
boxes = detection_graph.get_tensor_by_name('detection_boxes:0')
# Each score represent how level of confidence for each of the objects.
# Score is shown on the result image, together with the class label.
scores = detection_graph.get_tensor_by_name('detection_scores:0')
classes = detection_graph.get_tensor_by_name('detection_classes:0')
num_detections = detection_graph.get_tensor_by_name('num_detections:0')
# Actual detection.
(boxes, scores, classes, num_detections) = sess.run(
[boxes, scores, classes, num_detections],
feed_dict={image_tensor: image_np_expanded})
# Visualization of the results of a detection.
vis_util.visualize_boxes_and_labels_on_image_array(
image_np,
np.squeeze(boxes),
np.squeeze(classes).astype(np.int32),
np.squeeze(scores),
category_index,
use_normalized_coordinates=True,
line_thickness=8)
plt.figure(figsize=IMAGE_SIZE)
plt.imshow(image_np)
| 0.478529 | 0.910386 |
## MINI-PROJECT - SALES BONIFICATION: BUILDING A PROGRAM TO TRIGGER A WARNING MESSAGE VIA SMS WHEN AN EMPLOYEE BEATEN THE SALES GOAL
Imagine that your company has a database containing the names of employees and the total sales made in a specific month and you want to create a simple system for triggering messages, via SMS, when someone reaches the sales target. When an employee hits the sales target, he gets some bonus from the company. Using the Python language, we can build a simple, objective, and easily configure a system that allows the triggering of a warning via SMS when a condition is met. For the example of this mini-project, I defined that the hypothetical company's sales target is R$55,000. When the program identifies that an employee has reached the sale, a message will be triggered and an SMS will be sent to a specific number, which may be from the manager or area coordinator, for example. This type of automation can facilitate some processes within the company and can be adapted for different purposes, which can be extremely useful for decision-makers.
```
# Solution script
# 1- Open databases that are in excel spreadsheet format.
# 2- Check if any value in the sales column is greater than R$55,000.
# 3- If sales is greater than R$55,000 -> Sends an SMS with the name, month and total sales of the employee
# 4- If do not, do not take actions
# First, we need to install Twilio, the library that allows sending SMS via python
!pip install twilio
# Python integration with excel
!pip install openpyxl
# Importing necessary packages
import pandas as pd
from twilio.rest import Client
# Setting Twilio to send SMS
# Use your own account and token, get more information and create your account on https://www.twilio.com/console
# Your Account SID from twilio.com/console
account_sid = "AXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX"
# Your Auth Token from twilio.com/console
auth_token = "XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX"
client = Client(account_sid, auth_token)
# Creating a list of months
month_names = ["January",
"February",
"March",
"April",
"May",
"June"]
# Loading the datasets
for month in month_names:
print(month)
df_sales = pd.read_excel(f'{month}.xlsx')
print(df_sales)
# Verifying if any total sales value in the dataset is greater than R$55,000 and returning the employee's name and total sales
for month in month_names:
df_sales = pd.read_excel(f'{month}.xlsx')
if (df_sales['Sales'] > 55000).any():
employee = df_sales.loc[df_sales['Sales'] > 55000, 'Employee'].values[0]
sales = df_sales.loc[df_sales['Sales'] > 55000, 'Sales'].values[0]
print(f'In {month} someone hit the goal! Employee: {employee}, Total sales: {sales}')
# Setting message on twilio
message = client.messages.create(
to="+xxxxx", # Add the number to which the message should be sent
from_="+xxxxx", # Add your own Twilio number
body=f'In {month} someone hit the goal! Employee: {employee}, Total sales: {sales}')
print(message.sid)
```
from IPython.display import Image
Image("img/print.jpeg")
```
from IPython.display import Image
Image("img/print.jpeg")
```
Example of a message triggered via SMS. You can modify the content of the message and add any necessary content.
|
github_jupyter
|
# Solution script
# 1- Open databases that are in excel spreadsheet format.
# 2- Check if any value in the sales column is greater than R$55,000.
# 3- If sales is greater than R$55,000 -> Sends an SMS with the name, month and total sales of the employee
# 4- If do not, do not take actions
# First, we need to install Twilio, the library that allows sending SMS via python
!pip install twilio
# Python integration with excel
!pip install openpyxl
# Importing necessary packages
import pandas as pd
from twilio.rest import Client
# Setting Twilio to send SMS
# Use your own account and token, get more information and create your account on https://www.twilio.com/console
# Your Account SID from twilio.com/console
account_sid = "AXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX"
# Your Auth Token from twilio.com/console
auth_token = "XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX"
client = Client(account_sid, auth_token)
# Creating a list of months
month_names = ["January",
"February",
"March",
"April",
"May",
"June"]
# Loading the datasets
for month in month_names:
print(month)
df_sales = pd.read_excel(f'{month}.xlsx')
print(df_sales)
# Verifying if any total sales value in the dataset is greater than R$55,000 and returning the employee's name and total sales
for month in month_names:
df_sales = pd.read_excel(f'{month}.xlsx')
if (df_sales['Sales'] > 55000).any():
employee = df_sales.loc[df_sales['Sales'] > 55000, 'Employee'].values[0]
sales = df_sales.loc[df_sales['Sales'] > 55000, 'Sales'].values[0]
print(f'In {month} someone hit the goal! Employee: {employee}, Total sales: {sales}')
# Setting message on twilio
message = client.messages.create(
to="+xxxxx", # Add the number to which the message should be sent
from_="+xxxxx", # Add your own Twilio number
body=f'In {month} someone hit the goal! Employee: {employee}, Total sales: {sales}')
print(message.sid)
from IPython.display import Image
Image("img/print.jpeg")
| 0.445047 | 0.810216 |
## Diffusion preprocessing
Diffusion preprocessing typically comprises of a series of steps, which may vary depending on how the data is acquired. Some consensus has been reached for certain preprocessing steps, while others are still up for debate. The lesson will primarily focus on the preprocessing steps where consensus has been reached. Preprocessing is performed using a few well-known software packages (e.g. FSL, ANTs). For the purposes of these lessons, preprocessing steps requiring these software packages has already been performed for the dataset `ds000221` and the commands required for each step will be provided. This dataset contains single shell diffusion data with 7 b=0 s/mm^2 volumes (non-diffusion weighted) and 60 b=1000 s/mm^2 volumes. In addition, field maps (found in the `fmap` directory are acquired with opposite phase-encoding directions).
To illustrate what the preprocessing step may look like, here is an example preprocessing workflow from QSIPrep (Cieslak _et al_, 2020):

dMRI has some similar challenges to fMRI preprocessing, as well as some unique [ones](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3366862/).
Our preprocesssing of this data will consist of following steps and will make use of sub-010006:
1. Brainmasking the diffusion data
2. Applying FSL `topup` to correct for susceptibility induced distortions
3. FSL Eddy current distortion correction
4. Registration to T1w
### Brainmasking
The first step to the preprocessing workflow is to create an appropriate brainmask from the diffusion data! Start, by first importing the necessary modules. and reading the diffusion data! We will also grab the anatomical T1w image to use later on, as well as the second inversion from the anatomical acquisition for brainmasking purposes.
```
from bids.layout import BIDSLayout
layout = BIDSLayout("../../../data/ds000221", validate=False)
subj='010006'
# Diffusion data
dwi = layout.get(subject=subj, suffix='dwi', extension='nii.gz', return_type='file')[0]
# Anatomical data
t1w = layout.get(subject=subj, suffix='T1w', extension='nii.gz', return_type='file')[0]
import numpy as np
import nibabel as nib
dwi = nib.load(dwi)
dwi_affine = dwi.affine
dwi_data = dwi.get_fdata()
```
DIPY's `segment.mask` module will be used to create a brainmask from this. This module contains a function `median_otsu`, which can be used to segment the brain and provide a binary brainmask! Here, a brainmask will be created using the first non-diffusion volume of the data. We will save this brainmask to be used in our later future preprocessing steps. After creating the brainmask, we will start to correct for distortions in our images.
```
import os
from dipy.segment.mask import median_otsu
# vol_idx is a 1D-array containing the index of the first b0
dwi_brain, dwi_mask = median_otsu(dwi_data, vol_idx=[0])
# Create necessary folders to save mask
out_dir = '../../../data/ds000221/derivatives/uncorrected/sub-%s/ses-01/dwi/' % subj
# Check to see if directory exists, if not create one
if not os.path.exists(out_dir):
os.makedirs(out_dir)
img = nib.Nifti1Image(dwi_mask.astype(np.float32), dwi_affine)
nib.save(img, os.path.join(out_dir, "sub-%s_ses-01_brainmask.nii.gz" % subj))
```

### FSL [`topup`](https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/topup)
Diffusion images, typically acquired using spin-echo echo planar imaging (EPI), are sensitive to non-zero off-resonance fields. One source of these fields is from the susceptibilitiy distribution of the subjects head, otherwise known as susceptibility-induced off-resonance field. This field is approximately constant for all acquired diffusion images. As such, for a set of diffusion volumes, the susceptibility-induced field will be consistent throughout. This is mainly a problem due to geometric mismatches with the anatomical images (e.g. T1w), which are typically unaffected by such distortions.
`topup`, part of the FSL library, estimates and attempts to correct the susceptibility-induced off-resonance field by using 2 (or more) acquisitions, where the acquisition parameters differ such that the distortion differs. Typically, this is done using two acquisitions acquired with opposite phase-encoding directions, which results in the same field creating distortions in opposing directions.
Here, we will make use of the two opposite phase-encoded acquisitions found in the `fmap` directory of each subjet. These are acquired with a diffusion weighting of b = 0 s/mm^2. Alternatively, if these are not available, one can also extract and make use of the non-diffusion weighted images (assuming the data is also acquired with opposite phase encoding directions).
First, we will merge the two files so that all of the volumes are in 1 file.
```
%%bash
mkdir -p ../../../data/ds000221/derivatives/uncorrected_topup/sub-010006/ses-01/dwi/work
fslmerge -t ../../../data/ds000221/derivatives/uncorrected_topup/sub-010006/ses-01/dwi/work/sub-010006_ses-01_acq-SEfmapDWI_epi.nii.gz ../../data/ds000221/sub-010006/ses-01/fmap/sub-010006_ses-01_acq-SEfmapDWI_dir-AP_epi.nii.gz ../../data/ds000221/sub-010006/ses-01/fmap/sub-010006_ses-01_acq-SEfmapDWI_dir-PA_epi.nii.gz
```
Another file we will need to create is a text file containing the information about how the volumes were acquired. Each line in this file will pertain to a single volume in the merged file. The first 3 values of each line refers to the acquisition direction, typically along the y-axis (or anterior-posterior). The final value is the total readout time (from center of first echo to center of final echo), which can be determined from values contained within the json sidecar. Each line will look similar to `[x y z TotalReadoutTime]`. In this case, the file, which we created, is contained within the `pedir.txt` file in the derivative directory.
```
0 1 0 0.04914
0 1 0 0.04914
0 1 0 0.04914
0 -1 0 0.04914
0 -1 0 0.04914
0 -1 0 0.04914
```
With these two inputs, the next step is to make the call to `topup` to estimate the susceptibility-induced field. Within the call, a few parameters are used. Briefly:
* `--imain` specifies the previously merged volume
* `--datain` specifies the text file containing the information regarding the acquisition.
* `--config=b02b0.cnf` makes use of a predefined config file supplied with `topup`, which contains parameters useful to registering with good b=0 s/mm^2 images.
* `--out` defines the output files containing the spline coefficients for the induced field, as well as subject movement parameters
```
%%bash
topup --imain=../../../data/ds000221/derivatives/topup/sub-010006/ses-01/dwi/work/sub-010006_ses-01_acq-SEfmapDWI_epi.nii.gz --datain=../../data/ds000221/derivatives/topup/sub-010006/ses-01/dwi/work/pedir.txt --config=b02b0.cnf --out=../../data/ds000221/derivatives/topup/sub-010006/ses-01/dwi/work/topup
```
Next, we can apply the correction to the entire diffusion weighted volume by using `applytopup` Similar to `topup`, a few parameters are used. Briefly:
* `--imain` specifies the input diffusion weighted volume
* `--datain` again specifies the text file containing information regarding the acquisition - same file previously used
* `--inindex` specifies the index (comma separated list) of the input image to be corrected
* `--topup` name of field/movements (from previous topup step
* `--out` basename for the corrected output image
* `--method` (optional) jacobian modulation (jac) or least-squares resampling (lsr)
```
%%bash
applytopup --imain=../../../data/ds000221/sub-010006/ses-01/dwi/sub-010006_ses-01_dwi.nii.gz --datain=../../../data/ds000221/derivatives/topup/sub-010006/ses-01/dwi/work/pedir.txt --inindex=1 --topup=../../../data/ds000221/derivatives/topup/sub-010006/ses-01/dwi/work/topup --out=../../../data/ds000221/derivatives/topup/sub-010006/ses-01/dwi/dwi --method=jac
```

### FSL [`Eddy`](https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/eddy)
Another source of the non-zero off resonance fields is caused by the rapid switching of diffusion weighting gradients, otherwise known as eddy current-induced off-resonance fields. Additionally, the subject is likely to move during the diffusion protocol, which may be lengthy.
`eddy`, also part of the FSL library, attempts to correct for both eddy current-induced fields and subject movement by reading the gradient table and estimating the distortion volume by volume. This tool is also able to optionally detect and replace outlier slices.
Here, we will demonstrate the application of `eddy` following the `topup` correction step, by making use of both the uncorrected diffusion data, as well as distortion corrections from the previous step. Additionally, a text file, which maps each of the volumes to one of the corresponding acquisition directions from the `pedir.txt` file will have to be created. Finally, similar to `topup`, there are also a number of input parameters which have to be specified:
* `--imain` specifies the undistorted diffusion weighted volume
* `--mask` specifies the brainmask for the undistorted diffusion weighted volume
* `--acqp` specifies the the text file containing information regarding the acquisition that was previously used in `topup`
* `--index` is the text file which maps each diffusion volume to the corresponding acquisition direction
* `--bvecs` specifies the bvec file to the undistorted dwi
* `--bvals` similarily specifies the bval file to the undistorted dwi
* `--topup` specifies the directory and distortion correction files previously estimated by `topup`
* `--out` specifies the prefix of the output files following eddy correction
* `--repol` is a flag, which specifies replacement of outliers
```
%%bash
mkdir -p ../../../data/ds000221/derivatives/uncorrected_topup_eddy/sub-010006/ses-01/dwi/work
# Create an index file mapping the 67 volumes in 4D dwi volume to the pedir.txt file
indx=""
for i in `seq 1 67`; do
indx="$indx 1"
done
echo $indx > ../../../data/ds000221/derivatives/uncorrected_topup_eddy/sub-010006/ses-01/dwi/work/index.txt
eddy_openmp --imain=../../../data/ds000221/sub-010006/ses-01/dwi/sub-010006_ses-01_dwi.nii.gz --mask=../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/dwi/sub-010006_ses-01_brainmask.nii.gz --acqp=../../../data/ds000221/derivatives/uncorrected_topup/sub-010006/ses-01/dwi/work/pedir.txt --index=../../../data/ds000221/derivatives/uncorrected_topup_eddy/sub-010006/ses-01/dwi/work/index.txt --bvecs=../../../data/ds000221/sub-010006/ses-01/dwi/sub-010006_ses-01_dwi.bvec --bvals=../../../data/ds000221/sub-010006/ses-01/dwi/sub-010006_ses-01_dwi.bval --topup=../../../data/ds000221/derivatives/uncorrected_topup/sub-010006/ses-01/dwi/work/topup --out=../../../data/ds000221/derivatives/uncorrected_topup_eddy/sub-010006/ses-01/dwi/dwi --repol
```

### Registration with T1w
The final step to our diffusion processing is registration to an anatomical image (eg. T1-weighted). This is important because the diffusion data, typically acquired using echo planar imaging or EPI, enables faster acquisitions at the cost of lower resolution and introduction of distortions (as seen above). Registration with the anatomical image not only helps to correct for some distortions, it also provides us with a higher resolution, anatomical reference.
First, we will create a brainmask of the anatomical image using the second inversion. To do this, we will use FSL `bet` twice. The first call to `bet` will create a general skullstripped brain. Upon inspection, we can note that there is still some residual areas of the image which were included in the first pass. Calling `bet` a second time, we get a better outline of the brain and brainmask, which we can use for further processing.
```
%%bash
mkdir -p ../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/anat
bet ../../../data/ds000221/sub-010006/ses-01/anat/sub-010006_ses-01_inv-2_mp2rage.nii.gz ../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/anat/sub-010006_ses-01_space-T1w_broadbrain -f 0.6
bet ../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/anat/sub-010006_ses-01_space-T1w_broadbrain ../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/anat/sub-010006_ses-01_space-T1w_brain -f 0.4 -m
mv ../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/anat/sub-010006_ses-01_space-T1w_brain_mask.nii.gz ../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/anat/sub-010006_ses-01_space-T1w_brainmask.nii.gz
```

Note, we use `bet` here, as well as the second inversion of the anatomical image, as it provides us with a better brainmask. The `bet` command above is called to output only the binary mask and the fractional intensity threshold is also increased slightly (to 0.6) provide a smaller outline of the brain initially, and then decreased (to 0.4) to provide a larger outline. The flag `-m` indicates to the tool to create a brainmask in addition to outputting the extracted brain volume. Both the mask and brain volume will be used in our registration step.
Before we get to the registration, we will also update our DWI brainmask by performing a brain extraction using `dipy` on the eddy corrected image. Note that the output of `eddy` is not in BIDS format so we will include the path to the diffusion data manually. We will save both the brainmask and the extracted brain volume. Additionally, we will save a separate volume of only the first b0 to use for the registration.
```
from dipy.segment.mask import median_otsu
# Path of FSL eddy-corrected dwi
dwi = "../../../data/ds000221/derivatives/uncorrected_topup_eddy/sub-010006/ses-01/dwi/dwi.nii.gz"
# Load eddy-corrected diffusion data
dwi = nib.load(dwi)
dwi_affine = dwi.affine
dwi_data = dwi.get_fdata()
dwi_brain, dwi_mask = median_otsu(dwi_data, vol_idx=[0])
dwi_b0 = dwi_brain[:,:,:,0]
# Output directory
out_dir="../../../data/ds000221/derivatives/uncorrected_topup_eddy/sub-010006/ses-01/dwi"
# Save diffusion mask
img = nib.Nifti1Image(dwi_mask.astype(np.float32), dwi_affine)
nib.save(img, os.path.join(out_dir, "sub-010006_ses-01_dwi_proc-eddy_brainmask.nii.gz"))
# Save 4D diffusion volume
img = nib.Nifti1Image(dwi_brain, dwi_affine)
nib.save(img, os.path.join(out_dir, "sub-010006_ses-01_dwi_proc-eddy_brain.nii.gz"))
# Save b0 volume
img = nib.Nifti1Image(dwi_b0, dwi_affine)
nib.save(img, os.path.join(out_dir, "sub-010006_ses-01_dwi_proc-eddy_b0.nii.gz"))
```
To perform the registration between the diffusion volumes and T1w, we will make use of ANTs, specifically the `antsRegistrationSyNQuick.sh` script and `antsApplyTransform`. We will begin by registering the diffusion b=0 s/mm^2 volume to get the appropriate transforms to align the two images. We will then apply the inverse transformation to the T1w volume such that it is aligned to the diffusion volume
Here, we will constrain `antsRegistrationSyNQuick.sh` to perform a rigid and affine transformation (we will explain why in the final step). There are a few parameters that must be set:
* `-d` - Image dimension (2/3D)
* `-t` - Transformation type (`a` performs only rigid + affine transformation)
* `-f` - Fixed image (anatomical T1w)
* `-m` - Moving image (DWI b=0 s/mm^2)
* `-o` - Output prefix (prefix to be appended to output files)
```
%%bash
mkdir -p ../../../data/ds000221/derivatives/uncorrected_topup_eddy_regT1/sub-010006/ses-01/transforms
# Perform registration between b0 and T1w
antsRegistrationSyNQuick.sh -d 3 -t a -f ../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/anat/sub-010006_ses-01_space-T1w_brain.nii.gz -m ../../../data/ds000221/derivatives/uncorrected_topup_eddy/sub-010006/ses-01/dwi/sub-010006_ses-01_dwi_proc-eddy_b0.nii.gz -o ../../../data/ds000221/derivatives/uncorrected_topup_eddy_regT1/sub-010006/ses-01/transform/dwi_to_t1_
```
The transformation file should be created which we will use to apply the inverse transform with `antsApplyTransform` to the T1w volume. Similar to the previous command, there are few parameters that will need to be set:
* `-d` - Image dimension (2/3/4D)
* `-i` - Input volume to be transformed (T1w)
* `-r` - Reference volume (b0 of DWI volume)
* `-t` - Transformation file (can be called more than once)
* `-o` - Output volume in the transformed space.
Note that if more than 1 transformation file is provided, the order in which the transforms are applied to the volume is in reverse order of how it is inputted (e.g. last transform gets applied first).
```
%%bash
# Apply transform to 4D DWI volume
antsApplyTransforms -d 3 -i ../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/anat/sub-010006_ses-01_space-T1w_brain.nii.gz -r ../../../data/ds000221/derivatives/uncorrected_topup_eddy/sub-010006/ses-01/dwi/sub-010006_ses-01_dwi_proc-eddy_b0.nii.gz -t [../../../data/ds000221/derivatives/uncorrected_topup_eddy_regT1/sub-010006/ses-01/transform/dwi_to_t1_0GenericAffine.mat,1] -o ../../../data/ds000221/derivatives/uncorrected_topup_eddy_regT1/sub-010006/ses-01/anat/sub-010006_ses-01_space-dwi_T1w_brain.nii.gz
```

Following the transformation of the T1w volume, we can see that anatomical and diffusion weighted volumes are now aligned. It should be highlighted that as part of the transformation step, the T1w volume is resampled based on the voxel size of the ference volume (e.g. DWI).
### Preprocessing notes:
1. In this lesson, the T1w volume is registered to the DWI volume. This method minimizes the manipulation of the diffusion data. It is also possible to register the DWI volume to the T1w volume and would require the associated diffusion gradient vectors (bvec) to also be similarly rotated. If this step is not performed, one would have incorrect diffusion gradient directions relative to the registered DWI volumes. This also highlights a reason behind not performing a non-linear transformation for registration, as each individual diffusion gradient direction would also have to be subsequently warped. Rotation of the diffusion gradient vectors can be done by applying the affine transformation to each row of the file. Luckily, there are existing scripts that can do this. One such Python script was created by Michael Paquette: [`rot_bvecs_ants.py`](https://gist.github.com/mpaquette/5d59ad195778f9d984c5def42f53de6e).
2. We have only demonstrated the preprocessing steps where there is general consensus on how DWI data should be processed. There are also additional steps with certain caveats, which include denoising, unringing (to remove/minimize effects of Gibbs ringing artifacts), and gradient non-linearity correction (to unwarp distortions caused by gradient-field inhomogeneities using a vendor acquired gradient coefficient file.
3. Depending on how the data is acquired, certain steps may not be possible. For example, if the data is not acquired in two directions, `topup` may not be possible (in this situation, distortion correction may be better handled by registering with a T1w anatomical image directly.
4. There are also a number of tools available for preprocessing. In this lesson, we demonstrate some of the more commonly used tools alongside `dipy`.
### References
.. [Cieslak2020] M. Cieslak, PA. Cook, X. He, F-C. Yeh, T. Dhollander, _et al_, "QSIPrep: An integrative platform for preprocessing and reconstructing diffusion MRI", https://doi.org/10.1101/2020.09.04.282269
|
github_jupyter
|
from bids.layout import BIDSLayout
layout = BIDSLayout("../../../data/ds000221", validate=False)
subj='010006'
# Diffusion data
dwi = layout.get(subject=subj, suffix='dwi', extension='nii.gz', return_type='file')[0]
# Anatomical data
t1w = layout.get(subject=subj, suffix='T1w', extension='nii.gz', return_type='file')[0]
import numpy as np
import nibabel as nib
dwi = nib.load(dwi)
dwi_affine = dwi.affine
dwi_data = dwi.get_fdata()
import os
from dipy.segment.mask import median_otsu
# vol_idx is a 1D-array containing the index of the first b0
dwi_brain, dwi_mask = median_otsu(dwi_data, vol_idx=[0])
# Create necessary folders to save mask
out_dir = '../../../data/ds000221/derivatives/uncorrected/sub-%s/ses-01/dwi/' % subj
# Check to see if directory exists, if not create one
if not os.path.exists(out_dir):
os.makedirs(out_dir)
img = nib.Nifti1Image(dwi_mask.astype(np.float32), dwi_affine)
nib.save(img, os.path.join(out_dir, "sub-%s_ses-01_brainmask.nii.gz" % subj))
%%bash
mkdir -p ../../../data/ds000221/derivatives/uncorrected_topup/sub-010006/ses-01/dwi/work
fslmerge -t ../../../data/ds000221/derivatives/uncorrected_topup/sub-010006/ses-01/dwi/work/sub-010006_ses-01_acq-SEfmapDWI_epi.nii.gz ../../data/ds000221/sub-010006/ses-01/fmap/sub-010006_ses-01_acq-SEfmapDWI_dir-AP_epi.nii.gz ../../data/ds000221/sub-010006/ses-01/fmap/sub-010006_ses-01_acq-SEfmapDWI_dir-PA_epi.nii.gz
0 1 0 0.04914
0 1 0 0.04914
0 1 0 0.04914
0 -1 0 0.04914
0 -1 0 0.04914
0 -1 0 0.04914
%%bash
topup --imain=../../../data/ds000221/derivatives/topup/sub-010006/ses-01/dwi/work/sub-010006_ses-01_acq-SEfmapDWI_epi.nii.gz --datain=../../data/ds000221/derivatives/topup/sub-010006/ses-01/dwi/work/pedir.txt --config=b02b0.cnf --out=../../data/ds000221/derivatives/topup/sub-010006/ses-01/dwi/work/topup
%%bash
applytopup --imain=../../../data/ds000221/sub-010006/ses-01/dwi/sub-010006_ses-01_dwi.nii.gz --datain=../../../data/ds000221/derivatives/topup/sub-010006/ses-01/dwi/work/pedir.txt --inindex=1 --topup=../../../data/ds000221/derivatives/topup/sub-010006/ses-01/dwi/work/topup --out=../../../data/ds000221/derivatives/topup/sub-010006/ses-01/dwi/dwi --method=jac
%%bash
mkdir -p ../../../data/ds000221/derivatives/uncorrected_topup_eddy/sub-010006/ses-01/dwi/work
# Create an index file mapping the 67 volumes in 4D dwi volume to the pedir.txt file
indx=""
for i in `seq 1 67`; do
indx="$indx 1"
done
echo $indx > ../../../data/ds000221/derivatives/uncorrected_topup_eddy/sub-010006/ses-01/dwi/work/index.txt
eddy_openmp --imain=../../../data/ds000221/sub-010006/ses-01/dwi/sub-010006_ses-01_dwi.nii.gz --mask=../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/dwi/sub-010006_ses-01_brainmask.nii.gz --acqp=../../../data/ds000221/derivatives/uncorrected_topup/sub-010006/ses-01/dwi/work/pedir.txt --index=../../../data/ds000221/derivatives/uncorrected_topup_eddy/sub-010006/ses-01/dwi/work/index.txt --bvecs=../../../data/ds000221/sub-010006/ses-01/dwi/sub-010006_ses-01_dwi.bvec --bvals=../../../data/ds000221/sub-010006/ses-01/dwi/sub-010006_ses-01_dwi.bval --topup=../../../data/ds000221/derivatives/uncorrected_topup/sub-010006/ses-01/dwi/work/topup --out=../../../data/ds000221/derivatives/uncorrected_topup_eddy/sub-010006/ses-01/dwi/dwi --repol
%%bash
mkdir -p ../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/anat
bet ../../../data/ds000221/sub-010006/ses-01/anat/sub-010006_ses-01_inv-2_mp2rage.nii.gz ../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/anat/sub-010006_ses-01_space-T1w_broadbrain -f 0.6
bet ../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/anat/sub-010006_ses-01_space-T1w_broadbrain ../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/anat/sub-010006_ses-01_space-T1w_brain -f 0.4 -m
mv ../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/anat/sub-010006_ses-01_space-T1w_brain_mask.nii.gz ../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/anat/sub-010006_ses-01_space-T1w_brainmask.nii.gz
from dipy.segment.mask import median_otsu
# Path of FSL eddy-corrected dwi
dwi = "../../../data/ds000221/derivatives/uncorrected_topup_eddy/sub-010006/ses-01/dwi/dwi.nii.gz"
# Load eddy-corrected diffusion data
dwi = nib.load(dwi)
dwi_affine = dwi.affine
dwi_data = dwi.get_fdata()
dwi_brain, dwi_mask = median_otsu(dwi_data, vol_idx=[0])
dwi_b0 = dwi_brain[:,:,:,0]
# Output directory
out_dir="../../../data/ds000221/derivatives/uncorrected_topup_eddy/sub-010006/ses-01/dwi"
# Save diffusion mask
img = nib.Nifti1Image(dwi_mask.astype(np.float32), dwi_affine)
nib.save(img, os.path.join(out_dir, "sub-010006_ses-01_dwi_proc-eddy_brainmask.nii.gz"))
# Save 4D diffusion volume
img = nib.Nifti1Image(dwi_brain, dwi_affine)
nib.save(img, os.path.join(out_dir, "sub-010006_ses-01_dwi_proc-eddy_brain.nii.gz"))
# Save b0 volume
img = nib.Nifti1Image(dwi_b0, dwi_affine)
nib.save(img, os.path.join(out_dir, "sub-010006_ses-01_dwi_proc-eddy_b0.nii.gz"))
%%bash
mkdir -p ../../../data/ds000221/derivatives/uncorrected_topup_eddy_regT1/sub-010006/ses-01/transforms
# Perform registration between b0 and T1w
antsRegistrationSyNQuick.sh -d 3 -t a -f ../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/anat/sub-010006_ses-01_space-T1w_brain.nii.gz -m ../../../data/ds000221/derivatives/uncorrected_topup_eddy/sub-010006/ses-01/dwi/sub-010006_ses-01_dwi_proc-eddy_b0.nii.gz -o ../../../data/ds000221/derivatives/uncorrected_topup_eddy_regT1/sub-010006/ses-01/transform/dwi_to_t1_
%%bash
# Apply transform to 4D DWI volume
antsApplyTransforms -d 3 -i ../../../data/ds000221/derivatives/uncorrected/sub-010006/ses-01/anat/sub-010006_ses-01_space-T1w_brain.nii.gz -r ../../../data/ds000221/derivatives/uncorrected_topup_eddy/sub-010006/ses-01/dwi/sub-010006_ses-01_dwi_proc-eddy_b0.nii.gz -t [../../../data/ds000221/derivatives/uncorrected_topup_eddy_regT1/sub-010006/ses-01/transform/dwi_to_t1_0GenericAffine.mat,1] -o ../../../data/ds000221/derivatives/uncorrected_topup_eddy_regT1/sub-010006/ses-01/anat/sub-010006_ses-01_space-dwi_T1w_brain.nii.gz
| 0.446253 | 0.939471 |
# Uterine Corpus Endometrial Carcinoma (UCEC)
[Jump to the urls to download the GCT and CLS files](#Downloads)
**Authors:** Alejandra Ramos, Marylu Villa and Edwin Juarez
**Is this what you want your scientific identity to be?**
**Contact info:** Email Edwin at [[email protected]](mailto:[email protected]) or post a question in http://www.genepattern.org/help
This notebook provides the steps to download all the UCEC samples from The Cancer Genome Atlas (TCGA) contained in the Genomic Data Commons (GDC) Data portal. These samples can be downloaded as a GCT file and phenotype labels (primary tumor vs normal samples) can be downloaded as a CLS file. These files are compatible with other GenePattern Analyses.

# Overview
Endometrial cancer develops in the cells that form the inner lining of the uterus, or the endometrium, and is one of the most common cancers of the female reproductive system among American women
<p><img alt="Resultado de imagen para Uterine Corpus Endometrial Carcinoma" src="https://www.cancer.gov/images/cdr/live/CDR735233.jpg" style="width: 829px; height: 500px;" /></p>
# UCEC Statistics
In 2010, approximately 43,000 women in the United States were estimated to have been diagnosed and almost 8,000 to have died of endometrial cancer. This cancer occurs most commonly in women aged 60 years or older. About 69 percent of endometrial cancers are diagnosed at an early stage, and as a result about 83 percent of women will survive five years following the time of diagnosis.
<p><img alt="Imagen relacionada" src="https://www.safeworkaustralia.gov.au/sites/swa/files/indicator_chart_1.png" /></p>
https://www.safeworkaustralia.gov.au/statistics-and-research/statistics/disease-and-injuries/disease-and-injury-statistics-stateterritory
# Dataset's Demographic information
<p>TCGA contained 578 UCEC samples (543 primary cancer samples, and 35 normal tissue samples) from 555 people. Below is a summary of the demographic information represented in this dataset. If you are interested in viewing the complete study, as well as the files on the GDC Data Portal, you can follow <a href="https://portal.gdc.cancer.gov/repository?facetTab=cases&filters=%7B%22op%22%3A%22and%22%2C%22content%22%3A%5B%7B%22op%22%3A%22in%22%2C%22content%22%3A%7B%22field%22%3A%22cases.project.project_id%22%2C%22value%22%3A%5B%22TCGA-UVM%22%5D%7D%7D%2C%7B%22op%22%3A%22in%22%2C%22content%22%3A%7B%22field%22%3A%22files.analysis.workflow_type%22%2C%22value%22%3A%5B%22HTSeq%20-%20Counts%22%5D%7D%7D%2C%7B%22op%22%3A%22in%22%2C%22content%22%3A%7B%22field%22%3A%22files.experimental_strategy%22%2C%22value%22%3A%5B%22RNA-Seq%22%5D%7D%7D%5D%7D&searchTableTab=cases" target="_blank">this link.(these data were gathered on July 10th, 2018)</a></p>

# Login to GenePattern
<div class="alert alert-info">
<h3 style="margin-top: 0;"> Instructions <i class="fa fa-info-circle"></i></h3>
<ol>
<li>Login to the *GenePattern Cloud* server.</li>
</ol>
</div>
```
# Requires GenePattern Notebook: pip install genepattern-notebook
import gp
import genepattern
# Username and password removed for security reasons.
genepattern.display(genepattern.session.register("https://cloud.genepattern.org/gp", "", ""))
```
# Downloading RNA-Seq HTSeq Counts Using TCGAImporter
Use the TCGAImporter module to download RNA-Seq HTSeq counts from the GDC Data Portal using a Manifest file and a Metadata file
<p><strong>Input files</strong></p>
<ul>
<li><em>Manifest file</em>: a file containing the list of RNA-Seq samples to be downloaded.</li>
<li><em>Metadata file</em>: a file containing information about the files present at the GDC Data Portal. Instructions for downloading the Manifest and Metadata files can be found here: <a href="https://github.com/genepattern/TCGAImporter/blob/master/how_to_download_a_manifest_and_metadata.pdf" target="_blank">https://github.com/genepattern/TCGAImporter/blob/master/how_to_download_a_manifest_and_metadata.pdf</a></li>
</ul>
<p><strong>Output files</strong></p>
<ul>
<li><em>UCEC_TCGA.gct</em> - This is a tab delimited file that contains the gene expression (HTSeq counts) from the samples listed on the Manifest file. For more info on GCT files, look at reference <a href="#References">1</a><em> </em></li>
<li><em><em>UCEC_TCGA.cls</em> -</em> The CLS file defines phenotype labels (in this case Primary Tumor and Normal Sample) and associates each sample in the GCT file with a label. For more info on CLS files, look at reference <a href="#References">2</a></li>
</ul>
<div class="alert alert-info">
<h3 style="margin-top: 0;"> Instructions <i class="fa fa-info-circle"></i></h3>
<ol>
<li>Load the manifest file in **Manifest** parameter.</li>
<li>Load the metadata file in **Metadata** parameter.</li>
<li>Click **run**.</li>
</ol>
<p><strong>Estimated run time for TCGAImporter</strong> : ~ 10 minutes</p>
```
tcgaimporter_task = gp.GPTask(genepattern.session.get(0), 'urn:lsid:broad.mit.edu:cancer.software.genepattern.module.analysis:00369')
tcgaimporter_job_spec = tcgaimporter_task.make_job_spec()
tcgaimporter_job_spec.set_parameter("manifest", "https://cloud.genepattern.org/gp/users/marylu257/tmp/run2275209178994909933.tmp/UCEC_manifest.txt")
tcgaimporter_job_spec.set_parameter("metadata", "https://cloud.genepattern.org/gp/users/marylu257/tmp/run1507855829424081202.tmp/UCEC_metadata.json")
tcgaimporter_job_spec.set_parameter("output_file_name", "UCEC_TCGA")
tcgaimporter_job_spec.set_parameter("gct", "True")
tcgaimporter_job_spec.set_parameter("translate_gene_id", "False")
tcgaimporter_job_spec.set_parameter("cls", "True")
genepattern.display(tcgaimporter_task)
job35211 = gp.GPJob(genepattern.session.get(0), 35211)
genepattern.display(job35211)
collapsedataset_task = gp.GPTask(genepattern.session.get(0), 'urn:lsid:broad.mit.edu:cancer.software.genepattern.module.analysis:00134')
collapsedataset_job_spec = collapsedataset_task.make_job_spec()
collapsedataset_job_spec.set_parameter("dataset.file", "https://cloud.genepattern.org/gp/jobResults/31849/TCGA_dataset.gct")
collapsedataset_job_spec.set_parameter("chip.platform", "ftp://ftp.broadinstitute.org/pub/gsea/annotations/ENSEMBL_human_gene.chip")
collapsedataset_job_spec.set_parameter("collapse.mode", "Maximum")
collapsedataset_job_spec.set_parameter("output.file.name", "<dataset.file_basename>.collapsed")
genepattern.display(collapsedataset_task)
job32425 = gp.GPJob(genepattern.session.get(0), 32425)
genepattern.display(job32425)
```
# Downloads
<p>You can download the input and output files of TCGAImporter for this cancer type here:</p>
<p><strong>Inputs:</strong></p>
<ul>
<li><a href="https://datasets.genepattern.org/data/TCGA_HTSeq_counts/KIRP/KIRP_MANIFEST.txt" target="_blank">https://datasets.genepattern.org/data/TCGA_HTSeq_counts/UCEC/UCEC_MANIFEST.txt</a></li>
<li><a href="https://datasets.genepattern.org/data/TCGA_HTSeq_counts/KIRP/KIRP_METADATA.json" target="_blank">https://datasets.genepattern.org/data/TCGA_HTSeq_counts/UCEC/UCEC_METADATA.json</a></li>
</ul>
<p><strong>Outputs:</strong></p>
<ul>
<li><a href="https://datasets.genepattern.org/data/TCGA_HTSeq_counts/KIRP/KIRP_TCGA.gct" target="_blank">https://datasets.genepattern.org/data/TCGA_HTSeq_counts/UCEC/UCEC_TCGA.gct</a></li>
<li><a href="https://datasets.genepattern.org/data/TCGA_HTSeq_counts/KIRP/KIRP_TCGA.cls" target="_blank">https://datasets.genepattern.org/data/TCGA_HTSeq_counts/UCEC/UCEC_TCGA.cls</a></li>
</ul>
If you'd like to download similar files for other TCGA datasets, visit this link:
- https://datasets.genepattern.org/?prefix=data/TCGA_HTSeq_counts/
# References
[1] http://software.broadinstitute.org/cancer/software/genepattern/file-formats-guide#GCT
[2] http://software.broadinstitute.org/cancer/software/genepattern/file-formats-guide#CLS
[3] https://cancergenome.nih.gov/cancersselected/endometrial</p>
[4] https://www.google.com/search?q=Uterine+Corpus+Endometrial+Carcinoma&source=lnms&tbm=isch&sa=X&ved=0ahUKEwiu4e_s-IjcAhVoxVQKHeflBGAQ_AUICigB&biw=1366&bih=586#imgrc=S5Twtt5BPC9YTM:</p>
[5] https://www.google.com/search?q=Uterine+Corpus+Endometrial+Carcinoma+statistics&source=lnms&tbm=isch&sa=X&ved=0ahUKEwiYz9Th-YjcAhWI3lQKHVYHAU4Q_AUICigB&biw=1366&bih=586#imgdii=BivuaMT2MEQd8M:&imgrc=C_T7yrmM1NGcYM:</p>
|
github_jupyter
|
# Requires GenePattern Notebook: pip install genepattern-notebook
import gp
import genepattern
# Username and password removed for security reasons.
genepattern.display(genepattern.session.register("https://cloud.genepattern.org/gp", "", ""))
tcgaimporter_task = gp.GPTask(genepattern.session.get(0), 'urn:lsid:broad.mit.edu:cancer.software.genepattern.module.analysis:00369')
tcgaimporter_job_spec = tcgaimporter_task.make_job_spec()
tcgaimporter_job_spec.set_parameter("manifest", "https://cloud.genepattern.org/gp/users/marylu257/tmp/run2275209178994909933.tmp/UCEC_manifest.txt")
tcgaimporter_job_spec.set_parameter("metadata", "https://cloud.genepattern.org/gp/users/marylu257/tmp/run1507855829424081202.tmp/UCEC_metadata.json")
tcgaimporter_job_spec.set_parameter("output_file_name", "UCEC_TCGA")
tcgaimporter_job_spec.set_parameter("gct", "True")
tcgaimporter_job_spec.set_parameter("translate_gene_id", "False")
tcgaimporter_job_spec.set_parameter("cls", "True")
genepattern.display(tcgaimporter_task)
job35211 = gp.GPJob(genepattern.session.get(0), 35211)
genepattern.display(job35211)
collapsedataset_task = gp.GPTask(genepattern.session.get(0), 'urn:lsid:broad.mit.edu:cancer.software.genepattern.module.analysis:00134')
collapsedataset_job_spec = collapsedataset_task.make_job_spec()
collapsedataset_job_spec.set_parameter("dataset.file", "https://cloud.genepattern.org/gp/jobResults/31849/TCGA_dataset.gct")
collapsedataset_job_spec.set_parameter("chip.platform", "ftp://ftp.broadinstitute.org/pub/gsea/annotations/ENSEMBL_human_gene.chip")
collapsedataset_job_spec.set_parameter("collapse.mode", "Maximum")
collapsedataset_job_spec.set_parameter("output.file.name", "<dataset.file_basename>.collapsed")
genepattern.display(collapsedataset_task)
job32425 = gp.GPJob(genepattern.session.get(0), 32425)
genepattern.display(job32425)
| 0.382718 | 0.876423 |
# ChIPseq normalization based on binned spike-in local regression
## Core Functions
```
import itertools
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import tqdm
from scipy.interpolate import interp1d
from statsmodels.nonparametric import smoothers_lowess
def image_display(file):
from IPython.display import Image, display
display(Image(file))
def max_diff(df, combinations):
'''
Find the sample combination with the greatest average signal difference.
Parameters
----------
df : array-like of shape = [n_bins, n_samples]
combinations: list of non-repeating sample lists [(s1,s2), (s1,s3), (s2,s3)]
Returns
-------
list of samples with greatest mean signal differences
'''
mx = 0
comb = ['None', 'None']
for s1,s2 in combinations:
comb_max = abs((df[s1] - df[s2]).mean())
if comb_max > mx:
comb = [s1, s2]
mx = comb_max
return comb
def choice_idx_pmf(df, size=50000):
'''
Makes a pmf based on signal intensity and chooses random indicies based on pmf
Parameters
----------
df: dataframe with two columns
size: how many indicies to choose
Returns
-------
Values of indicies from the df based on the pmf
'''
difference = df.iloc[:,0] - df.iloc[:,1]
d2 = difference**2
total = d2.sum()
pmf2 = d2 / total
return np.random.choice(pmf2.index, size=size, replace=False, p=pmf2.values)
def get_subset_indicies(subset_df, combinations, size=50000):
'''
From spike dataframe, choose inidicies for modeling based on
the samples with the greatest mean signal difference.
Parameters
----------
df: dataframe of spike or subset signal values
combinations: list of non-repeating sample lists [(s1,s2), (s1,s3), (s2,s3)]
size: number of indicies to return
Returns
-------
list-like of chosen indicies
'''
comb = max_diff(subset_df, combinations)
return choice_idx_pmf(subset_df[comb], size=size)
def delta(difference, values, subset_indicies, num_samples):
'''
Makes a lowess model of the difference between samples based on the signal value
of the spike indicies (genomic bin) in that sample. Calcualte the differences over
the entire dataframe. (ie. Based on signal value at each index, calculate the
adjustment of the signal at that index in the direction that the spike in is
different to the other sample.)
Parameters
----------
difference: vector of calucated differences between two samples at spike-in indicies
values: signal values of one sample at spike-in indicies
num_samples: number of total samples in the experiment
Returns
-------
list of the correction values for that sample compared to one other sample
'''
low = smoothers_lowess.lowess(difference, values.loc[subset_indicies])
interp = interp1d(low[:,0], low[:,1], bounds_error=False,
fill_value=(difference[values.loc[subset_indicies].idxmin()],
"extrapolate")) # linear at high signal
return interp(values) / (num_samples - 1)
def normalize_lowess(df, spike_indicies, subset_size=50000, iterations=5, logit=True):
'''
Executes a minimization of the differences between spike-in samples based
on a non-parametric local regression model and applies the corrections to
all datapoints.
Parameters
----------
df: dataframe with sample names in columns. (binned genome signal means of IP and spike-in)
spike_indicies: list of spike-in indicies
subset_size: number of indicies to build model
iterations: number of iterations
logit: apply a log1p to the dataframe
Returns
-------
Normalized Dataframe
Dictionary of the max mean squared error at each iteration
'''
errors={}
d = df.apply(np.log1p).copy() if logit else df.copy()
samples = d.columns.tolist()
combinations = list(itertools.combinations(samples, 2))
for i in tqdm.tqdm(range(interations)):
#Initialize a change matrix
ddf = pd.DataFrame(0, columns=df.columns, index=d.index)
#choose spike in indicies for modeling
subset_index = get_subset_indicies(d.loc[spike_indicies], combinations, size=subset_size)
for s1, s2 in combinations:
#Calculate differences between two samples
difference = df.loc[subset_index,s1] - df.loc[subset_index,s2]
#Model the differences based on binned values and adjust the change matrix per comparison
ddf[s1] = ddf[s1] + delta(difference, df[s1], subset_index, len(samples))
ddf[s2] = ddf[s2] - delta(difference, df[s2], subset_index, len(samples))
#Make the iteration adjustments to the entire dataset
d = d - ddf
#errors[f'{i + 1}'] = ddf.loc[sub_index, samples].mean()
errors[f'{i + 1} MSE'] = ((ddf.loc[sub_index, samples])**2).mean()
MSE = {k: df.max() for k,df in errors.items() if 'MSE' in k}
normed_df = d.apply(np.expm1) if logit else d
return normed_df, MSE
```
## 1. Creating Binned Signal Matrix
```
import math
from multiprocessing import Pool
import pandas as pd
import pyBigWig as bw
import tqdm
from p_tqdm import p_map
d1 = 'H3K27me3_DMSO_Rep1.cpm.100bp.bw'
d2 = 'H3K27me3_DMSO_Rep2.cpm.100bp.bw'
D1 = bw.open(d1)
D2 = bw.open(d2)
D1.chroms()
bin_size = 100
start = []
end = []
ch = []
for chrom, size in tqdm.tqdm(D1.chroms().items()):
start = [*start, *(x for x in range(0,size,bin_size))]
ch = [*ch, *([chrom]*(math.ceil(size/bin_size)))]
end = [*end, *(x for x in range(bin_size,size,bin_size)), size]
df = pd.DataFrame({'chr':ch, 'start':start, 'end':end}, index=range(len(start)))
df[df.end % 100 > 0]
%%time
df['DMSO_1'] = df[['chr','start','end']].apply(lambda x: D1.stats(x[0], x[1], x[2])[0], axis=1)
%%time
df['DMSO_2'] = df2[['chr','start','end']].apply(lambda x: D2.stats(x[0], x[1], x[2])[0], axis=1)
E1 = bw.open('H3K27me3_EZH2i_Rep1.cpm.100bp.bw')
E2 = bw.open('H3K27me3_EZH2i_Rep2.cpm.100bp.bw')
P1 = bw.open('H3K27me3_PRMT5i_Rep1.cpm.100bp.bw')
P2 = bw.open('H3K27me3_PRMT5i_Rep2.cpm.100bp.bw')
%%time
df['EZH2i_1'] = df2[['chr','start','end']].progress_apply(lambda x: E1.stats(x[0], x[1], x[2])[0], axis=1)
df['EZH2i_2'] = df2[['chr','start','end']].progress_apply(lambda x: E2.stats(x[0], x[1], x[2])[0], axis=1)
df['PRMT5i_1'] = df2[['chr','start','end']].progress_apply(lambda x: P1.stats(x[0], x[1], x[2])[0], axis=1)
df['PRMT5i_2'] = df2[['chr','start','end']].progress_apply(lambda x: P2.stats(x[0], x[1], x[2])[0], axis=1)
df.to_csv('~/Desktop/norm_chip.txt', sep="\t")
```
## 2. Generate Normalized Average CPM driven by spike-in regression
```
np.random.seed(42)
sns.set(style='white')
norm_chip = pd.read_table('/Users/dlk41/Desktop/norm_chip1.txt', index_col=0)
fly_chr = {'2L_FLY', '2R_FLY', '3L_FLY', '3R_FLY', '4_FLY', 'X_FLY', 'Y_FLY'}
fly_df_index = norm_chip[norm_chip.chr.isin(fly_chr)].index
samples = norm_chip.columns[3:].tolist()
combinations = list(itertools.combinations(samples, 2))
iterations = 5
%%time
df = norm_chip[samples].copy()
errors = {}
for i in tqdm.tqdm(range(iterations)):
ddf = pd.DataFrame(0, columns=df.columns, index=df.index)
sub_index = get_subset_indicies(df.loc[fly_df_index], combinations, size=100000)
for s1, s2 in combinations:
difference = df.loc[sub_index,s1] - df.loc[sub_index,s2]
ddf[s1] = ddf[s1] + delta(difference, df[s1], sub_index, len(samples))
ddf[s2] = ddf[s2] - delta(difference, df[s2], sub_index, len(samples))
df = df - ddf
errors[f'{i + 1}'] = ddf.loc[sub_index, samples].mean()
errors[f'{i + 1} MSE'] = ((ddf.loc[sub_index, samples])**2).mean()
df[df < 0] = 0
print({k: df.max() for k,df in errors.items() if 'MSE' in k})
```
## Fly Tests
```
random_idx = norm_chip[norm_chip.index.isin(fly_df_index)].sample(100000).index
plt.scatter(np.log1p(norm_chip.loc[random_idx, 'DMSO_1']),
np.log1p(norm_chip.loc[random_idx, 'DMSO_2']),
s=1, alpha=.5, c='k')
plt.xlim(0,5)
plt.ylim(0,5)
plt.scatter(np.log1p(df.loc[random_idx, 'DMSO_1']),
np.log1p(df.loc[random_idx, 'DMSO_2']),
s=1, alpha=.5, c='k')
plt.scatter(np.log1p(norm_chip.loc[random_idx, 'EZH2i_1']),
np.log1p(norm_chip.loc[random_idx, 'EZH2i_2']),
s=1, alpha=.5, c='k')
plt.xlim(0,6)
plt.ylim(0,6)
plt.scatter(np.log1p(df.loc[random_idx, 'EZH2i_1']),
np.log1p(df.loc[random_idx, 'EZH2i_2']),
s=1, alpha=.5, c='k')
plt.scatter(np.log1p(norm_chip.loc[random_idx, 'PRMT5i_1']),
np.log1p(norm_chip.loc[random_idx, 'PRMT5i_2']),
s=1, alpha=.5, c='k')
plt.xlim(0,3.5)
plt.ylim(0,3.5)
plt.scatter(np.log1p(df.loc[random_idx, 'PRMT5i_1']),
np.log1p(df.loc[random_idx, 'PRMT5i_2']),
s=1, alpha=.5, c='k')
plt.scatter(np.log1p(norm_chip.loc[random_idx, 'EZH2i_1']),
np.log1p(norm_chip.loc[random_idx, 'DMSO_2']),
s=1, alpha=.5, c='k')
plt.xlim(0,6)
plt.ylim(0,6)
plt.scatter(np.log1p(df.loc[random_idx, 'EZH2i_1']),
np.log1p(df.loc[random_idx, 'DMSO_2']),
s=1, alpha=.5, c='k')
plt.xlim(0,5)
plt.ylim(0,5)
```
## Human test
```
random_h_idx = norm_chip[~norm_chip.index.isin(fly_df_index)].sample(1000000).index
plt.scatter(np.log1p(norm_chip.loc[random_h_idx, 'EZH2i_1']),
np.log1p(norm_chip.loc[random_h_idx, 'DMSO_1']),
s=1, alpha=.5, c='k')
plt.xlim(0,2.5)
plt.ylim(0,2.5)
plt.scatter(np.log1p(df.loc[random_h_idx, 'EZH2i_1']),
np.log1p(df.loc[random_h_idx, 'DMSO_1']),
s=1, alpha=.5, c='k')
plt.xlim(0,2)
plt.ylim(0,2)
```
## Export
```
df['chr'] = norm_chip.chr
df['start'] = norm_chip.start
df['end'] = norm_chip.end
human_index = df[~df.index.isin(fly_df_index)].index
for s in tqdm.tqdm(samples):
df.loc[human_index, ['chr', 'start', 'end', s]].to_csv(f'/Users/dlk41/Box/Norm_chip/updated/{s}.10it.100k.bedgraph', sep="\t", index=None, header=None)
fly_exp_df = df.loc[fly_df_index].copy()
fly_exp_df['chr'] = [x.split('_')[0] for x in fly_exp_df.chr.tolist()]
for s in tqdm.tqdm(samples):
fly_exp_df[['chr','start','end',s]].to_csv(f'/Users/dlk41/Box/Norm_chip/updated/{s}.10it.100k.bdgp6.bedgraph', sep="\t", index=None, header=None)
```
## Make bigwig files
```
import pyBigWig as bw
with bw.open('../H3K27me3_DMSO_Rep2.cpm.100bp.bw') as file:
chroms = file.chroms()
human_chroms = [(k,l) for k,l in chroms.items() if '_FLY' not in k]
fly_chroms = [(f"chr{k.split('_')[0]}",l) for k,l in chroms.items() if '_FLY' in k]
%%time
for sample in tqdm.tqdm(samples):
with bw.open(f'{sample}_adjusted.bw', 'w') as file:
file.addHeader(human_chroms)
for chromosome, _ in human_chroms:
file.addEntries(chromosome, 0, values=df[df.chr == chromosome][sample].values, span=100, step=100)
with bw.open(f'{sample}_adjusted_FLY.bw', 'w') as file:
file.addHeader(fly_chroms)
for fly_chrom, _ in fly_chroms:
file.addEntries(fly_chrom, 0, values=df[df.chr == fly_chrom][sample].values, span=100, step=100)
```
|
github_jupyter
|
import itertools
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import tqdm
from scipy.interpolate import interp1d
from statsmodels.nonparametric import smoothers_lowess
def image_display(file):
from IPython.display import Image, display
display(Image(file))
def max_diff(df, combinations):
'''
Find the sample combination with the greatest average signal difference.
Parameters
----------
df : array-like of shape = [n_bins, n_samples]
combinations: list of non-repeating sample lists [(s1,s2), (s1,s3), (s2,s3)]
Returns
-------
list of samples with greatest mean signal differences
'''
mx = 0
comb = ['None', 'None']
for s1,s2 in combinations:
comb_max = abs((df[s1] - df[s2]).mean())
if comb_max > mx:
comb = [s1, s2]
mx = comb_max
return comb
def choice_idx_pmf(df, size=50000):
'''
Makes a pmf based on signal intensity and chooses random indicies based on pmf
Parameters
----------
df: dataframe with two columns
size: how many indicies to choose
Returns
-------
Values of indicies from the df based on the pmf
'''
difference = df.iloc[:,0] - df.iloc[:,1]
d2 = difference**2
total = d2.sum()
pmf2 = d2 / total
return np.random.choice(pmf2.index, size=size, replace=False, p=pmf2.values)
def get_subset_indicies(subset_df, combinations, size=50000):
'''
From spike dataframe, choose inidicies for modeling based on
the samples with the greatest mean signal difference.
Parameters
----------
df: dataframe of spike or subset signal values
combinations: list of non-repeating sample lists [(s1,s2), (s1,s3), (s2,s3)]
size: number of indicies to return
Returns
-------
list-like of chosen indicies
'''
comb = max_diff(subset_df, combinations)
return choice_idx_pmf(subset_df[comb], size=size)
def delta(difference, values, subset_indicies, num_samples):
'''
Makes a lowess model of the difference between samples based on the signal value
of the spike indicies (genomic bin) in that sample. Calcualte the differences over
the entire dataframe. (ie. Based on signal value at each index, calculate the
adjustment of the signal at that index in the direction that the spike in is
different to the other sample.)
Parameters
----------
difference: vector of calucated differences between two samples at spike-in indicies
values: signal values of one sample at spike-in indicies
num_samples: number of total samples in the experiment
Returns
-------
list of the correction values for that sample compared to one other sample
'''
low = smoothers_lowess.lowess(difference, values.loc[subset_indicies])
interp = interp1d(low[:,0], low[:,1], bounds_error=False,
fill_value=(difference[values.loc[subset_indicies].idxmin()],
"extrapolate")) # linear at high signal
return interp(values) / (num_samples - 1)
def normalize_lowess(df, spike_indicies, subset_size=50000, iterations=5, logit=True):
'''
Executes a minimization of the differences between spike-in samples based
on a non-parametric local regression model and applies the corrections to
all datapoints.
Parameters
----------
df: dataframe with sample names in columns. (binned genome signal means of IP and spike-in)
spike_indicies: list of spike-in indicies
subset_size: number of indicies to build model
iterations: number of iterations
logit: apply a log1p to the dataframe
Returns
-------
Normalized Dataframe
Dictionary of the max mean squared error at each iteration
'''
errors={}
d = df.apply(np.log1p).copy() if logit else df.copy()
samples = d.columns.tolist()
combinations = list(itertools.combinations(samples, 2))
for i in tqdm.tqdm(range(interations)):
#Initialize a change matrix
ddf = pd.DataFrame(0, columns=df.columns, index=d.index)
#choose spike in indicies for modeling
subset_index = get_subset_indicies(d.loc[spike_indicies], combinations, size=subset_size)
for s1, s2 in combinations:
#Calculate differences between two samples
difference = df.loc[subset_index,s1] - df.loc[subset_index,s2]
#Model the differences based on binned values and adjust the change matrix per comparison
ddf[s1] = ddf[s1] + delta(difference, df[s1], subset_index, len(samples))
ddf[s2] = ddf[s2] - delta(difference, df[s2], subset_index, len(samples))
#Make the iteration adjustments to the entire dataset
d = d - ddf
#errors[f'{i + 1}'] = ddf.loc[sub_index, samples].mean()
errors[f'{i + 1} MSE'] = ((ddf.loc[sub_index, samples])**2).mean()
MSE = {k: df.max() for k,df in errors.items() if 'MSE' in k}
normed_df = d.apply(np.expm1) if logit else d
return normed_df, MSE
import math
from multiprocessing import Pool
import pandas as pd
import pyBigWig as bw
import tqdm
from p_tqdm import p_map
d1 = 'H3K27me3_DMSO_Rep1.cpm.100bp.bw'
d2 = 'H3K27me3_DMSO_Rep2.cpm.100bp.bw'
D1 = bw.open(d1)
D2 = bw.open(d2)
D1.chroms()
bin_size = 100
start = []
end = []
ch = []
for chrom, size in tqdm.tqdm(D1.chroms().items()):
start = [*start, *(x for x in range(0,size,bin_size))]
ch = [*ch, *([chrom]*(math.ceil(size/bin_size)))]
end = [*end, *(x for x in range(bin_size,size,bin_size)), size]
df = pd.DataFrame({'chr':ch, 'start':start, 'end':end}, index=range(len(start)))
df[df.end % 100 > 0]
%%time
df['DMSO_1'] = df[['chr','start','end']].apply(lambda x: D1.stats(x[0], x[1], x[2])[0], axis=1)
%%time
df['DMSO_2'] = df2[['chr','start','end']].apply(lambda x: D2.stats(x[0], x[1], x[2])[0], axis=1)
E1 = bw.open('H3K27me3_EZH2i_Rep1.cpm.100bp.bw')
E2 = bw.open('H3K27me3_EZH2i_Rep2.cpm.100bp.bw')
P1 = bw.open('H3K27me3_PRMT5i_Rep1.cpm.100bp.bw')
P2 = bw.open('H3K27me3_PRMT5i_Rep2.cpm.100bp.bw')
%%time
df['EZH2i_1'] = df2[['chr','start','end']].progress_apply(lambda x: E1.stats(x[0], x[1], x[2])[0], axis=1)
df['EZH2i_2'] = df2[['chr','start','end']].progress_apply(lambda x: E2.stats(x[0], x[1], x[2])[0], axis=1)
df['PRMT5i_1'] = df2[['chr','start','end']].progress_apply(lambda x: P1.stats(x[0], x[1], x[2])[0], axis=1)
df['PRMT5i_2'] = df2[['chr','start','end']].progress_apply(lambda x: P2.stats(x[0], x[1], x[2])[0], axis=1)
df.to_csv('~/Desktop/norm_chip.txt', sep="\t")
np.random.seed(42)
sns.set(style='white')
norm_chip = pd.read_table('/Users/dlk41/Desktop/norm_chip1.txt', index_col=0)
fly_chr = {'2L_FLY', '2R_FLY', '3L_FLY', '3R_FLY', '4_FLY', 'X_FLY', 'Y_FLY'}
fly_df_index = norm_chip[norm_chip.chr.isin(fly_chr)].index
samples = norm_chip.columns[3:].tolist()
combinations = list(itertools.combinations(samples, 2))
iterations = 5
%%time
df = norm_chip[samples].copy()
errors = {}
for i in tqdm.tqdm(range(iterations)):
ddf = pd.DataFrame(0, columns=df.columns, index=df.index)
sub_index = get_subset_indicies(df.loc[fly_df_index], combinations, size=100000)
for s1, s2 in combinations:
difference = df.loc[sub_index,s1] - df.loc[sub_index,s2]
ddf[s1] = ddf[s1] + delta(difference, df[s1], sub_index, len(samples))
ddf[s2] = ddf[s2] - delta(difference, df[s2], sub_index, len(samples))
df = df - ddf
errors[f'{i + 1}'] = ddf.loc[sub_index, samples].mean()
errors[f'{i + 1} MSE'] = ((ddf.loc[sub_index, samples])**2).mean()
df[df < 0] = 0
print({k: df.max() for k,df in errors.items() if 'MSE' in k})
random_idx = norm_chip[norm_chip.index.isin(fly_df_index)].sample(100000).index
plt.scatter(np.log1p(norm_chip.loc[random_idx, 'DMSO_1']),
np.log1p(norm_chip.loc[random_idx, 'DMSO_2']),
s=1, alpha=.5, c='k')
plt.xlim(0,5)
plt.ylim(0,5)
plt.scatter(np.log1p(df.loc[random_idx, 'DMSO_1']),
np.log1p(df.loc[random_idx, 'DMSO_2']),
s=1, alpha=.5, c='k')
plt.scatter(np.log1p(norm_chip.loc[random_idx, 'EZH2i_1']),
np.log1p(norm_chip.loc[random_idx, 'EZH2i_2']),
s=1, alpha=.5, c='k')
plt.xlim(0,6)
plt.ylim(0,6)
plt.scatter(np.log1p(df.loc[random_idx, 'EZH2i_1']),
np.log1p(df.loc[random_idx, 'EZH2i_2']),
s=1, alpha=.5, c='k')
plt.scatter(np.log1p(norm_chip.loc[random_idx, 'PRMT5i_1']),
np.log1p(norm_chip.loc[random_idx, 'PRMT5i_2']),
s=1, alpha=.5, c='k')
plt.xlim(0,3.5)
plt.ylim(0,3.5)
plt.scatter(np.log1p(df.loc[random_idx, 'PRMT5i_1']),
np.log1p(df.loc[random_idx, 'PRMT5i_2']),
s=1, alpha=.5, c='k')
plt.scatter(np.log1p(norm_chip.loc[random_idx, 'EZH2i_1']),
np.log1p(norm_chip.loc[random_idx, 'DMSO_2']),
s=1, alpha=.5, c='k')
plt.xlim(0,6)
plt.ylim(0,6)
plt.scatter(np.log1p(df.loc[random_idx, 'EZH2i_1']),
np.log1p(df.loc[random_idx, 'DMSO_2']),
s=1, alpha=.5, c='k')
plt.xlim(0,5)
plt.ylim(0,5)
random_h_idx = norm_chip[~norm_chip.index.isin(fly_df_index)].sample(1000000).index
plt.scatter(np.log1p(norm_chip.loc[random_h_idx, 'EZH2i_1']),
np.log1p(norm_chip.loc[random_h_idx, 'DMSO_1']),
s=1, alpha=.5, c='k')
plt.xlim(0,2.5)
plt.ylim(0,2.5)
plt.scatter(np.log1p(df.loc[random_h_idx, 'EZH2i_1']),
np.log1p(df.loc[random_h_idx, 'DMSO_1']),
s=1, alpha=.5, c='k')
plt.xlim(0,2)
plt.ylim(0,2)
df['chr'] = norm_chip.chr
df['start'] = norm_chip.start
df['end'] = norm_chip.end
human_index = df[~df.index.isin(fly_df_index)].index
for s in tqdm.tqdm(samples):
df.loc[human_index, ['chr', 'start', 'end', s]].to_csv(f'/Users/dlk41/Box/Norm_chip/updated/{s}.10it.100k.bedgraph', sep="\t", index=None, header=None)
fly_exp_df = df.loc[fly_df_index].copy()
fly_exp_df['chr'] = [x.split('_')[0] for x in fly_exp_df.chr.tolist()]
for s in tqdm.tqdm(samples):
fly_exp_df[['chr','start','end',s]].to_csv(f'/Users/dlk41/Box/Norm_chip/updated/{s}.10it.100k.bdgp6.bedgraph', sep="\t", index=None, header=None)
import pyBigWig as bw
with bw.open('../H3K27me3_DMSO_Rep2.cpm.100bp.bw') as file:
chroms = file.chroms()
human_chroms = [(k,l) for k,l in chroms.items() if '_FLY' not in k]
fly_chroms = [(f"chr{k.split('_')[0]}",l) for k,l in chroms.items() if '_FLY' in k]
%%time
for sample in tqdm.tqdm(samples):
with bw.open(f'{sample}_adjusted.bw', 'w') as file:
file.addHeader(human_chroms)
for chromosome, _ in human_chroms:
file.addEntries(chromosome, 0, values=df[df.chr == chromosome][sample].values, span=100, step=100)
with bw.open(f'{sample}_adjusted_FLY.bw', 'w') as file:
file.addHeader(fly_chroms)
for fly_chrom, _ in fly_chroms:
file.addEntries(fly_chrom, 0, values=df[df.chr == fly_chrom][sample].values, span=100, step=100)
| 0.654232 | 0.919643 |
# Programming in Python
## Session 1
### Aim of the Session
Learn/review the basics
- what is ...
- how to ...
### 'Hello World!'
```
# the culturally-expected introductory statement
# an example of what you can achieve with Python in just a few lines
```
### Literals
Values of a _type_, presented literally
```
# example name type designation
42 # integer int
2.016 # float float*
"Homo sapiens" # string str
```
- int: whole numbers e.g. 1, 1000, 6000000000
- float: 'floating point' non-whole numbers e.g. 1.9, 30.01, 10e3, 1e-3
- string: ordered sequence of characters, enclosed in quotation marks (single, double, _triple_)
```
# type conversions
```
#### Aside - Comments
Comments are preceded by a **#**, and are completely ignored by the python interpreter.
Comments can be on their own line or after a line of code.
Comments are an incredibly useful way to keep track of what you are doing in
your code. Use comments to document what you do as much as possible, it will
pay off in the long run.
### Exercises 1
```
# print some strings
# print some numbers (ints or floats)
# print multiple values of different types all at once
# (hints: use comma to separate values with a space, or + to join strings)
# print a string containing quote marks
```
### Operators & Operands
Using Python as a calculator: `+`, `-`, `/`, `*` etc are _operators_, the values/variables that they work on are _operands_.
```
# standard mathematical operations can be performed in Python
# and some less common ones
```
_Note: check out numpy, scipy, stats modules if you want to do a lot of maths_
### Variables
Store values (information) in memory, and (re-)use them. We give variables names (identifiers) so that we have a means of referring to the information on demand.
```
# variable assignment is done with '='
```
#### Variable naming
Rules:
- identifier lookup is case-sensitive
- `myname` & `MyName` are different
- must be unique in your working environment
- existing variable will be __over-written without warning__
- cannot start with a number, or any special symbol (e.g. $, %, @, -, etc...) except for "_" (underscore), which is OK.
- cannot have any spaces or special characters (except for "_" (underscore))
Conventions/good practice:
- identifiers (usually) begin with a lowercase letter
- followed by letters, numbers, underscores
- use a strategy to make reading easier
- `myName`
- `exciting_variable`
- long, descriptive > short, vague
### String Formatting
Create formatted strings, with variable values substituted in.
```
# two ways to do it in Python
name = 'Florence'
age = 73
print('%s is %d years old' % (name, age)) # common amongst many programming languages
print('{} is {} years old'.format(name, age)) # perhaps more consistent with stardard Python syntax
```
There is a long list of possible format options for numbers: https://pyformat.info/
### Data Structures
Programming generally requires building/working with much larger and more complex sets of data than the single values/words/sentences that we have looked at so far. In fact, finding ways to operate effectively (and efficiently) on complex structures in order to extract/produce information, _is_ (data) programming.
Python has two most commonly-used structures for storing multiple pieces of data - _lists_ and _dictionaries_. Let's look at these, and a few more, now.
#### Lists
```
# sequence of entries, in order and of any type
# accessing list entries
# adding/removing entries (remove/pop)
# length of list
# sets
```
#### Objects, Methods, and How To Get Help
In Python, everything is an _object_ - some value(s), packaged up with a set of things that can be done with/to it (___methods___), and pieces of information about it (___attributes___). This makes it very easy to perform the most commonly-needed operations for that/those type of value(s). The language has a standard syntax for accessing methods:
```
string_object = 'the cold never bothered me anyway'
# methods - object.something()
# more...
# dir() and help()
```
### Exercises 2
```
# add 'Sally' to the list of students' names
student_names = ['Sandy', 'Pete', 'Richard', 'Rebecca']
# access the fourth entry of the list
# join the list with a new list from another class
other_student_names = ['Sam', 'Fiona', 'Sarah', 'Richard', 'Sarah', 'Matthew']
```
#### Dictionaries
```
# collection of paired information - keys and values
student_marks = {'Alessio': 67, 'Nic': 48, 'Georg': 68}
empty_dict = {}
another_empty_dict = dict()
# accessing dict entries
# adding/changing/deleting entries
```
#### Mutable?
Object types can be divided into two categories - mutable & immutable. _Mutable_ objects can be changed 'in-place' - their value can be updated, added to, re-ordered etc without the need to create a whole new object every time. _Immutable_ types cannot be changed in place - once they have a value, this value cannot be altered. though, of course, it can __always__ be overwritten.
```
# lists are mutable
cities = ['Nairobi', 'Vancouver', 'Wellington', 'Beijing']
print(cities)
# strings are immutable
beatles = "I'd like to be under the sea"
```
### Looping
Time for some real programming. The biggest motivation for researches to learn a programming language is the opportunity to automate repetitive tasks and analyses.
For loops define a set of steps that will be carried out for all items in a sequence. The items in the sequence will be taken one-at-a-time, and the loop performed, until there are no more items to process.
```
for season in ['Spring', 'Summer', 'Autumn', 'Winter']:
print(season)
word = 'python'
for letter in word:
print(letter.upper())
print("Finished")
# range
# zip
# iterating through two lists simultaneously
# enumerate
```
## Exercise 3:
```
# calculate the mean of the elements in the list
list_of_numbers = [1, 2, 4, 8, 3, 6, 1, 9, 10, 5]
sum_of_values = ...
for ... in list_of_numbers:
... = ... + ...
mean_value = ... / len(...)
print(mean_value)
```
### Conditionals - if, elif, else
Looping allows you to perform a common set of operations on multiple pieces of data very quickly. But what if you want to treat the pieces differently, depending on some property or other of the objects?
This is the other central part of programming: testing for certain circumstances and changing the treatment of pieces of data accordingly. It is known as _flow control_, as you are controlling the flow of data through your script of operations.
#### if - elif - else
```
# use if statements to test for a condition (comparison, equality)
# use else to dictate what happens when the condition isn't met
# use elif to add more conditionals
# use more than one condition check
# list comprehensions with conditionals
```
|
github_jupyter
|
# the culturally-expected introductory statement
# an example of what you can achieve with Python in just a few lines
# example name type designation
42 # integer int
2.016 # float float*
"Homo sapiens" # string str
# type conversions
# print some strings
# print some numbers (ints or floats)
# print multiple values of different types all at once
# (hints: use comma to separate values with a space, or + to join strings)
# print a string containing quote marks
# standard mathematical operations can be performed in Python
# and some less common ones
# variable assignment is done with '='
# two ways to do it in Python
name = 'Florence'
age = 73
print('%s is %d years old' % (name, age)) # common amongst many programming languages
print('{} is {} years old'.format(name, age)) # perhaps more consistent with stardard Python syntax
# sequence of entries, in order and of any type
# accessing list entries
# adding/removing entries (remove/pop)
# length of list
# sets
string_object = 'the cold never bothered me anyway'
# methods - object.something()
# more...
# dir() and help()
# add 'Sally' to the list of students' names
student_names = ['Sandy', 'Pete', 'Richard', 'Rebecca']
# access the fourth entry of the list
# join the list with a new list from another class
other_student_names = ['Sam', 'Fiona', 'Sarah', 'Richard', 'Sarah', 'Matthew']
# collection of paired information - keys and values
student_marks = {'Alessio': 67, 'Nic': 48, 'Georg': 68}
empty_dict = {}
another_empty_dict = dict()
# accessing dict entries
# adding/changing/deleting entries
# lists are mutable
cities = ['Nairobi', 'Vancouver', 'Wellington', 'Beijing']
print(cities)
# strings are immutable
beatles = "I'd like to be under the sea"
for season in ['Spring', 'Summer', 'Autumn', 'Winter']:
print(season)
word = 'python'
for letter in word:
print(letter.upper())
print("Finished")
# range
# zip
# iterating through two lists simultaneously
# enumerate
# calculate the mean of the elements in the list
list_of_numbers = [1, 2, 4, 8, 3, 6, 1, 9, 10, 5]
sum_of_values = ...
for ... in list_of_numbers:
... = ... + ...
mean_value = ... / len(...)
print(mean_value)
# use if statements to test for a condition (comparison, equality)
# use else to dictate what happens when the condition isn't met
# use elif to add more conditionals
# use more than one condition check
# list comprehensions with conditionals
| 0.349311 | 0.975577 |
# 感知机模型
1、感知机是根据输入特征实例x进行二分类的线性分类模型:
$$
f(x)=\operatorname{sign}(w\cdot x+b)
$$
其中,$\operatorname{sign}$是符号函数,即
$$
\operatorname{sign}(x)=
\begin{cases}
+1, x\ge 0\\
-2, x<0\\
\end{cases}
$$
w为分离超平面的法向量,b为分离超平面的截距
2、数据集的线性可分性:
给定数据集
$$
T=\{(x_{1},y_{1}),(x_{2},y_{2}),...,(x_{n},y_{n})\}
$$
如果存在某个超平面S
$$
w\cdot x+b=0
$$
使得$y_{i}(w\cdot x_{i}+b)>0$,则称数据集T为线性可分数据集
## 损失函数
函数间隔损失函数
任意一点$x_{0}$到超平面S的距离为
$$
\frac{1}{\Arrowvert w \Arrowvert}\bracevert w\cdot x_{0}+b\bracevert
$$
对于误分类的数据$(x_{i}, y_{i})$来说,
$$
-y_{i}(w\cdot x_{i)}+b>0
$$
因此,误分类点$x_{i}$到超平面的距离为
$$
-\frac{1}{\Arrowvert w \Arrowvert}y_{i}(w\cdot x_{i}+b)
$$
这样,对于误分类点集合M,所有误分类点到超平面的距离之和为
$$
-\frac{1}{\Arrowvert w \Arrowvert}\sum_{x_{i}\in M} y_{i}(w\cdot x_{i}+b)
$$
不考虑$\frac{1}{\Arrowvert w \Arrowvert}$,得到损失函数
$$
L(w,b)=-\sum_{x_{i}\in M} y_{i}(w\cdot x_{i}+b)
$$
Q.
为什么这里不考虑$\frac{1}{\Arrowvert w \Arrowvert}$?
A.
1.超平面由法向量w和截距b确定,w的大小并不影响w的方向,$\frac{1}{\Arrowvert w \Arrowvert}$的大小并不影响分类的结果(这里也可以看作将$\Arrowvert w \Arrowvert$视为1)。
2.对于线性可分数据集,最终的损失函数值为0,$\frac{1}{\Arrowvert w \Arrowvert}$并不影响分类的最总结果。
3.采用梯度下降等算法时,不考虑$\frac{1}{\Arrowvert w \Arrowvert}$可以减少运算量。
## 感知机学习方法
### 1、感知机的学习目标
感知机的学习目标为最小化损失函数,即
$$
\min_{w,b}L(w,b)=-\sum_{x_{i}\in M} y_{i}(w\cdot x_{i}+b)
$$
### 2、采用梯度下降法进行学习
#### 2.1、梯度下降法
由泰勒公式:
$$
f(x + \Delta x)=f(x)+\sum_{i=1}^{\infty}\frac{1}{i!}f^{(i)}(x)(\Delta x)^i
$$
可得$f(x + \Delta x) = f(x)+f'(x)\Delta x + O(\Delta x)$,当$\Delta x*f'(x)>0$ 且 $\Delta x$值较小时,$f(x + \Delta x) > f(x)$,由此可构建一个递增的序列达到极大值,同理可达到极小值。
将该结论推广至高维情况,
$$
f(x + \Delta x)=f(x) + [\nabla f(x)]^T\cdot \Delta x + O(\Delta x)
$$
在$\Delta x$取值较小时,可以忽略$O(\Delta x)$,得到
$$
f(x + \Delta x)-f(x)=[\nabla f(x)]^T\cdot \Delta x=\Arrowvert \nabla f(x)\Arrowvert \Arrowvert\Delta x\Arrowvert \operatorname{cos}\theta
$$
其中$\operatorname{cos}\theta$为 $\nabla f(x)$和$\Delta x$夹角的余弦,当 $\theta = \pi$即增量方向与梯度方向相反时,下降速度最快。
P.S.在讨论时,我们要求$\Delta x$的值较小,在实现时一般添加一个较小的常数来进行限制称之为学习率。
P.S.S在这里我们只对损失函数进行一阶泰勒展开,高阶泰勒展开也是理论上可行的。
##### 2.1.1批梯度下降(BGD)
在计算梯度时,通过整个训练集的数据计算梯度。
优点是下降速度快;缺点是计算时间长,且不能在投入新数据时实时更新梯度。
##### 2.1.2随机梯度下降(SGD)
计算梯度时,只根据一个样本数据进行梯度计算。
优点是计算时间短且可以根据新数据实时更新梯度;缺点是噪音比较多,损失函数可能出现震荡。
##### 2.2.3小批量梯度下降(MBGD)
在计算梯度时,选取少量样本数据计算梯度。
优点可以降低更新时的方差,收敛更稳定;缺点是不能保证很好的收敛性,学习率如果选择的太小,收敛速度会很慢,如果太大,损失函数就会在极小值处不停地震荡甚至偏离。
P.S.考虑鞍点的情况,BGD会停留在鞍点,MBGD,SGD每次计算的方向会不同,陷入震荡。
P.S.S. 稀疏特征对应的参数在更新时的梯度会非常小几乎消失,所以在很多轮次后才可能得到稍微明显的更新,而稠密特征的参数则每次都会得到相对明显的更新。我们希望对于不同稠密度的特征对应的参数更新采取不同的学习率。对于稀疏的,我们希望调大他的学习率,让“难得”的一次更新步子更大,效果更加明显;对于稠密的,更新太过频繁,我们希望降低它的学习率,让每次的步子稍微小一点,不至于跳出全局最优点。
#### 2.2感知机学习算法原始形式
输入:训练集$T=\{(x_1, y_1),(x_2, y_2),(x_3, y_3),\cdots,(x_n, y_n)\}$, 学习率$\eta$
输出: w, b
(1)选取初值$w_0, b_0$
(2)在训练集选取数据$0(x_i, y_i)$
(3)若$y_{i}(w\cdot x_{i)}+b \le 0$
$$
w \gets w+\eta y_ix_i \\
b \gets b+\eta y_i
$$
(4)转至(2)直至训练集中没有误分类点
### 3、学习收敛性的证明
设训练数据集$T=\{(x_1, y_1),(x_2, y_2),(x_3, y_3),\cdots,(x_n, y_n)\}$是线性可分的,
(1)存在满足条件的 $\Arrowvert w_{opt}\Arrowvert =1$d的超平面$w_{opt}\cdot x+b_{opt}=0$将数据集完全正确分开且存在$\gamma >0$使得对所有$i=1,2,\cdots,n$
$$
y_i(w_{opt}\cdot x_i+b_{opt})\ge \gamma
$$
(2)令$R=\operatorname{max}_{1 \le i \le n} \Arrowvert x_i \Arrowvert$,则算法2.2的误分类次数k满足
$$
k \le \left( \frac{R}{\gamma} \right)^2
$$
证明:
(1)已知训练集线性可分,则存在w,b确定的超平面将训练集正确分开,令$w_{opt}=\frac{w}{\Arrowvert w \Arrowvert} \qquad b_{opt}=\frac{b}{\Arrowvert w \Arrowvert}$
已知对$i=0,1,\cdots,n$,
$$
y_i(w_{opt}\cdot x_i+b_{opt})> 0
$$
所以存在
$$
\gamma =\min_i\{ y_i(w_{opt}\cdot x_i+b_{opt})\}
$$
(2)令$\hat{w}_k=(w^T_{k},b_{k})^T \qquad \hat{x}=(x^T,1)^T$
对于第k个误分类点$(x_i, y_i)$,有
$$
w_k \gets w_{k-1}+\eta y_ix_i \\
b_k \gets w_{k-1}+\eta y_i
$$
即
$$
\hat{w_k} = \hat{w_{k-1}}+\eta y_i\hat{x_i}
$$
由此,两边同乘$\hat{w_{opt}}$得
$$
\hat{w_k}\cdot\hat{w_{opt}}=\hat{w_{k-1}}\cdot\hat{w_{opt}}+\eta y_i\hat{w_{opt}}\cdot\hat{x_i}\\
\ge\hat{w_{k-1}}\cdot\hat{w_{opt}}+\eta\gamma
$$
递推得到
$$
\hat{w_k}\cdot\hat{w_{opt}}\ge k\eta\gamma
$$
同时,可得到
$$
\Arrowvert\hat{w_k}\Arrowvert^2 = \Arrowvert\hat{w_{k-1}}\Arrowvert^2
+2\eta y_i\hat{w_{k-1}}\cdot\hat{x_i}+\eta^2\Arrowvert\hat{x_i}\Arrowvert^2 \\
\le\Arrowvert\hat{w_{k-1}}\Arrowvert^2 +\eta^2\Arrowvert\hat{x_i}\Arrowvert^2 \\
\le\Arrowvert\hat{w_{k-1}}\Arrowvert^2 +\eta^2R^2
\le k\eta^2R^2
$$
综上,可得
$$
k\eta\gamma\le\hat{w_k}\cdot\hat{w_{opt}}\le\Arrowvert\hat{w_k}\Arrowvert\Arrowvert\hat{w_{opt}}\Arrowvert\le\sqrt{k}\eta R\\
k^2\gamma^2\le kR^2
$$
即
$$
k \le \left( \frac{R}{\gamma} \right)^2
$$
### 4、感知机的对偶形式
#### 4.1对偶形式
对偶形式的基本想法是,将 w 和 b 表示为是咧 xi 和标记 yi的线性组合的形式,通过求解其系数而得到 w 和 b。
#### 4.2感知机的对偶形式
由算法可知,对误分类的点$(x_i, y_i)$,通过
$$
w\gets w+\eta y_ix_i\\
b\gets b+\eta y_i
$$
若修改n次,则最后学习到的w,b可以表示为
$$
w=\sum_{i=1}^N\alpha_iy_ix_i\\
b=\sum_{i=1}^N\alpha_iy_i
$$
这里$\alpha_i=k_i*\eta$,其中$k_i$为第i个实例由于误分而进行的更新次数
最后得到的模型为
$$
f(x)=\operatorname{sign}\left(\sum_{i=1}^N\alpha_iy_ix_i\cdot x+\sum_{i=1}^N\alpha_iy_i\right)
$$
相应地,对应的算法为:
(1)$\alpha\gets 0,\quad b\gets 0$
(2)在训练集选取数据$x_i, y_i$
(3)若$y_i\left(\sum_{i=1}^N\alpha_iy_ix_i\cdot x+\sum_{i=1}^N\alpha_iy_i\right)\le 0$
$$
\alpha_i\gets\alpha_i+\eta
\
$$
(4)转至(2)直至没有误分类数据
### 4.3对偶形式的实际意义
由上可见对偶形式与一般形式的算法本质没有区别,但如果预先将训练集中实例间的内积计算出来并以矩阵的形式存储下来,即Gram矩阵就可以减少训练时间
$$
G=[x_i\cdot x_j]_{N\times N}
$$
## 相关代码实现
```
#经典感知机
import numpy as np
class Perceptron:
def __init__(self, lr=0.01):
self.lr=lr
def fit(self, X, y):
self.w=np.random.randn(X.shape[1])
self.b=0
is_wrong=True
while is_wrong:
wrong_count=0
for X_,y_ in zip(X,y):
if y_*(self.sign(X_,self.w,self.b))<=0:
self.w=self.w+self.lr*np.dot(y_, X_)
self.b=self.b+self.lr*y_
wrong_count +=1
is_wrong = (wrong_count>0)
def sign(self,X,w,b):
return np.dot(X, w)+b
def predict(self,X):
return np.where((np.dot(X, self.w)+self.b)>=0.0, 1, -1)
#对偶形式感知机
class DualityPerceptron:
def __init__(self, lr=0.01):
self.lr=lr
def fit(self, X, y):
self.alpha=np.random.randn(X.shape[0])
self.b=0
G=np.dot(X, X.T)
is_wrong=True
while is_wrong:
wrong_count=0
for idx in range(X.shape[0]):
if y[idx]*(np.sum(self.alpha*y*G[idx])+self.b)<=0:
self.alpha+=self.lr
self.b+=self.lr*y[idx]
wrong_count +=1
is_wrong = (wrong_count>0)
self.w=np.sum(self.alpha*y*X.T, axis=1)
def predict(self, X):
return np.where((np.dot(X, self.w)+self.b)>=0.0, 1, -1)
import pandas as pd
df = pd.read_csv('http://archive.ics.uci.edu/ml/'
'machine-learning-databases/iris/iris.data',
header = None)
df.tail()
import matplotlib.pyplot as plt
import numpy as np
y = df.iloc[0: 100, 4].values
y = np.where(y == 'Iris-setosa', -1, 1)
X = df.iloc[0:100, [0,2]].values
plt.scatter(X[:50, 0], X[:50, 1],
color = 'red', marker = 'o',
label = 'setosa')
plt.scatter(X[50:100, 0], X[50:100, 1],
color = 'blue', marker = 'x',
label = 'versicolor')
plt.xlabel('sepal lenth [cm]')
plt.ylabel('prtap lenth [cm]')
plt.legend(loc = 'upper left')
plt.show()
from matplotlib.colors import ListedColormap
def plot_decision_regions(X, y, classifier, resolution = 0.02):
markers = ('s', 'x', 'o', '^', 'v')
colors = ('red', 'blue', 'lightgreen', 'gray', 'cyan')
cmap = ListedColormap(colors[:len(np.unique(y))])
# plot the decision surface
x1_min, x1_max = X[:, 0].min() - 1, X[:, 0].max() + 1
x2_min, x2_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx1, xx2 = np.meshgrid(np.arange(x1_min, x1_max, resolution),
np.arange(x2_min, x2_max, resolution))
Z = classifier.predict(np.array([xx1.ravel(), xx2.ravel()]).T)
Z = Z.reshape(xx1.shape)
plt.contourf(xx1, xx2, Z, alpha=0.4, cmap=cmap)
plt.xlim(xx1.min(), xx1.max())
plt.ylim(xx2.min(), xx2.max())
#plot class samples
for idx, cl in enumerate(np.unique(y)):
plt.scatter(x=X[y == cl, 0],
y=X[y == cl, 1],
alpha=0.8,
c=cmap(idx),
edgecolor='black',
marker=markers[idx],
label=cl)
ppn = Perceptron()
ppn.fit(X,y)
plot_decision_regions(X, y, classifier = ppn)
plt.xlabel('sepal lenth [cm]')
plt.ylabel('prtap lenth [cm]')
plt.legend(loc = 'upper left')
plt.tight_layout()
plt.show()
dppn = DualityPerceptron()
dppn.fit(X,y)
plot_decision_regions(X, y, classifier = dppn)
plt.xlabel('sepal lenth [cm]')
plt.ylabel('prtap lenth [cm]')
plt.legend(loc = 'upper left')
plt.tight_layout()
plt.show()
%timeit ppn.fit(X,y)
%timeit dppn.fit(X,y)
%timeit ppn.fit(X,y)
%timeit dppn.fit(X,y)
```
|
github_jupyter
|
#经典感知机
import numpy as np
class Perceptron:
def __init__(self, lr=0.01):
self.lr=lr
def fit(self, X, y):
self.w=np.random.randn(X.shape[1])
self.b=0
is_wrong=True
while is_wrong:
wrong_count=0
for X_,y_ in zip(X,y):
if y_*(self.sign(X_,self.w,self.b))<=0:
self.w=self.w+self.lr*np.dot(y_, X_)
self.b=self.b+self.lr*y_
wrong_count +=1
is_wrong = (wrong_count>0)
def sign(self,X,w,b):
return np.dot(X, w)+b
def predict(self,X):
return np.where((np.dot(X, self.w)+self.b)>=0.0, 1, -1)
#对偶形式感知机
class DualityPerceptron:
def __init__(self, lr=0.01):
self.lr=lr
def fit(self, X, y):
self.alpha=np.random.randn(X.shape[0])
self.b=0
G=np.dot(X, X.T)
is_wrong=True
while is_wrong:
wrong_count=0
for idx in range(X.shape[0]):
if y[idx]*(np.sum(self.alpha*y*G[idx])+self.b)<=0:
self.alpha+=self.lr
self.b+=self.lr*y[idx]
wrong_count +=1
is_wrong = (wrong_count>0)
self.w=np.sum(self.alpha*y*X.T, axis=1)
def predict(self, X):
return np.where((np.dot(X, self.w)+self.b)>=0.0, 1, -1)
import pandas as pd
df = pd.read_csv('http://archive.ics.uci.edu/ml/'
'machine-learning-databases/iris/iris.data',
header = None)
df.tail()
import matplotlib.pyplot as plt
import numpy as np
y = df.iloc[0: 100, 4].values
y = np.where(y == 'Iris-setosa', -1, 1)
X = df.iloc[0:100, [0,2]].values
plt.scatter(X[:50, 0], X[:50, 1],
color = 'red', marker = 'o',
label = 'setosa')
plt.scatter(X[50:100, 0], X[50:100, 1],
color = 'blue', marker = 'x',
label = 'versicolor')
plt.xlabel('sepal lenth [cm]')
plt.ylabel('prtap lenth [cm]')
plt.legend(loc = 'upper left')
plt.show()
from matplotlib.colors import ListedColormap
def plot_decision_regions(X, y, classifier, resolution = 0.02):
markers = ('s', 'x', 'o', '^', 'v')
colors = ('red', 'blue', 'lightgreen', 'gray', 'cyan')
cmap = ListedColormap(colors[:len(np.unique(y))])
# plot the decision surface
x1_min, x1_max = X[:, 0].min() - 1, X[:, 0].max() + 1
x2_min, x2_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx1, xx2 = np.meshgrid(np.arange(x1_min, x1_max, resolution),
np.arange(x2_min, x2_max, resolution))
Z = classifier.predict(np.array([xx1.ravel(), xx2.ravel()]).T)
Z = Z.reshape(xx1.shape)
plt.contourf(xx1, xx2, Z, alpha=0.4, cmap=cmap)
plt.xlim(xx1.min(), xx1.max())
plt.ylim(xx2.min(), xx2.max())
#plot class samples
for idx, cl in enumerate(np.unique(y)):
plt.scatter(x=X[y == cl, 0],
y=X[y == cl, 1],
alpha=0.8,
c=cmap(idx),
edgecolor='black',
marker=markers[idx],
label=cl)
ppn = Perceptron()
ppn.fit(X,y)
plot_decision_regions(X, y, classifier = ppn)
plt.xlabel('sepal lenth [cm]')
plt.ylabel('prtap lenth [cm]')
plt.legend(loc = 'upper left')
plt.tight_layout()
plt.show()
dppn = DualityPerceptron()
dppn.fit(X,y)
plot_decision_regions(X, y, classifier = dppn)
plt.xlabel('sepal lenth [cm]')
plt.ylabel('prtap lenth [cm]')
plt.legend(loc = 'upper left')
plt.tight_layout()
plt.show()
%timeit ppn.fit(X,y)
%timeit dppn.fit(X,y)
%timeit ppn.fit(X,y)
%timeit dppn.fit(X,y)
| 0.535098 | 0.936401 |
Breast cancer: Benign or Malignant
# **Data preprocessing**
```
#reading dataset from csv file
import pandas as pd
data=pd.read_csv("data.csv")
#view the data present
data
#drop first column having ids
data=data.iloc[:,1:32]
#convert the output column into numbers : 1 for 'M' , 2 for 'B'
match={'M':0,'B':1}
for i in range(0,len(data)):
data.loc[i,'diagnosis']=match[data.loc[i,'diagnosis']]
#data after adding diagnosis column to know if it benign or malignant
data
# split into input (X) and output (y) variables
X = data.iloc[:,1:]
y = data.iloc[:,0]
#converting the output column to int type from object type to feed into the model
y=y.astype('int')
#splitting the data into test and train sets
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42)
#scaling the input featuras they all are indifferent ranges and normalising them would ensure no input overshadows
# the other due to its high value.
from sklearn.preprocessing import StandardScaler
st_x= StandardScaler()
X_train= st_x.fit_transform(X_train)
X_test= st_x.transform(X_test)
```
# **Using KNN classifier**
```
#First alogorithm; KNN classifier
from sklearn.neighbors import KNeighborsClassifier
classifier= KNeighborsClassifier(n_neighbors=5)
#fitting the training data into the KNN model
classifier.fit(X_train, y_train)
#testing the model on the test set
y_pred= classifier.predict(X_test)
#testing out the metrics through confusion matrix
#inferred as:
# 63 values were outputted to 0 and were actually 0
# 117 values were outputted to 1 and were actually 1
# 4 values were outputted to 1 and were actually 0
# 4 values were outputted to 0 and were actually 1
from sklearn.metrics import confusion_matrix
print(confusion_matrix(y_test, y_pred))
#the model's accuracy was 96% i.e. it predicted 96% of the data correctly.
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
```
# **Using Logistic Regression**
```
#Second alogorithm; Logistic regression
from sklearn.linear_model import LogisticRegression
lm=LogisticRegression()
#fitting the training data into the KNN model
lm.fit(X_train,y_train)
#testing the model on the test set
y_pred=lm.predict(X_test)
#testing out the metrics through confusion matrix
#inferred as:
# 66 values were outputted to 0 and were actually 0
# 118 values were outputted to 1 and were actually 1
# 1 values were outputted to 1 and were actually 0
# 3 values were outputted to 0 and were actually 1
print(confusion_matrix(y_test, y_pred))
#the model's accuracy was 98% i.e. it predicted 98% of the data correctly.
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
```
|
github_jupyter
|
#reading dataset from csv file
import pandas as pd
data=pd.read_csv("data.csv")
#view the data present
data
#drop first column having ids
data=data.iloc[:,1:32]
#convert the output column into numbers : 1 for 'M' , 2 for 'B'
match={'M':0,'B':1}
for i in range(0,len(data)):
data.loc[i,'diagnosis']=match[data.loc[i,'diagnosis']]
#data after adding diagnosis column to know if it benign or malignant
data
# split into input (X) and output (y) variables
X = data.iloc[:,1:]
y = data.iloc[:,0]
#converting the output column to int type from object type to feed into the model
y=y.astype('int')
#splitting the data into test and train sets
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42)
#scaling the input featuras they all are indifferent ranges and normalising them would ensure no input overshadows
# the other due to its high value.
from sklearn.preprocessing import StandardScaler
st_x= StandardScaler()
X_train= st_x.fit_transform(X_train)
X_test= st_x.transform(X_test)
#First alogorithm; KNN classifier
from sklearn.neighbors import KNeighborsClassifier
classifier= KNeighborsClassifier(n_neighbors=5)
#fitting the training data into the KNN model
classifier.fit(X_train, y_train)
#testing the model on the test set
y_pred= classifier.predict(X_test)
#testing out the metrics through confusion matrix
#inferred as:
# 63 values were outputted to 0 and were actually 0
# 117 values were outputted to 1 and were actually 1
# 4 values were outputted to 1 and were actually 0
# 4 values were outputted to 0 and were actually 1
from sklearn.metrics import confusion_matrix
print(confusion_matrix(y_test, y_pred))
#the model's accuracy was 96% i.e. it predicted 96% of the data correctly.
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
#Second alogorithm; Logistic regression
from sklearn.linear_model import LogisticRegression
lm=LogisticRegression()
#fitting the training data into the KNN model
lm.fit(X_train,y_train)
#testing the model on the test set
y_pred=lm.predict(X_test)
#testing out the metrics through confusion matrix
#inferred as:
# 66 values were outputted to 0 and were actually 0
# 118 values were outputted to 1 and were actually 1
# 1 values were outputted to 1 and were actually 0
# 3 values were outputted to 0 and were actually 1
print(confusion_matrix(y_test, y_pred))
#the model's accuracy was 98% i.e. it predicted 98% of the data correctly.
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
| 0.449634 | 0.872021 |
# Analysis on correlation between earthquakes and surface mass loading
```
import xarray as xr
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
from global_land_mask import globe
import scipy.stats as stats
import os
import importlib
import geopandas as gpd
# file containing functions:
import loadquakes
importlib.reload(loadquakes)
```
### Load datasets (GRACE and declustered catalog)
```
dataset_path = '/data/fast1/surface-load/'
ds = xr.open_dataset(dataset_path + "GRCTellus.JPL.200204_202201.GLO.RL06M.MSCNv02CRI.nc")
# Events from 2002-04-16 to 2022-1-16 with magnitude >= 5.4
catalog = pd.read_csv('ind_and_bg_probs.csv')
catalog["time"] = catalog["time"].astype("datetime64")
# This is the catalog with all details from the USGS Earthquake Catalog
original_catalog = pd.read_csv('19970101-20220116.csv')
original_catalog = original_catalog[original_catalog['time']>'2002-04-16']
original_catalog = original_catalog[original_catalog['time']<'2022-01-16']
cat = original_catalog.reset_index()
depth = cat['depth']
catalog['depth'] = depth
catalog
```
### Filter events that that have less than 90% chance of being triggered
##### The mean probability of being triggered (P_triggered) for the catalog is 97.97% with a standard deviation of 9.25% . Here, we examine events that have less 90% probability of being triggered by another earthquake
```
mainshock = catalog[catalog['P_triggered']<.9]
mainshock.sort_values("magnitude")
```
### Add surface load to the earthquake dataframes
```
def get_mass(eq):
mass_change = ds.sel(lat=(eq["latitude"]),lon=(eq["longitude"]),time=(eq["time"]), method="nearest")
mass_change = mass_change["lwe_thickness"]
return mass_change.data
mainshock['dmass'] = mainshock.apply(lambda row: get_mass(row), axis=1)
catalog['dmass'] = catalog.apply(lambda row: get_mass(row), axis=1)
```
#### Are the mean/meadian values different between: 1. The whole dataset, 2. times with earthquakes, and 3. times with earthquakes that have low probability of being triggered?
The answer appears to be: no, not really. The Bayesian analysis explains why! It's because mainshocks are favored at large positive AND negative surface loads. The symmetry implies that the means don't change much.
```
print('Mainshock mean: %f'%mainshock['dmass'].mean())
print('Mainshock median: %f'%mainshock['dmass'].median())
print('Earthquake mean: %f'%catalog['dmass'].mean())
print('Earthquake median: %f'%catalog['dmass'].median())
print(ds.lwe_thickness.mean())
print(ds.lwe_thickness.median())
```
### Correlation between all events and surface mass loading
```
# takes a long time to run (has already been run for 2002-04-16 to 2022-01-16)
# loadquakes.calc_stats(np.array(catalog.dmass),np.array(ds['lwe_thickness'].data.flatten()))
importlib.reload(loadquakes)
def set_of_figures_load(all_time, earthquake_only,bayes_title,method):
fig,(ax1,ax2,ax3) = plt.subplots(3,1, figsize=(7,14))
plt.style.use('fivethirtyeight')
loadquakes.plot_hist(all_time, earthquake_only, ax1, ax2,
'a. Cumulative Distribution', 'b. Probability Density', method)
loadquakes.plot_bayes(all_time, earthquake_only, ax3, bayes_title,
method)
fig.tight_layout()
loadquakes.freedman_diaconis(ds['lwe_thickness'].data.flatten(), returnas = "bins")
set_of_figures_load(ds['lwe_thickness'].data.flatten(), catalog.dmass,
'c. Bayesian Probability of Event (n=12573)','fd')
set_of_figures_load(ds['lwe_thickness'].data.flatten(), catalog.dmass,
'c. Bayesian Probability of Event (n=12573)','Sturge')
```
## Correlation between mainshocks and surface mass loading
#### Both statistical tests produce p-values < 0.05, which indicate that the two samples are not drawn from the same distribution. This means that the distribution of surface mass load during earthquake occurrence is significantly different from background.
```
# takes a long time to run (has already been run for 2002-04-16 to 2022-01-16)
# loadquakes.calc_stats(np.array(mainshock.dmass),np.array(ds['lwe_thickness'].data.flatten()))
importlib.reload(loadquakes)
set_of_figures_load(ds['lwe_thickness'].data.flatten(), mainshock.dmass,
'c. Bayesian Probability of Event (n=537)','fd')
```
#### The conditional probability will change quantitatively depending on the bin size. However, the same qualitative distribution remains, where there is a higher relative conditional probability during periods of large loading and unloading.
#### Here, we calculate the Bayesian probability using two different bin sizes which are calculated via two methods:
##### a. Sturge's Rule
##### b. Freedman-Diaconis Rule
```
importlib.reload(loadquakes)
fig,(ax1,ax2) = plt.subplots(1,2, figsize=(17,6))
plt.style.use('fivethirtyeight')
loadquakes.plot_bayes(ds['lwe_thickness'].data.flatten(), mainshock.dmass, ax1,
"a. Bin size calculated via Sturge's Rule", method='Sturge')
loadquakes.plot_bayes(ds['lwe_thickness'].data.flatten(), mainshock.dmass, ax2,
"b. Bin size calculated via Freedman-Diaconis Rule", method='fd')
fig.tight_layout()
```
## Mapping earthquakes with high conditional probability
### First, calculate the conditional probability of each mainshock
```
conditional_probability = loadquakes.get_cond_probability(
np.array(ds['lwe_thickness'].data.flatten()),
np.array(mainshock.dmass), np.array(mainshock.dmass), 'fd')
mainshock['cp'] = conditional_probability
fig,ax=plt.subplots()
plt.hist(np.array(mainshock['cp']),11)
plt.title('Histogram of mainshock conditional probabilities')
plt.yscale('log')
from scipy.stats import iqr
iqr(mainshock['cp'])
np.median(mainshock.cp)
importlib.reload(loadquakes)
def all_figs(all_loads,full_catalog,declustered_catalog,bayes_title1,bayes_title2,method):
fig,((ax1,ax4),(ax2,ax5),(ax3,ax6)) = plt.subplots(3,2, figsize=(14,14))
plt.style.use('fivethirtyeight')
loadquakes.plot_hist(all_loads, full_catalog, ax1, ax2,
'a. Cumulative Distribution', 'b. Probability Density', method)
loadquakes.plot_bayes(all_loads, full_catalog, ax3, bayes_title1,
method)
loadquakes.plot_hist(all_loads, declustered_catalog, ax4, ax5,
'd. Cumulative Distribution (declustered catalog)',
'e. Probability Density (declustered catalog)',
method)
loadquakes.plot_bayes(all_loads, declustered_catalog, ax6, bayes_title2,
method)
fig.tight_layout()
all_figs(ds['lwe_thickness'].data.flatten(),
catalog.dmass,
mainshock.dmass,
'c. Bayesian Probability of Event (n=12573)',
'f. Bayesian Probability of Event (n=537)','fd')
np.min(mainshock.dmass)
```
### The relative conditional probability for the declustered catalog has a median of is 1.06 and an IQR of 0.68. We use the addition of these values as the threshold to categorize an event as a SLIQ.
```
sliq = mainshock[mainshock['cp']>1.74]
len(sliq)
sliq.sort_values('cp')
sliq.sort_values('magnitude')
from mpl_toolkits.axes_grid1 import make_axes_locatable
import matplotlib.cm as cm
# c = [0.5, 0.5, 0.5]
def probability_map_cb(events,color,label):
gdf = gpd.GeoDataFrame(events,
geometry=gpd.points_from_xy(events.longitude,
events.latitude))
world = gpd.read_file(gpd.datasets.get_path('naturalearth_lowres'))
ax = world.plot(color='white', edgecolor='black', figsize=(15,10))
divider = make_axes_locatable(ax)
cax = divider.append_axes("bottom", size="5%", pad=0.6)
ax.scatter(catalog.longitude, catalog.latitude, c="darkgrey", s=20,marker=".")
cmap = cm.get_cmap('viridis', 11) # 11 discrete colors
gdf.plot(ax=ax,cax=cax,alpha=0.5,column=color,cmap=cmap,legend=True,
edgecolor='k',
markersize = 1e-5*(events.magnitude)**10,
legend_kwds={'label': "Relative conditional probability of event",
'orientation': "horizontal"},
vmax=5,
vmin=1.74)
gdf.plot(ax=ax,facecolor="None",
edgecolor='k',
markersize = 1e-5*(events.magnitude)**10)
ax.set_xlabel('Longitude', fontsize = 15)
ax.set_ylabel("Latitude", fontsize = 15)
ax.set_title(label)
plt.show()
probability_map_cb(sliq,sliq.cp,
'SLIQs with relative conditional probability > 1.74')
montana = sliq[sliq['longitude']<-100]
montana
## Depth comparison sliq v declustered catalog
fig,(ax1,ax2) = plt.subplots(1,2, figsize=(15,5))
bins_depth = loadquakes.calculate_bin_sizes(mainshock.depth,'fd')
ax1.hist(sliq.depth,bins,density = True, cumulative=True, histtype='step',
label='SLIQs',linewidth=1.5)
ax1.hist(mainshock.depth, 50,density = True, cumulative=True,histtype='step',
label='Declustered catalog',linewidth=1.5)
yl = ax1.get_ylim()
ax1.set_ylim((-0.01,1.4*yl[1]))
xl = ax1.get_xlim()
ax1.set_xlim(xl[0],xl[1])
ax1.legend()
ax1.set_xlabel('Depth (km)', fontsize = 17)
ax1.set_ylabel("Cumulative probability", fontsize = 17)
ax1.set_title("Cumulative Distribution", fontsize = 17)
ax2.hist(sliq.depth, 50,density = True, cumulative=False, histtype='step',
label='SLIQs',linewidth=1.5)
ax2.hist(mainshock.depth, 50,density = True, cumulative=False,histtype='step',
label='Declustered catalog',linewidth=1.5)
yl = ax2.get_ylim()
ax2.set_ylim((-0.001,1.4*yl[1]))
xl = ax1.get_xlim()
ax2.set_xlim(xl[0],xl[1])
ax2.legend()
ax2.set_xlabel('Depth (km)', fontsize = 17)
ax2.set_ylabel("Probability", fontsize = 17)
ax2.set_title('Probability Density', fontsize = 17)
fig.tight_layout()
## Depth comparison sliq v all earthquakes
fig,(ax1,ax2) = plt.subplots(1,2, figsize=(15,5))
bins_depth = loadquakes.calculate_bin_sizes(catalog.depth,'fd')
ax1.hist(sliq.depth,bins,density = True, cumulative=True, histtype='step',
label='SLIQs',linewidth=1.5, color="mediumvioletred")
ax1.hist(catalog.depth, 50,density = True, cumulative=True,histtype='step',
label='All earthquakes',linewidth=1.5,color="royalblue")
yl = ax1.get_ylim()
ax1.set_ylim((-0.01,1.4*yl[1]))
xl = ax1.get_xlim()
ax1.set_xlim(xl[0],xl[1])
ax1.legend()
ax1.set_xlabel('Depth (km)', fontsize = 17)
ax1.set_ylabel("Cumulative probability", fontsize = 17)
ax1.set_title("Cumulative Distribution", fontsize = 17)
ax2.hist(sliq.depth, 50,density = True, cumulative=False, histtype='step',
label='SLIQs',linewidth=1.5,color="mediumvioletred")
ax2.hist(catalog.depth, 50,density = True, cumulative=False,histtype='step',
label='All earthquakes',linewidth=1.5,color="royalblue")
yl = ax2.get_ylim()
ax2.set_ylim((-0.001,1.4*yl[1]))
xl = ax1.get_xlim()
ax2.set_xlim(xl[0],xl[1])
ax2.legend()
ax2.set_xlabel('Depth (km)', fontsize = 17)
ax2.set_ylabel("Probability", fontsize = 17)
ax2.set_title('Probability Density', fontsize = 17)
fig.tight_layout()
```
### Depth statistics:
```
print('Catalog mean: %f'%catalog['depth'].mean())
print('Catalog median: %f'%catalog['depth'].median())
print('Declustered catalog mean: %f'%mainshock['depth'].mean())
print('Declustered catalog median: %f'%mainshock['depth'].median())
print('SLIQ mean: %f'%sliq['depth'].mean())
print('SLIQ median: %f'%sliq['depth'].median())
```
# Correlation between earthquakes and random data
##### *mean and std deviation of random array same as GRACE data
```
np.mean(ds['lwe_thickness'])
np.std(ds['lwe_thickness'])
random_load1 = np.random.normal(loc=-0.65387518, scale=25.49255185, size=(205, 360, 720))
ds_random = ds
ds_random = ds_random.assign(load=ds_random['lwe_thickness'] - ds_random['lwe_thickness'] + random_load1)
# earthquake catalog
mainshock_random = catalog[catalog['P_triggered']<.9]
def get_mass_random(eq):
mass_change = ds_random.sel(lat=(eq["latitude"]),lon=(eq["longitude"]),time=(eq["time"]), method="nearest")
mass_change = mass_change["load"]
return mass_change.data
mainshock_random['dmass'] = mainshock_random.apply(lambda row: get_mass_random(row), axis=1)
set_of_figures_load(ds_random['load'].data.flatten(), mainshock_random.dmass,'c. Bayesian Probability of Mainshock (n=537)')
stats.ks_2samp(mainshock_random.dmass,ds_random['load'].data.flatten())
```
|
github_jupyter
|
import xarray as xr
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
from global_land_mask import globe
import scipy.stats as stats
import os
import importlib
import geopandas as gpd
# file containing functions:
import loadquakes
importlib.reload(loadquakes)
dataset_path = '/data/fast1/surface-load/'
ds = xr.open_dataset(dataset_path + "GRCTellus.JPL.200204_202201.GLO.RL06M.MSCNv02CRI.nc")
# Events from 2002-04-16 to 2022-1-16 with magnitude >= 5.4
catalog = pd.read_csv('ind_and_bg_probs.csv')
catalog["time"] = catalog["time"].astype("datetime64")
# This is the catalog with all details from the USGS Earthquake Catalog
original_catalog = pd.read_csv('19970101-20220116.csv')
original_catalog = original_catalog[original_catalog['time']>'2002-04-16']
original_catalog = original_catalog[original_catalog['time']<'2022-01-16']
cat = original_catalog.reset_index()
depth = cat['depth']
catalog['depth'] = depth
catalog
mainshock = catalog[catalog['P_triggered']<.9]
mainshock.sort_values("magnitude")
def get_mass(eq):
mass_change = ds.sel(lat=(eq["latitude"]),lon=(eq["longitude"]),time=(eq["time"]), method="nearest")
mass_change = mass_change["lwe_thickness"]
return mass_change.data
mainshock['dmass'] = mainshock.apply(lambda row: get_mass(row), axis=1)
catalog['dmass'] = catalog.apply(lambda row: get_mass(row), axis=1)
print('Mainshock mean: %f'%mainshock['dmass'].mean())
print('Mainshock median: %f'%mainshock['dmass'].median())
print('Earthquake mean: %f'%catalog['dmass'].mean())
print('Earthquake median: %f'%catalog['dmass'].median())
print(ds.lwe_thickness.mean())
print(ds.lwe_thickness.median())
# takes a long time to run (has already been run for 2002-04-16 to 2022-01-16)
# loadquakes.calc_stats(np.array(catalog.dmass),np.array(ds['lwe_thickness'].data.flatten()))
importlib.reload(loadquakes)
def set_of_figures_load(all_time, earthquake_only,bayes_title,method):
fig,(ax1,ax2,ax3) = plt.subplots(3,1, figsize=(7,14))
plt.style.use('fivethirtyeight')
loadquakes.plot_hist(all_time, earthquake_only, ax1, ax2,
'a. Cumulative Distribution', 'b. Probability Density', method)
loadquakes.plot_bayes(all_time, earthquake_only, ax3, bayes_title,
method)
fig.tight_layout()
loadquakes.freedman_diaconis(ds['lwe_thickness'].data.flatten(), returnas = "bins")
set_of_figures_load(ds['lwe_thickness'].data.flatten(), catalog.dmass,
'c. Bayesian Probability of Event (n=12573)','fd')
set_of_figures_load(ds['lwe_thickness'].data.flatten(), catalog.dmass,
'c. Bayesian Probability of Event (n=12573)','Sturge')
# takes a long time to run (has already been run for 2002-04-16 to 2022-01-16)
# loadquakes.calc_stats(np.array(mainshock.dmass),np.array(ds['lwe_thickness'].data.flatten()))
importlib.reload(loadquakes)
set_of_figures_load(ds['lwe_thickness'].data.flatten(), mainshock.dmass,
'c. Bayesian Probability of Event (n=537)','fd')
importlib.reload(loadquakes)
fig,(ax1,ax2) = plt.subplots(1,2, figsize=(17,6))
plt.style.use('fivethirtyeight')
loadquakes.plot_bayes(ds['lwe_thickness'].data.flatten(), mainshock.dmass, ax1,
"a. Bin size calculated via Sturge's Rule", method='Sturge')
loadquakes.plot_bayes(ds['lwe_thickness'].data.flatten(), mainshock.dmass, ax2,
"b. Bin size calculated via Freedman-Diaconis Rule", method='fd')
fig.tight_layout()
conditional_probability = loadquakes.get_cond_probability(
np.array(ds['lwe_thickness'].data.flatten()),
np.array(mainshock.dmass), np.array(mainshock.dmass), 'fd')
mainshock['cp'] = conditional_probability
fig,ax=plt.subplots()
plt.hist(np.array(mainshock['cp']),11)
plt.title('Histogram of mainshock conditional probabilities')
plt.yscale('log')
from scipy.stats import iqr
iqr(mainshock['cp'])
np.median(mainshock.cp)
importlib.reload(loadquakes)
def all_figs(all_loads,full_catalog,declustered_catalog,bayes_title1,bayes_title2,method):
fig,((ax1,ax4),(ax2,ax5),(ax3,ax6)) = plt.subplots(3,2, figsize=(14,14))
plt.style.use('fivethirtyeight')
loadquakes.plot_hist(all_loads, full_catalog, ax1, ax2,
'a. Cumulative Distribution', 'b. Probability Density', method)
loadquakes.plot_bayes(all_loads, full_catalog, ax3, bayes_title1,
method)
loadquakes.plot_hist(all_loads, declustered_catalog, ax4, ax5,
'd. Cumulative Distribution (declustered catalog)',
'e. Probability Density (declustered catalog)',
method)
loadquakes.plot_bayes(all_loads, declustered_catalog, ax6, bayes_title2,
method)
fig.tight_layout()
all_figs(ds['lwe_thickness'].data.flatten(),
catalog.dmass,
mainshock.dmass,
'c. Bayesian Probability of Event (n=12573)',
'f. Bayesian Probability of Event (n=537)','fd')
np.min(mainshock.dmass)
sliq = mainshock[mainshock['cp']>1.74]
len(sliq)
sliq.sort_values('cp')
sliq.sort_values('magnitude')
from mpl_toolkits.axes_grid1 import make_axes_locatable
import matplotlib.cm as cm
# c = [0.5, 0.5, 0.5]
def probability_map_cb(events,color,label):
gdf = gpd.GeoDataFrame(events,
geometry=gpd.points_from_xy(events.longitude,
events.latitude))
world = gpd.read_file(gpd.datasets.get_path('naturalearth_lowres'))
ax = world.plot(color='white', edgecolor='black', figsize=(15,10))
divider = make_axes_locatable(ax)
cax = divider.append_axes("bottom", size="5%", pad=0.6)
ax.scatter(catalog.longitude, catalog.latitude, c="darkgrey", s=20,marker=".")
cmap = cm.get_cmap('viridis', 11) # 11 discrete colors
gdf.plot(ax=ax,cax=cax,alpha=0.5,column=color,cmap=cmap,legend=True,
edgecolor='k',
markersize = 1e-5*(events.magnitude)**10,
legend_kwds={'label': "Relative conditional probability of event",
'orientation': "horizontal"},
vmax=5,
vmin=1.74)
gdf.plot(ax=ax,facecolor="None",
edgecolor='k',
markersize = 1e-5*(events.magnitude)**10)
ax.set_xlabel('Longitude', fontsize = 15)
ax.set_ylabel("Latitude", fontsize = 15)
ax.set_title(label)
plt.show()
probability_map_cb(sliq,sliq.cp,
'SLIQs with relative conditional probability > 1.74')
montana = sliq[sliq['longitude']<-100]
montana
## Depth comparison sliq v declustered catalog
fig,(ax1,ax2) = plt.subplots(1,2, figsize=(15,5))
bins_depth = loadquakes.calculate_bin_sizes(mainshock.depth,'fd')
ax1.hist(sliq.depth,bins,density = True, cumulative=True, histtype='step',
label='SLIQs',linewidth=1.5)
ax1.hist(mainshock.depth, 50,density = True, cumulative=True,histtype='step',
label='Declustered catalog',linewidth=1.5)
yl = ax1.get_ylim()
ax1.set_ylim((-0.01,1.4*yl[1]))
xl = ax1.get_xlim()
ax1.set_xlim(xl[0],xl[1])
ax1.legend()
ax1.set_xlabel('Depth (km)', fontsize = 17)
ax1.set_ylabel("Cumulative probability", fontsize = 17)
ax1.set_title("Cumulative Distribution", fontsize = 17)
ax2.hist(sliq.depth, 50,density = True, cumulative=False, histtype='step',
label='SLIQs',linewidth=1.5)
ax2.hist(mainshock.depth, 50,density = True, cumulative=False,histtype='step',
label='Declustered catalog',linewidth=1.5)
yl = ax2.get_ylim()
ax2.set_ylim((-0.001,1.4*yl[1]))
xl = ax1.get_xlim()
ax2.set_xlim(xl[0],xl[1])
ax2.legend()
ax2.set_xlabel('Depth (km)', fontsize = 17)
ax2.set_ylabel("Probability", fontsize = 17)
ax2.set_title('Probability Density', fontsize = 17)
fig.tight_layout()
## Depth comparison sliq v all earthquakes
fig,(ax1,ax2) = plt.subplots(1,2, figsize=(15,5))
bins_depth = loadquakes.calculate_bin_sizes(catalog.depth,'fd')
ax1.hist(sliq.depth,bins,density = True, cumulative=True, histtype='step',
label='SLIQs',linewidth=1.5, color="mediumvioletred")
ax1.hist(catalog.depth, 50,density = True, cumulative=True,histtype='step',
label='All earthquakes',linewidth=1.5,color="royalblue")
yl = ax1.get_ylim()
ax1.set_ylim((-0.01,1.4*yl[1]))
xl = ax1.get_xlim()
ax1.set_xlim(xl[0],xl[1])
ax1.legend()
ax1.set_xlabel('Depth (km)', fontsize = 17)
ax1.set_ylabel("Cumulative probability", fontsize = 17)
ax1.set_title("Cumulative Distribution", fontsize = 17)
ax2.hist(sliq.depth, 50,density = True, cumulative=False, histtype='step',
label='SLIQs',linewidth=1.5,color="mediumvioletred")
ax2.hist(catalog.depth, 50,density = True, cumulative=False,histtype='step',
label='All earthquakes',linewidth=1.5,color="royalblue")
yl = ax2.get_ylim()
ax2.set_ylim((-0.001,1.4*yl[1]))
xl = ax1.get_xlim()
ax2.set_xlim(xl[0],xl[1])
ax2.legend()
ax2.set_xlabel('Depth (km)', fontsize = 17)
ax2.set_ylabel("Probability", fontsize = 17)
ax2.set_title('Probability Density', fontsize = 17)
fig.tight_layout()
print('Catalog mean: %f'%catalog['depth'].mean())
print('Catalog median: %f'%catalog['depth'].median())
print('Declustered catalog mean: %f'%mainshock['depth'].mean())
print('Declustered catalog median: %f'%mainshock['depth'].median())
print('SLIQ mean: %f'%sliq['depth'].mean())
print('SLIQ median: %f'%sliq['depth'].median())
np.mean(ds['lwe_thickness'])
np.std(ds['lwe_thickness'])
random_load1 = np.random.normal(loc=-0.65387518, scale=25.49255185, size=(205, 360, 720))
ds_random = ds
ds_random = ds_random.assign(load=ds_random['lwe_thickness'] - ds_random['lwe_thickness'] + random_load1)
# earthquake catalog
mainshock_random = catalog[catalog['P_triggered']<.9]
def get_mass_random(eq):
mass_change = ds_random.sel(lat=(eq["latitude"]),lon=(eq["longitude"]),time=(eq["time"]), method="nearest")
mass_change = mass_change["load"]
return mass_change.data
mainshock_random['dmass'] = mainshock_random.apply(lambda row: get_mass_random(row), axis=1)
set_of_figures_load(ds_random['load'].data.flatten(), mainshock_random.dmass,'c. Bayesian Probability of Mainshock (n=537)')
stats.ks_2samp(mainshock_random.dmass,ds_random['load'].data.flatten())
| 0.543348 | 0.894283 |
# 选择
## 布尔类型、数值和表达式

- 注意:比较运算符的相等是两个等到,一个等到代表赋值
- 在Python中可以用整型0来代表False,其他数字来代表True
- 后面还会讲到 is 在判断语句中的用发
```
2 ==True
```
## 字符串的比较使用ASCII值
```
'a' =='b'
# 90-A
# 80-90-B
number = eval(input('>>'))
if number>=90:
print('A')
elif 80 <= number < 90:
print('B')
elif 80 > number:
print('C')
```
## Markdown
- https://github.com/younghz/Markdown
## EP:
- <img src="../Photo/34.png"></img>
- 输入一个数字,判断其实奇数还是偶数
```
b1 = bool(4)
b1
```
## 产生随机数字
- 函数random.randint(a,b) 可以用来产生一个a和b之间且包括a和b的随机整数
```
import random
# number = eval(input('>>'))
a = random.randint(1,5)
while True:
number = eval(input('>>'))
if number == a:
print('OK')
break
elif number > a:
print('big')
elif number < a:
print('small')
```
## 其他random方法
- random.random 返回0.0到1.0之间前闭后开区间的随机浮点
- random.randrange(a,b) 前闭后开
## EP:
- 产生两个随机整数number1和number2,然后显示给用户,使用户输入数字的和,并判定其是否正确
- 进阶:写一个随机序号点名程序
```
import random
a = random.randint(1,5)
b = random.randint(1,5)
c = a + b
print('生成的数为:' + str(a) + ' 和 ' + str(b))
while True:
num = eval(input('>>'))
if num == c:
print('OK')
break
elif num > c:
print('big')
elif num < c:
print('small')
```
## if语句
- 如果条件正确就执行一个单向if语句,亦即当条件为真的时候才执行if内部的语句
- Python有很多选择语句:
> - 单向if
- 双向if-else
- 嵌套if
- 多向if-elif-else
- 注意:当语句含有子语句的时候,那么一定至少要有一个缩进,也就是说如果有儿子存在,那么一定要缩进
- 切记不可tab键和space混用,单用tab 或者 space
- 当你输出的结果是无论if是否为真时都需要显示时,语句应该与if对齐
```
```
## EP:
- 用户输入一个数字,判断其实奇数还是偶数
- 进阶:可以查看下4.5实例研究猜生日
## 双向if-else 语句
- 如果条件为真,那么走if内部语句,否则走else内部语句
## EP:
- 产生两个随机整数number1和number2,然后显示给用户,使用户输入数字,并判定其是否正确,如果正确打印“you‘re correct”,否则打印正确错误
## 嵌套if 和多向if-elif-else

## EP:
- 提示用户输入一个年份,然后显示表示这一年的动物

- 计算身体质量指数的程序
- BMI = 以千克为单位的体重除以以米为单位的身高

```
year = eval(input('输入年份:'))
a = year % 12
if a == 0:
print('你属猴')
elif a == 1:
print('你属鸡')
elif a == 2:
print('你属狗')
elif a == 3:
print('你属猪')
elif a == 4:
print('你属鼠')
elif a == 5:
print('你属牛')
elif a == 6:
print('你属虎')
elif a == 7:
print('你属兔')
elif a == 8:
print('你属龙')
elif a == 9:
print('你属蛇')
elif a == 10:
print('你属马')
else:
print('你属羊')
zhong = eval(input('输入体重:'))
hige = eval(input('输入身高:'))
BMI = zhong / (hige * hige)
if BMI < 18.5:
print('超轻')
elif 18.5 <= BMI < 25:
print('标准')
elif 25 <= BMI <30:
print('超重')
elif 30 <= BMI:
print('痴肥')
```
## 逻辑运算符

```
a = [1,2,3,4]
1 not in a
a = 19
a > 20
```


## EP:
- 判定闰年:一个年份如果能被4整除但不能被100整除,或者能被400整除,那么这个年份就是闰年
- 提示用户输入一个年份,并返回是否是闰年
- 提示用户输入一个数字,判断其是否为水仙花数
```
year = eval(input('输入年份:'))
if (year % 4 ==0) and (year % 100 !=0) or (year % 400 ==0):
print(str(year) + ' 是闰年')
else:
print(str(year) + ' 不是闰年')
import math
number = eval(input('输入数字:'))
a = number % 10
b = number // 10
c = b % 10
d = b // 10
number_ = math.pow(a,3) + math.pow(c,3) + math.pow(d,3)
if number == number_:
print(str(number) + ' 是水仙花数')
else:
print(str(number) + ' 不是水仙花数')
```
## 实例研究:彩票

```
import random
mod_1 = random.randint(0,9)
mod_2 = random.randint(0,9)
mod = str(mod_1) + str(mod_2)
print(str(mod))
num = input('>>')
if num == mod:
print('1等奖')
elif num[0] == mod[1] and num[1] == mod[0]:
print('2等奖')
elif num[0] == mod[1] or num[1] == mod[0] or num[0] == mod[0] or num[1] == mod[1]:
print('3等奖')
else:
print('谢谢惠顾')
```
# Homework
- 1

```
import math
a,b,c = eval(input('Enter a,b,c:'))
x = b * b - 4 * a * c
if x < 0:
print('The equation has no real roots')
elif x == 0:
r = (- b) / 2 * a
print('The root is ' + str(r))
elif x > 0:
y = math.sqrt(b * b - 4 * a * c)
r1 = ((- b) + y) / 2 * a
r2 = ((- b) - y) / 2 * a
print('The roots are ' + str(r1) + ' and ' + str(r2))
```
- 2

```
import random
a = random.randint(1,5)
b = random.randint(1,5)
c = a + b
print('生成的数为:' + str(a) + ' 和 ' + str(b))
num = eval(input('>>'))
if num == c:
print('真')
elif num !=c:
print('假')
```
- 3

```
31% 7
today = eval(input('Enter today day:'))
day = eval(input('Enter the number of days elapsed since taday:'))
b = (['Sunday','Monday','Tuesday','Wednesday','Thursday','Friday','Saturday'])
day_ = (day + today) % 7
print('Today is ' + str(b[today]) + ' and the future day is ' + str(b[day_]))
```
- 4

```
a = eval(input('输入第一个数字:'))
b = eval(input('输入第二个数字:'))
c = eval(input('输入第三个数字:'))
if a > b and b > c:
print('升序排列:' + str(c) + str(b) + str(a))
elif a > c and c > b:
print('升序排列:' + str(b) + str(c) + str(a))
elif b > c and c > a:
print('升序排列:' + str(a) + str(c) + str(b))
elif b > a and a > c:
print('升序排列:' + str(c) + str(a) + str(b))
elif c > a and a > b:
print('升序排列:' + str(b) + str(a) + str(c))
elif c > b and b > a:
print('升序排列:' + str(a) + str(b) + str(c))
```
- 5

```
weight_1,money_1 = eval(input('Enter weight and price for package 1 :'))
weight_2,money_2 = eval(input('Enter weight and price for package 2 :'))
pr_1 = money_1 / weight_1
pr_2 = money_2 / weight_2
if pr_1 > pr_2:
print('Package 2 has the better price')
else:
print('Package 1 has the better price')
```
- 6

```
mounth = eval(input('输入月份: '))
year = eval(input('输入年份: '))
day = [31,28,31,30,31,30,31,31,30,31,30,31]
if (year % 4 == 0) and (year % 100 != 0) or year % 400 == 0 :
day[1] = day[1] + 1
print(str(year) + '年' + str(mounth) + '月' + str(day[mounth-1]) + '天')
```
- 7

```
import random
a = random.randint(0,1)
x = eval(input('输入猜测值:'))
# a = [0,1]
# b = ['正面','反面']
# a= b
print(a)
if x==a:
print('OK')
else:
print('NOT OK')
import random
b=random.randint(0,1)
c=['正面','反面']
a = eval(input('输入猜测值:'))
if a==b:
print('the computer is',c[b],'You are',c[a],'too,it is a draw')
elif a>b and not(a==2 and b==0 or a==0 and b==2 ):
print('the computer is',c[b],'You are',c[a],'.You win')
else:
print('the computer is',c[b],'You are',c[a],'.You loss')
```
- 8

```
import random
a = eval(input('scissor(0)rock,(1), paper(2):'))
b = random.randint(0,2)
c = ['scissor','rock','paper']
if a==b:
print('the computer is ' + str(c[b]) + ',You are ' + str(c[a]) + 'too,it is a draw')
elif a>b and not(a==2 and b==0 or a==0 and b==2 ):
print('the computer is '+ str(c[b]) + ',You are ' + str(c[a]) + '.You win')
else:
print('the computer is ' + str(c[b]) + ',You are ' + str(c[a]) + '.You loss')
```
- 9

- 10

```
51% 13
52 // 13
import random
b = random.randint(0,52)
p = ['King','Ace','2','3','4','5','6','7','8','9','10','Jack','Queen']
h = ['Clubs','Diamonds','Hearts','Spades']
a = b % 13
c = b // 13
print('The card you picked is the ' + str(p[a-1]) + ' of ' + str(h[c]))
```
- 11

```
number = eval(input("Enter a three-digit integer: "))
number_ = (number % 10) * 100 + (number // 10 % 10) * 10 + (number // 100)
if number == number_:
print(str(number) + " is a palindrome")
else:
print(str(number) + " is not a palindrome")
```
- 12

```
a,b,c = eval(input('输入三边值:'))
if a + b > c and a + c > b and b + c > a:
s = a + b + c
print('该三角形周长为: ',s)
else:
print('这三边无法构成三角形')
```
|
github_jupyter
|
2 ==True
'a' =='b'
# 90-A
# 80-90-B
number = eval(input('>>'))
if number>=90:
print('A')
elif 80 <= number < 90:
print('B')
elif 80 > number:
print('C')
b1 = bool(4)
b1
import random
# number = eval(input('>>'))
a = random.randint(1,5)
while True:
number = eval(input('>>'))
if number == a:
print('OK')
break
elif number > a:
print('big')
elif number < a:
print('small')
import random
a = random.randint(1,5)
b = random.randint(1,5)
c = a + b
print('生成的数为:' + str(a) + ' 和 ' + str(b))
while True:
num = eval(input('>>'))
if num == c:
print('OK')
break
elif num > c:
print('big')
elif num < c:
print('small')
```
## EP:
- 用户输入一个数字,判断其实奇数还是偶数
- 进阶:可以查看下4.5实例研究猜生日
## 双向if-else 语句
- 如果条件为真,那么走if内部语句,否则走else内部语句
## EP:
- 产生两个随机整数number1和number2,然后显示给用户,使用户输入数字,并判定其是否正确,如果正确打印“you‘re correct”,否则打印正确错误
## 嵌套if 和多向if-elif-else

## EP:
- 提示用户输入一个年份,然后显示表示这一年的动物

- 计算身体质量指数的程序
- BMI = 以千克为单位的体重除以以米为单位的身高

## 逻辑运算符



## EP:
- 判定闰年:一个年份如果能被4整除但不能被100整除,或者能被400整除,那么这个年份就是闰年
- 提示用户输入一个年份,并返回是否是闰年
- 提示用户输入一个数字,判断其是否为水仙花数
## 实例研究:彩票

# Homework
- 1

- 2

- 3

- 4

- 5

- 6

- 7

- 8

- 9

- 10

- 11

- 12

| 0.167763 | 0.794624 |
# **Amazon Lookout for Equipment** - Getting started
*Part 1 - Data preparation*
## Initialization
---
This repository is structured as follow:
```sh
. lookout-equipment-demo
|
├── data/
| ├── interim # Temporary intermediate data are stored here
| ├── processed # Finalized datasets are usually stored here
| | # before they are sent to S3 to allow the
| | # service to reach them
| └── raw # Immutable original data are stored here
|
├── getting_started/
| ├── 1_data_preparation.ipynb <<< THIS NOTEBOOK <<<
| ├── 2_dataset_creation.ipynb
| ├── 3_model_training.ipynb
| ├── 4_model_evaluation.ipynb
| ├── 5_inference_scheduling.ipynb
| └── 6_cleanup.ipynb
|
└── utils/
└── lookout_equipment_utils.py
```
### Notebook configuration update
```
!pip install --quiet --upgrade tqdm tsia
```
### Imports
```
import boto3
import config
import os
import pandas as pd
from botocore.client import ClientError
```
Check the region and the availability of Amazon Lookout for Equipment in this region:
```
REGION_NAME = boto3.session.Session().region_name
try:
ssm_client = boto3.client('ssm')
response = ssm_client.get_parameters_by_path(
Path='/aws/service/global-infrastructure/services/lookoutequipment/regions',
)
available_regions = [r['Value'] for r in response['Parameters']]
if REGION_NAME not in available_regions:
raise Exception(f'Amazon Lookout for Equipment is only available in {available_regions}')
except ClientError as e:
print(f'This notebook does not have access to the SSM service to check the availability of Lookout for Equipment in the current region ({REGION_NAME}).')
print(f'If you are running this code in the following AWS region, it should be fine to continue: us-east-1, ap-northeast-2, eu-west-1')
print('Exception:')
print(' -', e)
```
### Parameters
Let's first check if the bucket name is defined, if it exists and if we have access to it from this notebook. If this notebook does not have access to the S3 bucket, you will have to update its permission:
```
BUCKET = config.BUCKET
PREFIX_TRAINING = config.PREFIX_TRAINING
PREFIX_LABEL = config.PREFIX_LABEL
if BUCKET == '<<YOUR_BUCKET>>':
raise Exception('Please update your Amazon S3 bucket name in the config.py file located at the root of this repository and restart the kernel for this notebook.')
else:
# Check access to the configured bucket:
try:
s3_resource = boto3.resource('s3')
s3_resource.meta.client.head_bucket(Bucket=BUCKET)
print(f'Bucket "{BUCKET}" exists')
# Expose error reason:
except ClientError as error:
error_code = int(error.response['Error']['Code'])
if error_code == 403:
raise Exception(f'Bucket "{BUCKET}" is private: access is forbidden!')
elif error_code == 404:
raise Exception(f'Bucket "{BUCKET}" does not exist!')
RAW_DATA = os.path.join('..', 'data', 'raw', 'getting-started')
TMP_DATA = os.path.join('..', 'data', 'interim', 'getting-started')
PROCESSED_DATA = os.path.join('..', 'data', 'processed', 'getting-started')
LABEL_DATA = os.path.join(PROCESSED_DATA, 'label-data')
TRAIN_DATA = os.path.join(PROCESSED_DATA, 'training-data')
INFERENCE_DATA = os.path.join(PROCESSED_DATA, 'inference-data')
os.makedirs(TMP_DATA, exist_ok=True)
os.makedirs(RAW_DATA, exist_ok=True)
os.makedirs(PROCESSED_DATA, exist_ok=True)
os.makedirs(LABEL_DATA, exist_ok=True)
os.makedirs(TRAIN_DATA, exist_ok=True)
os.makedirs(INFERENCE_DATA, exist_ok=True)
ORIGINAL_DATA = f's3://lookoutforequipmentbucket-{REGION_NAME}/datasets/getting-started/lookout-equipment-sdk-5min.zip'
```
## Downloading data
---
Downloading and unzipping the getting started dataset locally on this instance:
```
data_exists = os.path.exists(os.path.join(TMP_DATA, 'sensors-data', 'impeller', 'component2_file1.csv'))
raw_data_exists = os.path.exists(os.path.join(RAW_DATA, 'lookout-equipment.zip'))
if data_exists:
print('Dataset already available locally, nothing to do.')
print(f'Dataset is available in {TMP_DATA}.')
else:
if not raw_data_exists:
print('Raw data not found, downloading it')
!aws s3 cp $ORIGINAL_DATA $RAW_DATA/lookout-equipment.zip
print('Unzipping raw data...')
!unzip $RAW_DATA/lookout-equipment.zip -d $TMP_DATA
print(f'Done: dataset now available in {TMP_DATA}.')
```
## Preparing time series data
---
The time series data are available in the `sensors-data` directory. The industrial asset we are looking at is a [centrifugal pump](https://en.wikipedia.org/wiki/Centrifugal_pump). Such a pump is used to move a fluid by transfering the rotational energy provided by a motor to hydrodynamic energy:
<img src="assets/centrifugal_pump_annotated.png" alt="Centrifugal pump" style="width: 658px"/>
<div style="text-align: center"><i>Warman centrifugal pump in a coal preparation plant application</i>, by Bernard S. Janse, licensed under <a href="https://creativecommons.org/licenses/by/2.5/deed.fr">CC BY 2.5</a></div>
On a pump such as the one displayed in the photo above, the fluid enters at its axis (the black pipe arriving at the "eye" of the impeller. Measurements can be taken around the four main components of the centrifugal pump:
* The **impeller** (hidden into the round white casing above): this component consists of a series of curved vanes (blades)
* The drive **shaft** arriving at the impeller axis (the "eye")
* The **motor** connected to the impeller by the drive shaft (on the other end of the black pipe above)
* The **volute** chamber, offseted on the right compared to the impeller axis: this creates a curved funnel win a decreasing cross-section area towards the pump outlet (at the top of the white pipe above)
In the dataset provided, other sensors not located on one of these component are positionned at the **pump** level.
**Let's load the content of each CSV file (we have one per component) and build a single CSV file with all the sensors:** we will obtain a dataset with 10 months of data (spanning from `2019-01-01` to `2019-10-27`) for 30 sensors (`Sensor0` to `Sensor29`) with a 1-minute sampling rate:
```
%%time
# Loops through each subfolder of the original dataset:
sensor_df_list = []
tags_description_dict = dict()
for root, dirs, files in os.walk(os.path.join(TMP_DATA, 'sensors-data')):
# Reads each file and set the first column as an index:
for f in files:
print('Processing:', os.path.join(root, f))
df = pd.read_csv(os.path.join(root, f))
df['Timestamp'] = pd.to_datetime(df['Timestamp'])
df = df.set_index('Timestamp')
sensor_df_list.append(df)
component = root.split('/')[-1]
current_sensors = df.columns.tolist()
current_sensors = dict(zip(current_sensors, [component] * len(current_sensors)))
tags_description_dict = {**tags_description_dict, **current_sensors}
# Concatenate into a single dataframe:
equipment_df = pd.concat(sensor_df_list, axis='columns')
equipment_df = equipment_df.reset_index()
equipment_df['Timestamp'] = pd.to_datetime(equipment_df['Timestamp'])
equipment_df = equipment_df[[
'Timestamp', 'Sensor0', 'Sensor1', 'Sensor2', 'Sensor3', 'Sensor4',
'Sensor5', 'Sensor6', 'Sensor7', 'Sensor8', 'Sensor9', 'Sensor10',
'Sensor11', 'Sensor24', 'Sensor25', 'Sensor26', 'Sensor27', 'Sensor28',
'Sensor29', 'Sensor12', 'Sensor13', 'Sensor14', 'Sensor15', 'Sensor16',
'Sensor17', 'Sensor18', 'Sensor19', 'Sensor20', 'Sensor21', 'Sensor22',
'Sensor23'
]]
# Register a component for each sensor:
tags_description_df = pd.DataFrame.from_dict(tags_description_dict, orient='index')
tags_description_df = tags_description_df.reset_index()
tags_description_df.columns = ['Tag', 'Component']
print(equipment_df.shape)
equipment_df.head()
equipment_df['Timestamp'] = pd.to_datetime(equipment_df['Timestamp'])
equipment_df = equipment_df.set_index('Timestamp')
equipment_df
%%time
os.makedirs(os.path.join(TRAIN_DATA, 'centrifugal-pump'), exist_ok=True)
equipment_fname = os.path.join(TRAIN_DATA, 'centrifugal-pump', 'sensors.csv')
equipment_df.to_csv(equipment_fname)
```
Let's also persist the tags description file as it will be useful when analyzing the model results:
```
tags_description_fname = os.path.join(TMP_DATA, 'tags_description.csv')
tags_description_df.to_csv(tags_description_fname, index=None)
```
## Loading label data
---
This dataset contains synthetically generated anomalies over different periods of time. Labels are stored as time ranges with a start and end timestamp. Each label is a period of time where we know some anomalous behavior happen:
```
label_fname = os.path.join(TMP_DATA, 'label-data', 'labels.csv')
labels_df = pd.read_csv(label_fname, header=None)
labels_df.to_csv(os.path.join(PROCESSED_DATA, 'label-data', 'labels.csv'), index=None, header=None)
labels_df.columns = ['start', 'end']
labels_df.head()
```
## Uploading data to Amazon S3
---
Let's now load our training data and labels to Amazon S3, so that Lookout for Equipment can access them to train and evaluate a model.
```
train_s3_path = f's3://{BUCKET}/{PREFIX_TRAINING}centrifugal-pump/sensors.csv'
!aws s3 cp $equipment_fname $train_s3_path
label_s3_path = f's3://{BUCKET}/{PREFIX_LABEL}labels.csv'
!aws s3 cp $label_fname $label_s3_path
```
## (Optional) Data exploration
---
This section is optional and just aim at giving you a quick overview about what the data looks like:
```
import matplotlib.pyplot as plt
import numpy as np
import sys
import tsia
import warnings
sys.path.append('../utils')
import lookout_equipment_utils as lookout
%matplotlib inline
plt.style.use('Solarize_Light2')
plt.rcParams['lines.linewidth'] = 0.5
warnings.filterwarnings("ignore")
start = equipment_df.index.min()
end = equipment_df.index.max()
print(start, '|', end)
```
**Let's plot the first signal and the associated labels:** the `plot_timeseries` function is a utility function you can use to plot a signal and the associated labels on the same figure:
```
tag = 'Sensor0'
tag_df = equipment_df.loc[start:end, [tag]]
tag_df.columns = ['Value']
fig1, axes = lookout.plot_timeseries(
tag_df,
tag,
fig_width=20,
labels_df=labels_df,
custom_grid=False
)
```
**Run the following cell to get an overview of every signals in the dataset:** colors are allocated to each sensor according to the component it's associated to. This generates a big matplotlib picture in memory. On smaller instances, this can lead to some *out of memory* issues. Upgrade to a bigger instance, or clean up the memory of the instances if you have other notebooks running in parallel to this one:
```
df_list = []
features = equipment_df.columns.tolist()
for sensor in features:
df_list.append(equipment_df[[sensor]])
fig2 = tsia.plot.plot_multivariate_timeseries(
timeseries_list=df_list,
tags_list=features,
tags_description_df=tags_description_df,
tags_grouping_key='Component',
num_cols=3,
)
```
## Conclusion
---
In this notebook, you downloaded the getting started dataset and prepared it for ingestion in Amazon Lookout for Equipment.
You also had a quick overview of the dataset with basic timeseries visualization.
You uploaded the training time series data and the anomaly labels to Amazon S3: in the next notebook of this getting started, you will be acquainted with the Amazon Lookout for Equipment API to create your first dataset.
```
# Cleanup, might be necessary on smaller instances:
import gc
del fig1, fig2, equipment_df
gc.collect()
```
|
github_jupyter
|
. lookout-equipment-demo
|
├── data/
| ├── interim # Temporary intermediate data are stored here
| ├── processed # Finalized datasets are usually stored here
| | # before they are sent to S3 to allow the
| | # service to reach them
| └── raw # Immutable original data are stored here
|
├── getting_started/
| ├── 1_data_preparation.ipynb <<< THIS NOTEBOOK <<<
| ├── 2_dataset_creation.ipynb
| ├── 3_model_training.ipynb
| ├── 4_model_evaluation.ipynb
| ├── 5_inference_scheduling.ipynb
| └── 6_cleanup.ipynb
|
└── utils/
└── lookout_equipment_utils.py
!pip install --quiet --upgrade tqdm tsia
import boto3
import config
import os
import pandas as pd
from botocore.client import ClientError
REGION_NAME = boto3.session.Session().region_name
try:
ssm_client = boto3.client('ssm')
response = ssm_client.get_parameters_by_path(
Path='/aws/service/global-infrastructure/services/lookoutequipment/regions',
)
available_regions = [r['Value'] for r in response['Parameters']]
if REGION_NAME not in available_regions:
raise Exception(f'Amazon Lookout for Equipment is only available in {available_regions}')
except ClientError as e:
print(f'This notebook does not have access to the SSM service to check the availability of Lookout for Equipment in the current region ({REGION_NAME}).')
print(f'If you are running this code in the following AWS region, it should be fine to continue: us-east-1, ap-northeast-2, eu-west-1')
print('Exception:')
print(' -', e)
BUCKET = config.BUCKET
PREFIX_TRAINING = config.PREFIX_TRAINING
PREFIX_LABEL = config.PREFIX_LABEL
if BUCKET == '<<YOUR_BUCKET>>':
raise Exception('Please update your Amazon S3 bucket name in the config.py file located at the root of this repository and restart the kernel for this notebook.')
else:
# Check access to the configured bucket:
try:
s3_resource = boto3.resource('s3')
s3_resource.meta.client.head_bucket(Bucket=BUCKET)
print(f'Bucket "{BUCKET}" exists')
# Expose error reason:
except ClientError as error:
error_code = int(error.response['Error']['Code'])
if error_code == 403:
raise Exception(f'Bucket "{BUCKET}" is private: access is forbidden!')
elif error_code == 404:
raise Exception(f'Bucket "{BUCKET}" does not exist!')
RAW_DATA = os.path.join('..', 'data', 'raw', 'getting-started')
TMP_DATA = os.path.join('..', 'data', 'interim', 'getting-started')
PROCESSED_DATA = os.path.join('..', 'data', 'processed', 'getting-started')
LABEL_DATA = os.path.join(PROCESSED_DATA, 'label-data')
TRAIN_DATA = os.path.join(PROCESSED_DATA, 'training-data')
INFERENCE_DATA = os.path.join(PROCESSED_DATA, 'inference-data')
os.makedirs(TMP_DATA, exist_ok=True)
os.makedirs(RAW_DATA, exist_ok=True)
os.makedirs(PROCESSED_DATA, exist_ok=True)
os.makedirs(LABEL_DATA, exist_ok=True)
os.makedirs(TRAIN_DATA, exist_ok=True)
os.makedirs(INFERENCE_DATA, exist_ok=True)
ORIGINAL_DATA = f's3://lookoutforequipmentbucket-{REGION_NAME}/datasets/getting-started/lookout-equipment-sdk-5min.zip'
data_exists = os.path.exists(os.path.join(TMP_DATA, 'sensors-data', 'impeller', 'component2_file1.csv'))
raw_data_exists = os.path.exists(os.path.join(RAW_DATA, 'lookout-equipment.zip'))
if data_exists:
print('Dataset already available locally, nothing to do.')
print(f'Dataset is available in {TMP_DATA}.')
else:
if not raw_data_exists:
print('Raw data not found, downloading it')
!aws s3 cp $ORIGINAL_DATA $RAW_DATA/lookout-equipment.zip
print('Unzipping raw data...')
!unzip $RAW_DATA/lookout-equipment.zip -d $TMP_DATA
print(f'Done: dataset now available in {TMP_DATA}.')
%%time
# Loops through each subfolder of the original dataset:
sensor_df_list = []
tags_description_dict = dict()
for root, dirs, files in os.walk(os.path.join(TMP_DATA, 'sensors-data')):
# Reads each file and set the first column as an index:
for f in files:
print('Processing:', os.path.join(root, f))
df = pd.read_csv(os.path.join(root, f))
df['Timestamp'] = pd.to_datetime(df['Timestamp'])
df = df.set_index('Timestamp')
sensor_df_list.append(df)
component = root.split('/')[-1]
current_sensors = df.columns.tolist()
current_sensors = dict(zip(current_sensors, [component] * len(current_sensors)))
tags_description_dict = {**tags_description_dict, **current_sensors}
# Concatenate into a single dataframe:
equipment_df = pd.concat(sensor_df_list, axis='columns')
equipment_df = equipment_df.reset_index()
equipment_df['Timestamp'] = pd.to_datetime(equipment_df['Timestamp'])
equipment_df = equipment_df[[
'Timestamp', 'Sensor0', 'Sensor1', 'Sensor2', 'Sensor3', 'Sensor4',
'Sensor5', 'Sensor6', 'Sensor7', 'Sensor8', 'Sensor9', 'Sensor10',
'Sensor11', 'Sensor24', 'Sensor25', 'Sensor26', 'Sensor27', 'Sensor28',
'Sensor29', 'Sensor12', 'Sensor13', 'Sensor14', 'Sensor15', 'Sensor16',
'Sensor17', 'Sensor18', 'Sensor19', 'Sensor20', 'Sensor21', 'Sensor22',
'Sensor23'
]]
# Register a component for each sensor:
tags_description_df = pd.DataFrame.from_dict(tags_description_dict, orient='index')
tags_description_df = tags_description_df.reset_index()
tags_description_df.columns = ['Tag', 'Component']
print(equipment_df.shape)
equipment_df.head()
equipment_df['Timestamp'] = pd.to_datetime(equipment_df['Timestamp'])
equipment_df = equipment_df.set_index('Timestamp')
equipment_df
%%time
os.makedirs(os.path.join(TRAIN_DATA, 'centrifugal-pump'), exist_ok=True)
equipment_fname = os.path.join(TRAIN_DATA, 'centrifugal-pump', 'sensors.csv')
equipment_df.to_csv(equipment_fname)
tags_description_fname = os.path.join(TMP_DATA, 'tags_description.csv')
tags_description_df.to_csv(tags_description_fname, index=None)
label_fname = os.path.join(TMP_DATA, 'label-data', 'labels.csv')
labels_df = pd.read_csv(label_fname, header=None)
labels_df.to_csv(os.path.join(PROCESSED_DATA, 'label-data', 'labels.csv'), index=None, header=None)
labels_df.columns = ['start', 'end']
labels_df.head()
train_s3_path = f's3://{BUCKET}/{PREFIX_TRAINING}centrifugal-pump/sensors.csv'
!aws s3 cp $equipment_fname $train_s3_path
label_s3_path = f's3://{BUCKET}/{PREFIX_LABEL}labels.csv'
!aws s3 cp $label_fname $label_s3_path
import matplotlib.pyplot as plt
import numpy as np
import sys
import tsia
import warnings
sys.path.append('../utils')
import lookout_equipment_utils as lookout
%matplotlib inline
plt.style.use('Solarize_Light2')
plt.rcParams['lines.linewidth'] = 0.5
warnings.filterwarnings("ignore")
start = equipment_df.index.min()
end = equipment_df.index.max()
print(start, '|', end)
tag = 'Sensor0'
tag_df = equipment_df.loc[start:end, [tag]]
tag_df.columns = ['Value']
fig1, axes = lookout.plot_timeseries(
tag_df,
tag,
fig_width=20,
labels_df=labels_df,
custom_grid=False
)
df_list = []
features = equipment_df.columns.tolist()
for sensor in features:
df_list.append(equipment_df[[sensor]])
fig2 = tsia.plot.plot_multivariate_timeseries(
timeseries_list=df_list,
tags_list=features,
tags_description_df=tags_description_df,
tags_grouping_key='Component',
num_cols=3,
)
# Cleanup, might be necessary on smaller instances:
import gc
del fig1, fig2, equipment_df
gc.collect()
| 0.454714 | 0.839208 |
```
# Importing Libraries
import pandas as pd
import numpy as np
import math
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import log_loss
from tqdm import tqdm_notebook
from lightgbm import LGBMClassifier
from sklearn.decomposition import PCA
from functools import reduce
import warnings
warnings.filterwarnings('ignore')
# Loading files
df = pd.read_csv('radiant_pixels.csv')
sample_submission = pd.read_csv('SampleSubmission.csv')
train = df[df.crop_type != 0]
test = df[df.crop_type == 0]
target = train.crop_type
train.head()
```
### Bands
```
# Sentinel 2 band description
# Band 1 - Coastal aerosol 0.443 60
# Band 2 - Blue 0.490 10
# Band 3 - Green 0.560 10
# Band 4 - Red 0.665 10
# Band 5 - Vegetation Red Edge 0.705 20
# Band 6 - Vegetation Red Edge 0.740 20
# Band 7 - Vegetation Red Edge 0.783 20
# Band 8 - NIR 0.842 10
# Band 8A - Vegetation Red Edge 0.865 20
# Band 9 - Water vapour 0.945 60
# Band 10 - SWIR - Cirrus 1.375 60
# Band 11 - SWIR 1.610 20
# Band 12 - SWIR
# Getting the bands
B01, B02, B03, B04, B05 = df.filter(regex='._B01'), df.filter(regex='._B02'), df.filter(regex='._B03'), df.filter(regex='._B04'), df.filter(regex='._B05')
B06, B07, B08, B8A, B09 = df.filter(regex='._B06'), df.filter(regex='._B07'), df.filter(regex='._B08'), df.filter(regex='._B8A'), df.filter(regex='._B09')
B11, B12 = df.filter(regex='._B11'), df.filter(regex='._B12')
# Formulas for different vegetation indices
def aci(i): return B08.values[:, i] * (B04.values[:, i] + B03.values[:, i])
def avi (i): return (B08.values[:, i] * (1 - B04.values[:, i]) * (B08.values[:, i] - B04.values[:, i]))
def bai (i): return 1/((0.1 - B04.values[:, i]) ** 2 + (0.06 - B08.values[:, i]) ** 2)
def bgi (i): return B01.values[:, i] / B03.values[:, i]
def bri (i): return B01.values[:, i] / B05.values[:, i]
def ccci (i): return ((B08.values[:, i] - B05.values[:, i]) / (B08.values[:, i] + B05.values[:, i])) / ((B08.values[:, i] - B04.values[:, i]) / (B08.values[:, i] + B04.values[:, i]))
def cm (i): return B11.values[:, i] / B12.values[:, i]
def dyi (i): return B03.values[:, i] - B02.values[:, i]
def evi (i): return 2.5*(B08.values[:, i] - B04.values[:, i]) / (B08.values[:, i] + 6*B04.values[:, i] - 7.5*B02.values[:, i] + 1)
def exg (i): return 2 * B03.values[:, i] - B04.values[:, i] - B02.values[:, i]
def fidet (i): return B12.values[:, i] / (B8A.values[:, i] * B09.values[:, i])
def mcari (i): return ((B05.values[:, i] - B04.values[:, i]) - 2 * (B05.values[:, i] - B03.values[:, i])) * (B05.values[:, i] / B04.values[:, i])
def mi (i): return (B8A.values[:, i] - B11.values[:, i]) / (B8A.values[:, i] + B11.values[:, i])
def mrendvi (i): return (B06.values[:, i] - B05.values[:, i]) / (B06.values[:, i] + B05.values[:, i] - 2 * B01.values[:, i])
def mresr (i): return (B06.values[:, i] - B01.values[:, i]) / (B05.values[:, i] - B01.values[:, i])
def mtci (i): return (B06.values[:, i] - B05.values[:, i])/(B05.values[:, i] - B04.values[:, i])
def mtvi (i): return [1.5*(1.2 * (a - b) - 2.5 * (c - b))* math.sqrt((2 * a + 1)**2-(6 * a - 5 * math.sqrt(c)) - 0.5) for a, b, c in zip(B08.values[:, i], B03.values[:, i], B04.values[:, i])]
def nbr (i): return (B08.values[:, i] - B11.values[:, i]) / (B08.values[:, i]+ B11.values[:, i])
def ndsi (i): return (B03.values[:, i] - B11.values[:, i]) / (B03.values[:, i] + B11.values[:, i])
def nli (i): return (B08.values[:, i] **2 - B04.values[:, i]) / (B08.values[:, i] **2 + B04.values[:, i])
def ndmi (i): return (B08.values[:, i] - B11.values[:, i])/(B08.values[:, i] + B11.values[:, i])
def pvi (i): return (B08.values[:, i] - 0.3 * B04.values[:, i] - 0.5) / (math.sqrt(1 + 0.3 * 2))
def s2rep (i): return 705 + 35 * ((((B07.values[:, i] + B04.values[:, i])/2) - B05.values[:, i])/(B06.values[:, i] - B05.values[:, i]))
def si (i): return ((1 - B02.values[:, i]) * (1 - B03.values[:, i]) * (1 - B04.values[:, i]))
def sipi (i): return (B08.values[:, i] - B02.values[:, i]) / (B08.values[:, i] - B04.values[:, i])
def sr3 (i): return B05.values[:, i] / B04.values[:, i]
def tcari (i): return 3 * ((B05.values[:, i] - B04.values[:, i]) - 0.2 * (B05.values[:, i] - B03.values[:, i]) * (B05.values[:, i] / B04.values[:, i]))
def tvi (i): return (120 * (B06.values[:, i] - B03.values[:, i]) - 200 * (B04.values[:, i] - B03.values[:, i])) / 2
def vdvi (i): return (2 * B03.values[:, i] - B04.values[:, i] - B02.values[:, i]) / (2 * B03.values[:, i] + B04.values[:, i] + B02.values[:, i])
# Helper functions
num_dates = 25
bin_labels = list(range(3))
indices_df = []
# Function to convert continuos vegetation into a category via pca
def cat_indices(indice):
return pd.cut(PCA(1).fit_transform(pd.DataFrame(indice)).flatten(), len(bin_labels), labels = bin_labels)
# Calculating indices
def indice_calculator(func_name, indice_name):
holder = []
for i in range(num_dates):
holder.append(func_name(i))
indice_df = pd.DataFrame(np.array(holder).T, columns = [indice_name + '_date_' + str(i) for i in range(1, num_dates+1)])
differences_df = pd.DataFrame(indice_df.diff(axis = 1))
differences_df.columns = ['difference_'+indice_name +'_'+ str(i) for i in range(num_dates)]
indice_df['cat_' + indice_name] = cat_indices(np.nan_to_num(np.array(holder).T, nan=10, posinf=10, neginf=10))
indice_df = pd.merge(indice_df, differences_df, how = 'left', left_index=True, right_index=True)
return indice_df
# A list of all indices
indices = [(aci, 'aci'), (avi, 'avi'), (bai, 'bai'), (bgi, 'bgi'), (bri, 'bri'), (ccci, 'ccci'), (cm, 'cm'), (dyi, 'dyi'), (evi, 'evi'), (exg, 'exg'),\
(fidet, 'fidet'), (mcari, 'mcari'), (mi, 'mi'), (mrendvi, 'mrendvi'), (mresr, 'mresr'), (mtci, 'mtci'), (mtvi, 'mtvi'), (nbr, 'nbr'), \
(ndsi, 'ndsi'), (nli, 'nli'), (ndmi, 'ndmi'), (pvi, 'pvi'), (s2rep, 's2rep'), (si, 'si'), (sipi, 'sipi'), (sr3, 'sr3'), (tcari, 'tcari'),\
(tvi, 'tvi'), (vdvi, 'vdvi')]
# Calculating theindices
indices_dfs = []
for indice in indices:indices_dfs.append(indice_calculator(indice[0], indice[1]))
# Merging the indices dataframes
indices_df = reduce(lambda left,right: pd.merge(left,right,right_index=True, left_index=True,how='outer'), indices_dfs)
indices_df.head()
```
### Handling Categorical Features
```
# Transform categorical features into the appropriate type
categorical_features = [x for x in indices_df.columns if 'cat' in x]
for feature in categorical_features: indices_df[feature] = indices_df[feature].astype('category')
indices_df[[x for x in indices_df.columns if 'cat' in x]].dtypes
```
### Separating training and test sets
```
train_indices = indices_df.loc[train.index]
test_indices= indices_df.loc[test.index]
train_indices.head()
```
### Model training - Indices
```
# Model parameters
lgbm_params = {
'n_estimators': 50000,
'learning_rate': 0.05,
'colsample_bytree': 0.2,
'subsample': 0.2,
'reg_alpha': 10,
'reg_lambda': 5,
'num_leaves': 20,
'early_stopping_rounds': 300,
'n_jobs': -1,
'objective': 'multiclass',
'boosting': 'gbdt',
'feature_name': 'auto',
'categorical_features': 'auto'
}
# Training lgbm model
indices_oof_predictions, loss = [], []
for train_index, test_index in StratifiedKFold(n_splits=10).split(train_indices, target):
X_train, X_test, y_train, y_test = train_indices.iloc[train_index], train_indices.iloc[test_index], target[train_index], target[test_index]
lgbm = LGBMClassifier(**lgbm_params)
lgbm.fit(X_train, y_train, eval_set = [(X_test, y_test)], verbose = False)
indices_oof_predictions.append(lgbm.predict_proba(test_indices))
loss.append(log_loss(y_test, lgbm.predict_proba(X_test)))
print('Loss: ', log_loss(y_test, lgbm.predict_proba(X_test)))
print('Indices Cross Validation Loss: ', np.mean(loss))
```
### Submission file preparation
```
# Submission file preparation
submission_file = pd.DataFrame({'Field ID': test['Field ID']})
for i, j in enumerate(sample_submission.columns[1:]):
submission_file[j] = np.mean(indices_oof_predictions, 0)[:, i]
submission_file.to_csv('LGBM_SUB.csv', index = False)
submission_file.head()
```
|
github_jupyter
|
# Importing Libraries
import pandas as pd
import numpy as np
import math
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import log_loss
from tqdm import tqdm_notebook
from lightgbm import LGBMClassifier
from sklearn.decomposition import PCA
from functools import reduce
import warnings
warnings.filterwarnings('ignore')
# Loading files
df = pd.read_csv('radiant_pixels.csv')
sample_submission = pd.read_csv('SampleSubmission.csv')
train = df[df.crop_type != 0]
test = df[df.crop_type == 0]
target = train.crop_type
train.head()
# Sentinel 2 band description
# Band 1 - Coastal aerosol 0.443 60
# Band 2 - Blue 0.490 10
# Band 3 - Green 0.560 10
# Band 4 - Red 0.665 10
# Band 5 - Vegetation Red Edge 0.705 20
# Band 6 - Vegetation Red Edge 0.740 20
# Band 7 - Vegetation Red Edge 0.783 20
# Band 8 - NIR 0.842 10
# Band 8A - Vegetation Red Edge 0.865 20
# Band 9 - Water vapour 0.945 60
# Band 10 - SWIR - Cirrus 1.375 60
# Band 11 - SWIR 1.610 20
# Band 12 - SWIR
# Getting the bands
B01, B02, B03, B04, B05 = df.filter(regex='._B01'), df.filter(regex='._B02'), df.filter(regex='._B03'), df.filter(regex='._B04'), df.filter(regex='._B05')
B06, B07, B08, B8A, B09 = df.filter(regex='._B06'), df.filter(regex='._B07'), df.filter(regex='._B08'), df.filter(regex='._B8A'), df.filter(regex='._B09')
B11, B12 = df.filter(regex='._B11'), df.filter(regex='._B12')
# Formulas for different vegetation indices
def aci(i): return B08.values[:, i] * (B04.values[:, i] + B03.values[:, i])
def avi (i): return (B08.values[:, i] * (1 - B04.values[:, i]) * (B08.values[:, i] - B04.values[:, i]))
def bai (i): return 1/((0.1 - B04.values[:, i]) ** 2 + (0.06 - B08.values[:, i]) ** 2)
def bgi (i): return B01.values[:, i] / B03.values[:, i]
def bri (i): return B01.values[:, i] / B05.values[:, i]
def ccci (i): return ((B08.values[:, i] - B05.values[:, i]) / (B08.values[:, i] + B05.values[:, i])) / ((B08.values[:, i] - B04.values[:, i]) / (B08.values[:, i] + B04.values[:, i]))
def cm (i): return B11.values[:, i] / B12.values[:, i]
def dyi (i): return B03.values[:, i] - B02.values[:, i]
def evi (i): return 2.5*(B08.values[:, i] - B04.values[:, i]) / (B08.values[:, i] + 6*B04.values[:, i] - 7.5*B02.values[:, i] + 1)
def exg (i): return 2 * B03.values[:, i] - B04.values[:, i] - B02.values[:, i]
def fidet (i): return B12.values[:, i] / (B8A.values[:, i] * B09.values[:, i])
def mcari (i): return ((B05.values[:, i] - B04.values[:, i]) - 2 * (B05.values[:, i] - B03.values[:, i])) * (B05.values[:, i] / B04.values[:, i])
def mi (i): return (B8A.values[:, i] - B11.values[:, i]) / (B8A.values[:, i] + B11.values[:, i])
def mrendvi (i): return (B06.values[:, i] - B05.values[:, i]) / (B06.values[:, i] + B05.values[:, i] - 2 * B01.values[:, i])
def mresr (i): return (B06.values[:, i] - B01.values[:, i]) / (B05.values[:, i] - B01.values[:, i])
def mtci (i): return (B06.values[:, i] - B05.values[:, i])/(B05.values[:, i] - B04.values[:, i])
def mtvi (i): return [1.5*(1.2 * (a - b) - 2.5 * (c - b))* math.sqrt((2 * a + 1)**2-(6 * a - 5 * math.sqrt(c)) - 0.5) for a, b, c in zip(B08.values[:, i], B03.values[:, i], B04.values[:, i])]
def nbr (i): return (B08.values[:, i] - B11.values[:, i]) / (B08.values[:, i]+ B11.values[:, i])
def ndsi (i): return (B03.values[:, i] - B11.values[:, i]) / (B03.values[:, i] + B11.values[:, i])
def nli (i): return (B08.values[:, i] **2 - B04.values[:, i]) / (B08.values[:, i] **2 + B04.values[:, i])
def ndmi (i): return (B08.values[:, i] - B11.values[:, i])/(B08.values[:, i] + B11.values[:, i])
def pvi (i): return (B08.values[:, i] - 0.3 * B04.values[:, i] - 0.5) / (math.sqrt(1 + 0.3 * 2))
def s2rep (i): return 705 + 35 * ((((B07.values[:, i] + B04.values[:, i])/2) - B05.values[:, i])/(B06.values[:, i] - B05.values[:, i]))
def si (i): return ((1 - B02.values[:, i]) * (1 - B03.values[:, i]) * (1 - B04.values[:, i]))
def sipi (i): return (B08.values[:, i] - B02.values[:, i]) / (B08.values[:, i] - B04.values[:, i])
def sr3 (i): return B05.values[:, i] / B04.values[:, i]
def tcari (i): return 3 * ((B05.values[:, i] - B04.values[:, i]) - 0.2 * (B05.values[:, i] - B03.values[:, i]) * (B05.values[:, i] / B04.values[:, i]))
def tvi (i): return (120 * (B06.values[:, i] - B03.values[:, i]) - 200 * (B04.values[:, i] - B03.values[:, i])) / 2
def vdvi (i): return (2 * B03.values[:, i] - B04.values[:, i] - B02.values[:, i]) / (2 * B03.values[:, i] + B04.values[:, i] + B02.values[:, i])
# Helper functions
num_dates = 25
bin_labels = list(range(3))
indices_df = []
# Function to convert continuos vegetation into a category via pca
def cat_indices(indice):
return pd.cut(PCA(1).fit_transform(pd.DataFrame(indice)).flatten(), len(bin_labels), labels = bin_labels)
# Calculating indices
def indice_calculator(func_name, indice_name):
holder = []
for i in range(num_dates):
holder.append(func_name(i))
indice_df = pd.DataFrame(np.array(holder).T, columns = [indice_name + '_date_' + str(i) for i in range(1, num_dates+1)])
differences_df = pd.DataFrame(indice_df.diff(axis = 1))
differences_df.columns = ['difference_'+indice_name +'_'+ str(i) for i in range(num_dates)]
indice_df['cat_' + indice_name] = cat_indices(np.nan_to_num(np.array(holder).T, nan=10, posinf=10, neginf=10))
indice_df = pd.merge(indice_df, differences_df, how = 'left', left_index=True, right_index=True)
return indice_df
# A list of all indices
indices = [(aci, 'aci'), (avi, 'avi'), (bai, 'bai'), (bgi, 'bgi'), (bri, 'bri'), (ccci, 'ccci'), (cm, 'cm'), (dyi, 'dyi'), (evi, 'evi'), (exg, 'exg'),\
(fidet, 'fidet'), (mcari, 'mcari'), (mi, 'mi'), (mrendvi, 'mrendvi'), (mresr, 'mresr'), (mtci, 'mtci'), (mtvi, 'mtvi'), (nbr, 'nbr'), \
(ndsi, 'ndsi'), (nli, 'nli'), (ndmi, 'ndmi'), (pvi, 'pvi'), (s2rep, 's2rep'), (si, 'si'), (sipi, 'sipi'), (sr3, 'sr3'), (tcari, 'tcari'),\
(tvi, 'tvi'), (vdvi, 'vdvi')]
# Calculating theindices
indices_dfs = []
for indice in indices:indices_dfs.append(indice_calculator(indice[0], indice[1]))
# Merging the indices dataframes
indices_df = reduce(lambda left,right: pd.merge(left,right,right_index=True, left_index=True,how='outer'), indices_dfs)
indices_df.head()
# Transform categorical features into the appropriate type
categorical_features = [x for x in indices_df.columns if 'cat' in x]
for feature in categorical_features: indices_df[feature] = indices_df[feature].astype('category')
indices_df[[x for x in indices_df.columns if 'cat' in x]].dtypes
train_indices = indices_df.loc[train.index]
test_indices= indices_df.loc[test.index]
train_indices.head()
# Model parameters
lgbm_params = {
'n_estimators': 50000,
'learning_rate': 0.05,
'colsample_bytree': 0.2,
'subsample': 0.2,
'reg_alpha': 10,
'reg_lambda': 5,
'num_leaves': 20,
'early_stopping_rounds': 300,
'n_jobs': -1,
'objective': 'multiclass',
'boosting': 'gbdt',
'feature_name': 'auto',
'categorical_features': 'auto'
}
# Training lgbm model
indices_oof_predictions, loss = [], []
for train_index, test_index in StratifiedKFold(n_splits=10).split(train_indices, target):
X_train, X_test, y_train, y_test = train_indices.iloc[train_index], train_indices.iloc[test_index], target[train_index], target[test_index]
lgbm = LGBMClassifier(**lgbm_params)
lgbm.fit(X_train, y_train, eval_set = [(X_test, y_test)], verbose = False)
indices_oof_predictions.append(lgbm.predict_proba(test_indices))
loss.append(log_loss(y_test, lgbm.predict_proba(X_test)))
print('Loss: ', log_loss(y_test, lgbm.predict_proba(X_test)))
print('Indices Cross Validation Loss: ', np.mean(loss))
# Submission file preparation
submission_file = pd.DataFrame({'Field ID': test['Field ID']})
for i, j in enumerate(sample_submission.columns[1:]):
submission_file[j] = np.mean(indices_oof_predictions, 0)[:, i]
submission_file.to_csv('LGBM_SUB.csv', index = False)
submission_file.head()
| 0.361728 | 0.774935 |
```
import numba as nb
import numpy as np
def conv_kernel(x, w, rs, n, n_channels, height, width, n_filters, filter_height, filter_width, out_h, out_w):
for i in range(n):
for j in range(out_h):
for p in range(out_w):
window = x[i, ..., j:j+filter_height, p:p+filter_width]
for q in range(n_filters):
rs[i, q, j, p] += np.sum(w[q] * window)
@nb.jit(nopython=True)
def jit_conv_kernel(x, w, rs, n, n_channels, height, width, n_filters, filter_height, filter_width, out_h, out_w):
for i in range(n):
for j in range(out_h):
for p in range(out_w):
window = x[i, ..., j:j+filter_height, p:p+filter_width]
for q in range(n_filters):
rs[i, q, j, p] += np.sum(w[q] * window)
def conv(x, w, kernel, args):
n, n_filters = args[0], args[4]
out_h, out_w = args[-2:]
rs = np.zeros([n, n_filters, out_h, out_w], dtype=np.float32)
kernel(x, w, rs, *args)
return rs
def cs231n_conv(x, w, args):
n, n_channels, height, width, n_filters, filter_height, filter_width, out_h, out_w = args
shape = (n_channels, filter_height, filter_width, n, out_h, out_w)
strides = (height * width, width, 1, n_channels * height * width, width, 1)
strides = x.itemsize * np.asarray(strides)
x_cols = np.lib.stride_tricks.as_strided(x, shape=shape, strides=strides).reshape(
n_channels * filter_height * filter_width, n * out_h * out_w)
return w.reshape(n_filters, -1).dot(x_cols).reshape(n_filters, n, out_h, out_w).transpose(1, 0, 2, 3)
# 64 个 3 x 28 x 28 的图像输入(模拟 mnist)
x = np.random.randn(64, 3, 28, 28).astype(np.float32)
# 16 个 5 x 5 的 kernel
w = np.random.randn(16, x.shape[1], 5, 5).astype(np.float32)
n, n_channels, height, width = x.shape
n_filters, _, filter_height, filter_width = w.shape
out_h = height - filter_height + 1
out_w = width - filter_width + 1
args = (n, n_channels, height, width, n_filters, filter_height, filter_width, out_h, out_w)
print(np.linalg.norm((cs231n_conv(x, w, args) - conv(x, w, conv_kernel, args)).ravel()))
print(np.linalg.norm((cs231n_conv(x, w, args) - conv(x, w, jit_conv_kernel, args)).ravel()))
print(np.linalg.norm((conv(x, w, conv_kernel, args) - conv(x, w, jit_conv_kernel, args)).ravel()))
%timeit conv(x, w, conv_kernel, args)
%timeit conv(x, w, jit_conv_kernel, args)
%timeit cs231n_conv(x, w, args)
```
+ 注意:这里如果使用`np.allclose`的话会过不了`assert`;事实上,仅仅是将数组的`dtype`从`float64`变成`float32`、精度就会下降很多,毕竟卷积涉及到的运算太多
```
@nb.jit(nopython=True)
def jit_conv_kernel2(x, w, rs, n, n_channels, height, width, n_filters, filter_height, filter_width, out_h, out_w):
for i in range(n):
for j in range(out_h):
for p in range(out_w):
for q in range(n_filters):
for r in range(n_channels):
for s in range(filter_height):
for t in range(filter_width):
rs[i, q, j, p] += x[i, r, j+s, p+t] * w[q, r, s, t]
assert np.allclose(conv(x, w, jit_conv_kernel, args), conv(x, w, jit_conv_kernel, args))
%timeit conv(x, w, jit_conv_kernel, args)
%timeit conv(x, w, jit_conv_kernel2, args)
%timeit cs231n_conv(x, w, args)
```
+ 可以看到,使用`jit`和使用纯`numpy`进行编程的很大一点不同就是,不要畏惧用`for`;事实上一般来说,代码“长得越像 C”、速度就会越快
```
def max_pool_kernel(x, rs, *args):
n, n_channels, pool_height, pool_width, out_h, out_w = args
for i in range(n):
for j in range(n_channels):
for p in range(out_h):
for q in range(out_w):
window = x[i, j, p:p+pool_height, q:q+pool_width]
rs[i, j, p, q] += np.max(window)
@nb.jit(nopython=True)
def jit_max_pool_kernel(x, rs, *args):
n, n_channels, pool_height, pool_width, out_h, out_w = args
for i in range(n):
for j in range(n_channels):
for p in range(out_h):
for q in range(out_w):
window = x[i, j, p:p+pool_height, q:q+pool_width]
rs[i, j, p, q] += np.max(window)
@nb.jit(nopython=True)
def jit_max_pool_kernel2(x, rs, *args):
n, n_channels, pool_height, pool_width, out_h, out_w = args
for i in range(n):
for j in range(n_channels):
for p in range(out_h):
for q in range(out_w):
_max = x[i, j, p, q]
for r in range(pool_height):
for s in range(pool_width):
_tmp = x[i, j, p+r, q+s]
if _tmp > _max:
_max = _tmp
rs[i, j, p, q] += _max
def max_pool(x, kernel, args):
n, n_channels = args[:2]
out_h, out_w = args[-2:]
rs = np.zeros([n, n_filters, out_h, out_w], dtype=np.float32)
kernel(x, rs, *args)
return rs
pool_height, pool_width = 2, 2
n, n_channels, height, width = x.shape
out_h = height - pool_height + 1
out_w = width - pool_width + 1
args = (n, n_channels, pool_height, pool_width, out_h, out_w)
assert np.allclose(max_pool(x, max_pool_kernel, args), max_pool(x, jit_max_pool_kernel, args))
assert np.allclose(max_pool(x, jit_max_pool_kernel, args), max_pool(x, jit_max_pool_kernel2, args))
%timeit max_pool(x, max_pool_kernel, args)
%timeit max_pool(x, jit_max_pool_kernel, args)
%timeit max_pool(x, jit_max_pool_kernel2, args)
```
|
github_jupyter
|
import numba as nb
import numpy as np
def conv_kernel(x, w, rs, n, n_channels, height, width, n_filters, filter_height, filter_width, out_h, out_w):
for i in range(n):
for j in range(out_h):
for p in range(out_w):
window = x[i, ..., j:j+filter_height, p:p+filter_width]
for q in range(n_filters):
rs[i, q, j, p] += np.sum(w[q] * window)
@nb.jit(nopython=True)
def jit_conv_kernel(x, w, rs, n, n_channels, height, width, n_filters, filter_height, filter_width, out_h, out_w):
for i in range(n):
for j in range(out_h):
for p in range(out_w):
window = x[i, ..., j:j+filter_height, p:p+filter_width]
for q in range(n_filters):
rs[i, q, j, p] += np.sum(w[q] * window)
def conv(x, w, kernel, args):
n, n_filters = args[0], args[4]
out_h, out_w = args[-2:]
rs = np.zeros([n, n_filters, out_h, out_w], dtype=np.float32)
kernel(x, w, rs, *args)
return rs
def cs231n_conv(x, w, args):
n, n_channels, height, width, n_filters, filter_height, filter_width, out_h, out_w = args
shape = (n_channels, filter_height, filter_width, n, out_h, out_w)
strides = (height * width, width, 1, n_channels * height * width, width, 1)
strides = x.itemsize * np.asarray(strides)
x_cols = np.lib.stride_tricks.as_strided(x, shape=shape, strides=strides).reshape(
n_channels * filter_height * filter_width, n * out_h * out_w)
return w.reshape(n_filters, -1).dot(x_cols).reshape(n_filters, n, out_h, out_w).transpose(1, 0, 2, 3)
# 64 个 3 x 28 x 28 的图像输入(模拟 mnist)
x = np.random.randn(64, 3, 28, 28).astype(np.float32)
# 16 个 5 x 5 的 kernel
w = np.random.randn(16, x.shape[1], 5, 5).astype(np.float32)
n, n_channels, height, width = x.shape
n_filters, _, filter_height, filter_width = w.shape
out_h = height - filter_height + 1
out_w = width - filter_width + 1
args = (n, n_channels, height, width, n_filters, filter_height, filter_width, out_h, out_w)
print(np.linalg.norm((cs231n_conv(x, w, args) - conv(x, w, conv_kernel, args)).ravel()))
print(np.linalg.norm((cs231n_conv(x, w, args) - conv(x, w, jit_conv_kernel, args)).ravel()))
print(np.linalg.norm((conv(x, w, conv_kernel, args) - conv(x, w, jit_conv_kernel, args)).ravel()))
%timeit conv(x, w, conv_kernel, args)
%timeit conv(x, w, jit_conv_kernel, args)
%timeit cs231n_conv(x, w, args)
@nb.jit(nopython=True)
def jit_conv_kernel2(x, w, rs, n, n_channels, height, width, n_filters, filter_height, filter_width, out_h, out_w):
for i in range(n):
for j in range(out_h):
for p in range(out_w):
for q in range(n_filters):
for r in range(n_channels):
for s in range(filter_height):
for t in range(filter_width):
rs[i, q, j, p] += x[i, r, j+s, p+t] * w[q, r, s, t]
assert np.allclose(conv(x, w, jit_conv_kernel, args), conv(x, w, jit_conv_kernel, args))
%timeit conv(x, w, jit_conv_kernel, args)
%timeit conv(x, w, jit_conv_kernel2, args)
%timeit cs231n_conv(x, w, args)
def max_pool_kernel(x, rs, *args):
n, n_channels, pool_height, pool_width, out_h, out_w = args
for i in range(n):
for j in range(n_channels):
for p in range(out_h):
for q in range(out_w):
window = x[i, j, p:p+pool_height, q:q+pool_width]
rs[i, j, p, q] += np.max(window)
@nb.jit(nopython=True)
def jit_max_pool_kernel(x, rs, *args):
n, n_channels, pool_height, pool_width, out_h, out_w = args
for i in range(n):
for j in range(n_channels):
for p in range(out_h):
for q in range(out_w):
window = x[i, j, p:p+pool_height, q:q+pool_width]
rs[i, j, p, q] += np.max(window)
@nb.jit(nopython=True)
def jit_max_pool_kernel2(x, rs, *args):
n, n_channels, pool_height, pool_width, out_h, out_w = args
for i in range(n):
for j in range(n_channels):
for p in range(out_h):
for q in range(out_w):
_max = x[i, j, p, q]
for r in range(pool_height):
for s in range(pool_width):
_tmp = x[i, j, p+r, q+s]
if _tmp > _max:
_max = _tmp
rs[i, j, p, q] += _max
def max_pool(x, kernel, args):
n, n_channels = args[:2]
out_h, out_w = args[-2:]
rs = np.zeros([n, n_filters, out_h, out_w], dtype=np.float32)
kernel(x, rs, *args)
return rs
pool_height, pool_width = 2, 2
n, n_channels, height, width = x.shape
out_h = height - pool_height + 1
out_w = width - pool_width + 1
args = (n, n_channels, pool_height, pool_width, out_h, out_w)
assert np.allclose(max_pool(x, max_pool_kernel, args), max_pool(x, jit_max_pool_kernel, args))
assert np.allclose(max_pool(x, jit_max_pool_kernel, args), max_pool(x, jit_max_pool_kernel2, args))
%timeit max_pool(x, max_pool_kernel, args)
%timeit max_pool(x, jit_max_pool_kernel, args)
%timeit max_pool(x, jit_max_pool_kernel2, args)
| 0.428473 | 0.664867 |
# Using Tensorflow with H2O
This notebook shows how to use the tensorflow backend to tackle a simple image classification problem.
We start by connecting to our h2o cluster:
```
import sys, os
import h2o
from h2o.estimators.deepwater import H2ODeepWaterEstimator
import os.path
from IPython.display import Image, display, HTML
import pandas as pd
import numpy as np
import random
PATH=os.path.expanduser("~/h2o-3")
h2o.init(port=54321, nthreads=-1)
if not H2ODeepWaterEstimator.available(): exit
!nvidia-smi
%matplotlib inline
from IPython.display import Image, display, HTML
import matplotlib.pyplot as plt
```
## Image Classification Task
H2O DeepWater allows you to specify a list of URIs (file paths) or URLs (links) to images, together with a response column (either a class membership (enum) or regression target (numeric)).
For this example, we use a small dataset that has a few hundred images, and three classes: cat, dog and mouse.
```
frame = h2o.import_file(PATH + "/bigdata/laptop/deepwater/imagenet/cat_dog_mouse.csv")
print(frame.dim)
print(frame.head(5))
```
To build a LeNet image classification model in H2O, simply specify `network = "lenet"` and `backend="tensorflow"` to use the our pre-built TensorFlow lenet implementation:
```
model = H2ODeepWaterEstimator(epochs=500, network = "lenet", backend="tensorflow")
model.train(x=[0],y=1, training_frame=frame)
model.show()
model = H2ODeepWaterEstimator(epochs=100, backend="tensorflow",
image_shape=[28,28],
network="user",
network_definition_file=PATH + "/examples/deeplearning/notebooks/pretrained/lenet_28x28x3_3.meta",
network_parameters_file=PATH + "/examples/deeplearning/notebooks/pretrained/lenet-100epochs")
model.train(x=[0],y=1, training_frame=frame)
model.show()
```
# DeepFeatures
We can also compute the output of any hidden layer, if we know its name.
```
model.deepfeatures(frame, "fc1/Relu")
```
# Custom models
If you'd like to build your own Tensorflow network architecture, then this is easy as well.
In this example script, we are using the **Tensorflow** backend.
Models can easily be imported/exported between H2O and Tensorflow since H2O uses Tensorflow's format for model definition.
```
def simple_model(w, h, channels, classes):
import json
import tensorflow as tf
from tensorflow.python.framework import ops
# always create a new graph inside ipython or
# the default one will be used and can lead to
# unexpected behavior
graph = tf.Graph()
with graph.as_default():
size = w * h * channels
x = tf.placeholder(tf.float32, [None, size])
W = tf.Variable(tf.zeros([size, classes]))
b = tf.Variable(tf.zeros([classes]))
y = tf.matmul(x, W) + b
predictions = tf.nn.softmax(y)
# labels
y_ = tf.placeholder(tf.float32, [None, classes])
# train
cross_entropy = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=y, labels=y_))
train_step = tf.train.GradientDescentOptimizer(0.5).minimize(cross_entropy)
tf.add_to_collection(ops.GraphKeys.TRAIN_OP, train_step)
tf.add_to_collection("predictions", predictions)
# this is required by the h2o tensorflow backend
global_step = tf.Variable(0, name="global_step", trainable=False)
init = tf.global_variables_initializer()
tf.add_to_collection(ops.GraphKeys.INIT_OP, init.name)
tf.add_to_collection("logits", y)
saver = tf.train.Saver()
meta = json.dumps({
"inputs": {"batch_image_input": x.name, "categorical_labels": y_.name},
"outputs": {"categorical_logits": y.name},
"parameters": {"global_step": global_step.name},
})
print(meta)
tf.add_to_collection("meta", meta)
filename = "/tmp/lenet_tensorflow.meta"
tf.train.export_meta_graph(filename, saver_def=saver.as_saver_def())
return filename
filename = simple_model(28, 28, 3, classes=3)
model = H2ODeepWaterEstimator(epochs=500,
network_definition_file=filename, ## specify the model
image_shape=[28,28], ## provide expected (or matching) image size
channels=3,
backend="tensorflow",
)
model.train(x=[0], y=1, training_frame=frame)
model.show()
```
# Custom models with Keras
It is also possible to use libraries/APIs such as Keras to define the network architecture.
```
import tensorflow as tf
import json
from keras.layers.core import Dense, Flatten, Reshape
from keras.layers.convolutional import Conv2D
from keras.layers.pooling import MaxPooling2D
from keras import backend as K
from keras.objectives import categorical_crossentropy
from tensorflow.python.framework import ops
def keras_model(w, h, channels, classes):
# always create a new graph inside ipython or
# the default one will be used and can lead to
# unexpected behavior
graph = tf.Graph()
with graph.as_default():
size = w * h * channels
# Input images fed via H2O
inp = tf.placeholder(tf.float32, [None, size])
# Actual labels used for training fed via H2O
labels = tf.placeholder(tf.float32, [None, classes])
# Keras network
x = Reshape((w, h, channels))(inp)
x = Conv2D(20, (5, 5), padding='same', activation='relu')(x)
x = MaxPooling2D((2,2))(x)
x = Conv2D(50, (5, 5), padding='same', activation='relu')(x)
x = MaxPooling2D((2,2))(x)
x = Flatten()(x)
x = Dense(500, activation='relu')(x)
out = Dense(classes)(x)
predictions = tf.nn.softmax(out)
loss = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(labels=labels,logits=out))
train_step = tf.train.AdamOptimizer(1e-3).minimize(loss)
init_op = tf.global_variables_initializer()
# Metadata required by H2O
tf.add_to_collection(ops.GraphKeys.INIT_OP, init_op.name)
tf.add_to_collection(ops.GraphKeys.TRAIN_OP, train_step)
tf.add_to_collection("logits", out)
tf.add_to_collection("predictions", predictions)
meta = json.dumps({
"inputs": {"batch_image_input": inp.name,
"categorical_labels": labels.name},
"outputs": {"categorical_logits": out.name,
"layers": ','.join([m.name for m in tf.get_default_graph().get_operations()])},
"parameters": {}
})
tf.add_to_collection("meta", meta)
# Save the meta file with the graph
saver = tf.train.Saver()
filename = "/tmp/keras_tensorflow.meta"
tf.train.export_meta_graph(filename, saver_def=saver.as_saver_def())
return filename
filename = keras_model(28, 28, 3, classes=3)
model = H2ODeepWaterEstimator(epochs=50,
network_definition_file=filename, ## specify the model
image_shape=[28,28], ## provide expected (or matching) image size
channels=3,
backend="tensorflow",
)
model.train(x=[0], y=1, training_frame=frame)
model.show()
```
|
github_jupyter
|
import sys, os
import h2o
from h2o.estimators.deepwater import H2ODeepWaterEstimator
import os.path
from IPython.display import Image, display, HTML
import pandas as pd
import numpy as np
import random
PATH=os.path.expanduser("~/h2o-3")
h2o.init(port=54321, nthreads=-1)
if not H2ODeepWaterEstimator.available(): exit
!nvidia-smi
%matplotlib inline
from IPython.display import Image, display, HTML
import matplotlib.pyplot as plt
frame = h2o.import_file(PATH + "/bigdata/laptop/deepwater/imagenet/cat_dog_mouse.csv")
print(frame.dim)
print(frame.head(5))
model = H2ODeepWaterEstimator(epochs=500, network = "lenet", backend="tensorflow")
model.train(x=[0],y=1, training_frame=frame)
model.show()
model = H2ODeepWaterEstimator(epochs=100, backend="tensorflow",
image_shape=[28,28],
network="user",
network_definition_file=PATH + "/examples/deeplearning/notebooks/pretrained/lenet_28x28x3_3.meta",
network_parameters_file=PATH + "/examples/deeplearning/notebooks/pretrained/lenet-100epochs")
model.train(x=[0],y=1, training_frame=frame)
model.show()
model.deepfeatures(frame, "fc1/Relu")
def simple_model(w, h, channels, classes):
import json
import tensorflow as tf
from tensorflow.python.framework import ops
# always create a new graph inside ipython or
# the default one will be used and can lead to
# unexpected behavior
graph = tf.Graph()
with graph.as_default():
size = w * h * channels
x = tf.placeholder(tf.float32, [None, size])
W = tf.Variable(tf.zeros([size, classes]))
b = tf.Variable(tf.zeros([classes]))
y = tf.matmul(x, W) + b
predictions = tf.nn.softmax(y)
# labels
y_ = tf.placeholder(tf.float32, [None, classes])
# train
cross_entropy = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=y, labels=y_))
train_step = tf.train.GradientDescentOptimizer(0.5).minimize(cross_entropy)
tf.add_to_collection(ops.GraphKeys.TRAIN_OP, train_step)
tf.add_to_collection("predictions", predictions)
# this is required by the h2o tensorflow backend
global_step = tf.Variable(0, name="global_step", trainable=False)
init = tf.global_variables_initializer()
tf.add_to_collection(ops.GraphKeys.INIT_OP, init.name)
tf.add_to_collection("logits", y)
saver = tf.train.Saver()
meta = json.dumps({
"inputs": {"batch_image_input": x.name, "categorical_labels": y_.name},
"outputs": {"categorical_logits": y.name},
"parameters": {"global_step": global_step.name},
})
print(meta)
tf.add_to_collection("meta", meta)
filename = "/tmp/lenet_tensorflow.meta"
tf.train.export_meta_graph(filename, saver_def=saver.as_saver_def())
return filename
filename = simple_model(28, 28, 3, classes=3)
model = H2ODeepWaterEstimator(epochs=500,
network_definition_file=filename, ## specify the model
image_shape=[28,28], ## provide expected (or matching) image size
channels=3,
backend="tensorflow",
)
model.train(x=[0], y=1, training_frame=frame)
model.show()
import tensorflow as tf
import json
from keras.layers.core import Dense, Flatten, Reshape
from keras.layers.convolutional import Conv2D
from keras.layers.pooling import MaxPooling2D
from keras import backend as K
from keras.objectives import categorical_crossentropy
from tensorflow.python.framework import ops
def keras_model(w, h, channels, classes):
# always create a new graph inside ipython or
# the default one will be used and can lead to
# unexpected behavior
graph = tf.Graph()
with graph.as_default():
size = w * h * channels
# Input images fed via H2O
inp = tf.placeholder(tf.float32, [None, size])
# Actual labels used for training fed via H2O
labels = tf.placeholder(tf.float32, [None, classes])
# Keras network
x = Reshape((w, h, channels))(inp)
x = Conv2D(20, (5, 5), padding='same', activation='relu')(x)
x = MaxPooling2D((2,2))(x)
x = Conv2D(50, (5, 5), padding='same', activation='relu')(x)
x = MaxPooling2D((2,2))(x)
x = Flatten()(x)
x = Dense(500, activation='relu')(x)
out = Dense(classes)(x)
predictions = tf.nn.softmax(out)
loss = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(labels=labels,logits=out))
train_step = tf.train.AdamOptimizer(1e-3).minimize(loss)
init_op = tf.global_variables_initializer()
# Metadata required by H2O
tf.add_to_collection(ops.GraphKeys.INIT_OP, init_op.name)
tf.add_to_collection(ops.GraphKeys.TRAIN_OP, train_step)
tf.add_to_collection("logits", out)
tf.add_to_collection("predictions", predictions)
meta = json.dumps({
"inputs": {"batch_image_input": inp.name,
"categorical_labels": labels.name},
"outputs": {"categorical_logits": out.name,
"layers": ','.join([m.name for m in tf.get_default_graph().get_operations()])},
"parameters": {}
})
tf.add_to_collection("meta", meta)
# Save the meta file with the graph
saver = tf.train.Saver()
filename = "/tmp/keras_tensorflow.meta"
tf.train.export_meta_graph(filename, saver_def=saver.as_saver_def())
return filename
filename = keras_model(28, 28, 3, classes=3)
model = H2ODeepWaterEstimator(epochs=50,
network_definition_file=filename, ## specify the model
image_shape=[28,28], ## provide expected (or matching) image size
channels=3,
backend="tensorflow",
)
model.train(x=[0], y=1, training_frame=frame)
model.show()
| 0.516108 | 0.897381 |
# 8.1 Drawing random numbers
```
import random
random.random()
random.random()
```
## 8.1.1 The Seed
```
random.seed(3)
print(', '.join(['%.2f' % random.random() for i in range(11)]))
print(', '.join(['%.2f' % random.random() for i in range(11)]))
print(', '.join(['%.2f' % random.random() for i in range(11)]))
random.seed(3)
print(', '.join(['%.2f' % random.random() for i in range(11)]))
print(', '.join(['%.2f' % random.random() for i in range(11)]))
print(', '.join(['%.2f' % random.random() for i in range(11)]))
```
## 8.1.2 Uniformly Distributed Random Numbers
```
import matplotlib.pyplot as plt
plt.figure()
plt.plot([random.uniform(-1, 1) for i in range(300)], '+')
plt.show()
# normal distribution
plt.figure()
plt.plot([random.gauss(0, 1) for i in range(300)], '+')
plt.show()
```
## 8.1.3 Visualizing the Distribution
```
plt.figure()
plt.subplot(211)
plt.hist([random.random() for i in range(1000)], bins=50, normed=True)
plt.subplot(212)
plt.hist([random.random() for i in range(100000)], bins=50, normed=True)
plt.show()
```
# 8.1.4 Vectorized Drawing of random numbers
```
import numpy as np
np.random.random()
x = np.random.random(size=20)
x
```
# 8.1.5 Computing the mean and standard deviation
```
np.mean(x)
np.std(x)
np.sqrt(np.var(x))
```
# The Gaussian or normal distribution
```
random.gauss(0, 1)
np.random.normal()
# numpy random madness
# uniformly distributed numbers
print(np.random.uniform(-1, 1, (1, 3))) # here, the interval can be specified [low, high)
print(np.random.rand(1, 3)) # interval [0, 1), convenience function for uniform.
print(np.random.rand())
# uniform(a, b) is the same as (b - a) * rand() + a:
np.random.seed(0)
print(np.random.uniform(4, 5))
np.random.seed(0)
print((5 - 4) * np.random.rand() + 4) # however, this is longer and more confusing in this case!
# all identical, all like rand, but longer. (unuseful!)
print(np.random.random([1, 3]))
print(np.random.random_sample([1, 3]))
print(np.random.ranf([1, 3]))
print(np.random.sample([1, 3]))
# normally distributed numbers
print(np.random.normal(4, 2, (1, 3))) # here, mu and sigma can be specified
print(np.random.randn(1, 3)) # mu = 0, sigma = 1; convenience function for normal.
# normal(mu, sigma) is the same as sigma * randn() + mu:
np.random.seed(0)
print(np.random.normal(3, 0.5))
np.random.seed(0)
print(0.5 * np.random.randn() + 3) # however, this is longer and more confusing in this case!
# integers
print(np.random.randint(3, 9, size=(2, 2))) # draw int from [low, high)
# sample
print(np.random.choice([1, 2, 3, 4, 5], 3))
plt.figure()
plt.hist([np.random.randn() for i in range(1000)], bins=25, normed=True)
from scipy.stats import norm
x = np.linspace(-4, 4)
plt.plot(x, norm.pdf(x))
plt.show()
```
# 8.2 Drawing Integers
```
random.randint(2, 6) # upper limit is includet! (unlike in np.random.randint)
```
# 8.2.8 Drawing a random element from a list
```
a = np.arange(10)
random.choice(a)
np.random.choice(a)
random.shuffle(a) # changes the list (does not copy)
a
np.random.shuffle(a)
a
np.fromiter((np.random.rand() for i in range(20)), dtype=float) # turn generator into 1d array.
```
# 8.3 Computing Probabilities
```
random.choice("abc")
```
# 8.6 Random Walk in One Space Dimension
```
import numpy as np
import matplotlib.pyplot as plt
from math import sqrt
from scipy.stats import norm
n_steps = 10000
n_particles = 500
x = np.sum(np.random.choice((-1, 1), (n_steps, n_particles)), axis=1)
plt.figure()
plt.hist(x, bins=25, density=True)
plt.plot(x, np.zeros(x.shape), 'kx')
x = np.linspace(-2 * sqrt(n_steps), 2 * sqrt(n_steps))
plt.plot(x, norm.pdf(x, 0, sqrt(n_steps)))
plt.show()
# sigma obviously is not sqrt(n_steps!)
# -> find sigma using a monte carlo simulation!
x = np.sum(np.random.choice((-1, 1), (n_steps, n_particles)), axis=1) # random values
xm = x.mean()
variance = 1 / len(x) * np.sum((x - xm)**2)
sigma = sqrt(variance)
print(sigma, sqrt(n_steps)) # aha!
# again:
plt.figure()
plt.hist(x, bins=25, density=True)
plt.plot(x, np.zeros(x.shape), 'kx')
x_ = np.linspace(-sqrt(n_steps), sqrt(n_steps), 200)
plt.plot(x_, norm.pdf(x_, xm, sigma)) # xm does really approach 0.
plt.show()
```
|
github_jupyter
|
import random
random.random()
random.random()
random.seed(3)
print(', '.join(['%.2f' % random.random() for i in range(11)]))
print(', '.join(['%.2f' % random.random() for i in range(11)]))
print(', '.join(['%.2f' % random.random() for i in range(11)]))
random.seed(3)
print(', '.join(['%.2f' % random.random() for i in range(11)]))
print(', '.join(['%.2f' % random.random() for i in range(11)]))
print(', '.join(['%.2f' % random.random() for i in range(11)]))
import matplotlib.pyplot as plt
plt.figure()
plt.plot([random.uniform(-1, 1) for i in range(300)], '+')
plt.show()
# normal distribution
plt.figure()
plt.plot([random.gauss(0, 1) for i in range(300)], '+')
plt.show()
plt.figure()
plt.subplot(211)
plt.hist([random.random() for i in range(1000)], bins=50, normed=True)
plt.subplot(212)
plt.hist([random.random() for i in range(100000)], bins=50, normed=True)
plt.show()
import numpy as np
np.random.random()
x = np.random.random(size=20)
x
np.mean(x)
np.std(x)
np.sqrt(np.var(x))
random.gauss(0, 1)
np.random.normal()
# numpy random madness
# uniformly distributed numbers
print(np.random.uniform(-1, 1, (1, 3))) # here, the interval can be specified [low, high)
print(np.random.rand(1, 3)) # interval [0, 1), convenience function for uniform.
print(np.random.rand())
# uniform(a, b) is the same as (b - a) * rand() + a:
np.random.seed(0)
print(np.random.uniform(4, 5))
np.random.seed(0)
print((5 - 4) * np.random.rand() + 4) # however, this is longer and more confusing in this case!
# all identical, all like rand, but longer. (unuseful!)
print(np.random.random([1, 3]))
print(np.random.random_sample([1, 3]))
print(np.random.ranf([1, 3]))
print(np.random.sample([1, 3]))
# normally distributed numbers
print(np.random.normal(4, 2, (1, 3))) # here, mu and sigma can be specified
print(np.random.randn(1, 3)) # mu = 0, sigma = 1; convenience function for normal.
# normal(mu, sigma) is the same as sigma * randn() + mu:
np.random.seed(0)
print(np.random.normal(3, 0.5))
np.random.seed(0)
print(0.5 * np.random.randn() + 3) # however, this is longer and more confusing in this case!
# integers
print(np.random.randint(3, 9, size=(2, 2))) # draw int from [low, high)
# sample
print(np.random.choice([1, 2, 3, 4, 5], 3))
plt.figure()
plt.hist([np.random.randn() for i in range(1000)], bins=25, normed=True)
from scipy.stats import norm
x = np.linspace(-4, 4)
plt.plot(x, norm.pdf(x))
plt.show()
random.randint(2, 6) # upper limit is includet! (unlike in np.random.randint)
a = np.arange(10)
random.choice(a)
np.random.choice(a)
random.shuffle(a) # changes the list (does not copy)
a
np.random.shuffle(a)
a
np.fromiter((np.random.rand() for i in range(20)), dtype=float) # turn generator into 1d array.
random.choice("abc")
import numpy as np
import matplotlib.pyplot as plt
from math import sqrt
from scipy.stats import norm
n_steps = 10000
n_particles = 500
x = np.sum(np.random.choice((-1, 1), (n_steps, n_particles)), axis=1)
plt.figure()
plt.hist(x, bins=25, density=True)
plt.plot(x, np.zeros(x.shape), 'kx')
x = np.linspace(-2 * sqrt(n_steps), 2 * sqrt(n_steps))
plt.plot(x, norm.pdf(x, 0, sqrt(n_steps)))
plt.show()
# sigma obviously is not sqrt(n_steps!)
# -> find sigma using a monte carlo simulation!
x = np.sum(np.random.choice((-1, 1), (n_steps, n_particles)), axis=1) # random values
xm = x.mean()
variance = 1 / len(x) * np.sum((x - xm)**2)
sigma = sqrt(variance)
print(sigma, sqrt(n_steps)) # aha!
# again:
plt.figure()
plt.hist(x, bins=25, density=True)
plt.plot(x, np.zeros(x.shape), 'kx')
x_ = np.linspace(-sqrt(n_steps), sqrt(n_steps), 200)
plt.plot(x_, norm.pdf(x_, xm, sigma)) # xm does really approach 0.
plt.show()
| 0.295535 | 0.848157 |
# Pyspark 广播和累加器
对于并行处理,Apache Spark使用共享变量。当驱动程序将任务发送到集群上的执行程序时,共享变量的副本将在集群的每个节点上运行,以便可以将其用于执行任务。
Apache Spark支持两种类型的共享变量:
Broadcast
Accumulato
## 广播
Broadcast 广播变量用于跨所有节点保存数据副本。此变量缓存在所有计算机上,而不是在具有任务的计算机上发送。
```
from pyspark import SparkContext
sc = SparkContext("local","broadcast app")
word = sc.broadcast(["scala","java","python","spark"])
print(word.value) # Broadcast变量有一个名为value的属性,它存储数据并用于返回广播值。
print(word.value[2])
```
## 累加器
Accumulate 累加器变量用于通过关联和交换操作聚合信息。例如,您可以使用累加器进行求和操作或计数器(在MapReduce中)。
```
from pyspark import SparkContext
# sc = SparkContext("local","Accumulator app")
num = sc.accumulator(10)
def f(x):
global num
num+=x
rdd = sc.parallelize([20,30,40,50])
rdd.foreach(f)
print(num.value)
print(rdd.collect())
```
# PySpark SparkConf
以下是SparkConf最常用的一些属性
set(key,value) - 设置配置属性。
setMaster(value) - 设置主URL。
setAppName(value) - 设置应用程序名称。
get(key,defaultValue = None) - 获取密钥的配置值。
setSparkHome(value) - 在工作节点上设置Spark安装路径。
```
from pyspark import SparkConf, SparkContext
conf = SparkConf().setAppName("PySpark App").setMaster("local")
sc = SparkContext(conf=conf)
```
# PySpark SparkFiles
在Apache Spark中,您可以使用 sc.addFile 上传文件(sc是您的默认SparkContext),并使用 SparkFiles.get 获取工作者的路径。
SparkFiles解析通过 SparkContext.addFile() 添加的文件的路径。
SparkFiles包含以下类方法:
get(filename)
getrootdirectory()
```
from pyspark import SparkFiles
path = "/home/ace/Desktop/github/data_anasylsis/pyspark/5000_points.txt"
pathname = "500_points.txt"
sc.addFile(path)
print(SparkFiles.get(pathname))
print(SparkFiles.getRootDirectory())
```
# PySpark MLlib
mllib.classification - 支持二进制分类,多类分类和回归分析的各种方法。分类中一些最流行的算法是 随机森林,朴素贝叶斯,决策树 等。
mllib.clustering - 聚类是一种无监督的学习问题,您可以根据某些相似概念将实体的子集彼此分组。
mllib.fpm - 频繁模式匹配是挖掘频繁项,项集,子序列或其他子结构,这些通常是分析大规模数据集的第一步。 多年来,这一直是数据挖掘领域的一个活跃的研究课题。
mllib.linalg - 线性代数的MLlib实用程序。
mllib.recommendation - 协同过滤通常用于推荐系统。 这些技术旨在填写用户项关联矩阵的缺失条目。它目前支持基于模型的协同过滤,其中用户和产品由一小组可用于预测缺失条目的潜在因素描述。 spark.mllib使用交替最小二乘(ALS)算法来学习这些潜在因素。
mllib.regression - 线性回归属于回归算法族。 回归的目标是找到变量之间的关系和依赖关系。使用线性回归模型和模型摘要的界面类似于逻辑回归案例。
# PySpark Serializers
序列化用于Apache Spark的性能调优。通过网络发送或写入磁盘或持久存储在内存中的所有数据都应序列化。
Marshal Serializer 此序列化程序比PickleSerializer更快,但支持更少的数据类型。
Pickle Serializer 此序列化程序几乎支持任何Python对象,但可能不如更专业的序列化程序快
```
from pyspark.context import SparkContext
from pyspark.serializers import MarshalSerializer
sc.stop()
sc = SparkContext("local", "serialization app", serializer = MarshalSerializer()) # 使用MarshalSerializer序列化数据
print(sc.parallelize(list(range(1000))).map(lambda x: 2 * x).take(10))
sc.stop()
```
|
github_jupyter
|
from pyspark import SparkContext
sc = SparkContext("local","broadcast app")
word = sc.broadcast(["scala","java","python","spark"])
print(word.value) # Broadcast变量有一个名为value的属性,它存储数据并用于返回广播值。
print(word.value[2])
from pyspark import SparkContext
# sc = SparkContext("local","Accumulator app")
num = sc.accumulator(10)
def f(x):
global num
num+=x
rdd = sc.parallelize([20,30,40,50])
rdd.foreach(f)
print(num.value)
print(rdd.collect())
from pyspark import SparkConf, SparkContext
conf = SparkConf().setAppName("PySpark App").setMaster("local")
sc = SparkContext(conf=conf)
from pyspark import SparkFiles
path = "/home/ace/Desktop/github/data_anasylsis/pyspark/5000_points.txt"
pathname = "500_points.txt"
sc.addFile(path)
print(SparkFiles.get(pathname))
print(SparkFiles.getRootDirectory())
from pyspark.context import SparkContext
from pyspark.serializers import MarshalSerializer
sc.stop()
sc = SparkContext("local", "serialization app", serializer = MarshalSerializer()) # 使用MarshalSerializer序列化数据
print(sc.parallelize(list(range(1000))).map(lambda x: 2 * x).take(10))
sc.stop()
| 0.379263 | 0.869659 |
```
pip install wget
!python -m wget https://resources.lendingclub.com/LoanStats_2019Q1.csv.zip
!python -m wget https://resources.lendingclub.com/LoanStats_2019Q2.csv.zip
!python -m wget https://resources.lendingclub.com/LoanStats_2019Q3.csv.zip
!python -m wget https://resources.lendingclub.com/LoanStats_2019Q4.csv.zip
!python -m wget https://resources.lendingclub.com/LoanStats_2020Q1.csv.zip
import numpy as np
import pandas as pd
from pathlib import Path
from collections import Counter
from sklearn.model_selection import train_test_split
columns = [
"loan_amnt", "int_rate", "installment", "home_ownership", "annual_inc",
"verification_status", "pymnt_plan", "dti", "delinq_2yrs",
"inq_last_6mths", "open_acc", "pub_rec", "revol_bal", "total_acc",
"initial_list_status", "out_prncp", "out_prncp_inv", "total_pymnt",
"total_pymnt_inv", "total_rec_prncp", "total_rec_int",
"total_rec_late_fee", "recoveries", "collection_recovery_fee",
"last_pymnt_amnt", "collections_12_mths_ex_med", "policy_code",
"application_type", "acc_now_delinq", "tot_coll_amt", "tot_cur_bal",
"open_acc_6m", "open_act_il", "open_il_12m", "open_il_24m",
"mths_since_rcnt_il", "total_bal_il", "il_util", "open_rv_12m",
"open_rv_24m", "max_bal_bc", "all_util", "total_rev_hi_lim", "inq_fi",
"total_cu_tl", "inq_last_12m", "acc_open_past_24mths", "avg_cur_bal",
"bc_open_to_buy", "bc_util", "chargeoff_within_12_mths", "delinq_amnt",
"mo_sin_old_il_acct", "mo_sin_old_rev_tl_op", "mo_sin_rcnt_rev_tl_op",
"mo_sin_rcnt_tl", "mort_acc", "mths_since_recent_bc",
"mths_since_recent_inq", "num_accts_ever_120_pd", "num_actv_bc_tl",
"num_actv_rev_tl", "num_bc_sats", "num_bc_tl", "num_il_tl",
"num_op_rev_tl", "num_rev_accts", "num_rev_tl_bal_gt_0", "num_sats",
"num_tl_120dpd_2m", "num_tl_30dpd", "num_tl_90g_dpd_24m",
"num_tl_op_past_12m", "pct_tl_nvr_dlq", "percent_bc_gt_75",
"pub_rec_bankruptcies", "tax_liens", "tot_hi_cred_lim",
"total_bal_ex_mort", "total_bc_limit", "total_il_high_credit_limit",
"hardship_flag", "debt_settlement_flag",
"loan_status"
]
target = "loan_status"
# Load the data
df1 = pd.read_csv(Path(r'C:\Git\19-Supervised-Machine-Learning\Instructions\Resources\Generator\LoanStats_2019Q1.csv.zip'), skiprows=1)[:-2]
df2 = pd.read_csv(Path(r'C:\Git\19-Supervised-Machine-Learning\Instructions\Resources\Generator\LoanStats_2019Q2.csv.zip'), skiprows=1)[:-2]
df3 = pd.read_csv(Path(r'C:\Git\19-Supervised-Machine-Learning\Instructions\Resources\Generator\LoanStats_2019Q3.csv.zip'), skiprows=1)[:-2]
df4 = pd.read_csv(Path(r'C:\Git\19-Supervised-Machine-Learning\Instructions\Resources\Generator\LoanStats_2019Q4.csv.zip'), skiprows=1)[:-2]
df = pd.concat([df1, df2, df3, df4]).loc[:, columns].copy()
# Drop the null columns where all values are null
df = df.dropna(axis='columns', how='all')
# Drop the null rows
df = df.dropna()
# Remove the `Issued` loan status
issued_mask = df['loan_status'] != 'Issued'
df = df.loc[issued_mask]
# convert interest rate to numerical
df['int_rate'] = df['int_rate'].str.replace('%', '')
df['int_rate'] = df['int_rate'].astype('float') / 100
# Convert the target column values to low_risk and high_risk based on their values
x = {'Current': 'low_risk'}
df = df.replace(x)
x = dict.fromkeys(['Late (31-120 days)', 'Late (16-30 days)', 'Default', 'In Grace Period'], 'high_risk')
df = df.replace(x)
low_risk_rows = df[df[target] == 'low_risk']
high_risk_rows = df[df[target] == 'high_risk']
#df = pd.concat([low_risk_rows, high_risk_rows.sample(n=len(low_risk_rows), replace=True)])
df = pd.concat([low_risk_rows.sample(n=len(high_risk_rows), random_state=42), high_risk_rows])
df = df.reset_index(drop=True)
df = df.rename({target:'target'}, axis="columns")
df
df.to_csv('2019loans.csv', index=False)
# Load the data
validate_df = pd.read_csv(Path(r'C:\Git\19-Supervised-Machine-Learning\Instructions\Resources\Generator\LoanStats_2020Q1.csv.zip'), skiprows=1)[:-2]
validate_df = validate_df.loc[:, columns].copy()
# Drop the null columns where all values are null
validate_df = validate_df.dropna(axis='columns', how='all')
# Drop the null rows
validate_df = validate_df.dropna()
# Remove the `Issued` loan status
issued_mask = validate_df[target] != 'Issued'
validate_df = validate_df.loc[issued_mask]
# convert interest rate to numerical
validate_df['int_rate'] = validate_df['int_rate'].str.replace('%', '')
validate_df['int_rate'] = validate_df['int_rate'].astype('float') / 100
# Convert the target column values to low_risk and high_risk based on their values
x = dict.fromkeys(['Current', 'Fully Paid'], 'low_risk')
validate_df = validate_df.replace(x)
x = dict.fromkeys(['Late (31-120 days)', 'Late (16-30 days)', 'Default', 'In Grace Period', 'Charged Off'], 'high_risk')
validate_df = validate_df.replace(x)
low_risk_rows = validate_df[validate_df[target] == 'low_risk']
high_risk_rows = validate_df[validate_df[target] == 'high_risk']
validate_df = pd.concat([low_risk_rows.sample(n=len(high_risk_rows), random_state=37), high_risk_rows])
validate_df = validate_df.reset_index(drop=True)
validate_df = validate_df.rename({target:'target'}, axis="columns")
validate_df
validate_df.to_csv('2020Q1loans.csv', index=False)
```
|
github_jupyter
|
pip install wget
!python -m wget https://resources.lendingclub.com/LoanStats_2019Q1.csv.zip
!python -m wget https://resources.lendingclub.com/LoanStats_2019Q2.csv.zip
!python -m wget https://resources.lendingclub.com/LoanStats_2019Q3.csv.zip
!python -m wget https://resources.lendingclub.com/LoanStats_2019Q4.csv.zip
!python -m wget https://resources.lendingclub.com/LoanStats_2020Q1.csv.zip
import numpy as np
import pandas as pd
from pathlib import Path
from collections import Counter
from sklearn.model_selection import train_test_split
columns = [
"loan_amnt", "int_rate", "installment", "home_ownership", "annual_inc",
"verification_status", "pymnt_plan", "dti", "delinq_2yrs",
"inq_last_6mths", "open_acc", "pub_rec", "revol_bal", "total_acc",
"initial_list_status", "out_prncp", "out_prncp_inv", "total_pymnt",
"total_pymnt_inv", "total_rec_prncp", "total_rec_int",
"total_rec_late_fee", "recoveries", "collection_recovery_fee",
"last_pymnt_amnt", "collections_12_mths_ex_med", "policy_code",
"application_type", "acc_now_delinq", "tot_coll_amt", "tot_cur_bal",
"open_acc_6m", "open_act_il", "open_il_12m", "open_il_24m",
"mths_since_rcnt_il", "total_bal_il", "il_util", "open_rv_12m",
"open_rv_24m", "max_bal_bc", "all_util", "total_rev_hi_lim", "inq_fi",
"total_cu_tl", "inq_last_12m", "acc_open_past_24mths", "avg_cur_bal",
"bc_open_to_buy", "bc_util", "chargeoff_within_12_mths", "delinq_amnt",
"mo_sin_old_il_acct", "mo_sin_old_rev_tl_op", "mo_sin_rcnt_rev_tl_op",
"mo_sin_rcnt_tl", "mort_acc", "mths_since_recent_bc",
"mths_since_recent_inq", "num_accts_ever_120_pd", "num_actv_bc_tl",
"num_actv_rev_tl", "num_bc_sats", "num_bc_tl", "num_il_tl",
"num_op_rev_tl", "num_rev_accts", "num_rev_tl_bal_gt_0", "num_sats",
"num_tl_120dpd_2m", "num_tl_30dpd", "num_tl_90g_dpd_24m",
"num_tl_op_past_12m", "pct_tl_nvr_dlq", "percent_bc_gt_75",
"pub_rec_bankruptcies", "tax_liens", "tot_hi_cred_lim",
"total_bal_ex_mort", "total_bc_limit", "total_il_high_credit_limit",
"hardship_flag", "debt_settlement_flag",
"loan_status"
]
target = "loan_status"
# Load the data
df1 = pd.read_csv(Path(r'C:\Git\19-Supervised-Machine-Learning\Instructions\Resources\Generator\LoanStats_2019Q1.csv.zip'), skiprows=1)[:-2]
df2 = pd.read_csv(Path(r'C:\Git\19-Supervised-Machine-Learning\Instructions\Resources\Generator\LoanStats_2019Q2.csv.zip'), skiprows=1)[:-2]
df3 = pd.read_csv(Path(r'C:\Git\19-Supervised-Machine-Learning\Instructions\Resources\Generator\LoanStats_2019Q3.csv.zip'), skiprows=1)[:-2]
df4 = pd.read_csv(Path(r'C:\Git\19-Supervised-Machine-Learning\Instructions\Resources\Generator\LoanStats_2019Q4.csv.zip'), skiprows=1)[:-2]
df = pd.concat([df1, df2, df3, df4]).loc[:, columns].copy()
# Drop the null columns where all values are null
df = df.dropna(axis='columns', how='all')
# Drop the null rows
df = df.dropna()
# Remove the `Issued` loan status
issued_mask = df['loan_status'] != 'Issued'
df = df.loc[issued_mask]
# convert interest rate to numerical
df['int_rate'] = df['int_rate'].str.replace('%', '')
df['int_rate'] = df['int_rate'].astype('float') / 100
# Convert the target column values to low_risk and high_risk based on their values
x = {'Current': 'low_risk'}
df = df.replace(x)
x = dict.fromkeys(['Late (31-120 days)', 'Late (16-30 days)', 'Default', 'In Grace Period'], 'high_risk')
df = df.replace(x)
low_risk_rows = df[df[target] == 'low_risk']
high_risk_rows = df[df[target] == 'high_risk']
#df = pd.concat([low_risk_rows, high_risk_rows.sample(n=len(low_risk_rows), replace=True)])
df = pd.concat([low_risk_rows.sample(n=len(high_risk_rows), random_state=42), high_risk_rows])
df = df.reset_index(drop=True)
df = df.rename({target:'target'}, axis="columns")
df
df.to_csv('2019loans.csv', index=False)
# Load the data
validate_df = pd.read_csv(Path(r'C:\Git\19-Supervised-Machine-Learning\Instructions\Resources\Generator\LoanStats_2020Q1.csv.zip'), skiprows=1)[:-2]
validate_df = validate_df.loc[:, columns].copy()
# Drop the null columns where all values are null
validate_df = validate_df.dropna(axis='columns', how='all')
# Drop the null rows
validate_df = validate_df.dropna()
# Remove the `Issued` loan status
issued_mask = validate_df[target] != 'Issued'
validate_df = validate_df.loc[issued_mask]
# convert interest rate to numerical
validate_df['int_rate'] = validate_df['int_rate'].str.replace('%', '')
validate_df['int_rate'] = validate_df['int_rate'].astype('float') / 100
# Convert the target column values to low_risk and high_risk based on their values
x = dict.fromkeys(['Current', 'Fully Paid'], 'low_risk')
validate_df = validate_df.replace(x)
x = dict.fromkeys(['Late (31-120 days)', 'Late (16-30 days)', 'Default', 'In Grace Period', 'Charged Off'], 'high_risk')
validate_df = validate_df.replace(x)
low_risk_rows = validate_df[validate_df[target] == 'low_risk']
high_risk_rows = validate_df[validate_df[target] == 'high_risk']
validate_df = pd.concat([low_risk_rows.sample(n=len(high_risk_rows), random_state=37), high_risk_rows])
validate_df = validate_df.reset_index(drop=True)
validate_df = validate_df.rename({target:'target'}, axis="columns")
validate_df
validate_df.to_csv('2020Q1loans.csv', index=False)
| 0.542379 | 0.318644 |
# Comparative Linguistic Analysis of bioRxiv and PMC
```
%load_ext autoreload
%autoreload 2
from collections import defaultdict, Counter
import csv
import itertools
from pathlib import Path
import numpy as np
import pandas as pd
import pickle
import spacy
from scipy.stats import chi2_contingency
from tqdm import tqdm_notebook
from annorxiver_modules.corpora_comparison_helper import (
aggregate_word_counts,
dump_to_dataframe,
get_term_statistics,
KL_divergence,
)
```
# Full Text Comparison (Global)
## Gather Word Frequencies
```
biorxiv_count_path = Path("output/total_word_counts/biorxiv_total_count.tsv")
pmc_count_path = Path("output/total_word_counts/pmc_total_count.tsv")
nytac_count_path = Path("output/total_word_counts/nytac_total_count.tsv")
if not biorxiv_count_path.exists():
biorxiv_corpus_count = aggregate_word_counts(
list(Path("output/biorxiv_word_counts").rglob("*tsv"))
)
dump_to_dataframe(biorxiv_corpus_count, "output/biorxiv_total_count.tsv")
biorxiv_corpus_count.most_common(10)
if not pmc_count_path.exists():
pmc_corpus_count = aggregate_word_counts(
list(Path("../../pmc/pmc_corpus/pmc_word_counts").rglob("*tsv"))
)
dump_to_dataframe(pmc_corpus_count, "output/pmc_total_count.tsv")
pmc_corpus_count.most_common(10)
if not nytac_count_path.exists():
nytac_corpus_count = aggregate_word_counts(
list(Path("../../nytac/corpora_stats/output").rglob("*tsv"))
)
dump_to_dataframe(nytac_corpus_count, "output/nytac_total_count.tsv")
nytac_corpus_count.most_common(10)
biorxiv_total_count_df = pd.read_csv(biorxiv_count_path.resolve(), sep="\t")
pmc_total_count_df = pd.read_csv(pmc_count_path.resolve(), sep="\t")
nytac_total_count_df = pd.read_csv(nytac_count_path.resolve(), sep="\t")
biorxiv_sentence_length = pickle.load(open("output/biorxiv_sentence_length.pkl", "rb"))
pmc_sentence_length = pickle.load(
open("../../pmc/pmc_corpus/pmc_sentence_length.pkl", "rb")
)
nytac_sentence_length = pickle.load(
open("../../nytac/corpora_stats/nytac_sentence_length.pkl", "rb")
)
spacy_nlp = spacy.load("en_core_web_sm")
stop_word_list = list(spacy_nlp.Defaults.stop_words)
```
## Get Corpora Comparison Stats
```
biorxiv_sentence_len_list = list(biorxiv_sentence_length.items())
biorxiv_data = {
"document_count": len(biorxiv_sentence_length),
"sentence_count": sum(map(lambda x: len(x[1]), biorxiv_sentence_len_list)),
"token_count": biorxiv_total_count_df["count"].sum(),
"stop_word_count": (
biorxiv_total_count_df.query(f"lemma in {stop_word_list}")["count"].sum()
),
"avg_document_length": np.mean(
list(map(lambda x: len(x[1]), biorxiv_sentence_len_list))
),
"avg_sentence_length": np.mean(
list(itertools.chain(*list(map(lambda x: x[1], biorxiv_sentence_len_list))))
),
"negatives": (biorxiv_total_count_df.query(f"dep_tag =='neg'")["count"].sum()),
"coordinating_conjunctions": (
biorxiv_total_count_df.query(f"dep_tag =='cc'")["count"].sum()
),
"coordinating_conjunctions%": (
biorxiv_total_count_df.query(f"dep_tag =='cc'")["count"].sum()
)
/ biorxiv_total_count_df["count"].sum(),
"pronouns": (biorxiv_total_count_df.query(f"pos_tag =='PRON'")["count"].sum()),
"pronouns%": (biorxiv_total_count_df.query(f"pos_tag =='PRON'")["count"].sum())
/ biorxiv_total_count_df["count"].sum(),
"passives": (
biorxiv_total_count_df.query(
f"dep_tag in ['auxpass', 'nsubjpass', 'csubjpass']"
)["count"].sum()
),
"passive%": (
biorxiv_total_count_df.query(
f"dep_tag in ['auxpass', 'nsubjpass', 'csubjpass']"
)["count"].sum()
)
/ biorxiv_total_count_df["count"].sum(),
}
pmc_sentence_len_list = list(pmc_sentence_length.items())
pmc_data = {
"document_count": len(pmc_sentence_length),
"sentence_count": sum(map(lambda x: len(x[1]), pmc_sentence_len_list)),
"token_count": pmc_total_count_df["count"].sum(),
"stop_word_count": (
pmc_total_count_df.query(f"lemma in {stop_word_list}")["count"].sum()
),
"avg_document_length": np.mean(
list(map(lambda x: len(x[1]), pmc_sentence_len_list))
),
"avg_sentence_length": np.mean(
list(itertools.chain(*list(map(lambda x: x[1], pmc_sentence_len_list))))
),
"negatives": (pmc_total_count_df.query(f"dep_tag =='neg'")["count"].sum()),
"coordinating_conjunctions": (
pmc_total_count_df.query(f"dep_tag =='cc'")["count"].sum()
),
"coordinating_conjunctions%": (
pmc_total_count_df.query(f"dep_tag =='cc'")["count"].sum()
)
/ pmc_total_count_df["count"].sum(),
"pronouns": (pmc_total_count_df.query(f"pos_tag =='PRON'")["count"].sum()),
"pronouns%": (pmc_total_count_df.query(f"pos_tag =='PRON'")["count"].sum())
/ pmc_total_count_df["count"].sum(),
"passives": (
pmc_total_count_df.query(f"dep_tag in ['auxpass', 'nsubjpass', 'csubjpass']")[
"count"
].sum()
),
"passive%": (
pmc_total_count_df.query(f"dep_tag in ['auxpass', 'nsubjpass', 'csubjpass']")[
"count"
].sum()
)
/ pmc_total_count_df["count"].sum(),
}
nytac_sentence_len_list = list(nytac_sentence_length.items())
nytac_data = {
"document_count": len(nytac_sentence_length),
"sentence_count": sum(map(lambda x: len(x[1]), nytac_sentence_len_list)),
"token_count": nytac_total_count_df["count"].sum(),
"stop_word_count": (
nytac_total_count_df.query(f"lemma in {stop_word_list}")["count"].sum()
),
"avg_document_length": np.mean(
list(map(lambda x: len(x[1]), nytac_sentence_len_list))
),
"avg_sentence_length": np.mean(
list(itertools.chain(*list(map(lambda x: x[1], nytac_sentence_len_list))))
),
"negatives": (nytac_total_count_df.query(f"dep_tag =='neg'")["count"].sum()),
"coordinating_conjunctions": (
nytac_total_count_df.query(f"dep_tag =='cc'")["count"].sum()
),
"coordinating_conjunctions%": (
nytac_total_count_df.query(f"dep_tag =='cc'")["count"].sum()
)
/ nytac_total_count_df["count"].sum(),
"pronouns": (nytac_total_count_df.query(f"pos_tag =='PRON'")["count"].sum()),
"pronouns%": (nytac_total_count_df.query(f"pos_tag =='PRON'")["count"].sum())
/ nytac_total_count_df["count"].sum(),
"passives": (
nytac_total_count_df.query(f"dep_tag in ['auxpass', 'nsubjpass', 'csubjpass']")[
"count"
].sum()
),
"passive%": (
nytac_total_count_df.query(f"dep_tag in ['auxpass', 'nsubjpass', 'csubjpass']")[
"count"
].sum()
)
/ nytac_total_count_df["count"].sum(),
}
# This dataframe contains document statistics for each Corpus
# document count - the number of documents within the corpus
# Sentence count - the number of sentences within the corpus
# Token count - the number of tokens within the corpus
# Stop word counts - the number of stop words within the corpus
# Average document length - the average number of sentences within a document for a given corpus
# Average sentence length - the average number of words within a sentence for a given corpus
# Negatives - the number of negations (e.g. placing not in within a sentence) within a given corpus
# Coordinating Conjunctions - the number of coordinating conjunctions (and, but, for etc.) within a given corpus
# Pronouns - the number of pronouns within a given corpus
# Passive - the number of passive words within a given corpus
token_stats_df = pd.DataFrame.from_records(
[biorxiv_data, pmc_data, nytac_data], index=["bioRxiv", "PMC", "NYTAC"]
).T
token_stats_df.to_csv("output/figures/corpora_token_stats.tsv", sep="\t")
token_stats_df
```
## LogLikelihood + Odds Ratio + KL Divergence Calculations
The goal here is to compare word frequencies between bioRxiv and pubmed central. The problem when comparing word frequencies is that non-meaningful words (aka stopwords) such as the, of, and, be, etc., appear the most often. To account for this problem the first step here is to remove those words from analyses.
### Remove Stop words
```
biorxiv_total_count_df = (
biorxiv_total_count_df.query(f"lemma not in {stop_word_list}")
.groupby("lemma")
.agg({"count": "sum"})
.reset_index()
.sort_values("count", ascending=False)
)
biorxiv_total_count_df
pmc_total_count_df = (
pmc_total_count_df.query(f"lemma not in {stop_word_list}")
.groupby("lemma")
.agg({"count": "sum"})
.reset_index()
.sort_values("count", ascending=False)
.iloc[2:]
)
pmc_total_count_df
nytac_total_count_df = (
nytac_total_count_df.query(f"lemma not in {stop_word_list}")
.groupby("lemma")
.agg({"count": "sum"})
.reset_index()
.sort_values("count", ascending=False)
)
nytac_total_count_df
```
### Calculate LogLikelihoods and Odds ratios
```
biorxiv_vs_pmc = get_term_statistics(biorxiv_total_count_df, pmc_total_count_df, 100)
biorxiv_vs_pmc.to_csv(
"output/comparison_stats/biorxiv_vs_pmc_comparison.tsv", sep="\t", index=False
)
biorxiv_vs_pmc
biorxiv_vs_nytac = get_term_statistics(
biorxiv_total_count_df, nytac_total_count_df, 100
)
biorxiv_vs_nytac.to_csv(
"output/comparison_stats/biorxiv_nytac_comparison.tsv", sep="\t", index=False
)
biorxiv_vs_nytac
pmc_vs_nytac = get_term_statistics(pmc_total_count_df, nytac_total_count_df, 100)
pmc_vs_nytac.to_csv(
"output/comparison_stats/pmc_nytac_comparison.tsv", sep="\t", index=False
)
pmc_vs_nytac
```
## Calculate KL Divergence
```
term_grid = [100, 200, 300, 400, 500, 1000, 1500, 2000, 3000, 5000]
kl_data = []
for num_terms in tqdm_notebook(term_grid):
kl_data.append(
{
"num_terms": num_terms,
"KL_divergence": KL_divergence(
biorxiv_total_count_df, pmc_total_count_df, num_terms=num_terms
),
"comparison": "biorxiv_vs_pmc",
}
)
kl_data.append(
{
"num_terms": num_terms,
"KL_divergence": KL_divergence(
biorxiv_total_count_df, nytac_total_count_df, num_terms=num_terms
),
"comparison": "biorxiv_vs_nytac",
}
)
kl_data.append(
{
"num_terms": num_terms,
"KL_divergence": KL_divergence(
pmc_total_count_df, nytac_total_count_df, num_terms=num_terms
),
"comparison": "pmc_vs_nytac",
}
)
kl_metrics = pd.DataFrame.from_records(kl_data)
kl_metrics.to_csv(
"output/comparison_stats/corpora_kl_divergence.tsv", sep="\t", index=False
)
kl_metrics
```
# Preprint to Published View
```
mapped_doi_df = (
pd.read_csv("../journal_tracker/output/mapped_published_doi.tsv", sep="\t")
.query("published_doi.notnull()")
.query("pmcid.notnull()")
.groupby("preprint_doi")
.agg(
{
"author_type": "first",
"heading": "first",
"category": "first",
"document": "first",
"preprint_doi": "last",
"published_doi": "last",
"pmcid": "last",
}
)
.reset_index(drop=True)
)
mapped_doi_df.tail()
print(f"Total # of Preprints Mapped: {mapped_doi_df.shape[0]}")
print(f"Total % of Mapped: {mapped_doi_df.shape[0]/71118}")
preprint_count = aggregate_word_counts(
[
Path(f"output/biorxiv_word_counts/{Path(file)}.tsv")
for file in mapped_doi_df.document.values.tolist()
if Path(f"output/biorxiv_word_counts/{Path(file)}.tsv").exists()
]
)
preprint_count_df = (
pd.DataFrame.from_records(
[
{
"lemma": token[0],
"pos_tag": token[1],
"dep_tag": token[2],
"count": preprint_count[token],
}
for token in preprint_count
]
)
.query(f"lemma not in {stop_word_list}")
.groupby("lemma")
.agg({"count": "sum"})
.reset_index()
.sort_values("count", ascending=False)
)
preprint_count_df.head()
published_count = aggregate_word_counts(
[
Path(f"../../pmc/pmc_corpus/pmc_word_counts/{file}.tsv")
for file in mapped_doi_df.pmcid.values.tolist()
if Path(f"../../pmc/pmc_corpus/pmc_word_counts/{file}.tsv").exists()
]
)
published_count_df = (
pd.DataFrame.from_records(
[
{
"lemma": token[0],
"pos_tag": token[1],
"dep_tag": token[2],
"count": published_count[token],
}
for token in published_count
]
)
.query(f"lemma not in {stop_word_list}")
.groupby("lemma")
.agg({"count": "sum"})
.reset_index()
.sort_values("count", ascending=False)
)
published_count_df.head()
preprint_vs_published = get_term_statistics(preprint_count_df, published_count_df, 100)
preprint_vs_published.to_csv(
"output/comparison_stats/preprint_to_published_comparison.tsv",
sep="\t",
index=False,
)
preprint_vs_published
```
Main takeaways from this analysis:
1. On a global scale bioRxiv contains more field specific articles as top words consist of: neuron, gene, genome, network
2. "Patients" appear more correlated with PMC as most preprints involving patients are shipped over to medRxiv.
3. Many words associated with PMC are health related which ties back to the medRxiv note.
4. Citation styles change as preprints transition to published versions. Et Al. has a greater association within bioRxiv compared to PMC.
5. On a local scale published articles contain more statistical concepts (e.g., t-test) as well as quantitative measures (e.g. degree signs). (High associated lemmas are t, -, degree sign etc.)
6. Publish articles have a focus shift on mentioning figures, adding supplementary data etc compared to preprints.
7. Preprints have a universal way of citing published works by using the et al. citation. Hard to pinpoint if leading factor is because of peer review or journal style, but it will be an interesting point to discuss in the paper.
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
from collections import defaultdict, Counter
import csv
import itertools
from pathlib import Path
import numpy as np
import pandas as pd
import pickle
import spacy
from scipy.stats import chi2_contingency
from tqdm import tqdm_notebook
from annorxiver_modules.corpora_comparison_helper import (
aggregate_word_counts,
dump_to_dataframe,
get_term_statistics,
KL_divergence,
)
biorxiv_count_path = Path("output/total_word_counts/biorxiv_total_count.tsv")
pmc_count_path = Path("output/total_word_counts/pmc_total_count.tsv")
nytac_count_path = Path("output/total_word_counts/nytac_total_count.tsv")
if not biorxiv_count_path.exists():
biorxiv_corpus_count = aggregate_word_counts(
list(Path("output/biorxiv_word_counts").rglob("*tsv"))
)
dump_to_dataframe(biorxiv_corpus_count, "output/biorxiv_total_count.tsv")
biorxiv_corpus_count.most_common(10)
if not pmc_count_path.exists():
pmc_corpus_count = aggregate_word_counts(
list(Path("../../pmc/pmc_corpus/pmc_word_counts").rglob("*tsv"))
)
dump_to_dataframe(pmc_corpus_count, "output/pmc_total_count.tsv")
pmc_corpus_count.most_common(10)
if not nytac_count_path.exists():
nytac_corpus_count = aggregate_word_counts(
list(Path("../../nytac/corpora_stats/output").rglob("*tsv"))
)
dump_to_dataframe(nytac_corpus_count, "output/nytac_total_count.tsv")
nytac_corpus_count.most_common(10)
biorxiv_total_count_df = pd.read_csv(biorxiv_count_path.resolve(), sep="\t")
pmc_total_count_df = pd.read_csv(pmc_count_path.resolve(), sep="\t")
nytac_total_count_df = pd.read_csv(nytac_count_path.resolve(), sep="\t")
biorxiv_sentence_length = pickle.load(open("output/biorxiv_sentence_length.pkl", "rb"))
pmc_sentence_length = pickle.load(
open("../../pmc/pmc_corpus/pmc_sentence_length.pkl", "rb")
)
nytac_sentence_length = pickle.load(
open("../../nytac/corpora_stats/nytac_sentence_length.pkl", "rb")
)
spacy_nlp = spacy.load("en_core_web_sm")
stop_word_list = list(spacy_nlp.Defaults.stop_words)
biorxiv_sentence_len_list = list(biorxiv_sentence_length.items())
biorxiv_data = {
"document_count": len(biorxiv_sentence_length),
"sentence_count": sum(map(lambda x: len(x[1]), biorxiv_sentence_len_list)),
"token_count": biorxiv_total_count_df["count"].sum(),
"stop_word_count": (
biorxiv_total_count_df.query(f"lemma in {stop_word_list}")["count"].sum()
),
"avg_document_length": np.mean(
list(map(lambda x: len(x[1]), biorxiv_sentence_len_list))
),
"avg_sentence_length": np.mean(
list(itertools.chain(*list(map(lambda x: x[1], biorxiv_sentence_len_list))))
),
"negatives": (biorxiv_total_count_df.query(f"dep_tag =='neg'")["count"].sum()),
"coordinating_conjunctions": (
biorxiv_total_count_df.query(f"dep_tag =='cc'")["count"].sum()
),
"coordinating_conjunctions%": (
biorxiv_total_count_df.query(f"dep_tag =='cc'")["count"].sum()
)
/ biorxiv_total_count_df["count"].sum(),
"pronouns": (biorxiv_total_count_df.query(f"pos_tag =='PRON'")["count"].sum()),
"pronouns%": (biorxiv_total_count_df.query(f"pos_tag =='PRON'")["count"].sum())
/ biorxiv_total_count_df["count"].sum(),
"passives": (
biorxiv_total_count_df.query(
f"dep_tag in ['auxpass', 'nsubjpass', 'csubjpass']"
)["count"].sum()
),
"passive%": (
biorxiv_total_count_df.query(
f"dep_tag in ['auxpass', 'nsubjpass', 'csubjpass']"
)["count"].sum()
)
/ biorxiv_total_count_df["count"].sum(),
}
pmc_sentence_len_list = list(pmc_sentence_length.items())
pmc_data = {
"document_count": len(pmc_sentence_length),
"sentence_count": sum(map(lambda x: len(x[1]), pmc_sentence_len_list)),
"token_count": pmc_total_count_df["count"].sum(),
"stop_word_count": (
pmc_total_count_df.query(f"lemma in {stop_word_list}")["count"].sum()
),
"avg_document_length": np.mean(
list(map(lambda x: len(x[1]), pmc_sentence_len_list))
),
"avg_sentence_length": np.mean(
list(itertools.chain(*list(map(lambda x: x[1], pmc_sentence_len_list))))
),
"negatives": (pmc_total_count_df.query(f"dep_tag =='neg'")["count"].sum()),
"coordinating_conjunctions": (
pmc_total_count_df.query(f"dep_tag =='cc'")["count"].sum()
),
"coordinating_conjunctions%": (
pmc_total_count_df.query(f"dep_tag =='cc'")["count"].sum()
)
/ pmc_total_count_df["count"].sum(),
"pronouns": (pmc_total_count_df.query(f"pos_tag =='PRON'")["count"].sum()),
"pronouns%": (pmc_total_count_df.query(f"pos_tag =='PRON'")["count"].sum())
/ pmc_total_count_df["count"].sum(),
"passives": (
pmc_total_count_df.query(f"dep_tag in ['auxpass', 'nsubjpass', 'csubjpass']")[
"count"
].sum()
),
"passive%": (
pmc_total_count_df.query(f"dep_tag in ['auxpass', 'nsubjpass', 'csubjpass']")[
"count"
].sum()
)
/ pmc_total_count_df["count"].sum(),
}
nytac_sentence_len_list = list(nytac_sentence_length.items())
nytac_data = {
"document_count": len(nytac_sentence_length),
"sentence_count": sum(map(lambda x: len(x[1]), nytac_sentence_len_list)),
"token_count": nytac_total_count_df["count"].sum(),
"stop_word_count": (
nytac_total_count_df.query(f"lemma in {stop_word_list}")["count"].sum()
),
"avg_document_length": np.mean(
list(map(lambda x: len(x[1]), nytac_sentence_len_list))
),
"avg_sentence_length": np.mean(
list(itertools.chain(*list(map(lambda x: x[1], nytac_sentence_len_list))))
),
"negatives": (nytac_total_count_df.query(f"dep_tag =='neg'")["count"].sum()),
"coordinating_conjunctions": (
nytac_total_count_df.query(f"dep_tag =='cc'")["count"].sum()
),
"coordinating_conjunctions%": (
nytac_total_count_df.query(f"dep_tag =='cc'")["count"].sum()
)
/ nytac_total_count_df["count"].sum(),
"pronouns": (nytac_total_count_df.query(f"pos_tag =='PRON'")["count"].sum()),
"pronouns%": (nytac_total_count_df.query(f"pos_tag =='PRON'")["count"].sum())
/ nytac_total_count_df["count"].sum(),
"passives": (
nytac_total_count_df.query(f"dep_tag in ['auxpass', 'nsubjpass', 'csubjpass']")[
"count"
].sum()
),
"passive%": (
nytac_total_count_df.query(f"dep_tag in ['auxpass', 'nsubjpass', 'csubjpass']")[
"count"
].sum()
)
/ nytac_total_count_df["count"].sum(),
}
# This dataframe contains document statistics for each Corpus
# document count - the number of documents within the corpus
# Sentence count - the number of sentences within the corpus
# Token count - the number of tokens within the corpus
# Stop word counts - the number of stop words within the corpus
# Average document length - the average number of sentences within a document for a given corpus
# Average sentence length - the average number of words within a sentence for a given corpus
# Negatives - the number of negations (e.g. placing not in within a sentence) within a given corpus
# Coordinating Conjunctions - the number of coordinating conjunctions (and, but, for etc.) within a given corpus
# Pronouns - the number of pronouns within a given corpus
# Passive - the number of passive words within a given corpus
token_stats_df = pd.DataFrame.from_records(
[biorxiv_data, pmc_data, nytac_data], index=["bioRxiv", "PMC", "NYTAC"]
).T
token_stats_df.to_csv("output/figures/corpora_token_stats.tsv", sep="\t")
token_stats_df
biorxiv_total_count_df = (
biorxiv_total_count_df.query(f"lemma not in {stop_word_list}")
.groupby("lemma")
.agg({"count": "sum"})
.reset_index()
.sort_values("count", ascending=False)
)
biorxiv_total_count_df
pmc_total_count_df = (
pmc_total_count_df.query(f"lemma not in {stop_word_list}")
.groupby("lemma")
.agg({"count": "sum"})
.reset_index()
.sort_values("count", ascending=False)
.iloc[2:]
)
pmc_total_count_df
nytac_total_count_df = (
nytac_total_count_df.query(f"lemma not in {stop_word_list}")
.groupby("lemma")
.agg({"count": "sum"})
.reset_index()
.sort_values("count", ascending=False)
)
nytac_total_count_df
biorxiv_vs_pmc = get_term_statistics(biorxiv_total_count_df, pmc_total_count_df, 100)
biorxiv_vs_pmc.to_csv(
"output/comparison_stats/biorxiv_vs_pmc_comparison.tsv", sep="\t", index=False
)
biorxiv_vs_pmc
biorxiv_vs_nytac = get_term_statistics(
biorxiv_total_count_df, nytac_total_count_df, 100
)
biorxiv_vs_nytac.to_csv(
"output/comparison_stats/biorxiv_nytac_comparison.tsv", sep="\t", index=False
)
biorxiv_vs_nytac
pmc_vs_nytac = get_term_statistics(pmc_total_count_df, nytac_total_count_df, 100)
pmc_vs_nytac.to_csv(
"output/comparison_stats/pmc_nytac_comparison.tsv", sep="\t", index=False
)
pmc_vs_nytac
term_grid = [100, 200, 300, 400, 500, 1000, 1500, 2000, 3000, 5000]
kl_data = []
for num_terms in tqdm_notebook(term_grid):
kl_data.append(
{
"num_terms": num_terms,
"KL_divergence": KL_divergence(
biorxiv_total_count_df, pmc_total_count_df, num_terms=num_terms
),
"comparison": "biorxiv_vs_pmc",
}
)
kl_data.append(
{
"num_terms": num_terms,
"KL_divergence": KL_divergence(
biorxiv_total_count_df, nytac_total_count_df, num_terms=num_terms
),
"comparison": "biorxiv_vs_nytac",
}
)
kl_data.append(
{
"num_terms": num_terms,
"KL_divergence": KL_divergence(
pmc_total_count_df, nytac_total_count_df, num_terms=num_terms
),
"comparison": "pmc_vs_nytac",
}
)
kl_metrics = pd.DataFrame.from_records(kl_data)
kl_metrics.to_csv(
"output/comparison_stats/corpora_kl_divergence.tsv", sep="\t", index=False
)
kl_metrics
mapped_doi_df = (
pd.read_csv("../journal_tracker/output/mapped_published_doi.tsv", sep="\t")
.query("published_doi.notnull()")
.query("pmcid.notnull()")
.groupby("preprint_doi")
.agg(
{
"author_type": "first",
"heading": "first",
"category": "first",
"document": "first",
"preprint_doi": "last",
"published_doi": "last",
"pmcid": "last",
}
)
.reset_index(drop=True)
)
mapped_doi_df.tail()
print(f"Total # of Preprints Mapped: {mapped_doi_df.shape[0]}")
print(f"Total % of Mapped: {mapped_doi_df.shape[0]/71118}")
preprint_count = aggregate_word_counts(
[
Path(f"output/biorxiv_word_counts/{Path(file)}.tsv")
for file in mapped_doi_df.document.values.tolist()
if Path(f"output/biorxiv_word_counts/{Path(file)}.tsv").exists()
]
)
preprint_count_df = (
pd.DataFrame.from_records(
[
{
"lemma": token[0],
"pos_tag": token[1],
"dep_tag": token[2],
"count": preprint_count[token],
}
for token in preprint_count
]
)
.query(f"lemma not in {stop_word_list}")
.groupby("lemma")
.agg({"count": "sum"})
.reset_index()
.sort_values("count", ascending=False)
)
preprint_count_df.head()
published_count = aggregate_word_counts(
[
Path(f"../../pmc/pmc_corpus/pmc_word_counts/{file}.tsv")
for file in mapped_doi_df.pmcid.values.tolist()
if Path(f"../../pmc/pmc_corpus/pmc_word_counts/{file}.tsv").exists()
]
)
published_count_df = (
pd.DataFrame.from_records(
[
{
"lemma": token[0],
"pos_tag": token[1],
"dep_tag": token[2],
"count": published_count[token],
}
for token in published_count
]
)
.query(f"lemma not in {stop_word_list}")
.groupby("lemma")
.agg({"count": "sum"})
.reset_index()
.sort_values("count", ascending=False)
)
published_count_df.head()
preprint_vs_published = get_term_statistics(preprint_count_df, published_count_df, 100)
preprint_vs_published.to_csv(
"output/comparison_stats/preprint_to_published_comparison.tsv",
sep="\t",
index=False,
)
preprint_vs_published
| 0.275422 | 0.514095 |
# CS 109A/STAT 121A/AC 209A/CSCI E-109A: Homework 4
# Regularization, High Dimensionality, PCA
**Harvard University**<br/>
**Fall 2017**<br/>
**Instructors**: Pavlos Protopapas, Kevin Rader, Rahul Dave, Margo Levine
---
### INSTRUCTIONS
- To submit your assignment follow the instructions given in canvas.
- Restart the kernel and run the whole notebook again before you submit.
- Do not include your name(s) in the notebook even if you are submitting as a group.
- If you submit individually and you have worked with someone, please include the name of your [one] partner below.
---
Contributors: Ryan Janssen, Filip Michalsky
Enrollment Status (109A, 121A, 209A, or E109A): 209A
Import libraries:
```
import numpy as np
import pandas as pd
import matplotlib
import matplotlib.pyplot as plt
from sklearn.metrics import r2_score
import statsmodels.api as sm
from statsmodels.api import OLS
from sklearn.preprocessing import PolynomialFeatures
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Ridge
from sklearn.linear_model import Lasso
from sklearn.linear_model import RidgeCV
from sklearn.linear_model import LassoCV
from sklearn.decomposition import PCA
from scipy.stats import zscore
%matplotlib inline
# In this problem set, we are often iterating with small Lambda values that cause many warnings to appear.
# So, we'll turn off warnings for this set:
import warnings
warnings.filterwarnings('ignore')
```
# Continuing Bike Sharing Usage Data
In this homework, we will focus on multiple linear regression, regularization, dealing with high dimensionality, and PCA. We will continue to build regression models for the Capital Bikeshare program in Washington D.C. See Homework 3 for more information about the data.
Data Source: https://www.capitalbikeshare.com/system-data
*Note: please make sure you use all the processed data from HW 3 Part (a)...you make want to save the data set on your computer and reread the csv/json file here.
```
# Importing processed data from HW3:
test = pd.read_csv("df_test.csv", index_col=0)
train = pd.read_csv("df_train.csv", index_col=0)
x_train = train.drop('count', axis=1)
y_train = train['count']
x_test = test.drop('count', axis=1)
y_test = test['count']
```
## Part (f): Regularization/Penalization Methods
As an alternative to selecting a subset of predictors and fitting a regression model on the subset, one can fit a linear regression model on all predictors, but shrink or regularize the coefficient estimates to make sure that the model does not "overfit" the training set.
Use the following regularization techniques to fit linear models to the training set:
- Ridge regression
- Lasso regression
You may choose the shrikage parameter $\lambda$ from the set $\{10^{-5}, 10^{-4},...,10^{4},10^{5}\}$ using cross-validation. In each case,
- How do the estimated coefficients compare to or differ from the coefficients estimated by a plain linear regression (without shrikage penalty) in Part (b) fropm HW 3? Is there a difference between coefficients estimated by the two shrinkage methods? If so, give an explantion for the difference.
- List the predictors that are assigned a coefficient value close to 0 (say < 1e-10) by the two methods. How closely do these predictors match the redundant predictors (if any) identified in Part (c) from HW 3?
- Is there a difference in the way Ridge and Lasso regression assign coefficients to the predictors `temp` and `atemp`? If so, explain the reason for the difference.
We next analyze the performance of the two shrinkage methods for different training sample sizes:
- Generate random samples of sizes 100, 150, ..., 400 from the training set. You may use the following code to draw a random sample of a specified size from the training set:
```
#-------- sample
# A function to select a random sample of size k from the training set
# Input:
# x (n x d array of predictors in training data)
# y (n x 1 array of response variable vals in training data)
# k (size of sample)
# Return:
# chosen sample of predictors and responses
def sample(x, y, k):
n = x.shape[0] # No. of training points
# Choose random indices of size 'k'
subset_ind = np.random.choice(np.arange(n), k)
# Get predictors and reponses with the indices
x_subset = x[subset_ind, :]
y_subset = y[subset_ind]
return (x_subset, y_subset)
```
- Fit linear, Ridge and Lasso regression models to each of the generated sample. In each case, compute the $R^2$ score for the model on the training sample on which it was fitted, and on the test set.
- Repeat the above experiment for 10 random trials/splits, and compute the average train and test $R^2$ across the trials for each training sample size. Also, compute the standard deviation (SD) in each case.
- Make a plot of the mean training $R^2$ scores for the linear, Ridge and Lasso regression methods as a function of the training sample size. Also, show a confidence interval for the mean scores extending from **mean - SD** to **mean + SD**. Make a similar plot for the test $R^2$ scores.
How do the training and test $R^2$ scores compare for the three methods? Give an explanation for your observations. How do the confidence intervals for the estimated $R^2$ change with training sample size? Based on the plots, which of the three methods would you recommend when one needs to fit a regression model using a small training sample?
*Hint:* You may use `sklearn`'s `RidgeCV` and `LassoCV` classes to implement Ridge and Lasso regression. These classes automatically perform cross-validation to tune the parameter $\lambda$ from a given range of values. You may use the `plt.errorbar` function to plot confidence bars for the average $R^2$ scores.
```
# Fit ridge/lasso regressions to training set
alphas = np.logspace(-5,5, 11)
ridge = RidgeCV(alphas = alphas, fit_intercept=True, normalize=False)
ridge.fit(x_train, y_train)
lasso = LassoCV(alphas = alphas, fit_intercept=True, normalize=False)
lasso.fit(x_train, y_train)
# Get our linear regression parameters from HW3:
models_coefs = pd.read_csv("linear_params.csv",header = None, index_col=0).drop(['const'])
models_coefs.columns = ['Linear Regression']
models_coefs['Ridge Regression'] = ridge.coef_
models_coefs['Lasso Regression'] = lasso.coef_
# Compare parameters
print(models_coefs)
#alphas = np.logspace(-5,5, 11)
alphas = [10.0]
ridge = RidgeCV(alphas = alphas, fit_intercept=True, normalize=False)
lasso = LassoCV(alphas = alphas, fit_intercept=True, normalize=False)
linear = LinearRegression(fit_intercept=True)
ridge_subset_r2s, ridge_test_r2s, ridge_subset_stds, ridge_test_stds = [], [], [], []
lasso_subset_r2s, lasso_test_r2s, lasso_subset_stds, lasso_test_stds = [], [], [], []
linear_subset_r2s, linear_test_r2s, linear_subset_stds, linear_test_stds = [], [], [], []
trial_sizes = range(100, 450, 50)
for k in trial_sizes:
this_k_ridge_subset_r2s = []
this_k_ridge_test_r2s = []
this_k_lasso_subset_r2s = []
this_k_lasso_test_r2s = []
this_k_linear_subset_r2s = []
this_k_linear_test_r2s = []
for trial in range(10):
x_subset, y_subset = sample(x_train.values, y_train.values, k)
ridge.fit(x_subset, y_subset)
lasso.fit(x_subset, y_subset)
linear.fit(x_subset, y_subset)
this_k_ridge_subset_r2s.append(ridge.score(x_subset, y_subset))
this_k_ridge_test_r2s.append(ridge.score(x_test, y_test))
this_k_lasso_subset_r2s.append(lasso.score(x_subset, y_subset))
this_k_lasso_test_r2s.append(lasso.score(x_test, y_test))
this_k_linear_subset_r2s.append(linear.score(x_subset, y_subset))
this_k_linear_test_r2s.append(linear.score(x_test, y_test))
ridge_subset_r2s.append(np.array(this_k_ridge_subset_r2s).mean())
ridge_test_r2s.append(np.array(this_k_ridge_test_r2s).mean())
ridge_subset_stds.append(np.array(this_k_ridge_subset_r2s).std())
ridge_test_stds.append(np.array(this_k_ridge_test_r2s).std())
lasso_subset_r2s.append(np.array(this_k_lasso_subset_r2s).mean())
lasso_test_r2s.append(np.array(this_k_lasso_test_r2s).mean())
lasso_subset_stds.append(np.array(this_k_lasso_subset_r2s).std())
lasso_test_stds.append(np.array(this_k_lasso_test_r2s).std())
linear_subset_r2s.append(np.array(this_k_linear_subset_r2s).mean())
linear_test_r2s.append(np.array(this_k_linear_test_r2s).mean())
linear_subset_stds.append(np.array(this_k_linear_subset_r2s).std())
linear_test_stds.append(np.array(this_k_linear_test_r2s).std())
fig1 = plt.figure(figsize=(10,5))
fig1.tight_layout
ax1 = fig1.add_subplot(121)
ax1.plot(trial_sizes, ridge_subset_r2s, label = "ridge", color = 'purple')
ax1.plot(trial_sizes, lasso_subset_r2s, label = "lasso", color='green')
ax1.plot(trial_sizes, linear_subset_r2s, label = "linear", color = 'blue')
ax1.set_title("Sample size vs. model training R^2")
ax1.set_xlabel("trial sample size")
ax1.set_ylabel("train (subset) R^2")
ax1.legend()
ax2 = fig1.add_subplot(122)
ax2.plot(trial_sizes, ridge_test_r2s, label = "ridge", color='purple')
ax2.plot(trial_sizes, lasso_test_r2s, label = "lasso", color='green')
ax2.plot(trial_sizes, linear_test_r2s, label = "linear", color = 'blue')
ax2.set_title("Sample size vs. model test R^2")
ax2.set_xlabel("trial sample size")
ax2.set_ylabel("test R^2")
ax2.legend()
fig2 = plt.figure(figsize=(10,5))
fig2.tight_layout
ax3 = fig2.add_subplot(131)
ax4 = fig2.add_subplot(132)
ax5 = fig2.add_subplot(133)
ax3.errorbar(trial_sizes, ridge_subset_r2s, yerr=ridge_subset_stds, label = "ridge", color='purple')
ax4.errorbar(trial_sizes, lasso_subset_r2s, lasso_subset_stds, label = "lasso", color='green')
ax5.errorbar(trial_sizes, linear_subset_r2s, linear_subset_stds, label = "linear", color = 'blue')
ax3.set_title("Ridge regression - train")
ax3.set_xlabel("trial sample size")
ax3.set_ylabel("R^2 w/standard deviation")
ax3.set_ylim(0,0.8)
ax3.legend()
ax4.set_title("Lasso regression - train")
ax4.set_xlabel("trial sample size")
ax4.set_ylabel("R^2 w/standard deviation")
ax4.set_ylim(0,0.8)
ax4.legend()
ax5.set_title("Linear regression - train")
ax5.set_xlabel("trial sample size")
ax5.set_ylabel("R^2 w/standard deviation")
ax5.set_ylim(0,0.8)
ax5.legend()
fig3 = plt.figure(figsize=(10,5))
fig3.tight_layout
ax6 = fig3.add_subplot(131)
ax7 = fig3.add_subplot(132)
ax8 = fig3.add_subplot(133)
ax6.errorbar(trial_sizes, ridge_test_r2s, ridge_test_stds, label = "ridge", color='purple')
ax7.errorbar(trial_sizes, lasso_test_r2s, lasso_test_stds, label = "lasso", color='green')
ax8.errorbar(trial_sizes, linear_test_r2s, linear_test_stds, label = "linear", color = 'blue')
ax6.set_title("Ridge regression - test")
ax6.set_xlabel("trial sample size")
ax6.set_ylabel("R^2 w/standard deviation")
ax6.set_ylim(0,0.8)
ax6.legend()
ax7.set_title("Lasso regression - test")
ax7.set_xlabel("trial sample size")
ax7.set_ylabel("R^2 w/standard deviation")
ax7.set_ylim(0,0.8)
ax7.legend()
ax8.set_title("Linear regression - test")
ax8.set_xlabel("trial sample size")
ax8.set_ylabel("R^2 w/standard deviation")
ax8.set_ylim(0,0.8)
ax8.legend()
```
- How do the estimated coefficients compare to or differ from the coefficients estimated by a plain linear regression (without shrikage penalty) in Part (b) fropm HW 3? Is there a difference between coefficients estimated by the two shrinkage methods? If so, give an explantion for the difference.
** As we can see above, the larger coefficients from the linear regression are shrunk in both the Ridge and Lasso regressions. And the smaller coefficients from the linear regression are larger for the Ridge/Lasso.**
** The two shrinkage methods differ in that Lasso shrinks several coefficients to zero, whereas ridge doesn't shrink any coefficients to zero.**
- List the predictors that are assigned a coefficient value close to 0 (say < 1e-10) by the two methods. How closely do these predictors match the redundant predictors (if any) identified in Part (c) from HW 3?
** The Ridge coefficients never shrink to "close to zero" under these parameters. The Lasso coefficients [workingday, season_2.0, month_2.0, month_5.0, day_of_week_4.0, weather_2.0] all shrink to zero. **
** These were generally highly correlated coefficients from HW3 (c). For instance, workingday was correlated with all of the days of the week, and the respective months and seasons were correlated. **
- Is there a difference in the way Ridge and Lasso regression assign coefficients to the predictors `temp` and `atemp`? If so, explain the reason for the difference.
** For certain values of alpha, Lasso will assign temp a 0 coefficient, while Ridge will never do this. This is because Lasso is built to zero out certain redundant coefficients and there is a high degree of correlation between temp and atemp. **
** Interestingly, though, for most alphas (including the selected alpha above), even the Lasso regression will keep both temp and atemp. We believe this is because temp/atemp is some of the stronger predictors of count. **
- How do the training and test R2R2 scores compare for the three methods? Give an explanation for your observations. How do the confidence intervals for the estimated R2R2 change with training sample size? Based on the plots, which of the three methods would you recommend when one needs to fit a regression model using a small training sample?
** The training R^2 is generally decreasing with sample size for all 3 regressions, and the test R^2 is generally increasing for all 3 regressions. This is not surprising, since a larger training sample will help us to reduce overfitting. **
** Furthermore, the ridge regression generally performs the best on the test set, and the worst on the training set. We believe this is because the ridge model is a good balance between removing the higher-order predictors while still not "zeroing" out information as the lasso regression does. **
** Confidence intervals tend to generally decrease with increasing trial size (aka the train set size). Additionally, the Ridge regression seems to have the tightest confidence intervals. **
** For small sample sizes, ridge gains a bigger "advantage" on the test R^2 and a bigger "disadvantage" on the training R^2. Because our priority is maximizing out-of-sample predictive power, the best model for low sample sizes is using ridge regression. We believe this happens because ridge regression is effective for reducing collinearity. In smaller sample sizes, it's more difficult to separate the collinearity from the features, so ridge regression is useful here. **
## Part (g): Polynomial & Interaction Terms
Moving beyond linear models, we will now try to improve the performance of the regression model in Part (b) from HW 3 by including higher-order polynomial and interaction terms.
- For each continuous predictor $X_j$, include additional polynomial terms $X^2_j$, $X^3_j$, and $X^4_j$, and fit a multiple regression model to the expanded training set. How does the $R^2$ of this model on the test set compare with that of the linear model fitted in Part (b) from HW 3? Using a t-test, find out which of estimated coefficients for the polynomial terms are statistically significant at a significance level of 5%.
- Fit a multiple linear regression model with additional interaction terms $\mathbb{I}_{month = 12} \times temp$ and $\mathbb{I}_{workingday = 1} \times \mathbb{I}_{weathersit = 1}$ and report the test $R^2$ for the fitted model. How does this compare with the $R^2$ obtained using linear model in Part (b) from HW 3? Are the estimated coefficients for the interaction terms statistically significant at a significance level of 5%?
```
def add_poly_terms(x, selected_features, deg = 4):
"""
Adds polynomial terms of order deg, into ONLY selected_features.
Note that it does not include any interaction terms between the predictors as requested above.
"""
poly_fit = PolynomialFeatures(degree = deg, include_bias=False)
x_selected = x.copy()[selected_features]
x_out = x.drop(selected_features, axis = 1)
# Add each higher order term (doing these individually so we don't get interaction terms)
for i,feature in enumerate(selected_features):
for this_deg in range(deg):
poly_features = poly_fit.fit_transform(x_selected[feature].values.reshape(-1,1))
x_out[feature+"_deg_"+str(this_deg+1)] = poly_features[:,this_deg]
return x_out
continuous_predictors = ['temp', 'atemp', 'humidity', 'windspeed']
x_train_poly = add_poly_terms(x_train, continuous_predictors, deg = 4)
x_test_poly = add_poly_terms(x_test, continuous_predictors, deg = 4)
poly_ols = sm.OLS(y_train, sm.add_constant(x_train_poly))
poly_ols_fit = poly_ols.fit()
poly_ols_r2 = r2_score(y_test, poly_ols_fit.predict(sm.add_constant(x_test_poly)))
print("Polynomial regression train R^2 =", r2_score(y_train, poly_ols_fit.predict(sm.add_constant(x_train_poly))),"\n")
print("Polynomial regression test R^2 =", r2_score(y_test, poly_ols_fit.predict(sm.add_constant(x_test_poly))),"\n")
ttest=poly_ols_fit.pvalues
print("Model parameters of significance level > 5%:\n", ttest[ttest.values<0.05])
```
- For each continuous predictor $X_j$, include additional polynomial terms $X^2_j$, $X^3_j$, and $X^4_j$, and fit a multiple regression model to the expanded training set. How does the $R^2$ of this model on the test set compare with that of the linear model fitted in Part (b) from HW 3? Using a t-test, find out which of estimated coefficients for the polynomial terms are statistically significant at a significance level of 5%.
** The R^2 when polynomial terms is added increases to 0.29. (In HW3, the linear model, forward selection, and reverse selection had R^2 = 0.249, 0.274, and 0.273, respectively. **
** Using the t-test, the statistically significant polynomial features are:
** This adds a significant quadratic term for for temperature squared as expected. We also have significant linear terms for humidity and windspeed in the feature set. **
** This is as expected - for example, in HW3 we hypothesized there is a slightly quadratic reliationship with temperature. When it gets too hot people stop riding bikes. **
- Fit a multiple linear regression model with additional interaction terms $\mathbb{I}_{month = 12} \times temp$ and $\mathbb{I}_{workingday = 1} \times \mathbb{I}_{weathersit = 1}$ and report the test $R^2$ for the fitted model. How does this compare with the $R^2$ obtained using linear model in Part (b) from HW 3? Are the estimated coefficients for the interaction terms statistically significant at a significance level of 5%?
```
def copy_and_add_inter(x_train):
x_train_inter = x_train.copy(deep=True)
x_train_inter['month_12.0'] = 1 - x_train_inter['month_1.0'] - x_train_inter['month_2.0'] - x_train_inter['month_3.0']- x_train_inter['month_4.0'] - x_train_inter['month_5.0'] - x_train_inter['month_6.0'] - x_train_inter['month_7.0'] - x_train_inter['month_8.0']- x_train_inter['month_9.0']- x_train_inter['month_10.0']- x_train_inter['month_11.0']
x_train_inter['month12-temp'] = x_train_inter['month_12.0'] * x_train_inter['temp']
x_train_inter['wday1-weather1'] = x_train_inter['workingday'] * x_train_inter['weather_1.0']
return x_train_inter
#x_train.head(30)
x_train_inter = copy_and_add_inter(x_train)
x_test_inter = copy_and_add_inter(x_test)
inter_ols = sm.OLS(y_train, sm.add_constant(x_train_inter))
inter_ols_fit = inter_ols.fit()
inter_ols_r2_train = r2_score(y_train, inter_ols_fit.predict(sm.add_constant(x_train_inter)))
inter_ols_r2_test = r2_score(y_test, inter_ols_fit.predict(sm.add_constant(x_test_inter)))
print("Multilinear regression R^2 train set =", inter_ols_r2_train,"\n")
print("Multilinear regression R^2 test set =", inter_ols_r2_test,"\n")
ttest1=inter_ols_fit.pvalues
print("Model parameters of significance level > 5%:\n", ttest1[ttest1.values<0.05])
```
How does this compare with the R2
obtained using linear model in Part (b) from HW 3? Are the estimated coefficients for the interaction terms statistically significant at a significance level of 5%?
**The R2 on the test set (~28.1%) is higher than ~24.9% test R2 obtained in HW3. The interaction terms are not significant at a 5% significance level.**
## Part (h): PCA to deal with high dimensionality
We would like to fit a model to include all main effects, polynomial terms up to the $4^{th}$ order, and all interactions between all possible predictors and polynomial terms (not including the interactions between $X^1_j$, $X^2_j$, $X^3_j$, and $X^4_j$ as they would just create higher order polynomial terms).
- Create an expanded training set including all the desired terms mentioned above. What are the dimensions of this 'design matrix' of all the predictor variables? What are the issues with attempting to fit a regression model using all of these predictors?
- Instead of using the usual approaches for model selection, let's instead use principal components analysis (PCA) to fit the model. First, create the principal component vectors in python (consider: should you normalize first?). Then fit 5 different regression models: (1) using just the first PCA vector, (2) using the first two PCA vectors, (3) using the first three PCA vectors, etc... Briefly summarize how these models compare in the training set.
- Use the test set to decide which of the 5 models above is best to predict out of sample. How does this model compare to the previous models you've fit? What are the interpretations of this model's coefficients?
```
# First, let's generate a polynomial with all of the interation terms (this way, we don't get the higher-order interaction terms)
interaction_poly = PolynomialFeatures(degree = 2, interaction_only = True)
x_train_interaction = pd.DataFrame(interaction_poly.fit_transform(x_train), columns = interaction_poly.get_feature_names(x_train.columns))
x_test_interaction = pd.DataFrame(interaction_poly.fit_transform(x_test), columns = interaction_poly.get_feature_names(x_test.columns))
# Then, add in the polynomial terms used earlier
continuous_predictors = ['temp', 'atemp', 'humidity', 'windspeed']
x_big_train = add_poly_terms(x_train_interaction, continuous_predictors, deg = 4)
x_big_test = add_poly_terms(x_test_interaction, continuous_predictors, deg = 4)
print("Full expanded data set has",x_big_train.shape[1], "columns!")
#print(y_train.shape, sm.add_constant(x_big_train).shape)
big_ols = sm.OLS(y_train, sm.add_constant(x_big_train))
big_ols_fit = big_ols.fit()
big_ols_r2 = r2_score(y_train, big_ols_fit.predict(sm.add_constant(x_big_train)))
print("Our full feature training R^2 =",big_ols_r2)
big_ols_r2 = r2_score(y_test, big_ols_fit.predict(sm.add_constant(x_big_test)))
print("Our full feature test R^2 =",big_ols_r2,"\n")
# Fit PCA
num_comp = 5
pca = PCA(num_comp)
pca.fit(x_big_train)
x_train_pca = pca.transform(x_big_train)
x_test_pca = pca.transform(x_big_test)
print("Generating PCA with",num_comp, "components.\nExplained variance ratio:", pca.explained_variance_ratio_, "\n")
# Fit linear regression over sums of the first n components
lin_reg_model = LinearRegression(fit_intercept=True)
for n in range(1,5+1):
lin_reg_model.fit(x_train_pca[:,:n], y_train)
print("Training R^2 of first",n,"PCA components=", r2_score(y_train, lin_reg_model.predict(x_train_pca[:,:n])))
print("Test R^2 of first",n,"PCA components=", r2_score(y_test, lin_reg_model.predict(x_test_pca[:,:n])))
```
- Create an expanded training set including all the desired terms mentioned above. What are the dimensions of this 'design matrix' of all the predictor variables? What are the issues with attempting to fit a regression model using all of these predictors?
** This design matrix has a total of 452 predictors (and 330 data points). This is unfeasible for several reasons: **
** First, the model overfits very badly. The training R^2 is 0.91, but the test set R^2 is less than 0! In fact, we have more features than we do datapoints! **
** Additionally, it's very computationall expensive, in both time and space, to regress and manipulate this many features. **
- Instead of using the usual approaches for model selection, let's instead use principal components analysis (PCA) to fit the model. First, create the principal component vectors in python (consider: should you normalize first?). Then fit 5 different regression models: (1) using just the first PCA vector, (2) using the first two PCA vectors, (3) using the first three PCA vectors, etc... Briefly summarize how these models compare in the training set.
** The models perform quite poorly in the training set ($R^{2}<0.3$) for the first 3 components. The forth component adds a significant amount of predictive power, and the fifth term adds very little. We also see that by the fourth component, c.95% of the variance has been explained.**
** It's unfortunately difficult for us symbolize the meaning of each of the components (we just know they are orthogonal representations of the previous predictors. But it seems to us that a fair trade-off between computational power and model performance is to choose the first 4 components. **
- Use the test set to decide which of the 5 models above is best to predict out of sample. How does this model compare to the previous models you've fit? What are the interpretations of this model's coefficients?
** The training $R^{2}$ is highly non-linear versus the components added. The model has essentially no out-of-sample predictive power until the fourth principal component, when it becomes 0.22. This reinforces our hypothesis that at least the first four principal components are necessary. However, our model that uses fewer features (where PCA is not required) still performs better, so we would try to use a non-PCA-based solution with fewer interaction terms (if possible). **
** In this case, each of the coefficients are the weights of each respective principal component, which itself is an orthogonal representation of the previous features. **
## Part (i): Beyond Squared Error
We have seen in class that the multiple linear regression method optimizes the Mean Squared Error (MSE) on the training set. Consider the following alternate evaluation metric, referred to as the Root Mean Squared Logarthmic Error (RMSLE):
$$
\sqrt{\frac{1}{n}\sum_{i=1}^n (log(y_i+1) - log(\hat{y}_i+1))^2}.
$$
The *lower* the RMSLE the *better* is the performance of a model. The RMSLE penalizes errors on smaller responses more heavily than errors on larger responses. For example, the RMSLE penalizes a prediction of $\hat{y} = 15$ for a true response of $y=10$ more heavily than a prediction of $\hat{y} = 105$ for a true response of $100$, though the difference in predicted and true responses are the same in both cases.
This is a natural evaluation metric for bike share demand prediction, as in this application, it is more important that the prediction model is accurate on days where the demand is low (so that the few customers who arrive are served satisfactorily), compared to days on which the demand is high (when it is less damaging to lose out on some customers).
The following code computes the RMSLE for you:
```
#-------- rmsle
# A function for evaluating Root Mean Squared Logarithmic Error (RMSLE)
# of the linear regression model on a data set
# Input:
# y_test (n x 1 array of response variable vals in testing data)
# y_pred (n x 1 array of response variable vals in testing data)
# Return:
# RMSLE (float)
def rmsle(y, y_pred):
# Evaluate sqaured error, against target labels
# rmsle = \sqrt(1/n \sum_i (log (y[i]+1) - log (y_pred[i]+1))^2)
rmsle_ = np.sqrt(np.mean(np.square(np.log(y+1) - np.log(y_pred+1))))
return rmsle_
```
Use the above code to compute the training and test RMSLE for the polynomial regression model you fit in Part (g).
You are required to develop a strategy to fit a regression model by optimizing the RMSLE on the training set. Give a justification for your proposed approach. Does the model fitted using your approach yield lower train RMSLE than the model in Part (g)? How about the test RMSLE of the new model?
**Note:** We do not require you to implement a new regression solver for RMSLE. Instead, we ask you to think about ways to use existing built-in functions to fit a model that performs well on RMSLE. Your regression model may use the same polynomial terms used in Part (g).
```
lin_reg_model = LinearRegression(fit_intercept=True)
# Calculate train RMSLE on current model fit
lin_reg_model.fit(x_train_poly, y_train)
y_hat_train = lin_reg_model.predict(x_train_poly)
y_hat_test = lin_reg_model.predict(x_test_poly)
print("Train RMSLE on linear model fit =", rmsle(y_train, y_hat_train))
print("Test RMSLE on linear model fit =", rmsle(y_test, y_hat_test))
print("")
# Now, let's fit a new set on a revised response variable - specifically, log(y+1)
lin_reg_model.fit(x_train_poly, np.log1p(y_train))
y_hat_logit_train = np.expm1(lin_reg_model.predict(x_train_poly))
y_hat_logit_test = np.expm1(lin_reg_model.predict(x_test_poly))
print("Train RMSLE on RMSLE model fit =", rmsle(y_train, y_hat_logit_train))
print("Test RMSLE on RMSLE model fit =", rmsle(y_test, y_hat_logit_test))
```
Give a justification for your proposed approach. Does the model fitted using your approach yield lower train RMSLE than the model in Part (g)? How about the test RMSLE of the new model?
* ** We chose to train a linear model on a new y domain, denoted by $y_{new} = log(y + 1)$. By applying this domain transformation, our linear model will now minimize MSE in the $y_{new}$ domain, and therefore minimize RMSLE in the original $y$ domain. **
* ** As expected, this improves RMSLE in our training set. Training RMSLE improves from 0.311 to 0.291. Our test RMSLE actually results in higher RMSLE, but this does not invalidate our RMSLE fit. It just means that the model may be now overfitting for RMSLE (while it was overfitting for MSE previously). **
## Part (j): Dealing with Erroneous Labels
Due to occasional system crashes, some of the bike counts reported in the data set have been recorded manually. These counts are not very unreliable and are prone to errors. It is known that roughly 5% of the labels in the training set are erroneous (i.e. can be arbitrarily different from the true counts), while all the labels in the test set were confirmed to be accurate. Unfortunately, the identities of the erroneous records in the training set are not available. Can this information about presence of 5% errors in the training set labels (without details about the specific identities of the erroneous rows) be used to improve the performance of the model in Part (g)? Note that we are interested in improving the $R^2$ performance of the model on the test set (not the training $R^2$ score).
As a final task, we require you to come up with a strategy to fit a regression model, taking into account the errors in the training set labels. Explain the intuition behind your approach (we do not expect a detailed mathematical justification). Use your approach to fit a regression model on the training set, and compare its test $R^2$ with the model in Part (g).
**Note:** Again, we do not require you to implement a new regression solver for handling erroneous labels. It is sufficient that you to come up with an approach that uses existing built-in functions. Your regression model may use the same polynomial terms used in Part (g).
```
ridge1 = RidgeCV(alphas = alphas, fit_intercept=True, normalize=False)
ridge1.fit(x_train, y_train)
R2_init=r2_score(y_test, ridge1.predict(x_test))
print("Test R^2 from Part g)",r2_score(y_test, poly_ols_fit.predict(sm.add_constant(x_test_poly))))
print("Test R^2 from Ridge model with all train points",R2_init)
x_train_trim = x_train_poly.copy(deep = True)
y_train_trim = y_train.copy(deep = True)
#print(y_train_trim.shape)
Ridge_Test_R2s_after_drop = []
stop_constant = float("inf")
for i in range(int(len(y_train_trim)*0.05)):
ridge_reg_model = RidgeCV(alphas = alphas, fit_intercept=True, normalize=False)
ridge_reg_model.fit(x_train_trim, y_train_trim)
y_prec_train = ridge_reg_model.predict(x_train_trim)
y_prec_test = ridge_reg_model.predict(x_test_poly)
#print("Train R-score is", r2_score(y_train_trim,y_prec_train))
#print("Test R-score is",r2_score(y_test,y_prec_test))
#index of the worst value
y_worst = np.argmax(np.abs(y_train_trim - y_prec_train))
#worst value
worst_error =np.max(np.abs(y_train_trim - y_prec_train))
#print("Dropping the worst y", worst_error, "at the index", y_train_trim.index[y_worst],"\n")
if worst_error < 2600:
print("BREAK")
break
x_train_trim = x_train_trim.drop(x_train_trim.index[y_worst])
y_train_trim = y_train_trim.drop(y_train_trim.index[y_worst])
Ridge_Test_R2s_after_drop.append(r2_score(y_test,y_prec_test))
print("R^2 after removing ~5% of data points", Ridge_Test_R2s_after_drop[-1])
```
We use the Ridge regression to fit our training data and we iteratively remove the ~5% of the data points with the highest residuals when predicting on our train data. We see that our test R^2 from Ridge goes up from 27.4% to 30.2% after removing these points and this is higher than test R^2 of 29.4% from Part g).
We chose this method because we have no a priori information about how the error in the 5% of "bad" counts is skewed (ie, we don't know if it's too high, too low, completely random, etc). So in our opinion, the best remaining assumption that the outliers with the higher residual error are the most likely to be erroneous.
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib
import matplotlib.pyplot as plt
from sklearn.metrics import r2_score
import statsmodels.api as sm
from statsmodels.api import OLS
from sklearn.preprocessing import PolynomialFeatures
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Ridge
from sklearn.linear_model import Lasso
from sklearn.linear_model import RidgeCV
from sklearn.linear_model import LassoCV
from sklearn.decomposition import PCA
from scipy.stats import zscore
%matplotlib inline
# In this problem set, we are often iterating with small Lambda values that cause many warnings to appear.
# So, we'll turn off warnings for this set:
import warnings
warnings.filterwarnings('ignore')
# Importing processed data from HW3:
test = pd.read_csv("df_test.csv", index_col=0)
train = pd.read_csv("df_train.csv", index_col=0)
x_train = train.drop('count', axis=1)
y_train = train['count']
x_test = test.drop('count', axis=1)
y_test = test['count']
#-------- sample
# A function to select a random sample of size k from the training set
# Input:
# x (n x d array of predictors in training data)
# y (n x 1 array of response variable vals in training data)
# k (size of sample)
# Return:
# chosen sample of predictors and responses
def sample(x, y, k):
n = x.shape[0] # No. of training points
# Choose random indices of size 'k'
subset_ind = np.random.choice(np.arange(n), k)
# Get predictors and reponses with the indices
x_subset = x[subset_ind, :]
y_subset = y[subset_ind]
return (x_subset, y_subset)
# Fit ridge/lasso regressions to training set
alphas = np.logspace(-5,5, 11)
ridge = RidgeCV(alphas = alphas, fit_intercept=True, normalize=False)
ridge.fit(x_train, y_train)
lasso = LassoCV(alphas = alphas, fit_intercept=True, normalize=False)
lasso.fit(x_train, y_train)
# Get our linear regression parameters from HW3:
models_coefs = pd.read_csv("linear_params.csv",header = None, index_col=0).drop(['const'])
models_coefs.columns = ['Linear Regression']
models_coefs['Ridge Regression'] = ridge.coef_
models_coefs['Lasso Regression'] = lasso.coef_
# Compare parameters
print(models_coefs)
#alphas = np.logspace(-5,5, 11)
alphas = [10.0]
ridge = RidgeCV(alphas = alphas, fit_intercept=True, normalize=False)
lasso = LassoCV(alphas = alphas, fit_intercept=True, normalize=False)
linear = LinearRegression(fit_intercept=True)
ridge_subset_r2s, ridge_test_r2s, ridge_subset_stds, ridge_test_stds = [], [], [], []
lasso_subset_r2s, lasso_test_r2s, lasso_subset_stds, lasso_test_stds = [], [], [], []
linear_subset_r2s, linear_test_r2s, linear_subset_stds, linear_test_stds = [], [], [], []
trial_sizes = range(100, 450, 50)
for k in trial_sizes:
this_k_ridge_subset_r2s = []
this_k_ridge_test_r2s = []
this_k_lasso_subset_r2s = []
this_k_lasso_test_r2s = []
this_k_linear_subset_r2s = []
this_k_linear_test_r2s = []
for trial in range(10):
x_subset, y_subset = sample(x_train.values, y_train.values, k)
ridge.fit(x_subset, y_subset)
lasso.fit(x_subset, y_subset)
linear.fit(x_subset, y_subset)
this_k_ridge_subset_r2s.append(ridge.score(x_subset, y_subset))
this_k_ridge_test_r2s.append(ridge.score(x_test, y_test))
this_k_lasso_subset_r2s.append(lasso.score(x_subset, y_subset))
this_k_lasso_test_r2s.append(lasso.score(x_test, y_test))
this_k_linear_subset_r2s.append(linear.score(x_subset, y_subset))
this_k_linear_test_r2s.append(linear.score(x_test, y_test))
ridge_subset_r2s.append(np.array(this_k_ridge_subset_r2s).mean())
ridge_test_r2s.append(np.array(this_k_ridge_test_r2s).mean())
ridge_subset_stds.append(np.array(this_k_ridge_subset_r2s).std())
ridge_test_stds.append(np.array(this_k_ridge_test_r2s).std())
lasso_subset_r2s.append(np.array(this_k_lasso_subset_r2s).mean())
lasso_test_r2s.append(np.array(this_k_lasso_test_r2s).mean())
lasso_subset_stds.append(np.array(this_k_lasso_subset_r2s).std())
lasso_test_stds.append(np.array(this_k_lasso_test_r2s).std())
linear_subset_r2s.append(np.array(this_k_linear_subset_r2s).mean())
linear_test_r2s.append(np.array(this_k_linear_test_r2s).mean())
linear_subset_stds.append(np.array(this_k_linear_subset_r2s).std())
linear_test_stds.append(np.array(this_k_linear_test_r2s).std())
fig1 = plt.figure(figsize=(10,5))
fig1.tight_layout
ax1 = fig1.add_subplot(121)
ax1.plot(trial_sizes, ridge_subset_r2s, label = "ridge", color = 'purple')
ax1.plot(trial_sizes, lasso_subset_r2s, label = "lasso", color='green')
ax1.plot(trial_sizes, linear_subset_r2s, label = "linear", color = 'blue')
ax1.set_title("Sample size vs. model training R^2")
ax1.set_xlabel("trial sample size")
ax1.set_ylabel("train (subset) R^2")
ax1.legend()
ax2 = fig1.add_subplot(122)
ax2.plot(trial_sizes, ridge_test_r2s, label = "ridge", color='purple')
ax2.plot(trial_sizes, lasso_test_r2s, label = "lasso", color='green')
ax2.plot(trial_sizes, linear_test_r2s, label = "linear", color = 'blue')
ax2.set_title("Sample size vs. model test R^2")
ax2.set_xlabel("trial sample size")
ax2.set_ylabel("test R^2")
ax2.legend()
fig2 = plt.figure(figsize=(10,5))
fig2.tight_layout
ax3 = fig2.add_subplot(131)
ax4 = fig2.add_subplot(132)
ax5 = fig2.add_subplot(133)
ax3.errorbar(trial_sizes, ridge_subset_r2s, yerr=ridge_subset_stds, label = "ridge", color='purple')
ax4.errorbar(trial_sizes, lasso_subset_r2s, lasso_subset_stds, label = "lasso", color='green')
ax5.errorbar(trial_sizes, linear_subset_r2s, linear_subset_stds, label = "linear", color = 'blue')
ax3.set_title("Ridge regression - train")
ax3.set_xlabel("trial sample size")
ax3.set_ylabel("R^2 w/standard deviation")
ax3.set_ylim(0,0.8)
ax3.legend()
ax4.set_title("Lasso regression - train")
ax4.set_xlabel("trial sample size")
ax4.set_ylabel("R^2 w/standard deviation")
ax4.set_ylim(0,0.8)
ax4.legend()
ax5.set_title("Linear regression - train")
ax5.set_xlabel("trial sample size")
ax5.set_ylabel("R^2 w/standard deviation")
ax5.set_ylim(0,0.8)
ax5.legend()
fig3 = plt.figure(figsize=(10,5))
fig3.tight_layout
ax6 = fig3.add_subplot(131)
ax7 = fig3.add_subplot(132)
ax8 = fig3.add_subplot(133)
ax6.errorbar(trial_sizes, ridge_test_r2s, ridge_test_stds, label = "ridge", color='purple')
ax7.errorbar(trial_sizes, lasso_test_r2s, lasso_test_stds, label = "lasso", color='green')
ax8.errorbar(trial_sizes, linear_test_r2s, linear_test_stds, label = "linear", color = 'blue')
ax6.set_title("Ridge regression - test")
ax6.set_xlabel("trial sample size")
ax6.set_ylabel("R^2 w/standard deviation")
ax6.set_ylim(0,0.8)
ax6.legend()
ax7.set_title("Lasso regression - test")
ax7.set_xlabel("trial sample size")
ax7.set_ylabel("R^2 w/standard deviation")
ax7.set_ylim(0,0.8)
ax7.legend()
ax8.set_title("Linear regression - test")
ax8.set_xlabel("trial sample size")
ax8.set_ylabel("R^2 w/standard deviation")
ax8.set_ylim(0,0.8)
ax8.legend()
def add_poly_terms(x, selected_features, deg = 4):
"""
Adds polynomial terms of order deg, into ONLY selected_features.
Note that it does not include any interaction terms between the predictors as requested above.
"""
poly_fit = PolynomialFeatures(degree = deg, include_bias=False)
x_selected = x.copy()[selected_features]
x_out = x.drop(selected_features, axis = 1)
# Add each higher order term (doing these individually so we don't get interaction terms)
for i,feature in enumerate(selected_features):
for this_deg in range(deg):
poly_features = poly_fit.fit_transform(x_selected[feature].values.reshape(-1,1))
x_out[feature+"_deg_"+str(this_deg+1)] = poly_features[:,this_deg]
return x_out
continuous_predictors = ['temp', 'atemp', 'humidity', 'windspeed']
x_train_poly = add_poly_terms(x_train, continuous_predictors, deg = 4)
x_test_poly = add_poly_terms(x_test, continuous_predictors, deg = 4)
poly_ols = sm.OLS(y_train, sm.add_constant(x_train_poly))
poly_ols_fit = poly_ols.fit()
poly_ols_r2 = r2_score(y_test, poly_ols_fit.predict(sm.add_constant(x_test_poly)))
print("Polynomial regression train R^2 =", r2_score(y_train, poly_ols_fit.predict(sm.add_constant(x_train_poly))),"\n")
print("Polynomial regression test R^2 =", r2_score(y_test, poly_ols_fit.predict(sm.add_constant(x_test_poly))),"\n")
ttest=poly_ols_fit.pvalues
print("Model parameters of significance level > 5%:\n", ttest[ttest.values<0.05])
def copy_and_add_inter(x_train):
x_train_inter = x_train.copy(deep=True)
x_train_inter['month_12.0'] = 1 - x_train_inter['month_1.0'] - x_train_inter['month_2.0'] - x_train_inter['month_3.0']- x_train_inter['month_4.0'] - x_train_inter['month_5.0'] - x_train_inter['month_6.0'] - x_train_inter['month_7.0'] - x_train_inter['month_8.0']- x_train_inter['month_9.0']- x_train_inter['month_10.0']- x_train_inter['month_11.0']
x_train_inter['month12-temp'] = x_train_inter['month_12.0'] * x_train_inter['temp']
x_train_inter['wday1-weather1'] = x_train_inter['workingday'] * x_train_inter['weather_1.0']
return x_train_inter
#x_train.head(30)
x_train_inter = copy_and_add_inter(x_train)
x_test_inter = copy_and_add_inter(x_test)
inter_ols = sm.OLS(y_train, sm.add_constant(x_train_inter))
inter_ols_fit = inter_ols.fit()
inter_ols_r2_train = r2_score(y_train, inter_ols_fit.predict(sm.add_constant(x_train_inter)))
inter_ols_r2_test = r2_score(y_test, inter_ols_fit.predict(sm.add_constant(x_test_inter)))
print("Multilinear regression R^2 train set =", inter_ols_r2_train,"\n")
print("Multilinear regression R^2 test set =", inter_ols_r2_test,"\n")
ttest1=inter_ols_fit.pvalues
print("Model parameters of significance level > 5%:\n", ttest1[ttest1.values<0.05])
# First, let's generate a polynomial with all of the interation terms (this way, we don't get the higher-order interaction terms)
interaction_poly = PolynomialFeatures(degree = 2, interaction_only = True)
x_train_interaction = pd.DataFrame(interaction_poly.fit_transform(x_train), columns = interaction_poly.get_feature_names(x_train.columns))
x_test_interaction = pd.DataFrame(interaction_poly.fit_transform(x_test), columns = interaction_poly.get_feature_names(x_test.columns))
# Then, add in the polynomial terms used earlier
continuous_predictors = ['temp', 'atemp', 'humidity', 'windspeed']
x_big_train = add_poly_terms(x_train_interaction, continuous_predictors, deg = 4)
x_big_test = add_poly_terms(x_test_interaction, continuous_predictors, deg = 4)
print("Full expanded data set has",x_big_train.shape[1], "columns!")
#print(y_train.shape, sm.add_constant(x_big_train).shape)
big_ols = sm.OLS(y_train, sm.add_constant(x_big_train))
big_ols_fit = big_ols.fit()
big_ols_r2 = r2_score(y_train, big_ols_fit.predict(sm.add_constant(x_big_train)))
print("Our full feature training R^2 =",big_ols_r2)
big_ols_r2 = r2_score(y_test, big_ols_fit.predict(sm.add_constant(x_big_test)))
print("Our full feature test R^2 =",big_ols_r2,"\n")
# Fit PCA
num_comp = 5
pca = PCA(num_comp)
pca.fit(x_big_train)
x_train_pca = pca.transform(x_big_train)
x_test_pca = pca.transform(x_big_test)
print("Generating PCA with",num_comp, "components.\nExplained variance ratio:", pca.explained_variance_ratio_, "\n")
# Fit linear regression over sums of the first n components
lin_reg_model = LinearRegression(fit_intercept=True)
for n in range(1,5+1):
lin_reg_model.fit(x_train_pca[:,:n], y_train)
print("Training R^2 of first",n,"PCA components=", r2_score(y_train, lin_reg_model.predict(x_train_pca[:,:n])))
print("Test R^2 of first",n,"PCA components=", r2_score(y_test, lin_reg_model.predict(x_test_pca[:,:n])))
#-------- rmsle
# A function for evaluating Root Mean Squared Logarithmic Error (RMSLE)
# of the linear regression model on a data set
# Input:
# y_test (n x 1 array of response variable vals in testing data)
# y_pred (n x 1 array of response variable vals in testing data)
# Return:
# RMSLE (float)
def rmsle(y, y_pred):
# Evaluate sqaured error, against target labels
# rmsle = \sqrt(1/n \sum_i (log (y[i]+1) - log (y_pred[i]+1))^2)
rmsle_ = np.sqrt(np.mean(np.square(np.log(y+1) - np.log(y_pred+1))))
return rmsle_
lin_reg_model = LinearRegression(fit_intercept=True)
# Calculate train RMSLE on current model fit
lin_reg_model.fit(x_train_poly, y_train)
y_hat_train = lin_reg_model.predict(x_train_poly)
y_hat_test = lin_reg_model.predict(x_test_poly)
print("Train RMSLE on linear model fit =", rmsle(y_train, y_hat_train))
print("Test RMSLE on linear model fit =", rmsle(y_test, y_hat_test))
print("")
# Now, let's fit a new set on a revised response variable - specifically, log(y+1)
lin_reg_model.fit(x_train_poly, np.log1p(y_train))
y_hat_logit_train = np.expm1(lin_reg_model.predict(x_train_poly))
y_hat_logit_test = np.expm1(lin_reg_model.predict(x_test_poly))
print("Train RMSLE on RMSLE model fit =", rmsle(y_train, y_hat_logit_train))
print("Test RMSLE on RMSLE model fit =", rmsle(y_test, y_hat_logit_test))
ridge1 = RidgeCV(alphas = alphas, fit_intercept=True, normalize=False)
ridge1.fit(x_train, y_train)
R2_init=r2_score(y_test, ridge1.predict(x_test))
print("Test R^2 from Part g)",r2_score(y_test, poly_ols_fit.predict(sm.add_constant(x_test_poly))))
print("Test R^2 from Ridge model with all train points",R2_init)
x_train_trim = x_train_poly.copy(deep = True)
y_train_trim = y_train.copy(deep = True)
#print(y_train_trim.shape)
Ridge_Test_R2s_after_drop = []
stop_constant = float("inf")
for i in range(int(len(y_train_trim)*0.05)):
ridge_reg_model = RidgeCV(alphas = alphas, fit_intercept=True, normalize=False)
ridge_reg_model.fit(x_train_trim, y_train_trim)
y_prec_train = ridge_reg_model.predict(x_train_trim)
y_prec_test = ridge_reg_model.predict(x_test_poly)
#print("Train R-score is", r2_score(y_train_trim,y_prec_train))
#print("Test R-score is",r2_score(y_test,y_prec_test))
#index of the worst value
y_worst = np.argmax(np.abs(y_train_trim - y_prec_train))
#worst value
worst_error =np.max(np.abs(y_train_trim - y_prec_train))
#print("Dropping the worst y", worst_error, "at the index", y_train_trim.index[y_worst],"\n")
if worst_error < 2600:
print("BREAK")
break
x_train_trim = x_train_trim.drop(x_train_trim.index[y_worst])
y_train_trim = y_train_trim.drop(y_train_trim.index[y_worst])
Ridge_Test_R2s_after_drop.append(r2_score(y_test,y_prec_test))
print("R^2 after removing ~5% of data points", Ridge_Test_R2s_after_drop[-1])
| 0.69233 | 0.937555 |
# A - 2 - Full Texts Acquisition
## Description
**Process aim:**
This process aims at adding a field containing the full text of the resources to the metadata dataframe.
**Input:** A csv files containing metadata including at least one columns with URLs
**Sub-processes**:
1. Import metadata
2. Get and save PDF files
3. Extract full texts from PDF files
4. Add full text to dataset and save
**Output:** a CSV file
## 1. Import metadata
```
import pandas as pd
import requests
import textract
import logging
logging.basicConfig(level=logging.INFO)
logger = logging.getLogger(__name__)
```
Write the name of the fields to keep. These might be either
* identifiers (ie. record_id)
* target labels: field that are intended to be predicted in other world automatically generated (i.e. subjects_geo, subjects_topics, etc)
* url of the resource in English (i.e. url_English)
* features: field that includes characteristics of the text that will be used to predict the labels (i.e. title)
* any other field that you want to keep to analyze your dataset
```
# Columns of the dataset to keep
columns = ['record_id','body', 'date', 'session', 'subjects_geo','subjects_primary', 'subjects_topics', 'symbol',
'title', 'type','url_English']
# Load the dataset and create a Pandas dataframe
dataset = (pd.read_csv('data/0_input_data/metadata/input/doc_2000_2017.csv',usecols=columns, index_col='record_id', dtype='str'))
```
## 2. Get and save PDF files
We need the full text of the resources described in the MARC XML that will be used later to infer some metadata. In this case we will focus on English texts only.
For this step start by creating a list containing for each record that has an English url, the record id, and the url. We then use the function save_files() to get the files using the url and save them in the the folder data/acquisition.
```
def save_files(files_list, save_path, file_extension):
'''
Takes a list of of lists (record_id and url), the path of the location where the files
will be saved, and the extension of the file type. Get the files through htpp requests
and save them. Returns a list of record_id, corresponding to files that could not be
saved.
'''
errors = []
for item in files_list:
save_as = save_path + str(item[0]) + file_extension
file_url = item[1]
response = requests.get(file_url)
if response.status_code == requests.codes.ok:
with open(save_as, 'wb') as f:
f.write(response.content)
else:
errors.append(item)
return errors
def last_saved_file(record_id,file_list):
'''
Print the index of corresponding to the record_id in file_list
To use if save_files stops in order to restart the downloads where it stoped.
'''
i = 0
for item in file_list:
if record_id in item:
print('{} : {}'.format(i,item))
i +=1
# Create a list of record_id and url for all record that have an url
en_list = (dataset.reset_index()[['record_id', 'url_English']]
.dropna() # filter out if non values
.values.tolist())
# Output the length of the list
len(en_list)
# Get the files and save them in pdf
save_files(en_list, 'data/A_input_data/files/', '.pdf')
```
If the script stops running before the list is completed, get the latest saved file, use last_saved_file to get the index number and restart at index number:
```
# last_record_id = # past the record_id of the latest file saved
# last_saved_file(last_record_id,en_list)
# Restart at index, replace *** by the index number
# save_files(en_list[***:], 'data/A_input_data/files/', '.pdf')
```
## 3. Extract full text from PDF files
Using the same files list we then use the convert_to_pdf_function, to get the content of the PDFs, convert it to a string of text, and store this as a third column in the initial list. Note that if a page cannot be processed, then it will be skipped altogether.
```
def convert_pdf_to_text(files_list, path):
'''
Takes a list of list, and a path to files in pdf. Read each files and convert to text.
Append the resulting texts to the initial list and return the list.
'''
new_list = []
i = 1
for file in files_list:
file_path = path + str(file[0]) + '.pdf'
full_text = ""
try:
full_text = textract.process(file_path)
full_text = full_text.decode() # convert unicode bytes
except:
logger.exception("record {}: could not convert pdf to text".format(file[0]))
file.append(full_text)
new_list.append(file)
i +=1
return new_list
# Add a column with the full text of the pdf
en_list = convert_pdf_to_text(en_list,'data/A_input_data/files/')
```
## 4. Add full texts to the metadata and save the output
To finish, we create a new dataframe with only the record_id and the full text. As they have the same index, the record-id, we can join it to the metadata dataframe and easily save the result as a CSV.
```
# Create a new dataframe withg the record_id and the full text
full_text = (pd.DataFrame(en_list, columns=['record_id','url','text'])
.drop('url', axis=1)
.set_index('record_id')
)
# Join the result to the metadta dataset
dataset = dataset.join(full_text)
# Check the result
dataset.info()
# Save the content of the dataset in data/pre-processing/
dataset.to_csv('data/A_input_data/metadata/output/doc_2000_2017.csv')
```
|
github_jupyter
|
import pandas as pd
import requests
import textract
import logging
logging.basicConfig(level=logging.INFO)
logger = logging.getLogger(__name__)
# Columns of the dataset to keep
columns = ['record_id','body', 'date', 'session', 'subjects_geo','subjects_primary', 'subjects_topics', 'symbol',
'title', 'type','url_English']
# Load the dataset and create a Pandas dataframe
dataset = (pd.read_csv('data/0_input_data/metadata/input/doc_2000_2017.csv',usecols=columns, index_col='record_id', dtype='str'))
def save_files(files_list, save_path, file_extension):
'''
Takes a list of of lists (record_id and url), the path of the location where the files
will be saved, and the extension of the file type. Get the files through htpp requests
and save them. Returns a list of record_id, corresponding to files that could not be
saved.
'''
errors = []
for item in files_list:
save_as = save_path + str(item[0]) + file_extension
file_url = item[1]
response = requests.get(file_url)
if response.status_code == requests.codes.ok:
with open(save_as, 'wb') as f:
f.write(response.content)
else:
errors.append(item)
return errors
def last_saved_file(record_id,file_list):
'''
Print the index of corresponding to the record_id in file_list
To use if save_files stops in order to restart the downloads where it stoped.
'''
i = 0
for item in file_list:
if record_id in item:
print('{} : {}'.format(i,item))
i +=1
# Create a list of record_id and url for all record that have an url
en_list = (dataset.reset_index()[['record_id', 'url_English']]
.dropna() # filter out if non values
.values.tolist())
# Output the length of the list
len(en_list)
# Get the files and save them in pdf
save_files(en_list, 'data/A_input_data/files/', '.pdf')
# last_record_id = # past the record_id of the latest file saved
# last_saved_file(last_record_id,en_list)
# Restart at index, replace *** by the index number
# save_files(en_list[***:], 'data/A_input_data/files/', '.pdf')
def convert_pdf_to_text(files_list, path):
'''
Takes a list of list, and a path to files in pdf. Read each files and convert to text.
Append the resulting texts to the initial list and return the list.
'''
new_list = []
i = 1
for file in files_list:
file_path = path + str(file[0]) + '.pdf'
full_text = ""
try:
full_text = textract.process(file_path)
full_text = full_text.decode() # convert unicode bytes
except:
logger.exception("record {}: could not convert pdf to text".format(file[0]))
file.append(full_text)
new_list.append(file)
i +=1
return new_list
# Add a column with the full text of the pdf
en_list = convert_pdf_to_text(en_list,'data/A_input_data/files/')
# Create a new dataframe withg the record_id and the full text
full_text = (pd.DataFrame(en_list, columns=['record_id','url','text'])
.drop('url', axis=1)
.set_index('record_id')
)
# Join the result to the metadta dataset
dataset = dataset.join(full_text)
# Check the result
dataset.info()
# Save the content of the dataset in data/pre-processing/
dataset.to_csv('data/A_input_data/metadata/output/doc_2000_2017.csv')
| 0.483892 | 0.846324 |
# Classifying Fashion-MNIST
Now it's your turn to build and train a neural network. You'll be using the [Fashion-MNIST dataset](https://github.com/zalandoresearch/fashion-mnist), a drop-in replacement for the MNIST dataset. MNIST is actually quite trivial with neural networks where you can easily achieve better than 97% accuracy. Fashion-MNIST is a set of 28x28 greyscale images of clothes. It's more complex than MNIST, so it's a better representation of the actual performance of your network, and a better representation of datasets you'll use in the real world.
<img src='assets/fashion-mnist-sprite.png' width=500px>
In this notebook, you'll build your own neural network. For the most part, you could just copy and paste the code from Part 3, but you wouldn't be learning. It's important for you to write the code yourself and get it to work. Feel free to consult the previous notebooks though as you work through this.
First off, let's load the dataset through torchvision.
```
import torch
from torchvision import datasets, transforms
import helper
# Define a transform to normalize the data
transform = transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.5,), (0.5,))])
# Download and load the training data
trainset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)
# Download and load the test data
testset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=False, transform=transform)
testloader = torch.utils.data.DataLoader(testset, batch_size=64, shuffle=True)
```
Here we can see one of the images.
```
type(trainset)
image, label = next(iter(trainloader))
helper.imshow(image[0,:]);
image[0].shape
```
## Building the network
Here you should define your network. As with MNIST, each image is 28x28 which is a total of 784 pixels, and there are 10 classes. You should include at least one hidden layer. We suggest you use ReLU activations for the layers and to return the logits or log-softmax from the forward pass. It's up to you how many layers you add and the size of those layers.
```
# TODO: Define your network architecture here
from torch import nn
from torch import optim
import torch.nn.functional as F
class Network(nn.Module):
def __init__(self):
super().__init__()
self.fc1 = nn.Linear(784, 128)
self.fc2 = nn.Linear(128, 64)
self.fc3 = nn.Linear(64, 10)
def forward(self, x):
x = x.view(x.shape[0], -1)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = F.log_softmax(self.fc3(x), dim=1)
return x
```
# Train the network
Now you should create your network and train it. First you'll want to define [the criterion](http://pytorch.org/docs/master/nn.html#loss-functions) ( something like `nn.CrossEntropyLoss`) and [the optimizer](http://pytorch.org/docs/master/optim.html) (typically `optim.SGD` or `optim.Adam`).
Then write the training code. Remember the training pass is a fairly straightforward process:
* Make a forward pass through the network to get the logits
* Use the logits to calculate the loss
* Perform a backward pass through the network with `loss.backward()` to calculate the gradients
* Take a step with the optimizer to update the weights
By adjusting the hyperparameters (hidden units, learning rate, etc), you should be able to get the training loss below 0.4.
```
# TODO: Create the network, define the criterion and optimizer
model = Network()
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.parameters(), lr=0.01)
# TODO: Train the network here
epochs = 5
for e in range(epochs):
running_loss = 0
for images, labels in trainloader:
# forward pass
output = model(images)
# loss counting
loss = criterion(output, labels)
# optimizer reset
optimizer.zero_grad()
# backward pass
loss.backward()
# applying optimizer
optimizer.step()
running_loss += loss.item()
else:
print(f"Training loss: {running_loss/len(trainloader)}")
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import helper
# Test out your network!
dataiter = iter(testloader)
images, labels = dataiter.next()
img = images[0]
# Convert 2D image to 1D vector
img = img.resize_(1, 784)
# TODO: Calculate the class probabilities (softmax) for img
ps = torch.exp(model(img))
# Plot the image and probabilities
helper.view_classify(img.resize_(1, 28, 28), ps, version='Fashion')
```
|
github_jupyter
|
import torch
from torchvision import datasets, transforms
import helper
# Define a transform to normalize the data
transform = transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.5,), (0.5,))])
# Download and load the training data
trainset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)
# Download and load the test data
testset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=False, transform=transform)
testloader = torch.utils.data.DataLoader(testset, batch_size=64, shuffle=True)
type(trainset)
image, label = next(iter(trainloader))
helper.imshow(image[0,:]);
image[0].shape
# TODO: Define your network architecture here
from torch import nn
from torch import optim
import torch.nn.functional as F
class Network(nn.Module):
def __init__(self):
super().__init__()
self.fc1 = nn.Linear(784, 128)
self.fc2 = nn.Linear(128, 64)
self.fc3 = nn.Linear(64, 10)
def forward(self, x):
x = x.view(x.shape[0], -1)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = F.log_softmax(self.fc3(x), dim=1)
return x
# TODO: Create the network, define the criterion and optimizer
model = Network()
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.parameters(), lr=0.01)
# TODO: Train the network here
epochs = 5
for e in range(epochs):
running_loss = 0
for images, labels in trainloader:
# forward pass
output = model(images)
# loss counting
loss = criterion(output, labels)
# optimizer reset
optimizer.zero_grad()
# backward pass
loss.backward()
# applying optimizer
optimizer.step()
running_loss += loss.item()
else:
print(f"Training loss: {running_loss/len(trainloader)}")
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import helper
# Test out your network!
dataiter = iter(testloader)
images, labels = dataiter.next()
img = images[0]
# Convert 2D image to 1D vector
img = img.resize_(1, 784)
# TODO: Calculate the class probabilities (softmax) for img
ps = torch.exp(model(img))
# Plot the image and probabilities
helper.view_classify(img.resize_(1, 28, 28), ps, version='Fashion')
| 0.813424 | 0.99031 |
# Explore the data
We have downloaded the ICESat-2 data and saved them as `./download/processed_ATL03_20190805232841_05940403_004_01.h5`. Before diving into a specific analysis routine, let us see if we can have a general overview about the data using the Jupyter tools. For this stage, we want tools that provide quick access to data, preferably in many ways. And we also want to have some funcionaility to manually navigate to different part of the data. Jupyter's language-agnostic nature (i.e., not bonded to any specific programming languages) and support to interactive plotting widgets are designed to address these needs.
```{admonition} Goals
:class: tip
Explore the data file, including its data structure, size, geospatial information, and so on.
```
## Steps
### Check data structure
Since the data are stored using the HDF5 format (as indicated by the file extension), we will need certain tools to read them. For example, we can use the `h5ls` command-line tool to have a quick look of the data structure. On Jupyter notebook, we can use the `!` character to use any command-line tool, and we can even pass the variable defined in the other cell (Python or Shell block) to the `h5ls` command.
```
filename = 'download/processed_ATL03_20190805232841_05940403_004_01.h5'
```
This string variable is now shared by the Python kernel and the shell environment. Note that we also use `grep` here because the full, nested data structure is very long, and we are only interested in the gt1l beam (which {cite}`Herzfeld2021` use in their study).
```
!h5ls -r $filename | grep ^/gt1l/heights
!h5ls -r $filename | grep ^/gt1l/geolocation/segment
```
### Load the data
Now we use h5py (the python library for working with HDF5) and numpy to open the file and load the data we want.
```
import h5py
import numpy as np
with h5py.File(filename, 'r') as f:
lon_ph = f['gt1l']['heights']['lon_ph'][:] # photon longitude (x)
lat_ph = f['gt1l']['heights']['lat_ph'][:] # photon latitude (y)
h_ph = f['gt1l']['heights']['h_ph'][:] # photon elevation (z), in m
dist_ph = f['gt1l']['heights']['dist_ph_along'][:] # photon horizontal distance from the beginning of the parent segment, in m
seg_length = f['gt1l']['geolocation']['segment_length'][:] # horizontal of each segment, in m
seg_ph_count = f['gt1l']['geolocation']['segment_ph_cnt'][:] # photon count in each segment, in m
```
We can easily check the content and statistical information of each variable. For example:
```
print(sum(seg_ph_count))
print(h_ph.shape[0]) # this should equal to the summation of the photon count.
```
### Prepare the data
We often need to apply a few steps to the raw data before visualizing or analyzing them. Take these ICESat-2 data for example: if we want to plot the elevation along this track, we need the distance along the track as x values, which is not provided but can be calculated on our own. Using Jupyter Notebook, we can quickly design a Python function for this variable.
```
def make_dist_alongtrack(ph_count, seg_length, dist_ph):
'''
For detailed explanation of each variable and reasoning of the code, see ICESat-2 ATL03 documentation.
'''
repeat_num = np.cumsum(seg_length) - seg_length[0]
dist_alongtrack = np.repeat(repeat_num, ph_count)
dist_alongtrack += dist_ph
return dist_alongtrack
dist_alongtrack = make_dist_alongtrack(seg_ph_count, seg_length, dist_ph) # distance along track for each photon, in m
```
(sec:explore-plot)=
### Plot the data
Plotting the data is a great way to obtain a brief overview. Using interactive matplotlib figures powered by `ipympl`, we can speed up the exploration and quickly focus on the key elements of our data.
Use the following Notebook command to activate interactive matplotlib environment:
```
%matplotlib widget
```
And then we import matplotlib.
```
import matplotlib.pyplot as plt
```
Now every single figure will come with a control panel, and we can use the buttons to pan, zoom, and save the figure. This is especially very helpful in our case because crevasses are small scale features and do not show everywhere along the track. We have to really zoom in to a certain area in order to see them.
```
fig, ax = plt.subplots(1, 1, figsize=(7, 3))
ax.plot(dist_alongtrack, h_ph, '.', markersize=1)
```
After a careful check up, we are able to locate the segment where [Figure 6a](sec:ideas-goals) of {cite}`Herzfeld2021` uses. (Note that the along-track distance readings are different because we are using a subset of the original data granule .)
```
fig, ax2 = plt.subplots(1, 1, figsize=(7, 3))
ax2.plot(dist_alongtrack, h_ph, '.', markersize=1)
ax2.set_xlim(15090, 16090)
ax2.set_ylim(320, 363)
```
```{admonition} Summary
The Jupyter ecosystem provides multiple and interactive approaches to access and explore the data.
```
```{bibliography}
:filter: docname in docnames
```
|
github_jupyter
|
## Steps
### Check data structure
Since the data are stored using the HDF5 format (as indicated by the file extension), we will need certain tools to read them. For example, we can use the `h5ls` command-line tool to have a quick look of the data structure. On Jupyter notebook, we can use the `!` character to use any command-line tool, and we can even pass the variable defined in the other cell (Python or Shell block) to the `h5ls` command.
This string variable is now shared by the Python kernel and the shell environment. Note that we also use `grep` here because the full, nested data structure is very long, and we are only interested in the gt1l beam (which {cite}`Herzfeld2021` use in their study).
### Load the data
Now we use h5py (the python library for working with HDF5) and numpy to open the file and load the data we want.
We can easily check the content and statistical information of each variable. For example:
### Prepare the data
We often need to apply a few steps to the raw data before visualizing or analyzing them. Take these ICESat-2 data for example: if we want to plot the elevation along this track, we need the distance along the track as x values, which is not provided but can be calculated on our own. Using Jupyter Notebook, we can quickly design a Python function for this variable.
(sec:explore-plot)=
### Plot the data
Plotting the data is a great way to obtain a brief overview. Using interactive matplotlib figures powered by `ipympl`, we can speed up the exploration and quickly focus on the key elements of our data.
Use the following Notebook command to activate interactive matplotlib environment:
And then we import matplotlib.
Now every single figure will come with a control panel, and we can use the buttons to pan, zoom, and save the figure. This is especially very helpful in our case because crevasses are small scale features and do not show everywhere along the track. We have to really zoom in to a certain area in order to see them.
After a careful check up, we are able to locate the segment where [Figure 6a](sec:ideas-goals) of {cite}`Herzfeld2021` uses. (Note that the along-track distance readings are different because we are using a subset of the original data granule .)
| 0.810216 | 0.987351 |
# Text search/indexing using prime number factorisation
---
## Abstract
This report details the proof of concept for using prime numbers for indexing text with the intention of searching for words within a data set. This technique is shown to significantly out perform regular string comparison in all the aspect that were tested i.e number of records, lengths of records, length of dictionary, and number of words searched for.
The targeted use case for this algorithm is in exploratory text analysis by finding the frequency that specific word combinations appear together within the same text record.
---
## Background knowledge
This technique utilises the knowledge described by the unique-prime-factorization theorem where it is understood that every integer greater than 1 can be broken down into a unique list of its prime factors.
---
## Technique theory
The process is broken down into preprocessing and searching:
#### Preprocessing
* Defining the dictionary of the data set
* Index the dictionary by storing a unique prime number for each word in the dictionary
* Index the text records by storing the product of the prime numbers that make up each text record
#### Searching
* Fetch the unique prime number of the word to search for from the indexed dictionary
* Find the modulo of each indexed text record against the value of the indexed word being searched for
* If the modulo calculation equals 0 then the search word exists in that record
#### Example
Take the data set of text:
* "black cat on mat",
* "black hat for you",
* "cat sat on you"
The dictionary of this data set is:
* "black",
* "cat",
* "on",
* "mat",
* "hat",
* "for",
* "you",
* "sat"
Now assign prime number to each word in dictionary:
* "black": 2,
* "cat" : 3,
* "on" : 5,
* "mat" : 7,
* "hat" : 11,
* "for" : 13,
* "you" : 17,
* "sat" : 19
Now turn each text record into the product of its prime numbers:
* "black cat on mat" = "black(2) cat(3) on(5) mat(7)" = 2 x 3 x 5 x 7 = **210**
* "black hat for you" = black(2) hat(11) for(13) you(17) = 2 x 11 x 13 x 17 = **4862**
* "cat sat on you" = "cat(3) sat(19) on(5) you(17)" = 3 x 19 x 5 x 17 = **4845**
Now the indexed data set is:
* 210,
* 4862,
* 4845
To search for the word **"sat"**(prime equvilant 19) in the data set we calculate the modulo of its equvilate prime across the data set and where the result is '0' the word exists within that text record:
* 210 % 19 = 1 (**not a factor**)
* 4862 % 19 = 17 (**not a factor**)
* 4845 % 19 = 0 (**is a factor** "cat **sat** on you")
Another example, search for **"you"** (prime equivilant 17):
* 210 % 17 = 6 (**not a factor**)
* 4862 % 17 = 0 (**is a factor** "black hat for **you**")
* 4845 % 17 = 0 (**is a factor** "cat sat on **you**")
#### Searching for multiple words
Because of the associative property of factors, to search for multiple words you do the same as before except calculate the modulo using the product of the words that you are searching for:
Examples:
---
Search for "you" and "cat"
"you"(17) x "cat"(3) = 51
* 210 % 51 = 6 (**not a factor**)
* 4862 % 51 = 17 (**not a factor**)
* 4845 % 51 = 0 (**is a factor** "**cat** sat on **you**")
---
Search for "cat" and "on"
"cat"(3) x "on"(5) = 15
* 210 % 15 = 0 (**is a factor** "black **cat** **on** mat")
* 4862 % 15 = 2 (**not a factor**)
* 4845 % 15 = 0 (**is a factor** "**cat** sat **on** you")
---
Search for "black", "hat" and "you"
"black"(2) x "hat"(11) x you(17) = 374
* 210 % 374 = 210 (**not a factor**)
* 4862 % 374 = 0 (**is a factor** "**black** **hat** for **you**")
* 4845 % 374 = 231 (**not a factor**)
---
# Implimentation and testing
This section shows an implimentation of the technique along side a standard string comparision implimentation with performance camapaisons made between the two techniques.
Steps in this section are:
1. Generate synthetic data set to test with
2. Impliment standard string comparision serach functions (base line technique to compare against)
3. Impliment prime factorisation index/serach function
4. Compare performance
```
%matplotlib inline
import random
import time
import copy
import numpy as np
import numpy.core.defchararray as npstr
import matplotlib.pyplot as plt
```
---
## Synthetic test data generation
First step is developing convienent functions that allow for control over different varibles concerning the data set. This makes comparing performance more informative and reproducable.
```
# generates a random string of letters for a given length
def generateWord(length):
abc = 'abcdefghijklmnopqrstuvwxyz'
word = ''
for i in range(length):
word += random.choice(abc)
return word
# generates a normally distributed list of numbers with a given length, mean and diviation
def generateLengths(count,mean, dist, showGraph = False):
s = np.random.normal(mean, dist, count)
s = np.round(s,0).astype(int)
lessThanOne = s < 1
s[lessThanOne] = 1
if showGraph:
plt.hist(s, 30, normed=True)
plt.title("Histogram of lengths")
plt.show()
return s
# generates a random dictionary of words whose lengths are normally distributed
def generateDictionary(count):
currentDict = []
wordLengths = generateLengths(count,5,1)
for length in wordLengths:
oldLen = len(currentDict)
while(len(currentDict) == oldLen):
word = generateWord(length)
if word not in currentDict:
currentDict.append(word)
return np.asarray(currentDict)
# Selects random words from a given dictionary that matches given length requirments
def pickRandomWordsFromDict(dictionary, minLength, maxLength, count):
output = []
for word in dictionary:
if maxLength >= len(word) >= minLength:
output.append(word)
if len(output)==count:
return output
return ValueError('Dictionary did not conatin enough words of your min length')
# generates a list of randomy generated strings of text made of of words from a given dictionary
# and where the number of words used in the text are normally distributed to a given mean and diviation
def generateComments(count,dictionary,mean,dist,showGraph = False):
comments = []
lengths = generateLengths(count,mean,dist,showGraph)
for length in lengths:
comment = ''
for i in range(length):
comment += random.choice(dictionary) + ' '
comments.append(comment.rstrip())
return np.asarray(comments)
```
## Testing syntehtic data functions
```
generateWord(3)
generateLengths(100,5,1,True)
dictionary = generateDictionary(100)
dictionary [1:5]
pickRandomWordsFromDict(dictionary,3,8,3)
comments = generateComments(1000000,dictionary,4,1,True)
comments[1:5]
```
---
# Implimenting word search using string comparison (base line technique)
```
# Finds records that conatin specific words using numpy's string comparitor function
def findCommentsWithWords(comments,words):
currentVocab = copy.deepcopy(comments)
for w in words:
currentVocab = currentVocab[(npstr.find(currentVocab, w) != -1)]
return currentVocab
#Returns number of records that contain specific words
def numberOfMatches(comments, words):
result = findCommentsWithWords(comments,words)
return len(result)
```
### Test speed of finding number of records with specific words
```
randoWords = pickRandomWordsFromDict(dictionary,3,8,2)
randoWords
start = time.time()
print(str(numberOfMatches(comments,randoWords)) + ' number of records found containing ' + str(randoWords))
end = time.time()
print(str(end - start)+' seconds')
```
---
# Implimenting word indexing/search by prime factorisation
### Preprocessing/Indexing
```
# Assigns a prime number to each word in a given dictionary and returns a dict object
def createIndexSchema(dictionary):
primes = np.genfromtxt ('primes.csv', delimiter=",").astype(int)
primeFit = primes[1:len(dictionary)+1,1]
index = dict(np.c_[dictionary,primeFit])
return index
# Converts a list of strings into the products of the words in the string's prime number indexes
def indexComments(comments,indexSchemal):
output = []
for comment in comments:
prod = 1
words = comment.split(' ')
for word in words:
if word in indexSchemal:
prod *= int(indexSchemal[word])
output.append(prod)
return output
# Return the indexed dictionary and text data for a given normal dictionary of words and list of text data
def preprocessPrimeComments(dictionary,comments):
indexSchemal = createIndexSchema(dictionary)
indexedComments = indexComments(comments,indexSchemal)
return [indexSchemal,indexedComments]
```
### Searching
```
# converts given words into their indexed product
def convertWordsToProduct(indexSchema, words):
output = 1
for word in words:
output *= int(indexSchema[word])
return output
# calculates the modulo of the indexed text data against the given search word product
# and return a boolean list of which records match the search
def searchByPrimeFact(indexedComments,serachProduct):
return (np.mod(indexedComments, serachProduct) == 0)
# returns a boolean list of which of the given indexed records conatain
# the given words using the given indexed dictionary
def findCommentsUsingPrimeFact(indexedComments,index, words):
prod = convertWordsToProduct(index, words)
return searchByPrimeFact(indexedComments,prod)
```
### Test speed of finding numberof records with specific words
```
[index, indexedComments] = preprocessPrimeComments(dictionary,comments)
start = time.time()
print(str(findCommentsUsingPrimeFact(indexedComments,index,randoWords).sum())+ ' number of records found containing ' + str(randoWords) )
end = time.time()
print(str(end - start)+' seconds')
```
---
# Compare Performance
The different varibles that will be tested are:
* Number of comments
* Number of query words
* Length of comments
* Dictionary length
```
# times the execution fo a function returning the seconds it took and the output of the function
def timeFunction(func, *params):
start = time.time()
output = func(*params)
end = time.time()
return[output, (end - start)]
# generate text records using the previously defined synthetic data function
def generateCommentData(dictLength, numOfComments, commentLengthMean, commentLengthDist, graph = False):
random.seed(50)
np.random.seed(15)
dictionary = generateDictionary(dictLength)
comments = generateComments(numOfComments,dictionary,commentLengthMean,commentLengthDist,graph)
return [dictionary, comments]
[d,c] = generateCommentData(300,100000,7,1, True)
```
## Performance against number of comments to search through
```
[d,c] = generateCommentData(300,100000,7,1, False)
wordsToserachFor = pickRandomWordsFromDict(d,3,7,3)
[indx,indxCom] = preprocessPrimeComments(d,c)
stringCompTimings1 = []
primeTimings1 = []
xIncrement1 = []
for n in range(100,100000,1000):
xIncrement1.append(n)
[o,t] = timeFunction(numberOfMatches, c[:n], wordsToserachFor)
stringCompTimings1.append(t)
[o,t] = timeFunction(findCommentsUsingPrimeFact, indxCom[:n],indx, wordsToserachFor)
primeTimings1.append(t)
plt.figure()
plt.title("Plot of speed of search based on number of commenst to search through")
plt.ylabel("Time taken (seconds)")
plt.xlabel("Number of comments to search through")
strCompPlot, = plt.plot(xIncrement1,stringCompTimings1, label="Numpy String Compare")
primeFactPlot, = plt.plot(xIncrement1,primeTimings1, label="My Prime Factorisation")
plt.legend(handles=[strCompPlot,primeFactPlot],loc=2)
plt.show()
```
# Performance against number of words searched for
```
stringCompTimings2 = []
primeTimings2 = []
xIncrement2 = []
for n in range(1,100):
xIncrement2.append(n)
randWords2 = pickRandomWordsFromDict(d,2,7,n)
[o,t] = timeFunction(numberOfMatches, c[:100000], randWords2)
stringCompTimings2.append(t)
[o,t] = timeFunction(findCommentsUsingPrimeFact, indxCom[:10000],indx, randWords)
primeTimings2.append(t)
plt.figure()
plt.title("Plot of speed of search based on number of words searched for")
plt.ylabel("Time taken (seconds)")
plt.xlabel("Number of words searched for")
strCompPlot, = plt.plot(xIncrement2,stringCompTimings2, label="Numpy String Compare")
primeFactPlot, = plt.plot(xIncrement2,primeTimings2, label="My Prime Factorisation")
plt.legend(handles=[strCompPlot,primeFactPlot],loc=5)
plt.show()
```
# Performance against length of comments to search through
```
stringCompTimings3 = []
primeTimings3 = []
xIncrement3 = []
for n in range(5,500,10):
xIncrement3.append(n)
[d3,c3] = generateCommentData(30,1000,n,1)
randWords3 = pickRandomWordsFromDict(d3,2,7,4)
[o,t] = timeFunction(numberOfMatches, c3, randWords3)
stringCompTimings3.append(t)
[indexSchema3, indxCom3] = preprocessPrimeComments(d3,c3)
[o,t] = timeFunction(findCommentsUsingPrimeFact, indxCom3,indexSchema3, randWords3)
primeTimings3.append(t)
plt.figure()
plt.title("Plot of speed of search based on length of comments to search through")
plt.ylabel("Time taken (seconds)")
plt.xlabel("Mean length of comments searched through")
strCompPlot, = plt.plot(xIncrement3,stringCompTimings3, label="Numpy String Compare")
primeFactPlot, = plt.plot(xIncrement3,primeTimings3, label="My Prime Factorisation")
plt.legend(handles=[strCompPlot,primeFactPlot],loc=2)
plt.show()
```
# Performance against length of dictionary
```
stringCompTimings4 = []
primeTimings4 = []
xIncrement4 = []
for n in range(10,1000,10):
xIncrement4.append(n)
[d4,c4] = generateCommentData(n,2000,10,1)
randWords4 = pickRandomWordsFromDict(d4,2,7,3)
[o,t] = timeFunction(numberOfMatches, c4, randWords4)
stringCompTimings4.append(t)
[indexSchema4, indxCom4] = preprocessPrimeComments(d4,c4)
[o,t] = timeFunction(findCommentsUsingPrimeFact, indxCom4,indexSchema4, randWords4)
primeTimings4.append(t)
plt.figure()
plt.title("Plot of speed of search based on length of dictionary")
plt.ylabel("Time taken (seconds)")
plt.xlabel("Number of unique words in text (dictionary length)")
strCompPlot, = plt.plot(xIncrement4,stringCompTimings4, label="Numpy String Compare")
primeFactPlot, = plt.plot(xIncrement4,primeTimings4, label="My Prime Factorisation")
plt.legend(handles=[strCompPlot,primeFactPlot],loc=1)
plt.show()
```
---
## Conclusion
|
github_jupyter
|
%matplotlib inline
import random
import time
import copy
import numpy as np
import numpy.core.defchararray as npstr
import matplotlib.pyplot as plt
# generates a random string of letters for a given length
def generateWord(length):
abc = 'abcdefghijklmnopqrstuvwxyz'
word = ''
for i in range(length):
word += random.choice(abc)
return word
# generates a normally distributed list of numbers with a given length, mean and diviation
def generateLengths(count,mean, dist, showGraph = False):
s = np.random.normal(mean, dist, count)
s = np.round(s,0).astype(int)
lessThanOne = s < 1
s[lessThanOne] = 1
if showGraph:
plt.hist(s, 30, normed=True)
plt.title("Histogram of lengths")
plt.show()
return s
# generates a random dictionary of words whose lengths are normally distributed
def generateDictionary(count):
currentDict = []
wordLengths = generateLengths(count,5,1)
for length in wordLengths:
oldLen = len(currentDict)
while(len(currentDict) == oldLen):
word = generateWord(length)
if word not in currentDict:
currentDict.append(word)
return np.asarray(currentDict)
# Selects random words from a given dictionary that matches given length requirments
def pickRandomWordsFromDict(dictionary, minLength, maxLength, count):
output = []
for word in dictionary:
if maxLength >= len(word) >= minLength:
output.append(word)
if len(output)==count:
return output
return ValueError('Dictionary did not conatin enough words of your min length')
# generates a list of randomy generated strings of text made of of words from a given dictionary
# and where the number of words used in the text are normally distributed to a given mean and diviation
def generateComments(count,dictionary,mean,dist,showGraph = False):
comments = []
lengths = generateLengths(count,mean,dist,showGraph)
for length in lengths:
comment = ''
for i in range(length):
comment += random.choice(dictionary) + ' '
comments.append(comment.rstrip())
return np.asarray(comments)
generateWord(3)
generateLengths(100,5,1,True)
dictionary = generateDictionary(100)
dictionary [1:5]
pickRandomWordsFromDict(dictionary,3,8,3)
comments = generateComments(1000000,dictionary,4,1,True)
comments[1:5]
# Finds records that conatin specific words using numpy's string comparitor function
def findCommentsWithWords(comments,words):
currentVocab = copy.deepcopy(comments)
for w in words:
currentVocab = currentVocab[(npstr.find(currentVocab, w) != -1)]
return currentVocab
#Returns number of records that contain specific words
def numberOfMatches(comments, words):
result = findCommentsWithWords(comments,words)
return len(result)
randoWords = pickRandomWordsFromDict(dictionary,3,8,2)
randoWords
start = time.time()
print(str(numberOfMatches(comments,randoWords)) + ' number of records found containing ' + str(randoWords))
end = time.time()
print(str(end - start)+' seconds')
# Assigns a prime number to each word in a given dictionary and returns a dict object
def createIndexSchema(dictionary):
primes = np.genfromtxt ('primes.csv', delimiter=",").astype(int)
primeFit = primes[1:len(dictionary)+1,1]
index = dict(np.c_[dictionary,primeFit])
return index
# Converts a list of strings into the products of the words in the string's prime number indexes
def indexComments(comments,indexSchemal):
output = []
for comment in comments:
prod = 1
words = comment.split(' ')
for word in words:
if word in indexSchemal:
prod *= int(indexSchemal[word])
output.append(prod)
return output
# Return the indexed dictionary and text data for a given normal dictionary of words and list of text data
def preprocessPrimeComments(dictionary,comments):
indexSchemal = createIndexSchema(dictionary)
indexedComments = indexComments(comments,indexSchemal)
return [indexSchemal,indexedComments]
# converts given words into their indexed product
def convertWordsToProduct(indexSchema, words):
output = 1
for word in words:
output *= int(indexSchema[word])
return output
# calculates the modulo of the indexed text data against the given search word product
# and return a boolean list of which records match the search
def searchByPrimeFact(indexedComments,serachProduct):
return (np.mod(indexedComments, serachProduct) == 0)
# returns a boolean list of which of the given indexed records conatain
# the given words using the given indexed dictionary
def findCommentsUsingPrimeFact(indexedComments,index, words):
prod = convertWordsToProduct(index, words)
return searchByPrimeFact(indexedComments,prod)
[index, indexedComments] = preprocessPrimeComments(dictionary,comments)
start = time.time()
print(str(findCommentsUsingPrimeFact(indexedComments,index,randoWords).sum())+ ' number of records found containing ' + str(randoWords) )
end = time.time()
print(str(end - start)+' seconds')
# times the execution fo a function returning the seconds it took and the output of the function
def timeFunction(func, *params):
start = time.time()
output = func(*params)
end = time.time()
return[output, (end - start)]
# generate text records using the previously defined synthetic data function
def generateCommentData(dictLength, numOfComments, commentLengthMean, commentLengthDist, graph = False):
random.seed(50)
np.random.seed(15)
dictionary = generateDictionary(dictLength)
comments = generateComments(numOfComments,dictionary,commentLengthMean,commentLengthDist,graph)
return [dictionary, comments]
[d,c] = generateCommentData(300,100000,7,1, True)
[d,c] = generateCommentData(300,100000,7,1, False)
wordsToserachFor = pickRandomWordsFromDict(d,3,7,3)
[indx,indxCom] = preprocessPrimeComments(d,c)
stringCompTimings1 = []
primeTimings1 = []
xIncrement1 = []
for n in range(100,100000,1000):
xIncrement1.append(n)
[o,t] = timeFunction(numberOfMatches, c[:n], wordsToserachFor)
stringCompTimings1.append(t)
[o,t] = timeFunction(findCommentsUsingPrimeFact, indxCom[:n],indx, wordsToserachFor)
primeTimings1.append(t)
plt.figure()
plt.title("Plot of speed of search based on number of commenst to search through")
plt.ylabel("Time taken (seconds)")
plt.xlabel("Number of comments to search through")
strCompPlot, = plt.plot(xIncrement1,stringCompTimings1, label="Numpy String Compare")
primeFactPlot, = plt.plot(xIncrement1,primeTimings1, label="My Prime Factorisation")
plt.legend(handles=[strCompPlot,primeFactPlot],loc=2)
plt.show()
stringCompTimings2 = []
primeTimings2 = []
xIncrement2 = []
for n in range(1,100):
xIncrement2.append(n)
randWords2 = pickRandomWordsFromDict(d,2,7,n)
[o,t] = timeFunction(numberOfMatches, c[:100000], randWords2)
stringCompTimings2.append(t)
[o,t] = timeFunction(findCommentsUsingPrimeFact, indxCom[:10000],indx, randWords)
primeTimings2.append(t)
plt.figure()
plt.title("Plot of speed of search based on number of words searched for")
plt.ylabel("Time taken (seconds)")
plt.xlabel("Number of words searched for")
strCompPlot, = plt.plot(xIncrement2,stringCompTimings2, label="Numpy String Compare")
primeFactPlot, = plt.plot(xIncrement2,primeTimings2, label="My Prime Factorisation")
plt.legend(handles=[strCompPlot,primeFactPlot],loc=5)
plt.show()
stringCompTimings3 = []
primeTimings3 = []
xIncrement3 = []
for n in range(5,500,10):
xIncrement3.append(n)
[d3,c3] = generateCommentData(30,1000,n,1)
randWords3 = pickRandomWordsFromDict(d3,2,7,4)
[o,t] = timeFunction(numberOfMatches, c3, randWords3)
stringCompTimings3.append(t)
[indexSchema3, indxCom3] = preprocessPrimeComments(d3,c3)
[o,t] = timeFunction(findCommentsUsingPrimeFact, indxCom3,indexSchema3, randWords3)
primeTimings3.append(t)
plt.figure()
plt.title("Plot of speed of search based on length of comments to search through")
plt.ylabel("Time taken (seconds)")
plt.xlabel("Mean length of comments searched through")
strCompPlot, = plt.plot(xIncrement3,stringCompTimings3, label="Numpy String Compare")
primeFactPlot, = plt.plot(xIncrement3,primeTimings3, label="My Prime Factorisation")
plt.legend(handles=[strCompPlot,primeFactPlot],loc=2)
plt.show()
stringCompTimings4 = []
primeTimings4 = []
xIncrement4 = []
for n in range(10,1000,10):
xIncrement4.append(n)
[d4,c4] = generateCommentData(n,2000,10,1)
randWords4 = pickRandomWordsFromDict(d4,2,7,3)
[o,t] = timeFunction(numberOfMatches, c4, randWords4)
stringCompTimings4.append(t)
[indexSchema4, indxCom4] = preprocessPrimeComments(d4,c4)
[o,t] = timeFunction(findCommentsUsingPrimeFact, indxCom4,indexSchema4, randWords4)
primeTimings4.append(t)
plt.figure()
plt.title("Plot of speed of search based on length of dictionary")
plt.ylabel("Time taken (seconds)")
plt.xlabel("Number of unique words in text (dictionary length)")
strCompPlot, = plt.plot(xIncrement4,stringCompTimings4, label="Numpy String Compare")
primeFactPlot, = plt.plot(xIncrement4,primeTimings4, label="My Prime Factorisation")
plt.legend(handles=[strCompPlot,primeFactPlot],loc=1)
plt.show()
| 0.594904 | 0.973139 |
# Module 4: Measuring plant phenotypes with PlantCV - Multiple plants
[PlantCV homepage](https://plantcv.danforthcenter.org/)
[PlantCV documentation](https://plantcv.readthedocs.io/en/stable/)
```
%matplotlib widget
import matplotlib
from plantcv import plantcv as pcv
import numpy as np
import cv2
matplotlib.rcParams["figure.max_open_warning"] = False
pcv.params.debug = "plot"
pcv.params.text_size = 10
pcv.params.text_thickness = 10
pcv.params.line_thickness = 10
pcv.__version__
```
## Refresher: plant segmentation
```
# Open image file
img, imgpath, imgname = pcv.readimage(filename="images/10.9.1.244_pos-165-002-009_2020-02-29-20-05.jpg")
# Convert the RGB image into a grayscale image by choosing one of the HSV or LAB channels
gray_img = pcv.rgb2gray_lab(rgb_img=img, channel="a")
# Instead of setting a manual threshold, try an automatic threshold method such as Otsu
bin_img = pcv.threshold.otsu(gray_img=gray_img, max_value=255, object_type="dark")
# Remove "salt" noise from the binary image
filter_bin = pcv.fill(bin_img=bin_img, size=100)
```
## Measuring the shape and color of objects in digital image
At this stage we have a binary mask that labels plant pixels (white) and background pixels (black). There are multiple plants but we cannot tell which pixels belong to each plant
```
# Identify connected components (contours) using the binary image
cnt, cnt_str = pcv.find_objects(img=img, mask=filter_bin)
# Plot each contour to see where they are
pcv.params.color_sequence = "random"
cp = img.copy()
for i in range(0, len(cnt)):
cv2.drawContours(cp, cnt, i, pcv.color_palette(num=100, saved=False)[0], thickness=-1, hierarchy=cnt_str)
pcv.plot_image(cp)
```
We have distinct contours for each plant and some (most) plants are composed of multiple contours, how do we assign these to individual plants?
```
# Create a region of interest (ROI) for one plant
roi, roi_str = pcv.roi.circle(img=img, x=1460, y=1400, r=100)
# Filter the contours using the ROI
plant_cnt, plant_str, plant_mask, plant_area = pcv.roi_objects(img=img, roi_contour=roi, roi_hierarchy=roi_str, object_contour=cnt, obj_hierarchy=cnt_str)
# Flatten contours into a single object
# Combine the contours into a single plant object
plant, mask = pcv.object_composition(img=img, contours=plant_cnt, hierarchy=plant_str)
# Measure the size and shape of the plant
shape_img = pcv.analyze_object(img=img, obj=plant, mask=mask)
# Output measurements
print(f"Leaf area = {pcv.outputs.observations['default']['area']['value']} pixels")
print(f"Convex hull area = {pcv.outputs.observations['default']['convex_hull_area']['value']} pixels")
print(f"Solidity = {pcv.outputs.observations['default']['solidity']['value']}")
print(f"Perimeter = {pcv.outputs.observations['default']['perimeter']['value']} pixels")
print(f"Width = {pcv.outputs.observations['default']['width']['value']} pixels")
print(f"Height = {pcv.outputs.observations['default']['height']['value']} pixels")
print(f"Center of mass = {pcv.outputs.observations['default']['center_of_mass']['value']}")
print(f"Convex hull vertices = {pcv.outputs.observations['default']['convex_hull_vertices']['value']}")
print(f"Plant in frame = {pcv.outputs.observations['default']['object_in_frame']['value']}")
print(f"Bounding ellipse center = {pcv.outputs.observations['default']['ellipse_center']['value']}")
print(f"Bounding ellipse center major axis length = {pcv.outputs.observations['default']['ellipse_major_axis']['value']} pixels")
print(f"Bounding ellipse center minor axis length = {pcv.outputs.observations['default']['ellipse_minor_axis']['value']} pixels")
print(f"Bounding ellipse angle of rotation = {pcv.outputs.observations['default']['ellipse_angle']['value']} degrees")
print(f"Bounding ellipse eccentricity = {pcv.outputs.observations['default']['ellipse_eccentricity']['value']}")
# Measure the color properties of the plant
color_hist = pcv.analyze_color(rgb_img=img, mask=mask, colorspaces="hsv")
```

```
# Output measurements
print(f"Hue circular mean = {pcv.outputs.observations['default']['hue_circular_mean']['value']} degrees")
print(f"Hue circular mean standard deviation = {pcv.outputs.observations['default']['hue_circular_std']['value']} degrees")
print(f"Hue median = {pcv.outputs.observations['default']['hue_median']['value']} degrees")
```
|
github_jupyter
|
%matplotlib widget
import matplotlib
from plantcv import plantcv as pcv
import numpy as np
import cv2
matplotlib.rcParams["figure.max_open_warning"] = False
pcv.params.debug = "plot"
pcv.params.text_size = 10
pcv.params.text_thickness = 10
pcv.params.line_thickness = 10
pcv.__version__
# Open image file
img, imgpath, imgname = pcv.readimage(filename="images/10.9.1.244_pos-165-002-009_2020-02-29-20-05.jpg")
# Convert the RGB image into a grayscale image by choosing one of the HSV or LAB channels
gray_img = pcv.rgb2gray_lab(rgb_img=img, channel="a")
# Instead of setting a manual threshold, try an automatic threshold method such as Otsu
bin_img = pcv.threshold.otsu(gray_img=gray_img, max_value=255, object_type="dark")
# Remove "salt" noise from the binary image
filter_bin = pcv.fill(bin_img=bin_img, size=100)
# Identify connected components (contours) using the binary image
cnt, cnt_str = pcv.find_objects(img=img, mask=filter_bin)
# Plot each contour to see where they are
pcv.params.color_sequence = "random"
cp = img.copy()
for i in range(0, len(cnt)):
cv2.drawContours(cp, cnt, i, pcv.color_palette(num=100, saved=False)[0], thickness=-1, hierarchy=cnt_str)
pcv.plot_image(cp)
# Create a region of interest (ROI) for one plant
roi, roi_str = pcv.roi.circle(img=img, x=1460, y=1400, r=100)
# Filter the contours using the ROI
plant_cnt, plant_str, plant_mask, plant_area = pcv.roi_objects(img=img, roi_contour=roi, roi_hierarchy=roi_str, object_contour=cnt, obj_hierarchy=cnt_str)
# Flatten contours into a single object
# Combine the contours into a single plant object
plant, mask = pcv.object_composition(img=img, contours=plant_cnt, hierarchy=plant_str)
# Measure the size and shape of the plant
shape_img = pcv.analyze_object(img=img, obj=plant, mask=mask)
# Output measurements
print(f"Leaf area = {pcv.outputs.observations['default']['area']['value']} pixels")
print(f"Convex hull area = {pcv.outputs.observations['default']['convex_hull_area']['value']} pixels")
print(f"Solidity = {pcv.outputs.observations['default']['solidity']['value']}")
print(f"Perimeter = {pcv.outputs.observations['default']['perimeter']['value']} pixels")
print(f"Width = {pcv.outputs.observations['default']['width']['value']} pixels")
print(f"Height = {pcv.outputs.observations['default']['height']['value']} pixels")
print(f"Center of mass = {pcv.outputs.observations['default']['center_of_mass']['value']}")
print(f"Convex hull vertices = {pcv.outputs.observations['default']['convex_hull_vertices']['value']}")
print(f"Plant in frame = {pcv.outputs.observations['default']['object_in_frame']['value']}")
print(f"Bounding ellipse center = {pcv.outputs.observations['default']['ellipse_center']['value']}")
print(f"Bounding ellipse center major axis length = {pcv.outputs.observations['default']['ellipse_major_axis']['value']} pixels")
print(f"Bounding ellipse center minor axis length = {pcv.outputs.observations['default']['ellipse_minor_axis']['value']} pixels")
print(f"Bounding ellipse angle of rotation = {pcv.outputs.observations['default']['ellipse_angle']['value']} degrees")
print(f"Bounding ellipse eccentricity = {pcv.outputs.observations['default']['ellipse_eccentricity']['value']}")
# Measure the color properties of the plant
color_hist = pcv.analyze_color(rgb_img=img, mask=mask, colorspaces="hsv")
# Output measurements
print(f"Hue circular mean = {pcv.outputs.observations['default']['hue_circular_mean']['value']} degrees")
print(f"Hue circular mean standard deviation = {pcv.outputs.observations['default']['hue_circular_std']['value']} degrees")
print(f"Hue median = {pcv.outputs.observations['default']['hue_median']['value']} degrees")
| 0.727492 | 0.921358 |
## 说明
请按照填空顺序编号分别完成 参数优化,不同基函数的实现
```
import numpy as np
import matplotlib.pyplot as plt
def load_data(filename):
"""载入数据。"""
xys = []
with open(filename, 'r') as f:
for line in f:
xys.append(map(float, line.strip().split()))
xs, ys = zip(*xys)
return np.asarray(xs), np.asarray(ys)
```
## 不同的基函数 (basis function)的实现 填空顺序 2
请分别在这里实现“多项式基函数”以及“高斯基函数”
其中以及训练集的x的范围在0-25之间
```
def identity_basis(x):
ret = np.expand_dims(x, axis=1)
return ret
def multinomial_basis(x, feature_num=10):
'''多项式基函数'''
x = np.expand_dims(x, axis=1) # shape(N, 1)
#==========
#todo '''请实现多项式基函数'''
ret = [x]
for i in range(2,feature_num+1):
ret.append(x**i)
ret = np.concatenate(ret, axis=1)
#==========
return ret
def gaussian_basis(x, feature_num=10):
'''高斯基函数'''
#==========
#todo '''请实现高斯基函数'''
#print(x.shape)
centers = np.linspace(0, 25, feature_num)
width = 1.0 * (centers[1] - centers[0])
x = np.expand_dims(x, axis=1)
#print(x.shape)
#print([x]*feature_num)
x = np.concatenate([x]*feature_num, axis=1)
print(x.shape)
out = (x-centers)/width
ret = np.exp(-0.5 * out ** 2)
return ret
#==========
```
## 返回一个训练好的模型 填空顺序 1 用最小二乘法进行模型优化
## 填空顺序 3 用梯度下降进行模型优化
> 先完成最小二乘法的优化 (参考书中第二章 2.3中的公式)
> 再完成梯度下降的优化 (参考书中第二章 2.3中的公式)
在main中利用训练集训练好模型的参数,并且返回一个训练好的模型。
计算出一个优化后的w,请分别使用最小二乘法以及梯度下降两种办法优化w
```
def main(x_train, y_train):
"""
训练模型,并返回从x到y的映射。
"""
basis_func = gaussian_basis
phi0 = np.expand_dims(np.ones_like(x_train), axis=1)
phi1 = basis_func(x_train)
phi = np.concatenate([phi0, phi1], axis=1)
w = np.dot(np.linalg.pinv(phi), y_train)
#==========
#todo '''计算出一个优化后的w,请分别使用最小二乘法以及梯度下降两种办法优化w'''
#==========
def f(x):
phi0 = np.expand_dims(np.ones_like(x), axis=1)
phi1 = basis_func(x)
phi = np.concatenate([phi0, phi1], axis=1)
y = np.dot(phi, w)
return y
return f
```
## 评估结果
> 没有需要填写的代码,但是建议读懂
```
def evaluate(ys, ys_pred):
"""评估模型。"""
std = np.sqrt(np.mean(np.abs(ys - ys_pred) ** 2))
return std
# 程序主入口(建议不要改动以下函数的接口)
if __name__ == '__main__':
train_file = 'train.txt'
test_file = 'test.txt'
# 载入数据
x_train, y_train = load_data(train_file)
x_test, y_test = load_data(test_file)
print(x_train.shape)
print(x_test.shape)
# 使用线性回归训练模型,返回一个函数f()使得y = f(x)
f = main(x_train, y_train)
y_train_pred = f(x_train)
std = evaluate(y_train, y_train_pred)
print('训练集预测值与真实值的标准差:{:.1f}'.format(std))
# 计算预测的输出值
y_test_pred = f(x_test)
# 使用测试集评估模型
std = evaluate(y_test, y_test_pred)
print('预测值与真实值的标准差:{:.1f}'.format(std))
#显示结果
plt.plot(x_train, y_train, 'ro', markersize=3)
# plt.plot(x_test, y_test, 'k')
plt.plot(x_test, y_test_pred, 'k')
plt.xlabel('x')
plt.ylabel('y')
plt.title('Linear Regression')
plt.legend(['train', 'test', 'pred'])
plt.show()
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
def load_data(filename):
"""载入数据。"""
xys = []
with open(filename, 'r') as f:
for line in f:
xys.append(map(float, line.strip().split()))
xs, ys = zip(*xys)
return np.asarray(xs), np.asarray(ys)
def identity_basis(x):
ret = np.expand_dims(x, axis=1)
return ret
def multinomial_basis(x, feature_num=10):
'''多项式基函数'''
x = np.expand_dims(x, axis=1) # shape(N, 1)
#==========
#todo '''请实现多项式基函数'''
ret = [x]
for i in range(2,feature_num+1):
ret.append(x**i)
ret = np.concatenate(ret, axis=1)
#==========
return ret
def gaussian_basis(x, feature_num=10):
'''高斯基函数'''
#==========
#todo '''请实现高斯基函数'''
#print(x.shape)
centers = np.linspace(0, 25, feature_num)
width = 1.0 * (centers[1] - centers[0])
x = np.expand_dims(x, axis=1)
#print(x.shape)
#print([x]*feature_num)
x = np.concatenate([x]*feature_num, axis=1)
print(x.shape)
out = (x-centers)/width
ret = np.exp(-0.5 * out ** 2)
return ret
#==========
def main(x_train, y_train):
"""
训练模型,并返回从x到y的映射。
"""
basis_func = gaussian_basis
phi0 = np.expand_dims(np.ones_like(x_train), axis=1)
phi1 = basis_func(x_train)
phi = np.concatenate([phi0, phi1], axis=1)
w = np.dot(np.linalg.pinv(phi), y_train)
#==========
#todo '''计算出一个优化后的w,请分别使用最小二乘法以及梯度下降两种办法优化w'''
#==========
def f(x):
phi0 = np.expand_dims(np.ones_like(x), axis=1)
phi1 = basis_func(x)
phi = np.concatenate([phi0, phi1], axis=1)
y = np.dot(phi, w)
return y
return f
def evaluate(ys, ys_pred):
"""评估模型。"""
std = np.sqrt(np.mean(np.abs(ys - ys_pred) ** 2))
return std
# 程序主入口(建议不要改动以下函数的接口)
if __name__ == '__main__':
train_file = 'train.txt'
test_file = 'test.txt'
# 载入数据
x_train, y_train = load_data(train_file)
x_test, y_test = load_data(test_file)
print(x_train.shape)
print(x_test.shape)
# 使用线性回归训练模型,返回一个函数f()使得y = f(x)
f = main(x_train, y_train)
y_train_pred = f(x_train)
std = evaluate(y_train, y_train_pred)
print('训练集预测值与真实值的标准差:{:.1f}'.format(std))
# 计算预测的输出值
y_test_pred = f(x_test)
# 使用测试集评估模型
std = evaluate(y_test, y_test_pred)
print('预测值与真实值的标准差:{:.1f}'.format(std))
#显示结果
plt.plot(x_train, y_train, 'ro', markersize=3)
# plt.plot(x_test, y_test, 'k')
plt.plot(x_test, y_test_pred, 'k')
plt.xlabel('x')
plt.ylabel('y')
plt.title('Linear Regression')
plt.legend(['train', 'test', 'pred'])
plt.show()
| 0.279828 | 0.848345 |
```
!pip install pydgraph
import json
import pydgraph
from pydgraph import DgraphClient, DgraphClientStub
def set_process_schema(client):
schema = """node_key: string @upsert @index(hash) .
pid: int @index(int) .
created_time: int @index(int) .
asset_id: string @index(hash) .
terminate_time: int @index(int) .
image_name: string @index(exact, hash, trigram, fulltext) .
process_name: string @index(exact, hash, trigram, fulltext) .
arguments: string @index(fulltext) @index(trigram) .
bin_file: uid @reverse .
children: uid @reverse .
created_files: uid @reverse .
deleted_files: uid @reverse .
read_files: uid @reverse .
wrote_files: uid @reverse .
created_connection: uid @reverse .
bound_connection: uid @reverse ."""
op = pydgraph.Operation(schema=schema)
client.alter(op)
def set_file_schema(client):
schema = """
node_key: string @upsert @index(hash) .
file_name: string @index(exact, hash, trigram, fulltext) .
asset_id: string @index(exact, hash, trigram, fulltext) .
file_path: string @index(exact, hash, trigram, fulltext) .
file_extension: string @index(exact, hash, trigram, fulltext) .
file_mime_type: string @index(exact, hash, trigram, fulltext) .
file_size: int @index(int) .
file_version: string @index(exact, hash, trigram, fulltext) .
file_description: string @index(exact, hash, trigram, fulltext) .
file_product: string @index(exact, hash, trigram, fulltext) .
file_company: string @index(exact, hash, trigram, fulltext) .
file_directory: string @index(exact, hash, trigram, fulltext) .
file_inode: int @index(int) .
file_hard_links: string @index(exact, hash, trigram, fulltext) .
md5_hash: string @index(exact, hash, trigram, fulltext) .
sha1_hash: string @index(exact, hash, trigram, fulltext) .
sha256_hash: string @index(exact, hash, trigram, fulltext) .
"""
op = pydgraph.Operation(schema=schema)
client.alter(op)
def set_outbound_connection_schema(client, engagement=False):
schema = """
create_time: int @index(int) .
terminate_time: int @index(int) .
last_seen_time: int @index(int) .
ip: string @index(exact, trigram, hash) .
port: string @index(exact, trigram, hash) .
"""
op = pydgraph.Operation(schema=schema)
client.alter(op)
def set_external_ip_schema(client, engagement=False):
schema = """
external_ip: string @index(exact, trigram, hash) .
"""
op = pydgraph.Operation(schema=schema)
client.alter(op)
mclient = DgraphClient(DgraphClientStub('alpha0.mastergraphcluster.grapl:9080'))
eclient = DgraphClient(DgraphClientStub('alpha0.engagementgraphcluster.grapl:9080'))
# master nodes
set_process_schema(mclient)
set_file_schema(mclient)
set_outbound_connection_schema(mclient)
set_external_ip_schema(mclient)
# engagement nodes
set_process_schema(eclient)
set_file_schema(eclient)
set_outbound_connection_schema(eclient)
set_external_ip_schema(eclient)
```
|
github_jupyter
|
!pip install pydgraph
import json
import pydgraph
from pydgraph import DgraphClient, DgraphClientStub
def set_process_schema(client):
schema = """node_key: string @upsert @index(hash) .
pid: int @index(int) .
created_time: int @index(int) .
asset_id: string @index(hash) .
terminate_time: int @index(int) .
image_name: string @index(exact, hash, trigram, fulltext) .
process_name: string @index(exact, hash, trigram, fulltext) .
arguments: string @index(fulltext) @index(trigram) .
bin_file: uid @reverse .
children: uid @reverse .
created_files: uid @reverse .
deleted_files: uid @reverse .
read_files: uid @reverse .
wrote_files: uid @reverse .
created_connection: uid @reverse .
bound_connection: uid @reverse ."""
op = pydgraph.Operation(schema=schema)
client.alter(op)
def set_file_schema(client):
schema = """
node_key: string @upsert @index(hash) .
file_name: string @index(exact, hash, trigram, fulltext) .
asset_id: string @index(exact, hash, trigram, fulltext) .
file_path: string @index(exact, hash, trigram, fulltext) .
file_extension: string @index(exact, hash, trigram, fulltext) .
file_mime_type: string @index(exact, hash, trigram, fulltext) .
file_size: int @index(int) .
file_version: string @index(exact, hash, trigram, fulltext) .
file_description: string @index(exact, hash, trigram, fulltext) .
file_product: string @index(exact, hash, trigram, fulltext) .
file_company: string @index(exact, hash, trigram, fulltext) .
file_directory: string @index(exact, hash, trigram, fulltext) .
file_inode: int @index(int) .
file_hard_links: string @index(exact, hash, trigram, fulltext) .
md5_hash: string @index(exact, hash, trigram, fulltext) .
sha1_hash: string @index(exact, hash, trigram, fulltext) .
sha256_hash: string @index(exact, hash, trigram, fulltext) .
"""
op = pydgraph.Operation(schema=schema)
client.alter(op)
def set_outbound_connection_schema(client, engagement=False):
schema = """
create_time: int @index(int) .
terminate_time: int @index(int) .
last_seen_time: int @index(int) .
ip: string @index(exact, trigram, hash) .
port: string @index(exact, trigram, hash) .
"""
op = pydgraph.Operation(schema=schema)
client.alter(op)
def set_external_ip_schema(client, engagement=False):
schema = """
external_ip: string @index(exact, trigram, hash) .
"""
op = pydgraph.Operation(schema=schema)
client.alter(op)
mclient = DgraphClient(DgraphClientStub('alpha0.mastergraphcluster.grapl:9080'))
eclient = DgraphClient(DgraphClientStub('alpha0.engagementgraphcluster.grapl:9080'))
# master nodes
set_process_schema(mclient)
set_file_schema(mclient)
set_outbound_connection_schema(mclient)
set_external_ip_schema(mclient)
# engagement nodes
set_process_schema(eclient)
set_file_schema(eclient)
set_outbound_connection_schema(eclient)
set_external_ip_schema(eclient)
| 0.452778 | 0.183265 |
# Keras LSTM Text Generation
Text generation is a important nlp problem which can enable computers to write.
<table align="left"><td>
<a target="_blank" href="https://colab.research.google.com/github/TannerGilbert/Tutorials/blob/master/Keras-Tutorials/4.%20LSTM%20Text%20Generation/Keras%20LSTM%20Text%20Generation.ipynb">
<img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab
</a>
</td><td>
<a target="_blank" href="https://github.com/TannerGilbert/Tutorials/blob/master/Keras-Tutorials/4.%20LSTM%20Text%20Generation/Keras%20LSTM%20Text%20Generation.ipynb">
<img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td></table>
```
from __future__ import print_function
from tensorflow.keras.callbacks import LambdaCallback
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation
from tensorflow.keras.layers import LSTM
from tensorflow.keras.optimizers import RMSprop
import numpy as np
import random
import sys
!wget -O sherlock_homes.txt http://www.gutenberg.org/files/1661/1661-0.txt
text = open('sherlock_homes.txt', 'r').read().lower()
print('text length', len(text))
print(text[:1000])
```
## Map chars to integers
```
chars = sorted(list(set(text)))
print('total chars: ', len(chars))
char_indices = dict((c, i) for i, c in enumerate(chars))
indices_char = dict((i, c) for i, c in enumerate(chars))
```
## Split up into subsequences
```
maxlen = 40
step = 3
sentences = []
next_chars = []
for i in range(0, len(text) - maxlen, step):
sentences.append(text[i: i + maxlen])
next_chars.append(text[i + maxlen])
print('nb sequences:', len(sentences))
print(sentences[:3])
print(next_chars[:3])
x = np.zeros((len(sentences), maxlen, len(chars)), dtype=bool)
y = np.zeros((len(sentences), len(chars)), dtype=bool)
for i, sentence in enumerate(sentences):
for t, char in enumerate(sentence):
x[i, t, char_indices[char]] = 1
y[i, char_indices[next_chars[i]]] = 1
print(x[:3])
print(y[:3])
```
## Building Model
In this notebook a small recurrent neural networks is used for both simplicity and because of the training time but if you want to train a more sophisticated model you can increase the size of the network. You can also use a model pretrained on some other text like wikipedia text to both speed up the training process and get better results.
```
model = Sequential()
model.add(LSTM(128, input_shape=(maxlen, len(chars))))
model.add(Dense(len(chars)))
model.add(Activation('softmax'))
optimizer = RMSprop(learning_rate=0.01)
model.compile(loss='categorical_crossentropy', optimizer=optimizer)
```
## Helper Functions
These helper functions are taken from the [official Keras text generation notebook](https://github.com/keras-team/keras/blob/master/examples/lstm_text_generation.py).
```
def sample(preds, temperature=1.0):
# helper function to sample an index from a probability array
preds = np.asarray(preds).astype('float64')
preds = np.log(preds) / temperature
exp_preds = np.exp(preds)
preds = exp_preds / np.sum(exp_preds)
probas = np.random.multinomial(1, preds, 1)
return np.argmax(probas)
def on_epoch_end(epoch, logs):
# Function invoked at end of each epoch. Prints generated text.
print()
print('----- Generating text after Epoch: %d' % epoch)
start_index = random.randint(0, len(text) - maxlen - 1)
for diversity in [0.2, 0.5, 1.0, 1.2]:
print('----- diversity:', diversity)
generated = ''
sentence = text[start_index: start_index + maxlen]
generated += sentence
print('----- Generating with seed: "' + sentence + '"')
sys.stdout.write(generated)
for i in range(400):
x_pred = np.zeros((1, maxlen, len(chars)))
for t, char in enumerate(sentence):
x_pred[0, t, char_indices[char]] = 1.
preds = model.predict(x_pred, verbose=0)[0]
next_index = sample(preds, diversity)
next_char = indices_char[next_index]
generated += next_char
sentence = sentence[1:] + next_char
sys.stdout.write(next_char)
sys.stdout.flush()
print()
print_callback = LambdaCallback(on_epoch_end=on_epoch_end)
```
## Defining callbacks
```
from tensorflow.keras.callbacks import ModelCheckpoint
filepath = "weights.hdf5"
checkpoint = ModelCheckpoint(filepath, monitor='loss',
verbose=1, save_best_only=True,
mode='min')
from tensorflow.keras.callbacks import ReduceLROnPlateau
reduce_lr = ReduceLROnPlateau(monitor='loss', factor=0.2,
patience=1, min_lr=0.001)
callbacks = [print_callback, checkpoint, reduce_lr]
```
## Training the model
```
model.fit(x, y, batch_size=128, epochs=5, callbacks=callbacks)
```
## Testing the model
Now that we have a trained network we can test it using a method simular to the ``on_epoch_end`` method above.
```
def generate_text(length, diversity):
# Get random starting text
start_index = random.randint(0, len(text) - maxlen - 1)
generated = ''
sentence = text[start_index: start_index + maxlen]
generated += sentence
for i in range(length):
x_pred = np.zeros((1, maxlen, len(chars)))
for t, char in enumerate(sentence):
x_pred[0, t, char_indices[char]] = 1.
preds = model.predict(x_pred, verbose=0)[0]
next_index = sample(preds, diversity)
next_char = indices_char[next_index]
generated += next_char
sentence = sentence[1:] + next_char
return generated
print(generate_text(500, 0.2))
```
|
github_jupyter
|
from __future__ import print_function
from tensorflow.keras.callbacks import LambdaCallback
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation
from tensorflow.keras.layers import LSTM
from tensorflow.keras.optimizers import RMSprop
import numpy as np
import random
import sys
!wget -O sherlock_homes.txt http://www.gutenberg.org/files/1661/1661-0.txt
text = open('sherlock_homes.txt', 'r').read().lower()
print('text length', len(text))
print(text[:1000])
chars = sorted(list(set(text)))
print('total chars: ', len(chars))
char_indices = dict((c, i) for i, c in enumerate(chars))
indices_char = dict((i, c) for i, c in enumerate(chars))
maxlen = 40
step = 3
sentences = []
next_chars = []
for i in range(0, len(text) - maxlen, step):
sentences.append(text[i: i + maxlen])
next_chars.append(text[i + maxlen])
print('nb sequences:', len(sentences))
print(sentences[:3])
print(next_chars[:3])
x = np.zeros((len(sentences), maxlen, len(chars)), dtype=bool)
y = np.zeros((len(sentences), len(chars)), dtype=bool)
for i, sentence in enumerate(sentences):
for t, char in enumerate(sentence):
x[i, t, char_indices[char]] = 1
y[i, char_indices[next_chars[i]]] = 1
print(x[:3])
print(y[:3])
model = Sequential()
model.add(LSTM(128, input_shape=(maxlen, len(chars))))
model.add(Dense(len(chars)))
model.add(Activation('softmax'))
optimizer = RMSprop(learning_rate=0.01)
model.compile(loss='categorical_crossentropy', optimizer=optimizer)
def sample(preds, temperature=1.0):
# helper function to sample an index from a probability array
preds = np.asarray(preds).astype('float64')
preds = np.log(preds) / temperature
exp_preds = np.exp(preds)
preds = exp_preds / np.sum(exp_preds)
probas = np.random.multinomial(1, preds, 1)
return np.argmax(probas)
def on_epoch_end(epoch, logs):
# Function invoked at end of each epoch. Prints generated text.
print()
print('----- Generating text after Epoch: %d' % epoch)
start_index = random.randint(0, len(text) - maxlen - 1)
for diversity in [0.2, 0.5, 1.0, 1.2]:
print('----- diversity:', diversity)
generated = ''
sentence = text[start_index: start_index + maxlen]
generated += sentence
print('----- Generating with seed: "' + sentence + '"')
sys.stdout.write(generated)
for i in range(400):
x_pred = np.zeros((1, maxlen, len(chars)))
for t, char in enumerate(sentence):
x_pred[0, t, char_indices[char]] = 1.
preds = model.predict(x_pred, verbose=0)[0]
next_index = sample(preds, diversity)
next_char = indices_char[next_index]
generated += next_char
sentence = sentence[1:] + next_char
sys.stdout.write(next_char)
sys.stdout.flush()
print()
print_callback = LambdaCallback(on_epoch_end=on_epoch_end)
from tensorflow.keras.callbacks import ModelCheckpoint
filepath = "weights.hdf5"
checkpoint = ModelCheckpoint(filepath, monitor='loss',
verbose=1, save_best_only=True,
mode='min')
from tensorflow.keras.callbacks import ReduceLROnPlateau
reduce_lr = ReduceLROnPlateau(monitor='loss', factor=0.2,
patience=1, min_lr=0.001)
callbacks = [print_callback, checkpoint, reduce_lr]
model.fit(x, y, batch_size=128, epochs=5, callbacks=callbacks)
def generate_text(length, diversity):
# Get random starting text
start_index = random.randint(0, len(text) - maxlen - 1)
generated = ''
sentence = text[start_index: start_index + maxlen]
generated += sentence
for i in range(length):
x_pred = np.zeros((1, maxlen, len(chars)))
for t, char in enumerate(sentence):
x_pred[0, t, char_indices[char]] = 1.
preds = model.predict(x_pred, verbose=0)[0]
next_index = sample(preds, diversity)
next_char = indices_char[next_index]
generated += next_char
sentence = sentence[1:] + next_char
return generated
print(generate_text(500, 0.2))
| 0.571288 | 0.916857 |
# hicetnunc.xyz open dataset and parsers
<a href="https://creativecommons.org/licenses/by/4.0/"><img src="https://img.shields.io/badge/license-CC%20BY-green" /></a> <img src="https://img.shields.io/badge/python-3.6-yellow"/> <a href="https://hub.docker.com/repository/docker/pallada92/hicetnunc-dataset"><img src="https://img.shields.io/badge/docker%20hub-pallada92%2Fhicetnunc--dataset-blue" /></a>
[Hic et nunc](http://hicetnunc.xyz) is a new eco-friendly [NFT](https://en.wikipedia.org/wiki/Non-fungible_token) marketplace, built on top of [Tezos](https://en.wikipedia.org/wiki/Tezos) blockchain.
It is especially popular in generative graphics and data viz community, so I've decided to share data and all scripts that I've made for https://hashquine.github.io/hicetnunc rating.
It is published under [CC BY](https://creativecommons.org/licenses/by/4.0/) license, so that it is even possible to sell NFTs that use that data (or modified scripts) as long as there is the following phrase somewhere in the token description: `based on @hashquine dataset`.
Since hicetnunc servers are already under an extreme load due to quick growth, I've reorganized code, so that all data is taken from Tezos blockchain and IPFS **without any calls** to the [hicetnunc.xyz](http://hicetnunc.xyz) website or API.
## Data sources
* Blockchain transactions by [TzStats API](https://tzstats.com/docs/api#tezos-api) ([better-call.dev](https://better-call.dev) was not used in order not to interfere with hicetnunc backend).
* [IPFS](https://ru.wikipedia.org/wiki/IPFS) by [cloudflare-ipfs.com](https://cloudflare-ipfs.com/) and [ipfs.io](https://ipfs.io/) depending on mime type (same sources as in hicetnunc frontend).
* Wallet address owner metadata (name, Twitter etc.) from [api.tzkt.io](https://api.tzkt.io/#operation/Accounts_GetMetadata) (same source as in hicetnunc frontend).
## What data is available
* Money data: list of all purchases, prices and commissions.
* All NFTs raw files, their previews and thumbnails, although 3d files and interactive SVG/HTML files are not yet processed properly.
* Authors metadata [verified via tzkt.io](https://github.com/hicetnunc2000/hicetnunc/blob/main/FAQ.md#how-to-get-verified) like Twitter account address.
* Token transfers: list of changes of tokens owners including burns and direct transfers.
* All metadata available for tokens.
* Swaps and mints.
Data not available:
* Everything connected with [hDAO tokens](https://github.com/hicetnunc2000/hicetnunc/blob/main/FAQ.md#what-are-those-little-circles-on-each-post-hdao-what-is-that) and [hDAO feed](https://www.hicetnunc.xyz/hdao). Although all related transactions are already being collected, they are not analysed yet.
* Twitter statistics like the number of followers.
* Direct money transfers between users, when NFT tokens are not transferred in the same transaction.
## Dataset schema
The goal was to simplify data analysis and visualization with a wide range of existing tools, so there are lots of redundant fields, which contain precalculated aggregations and different representations of the same data.
All files have two equivalent versions: JSON and CSV.
* JSON files are dictionary of dictionaries with rows of CSV files are indexed by the `*_id` field.
* CSV files have commas as delimiters.
* Fields values are ether numbers or strings, empty values represented by `-1` or `""`.
* All identifiers are strings.
Any field, which references some event in the blockchain (for example, mint time) have 4 representations:
* `mint_iso_date` — string with UTC date and time: `"2021-03-01T15:00:00Z"`,
* `mint_stamp` — integer Unix timestamp in seconds: `1614610800`,
* `mint_hash` — string with transaction hash, where event occurred: `"oom5Ju6X9nYpBCi..."`,
* `mint_row_id` — integer with global unique operation id (internal to TzStats) with that event: `42181049`
Any field, which references a set of values (like the set of prices of sold works), have following aggregations:
* `sold_count` — values count,
* `sold_nonzero_count` — number of positive values,
* `sold_zero_count` — number of zeros,
* `sold_price_min` — minimum value (excl. zeros),
* `sold_price_max` — maximum value,
* `sold_price_sum` — sum of values,
* `sold_price_avg` — average value (sum divided by count excl. zeros).
## Essential information about contracts logic
There are 3 Tezos addresses, which are common to most of hicetnunc transactions:
* **"NFT" contract**: [KT1RJ6PbjHpwc3M5rw5s2Nbmefwbuwbdxton](https://tzstats.com/KT1RJ6PbjHpwc3M5rw5s2Nbmefwbuwbdxton).
* This is the registry of all owners of all NFT tokens.
* This is the typical way how most of NFTs works on Ethereum or on Tezos.
* Click on "Bigmap #511" tab in Tezos explorer to see registry of token owners.
* This contract is the single "source of truth" about current owners of all NFT tokens issued by hicetnunc.
If there is no information about token owner in the registry, than that person doesn't own any tokens.
* This contract also the registry of tokens infos (metadata).
* Token info is a small JSON structure stored on on IPFS.
* [Here is an example](https://ipfs.tzstats.com/ipfs/Qme3LXQF2UaqCx1ksDtHSSFTmuER8AehoayHKoTvfT9rQ6) of such structure.
* It contains link to IPFS with NFT binary contents (some image, for example).
* It also contains title, description, creator and tags.
* It **does not** contain price or related information.
* Only link to IPFS is stored on the blockchain.
* Note, that, however, there is no way to alter token metadata after minting.
* Click on "Bigmap #514" tab in Tezos explorer to see mapping from tokens to IPFS urls.
* Every token owner can call "transfer" method of the contract to send tokens to other address.
* This contract can't do any money related operations.
Money logic should be implemented in other contracts, which call "NFT" contract as a part of transaction operation.
* There is also a "mint" method in this contract, but it can only be called by the "Art house" contract.
* **"Art house" contract**: [KT1Hkg5qeNhfwpKW4fXvq7HGZB9z2EnmCCA9](https://tzstats.com/KT1Hkg5qeNhfwpKW4fXvq7HGZB9z2EnmCCA9).
* This contract implements money related operations on hicetnunc.
* It's main structure is a swap. It is some amount of tokens, which are available of sale for specific price.
* Click on "Bigmap #523" tab in Tezos explorer to see all current swaps.
* Note, that there may be other contracts implementing swap mechanism. These contracts may decide not to pay comission or royalties.
* Objects can be minted only with this contract by calling "mint_OBJKT" method.
* This contract keeps track of royalties and assigns tokens ids.
* **Comission wallet:** [tz1UBZUkXpKGhYsP5KtzDNqLLchwF4uHrGjw](https://tzstats.com/tz1UBZUkXpKGhYsP5KtzDNqLLchwF4uHrGjw).
* 2.5% of every purchase via "Art house" contract swaps is sent to this wallet.
There are several other contracts related to [curation](https://tzstats.com/KT1TybhR7XraG75JFYKSrh7KnxukMBT5dor6) and [hDAO](https://tzstats.com/KT1AFA2mwNUMNd4SsujE1YYp29vd8BZejyKW) mechanisms, which are independent from the contracts mentioned above.
Actually, hicetnunc was not created just as another NFT marketplace, it has much broader mission as hDAO (hicetnunc [DAO](https://en.wikipedia.org/wiki/The_DAO_(organization))). You can get the idea of the creators vision [on hicetnunc blog](https://hicetnunc2000.medium.com/). As a result, only a small subset of contract's logic is actually used during hicetnunc website operation.
### Official and external swap mechanisms
* Official swap mechanism by hicetnunc "Art house" contract.
* Any token bought on hicetnunc website is a part of some swap.
* Swap is just some amount of tokens, which are offered for sale by specific price.
* When swap is created, the seller sends all offered tokens to the "Art house" smart contract.
* Then anybody can send the required amount of money to the "collect" method of contract and get tokens in return.
* 2.5% of comission is transferred to hicetnunc comission wallet.
* 10% of royalties (this parameter is configurable in general) is transferred to token author.
* rest of money is sent to swap creator.
* proportional amount of hDAO tokens are also sent to buyer, seller, token author and comission wallet.
* Seller can cancel swap any time and get unsold tokens back.
* Swaps can be created by any token owner any number of times.
* In this dataset official swaps are treated as `author->swap`, `other->swap`, `swap->author`, `swap->other` transfers.
* External swap mechanisms.
* Since "transfer" method of "NFT" contract can be called by any token owner directly, it is possible to make custom smart contracts, which implement any desired logic.
* These custom contracts are not required not pay comission or royalties to hicetnunc.
* In general, swap contracts can be used to exchange any types entities.
* Example: https://quipuswap.com/swap
* In this dataset external swaps are treated as `other->other` transfers. The related price is guessed heuristically (as half of money transferred in all operations) and may not be always correct.
### Token lifecycle
In contrast to NFT definition, each NFT artwork in hicetnunc can have multiple copies, which are fungible. The NFT contract only tracks the amount of copies owned by each address. This means, that there is no way (even in theory) to track history of single copy like [it can be done on OpenSea](https://opensea.io/assets/0x06012c8cf97bead5deae237070f9587f8e7a266d/1864227), for example.
It is possible, however, to track history of token groups to some extent. Here is a list of possible owner types in this dataset:
* `author` — the person, who created the tokens during mint.
* `user` — any other hicetnunc user.
* `ext` — any external contract (external swap mechanism, for example).
* `burn` — reserved address for burning tokens.
* `swap` — when tokens are offered on sale in official swaps.
List of possible transitions:
* `mint->author`, `mint->user` First, every token should be minted.
* For each token type there may exist only single mint operation. It is impossible to mint additional tokens later.
* The only way to mint a token is to call "mint_OBJKT" method in "Art house" contract.
* [Here is](https://tzstats.com/ooVQqSXkhKHKi6ZDbT5tUxftLYNvC3zpuPrb8qWBEyjwy1hASLv) a typical mint transaction
* Internally it calls "mint" method in "NFT" contract.
* In dataset the sender is empty for mint operations.
* As result of mint operation, all tokens are transferred to some address. In 99% of cases this is the transaction sender, but sometimes it is different.
* Royalties are always sent to the mint transaction sender.
* `author->swap`, `user->swap` Any token owner can create official swap.
* Hicetnunc swap is created by calling "swap" method in "Art house" contract.
* [Here is](https://tzstats.com/opYXNWa6Cs8LoFvsguVpKocmQ6JpksSuTRWcACSfCkuY4UTNkhC) a typical swap creation transaction by author.
* Internally tokens are transferred to the "Art house" address.
* `swap->author`, `swap->other` There are two situations, when tokens may be transferred from a swap.
1. Purchase
* When token is purchased on hicetnunc website, it is transferred to the buyer. This is the main operation on hicetnunc.
* Buyer should call "collect" method of "Art house" contract and send required amount of money with it.
* [Here is](https://tzstats.com/op7ft9rqdYvbctZ5NFw2wPDmioBx29nPREgeZgxmdypxL5nxyAk) an example of "collect" transaction.
* First 3 internal operations send money to token creator (royalties), hicetnunc wallet (comission) and to the seller (which is the same as token creator in some cases) in that order.
* Fourth operation creates hDAO tokens and sends them to the buyer, seller and hicetnunc wallet. These tokens have special meaning and are not tracked in this dataset.
* Last internal operation does the actual token transfer.
* Note, that case of zero price is handled differently.
* [Here is](https://tzstats.com/ooKbTDkkT9fHoXxrkN5cAEFfbrXnd47YZuGHR991YzEtqneeGrQ) an example of purchase with zero price.
2. Swap cancel
* When swap creator decides to cancel swap, all remaining tokens are transferred back to him.
* `author->user`, `user->user` Any token owner can transfer tokens directly to other users for free by calling "transfer" method of "NFT" contract.
* [Here is](https://tzstats.com/ooDEeiWKwk7eL4DgUELErf6qkycYisbehWZsU3R1M2XWA5DKW2P) an example of direct transfer transaction from author to other user.
* `author->ext`, `user->ext`, `ext->user`, `ext->author` — external swaps
* [Here is](https://tzstats.com/ooF1bszbutpvvb5LWrcmd5A1WoqSKGicB2wr7SsVruKbWoaDasD) an example of external swap.
* `author->burn`, `other->burn` Any token owner can transfer tokens to burn address <code>tz1burnburnburnburnburnburnburjAYjjX</code>.
* [Here is](https://tzstats.com/ooDEeiWKwk7eL4DgUELErf6qkycYisbehWZsU3R1M2XWA5DKW2P) and example of burn transfer from author.
* Tokens can never be transferred from burn address since it is impossible to retrieve its private key (similar to how it is impossible to reverse hash containing all zeros).
## Details about edge cases
### How to define the author of the token
1. `mint_sender` The address of the sender of the "mint" transaction.
* This is the person, who receives royalties in hicetnunc.
* In this dataset token author this is equivalent to token author.
2. `issuer` The address of the receiver of tokens after the mint transaction.
* It is also the first parameter of the "mint" call in "Art house" contract.
* [Here is](https://tzstats.com/ooVQqSXkhKHKi6ZDbT5tUxftLYNvC3zpuPrb8qWBEyjwy1hASLv) an example of mint, where transaction sender and token issuer are different.
3. `info_creator` Field "creators" in JSON in token metadata.
* [Here is](https://tzstats.com/onu5q4QMQRFD7NsFDWjk5WaeBk8PR2orHhmN6M7qntaTFyMGjJD) an example of mint, where metadata creator field is different from transaction sender and issuer.
* As of 4th of April, it always has single entry.
* Sometimes it is empty.
* [Here is](https://tzstats.com/op8uvfPYcy1Yofn9eCgjrfNvFJkpZfjT6wpP8yezczXxPJcc8Pa) an example of mint with empty metadata creator field.
* Note, that [corresponding token page](https://www.hicetnunc.xyz/objkt/12123) has a bug, that it shows token owner controls on token page.
### Hicetnunc core addresses can own NFTs as regular users
* Any user can send any NFT tokens to "NFT" or "Art house contract"
* Technically, it has the same effect as sending this tokens to burn address, since contracts were not programmed to send their own NFTs (except from swap mechanism) under any circumstances.
* Comission wallet sometimes mint NFTs and buys them from other users.
* Since it is not a contract and manipulated by a real person (hicetnunc creator).
### Void transactions
* It is possible to send 0 tokens. [Example](https://tzstats.com/ooXTr2AJBN95EiN3u7NcUg5K7Pkd8nRHNRxa8CbxRNhQZEW4QLN).
* Sender and receiver can be the same. [Example](https://tzstats.com/opUVg6edpbHtJ94VgHTcwbDnodoegKKwmQ8C6iC9avVX6vZPQd4)
### Strange issue with negative swap count:
* https://tzstats.com/opYoTN4LUvNq7F5oiq7a6frW1Q5UubQuXSrDmrqrJGJzHnKaJQQ/47335724
* https://tzstats.com/oo7Sr2daVXamKTDtajRRvJGmTmYAantjJeLoPkTD6hraVHXQR1B/47335862
## How to update dataset
Note, that the code is still experimental and may require substantial changes to make it run. But you can read introduction in [HACKING.md](./HACKING.md)
```
%run -i ../../src/reload.py
import src
import config
import json
from pathlib import Path
nb_json = src.utils.read_json(Path('./make_readme.ipynb'))
assert nb_json['cells'][0]['cell_type'] == 'markdown'
readme_intro = ''.join(nb_json['cells'][0]['source'])
readme_outro = ''.join(nb_json['cells'][1]['source'])
fields_list = src.utils.read_json(config.datasets_fields_file)
tokens_ds_fields = fields_list['tokens']
addrs_ds_fields = fields_list['addrs']
sells_ds_fields = fields_list['sells']
transfers_ds_fields = fields_list['transfers']
swaps_ds_fields = fields_list['swaps']
Path('../../README.md').write_text(readme_intro + f'''
### [tokens.json](./dataset/tokens.json) and [tokens.csv](./dataset/tokens.csv) — of all NFTs tokens
There is a confusing fact, that in hicetnunc each NFT can have multiple identical instances, which are fungible.
In this document term "token" refers to the set of all that instances.
There are following invariants:
<pre>mint_count = author_owns_count + available_count + available_zero_count + other_own_count + burn_count
author_sent_count <= other_own_count + available_count + available_zero_count</pre>
{src.formatters.md_fields_schema.db_fields_schema_to_md(tokens_ds_fields)}
### [addrs.json](./dataset/addrs.json) and [addrs.csv](./dataset/addrs.csv) — of all hicetnunc users
All users, who ever created or owned NFT token.
{src.formatters.md_fields_schema.db_fields_schema_to_md(addrs_ds_fields)}
### [sells.json](./dataset/sells.json) and [sells.csv](./dataset/sells.csv) — of all purchases via "official" hicetnunc swaps
There is the following invariant:
<pre>price * count = total_royalties + total_comission + total_seller_income</pre>
{src.formatters.md_fields_schema.db_fields_schema_to_md(sells_ds_fields)}
### [transfers.json](./dataset/transfers.json) and [transfers.csv](./dataset/transfers.csv) — all token transfers
{src.formatters.md_fields_schema.db_fields_schema_to_md(transfers_ds_fields)}
### [swaps.json](./dataset/swaps.json) and [swaps.csv](./dataset/swaps.csv) — all "official" hicetnunc swaps ever created
{src.formatters.md_fields_schema.db_fields_schema_to_md(swaps_ds_fields)}
''' + readme_outro, 'utf-8')
total_cols = 0
total_rows = 0
for ds_id, ds in fields_list.items():
ds_json = src.utils.read_json(config.dataset_dir / (ds_id + '.json'))
total_rows += len(ds_json)
for field_group in ds.values():
total_cols += len(field_group)
print(len(fields_list), 'tables', total_cols, 'columns', total_rows, 'rows')
```
|
github_jupyter
|
%run -i ../../src/reload.py
import src
import config
import json
from pathlib import Path
nb_json = src.utils.read_json(Path('./make_readme.ipynb'))
assert nb_json['cells'][0]['cell_type'] == 'markdown'
readme_intro = ''.join(nb_json['cells'][0]['source'])
readme_outro = ''.join(nb_json['cells'][1]['source'])
fields_list = src.utils.read_json(config.datasets_fields_file)
tokens_ds_fields = fields_list['tokens']
addrs_ds_fields = fields_list['addrs']
sells_ds_fields = fields_list['sells']
transfers_ds_fields = fields_list['transfers']
swaps_ds_fields = fields_list['swaps']
Path('../../README.md').write_text(readme_intro + f'''
### [tokens.json](./dataset/tokens.json) and [tokens.csv](./dataset/tokens.csv) — of all NFTs tokens
There is a confusing fact, that in hicetnunc each NFT can have multiple identical instances, which are fungible.
In this document term "token" refers to the set of all that instances.
There are following invariants:
<pre>mint_count = author_owns_count + available_count + available_zero_count + other_own_count + burn_count
author_sent_count <= other_own_count + available_count + available_zero_count</pre>
{src.formatters.md_fields_schema.db_fields_schema_to_md(tokens_ds_fields)}
### [addrs.json](./dataset/addrs.json) and [addrs.csv](./dataset/addrs.csv) — of all hicetnunc users
All users, who ever created or owned NFT token.
{src.formatters.md_fields_schema.db_fields_schema_to_md(addrs_ds_fields)}
### [sells.json](./dataset/sells.json) and [sells.csv](./dataset/sells.csv) — of all purchases via "official" hicetnunc swaps
There is the following invariant:
<pre>price * count = total_royalties + total_comission + total_seller_income</pre>
{src.formatters.md_fields_schema.db_fields_schema_to_md(sells_ds_fields)}
### [transfers.json](./dataset/transfers.json) and [transfers.csv](./dataset/transfers.csv) — all token transfers
{src.formatters.md_fields_schema.db_fields_schema_to_md(transfers_ds_fields)}
### [swaps.json](./dataset/swaps.json) and [swaps.csv](./dataset/swaps.csv) — all "official" hicetnunc swaps ever created
{src.formatters.md_fields_schema.db_fields_schema_to_md(swaps_ds_fields)}
''' + readme_outro, 'utf-8')
total_cols = 0
total_rows = 0
for ds_id, ds in fields_list.items():
ds_json = src.utils.read_json(config.dataset_dir / (ds_id + '.json'))
total_rows += len(ds_json)
for field_group in ds.values():
total_cols += len(field_group)
print(len(fields_list), 'tables', total_cols, 'columns', total_rows, 'rows')
| 0.302803 | 0.838878 |
# EDA on Stroke data
Link to dataset: [Stroke Prediction Dataset](https://www.kaggle.com/fedesoriano/stroke-prediction-dataset)
## Import
Import necessary libraries and read the dataset.
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
import warnings
warnings.filterwarnings('ignore')
```
Explore the dataset and check for missing data.
```
strokeDF = pd.read_csv('healthcare-dataset-stroke-data.csv') # Read CSV file into dataframe.
strokeDF.head() # Get and print the first 5 rows.
strokeDF.info() # Getting information about the dataframe.
```
There are missing data in the 'bmi' column.
## Exploratory Data Analysis
```
print(strokeDF.shape) # Get rows and columns from dataframe.
print('The dataset has {} rows and {} columns.'.format(strokeDF.shape[0], strokeDF.shape[1]))
strokeDF.describe() # Generate descriptive statistics on the dataframe.
strokeDF.describe(include='object') # Generate descriptive statistics including object types.
```
### Summary Statistics
```
# Calculate mean for each numerical field.
strokeDF.mean()
# Calculate median for each numerical field.
strokeDF.median()
# Calculate standard deviation for each numerical field.
strokeDF.std()
# Calculate variance for each numerical field.
strokeDF.var()
# Data distribution
strokeDF.hist(figsize=(20,20), color='red')
plt.show()
# Use seaborn to lot a histogram to compare count of stroke.
sns.factorplot('stroke',data=strokeDF,kind='count')
print("Number of patient with stroke: " + str(len(strokeDF[strokeDF['stroke'] == 1])))
print("Number of patient without stroke: " + str(len(strokeDF[strokeDF['stroke'] == 0])))
```
We have more patients with no stroke than stroke in this dataset.
```
plt.figure(figsize = (10,6))
plt.scatter(x=strokeDF['age'], y=strokeDF['avg_glucose_level'], alpha=0.5)
plt.ylabel('avg_glucose_level')
plt.xlabel('age')
plt.colorbar()
plt.show()
```
From the scatter plot above, it is observed that old-aged patient has higher average glocose level generally.
```
sns.lmplot('age', 'avg_glucose_level', data=strokeDF, fit_reg= False, aspect=2, size =4, hue='stroke')
```
Old-aged patient has higher chance of getting stroke regardless of their average glucose level.
```
groupedDF = strokeDF[['gender','work_type','smoking_status','stroke']]
summary = pd.concat([pd.crosstab(groupedDF[x], groupedDF.stroke) for x in groupedDF.columns[:-1]], keys=groupedDF.columns[:-1])
summary
plt.figure(figsize=(10,5))
strok=strokeDF.loc[strokeDF['stroke']==1]
sns.countplot(data=strok,x='ever_married');
```
From the hsitogram above, it is concluded that getting married increase chances of getting stroke. 😂
```
plt.figure(figsize=(10,5))
sns.countplot(data=strok, x='work_type');
```
Working at private sector has higher chance of getting you a stroke.
This simple EDA on the dataset is performed as part of a university assignment.
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
import warnings
warnings.filterwarnings('ignore')
strokeDF = pd.read_csv('healthcare-dataset-stroke-data.csv') # Read CSV file into dataframe.
strokeDF.head() # Get and print the first 5 rows.
strokeDF.info() # Getting information about the dataframe.
print(strokeDF.shape) # Get rows and columns from dataframe.
print('The dataset has {} rows and {} columns.'.format(strokeDF.shape[0], strokeDF.shape[1]))
strokeDF.describe() # Generate descriptive statistics on the dataframe.
strokeDF.describe(include='object') # Generate descriptive statistics including object types.
# Calculate mean for each numerical field.
strokeDF.mean()
# Calculate median for each numerical field.
strokeDF.median()
# Calculate standard deviation for each numerical field.
strokeDF.std()
# Calculate variance for each numerical field.
strokeDF.var()
# Data distribution
strokeDF.hist(figsize=(20,20), color='red')
plt.show()
# Use seaborn to lot a histogram to compare count of stroke.
sns.factorplot('stroke',data=strokeDF,kind='count')
print("Number of patient with stroke: " + str(len(strokeDF[strokeDF['stroke'] == 1])))
print("Number of patient without stroke: " + str(len(strokeDF[strokeDF['stroke'] == 0])))
plt.figure(figsize = (10,6))
plt.scatter(x=strokeDF['age'], y=strokeDF['avg_glucose_level'], alpha=0.5)
plt.ylabel('avg_glucose_level')
plt.xlabel('age')
plt.colorbar()
plt.show()
sns.lmplot('age', 'avg_glucose_level', data=strokeDF, fit_reg= False, aspect=2, size =4, hue='stroke')
groupedDF = strokeDF[['gender','work_type','smoking_status','stroke']]
summary = pd.concat([pd.crosstab(groupedDF[x], groupedDF.stroke) for x in groupedDF.columns[:-1]], keys=groupedDF.columns[:-1])
summary
plt.figure(figsize=(10,5))
strok=strokeDF.loc[strokeDF['stroke']==1]
sns.countplot(data=strok,x='ever_married');
plt.figure(figsize=(10,5))
sns.countplot(data=strok, x='work_type');
| 0.58948 | 0.909425 |
# Plotting Depth and Cyanobacteria (phylum) reads, bacterias that obtain their energy through photosynthesis, normalised by number of InterPro annotations, for Tara Oceans project PRJEB1787.
The following task shows how to analysie metadata and annotations retrieved from the EMG API and combined on the fly to generate the visualisations.
```
import copy
try:
from urllib import urlencode
except ImportError:
from urllib.parse import urlencode
from pandas import DataFrame
import matplotlib.pyplot as plt
import numpy as np
from jsonapi_client import Session, Filter
API_BASE = 'https://www.ebi.ac.uk/metagenomics/api/latest/'
```
List all runs
https://www.ebi.ac.uk/metagenomics/api/latest/runs?experiment_type=metagenomic&study_accession=PRJEB1787
```
def find_metadata(metadata, key):
"""
Extract metadata value for given key
"""
for m in metadata:
if m['key'].lower() == key.lower():
return m['value']
return None
metadata_key = 'geographic location (depth)'
normilize_key = 'Predicted CDS with InterProScan match'
# map GO terms to the temperature
result = {}
with Session(API_BASE) as s:
# temporary dict to store accession and metadata
metadata_map = {}
# list of runs missing metadata
missing_meta = list()
print('Loading data from API.', end='', flush=True)
# preparing url
params = {
'experiment_type': 'metagenomic',
'study_accession': 'ERP001736',
'page_size': 100,
'include': 'sample',
}
f = Filter(urlencode(params))
# list runs
for anls in s.iterate('analyses', f):
print('.', end='', flush=True)
# find temperature for each run
try:
m_value = float(find_metadata(anls.sample.sample_metadata, metadata_key))
except:
m_value = None
if m_value is not None:
metadata_map[anls.accession] = m_value
else:
# missing value, skip run!
missing_meta.append(anls.accession)
continue
_pcds = int(find_metadata(anls.analysis_summary, normilize_key))
if _pcds is None:
# missing value, skip run!
continue
_temperature = metadata_map[anls.accession]
try:
result[_temperature]
except KeyError:
result[_temperature] = {}
# list a summary of GO terms derived from InterPro matches
for ann in anls.taxonomy:
try:
ann.hierarchy['phylum']
except KeyError:
continue
if len(ann.hierarchy['phylum']) > 0:
l = "{}:{}".format(ann.hierarchy['kingdom'], ann.hierarchy['phylum'])
try:
result[_temperature][l]
except KeyError:
result[_temperature][l] = list()
# normalize annotation counts, adjusting value
_norm = int(ann.count)/_pcds
# assign value
result[_temperature][l].append(_norm)
print("DONE")
# print("Missing: ", missing_meta)
```
### Clean up data
```
# remove invalid temperatures
for k in copy.deepcopy(list(result.keys())):
if k > 2000:
del result[k]
# average value of the same temperature
for k in result:
for k1 in result[k]:
result[k][k1] = np.mean(result[k][k1])
```
### Calculate correlation
```
from scipy.stats import spearmanr
df = DataFrame(result)
df_go = df.T[['Bacteria:Cyanobacteria']].copy()
x = df_go.index.tolist()
correl = []
correl_p = []
for k in df_go.keys():
y = list(df_go[k])
rho, p = spearmanr(x, y)
correl.append(rho)
correl_p.append(p)
df_go.loc['rho'] = correl
df_go.loc['p'] = correl_p
df_go
```
### Plot
```
df = DataFrame(result)
df_go_plot = df.T[['Bacteria:Cyanobacteria']].copy()
pl = df_go_plot.plot(
y=['Bacteria:Cyanobacteria'], use_index=True, style='o', figsize=(8,5),
title='Depth and Cyanobacteria (phylum) reads, bacterias that obtain their energy through photosynthesis, normalised by number of InterPro annotations, for Tara Oceans project PRJEB1787',
)
pl.set_xlabel("Depth m")
pl.set_ylabel("Relative abundance")
plt.show()
```
|
github_jupyter
|
import copy
try:
from urllib import urlencode
except ImportError:
from urllib.parse import urlencode
from pandas import DataFrame
import matplotlib.pyplot as plt
import numpy as np
from jsonapi_client import Session, Filter
API_BASE = 'https://www.ebi.ac.uk/metagenomics/api/latest/'
def find_metadata(metadata, key):
"""
Extract metadata value for given key
"""
for m in metadata:
if m['key'].lower() == key.lower():
return m['value']
return None
metadata_key = 'geographic location (depth)'
normilize_key = 'Predicted CDS with InterProScan match'
# map GO terms to the temperature
result = {}
with Session(API_BASE) as s:
# temporary dict to store accession and metadata
metadata_map = {}
# list of runs missing metadata
missing_meta = list()
print('Loading data from API.', end='', flush=True)
# preparing url
params = {
'experiment_type': 'metagenomic',
'study_accession': 'ERP001736',
'page_size': 100,
'include': 'sample',
}
f = Filter(urlencode(params))
# list runs
for anls in s.iterate('analyses', f):
print('.', end='', flush=True)
# find temperature for each run
try:
m_value = float(find_metadata(anls.sample.sample_metadata, metadata_key))
except:
m_value = None
if m_value is not None:
metadata_map[anls.accession] = m_value
else:
# missing value, skip run!
missing_meta.append(anls.accession)
continue
_pcds = int(find_metadata(anls.analysis_summary, normilize_key))
if _pcds is None:
# missing value, skip run!
continue
_temperature = metadata_map[anls.accession]
try:
result[_temperature]
except KeyError:
result[_temperature] = {}
# list a summary of GO terms derived from InterPro matches
for ann in anls.taxonomy:
try:
ann.hierarchy['phylum']
except KeyError:
continue
if len(ann.hierarchy['phylum']) > 0:
l = "{}:{}".format(ann.hierarchy['kingdom'], ann.hierarchy['phylum'])
try:
result[_temperature][l]
except KeyError:
result[_temperature][l] = list()
# normalize annotation counts, adjusting value
_norm = int(ann.count)/_pcds
# assign value
result[_temperature][l].append(_norm)
print("DONE")
# print("Missing: ", missing_meta)
# remove invalid temperatures
for k in copy.deepcopy(list(result.keys())):
if k > 2000:
del result[k]
# average value of the same temperature
for k in result:
for k1 in result[k]:
result[k][k1] = np.mean(result[k][k1])
from scipy.stats import spearmanr
df = DataFrame(result)
df_go = df.T[['Bacteria:Cyanobacteria']].copy()
x = df_go.index.tolist()
correl = []
correl_p = []
for k in df_go.keys():
y = list(df_go[k])
rho, p = spearmanr(x, y)
correl.append(rho)
correl_p.append(p)
df_go.loc['rho'] = correl
df_go.loc['p'] = correl_p
df_go
df = DataFrame(result)
df_go_plot = df.T[['Bacteria:Cyanobacteria']].copy()
pl = df_go_plot.plot(
y=['Bacteria:Cyanobacteria'], use_index=True, style='o', figsize=(8,5),
title='Depth and Cyanobacteria (phylum) reads, bacterias that obtain their energy through photosynthesis, normalised by number of InterPro annotations, for Tara Oceans project PRJEB1787',
)
pl.set_xlabel("Depth m")
pl.set_ylabel("Relative abundance")
plt.show()
| 0.366363 | 0.886617 |
```
%matplotlib inline
from qcodes.dataset.experiment_container import new_experiment
from qcodes.dataset.experiment_container import load_by_id
from qcodes.dataset.plotting import plot_by_id
import nanotune as nt
from nanotune.model.capacitancemodel import CapacitanceModel
```
## Capacitance model for quantum dots
The system of electrostatic gates, dots and reservoirs is represented by a system of conductors connected via resistors and capacitors. Albeit classical and simple, the capacitance model explains and qualitatively reproduces relevant transport features of gate-defined quantum dots.
The capacitance model is defined by a capacitance matrix $\mathbf{C}$, whose elements $C_{ij}$ are the capacitances between individual elements $i$ and $j$. We distinguish between two different types of elements, the charge and the voltage nodes, representing quantum dots and electrostatic gates respectively. Each node $i$ is defined by its charge $Q_{i}$ and electrical potential $V_{i}$. For simplicity, we write charges and potentials on all nodes of the system in vector notation. We denote charges on charge and voltages nodes by $\vec{Q_{c}}$ and $\vec{Q_{v}}$ respectively, and electrical potentials by $\vec{V}_{c}$ and $\vec{V}_{v}$. The capacitance model allows to calculate potentials on voltage nodes resulting in the desired number of charges on charge nodes. We consider a system of $N_{c}$ charge nodes and $N_{v}$ voltage nodes.
The capacitor connecting node $j$ and node $k$ has a capacitance $C_{jk}$ and stores a charge $q_{jk}$. The total charge on node $j$ is the sum of the charges of all capacitors connected to it,
\begin{equation}
Q_{j} = \sum_{k} q_{jk} = \sum_{k} C_{jk} (V_{j} - V_{k}).
\end{equation}
Using the vector notation for charges and electrical potentials introduced above, this relation can be expressed using the capacitance matrix, $\vec{Q} = \mathbf{C} \vec{V}$. Distinguishing between charge and voltage node sub-systems, this relation becomes
\begin{equation}
\begin{pmatrix} \vec{Q_{c}} \\ \vec{Q_{v}} \end{pmatrix} =
\begin{pmatrix}
\mathbf{C_{cc}} & \mathbf{C_{cv}} \\
\mathbf{C_{vc}} & \mathbf{C_{vv}}
\end{pmatrix}
\begin{pmatrix}
\vec{V_{c}} \\
\vec{V}_{v}
\end{pmatrix}.
\end{equation}
Diagonal elements of the capacitance matrix, $C_{jj}$, are total capacitances of each node and carry the opposite sign of the matrix's off-diagonal elements.
The off-diagonal elements of $\mathbf{\mathbf{C_{cc}}}$ are capacitances between charge nodes, while the off-diagonal elements of $\mathbf{\mathbf{C_{vv}}}$ are capacitances between voltage nodes. The elements of $\mathbf{\mathbf{C_{cv}}}$ are capacitances between voltage and charge nodes, and allow to calculate so-called virtual gate coefficients - useful knobs in semiconductor qubit experiments.
Illustration of capacitances between gates and gates and dots:
Each gate voltage $V_{i}$ will tune the number of charges on each dot. The capacitance of the dots, $C_{A}$ and $C_{B}$, are sums of all capacitances connects to $A$ and $B$ respectively. Gates located further away will have a smaller capacitive coupling. Most labels of capacitance between gates are omitted for readability.
<img src="../quantum_dots/quantum_dots-08.svg" width="300"/>
Names and layout indices of gates:
Six electrostatic gates, three barriers and two plungers, are used to define two, possibly coupled, quantum dots. Barrier gates are primarily used to create potential barriers, while plungers are used to tune electron density and thus the dot's electrochemical potentials.
<img src="../quantum_dots/quantum_dots-09.svg" width="200"/>
Initiate or set database where data should be saved. When initializing a database a new qcodes experiment needs to be created as well.
```
exp_name = 'capacitance_extraction'
sample_name = 'capa_model'
db_name = 'capa_model_test.db'
nt.new_database(db_name, '.')
new_experiment(exp_name, sample_name)
```
Initialize an instance of CapacitanceModel with six gates and two dots
```
voltage_nodes = {
0: 'top_barrier',
1: 'left_barrier',
2: 'left_plunger',
3: 'central_barrier',
4: 'right_plunger',
5: 'right_barrier',
}
charge_nodes = {
0: 'A',
1: 'B'
}
qdot = CapacitanceModel(
'qdot',
charge_nodes=charge_nodes,
voltage_nodes=voltage_nodes,
db_name=db_name,
db_folder='.'
)
```
Set voltages, define capacitance matrix and calculate ground state dot occupation
```
qdot.V_v([
-1, # top_barrier
-1, # left_barrier
-0.5, # left_plunger
-1, # central_barrier
-0.1, # right_plunger
-1, # right_barrier
])
# off-diagonal entries of C_cc, ie.e inter-dot capacitance
qdot.C_cc([[-2]])
# # capacitances between gates and dots
# top_b left_b left_p center right_p right_b
qdot.C_cv([[-0.6, -0.6, -0.5, -0.5, -0.1, -0.1], # A
[-0.5, -0.3, -0.2, -0.8, -0.5, -0.9]]) # B
print(qdot.determine_N())
```
Define which dot occupancies to probe and calculate respective voltage ranges of two gates to sweep.
```
N_limits = [(0, 2), (0, 2)]
sweep_ranges = qdot.determine_sweep_voltages([2, 4], N_limits=N_limits)
print(sweep_ranges)
%%time
dataid = qdot.sweep_voltages(
[2, 4],
sweep_ranges,
n_steps=[40,40],
target_snr_db=100,
add_noise=False,
add_charge_jumps=False,
normalize=False,
)
plot_by_id(dataid)
```
Sweep one voltage to show Coulomb oscillations
```
qdot.V_v([
-1, # top_barrier
-1, # left_barrier
3.2, # left_plunger
-1, # central_barrier
-0.1, # right_plunger
-1, # right_barrier
])
dataid = qdot.sweep_voltage(
4,
sweep_ranges[1],
n_steps=200,
target_snr_db=100,
normalize=False,
)
plot_by_id(dataid)
```
|
github_jupyter
|
%matplotlib inline
from qcodes.dataset.experiment_container import new_experiment
from qcodes.dataset.experiment_container import load_by_id
from qcodes.dataset.plotting import plot_by_id
import nanotune as nt
from nanotune.model.capacitancemodel import CapacitanceModel
exp_name = 'capacitance_extraction'
sample_name = 'capa_model'
db_name = 'capa_model_test.db'
nt.new_database(db_name, '.')
new_experiment(exp_name, sample_name)
voltage_nodes = {
0: 'top_barrier',
1: 'left_barrier',
2: 'left_plunger',
3: 'central_barrier',
4: 'right_plunger',
5: 'right_barrier',
}
charge_nodes = {
0: 'A',
1: 'B'
}
qdot = CapacitanceModel(
'qdot',
charge_nodes=charge_nodes,
voltage_nodes=voltage_nodes,
db_name=db_name,
db_folder='.'
)
qdot.V_v([
-1, # top_barrier
-1, # left_barrier
-0.5, # left_plunger
-1, # central_barrier
-0.1, # right_plunger
-1, # right_barrier
])
# off-diagonal entries of C_cc, ie.e inter-dot capacitance
qdot.C_cc([[-2]])
# # capacitances between gates and dots
# top_b left_b left_p center right_p right_b
qdot.C_cv([[-0.6, -0.6, -0.5, -0.5, -0.1, -0.1], # A
[-0.5, -0.3, -0.2, -0.8, -0.5, -0.9]]) # B
print(qdot.determine_N())
N_limits = [(0, 2), (0, 2)]
sweep_ranges = qdot.determine_sweep_voltages([2, 4], N_limits=N_limits)
print(sweep_ranges)
%%time
dataid = qdot.sweep_voltages(
[2, 4],
sweep_ranges,
n_steps=[40,40],
target_snr_db=100,
add_noise=False,
add_charge_jumps=False,
normalize=False,
)
plot_by_id(dataid)
qdot.V_v([
-1, # top_barrier
-1, # left_barrier
3.2, # left_plunger
-1, # central_barrier
-0.1, # right_plunger
-1, # right_barrier
])
dataid = qdot.sweep_voltage(
4,
sweep_ranges[1],
n_steps=200,
target_snr_db=100,
normalize=False,
)
plot_by_id(dataid)
| 0.440229 | 0.984694 |
http://preview.d2l.ai/d2l-en/master/chapter_generative-adversarial-networks/gan.html
```
%matplotlib inline
from d2l import tensorflow as d2l
import tensorflow as tf
```
https://www.tensorflow.org/api_docs/python/tf/random/normal
https://www.tensorflow.org/api_docs/python/tf/norm
```
X = d2l.normal([1000, 2], 0.0, 1, tf.float32)
A = d2l.tensor([[1, 2], [-0.1, 0.5]], tf.float32)
b = d2l.tensor([1, 2], tf.float32)
data = d2l.matmul(X, A) + b
d2l.set_figsize()
d2l.plt.scatter(d2l.numpy(data[:100, 0]), d2l.numpy(data[:100, 1]));
print(f'The covariance matrix is\n{d2l.matmul(tf.transpose(A), A)}')
```
https://github.com/zxjzxj9/deeplearning/blob/master/gan_mnist/gan_mnist.py
https://www.tensorflow.org/api_docs/python/tf/convert_to_tensor
```
batch_size = 8
data_iter = d2l.load_array((data,), batch_size).convert_to_tensor()
```
https://learning.oreilly.com/library/view/hands-on-generative-adversarial/9781789538205/e5a3d881-308a-4971-a086-9ae66bcdab38.xhtml
```
net_G = tf.keras.Sequential()
net_G.add(tf.keras.layers.Dense(2))
```
tf.keras.activations.tanh(a)
https://www.tensorflow.org/api_docs/python/tf/keras/activations/tanh
```
net_D = tf.keras.Sequential()
net_D.add(tf.keras.layers.Dense(5, activation='tanh'))
net_D.add(tf.keras.layers.Dense(3, activation='tanh'))
net_D.add(tf.keras.layers.Dense(1, activation='sigmoid'))
```
with tf.GradientTape() as t:
http://preview.d2l.ai/d2l-en/master/chapter_preliminaries/autograd.html
t.gradient(y, x)
t.gradient(loss_D, trainer_D)
.apply_gradients(zip(grads, self.generator.trainable_weights))
https://keras.io/examples/generative/dcgan_overriding_train_step/
```
#@save
def update_D(X, Z, net_D, net_G, loss, trainer_D):
"""Update discriminator."""
batch_size = tf.shape(X)[0]
ones = tf.ones((batch_size, ), tf.float32)
zeros = tf.zeros((batch_size, ), tf.float32)
with tf.GradientTape() as t:
real_Y = net_D(tf.convert_to_tensor(X))
fake_X = net_G(tf.convert_to_tensor(Z))
# Do not need to compute gradient for `net_G`, detach it from
# computing gradients.
fake_Y = net_D(tf.stop_gradient(fake_X))
loss_D = (loss(real_Y, ones) + loss(fake_Y, zeros)) / 2
grads = t.gradient(loss_D, net_D.trainable_variables)
trainer_D.apply_gradients(zip(grads, net_D.trainable_weights))
return float(loss_D)
```
https://www.tensorflow.org/api_docs/python/tf/reshape
```
#@save
def update_G(Z, net_D, net_G, loss, trainer_G):
"""Update generator."""
batch_size = tf.shape(Z)[0]
ones = tf.ones((batch_size, ), tf.float32)
with tf.GradientTape() as t:
# We could reuse `fake_X` from `update_D` to save computation
fake_X = net_G(tf.convert_to_tensor(Z))
# Recomputing `fake_Y` is needed since `net_D` is changed
fake_Y = net_D(fake_X)
loss_G = loss(fake_Y, ones)
grads = t.gradient(loss_G, net_G.trainable_variables)
trainer_G.apply_gradients(zip(grads, net_G.trainable_weights))
return float(loss_G)
```
https://www.google.com/search?q=numpy()+tensorflow&rlz=1C1GCEA_enJP909HK909&oq=numpy()+tensorflow&aqs=chrome..69i57j0l7.2792j0j1&sourceid=chrome&ie=UTF-8
https://www.tensorflow.org/api_docs/python/tf/keras/optimizers/Adam
tf.keras.optimizers.Adam(learning_rate=0.1)
m.compile
m = tf.keras.models.Sequential([tf.keras.layers.Dense(10)])
m.compile(opt, loss='mse')
https://stackoverflow.com/questions/55683729/bcewithlogitsloss-in-keras
https://www.tensorflow.org/api_docs/python/tf/keras/losses/BinaryCrossentropy
def weight_variable(shape, w=0.1):
initial = tf.truncated_normal(shape, stddev=w)
return tf.Variable(initial)
https://stackoverflow.com/questions/43489697/tensorflow-weight-initialization
The values generated are similar to values from a tf.keras.initializers.RandomNormal initializer except that values more than two standard deviations from the mean are discarded and re-drawn.
https://keras.io/api/layers/initializers/
tf.Variable(initial_value=w_init(
shape=(input_dim, unit), dtype=tf.float32), trainable=True)
https://zhuanlan.zhihu.com/p/59481536
https://keras.io/api/layers/initializers/
numpy.array
https://stackoverflow.com/questions/41198144/attributeerror-tuple-object-has-no-attribute-shape
https://github.com/tensorflow/tensorflow/issues/29972
tf.stop_gradient(y)
http://preview.d2l.ai/d2l-en/master/chapter_preliminaries/autograd.html?highlight=detach
from_logits=True 9.2.4 TensorFlow 2.0 神经网络实践
https://github.com/czy36mengfei/tensorflow2_tutorials_chinese/blob/master/025-GAN/002-DCGAN.ipynb
```
def train(net_D, net_G, data_iter, num_epochs, lr_D, lr_G, latent_dim, data):
loss = tf.keras.losses.BinaryCrossentropy(reduction=
tf.keras.losses.Reduction.SUM)
tf.keras.initializers.RandomNormal(net_D, stddev=0.02)
tf.keras.initializers.RandomNormal(net_G, stddev=0.02)
trainer_D = tf.keras.optimizers.Adam(learning_rate=lr_D)
trainer_G = tf.keras.optimizers.Adam(learning_rate=lr_G)
net_D.compile(optimizer=trainer_D)
net_G.compile(optimizer=trainer_G)
animator = d2l.Animator(xlabel='epoch', ylabel='loss',
xlim=[1, num_epochs], nrows=2, figsize=(5, 5),
legend=['discriminator', 'generator'])
animator.fig.subplots_adjust(hspace=0.3)
for epoch in range(num_epochs):
# Train one epoch
timer = d2l.Timer()
metric = d2l.Accumulator(3) # loss_D, loss_G, num_examples
for X in data_iter:
batch_size = tf.shape(X)[0]
Z = d2l.normal([batch_size, latent_dim], 0, 1, tf.float32)
metric.add(update_D(X, Z, net_D, net_G, loss, trainer_D),
update_G(Z, net_D, net_G, loss, trainer_G),
batch_size)
# Visualize generated examples
Z = d2l.normal([100, latent_dim], 0, 1, tf.float32)
fake_X = net_G(tf.stop_gradient(Z)).numpy()
animator.axes[1].cla()
animator.axes[1].scatter(data[:, 0], data[:, 1])
animator.axes[1].scatter(fake_X[:, 0], fake_X[:, 1])
animator.axes[1].legend(['real', 'generated'])
# Show the losses
loss_D, loss_G = metric[0]/metric[2], metric[1]/metric[2]
animator.add(epoch + 1, (loss_D, loss_G))
print(f'loss_D {loss_D:.3f}, loss_G {loss_G:.3f}, '
f'{metric[2] / timer.stop():.1f} examples/sec')
lr_D, lr_G, latent_dim, num_epochs = 0.05, 0.005, 2, 20
train(net_D, net_G, data_iter, num_epochs, lr_D, lr_G,
latent_dim, d2l.numpy(data[:100]))
```
https://www.tensorflow.org/api_docs/python/tf/ones
|
github_jupyter
|
%matplotlib inline
from d2l import tensorflow as d2l
import tensorflow as tf
X = d2l.normal([1000, 2], 0.0, 1, tf.float32)
A = d2l.tensor([[1, 2], [-0.1, 0.5]], tf.float32)
b = d2l.tensor([1, 2], tf.float32)
data = d2l.matmul(X, A) + b
d2l.set_figsize()
d2l.plt.scatter(d2l.numpy(data[:100, 0]), d2l.numpy(data[:100, 1]));
print(f'The covariance matrix is\n{d2l.matmul(tf.transpose(A), A)}')
batch_size = 8
data_iter = d2l.load_array((data,), batch_size).convert_to_tensor()
net_G = tf.keras.Sequential()
net_G.add(tf.keras.layers.Dense(2))
net_D = tf.keras.Sequential()
net_D.add(tf.keras.layers.Dense(5, activation='tanh'))
net_D.add(tf.keras.layers.Dense(3, activation='tanh'))
net_D.add(tf.keras.layers.Dense(1, activation='sigmoid'))
#@save
def update_D(X, Z, net_D, net_G, loss, trainer_D):
"""Update discriminator."""
batch_size = tf.shape(X)[0]
ones = tf.ones((batch_size, ), tf.float32)
zeros = tf.zeros((batch_size, ), tf.float32)
with tf.GradientTape() as t:
real_Y = net_D(tf.convert_to_tensor(X))
fake_X = net_G(tf.convert_to_tensor(Z))
# Do not need to compute gradient for `net_G`, detach it from
# computing gradients.
fake_Y = net_D(tf.stop_gradient(fake_X))
loss_D = (loss(real_Y, ones) + loss(fake_Y, zeros)) / 2
grads = t.gradient(loss_D, net_D.trainable_variables)
trainer_D.apply_gradients(zip(grads, net_D.trainable_weights))
return float(loss_D)
#@save
def update_G(Z, net_D, net_G, loss, trainer_G):
"""Update generator."""
batch_size = tf.shape(Z)[0]
ones = tf.ones((batch_size, ), tf.float32)
with tf.GradientTape() as t:
# We could reuse `fake_X` from `update_D` to save computation
fake_X = net_G(tf.convert_to_tensor(Z))
# Recomputing `fake_Y` is needed since `net_D` is changed
fake_Y = net_D(fake_X)
loss_G = loss(fake_Y, ones)
grads = t.gradient(loss_G, net_G.trainable_variables)
trainer_G.apply_gradients(zip(grads, net_G.trainable_weights))
return float(loss_G)
def train(net_D, net_G, data_iter, num_epochs, lr_D, lr_G, latent_dim, data):
loss = tf.keras.losses.BinaryCrossentropy(reduction=
tf.keras.losses.Reduction.SUM)
tf.keras.initializers.RandomNormal(net_D, stddev=0.02)
tf.keras.initializers.RandomNormal(net_G, stddev=0.02)
trainer_D = tf.keras.optimizers.Adam(learning_rate=lr_D)
trainer_G = tf.keras.optimizers.Adam(learning_rate=lr_G)
net_D.compile(optimizer=trainer_D)
net_G.compile(optimizer=trainer_G)
animator = d2l.Animator(xlabel='epoch', ylabel='loss',
xlim=[1, num_epochs], nrows=2, figsize=(5, 5),
legend=['discriminator', 'generator'])
animator.fig.subplots_adjust(hspace=0.3)
for epoch in range(num_epochs):
# Train one epoch
timer = d2l.Timer()
metric = d2l.Accumulator(3) # loss_D, loss_G, num_examples
for X in data_iter:
batch_size = tf.shape(X)[0]
Z = d2l.normal([batch_size, latent_dim], 0, 1, tf.float32)
metric.add(update_D(X, Z, net_D, net_G, loss, trainer_D),
update_G(Z, net_D, net_G, loss, trainer_G),
batch_size)
# Visualize generated examples
Z = d2l.normal([100, latent_dim], 0, 1, tf.float32)
fake_X = net_G(tf.stop_gradient(Z)).numpy()
animator.axes[1].cla()
animator.axes[1].scatter(data[:, 0], data[:, 1])
animator.axes[1].scatter(fake_X[:, 0], fake_X[:, 1])
animator.axes[1].legend(['real', 'generated'])
# Show the losses
loss_D, loss_G = metric[0]/metric[2], metric[1]/metric[2]
animator.add(epoch + 1, (loss_D, loss_G))
print(f'loss_D {loss_D:.3f}, loss_G {loss_G:.3f}, '
f'{metric[2] / timer.stop():.1f} examples/sec')
lr_D, lr_G, latent_dim, num_epochs = 0.05, 0.005, 2, 20
train(net_D, net_G, data_iter, num_epochs, lr_D, lr_G,
latent_dim, d2l.numpy(data[:100]))
| 0.870253 | 0.888227 |
```
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
data_file_listings_path = "/Users/Edo/Resources/listings.csv"
data_file_listings_path
#!pwd to check the working directory
raw_df = pd.read_csv(data_file_listings_path)
print(f"The dataset contains {len(raw_df)} Airbnb listings")
pd.set_option('display.max_columns', len(raw_df.columns)) # To view all columns
pd.set_option('display.max_rows', 100)
raw_df.head(3)
cols_to_drop = ['listing_url', 'scrape_id', 'last_scraped', 'name','description', 'neighborhood_overview',
'picture_url', 'host_id', 'host_url', 'host_name', 'host_location', 'host_about',
'host_thumbnail_url', 'host_picture_url','number_of_reviews_l30d',
'host_neighbourhood', 'host_verifications', 'calendar_last_scraped']
df = raw_df.drop(cols_to_drop, axis=1)
df.isna().sum()
#Columns with several NULL entries are dropped too.
df.drop(['bathrooms', 'host_acceptance_rate', 'neighbourhood_group_cleansed',
'calendar_updated', 'license'], axis=1, inplace=True)
df.set_index('id', inplace=True) # ID as index
df.isna().sum()
print(sum((df.host_listings_count == df.host_total_listings_count) == False))
df.loc[((df.host_listings_count == df.host_total_listings_count) == False)][:5]
# host_listings_count and host_total_listings_count are the same in all but 248 cases.
#These cases are those where the value is NaN. Therefore one of these columns can be dropped.
#Other columns which split these into type of property will also be dropped,
#as they will be highly correlated (one will be the total of the others).
df.drop(['host_total_listings_count', 'calculated_host_listings_count',
'calculated_host_listings_count_entire_homes', 'calculated_host_listings_count_private_rooms',
'calculated_host_listings_count_shared_rooms'], axis=1, inplace=True)
#Columns for property location can be dropped because one column for area will be kept, neighboorhood_cleansed
df.drop(['latitude', 'longitude', 'neighbourhood'], axis=1, inplace=True)
sum((df.minimum_nights == df.minimum_minimum_nights) == False)
df.drop(['minimum_minimum_nights', 'maximum_minimum_nights', 'minimum_maximum_nights',
'maximum_maximum_nights', 'minimum_nights_avg_ntm', 'maximum_nights_avg_ntm'], axis=1, inplace=True)
# Replacing columns with f/t with 0/1
df.replace({'f': 0, 't': 1}, inplace=True)
# Plotting the distribution of numerical and boolean categories
df.hist(figsize=(20,20));
df.drop(['has_availability', 'host_has_profile_pic'], axis=1, inplace=True)
df.isna().sum()
```
Cleaning each of the columns
host_since
This is a datetime column, and will be converted into a measure of the number of days that a host has been on the platform, measured from the date that the data was scraped (July , 2021). The original column will be left in initially for EDA, and dropped later.
host_response_time
About a third of rows don't have a value for host_response_time, and the majority of these have also not yet been reviewed.
```
print("Null values:", df.host_response_time.isna().sum())
print(f"Proportion: {round((df.host_response_time.isna().sum()/len(df))*100, 1)}%")
# Number of rows without a value for host_response_time which have also not yet had a review
len(df[df.loc[ :,['host_response_time', 'first_review'] ].isnull().sum(axis=1) == 2])
df.host_response_time.fillna("unknown", inplace=True)
df.host_response_time.value_counts(normalize=True)
df.host_response_time.value_counts(normalize=True)
```
host_response_rate
The same story for host_response_rate, with about a third of values being null. This will also be kept as its own category. Because about 75% of hosts respond 100% of the time, this will be kept as its own category, and other values will be grouped into bins.
```
print("Null values:", df.host_response_rate.isna().sum())
print(f"Proportion: {round((df.host_response_rate.isna().sum()/len(df))*100, 1)}%")
# Removing the % sign from the host_response_rate string and converting to an integer
df.host_response_rate = df.host_response_rate.str[:-1].astype('float64')
print("Mean host response rate:", round(df['host_response_rate'].mean(),0))
print("Median host response rate:", df['host_response_rate'].median())
print(f"Proportion of 100% host response rates: {round(((df.host_response_rate == 100.0).sum()/df.host_response_rate.count())*100,1)}%")
# Bin into four categories
df.host_response_rate = pd.cut(df.host_response_rate,
bins=[0, 50, 90, 99, 100],
labels=['0-49%', '50-89%', '90-99%', '100%'],
include_lowest=True)
# Converting to string
df.host_response_rate = df.host_response_rate.astype('str')
# Replace nulls with 'unknown'
df.host_response_rate.replace('nan', 'unknown', inplace=True)
# Category counts
df.host_response_rate.value_counts()
```
host_is_superhost
There are 21 rows NA values for each of five different host-related features. These rows will be dropped.
```
# Number of rows without a value for multiple host-related columns
len(df[df.loc[ :,['host_since', 'host_is_superhost', 'host_listings_count',
'host_identity_verified'] ].isnull().sum(axis=1) == 5])
df.dropna(subset=['host_since'], inplace=True)
```
property_type
Some cleaning of property types is required as there are a large number of categories with only a few listings. The categories Apartment, House and Other will be used, as most properties can be classified as either apartment or house.
```
df.property_type.value_counts()
# Replacing categories that are types of houses or apartments
df.property_type.replace({
'Entire apartment': 'Apartment',
'Private room in apartment': 'Apartment',
'Private room in house': 'House',
'Entire condominium': 'House',
'Entire loft': 'Apartment',
'Private room in condominium': 'Apartment',
'Entire house': 'House',
'Private room in guest suite': 'Apartment',
'Entire serviced apartment': 'Apartment',
'Entire guest suite': 'House',
'Private room in serviced apartment': 'Apartment',
'Private room in loft': 'Apartment',
'Private room in guesthouse': 'House',
'Shared room in apartment': 'Apartment',
'Shared room in house': 'House',
'Room in serviced apartment': 'Apartment',
'Entire guesthouse': 'House',
'Private room in townhouse': 'House',
'Private room in tiny house': 'House',
'Entire townhouse': 'House',
'Private room': 'House',
'Tiny house': 'House',
'Shared room in guesthouse': 'House',
'Casa particular (Cuba)': 'House',
'Private room in farm stay': 'House',
'Private room in villa': 'House',
'Private room in hut': 'House',
'Private room in dome house': 'House',
'Private room in bungalow': 'House',
'Entire villa': 'House',
'Room in casa particular': 'House',
'Entire villa': 'House',
'Shared room in serviced apartment': 'Apartment',
'Dome house': 'House'
}, inplace=True)
# Replacing other categories with 'other'
df.loc[~df.property_type.isin(['House', 'Apartment']), 'property_type'] = 'Other'
```
bathrooms, bedrooms and beds
Missing values will be replaced with the median (to avoid strange fractions).
```
for col in ['bedrooms', 'beds']:
df[col].fillna(df[col].median(), inplace=True)
```
amenities
Amenities is a list of additional features in the property, i.e. whether it has a TV or parking. Examples are below:
```
# Example of amenities listed
df.amenities[:1].values
# Creating a set of all possible amenities
amenities_list = list(df.amenities)
amenities_list_string = " ".join(amenities_list)
amenities_list_string = amenities_list_string.replace('{', '')
amenities_list_string = amenities_list_string.replace('}', ',')
amenities_list_string = amenities_list_string.replace('"', '')
amenities_set = [x.strip() for x in amenities_list_string.split(',')]
amenities_set = set(amenities_set)
amenities_set
df.loc[df['amenities'].str.contains('24-hour check-in'), 'check_in_24h'] = 1
df.loc[df['amenities'].str.contains('Air conditioning|Central air conditioning'), 'air_conditioning'] = 1
df.loc[df['amenities'].str.contains('Amazon Echo|Apple TV|Game console|Netflix|Projector and screen|Smart TV'), 'high_end_electronics'] = 1
df.loc[df['amenities'].str.contains('BBQ grill|Fire pit|Propane barbeque'), 'bbq'] = 1
df.loc[df['amenities'].str.contains('Balcony|Patio'), 'balcony'] = 1
df.loc[df['amenities'].str.contains('Beach view|Beachfront|Lake access|Mountain view|Ski-in/Ski-out|Waterfront'), 'nature_and_views'] = 1
df.loc[df['amenities'].str.contains('Bed linens'), 'bed_linen'] = 1
df.loc[df['amenities'].str.contains('Breakfast'), 'breakfast'] = 1
df.loc[df['amenities'].str.contains('TV'), 'tv'] = 1
df.loc[df['amenities'].str.contains('Coffee maker|Espresso machine'), 'coffee_machine'] = 1
df.loc[df['amenities'].str.contains('Cooking basics'), 'cooking_basics'] = 1
df.loc[df['amenities'].str.contains('Dishwasher|Dryer|Washer'), 'white_goods'] = 1
df.loc[df['amenities'].str.contains('Elevator'), 'elevator'] = 1
df.loc[df['amenities'].str.contains('Exercise equipment|Gym|gym'), 'gym'] = 1
df.loc[df['amenities'].str.contains('Family/kid friendly|Children|children'), 'child_friendly'] = 1
df.loc[df['amenities'].str.contains('parking'), 'parking'] = 1
df.loc[df['amenities'].str.contains('Garden|Outdoor|Sun loungers|Terrace'), 'outdoor_space'] = 1
df.loc[df['amenities'].str.contains('Host greets you'), 'host_greeting'] = 1
df.loc[df['amenities'].str.contains('Hot tub|Jetted tub|hot tub|Sauna|Pool|pool'), 'hot_tub_sauna_or_pool'] = 1
df.loc[df['amenities'].str.contains('Internet|Pocket wifi|Wifi'), 'internet'] = 1
df.loc[df['amenities'].str.contains('Long term stays allowed'), 'long_term_stays'] = 1
df.loc[df['amenities'].str.contains('Pets|pet|Cat(s)|Dog(s)'), 'pets_allowed'] = 1
df.loc[df['amenities'].str.contains('Private entrance'), 'private_entrance'] = 1
df.loc[df['amenities'].str.contains('Safe|Security system'), 'secure'] = 1
df.loc[df['amenities'].str.contains('Self check-in'), 'self_check_in'] = 1
df.loc[df['amenities'].str.contains('Smoking allowed'), 'smoking_allowed'] = 1
df.loc[df['amenities'].str.contains('Step-free access|Wheelchair|Accessible'), 'accessible'] = 1
df.loc[df['amenities'].str.contains('Suitable for events'), 'event_suitable'] = 1
# Replacing nulls with zeros for new columns
cols_to_replace_nulls = df.iloc[:,41:].columns
df[cols_to_replace_nulls] = df[cols_to_replace_nulls].fillna(0)
# Produces a list of amenity features where one category (true or false) contains fewer than 10% of listings
infrequent_amenities = []
for col in df.iloc[:,41:].columns:
if df[col].sum() < len(df)/10:
infrequent_amenities.append(col)
print(infrequent_amenities)
# Dropping infrequent amenity features
df.drop(infrequent_amenities, axis=1, inplace=True)
# Dropping the original amenity feature
df.drop('amenities', axis=1, inplace=True)
df.columns[40:]
```
price
Price will be converted to an integer. Currently it is a string because there is a currency sign.
```
df.price = df.price.str[1:-3]
df.price = df.price.str.replace(",", "")
df.price = df.price.astype('int64')
```
availability
There are multiple different measures of availability, which will be highly correlated with each other. Only one will be retained, availability for 90 days (availability_90)
```
df.drop(['availability_30', 'availability_60', 'availability_365'], axis=1, inplace=True)
```
first_review and last_review¶
```
print(f"Null values in 'first_review': {round(100*df.first_review.isna().sum()/len(df),1)}%")
print(f"Null values in 'review_scores_rating': {round(100*df.review_scores_rating .isna().sum()/len(df),1)}%")
df.first_review = pd.to_datetime(df.first_review) # Converting to datetime
# Calculating the number of days between the first review and the date the data was scraped
df['time_since_first_review'] = (pd.datetime(2019, 4, 9) - df.first_review).astype('timedelta64[D]')
# Distribution of the number of days since first review
df.time_since_first_review.hist(figsize=(15,5), bins=30);
def bin_column(col, bins, labels, na_label='unknown'):
"""
Takes in a column name, bin cut points and labels, replaces the original column with a
binned version, and replaces nulls (with 'unknown' if unspecified).
"""
df[col] = pd.cut(df[col], bins=bins, labels=labels, include_lowest=True)
df[col] = df[col].astype('str')
df[col].fillna(na_label, inplace=True)
# Binning time since first review
bin_column('time_since_first_review',
bins=[0, 182, 365, 730, 1460, max(df.time_since_first_review)],
labels=['0-6 months',
'6-12 months',
'1-2 years',
'2-3 years',
'4+ years'],
na_label='no reviews')
df.last_review = pd.to_datetime(df.last_review) # Converting to datetime
# Calculating the number of days between the most recent review and the date the data was scraped
df['time_since_last_review'] = (pd.datetime(2019, 4, 9) - df.last_review).astype('timedelta64[D]')
# Distribution of the number of days since last review
df.time_since_last_review.hist(figsize=(15,5), bins=30);
# Binning time since last review
bin_column('time_since_last_review',
bins=[0, 14, 60, 182, 365, max(df.time_since_last_review)],
labels=['0-2 weeks',
'2-8 weeks',
'2-6 months',
'6-12 months',
'1+ year'],
na_label='no reviews')
# Dropping last_review - first_review will be kept for EDA and dropped later
df.drop('last_review', axis=1, inplace=True)
```
review ratings columns
As above, listings without reviews will be kept and replaced with unknown. Other ratings will be grouped into bins. The histograms below were produced in order to decide on useful bins. The majority of ratings are 9 or 10 out of 10, as also seen in the value count below. Therefore for these columns, 9/10 and 10/10 will be kept as separate groups, and 1-8/10 will be binned together (as this is, by Airbnb standards, a 'low' rating).
```
# Checking the distributions of the review ratings columns
variables_to_plot = list(df.columns[df.columns.str.startswith("review_scores") == True])
fig = plt.figure(figsize=(12,8))
for i, var_name in enumerate(variables_to_plot):
ax = fig.add_subplot(3,3,i+1)
df[var_name].hist(bins=10,ax=ax)
ax.set_title(var_name)
fig.tight_layout()
plt.show()
# Creating a list of all review columns that are scored out of 10
variables_to_plot.pop(0)
# Binning for all columns scored out of 10
for col in variables_to_plot:
bin_column(col,
bins=[0, 8, 9, 10],
labels=['0-8/10', '9/10', '10/10'],
na_label='no reviews')
# Binning column scored out of 100
bin_column('review_scores_rating',
bins=[0, 80, 95, 100],
labels=['0-79/100', '80-94/100', '95-100/100'],
na_label='no reviews')
```
number_of_reviews_ltm and reviews_per_month¶
```
df.drop(['number_of_reviews_ltm', 'reviews_per_month'], axis=1, inplace=True)
df.head()
df.dtypes
# Save cleaned dataset
listings_cleaned = df.to_csv(r'/Users/Edo/Resources/listings_cleaned.csv', index=id, header=True)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
data_file_listings_path = "/Users/Edo/Resources/listings.csv"
data_file_listings_path
#!pwd to check the working directory
raw_df = pd.read_csv(data_file_listings_path)
print(f"The dataset contains {len(raw_df)} Airbnb listings")
pd.set_option('display.max_columns', len(raw_df.columns)) # To view all columns
pd.set_option('display.max_rows', 100)
raw_df.head(3)
cols_to_drop = ['listing_url', 'scrape_id', 'last_scraped', 'name','description', 'neighborhood_overview',
'picture_url', 'host_id', 'host_url', 'host_name', 'host_location', 'host_about',
'host_thumbnail_url', 'host_picture_url','number_of_reviews_l30d',
'host_neighbourhood', 'host_verifications', 'calendar_last_scraped']
df = raw_df.drop(cols_to_drop, axis=1)
df.isna().sum()
#Columns with several NULL entries are dropped too.
df.drop(['bathrooms', 'host_acceptance_rate', 'neighbourhood_group_cleansed',
'calendar_updated', 'license'], axis=1, inplace=True)
df.set_index('id', inplace=True) # ID as index
df.isna().sum()
print(sum((df.host_listings_count == df.host_total_listings_count) == False))
df.loc[((df.host_listings_count == df.host_total_listings_count) == False)][:5]
# host_listings_count and host_total_listings_count are the same in all but 248 cases.
#These cases are those where the value is NaN. Therefore one of these columns can be dropped.
#Other columns which split these into type of property will also be dropped,
#as they will be highly correlated (one will be the total of the others).
df.drop(['host_total_listings_count', 'calculated_host_listings_count',
'calculated_host_listings_count_entire_homes', 'calculated_host_listings_count_private_rooms',
'calculated_host_listings_count_shared_rooms'], axis=1, inplace=True)
#Columns for property location can be dropped because one column for area will be kept, neighboorhood_cleansed
df.drop(['latitude', 'longitude', 'neighbourhood'], axis=1, inplace=True)
sum((df.minimum_nights == df.minimum_minimum_nights) == False)
df.drop(['minimum_minimum_nights', 'maximum_minimum_nights', 'minimum_maximum_nights',
'maximum_maximum_nights', 'minimum_nights_avg_ntm', 'maximum_nights_avg_ntm'], axis=1, inplace=True)
# Replacing columns with f/t with 0/1
df.replace({'f': 0, 't': 1}, inplace=True)
# Plotting the distribution of numerical and boolean categories
df.hist(figsize=(20,20));
df.drop(['has_availability', 'host_has_profile_pic'], axis=1, inplace=True)
df.isna().sum()
print("Null values:", df.host_response_time.isna().sum())
print(f"Proportion: {round((df.host_response_time.isna().sum()/len(df))*100, 1)}%")
# Number of rows without a value for host_response_time which have also not yet had a review
len(df[df.loc[ :,['host_response_time', 'first_review'] ].isnull().sum(axis=1) == 2])
df.host_response_time.fillna("unknown", inplace=True)
df.host_response_time.value_counts(normalize=True)
df.host_response_time.value_counts(normalize=True)
print("Null values:", df.host_response_rate.isna().sum())
print(f"Proportion: {round((df.host_response_rate.isna().sum()/len(df))*100, 1)}%")
# Removing the % sign from the host_response_rate string and converting to an integer
df.host_response_rate = df.host_response_rate.str[:-1].astype('float64')
print("Mean host response rate:", round(df['host_response_rate'].mean(),0))
print("Median host response rate:", df['host_response_rate'].median())
print(f"Proportion of 100% host response rates: {round(((df.host_response_rate == 100.0).sum()/df.host_response_rate.count())*100,1)}%")
# Bin into four categories
df.host_response_rate = pd.cut(df.host_response_rate,
bins=[0, 50, 90, 99, 100],
labels=['0-49%', '50-89%', '90-99%', '100%'],
include_lowest=True)
# Converting to string
df.host_response_rate = df.host_response_rate.astype('str')
# Replace nulls with 'unknown'
df.host_response_rate.replace('nan', 'unknown', inplace=True)
# Category counts
df.host_response_rate.value_counts()
# Number of rows without a value for multiple host-related columns
len(df[df.loc[ :,['host_since', 'host_is_superhost', 'host_listings_count',
'host_identity_verified'] ].isnull().sum(axis=1) == 5])
df.dropna(subset=['host_since'], inplace=True)
df.property_type.value_counts()
# Replacing categories that are types of houses or apartments
df.property_type.replace({
'Entire apartment': 'Apartment',
'Private room in apartment': 'Apartment',
'Private room in house': 'House',
'Entire condominium': 'House',
'Entire loft': 'Apartment',
'Private room in condominium': 'Apartment',
'Entire house': 'House',
'Private room in guest suite': 'Apartment',
'Entire serviced apartment': 'Apartment',
'Entire guest suite': 'House',
'Private room in serviced apartment': 'Apartment',
'Private room in loft': 'Apartment',
'Private room in guesthouse': 'House',
'Shared room in apartment': 'Apartment',
'Shared room in house': 'House',
'Room in serviced apartment': 'Apartment',
'Entire guesthouse': 'House',
'Private room in townhouse': 'House',
'Private room in tiny house': 'House',
'Entire townhouse': 'House',
'Private room': 'House',
'Tiny house': 'House',
'Shared room in guesthouse': 'House',
'Casa particular (Cuba)': 'House',
'Private room in farm stay': 'House',
'Private room in villa': 'House',
'Private room in hut': 'House',
'Private room in dome house': 'House',
'Private room in bungalow': 'House',
'Entire villa': 'House',
'Room in casa particular': 'House',
'Entire villa': 'House',
'Shared room in serviced apartment': 'Apartment',
'Dome house': 'House'
}, inplace=True)
# Replacing other categories with 'other'
df.loc[~df.property_type.isin(['House', 'Apartment']), 'property_type'] = 'Other'
for col in ['bedrooms', 'beds']:
df[col].fillna(df[col].median(), inplace=True)
# Example of amenities listed
df.amenities[:1].values
# Creating a set of all possible amenities
amenities_list = list(df.amenities)
amenities_list_string = " ".join(amenities_list)
amenities_list_string = amenities_list_string.replace('{', '')
amenities_list_string = amenities_list_string.replace('}', ',')
amenities_list_string = amenities_list_string.replace('"', '')
amenities_set = [x.strip() for x in amenities_list_string.split(',')]
amenities_set = set(amenities_set)
amenities_set
df.loc[df['amenities'].str.contains('24-hour check-in'), 'check_in_24h'] = 1
df.loc[df['amenities'].str.contains('Air conditioning|Central air conditioning'), 'air_conditioning'] = 1
df.loc[df['amenities'].str.contains('Amazon Echo|Apple TV|Game console|Netflix|Projector and screen|Smart TV'), 'high_end_electronics'] = 1
df.loc[df['amenities'].str.contains('BBQ grill|Fire pit|Propane barbeque'), 'bbq'] = 1
df.loc[df['amenities'].str.contains('Balcony|Patio'), 'balcony'] = 1
df.loc[df['amenities'].str.contains('Beach view|Beachfront|Lake access|Mountain view|Ski-in/Ski-out|Waterfront'), 'nature_and_views'] = 1
df.loc[df['amenities'].str.contains('Bed linens'), 'bed_linen'] = 1
df.loc[df['amenities'].str.contains('Breakfast'), 'breakfast'] = 1
df.loc[df['amenities'].str.contains('TV'), 'tv'] = 1
df.loc[df['amenities'].str.contains('Coffee maker|Espresso machine'), 'coffee_machine'] = 1
df.loc[df['amenities'].str.contains('Cooking basics'), 'cooking_basics'] = 1
df.loc[df['amenities'].str.contains('Dishwasher|Dryer|Washer'), 'white_goods'] = 1
df.loc[df['amenities'].str.contains('Elevator'), 'elevator'] = 1
df.loc[df['amenities'].str.contains('Exercise equipment|Gym|gym'), 'gym'] = 1
df.loc[df['amenities'].str.contains('Family/kid friendly|Children|children'), 'child_friendly'] = 1
df.loc[df['amenities'].str.contains('parking'), 'parking'] = 1
df.loc[df['amenities'].str.contains('Garden|Outdoor|Sun loungers|Terrace'), 'outdoor_space'] = 1
df.loc[df['amenities'].str.contains('Host greets you'), 'host_greeting'] = 1
df.loc[df['amenities'].str.contains('Hot tub|Jetted tub|hot tub|Sauna|Pool|pool'), 'hot_tub_sauna_or_pool'] = 1
df.loc[df['amenities'].str.contains('Internet|Pocket wifi|Wifi'), 'internet'] = 1
df.loc[df['amenities'].str.contains('Long term stays allowed'), 'long_term_stays'] = 1
df.loc[df['amenities'].str.contains('Pets|pet|Cat(s)|Dog(s)'), 'pets_allowed'] = 1
df.loc[df['amenities'].str.contains('Private entrance'), 'private_entrance'] = 1
df.loc[df['amenities'].str.contains('Safe|Security system'), 'secure'] = 1
df.loc[df['amenities'].str.contains('Self check-in'), 'self_check_in'] = 1
df.loc[df['amenities'].str.contains('Smoking allowed'), 'smoking_allowed'] = 1
df.loc[df['amenities'].str.contains('Step-free access|Wheelchair|Accessible'), 'accessible'] = 1
df.loc[df['amenities'].str.contains('Suitable for events'), 'event_suitable'] = 1
# Replacing nulls with zeros for new columns
cols_to_replace_nulls = df.iloc[:,41:].columns
df[cols_to_replace_nulls] = df[cols_to_replace_nulls].fillna(0)
# Produces a list of amenity features where one category (true or false) contains fewer than 10% of listings
infrequent_amenities = []
for col in df.iloc[:,41:].columns:
if df[col].sum() < len(df)/10:
infrequent_amenities.append(col)
print(infrequent_amenities)
# Dropping infrequent amenity features
df.drop(infrequent_amenities, axis=1, inplace=True)
# Dropping the original amenity feature
df.drop('amenities', axis=1, inplace=True)
df.columns[40:]
df.price = df.price.str[1:-3]
df.price = df.price.str.replace(",", "")
df.price = df.price.astype('int64')
df.drop(['availability_30', 'availability_60', 'availability_365'], axis=1, inplace=True)
print(f"Null values in 'first_review': {round(100*df.first_review.isna().sum()/len(df),1)}%")
print(f"Null values in 'review_scores_rating': {round(100*df.review_scores_rating .isna().sum()/len(df),1)}%")
df.first_review = pd.to_datetime(df.first_review) # Converting to datetime
# Calculating the number of days between the first review and the date the data was scraped
df['time_since_first_review'] = (pd.datetime(2019, 4, 9) - df.first_review).astype('timedelta64[D]')
# Distribution of the number of days since first review
df.time_since_first_review.hist(figsize=(15,5), bins=30);
def bin_column(col, bins, labels, na_label='unknown'):
"""
Takes in a column name, bin cut points and labels, replaces the original column with a
binned version, and replaces nulls (with 'unknown' if unspecified).
"""
df[col] = pd.cut(df[col], bins=bins, labels=labels, include_lowest=True)
df[col] = df[col].astype('str')
df[col].fillna(na_label, inplace=True)
# Binning time since first review
bin_column('time_since_first_review',
bins=[0, 182, 365, 730, 1460, max(df.time_since_first_review)],
labels=['0-6 months',
'6-12 months',
'1-2 years',
'2-3 years',
'4+ years'],
na_label='no reviews')
df.last_review = pd.to_datetime(df.last_review) # Converting to datetime
# Calculating the number of days between the most recent review and the date the data was scraped
df['time_since_last_review'] = (pd.datetime(2019, 4, 9) - df.last_review).astype('timedelta64[D]')
# Distribution of the number of days since last review
df.time_since_last_review.hist(figsize=(15,5), bins=30);
# Binning time since last review
bin_column('time_since_last_review',
bins=[0, 14, 60, 182, 365, max(df.time_since_last_review)],
labels=['0-2 weeks',
'2-8 weeks',
'2-6 months',
'6-12 months',
'1+ year'],
na_label='no reviews')
# Dropping last_review - first_review will be kept for EDA and dropped later
df.drop('last_review', axis=1, inplace=True)
# Checking the distributions of the review ratings columns
variables_to_plot = list(df.columns[df.columns.str.startswith("review_scores") == True])
fig = plt.figure(figsize=(12,8))
for i, var_name in enumerate(variables_to_plot):
ax = fig.add_subplot(3,3,i+1)
df[var_name].hist(bins=10,ax=ax)
ax.set_title(var_name)
fig.tight_layout()
plt.show()
# Creating a list of all review columns that are scored out of 10
variables_to_plot.pop(0)
# Binning for all columns scored out of 10
for col in variables_to_plot:
bin_column(col,
bins=[0, 8, 9, 10],
labels=['0-8/10', '9/10', '10/10'],
na_label='no reviews')
# Binning column scored out of 100
bin_column('review_scores_rating',
bins=[0, 80, 95, 100],
labels=['0-79/100', '80-94/100', '95-100/100'],
na_label='no reviews')
df.drop(['number_of_reviews_ltm', 'reviews_per_month'], axis=1, inplace=True)
df.head()
df.dtypes
# Save cleaned dataset
listings_cleaned = df.to_csv(r'/Users/Edo/Resources/listings_cleaned.csv', index=id, header=True)
| 0.464173 | 0.681594 |
```
from opf_python import sc
from opf_python import fcc
from opf_python import bcc
from opf_python import rhom
from opf_python import stet
from opf_python import hx
from opf_python import body_tet
from opf_python import so
from opf_python import base_ortho
from opf_python import face_ortho
from opf_python import body_ortho
from opf_python import sm
from opf_python import base_mono
import time
sc_res = []
count = 0
start = time.time()
for n in range(1,100001):
temp = sc.sc_3(n)
count += len(temp)
if n%1000==0:
end = time.time()
interval = end-start
sc_res.append([n,interval,count])
print("n: ",n," time: ",interval," count: ",count)
fcc_res = []
count = 0
start = time.time()
for n in range(1,100001):
temp = fcc.fcc_1(n)
count += len(temp)
if n%1000==0:
end = time.time()
interval = end-start
fcc_res.append([n,interval,count])
print("n: ",n," time: ",interval," count: ",count)
fcc_res2 = []
count = 0
start = time.time()
for n in range(1,47):
for m in [1,4,16]:
if (n*n*n*m)<100000:
temp = fcc.fcc_1(n*n*n*m)
count += len(temp)
end = time.time()
interval = end-start
fcc_res2.append([n*n*n*m,interval,count])
print("n: ",n*n*n*m," time: ",interval," count: ",count)
so_res = []
count = 0
start = time.time()
for n in range(1,100001):
temp = so.so_32(n)
count += len(temp)
if n%1000==0:
end = time.time()
interval = end-start
so_res.append([n,interval,count])
print("n: ",n," time: ",interval," count: ",count)
fo_res = []
count = 0
start = time.time()
for n in range(1,100001):
temp = face_ortho.face_ortho_26(n)
count += len(temp)
if n%1000==0:
end = time.time()
interval = end-start
fo_res.append([n,interval,count])
print("n: ",n," time: ",interval," count: ",count)
import matplotlib.pyplot as plt
xf = []
tf = []
nf = []
for f in fo_res:
xf.append(f[0])
tf.append(f[1])
nf.append(f[2])
xs = []
ts = []
ns = []
for s in so_res:
xs.append(s[0])
ts.append(s[1])
ns.append(s[2])
xf1 = []
tf1 = []
nf1 = []
for f in fcc_res:
xf1.append(f[0])
tf1.append(f[1])
nf1.append(f[2])
xf2 = []
tf2 = []
nf2 = []
for f in fcc_res2:
xf2.append(f[0])
tf2.append(f[1])
nf2.append(f[2])
plt.plot(xf,tf,label="fco")
plt.plot(xs,ts,label="so")
plt.plot(xf1,tf1,label="fcc all n")
plt.plot(xf2,tf2,label="fcc relevant n")
plt.xscale('log')
plt.yscale('log')
plt.title("Comulative Time scale for generating srHNFs")
plt.ylabel("Time (sec)")
plt.xlabel("Volume factor (n)")
plt.legend()
plt.savefig("srHNF_algorithm_timing.pdf")
plt.show()
plt.plot(xf,nf,label="fco")
plt.plot(xs,ns,label="so")
plt.plot(xf1,nf1,label="fcc all n")
plt.plot(xf2,nf2,label="fcc relevant n")
plt.xscale('log')
plt.yscale('log')
plt.title("Comulative number of srHNFs")
plt.ylabel("Time (sec)")
plt.xlabel("Volume factor (n)")
plt.legend()
plt.savefig("srHNFs_number.pdf")
plt.show()
```
|
github_jupyter
|
from opf_python import sc
from opf_python import fcc
from opf_python import bcc
from opf_python import rhom
from opf_python import stet
from opf_python import hx
from opf_python import body_tet
from opf_python import so
from opf_python import base_ortho
from opf_python import face_ortho
from opf_python import body_ortho
from opf_python import sm
from opf_python import base_mono
import time
sc_res = []
count = 0
start = time.time()
for n in range(1,100001):
temp = sc.sc_3(n)
count += len(temp)
if n%1000==0:
end = time.time()
interval = end-start
sc_res.append([n,interval,count])
print("n: ",n," time: ",interval," count: ",count)
fcc_res = []
count = 0
start = time.time()
for n in range(1,100001):
temp = fcc.fcc_1(n)
count += len(temp)
if n%1000==0:
end = time.time()
interval = end-start
fcc_res.append([n,interval,count])
print("n: ",n," time: ",interval," count: ",count)
fcc_res2 = []
count = 0
start = time.time()
for n in range(1,47):
for m in [1,4,16]:
if (n*n*n*m)<100000:
temp = fcc.fcc_1(n*n*n*m)
count += len(temp)
end = time.time()
interval = end-start
fcc_res2.append([n*n*n*m,interval,count])
print("n: ",n*n*n*m," time: ",interval," count: ",count)
so_res = []
count = 0
start = time.time()
for n in range(1,100001):
temp = so.so_32(n)
count += len(temp)
if n%1000==0:
end = time.time()
interval = end-start
so_res.append([n,interval,count])
print("n: ",n," time: ",interval," count: ",count)
fo_res = []
count = 0
start = time.time()
for n in range(1,100001):
temp = face_ortho.face_ortho_26(n)
count += len(temp)
if n%1000==0:
end = time.time()
interval = end-start
fo_res.append([n,interval,count])
print("n: ",n," time: ",interval," count: ",count)
import matplotlib.pyplot as plt
xf = []
tf = []
nf = []
for f in fo_res:
xf.append(f[0])
tf.append(f[1])
nf.append(f[2])
xs = []
ts = []
ns = []
for s in so_res:
xs.append(s[0])
ts.append(s[1])
ns.append(s[2])
xf1 = []
tf1 = []
nf1 = []
for f in fcc_res:
xf1.append(f[0])
tf1.append(f[1])
nf1.append(f[2])
xf2 = []
tf2 = []
nf2 = []
for f in fcc_res2:
xf2.append(f[0])
tf2.append(f[1])
nf2.append(f[2])
plt.plot(xf,tf,label="fco")
plt.plot(xs,ts,label="so")
plt.plot(xf1,tf1,label="fcc all n")
plt.plot(xf2,tf2,label="fcc relevant n")
plt.xscale('log')
plt.yscale('log')
plt.title("Comulative Time scale for generating srHNFs")
plt.ylabel("Time (sec)")
plt.xlabel("Volume factor (n)")
plt.legend()
plt.savefig("srHNF_algorithm_timing.pdf")
plt.show()
plt.plot(xf,nf,label="fco")
plt.plot(xs,ns,label="so")
plt.plot(xf1,nf1,label="fcc all n")
plt.plot(xf2,nf2,label="fcc relevant n")
plt.xscale('log')
plt.yscale('log')
plt.title("Comulative number of srHNFs")
plt.ylabel("Time (sec)")
plt.xlabel("Volume factor (n)")
plt.legend()
plt.savefig("srHNFs_number.pdf")
plt.show()
| 0.223631 | 0.202266 |
## 0. Imports
```
import numpy as np
import matplotlib.pyplot as plt
plt.style.use('seaborn-whitegrid')
from ipywidgets import interactive
import matplotlib as mpl
%matplotlib widget
# %matplotlib notebook
```
## 1. Daten generieren
Generiere Trainings- und Testdaten mit einem Merkmal und einer kontinuierlichen Zielvariablen.
Zwischen den Variabeln herrscht ein logistischer Zusammenhang.
```
# der wahre funktionale Zusammenhang zwischen
def true_function(x):
f = 5 / (1 + np.exp(-x + 2))
return f
rng = np.random.RandomState(1)
# Daten - Beobachtungen / Merkmale
x_train = 10 * rng.rand(20)
x_test = 10 * rng.rand(20)
# Daten - Zielvariablen
y_train = true_function(x_train) + 0.5 * rng.randn(20)
y_test = true_function(x_test) + 0.5 * rng.randn(20)
# Zum Plotten
xx = np.linspace(0, 10)
ff = true_function(xx)
```
## 1.1. Visualisierung der Daten
```
plt.figure()
# plt.plot(xx, ff)
plt.scatter(x_train, y_train, alpha=0.7)
plt.scatter(x_test, y_test, alpha=0.7)
```
## 1.2. Visualisierung der Verlustfunktion
```
# convenience functions and variables zur Darstellung der Verlustfunktion
def empirical_risk(w, b, x_sample, y_sample):
# makes heavy use of broadcasting
W = np.repeat(w[..., np.newaxis], x_sample.shape[0], axis=-1)
B = np.repeat(b[..., np.newaxis], x_sample.shape[0], axis=-1)
Y_pred = W * x_sample + B
loss = np.mean((Y_pred - y_sample)**2, axis=-1)
return loss
def weight_norm(W, B):
return W**2 + B**2
def L1_norm(W, B):
return np.abs(W) + np.abs(B)
ws = np.linspace(-10, 10, 1000)
bs = np.linspace(-10, 10, 1000)
def get_argmin(L):
argmin = np.argmin(L)
argmin = np.unravel_index(argmin, L.shape)
return ws[argmin[0]], bs[argmin[1]]
W, B = np.meshgrid(ws, bs)
L = empirical_risk(W, B, x_train, y_train)
L_reg = weight_norm(W, B)
L_reg_l1 = L1_norm(W, B)
L_test = empirical_risk(W, B, x_test, y_test)
L_min, L_max = L.min(), L.max()
```
### 1.2.1. Empirsche Verlustfunktion ohne Regularisierung
```
from mpl_toolkits import mplot3d
fig = plt.figure(figsize=(8, 6))
ax = plt.axes(projection="3d")
ax.contour3D(W, B, L, 50, cmap="viridis")
ax.set_xlabel(r"$w$")
ax.set_ylabel(r"$b$")
ax.set_zlabel(r"$\mathcal{L}_{E}$");
ax.view_init(30, 75)
```
### 1.2.2. Regularisierungsfunktion
```
plt.close("all")
fig = plt.figure(figsize=(8, 6))
ax = plt.axes(projection="3d")
ax.contour3D(W, B, L_reg, 50, cmap="Greens")
# ax.contour3D(W, B, lambda_*L_reg, 50, cmap="Greens")
# ax.contour3D(W, B, L_reg_l1, 50, cmap="Greens")
ax.set_xlabel(r"$w$")
ax.set_ylabel(r"$b$")
ax.set_zlabel(r"$\mathcal{L}_{reg}$");
ax.view_init(30, 75)
```
### 1.2.3. Empirische Verlustfunktion + Regularisierungsfunktion
```
plt.close("all")
alpha = 1.0
fig = plt.figure(figsize=(8, 6))
ax = plt.axes(projection="3d")
ax.contour3D(W, B, L + alpha * L_reg, 50, cmap="rainbow")
ax.set_xlabel(r"$w$")
ax.set_ylabel(r"$b$")
ax.set_zlabel(r"$\mathcal{L}_E + \mathcal{L}_{reg}$");
ax.view_init(30, 75)
fig = plt.figure(figsize=(8, 6))
ax = plt.axes(projection="3d")
ax.contour3D(W, B, L, 50, cmap="viridis")
ax.set_xlabel(r"$w$")
ax.set_ylabel(r"$b$")
ax.set_zlabel(r"$\mathcal{L}_E$");
ax.view_init(30, 75)
```
### 1.2.4 Empirische Verlustfunktion vs Test-Verlustfunktion
```
alpha = 5.0
plt.close("all")
fig = plt.figure(figsize=(12, 6))
plt.subplot(131)
plt.contour(W, B, L, 50, cmap="viridis")
plt.title(r"$\mathcal{L}_E$")
plt.subplot(132)
plt.contour(W, B, L + alpha*L_reg , 50, cmap="rainbow")
plt.title(r"$\mathcal{L}_E + \alpha \cdot \mathcal{L}_{reg}$")
plt.subplot(133)
plt.contour(W, B, L_test, 50, cmap="seismic")
plt.title(r"$\mathcal{L}_T$");
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
plt.style.use('seaborn-whitegrid')
from ipywidgets import interactive
import matplotlib as mpl
%matplotlib widget
# %matplotlib notebook
# der wahre funktionale Zusammenhang zwischen
def true_function(x):
f = 5 / (1 + np.exp(-x + 2))
return f
rng = np.random.RandomState(1)
# Daten - Beobachtungen / Merkmale
x_train = 10 * rng.rand(20)
x_test = 10 * rng.rand(20)
# Daten - Zielvariablen
y_train = true_function(x_train) + 0.5 * rng.randn(20)
y_test = true_function(x_test) + 0.5 * rng.randn(20)
# Zum Plotten
xx = np.linspace(0, 10)
ff = true_function(xx)
plt.figure()
# plt.plot(xx, ff)
plt.scatter(x_train, y_train, alpha=0.7)
plt.scatter(x_test, y_test, alpha=0.7)
# convenience functions and variables zur Darstellung der Verlustfunktion
def empirical_risk(w, b, x_sample, y_sample):
# makes heavy use of broadcasting
W = np.repeat(w[..., np.newaxis], x_sample.shape[0], axis=-1)
B = np.repeat(b[..., np.newaxis], x_sample.shape[0], axis=-1)
Y_pred = W * x_sample + B
loss = np.mean((Y_pred - y_sample)**2, axis=-1)
return loss
def weight_norm(W, B):
return W**2 + B**2
def L1_norm(W, B):
return np.abs(W) + np.abs(B)
ws = np.linspace(-10, 10, 1000)
bs = np.linspace(-10, 10, 1000)
def get_argmin(L):
argmin = np.argmin(L)
argmin = np.unravel_index(argmin, L.shape)
return ws[argmin[0]], bs[argmin[1]]
W, B = np.meshgrid(ws, bs)
L = empirical_risk(W, B, x_train, y_train)
L_reg = weight_norm(W, B)
L_reg_l1 = L1_norm(W, B)
L_test = empirical_risk(W, B, x_test, y_test)
L_min, L_max = L.min(), L.max()
from mpl_toolkits import mplot3d
fig = plt.figure(figsize=(8, 6))
ax = plt.axes(projection="3d")
ax.contour3D(W, B, L, 50, cmap="viridis")
ax.set_xlabel(r"$w$")
ax.set_ylabel(r"$b$")
ax.set_zlabel(r"$\mathcal{L}_{E}$");
ax.view_init(30, 75)
plt.close("all")
fig = plt.figure(figsize=(8, 6))
ax = plt.axes(projection="3d")
ax.contour3D(W, B, L_reg, 50, cmap="Greens")
# ax.contour3D(W, B, lambda_*L_reg, 50, cmap="Greens")
# ax.contour3D(W, B, L_reg_l1, 50, cmap="Greens")
ax.set_xlabel(r"$w$")
ax.set_ylabel(r"$b$")
ax.set_zlabel(r"$\mathcal{L}_{reg}$");
ax.view_init(30, 75)
plt.close("all")
alpha = 1.0
fig = plt.figure(figsize=(8, 6))
ax = plt.axes(projection="3d")
ax.contour3D(W, B, L + alpha * L_reg, 50, cmap="rainbow")
ax.set_xlabel(r"$w$")
ax.set_ylabel(r"$b$")
ax.set_zlabel(r"$\mathcal{L}_E + \mathcal{L}_{reg}$");
ax.view_init(30, 75)
fig = plt.figure(figsize=(8, 6))
ax = plt.axes(projection="3d")
ax.contour3D(W, B, L, 50, cmap="viridis")
ax.set_xlabel(r"$w$")
ax.set_ylabel(r"$b$")
ax.set_zlabel(r"$\mathcal{L}_E$");
ax.view_init(30, 75)
alpha = 5.0
plt.close("all")
fig = plt.figure(figsize=(12, 6))
plt.subplot(131)
plt.contour(W, B, L, 50, cmap="viridis")
plt.title(r"$\mathcal{L}_E$")
plt.subplot(132)
plt.contour(W, B, L + alpha*L_reg , 50, cmap="rainbow")
plt.title(r"$\mathcal{L}_E + \alpha \cdot \mathcal{L}_{reg}$")
plt.subplot(133)
plt.contour(W, B, L_test, 50, cmap="seismic")
plt.title(r"$\mathcal{L}_T$");
| 0.600891 | 0.929184 |
# Descarga de históricos de precios
<img style="float: right; margin: 0px 0px 15px 15px;" src="https://upload.wikimedia.org/wikipedia/commons/7/7d/Copper_Price_History_USD.png" width="600px" height="400px" />
> Entonces, en la clase anterior vimos que podemos caracterizar la distribución de rendimientos de un activo mediante una medida de tendencia central (media: rendimiento esperado) y una medida de dispersión (desviación estándar: volatilidad).
> Estas medidas se pueden calcular cuando tenemos escenarios probables de la economía y conocemos sus probabilidades de ocurrencia. Ahora, si no conocemos dichos escenarios, ¿qué podemos hacer?
Como no hay naide que nos diga las probabilidades de los sucesos debemos de tomar los hostóricos que necesitamos y analizarlos.
*Objetivos:*
- Aprender a importar datos desde archivos separados por comas (extensión `.csv`).
- Descargar el paquete `pandas-datareader`.
- Aprender a descargar datos desde fuentes remotas.
**Referencias:**
- http://pandas.pydata.org/
- https://pandas-datareader.readthedocs.io/en/latest/
## 1. Importar datos desde archivos locales
<img style="float: left; margin: 0px 0px 15px 15px;" src="https://upload.wikimedia.org/wikipedia/commons/8/86/Microsoft_Excel_2013_logo.svg" width="300px" height="125px" />
<img style="float: right; margin: 0px 0px 15px 15px;" src="https://upload.wikimedia.org/wikipedia/commons/0/0a/Python.svg" width="300px" height="125px" />
### 1.1. ¿Porqué?
- Muchas veces tenemos bases de datos proporcionadas como archivos locales.
- Para poder analizar, procesar y tomar decisiones con estos datos, es necesario importarlos a python.
- Ejemplos de archivos donde comúnmente se guardan bases de datos son:
- `.xls` o `.xlsx`
- `.cvs`
- Excel es ampliamente usado en distintos campos de aplicación en todo el mundo.
- Nos guste o no, esto también aplica a ciencia de datos (ingeniería financiera).
- Muchos de ustedes en su futuro académico y profesional tendrán que trabajar con estas hojas de cálculo, pero no siempre querrán trabajar directamente con ellas si tienen que hacer un análisis un poco más avanzado de los datos.
- Por eso en Python se han implementado herramientas para leer, escribir y manipular este tipo de archivos.
En esta clase veremos cómo podemos trabajar con Excel y Python de manera básica utilizando la librería *pandas*.
### 1.2. Reglas básicas para antes de leer hojas de cálculo
Antes de comenzar a leer una hoja de cálculo en Python (o cualquier otro programa), debemos considerar el ajustar nuestro archivo para cumplir ciertos principios, como:
- La primer fila de la hoja de cálculo se reserva para los títulos, mientras que la primer columna se usa para identificar la unidad de muestreo o indización de los datos (tiempo, fecha, eventos...)
- Evitar nombres, valores o campos con espacios en blanco. De otra manera, cada palabra se interpreta como variable separada y resultan errores relacionados con el número de elementos por línea.
- Los nombres cortos se prefieren sobre nombre largos.
- Evite símbolos como ?, $, %, ^, &, *, (,),-,#, ?, ,,<,>, /, |, \, [ ,] , {, y }.
- Borre cualquier tipo de comentario que haya hecho en su archivo para evitar columnas extras.
- Asegúrese de que cualquier valor inexistente esté indicado como NA.
Si se hizo algún cambio, estar seguro de guardarlo.
Si estás trabajando con Microsoft Excel, verás que hay muchas opciones para guardar archivos, a parte de las extensiones por defecto .xls or .xlsx. Para esto ir a “Save As” y seleccionar una de las extensiones listadas en “Save as Type”.
La extensión más común es .csv (archivos de texto separados por comas).
**Actividad.** Descargar precios de acciones de Apple (AAPL) de Yahoo Finance, con una ventana de tiempo desde el 01-01-2015 al 31-12-2017 y frecuencia diaria.
- Ir a https://finance.yahoo.com/.
- Buscar cada una de las compañías solicitadas.
- Dar click en la pestaña *'Historical Data'*.
- Cambiar las fechas en *'Time Period'*, click en *'Apply'* y, finalmente, click en *'Download Data'*.
- **¡POR FAVOR! GUARDAR ESTOS ARCHIVOS EN UNA CARPETA LLAMADA precios EN EL MISMO DIRECTORIO DONDE TIENEN ESTE ARCHIVO**.
### 1.3. Carguemos archivos .csv como ventanas de datos de pandas
Ahora podemos comenzar a importar nuestros archivos.
Una de las formas más comunes de trabajar con análisis de datos es en pandas. Esto es debido a que pandas está construido sobre NumPy y provee estructuras de datos y herramientas de análisis fáciles de usar.
```
# Importamos pandas
import pandas as pd
```
Para leer archivos `.csv`, utilizaremos la función `read_csv` de pandas:
```
# Función read_csv
help(pd.read_csv)
# Cargamos hoja de calculo en un dataframe
aapl = pd.read_csv('precios/AAPL.csv')
aapl
```
#### Anotación #1
- Quisieramos indizar por fecha.
```
# Cargamos hoja de calculo en un dataframe
aapl = pd.read_csv('precios/AAPL.csv', index_col='Date')
aapl
%matplotlib inline
# Graficar precios de cierre y precios de cierre ajustados
#Debemos de colocar doble corchete para acceder correctamente
aapl[['Close','Adj Close']].plot(grid=True)
```
#### Anotación #2
- Para nuestra aplicación solo nos interesan los precios de cierre de las acciones (columna Adj Close).
```
# Cargamos hoja de calculo en un dataframe
aapl = pd.read_csv('precios/AAPL.csv',
index_col ='Date',
usecols =['Date','Adj Close']
)
aapl
```
**Actividad.** Importen todos los archivos .csv como acabamos de hacerlo con el de apple. Además, crear un solo DataFrame que cuyos encabezados por columna sean los nombres respectivos (AAPL, AMZN,GFNORTEO.MX,GFINBURO.MX) y contengan los datos de precio de cierre.
```
aapl = pd.read_csv('precios/AAPL.csv',index_col ='Date',usecols =['Date','Adj Close'])
amzn = pd.read_csv('precios/AMZN.csv',index_col ='Date',usecols =['Date','Adj Close'])
gfn = pd.read_csv('precios/GFNORTEO.MX.csv',index_col ='Date',usecols =['Date','Adj Close'])
gfi = pd.read_csv('precios/GFINBURO.MX.csv',index_col ='Date',usecols =['Date','Adj Close'])
closes=pd.DataFrame(data={'AAPL':aapl['Adj Close'],
'AMZN':amzn['Adj Close'],
'GFNORTEO.MX':gfn['Adj Close'],
'GFINBURO.MX':gfi['Adj Close']})
closes
```
## 2. Descargar los datos remotamente
Para esto utilizaremos el paquete *pandas_datareader*.
**Nota**: Usualmente, las distribuciones de Python no cuentan, por defecto, con el paquete *pandas_datareader*. Por lo que será necesario instalarlo aparte:
- buscar en inicio "Anaconda prompt" y ejecutarlo como administrador;
- el siguiente comando instala el paquete en Anaconda: *conda install pandas-datareader*;
- una vez finalice la instalación correr el comando: *conda list*, y buscar que sí se haya instalado pandas-datareader
tiene que ser del 7 para arriba
```
# Importar el modulo data del paquete pandas_datareader. La comunidad lo importa con el nombre de web
import pandas_datareader.data as web
```
El módulo data del paquete pandas_datareader contiene la funcion `DataReader`:
```
# Función DataReader
help(web.DataReader)
```
- A esta función le podemos especificar la fuente de los datos para que se use la api específica para la descarga de datos de cada fuente.
- Fuentes:
- Google Finance: su api ya no sirve.
- Quandl: necesita registrarse para obtener un api key.
- IEX: los datos tienen antiguedad máxima de 5 años y de equities estadounidenses.
- Yahoo! Finance: su api ha tenido cambios significativos y ya no es posible usarla desde DataReader. Sin embargo permite obtener datos de distintas bolsas (incluida la mexicana), por eso le haremos la luchita.
```
# Ejemplo google finance
aapl= web.DataReader(name='AAPL',
data_source='google',
start='2015-01-01')
aapl
# Ejemplo iex
aapl= web.DataReader(name='AAPL',
data_source='iex',
start='2015-01-01')
# Ejemplo yahoo
aapl= web.DataReader(name='AAPL',
data_source='yahoo',
start='2015-01-01')
aapl
```
Sin embargo no se pueden descargar varios a la vez. Intentémoslo hacer nosotros así sea de manera rudimentaria:
```
# Función para descargar precios de cierre ajustados:
def get_adj_closes(tickers, start_date=None, end_date=None):
# Fecha inicio por defecto (start_date='2010-01-01') y fecha fin por defecto (end_date=today)
# Descargamos DataFrame con todos los datos
closes = web.DataReader(name=tickers, data_source='yahoo', start=start_date, end=end_date)
# Solo necesitamos los precios ajustados en el cierre
closes = closes['Adj Close']
# Se ordenan los índices de manera ascendente
closes.sort_index(inplace=True)
return closes
```
**Nota**: Para descargar datos de la bolsa mexicana de valores (BMV), el ticker debe tener la extensión MX.
Por ejemplo: *MEXCHEM.MX*, *LABB.MX*, *GFINBURO.MX* y *GFNORTEO.MX*.
```
# Ejemplo: 'AAPL', 'MSFT', 'NVDA', '^GSPC'
closes= get_adj_closes(tickers=['AAPL','MSFT','NVDA','^GSPC'])
closes
# Gráfico
closes.plot(figsize=(10,8),grid=True)
```
**Conclusión**
- Aprendimos a importar datos desde archivos locales.
- Aprendimos a importar datos remotamente con el paquete pandas_datareader. Queda pendiente Infosel (después vemos esto).
¿Ahora qué? Pues con estos históricos, obtendremos los rendimientos y ellos nos servirán como base para caracterizar la distribución de rendimientos...
### ¡Oh, si!
<script>
$(document).ready(function(){
$('div.prompt').hide();
$('div.back-to-top').hide();
$('nav#menubar').hide();
$('.breadcrumb').hide();
$('.hidden-print').hide();
});
</script>
<footer id="attribution" style="float:right; color:#808080; background:#fff;">
Created with Jupyter by Esteban Jiménez Rodríguez.
</footer>
|
github_jupyter
|
# Importamos pandas
import pandas as pd
# Función read_csv
help(pd.read_csv)
# Cargamos hoja de calculo en un dataframe
aapl = pd.read_csv('precios/AAPL.csv')
aapl
# Cargamos hoja de calculo en un dataframe
aapl = pd.read_csv('precios/AAPL.csv', index_col='Date')
aapl
%matplotlib inline
# Graficar precios de cierre y precios de cierre ajustados
#Debemos de colocar doble corchete para acceder correctamente
aapl[['Close','Adj Close']].plot(grid=True)
# Cargamos hoja de calculo en un dataframe
aapl = pd.read_csv('precios/AAPL.csv',
index_col ='Date',
usecols =['Date','Adj Close']
)
aapl
aapl = pd.read_csv('precios/AAPL.csv',index_col ='Date',usecols =['Date','Adj Close'])
amzn = pd.read_csv('precios/AMZN.csv',index_col ='Date',usecols =['Date','Adj Close'])
gfn = pd.read_csv('precios/GFNORTEO.MX.csv',index_col ='Date',usecols =['Date','Adj Close'])
gfi = pd.read_csv('precios/GFINBURO.MX.csv',index_col ='Date',usecols =['Date','Adj Close'])
closes=pd.DataFrame(data={'AAPL':aapl['Adj Close'],
'AMZN':amzn['Adj Close'],
'GFNORTEO.MX':gfn['Adj Close'],
'GFINBURO.MX':gfi['Adj Close']})
closes
# Importar el modulo data del paquete pandas_datareader. La comunidad lo importa con el nombre de web
import pandas_datareader.data as web
# Función DataReader
help(web.DataReader)
# Ejemplo google finance
aapl= web.DataReader(name='AAPL',
data_source='google',
start='2015-01-01')
aapl
# Ejemplo iex
aapl= web.DataReader(name='AAPL',
data_source='iex',
start='2015-01-01')
# Ejemplo yahoo
aapl= web.DataReader(name='AAPL',
data_source='yahoo',
start='2015-01-01')
aapl
# Función para descargar precios de cierre ajustados:
def get_adj_closes(tickers, start_date=None, end_date=None):
# Fecha inicio por defecto (start_date='2010-01-01') y fecha fin por defecto (end_date=today)
# Descargamos DataFrame con todos los datos
closes = web.DataReader(name=tickers, data_source='yahoo', start=start_date, end=end_date)
# Solo necesitamos los precios ajustados en el cierre
closes = closes['Adj Close']
# Se ordenan los índices de manera ascendente
closes.sort_index(inplace=True)
return closes
# Ejemplo: 'AAPL', 'MSFT', 'NVDA', '^GSPC'
closes= get_adj_closes(tickers=['AAPL','MSFT','NVDA','^GSPC'])
closes
# Gráfico
closes.plot(figsize=(10,8),grid=True)
| 0.30054 | 0.955068 |
```
# Run this!
import warnings
warnings.filterwarnings("ignore")
import tensorflow as tf
import keras
print('keras using %s backend'%keras.backend.backend())
import pandas as pd
import numpy as np
import matplotlib.pyplot as graph
%matplotlib inline
graph.rcParams['figure.figsize'] = (15,5)
graph.rcParams["font.family"] = 'DejaVu Sans'
graph.rcParams["font.size"] = '12'
graph.rcParams['image.cmap'] = 'rainbow'
# This loads the dataset
dataset = pd.read_csv('Data/dog_data.csv')
print(dataset.head())
# This tells us the shape of the data set
print("Shape of data set:", dataset.shape)
# Defines the feature dataframe
features = dataset.drop(['breed'], axis = 1)
from sklearn.preprocessing import OneHotEncoder
# This sets the labels (numerical)
labels = np.array(dataset['breed'])
onehot = OneHotEncoder(sparse = False).fit_transform(np.transpose([labels]))
print(onehot[:5])
```
### Modelo a entrenar con 160 datos y testear con 40
```
# Run this! This sets up our training and test sets.
# This takes the first 160 examples for our training set
train_X = features.values[:160]
train_Y = onehot[:160]
# This takes the last 40 examples of the 200 for our test set
test_X = features.values[160:]
test_Y = onehot[160:]
```
### Primera capa - capa de entradas con 3 nodos
### Segunda " - 4 nodos
### Tercera " - 2 nodos
### Cuarta " - capa de salidas con 3 nodos
```
# Set a randomisation seed for replicatability.
np.random.seed(6)
# This creates our base model for us to add to
model = keras.models.Sequential()
structure = [3, 4, 2, 3]
# Input layer + hidden layer 1
model.add(keras.layers.Dense(units=structure[1], input_dim = structure[0], activation = 'relu'))
# Hidden layer 2
model.add(keras.layers.Dense(units=structure[2], activation = 'relu'))
# Output layer - note that the activation function is softmax
# Softmax will predict a category and provide a value for how likely this is the correct prediction.
model.add(keras.layers.Dense(units=structure[3], activation = tf.nn.softmax))
print("Layer structure:", structure)
# Let's compile the model
model.compile(loss = 'categorical_crossentropy', optimizer = 'sgd', metrics = ['accuracy'])
# Time to fit the model
print('Starting training')
training_stats = model.fit(train_X, train_Y, batch_size = 1, epochs = 24, verbose = 0)
print('Training finished')
print('Training Evaluation: loss = %0.3f, accuracy = %0.2f%%'
%(training_stats.history['loss'][-1], 100 * training_stats.history['accuracy'][-1]))
# Run this!
accuracy, = graph.plot(training_stats.history['accuracy'],label = 'Accuracy')
training_loss, = graph.plot(training_stats.history['loss'],label = 'Training Loss')
graph.legend(handles = [accuracy,training_loss])
loss = np.array(training_stats.history['loss'])
xp = np.linspace(0, loss.shape[0], 10 * loss.shape[0])
graph.plot(xp, np.full(xp.shape, 1), c = 'k', linestyle = ':', alpha = 0.5)
graph.plot(xp, np.full(xp.shape, 0), c = 'k', linestyle = ':', alpha = 0.5)
graph.show()
evaluation = model.evaluate(test_X, test_Y, verbose=0)
print('Test Set Evaluation: loss = %0.6f, accuracy = %0.2f' %(evaluation[0], 100*evaluation[1]))
# [age, weight, height]
new_sample = [9, 7, 7]
graph.plot(new_sample[0], new_sample[1], 'ko', marker='x')
graph.scatter(train_X[:,0], train_X[:,1], c = labels[:160])
graph.title('samples by age and weight')
graph.xlabel('age')
graph.ylabel('weight')
graph.show()
# Plot out the age-height relationship
graph.plot(new_sample[0], new_sample[2], 'ko', marker='x')
graph.scatter(train_X[:,0], train_X[:,2], c = labels[:160])
graph.title('samples by age and height')
graph.xlabel('age')
graph.ylabel('height')
graph.show()
predicted = model.predict(np.array([new_sample]))
print('Breed prediction for %s:' %(new_sample))
print(np.around(predicted[0],2))
print('Breed %s, with %i%% certainty.' %(np.argmax(predicted), np.round(100 * predicted[:, np.argmax(predicted)][0])))
```
|
github_jupyter
|
# Run this!
import warnings
warnings.filterwarnings("ignore")
import tensorflow as tf
import keras
print('keras using %s backend'%keras.backend.backend())
import pandas as pd
import numpy as np
import matplotlib.pyplot as graph
%matplotlib inline
graph.rcParams['figure.figsize'] = (15,5)
graph.rcParams["font.family"] = 'DejaVu Sans'
graph.rcParams["font.size"] = '12'
graph.rcParams['image.cmap'] = 'rainbow'
# This loads the dataset
dataset = pd.read_csv('Data/dog_data.csv')
print(dataset.head())
# This tells us the shape of the data set
print("Shape of data set:", dataset.shape)
# Defines the feature dataframe
features = dataset.drop(['breed'], axis = 1)
from sklearn.preprocessing import OneHotEncoder
# This sets the labels (numerical)
labels = np.array(dataset['breed'])
onehot = OneHotEncoder(sparse = False).fit_transform(np.transpose([labels]))
print(onehot[:5])
# Run this! This sets up our training and test sets.
# This takes the first 160 examples for our training set
train_X = features.values[:160]
train_Y = onehot[:160]
# This takes the last 40 examples of the 200 for our test set
test_X = features.values[160:]
test_Y = onehot[160:]
# Set a randomisation seed for replicatability.
np.random.seed(6)
# This creates our base model for us to add to
model = keras.models.Sequential()
structure = [3, 4, 2, 3]
# Input layer + hidden layer 1
model.add(keras.layers.Dense(units=structure[1], input_dim = structure[0], activation = 'relu'))
# Hidden layer 2
model.add(keras.layers.Dense(units=structure[2], activation = 'relu'))
# Output layer - note that the activation function is softmax
# Softmax will predict a category and provide a value for how likely this is the correct prediction.
model.add(keras.layers.Dense(units=structure[3], activation = tf.nn.softmax))
print("Layer structure:", structure)
# Let's compile the model
model.compile(loss = 'categorical_crossentropy', optimizer = 'sgd', metrics = ['accuracy'])
# Time to fit the model
print('Starting training')
training_stats = model.fit(train_X, train_Y, batch_size = 1, epochs = 24, verbose = 0)
print('Training finished')
print('Training Evaluation: loss = %0.3f, accuracy = %0.2f%%'
%(training_stats.history['loss'][-1], 100 * training_stats.history['accuracy'][-1]))
# Run this!
accuracy, = graph.plot(training_stats.history['accuracy'],label = 'Accuracy')
training_loss, = graph.plot(training_stats.history['loss'],label = 'Training Loss')
graph.legend(handles = [accuracy,training_loss])
loss = np.array(training_stats.history['loss'])
xp = np.linspace(0, loss.shape[0], 10 * loss.shape[0])
graph.plot(xp, np.full(xp.shape, 1), c = 'k', linestyle = ':', alpha = 0.5)
graph.plot(xp, np.full(xp.shape, 0), c = 'k', linestyle = ':', alpha = 0.5)
graph.show()
evaluation = model.evaluate(test_X, test_Y, verbose=0)
print('Test Set Evaluation: loss = %0.6f, accuracy = %0.2f' %(evaluation[0], 100*evaluation[1]))
# [age, weight, height]
new_sample = [9, 7, 7]
graph.plot(new_sample[0], new_sample[1], 'ko', marker='x')
graph.scatter(train_X[:,0], train_X[:,1], c = labels[:160])
graph.title('samples by age and weight')
graph.xlabel('age')
graph.ylabel('weight')
graph.show()
# Plot out the age-height relationship
graph.plot(new_sample[0], new_sample[2], 'ko', marker='x')
graph.scatter(train_X[:,0], train_X[:,2], c = labels[:160])
graph.title('samples by age and height')
graph.xlabel('age')
graph.ylabel('height')
graph.show()
predicted = model.predict(np.array([new_sample]))
print('Breed prediction for %s:' %(new_sample))
print(np.around(predicted[0],2))
print('Breed %s, with %i%% certainty.' %(np.argmax(predicted), np.round(100 * predicted[:, np.argmax(predicted)][0])))
| 0.829216 | 0.791741 |
# Data Gathering
This recipe shows how we will be accessing the datasets necessary for the rest of the book.
We start by loading the necessary libraries and resetting the computational graph.
```
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
from tensorflow.python.framework import ops
ops.reset_default_graph()
```
## The Iris Dataset (R. Fisher / Scikit-Learn)
One of the most frequently used ML datasets is the iris flower dataset. We will use the easy import tool, `datasets` from scikit-learn. You can read more about it here: http://scikit-learn.org/stable/modules/generated/sklearn.datasets.load_iris.html#sklearn.datasets.load_iris
```
from sklearn.datasets import load_iris
iris = load_iris()
print(len(iris.data))
print(len(iris.target))
print(iris.data[0])
print(set(iris.target))
```
## Low Birthrate Dataset (Hosted on Github)
The 'Low Birthrate Dataset' is a dataset from a famous study by Hosmer and Lemeshow in 1989 called, "Low Infant Birth Weight Risk Factor Study". It is a very commonly used academic dataset mostly for logistic regression. We will host this dataset on the public Github here:
https://github.com/nfmcclure/tensorflow_cookbook/raw/master/01_Introduction/07_Working_with_Data_Sources/birthweight_data/birthweight.dat
```
import requests
birthdata_url = 'https://github.com/nfmcclure/tensorflow_cookbook/raw/master/01_Introduction/07_Working_with_Data_Sources/birthweight_data/birthweight.dat'
birth_file = requests.get(birthdata_url)
birth_data = birth_file.text.split('\r\n')
birth_header = birth_data[0].split('\t')
birth_data = [[float(x) for x in y.split('\t') if len(x)>=1] for y in birth_data[1:] if len(y)>=1]
print(len(birth_data))
print(len(birth_data[0]))
```
## Housing Price Dataset (UCI)
We will also use a housing price dataset from the University of California at Irvine (UCI) Machine Learning Database Repository. It is a great regression dataset to use. You can read more about it here:
https://archive.ics.uci.edu/ml/datasets/Housing
```
import requests
housing_url = 'https://archive.ics.uci.edu/ml/machine-learning-databases/housing/housing.data'
housing_header = ['CRIM', 'ZN', 'INDUS', 'CHAS', 'NOX', 'RM', 'AGE', 'DIS', 'RAD', 'TAX', 'PTRATIO', 'B', 'LSTAT', 'MEDV']
housing_file = requests.get(housing_url)
housing_data = [[float(x) for x in y.split(' ') if len(x)>=1] for y in housing_file.text.split('\n') if len(y)>=1]
print(len(housing_data))
print(len(housing_data[0]))
```
## MNIST Handwriting Dataset (Yann LeCun)
The MNIST Handwritten digit picture dataset is the `Hello World` of image recognition. The famous scientist and researcher, Yann LeCun, hosts it on his webpage here, http://yann.lecun.com/exdb/mnist/ . But because it is so commonly used, many libraries, including TensorFlow, host it internally. We will use TensorFlow to access this data as follows.
If you haven't downloaded this before, please wait a bit while it downloads
```
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("MNIST_data/", one_hot=True)
print(len(mnist.train.images))
print(len(mnist.test.images))
print(len(mnist.validation.images))
print(mnist.train.labels[1,:])
```
## CIFAR-10 Data
The CIFAR-10 data ( https://www.cs.toronto.edu/~kriz/cifar.html ) contains 60,000 32x32 color images of 10 classes collected by Alex Krizhevsky, Vinod Nair, and Geoffrey Hinton. Alex Krizhevsky maintains the page referenced here. This is such a common dataset, that there are built in functions in TensorFlow to access this data (the keras wrapper has these commands). Note that the keras wrapper for these functions automatically splits the images into a 50,000 training set and a 10,000 test set.
```
from PIL import Image
# Running this command requires an internet connection and a few minutes to download all the images.
(X_train, y_train), (X_test, y_test) = tf.contrib.keras.datasets.cifar10.load_data()
```
The ten categories are (in order):
<ol start="0">
<li>Airplane</li>
<li>Automobile</li>
<li>Bird</li>
<li>Car</li>
<li>Deer</li>
<li>Dog</li>
<li>Frog</li>
<li>Horse</li>
<li>Ship</li>
<li>Truck</li>
</ol>
```
X_train.shape
y_train.shape
y_train[0,] # this is a frog
# Plot the 0-th image (a frog)
%matplotlib inline
img = Image.fromarray(X_train[0,:,:,:])
plt.imshow(img)
```
## Ham/Spam Texts Dataset (UCI)
We will use another UCI ML Repository dataset called the SMS Spam Collection. You can read about it here: https://archive.ics.uci.edu/ml/datasets/SMS+Spam+Collection . As a sidenote about common terms, when predicting if a data point represents 'spam' (or unwanted advertisement), the alternative is called 'ham' (or useful information).
This is a great dataset for predicting a binary outcome (spam/ham) from a textual input. This will be very useful for short text sequences for Natural Language Processing (Ch 7) and Recurrent Neural Networks (Ch 9).
```
import requests
import io
from zipfile import ZipFile
# Get/read zip file
zip_url = 'http://archive.ics.uci.edu/ml/machine-learning-databases/00228/smsspamcollection.zip'
r = requests.get(zip_url)
z = ZipFile(io.BytesIO(r.content))
file = z.read('SMSSpamCollection')
# Format Data
text_data = file.decode()
text_data = text_data.encode('ascii',errors='ignore')
text_data = text_data.decode().split('\n')
text_data = [x.split('\t') for x in text_data if len(x)>=1]
[text_data_target, text_data_train] = [list(x) for x in zip(*text_data)]
print(len(text_data_train))
print(set(text_data_target))
print(text_data_train[1])
```
## Movie Review Data (Cornell)
The Movie Review database, collected by Bo Pang and Lillian Lee (researchers at Cornell), serves as a great dataset to use for predicting a numerical number from textual inputs.
You can read more about the dataset and papers using it here:
https://www.cs.cornell.edu/people/pabo/movie-review-data/
```
import requests
import io
import tarfile
movie_data_url = 'http://www.cs.cornell.edu/people/pabo/movie-review-data/rt-polaritydata.tar.gz'
r = requests.get(movie_data_url)
# Stream data into temp object
stream_data = io.BytesIO(r.content)
tmp = io.BytesIO()
while True:
s = stream_data.read(16384)
if not s:
break
tmp.write(s)
stream_data.close()
tmp.seek(0)
# Extract tar file
tar_file = tarfile.open(fileobj=tmp, mode="r:gz")
pos = tar_file.extractfile('rt-polaritydata/rt-polarity.pos')
neg = tar_file.extractfile('rt-polaritydata/rt-polarity.neg')
# Save pos/neg reviews
pos_data = []
for line in pos:
pos_data.append(line.decode('ISO-8859-1').encode('ascii',errors='ignore').decode())
neg_data = []
for line in neg:
neg_data.append(line.decode('ISO-8859-1').encode('ascii',errors='ignore').decode())
tar_file.close()
print(len(pos_data))
print(len(neg_data))
print(neg_data[0])
```
## The Complete Works of William Shakespeare (Gutenberg Project)
For training a TensorFlow Model to create text, we will train it on the complete works of William Shakespeare. This can be accessed through the good work of the Gutenberg Project. The Gutenberg Project frees many non-copyright books by making them accessible for free from the hard work of volunteers.
You can read more about the Shakespeare works here:
http://www.gutenberg.org/ebooks/100
```
# The Works of Shakespeare Data
import requests
shakespeare_url = 'http://www.gutenberg.org/cache/epub/100/pg100.txt'
# Get Shakespeare text
response = requests.get(shakespeare_url)
shakespeare_file = response.content
# Decode binary into string
shakespeare_text = shakespeare_file.decode('utf-8')
# Drop first few descriptive paragraphs.
shakespeare_text = shakespeare_text[7675:]
print(len(shakespeare_text))
```
## English-German Sentence Translation Database (Manythings/Tatoeba)
The Tatoeba Project is also run by volunteers and is set to make the most bilingual sentence translations available between many different languages. `Manythings.org` compiles the data and makes it accessible.
http://www.manythings.org/corpus/about.html#info
More bilingual sentence pairs: http://www.manythings.org/bilingual/
```
# English-German Sentence Translation Data
import requests
import io
from zipfile import ZipFile
sentence_url = 'http://www.manythings.org/anki/deu-eng.zip'
r = requests.get(sentence_url)
z = ZipFile(io.BytesIO(r.content))
file = z.read('deu.txt')
# Format Data
eng_ger_data = file.decode()
eng_ger_data = eng_ger_data.encode('ascii',errors='ignore')
eng_ger_data = eng_ger_data.decode().split('\n')
eng_ger_data = [x.split('\t') for x in eng_ger_data if len(x)>=1]
[english_sentence, german_sentence] = [list(x) for x in zip(*eng_ger_data)]
print(len(english_sentence))
print(len(german_sentence))
print(eng_ger_data[10])
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
from tensorflow.python.framework import ops
ops.reset_default_graph()
from sklearn.datasets import load_iris
iris = load_iris()
print(len(iris.data))
print(len(iris.target))
print(iris.data[0])
print(set(iris.target))
import requests
birthdata_url = 'https://github.com/nfmcclure/tensorflow_cookbook/raw/master/01_Introduction/07_Working_with_Data_Sources/birthweight_data/birthweight.dat'
birth_file = requests.get(birthdata_url)
birth_data = birth_file.text.split('\r\n')
birth_header = birth_data[0].split('\t')
birth_data = [[float(x) for x in y.split('\t') if len(x)>=1] for y in birth_data[1:] if len(y)>=1]
print(len(birth_data))
print(len(birth_data[0]))
import requests
housing_url = 'https://archive.ics.uci.edu/ml/machine-learning-databases/housing/housing.data'
housing_header = ['CRIM', 'ZN', 'INDUS', 'CHAS', 'NOX', 'RM', 'AGE', 'DIS', 'RAD', 'TAX', 'PTRATIO', 'B', 'LSTAT', 'MEDV']
housing_file = requests.get(housing_url)
housing_data = [[float(x) for x in y.split(' ') if len(x)>=1] for y in housing_file.text.split('\n') if len(y)>=1]
print(len(housing_data))
print(len(housing_data[0]))
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("MNIST_data/", one_hot=True)
print(len(mnist.train.images))
print(len(mnist.test.images))
print(len(mnist.validation.images))
print(mnist.train.labels[1,:])
from PIL import Image
# Running this command requires an internet connection and a few minutes to download all the images.
(X_train, y_train), (X_test, y_test) = tf.contrib.keras.datasets.cifar10.load_data()
X_train.shape
y_train.shape
y_train[0,] # this is a frog
# Plot the 0-th image (a frog)
%matplotlib inline
img = Image.fromarray(X_train[0,:,:,:])
plt.imshow(img)
import requests
import io
from zipfile import ZipFile
# Get/read zip file
zip_url = 'http://archive.ics.uci.edu/ml/machine-learning-databases/00228/smsspamcollection.zip'
r = requests.get(zip_url)
z = ZipFile(io.BytesIO(r.content))
file = z.read('SMSSpamCollection')
# Format Data
text_data = file.decode()
text_data = text_data.encode('ascii',errors='ignore')
text_data = text_data.decode().split('\n')
text_data = [x.split('\t') for x in text_data if len(x)>=1]
[text_data_target, text_data_train] = [list(x) for x in zip(*text_data)]
print(len(text_data_train))
print(set(text_data_target))
print(text_data_train[1])
import requests
import io
import tarfile
movie_data_url = 'http://www.cs.cornell.edu/people/pabo/movie-review-data/rt-polaritydata.tar.gz'
r = requests.get(movie_data_url)
# Stream data into temp object
stream_data = io.BytesIO(r.content)
tmp = io.BytesIO()
while True:
s = stream_data.read(16384)
if not s:
break
tmp.write(s)
stream_data.close()
tmp.seek(0)
# Extract tar file
tar_file = tarfile.open(fileobj=tmp, mode="r:gz")
pos = tar_file.extractfile('rt-polaritydata/rt-polarity.pos')
neg = tar_file.extractfile('rt-polaritydata/rt-polarity.neg')
# Save pos/neg reviews
pos_data = []
for line in pos:
pos_data.append(line.decode('ISO-8859-1').encode('ascii',errors='ignore').decode())
neg_data = []
for line in neg:
neg_data.append(line.decode('ISO-8859-1').encode('ascii',errors='ignore').decode())
tar_file.close()
print(len(pos_data))
print(len(neg_data))
print(neg_data[0])
# The Works of Shakespeare Data
import requests
shakespeare_url = 'http://www.gutenberg.org/cache/epub/100/pg100.txt'
# Get Shakespeare text
response = requests.get(shakespeare_url)
shakespeare_file = response.content
# Decode binary into string
shakespeare_text = shakespeare_file.decode('utf-8')
# Drop first few descriptive paragraphs.
shakespeare_text = shakespeare_text[7675:]
print(len(shakespeare_text))
# English-German Sentence Translation Data
import requests
import io
from zipfile import ZipFile
sentence_url = 'http://www.manythings.org/anki/deu-eng.zip'
r = requests.get(sentence_url)
z = ZipFile(io.BytesIO(r.content))
file = z.read('deu.txt')
# Format Data
eng_ger_data = file.decode()
eng_ger_data = eng_ger_data.encode('ascii',errors='ignore')
eng_ger_data = eng_ger_data.decode().split('\n')
eng_ger_data = [x.split('\t') for x in eng_ger_data if len(x)>=1]
[english_sentence, german_sentence] = [list(x) for x in zip(*eng_ger_data)]
print(len(english_sentence))
print(len(german_sentence))
print(eng_ger_data[10])
| 0.393618 | 0.975414 |
```
import os
import glob
import dask
from dask.distributed import Client, progress, LocalCluster
import geopandas as gpd
import pandas as pd
from pyFIRS.wrappers import lastools
from pyFIRS.wrappers import fusion
from pyFIRS.utils import validation_summary, move_invalid_tiles, fname, PipelineError, inspect_failures
# data handling directories
WORKDIR = os.path.abspath('F:/willamette-valley_2009/')
TARGET_EPSG = 6339 # utm 10N, NAD83_2011
# TARGET_EPSG = 6340 # utm 11N, NAD83_2011
SRC = os.path.join(WORKDIR, 'src')
src_tiles = glob.glob(os.path.join(SRC, '*.laz'))
# src_tiles = glob.glob(os.path.join(SRC, '*.las'))
# where we're going to put processed source tiles
RAW = os.path.join(WORKDIR, 'raw')
print('Found {:,d} tiles in source directory:\n'
' {}'.format(len(src_tiles), SRC))
```
# Enough already, let's get to work with some lidar data
We'll define where we can find the binary executables for LAStools and FUSION command line tools.
```
las = lastools.useLAStools('C:/Program Files/LAStools/bin')
fus = fusion.useFUSION('C:/Program Files/FUSION/')
# take a peak at info from a lidar source tile
info_proc = las.lasinfo(i=src_tiles[0],
echo=True)
```
### Setting up parallel computing using `dask.distributed`
`LAStools` offers native multi-core processing as an optional argument (`cores`) supplied to its command-line tools. `FUSION` command line tools do not. To enable parallel processing of `FUSION` commands, we'll use `dask.distributed` to schedule the processing of tiles in asynchronous parallel batches. This approach also offers us the ability to track progress using a progress bar.
You'll first need to launch a parallel computing cluster.
```
cluster=LocalCluster()#(scheduler_port=7001, dashboard_address=7002)
c = Client(cluster)
```
At this point, you should also be able to view an interactive dashboard on port 7002. If you're executing this on a remote server, you'll need to set up port forward so you can view the dashboard on your local machine's browser. Once you've done that, or if you're processing on your own machine, you can view the dashboard at [http://localhost:7002/status](http://localhost:7002/status).
```
# push our working directories and wrapper classes to the workers on the cluster as well
c.scatter([WORKDIR, SRC, RAW,
las, fus,
TARGET_EPSG],
broadcast=True);
def log_error(tile_id, process, error_msg):
logfile = os.path.join(RAW, 'failed', tile_id + '.txt')
os.makedirs(os.path.dirname(logfile), exist_ok=True)
with open(logfile, '+w') as f:
f.write('{} | {}: {}'.format(tile_id, process, error_msg))
return
def has_error(tile_id):
errors = glob.glob(os.path.join(RAW, 'failed', '*.txt'))
tiles_with_errors = [fname(error) for error in errors]
if tile_id in tiles_with_errors:
return True
else:
return False
```
## Get the raw data into our working directory
First, move the tiles over to our working directory.
When we define functions using the `dask.delayed` decorator, the function will have 'lazy' instead of 'eager' execution. We can map the function to a list of inputs and it will not execute for any of them until we ask for results to be computed. When we use the `compute()` method for the client managing the scheduler that sends jobs to the workers, it then starts running the jobs.
```
@dask.delayed
def import_tile(tile_id):
INFILE = os.path.join(SRC, tile_id + '.laz')
# INFILE = os.path.join(SRC, tile_id + '.las')
OUTFILE = os.path.join(RAW, tile_id + '.laz')
if os.path.exists(OUTFILE):
pass
else:
try:
proc_import = las.las2las(i=INFILE,
drop_withheld=True,
drop_class=(7,18), # classified as noise
# epsg=32149, # specify the source lidar projection, washington state plane south
# epsg=2927, # specify the source lidar projection, washington state plane south
longlat=True, # original data is in geographic coordinates
# elevation_surveyfeet=True,
# survey_feet=True,
# nad83_2011=True, # original data in nad83_2011 datum
nad83_harn=True, # original data in nad83_harn datum
target_epsg=TARGET_EPSG, # reproject
dont_remove_empty_files=True,
cpu64=True,
odir=RAW,
olaz=True)
except PipelineError as e:
log_error(tile_id, 'import_tile', e.message)
return tile_id
```
Next, validate that the data match LAS specifications and have not been corrupted.
```
@dask.delayed
def validate(tile_id):
INFILE = os.path.join(RAW, tile_id + '.laz')
OUTFILE = os.path.join(RAW, tile_id + '.xml')
if os.path.exists(OUTFILE):
pass
else:
try:
proc_validate = las.lasvalidate(i=INFILE,
o=OUTFILE)
except PipelineError as e:
log_error(tile_id, 'validate', e.message)
return tile_id
```
Next, create spatial indexes for the input files to allow fast spatial queries (which are used, for example, when retiling and adding buffers).
```
@dask.delayed
def make_index(tile_id):
INFILE = os.path.join(RAW, tile_id + '.laz')
OUTFILE = os.path.join(RAW, tile_id + '.lax')
if os.path.exists(OUTFILE):
pass
else:
try:
proc_index = las.lasindex(i=INFILE,
cpu64=True)
except PipelineError as e:
log_error(tile_id, 'make_index', e.message)
return tile_id
@dask.delayed
def make_boundary(tile_id):
INFILE = os.path.join(RAW, tile_id + '.laz')
OUTFILE = os.path.join(RAW, tile_id + '.shp')
if os.path.exists(OUTFILE):
pass
else:
try:
proc_bnd = las.lasboundary(i=INFILE,
o=OUTFILE,
disjoint=True,
labels=True,
use_lax=True,
cpu64=True)
except PipelineError as e:
log_error(tile_id, 'make_index', e.message)
return tile_id
```
## Hand-build the computational graph
Define the recipe for computations.
```
tile_ids = [fname(tile) for tile in src_tiles]
get_data = {}
for tile in tile_ids:
get_data['import-{}'.format(tile)]=(
import_tile,
tile)
get_data['validate-{}'.format(tile)]=(
validate,
'import-{}'.format(tile))
get_data['index-{}'.format(tile)]=(
make_index,
'validate-{}'.format(tile))
get_data['boundary-{}'.format(tile)]=(
make_boundary,
'index-{}'.format(tile))
# this empty function will be added to recipe for computations
# it will be defined to depend upon all previous steps being completed
@dask.delayed
def done_importing(*args, **kwargs):
return
get_data['done_importing']=(
done_importing,
['boundary-{}'.format(tile) for tile in tile_ids])
get_data_graph = c.get(get_data, 'done_importing') # build the computational graph
get_data_results = c.persist(get_data_graph) # start executing it
progress(get_data_results) # progress bars
# c.cancel(get_data_results)
inspect_failures(os.path.join(RAW, 'failed'))
validation_summary(xml_dir=RAW, verbose=False)
# move_invalid_tiles(xml_dir=RAW, dest_dir=os.path.join(RAW, 'invalid'))
```
## Merge all the individual tile boundaries into a tile index
```
boundaries = glob.glob(os.path.join(RAW, '*.shp'))
# set up a lazy read_file function so we can read in files in parallel
gdfs = [dask.delayed(gpd.read_file)(shp) for shp in boundaries]
# and a lazy concatenation
gather_tiles = dask.delayed(pd.concat)(gdfs, axis=0, ignore_index=True)
# now execute the read and concatenate with the cluster
tileindex = gather_tiles.compute()
tileindex.crs = "EPSG:{}".format(TARGET_EPSG)
tileindex.head()
tileindex.to_file(os.path.join(RAW, 'raw_tileindex.shp'), index=False)
# c.close()
# cluster.close()
```
|
github_jupyter
|
import os
import glob
import dask
from dask.distributed import Client, progress, LocalCluster
import geopandas as gpd
import pandas as pd
from pyFIRS.wrappers import lastools
from pyFIRS.wrappers import fusion
from pyFIRS.utils import validation_summary, move_invalid_tiles, fname, PipelineError, inspect_failures
# data handling directories
WORKDIR = os.path.abspath('F:/willamette-valley_2009/')
TARGET_EPSG = 6339 # utm 10N, NAD83_2011
# TARGET_EPSG = 6340 # utm 11N, NAD83_2011
SRC = os.path.join(WORKDIR, 'src')
src_tiles = glob.glob(os.path.join(SRC, '*.laz'))
# src_tiles = glob.glob(os.path.join(SRC, '*.las'))
# where we're going to put processed source tiles
RAW = os.path.join(WORKDIR, 'raw')
print('Found {:,d} tiles in source directory:\n'
' {}'.format(len(src_tiles), SRC))
las = lastools.useLAStools('C:/Program Files/LAStools/bin')
fus = fusion.useFUSION('C:/Program Files/FUSION/')
# take a peak at info from a lidar source tile
info_proc = las.lasinfo(i=src_tiles[0],
echo=True)
cluster=LocalCluster()#(scheduler_port=7001, dashboard_address=7002)
c = Client(cluster)
# push our working directories and wrapper classes to the workers on the cluster as well
c.scatter([WORKDIR, SRC, RAW,
las, fus,
TARGET_EPSG],
broadcast=True);
def log_error(tile_id, process, error_msg):
logfile = os.path.join(RAW, 'failed', tile_id + '.txt')
os.makedirs(os.path.dirname(logfile), exist_ok=True)
with open(logfile, '+w') as f:
f.write('{} | {}: {}'.format(tile_id, process, error_msg))
return
def has_error(tile_id):
errors = glob.glob(os.path.join(RAW, 'failed', '*.txt'))
tiles_with_errors = [fname(error) for error in errors]
if tile_id in tiles_with_errors:
return True
else:
return False
@dask.delayed
def import_tile(tile_id):
INFILE = os.path.join(SRC, tile_id + '.laz')
# INFILE = os.path.join(SRC, tile_id + '.las')
OUTFILE = os.path.join(RAW, tile_id + '.laz')
if os.path.exists(OUTFILE):
pass
else:
try:
proc_import = las.las2las(i=INFILE,
drop_withheld=True,
drop_class=(7,18), # classified as noise
# epsg=32149, # specify the source lidar projection, washington state plane south
# epsg=2927, # specify the source lidar projection, washington state plane south
longlat=True, # original data is in geographic coordinates
# elevation_surveyfeet=True,
# survey_feet=True,
# nad83_2011=True, # original data in nad83_2011 datum
nad83_harn=True, # original data in nad83_harn datum
target_epsg=TARGET_EPSG, # reproject
dont_remove_empty_files=True,
cpu64=True,
odir=RAW,
olaz=True)
except PipelineError as e:
log_error(tile_id, 'import_tile', e.message)
return tile_id
@dask.delayed
def validate(tile_id):
INFILE = os.path.join(RAW, tile_id + '.laz')
OUTFILE = os.path.join(RAW, tile_id + '.xml')
if os.path.exists(OUTFILE):
pass
else:
try:
proc_validate = las.lasvalidate(i=INFILE,
o=OUTFILE)
except PipelineError as e:
log_error(tile_id, 'validate', e.message)
return tile_id
@dask.delayed
def make_index(tile_id):
INFILE = os.path.join(RAW, tile_id + '.laz')
OUTFILE = os.path.join(RAW, tile_id + '.lax')
if os.path.exists(OUTFILE):
pass
else:
try:
proc_index = las.lasindex(i=INFILE,
cpu64=True)
except PipelineError as e:
log_error(tile_id, 'make_index', e.message)
return tile_id
@dask.delayed
def make_boundary(tile_id):
INFILE = os.path.join(RAW, tile_id + '.laz')
OUTFILE = os.path.join(RAW, tile_id + '.shp')
if os.path.exists(OUTFILE):
pass
else:
try:
proc_bnd = las.lasboundary(i=INFILE,
o=OUTFILE,
disjoint=True,
labels=True,
use_lax=True,
cpu64=True)
except PipelineError as e:
log_error(tile_id, 'make_index', e.message)
return tile_id
tile_ids = [fname(tile) for tile in src_tiles]
get_data = {}
for tile in tile_ids:
get_data['import-{}'.format(tile)]=(
import_tile,
tile)
get_data['validate-{}'.format(tile)]=(
validate,
'import-{}'.format(tile))
get_data['index-{}'.format(tile)]=(
make_index,
'validate-{}'.format(tile))
get_data['boundary-{}'.format(tile)]=(
make_boundary,
'index-{}'.format(tile))
# this empty function will be added to recipe for computations
# it will be defined to depend upon all previous steps being completed
@dask.delayed
def done_importing(*args, **kwargs):
return
get_data['done_importing']=(
done_importing,
['boundary-{}'.format(tile) for tile in tile_ids])
get_data_graph = c.get(get_data, 'done_importing') # build the computational graph
get_data_results = c.persist(get_data_graph) # start executing it
progress(get_data_results) # progress bars
# c.cancel(get_data_results)
inspect_failures(os.path.join(RAW, 'failed'))
validation_summary(xml_dir=RAW, verbose=False)
# move_invalid_tiles(xml_dir=RAW, dest_dir=os.path.join(RAW, 'invalid'))
boundaries = glob.glob(os.path.join(RAW, '*.shp'))
# set up a lazy read_file function so we can read in files in parallel
gdfs = [dask.delayed(gpd.read_file)(shp) for shp in boundaries]
# and a lazy concatenation
gather_tiles = dask.delayed(pd.concat)(gdfs, axis=0, ignore_index=True)
# now execute the read and concatenate with the cluster
tileindex = gather_tiles.compute()
tileindex.crs = "EPSG:{}".format(TARGET_EPSG)
tileindex.head()
tileindex.to_file(os.path.join(RAW, 'raw_tileindex.shp'), index=False)
# c.close()
# cluster.close()
| 0.247441 | 0.654039 |
# Guerrieri and Lorenzoni (2017)
# Credit Crises, Precautionary Savings, and The Liquidity Trap
Notebook created by William Du and Tung Sheng Hsieh
This notebook uses the Econ-ARK/HARK toolkit to replicate the results of Guerrieri and Lorenzoni (2017). We create a new AgentType, GLConsumerType, that inherits the IndShockConsumerType and a Solver, GLSolver, that inherits the ConsIndShockSolver.
We managed to closely replicate the initial Optimal Consumption and Labor Supply Steady States found Figure 1 of the paper.
## Summary
This paper uses a heterogeneous agents model with incomplete markets and endogenous labor supply to analyze the effects of a credit crunch on consumer spending.
Main Findings:
(i) A credit crunch leads to a fall in consumption and real interest rates due to a forced deleveraging and an increase in precautionary savings.
(ii) Adding nominal rigidities to the baseline model may exacerbate the effects on output as the zero lower bound may prevent the real interest rate from falling sufficiently in order to attain the flexible price equilibrium.
(iii) Adding durable goods to the baseline model does not fundamentally alter the baseline result mentioned in (i). In this extension, there is a fall in consumption in both non durables and durables for credit constrained consumers and an increase in precautionary savings through durables and bonds for net lenders.
## Non-Technical Overview
The authors consider a heterogeneous agents model where there is a continuum of infinitely lived households with idiosyncratic uncertainty to their labor productivity and subject to a borrowing limit. In order to understand the effects of a credit crunch, the authors analyze how a shock to the borrowing limit (a tightening in credit) influences each household's consumption decision and the resulting interest rate dynamics.
## Baseline Model
#### Households/Producers
There is a continuum of infinitely lived households with preferences represented by
the utility function: $$
\mathrm{E}\left[\sum_{t=0}^{\infty} \beta^{t} U\left(c_{i t}, n_{i t}\right)\right]
$$
where $U(c, n)=\frac{c^{1-\gamma}}{1-\gamma}+\psi \frac{(1-n)^{1-\eta}}{1-\eta}$
Each household \textit{i} chooses $c_{it}$ and $n_{it}$ to maximize their lifetime expected utility subject to their household budget constraint described below. Production is dependent on the choice of $n_{it}$
$$
y_{i t}=\theta_{i t} n_{i t}
$$
where $ \theta_{i t}$ is an idiosyncratic shock to the labor productivity of household \textit{i}, which follows a Markov chain on the space $\left\{\theta^{1}, \ldots, \theta^{S}\right\}$. Let $\theta^{1} =0$.
#### Household Budget Constraint
$$
q_{t} b_{i t+1} +c_{i t}+\tilde{\tau}_{i t} \leq b_{i t}+y_{i t}
$$
where $q_{t}$ is the bond price, $\tilde{\tau}_{i t}$ are taxes such that $\tilde{\tau}_{i t}=\tau_{t}$ if $\theta_{i t}>0$ and $\tilde{\tau}_{i t}=\tau_{t}-v_{t}$ if $\theta_{i t}=0$ where $v_{t}$ is unemployment insurance. $b_{i t+1}$ are bond holdings.
Household debt is bounded below by an exogenous limit $\phi > 0$. That is,
$$b_{i t+1} \geq-\phi$$
A credit crunch is equivalent to lowering the value of $\phi$.
#### Government
The government chooses the aggregate supply of bonds $B_{t}$, the unemployment benefit $v_{t}$ and the lump sum tax $\tau_{t}$ so as to satisfy the budget constraint
$$B_{t}+v_{t} u=q_{t} B_{t+1}+\tau_{t}$$
where where u = Pr ($\theta_{it}$ = 0) is the fraction of unemployed agents in the population.
We assume that the supply of government bonds $B$ and unemployment insurance $v$ are kept constant.
Taxes $\tau$ adjust to ensure the government budget balances.
### Dynamic Program
For household i, the Bellman equation is
$$
\begin{aligned}
V_{it}(b_{it}, \theta_{it})=\max _{c_{it}, n_{it}, b_{it+1}} & U(c_{it}, n_{it})+\beta E\left[V\left(b_{it+1}, \theta_{it+1}\right) \mid \theta_{it}\right] \\
\text { s.t. } & b_{it}+\theta_{it} n_{it} -\tau(\theta_{it}) \geq q_{t} b_{it+1}+c_{it}, \\
& b_{it+1}+\phi \geq 0
\end{aligned}
$$
## Calibration Summary
The table below indicates the calibrations for the initial steady state.
| Parameter | MATLAB | HARK | Value | Description |
|:---------:|:---------:|:----:|:--------|:-----------|
| $\gamma$ | `gam` | `CRRA` | $4$ | Coefficient of relative risk aversion |
| $r$ |`r` | `Rfree-1` | $0.00625$| Quarterly interest rate |
| $\eta$ | `eta` | `eta` | $1.5$ | Curvature of utility from leisure |
| $\beta$ | `bet` | `DiscFac` | $0.9457$ | Discount factor |
| $v$ | `nu` | `nu` | $0.16$ | UI benefits |
| $B$ | `B` | `B` | $2.56$ | Net supply of bonds |
| $\psi$ | `pssi` | `pssi` | $18.154609$ | Disutility from labor |
| $\phi$ | `phi` | `-BoroCnstArt` | $1.60$ | Borrowing limit|
| $\pi_{e,u}$ | `sep` | `sep` | $0.0573$ | Separation probability|
| $\pi_{u,e}$ | `fin` | `fin` | $0.8820$ | Job-finding probability |
| $\rho$ | | | $0.967$ | Persistence of wage process |
| $\sigma^2_\varepsilon$|| | $0.017$| Variance of wage process |
The last 4 parameters are used to construct the Markov Transition matrix.
The wage process defined by $\rho$ and $\sigma^2_\varepsilon$ is approximated by a 12 state markov chain following the approach in Tauchen (1986). The authors perform this approximation outside of the code provided and load the resulting 12 x 12 markov array from inc_process.mat which will be seen below when the complete 12 x 13 markov transition Matrix is constructed.
The state of unemployment ($\theta_{it}=0$) is then added to this 12 state markov chain to create a 13 x 13 Markov Transition Matrix.
## Results
#### Baseline Model
Consumers who's borrowing constraint was slack are forced to deleverage when the borrowing limit falls. These consumers both increase their labor supply and reduce their consumption. Furthermore, this deleveraging requires in an increase in the demand for bonds to a fall in the real interest rate.
There is an increase in precautionary savings for non-constrained consumers (who are not at the right tail of the initial bond distribution) in order to buffer themselves against future shocks. Increasing their precautionary motive requires an increase in the demand for bonds leading the real interest rate to fall.
Only highly productive consumers (those who did not experience a significant negative productivity shock) will decumulate bonds and increase consumption from lowered interest rates.
# Replication
```
from time import process_time
from copy import deepcopy, copy
import numpy as np
from HARK.ConsumptionSaving.GLModel import GLConsumerType
from HARK.distribution import DiscreteDistribution
from scipy.io import loadmat
from HARK.utilities import plotFuncs
from time import time
mystr = lambda number: "{:.4f}".format(number)
```
### Construct Markov Transition Matrix
```
#Import income process to build transition matrix
Matlabdict = loadmat('inc_process.mat')
data = list(Matlabdict.items())
data_array=np.asarray(data)
Pr=data_array[4,1]
pr = data_array[5,1]
fin = 0.8820 #job-finding probability
sep = 0.0573 #separation probability
#constructing transition Matrix
G=np.array([1-fin]).reshape(1,1)
A = np.concatenate((G, fin*pr), axis=1)
K= sep**np.ones(12).reshape(12,1)
D=np.concatenate((K,np.multiply((1-sep),Pr)),axis=1)
MrkvArray = np.concatenate((A,D))
```
### Dictionary for Parameters
```
GLDict={
# Parameters shared with the perfect foresight model
"CRRA": 4.0, # Coefficient of relative risk aversion
"Rfree": 1.00625*np.ones(13), # Interest factor on assets
"DiscFac": .9457, # Intertemporal discount factor
"LivPrb" : [1.00*np.ones(13)], # Survival probability
"PermGroFac" :[1.00*np.ones(13)], # Permanent income growth factor
# Parameters that specify the income distribution over the lifecycle
"PermShkStd" : [0.0], # Standard deviation of log permanent shocks to income
"PermShkCount" : 1, # Number of points in discrete approximation to permanent income shocks
"TranShkStd" : [0.0], # Standard deviation of log transitory shocks to income
"TranShkCount" : 1, # Number of points in discrete approximation to transitory income shocks
"UnempPrb" : 0, # Probability of unemployment while working
"IncUnemp" : 0, # Unemployment benefits replacement rate
"UnempPrbRet" : 0, # Probability of "unemployment" while retired
"IncUnempRet" : 0, # "Unemployment" benefits when retired
"T_retire" : 0, # Period of retirement (0 --> no retirement)
"tax_rate" : 0.0, # Flat income tax rate (legacy parameter, will be removed in future)
# Parameters for constructing the "assets above minimum" grid
"aXtraMin" : .000006, # Minimum end-of-period "assets above minimum" value
"aXtraMax" : 50, # Maximum end-of-period "assets above minimum" value
"aXtraCount" : 182, # Number of points in the base grid of "assets above minimum"
"aXtraNestFac" : 3, # Exponential nesting factor when constructing "assets above minimum" grid
"aXtraExtra" : [None], # Additional values to add to aXtraGrid
# A few other paramaters
"BoroCnstArt" : -1.6, # Artificial borrowing constraint; imposed minimum level of end-of period assets
"vFuncBool" : True, # Whether to calculate the value function during solution
"CubicBool" : False, # Preference shocks currently only compatible with linear cFunc
"T_cycle" : 1, # Number of periods in the cycle for this agent type
# Parameters only used in simulation
"AgentCount" : 10000, # Number of agents of this type
"T_sim" : 120, # Number of periods to simulate
"aNrmInitMean" : -6.0, # Mean of log initial assets
"aNrmInitStd" : 1.0, # Standard deviation of log initial assets
"pLvlInitMean" : 0.0, # Mean of log initial permanent income
"pLvlInitStd" : 0.0, # Standard deviation of log initial permanent income
"PermGroFacAgg" : 1.0, # Aggregate permanent income growth factor
"T_age" : None, # Age after which simulated agents are automatically killed
# Additional Parameters specific to Guerrieri et Lorenzoni
"MrkvArray" : [MrkvArray], # Markov Transition Matrix
'eta': 1.5, # Curvature of utility from leisure
'nu': 0.16, # UI benefits
'pssi': 18.154609, # Coefficient on leisure in utility
'B': 2.56, # Bond Supply
}
```
### Construct degenerate Income Distribution
```
IncomeDstnReg = DiscreteDistribution(np.ones(1), [np.ones(1), np.zeros(1)])
IncomeDstn = 13*[IncomeDstnReg]
```
Since the GLConsumerType is a subclass of the IndShockConsumerType , we must specify an income distribution.
However, because there are no transitory nor permanent shocks to income in the model, the Income distribution is degenerate.
### Create an Instance of the GLConsumerType
```
GLexample = GLConsumerType(**GLDict)
GLexample.IncomeDstn = [IncomeDstn]
```
### Solve the Agent's Problem
```
start_time = time()
GLexample.solve()
end_time = time()
print(
"Solving a GLexample took "
+ mystr(end_time - start_time)
+ " seconds."
)
```
## Initial Steady States
### Consumption Functions for $ \theta_{it} = \theta^{2} , \theta^{8}$
```
plotFuncs([GLexample.solution[0].cFunc[1],
GLexample.solution[0].cFunc[7]
], -2, 12)
#note due to the fact that python's index begins at 0, cFunc[i] is the consumption function for when \theta^{i+1
```
It is apparent that consumption varies with bond holdings. At high levels of bond holdings, the consumer's behavior is similar to that of the Permanent Income Hypothesis. The consumption function is concave due to the precautionary motive as there is positive probability that the agent will be unemployed.
### Labor Supply Functions for $ \theta_{it} = \theta^{2} , \theta^{8}$
```
plotFuncs([GLexample.solution[0].LFunc[1],
GLexample.solution[0].LFunc[7]
], -2, 12)
```
The labor supply functions are convex as higher levels of bond holdings capture an income effect that in turn lowers the amount of labor supplied.
## Figures in Paper

The figures produced in our code closely resemble those found in the paper. The only notable difference is the labor supply function when $ \theta_{it} = \theta^{8}$.
## Optimal Consumption and Labor Supply for All $\theta_{it}$
```
plotFuncs(GLexample.solution[0].cFunc[1:12], -2, 12)
plotFuncs(GLexample.solution[0].LFunc[1:12],-2, 12)
```
|
github_jupyter
|
from time import process_time
from copy import deepcopy, copy
import numpy as np
from HARK.ConsumptionSaving.GLModel import GLConsumerType
from HARK.distribution import DiscreteDistribution
from scipy.io import loadmat
from HARK.utilities import plotFuncs
from time import time
mystr = lambda number: "{:.4f}".format(number)
#Import income process to build transition matrix
Matlabdict = loadmat('inc_process.mat')
data = list(Matlabdict.items())
data_array=np.asarray(data)
Pr=data_array[4,1]
pr = data_array[5,1]
fin = 0.8820 #job-finding probability
sep = 0.0573 #separation probability
#constructing transition Matrix
G=np.array([1-fin]).reshape(1,1)
A = np.concatenate((G, fin*pr), axis=1)
K= sep**np.ones(12).reshape(12,1)
D=np.concatenate((K,np.multiply((1-sep),Pr)),axis=1)
MrkvArray = np.concatenate((A,D))
GLDict={
# Parameters shared with the perfect foresight model
"CRRA": 4.0, # Coefficient of relative risk aversion
"Rfree": 1.00625*np.ones(13), # Interest factor on assets
"DiscFac": .9457, # Intertemporal discount factor
"LivPrb" : [1.00*np.ones(13)], # Survival probability
"PermGroFac" :[1.00*np.ones(13)], # Permanent income growth factor
# Parameters that specify the income distribution over the lifecycle
"PermShkStd" : [0.0], # Standard deviation of log permanent shocks to income
"PermShkCount" : 1, # Number of points in discrete approximation to permanent income shocks
"TranShkStd" : [0.0], # Standard deviation of log transitory shocks to income
"TranShkCount" : 1, # Number of points in discrete approximation to transitory income shocks
"UnempPrb" : 0, # Probability of unemployment while working
"IncUnemp" : 0, # Unemployment benefits replacement rate
"UnempPrbRet" : 0, # Probability of "unemployment" while retired
"IncUnempRet" : 0, # "Unemployment" benefits when retired
"T_retire" : 0, # Period of retirement (0 --> no retirement)
"tax_rate" : 0.0, # Flat income tax rate (legacy parameter, will be removed in future)
# Parameters for constructing the "assets above minimum" grid
"aXtraMin" : .000006, # Minimum end-of-period "assets above minimum" value
"aXtraMax" : 50, # Maximum end-of-period "assets above minimum" value
"aXtraCount" : 182, # Number of points in the base grid of "assets above minimum"
"aXtraNestFac" : 3, # Exponential nesting factor when constructing "assets above minimum" grid
"aXtraExtra" : [None], # Additional values to add to aXtraGrid
# A few other paramaters
"BoroCnstArt" : -1.6, # Artificial borrowing constraint; imposed minimum level of end-of period assets
"vFuncBool" : True, # Whether to calculate the value function during solution
"CubicBool" : False, # Preference shocks currently only compatible with linear cFunc
"T_cycle" : 1, # Number of periods in the cycle for this agent type
# Parameters only used in simulation
"AgentCount" : 10000, # Number of agents of this type
"T_sim" : 120, # Number of periods to simulate
"aNrmInitMean" : -6.0, # Mean of log initial assets
"aNrmInitStd" : 1.0, # Standard deviation of log initial assets
"pLvlInitMean" : 0.0, # Mean of log initial permanent income
"pLvlInitStd" : 0.0, # Standard deviation of log initial permanent income
"PermGroFacAgg" : 1.0, # Aggregate permanent income growth factor
"T_age" : None, # Age after which simulated agents are automatically killed
# Additional Parameters specific to Guerrieri et Lorenzoni
"MrkvArray" : [MrkvArray], # Markov Transition Matrix
'eta': 1.5, # Curvature of utility from leisure
'nu': 0.16, # UI benefits
'pssi': 18.154609, # Coefficient on leisure in utility
'B': 2.56, # Bond Supply
}
IncomeDstnReg = DiscreteDistribution(np.ones(1), [np.ones(1), np.zeros(1)])
IncomeDstn = 13*[IncomeDstnReg]
GLexample = GLConsumerType(**GLDict)
GLexample.IncomeDstn = [IncomeDstn]
start_time = time()
GLexample.solve()
end_time = time()
print(
"Solving a GLexample took "
+ mystr(end_time - start_time)
+ " seconds."
)
plotFuncs([GLexample.solution[0].cFunc[1],
GLexample.solution[0].cFunc[7]
], -2, 12)
#note due to the fact that python's index begins at 0, cFunc[i] is the consumption function for when \theta^{i+1
plotFuncs([GLexample.solution[0].LFunc[1],
GLexample.solution[0].LFunc[7]
], -2, 12)
plotFuncs(GLexample.solution[0].cFunc[1:12], -2, 12)
plotFuncs(GLexample.solution[0].LFunc[1:12],-2, 12)
| 0.560974 | 0.993505 |
```
import pandas as pd
import seaborn as sns
sns.set()
!ls ../*.csv
df = pd.read_csv("../csvs/c2x2-paper-d200-t1.csv")
df_fix = pd.read_csv("../csvs/c2x2-paper-d200-t1-v2.csv")
sns.lineplot('depth', 'duration', data=df);
sns.lineplot('depth', 'duration', data=df_fix);
sns.lineplot('depth', 'is_solved', data=df);
sns.lineplot('depth', 'is_solved', data=df_fix);
sns.lineplot('depth', 'solve_steps', data=df);
sns.lineplot('depth', 'solve_steps', data=df_fix);
sns.lineplot('depth', 'solve_steps', data=df[df.is_solved == 1]);
sns.lineplot('depth', 'solve_steps', data=df_fix[df_fix.is_solved == 1]);
sns.lineplot('depth', 'solve_steps', data=df[df.is_solved == 0]);
sns.lineplot('depth', 'solve_steps', data=df_fix[df_fix.is_solved == 0]);
df_fix[df_fix.is_solved == 0].head()
```
# Cube 2x2 zero goal fix
```
df_c2_zg = pd.read_csv("../csvs/c2x2-zero-goal-d200-t1.csv")
df_c2_zg_fix = pd.read_csv("../csvs/c2x2-zero-goal-d200-t1-v2.csv")
sns.lineplot('depth', 'duration', data=df_c2_zg);
sns.lineplot('depth', 'duration', data=df_c2_zg_fix);
sns.lineplot('depth', 'is_solved', data=df_c2_zg);
sns.lineplot('depth', 'is_solved', data=df_c2_zg_fix);
sns.lineplot('depth', 'is_solved', data=df_fix);
sns.lineplot('depth', 'is_solved', data=df_c2_zg_fix);
sns.lineplot('depth', 'solve_steps', data=df_c2_zg);
sns.lineplot('depth', 'solve_steps', data=df_c2_zg_fix);
sns.lineplot('depth', 'solve_steps', data=df_fix);
sns.lineplot('depth', 'solve_steps', data=df_c2_zg_fix);
sns.lineplot('depth', 'solve_steps', data=df_fix[df_fix.is_solved==1]);
sns.lineplot('depth', 'solve_steps', data=df_c2_zg_fix[df_c2_zg_fix.is_solved==1]);
```
Significant improvement with the fix.
# Cube 3x3, paper method
```
df_c3_p = pd.read_csv("../csvs/c3x3-paper-d200-t1.csv")
df_c3_p_fix = pd.read_csv("../csvs/c3x3-paper-d200-t1-v2.csv")
sns.lineplot('depth', 'duration', data=df_c3_p);
sns.lineplot('depth', 'duration', data=df_c3_p_fix);
sns.lineplot('depth', 'is_solved', data=df_c3_p);
sns.lineplot('depth', 'is_solved', data=df_c3_p_fix);
sns.lineplot('depth', 'solve_steps', data=df_c3_p);
sns.lineplot('depth', 'solve_steps', data=df_c3_p_fix);
```
No improvement
# Cube 3x3, zero goal
Compare previous results with fixed (which gave more steps to solve for each cube)
```
df_c3_zg = pd.read_csv("../csvs/c3x3-zero-goal-d200-t1.csv")
df_c3_zg_fix = pd.read_csv("../csvs/c3x3-zero-goal-d200-t1-v2.csv")
sns.lineplot('depth', 'duration', data=df_c3_zg);
sns.lineplot('depth', 'duration', data=df_c3_zg_fix);
sns.lineplot('depth', 'is_solved', data=df_c3_zg);
sns.lineplot('depth', 'is_solved', data=df_c3_zg_fix);
sns.lineplot('depth', 'solve_steps', data=df_c3_zg);
sns.lineplot('depth', 'solve_steps', data=df_c3_zg_fix);
sns.lineplot('depth', 'solve_steps', data=df_c3_zg[df_c3_zg.is_solved==1]);
sns.lineplot('depth', 'solve_steps', data=df_c3_zg_fix[df_c3_zg_fix.is_solved==1]);
```
No improvement versus old version
# Cube 3x3, paper method versus zero goal
Compare fixed results on paper with fixed in my method
```
sns.lineplot('depth', 'duration', data=df_c3_p_fix);
sns.lineplot('depth', 'duration', data=df_c3_zg_fix);
sns.lineplot('depth', 'is_solved', data=df_c3_p_fix);
sns.lineplot('depth', 'is_solved', data=df_c3_zg_fix);
sns.lineplot('depth', 'solve_steps', data=df_c3_p_fix);
sns.lineplot('depth', 'solve_steps', data=df_c3_zg_fix);
```
No much improvements. Probably, steps limit is too small.
# Cube 3x3, zero goal trained with decay and without
Compare policies obtained from training with LR decay and without it
```
df_c3_nodecay = pd.read_csv("../csvs/c3x3-zero-goal-d200-no-decay-v2.csv")
df_c3_decay = pd.read_csv("../csvs/c3x3-zero-goal-d200-t1-v2.csv")
sns.lineplot('depth', 'duration', data=df_c3_nodecay);
sns.lineplot('depth', 'duration', data=df_c3_decay);
sns.lineplot('depth', 'is_solved', data=df_c3_nodecay);
sns.lineplot('depth', 'is_solved', data=df_c3_decay);
sns.lineplot('depth', 'solve_steps', data=df_c3_nodecay);
sns.lineplot('depth', 'solve_steps', data=df_c3_decay);
```
Almost the same
# Final conclusions
After fix, on 2x2 cube, 'zero goal' method solves much more cubes than trained on paper.
Cubes 3x3 has almost zero improvement, probably due to untuned MCTS search and small amount of searches performed.
Future directions:
* Experiment with MCTS parameters, check exploration constants effect
* Give more steps to cube 3x3
* Optimize performance of MCTS (parallelization, etc) to be able to test deeper searches.
|
github_jupyter
|
import pandas as pd
import seaborn as sns
sns.set()
!ls ../*.csv
df = pd.read_csv("../csvs/c2x2-paper-d200-t1.csv")
df_fix = pd.read_csv("../csvs/c2x2-paper-d200-t1-v2.csv")
sns.lineplot('depth', 'duration', data=df);
sns.lineplot('depth', 'duration', data=df_fix);
sns.lineplot('depth', 'is_solved', data=df);
sns.lineplot('depth', 'is_solved', data=df_fix);
sns.lineplot('depth', 'solve_steps', data=df);
sns.lineplot('depth', 'solve_steps', data=df_fix);
sns.lineplot('depth', 'solve_steps', data=df[df.is_solved == 1]);
sns.lineplot('depth', 'solve_steps', data=df_fix[df_fix.is_solved == 1]);
sns.lineplot('depth', 'solve_steps', data=df[df.is_solved == 0]);
sns.lineplot('depth', 'solve_steps', data=df_fix[df_fix.is_solved == 0]);
df_fix[df_fix.is_solved == 0].head()
df_c2_zg = pd.read_csv("../csvs/c2x2-zero-goal-d200-t1.csv")
df_c2_zg_fix = pd.read_csv("../csvs/c2x2-zero-goal-d200-t1-v2.csv")
sns.lineplot('depth', 'duration', data=df_c2_zg);
sns.lineplot('depth', 'duration', data=df_c2_zg_fix);
sns.lineplot('depth', 'is_solved', data=df_c2_zg);
sns.lineplot('depth', 'is_solved', data=df_c2_zg_fix);
sns.lineplot('depth', 'is_solved', data=df_fix);
sns.lineplot('depth', 'is_solved', data=df_c2_zg_fix);
sns.lineplot('depth', 'solve_steps', data=df_c2_zg);
sns.lineplot('depth', 'solve_steps', data=df_c2_zg_fix);
sns.lineplot('depth', 'solve_steps', data=df_fix);
sns.lineplot('depth', 'solve_steps', data=df_c2_zg_fix);
sns.lineplot('depth', 'solve_steps', data=df_fix[df_fix.is_solved==1]);
sns.lineplot('depth', 'solve_steps', data=df_c2_zg_fix[df_c2_zg_fix.is_solved==1]);
df_c3_p = pd.read_csv("../csvs/c3x3-paper-d200-t1.csv")
df_c3_p_fix = pd.read_csv("../csvs/c3x3-paper-d200-t1-v2.csv")
sns.lineplot('depth', 'duration', data=df_c3_p);
sns.lineplot('depth', 'duration', data=df_c3_p_fix);
sns.lineplot('depth', 'is_solved', data=df_c3_p);
sns.lineplot('depth', 'is_solved', data=df_c3_p_fix);
sns.lineplot('depth', 'solve_steps', data=df_c3_p);
sns.lineplot('depth', 'solve_steps', data=df_c3_p_fix);
df_c3_zg = pd.read_csv("../csvs/c3x3-zero-goal-d200-t1.csv")
df_c3_zg_fix = pd.read_csv("../csvs/c3x3-zero-goal-d200-t1-v2.csv")
sns.lineplot('depth', 'duration', data=df_c3_zg);
sns.lineplot('depth', 'duration', data=df_c3_zg_fix);
sns.lineplot('depth', 'is_solved', data=df_c3_zg);
sns.lineplot('depth', 'is_solved', data=df_c3_zg_fix);
sns.lineplot('depth', 'solve_steps', data=df_c3_zg);
sns.lineplot('depth', 'solve_steps', data=df_c3_zg_fix);
sns.lineplot('depth', 'solve_steps', data=df_c3_zg[df_c3_zg.is_solved==1]);
sns.lineplot('depth', 'solve_steps', data=df_c3_zg_fix[df_c3_zg_fix.is_solved==1]);
sns.lineplot('depth', 'duration', data=df_c3_p_fix);
sns.lineplot('depth', 'duration', data=df_c3_zg_fix);
sns.lineplot('depth', 'is_solved', data=df_c3_p_fix);
sns.lineplot('depth', 'is_solved', data=df_c3_zg_fix);
sns.lineplot('depth', 'solve_steps', data=df_c3_p_fix);
sns.lineplot('depth', 'solve_steps', data=df_c3_zg_fix);
df_c3_nodecay = pd.read_csv("../csvs/c3x3-zero-goal-d200-no-decay-v2.csv")
df_c3_decay = pd.read_csv("../csvs/c3x3-zero-goal-d200-t1-v2.csv")
sns.lineplot('depth', 'duration', data=df_c3_nodecay);
sns.lineplot('depth', 'duration', data=df_c3_decay);
sns.lineplot('depth', 'is_solved', data=df_c3_nodecay);
sns.lineplot('depth', 'is_solved', data=df_c3_decay);
sns.lineplot('depth', 'solve_steps', data=df_c3_nodecay);
sns.lineplot('depth', 'solve_steps', data=df_c3_decay);
| 0.144239 | 0.682064 |
```
import batoid
import numpy as np
import matplotlib.pyplot as plt
from ipywidgets import interact
telescope = batoid.Optic.fromYaml("LSST_r_baffles.yaml")
def pupil(thx, thy, nside=512, dx=0):
scope = telescope.withGloballyShiftedOptic("M2",(dx,0,0))
rays = batoid.RayVector.asGrid(
optic=scope, wavelength=620e-9,
theta_x=thx, theta_y=thy,
nx=nside, ny=nside
)
rays2 = rays.copy()
scope.stopSurface.interact(rays2)
scope.trace(rays)
w = ~rays.vignetted
return rays2.x[w], rays2.y[w]
def drawCircle(ax, cx, cy, r, **kwargs):
t = np.linspace(0, 2*np.pi, 1000)
x = r*np.cos(t)+cx
y = r*np.sin(t)+cy
ax.plot(x, y, **kwargs)
def drawRay(ax, cx, cy, width, theta, **kwargs):
R = np.array([[np.cos(theta), -np.sin(theta)], [np.sin(theta), np.cos(theta)]])
dx = np.linspace(0, 4.1, 1000)
dy = np.ones_like(dx)*width/2
bx = np.copy(dx)
by = -dy
dx, dy = R.dot(np.vstack([dx, dy]))
bx, by = R.dot(np.vstack([bx, by]))
dx += cx
dy += cy
bx += cx
by += cy
ax.plot(dx, dy, **kwargs)
ax.plot(bx, by, **kwargs)
def drawRectangle(ax, cx, cy, width, height, **kwargs):
x = width/2*np.array([-1,-1,1,1,-1])
y = height/2*np.array([-1,1,1,-1,-1])
x += cx
y += cy
ax.plot(x, y, **kwargs)
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:100% !important; }</style>"))
@interact(thx=(-2.0, 2.0, 0.01), thy=(-2.0, 2.0, 0.01), dx=(-10, 10, 1))
def modelPlot(thx=1.67, thy=0.0, dx=0.0):
thx = np.deg2rad(thx)
thy = np.deg2rad(thy)
fig, ax = plt.subplots(1, 1, figsize=(24, 24))
ax.scatter(*pupil(thx,thy,dx=dx*1e-2), s=0.1, c='k')
ax.set_aspect('equal')
# Primary mirror
drawCircle(ax, 0, 0, 4.18, c='r')
drawCircle(ax, 0, 0, 2.55, c='r')
scales = {
'M2_inner': 2.3698999752679404,
'M2_outer': 4.502809953009087,
'M3_inner': 1.1922312943631603,
'M3_outer': 5.436574702296011,
# 'L1_entrance': 7.697441260764198,
# 'L1_exit': 8.106852624652701,
# 'L2_entrance': 10.748915941599885,
# 'L2_exit': 11.5564127895276,
# 'Filter_entrance': 28.082220873785978,
# 'Filter_exit': 30.91023954045243,
# 'L3_entrance': 54.67312185149621,
# 'L3_exit': 114.58705556485711
}
speeds = {
'M2_inner': 16.8188788239707,
'M2_outer': 16.8188788239707,
'M3_inner': 53.22000661238318,
'M3_outer': 53.22000661238318,
# 'L1_entrance': 131.76650078100135,
# 'L1_exit': 137.57031952814913,
# 'L2_entrance': 225.6949885074127,
# 'L2_exit': 237.01739037674315,
# 'Filter_entrance': 802.0137451419788,
# 'Filter_exit': 879.8810309773828,
# 'L3_entrance': 1597.8959863335774,
# 'L3_exit': 3323.60145194633
}
for k in scales:
drawCircle(ax, -speeds[k]*thx, -speeds[k]*thy, scales[k], c='r')
# ax.set_xlim(-50,50)
# ax.set_ylim(-50,50)
ax.set_xlim(-5,5)
ax.set_ylim(-5,5)
ax.axvline(c='k')
ax.axhline(c='k')
fig.show()
```
|
github_jupyter
|
import batoid
import numpy as np
import matplotlib.pyplot as plt
from ipywidgets import interact
telescope = batoid.Optic.fromYaml("LSST_r_baffles.yaml")
def pupil(thx, thy, nside=512, dx=0):
scope = telescope.withGloballyShiftedOptic("M2",(dx,0,0))
rays = batoid.RayVector.asGrid(
optic=scope, wavelength=620e-9,
theta_x=thx, theta_y=thy,
nx=nside, ny=nside
)
rays2 = rays.copy()
scope.stopSurface.interact(rays2)
scope.trace(rays)
w = ~rays.vignetted
return rays2.x[w], rays2.y[w]
def drawCircle(ax, cx, cy, r, **kwargs):
t = np.linspace(0, 2*np.pi, 1000)
x = r*np.cos(t)+cx
y = r*np.sin(t)+cy
ax.plot(x, y, **kwargs)
def drawRay(ax, cx, cy, width, theta, **kwargs):
R = np.array([[np.cos(theta), -np.sin(theta)], [np.sin(theta), np.cos(theta)]])
dx = np.linspace(0, 4.1, 1000)
dy = np.ones_like(dx)*width/2
bx = np.copy(dx)
by = -dy
dx, dy = R.dot(np.vstack([dx, dy]))
bx, by = R.dot(np.vstack([bx, by]))
dx += cx
dy += cy
bx += cx
by += cy
ax.plot(dx, dy, **kwargs)
ax.plot(bx, by, **kwargs)
def drawRectangle(ax, cx, cy, width, height, **kwargs):
x = width/2*np.array([-1,-1,1,1,-1])
y = height/2*np.array([-1,1,1,-1,-1])
x += cx
y += cy
ax.plot(x, y, **kwargs)
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:100% !important; }</style>"))
@interact(thx=(-2.0, 2.0, 0.01), thy=(-2.0, 2.0, 0.01), dx=(-10, 10, 1))
def modelPlot(thx=1.67, thy=0.0, dx=0.0):
thx = np.deg2rad(thx)
thy = np.deg2rad(thy)
fig, ax = plt.subplots(1, 1, figsize=(24, 24))
ax.scatter(*pupil(thx,thy,dx=dx*1e-2), s=0.1, c='k')
ax.set_aspect('equal')
# Primary mirror
drawCircle(ax, 0, 0, 4.18, c='r')
drawCircle(ax, 0, 0, 2.55, c='r')
scales = {
'M2_inner': 2.3698999752679404,
'M2_outer': 4.502809953009087,
'M3_inner': 1.1922312943631603,
'M3_outer': 5.436574702296011,
# 'L1_entrance': 7.697441260764198,
# 'L1_exit': 8.106852624652701,
# 'L2_entrance': 10.748915941599885,
# 'L2_exit': 11.5564127895276,
# 'Filter_entrance': 28.082220873785978,
# 'Filter_exit': 30.91023954045243,
# 'L3_entrance': 54.67312185149621,
# 'L3_exit': 114.58705556485711
}
speeds = {
'M2_inner': 16.8188788239707,
'M2_outer': 16.8188788239707,
'M3_inner': 53.22000661238318,
'M3_outer': 53.22000661238318,
# 'L1_entrance': 131.76650078100135,
# 'L1_exit': 137.57031952814913,
# 'L2_entrance': 225.6949885074127,
# 'L2_exit': 237.01739037674315,
# 'Filter_entrance': 802.0137451419788,
# 'Filter_exit': 879.8810309773828,
# 'L3_entrance': 1597.8959863335774,
# 'L3_exit': 3323.60145194633
}
for k in scales:
drawCircle(ax, -speeds[k]*thx, -speeds[k]*thy, scales[k], c='r')
# ax.set_xlim(-50,50)
# ax.set_ylim(-50,50)
ax.set_xlim(-5,5)
ax.set_ylim(-5,5)
ax.axvline(c='k')
ax.axhline(c='k')
fig.show()
| 0.482917 | 0.61422 |
# Yolov5 Pytorch 1.7 for Edge Devices with Amazon SageMaker
Amazon SageMaker is a fully managed machine learning service. With SageMaker, data scientists and developers can quickly and easily build and train machine learning models, and then directly deploy them into a production-ready hosted environment. It provides an integrated Jupyter authoring notebook instance for easy access to your data sources for exploration and analysis, so you don't have to manage servers. It also provides common machine learning algorithms that are optimized to run efficiently against extremely large data in a distributed environment. With native support for bring-your-own-algorithms and frameworks, SageMaker offers flexible distributed training options that adjust to your specific workflows.
SageMaker also offers capabilities to prepare models for deployment at the edge. [SageMaker Neo](https://docs.aws.amazon.com/sagemaker/latest/dg/neo.html) is a capability of Amazon SageMaker that enables machine learning models to train once and run anywhere in the cloud and at the edge and [Amazon SageMaker Edge Manager](https://docs.aws.amazon.com/sagemaker/latest/dg/edge.html) provides model management for edge devices so you can optimize, secure, monitor, and maintain machine learning models on fleets of edge devices such as smart cameras, robots, personal computers, and mobile devices.
In this notebook we'll train a [**Yolov5**](https://github.com/ultralytics/yolov5) model on Pytorch using Amazon SageMaker to draw bounding boxes around images and then, compile and package it so that it can be deployed on an edge device(in this case, a [Jetson Xavier](https://developer.nvidia.com/jetpack-sdk-441-archive)).
### Pre-requisites
Let us start with setting up the pre-requisites for this notebook. First, we will sagemaker and other related libs and then set up the role and buckets and some variables. Note that, we are also specifying the size of the image and also the model size taken as Yolov5s where s stand for small. Check out the [github doc of yolov5](https://github.com/ultralytics/yolov5) to understand how model sizes differ.
```
import sagemaker
import numpy as np
import glob
import os
from sagemaker.pytorch.estimator import PyTorch
role = sagemaker.get_execution_role()
sagemaker_session=sagemaker.Session()
bucket_name = sagemaker_session.default_bucket()
img_size=640
model_type='s' # s=small,m=medium,l=large,x=xlarge, s6,m6,l6,x6
model_name=f"yolov5-{model_type}"
```
### Download the official Yolov5 repo - version 5.0
Now, we will download the official Yolov5 version from the [official repository](https://github.com/ultralytics/yolov5). We will place it in a local directory `yolov5`
```
if not os.path.isdir('yolov5'):
!git clone https://github.com/ultralytics/yolov5 && \
cd yolov5 && git checkout v5.0 && \
git apply ../yolov5_inplace.patch
!echo 'tensorboard' > yolov5/requirements.txt
!echo 'seaborn' >> yolov5/requirements.txt
```
### Prepare a Python script that will be the entrypoint of the training process
Now, we will create a training script to train the Yolov5 model. The training script will wrap the original training scripts and expose the parameters to SageMaker Estimator. The script accepts different arguments which will control the training process.
```
%%writefile yolov5/sagemaker_train.py
import sys
import subprocess
## We need to remove smdebug to avoid the Hook bug https://github.com/awslabs/sagemaker-debugger/issues/401
subprocess.check_call([sys.executable, "-m", "pip", "uninstall", "-y", "smdebug"])
import os
import yaml
import argparse
import torch
import shutil
import models
import torch.nn as nn
from utils.activations import Hardswish, SiLU
if __name__ == '__main__':
os.environ['WANDB_MODE'] = 'disabled' # disable weights and bias
parser = argparse.ArgumentParser()
parser.add_argument('--num-classes', type=int, default=80, help='Number of classes')
parser.add_argument('--img-size', type=int, default=640, help='Size of the image')
parser.add_argument('--epochs', type=int, default=1, help='Number of epochs')
parser.add_argument('--batch-size', type=int, default=16, help='Batch size')
parser.add_argument('--model-dir', type=str, default=os.environ["SM_MODEL_DIR"], help='Trained model dir')
parser.add_argument('--train', type=str, default=os.environ["SM_CHANNEL_TRAIN"], help='Train path')
parser.add_argument('--validation', type=str, default=os.environ["SM_CHANNEL_VALIDATION"], help='Validation path')
parser.add_argument('--model-type', type=str, choices=['s', 'm', 'l', 'x', 's6', 'm6', 'l6', 'x6'], default="s", help='Model type')
# hyperparameters
with open('data/hyp.scratch.yaml', 'r') as f:
hyperparams = yaml.load(f, Loader=yaml.FullLoader)
for k,v in hyperparams.items():
parser.add_argument(f"--{k.replace('_', '-')}", type=float, default=v)
args,unknown = parser.parse_known_args()
base_path=os.path.dirname(__file__)
project_dir = os.environ["SM_OUTPUT_DATA_DIR"]
# prepare the hyperparameters metadata
with open(os.path.join(base_path,'data', 'hyp.custom.yaml'), 'w' ) as y:
y.write(yaml.dump({h:vars(args)[h] for h in hyperparams.keys()}))
# prepare the training data metadata
with open(os.path.join(base_path,'data', 'custom.yaml'), 'w') as y:
y.write(yaml.dump({
'names': [f'class_{i}' for i in range(args.num_classes)],
'train': args.train,
'val': args.validation,
'nc': args.num_classes
}))
# run the training script
train_cmd = [
sys.executable, os.path.join(base_path,'train.py'),
"--data", "custom.yaml",
"--hyp", "hyp.custom.yaml",
"--weights", f"yolov5{args.model_type}.pt",
"--img", str(args.img_size),
"--batch", str(args.batch_size),
"--epochs", str(args.epochs),
"--project", project_dir
]
subprocess.check_call(train_cmd)
# run the export script
export_cmd = [
sys.executable, os.path.join(base_path,'models', 'export.py'),
"--weights", os.path.join(project_dir, 'exp', 'weights', 'best.pt'),
"--batch-size", "1",
"--img-size", str(args.img_size)
]
# tracing and saving the model
inp = torch.rand(1, 3, args.img_size, args.img_size).cpu()
model = torch.load(os.path.join(project_dir, 'exp', 'weights', 'best.pt'), map_location='cpu')['model']
model.eval().float()
model(inp)
# Update model
for k, m in model.named_modules():
if isinstance(m, models.common.Conv): # assign export-friendly activations
if isinstance(m.act, nn.Hardswish):
m.act = Hardswish()
elif isinstance(m.act, nn.SiLU):
m.act = SiLU()
model_trace = torch.jit.trace(model, inp, strict=False)
model_trace.save(os.path.join(args.model_dir, 'model.pth'))
```
### Preparing the dataset
Here we'll download a sample dataset [coco128](https://github.com/ultralytics/yolov5/releases/download/v1.0/coco128.zip). We can also replace this step with any other dataset.
Just take a look on the labels format and create your dataset scructure following the same standard (COCO).
```
import os
if not os.path.exists('coco128'):
!wget -q https://github.com/ultralytics/yolov5/releases/download/v1.0/coco128.zip
!unzip -q coco128.zip && rm -f coco128.zip
print('BBoxes annotation')
print('class x_center y_center width height')
!head coco128/labels/train2017/000000000009.txt
```
### Upload the dataset to S3
Once the dataset has been downloaded locally, we'll upload the dataset to an S3 bucket created earlier. We are setting up the training and validation dataset s3 locations here.
```
prefix='data/coco128'
!rm -f coco128/labels/train2017.cache
train_path = sagemaker_session.upload_data('coco128', key_prefix=f'{prefix}/train')
val_path = sagemaker_session.upload_data('coco128', key_prefix=f'{prefix}/val')
print(train_path, val_path)
```
### Prepare the SageMaker Estimator to train the model
Now it's time to create an Estimater and train the model with the training script created in earlier step. We are using Pytorch estimator and supplying other arguments in the estimator. Note that we are supplying the `source_dir` so that sagemaker can pick up the training script and other related files from there. Once the estimator is ready, we start the training using the `.fit()` method.
```
estimator = PyTorch(
'sagemaker_train.py',
source_dir='yolov5',
framework_version='1.7',
role=role,
sagemaker_session=sagemaker_session,
instance_type='ml.p3.2xlarge',
instance_count=1,
py_version='py3',
hyperparameters={
'epochs': 2,
'batch-size': 16,
'lr0': 0.0001,
'num-classes': 80,
'img-size': img_size,
'model-type': model_type
}
)
estimator.fit({'train': train_path, 'validation': val_path})
```
### Compile your trained model for the edge device
Once the model has been traied, we need to compile the model using SageMaker Neo. This step is needed to compile the model for the specific hardware on which this model will be deployed.
In this notebook, we will compile a model for [Jetson Xavier Jetpack 4.4.1](https://developer.nvidia.com/jetpack-sdk-441-archive).
In case, you want to compile for a different hardware platform, just change the parameters bellow to adjust the target to your own edge device. Also, note, that if you dont have GPU available on the hardware device, then you can comment the `Accelerator` key:value in the `OutputConfig`.
The below cell also calls the `describe_compilation_job` API in a loop to wait for the compilation job to complete. In actual applications, it is advisable to setup a cloudwatch event which can notify OR execute the next steps once the compilation job is complete.
```
import time
import boto3
sm_client = boto3.client('sagemaker')
compilation_job_name = f'{model_name}-pytorch-{int(time.time()*1000)}'
sm_client.create_compilation_job(
CompilationJobName=compilation_job_name,
RoleArn=role,
InputConfig={
'S3Uri': f'{estimator.output_path}{estimator.latest_training_job.name}/output/model.tar.gz',
'DataInputConfig': f'{{"input": [1,3,{img_size},{img_size}]}}',
'Framework': 'PYTORCH'
},
OutputConfig={
'S3OutputLocation': f's3://{sagemaker_session.default_bucket()}/{model_name}-pytorch/optimized/',
'TargetPlatform': {
'Os': 'LINUX',
'Arch': 'X86_64',
},
},
StoppingCondition={ 'MaxRuntimeInSeconds': 900 }
)
#check for the compilation job to complete
while True:
resp = sm_client.describe_compilation_job(CompilationJobName=compilation_job_name)
if resp['CompilationJobStatus'] in ['STARTING', 'INPROGRESS']:
print('Running...')
else:
print(resp['CompilationJobStatus'], compilation_job_name)
break
time.sleep(5)
```
### Finally create a SageMaker Edge Manager packaging job
Once the model has been compiled, it is time to create an edge manager packaging job. Packaging job take SageMaker Neo–compiled models and make any changes necessary to deploy the model with the inference engine, Edge Manager agent.
We need to provide the name used for the Neo compilation job, a name for the packaging job, a role ARN, a name for the model, a model version, and the Amazon S3 bucket URI for the output of the packaging job. Note that Edge Manager packaging job names are case-sensitive.
The below cell also calls the `describe_edge_packaging_job` API in a loop to wait for the packaging job to complete. In actual applications, it is advisable to setup a cloudwatch event which can notify OR execute the next steps once the compilation job is complete.
```
import time
model_version = '1.0'
edge_packaging_job_name=f'{model_name}-pytorch-{int(time.time()*1000)}'
resp = sm_client.create_edge_packaging_job(
EdgePackagingJobName=edge_packaging_job_name,
CompilationJobName=compilation_job_name,
ModelName=model_name,
ModelVersion=model_version,
RoleArn=role,
OutputConfig={
'S3OutputLocation': f's3://{bucket_name}/{model_name}'
}
)
while True:
resp = sm_client.describe_edge_packaging_job(EdgePackagingJobName=edge_packaging_job_name)
if resp['EdgePackagingJobStatus'] in ['STARTING', 'INPROGRESS']:
print('Running...')
else:
print(resp['EdgePackagingJobStatus'], compilation_job_name)
break
time.sleep(5)
```
### Done !!
And we are done with all the steps needed to prepare the model for deploying to edge. The model package is avaialble in S3 and can be taken from there to deploy it to edge device. Now you need to move over to your edge device and download and setup edge manager agent(runtime), model and other related artifacts on the device. Please check out the [documentation](https://docs.aws.amazon.com/sagemaker/latest/dg/edge.html) for detailed steps.
|
github_jupyter
|
import sagemaker
import numpy as np
import glob
import os
from sagemaker.pytorch.estimator import PyTorch
role = sagemaker.get_execution_role()
sagemaker_session=sagemaker.Session()
bucket_name = sagemaker_session.default_bucket()
img_size=640
model_type='s' # s=small,m=medium,l=large,x=xlarge, s6,m6,l6,x6
model_name=f"yolov5-{model_type}"
if not os.path.isdir('yolov5'):
!git clone https://github.com/ultralytics/yolov5 && \
cd yolov5 && git checkout v5.0 && \
git apply ../yolov5_inplace.patch
!echo 'tensorboard' > yolov5/requirements.txt
!echo 'seaborn' >> yolov5/requirements.txt
%%writefile yolov5/sagemaker_train.py
import sys
import subprocess
## We need to remove smdebug to avoid the Hook bug https://github.com/awslabs/sagemaker-debugger/issues/401
subprocess.check_call([sys.executable, "-m", "pip", "uninstall", "-y", "smdebug"])
import os
import yaml
import argparse
import torch
import shutil
import models
import torch.nn as nn
from utils.activations import Hardswish, SiLU
if __name__ == '__main__':
os.environ['WANDB_MODE'] = 'disabled' # disable weights and bias
parser = argparse.ArgumentParser()
parser.add_argument('--num-classes', type=int, default=80, help='Number of classes')
parser.add_argument('--img-size', type=int, default=640, help='Size of the image')
parser.add_argument('--epochs', type=int, default=1, help='Number of epochs')
parser.add_argument('--batch-size', type=int, default=16, help='Batch size')
parser.add_argument('--model-dir', type=str, default=os.environ["SM_MODEL_DIR"], help='Trained model dir')
parser.add_argument('--train', type=str, default=os.environ["SM_CHANNEL_TRAIN"], help='Train path')
parser.add_argument('--validation', type=str, default=os.environ["SM_CHANNEL_VALIDATION"], help='Validation path')
parser.add_argument('--model-type', type=str, choices=['s', 'm', 'l', 'x', 's6', 'm6', 'l6', 'x6'], default="s", help='Model type')
# hyperparameters
with open('data/hyp.scratch.yaml', 'r') as f:
hyperparams = yaml.load(f, Loader=yaml.FullLoader)
for k,v in hyperparams.items():
parser.add_argument(f"--{k.replace('_', '-')}", type=float, default=v)
args,unknown = parser.parse_known_args()
base_path=os.path.dirname(__file__)
project_dir = os.environ["SM_OUTPUT_DATA_DIR"]
# prepare the hyperparameters metadata
with open(os.path.join(base_path,'data', 'hyp.custom.yaml'), 'w' ) as y:
y.write(yaml.dump({h:vars(args)[h] for h in hyperparams.keys()}))
# prepare the training data metadata
with open(os.path.join(base_path,'data', 'custom.yaml'), 'w') as y:
y.write(yaml.dump({
'names': [f'class_{i}' for i in range(args.num_classes)],
'train': args.train,
'val': args.validation,
'nc': args.num_classes
}))
# run the training script
train_cmd = [
sys.executable, os.path.join(base_path,'train.py'),
"--data", "custom.yaml",
"--hyp", "hyp.custom.yaml",
"--weights", f"yolov5{args.model_type}.pt",
"--img", str(args.img_size),
"--batch", str(args.batch_size),
"--epochs", str(args.epochs),
"--project", project_dir
]
subprocess.check_call(train_cmd)
# run the export script
export_cmd = [
sys.executable, os.path.join(base_path,'models', 'export.py'),
"--weights", os.path.join(project_dir, 'exp', 'weights', 'best.pt'),
"--batch-size", "1",
"--img-size", str(args.img_size)
]
# tracing and saving the model
inp = torch.rand(1, 3, args.img_size, args.img_size).cpu()
model = torch.load(os.path.join(project_dir, 'exp', 'weights', 'best.pt'), map_location='cpu')['model']
model.eval().float()
model(inp)
# Update model
for k, m in model.named_modules():
if isinstance(m, models.common.Conv): # assign export-friendly activations
if isinstance(m.act, nn.Hardswish):
m.act = Hardswish()
elif isinstance(m.act, nn.SiLU):
m.act = SiLU()
model_trace = torch.jit.trace(model, inp, strict=False)
model_trace.save(os.path.join(args.model_dir, 'model.pth'))
import os
if not os.path.exists('coco128'):
!wget -q https://github.com/ultralytics/yolov5/releases/download/v1.0/coco128.zip
!unzip -q coco128.zip && rm -f coco128.zip
print('BBoxes annotation')
print('class x_center y_center width height')
!head coco128/labels/train2017/000000000009.txt
prefix='data/coco128'
!rm -f coco128/labels/train2017.cache
train_path = sagemaker_session.upload_data('coco128', key_prefix=f'{prefix}/train')
val_path = sagemaker_session.upload_data('coco128', key_prefix=f'{prefix}/val')
print(train_path, val_path)
estimator = PyTorch(
'sagemaker_train.py',
source_dir='yolov5',
framework_version='1.7',
role=role,
sagemaker_session=sagemaker_session,
instance_type='ml.p3.2xlarge',
instance_count=1,
py_version='py3',
hyperparameters={
'epochs': 2,
'batch-size': 16,
'lr0': 0.0001,
'num-classes': 80,
'img-size': img_size,
'model-type': model_type
}
)
estimator.fit({'train': train_path, 'validation': val_path})
import time
import boto3
sm_client = boto3.client('sagemaker')
compilation_job_name = f'{model_name}-pytorch-{int(time.time()*1000)}'
sm_client.create_compilation_job(
CompilationJobName=compilation_job_name,
RoleArn=role,
InputConfig={
'S3Uri': f'{estimator.output_path}{estimator.latest_training_job.name}/output/model.tar.gz',
'DataInputConfig': f'{{"input": [1,3,{img_size},{img_size}]}}',
'Framework': 'PYTORCH'
},
OutputConfig={
'S3OutputLocation': f's3://{sagemaker_session.default_bucket()}/{model_name}-pytorch/optimized/',
'TargetPlatform': {
'Os': 'LINUX',
'Arch': 'X86_64',
},
},
StoppingCondition={ 'MaxRuntimeInSeconds': 900 }
)
#check for the compilation job to complete
while True:
resp = sm_client.describe_compilation_job(CompilationJobName=compilation_job_name)
if resp['CompilationJobStatus'] in ['STARTING', 'INPROGRESS']:
print('Running...')
else:
print(resp['CompilationJobStatus'], compilation_job_name)
break
time.sleep(5)
import time
model_version = '1.0'
edge_packaging_job_name=f'{model_name}-pytorch-{int(time.time()*1000)}'
resp = sm_client.create_edge_packaging_job(
EdgePackagingJobName=edge_packaging_job_name,
CompilationJobName=compilation_job_name,
ModelName=model_name,
ModelVersion=model_version,
RoleArn=role,
OutputConfig={
'S3OutputLocation': f's3://{bucket_name}/{model_name}'
}
)
while True:
resp = sm_client.describe_edge_packaging_job(EdgePackagingJobName=edge_packaging_job_name)
if resp['EdgePackagingJobStatus'] in ['STARTING', 'INPROGRESS']:
print('Running...')
else:
print(resp['EdgePackagingJobStatus'], compilation_job_name)
break
time.sleep(5)
| 0.38549 | 0.951504 |
```
import keras
import keras.backend as K
from keras.datasets import mnist
from keras.models import Sequential, Model, load_model
from keras.layers import Dense, Dropout, Activation, Flatten, Input, Lambda
from keras.layers import Conv2D, MaxPooling2D, AveragePooling2D, Conv1D, MaxPooling1D, LSTM, ConvLSTM2D, GRU, CuDNNLSTM, CuDNNGRU, BatchNormalization, LocallyConnected2D, Permute, TimeDistributed, Bidirectional
from keras.layers import Concatenate, Reshape, Conv2DTranspose, Embedding, Multiply, Activation
from functools import partial
from collections import defaultdict
import os
import pickle
import numpy as np
import scipy.sparse as sp
import scipy.io as spio
import isolearn.io as isoio
import isolearn.keras as isol
import matplotlib.pyplot as plt
import tensorflow as tf
from keras.backend.tensorflow_backend import set_session
import pandas as pd
def contain_tf_gpu_mem_usage() :
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
sess = tf.Session(config=config)
set_session(sess)
contain_tf_gpu_mem_usage()
class MySequence :
def __init__(self) :
self.dummy = 1
keras.utils.Sequence = MySequence
import isolearn.keras as iso
from sequence_logo_helper_protein import plot_protein_logo, letterAt_protein
class IdentityEncoder(iso.SequenceEncoder) :
def __init__(self, seq_len, channel_map) :
super(IdentityEncoder, self).__init__('identity', (seq_len, len(channel_map)))
self.seq_len = seq_len
self.n_channels = len(channel_map)
self.encode_map = channel_map
self.decode_map = {
val : key for key, val in channel_map.items()
}
def encode(self, seq) :
encoding = np.zeros((self.seq_len, self.n_channels))
for i in range(len(seq)) :
if seq[i] in self.encode_map :
channel_ix = self.encode_map[seq[i]]
encoding[i, channel_ix] = 1.
return encoding
def encode_inplace(self, seq, encoding) :
for i in range(len(seq)) :
if seq[i] in self.encode_map :
channel_ix = self.encode_map[seq[i]]
encoding[i, channel_ix] = 1.
def encode_inplace_sparse(self, seq, encoding_mat, row_index) :
raise NotImplementError()
def decode(self, encoding) :
seq = ''
for pos in range(0, encoding.shape[0]) :
argmax_nt = np.argmax(encoding[pos, :])
max_nt = np.max(encoding[pos, :])
if max_nt == 1 :
seq += self.decode_map[argmax_nt]
else :
seq += "0"
return seq
def decode_sparse(self, encoding_mat, row_index) :
encoding = np.array(encoding_mat[row_index, :].todense()).reshape(-1, 4)
return self.decode(encoding)
class NopTransformer(iso.ValueTransformer) :
def __init__(self, n_classes) :
super(NopTransformer, self).__init__('nop', (n_classes, ))
self.n_classes = n_classes
def transform(self, values) :
return values
def transform_inplace(self, values, transform) :
transform[:] = values
def transform_inplace_sparse(self, values, transform_mat, row_index) :
transform_mat[row_index, :] = np.ravel(values)
#Re-load cached dataframe (shuffled)
dataset_name = "coiled_coil_binders"
experiment = "baker_big_set_5x_negatives"
pair_df = pd.read_csv("pair_df_" + experiment + "_in_shuffled.csv", sep="\t")
print("len(pair_df) = " + str(len(pair_df)))
print(pair_df.head())
#Generate training and test set indexes
valid_set_size = 0.0005
test_set_size = 0.0995
data_index = np.arange(len(pair_df), dtype=np.int)
train_index = data_index[:-int(len(pair_df) * (valid_set_size + test_set_size))]
valid_index = data_index[train_index.shape[0]:-int(len(pair_df) * test_set_size)]
test_index = data_index[train_index.shape[0] + valid_index.shape[0]:]
print('Training set size = ' + str(train_index.shape[0]))
print('Validation set size = ' + str(valid_index.shape[0]))
print('Test set size = ' + str(test_index.shape[0]))
#Sub-select smaller dataset
n_train_pos = 20000
n_train_neg = 20000
n_test_pos = 2000
n_test_neg = 2000
orig_n_train = train_index.shape[0]
orig_n_valid = valid_index.shape[0]
orig_n_test = test_index.shape[0]
train_index_pos = np.nonzero((pair_df.iloc[train_index]['interacts'] == 1).values)[0][:n_train_pos]
train_index_neg = np.nonzero((pair_df.iloc[train_index]['interacts'] == 0).values)[0][:n_train_neg]
train_index = np.concatenate([train_index_pos, train_index_neg], axis=0)
np.random.shuffle(train_index)
test_index_pos = np.nonzero((pair_df.iloc[test_index]['interacts'] == 1).values)[0][:n_test_pos] + orig_n_train + orig_n_valid
test_index_neg = np.nonzero((pair_df.iloc[test_index]['interacts'] == 0).values)[0][:n_test_neg] + orig_n_train + orig_n_valid
test_index = np.concatenate([test_index_pos, test_index_neg], axis=0)
np.random.shuffle(test_index)
print('Training set size = ' + str(train_index.shape[0]))
print('Test set size = ' + str(test_index.shape[0]))
#Calculate sequence lengths
pair_df['amino_seq_1_len'] = pair_df['amino_seq_1'].str.len()
pair_df['amino_seq_2_len'] = pair_df['amino_seq_2'].str.len()
#Initialize sequence encoder
seq_length = 81
residue_map = {'D': 0, 'E': 1, 'V': 2, 'K': 3, 'R': 4, 'L': 5, 'S': 6, 'T': 7, 'N': 8, 'H': 9, 'A': 10, 'I': 11, 'G': 12, 'P': 13, 'Q': 14, 'Y': 15, 'W': 16, 'M': 17, 'F': 18, '#': 19}
encoder = IdentityEncoder(seq_length, residue_map)
#Construct data generators
class CategoricalRandomizer :
def __init__(self, case_range, case_probs) :
self.case_range = case_range
self.case_probs = case_probs
self.cases = 0
def get_random_sample(self, index=None) :
if index is None :
return self.cases
else :
return self.cases[index]
def generate_random_sample(self, batch_size=1, data_ids=None) :
self.cases = np.random.choice(self.case_range, size=batch_size, replace=True, p=self.case_probs)
def get_amino_seq(row, index, flip_randomizer, homodimer_randomizer, max_seq_len=seq_length) :
is_flip = True if flip_randomizer.get_random_sample(index=index) == 1 else False
is_homodimer = True if homodimer_randomizer.get_random_sample(index=index) == 1 else False
amino_seq_1, amino_seq_2 = row['amino_seq_1'], row['amino_seq_2']
if is_flip :
amino_seq_1, amino_seq_2 = row['amino_seq_2'], row['amino_seq_1']
if is_homodimer and row['interacts'] < 0.5 :
amino_seq_2 = amino_seq_1
return amino_seq_1, amino_seq_2
flip_randomizer = CategoricalRandomizer(np.arange(2), np.array([0.5, 0.5]))
homodimer_randomizer = CategoricalRandomizer(np.arange(2), np.array([0.95, 0.05]))
batch_size = 32
data_gens = {
gen_id : iso.DataGenerator(
idx,
{ 'df' : pair_df },
batch_size=(idx.shape[0] // batch_size) * batch_size,
inputs = [
{
'id' : 'amino_seq_1',
'source_type' : 'dataframe',
'source' : 'df',
#'extractor' : lambda row, index, flip_randomizer=flip_randomizer, homodimer_randomizer=homodimer_randomizer: (get_amino_seq(row, index, flip_randomizer, homodimer_randomizer)[0] + "#" * seq_length)[:seq_length],
'extractor' : lambda row, index, flip_randomizer=flip_randomizer, homodimer_randomizer=homodimer_randomizer: get_amino_seq(row, index, flip_randomizer, homodimer_randomizer)[0],
'encoder' : IdentityEncoder(seq_length, residue_map),
'dim' : (1, seq_length, len(residue_map)),
'sparsify' : False
},
{
'id' : 'amino_seq_2',
'source_type' : 'dataframe',
'source' : 'df',
#'extractor' : lambda row, index, flip_randomizer=flip_randomizer, homodimer_randomizer=homodimer_randomizer: (get_amino_seq(row, index, flip_randomizer, homodimer_randomizer)[1] + "#" * seq_length)[:seq_length],
'extractor' : lambda row, index, flip_randomizer=flip_randomizer, homodimer_randomizer=homodimer_randomizer: get_amino_seq(row, index, flip_randomizer, homodimer_randomizer)[1],
'encoder' : IdentityEncoder(seq_length, residue_map),
'dim' : (1, seq_length, len(residue_map)),
'sparsify' : False
},
{
'id' : 'amino_seq_1_len',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : lambda row, index, flip_randomizer=flip_randomizer, homodimer_randomizer=homodimer_randomizer: len(get_amino_seq(row, index, flip_randomizer, homodimer_randomizer)[0]),
'encoder' : lambda t: t,
'dim' : (1,),
'sparsify' : False
},
{
'id' : 'amino_seq_2_len',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : lambda row, index, flip_randomizer=flip_randomizer, homodimer_randomizer=homodimer_randomizer: len(get_amino_seq(row, index, flip_randomizer, homodimer_randomizer)[1]),
'encoder' : lambda t: t,
'dim' : (1,),
'sparsify' : False
}
],
outputs = [
{
'id' : 'interacts',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : lambda row, index: row['interacts'],
'transformer' : NopTransformer(1),
'dim' : (1,),
'sparsify' : False
}
],
randomizers = [flip_randomizer, homodimer_randomizer],
shuffle = True
) for gen_id, idx in [('train', train_index), ('valid', valid_index), ('test', test_index)]
}
#Load data matrices
[x_1_train, x_2_train, l_1_train, l_2_train], [y_train] = data_gens['train'][0]
[x_1_test, x_2_test, l_1_test, l_2_test], [y_test] = data_gens['test'][0]
print("x_1_train.shape = " + str(x_1_train.shape))
print("x_2_train.shape = " + str(x_2_train.shape))
print("x_1_test.shape = " + str(x_1_test.shape))
print("x_2_test.shape = " + str(x_2_test.shape))
print("l_1_train.shape = " + str(l_1_train.shape))
print("l2_train.shape = " + str(l_2_train.shape))
print("l_1_test.shape = " + str(l_1_test.shape))
print("l2_test.shape = " + str(l_2_test.shape))
print("y_train.shape = " + str(y_train.shape))
print("y_test.shape = " + str(y_test.shape))
#Define sequence templates
sequence_templates = [
'$' * i + '@' * (seq_length - i)
for i in range(seq_length+1)
]
sequence_masks = [
np.array([1 if sequence_templates[i][j] == '$' else 0 for j in range(len(sequence_templates[i]))])
for i in range(seq_length+1)
]
#Load cached dataframe (shuffled)
dataset_name = "coiled_coil_binders"
experiment = "coiled_coil_binders_alyssa"
data_df = pd.read_csv(experiment + ".csv", sep="\t")
print("len(data_df) = " + str(len(data_df)))
test_df = data_df.copy().reset_index(drop=True)
batch_size = 32
test_df = test_df.iloc[:(len(test_df) // batch_size) * batch_size].copy().reset_index(drop=True)
print("len(test_df) = " + str(len(test_df)))
print(test_df.head())
#Construct test data
batch_size = 32
test_gen = iso.DataGenerator(
np.arange(len(test_df), dtype=np.int),
{ 'df' : test_df },
batch_size=(len(test_df) // batch_size) * batch_size,
inputs = [
{
'id' : 'amino_seq_1',
'source_type' : 'dataframe',
'source' : 'df',
#'extractor' : lambda row, index: (row['amino_seq_1'] + "#" * seq_length)[:seq_length],
'extractor' : lambda row, index: row['amino_seq_1'],
'encoder' : IdentityEncoder(seq_length, residue_map),
'dim' : (1, seq_length, len(residue_map)),
'sparsify' : False
},
{
'id' : 'amino_seq_2',
'source_type' : 'dataframe',
'source' : 'df',
#'extractor' : lambda row, index: row['amino_seq_2'] + "#" * seq_length)[:seq_length],
'extractor' : lambda row, index: row['amino_seq_2'],
'encoder' : IdentityEncoder(seq_length, residue_map),
'dim' : (1, seq_length, len(residue_map)),
'sparsify' : False
},
{
'id' : 'amino_seq_1_len',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : lambda row, index: len(row['amino_seq_1']),
'encoder' : lambda t: t,
'dim' : (1,),
'sparsify' : False
},
{
'id' : 'amino_seq_2_len',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : lambda row, index: len(row['amino_seq_2']),
'encoder' : lambda t: t,
'dim' : (1,),
'sparsify' : False
}
],
outputs = [
{
'id' : 'interacts',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : lambda row, index: row['interacts'],
'transformer' : NopTransformer(1),
'dim' : (1,),
'sparsify' : False
}
],
randomizers = [],
shuffle = False
)
#Load data matrices
[x_1_test, x_2_test, l_1_test, l_2_test], [y_test] = test_gen[0]
print("x_1_test.shape = " + str(x_1_test.shape))
print("x_2_test.shape = " + str(x_2_test.shape))
print("l_1_test.shape = " + str(l_1_test.shape))
print("l_2_test.shape = " + str(l_2_test.shape))
print("y_test.shape = " + str(y_test.shape))
#Load predictor model
def get_shared_model() :
gru_1 = Bidirectional(CuDNNGRU(64, return_sequences=False), merge_mode='concat')
drop_1 = Dropout(0.25)
def shared_model(inp) :
gru_1_out = gru_1(inp)
drop_1_out = drop_1(gru_1_out)
return drop_1_out
return shared_model
shared_model = get_shared_model()
#Inputs
res_1 = Input(shape=(seq_length, 19 + 1))
res_2 = Input(shape=(seq_length, 19 + 1))
#Outputs
true_interacts = Input(shape=(1,))
#Interaction model definition
dense_out_1 = shared_model(res_1)
dense_out_2 = shared_model(res_2)
layer_dense_pair_1 = Dense(128, activation='relu')
dense_out_pair = layer_dense_pair_1(Concatenate(axis=-1)([dense_out_1, dense_out_2]))
pred_interacts = Dense(1, activation='sigmoid', kernel_initializer='zeros')(dense_out_pair)
predictor = Model(
inputs=[
res_1,
res_2
],
outputs=pred_interacts
)
predictor.load_weights('saved_models/ppi_rnn_baker_big_set_5x_negatives_classifier_symmetric_drop_25_5x_negatives_balanced_partitioned_data_epoch_10.h5', by_name=True)
predictor.trainable = False
predictor.compile(
optimizer=keras.optimizers.SGD(lr=0.1),
loss='mean_squared_error'
)
#Generate (original) predictions
pred_train = predictor.predict([x_1_train[:, 0, ...], x_2_train[:, 0, ...]], batch_size=32)
pred_test = predictor.predict([x_1_test[:, 0, ...], x_2_test[:, 0, ...]], batch_size=32)
pred_train = np.concatenate([1. - pred_train, pred_train], axis=1)
pred_test = np.concatenate([1. - pred_test, pred_test], axis=1)
#Make two-channel targets
y_train = np.concatenate([1. - y_train, y_train], axis=1)
y_test = np.concatenate([1. - y_test, y_test], axis=1)
from keras.layers import Input, Dense, Multiply, Flatten, Reshape, Conv2D, MaxPooling2D, GlobalMaxPooling2D, Activation
from keras.layers import BatchNormalization
from keras.models import Sequential, Model
from keras.optimizers import Adam
from keras import regularizers
from keras import backend as K
import tensorflow as tf
import numpy as np
from keras.layers import Layer, InputSpec
from keras import initializers, regularizers, constraints
class InstanceNormalization(Layer):
def __init__(self, axes=(1, 2), trainable=True, **kwargs):
super(InstanceNormalization, self).__init__(**kwargs)
self.axes = axes
self.trainable = trainable
def build(self, input_shape):
self.beta = self.add_weight(name='beta',shape=(input_shape[-1],),
initializer='zeros',trainable=self.trainable)
self.gamma = self.add_weight(name='gamma',shape=(input_shape[-1],),
initializer='ones',trainable=self.trainable)
def call(self, inputs):
mean, variance = tf.nn.moments(inputs, self.axes, keep_dims=True)
return tf.nn.batch_normalization(inputs, mean, variance, self.beta, self.gamma, 1e-6)
def bernoulli_sampling (prob):
""" Sampling Bernoulli distribution by given probability.
Args:
- prob: P(Y = 1) in Bernoulli distribution.
Returns:
- samples: samples from Bernoulli distribution
"""
n, x_len, y_len, d = prob.shape
samples = np.random.binomial(1, prob, (n, x_len, y_len, d))
return samples
class INVASE():
"""INVASE class.
Attributes:
- x_train: training features
- y_train: training labels
- model_type: invase or invase_minus
- model_parameters:
- actor_h_dim: hidden state dimensions for actor
- critic_h_dim: hidden state dimensions for critic
- n_layer: the number of layers
- batch_size: the number of samples in mini batch
- iteration: the number of iterations
- activation: activation function of models
- learning_rate: learning rate of model training
- lamda: hyper-parameter of INVASE
"""
def __init__(self, x_train, y_train, model_type, model_parameters):
self.lamda = model_parameters['lamda']
self.actor_h_dim = model_parameters['actor_h_dim']
self.critic_h_dim = model_parameters['critic_h_dim']
self.n_layer = model_parameters['n_layer']
self.batch_size = model_parameters['batch_size']
self.iteration = model_parameters['iteration']
self.activation = model_parameters['activation']
self.learning_rate = model_parameters['learning_rate']
#Modified Code
self.x_len = x_train.shape[1]
self.y_len = x_train.shape[2]
self.dim = x_train.shape[3]
self.label_dim = y_train.shape[1]
self.model_type = model_type
optimizer = Adam(self.learning_rate)
# Build and compile critic
self.critic = self.build_critic()
self.critic.compile(loss='categorical_crossentropy',
optimizer=optimizer, metrics=['acc'])
# Build and compile the actor
self.actor = self.build_actor()
self.actor.compile(loss=self.actor_loss, optimizer=optimizer)
if self.model_type == 'invase':
# Build and compile the baseline
self.baseline = self.build_baseline()
self.baseline.compile(loss='categorical_crossentropy', optimizer=optimizer, metrics=['acc'])
def actor_loss(self, y_true, y_pred):
"""Custom loss for the actor.
Args:
- y_true:
- actor_out: actor output after sampling
- critic_out: critic output
- baseline_out: baseline output (only for invase)
- y_pred: output of the actor network
Returns:
- loss: actor loss
"""
y_pred = K.reshape(y_pred, (K.shape(y_pred)[0], self.x_len*self.y_len*1))
y_true = y_true[:, 0, 0, :]
# Actor output
actor_out = y_true[:, :self.x_len*self.y_len*1]
# Critic output
critic_out = y_true[:, self.x_len*self.y_len*1:(self.x_len*self.y_len*1+self.label_dim)]
if self.model_type == 'invase':
# Baseline output
baseline_out = \
y_true[:, (self.x_len*self.y_len*1+self.label_dim):(self.x_len*self.y_len*1+2*self.label_dim)]
# Ground truth label
y_out = y_true[:, (self.x_len*self.y_len*1+2*self.label_dim):]
elif self.model_type == 'invase_minus':
# Ground truth label
y_out = y_true[:, (self.x_len*self.y_len*1+self.label_dim):]
# Critic loss
critic_loss = -tf.reduce_sum(y_out * tf.log(critic_out + 1e-8), axis = 1)
if self.model_type == 'invase':
# Baseline loss
baseline_loss = -tf.reduce_sum(y_out * tf.log(baseline_out + 1e-8),
axis = 1)
# Reward
Reward = -(critic_loss - baseline_loss)
elif self.model_type == 'invase_minus':
Reward = -critic_loss
# Policy gradient loss computation.
custom_actor_loss = \
Reward * tf.reduce_sum(actor_out * K.log(y_pred + 1e-8) + \
(1-actor_out) * K.log(1-y_pred + 1e-8), axis = 1) - \
self.lamda * tf.reduce_mean(y_pred, axis = 1)
# custom actor loss
custom_actor_loss = tf.reduce_mean(-custom_actor_loss)
return custom_actor_loss
def build_actor(self):
"""Build actor.
Use feature as the input and output selection probability
"""
actor_model = Sequential()
actor_model.add(Conv2D(self.actor_h_dim, (1, 3), padding='same', activation='linear'))
actor_model.add(InstanceNormalization())
actor_model.add(Activation(self.activation))
for _ in range(self.n_layer - 2):
actor_model.add(Conv2D(self.actor_h_dim, (1, 3), padding='same', activation='linear'))
actor_model.add(InstanceNormalization())
actor_model.add(Activation(self.activation))
actor_model.add(Conv2D(1, (1, 1), padding='same', activation='sigmoid'))
feature = Input(shape=(self.x_len, self.y_len, self.dim), dtype='float32')
selection_probability = actor_model(feature)
return Model(feature, selection_probability)
def build_critic(self):
"""Build critic.
Use selected feature as the input and predict labels
"""
critic_model = Sequential()
critic_model.add(Conv2D(self.critic_h_dim, (1, 3), padding='same', activation='linear'))
critic_model.add(InstanceNormalization())
critic_model.add(Activation(self.activation))
for _ in range(self.n_layer - 2):
critic_model.add(Conv2D(self.critic_h_dim, (1, 3), padding='same', activation='linear'))
critic_model.add(InstanceNormalization())
critic_model.add(Activation(self.activation))
critic_model.add(Flatten())
critic_model.add(Dense(self.critic_h_dim, activation=self.activation))
#critic_model.add(Dropout(0.2))
critic_model.add(Dense(self.label_dim, activation ='softmax'))
## Inputs
# Features
feature = Input(shape=(self.x_len, self.y_len, self.dim), dtype='float32')
# Binary selection
selection = Input(shape=(self.x_len, self.y_len, 1), dtype='float32')
# Element-wise multiplication
critic_model_input = Multiply()([feature, selection])
y_hat = critic_model(critic_model_input)
return Model([feature, selection], y_hat)
def build_baseline(self):
"""Build baseline.
Use the feature as the input and predict labels
"""
baseline_model = Sequential()
baseline_model.add(Conv2D(self.critic_h_dim, (1, 3), padding='same', activation='linear'))
baseline_model.add(InstanceNormalization())
baseline_model.add(Activation(self.activation))
for _ in range(self.n_layer - 2):
baseline_model.add(Conv2D(self.critic_h_dim, (1, 3), padding='same', activation='linear'))
baseline_model.add(InstanceNormalization())
baseline_model.add(Activation(self.activation))
baseline_model.add(Flatten())
baseline_model.add(Dense(self.critic_h_dim, activation=self.activation))
#baseline_model.add(Dropout(0.2))
baseline_model.add(Dense(self.label_dim, activation ='softmax'))
# Input
feature = Input(shape=(self.x_len, self.y_len, self.dim), dtype='float32')
# Output
y_hat = baseline_model(feature)
return Model(feature, y_hat)
def train(self, x_train, y_train):
"""Train INVASE.
Args:
- x_train: training features
- y_train: training labels
"""
for iter_idx in range(self.iteration):
## Train critic
# Select a random batch of samples
idx = np.random.randint(0, x_train.shape[0], self.batch_size)
x_batch = x_train[idx,:]
y_batch = y_train[idx,:]
# Generate a batch of selection probability
selection_probability = self.actor.predict(x_batch)
# Sampling the features based on the selection_probability
selection = bernoulli_sampling(selection_probability)
# Critic loss
critic_loss = self.critic.train_on_batch([x_batch, selection], y_batch)
# Critic output
critic_out = self.critic.predict([x_batch, selection])
# Baseline output
if self.model_type == 'invase':
# Baseline loss
baseline_loss = self.baseline.train_on_batch(x_batch, y_batch)
# Baseline output
baseline_out = self.baseline.predict(x_batch)
## Train actor
# Use multiple things as the y_true:
# - selection, critic_out, baseline_out, and ground truth (y_batch)
if self.model_type == 'invase':
y_batch_final = np.concatenate((np.reshape(selection, (y_batch.shape[0], -1)),
np.asarray(critic_out),
np.asarray(baseline_out),
y_batch), axis = 1)
elif self.model_type == 'invase_minus':
y_batch_final = np.concatenate((np.reshape(selection, (y_batch.shape[0], -1)),
np.asarray(critic_out),
y_batch), axis = 1)
y_batch_final = y_batch_final[:, None, None, :]
# Train the actor
actor_loss = self.actor.train_on_batch(x_batch, y_batch_final)
if self.model_type == 'invase':
# Print the progress
dialog = 'Iterations: ' + str(iter_idx) + \
', critic accuracy: ' + str(critic_loss[1]) + \
', baseline accuracy: ' + str(baseline_loss[1]) + \
', actor loss: ' + str(np.round(actor_loss,4))
elif self.model_type == 'invase_minus':
# Print the progress
dialog = 'Iterations: ' + str(iter_idx) + \
', critic accuracy: ' + str(critic_loss[1]) + \
', actor loss: ' + str(np.round(actor_loss,4))
if iter_idx % 100 == 0:
print(dialog)
def importance_score(self, x):
"""Return featuer importance score.
Args:
- x: feature
Returns:
- feature_importance: instance-wise feature importance for x
"""
feature_importance = self.actor.predict(x)
return np.asarray(feature_importance)
def predict(self, x):
"""Predict outcomes.
Args:
- x: feature
Returns:
- y_hat: predictions
"""
# Generate a batch of selection probability
selection_probability = self.actor.predict(x)
# Sampling the features based on the selection_probability
selection = bernoulli_sampling(selection_probability)
# Prediction
y_hat = self.critic.predict([x, selection])
return np.asarray(y_hat)
#Concatenate input binder pairs
x_train = np.concatenate([x_1_train, x_2_train], axis=2)
x_test = np.concatenate([x_1_test, x_2_test], axis=2)
#Execute INVASE
mask_penalty = 0.05
hidden_dims = 32
n_layers = 4
epochs = 50
batch_size = 128
model_parameters = {
'lamda': mask_penalty,
'actor_h_dim': hidden_dims,
'critic_h_dim': hidden_dims,
'n_layer': n_layers,
'batch_size': batch_size,
'iteration': int(x_train.shape[0] * epochs / batch_size),
'activation': 'relu',
'learning_rate': 0.0001
}
invase_model = INVASE(x_train, pred_train, 'invase', model_parameters)
invase_model.train(x_train, pred_train)
importance_scores_test = invase_model.importance_score(x_test)
importance_scores_1_test, importance_scores_2_test = importance_scores_test[:, :, :seq_length, :], importance_scores_test[:, :, seq_length:, :]
#Evaluate INVASE model on train and test data
invase_pred_train = invase_model.predict(x_train)
invase_pred_test = invase_model.predict(x_test)
print("Training Accuracy = " + str(np.sum(np.argmax(invase_pred_train, axis=1) == np.argmax(pred_train, axis=1)) / float(pred_train.shape[0])))
print("Test Accuracy = " + str(np.sum(np.argmax(invase_pred_test, axis=1) == np.argmax(pred_test, axis=1)) / float(pred_test.shape[0])))
#Gradient saliency/backprop visualization
import matplotlib.collections as collections
import operator
import matplotlib.pyplot as plt
import matplotlib.cm as cm
import matplotlib.colors as colors
import matplotlib as mpl
from matplotlib.text import TextPath
from matplotlib.patches import PathPatch, Rectangle
from matplotlib.font_manager import FontProperties
from matplotlib import gridspec
from matplotlib.ticker import FormatStrFormatter
def plot_protein_logo(residue_map, pwm, sequence_template=None, figsize=(12, 3), logo_height=1.0, plot_start=0, plot_end=164) :
inv_residue_map = {
i : sp for sp, i in residue_map.items()
}
#Slice according to seq trim index
pwm = pwm[plot_start: plot_end, :]
sequence_template = sequence_template[plot_start: plot_end]
entropy = np.zeros(pwm.shape)
entropy[pwm > 0] = pwm[pwm > 0] * -np.log2(np.clip(pwm[pwm > 0], 1e-6, 1. - 1e-6))
entropy = np.sum(entropy, axis=1)
conservation = np.log2(len(residue_map)) - entropy#2 - entropy
fig = plt.figure(figsize=figsize)
ax = plt.gca()
height_base = (1.0 - logo_height) / 2.
for j in range(0, pwm.shape[0]) :
sort_index = np.argsort(pwm[j, :])
for ii in range(0, len(residue_map)) :
i = sort_index[ii]
if pwm[j, i] > 0 :
nt_prob = pwm[j, i] * conservation[j]
nt = inv_residue_map[i]
color = None
if sequence_template[j] != '$' :
color = 'black'
if ii == 0 :
letterAt_protein(nt, j + 0.5, height_base, nt_prob * logo_height, ax, color=color)
else :
prev_prob = np.sum(pwm[j, sort_index[:ii]] * conservation[j]) * logo_height
letterAt_protein(nt, j + 0.5, height_base + prev_prob, nt_prob * logo_height, ax, color=color)
plt.xlim((0, plot_end - plot_start))
plt.ylim((0, np.log2(len(residue_map))))
plt.xticks([], [])
plt.yticks([], [])
plt.axis('off')
plt.axhline(y=0.01 + height_base, color='black', linestyle='-', linewidth=2)
for axis in fig.axes :
axis.get_xaxis().set_visible(False)
axis.get_yaxis().set_visible(False)
plt.tight_layout()
plt.show()
def plot_importance_scores(importance_scores, ref_seq, figsize=(12, 2), score_clip=None, sequence_template='', plot_start=0, plot_end=96, save_figs=False, fig_name=None) :
end_pos = ref_seq.find("#")
fig = plt.figure(figsize=figsize)
ax = plt.gca()
if score_clip is not None :
importance_scores = np.clip(np.copy(importance_scores), -score_clip, score_clip)
max_score = np.max(np.sum(importance_scores[:, :], axis=0)) + 0.01
for i in range(0, len(ref_seq)) :
mutability_score = np.sum(importance_scores[:, i])
letterAt_protein(ref_seq[i], i + 0.5, 0, mutability_score, ax, color=None)
plt.sca(ax)
plt.xlim((0, len(ref_seq)))
plt.ylim((0, max_score))
plt.axis('off')
plt.yticks([0.0, max_score], [0.0, max_score], fontsize=16)
for axis in fig.axes :
axis.get_xaxis().set_visible(False)
axis.get_yaxis().set_visible(False)
plt.tight_layout()
if save_figs :
plt.savefig(fig_name + ".png", transparent=True, dpi=300)
plt.savefig(fig_name + ".eps")
plt.show()
np.sum(importance_scores_1_test[0, 0, :, 0] + importance_scores_2_test[0, 0, :, 0])
np.max(importance_scores_1_test[0, 0, :, 0])
#Visualize importance for binder 1
for plot_i in range(0, 5) :
print("Test sequence " + str(plot_i) + ":")
sequence_template = sequence_templates[l_1_test[plot_i, 0]]
plot_protein_logo(residue_map, x_1_test[plot_i, 0, :, :], sequence_template=sequence_template, figsize=(12, 1), plot_start=0, plot_end=81)
plot_importance_scores(importance_scores_1_test[plot_i, 0, :, :].T, encoder.decode(x_1_test[plot_i, 0, :, :]), figsize=(12, 1), score_clip=None, sequence_template=sequence_template, plot_start=0, plot_end=81)
#Visualize importance for binder 2
for plot_i in range(0, 5) :
print("Test sequence " + str(plot_i) + ":")
sequence_template = sequence_templates[l_2_test[plot_i, 0]]
plot_protein_logo(residue_map, x_2_test[plot_i, 0, :, :], sequence_template=sequence_template, figsize=(12, 1), plot_start=0, plot_end=81)
plot_importance_scores(importance_scores_2_test[plot_i, 0, :, :].T, encoder.decode(x_2_test[plot_i, 0, :, :]), figsize=(12, 1), score_clip=None, sequence_template=sequence_template, plot_start=0, plot_end=81)
#Save predicted importance scores
model_name = "invase_" + dataset_name + "_conv" + "_zeropad_no_drop_penalty_005"
np.save(model_name + "_importance_scores_1_test", importance_scores_1_test)
np.save(model_name + "_importance_scores_2_test", importance_scores_2_test)
#Binder DHD_154
seq_1 = "TAEELLEVHKKSDRVTKEHLRVSEEILKVVEVLTRGEVSSEVLKRVLRKLEELTDKLRRVTEEQRRVVEKLN"[:81]
seq_2 = "DLEDLLRRLRRLVDEQRRLVEELERVSRRLEKAVRDNEDERELARLSREHSDIQDKHDKLAREILEVLKRLLERTE"[:81]
print("Seq 1 = " + seq_1)
print("Seq 2 = " + seq_2)
encoder = IdentityEncoder(81, residue_map)
test_onehot_1 = np.tile(np.expand_dims(np.expand_dims(encoder(seq_1), axis=0), axis=0), (batch_size, 1, 1, 1))
test_onehot_2 = np.tile(np.expand_dims(np.expand_dims(encoder(seq_2), axis=0), axis=0), (batch_size, 1, 1, 1))
test_len_1 = np.tile(np.array([[len(seq_1)]]), (batch_size, 1))
test_len_2 = np.tile(np.array([[len(seq_2)]]), (batch_size, 1))
pred_interacts = predictor.predict(x=[test_onehot_1[:, 0, ...], test_onehot_2[:, 0, ...]])[0, 0]
print("Predicted interaction prob = " + str(round(pred_interacts, 4)))
x_1_test = test_onehot_1[:1]
x_2_test = test_onehot_2[:1]
#Concatenate input binder pairs
x_test = np.concatenate([x_1_test, x_2_test], axis=2)
#Execute INVASE
mask_penalty = 0.05
hidden_dims = 32
n_layers = 4
epochs = 50
batch_size = 128
model_parameters = {
'lamda': mask_penalty,
'actor_h_dim': hidden_dims,
'critic_h_dim': hidden_dims,
'n_layer': n_layers,
'batch_size': batch_size,
'iteration': int(x_train.shape[0] * epochs / batch_size),
'activation': 'relu',
'learning_rate': 0.0001
}
invase_model = INVASE(x_train, pred_train, 'invase', model_parameters)
invase_model.train(x_train, pred_train)
importance_scores_test = invase_model.importance_score(x_test)
importance_scores_1_test, importance_scores_2_test = importance_scores_test[:, :, :seq_length, :], importance_scores_test[:, :, seq_length:, :]
save_figs = True
model_name = "invase_" + dataset_name + "_conv" + "_zeropad_no_drop_penalty_005"
pair_name = "DHD_154"
#Visualize importance for binder 1
for plot_i in range(0, 1) :
print("Test sequence " + str(plot_i) + ":")
sequence_template = sequence_templates[l_1_test[plot_i, 0]]
plot_protein_logo(residue_map, x_1_test[plot_i, 0, :, :], sequence_template=sequence_template, figsize=(12, 1), plot_start=0, plot_end=81)
plot_importance_scores(importance_scores_1_test[plot_i, 0, :, :].T, encoder.decode(x_1_test[plot_i, 0, :, :]), figsize=(12, 1), score_clip=None, sequence_template=sequence_template, plot_start=0, plot_end=81, save_figs=save_figs, fig_name=model_name + "_scores_" + pair_name + "_binder_1")
#Visualize importance for binder 2
for plot_i in range(0, 1) :
print("Test sequence " + str(plot_i) + ":")
sequence_template = sequence_templates[l_2_test[plot_i, 0]]
plot_protein_logo(residue_map, x_2_test[plot_i, 0, :, :], sequence_template=sequence_template, figsize=(12, 1), plot_start=0, plot_end=81)
plot_importance_scores(importance_scores_2_test[plot_i, 0, :, :].T, encoder.decode(x_2_test[plot_i, 0, :, :]), figsize=(12, 1), score_clip=None, sequence_template=sequence_template, plot_start=0, plot_end=81, save_figs=save_figs, fig_name=model_name + "_scores_" + pair_name + "_binder_2")
```
|
github_jupyter
|
import keras
import keras.backend as K
from keras.datasets import mnist
from keras.models import Sequential, Model, load_model
from keras.layers import Dense, Dropout, Activation, Flatten, Input, Lambda
from keras.layers import Conv2D, MaxPooling2D, AveragePooling2D, Conv1D, MaxPooling1D, LSTM, ConvLSTM2D, GRU, CuDNNLSTM, CuDNNGRU, BatchNormalization, LocallyConnected2D, Permute, TimeDistributed, Bidirectional
from keras.layers import Concatenate, Reshape, Conv2DTranspose, Embedding, Multiply, Activation
from functools import partial
from collections import defaultdict
import os
import pickle
import numpy as np
import scipy.sparse as sp
import scipy.io as spio
import isolearn.io as isoio
import isolearn.keras as isol
import matplotlib.pyplot as plt
import tensorflow as tf
from keras.backend.tensorflow_backend import set_session
import pandas as pd
def contain_tf_gpu_mem_usage() :
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
sess = tf.Session(config=config)
set_session(sess)
contain_tf_gpu_mem_usage()
class MySequence :
def __init__(self) :
self.dummy = 1
keras.utils.Sequence = MySequence
import isolearn.keras as iso
from sequence_logo_helper_protein import plot_protein_logo, letterAt_protein
class IdentityEncoder(iso.SequenceEncoder) :
def __init__(self, seq_len, channel_map) :
super(IdentityEncoder, self).__init__('identity', (seq_len, len(channel_map)))
self.seq_len = seq_len
self.n_channels = len(channel_map)
self.encode_map = channel_map
self.decode_map = {
val : key for key, val in channel_map.items()
}
def encode(self, seq) :
encoding = np.zeros((self.seq_len, self.n_channels))
for i in range(len(seq)) :
if seq[i] in self.encode_map :
channel_ix = self.encode_map[seq[i]]
encoding[i, channel_ix] = 1.
return encoding
def encode_inplace(self, seq, encoding) :
for i in range(len(seq)) :
if seq[i] in self.encode_map :
channel_ix = self.encode_map[seq[i]]
encoding[i, channel_ix] = 1.
def encode_inplace_sparse(self, seq, encoding_mat, row_index) :
raise NotImplementError()
def decode(self, encoding) :
seq = ''
for pos in range(0, encoding.shape[0]) :
argmax_nt = np.argmax(encoding[pos, :])
max_nt = np.max(encoding[pos, :])
if max_nt == 1 :
seq += self.decode_map[argmax_nt]
else :
seq += "0"
return seq
def decode_sparse(self, encoding_mat, row_index) :
encoding = np.array(encoding_mat[row_index, :].todense()).reshape(-1, 4)
return self.decode(encoding)
class NopTransformer(iso.ValueTransformer) :
def __init__(self, n_classes) :
super(NopTransformer, self).__init__('nop', (n_classes, ))
self.n_classes = n_classes
def transform(self, values) :
return values
def transform_inplace(self, values, transform) :
transform[:] = values
def transform_inplace_sparse(self, values, transform_mat, row_index) :
transform_mat[row_index, :] = np.ravel(values)
#Re-load cached dataframe (shuffled)
dataset_name = "coiled_coil_binders"
experiment = "baker_big_set_5x_negatives"
pair_df = pd.read_csv("pair_df_" + experiment + "_in_shuffled.csv", sep="\t")
print("len(pair_df) = " + str(len(pair_df)))
print(pair_df.head())
#Generate training and test set indexes
valid_set_size = 0.0005
test_set_size = 0.0995
data_index = np.arange(len(pair_df), dtype=np.int)
train_index = data_index[:-int(len(pair_df) * (valid_set_size + test_set_size))]
valid_index = data_index[train_index.shape[0]:-int(len(pair_df) * test_set_size)]
test_index = data_index[train_index.shape[0] + valid_index.shape[0]:]
print('Training set size = ' + str(train_index.shape[0]))
print('Validation set size = ' + str(valid_index.shape[0]))
print('Test set size = ' + str(test_index.shape[0]))
#Sub-select smaller dataset
n_train_pos = 20000
n_train_neg = 20000
n_test_pos = 2000
n_test_neg = 2000
orig_n_train = train_index.shape[0]
orig_n_valid = valid_index.shape[0]
orig_n_test = test_index.shape[0]
train_index_pos = np.nonzero((pair_df.iloc[train_index]['interacts'] == 1).values)[0][:n_train_pos]
train_index_neg = np.nonzero((pair_df.iloc[train_index]['interacts'] == 0).values)[0][:n_train_neg]
train_index = np.concatenate([train_index_pos, train_index_neg], axis=0)
np.random.shuffle(train_index)
test_index_pos = np.nonzero((pair_df.iloc[test_index]['interacts'] == 1).values)[0][:n_test_pos] + orig_n_train + orig_n_valid
test_index_neg = np.nonzero((pair_df.iloc[test_index]['interacts'] == 0).values)[0][:n_test_neg] + orig_n_train + orig_n_valid
test_index = np.concatenate([test_index_pos, test_index_neg], axis=0)
np.random.shuffle(test_index)
print('Training set size = ' + str(train_index.shape[0]))
print('Test set size = ' + str(test_index.shape[0]))
#Calculate sequence lengths
pair_df['amino_seq_1_len'] = pair_df['amino_seq_1'].str.len()
pair_df['amino_seq_2_len'] = pair_df['amino_seq_2'].str.len()
#Initialize sequence encoder
seq_length = 81
residue_map = {'D': 0, 'E': 1, 'V': 2, 'K': 3, 'R': 4, 'L': 5, 'S': 6, 'T': 7, 'N': 8, 'H': 9, 'A': 10, 'I': 11, 'G': 12, 'P': 13, 'Q': 14, 'Y': 15, 'W': 16, 'M': 17, 'F': 18, '#': 19}
encoder = IdentityEncoder(seq_length, residue_map)
#Construct data generators
class CategoricalRandomizer :
def __init__(self, case_range, case_probs) :
self.case_range = case_range
self.case_probs = case_probs
self.cases = 0
def get_random_sample(self, index=None) :
if index is None :
return self.cases
else :
return self.cases[index]
def generate_random_sample(self, batch_size=1, data_ids=None) :
self.cases = np.random.choice(self.case_range, size=batch_size, replace=True, p=self.case_probs)
def get_amino_seq(row, index, flip_randomizer, homodimer_randomizer, max_seq_len=seq_length) :
is_flip = True if flip_randomizer.get_random_sample(index=index) == 1 else False
is_homodimer = True if homodimer_randomizer.get_random_sample(index=index) == 1 else False
amino_seq_1, amino_seq_2 = row['amino_seq_1'], row['amino_seq_2']
if is_flip :
amino_seq_1, amino_seq_2 = row['amino_seq_2'], row['amino_seq_1']
if is_homodimer and row['interacts'] < 0.5 :
amino_seq_2 = amino_seq_1
return amino_seq_1, amino_seq_2
flip_randomizer = CategoricalRandomizer(np.arange(2), np.array([0.5, 0.5]))
homodimer_randomizer = CategoricalRandomizer(np.arange(2), np.array([0.95, 0.05]))
batch_size = 32
data_gens = {
gen_id : iso.DataGenerator(
idx,
{ 'df' : pair_df },
batch_size=(idx.shape[0] // batch_size) * batch_size,
inputs = [
{
'id' : 'amino_seq_1',
'source_type' : 'dataframe',
'source' : 'df',
#'extractor' : lambda row, index, flip_randomizer=flip_randomizer, homodimer_randomizer=homodimer_randomizer: (get_amino_seq(row, index, flip_randomizer, homodimer_randomizer)[0] + "#" * seq_length)[:seq_length],
'extractor' : lambda row, index, flip_randomizer=flip_randomizer, homodimer_randomizer=homodimer_randomizer: get_amino_seq(row, index, flip_randomizer, homodimer_randomizer)[0],
'encoder' : IdentityEncoder(seq_length, residue_map),
'dim' : (1, seq_length, len(residue_map)),
'sparsify' : False
},
{
'id' : 'amino_seq_2',
'source_type' : 'dataframe',
'source' : 'df',
#'extractor' : lambda row, index, flip_randomizer=flip_randomizer, homodimer_randomizer=homodimer_randomizer: (get_amino_seq(row, index, flip_randomizer, homodimer_randomizer)[1] + "#" * seq_length)[:seq_length],
'extractor' : lambda row, index, flip_randomizer=flip_randomizer, homodimer_randomizer=homodimer_randomizer: get_amino_seq(row, index, flip_randomizer, homodimer_randomizer)[1],
'encoder' : IdentityEncoder(seq_length, residue_map),
'dim' : (1, seq_length, len(residue_map)),
'sparsify' : False
},
{
'id' : 'amino_seq_1_len',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : lambda row, index, flip_randomizer=flip_randomizer, homodimer_randomizer=homodimer_randomizer: len(get_amino_seq(row, index, flip_randomizer, homodimer_randomizer)[0]),
'encoder' : lambda t: t,
'dim' : (1,),
'sparsify' : False
},
{
'id' : 'amino_seq_2_len',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : lambda row, index, flip_randomizer=flip_randomizer, homodimer_randomizer=homodimer_randomizer: len(get_amino_seq(row, index, flip_randomizer, homodimer_randomizer)[1]),
'encoder' : lambda t: t,
'dim' : (1,),
'sparsify' : False
}
],
outputs = [
{
'id' : 'interacts',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : lambda row, index: row['interacts'],
'transformer' : NopTransformer(1),
'dim' : (1,),
'sparsify' : False
}
],
randomizers = [flip_randomizer, homodimer_randomizer],
shuffle = True
) for gen_id, idx in [('train', train_index), ('valid', valid_index), ('test', test_index)]
}
#Load data matrices
[x_1_train, x_2_train, l_1_train, l_2_train], [y_train] = data_gens['train'][0]
[x_1_test, x_2_test, l_1_test, l_2_test], [y_test] = data_gens['test'][0]
print("x_1_train.shape = " + str(x_1_train.shape))
print("x_2_train.shape = " + str(x_2_train.shape))
print("x_1_test.shape = " + str(x_1_test.shape))
print("x_2_test.shape = " + str(x_2_test.shape))
print("l_1_train.shape = " + str(l_1_train.shape))
print("l2_train.shape = " + str(l_2_train.shape))
print("l_1_test.shape = " + str(l_1_test.shape))
print("l2_test.shape = " + str(l_2_test.shape))
print("y_train.shape = " + str(y_train.shape))
print("y_test.shape = " + str(y_test.shape))
#Define sequence templates
sequence_templates = [
'$' * i + '@' * (seq_length - i)
for i in range(seq_length+1)
]
sequence_masks = [
np.array([1 if sequence_templates[i][j] == '$' else 0 for j in range(len(sequence_templates[i]))])
for i in range(seq_length+1)
]
#Load cached dataframe (shuffled)
dataset_name = "coiled_coil_binders"
experiment = "coiled_coil_binders_alyssa"
data_df = pd.read_csv(experiment + ".csv", sep="\t")
print("len(data_df) = " + str(len(data_df)))
test_df = data_df.copy().reset_index(drop=True)
batch_size = 32
test_df = test_df.iloc[:(len(test_df) // batch_size) * batch_size].copy().reset_index(drop=True)
print("len(test_df) = " + str(len(test_df)))
print(test_df.head())
#Construct test data
batch_size = 32
test_gen = iso.DataGenerator(
np.arange(len(test_df), dtype=np.int),
{ 'df' : test_df },
batch_size=(len(test_df) // batch_size) * batch_size,
inputs = [
{
'id' : 'amino_seq_1',
'source_type' : 'dataframe',
'source' : 'df',
#'extractor' : lambda row, index: (row['amino_seq_1'] + "#" * seq_length)[:seq_length],
'extractor' : lambda row, index: row['amino_seq_1'],
'encoder' : IdentityEncoder(seq_length, residue_map),
'dim' : (1, seq_length, len(residue_map)),
'sparsify' : False
},
{
'id' : 'amino_seq_2',
'source_type' : 'dataframe',
'source' : 'df',
#'extractor' : lambda row, index: row['amino_seq_2'] + "#" * seq_length)[:seq_length],
'extractor' : lambda row, index: row['amino_seq_2'],
'encoder' : IdentityEncoder(seq_length, residue_map),
'dim' : (1, seq_length, len(residue_map)),
'sparsify' : False
},
{
'id' : 'amino_seq_1_len',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : lambda row, index: len(row['amino_seq_1']),
'encoder' : lambda t: t,
'dim' : (1,),
'sparsify' : False
},
{
'id' : 'amino_seq_2_len',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : lambda row, index: len(row['amino_seq_2']),
'encoder' : lambda t: t,
'dim' : (1,),
'sparsify' : False
}
],
outputs = [
{
'id' : 'interacts',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : lambda row, index: row['interacts'],
'transformer' : NopTransformer(1),
'dim' : (1,),
'sparsify' : False
}
],
randomizers = [],
shuffle = False
)
#Load data matrices
[x_1_test, x_2_test, l_1_test, l_2_test], [y_test] = test_gen[0]
print("x_1_test.shape = " + str(x_1_test.shape))
print("x_2_test.shape = " + str(x_2_test.shape))
print("l_1_test.shape = " + str(l_1_test.shape))
print("l_2_test.shape = " + str(l_2_test.shape))
print("y_test.shape = " + str(y_test.shape))
#Load predictor model
def get_shared_model() :
gru_1 = Bidirectional(CuDNNGRU(64, return_sequences=False), merge_mode='concat')
drop_1 = Dropout(0.25)
def shared_model(inp) :
gru_1_out = gru_1(inp)
drop_1_out = drop_1(gru_1_out)
return drop_1_out
return shared_model
shared_model = get_shared_model()
#Inputs
res_1 = Input(shape=(seq_length, 19 + 1))
res_2 = Input(shape=(seq_length, 19 + 1))
#Outputs
true_interacts = Input(shape=(1,))
#Interaction model definition
dense_out_1 = shared_model(res_1)
dense_out_2 = shared_model(res_2)
layer_dense_pair_1 = Dense(128, activation='relu')
dense_out_pair = layer_dense_pair_1(Concatenate(axis=-1)([dense_out_1, dense_out_2]))
pred_interacts = Dense(1, activation='sigmoid', kernel_initializer='zeros')(dense_out_pair)
predictor = Model(
inputs=[
res_1,
res_2
],
outputs=pred_interacts
)
predictor.load_weights('saved_models/ppi_rnn_baker_big_set_5x_negatives_classifier_symmetric_drop_25_5x_negatives_balanced_partitioned_data_epoch_10.h5', by_name=True)
predictor.trainable = False
predictor.compile(
optimizer=keras.optimizers.SGD(lr=0.1),
loss='mean_squared_error'
)
#Generate (original) predictions
pred_train = predictor.predict([x_1_train[:, 0, ...], x_2_train[:, 0, ...]], batch_size=32)
pred_test = predictor.predict([x_1_test[:, 0, ...], x_2_test[:, 0, ...]], batch_size=32)
pred_train = np.concatenate([1. - pred_train, pred_train], axis=1)
pred_test = np.concatenate([1. - pred_test, pred_test], axis=1)
#Make two-channel targets
y_train = np.concatenate([1. - y_train, y_train], axis=1)
y_test = np.concatenate([1. - y_test, y_test], axis=1)
from keras.layers import Input, Dense, Multiply, Flatten, Reshape, Conv2D, MaxPooling2D, GlobalMaxPooling2D, Activation
from keras.layers import BatchNormalization
from keras.models import Sequential, Model
from keras.optimizers import Adam
from keras import regularizers
from keras import backend as K
import tensorflow as tf
import numpy as np
from keras.layers import Layer, InputSpec
from keras import initializers, regularizers, constraints
class InstanceNormalization(Layer):
def __init__(self, axes=(1, 2), trainable=True, **kwargs):
super(InstanceNormalization, self).__init__(**kwargs)
self.axes = axes
self.trainable = trainable
def build(self, input_shape):
self.beta = self.add_weight(name='beta',shape=(input_shape[-1],),
initializer='zeros',trainable=self.trainable)
self.gamma = self.add_weight(name='gamma',shape=(input_shape[-1],),
initializer='ones',trainable=self.trainable)
def call(self, inputs):
mean, variance = tf.nn.moments(inputs, self.axes, keep_dims=True)
return tf.nn.batch_normalization(inputs, mean, variance, self.beta, self.gamma, 1e-6)
def bernoulli_sampling (prob):
""" Sampling Bernoulli distribution by given probability.
Args:
- prob: P(Y = 1) in Bernoulli distribution.
Returns:
- samples: samples from Bernoulli distribution
"""
n, x_len, y_len, d = prob.shape
samples = np.random.binomial(1, prob, (n, x_len, y_len, d))
return samples
class INVASE():
"""INVASE class.
Attributes:
- x_train: training features
- y_train: training labels
- model_type: invase or invase_minus
- model_parameters:
- actor_h_dim: hidden state dimensions for actor
- critic_h_dim: hidden state dimensions for critic
- n_layer: the number of layers
- batch_size: the number of samples in mini batch
- iteration: the number of iterations
- activation: activation function of models
- learning_rate: learning rate of model training
- lamda: hyper-parameter of INVASE
"""
def __init__(self, x_train, y_train, model_type, model_parameters):
self.lamda = model_parameters['lamda']
self.actor_h_dim = model_parameters['actor_h_dim']
self.critic_h_dim = model_parameters['critic_h_dim']
self.n_layer = model_parameters['n_layer']
self.batch_size = model_parameters['batch_size']
self.iteration = model_parameters['iteration']
self.activation = model_parameters['activation']
self.learning_rate = model_parameters['learning_rate']
#Modified Code
self.x_len = x_train.shape[1]
self.y_len = x_train.shape[2]
self.dim = x_train.shape[3]
self.label_dim = y_train.shape[1]
self.model_type = model_type
optimizer = Adam(self.learning_rate)
# Build and compile critic
self.critic = self.build_critic()
self.critic.compile(loss='categorical_crossentropy',
optimizer=optimizer, metrics=['acc'])
# Build and compile the actor
self.actor = self.build_actor()
self.actor.compile(loss=self.actor_loss, optimizer=optimizer)
if self.model_type == 'invase':
# Build and compile the baseline
self.baseline = self.build_baseline()
self.baseline.compile(loss='categorical_crossentropy', optimizer=optimizer, metrics=['acc'])
def actor_loss(self, y_true, y_pred):
"""Custom loss for the actor.
Args:
- y_true:
- actor_out: actor output after sampling
- critic_out: critic output
- baseline_out: baseline output (only for invase)
- y_pred: output of the actor network
Returns:
- loss: actor loss
"""
y_pred = K.reshape(y_pred, (K.shape(y_pred)[0], self.x_len*self.y_len*1))
y_true = y_true[:, 0, 0, :]
# Actor output
actor_out = y_true[:, :self.x_len*self.y_len*1]
# Critic output
critic_out = y_true[:, self.x_len*self.y_len*1:(self.x_len*self.y_len*1+self.label_dim)]
if self.model_type == 'invase':
# Baseline output
baseline_out = \
y_true[:, (self.x_len*self.y_len*1+self.label_dim):(self.x_len*self.y_len*1+2*self.label_dim)]
# Ground truth label
y_out = y_true[:, (self.x_len*self.y_len*1+2*self.label_dim):]
elif self.model_type == 'invase_minus':
# Ground truth label
y_out = y_true[:, (self.x_len*self.y_len*1+self.label_dim):]
# Critic loss
critic_loss = -tf.reduce_sum(y_out * tf.log(critic_out + 1e-8), axis = 1)
if self.model_type == 'invase':
# Baseline loss
baseline_loss = -tf.reduce_sum(y_out * tf.log(baseline_out + 1e-8),
axis = 1)
# Reward
Reward = -(critic_loss - baseline_loss)
elif self.model_type == 'invase_minus':
Reward = -critic_loss
# Policy gradient loss computation.
custom_actor_loss = \
Reward * tf.reduce_sum(actor_out * K.log(y_pred + 1e-8) + \
(1-actor_out) * K.log(1-y_pred + 1e-8), axis = 1) - \
self.lamda * tf.reduce_mean(y_pred, axis = 1)
# custom actor loss
custom_actor_loss = tf.reduce_mean(-custom_actor_loss)
return custom_actor_loss
def build_actor(self):
"""Build actor.
Use feature as the input and output selection probability
"""
actor_model = Sequential()
actor_model.add(Conv2D(self.actor_h_dim, (1, 3), padding='same', activation='linear'))
actor_model.add(InstanceNormalization())
actor_model.add(Activation(self.activation))
for _ in range(self.n_layer - 2):
actor_model.add(Conv2D(self.actor_h_dim, (1, 3), padding='same', activation='linear'))
actor_model.add(InstanceNormalization())
actor_model.add(Activation(self.activation))
actor_model.add(Conv2D(1, (1, 1), padding='same', activation='sigmoid'))
feature = Input(shape=(self.x_len, self.y_len, self.dim), dtype='float32')
selection_probability = actor_model(feature)
return Model(feature, selection_probability)
def build_critic(self):
"""Build critic.
Use selected feature as the input and predict labels
"""
critic_model = Sequential()
critic_model.add(Conv2D(self.critic_h_dim, (1, 3), padding='same', activation='linear'))
critic_model.add(InstanceNormalization())
critic_model.add(Activation(self.activation))
for _ in range(self.n_layer - 2):
critic_model.add(Conv2D(self.critic_h_dim, (1, 3), padding='same', activation='linear'))
critic_model.add(InstanceNormalization())
critic_model.add(Activation(self.activation))
critic_model.add(Flatten())
critic_model.add(Dense(self.critic_h_dim, activation=self.activation))
#critic_model.add(Dropout(0.2))
critic_model.add(Dense(self.label_dim, activation ='softmax'))
## Inputs
# Features
feature = Input(shape=(self.x_len, self.y_len, self.dim), dtype='float32')
# Binary selection
selection = Input(shape=(self.x_len, self.y_len, 1), dtype='float32')
# Element-wise multiplication
critic_model_input = Multiply()([feature, selection])
y_hat = critic_model(critic_model_input)
return Model([feature, selection], y_hat)
def build_baseline(self):
"""Build baseline.
Use the feature as the input and predict labels
"""
baseline_model = Sequential()
baseline_model.add(Conv2D(self.critic_h_dim, (1, 3), padding='same', activation='linear'))
baseline_model.add(InstanceNormalization())
baseline_model.add(Activation(self.activation))
for _ in range(self.n_layer - 2):
baseline_model.add(Conv2D(self.critic_h_dim, (1, 3), padding='same', activation='linear'))
baseline_model.add(InstanceNormalization())
baseline_model.add(Activation(self.activation))
baseline_model.add(Flatten())
baseline_model.add(Dense(self.critic_h_dim, activation=self.activation))
#baseline_model.add(Dropout(0.2))
baseline_model.add(Dense(self.label_dim, activation ='softmax'))
# Input
feature = Input(shape=(self.x_len, self.y_len, self.dim), dtype='float32')
# Output
y_hat = baseline_model(feature)
return Model(feature, y_hat)
def train(self, x_train, y_train):
"""Train INVASE.
Args:
- x_train: training features
- y_train: training labels
"""
for iter_idx in range(self.iteration):
## Train critic
# Select a random batch of samples
idx = np.random.randint(0, x_train.shape[0], self.batch_size)
x_batch = x_train[idx,:]
y_batch = y_train[idx,:]
# Generate a batch of selection probability
selection_probability = self.actor.predict(x_batch)
# Sampling the features based on the selection_probability
selection = bernoulli_sampling(selection_probability)
# Critic loss
critic_loss = self.critic.train_on_batch([x_batch, selection], y_batch)
# Critic output
critic_out = self.critic.predict([x_batch, selection])
# Baseline output
if self.model_type == 'invase':
# Baseline loss
baseline_loss = self.baseline.train_on_batch(x_batch, y_batch)
# Baseline output
baseline_out = self.baseline.predict(x_batch)
## Train actor
# Use multiple things as the y_true:
# - selection, critic_out, baseline_out, and ground truth (y_batch)
if self.model_type == 'invase':
y_batch_final = np.concatenate((np.reshape(selection, (y_batch.shape[0], -1)),
np.asarray(critic_out),
np.asarray(baseline_out),
y_batch), axis = 1)
elif self.model_type == 'invase_minus':
y_batch_final = np.concatenate((np.reshape(selection, (y_batch.shape[0], -1)),
np.asarray(critic_out),
y_batch), axis = 1)
y_batch_final = y_batch_final[:, None, None, :]
# Train the actor
actor_loss = self.actor.train_on_batch(x_batch, y_batch_final)
if self.model_type == 'invase':
# Print the progress
dialog = 'Iterations: ' + str(iter_idx) + \
', critic accuracy: ' + str(critic_loss[1]) + \
', baseline accuracy: ' + str(baseline_loss[1]) + \
', actor loss: ' + str(np.round(actor_loss,4))
elif self.model_type == 'invase_minus':
# Print the progress
dialog = 'Iterations: ' + str(iter_idx) + \
', critic accuracy: ' + str(critic_loss[1]) + \
', actor loss: ' + str(np.round(actor_loss,4))
if iter_idx % 100 == 0:
print(dialog)
def importance_score(self, x):
"""Return featuer importance score.
Args:
- x: feature
Returns:
- feature_importance: instance-wise feature importance for x
"""
feature_importance = self.actor.predict(x)
return np.asarray(feature_importance)
def predict(self, x):
"""Predict outcomes.
Args:
- x: feature
Returns:
- y_hat: predictions
"""
# Generate a batch of selection probability
selection_probability = self.actor.predict(x)
# Sampling the features based on the selection_probability
selection = bernoulli_sampling(selection_probability)
# Prediction
y_hat = self.critic.predict([x, selection])
return np.asarray(y_hat)
#Concatenate input binder pairs
x_train = np.concatenate([x_1_train, x_2_train], axis=2)
x_test = np.concatenate([x_1_test, x_2_test], axis=2)
#Execute INVASE
mask_penalty = 0.05
hidden_dims = 32
n_layers = 4
epochs = 50
batch_size = 128
model_parameters = {
'lamda': mask_penalty,
'actor_h_dim': hidden_dims,
'critic_h_dim': hidden_dims,
'n_layer': n_layers,
'batch_size': batch_size,
'iteration': int(x_train.shape[0] * epochs / batch_size),
'activation': 'relu',
'learning_rate': 0.0001
}
invase_model = INVASE(x_train, pred_train, 'invase', model_parameters)
invase_model.train(x_train, pred_train)
importance_scores_test = invase_model.importance_score(x_test)
importance_scores_1_test, importance_scores_2_test = importance_scores_test[:, :, :seq_length, :], importance_scores_test[:, :, seq_length:, :]
#Evaluate INVASE model on train and test data
invase_pred_train = invase_model.predict(x_train)
invase_pred_test = invase_model.predict(x_test)
print("Training Accuracy = " + str(np.sum(np.argmax(invase_pred_train, axis=1) == np.argmax(pred_train, axis=1)) / float(pred_train.shape[0])))
print("Test Accuracy = " + str(np.sum(np.argmax(invase_pred_test, axis=1) == np.argmax(pred_test, axis=1)) / float(pred_test.shape[0])))
#Gradient saliency/backprop visualization
import matplotlib.collections as collections
import operator
import matplotlib.pyplot as plt
import matplotlib.cm as cm
import matplotlib.colors as colors
import matplotlib as mpl
from matplotlib.text import TextPath
from matplotlib.patches import PathPatch, Rectangle
from matplotlib.font_manager import FontProperties
from matplotlib import gridspec
from matplotlib.ticker import FormatStrFormatter
def plot_protein_logo(residue_map, pwm, sequence_template=None, figsize=(12, 3), logo_height=1.0, plot_start=0, plot_end=164) :
inv_residue_map = {
i : sp for sp, i in residue_map.items()
}
#Slice according to seq trim index
pwm = pwm[plot_start: plot_end, :]
sequence_template = sequence_template[plot_start: plot_end]
entropy = np.zeros(pwm.shape)
entropy[pwm > 0] = pwm[pwm > 0] * -np.log2(np.clip(pwm[pwm > 0], 1e-6, 1. - 1e-6))
entropy = np.sum(entropy, axis=1)
conservation = np.log2(len(residue_map)) - entropy#2 - entropy
fig = plt.figure(figsize=figsize)
ax = plt.gca()
height_base = (1.0 - logo_height) / 2.
for j in range(0, pwm.shape[0]) :
sort_index = np.argsort(pwm[j, :])
for ii in range(0, len(residue_map)) :
i = sort_index[ii]
if pwm[j, i] > 0 :
nt_prob = pwm[j, i] * conservation[j]
nt = inv_residue_map[i]
color = None
if sequence_template[j] != '$' :
color = 'black'
if ii == 0 :
letterAt_protein(nt, j + 0.5, height_base, nt_prob * logo_height, ax, color=color)
else :
prev_prob = np.sum(pwm[j, sort_index[:ii]] * conservation[j]) * logo_height
letterAt_protein(nt, j + 0.5, height_base + prev_prob, nt_prob * logo_height, ax, color=color)
plt.xlim((0, plot_end - plot_start))
plt.ylim((0, np.log2(len(residue_map))))
plt.xticks([], [])
plt.yticks([], [])
plt.axis('off')
plt.axhline(y=0.01 + height_base, color='black', linestyle='-', linewidth=2)
for axis in fig.axes :
axis.get_xaxis().set_visible(False)
axis.get_yaxis().set_visible(False)
plt.tight_layout()
plt.show()
def plot_importance_scores(importance_scores, ref_seq, figsize=(12, 2), score_clip=None, sequence_template='', plot_start=0, plot_end=96, save_figs=False, fig_name=None) :
end_pos = ref_seq.find("#")
fig = plt.figure(figsize=figsize)
ax = plt.gca()
if score_clip is not None :
importance_scores = np.clip(np.copy(importance_scores), -score_clip, score_clip)
max_score = np.max(np.sum(importance_scores[:, :], axis=0)) + 0.01
for i in range(0, len(ref_seq)) :
mutability_score = np.sum(importance_scores[:, i])
letterAt_protein(ref_seq[i], i + 0.5, 0, mutability_score, ax, color=None)
plt.sca(ax)
plt.xlim((0, len(ref_seq)))
plt.ylim((0, max_score))
plt.axis('off')
plt.yticks([0.0, max_score], [0.0, max_score], fontsize=16)
for axis in fig.axes :
axis.get_xaxis().set_visible(False)
axis.get_yaxis().set_visible(False)
plt.tight_layout()
if save_figs :
plt.savefig(fig_name + ".png", transparent=True, dpi=300)
plt.savefig(fig_name + ".eps")
plt.show()
np.sum(importance_scores_1_test[0, 0, :, 0] + importance_scores_2_test[0, 0, :, 0])
np.max(importance_scores_1_test[0, 0, :, 0])
#Visualize importance for binder 1
for plot_i in range(0, 5) :
print("Test sequence " + str(plot_i) + ":")
sequence_template = sequence_templates[l_1_test[plot_i, 0]]
plot_protein_logo(residue_map, x_1_test[plot_i, 0, :, :], sequence_template=sequence_template, figsize=(12, 1), plot_start=0, plot_end=81)
plot_importance_scores(importance_scores_1_test[plot_i, 0, :, :].T, encoder.decode(x_1_test[plot_i, 0, :, :]), figsize=(12, 1), score_clip=None, sequence_template=sequence_template, plot_start=0, plot_end=81)
#Visualize importance for binder 2
for plot_i in range(0, 5) :
print("Test sequence " + str(plot_i) + ":")
sequence_template = sequence_templates[l_2_test[plot_i, 0]]
plot_protein_logo(residue_map, x_2_test[plot_i, 0, :, :], sequence_template=sequence_template, figsize=(12, 1), plot_start=0, plot_end=81)
plot_importance_scores(importance_scores_2_test[plot_i, 0, :, :].T, encoder.decode(x_2_test[plot_i, 0, :, :]), figsize=(12, 1), score_clip=None, sequence_template=sequence_template, plot_start=0, plot_end=81)
#Save predicted importance scores
model_name = "invase_" + dataset_name + "_conv" + "_zeropad_no_drop_penalty_005"
np.save(model_name + "_importance_scores_1_test", importance_scores_1_test)
np.save(model_name + "_importance_scores_2_test", importance_scores_2_test)
#Binder DHD_154
seq_1 = "TAEELLEVHKKSDRVTKEHLRVSEEILKVVEVLTRGEVSSEVLKRVLRKLEELTDKLRRVTEEQRRVVEKLN"[:81]
seq_2 = "DLEDLLRRLRRLVDEQRRLVEELERVSRRLEKAVRDNEDERELARLSREHSDIQDKHDKLAREILEVLKRLLERTE"[:81]
print("Seq 1 = " + seq_1)
print("Seq 2 = " + seq_2)
encoder = IdentityEncoder(81, residue_map)
test_onehot_1 = np.tile(np.expand_dims(np.expand_dims(encoder(seq_1), axis=0), axis=0), (batch_size, 1, 1, 1))
test_onehot_2 = np.tile(np.expand_dims(np.expand_dims(encoder(seq_2), axis=0), axis=0), (batch_size, 1, 1, 1))
test_len_1 = np.tile(np.array([[len(seq_1)]]), (batch_size, 1))
test_len_2 = np.tile(np.array([[len(seq_2)]]), (batch_size, 1))
pred_interacts = predictor.predict(x=[test_onehot_1[:, 0, ...], test_onehot_2[:, 0, ...]])[0, 0]
print("Predicted interaction prob = " + str(round(pred_interacts, 4)))
x_1_test = test_onehot_1[:1]
x_2_test = test_onehot_2[:1]
#Concatenate input binder pairs
x_test = np.concatenate([x_1_test, x_2_test], axis=2)
#Execute INVASE
mask_penalty = 0.05
hidden_dims = 32
n_layers = 4
epochs = 50
batch_size = 128
model_parameters = {
'lamda': mask_penalty,
'actor_h_dim': hidden_dims,
'critic_h_dim': hidden_dims,
'n_layer': n_layers,
'batch_size': batch_size,
'iteration': int(x_train.shape[0] * epochs / batch_size),
'activation': 'relu',
'learning_rate': 0.0001
}
invase_model = INVASE(x_train, pred_train, 'invase', model_parameters)
invase_model.train(x_train, pred_train)
importance_scores_test = invase_model.importance_score(x_test)
importance_scores_1_test, importance_scores_2_test = importance_scores_test[:, :, :seq_length, :], importance_scores_test[:, :, seq_length:, :]
save_figs = True
model_name = "invase_" + dataset_name + "_conv" + "_zeropad_no_drop_penalty_005"
pair_name = "DHD_154"
#Visualize importance for binder 1
for plot_i in range(0, 1) :
print("Test sequence " + str(plot_i) + ":")
sequence_template = sequence_templates[l_1_test[plot_i, 0]]
plot_protein_logo(residue_map, x_1_test[plot_i, 0, :, :], sequence_template=sequence_template, figsize=(12, 1), plot_start=0, plot_end=81)
plot_importance_scores(importance_scores_1_test[plot_i, 0, :, :].T, encoder.decode(x_1_test[plot_i, 0, :, :]), figsize=(12, 1), score_clip=None, sequence_template=sequence_template, plot_start=0, plot_end=81, save_figs=save_figs, fig_name=model_name + "_scores_" + pair_name + "_binder_1")
#Visualize importance for binder 2
for plot_i in range(0, 1) :
print("Test sequence " + str(plot_i) + ":")
sequence_template = sequence_templates[l_2_test[plot_i, 0]]
plot_protein_logo(residue_map, x_2_test[plot_i, 0, :, :], sequence_template=sequence_template, figsize=(12, 1), plot_start=0, plot_end=81)
plot_importance_scores(importance_scores_2_test[plot_i, 0, :, :].T, encoder.decode(x_2_test[plot_i, 0, :, :]), figsize=(12, 1), score_clip=None, sequence_template=sequence_template, plot_start=0, plot_end=81, save_figs=save_figs, fig_name=model_name + "_scores_" + pair_name + "_binder_2")
| 0.77552 | 0.424412 |
```
# -*- coding: utf-8 -*-
# This work is part of the Core Imaging Library (CIL) developed by CCPi
# (Collaborative Computational Project in Tomographic Imaging), with
# substantial contributions by UKRI-STFC and University of Manchester.
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
# http://www.apache.org/licenses/LICENSE-2.0
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# Copyright 2021 UKRI-STFC, Technical University of Denmark
# Authored by: Jakob S. Jørgensen (DTU)
```
# Additional open-ended exercises, datasets and resources
To further experiment with tomographic reconstruction and data processing using CIL we provide a few additional data sets, suggestions for experiments and links to CIL resources including publications, documentation and demos.
## Data sets
The following data sets have been downloaded and are available on the STFC Cloud shared drive (read only).
Most of the data sets provided are 3D. For faster execution times in the experiments we suggest to extract a single slice and work only with such a 2D data set. From the loaded 3D `AcquisitionData` object, `data3D`, a 2D slice `AcquisitionData` can be extracted using the `get_slice` method, for example
`data2D = data3D.get_slice(vertical=27)`
will extract vertical slice number 27 as a new 2D `AcquisitionData` object, `data2D`.
For Cone3D cone-beam datasets only the central slice can be extracted as a Cone2D (fan-beam) dataset, this is done using
`data2D = data3D.get_slice(vertical='centre')`
If an even number of slices are available in `data3D`, then the above command will produce a slice that is interpolated from the two central slices.
Nikon datasets can be loaded using the CIL `NikonDataReader` in the `cil.io` module.
Zeiss datasets can be loaded using the CIL `TXRMDataReader` in the `cil.io` module.
**Crystals in clay:**
Cone-beam Nikon data set of crystals in clay from, described at
https://zenodo.org/record/4912635
**SophiaBeads:**
Cone-beam Nikon data set of glass beads. Data sets with different trade-offs between numbers of projections and exposure time are available, the 256-projection data set is available on the STFC Cloud. Description at
https://zenodo.org/record/16474
**LEGO laminography dataset:**
A rotary laminography tomography dataset of a sample of LEGO brick acquired on a Nikon micro-CT instrument, described at
https://zenodo.org/record/2540509
**HDTomo datasets:**
Six Zeiss cone-beam data sets including the walnut dataset, as well as Kinder Surprise chocolate eggs, a USB stick and more, with descriptions at
https://zenodo.org/record/4822516
**Sandstone:**
The parallel-beam sandstone synchrotron data set, both a small extracted data set and the full projections are available from
https://zenodo.org/record/4912435
#### Additional tomography data resources NOT on the STFC Cloud
FIPS data: A collection of X-ray CT data sets is available from the Finnish Inverse Problems Society:
https://www.fips.fi/dataset.php
## Suggestions for experiments
**Try reconstructing different real data sets:**
Choose one or more of the datasets listed above. Load the dataset, determine and carry out any preprocessing required, and compute an FBP or FDK reconstruction. Try also to reconstruct using your favourite iterative/regularized reconstruction method. Apply CIL processors to preprocess data as necessary, possibly after having added noise, artifacts or reduced the datasets yourself.
*Relevant notebooks:* 01_optimisation_gd_fista.ipynb, Week1/01_intro_walnut_conebeam.ipynb, Week1/02_intro_sandstone_parallel_roi.ipynb
**Synthetic data:**
Try out loading and generating simulated data from phantoms provided by CIL (cil.utilities.dataexample) or the TomoPhantom CIL plugin (cil.plugins.TomoPhantom). Choose either full data or incomplete data of your choice, add noise, and reconstruct first using FBP and then using regularised reconstruction methods.
*Relevant notebooks:* 01_optimisation_gd_fista.ipynb, 02_tikhonov_block_framework.ipynb, Week1/03_preprocessing.ipynb
**Reduced data reconstruction:**
Use CIL processors (e.g. Slicer or Binner or Masker/MaskGenerator) to remove parts of or downsample data sets, for example to obtain a reduced number of projections, a limited angle problem, truncated projections (region of interest data), exterior problem, etc. Compare for example different regularised reconstruction methods at increasingly few projections.
*Relevant notebooks:* 01_optimisation_gd_fista.ipynb, 03_preprocessing.ipynb
**Denoising, deblurring, inpainting:**
CIL is developed for tomography but can handle general (at present, linear) inverse problems. Using IdentityOperator, BlurringOperator and MaskOperator provided by CIL it is possible to set up denoising, deblurring and inpainting problems. Choose one or more of these problems and a test image, simulate some data, choose a regularised recontruction problem and compute reconstructions.
*Relevant notebooks:* Week3/01_Color_Processing.ipynb
**Effect of regularisation parameter:**
Run Tikhonov and TV-regularized reconstruction with a wide range of different values for the regularisation parameter to see the effect on the reconstruction, ranging from under- to over-regularised.
*Relevant notebooks:* 01_optimisation_gd_fista.ipynb
**Anisotropic regularisation:**
Normally we use the same regularisation in all spatial dimensions. Sometimes we may have an image with different behaviour in different dimensions, for example smooth in the y-dimension but edges in the x-direction. Using CIL BlockOperators it is possible to use different regularisers in different dimensions, for example a FiniteDifferenceOperator in y and an IndentityOperator (or no regularisation) in x, or FiniteDifferenceOperators in both x and y but having different regularisation parameters. Implement such anisotropic regularisation in a Tikhonov formulation and demonstrate the effect on a synthetic data reconstruction problem of your choice.
*Relevant notebooks:* 01_optimisation_gd_fista.ipynb
**Verify algorithms against each other:**
Compare FISTA and PDHG for solving the same problem, such as TV-regularised or L1-norm regularised least squares. As the same optimisation problem is specified, the different algorithms should produce the same solution, when converged. Try to confirm whether they produce the same solution. You may need to run a large number of iterations. You can also compare with the smoothed TV regulariser, in which case the optimisation problem is smooth and so can be solved using the gradient descent algorithm.
*Relevant notebooks:* 01_optimisation_gd_fista.ipynb
**Compare convergence speed of PDHG using different step sizes:**
The sigma and tau step sizes in PDHG can have a dramatic influence on the convergence speed. Experiment with different choices (that must satisfy the constraint specified) and compare the convergence speed. Try on different test problems, including synthetic and real data, and see if there is a trend for the best choice of step sizes across data sets, or it is data set dependent.
*Relevant notebooks:* 03_PDHG.ipynb
**SPDHG subsets and probabilities:**
In SPDHG we need to specify the number of subsets to use and the probabilities with which to choose each subset and the regulariser. Experiment with different numbers of subsets and probabilities and compare the effect on reconstruction quality and speed.
*Relevant notebooks:* 04_SPDHG.ipynb
**Other algorithms, operators and functions:**
Explore other tools offered by CIL such as the LADMM algorithm, TGV regularisation and weighted least squares and Kullback-Leibler divergence data fidelities, set up test problems and try out new algorithms and optimisation problems and compare the results with problems previously solved.
*Relevant notebooks:* Week3/01_Color_Processing.ipynb
## Resources
**CIL documentation**
https://tomographicimaging.github.io/CIL/
**Main CIL GitHub repository**
https://github.com/TomographicImaging/CIL
**CIL demos and training material repository**
https://github.com/TomographicImaging/CIL-Demos
**Core Imaging Library -- Part I: a versatile Python framework for tomographic imaging**
by Jakob S. Jørgensen, Evelina Ametova, Genoveva Burca, Gemma Fardell, Evangelos Papoutsellis, Edoardo Pasca, Kris Thielemans, Martin Turner, Ryan Warr, William R. B. Lionheart, Philip J. Withers
https://arxiv.org/abs/2102.04560
**Core Imaging Library -- Part II: Multichannel reconstruction for dynamic and spectral tomography**
by Evangelos Papoutsellis, Evelina Ametova, Claire Delplancke, Gemma Fardell, Jakob S. Jørgensen, Edoardo Pasca, Martin Turner, Ryan Warr, William R. B. Lionheart, and Philip J. Withers
https://arxiv.org/abs/2102.06126
**Enhanced hyperspectral tomography for bioimaging by spatiospectral reconstruction**
by Ryan Warr, Evelina Ametova, Robert J. Cernik, Gemma Fardell, Stephan Handschuh, Jakob S. Jørgensen, Evangelos Papoutsellis, Edoardo Pasca, and Philip J. Withers
https://arxiv.org/abs/2103.04796
**Crystalline phase discriminating neutron tomography using advanced reconstruction methods**
by Evelina Ametova, Genoveva Burca, Suren Chilingaryan, Gemma Fardell, Jakob S. Jørgensen, Evangelos Papoutsellis, Edoardo Pasca, Ryan Warr, Martin Turner, William R B Lionheart, and Philip J Withers
https://doi.org/10.1088/1361-6463/ac02f9
|
github_jupyter
|
# -*- coding: utf-8 -*-
# This work is part of the Core Imaging Library (CIL) developed by CCPi
# (Collaborative Computational Project in Tomographic Imaging), with
# substantial contributions by UKRI-STFC and University of Manchester.
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
# http://www.apache.org/licenses/LICENSE-2.0
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# Copyright 2021 UKRI-STFC, Technical University of Denmark
# Authored by: Jakob S. Jørgensen (DTU)
| 0.274643 | 0.858955 |
#### Copyright 2020 DeepMind Technologies Limited. All Rights Reserved.
#### Licensed under the Apache License, Version 2.0 (the "License");
#### Full license text
```
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# A (very) basic GAN for MNIST in JAX/Haiku
Based on a TensorFlow tutorial written by Mihaela Rosca.
Original GAN paper: https://papers.nips.cc/paper/5423-generative-adversarial-nets.pdf
## Imports
```
# Uncomment the line below if running on colab.research.google.com.
# !pip install dm-haiku
import functools
from typing import Any, NamedTuple
import haiku as hk
import jax
import optax
import jax.numpy as jnp
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
import tensorflow as tf
import tensorflow_datasets as tfds
```
## Define the dataset
```
# Download the data once.
mnist = tfds.load("mnist")
def make_dataset(batch_size, seed=1):
def _preprocess(sample):
# Convert to floats in [0, 1].
image = tf.image.convert_image_dtype(sample["image"], tf.float32)
# Scale the data to [-1, 1] to stabilize training.
return 2.0 * image - 1.0
ds = mnist["train"]
ds = ds.map(map_func=_preprocess,
num_parallel_calls=tf.data.experimental.AUTOTUNE)
ds = ds.cache()
ds = ds.shuffle(10 * batch_size, seed=seed).repeat().batch(batch_size)
return iter(tfds.as_numpy(ds))
```
## Define the model
```
class Generator(hk.Module):
"""Generator network."""
def __init__(self, output_channels=(32, 1), name=None):
super().__init__(name=name)
self.output_channels = output_channels
def __call__(self, x):
"""Maps noise latents to images."""
x = hk.Linear(7 * 7 * 64)(x)
x = jnp.reshape(x, x.shape[:1] + (7, 7, 64))
for output_channels in self.output_channels:
x = jax.nn.relu(x)
x = hk.Conv2DTranspose(output_channels=output_channels,
kernel_shape=[5, 5],
stride=2,
padding="SAME")(x)
# We use a tanh to ensure that the generated samples are in the same
# range as the data.
return jnp.tanh(x)
class Discriminator(hk.Module):
"""Discriminator network."""
def __init__(self,
output_channels=(8, 16, 32, 64, 128),
strides=(2, 1, 2, 1, 2),
name=None):
super().__init__(name=name)
self.output_channels = output_channels
self.strides = strides
def __call__(self, x):
"""Classifies images as real or fake."""
for output_channels, stride in zip(self.output_channels, self.strides):
x = hk.Conv2D(output_channels=output_channels,
kernel_shape=[5, 5],
stride=stride,
padding="SAME")(x)
x = jax.nn.leaky_relu(x, negative_slope=0.2)
x = hk.Flatten()(x)
# We have two classes: 0 = input is fake, 1 = input is real.
logits = hk.Linear(2)(x)
return logits
def tree_shape(xs):
return jax.tree_map(lambda x: x.shape, xs)
def sparse_softmax_cross_entropy(logits, labels):
one_hot_labels = jax.nn.one_hot(labels, logits.shape[-1])
return -jnp.sum(one_hot_labels * jax.nn.log_softmax(logits), axis=-1)
class GANTuple(NamedTuple):
gen: Any
disc: Any
class GANState(NamedTuple):
params: GANTuple
opt_state: GANTuple
class GAN:
"""A basic GAN."""
def __init__(self, num_latents):
self.num_latents = num_latents
# Define the Haiku network transforms.
# We don't use BatchNorm so we don't use `with_state`.
self.gen_transform = hk.without_apply_rng(
hk.transform(lambda *args: Generator()(*args)))
self.disc_transform = hk.without_apply_rng(
hk.transform(lambda *args: Discriminator()(*args)))
# Build the optimizers.
self.optimizers = GANTuple(gen=optax.adam(1e-4, b1=0.5, b2=0.9),
disc=optax.adam(1e-4, b1=0.5, b2=0.9))
@functools.partial(jax.jit, static_argnums=0)
def initial_state(self, rng, batch):
"""Returns the initial parameters and optimize states."""
# Generate dummy latents for the generator.
dummy_latents = jnp.zeros((batch.shape[0], self.num_latents))
# Get initial network parameters.
rng_gen, rng_disc = jax.random.split(rng)
params = GANTuple(gen=self.gen_transform.init(rng_gen, dummy_latents),
disc=self.disc_transform.init(rng_disc, batch))
print("Generator: \n\n{}\n".format(tree_shape(params.gen)))
print("Discriminator: \n\n{}\n".format(tree_shape(params.disc)))
# Initialize the optimizers.
opt_state = GANTuple(gen=self.optimizers.gen.init(params.gen),
disc=self.optimizers.disc.init(params.disc))
return GANState(params=params, opt_state=opt_state)
def sample(self, rng, gen_params, num_samples):
"""Generates images from noise latents."""
latents = jax.random.normal(rng, shape=(num_samples, self.num_latents))
return self.gen_transform.apply(gen_params, latents)
def gen_loss(self, gen_params, rng, disc_params, batch):
"""Generator loss."""
# Sample from the generator.
fake_batch = self.sample(rng, gen_params, num_samples=batch.shape[0])
# Evaluate using the discriminator. Recall class 1 is real.
fake_logits = self.disc_transform.apply(disc_params, fake_batch)
fake_probs = jax.nn.softmax(fake_logits)[:, 1]
loss = -jnp.log(fake_probs)
return jnp.mean(loss)
def disc_loss(self, disc_params, rng, gen_params, batch):
"""Discriminator loss."""
# Sample from the generator.
fake_batch = self.sample(rng, gen_params, num_samples=batch.shape[0])
# For efficiency we process both the real and fake data in one pass.
real_and_fake_batch = jnp.concatenate([batch, fake_batch], axis=0)
real_and_fake_logits = self.disc_transform.apply(disc_params,
real_and_fake_batch)
real_logits, fake_logits = jnp.split(real_and_fake_logits, 2, axis=0)
# Class 1 is real.
real_labels = jnp.ones((batch.shape[0],), dtype=jnp.int32)
real_loss = sparse_softmax_cross_entropy(real_logits, real_labels)
# Class 0 is fake.
fake_labels = jnp.zeros((batch.shape[0],), dtype=jnp.int32)
fake_loss = sparse_softmax_cross_entropy(fake_logits, fake_labels)
return jnp.mean(real_loss + fake_loss)
@functools.partial(jax.jit, static_argnums=0)
def update(self, rng, gan_state, batch):
"""Performs a parameter update."""
rng, rng_gen, rng_disc = jax.random.split(rng, 3)
# Update the discriminator.
disc_loss, disc_grads = jax.value_and_grad(self.disc_loss)(
gan_state.params.disc,
rng_disc,
gan_state.params.gen,
batch)
disc_update, disc_opt_state = self.optimizers.disc.update(
disc_grads, gan_state.opt_state.disc)
disc_params = optax.apply_updates(gan_state.params.disc, disc_update)
# Update the generator.
gen_loss, gen_grads = jax.value_and_grad(self.gen_loss)(
gan_state.params.gen,
rng_gen,
gan_state.params.disc,
batch)
gen_update, gen_opt_state = self.optimizers.gen.update(
gen_grads, gan_state.opt_state.gen)
gen_params = optax.apply_updates(gan_state.params.gen, gen_update)
params = GANTuple(gen=gen_params, disc=disc_params)
opt_state = GANTuple(gen=gen_opt_state, disc=disc_opt_state)
gan_state = GANState(params=params, opt_state=opt_state)
log = {
"gen_loss": gen_loss,
"disc_loss": disc_loss,
}
return rng, gan_state, log
```
## Train the model
```
#@title {vertical-output: true}
num_steps = 20001
log_every = num_steps // 100
# Let's see what hardware we're working with. The training takes a few
# minutes on a GPU, a bit longer on CPU.
print(f"Number of devices: {jax.device_count()}")
print("Device:", jax.devices()[0].device_kind)
print("")
# Make the dataset.
dataset = make_dataset(batch_size=64)
# The model.
gan = GAN(num_latents=20)
# Top-level RNG.
rng = jax.random.PRNGKey(1729)
# Initialize the network and optimizer.
rng, rng1 = jax.random.split(rng)
gan_state = gan.initial_state(rng1, next(dataset))
steps = []
gen_losses = []
disc_losses = []
for step in range(num_steps):
rng, gan_state, log = gan.update(rng, gan_state, next(dataset))
# Log the losses.
if step % log_every == 0:
# It's important to call `device_get` here so we don't take up device
# memory by saving the losses.
log = jax.device_get(log)
gen_loss = log["gen_loss"]
disc_loss = log["disc_loss"]
print(f"Step {step}: "
f"gen_loss = {gen_loss:.3f}, disc_loss = {disc_loss:.3f}")
steps.append(step)
gen_losses.append(gen_loss)
disc_losses.append(disc_loss)
```
## Visualize the losses
Unlike losses for classifiers or VAEs, GAN losses do not decrease steadily, instead going up and down depending on the training dynamics.
```
sns.set_style("whitegrid")
fig, axes = plt.subplots(1, 2, figsize=(20, 6))
# Plot the discriminator loss.
axes[0].plot(steps, disc_losses, "-")
axes[0].plot(steps, np.log(2) * np.ones_like(steps), "r--",
label="Discriminator is being fooled")
axes[0].legend(fontsize=20)
axes[0].set_title("Discriminator loss", fontsize=20)
# Plot the generator loss.
axes[1].plot(steps, gen_losses, '-')
axes[1].set_title("Generator loss", fontsize=20);
```
## Visualize samples
```
#@title {vertical-output: true}
def make_grid(samples, num_cols=8, rescale=True):
batch_size, height, width = samples.shape
assert batch_size % num_cols == 0
num_rows = batch_size // num_cols
# We want samples.shape == (height * num_rows, width * num_cols).
samples = samples.reshape(num_rows, num_cols, height, width)
samples = samples.swapaxes(1, 2)
samples = samples.reshape(height * num_rows, width * num_cols)
return samples
# Generate samples from the trained generator.
rng = jax.random.PRNGKey(12)
samples = gan.sample(rng, gan_state.params.gen, num_samples=64)
samples = jax.device_get(samples)
samples = samples.squeeze(axis=-1)
# Our model outputs values in [-1, 1] so scale it back to [0, 1].
samples = (samples + 1.0) / 2.0
plt.gray()
plt.axis("off")
samples_grid = make_grid(samples)
plt.imshow(samples_grid);
```
|
github_jupyter
|
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# Uncomment the line below if running on colab.research.google.com.
# !pip install dm-haiku
import functools
from typing import Any, NamedTuple
import haiku as hk
import jax
import optax
import jax.numpy as jnp
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
import tensorflow as tf
import tensorflow_datasets as tfds
# Download the data once.
mnist = tfds.load("mnist")
def make_dataset(batch_size, seed=1):
def _preprocess(sample):
# Convert to floats in [0, 1].
image = tf.image.convert_image_dtype(sample["image"], tf.float32)
# Scale the data to [-1, 1] to stabilize training.
return 2.0 * image - 1.0
ds = mnist["train"]
ds = ds.map(map_func=_preprocess,
num_parallel_calls=tf.data.experimental.AUTOTUNE)
ds = ds.cache()
ds = ds.shuffle(10 * batch_size, seed=seed).repeat().batch(batch_size)
return iter(tfds.as_numpy(ds))
class Generator(hk.Module):
"""Generator network."""
def __init__(self, output_channels=(32, 1), name=None):
super().__init__(name=name)
self.output_channels = output_channels
def __call__(self, x):
"""Maps noise latents to images."""
x = hk.Linear(7 * 7 * 64)(x)
x = jnp.reshape(x, x.shape[:1] + (7, 7, 64))
for output_channels in self.output_channels:
x = jax.nn.relu(x)
x = hk.Conv2DTranspose(output_channels=output_channels,
kernel_shape=[5, 5],
stride=2,
padding="SAME")(x)
# We use a tanh to ensure that the generated samples are in the same
# range as the data.
return jnp.tanh(x)
class Discriminator(hk.Module):
"""Discriminator network."""
def __init__(self,
output_channels=(8, 16, 32, 64, 128),
strides=(2, 1, 2, 1, 2),
name=None):
super().__init__(name=name)
self.output_channels = output_channels
self.strides = strides
def __call__(self, x):
"""Classifies images as real or fake."""
for output_channels, stride in zip(self.output_channels, self.strides):
x = hk.Conv2D(output_channels=output_channels,
kernel_shape=[5, 5],
stride=stride,
padding="SAME")(x)
x = jax.nn.leaky_relu(x, negative_slope=0.2)
x = hk.Flatten()(x)
# We have two classes: 0 = input is fake, 1 = input is real.
logits = hk.Linear(2)(x)
return logits
def tree_shape(xs):
return jax.tree_map(lambda x: x.shape, xs)
def sparse_softmax_cross_entropy(logits, labels):
one_hot_labels = jax.nn.one_hot(labels, logits.shape[-1])
return -jnp.sum(one_hot_labels * jax.nn.log_softmax(logits), axis=-1)
class GANTuple(NamedTuple):
gen: Any
disc: Any
class GANState(NamedTuple):
params: GANTuple
opt_state: GANTuple
class GAN:
"""A basic GAN."""
def __init__(self, num_latents):
self.num_latents = num_latents
# Define the Haiku network transforms.
# We don't use BatchNorm so we don't use `with_state`.
self.gen_transform = hk.without_apply_rng(
hk.transform(lambda *args: Generator()(*args)))
self.disc_transform = hk.without_apply_rng(
hk.transform(lambda *args: Discriminator()(*args)))
# Build the optimizers.
self.optimizers = GANTuple(gen=optax.adam(1e-4, b1=0.5, b2=0.9),
disc=optax.adam(1e-4, b1=0.5, b2=0.9))
@functools.partial(jax.jit, static_argnums=0)
def initial_state(self, rng, batch):
"""Returns the initial parameters and optimize states."""
# Generate dummy latents for the generator.
dummy_latents = jnp.zeros((batch.shape[0], self.num_latents))
# Get initial network parameters.
rng_gen, rng_disc = jax.random.split(rng)
params = GANTuple(gen=self.gen_transform.init(rng_gen, dummy_latents),
disc=self.disc_transform.init(rng_disc, batch))
print("Generator: \n\n{}\n".format(tree_shape(params.gen)))
print("Discriminator: \n\n{}\n".format(tree_shape(params.disc)))
# Initialize the optimizers.
opt_state = GANTuple(gen=self.optimizers.gen.init(params.gen),
disc=self.optimizers.disc.init(params.disc))
return GANState(params=params, opt_state=opt_state)
def sample(self, rng, gen_params, num_samples):
"""Generates images from noise latents."""
latents = jax.random.normal(rng, shape=(num_samples, self.num_latents))
return self.gen_transform.apply(gen_params, latents)
def gen_loss(self, gen_params, rng, disc_params, batch):
"""Generator loss."""
# Sample from the generator.
fake_batch = self.sample(rng, gen_params, num_samples=batch.shape[0])
# Evaluate using the discriminator. Recall class 1 is real.
fake_logits = self.disc_transform.apply(disc_params, fake_batch)
fake_probs = jax.nn.softmax(fake_logits)[:, 1]
loss = -jnp.log(fake_probs)
return jnp.mean(loss)
def disc_loss(self, disc_params, rng, gen_params, batch):
"""Discriminator loss."""
# Sample from the generator.
fake_batch = self.sample(rng, gen_params, num_samples=batch.shape[0])
# For efficiency we process both the real and fake data in one pass.
real_and_fake_batch = jnp.concatenate([batch, fake_batch], axis=0)
real_and_fake_logits = self.disc_transform.apply(disc_params,
real_and_fake_batch)
real_logits, fake_logits = jnp.split(real_and_fake_logits, 2, axis=0)
# Class 1 is real.
real_labels = jnp.ones((batch.shape[0],), dtype=jnp.int32)
real_loss = sparse_softmax_cross_entropy(real_logits, real_labels)
# Class 0 is fake.
fake_labels = jnp.zeros((batch.shape[0],), dtype=jnp.int32)
fake_loss = sparse_softmax_cross_entropy(fake_logits, fake_labels)
return jnp.mean(real_loss + fake_loss)
@functools.partial(jax.jit, static_argnums=0)
def update(self, rng, gan_state, batch):
"""Performs a parameter update."""
rng, rng_gen, rng_disc = jax.random.split(rng, 3)
# Update the discriminator.
disc_loss, disc_grads = jax.value_and_grad(self.disc_loss)(
gan_state.params.disc,
rng_disc,
gan_state.params.gen,
batch)
disc_update, disc_opt_state = self.optimizers.disc.update(
disc_grads, gan_state.opt_state.disc)
disc_params = optax.apply_updates(gan_state.params.disc, disc_update)
# Update the generator.
gen_loss, gen_grads = jax.value_and_grad(self.gen_loss)(
gan_state.params.gen,
rng_gen,
gan_state.params.disc,
batch)
gen_update, gen_opt_state = self.optimizers.gen.update(
gen_grads, gan_state.opt_state.gen)
gen_params = optax.apply_updates(gan_state.params.gen, gen_update)
params = GANTuple(gen=gen_params, disc=disc_params)
opt_state = GANTuple(gen=gen_opt_state, disc=disc_opt_state)
gan_state = GANState(params=params, opt_state=opt_state)
log = {
"gen_loss": gen_loss,
"disc_loss": disc_loss,
}
return rng, gan_state, log
#@title {vertical-output: true}
num_steps = 20001
log_every = num_steps // 100
# Let's see what hardware we're working with. The training takes a few
# minutes on a GPU, a bit longer on CPU.
print(f"Number of devices: {jax.device_count()}")
print("Device:", jax.devices()[0].device_kind)
print("")
# Make the dataset.
dataset = make_dataset(batch_size=64)
# The model.
gan = GAN(num_latents=20)
# Top-level RNG.
rng = jax.random.PRNGKey(1729)
# Initialize the network and optimizer.
rng, rng1 = jax.random.split(rng)
gan_state = gan.initial_state(rng1, next(dataset))
steps = []
gen_losses = []
disc_losses = []
for step in range(num_steps):
rng, gan_state, log = gan.update(rng, gan_state, next(dataset))
# Log the losses.
if step % log_every == 0:
# It's important to call `device_get` here so we don't take up device
# memory by saving the losses.
log = jax.device_get(log)
gen_loss = log["gen_loss"]
disc_loss = log["disc_loss"]
print(f"Step {step}: "
f"gen_loss = {gen_loss:.3f}, disc_loss = {disc_loss:.3f}")
steps.append(step)
gen_losses.append(gen_loss)
disc_losses.append(disc_loss)
sns.set_style("whitegrid")
fig, axes = plt.subplots(1, 2, figsize=(20, 6))
# Plot the discriminator loss.
axes[0].plot(steps, disc_losses, "-")
axes[0].plot(steps, np.log(2) * np.ones_like(steps), "r--",
label="Discriminator is being fooled")
axes[0].legend(fontsize=20)
axes[0].set_title("Discriminator loss", fontsize=20)
# Plot the generator loss.
axes[1].plot(steps, gen_losses, '-')
axes[1].set_title("Generator loss", fontsize=20);
#@title {vertical-output: true}
def make_grid(samples, num_cols=8, rescale=True):
batch_size, height, width = samples.shape
assert batch_size % num_cols == 0
num_rows = batch_size // num_cols
# We want samples.shape == (height * num_rows, width * num_cols).
samples = samples.reshape(num_rows, num_cols, height, width)
samples = samples.swapaxes(1, 2)
samples = samples.reshape(height * num_rows, width * num_cols)
return samples
# Generate samples from the trained generator.
rng = jax.random.PRNGKey(12)
samples = gan.sample(rng, gan_state.params.gen, num_samples=64)
samples = jax.device_get(samples)
samples = samples.squeeze(axis=-1)
# Our model outputs values in [-1, 1] so scale it back to [0, 1].
samples = (samples + 1.0) / 2.0
plt.gray()
plt.axis("off")
samples_grid = make_grid(samples)
plt.imshow(samples_grid);
| 0.955382 | 0.786828 |
# Воркшоп по AI для архитекторов: Практикa
## Датасет
Мы уже определились с тем, какую задачу мы будем решать. Нужно выбрать подходящий набор данных! Мы предподготовили для воркшопа четыре набора размеченных данных:
* **green** - леса и парки
* **water** - водные объекты
* **residential** - жилые здания
* **non-residential** - нежилые здания
Каждый набор данных содержит две папки –– `train` (на данных из этой папки мы будем обучать нашу нейросеть) и `test` (на данных из этой папки мы будем проверять то, как работает наша нейросеть).
В свою очередь, каждая из этих папок содержит две подпапки –– `tile` и `mask`. В папке `tile` находятся тайлы (кусочки) спутниковых снимков, а в папке `mask` для каждого такого снимка есть *маска* интересующих нас объектов с тайла.
#### Упражненьице
Нужно правильно прописать путь к папке `train`, чтобы запустить обучение на нужном датасете
```
dataset_path = '/Users/slobanova/ipynbs/workshop/datasets/water/train/'
```
Посмотрим на картинки из датасета (попробуйте понажимать `Tab` в процессе написания пути):
```
from PIL import Image
Image.open('/Users/slobanova/ipynbs/workshop/datasets/water/train/tile/0.79166.41078.17.sat.tile.jpg')
Image.open('/Users/slobanova/ipynbs/workshop/datasets/water/train/mask/0.79166.41078.17.mask.tile.png')
```
С такими парами изображений мы и будем работать. Будем пытаться находить на спутниковых снимках объекты, маски которых мы скормим нейросети.
Чтобы в дальнейшем нам не приходилось каждый раз прописывать путь полностью (абсолютный путь), напишем функцию, которая будет возвращать нам путь к папке с текущим проектом. Тогда мы сможем использовать относительные, а не абсолютные пути:
```
import os
def get_root():
return os.path.abspath('')
# Получим путь к папке проекта:
root = get_root()
# Выведем путь
root
```
А тут давайте укажем название датасета, с которым хотим работать. Это понадобится нам в дальнейшем как составная часть относительного пути:
```
DATASET = 'water'
```
### Класс датасета
В библиотеке `torch`, которой мы собираемся пользоваться, для работы с данными, необходимо их описать особым образом. Этот способ описания позволяет привести данные к единому виду, а библиотеке обращаться к разным датасетам одного типа одинаково.
```
# Здесь мы импортируем нужные для работы кода библиотеки
import numpy as np
import torch
from torchvision import transforms as T
# Здесь мы описываем атрибуты и способы работы с нашим датасетом
class MaskDataset(object):
# Инициализация датасета
def __init__(self, root):
self.root = root
# Загрузка масок из папки
masks = list(sorted(os.listdir(os.path.join(root, "mask"))))
self.masks = []
self.imgs = []
# Для каждой маски мы находим соответствующий спутниковый снимок
for mask_file in masks:
img_mask_path = os.path.join(root, 'mask', mask_file)
img_file = mask_file.replace('.mask.', '.sat.').replace('.png', '.jpg')
img_mask = Image.open(img_mask_path).quantize(colors=256, method=2)
img_mask = np.array(img_mask)
if np.min(img_mask) == np.max(img_mask):
continue
self.masks.append(mask_file)
self.imgs.append(img_file)
# Обработка значений минимума и максимума для ббоксов (нужна для пограничных случаев)
@staticmethod
def _normalize_min_max(min_, max_):
if min_ == max_:
if max_ == 255:
min_ -= 1
else:
max_ += 1
elif min_ > max_:
min_, max_ = max_, min_
return min_, max_
# Этот метод описывает получение объекта (пара "снимок+маска") и его свойств
def __getitem__(self, idx):
# Загружаем снимки и маски
img_path = os.path.join(self.root, "tile", self.imgs[idx])
mask_path = os.path.join(self.root, "mask", self.masks[idx])
img_mask = Image.open(mask_path).quantize(colors=256, method=2)
img_mask = np.array(img_mask)
# Уникальный цвет на маске соответствует уникальному типу объекта
obj_ids = np.unique(img_mask)
# Первый цвет в списке - цвет фона, так что мы убираем его из списка цветов объектов
obj_ids = obj_ids[1:]
# Собираем бинарную маску, в которой для каждого пикселя будет говориться, есть ли на нем искомый объект
masks = img_mask == obj_ids[:, None, None]
masks = np.bitwise_not(masks)
# Получаем ббоксы для каждого снимка
num_objs = len(obj_ids)
boxes = []
try:
for i in range(num_objs):
pos = np.where(masks[i])
xmin = np.min(pos[1])
xmax = np.max(pos[1])
ymin = np.min(pos[0])
ymax = np.max(pos[0])
xmin, xmax = self._normalize_min_max(xmin, xmax)
ymin, ymax = self._normalize_min_max(ymin, ymax)
boxes.append([xmin, ymin, xmax, ymax])
except IndexError as e:
print(e)
print(img_path)
print(mask_path)
raise
# Конвертируем полученные ббоксы в тензор
boxes = torch.as_tensor(boxes, dtype=torch.float32)
# У нас только один класс - вешаем ярлыки-единички на все объекты на снимке
labels = torch.ones((num_objs,), dtype=torch.int64)
masks = torch.as_tensor(masks, dtype=torch.uint8)
image_id = torch.tensor([idx])
if boxes.size()[0] > 0:
area = (boxes[:, 3] - boxes[:, 1]) * (boxes[:, 2] - boxes[:, 0])
else:
area = torch.as_tensor(0)
# Здесь описываем атрибуты объекта в нашем датасете
target = {}
target["boxes"] = boxes
target["area"] = area
target["labels"] = labels
target["masks"] = masks
target["image_id"] = image_id
# Применяем преобразования к снимкам
transforms = self.get_transform()
img_tensor = transforms(Image.open(img_path).convert("RGB"))
return img_tensor, target
# С помощью этого метода мы сможем получать количество объектов в датасете
def __len__(self):
return len(self.imgs)
# С помощью этого метода мы получаем список трансформаций - преобразований исходных элементов датасета
def get_transform(self):
transforms = list()
# Наше единственное преобразование - перевод изображения в формат тензора (библиотека pytorch работает только с тензорами)
transforms.append(T.ToTensor())
return T.Compose(transforms)
```
Теперь мы можем загрузить данные из папки `train` в описанную структуру:
```
# Здесь мы определим размер валидационной выборки
val_subset_number = 10
# Загружаем данные из папки
whole_dataset = MaskDataset(os.path.join(root, 'datasets', DATASET, 'train'))
# Создадим список перемешанных номеров элементов датасета
indices = torch.randperm(len(whole_dataset)).tolist()
# Переопределим датасеты - теперь данные в них перемешаны
train_dataset = torch.utils.data.Subset(whole_dataset, indices[:-val_subset_number])
val_dataset = torch.utils.data.Subset(whole_dataset, indices[-val_subset_number:])
```
В `torch` для того, чтобы получать батчи данных из датасета, используется объект типа [DataLoader](https://pytorch.org/docs/stable/_modules/torch/utils/data/dataloader.html):
```
import utils
# Создадим DataLoader для тренировочного датасета:
train_loader = torch.utils.data.DataLoader(
train_dataset, # Здесь мы определяем датасет, для которого создается лодер
batch_size=2, # Определим количество элементов в батче
shuffle=True, # Будем перемешивать данные внутри датасета каждую эпоху
collate_fn=utils.collate_fn # Вспомогательная функция, приводит данные в батче к определенному виду
)
# Создадим DataLoader для валидационного датасета:
val_loader = torch.utils.data.DataLoader(
val_dataset,
batch_size=1,
shuffle=False,
collate_fn=utils.collate_fn
)
```
Ура! Мы разобрались с датасетом!
## Выбор модели и загрузка весов
Теперь необходимо определиться с архитектурой сети, которую мы собираемся использовать в качестве backbone. В `torch` реализовано много [моделей](https://pytorch.org/docs/stable/torchvision/models.html), мы же будем работать со следующими:
* [ResNet](https://arxiv.org/abs/1512.03385)
* [MobileNetV2](https://arxiv.org/abs/1801.04381)
Вы должны определиться с тем, какую архитектуру вы будете использовать в этом проекте –– укажите `resnet` или `mobilenet` в поле:
```
NETWORK = 'resnet'
```
Следующая функция возвращает нам выбранную модель:
```
def get_model(network='resnet', num_classes = 2):
import torchvision
from torchvision.models.detection import FasterRCNN, MaskRCNN
from torchvision.models.detection.rpn import AnchorGenerator
from torchvision.models.detection.faster_rcnn import FastRCNNPredictor
if network == 'resnet':
# Загружаем "чистую" модель
model = torchvision.models.detection.maskrcnn_resnet50_fpn(pretrained=False)
# В качестве предиктора ббоксов выберем FastRCNNPredictor, меняем количество выходных нейронов на количество классов = 2
in_features = model.roi_heads.box_predictor.cls_score.in_features
model.roi_heads.box_predictor = FastRCNNPredictor(in_features, num_classes)
return model
if network == 'mobilenet':
# Загружаем "чистую" модель
backbone = torchvision.models.mobilenet_v2(pretrained=False).features
backbone.out_channels = 1280
# Инициализируем генератор окон разных размеров
anchor_generator = AnchorGenerator(sizes=((32, 64, 128, 256, 512),),
aspect_ratios=((0.5, 1.0, 2.0),))
# Здесь мы инициализируем "голову" сети (предсказатель предполагаемых объектов)
roi_pooler = torchvision.ops.MultiScaleRoIAlign(featmap_names=[0], # Ищем единственный объект
output_size=7, # Размер выходного RoI
sampling_ratio=2) # Количество опорных точек
# Собираем MaskRCNN модель из частей
model = MaskRCNN(backbone,
num_classes=2,
rpn_anchor_generator=anchor_generator,
box_roi_pool=roi_pooler)
return model
else:
print('Неправильная модель, попробуй "resnet" и "mobilenet"')
return
```
Загрузим модель с помощью функции `get_model()`:
```
model = get_model(NETWORK)
```
Вспомогательная функция для формирования названия файла с весами (нужна будет для сохранения и загрузки весов):
```
def get_weights_filepath(epoch, dataset, network, is_train=False):
file_segments = []
if is_train:
file_segments.append('train')
if epoch is not None:
file_segments.append('ep{}'.format(epoch))
file_segments.append(dataset)
file_segments.append(network)
root = get_root()
file_name = '_'.join(file_segments) + '.pt'
return os.path.join(root, 'weights', file_name)
```
Одна из особенностей библиотеки PyTorch - возможность работы с GPU. Это позволит нам быстрее работать с моделью: тренировать ее и получать предсказания. Чтобы узнать, можем ли мы использовать GPU, запустим ячейку:
```
device = torch.device('cuda:0') if torch.cuda.is_available() else torch.device('cpu')
device.type
```
Если вы увидели `'cpu'` - чтож, обучать модель вам будет дольше по времени. Остальные могут выдохнуть. Да-да, это несправделивый мир машинного обучения, в котором вычислительные мощности решают!
#### Загрузка весов
В нашем воркшопе мы используем предподготовленные веса. Это значит, что мы будем инициализировать веса в нашей модели не случайным образом, а загрузим "сейв", который кто-то когда-то сделал для этой же архитектуры.
Все веса расположены в папке `weights`. Для каждого набора данных мы обучали сети обеих архитектур на протяжении 35-45 эпох. Результаты работы после такого обучения не идеальны, но уже гораздо лучше, чем после случайной инициализации. В этом модуле мы загрузим веса в модель:
```
weights_file = DATASET + '_' + NETWORK + '_' + 'initial_weights.pt'
WEIGHTS = os.path.join(root, 'weights', weights_file)
if not os.path.isfile(WEIGHTS):
print('Нет таких весов!')
# Загружаем веса
model.load_state_dict(torch.load(WEIGHTS))
# model.load_state_dict(torch.load(WEIGHTS, map_location=torch.device('cpu')))
# Загружаем модель в память устройства
model.to(device);
```
### Оптимизатор и гиперпараметры
_<..Вставьте еще одну лекцию и сто презентаций..>_
Оптимизатор - движущая сила всего процесса обучения. Именно он занимается обновлением весов модели, поэтому от того, как мы его зададим, зависит то, насколько быстро наша модель станет давать приемлемые предсказания, станет ли она их давать вообще и в принципе весь результат.
Вот [тут](https://pytorch.org/docs/stable/optim.html) можно посмотреть список всех методов оптимизации, зашитых в библиотеку PyTorch. Вы можете использовать любой по желанию, если разберетесь. А если нет - в ячейке ниже уже описана парочка таких методов (я надеюсь, что к этому моменту мы уже рассказали вам про них):
* [SGD](https://pytorch.org/docs/stable/_modules/torch/optim/sgd.html)
* [Adam](https://pytorch.org/docs/stable/_modules/torch/optim/adam.html)
```
# Создаем список всех весов в модели
params = [p for p in model.parameters() if p.requires_grad]
# Выбираем оптимизатор и указываем его параметры
# Градиентный спуск
optimizer = torch.optim.SGD(params,
lr=0.005, # Коэффициент скорости обучения, должен быть не слишком большим и не слишком маленьким
momentum=0.9, # Коэффициент "ускорения"
weight_decay=0.0005 # Коэффициент устойчивости нестабильных весов
)
# Adam
optimizer = torch.optim.Adam(params,
lr=0.001,
weight_decay=0.0005
)
```
### Обучение модели
Ну что, перейдем к самой ответственной части? Нужно собрать все вместе: датасет, модель, оптимизатор.. И обучать модель. Как обычно, параметры `NUM_EPOCHS` и `EPOCH_SAVING_RATE` останутся на ваше усмотрение, но мы настоятельно рекомендуем сохраняться не реже раза в 5 эпох, а количество эпох для обучения оценивать трезво - модель должна успеть доучиться до завтра, но при этом не __пере__обучиться.
```
# Добавляем функцию для динамического обновления коэффициента скорости обучения
lr_scheduler = torch.optim.lr_scheduler.StepLR(optimizer, # будем обновлять коэффициент в нашем оптимизаторе
step_size=3, # каждые N эпох коэффициент будет обновляться
gamma=0.1 # как будет изменяться коэффициент (new_lr = lr * gamma)
)
# Количество эпох
NUM_EPOCHS = 2
# Будем сохранять веса каждые N эпох:
EPOCH_SAVING_RATE = 5
import time
from engine import train_one_epoch, evaluate
print('Начинаем обучать модель: {}'.format(time.asctime()))
print('Устройство: {}'.format(device))
print('Датасет: {}'.format(DATASET))
print('Модель: {}'.format(NETWORK))
print('Эпохи: {}'.format(NUM_EPOCHS))
print('Загруженные веса: {}'.format(WEIGHTS))
print('--- -- -- -- -- -- ---')
# Список для хранения времени обучения за эпоху
learning_ts_list = list()
loss_acc_list = []
val_accuracy_list = []
for epoch in range(NUM_EPOCHS):
epoch_learning_ts = time.time()
try:
# Тренируем одну эпоху, выводим информацию каждые 10 батчей
loss_acc_dict = train_one_epoch(model, optimizer, train_loader, device, epoch, print_freq=10)
# Обновим коэффициент скорости обучения оптимизатора:
lr_scheduler.step()
# Оценка на валидационном датасете
val_accuracies = evaluate(model, val_loader, device=device)
mean_acc = float(np.mean(val_accuracies))
print('Эпоха {} окончена, средняя accuracy на обучении {}%'.format(epoch, mean_acc * 100))
# Сохраним для истории loss и accuracy
with torch.no_grad():
loss_acc_list.append(loss_acc_dict)
val_accuracy_list.append(mean_acc)
# Сохраняем веса в отдельный файл каждые EPOCH_SAVING_RATE эпох
if epoch >= EPOCH_SAVING_RATE and epoch % EPOCH_SAVING_RATE == 0:
train_weights_file_path = get_weights_filepath(epoch=epoch, dataset=DATASET, network=NETWORK, is_train=True)
torch.save(model.state_dict(), train_weights_file_path)
# Если произойдет какая-либо ошибка, мы хотим ее вывести и сохранить текущие веса
except Exception as e:
import traceback
print(e)
print(traceback.format_exc())
# Сохраняем текущие веса
train_weights_file_path = get_weights_filepath(epoch=epoch, dataset=DATASET, network=NETWORK, is_train=True)
torch.save(model.state_dict(), train_weights_file_path)
# Записываем, сколько времени мы потратили на эту эпоху (интересно и познавательно)
epoch_learning_ts = time.time() - epoch_learning_ts
learning_ts_list.append(epoch_learning_ts)
avg_learn_time = np.mean(learning_ts_list)
print('Время обучения: {} сек'.format(int(epoch_learning_ts)))
print('Среднее время обучения: {} сек'.format(int(avg_learn_time)))
# Сохраняем конечные веса модели после обучения
weights_file_path = get_weights_filepath(epoch=None, dataset=DATASET, network=NETWORK, is_train=False)
torch.save(model.state_dict(), weights_file_path)
print("Вот и всё!")
# Преобразуем словаль со статистикой по обучению в отдельные переменные
tr_accuracy_list = [np.mean(x['tr_accuracy_list']) for x in loss_acc_list]
loss_aggregate = [np.mean(x['loss_aggregate']) for x in loss_acc_list]
loss_classifier = [np.mean(x['loss_classifier']) for x in loss_acc_list]
loss_box_reg = [np.mean(x['loss_box_reg']) for x in loss_acc_list]
loss_mask = [np.mean(x['loss_mask']) for x in loss_acc_list]
```
### Построим график loss и метрики качества
Чтобы диагностировать проблемы при обучении, полезно иногда смотреть на то, как изменяется со временем loss и accuracy на тренировочном и валидационном датасетах. Если грубо говорить, _loss_ должна убывать со временем, а _accuracy_ расти.
```
from matplotlib import pyplot as plt
%matplotlib inline
fig, ax = plt.subplots(2, 1, figsize=(15, 20))
# Нарисуем первый график, он будет показывать, как изменялись loss'ы по эпохам на трейне и на валидации
ax[0].plot(list(range(len(loss_classifier))), loss_classifier, label='Classifier loss', c='c')
ax[0].plot(list(range(len(loss_box_reg))), loss_box_reg, label='Bbox regression loss', c='m')
ax[0].plot(list(range(len(loss_mask))), loss_mask, label='Mask loss', c='y')
ax[0].plot(list(range(len(loss_aggregate))), loss_aggregate, label='Aggregated loss', linewidth=3, c='0.5')
ax[0].set_xlabel('Epoch')
ax[0].set_ylabel('Losses')
ax[0].legend(loc='upper right')
# Нарисуем второй график, он будет показывать, как изменялись аккураси и совокупный loss по эпохам на трейне и на валидации
ax[1].plot(list(range(len(tr_accuracy_list))), tr_accuracy_list, label='Train accuracy', linestyle='--', c='y')
ax[1].plot(list(range(len(val_accuracy_list))), val_accuracy_list, label='Validation accuracy', c='y')
ax[1].plot(list(range(len(loss_aggregate))), loss_aggregate, label='Training aggregated loss', linewidth=3, linestyle='--', c='0.5')
ax[1].set_xlabel('Epoch')
ax[1].set_ylabel('Accuracy & aggregated loss')
ax[1].legend(loc='upper right')
plt.show()
```
## Посмотрим, что же у нас вышло
Ну что, наверное хочется посмотреть, как работает наш алгоритм? Это можно сделать, посмотрев на предсказание, полученное моделью `pred = model(img)`, однако его не очень приятно читать и довольно сложно соотнести с реальным изображением. Чтобы было проще узнать, что и где нашла наша сеть, используем пару новых функций и сохраним изображения в папку `result`.
Для начала разберемся, с каким датасетом мы будем работать, и куда сохраним результаты. Если предыдущие части кода вы запускали относительно давно, то компьютеру нужно будет освежить память и задать нужные нам переменные заново.
```
# Это секция с закоментированными строками может пригодиться, если в памяти компьютера не осталось переменных с обучения
# weights_file_path = '/Users/slobanova/ipynbs/workshop/weights/water_resnet_initial_weights.pt'
# model = get_model('mobilenet')
# model.load_state_dict(torch.load(weights_file_path, map_location=torch.device('cpu')))
# model.to(device)
# DATASET = 'water'
# Указываем путь к папке со снимками для проверки
dataset_path = os.path.join(root, 'datasets', DATASET, 'train')
# Указываем путь для сохранения итоговых изображений
result_path = os.path.join(root, 'result')
# Задаем названия лейблов для ббоксов
OBJECT_LABELS = [
'__background__', DATASET
]
```
Опишем функцию, которая будет получать предсказание от модели и в зависимости от вероятности наличия объекта (вероятность больше или меньше порогового значения `threshold`) будет передавать объекты в функцию для рисования:
```
# Функция для получения списка преобразований
def get_transform(train):
transforms = [
T.ToTensor(),
]
if train:
transforms.append(T.RandomHorizontalFlip(0.5))
return T.Compose(transforms)
# Функция для получения предсказания для спутникого снимка
def get_prediction(img_path, threshold):
# Переводим модель в режим оценки
model.eval()
img = Image.open(img_path) # Открываем картинку
transform = T.Compose([T.ToTensor()])
img = transform(img) # Применяем к ней трансформации
pred = model([img]) # Получаем предсказание модели по снимку
pred_class = [OBJECT_LABELS[i] for i in list(pred[0]['labels'].cpu().numpy())] # Получаем классы распознанных объектов - лейблы
pred_boxes = [[(i[0], i[1]), (i[2], i[3])] for i in list(pred[0]['boxes'].detach().cpu().numpy())] # Получаем ббоксы объектов
pred_score = list(pred[0]['scores'].detach().cpu().numpy()) # Получаем вероятности для объектов
pred_masks = list(pred[0]['masks'].detach().cpu().numpy()) # Маски объектов
print(os.path.basename(img_path))
# Здесь мы выбираем объекты, вероятность которых > threshold
pred_selected = [pred_score.index(x) for x in pred_score]
if len(pred_selected) == 0:
return [], [], [], []
pred_filtered_values = [x for x in pred_score if x > threshold]
if len(pred_filtered_values) == 0:
return [], [], [], []
print("Вероятности для всех найденных объектов: {}.".format(pred_score))
pred_selected = [pred_score.index(x) for x in pred_score if x > threshold]
pred_boxes = [pred_boxes[idx] for idx in pred_selected]
pred_class = [pred_class[idx] for idx in pred_selected]
pred_score = [pred_score[idx] for idx in pred_selected]
pred_masks = [pred_masks[idx] for idx in pred_selected]
return pred_boxes, pred_class, pred_score, pred_masks
```
Напишем функцию для отрисовки найденных объектов:
```
import cv2
# Функция рисования результатов обработки, сохраняет снимок с нанесенными ббоксами и отдельно предсказанную маску
def object_detection_api(img_path, threshold=0.15, rect_th=1, text_size=0.4, text_th=3):
boxes, pred_cls, scores, masks = get_prediction(img_path, threshold) # Получим данные о найденных объектах на снимке
img = cv2.imread(img_path) # Читаем изображение
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB) # Конвертируем цвета (особенность работы с cv2)
# Рисуем ббокс
for i in range(len(boxes)):
cv2.rectangle( # Добавляем прямоугольник ббокса на картинку
img,
boxes[i][0],
boxes[i][1],
color=(0, 255, 0),
thickness=rect_th
)
cv2.putText( # Добавляем подпись к прямоугольнику (вероятность)
img,
str(scores[i]),
boxes[i][0],
cv2.FONT_HERSHEY_SIMPLEX,
text_size,
color=(0, 255, 0),
thickness=1
)
height, width, _ = img.shape
# Сделаем пустой холст для рисования маски
heatmap_mask = np.zeros((256,256), dtype=np.uint8)
# Накладываем единичные маски друг на друга
for i in range(len(masks)):
the_mask = masks[i][0]
heatmap_mask = np.uint8(255 * the_mask) + heatmap_mask
# Сохраняем изображение с ббоксами
plt.imshow(img)
saving_file = os.path.join(result_path, os.path.basename(img_path).replace('.sat.', '.bbox.'))
plt.savefig(saving_file)
# Сохраняем изображение маски
plt.imshow(heatmap_mask)
saving_file = os.path.join(result_path, os.path.basename(img_path).replace('.sat.', '.heat.'))
plt.savefig(saving_file)
```
Теперь мы можем запустить каскад функций (параметр `threshold` можно и нужно менять):
```
data = os.scandir(os.path.join(dataset_path, 'tile'))
for i, item in enumerate(data):
object_detection_api(item.path, threshold = 0.5)
# Будем смотреть первые 20 снимков
if i == 19:
break
```
Теперь можно посмотреть на эти картинки в папке. Погнали!
## Запуск на новых данных
Если вы дошли до этого этапа, то с большой вероятностью вы смогли выполнить предыдущий шаг. Сейчас мы попробуем получить результаты алгоритма на снимках Санкт-Петербурга. Эти снимки хранятся в папке `test`. Мы специально не трогали этот набор данных до последнего: сейчас мы будем смотреть, как же в реальности работает наш алгоритм на данных, которых он раньше не видел.
#### Упражненьице
* Создайте новую папку для сохранения результатов. Например, `spb_result`
* Получите обработанные сетью картинки и сохраните их в эту папку
_Не стесняйтесь спрашивать, если что-то не понятно!_
|
github_jupyter
|
dataset_path = '/Users/slobanova/ipynbs/workshop/datasets/water/train/'
from PIL import Image
Image.open('/Users/slobanova/ipynbs/workshop/datasets/water/train/tile/0.79166.41078.17.sat.tile.jpg')
Image.open('/Users/slobanova/ipynbs/workshop/datasets/water/train/mask/0.79166.41078.17.mask.tile.png')
import os
def get_root():
return os.path.abspath('')
# Получим путь к папке проекта:
root = get_root()
# Выведем путь
root
DATASET = 'water'
# Здесь мы импортируем нужные для работы кода библиотеки
import numpy as np
import torch
from torchvision import transforms as T
# Здесь мы описываем атрибуты и способы работы с нашим датасетом
class MaskDataset(object):
# Инициализация датасета
def __init__(self, root):
self.root = root
# Загрузка масок из папки
masks = list(sorted(os.listdir(os.path.join(root, "mask"))))
self.masks = []
self.imgs = []
# Для каждой маски мы находим соответствующий спутниковый снимок
for mask_file in masks:
img_mask_path = os.path.join(root, 'mask', mask_file)
img_file = mask_file.replace('.mask.', '.sat.').replace('.png', '.jpg')
img_mask = Image.open(img_mask_path).quantize(colors=256, method=2)
img_mask = np.array(img_mask)
if np.min(img_mask) == np.max(img_mask):
continue
self.masks.append(mask_file)
self.imgs.append(img_file)
# Обработка значений минимума и максимума для ббоксов (нужна для пограничных случаев)
@staticmethod
def _normalize_min_max(min_, max_):
if min_ == max_:
if max_ == 255:
min_ -= 1
else:
max_ += 1
elif min_ > max_:
min_, max_ = max_, min_
return min_, max_
# Этот метод описывает получение объекта (пара "снимок+маска") и его свойств
def __getitem__(self, idx):
# Загружаем снимки и маски
img_path = os.path.join(self.root, "tile", self.imgs[idx])
mask_path = os.path.join(self.root, "mask", self.masks[idx])
img_mask = Image.open(mask_path).quantize(colors=256, method=2)
img_mask = np.array(img_mask)
# Уникальный цвет на маске соответствует уникальному типу объекта
obj_ids = np.unique(img_mask)
# Первый цвет в списке - цвет фона, так что мы убираем его из списка цветов объектов
obj_ids = obj_ids[1:]
# Собираем бинарную маску, в которой для каждого пикселя будет говориться, есть ли на нем искомый объект
masks = img_mask == obj_ids[:, None, None]
masks = np.bitwise_not(masks)
# Получаем ббоксы для каждого снимка
num_objs = len(obj_ids)
boxes = []
try:
for i in range(num_objs):
pos = np.where(masks[i])
xmin = np.min(pos[1])
xmax = np.max(pos[1])
ymin = np.min(pos[0])
ymax = np.max(pos[0])
xmin, xmax = self._normalize_min_max(xmin, xmax)
ymin, ymax = self._normalize_min_max(ymin, ymax)
boxes.append([xmin, ymin, xmax, ymax])
except IndexError as e:
print(e)
print(img_path)
print(mask_path)
raise
# Конвертируем полученные ббоксы в тензор
boxes = torch.as_tensor(boxes, dtype=torch.float32)
# У нас только один класс - вешаем ярлыки-единички на все объекты на снимке
labels = torch.ones((num_objs,), dtype=torch.int64)
masks = torch.as_tensor(masks, dtype=torch.uint8)
image_id = torch.tensor([idx])
if boxes.size()[0] > 0:
area = (boxes[:, 3] - boxes[:, 1]) * (boxes[:, 2] - boxes[:, 0])
else:
area = torch.as_tensor(0)
# Здесь описываем атрибуты объекта в нашем датасете
target = {}
target["boxes"] = boxes
target["area"] = area
target["labels"] = labels
target["masks"] = masks
target["image_id"] = image_id
# Применяем преобразования к снимкам
transforms = self.get_transform()
img_tensor = transforms(Image.open(img_path).convert("RGB"))
return img_tensor, target
# С помощью этого метода мы сможем получать количество объектов в датасете
def __len__(self):
return len(self.imgs)
# С помощью этого метода мы получаем список трансформаций - преобразований исходных элементов датасета
def get_transform(self):
transforms = list()
# Наше единственное преобразование - перевод изображения в формат тензора (библиотека pytorch работает только с тензорами)
transforms.append(T.ToTensor())
return T.Compose(transforms)
# Здесь мы определим размер валидационной выборки
val_subset_number = 10
# Загружаем данные из папки
whole_dataset = MaskDataset(os.path.join(root, 'datasets', DATASET, 'train'))
# Создадим список перемешанных номеров элементов датасета
indices = torch.randperm(len(whole_dataset)).tolist()
# Переопределим датасеты - теперь данные в них перемешаны
train_dataset = torch.utils.data.Subset(whole_dataset, indices[:-val_subset_number])
val_dataset = torch.utils.data.Subset(whole_dataset, indices[-val_subset_number:])
import utils
# Создадим DataLoader для тренировочного датасета:
train_loader = torch.utils.data.DataLoader(
train_dataset, # Здесь мы определяем датасет, для которого создается лодер
batch_size=2, # Определим количество элементов в батче
shuffle=True, # Будем перемешивать данные внутри датасета каждую эпоху
collate_fn=utils.collate_fn # Вспомогательная функция, приводит данные в батче к определенному виду
)
# Создадим DataLoader для валидационного датасета:
val_loader = torch.utils.data.DataLoader(
val_dataset,
batch_size=1,
shuffle=False,
collate_fn=utils.collate_fn
)
NETWORK = 'resnet'
def get_model(network='resnet', num_classes = 2):
import torchvision
from torchvision.models.detection import FasterRCNN, MaskRCNN
from torchvision.models.detection.rpn import AnchorGenerator
from torchvision.models.detection.faster_rcnn import FastRCNNPredictor
if network == 'resnet':
# Загружаем "чистую" модель
model = torchvision.models.detection.maskrcnn_resnet50_fpn(pretrained=False)
# В качестве предиктора ббоксов выберем FastRCNNPredictor, меняем количество выходных нейронов на количество классов = 2
in_features = model.roi_heads.box_predictor.cls_score.in_features
model.roi_heads.box_predictor = FastRCNNPredictor(in_features, num_classes)
return model
if network == 'mobilenet':
# Загружаем "чистую" модель
backbone = torchvision.models.mobilenet_v2(pretrained=False).features
backbone.out_channels = 1280
# Инициализируем генератор окон разных размеров
anchor_generator = AnchorGenerator(sizes=((32, 64, 128, 256, 512),),
aspect_ratios=((0.5, 1.0, 2.0),))
# Здесь мы инициализируем "голову" сети (предсказатель предполагаемых объектов)
roi_pooler = torchvision.ops.MultiScaleRoIAlign(featmap_names=[0], # Ищем единственный объект
output_size=7, # Размер выходного RoI
sampling_ratio=2) # Количество опорных точек
# Собираем MaskRCNN модель из частей
model = MaskRCNN(backbone,
num_classes=2,
rpn_anchor_generator=anchor_generator,
box_roi_pool=roi_pooler)
return model
else:
print('Неправильная модель, попробуй "resnet" и "mobilenet"')
return
model = get_model(NETWORK)
def get_weights_filepath(epoch, dataset, network, is_train=False):
file_segments = []
if is_train:
file_segments.append('train')
if epoch is not None:
file_segments.append('ep{}'.format(epoch))
file_segments.append(dataset)
file_segments.append(network)
root = get_root()
file_name = '_'.join(file_segments) + '.pt'
return os.path.join(root, 'weights', file_name)
device = torch.device('cuda:0') if torch.cuda.is_available() else torch.device('cpu')
device.type
weights_file = DATASET + '_' + NETWORK + '_' + 'initial_weights.pt'
WEIGHTS = os.path.join(root, 'weights', weights_file)
if not os.path.isfile(WEIGHTS):
print('Нет таких весов!')
# Загружаем веса
model.load_state_dict(torch.load(WEIGHTS))
# model.load_state_dict(torch.load(WEIGHTS, map_location=torch.device('cpu')))
# Загружаем модель в память устройства
model.to(device);
# Создаем список всех весов в модели
params = [p for p in model.parameters() if p.requires_grad]
# Выбираем оптимизатор и указываем его параметры
# Градиентный спуск
optimizer = torch.optim.SGD(params,
lr=0.005, # Коэффициент скорости обучения, должен быть не слишком большим и не слишком маленьким
momentum=0.9, # Коэффициент "ускорения"
weight_decay=0.0005 # Коэффициент устойчивости нестабильных весов
)
# Adam
optimizer = torch.optim.Adam(params,
lr=0.001,
weight_decay=0.0005
)
# Добавляем функцию для динамического обновления коэффициента скорости обучения
lr_scheduler = torch.optim.lr_scheduler.StepLR(optimizer, # будем обновлять коэффициент в нашем оптимизаторе
step_size=3, # каждые N эпох коэффициент будет обновляться
gamma=0.1 # как будет изменяться коэффициент (new_lr = lr * gamma)
)
# Количество эпох
NUM_EPOCHS = 2
# Будем сохранять веса каждые N эпох:
EPOCH_SAVING_RATE = 5
import time
from engine import train_one_epoch, evaluate
print('Начинаем обучать модель: {}'.format(time.asctime()))
print('Устройство: {}'.format(device))
print('Датасет: {}'.format(DATASET))
print('Модель: {}'.format(NETWORK))
print('Эпохи: {}'.format(NUM_EPOCHS))
print('Загруженные веса: {}'.format(WEIGHTS))
print('--- -- -- -- -- -- ---')
# Список для хранения времени обучения за эпоху
learning_ts_list = list()
loss_acc_list = []
val_accuracy_list = []
for epoch in range(NUM_EPOCHS):
epoch_learning_ts = time.time()
try:
# Тренируем одну эпоху, выводим информацию каждые 10 батчей
loss_acc_dict = train_one_epoch(model, optimizer, train_loader, device, epoch, print_freq=10)
# Обновим коэффициент скорости обучения оптимизатора:
lr_scheduler.step()
# Оценка на валидационном датасете
val_accuracies = evaluate(model, val_loader, device=device)
mean_acc = float(np.mean(val_accuracies))
print('Эпоха {} окончена, средняя accuracy на обучении {}%'.format(epoch, mean_acc * 100))
# Сохраним для истории loss и accuracy
with torch.no_grad():
loss_acc_list.append(loss_acc_dict)
val_accuracy_list.append(mean_acc)
# Сохраняем веса в отдельный файл каждые EPOCH_SAVING_RATE эпох
if epoch >= EPOCH_SAVING_RATE and epoch % EPOCH_SAVING_RATE == 0:
train_weights_file_path = get_weights_filepath(epoch=epoch, dataset=DATASET, network=NETWORK, is_train=True)
torch.save(model.state_dict(), train_weights_file_path)
# Если произойдет какая-либо ошибка, мы хотим ее вывести и сохранить текущие веса
except Exception as e:
import traceback
print(e)
print(traceback.format_exc())
# Сохраняем текущие веса
train_weights_file_path = get_weights_filepath(epoch=epoch, dataset=DATASET, network=NETWORK, is_train=True)
torch.save(model.state_dict(), train_weights_file_path)
# Записываем, сколько времени мы потратили на эту эпоху (интересно и познавательно)
epoch_learning_ts = time.time() - epoch_learning_ts
learning_ts_list.append(epoch_learning_ts)
avg_learn_time = np.mean(learning_ts_list)
print('Время обучения: {} сек'.format(int(epoch_learning_ts)))
print('Среднее время обучения: {} сек'.format(int(avg_learn_time)))
# Сохраняем конечные веса модели после обучения
weights_file_path = get_weights_filepath(epoch=None, dataset=DATASET, network=NETWORK, is_train=False)
torch.save(model.state_dict(), weights_file_path)
print("Вот и всё!")
# Преобразуем словаль со статистикой по обучению в отдельные переменные
tr_accuracy_list = [np.mean(x['tr_accuracy_list']) for x in loss_acc_list]
loss_aggregate = [np.mean(x['loss_aggregate']) for x in loss_acc_list]
loss_classifier = [np.mean(x['loss_classifier']) for x in loss_acc_list]
loss_box_reg = [np.mean(x['loss_box_reg']) for x in loss_acc_list]
loss_mask = [np.mean(x['loss_mask']) for x in loss_acc_list]
from matplotlib import pyplot as plt
%matplotlib inline
fig, ax = plt.subplots(2, 1, figsize=(15, 20))
# Нарисуем первый график, он будет показывать, как изменялись loss'ы по эпохам на трейне и на валидации
ax[0].plot(list(range(len(loss_classifier))), loss_classifier, label='Classifier loss', c='c')
ax[0].plot(list(range(len(loss_box_reg))), loss_box_reg, label='Bbox regression loss', c='m')
ax[0].plot(list(range(len(loss_mask))), loss_mask, label='Mask loss', c='y')
ax[0].plot(list(range(len(loss_aggregate))), loss_aggregate, label='Aggregated loss', linewidth=3, c='0.5')
ax[0].set_xlabel('Epoch')
ax[0].set_ylabel('Losses')
ax[0].legend(loc='upper right')
# Нарисуем второй график, он будет показывать, как изменялись аккураси и совокупный loss по эпохам на трейне и на валидации
ax[1].plot(list(range(len(tr_accuracy_list))), tr_accuracy_list, label='Train accuracy', linestyle='--', c='y')
ax[1].plot(list(range(len(val_accuracy_list))), val_accuracy_list, label='Validation accuracy', c='y')
ax[1].plot(list(range(len(loss_aggregate))), loss_aggregate, label='Training aggregated loss', linewidth=3, linestyle='--', c='0.5')
ax[1].set_xlabel('Epoch')
ax[1].set_ylabel('Accuracy & aggregated loss')
ax[1].legend(loc='upper right')
plt.show()
# Это секция с закоментированными строками может пригодиться, если в памяти компьютера не осталось переменных с обучения
# weights_file_path = '/Users/slobanova/ipynbs/workshop/weights/water_resnet_initial_weights.pt'
# model = get_model('mobilenet')
# model.load_state_dict(torch.load(weights_file_path, map_location=torch.device('cpu')))
# model.to(device)
# DATASET = 'water'
# Указываем путь к папке со снимками для проверки
dataset_path = os.path.join(root, 'datasets', DATASET, 'train')
# Указываем путь для сохранения итоговых изображений
result_path = os.path.join(root, 'result')
# Задаем названия лейблов для ббоксов
OBJECT_LABELS = [
'__background__', DATASET
]
# Функция для получения списка преобразований
def get_transform(train):
transforms = [
T.ToTensor(),
]
if train:
transforms.append(T.RandomHorizontalFlip(0.5))
return T.Compose(transforms)
# Функция для получения предсказания для спутникого снимка
def get_prediction(img_path, threshold):
# Переводим модель в режим оценки
model.eval()
img = Image.open(img_path) # Открываем картинку
transform = T.Compose([T.ToTensor()])
img = transform(img) # Применяем к ней трансформации
pred = model([img]) # Получаем предсказание модели по снимку
pred_class = [OBJECT_LABELS[i] for i in list(pred[0]['labels'].cpu().numpy())] # Получаем классы распознанных объектов - лейблы
pred_boxes = [[(i[0], i[1]), (i[2], i[3])] for i in list(pred[0]['boxes'].detach().cpu().numpy())] # Получаем ббоксы объектов
pred_score = list(pred[0]['scores'].detach().cpu().numpy()) # Получаем вероятности для объектов
pred_masks = list(pred[0]['masks'].detach().cpu().numpy()) # Маски объектов
print(os.path.basename(img_path))
# Здесь мы выбираем объекты, вероятность которых > threshold
pred_selected = [pred_score.index(x) for x in pred_score]
if len(pred_selected) == 0:
return [], [], [], []
pred_filtered_values = [x for x in pred_score if x > threshold]
if len(pred_filtered_values) == 0:
return [], [], [], []
print("Вероятности для всех найденных объектов: {}.".format(pred_score))
pred_selected = [pred_score.index(x) for x in pred_score if x > threshold]
pred_boxes = [pred_boxes[idx] for idx in pred_selected]
pred_class = [pred_class[idx] for idx in pred_selected]
pred_score = [pred_score[idx] for idx in pred_selected]
pred_masks = [pred_masks[idx] for idx in pred_selected]
return pred_boxes, pred_class, pred_score, pred_masks
import cv2
# Функция рисования результатов обработки, сохраняет снимок с нанесенными ббоксами и отдельно предсказанную маску
def object_detection_api(img_path, threshold=0.15, rect_th=1, text_size=0.4, text_th=3):
boxes, pred_cls, scores, masks = get_prediction(img_path, threshold) # Получим данные о найденных объектах на снимке
img = cv2.imread(img_path) # Читаем изображение
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB) # Конвертируем цвета (особенность работы с cv2)
# Рисуем ббокс
for i in range(len(boxes)):
cv2.rectangle( # Добавляем прямоугольник ббокса на картинку
img,
boxes[i][0],
boxes[i][1],
color=(0, 255, 0),
thickness=rect_th
)
cv2.putText( # Добавляем подпись к прямоугольнику (вероятность)
img,
str(scores[i]),
boxes[i][0],
cv2.FONT_HERSHEY_SIMPLEX,
text_size,
color=(0, 255, 0),
thickness=1
)
height, width, _ = img.shape
# Сделаем пустой холст для рисования маски
heatmap_mask = np.zeros((256,256), dtype=np.uint8)
# Накладываем единичные маски друг на друга
for i in range(len(masks)):
the_mask = masks[i][0]
heatmap_mask = np.uint8(255 * the_mask) + heatmap_mask
# Сохраняем изображение с ббоксами
plt.imshow(img)
saving_file = os.path.join(result_path, os.path.basename(img_path).replace('.sat.', '.bbox.'))
plt.savefig(saving_file)
# Сохраняем изображение маски
plt.imshow(heatmap_mask)
saving_file = os.path.join(result_path, os.path.basename(img_path).replace('.sat.', '.heat.'))
plt.savefig(saving_file)
data = os.scandir(os.path.join(dataset_path, 'tile'))
for i, item in enumerate(data):
object_detection_api(item.path, threshold = 0.5)
# Будем смотреть первые 20 снимков
if i == 19:
break
| 0.310485 | 0.966505 |
# Creating a Real-Time Inferencing Service
After training a predictive model, you can deploy it as a real-time service that clients can use to get predictions from new data.
## Before You Start
Before you start this lab, ensure that you have completed the *Create an Azure Machine Learning Workspace* and *Create a Compute Instance* tasks in [Lab 1: Getting Started with Azure Machine Learning](./labdocs/Lab01.md). Then open this notebook in Jupyter on your Compute Instance.
## Connect to Your Workspace
The first thing you need to do is to connect to your workspace using the Azure ML SDK.
> **Note**: If the authenticated session with your Azure subscription has expired since you completed the previous exercise, you'll be prompted to reauthenticate.
```
import azureml.core
from azureml.core import Workspace
# Load the workspace from the saved config file
ws = Workspace.from_config()
print('Ready to use Azure ML {} to work with {}'.format(azureml.core.VERSION, ws.name))
```
## Train and Register a Model
Now let's train and register a model.
```
from azureml.core import Experiment
from azureml.core import Model
import pandas as pd
import numpy as np
import joblib
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import roc_auc_score
from sklearn.metrics import roc_curve
# Create an Azure ML experiment in your workspace
experiment = Experiment(workspace = ws, name = "diabetes-training")
run = experiment.start_logging()
print("Starting experiment:", experiment.name)
# load the diabetes dataset
print("Loading Data...")
diabetes = pd.read_csv('data/diabetes.csv')
# Separate features and labels
X, y = diabetes[['Pregnancies','PlasmaGlucose','DiastolicBloodPressure','TricepsThickness','SerumInsulin','BMI','DiabetesPedigree','Age']].values, diabetes['Diabetic'].values
# Split data into training set and test set
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=0)
# Train a decision tree model
print('Training a decision tree model')
model = DecisionTreeClassifier().fit(X_train, y_train)
# calculate accuracy
y_hat = model.predict(X_test)
acc = np.average(y_hat == y_test)
print('Accuracy:', acc)
run.log('Accuracy', np.float(acc))
# calculate AUC
y_scores = model.predict_proba(X_test)
auc = roc_auc_score(y_test,y_scores[:,1])
print('AUC: ' + str(auc))
run.log('AUC', np.float(auc))
# Save the trained model
model_file = 'diabetes_model.pkl'
joblib.dump(value=model, filename=model_file)
run.upload_file(name = 'outputs/' + model_file, path_or_stream = './' + model_file)
# Complete the run
run.complete()
# Register the model
run.register_model(model_path='outputs/diabetes_model.pkl', model_name='diabetes_model',
tags={'Training context':'Inline Training'},
properties={'AUC': run.get_metrics()['AUC'], 'Accuracy': run.get_metrics()['Accuracy']})
print('Model trained and registered.')
```
## Deploy a Model as a Web Service
You have trained and registered a machine learning model that classifies patients based on the likelihood of them having diabetes. This model could be used in a production environment such as a doctor's surgery where only patients deemed to be at risk need to be subjected to a clinical test for diabetes. To support this scenario, you will deploy the model as a web service.
First, let's determine what models you have registered in the workspace.
```
from azureml.core import Model
for model in Model.list(ws):
print(model.name, 'version:', model.version)
for tag_name in model.tags:
tag = model.tags[tag_name]
print ('\t',tag_name, ':', tag)
for prop_name in model.properties:
prop = model.properties[prop_name]
print ('\t',prop_name, ':', prop)
print('\n')
```
Right, now let's get the model that we want to deploy. By default, if we specify a model name, the latest version will be returned.
```
model = ws.models['diabetes_model']
print(model.name, 'version', model.version)
```
We're going to create a web service to host this model, and this will require some code and configuration files; so let's create a folder for those.
```
import os
folder_name = 'diabetes_service'
# Create a folder for the web service files
experiment_folder = './' + folder_name
os.makedirs(experiment_folder, exist_ok=True)
print(folder_name, 'folder created.')
# Set path for scoring script
script_file = os.path.join(experiment_folder,"score_diabetes.py")
```
The web service where we deploy the model will need some Python code to load the input data, get the model from the workspace, and generate and return predictions. We'll save this code in an *entry script* (often called a *scoring script*) that will be deployed to the web service:
```
%%writefile $script_file
import json
import joblib
import numpy as np
from azureml.core.model import Model
# Called when the service is loaded
def init():
global model
# Get the path to the deployed model file and load it
model_path = Model.get_model_path('diabetes_model')
model = joblib.load(model_path)
# Called when a request is received
def run(raw_data):
# Get the input data as a numpy array
data = np.array(json.loads(raw_data)['data'])
# Get a prediction from the model
predictions = model.predict(data)
# Get the corresponding classname for each prediction (0 or 1)
classnames = ['not-diabetic', 'diabetic']
predicted_classes = []
for prediction in predictions:
predicted_classes.append(classnames[prediction])
# Return the predictions as JSON
return json.dumps(predicted_classes)
```
The web service will be hosted in a container, and the container will need to install any required Python dependencies when it gets initialized. In this case, our scoring code requires **scikit-learn**, so we'll create a .yml file that tells the container host to install this into the environment.
```
from azureml.core.conda_dependencies import CondaDependencies
# Add the dependencies for our model (AzureML defaults is already included)
myenv = CondaDependencies()
myenv.add_conda_package('scikit-learn')
# Save the environment config as a .yml file
env_file = os.path.join(experiment_folder,"diabetes_env.yml")
with open(env_file,"w") as f:
f.write(myenv.serialize_to_string())
print("Saved dependency info in", env_file)
# Print the .yml file
with open(env_file,"r") as f:
print(f.read())
```
Now you're ready to deploy. We'll deploy the container a service named **diabetes-service**. The deployment process includes the following steps:
1. Define an inference configuration, which includes the scoring and environment files required to load and use the model.
2. Define a deployment configuration that defines the execution environment in which the service will be hosted. In this case, an Azure Container Instance.
3. Deploy the model as a web service.
4. Verify the status of the deployed service.
> **More Information**: For more details about model deployment, and options for target execution environments, see the [documentation](https://docs.microsoft.com/azure/machine-learning/how-to-deploy-and-where).
Deployment will take some time as it first runs a process to create a container image, and then runs a process to create a web service based on the image. When deployment has completed successfully, you'll see a status of **Healthy**.
```
from azureml.core.webservice import AciWebservice
from azureml.core.model import InferenceConfig
# Configure the scoring environment
inference_config = InferenceConfig(runtime= "python",
entry_script=script_file,
conda_file=env_file)
deployment_config = AciWebservice.deploy_configuration(cpu_cores = 1, memory_gb = 1)
service_name = "diabetes-service"
service = Model.deploy(ws, service_name, [model], inference_config, deployment_config)
service.wait_for_deployment(True)
print(service.state)
```
Hopefully, the deployment has been successful and you can see a status of **Healthy**. If not, you can use the following code to check the status and get the service logs to help you troubleshoot.
```
print(service.state)
print(service.get_logs())
# If you need to make a change and redeploy, you may need to delete unhealthy service using the following code:
#service.delete()
```
Take a look at your workspace in [Azure ML Studio](https://ml.azure.com) and view the **Endpoints** page, which shows the deployed services in your workspace.
You can also retrieve the names of web services in your workspace by running the following code:
```
for webservice_name in ws.webservices:
print(webservice_name)
```
## Use the Web Service
With the service deployed, now you can consume it from a client application.
```
import json
x_new = [[2,180,74,24,21,23.9091702,1.488172308,22]]
print ('Patient: {}'.format(x_new[0]))
# Convert the array to a serializable list in a JSON document
input_json = json.dumps({"data": x_new})
# Call the web service, passing the input data (the web service will also accept the data in binary format)
predictions = service.run(input_data = input_json)
# Get the predicted class - it'll be the first (and only) one.
predicted_classes = json.loads(predictions)
print(predicted_classes[0])
```
You can also send multiple patient observations to the service, and get back a prediction for each one.
```
import json
# This time our input is an array of two feature arrays
x_new = [[2,180,74,24,21,23.9091702,1.488172308,22],
[0,148,58,11,179,39.19207553,0.160829008,45]]
# Convert the array or arrays to a serializable list in a JSON document
input_json = json.dumps({"data": x_new})
# Call the web service, passing the input data
predictions = service.run(input_data = input_json)
# Get the predicted classes.
predicted_classes = json.loads(predictions)
for i in range(len(x_new)):
print ("Patient {}".format(x_new[i]), predicted_classes[i] )
```
The code above uses the Azure ML SDK to connect to the containerized web service and use it to generate predictions from your diabetes classification model. In production, a model is likely to be consumed by business applications that do not use the Azure ML SDK, but simply make HTTP requests to the web service.
Let's determine the URL to which these applications must submit their requests:
```
endpoint = service.scoring_uri
print(endpoint)
```
Now that you know the endpoint URI, an application can simply make an HTTP request, sending the patient data in JSON (or binary) format, and receive back the predicted class(es).
```
import requests
import json
x_new = [[2,180,74,24,21,23.9091702,1.488172308,22],
[0,148,58,11,179,39.19207553,0.160829008,45]]
# Convert the array to a serializable list in a JSON document
input_json = json.dumps({"data": x_new})
# Set the content type
headers = { 'Content-Type':'application/json' }
predictions = requests.post(endpoint, input_json, headers = headers)
predicted_classes = json.loads(predictions.json())
for i in range(len(x_new)):
print ("Patient {}".format(x_new[i]), predicted_classes[i] )
```
You've deployed your web service as an Azure Container Instance (ACI) service that requires no authentication. This is fine for development and testing, but for production you should consider deploying to an Azure Kubernetes Service (AKS) cluster and enabling authentication. This would require REST requests to include an **Authorization** header.
## Delete the Service
When you no longer need your service, you should delete it to avoid incurring unecessary charges.
```
service.delete()
print ('Service deleted.')
```
For more information about publishing a model as a service, see the [documentation](https://docs.microsoft.com/azure/machine-learning/how-to-deploy-and-where)
|
github_jupyter
|
import azureml.core
from azureml.core import Workspace
# Load the workspace from the saved config file
ws = Workspace.from_config()
print('Ready to use Azure ML {} to work with {}'.format(azureml.core.VERSION, ws.name))
from azureml.core import Experiment
from azureml.core import Model
import pandas as pd
import numpy as np
import joblib
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import roc_auc_score
from sklearn.metrics import roc_curve
# Create an Azure ML experiment in your workspace
experiment = Experiment(workspace = ws, name = "diabetes-training")
run = experiment.start_logging()
print("Starting experiment:", experiment.name)
# load the diabetes dataset
print("Loading Data...")
diabetes = pd.read_csv('data/diabetes.csv')
# Separate features and labels
X, y = diabetes[['Pregnancies','PlasmaGlucose','DiastolicBloodPressure','TricepsThickness','SerumInsulin','BMI','DiabetesPedigree','Age']].values, diabetes['Diabetic'].values
# Split data into training set and test set
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=0)
# Train a decision tree model
print('Training a decision tree model')
model = DecisionTreeClassifier().fit(X_train, y_train)
# calculate accuracy
y_hat = model.predict(X_test)
acc = np.average(y_hat == y_test)
print('Accuracy:', acc)
run.log('Accuracy', np.float(acc))
# calculate AUC
y_scores = model.predict_proba(X_test)
auc = roc_auc_score(y_test,y_scores[:,1])
print('AUC: ' + str(auc))
run.log('AUC', np.float(auc))
# Save the trained model
model_file = 'diabetes_model.pkl'
joblib.dump(value=model, filename=model_file)
run.upload_file(name = 'outputs/' + model_file, path_or_stream = './' + model_file)
# Complete the run
run.complete()
# Register the model
run.register_model(model_path='outputs/diabetes_model.pkl', model_name='diabetes_model',
tags={'Training context':'Inline Training'},
properties={'AUC': run.get_metrics()['AUC'], 'Accuracy': run.get_metrics()['Accuracy']})
print('Model trained and registered.')
from azureml.core import Model
for model in Model.list(ws):
print(model.name, 'version:', model.version)
for tag_name in model.tags:
tag = model.tags[tag_name]
print ('\t',tag_name, ':', tag)
for prop_name in model.properties:
prop = model.properties[prop_name]
print ('\t',prop_name, ':', prop)
print('\n')
model = ws.models['diabetes_model']
print(model.name, 'version', model.version)
import os
folder_name = 'diabetes_service'
# Create a folder for the web service files
experiment_folder = './' + folder_name
os.makedirs(experiment_folder, exist_ok=True)
print(folder_name, 'folder created.')
# Set path for scoring script
script_file = os.path.join(experiment_folder,"score_diabetes.py")
%%writefile $script_file
import json
import joblib
import numpy as np
from azureml.core.model import Model
# Called when the service is loaded
def init():
global model
# Get the path to the deployed model file and load it
model_path = Model.get_model_path('diabetes_model')
model = joblib.load(model_path)
# Called when a request is received
def run(raw_data):
# Get the input data as a numpy array
data = np.array(json.loads(raw_data)['data'])
# Get a prediction from the model
predictions = model.predict(data)
# Get the corresponding classname for each prediction (0 or 1)
classnames = ['not-diabetic', 'diabetic']
predicted_classes = []
for prediction in predictions:
predicted_classes.append(classnames[prediction])
# Return the predictions as JSON
return json.dumps(predicted_classes)
from azureml.core.conda_dependencies import CondaDependencies
# Add the dependencies for our model (AzureML defaults is already included)
myenv = CondaDependencies()
myenv.add_conda_package('scikit-learn')
# Save the environment config as a .yml file
env_file = os.path.join(experiment_folder,"diabetes_env.yml")
with open(env_file,"w") as f:
f.write(myenv.serialize_to_string())
print("Saved dependency info in", env_file)
# Print the .yml file
with open(env_file,"r") as f:
print(f.read())
from azureml.core.webservice import AciWebservice
from azureml.core.model import InferenceConfig
# Configure the scoring environment
inference_config = InferenceConfig(runtime= "python",
entry_script=script_file,
conda_file=env_file)
deployment_config = AciWebservice.deploy_configuration(cpu_cores = 1, memory_gb = 1)
service_name = "diabetes-service"
service = Model.deploy(ws, service_name, [model], inference_config, deployment_config)
service.wait_for_deployment(True)
print(service.state)
print(service.state)
print(service.get_logs())
# If you need to make a change and redeploy, you may need to delete unhealthy service using the following code:
#service.delete()
for webservice_name in ws.webservices:
print(webservice_name)
import json
x_new = [[2,180,74,24,21,23.9091702,1.488172308,22]]
print ('Patient: {}'.format(x_new[0]))
# Convert the array to a serializable list in a JSON document
input_json = json.dumps({"data": x_new})
# Call the web service, passing the input data (the web service will also accept the data in binary format)
predictions = service.run(input_data = input_json)
# Get the predicted class - it'll be the first (and only) one.
predicted_classes = json.loads(predictions)
print(predicted_classes[0])
import json
# This time our input is an array of two feature arrays
x_new = [[2,180,74,24,21,23.9091702,1.488172308,22],
[0,148,58,11,179,39.19207553,0.160829008,45]]
# Convert the array or arrays to a serializable list in a JSON document
input_json = json.dumps({"data": x_new})
# Call the web service, passing the input data
predictions = service.run(input_data = input_json)
# Get the predicted classes.
predicted_classes = json.loads(predictions)
for i in range(len(x_new)):
print ("Patient {}".format(x_new[i]), predicted_classes[i] )
endpoint = service.scoring_uri
print(endpoint)
import requests
import json
x_new = [[2,180,74,24,21,23.9091702,1.488172308,22],
[0,148,58,11,179,39.19207553,0.160829008,45]]
# Convert the array to a serializable list in a JSON document
input_json = json.dumps({"data": x_new})
# Set the content type
headers = { 'Content-Type':'application/json' }
predictions = requests.post(endpoint, input_json, headers = headers)
predicted_classes = json.loads(predictions.json())
for i in range(len(x_new)):
print ("Patient {}".format(x_new[i]), predicted_classes[i] )
service.delete()
print ('Service deleted.')
| 0.595022 | 0.964556 |
```
import matplotlib.pyplot as plt
import matplotlib.cm as cmap
%matplotlib inline
import numpy as np
np.random.seed(206)
import theano
import theano.tensor as tt
import pymc3 as pm
```
# Mean and Covariance Functions
A large set of mean and covariance functions are available in PyMC3. It is relatively easy to define custom mean and covariance functions. Since PyMC3 uses Theano, their gradients do not need to be defined by the user.
## Mean functions
The following mean functions are available in PyMC3.
- `gp.mean.Zero`
- `gp.mean.Constant`
- `gp.mean.Linear`
All follow a similar usage pattern. First, the mean function is specified. Then it can be evaluated over some inputs. The first two mean functions are very simple. Regardless of the inputs, `gp.mean.Zero` returns a vector of zeros with the same length as the number of input values.
### Zero
```
zero_func = pm.gp.mean.Zero()
X = np.linspace(0, 1, 5)[:, None]
print(zero_func(X).eval())
```
The default mean functions for all GP implementations in PyMC3 is `Zero`.
### Constant
`gp.mean.Constant` returns a vector whose value is provided.
```
const_func = pm.gp.mean.Constant(25.2)
print(const_func(X).eval())
```
As long as the shape matches the input it will receive, `gp.mean.Constant` can also accept a Theano tensor or vector of PyMC3 random variables.
```
const_func_vec = pm.gp.mean.Constant(tt.ones(5))
print(const_func_vec(X).eval())
```
### Linear
`gp.mean.Linear` is a takes as input a matrix of coefficients and a vector of intercepts (or a slope and scalar intercept in one dimension).
```
beta = np.random.randn(3)
b = 0.0
lin_func = pm.gp.mean.Linear(coeffs=beta, intercept=b)
X = np.random.randn(5, 3)
print(lin_func(X).eval())
```
## Defining a custom mean function
To define a custom mean function, subclass `gp.mean.Mean`, and provide `__call__` and `__init__` methods. For example, the code for the `Constant` mean function is
```python
import theano.tensor as tt
class Constant(pm.gp.mean.Mean):
def __init__(self, c=0):
Mean.__init__(self)
self.c = c
def __call__(self, X):
return tt.alloc(1.0, X.shape[0]) * self.c
```
Remember that Theano must be used instead of NumPy.
## Covariance functions
PyMC3 contains a much larger suite of built-in covariance functions. The following shows functions drawn from a GP prior with a given covariance function, and demonstrates how composite covariance functions can be constructed with Python operators in a straightforward manner. Our goal was for our API to follow kernel algebra (see Ch.4 of Rassmussen + Williams) as closely as possible. See the main documentation page for an overview on their usage in PyMC3.
### Exponentiated Quadratic
$$
k(x, x') = \mathrm{exp}\left[ -\frac{(x - x')^2}{2 \ell^2} \right]
$$
```
lengthscale = 0.2
eta = 2.0
cov = eta**2 * pm.gp.cov.ExpQuad(1, lengthscale)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
```
### Two (and higher) Dimensional Inputs
#### Both dimensions active
It is easy to define kernels with higher dimensional inputs. Notice that the ```ls``` (lengthscale) parameter is an array of length 2. Lists of PyMC3 random variables can be used for automatic relevance determination (ARD).
```
x1, x2 = np.meshgrid(np.linspace(0,1,10), np.arange(1,4))
X2 = np.concatenate((x1.reshape((30,1)), x2.reshape((30,1))), axis=1)
ls = np.array([0.2, 1.0])
cov = pm.gp.cov.ExpQuad(input_dim=2, ls=ls)
K = theano.function([], cov(X2))()
m = plt.imshow(K, cmap="inferno", interpolation='none'); plt.colorbar(m);
```
#### One dimension active
```
ls = 0.2
cov = pm.gp.cov.ExpQuad(input_dim=2, ls=ls, active_dims=[0])
K = theano.function([], cov(X2))()
m = plt.imshow(K, cmap="inferno", interpolation='none'); plt.colorbar(m);
```
#### Product of covariances over different dimensions
Note that this is equivalent to using a two dimensional `ExpQuad` with separate lengthscale parameters for each dimension.
```
ls1 = 0.2
ls2 = 1.0
cov1 = pm.gp.cov.ExpQuad(2, ls1, active_dims=[0])
cov2 = pm.gp.cov.ExpQuad(2, ls2, active_dims=[1])
cov = cov1 * cov2
K = theano.function([], cov(X2))()
m = plt.imshow(K, cmap="inferno", interpolation='none'); plt.colorbar(m);
```
### White Noise
$$
k(x, x') = \sigma^2 \mathrm{I}_{xx}
$$
```
sigma = 2.0
cov = pm.gp.cov.WhiteNoise(sigma)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
```
### Constant
$$
k(x, x') = c
$$
```
c = 2.0
cov = pm.gp.cov.Constant(c)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
```
### Rational Quadratic
$$
k(x, x') = \left(1 + \frac{(x - x')^2}{2\alpha\ell^2} \right)^{-\alpha}
$$
```
alpha = 0.1
ls = 0.2
tau = 2.0
cov = tau * pm.gp.cov.RatQuad(1, ls, alpha)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
```
### Exponential
$$
k(x, x') = \mathrm{exp}\left[ -\frac{||x - x'||}{2\ell^2} \right]
$$
```
inverse_lengthscale = 5
cov = pm.gp.cov.Exponential(1, ls_inv=inverse_lengthscale)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
```
### Matern 5/2
$$
k(x, x') = \left(1 + \frac{\sqrt{5(x - x')^2}}{\ell} +
\frac{5(x-x')^2}{3\ell^2}\right)
\mathrm{exp}\left[ - \frac{\sqrt{5(x - x')^2}}{\ell} \right]
$$
```
ls = 0.2
tau = 2.0
cov = tau * pm.gp.cov.Matern52(1, ls)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
```
### Matern 3/2
$$
k(x, x') = \left(1 + \frac{\sqrt{3(x - x')^2}}{\ell}\right)
\mathrm{exp}\left[ - \frac{\sqrt{3(x - x')^2}}{\ell} \right]
$$
```
ls = 0.2
tau = 2.0
cov = tau * pm.gp.cov.Matern32(1, ls)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
```
### Cosine
$$
k(x, x') = \mathrm{cos}\left( 2 \pi \frac{||x - x'||}{ \ell^2} \right)
$$
```
period = 0.5
cov = pm.gp.cov.Cosine(1, period)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
```
### Linear
$$
k(x, x') = (x - c)(x' - c)
$$
```
c = 1.0
tau = 2.0
cov = tau * pm.gp.cov.Linear(1, c)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
```
### Polynomial
$$
k(x, x') = [(x - c)(x' - c) + \mathrm{offset}]^{d}
$$
```
c = 1.0
d = 3
offset = 1.0
tau = 0.1
cov = tau * pm.gp.cov.Polynomial(1, c=c, d=d, offset=offset)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
```
### Multiplication with a precomputed covariance matrix
A covariance function ```cov``` can be multiplied with numpy matrix, ```K_cos```, as long as the shapes are appropriate.
```
# first evaluate a covariance function into a matrix
period = 0.2
cov_cos = pm.gp.cov.Cosine(1, period)
K_cos = theano.function([], cov_cos(X))()
# now multiply it with a covariance *function*
cov = pm.gp.cov.Matern32(1, 0.5) * K_cos
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
```
### Applying an arbitary warping function on the inputs
If $k(x, x')$ is a valid covariance function, then so is $k(w(x), w(x'))$.
The first argument of the warping function must be the input ```X```. The remaining arguments can be anything else, including random variables.
```
def warp_func(x, a, b, c):
return 1.0 + x + (a * tt.tanh(b * (x - c)))
a = 1.0
b = 5.0
c = 1.0
cov_m52 = pm.gp.cov.ExpQuad(1, 0.2)
cov = pm.gp.cov.WarpedInput(1, warp_func=warp_func, args=(a,b,c), cov_func=cov_m52)
X = np.linspace(0, 2, 400)[:,None]
wf = theano.function([], warp_func(X.flatten(), a,b,c))()
plt.plot(X, wf); plt.xlabel("X"); plt.ylabel("warp_func(X)");
plt.title("The warping function used");
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
```
### Constructing `Periodic` using `WarpedInput`
The `WarpedInput` kernel can be used to create the `Periodic` covariance. This covariance models functions that are periodic, but are not an exact sine wave (like the `Cosine` kernel is).
The periodic kernel is given by
$$
k(x, x') = \exp\left( -\frac{2 \sin^{2}(\pi |x - x'|\frac{1}{T})}{\ell^2} \right)
$$
Where T is the period, and $\ell$ is the lengthscale. It can be derived by warping the input of an `ExpQuad` kernel with the function $\mathbf{u}(x) = (\sin(2\pi x \frac{1}{T})\,, \cos(2 \pi x \frac{1}{T}))$. Here we use the `WarpedInput` kernel to construct it.
The input `X`, which is defined at the top of this page, is 2 "seconds" long. We use a period of $0.5$, which means that functions
drawn from this GP prior will repeat 4 times over 2 seconds.
```
def mapping(x, T):
c = 2.0 * np.pi * (1.0 / T)
u = tt.concatenate((tt.sin(c*x), tt.cos(c*x)), 1)
return u
T = 0.6
ls = 0.4
# note that the input of the covariance function taking
# the inputs is 2 dimensional
cov_exp = pm.gp.cov.ExpQuad(2, ls)
cov = pm.gp.cov.WarpedInput(1, cov_func=cov_exp,
warp_func=mapping, args=(T, ))
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
```
### Periodic
There is no need to construct the periodic covariance this way every time. A more efficient implementation of this covariance function is built in.
```
period = 0.6
ls = 0.4
cov = pm.gp.cov.Periodic(1, period=period, ls=ls)
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
```
### Gibbs
The Gibbs covariance function applies a positive definite warping function to the lengthscale. Similarly to ```WarpedInput```, the lengthscale warping function can be specified with parameters that are either fixed or random variables.
```
def tanh_func(x, ls1, ls2, w, x0):
"""
ls1: left saturation value
ls2: right saturation value
w: transition width
x0: transition location.
"""
return (ls1 + ls2) / 2.0 - (ls1 - ls2) / 2.0 * tt.tanh((x - x0) / w)
ls1 = 0.05
ls2 = 0.6
w = 0.3
x0 = 1.0
cov = pm.gp.cov.Gibbs(1, tanh_func, args=(ls1, ls2, w, x0))
wf = theano.function([], tanh_func(X, ls1, ls2, w, x0))()
plt.plot(X, wf); plt.ylabel("tanh_func(X)"); plt.xlabel("X"); plt.title("Lengthscale as a function of X");
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
```
### Defining a custom covariance function
Covariance function objects in PyMC3 need to implement the `__init__`, `diag`, and `full` methods, and subclass `gp.cov.Covariance`. `diag` returns only the diagonal of the covariance matrix, and `full` returns the full covariance matrix. The `full` method has two inputs `X` and `Xs`. `full(X)` returns the square covariance matrix, and `full(X, Xs)` returns the cross-covariances between the two sets of inputs.
For example, here is the implementation of the `WhiteNoise` covariance function:
```python
class WhiteNoise(pm.gp.cov.Covariance):
def __init__(self, sigma):
super(WhiteNoise, self).__init__(1, None)
self.sigma = sigma
def diag(self, X):
return tt.alloc(tt.square(self.sigma), X.shape[0])
def full(self, X, Xs=None):
if Xs is None:
return tt.diag(self.diag(X))
else:
return tt.alloc(0.0, X.shape[0], Xs.shape[0])
```
If we have forgotten an important covariance or mean function, please feel free to submit a pull request!
|
github_jupyter
|
import matplotlib.pyplot as plt
import matplotlib.cm as cmap
%matplotlib inline
import numpy as np
np.random.seed(206)
import theano
import theano.tensor as tt
import pymc3 as pm
zero_func = pm.gp.mean.Zero()
X = np.linspace(0, 1, 5)[:, None]
print(zero_func(X).eval())
const_func = pm.gp.mean.Constant(25.2)
print(const_func(X).eval())
const_func_vec = pm.gp.mean.Constant(tt.ones(5))
print(const_func_vec(X).eval())
beta = np.random.randn(3)
b = 0.0
lin_func = pm.gp.mean.Linear(coeffs=beta, intercept=b)
X = np.random.randn(5, 3)
print(lin_func(X).eval())
import theano.tensor as tt
class Constant(pm.gp.mean.Mean):
def __init__(self, c=0):
Mean.__init__(self)
self.c = c
def __call__(self, X):
return tt.alloc(1.0, X.shape[0]) * self.c
lengthscale = 0.2
eta = 2.0
cov = eta**2 * pm.gp.cov.ExpQuad(1, lengthscale)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
x1, x2 = np.meshgrid(np.linspace(0,1,10), np.arange(1,4))
X2 = np.concatenate((x1.reshape((30,1)), x2.reshape((30,1))), axis=1)
ls = np.array([0.2, 1.0])
cov = pm.gp.cov.ExpQuad(input_dim=2, ls=ls)
K = theano.function([], cov(X2))()
m = plt.imshow(K, cmap="inferno", interpolation='none'); plt.colorbar(m);
ls = 0.2
cov = pm.gp.cov.ExpQuad(input_dim=2, ls=ls, active_dims=[0])
K = theano.function([], cov(X2))()
m = plt.imshow(K, cmap="inferno", interpolation='none'); plt.colorbar(m);
ls1 = 0.2
ls2 = 1.0
cov1 = pm.gp.cov.ExpQuad(2, ls1, active_dims=[0])
cov2 = pm.gp.cov.ExpQuad(2, ls2, active_dims=[1])
cov = cov1 * cov2
K = theano.function([], cov(X2))()
m = plt.imshow(K, cmap="inferno", interpolation='none'); plt.colorbar(m);
sigma = 2.0
cov = pm.gp.cov.WhiteNoise(sigma)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
c = 2.0
cov = pm.gp.cov.Constant(c)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
alpha = 0.1
ls = 0.2
tau = 2.0
cov = tau * pm.gp.cov.RatQuad(1, ls, alpha)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
inverse_lengthscale = 5
cov = pm.gp.cov.Exponential(1, ls_inv=inverse_lengthscale)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
ls = 0.2
tau = 2.0
cov = tau * pm.gp.cov.Matern52(1, ls)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
ls = 0.2
tau = 2.0
cov = tau * pm.gp.cov.Matern32(1, ls)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
period = 0.5
cov = pm.gp.cov.Cosine(1, period)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
c = 1.0
tau = 2.0
cov = tau * pm.gp.cov.Linear(1, c)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
c = 1.0
d = 3
offset = 1.0
tau = 0.1
cov = tau * pm.gp.cov.Polynomial(1, c=c, d=d, offset=offset)
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
# first evaluate a covariance function into a matrix
period = 0.2
cov_cos = pm.gp.cov.Cosine(1, period)
K_cos = theano.function([], cov_cos(X))()
# now multiply it with a covariance *function*
cov = pm.gp.cov.Matern32(1, 0.5) * K_cos
X = np.linspace(0, 2, 200)[:,None]
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
def warp_func(x, a, b, c):
return 1.0 + x + (a * tt.tanh(b * (x - c)))
a = 1.0
b = 5.0
c = 1.0
cov_m52 = pm.gp.cov.ExpQuad(1, 0.2)
cov = pm.gp.cov.WarpedInput(1, warp_func=warp_func, args=(a,b,c), cov_func=cov_m52)
X = np.linspace(0, 2, 400)[:,None]
wf = theano.function([], warp_func(X.flatten(), a,b,c))()
plt.plot(X, wf); plt.xlabel("X"); plt.ylabel("warp_func(X)");
plt.title("The warping function used");
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
def mapping(x, T):
c = 2.0 * np.pi * (1.0 / T)
u = tt.concatenate((tt.sin(c*x), tt.cos(c*x)), 1)
return u
T = 0.6
ls = 0.4
# note that the input of the covariance function taking
# the inputs is 2 dimensional
cov_exp = pm.gp.cov.ExpQuad(2, ls)
cov = pm.gp.cov.WarpedInput(1, cov_func=cov_exp,
warp_func=mapping, args=(T, ))
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
period = 0.6
ls = 0.4
cov = pm.gp.cov.Periodic(1, period=period, ls=ls)
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
def tanh_func(x, ls1, ls2, w, x0):
"""
ls1: left saturation value
ls2: right saturation value
w: transition width
x0: transition location.
"""
return (ls1 + ls2) / 2.0 - (ls1 - ls2) / 2.0 * tt.tanh((x - x0) / w)
ls1 = 0.05
ls2 = 0.6
w = 0.3
x0 = 1.0
cov = pm.gp.cov.Gibbs(1, tanh_func, args=(ls1, ls2, w, x0))
wf = theano.function([], tanh_func(X, ls1, ls2, w, x0))()
plt.plot(X, wf); plt.ylabel("tanh_func(X)"); plt.xlabel("X"); plt.title("Lengthscale as a function of X");
K = cov(X).eval()
plt.figure(figsize=(14,4))
plt.plot(X, pm.MvNormal.dist(mu=np.zeros(K.shape[0]), cov=K).random(size=3).T);
plt.xlabel("X");
class WhiteNoise(pm.gp.cov.Covariance):
def __init__(self, sigma):
super(WhiteNoise, self).__init__(1, None)
self.sigma = sigma
def diag(self, X):
return tt.alloc(tt.square(self.sigma), X.shape[0])
def full(self, X, Xs=None):
if Xs is None:
return tt.diag(self.diag(X))
else:
return tt.alloc(0.0, X.shape[0], Xs.shape[0])
| 0.528777 | 0.983486 |
# Machine Learning Engineer Nanodegree
## Model Evaluation & Validation
## Project: Predicting Boston Housing Prices
## Getting Started
In this project, you will evaluate the performance and predictive power of a model that has been trained and tested on data collected from homes in suburbs of Boston, Massachusetts. A model trained on this data that is seen as a *good fit* could then be used to make certain predictions about a home — in particular, its monetary value. This model would prove to be invaluable for someone like a real estate agent who could make use of such information on a daily basis.
The dataset for this project originates from the [UCI Machine Learning Repository](https://archive.ics.uci.edu/ml/datasets/Housing). The Boston housing data was collected in 1978 and each of the 506 entries represent aggregated data about 14 features for homes from various suburbs in Boston, Massachusetts. For the purposes of this project, the following preprocessing steps have been made to the dataset:
- 16 data points have an `'MEDV'` value of 50.0. These data points likely contain **missing or censored values** and have been removed.
- 1 data point has an `'RM'` value of 8.78. This data point can be considered an **outlier** and has been removed.
- The features `'RM'`, `'LSTAT'`, `'PTRATIO'`, and `'MEDV'` are essential. The remaining **non-relevant features** have been excluded.
- The feature `'MEDV'` has been **multiplicatively scaled** to account for 35 years of market inflation.
```
# Import libraries necessary for this project
from sklearn.model_selection import GridSearchCV, ShuffleSplit, train_test_split
from sklearn.tree import DecisionTreeRegressor
from sklearn.metrics import make_scorer, r2_score
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
# Import supplementary visualizations code visuals.py
import visuals as vs
# Pretty display for notebooks
%matplotlib inline
# Load the Boston housing dataset
data = pd.read_csv('housing.csv')
prices = data['MEDV']
features = data.drop('MEDV', axis = 1)
```
### Implementation: Calculate Statistics
In the code cell below, you will need to implement the following:
- Calculate the minimum, maximum, mean, median, and standard deviation of `'MEDV'`, which is stored in `prices`.
- Store each calculation in their respective variable.
```
minimum_price = min(prices)
maximum_price = max(prices)
mean_price = prices.mean()
median_price = prices.median()
std_price = np.std(prices)
# Show the calculated statistics
print("Statistics for Boston housing dataset:\n")
print("Minimum price: ${}".format(minimum_price))
print("Maximum price: ${}".format(maximum_price))
print("Mean price: ${:.2f}".format(mean_price))
print("Median price ${}".format(median_price))
print("Standard deviation of prices: ${:.2f}".format(std_price))
```
### Question 1 - Feature Observation
As a reminder, we are using three features from the Boston housing dataset: `'RM'`, `'LSTAT'`, and `'PTRATIO'`. For each data point (neighborhood):
- `'RM'` is the average number of rooms among homes in the neighborhood.
- `'LSTAT'` is the percentage of homeowners in the neighborhood considered "lower class" (working poor).
- `'PTRATIO'` is the ratio of students to teachers in primary and secondary schools in the neighborhood.
** Using your intuition, for each of the three features above, do you think that an increase in the value of that feature would lead to an **increase** in the value of `'MEDV'` or a **decrease** in the value of `'MEDV'`? Justify your answer for each.**
**Answer:
1) An increase in the value of the feature "RM" will increase the worth of the house, this because we have to suppose that having more rooms means having a bigger house.
Even if this is not necessarly true, without having the square meters as a feature we have to rely on the number of rooms.
2) By increasing the value of "LSTAT" the worth of the house will decrease, because this means that the house is not in a rich area.
3) An increase in the value of "PTRATIO" will decrease the worth of the house, because this means having less schools or smaller schools, consequently school's rooms will be overcrowded.
As confirmation to my hypothesis the graph below, shows the price for every feature variation.
**
```
# code to visualize my theory on question 1
def show_price_on_feature_variation():
plt.figure(figsize=(15, 5))
for i, col in enumerate(features.columns):
plt.subplot(1, 3, i+1)
plt.plot(data[col], prices, 'x')
plt.title('%s x MEDV' % col)
plt.xlabel(col)
plt.ylabel('MEDV')
show_price_on_feature_variation()
```
### Implementation: Shuffle and Split Data
For the code cell below, you will need to implement the following:
- Use `train_test_split` from `sklearn.cross_validation` to shuffle and split the `features` and `prices` data into training and testing sets.
- Split the data into 80% training and 20% testing.
- Set the `random_state` for `train_test_split` to a value of your choice. This ensures results are consistent.
- Assign the train and testing splits to `X_train`, `X_test`, `y_train`, and `y_test`.
```
X_train, X_test, y_train, y_test = train_test_split(features, prices, test_size=0.2, random_state=42)
```
### Question 2 - Training and Testing
* What is the benefit to splitting a dataset into some ratio of training and testing subsets for a learning algorithm?
**Answer:
To train the model we must use only the training set and leave some data as testing set, otherwise if we use the whole data available, we don't have a way to verify how the model performs with unseen data.
For example if we train our model without testing it we might get a really good fit with high score ratio, but when we test it with some other data it is not predicting as good as we were expecting.
In this case we talk about overfitting which means the model is not generalizing well but instead it memorizes the data.
In case of overfitting then, the only way to detect it is by looking how the model reacts with data it has never seen before (testing data).
For all these reasons we need to split our data, so we can see how the model reacts with data it has never seen before and its capability to generalize them.**
----
## Analyzing Model Performance
### Learning Curves
The following code cell produces four graphs for a decision tree model with different maximum depths. Each graph visualizes the learning curves of the model for both training and testing as the size of the training set is increased. Note that the shaded region of a learning curve denotes the uncertainty of that curve (measured as the standard deviation). The model is scored on both the training and testing sets using R<sup>2</sup>, the coefficient of determination.
```
# Produce learning curves for varying training set sizes and maximum depths
vs.ModelLearning(features, prices)
```
### Question 3 - Learning the Data
* Choose one of the graphs above and state the maximum depth for the model.
* What happens to the score of the training curve as more training points are added? What about the testing curve?
* Would having more training points benefit the model?
**Answer:
Maximum depth = 3
By increasing the number of training points, the score of the training curve decreases, more gradually after 50 training points.
About the testing curve, it's score has a big increase during the first 50 training points, after them it increases gradually.
The two curves are converging in a good score ratio by increasing the number of training points, but as we can see from the graph, after 300 training points the convergence between the 2 curves is imperceptible, so in conclusion having more training points will not significantly benefit the model.
**
### Complexity Curves
The following code cell produces a graph for a decision tree model that has been trained and validated on the training data using different maximum depths. The graph produces two complexity curves — one for training and one for validation. Similar to the **learning curves**, the shaded regions of both the complexity curves denote the uncertainty in those curves, and the model is scored on both the training and validation sets using the `performance_metric` function.
```
vs.ModelComplexity(X_train, y_train)
```
### Question 4 - Bias-Variance Tradeoff
* When the model is trained with a maximum depth of 1, does the model suffer from high bias or from high variance?
* How about when the model is trained with a maximum depth of 10? What visual cues in the graph justify your conclusions?
**Answer:
1) It suffers from high bias.
2) At maximum depth of 10 the model suffers from high variance, because the gap between the training score and the validation score is high, which means the model is just memorizing the training data and is not generalizing well the data.**
### Question 5 - Best-Guess Optimal Model
* Which maximum depth do you think results in a model that best generalizes to unseen data?
* What intuition lead you to this answer?
**Answer:
The model with maximum depth of 4 generalizes the best, this because the score of training and validation are high but also not distant from each other (validation score with maximum depth of 3 is a bit less).**
-----
## Evaluating Model Performance
In this final section of the project, you will construct a model and make a prediction on the client's feature set using an optimized model from `fit_model`.
### Question 6 - Grid Search
* What is the grid search technique?
* How it can be applied to optimize a learning algorithm?
**Answer:
The grid search technique is a technique that tests differrent hyperparameters to find out which is the best based on the score ratio.
An example would be using the maximum depth with value 1, 3, 7 and 10 for a decision tree.
The grid search technique will tests each one of these values and will pick the best based on the score ratio.
It is very usefull because we can automatize the process instead of changing the parameters to see which configuration fits the best the data.
**
### Question 7 - Cross-Validation
* What is the k-fold cross-validation training technique?
* What benefit does this technique provide for grid search when optimizing a model?
**Answer:
The k-fold cross-validation technique is a technique that divides all the training data into groups, called folds and "K" is the number of folds.
Each fold is constituted by two arrays, the first one is related to the training set, and the second one to the test set.
So for example this technique will divide only the training data into 4 (K=4) number of folds and it will return 4 training sets and 4 testing sets.
K-fold technique provides a big benefit to grid search technique because it generate different training and testing sets which are used from the grid search technique to pick up the best configuration for the model.
In this way we can test the model with a large variety of data which help us to generalize it better.
In addiction, K-fold helps to prevent overfitting by providing test sets from the training set.
**
### Implementation: Fitting a Model
The `ShuffleSplit()` implementation below will create 10 (`'n_splits'`) shuffled sets, and for each shuffle, 20% (`'test_size'`) of the data will be used as the *validation set*.
For the `fit_model` function in the code cell below, you will need to implement the following:
- Use [`DecisionTreeRegressor`](http://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeRegressor.html) from `sklearn.tree` to create a decision tree regressor object.
- Assign this object to the `'regressor'` variable.
- Create a dictionary for `'max_depth'` with the values from 1 to 10, and assign this to the `'params'` variable.
- Use [`make_scorer`](http://scikit-learn.org/stable/modules/generated/sklearn.metrics.make_scorer.html) from `sklearn.metrics` to create a scoring function object.
- Pass the `r2_score` function as a parameter to the object.
- Assign this scoring function to the `'scoring_fnc'` variable.
- Use [`GridSearchCV`](http://scikit-learn.org/0.17/modules/generated/sklearn.grid_search.GridSearchCV.html) from `sklearn.grid_search` to create a grid search object.
- Pass the variables `'regressor'`, `'params'`, `'scoring_fnc'`, and `'cv_sets'` as parameters to the object.
- Assign the `GridSearchCV` object to the `'grid'` variable.
```
def fit_model(X, y):
""" Performs grid search over the 'max_depth' parameter for a
decision tree regressor trained on the input data [X, y]. """
cv_sets = ShuffleSplit(X.shape[0], test_size = 0.20, random_state = 0)
regressor = DecisionTreeRegressor()
params = {'max_depth':[1,2,3,4,5,6,7,8,9,10]}
scoring_fnc = make_scorer(r2_score)
grid = GridSearchCV(regressor, params, scoring=scoring_fnc, cv=cv_sets)
grid = grid.fit(X, y)
return grid.best_estimator_
```
### Making Predictions
### Question 8 - Optimal Model
* What maximum depth does the optimal model have? How does this result compare to your guess in **Question 5**?
```
# Fit the training data to the model using grid search
reg = fit_model(X_train, y_train)
# Print the value for 'max_depth'
print("Parameter 'max_depth' is {} for the optimal model.".format(reg.get_params()['max_depth']))
```
**Answer:
The optimal model has the value of maximum depth of 4.
### Question 9 - Predicting Selling Prices
Imagine that you were a real estate agent in the Boston area looking to use this model to help price homes owned by your clients that they wish to sell. You have collected the following information from three of your clients:
| Feature | Client 1 | Client 2 | Client 3 |
| :---: | :---: | :---: | :---: |
| Total number of rooms in home | 5 rooms | 4 rooms | 8 rooms |
| Neighborhood poverty level (as %) | 17% | 32% | 3% |
| Student-teacher ratio of nearby schools | 15-to-1 | 22-to-1 | 12-to-1 |
* What price would you recommend each client sell his/her home at?
* Do these prices seem reasonable given the values for the respective features?
```
# Produce a matrix for client data
client_data = [[5, 17, 15], # Client 1
[4, 32, 22], # Client 2
[8, 3, 12]] # Client 3
# Show predictions
for i, price in enumerate(reg.predict(client_data)):
print("Predicted selling price for Client {}'s home: ${:,.2f}".format(i+1, price))
```
**Answer:
Client1 = $403,025.00.
Client2 = $237,478.72.
Client3 = $931,636.36.
By looking at the output above we can see the minimum, median and maximum value of the price and every feature in our dataset.
The client number 2 has only 4 rooms, Neighborhood poverty level at 32 percent and Student-teacher ratio of 22 to 1.
As we can see the number of rooms is close to the "RM min" value of our dataset and the other 2 features are close to the respective max value.
As we saw in the Data Exploration graph, we were expecting a low price of the house with these features and as we can see, the price is not far from the minimum price in our dataset.
The client number 1 has features more in the average of the dataset, infact the price of his house is in the average of our data.
Same thing for the client number 3 who has 8 rooms and really low level of Neighborhood poverty and Student-teacher ratio, with these features we are expecting a really high price of the house infact it is close to the highest price of our dataset.
As we saw in the Data Exploration section, by increasin the number of rooms the price gets higher and by increasing Neighborhood poverty level or Student-teacher ratio, the price of the house decrease.
In conclusion we can confirm that these prices seem reasonable, given the values for the respective features, because are in the range of our dataset and also preaty close to our minimum, median and maximum value of our price dataset.
**
### Sensitivity
An optimal model is not necessarily a robust model. Sometimes, a model is either too complex or too simple to sufficiently generalize to new data. Sometimes, a model could use a learning algorithm that is not appropriate for the structure of the data given. Other times, the data itself could be too noisy or contain too few samples to allow a model to adequately capture the target variable — i.e., the model is underfitted.
**Run the code cell below to run the `fit_model` function ten times with different training and testing sets to see how the prediction for a specific client changes with respect to the data it's trained on.**
```
vs.PredictTrials(features, prices, fit_model, client_data)
```
### Question 10 - Applicability
* In a few sentences, discuss whether the constructed model should or should not be used in a real-world setting.
- How relevant today is data that was collected from 1978? How important is inflation?
- Are the features present in the data sufficient to describe a home? Do you think factors like quality of apppliances in the home, square feet of the plot area, presence of pool or not etc should factor in?
- Is the model robust enough to make consistent predictions?
- Would data collected in an urban city like Boston be applicable in a rural city?
- Is it fair to judge the price of an individual home based on the characteristics of the entire neighborhood?
**Answer:
This model unfortunatly can not be used in a real world setting.
The collected data are too old and the inflation is a crucial point to consider.
As i said in the question 1 the features are not really enough, for example we have the number of rooms but not the square meters of the area, are missing features like presence of garden, pool, balcony etc..
The data is collected only in Boston, so it can't be used for other countries neither other cities.
The features describe the neighborhood more than the house itself, so for all these reasons the model can not be considered as useful in a real scenario.
**
|
github_jupyter
|
# Import libraries necessary for this project
from sklearn.model_selection import GridSearchCV, ShuffleSplit, train_test_split
from sklearn.tree import DecisionTreeRegressor
from sklearn.metrics import make_scorer, r2_score
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
# Import supplementary visualizations code visuals.py
import visuals as vs
# Pretty display for notebooks
%matplotlib inline
# Load the Boston housing dataset
data = pd.read_csv('housing.csv')
prices = data['MEDV']
features = data.drop('MEDV', axis = 1)
minimum_price = min(prices)
maximum_price = max(prices)
mean_price = prices.mean()
median_price = prices.median()
std_price = np.std(prices)
# Show the calculated statistics
print("Statistics for Boston housing dataset:\n")
print("Minimum price: ${}".format(minimum_price))
print("Maximum price: ${}".format(maximum_price))
print("Mean price: ${:.2f}".format(mean_price))
print("Median price ${}".format(median_price))
print("Standard deviation of prices: ${:.2f}".format(std_price))
# code to visualize my theory on question 1
def show_price_on_feature_variation():
plt.figure(figsize=(15, 5))
for i, col in enumerate(features.columns):
plt.subplot(1, 3, i+1)
plt.plot(data[col], prices, 'x')
plt.title('%s x MEDV' % col)
plt.xlabel(col)
plt.ylabel('MEDV')
show_price_on_feature_variation()
X_train, X_test, y_train, y_test = train_test_split(features, prices, test_size=0.2, random_state=42)
# Produce learning curves for varying training set sizes and maximum depths
vs.ModelLearning(features, prices)
vs.ModelComplexity(X_train, y_train)
def fit_model(X, y):
""" Performs grid search over the 'max_depth' parameter for a
decision tree regressor trained on the input data [X, y]. """
cv_sets = ShuffleSplit(X.shape[0], test_size = 0.20, random_state = 0)
regressor = DecisionTreeRegressor()
params = {'max_depth':[1,2,3,4,5,6,7,8,9,10]}
scoring_fnc = make_scorer(r2_score)
grid = GridSearchCV(regressor, params, scoring=scoring_fnc, cv=cv_sets)
grid = grid.fit(X, y)
return grid.best_estimator_
# Fit the training data to the model using grid search
reg = fit_model(X_train, y_train)
# Print the value for 'max_depth'
print("Parameter 'max_depth' is {} for the optimal model.".format(reg.get_params()['max_depth']))
# Produce a matrix for client data
client_data = [[5, 17, 15], # Client 1
[4, 32, 22], # Client 2
[8, 3, 12]] # Client 3
# Show predictions
for i, price in enumerate(reg.predict(client_data)):
print("Predicted selling price for Client {}'s home: ${:,.2f}".format(i+1, price))
vs.PredictTrials(features, prices, fit_model, client_data)
| 0.880052 | 0.994672 |
For those who decide to implement and experiment with convolutional layers for the second coursework, below a skeleton class and associated test functions for the `fprop`, `bprop` and `grads_wrt_params` methods of the class are included.
The test functions assume that in your implementation of `fprop` for the convolutional layer, outputs are calculated only for 'valid' overlaps of the kernel filters with the input - i.e. without any padding.
It is also assumed that if convolutions with non-unit strides are implemented the default behaviour is to take unit-strides, with the test cases only correct for unit strides in both directions.
```
import mlp.layers as layers
import mlp.initialisers as init
class ConvolutionalLayer(layers.LayerWithParameters):
"""Layer implementing a 2D convolution-based transformation of its inputs.
The layer is parameterised by a set of 2D convolutional kernels, a four
dimensional array of shape
(num_output_channels, num_input_channels, kernel_dim_1, kernel_dim_2)
and a bias vector, a one dimensional array of shape
(num_output_channels,)
i.e. one shared bias per output channel.
Assuming no-padding is applied to the inputs so that outputs are only
calculated for positions where the kernel filters fully overlap with the
inputs, and that unit strides are used the outputs will have spatial extent
output_dim_1 = input_dim_1 - kernel_dim_1 + 1
output_dim_2 = input_dim_2 - kernel_dim_2 + 1
"""
def __init__(self, num_input_channels, num_output_channels,
input_dim_1, input_dim_2,
kernel_dim_1, kernel_dim_2,
kernels_init=init.UniformInit(-0.01, 0.01),
biases_init=init.ConstantInit(0.),
kernels_penalty=None, biases_penalty=None):
"""Initialises a parameterised convolutional layer.
Args:
num_input_channels (int): Number of channels in inputs to
layer (this may be number of colour channels in the input
images if used as the first layer in a model, or the
number of output channels, a.k.a. feature maps, from a
a previous convolutional layer).
num_output_channels (int): Number of channels in outputs
from the layer, a.k.a. number of feature maps.
input_dim_1 (int): Size of first input dimension of each 2D
channel of inputs.
input_dim_2 (int): Size of second input dimension of each 2D
channel of inputs.
kernel_dim_x (int): Size of first dimension of each 2D channel of
kernels.
kernel_dim_y (int): Size of second dimension of each 2D channel of
kernels.
kernels_intialiser: Initialiser for the kernel parameters.
biases_initialiser: Initialiser for the bias parameters.
kernels_penalty: Kernel-dependent penalty term (regulariser) or
None if no regularisation is to be applied to the kernels.
biases_penalty: Biases-dependent penalty term (regulariser) or
None if no regularisation is to be applied to the biases.
"""
self.num_input_channels = num_input_channels
self.num_output_channels = num_output_channels
self.input_dim_1 = input_dim_1
self.input_dim_2 = input_dim_2
self.kernel_dim_1 = kernel_dim_1
self.kernel_dim_2 = kernel_dim_2
self.kernels_init = kernels_init
self.biases_init = biases_init
self.kernels_shape = (
num_output_channels, num_input_channels, kernel_dim_1, kernel_dim_2
)
self.inputs_shape = (
None, num_input_channels, input_dim_1, input_dim_2
)
self.kernels = self.kernels_init(self.kernels_shape)
self.biases = self.biases_init(num_output_channels)
self.kernels_penalty = kernels_penalty
self.biases_penalty = biases_penalty
def fprop(self, inputs):
"""Forward propagates activations through the layer transformation.
For inputs `x`, outputs `y`, kernels `K` and biases `b` the layer
corresponds to `y = conv2d(x, K) + b`.
Args:
inputs: Array of layer inputs of shape
(batch_size, num_input_channels, input_dim_1, input_dim_2).
Returns:
outputs: Array of layer outputs of shape
(batch_size, num_output_channels, output_dim_1, output_dim_2).
"""
raise NotImplementedError()
def bprop(self, inputs, outputs, grads_wrt_outputs):
"""Back propagates gradients through a layer.
Given gradients with respect to the outputs of the layer calculates the
gradients with respect to the layer inputs.
Args:
inputs: Array of layer inputs of shape
(batch_size, num_input_channels, input_dim_1, input_dim_2).
outputs: Array of layer outputs calculated in forward pass of
shape
(batch_size, num_output_channels, output_dim_1, output_dim_2).
grads_wrt_outputs: Array of gradients with respect to the layer
outputs of shape
(batch_size, num_output_channels, output_dim_1, output_dim_2).
Returns:
Array of gradients with respect to the layer inputs of shape
(batch_size, num_input_channels, input_dim_1, input_dim_2).
"""
raise NotImplementedError()
def grads_wrt_params(self, inputs, grads_wrt_outputs):
"""Calculates gradients with respect to layer parameters.
Args:
inputs: array of inputs to layer of shape (batch_size, input_dim)
grads_wrt_to_outputs: array of gradients with respect to the layer
outputs of shape
(batch_size, num_output_channels, output_dim_1, output_dim_2).
Returns:
list of arrays of gradients with respect to the layer parameters
`[grads_wrt_kernels, grads_wrt_biases]`.
"""
raise NotImplementedError()
def params_penalty(self):
"""Returns the parameter dependent penalty term for this layer.
If no parameter-dependent penalty terms are set this returns zero.
"""
params_penalty = 0
if self.kernels_penalty is not None:
params_penalty += self.kernels_penalty(self.kernels)
if self.biases_penalty is not None:
params_penalty += self.biases_penalty(self.biases)
return params_penalty
@property
def params(self):
"""A list of layer parameter values: `[kernels, biases]`."""
return [self.kernels, self.biases]
@params.setter
def params(self, values):
self.kernels = values[0]
self.biases = values[1]
def __repr__(self):
return (
'ConvolutionalLayer(\n'
' num_input_channels={0}, num_output_channels={1},\n'
' input_dim_1={2}, input_dim_2={3},\n'
' kernel_dim_1={4}, kernel_dim_2={5}\n'
')'
.format(self.num_input_channels, self.num_output_channels,
self.input_dim_1, self.input_dim_2, self.kernel_dim_1,
self.kernel_dim_2)
)
```
The three test functions are defined in the cell below. All the functions take as first argument the *class* corresponding to the convolutional layer implementation to be tested (**not** an instance of the class). It is assumed the class being tested has an `__init__` method with at least all of the arguments defined in the skeleton definition above. A boolean second argument to each function can be used to specify if the layer implements a cross-correlation or convolution based operation (see note in [seventh lecture slides](http://www.inf.ed.ac.uk/teaching/courses/mlp/2016/mlp07-cnn.pdf)).
```
import numpy as np
def test_conv_layer_fprop(layer_class, do_cross_correlation=False):
"""Tests `fprop` method of a convolutional layer.
Checks the outputs of `fprop` method for a fixed input against known
reference values for the outputs and raises an AssertionError if
the outputted values are not consistent with the reference values. If
tests are all passed returns True.
Args:
layer_class: Convolutional layer implementation following the
interface defined in the provided skeleton class.
do_cross_correlation: Whether the layer implements an operation
corresponding to cross-correlation (True) i.e kernels are
not flipped before sliding over inputs, or convolution
(False) with filters being flipped.
Raises:
AssertionError: Raised if output of `layer.fprop` is inconsistent
with reference values either in shape or values.
"""
inputs = np.arange(96).reshape((2, 3, 4, 4))
kernels = np.arange(-12, 12).reshape((2, 3, 2, 2))
if do_cross_correlation:
kernels = kernels[:, :, ::-1, ::-1]
biases = np.arange(2)
true_output = np.array(
[[[[ -958., -1036., -1114.],
[-1270., -1348., -1426.],
[-1582., -1660., -1738.]],
[[ 1707., 1773., 1839.],
[ 1971., 2037., 2103.],
[ 2235., 2301., 2367.]]],
[[[-4702., -4780., -4858.],
[-5014., -5092., -5170.],
[-5326., -5404., -5482.]],
[[ 4875., 4941., 5007.],
[ 5139., 5205., 5271.],
[ 5403., 5469., 5535.]]]]
)
layer = layer_class(
num_input_channels=kernels.shape[1],
num_output_channels=kernels.shape[0],
input_dim_1=inputs.shape[2],
input_dim_2=inputs.shape[3],
kernel_dim_1=kernels.shape[2],
kernel_dim_2=kernels.shape[3]
)
layer.params = [kernels, biases]
layer_output = layer.fprop(inputs)
assert layer_output.shape == true_output.shape, (
'Layer fprop gives incorrect shaped output. '
'Correct shape is \n\n{0}\n\n but returned shape is \n\n{1}.'
.format(true_output.shape, layer_output.shape)
)
assert np.allclose(layer_output, true_output), (
'Layer fprop does not give correct output. '
'Correct output is \n\n{0}\n\n but returned output is \n\n{1}.'
.format(true_output, layer_output)
)
return True
def test_conv_layer_bprop(layer_class, do_cross_correlation=False):
"""Tests `bprop` method of a convolutional layer.
Checks the outputs of `bprop` method for a fixed input against known
reference values for the gradients with respect to inputs and raises
an AssertionError if the returned values are not consistent with the
reference values. If tests are all passed returns True.
Args:
layer_class: Convolutional layer implementation following the
interface defined in the provided skeleton class.
do_cross_correlation: Whether the layer implements an operation
corresponding to cross-correlation (True) i.e kernels are
not flipped before sliding over inputs, or convolution
(False) with filters being flipped.
Raises:
AssertionError: Raised if output of `layer.bprop` is inconsistent
with reference values either in shape or values.
"""
inputs = np.arange(96).reshape((2, 3, 4, 4))
kernels = np.arange(-12, 12).reshape((2, 3, 2, 2))
if do_cross_correlation:
kernels = kernels[:, :, ::-1, ::-1]
biases = np.arange(2)
grads_wrt_outputs = np.arange(-20, 16).reshape((2, 2, 3, 3))
outputs = np.array(
[[[[ -958., -1036., -1114.],
[-1270., -1348., -1426.],
[-1582., -1660., -1738.]],
[[ 1707., 1773., 1839.],
[ 1971., 2037., 2103.],
[ 2235., 2301., 2367.]]],
[[[-4702., -4780., -4858.],
[-5014., -5092., -5170.],
[-5326., -5404., -5482.]],
[[ 4875., 4941., 5007.],
[ 5139., 5205., 5271.],
[ 5403., 5469., 5535.]]]]
)
true_grads_wrt_inputs = np.array(
[[[[ 147., 319., 305., 162.],
[ 338., 716., 680., 354.],
[ 290., 608., 572., 294.],
[ 149., 307., 285., 144.]],
[[ 23., 79., 81., 54.],
[ 114., 284., 280., 162.],
[ 114., 272., 268., 150.],
[ 73., 163., 157., 84.]],
[[-101., -161., -143., -54.],
[-110., -148., -120., -30.],
[ -62., -64., -36., 6.],
[ -3., 19., 29., 24.]]],
[[[ 39., 67., 53., 18.],
[ 50., 68., 32., -6.],
[ 2., -40., -76., -66.],
[ -31., -89., -111., -72.]],
[[ 59., 115., 117., 54.],
[ 114., 212., 208., 90.],
[ 114., 200., 196., 78.],
[ 37., 55., 49., 12.]],
[[ 79., 163., 181., 90.],
[ 178., 356., 384., 186.],
[ 226., 440., 468., 222.],
[ 105., 199., 209., 96.]]]])
layer = layer_class(
num_input_channels=kernels.shape[1],
num_output_channels=kernels.shape[0],
input_dim_1=inputs.shape[2],
input_dim_2=inputs.shape[3],
kernel_dim_1=kernels.shape[2],
kernel_dim_2=kernels.shape[3]
)
layer.params = [kernels, biases]
layer_grads_wrt_inputs = layer.bprop(inputs, outputs, grads_wrt_outputs)
assert layer_grads_wrt_inputs.shape == true_grads_wrt_inputs.shape, (
'Layer bprop returns incorrect shaped array. '
'Correct shape is \n\n{0}\n\n but returned shape is \n\n{1}.'
.format(true_grads_wrt_inputs.shape, layer_grads_wrt_inputs.shape)
)
assert np.allclose(layer_grads_wrt_inputs, true_grads_wrt_inputs), (
'Layer bprop does not return correct values. '
'Correct output is \n\n{0}\n\n but returned output is \n\n{1}'
.format(true_grads_wrt_inputs, layer_grads_wrt_inputs)
)
return True
def test_conv_layer_grad_wrt_params(
layer_class, do_cross_correlation=False):
"""Tests `grad_wrt_params` method of a convolutional layer.
Checks the outputs of `grad_wrt_params` method for fixed inputs
against known reference values for the gradients with respect to
kernels and biases, and raises an AssertionError if the returned
values are not consistent with the reference values. If tests
are all passed returns True.
Args:
layer_class: Convolutional layer implementation following the
interface defined in the provided skeleton class.
do_cross_correlation: Whether the layer implements an operation
corresponding to cross-correlation (True) i.e kernels are
not flipped before sliding over inputs, or convolution
(False) with filters being flipped.
Raises:
AssertionError: Raised if output of `layer.bprop` is inconsistent
with reference values either in shape or values.
"""
inputs = np.arange(96).reshape((2, 3, 4, 4))
kernels = np.arange(-12, 12).reshape((2, 3, 2, 2))
biases = np.arange(2)
grads_wrt_outputs = np.arange(-20, 16).reshape((2, 2, 3, 3))
true_kernel_grads = np.array(
[[[[ -240., -114.],
[ 264., 390.]],
[[-2256., -2130.],
[-1752., -1626.]],
[[-4272., -4146.],
[-3768., -3642.]]],
[[[ 5268., 5232.],
[ 5124., 5088.]],
[[ 5844., 5808.],
[ 5700., 5664.]],
[[ 6420., 6384.],
[ 6276., 6240.]]]])
if do_cross_correlation:
kernels = kernels[:, :, ::-1, ::-1]
true_kernel_grads = true_kernel_grads[:, :, ::-1, ::-1]
true_bias_grads = np.array([-126., 36.])
layer = layer_class(
num_input_channels=kernels.shape[1],
num_output_channels=kernels.shape[0],
input_dim_1=inputs.shape[2],
input_dim_2=inputs.shape[3],
kernel_dim_1=kernels.shape[2],
kernel_dim_2=kernels.shape[3]
)
layer.params = [kernels, biases]
layer_kernel_grads, layer_bias_grads = (
layer.grads_wrt_params(inputs, grads_wrt_outputs))
assert layer_kernel_grads.shape == true_kernel_grads.shape, (
'grads_wrt_params gives incorrect shaped kernel gradients output. '
'Correct shape is \n\n{0}\n\n but returned shape is \n\n{1}.'
.format(true_kernel_grads.shape, layer_kernel_grads.shape)
)
assert np.allclose(layer_kernel_grads, true_kernel_grads), (
'grads_wrt_params does not give correct kernel gradients output. '
'Correct output is \n\n{0}\n\n but returned output is \n\n{1}.'
.format(true_kernel_grads, layer_kernel_grads)
)
assert layer_bias_grads.shape == true_bias_grads.shape, (
'grads_wrt_params gives incorrect shaped bias gradients output. '
'Correct shape is \n\n{0}\n\n but returned shape is \n\n{1}.'
.format(true_bias_grads.shape, layer_bias_grads.shape)
)
assert np.allclose(layer_bias_grads, true_bias_grads), (
'grads_wrt_params does not give correct bias gradients output. '
'Correct output is \n\n{0}\n\n but returned output is \n\n{1}.'
.format(true_bias_grads, layer_bias_grads)
)
return True
```
An example of using the test functions if given in the cell below. This assumes you implement a convolution (rather than cross-correlation) operation. If the implementation is correct
```
all_correct = test_conv_layer_fprop(ConvolutionalLayer, False)
all_correct &= test_conv_layer_bprop(ConvolutionalLayer, False)
all_correct &= test_conv_layer_grad_wrt_params(ConvolutionalLayer, False)
if all_correct:
print('All tests passed.')
```
|
github_jupyter
|
import mlp.layers as layers
import mlp.initialisers as init
class ConvolutionalLayer(layers.LayerWithParameters):
"""Layer implementing a 2D convolution-based transformation of its inputs.
The layer is parameterised by a set of 2D convolutional kernels, a four
dimensional array of shape
(num_output_channels, num_input_channels, kernel_dim_1, kernel_dim_2)
and a bias vector, a one dimensional array of shape
(num_output_channels,)
i.e. one shared bias per output channel.
Assuming no-padding is applied to the inputs so that outputs are only
calculated for positions where the kernel filters fully overlap with the
inputs, and that unit strides are used the outputs will have spatial extent
output_dim_1 = input_dim_1 - kernel_dim_1 + 1
output_dim_2 = input_dim_2 - kernel_dim_2 + 1
"""
def __init__(self, num_input_channels, num_output_channels,
input_dim_1, input_dim_2,
kernel_dim_1, kernel_dim_2,
kernels_init=init.UniformInit(-0.01, 0.01),
biases_init=init.ConstantInit(0.),
kernels_penalty=None, biases_penalty=None):
"""Initialises a parameterised convolutional layer.
Args:
num_input_channels (int): Number of channels in inputs to
layer (this may be number of colour channels in the input
images if used as the first layer in a model, or the
number of output channels, a.k.a. feature maps, from a
a previous convolutional layer).
num_output_channels (int): Number of channels in outputs
from the layer, a.k.a. number of feature maps.
input_dim_1 (int): Size of first input dimension of each 2D
channel of inputs.
input_dim_2 (int): Size of second input dimension of each 2D
channel of inputs.
kernel_dim_x (int): Size of first dimension of each 2D channel of
kernels.
kernel_dim_y (int): Size of second dimension of each 2D channel of
kernels.
kernels_intialiser: Initialiser for the kernel parameters.
biases_initialiser: Initialiser for the bias parameters.
kernels_penalty: Kernel-dependent penalty term (regulariser) or
None if no regularisation is to be applied to the kernels.
biases_penalty: Biases-dependent penalty term (regulariser) or
None if no regularisation is to be applied to the biases.
"""
self.num_input_channels = num_input_channels
self.num_output_channels = num_output_channels
self.input_dim_1 = input_dim_1
self.input_dim_2 = input_dim_2
self.kernel_dim_1 = kernel_dim_1
self.kernel_dim_2 = kernel_dim_2
self.kernels_init = kernels_init
self.biases_init = biases_init
self.kernels_shape = (
num_output_channels, num_input_channels, kernel_dim_1, kernel_dim_2
)
self.inputs_shape = (
None, num_input_channels, input_dim_1, input_dim_2
)
self.kernels = self.kernels_init(self.kernels_shape)
self.biases = self.biases_init(num_output_channels)
self.kernels_penalty = kernels_penalty
self.biases_penalty = biases_penalty
def fprop(self, inputs):
"""Forward propagates activations through the layer transformation.
For inputs `x`, outputs `y`, kernels `K` and biases `b` the layer
corresponds to `y = conv2d(x, K) + b`.
Args:
inputs: Array of layer inputs of shape
(batch_size, num_input_channels, input_dim_1, input_dim_2).
Returns:
outputs: Array of layer outputs of shape
(batch_size, num_output_channels, output_dim_1, output_dim_2).
"""
raise NotImplementedError()
def bprop(self, inputs, outputs, grads_wrt_outputs):
"""Back propagates gradients through a layer.
Given gradients with respect to the outputs of the layer calculates the
gradients with respect to the layer inputs.
Args:
inputs: Array of layer inputs of shape
(batch_size, num_input_channels, input_dim_1, input_dim_2).
outputs: Array of layer outputs calculated in forward pass of
shape
(batch_size, num_output_channels, output_dim_1, output_dim_2).
grads_wrt_outputs: Array of gradients with respect to the layer
outputs of shape
(batch_size, num_output_channels, output_dim_1, output_dim_2).
Returns:
Array of gradients with respect to the layer inputs of shape
(batch_size, num_input_channels, input_dim_1, input_dim_2).
"""
raise NotImplementedError()
def grads_wrt_params(self, inputs, grads_wrt_outputs):
"""Calculates gradients with respect to layer parameters.
Args:
inputs: array of inputs to layer of shape (batch_size, input_dim)
grads_wrt_to_outputs: array of gradients with respect to the layer
outputs of shape
(batch_size, num_output_channels, output_dim_1, output_dim_2).
Returns:
list of arrays of gradients with respect to the layer parameters
`[grads_wrt_kernels, grads_wrt_biases]`.
"""
raise NotImplementedError()
def params_penalty(self):
"""Returns the parameter dependent penalty term for this layer.
If no parameter-dependent penalty terms are set this returns zero.
"""
params_penalty = 0
if self.kernels_penalty is not None:
params_penalty += self.kernels_penalty(self.kernels)
if self.biases_penalty is not None:
params_penalty += self.biases_penalty(self.biases)
return params_penalty
@property
def params(self):
"""A list of layer parameter values: `[kernels, biases]`."""
return [self.kernels, self.biases]
@params.setter
def params(self, values):
self.kernels = values[0]
self.biases = values[1]
def __repr__(self):
return (
'ConvolutionalLayer(\n'
' num_input_channels={0}, num_output_channels={1},\n'
' input_dim_1={2}, input_dim_2={3},\n'
' kernel_dim_1={4}, kernel_dim_2={5}\n'
')'
.format(self.num_input_channels, self.num_output_channels,
self.input_dim_1, self.input_dim_2, self.kernel_dim_1,
self.kernel_dim_2)
)
import numpy as np
def test_conv_layer_fprop(layer_class, do_cross_correlation=False):
"""Tests `fprop` method of a convolutional layer.
Checks the outputs of `fprop` method for a fixed input against known
reference values for the outputs and raises an AssertionError if
the outputted values are not consistent with the reference values. If
tests are all passed returns True.
Args:
layer_class: Convolutional layer implementation following the
interface defined in the provided skeleton class.
do_cross_correlation: Whether the layer implements an operation
corresponding to cross-correlation (True) i.e kernels are
not flipped before sliding over inputs, or convolution
(False) with filters being flipped.
Raises:
AssertionError: Raised if output of `layer.fprop` is inconsistent
with reference values either in shape or values.
"""
inputs = np.arange(96).reshape((2, 3, 4, 4))
kernels = np.arange(-12, 12).reshape((2, 3, 2, 2))
if do_cross_correlation:
kernels = kernels[:, :, ::-1, ::-1]
biases = np.arange(2)
true_output = np.array(
[[[[ -958., -1036., -1114.],
[-1270., -1348., -1426.],
[-1582., -1660., -1738.]],
[[ 1707., 1773., 1839.],
[ 1971., 2037., 2103.],
[ 2235., 2301., 2367.]]],
[[[-4702., -4780., -4858.],
[-5014., -5092., -5170.],
[-5326., -5404., -5482.]],
[[ 4875., 4941., 5007.],
[ 5139., 5205., 5271.],
[ 5403., 5469., 5535.]]]]
)
layer = layer_class(
num_input_channels=kernels.shape[1],
num_output_channels=kernels.shape[0],
input_dim_1=inputs.shape[2],
input_dim_2=inputs.shape[3],
kernel_dim_1=kernels.shape[2],
kernel_dim_2=kernels.shape[3]
)
layer.params = [kernels, biases]
layer_output = layer.fprop(inputs)
assert layer_output.shape == true_output.shape, (
'Layer fprop gives incorrect shaped output. '
'Correct shape is \n\n{0}\n\n but returned shape is \n\n{1}.'
.format(true_output.shape, layer_output.shape)
)
assert np.allclose(layer_output, true_output), (
'Layer fprop does not give correct output. '
'Correct output is \n\n{0}\n\n but returned output is \n\n{1}.'
.format(true_output, layer_output)
)
return True
def test_conv_layer_bprop(layer_class, do_cross_correlation=False):
"""Tests `bprop` method of a convolutional layer.
Checks the outputs of `bprop` method for a fixed input against known
reference values for the gradients with respect to inputs and raises
an AssertionError if the returned values are not consistent with the
reference values. If tests are all passed returns True.
Args:
layer_class: Convolutional layer implementation following the
interface defined in the provided skeleton class.
do_cross_correlation: Whether the layer implements an operation
corresponding to cross-correlation (True) i.e kernels are
not flipped before sliding over inputs, or convolution
(False) with filters being flipped.
Raises:
AssertionError: Raised if output of `layer.bprop` is inconsistent
with reference values either in shape or values.
"""
inputs = np.arange(96).reshape((2, 3, 4, 4))
kernels = np.arange(-12, 12).reshape((2, 3, 2, 2))
if do_cross_correlation:
kernels = kernels[:, :, ::-1, ::-1]
biases = np.arange(2)
grads_wrt_outputs = np.arange(-20, 16).reshape((2, 2, 3, 3))
outputs = np.array(
[[[[ -958., -1036., -1114.],
[-1270., -1348., -1426.],
[-1582., -1660., -1738.]],
[[ 1707., 1773., 1839.],
[ 1971., 2037., 2103.],
[ 2235., 2301., 2367.]]],
[[[-4702., -4780., -4858.],
[-5014., -5092., -5170.],
[-5326., -5404., -5482.]],
[[ 4875., 4941., 5007.],
[ 5139., 5205., 5271.],
[ 5403., 5469., 5535.]]]]
)
true_grads_wrt_inputs = np.array(
[[[[ 147., 319., 305., 162.],
[ 338., 716., 680., 354.],
[ 290., 608., 572., 294.],
[ 149., 307., 285., 144.]],
[[ 23., 79., 81., 54.],
[ 114., 284., 280., 162.],
[ 114., 272., 268., 150.],
[ 73., 163., 157., 84.]],
[[-101., -161., -143., -54.],
[-110., -148., -120., -30.],
[ -62., -64., -36., 6.],
[ -3., 19., 29., 24.]]],
[[[ 39., 67., 53., 18.],
[ 50., 68., 32., -6.],
[ 2., -40., -76., -66.],
[ -31., -89., -111., -72.]],
[[ 59., 115., 117., 54.],
[ 114., 212., 208., 90.],
[ 114., 200., 196., 78.],
[ 37., 55., 49., 12.]],
[[ 79., 163., 181., 90.],
[ 178., 356., 384., 186.],
[ 226., 440., 468., 222.],
[ 105., 199., 209., 96.]]]])
layer = layer_class(
num_input_channels=kernels.shape[1],
num_output_channels=kernels.shape[0],
input_dim_1=inputs.shape[2],
input_dim_2=inputs.shape[3],
kernel_dim_1=kernels.shape[2],
kernel_dim_2=kernels.shape[3]
)
layer.params = [kernels, biases]
layer_grads_wrt_inputs = layer.bprop(inputs, outputs, grads_wrt_outputs)
assert layer_grads_wrt_inputs.shape == true_grads_wrt_inputs.shape, (
'Layer bprop returns incorrect shaped array. '
'Correct shape is \n\n{0}\n\n but returned shape is \n\n{1}.'
.format(true_grads_wrt_inputs.shape, layer_grads_wrt_inputs.shape)
)
assert np.allclose(layer_grads_wrt_inputs, true_grads_wrt_inputs), (
'Layer bprop does not return correct values. '
'Correct output is \n\n{0}\n\n but returned output is \n\n{1}'
.format(true_grads_wrt_inputs, layer_grads_wrt_inputs)
)
return True
def test_conv_layer_grad_wrt_params(
layer_class, do_cross_correlation=False):
"""Tests `grad_wrt_params` method of a convolutional layer.
Checks the outputs of `grad_wrt_params` method for fixed inputs
against known reference values for the gradients with respect to
kernels and biases, and raises an AssertionError if the returned
values are not consistent with the reference values. If tests
are all passed returns True.
Args:
layer_class: Convolutional layer implementation following the
interface defined in the provided skeleton class.
do_cross_correlation: Whether the layer implements an operation
corresponding to cross-correlation (True) i.e kernels are
not flipped before sliding over inputs, or convolution
(False) with filters being flipped.
Raises:
AssertionError: Raised if output of `layer.bprop` is inconsistent
with reference values either in shape or values.
"""
inputs = np.arange(96).reshape((2, 3, 4, 4))
kernels = np.arange(-12, 12).reshape((2, 3, 2, 2))
biases = np.arange(2)
grads_wrt_outputs = np.arange(-20, 16).reshape((2, 2, 3, 3))
true_kernel_grads = np.array(
[[[[ -240., -114.],
[ 264., 390.]],
[[-2256., -2130.],
[-1752., -1626.]],
[[-4272., -4146.],
[-3768., -3642.]]],
[[[ 5268., 5232.],
[ 5124., 5088.]],
[[ 5844., 5808.],
[ 5700., 5664.]],
[[ 6420., 6384.],
[ 6276., 6240.]]]])
if do_cross_correlation:
kernels = kernels[:, :, ::-1, ::-1]
true_kernel_grads = true_kernel_grads[:, :, ::-1, ::-1]
true_bias_grads = np.array([-126., 36.])
layer = layer_class(
num_input_channels=kernels.shape[1],
num_output_channels=kernels.shape[0],
input_dim_1=inputs.shape[2],
input_dim_2=inputs.shape[3],
kernel_dim_1=kernels.shape[2],
kernel_dim_2=kernels.shape[3]
)
layer.params = [kernels, biases]
layer_kernel_grads, layer_bias_grads = (
layer.grads_wrt_params(inputs, grads_wrt_outputs))
assert layer_kernel_grads.shape == true_kernel_grads.shape, (
'grads_wrt_params gives incorrect shaped kernel gradients output. '
'Correct shape is \n\n{0}\n\n but returned shape is \n\n{1}.'
.format(true_kernel_grads.shape, layer_kernel_grads.shape)
)
assert np.allclose(layer_kernel_grads, true_kernel_grads), (
'grads_wrt_params does not give correct kernel gradients output. '
'Correct output is \n\n{0}\n\n but returned output is \n\n{1}.'
.format(true_kernel_grads, layer_kernel_grads)
)
assert layer_bias_grads.shape == true_bias_grads.shape, (
'grads_wrt_params gives incorrect shaped bias gradients output. '
'Correct shape is \n\n{0}\n\n but returned shape is \n\n{1}.'
.format(true_bias_grads.shape, layer_bias_grads.shape)
)
assert np.allclose(layer_bias_grads, true_bias_grads), (
'grads_wrt_params does not give correct bias gradients output. '
'Correct output is \n\n{0}\n\n but returned output is \n\n{1}.'
.format(true_bias_grads, layer_bias_grads)
)
return True
all_correct = test_conv_layer_fprop(ConvolutionalLayer, False)
all_correct &= test_conv_layer_bprop(ConvolutionalLayer, False)
all_correct &= test_conv_layer_grad_wrt_params(ConvolutionalLayer, False)
if all_correct:
print('All tests passed.')
| 0.945576 | 0.990112 |
```
"""
This notebook is to demonstrate multi-class classification using PyTorch
The dataset used for the task is GTSRB - German Traffic Sign Recognition Database. This datset consists of 39209 training examples and 12631 test images, belonging to 43 classes
This notebook is used to perform inference using the saved model. Training and generating the model is done in a separate notebook
"""
# Few imports
import torch
import torchvision
from torchvision import transforms
import torch.utils.data as data
import time
import numpy as np
import os
import pandas as pd
import matplotlib.pyplot as plt
from PIL import Image
from sklearn.metrics import confusion_matrix
from sklearn.metrics import classification_report
from sklearn.metrics import ConfusionMatrixDisplay
# Define transformations
test_transforms = transforms.Compose([
transforms.Resize([112, 112]),
transforms.ToTensor()
])
# Define path of test data
test_data_path = "../GTSRB/Test"
test_data = torchvision.datasets.ImageFolder(root = test_data_path, transform = test_transforms)
test_loader = data.DataLoader(test_data, batch_size=1, shuffle=False)
# Define hyperparameters
numClasses = 43
# Generating labels of classes
num = range(numClasses)
labels = []
for i in num:
labels.append(str(i))
labels = sorted(labels)
for i in num:
labels[i] = int(labels[i])
print("List of labels : ")
print("Actual labels \t--> Class in PyTorch")
for i in num:
print("\t%d \t--> \t%d" % (labels[i], i))
# Read the image labels from the csv file
# Note: The labels provided are all numbers, whereas the labels assigned by PyTorch dataloader are strings
df = pd.read_csv("../GTSRB/Test.csv")
numExamples = len(df)
labels_list = list(df.ClassId)
# Load the saved model
from class_alexnetTS import AlexnetTS
MODEL_PATH = "../Model/pytorch_classification_alexnetTS.pth"
model = AlexnetTS(numClasses)
model.load_state_dict(torch.load(MODEL_PATH))
model = model.cuda()
# Perform classification
y_pred_list = []
corr_classified = 0
with torch.no_grad():
model.eval()
i = 0
for image, _ in test_loader:
image = image.cuda()
y_test_pred = model(image)
y_pred_softmax = torch.log_softmax(y_test_pred[0], dim=1)
_, y_pred_tags = torch.max(y_pred_softmax, dim=1)
y_pred_tags = y_pred_tags.cpu().numpy()
y_pred = y_pred_tags[0]
y_pred = labels[y_pred]
y_pred_list.append(y_pred)
if labels_list[i] == y_pred:
corr_classified += 1
i += 1
print("Number of correctly classified images = %d" % corr_classified)
print("Number of incorrectly classified images = %d" % (numExamples - corr_classified))
print("Final accuracy = %f" % (corr_classified / numExamples))
# Print classification report
print(classification_report(labels_list, y_pred_list))
# Print confusion matrix
def plot_confusion_matrix(labels, pred_labels, classes):
fig = plt.figure(figsize = (20, 20));
ax = fig.add_subplot(1, 1, 1);
cm = confusion_matrix(labels, pred_labels);
cm = ConfusionMatrixDisplay(cm, display_labels = classes);
cm.plot(values_format = 'd', cmap = 'Blues', ax = ax)
plt.xticks(rotation = 20)
labels_arr = range(0, numClasses)
plot_confusion_matrix(labels_list, y_pred_list, labels_arr)
print(y_pred_list[:20])
print(labels_list[:20])
# Display first 30 images, along with the actual and predicted classes
fig, axs = plt.subplots(6,5,figsize=(50,75))
#fig.tight_layout(h_pad = 50)
for i in range(30):
row = i // 5
col = i % 5
imgName = '../GTSRB/Test/' + df.iloc[i].Path
img = Image.open(imgName)
axs[row, col].imshow(img)
title = "Pred: %d, Actual: %d" % (y_pred_list[i], labels_list[i])
axs[row, col].set_title(title, fontsize=50)
plt.savefig("predictions.png", bbox_inches = 'tight', pad_inches=0.5)
```
|
github_jupyter
|
"""
This notebook is to demonstrate multi-class classification using PyTorch
The dataset used for the task is GTSRB - German Traffic Sign Recognition Database. This datset consists of 39209 training examples and 12631 test images, belonging to 43 classes
This notebook is used to perform inference using the saved model. Training and generating the model is done in a separate notebook
"""
# Few imports
import torch
import torchvision
from torchvision import transforms
import torch.utils.data as data
import time
import numpy as np
import os
import pandas as pd
import matplotlib.pyplot as plt
from PIL import Image
from sklearn.metrics import confusion_matrix
from sklearn.metrics import classification_report
from sklearn.metrics import ConfusionMatrixDisplay
# Define transformations
test_transforms = transforms.Compose([
transforms.Resize([112, 112]),
transforms.ToTensor()
])
# Define path of test data
test_data_path = "../GTSRB/Test"
test_data = torchvision.datasets.ImageFolder(root = test_data_path, transform = test_transforms)
test_loader = data.DataLoader(test_data, batch_size=1, shuffle=False)
# Define hyperparameters
numClasses = 43
# Generating labels of classes
num = range(numClasses)
labels = []
for i in num:
labels.append(str(i))
labels = sorted(labels)
for i in num:
labels[i] = int(labels[i])
print("List of labels : ")
print("Actual labels \t--> Class in PyTorch")
for i in num:
print("\t%d \t--> \t%d" % (labels[i], i))
# Read the image labels from the csv file
# Note: The labels provided are all numbers, whereas the labels assigned by PyTorch dataloader are strings
df = pd.read_csv("../GTSRB/Test.csv")
numExamples = len(df)
labels_list = list(df.ClassId)
# Load the saved model
from class_alexnetTS import AlexnetTS
MODEL_PATH = "../Model/pytorch_classification_alexnetTS.pth"
model = AlexnetTS(numClasses)
model.load_state_dict(torch.load(MODEL_PATH))
model = model.cuda()
# Perform classification
y_pred_list = []
corr_classified = 0
with torch.no_grad():
model.eval()
i = 0
for image, _ in test_loader:
image = image.cuda()
y_test_pred = model(image)
y_pred_softmax = torch.log_softmax(y_test_pred[0], dim=1)
_, y_pred_tags = torch.max(y_pred_softmax, dim=1)
y_pred_tags = y_pred_tags.cpu().numpy()
y_pred = y_pred_tags[0]
y_pred = labels[y_pred]
y_pred_list.append(y_pred)
if labels_list[i] == y_pred:
corr_classified += 1
i += 1
print("Number of correctly classified images = %d" % corr_classified)
print("Number of incorrectly classified images = %d" % (numExamples - corr_classified))
print("Final accuracy = %f" % (corr_classified / numExamples))
# Print classification report
print(classification_report(labels_list, y_pred_list))
# Print confusion matrix
def plot_confusion_matrix(labels, pred_labels, classes):
fig = plt.figure(figsize = (20, 20));
ax = fig.add_subplot(1, 1, 1);
cm = confusion_matrix(labels, pred_labels);
cm = ConfusionMatrixDisplay(cm, display_labels = classes);
cm.plot(values_format = 'd', cmap = 'Blues', ax = ax)
plt.xticks(rotation = 20)
labels_arr = range(0, numClasses)
plot_confusion_matrix(labels_list, y_pred_list, labels_arr)
print(y_pred_list[:20])
print(labels_list[:20])
# Display first 30 images, along with the actual and predicted classes
fig, axs = plt.subplots(6,5,figsize=(50,75))
#fig.tight_layout(h_pad = 50)
for i in range(30):
row = i // 5
col = i % 5
imgName = '../GTSRB/Test/' + df.iloc[i].Path
img = Image.open(imgName)
axs[row, col].imshow(img)
title = "Pred: %d, Actual: %d" % (y_pred_list[i], labels_list[i])
axs[row, col].set_title(title, fontsize=50)
plt.savefig("predictions.png", bbox_inches = 'tight', pad_inches=0.5)
| 0.907268 | 0.912903 |
## Steel Surface defect detection
An Automatic real-time Optical Inspection System for classifying and detecting defects in steel surfaces. Designed and trained a CNN framework using EfficientNet with U-Net Architecture and applying Mask R-CNN, to build an efficient search-heuristics method for feature extraction and representations from raw pixel and segmenting a masks over the defects for industrial applications.
## Importing Libraries
```
import os
import cv2
import pdb
import time
import warnings
import random
import numpy as np
import pandas as pd
from tqdm import tqdm_notebook as tqdm
from torch.optim.lr_scheduler import ReduceLROnPlateau
from sklearn.model_selection import train_test_split
import torch
import torch.nn as nn
from torch.nn import functional as F
import torch.optim as optim
import torch.backends.cudnn as cudnn
from torch.utils.data import DataLoader, Dataset, sampler
from matplotlib import pyplot as plt
from albumentations import (HorizontalFlip, ShiftScaleRotate, Normalize, Resize, Compose, GaussNoise)
from albumentations.pytorch import ToTensor
from model import Unet
warnings.filterwarnings("ignore")
seed = 69
random.seed(seed)
os.environ["PYTHONHASHSEED"] = str(seed)
np.random.seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
```
## RLE-Mask utility functions
```
def mask2rle(img):
'''
img: numpy array, 1 -> mask, 0 -> background
Returns run length as string formated
'''
pixels= img.T.flatten()
pixels = np.concatenate([[0], pixels, [0]])
runs = np.where(pixels[1:] != pixels[:-1])[0] + 1
runs[1::2] -= runs[::2]
return ' '.join(str(x) for x in runs)
def make_mask(row_id, df):
'''Given a row index, return image_id and mask (256, 1600, 4) from the dataframe `df`'''
fname = df.iloc[row_id].name
labels = df.iloc[row_id][:4]
masks = np.zeros((256, 1600, 4), dtype=np.float32) # float32 is V.Imp
# 4:class 1~4 (ch:0~3)
for idx, label in enumerate(labels.values):
if label is not np.nan:
label = label.split(" ")
positions = map(int, label[0::2])
length = map(int, label[1::2])
mask = np.zeros(256 * 1600, dtype=np.uint8)
for pos, le in zip(positions, length):
mask[pos:(pos + le)] = 1
masks[:, :, idx] = mask.reshape(256, 1600, order='F')
return fname, masks
```
## Dataloader
```
class SteelDataset(Dataset):
def __init__(self, df, data_folder, mean, std, phase):
self.df = df
self.root = data_folder
self.mean = mean
self.std = std
self.phase = phase
self.transforms = get_transforms(phase, mean, std)
self.fnames = self.df.index.tolist()
def __getitem__(self, idx):
image_id, mask = make_mask(idx, self.df)
image_path = os.path.join(self.root, "train_images", image_id)
img = cv2.imread(image_path)
augmented = self.transforms(image=img, mask=mask)
img = augmented['image']
mask = augmented['mask'] # 1x256x1600x4
mask = mask[0].permute(2, 0, 1) # 4x256x1600
return img, mask
def __len__(self):
return len(self.fnames)
def get_transforms(phase, mean, std):
list_transforms = []
if phase == "train":
list_transforms.extend(
[
HorizontalFlip(p=0.5), # only horizontal flip as of now
]
)
list_transforms.extend(
[
Normalize(mean=mean, std=std, p=1),
ToTensor(),
]
)
list_trfms = Compose(list_transforms)
return list_trfms
def provider(
data_folder,
df_path,
phase,
mean=None,
std=None,
batch_size=8,
num_workers=4,
):
'''Returns dataloader for the model training'''
df = pd.read_csv(df_path)
# https://www.kaggle.com/amanooo/defect-detection-starter-u-net
df['ImageId'], df['ClassId'] = zip(*df['ImageId_ClassId'].str.split('_'))
df['ClassId'] = df['ClassId'].astype(int)
df = df.pivot(index='ImageId',columns='ClassId',values='EncodedPixels')
df['defects'] = df.count(axis=1)
train_df, val_df = train_test_split(df, test_size=0.2, stratify=df["defects"], random_state=69)
df = train_df if phase == "train" else val_df
image_dataset = SteelDataset(df, data_folder, mean, std, phase)
dataloader = DataLoader(
image_dataset,
batch_size=batch_size,
num_workers=num_workers,
pin_memory=True,
shuffle=True,
)
return dataloader
```
## Some more utility functions
Dice and IoU metric implementations, metric logger for training and validation.
```
def predict(X, threshold):
'''X is sigmoid output of the model'''
X_p = np.copy(X)
preds = (X_p > threshold).astype('uint8')
return preds
def metric(probability, truth, threshold=0.5, reduction='none'):
'''Calculates dice of positive and negative images seperately'''
'''probability and truth must be torch tensors'''
batch_size = len(truth)
with torch.no_grad():
probability = probability.view(batch_size, -1)
truth = truth.view(batch_size, -1)
assert(probability.shape == truth.shape)
p = (probability > threshold).float()
t = (truth > 0.5).float()
t_sum = t.sum(-1)
p_sum = p.sum(-1)
neg_index = torch.nonzero(t_sum == 0)
pos_index = torch.nonzero(t_sum >= 1)
dice_neg = (p_sum == 0).float()
dice_pos = 2 * (p*t).sum(-1)/((p+t).sum(-1))
dice_neg = dice_neg[neg_index]
dice_pos = dice_pos[pos_index]
dice = torch.cat([dice_pos, dice_neg])
# dice_neg = np.nan_to_num(dice_neg.mean().item(), 0)
# dice_pos = np.nan_to_num(dice_pos.mean().item(), 0)
# dice = dice.mean().item()
num_neg = len(neg_index)
num_pos = len(pos_index)
return dice, dice_neg, dice_pos, num_neg, num_pos
class Meter:
'''A meter to keep track of iou and dice scores throughout an epoch'''
def __init__(self, phase, epoch):
self.base_threshold = 0.5 # <<<<<<<<<<< here's the threshold
self.base_dice_scores = []
self.dice_neg_scores = []
self.dice_pos_scores = []
self.iou_scores = []
def update(self, targets, outputs):
probs = torch.sigmoid(outputs)
dice, dice_neg, dice_pos, _, _ = metric(probs, targets, self.base_threshold)
self.base_dice_scores.extend(dice.tolist())
self.dice_pos_scores.extend(dice_pos.tolist())
self.dice_neg_scores.extend(dice_neg.tolist())
preds = predict(probs, self.base_threshold)
iou = compute_iou_batch(preds, targets, classes=[1])
self.iou_scores.append(iou)
def get_metrics(self):
dice = np.nanmean(self.base_dice_scores)
dice_neg = np.nanmean(self.dice_neg_scores)
dice_pos = np.nanmean(self.dice_pos_scores)
dices = [dice, dice_neg, dice_pos]
iou = np.nanmean(self.iou_scores)
return dices, iou
def epoch_log(phase, epoch, epoch_loss, meter, start):
'''logging the metrics at the end of an epoch'''
dices, iou = meter.get_metrics()
dice, dice_neg, dice_pos = dices
print("Loss: %0.4f | IoU: %0.4f | dice: %0.4f | dice_neg: %0.4f | dice_pos: %0.4f" % (epoch_loss, iou, dice, dice_neg, dice_pos))
return dice, iou
def compute_ious(pred, label, classes, ignore_index=255, only_present=True):
'''computes iou for one ground truth mask and predicted mask'''
pred[label == ignore_index] = 0
ious = []
for c in classes:
label_c = label == c
if only_present and np.sum(label_c) == 0:
ious.append(np.nan)
continue
pred_c = pred == c
intersection = np.logical_and(pred_c, label_c).sum()
union = np.logical_or(pred_c, label_c).sum()
if union != 0:
ious.append(intersection / union)
return ious if ious else [1]
def compute_iou_batch(outputs, labels, classes=None):
'''computes mean iou for a batch of ground truth masks and predicted masks'''
ious = []
preds = np.copy(outputs) # copy is imp
labels = np.array(labels) # tensor to np
for pred, label in zip(preds, labels):
ious.append(np.nanmean(compute_ious(pred, label, classes)))
iou = np.nanmean(ious)
return iou
```
## Model Initialization
```
model = Unet("efficientnet-b3", encoder_weights="imagenet", classes=4, activation=None)
```
### Training and Validation
```
class Trainer(object):
'''This class takes care of training and validation of our model'''
def __init__(self, model):
self.num_workers = 6
self.batch_size = {"train": 4, "val": 4}
self.accumulation_steps = 32 // self.batch_size['train']
self.lr = 5e-4
self.num_epochs = 20
self.best_loss = float("inf")
self.phases = ["train", "val"]
self.device = torch.device("cuda:0")
torch.set_default_tensor_type("torch.cuda.FloatTensor")
self.net = model
self.criterion = torch.nn.BCEWithLogitsLoss()
self.optimizer = optim.Adam(self.net.parameters(), lr=self.lr)
self.scheduler = ReduceLROnPlateau(self.optimizer, mode="min", patience=3, verbose=True)
self.net = self.net.to(self.device)
cudnn.benchmark = True
self.dataloaders = {
phase: provider(
data_folder=data_folder,
df_path=train_df_path,
phase=phase,
mean=(0.485, 0.456, 0.406),
std=(0.229, 0.224, 0.225),
batch_size=self.batch_size[phase],
num_workers=self.num_workers,
)
for phase in self.phases
}
self.losses = {phase: [] for phase in self.phases}
self.iou_scores = {phase: [] for phase in self.phases}
self.dice_scores = {phase: [] for phase in self.phases}
def forward(self, images, targets):
images = images.to(self.device)
masks = targets.to(self.device)
outputs = self.net(images)
loss = self.criterion(outputs, masks)
return loss, outputs
def iterate(self, epoch, phase):
meter = Meter(phase, epoch)
start = time.strftime("%H:%M:%S")
print(f"Starting epoch: {epoch} | phase: {phase} | ⏰: {start}")
batch_size = self.batch_size[phase]
self.net.train(phase == "train")
dataloader = self.dataloaders[phase]
running_loss = 0.0
total_batches = len(dataloader)
# tk0 = tqdm(dataloader, total=total_batches)
self.optimizer.zero_grad()
for itr, batch in enumerate(dataloader): # replace `dataloader` with `tk0` for tqdm
images, targets = batch
loss, outputs = self.forward(images, targets)
loss = loss / self.accumulation_steps
if phase == "train":
loss.backward()
if (itr + 1 ) % self.accumulation_steps == 0:
self.optimizer.step()
self.optimizer.zero_grad()
running_loss += loss.item()
outputs = outputs.detach().cpu()
meter.update(targets, outputs)
# tk0.set_postfix(loss=(running_loss / ((itr + 1))))
epoch_loss = (running_loss * self.accumulation_steps) / total_batches
dice, iou = epoch_log(phase, epoch, epoch_loss, meter, start)
self.losses[phase].append(epoch_loss)
self.dice_scores[phase].append(dice)
self.iou_scores[phase].append(iou)
torch.cuda.empty_cache()
return epoch_loss
def start(self):
for epoch in range(self.num_epochs):
self.iterate(epoch, "train")
state = {
"epoch": epoch,
"best_loss": self.best_loss,
"state_dict": self.net.state_dict(),
"optimizer": self.optimizer.state_dict(),
}
with torch.no_grad():
val_loss = self.iterate(epoch, "val")
self.scheduler.step(val_loss)
if val_loss < self.best_loss:
print("******** New optimal found, saving state ********")
state["best_loss"] = self.best_loss = val_loss
torch.save(state, "./model.pth")
print()
train_df_path = 'steel-defect-detection/train.csv'
data_folder = "steel-defect-detection/"
test_data_folder = "steel-defect-detection/test_images"
model_trainer = Trainer(model)
model_trainer.start()
# PLOT TRAINING
losses = model_trainer.losses
dice_scores = model_trainer.dice_scores # overall dice
iou_scores = model_trainer.iou_scores
def plot(scores, name):
plt.figure(figsize=(15,5))
plt.plot(range(len(scores["train"])), scores["train"], label=f'train {name}')
plt.plot(range(len(scores["train"])), scores["val"], label=f'val {name}')
plt.title(f'{name} plot'); plt.xlabel('Epoch'); plt.ylabel(f'{name}');
plt.legend();
plt.show()
plot(losses, "BCE loss")
plot(dice_scores, "Dice score")
plot(iou_scores, "IoU score")
```
|
github_jupyter
|
import os
import cv2
import pdb
import time
import warnings
import random
import numpy as np
import pandas as pd
from tqdm import tqdm_notebook as tqdm
from torch.optim.lr_scheduler import ReduceLROnPlateau
from sklearn.model_selection import train_test_split
import torch
import torch.nn as nn
from torch.nn import functional as F
import torch.optim as optim
import torch.backends.cudnn as cudnn
from torch.utils.data import DataLoader, Dataset, sampler
from matplotlib import pyplot as plt
from albumentations import (HorizontalFlip, ShiftScaleRotate, Normalize, Resize, Compose, GaussNoise)
from albumentations.pytorch import ToTensor
from model import Unet
warnings.filterwarnings("ignore")
seed = 69
random.seed(seed)
os.environ["PYTHONHASHSEED"] = str(seed)
np.random.seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
def mask2rle(img):
'''
img: numpy array, 1 -> mask, 0 -> background
Returns run length as string formated
'''
pixels= img.T.flatten()
pixels = np.concatenate([[0], pixels, [0]])
runs = np.where(pixels[1:] != pixels[:-1])[0] + 1
runs[1::2] -= runs[::2]
return ' '.join(str(x) for x in runs)
def make_mask(row_id, df):
'''Given a row index, return image_id and mask (256, 1600, 4) from the dataframe `df`'''
fname = df.iloc[row_id].name
labels = df.iloc[row_id][:4]
masks = np.zeros((256, 1600, 4), dtype=np.float32) # float32 is V.Imp
# 4:class 1~4 (ch:0~3)
for idx, label in enumerate(labels.values):
if label is not np.nan:
label = label.split(" ")
positions = map(int, label[0::2])
length = map(int, label[1::2])
mask = np.zeros(256 * 1600, dtype=np.uint8)
for pos, le in zip(positions, length):
mask[pos:(pos + le)] = 1
masks[:, :, idx] = mask.reshape(256, 1600, order='F')
return fname, masks
class SteelDataset(Dataset):
def __init__(self, df, data_folder, mean, std, phase):
self.df = df
self.root = data_folder
self.mean = mean
self.std = std
self.phase = phase
self.transforms = get_transforms(phase, mean, std)
self.fnames = self.df.index.tolist()
def __getitem__(self, idx):
image_id, mask = make_mask(idx, self.df)
image_path = os.path.join(self.root, "train_images", image_id)
img = cv2.imread(image_path)
augmented = self.transforms(image=img, mask=mask)
img = augmented['image']
mask = augmented['mask'] # 1x256x1600x4
mask = mask[0].permute(2, 0, 1) # 4x256x1600
return img, mask
def __len__(self):
return len(self.fnames)
def get_transforms(phase, mean, std):
list_transforms = []
if phase == "train":
list_transforms.extend(
[
HorizontalFlip(p=0.5), # only horizontal flip as of now
]
)
list_transforms.extend(
[
Normalize(mean=mean, std=std, p=1),
ToTensor(),
]
)
list_trfms = Compose(list_transforms)
return list_trfms
def provider(
data_folder,
df_path,
phase,
mean=None,
std=None,
batch_size=8,
num_workers=4,
):
'''Returns dataloader for the model training'''
df = pd.read_csv(df_path)
# https://www.kaggle.com/amanooo/defect-detection-starter-u-net
df['ImageId'], df['ClassId'] = zip(*df['ImageId_ClassId'].str.split('_'))
df['ClassId'] = df['ClassId'].astype(int)
df = df.pivot(index='ImageId',columns='ClassId',values='EncodedPixels')
df['defects'] = df.count(axis=1)
train_df, val_df = train_test_split(df, test_size=0.2, stratify=df["defects"], random_state=69)
df = train_df if phase == "train" else val_df
image_dataset = SteelDataset(df, data_folder, mean, std, phase)
dataloader = DataLoader(
image_dataset,
batch_size=batch_size,
num_workers=num_workers,
pin_memory=True,
shuffle=True,
)
return dataloader
def predict(X, threshold):
'''X is sigmoid output of the model'''
X_p = np.copy(X)
preds = (X_p > threshold).astype('uint8')
return preds
def metric(probability, truth, threshold=0.5, reduction='none'):
'''Calculates dice of positive and negative images seperately'''
'''probability and truth must be torch tensors'''
batch_size = len(truth)
with torch.no_grad():
probability = probability.view(batch_size, -1)
truth = truth.view(batch_size, -1)
assert(probability.shape == truth.shape)
p = (probability > threshold).float()
t = (truth > 0.5).float()
t_sum = t.sum(-1)
p_sum = p.sum(-1)
neg_index = torch.nonzero(t_sum == 0)
pos_index = torch.nonzero(t_sum >= 1)
dice_neg = (p_sum == 0).float()
dice_pos = 2 * (p*t).sum(-1)/((p+t).sum(-1))
dice_neg = dice_neg[neg_index]
dice_pos = dice_pos[pos_index]
dice = torch.cat([dice_pos, dice_neg])
# dice_neg = np.nan_to_num(dice_neg.mean().item(), 0)
# dice_pos = np.nan_to_num(dice_pos.mean().item(), 0)
# dice = dice.mean().item()
num_neg = len(neg_index)
num_pos = len(pos_index)
return dice, dice_neg, dice_pos, num_neg, num_pos
class Meter:
'''A meter to keep track of iou and dice scores throughout an epoch'''
def __init__(self, phase, epoch):
self.base_threshold = 0.5 # <<<<<<<<<<< here's the threshold
self.base_dice_scores = []
self.dice_neg_scores = []
self.dice_pos_scores = []
self.iou_scores = []
def update(self, targets, outputs):
probs = torch.sigmoid(outputs)
dice, dice_neg, dice_pos, _, _ = metric(probs, targets, self.base_threshold)
self.base_dice_scores.extend(dice.tolist())
self.dice_pos_scores.extend(dice_pos.tolist())
self.dice_neg_scores.extend(dice_neg.tolist())
preds = predict(probs, self.base_threshold)
iou = compute_iou_batch(preds, targets, classes=[1])
self.iou_scores.append(iou)
def get_metrics(self):
dice = np.nanmean(self.base_dice_scores)
dice_neg = np.nanmean(self.dice_neg_scores)
dice_pos = np.nanmean(self.dice_pos_scores)
dices = [dice, dice_neg, dice_pos]
iou = np.nanmean(self.iou_scores)
return dices, iou
def epoch_log(phase, epoch, epoch_loss, meter, start):
'''logging the metrics at the end of an epoch'''
dices, iou = meter.get_metrics()
dice, dice_neg, dice_pos = dices
print("Loss: %0.4f | IoU: %0.4f | dice: %0.4f | dice_neg: %0.4f | dice_pos: %0.4f" % (epoch_loss, iou, dice, dice_neg, dice_pos))
return dice, iou
def compute_ious(pred, label, classes, ignore_index=255, only_present=True):
'''computes iou for one ground truth mask and predicted mask'''
pred[label == ignore_index] = 0
ious = []
for c in classes:
label_c = label == c
if only_present and np.sum(label_c) == 0:
ious.append(np.nan)
continue
pred_c = pred == c
intersection = np.logical_and(pred_c, label_c).sum()
union = np.logical_or(pred_c, label_c).sum()
if union != 0:
ious.append(intersection / union)
return ious if ious else [1]
def compute_iou_batch(outputs, labels, classes=None):
'''computes mean iou for a batch of ground truth masks and predicted masks'''
ious = []
preds = np.copy(outputs) # copy is imp
labels = np.array(labels) # tensor to np
for pred, label in zip(preds, labels):
ious.append(np.nanmean(compute_ious(pred, label, classes)))
iou = np.nanmean(ious)
return iou
model = Unet("efficientnet-b3", encoder_weights="imagenet", classes=4, activation=None)
class Trainer(object):
'''This class takes care of training and validation of our model'''
def __init__(self, model):
self.num_workers = 6
self.batch_size = {"train": 4, "val": 4}
self.accumulation_steps = 32 // self.batch_size['train']
self.lr = 5e-4
self.num_epochs = 20
self.best_loss = float("inf")
self.phases = ["train", "val"]
self.device = torch.device("cuda:0")
torch.set_default_tensor_type("torch.cuda.FloatTensor")
self.net = model
self.criterion = torch.nn.BCEWithLogitsLoss()
self.optimizer = optim.Adam(self.net.parameters(), lr=self.lr)
self.scheduler = ReduceLROnPlateau(self.optimizer, mode="min", patience=3, verbose=True)
self.net = self.net.to(self.device)
cudnn.benchmark = True
self.dataloaders = {
phase: provider(
data_folder=data_folder,
df_path=train_df_path,
phase=phase,
mean=(0.485, 0.456, 0.406),
std=(0.229, 0.224, 0.225),
batch_size=self.batch_size[phase],
num_workers=self.num_workers,
)
for phase in self.phases
}
self.losses = {phase: [] for phase in self.phases}
self.iou_scores = {phase: [] for phase in self.phases}
self.dice_scores = {phase: [] for phase in self.phases}
def forward(self, images, targets):
images = images.to(self.device)
masks = targets.to(self.device)
outputs = self.net(images)
loss = self.criterion(outputs, masks)
return loss, outputs
def iterate(self, epoch, phase):
meter = Meter(phase, epoch)
start = time.strftime("%H:%M:%S")
print(f"Starting epoch: {epoch} | phase: {phase} | ⏰: {start}")
batch_size = self.batch_size[phase]
self.net.train(phase == "train")
dataloader = self.dataloaders[phase]
running_loss = 0.0
total_batches = len(dataloader)
# tk0 = tqdm(dataloader, total=total_batches)
self.optimizer.zero_grad()
for itr, batch in enumerate(dataloader): # replace `dataloader` with `tk0` for tqdm
images, targets = batch
loss, outputs = self.forward(images, targets)
loss = loss / self.accumulation_steps
if phase == "train":
loss.backward()
if (itr + 1 ) % self.accumulation_steps == 0:
self.optimizer.step()
self.optimizer.zero_grad()
running_loss += loss.item()
outputs = outputs.detach().cpu()
meter.update(targets, outputs)
# tk0.set_postfix(loss=(running_loss / ((itr + 1))))
epoch_loss = (running_loss * self.accumulation_steps) / total_batches
dice, iou = epoch_log(phase, epoch, epoch_loss, meter, start)
self.losses[phase].append(epoch_loss)
self.dice_scores[phase].append(dice)
self.iou_scores[phase].append(iou)
torch.cuda.empty_cache()
return epoch_loss
def start(self):
for epoch in range(self.num_epochs):
self.iterate(epoch, "train")
state = {
"epoch": epoch,
"best_loss": self.best_loss,
"state_dict": self.net.state_dict(),
"optimizer": self.optimizer.state_dict(),
}
with torch.no_grad():
val_loss = self.iterate(epoch, "val")
self.scheduler.step(val_loss)
if val_loss < self.best_loss:
print("******** New optimal found, saving state ********")
state["best_loss"] = self.best_loss = val_loss
torch.save(state, "./model.pth")
print()
train_df_path = 'steel-defect-detection/train.csv'
data_folder = "steel-defect-detection/"
test_data_folder = "steel-defect-detection/test_images"
model_trainer = Trainer(model)
model_trainer.start()
# PLOT TRAINING
losses = model_trainer.losses
dice_scores = model_trainer.dice_scores # overall dice
iou_scores = model_trainer.iou_scores
def plot(scores, name):
plt.figure(figsize=(15,5))
plt.plot(range(len(scores["train"])), scores["train"], label=f'train {name}')
plt.plot(range(len(scores["train"])), scores["val"], label=f'val {name}')
plt.title(f'{name} plot'); plt.xlabel('Epoch'); plt.ylabel(f'{name}');
plt.legend();
plt.show()
plot(losses, "BCE loss")
plot(dice_scores, "Dice score")
plot(iou_scores, "IoU score")
| 0.787646 | 0.899387 |
```
from selenium import webdriver
driver = webdriver.Firefox()
driver.get("http://www.naver.com/")
```
# 다나와 사이트 크롤링
```
driver.get("http://prod.danawa.com/list/?cate=112758&searchOption=btnAllOptUse=true/searchMaker%5B%5D=702/innerSearchKeyword=")
import pandas as pd
import pickle
post= driver.find_elements_by_css_selector("#productListArea")[0].find_elements_by_css_selector("div.main_prodlist.main_prodlist_list")[0]
post_elements = post.find_elements_by_css_selector("div.prod_main_info")
a = post_elements[0].text.strip()
a
import re
pattern = re.compile("[SSDDRLmTAM23.]{3,5} / [0-9]{1,3}[G,T]{1}B")
pattern2 = re.compile("[0-9]?[.]?[0-9]{1,3}[ K]{0,2}?g")
pattern2.findall(a)
df = pd.DataFrame(columns=["CPU" , "CPU2", "CPU3", "Ram" , "Memory_type" ,"Memory_size", "Weight" ,"Size", "Price"])
import re
def crawling():
for post_element in post_elements:
spec = post_element.find_element_by_css_selector("div.spec_list").text.strip()
price = post_element.find_element_by_css_selector("div.prod_pricelist").text.strip()
cpu_pattern = re.compile("코어i[0-9]{1}")
cpu2_pattern = re.compile("[0-9]{1}세대")
cpu3_pattern = re.compile("[0-9 ]{4}[A-Z]{1,2}")
ram_pattern = re.compile(" [0-9]{1,2}GB")
memory_pattern = re.compile("[SSDDRLmTAM234.]{3,5} / [0-9]{1,3}[G,T]{1}B")
weight_pattern = re.compile("[0-9]?[.]?[0-9]{1,3}[ K]{0,2}?g")
size_pattern = re.compile("[]?{1,2}[0-9]{1,2}[.]?[0-9 ]{1,3}인치")
price_pattern = re.compile("[0-9]?[,]?[0-9]{2,3}[,][0-9]{3}원")
cpu = cpu_pattern.findall(spec)[0]
cpu2 = cpu2_pattern.findall(spec)[0]
cpu3 = cpu3_pattern.findall(spec)[0]
ram = ram_pattern.findall(spec)[0]
memory = memory_pattern.findall(spec)
if memory:
memory_type = memory_pattern.findall(spec)[0].split("/")[0]
else:
memory = "HDD"
memory_size = memory_pattern.findall(spec)[0].split("/")[1]
weight = weight_pattern.findall(spec)
if weight:
weight = weight_pattern.findall(spec)[0]
else:
weight = "None"
size = size_pattern.findall(spec)
if size:
size = size_pattern.findall(spec)[0]
else:
size = "None"
try:
price = price_pattern.findall(price)[0]
data = {"CPU" : cpu , "CPU2": cpu2 , "CPU3":cpu3 , "Ram" : ram , "Memory_type" : memory_type ,"Memory_size": memory_size, "Weight" : weight , "Size":size , "Price" : price}
df.loc[len(df)] = data
except:
pass
return df
# 자바스크립트 페이지가져오기 (movePage)
url = driver.find_elements_by_css_selector("div.number_wrap")[0].find_elements_by_css_selector("a")[0].get_attribute("href")
driver.get(url)
import time
for i in range(1,18):
time.sleep(3)
driver.execute_script("movePage({i})".format(i = i))
time.sleep(3)
post= driver.find_elements_by_css_selector("#productListArea")[0].find_elements_by_css_selector("div.main_prodlist.main_prodlist_list")[0]
time.sleep(3.6)
post_elements = post.find_elements_by_css_selector("div.prod_main_info")
time.sleep(3)
crawling()
```
# 주의:
Memory_type에 보조기억장치(SSD,M.2..)가 아닌 DDR3L이 적혀있을 경우 HDD임을 뜻한다.
이 점은 나중에 전처리를 할 예정!!
### SSD, M.2 -> SSD로 통일
### DDR3L => SSD가 없어 Ram타입이나온것으로, HDD로 전처리 할 예정
```
df
#df.to_pickle("DanawaLg.pkl")
import pickle
pickle.load(open("DanawaLg.pkl","rb"))
#lg_df.to_csv("DanawaLg.csv")
```
|
github_jupyter
|
from selenium import webdriver
driver = webdriver.Firefox()
driver.get("http://www.naver.com/")
driver.get("http://prod.danawa.com/list/?cate=112758&searchOption=btnAllOptUse=true/searchMaker%5B%5D=702/innerSearchKeyword=")
import pandas as pd
import pickle
post= driver.find_elements_by_css_selector("#productListArea")[0].find_elements_by_css_selector("div.main_prodlist.main_prodlist_list")[0]
post_elements = post.find_elements_by_css_selector("div.prod_main_info")
a = post_elements[0].text.strip()
a
import re
pattern = re.compile("[SSDDRLmTAM23.]{3,5} / [0-9]{1,3}[G,T]{1}B")
pattern2 = re.compile("[0-9]?[.]?[0-9]{1,3}[ K]{0,2}?g")
pattern2.findall(a)
df = pd.DataFrame(columns=["CPU" , "CPU2", "CPU3", "Ram" , "Memory_type" ,"Memory_size", "Weight" ,"Size", "Price"])
import re
def crawling():
for post_element in post_elements:
spec = post_element.find_element_by_css_selector("div.spec_list").text.strip()
price = post_element.find_element_by_css_selector("div.prod_pricelist").text.strip()
cpu_pattern = re.compile("코어i[0-9]{1}")
cpu2_pattern = re.compile("[0-9]{1}세대")
cpu3_pattern = re.compile("[0-9 ]{4}[A-Z]{1,2}")
ram_pattern = re.compile(" [0-9]{1,2}GB")
memory_pattern = re.compile("[SSDDRLmTAM234.]{3,5} / [0-9]{1,3}[G,T]{1}B")
weight_pattern = re.compile("[0-9]?[.]?[0-9]{1,3}[ K]{0,2}?g")
size_pattern = re.compile("[]?{1,2}[0-9]{1,2}[.]?[0-9 ]{1,3}인치")
price_pattern = re.compile("[0-9]?[,]?[0-9]{2,3}[,][0-9]{3}원")
cpu = cpu_pattern.findall(spec)[0]
cpu2 = cpu2_pattern.findall(spec)[0]
cpu3 = cpu3_pattern.findall(spec)[0]
ram = ram_pattern.findall(spec)[0]
memory = memory_pattern.findall(spec)
if memory:
memory_type = memory_pattern.findall(spec)[0].split("/")[0]
else:
memory = "HDD"
memory_size = memory_pattern.findall(spec)[0].split("/")[1]
weight = weight_pattern.findall(spec)
if weight:
weight = weight_pattern.findall(spec)[0]
else:
weight = "None"
size = size_pattern.findall(spec)
if size:
size = size_pattern.findall(spec)[0]
else:
size = "None"
try:
price = price_pattern.findall(price)[0]
data = {"CPU" : cpu , "CPU2": cpu2 , "CPU3":cpu3 , "Ram" : ram , "Memory_type" : memory_type ,"Memory_size": memory_size, "Weight" : weight , "Size":size , "Price" : price}
df.loc[len(df)] = data
except:
pass
return df
# 자바스크립트 페이지가져오기 (movePage)
url = driver.find_elements_by_css_selector("div.number_wrap")[0].find_elements_by_css_selector("a")[0].get_attribute("href")
driver.get(url)
import time
for i in range(1,18):
time.sleep(3)
driver.execute_script("movePage({i})".format(i = i))
time.sleep(3)
post= driver.find_elements_by_css_selector("#productListArea")[0].find_elements_by_css_selector("div.main_prodlist.main_prodlist_list")[0]
time.sleep(3.6)
post_elements = post.find_elements_by_css_selector("div.prod_main_info")
time.sleep(3)
crawling()
df
#df.to_pickle("DanawaLg.pkl")
import pickle
pickle.load(open("DanawaLg.pkl","rb"))
#lg_df.to_csv("DanawaLg.csv")
| 0.179531 | 0.304307 |
<a href="https://colab.research.google.com/github/diegosaldiasq/al-vectores/blob/main/Analisis_de_sentimiento.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import numpy as np
import pandas as pd
def sentimiento(Tweet):
#lower() = conviente todas la letras de la palabras en minusculas
#split(" ") = separa la cadena por cada palabra para formar el vector separado por sus palabras
tweet = Tweet.replace("!","").replace(",","").replace(".","").lower().split(" ")
palabras = ['muerte','pérdida','luto','excelente','gran','positivo','vaya','ignorante','aprender','platzi','asesorarme','quiero']
palabras_positivas = ['excelente','gran','positivo','platzi']
palabras_neutras = ['pérdida','aprender','asesorarme','quiero']
palabras_negativas = ['muerte','luto','ignorante','vaya']
w = []
positivas = 0
neutras = 0
negativas = 0
for i in palabras:
w.append(tweet.count(i))
if i in tweet and i in palabras_positivas:
positivas += 1
elif i in tweet and i in palabras_neutras:
neutras += 1
elif i in tweet and i in palabras_negativas:
negativas +=1
s = np.array([positivas,neutras,negativas])
w = np.array(w)
calidad = (np.ones(w.size)/w.size).T@w
puntaje = (np.ones(s.size)/s.size).T@s
V_score = np.array([1,0,-1])
score = V_score.T@s
p_s = s/(s[0]+s[1]+s[2])
return Tweet,calidad,puntaje,score,p_s[0],p_s[1],p_s[2],w,s
tweet1 = "Gran mexicano y excelente en su área, su muerte es una enorme perdida y debería ser luto nacional!!!"
tweet2 = "Vaya señora que bueno que se asesora por alguien inteligente no por el ignorante del Gatt."
tweet3 = "Se me ocurre y sin ver todos los videos de Plazti que me informéis por dónde empiezo. Entiendo que os tendría que decir quién soy y que quiero, vamos conocerme para asesorarme bien. Un saludo"
tweet4 = "Soy docente universitario, estoy intentando preparar mis clases en modo platzi bien didáctico, (le llamo modo noticiero), descargue una plataforma gratuita de grabación y transmisión de vídeo, se llama Obs estudio!bueno la sigo remando con sus funciones pero sé que saldrá algo!"
tweets = [tweet1,tweet2,tweet3,tweet4]
resultados = []
positivos = []
negativos = []
c_p = 0
n_p = 0
for j in tweets:
resultados.append(sentimiento(j))
positivos.append(sentimiento(j)[0:5:4])
negativos.append(sentimiento(j)[0:7:6])
c_p += sentimiento(j)[1]
n_p += 1
#Tweet mas positivo
po2 = []
ne2 = []
po = np.array(positivos)
ne = np.array(negativos)
for ax, i in enumerate(po):
po2.append(po[ax][1])
po2 = max(sorted(po2))
val_m = 0
for i,value in po:
if po2 == value:
val_m = i
cout = 0
for i,value in positivos:
cout +=1
if val_m == i:
print(f'El Tweet mas positivo es: el tweet{cout}\n --> {i}\n')
#Tweet mas negativo
for bx, j in enumerate(ne):
ne2.append(ne[bx][1])
ne2 = max(sorted(ne2))
val_n = 0
for i,value in ne:
if ne2 == value:
val_n = i
cout2 = 0
for i,value in negativos:
cout2 += 1
if val_n == i:
print(f'El Tweet mas negativo es: el tweet{cout2}\n --> {i}\n')
#Calidad promedio
calidad_promedio = c_p/n_p
print(f'La calidad promedio es: {round(calidad_promedio,2)}')
df = pd.DataFrame(resultados, columns=['Tweet','Calidad','Promedio sentimiento','Score','P_positiva','P_neutra','P_negativa','Vector veces','Vector sensibilidad'])
df
```
|
github_jupyter
|
import numpy as np
import pandas as pd
def sentimiento(Tweet):
#lower() = conviente todas la letras de la palabras en minusculas
#split(" ") = separa la cadena por cada palabra para formar el vector separado por sus palabras
tweet = Tweet.replace("!","").replace(",","").replace(".","").lower().split(" ")
palabras = ['muerte','pérdida','luto','excelente','gran','positivo','vaya','ignorante','aprender','platzi','asesorarme','quiero']
palabras_positivas = ['excelente','gran','positivo','platzi']
palabras_neutras = ['pérdida','aprender','asesorarme','quiero']
palabras_negativas = ['muerte','luto','ignorante','vaya']
w = []
positivas = 0
neutras = 0
negativas = 0
for i in palabras:
w.append(tweet.count(i))
if i in tweet and i in palabras_positivas:
positivas += 1
elif i in tweet and i in palabras_neutras:
neutras += 1
elif i in tweet and i in palabras_negativas:
negativas +=1
s = np.array([positivas,neutras,negativas])
w = np.array(w)
calidad = (np.ones(w.size)/w.size).T@w
puntaje = (np.ones(s.size)/s.size).T@s
V_score = np.array([1,0,-1])
score = V_score.T@s
p_s = s/(s[0]+s[1]+s[2])
return Tweet,calidad,puntaje,score,p_s[0],p_s[1],p_s[2],w,s
tweet1 = "Gran mexicano y excelente en su área, su muerte es una enorme perdida y debería ser luto nacional!!!"
tweet2 = "Vaya señora que bueno que se asesora por alguien inteligente no por el ignorante del Gatt."
tweet3 = "Se me ocurre y sin ver todos los videos de Plazti que me informéis por dónde empiezo. Entiendo que os tendría que decir quién soy y que quiero, vamos conocerme para asesorarme bien. Un saludo"
tweet4 = "Soy docente universitario, estoy intentando preparar mis clases en modo platzi bien didáctico, (le llamo modo noticiero), descargue una plataforma gratuita de grabación y transmisión de vídeo, se llama Obs estudio!bueno la sigo remando con sus funciones pero sé que saldrá algo!"
tweets = [tweet1,tweet2,tweet3,tweet4]
resultados = []
positivos = []
negativos = []
c_p = 0
n_p = 0
for j in tweets:
resultados.append(sentimiento(j))
positivos.append(sentimiento(j)[0:5:4])
negativos.append(sentimiento(j)[0:7:6])
c_p += sentimiento(j)[1]
n_p += 1
#Tweet mas positivo
po2 = []
ne2 = []
po = np.array(positivos)
ne = np.array(negativos)
for ax, i in enumerate(po):
po2.append(po[ax][1])
po2 = max(sorted(po2))
val_m = 0
for i,value in po:
if po2 == value:
val_m = i
cout = 0
for i,value in positivos:
cout +=1
if val_m == i:
print(f'El Tweet mas positivo es: el tweet{cout}\n --> {i}\n')
#Tweet mas negativo
for bx, j in enumerate(ne):
ne2.append(ne[bx][1])
ne2 = max(sorted(ne2))
val_n = 0
for i,value in ne:
if ne2 == value:
val_n = i
cout2 = 0
for i,value in negativos:
cout2 += 1
if val_n == i:
print(f'El Tweet mas negativo es: el tweet{cout2}\n --> {i}\n')
#Calidad promedio
calidad_promedio = c_p/n_p
print(f'La calidad promedio es: {round(calidad_promedio,2)}')
df = pd.DataFrame(resultados, columns=['Tweet','Calidad','Promedio sentimiento','Score','P_positiva','P_neutra','P_negativa','Vector veces','Vector sensibilidad'])
df
| 0.068928 | 0.876423 |
# How do starspot contrasts vary with stellar effective temperature in the Gaia $G$ bandpass?
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from astropy.io import ascii
import astropy.units as u
from astropy.modeling.blackbody import blackbody_lambda
```
Use starspot properties from Table 5 of [Berdyugina 2005](http://adsabs.harvard.edu/abs/2005LRSP....2....8B):
```
table5 = ascii.read('data/berdyugina2005.csv')
```
Use the $G$ band transmittance curve from Figure 3 of [Jordi et al. 2010](https://arxiv.org/abs/1008.0815), accessed via the [SVO Filter Profile Service](http://svo2.cab.inta-csic.es/svo/theory/fps/index.php?id=GAIA/GAIA0.G&&mode=browse&gname=GAIA&gname2=GAIA0#filter):
```
gaia_wl, gaia_transmissivity = np.loadtxt('data/gaiagband_svo.txt', unpack=True)
gaia_wl *= u.Angstrom
kepler_wl, kepler_transmissivity = np.loadtxt('data/keplerband_svo.txt', unpack=True)
kepler_wl *= u.Angstrom
tess_wl, tess_transmissivity = np.loadtxt('data/tess.csv', delimiter=',', unpack=True)
tess_wl *= u.nm
tess_wl = tess_wl.to(u.Angstrom)
plt.plot(gaia_wl, gaia_transmissivity, label='Gaia')
plt.plot(kepler_wl, kepler_transmissivity/kepler_transmissivity.max(), label='Kepler')
plt.plot(tess_wl, tess_transmissivity, label='TESS')
plt.legend()
plt.gca().set(xlabel='Wavelength [Angstrom]', ylabel='Transmissivity');
#plt.plot(table5['Teff K'], table5['deltaT K'], 'k.')
weighted_sunspot_temp_diff = (5770-5000)/5 + (5770-4200)*4/5
x = np.concatenate([table5['Teff K'].data.data, [5770]])
y = np.concatenate([table5['deltaT K'].data.data,
[weighted_sunspot_temp_diff]])
x = x[y != 0]
y = y[y != 0]
yerr = 300 * np.ones_like(x)
yerr[-1] = 100
plt.plot(x, y, 'k.')
p, V = np.polyfit(x, y, 2, cov=True, w=1/yerr)
y_fit = np.polyval(p, x)
a0_err = np.sqrt(V[0, 0])
a1_err = np.sqrt(V[1, 1])
a2_err = np.sqrt(V[2, 2])
sort = np.argsort(x)
p_plus = p + np.array([a0_err, a1_err, a2_err])/20
p_minus = p - np.array([a0_err, a1_err, a2_err])/20
y_fit_plus = np.polyval(p_plus, x)
y_fit_minus = np.polyval(p_minus, x)
fit_params = [p_plus, p, p_minus]
x_range = np.linspace(x.min(), x.max(), 100)
np.savetxt('data/spot_temp_contrast.txt',
np.vstack([x_range, np.polyval(p, x_range)]).T)
plt.plot(x[sort], y_fit_plus[sort])
plt.plot(x[sort], y_fit[sort])
plt.plot(x[sort], y_fit_minus[sort])
plt.show()
```
Plot the spot and photosphere temperatures, along with the spot contrasts integrated in the Gaia bandpass.
```
import sys
sys.path.insert(0, '../')
from mrspoc import get_table_ms
from mrspoc.tgas import mamajek_path
mamajek = ascii.read(mamajek_path, format='commented_header')
effective_temperatures = {spt[:-1]: teff for spt, teff in
zip(mamajek['SpT'].data, mamajek['Teff'].data)}
table = get_table_ms()
fontsize = 13
temp_phots = np.linspace(x.min(), x.max(), 50)
labels = ['upper', 'best', 'lower']
linestyles = ['--', '-', '-.']
teff_bounds = [3000, 6000]
fig, ax = plt.subplots(3, 1, figsize=(3.5, 10))
gaia_best_contrasts = None
for fit_p, ls, label in zip(fit_params, linestyles, labels):
get_temp_diff = lambda tphot: np.polyval(fit_p, tphot)
ax[0].plot(temp_phots, get_temp_diff(temp_phots), label=label, ls=ls, lw=1.5)
gaia_contrasts = []
for temp in temp_phots:
phot_irr = blackbody_lambda(gaia_wl, temp)
spot_temp = temp - get_temp_diff(temp)
spot_irr = blackbody_lambda(gaia_wl, spot_temp)
contrast = (1 - np.trapz(spot_irr*gaia_transmissivity*gaia_wl) /
np.trapz(phot_irr*gaia_transmissivity*gaia_wl))
gaia_contrasts.append(contrast)
ax[1].plot(temp_phots, gaia_contrasts, ls=ls, lw=1.5, label=label)
if label == 'best':
gaia_best_contrasts = gaia_contrasts
kepler_contrasts = []
for temp in temp_phots:
phot_irr = blackbody_lambda(kepler_wl, temp)
spot_temp = temp - get_temp_diff(temp)
spot_irr = blackbody_lambda(kepler_wl, spot_temp)
contrast = (1 - np.trapz(spot_irr*kepler_transmissivity*kepler_wl) /
np.trapz(phot_irr*kepler_transmissivity*kepler_wl))
kepler_contrasts.append(contrast)
ax[2].plot(temp_phots, kepler_contrasts, ls=ls,
lw=1.5, label='Kepler', color='#9e0303')
tess_contrasts = []
for temp in temp_phots:
phot_irr = blackbody_lambda(tess_wl, temp)
spot_temp = temp - get_temp_diff(temp)
spot_irr = blackbody_lambda(tess_wl, spot_temp)
contrast = (1 - np.trapz(spot_irr*tess_transmissivity*tess_wl) /
np.trapz(phot_irr*tess_transmissivity*tess_wl))
tess_contrasts.append(contrast)
ax[2].plot(temp_phots, tess_contrasts, ls='--',
lw=1.5, label='TESS', color='#9f41fc')
ax[0].legend(loc='upper left', fontsize=8)
ax[1].legend(loc='lower right')
ax[2].legend(loc='lower right')
ax[0].plot(x[:-1], y[:-1], 'k.')
ax0_upper = ax[0].twiny()
label_sptypes = ['M4', 'M0', 'K4', 'K0', 'G2']
label_teffs = [effective_temperatures[l] for l in label_sptypes]
ax0_upper.set_xlim(teff_bounds)
ax0_upper.set_xticks(label_teffs)
ax0_upper.set_xticklabels(label_sptypes)
sunspot_contrasts = []
for temp_diff, temp_phot, label in zip([5770-5000, 5770-4200],
[5770, 5770],
['umbra', 'penumbra']):
phot_irr = blackbody_lambda(gaia_wl, temp_phot)
spot_temp = temp_phot - temp_diff
spot_irr = blackbody_lambda(gaia_wl, spot_temp)
contrast = (1 - np.trapz(spot_irr*gaia_transmissivity*gaia_wl) /
np.trapz(phot_irr*gaia_transmissivity*gaia_wl))
sunspot_contrasts.append(contrast.value)
ax[0].scatter(temp_phot, weighted_sunspot_temp_diff,
color='k', marker='$\odot$',
label='sun', s=150)
area_weighted_sunspot_contrast = np.dot(sunspot_contrasts, [1/5, 4/5])
ax[1].set_ylabel('Spot contrast\nin Gaia $G$ band', fontsize=fontsize)
ax[2].set_ylabel('Spot contrast\n(other missions)', fontsize=fontsize)
ax[-1].set_xlabel("$T_{\mathrm{phot}}$ [K]", fontsize=fontsize)
ax[0].set_ylabel("$T_{\mathrm{phot}} - T_{\mathrm{spot}}$ [K]",
fontsize=fontsize)
xticks = np.arange(3000, 7000, 1000)
for axis in ax:
axis.grid(ls='--')
axis.set_xlim(teff_bounds)
axis.set_xticks(xticks)
for i in range(3):
ax[i].set_xticklabels([])
for i in range(1, 3):
#ax[i].set_xticks(ax[0].get_xticks())
ax[i].set_ylim([0, 1])
ax[i].scatter(5770, area_weighted_sunspot_contrast, color='k', marker='$\odot$',
label='sun', s=100)
ax[-1].set_xticklabels(xticks)
#ax[3].hist(table['Teff'], range=teff_bounds, color='k')
#ax[3].set_yticks([])
#ax[3].set_ylabel('Relative number\nof TGAS stars', fontsize=fontsize)
fig.subplots_adjust(hspace=0.08)
fig.savefig('contrasts.pdf', bbox_inches='tight')
```
*Upper*: The spot temperature contrasts scale quadratically with photosphere effective temperatures. The mean, area-weighted sunspot contrast is labeled $\odot$ (including both the umbra and penumbra).
*Lower*: Spot flux contrasts, approximated by integrating blackbody radiance curves with the temperatures of the photosphere and spot, convolved with the Gaia $G$ bandpass. The area-weighted mean sunspot contrast is 0.7, marked with $\odot$.
```
# plt.plot(temp_phots, gaia_best_contrasts)
# plt.plot(temp_phots, kepler_contrasts)
# plt.plot(temp_phots, tess_contrasts)
diffs = [np.array(kepler_contrasts) - np.array(gaia_best_contrasts),
np.array(tess_contrasts) - np.array(gaia_best_contrasts)]
plt.plot(temp_phots, np.mean(np.abs(diffs)/np.array(gaia_best_contrasts), axis=0))
plt.ylabel('max fractional err between missions')
plt.show()
np.count_nonzero((table['Teff'].data < 5770) & (table['Teff'].data > 3550))/len(table)
len(table)
```
|
github_jupyter
|
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from astropy.io import ascii
import astropy.units as u
from astropy.modeling.blackbody import blackbody_lambda
table5 = ascii.read('data/berdyugina2005.csv')
gaia_wl, gaia_transmissivity = np.loadtxt('data/gaiagband_svo.txt', unpack=True)
gaia_wl *= u.Angstrom
kepler_wl, kepler_transmissivity = np.loadtxt('data/keplerband_svo.txt', unpack=True)
kepler_wl *= u.Angstrom
tess_wl, tess_transmissivity = np.loadtxt('data/tess.csv', delimiter=',', unpack=True)
tess_wl *= u.nm
tess_wl = tess_wl.to(u.Angstrom)
plt.plot(gaia_wl, gaia_transmissivity, label='Gaia')
plt.plot(kepler_wl, kepler_transmissivity/kepler_transmissivity.max(), label='Kepler')
plt.plot(tess_wl, tess_transmissivity, label='TESS')
plt.legend()
plt.gca().set(xlabel='Wavelength [Angstrom]', ylabel='Transmissivity');
#plt.plot(table5['Teff K'], table5['deltaT K'], 'k.')
weighted_sunspot_temp_diff = (5770-5000)/5 + (5770-4200)*4/5
x = np.concatenate([table5['Teff K'].data.data, [5770]])
y = np.concatenate([table5['deltaT K'].data.data,
[weighted_sunspot_temp_diff]])
x = x[y != 0]
y = y[y != 0]
yerr = 300 * np.ones_like(x)
yerr[-1] = 100
plt.plot(x, y, 'k.')
p, V = np.polyfit(x, y, 2, cov=True, w=1/yerr)
y_fit = np.polyval(p, x)
a0_err = np.sqrt(V[0, 0])
a1_err = np.sqrt(V[1, 1])
a2_err = np.sqrt(V[2, 2])
sort = np.argsort(x)
p_plus = p + np.array([a0_err, a1_err, a2_err])/20
p_minus = p - np.array([a0_err, a1_err, a2_err])/20
y_fit_plus = np.polyval(p_plus, x)
y_fit_minus = np.polyval(p_minus, x)
fit_params = [p_plus, p, p_minus]
x_range = np.linspace(x.min(), x.max(), 100)
np.savetxt('data/spot_temp_contrast.txt',
np.vstack([x_range, np.polyval(p, x_range)]).T)
plt.plot(x[sort], y_fit_plus[sort])
plt.plot(x[sort], y_fit[sort])
plt.plot(x[sort], y_fit_minus[sort])
plt.show()
import sys
sys.path.insert(0, '../')
from mrspoc import get_table_ms
from mrspoc.tgas import mamajek_path
mamajek = ascii.read(mamajek_path, format='commented_header')
effective_temperatures = {spt[:-1]: teff for spt, teff in
zip(mamajek['SpT'].data, mamajek['Teff'].data)}
table = get_table_ms()
fontsize = 13
temp_phots = np.linspace(x.min(), x.max(), 50)
labels = ['upper', 'best', 'lower']
linestyles = ['--', '-', '-.']
teff_bounds = [3000, 6000]
fig, ax = plt.subplots(3, 1, figsize=(3.5, 10))
gaia_best_contrasts = None
for fit_p, ls, label in zip(fit_params, linestyles, labels):
get_temp_diff = lambda tphot: np.polyval(fit_p, tphot)
ax[0].plot(temp_phots, get_temp_diff(temp_phots), label=label, ls=ls, lw=1.5)
gaia_contrasts = []
for temp in temp_phots:
phot_irr = blackbody_lambda(gaia_wl, temp)
spot_temp = temp - get_temp_diff(temp)
spot_irr = blackbody_lambda(gaia_wl, spot_temp)
contrast = (1 - np.trapz(spot_irr*gaia_transmissivity*gaia_wl) /
np.trapz(phot_irr*gaia_transmissivity*gaia_wl))
gaia_contrasts.append(contrast)
ax[1].plot(temp_phots, gaia_contrasts, ls=ls, lw=1.5, label=label)
if label == 'best':
gaia_best_contrasts = gaia_contrasts
kepler_contrasts = []
for temp in temp_phots:
phot_irr = blackbody_lambda(kepler_wl, temp)
spot_temp = temp - get_temp_diff(temp)
spot_irr = blackbody_lambda(kepler_wl, spot_temp)
contrast = (1 - np.trapz(spot_irr*kepler_transmissivity*kepler_wl) /
np.trapz(phot_irr*kepler_transmissivity*kepler_wl))
kepler_contrasts.append(contrast)
ax[2].plot(temp_phots, kepler_contrasts, ls=ls,
lw=1.5, label='Kepler', color='#9e0303')
tess_contrasts = []
for temp in temp_phots:
phot_irr = blackbody_lambda(tess_wl, temp)
spot_temp = temp - get_temp_diff(temp)
spot_irr = blackbody_lambda(tess_wl, spot_temp)
contrast = (1 - np.trapz(spot_irr*tess_transmissivity*tess_wl) /
np.trapz(phot_irr*tess_transmissivity*tess_wl))
tess_contrasts.append(contrast)
ax[2].plot(temp_phots, tess_contrasts, ls='--',
lw=1.5, label='TESS', color='#9f41fc')
ax[0].legend(loc='upper left', fontsize=8)
ax[1].legend(loc='lower right')
ax[2].legend(loc='lower right')
ax[0].plot(x[:-1], y[:-1], 'k.')
ax0_upper = ax[0].twiny()
label_sptypes = ['M4', 'M0', 'K4', 'K0', 'G2']
label_teffs = [effective_temperatures[l] for l in label_sptypes]
ax0_upper.set_xlim(teff_bounds)
ax0_upper.set_xticks(label_teffs)
ax0_upper.set_xticklabels(label_sptypes)
sunspot_contrasts = []
for temp_diff, temp_phot, label in zip([5770-5000, 5770-4200],
[5770, 5770],
['umbra', 'penumbra']):
phot_irr = blackbody_lambda(gaia_wl, temp_phot)
spot_temp = temp_phot - temp_diff
spot_irr = blackbody_lambda(gaia_wl, spot_temp)
contrast = (1 - np.trapz(spot_irr*gaia_transmissivity*gaia_wl) /
np.trapz(phot_irr*gaia_transmissivity*gaia_wl))
sunspot_contrasts.append(contrast.value)
ax[0].scatter(temp_phot, weighted_sunspot_temp_diff,
color='k', marker='$\odot$',
label='sun', s=150)
area_weighted_sunspot_contrast = np.dot(sunspot_contrasts, [1/5, 4/5])
ax[1].set_ylabel('Spot contrast\nin Gaia $G$ band', fontsize=fontsize)
ax[2].set_ylabel('Spot contrast\n(other missions)', fontsize=fontsize)
ax[-1].set_xlabel("$T_{\mathrm{phot}}$ [K]", fontsize=fontsize)
ax[0].set_ylabel("$T_{\mathrm{phot}} - T_{\mathrm{spot}}$ [K]",
fontsize=fontsize)
xticks = np.arange(3000, 7000, 1000)
for axis in ax:
axis.grid(ls='--')
axis.set_xlim(teff_bounds)
axis.set_xticks(xticks)
for i in range(3):
ax[i].set_xticklabels([])
for i in range(1, 3):
#ax[i].set_xticks(ax[0].get_xticks())
ax[i].set_ylim([0, 1])
ax[i].scatter(5770, area_weighted_sunspot_contrast, color='k', marker='$\odot$',
label='sun', s=100)
ax[-1].set_xticklabels(xticks)
#ax[3].hist(table['Teff'], range=teff_bounds, color='k')
#ax[3].set_yticks([])
#ax[3].set_ylabel('Relative number\nof TGAS stars', fontsize=fontsize)
fig.subplots_adjust(hspace=0.08)
fig.savefig('contrasts.pdf', bbox_inches='tight')
# plt.plot(temp_phots, gaia_best_contrasts)
# plt.plot(temp_phots, kepler_contrasts)
# plt.plot(temp_phots, tess_contrasts)
diffs = [np.array(kepler_contrasts) - np.array(gaia_best_contrasts),
np.array(tess_contrasts) - np.array(gaia_best_contrasts)]
plt.plot(temp_phots, np.mean(np.abs(diffs)/np.array(gaia_best_contrasts), axis=0))
plt.ylabel('max fractional err between missions')
plt.show()
np.count_nonzero((table['Teff'].data < 5770) & (table['Teff'].data > 3550))/len(table)
len(table)
| 0.321993 | 0.911219 |
<a href="https://colab.research.google.com/github/https-deeplearning-ai/tensorflow-1-public/blob/master/C3/W2/ungraded_labs/C3_W2_Lab_1_imdb.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Ungraded Lab: Training a binary classifier with the IMDB Reviews Dataset
In this lab, you will be building a sentiment classification model to distinguish between positive and negative movie reviews. You will train it on the [IMDB Reviews](http://ai.stanford.edu/~amaas/data/sentiment/) dataset and visualize the word embeddings generated after training.
Let's get started!
## Download the Dataset
First, you will need to fetch the dataset you will be working on. This is hosted via [Tensorflow Datasets](https://www.tensorflow.org/datasets), a collection of prepared datasets for machine learning. If you're running this notebook on your local machine, make sure to have the [`tensorflow-datasets`](https://pypi.org/project/tensorflow-datasets/) package installed before importing it. You can install it via `pip` as shown in the commented cell below.
```
# Install this package if running on your local machine
# !pip install -q tensorflow-datasets
```
The [`tfds.load`](https://www.tensorflow.org/datasets/api_docs/python/tfds/load) method downloads the dataset into your working directory. You can set the `with_info` parameter to `True` if you want to see the description of the dataset. The `as_supervised` parameter, on the other hand, is set to load the data as `(input, label)` pairs.
```
import tensorflow_datasets as tfds
# Load the IMDB Reviews dataset
imdb, info = tfds.load("imdb_reviews", with_info=True, as_supervised=True)
# Print information about the dataset
print(info)
```
As you can see in the output above, there is a total of 100,000 examples in the dataset and it is split into `train`, `test` and `unsupervised` sets. For this lab, you will only use `train` and `test` sets because you will need labeled examples to train your model.
## Split the dataset
If you try printing the `imdb` dataset that you downloaded earlier, you will see that it contains the dictionary that points to [`tf.data.Dataset`](https://www.tensorflow.org/api_docs/python/tf/data/Dataset) objects. You will explore more of this class and its API in Course 4 of this specialization. For now, you can just think of it as a collection of examples.
```
# Print the contents of the dataset you downloaded
print(imdb)
```
You can preview the raw format of a few examples by using the [`take()`](https://www.tensorflow.org/api_docs/python/tf/data/Dataset#take) method and iterating over it as shown below:
```
# Take 2 training examples and print its contents
for example in imdb['train'].take(2):
print(example)
```
You can see that each example is a 2-element tuple of tensors containing the text first, then the label (shown in the `numpy()` property). The next cell below will take all the `train` and `test` sentences and labels into separate lists so you can preprocess the text and feed it to the model later.
```
import numpy as np
# Get the train and test sets
train_data, test_data = imdb['train'], imdb['test']
# Initialize sentences and labels lists
training_sentences = []
training_labels = []
testing_sentences = []
testing_labels = []
# Loop over all training examples and save the sentences and labels
for s,l in train_data:
training_sentences.append(s.numpy().decode('utf8'))
training_labels.append(l.numpy())
# Loop over all test examples and save the sentences and labels
for s,l in test_data:
testing_sentences.append(s.numpy().decode('utf8'))
testing_labels.append(l.numpy())
# Convert labels lists to numpy array
training_labels_final = np.array(training_labels)
testing_labels_final = np.array(testing_labels)
```
## Generate Padded Sequences
Now you can do the text preprocessing steps you've learned last week. You will tokenize the sentences and pad them to a uniform length. We've separated the parameters into its own code cell below so it will be easy for you to tweak it later if you want.
```
# Parameters
vocab_size = 10000
max_length = 120
embedding_dim = 16
trunc_type='post'
oov_tok = "<OOV>"
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
# Initialize the Tokenizer class
tokenizer = Tokenizer(num_words = vocab_size, oov_token=oov_tok)
# Generate the word index dictionary for the training sentences
tokenizer.fit_on_texts(training_sentences)
word_index = tokenizer.word_index
# Generate and pad the training sequences
sequences = tokenizer.texts_to_sequences(training_sentences)
padded = pad_sequences(sequences,maxlen=max_length, truncating=trunc_type)
# Generate and pad the test sequences
testing_sequences = tokenizer.texts_to_sequences(testing_sentences)
testing_padded = pad_sequences(testing_sequences,maxlen=max_length)
```
## Build and Compile the Model
With the data already preprocessed, you can proceed to building your sentiment classification model. The input will be an [`Embedding`](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Embedding) layer. The main idea here is to represent each word in your vocabulary with vectors. These vectors have trainable weights so as your neural network learns, words that are most likely to appear in a positive tweet will converge towards similar weights. Similarly, words in negative tweets will be clustered more closely together. You can read more about word embeddings [here](https://www.tensorflow.org/text/guide/word_embeddings).
After the `Embedding` layer, you will flatten its output and feed it into a `Dense` layer. You will explore other architectures for these hidden layers in the next labs.
The output layer would be a single neuron with a sigmoid activation to distinguish between the 2 classes. As is typical with binary classifiers, you will use the `binary_crossentropy` as your loss function while training.
```
import tensorflow as tf
# Build the model
model = tf.keras.Sequential([
tf.keras.layers.Embedding(vocab_size, embedding_dim, input_length=max_length),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(6, activation='relu'),
tf.keras.layers.Dense(1, activation='sigmoid')
])
# Setup the training parameters
model.compile(loss='binary_crossentropy',optimizer='adam',metrics=['accuracy'])
# Print the model summary
model.summary()
```
## Train the Model
Next, of course, is to train your model. With the current settings, you will get near perfect training accuracy after just 5 epochs but the validation accuracy will plateau at around 83%. See if you can still improve this by adjusting some of the parameters earlier (e.g. the `vocab_size`, number of `Dense` neurons, number of epochs, etc.).
```
num_epochs = 10
# Train the model
model.fit(padded, training_labels_final, epochs=num_epochs, validation_data=(testing_padded, testing_labels_final))
```
## Visualize Word Embeddings
After training, you can visualize the trained weights in the `Embedding` layer to see words that are clustered together. The [Tensorflow Embedding Projector](https://projector.tensorflow.org/) is able to reduce the 16-dimension vectors you defined earlier into fewer components so it can be plotted in the projector. First, you will need to get these weights and you can do that with the cell below:
```
# Get the embedding layer from the model (i.e. first layer)
embedding_layer = model.layers[0]
# Get the weights of the embedding layer
embedding_weights = embedding_layer.get_weights()[0]
# Print the shape. Expected is (vocab_size, embedding_dim)
print(embedding_weights.shape)
```
You will need to generate two files:
* `vecs.tsv` - contains the vector weights of each word in the vocabulary
* `meta.tsv` - contains the words in the vocabulary
For this, it is useful to have `reverse_word_index` dictionary so you can quickly lookup a word based on a given index. For example, `reverse_word_index[1]` will return your OOV token because it is always at index = 1. Fortunately, the `Tokenizer` class already provides this dictionary through its `index_word` property. Yes, as the name implies, it is the reverse of the `word_index` property which you used earlier!
```
# Get the index-word dictionary
reverse_word_index = tokenizer.index_word
```
Now you can start the loop to generate the files. You will loop `vocab_size-1` times, skipping the `0` key because it is just for the padding.
```
import io
# Open writeable files
out_v = io.open('vecs.tsv', 'w', encoding='utf-8')
out_m = io.open('meta.tsv', 'w', encoding='utf-8')
# Initialize the loop. Start counting at `1` because `0` is just for the padding
for word_num in range(1, vocab_size):
# Get the word associated at the current index
word_name = reverse_word_index[word_num]
# Get the embedding weights associated with the current index
word_embedding = embedding_weights[word_num]
# Write the word name
out_m.write(word_name + "\n")
# Write the word embedding
out_v.write('\t'.join([str(x) for x in word_embedding]) + "\n")
# Close the files
out_v.close()
out_m.close()
```
When running this on Colab, you can run the code below to download the files. Otherwise, you can see the files in your current working directory and download it manually.
```
# Import files utilities in Colab
try:
from google.colab import files
except ImportError:
pass
# Download the files
else:
files.download('vecs.tsv')
files.download('meta.tsv')
```
Now you can go to the [Tensorflow Embedding Projector](https://projector.tensorflow.org/) and load the two files you downloaded to see the visualization. You can search for words like `worst` and `fantastic` and see the other words closely located to these.
## Wrap Up
In this lab, you were able build a simple sentiment classification model and train it on preprocessed text data. In the next lessons, you will revisit the Sarcasm Dataset you used in Week 1 and build a model to train on it.
|
github_jupyter
|
# Install this package if running on your local machine
# !pip install -q tensorflow-datasets
import tensorflow_datasets as tfds
# Load the IMDB Reviews dataset
imdb, info = tfds.load("imdb_reviews", with_info=True, as_supervised=True)
# Print information about the dataset
print(info)
# Print the contents of the dataset you downloaded
print(imdb)
# Take 2 training examples and print its contents
for example in imdb['train'].take(2):
print(example)
import numpy as np
# Get the train and test sets
train_data, test_data = imdb['train'], imdb['test']
# Initialize sentences and labels lists
training_sentences = []
training_labels = []
testing_sentences = []
testing_labels = []
# Loop over all training examples and save the sentences and labels
for s,l in train_data:
training_sentences.append(s.numpy().decode('utf8'))
training_labels.append(l.numpy())
# Loop over all test examples and save the sentences and labels
for s,l in test_data:
testing_sentences.append(s.numpy().decode('utf8'))
testing_labels.append(l.numpy())
# Convert labels lists to numpy array
training_labels_final = np.array(training_labels)
testing_labels_final = np.array(testing_labels)
# Parameters
vocab_size = 10000
max_length = 120
embedding_dim = 16
trunc_type='post'
oov_tok = "<OOV>"
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
# Initialize the Tokenizer class
tokenizer = Tokenizer(num_words = vocab_size, oov_token=oov_tok)
# Generate the word index dictionary for the training sentences
tokenizer.fit_on_texts(training_sentences)
word_index = tokenizer.word_index
# Generate and pad the training sequences
sequences = tokenizer.texts_to_sequences(training_sentences)
padded = pad_sequences(sequences,maxlen=max_length, truncating=trunc_type)
# Generate and pad the test sequences
testing_sequences = tokenizer.texts_to_sequences(testing_sentences)
testing_padded = pad_sequences(testing_sequences,maxlen=max_length)
import tensorflow as tf
# Build the model
model = tf.keras.Sequential([
tf.keras.layers.Embedding(vocab_size, embedding_dim, input_length=max_length),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(6, activation='relu'),
tf.keras.layers.Dense(1, activation='sigmoid')
])
# Setup the training parameters
model.compile(loss='binary_crossentropy',optimizer='adam',metrics=['accuracy'])
# Print the model summary
model.summary()
num_epochs = 10
# Train the model
model.fit(padded, training_labels_final, epochs=num_epochs, validation_data=(testing_padded, testing_labels_final))
# Get the embedding layer from the model (i.e. first layer)
embedding_layer = model.layers[0]
# Get the weights of the embedding layer
embedding_weights = embedding_layer.get_weights()[0]
# Print the shape. Expected is (vocab_size, embedding_dim)
print(embedding_weights.shape)
# Get the index-word dictionary
reverse_word_index = tokenizer.index_word
import io
# Open writeable files
out_v = io.open('vecs.tsv', 'w', encoding='utf-8')
out_m = io.open('meta.tsv', 'w', encoding='utf-8')
# Initialize the loop. Start counting at `1` because `0` is just for the padding
for word_num in range(1, vocab_size):
# Get the word associated at the current index
word_name = reverse_word_index[word_num]
# Get the embedding weights associated with the current index
word_embedding = embedding_weights[word_num]
# Write the word name
out_m.write(word_name + "\n")
# Write the word embedding
out_v.write('\t'.join([str(x) for x in word_embedding]) + "\n")
# Close the files
out_v.close()
out_m.close()
# Import files utilities in Colab
try:
from google.colab import files
except ImportError:
pass
# Download the files
else:
files.download('vecs.tsv')
files.download('meta.tsv')
| 0.884962 | 0.990866 |
# Determinando força do concreto com modelos de regressão
Dataset do Kaggle: https://www.kaggle.com/pavanraj159/concrete-compressive-strength-data-set
Importando bibliotecas para manipulação e visualização dos dados
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
```
Lendo Arquivo CSV
```
dados = pd.read_csv('compresive_strength_concrete.csv')
```
Exibindo cinco primeiras linhas
```
dados.head()
dados.columns = ['Cimento','Blast Furnace Slag','Fly Ash','Water','Superplasticizer','Coarse Aggregate','Fine Aggregate','Age','Compressive strength']
dados.head()
```
Exibindo informações dos dados
```
dados.info()
```
Verificando se existem NaNs
```
print(dados.isna().sum())
```
Visualização dos dados
```
plt.figure(figsize=(10,10))
plt.subplot(3,3,1)
dados[dados.columns[0]].plot(kind='hist')
plt.xlabel(dados.columns[0])
plt.subplot(3,3,2)
dados[dados.columns[1]].plot(kind='hist')
plt.xlabel(dados.columns[1])
plt.subplot(3,3,3)
dados[dados.columns[2]].plot(kind='hist')
plt.xlabel(dados.columns[2])
plt.subplot(3,3,4)
dados[dados.columns[3]].plot(kind='hist')
plt.xlabel(dados.columns[3])
plt.subplot(3,3,5)
dados[dados.columns[4]].plot(kind='hist')
plt.xlabel(dados.columns[4])
plt.subplot(3,3,6)
dados[dados.columns[5]].plot(kind='hist')
plt.xlabel(dados.columns[5])
plt.subplot(3,3,7)
dados[dados.columns[6]].plot(kind='hist')
plt.xlabel(dados.columns[6])
plt.subplot(3,3,8)
dados[dados.columns[7]].plot(kind='hist')
plt.xlabel(dados.columns[7])
plt.subplot(3,3,9)
dados[dados.columns[8]].plot(kind='hist')
plt.xlabel(dados.columns[8])
plt.tight_layout()
```
Verificando correlação entre as variáveis
```
corr = dados.corr()
sns.heatmap(corr)
```
Normalizando valores no intervalo entre 0 e 1
```
from sklearn.preprocessing import MinMaxScaler
colunas = dados.columns
for col in colunas:
scaler = MinMaxScaler(feature_range=(0, 1))
dados[col] = scaler.fit_transform(dados[col].values.reshape(-1, 1))
dados.head()
```
Determinando variáveis X e Y
```
X = dados[colunas[0:-1]].values
Y = dados[dados.columns[-1]].values
```
Criando amostras de treino e teste
```
from sklearn.model_selection import train_test_split
X_treino, X_teste, Y_treino, Y_teste = train_test_split(X, Y, test_size=0.25, random_state=42)
```
Importando bibliotecas para cálculo dos erros dos modelos
```
from sklearn.metrics import mean_absolute_error,mean_squared_error
```
Criando função para cálculo do erro percentual absoluto médio
```
def mean_absolute_percentage_error(y_true, y_pred):
y_true, y_pred = np.array(y_true), np.array(y_pred)
return np.mean(np.abs((y_true - y_pred) / y_true)) * 100
```
Modelo 1: Regressão Linear
```
from sklearn.linear_model import LinearRegression
linreg = LinearRegression()
linreg.fit(X_treino,Y_treino)
Y_pred_linreg = linreg.predict(X_teste)
MAE_linreg = mean_absolute_error(Y_teste,Y_pred_linreg)
MSE_linreg = mean_squared_error(Y_teste,Y_pred_linreg)
RMSE_linreg = np.sqrt(MSE_linreg)
MAPE_linreg = mean_absolute_percentage_error(Y_teste,Y_pred_linreg)
print("MAE = {:0.2f}".format(MAE_linreg))
print("MAPE = {:0.2f}%".format(MAPE_linreg))
print("MSE = {:0.2f}".format(MSE_linreg))
print("RMSE = {:0.2f}".format(RMSE_linreg))
```
Modelo 2: Support Vector Regressor
```
from sklearn.svm import SVR
svr = SVR()
svr.fit(X_treino,Y_treino)
Y_pred_svr = svr.predict(X_teste)
MAE_svr = mean_absolute_error(Y_teste,Y_pred_svr)
MSE_svr = mean_squared_error(Y_teste,Y_pred_svr)
RMSE_svr = np.sqrt(MSE_svr)
MAPE_svr = mean_absolute_percentage_error(Y_teste,Y_pred_svr)
print("MAE = {:0.2f}".format(MAE_svr))
print("MAPE = {:0.2f}%".format(MAPE_svr))
print("MSE = {:0.2f}".format(MSE_svr))
print("RMSE = {:0.2f}".format(RMSE_svr))
```
Modelo 3: Decision Tree Regressor
```
from sklearn.tree import DecisionTreeRegressor
dte = DecisionTreeRegressor()
dte.fit(X_treino,Y_treino)
Y_pred_dte = dte.predict(X_teste)
MAE_dte = mean_absolute_error(Y_teste,Y_pred_dte)
MSE_dte = mean_squared_error(Y_teste,Y_pred_dte)
RMSE_dte = np.sqrt(MSE_dte)
MAPE_dte = mean_absolute_percentage_error(Y_teste,Y_pred_dte)
print("MAE = {:0.2f}".format(MAE_dte))
print("MAPE = {:0.2f}%".format(MAPE_dte))
print("MSE = {:0.2f}".format(MSE_dte))
print("RMSE = {:0.2f}".format(RMSE_dte))
```
Modelo 4: Random Forest Regressor
```
from sklearn.ensemble import RandomForestRegressor
rfr = RandomForestRegressor()
rfr.fit(X_treino,Y_treino)
Y_pred_rfr = rfr.predict(X_teste)
MAE_rfr = mean_absolute_error(Y_teste,Y_pred_rfr)
MSE_rfr = mean_squared_error(Y_teste,Y_pred_rfr)
RMSE_rfr = np.sqrt(MSE_rfr)
MAPE_rfr = mean_absolute_percentage_error(Y_teste,Y_pred_rfr)
print("MAE = {:0.2f}".format(MAE_rfr))
print("MAPE = {:0.2f}%".format(MAPE_rfr))
print("MSE = {:0.2f}".format(MSE_rfr))
print("RMSE = {:0.2f}".format(RMSE_rfr))
```
Modelo 5: AdaBoost Regressor
```
from sklearn.ensemble import AdaBoostRegressor
ada = AdaBoostRegressor()
ada.fit(X_treino,Y_treino)
Y_pred_ada = ada.predict(X_teste)
MAE_ada = mean_absolute_error(Y_teste,Y_pred_ada)
MSE_ada = mean_squared_error(Y_teste,Y_pred_ada)
RMSE_ada = np.sqrt(MSE_ada)
MAPE_ada = mean_absolute_percentage_error(Y_teste,Y_pred_ada)
print("MAE = {:0.2f}".format(MAE_ada))
print("MAPE = {:0.2f}%".format(MAPE_ada))
print("MSE = {:0.2f}".format(MSE_ada))
print("RMSE = {:0.2f}".format(RMSE_ada))
modelo = ["Regressão linear","SVR","Decion Tree","Random Forest","Ada Boost"]
MAE = [MAE_linreg,MAE_svr,MAE_dte,MAE_rfr,MAE_ada]
MAPE = [MAPE_linreg,MAPE_svr,MAPE_dte,MAPE_rfr,MAPE_ada]
MSE = [MSE_linreg,MSE_svr,MSE_dte,MSE_rfr,MSE_ada]
RMSE = [RMSE_linreg,RMSE_svr,RMSE_dte,RMSE_rfr,RMSE_ada]
dici = {"Modelo" : modelo, "MAE" : MAE, "MAPE" : MAPE, "MSE" : MSE, "RMSE" : RMSE}
pd_dici = pd.DataFrame(dici).sort_values(by="MAPE")
pd_dici
```
Modelo Random Forest apresentou melhor precisão em comparação aos demais modelos
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
dados = pd.read_csv('compresive_strength_concrete.csv')
dados.head()
dados.columns = ['Cimento','Blast Furnace Slag','Fly Ash','Water','Superplasticizer','Coarse Aggregate','Fine Aggregate','Age','Compressive strength']
dados.head()
dados.info()
print(dados.isna().sum())
plt.figure(figsize=(10,10))
plt.subplot(3,3,1)
dados[dados.columns[0]].plot(kind='hist')
plt.xlabel(dados.columns[0])
plt.subplot(3,3,2)
dados[dados.columns[1]].plot(kind='hist')
plt.xlabel(dados.columns[1])
plt.subplot(3,3,3)
dados[dados.columns[2]].plot(kind='hist')
plt.xlabel(dados.columns[2])
plt.subplot(3,3,4)
dados[dados.columns[3]].plot(kind='hist')
plt.xlabel(dados.columns[3])
plt.subplot(3,3,5)
dados[dados.columns[4]].plot(kind='hist')
plt.xlabel(dados.columns[4])
plt.subplot(3,3,6)
dados[dados.columns[5]].plot(kind='hist')
plt.xlabel(dados.columns[5])
plt.subplot(3,3,7)
dados[dados.columns[6]].plot(kind='hist')
plt.xlabel(dados.columns[6])
plt.subplot(3,3,8)
dados[dados.columns[7]].plot(kind='hist')
plt.xlabel(dados.columns[7])
plt.subplot(3,3,9)
dados[dados.columns[8]].plot(kind='hist')
plt.xlabel(dados.columns[8])
plt.tight_layout()
corr = dados.corr()
sns.heatmap(corr)
from sklearn.preprocessing import MinMaxScaler
colunas = dados.columns
for col in colunas:
scaler = MinMaxScaler(feature_range=(0, 1))
dados[col] = scaler.fit_transform(dados[col].values.reshape(-1, 1))
dados.head()
X = dados[colunas[0:-1]].values
Y = dados[dados.columns[-1]].values
from sklearn.model_selection import train_test_split
X_treino, X_teste, Y_treino, Y_teste = train_test_split(X, Y, test_size=0.25, random_state=42)
from sklearn.metrics import mean_absolute_error,mean_squared_error
def mean_absolute_percentage_error(y_true, y_pred):
y_true, y_pred = np.array(y_true), np.array(y_pred)
return np.mean(np.abs((y_true - y_pred) / y_true)) * 100
from sklearn.linear_model import LinearRegression
linreg = LinearRegression()
linreg.fit(X_treino,Y_treino)
Y_pred_linreg = linreg.predict(X_teste)
MAE_linreg = mean_absolute_error(Y_teste,Y_pred_linreg)
MSE_linreg = mean_squared_error(Y_teste,Y_pred_linreg)
RMSE_linreg = np.sqrt(MSE_linreg)
MAPE_linreg = mean_absolute_percentage_error(Y_teste,Y_pred_linreg)
print("MAE = {:0.2f}".format(MAE_linreg))
print("MAPE = {:0.2f}%".format(MAPE_linreg))
print("MSE = {:0.2f}".format(MSE_linreg))
print("RMSE = {:0.2f}".format(RMSE_linreg))
from sklearn.svm import SVR
svr = SVR()
svr.fit(X_treino,Y_treino)
Y_pred_svr = svr.predict(X_teste)
MAE_svr = mean_absolute_error(Y_teste,Y_pred_svr)
MSE_svr = mean_squared_error(Y_teste,Y_pred_svr)
RMSE_svr = np.sqrt(MSE_svr)
MAPE_svr = mean_absolute_percentage_error(Y_teste,Y_pred_svr)
print("MAE = {:0.2f}".format(MAE_svr))
print("MAPE = {:0.2f}%".format(MAPE_svr))
print("MSE = {:0.2f}".format(MSE_svr))
print("RMSE = {:0.2f}".format(RMSE_svr))
from sklearn.tree import DecisionTreeRegressor
dte = DecisionTreeRegressor()
dte.fit(X_treino,Y_treino)
Y_pred_dte = dte.predict(X_teste)
MAE_dte = mean_absolute_error(Y_teste,Y_pred_dte)
MSE_dte = mean_squared_error(Y_teste,Y_pred_dte)
RMSE_dte = np.sqrt(MSE_dte)
MAPE_dte = mean_absolute_percentage_error(Y_teste,Y_pred_dte)
print("MAE = {:0.2f}".format(MAE_dte))
print("MAPE = {:0.2f}%".format(MAPE_dte))
print("MSE = {:0.2f}".format(MSE_dte))
print("RMSE = {:0.2f}".format(RMSE_dte))
from sklearn.ensemble import RandomForestRegressor
rfr = RandomForestRegressor()
rfr.fit(X_treino,Y_treino)
Y_pred_rfr = rfr.predict(X_teste)
MAE_rfr = mean_absolute_error(Y_teste,Y_pred_rfr)
MSE_rfr = mean_squared_error(Y_teste,Y_pred_rfr)
RMSE_rfr = np.sqrt(MSE_rfr)
MAPE_rfr = mean_absolute_percentage_error(Y_teste,Y_pred_rfr)
print("MAE = {:0.2f}".format(MAE_rfr))
print("MAPE = {:0.2f}%".format(MAPE_rfr))
print("MSE = {:0.2f}".format(MSE_rfr))
print("RMSE = {:0.2f}".format(RMSE_rfr))
from sklearn.ensemble import AdaBoostRegressor
ada = AdaBoostRegressor()
ada.fit(X_treino,Y_treino)
Y_pred_ada = ada.predict(X_teste)
MAE_ada = mean_absolute_error(Y_teste,Y_pred_ada)
MSE_ada = mean_squared_error(Y_teste,Y_pred_ada)
RMSE_ada = np.sqrt(MSE_ada)
MAPE_ada = mean_absolute_percentage_error(Y_teste,Y_pred_ada)
print("MAE = {:0.2f}".format(MAE_ada))
print("MAPE = {:0.2f}%".format(MAPE_ada))
print("MSE = {:0.2f}".format(MSE_ada))
print("RMSE = {:0.2f}".format(RMSE_ada))
modelo = ["Regressão linear","SVR","Decion Tree","Random Forest","Ada Boost"]
MAE = [MAE_linreg,MAE_svr,MAE_dte,MAE_rfr,MAE_ada]
MAPE = [MAPE_linreg,MAPE_svr,MAPE_dte,MAPE_rfr,MAPE_ada]
MSE = [MSE_linreg,MSE_svr,MSE_dte,MSE_rfr,MSE_ada]
RMSE = [RMSE_linreg,RMSE_svr,RMSE_dte,RMSE_rfr,RMSE_ada]
dici = {"Modelo" : modelo, "MAE" : MAE, "MAPE" : MAPE, "MSE" : MSE, "RMSE" : RMSE}
pd_dici = pd.DataFrame(dici).sort_values(by="MAPE")
pd_dici
| 0.572842 | 0.877844 |
## Statistical Modeling on Past vs Current Year Weather Data
My Spring 2017 study to determining if the highs and lows of this year are more severe than those of last year using statistical methods.
The raw weather data can be accessed [here](http://www.georgiaweather.net/index.php?variable=HI&site=WATHORT) and is collected at the University of Georgia [Horticulture Research Farm](http://www.caes.uga.edu/departments/horticulture/about/facilities/farm.html) in Watkinsville, Georgia.
```
import bs4 as bs
import urllib
import urllib2
import numpy as np
import scipy.stats as stats
import math
def scrape_temperatures(fromMonth, fromDay, fromYear, toMonth, toDay, toYear):
# request website
url = 'http://www.georgiaweather.net/index.php?variable=HI&site=WATHORT'
action_url = 'index.php?variable=HI&site=WATHORT'
# get source
source = urllib2.urlopen(url)
# submitting form
print('Retrieving daily temperatures from %s-%s-%s to %s-%s-%s' %(fromMonth, fromDay, fromYear, toMonth, toDay, toYear))
req = urllib2.Request(url,
data=urllib.urlencode({'fromMonth': fromMonth,
'toMonth': toMonth,
'fromDay': fromDay,
'toDay': toDay,
'fromYear': fromYear,
'toYear': toYear}),
headers={'User-Agent': 'Mozilla something',
'Cookie': 'name=value; name2=value2'})
response = urllib2.urlopen(req)
# get new source
soup = bs.BeautifulSoup(response, "lxml", from_encoding="utf-8")
# retrieve table data
td = []
for table in soup.findAll('table', {'class': 'tableBackground', 'width': '90%'}):
td = table.findAll('td', {'class': 'tdClass'})
# Organize table data
date = []
max = []
min = []
rain = []
temp_range = []
# separates data by date, max temp, min temp, and rain
for i in range(len(td) / 4):
# unused attributes
date.append(str(td[i * 4].text))
max.append(float(td[(i * 4) + 1].text))
min.append(float(td[(i * 4) + 2].text))
rain.append(float(td[(i * 4) + 3].text))
# value to return
temp_range.append([max[i], min[i]])
return temp_range
# retrieve daily temperatures
X = scrape_temperatures(fromMonth='January', fromDay='1', fromYear='2017', toMonth='January', toDay='31', toYear='2017')
Y = scrape_temperatures(fromMonth='January', fromDay='1', fromYear='2016', toMonth='January', toDay='31', toYear='2016')
# separate max & min values
xmax = (np.array(X)[:,0])
xmin = (np.array(X)[:,1])
ymax = (np.array(Y)[:,0])
ymin = (np.array(Y)[:,1])
def paired_ttest(X, Y, alph):
# difference
d = X - Y
# mean of difference
d_bar = np.mean(d)
N = len(X)
df = len(X)+len(Y) - 2
# np.std divides the statistic by N - ddof
s = np.std(d, ddof=2)
# critical value: 1-alpha/2 because it is a two sided test
cval = stats.t.ppf(1 - alph/2, df)
print "critical value=", cval
# t-statistic
t = (d_bar - 0) / (s / math.sqrt(N))
# p-value: multiplying by 2 because its a two tailed test
pval = 2.0 * (1 - stats.t.cdf(t, df))
print "pval=", pval
# critical value and pvalue
return [cval, pval]
alph = 0.05
# calculate critical and p values
highs = paired_ttest(xmax, ymax, alph)
lows = paired_ttest(xmin, ymin, alph)
# hypothesis testing using alpha and the calculated p-value
if highs[1] < alph:
print 'Based on the p-value of the daily highs we Reject Null Hypothesis'
else:
print 'Based on the p-value of the daily highs we Fail to Reject Null Hypothesis'
# hypothesis testing using alpha and the calculated p-value
if lows[1] < alph:
print 'Based on the p-value of the daily lows we Reject Null Hypothesis'
else:
print 'Based on the p-value of the daily lows we Fail to Reject Null Hypothesis'
```
|
github_jupyter
|
import bs4 as bs
import urllib
import urllib2
import numpy as np
import scipy.stats as stats
import math
def scrape_temperatures(fromMonth, fromDay, fromYear, toMonth, toDay, toYear):
# request website
url = 'http://www.georgiaweather.net/index.php?variable=HI&site=WATHORT'
action_url = 'index.php?variable=HI&site=WATHORT'
# get source
source = urllib2.urlopen(url)
# submitting form
print('Retrieving daily temperatures from %s-%s-%s to %s-%s-%s' %(fromMonth, fromDay, fromYear, toMonth, toDay, toYear))
req = urllib2.Request(url,
data=urllib.urlencode({'fromMonth': fromMonth,
'toMonth': toMonth,
'fromDay': fromDay,
'toDay': toDay,
'fromYear': fromYear,
'toYear': toYear}),
headers={'User-Agent': 'Mozilla something',
'Cookie': 'name=value; name2=value2'})
response = urllib2.urlopen(req)
# get new source
soup = bs.BeautifulSoup(response, "lxml", from_encoding="utf-8")
# retrieve table data
td = []
for table in soup.findAll('table', {'class': 'tableBackground', 'width': '90%'}):
td = table.findAll('td', {'class': 'tdClass'})
# Organize table data
date = []
max = []
min = []
rain = []
temp_range = []
# separates data by date, max temp, min temp, and rain
for i in range(len(td) / 4):
# unused attributes
date.append(str(td[i * 4].text))
max.append(float(td[(i * 4) + 1].text))
min.append(float(td[(i * 4) + 2].text))
rain.append(float(td[(i * 4) + 3].text))
# value to return
temp_range.append([max[i], min[i]])
return temp_range
# retrieve daily temperatures
X = scrape_temperatures(fromMonth='January', fromDay='1', fromYear='2017', toMonth='January', toDay='31', toYear='2017')
Y = scrape_temperatures(fromMonth='January', fromDay='1', fromYear='2016', toMonth='January', toDay='31', toYear='2016')
# separate max & min values
xmax = (np.array(X)[:,0])
xmin = (np.array(X)[:,1])
ymax = (np.array(Y)[:,0])
ymin = (np.array(Y)[:,1])
def paired_ttest(X, Y, alph):
# difference
d = X - Y
# mean of difference
d_bar = np.mean(d)
N = len(X)
df = len(X)+len(Y) - 2
# np.std divides the statistic by N - ddof
s = np.std(d, ddof=2)
# critical value: 1-alpha/2 because it is a two sided test
cval = stats.t.ppf(1 - alph/2, df)
print "critical value=", cval
# t-statistic
t = (d_bar - 0) / (s / math.sqrt(N))
# p-value: multiplying by 2 because its a two tailed test
pval = 2.0 * (1 - stats.t.cdf(t, df))
print "pval=", pval
# critical value and pvalue
return [cval, pval]
alph = 0.05
# calculate critical and p values
highs = paired_ttest(xmax, ymax, alph)
lows = paired_ttest(xmin, ymin, alph)
# hypothesis testing using alpha and the calculated p-value
if highs[1] < alph:
print 'Based on the p-value of the daily highs we Reject Null Hypothesis'
else:
print 'Based on the p-value of the daily highs we Fail to Reject Null Hypothesis'
# hypothesis testing using alpha and the calculated p-value
if lows[1] < alph:
print 'Based on the p-value of the daily lows we Reject Null Hypothesis'
else:
print 'Based on the p-value of the daily lows we Fail to Reject Null Hypothesis'
| 0.238994 | 0.849784 |
```
import astropy.coordinates as coord
import astropy.units as u
from astropy.table import Table, join, vstack
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
%matplotlib inline
from astropy.io import ascii
import gala.coordinates as gc
import gala.dynamics as gd
from gala.units import galactic
from pyia import GaiaData
xmatch = Table.read('/Users/adrian/data/APOGEE_DR15beta/gaiadr2_allStar-t9-l31c-58158.fits')
xmatch.rename_column('apstar_id', 'APSTAR_ID')
apogee = Table.read('/Users/adrian/data/APOGEE_DR15beta/allStar-t9-l31c-58158.fits')
stream_mask = (apogee['APOGEE2_TARGET1'] & (2**18 + 2**19)) != 0
stream_mask.sum()
apogee_gaia = join(apogee[stream_mask], xmatch, keys='APSTAR_ID')
ndim_cols = [x for x in apogee_gaia.columns if apogee_gaia[x].ndim == 1]
apogee_gaia = apogee_gaia[ndim_cols]
# select distance stuff
apogee_gaia = apogee_gaia[(apogee_gaia['parallax'] < 1.) | ((apogee_gaia['parallax'] - apogee_gaia['parallax_error']) < 1.)]
g = GaiaData(apogee_gaia)
len(g)
np.unique(apogee_gaia['FIELD'])
gd1_mask = np.array(['GD1' in f for f in apogee_gaia['FIELD']])
orp_mask = np.array(['ORPHAN' in f for f in apogee_gaia['FIELD']])
pal5_mask = np.array(['PAL5' in f for f in apogee_gaia['FIELD']])
tri_mask = np.array(['TRIAND' in f for f in apogee_gaia['FIELD']])
gd1_mask.sum(), orp_mask.sum(), pal5_mask.sum(), tri_mask.sum()
fig, axes = plt.subplots(1, 3, figsize=(12, 4), sharex=True, sharey=True)
for i, name, mask in zip(range(3),
['GD-1', 'Orp', 'Pal5'],
[gd1_mask, orp_mask, pal5_mask]):
ax = axes.flat[i]
ax.plot(apogee_gaia['TEFF'][mask], apogee_gaia['LOGG'][mask],
marker='.', linestyle='none')
ax.set_title(name)
ax.set_xlabel('Teff')
ax.set_xlim(6500, 3500)
ax.set_ylim(4, 0)
axes[0].set_ylabel('logg')
fig.tight_layout()
fig, axes = plt.subplots(1, 3, figsize=(12, 4), sharex=True, sharey=True)
for i, name, mask in zip(range(3),
['GD-1', 'Orp', 'Pal5'],
[gd1_mask, orp_mask, pal5_mask]):
ax = axes.flat[i]
ax.plot(apogee_gaia['RA'][mask], apogee_gaia['DEC'][mask],
marker='.', linestyle='none')
ax.set_title(name)
ax.set_xlabel('RA')
fig.tight_layout()
ax.set_xlim(360, 0)
ax.set_ylim(-30, 90)
axes[0].set_ylabel('Dec')
fig.tight_layout()
pm_poly = np.load('../../gd1-dr2/output/pm_poly.npy')
fig, axes = plt.subplots(2, 3, figsize=(12, 8), sharex='row', sharey='row')
fig2, axes2 = plt.subplots(1, 3, figsize=(12, 4.5), sharex=True)
for line in zip(range(3),
['GD-1', 'Orp', 'Pal5'],
[gd1_mask, orp_mask, pal5_mask],
[gc.GD1(), gc.Orphan(), gc.Pal5()],
[8.*u.kpc, 30*u.kpc, 22*u.kpc]):
i, name, mask, frame, dist = line
lon, lat, _ = frame.get_representation_component_names().keys()
stream_g = g[mask]
c = stream_g.get_skycoord(distance=dist,
radial_velocity=np.array(stream_g.VHELIO_AVG)*u.km/u.s)
c = gc.reflex_correct(c.transform_to(frame))
pm1 = getattr(c, 'pm_{0}_cos{1}'.format(lon, lat)).to(u.mas/u.yr)
pm2 = getattr(c, 'pm_{0}'.format(lat)).to(u.mas/u.yr)
# top row
ax = axes[0, i]
ax.scatter(pm1, pm2,
c=stream_g.FE_H, vmin=-2., vmax=0,
marker='o', alpha=0.7, linewidth=0)
if name == 'GD-1':
ax.add_patch(mpl.patches.Polygon(pm_poly, facecolor='none', edgecolor='k', zorder=-10))
ax.set_title(name)
ax.axhline(0, zorder=-100, color='#cccccc')
ax.set_xlabel('pm1')
# bottom row
ax = axes[1, i]
ax.scatter(getattr(c, lon).wrap_at(180*u.deg),
c.radial_velocity.to(u.km/u.s),
c=stream_g.FE_H, vmin=-2., vmax=-1,
marker='o', alpha=0.7, linewidth=0)
ax.set_xlabel('phi1')
# metallicity
axes2[i].hist(stream_g.FE_H, bins=np.linspace(-2.5, 0.5, 32));
axes2[i].set_xlabel('[Fe/H]')
fig.tight_layout()
axes[0, 0].set_xlim(-15, 15)
axes[0, 0].set_ylim(-15, 15)
axes[1, 0].set_xlim(-120, 60)
axes[1, 0].set_ylim(-300, 300)
axes[0, 0].set_ylabel('pm2')
axes[1, 0].set_ylabel('RV')
fig.tight_layout()
fig2.tight_layout()
```
---
```
kop_vr = ascii.read("""phi1 phi2 vr err
-45.23 -0.04 28.8 6.9
-43.17 -0.09 29.3 10.2
-39.54 -0.07 2.9 8.7
-39.25 -0.22 -5.2 6.5
-37.95 0.00 1.1 5.6
-37.96 -0.00 -11.7 11.2
-35.49 -0.05 -50.4 5.2
-35.27 -0.02 -30.9 12.8
-34.92 -0.15 -35.3 7.5
-34.74 -0.08 -30.9 9.2
-33.74 -0.18 -74.3 9.8
-32.90 -0.15 -71.5 9.6
-32.25 -0.17 -71.5 9.2
-29.95 -0.00 -92.7 8.7
-26.61 -0.11 -114.2 7.3
-25.45 -0.14 -67.8 7.1
-24.86 0.01 -111.2 17.8
-21.21 -0.02 -144.4 10.5
-14.47 -0.15 -179.0 10.0
-13.73 -0.28 -191.4 7.5
-13.02 -0.21 -162.9 9.6
-12.68 -0.26 -217.2 10.7
-12.55 -0.23 -172.2 6.6""")
w = np.load('../../gd1-dr2/data/stream_model.npy')
stream_w = gd.PhaseSpacePosition(pos=w[:, :3].T*u.kpc,
vel=w[:, 3:].T*u.km/u.s)
model_c = stream_w.to_coord_frame(gc.GD1)
name = 'GD-1'
mask = gd1_mask
frame = gc.GD1()
dist = 10*u.kpc
stream_g = g[mask]
c = stream_g.get_skycoord(distance=dist,
radial_velocity=np.array(stream_g.VHELIO_AVG)*u.km/u.s)
c = gc.reflex_correct(c.transform_to(frame))
pm1 = c.pm_phi1_cosphi2
pm2 = c.pm_phi2
fig, ax = plt.subplots(1, 1, figsize=(8, 6))
ax.scatter(c.phi1.wrap_at(180*u.deg),
stream_g.VHELIO_AVG,
c='k', marker='o', alpha=0.5, linewidth=0)
# ax.scatter(kop_vr['phi1'], kop_vr['vr'])#, kop_vr['err']
ax.scatter(model_c.phi1.wrap_at(180*u.deg),
model_c.radial_velocity.to(u.km/u.s),
zorder=-100, color='#aaaaaa')
ax.set_xlabel('phi1')
ax.set_ylabel('RV')
ax.set_xlim(-100, 30)
ax.set_ylim(-320, 320)
fig.tight_layout()
phi1 = c.phi1.wrap_at(180*u.deg).degree
rv = stream_g.VHELIO_AVG
rv_mask = (((phi1 > 0) & (rv < -220) & (rv > -300)) |
((phi1 < -20) & (phi1 > -40) & (rv < -95) & (rv > -200)) |
((phi1 > -60) & (phi1 < -50) & (rv > 90) & (rv < 150)))
feh_mask = (stream_g.FE_H > -1.9) & (stream_g.FE_H < -1.4)
```
|
github_jupyter
|
import astropy.coordinates as coord
import astropy.units as u
from astropy.table import Table, join, vstack
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
%matplotlib inline
from astropy.io import ascii
import gala.coordinates as gc
import gala.dynamics as gd
from gala.units import galactic
from pyia import GaiaData
xmatch = Table.read('/Users/adrian/data/APOGEE_DR15beta/gaiadr2_allStar-t9-l31c-58158.fits')
xmatch.rename_column('apstar_id', 'APSTAR_ID')
apogee = Table.read('/Users/adrian/data/APOGEE_DR15beta/allStar-t9-l31c-58158.fits')
stream_mask = (apogee['APOGEE2_TARGET1'] & (2**18 + 2**19)) != 0
stream_mask.sum()
apogee_gaia = join(apogee[stream_mask], xmatch, keys='APSTAR_ID')
ndim_cols = [x for x in apogee_gaia.columns if apogee_gaia[x].ndim == 1]
apogee_gaia = apogee_gaia[ndim_cols]
# select distance stuff
apogee_gaia = apogee_gaia[(apogee_gaia['parallax'] < 1.) | ((apogee_gaia['parallax'] - apogee_gaia['parallax_error']) < 1.)]
g = GaiaData(apogee_gaia)
len(g)
np.unique(apogee_gaia['FIELD'])
gd1_mask = np.array(['GD1' in f for f in apogee_gaia['FIELD']])
orp_mask = np.array(['ORPHAN' in f for f in apogee_gaia['FIELD']])
pal5_mask = np.array(['PAL5' in f for f in apogee_gaia['FIELD']])
tri_mask = np.array(['TRIAND' in f for f in apogee_gaia['FIELD']])
gd1_mask.sum(), orp_mask.sum(), pal5_mask.sum(), tri_mask.sum()
fig, axes = plt.subplots(1, 3, figsize=(12, 4), sharex=True, sharey=True)
for i, name, mask in zip(range(3),
['GD-1', 'Orp', 'Pal5'],
[gd1_mask, orp_mask, pal5_mask]):
ax = axes.flat[i]
ax.plot(apogee_gaia['TEFF'][mask], apogee_gaia['LOGG'][mask],
marker='.', linestyle='none')
ax.set_title(name)
ax.set_xlabel('Teff')
ax.set_xlim(6500, 3500)
ax.set_ylim(4, 0)
axes[0].set_ylabel('logg')
fig.tight_layout()
fig, axes = plt.subplots(1, 3, figsize=(12, 4), sharex=True, sharey=True)
for i, name, mask in zip(range(3),
['GD-1', 'Orp', 'Pal5'],
[gd1_mask, orp_mask, pal5_mask]):
ax = axes.flat[i]
ax.plot(apogee_gaia['RA'][mask], apogee_gaia['DEC'][mask],
marker='.', linestyle='none')
ax.set_title(name)
ax.set_xlabel('RA')
fig.tight_layout()
ax.set_xlim(360, 0)
ax.set_ylim(-30, 90)
axes[0].set_ylabel('Dec')
fig.tight_layout()
pm_poly = np.load('../../gd1-dr2/output/pm_poly.npy')
fig, axes = plt.subplots(2, 3, figsize=(12, 8), sharex='row', sharey='row')
fig2, axes2 = plt.subplots(1, 3, figsize=(12, 4.5), sharex=True)
for line in zip(range(3),
['GD-1', 'Orp', 'Pal5'],
[gd1_mask, orp_mask, pal5_mask],
[gc.GD1(), gc.Orphan(), gc.Pal5()],
[8.*u.kpc, 30*u.kpc, 22*u.kpc]):
i, name, mask, frame, dist = line
lon, lat, _ = frame.get_representation_component_names().keys()
stream_g = g[mask]
c = stream_g.get_skycoord(distance=dist,
radial_velocity=np.array(stream_g.VHELIO_AVG)*u.km/u.s)
c = gc.reflex_correct(c.transform_to(frame))
pm1 = getattr(c, 'pm_{0}_cos{1}'.format(lon, lat)).to(u.mas/u.yr)
pm2 = getattr(c, 'pm_{0}'.format(lat)).to(u.mas/u.yr)
# top row
ax = axes[0, i]
ax.scatter(pm1, pm2,
c=stream_g.FE_H, vmin=-2., vmax=0,
marker='o', alpha=0.7, linewidth=0)
if name == 'GD-1':
ax.add_patch(mpl.patches.Polygon(pm_poly, facecolor='none', edgecolor='k', zorder=-10))
ax.set_title(name)
ax.axhline(0, zorder=-100, color='#cccccc')
ax.set_xlabel('pm1')
# bottom row
ax = axes[1, i]
ax.scatter(getattr(c, lon).wrap_at(180*u.deg),
c.radial_velocity.to(u.km/u.s),
c=stream_g.FE_H, vmin=-2., vmax=-1,
marker='o', alpha=0.7, linewidth=0)
ax.set_xlabel('phi1')
# metallicity
axes2[i].hist(stream_g.FE_H, bins=np.linspace(-2.5, 0.5, 32));
axes2[i].set_xlabel('[Fe/H]')
fig.tight_layout()
axes[0, 0].set_xlim(-15, 15)
axes[0, 0].set_ylim(-15, 15)
axes[1, 0].set_xlim(-120, 60)
axes[1, 0].set_ylim(-300, 300)
axes[0, 0].set_ylabel('pm2')
axes[1, 0].set_ylabel('RV')
fig.tight_layout()
fig2.tight_layout()
kop_vr = ascii.read("""phi1 phi2 vr err
-45.23 -0.04 28.8 6.9
-43.17 -0.09 29.3 10.2
-39.54 -0.07 2.9 8.7
-39.25 -0.22 -5.2 6.5
-37.95 0.00 1.1 5.6
-37.96 -0.00 -11.7 11.2
-35.49 -0.05 -50.4 5.2
-35.27 -0.02 -30.9 12.8
-34.92 -0.15 -35.3 7.5
-34.74 -0.08 -30.9 9.2
-33.74 -0.18 -74.3 9.8
-32.90 -0.15 -71.5 9.6
-32.25 -0.17 -71.5 9.2
-29.95 -0.00 -92.7 8.7
-26.61 -0.11 -114.2 7.3
-25.45 -0.14 -67.8 7.1
-24.86 0.01 -111.2 17.8
-21.21 -0.02 -144.4 10.5
-14.47 -0.15 -179.0 10.0
-13.73 -0.28 -191.4 7.5
-13.02 -0.21 -162.9 9.6
-12.68 -0.26 -217.2 10.7
-12.55 -0.23 -172.2 6.6""")
w = np.load('../../gd1-dr2/data/stream_model.npy')
stream_w = gd.PhaseSpacePosition(pos=w[:, :3].T*u.kpc,
vel=w[:, 3:].T*u.km/u.s)
model_c = stream_w.to_coord_frame(gc.GD1)
name = 'GD-1'
mask = gd1_mask
frame = gc.GD1()
dist = 10*u.kpc
stream_g = g[mask]
c = stream_g.get_skycoord(distance=dist,
radial_velocity=np.array(stream_g.VHELIO_AVG)*u.km/u.s)
c = gc.reflex_correct(c.transform_to(frame))
pm1 = c.pm_phi1_cosphi2
pm2 = c.pm_phi2
fig, ax = plt.subplots(1, 1, figsize=(8, 6))
ax.scatter(c.phi1.wrap_at(180*u.deg),
stream_g.VHELIO_AVG,
c='k', marker='o', alpha=0.5, linewidth=0)
# ax.scatter(kop_vr['phi1'], kop_vr['vr'])#, kop_vr['err']
ax.scatter(model_c.phi1.wrap_at(180*u.deg),
model_c.radial_velocity.to(u.km/u.s),
zorder=-100, color='#aaaaaa')
ax.set_xlabel('phi1')
ax.set_ylabel('RV')
ax.set_xlim(-100, 30)
ax.set_ylim(-320, 320)
fig.tight_layout()
phi1 = c.phi1.wrap_at(180*u.deg).degree
rv = stream_g.VHELIO_AVG
rv_mask = (((phi1 > 0) & (rv < -220) & (rv > -300)) |
((phi1 < -20) & (phi1 > -40) & (rv < -95) & (rv > -200)) |
((phi1 > -60) & (phi1 < -50) & (rv > 90) & (rv < 150)))
feh_mask = (stream_g.FE_H > -1.9) & (stream_g.FE_H < -1.4)
| 0.429429 | 0.504516 |
# Tutorial: Import and export
<figure style="display: table; text-align:center; margin-left: auto; margin-right:auto">
[](https://mybinder.org/v2/gh/simphony/docs/master?filepath=docs%2Fsource%2Fjupyter%2Fimport_export.ipynb "Click to run the tutorial yourself!")
</figure>
In this tutorial we will be covering the import and export capabilities of OSP-core. The utility functions that provide these functionalities are `import_cuds` and `export_cuds`, respectively.
<div class="admonition important">
<div class="admonition-title" style="font-weight: bold"><div style="display: inline-block">Tip</div></div>
The full API specifictions of the import and export functions can be found in the
[utilities API reference page](../api_ref.html#osp.core.utils.export_cuds).
</div>
</div>
For our running example, we'll be using the *city ontology* that was already introduces in the [cuds API tutorial](./cuds_api.html). First, make sure the city ontology is installed. If not, run the following command:
```
!pico install city
```
Next we create a few CUDS objects:
```
from osp.core.namespaces import city
c = city.City(name="Freiburg", coordinates=[47, 7])
p1 = city.Citizen(name="Peter")
p2 = city.Citizen(name="Anne")
c.add(p1, rel=city.hasInhabitant)
c.add(p2, rel=city.hasInhabitant)
```
Now we can use the `export_cuds` methods to export the data into a file:
```
from osp.core.utils import export_cuds
export_cuds(c, file='./data.ttl', format='turtle')
```
This will create the file `data.ttl` with the following content:
```
from sys import platform
if platform == 'win32':
!more data.ttl
else:
!cat data.ttl
```
You can change the format by entering a different value for the parameter `format`. The supported formats are “xml”, “n3”, “turtle”, “nt”, “pretty-xml”, “trix”, “trig” and “nquads”.
To import data, we can use the `import` method. Let's assume we wish to import data into an SQLite session. The following code will help us to achieve our aim:
```
from osp.wrappers.sqlite import SqliteSession
from osp.core.utils import import_cuds
with SqliteSession("test.db") as session:
wrapper = city.CityWrapper(session=session)
c = import_cuds('./data.ttl')
wrapper.add(c)
session.commit()
```
Now we can verify the data was indeed imported:
```
from osp.core.utils import pretty_print
with SqliteSession("test.db") as session:
wrapper = city.CityWrapper(session=session)
pretty_print(wrapper)
```
<div class="admonition important">
<div class="admonition-title" style="font-weight: bold"><div style="display: inline-block">Notes</div></div>
1. The format is automatically inferred from the file extension. To specify it explicitly, you can add the `format` parameter, like so: `import_cuds('./data.ttl', format='turtle')`.
1. The `session` parameter is optional and inferred automatically from the context that created by the `with` statement (see the [tutorial on multiple wrappers](./multiple_wrappers.html) for more information). You can specify the session explicitly like so: `import_cuds('./data.ttl', session=session)`.
</div>
|
github_jupyter
|
!pico install city
from osp.core.namespaces import city
c = city.City(name="Freiburg", coordinates=[47, 7])
p1 = city.Citizen(name="Peter")
p2 = city.Citizen(name="Anne")
c.add(p1, rel=city.hasInhabitant)
c.add(p2, rel=city.hasInhabitant)
from osp.core.utils import export_cuds
export_cuds(c, file='./data.ttl', format='turtle')
from sys import platform
if platform == 'win32':
!more data.ttl
else:
!cat data.ttl
from osp.wrappers.sqlite import SqliteSession
from osp.core.utils import import_cuds
with SqliteSession("test.db") as session:
wrapper = city.CityWrapper(session=session)
c = import_cuds('./data.ttl')
wrapper.add(c)
session.commit()
from osp.core.utils import pretty_print
with SqliteSession("test.db") as session:
wrapper = city.CityWrapper(session=session)
pretty_print(wrapper)
| 0.195364 | 0.946941 |
# Examples
```
import numpy as np
import pandas as pd
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
```
## Ticks
```
plt.figure(figsize=(6, 3))
plt.plot([2, 4, 6, 8], 'o', [1, 5, 9, 13], 's')
plt.xticks(ticks=np.arange(4))
plt.show()
plt.figure(figsize=(6, 3))
plt.plot([2, 4, 6, 8], 'o', [1, 5, 9, 13], 's')
plt.xticks(ticks=np.arange(4), labels=['January', 'February', 'March', 'April'], rotation=20)
plt.show()
```
## Basic Text and Legend Functions
```
fig = plt.figure()
plt.plot(4, 2, 'o')
plt.xlim([0, 10])
plt.ylim([0, 10])
fig.suptitle('Suptitle', fontsize=10, fontweight='bold')
ax = plt.gca()
ax.set_xlabel('X Label')
ax.set_ylabel('Y Label')
ax.text(4, 6, 'Text in Data Coords', bbox={'facecolor': 'yellow', 'alpha':0.5, 'pad':10})
ax.annotate('Example of Annotate', xy=(4,2), xytext=(8,4), arrowprops=dict(facecolor='green', shrink=0.05))
plt.show()
```
## Legends
```
plt.figure()
plt.plot([1, 2, 3], label='Label 1')
plt.plot([2, 4, 3], label='Label 2')
plt.legend()
plt.show()
```
## Bar Chart
```
plt.figure()
plt.bar(['A', 'B', 'C', 'D'], [20, 25, 40, 10])
plt.show()
plt.figure()
labels = ['A', 'B', 'C', 'D']
x = np.arange(len(labels))
width = 0.4
plt.bar(x - width / 2, [20, 25, 40, 10], width=width)
plt.bar(x + width / 2, [30, 15, 30, 20], width=width)
# Ticks and tick labels must be set manually
plt.xticks(x)
ax = plt.gca()
ax.set_xticklabels(labels)
plt.show()
```
## Pie Chart
```
plt.figure()
plt.pie([0.4, 0.3, 0.2, 0.1], explode=(0.1, 0, 0, 0), labels=['A', 'B', 'C', 'D'])
plt.show()
```
## Stacked Bar Chart
```
plt.figure()
labels = ['A', 'B', 'C']
x = np.arange(len(labels))
bars1 = [10, 20, 30]
bars2 = [15, 10, 20]
bars3 = [20, 10, 10]
plt.bar(x, bars1)
plt.bar(x, bars2, bottom=bars1)
plt.bar(x, bars3, bottom=np.add(bars1, bars2))
plt.xticks(x, labels)
plt.show()
```
## Stacked Area Chart
```
plt.figure()
plt.stackplot([1, 2, 3, 4], [2, 4, 5, 8], [1, 5, 4, 2])
plt.show()
```
## Histogram
```
plt.figure()
x = np.random.normal(0, 1, size=1000)
plt.hist(x, bins=30, density=True)
plt.show()
```
## Box Plot
```
plt.figure()
x1 = np.random.normal(0, 1, size=100)
x2 = np.random.normal(0, 1, size=100)
plt.boxplot([x1, x2], labels=['A', 'B'])
plt.show()
```
## Scatter Plot
```
x = np.random.normal(0, 1, size=100)
y = x + np.random.normal(0, 1, size=100)
plt.figure()
plt.scatter(x, y)
plt.show()
```
## Bubble Plot
```
x = np.random.normal(0, 1, size=100)
y = np.random.normal(0, 1, size=100)
z = np.random.normal(0, 1, size=100)
c = np.random.normal(0, 1, size=100)
plt.figure()
plt.scatter(x, y, s=z*500, c=c, alpha=0.5)
plt.colorbar()
plt.show()
```
## Subplots
```
series = [np.cumsum(np.random.normal(0, 1, size=100)) for i in range(4)]
fig, axes = plt.subplots(2, 2)
axes = axes.ravel()
for i, ax in enumerate(axes):
ax.plot(series[i])
plt.show()
fig, axes = plt.subplots(2, 2, sharex=True, sharey=True)
axes = axes.ravel()
for i, ax in enumerate(axes):
ax.plot(series[i])
plt.show()
```
## Tight Layout
```
fig, axes = plt.subplots(2, 2)
axes = axes.ravel()
for i, ax in enumerate(axes):
ax.plot(series[i])
ax.set_title('Subplot ' + str(i))
plt.show()
fig, axes = plt.subplots(2, 2)
axes = axes.ravel()
for i, ax in enumerate(axes):
ax.plot(series[i])
ax.set_title('Subplot ' + str(i))
plt.tight_layout()
plt.show()
```
## GridSpec
```
plt.figure()
gs = matplotlib.gridspec.GridSpec(3, 4)
ax1 = plt.subplot(gs[:3, :3])
ax2 = plt.subplot(gs[0, 3])
ax3 = plt.subplot(gs[1, 3])
ax4 = plt.subplot(gs[2, 3])
ax1.plot(series[0])
ax2.plot(series[1])
ax3.plot(series[2])
ax4.plot(series[3])
plt.tight_layout()
plt.show()
```
## Images
```
import matplotlib.image as mpimg
import os
img_filenames = sorted(os.listdir('../../Datasets/images'))
imgs = [mpimg.imread(os.path.join('../../Datasets/images', img_filename)) for img_filename in img_filenames]
gray = lambda rgb : np.dot(rgb[... , :3] , [0.299 , 0.587, 0.114])
img = gray(imgs[0])
plt.figure()
plt.imshow(img, cmap='jet')
plt.colorbar()
plt.show()
plt.figure()
plt.hist(img.ravel(), bins=256, range=(0, 255))
plt.show()
fig, axes = plt.subplots(1, 2)
labels = ['coast', 'beach']
for i in range(2):
axes[i].imshow(imgs[i])
axes[i].set_xticks([])
axes[i].set_yticks([])
axes[i].set_xlabel(labels[i])
plt.show()
```
## Writing Mathematical Expressions
```
plt.figure()
x = np.arange(0, 10, 0.1)
y = np.cos(x)
plt.plot(x, y)
plt.xlabel('$x$')
plt.ylabel('$\cos(x)$')
plt.show()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
plt.figure(figsize=(6, 3))
plt.plot([2, 4, 6, 8], 'o', [1, 5, 9, 13], 's')
plt.xticks(ticks=np.arange(4))
plt.show()
plt.figure(figsize=(6, 3))
plt.plot([2, 4, 6, 8], 'o', [1, 5, 9, 13], 's')
plt.xticks(ticks=np.arange(4), labels=['January', 'February', 'March', 'April'], rotation=20)
plt.show()
fig = plt.figure()
plt.plot(4, 2, 'o')
plt.xlim([0, 10])
plt.ylim([0, 10])
fig.suptitle('Suptitle', fontsize=10, fontweight='bold')
ax = plt.gca()
ax.set_xlabel('X Label')
ax.set_ylabel('Y Label')
ax.text(4, 6, 'Text in Data Coords', bbox={'facecolor': 'yellow', 'alpha':0.5, 'pad':10})
ax.annotate('Example of Annotate', xy=(4,2), xytext=(8,4), arrowprops=dict(facecolor='green', shrink=0.05))
plt.show()
plt.figure()
plt.plot([1, 2, 3], label='Label 1')
plt.plot([2, 4, 3], label='Label 2')
plt.legend()
plt.show()
plt.figure()
plt.bar(['A', 'B', 'C', 'D'], [20, 25, 40, 10])
plt.show()
plt.figure()
labels = ['A', 'B', 'C', 'D']
x = np.arange(len(labels))
width = 0.4
plt.bar(x - width / 2, [20, 25, 40, 10], width=width)
plt.bar(x + width / 2, [30, 15, 30, 20], width=width)
# Ticks and tick labels must be set manually
plt.xticks(x)
ax = plt.gca()
ax.set_xticklabels(labels)
plt.show()
plt.figure()
plt.pie([0.4, 0.3, 0.2, 0.1], explode=(0.1, 0, 0, 0), labels=['A', 'B', 'C', 'D'])
plt.show()
plt.figure()
labels = ['A', 'B', 'C']
x = np.arange(len(labels))
bars1 = [10, 20, 30]
bars2 = [15, 10, 20]
bars3 = [20, 10, 10]
plt.bar(x, bars1)
plt.bar(x, bars2, bottom=bars1)
plt.bar(x, bars3, bottom=np.add(bars1, bars2))
plt.xticks(x, labels)
plt.show()
plt.figure()
plt.stackplot([1, 2, 3, 4], [2, 4, 5, 8], [1, 5, 4, 2])
plt.show()
plt.figure()
x = np.random.normal(0, 1, size=1000)
plt.hist(x, bins=30, density=True)
plt.show()
plt.figure()
x1 = np.random.normal(0, 1, size=100)
x2 = np.random.normal(0, 1, size=100)
plt.boxplot([x1, x2], labels=['A', 'B'])
plt.show()
x = np.random.normal(0, 1, size=100)
y = x + np.random.normal(0, 1, size=100)
plt.figure()
plt.scatter(x, y)
plt.show()
x = np.random.normal(0, 1, size=100)
y = np.random.normal(0, 1, size=100)
z = np.random.normal(0, 1, size=100)
c = np.random.normal(0, 1, size=100)
plt.figure()
plt.scatter(x, y, s=z*500, c=c, alpha=0.5)
plt.colorbar()
plt.show()
series = [np.cumsum(np.random.normal(0, 1, size=100)) for i in range(4)]
fig, axes = plt.subplots(2, 2)
axes = axes.ravel()
for i, ax in enumerate(axes):
ax.plot(series[i])
plt.show()
fig, axes = plt.subplots(2, 2, sharex=True, sharey=True)
axes = axes.ravel()
for i, ax in enumerate(axes):
ax.plot(series[i])
plt.show()
fig, axes = plt.subplots(2, 2)
axes = axes.ravel()
for i, ax in enumerate(axes):
ax.plot(series[i])
ax.set_title('Subplot ' + str(i))
plt.show()
fig, axes = plt.subplots(2, 2)
axes = axes.ravel()
for i, ax in enumerate(axes):
ax.plot(series[i])
ax.set_title('Subplot ' + str(i))
plt.tight_layout()
plt.show()
plt.figure()
gs = matplotlib.gridspec.GridSpec(3, 4)
ax1 = plt.subplot(gs[:3, :3])
ax2 = plt.subplot(gs[0, 3])
ax3 = plt.subplot(gs[1, 3])
ax4 = plt.subplot(gs[2, 3])
ax1.plot(series[0])
ax2.plot(series[1])
ax3.plot(series[2])
ax4.plot(series[3])
plt.tight_layout()
plt.show()
import matplotlib.image as mpimg
import os
img_filenames = sorted(os.listdir('../../Datasets/images'))
imgs = [mpimg.imread(os.path.join('../../Datasets/images', img_filename)) for img_filename in img_filenames]
gray = lambda rgb : np.dot(rgb[... , :3] , [0.299 , 0.587, 0.114])
img = gray(imgs[0])
plt.figure()
plt.imshow(img, cmap='jet')
plt.colorbar()
plt.show()
plt.figure()
plt.hist(img.ravel(), bins=256, range=(0, 255))
plt.show()
fig, axes = plt.subplots(1, 2)
labels = ['coast', 'beach']
for i in range(2):
axes[i].imshow(imgs[i])
axes[i].set_xticks([])
axes[i].set_yticks([])
axes[i].set_xlabel(labels[i])
plt.show()
plt.figure()
x = np.arange(0, 10, 0.1)
y = np.cos(x)
plt.plot(x, y)
plt.xlabel('$x$')
plt.ylabel('$\cos(x)$')
plt.show()
| 0.526099 | 0.945197 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.